Note: Descriptions are shown in the official language in which they were submitted.
CA 02314357 2000-06-12
WO 99/31969 PCT1US98/27056
TRANSGENIC ANIMAL MODEL FOR
DEGENERATIVE DISEASES OF CARTILAGE
Field of the Invention
The present invention pertains to transgenic mammals that express
recombinant matrix-degading enzymes in a temporally and spatially regulated
manner.
The invention further pertains to model systems incorporating such transgenic
mammals
for studying degenerative joint diseases, including systems for identifying
therapeutic
agents and treatment regimens.
Background of the Invention
Degenerative diseases of cartilage, including joint and disc diseases such
as osteoarthritis, rheumatoid arthritis, and osteochondrodysplasias, are
widespread,
1 S particularly in the elderly. Early symptoms common to these diseases
include progressive
loss of proteoglycans in the joint (as evidenced by loss of metachromasia);
collagen
degadation; fibrillation of the cartilage surface; and, ultimately, loss of
cartilage (which
is evidenced radiologically as joint space narrowing).
One of the primary targets affected by these diseases is type II collagen,
the major structural collagen found in articular cartilage. There is a balance
between the
production of type II collagen and catabolic enzymes that degrade type II
collagen during
normal remodeling of cartilage and bone. Pathological conditions such as,
e.g.,
degenerative joint diseases, may result when this balance is disrupted.
Among the enzymes that degade extracellular matrix components are
matrix metalloproteinases (M1V)Ps), a family of zinc-dependent enzymes, and
aggecanase
(Table 1).
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
2
Table 1
Matr~-De=riding
Enzymes
SUBSTRATES
CollagenGelatinProttoglyeanFibrantainLanystinF.lastinOther
S I. M~alloprotdnases
Collagmases
MMP-I I,
II,
II1,
(intestinal VII,
collagenase) X
MMP-8 I,
II,
III
1 (neutrophil
~ collagenase)
MMP-13 I,
II,
III
(collagenase
3)
Gelatlnases
MMP-2 IV, J .l J
V,
VII,
1 (gelatinise XI
S A)
MMP-9 IV, ./
V
(gelatinise
B)
Stromelysins
MMP-3 ./ ./ ,/ ,/ activate
ZO (stromelysin t
1)
zymogens
MMP-7 IV ./ .~ .~ .~ d
(matrilysin)
MMP-10 IV, ./ ./ / activates
V,
IX
(stromelysin
2) MIHP
zymogrns
ZS MMP-11 IV .~ .~ activates
(stromelysin serpins
3)
Other
MMP-12 ,/
(metalloelastase)
3~ MMP-14 d proMMP-
2,
proMMP-
13
MMP-15
MMP-16 proMMP-
2
MMP-17 I
II. Aggreamaae ,/
3S
MMPs are synthesized in articulating joints by chondrocytes, which, in
mature articular cartilage, are terminally differentiated cells that maintain
the cartilage-
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
3
specific matrix phenotype. Overexpression of MMPs relative to endogenous MMP
inhibitors, as occurs in degenerative joint diseases, may result in cartilage
degradation.
For example, Type II collagen is a substrate for MMP-13 and MMP-1 (Knauper et
al., J.
Biol. Chem. 271:1544, 1996) and both MMP-1 and MMP-13 proteins can be detected
S immunohistochemically in human osteoarthritic tissues. In some cases, MMP-13
and its
cleavage products are found at higher levels than ~-1 (Billinghurst et al., J.
Clin.
Inves. 99:1534, 1997). Thus, MMP-13 may play an important role in cartilage
degradation
associated with osteoarthritis and other degenerative joint diseases (Mitchell
et al., J. Clin.
Inves. 97:761, 1996).
Animal models for osteoarthritis-related syndromes have been described in
guinea pigs (Watson et al., Arth. Rheum. 39:1327, 1996) and in the inbred
STR/ORT strain
of mice (Das-Gupta et al., Int. J. Fxp. Path. 74:627, 1993). In guinea pigs,
spontaneous
osteoarthritis has a long course of development (six months or more), and only
certain
sublines of STR/ORT mice consistently develop degenerative joint disease.
Thus, the
duration and/or variability of these models renders them less applicable to
drug discovery
studies.
Other osteoarthritis-related models include surgically-induced joint
destabilization, e.g., anterior cruciate ligament transection and/or partial
meniscectomy in
rabbits and dogs, which stimulates cartilage degradation (Hulth et al., Acta
Orthop. Scand
41:522, 1970). Another model employs injection of bacterial collagenase into
the joints of
an animal to induce a biochemical ligament transection (Van der Kraan et al.,
J. Exp.
Pathol. 71:19, 1990). Because (i) surgical or other manipulation of individual
animals is
required; (ii) the animals are large and expensive; and/or (iii) the course of
disease is not
consistent, these models cannot easily be used in large-scale studies,
including drug
screening.
Transgenic animal models, in principle, can provide the opportunity for a
reproducible animal model system for degenerative joint diseases. However,
previous
attempts to engineer transgenic animals expressing MMPs such as MMP-1 and
stromelysin have not resulted in an observable joint degeneration phenotype in
the
transgenic animals. This could be due to embryonic lethality caused by
constitutive
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/2?056
4
expression of these enzymes. Witty et al., Mol.Biol. Cell 6:1287, 1995, have
created
transgenic animals that constitutively express M1V>P-1 and stromelysin in
mammary tissue,
but these animals do not exhibit symptoms of osteoarthritis. D'Armiento et
al., Cell
71:955, 1992, disclose transgenic mice that express human interstitial
collagenase in the
lung. Liu et al., J. Cell Biol. 130:227, 1995, disclose transgenic animals
that overexpress
mutated type II collagen, resulting in connective tissue defects but not
osteoarthritis. None
of these transgenic animal systems provides a useful animal model for
osteoarthritis
(Khokha et al., Cancer arid Metastasis Rev. 14:97, 1995; Shapiro, Matrix Biol.
15:527,
1997).
Thus, there is a need in the art for animal model systems that mimic human
degenerative joint diseases such as, e.g., osteoarthritis, rheumatoid
arthritis, and
chondrodysplasias. Transgenic animals containing regulatable heterologous
genes whose
expression results in cartilage degeneration are particularly advantageous in
providing
reproducible experimental control over the timing and the level of expression
of the
transgenes and, thereby, over the pathological syndrome itself. Such animals
can be used
to determine what level of expression of the transgene is required to cause
disease and,
importantly, can be used for drug discovery and optimization of treatment
regimens. In
particular, such transgenic animals can be used to further define the role of
matrix-
degrading enzymes in cartilage degradation and as an in vivo screen to
identify compounds
that modulate these enzymes or compounds that inhibit the progression of
degenerative
joint diseases.
Summary of the Invention
The present invention provides transgenic non-human animals or the
progeny thereof whose somatic and germline cells contain, in stably integrated
form, one
or more heterologous or recombinant genes encoding polypeptides comprising
enzymatically active matrix-degrading enzymes (MDEs), preferably MMPs. MMPs
for
use in the invention comprise one or more of MNIP-1, MMP-2, MIVII'-3, NIMP-7,
MMP-8,
NI1VIP-9, M1V11'-10, MIvv)P-11, MMP-12, M1VVIP-13, MIv)I'-14, M1VVIP-15,
M1VVIP-16, and
MMP-17; preferably one or more of MMP-1, MNIP-3, I~~VIP-8, and NINIP-13; and
most
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/Z7056
preferably one or more of M1VVIP-1 and MMP-13; and include enzymatically
active
variants, fragments, and combinations of these polypeptides. Other matrix-
degrading
enzymes can also be used, including, e.g., aggecanase. The MDEs may be derived
from
any species, preferably human. In preferred embodiments, the recombinant MDE-
5 encoding genes are selectively expressed in articular chondrocytes of the
transgenic animal
and expression results in pathological symptoms characteristic of degenerative
joint
disease.
In one aspect, the invention provides a transgenic animal or the progeny
thereof whose somatic and germline cells contain a stably integrated first
recombinant
gene encoding an 1V1DE or an enzymatically active derivative or variant
thereof, preferably
a constitutively active proMMf-13 variant (designated MMP-13*) comprising the
sequence depicted in SEQ ID NO: 4. Preferably, the first recombinant gene is
under the
control of a first regulatable promoter; most preferably, the first
regulatable promoter
comprises a tet07 sequence, such as, e.g., the promoter depicted in SEQ 1D NO:
5. The
transgenic animal may further comprise a second recombinant gene encoding a
polypeptide that regulates the first regulatable promoter and is preferably a
tTA
polypeptide. In these embodiments, the second recombinant gene is under the
control of a
second regulatable promoter, preferably one that comprises sequences derived
from a
joint-specific promoter, and most preferably a type II collagen promoter, such
as, e.g., the
promoter depicted in SEQ B~ NO: 6. Selective expression of the second
recombinant gene
in joint tissues thus results in regulated joint-specific expression of the
recombinant MDE.
In another aspect, the invention provides isolated nucleic acids encoding
enzymatically active M1VIP variants, preferably human proMIvvlp-13 variants,
and most
preferably ~-13*. The invention also encompasses recombinant cloning vectors
comprising these nucleic acids; cells comprising the vectors; methods for
producing
M1VVIP-13-derived polypeptides comprising culturing the cells under conditions
appropriate
for MIV>I'-13 expression; and isolated MIV)P-13-derived polypeptides.
In yet another aspect, the invention provides methods for producing
phenotypic changes characteristic of cartilage-degenerative disease in a
mammal, which
comprise exposing the transgenic animals of the invention to conditions that
result in
CA 02314357 2000-06-12
WO 99/31969 PCT/US98rt7056
6
expression of the MDEs encoded by the transgenes. In a preferred embodiment, a
transgenic animal comprising a first recombinant gene encoding MMP-13*
operably
linked to a tet07 promoter and a second recombinant gene encoding a tTA
protein
operably linked to a type II collagen promoter is maintained in the presence
of tetracycline
or a tetracycline analogue. When it is desired to induce expression of MIVB'-
13 *,
tetracycline or the tetracycline analogue is withdrawn, MMP-13 * is
selectively expressed
in joint tissues, and phenotypic changes characteristic of cartilage-
degenerative disease
result.
In yet another aspect, the invention provides methods for determining the
potential of a composition to counteract cartilage-degenerative disease. The
methods are
carried out by administering a known dose of the composition to the transgenic
animals of
the invention, either before or after phenotypic indicators of cartilage-
degenerative disease
have developed; monitoring the indicators for a predetermined time following
administration of the composition; and comparing the extent of the indicators
in the animal
to which the composition was administered relative to a control transgenic
animal that had
not been exposed to the composition. Any diiTerence in (i) the nature or
extent of
phenotypic indicators of cartilage-degenerative disease, {ii) the time
required for the
indicators to develop, or (iii) the need for other ameliorative treatments
indicates the
potential of the composition to counteract cartilage-degenerative disease.
Brief Description of the Drawings
Figure lA and 1B. (A) Schematic illustration of the structure of human
Nllvlp-13 (collagenase-3). The first box (corresponding to the extreme
aminoterrninus)
represents the pre domain (signal peptide) that targets nascent proMMP-13 for
secretion.
The second box represents the pro domain, which is involved in maintaining the
latency of
the enzyme. A conserved sequence (SEQ 1D NO:1) within the pro domain that is
important for maintaining enzyme latency is shown. The third box represents
the 170-
amino acid catalytic domain, which contains a conserved region (shown; SEQ 1D
N0:2)
that is important for catalytic activity. The fourth box represents the 200-
amino acid
carboxyterminal domain. (B) Illustration of the nucleic acid sequence (SEQ ID
N0:3)
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
7
encoding a constitutively active variant of human pro MMP-13, designated MMP-
13 *, and
the amino acid sequence of MMP-13 * (SEQ ID N0:4). The residues that are,
mutated
relative to wild-type MMP-13, which are depicted in larger type, are GTC at
nucleotide
positions 299-301.
Figures 2A and 2B. Binary strategy used to obtain chondrocyte specific,
doxycycline (DOX) regulated MMP 13 * expression. (A) The first construct
illustrates the
rat type II collagen promoter driving expression of the tetracycline repressor
- VP16
activator fusion protein (TA), followed by an SV40 splice and polyadenylation
signal (this
construct is referred to as CPE - TA). (B) The second construct illustrates
the Tet07
promoter (Gossen and Bujard, Proc. Natl. Acad Sci. USA, 895547, 1992), driving
expression of a constitutively active human MMP 13 protein, followed by an
SV40 splice
and polyadenylation signal (construct referred to as Tet07 - MMP13*). These
two
independent transgene constructs were co-microinejected into fertilized mouse
embryos to
generate a double transgenic harboring both genes. In the presence of DOX,
transgene
expression is off; when DOX is removed, transgene expression is turned on. An
arrow
denotes the transcription start site, while the asterisk indicates a
constitutively active
mutation in the MMP 13 transgene.
Figure 3A and 3B. The type II collagen gene promoter directs expression
to the joints of tester transgenic mice expressing ~i-galactosidase under the
control of the
rat type Ii collagen promoter. (A) Diagram of the CPE -lacZ construct. The rat
type II
collagen promoter (first box) drives expression of the (3-galactosidase (lacZ)
gene (second
box), which is followed by a ~-globin splice and polyadenylation signal (final
boxes
joined by splice symbol). (B) Whole mount staining for (3-galactosidase
activity on E16
embryos. The embryo on the left shows the staining (arrows) of the transgenic
compared
to a wild-type embryo. (C) Higher magnification of a transgenic elbow and
front paw;
~-gal staining is visible (arrows). Figures 3B and 3C more dramatically
illustrate the
invention when rendered in color, although black and white rendering is
sufficient for
understanding the invention.
Figure 4A and 4B. Expression profile of the TA and MMP 13 RNA by
RT-PCR. (A) Amplification of TA cDNA from total RNA. (B) Amplification of
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
8
MMP 13 * cDNA from total RNA. Lane 1.: ~x 164 Hae III MW markers; Lane Z: PCR
amplification of transgenic (line 6) genomic DNA; Lane 3: PCR amplification of
non-transgenic genomic DNA; Lane 4: a wild-type mouse maintained on DOX; Lane
5: a
wild-type mouse off DOX; Lanes 6-7: transgenic mice (four months) maintained
on
DOX; Lanes 8-9: transgenic mice (four months) removed from DOX at birth. The
arrows la and lb indicate a 648 by MMP13* specific fragment and a 859 by
specific
fragment, respectively. Each reaction was run using c-fos primers as an
internal control,
spliced mRNA yielding 187 by (arrow 3) and unspliced mRNA 303 by (arrow 2). No
bands were detected in corresponding lanes containing RNA for PCR that was not
treated
with RT (data not shown).
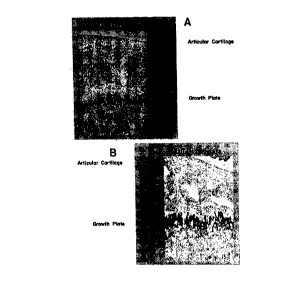
Figure 5A and 5B. Photographic illustrations of immunohistochemical
localization of type II collagen cleavage products in the growth plate and
articular
cartilage of transgenic mice expressing the transgenes shown in Figure 2. The
tissues
were stained with an antibody that recognizes cleavage products of type II
collagen. (A)
Tissue derived from a mouse that had been maintained on doxycycline to repress
MMP-
13* expression. (B) Tissue derived from a mouse that had been withdrawn from
doxycycline, allowing expression of MMP-13*, for 30 days at 3 months of age.
Figures
SA and B more dramatically illustrate the invention when rendered in color,
although
black and white rendering is sufficient for understanding the invention.
ZO Figure 6A, 6B, and 6C. A color photographic illustration of Safranin O
staining of the articular cartilage and growth plate of the patella of double
transgenic
mice. (A) Tissue derived from a mouse maintained on doxycycline. (B) Tissue
derived
from a mouse 7 days after withdrawal from doxycycline. (C) Tissue derived from
a
mouse 14 days after withdrawal from doxycycline. Figures 6A-C more
dramatically
illustrate the invention when rendered in color, although black and white
rendering is
sufficient for understanding the invention.
Figures 7A, 7B, 7C, and 7D. Longitudinal section through the hind knee
joints (A and B) and synovium (C and D). (A and C) Age match littermate
control and
(B and D) line 6 removed from DOX. Abbreviations: L, lesion; AC, articular
cartilage;
AG, angiogenesis, IH, infiltration hyperplasia; BM, bone marrow; and, PL,
patella
CA 02314357 2000-06-12
WO 99/31969 PCTNS98/Z7056
9
ligament. The location of the femur and patella are labelled on the figure.
Figures 7A-D
more dramatically illustrate the invention when rendered in color, although
black and
white rendering is sufficient for understanding the invention.
Detailed Description of the Invention
The present invention is based on the discovery regulated expression of
matrix-degrading enzymes in cartilage in transgenic mice results in
characteristic
phenotypic changes associated with matrix degenerative diseases of the joints
and
intervertebral discs. The animal models of the invention provide novel model
systems for
matrix degenerative disease syndromes which can be used for detailed
characterization of
human joint and intervertebral disc pathologies as well as for drug discovery
and
optimization of treatment regimens.
A transgenic animal according to the invention is an animal having cells
that contain a transgene which was introduced into the animal or an ancestor
of the animal
at a prenatal (embryonic) stage. A transgenic animal can be created, for
example, by
introducing the gene of interest into the male pronucleus of a fertilized
oocyte by, e.g.,
microinjection, and allowing the oocyte to develop in a pseudopregnant female
foster
animal. The gene of interest may include appropriate promoter sequences, as
well as
intronic sequences and polyadenylation signal sequences. Methods for producing
transgenic animals are disclosed in, e.g., U.S. Patent Nos. 4,736,866 and
4,870,009 and
Hogan et al., A LaboratoryManual, Cold Spring Harbor Laboratory, 1986. A
transgenic
founder animal can be used to breed additional animals carrying the transgene.
A
transgenic anima! carrying one transgene can also be bred to another
transgenic animal
carrying a second transgene to create a "double transgenic" animal carrying
two
transgenes. Alternatively, two transgenes can be co-microinjected to produce a
double
transgenic animal. Animals carrying more than two transgenes are also
possible.
Furthermore, heterozygous transgenic animals, i.e., animals carrying one copy
of a
transgene, can be bred to a second animal heterozygous for the same transgene
to produce
homozygous animals carrying two copies of the transgene.
The present invention encompasses transgenic animals, preferably
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
mammals, which express MDEs, particularly MMPs, and most particularly those
MMPs
having collagenase activity, from a recombinant gene. MDFs for use in the
invention
include without limitation MMPs and aggrecanase. Useful MMPs include without
limitation the collagenases designated MMP-1, MMP-8 and MMP-13; the
stromelysins
5 designated MMP-3, MMP-10, and MMP-11; the gelatinases designated MMP-2 and
MMP-9; the metalloelastase designated MMP-12; and membrane-type MMPs
designated
MMP-14, MMP-15, MMP-16, and MMP-17 ( Matrisian, BioEssays, 14:455, 1992).
Matrix-degrading activity as used herein refers to the proteolytic degradation
of matrix
components, including, e.g., collagen, particularly type II collagen and most
particularly
10 the triple helical form of type II collagen. Any polypeptide exhibiting
matrix-degrading
activity may be used in practicing the invention, including enzymatically
active fragments
of the above-described enzymes. Preferably, MMP-13 enzymatic activity is
expressed.
MMP-13 enzymatic activity as used herein refers to the proteolytic degradation
of type II
collagen. Any MMP-13 polypeptide or fragment or derivative thereof that
exhibits MMP-
13 enzymatic activity may be used. The enzymes may be derived from any animal
species, including without limitation human, mouse, rat, rabbit, pig, cow, or
non-human
primate, or combinations thereof. Preferably, the MMP-13 or derivative thereof
is of
human origin.
Normally, MMPs are synthesized as precursors (i.e., zymogens or
proenzymes) whose enzymatic activity is latent; proteolytic removal of the pro
region after
secretion produces the enzymatically active protein. In preferred embodiments
of the
invention, the need for proteolytic processing is circumvented by the use of
enzyme or
proenzyme variants that are enzymatically active even when uncleaved. Such
variants can
be produced using conventional techniques for site-directed or random
mutagenesis
coupled with analysis of collagenase enzymatic activity (see below). In this
manner,
modifications (including, e.g., insertions, deletions, and substitutions), may
be introduced
into a proenzyme sequence, particularly within the pro region or near the pro
region
cleavage site, to produce a constitutively active polypeptide which does not
require
proteolytic processing for activation. Alternatively, the pro region may be
deleted
entirely. Furthermore, recombinant genes may be used in which the sequence
encoding
CA 02314357 2000-06-12
WO 99/31969 PCT/US98I27056
11
the native signal peptide is replaced by a heterologous sequence that
functions as a signal
peptide, i.e., promotes secretion. The use of genes encoding any such modified
MMP
polypeptides is encompassed by the invention.
Preferably, a constitutively active MMP-13 variant is used in practicing the
invention. Most preferably, the MMP-13 variant comprises a sequence containing
a
mutation in the sequence encoding the PRCGVPDV region, SEQ ID N0:7,
specifically a
substitution of Prop' to Val; the sequence of this polypeptide is depicted in
SEQ ID NO: 4
and this polypeptide is designated MMP-13*. In another embodiment, the
constitutively
active MMP-13 variant comprises a substitution of Va198 to Gly.
The transgenic animals of the invention preferably express MMP activity in
a regulated manner. Regulated expression as used herein refers to temporal
andlor spatial
control. Temporal control refers to the ability to repress expression of MMP
activity until
a predetermined time in the development of the transgenic animal, after which
MMP
expression may be activated and maintained for as long as desired. Preferably,
MMP
expression is repressed throughout embryonic development and activated in the
adult
animal. Spatial control refers to the ability to selectively express MMP
activity in
particular tissues. Preferably, MMP activity is selectively expressed in joint
tissues, most
preferably in articular chondrocytes.
Temporal control of MMP expression is achieved by use of one or more
polypeptides comprising a transcriptional repressor, a transcriptional
activator or enhancer,
or combinations thereof, in conjunction with a promoter responsive to the
transcriptional
repressor/activator used to which the MMP-encoding sequence is operably
linked. In one
set of embodiments, temporal control of MMP expression is achieved by (i)
expression in
the transgenic animal of a repressor polypeptide operably linked to a
polypeptide that
directly or indirectly activates transcription in eucaryotic cells, creating a
repressor-
activator fusion polypeptide; and (ii) the coupled use of a target promoter
operably linked
to an MMP-encoding sequence whose transcriptional activity is responsive to
the
repressor-activator fusion polypeptide. Typically, nucleotide sequences
encoding the
repressor polypeptide are ligated in-frame to sequences encoding the
transcriptional
activator polypeptide to create a chimeric gene encoding a fusion protein.
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
12
Useful repressor polypeptides include without limitation polypeptides
comprising sequences derived from bacterial repressors, including without
limitation
tetracycline repressor, LacR repressor, KRAB domain, and lambda repressor (cro
and cI),
as well as eukaryotic repressors, including without limitation those involved
in amino acid
or sugar synthesis. Useful direct transcriptional activator polypeptides
include without
limitation herpes simplex virus protein 16 (VP16); yeast GAL14; yeast STAT;
steroid
receptors such as, e.g., progesterone receptor and estrogen receptor; and
constitutive
activators such as, e.g., c-fos, c-jun, and SP-1. Alternatively, the repressor
polypeptide
may be linked to a polypeptide that indirectly activates transcription by
recruiting a
transcriptional activator to interact with the repressor-activator fusion
protein; such
indirect activator polypeptides include without limitation TATA Box Binding
Protein
(TBP) and basic transcription factors, including, e.g., basic transcription
factor D.
According to the invention, each repressor-activator fusion protein is used
in conjunction with a target promoter that is responsive to the particular
fusion protein and
that regulates transcription of an MDE-encoding sequence. Typically, the
promoter
comprises at least one operator sequence responsive to the repressor component
of the
repressor-activator fusion polypeptide, which is operably linked to at least a
minimal
promoter that supports transcription in eucaryotic cells. Examples of suitable
repressor-
responsive operator sequences include without limitation sequences derived
from the
tetracycline resistance operon encoded in TnlO in E toll, the lambda repressor
operon,
and the yeast GAL repressor operon. Examples of suitable eucaryotic promoters
from
which minimal promoters may be derived include without limitation the
cytomegalovizus
(CMS IE promoter, PtK-1 (thymidine kinase) promoter, HSP (heat shock protein)
promoter, and any eukaryotic promoter containing a TATA box. Minimal promoter
sequences may be derived from these promoters by (i) creating deletion mutants
using
conventional methods and (ii) testing the ability of the resulting sequences
to activate
transcription in a cell line. U.S. Patent No. 5,650,298 discloses a repressor-
activator
fusion protein comprised of sequences derived from the tetracycline repressor
fused to
VP16 sequences, which is designated tTA, and a tTA-responsive promoter,
designated
tet07, which comprises a TnlO-derived sequence linked to a portion of the CMV
IE
CA 02314357 2000-06-12
WO 99/31969 PGT/US98n7056
13
promoter.
Alternatively, temporal control is achieved by (i) expression in the
transgenic animal of a heterologous or recombinant transcriptional activator
polypeptide or
polypeptides and (ii) the coupled use of a target promoter operably linked to
an MMP-
encoding sequence whose transcriptional activity is responsive to the
heterologous or
recombinant transcriptional activator. Useful transcriptional activators
include without
limitation a modified ecdysone receptor, in which a VP16 transactivation
domain linked to
the aminotenminal transactivation domain of the glucocorticoid receptor is
fused to the
ligand-binding domain and carboxyterminal sequence of the ecdysone receptor
(No et al.,
Proc. Natl. Acad Sci. USA 93:3346, 1996); a chimeric protein, designated pGL-
VP,
comprising VP16 activator sequences, GAL4 activation sequences, and a mutated
human
progesterone receptor ligand-binding domain (Wang et al., Proc. Natl. Acad
Sci. USA
91:8180, 1994; Wang et al., Gene Therapy 4:432, 1997); and chimeric proteins
comprising
transcriptionai activators fused to estrogen (or other steroid) binding
domains (Mattioni et
1 S al., Meih. Cell Biol. 43:335, 1994). The ecdysone receptor system utilizes
retinoid X
receptor (RXR) to form heterodimers with the chimeric receptor, and responds
to
ecdysone, muristerone (an ecdysone analogue) or dexamethasone. The pGL-VP
system is
responsive to mifepristone (RU486). Chimeric receptors containing an estrogen
binding
domain respond to hydroxytamoxifen (an estrogen analogue).
Spatial control of 1VIDE expression is achieved by the use of transcriptional
promoters that direct transcription selectively in joint tissues. Joint-
specific expression as
used herein refers to expression that is greater in joints than in other
cells; typically, the
level of expression in non-joint tissues is less than 10% of the level of
expression in joints.
Preferably, expression in non joint tissues is undetectable. Useful promoter
sequences that
confer joint-specific expression on a sequence to which they are operably
linked include
without limitation sequences derived from the collagen type II promoter. It
will be
understood that a joint-specific promoter according to the invention may
comprise one or
more copies of particular sequences or sub-sequences, and these sequences may
be in
direct or inverted orientation relative to each other and relative to the
sequence whose
expression is regulated by the promoter.
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/Z7056
14
Coordinated spatial and temporal control of MDE expression is preferably
achieved by (i) placing expression of the repressor-activator fusion
polypeptide or the
transcriptional activator polypeptide under the control of a joint-specific
promoter; (ii)
placing the expression of the MDE or a derivative thereof under the control of
a promoter
responsive to the repressor-activator fusion polypeptide or the
transcriptional activator
polypeptide; and (iii) maintaining the transgenic animal during fetal
development and
early life under conditions in which MDE expression is repressed.
The method by which transgenic animals are maintained during fetal and
early post-natal development so that MDE expression is repressed will depend
on the
particular transgenes being expressed. When a repressor-activator fusion
polypeptide is
used, repression is achieved by providing the animal with an agent that binds
to the
repressor-activator fusion protein and results in repression of transcription
of the target
MDE gene. In animals comprising a transgene encoding a repressor-activator
fusion
polypeptide containing tet repressor sequences, repression is achieved by
providing
tetracycline or a tetracycline analogue in the food or drinking water of the
mother and,
following birth, of the progeny. Tetracycline or an analogue may also be
provided using
surgically implanted subcutaneous time-release pellets (Innovative Research of
America,
Inc., Sarasota FL). In this case, binding of tetracycline or a tetracycline
analogue to the
repressor-activator fusion protein prevents the fusion protein from binding
to, and
activating transcription of, the cognate promoter. Tetracycline analogues are
compounds
closely related to tetracycline which bind to the tet repressor with a Ka of
at least about
106M'', preferably with an affinity of about IO~IvI'' or greater. Useful
tetracycline
analogues include without limitation doxycycline, anhdryrotetracycline,
chlortetracycline,
epioxytetracycline, and the like. The dosage used is one that will result in
substantial
repression of MMP expression. Typically, tetracycline or a tetracycline
analogue is
administered in the animal's drinking water at a dosage of about 1 mglml. When
it is
desired that MMPs be expressed, the tetracycline or analogue thereof is
withheld.
In other embodiments, repression is achieved by withholding from
the animal an agent required for activity of the transcriptional activator
polypeptide. For
example, if the transcriptional activator is a modified ecdysone receptor, the
animals are
CA 02314357 2000-06-12
WO 99/31969 PCT1US98/27056
maintained in the absence of ecdysone or an ecdysone analogue throughout fetal
and early
post-natal development. Ecdysone analogues are compounds closely related to
ecdysone
which bind to the modified ecdysone receptor with a Ka of at least about
106M''. Useful
ecdysone analogues include without limitation muristerone A. When it is
desired that
S MDEs be expressed, the animals are given, e.g., ecdysone or muristerone A
via
intraperitoneal injections at dosages of between about 10 mg and about 20
mg/animal.
Similarly, when pGL-VP is used, activation is achieved by providing
mifepristone.
In a preferred embodiment of the invention, a transgenic animal is
constructed whose somatic and germline cells contain in stably integrated form
two
10 recombinant genes: (i) a first recombinant gene comprising a sequence
encoding MMP-
13 *, wherein the sequence is operably linked to a tet07 promoter; and (ii) a
second
recombinant gene encoding a tTA protein operatively linked to a collagen type
II
promoter. In this embodiment, animals are maintained in the presence of
tetracycline or a
tetracycline analogue throughout fetal and early post-natal development to
repress the
15 gene. Afterwards, tetracycline or the tetracycline analogue is withdrawn,
and MMP-13
enzymatic activity is selectively expressed in joint tissues.
Animal Models for Cartilage-Deuenerative Diseases
The present invention provides animal model systems in which phenotypic
changes characteristic of cartilage-degenerative diseases, such as, e.g.,
joint or disc
disease, are reproducibly exhibited. These diseases include without limitation
osteoarthritis, rheumatoid arthritis, chondrodysplasias, and degenerative
intervertebral disc
diseases. The model systems of the invention exhibit one or more phenotypic
indicators
common to these diseases, which include without limitation loss of
proteoglycan (as
indicated by, e.g., loss of Safranin O staining) and cleavage of type II
collagen in the
affected tissues. The systems encompass the transgenic animals described
above, in which
recombinant or heterologous MDEs, particularly MMPs, are expressed in
cartilage at a
predetermined time in the life of the transgenic animal. The timing of the
appearance of
cartilage-degenerative indicators is determined by activating MDE expression
and
monitoring the effects on cartilage (see below). Preferably, one or more MDEs
are
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
16
expressed after birth, most preferably after the animal has reached adulthood.
Expression of the transgenes is typically monitored by extracting mRNA
from different tissues and subjecting the extracted mRNA to one or more of the
following:
(i) reverse transcriptase-polymerase chain reaction (RT-PCR), using primers
homologous
to the transgene; (ii) RNAase protection; and (iii) Northern blot analysis.
Alternatively, in
situ hybridization may used.
The physiological effects of MDE expression on articular cartilage are
monitored in test animals by sacrificing the animals and subjecting paraffin-
embedded
decalcified cartilage to staining with (i) hematoxylin and eosin (using
conventional
techniques) followed by double staining with (ii) Safranin O and fast green
(Peter et al., J.
Fxp. Pathol. 71:19, 1990). Alternatively, frozen sections may be obtained and
stained
with antibodies that are specific for cleavage fragments derived from type II
collagen
(Billinghurst et al., J. Clin. Invest. 99:1534, 1997). Typically, expression
of the MMP
transgene(s) for at least about 7 days results in detectable loss of
proteoglycan and changes
in growth plate morphology (see, e.g., Example 5 below). Animal models in
which
expression of lV~Es, particularly MMPs, and most particularly an enzymatically
active
form of MMP-13, results in proteoglycan loss and/or cleavage of type II
collagen are
within the scope of the invention.
Other phenotypic indicators of cartilage-degenerative disease which can be
monitored in transgenic animals produced according to the invention include
without
limitation gross observations of changes in joint function and histological
evidence of (i)
fibrillation and loss of articular cartilage and (ii) osteophyte formation.
Syndromes for which the transgenic animals of the invention provide useful
models include without limitation any pathological condition that manifests a
disturbance
in the composition, morphology, and/or function of cartilage, including
osteoarthritis;
rheumatoid arthritis; degenerative intervertebral disc diseases;
chondrodysplasias,
including, e.g., Kniest dysplasia, achondrogenesis, and hypophosphatasia; and
proteoglycan-mediated disorders, such as occur, e.g., in brachymorphic animals
(Hall et
al., Cartilage: Molecular Aspects, CRC Press, 1991, pp. 201-203).
In further embodiments of the invention, the transgenic animals can be
CA 02314357 2000-06-12
WO 99/31969 PCTNS98/27056
17
subjected to additional treatments to modulate the cartilage-degenerative
indicators and/or
to supplement the animals' disease phenotype with additional physiological
effects such
as, e.g., those associated with a particular disease. For example, the
transgenic animals
may be further treated with inflammatory mediators to augment collagen
degradation
and/or induce loss of proteoglycan (see, e.g., Example 6 below). Furthermore,
the timing
and extent of MDE induction, with or without additional treatments, can be
adapted to
replicate the symptomatology of a particular disease or syndrome.
Methods for Evaluating Dm~s that Modulate Degenerative Diseases of Cartilagg
The present invention encompasses methods for discovery and evaluation
of drugs and therapies for their efficacy against degenerative diseases of
cartilage,
particularly degenerative joint diseases. In one embodiment of the invention,
the
transgenic animals of the invention are maintained under conditions in which
expression
of one or more MDEs results in one or more phenotypic indicators of cartilage-
t5 degenerative disease. Once the symptoms have developed, the potential of a
composition
to counteract cartilage-degenerative disease can be evaluated by administering
a known
dose of the composition to the animal in which the symptoms have developed;
monitoring
the phenotypic indicators for a predetermined time following administration of
the
composition; and comparing the extent of the phenotypic indicators in the
animal to which
the composition was administered relative to a control animal. Control animals
comprise
age- and sex-matched transgenic animals that are maintained under an identical
regimen
(i.e., express the transgenes) but which do not receive the composition. Any
statistically
significant difference in the extent or nature of the phenotypic indicators
indicates the
potential of the composition to counteract cartilage-degenerative disease. As
used herein,
phenotypic indicators of cartilage-degenerative disease refer to proteoglycan
loss, joint
space narrowing, collagen degradation, and destruction of cartilage.
In another embodiment of the invention, the potential of a composition to
counteract degenerative diseases of cartilage, particularly degenerative joint
disease, is
evaluated by administering to a transgenic animal a known dose of the
composition before
and/or simultaneous with the induction of MDE expression in the transgenic
animal;
CA 02314357 2000-06-12
WO 99131969 PCT/US98/27056
18
monitoring phenotypic indicators of cartilage-degenerative disease for a
predetermined
time following administration of the composition and MDE induction; and
comparing the
extent of the phenotypic indicators andlor disease in the animal to which the
composition
was administered relative to a control animal that had not been exposed to the
composition. In this embodiment, any statistically significant difference in
the extent or
nature of the phenotypic indicators and/or disease, or any statistically
significant delay in
appearance of the phentoypic indicators or disease, indicates the potential of
the
composition to counteract cartilage-degenerative disease.
A further indication of the potential of a composition to counteract
cartilage-degenerative disease is the ability of the composition to cause any
reduction in
the extent or duration of other treatments, including, e.g., the dosage and
timing of
administration of other therapeutic agents used to alleviate symptoms of the
disease.
Compounds that may be tested for anti-cartilage-degenerative disease
potential may be found in, for example, natural product libraries,
fermentation libraries
1 S (encompassing plants and microorganisms), combinatorial libraries,
compound files,
synthetic compound libraries, and compounds resulting from directed rational
drug design
and synthesis. For example, synthetic compound libraries are commercially
available
from Maybridge Chemical Co. (Trevillet, Cornwall, UK), Comgenex (Princeton,
Nn,
Brandon Associates (Merrimack, NH), and Microsource (New Milford, CT). A rare
chemical library is available from Aldrich Chemical Company, Inc. (Milwaukee,
WI).
Alternatively, libraries of natural compounds in the form of bacterial,
fungal, plant and
animal extracts are available from, for example, Pan Laboratories (Bothell,
WA) or
MycoSearch (NC), or are readily producible. Additionally, natural and
synthetically
produced libraries and compounds are readily modif'led through conventional
chemical,
physical, and biochemical means (Blondelle et al., TibTech 14:60, 1996).
Transgertic animals
Transgenic animals as used herein refers to animals into which one or more
heterologous and/or recombinant genes have been introduced. The transgenes may
be
from a different species, or from the same species as the transgenic animal
but are not
CA 02314357 2000-06-12
WO 99/31969 PC'T/US98/27056
19
naturally found in the animal in the configuration and/or at the chromosomal
locus
conferred by the transgene. Transgenes may comprise foreign DNA sequences,
i.e.,
sequences not normally found in the genome of the host animal. Alternatively
or
additionally, transgenes may comprise endogenous DNA sequences that have been
rearranged or mutated in vitro in order to alter the normal in vivo pattern of
expression of
the gene, or to alter or eliminate the biological activity of an endogenous
gene product
encoded by the gene. Also encompassed by the invention are DNA fragments that
are
introduced into a pre-existing gene to, e.g., change patterns of expression or
to provide
additional means of regulating the expression of the gene (Watson et al., "The
Introduction
of Foreign Genes Into Mice," in Recombinant DNA, 2d Ed., W.H. Freeman & Co.,
New
York, 1992, pp. 255-272; Gordon, J.W., Intl. Rev. Cytol. 115:171,1989;
Jaenisch, Science
240:1468, 1989; Rossant, Neuron 2:323, 1990).
The transgenic non-human animals of the invention are produced by
introducing transgenes into the germline of the non-human animal. Embryonal
target cells
at various developmental stages are used to introduce the transgenes of the
invention.
Different methods are used depending on the stage of development of the
embryonal target
cell(s). Such methods include, but are not limited to, microinjection of
zygotes, viral
integration, and transformation of embryonic stem cells as described below.
Microinjection of zygotes is the preferred method for incorporating
transgenes into animal genomes. A zygote, which is a fertilized owm that has
not
undergone pronuclei fusion or subsequent cell division, is the preferred
target cell for
microinjection of transgenic DNA sequences. The murine male pronucleus reaches
a size
of approximately 20 micrometers in diameter, a feature which allows for the
reproducible
injection of 1-2 picoliters of a solution containing transgenic DNA sequences.
The use of
a zygote for introduction of transgenes has the advantage that, in most cases,
the injected
transgenic DNA sequences will be incorporated into the host animal's genome
before the
first cell division (Brinster et al., Proc. Natl. Acad. Sci. USA 82:4438,
1985). As a
consequence, all cells of the resultant transgenic animals (founder animals)
stably carry an
incorporated transgene at a particular genetic locus, referred to as a
transgenic allele. The
transgenic allele demonstrates Mendelian inheritance, i.e., half of the
offspring resulting
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
from the cross of a tiansgenic animal with a non-transgenic animal will
inherit the
transgenic allele, in accordance with Mendel's rules of random assortment.
2. Viral integation can also be used to introduce the transgenes of the
invention into an animal. The developing embryos are cultured in vitro to the
blastocyte
5 developmental stage. The blastomeres may be infected with appropriate
retroviruses
(Jaenich, Proc. Natl. Acac~ Sci. USA 73:1260). Infection of the blastomeres is
enhanced by
enzymatic removal of the zona pellucida. Transgenes are introduced via viral
vectors
which are typically replication-defective but which remain competent for
integration of
viral-associated DNA sequences, including transgenic DNA sequences linked to
such viral
10 sequences, into the host animal's genome. Transfection is easily and
efficiently obtained
by culture of blastomeres on a monolayer of cells producing the transgene-
containing viral
vector. Alternatively, infection may be performed using cells at a later
developmental
stage, such as blastocoeles. In any event, most transgenic founder animals
produced by
viral integration will be mosaics for the transgenic allele; that is, the
transgene is
15 incorporated into only a subset of all the cells that form the transgenic
founder animals.
Moreover, multiple viral integration events may occur in a single founder
animal,
generating multiple transgenic alleles which will segregate in future
generations of
offspring. Introduction of transgenes into gernlline cells by this method is
possible but
probably occurs at a low frequency. However, once a transgene has been
introduced into
20 germline cells by this method, offspring may be produced in which the
transgenic allele is
present in all of the animal's cells, i.e., in both somatic and germline
cells.
3. Embryonal stem (ES) cells can also serve as target cells for
introduction of the transgenes of the invention into animals. ES cells are
obtained from
pre-implantation embryos that are cultured in vitro (Evans et al., Nature
292:154, I981).
ES cells that have been transformed with a transgene can be combined with an
animal
blastocyst, after which the ES cells colonize the embryo and contribute to the
germline of
the resulting animal (which is a chimera, i.e., composed of cells derived from
two or more
animals). Again, once a transgene has been introduced into germline cells by
this method,
offspring may be produced in which the transgenic allele is present in all of
the animal's
cells, i.e., in both somatic and germline cells.
CA 02314357 2000-06-12
WO 99/31969 PC'f/US98/27056
21
Although the initial introduction of a transgene is a Lamarckian (non--
Mendelian) event, the transgenes of the invention may be stably integrated
into germ line
cells and transmitted to offspring of the transgenic animal as Mendelian loci.
Other
transgenic techniques result in mosaic transgenic animals, in which some cells
carry the
transgenes and other cells do not. In mosaic transgenic animals in which germ
line cells
do not carry the transgenes, transmission of the transgenes to offspring does
not occur.
Nevertheless, mosaic transgenic animals are capable of demonstrating
phenotypes
associated with the transgenes.
In practicing the invention, animals of the transgenic maintenance line are
crossed with animals having a genetic background in which expression of the
transgene
results in symptoms of cartilage-degenerative disease. Offspring that have
inherited the
transgenes of the invention are distinguished from littermates that have not
inherited
transgenes by analysis of genetic material from the offspring for the presence
of nucleic
acid sequences derived from the transgenes of the invention. For example,
biological
fluids that contain polypeptides uniquely encoded by the transgenes of the
invention may
be immunoassayed for the presence of the polypeptides. A simpler and more
reliable
means of identifying transgenic offspring comprises obtaining a tissue sample
from an
extremity of an animal, such as, for example, a tail, and analyzing the sample
for the
presence of nucleic acid sequences corresponding to the DNA sequence of a
unique
portion or portions of the transgenes of the invention. The presence of such
nucleic acid
sequences may be determined by, e.g., hybridization ("Southern") analysis with
DNA
sequences corresponding to unique portions of the transgene, analysis of the
products of
PCR reactions using DNA sequences in a sample as substrates, oligonucleotides
derived
from the transgene's DNA sequence, and the like.
Nucleic Acids. Vectors. Expression Systems. and Polypeptides
The present invention encompasses isolated nucleic acids encoding MDFs,
particularly MMPs, and enzymatically active fragments derived therefrom, as
well as
constitutively active MMP variants and enzymatically active fragments derived
therefrom.
The invention also encompasses complements of the above nucleic acids; vectors
CA 02314357 2000-06-12
WO 99/31969 PC'f/US98I27056
22
comprising the nucleic acids; cells comprising the vectors; and isolated
polypeptides
encoded by the nucleic acids.
Many techniques in molecular biology, microbiology, recombinant DNA, and
protein biochemistry are used in practicing the present invention, such as
those explained
in, for example, Sambrook et al., 1989, Molecular Cloning: A Laboratory
Manual,
Second Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New
York;
DNA Cloning: A Practical Approach, Volumes I and II, 1985 (D.N. Glover ed.);
Oligonucleotide Synthesis, 1984, (M.L. Gait ed.); Transcription and
Translation, 1984
(Names and Higgins eds.); A Practical Guide to Molecular Cloning; the series,
Methods in
Enzymology (Academic Press, Inc.); and Protein Pur~cation: Principles and
Practice,
Second Edition (Springer-Verlag, N.Y.).
"Nucleic acid" or "polynucleotide" as used herein refers to purine- and
pyrimidine-containing polymers of any length, either polyribonucleotides or
polydeoxyribonucleotides or mixed polyribo-polydeoxyribo nucleotides. This
includes
single- and double-stranded molecules, such as, for example, DNA-DNA, DNA-RNA
and
RNA-RNA hybrids, as well as "protein nucleic acids" (PNA) formed by
conjugating bases
to an amino acid backbone. This also includes nucleic acids containing
modified bases.
A "coding sequence" or a "protein-coding sequence" is a polynucleotide
sequence capable of being transcribed into mRNA and/or capable of being
translated into a
polypeptide. The boundaries of the coding sequence are typically determined by
a
translation start codon at the 5'-terminus and a translation stop codon at the
3'-terminus.
A "complement" of a nucleic acid sequence as used herein refers to the
"antisense" sequence that participates in Watson-Crick base-pairing with the
original
sequence.
An "isolated" nucleic acid or polypeptide as used herein refers to a component
that is removed from its original environment (for example, its natural
environment if it is
naturally occurring). An isolated nucleic acid or polypeptide typically
contains less than
about 50%, preferably less than about 75%, and most preferably less than about
90%, of
the cellular components with which it was originally associated.
A nucleic acid or polypeptide sequence that is "derived from" a designated
CA 02314357 2000-06-12
WO 99/31969 PCTNS98/27056
23
sequence refers to a sequence that corresponds to a region of the designated
sequence. For
nucleic acid sequences, this encompasses sequences that are homologous or
complementary to the sequence, as well as "sequence-conservative variants" and
"function-conservative variants." For polypeptide sequences, this encompasses
"function-
s conservative variants." Sequence-conservative variants are those in which a
change of
one or more nucleotides in a given codon position results in no alteration in
the amino acid
encoded at that position. Function-conservative variants are those in which a
given amino
acid residue in a polypeptide has been changed without altering the overall
conformation
and function of the native polypeptide, including, but not limited to,
replacement of an
amino acid with one having similar physico-chemical properties (such as, for
example,
acidic, basic, hydrophobic, and the like). "Function-conservative" variants
also include
any polypeptides that have the ability to elicit antibodies specific to a
designated
polypeptide.
Nucleic acids comprising any of the sequences disclosed herein or
subsequences thereof can be prepared by conventional methods. For example, DNA
can
be chemically synthesized using, e.g., the phosphoramidite solid support
method of
Matteucci et al., 1981, J. Am. Chem. Soc. 103:3185, the method of Yoo et al.,
1989, J.
Biol. Chem. 764:17078, or other well known methods. This can be performed by
sequentially linking a series of oligonucleotide cassettes comprising pairs of
synthetic
oligonucleotides.
Due to the degeneracy of the genetic code, many different nucleotide
sequences can encode polypeptides having the amino acid sequences defined
herein or
subsequences thereof. The codons can be selected for optimal expression in
prokaryotic or
eukaryotic systems. Such degenerate variants are also encompassed by this
invention.
The nucleic acids may also be modified by many means known in the art.
Non-limiting examples of such modifications include methylation, "caps",
substitution of
one or more of the naturally occurring nucleotides with an analog,
internucleotide
modifications such as, for example, those with uncharged linkages (e.g.,
methyl
phosphonates, phosphotriesters, phosphoroamidates, carbamates, etc.) and with
charged
linkages (e.g., phosphorothioates, phosphorodithioates, etc.). Nucleic acids
may contain
CA 02314357 2000-06-12
WO 99/31969 PCT/U898/27056
24
one or more additional covalently linked moieties, such as, for example,
proteins (e.g.,
nucleases, toxins, antibodies, signal peptides, poly-L-lysine, etc.),
intercalators (e.g.,
acridine, psoralen, etc.), chelators (e.g., metals, radioactive metals, iron,
oxidative metals,
etc.), and alkylators. PNAs are also encompassed by the term "nucleic acid".
The nucleic
acid may be derivatized by formation of a methyl or ethyl phosphotriester or
an alkyl
phosphoramidate linkage. Furthermore, the nucleic acid sequences of the
present
invention may also be modified with a label capable of providing a detectable
signal,
either directly or indirectly. Exemplary labels include radioisotopes,
fluorescent
molecules, biotin, and the like.
The polypeptides of the invention may be expressed by using many known
vectors, such as pUC plasmids, pET plasmids (Novagen, Inc., Madison, WI), or
pRSET or
pREP plasmids (Invitrogen, San Diego, CA), and many appropriate host cells,
using
methods disclosed or cited herein or otherwise known to those skilled in the
relevant art.
The particular choice of vector/host is not critical to the practice of the
invention.
Recombinant cloning vectors will often include one or more replication systems
for
cloning or expression; one or more markers for selection in the host, such as,
for example,
antibiotic resistance; and one or more expression cassettes. The inserted
coding sequences
may be synthesized by standard methods, isolated from natural sources,
prepared as
hybrids, or the like. Ligation of the coding sequences to transcriptional
regulatory
elements and/or to other amino acid coding sequences may be achieved by known
methods. Suitable host cells may be transformed/transfected/infected as
appropriate by
any suitable method including electroporation, CaCl2 mediated DNA uptake,
fungal
infection, microinjection, microprojectile, or other established methods.
Appropriate host cells include bacteria, archebacteria, fungi, yeast, plant,
and
animal cells, and especially mammalian cells. Of particular interest are E.
coli, S aureus,
B. subtilis, Saccharomyces cerevisiae, Saccharomyces carlsbergensis,
Schizosaccharomyces pombi, SF9 cells, C 129 cells, 293 cells, Neurospora, CHO
cells,
COS cells, HeLa cells, and immortalized mammalian myeloid and lymphoid cell
lines.
Preferred replication systems include M13, ColEl, SV40, baculovirus, lambda,
adenovirus, cytomegalovirus, and the like. A large number of transcription
initiation and
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
termination regulatory regions have been isolated and are effective in the
transcription and
translation of heterologous proteins in the various hosts. Examples of these
regions,
methods of isolation, manner of manipulation, etc. are known in the art
(Ausubel et al.,
Current Protocols in Molecular Biology, John Wiley, 1997). Under appropriate
5 expression conditions, host cells can be used as a source of recombinantly
produced
peptides and polypeptides.
The MDEs of the present invention, including function-conservative variants,
may be isolated from native or heterologous organisms or cells (including, but
not limited
to, bacteria, fungi, insect, plant, and mammalian cells) into which the
protein-coding
10 sequence has been introduced and expressed. Alternatively, these
polypeptides may be
produced in cell-free protein synthesis systems, which may additionally be
supplemented
with microsomal membranes to achieve glycosylation and signal peptide
processing of
preprocollagenases. Furthermore, the polypeptides may be chemically
synthesized by
commercially available automated procedures, including, without limitation,
exclusive
15 solid phase synthesis, partial solid phase methods, fragment condensation,
or classical
solution synthesis.
Methods for polypeptide purification are well-known in the art, including,
without limitation, preparative disc-gel electrophoresis, isoelectric
focusing, HPLC,
reversed-phase HPLC, gel filtration, ion exchange and partition
chromatography, and
20 countercunrent distribution. For some purposes, it is preferable to produce
the polypeptide
in a recombinant system in which the protein contains an additional sequence
tag that
facilitates purification, such as, but not limited to, a polyhistidine
sequence. The
polypeptide can then be purified from a crude lysate of the host cell by
chromatography on
an appropriate solid-phase matrix. Alternatively, antibodies produced against
the protein
25 or against peptides derived therefrom can be used as purification reagents.
Other
purification methods are possible.
The construction and analysis of MMP variants and derivatives that exhibit
enzymatic activity, and preferably constitutive enzymatic activity, can be
achieved by
routine application of conventional methods. First, a nucleic acid encoding an
MMP is
modified either by site-directed or random mutagenesis, or is used in a
construction
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
26
scheme as one segment of a fusion gene. Preferably, the procedure results in a
modification either contained within the sequence encoding the pro region or
near the pro
region cleavage site; this includes deleting the pro region entirely.
Alternatively,
sequences may be constructed that encode fusion proteins either between
enzymatically
S active MMP domains and other polypeptides, or between different MIvJQ's. The
modified
nucleic acid is then used to program synthesis of a variant MlVI~', either in
a cell-free
system, in intact cells {including permeabilized cells), or in a transgenic
animal.
Preferably, either a cell-free system or a cell culture system is used to
express the MIVIP
variant or derivative. The extent of pro region cleavage is assessed by
metabolic labelling
and resolution of the NllVIP product by SDS-PAGE. Finally, M1V)I' enzymatic
activity is
measured using conventional assays, such as, by quantifying the cleavage of
natural
substrates or model peptides, as disclosed, e.g., in Weingarten et al.,
Biochem. 24:6730,
1985; Woessner et al., J. Biol.Chem., 263:16918, 1988, and Knight et al.,
FEBSLetts.,
296:263, 1992. In this manner, a large number of MMP variants and derivatives,
including, e.g., function-conservative variants of MNNIP-13*, can be created
routinely and
assayed for MMf' enzymatic activity.
Description of the Preferred Embodiments
The following examples are intended to illustrate the present invention
without
limitation.
Example 1: Construction of a Gene Encoding a Modified, Constitutively Active
ProMMP-13
The following experiments were performed to create a gene encoding a
procollagenase derived from M1VVIP-13 that is enzymatically active in the
absence of pro
region cleavage. The sequence of this proMIVVIP-13 variant, designated MMP 13
*, is
shown in SEQ ID N0:4.
Site-directed mutagenesis to modify MMP-13 cDNA
A cDNA fragment encoding proMlV)~' was obtained by digesting plasmid
pNot3A (Freije et al., J. Biol. Chem. 269:16766, 1994; GENBANK accession
number
CA 02314357 2000-06-12
WO 99/31969 PG"fNS98/27056
27
X75308) with XbaI and HindIII and purifying the resulting ~ 1 S 15 by
fragment. This
fragment was subcloned into the Tet-resistant/Amp-sensitive pAlter plasmid
(Promega,
Madison, WI) that had been digested with XbaI and HindIII.
Site-directed mutagenesis was performed using the Altered Sites II in vitro
Mutagenesis System (Promega, Madison, WI). Briefly, phagemid single-stranded
DNA
was purified from cultures containing the helper phage 8408 (Promega). In
addition to the
Amp repair - Tet knock-out conversion oligos (Promega), an oligonucleotide
having the
sequence 5'-AAGCCAAGATGCGGGGTTGTCGATGTGGGTGAATACAAT-3', SEQ
ID N0:8, was phosphorylated and annealed to the single-stranded DNA, followed
by
mutant strand synthesis. The reaction mixture was then used to transform the
repair-minus
E coli strain ES 1301 mutS, and the culture was grown in ampicillin selective
media.
Plasmid DNA was isolated from isolated clones and transformed into JM 109
cells, which
were then plated on LB plates containing 120 pg/ml ampicillin.
The above procedure resulted in a proline-to-valine substitution at amino acid
IS 99. The modified proMMP was designated MMP13* (SEQ ID N0:4).
Using a similar technique, site-directed mutagenesis was also used to
introduce a valine to glycine mutation at amino acid 98. A mutagenic
oligonucleotide
having the sequence 5'-
GAAAAAGCCAAGATGCGGGGGTCCTGATGTGGGTGAATAC-5', SEQ ID N0:9
was used as described above. This procedure resulted in a valine-to-glycine
substitution at
amino acid 98.
After confirmation of the above mutations by direct sequencing, cDNA
encoding MMP 13 * cDNA was excised from the pAlter-MMP 13 * vector by
digestion with
EcoRI and HindiII.
Example 2: Determination of the Enzymatic Activity of MMP-13*
Materials and Methods
cDNAs encoding both mutant forms of MMP13 and wild-type MMP-13 were
subcloned into a BS(SK-) vector (Stratagene) containing the CMV promoter (Xho
I - Eco
RI) and the SV40 splice poly (A)n (Xba I - Nco I). Duplicate cultures of Hela
cells (10 cm
CA 02314357 2000-06-12
WO 99/31969 PGT/US98IZ7056
28
dishes) were transfected with 50 ~g of these plasmids using the CaP04
precipitation
method (Promega). Five hours later, cells were subjected to a 1-minute
glycerol shock
using a solution containing an equal volume of 2 X HBS + 30 % glycerol. This
procedure
is described in the Profection Mammalian Transfection Systems technical manual
S (Promega).
Twenty-four hours following transfection, the culture medium (D-MEM
containing 10 % fetal bovine serum) was replaced with D-MEM containing no
serum and
pM CGS-27023A (Ciba-Geigy), an MMP inhibitor. It is believed that, in the
absence
of an added MMP inhibitor, MMPs produced by the culture autodigest; thus,
addition of
10 an MMP inhibitor to the culture medium resulted in a detectable MMP13 band.
Forty-eight hours after the addition of serum-free medium containing the
MMP inhibitor, 10 ml of supernatant were collected and concentrated about 200-
fold
using Centriprep-30 and Centricon-10 concentrators (Amicon), after which an
equal
volume of 2X Tris-glycine SDS running buffer was added to each sample. The
samples
were then applied to a 4-16% pre-stained beta-casein zymogram SDS
polyacrylamide gel
(Novex). After electrophoresis, the gels were renatured in renaturing buffer
(Novex) for
30 minutes at room temperature, followed by overnight incubation at
37°C in zymogram
developing buffer (Novex).
Results
Normally, MMPs are synthesized as precursors (i.e., procollagenases or
zymogens) whose enzymatic activity is latent; proteolytic removal of the
proregion after
secretion produces the enzymatically active protein. The need for proteolytic
processing is
circumvented by the use of a procollagenase variant that is enzymatically
active even
when uncleaved. The constitutively active MMP 13 variant used as the transgene
contains
a proline to valine substitution in the sequence encoding the PRCGVPDV {SEQ ID
N0:7)
region, which is highly conserved among MMPs and important for controlling
enzyme
latency.
The activity of the altered MMP 13 protein, was determined on a casein
zymogram {data not shown). Three MMP species are secreted from control HeLa
cells.
One runs at the correct molecular weight for the 92 kDa MMP9 enzyme, also
referred to as
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
29
gelatinase B. Cleavage of the prodomain of MMPs during enzyme activation
results in a
loss of approximately 10 kDa. The molecular weight of a second band is
consistent with
the processed form of gelatinase B. Stromelysin-1 (MMP3) and stromelysin-2
(MMP10)
are both 54 kDa and one or both may represent the fourth band. Pro-MIVll' 13
is expected
to migrate ~60 kDa, which is observed in lane 1 containing uncleaved
recombinant
MIVll' 13. hIeLa cells were transfected with constructs expressing both
parental and mutant
MNNIP 13. Only the ~60-kDa pro-MMP 13 form was detected from cells expressing
both
constructs, indicating that autoproteolysis of the proenzyme to the mature
form is not
likely to occur in an exogenous system. These results showed that the proline
to valine
substitution did not interfere with its native NIIVVIP13 activity or substrate
specificity.
This method provides a rapid screen for M1VVIP 13 variants that retain MMP 13
enzymatic activity.
Example 3: Construction of Transgenic Vectors
~'onstruction of MlVlp-13* Linked to tet07
In a further step, cDNA encoding M1V»'-13* was operably linked to a
transcriptional regulatory sequence derived from the tet07 promoter as
follows:
The B S(SK-) vector (Stratagene) was digested with Kpnl and NotI. A
synthetic duplex oligonucleotide having the following sequence was digested
with KpnI
and Not I and ligated to the vector:
5'-GGTACCACTAGTAAGCTTAGATCTCATATGGTCGACCCCGGGGAATTCCTGC
AGGGATCCTCTAGAAGTACTCCATGGGTATACATCGATGCGGCCGC-3',SEQID
NO:10
The B S(SK-) vector as modified above was digested with XbaI and NcoI. A
745 by fragment containing the SV40 splice site and polyadenylation signal,
which was
obtained by digesting pcDNAI/Amp (Invitrogen, Carlsbad, CA) with XbaI and
NcoI, was
ligated to this vector.
The resulting vector was linearized by digestion with XhoI and EcoRI and
ligated to a 460 by Xhol-EcoRI fragment containing the tet07 promoter region
from
pLTI~ 10-3 (Gossen et al., Proc. Natl. Acac~ Sci. USA 89:5547, 1992). This
vector was
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/2~056
then digested with SpeI, blunt-ended with Klenow polymerise, and digested with
EcoRI.
pAlter-MMP13* was digested with HindIII, blunt-ended with Klenow polymerise,
and
digested with EcoRI to obtain an MMP 13 *-encoding fragment. The MMP 13 *
EcoRI
fragment was cloned into the EcoRI digested vector obtained above.
5 The 2792 by transgene, SEQ ID NO:11 (Figure 2B), was excised by digestion
with XhoI and Notl and purified using CsCI gradient centrifugation prior to
microinjection
into fertilized eggs.
Construction of a Collagen Tyne II-Promoter-Linked tTa Gene
A gene encoding a tTA repressor-activator fusion protein was operably linked
10 to a joint-specific (type II collagen) promoter.
The modified BS(SK-) vector containing the SV40 splice site and
polyadenylation signal as described in Example 1 above was digested with Ndel
and Sma I
and ligated to a 1897 by fragment containing the collagen II promoter and
enhancer. This
fragment was obtained by digesting plasmid PBSAH1 with HindIII, after which it
was
15 blunt-ended with Klenow and digested with NdeI.
The plasmid was then digested with EcoRI and BamHI and ligated to a 1025
by fragment encoding the tetracycline/VP16 repressor-activator fusion protein
that had
been excised from the pUHGlS-1 plasmid (Gossen et al., supra) using EcoRI and
BamHI.
The plasmid was linearized by digestion with BgIII, dephosphorylated using
calf intestinal
20 phosphatase, and ligated to a 1554 BamHI enhancer fragment obtained from
plasmid
PBS~Hl .
Finally, the 5276 by transgene, SEQ ID N0:12 (Figure 2A), was excised from
the vector by digestion with KpnI and NotI, gel purified, purified by CsCI
gradient
centrifugation, dialyzed against microinjection buffer (5 mM Tris-HCl pH 7.4,
0.1 mM
25 EDTA pH 8.0) and used for microinjection.
Tvpe II collagen promoter-~3-galactosidase serve
A reporter gene, suitable for assessing the tissue-specific expression
conferred
by the type II collagen promoter, was operably linked to the type II collagen
promoter.
A 4179 by BamHI-BgIII fragment containing the ~i-galactosidase gene fused
30 to the ~-globin splice sequence and polyadenylation signal was excised from
plasmid
CA 02314357 2000-06-12
WO 99/31969 PCT/US98I27056
31
pUGHl6-3 (Gossen et al., supra) and cloned into the BamHI site of unmodified
BS(SK-)
(Stratagene). This plasmid was digested with EcoRI and HindIII and ligated to
a 655 by
Hind III-Eco RI fragment containing the type II collagen promoter sequence,
which was
excised from the plasmid described above. The plasmid was then digested with
EcoRI and
S ligated to a 2807 by Eco RI fragment which had been excised from the type II
collagen
promoter plasmid described above. Restriction mapping was used to verify the
orientation
of each insert. The 7664 by transgene, SEQ ID N0:13 (Figure 3A), was excised
by
digestion with HindIil and Notl, gel purified, purified by CsCI gradient
centrifugation,
dialyzed against microinjection buffer (5 mM Tris pH 7.4, 0.1 mM EDTA pH 8.0),
and
used for microinjection into mouse embryos.
Results
Figure 2A and 2B shows a schematic diagram of the synthetic genes
generated to achieve regulated expression of MMP13 in chondrocytes. Inducible
expression of the transgene was accomplished using the tetracycline
regulatable gene
1 S expression system (Gossen et al., supra; Furth et al., Proc. Natl. Acac~
Sci. USA,
91:9302-9306, 1994). The first construct shown in Figure 2A places expression
of the
tetracycline-controlled VP 16 transactivator fusion protein under the control
of the type II
collagen gene promoter. This construct directs expression of the VP16 fusion
protein to
chondrocytes. The second construct shown in Figure 2B places expression a cDNA
encoding a modified version of MMP 13 protein (MMP 13 *) under the direction
of the
VP 16 fusion protein. In the presence of doxycycline, a tetracycline analog
provided in the
drinking water, the VP 16 fusion protein does not bind to the Tet07 promoter
of the
synthetic MMP13* gene and the gene is silent. Upon removal of doxycycline, the
transactivator stimulates transcription of the human MMP 13 * cDNA and the
production of
the modified protein product.
In addition to verifying the MMP 13 * activity, prior to microinjection, both
transgene constructs shown in Figure 2A and 2B were tested in primary bovine
chondrocytes and in embryonic mouse fibroblast. The results showed the ability
of the
collagen II promoter to induce expression of a second construct containing
either the
Tet07-luciferase or the Tet07-MMP13* (data not shown).
CA 02314357 2000-06-12
WO 99/31969 PCTNS98/27056
32
Figure 3A is a schematic illustration of a transgene used to assess
tissue-specific regulation conferred by type II collagen promoter. The nucleic
acid
construct comprises: (i) sequences derived from a rate type II collagen
promoter; (ii)
sequences encoding bacterial (3-galactosidase (Lac Z); and (iii) sequences
comprising an
(3-globin splice and polyadenylation signal.
Example 4: Production and Characterization of Transgenic Mice Expressing
Tetracycline-Regulated LacZ or MMP-13* in Joint Tissues
The following experiments were performed to produce transgenic mice
expressing MMP-13* or a LacZ (~i-galactosidase) reporter gene.
Materials and Methods
Preparation aced testing of transgene mice. To produce mice expressing
MMP-13* under tetracycline regulation, an XhoI-Notl tet07-IvM'-13* DNA
fragment of
about 2784 base pairs (Figure 2B) and a Kpnl - NotI CPE-tTA DNA fragment of
about
5265 base pairs (Figure 2A) were co-microinjected into fertilized mouse
embryos in
equimolar amounts. To produce mice expressing the reporter gene, a HindIII-
NotI LacZ
7641 base pairs fragment (Figure 3A) was injected into (FVB/N) fertilized eggs
as
described (Hogan et al., Manipulating the Mouse Embryo, Cold Spring Harbor
Laboratories, 1996). (The size of the restriction fragments is smaller than
the transgenes
described in Example 3 because the endonuclease cleavage sites are internal to
the
transgenes.)
Founder animals were first identified by PCR as follows. The tTA-encoding
transgene was identified using a primer corresponding to the tTA sequence (5'-
CGAGGGCCTGCTCGATCTCC-3', SEQ ID N0:14) and a primer corresponding to a 3'
untranslated sequence (5'-GGCATTCCACCACTGCTCCC-3', SEQ ID NO:15). The
resulting PCR product was 584 by in size. The NM' 13 * -encoding transgene was
identified using primers corresponding to sequences encoding M1V>I'13* (5'-
GAGCACCCTTCTCATGACCTC-3', SEQ ID N0:16) and the 3' untranslated region,
respectively. The resulting PCR product was 731 by in size.
The LacZ-encoding transgene was identified using primers corresponding to
CA 02314357 2000-06-12
WO 99/31969 PCTNS98/27056
33
the nuclear localization signal of the ~-galactosidase gene (5'-
GTTGGTGTAGATGGGCGCATCG-3', SEQ ID N0:17) and the collagen promoter (5'-
GCGGGGTCTCAGGTTACAGCC-3', SEQ ID N0:18). The resulting PCR product was
673 by in size.
Southern blot analysis of tail DNA digested with BamHI/NcoI or PwIIINcoI
and hybridized to the 3' untranslated region under high stringency conditions
was
performed to confirm the results obtained using PCR. The number of copies of
transgene
DNA that integrated into the genome was determined by comparing the relative
intensity
of the hybridization signal from transgenic mice with that obtained using
control DNAs
containing 10 and 100 genome equivalents of the same DNA that was injected.
Copy number was confirmed using Taqman quantitative PCR according to
manufacturer's specifications (Perkin Elmer). Transgenic lines were generated
by mating
founder animals to FVB/N wild-type mice and the subsequent generations were
identified
by PCR.
All mice were administered doxycycline (Sigma Chemical Co., St. Louis MO)
prepared as a 100 mg/ml stock solution in 50% ethanol, and diluted to a final
concentration of 1.0 mg/ml in acidic drinking water, which was changed on a
daily basis
(Schultze et al., Nature Biotech. 14:499, 1996).
Whole Embryo Staining for /~ Galactosidase (lack Activity. Wild-type
females were mated with transgenic males harboring the CPE-lacZ construct. On
embryonic day 16 (E16), the females were sacrificed, and the embryos stained
for
~-galactosidase activity as described (Hogan et al., supra, 1996). Prior to
fixation, tails
from the E16 embryos were removed, digested and analyzed for transgene
transmission by
PCR.
Expression Analysis via RT PCR Transgene expression was assayed by
RT-PCR. Total RNA was isolated from tissues following homogenization in Trizol
(Life
Technologies). First strand synthesis was generated using the Superscript
preamplification
kit by Gibco/BRL. Briefly, Spg of total RNA was treated with DNase I for 15
minutes at
RT, then inactivate by the addition of 2 gl of 25 mM EDTA and heated to
65°C for fifteen
minutes. Following, the RNA was annealed to 0.5 pg oligo dT and reverse
transcribed
CA 02314357 2000-06-12
WO 99/31969 PCT/US98127056
34
according to manufacturer's specifications. PCR analysis of the cDNA was done
using the
following primers sets (5' to 3' sequences): tet activator:
CGCCCAGAAGCTAGGTGTAGAG (SEQ D7 N0:19) and
CGGCCATATCCAGAGCGCCG (SEQ ID N0:20); MMP13*:
GCCCTCTGGCCTGCTGGCTCATG (SEQ ID N0:21) and
CAGGAGAGTCTTGCCTGTATCCTC (SEQ >D N0:22). The resulting PCR products
are 859 by and 648 bp, respectively. To test the integrity of the mRNA and the
efficiency
of the reverse transcriptase, each PCR reaction also contained the following c-
fos primer
set: 5'-AGGAGGGAGCTGACAGATACACTCC-3' (SEQ ID N0:23) and
5'-AGGCCACAGACATCTCCTCTGG-3' (SEQ m N0:24). PCR analysis was
performed on the cDNA using Taq-gold (Perkin Elmer) for 10 minutes at
95°C, followed
by 35 cycles of 60 seconds at 96°C, 90 seconds at 67°C, and 60
seconds at 72°C. A final
twelve minute extension was done at 72°C. Reaction products were run on
2.0% agarose
gels and visualized by ethidium bromide staining.
Embryonic FibroblastlRT PCR Tet07-lacZ females were mated to
transgenic males harboring both the CPE-tTA and the Tet07-MMP 13 * transgenes.
On
E15, the females were sacrificed, and fibroblast prepared from the embryos
(Graham et al.,
Virology, 52:456, 1973; Lopata et al., Nucl. Acidr Res., 12:5707, 1984). The
fibroblast
were cultured in DMEM (GibcoBRL) containing 10% FBS (GibcoBRL). The fibroblast
were subsequently transfected via the calciumphosphate precipitation method
with the
desired expression plasmids. Forty-eight hours after transfection, total RNA
was prepared
from the transfected cells using the Trizol method (GibcoBRL). First strand
cDNA
synthesis was prepared using the Superscript preamplification system
(GibcoBRL).
MMP13* expression was identified using primers specific for human MMP13
(GCCCTCTGGCCTGCTGGCTCATG) (SEQ )D NO:21) and
(CAGGAGAGTCTTGCCTGTATCCTC) (SEQ ID N0:22). The resulting PCR was 648
by in size.
Immunohistochemistry. Expression of MMP 13 * in the double transgenic
lines was further analyzed by immunohistochemistry, using antibodies specific
for MMP-
13-derived type II collagen cleavage fragments. For this purpose, joints were
fixed in 4%
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
paraformaldehyde in PBS at neutral pH for 60 minutes at room temperature. They
were
then rinsed twice in PBS, incubated in O.1M Tris-HCI, pH 7.4, overnight, and
partially
decalcified in 0.2M EDTA at neutral pH. The samples were transferred to TOC
medium
and 6-mm frozen sections were obtained using a HackerBright cryostat. The
sections
5 were stained with an antibody that recognizes an epitope present in a
degradation product
of type II collagen, specifically, in the TC" degradation product, which is
also designated
the 3/4 -piece. Billinghurst et al., J. Clin. Invest. 99:1534, 1997.
Results
Tetracycline and their analogues are known inhibitors of MMP activity. As a
10 result, we compared the serum levels of DOX when 1 mg/ml was added to the
drinking
water and the in vitro ICso, In a MCA fluorescent assay the ICso = 59.1 ~M,
whereas the
serum levels measured 2.64 ~M using a zone of inhibition assay. These data
show that the
amount of DOX in the serum is 22.4 fold below the level at which 50% of MMP
activity
could be inhibited. Thus, it is unlikely that there is a significant
inhibition due to the
15 DOX.
Figure 3B is a photographic illustration of whole mount staining for
j3-galactosidase activity of embryonic day 16 transgenic mouse embryos
expressing the
transgene illustrated in Figure 3A. Blue staining (arrows) is evident in the
joints
throughout the body of the transgenic animal, while no staining is observed in
the
20 non-transgenic, wild-type littermate. Specifically, joints including the
ankles, knees, hips,
phalanges, wrists, elbows, shoulders, and vertebrae showed expression of the
transgene.
In addition to the cartilage of the joints, cartilage that has not ossified to
the bone at this
stage of development, i.e., some of the facial, skull, and rib bones, also
stained blue.
Figure 3C shows an enlargement of the elbow and paw. These data confirm the
25 expression abilities of the type II collagen promoter, and are useful in
determining those
tissues (joints) that will express the MMP13* transgene.
The constructs shown in Figure 2A and 2B were co-microinjected into
fertilized mouse embryos. Out of 112 newborn mice, 7 transgenic founders
harboring
both transgenes were identified, however, only four of these transgenic lines
were capable
30 of breeding. The transgenes were identified by PCR and verified by Southern
blot
CA 02314357 2000-06-12
WO 99!31969 PCT/US98/27056
36
analysis using a transgene-specific probe '(data not shown). The copy number
for each of
the four transgenics was further accessed using Taqman quantitative PCR (data
not
shown). Briefly, transgene copy number ranged from 1-32 and 1-20 for the tet
activator
and MMP13*, respectively. Specifically, line 6 contained ~8 copies of the tet
activator
and ~3 copies of the MMP 13 * transgene.
Expression of the TA and MMP 13 * transgenes were initially evaluated in the
hind-knee joints of four-month old mice (line 6). Amplification of the c-fos
endogenous
cDNA was used as a control to verify the efficacy of each reaction. Figure 4
shows
amplification of an 890 by fragment resulting from a TA-specific primer set.
RT-PCR
showed the TA transgene to be expressed in transgenic mice, both on and off
DOX, but
was not expressed in the non-transgenic controls (lanes 4-5). Constitutive
expression of
the TA is expected since it is driven by a constitutively active promoter.
Moreover,
expression of the TA is limited to the joints and was not observed by the RT-
PCR in other
tissues including brain, heart, liver, kidney, spleen, or skeletal muscle
(data not shown).
Figure 4B shows amplification of a b45 by fragment resulting from an
MMP 13 * specific primer set. Note, the MMP 13 * primer set is specific for
human
MMP13 and does not react with its endogenous mouse homologue, collagenase-1.
RT-PCR showed that MMP 13 * was not expressed in the non-transgenic controls
(lanes
4-5). Lanes 6-7 show that there is expression of the MMP 13 * transgene in
mice
maintained on DOX. Removal of DOX from the drinking water induces a
significant
amount of expression (lanes 8-9). We estimate a 5-10 fold induction.
Furthermore,
following gel electrophoresis, PCR fragments were transferred to a nylon
membrane and
hybridized to a TA or MMP 13 * specific probe to verify the identity of the
PCR product
(data not shown).
Fibroblasts from several transgenic lines (such as, e.g., lines 8 and 42) were
capable of expressing MMP 13 *, as evidenced by the appearance of a PCR
product of the
predicted size. No MMP13* RT-PCR band was detected from cells transfected with
vehicle alone. These results indicated that, in these mice, the MMP13*
transgene is
integrated into a transcriptionally active region of the chromatin.
~As early as 3 days after removal of the mice from doxycycline, MMP-13
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/Z7056
37
cleavage products could be detected immunologically. After 30 days without
doxycycline,
a substantial increase in staining in the growth plate and in the articuiar
cartilage could be
seen (Figure $), but the results differed among different lines of mice (see
Table 1 below).
Table 1
$ ~ Immuno6istochemistry
Fl Days Type II Collagen Loss of
Anima! off DOX hMMPl3 Ab Cleavage FragmentsSafranin
Ab O Stain
Line 8 wt - - not reanarkable
Line 8 0 d - - "
3d + +
7 d ++ ++ Mild
14 d +++ +++ Moderate
- - not remarkable
Line 6 30 d Moderate
1$ Line 8 30 d +++ +++ Moderate
Line 42 30 d + - not remarkable
Example 5: Phenotype of MMP13* Transgenic Mice
Materials and Methods
To study the effect of MMP 13 * activity on cartilage in adult transgenic
animals, mice were withdrawn from doxycycline for increasing times, after
which they
were sacrificed. Paraffin-embedded formaldehyde-fixed sections of decalcified
cartilage
were sectioned and stained with (i) hematoxylin and eosin (H&E) and (ii)
Safranin O
2$ followed by fast green (American Histo Labs, Gaithersburg MD). Staining
techniques in
articular cartilage have been described (Peter et al., J. Exp. Pathol. 71:19,
1990).
Results
Control transgenic animals that lack MIVVIP 13 * expression retain a
significant
amount of safranin O stain in both the articular cartilage as well as the
growth plate of
their patella (Figure 6A). By contrast, transgenic animals from line 8 show a
substantial
loss of safranin O staining in their joints following doxycyline withdrawal.
After seven
days, a mild reduction of safranin O staining is observed in the articular
cartilage of the
patella (Figure 6B), which progresses by day 14 to moderate loss of stain in
articular
CA 02314357 2000-06-12
WO 99/31969 PCT/US98/27056
38
cartilage as well as the growth plate (Figure 6C). A significant loss of
safranin O stain
was also observed in the other joints including the cartilage of the tarsus
and femur, as
well as wrist and knuckle, indicating a reduced proteoglycan concentration in
these areas
compared to controls.
To access any changes in the articular cartilage due to transgene expression,
mice from line 6 were maintained or removed from DOX for 114 days, and their
joints
were sectioned and stained with H&E. When compared with an age matched
littermate,
control the transgenic removed from DOX developed a pathology reminiscent of
osteoarthritis. Shown in Figure 7A, the control animal showed no lesions or
other
osteoarthritis pathologies, whereas the transgenic animal shows the formation
of lesions in
its articular cartilage (Figure 7B). More specifically, the H&E sections show
considerable
loss of cartilage, focal erosions, erosions that extend into the bone, and an
inflamed
synovium. Within the synovium, there is evidence of fibroid necrosis,
metaplasia, and
synovial cell hyperplasia. In addition to these symptoms of osteoarthritis,
some changes
1 S observed are more characteristic of rheumatoid arthritis. These changes
include
angiogenesis as seen by an infiltration of red blood cells, monocytes, and
macrophages.
Figures 7C and 7D show the synovium at a higher magnification.
Discussion
Transgenic mice expressing a constitutively active human MMP13 protein in
the articular cartilage of adult animals have been successfully created. We
showed that a
unique combination of technologies, i.e., the tetracycline regulatable gene
expression
system and chondrocyte specific expression of a constitutively active MMP
protein, has
enabled us to develop a transgenic model resulting in lesion formation and
other
osteoarthritis pathologies. Using a regulatable/inducible system enabled us to
bypass
deleterious embryonic effects, while allowing significant hMiVl:Pl3*
expression in the
adult mouse. The Lac Z staining shown in Figure 3, demonstrates that type II
collagen
expression occurs during development at about stage E16 and that type II
collagen is
involved during the formation of the skeleton. Type II collagen, as well as
type I collagen
are known to be substrates for proteolytic cleavage by MMP13. Therefore, we
predicted
that unregulated expression of MMP13 during embryogenesis would be lethal.
CA 02314357 2000-06-12
WO 99/31969 PCT/US98I2'1056
39
Four transgenic lines containing both transgenes were established. In the two
lines that expressed hMMP 13 *, one expressed at very low levels, whereas the
other line
expressed significant amounts of hMMPl3*. The line that expressed only
marginal
amounts of transgene displayed histological evidence of proteoglycan loss.
However, after
six months off DOX no lesions were observed. Line 6, which expressed
significantly
greater amounts of hMIVV>P13*, showed osteoarthritis pathologies including
lesion
formation, cartilage degradation, and an inflamed synovium after four months
off DOX.
Moreover, expression of hMIVVIP13* in line 6 can be controlled, i.e., turned
on and offby
the investigator. This result provides additional evidence that the phenotype
is due to
expression of hMMPl3* and not the result of the integration site in the
chromatin.
As observed in line 6, the joint destruction/erosion, lesions, fibroid
necrosis,
metaplasia, synovial cell hyperplasia and an inflamed synovium (in the absence
of T-cells)
are among pathologies observed in patients with osteoarthritis. However, not
all of the
pathologies observed in the transgenics are reminiscent of osteoarthritis. For
example,
angiogenesis and infiltration of monocytes and macrophages are pathologies
observed
during the inflammation process associated with rheumatoid arthritis. Note,
the absence of
neutrophils in the synovial fluid. Migration of neutrophils to the site of
inflammation is a
hallmark pathology of rheumatoid arthritis.
The data presented in this paper provides direct evidence that MMP13 is a
critical player in the development of osteoarthritis. Moreover, the
transgenics described in
this paper provide an animal model to test the efficacy of therapeutics.
Compounds that
modulate the activity of MMP13 or inhibit progression of osteoarthritis can be
monitored
by determining lesion formation and other OA pathologies at various times
during the
progression of the disease. The ability to turn on and off hMMP 13 *
expression, thus
controlling production/timing of lesion formation will be advantageous to
determining the
compound efficacy.
This OA-like transgenic model can also be used to answer a growing list of
biological questions. In addition to MMP13, interstitial collagenase (MMP1)
and
neutrophil collagenase (MMPB) have been shown to cleave type II collagen.
Thus,
transgenic mice expressing constitutively active MMP1 or MMP8 would be
expected to
CA 02314357 2000-06-12
WO 99/31969 PCTNS98/29056
show a similar phenotype in their articular cartilage.
Ezample 6: Augmentation of the Development of Symptoms of Joint
Degenerative Disease in MMP-13 Transgenic Mice
The following treatments are performed to enhance the symptoms of joint
degeneration exhibited by the transgenic animals of the invention.
A group of transgenic mice are treated to induce expression of the transgenes
at 4-12 weeks of age. Two to six weeks after induction, the mice are injected
10 intraperitonealy with an inflammatory agent, including without limitation,
lipopolysaccharide (10-100 gg), zymosan (1-10 mg), the superantigen
Staphylococcal
Enterotoxin B (1-100 gg), or TGF-(3 (1-10 fig). Alternatively, the animals are
injected
intraarticularly with an inflammatory or chondrocyte function-modulating
agent, including
without limitation, lipopolysaccharide (1-100 ng), zymosan (50-250 fig),
papain (10-100
15 fig), TGF-~i (0.01-1 wg), Bone Morphogenic Protein -2 (2-1000 ng), IL-1 (1-
100 ng), TGF-
a (10-200 ng), IGF (0.01-1 pg), or FGF (0.01-1 pg). Age- and sex-matched
transgenic
mice maintained under a regimen in which the transgenes are not expressed
receive the
same treatment and serve as controls.
The development of symptoms of degenerative joint disease is monitored by
20 gross observation of joint swelling and function, and by histological
evaluation of the joint
at selected timepoints after exposure to the inflammatory agent.
The agents will induce an acute inflammatory response and/or transient loss of
proteoglycan with a duration of less than one week. The acute inflammatory
response
and/or transient cartilage changes will upregulate gene expression in the
chondrocytes,
25 enhancing the expression of the transgene and increasing the levels of MMP-
13 produced.
All patents, applications, articles, publications, and test methods mentioned
above are hereby incorporated by reference in their entirety.
Many variations of the present invention will suggest themselves to those
skilled in the art in light of the above detailed description. Such obvious
variations are
30 within the full intended scope of the appended claims.
CA 02314357 2000-06-12
WO 99/31969 PCT/US98IZ7056
1
SEQUENCE LISTING
<110> Neuhold, Lisa
Killar, Loran
<120> TRANSGENIC ANIMAL MODEL FOR DEGENERATIVE
DISEASES OF CARTILAGE
<130> AHP-97285PCT
<140> TBA
<141>
<150> 60/068,312
<151> 1997-12-19
<150> 08/994,689
<151> 1997-12-19
<160> 24
<170> FastSEQ for Windows Version 3.0
<210> 1
<211> 7
<212> PRT
<213> Homo Sapiens
<400> 1
Pro Arg Cys Gly Xaa Pro Asp
1 5
<210> 2
<211> 12
<212> PRT
<213> Homo Sapiens
<400> 2
His Glu Xaa Gly His Xaa Xaa Xaa Xaa Xaa His Ser
1 5 10
<210> 3
<211> 1521
<212> DNA
<213> Homo sapiens
<400> 3
caagatgcatccaggggtcctggctgccttcctcttcttgagctggactcattgtcgggc 60
cctgccccttcccagtggtggtgatgaagatgatttgtctgaggaagacctccagtttgc 120
agagcgctacctgagatcatactaccatcctacaaatctcgcgggaatcctgaaggagaa 180
tgcagcaagctccatgactgagaggctccgagaaatgcagtctttcttcggcttagaggt 240
gactggcaaacttgacgataacaccttagatgtcatgaaaaagccaagatgcggggttgt 300
cgatgtgggtgaatacaatgttttccctcgaactcttaaatggtccaaaatgaatttaac 360
ctacagaattgtgaattacacccctgatatgactcattctgaagtcgaaaaggcattcaa 420
aaaagccttcaaagtttggtccgatgtaactcctctgaattttaccagacttcacgatgg 480
cattgctgacatcatgatctcttttggaattaaggagcatggcgacttctacccatttga 540
tgggccctctggcctgctggctcatgcttttcctcctgggccaaattatggaggagatgc 600
ccattttgatgatgatgaaacctggacaagtagttccaaaggctacaacttgtttcttgt 660
tgctgcgcatgagttcggccactccttaggtcttgaccactccaaggaccctggagcact 720
catgtttcctatctacacctacaccggcaaaagccactttatgcttcctgatgacgatgt 780
SUBSTITUTE SHEET (RULE 26)
CA 02314357 2000-06-12
WO 99/31969 PCTNS98~Z7056
2
acaagggatccagtctctctatggtccaggagatgaagaccccaaccctaaacatccaaa840
aacgccagacaaatgtgacccttccttatcccttgatgccattaccagtctccgaggaga900
aacaatgatctttaaagacagattcttctggcgcctgcatcctcagcaggttgatgcgga960
gctgtttttaacgaaatcattttggccagaacttcccaaccgtattgatgctgcatatga1020
gcacccttctcatgacctcatcttcatcttcagaggtagaaaattttgggctcttaatgg1080
ttatgacattctggaaggttatcccaaaaaaatatctgaactgggtcttccaaaagaagt1140
taagaagataagtgcagctgttcactttgaggatacaggcaagactctcctgttctcagg1200
aaaccaggtctggagatatgatgatactaaccatattatggataaagactatccgagact1260
aatagaagaagacttcccaggaattggtgataaagtagatgctgtctatgagaaaaatgg1320
ttatatctattttttcaacggacccatacagtttgaatacagcatctggagtaaccgtat1380
tgttcgcgtcatgccagcaaattccattttgtggtgttaagtgtctttttaaaaattgtt1440
atttaaatcctgaagagcatttggggtaatacttccagaagtgcggggtaggggaagaag1500
agctatcaggagaaagcttgg 1521
<210> 4
<211> 471
<212> PRT
<213> Homo sapiens
<400> 4
Met His Pro Gly Val Leu Ala Ala Phe Leu Phe Leu Ser Trp Thr His
1 5 10 15
Cys Arg Ala Leu Pro Leu Pro Ser Gly Gly Asp Glu Asp Asp Leu Ser
20 25 30
Glu Glu Asp Leu Gln Phe Ala Glu Arg Tyr Leu Arg Ser Tyr Tyr His
35 40 45
Pro Thr Asn Leu Ala Gly Ile Leu Lys Glu Asn Ala Ala Ser Ser Met
50 55 60
Thr Glu Arg Leu Arg Glu Met Gln Ser Phe Phe Gly Leu Glu Val Thr
65 70 75 80
Gly Lys Leu Asp Asp Asn Thr Leu Asp Val Met Lys Lys Pro Arg Cys
85 90 95
Gly Val Val Asp Val Gly Glu Tyr Asn Val Phe Pro Arg Thr Leu Lys
100 105 110
Trp Ser Lys Met Asn Leu Thr Tyr Arg Ile Val Asn Tyr Thr Pro Asp
115 120 125
Met Thr His.Ser Glu Val Glu Lys Ala Phe Lys Lys Ala Phe Lys Val
130 135 140
Trp Ser Asp Val Thr Pro Leu Asn Phe Thr Arg Leu His Asp Gly Ile
145 150 155 160
Ala Asp Ile Met Ile Ser Phe Gly Ile Lys Glu His Gly Asp Phe Tyr
165 170 175
Pro Phe Asp Gly Pro Ser Gly Leu Leu Ala His Ala Phe Pro Pro Gly
180 185 190
Pro Asn Tyr Gly Gly Asp Ala His Phe Asp Asp Asp Glu Thr Trp Thr
195 200 205
Ser Ser Ser Lys Gly Tyr Asn Leu Phe Leu Val Ala Ala His Glu Phe
210 215 220
Gly His Ser Leu Gly Leu Asp His Ser Lys Asp Pro Gly Ala Leu Met
225 230 235 240
Phe Pro Ile Tyr Thr Tyr Thr Gly Lys Ser His Phe Met Leu Pro Asp
245 250 255
Asp Asp Val Gln Gly Ile Gln Ser Leu Tyr Gly Pro Gly Asp Glu Asp
260 265 270
Pro Asn Pro Lys His Pro Lys Thr Pro Asp Lys Cys Asp Pro Ser Leu
275 280 285
Ser Leu Asp Ala Ile Thr Ser Leu Arg Gly Glu Thr Met Ile Phe Lys
290 295 300
Asp Arg Phe Phe Trp Arg Leu His Pro Gln Gln Val Asp Ala Glu Leu
305 310 315 320
Phe Leu Thr Lys Ser Phe Trp Pro Glu Leu Pro Asn Arg Ile Asp Ala
SUBSTITUTE SHEET (RULE 26)
CA 02314357 2000-06-12
WO 99/31969 3 PCTNS98/Z7056
325 330 335
Ala Tyr Glu His Pro Ser His Asp Leu Ile Phe Ile Phe Arg Gly Arg
340 345 350
Lys Phe Trp Ala Leu Asn Gly Tyr Asp Ile Leu Glu Gly Tyr Pro Lys
355 360 365
Lys Ile Ser Glu Leu Gly Leu Pro Lys Glu Val Lys Lys Ile Ser Ala
370 375 380
Ala Val His Phe Glu Asp Thr Gly Lys Thr Leu Leu Phe Ser Gly Asn
385 390 395 400
Gln Val Trp Arg Tyr Asp Asp Thr Asn His Ile Met Asp Lys Asp Tyr
405 410 415
Pro Arg Leu Ile Glu Glu Asp Phe Pro Gly Ile Gly Asp Lys Val Asp
420 425 430
Ala Val Tyr Glu Lys Asn Gly Tyr Ile Tyr Phe Phe Asn Gly Pro Ile
435 440 445
Gln Phe Glu Tyr Ser Ile Trp Ser Asn Arg Ile Val Arg Val Met Pro
450 455 460
Ala Asn Ser Ile Leu Trp Cys
465 470
<210> 5
<211> 470
<212> DNA
<213> Artificial Sequence
<220>
<223> Tet promoter cloned into cytomegaloviral promoter,
"tet 07 promoter"
<400> 5
ctcgagtttaccactccctatcagtgatagagaaaagtgaaagtcgagtttaccactccc 60
tatcagtgatagagaaaagtgaaagtcgagtttaccactccctatcagtgatagagaaaa 120
gtgaaagtcgagtttaccactccctatcagtgatagagaaaagtgaaagtcgagtttacc 180
actccctatcagtgatagagaaaagtgaaagtcgagtttaccactccctatcagtgatag 240
agaaaagtgaaagtcgagtttaccactccctatcagtgatagagaaaagtgaaagtcgag 300
ctcggtacccgggtcgagtaggcgtgtacggtgggaggcctatataagcagagctcgttt 360
agtgaaccgtcagatcgcctggagacgccatccacgctgttttgacctccatagaagaca 420
ccgggaccgatccagcctccgcggccccgaattagcttgatatcgaattc 470
<210> 6
<211> 3479
<212> DNA
<213> Unknown
<220>
<223> Sprague Dawiey rat type II collagen promoter.
<400> 6
ggtaccactagtaagcttagatccactgtctgggattatatcaggacaaccgaagcctgg 60
aaagtgtattaggtagagcattttcttccacgtgtttgggcacgtttccgacagctagga 120
ttccagctctgtctttgtatgttacagactgtaaatcaatcgcaggtgaaactgtttgga 180
cagtaggtggggatcaaagaccctccgcccgtgagactctaggcgctttcccctgccacc 240
agcctgtctccagagatgctctggaaggaggcgggcccgggcggtctttctgctctttag 300
cgtggcggacgcggcggcgggggcagggctggagcagagagcgctgcagtgatagaactt 360
tctgaccccgctgcgcagggcggcagggtggcagggtggcagggtggcgagctaagccag 420
agccgaacgctggagctctgggaggaacatcgaaggtttgtatgtggtctgagatcggcc 480
tgactatatttttttgtcctaaatttgcaagcacacacccacaaagctgcggtcttgacc 540
ggtattctttatagagcgcaatggagtgagctgagtgtctaaacgatttccctaattcat 600
ctgatagcagaggcgctctcctaattggcgaagagctgcctcatgtccgcaactttttgg 660
cagagtgaattccacagctttgtgtgtgtgtgtgggggggggtgtaaggggtgtctaaaa 720
ctttcggtctcctactattctgtatctcgaccggttggttttacaccccggctcatctca 780
SUBSTITUTE SHEET (RULE 28)
CA 02314357 2000-06-12
WO 99131969 4 ' PCTNS98I27056
tcaacgcaaacacccccactctcctatggacccaaggacctgacgtgggggaaggtggac840
attaggaatgtcagaaacctagagtccacgctcctcctctccatctttccacgagtttgg900
gaaacttcttggctgcgaagactttgacccacatctgcatttctcagccccagcttccaa960
aagtgctgcaggttcgggaggggagacctcagtcctcctttgtgaggcttgtttgcgttg1020
agggattggcagcgatggcttccagatgggctgaaaccctgcccgtatttatttaaactg1080
gttcctcgtggagagctgtgaatcgggctctgtatgcgctcgagaaaagccccattcatg1140
agaggcaaggcccagtgggtccccccgactccccgacccccctctcccacaatatatccc1200
ccctccctgtgcccgcctgccgccacctcccgggctccggccccgcgcgcagcggcgacg1260
aagcaacacagttccccgaaagaggtagctttttaattggccagccacaaagaatcactt1320
atgccgcacggcggtaacgaggggaaccggatcgggcggccaggatgctatctgtgtagc1380
ccttttcgtgccacaattagggtggtgctggcttcctccgaccgcacctaggcgatctgg1440
ttacactgttggctcctttcttgggcagtcatttaatcctactttttactctacgaatgt1500
ctgtctgatggagggctgtgtccggagccccatccacaaagagtcagccagcagctctca1560
cacccggctggatctcatatggtgcactctcagtacaatctgctctgatgccgcatagtt1620
aagccagccaagctagcttgcgcaagctagcttgcgatccgtaaaaatgtgtgagagtta1680
caaaatgtcttccgggctaagatccgacagccatggtccaaagaagacttcggcactgca1740
gacttaaaaccagctttctagcagaggcagaaggatctagagccaaaggcaaagacttga1800
ataggctgggaagatgcaagaatggcattttacataaagaacactctctccttttccagc1860
cagcacacttgcatagaaattaagttttacacttgaagttctttgtttccatcctgagaa1920
gctccaaagtctgaggtggtgtggtatgctgggtaattctccccaccccccaacattccc1980
tgggggttccatgggggtagcttctcccaaggacttccagcggcaacacagaaatcccac2040
ttcgagacaaaggagttactgcttaaatcaggccctaatttccaaggttccctttgctta2100
aagttccctagaggaccatctcacttctaaagaaaaggtgtattcggggacccatcctca2160
acctccttgttatggaaggagacttcgggaacagagcaagggctgagcctccggcagttt2220
ggggtaaggttggggttggggggagcaaggaaggcaagtgaggctggaggcccagggata2280
ggggaagatgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtctcggggatgg2340
tggtggtggacaactaggaaactctggcgctttctcctcccctcacaaaactgagtccag2400
ctggagccgcctccagactctctggccagggcctcagagtggtcaacagtccctggccag2460
cgttgctctctccaggctaagggcacccactcccctggagattcctgaacctgggccagg2520
aagagccgaattagacaagtgtctccaatccggctgcgtgcggattttgttgcggtgtcc2580
ctcggttgtctgcagttcctttagtcccttccctggcctgccccttacacctccacacag2640
gtccccctctgtgtaggaatacaccagaccctctcttagccacacacacctccagtcccc2700
cgtctacctagatttttttcatagctagttggatgggggatgggttagggaggctgggtt2760
tgcgagcctccaggtgggagttcaccgacaggtactccgcaaaggagctggaaggcaggt2820
ctggaaaactgtcccccagatttaggattctgggcagcttccatcagcttatactttggc2880
tcccccgccccctaaactccccatccccaccttcctttctcccgttacttcgtcctccct2940
cgcctttccagccttgagtctaaagctccatgcttatgcctctgcaaacaaccccctccc3000
ttctaaccccagcagaactccgaggaaaggggccggaggccccccttctcgcctgtggtt3060
agagggggcagtgtggcagtcccaagtgggggcgaccggaggccgtctcggtgccccgcc3120
cgatcaggccactgggcacatcgggggcgggaagctgggctcaccaaaggggcgactggc3180
cttggcaggtgtgggctctggtccggcctgggcaggctccgggggcggggtctcaggtta3240
cagccccgcggggggctggggggcggcccgcggtttgggctggtttgccagcctttggag3300
cgaccgggagcatataaccggagcctctgctgggagaagacgcagagcgccgctgggctg3360
ccgggtctcctgcctcctcctcctgctcctagagcctcctgcatgagggcgcggtagaga3420
cccggacccgctccgtgctctgccgcctcgccgagcttcgcccgcaagctggggaattc 3479
<210> 7
<211> 8
<212> PRT
<213> Homo sapiens
<400> 7
Pro Arg Cars Gly Val Pro Asp Val
<210> 8
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
suesTrtvrs sH~r f RULE 2s)
CA 02314357 2000-06-12
WO 99/31969 p~yVgggn~p~
<223> MMP13* Mutagenic Oligonucleotide
<400> 8
aagccaagat gcggggttgt cgatgtgggt gaatacaat 39
<210> 9
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> MMP13* Mutagenic Oligonucleotide
<400> 9
gaaaaagcca agatgcgggg gtcctgatgt gggtgaatac 40
<210> 10
<211> 98
<212> DNA
<2i3> Artificial Sequence
<220>
<223> HS(SK-) vector
<400> 10
ggtaccacta gtaagcttag atctcatatg gtcgaccccg gggaattcct gcagggatcc 60
tctagaagta ctccatgggt atacatcgat gcggccgc 98
<210> 11
<211> 2792
<212> DNA
<213> Artificial Sequence
<220>
<223> tet-07/MMP13* transgene
<400> 11
ctcgagtttaccactccctatcagtgatagagaaaagtgaaagtcgagtttaccactccc60
tatcagtgatagagaasagtgaaagtcgagtttaccactccctatcagtgatagagaaaa120
gtgaaagtcgagtttaccactccctatcagtgatagagaaaagtgaaagtcgagtttacc180
actccctatcagtgatagagaasagtgaaagtcgagtttaccactccctatcagtgatag240
agaaaagtgaaagtcgagtttaccactccctatcagtgatagagaaaagtgaaagtcgag300
ctcggtacccgggtcgagtaggcgtgtacggtgggaggcctatataagcagagctcgttt360
agtgaaccgtcagatcgcctggagacgccatccacgctgttttgacctccatagaagaca420
ccgggaccgatccagcctccgcggccccgaattagcttgatatcgaattcgagctcggta480
cccggggatcctctagacaagatgcatccaggggtcctggctgccttcctcttcttgagc540
tggactcattgtcgggccctgccccttcccagtggtggtgatgaagatgatttgtctgag600
gaagacctccagtttgcagagcgctacctgagatcatactaccatcctacaaatctcgcg660
ggaatcctgaaggagaatgcagcaagctccatgactgagaggctccgagaaatgcagtct720
ttcttcggcttagaggtgactggcaaacttgacgataacaccttagatgtcatgaaaaag780
ccaagatgcggggttgtcgatgtgggtgaatacaatgttttccctcgaactcttaaatgg840
tccaaaatgaatttaacctacagaattgtgaattacacccctgatatgactcattctgaa900
gtcgaaaaggcattcaaaaaagccttcaaagtttggtccgatgtaactcctctgaatttt960
accagacttcacgatggcattgctgacatcatgatctcttttggaattaaggagcatggc1020
gacttctacccatttgatgggccctctggcctgctggctcatgcttttcctcctgggcca1080
aattatggaggagatgcccattttgatgatgatgaaacctggacaagtagttccaaaggc1140
tacaacttgtttcttgttgctgcgcatgagttcggccactccttaggtcttgaccactcc1200
aaggaccctggagcactcatgtttcctatctacacctacaccggcaaaagccactttatg1260
cttcctgatgacgatgtacaagggatccagtctctctatggtccaggagatgaagacccc1320
aaccctaaacatccaaaaacgccagacaaatgtgacccttccttatcccttgatgccatt1380
accagtctccgaggagaaacaatgatctttaaagacagattcttctggcgcctgcatcct1440
SUBSTITUTE SHEET (RULE Z6)
CA 02314357 2000-06-12
WO 99/31969 6 PCT/US98/27056
cagcaggttgatgcggagctgtttttaacgaaatcattttggccagaacttcccaaccgt150U
attgatgctgcatatgagcacccttctcatgacctcatcttcatcttcagaggtagaaaa1560
ttttgggctcttaatggttatgacattctggaaggttatcccaaaaaaatatctgaactg1620
ggtcttccaaaagaagttaagaagataagtgcagctgttcactttgaggatacaggcaag1680
actctcctgttctcaggaaaccaggtctggagatatgatgatactaaccatattatggat1740
aaagactatccgagactaatagaagaagacttcccaggaattggtgataaagtagatgct1800
gtctatgagaaaaatggttatatctattttttcaacggacccatacagtttgaatacagc1860
atctggagtaaccgtattgttcgcgtcatgccagcaaattccattttgtggtgttaagtg1920
tctttttaaaaattgttatttaaatcctgaagagcatttggggtaatacttccagaagtg1980
cggggtaggggaagaagagctatcaggagaaagctctagttctagagggccctattctat2040
agtgtcacctaaatgctagaggatctttgtgaaggaaccttacttctgtggtgtgacata2100
attggacaaactacctacagagatttaaagctctaaggtaaatataaaatttttaagtgt2160
ataatgtgttaaactactgattctaattgtttgtgtattttagattccaacctatggaac2220
tgatgaatgggagcagtggtggaatgcctttaatgaggaaaacctgttttgctcagaaga2280
aatgccatctagtgatgatgaggctactgctgactctcaacattctactcctccaaaaaa2340
gaagagaaaggtagaagaccccaaggactttccttcagaattgctaagttttttgagtca2400
tgctgtgtttagtaatagaactcttgcttgctttgctatttacaccacaaaggaaaaagc2460
tgcactgctatacaagaaaattatggaaaaatatttgatgtatagtgccttgactagaga2520
tcataatcagccataccacatttgtagaggttttacttgctttaaaaaacctcccacacc2580
tccccctgaacctgaaacataaaatgaatgcaattgttgttgttaacttgtttattgcag2640
cttataatggttacaaataaagcaatagcatcacaaatttcacaaataaagcattttttt2700
cactgcattctagttgtggtttgtccaaactcatcaatgtatcttatcatgtctggatca2760
tcccgccatgggtatacatcgatgcggccgcc
2792
<210> 12
<211> 5276
<212> DNA
<213> Artificial Sequence
<220>
<223> tetracycline/VP16 repressor-activator fusion
linked to a type II collagen prort~oter and
enhancer, "CPE-tTA transgene"
<400>
12
ggtaccactagtaagcttagatccactgtctgggattatatcaggacaaccgaagcctgg60
aaagtgtattaggtagagcattttcttccacgtgtttgggcacgtttccgacagctagga120
ttccagctctgtctttgtatgttacagactgtaaatcaatcgcaggtgaaactgtttgga180
cagtaggtggggatcaaagaccctccgcccgtgagactctaggcgctttcccctgccacc240
agcctgtctccagagatgctctggaaggaggcgggcccgggcggtctttctgctctttag300
cgtggcggacgcggcggcgggggcagggctggagcagagagcgctgcagtgatagaactt360
tctgaccccgctgcgcagggcggcagggtggcagggtggcagggtggcgagctaagccag420
agccgaacgctggagctctgggaggaacatcgaagtgtttgtatgtggtctgagatcggc480
ctgactatatttttttgtcctaaatttgcaagcacacacccacaaagctgcggtcttgac540
cggtattctttatagagcgcaatggagtgagctgagtgtctaaacgatttccctaattca600
tctgatagcagaggcgctctcctaattggcgaagagctgcctcatgtccgcaactttttg660
gcagagtgaattccacagctttgtgtgtgtgtgtgggggggggtgtaaggggtgtctaaa720
actttcggtctcctactattctgtatctcgaccggttggttttacaccccggctcatctc780
atcaacgcaaacacccccactctcctatggacccaaggacctgacgtgggggaaggtgga840
cattaggaatgtcagaaacctagagtccacgctcctcctctccatctttccacgagtttg900
ggaaacttcttggctgcgaagactttgacccacatctgcatttctcagccccagcttcca960
asagtgctgcaggttcgggaggggagacctcagtcctcctttgtgaggcttgtttgcgtt1020
gagggattggcagcgatggcttccagatgggctgaaaccctgcccgtatttatttaaact1080
ggttcctcgtggagagctgtgaatcgggctctgtatgcgctcgagaaaagccccattcat1140
gagaggcaaggcccagtgggtccccccgactccccgacccccctctcccacaatatatcc1200
cccctccctgtgcccgcctgccgccacctcccgggctccggccccgcgcgcagcggcgac1260
gaagcaacacagttccccgaaagaggtagctttttaattggccagccacaaagaatcact1320
tatgccgcacggcggtaacgaggggaaccggatcgggcggccaggatgctatctgtgtag1380
cccttttcgtgccacaattagggtggtgctggcttcctccgaccgcacctaggcgatctg1440
gttacactgttggctcctttcttgggcagtcatttaatcctactttttactctacgaatg1500
tctgtctgatggagggctgtgtccggagccccatccacaaagagtcagccagcagctctc1560
SUBSTITUTE SHEET (RULE 26)
CA 02314357 2000-06-12
WO 99/31969 ~ PCT/US98l27056
aatgtatctt atcatgtctg gatcatcccg ccatgggtat acatcgatgc ggccgc 5276
<210> 13
<211> 7664
<212> DNA
<213> Artificial Sequence
<220>
<223> beta-galactosidase (lacZ> gene linked to
beta-globin splice sequence and polyadenylation
signal, and linked to type II collagen pro~ter
sequence
<400> 13
ggtaccacta gtaagcttag tcaggacaaccgaagcctgg60
atccactgtc tgggattata
aaagtgtatt aggtagagca cacgtttccgacagctagga120
ttttcttcca cgtgtttggg
ttccagctct gtctttgtat cgcaggtgaaactgtttgga180
gttacagact gtaaatcaat
cagtaggtgg ggatcaaaga aggcgctttcccctgccacc240
ccctccgccc gtgagactct
agcctgtctc cagagatgctctggaaggag gcggtctttctgctctttag300
gcgggcccgg
cgtggcggac gcggcggcgg gcgctgcagtgatagaactt360
gggcagggct ggagcagaga
tctgaccccg ctgcgcagggcggcagggtggcagggtggcagggtggcgagctaagccag420
agccgaacgc tggagctctgggaggaacatcgaagtgtttgtatgtggtctgagatcggc480
ctgactatat ttttttgtcctaaatttgcaagcacacacccacaaagctgcggtcttgac540
cggtattctt tatagagcgcaatggagtgagctgagtgtctaaacgatttccctaattca600
tctgatagca gaggcgctctcctaattggcgaagagctgcctcatgtccgcaactttttg660
gcagagtgaa ttccacagctttgtgtgtgtgtgtgggggggggtgtaaggggtgtctaaa720
actttcggtc tcctactattctgtatctcgaccggttggttttacaccccggctcatctc780
atcaacgcaa acacccccactctcctatggacccaaggacctgacgtgggggaaggtgga840
cattaggaat gtcagaaacctagagtccacgctcctcctctccatctttccacgagtttg900
ggaaacttct tggctgcgaagactttgacccacatctgcatttctcagccccagcttcca960
aaagtgctgc aggttcgggaggggagacctcagtcctcctttgtgaggcttgtttgcgtt1020
gagggattgg cagcgatggcttccagatgggctgaaaccctgcccgtatttatttaaact1080
ggttcctcgt ggagagctgtgaatcgggctctgtatgcgctcgagaaaagccccattcat1140
gagaggcaag gcccagtgggtccccccgactccccgacccccctctcccacaatatatcc1200
cccctccctg tgcccgcctgccgccacctcccgggctccggccccgcgcgcagcggcgac1260
gaagcaacac agttccccgaaagaggtagctttttaattggccagccacaaagaatcact1320
tatgccgcac ggcggtaacgaggggaaccggatcgggcggccaggatgctatctgtgtag1380
cccttttcgt gccacaattagggtggtgctggcttcctccgaccgcacctaggcgatctg1440
gttacactgt tggctcctttcttgggcagtcatttaatcctactttttactctacgaatg1500
tctgtctgat ggagggctgtgtccggagccccatccacaaagagtcagccagcagctctc1560
acacccggct ggatctcatatggtgcactctcagtacaatctgctctgatgccgcatagt1620
taagccagcc aagctagcttgcgcaagctagcttgcgatccgtaaaaatgtgtgagagtt1680
acaaaatgtc ttccgggctaagatccgacagccatggtccaaagaagacttcggcactgc1740
agacttaaaa ccagctttctagcagaggcagaaggatctagagccaaaggcaaagacttg1800
aataggctgg gaagatgcaagaatggcattttacataaagaacactctctccttttccag1860
ccagcacact tgcatagaaattaagttttacacttgaagttctttgtttccatcctgaga1920
agctccaaag tctgaggtggtgtggtatgctgggtaattctccccaccccccaacattcc1980
ctgggggttc catgggggtagcttctcccaaggacttccagcggcaacacagaaatccca2040
cttcgagaca aaggagttactgcttasatcaggccctaatttccaaggttccctttgctt2100
aaagttccct agaggaccatctcacttctaaagaaaaggtgtattcggggacccatcctc2160
aacctccttg ttatggaaggagacttcgggaacagagcaagggctgagcctccggcagtt2220
tggggtaagg ttggggttggggggagcaaggaaggcaagtgaggctggag 2280
gcccagggat
aggggaagat gtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtctcggggatg2340
gtggtggtgg acaactaggaaactctggcgctttctcctcccctcacaaa 2400
actgagtcca
gctggagccg cctccagactctctggccagggcctcagag 2460
tggtcaacag
tccctggcca
gcgttgctct ctccaggctaagggcacccactcccctgga 2520
gattcctgaa
cctgggccag
gaagagccga attagacaagtgtctccaatccggctgcgt 2580
gcggattttg
ttgcggtgtc
cctcggttgt ctgcagttcctttagtcccttccctggcct 2640
gccccttaca
cctccacaca
ggtccccctc tgtgtaggaa cctctcttag 2700
tacaccagac ccacacacac
ctccagtccc
SUBSTITUTE SHEET (RULE 26~
CA 02314357 2000-06-12
WO 99/31969 8 PCT/US98/29056
acacccggct ggatctcata tggtgcactc tcagtacaat ctgctctgat 1620
gccgcatagt
taagccagcc aagctagctt gcgcaagcta gcttgcgatc cgtaaaaatg 1680
tgtgagagtt
acaaaatgtc ttccgggcta agatccgaca gccatggtcc aaagaagact 1740
tcggcactgc
agacttaaaa ccagctttct agcagaggca gaaggatcta gagccaaagg 1800
caaagacttg
aataggctgg gaagatgcaa gaatggcatt ttscataaag aacactctct 1860
ccttttccag
ccagcacact tgcatagaaa ttaagtttta cacttgaagt tctttgtttc 1920
catcctgaga
agctccaaag tctgaggtgg tgtggtatgc tgggtaattc tccccacccc 1980
ccaacattcc
ctgggggttc catgggggta gcttctccca aggacttcca gcggcascac 2040
agaaatccca
cttcgagaca aaggagttac tgcttaaatc aggccctaat ttccaaggtt 2100
ccctttgctt
aaagttccct agaggaccat ctcacttcta aagaaaaggt gtattcgggg 2160
acccatcctc
aacctccttg ttatggaagg agacttcggg aacagagcaa gggctgagcc 2220
tccggcagtt
tggggtaagg ttggggttgg ggggagcaag gaaggcaagt gaggctggag 2280
gcccagggat
aggggaagat gtgtgtgtgt gtgtgtgtgt gtgtgtgtgt gtgtgtgtgt 2340
ctcggggatg
gtggtggtgg acaactagga aactctggcg ctttctcctc ccctcacaaa 2400
actgagtcca
gctggagccg cctccagact ctctggccag ggcctcagag tggtcaacag 2460
tccctggcca
gcgttgctct ctccaggcta agggcaccca ctcccctgga gattcctgaa 2520
cctgggccag
gaagagccga attagacaag tgtctccaat ccggctgcgt gcggattttg 2580
ttgcggtgtc
cctcggttgt ctgcagttcc tttagtccct tccctggcct gccccttaca 2640
cctccacaca
ggtccccctc tgtgtaggaa tacaccagac cctctcttag ccacacacac 2700
ctccagtccc
ccgtctacct agattttttt catagctagt tggatggggg atgggttagg 2760
gaggctgggt
ttgcgagcct ccaggtggga gttcaccgac aggtactccg caaaggagct 2820
ggaaggcagg
tctggaaaac tgtcccccag atttaggatt ctgggcagct tccatcagct 2880
tatactttgg
ctcccccgcc ccctaaactc cccatcccca ccttcctttc tcccgttact 2940
tcgtcctccc
tcgcctttcc agccttgagt ctaaagctcc atgcttatgc ctctgcaaac 3000
aaccccctcc
cttctaaccc cagcagaact ccgaggaaag gggccggagg ccccccttct 3060
cgcctgtggt
tagagggggc agtgtggcag tcccaagtgg gggcgaccgg aggccgtctc 3120
ggtgccccgc
ccgatcaggc cactgggcac atcgggggcg ggaagctggg ctcaccaaag 3180
gggcgactgg
ccttggcagg tgtgggctct ggtccggcct gggcaggctc cgggggcggg 3240
gtctcaggtt
acagccccgc ggggggctgg ggggcggccc gcggtttggg ctggtttgcc 3300
agcctttgga
gcgaccggga gcatataacc ggagcctctg ctgggagaag acgcagagcg 3360
ccgctgggct
gccgggtctc ctgcctcctc ctcctgctcc tagagcctcc tgcatgaggg 3420
cgcggtagag
acccggaccc gctccgtgct ctgccgcctc gccgagcttc gcccgcaagc 3480
tggggaattc
atatgtctag attagataaa agtaaagtga ttaacagcgc attagagctg 3540
cttaatgagg
tcggaatcga aggtttaaca acccgtaaac tcgcccagaa gctaggtgta 3600
gagcagccta
cattgtattg gcatgtaaaa aataagcggg ctttgctcga cgccttagcc 3660
attgagatgt
tagataggca ccatactcac ttttgccctt tagaagggga aagctggcaa 3720
gattttttac
gtaataacgc taaaagtttt agatgtgctt tactaagtca tcgcgatgga 3780
gcaaaagtac
atttaggtac acggcctaca gaaaaacagt atgaaactct cgaaaatcaa 3840
ttagcctttt
tatgccaaca aggtttttca ctagagaatg cattatatgc actcagcgct 3900
gtggggcatt
ttactttagg ttgcgtattg gaagatcaag agcatcaagt cgctaaagaa 3960
gaaagggaaa
cacctactac tgatagtatg ccgccattat tacgacaagc tatcgaatta 4020
tttgatcacc
aaggtgcaga gccagccttc ttattcggcc ttgaattgat catatgcgga 4080
ttagaaaaac
aacttaaatg tgaaagtggg tccgcgtaca gccgcgcgcg tacgaaaaac 4140
aattacgggt
ctaccatcga gggcctgctc gatctcccgg acgacgacgc ccccgaagag 4200
gcggggctgg
cggctccgcg cctgtccttt ctccccgcgg gacacacgcg cagactgtcg 4260
acggcccccc
cgaccgatgt cagcctgggg gacgagctcc acttagacgg cgaggacgtg 4320
gcgatggcgc
atgccgacgc gctagacgat ttcgatctgg acatgttggg ggacggggat 9380
tccccgggtc
cgggatttac cccccacgac tccgccccct acggcgctct ggatatggcc 4440
gacttcgagt
ttgagcagat gtttaccgat gcccttggaa ttgacgagta cggtgggtag 4500
ggggcgcgag
gatcctctag agggccctat tctatagtgt cacctaaatg ctagaggatc 4560
tttgtgaagg
aaccttactt ctgtggtgtg acataattgg acaaactacc tacagagatt 4620
taaagctcta
aggtaaatat aaaattttta agtgtataat gtgttaaact actgattcta 4680
attgtttgtg
tattttagat tccaacctat ggaactgatg aatgggagca gtggtggaat 4740
gcctttaatg
aggaaaacct gttttgctca gaagaaatgc catctagtga tgatgaggct 4800
actgctgact
ctcaacattc tactcctcca aaaaagaaga gaaaggtaga agaccccaag 4860
gactttcctt
cagaattgct aagttttttg agtcatgctg tgtttagtaa tagaactctt 4920
gcttgctttg
ctatttacac cacaaaggaa aaagctgcac tgctatacaa gaaaattatg 4980
gaaaaatatt
tgatgtatag tgccttgact agagatcata atcagccata ccacatttgt 5040
agaggtttta
cttgctttaa aaaacctccc acacctcccc ctgaacctga aacataaaat 5100
gaatgcaatt
gttgttgtta acttgtttat tgcagcttat aatggttaca aataaagcaa 5160
tagcatcaca
aatttcacaa ataaagcatt tttttcactg cattctagtt gtggtttgtc 5220
caaactcatc
SUBSTIME SHEET (RULE 2~
CA 02314357 2000-06-12
WO 99/31969 g PGT/US98/Z7056
ccgtctacct agattttttt catagctagt tggatggggg atgggttagg2760
gaggctgggt
ttgcgagcct ccaggtggga gttcaccgac aggtactccg caaaggagct2820
ggaaggcagg
tctggaaaac tgtcccccag atttaggatt ctgggcagct tccatcagct2880
tatactttgg
ctcccccgcc ccctaaactc cccatcccca ccttcctttc tcccgttact2940
tcgtcctccc
tcgcctttcc agccttgagt ctaaagctcc atgcttatgc ctctgcaaac3000
aaccccctcc
cttctaaccc cagcagaact ccgaggaaag gggccggagg ccccccttct3060
cgcctgtggt
tagagggggc agtgtggcag tcccaagtgg gggcgaccgg aggccgtctc3120
ggtgccccgc
ccgatcaggc cactgggcac atcgggggcg ggaagctggg ctcaccaaag3180
gggcgactgg
ccttggcagg tgtgggctct ggtccggcct gggcaggctc cgggggcggg3240
gtctcaggtt
acagccccgc ggggggctgg ggggcggccc gcggtttggg ctggtttgcc3300
agcctttgga
gcgaccggga gcatataacc ggagcctctg ctgggagaag acgcagagcg3360
ccgctgggct
gccgggtctc ctgcctcctc ctcctgctcc tagagcctcc tgcatgaggg3420
cgcggtagag
acccggaccc gctccgtgct ctgccgcctc gccgagcttc gcccgcaagc3480
tggggaattc
ggatccccgg gatcgaaaga gcctgctaaa gcaaaaaaga agtcaccatg3540
tcgtttactt
tgaccaacaa gaacgtgatt ttcgttgccg gtctgggagg cattggtctg3600
gacaccagca
aggagctgct caagcgcgat cccgtcgttt tacaacgtcg tgactgggaa3660
aaccctggcg
ttacccaact taatcgcctt gcagcacatc cccctttcgc cagctggctt3720
tatagcgaag
aggcccgcac cgatcgccct tcccaacagt tgcgcagcct gaatggcgaa3780
tggcgctttg
cctggtttcc ggcaccagaa gcggtgccgg aaagctggct ggagtgcgat3840
cttcctgagg
ccgatactgt cgtcgtcccc tcaaactggc agatgcacgg ttacgatgcg3900
cccatctaca
ccaacgtaac ctattccatt acggtcaatc cgccgtttgt tcccacggag3960
aatccgacgg
gttgttactc gctcacattt aatgttgatg aaagctggct acaggaaggc4020
cagacgcgaa
ttatttttga tggcgttaac ttggcgtttc atctgtggtg caacgtgcgc4080
tgggtcggtt
acggccagga cagtcgtttg ccgtctgaat ttgacctgag cgcattttta4140
cgcgccggag
aaaaccgcct cgcggtgatg gtgctgcgtt ggagtgacgg cagttatctg4200
gaagatcagg
atatgtggcg gatgagcggc attttccgtg acgtctcgtt gctgcataaa4260
ccgactacac
aaatcagcga tttccatgtt gccactcgct ttaatgatga tttcagccgc4320
gctgaactgg
aggctgaagt tcagatgtgc ggcgagttgc gtgactacct acgggtaaca4380
gtttctttat
ggcagggtga aacgcaggtc gccagcggca ccgcgccttt cggcggtgaa4440
attatcgatg
agcgtggtgg ttatgccgat cgcgtcacac tacgtctgaa cgtcgaaasc4500
ccgaaactgt
ggagcgccga aatcccgaat ctctatcgtg cggtggttga actgcacacc4560
gccgacggca
cgctgattga agcagaagcc tgcgatgtcg gtttccgcga ggtgcggatt4620
gaaaatggtc
tgctgctgct gaacggcaag ccgttgctga ttcgaggcgt taaccgtcac4680
gagcatcatc
ctctgcatgg tcaggtcatg gatgagcaga cgatggtgca ggatatcctg4740
ctgatgaagc
agaacaactt taacgccgtg cgctgttcgc attatccgaa ccatccgctg4800
tggtacacgc
tgtgcgaccg ctacggcctg tatgtggtgg atgaagccaa tattgaaacc4860
cacggcatgg
tgccaatgaa tctgctgacc gatgatccgc gctggctacc ggcgatgagc4920
gaacgcgtaa
cgcgaatggt gcagcgcgat cgtaatcacc cgagtgtgat catctggtcg4980
ctggggaatg
aatcaggcca cggcgctaat cacgacgcgc tgtatcgctg gatcaaatct5040
gtcgatcctt
cccgcccggt gcagtatgaa ggcggcggag ccgacaccac ggccaccgat5100
attatttgcc
cgatgtacgc gcgcgtggat gaagaccagc ccttcccggc tgtgccgaaa5160
tggtccatca
aaaaatggct ttcgctacct ggagagacgc gcccgctgat cctttgcgaa5220
tacgcccacg
cgatgggtaa cagtcttggc ggtttcgcta aatactggca ggcgtttcgt5280
cagtatcccc
gtttacaggg cggcttcgtc tgggactggg tggatcagtc gctgattaaa5340
tatgatgaaa
acggcaaccc gtggtcggct tacggcggtg attttggcga tacgccgaac5400
catcgccagt
tctgtatgaa cggtctggtc tttgccgacc gcacgccgca tccagcgctg5460
acggaagcaa
aacaccagca gcagtttttc cagttccgtt tatccgggca aaccatcgaa5520
gtgaccagcg
aatacctgtt ccgtcatagc gataacgagc tcctgcactg gatggtggcg5580
ctggatggta
agccgctggc aagcggtgaa gtgcctctgg atgtcgctcc acaaggtaaa5640
cagttgattg
aactgcctga actaccgcag ccggagagcg ccgggcaact ctggctcaca5700
gtacgcgtag
tgcaaccgaa cgcgaccgga tggtcagaag ccgggcacat cagcgcctgg5760
cagcagtggc
gtctggcgga aaacctcagt gtgacgctcc ccgccgcgtc ccacgccatc5820
ccgcatctga
ccaccagcga aatggatttt tgcatcgagc tgggtaataa gcgttggcaa5880
tttaaccgcc
agtcaggctt tctttcacag ctgtggattg gcgataaaaa acaactgctg5940
acgccgctgc
gcgatcagtt cacccgtgca ccgctggata acgacattgg cgtaagtgaa6000
gcgacccgca
ttgaccctaa cgcctgggtc gaacgctgga aggcggcggg ccattaccag6060
gccgaagcag
cgttgttgca gtgcacggca gatacacttg ctgatgcggt gctgattacg6120
accgctcacg
cgtggcagca tcaggggaaa accttattta tcagccggaa aacctaccgg6180
attgatggta
gtggtcaaat ggcgattacc gttgatgttg aagtggcgag cgatacaccg6240
catccggcgc
ggattggcct gaactgccag ctggcgcagg tagcagagcg ggtaaactgg6300
ctcggattag
ggccgcaaga aaactatccc gaccgcctta ctgccgcctg ttttgaccgc6360
tgggatctgc
SU6STITUTE SHEET fRUIE 2~
CA 02314357 2000-06-12
WO 99/31969 PCTNS98/Z9056
cattgtcagacatgtataccccgtacgtcttcccgagcgaaaacggtctgcgctgcggga6420
cgcgcgaattgaattatggcccacaccagtggcgcggcgacttccagttcaacatcagcc6480
gctacagtcaacagcaactgatggaaaccagccatcgccatctgctgcacgcggaagaag6540
gcacatggctgaatatcgacggtttccatatggggattggtggcgacgactcctggagcc6600
cgtcagtatcggcggaattacagctgagcgccggtcgctaccattaccagttggtctggt6660
gtcaaaaataataataaccggcaggccatgtctgaaagtattcgcgtaaggaaatccatt6720
atgtactatttaaaaaacacaaacttttggatgttcggtttattctttttcttttacttt6780
tttatcatgggagcctacttcccgtttttcccgatttggctacatgacatcaaccatatg6840
agcaaaagtgatacgggtattatttttgccgctatttctctgttgtcgctattattccaa6900
ccgctgttggtctgctttctgacaaactcggcctcgactctagactgagaacttcagggt6960
gagtttggggacccttgattgttctttctttttcgctattgaaaaattcatgttatatgg7020
agggggcaaagttttcagggtgttgtttagaatgggaagatgtcccttgtatcaccatgg7080
accctcatgataattttgtttctttcactttctactctgttgacaaccattgtctcctct7140
tattttcttttcattttctgtaacttttttcgttaaactttagcttgcatttgtaacgaa7200
tttttaaattcactttcgtttatttgtcagattgtaagtactttctctaatcactttttt7260
ttcaaggcaatcagggtaattatattgtacttcagcacagttttagagaacaattgttat7320
aattsaatgataaggtagaatatttctgcatataaattctggctggcgtggaaatattct7380
tattggtagaaacaactacatcctggtaatcatcctgcctttctctttatggttacaatg7440
atatacactgtttgagatgaggataaaatactctgagtccaaaccgggcccctctgctaa7500
ccatgttcatgccttcttctttttcctacagctcctgggcaacgtgctggttgttgtgct7560
gtctcatcattttggcaaagaattcactcctcaggtgcaggctgcctatcagaaggtggt7620
ggctggtgtggccaatgccctggctcacaaataccactgagatc 7664
<210> 14
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> primer corresponding to tTA sequence
<400> 14
cgagggcctg ctcgatctcc 20
<210> 15
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> primer corresponding to 3' untranslated region of
tTA sequence
<400> 15
ggcattccac cactgctccc 20
<210> 16
<211> 21
<212> DNA
<213> Homo sapiens
<400> 16
gagcaccctt ctcatgacct c 21
<210> 17
<211> 22
<212> DNA
<213> Escherichia coli
suesmurs sH~r ~RUC.s 26)
CA 02314357 2000-06-12
WO 99/31969 PCTNS98/Z9056
11
<400> 17
gttggtgtag atgggcgcat cg 22
<210> 18
<211> 21
<212> DNA
<213> Escherichia coli
<400> 18
gcggggtctc aggttacagc c 21
<210> 19
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> PCR primer corresponding to tet activator
<400> 19
cgcccagaag ctaggtgtag ag 22
<210> 20
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> PCR primer for tet activator
<400> 20
cggccatatc cagagcgccg 20
<210> 21
<211> 23
<212> DNA
<213> Homo sapiens
<400> 21
gccctctggc ctgctggctc atg 23
<210> 22
<211> 24
<212> DNA
<213> Homo sapiens
<400> 22
caggagagtc ttgcctgtat cctc 24
<210> 23
<211> 25
<212> DNA
<213> Mus muscalis
<400> 23
aggagggagc tgacagatac actcc 25
<210> 24
<211> 22
<212> DNA
<213> Mus muscalis
SU9SmUTE SHEET (RULE 26)
CA 02314357 2000-06-12
WO 99/31969 12 PCTNS98/27056
<400> 24
aggccacaga catctcctct gg 22
SU6STITUTE SHEET (RULE 2~