Note: Descriptions are shown in the official language in which they were submitted.
CA 02008735 1999-12-20
SUr~IARY OF THE INVENTION
The control of human cell growth is of medical
importance. As a result, growth factors, such as epidermal
growth factor (EGF), are of interest, as cell growth is
regulated by the interaction between growth factors and their
receptors in the cell. Here, we disclose the invention of
monoclonal antibodies that are useful because they block the
interaction of EGF with human cell receptors and inhibit cell
growth. They are also useful because they are analytical
reagents of great specificity. Also disclosed are segments of
the EGF receptor of value as monoclonal antibody targets and
immunogens or immunogen components.
The monoclonal antibodies we discovered are unusual
anti-EGF receptor antibodies in that they also recognize
deglycosylated and denatured EGF receptors. These properties
were essential for us to determine the amino acid sequence of
the epitope where the antibodies bind. Taking advantage of
these properties and using cyanogen bromide fragments of a
truncated form of the EGF receptor secreted by A431 cells (26)
and synthetic peptides, we determined that all three antibodies
recognize epitopes encompassed by 14 amino acids of the EGF
receptor. This region of the receptor is located between the
extracellular cystein-rich domains and includes an Arg-Gly-Asp-
Ser recognition sequence for cell adhesion receptors (27).
- 1 -
._ % 2008735
In additional experiments we allowed a truncated from
of the human EGF receptor to bind to mouse EGF and then
crosslinked the two entities with an amine-reactive
crosslinking reagent, disuccinimidyl succinate (DSS). (Mouse
EGF, a commonly used substitute for human EGF, was the form of
EGF used in all experiments in this application.) This allowed
us to pinpoint a single amino acid in the receptor that was
linked to the EGF, indicating that the amino acid was in the
EGF binding site. Combining the knowledge of the locations of
that amino acid with the knowledge of the locations of the
three antibody epitopes, as well a;s other information on the
structure of the receptor, allowed us to the identify regions
of the receptor of particular value as monoclonal antibody
targets.
In accordance with an Eambodiment of the present
invention there is provided a monoclonal antibody that competes
with EGF for binding to the natural EGF receptor and binds to
an epitope between residues Ala-351 and Asp-364 of the natural
EGF receptor, which epitope remains immunogenically active
after denaturation.
In accordance with another embodiment of the present
invention there is provided a chemical compound containing an
amino acid sequence consisting of al.l or part of the human EGF
receptor segment that extends from residue Phe-321 to residue
Glu-367 in natural, denatured or synthetic conformation, said
sequence having a length effectivs~ to raise antibodies that
compete with EGF binding.
Yet another embodiment of the present invention
provides a hybridoma selected from the group consisting of
hybridoma ATCC HB 10342, hybridoma A,TCC HB 10343, and hybridoma
ATCC HB 10344.
Among the known growth factors and receptors (1),
- 2 -
'~200g~'35
intracellular signal transduction initiated by interactions
between epidermal growth factor (EGF') and its receptor has been
particularly well studied (2-5)" EGF is a 53-residue
polypeptide that stimulates the growth of a variety of cell
types and is synthesized in the form of an M,. 128,000 precursor
molecule. The mature EGF receptor is an M,. 170,000
glycoprotein with intrinsic tyrosine-specific protein kinase
activity and contains two functional domains linked by a
transmembrane region (11-14). Th;e extracellular domain is
heavily glycosylated and possesses a single EGF-binding site,
and the intracellular portion of the receptor includes a
tyrosine protein kinase domain with three
30
- 2a -
CA 02008735 1999-12-20
COOH-terminal autophosphorylation sites at residues 1068, 1148,
and 1173 (14-19). The extracellular domain of the EGF receptor
contains a large number of cysteine residues, 51 in total,
which are clustered in two regions (14). These cysteine
residues are conserved in the extracelluar domain of the
chicken EGF receptor which is 75% identical to that of the
human receptor (20). Similar clusters of cysteine residues have
been found in the extra-cellular portions of the receptors for
insulin, insulin-like growth factor I, nerve growth factor, and
low density lipoprotein (2). Although these cysteine-rich
regions probably contribute to receptor tertiary structure
through the formation of intramolecular disulfide bridges, the
role of these regions in receptor function has not been
elucidated.
Very little is known about the structural features
of the EGF receptor that are involved in the binding of EGF.
Carpenter and Cohen observed that several lectins reversibly
inhibited the binding of radiolabeled EGF to human fibroblasts
(21). However, the antigenic specificities of mono monoclonal
antibodies to EGF receptors of A431 human epidermoid carcinoma
cells indicate that antibodies which block EGF binding do not
recognize carbohydrate determinants while those with
carbohydrate specificities do not inhibit EGF binding (22, 23).
These results suggest that N-linked oligosaccharides, which
comprise 40% of the mass of the EGF receptor extracelluar
domain (2) are not directly involved in EGF binding. In
addition, Slieker et al. found that oligosaccharide addition
to pro-EGF receptors was necessary to acquire the ability to
bind EGF but that deglycosylated receptors retained binding
- 3 -
CA 02008735 1999-12-20
activity (24). Lax et al. have cleaved with cyanogen bromide
human EGF receptors cross-linked to l2sl-EGF and have identified
with anti-peptide antibodies a labeled Mr 50,000 receptor
fragment spanning residues 294-543, which is largely located
between the two cysteine-rich regions of the extracellular
domain (25). Thus, the EGF-binding site of the receptor
resides within a 250-residue region of the extracellular
domain. The location of the EGF binding site was not
characterized in further detail.
BRIEF DESCRIPTION OF THE DRAWINGS
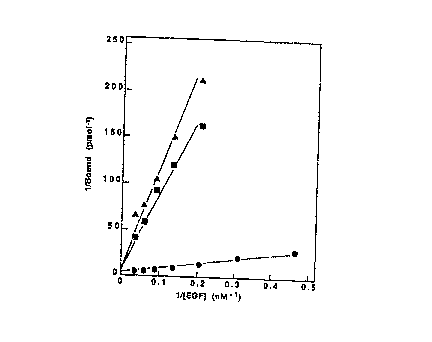
FIG. 1. Effect of monoclonal antibodies on l2sl-EGF
binding to A431 cells. Duplicate cultures of live A431 cells
were incubated at 4°C with increasing concentrations of l2sI-EGF
in the absence (filled in circles) or presence of 20 nM LA22
IgG (filled in triang,es) or 20 nM LA58 IgG (filled in squares)
as described under "Experimental Procedures". The data are
shown in a double reciprocal Lineweaver-Burk plot.
FIG. 2. Immunoreactivity of intact ERRP and
deglycosylated ERRP. Full length A431 EGF receptor (1~ 1),
ERRP (lanes 2, 4, 6, and 8) and ERRP treated with
endoglycosidase F (lanes 3, 5, 7, and 9) were electrophoresed
in a 10% polyacrylamide gel under denaturing conditions and
electrophoretically transferred to nitrocellulose. The membrane
was reacted with monoclonal antibodies LA90 (1-3), LA58
(lanes 4 and 5), 455 IgG (lanes 6 and 7), and LA22 (8 and
9). Bound antibody was detected with l2sl-protein A and
autoradiography.
- 4 -
CA 02008735 1999-12-20
FIG. 3. Reverse phase HPLC chromatography of ERRP
fragments after CNBr cleavage. (A) CNBr-generated ERRP
fragments were fractionated on a C-4 reverse phase column using
a 20-60% gradient of acetonitrile in aqueous 0.1% TFA. (B) A
l0 ~1 aliquot of each column fraction was reacted with LA22 in
a dot blot assay.
FIG. 4. LA22-reactive Mr 43,000 and Mr 37,000 ERRP
fragments. CNBr fragments in fraction 19 (Fig. 3) were
rechromatographed on a C-4 reverse phase HPLC column. The main
absorbance peak was analyzed by N-terminal sequencing and
electrophoresis. Edman degradation yielded a major amino acid
sequence of XEDGV and a minor sequence of XXNPE. Mr 43,000 and
Mr 37,000 bands were detected after SDS-PAGE in a 15%
polyacrylamide gel by silver staining (inset, right lane) and
western blotting with LA22 antibodies(inset, left lane).
FIG. 5. Schematic representations of the ERRP of A431
cells. The cysteine-rich regions from Cys-133 to Cys-313 and
Cys-446 to Cys-612 are hatched. The amino acid sequence
deduced from ERRP cDNA (14) for residues 295 through 370,
denoted by the ~, is presented below. Consensus sequences
for potential asparagine-linked glycosylation sites are
overlined, the Arg-Gly-Asp-Ser adhesion molecule recognition
site (27) is , and the 14-amino acid peptide recognized
by LA22, LA58, and LA90 is . Amino acid differences
in the chicken EGF receptor sequence homologous to residues 295
through 370 of the human receptor (20) are indicated.
- 5 -
CA 02008735 1999-12-20
FIG. 6. LA22-reactive Mr 17,000 ERRP fragment. CNBr
fragments in fraction 21 (Fig. 3) were rechromatographed on a
C-4 reverse phase HPLC column. Peak C was analyzed by N
terminal sequencing and electrophoresis. Edman degradation
yielded an amino acid sequence of EEDGV. The Mr 17,000 ERRP
fragment was detected after SDS-PAGE by silver staining (inset,
right lane) and western blotting with LA22 antibodies (inset,
left lane) .
FIG. 7. Reverse phase HPLC chromatography of acid-
cleaved Mr 43, 000 and Mr 37, 000 CNBr fragments of ERRP. The Mr
43,000 and Mr 37,000 CNBr-generated ERRP fragments (Fig. 4)
were cleaved with 0.1% trifluoracetic acid (28) and were
chromatographed on a C-4 reverse phase column. Peak D, which
reacted with LA22 antibodies in a dot blot assay, was analyzed
by N-terminal sequencing and electrophoresis. Edman
degradation yielded a major amino acid sequence of EEDGVR and
a minor sequence of DVNPEG. This peak was resolved by
electrophoresis into bands of Mr 23,000 and Mr 17,000 (inset,
left lane) that reacted with L22 antibodies (inset, middle
lane). These bands were converted to LA22-reactive bands of
lower molecular weight by endoglycosidase F (inset, right
lane).
FIG. 8. S. aureus V8 protease digestion of LA22-
reactive Mr 43,000 and Mr 37,000 ERRP fragments. The Mr 43,000
and Mr CNBr-generated ERRP fragments (Fig. 4) were digested
with S. aureus V8 protease at pH 8.0 (29), and the digestion
products were chromatographed in a C-4 reverse phase HPLC
column. Peak E, which reacted with L22 antibodies in a dot
blot assay, was analysed by N-terminal sequencing and
electrophoresis. Edman degradation yielded a single sequence
- 6 -
CA 02008735 1999-12-20
of FKDSLSIXATNIKHFKXCTSI. By SDS-PAGE, peak I was resolved
into a single silver-stained band of Mr 15,000 (inset, right
lane) that reacted with LA22 antibodies (inset, left lane).
FIG. 9. Inhibition of antibody binding to ERRP by
synthetic peptides. Synthetic peptides of 24 residues (A) and
14 residues (B) corresponding to EGF receptor sequences Asp-344
to Glu-367 and Ala-351 to Asp-364 (Fig. 4) (14), respectively,
were used to inhibit the binding of purified LA22 (A,
triangles; B, Qnen circles), LA58 (A, circles; B, ~, and
LA90 (A; sr~,al res, B, trian~~les) to immobilize ERRP. The
inhibition of LA22 binding by purified ERRP (filled in circles)
is presented in panel B.
FIG. 10. Immunoprecipitation of the cross-linked
fragments produced from [l2sl]EGF-ERRP complexes by Endo-Glu-C.
The reduced and carboxymethylated cross-linking complex was
digested by Endo Glu-C and immunoprecipitated by immobilized
LA22. The immunoprecipitated fragments was either run on 15%
SDS-PAGE (lane 1) or subjected to further treatment with Endo
Glu-C (lane 2) or Endo-F (lane 3) prior to electrophoresis.
The dried gel was exposed to X-ray film.
FIG. 11. Cleavage of [lzsI]EGF-ERRP complexes by Endo
Lys-C. Reduced and carboxymethylated cross-linked complexes
were subjected to cleavage by Endo Lys-C. The resulting
fragments were immunoprecipitated by immobilized LA22, and the
immunoprecipitate was either directly run on 15% SDS-PAGE (lane
A), or further treated with Endo-F (lane B) or Endo Glu-C (lane
C), respectively, before electrophoresis. The dried gel was
exposed to X-ray film.
-
CA 02008735 1999-12-20
FIG. 12. Inhibition of A431 growth by EGF receptor
monoclonal antibodies. A431 cells were seeded in 24-well
plates at 5 x 103 cells/ml in DME/F12 medium, and they were
cultured in the presence of increasing concentrations of
monoclonal antibodies LA22 (filled in circles) , LA58 (f~
in scruares), and LA90 (filled in triangles) for 6 days at 37°C.
Cells in duplicate wells were harvested by trypsinization and
counted with a Coulter particle counter.
DETAILED DESCRIPTION OF PREFERRED EMBODIMENTS
The invention is, in its first aspect, a monoclonal
antibody that competes with EGF binding to the natural human
EGF receptor and binds to an epitope residing on the human EGF
receptor between residues Cys-313 and Cys-446, which epitope
remains immunologically active after denaturation. For the
purpose of the present specification and claims, "denaturation"
means an alteration of the three-dimensional conformation from
that naturally occurring, including deglycosylation and
reduction of disulfide bonds, such as, for example, by boiling,
treatment with SDS or other strong detergent, and/or treatment
with reducing agents, such as ~i-mercaptoethanol. Of course,
"denaturation" as used herein does not comprehend conditions so
extreme as to cause splitting of the peptide bonds or
destruction of the amino acids. Cys-313 and Cys-446 are two
residues that mark the limits of two cysteine-rich regions and
also encompass the receptor segment that contains both the
newly discovered antigenic determinant extending from residue
Ala-351 to Asp-364 and the residue which can be cross-linked
_ g -
CA 02008735 1999-12-20
to bound EGF, Lys-336: any monoclonal antibody with the
foregoing properties is considered part of the invention. In
preferred embodiments the invention is either (1) a monoclonal
antibody that reacts with an epitope residing on the receptor
between residues Cys-113 and Asp-364, (2) one that reacts with
an epitope between residues Lys-336 and Asp-364, (3) one that
reacts with an epitope residing between residues Ala-351 and
Asp-364, (4) one that reacts with the same epitope as a
monoclonal antibody produced by a hybridoma selected from the
group hybridoma ATCC HB 10342, hybridoma ATCC HB 10343, and
hybridoma ATCC HB 10344, or (5) one that is produced by one of
those three hybridomas.
In a second aspect, the invention is a chemical
compound which includes an amino acid sequence comprising all
or part of the human EGF receptor segment that extends from
residue Phe-321 to residue Glu-367, said segment being
substantially free of the remainder of the receptor. The
receptor has a length effective to raise antibodies that
compete with EGF binding. Usually, the minimum amino acid
length to provide such immur~ogenicity is about ten amino acids.
In preferred embodiments, the chemical compound includes all
or part of that portion of the receptor segment that extends
from (1) residue Lys-336 to residue Asp-364, or (2) residue
Ala-351 to residue Asp-364, in each preferred embodiment said
portion or part thereof being substantially free of the
remainder of the receptor and other components of human cells.
The segment may be one isolated from human cells by techniques
which may or may not cause denaturation of the segment. It may
also be one synthesized by known techniques. For those
- g -
CA 02008735 1999-12-20
epitopes which bind to antibodies in both the natural or
denatured form of the segment bearing the epitopes, a linear
synthesized segment will also generate the desired antibodies,
regardless of its natural conformation or glycosylation. Of
course, the synthetic sequence may comprise segments which
contain glycosylated structures or are otherwise manipulated
in the laboratory to emulate the natural conformation and
structure of the receptor.
The amino acid sequence present in the chemical
compound of the present invention is a shortened segment of the
entire human EGF receptor and as such does not comprehend the
entire native receptor. However, the compound may contain
other moieties or sequences for specified purposes, such as,
for example, a carrier protein to enhance the immunogenicity
of the compound.
Receptor fragments of the present invention may also
be produced by recombinant DNA procedures. The DNA encoding
the receptor fragment may be synthetic DNA, isolated genomic
DNA, cDNA, or a combination thereof. The DNA can then be
inserted into any appropriate vector, such as a plasmid or
virus, and introduced into an appropriate host cell, either
prokaryotic or eukaryotic. Such techniques are set forth, for
example, in Sambrook et al, "Molecular Cloning: A Laboratory
Manual", Second Edition, Cold Spring Harbor Laboratory Press,
1989. The present invention also comprehends such DNA, vectors
and host cells.
In a further aspect, the invention is a process of
inhibiting the growth of human cells which are EGF-responsive,
which comprises allowing such cells to come into contact with
a monoclonal antibody of the present invention.
- 10 -
2008735
In another aspect, the invention is the use of compounds
including EGF sequences of the present invention in the known
procedures of obtaining polyclonal or monoclonal antibodies by
subjecting an animal to an immunogen and obtaining the antibodies,
or the cells which produce the antibodies, raised in such animals.
The cells may be used to form monoclonal hybridoma cell lines
which produce a monoclonal antibody of the present invention.
In yet a further aspect, the present invention is a
hybridoma capable of producing a monoclonal antibody in accordance
with the present invention, and preferably one of hybridoma ATCC
HB 10342, hybridoma ATCC HB 10343, and hybridoma ATCC HB 10344.
4Aith respect to receptor segments, the segment between
residues 321 and 367 is disclosed in this application. The resi-
dues from 321 to 336 can be split off with Lys-C endoproteinase.
The linkage between residues 364 anal 365 is acid labile; that
property can be used to split off residues 365 to 367. The l4mer
between residues 351 and 364 can be: synthesized as described in
this application. These methods, i.n combination with other meth-
ods described here, allow the production of a segment substantial-
ly free of the remainder of the receptor and other components of
human cells as indicated by chromatographic and/or electorphoretic
analysis.
MATERIALS AND METHODS
Abbreviations: EGF, epidermal growth factor; ERRP, EGF
receptor-related protein; HPLC, high performance liquid chromatog-
raphy; SDS, sodium dodecyl sulfate; DME, Dulbecco's Modified
3
- 11 "
~. ~ 2008735
Eagle's medium; BSA bovine serum albumin; ELISA, enzyme-linked
immunosorbant assay.
Production of hybridomas and monoclonal antibodies.
Hybridomas were produced by fusing X63-Ag8.653 mouse myeloma
cells (30), obtained from the American Type Culture Collection
(Rockvile, MD), with splenocytes from Balb/c mice immunized
with A431 human epidermoid carcinoma cells (22) as described
previously (31,32). The hybridomas were selected in HAT-
supplemented RD+5F (factor) medium, (33) containing 5% fetal
calf serum (FCS; Hyclone, Logam, UT), and they were
subsequently cloned and maintained in RD+5F medium. The
monoclonal antibodies LA22, IgG2a, LA58 IgGi, and LA90 IgG2a
were selected for their ability t:o inhibit the binding of
[l2sl]EGF (receptor grade; Collaborative Research, Bedford, MA)
to intact A431 cells (L. Wang and J.D. Sato, unpublished
results) as described previously (31,34). Monoclonal
antibodies were purified from serum-free hybridoma media by
chromatography on protein A-*Ssaphrose (Pharmacia Inc.,
Piscataway, N.J.) (35). Purified antibodies were dialyzed
against phosphate-buffered saline and filter-sterilized.
Monoclonal antibody LA22 l:gG2a, also referred to here
as monoclonal antibody LA22, was deposited on January 24, 1990,
at the American Type Culture Collection, Rockville, Maryland
20852, and was given the ATCC accession number HB 10342.
Monoclonal antibody LA58 IgGl, also referred to here as
monoclonal antibody LA58, was deposited on January 24, 1990,
at the American Type Culture Collection, Rockville, Maryland
20852, and was given the ATCC accession number HB 10343.
Monoclonal antibody LA90 IgG2a, also referred to here as
monoclonal antibody LA90, was deposited on January 24, 1990,
at the American Type Culture Collection, Rockville, Maryland
20852, and was given the ATGC acce:~sion number HB 10344.
*Trade-mark
- 12 -
~
2008735
Purification of EGF rece~ator-related protein ~ ERRPI
from conditioned medium. The truncated form of the EGF
receptor (ERRP: 26) secreted by A431 cells was purified from
unsupplemented DME/F12 medium (Gibco, Grand Island, NY)
conditioned by A431 cells in ro:Ller bottle cultures. The
conditioned medium was passed over a column of 528IgG EGF
receptor monoclonal antibody (31, 3~t) immobilized on *Affi-gel
(BioRad, Richmond, CA). Bound ERRP was eluted from the
column in 0.1 M citrate buffer (pH 4.0), and the elute was
10 dialyzed against distilled water and lyophilized.
Binding assays with EGF and monoclonal antibodies.
The binding assays with EGF and monoclonal antibodies were done
with paraformaldehyde-fixed A431 ce7lls in punctured microtiter
wells containing glass fiber filters (V and P Enterprises, San
Diego, CA). EGF, LA22 IgG, LA58 IgG and LA90 IgG were labeled
with Na[lzsl] (Amersham, Arlington Heights, IL) by the
chloramine T method. 2x104 A431 cells were incubated in
duplicate wells with 0.3 - 0.5 nM l2sl-ligand and increasing
concentrations of unlabeled competing ligand in 50 ~1 DME/F12
medium containing 0.2% BSA. After two hours at room
temperature the cells were washed
*Trade-mark
- 13 -
CA 02008735 1999-12-20
four times with PBS containing 0.25% gelatin and 5% newborn
calf serum. Cell-bound radioactivity was measured with a model
1274 LKB gamma counter. Maximum inhibition of binding was
measured in the presence of a 200-fold excess of unlabeled
homologous ligand.
The competitive nature of the antibody-mediated
inhibition of 1251[EGF]-receptor binding was determined with
confluent microtiter cultures of live A431 cells. Cells in
duplicate cultures were incubated with increasing
concentrations of [1251]EGF (2.0x105 cpm/ng) in the presence or
absence of 20 nM monoclonal antibody in DME/F12 medium
containing 0.5% BSA. After two hours at 4°C the cells were
washed three times with 0.5% BSA in DME/F12 medium and were
solubilized in 0.1 N NaOH. Cell-bound radioactivity was
corrected for non-specific binding in the presence of a 200-
fold excess of unlabeled EGF. The results were analyzed in a
double-reciprocal Lineweaver-Burk plot.
Zodetection methods with ,purified monoclonal
For Western blotting ( 3 6 ) 2 E.cg each of intact
A431 EGF receptors, ERRP, ERRP deglycosylated with
endoglycosidase F containing peptide N-glycosidase (Boehringer-
Mannheim, Indianapolis, IN) or ERRP peptides were
electrophoresed in a SDS-polyacrylamide gel (SDS-PAGE) (37)
using a miniprotean II apparatus (BioRad). Molecular weight
markers were purchased from BioRad. The proteins were
electrophoretically transferred with a mini-transblot assembly
(BioRad) to 0.45 ~cm or 0.2 ~cm nitrocellulose (Schleicher and
Schuell, Kenne, NH) in 25 nM Tris buffer (pH 8.3) containing
192 mM glycine and 20% (v/v) methanol. The nitrocellulose
membranes were incubated with a 5% solution of non-fat dry milk
- 14 -
CA 02008735 1999-12-20
for 1 hr and rinsed twice with TBS (20 nM Tris/500 nM NaCl/pIi
7.5) to block non-specific binding sites. The nitrocellulose-
bound proteins were incubated overnight at 4°C with purified
monoclonal antibodies at 1 fcg/ml in a 1% solution of fraction
V BSA (Sigma Chemical Co., St. Louis, MO). The membranes were
rinsed with TBS and incubated for 1 hr in a 1:1000 dilution of
affinity purified rabbit anti-mouse Ig (Cappel, Malvern, PA)
n TBS. After two washes in TBS the nitrocellulose membranes
were incubated for 1 hr in a 1:1000 dilution of l2sl-protein A
(50-100 ~CCi/~g; New England Nuclear, Boston, MAO in TBS. TBS-
washed and air-dried membranes were exposed to XAR5 film
(Kodak, Rochester, NY) and bound antibody was detected by
autoradiography. For dot-blotting peptides and protein were
adsorbed to 0.2 ~m nitrocellulose membranes (Schleicher and
Schuell) in a 96-well dot-blot apparatus (BioRad), and the
membranes were reacted with purified monoclonal antibodies and
processed as described above.
Selective fragmentation of ERRP and yep
purification. Affinity-purified ERRP was reduced with 100 mM
dithiothreitol (Sigma Chemical Co.) and carboxymethylated with
iodoacetic acid (Sigma Chemical Co.). Alkylated ERRP was
dialyzed against 70% formic acid (Aldrich Chemical Co.,
Milwaukee, WI) and was cleaved with cyanogen bromide (CNBr:
Aldrick Chem. Co.) in the dark at 20°C for 20 hr. Acid-
sensitive aspartyl-prolyl peptide bonds (38,39) were cleaved
at 100°C for 30 min at 0.1 % trifluoracetic acid (Pierce
Chemical Co., Rockford, IL) as described by
- 15 -
'~200873'~
Tarr (28). For cleavage at glut;amic acid residues CNBr-
fragments of ERRP were digested with S. aureus V8 protease
(Boehringer-Mannheim) at a 10:1 (w/w) ratio in 1% ammonium
bicarbonate, pH 8, at 37°C for 24 hr. ERRP was deglycosylated
with endoglycosidase F (Boehringer-Mannheim) (6mU/~.cg ERRP) for
12 hr at 37°C in 50 ~.cl 20 mm potassium phosphate (pH 7.0)
containing 25 mM EDTA and 0.1% SDS. ERRP peptides were
purified by reverse phase HPLC on *Vydac columns (The
Separations Group, Hesperia, CA) in aqueous solvents of 0.1%
trifluoracetic acid (Pierce Chemical Co.) and acetonitrile
(Burdick and Jackson, Muskeogon, MI) using a Beckman model 344
HPLC gradient system (Beckman Instruments, Inc., San Ramon,
CA).
Competitive binding assavs using synthetic ~e_ptides
96-well ELISA plates (Falcon, Oxnard, CA) were incubated
overnight at 4°C with 1 ~g/ml purified ERRP in 50 mM ammonium
bicarbonate, pH 9.6. Non-specific binding sites were blocked
with 5% non-fat dry milk for 1 hr, and the plates were washed
twice with 1% fraction V BSA (Sigma chemical Co.) in 20 mM Tris
buffer, pH 8.0 (EIA buffer). Purified monoclonal antibodies at
10 nM in EIA buffer were incubated with increasing
concentrations of synthetic 24-residue or 14-residue ERRP
peptides (see EXAMPLES) or purified :ERRP for 1 hr at 20°C. The
plates were washed three times with EIA buffer, and rabbit and
anti-mouse IgG antibodies (1:1000 dilution in EIA buffer;
Cappel) were added. After 1 hr the plates were washed with EIA
buffer and lzsl-protein A (New England Nuclear) in EIA buffer
was added for a further hr. After a final
*Trade-mark
- 16 -
CA 02008735 1999-12-20
series of washes, the plates were air-dried, and the wells were
counted with a model 1274 LKB gamma counter.
Amino acid analysis,. Edman degradation, and yeytide
~vnthesis. Phenylthiocarbamyl amino acid analysis and
microsequence analysis were done using a model 420A PTC amino
acid analyzer and a model 470 gas phase sequences (both from
Applied Biosystems, Inc.), respectively, as described by Crabb
et al. (40). All sequences reagents and solvents were from
Applied Biosystems. With Beta-lactoglobulin A as a test
substrate the sequences gave initial yields of 40% and
repetitive yields in excess of 92%. The N-terminal sequences
of ERRP fragments were localized by comparison with the primary
receptor structure deduced from cloned cDNA (14). Solid phase
peptide synthesis was carried out essentially as described by
Merrifield (41) using an Applied Biosystems Model 430A
automatic peptide synthesizer. A 24 residue peptide
(DLHILPVAFRGDSFTHTPPLDPQD), a 14 residue peptide
(AFRGDSFTHTPPLD), and a 4 residue peptide (RGDS) were
synthesized using t-butyloxycarbonyl (tBOC-N°lp~"-protected amino
acids and "Pam" resins (Applied Biosystems). Side chain
protecting groups used were Asp(O Bzl), Ser(Bzl), Thr(Bzl),
Arg (NalphaTOS) and His (Bom) where Bzl = benzyl, Tos = toluene
sulfoxyl, and Bom = benzyloxymethyl. The remaining amino acids
had no side chain protecting groups. All amino acids were
obtained from Peninsula Laboratories except tBoc-His(Bom),
which was from Bachem. After assembly the protecting groups
were removed and the peptide-resin anchoring bond was cleaved
with 80-90% (v/v) anhydrous hydrogen fluoride in the presence
- 17 -
CA 02008735 1999-12-20
of anisole and dimethylsulfide. Before use synthetic peptides
were purified by reverse phase HPLC and were subjected to amino
acid analysis and Edman degradation.
Table I
Amino Acid Analysis
Peptide Ea ERRPb
Analysis 1 Analysis 2 (Residues 321-367)
Asx 6.7 6.5 7
Glx 2.1 2.5 2
Ser 4.3 4.7 5
Gly 2.4 3.2 2
His 3.1 3.4 3
Arg 1.0 1.1 1
Thr 3.9 3.7 4
Ala 2.1 2.3 2
Pro 3.8 3.6 4
Met 0 0 0
Tyr 0 0 0
Val 1.0 1.1 1
Ile 3.8 3.5 4
Leu 4.1 3.8 4
Phe 4.0 3.4 4
Lys 3.2 3.0 3
Total
Residues 46 46 46
*Numbers of amino acid residues were calculated assuming a
peptide length of 46 residues. Peptide E was generated by V8
protease cleavage of ERRP and was purified by reverse phase
HPLC as shown in Fig. 8. 21 and 40 pmol of peptide E were
subjected to amino acid analysis.
HComposition inferred from cDNA sequence (14).
- 18 -
i i
CA 02008735 1999-12-20
~ Iodination of EGF and TGF-alb. Receptor grade EGF (Upstate
Biotechnology, Inc. Lake Placid, NY) and synthetic rat TGF-
alpha (Peninsula Lab, Belmont, CA) were iodinated with Na[125I]
(Amersham, Arlington Heights, IL) by the chloramine T methid
(42) to specific activities of 4.0 x 105 cpm/ng and 3.5 x 105
cpm/ng, respectively.
Cross-linkingv and enzymatic digestion. ERRP was incubated with
[ 125I ] EGF or [ 125I ] TGF-alpha at a ratio of 1000 :1 (w/w) for 2 hr
at room temperature. Disuccinimidyl suberate (DSS) (Pierce
Chemical Co., Rockford, IL) was added to the final
concentration of 1 mM. After 20 min, the reaction was stopped
by the addition of reduction and alkylation buffer (1 M Tris,
pH 8.4, 6 M GnHCl and 5 M EDTA). The detailed procedures for
reduction and alkylation were described previously (40). The
alkylated cross-linking material was dialyzed in either
endoproteinase Glu-C digestion buffer (0.05 M NH4HC0~, pH 7.8)
or endoproteinase Lys-C digestion buffer (0.1 M NH,HCO3, pH
9.0). Cleavage by endoproteinase Glu-C (Boehringer-Mannheim,
Indianapolis, IN) was carried out at an enzyme-protein ratio
of 1:40 (w/w) for 14 hours at room temperature, and cleavage
by endoproteinase Lys-C (Boehringer-Mannheim) was performed at
a ratio of 1:40 (w/w) for 10 hr at 37°C.
Immunoyrecipitation of ERRP fragments. The protease activity
in digests was inactivated by boiling for 2 min and
immunoprecipitation was performed by addition of LA22 IgG2 a
(43) conjugated to Affi-Gel 10 (BioRad, Richmond, CA) (1 mg
antibody/ml gel). After
- 19 -
2008735
a 1;~ hr incubation at 22°C, the sub>ernatant was removed, and the
gel was washed twice with 1% BSA in PBS and twice by PBS or, for
further cleavage, by either Endo Gl.u-C digestion buffer or Endo
Lys-C digestion buffer. The gels washed with digestion buffers
were boiled for 2 min to release pe=ptides from the antibodies.
Proteases were then added to the ge=l slurry and incubated for 10
hrs at room temperature for Endo GI_u-C and at 37°C for Endo Lys-
C.
Inhibition or growth of a human tumor cell line A431 cells (22)
were plated in 24-well plates (Cost=ar, Cambridge, MA) at 5 x 103
cells/ml in DME/F12 medium. Increasing concentrations of protein
A-purified monoclonal antibodies LA22, LA58 and LA90 were added to
duplicate wells as indicated in Fic~. 12. After a six day incuba-
tion at 37°C, the cells were trypsinized and counted with a
Coulter Particle counter (Coulter F'slectronics).
EXAMPLES
Interaction between monoclonal ant_Lbodies and the EGF-binding
domain of the EGF receptor
Three monoclonal antibod_Les, LA22, LA58, and LA90 were
used to study the interaction betwe=en EGF and the ligand-binding
domain of the human EGF receptor. All three of the antibodies
inhibited the binding of lzsl-EGF i.o A431 human epidermoid carci-
noma cells (22) (Table II). In addition, each antibody completely
inhibited the binding of the other two antibodies to A431 cells,
and the binding of each antibody w<~s inhibited up to 87% by EGF.
''.. .;: _ I
i - 20 _
i ~
CA 02008735 1999-12-20
Furthermore, the binding of all three antibodies was blocked
by the EGF-inhibiting monoclonal antibody 528 IgG (33, 34) but
not by the oligosaccharide-specific EGF receptor antibody 455
IgG (32) . Lineweaver-Burk plots of l2sl-EGF binding to live
A431 cells in the presence of 20 nM LA22 or 20 nM LA58
indicated that the antibodies did not detectably alter B"~X
(Fig. 1). Similar results were obtained with l2sl-EGF binding
to fixed A431 cells in the presence of antibodies at 66 nM
(results not shown). Thus, these antibodies, like 528 IgG (44)
mainly inhibited EGF receptor interactions in a competitive
manner. These results suggested that the monoclonal antibodies
recognized spatially related epitopes and that these antigenic
determinants were within, or immediately adjacent to, the
receptor EGF-binding site.
30
- 21 -
i
CA 02008735 1999-12-20
TABLE II
Mutually competitive binding of EGF and
monoclonal antibodies
Competitive binding assays were done using
paraformaldehyde-fixed A431 cells as described under
"Experimental Procedures". The concentrations and specific
activities of lzSl_labeled ligands were: EGF (0.5 nM, 4.8 x 105
cpm/ng): LA22 (0.3 nM, 1.5 x 105 cpm/ng); LA58 (0.4 nM, 8.0 x
104 cpm/ng) : and LA90 (0.3 nM, 9.3 x 104 cpm/ng) . Competing
ligands were added to saturating concentrations. The results
are expressed as percent maximum inhibition.
Competing ligand
125I-Ligand
EGF LA22 LA58 LA90 528 455
EGF 97 87 89 85 97 0
LA22 87 100 100 100 100 0
LA58 84 100 100 100 100 0
LA90 85 100 97 100 98 0
- 22 -
CA 02008735 1999-12-20
~ Tntorac-tinn of monoclonal antibodies with deylyco~ylated
receptor segments
Western analysis was used to determine whether the
antibodies recognized continuous epitopes as opposed to
conformational assembly or carbohydrate determinants on EGF
receptors. The antibodies were reacted within tact or
deglycosylated A431 EGF receptor-related protein (ERRP), a
truncated form of EGF receptor that is secreted by A431 cells
(26). The receptor-related protein is a Mr 105,000
glycoprotein that binds EGF and includes the entire
extracellular domain of the EGF receptor ( 14 ) . As shown in
Fig. 2, all three monoclonal antibodies bound to both the
intact and deglycosylated forms of ERRP. In contrast, 455 IgG
(31), an EGF receptor monoclonal antibody that recognizes a
blood group A-related oligosaccharide epitope (45), bound to
intact but not to deglycosylated ERRP (Fig. 2, 6 and 7).
These results indicated that LA22, LA90, and LA58 did not
recognize endoglycosidase F-sensitive N-linked glycans, and
they indicated that the antibodies recognized continuous
peptide epitopes.
Fmrt_h_o__r localization of theepitones recogw~i ~.~d by the
monoclonal antibodies
The ability of the three monoclonal antibodies to
react with denatured ERRP was exploited to define the epitopes
of these EGF competitive antibodies. Cyanogen bromide (CNBr)-
generated fragments of reduced and carboxymethylated ERRP were
resolved by reverse-phase HPLC (Fig. 3A), and each fraction was
tested for immunoreactivity to LA22 (Fig. 3B). Inspection of
the deduced amino acid sequence of ERRP (14) predicted that
-23 -
IgG (32) . Lineweaver-Burk
CA 02008735 1999-12-20
complete cleavage would generate 11 CNBr fragments ranging in
size from 9 to 250 amino acids. As shown in Fig. 3, LA22 IgG
reacted most strongly with fractions 19-22 which corresponded
to an absorbance peak eluting from the HPLC column between 40-
50% acetonitrile. Fraction 19 consisted of a broad peak on
reverse-phase HPLC (Fig. 4) which was resolved by SDS-
polyacrylamide gel electrophoresis into two immunoreactive
bands of Mr 370,000 and 43,000 (Fig. 4, inset). When subjected
to amino acid sequencing, this HPLC peak yielded a major
sequence (initial yield 38 pmol) staring at Glu-295 (XEDGV ...,
Fig. 5) and a minor sequence (initial yield 7 pmol) starting
with Asp-254 (XXNPE...) (14). Fraction 21 (Fig. 3) yielded an
additional immunoreactive CNBr peptide of Mr 170,000 (C,
Fig. 6), which also started with Glu-295 (EEDGV ... , peptide
C; initial yield 28 pmol).
The presence of an acid-labile peptide bond between
Asp-364 and Pro-365 (14) suggested that Asp-364 was the COOH-
terminal amino acid of the Mr 17,000 CNBr peptide. This
possibility was supported by the observation that the Mr 43,000
and 37,000 CNBr fragments (Fig. 4) were converted by acid
treatment to Mr 23,000 and 17,000 LA22-reactive polypeptides
with conserved NH2-terminal amino acid sequences (Fig. 7).
Together these results suggested that the epitope recognized .
by LA22 resided within the 70 amino acids between Glu-295 and
Asp-364 (Fig. 5). This 70-residue peptide had two potential
N-linked glycosylation sites at Asn-328 and Asn-33? (14). The
difference between the apparent molecular weight of this 70-
amino acid peptide (Mr 170,000) and that deduced from its
- 24 -
CA 02008735 1999-12-20
primary structure (Mr 9,200) suggested that the peptide was
modified at one or both glycosylation sites. Endoglycosidase
F digestion of the Mr 23,000 and 17,000 immunoreactive peptides
resulted in molecular weight decreases in both peptides (Fig.
7, inset) indicating that these peptides were indeed
glycosylated.
To further localize the epitope recognized by LA22,
the Mr 43,000 and 37,000 CNBr peptides (Fig. 4, inset) were
cleaved with V8 protease at glutamyl residues (29) generating
an Mr 15,000 immunoreactive fragment (~ntide E, Fig. 8).
Edman degradation of this peptide yielded a sequence starting
with Phe-321 (FKDSLSIXATNIKHFKXCTSI..., initial yield 100
pmol). The first 21 amino acids of this peptide were identical
to the corresponding amino acid sequence deduced from cloned
EGF receptor cDNA (14) (Fig. 5) with the exception that no
detectable PTH yields were obtained for Asn-329 and Asn-337.
These results further suggested that both asparagine residues
were glycosylated. Based on the cleavage specificity of V8
protease and the molecular weight of peptide E, the COOH-
terminal amino acid of this peptide was predicted to be Glu-
367. This prediction was confirmed by amino acid analysis
(Table I): the amino acid composition of the Mr 15,000 peptide
was virtually identical to the known amino acid composition of
the 47-residue sequence from Phe-321 to Glu-367 (14) (Fig. 5).
Analyses of peptides C and E suggested that the
epitope for LA22 resided within the 44 amino acids from Phe-321
to Asp-364 (Fig. 5). Preliminary experiments indicated that
the reactivity of LA22 for ERRP was abolished by Arg-C
endoproteinase (data not shown). Since erg-353 was the sole
- 25 -
CA 02008735 1999-12-20
arginine residue between Phe-321 and Asp-364 (14) (Fig. 5), it
was likely that the epitope for LA22 was within the COOH-
terminal portion of this 44-residue amino acid sequence.
Therefore, a 24-residue peptide was synthesized based upon the
receptor sequence from Asp-344 to Glu-367, which included the
Arg-Gly-Asp-Ser adhesion molecule receptor recognition sequence
(27) at residues 353-356. The synthetic 24-mer inhibited the
binding of antibodies LA22, LA58, and LA90 to immobilized ERRP
(Fig. 9A): the concentration of the 24-mer needed to inhibit
antibody binding by 50% ranged from 0.7 to 2 ,ccM. These results
demonstrated that these three mutually competitive antibodies
(Table II) recognized closely spaced and perhaps identical
antigenic determinants. A synthetic 14-residue peptide, Ala-
351 to Asp-364 (Fig. 5), within the 24-mer sequence also
effectively inhibited the binding of all three antibodies to
ERRP (Fig. 9B) with half-maximal inhibition occurring at
concentrations of 1-3 ~.cM. Both synthetic peptides were 10- to
50-fold less effective than intact ERRP in inhibiting antibody
binding. Although both peptides included the recognition
sequence for adhesion molecule receptors, a synthetic Arg-Gly-
Asp-Ser tetramer had no effect on antibody binding to ERRP
(data not shown). From these results the epitopes for the EGF
competitive antibodies LA22, LA58, and LA90 were assigned to
a region of the EGF receptor extracellular domain not larger
than 14 amino acids, which was flanked by the two cysteine-rich
sequences at residues 134-313 and 446-612 (14). The antibody
reactive synthetic peptides neither bound lzsl-EGF when
immobilized on a plastic substrate nor inhibited l2sl-EGF
binding to A 431 cells (results not shown). Thus, the epitopes
- 26 -
CA 02008735 1999-12-20
for antibodies LA22, LA58, and LA90 did not comprise the entire
ligand-binding region of the human EGF receptor.
Discussion of the foregoing' results
We have used the truncated Mr 105,000 form of the EGF
receptor secreted by A431 human epidermoid carcinoma cells (26)
to define the epitopes of three EGF competitive monoclonal
antibodies. The truncated receptor (ERRP) is identical in
sequence to the extracellular domain of the full length human
EGF receptor, but it includes 17 nonidentical COOH-terminal
amino acids starting from residue 617 (14). This form of the
EGF receptor is encoded by an overexpressed 2.8-kb mRNA in A431
cells (14).
The monoclonal antibodies under study were raised
against intact A431 cells and A431 membrane preparations, and
they were selected for the ability to inhibit l2sl-EGF binding
to A431 cells. The antibodies LA22, LA58, and LA90 were
mutually inhibitory, and they were largely inhibited by EGF in
binding to A431 cells (Table II). These three antibodies were
therefore, to that extent, similar in their binding properties
to the EGF receptor monoclonal antibodies 528, 225, and 579
(31, 34) and distinct from receptor antibody 455 which bound
to an oligosaccharide determinant without inhibiting EGF
binding (31,45). The binding of LA22, LA58, and LA90 to A431
cells was completely inhibited by 528 IgG but was not affected
by 455 IgG (Table II). Like 528 IgG (44), these antibodies
were competitive inhibitors of EGF receptor interactions (Fig.'
1) .
- 27 -
CA 02008735 1999-12-20
Unlike previously reported EGF competitive monoclonal
antibodies (23,31,46), LA22, LA58, and LA90 recognized both
denatured and deglycosylated forms of the EGF receptor (Fig.
2). These properties enabled us to analyze the binding of
these antibodies to CNB4- and protease-generated ERRP peptides.
Based on the recognition by LA22 of ERRP peptide C, a 70-amino
acid fragment, and peptide E, a 47-amino acid V8 protease-
generated ERRP peptide, the epitope for LA22 was localized to
the 44 amino acids between Phe-321 and Asp-364 (Fig. 5).
Inhibition of the binding of LA22, LA58, and LA90 to ERRP by
synthetic peptides (Fig. 9) placed the epitopes for these
antibodies within the 14 amino acids from Ala-351 to Asp-364
(Fig. 5). Thus, these three EGF competitive antibodies
recognized closely spaced amino acid determinants within a very
limited region of the extracellular domain of the EGF receptor.
This region of the receptor is located between the two
cysteine-rich regions that span residues 134-313 and residues
446-612 (14).
Although antibodies LA22, LA58, and LA90 bound to a
short region of the EGF receptor, it remained possible that
they recognize adjacent or overlapping epitopes (47). Studies
with synthetic peptide immunogens have indicated that peptides
of fewer than 10 amino acids are generally poor immunogens and
that the optimum length for immunogenic peptides is 10-15 amino
acids (48,49). These findings suggested that the 14-amino acid
sequence of the EGF receptor that bound all three antibodies
comprised but a single antigenic determinant and that the
antibodies had identical antigenic specificities. An Arg-Gly-
Asp-Ser recognition site for adhesion molecule receptors
- 28 -
CA 02008735 1999-12-20
~ occurred with the NH2-terminal half of the antibody-binding 14-
mer at receptor residues 353-356 (14). Whether or not this
sequence of amino acids can mediate the binding of receptors
for fibronectin or other adhesion molecules to EGF receptors
is unknown (27). The Arg-Gly-Asp-Ser tetramer was reported to
inhibit the attachment of normal rat kidney cells to
fibronectin-coated plastic (50), but it did not inhibit the
binding of any of the antibodies to immobilized ERRP. Thus,
this tetramer does not by itself constitute the epitope
recognized by these antibodies. However, we cannot rule out
the possibility that it is an integral part of the epitope.
The 14-residue human EGF receptor epitope described
here is 71.4% identical to the homologous region of the chicken
EGF receptor (Ala-352 to Asp-365) (20). Of the four amino acid
differences between the two sequences, two occur in the
adhesion receptor recognition tetramer such that the human Arg-
Gly-Asp-Ser sequence corresponds to Leu-Gly-Asp-Ala in the
chicken receptor (20). The remaining two amino acid
differences occur at residues 359 (His to Lys) and 361 (Pro to
Leu) of the human receptor (14,20) (Fig. 5). These amino acid
alterations may help to explain the 100-fold difference in the
of f inities of human and chicken EGF receptors of murine EGF
(20) .
Our results with EGF competitive antibodies are
consistent with and extend those of Lax et al (25) who reported
that EGF could be covalently cross-linked to the EGF receptor
between Met-294 and Asn-544. We have further found, as
described below that immobilize LA22 IgG precipitated a Mr
- 29 -
CA 02008735 1999-12-20
a
18,000 V8 protease-generated ERRP fragment cross-linked to l2sl-
EGF which suggested that the epitope for the monoclonal
antibodies and the receptor residues) covalently linked to EGF
colocalized to the 47 amino acids of ERRP peptide E (Fig. 8)
(51). The locations of the cysteine-rich regions in the
primary structure of the EGF receptor ( 14 ) suggest that the
native conformation of the extracellular domain is of vital
importance for the binding of EGF (25) and most EGF competitive
antibodies (23). The smallest antibody-reactive synthetic
peptide described here does not bind EGF. However, the
mutually competitive binding properties of LA22, LA58, LA90,
and EGF strongly imply that the 14-amino acid region of the
receptor between Ala-351 and Asp-364 participates in the
formation of the EGF-binding site. This conclusion supports
a model of EGF receptor structure in which a ligand-binding
cleft is formed between the two cysteine-rich regions of the
extracellular domain.
ERRP secreted by A431 cells contains the entire
extracellular domain of the EGF receptor (14). To identify
receptor cross-linking sites, [lasl]EGF was covalently cross-
linked to ERRP with DSS, and the [l2sl]EGF-ERRP complex was
cleaved by proteases. The reduced and alkylated [lzsl]EGF-ERRP
complex was first digested with endoproteinase Glu-C (Endo Glu-
C), an enzyme that specifically cleaves at the carboxyl side
of glutamyl residues (29). The enzyme was then heat-
inactivated and the cleavage products were immunoprecipitated
- 30 -
CA 02008735 1999-12-20
' with immobilized LA22. Bound peptides were released by boiling
in SDS-PAGE sample buffer and were analyzed by electrophoresis
and autoradiography. A single radiolabeled Mr 18,000 band was
detected (Fig. 10, lane 1). This indicated that the epitope
of LA22 was in close proximity to the cross-linked receptor
residue(s). To ensure complete cleavage, the Mr 18,000
fragment was treated again with Endo Glu-C. No further shift
in mobility or decrease in the intensity of the band was
observed (Fig. 10, lane 2).
As noted above, the smallest Endo Glu-C ERRP fragment
which contained the LA22 epitope consisted of the 47 amino
acids from Phe-321 to Glu-367; this Mr 15,000 peptide included
about Mr 9,000 in N-linked carbohydrates distributed between
Asn-328 and Asn-337. The Mr 18,000 fragment observed here most
likely consisted of this 47 amino acid peptide with a further
Mr 3,000 contributed by EGF residues Asn-1 to Glu-24. To
confirm this idea, the Mr 18,000 fragment was treated with a
mixture of endoglycosidase F and N-glycosidase F (Endo-F).
Endo-F treatment reduced molecular weight of the radiolabeled
fragment by about Mr 9,000 (Fig. 10, lane 3). This result was
consistent with our previous observation that the Endo Glu-C
fragment was shifted from Mr 15,000 to Mr 6,000 by Endo-F
treatment (43).
According to the amino acid sequence deduced from
cloned EGF receptor cDNA (14), this 47 amino acid fragment
includes only three lysine residues, Lys-332, Lys-333 and Lys-
336, which are potentially available to react with the amine-
specific cross-linker DSS. To determine which of these
residues were cross-linked to EGF, [l2sl]EGF-ERRP complexes were
- 31 -
CA 02008735 1999-12-20
cleaved with endoproteinase Lys-C (Endo Lys-C), which
specifically cleaves peptide bonds on the carboxyl side of
lysyl residues (52). At least five LA22-reactive radiolabeled
fragments were detected (Fig. 11, lane A), which indicated that
the digestion was incomplete. The two smallest Endo Lys-C
fragments immunoprecipitated by immobilized LA22 antibodies
were a peptide of approximately Mr 12,500 and a prominent
radiolabeled Mr 18,000 peptide. When deglycosylated with Endo
F, both bands shifted in molecular weight by approximately Mr
1-2,000 (Fig. 11 lane B). As this shift in molecular weight
did not eqixal or exceed Mr 9,000, it was evident that both ERRP
fragments contained only a single glycosylated residue, Asn-
337, which was the most proximal to the LA22 epitope (Ala-351
to Asp-364) of the two glycosylation sites between Phe-321 and
Glu-367. This experiment further indicated that the
oligosaccharides N-linked to Asn-328 and Asn-337 contributed
to Mr 7-8,000 and Mr 1-2,000, respectively, to the mass of
ERRP.
Since the NH2-termini of the Mr 12, 500 and the Mr
18,000 ERRP fragments resided between Asn-328 and Asn-337, the
cleavage specificity of Endo Lys-C indicated that His-334 was
the NHZ-terminal residue of both fragments and that Lys-336 was
the sole receptor residue that could be cross-linked by DSS to
EGF. As EGF possesses no lysyl residues, it is most likely
cross-linked to ERRP via the amino group of Asn-1. Although
the COOH-terminal residues of the ERRP fragments could not be
identified unambiguously, the molecular masses of the [l2sl]EGF-
peptide complexes were consistent with the Mr 12,500 fragment
terminating with Lys-372 or Lys-375 and the Mr 18,000 fragment
- 32 -
CA 02008735 1999-12-20
ending at Lys-407. The existence of the radiolabeled Mr 18,000
fragment suggested that the cleavage of ERRP by Endo Lys-C at
residues 372 and 375 was very inefficient.
To eliminate the possibility that cleavage by Endo
Lys-C was occurring at residues other than lysine, the
immunoprecipitated fragments generated with Endo Lys-C were
digested with Endo Glu-C. Three radiolabeled fragments were
expected if Endo Lys-C were acting in a specific manner: an Mr
18,000 band corresponding to receptor fragments from either
Phe-321 or Asp-323 to Glu-367, which would result from any
fragment bearing the LA22 epitope terminating N-terminal to
Lys-333 and C-terminal to Lys-372: and Mr 9,000 receptor
fragment corresponding to His-334 to Glu-367, which would be
produced from fragments with His-334 as the NH2-tenainal amino
acid and any lysyl residue at the COOH-terminus; and a Mr 3,000
peptide representing EGF residues 25 to 53 that would be
released by the protease. The Endo Glu-C digest was resolved
by SDS-PAGE into three bands of Mr 18,000, Mr 9,500 and Mr 3,000
(Fig. 11, lane C). Thus, the original Endo Lys-C cleavages
occurred specifically at lysyl residues even though the
reaction did not go to completion.
We applied the same strategy to investigate the EGF
receptor binding site for TGF-alpha. After cross-linking
[l2sl]TGF-alpha to ERRP with DSS, the complex was subjected to
reduction and alkylation and Endo Glu-C cleavage. An Mr 18,500
band was detected by autoradiography after immunoprecipitation
with LA22 antibodies. Complete cleavage was confirmed by the
- 33 -
CA 02008735 1999-12-20
lack of further digestion with ,additional Endo Glu-C. This
antibody-reactive Endo Glu-C-generated ERRP fragment was most
likely the same as that cross-linked with EGF. That is, the
band was composed of the Mr 15,000 forty-seven residue receptor
fragment Phe-321 to Gln-367 with a Mr X3,000 TGF-alpha fragment
from Val-1 to Gln-26 or Lys-29 to Ala-50. These data indicated
that the receptor site cross-linked with TGF-alpha was also in
close proximity to the LA22 epitope and was included within the
same 47 amino acid ERRP fragment.
r"t,_i_hit~~n of growth of a human tumor cell line
The EGF receptor monoclonal antibodies LA22, LA58 and
LA90 were tested for the ability to inhibit the growth in vitro
of A431 human epidermoid carcinoma cells (22), which contain
a large number of EGF receptors. After a 6 day incubation
period at 37°C, all three antibodies at concentrations greater
than 1 ~Cg/ml severely inhibited A431 proliferation (Fig. 12).
In the presence of 20 ~g/ml of each antibody there was
essentially no increase in cell number.
EGF receptor antibodies seem to be most effective
against tumor cells that express an abnormally high number of
EGF receptors. This situation occurs frequently, but not
always, in squamous cell carcinomas, non-neuronal brain tumors,
breast cancers, and gastric carcinomas. Such antibodies are
not, however, effective against all tumors that express EGF
receptors.
Monoclonal antibodies can be delivered to a patient
through either the blood stream or peritoneal cavity and can
- 34 -
CA 02008735 1999-12-20
' be delivered directly to the tumor site. A preferred
embodiment would be chimeric antibodies in which the variable
region of the mouse heavy and light chains are spliced through
recombinant DNA technology to C regions of human heavy and
light chains.
20
- 35 -
CA 02008735 1999-12-20
' REFERENCES
1. James, R., and Bradshaw, R.A. (1984) Annu. Rev. Biochem
259-292.
2. Carpenter, G. (1987) Annu. Rev. Biochem, ~, 881-914.
3. Gill, G.N., Bertices, P.J., and Stanton, J.B. (1987) Mol.
Cell. Endocrinol. ~, 169-186.
4. Yarden, Y., and Ullrich, A. (1988) Biochemistry ~, 3113-
3119.
5. Schlessinger, J. (1988) Biochemistry ~, 3119-3123.
6. Savage, C.R., Jr., Inagami, T., and Cohen, S. (1972) J.
Biol. Chem. ~,, 7612-7621.
7. Savage, C.R. , Jr. , Hash, J.H. , and Cohen, S. (1973)
J.
Biol. Chem. 248, 7669-7672,
8. Carpenter, G., and Cohen, S. (1979) Annu. Rev. Biochem.
g$, 193-216.
9. Gray, A., Dull, T.J., and Ullrich, A. (1983) Nature 303,
722-725.
10. Scott, J., Urdea, M., Quiroga, M., Sanchez-Pescador, R.,
Fong, N., Selby, M., Rutter, W.J., and Bell, G.I. (1983)
Science ~, 236-240.
11. Cohen, S., Carpenter, G., and King, L., Jr. (198) J.
Biol. Chem. 255, 4834-4842.
12. Ushiro, H., and Cohen, S. (1980) J. Biol. Chem 255, 8363-
8365.
13. Buhrow, S.A., Cohen, S., and Staros, J.V. (1982) J. Biol.
Chem. 257, 4019-4022.
- 36 -
CA 02008735 1999-12-20
14. Ullrich, A., Coussens, L., Hayflick, J.S., Dull, T.J.,
Gray, A., Tem, A.W., Lee, J., Yarden, Y., Liberman, T.A.,
Schlessinger, J., Downward, J., Mayes, E.L.V., Whittle,
N., Waterfield, M.D., and Seeburg, P.H. (1984) Nature
309, 418-425.
15. Mayes, E., and Waterfield, M.D. (1984) EMBO J. $, 531-
537.
16. Childs, R.A., Gregoriou, M., Scudder, P., Thorpe, S.J.,
Rees, A., and Feizi, T. (1984) EMBO J. $,2227-2233.
17. Cummings, R.D., Soderquist, A.M. and Carpenter, G. (1985)
J. Biol. Chem. 260, 11944-11952.
18. Weber, W., Bertices, P.J., and Gill, G.N. (1984) J. Biol.
Chem. 259, 14631-14636
19. Downward, J., Parker, P., and Waterfield, M.D. (1984)
Nature $11, 483-485.
20. Lax, I., Johnson, A., Howk, R., Sap, J., Bellot,F.,
Winkler, M., Ullrich, A., Vennstrom, B., Schlessinger,
J., and Givol, D. (1988) Moll Cell Biol. $, 1970-1978.
21. Carpenter, G., and Cohen, S. (1977) Biochem. Biophys.
Res. Commun. ]~, 545-552.
22. Fabricant, R.N., DeLarco, J.E., and Todaro, G. (1977)
Proc. Natl. Acad. Sci. U.S.A. fig, 565-569.
23. Sato, J.D., Le, A., and Kawamoto, T. (1987) Methods
Enzymol. 146, 63-81.
24. Slieker, L.J., Martensen, T.M., and Lane, M.D. (1986)
J.
Biol. Chem. ?~,1, 15233-15241.
- 37 -
CA 02008735 1999-12-20
25. Lax, I., Burgess, W.H., Bellot, F., Ullrich, A.,
Schlessinger, J., and Givol, D. (1988) Mol. Cell. Biol.
$, 1831-1834.
26. Weber, W., Gill, G.N., and Spiess, J. (1984) Science 224,
294-297.
27. Rouslahti, E., and Pierschbacher, M.D. (1986) Cell 4.4.
517-518.
28. Tarr, G.E. (1986) Methods of Protein
Microcharacterization; A Practical Handbook (Shively,
J.E., ed.) pp. 155-194, Humana Press, Clifton, N.J.)
29. Houmard, J. and Drapeau, G.R. (1972) Proc. Natl. Acad.
Sci. x:3506-3509.
30. Kearney, J.F., Radbruch, A., Liesgang, B., and Rajewsky,
K. (1979) J. Immunol. 123, 1548-1550.
31. Sato, J.D., Kawamoto, T., Le, A., Mendelsohn, J.,
Polikoff, J., and Sato, G.H. (1983) Mol. Bio. Med. ~,
511-529.
32. Kawamoto, T., Sato, J.D., Le, A., McClure, D.B., and
Sato, G.H. (1983) Anal. Biochem. 130, 445-453
33. Sato, J.D., Kawamoto, T., and Okamoto, T. (1987) J. Exp.
Med. 165, 1761-1766
34. Kawamoto, T., Sato, J.D., Le, A., Polikoff, J., Sato,
G.H., and Mendelsohn, J. (1983) Proc. Natl. Acad. Sci.
USA $Q:1337-1341.
35. Ey, P.L., Prowse, S.J., and Jenkin, C.R. (1978).
Immunochem. x:429-436
36. Burnette, W.N. (1981) Anal. Biochem. ~i~:195-203.
- 38 -
CA 02008735 1999-12-20
r
' 37. Laemli, U.K. (1970) Nature 227:680-685.
38. Janregni=Adell, J., and Marti, J. (1975) Anal. Biochem.
x:468-473.
39. Mole, J.E., Bhown, A.S. and Bennett, J.C. (1977) J.
Immunol. 118: 67-70.
40. Crabb, J.W., Johnson, C.W., Carr, S.A., Armes, L.G., and
Saari, J.C. (1988) J. Biol. Chem. 263: 18678-18687.
41. Merrifield, R.B. (1963) J. Am. Chem. Soc. $,~: 2149-2154
42. Hunter, W.M., and Greenwood, F.C. (1962) Nature 194: 495-
496.
43. Wu, D., Wang, L., Sato, G.H., West, K.A., Harris, W.R.,
Crabb, J.W. and Sato, J.D. (1989) J. Biol. Chem. 264:
17469-17575.
44. Gill, G.N., Kawamoto, T., Cochet, C., Le, A., Sato, J.D.,
Masui, H., McLeod, C., and Mendelsohn, J. (1984) J. Biol.
Chem. 259: 7755-7760.
45. Gooi, H.C., Hounsell, E.F., Lax, I., Kris, R.M.,
Libermann, T.A., Schlessinger, J., Sato, J.D., Kawamoto,
T., Mendelsohn, J., and Feizi, T. (1985) Bioci. Rep. ~:
83-94.
- 39 -
CA 02008735 1999-12-20
A
46. Murthy U., Basu, A., Rodeck, U., Herlyn, M., Ross, A.H.,
and Das, M. (1987). Arch Biochem. Biophys. 252:549-560.
47. Benjamin, D.C., Berzofsky, J.A., East, I.J., Gurd,
F.R.N:, Hannum, C., Leach, S.J., Margoliash, E., Michael,
J.G., Miller, A., Prager, E.M., Reichlin, M., Sercarz,
E.E., Smith-Gill, S.J., Todd, P.E., and Wilson, A.C.
(1984) Annu. Rev. Immunol. 2,:67-101.
48. Palfreyman, J.W., Aitchenson, T.C., and Taylor, P. (1984)
J. Immunol. Methods $$:149-161.
49. Walter, G. (1986). J. Immunol. Methods $$:149-161.
50. Pierschbacher, M.D. and Ruoslahti, E. (1984) Nature
309:30-33.
51. Wu, D., Wang L., Chi, Y., Sato, G.H., and Sato, J.D.
(1990) Submitted for publication
52. Jekel, P.A., Weijer, and Beinteme, J. (1983) Anal.
Biochem, 134:347-354.
30
- 40 -