Note: Descriptions are shown in the official language in which they were submitted.
WOg2/20796 . PCT/US92/~146 ::
2103161 :
D-TYPE CYCLIN AND USES RELATED THERETO
__________________________ _ _ .
~ Descri~tion
__ ____ _ ___
~ Back~round of the Invention
____ ______________________
A typical cell cycle of a eukaryotic cell includes
05 the M phase, which includes nuclear division (mitosis)
and cytoplasmic division or cytokinesis and interphase,
which begins with the Gl phase, proceeds into the S phase
and ends with the G2 phase, which continues until mitosis
begins, initiating the next M phase. .In the S phase, DNA
~0 replication and histone synthesis occurs, while in the Gl
and G2 phases, no net DNA synthesis occurs, although
damaged DNA can be repaired. There are several key
changes which occur during the cell cycle, including a
critical point in the Gl phase called the restriction
15 point or start, beyond which a cell is committed to
completing the S, G2 and M phases.
Onset of the M phase appears to be regulated by a
common mechanism in all eukaryotic cells. A key element
of this mechanism is the protein kinase p84cdC2, whose
20 activation requires changes in phosphorylation and
interaction with proteins referred to as cyclins, which
also have an ongoing role in the M phase after acti-
vation.
Cyclins are proteins that were discovered due to
25 their intense synthesis following the fertilization o
marine invertebrate eggs ~Rosenthal, E.T. et _1., Cell
20:487-494 (1980)). It was subsequently observed that
the abundance of two types of cyclin, A and B, oscillated
WOg2/20796 PCT/US92/04146
-2-
2103161
during ~he early cleavage divisions due to abrupt proteo-
lytic degradation of the polypeptides at ~itosis and
thus, they derived their name (E~ans, T. et al., Cell
33:389-396 (1983); Swenson, K.I. et al., Cell 47:867-870
05 (1986); Standart, N. et al., Dev. Biol. 124:248 258
(1987)).
Active rather than passive involvement of cyclins in
regulation of cell di~ision became apparent with the
obser~ation that a clam cyclin mRNA could cause acti-
vation of frog oocytes and entry of these cells into Mphase (Swenson, K.I. et al., Cell 47:867-870 (1986)).
Activation of frog oocytes is associated with elaboration
of an M phase inducing factor known as MPF (Masui, Y. and
C.L. Markert, J. Ex~. Zool. 177:129 146 (1971); Smith,
L.D. and R.E. Ecker, Dev. Biol. 25:232-247 (1971)). MPF
is a protein kinase in which the catalytic s~bunit is the
fro~ homolog of the cdc2 protein kinase (Dunphy, ~.G. et
al., Cell 54:423-431 (1988); Gautier, J. et al., Cell
54:433-439 (1988); Arion, D. et al., Cell 55:371-378
(1988))
Three types of classes of cyclins have been identi-
fied to date: B, A and CLN cyclins. The B-type cyclin
has been shown to act in mitosis by serving as an
integral subunit of the cdc2 protein kinase (Booher, R.
and D. Beach, EMB0 J. 6:3441-3447 (1987); Draetta, G. et
al., Cell 56:829-838 (1989); Labbe, J.C. et al., Cell
57:253-263 (1989); Labbe, J.C. et al., EMBO_J. 8:3053-
3058 (1989); Mei~er, L. et al., EMBO_J. 8:2275-2282
(1989); Gautier, J. e_ al., Cell 60:487-494 (1990)). The
A-type cyclin also independently associates with the cdc2
W092/20796 PCT/US92/~146
~3~ 2103161
kinase, forming an enzyme that appears to act earlier in
the division cycle than mitosis (Draetta, G. et al., Cell
56:829-838 (1989); Minshull, J. et al., EMB0 J. 9:2865-
2875 (1990); Giordano, A. et al., Cell 58:981-990 (1989);
05 Pines, J. and T. Hunter, Nature 346:760-763 (1990)). The
functional difference between these two classes of
cyclins is not yet fully understood.
Cellular and molecular studies of cyclins in
invertebrate and vertebrate embryos ha~e been accompanied
by genetic studies, particularly in ascomycete yeasts.
In the fission yeast, the cdc13 gene encodes a B-type
cyclin that acts in cooperation with cdc2 to regulate
entry into mitosis (Booher, R. and D. Beach, ~MB0 J.,
6:3441-3447 (1987); Booher, R. and D. Beach, EMB0 J.
7:2321-2327 (1988); Hagan, I. et al., J. Cell Sci.
91:587-595 (1988); Solomon, M., Cell 54:738-740 (1988);
Goebl, M. and B. Byers, Cell 54:433-439 (1988); Booher,
R.N. et al., Cell 58:485-497 (1989)).
__ ___ ____ __
Genetic studies in both the budding yeast and
fission yeast have revealed that cdc2 (or CDC28 in
budding yeast) acts at two independent points in the cell
cycle: mitosis and the so-called cell cycle "start"
(Hartwell, L.H., J. Mol. Biol., 104:803-817 ~1971);
_____________ ___
Nurse, P. and Y. Bissett, Nature 292:558-560 {1981~;
Piggot, J.R. et al., Nature 298:391-393 (1982); Reed,
S.I. and C. Wittenberg, Proc._Nat._Acad._Sci._USA
87:5697-5701 (1990)).
In budding yeast, the start function of the CDC28
protein also requires association of the-catalytic
3~ subunit of the protein kinase with ancillary proteins
WO 92/20796 PCI`/USg2J04146
~ ?
2103161 -4-
that are structurally related to A and B-type cyclins.
This third class of cyclin has been called the Cln class,
and three genes comprising a partially redundant gene
family have been described (Nash, R. et al., EMB0 J.
05 7:4335-4346 (1988); Hadwiger, J.A. et al., Prot._Natl.
Acad._Sci. USA 86:6255-6259 (1989); Richardson, H.E. et
_1., Cell 59:1127-1133 (1989)). The CLN genes are_ ____ __
essential for execution of start and in their absence,
cells become arrested in the Gl phase of the cell cycle.
10 T~e CLNl and CLN2 transcripts oscillate in abundance
through the cell cycle, but the CLN3 transcript does not.
~' In addition, the Cln2 protein has been shown to oscillate
in parallel with its mRNA (Nash, R. et al., EMBO_J.
7:4335-4346 (1988); Cross, F.R., Mol._Cell._Biol.
8:4675-4684 (1988); Richardson, H.E. et al., Cell
59:1127-1133 (1988); Wittenberg, et al., l9gO)).
Although the precise biochemical properties con-
ferred on cdc2/CDC28 by association with different
cyclins have not bee~ fully elaborated, genetic studies
of cyclin mutants clearly establishes that they confer
"Gl" and "G2" properties on the catalytic subunit
(Booher, R. and D. Beach, EMB0 J. 6:3441-3447 (1987);
Nash, R. et al., EMBO_J. 7:4335-4346 (1988); Richardson,
H.E. et al., Cell 56:1127-1133 (1989)).
cdc2 and cyclins have been found not only in embryos
and yeasts, but also in somatic human cells. The
function of the cdc2/cyclin B enzyme appears to be the
same in human cells as in other cell types (Riabowol, K.
et al., Cell 57:393-401 (1989)). A human A type cyclin
30 has also been found in association with cdc2. No CLN
W092/20796 PCT/US92/04146
, ,?,1031~il
5 ,;
type cyclin has yet been described in mammalian cells. A
better unders~anding of the elements involved in cell
cycle regulation and of their interactions would con-
tribute to a better understancing of cell replication and
05 perhaps even alter or control the process.
Summary_of_the_Inve tion
The present invention relates to a novel class of
cyclins, referred to as D-type cyclins, which are of
mammalian origin and are a n~w family of cyelins related
10 to, but distinct from, previously described A, B or CLN
type cyclins. In particulsr, it relates to human
cyclins, encoded by genes shown to be able to replace a
CLN-type gene essential for cell cycle star~ in yeast,
which complement a deficiency of a protein essential for
15 cell cycle start and which, on the basis of protein
structure, are on a different branch of the evolutionary
tree from A, B or GLN type cyclins. Three members of the x
new family of D-type cyclins, referred to as the human
D-type gene family, are described herein. They encode
20 small (33-34 KDa) proteins which share an average of 57%
identity over the entire coding region and 78~ in the
cyclin box. One member of this new cyclin family, cyclin
Dl or CCNDl, is 295 amino acld residues and has an
estimated molecular weight of 33,670 daltons (Da). A ~-
25 second member, cyclin D2 or CCND2, is 289 amino acid
residues and has an estimated molecular weight of 33,045
daltons. It has been mapped to chromosome 12p band pl3.
A third member, cyclin D3 or CCND3, is 292 amino acid
residues and has an estimated molecular weight of
30 approximately 32,482 daltons. It has been mapped to
W092/~0796 PCT/US92/04146
2103161 -6-
chromosome 6p band p21. The D-type cyclins described
herein are the smallest cyclin proteins identified to
date. All three cyclin genes described herein are
interrupted by an intron at the same position. D-type
05 cyclins of the present invention can be produced using
recombinant techniques, can be synthesized chemically or
can be isolated or purified from sources in which they
occur naturally. Thus, the present invention includes
reccmbinant D-type cyclins, isolated or'~urified D-type
cyclins and ~ynthetic D-type cyclins.
The present invention also relates to DNA or RNA
encoding a D-eype cyclin of mammalian origin, particu-
larly of human origin, as well as to antibodies, both
polyclonal and monoclonal, specific for a D^type cyclin
Of mammalian, particularly human, origin.
The present i~vention further relates to a method of
isolating genes encoding other cyclins, such as other
D-type cyclins and related (but non-D type~ cyclins. It
also has diagnostic and therspeutic aspects. For ex-
20 ample, it relates to a method in which the presenceand/or quantity of a D-type cyclin (or cyclins) in
tissues or biological ~amples, such as blood, urine,
feces, mucous or saliva, is determined, using a nucleic
acid probe based on a D-type cyclin gene or genes des-
25 cribed herein or an antibody specific for a D-type
cyclin. This embodiment can be used to predict whether
cells are likely to undergo cell division at an ab-
normally high rate (i.e., if cells are likely to be
cancero~s), by determining whether their cyclin levels or
30 activity are elevated (elevated level of activity being
- indicative of an increased probability that cells will
VWD92/207g6 PCT/US9~/~146
210316i ``
- 7 - ; à
undergo an abnormally high rate of division). The
present method also relates to a diagnostic method in
which the occurrence of cell division at an abnormally
high rate is assessed based on abnormally high levels of
05 a D-type cyclin(s), a gene(s) encoding a D-type cyclin(s)
or a transcription product(s) (RNA).
In addition, the present invention relates to a
method of modulating (decreasing or enhancing) cell
division by altering the activity of at least one D-type
cyclin, such as D2, D2 or D3 in cells. The present
invention particularly relates to a method of inhibiting
' increased cell division by interfering with the act:ivity -~
or function of a D-type cyclin(s). In this therapeutic ~
method, function of D-type cyclin(s) is blocked (totally ~`
15 or partially) by interfering with its ability to activate -
the protein kinase it would otherwise (normally) activate
(e.g., p34 or a related protein kinase), by means of ~`~
agents which interfere with D-~ype cyclin activity, -;~
either directly or indirectly. Such agents include
anti-sense sequences or other transcriptional modulators
which bind D cyclin-encoding DNA or RNA; antibodies which `~
bind either the D-type cyclin or a ~olecule with which a ~-~
D-type cyclin must interact or bind in order to carry out
its role in cell cycle start; substances which bind the
D-type cyclin(s); agents (e.g., proteases) which degrade
or otherwise inactivate the D-type cyclin(s); or agents
(e.g., small organic molecules~ which interfere with
association of the D-type cyclin with the catalytic
subunit of the kinase. The subject invention also
relates to agents (e.g., oligonucleotides, antibodies,
peptides) useful in the isolation, diagnostic or thera-
peutic ~ethods described.
~"~" ~ ` r~3
W092/20796 PCT/USg2/04146
. ~, . . . ~ . -
21031~ 8-
Brief Descri~tion of the Fi~ures
____________ ______________ ____
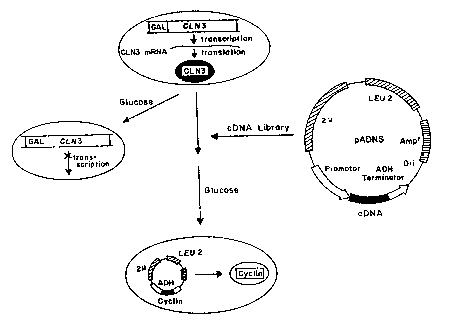
Figure 1 is a schematic representation of a genetic
screen for human cyclin genes.
Figure 2 is the human cyclin Dl nucleic acid
05 sequence (SEQ ID No. l) and amino acid sequence (SEQ ID
No. 2), in which nucleotide numbers and amino acid
numbers are on the right, amino acid numbers are given
with the initiation methionine as number one and the stop
codon is indicated by an asterisk.
Fig~re 3 is the human cyclin D2 nucleic acid
sequence (SEQ ID No. 3) and amino acid sequence (SEQ ID
~' No. 4) in which nucleotide numbers and aminc acid nu~bers
are on the right, amino acid numbers are gi~en with the
initiation methionine as number one and the stop codon is
indicated by an asterisk.
Figure 4 is the human cyclin D3 nucleic acid
sequence (SEQ ID No. 5) and a~ino acid sequence ~SEQ ID
No. 6), in which nucleotide numbers and amino acid -
numbers are on the right, amino acid numbers are given
with the initiation methionine as number one and the stop
codon is indicated by an asterisk.
Figure 5 shows the cyclin gene family.
Figure SA shows the amino acid sequence alignment of
seven cyclin genes (CYCDl-Hs, SEQ ID No. 7; CYCA-Hs, SEQ
ID No. 8; CYCA-Dm, SEQ ID No. 9; CYCBl-Hs, SEQ ID No. 10;
CDC13-Sp, SEQ ID No. 11; CLNl-Sc, SE~ ID No. 12; CLN3-Sc,
SEQ ID No. 13), in which numbers within certain sequences
indicate the number of amino acid residues omitted from
the sequence as the result of insertion.
Figure 5B is a schematic representation of the
evolutionary tree of the cyclin family, constructed using
the Neighbor-Joining method; the length of horizontal
line reflects the divergence.
W092/20796 - PCT/US92/04146
` 2103161
g , .
Figure 6 shows alternative polyadenylation of the
cyclin Dl gene transcript.
Figure 6A is a comparison of several cDNA clones
isolated from different cell lines. Open boxes represent
05 the 1.7 kb small transcript containing the coding region
of cyclin Dl gene. Shadowed boxes represent the 3'
fragment present in the 4.8 kb long transcript.
Restriction sites are given above each cDNA clone to ~-
indicate the alignment of these clones.
Figure 6B shows the nucleotide sequence surrou~ding
the first polyadenylation site for several c~NA clones -
(CYCDl-21, SEQ ID No. 14; CYCDl-H12, SEQ ID No. 15; -
CYCDl-HO34, SEQ ID No. 16; CYCDl-T078, SEQ ID No. 17 and
a genomic clone; CYCDl-GO68, SEQ ID No. 18).
Figure 6C is a summary of the structure and alter-
native polyadenylation of the cyclin Dl gene. Open boxes
represent the small transcript, the shadowed box repre-
sents the 3' sequence in the large transcript and the
filled boxes indicate the coding regions.
Figure 7 shows the protein sequence comparison of
eleven mammalian cyclins (CYCDl-Hs, SEQ ID No. 19; ~-
CYLl -Mm r SEQ ID No . 20; CYCD2-Hs, SEQ ID No. 21;
CYCL2-Mm, SEQ ID No. 22; CYCD3-Hs, SEQ ID No. 23;
CYL3-Mm, SEQ ID No. 24; CYCA-Hs, SEQ ID No. 25; CYCRl-Hs,
SEQ ID No. 26; CYCB2-Hs, SEQ ID No. 27; CYCC-Hs, SEQ ID
No. 28; CYCE-Hs, SEQ ID No. 29).
Figure 8 is a schematic representation of the
genomic structure of human cyclin D genes, in which each
diagram represents one restriction fragment from each
cyclin D gene that has been completely sequenced. Solid
WOg2/20796 PCT/US92/~146
2i03161 -lO-
boxes indicate exon sequences, open boxes indicate intron
or 5' and 3' untranslated sequences and hatched boxes
represent pseudogenes. The positions of certain
restriction sites, ATG and stop codons are indicated at
05 the top of each clone.
Figure 9 is the nucleic acid sequence (SEQ ID No.
30) and amino acid sequence (S~Q ID No. 31) of a cyclin
D2 pseudogene.
Figure 10 is the nucleic acid se~uence (SEQ ID No.
32) and the amino acid sequence (SEQ ID No. 33) of a
cyclin D3 pseudogene. `
~, Figure 11 is the nucleic acid sequence (SEQ ID ~o.
34) of 1.3 kb of human cyclin Dl promoter; the sequence
ends at initiation ATG codon and transcription starts at
approximately nucleotide -160.
Figure 12 is the nucleotide sequence (SEQ ID No. 35)
of 1.6 kb of human cyclin D2 promoter; the sequence ends
at initiation ATG codon and transcription starts at
approximately nucleotide -170.
Figure 13 is the nucleotide sequence (SEQ ID No. 36)
of 3.2 kb of human cyclin D3 promoter; the sequence ends
at initiation ATG codon and txanscription starts at
approximately nucleotide -160.
Detailed Descri~tion of the Invention
_______________ _____________________
As described herein, a new class of mammalian cyclin
proteins, designated D-type cyclins, has been identified,
isolated and shown to serve as a control element for the
cell cycle start, in ~hat they fill the role of a known
cyclin protein by activatin~, a protein kinase whose
30 activation is essential for cell cycle start, an event in
W092/20796 PCT/US92/~1~
3 1 6 1
the Gl phase at which a cell becomes committed to cell
division. Specifically, human D-type cyclin proteins, as
well as the genes which encode them, have been identi-
fied, isolated and shown to be able to replace CLN type
05 cyclin known to be essential for cell cycle start in
yeast. The chromosomal locations of CCND2 and CCND3 have
also been mapped.
As a result, a new class of cyclins (D type) is
available, as are DNA and RNA encoding the novel D-type ~;
cyclins, antibodies specific for (which bind to) D-type
cyclins and methods of their use in the identification of
additional cyclins, the detection of such proteins and
oligonucleotides in biological samples, the inhibition of
abnormally increased rates of cell division and the
identification of inhibitor~ of cyclins.
The following is a description of the identification
and characterization of human D-type cyclins and of the
uses of these no~el cyclins and related products.
'
Isolation and_Characterization_of Human Cyclin
D2 and D3
_ __ ____ .
As represented schematically in Figure 1 and des-
cribed in detail in Example 1, a mutant yeast strain in
which two of the three CLN genes (CLNl and CLN2) were
inactive and expression of the third was conditional, was
25 used to identify human cDNA clones which rescue yeast
from CLN deficiency. A human glioblastoma cDNA library
carried in a yeast expression ~ector (pADNS) was intro-
duced into the mutant yeast strain. Two yeast transform-
ants (pCYCDl-21 and pCYCDl-19) which ~rew despite the
WO92!20796 PCT/US92/04146
!, L~
2103161 -12-
lack of function of all three CLN genes and were not
revertants, were identified and recovered in E. coll.
Both rescued the mutant (CLN deficient) strain when
reintroduced into yeast, although rescue was inefficient `~:
05 and the rescued strain grew relatively poorly.
pCYCDl-l9 and pCYCDl-21 were shown, by restriction
mapping and partial DNA sequence analysis, to be inde~
pendent clones representing the same gene. A HeLa cDNA
library was screened for a full length cDNA clone, using
the 1.2 kb insert of pCYCDl-21 as probe. Complete
sequencing was done of the longest of~nine posi~i~e
clones identified in this manner (pCYCDl-H12; 1325 bp).
The sequence of the 1.2 kb insert is presented in Figure
2; the predicted protein product of the gene is of
approximate molecular weight 34,000 daltons.
Cyclin D2 and cyclin D3 cDNAs were isolated using
the polymerase chain reaction and three oligonucleotlde
probes derived from three highly conserved regions of
D-type cyclins, as descrihed in Example 4. As described,
two 5' oligonucleotides and one 3' degenerate oligonucle-
otide were used for this purpose. The nucleotide and
- amino acid sequences of the CCND2 gene and encoded D2
cyclin protein are represented in Figure 3 and of the
CCND3 gene and encoded D3 cyclin protein are represented
in Figure 4. A deposit of plasmid pCYC-D3 was made with
the American Type Culture Collection (Rockville, MD) on
May 14, lg91, under the terms of the Budapest Treaty.
Accession number 68620 has been assigned to the deposit.
Comparison of the CYCDl-H12-encoded protein se~uence
with that of known cyclins (see Figure SA) showed that
W092/20796 PCT/US92/~146
-13- ~103161
. . ., ~,
there was homology between the new cyclin and A, B and
CLN type cyclins, but also made it clear that CYCDl
- differs from these existing classes.
An assessment of how this new cyclin gene and its
05 product might be related in an e~olutionary sense to
other cyclin genes was carried out by a comprehensive
comparison of the amino acid sequences of all known
cyclins (Figure 5B and Example 1). Results of this
comparison showed that CYCDl represents a new class of
cyclin, designated herein cyclin D.
Expression of cyclin Dl gene in human cells was
' studied using Northern analysis, as described in Example
2. Results showed that levels of cyclin Dl expression
were very low in se~eral cell lines. The entire coding
region of the CYCDl gene was used to probe poly~A)+ RNA
from HeLa cells and demonstrated the presence of two
major transcripts, one approximately 4.8 kb and the other
approximately 1.7 kb, with the higher molecular weight
form being the more abunda~t. Most of the cDNA clonas
isolated from various cDNA l~braries proved to be very
si~ilar to clone ~CYCDl-H12 and, thus, it appears that `-
the 1.7 kb transcript detected in ~orthern blots corres-
ponds to the nucleo~ide sequenee of Figure 2. The origin
of the larger (4.8 kb) transcript was unclear. As
described in Example 2, it appears that the two mRNAs
detected (4.8 kb and 1.7 kb) arose by differential
polyadenylation of CYCDl (Figure 6).
Differential expression of cyclin Dl in different
tissues and cell lines was also assessed, as described in
Example 3. Screening of cDNA libraries to obtain full
length CYCDl clones had demonstrated that the cDNA
W092/20796 - PCT/US92/04146
~ 14-
2103161
library from the human glioblastoma cell line (U118 MG) ;
used to produce yeast transformants produced many more
positives than the other three cDNA libraries (human HeLa
cell cDNA, human T cell cDNA, human teratocarcinoma cell
05 cDNA). Northern and Western blotting were carried out to
determine whether cyclin Dl is differentially expressed.
Results showed (Example 3) that the level of transcript
is 7 to 10 fold higher in the glioblastoma (U118 MG)
cells than in HeLa cells, and that in both HeLa and U118
MG cells, the high and low molecular weight transcripts
occurred. Uestern blotting using anti-CYLl antibody
readily detected the presence of a 34kd polypept~de in
the glioblas~oma cells and demonstrated tha~ the protein
is far less abundant in HeLa cells and not detectable in
the 293 cells. The molecular weight of the anti-CYCLl
cross reactive material identified in U118 MG and HeLa
cells is exactly that of the human CYCDl protein ex-
pressed in E. coli. Thus, results demonstrated differ-
ential occurrence of the cyclin Dl in the cell types
analyzed, with the highest levels being in cells of
neural origin.
As also described herein (Example 6), human genomic
libraries were screened using cDNA probes and genomic
clones of human D-type cyclins, specifically Dl, D2 and
D3, have been isolated and characterized. Nucleic acid
sequences of cyclin Dl, D2 and D3 promoters are repre-
sented in Figures 11-13. Specifically, the entire 1.3 kb
cyclin Dl cDNA clone was used as a probe to screen a
normal human liver genomic library, resulting in identi-
fication of three positive clones. One of these clones(G6) contained a DNA insert shown to contain 1150 bp of
-W092/20796 PCT/US92/~146
-15- 21031~1 ~
upstream promoter sequence and a 198 bp exon, followed by
an intron. Lambda genomic clones corresponding to the
human cyclin D2 and lambda genomic clones corresponding
to the human cyclin D3 were also isolated and character-
- 05 ized, using a similar approach. One clone (~D2-G4) was
shown to contain (Figure 8B) a 2.7 kb SacI SmaI fragment
which includes 1620 bp of sequence 5' to the presumptive
initiating methionine codon identified in D2 cDNA (Figure
3) and a 195 bp exon followed by a 907 bp intervening
sequence. One clone (G9) was shown to contain (Flgure
8C) 1.8 kb of sequence 5' to the presumptive initiating
methionine codon identified in D3 cDNA (Figure 4), a 198
bp exon 1, a 684 bp exon 2 and a 870 bp intron.
Thus, as a result of the work described herein, a
novel class of mammalian cyclins, designated cyclin D or
D-type cyclin, has been identified and shown to be dis- -
tinct, on the basis of structure of the gene (protein) ~
product, from previously-identified cyclins. Three ;
members of this new class, designated cyclin Dl or CCNDl, ~`
zo cyclin D2 or CCND2 and cyclin D3 or CCND3, have been -~
isolated and sequenced. They have been shown to fulfill
the role of snother cyclin (CLN type) in activation of
the protein kinase (CDC28) which is essential for cell
cycle start in yeast. It has also been shown that the
25 cyclin Dl gene is expressed differentially in different
cell types, with expression being highest in cells of
neural origin.
Uses of the Invention
It is possible, using the methods and materials
30 described herein, to identify genes (DNA or RNA) which
W092/20796 PCT/US92/04146
~`1 0 ~1`6-1 - 16-
encode other cyclins (DNA or RNA which replaces a gene
essential for cell cycle start). This method can be used
to identify additional members of the cyclin D class or
other (non-D type) cyclins of either human or nonhuman
05 origin. This can be done, for example, by screening
other cDNA libraries using the budding yeast strain
conditional for CLN cyclin expression, described in
Example 1, or another mutsnt in which the ability of a
gene to replace cyclin expression can be assessed and
used to identify cyclin homologues. This method is
carried out as described herein, particularly in Example
~' 1 and as represented in Figure 1. A cDNA library carried
in an appropriate yeast vector (e.g., pADNS) is intro-
duced into a mutant yeast strain, such as the strain
described herein (Example 1 and Experimental Procedures).
The strain used contains altered CLN genes. In the case
of the specific strain descr~bed herein, insertional `~
mutations in the CLNl and CLN2 genes rendered them
inactive and alteration of the CLN3 gene allowed for its ;~
conditional expression from a galactose-inducible,
glucose-repressible promoter; as exemplified, this
promoter is a galactose-inducible, glucose-repressible-
promoter but others can be used.
Mutant yeast transformed with ~he cDNA library in
the expression vector are screened for their ability to
grow on glucose-containing medium. In medium containing
galactose, the CLN3 gene is expressed and cell viability
is maintained, despite the absence of CLNl and CLN2. In
medium containing glucose, all CLN function is lost and
the yeast cells arrest in the Gl phase of the cell cycle.
- Thus, the ability of a yeast transformant to grow on
W092/20796 - PCT/US9~/04146
-17- ~103161
glucose-containing m~dium is an indication of the pre-
sence in the transformant of DNA able to replace the
function of a gene essential for cell cycle start.
Although not required, this can be confirmed by use of an
05 expression vector, such as pADNS, which contains a
selectable marker (the LEU2 marker is present in pADNS).
Assessment of the plasmid stability shows whether the
ability to grow on glucose-containing medium is the
result of reversion or the presence of DNA function
(~ntroduction of DNA which replaces the unexpressed or
nonfunctional yeast gene(s) essential for cell cycle
start). ~sing this method, cyclins of all types (D type,
non-D type) can be identified by their ability to replace .
CLN3 function when transformants are grown on glucose.
15Screening of additional cDNA or genomic libraries eo
identify other cyclin genes can be carried out using all
or a portion of the human D-type cyclin DNAs disclosed
herein as probes; for example, all or a portion of the
Dl, D2 or D3 cDNA sequences of Figures 2-4, respecti~ely,
20 or all or a portion of the corresponding genomic sequen- -
ces described herein can be used as probes. The hybrid-
ization conditions can be varied as desired and, as a
result, the sequences identified will be of greater or
les~er complementarity to the probe sequence (i.e., if
higher or lower stringency conditions are used~. Addi-
tionally, an anti-D type cyclin antibody, such as CYLl or
another raised against Dl or D3 or other human D-type
cyclin, can be used to detect other recombinant D-type
cyclins produced in appropriate host cells transformed
with a vector containing DNA thought to encode a cyclin.
W092/20796 PCT/US92/04146
-18-
~103161
Based on work described herein, it is possible to
detect altered expression of a D-type cyclin or increased
rates of cell division in cells obtained from a tissue or
biological sample, such as blood, urine, feces, mucous or
05 saliva. This has poeential for use for diagnostic and
prognostic purposes since, for example, there appears to
be a link between alteration of a cyclin gene expression
and cellular transformation or abnormal cell prolifer-
ation. For example, several previous reports have
suggested the oncogenic potential of altered human cyclin
A function. The human cyclin A gene was found to be a
~, target for hepatitis B virus integration in a hepato-
cellular carcinoma (Uand, J. et al., Nature 343:555-557
(1990)). Cyclin A has also been shown to associate with
adenovirus ElA in virally infected cells (Giordano, A et
al., Cell 58:981-990 (1989); Pines, J. and T. Hunter,
Nature 34~-760-763 (1990)). Further, the PRADl gene,
______ ___
which has the same sequence as the cyclin Dl gene, may
play an important role in the development of various
tumors (e.g., non-parathyroid neoplasis, human breast
carcinomas and squamous cell carcinomas) with abnormal-
ities in chromosome llql3. In particular, identification
of CCNDl (PRADl) as a candidate BCLl oncogene provides
the most direct evidence for the oncogenic potential of
cyclin genes. This also suggests that other members of
the D-type cyclin family may be involved in oncogenesis.
In this context, the chromosomal locations of the CC~D2
and CCND3 genes have been mapped to 12pl3 and 6p21,
respectively. Region 12pl3 contains sites of several
translocations that are associated with specific immuno-
phenotypes of disease, such as acute lymphoblastic
W092/20796 PCT/US92/~146
2103161
- 19 - ~ .
leukemia, chronic myelomoncytic leukemia, and acute
myeloid leukemia. Particularly, the isochromosome of the
short arm of chromosome 12 [1(12p)] is one of a few known
consistent chromosomal abnormalities in human solid
05 tumors and is seen in 90% of adult testicular germ cell
tumors. Region 6p21, on the other hand, has been impli-
cated in the manifestation of chronic lymphoproliferative
disorder and leiomyoma. Region tp21, the locus of ~LA
complex, is also one of the best characterized regions of
the human genome. Many diseases have been previously
linked to the KLA complex, but the etiology of few of
~- these diseases is fully understood. Molecular cloning ;~
and chromosomal localization of cyclins D2 and D3 should
make it possible to determine whether they are directly
involved in these translocations, and if so, whether they
are activated. If they prove to be involved, diagnostic
and therapeutic methods described herein can be used to
assess an individual's disease state or probability of
developing a condition associated with or caused by such
translocations, to monitor therapy effec~iveness (by
assessing the effect of a drug or drugs on cell
proliferation) and to provide treatment. ~`
The present invention includes a diagnostic method
to detect altered expression of a cyclin gene, such as
cyclin Dl, D2, D3 or another D-type cyclin. The method
can be carried out to detect altered expression in cells
or in a biological sample. As shown herein, there is
high sequence similarity among cyclin D genes, which
indicates that different members of D-type cyclins ~ay
use similar mechanisms in regulating the cell cycle
(e.g., association with the same catalytic subunit and
W092/20796 PCT/US92/~146
~ -20-
acting upon the same substrates). The fact that there is
cell-type-specific differential expression, in both mouse
and human cells, makes it reasonable to suggest that
different cell lineages or d~fferent tissues may use
05 different D-type cyclins to perform very similar func-
tions and that altered tissue-specific expression of
cyclin D genes as a result of translocation or other
mutational events may contribute to abnormal cell
proliferation. As described herein, cyclin Dl is ex-
pressed differentially in tissues analyzed; in particu-
lar, it has been shown to be expressed at the highest
~' levels in cells of neural origin (e.g., glioblastoma
cells).
As a result of the work described herein, D-type
cyclin expression can be detected and/or quantitated and
results used as an indicator of normal or abnormal (e.g.,
abnormally high rate of) cell division. Differential
expression (either expression in ~arious cell types or of
one or more of the types of D cyclins) can also be
determined.
In a diagnostic method of ehe present invention,
cells obtained from an individusl are processed in order
to render nucleic acid sequences in them available for
hybridization with complementary nucleic acid se~uences.
25 All or a portion of the Dl, D2 and/or D3 cyclin (or other
D-type cyclin gene) sequences can be used as a probe(s).
Such probes can be a portion of a D-type cyclin gene;
such a portion must be of sufficient length to hybridize
to complementary sequences in a sample and remain hybri-
30 dized under the conditions used and will generally be atleast six nucleotides long. Hybridization is detected
using known techniques (e.g., measurement of labeled
W092/20796 PCT/US~2/~1~
-21- ~ 3 1 ~ I
hybridization complexes, if radiolabeled or fluorescently
labeled oligonucleotide probed are used). The extent to
which hybridi~ation occurs is quantitated; increased
levels of the D-type cyclin gene is indicative of
05 increased potential for cell di~ision.
Alternatively, the extent to which a D-type cyclin ~;~
(or cyclins) is present in cells, in a specific cell type
or in a body fluid can be determined using known tech-
nlques and an antibody specific for the D-type cyclin(s).
10 In a third type of diagnostic method, complex formation
between the D-type cyclin and the protein kinase with
which it normally or typically complexes is assessed,
using exogenous substrate, such as histone H1, as a
substrate. Arion, D. et al., Cell, 55:371-378 (1988~.
15 In each diagnostic method, comparison of results obtained
from cells or a body fluid being analyzed with results
obtained from an appropriate control ~e.g., cells of the
same type known to have normal D-type cyclin levels
and/or activity or the same body fluid obtained from an
20 individual known to have normal D-type cyclin levels
and/or activity) is carried out. Increased D-type cyclin
levels and/or activity may be indicative of an increased
probability of abnormal cell prolifera~ion or oncogenesis
or of the actual occurrence of abnoxmal prolifera~ion or
25 oncogenesis. It is also possible to detect more than one
type of cyclin (e.g., A, B, and/or D) in a cell or tissue
sample by using a set of probes (e.g., a set of nucleic
acid probes or a set of antibodies), the members of which
each recognize and bind to a selected cyclin and col-
30 lectively provide information about twv or more cyclinsin the tissues or cells analyzed. Such probes are also
W092/20796 - PCT/US92/04146
~ 22-
2~03161
the sub;ect of the present invention; they will generally
be detectably labelled (e.g., with a radioactive label, a
fluorescent material, biotin or another member of a
binding pair or an enzyme).
05 A method of inhibiting cell division, particularly
cell division which would otherwise occur at an abnormal-
ly high rate, is also possible. For example, increased
cell division is reduced or prevented by introducing into
cells a drug or other agent which can block, directly or
indirectly, formation of the protein kinase-D type cyclin
complex and, thus, block activation of the enzyme. In
~' o~e embodiment, complex formation is prevented in an
indirect manner, such as by preventing transcription
and/or translation of the D-type cyclin DNA and~or RNA.
15 This can be carried out by introducing antisense oligo- -
nucleotides into cells, in which they hybridize to the
'cyclin-encoding nucleic acid sequences, preventing their
further processing. It is also possible to inhibit
expression of the cyclin by interfering with an essential
D-type transcription factor. There are reasons to
believe that the regulation of cyclin gene transcription
may play an important role in regulating the cell cycle
and cell growth and oscillations of cyclin mRNA levels
are critical in controlling cell division. The Gl phase
25 is the time at which cells commit to a new round of
division in response to external and internal sequences
and, thus, trar.scription factors which regulate expres-
sion of Gl cyclins are surely important in controlling
cell proliferation. Modulation of the transcription
30 factors is one route by which D-type cyclin activity can
be influenced, resulting, in the case of inhibition or
W092/20796 PCT/US92/~l~
-23- ~1~3161
prevention of function of the transcription factor(s), in
reduced D-type cyclin activity. Alternatively, complex
formation can be prevented indirectly by degrading the D-
type cyclin(s), such as by in~roducing a protease or
05 substance which enhances cyclin breakdown into celIs. In
either case, the effect is indirect in that less D-type
cyclin is available than would otherwise be the case.
In another embodiment, protein kinase-D type cyclin
complèx formation is prevented in a more direct manner
10 by, for example, introducing into cells a drug or other -~
agent which binds the protein kinase or the D-type cyclin
or otherwise interferes with the physical association
between the cyclin and the protein kinase it activates
(e.g., by intercalation) or disrupts the catalytic
acti~lty of the enzyme. This can be effected by means of
antibodies which bind the kinase or the cyclin or a
peptide or low molecular weight organlc compound which,
like the endogenous D-type cyclin, b~nds the protein
kinase, but whose binding does not result in activation
of the enzyme or results in its being disabled or de-
graded. Peptides and small organic compounds to be used
for this purpose can be designed, based on analysis of
the amino acid sequences of D-type cyclins, to include
residues necessary for binding and to exclude residues
25 whose presence results in activation. This can be done,
for example, by systematically mapping the binding
site(s) and designing molecules which recognize or
otherwise associate with the site(s~ necessary for
activation, but do not cause activation. As described
30 herein, there is differential expression in tissues of
W092/2~796 ~CT/US92/~146
~ 03161 -24-
D-type cyclins. Thus, it is possible to selectively
decrease mitotic capability of cells by the use of an
a~ent (e.g., an antibody or anti-sense or other nucleic
acid molecule) which is designed to interfere with
05 (inhibit) the activity and/or level of expression of a
selccted type (or types) of D cyclin. For example, in
treating tumors in~olving the central nervous system or
other non-hemotopoietic tissues, agents which selectively
inhibit cyclin Dl might be expected to be particularly
10 useful, since Dl has been sh~wn to be differentially
expressed (expressed at particularly high levels in cells
of neural origin).
Antibodies specifically reacti~e with D-type cyclins
of the present ~nvention can also be produc~d, using
15 known methods. For example, anti-D type cyclin antisera
can be produced by injecting an appropriate host (e.g.,
rabbits, mice, rats, pigs) with the D-type cyclin against
which anti sera is desired and withdrawing blood from the
ho.st animal after sufficient time for an~ibodies to have
20 been formed. Monoclonal antibodies can also be produced
using known techniques. Sambrook, J. et al., Molecular
Clonin~. _A_Laboratory Manual, Cold Spring ~arbor
Laboratory, Cold Spring Harbor, NY (1989).
The present invention also includes a method of
25 screening compounds or molecules for their ability to
inhibit or suppress the function of a cyclin, particu-
larly a D-type cyclin. For example, mutant cells as
described herein, in which a D-type cyclin such as Dl or
D3, is expressed, can be used. A compound or molecule to
30 be assessed for its ability to inhibit a D-type cyclin is
contacted with the cells, under conditions appropriate
W092/20796 - PCT/US92/041~
-25- 21031Gl
for entry of the compound or molecule into the cells.
Inhibition of the cyclin will result in arrest of the
cells or a reduced rate of cell division. Comparison of
the rate or extent of cell di~ision in the presence of
05 the compound or ~olecule being assessed with cell
division of an appropriate control (e.g., the same type
of cells without added test drug> will demonstrate the
ability or inability of the compound or molecule to
inhibit the cyclin. Existing compounds or molecules
(e.g., those present in a fermentation broth or a
chemical "library"~ or those developed to inhibit the
~' cyclin activation of its protein kinase can be screened
for their effectiveness using this method. Dr~gs which
inhibit D-type cyclin are also the subject of this
invention.
The present invention will now be illustrated by the
following examples, which are not ~ntended to be limiting -
in any way.
_XAMPLES______
Experimental procedurPs for Examples 1-3 are pre-
sented after Example 3.
EXAMPLE 1 Identification of Human cDNA Clones that
_.________ ________________________________________
Rescue CLN Deficiency
____________________
In S. cerevisiae, there are three Cln proteins.
_____________
25 Disruption of any one CLN gene has little effect on
growth, but if all three CLN genes are disrupted, ~he
cells arrest in Gl (Richardson, H.E. et al , Cell
59:1127-1133 (1989)). A yeast strain was constructed, as
W092~20796 PCT/US92/~l~
;: -26-
2103161
described below, which contained insertional mutations in
the CLN1 and CLN2 genes to render them inactive. The
remaining CLN3 gene was further altered to allow for
conditional expression from the galactose-inducible,
05 glucose-repressible promoter GALl (see Figure 1). The
strain is designated 305-15d #21. In medium containing
galsctose the CLN3 gene is expressed and despite the
absence of both CLNl and CLN2, cell viability is retaîned
(Fig. 1). In a medium containing glucose, all CLN
1~ function is lost and the cells arrest in the Gl phase of
the cell cycle.
~' A human glioblastoma cDNA 11brary carried in the
yeast expression vector pADNS (Colicelli, J. et al., Pro.
Natl. Acad. Sci. USA 86:3599-3603 (1989)) was introduced
____________________ __
15 into the yeast. The vector pADNS has the LEU2 marker,
the 2~ replication origin~ and the promoter and ter-
minator sequences from the yeast alcohol dehydrogenase
gene (Figure 1). Approximately 3 x 10 transformants
were screened for the ability to grow on glucose con-
20 taining medium. After 12 days of incubation, twelvecolonies were obtained. The majority of these proved to
be re~ertants. However, in two cases, the ability to
grow on glucose correlated with the maintenance of the
LEU2 marker as assessed by plasmid stability tests.
25 These two yeast transformants carried plasmids designated
pCYCDl-21 and pCYCD1-19 (see below). Both were recovered
in E._coli. Upon reintroduction into yeast, the plasmids
rescued the CLN deficient strain, although the rescue was
inefficient and the rescued strain grew relatively
30 poorly.
W092/20796 PCT/US92/~146
-27- ~ 10~31 6 1
The restriction map and partial DNA sequence analy-
sis revealed that pCYCDl-19 and pCYCDl-21 were indepen-
dent clones representing the same gene. The 1.2 kb
insert of pCYCDl-21 was used as probe to screen a human
05 HeLa cDNA library for a full length cDNA clone. Approxi-
mately 2 million cDNA clones were screened and 9 posi-
tives were obtained. The longest one of these clones,
pCYCDl-Hl2 (1325 bp), was completely sequenced (Figure ~-
2). The sequence exhiblts a very high GC content within
the coding region (61%) and contains a poly A tail (69 A
residues). The estimated molecular weight of the predic-
ted protein product of the gene is 33,670 daltons start-
ing from the first in-frame AUG codon at nucleotide 145 H~
(Figure 2). The predicted protein is related to other
syclins (see below) and has an unusually low pI of 4.9
(compared to 6.4 of human cyclin A, 7.7 of human cyclin B
and 5.6 of CLNl), largely contributed by the high concen-
tration of acidic residues at its C-terminus.
There are nelther methionine nor s~op codons 5' to
the predicted initiating methionine at nucleotide 145.
Because of this and also because of the apparent N-
terminal ~runcation of CYCDl with respect to other
cyclins (see below for more detail), four additional
human cD~A libraries were further screened to see if the
~CYCDl-H12 clone might lack the full 5' region of the
cDNA. Among more than 100 cDNA clones isolated from
these screens, none was found that had a more extensive
5' region than that of ~CYCDl-Hl2. The full length
coding capacity of clone H12 was later confirmed by
Uestern blot analysis (see below).
092~20796 ~ PCT/US92/~146
r ; - 2 8 ~
CYCDl encodes the smallest (34 kd) cyclin protein
identified so far, compared to the 49 kd human cyclin A,
50 kd human cyclin B and 62 kd S._cerevisi_e CLN1. By
comparison with A and B type cyclins, the difference is
05 due to the lack of almost the entire N-terminal segment
that contains the so called "destruction box" identified
in both A and B type cyclins (Glotzer, M. et al., Nat_re
349:132-138 (1~91)).
Sequence_AnalysiS_of_Dl_an _ComEarison with Other
Cyclins
Sequence analysis revealed homology between the
CYCDl-H12 encoded protein and other cyclins, How-
e~er, it is clear that CYCD1 differs fro~ the three
existing ciasses of cyclins, A, B and CLN. To examine
how this new cyclin gene might be evolutionary related to
other cyclins, a comprehensive amino acid sequence
comparison of all cyclin genes was conducted. Fifteen
previously publishea cyclin sequences as well as CYCDl
were first aligned using a strategy described in detail
by Xiong and Eickbush (Xiong, Y. and T.H. Eickbush, EMBO
J. 9:3353-3362 (1990)). Effort was made to reach the
maximum similarity between sequences with the minimum
introduction of insertion/deletions and to include as
much sequence as possible. With the exception of CLN
cyclins, this alignment contains about 200 amino acids
residues which occupies more than 70% of total coding
region of CYCD1 (Figure 5A). There is a conserved domain
and some scattered similarities between members of A and
B type cyclins N-terminal to the aligned region (Glotzer,
M. _t al., Nature 349:132-138 (1991)), but this is not___ ______ ___
W092/20796 PCT/US92/~l~
-29- 2103161
present in either CLN cyclins or CYCDl and CYLl and so
they were not included in the alignment.
The percent divergence for all pairwise comparisons
of the 17 aligned sequences was calculated and used to
05 construct an evolutionary tree of cyclin gene family
using the Neighbor-Joining method (Saitou, N. and M. Nei,
_ol. Biol. Evol. 4:406-425 (1987) and Experimental______________ _
Procedures~. Because of the lowest similarity of CLN
cyclins to ~he other three classes, the tree (Figure 5B)
was rooted at the connection between the CLN cyclins and
the others. It is ~ery clear from this evolutionary tree
that CYC~Dl, CYCD2 and CYCD3 represent a distinct new
class of cyclin, designated cyclin D.
EXAMPLE_2 Ex~ression_of_the Cyclin_Dl_Ge_e_i__Human
Cells
Express~on of cyclin Dl gene in human cells was
studied by Northern analysis. Initial studies indicated
that the level of cyclin Dl expression was very low in
several cell lines. Poly (A)~RNA was prepared from HeLa
cells and probed with the entire coding region of CYCDl
gene. Two major transcripts of 4.8 kb and 1.7 kb were
detected. The high molecular weight form was the most
abundant. With the exception of a few cDNA clones, which
were truncated at either the 5' or 3' ends, most of the
cDNA clones isolated from various different cDNA librar-
ies are very similar to the clone ~CYCDl-H12 (Ei~ure 2).
Thus, it appears that the 1.7 kb transcript detected in
Northern blots corresponds to nucleotide sequence in
Figure 2.
W092~20796 - PCT/US92/04146
2103 161 - 30-
To understand the origin of the larger 4.8 kb
transcript, both 5' and 3' end sub-fragments of the
~CYCDl-H12 clone were used to screen both cDNA and
genomic libraries, to test whether there might be alter-
05 na~ive transcription initiation, polyadenylstion and/ormRNA splicing. Two longer cDNA clones, ~CYCDl-H034 (1.7
kb) from HeLa cells and ~DYDCl-T078 (4.1 kb) from human
teratocarcinoma cells, as well as several genomic clones
were isolated and partially sequenced. Both ~CYCDl-H034
and ~CYCDl-T078 have identical sequences to ~CYCDl-H12
clone from their 5' ends (Figure 6). Both differ from
~CYCDl-H12 in having additional sequences at the 3' end,
after the site of polyadenylation. These 3' sequences
are the same in ~CYCDl-H034 and ~CYCDl-T078, but extend
further in the latter clone (Figure 6). Nucleotide
sequencing of a genomic clone within this reglon revealed
colinearity between the cDNAs and the genomic D~A (Figure
6). There is a single base deletion (an A residue) in
~CYCDl-T078 cDNA clone. This ~ay be the result of
polymorphis~, although it is not possible to exclude the
possibility that some other mechanism is involved. The
same 4.8 kb transcript, but not the 1.7 kb transcript,
was detected using the 3' end extra fragment from clone
T078 as a probe.
It appears that the two mRNAs detected in Northern
blots arise by differential polyadenylation (Figure 6).
Strangely, there is no recognizable polyadenylation
sequence (AAUAAA) anywhere within the sequence of clone
~CYCDl-H12, even though polyadenylation has clearly
occurred (Figure 2). There is also no close variant of
AAUAAA (nothing with less than two mismatches).
- W092/20796 PCT/US92/04146
2l~)3l6l :
EXAMPLE 3 Differen~ial Ex~ression of Cyclin Dl Gene
_________ _______________ ____________ ____________
i__Different_Cell Ty~es
During the screening of cDNA libraries to obtain
full length clones of CYCDl, it became evident that the
05 cDNA library derived from the human glioblastoma cell
line (U118 MG) from which the yeast transformants were
obtained gave rise to many more positi~es than the other
four cDNA libraries. Northern and Western blotting were
carried out to explore the possibility that cyclin Dl
might be differentially expressed in different tissues or
cell lines. Total RNA was isolated from U118 MG cells`
~-~ and analyzed by Northern blot using the CYCDl gene coding ~
region as probe. The level of transcript is 7 to 10 fold
higher in the glioblastoma cells, compared to HeLa cells.
In both HeLa and U118 MG cells, both high and low
molecular weight transcripts are observed.
To investigate whether the abundant CYCDl message in
the U118 MG cell line is reflected at the protein level,
cell extracts were prepared and Western blotting was `~
performed using anti-CYLl prepared against mouse CYLl
(provided by Matsushime, H. et al.). This anti-CYLl
antibody was able to detect nanogrsm quantities of
recombinant CYCDl on Uestern blots ~data not shown), and
was also able to detect CYCDl in the original yeast
transformants ~y immunoprecipitation and Wes~ern
analysis. Initial experiments using total cell extracts,
from HeLa, 293 or U118 MG cells failed to detect any
signal. However, if the cell extracts were immuno-
precipitated with the serum before being subjected to
30 SDS-PAGE and immunoblotting, a 34 kd polypeptide was
readily detected in Ull~ MG cells. The protein is far
W092/20796 PCT/US92/~146
-32-
2103161
less abundant in HeLa cells and was not detectable in 293
cells. The molecular weight of the anti-CYCLl cross-
reactive material from U118 MG and HeLa is exactly that
of the h~man CYCDl protein expressed in E. coli. This
05 arg~es that the sequenced cDNA clones contain the entire
open reading frame.
EXPERIMENTAL PROCEDURES
Strain Construction
The parental strain was BF305-lSd (MATa leu2-3
leu2-112 his3-11 his3-lS ura3-52 trpl adel metl4 arg5,6)
(Futcher, B. and J. Carbon, Mol._Cell. Biol. 6:2213-2222
(1986)). The strain was converted into a conditional
cln- strain in three steps. First, the chromosomal CLN3
gene was placed under control of the GALl promoter. A
15 0.75 kb Eco~I-BamHI fragment containing the bidirectional -~
GAL10-GALl promoters was fused to the 5' end of the CLN3
gene, such that the BamHI (GALl) end was attached 110
nucleotides upstream of the CLN3 start codon. An EcoRI
fragment stretching from the GAL10 promoter to the middle
of CLN3 (Nash, R. e_ al., EMBO J. 7:4335-4346 (198B)) was
then subcloned between the XhoI and EcoRI sites of pBF30
(Nash, R. et al., _MBO J. 7:4335-4346 (1988)). The
ligation of the XhoI end to the EcoRI end was accom-
plished by filling in the ends with Klenow, and blunt-end
25 ligating (destroying the EcoRI site). As a result, the
GALl promoter had replaced the DNA normally found between
-110 and -411 upstream of CLN3. Next, an EcoRI to SphI
fragment was excised from this new pBF30 derivative.
This fragment had extensive 5' and 3' homology to the
W092/20796 - PCT/US92/04146
33 2103161
CLN3 region, but contained the GALl promoter and a URA3
marker just upstream of CLN3. Strain BF305-15d was
transformed with this fragment and Ura+ transformants
were selected. These were checked by Southern analysis.
05 In addition, average cell size was measured when the GAL1
promoter was induced or uninduced. Uhen the GALl
promoter was induced by growing the cells in 1% raffinose
and 1~ galactose, mode cell volume was about 25~m3 (com-
pared to a mode volume of about 40~m3 for the parental
strain) whereas when the promoter was not induced
(raffinose alone), or was repressed by the presence of
glucose, cell volume was much larger than for the wild-
type strain. These experiments showed that CLN3 had been
placed under control of the GAL1 pro~oter. It is ~m-
portant to note that this GALl-controlled, glucose
repressible gene is the only source of CLN3 protein in
the cell.
Second, the CLNl gene was disrupted. A fragment of
CLNl was obtained from I. Fitch, and used to obtain a ;~
full length clone of CLNl by hybridization, and this was
subcloned into a pUC plasmid. A BamHI fragment carrying
the-HIS3 gene was inserted into an NcoI site in the CLN1
open reading frame. A large EcoRI fragment with ex-
tensive 5' and 3' homology to the CLN1 region was then
excised, and used to transform the BF305-15d GAL-CLN3
strain described above. Transformation was done on
YNB-his raffinose galactose plates. His~ clones were
selected, and checked by Southern analysis.
Finally, the CLN2 gene was disrupted. A fragment of
CLN2 was obtained from I. Fitch, and used to obtain a
full length clone of CLN2 by hybridization, and this was
W092/20796 PCT/USg2/~146
A
-34-
2103161
subcloned into a pUC plasmid. An EcoRI fragment carrying
the TRPl gene was inserted into an SpeI site in the CLN2
open reading frame. A BamHI-KpnI fragment was excised
and used to trsnsform the BF305-lSd GAL-CLN3 HIS3::clnl
05 strain described above. Transformation was done on
YNB-trp raffinose galactose plates. Trp+ clones were
selected. In this case, because the TRPl fragment
included an ARS, many of the transformants contained ~
autonomously replicatlng plasmid rather than a disrupted ~;
CLN2 gene. Howe~er, several percent of the transformants
were simple TRPl::cln2 disruptants, as shown by pheno-
~' typic and Southern analysis.
One particular 305-15d GALl-CLN3 HIS3::clnl
TRPl::cln2 transformant called clone #21 (referred to
hereafter as 305-15d #21) was analyzed extensively. When
grown in 1% raffinose and 1~ galactose, it had a doubling
time indistinguishable from the CLN wild-type parental
strain. However, it displayed a moderate Wee phenotype
(small cell volume), as expected for a CLN3 over-
expressor. When glucose was added, or when galactose wasremoved, cells accumulated in Gl phase, and cell division
ceased, though cells continued to increase in mass and
volume. After overnight incubation in the Gl-arrested
state, essentially no budded cells were seen, and a large
proportion of the cells had lysed due to their uncon-
trolled increase in size.
When 305-15d #21 was spread on glucose plates,
revertant colonies arose at a frequency of about 10-7.
The nature of these glucose-resistant, galactose-
independent mutants was not investigated.
W092/2079~ PCT/US9~/~146
~103161
-35-
Yeast_S~hero~lasts_Transformation
S. cerevisae spheroplasts transformation was carried
______.______
out according to Burgers and Pe~ci~al and Allshire
(Burgers, P.M.J. and K.J. Percival, Anal. Biochem.
05 163:391 397 (1987); Allshire, R.C., Proc._Natl._Acad.
Sci. USA 87:4043-4047 (1990)).
Cell Culture -
__..__________
HeLa and 293 cells were cultured at 37-C either on
plates or in suspension in Dulbecco's modified Eagle's
10 medium (DMEM) supplemented with 10% fetal calf serum. -~
~lioblastoma U118 MG cells were cultured on plates in
DMEM supplemented with 15% fetal bovine serum and 0.1 mM
non-essential amino acid (GIBCO~.
Nucleic Acid Procedures
Most molecular biology techniques were essentially
the ~ame as described by Sambrook et al. (Sambrook, J. et
al., Molecular Clonin~ A Laborat~ry Manual Cold Spring
Harbor Laboratory, Cold Spring Harbor, NY (1989)).
~hagmid vectorc pUC118 or pUCll9 (Vieira, J. and 3.
Messing, Meth. E_z~mol. 153:3 11 (1987)) or pBlueScript
(Stratagene) were used as cloning vectors. DNA sequences
were determined either by a chain termination method
(Sanger, F. et al., Proc. Natl. Acad. Sci._USA 74:5463-
5467 (1977)) using Sequenase Kit (United States Biochem-
ical) or on an Automated Sequencing System (373A, AppliedBiosystems).
Human HeLa cell cDNA library in ~ZAP II was pur-
chased from Stratagene. Human T cell cDNA library in
~gtlO was a gift of M. Gillman (Cold Spring Harbor
W092/20796- PCT/US92~04146
~ -36-
w103161
Laboratory). Human glioblastoma U118 MG and glioblastoma
SW1088 cell cDNA libraries in ~ZAP II were gifts of M.
Wigler (Cold Spring Harbor Laboratory). Human terato- ~
carcinoma cell cDNA library ~gtlO was a gift of Skow- `
05 ronski (Cold Spring Harbor Laboratory). Normal human
liver genomic library ~GEM-ll was purchased from Promega.
Total RNA from cell culture was extracted exactly
according to Sambrook et al. (Sambrook, J. et al.,
Molecular Clonin~: A Laboratory Manual Cold Spring
________________ ______________ _______
Harbor Laboratory, Cold Spring Harbor, NY (1939)) using
guanidium thiocyanate followed by centrirugation in CsCl
~, solution. Poly(A)+RNA was isolated from total RNA
preparation using Poly (A)~Quick push columns (Strata-
gene). RNA samples were separated on a 1% agarose-
15 formaldehyde-MOPS gel and transferred to a nitrocellulose
filter. Northern hybridizations (as well as library
screening) were carried out at 68-C in a solution con-
taining 5 x Denhardt's solution, 2 x SSC, 0.1% SDS, 100
~g/ml denatured Salmon sperm DNA, 25 ~M NaP04 (pH7.0) and
10% dextran sulfate. Probes were labelled by the random
priming labelling method (Feinberg, A. and ~. Vogelstein,
Anal. Biochem. 132:6-13 (1983)). A 1.3 kb Hind III
______________ ___
fragment of cDNA clone pCYCDl-Hl2 was used as codin~
region probe for Northern hybridization and genomic
library screening, a 1.7 kb Hind III-EcoRI fragment from
cDNA clone pCYCDl-T073 ~as used as 3' fragment probe.
To express human cyclin Dl gene in bacteria, a 1.3
kb Nco I-Hind II fra~ment of pCYCDl-H12 containing the
entire CYCDl open reading frame was subcloned into a T7
expression vector (pET3d, Studier, F.W. e_ al., Methods
in Enzymolo~y 185:60-89 (1990)). Induction of E. coli
______ ____ ___ __ ____
W092/20796 PCT/US9~/04146
~37~ 2103161
strain BL21 (DE3) harboring the expression construct was
according to Studier (Studier~ F.W. et al., Methods in
Enzymolo~y 185:60-89 (1990)). Bacterial culture was
lysed by sonication in a lysis buffer (5 mM EDTA, 10%
glycerol, 50 mM Tris-HCL, pH 8.0, 0.005% Triton X-100)
containing 6 M urea (CYCDl encoded p34 is only partial
soluble in 8 M urea), centrifuged for 15 minutes at
20,000 g force. The pellet was washed once in the lysis
buffer with 6 M urea, pelleted again, resuspended in
lysis buffer containing 8 M urea, and centrifuged. The
supernatant which enriched the 34 kd CYCDl protein was
loaded on a 10% polyacrymide gel. The 34 kd band was cut
from the gel and eluted with PBS containing 0.1~ SDS.
Se~uence Ali~nment and Formation of an Evolutionary Tree
__ _________ __ --____ _ :
Protein sequence alignment was conducted vixtually
by eye according to the methods described and discussed
in detail by Xiong and Eickbush (Xiong, Y. and T.H. .
Eickbush, EMB0 J. 9:3353-3362 (1990)). Numbers within
certain sequenc~s indicate the number of amino acid
20 residues omitted from the sequence as the result of
insertion.
Numbers within certain sequences indicate the number
of amino acid residues omitted from the sequence as the
result of insertion (e.g., for CLNl, ...TUG25RLS...-
25 indicates that 25 amino acids ha~e been omitted between Gand R). Sources for each sequence used in this alignment
and in the construction of an evolutionary tree (Figure
5B) are as follows: C~CA-Hs, human A type cyclin (Wang,
J. et al., Nature 343:555-557 (1990)); CYCA-Xl, Xenopus
30 A-type cyclin (Minshull, J. et al., EMBO_J. 9:2865-2875
W092/20796 - PCT/US92/~l~
2103161 -38-
~1990)); CYCA-Ss, clam A-type cyclin (Swenson, K.I. et
al., Cell 47:867-870 (1986); CYCA-Dm, Drosophila A-type
cyclin (Lehner, C.F. and P.H. O'Farrell, Cell 56:957-968
(1989)); CYCBl-Hs, human Bl-type cyclin (Pines, J. and T.
05 Hunter, Cell 58:833-846 (1989)); CYCBl-Xl and CYCB2-Xl,
Xenopus Bl- and B2-type cyclin (Minshull, J. et al., Cell
56:947-956 (1989)); CYCB-Ss, clam B-type cyclin
(Westendorf, J.M et al., J. Cell Biol., 108:1431-1444
(1989)); CYCB-Asp, starfish B-type cyclin (Tachibana, K.
et al., Dev. Biol. 140:241-252 (1990)); CYCB-Arp, sea
urchin B-type cyclin (Pines, J. and T. Hunter, EMB0 J.
, S:2987-2995 (1987)); CYCB-Dm, Drosophila B-type cyclin
(Lehner, C.F. and P.H. O'Farrell, Cell 61:535-547
(1990)); CDC13-Sp, S. po~be CDC13 (Booher, R. and D.
15 Beach, EMBO_J. 7:2321-2327 (1988)); CLNl-Sc and CLN2-Sc,
S. cerevisiae cyclin 1 and 2 (Hadwiger, J.A. et al.,
Proc. Natl. Acad. Sci. USA 86:6255-6259 (1989)); CLN3-Sc,
S. cerevisiae cyclin 3 (Nash, R. et al., EMB0 J. 7:4335-
_____________ :
4346 (1988)).
A total of 17 cyclin sequences were aligned and two
representive sequences from each class are presented in
Figure 5A.
Percent divergence of all pairwise comparison of 17
sequences were calculated from 154 amino acid residues
25 common to all 17 sequences, which does no`t include the 50
residue segments loca~ed at N-terminal part of A, B and
D-type cyclins because of its absence from CLN type
cyclins. A ~ap/insertion was counted as one mismatch
regardless of its size. Before tree construction, all
30 values were changed to distance with Poisson correction
(d ~ -log S, where the S = sequence similarity (Nei, M.,
- W092/20796 PCT/US92i~146
~,103161 ~
-39- :
Molecular Evolutionary Genetics pp. 287-326 Columbia
________ ____________ _________ ~
University Press, NY (1987)). Calculation of pairwise
comparison and Poisson correction were conducted using
computer programs developed at University of Rochester.
Evolutionary treec of cyclin gene fam~ly was generated by
05 the Neighbor-Joining program tSaitou, N. and M. Nei, Mol.
Biol. Evol. 4:406-567 (1987)). All calculations were
___________ _
conducted on VAX computer MicroVMS V4.4 of Cold Spring
Harbor Laboratory. The reliability of the tree was
e~aluated by using a subset sequence (e.g., A, B and
D-type cyclins), including ~ore residues (e.g., the 50-
~- residue segment located at C-terminal of A, B and D-type
cyclins, ~igure 5A) or adding several other unpublished
cyclin sequences. They all gave rise to the tree with
the same topology as the one presented in Figure SB.
Immunopreci~itatio__and_Western_Blots
Cells from 60 to 80% confluent 100 mm dish were
lysed in 1 ml of lysis buffer (50 mM Tris-HCl, pH 7.4,
150 mM NaCl, 20 mM EDTA, 0.5% NP-40, 0.5~ Nadeoxycholate,
1 mM PMSF) for 30 minutes on ice. Immunoprecipi~ation
was carried out using 1 mg protein from each cell lysate
at 4C for overnight. After equilibrated with the lysis
buffer, 60 ~1 of Protein A-agarose (PIERCE) was added to
each immunoprecipitation and incubated at 4C for 1 hour
with constant rotating. The immunoprecipitate was washed
three times with the lysis buffer and final resuspended
in 50 ~1 2 x SDS protein sample buffer, boiled for 5
minutes and loaded onto a 10% polyacrymide gel. Proteins
were transferred to a nitrocellulose filter using a SDE
Electroblotting System (Millipore) for 45 minutes at a
W092r20796 PCT/US92/~1~
~103161
constant current of 400 mA. The filter was blocked for 2
to 6 hours with 1 x PBS, 3% BSA and 0.1% sodium azide,
washed 10 minutes each time and 6 times with NET gel
buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1~ NP-40,
05 1 mM EDTA, 0.25% gelatin and 0.02 sodium azide), radio-
labelled with l25I-Protein A for l hour in blocking
solution with shaking. The blot was then washed 10
minutes each time and 6 times with the NET gel buffer
before autoradiography.
The tree was constructed using the Neighbor-Joining
method (Saitou, N. and M. Nei, Mol. Biol. Evol., 4:406-
,, 425 (1987). The length of horizon~al line reflects the
divergence. The branch len~th between the node con-
necting the CLN cyclins and other cyclins was arbitrary
15 divided~
MATERIALS_AND NETHODS
The following m~terials and methods were used in the
work described in Examples 4-6.
Molecular Clonin~
The human HeLa cell cDNA library, the human glio-
blastoma cell U118 MG cDNA library, the normal human
liver ~enomic library, and the hybridization buffer were
the same as those described above. A human hippocampus
cDNA library was purchased from Stratagene, Inc. High-
and low-stringency hybridizations were carried ou~ at 68
and 50C, respectively. To prepare template DNA for PCR
reactions, approximately 2 million lambda phages from
each cDNA library were plated at a density of 105 PFU/
150-mm plate, and DNA was prepared from the plate lysate
W092t20796 - PCT/US92/04146
21031~1
-41-
according to Sambrook, J. et _1., _olecul_r_Clo_in~. _A
L_borator~ _anual, 2nd ed., Cold Spring Harbor Labora-
tory, Cold Spring Harbor, NY, 1989.
EXAMPLE_4 Isolation of Human_Cyclin D2 an D3 cDNAs
05 To isolate human cyclin D2 and D3 cDNAs, two 5'
oligonucleotides and one 3' degenerate oligonucleotide
were deri~ed from three highly conserved regions of human
CCNDl, mouse cyll, cyl2, and cyl3 D-type cyclins (Matsu-
shime, H. et al., Cell 65:701-713 (1991); Xiong, Y.
et_al., Cell 65:691-699; Fig. 8). The first 5' oligo-
nucleotide primer, HCNDll, is a 8192-fold degenerate
38-mer (TGGATG[T/C]TNGA[A/G]GTNTG[T/C]GA[A/C]GA[A/G~CA-
lA/GlAAlA/G~TGlT/C)GAlA/G]~A)(SEQ ID No. 37), encoding 13
amino acids (UMLEVCEEQKCEE)(SEQ ID No. 33). The second
5' oligonucleotide primer, HCN~12, is a 8192-fold degen-
erate 29-~er (GTNTT[T/C~CCN[T/C~TNGCNATGAA[T/C]TA[T/C]- ;
TNGA)(SEQ ID No. 39), encoding 10 amino acids
(VFPLAMNYLD)(SEQ ID No. 40). The 3' primer, HCND13, is a
3072~fold degenerate 24-mer ([A/GlTCNGT[A/G]TA~A/G/T]AT-
[A/G]CANA[A/G][T/C]TT-[T/C]TC)(SEQ ID No. 41), encoding 8
amino acids (EKLCIYTD)~SEQ ID No. 42). The PCR reactions
were carried out for 30 cycles at 94~C for 1 min, 48C
for 1 min, and 72C for 1 min. The reactions contained
50 ~M KCl, 10 mM Tris-HCl(pH 8.3), 1~5 mM MgC12, 0.01~
gelatin, 0.2 mM each of dATP, dGTP, dCTP, and dTTP, 2.5
units of Taq polymerase, 5 ~M of oligonucleotide, and
2-10 ~g of template DNA. PCR products generated by
HCNDll and HCND13 were verified in a second-round PCT
reaction using HCND12 and HCND13 as the primers. After
resolution on a 1.2% agarose gel, DNA fragments ~ith the
. ` ' ! !: ! ' '.( ' ' !; .,` .
W092/20796 PCT/US92tO414
~ J
-42-
2103161
expec~ed size (200 bp between primer HCNDll and HCND13)
were purified and subcloned into the SmaI site of phagmid
vector pUC118 for sequencing.
To isolate full-length cyclin D3 cDNA, the 201-bp
05 fragment of the D3 PCR product was labeled with oligo-
nucleotide primers HCNDll and HCND13 using a random-
primed labeling technique (Feinberg, A. P. et _1., A__l.
Biochem. 132:6-13 (1983)) and used to screen a human HeLa
________ ___
cell cDNA library. The probe used to screen the human
genomic library for the CCND3 gene was a 2-kb EcoRI
fragment derived from cDNA clone ~D3-H34. All hybridi-
~, zations for the screen of human cyclin D3 were carriedout at high str~ngency. The PCR clones corresponding to CCNDl and CCND3 have
been repeatedly isolated from both cDNA libraries; CCND2
has not. To isolate cyclin D2, a l-kb EcoRI fragment
derived fro~ mouse c~12 cDNA was used as a probe to
screen a human genomic library. Under low-strin&ency
conditions, this probe hybridized to both human cyclins
Dl and D2. The cyclin Dl elones were eliminated through
another hybridization with a human cyclin D1 probe at ~;
high stringency. Human CCND2 genomic clones were sub-
sequently identified by partial sequencing and by com-
paring the predicted protein sequence with ~hat of human
cyclins Dl and D3 as well as mouse cyl2.
As described above, human CCNDl (cyclin Dl) was
isolated by rescuing a triple Cl_ deficiency mutant of
Saccharomyces cerevisiae using a genetic complementation
_________ ___ __________
screen. Evolutionary proximity between hu~an and mouse,
and the high sequence similarity among cyll, cyl2, and
cyl3, suggested the existence of two additional D-type
W092/20796 PCT/US92/~146
2103161
-43-
cyclin genes in the human genome. The PCR technique was
first used to isolate the putative human cyclin D2 and D3
genes. Three degenerate oligonucleotide primers were
deri~ed from highly conserved regions of human CCNDl,
05 mouse c~ll, c~12, and cyl3. Using these primers, cyclin
Dl and a 200-bp DNA fragment that appeared to be the
human homolog of mouse _yl3 from both human HeLa cell and
glioblastoma cell cDNA libraries was isolated. A human
HeLa cell cDNA library was screened wlth this PCR product
as probe to obtain a full-length D3 clone. Some 1.2
million cDNA clones were screened, and six positives were
~' obtained. The longest cDNA clone from this screen,
AD3-H34 (196~ bp), was completely sequenced ~Figure 4).
Because a putative human cyclin D2 cD~A was not
detected by PCR, mouse cyl2 cDNA was used as a heterolo-
gous probe to screen a human c~NA library at low str~n-
gency. This resulted, initially, in isolation of 10
clones from the HeLa cell cDNA library, but all corres-
ponded to the human cyclin D1 gene on the basis of
restriction mapping. Presumably, this was because cyclin
D2 in HeLa cells is expressed at very low levels. Thus,
the same probe was used to screen a human genomic
library, based on the assumption that the representation
of Dl and D2 should be approximately equal. Of the 18
positives obtained, 10 corresponded to human cyclin Dl
and 8 appeared to contain human cyclin D2 sequences (see
bel~w). A 0.4-kb BamHI restriction fragment derived from
AD2-G1 1 of the 8 putative cyclin D2 clones, was ~hen
used as probe to screen a human hippocampus cDNA library
at high stringency to search for a full-length cDNA clone
of the cyclin D2 gene. Nine positives were obtained
wo 92~20796 - P~r/us92to4t46
210'~161
after screening of approximately 1 million cDNA clones.
The longest cDNA clone, ~D2-P3 (19ll bp), was completely
sequenced (Figure 3). Neither ~D2-P3 nor ~D3-H34 con-
tains a poly(A) sequence, suggesting that part of the 3'
05 untranslated region might be missing.
The DNA sequence of AD2-P3 revealed an open reading
frame that could encode a 289-amino-acid protein with a
33,045-Da calculated molecular weight. A similar analy-
sis of ~D3-H34 revealed a 292-amino-acid open reading
frame encoding a protein with a 32,482-Da calculated
molecular weight. As in the case of human cyclin Dl,
there is neither methionine nor stop codons 5' to the
presumptive initiating methionine codon for both ~D2-P3 ~,
(nucleotide position 22, Figure 3) and ~D3-H34 (nucleo-
t$de position 101, Figure 4). On the basis of the
protein sequence comparison with human cyclin Dl and
mouse c~ll (Figure 7) and preliminary results of the
RNase protection experiment, both ~D2-P3 and ~D3-H34 are
believed to contain full-length coding regions.
The protein sequence of all ll mAmmalian cyclins
identified to date were compared to assess their struc-
tural and evolutionary relationships. This includes `~
cyclin A, cyclins Bl and B2, 5iX D-type cyclins (three
from human and three from mouse), and the recently
identified cyclins E and C (Figure 7). Several features
concerning D-type cyclins can be seen from this compari-
son. First, as noted pre~iously for cyclin D1, all three
cyclin D genes encode a similar small size protein
ranging from 289 to 295 amino acid residues, the shortest
cyclins found so far. Second, they all lack the so-
called "destruction box" identified in the N-terminus of
W092/20796 PCT/US92/~146
~1i)3161
-45-
both A- and B-type cyclins, which targets it for ubi-
quitin-dependent degradation (Glotzer, M. et al., Nature
349:132-138 (1991)). This sug~ests either that the
D-type cyclins have evolved a different mechanism to
05 govern their periodic degradation during each cell cycle
or that they do not undergo such destruction. Third, the
three human cyclin D genes share ~ery high similarity
over their entire coding re~ion:60~ between Dl and D2,
60~ between D2 and D3, snd 52~ between Dl and D3.
Fourth, members of the D-type cyclins are more closely
related to each other tha~ are members of the B-type
cyclins, averaging 78% for three cyclin D genes in the
cyclin box versus 57% for two cyclin B genes. This
suggests that the separation (emergence) of D-type
1~ cyclins occurred after that of cyclin Bl from B2.
Finally, using th~ well-characterized mitotic B-type
cyclin as an index, the most closely related genes are
cyclin A (average 51~), followed by the E-type (40~),
D-type (2g~), and C-type cyclins (20%).
EXAMPLE_5 Chromosome Localization_of_CCND2_and
CCND3
_____
The chromosome localization of CCND2 and CCND3 was
determined by fluoresoence in situ hybridization.
Chromosome i_ situ suppression hybridization and in situ
~5 hybridization banding were performed as described pre-
viously (Lichter, T. et al., Science 247-64-69 (1990);
Baldini, A. et al., Genomics 9:770-774 (1991)). Briefly
__ ___ ________ _
~D2-G4 and ~D3-~9 lambda genomic DNAs containing inserts
of 15 and 16 kb, respectively, were labeled with biotin-
ll-dUTP (Sigma) by nick-translation (Brigatti, D. J. et
W092/20796 - PCT/US92/04146
~,"
46-
2lo3l6l
al., Urolo~y 126:32 50 (1983); Boyle, A. L., In Current
Protocols in Molecular Biolo~y, Wiley, New York, 1991).
Probe size ranged between 200 and 400 nucleotides, and
unincorporated nucleotides were separated from probes
05 using Sephadex G-50 spin columns (Sambrook, J. et al.,
Molecular Clonin~: A Laboratory Manual, 2nd ed., Cold
________________ ______________ _______
Spring Harbor Laboratory, Cold Spring Harbor, NY, 1989).
Metaphase chro~oso~e spreads prepared by the standard
technique (Lichter, T. et al., Science 247:64-69 (1990))
were hybridized in situ with biotin-labeled D2-G4 or
D3-G9. Denaturation and preannealing of 5 ~g of DNase-
treated human placental DNA, 7 ~g of DNased salmon sperm
DNA, and 100 ng of labeled probe were performed before
the cocktail was applied to Alu prehybridized slides.
is ,he in situ hybridization banding pattern used for
__ ____
chromosome identification and visual localization of the ;~
probe was generated by cohybridizing the spreads with 40
ng of an Alu 48-mer oligonucleotide. This Alu oligo was
chemically labeled with digoxigenin-ll-dUTP (Boehringer-
20 Mannheim) and denatured before being applied to denatured
chromosomes. Following 16-18 h of incubation at 37C and
posthybridization wash, slides were incubated with
blocking solution and detection reagent (Lichter, T. et
al., Science 247:64-6g (1990)). Biotin-labeled DNA was
__ _ ___ ____ __ _
25 detected using fluorescence isothiocyanate (FITC)-
conjugated avidin DCS (S ~g/ml) (Vector Laboratories);
digoxigenin-labeled DNA was detected using a rhodamine~
conjugated anti-digoxigenin antibody (Boehringer-
Mannheim). Fluorescence signals were imaged separately
30 using a Zeiss Axioskop-20 epifluorescence microscope
equipped with a cooled CCD camera (Photometrics CH220).
- W092/20796 PCT/US9~/04146
-47- 2103161
Camera control and image acquisition were performed using
an Apple Macintosh IIX computer. The gray scale images
were pseudocolored and merged electronically as described
pre~iously (Baldini, A. et al., Ge_o_ics 9:770-774
- 05 (1991)~. Image processing was done on a.Macintosh IIci
computer using Gene Join Maxpix (software by Tim Rand in
the laboratory of n . Uard, Yale~ to merge FITC and
rhodamine images. Photographs were taken directly from
the computer monitor.
Chromosomal fluorescence in sit_ hybridization was
used to localize D2-G4 and D3-G9. The cytogenetic
location of D2-G4 on chromosome 12p band 13 and that of
D3-G9 on chromosome 6p band 21 were determined by direct
visualization of the two-color fluorescence in situ
hybr~dization using the biotin-labeled probe and the
digoxigen-labeled Alu 48-mer oligonucleotide (Fig. 5).
The Al_ 48-mer R-bands, consistent with the con-
ventional R-banding pattern, were imaged and merged with
images genPrated from the D2-G4 and D3-G9 hybridized
probes. The loci of D2-G4 and D3-G9 were visualized
against the Al_ banding by merging the corresponding FITC
and rhodamine images. This merged image allows the
direct visualization of D2-G4 and D3-G9 on chromosomes 12
and 6, respectively. The D2-G4 probe lies on the posi-
tive R-band 12pl3, while D3-G9 lies on the positive
R-band 6p21.
Cross-hybridization W25 not detected with either
pseudogene cyclin D2 or D3, presumably because the
potentially cross-hybridizing sequence represents only a
sufficiently small proportion of the 15- and 16-kb
genomic fragments (nonsuppressed) used as probe, and the
W092/20796 PCT/US~2/~146
~ 48-
;~1 03161
nucleotide sequences of pseudogenes have diverged from
their ancestral active ~enes. .
EXAMPLE_6 Isolatio_ an_ Characteriza_io_ of Ge_omic
Clones of Human D-Ty~e Cyclins
05 Genomic clones of human D-type cyclins were isolated
and characterized to study the genomic structure and to ~
obtaln probes for chromosomal mapping. The entire 1.3-kb ~;
cyclin Dl cDNA clone was used as probe to screen a normal
human liver genomic library. Five million lambda clones
10 were screened, and three positives were obtained. After
~, initial restriction mapping and hybridizations, lambda ;~
clone G6 was chosen for further analysis. A 1.7-kb BamHI
restriction fragment ~f ~Dl-G6 was subcloned into pUC118
and completely sequenced. Comparison with the cDNA
15 clones pre~iously isolated and RNase protection experi-
ment results (~ithers, D.A. et al., Mol._Cell._Biol.
11:4846 4853 (1991)) indicated that this fragment corres-
ponds to the 5' part of the cyclin Dl gene. As shown in
Figure 8A, it contains 1150 bp of upstream promoter
20 sequence and a 198-bp exon followed by an intron.
Eighteen lambda genomic clones were isolated from a
similar screening using mouse cyl2 cDNA as a probe under
low-stringency hybridization conditions, as described
above (Example 4). Because it was noted in previous cDNA
25 library screening that the mouse cyl2 cDNA probe can
cross-hybridize with the hu~an Dl gene at low stringency,
a dot-blot hybridization at high stringency was carried
out, using the human Dl cDNA probe. Ten of the 18 clones
hybridized with the human Dl probe and 8 did not. On the
30 basis of the restriction digestion analysis, the 8 lambda
W092/20796 - PCT/US92/~146
49 21 03161
clones that did not hybridize with the human Dl probe at
high stringency fall into three classes respresented by
~D2-Gl, ~D2-G2, and ~D2-G4, respectively. These three
lambda clones were subcloned into a pUC plasmid vector,
05 and small restriction fragments containing coding region
were identified by Southern hybridization using a mouse
cyl2 cDNA probe. A 0.4-kb BamHI fragment derived from
~D2-Gl was subsequently used as a probe to screen a human
hippocampus cell cDNA library at high stringency. ~`
10 Detailed restriction mapping and partial sequencing
indicated that ~D2-Gl and ~D2-G2 were two different
clones corresponding to the same gene, whereas ~D2-G4
appeared to correspond to a different gene. A 2.7-kb
SacI-S~aI fragment from ~D2-G4 and 1.5-kb BclI-B~lII
15 fragment from ~D2-Gl have been completely sequenced.
Nucleotide sequence comparison revealed that the clone
~D2-G4 corresponds ~o the D2 cD~A clone ~D2-P3 (Figure
3). As shown in Figure 8A, the 2.7-kb SacI-SmaI fragment
contains 1620 bp of sequence 5' to the presumptive
20 initiating methionine codon identified in D2 cDNA (Figure
3~ and a 195-bp exon followed by a 907-bp intervening
sequence.
Lambda genomic clones corresponding to the human
cyclin D3 were isolated from the same genomic library
25 using human D3 cDNA as a probe. Of four million clones
screened, nine were positives. Two classes of clones,
represented by lD3-G4 and ~D3-G9, were distinguished by
restriction digestion analysis. A 2.0-kb Hi_dIII-Sc_I
restriction fragment from ~D3-G5 and a 3.7-kb SacI-
30 Hi_dIII restriction fragment from ~D3-G9 were further
subcloned into a p~C plasmid vector for more detailed
W092/20796 PCT/US92/04146
~103161
restriction mapping and complete sequencing, as they both ;
hybridized to the 5' cyclin D3 cDNA probe. As presented
in Figure 9C, the 3.7-kb fragment from clone G9 contains
1.8 kb of sequence 5' to the presumptive initiating
05 methionine codon identified in D3 cDNA (Figure 4), a
198-bp exon 1, a 684-bp exon 2, and a 870-bp intron. ~-
Comparison of the genomic clones of cyclins Dl, D2,
and D3 revealed that the coding regions of all three
human CCND genes are interrupted at the same position by
10 an intron (indicated by an arrow in Figure 8). This
indicated that the intron occurred be~ore the separation
of cyclin D genes.
EXAMPLE 7 Isolation and Characterizatio_ of T_o
Cyclin D Pseudo~enes
______ ______ ____
The 1.5-kb BclI-B~lII fragment subcloned from clone
~2-Gl has been completely sequenced and compared with
cyclin D2 c~NA clone ~D2-P3. As shown in Figure 10, it
contains three internal stop codons (nucleotide positions
495, 956, and 1310, indicated by asterisks), two frame-
20 shifts (position 1188 and 1291, slash lines), one
insertion, and one deletion. It has also accumulated
many missense nucleotide substitutions, some of which
occurred at the positions that are conserved in all
cyclins. For example, triplet CGT at position 277 to 279
2~ of D2 cDNA (Fi~ure 3) encodes amino acid Arg, which is an
invariant residue in all cyclins ~see Figure 8). A
nucleotide change from C to T at the corresponding
position (nucleotide 731) in clone D2-Gl (Figure 10) gave
rise to a triplet TGT encoding Cys instead of Arg.
30 Sequencing of the 2.0-kb Hi_dIII-ScaI fragment from clone
WOg2/20796 PCT/US~2/04t46
-51~ 3161
~D3-G5 re~ealed a cyclin D3 pseudogene (Figure 11). In
addition to a nonsense mutation (nucleotide position
1265), two frameshifts (position 1210 and 1679), a 15-bp
internal duplication (underlined region from position
05 1361 to 1376?, and many missense mutations, a nucleotide
change from A to G at position 1182 resulted in an amino ;~
acid change from the presumptive initiating methionine
codon ATG to GTG encoding Val. On the basis of these
analyses, we conclude that clones ~D2-Gl and ~D3-G5
10 contain pseudogenes of cyclins D2 and D3, respectively.
E~UIVALENTS
Those skilled in the art will recognize, or be able
to ascertain using no more than routine experimentation,
many equi~alents to the specific embodiments of the
15 invention described herein. Such equi~alents are in-
tended to be encompassed by the following claims.