Note: Descriptions are shown in the official language in which they were submitted.
~i t i!
~o 93/17109 ~ PCT/GB93~00332
CA2 1 i 7468
IBV SPIKE PROTEIN (2)
Backqro~d of the inventlon
1. Fi~ld of thQ inventiQn
This invention relates to the spike protein of infectious
brsnchitis virus (IBV) and to a recombinant DNA method of
preparing lt. IBV is a virus which causes respiratory disease in
the fowl, and is of particular importance in relation to poultry.
2. Pescris~iQn of the Drior art
I8V is a virus of the type Coronaviridae. It has a single-
stranded RNA genome, approximately 20 kb in length, of positive
polarity, which specifies the production of three major
structural proteins: nucleocapsid protein, membrane
glycoprotein, and spike glycoprotein. The spike glycoprotein is
so called because it is present in the teardrop-shaped surface
projections or spikes protruding from the lipid membrane of the
virus~ The spike protein is believed likely to be responsible
for immunogen~city of the virus, partly by analogy with the spike
prote~ns of other coronaviruses and partly by 7n v~tro
neutralisation experiments, see, for example, D. Cav~nagh et al.,
Av~an Pathology 13, 573-583 (1984). Although the term "spike
protein" is used to refer to the glycoproteinaceous material of
the sp~ke, it has been characterised by D. Cavanagh, Journal.
General Virology 64, 1187-1191; 1787-1791; and 2577-2583 (1983)
as comprising two or three copies each of two glycopolypept~des,
Sl ~90,000 daltons) and S2 (84,000 daltons). The polypeptide
components of the glycopolypept~des Sl and S2 have been estimated
after enzymatic removal of oligosacchar1des to have a co~btned
molecular weight of approx~mately 125,000 daltons. It appears
that the spike protein is attached to the viral membrane by the
S2 polypeptide. Thus, the protein comprises an extra cellular
domain, a transmembrane domain and a cytoplasmic anchor domain.
European Patent Application Publication No. 218625A NRDC ~and
equiva1ent US Patent S,032,520 and corresponding application in
3~ Japan) d7scloses the clon~ng of cDNA sequences cod~ng for the
spike protein ~recursor as well as sequences coding specifically
SU13STITUTE SHEET
r~liG~,31~0332
CA2117468 1; ~ i ,9~
for the Sl and S2 polypept~des. Such a DNA mole~ule wh1ch codes
for an IBV sp1ke prote1n will hereinafter be referred to as "spike
DNA" for brev1ty. The d~sclosed splke DNA codes for the whole
spike prote1n, i.e. all 3 domains.
Summary of the ~nven~ion
It has now been found that it is unnecessary to clone the
whole sp~ke cDNA dlsclosed in EP 21862~A 1n order to obta~n an
~mmunological response: a truncated "sp1ke DNA" cons~derably
shorter ~n length may be cloned and expressed to produce a
polypept~de that w111 generate an immunolog~cal response. Thus,
the present ~nvent10n relates to a DNA molecule whlch codes
substantially for a truncated IBV spike protein polypeptide. The
truncated IBV sp~ke protein polypept~de produced as a resu1t of
the clon~ng and expression of a DNA molecule of the present
lnvent~on 1s characterised in lacking the transmembrane doma~n and
cytoplasmic anchor reg10n of the nat1ve IBV S2 spike protein
polypeptide, but otherwise encodtng the whole of the Sl and
rema1nder of the S2 polypept1de except that the Sl polypeptide can
lack up to lO amino ac~ds at the N-term~nal end and the truncated
S2 polypeptide can lack up to 10 am1no ac~ds at ~ts truncated end.
A DNA molecule according to the 1nventlon ~s shown in the
Sequence L~st1ng (SEQ ID N0: 1). Th~s DNA ~olecule was obta~ned
as a result of research on ~he M41 stra1n o~ IBV, but ~b~-~ s
expected that slmllarly truncated sptke pro~e~n of cDNA of other
IBV serotypes and stra~ns such as Beaudette, M~2, ~l~2,
Connect1cut 1solate A5968, Arkansas and Holland stralns H120, H5~,
Ma5, D207, D212, D3128 and D3896, whether or not exh~b~t~ng a h~gh
degree of homology w~th M41, wil1 express I~V sp1ke prote1n.
In referr~ng to a DNA molecule def~nëd as codlng substantially
for a truncated IBV sp1ke prote~n ~t w111 be appreclated that it
~s 1ntended not to exclude DNA flanklng sequences, wh~ch may be,
for example, cDNA to flank~ng sequences ~n the IBV RNA
genome (other than transmembrane sequences) or may be fore~gn
sequences der1ved from other genes, such as leader sequences
that may ass1st ~n dr1v1ng expresslon of the truncated polypept~de
or may be a short sequence of plasm~d DNA. Also, ~t ~s not
~ntended that the DNA molecule should necessar11y code for
~, 1..., r ,~
_ . _ .. _ , . ...
CA21 1 7468
~ 93~171Q~ PCT/GB93/00332
_ ~ _
amino acids extending right up to the 5'- term~nus or 3'-
truncated end. It may be possible to obtain expression of the
truncated sp~ke protein lacking say, up to 5 or eYen 10 of the
amino acids (30 nucleotides) at either end.
The invent~on also includes a vector containing the above
defined DNA molecule, including a cloning vector sllch as a
plasmid or phage or expression vector, preferably a pox virus
vector, and a host contalning the vector. Mammalian cells
contain~ng the above-defined DNA molecule, whether as naked DNA
or contained in a vector, are also included. Further, the
invention includes isolated biosynthetic truncated spike protein
polypeptide and ~ts expression from mammalian cells.
Brief descriDtion of the drawinas
Figures 1-17 show plasmid constructs of use in the
prepara~ion of DNA molecules of the present invention.
DescriDtion of the preferred embodiments
SEQ ID NO: 1 shows the complete nucleotide sequence of a cDNA
molecule of the invention obtained from IBV genomic RNA M41
strain. The IBV RNA of other strains is believed to be fairly
s~milar to that of M41, and therefore oligonucleotides derived
from DNA of the present invention can be used as primers for
sequencing RNA of other serotypes thus enabling truneated cDNA
for all or virtually all other serotypes to be prepared usin~-
~methods described hereinafter. Obviously, those serotypes in
wh kh the ent~re IBV spike protein cDNA has a h~gh degree of
nucleotide sequence homology with IBV M41 stra~n are slightly
pre~erred, as giving a w~der choice of potent~al oligonucleot~des.
The vectors included in the invention are clon~ng and
expression vectors. The DNA molecule of the present invention is
conv~niently multiplied by insertion in a prokaryotic vector, for
example pBR322, and cloning in an appropriate host such as a
bacterial host, especially E. coli. Alternatively, using
appropriate dlfferent vectors it could be multiplied in (say~
8acillus species, or a yeast. For expression, mammalian cells
can be transfectod by the calcium phosphate precip~tat~on method
or transformed by a viral vector. Viral vectors include
CA21 1 7468
WO 93/17109 PCI/GB93/003~2
-- 4 --
retroviruses and poxviruses such as fowlpox virus or vaccinia
virus.
A DNA molecule of the present invention may be prepared by
first obtaining full length IBV spike DNA in a suitable plasmid.
European Patent 218625A NRDC predicts the probable transmembrane
domain of the spike protein and indieates the region of DNA
coding for it. A suitable endonuclease restriction site near the
beginning of the DNA sequence coding for the transmembrane
domain, can ~hen be ident~fied. Uslng the des~red endonuclease,
the IBV spike DNA may be cleaved and the truncated DNA molecule
coding for the extracellular domain, introduced into a viral
vector as described below. Care is needed to ensure either that
the chosen restriction site is a unique one in the spike DNA. or
that a cloning procedure such as described in the Example is
devised to compensate. In the Example, a two-step cloning
~ proeess was used to overcome a second Styl site in the M41 spike
- DNA molecule. Alternatively, once it is known where the sequencecoding for the transmembrane domain begins, the truncat1On can be
brought about uslng Polymerase Chain Reaction (PCR) cloning or by
using ollgonucl20tide site-dlrected mutagenesis. In the latter
method, a stop codon is ~nserted at the desired positlon.
The truncated IBV sp~ke DNA can be introduced into the viral
vector as ~ollows. The DNA is inserted into a plasmid containi~
an appropriate non-essential region of poxvirus DNA, such as the
thymldine kinase gene of vacc1nia virus or ~nto any su~table
non-essent~al region of fowlpox virus, e.g. as described in
European Patent 353851A, so that the insert interrupts the NER
sequence. A poxvirus promoter, e.g. the vaccinia virus p7.5K
promoter, which is usable in vaccinia virus or avipoxviruses. or
a fowlpox virus promoter as described in our prior patent
appllcations publication Nos. W089/03879, W090104638 and
W091/02072, is also introduced into the NER sequence in such a
pos~tion that it will operate on the inserted truncated spike DNA
sequence. When an intergenic NER is used a "marker" gene w~th
its own promoter e.g. the lac Z gene will be inser~ed along with
the sequense coding for the truncated spike protein. When the
SU13STITUTE SHE~ET
CA21 1 7468
~o 93/17109 PCT/GB93/00332
poxvirus and the plasmid recombinant DNA are co-transfected into
a mammalian cel1, homologous recombination takes place between
the poxvirus NER, such as TK in vacclnia virus, or a said
non-essential region of fowlpox virus and the same gene or region
S present in the plasmid. Since the truncated IBV spike DNA has
thereby interrupted the poxvirus gene, viruses lacking the gene
expression product, such as TK, are selected. If the NER used is
an intergenic region, viruses expressing the truncated spike
protein ~11 be identified by the co-expression of the "marker"
gene e.g. blue plaques colonies if lac Z ~s the marker gene.
Once such a recombinant virus vector has been thus constructed it
can be used to introduce the truncated IBV spike DNA directly
into the desired host cells without the need for any separate
step of transfec~ing plasmid recsmbinant DNA into the cells.
In order to improve the expression of the truncated spike
protein it may be preferable to replace part or all of the
untranslated leader sequence upstream of the spike gene. The
leader sequence is the region between the TAATTATT of the
promoter sequence and the ATG initiation codon of the gene. By
replacing part or all of the native FPV IBV leader sequence wlth
leader sequences derived from other related vlruses such as
poxviruses it may be poss1ble to initiate stronger translation in
FPV.
Such leader sequences could be deriYed from: (1) part or all
of the sequences found downstream of other poxvlral promoters
e~g. the vaccinia v1rus p7.5 pro0oter (li) part or all of the
leader sequences from foreign gsnes that have been shown to be
well expressed in sells infected by the appropriate recombinant
poxviruses or (~ synthetlc sequences shown to promote
efficient translation in poxvirus-infected cells. The
replacement of appropriate sequences can be accomplished using
PCR cloning or by inserting synthetic oligonucleotides. The
choice of leader sequence to be used and the method of inser~ion
is well within the ability of skilled man. The Example 2
hereinafter illustrates how the procedure could be performed.
SUBSTITUTE SHEET
CA 2 1 1 7468
W O 93~17109 PCT/GB93/00
The invention therefore further relates to a vector wherein
containing part or all of a sequence found downstream of a
poxvirus promoter, not being the poxvirus promoter of use in the
vector, between the promoter and the IBV DNA Molecule.
With a view ultimately to obtaining expression of the
recombinant virus in ViYo, the preferred poxvirus is fowlpox
virus. It may be that the inserted truncated IBV DNA contains a
sequence, which, in the fowlpox vector, leads to premature
termination of transcription. In this case, the truncated sp~ke
DNA would have to be mod~fied slightly by one or two nucleotides,
thereby to allow transcription to proceed along the full length
of the gene.
The vector can be introduced into any appropriate host by any
method known in recombinant DNA technology. Hosts include
E. coli, Bacillus 5pp, animal cells such as avian or mammalian
cells and yeasts. The method of introduction can be transformed
by a plasmid or cosmid vector, or infection by a phage or viral
vector etc. as known in recombinant DNA technology.
The following Examples illustrate the invention. All
temperatures are in C.
EXAMPLE 1
I. Prepara~ion ~f a full lenqth IBV s~ike prot~n cDNA from M41
~tra~n
Example 2 of European Patent Application Publicat~on
No. 218625 (NRDC) describes the preparation of cDNA coding for
the spike protein precursor of IBV strain M41. It describes
therein the preparat~on of plasmids pMB276 and pMB2~ containing
the entire M41 sp~ke protein cDNA sequence.
An initial step in the preparation of a DNA molecule encoding
a truncated IBV spike protein was to join pMB276 and pMB250 to
produce a full length clone of the IBV M41 spike gene.
1. Joinlnq pMB276 and pMB250 a~ a shared Ndel site to
produce a ~ull lenath çLone of the IBV M41 s~ike aene ~in DMB374)
Plasmids pMB276 and pMB250 were digested w~th Ndel (20 units)
in 50mM tris-HCl pH 8.0, lOmM MgC12, SOmM NaCl, final volume 20~1.
SUBSTITU I E SHEET
CA 2 1 1 74 68
YYO 93/1710g P~T/GB93/00332
The digested DNA was then phenol-extracted with an equal volume
of TE-saturated phenol, ether extracted twice with an equal
volume o~ water-saturated ether, then ethanol- precip~tated. The
precip~tated DNA was resuspended in 15~1 water. Then 2.5~1 of
each digest were ligated together in a total volume of 10~1 in
50mM tris~HCl pH 7.5, lOmM MgC12, lOmM ~TT, lmM ATP, 1 unit T4
DNA ligase at 4C overn~ght. Ligated DNA, in 1~1 of the ligation
mix, was transformed into competent E. col1 DH5 and transformed
bacteria were selected on agar plates containing tetracycline.
Transformant colonies were grown in L broth plus tetracycline
and DNA was isolated therefrom using a standard procedure
described by Holmes and Quigley (1981), Analytical Biochemistry
114: 193-197. Following digestion of the isolated DNA with Ndel
and agarose gel electrophoresis, i~ was apparent that, of 48
clones screened, one Cno. 17) had ~nher~ted the desired fragments
from the parental plasmids, viz. a fragment of circa 6kbp from
pM3276, Fig. 1 and a fragment of about 4kbp from pMB 250, F~g. 2.
The desired recombinant plasmid would also have a fragment,
following Pstl d1gestion, equivalent to the length of pBR322
(~stl sites flank the M41 spike cDNA). Analysis of clone 17
: showed that ~t d~d not have a pBR322-s~zed Pstl fragment,
:~ indicat7ng that the two Ndel fragmen~s had tlgated together in
the wrong relative orientaff 0n. Clone 17 DNA was therefor~
digested with Ndel and religated (using procedures -described
above) to allow isolation of recomblnan~s with the two Ndel
fragments in the correct or3entat~on. Analys~s of Pstl-digested
DNA from a number of clones showed that about 50X had religated
to glve the correct or3entat30n. One of these clones was saved,
as pMB374, Fig. 3.
2. Clonlna _the IBV M41 sPike Qene under cQntrJ2L~ hp
vacc1nia v1rus ~7.5 Dromoter to make DGSS2
The IBV M41 spike protein gene was cut out of pMB374 by
digest10n of the plasmid with Tthlll 1, see Fig. 3, in lOmM
trls-Hcl pH 7.4, lOmM MgC12~ 50mM NaCl, lOmM ~-mercaptoethanol,
at 65C ln a final volumæ o~ 20~1. The DNA was made blunt-ended
SUE~STITIJTE SHEET
CA21 17468
W O 93/17109 PCT/GB93/003
by the addition of 0.025mM dATP, dCTP, dGTP, dTTP and 5 units of
Klenow polymerase, followed by ineubation for lh at room
temperature. The digestion products were electrophore~ed on an
agarose gel using standard procedures as described by Maniatis
et al., (1982) in "Molecu1ar cloning: a laboratory manual" ~Cold
Spring Harbor Laboratory) and a Skb fragment, containing the
sp~ke gene was purified using "Geneclean II" (Blo lOl~ as per
supplier's ~nstructions. The purified DNA was then cloned into
the ~m~l site of pGS20 (from Dr. G. L. Smith, Sir William Dunn
School of Pathology, University of Oxford, South Parks Road,
Oxford OXl 3RE, as described in Mackett, Smith & Moss (l984),
J. V~rol. 49, 857-B64) to make pGSS2, see Fig. 4.
II. TruncatiQn o~ M41 spike gene
It was desired to truncate the spike protein gene so that the
lS protein would not carry a transmembrane segment and a cytoplasmic
domain. This could be conveniently achieved by cutting the gene
; at the ~y1 site (position 4384 in pGSS2). As there is another
StYl s1te within the gene, a two step process was devised. This
involved the transfer of the spike prote~n gene sequences
(wlthout the p7.5 promoter~ to pUCl9 (Yanlsch-Perron, Vie1ra &
Messing~ 1985, Gene 33, 103-ll9). Finally the truncated spike
gene was transferred to the fowlpoxvirus expression plasmid,
pEFL29. These steps are descr1bed below. .
1. Transfer of the ~yl fra~ment within the spike qene frQm
D~SSZ to pU~l9 to make pUC/M4lStv
pGSS2 was digested with Stvl, the DNA was made blunt-ended
with Klenow polymerase and the 1.95kb fragment (2430-43843 was
; recovered and pur~f~ed. This fragment was ligated into pUCl9
di gested wi th ~m~l- Recombi nants carrying the inserted fragment
were isolated and the orientation of the inserted fragment was
checked by digestion of their plasmid DNA with Mlul and BamHl.
The required recombinant had a small Mlul/BamHl fragment of 480
bp (and not 1480bp) and was given the title pUC/M41Sty, Fig. 5
(note that ligation of the blunt-ended Styl fragment into the
~m~l site restores the Stvl sites but not the Smal site~.
- SUBSTITUl-E SWEEi~l'
` CA2117468
~0 93/17109 PCr/GB93/00332
2. Cloninq ~he N-terminal part of the sDike qene from DG~S2
intQ pUC/M41 S~y ~o g~ve pU~/M41 Bam-Stv (conta1ninq spike
se~u~nces ~ mm the N-terminus to the C-terminal ~tyl site)
Plasmids pGSS2, Fig. 4, and pUC/M41Sty, Fig. 5, were both
d~gested with BamHl and Afl2 and fragments of 2.2kb and 3.8kb,
respectlvely, were recovered. The purified fra~ments were
liyated together and recombtnants were isolated. The required
recombinant (tltled pUCJM41 Bam-Sty3, Fig. 6, had the 0.85kb
BamHllAfl2 fragment of pUC/M41Sty replaced by a 2.2kb fragment
from pGSS2, Fig. 6.
3. Tr~n~fer of ~he truncated s~ik~ qene from pUC/M41
Bam-~y into the f~wlDoxvirus exDression vectQr. ~EFL29. to aive
uEFS17
The entire truncated spike gene sequences were cut out of
pUC/M41 Bam-Sty using ~mHl and E~oRl. Followlng repair of the
ends of the DNA w~th Klenow polymerase, the 3.3kb fragment was
isola~ed. purified and blunt-end liga~ed ~nto pEFL29, F~g. 7,
digested with ~m~l- Recomb1nants were screened by digestion with
BamHl/B~12 to check that the sp~ke gene insert was in the correct
or~entation relat~ve to the p7.5 promoter in pEFL29. Correct
recombinants were titled pEFSl7, Fig. 8. (The derivation of
pEFL2~ ~s descr7bed below).
4. Derivatlon of pEFL29 _~
A DNA fragment containing the fowlpoxvirus 4b- promoter
dr1vlng a l~Z reporter gene was cut sut of plasmid pNM~b30 (see
the releYant fowlpox virus promoter patent spec~f~cat~on
(W089l03879), page 35, Table 2~ using ~QRl and ~L~l. The
fragment was end-repaired and ~as then blunt-end ligated ~nto the
end-repaired B~12 s~te of a plasmid containiny part of the
terminal ~mHl fragment of fowlpoxvirus (p83ME, described in
Boursnell et al., l990, J. Gen. Virol. 71, 621-628) to create
plasmid pEFLlO.
The vacc~nia virus p7.5 promoter was then introduced, on
a 300bp çQRl (end-repaired) DNA fragment from pGSZ0 (see above),
into the ~ç~l site of pEFLlO. A recombinant with the
5UBSTITUTE SHE~T
CA21 1 7468
w o 93~17109 PCT/GBs3/003.~?~
-- 10 --
p7.5 promoter in such an orientation that transcript~on from it
is initiated in the opposite direction to that from the
fowlpoxvirus 4b promoter, identified by restriction analysis
using BamHl, was titled pEFL29.
5. Isolation of recombinant fowlpoxvirus exDressina the
truncated IBV M41 spike gene
Chick embryo f~broblasts (CEFs), at 80% confluence, were
infected with the Duphar "Poxine" strain of fowlpoxvirus at a
~ultiplic~ty of infection (m.o.i.) of 1. At 4h post-infection,
pEFS17 DNA (10~9 per 25cm2 flask) was introduced to the cells
using the 'Lipofectin' method ~8RL) under manufacturer~s
instructions. Five days post-infection, when there was complete
cytopathic effect, the cells were harvested. Virus, released
from the cells by freeze/thawing three times, was used at various
dilutions to infect CEFs which were then overlaid w1th agarose to
allow plaques to form. When plaques were vis1ble the plates were
overlaid with X-gal agarose. Two days la~er, blue plaques were
p~cked and virus was released by freeze/thawing. The virus was
titrated again, overlaid with X-gal agarose and blue plaques were
picked again. This procedure was repeated three more times.
Finally two plaques (fpl74Pxllll and fpl74Px1121) were chosen for
further characterisation.
III. Characterisat~on of the fowlpoxvirus/EFS17 recombinant
~1!~
Z~ 1. (a) Plaque hybridisation analysls
These viruses were propagated and plaques on CEFs were onee
more obtai ned . The agarose overlay was removed and a
nitrocellulose f~lter was applied to the cell sheet. A piece
of 3MM filter paper, soaked in 20X SSC, was applied to the
nitrocellu1Ose filter. The nitrocellulose filter was then
removed and baked at 80C in a vacuum oven. The filters were
then probed with a 32P-radiolabelled probe speci~ic ~or the IBV
M41 spike protein gene, to verify that the recombinan~ fowlpox
viruses carried the IBV M41 spike protein gene.
SUBSTITUTE SH8~ET
CA2i i 7468
YVO 93/17109 pcT/GBs3/oo332
2. (b) Radio-immunoprecipitation assay (RIPA)
CEFs were infected with the fpEFSl7 recombinant viruses (or
with a control 'poxine'/lacZ recombinant virus or mock-infected)
at a m.o.i. of 10. At 24h post-infection the tissue culture
medium was replaced with methionine-free medium to 'starve' the
cells (i.e. to deplete the cells of their intracellular
methion~ne pool~ for lh. The cells were then labelled with
35S-methionine (lOO~Ci) for 3h. Then they were harvested, washed
and lysed ~n RIPA buffer (the RIPA procedures used are described
in detail in "Antibodies: a laboratory manual", Harlow and Lane
~1988), Cold Spring Harbor Laboratory, New York). A polyclonal
serum raised in rabbits against purified IBV M41 spike protein
was added to the clarified extracts and immune complexes were
precipitated with protein-A/Sepharose.
The protein-A/Sepharose was washed thrice with RIPA buffer, then
resuspended in SDS-PAGE sample buffer and boiled for 3 min. The
samples were then applied to a 5-lOZ gradient SDS-PAGE gel and
electrophoresed. The gel was fixed and exposed by fluorography.
RIPA analysls sho~ed that cells infected with the fpEFSl7
recomb~nant v~ruses, but not those infected ~th control
'pox~ne'/lacZ recombinant nor uninfected cells, synthesised a new
prote~n (apparent molecular we~ght about 160K). The band
appeared 'fuzzy', characteristic of an extensively glycosylate~.
prote~n sueh as the spike protein. When the infected eells were
'starved' and labelled in the presence of tunicamycin, an
~nhibitor of N-11nked glycosylation, a fa~nt band was seen at
about 120K (the predicted size of the unmodified primary
translation product) but most of the new product appeared as two
closely migrating b~nds of 90-9SK, suggesff ng that the
30 unglycosylated protein was unstable and was being cleaved by
protease activity.
SUBSTITUTE SHEET
CA21 1 7468
WO 93/17109 PCl/GB93/003
- 12 -
EXAMPLE 2
The Example below describes the replacement of the
untranslated IBV spike sequences with sequences derived from part
of the leader downstream of the p7.5 promoter, by cloning
S synthetic oligonucleotides between the BamHI site in the leader
and a Spel site near the 5' end of the I~V spike coding
sequence. The complete leader is then cloned upstream of the
truncated IBV spike gene from pEFS 17 to give pEFS 20.
In summary the 83 base pair BamHI-Spel fragment (SEQ ID N0 3)
10in pEFS17 i 5 replaced with a synthetic leader based on p7.5
(SEQ ID N0 4) using the oligonucleotides MAS-H7 and MAS-H8
(SEQ ID 5 and 6 respectively).
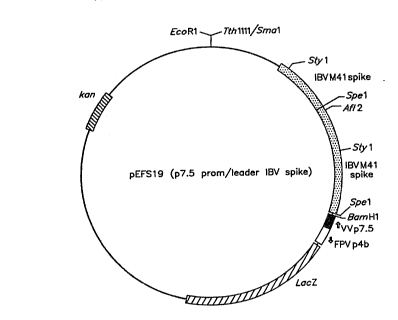
1) Replacinq non-translated leader from IBV in_pGSS2 with leader
sequences from the va~cinia virus D7.5 promoter
15Plasmid pGSS2 (Fig. 4) was digested with BamHI (1059) and
Spel (3358), and fragments of lOkb and 2.2kb (Fig. 9 were
recovered. To anneal synthetic oligonucleotides MAS-H7 and
MAS-H8, 50 pmol of each were mixed in 10~1 water. They were then
bolled for 3 minutes and allowed to cool slowly to room
temperature. The annealed oligonucleotide duplex (0.2 to 5 pmol)
was then ligated to the 10 kb BamHI-Spel fragment from pGSS2.
The required recombinant, pGSS3 (~ig. 10), had retained the BamHI
and Spel sites but had deleted a 2.2 kb Spel fragment relative t~
pGSS2.
2) Replasina the ~eleted 2.2 kb S~el fraament from vGSS2 into
DGS~3 ~o make p~SS4
The 2.2 kb Spel fragment from pGSS2 (Fig 11) was ligated into
pGSS3 11nearised w~th Spel. The presence and orientation of the
inserted Spel fragment in the resultant recombinant, pGSS4
~Fig. 12), was verified by digestion ~ith Spel, Aflll, Mlul or
BamHI/Sall.
5UBSTITUTE SHEET
~A2 1 1 ~4 68
~0 93/17109 PCT/GB93/00332
3) Cloninq_~he IBV s~ike qene with the new leader from pGSS4
into the eX~ressiQn vector. pEFL29
Plasmid pGSS4 was digested with BamHI and EcoRI, repaired
with Klenow polymerase then a 4.~ kb fragment ~Fig. l3) was
recovered and ligated into pEFL29 (Fig. 7) digested with Smal.
The presence and orientation of the spike gene insert in the
desired recombinant, pEFSl9 (Fig. 14), was checked by digestion
with BamHl, EcoRI, Styl or BamHI/Styl.
4) Comb~ninq the new leader and 5'-terminus of the I~V spike
qene (fr~m DEF~l9) with the ~-terminus nf the truncated sDike
qene (from pEF~17)
Plasmids pEFS17 and pEFSl9 were digested with Ncol and BglII
then 3 kb (Fig. l5) and ll.8 kb (Fig. 16) fragments,
respectively, were recovered and ligated together. The required
lS recombinant pEFS20 (Fig. 17) was checked by digestion with Kpnl,
BamHI, Styl and BamHI/Styl.
Recomb~nant fowlpox viruses were derived, using pEFS20, and
analysed as described above in Example l.III for pEFSl7.
5LIBE;TITUTE SHEET
CA21 1 7468
WO93/17109 PCT/GB93/00~
_ ",,~ _
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: British, Technology Group Ltd
(ii) TITLE OF INVENTION: IBV Spike Protein (2)
(iii) NUMBER OF SEQUENCES~ 6
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: British Technology Group Ltd
(B) STREET: l0l Newington Causeway
(C) CITY: London
(E) COUNTRY: U.K.
(F) ZIP: SEl 6BU
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy ~isk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: PatentIn Release #l.0, Version ~l.25
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER:
(B~ FI~ING DATE:
(C) CLASSIFICATION:
(vii~ PRIOR ~PPLICATION DATA:
(A) APPLICATION NUMBER: GB 9203509.6
~B) FILING DATE: l9-FEB-1992
tviii) ATTORNEY/AGENT INFORMATION:
t~) NAM~: Percy, R K
(C3 REFERENCE/DOCKET NUMBER: 135324 ~
(ix) TELECO~MUNICATION INFORMATION:
(A) TELEPHONE: 017 403 6666
(B) TELEFAX: 071 403 7568
(2~ INFORMATION FOR SEQ ID NO:l:
(i) SEQUENCE CHARACTERISTICS:
(A) LEN~TH: 3281 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii~ MOLECULE TYPE: cDNA
(iv) ANTI-SENSE: NO
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Infectious bronchitis virus
(B) STRAI~: M4l
SUBSTITU I t SHEET
CA21 1 7468
93~17109 PCT/GB~3/00332
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: l..3281
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:l:
ATG TTG GTA ACA CCT CTT TTA CTA GTG ACT CTT TTG TGT GTA CTA TGT
48
Met Leu Val Thr Pro Leu Leu Leu Val Thr Leu Leu Cys Val Leu Cys
l 5 lO 15
AGT GCT GCT TTG T~T CAC AGT AGT TCT TAC GTT TAC TAC TAC CAA AGT
96
Ser Ala Ala Leu Tyr A~p Ser Ser Ser Tyr Val Tyr Tyr Tyr Gln Ser
GCC TTT AGA C~A CCT AAT GGT TGG CAT TTA CAC GGG GGT GCT TAT GCG
144
Ala Phe Arg Pro Pro Asn Gly Trp His Leu His Gly Gly Ala Tyr Ala
GTA GTT AAT ATT TCT AGC GAA TCT AAT AAT GCA GGC TCT TCA CCT GGG
192
Val Val Asn Ile S~r Ser Glu Ser Asn Asn Ala Gly Ser Ser Pro Gly
TGT ATT GTT GGT ACT ATT CAT GGT GGT CGT GTT GTT AAT GCT TCT T
240
Cys Ile Val Gly Thr Ile His Gly Gly Arg Val Val ~sn Al~ Ser Ser
6~ 70 75 ~9
ATA GCT ATG ACG GCA CCG TCA TCA GGT ATG GCT TGG TCT AGC AGT CAG
288
Ile Ala Met Thr Ala Pro Ser Ser Gly Met Ala Trp Ser Ser Ser Gln
go 95
TTT TGT ACT GCA CAC TGT AAC TTT TCA GAT ACT ACA GTG TTT GTT ACA
336
Phe Cys Thr Ala His Cys Asn Phe Ser Asp Thr Thr Val Phe Val Thr
lO0 105 lt0
CAT TGT TAT AAA TAT GAT GGG TGT CCT ATA ACT GGC ATG CGT CAA AAG
3~4
SUBSTITUTE 5HEET
CA~ I 1 7468
W093/17l09 PCT/GB93/00~
His Cys Tyr Lys Tyr Asp Gly Cys Pr~ Ile Thr Gly Met Arg Gln Lys
115 lZ0 125
AAT TTT TTA CGT GTT TCT GCT ATG AAA AAT GGC CAG CTT TTC TAT AAT
432
Asn Phe Leu Arg Val Ser Ala Met Lys Asn Gly Gln Leu Phe Tyr Asn
130 135 140
TTA ACA GTT AGT ~TA GCT AAG TAC CCT ACT TTT AAA TCA TTT CAG TGT
48~
Leu Thr Val Ser Val Ala Lys Tyr Pro Thr Phe Lys Ser Phe Gln Cys
145 150 155 160
GTT AAT AAT TTA ACA TCC GTA TAT TTA AAT GGT GAT CTT GTT TAC ACC
528
Val Asn Asn Leu Thr Ser Val Tyr Leu Asn Gly Asp Leu Val Tyr Thr
165 170 175
TCT AAT GAG ACC ACA GAT GTT ACA TCT GCA GGT GTT TAT TTT AAA GCT
576
Ser Asn Glu Thr Thr Asp Val Thr Ser Ala Gly Val Tyr Phe Lys Ala
1~0 185 190
GGT GGA CCT ATA ACT TAT AAA GTT ATG AGA G~A GTT AAA GCC CTG GCT
624
Gly Gly P~o Ile Thr Tyr Lys Val Met Arg Glu Val Lys Ala Leu A~
195 20~ 205
TAT TTT GTT AAT GGT ACT GCA CAA GAT GTT ATT TTG TGT GAT GGA TCA
672
Tyr Phe Val Asn Gly Thr Ala Gln Asp Val Ile Leu Cys Asp Gly Ser
210 215 220
CCT AGA GGC TTG TTA GCA TGC CAG TAT AAT ACT GGC AAT TTT TCA GAT
720
Pro Arg Gly Leu Leu Ala Cys Gln Tyr Asn Thr Gly Asn Phe Ser Asp
225 230 235 240
~GC TTT TAT CCT TTT ATT AAT AGT AGT TTA GTT AAG CAG AAG TTT ATT
76~
Gly Phe Tyr Pro Phe Ile Asn Ser Ser Leu Val Lys Gln Lys Phe Ile
~;UE~STITUTE SHEET
CA2i 1 7468
.~O 93/17l09 PCr/GE~93/00332
245 250 255
GTC TAT CGT GAA AAT AGT GTT AAT ACT ACT TTT ACG TTA CAC AAT TTC
816
Val Tyr Arg Glu Asn Ser Val Asn Thr Thr Phe Thr Leu His Asn Phe
260 265 270
ACT TTT CAT AAT GAG ACT GGC GC:C AAC CCT AAT CCT AGT GGT GTT CAG
864
Thr Phe His Asn Glu Thr Gly Ala Asn Pro Asn Pro Ser Gly Val Gln
275 280 285
AAT ATT CA~ ACT TAC CAA ACA CAA ACA GCT CAG AGT GGT TAT TAT AAT
gl2
Asn ïle. Gln Thr Tyr Gln Thr Gln Thr Ala Gln Ser Gly Tyr Tyr Asn
290 295 300
TTT AAT TTT TCC TTT CTG AGT AGT TTT GTT TAT AAG GAG TCT AAT TTT
960
Phe Asn Phe Ser Phe Leu Ser Ser Phe Val Tyr Lys Glu Ser Asn Phe
3~5 310 315 320
ATG TAT GGA TCT TAT CAC CC}~ AGT TGT AAT TTT AGA CTA GAA ACT ATT
10~8
Met Tyr Gly Ser Tyr His Pro Ser Cys Asn Phe Arg Leu Glu Thr I le
325 330 335_.<,
AAT A~T GGC TTG TGG TTT AAT TCA CTT TCA GTT TCA ATT GCT TAC GGT
1056
Asn Asn Gly Leu Trp Phe Asn Sor Leu Ser Val Ser Ile Ala Tyr ~;ly
340 345 350
CCT CTT CAA GGT GGT TGC AAG CAA TCT GTC TTT AGT GGT AGA GCA ACT
1104
Pro Leu Gln Gly Gly Cys Lys Gln Ser Val Phe Ser Gly Arg Ala Thr
3S5 360 365
TGT TGT TAT GCT TAT TCA TAT GGA GGT CCT TCG CTG TGT AAA GGT GTT
1152
Cys Cys Tyr Ala Tyr Ser Tyr Gly Gly Pro Ser Leu Cys Lys Gly Val
37~ 375 380
SUBSTITUTE SHEET
~;A~ i /46~
WO 93/171Og ~5 PCI~GB93/003?^`
TAT TCA GGT GAG TTA GAT CTT AAT TTT GAA TGT GGA CTG TTA GTT TAT
1200
Tyr Ser Gly Glu Leu Asp Leu Asn Phe Glu Cys Gly Leu Leu V~l Tyr
38~ 390 395 40~
GTT ACT AAG AGC GGT GGC TCT CGT ATA CAA ACA GCC ACT GAA CCG CCA
1248
Val Thr Lys Ser Gly Gly Ser Arg Ile Gln Thr Ala Thr Glu Pro Pro
405 410 41S
GTT ATA ACT CGA CAC AAT TAT ~AT AAT ATT ACT TTA AAT ACT TGT GTT
I296 .
Val Ile Thr Arg His Asn Tyr Asn Asn Ile Thr Leu Asn Thr Cys Val
. 420 425 430
GAT TAT AAT ATA TAT GGC AGA ACT GGC CAA GGT TTT ATT ACT AAT GTA
1344
Asp Tyr Asn Ile Tyr Gly Arg Thr Gly Gln Gly Phe Ile Thr Asn Val
435 440 445
ACC ~AC TCA GCT GTT AGT TAT AAT TAT CTA GCA GAC GCA GGT TTG GCT
13g2
Thr Asp Ser Ala Val Ser Tyr Asn Tyr Leu Ala ~sp Ala Gly Leu Ala
450 455 460
ATT ~T~ GAT ACA TCT GGT TCC ATA GAC ATC TTT GTT GTA CAA GGT GAA~
1440
Ile Leu Asp Thr Ser Gly Ser Ile Asp Ile Phe Val Val `Gln Gly Glu
465 470 475 ~80
TAT GGT CTT ACT TAT TAT AAG GTT AAC CCT TGC GAA GAT GTC AAC CAG
1488
Tyr ~ly Leu Thr Tyr Tyr Lys Val Asn Pro Cys Glu Asp Val Asn Gln
485 490 495
CAG TTT GT;~ GTT TCT GGT GGT ~ TTA GTA GGT ATT CTT ACT TCA CGT
153~
Gln Phe Val Val Ser Gly Gly Lys Leu Val Gly Ile Leu Thr Ser Arg
00 505 S10
~AT GAG ACT GGT TCT CAG CTT CTT GAG AAC CAG TTT TAC ATT AAA ATC
158 4
SU13STITUTE SHEET
CA27 i 7468
~v~93~17109 ~q_ PCTJGB93/00332
Asn Glu Thr Gly Ser Gln Leu Leu Glu Asn Gln Phe Tyr Ile Lys Ile
515 520 525
ACT AAT GGA ACA CGT CGT TTT AGA CGT TCT ATT ACT GAA AAT GTT GCA
1632
Thr Asn Gly Thr Arg Arg Phe Arg Arg Ser Ile Thr Glu Asn Val Ala
530 535 540
AAT TGC CCT TAT GTT AGT TAT GGT AAG TTT TGT ATA AAA CCT GAT GGT
1680
Asn Cys Pro Tyr Val Ser Tyr Gly Lys PhP Cys Ile Lys Pro Asp Gly
545 550 555 560
TCA ATT GCC ACA ATA GTA CCA AAA CAA TTG GAA CAG TTT GTG GCA CCT
1728
Ser Ile Ala Thr Ile Val Pro Lys Gln Leu Glu Gln Phe Val Ala Pro
565 ~70 575
TTA CTT AAT GTT ACT GAA AAT GTG CTC ATA ~CT AAC AGT TTT AAT TTA
1776
Leu Leu Asn Val Thr Glu Asn Val Leu Ile Pro Asn Ser Phe Asn Leu
5~0 585 590
ACT GTT ACA GAT GA& TAC ATA CAA ACG CGT ATG GAT AAG GTC CAA ATT
1~24
Thr Yal Thr Asp Glu Tyr Ile Gln Thr Arg Met Asp Lys Val Gln ~e
595 600 60~ ~
AAT TGT CTG CAG TAT GTT TGT GGC AAT TCT CTG ~AT TGT AGA GAT TTG
1872
Asn Cy5 Leu Gln Tyr Val Cys Gly A~n Ser Leu Asp Cy5 Arg Asp Leu
610 6lS 620
TTT CAA CAA TAT GGG CCT GTT TGT GAC AAC ATA TTG TCT GTA GTA AAT
1920
Phe Gln Gln Tyr Gly Pro Val Cys Asp Asn Ile Leu Ser Val Val Asn
6~5 630 635 640
AGT ATT GGT CAA AAA GAA GAT ATG GAA CTT TTG AAT TTC TAT TCT TCT
1968
Ser Ile Gly Gln Lys Glu Asp Met Glu Leu Leu Asn Phe Tyr Ser Ser
~:IJR~:TITII~ ~ F F T
CA21 1 7468
WOg3/l7l09 PCr/GB93/00
_ 2O-
645 65~ 655
ACT AAA CCG GCT GGT TTT AAT ACA CCA TTT CTT AGT AAT GTT AGC ACT
2016
Thr Lys Pro Ala Gly Phe Asn Thr Pro Ph Leu Ser Asn Val Ser Thr
660 665 670
GGT GAG TTT AAT ATT TCT CTT CTG TTA ACA ACT CCT AGT AGT CCT AGA
2064
Gly Glu Phe Asn Ile Ser Leu Leu Leu Thr Thr Pro Ser Ser Pro Arg
675 6~0 685
AGG CGT TCT TTT ATT GAA GAC CTT CTA TTT ACA AGC GTT GAA TCT GTT
2112
Arg Arg Ser Phe Ile Glu Asp Leu Leu Phe Thr Ser Yal Glu Ser Val
690 ~95 700
GGA TTA CCA ACA GAT GAC GCA TAC AAA AAT TGC ACT GCA GGA CCT TTA
2160
Gly Leu Pro Thr Asp Asp Ala Tyr Lys Asn Cys Thr Ala Gly Pro Leu
705 710 715 ~20
GGT TTT CTT AAG GAC CTT GCG T&T GCT CGT GAA TAT AAT GGT TTG CTT
2208
Gly Phe Leu Lys Asp Leu Ala Cys Ala Arg Glu Tyr Asn Gly Leu Leu
725 730 735 ~,
GTG TTG CCT CCC ATT ATA ACA GCA GAA ATG CAA ACT TTG TAT ACT AGT
2256
Val Leu Pro Pro Ile Ile Thr Ala Glu Met Gln Thr Leu Tyr Thr Ser
740 745 75~
TCT CTA GTA GCT TCT ATG GCT TTT GGT GGT ATT ACT GCA GCT GGT GCT
~304
Ser Leu Val Ala Ser Met Ala Phe Çly Gly Ile Thr Ala Ala Gly Ala
755 760 765
ATA CCT TTT GCC ACA CAA CTG CAG GCT AGA ATT AAT CAC TTG GGT ATT
235~
Ile Pro Phe Ala Thr Gln Leu Gln Ala Arg Ile Asn His Leu Gly Ile
770 77~ 780
SUBSTITUTE 5HEE~T
CA-2 i i 74h8
~'~93/17109 PCT/GB93~00332
ACC CAG TCA CTT TT~ TTG AAG AAT CAA GAA AAA ATT GCT GCT TCC TTT
2400
Thr Gln Ser Leu Leu Leu Lys Asn Gln Glu Lys Ile Ala Ala Ser Phe
785 ?90 795 800
AAT AAG GCC ATT GGT CGT ATG CAG GAA GGT TTT AGA AGT ACA TCT CTA
2448
Asn Lys Ala Ile Gly Arg Met Gln Glu Gly Phe Arg Ser Thr Ser Leu
805 810 815
GCA TTA CAA CAA ATT CAA GAT GTT GTT AAT AAG CAG AGT GCT ATT CTT
24g6
Ala Leu Gln Gln Ile Gln Asp Val Val Asn Lys Gln Ser Ala Ile Leu
820 825 830
ACT GAG ACT ATG GCA TCA CTT AAT AAA AAT TTT GGT GCT ATT TCT TCT
2544
Thr Glu Thr Met ~la Ser Leu Asn Lys Asn Phe Gly Ala Ile Ser Ser
835 840 845
GTG ATT CAA G~A ATC TAC CAG CAA CTT GAC GCC ATA CAA GCA AAT GCT
25~2
Val Ile Gln Glu Ile Tyr Gln Gln Leu Asp Ala Ile Gln Ala Asn Ala
8~0 855 860
CAA GTG GAT CGT CTT ATA ACT GGT AGA TTG TCA TCA CTT TCT GTT T~
2640
Gln Val A~p Arg ~eu Ile Thr Gly Arg Leu Ser S~r Leu Ser Val Leu
8~5 870 ~75 880
GCA TCT GCT AAG CAG GCG GAG CAT ATT AGA GTG TCA CA~ CAG CGT GAG
2688
Ala 5er ~la ~ys Gln Ala Glu His Ile Arg Val Ser Gln Gln Arg Glu
885 890 ~95
TTA GCT ACT CAG AAA ATT AAT GAG TGT GTT AAG TCA CAG TCT ATT AGG
2736
Leu Ala Thr Gln Lys Ile Asn Glu Cys Val Lys Ser Gln Ser Ile Arg
9~0 905 9lO
TAC TCC TTT TGT GGT AAT G~A CGA CAT GTT CTA ACC ATA CCG CAA AAT
27~4
SUBSTITIJTE SHEET
C A 2 i ~ 7 4 ~
WO93~7109 -~2- PCT/GB93/00~
Tyr `Ser Phe Cys Gly Asn Gly Arg His Val Leu Thr Ile Pro Gln Asn
915 920 925
GCA CCT AAT GGT ATA GTG TTT ATA CAC TTT TCT TAT ACT CCA GAT AGT
2~32
Ala Pro Asn Gly Ile Val Phe Ile His Phe Ser Tyr Thr Pro Asp Ser
930 935 940
TTT GTT AAT GTT ACT GCA ATA GTG GGT TTT TGT GTA ~AG CCA GCT AAT
28~0
Phe Val Asn Val Thr Ala Ile Val Gly Phe Cys Val Lys Pro Ala Asn
945 950 955 960
GCT AGT CAG TAT GCA ATA GTA CCC GCT AAT GGT AGG GGT ATT TTT ATA
2928
Ala Ser Gln Tyr Ala Ile Val Pro Ala Asn ~ly Arg Gly Ile Phe Ile
965 970 975
CAA GTT AAT GGT P~GT TAC TAC ATC ACA GCA CGA GAT ATG TAT ATG CCA
29~6
Gln Val ~sn Gly Ser Tyr Tyr Ile Thr Ala Arg Asp Met Tyr Met Pro
980 985 990
AGA GCT ATT ACT GCA GGA GAT ATA GTT ACG CTT ACT TCT TGT CAA GCA
3024
Arg Ala Ile Thr Ala Gly Asp Ile Val Thr Leu Thr Ser Cys Gln Ala~
9~5 1000 1005 -
AAT TAT GTA AGT GTA AAT AAG ACC GTC ATT ACT ACA TTC GTA GAC AAT
3072
Asn Tyr Val Ser Val Asn Lys Thr Val Ile Thr Thr Phe Val Asp Asn
1010 1015 1020
GAT GAT TTT GAT TTT AP~T GAC GAA TTG TCA AAA TGG TGG AAT GAC ACT
3120
Asp Asp Phe Asp Phe Asn Asp Glu Leu Ser Lys Trp Trp Asn Asp Thr
1~25 1030 1035 1040
AAG CAT GAC~ CTA CCA GAC TTT GAC AAA TTC AAT TAC ACA GTA CCT ATA
3 168
Ly5 His Glu Leu Pro Asp Phe Asp Lys Phe Asn Tyr Thr Val Pro Ile
1045 1050 10~5
SUE~STITLJTE SHEET
C~2i 1 7468
`V093/17109 PCT/GB93/00332
-~3-
CTT GAC ATT GAT AGT GAA ATT GAT CGT ATT CAA GGC GTT ATA CAG GGT
3216
Leu Asp Ile Asp Ser Glu Ile Asp Arg Ile Gln Gly Val Ile Gln Gly
1060 1065 1070
CTT AAT GAC TCT TTA ATA GAC CTT GAA AAA CTT TCA ATA CTC AAA ACT
3264
Leu Asn Asp Ser Leu Ile Asp ~u Glu Lys Leu Ser Ile Leu Lys Thr
1075 1080 1085
TAT ATT A~G TGG CCA AG
3281
Tyr Ile Lys Trp Pro
1090
(2) INFORMATION FOR SEQ ID NO:2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: lO93 amino acids
(B) TYPE: amino acid
~D) TOPOLOGY: linear
( ii ) MOLECIJLE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2:
Met Leu Val Thr Pro Leu Leu Leu Val Thr Leu Leu Cys Val Leu Cys
Ser Ala Ala Leu Tyr Asp Ser Ser Ser Tyr Val Tyr Tyr Tyr Gln Ser
Ala Phe Arg Pro Pro Asn Gly Trp His Leu His Gly &ly Ala Tyr Ala
Val Val Asn Ile Ser Ser Glu Ser Asn Asn Ala Gly Ser Ser Pro Gly
Cys Ile Val Gly Thr Ile His Gly Gly Arg Val Val Asn Ala Ser Ser
Ile Ala Met Thr Ala Pro Ser Ser Gly Met Ala Trp Sar Ser Ser Gln
Phe Cys Thr Ala His Cys Asn Phe Ser Asp Thr Thr Val Phe Val Thr
lOO . 105 llO
SIJBSTITUT~ SHEET
CA2i 1 7468
WO93/17l09 PCT/GB93/003.
His Cys Tyr Lys Tyr Asp Gly Cys Pro Ile Thr Gly Met Arg Gln Lys
115 120 125
Asn Phe Leu ~rg Val Ser Ala Met ~ys Asn Gly Gln Leu Phe Tyr Asn
130 135 140
Leu Thr Val Ser Val Ala Lys Tyr Pro Thr Phe Lys Ser Phe Gln Cys
145 150 155 160
~al Asn Asn Leu Thr Ser Val Tyr Leu Asn Gly Asp Leu Val Tyr Thr
165 170 175
Ser Asn Glu Thr Thr Asp Val Thr Ser Ala Gly Val Tyr Phe Lys Ala
180 185 190
Gly Gly Pro Ile Thr Tyr Lys Val Met Arg Glu Val Lys Ala Leu Ala
195 200 205
; Tyr Phe Val Asn Gly Thr Ala Gln Asp Val Ile Leu Cys ~sp Gly Ser
210 215 220
Pro Arg Gly Leu Leu Ala Cys Gln Tyr Asn Thr Gly Asn Phe Ser Asp
225 230 235 240
Gly Phe Tyr Pro Phe Ile Asn Ser Ser Leu Val Lys Gln Lys Phe Ile
245 250 255
Val Tyr Arg Glu Asn Ser Val Asn Thr Thr Phe Thr Leu His Asn Phe
260 265 270
Thr Phe His Asn GlU Thr Gly Ala Asn Pro Asn Pro Ser Gly Val Gln
275 280 285
Asn Ile Gln Thr Tyr Gln Thr Gln Thr Ala Gln Ser Gly Tyr Tyr Asn
290 295 300
Phe Asn Phe Ser Phe Leu Ser Ser Phe Val Tyr Lys Glu Ser Asn Phe
3~5 310 315 320
Met Tyr Gly Ser Tyr His Pro Ser Cys Asn Phe Arg Leu Glu Thr Ile
325 330 335
Asn Asn Gly Leu Trp Phe Asn Ser Leu Ser Val Ser Ile Ala Tyr Gly
340 345 350
SUE~STITUTE 5HEET
CA2i 1 7468
~093/17109 PCT/GB93/00332
_2S-
Pro Leu Gln Gly Gly Cys Lys Gln Ser Val Phe Ser Gly Arg Ala Thr
355 360 365
Cys Cys Tyr Ala Tyr Ser Tyr Gly Gly Pro Ser Leu Cys Lys Gly Val
370 37S 380
Tyr Ser Gly Glu Leu Asp Leu Asn Phe Glu Cys Gly Leu Leu Val Tyr
385 390 395 400
Val Thr Lys Ser Gly Gly Ser Arg Ile Gln Thr Ala Thr Glu Pro Pro
405 410 415
.~.
Val Ile Thr Arg His Asn Tyr Asn Asn Ile Thr Leu Asn Thr Cys Val
420 425 430
Asp Tyr Asn Ile Tyr Gly Arg Thr Gly Gln Gly Phe Ile Thr Asn Val
435 440 445
Thr Asp Ser Ala Val Ser Tyr Asn Tyr Leu Ala Asp Ala Gly Leu Ala
450 455 460
.
:i:le Leu A5p Thr Ser Gly Ser Ile Asp Ile Phe Val Val Gln Gly Glu
465 470 475 480
Tyr Gly Leu Thr Tyr Tyr Lys Val Asn Pro Cys Glu Asp Val Asn Gln
485 490 495
Gln Phe Val Val Ser Gly Gly Lys Leu Val Gly Ile Leu Thr Ser Arg
500 505 51~
-
Asn Glu Thr Gly Ser Gln Leu Leu Glu Asn ~;ln Phe Tyr Ile I,ys Ile
515 520 525
Thr Asn Gly Thr Arg Arg Phe Arg Arg Ser Ile Thr Glu Asn Val Ala
530 535 S40
Asn Cys Pro Tyr Val Ser Tyr Gly Lys Phe Cys Ile ~ys Pro Asp ~ly
545 550 555 560
er Ile Ala Thr Ile Val Pro Lys ~ln Leu Glu Gln Phe Yal Ala Pro
565 570 575
eu Leu Asn Val Thr Glu Asn Val Leu Ile Pro Asn Ser Ph~ Asn Leu
580 585 530
Thr Val Thr Asp Glu Tyr Ile Gln Thr Arg Met Asp Lys Yal Gln Ile
SIJE~8TITUTE SHEET
CA21 1 7468
WO93J17109 PCT/GB93/003
~2~-
595 600 605
Asn Cys Leu Gln Tyr Val Cys Gly Asn Ser Leu Asp Cys Arg Asp Leu
610 615 620
Phe Gln Gln Tyr Gly Pro Val Cys Asp Asn Ile Leu Ser Val Val Asn
625 630 635 640
Ser Ile Gly Gln Lys Glu Asp Met Glu Leu Leu Asn Phe Tyr Ser Ser
645 650 655
Thr Lys Pro Ala Gly Phe Asn Thr Pro Phe Leu Ser Asn Val Ser Thr
660 655 670
Gly Glu Phe Asn Ile Ser Leu Leu Leu Thr Thr Pro Ser Ser Pro Arg
675 680 685
Arg Arg Ser Phe Ile Glu Asp Leu Leu Phe Thr Ser Val Glu Ser Val
690 695 700
Gly Leu Pro Thr Asp Asp Ala Tyr Lys Asn Cys Thr Ala Gly Pro Leu
70S 710 715 720
Gly Phe Leu Lys Asp Leu Ala Cys Ala Arg Glu Tyr Asn Gly Leu Leu
725 730 735
Val Leu Pro Pro Ile Ile Thr Ala Glu Met GIn Thr Leu Tyr Thr Ser
740 745 750
. ,-~
Ser Leu Val Ala Ser Met Ala Phe Gly Gly Ile Thr Ala Ala Gly Ala
755 760 765
Ile Pro Phe Ala Thr Gln Leu Gln Ala Arg Ile Asn His Leu ~ly Ile
770 775 780
Thr Gln Ser Leu Leu Leu Lys Asn Gln Glu Lys Ile Ala Ala Ser Phe
785 790 795 800
Asn Lys Ala Ile Gly Arg Met Gln Glu Gly Phe Arg Ser Thr Ser Leu
895 810 815
Ala Leu G1n Gln Ile Gln ~sp Val Val Asn Lys Gln Ser Ala Ile Leu
8~0 825 830
Thr Glu Thr Met Ala Ser Leu Asn Lys Asn Phe Gly Ala Ile Ser SPr
835 ~40 845
5UBSTITUTE SHEET
CA2i`1 7468
`~'~ 93/17109 PCT/GB93/0033.
Val Ile Gln Glu Ile Tyr Gln Gln Leu Asp Ala Ile Gln Ala Asn Ala
8~0 855 860
Gln Val Asp Arg Leu Ile Thr Gly Arg Leu Ser Ser Leu Ser Val Leu
865 870 875 880
Ala Ser Ala Lys Gln Ala Glu His Ile Arg Val Ser Gln Gln Arg Glu
885 890 895
Leu Ala Thr Gln Lys Ile Asn Glu Cys Val Lys Ser Gln Ser Il~ Arg
900 905 910
Tyr Ser Phe Cys Gly Asn Gly Arg His Val Leu Thr Ile Pro Gln Asn
915 920 925
Ala Pro Asn ~ly Ile Val Phe Ile His Phe Ser Tyr Thr Pro Asp Ser
930 935 940
Phe Val Asn Val Thr Ala Ile Val Gly Phe Cys Val Lys Pro Ala Asn
945 950 955 g6~
Ala Ser Gln Tyr Ala ~le Val Pro Ala Asn Gly Arg Gly Ile Phe Ile
965 970 975
Gln Val Asn Gly Ser Tyr Tyr Ile Thr Ala Arg Asp Met Tyr Met Pro
980 985 990
Arg Ala Ile Thr Ala ~ly Asp Ile Val Thr Leu Thr Ser Cys Gln A
99S - 1000 1005-
Asn Tyr Val Ser Val Asn Lys Thr Val Ile Thr Thr Phe Val Asp Asn
1010 1015 102~
Asp Asp ~he Asp Phe Asn Asp Glu Leu Ser Lys Trp Trp Asn Asp Thr
1025 1030 1035 1040
Lys His Glu Leu Pro Asp Phe Asp Lys Phe Asn Tyr Thx Val Pro Ile
1045 1050 1055
Leu Asp Ile Asp Ser Glu Ile Asp Arg Ile Gln Gly Val Ile Gln Gly
1060 1065 1070
Leu Asn Asp Ser Leu Ile Asp Leu Glu Lys Leu Ser Ile Leu Lys Thr
1075 1080 1085
.
SUBSTlTlJTE SHEET
CA 2 i 1 74 68
W093/l7l09 PCT/GB93/003~-
Tyr Ile Lys Trp Pro
10~0
(2) INFORMATION FQR SEQ ID NO:3:
(i~ SEQUENCE CHARACTERISTICS:
(A~ LENGTH: 83 base pairs
(B) TYPE: nucleic acid
(C~ STRANDEDNESS: single
~D) TOPOLOGY: linear
(ii3 MOLECULE TYPE: cDNA
(vi~ ORIGINAL SOURCE:
(A~ ORGANICM: Infectious bronchitis virus
(B) STRAIN: M41
(ix) FEATURE:
(A) NAME/KEY: misc_feature
(B) LOCATION: 7..57
~D) OTHER INFORMATION: /function= "IBV LEADER SEQUENCE"
(ix) FEATURE:
(A) NAME/KEY: misc feature
(B) LOCATION: 58..83
(D) OTHER INFORMATION: /function= "IBV SPIKE CODING
SEQUENCE"
(xi~ SEQUENCE DESCRIPTION: SEQ ID NO:3:
GGATCCCCGA TCCCCTAGTCTTTAATTTAA TTAAGTGTGG TAAGTTACTG GTAAGAGATG
TTGGTAACAC CTCTTTTACT AGT
83
(2) INFORMATION FOR SEQ ID NO:4:
(i) SE~UENCE CHARACTERISTICS:
(A) LENGTH: 46 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
(~i) ORIGINAL SOURCE:
(A) ORGANISM: Infectious bronchitis virus
(B) STRAIN: M41
~ix~ FEAT~RE:
~A) NAME/KEY: misc_feature
(B) LOCATION: 7..20
(D) OTHER INFORMATION: /function= "VACCINIA P7.5 LEADER
SEQ~JENCE"
(ix) FEATURE:
SUBSTITUTE SHEET
CA 2 1 1 7468
~'~ 93~17109 ~ ; PCI/GB93J00332
(A) NAME/KEY: misc _ feat~e
(B) LOCATION: 2l. . 46
(D) OTHER INFORMATION: /function= "IBV SPIKE CODING
SEQUENCE"
(xi) SEQUENCE r)ESCRIPTION: SEQ ID NO: 4:
GGATCCAATC: AATAGCAATC ATGTTGGTAA CACCTCTTTT ACTAGT
46
2 ) INFORMATION FOPc SEQ ID NO: 5:
( i ) SEQUENCE CH~RACTERISTICS:
(A) LENGTH: 4 O base pairs
(B) TYPE: nucleic acid
t C3 ST~A~DEDNESS: single
( D ~ TOPOLOGY: l inear
MC~LECULE TYPE: cDNA
(xi) SEQUENCE DESCRIPTIO~I: SEQ ID NO:5:
GATCCAATCA ATAGCAATCA TGTTGGTAP,C ACCTCTTTTA
( 2 ) INFORMATION FOR S~Q ID NO: 6:
( i 3 SEQUENOE CHARACThRI STICS:
(A) LENGTH: 4 0 base pairs
(B) TYPE:: nucleic acid
(C~ STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
(xi) SEQUENCE DESC~IPTION: SEQ ID NO:6:
CTAGTAAAAG AGGTGTT~CC AACATGATTG CTATTGATTG
~....
,
SUBSTITU I ~ SHE~E~T