Note: Descriptions are shown in the official language in which they were submitted.
~Lll'7D9
o.z. 0050/42623
The specific genetic modification of Ashbya qossypii
The present invention relates to a method for thespeci~ic genetic modification of Ashbya gossypii by
homologous recombination, and to Ashbya gossypii strains
modified by this method.
The filamentous hemiascomycete Ashbya gossypii is
of great interest, for example, as riboflavin-producing
microorganism.
A method for specific genetic modification by
homologous recombination for the yeast Saccharomyces
cerevisiae has been described by Struhl et al. tProc.
Natl. Acad. Sci. USA, 76 (1979) 1035-1039) and later
extended by Rothstein (Meth. Enzymology Vol. 194, p.281
et seq.).
Homologous recombination has been observed in
filamentous ascomycetes such as Aspergillus or
Neurospora; howe~er, the proportion of homologous
-~ recombination is often low compared with non-homologous
recombination. In addition, reliable parameters leading
preferentially or almost exclusively to homologous
recombination are not known (Microbiological Reviews, 53
;~ (1989) 148-170).
It is an object of the present in~ention to
provide a~method for the specific genetic modification of
~,: : .
Ashbya gossyp1i by homologous recombination.
We have found that this object is achieved
particularly effectively when Ashbya gossypii is
tran~formed with vectors which contain the DNA intended
for recombination flanked by one or moxe gene regions of
Ash~ya gossypii. For the method according to the
invention, preference is given to using as gene region of
Ashbya gossypii DNA sequences in the region of the gene
locus of the translation elongation factor EF-1~ (TEF
, ~
gene)
The gene of the translation elongation factor EF-
1~ (TEF) of Ashbya gossypii is con~ained in a 4.6 kb
EcoRI-EcoRI fragment and a 6.3 kb BamHI-BamHI fragment
,
2ii ~7~
2 - O.Z. 0050/42623
which can be isolated from Ashbya DNA gene banks by
hybridization with a TEF probe from Saccharomyces
cerevisiae (EM~O J.3, (1984) 3311-3315) (Fig. 12).
The coding part of the TEF gene is located within
the overlapping 2.1 kb EcoRI-BamHI fragment whose
sequence is shown in DNA Sequence Listing SEQ ID NO:l
together with the adjacent sequences.
We have also found that the Ashbya gossypii
strains modified by this method have valuable properties.
By transformation of Ashbya gossypii is meant the
~,
~ transfer of recombinant DNA into Ashbya gossypii nuclei
; or other DNA-bearing organelles by various methods.
, .
P~rticularly suitable transfer methods are protoplast
transformation and~electroporation.
;~ 15 The protoplast transformation is expediently
;~ carried out by preparing protoplasts from the mycelium
using enzymes~such ~as zymolase. The protoplasts are
suspended in an aqueous calcium-containing buffer in a
concentration of from l`to 10 x lOa/ml, preferably from 3
2;0 ~ to 5 x 10~/ml, and incubated with the purified DNA. The
purified DNA is~added in an amount of from 1 to 50 ~g,
preferably from 10 to 25 ~g, per 100 ~l of protoplast
suspension. The~transformed Ashbya gossypii cells are
ncubated in nutrient medium and streaked onto agar
25~ plates.~ When ~ a~ ;selection marker, for example an
antibiotic-resistance gene, is used the successfully
t}ansformed cells ca~be identified particularly easily
by~ growth ~ on antibiotic-containing nutrient media.
Subsequent- clonaI purification via spore isolation is
expedient.
Used as vectors for the transformation are recom-
binant DNA constructs which contain the DNA intended for
recombination ~lanked by one or more gene regions of
Ashbya gossypii. A flanking gene region particularly
suitable for homologous recombination is the TEF gene
region. The DNA intended for recombination is, for
~; example, flanked as represented in the plasmids pAG-102
~:
7 ~ ~
-~ - 3 - o.z. 0050/42623
and pAG-145 (Figs. 4 and 11).
The DNA for recombination can likewise originate
from Ashbya gossypii; however, it is also possible to use
DNA from other organisms, both of prokaryotic and of
eukaryotic origin, or synthetic DNA.
When DNA from Ashbya gossypii is used as DNA for
recombination, it is possible, for example, to place
weakly expressed Ashbya gossypii genes under the control
of a strong promoter (eg. TEF promoter) by integration
which is base-pair accurate, and thus to increase
expression without needing to use heterologous expression
signals.
It is also possible with the method according to
the invention for DNA from other organisms than Ashbya
gossypii, for example from the genetically better defined
yeast Saccharomyces cerevisiae or the bacteria sacillus
subtilis or Escherichia coli, to be introduced
specifically into Ashbya gossypii cells and to replicate
stably and, where appropriate, to be expressed therein.
The DNA for recombination can be coding or non-
coding DNA, and the use of coding DNA is preferred.
` The vectors used in the method according to the
invention can, furthermore, contain other DNA sequences
such as selection markers. Shuttle vectors which both
replicate in bacteria and are used for the transformation
of Ashbya gos~ypii are preferably used. For this purpose
these vector~ usually have a bacterial DNA replication
origin and one or more antibiotic-resistance genes. The
plasmids pAG-102 and pAG-145 or derivatives thereof are
par~icularly preferred as vectors (Figs. 4 and 11). By
derivatives are meant those plasmids which contain other
DNA sequences flanked by A. gossypii DN~ in addition to
or in place of the G418-resistance-gene.
When circular DNA is used as vector, this is
linearized, before the transformation of Ashbya gossypii,
so as to result in fragments which ha~e a terminal A.
gossypii sequence and which initiate the homologous
- 4 - o.z. 0050/42623
recombination.
The Ashbya gossypii strains which have been
specifically modified by this method can be identified by
the fact that integration of vector DNA h~s taken place
at the predetermined sites of the homology region (eg.
TEF gene locus). This can be detected, for example, by
Southern blotting with a vector probe. It is particularly
straightforward to isolate modified Ashbya gossypii
strains when the vector DNA contains a selection marker.
It is then possible to isolate exclusively the modified
~; strains under selective conditions, for example by
:~ antibio~ic selection.
The strains genetically modified by homologous
recombination with ~he plasmid pAG-102 are called LU8334
to LU8341. These strains have been deposited at the
: Deutsche Sammlung von Mikroorganismen und Zellkulturen
GmbH, Braunschweig, under the following numbers:
U 8334: DSM 6661
` : LU 8335: DSM 6662
LU 8336: DSM 6663
:; :LU 8337: DSM 6664
LU 8338: DSM 6665
LU 8339: DSM 6666
: L~ 8340: DSM 6667
:` 25 :~ ~U:8341: DSM 6668
: The method according to the invention makes it
: possible for specific genetic modifications, such as
insertions, deletions and substitutions which are base-
: pair accurate, to be effected straightforwardly and
genetically stably~or unstably as desired. This means
that the hemiascomycete Ashbya gossypii is available for
: methods of génetic manipulation which aim at, for
example, genetically stable overexpression through
homologous promoters to an extent similar to the single-
cell yeast S. cerevisiae.
: The following Examples illustrate the invention.
..
- 5 - o.Z. 0050/42623
EXAMPLE 1
Isolation of the Ashbya gossypii TEF gene region
DNA isolated from A.gossypii mycelium was cut
with the restriction endonucleases EcoRI and BamHI. DNA
fragments which harbor the TEF gene or parts thereof were
identified after separation of the restriction fragments
according to size in an agarose gel electrophoresis and
subsequent hybridization with a 32P-labeled heterologous
TEF gene probe. The TEF gene probe comprises nucleotides
363 to 1235 of the 1377 bp-long open reading frame of the
S. cerevisiae TEF2 gene (Schirmaier and Philippsen, EMBO
J. 3 (1984), 3311-3315). A 4.6 kb-long EcoRI fragment and
a 6.3 kb-long BamHI fragment hybridized with the hetero-
logous TEF gene probe. Fragments with lengths in this
range were eluted from agarose gels, cloned into the
vector pUC8 (Vieira and Messing, Gene 19 (1982), 259-268)
which has been cut with EcoRI and BamHI, and transformed
into E.coli. The clones with TEF DNA were identified by
colony hybridization (Grunstein and Hogness, Proc. Natl.
Acad. Sci. USA 72 (1975), 3961-3~65) using the 32P-labeled
heterologous probe. The positive clones contained either
the 4.6 kb-long EcoRI fragment or the 6.3 kb-long BamHI
fragment. The two clones overlap in a region of 2.1 kb
which harbors the homology with the TEF gene probe and
which was sequenced. The sequence of the 2.i kb fragment
and adjacent regions is ~ontained in SEQ ID NO: 1.
SEQ ID NO: 1 contains the open reading frame of 1377 bp
(SEQ ID NO: 2), 436 bp of the 5' non-coding region and
686 bp of the 3' non-coding region. Subsequently the
promoter region was isolated as 403 bp-long HindIII/
HincII fragment which, besides the 379 bp in front of the
start codon, also harbors the first 24 bp of the open
reading frame of the TEF gene, and was employed for
pAG100 constructions.
3S EXAMPLE 2
Plasmid constructions
a) The vector pAG-1 (Fig. 1) (deposited as DSM 6010),
~I~i7a~
- 6 - O.Z. 0~50/42623
a derivative of the vector pEX4, was prepared as
described by Ernst and Chan, J.Bacteriol. 163
tl985), 8-14. pAG-l contains a 1.7 kb SalI fragment
with the G418-resistance gene (G41~r), which codes
for the aminoglycoside phosphotransferase
(APH(3')I), of the transposon Tn903~ In the original
pEX4 construct, initially the 1695 bp PvuII fragment
of Tn903 (Oka et al., J.Mol.Biol. 147 (1981), 217-
226) was ligated into a plasmid with filled-in SalI
cleavage sites. The SalI cleavage sites were
retain~d during this, and the resistance gene can be
isolated as 1.7 kb SalI fragment. pAG-l contains the
Saccharomyces cerevisiae ARS elements ARSl and 2~
ARS and undergoes autonomous replication in Ashbya
gossypii.
b) pAG-2 ~Fig. 2). The 1.7 kb SalI fragment with the
G418-resistance gene was cut out of pAG-l and
~; inserted into the SalI cleavage site of the SO
cere~isiae-E. coli shuttle vector YEp24 (Botstein et
al., Gene 8 (1979), 17-24; New England Biolabs Inc.,
Beverly, M~, USA, 1988-1989 Catalog, 112-113). The
structure of the new plasmid - pAG-2 - was checked
by restriction endonuclease mapping, using the XhoI
~leavage site located in the 1.7 kb SalI fragment to
check the insert orientation. pAG-2 contains the
Saccharomyces cerevisiae ARS element 2~ ARS and
undergoes autonomous replication in Ashbya gossypii
(Wright and Philippsen, Gene 109 (1991), 99-105).
;, c) pAG-100 (Fig. 3). A 403 bp long HindIII/HincII
fragment which contains the promoter region and the
first 24 bp of the open reading frame of the gene
for the translation elongation factor EF-l~ (TEF
gene) from A.gossypii was inserted into the XhoI
cleavage site of pAG-2, which is located 30 bp in
the 3' direction behind the start of translation of
,
- 7 - o.z. ooso/42623
the G418-resistance gene, after the protruding ends
had been filled in. The orientation of the fragment
in the resulting plasmid pAG-100 was checked by
restriction endonuclease mapping with HindIIIO
Insertion of the 403 bp-long fragment resulted in
the 10 N-terminal amino acids of APH(3')I being
replaced by the first 8 amino acids of the
A.gossypii translation elongation factor EF~
Deletion or replacement of the first 19 amino acids
of APH(3')I by other amino acids does not result in
loss of activity (Chen and Fukuhara, Gene 69 (1988),
181-192). pAG-100 contains the Saccharomyces
cerevisiae ~RS element 2~ ARS and undergoes
autonomous replication in Ashbya gossypii.
d) pAG-102 ~Fig. 4). The starting vector is pUC19 in
which the BamHI site has been inactivated by cutt-
ingr filling in and religation. The 4.6 kb EcoRI
fragment of the TEF region was cloned into the EcoRI
site of pUCl9 (arrow = open reading frame of the TEF
gene). Subsequently, the 2.1 kb SalI fragment from
~;~ p~G100 was, after the ends had been filled in,
cloned into the SspI site (thin arrow = open reading
frame of the G418-resistance gene; thick white arrow
= TEF promoter HindI I I -HincII fragment).
;~ .
e) Constxuction of pAG-103 (Fig. 10)
The 6.3 kb BamHI fragment with the TEF gene from A.
gossypii was cloned into the Bam~I cleavage site of
the plasmid pBluescript II SK (Alting-Mees and
Short, Nucl. Acids Res., 17 (1989) 9494). The
orientation of this fragment was checked by
restriction cleavages with EcoRI, HincII and HindIII
(TEF = TEF gene from A. gossypii, white arrow on
black ground = TEF promoter).
,' D 9
- 8 - o.z. 0050/42623
f) Construction of pAG-121
A 669 bp SalI/HindIII fragment which harbors the 3'
end of the G418-resistance gene was isolated from
pAG-100 and inserted into the SalI/HindIII-cut
plasmid pBluescript II SK + (Alting-Mees and Short,
1989).
g) Construction of pAG-122
A 940 bp fragment which harbors the completing 5'
end of the G418-resistance gene fused to the TEF
promoter was inserted into the HindIII cleavage site
of pAG-121. This fragment was obtained by cleavage
of pAG-loO with HindIII. The insertion of this
fragment results in a complete G418-resistance gene.
h) Construction of pAG-145 (Fig. 11)
pAG-103 was partially cleaved with HindIII, and the
linearized plasmid was isolated from an agarose gel.
The protruding ends were converted into blunt ends
by filling in using the Klenow fragment of DNA-
polymerase I, and a 1.64 kb BamHI/SalI-fragment from
pAG-122 with the kanamycin-resistance gene under the
control of the TEF promoter was in~erted after the
protruding ends ~ad been filled in. The position and
orientation of the G418-resistance gene was checked
by ~estriction cleavages (white or shaded arrow on
~ black ground = TEF promoter (HindIII/HincII
fragment), shaded = G418-resistance gene). The
fusion of the filled-in SalI and HindIII cleavage
sites restores HindIII cleavage sites.
EXAMPLE 3
Transformation of A.gossypii with TEF gene region vectors
The transformations were carried out as outlined
below:
a~
- ~ - o.z. 0050/42623
inoculate 200 ml of MA2 with about 1-2x107 spores
incubate in baffle flasks at 27C and 350 rpm for
32-40 h
filter off mycelium with suction and wash lx in
30 ml of H20
determine fresh weight (about 2-3 g)
suspend mycelium in 30 ml of SD and incubate at 30C
in a shaker for 30 min
suspend mycelium in 5-10 ml of SPEZ per g fresh
weight
incubate in a shaking waterbath at 30C, check for
protoplast formation under the microscope (proto
plast formation should have reached more than 90
after 30 min)
filter protoplast suspension through glass ilter
(Schott, porosity 1)
centrifuge filtrate for 5 min (Sorvall SM24 rotor,
1800 rpm)
wash sediment lx in 20 ml of ST and lx in 20 ml of
STC
uspend protoplasts in 20 ml of STC and determine
titer in a counter
after cen~rifugation, resuspend protoplasts at a
density of 4xlO~/ml in STC
add 100 ~l of protoplast suspension to DNA ln a
maximum of 15 ~l of TE and mix (amounts of DNA for
integrative transformat on with linearized TEF gene
region vectors: 15-20 ~g)
incub-ate at room temperature for 15 min
cautiously add l ml of PTC40 and mix by inversion
centrifuge for 5 min (Heraeus Biofuge A, 1500 rpm)
carefully remove supernatant, and suspend sediment
in l ml of SMTCI
incubate at 27~C for 3 h, mix by inversion about
every 45 min
after centrifugation, suspend sediments in 1 ml of SM
mix suspension with 9 ml of SMA2 top layer and add
~ i L i 7 3 ~
- 10 - O. Z . 0050/42623
~o SMA2 plate (20 ml of SMA2 agar per plate)
- incubate plates at 27C for 18 h
- cover plates with G418 (0.54 ml of G418 stock
solution + 0.46 ml of H2O + 6 ml of 0.5~ agarose (in
H2O, preheated to 42C))
- incubate plates further at 27C, transformants are
visible after 3-6 days.
Media and solutions
Media: MA2: peptone (Gibco casein
hydrolysate, No. 140) : 10 g/l
yeast extract (Gibco) : 1 g/1
glucose 10 g/1
myo-inositol 0.3 g/1
SMA2 agar: sorbitol : 1 M
peptone : 10 g/l
yeast extract : 1 g/l
glucose 20 g/l
myo-inositol 0.3 g/l
agar (Gibco) 12 g/l
: 20 SMA2 top layer: as SMA2 agar with 0.8% agarose in
: place of agar
Solutions: SD: lM sorbitol; 50 mM dithiothreitol
:~ SPEZ: lM sorbitol; 10 mM Na phosphate buffer
pH 5.8; 10 mM EDTA; 2 mg/ml Zymolyase
~:~ 25 20 T (Seikagaku Kogyo Co., Tokyo)
ST: lM sorbitol; 10 mM tris-HC1 pH 8
STC: lM sorbitol; 10 mM tris-HCl pH 8;
10 mM CaCl2
TE: 10 mM tris-HCl; 1 mM EDTA
. EDTA: ethylenediaminetetraacetic acid
Tris: tris(hydroxyethyl)aminomethane
SDS: sodium lauryl sulfate
ddH2O: double-distilled water
TBE: 100 mM tris, 100 mM boric acid, 2 mM
EDTA, pH = 8.0
pTC40: 40% (w/v) polyethylene glycol 4000
(Merck); 10 mM tris-Cl pH 8; 10 mM
ù 9
~ o z. 0050/42623
CaCl2
SMTCI: 50~ SM (see below); 50% STCi 0.03 g/l
myo-inositol
SM: 50~ 2M sorbitol; 50% MA2
S G418 stock solution: 20 mg/ml G418 (Geneticin,
Gibco) in H2O
EXAMPLE 4
Transformation of A. gossypii with pAG-102
The plasmid pAG-102 (Figure 4) contains a 4.6 kb
EcoRI fragment from the Ashbya gossypii TEF region (TEF
= gene of the translation elongation factor EF-1~. The
plasmid also harbors a G418-resistance gene under the
control of the TEF promoter, which confers resistance to
the aminoglycoside G418 on Ashbya gossypii transformants.
The plasmid has no signals for autonomous replication in
Ashbya gossypii. pAG-102 DNA was cut within the TEF
homolo~y region (3' end of the TEF gene) at the BamHI
ite and used for the transformation of Ashbya gossypii
protoplasts as in Example 3. This allows homologous
recombination 3' of the TEF gene to be induced. The eight
independently obtained G418-resistant transformants
(hU8334-LU8341, deposited at the DSM, Braunschweig)
retained their G418 resistance after clonal purification
and without selection pressure. The pAG-102 DN~ had been
~tably integrated into the Ashbya gossypii genome.
Figure 5a shows a chromosome fractionation of t~e
eight transformants and of the wild-type strain (middle
lane). The TEF gene is located on the largest of the five
sible chromosomes, and the plasmid had also been
integrated into this in all eight cases (Fig. 5b).
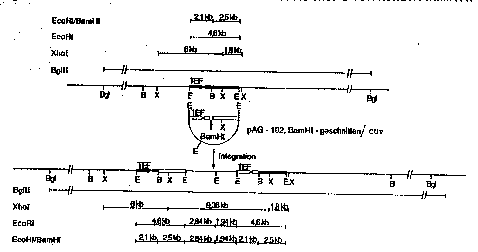
These integrations took place exclusively at the
TEF locus, since the BglII fragment which contains the
TEF locus is about 10 kb longer in all eight transform-
ants than in the wild-type BglII fra~ment (Fig. 6).
Homologous recombination ought, as shown in Figure 7,
result in a precise doubling of the 4.6 kb TEF region.
This is the case as is shown by analysis of the
~ t ~ V 3
- 12 - o.z. 0050/42623
transformants and wild-type DNA after cleavage with BamHI
and EcoRI (Fig. 8a,b).
To check, DNA frayments which con~ain the novel
joins between chromosomal and plasmid DNA were cloned
from three of the eight transformants. Sequence analysis
in the region of the BamHI sites showed no change from
the wild~type DNA.
After storage at -70 for one year, the eight
transformants were reanalyzed after renewed mycelial
1~ growth and sporulation. This revealed evîdence of
amplification since three of the analyzed clones harbored
two copies of the plasmid, and two in fact haxbored three
copies (Fig. 9), in some cases with sequence overlaps.
One of these overlaps probably led to tandem duplication
lS of the G418-resistance gene. The reason for these
overlaps is perhaps the presence of two TEF promoter
regions per copy of the plasmid ~see Fig. 4).
EXAMPLE 5
Transformation of A. gossypii with pAG-145
A. gossypii protoplasts were transformed with
BamHI-cut plasmid pAG-14S. This makes it po~sible
simultaneously to induce homologous recombination 5' and
3' from the TEF gene. Four transformants were subjected
to clonal purification and investigated by Southern
analysis for integration of the kanamycin-resistance
gene. The analyses of EcoRI-cleaved DNA were carried out
wi~h two clonally purified strains of each ~ransformant,
and the analyses of BglII-cleaved DNA were carried out
with one ~lonally purified strain of each transformant.
The results of -these analyses are shown in Fig. 13 and
Fig. 14, and interpretation of the data based on the
model is carried out in Fig. 15.
All the transformants show two bands in the BglII
Southern: the larger band with a length of about 18.6 kb
does not occur in the untransformed wild type. It
corresponds to the TEF locus after integration of the
G418-resistance gene (see Fig. 15b). The smaller band
~, - 13 - o.z. 0050/42623
with a length of about 17 kb also occurs in the wild type
and corresponds to the TEF locus without integrated G418-
resistance gene. It can be formed either by a wild-type
copy of the TEF locus (see Fig. 15a), or by a secondary
event after the integration, namely a deletion of the
G418-resistance gene by homologous recombination between
the two TEF promoter copies (see Fig. 15c).
This would be advantageous, for example, if the
intention is to delete an integrated heterologous marker
after integration of a sequence signal or of a gene.
The analysis with EcoRI-cleaved DNA was carried
out in order to investigate this. The TEF promoter
fragment of the Ç418-resistance gene is preceded by an
additional EcoRI cleavage site which derives from the
lS polylinker of pAG-122 and therefore it does not occur in
front of the TEF promoter fragment of the TEF gene. If
the 17 kb BglII fra~ment in the transformants had been
; produced by integration and subsequent deletion, the
a~ditional EcoRI cleavage site ought to be detectable
- 20 because it is located in front of the duplicated region
and ough~ therefore not to be affected by the deletion
(see Fig. lSc). In this case, the transformants should no
~onger have the 1.55 kb EcoRI fragment of the wild-type
locus but, instead, a 1.32 kb fragment and a 0.27 kb
fragment. The absence of the 1.55 kb fragment and the
pre~ence of the 1.32 kb fragment were shown for
transformants 1, 3, 4 and 5 ~Fig. 14).
A stability test was carried out with the
clonally purified transformants 1 to 5. Mycelium was
incubated on non-selective medium for six days and
subsequently mycelium from the edge of each colony was
transferred to a new plate of non-selective medium and to
a plate containing selective medium. Ater incubation on
non-selective medium for a further three days, mycelium
was again transferred to new plates. After six days of
non-selective growth, all the transformants were still
G418-resistant. After nine days, transformants 1, 2 and
ïd~
- 14 - O.Z. 0050/42623
3 had lost their resistance to G418. The Southern data
indicate that the loss is caused by a homologous
recombination between the TEF promoter fragments. The
relatively high frequency of deletion of the heterologous
marker (G418-resistance gene) observed with this
construction is in some circumstances ad~-antageous for
cotransformation.
a) Cultivation of Ashbya gossypii
Cultivation in a liquid culture or on a plate is
achieved by inoculation with mycelium or spores.
Medium: 1 liter MA-2: 1 g peptone
1 g yeast extract
10 g glucose
12 g agar
Selective medium: MA-2-G418: MA-2 ~ 200 mg G418/ml
b) DNA isolation
1. Make up liquid culture in MA-2 medium in a
baffle flask. 200 ml of medium produce about
1.5 g of mycelium. Incubation at 27C for 48 h.
2. Wash mycelium: filter off mycelium with suc-
tion
take up mycelium in 30 ml of
ddH2O
repeat 2-3 times
25 ~ 3. Then take up the mycelium (about 1.5 g) in
30 ml of SCE buffer
SCE: 1 M sorbitol
0.1 M sodium citrate
. 60 mM EDTA pH 8
4. Addition of 30 ml of 2-mercaptoethanol (14 M)
, Addition of 5 mg of zymolase per 1.5 g of
mycelium
~;~ 5. Incubation at 37C for 1 h
6. Check protoplast formation under the microscope
and, if necessary, prolong incubation time
7. Addition of 3 ml of 0.5 M EDTA pH 7.5
Addition of 3 ml of 10% SDS
~11i 7~3
- 15 - o.z. 0050/42623
8. Incubation at room temperature for 5 min
9. Addition of 5 mg of proteinase K per 1.5 g of
mycelium
10. Incubation at 37C for 1 h
11. Determination of the volume
Addition of the same volume of 5M ammonium
acetate solution
12. Centrifugation at 15,000 rpm for 15 min
13. Discard pellet
14. Ethanol precipitation with 2.5 times the volume
15. Centrifugation at 10,000 rpm for 10 min
16. Wash pellet in 70% ethanol, dry and resuspend
in 4 ml of TE 10:1
17. Addition of 0.5 mg of RNAse
18. Incubation at 37C for 1 h
19. Phenol extraction
20. Phenol/chloroform extraction
21. Determination of the volume
22. DNA precipitation with the ~ame volume of 5M
ammonium acetate solution and 2.5 time3 the
~: volume of ethanol
: 23. Resuspend DNA in 1 ml of TE 10:1.
c) DN~ cleavage for Southern analyses
About 1 ~g of DNA were cleaved per mixture.
: 25 EcoRI/BamHI double digestion:
: DNA 20 ~1
, ~
10 x MS buffer 20 ~1
ddH2O 159 ~1 -
BamHI 10 units
200 ~1
Incubation at 37C overnight
Addition of 10 ~1 of AS buffer
Addition of 10 units EcoRI
Incubation at 37C overnight
BglII digestion:
DN~ 20 ~1
10 x MS buffer 20 ~1
6 - o.z. 0050/42623
ddH2O 159 ~1
BglII 10 units
200 ~l
Incubation at 37C overnight
Subsequently:
DNA precipitation with the same volume of 5M ammonium
acetate solution and 2.5 times the volume of ethanol
Centrifugation at 15,000 rpm for 15 min
Wash pellet in 70~ ethanol
Dry pellet and resuspend in 2Q ~l of TE 10:1.
Buffer: lxMS buffer: 50 mM NaCl
10 mM tris-HCl pH 7.5
- ' 10 mM MgCl 2
~; 1 mM dithiothreitol
AS buffer: 500 mM NaCl
400 mM tris-HCl pH 7.5
: Running conditions:
For Eco/Bam digestion: 0.8~ agarose gel
3 V/cm
8 h running time
For BylII digestion: 0.6~ agarose gel
: 1 V/cm
66 h running time
The gel obtained in this way were transferred to
25 ~ Hybond N membranes by Southern transfer and baked at
80C for 2 h. Prehyb~idization, h~bridiza~ion and
subsequent detection were carried out non-radioac-
: ti~ely under stringent conditions ('Non-radioactive
Label~ng and Detection' Applications Manual, Boehrin-
~ ger Mannheiml Order Number: 1093 657).
~, d) OFAGE preparation
1. Determine fresh weight of the mycelium by filter-
: ing with suction an MA-2 liquid culture
2. Resuspend 0.5 g of mycelium in 2 ml of 50 mM EDTA
pH 7.5
~; 3. Addition of buffer I
: 4. Cautious addition of 5 ml of low melting agarose
~ iii7 ~3
7 - o.z. 0050/42623
(40C)
5. Place mixture in a Petri dish t5 cm diameter) and
leave to solidify
6. Co~er with buffer II
7. Incubation at 37C overnight
8. Remove buffer II by aspiration and add buffer III
9. Incubation at 55C overnight
10. For preservation of the preparation, remove
buffer III by aspiration and replace by 5 ml of
0.5 M EDTA pH 8.5, and store Petri dish at 4C.
Buffers: I: 3 mg of Zymolyase
320 ~1 of 0.5M dithiothreitol
0.9 ml of SCE
4.5 ml of 0.5M EDTA pH 8.5
50 ~l of lM tris-HCl pH 8
200 ~l of 0.5M dithiothreitol
250 ~l of ddH20
III: 5 mg of protein~se K
50 mg of N-lauroyl sarcosinate
4.5 ml of 0.5M EDTA pH 8
50 ~l of lM tris:HCl pH 8
450 ~l of ddH20
: Low melting agarose: 1~ in 0.125 M EDTA pH 7.5
e)~ OFAGE run (apparatus: Rotaphor , Biometra)
: 1. Cut small block out of OFAGE preparation
: 2. Wash 3 times for 1 h at room temperature in 5 ml
~: of 50 mM EDT~ pH 7.5
3. Place blocks in the pocket~ in the gel
; Gel: 1~ agarose gel in 0.4x TBE
4. Running conditions: 20 h running time
~, 48 ~ pulse duration
- 4-9C buffer temperature
0.4x TBE running buffer
300 V voltage
80-90 mA current
: Legends for Figures 5 to 9 and 12 to 14
Fig. 5 a: Chromosome separation wi~h OFAGE.
a 3
18 - O.Z. 0050/42623
From left to right: LU8334, LU8335, LU8336,
LU8337, ATCC 10985, LU8338, LU8339, LU8340,
LU8341.
b: Hybridization of the separated chromosomes
with a pUCl9 DNA probe
Fig. 6 Fractionation of BglII-cleaved DNA of the
strains (from left to right) LU8334, LU8335,
LU8335, LU8337, ATCC 10985, LU8338, LU8339,
LU8340, ~U834~ and hybridization with a pAG-102
DNA probe
: Fig. 7 Diagram of the integration of the plasmid
~: p~G-102 by homologous recombination. Chromosom-
al DNA is indicated by thick lines, and the
~: plasmid DNA by thin or double lines.
Arrows show the position of the TEF reading
frame. B=BamHI, E=EcoRI, Bgl=BglII
Fig. 8 a: Fractionation of BamHI plus EcoRI-cleaved
pAG-102 and Ashbya gossypii DNA, and HindIII-
and EcoRI-cut A DNA. From left to right: A,
pAG-102, ATCC 10895, LU8334 to LU8341, A.
b: Hybridization with a pAG-102 DNA probe.
: c: Und~rexpo~ure of the pAG-102 lane.
Fig. 9: Analysis of BglII-cleaved DNA of the pAG-102
transformants 1 to 8 after double clonal
~ purification
DNA of pAG-102 transformants after double
clonal purification and of the untransformed
; wild-type was cleaved with BglII, fractionated
~:; on a 0.4% agarose g~.l and hybridized in a
. Southern experiment with radio-labeled pAG-102
DNA. The fragment about 17 kb in size which
corresponds to the wild-type copy occurred in
none of the transformants. The bands which are
present instead indicate single integration
: 35 into transformants 2, 3 and 6 and double and
~ triple integrations into transformants 1, 4, 5,
: 7 and 8, which is confirmed by further
J
. ~ - 19 - o.z. 0050/42623
cleavages. Transformants 1 to 8 are derived
from LU8334 to LU8341. By comparison with these
strains, they have been clonally purified one
further time (wt: wild ~ype). The length data
are approximate.
Fig. 12 Comparison of the homologous regions used for
the integration of pAG-102 and pAG-145.
a: Homologous region of pAG-1~2: 4.6 kb EcoRI
fragment
b: Homologous region of pAG-145: 6.3 kb BamHI
fragment
The arrow indicates the point in the genome in
which the foreign DNA is integrated after
homologous recombination.
Fig. 13: Southern with BglII-cleaved DNA from five pAG-
145 transformants
Genomic DNA from five clonally purified
transformants of the plasmid pAG-145 and of the
untransformed initial strain was cleaved wi~h
BglII, fractionated on a 0.4~ agarose gel and
hybridized in a Southern experiment with radio-
labeled pAG-145 D~A. All the transformants
(lanes 1 to 5), show a signal with a length of
abou~ 17 kb which also occurs in the wild-type
DN~ (lane 6~. In addition, all the
~ransformants possess a fragment wi~h a length
of a~out 18.6 kb, which indicates integration
of the G418-resistance gene into the 17 kb
BglII fragment.
Fig. 14: Southern with EcoRI-cleaved DNA from five pAG-
145 transformants
Genomic DNA from in each case two clonally
^?~
.~ - 20 - o.z. 0050/42623
purified strains of five transformants of the
plasmid pAG-145 and of the untransformed
initial strain was cleaved with EcoRI,
fractionated on a 0.8~ agarose gel and
hybridized in a Southern experiment with radio-
labeled pAG-103 DNA. Transformants 1 (lanes 1,
2)~ 3 (lanes 5, 6), 4 (lanes 7, 8) and 5 (lanes
9, 10) no longer have the 1.55 kb fragment
which occurs in the wild-type DNA (11) but
instead have a 1.32 kb fragment and, in
addition, a 1.6 kb fragment which corresponds
to the G418-resistance gene. Fragments of this
length also occur in the lane with EcoRI-
cleaved pAG-14S DNA ~lanes 12, 13).
Transformant 2 (lanes 3, 4) shows the 1.6 kb
fragment (G418-resistance gene) and a 1.55 kb
fragment, which indicates diploidization or a
gene conversion. For this reason, OFAGE
(orthogonal field alternation gel
electrophoresis), was used to investigate
whether deploidization had taken place in
respect of the largest chromosome on which the
homolo~y region is located. No evidence was
found of this. The presence of the 5.15 kb and
~ ~ 4.~ kb fragments and the absence of other
signals prove the homologous recombination in
` ; all the transformants.
Fig. lS: -Model o~ the integration of the ~amHI fragment
from pAG-145 into the genome of A. gos~ypii and
of the subsequent deletion o the G418-
resistance gene by homologous recombination.
Black bars: TEF promoter ~HindIII/HincII
fragment), G418r: G418-resistance gene, TEF: A.
gossypii TEF-gene, Arrows: direction of
transcription.
O.~. 0050/42S23 ~ 9
.
21
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT:
(A) NAME: BASF Aktiengesellschaft
(B) STREET: Carl-Bosch-Strasse 38
(C) CITY: Ludwigshafen
(E) COUNTRY: Bundesrepublik Deutschland
(F) POSTAL CODE (ZIP): D-6700
(G) TELEPHONE: 0621/6048526
(H) TELEFAX: 0621/6043123
(I) TELEX: 1762175170
(ii) TIT~.E OF INVENTION: Verfahren zur gezielten genetischen
Veraenderung von Ashbya gossypii
: (iii) NUMBER OF SEQUENCES: 2
(iv) COMPUTER READABLE FORM:
Not Applicable
(v) CURRENT APPLICATION DATA:
APPLICATION NUMBER:
(2) INFORMATION FOR SEQ ID NO:l:
: ~i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2496 base pairs
(B) TYPE: nucleic acid
(C) STRAN~EDNESS: single
(D) TOPOLOGY: linear
: ~ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Ashbya gossypii
(ix) FEATURE:
(A) NAMEfKEY: CDS
~B) LOCATION: 437..1810
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:l:
:: TGGGCGGCCC GCACTCGTGC CTTCTTCGCA CGGGCCGCCG AACCCCGCCA GGCCATCAAG 60
: CTTGCCTCGT CCCGCCGGGT CACCCGGCCA GCGACATGGA GGCCCAGAAT ACCCTCCTTG 120
ACAGTCTTGA CGTGCGCAGC TCAGGGGCAT GATGTGACTG TCGCCCGTAC ATTTAGCCCA 180
TACATCCCCA TGTATAATCA TTTGCATCCA TACATTTTGA TGGCCGCACG GCGCGAAGCA 240
AAAATTACGG CTCCTCGCTG CAGACCTGCG AGCAGGGAAA CGCTCCCCTC ACAGACGCGT 300
TGAATTCTCC CCACGGCGCG CCCCTGTAGA CAAATATAAA AGGTTAGGAT TTGCCACTGA 360
GGTTCTTCTT TCATATACTT CCTTTTAAAA TCTTGCTAGG ATACAGTTCT CACATCACAT 420
CCGAACATAA ACAAAA ATG GGT AAG GAA AAG ACT CAC GTT AAC GTT GTC 469
Met Gly Lys Glu Lys Thr ~is Val Asn Val Val
1 5 10
GTC ATC GGT CAC GTC GAC TCT GGT AAG TCT ACT ACC ACC GGT CAC TTG 517
Val Ile Gly His Val Asp Ser Gly Lys Ser Thr Thr Thr Gly His Leu
O.Z. 0050/42623
2ill7~3
22
ATC TAC AAG TGT GGT GGT ATT GAC AAG AGA ACC ATC GAG AAG TTC GAG 565
Ile Tyr Lys Cys Gly Gly Xle Asp Lys Arg Thr Ile Glu Lys Phe Glu
30 35 40
AAG GAG GCT GCC GAG TTG GGT AAG GGT TCT TTC AAG TAC GCC TGG GTT 613
Lys Glu P~la Ala Glu Leu Gly Lys Gly Ser Phe Lys ryr Ala Trp Val
45 50 55
TTG GAC AAA TTG AAG GCT GAG AGA GAG AGA GGT ATC ACC ATC GAC ATT 661
Leu Asp Lys Leu Lys Ala Glu Arg Glu Arg Gly Ile Thr Ile Asp Ile
60 65 70 75
GCG TTG TGG AAG TTC GAG ACT CCA AAG TAC CAC GTC ACT GTC ATT GAC 709
Ala Leu Trp Lys Phe Glu Thr Pro Lys Tyr His Val Thr Val Ile Asp
80 85 90
CCC CCA GGC CAC AGA GAC TTC ATC AAG AAC ATG ATT ACC GGT ACT TCT 757
Pro Pro Gly His Arg Asp Ph~ Ile Lys Asn Met Ile Thr Gly Thr Ser
95 100 105
CAA GCT GAC TGT GCC ATC TTG ATC ATT GCT GGT GGT GTC GGT GAG TTC 805
Gln Ala Asp Cys Ala Ile Leu Ile Ile Ala Gly Gly Val Gly Glu Phe
110 115 120
GAG GCT GGT ATC TCC AAG GAC GGT CAG ACC AGA GAG CAC GCT TTG TTG 853
Glu Ala Gly Ile Ser Lys ASp Gly Gln Thr Arg Glu His ~la Leu Leu
125 130 135
GCT TAC ACC TTG GGT GTC AAG CAG TTG ATC GTT GCC ATC AAC AAG ATG 901
Ala Tyr Thr Leu Gly Val Lys Gln Leu Ile Val Ala Ile Asn Lys Met
140 145 150 155
GAC TCC GTC AAG TGG GAC GAG TCC AGA TAC CAG GAG ATT GTC AAG GAG 94g
Asp~ Ser Val Lys Trp Asp Glu Ser Arg Tyr Gln Glu Ile Val Lys Glu
160: 165 170
ACC TCC AAC TTC ATC AAG AAG GTC GGT TAC AAC CCT AAG ACT GTT CCA 997
Thr Ser Asn Phe Ile Lys Lys Val Gly Tyr Asn Pro Lys Thr Val Pro
175 180 185
TTC GTT CCA ATC TCC GGC TGG AAC GGT GAC AAC ATG ATT GAG GCC ACC 1045
Phe Val Pro Ile Ser Gly Trp Asn Gly Asp Asn Met Ile Glu Ala Thr
190 195 200
A~C AAC GCC CC~ TGG TAC AAG GGC TGG GAG AAG GAG ACC AAG GCT GGT 1093
Thr Asn Ala Pro Trp Tyr Lys Gly Trp Glu Lys Glu Thr Lys Ala Gly
205 210 215
GCC GTC AAG GGT AAG ACC TTG TTG GAG GCC ATT GAC GCC ATT GAG CCA 1141
Ala Val Lys Gly Lys Thr Leu Leu Glu Ala Ile Asp Ala Ile Glu Pro
220 225 230 235
CCT GTC AGA CCA ACT GAC AAG GCA TTG AGA TTG CCA TTG CAG GAT GTC 1189
Pro Val Arg Pro Thr Asp Lys Ala Leu Arg Leu Pro Leu Gln Asp Val
240 245 250
TAC AAG ATC GGT GGT ATT GGT ACG GTT CCA GTC GGC AGA GTC GAG ACC 1237
Tyr Lys Ile Gly Gly Ile Gly Thr Val Pro Val Gly Arg Val Glu Thr
255 260 265
GGT GTC ATC AAG CCA GGT ATG GTT GTT ACC TTC GCC CCA TCC GGT GTC 1285
Gly Val Ile Lys Pro Gly Met Val Val Thr Phe Ala Pro Ser Gly Val
270 275 280
O.~. 0~50/4~i623
3~ 3
:: 23
ACC ACT GAA GTC AAG TCC GTC GAG ATG CAC CAC GAG CAA TTG GAG GAG 1333
Thr Thr Glu Val Lys Ser Val Glu Met His His Glu Gln Leu Glu Glu
~85 290 295
GGT GTC CCA GGT GAC AAC GTT GGT TTC AAC GTC AAG AAC GTC TCC GTC 1381
Gly Val Pro Gly Asp Asn Val Gly Phe Asn Val Lys Asn Val Ser Val
300 305 310 315
AAG GAG ATC AGA AGA GGT AAC GTT TGC GGT GAC TCC AAG AAC GAC CCA 142g
Lys Glu Ile Arg Arg Gly Asn Val Cys Gly Asp Ser Lys Asn Asp Pro
320 3~i5 330
CCA AAG GCT GCT GAG TCC TTC AAC GCT ACC GTC ATT GTC TTG AAC CAC 1477
Pro Lys Ala Ala Glu Ser Phe Asn Ala Thr Val Ile Val Leu Asn His
335 340 345
CCA GGT CAA ATC TCT GCC GGT TAC TCT CCA GTC TTG GAC TGT CAC ACT 1525
Pro Gly Gln Ile Ser Ala Gly Tyr Ser Pro Val Leu Asp Cys His Thr
350 355 360
GCC CAC ATT GCT TGT AAG TTC GAC GAG TTG TTG GAG AAG AAC GAC AGA 1573
Ala His Ile Ala Cys Lys Phe Asp Glu Leu 1eu Glu Lys Asn Asp Arg
365 370 375
AGA ACC GGT AAG AAG TTG GAA GAC TCT CCA AAG TTC CTA AAG GCC GGT 1621
Arg Thr Gly Lys Lys Leu Glu Asp Ser Pro Lys Phe Leu Lys Ala Gly
380 385 390 395
GAC GCT GCC ATG GTC AAG TTT GTC CCA TCC AAG CCA ATG TGT GTT GAG 1669
Asp Ala Ala Met Val Lys Phe Val Pro Ser Lys Pro ~et Cys Val Glu
400 405 410
GCT TTC ACC GAC TAC CCA CCA TTG GGT AGA TTC GCT GTC AGA GAC ATG 1717
Ala:Phe Thr Asp Tyr Pro Pro Leu Gly Arg Phe Ala Val Arg Asp Met
15 : 420 425
AGA CAG ACC GTT GCT GTC GGT GTC ATC AAG TCT GTT GTC AAG TCC GAC 1765
Arg Gln Thr Val Ala ~Val Gly Val Ile Lys Ser Val Val Lys Ser Asp
430 : 435 440
AAG GCT GGT AAG GTC ACC AAG GCC GCC CAA AAG GCT GGT AAG AAA 1810
Lys~Ala Gly Lys Val Thr L~s Ala Ala Gln Lys Ala Gly Lys Lys
445~ 450 455
TAGAGTAACT GACAATA~AA AGATTCTTGT TTTCAAGAAC TTGTCATTTG TATAGTTTTT 1870
TTATATTGTA GTTGTTCTAT TTTAATCAAA TGTTAGCGTG ATTTATATTT TTTTTGCCTC 1930
GACATCATCT GCCCAGATGC~GAAGTTAAGT GCGCAGAAAG TAATATCATG CGTCAATCGT 1990
ATGTGAATGC TGGTCGCTAT ACTGCTGTCG ATTCGATACT AACGCCGCCA TCCAGTGTCT 2050
ACCTGTCAAA TTTGCCAGCG TCAAATGCCT CCAGGATAGA ATATGCTCGA CAACTGTTGA 2110
AGTCCATCAA CAAGGATAAC CCATATGCTC TATCGGCGGA GAAAACGTTG CCAGAGCCGC 2170
TTCCTTCCGC AGACGTGCCC CTTCCACTGC TAGATGAGAA GTACGGGGTA GTTAGTGTTT 2230
CCAGGCCTCG TAAATGCCGC AATAAATGCT TCCTTGGGTT CGCTACGCCA TCTCAGGCAG 2290
ACGAGTTTCT ACAAAACTTC AAGGACCGCC TTTTCATATA TGGCCACCAG GTCAATATAG 2350
AGCCAGCGAA GCATGATGCA TTCTGGTATA TTGAACGCGA GGATCCGCGA GCATGCCAAC 2410
GCGTGCGCAA ACACCGAGCG CCAGTTGCTG ATCCAGAAGC CAAGTTGCGT GTTCGTAGCA 2470
ACCGCCGCCT GGCCAGGTTA CGAAGC 2496
(2) INFORMATION FOR SEQ ID NO:2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 458 amino acids
-
O.Z. 0050/4~623
~ ~ ~ i 7 `~ .3
. 24
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SE~UENCE DESCRIPTION: SEQ ID NO:2:
Met Gly Lys Glu Lys Thr His Val Asn Val Val Val Ile Gly His Val
1 5 10 15sp Ser Gly Lys Ser Thr Thr Thr Gly His Leu Ile Tyr Lys Cys Gly
Gly Ile Asp Lys Arg Thr Ile Glu Lys Phe Glu Lys Glu Ala Ala Glu
Leu Gly Lys Gly Ser Phe Lys Tyr Ala Trp Val Leu Asp Lys Leu Lys
Ala Glu Arg Glu Arg Gly Ile Thr Ile Asp Ile Ala Leu Trp Lys Phe
80lu Thr Pro Lys Tyr His Val Thr Val Ile Asp Pro Pro Gly His Arg
95sp Phe Ile Lys Asn Met Ile Thr Gly Thr Ser Gln Ala Asp Cys Ala
100 105 110
Ile Leu Ile Ile Ala Gly Gly Val Gly Glu Phe Glu Ala Gly Ile Ser
llS 120 125
1ys Asp Gly Gln Thr Arg Glu His Ala Leu Leu Ala Tyr Thr Leu Gly
130 135 140
Val Lys Gln Leu Ile Val Ala Ile Asn Lys Met Asp Ser Val Lys Trp
145 150 155 160sp Glu Ser Arg Tyr Gln Glu Ile Val Lys Glu Thr Ser Asn Phe Ile
165 170 175ys Lys Val Gly Tyr Asn Pro Lys Thr Val Pro Phe Val Pro Ile Ser
180 185 190
Gly Trp Asn Gly Asp Asn Met Ile Glu Ala Thr Thr ~sn Ala Pro Trp
195 200 205
Tyr Lys Gly Trp Glu Lys Glu Thr Lys Ala Gly Ala Val Lys Gly Lys
210 ~15 220
Thr Leu Leu Glu Ala Ile Asp Ala Ile Glu Pro Pro Val Arg Pro Thr
225 230 235 240sp Lys Ala Leu Arg Leu Pro Leu Gln Asp Val Tyr Lys Ile Gly Gly
245 250 255'le Gly Thr Val Pro Val Gly Arg Val Glu Thr Gly Val Ile Lys Pro
260 265 270
Gly Met Val Val Thr Phe Ala Pro Ser Gly Val Thr Thr Glu Val Lys
275 280 285
Ser Val Glu Met His His Glu Gln 1eu Glu Glu Gly Val Pro Gly Asp
290 295 300
Asn Val Gly Phe Asn Val Lys Asn Val Ser Val Lys Glu Ile Arg Arg
305 310 315 320ly Asn Val Gys Gly Asp Ser Lys Asn Asp Pro Pro Lys Ala Ala Glu
325 330 335er Phe Asn Ala Thr Val Ile Val Leu Asn His Pro Gly Gln Ile Ser
340 345 350
O . Z . O O S O / 42 62 3 ~ i 1 i 7 a
.. 25
Ala Gly Tyr Ser Pro Val Leu Asp Cys His Thr Ala His I le Ala Cys
355 360 365
Lys Phe Asp Glu Leu Leu Glu Lys Asn Asp Arg Arg Thr Gly Lys Lys
370 375 380
Leu Glu Asp Ser Pro Lys Phe Leu Lys Ala Gly Asp Ala Ala Met Val
385 390 395 400ys Phe Val Pro Ser Lys Pro Met Cys Val Glu Ala Phe Thr Asp Tyr
405 410 415ro Pro Leu Gly Arg Phe Ala Val Arg Asp Met Arg Gln Thr Val Ala
420 425 ~30
Val Gly Val Ile Lys Ser Val Val Lys Ser Asp Lys Ala Gly Lys Val
435 440 445
Thr Lys Ala Ala Gln Lys Ala Gly Lys Lys
450 455
.