Note: Descriptions are shown in the official language in which they were submitted.
WO 93~1213 PCT/US~2/0~661
21i291t,
A~LERGENIC PROTEINS AND PEPTIDES FRO~ JAPANESE CEDAR
POLLEN
S
~ack~round oî the Invention
Genetically predisposed individuals, who make up about 10% of the
population, become hypersensitized (allergic) to antigens from a variety of
environmental sources to which they ar~ exposed. Those an~igens that can induce
Oimmediate and/or delayed Iypes of hypersensi~vity are known as allergens. (King,
T.P., Adv. Imml~nol. ~: 77-105, (1976)). Anaphylaxis or atopy, which includes the
symptoms of hay fever, asthma, and hives, is one form of immcdiate allergy. It can
~e caused by a vanety of atopic allergens, such as products of grasses, ~ees, weeds,
animal dander, insects, food. drugs, and chemicals.
l~e antibodies involved in atopic allergy belong primarily to the lgE
class of immlmoglobulins. IgE binds to mast cells and basophils. Upon
c ombinatio~ of a specifilc allergen with IgE bo~md to mast cells or basophils, the IgE
may be cross~ ed on the cçll surface, resul~ing in ~e physiological ef~ects of IgE-
antigen interachon. These physiological effects include the release of, among other
J.O substanses, histamine, serotonin, heparin, a chemotactis factor for eosinophilic
leukocy~s andlor the leuko~enes, C4, D4, and E4, wbich cause prolonged
c onst~icl:ion of bronchial smooth muscle ~ells (Hood, L.E. et al. ~ nology (2nded.), llle Benjamin/Cumming Publishing (~o., Inc. (1984)). Thcse release,~
~ ~ substances are the me~iators which result in allergiG symp~oms caused by a
!5 : combina~on of I~E with:a specific allergen. Through ~em, the effects ~ an
llergen are manifes~ed. Such effects may be systemic or local in na~, depending
on ~e routc by which the an~en entered ~e body and the pattem of deposi~ion of
IgE on mast cells or basophils. Local manifesta~sns ~enerally o~ur on epithelialsurfaces at the location at which the aller~cn cntered ~e body. Systemic effects can
~30 include anaphylaxis (anaphylactic shock), which is the result of an IgE-basophil
response to circulating (intravascular) antigen.
~: ~ Japanese cedar (Sugi; Cryp~orneria japonica) pollinosis is one of ~he
mos~ important aller~ic diseases in Japan. The nurnber of patients suffering from
this disease is on the increase and in some areas, more than 10% of the population
~YO 93/~213 Pcr/uss2/o~66
'2
?.9~"
are affected. Treatment of Japanese cedar pollinosis by administra~ion of Japanese
cedar pollen extract to effect hyposensitization to the aller~en has been attempted.
Hyposensitization usin~ Japanese cedar pollen extract. however~ has drawbaclcs in
that it can~ elicit anaphylaxis if hi~h doses are used~ whereas when low doses are used
to avoid anaphylaxis, treat nent must be continued for several years to build up a
tolerance for thé extract.
The major aller~en from Japanese cedar pollen has been puri~led and
desi~nated as Sugi basic protein (SBP) or C~ j 1. This protein is reported to be a
basic protein with a molecular weight of 41-50 kDa and a pI of 8.~. There appear to
be multiple isoforms of the allergen, apparently due in part to differential
glycosylation (Yasueda et al. (1983) J. Allerg~ Clin. Immlmol. ?1: 77-86; and Taniai
et al. (198~) F~B3 Lerters ~: 329-332. The sequence of the first twenty amino
acids at d~e N-terminal end of Cry j I and a sixteen ~nino acid internal sequence
have been determined (Taniai supr~).
S A second allergen from Japanese cedar pollen having a molecular
weigh~ of about 37 kDa known as CJ y j II has also been repor~d (Sakaguchi e~ al.
(1990) Allergy 45: 309-~12). This aller~en was found to have no imm~unolo~ical
cross- reacdvity with Cry j I. Most patients widl Japanese cedar pollinosis werefound to have IgE antibodies to both C7y j I and Gy j II, however, sera from some
~0 ~ ~ paaents reac~ed with only C~ j I or Cryj II.
In addition to hyposensitiza~don of Japanese cedar pollinosis patients
with low doses of Japanese cedar pollen extract, U.S. patent 4,93~,239 issued July 3,
1990 to Matsuhashi et al. discloses a hyposensitizaaon agent comprising a~
saccharide covalendy~ lced to a Japanese cedar pollen allergen for
hyposensitization of persons serlsitive to 3apanese cedar pollen. l~is
hy~osensitizadon agent is repor~d to enhance the produc~ion of Ig~3 and IgM
an~ibodies, but reduce production of IgE antibodies which are speci~lc to the aller~en
-- ~ and responsible for anaphylaxis and allergy. The allergens used in the
hyposensitîzation agent preferably have an NH2~ minal amino acid sequenee of
Asp-Asn-Pro-Ile-As~Ser-X-Trp-Arg-Gly-As~Ser-Asn-Trp-Ala-Gln-Asn-Arg-Met-
Lys-, wherein X is Ser, Cys, Thr, or His (SEQ ID NO: 18). Addi~ionally, Usui et àl.
(1990) Int. Arch. Allergy Appl. Irr~rnunol. 91: 74-79 reported that the ability of a Sugi
basic protein (i.e., Cry j I~-pullulan conju~ate to elicit the Ar~us reac~ion wa~
markedly reduced, about 1.000 times lower ~han that of nalive Su~i basic protein and
-
WO 93/01213 Pcrfus92/05661
2112913
suggested that the Sugi basic protein-pullulan conjugate would be a ~ood candidate
for desensi~ization therapy a~ainst cedar pollinosis.
The Cr j I aller~en found in C'nplomeria jap~nica has also been
found to be cross-reactive with aller~ens in the p~llen from other species of trees,
s including Cupressus sempervirens. Panzani et al. (Annals of Allerg~ ~7: 26-3~J
(1986)) reported that cross reactivity was de~ected between aller~ens in the pollens
of Cupressus sempervirens and Cr~ptorneria japonica in s~in testing~ RAST and
RAST inhibition. A 50 kDa allergen isolated from Mountain Cedar (Juniperus
sabinotdes) has the NH2-terminal sequence AspAsnProIleAsp (SEQ ID NO: 25)
~0~ (Gross et al, (1978) Scand. J. Immunol. ~: 437-441) which is the same sequence as
the fi~st five amino acids of the NH-2 terminal end of the Cry j 1 allergen. The Cr~ j
I allergen has also been found to be allergenically cross-reactive with the following
species of trees: G~pressl~s arizonica, Cupressus m~tcrocarpa, Junip~rus virginiana,
Junipe~us cornmlmis, Thuyct orien~alis, and Chamf ecyparis obtusa.
S Despite the attention Japanese cedar pollinosis allergens have
received, defini~on or characteriz~tion of the allergens responsible ~or its adverse
effects on people is far from cs)mplete. Current desensitization therapy involves
trea~nen~ with pollen ~xtract widl its attendaQt risks of anaphylaxîs if high doses of
pollen extract are administered~ or long desensi~iza~ion times when low doses of.0~ pollen cxtract are administered.
,
SummaryQf~heInvenaon
The presen~ invention provides nucleic acid sequences coding for the
ryp~meria japonica ~major po}ler~ allergen Cry j I and fra~ments thereof. The
present invention also p~ovides purified C)y j I and at least one fragment thereof
prcduced in a host c~ll transfonned wi~ a nucleic acid se~uence coding for Cr y j I
or at least one fragment thereof and firagments of C~y j I prepared syndle~cally. As
used herein, a fragment of the nucldc acid sequence coding for the entire amino acid
uence of Gy j I refers to a nucleotide sequence having fewer bases than the
~io ~ : nucleotide sequence coding for the en~ire amino acid sequence of Cry J I and/or
mature Gy j I. Gy j I and fragments thereof are useful for dia~nosing. treating, and
preventing Japanese cedar pollinosis. Tlli5 invention is more par~icul~rly described
in the appended claims arld is described in its preferred embodiments in the
following description.
,
wO 93/Vl~l~ Pcr/us~2/05661
3~
Brief ~ptiQn of th~ Dr~wings
"~
Fign la is a ~raphic represen~uon of affinity purified Cry j I on
Superdex 75 (2.6 by 60 cm) equilibrated with 1() mM sodium acetate (pH 5.()) and0.15 M NaCl;
Fi~. Ib shows an SDS-PAGE (12.5~) analysis of the fractions from
the major peak shvwn in Fig la;
Fig. 2 shows a Wes~rn blot of isoforms of purified native Cry j 1
O proteins separated by SDS-PAGE and probed with mAB CBF2;
Fig. 3 is a graphic representation of allergic sera titration of different
purified frac~ions of purified na~ive Cr~ j I usin~ plasma from a pQOl of fifteen
allergic patients;
Figs. 4a-b show ~e composite nucleic acid sequence from the two
S overlapping clones JC 71.6 and pUC19JC9lA eoding for Cry j I. The complete
cDNA æquenee f~r Gy j I is composed of 1312 nucleotides. including 66
nucleo~ides of ~' un~anslated sequence, an open reading frame starting with the
codon for an ~ni~a~ng methionine, of 1122 nucleo~ides. and a 3' untranslated region.
: Figs. 4a-b also show the deduced amino acid sequence of C yj I;
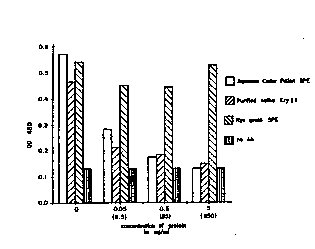
.0 . Fig. 5a is a graphic representa~ion of the results of IgE binding
reactivity wherein ~e coating an~igen is soluble pollen extract ~SPE~ from Japanese
cedar pollen;
~: Fig. Sb is a graphic lepresentahon of the results vf IgE binding
; ~ rea~iYity wherein ~e coating an~gen is purified na~ive Gy; I;
~: : Fig. 6 is ~a graphic representa~on of the results of a compe~i~ion
El,ISA ~vith pooled human pla~ma (PHP) from 15 patients wherein ~e coa~ng
antigen is soluble poll~n extract (SPE) from Japanese cedar pollen;
Fig. 7 is a graphic representaaon of ~he r~sults of a compe~ition
ELISA using plasma from individual patients (indica~d by patient numbers)
wherein ~e coa~ing antigen is soluble pollen extract (SPE) from Japanese cedar
pollen and the competing antigen is punfied native Cry j I;
Fig. 8a is a graphic representation of the results ~rsm a direct binding
ELISA using plasma from seven individual patients (indicated by patient numbers~wherein the coating antigen is soluble pollen ex~ract (SPE~) from Japanese cedar
WC:~ 93/01213 Pcr/us92/05661
21:L29 l3
pollen:
Fi~. 8b is a ~raphic represen~uon of the results ~rom ~ direct bindin~
ELISA using plasma from seven individual pauents (indicated by patient numbers)
wherein the coating an~gen is denatured soluble pollen extract which has been
dena~ured by boiling in the presence of a reducing agent. DlT;
Fig. 9 is a graphic lepresentaion of a direct ELISA where the wells
`: ~ were coated with recombinant C y j I (rCry j I) and IgE binding was assayed on
: individual patients;
Fi~. lOa is a graphic representation of the ~esulss of a capture ELISA
using pooled human plasma from fif~en patients wherein the wells were coated with
CBF2 (IgG) mAb, PBS was used as a negative anugen con~ol. and the antigen was
purified rccombinant Cr~
Fig. lOb is a graphic representation of the results of a capnlre ELISA
using rabbit an~i-Amb al arld II, wherein the wells were coated with 20 llg/ml CBF2
(IgG~, PBS was uscd as a negative antigen con~rol and the antigen was purified
recombinant Gy j I:
Fig. ll is a graphic ~ep~esentauon of a histamine release assay
rmed on one Japanese cedar pollen allcrgic patien~ using SPE from Japanese
; cedar pollen, purified na~ive Gy j I and recombinant Cry j I as the added antigens:
o ~ and ~
hg. 12 Is a graphic represen~on of the ~esults of-a T cell
proliferadon ass~y using blood from pa~ient ~999 wherein the antigen is recombislant
Cry j I protein, punfied na~ive C~y j I protein, or recombin~nt Amb a l . l.
The present inven~ion provides nucleic acid sequences coding for C~
j 1, the major allergen found in lapanese cedar pollen. The nucleic ~cid sequence
codinP for CrY j I preferably has the sequence shown in Fi~s. ~a and 4b (SEQ ID
NO: l). llle nucleic ~cid sequence coding ~or Cr~ j I shown in Figs. 4;1 and ~b
:3~) (SEQ lD NO: 1! cont~ins ~ ~1 ~nino acid leader sequence from b;lse ~6 ~hrough
base 1''8 This le~der se~uence is cleaved ~rom the mature protein which is encoded
- ~ by bases 129 through 1187. The deduced amino acid sequence'o~ Cry j 1 is also
shown in Figs. 4~ and 4b (SEQ ID NO: ''). The nucleic acid sequence of the
invention codes tor a protein havin_ ~ predicted molecul;lr wei~ht of 38.5 kDa. with
WO 93/0121~ pcr/uss2/Q~66l
?.~-~g~3
~.
a pI of 7.8 and five potential N-linked glycosylation sites. Utiliza~ion o~ these
glycosylation sites will increase the rnolecular wei~ht and atfect ~he pI of the mature
pro~in. The deduced amino acid sequence for the mature protein encoded by the
nucleic acid sequence of the invention is identical with the known NH )-terminal ~d
in~ernal amino acid sequences reported by Tanicu et al.. supra. The NH2-terminal
end of C~y J I r~ported by Taniai et al.. supra has the sequence shown in SEQ IDNO: 18. The intemal sequence reported by Taniai et al.. supra has the sequence
~- GluAlaPheAsnValGluAsnGlyAAsnAlaThrProGlnLeuThrLys (SEQ ID NO: 19).There are sequence polymorphisms observed in the nucleic acid sequence of the
O invention. For example. single independent nuleotide subs~tutions at the codons
e~coding amino acids 38~ 5l cmd 74 (GGA vs. G~A. GTG vs. GCG. ~d GGG vs.
GAG, respectively) of SEQ ID #l may result in amino acid polymorphisms (G vs. E,V vs. A, and G vs. E. respecnvçly) at these sites. In add;~tion. a single nucleotide
subsitudon has been detected in one cDNA clone dedved frvm Cryptomena
~15 japonica polleo collected in ~apan. ~his subs~i~ution in the codon for amino acid 60
AT vs. CAT) of SEQ ID #l may result in an amino acid polymolphism (Y vs. H)
at this~site. A~dicional silent nucleoud~ subs~itutions have been de~cted. It isexpected thas ~erc arc additional scquence polymorphisms. and it will be
appreaatcd by one s~ led in the art that one or more nucleotides (up to about 1% of
~0 thc nuclco~idcs) in the nucleic acid sequence coding for Gy j I may vary among
:
i ndividual C~ryptomeria japoni~a plants due to natural allelic variation. ~ny and all
such nucleotide variations and r~sullting amino acid polymorphisms are within ~he
scop~ of ~he invennon. Furtbermore. ther~ may be one or more family members of
ry j 1. Such ~amily mçmbers are defined as proteins related in function and ~nino
~5 acid sequence to Gy~ I bu~ encoded by genes a~ separa~e gcneuc loci.
Fragments of the nucleic acid scquence codin~ for fragments of Cry j
se also within the scope of the inYention. Fr~gments within the scope of the
invention include those coding for parts of C~ j I which induce an immune response
;- in marnmals. preférably humans. such as stimulation of minimal ~nounts of IgE;
31) binding of IgE; eliciong the production of IgG and IgM antibodies: or the elicitin~
of a T cell response such as proliferation and/or lymphokine secretion ;md/or the
induclion of T cell anergy. The foregoing fragments of ~ry j I ~e referred to herein
as antigenic fra men~. Fragments within the scope of the invention also include
those capable o~ hybndizin~ with nucleic acid from other plant specles for u~e in
Wo 43/01213 pcr~us92/o~661
21;~913
screening protocols to de~ct aller. ens th~lt are cross-re~cuve with Cr j 1 As used
herein~ a fragment of the nucleic acid sequence coding for Cr~ j I refers to a
nucleo~ide sequence having fewer bases than the nucleo~ide sequence coding for the
entire amino acid sequence of C~ j I and/or ma~ure Cry j I. Generally. the nucleic
acid sequence coding for the ~ragment or fra~ments of Cry j I will be selected from
the bases coding for the mature pro~ein. however. in some inst~nces it may be
desirable eo select all or a part of a fragment or fragments from the leader sequence
portion of the nucleic acid sequence of the invention. The nucleic acid sequence:of
the invention may also con~ain linker sequences~ modified res~ic~ion endonucle~esites and other sequences useful for cloning, expression or punfication of Gy j I or
fragments thereof.
A nucleic acid sequence coding for C~y j I may be obtained ~rom
Cryptomeria japonica plants. However, Appl~;cants have ~ound that mRNA coding
for CJY j I could not ~e obtain~d from commercially available Cryptom~na japonica
~5~ pollen. Th;s ~nability to obtain mRNA ~rom the pollen may be due to problems with
storage or tral~spor~ation of commercially available pollen. Applican~s have found
that fE~sh pollen and s~in~ cones are a good source of Gy j I mRNA. It may
iso be possiUe to obtain thc nudeic acid sequence coding for Cry j I ~rom genomic
DNA. C~ypto~r~ria japoni~a iS a well-lcnown species of cedar. and plant ma~nal
~o ~ m~f be obtained from wild, cul~ivated, or ornamental plants. The nucleic acid~
sequcnce coding for Gy^; I may be ~bt~ined using ~e method disclosed- herein or
any other suitablc techniques for isolation and clonin~ of genes. The nucleic acid
sequcnce of the invention may be DNA or RNA.
nle present invention provides expression vectors and hos~ cells transfonned
5~ to exprcss the nucleic acid sequences of the invention. Nucleic a~id codin~, for C~;
I. or at least one fragment thereof may be expresscd in bacterial cells such as E. cofi.
insect cells (baculovims), ye2st, or mammalian cells such as Chinese hamster ovary
cells ~CHO). Suitable exprcssion vectors. promoters, enhancers. and other
expression control elements may be found in Sarnbrook et al. MolYcular Clonin~
o L~boratorv Manu~ll. sccond edition. Cold Sprin~ Harbor Labor~tory Prçss. Co1~1
Spnn~ Harbor. New York (1989). Othersuitable expression veclors. promnters.
enhanccrs, and other expression elements are known to those skilled in the art.
Expression in marnmalian. yeast or insect cells leads to partial or complete
glycosylat1un o~ the recombinant material and ~onnation ot any inler- or intra-chain
WO ~3/01213 pcr/vs92/o~661
9~ 3
disulfide bonds. Suitable vectors for expression in yeas$ include YepSecl (Bald~ri
et al. (19873 Embo J. ~: 229-234); pMFa (Ku~an and Herskowitz (1982) Cell 3n
933-943); JRY8g (Schultz et al. (1987) Gene 54: 113-123) and pYl~S2 (Invitrogen
Corporation~ San Diego, CA)~ These veclors are freely available. Baculovirus andS mammalian expression systems are also available. For example, a baculovi~us
sys~em is commercially available (PharMingen, San Diego. CA) for expression in
msec~ cells while the pMSG veetor is commerically available (Pharmacia,
Piscataway, NJl for e~cpression in mammalian cells.
For expression in E. coli, suitable expression vec~ors in~lude, arnong others,
~0~ ~ pll~C (Amann et al. ~1988) Gene ~: 301-315); pG~X (~rad C arp., Melbourne.
Australia); pMAL (N.E. Biolabs, Beverly, MA); pRlT5 (Phannacia. Piscataway,
NJ); pET-l ld (Novagcn, Madison, WI) Jameel ct al., (1990~ J. Virol. 64:3963-
3966; and pSEM (Knapp et al. (1990) BioTechniques 8: 28Q-281). The use of
~, and pET- l ld, ~or example. will lead to the exp~ession of unfused protein.
~15~ The u~e of pMAI" pRlI`5 pSEM and ~EX will lcad to thc exprcssion of allergen
fused to maltose E binding protein (pMAL), protein A (pRlT5), truncated 13-
gala~tosidase (PSEM). or glutadlione S-transferase ~pGEX). When C7y j I,
agment. or fragmellts ~e~eof is expressed as a fusion pro~in, it is par~cularly
ageous to introduce an enzymatic cleavag? sise at the fusion junction between
the camor protein and Cr~ j I or fragm~nt thereof. Cry; I ~r f~agment the~of maythen be recove~ed from the fusion protein ~rough enzymatic cl~avagc a~ the
enyma~ic sitc and biochemical purification using conv&ndonal ~echniques for
purifica~i~n of proteins and peptides. Suitable enz~rmatic cleaYage sites include
thos~ for blood clot~ing Fa~tor Xa or thrombin for which the appropriate enzymesand protocols for cleavage are commcrcially available from for example Sigma
Chemic~l Company. St. Louis, MO and N.E. Biolabs, Beverly, MA. The diffe~ent
vectors also have di~ferent promoaer regions allowing constitu~ive or inducible
expression with, for example. IPIG induction (PRTC. Am~nn et al.~ ( 1988) supra;pET- 1 ld. Novagen, Madison. WI) or temperature induction (pRIT5, Pharmaci;l.
3 n Piscasaway, NJ) . It may also be appropriate to express recombinant Cr~ j 1 in
different E. coli hosts that have an altered capacity to de~rade recombln~ntly
expressed proteins (e.g. U.S. pa~nt 4,758,512). Alternatively, it may be
~dvan~ageous to alter the nucleic acid sequence to use codons preferentially utilized
by E. coli. where such nucleic acid alleration would not affect the amino acid
l ; ~
WO 93~01213 PCr/US92/05661
21129~
sequence of the expressed protein.
Host cells can be transformed to express the nucleic acid sequences of
the invention usin~ conventional techniques such as calcium phosphate or calciumchloride co-precipitation, DEAE-dextran-mediated transfection. or electroporation.
Suitable melhods for transfonnin~ the host cells may be found in Sarnbroo~c et al.
supr~, and other laboratory textbooks.
The nucleic acid sequences of the inven~ion may also be synthesized
using standard techniques.
The present invention also provides a method of producing purified
3apanese cedar pollen allergen C~ j I or at least one fragment thereof comprising the
steps of culturing a host cell transformed with a DNA sequence encoding Japanesecedar pollen allergen Gy j I or at least one fragment thereof in an appropriate
medium to produce a mixture of cells and medium containing said Japanese cedar
pollen allergen C~y j I or at least one fragment thereof; and punfying the mixture to
S produce substantially pure Japanese cedar pollen allergen Gy j I or at least one
~ragment thereo Host cells transformed with an expression vector ~ontaining DNAc~ng for C~y j I or at least one fragment dlereof are cultured in a suitable medium
for ~e host cell. Gy j I protein and pep~des can be pu~ed from cell culture
medium, host cells, or bo~ using techniques known in ~ art for purifying peptides
O ~ and proteins including ion-cxehange ehromatography~ gel filtraaon chromatography,
ult~afLltra~on, electrophoresis and immunopurification with andbodies specific for
Cry j I or fragments thereof. lhe tenns isolated and purified are used
ihterchangcably herein and refer to peptides, protein, proteir. fragments, and nucleic
id sequences substantially free of cellular ma~rial or culture med;um when
- produced by recombinant DNA ~chniques, or chcmical precursors or other
chemicals when syn~esized chemically.
Ano~er aspect of ~e invention provides prepara~ions comprising
Japanesc ccdar pollen allergen Gy j I or at least one ~agment thereof synthesized in
a host cdl ~ansfonned with a DNA sequence encoding all or a por~ion of Japanese
~0 ' cedar pollen allergen Cry j I, or chemically synthesized, and purified Japanese cedar
pollen allergen ~y j I protein, or at least one antigenic fragment thereof produced in
a bost cell transformed with a nucleic acid sequence of the invention, or chemically
synthesiæd. In preferred embodiments of the invention ~he C)y j I protein is
produced in a host cell transformed with the nucleic acid sequence coding for at least
WO 93/01213 ~ Pcr/US92/05~61
the mature C~y j I protein.
Fra~ments of an allergen from Japanese cedar pollen, preferably Cr~ j
_,
I, eliciting a desired antigenic response (referred to herein as antigenic fra~ments)
may be obtained. for exarnple. by screening peptides recombinantly produced fromS the corresponding fra~ment of the nucleic acid sequence of the invention codin~ for
such peptides, synthesized chemically using techniques known in the art. or
produced by chemical cleavage of the allergen, the allergen may be arbitrarily
divided into fragments of a desired length with no overlap of the peptides~ or
preferably divided into fragments of a desired length with no overlap of the peptides.
1 10 or preferably divided into overlapping fragments of a desired length. The fragments
are tested to determine their an~igenicity (e.g. the ability of the fragment to induce an
immune response). If fragments of Japanese cedar pollen allergen. e.g.C~ j, I are ~o
be used for therapeutic purposes, then the fragments of Japanese cedar pollen
allergen which are capable of eliciting a T cell rcsponse such as stimulation (i.e.~
lS proliferation or lymphokine secre~ion) and/or are capable of inducing T cell anergy
are particularly desirable and fragments of Japanese cedar pollen which have
minimal IgE s~mulating activity are also desirable. Addi~ionally, for ~erapeuticpurposes, purifed Japanese cedar pollen allergerls, e.g. Cry j I, and fragments thereof
preferably do not bind IgE specific for Japanese cedar pollen or bind such ~gE to a
~0 substantially lesser extent dlan the purified native Japanese cedar pollen allergen
binds such IgE. I~ ~e purified Japanese cedar pollen allergen or fragment or
*agments ~ereof bind IgE, it is preferable that such binding does not result in ~he
release of mediators (e.g. histamines) from m~ ceL~s or basophils. Minimal IgE
;; ~ stimula~ng ac~ r refers to IgE s~imula~ng activi~ that is less than ~e amoun~ o~
IgE producdon seimulated by thc native C yj I protein.
; ~ Purified protein allergens from Japanese cedar pollen or prefe~ed
antigenic ~rag~en~s ~ereof, when administered to a Japanese cedar pollen-sensitive
individual, or an individual allergic to an allergen cross-reac~ive with Japanese cedar
pollen allergcn, such as allergen from the pollen of Cupressus semper~irens or
Juniperus sabinoides etc. (discussed previously) are capable of modifying ~e
allergic response of the individual to Japanese cedar pollen or such cross-reac~ive
allergen of the individual, and preferably are capable of modifying the B-cell
response, T-cell ~esponse or both the B-cell and the T-cell response of the individual
to the allergen. As used herein, modification of the allergic response of an
3~10 g3/01213 Pcr/l-lS92/05661
21:1291:~
individual sensitive to a Japanese cedar pollen allergen can be defined as non-
responsiveness or diminution in symptoms to the aller~en. s determined by standard
clinical procedures (See e.~. Varney et al. Brirish Medical Journal. 302:2~5-269(1990)~.
The purified Cry j 1 protein or fragments thereof are preferably tested
in mammalian rnodels of Japanese cedar pollinosis such as the mouse model
disc!osed in Ta m ura et al. (1986) Microbiol. Imml~nol. ~: 883-896, or U.S. patent
4~39,239; or the primate model disclosed in Chiba et al. (1990) In~. Arch. Allergy
Itr~nol. 2~: 83-88. Ini~ial screening ~or IgE binding to ~he protein or fragments
O thereof may be performed by scratch tests or intradermal slcin tests on laboratory
anima1s or human volunteers, or in in vitro syste m s such a3 R A S T
(radioallergosorbent test), ~A S T inhibition, E LIS A assay, radioimmunoassay (RlA),
or histamine release (see ~xamples 7 and 8).
Aneigenic firagments of ~e present invention which have T cell
S s~imula~ng activi~y, and thus comprise at least one T cell epitope a~e par~icularly
desirable. T cell epitopes are believed eo be involved in initiation and perpetuation
of ~e immune r~sponse to a protein allergen which is responsible for the clinieal
symptoms of allergy. l~e~ T cell cpitopes are thought to trigger early events at the
level of d~e T helper cell by binding to an appropriate HLA molecule on the sur~aee
~0 of an an~gen presenting cell and s~mula~ing ~he relevant T cell subpopulation.
` ~ Thcse e~nts lead to T cell proliferation, l~nphokine secretion, local inflammatory
reac~nons, recn~i~nent of additional ~nmune cells to dle site. and activa~ion of the B
~ cdl cascads le~g to produ~ion of andbodies. One iso~ype of these an~ibodies,
; ~ IgE, is fundamentally important to the development of allergic symptoms and its
:
~5 ~uction is influen~ed carly in ~c cascade o~ events, at the level of the T helper
cell, by ~he natl~re of the lymphokines secre~d. A T cell epitope is ~e basic element
~- or smallest unit of recognition by a T cell receptor, where the epitope eomprises
~- amino a~ids essential to reccptor recognidon. Amino acid sequen~es which mimic
dlose of ~e T cell epitopes and which modify the allergic response to protein
30' allergens are wi~in the scope of this inven~ion.
Exposure of patients to purified protein allergens of the present
invention sr to ~he antigenic firagments of the present invention which comprise at
least one T cell epitope and are denved from protein allergens may tolerize or
anergize appropriate T cell subpopulations such that they become unresponsive to
. I]
WO 93/0121~ ,~ PCI/US92/056~1
the protein allergen and do not par~icipate in stimulating an immune response upon
such exposure. In addition, administration of the protein aller~en of the invention or
an anti~enic fra~ment of the present invemion which comprises at least one T cell
epitope may modify the lympholcine secre~ion profile as compared with exposure to
the naturally-occurring protein allergen or portion thereof (e.~. result in a decrease
of IL-4 and/or an increase in IL-2). Furthermore. exposure to such anh~enic
fragment or protein allergen may influence T cell subpopulations which normally
participate in the response to the allergen such that these T cells are drawn away
from the site(s~ of normal exposure to the allergen (e.g.. nasal mucosa, skin, and
10lung) towards the si~(s) of therapeutic administration of the fragment or protein
allergen. This redistribution of T cell su~populations may ameliorate or reduce the
ability of an individual's immune system to stimulate the usual immune respo~ise at
the site of normal exposure to the allergen, resulting in a dimunu~ion in allergic
symptoms.
15The purified Gy j I protein, and fragments or portions derived
`;therefrom (peptides~ can be used in methods of diagnosing, treating and preventing
allergic reactions to Japanese cedar pollen allergen or a cross reactive proteinaller~en. Thus the present invention provides therapeutic compositions comprisinp,
purified Japanese cedar pollen allergen Gy j I or at least one fragment thereof
~20 ~ ~ produced in a host cell ~ansformed to express C~y J I or at least one ~ragment
~ereof, and a pha~naceutically acceptable ca~ier or diluent. The therapeu~ic
compositions of the invention may also comprise synthetically prepared C~y j I or at
least one fragment ~hereof and a pharmaceutically accepeable carrier or diluent._-t
Admi~istration of the. ~erapelltic compositions of dle present inven~ion ~o an
indi~idual to be desensi~ed can be carried out using lcnown techniqucs. Cry j 1
prot~in or at lcast one fragment ~ereof may be administered ~o an individual in
.~
combinadon with, for example, an appropriate diluent, a calTier andlor an adjuvant.
Pharmaceutically acceptable diluents include saLine and aqueous buffer solutions.
Phannaceutically acceptable carners include polyethylene glycol (Wie et al. (1981)
Int. Arch. Allergy Appl Irnmunol. :8~99) and liposornes (Strejan el al. (1984~ J.
Neuroimm~nol 1: 27). For purposes of inducing T cell anergy, the therapeutic
composition is preferably administered in nonimmunogenic form, e.g. it does not
contain adjuvant. Such c~mpositions will ~enerally be administered by injection
(suhcut~meous, mrravenous. etc.). oral administrauon, inhalation, transderma]
-
WO ~1/01 ~11 PCI`/U692/0:,661
` 21i29:~3
application or rectal administration. The therapeutic compositions of the invention
_r are administered to Japanese cedar pollen-sensitive individuals at dosages and for
lengths of time effective to reduce sensitivity (i.e~ reduce the aller~ic response~ of
the individual to Japanese cedar pollen. Effec~ive amounts of the therapeulic
composi~ions will vary according to factors suchras the degree of sensitivity of the
individual to Japanese cedar pollen, the age, sex. and weight of the individual. and
the abili~y of the Gy j 1 pro~ein or fragment thereof to elicit an an~i~enic response in
the individual.
~ _
The Cry j I cDNA (or the mRNA from which it was transcribed) or a
O por~ion thereof can be used to identify similar sequences in any variety or type of
plant and thus, to identify or "pull out" sequences which have sufficient homology to
hybridiæ to the Cry j l cDNA or mRNA or por~on thereof, for example, DNA from
` ~ allergens of G pressus sem~en~irens, Juniperus sabinoides etc., under condiuons of
low stringency. Those sequences which have sufficient homology (generally greater
than 40%) san be selectcd for further assessment using the method described herein.
tematiYely~ hlgh S*ingency conditions can be used. In ~is manner, DNA of ~he
present inYention can be used to iden~y, in o~er ~pes of plants, preferably related
families, genera, or species su~h aS Jurliperus, or Cupre~sus, sequences encoding
polypepddes having amino a~id sequenccs similar to ~hat of Japanese cedar pollenallergcn Cr~ j I, and thus to identify allergens in other species. Thus, the present
invention includes not: only CJy j I, but also o~er allergens encoded by DNA which
hybndizes to DNA of ~e p~esent inYcn~om The invenoon further includes isolated
allergenic proteins or fragments ~ercof dlat are immunologically related to ~ry i
or~ agments ~ereof. such as by antibody croSs-Dac~Yity wherein the isolated
geI~ic proteins or fragments thercof are capable of binding to antibodies specific
for~e ~ro~in and pcptides of the invention, or by T cdl cross-reactivity whereinthe ~isolated allergenic pro~ins or fragmcnts ther~of arc capable of sdmulati~g T
: cells SpCCiflC for the pr tein and peptides ~ ~is inven~don.
Proteins or peptides encoded by the cDNA of the present invention
3~ ~ can be used. for example as "purified" allergens. Such purified allergens are useful
in ~e standardization of allergen ex~acts which are key reagents for the diagnosis
; ~; and t~eatment of Japanese cedar pollinosis. Furthermore. by using peptides based on
the nucleic acid sequences of G~ j I, anti-peptide an~isera or monoclonal antibodies
can be made using standard methods. These sera or monoclonal antibodies can be
:
~ 13
WO ~3/0121~ Pcr/Us92/0~661
used to standardi~e allergen extracts.
_,~ Through use of the pep~ides and prt)tein of the present inven~ion,
preparations of consistent. well-defined composition and bic)lo~gical activity can be
made ar;d administered for therapeutic purposes (e.g. t() modify the aller~ie response
of a Japanese cedar sensi~ive individual to pollen~of such trees). Administration of
such peptides or protein ~may, for example, modify B-cell response to Cr) j I
allergen, T-cell response to Cr~ j I allergen or both responses. Purified peptides can
also be used to study the m echanism of immuno~erapy of Crvptomeria japonica
allergy and to design modified deriYatives or analogues useful in immunotherapy.a Work by others has shown that hi~h doses of allergens ~enerally
produce ~he best results (i.e., best symptom relief). However~ many people are
~- unable to tolerate large doses of allergens because of aller~ic reachonS to the
allergens. Modification of naturally-occurring allergens can be designed in such a
manner ~t modified peptides or modified allergens which have the same or
- 5 enhanced therapeutic proper~es as ~e corresponding naturally-occurring allergen
but have reduced side ef~ec~. (especially anaphylactic reactions) can be produced.
These can be, for ex~nple, a protein or pep~ide of the present invention (e.g., one
hanng ~11 or a portion of the amino a~id sequence of C?y j I), or a modified protein
or peptide, or protein or peptide analogue. It is possible to modify ~he structure of a
0 pro~in or pep~de of ~he invenaon for such purposes as increasing solubility.
enhancing dlerapeutic or preventive efficacy, or stability (e.~., shelf lif~ ~x ~, and
resistance to proteoly~ic degradation in ViVQ). A modified protein or peptide can be
produced în wh;ch the amino acid sequence has been altered, such as by amino acid
subs~ on~ deletion, or addi~on, tO modify immunogenicity and/or r~duce
allergenicity, or to which a componcn~ has been added for ~he same purpose. For
example, ~e amino acsd Iesiducs essential to T cell epitope func~on can be
detennined using known techniques (e.g., substitu~ion of each residue and
determillation of thc prescnce or absence of T cell reactivity). Those residues shown
~o be essential can be modified (e.g., replaced by another amino acid whose presence
is shown to enhance T cell reactivity), as can those which are not required for T cell
reactivity (e.g., by being replaced by another amino acid whose incorpora~ion
enhances T cell reactivity but does not diminish binding to relevant MHC). Another
exarnple of a modific tion of protein or peptides is substitlltion of cysteine residues
preferab}y with alanine, serine. ~hreonine, leucine or ~lutamic acid to minimize
14
WO 93/0121~ Pcr/us92/o566 1
2112913
dimerization via disulfide linkages. Another example of modification of the
peptides of the invention is by chemical modification of amino acid side chains or
.~,
cyclization of the peptide.
ln order tO enhance stability andlor reactivity. the pro~in or peptides
of the inven~ion can also be modified lo incorporate one nr more polymorphisms in
the amino acid sequence of ~e protein aller~en resulting from natural allelic
variation. Additionally, D-amino acids, non-natural amino aeids or non-amino acid
analo~ues can be substituted or added tO produce a modified protein or peptide
within the scope of thas inven~ion. Furtherrnor! ~ proteins or peptides of the present
O invention can be modi~aed using the polyethylene glycol ~PEG) method of A. Sehon
and co-workers (Wie et al. supra) to produce a protein or peptide conjugated with
P~G. In addition, PEG can be added during chemical synthesis of a pro~in or
pep~ide of ~he invention. Modifica~ons of proteins or pep~ides or por~ions thereof
can also include reduction/ alyklation (Tarr in: ~ethods of Protein
l~icrocharacterization, J.E. SilYer ed. Humana Press, Clifton. NJ, pp 15S-194
(19863); acylation (Tarr, supra) chemical coupling to an appropriate carrier (Mishell
-~ and Shiigi, eds, Selected Methods in Cellular l~ nology, WH F~eman, San
Francisco, CA (19803; U.S. Pa~ent 4~939,239; or mild formalin trea~nent ~Marsh
Internanonal Archives of Aller~y and Applied Inu7luJwlogy, 41:199-215 (1971)).
~0 To facilitate purifica~on and poten~ally increase solubility of proteins
or pep~des of ~e invendon, i~ is possible to add reporter group~s) to the pep~ide
backbone. For cxample, poly~ dine can be added to a pep~ide to purify the
peptide Oll ~nmobiliæd metal ion aff~nity chromatography (Hochuli, E. et al;
Bi~/~echnology, 6:1321-1325 (1988)). In addition, specific endoprotease cleava~esi~s can be introduced, if ~esi~d, ben~een a reporter group and amino acid
`s~u~nces of a peptide to facilitate isolation of peptides free o irrelevant sequences.
In order to successfi~lly desensitize an individual to a pro~ein antigen. it may be
necessa~ to incr~ e solubi3ity of a protein or pcp~ide by adding functional
groups to the peptide or by not including hydrophobic T cell epitopes or regions30' containing hydrophobic epi~opes in the pep~ides or hydrophobic regions of the
pro~ein or pep~ide.
To potentially aid proper antigen processing of T cell epitopes within
a peptide. c~nonical pro~ease sensitive sites can be recombinantly or synthetically
engineered between regions, each comprising at least one T cell epitope. For
WO 93/01213 g,~ ,} PcrtUS92/0~66
example, charged amino acid pairs, such as KK or RR, can be introduced between
regions within a peptide during recombinant construction of the peptide. The
resultin~ peptide can be rendered sensitive to cathepsin andlor other trypsin-like
enzymes cleavage to ~enerate portions of ~he peptide containin~ one or more T cell
epitopes. ln addition~ such charged amino acid residues can result in an increase in
solubility of a peptide.
Si~-directed mutagenesis of DNA encoding a peptide or protein of
the invention (e.g. Cr~ j I or a fragment thereof) can be used to modify the structure
of ~he peptide or protein by methods Icnown in the art. Such methods may, among
0 others, include PCR with de~enerate oligonucleotides (Ho et al., Gene, 77:51-59
(1989)) or total synthesis of mutated genes (Hostomsky, Z. et al., Biochem. Bioph~s,
: Res. Comm.,~:~0$6-1063 (l989)). To enhance bacterial expression, the
aforementioned methods can be used in conjunction with other procedures to chan~e
the eucaryotic codons in DNA cons~ucts encoding protein or peptides of the
invention to ones preferentially used in E. coli, yeast, mammalian cells, or other
eukaryo~ic cells.
Using the structural information now ~vailable, it is possible to design
Cry j T peptides which, when administered to a Japanese cedar pollen sensi~ive
individual in sufficient ~uantities, will modify the individual's allergic response to
Japanese cedar pollen. ~is can be done, for example, by examining ~e struc~ure of
C)y j 1, producing pepddes (via an expression system, synthe~cally or otherwise~ to
be examined for their ability to influence B-cell and/or T-cell ~sponses iI~ Japanese
c ~dar pollen sensitiv~ individuals and sdec~ing appropriate peptides which con~in
epitopes recognized by the cells. In referring to an epitope, the epitope will be the
basic element or smallest unit of rccogni~on by a receptor, particularly
immunoglobulins, histocompa~bility antigens and T cell receptors where the epi~ope
comp~ises almino a~ids essential to receptor recogni~on. Amino acid sequences
which mimic those of ~e epitopes and which are capable of down re~ula~ing allergic
r~sponse to ~j I can also be used,
It is now also possible to design an agent or a drug capable of
blocking or inhibiting the ability of Japanese cedar pollen allergen to induce an
al}ergic reaction in Japanese cedar pollen sensitive individuals. Such a~ents could be
designed. for exarnple, in such a manner that they would bind to relevant anti-Cr~ j I
I~Es. thus preventing I E-allergen binding and subsequent mast cell degranulation.
16
:
W~ 93/01213 2 1 1 2 9 1 3 P(~r/US92/O~S61
Altematively, such agents could bind to cellular components of the immune syslem,
resultin~ in suppression or desensitization of the aller~ic response to C)ypromeria
.~,~
japonica pollen aller~ens. A non-restrictive example of this is the use of appropriate
B- and T-cell epitope peptides, or modifications thereof. based on the cDNA/protein
S structures of the present invention to suppress the allergic response to Japanese cedar
pollen. This can be canied out by defiming the structures of B- and T-cell epitope
peptides which affect B- and T-cell function in in vitro studies with blood
components from Japanese cedar pollen sensitive individuals.
Protein, peptides or antibodies of the present invention can also be
O used for detecting and diagnosing Japanese cedar pollinosis. For example. this could
be done ~y combining blood or blood products obtained from an individual to be
assessed for sensi~ivity to Japanese cedar pollen wi~h an isolated antigenic pepti~e or
peptides of Cty j I, or isolated Gy j I protein, under condiiQns appropriate forbinding of components in the blood (e.g., antibodies, T-cells, B- cells) with the
I S peptide(s) or protein and determi~g the extent to which such binding occurs.
The present invention also provides a method of producing CrY j I or
fragmen~ ~ereof compr;ising culturîng a host cell containing an expression ./ec~or
which contains DNA encoding all or at least one fra~ment of Gy j I under
conditions appropriate for expression of Gy i I or a~ least one fragment. The
~'.0 expressed product is ~en recovered, using lalo~m techniques. Alterna~vely9 C~y j I
~; or ~gment thereof can be synthesized using known mechal~ical or chemical techniques.
The DNA used in any embodiment of this invention can be cDNA~
obtained as descnbed herein, or alternadvely, can be any oligodeoxynuGleohde
t~ sequence having aJl or a portion of a sequencc represented herein, or d~dr functional
equivalen~s. Such oligodeoxynucleotide sequences can be produced chemically or
enzymatically, using known ecchniques. A ~unc~onal equivalent of an
oligonucleohde sequence is one which is l) a sequencc capable of hybridizing to a
complementary oligonucleotide to which the sequcnce (or corresponding sequence
portions3 of SEQ ID NO: 1 or fragments thereof hybridizes, or 2) the sequence (or
corresponding sequence por~on) complemçntary to SEQ ID NO: 1, and/or 3) a
sequence which encodes a product (e.g., a polypeptide or pep~de) having the samefunctional characteristics of the product encoded by the ~sequence (or correspondin~
sequence portion) of SEQ ID NO: l. Whether a functional equivalent must meet
W093/~12~3 ~3 Pcr/US92/~66
one or both criteria will depend on i~s use (e.g.. if it is to be used only as an
oligoprobe, it need meet only the first or second criteria and if it is to be used to
produce a Cr~ j 1 aller~en. it need only meet the third criterion).
The invention is further illustrated by the following non-limitin~
; exarnples.
Example 1
riScatiQn of Na~ e .1~~edar Pollen A11er~en (Cr~Ll
The ~ollowing is a description of the work done to biochemically
O purify the major allergen, C~Y j I in the native fo~n. The purification was modi~led
from published procedures (Yasueda et al., J. Allergy Clin. Irnmunol. 71:77. 1983).
lOOg of Japanese cedar pollen obt~ined from Japan (Hollister-~tier,
Spokane, WA) was defatted in 1 L diethyl ether three ~imes, the pollen was collected
after filtration and ~e ether was dried off ~ a vacuum.
The defatted pollen was e~ctracted at 4C overnight in 2 L extrac~ion
bu~er containing ~0 mM tris-HCI, pH 7.8t 0.2 M NaCI and protease inhibitors in
final concentrations: soybean trypsin inhibitor (2 ~g~ml), leupeptin (1 llg/ml),pepstatin A (lf llgJml3 and phenyl methyl sulfonyl fluoride (0.17 mg/ml). The
insoluble ma~rial was r~ex~acted with 1.2 L extrac~on buffer at 4C overnight and
7fl~ bo~ ex~cts were combined toge~er and depigmented by batch absorption with
Wha~an DE-52 DEAE cellulose (2~0 g dry weight) equilibrated with'~he extraction
~ buffer.
;~ e depigmented material was then fractionated by ammonium
~ sulf~te precipitation at 80% saturation ~4C), which removed much of ~e lower
molecular weight material. The resultant partially purified C?y j i was ei~her
dialyzed in PBS bu~fer and used in T cell studies (see Example 6) or subjected to
further purifica~on as descr~bed below.
The cnriched ~Jy j I material was then dialyzed against 50 mM Na-acetate,
pH 5.0 at 4C with 50 mM Na-acetate, pH 5.0 with protease inhibitors. The unbound
material ~basic proteins) was then applied ~o a 50 ml caaon exchange column ~Wha~rnan
CM-52) which was equilibrated at 4C wi~h 10 mM Na-acetate, pH 5.0 with proteaseinhibi~ors. C~y j I was eluted in the early fraclions of a linear ~radient 0.3 M NaCl.
The enriched Cr~ j I material was lyophiliud and was then purified by FPLC over a 300
rnl Superdex 75 column (Pharmacia) at a flow rate of 30 ml/h in lO mM Na-acetate. pH
W~ 93J01213 Pcr/uss2/o~66l
21i2913
5.0 at 25C.
The purified Cr~ j I was further applied to FPLC S-Sepharose 16110 column
chromato~raphy (PhaImacia) with a linear ~radient of 0 - 1 M NaCI at 25C. Cr~ j 1
eluted as the major pealc was subjected to a second ~el filtration chromato~raphy. FPLC
Superdex 75 column (2.6 by 60 cm)(Phannacia, Piscataway, NJ) was eluted with a
downward flow of I0 mM Na-acetate, pH 5.0 with 0.15 M NaCI at a flow rate of 30
ml/h at 25C. Fig. la shows the chromato~raphy on gel filtration. Only C~ j 1 was
detected (Fig. lb, lane 2 to lane 8). Gy j I was fractionated into 3 bands as analyzed by
SDS-PAGE using silver suining (Fig. lb) As shown in Fig. lb, SDS PAG~ (12.5%j
0~ analysis of the fractions from the major peak shown in Fig. la was perforrned under
reducing~conditions. The gel was siiver stained using the silver staining kit from Bio-
`~ Rad. The samples in each lane were as follows: Lane l, pres~ained standard pr~teins
(Gibco BRL) ~ncluding ova!bumin (43,000 kD), carbonic anhydrase (29,000 kD), anda-lactoglobulin ~18,400 kD); lane 2, fraction 36; lane 3 fraction 37; lane 4 fraction 38;
5~ ~ ~ lane 5 fraction 39; lanc 6 frac~ion 41, lane 7 fracaon 43; and lane 8 frac~ion 44. All
, ~ fractions areshownin~Fi~. la.
` ~ ese proteins were also analyzed by Western blot~ing using mouse
` ~ monoclon 1 an~body CBF~ (E;ig. 2). As shown in Fig. 2, an ali~uot of fraction 36 (lane
; 1), frac~don 39, (lane 2) and fracdon 43 ~lane 3) purified from the Superdex 75 as shown
20~ in Fig. 1 ~was scpara~d by SDS-P~GE, clectroblo~ed onto nitrocelluslose and probcd
wi~mAB CBF2. Biot;nlylatcd goat and-mouse Ig was used for ~e second an~ibody
and~bound~andbody was; re~voaled by l25I-sheptavidin. Ibe monoclonal CBF2 was
r ai~ ag~t ragweed allergenAmb a I by Dr. D. Klapper~(Chapel Hill, N. Carolina~.~ Because ~of ~e homoIogy ~wecn ~e Amb a l and CJY j I sequences, a number of
25 ~ andbodies raised against Amb a I weretested for reac~ivity with Gy; I. The results
showed ~at CB~ rccognize-d denatur~d Cr~j I as detect~d by ELISA and Western
blot~ng.~ ~addition,~Wcstern blotdng also demonstrated ~at no other bands were
deicctcd~ by CBP2, other than Cry j I in ~e expccted molecular wdght range (Fig. 2~.
T hesc results were consistent with the findings from protein sequcncing. When fraction
44 and frac~ion 39 (Figilb) were subjected to N-tenninal sequencing, only Crvj Isequencewasdetected. ~ ~
In summary, ~ree C~y j I isoforms of different molecular wei~ht were
:: :
pur;fied from pollen e%tract. The molecular weights es~mated by SDS-PAGE ran~ed
from 40-35 kD under both reducing and non-reducing condi~ions. The isoelectric point
,~
~::: : :
.
~ ,
WO 93/01213
Pcr/US92/0~661
~ 9~3
of these isoforms is approximately 9.5-8.6. with an average pl of 9Ø The N-terminal
20 amino acid sequence was the same in these 3 bands and was identical to previously
published Cn~j I sequence (Taniai et al. supra). The 3 isofonns are all recognized by
monoclonal antibody CBF2 as shown in the alleroic sera titration of different puri~led
subfractions of Cry j I usin~ a pool of fifteen allergic patient plasma. They all bind
aller~ic patient IgE (Fi~. 3). The difference in molecular weight and isoelectric point
mi~ht in part be due to post-translational modification, e.g. glycosyJation.
phosphorylation or lipid content might be different in these iso~orms. The possibility
that these different iso~omls might be due to protease degradation cannot be ruled out at
0 present even though it is unlikely due to the fact that four different protease inhibitors
were used during extraction and purification. The other possibility could be due to
polymorphism in the gene or alterna~e splicing in the mRNA though only one majorform of Gy j I protein has been detected in cDNA cloning studies (see Example 4).
Ano~er approach which may be used to purify na~ive C~y j 1 or
recombinant Gy j I is immunoaffinity chromatography. This technique provides a
vcry selective protein purification due to the specificity of the interachon between
monocl~nal an~bodies and antigen. For the purpose of producing C~ reac~ive
monoclonaI antibodies, female Balbl/c mice were obtained from Jackson Labs. Eachmouse was initially immunized intraperitoneally with 7~100 llg purified native Cry;
99% puri~y lower band, as shown in Fig. lb), emulsified in Freund's complete
ad5uvan~ One fur~er intravenous injection of 10 ~g purified native C~y j I in PBS
was given ~4 days ~er the inidal injection. The spleen was removed 3 days later
and myeloma fusion was conducted as described ~Current Protocols in Immunology,
l991, Coligan et al, eds.) using ~be myeloma line SP2Ø The cells were cultured in
- 25 10% ~etal caL~ serum (HybIimax), hypoxanthinc and azaserine and wdls containing
colonies of hybridoma cclls were screened for an~ibod~r production using an~igen-
binding EUSA.
Cells from positive wells wer~ cloncd at three-tendls cell/well in 10% fetal
calf seram (Hybnmax), hypoxanthine and positivc clones were subcloned one more time
3 oi in hypoxanthine medium . Capture ELISA (see Example 7) was used ~or secondary and
ertialy screening. This assay offers the advantage that a clone that recognizes the na~ive
protein may be selected and thus may be useful for immunoaffinity purification. Thus~
~ ~ lhe mAbs will provide a useful tool in purification of C~ j 1 from pollen extracts.
-~ Similarly, monoclonal antibodies that bind to recombinant C~ j I can also be used for
:
WO 93/01213 Pcr/US~2/0~661
21~913
immunoaffinity chromatography. In addition, the monoclonal antibodies generated may
~,~ be useful for diagnosic purposes. It may also be possible to raise different mAbs that
show some specificity towards these different isoforms of Cr~ j I and thus wou}dprovide a useful tool to characterize these isoforms.
;
Example 2
Attempted Extracffon of RNA From Japanese Cedar Pollen
Mul2iple attempts were made to obtain RNA ~rom commercially-
3 available, non-defatted, Cryp~om~ria japonica (Japanese cedar) pollcn (Hollister
S~ier, Sea~le, WA). Ini~ially, the method of Sambrook et al., Molecular Cloning. A
Labor~tory Manual, ~old Spring Harbor Laboratory P~ss, Cold Spring Harbor,
New York (19B9) was used in which the sample was suspended and lysed in 4 M
guanidine buffer, ground under liquid nitrogen, and pelleted through 5.7 M cesium
chloridc by ultracen~ifuga~don. Various amounts (3, 5 and lO g~ of pollen in
varying amounts of guanidine Iysis buffer (lO and 25 ml) weFe ¢ied. Centrifugation
~-~ ; through cesium resulted in viscous material in the bottom of ~e tube, from which it
was not possible to recover an RNA pelle~ Although it was possible to obtain RNA- from de~a~ed Ambrosia ar~ènusiifolia (ragweed) pollen (Greer Laboratories, Lenior,
~: û NC) using ~is protocol, defatt;ng the Cryptorneria japonica p~len wi~ acetone
before guanidine cxtraction also did not yidd any RNA, as dletennined by
absorbanCe at A2~
~ : `
An acid pb~nol extrac~n of RNA according to the me~od yl,
S~k et al., suprd was ~cmpted from Gyptomeria Japolu~a pollen. The
pollen was grolmd and shcarcd in 45 ~ guanidine soludon, acidified by addition of
2 M sodium aceltate, and extrac~cd wi~ watc~ rated phenol plus chloroform.
~r pr~cipitation, ~e pelle~ was washed wi~ 4 M lithium chloride, rcdissolved in
10 mM Tris/5 mM EDTA11% SDS, chloroform cxtræted, and re-pr~cipita~d with
NaCI and absolute e~anol. It was possible ~o extract Am~rosia ar~enusiifolia but not
ryp~orr~ria j~ponica RNA with this procedure.
Next, 4 g of Cryptornerul japonica pollen was suspended in lO ml
extrac~on buffer (50 mM Tris, pH 9.0, 0.2 M NaCl, lO mM Mg acetate and
dicthylpyrocarbonate (DEPC) to 0.1%), ground in a mortar and pestle on d~y ice,
transfelTed to a cen~ifuge tube wi~h 1% SDS, lO mM EDTA and O.5~o N- lauroyl
'~1
WO 93J01213 Pcr/us92/056~1
` ~, 3
sarcosine, and the mixture was extracted five ~imes with warm phenol. The aqueous
phase was recovered after the final centrifu~ation, 2.5 vol. absolute ethanol was
..~,~
added, and the mixture was incubated overni~ht at 4C. The pellet was recovered by
centrifugahon, resuspended in 1 ml dH20 by heating to 65C. and reprecipitated by
the addition of 0.1 vol. 3 M Na aceta~e and 2.0 vol. of ethanol. No detectable RNA
was recovered in the pellet as judged by absorbance at A260 an~ gel ~lectrophoresis.
Finally, 500 mg of Cry~ptomeria japonica pollen was ~round by
mortar and pes~le on d~ ice and suspended in S ml of 50 mM Tris pH 9.0 with ~.2
M NaCl, 1 mM EDTA, 1~ SDS that had been Ireated overnight with 0.1% DEPC.
~10 as previously described in Frankis and Mascarhenas (1980) Ann. BOI. 4'2: ~95- 599.
After five extractions with phenol/chloroform/lsoamyl alcohol (mixed at 25:24:1),
material was p~ecipitated ~rom ~he aqueous phase wi~h 0.1 volume 3 M sodium
acetate and 2 volumes ethanol. The pellet was recovered by centrifuga~ion,
resuspended in dH20 and heated to 65C to solubilize the precipitated material.
Further precipita~ons with lithium chloride were not done. There was no detectable
A recovered, as detennined by absorbance at A26() and gel elec~ophoresis.
In summary, it has not been possible to recover RNA frorn the
commercial pollen. It is not known whether the RNA has been degraded d-tring
storage or shipmen~, or whe~er the protocols used in this example did not allow
~2û recovcry of cxtant RNA. However, RNA was recovered from ~resh Cryptomeria
faponic~;pollen and staminate cone samples. (See Example 3.)
~ Example 3
-~25 ~ Extr~ction of RNA From Japanese Cedar Pollen and Staminate Cones and
: ~ :
/CIoning o~ Cry jI
F~esh pollen and staminate cone samples~ collected from a single
:: ~ C1yptomeria ja~onica (Japanese cedar) tree at the Ar~old ~rbor~tum (Boston, MA),
were ~rozen immedia~ely on dry ice. RNA was prepared from ~00 mg of eaeh
~30 ~ sample, essentially as described by Frankis and Mascarenhas, supra. The samples
were ground by mortar and pestle on dry ice and suspended in S ml of 50 mM Tns
pH 9.0 with 0.2 M NaCI, l mM EDTA, 1% SDS that had been treated overnight
with 0.1% DEPC. After five extractions with phenoVchloroforrn/isoamyl alcohol
(mixed at 25:24:1), the RNA was precipitated from the aqueous phase with O.l
.
__
WO 93/01~l3 PCI/US92/0~661
211~13
volume 2 M sodium acetate and 2 volumes ethanol. The pellets were recovered by
centrifuga~ion, resuspended in dH20 and heated to 65C for ~ min. Two ml of 4 M
lithium chloride were added to the RNA preparations and they were incubated
overnight at 0C. The RNA pellets were recovered by centrifu~ation. resuspended
in 1 ml dH20. and again precipitated with 3 M sodium acetate and ethanol overnight.
The fin~l pellets were resuspended in 100 ~11 dH20 and stored at -80C.
First strand cDNA was synthesized from 8 llg flowerhead and 4 ~ug
pollen RNA using a commercially available Icit (cDNA synthesis systems kit, BRL,Gaith~rsburg, MD) with oligo dT priming according to the method of Gubler and
0 Hoffman (1~83) ~ene 2~: 263-269. An attempt was made to amplify cDNA
encoding C~ j I using the degenerate oligonucleotide CP- 1 (which has the sequence
5'-GAT~AATC~CGATAGA~AG-3', wherein T a~ posihon 3 ean also be C; T at
position 6 can also bc C; C; at position 9 can also bc A,T, or C; A at posi~ion 12 can
also be T, or C; T at posi~ion 15 can also be C; A at position 16 can also be T; and G
.5 at position 17 can also be C; SEQ ID NO: 3) and plimers EDT and ED. P~imer EDT
has the sequence ~'-GGAAlTCTCT~GACTGCAGG~3'~SEQ
II) NO: 24~. Primer EI) has dle sequence 5~-GGAAT~ TAGAcTGcAGGT-3~
(SEQ ID NO: 23). CP-l is ~e degenerate oligomlcleotide s~uence encoding the
first six amino acids o~ the amino terminus (AspAsnProIleAspSer, amino acids 1-6of SE3Q IO NO: 1) of Cry j I. EDT will hybridize with the poly A tail o~ the gene.
All oligonucleo~ides were synthesized by Resear~h Genedcs, Inc. Huntsville, AL.
Polymerase chain reac~ons (PCR) were carried out using a commercially available
kit ~(}ene~p DNA ~mplifiua~on kit, Perkin Elmer Cetus, NorwaLk. CT) whereby
~ ~10 ~1 lOx buffer containing dNTPs was mixed wi~ 1 llg of CP- 1 and 1 ~lg of
~ ED/EDT p~ners (ED:E3DT in a 3:1 M ratio), cDNA (3-~ ~1 of a 20 ~1 fi~st strand
cDNA rcacdon mix), 0.5 ~1 Amplitaq DNA polymerase, and distilled wa~er to 100
The samples were amplified with a programmable thennai controller
(MJ Research, Inc., Cambridge, MA). The first S rounds of amplification consisted
~30 of denaturation at 94C ~or 1 minute, annealing of primers to the template at 4~C
for 1.5 minutes, and chain elongation at 70C for 2 minutes. The final 20 rounds of
amplificadon consis~ed of denaturation as above, annealillg at 55C ~or 1.5 minutes,
and elongation as above. Five percent (5 Jll) of this initial amplification was then
used in a secondaly arnplification with 1 llg each of CP-2 (which has ~he se~uence
WO 93/01213 Pcr/US~2/05661
C~
5'- G&GAAl~CAAlTG{iGCGGAGAATGG-3' wherein T at position 11 can also
be C; G at position 17 can also be A, T, or C: G at position 20 can also be A; T at
position 23 can also be C; and G at position 24 can also be C) (SEQ ID NO: 4). anested primer, and ED, as above. The sequence ~'-GGGAAl-rC-3' (bases I throu h
S 8 of SE(2 ID NO: 4) in primer CP-2 represents~an Eco Rl si~e added for clonine
pulposes; the aemaining degenera~e oligonucleotide sequence encodes arnino acids13-18 of Cry j 1 (AsnTrpAlaGlnAsnArg, arnino acids 13 through 18 of SEQ ID NO:
1). Multiple DNA bands were resolved on a 1% GTG agarose ~el (FMC, Rockport,
ME~, none of which hybridized with 32p end- labeled probe CP-3 (SEQ ID NO: 5)
in a Southern blot performed according to the method in Sarnbrook et al. supra.
Therefore, it was not possible to select a specific Gy j I DNA band and this
approach was not pursued. CP-3 has the sequence. 5'-
CTGcAGccATmcIAcATIAAA 3~ wherein A at posi~ion 9 can also be G; T at
posiion 12 can also be C; A at posi~ion 18 can also be G; and A at posi~ion 21 can
also be G~ (SEQ ID NO: 5). Inosine (I) is used at posi~ion 15 in place of G or A or
. T or C to reduce degener~y (Kno~h et al. (1988) Nucleic Acids ~es. ~: 10932).
~he sequence 5'-CTGCAG-3' (bases 1 through 6 of SEQ ID NO: 5~ in primer CP-3
represent a Pst I ~te added for ~loning purposes; the remaining degenerate
oligonucleotide sequcnce is the non-coding strand sequence co~responding to coding
: 2Q strand sequence encoding amino acids PheAsnValGluAsnCily (~mino acids 327
: ~ ~rough 332 of SEQ ID NO: 1) from ~e internal sequence of C~j I.~; A primary PCR was also perfolmed on first-strand cDNA using CP- l
(SEQ ID NO: 3) and CP-3 (SEQ ID NO: ~), as above. A secondary PCR was
perfo~ed using 5% of ~e primaly reac~on ~g CP-2 (SEQ ID NO.. 4) and CP-3
(SEQ ID NO: 5). Again, mul~ple bands were observed, none of whi~h could be
specifically identi~lsd in a Southern blot as C y j I, and this approach was also not
pu~sued.
: ~ Double-s~anded cDNA was dlen synthesiæd from approximately 4
llg (pollen) or 8 llg (flowerhead) RNA using a commercially available kît (cDNA
3~ Synthesis System kit, BRL, Gaithersburg, MD). After a phenol extraction and
ethanol precipitation, the cDNA was blunted with T4 DNA polymerase (Promega~
Madison, WI~, and ligated to ethanol precipieated, self-annealed. AT (SEQ ID NO:20) and AL (SEQ ID NO: 22) oligonucleotides for use in a modi~led Anchored PCR
reaction~ according to the method in Rafnar et al. (1991) J. Biol. Chem. ~: 1229^
~4
WO 93/0121~ PCl /US~2/0~661
21i291~
1236; Frohman et al. (l990) Proc. Na~l. Acad. Sci. USA ~: 8998-9002; and Roux e~al. (1990) BioTech. 8: 48-57. Oli~onucleotide AT has the sequence 5'-
GGGTcTAGAGGrAccGTccGATcGATcATI-3~(sEQ ID NO: 20) (Rafnar et al.
supra ). Oligonucleotide AL has the sequence 5'-AATGATCGATGCT-3' (SEQ lD
NO: 22) (Rafnar et al. 3wpra. The amino terminus of Cr~ j 1 was amplified from the
linkered cDNA (3 ul from a 20 ~I reaction~ with 1 ~lg each of oligonucleotides AP
(SEQ ID NO: 21) and degenerate ~ry j 1 primer CP-7 (which has the sequence 5'-
ITCATICGATICTGGGCCCA-3' wherein G at posi~ion 8 can also be T; A at
position 9 can also be G; C at position 12 can also be T; and G at posi~ion 15 can
~0 also be A, T, or C)(SEQ ID NO: 6). lnosine (I) is used at posi~ion 6 in place of G or
A or T or C to reduce de~eneracy (Knoth et al. supra). The degenerate
oligonucleotide CP-7 (SEQ ID NO: 6) is the non-coding strand sequence
corresponding to coding strand sequence encoding amino acids 14-20
(1 rpAlaGlnAsnArgMetLys) from ~he asnino tenninus of Cry j I (amino acids 14-20
:1~ of SEQ ID NO: l~. Oligonucleo~de AP has ~e sequence 5'-
GGGTCTAGAG~}TACCGTCCG-3' (SEQ ID NO- 2l3.
The p~nary PCR reaction was carried out as described herein. Five
percent (5 ~1) of ~is ini~dal amplifica~on was ~en used in a secondary amplificahon
g e~ch of AP (SEQ ID NC): 21) and degenerate C~y j I p~ner CP-8 (SEC~
ID NO: 7) an intemally nested Cry j I oligonucleotide primer, as described herein.
Primer CP-8 has the ~ sequence S'~CIGCAGCGATICT~G&CCCAAAl~-3'
-:: wherein G at position 9 can also be T; A at posi~on l0 can also be G; C at posi~on
13~ can ~so be T; G at posieion 16 can also be A, T, or C; and A at posi~ion 23 c~
~: also be G)(SEQ ID NO: 7). Ihc nucleotides ~'-CCIG~AG-3' (bases l.through 7 of
SEQ ID NO: 7) r~present a Ps~ I res~ic~ion site added for cloning purp~ses. The
remaining degenerate oligonucleotide sequence is the non-coding s~and seq~ence
conespo~ding to coding strand sequence encoding amino acids 13-18 of Cy j I
(AsnTrpAlaGlnAsnArg, amino acids 13-18 of SEQ ID NO:: l) from the amino
terminus of Gy j I. The dominant amplified product was a DNA band of
approximately 193 base pairs, as visualized on an ethidium bromide (EtBr)-stained
3% GTG agarose gel.
Amplified DNA was recovered by sequential chloroform. phenol. and
chloroform extractions, followed by precipitation at -20C with 0.5 volumes of 7.5
ammonium acetate and l.5 volumes of isopropanol. After precipitation and washin~
~5
W~ 93/01213 PC~/US92~056
~ ,9~-'t~`~
with 70% ethanol, the DNA was simultaneously digested with Xba I and Pst I in a
15 111 reaction and electrophoresed throuth a prepa~a~ive 3% GTG NuSievç low melt
gel (F'MC, Rockport~ ME). The appropriate sized DNA band was visualiæd by EtBr
staining, excised, and ligated into appropriately digested Ml3mpl8 for sequencing
; by the dideoxy chain termination method ~San~er et al. (1977) Proc. Natl Acad Sci.
U~A 74: ~463-5476) using a commercial}y available sequencing kit (Sequenase kil.U.S. Biochemicals, Cleveland, OH). It was initially thought that ligatable material
could on}y be derived from staminate cone-derived RNA. However, upon
subsequent examination, it was shown that li~atable material could be recovered
O ~rom PCR product generated from pollen-derived RNA, and from st~ninate cone-
derived RNA.
The clone designated ~C71.6 was found to contain a par~ial sequence
: of Cry j I. This was confirmed as an authentic clone of CJy j I by ha~ng complete
identity to ~e disclosed NH2-tcnninal sequence of Cry j I (Tania~ et al. supra). The
amino acid at posilion 7 was determined to be cysteine (Cys) in agreement with the
; sequence disclosed in U.S. patlent 4, 939,239. Amino acid numbering is based on dle
sequence of the mature pro~in; amino acid 1 corresponds to the aspar~ic acid (Asp)
disclosed as the NH2~ Tninus of Gy j I ~Taniai et al. supr~) The initia~ing
methionine was found to be amino acid -21 relative to the first amino acid of the
~D~ mature protein. The posidon of the initiating methionine was supported by the
presence of ups~eaTn in~ rne-stop codons and ~ by 78% homology of the
surrounding nucleotide sequence with the plant consensus sequence that
encompasses the ini~ia~ng methionine, as r~ported by Lutcke et al. (1987) EMBO J
.
6:43~8.
The cDNA cncoding dl~ remainder of Cry j I gene was cloned from
~e linkered cDNA by using oligonucleo~des CP-9 (which has the sequence 5'-
ATGGAITCCCCITGCI~A-3'~(SEQ ID NO: g) and AP (SEQ ID NO: 21) in the
:pnmary PCR reac~on. Oligonucleo~ide CP-9 (SEQ ID NO: 8) encodes amino acids
MetAspSerProCysLeu of ~ y j I (amino acids -21 through -16 of SEQ ID NO: i)
;30 from the leader seguence of C~y j I, and is based on the nuc~eotide sequence
d termined for the partial C7 yj I clone JC76.1.
A secondary PCR reaction was perfonned on 5% of the initial
amplification mixture, with 1 ~g each of AP (SEQ ID NO: 21) and {:P-10 (which
has the sequence 5'-GGGAATTCGATAATCCCATAGACAGC-3')(SEQ ID NO: 9).
WO 93/01213 PCl /US92/0~661
` 21i2913
the nested primer. l~e nucleotide sequence ~'-GGGAAl-rC-3' of primer CP- lO
.~r ~bases 1 throu~h 8 of SEQ ID NO: 9) represent an Eco Rl restric~ion site added fnr
cloning purposes. The remaining oligonuclec)tide sequence encodes amino acids 3-6
of Cr v j I (AspAsnProIleAspSer) (arnino acids 1 through 6 of SEQ lD NO: 1). and is
based on the nucleo~ide sequence determined for the partial Cr~ j 1 clone JC76.1.
The amplified DNA product was puri~ed and precipitated as above, followed by
digestion with Eco RI and Xba I and electrophoresis through a preparahve 1% low
rnelt gel. The dominant DNA band was excised and ligated into M13mpl9 and
pUC19 for sequencing. Again, ligatable material was recovered from cDNA
generated from pollen-derived RNA, and from staminate cone-derived RNA. Two
clones, designated pUC19JC9la and pUC19JC9ld. were selected for full-length
sequencing. They were subsequently found to have identical sequences.
` ~ ~ DNA was sequenced by d~e dideoxy chain ~rmina~ion method
(Sanger et al. supr~) using a commercially available kit (sequenase kit (U.S.
Biochemicals, Cleveland, oHj. Both strands were completely sequenced using M13
orward and reverse primers (N.E. Biolabs, Beverly, MA) aDd internal sequencing
p~ers CP-13 (SEQ ID NO: 10), CP-14 (SEQ ID NO: 11), CP~ SEQ II:) NO:
12), CP-16 (SEQ ID NO: 13), CP-18 (SEQ ID NO: 15)~ CP-19 (SEQ ID NO: 16),
and CP-20 (SEQ II) NO: 17). CP-13 ha~ the sequence 5'-
~20 A~CTAT TACATTGC-3'(SEQIDNO:10). CP-13(SEQIONO:10)encodes
amino acids 82-87 of C~y j I (Me~roMetTyrIleAla, amino acids 82 through 87 of
SEQ ID NO: 1). CP-14 has the sequence 5'-GCAATGTACATAGGCAT-3' (SEQ
ID NO: 11) and corresponds to the non-coding strand sequence of CP-13 SEQ I~
NO: ~10). CP-15 has ~e sequenc~ 5'- TCCAAlTClTCI'C~ATGGI~-3' (~SEQ ID
NO: ~ 12) Yvhich cncodes amino acids 169-174 of C7y j I (SerAsnSerSerAspGly,
amino a~ds 169 ~rough 174 of SEQ II) NO: 1~. CP-16 has the sequence 5'-
TITTGTCAAITGAGGAGT-3' (SEQ ID NO: 13) which is the non-coding strand
~; sequence which cor~Esponds to coding strasld sequence encoding amino acids 335-
. 340 of Cr~ j I (l~rP~oGlnLeuThrLys, amino acids 335 through 340 of SEQ ID NO:
~ 30 ~ 1). CP-18 has the sequence 5'-TAGCAACI~CCAGTCG~GT-3' (SEQ ID NO: 15
which is the non-coding strand sequence which substantially corresponds to codin~
s~and sequence encoding amino acids l 81 through 186 of Cr~ j 1
(ThrSerThrGlyValThr, amino acids lB1 through 186 of SEQ ID NO: l) except that
the fourth nucleotide of CP-18 (SEQ ID NO: 1~) was synthesiæd as a C rather than
~7
1:
WO ~3/01213 ~ PCr/USg2/05661
the correct nucleo~ide, T. CP-l9 which has the sequence 5'-
TAGCTCTCAmGGTGC-3' (SEQ ID NO: 16) is the non-coding strand sequence
which corresponds to codin~ strand sequence encodin~ amino acids 270 through 275of Cry j 1 (AlaProAsnGluSerTyr~ amino acids 270 through 275 nf SEQ ID NO: 1).
CP-20 has the sequence ~'- TATGCAAl~GGl~GGGAGT-3' (SEQ lD NO: 17)
- which is ~e coding strand sequence for arnino acids 251-256 of Cr~ j I
(TyrAlaIleGlyGlySer, amino acids 251 through 256 of SEQ ID NO: 1). The
sequenced DNA was found to have the sequence shown in Figs. 4a and 4b (SEQ ID
NO: 1). This is a composite sequence from the two o~erlapping clones JC 71.6 andO pUC19J9lA. The comple~ cDNA sequence for Cy j I is composed of 1312
nucleo~ides, including 66 nucleo~ides of S untranslated sequence, an open reading
frame s~ing with the co~on ~or an initiating me~hionine, of 112~ nucleotides, and a
3' untranslated region. ~cre is a consensus polyadenyla~ion signal sequence in the
3' unt~nslated r~gion 2~ nucleo~des 5' to ~he poly A $ail. The position of the
ini~aa~ing methionine is confLrmed by the presence of in- frame upstream stop codons
:: and by 78% homology wi~ the plant consensus sequence that encompasses ~e
~- initiating methior~ine (AAAA~ GA (bases 62 through 70 of SEQ ID NO: 1~
found in C~y j I compa~ed with the AAcA~gGc consensus sequence ~or plants,
Lutcke et al. (1987~ EMBO J. ~: 43-48). The open reading frame encodes a protein20: of 374 amino acids of which the first 21 ~nino acids compdse a leader scquence that
is cle~ved from the mature protein. The amino terminus of the mature protein wasiden~fied by compansorl widl the pu~lished NH2-terminal sequence (Taniai et al.
I
(1988) supra) and wi~ scquence determined by direct amino acid analysis of
pu~ifi~ nadve ~ Thc deduced amino acid sequence of the mature protein,
~5 - comprised of 353 amino acids has complete sequence identity with the published
prote~n sequence for C7y j I (Taniai et al. supra3, including the fis~it twenty arninG
acids for thc NH2-terminal and sixtecn contiguous internal amino acids. The mature
pr~tein also contains five potendial M-linked glycosylation sites corresponding to ~e
consensus sequence N-X-SIT.
: 30 :
Example 4
Extrac~don o~ IA from Japanese Cediar Pollen Co}lected in Japan
.
28
WO 93/0121~ PCr/US92/05661
.
21129~3
Fresh pollen collected from a pool of Cryptorneria japonica (Japanese
cedar) trees in Japan was frozen immediately on dry ice. RNA was prepared from
500 mg of the pollen, essentially as described by Frankis and Mascarenhas Ann. Bot.
45:595-599. The samples were ~round by mortar and pesLle on dry ice and
S suspended in 5 ml of 50 mM Tris pH 9.0 with 0.2 M NaCI. 1 mM EDTA, 1~ SDS
that had been treated overni~ht with 0.1% DEPC. After five extractions with
phenoVchloroform/isoamyl alcohol ~mixed at 25:24:1). the RNA was precipitated
from the a~ueous phase with 0.1 volume 3 M sodium ace~ate and 2 volumes ethanol.The pellets were recovered by centrifugation. resuspended in dH~0 and heated to
O 65~ for S minutes. Two ml of 4 M lithium chloride were added to the RNA
preparations and they were incubated o~ernight at 9C. The RNA pellets were
recovered by centrifuga~ion, ~suspended in 1 ml dH20, and a~ain precipitated with
3 M sodium acetate and ethanol overnight. The final pellets were resuspended in
100 ,ul dH20 and stored at -80C.
Double stranded cDNA was synthesized from 8 ilg pollen RNA using
the ~DNA Synthesis Systems kit (BRL) wi~ oligo dT priming according to the
method of Gubler and Hoffman (1983) Gene 25:263-269. Polymerase chain
reac~ons (PCR) were carried out using the GeneAmp DNA Amplifica~ion kit
(Per~in Elmer Cehls) whereby 10 111 lOx buffer con~ing dNl Ps was mixed with
-~0 ~ 1~ pmol each of a sense oligonucleo~ide and an an~-sense oligonucleotide, (lû 111
~ of a 400 ~1 double stranded cDNA reac~ion mix), 0.~ ~ Amplitaq DNA polymerase.
,
and dis~lled water to 100
The samples were amplified with a programmable thermal control~ç~
~ ~ ~ from M~ Research, Inc. (C~ambndge, MA). The ~Irst 5 rounds of amplification
; ~ Z~ consisted OI den~tura~on at 94~ for 1 minute, annealing of primers to the template
at 45C for 1 minute, and chain elongadon at 72~ for 1 minute. The final 20
; ~ rounds of amplificadon consisted of denatura~on as above, annealing at 55C for 1
minute, and elon~adon as aboYe.
~ Seven different Cry j I pnmer pairs were used to amplify the double stranded
cDNA as follows: CP-9 (SEQ. ID #8) and CP-17 (SEQ. ID #14). CP-10 (SEQ. ID
9? and CP-17 (SEQ. ID #14), CP-10 (SEQ. ID #9) and CP-16 (SEQ. ID #13), CP-
- 10 ~SEQ. ID #9) and CP-19 (SEQ. ID ~16), CP-10 ~SEQ. ID #9) and CP-18 ~SEQ.
ID #15), CP-13 (SEQ. ID #10) and CP-17 (SEQ. Il) #14), and CP-13 (SEQ. ID
#10) and ~P-19 (SEQ. ID ~16). CP-17 (SEQ. ID #14) has the sequence 5'-
I ~
,9
:
WO g3/01213
PCr/USg2/0~661
?.~ 9~-~
CCTGCAGAAGCl-rC~TCAACAACGTTTAGA-3' and corresponds to non-coding
strand sequence that corresponds to codin~ strand sequence encodin~ amino acids
SKRC$ (amino acids 350-3S3 and the stop codon of SEQ. ID #1~. The nucleotide
sequence 5'-CCTGCAGAAGCl'r-3' (bases 1 through 13 of SEQ. ID # 14)
S represents Pst I and Hin dIII restriction sites added for cloning purposes. The
nucleotide sequence 5'-TCA-3' (bases 13 throu~h 15 of SEQ. ID # 14) correspond IO
the non-coding strand sequence of a stop codon. All of the amplifications yielded
products of the expected size when viewed on ethidium bromide (EtBr)-stained
~ ~ .
`~ ~ ~ agarose gels. Two of these~primer pairs were used in amplifilcations whose products
-~ 0 were cloned into pUC19 for full-length sequencing. The PCR re~ction with CP-I0
(SEQ. lD #9) and CP-16 (S~Q. ID #13) on the double stranded ~DNA yielded a
band of approximately l.l kb, and was called JC130. A separate first strand cD~Areac~on was done with 8 ~g pollen RNA as described above and amplified with
oligonucleotide primers CP-10 (SEQ. II~ ~9) and CP-17 (SEQ. ID #14). This
amplificadon yiclded a full-lcngtb cDNA. named JC13~, from the amino tenninus ofthe mature protein to dle stop codon.
Amplified DNA was Fecovered by sequential chloroform, phenol, and
chloroforrrl extractions. followcd by precipitadon at -20C with 0.5 volumes of 7.5
~ ammonium acetate and 1.5 volumes of isopropanol. After precipita~on and washing
- W ~ widl~ 70% cthanol. the DNA was blunted with T4 polymerase followed by digestion
Eco ~, in thc case~of JC130, or simultancously digested with Eco RI and Ps
L iI~ thc case of JCl35, in a 15 1~1 reaction and electrophoresed ~rough a preparative
1%~ScaPlaque low mdt gel (FMC). Appropriatc sized DNA bands were visualized
by~ÉtBrstaining, a~cis~d, and ligated into appropriatcly digested pUCl9 for dideoxy
` 25 ~ D~A sequencing by ~e~didcoxy chaIn termination method tSanger et al. (1977)
Proc.; N~tl. Acad. ScL ~ USA 74:5463-5476) using a commcrcially available
sequencing kit (Sequenase kit, U.S. Biochcmicals, Clevcland, OH).
Both suands werc sequenccd using M13 forward and revcrse primers ~N.E.
Biolabs, Bevcrly, ~A) and internal sequcncing primers CP-13 (SEQ. ID #10). CP-
~. , .
15 (SEQ. ID #12), CP-16 (SEQ ID #13), CP-18 (SEQ. ID ~15), CP-l9 (SEQ. ID
#i6) and CP-20 (SEQ. ID #17). Two clones from amplification JC130 ~JC130a and
JC130b) and one clone from amplification JC135 ~JC135g) were found to be Cn j 1
clones upon sequencing. The nucleotide and deduced amino acid sequences of
clones JC130a and JC135g were identical to previously known C~ j I sequence
~ ' :
WO 93J01213 Pcr/US92/OS~61
21129l3
(SEQ. ID #l). Clone JC130b was found to contain a sin~le nucleotide difference
~,~ from the previously known Cr~j I sequence (SEQ. ID ~1). Clone JCl30b had a T at
nucleotide position 306 of Seq. ID ~1 rather than the previously described C. -This
nucleotide chan~e results in a predicted amino acid chan~e from a Tyr to a His at
amino acid 60 of the mature C~ j 1 protein. This polymolphism has not yet been
conf~rmed in an independently-derived PCR clone or by direct amino acid
sequencing. However, such polymorphisms in primary nucleotide and amino acid
sequences are expected.
Examp1e ~
Expr~ssion of Cry j I
E~pression of Cry j I was performed as follows. Ten ~1, of pUCl9JC9la was
diges~d wi~ Xba I, precipitated, then blun~ed with T4 polymerase. Bam HI linkers(N.E. Biolabs, Beverly, MA) were blunt-end ligated to pUCl9JC9la overnight and
excess linkers were removed by filtra~ion ~rough a NACS ion exchange
minicolumn (BRL, Gaithersburg, MD). The linkered cDNA was ~en digested
simultaneously with Eco RI and Bam F~. The ~ryj I insert (extending from the
~0 nucleo~ides encoding the amino terminus of the mature protein through the stop
~; ~ c~on) was isolated by dectrophorcsis of this digest through a 1% SeaPlaque low
mdt agarose gel. The insert was then ligated into the appropriately digested
expression vector pET-1 ld (Novagen, Madison, WI; Jameel et al. (1990) J. Varol.:3963-3966) modified to con~ain a sequence encoding 6 histidines (His 6) . _-~
2~ immediately 3' of ~e AIG initiation codon followed by a unique Eco RI
endonuclease restricdon site. A second Eco RI endonuclease restriction site in the
vector, along with ndghbonng Cla I and Hind m endonuclease restriction sites, had
p~eviously b~en removed by digestion with Eco Rl and Hind ~Jl, blun~ed and
~eligated. The his~dine (His6) sequence was added for a~finity purification of the
~30, recombinant protein (Cry j I) on a Ni2+ chelating column (Hochuli et al. (1987) J.
Chromatog. 411:177-184; Hochuli et al. (1988) Bio/Tech. 6:1321-1325.). A
recombinant clone was used to transform Esc~terichia coli strain BL21-DE3 which
harbors a plasmid that has an isopropyl-l~D-thiogalactopyranoside (IPTG)-inducible
promoter preceding the gene encoding T7 polymerase. Induction with IPTG leads tohigh levels of 17 polymerase expression, which is necessary for expression of the
31
WO 93/û121~
PCr/US92/05661
?,~ 3
recombinant protein in pET-l ld, which has a T7 promoter. Clone pET-l ld~
HRhis6JC9la.d was confirmed by dideoxy sequencing (Sanger et al. Supra) with
CP-14 (SEQ. ID #11) to be a Cr~ j I clone in the correct reading frame for
expression.
; Expression of the recombinant protein was-~onfirmed in an iniual small
culture (~0 ml). An overnight culture of clone pET-} ld~HRhis6JC9la.d was used
to inoculate 50 ml of media (13rain Heart lnfusion Media~ Difco) containing
ampicillin (200 ~lglml), grown to an A600 = 1.0 and then induced with IPTG ( I
mM, final concentration) for 2 hrs. One ml aliquots of the bacteria were collected
) before and after induction, pelleted by centrifugation, and crude cell Iysates prepared
by boiling the pellets for 5 minutes in 50 mM Tris HCl, pH 6.8, 2 mM EDTA, 1%
SDS, 1% ~mercaptoethanol, 10% glycerol, 0.25% bromophenol blue (Studier et:al.,
(1990l A~e~hods in Enzyrn~logy 185:6~89). Recombinant protein expression was
visuali~ed as a band with the predic~ed molecular weight of approximately 38 kDaon a Coomassie blue-stained SDS-PAGE gel, according to the method in Sambrook
- et al.~ supra, on which 40 111 of the crude lysate was loaded. A negative con~ol
consisted of crude lysates from uninduced bactcria containing the plasmid Wit}l C~y j
I ~nd an induced lysa~ ~rom bacteria carrying no plasmid.
The pET-l ld~ HRhis6JC9lad clone was then grown on a large scale ~or
.0~- ~ecombinant protein expression and pu~ification. A 2 ml cul~re bacteria con~aining
the rccombinant plasmid was grown for 8 hr, ~en streaked onto solid media (e.g. 6
petri plates ~100 x 15 mm) with 1.5% agarose in LB medium (G;~BRl.
Gaithersburg, MD) containîng 200 Il~/ml ampicillin), grown to confluence
oven~ight7 then scraped into 9 L of liquid media (Brain Heart ~fusion media, Difc~
containing ampicillin (200 llg/ml). The ca~lture was grown undl ~he A600 is l.û,D~G added (l mM final concentration), and the culture grown i~or an additional 2hours.
Bac~eria was recovered by centrifugation (7,930 x g, 10 min~, and lysed in g0
ml of 6M Guanidine-HCl, 0.1M Na2HP04, pH 8.0 for 1 hour with v;gorous
30' shaking. Insoluble material was removed by cen~i~ugation (11,000 x g. 10 min, 4
C). The pH of the lysate was adjusted to pH 8.0, and the Iysate applied to an 80 ml
Nickel NTA agarose column (Qiagen) that had been equilibrated with 6 M
Guanidine HCl, 100 mM Na2HP04, pH 8Ø 1 he column was sequen~ially washed
with 6 M Guanidine HCl, 100 mM Ma2HPO4, 10 mM Tris-HCl, pH 8.0, then 8 M
.
3~
WO 93/01213 Pcr/US92/05661
21~2913
urea, 100 mM Na2HP04, pH 8.0, and finally 8 M urea, 100 mM sodium acetate, 10
mM Tris-HCI, pH 6.3. The column was washed with each buffer until the flow
throu~h has an A28Q~ 0-05
llle recombinant protein. Cry j 1, was eluted with 8 M urea. 100 mM sodium
acetate, 10 mM Tris-HCl, pH 4.5, and eollected inrlO ml aliquots. Tne protein
concentra~ion of each fraction was determined by A280 and the peak fractions
pooled. An aliquot of the collected recombinant protein was analyzed on SDS-
PAGE according to the method in Sambrook et al., supra.
The first 9 L prep, JCpET-1, yielded 30 mg of Cryj I wi~ approximately
O 78% purity, as detennined by densitometry (Shimadzu Flying Spot Scanner,
Shimadzu SCiGIl~i~lC Instrumen~, Inc., Braintree, MA) of the Coomassie-blue stained
SDS-PAGE; gel. A second 9 L prep prepared the same way, JCpET-2, yielded 41
mg of Cryj I wi~ approximately 77% purity.
,s Example 6
la~ e Cedar Pollen ~ller~ic Patie~ T CeU Studies witll C~ j I ~ P marv
edar Poll~n An'd~en.
~ ~0
T Cell Responses to Cedar Pollen Anti~en Peptides
Peripheral blood mononuclear cells (PBMC) were purified by lymphocyte
separa~on medium (LSM) centrifugation of 60 ml of heparinized blood from ~ '
~2S : ~ ~ Japanese cedar pollen-allergic patients who exhibited clinical symptoms of seasonal
rhinitis and were MAST and/or skin test posi~ve for Japanese cedar pollen. Long
~n T cell lines were established by stimulation of 2 X lO~ PBL/ml in buLk cultures
of comple~ mcdium (RPMI-1640, 2 mM L-glutamine, 100 U/ml penicillin/strepto-
~: mycin, sxlo~5M 2-mercaptoethanol, and lO mM HEPES supplemented with ~2O
: 30, heat inactivated human AB serum) with 20 llg/ml of par~ally purified native Cry j I
(75% puri~ containing three bands similar to ~he three bands in ~;ig. 2) for 7 days at
~; ~ 37C in a hwnidified 5% CO~ incubator to select for Gy j I reactive T cells. This
amount of priming antigen was determined to be optimal for the activation of T cells
from most cedar pollen allergic patients. Viable cells were purified ~y LSM
centrifugation and cultured in complete medium supplemented with ~ units
33
WO 93/0l~l3
PCl /US92/0~66 1
9 ~
recombinant human IL-2/ml and ~ units recombinant human lL-4/ml for up to three
weeks until the cells no longer responded to lymphokines and were considered
"rested". The ability of the T cells to proliferate to recombinant Cry j I (rCrY j I).
purified native Cr~ j 1, or recombinant Amb a 1.1 (rAmb al. 1 ) was then assessed.
For assay, 2 X 104 rested cells were restimulated in the presence of 4 X 104
autologous Epstein-Barr virus (EBV)-transformed B cells (prepared as deseribed
below~ (gamma-i~Tadiated with 25,000 RADS) wi~h 2-50 ~ /ml of rGy; I, puri~led
native Cr~ j I or rAmb a I. 1, in a volume of 200 ~I complete medium in duplicate or
~iplicate wells in 96-well round bonom plates for 2-4 days. The op~imal incubation
was found to be 3 days. Each well then received 1 ,uCi tritiated thymidine for 16-20
hours. The counts incorporated were collected onto glass fiber filter mats and
processed for liquid scintillation counting. Fig. 12 shows the e~fect of vary~ng :
antigen dose in assays with recombinant Cry j I, purified native Cry j I, and
recombinant Amb a I. 1. The results shown in Fig. 12 demonstrate that patient #999
S ~esponds well to recombinan~ Cry j I, and purified native Cry j I, but not to
reeombinant Amb a I. 1. Th}s indica~es that Cryj I T cell epitopes are recognized by
T cells ~rom this particular allergic patient and that rGy j I contains such T cell
epitopes.
0 Prepsra~on ~f (EBV3-trans~ormed B Cells for
Use as Antigen Presenting Cells
Autologous EBV-~ansformed cell lines were y-irradiated with 25,000 Rad
and used as an~igen presenting ceLls in secondary proliferation assays and secondary
~25 butk stimulations. These cell lines wcre also usul as a control in the immuno-
fluorescencc flow cytometry analysis. These EBV-~ansformed cell lines were made
by mcubating S X 106 PBL with 1 ml of }~-59/8 Mannoset cell line (ATCC
~; CRL 1612, American Type Culture Collec~ion, Rockville, MD) condi~ioned medium
in the presence of 1 ~lg/ml phorbol 12-myristate 13-acetate (PMA) at 37C for 60minutes in 12 X 75 mm polypropylene round-bottom Falcon snap cap n~bes (Becton
Dickinson LAbware, Lincoln Park. Nn. These cells were then diluted to 1.25 X 1()6
cells/ml in RPMI-l~0 as described above except supplemented with 10% heat-
inactivated fetal bovine serum and cultured in 200 ~l aliquots in fla~ bottom culture
plates until visible colonies were detected. l hey were then trans~lTed to lar~er
34
WO g3/01213 P~r/us92/05661
2:~12913
wells until the cell lines were established.
Examp1e 7
S
~:~ C~yj I as the MaJor Cedar Pollen Allergen
~ .
To examine the importance of Cr~j I, reported as the major allergen of
~ ~ Japanese cedar pollen, both direct and competihon ELISA assays were perfonned.
- ~0 ~ For the direct ELISA assays, wells were coated with either soluble pollen ex~ct
(SPE) for Japanese cedar pollen or punfied native C~j I (assayed at 90% purity by
protein sequencing) and human lgE antibody binding to these antigens was analyzed.
Pooled human plasma. consisting of an equal volume of plasma from 15 patients
with a Japanese cedar pollen M.AST score of 2.5 or grea~r. and two individual
13 patient plasma samples were compared in this assay. Fig. 5 shows the resul~ of the
~inding seactivity with thcse two antigens. The overall pat~m of binding is verysimilar whe~er ~e coating antigen is SPE (Flg. 5a) or purified native Cry j I (Fig.
Sb)-
In the Gompe~idon æsay, ELISA wells were coated with Japanese cedar
pollen SPE and then allergic paaen~ IgE bi~ding was measured in the presence of
comp~ng purifi~ native Cry; I in solution. The source of allergie IgE in these
; assays was either ~e pool of plasma from lS patients (denoted PHP) or seven
individual plasma samples from patients wi~ a Japanese cedar MAST score of 2.5
or grea~r. The compe~ion assay using the pooled human plasma samples compares
25~ e comp~tihve binding capacity of purifted nadve Cryj I to Japanese cedar pollen
SPE and an i~e]evant aliergen source, Iye grass SPE. Pig. 6 shows dle graphed
r esults of the competition ELISA with pooled human plasma. The concen~ation of
protein present in ~he Japanesc cedar pollen SPE is approximately 170 times greater
at oach competing point than is the purified na~ive Gy; I . From th~s analysis it is
~ 30i clear ~lat the purified native Cryj I competes vely well for lgE binding to the~whole
- ~ ~ range of proteins present in the Japianese cedar pollen soluble pollen extract. This
~ ,~
implies that mos~ of the an~-Cry j I IgE reac~ivi~ is direc~d a~ainst purified native
Cr~j I . The negative control shows no specific competitive ac~ivity and the
competing SPE in solution can completely remove binding to the coated wells. This
WO 93/0121~ PCI/llS92/05661
assay was repeated with individual patients as a measure of the range of the lgEresponse within the allergic population. Fi~. 7 shows this ~esult where the
competi~ion of binding to SPE was perfo~ned with purified native Cr~ j I . The
results demonstrate that a1though the patients show different dose response to
i Japanese cedar pollen SPE, each of the seven patients' IgE bindin~ to ~apanese cedar
pollen SPE could be competed with purified native Cry; I. The implications of
these dat~ are that for each patient the IgE reactivity directed a~ainst Cry j I is
predominant but that there is variation in this reactivity be~ween patients. Theoverall conclusion is that these data support the previous findings (Yasueda et al
: ) (1988) supra) that C~yj I is the major allergen of Japanese cedar pollen.
The reactivity of IgE from cedar pollen allergic patients to the pollen proteinsis dramatically reduced when these proteins are denatured. One method of analyiing
~is property is through direct binding ELISA where the coating antigen is the
Japanese cedar pollen SPE or denatured Japanese cedar pollen SPE which has been
S denatured by boiling in the presence of a reducing agent Dl r. This is ~en
examined with allergic patient plasma for IgE binding reactivity. Fig. 8a, shows the
;~ direct binding assay to the SPE wilth seven individual plasma samples. ~ Fig. 8b,
the binding results with the denatured SPE demonstrates the marked decrease in
reactivity following ~is trea~nent. To determine ~e extent of Gryj I binding to the
~0 ELISA wclls, Gy; I was detected with a rabbit polyclonal an~sera against the Amb
a I & II pro~ amily. Tbese proteins have high sequence idend~y (46%) with
d d~is antisera can be used as a cross reactive antibody de~ection system. In
conclusion, ~ese data demonstra~ a marked loss in IgE reac~ivity following
denatura~on of the SPE.
: :
~7,~
;nple 8
IgE Rea~ r and Histam~ne Release Analysis
i0 ' The recombinant C~y; I protein (rCr~ j I), expressed in bacteria and then
purified (as described in Example 5), has been examined for IgE reac~ivity. The first
method applied to ~is examination was direct ELISA where wells were coated with
the recombinant Cryj I and IgE binding was assayed on in-1ividual patients. Fig. 9
is the graphic r~presenta~ion of this direct ELISA. The only positive signals on this
36
W~ J(~121.~ 1~Cr/lJS92~05~6l
~ l I 2 !~
data ~c~ are from thc ~wc) control antisera rabbit polyclonal anti-Amb a I & II (Rabbit
smti-Amh a I ~ Il) nncl C~T3P2, a monQclonal antibody rai~cd again~t An~7 ~ I that
.~,~
cro~ rellcl~ with ~r~j I . By lhi~ melho(l all patients ~stul ~howcd no I~E
reactiYity with the rec()mbitlant Cr~
i Another meth()(J of Imalysis that was applied lO the ~x~mination of I~E
r~activity to the r~combinant Cryj I wns a capture ELISA. This analysis relics on
th~ u~e of a deflned anitibl)dy~ in this cu~;e CBF2 to bind the anti~en and allow fnr
th~ b~n~lin~ of antib~)llies to other epitope si~es rhe f(~nnat of this capture ELISA is
1~ we}ls arc ~oatcd with MA~ CBF2, ~) andgcn or PBS (as on~ type of ne~ative
ct)n~ol~ i~ add~d and capturcd by specific interaction with the coated MAb, 3) either
the contr~l smtibody anti-Amb ~ Fig. IOb) or human ~ rgic plasma (Fig.
~ lOa) is added as thc detecting ~ntibody. and 4~ d~tcction of antibody binding is
-~:: as~nycd. ~ a and 1(3b are thc graphed ~sul¢s of these assays. For the IgE
lysi~, tha pooled human pl~sma (15 patients) was uscd. The conclusion from
: ~ lhcs~ ~sul~ i~ that therc i~ no indication of any s~ciflc binding of human allergic
~ r~ry; I by ~his mcth~d of analysis. Howcver. thc ~apturc of rCryj 1 works
ev(dGn~¢d by the cont~l antibo~y binding curve, shown in Fi~. lOb l~he lack of
bindin~ to rCryj I may be due to absencs of carbohydra~ or an~r other post-
~ n~la~onal rnodification and/or tha~ the major~ty of I~E c2rlnot react with
O d~n~tu~ Cry r I. ~S~, comp~tidon ELISA and Wcstcrn blotung da~ also
dt~ OnS~l~eS no ~peclf~c IgE rc~civity to the rCr~ da~ not shown).
A histaminc rele~s~ assay was performed on one Japan~se cedar pollen
pad~nt us~ng Japaslcse ~dar pollcn SPE. pur~lçd nativa Cry j I and rCr~ j I
addcd antl~cnsO This assay is fl mcssur~ of I~E ~activ~ty sh~u~h hum~n
basophil m~di~tor s~lease. T~e r~ul~s of this assay~ sh~w~ in Fi~. 11, demonstratc
:~ ~n~ h~stamine r~lea~ wi~ both purifiçd na~ve Cry; I and the Japanesc c~ar
llen SPE over a wido conccntration range. Thc only point whes~ re is any
m~urable histamino rcleasc with the C~; I is at ~ hi~hest COnCentratiQn, 50
/ml. Two possible explanations for this ~lcaso by thc r~ spacific
lo ~c~tivity with a vcry low propor~ion of the and C~ j 1 I~E capable of ~cognizin~
~he resombinant form of ~ryj I, or 23 non-spccific releasc ~aused by l~w abundance
of bac~rial conl~rninants ob~erved only at the hi~hcst an~i~en conccntration~ Thus
~ar, this result has only becn shown in a single paticnt. In additi~n. the da~ shown
rom sin~le da~ pc3ints at c~eh protcin conc~ntration
?I?
W~ 93/01213 ,~ PCr/US9~/0~661
c~ 9~'
lt may be possible to use this recombinantly expressed cr~ j 1 protein for
immunotherapy as ~. coli exp~essed material has T cell reactivity (Exarnple 6). but
does not appear to bind I~E from Cr~tpomeria japonicQ atopes nor cause histaminerelease from the mast cells and basophils of such atopes in l~itro. Expression of rCr~
i j 1 which is capable of binding I~E could be achie~ed in yeast. insect (baculoYirus)
or mammalian cells (e.g. CHO. human and mouse). A rcr)~j I capable of actively
binding Igl~ may be important for the use of recombinant ma~erial for diagnosticpurposes.
Although the invention has been described with reference to its preferred
embodiments, other embodiments, can achieve the sarne results. Variations and
modifications to the present invention will be obvious to those skilled in the art and
it is intended to cover in the appended claims all such modification and equivaler ts
and follow in the true spirit and scope of this invention.
;,
WO 93/01213 PCr~US92/0~661
2112~ 13
SEQUE.'~1CE LlSTlN'G
I l ! GE~ERAL ~ FOR~1 ATlO~:
i) APPLICAl\T: Griffith~ Irv~in J.
P()llt)~ J()anne. B()nd Julian
(ii) TITLE OF INVENTiON: Aller~enic Prntein.c An~ Peptide~ From
Japanese Cedar Pollen
(iii) NUMBER OF SEQUENCES: 25
(i~ ) CORRESPONDENCE ~DDRESS:
~A) ADDRESSEE: ImmuLo~ic Pharm~ceutical Corporation
(B) STREET: One Kendall Square, Building 600
(C) ClTY: Boston
(1:)) STATE: MA
(E) COUNTRY: USA
~0 (F) ~;IP: 0213~
, ~ ~
(v~ COMPIJlER READABLE FORM:
(A) MEDrllM lYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
'5 (C) OPERATINC~ SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: PatentIn R~lease #1.0, Vcrsion ~1.''~
(vi) CU~NT APPLICATION DATA:
(A) APPLICATION NUMBER:
~` ~ 30 (B) FILINGDATE: ._.
C) CLASSIFICATION:
(viii) ATTORNEY/AGEN-r ~FORMATION:
:~: : : (A~NA~:StaceyL. Channing
(B) REGISTRATION NUMBER: 31,095
C) REFERENCE/DOCKETNUMBER: I:PC-02~CPCT
~ix) TELECOMMUNICATION ~FORMATION:
' (A) lELEPHONE: (617) 494-0060
~B) TELEFAX: (617) 494-4964
:
lFORMATION FO~ SEQ l:D NO:I:
WO ~3/01213 Pcr/US92/05661
9~3
(i) SEQUENCE CHARACTERlSTICS:
(A) LENGTH: 1337 base p~ir~
(B) TYPE: nucleic acid
(C? STRANDEDNESS: ~in~le
s (D) TOPOLOGY: line~r
~ii) MOLECVLE TYPE: cDNA tO mRNA
(vi) ORlGINAL SOURCE:
In (A) ORGANISM: Crvtpomeria japonica
(ix) FEATURE:
(A) NAME/KEY: CDS
~: ` (B) LOCATION: 66.. 1 187
(ix) FEATURE~:
(A) NAME/KEY: ma~_pephde
(B) LOCATION: 1 29.. 11 87
20 -
~ (xi ) SEQUENCE DESCRIPTION: SEQ ID NO :1:
,.
AGTCAATCTG CTCATAATCA T~GCATAGCC GTATAGAAAG AAATTCTACA CTCTGCTACC 60
25 AAAAA ATG GAT TCC CCT TGC TTA GTA GCA TTA CTG GTT TTC TCT TTT 107
~et Asp Ser Pro Cys Leu Val Ala Leu Leu Val Phe Ser Phe
21 -20 -15 -10
GTA ATT GGA TCT TGC TTT TCT GAT AAT CCC ATA GAC AGC ,GC TGG AGA 155
30 Val Ile'~ly Ser Cys Phe Ser Asp Asn Pro Ile Asp Ser Cys Trp Arg
-5 1 5
:G5A GAC TCA AAC TGG GCC CAA AAT AGA ATG AAG CTC GCA GAT TGT GCA 2G~
Gly Asp Ser Asn Trp Ala Gln Asn Arg Mgt Lys Leu Ala Asp Cys Ala
10~ : 15 20 25
TG GGC TTC GGA AGC TCC ACC ATG GGA GGC AAG GGA GGA ~A~ CTT TAT 251
Val Gly Phe Gly Ser Ser Thr Met Gly Gly Lys Gly Gly Asp Leu Tyr
~-: 30 35 q
ACG:~GTC ACG AAC TCA GAT GAC GAC CCT GTG AAT CCT GCA CCA GGA ACT 29
Thr Val Thr Asn S~r Asp Asp Asp Pro Val Asn Pro Ala Pro Gly Thr
45 CTG CGC TAT GGA GCP ~CC CGA GAT AGG CCC CTG TGG ..T~. ATT TTC AGT 347
Leu Arg Tyr Gly ~la Thr Arg Asp ~rg Pro Leu Trp Ile Ile Phe Ser
GGG AAT ATG AAT ~T~. AAG CTC AAA ATG CCT ATG TAC ATT GCT GGG T~.T 395
50 Gly Asn Met Asn Ile Lys Leu L~ys Met Pro Met T~r Ile .~la Gly Tvr
: 75 ~0 ~5
A~G hCT TTT GP.T GGC A.-G GG.} GCA CA~ GTT TAT ATT GG~ .T GGC GGT 44
Lys Thr Phe ~sp Gly ~rg Gly .la Gln -~;21 ~j~- Ile Gly Asn Gly Gl~
4()
WO 93/1~1213 PCl/US92/0~661
` 2~ ~291:~
gQ 95 l~G ^~
CCC TGT GTG TTT ATC AAG AGA GTT AGC A~T GTT ATC ATA CAC GGT TTG 491
~F ~s Val Phe Ile Lys Arg Val Ser Asn Val Tle Ile His Gly Le~
llO 115 12G
TAT CTG TAC GGC TGT AGT Ar T AGT GTT TTG GGG ~T GTT TTG ATk AkC S39
T~r Leu ~r Gly Cys Ser Thr Ser Val Leu Gly Asn Val Leu Ile Asn
125 130 '~5
GAG AGT TTT GGG GTG GAG CCT GTT CAT CCT CAG GAT GGC G~T GCT CTT 587
Glu Ser Phe Gly Val Glu Pro Val His Pro Gln Asp Gly Asp ~la Leu
140 145 150
15 ACT CTG CGC ACT GCT ACA AAT ATT T~G ATT GAT C~.T AAT TC* TTC TCC 635
Thr Leu Arg Tnr Ala Thr Asn Ile Trp Ile Asp His Asn Ser Phe Ser
155 160 165
AAT TCT TCT GAT GGT CTG GTC GAT GTC ACT CTT ACT TCG ACT GGA GTT 683
20 Asn Ser Ser Asp Gly Leu Val Asp Val Thr Leu Thr Ser Thr Gly Val
170 175 180 185
ACT ATT TCA AAC AAT CTT TTT TTC AAC CAT CAT A~A GTG ATG TTG TTA 731
Thr Ile Ser Asn Asn Leu Phe Phe Asn His His Lys ~al Met Leu Leu
23 190 19~ 200
GGG CAT GAT GAT GCA TAT AGT GAT GAC AAA TCC ATG AAG GTG ACA GTG 779
Gly His Asp Asp Ala Tyr Ser Asp Asp Lys Ser Met Lys Val Thr Val
205 210 215
~: GCG TTC AAT C~A TTT GGA CCT AAC TGT GGA CAA AGA ATG CCC AGG GCA 827
Ala Phe Asn Gln Phe Gly Pro Asn Cys Gly Gln Arg Met Pro Arg Ala
220 225 230
35 CGA TAT GGA CTT GTA CAT GTT GCA AAC AAT AAT TAT GAC CCA TGG ACT 875
Arg Tyr Gly Leu Val His Val Ala Asn Asn Asn Tyr Asp Pro Trp Thr
235 240 245
ATA TAT GCA ATT GGT GGG AGT TCA AAT CCA ACC ATT CTA AGT G~A GGG 923
40 Ile Tyr Ala Ile Gly Gly Ser Ser Asn Pro Thr Ile Leu Ser Glu Gly
250 : ~ 255 260 265
AAT AGT TTC ACT GCA CCA AAT GAG AGC TAC AAG AAG CAA GTA ACC ATA 971
A~n:Ser Phe Thr Ala Pro Asn Glu Ser Tyr Lys Lys Gln Val Thr Ile
270 275 280
CGT ATT GGA TGC AAA ACA TCA TCA TCT TGT TCA AAT TGG GTG TGG CAA 1019
: Arg Ile Gly Cys Lys Thr Ser Ser Ser Cys Ser Asn Trp Val Trp Gln
: : 285 290 295
5~ :
TCT ACA CAA GAT GTT TTT TAT AAT GGA GCT TAT TTT GTA TCA TCA GGG 1067
Ser Thr Gln Asp Val Phe Tyr Asn Gly Ala Tyr Phe Val Ser Ser Gly
300 30~ 310
55 AAA TAT GAA GGG GGT AAT ATA TAC ACA AAG AAÆ GAA GCT TTC PP.T GTT l11S
Lys Tyr Glu Gly Gly Asn Ile Tyr Thr Lys Lys Glu Ala Phe Asn Val
315 320 325
GAG hAT GGG ~ÆT GCA ACT CCT C~ TTG ACA ~ AT GCT GGG GTT TTA 1163
60 Glu Asn Gly Asn ~la Thr Pro ~ln L~u Thr Lys ~.sn Ala Gly Val Leu
330 335 34C 3
~1
WO 93/01213 PCT/~JS92/05~61
9~ ~
hCr T~7C TCT CTC TCT ~ ~ CGT TGT TG~TGhTGC~ T~T.7.TTCTrG CATGTTCTA~ 121
Thr Cys Ser Leu Ser Lys Arg Cys
350
T-'-m--Tr '.TT . ~C' ~17~_r~r; ,b~G,~ ,",,?, T~ ,mJ.~,T~ m--mr.7 ~
~TA~7iTG T.~T-TTTT~C TP.TT;rr~'_:. r'__'_ T~.T~ ~ T^GG _G^. T..C^T_TAG.. ~
In
(2) I~ORMATION FOR SEQ I~ NO:2:
(i) SEQUENCE CHARACTERISTICS:
15(A) LENGTH: 374 amino acids
(B) TYPE: amino acid
~D) TOPOLOGY: linear
(ii) MOL~CULE TYPE: protein
(xi) SEQUENCE ~ESCRIPTION: SEQ ID NO:2:
Met Asp Ser Pro Cys Leu Val Ala Leu Leu V21 Phe Ser Phe V~l Iie
-21 -20 -15 -10
Gly Ser Cys Phe Ser Asp Asn Pro Ile Asp Ser Cys Trp Arg Gly Asp
-5 1 5 10
o Ser Asn Trp Ala Gln Asn Arg Met Lys Leu Ala Asp Cys Ala Val Gly
3015 20 25
Phe Gly Ser Ser ~,hr Met Gly Gly Lys Gly Gly Asp Leu Tyr Thr Val
35 Thr Asn Ser Asp Asp Asp Pro Val Asn Pro Ala Pro Gly Thr Leu Arg
: Tyr Gly Ala Thr Arg Asp Arg Pro Leu Trp Ile Ile Phe Ser Gly Asn
60 65 70 '5
Met~Asn Ile Lys Leu Lys Met Pro Met Tyr Ile Ala Gly Tyr Lys Thr
80 85 90 .
Phe A;sp Gly Arg Gly Ala Gln Val Tyr Ile Gly Asn Gly Gly Pro Cys
4~95 100 105
Val Phe }le Lys Arg Val Ser Asn Val Ile Ile ~is Gly Leu ~yr Leu
110 115 12~
5~ Tyr Gly Cys Ser Thr Ser Val Leu Gly Asn Val Leu Ile Asn Glu Ser
lZ5 130 135
Phe Gly Val ~lu Pro Val His Pro ~ln ~sp Gly .~.sp Ala Leu Thr Leu
~5 140 145 150 155
Arg Tnr ~la Thr Asn Ile Trp I1e Asp His Asn Ser Phe Ser Asn Ser
- 160 165 1/0
Ser hsp Gly Leu val aSp Val Tr.r Leu Thr Ser ~hr Gl~f Val Tn~ Ile
60 1~ ~80 1~5
WO93/0121~i PCT/US92/0566l
21i29:13
Ser As.~ Asn Leu Phe Phe ~sn ~lS Hic ' j'S ~ . Leu Leu Gl~ s
190 195 ~00
~,~ ~sp .hsp ~la Tyr Ser A_p ~.sF Lys Se- M~ c ;~al Trr ~J~l .'.la ~;r~C
205 10 ,;~
~sn Gln ?he Gl~ Pro ~.sr. C;~s Glj~ Gl.. .~ t Pr~ .-.r ~.ia ~rg ~ r
~ -- ~ 230 _~5
10 Gl)~ Leu Val His Val Ala Asn Asn ~s~ ~r Asp Pro Trp Thr Ile ~r
24Q 245 250
Ala Ile Gly Gly Ser Ser Asn Pr~ ~hr Ile Leu Ser Glu Gly Acn Ser
255 260 265
Phe Thr ~la Pro Asn Glu Ser ~fr Lys Lys Gln Val Thr Ile Arg Ile
270 275 280
Gly Cys Lys Thr Ser Ser Ser Cys Ser Asn Trp Val Trp Gln Ser Thr
285 29~ 295
Gln ASD Val Phe Tyr Asn Gly Ala Tyr Phe Val Ser Ser Gly Lys ~yr
300 305 310 315
25 Glu Gly Gly Asn Ile Tyr Thr Lys Lys Glu Ala Phe Asn Val Glu Asn
320 325 330
: ~ Gly Asn Ala Thr Pro Gln Leu Thr Lys Asn Ala Gly Val ~eu Thr Cys
335 340 345
~ Ser Leu Ser Lys Arg Cys
:: 3~0
::
(2) INFORMATION FOR SEQ ID NO:3:
:~ 35
(i) SEQUENCE CHARACTERISTICS:
tA) LENGTH: 17 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
~ (D) TOPOLOGY: linear
: ~X1) SEQUENCE DESCRIPTION: SEQ ID NO:~:
` 45.
GAYAAYCCNA THGAYWS
(2) INFO~MATION FOR SEQ ID NO:4:
(i) SEQUENCE CHARACTERISTICS:
~A) LENGTH: 25 base pairs
(B) TYPE: nucleic acid
: (C) STRANDEDNESS: single
: (D) TOPOLOGY: linear
~ ~ 55
4~i
WO93,'0121~ PCT/US92/05661
J~
(~i) SEQ'JENCE DESCRI?TIOI;: SE2 lv I~O:~:
GGGAATTCAA YTGGGCNCAR AAYSG 25
_,~
~) INFORM~.TION FOR SEQ I~ NC~
(i) SEQUENCE CHAR~:CTERISTTCS-
(A) LENGTH: 23 base pairs
lB) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
: (ix) FEATURE:
(A) NAME/KEY: modified_base
(B) LOCATION: l5
(D) OTHER INFORMATION: /mod_base= i
.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:5:
CTGCAGCCRT TYTCNACRTT RAA 23
(2) INFORMATION FOR SEQ ID NO:6:
25:
- ~i) SEQUE~CE CHARACTERISTICS:
(A) LENGTH: 20 base pairs
(B) TYPE: nucleic acid
~C) STRANDEDNESS: single
(D) TOPOLOGY: linear
: ~ ~ix) FEATURE:
~ : : (A) NAME/KEY: modified_base
- 35 ~B) LOCATION: 6 ~.
~D) OTHER INFO~M~TION: /mod_base= i
(xi~ SEQUENCE DESCRIPTION: SEQ ID NO:6:
:~, ~
;~ TTCATNCKRT TYTGNGCCCA 20
: - :
12~ INFORM~TION FOR SEQ ID NO:7:
:~ 45 . (i~ SEQUENCE CHARACTERISTICS:
(A) LENGTH: 25 base pairs
(B) TYPE: nucleic acid
(C~ STRANDEDNESS: single
~D) TOPOLOGY: line~r
;
W~9~/01213 PCT/~S92/036~1
2112913
(xi) SEQUENCE ~E~C~IPTIOl~: SEQ I~ :7:
CCTGCAGCKR TTYTGNGCCC ~ARTT 25
~,~
t2i INFORMATION FOR SEQ ID NO:8:
(i) SEQUENCE CHAR~ TE~ISTI.~S:
IA) LENGT~: 18 bâse pairs
(B) TYPr.: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:8:
ATGGATTCCC CTTGCTTA l8
(2) INFORMATION FOR SEQ ID NO:9:
~0
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 26 base pairs
~B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:9:
: GGGAATTCGA TAATCCCAT~ GACAGC 26
~ (2) I~FORMATION FOR SEQ ID NO:l0:
;~: 35 (i) SEQUENCE CHARACTERISTICS: .~--
` (A)-LENGTH: 17 base pairs
(B) TYPE: nucleic acid
; ~C) Sl1RANDEDNESS: single
tD) TOPOLOGY: lin~ar
~xi) SEQUENCE DESCRIPTION: SEQ ID NO:l0-
45 ATGCCTATGT ACATTGC l7
(2) INFORM~TION FOR SEQ ID NO:ll:
(i) SEQUENCE CHA~ACTERISTICS:
(A) LENGTH: 1? base pairs
(B) TYPE: nucleic acid
(C) STR~D~DNESS: single
WO93/01213 PCr/US92~0~661
19~
~D) TOPOLOG~: iinear
_r
~Xi) SEQUENCE DESCRIPTION: ~EQ ILI~
G^~h.TGT~CA TAGGCPT
~2) INFORMATION FOR SEQ ID NO:12:
(i) SEQUENCE CHARACTERISTICS:
~A) LENGTH: 18 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
1S (D ! TOPOLOGY: linear
(Xi) SEQUÉNCE DESCRIPTION: SEQ ID NO:12:
TCCAATTCTT CTGATGGT
(2) INFORMATION FOR SEQ ID NO:13:
25 ~ ~i) SEQUENCE CHARACTERISTIC~:
: (A) LENGTH:~18 base pairs
(B) TYPE: nucleic acid
M : ~ ) STRANDEDNESS: single
(D) TOPOLOGY: linear
: ~ ~
~ ~ ,
: (Xi) SEQUENCE~DESCRIPTION: SEQ ID NO 13
35 TTTTGTAAT:TGAGGAGT ~
(2~) INFORMATION~FOR SEQ ID NO:14:
):SEQ~ENCE: CHARACTE~ISTICS:
: (A)~ LEN~TH::: 3Q base pairs
B) :TYPE~:~nucleic acid
; ; (C:):~STRANDED~ESS: single ~ :
D) TOPOLOGY: linear
` ~ ;45
: , : ~ : :: :
~ ::(xi) SEQUENCE~DESCRIPTION: SEQ ID NO:14: :
:
,
CCTGCAGAAG CTTCATCAAC ~CGTTTAGA ~:
50~
2~ INrORMATION FOR SEQ ID NO:15:
.
~ 46
:
:
WO~3~01213 PCT~S~2~Q~661
2112~13
(i) SEQUENCE C~ARACTERISTICS:
(A) LENGT~: l9 base pairs
(B) TYPE: nucleic acid
~C) ST~NDEDNESS: single
~) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:15:
TAGCAACTC AGTCGAAGT
i2) INFORMATION FOR SEQ ID NO:16:
::
; ~ ~ 1:5 ~ (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 17 base pairs
` ~B) TYPE: nucleic acid
C1 STRANDEDNESS: single
(D): TOPOLOGY: linear
: 20~
xi) ~SEQUENCE~DESCRIPTION: SEQ ID NO:16:
T~CTCTCAT TTGGTGC~
(;2) INFO~MATIO~FOR~SEQ ~D NO:17:
(i) SEQUENCE~;CH~RACTERISTICS:
30:~ :(A~ LENGTH: ~8:base pairs
:(B~: TYPE:::nucleic acid
:(C)~S ~ EDNESS: single :~
:(D~ TOPOLOGY: linear:~ ~
(x~ ;SEQ~ENCE~DESCRIPTION~ SEQ:ID~NO:~17.
TATGCP~ATTG ~G~GGGAGT~
(2~)~INFoRMATIoN~:FoR~sEQ ID~NO:18
i1 SEQUENCE~CHAR~CTERISTICS ~
r ~ ENGTH 20 amino acids
:45 ~ ; (B~ TYPE: amino acid
D)~TOPOLOGY~: ~linear~
MOLECULE;~TYPE- peptlde~
50~ (:v) FRAGMENT~TYPE: N-te~rmlnal
(Vl ): ORIGINAL~SOURCE~
W O ~3~01~1~ PC~/US92/05661
IA) ORG~'IS~.: Cr~ptom~rl â ~z~o~.icc
~ ix ) ~E.~ 'rrJp~ - -
_3_.-.~lC.
7 ~^ Ser, Cyc, ?.~ r ~lc~
..
: 10 (xi) SEQUrI~C~ DESCRIPTIr~1;: SE~ 8:
Asp Asn Pro 11e ~.sp Ser ~:aa Tr~ ~.rg Gly ~.sp ~ sn Trp ~1~ Gln
Asn Arg Met Lys
: ~j 70
) INFORMATION FOR SEQ ID NO:l9:
~i) SEQUENCE CHARACTERISTICS:
: IA) LENGTH: 16 amino acids
(B) TY?E: amino acid
D)~ TOPOLOGY:~:linear
(ii) MOLECULE;TYPE: peptide
(v) FRAGMENT TYPE: internal
Ivi) ORIGINAL SOURCE:
30~ (A) ORGANISM:~Cryptomeriâ japonica
(xi3 SEQUENCE~D SCRIPTION: SEQ ID NO:l9:
:35~ Glu Ala Phe Asn~Val Glu Asn Gly Asn Ala ~.hr Pro Gln Leu Thr Lys
2:) INFOF~ATIQN FOR~SEQ ID NO.20:
SEQUENCE~CXARACT RISTICS:
A)~ ~ENGT~ 30~ase pairs ~ ~ : :; : -
B)~: TYPE::~ nucl~ic acid :~
C)-~STR~NDEDNESS: single
45~ D)~TOPOLOGY:~1i~e~
(xi)~ S~QUEI~CE ~SCRI~PTION~: SSQ ~D~MO:~20
CGG~CT~.GAGIGT~CCGT~CG ATCGATC~TT
W~93/01213 PCT/US92~661
2~12~1 3
(2) INFORMATION FOR SEQ ID NO:2l:
-~ (i) SEQUENCE CHARACTERISTICS:
(A~ LENGTH: 20 base pairs
: 5 (B) TYPE: nucleic acid
(C~ STR~NDEDNESS: single
(D~ TOPOLOGY: linear
(xi) SEQUE~C~ DESCRIPTION: SEQ ID NO:21:
GGGTCTAGAG GTACCGTCCG
15 (2) INFORMRTION FOR SEQ ID NO:22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 base pairs
: :- (8~ TYPE: nucleic acid
~C) STRANDÉDNESS: single
. (D) TOPOLOGY: linear
, :
xi3 SEQUXNCE DESCRIPTION: SEQ ID NO.22:
AAT~ATCGAT GCT
(2) INFORNATION FOR SEQ ID NO:23:
30~
(i) SEQUENCE C ~ CT RISTICS:
tA) LENGTH:: 21 base pairs
: (B) TYPE:: nucleic acid
(C~ STRANDEDNESS: single
;35~ D) TOPOLOGY. llnear
; (xi:)~SEQUENCE DESCRIPTION: SEQ ID NO:23:
GGAATTCT~T AG~TGCAGG T :
(2) INFORM~TION FOR SEQ ID NO:24:
: 45 ~i) SE5~UENCE ~CHARACTERISTICS:
(A) LENGTH. 35 base pairs:
B ) TYPE:: nucleic acid
C) ST~ANDEDNESS: sing~ e
D ) TOPOLOGY: l inear
: ~ ~
::
:: : : :
~ 49
: :
W~ 93/01213 PCI/US92/OS661
(xi ) SEQUENCE DESCRIPTION: SEQ ID NO: 24:
GG~.TTCTCT .Z~G~CTGCAG'' T. TTTTT T T~ TTTT~
2 ) INFORMATTON FOR SEr) IO l~C: ' 5:
( i ) SEQUENCE CHP.RZ~CTERISTICS:
(A) LENGTH: 5 amino acids
(B~ TYPE: amino acid
; 10 ~D) TOPOLQGY: l1near
( i i ) MOLECULE TYPE: peptide
~ ~ (v) FRAGMENT TYPE: N-terminal
: 15
~vi ) ORIGINAL SOURCE:
; : (A) ORGANISM: Juniperus sablnoldes
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 25:
,
~ : Asp ~sn Pro:Ile:~Asp
: