Note: Descriptions are shown in the official language in which they were submitted.
-
. CA 02213622 1997-08-22
W O 96127026 1~~ .'02380
DETECTION OF BIOMOLECULES
FIELD OF THE INVENTION
The present invention concerns a mcthocl and kit for thc detection
of the presence of catalytically active ribozymes in a medium. The method
and kit of the invention may be useful within the framework of a method
5 and kit for the detection of the presence of specific biomolecules in a test
sample.
BACKGROUND OF THE INVENTION
Detection of the presence of specific biomolecules, such as DNA
10 or RNA sequences, proteins, anti~ens, antibodies, etc., in a sample is
required for a variety of expcrimental, diagnostic and therapeutic purposes.
A multitudc of assays are available for detecting proteinaceous biomolecules
such as gel electrophoretogram, HPLC, affinity chromatography, as well as
other assays which are performcd by use of an appropriately labelled probe.
1~ While such assays are satisfactory where the proteinaceous biomolecule to
be detectcd is presellt in sufficicntly large quantitics, they are at times not
sensitive enough to allow dCtCCtiOII of minute quantities of biomolecules.
SUBS 111 UTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96/27026 PCTnUS96102380
DNA or RNA sequences can be detected by the use of a labelled
probe. Where the DNA or RNA sequences to be detected present in only
very small amounts, they have to be amplified by methods such as LCR
(ligase chain reaction), SSR (self-sustained sequence replication) or PCR
5 (polymerase chain-reaction).
Although amplification methods such as PCR have had an
extremely high impact on basic research, they have been slow in making the
transition to the clinical setting. The primary reason for this is that the
requirement for automation combined with the clinical environment of the
10 samples, have yielded processes that are complex, slow and expensive. The
need for protein enzymes with their high sensitivity to environmental factors
necessitates a very controlled environment in which they are to operate.
Typically, a clinical sample contains many components that can interfere
with the enzyme's ability to perform its catalytic activity. In addition, the
15 standard methods that are used for sample preparation to release the nucleic
acids, such as Guanidine thiocyanate or Phenol extraction are unsuitable for
protein bascd enzymatic activity and it is thcrcforc nccessary to rcmove the
target nucleic acid from sample preparation.
Ribozymes are typically RNA molecules having enzyme-like
20 catalytic activities that are usually of cleavage, splicing or ligation of nucleic
acid sequences. The known substrates for ribozymes are RNA molecules
although there have been some indications that ribozymes may act on DNA
molecules and on proteins.
Natural ribozymes which participate in intracellular reaction work
25 in cis, catalyzing only a single turnover, and are usually self-modified
during the reaction. However, ribozymes can be engineered to act in trans,
in a truly catalytic manner, with a turnover greater than one and without
being self-modified. Two distinct regions can be identified in a ribozyme:
the binding region which givcs the ribozyme its specificity through
SUBSTITUTE SHEET (RULE 26)
. CA 02213622 1997-08-22
W O 96n7026 PCTAUS96/02380 - 3 -
hybridization to a specific nucleic acid sequence (and possibly also to
specific proteins), and a catalytic region which gives the ribozyme the
activity of cleavage, ligation or splicing. Each class of ribozymes cleaves
a dirr~ t sequence of nucleotides using a ~ tinct mech~ m of action.
S Each class is further distinguished by the number of nucleotide bases that
are essential for its catalytic activity and by the degree of the specificity ofthe ribozyme and the target sequence (Robert H. Simons, Annual Revie~
of Bioche~zistry, 61, pp. 641-671, (1992)).
It has recently been proposed to use ribozymes in order to treat
10 ~ e~ces or genetic disorders by cleaving a target RNA, such as viral RNA
or messengcr RNA transcribcd from gcncs that should bc turned off. This
method is proposed as an alternative to blockage of the RNA transcript by
the use of antisense sequences. Owing to the catalytic nature of the
ribozyme, a single ribozyme molecule cleaves many molecules of target
15 RNA and therefore therapeutical activity is achieved in relatively lower
concentrations than those required in an antisense treatment (WO 96/ 3569).
The use of ribozymes for diagnostic purposes has been only
seldomly mentioned. WO 94/13833 describes a method for detecting
nucleic acid molecules in a solution by tailoring a specific ribozyme
20 molecule having two regions, one complementary to the nucleic acid
sequence to be detected, and the other complementary to a co-target
molecule bearing a detectable label. The ribozyme is able to specifically
and reversibly bind both a selected target nucleic acid sequence and to the
labelled co-target. When both the target and the co-target are bound, the
25 ribozyme undergoes a conformational change which renders it active and
able to cleave the label off the co-target, and the free label can then be
detected. Upon cleavage of the co-target, the ribozyme is able to re-
associate with an additional co-target, cleaving more label and producing
more detectable signals.
SU B STITUTE SH EET (RU LE 26)
CA 02213622 1997-08-22
W O 96127026 PCTnUS96/02380
_ 4 _
Although the inventors of WO 94/13833 termed their invention
"amplification of signal" there is actually no amplification in the number
of ribozymes produced, but rather the reaction is purely an enzymatic
reaction, wherein the catalytic substance (in this case the ribozyme) cleaves
5 the substrate (in this case the co-target) and then disassociate and cleave
another substrate. There is no true amplification of the number of active
ribozymes involved in the reaction occurs.
GLOSSARY
Below is a glossary of terms which are used in the following
description and claims. However, this glossary should not be considered
separately and for full comprehension of the various terms and the meaning
in which these terms havc in the context of the invention, the glossary
should be read in conjwlctioll with thc remain~lcr of the disclosure herein.
Ribozyme -- a nucleic acid molecule which possesses an enzyme-like
catalytic acivity. The term ~'ri~o7y~7te" as used in the art generally refers
to RNA molecules having a catalytic activity although in the context of the
present invention this term is used to denote a catalytically active (enzyme-
20 like) oligonucleotide in gcneral. The ribozyme of the invention may thusbe an RNA molecule, may be an oligonucleotide comprising dNTPs or
composed completely of dNTPs and may also comprise a variety of non-
naturally occurring nucleotides such as IsoG or IsoC 5'-0-(1-thio-
triphosphate) nucleosites and 5-0-methyl nucleotides. The ribozyme which
25 may be used in accordance with the present invention may be comprised
exclusively of nucleic acids as described above or may require a co-factor
for their catalytic activity. The ribozymes may have a catalytic activity of
cleavage, ligation, splicing or splicing-out (removal) of oligonucleotide
SUBSTITUTE SHEET (RULE 26)
-
- CA 02213622 1997-08-22
W O 96n7026 r~l/U~96/02380
sequence, addition of groups to oligonucleotides, rearrangèment of nucleic
acid sequences, etc.
Assayed biomolecule - a molecule the presence of which in the test sample
S is to be detected. It can be an oligonucleotide or a member of a recognition
pair such as receptor/ligand, antibody/antigen, lectin/glycoprotein, etc.
Initiation ribozyme - The ribozyme which initiates the reaction where
more ribozymes arc produccd, evcntually Icacling to the gencration of a
10 detectable label. Where the method of the invention is used to detect
biomolecules, the initiation ribozyme is part of the detection system (see
below), and serves as a reporter for the presence of the assayed biomolecule,
since only in the presence of said assayed biomolecules it is either generated
or it becomes catalytically active. The prcsence of an active initiation
1~ ribozymc activates thc catalytic system (scc bclow).
Detection system - the combination of molecules and reagents that enable
the production or activation of a catalytically active initiation ribozyme,
which serves as a reporter for the presence of the assayed biomolecules in
20 the test sample. In other words, in the presence of the assayed
biomolecules, following a reaction or a cascade of reactions, a catalytically
active initiation ribozyme is eventually generated. The presence of the
initiation ribozyme is verified in the catalytic system (see below) where it
brings to the generation of more active ribozymes in an amplificatory
5 manner (the ribozymes themselves or a product of their catalytic activity,
e.g. a free label is then detected at the final stage of the assay).
Inactive ribozyme - a potentially catalytically active ribozyme which
cannot exert its catalytic activity (cleavage, splicing, ligation, etc.) until it
SUBSTITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96/27026 ~ 6/02380
has been modified, or until the conditions in the medium have not been
amended to such in which it becomes active.
Activation - rendering an inactive ribozyme catalytically active by some
5 kind of catalytic action (cleavage, splicing, ligation, addition of groups,
rearrangement), or by change of external conditions (such as addition of
magnesium ions).
Inhibitory moiety - a moiety which may at tim~s be present in the second
10 complex molecule (see below) and which when present renders the
initiation ribozyme inactivc. The inhibitory effcct of the inhibitory moiety
can be terminated by its modification or removal from the complex
molecule.
15 Complex molecule - a molecule which forms part of the detection system
in accordance with an embodiment of thc invention referred to herein as the
"activation embodiment". In one mode of carrying out the activation
embodiment, a '~irst comple~- molecule" is being used, which comprises an
initiation ribozyme which is a priori, catalytically inactive (for example, due
20 to lack of magnesium ions in the medium) linked to a sequence capable of
being cleaved by an active initiation ribozyme and comprising in addition
a recognition biomolecule (see below). In another mode of carrying out
the activation embodiment, a "second complex molecule" is being used,
which comprises an initiation ribozyme which is a prioli, catalytically
25 inactive and is linked to a sequence capable of being cleaved by an active
initiation ribozyme and comprising in addition a recognition biomolecule
(see below). The'second complcx molccule further comprises an inhibitory
moiety.
SUBSTITUTE SHEET (RULE 26)
=
. CA 02213622 1997-08-22
W O 96127026 1~1~ 02380
-- 7 --
Recognition biomolecule - a molecule capable of specifically recogni7~ing
and binding to the assayed biomolecule. Where the assayed biomolecule is
an oligonucleotide sequence, the recognition biomolecule is the complemen-
tary sequence. Where the assayed biomolecule is a member of a recognition
5 pair (such as antigen-antibody) the recognition biomolecule is the other
member of the pair.
First oligonucleotide - an oligonucleotide, typically a DNA molecule,
which comprises from 3'-5': a double-stranded functional promotor, a
10 single-stranded sequence that codes for a sequence complementary to the
sequence of the initiation ribozymes, a single-stranded sequence identical
(with the necess~ry U-T replacements) with an RNA sequence capable of
being cleaved by a catalytically active initiation ribozyme, and a single-
stranded sequence complementary to the ~ '-part of the oligonucleotide
15 sequence to be detected.
Second oligonucleotide - an oligonucleotide, typically a DNA molecule,
which comprises from 3'--~': a single-stranded 3'--part complementary
to the oligonucleotide sequence to be detected, and a triggering oligonu-
20 cleotide template (see below).
Triggering oligonucleotide template - an oligonucleotide sequence which
is part of the second oligonuclcotide, thc transcription product of which is
capable of hybridizing with the back promotor construct (see below).
Triggering oligonucleotide sequence - the transcriptional product of the
triggering oligonucleotide template, capable of hybridizing with the back
promotor construct (xee below) and after the back promotor has been
SUBS 1~1 UTE SHLET tRULE 2~)
CA 02213622 1997-08-22
W 096n7026 ~ 02380
- 8 -
completed, can bring to transcription of an oligonucleotide sequence to
which it is attached.
Non-template strand oligonucleotide - the transcription product of the
S oligonucleotide hybrid obtained by hybridization of the assayed nucleic acid
sequence, the first oligonucleotide and the second oligonucleotide, and which
comprises from 3'-5': a triggering oligonucleotide sequence, a sequence
complementary to the assayed biomolecule, a sequence complementary to a
sequence which can be cleaved by the initiation ribozyme, and a sequence
10 complementary to the initiation ribozyme.
Back promotor construct - a single-stranded promotor sequence attached
to a single-stranded sequence capable of hybridizing with the triggering
oligonucleotide sequencc. Aftcr hybridization with the triggering oligonu-
lS cleotide sequence, and upon action of a suitable DNA polymerase, afunctional double-stranded promotor is created, which in the presence of a
transcri~tion Systf~7~ (s;cc below) is capable of producing the final
oligonucleotide transcriI)t (sec below).
20 Final oligonucleotide transcript - the transcriptional product of the
oligonucleotide hybrids obtained following hybridization of the back
promotor construct, non-template strand oligonuclcotide (after the promotor
has been completed by a suitable DNA polymerase to a double-stranded
functional promoter), and comprises from ~'-3': an initiation ribozyme
25 sequence, a sequence capable of being cleaved by said initiation ribozyme,
a sequence that codes for thc complcment of the detected sequence on the
assayed biomolecule, and a sequence that codes for the complement of the
triggering oligonucleotide template. The initiation ribozyme in said final
SUBSTITUTE SH EET (RULE 26~
- CA 02213622 1997-08-22
W O 96/27026 ~-liU'r.'v23~C
_ 9 _
oligonucleotide transcript can cleave its adjacent sequence thus freeing itself
and yielding a free, fully active initiation ribozyme.
Third oligonucleotide sequence - a nucleic acid sequence complementary
S to the S'-part of the sequcnce to be dctectcd in thc assayed biomolccule.
Third composite molecule - a molecule used in the "assembly embodi-
ment" of the invention, and which comprises the third oligonucleotide
sequence linked to part of the initiation ribozyme. It optionally also
10 comprises a sequence cleavable by an active initiation ribozyme.
Fourth oligonucleotide sequence - an oligonucleotide sequence comple-
mentary to the 3 ' -part of the sequence to be detected in the assayed
biomolecule.
Fourth composite molecule - a molecule used in the "assembly embodi-
ment" of the invention which comprises the fourth oligonucleotide sequence
linked to part of the initiation ribozyme required to complete the part present
in the third composite to obtain a complete catalytically active ribozyme.
20 It optionally also comprises a sequence cleavable by an active initiation
ribozyme.
Catalytic system - An ensemble of molecules and reaction mixtures which
in the presence of a catalytically active initiation ribozyme produces a
- S detectable signal. This enscmble of moleculcs comprises a combination of
composite molecules comprising a ribozyme which is a priori inactive. By
one embodiment, the catalytic system comprises reagents and a combination
of a first composite molecule and a second composite molecule (see below),
comprising a first ancl SCCOlld ribozyme (inactive), respectively. The first
SUBSTI I UTE SHEE~ (RULE 26)
CA 02213622 1997-08-22
W 096/27026 ~ g6tO2380
-- 10-- .
and second ribozymes arc a priori inactive and either or both can be
activated by a catalytically active initiation ribozyme. Active first ribozyme
may activate inactive second ribozyme molecules, and active second
ribozymes may activate inactive first ribozyme to cause amplification of the
5 number of active ribozymes in a positive feedback manner. Alternatively,
the first and second ribozymes may be immobilized or spatially separated
from each other and cleavage of one or both by the initiation ribozyme
causes their release to the medium. Released, free ribozymes can free
immobilized second ribozymes (for example by cleavage) and free second
10 ribozymes can in turn release and free first ribozyme, thus giving rise to self
amplifying reaction cascade which rapidly yields an amplification in the
number of active ribozymes in a positive feedback manner.
The catalytic systcm may also comprise in accordance with
another emboidment, only one species of composite molecules which are
15 immobilized or spatially separated from each other in the reaction vessel or
which are a priori inactive. An initiation ribozyme activates the inactive
ribozyme or rcleases thc ribozymc from thc compositc molccule and active
or released ribozymes then act to respectively activate inactive or free
immobilized other ribozymes, then giving rise to a self amplifying reaction
20 cascade which rapidly yields an amplfication in the number of active
ribozymes in a positivc fecdback manncr.
As a result of ribozyme activation or release, a detectable signal
is produced which signal is indicative to the presencc of the initiation
ribozyme in the medium.
First composite molecule - comprises a fiJst ribo~y~7le (see below),
optionally labelled, linked to second nucleic acid se4uence (see below).
SU8 STlTUTE SH EET (RULE 26)
. CA 02213622 1997-08-22
W 0 96r27026 ~~ 02380
Second composite molecule - eomprises a seco)ld ~ o~y~ (see below),
optionally labelled, linked to afirst n~lcleic acid seq~lence (see below).
First nucleic acid sequence - an oligonueleotide sequenee whieh is part of
5 the seeond eomposite moleeule and whieh is a target for the eatalytie
aetivity of the first ribozyme (see below). Following the eatalytic aetivity
of the first ribozyme on the first nucleic acid sequenee, the seeoncl ribozyme
(see below) is either released into the medium or becomes eatalytically
active.
Second nucleic acid sequence - an oligonucleotide sequence which is part
of the first composite molecule and which is a target for the catalytic
activity of the second ribozymc (sec bclow). Followillg thc catalytic activity
of the second ribozyme On the second nucleic acid sequence, the first
15 ribozyme (see below) is either relcase(l into the medium or becomes
catalytically active.
First ribozyme - part of the first composite molecule -- is capable of
cleavinL~ thc first IlUClCiC acid scqucllcc and is i~lcntical in its catalytic
20 activity to the initiation ribozyme. It is optionally labelled.
Second ribozyme - part of the second composite molecule - is capable of
cleaving the seeond nucleie aeid sequenee and is optionally labelled.
25 Third ribozyme - a ribozyme whieh may form part of the catalytic system
in accordanee with another embodiment thereof. The third ribozymes are
initially inaetive. Initiation ribozyme acts to activate the third ribozyme by
exerting itsi eatalytic activity thereon (in a manller to be explained below)
SUBSmUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96/27026 . ~11~_./02380
and the aetivated ribozymes ean then aet, to aetivate other third ribozymes
in the eatalytie system.
Transcription system - ensemble of oligonueleotide, nueleotides, RNA
5 polymerase and reagents whieh, in the presenee of an oligonueleotide
template bring to the transeription of an oligonueleotide transeript.
Fourth ribozyme - a ribozyme whieh is part of the eatalytic ~;ystem in
aeeordanee with an embodiment thereof, and whieh onee catalytically aetive
10 ean ligate two parts of the fifth ribozyme (see below) to produee a
eatalytieally aetive fifth ribozyme. The fourth ribozyme is eomposed of at
least two eomponents, whieh are initially separated, and whieh are ligated
together by Ihe fifth ribozyme (when eatalytieally aetive). Following sueh
ligation, the fourth ribozyme beeomes eatalytieally aetive.
Fifth ribozyme - a ribozyme whieh is part of the eatalytie system
eomprising the fourth ribozyme and which once catalytically active ean
ligate together two parts of the fourth ribozyme to produee a catalytically
aetive fourth ribozyme. The fifth ribozyme is composed of at least two
20 eomponents, which are initially separated and whieh are ligated together by
the fourth ribozyme. Following such ligation, the fifth ribozyme becomes
catalytically active.
Sixth ribozyme - a speeific example of the third ribozyme wherein the
5 inactive ribozyme carries an extra nucleic acid sequence, and is activated
upon eleavage or splicillg-out (i.e. removal) of this sequence.
SUBSTlTUTE SH EET (RULE 26)
- CA 02213622 1997-08-22
W O 96127026 PCTnUS96102380
- 13 -
Seventh ribozyme - a ribozyme whcrcin the assayed llucleic acid sequence
completes a mi~in~ portion essential for its catalytic activity, and thus once
combined with the assayed sequence it becomes catalytically active.
5 SUMMARY OF THE INVENTION
The present invention provides a ribozyme-based signal-
amplification method which is relatively simple to perform, is rapid and
inexpensive. Unlike hitherto available detection-amplification methods, the
method of the invention is also suitable for a point-of-care (POC) testing.
1() One a~va!lta~c of thc mcthod of the invention is in that ribozymes
are active under conditions foulld in thc clinical environmcnt, e.L~. in
bioligical fluids. Furthermore, as will be shown further below, the signal-
amplification method in accordance with the invention does not require the
observance of specific collcJitiolls in order to ensure specificity (strict
observance of conditions is a must in prior art si~nal-amplification
methods). Additionally, ribozymes are functional in various sample
preparation cocktails, e.g. 1 M Guanadine thiocyanate as well as in a
saturated phenol preparation, which usually inhibits function of other
detection-amplification systems.
Ribozymes are composed of nucleic acid sequences and thus assay
and probe sequences can be included in the ribozyme molecule. Further-
more, it is possible to increase the specificity of the amplification process
by engineering the ribozyme such that part of the assayed sequence itself is
required for the ribozyme to exert its catalytic activity.
A very powerful technique, termed in the art as "in vitro
evolution " has been successfully applied to ribozymes to produce a
ribozyme with various catalytic activities and specificities. By such
techniques, a vast array of potcntial ribozymc~ arc screcned for activity.
Those ribozymes that show activity are purificd for further rounds of
SUBSTmJTE SHEET (RULE Z6)
CA 02213622 1997-08-22
W 096/27026 1~l~ 02380
- 14 -
selection and after repeated rounds only the most potent candidates remain.
In traditional amplification techniques, following a choice of the enzyme, the
environment where the enzyme is to operate, i.e. the sample-comprising
medium, has to be modified to allow proper activity of the enzyme. In the
case of ribozymes, using in vitro evolution, it is possible to select a
ribozyme which is highly active in a dcsired clinical (biulogical) medium,
by performing the in vct)-o evolution in a selcction medium which is
identical in its composition to the clinical sample.
The present invention provides a sensitive method for the
10 detection of a cataytically active ribozyme (referred to herein as "initiation
ribo7yme") in a medium. Detection of the presence of a catalytically active
initiation ribozyme may be a goal by itself, although usually the catalytically
active initiation ribozyme serves as reporter for the presence of other
biomolecules in the tcst samplc. Once a mcdium comprising an active
15 initiation ribozyme is introduced into a catalytic system in accordance with
the invention, there results a catalytic reaction cascade which gives rise to
an exponential amplification in the number of active ribozymes. The
catalytic system comprises ribozymes which are either inactive or spatially
separated from one another such that they camlot exert their catalytic
20 activity; the initiation ribozymc frecs or activatcs ribozymes of the catalytic
system which in turn free or activate respectively, further ribozymes of the
system. The ribozymes either carry a detectable label or the catalytic
activity causes generation of a detectable label, which then serves as an
indication of the catalytic cascade which occurred in the system.
According to embodiments of the present invnetion in which the
ribozymes are initially immobilized, the presence of free ribozymes in the
reaction medium may serve as a dctectable signal by itself. In accordance
with another embodiment, each active ribozyme is made to carry or produce
a detectable label and thcse thcn serve as the dctcctable signal.
SUBSTmJTE SHEET (RULE 26)
- CA 02213622 1997-08-22
W O 96/27026 PCTAUS96/02380
Accor~ling to the method of the invelltioll there is very little false
positive signal, i.e. Iow noise Ievel; furthermore the method of the invention
enables the detection of several biomolecules in a single assay system.
The present invention thus provides a method for detecting the
5 presence of a catalytically active initiation ribozyme in a medium, compris-
ing the steps of:
(a) providing a catalytic system comprising
(aa) ribozymes which are a pri~ri catalytically inactive or
spatially confined such that they cannot exert their catalytic
activity on their target; the target of the ribozymes being other
ribozymes of the catalytic system, their catalytic activity on such
other ribozymes causing either:
(i) activation of inactive ribozymes,
(ii) release of spatially confined ribozymes to allow them
to reach their targets;
at least some of the ribozymes of the catalytic system being a target of
the catalytic activity of the initiation ribozyme, the catalytic activity of theinitiation ribozyme on said some of the ribozymes being that of(i) or (ii)
above; ~nd comprising
(ab) a detectable label having detectable properties such that the
catalytic activity of the ribozymes causes a change in the
detectable properties;
(b) contacting the medium with said catalytic system;
(c) providing conditions permitting said catalytically active initiation
ribozyme and catalytically active ribozymes of the catalytic system to exert
their catalytic activity, whereby the presence of a catalytically active
initiation ribozyme gives rise to a reaction cascade in which ribozymes of
the catalytic system are activated or freed into the medium; and
SU BSTlllJTE SHEET(RULE 26)
CA 022l3622 1997-08-22
W O 96/27026 l~l1~ 2380 - 16 -
(d) detecting said detectable properties, a change in said properties
being an indication of the presence of an active initiation ribozyme in said
medlum.
The prime utility of the ribozyme detection method of the
5 invention, is within the framework of an assay designed to detect the
presence of a biomolecule such as: a specific nucleic acid sequence, a
member of a binding couple such as antibody-antigcn, sugar-lectin, etc., in
a biological sample. Such an assay may conceptually be thought of as
comprising two distinct components (although these components may be
10 included physically together in a single reaction vessel): a detection system and a catalytic system. In such an assay, the presence of the assayed
biomolecule brings to the production, in a manner to be described further
below, of a catalytically active initiation ribozyme in the medium. The
catalytically active initiation ribozyme then acts as a reporter molecule in the15 catalytic system, giving rise, following a reaction cascade which amplifies
the number of active ribozymcs, to the appearance of a detectable label in
the reaction medium as generally described above. The appearance of such
a detectable labcl in the mcdium of the catalytic system thus indicates the
presence of the assayed biomolecule in the original assayed biological
0 sample.
DETAILED DESCRIP~ION OF THE INVENTION
In accordance with the invelltion, novel use is made of ribozymes.
In the most gcneralizcd scnsc, a catalytic system comprising ribozymes is
5 used for the detection of thc prescnce of a catalytically active initiation
ribozyme in an assayed medium. Detection of the presence of a catalytically
active initiation ribozyme in all assayed medium may be a goal by itself, for
cxamplc, in thc proccss of prcparation of ribozymes by in vitro evolution.
In addition, in accordance with a prcfcrrcd embodiment of the invention, the
SUBSTITUTE SH E~T (RULE 26)
=
- CA 02213622 1997-08-22
W O 96127026 PCTnUS96/02380
- 17 -
catalytically active initiation ribozyme serves as a reporter ~or the presence
of an assayed biomolecule (other than the initiation ribozyme) in an assayed
biological sample.
The ribozymes used in accordance with the invention may be
S comprised entirely of RNA. At time it is possible also to replace some of
the ribonucleotides ( "rNTPs ") in the RNA with deoxy nucleotides
("dNTPs") or some other naturally or non-naturally occurring nucleotides
such as IsoG or IsoC 5'-0-(1-thiotriphosphate) nucleotsites and
5-0-methyl nucleotides. Such replacement is at times desired, for example,
10 to increase stability of the ribozyme to RNAase which are present in almost
all biological samples. While this will not specifically be mentioned at each
time, it is understood that thc term "ribo~yme" means to denote catalytic
nucleotides composed entirely of rNTPs or catalytic oligonucleotides
wherein some of the rNTPs have been replaced by dNTPs or other
15 nucleotides. The ribozyme may also be entirely composed of DNA (Breaker
et al., Chenzishy and Bcology, 1(4):2~3-9, 1994).
The ribozymcs of the invention may comprise of nuclcic acid
sequences as described above complexed with a non-nucleic acid molecule
such as a protein, polypeptide, fatty-acid, dye, antibiotic, or a carbohydrate.
20 The non-nucleic acid moiety complexed with the ribozyme may serve as a
co-factor for the ribozyme's catalytic activity.
In the following, usc of thc term "oligon~cleotide" will be made.
The oligonucleotides may, clepending on the context, be a DNA oligo-
nucleotide (consisting entirely of dNTPs) or a RNA oligonucleotide
25 (consisting entirely of rNTPs). However, specifically in the case of RNA
oligonucleotides, it is at times desired to replace some or all of the rNTPs
with dNTPs or other naturally and non-naturally occurring nucleotides.
The present invention provides, in its broadest sense, a method for
the detection of the presence in a tested medium of a catalytically active
SUBSTITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W 096127026 ~1/~ 02380
initiation ribozyme. By "catalytically active", it is meant that a ribozyme
is capable of carrying out a catalytic reaction such as cleavage, splicing,
ligation, addition of specific groups such as phosphate to molecules,
rearrangement of nucleic acid sequences and the like.
S The catalytic system in accordance with one embodiment for
carrying out the invention comprises two species of ribozymes which are a
priori inactive, but become catalytically active as a result of the exertion of
a catalytic activity thereon. For example, each ribozyme may have a nick
or break in a portion essential for its activity and prior ligation of this nickor break is thus required for its activation. The catalytic system in this case
compriscs two spccies of ribozymcs, each spccics a pJ-i~l-i broken into two
components and thus initially inactive. An active ribozyme of one species
is capable of ligating the two components of thc second species of ribozyme
thus rendering it active, and an active ribozyme of the second species of
ribozyme is capable of ligating the two componcnts of the first species of
ribozymes, thereby rendering it active. Then the activation proceeds by
cross-ligation in a positive feedback amplificatory manner.
The first active ribozyme of one species may be produced by the
initiation ribozyme which is a product of the ~Ictection system, in one of two
routes. According to one routc, some of thc ribozymes of the first species
are a priori fully assembled but cannot ligate the parts of the second species
of ribozyme, since they are spatially separated therefrom, for example, as a
result of being immobilizcd by means of a porous membrane, etc. The
initiation ribozyme cleaves thc molecules of the immobilized fully assembled
first species of ribozyme, and the free first species of ribozyme then ligates
the second spccies of ribozymcs which Call in tum ligate members of the
first species of the ribozyme which are not, a priori, assembled and so on.
According to thc sccond route, the initiation ribozyme is itself a
ligating ribozymc which ligates from its parts, at least one species of the two
SUBSTITUTE SHEET (RULE 26)
- CA 02213622 1997-08-22
W O 96/27026 ~ U~ 02380
- 14 -
ribozyme of the catalytic systcm, thus initiating the cross-ligation cascade.
In such a case there is no need to spatially separate various members of the
catalytic system since until the initiation ribozyme is introduced to the
reaction mixture no catalytic process can begin.
Another example are ribozymes which have a redundant sequence
which renders the ribozyme inactive and it thus needs to be either cleaved
or spliced-out for the ribozyme's activation. Further example are ribozymes
which require sequence rearrangement or addition of specific groups for
activation. Yet another examplc arc ribozymes which require reverse exon
splicing, i.e. addition of a sequence internal to the ribozyme.
One species of ribozymes when in the active form, may activate
inactive ribozymes of the SCCOlld species and vice velsa, as it possesses the
catalytic properties (ligation, cleavage, splicing, rearrangement, etc.) required
to moclify thc othcr spccics of ribozymc from an inaclivc to an activc form.
The two ribozymes may potentially possess the same type of catalytic
activity (e.g. both are ligating rizoymes or both cleaving ribozymes, etc.) or
may possess different types of catalytic activities.
Onc or both spccics of thc c~ pr~ inactivc ribozymcs is activated
by a catalytically active initiation ribozyme. Prior to introduction of the
initiation ribozyme to the medium, the catalytic system is çcsenti~lly silent
as no catalytic activity takes place. In the presence of such an initiation
ribozyme, a ribozyme amplification cascade begins since each active
ribozyme generates in turn more active ribozymes in a positive feedback
manner. Active ribozymes give rise to a signal which can be detected in a
manner as described hereinbelow and such a signal is indicative to the
presence of the original catalytically active initiation ribozyme in the
medium.
The catalytic system in accordance with another embodiment for
carrying out the invention compriscs two species of composite molecules,
SUBSTITVTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96n7026 ~ 23~0
-- ')O --
each comprising a ribozyme linked to a cleavable nucleic acid sequence.
The ribozyme in one species of composite molecules is capable of cleaving
the nucleic acid sequence in the other species of composite molecules so that
cross-cleavage between the two species of composite molecules is, in
S principle, possible, while self-cleavage is avoided. However, prior to
introduction of the initiation ribozyme to the medium, cross-cleavage does
not result since the two specics of composite molecules are constructed so
as to prevent mutual interaction between them, while cleaved ribozymes are
able to interact with other composite molecules in the test vessel and further
10 release ribozymes to thc medium in a positive feedback manner.
Prevention of mutual interaction can be done, for example, by
immobilizing each species of composite molecules to opposite sides of the
test vessels; by linking each species of the composite molecule to different
beads or different colloid particles having properties, e.g. size or other
1~ properties, e.g. the same electric charge (which repels the beads from one
another) which prevents any kind of interaction between molecules attached
to one with moleculcs attachcd to anothcr; by linking the composite
molecules to moieties having the same electric charge, so that the electrical
rejection between said moictics will prevent any interaction between the two
~0 composite molecules; by placing each species of composite molecules at
opposite sides of a porous membrane which does not allow permeation
therethrough of the full composite molecule, but allows free passage of
cleaved ribozyme.
A catalytically active initiation ribozyme, either present in the test
''S medium a priori, or produced as a result of the presence of another
biomolecule in a biological sample (such a ribozyme being in this case) ("a
reporter- ribo~ynle") is ablc to clcave a spccific nucleic acid present in one
or both species of composite molecules thus freeing the ribozymes of the
catalytic system. The cleaved ribozymes are able to interact freely in the
SUBSTlTUTE SH EET (RULE 26)
. CA 02213622 1997-08-22
W O 96/27026 ~1l~','~02380
reaction vessel with the ribozymes from the other species of composite
molecules, which in turn, can again cleave the ribozymes from the first
species of composite molecules, thus creating a "ping-pong" cross-cleavage
of ribozymes. Such cross-cleavage of ribozymes acts in a positive feedback
manner, c~ in~ substantial amplification of the reaction. Either or both
species of ribozymes typically bcars dctectablc labcls. Thc detcction of
cleaved labels, indicates the presence of the catalytically active initiation
ribozyme in the reaction mixture. Where the initiation ribozyme is a
reporter ribozyme, detection of a frce label indicates thc presence of an
10 assayed biomolecule in the reaction mixture.
The catalytic system may comprise, in accordance with other
embodiment, only OllC ~ipCCiCS of inactivc ribozymes or onc specics of
composite moleculcs comprisill~ a ribozymc and a llUCICiC acid scqucncc
cleaved by the ribozyme when converted into a free or active form. In an
15 analogous manner to that dcscribed above for the said one embodiment, each
single molecule of the ribozyme is inactive until the catalytically active
initiation ribozyme is introduce<l to the medium: for example, each inactive
molecule is in the form of a closed-circle which may be opcned by cleavage
or splicir,g-out of a strctch of nucleotides, by a catalytically active initiation
0 ribozyme. Activated (open) ribozymes thcn opcn and activate other such
closed-circle moleculcs of the catalytic system.
In an analogous manner to that described above for the said
another above second cmbodiment, each single species of the composite
molecule may comprise a ribozyme positioned in an orientation which
25 prevents self-cleavage of the adjacent cleavable nucleic acid sequence, for
example, by placing thc sequcnce immediately adjacent to the ribozyme.
The fact that there are no intcrvening sequences between the ribozyme and
the cleavable scqucncc stcarically inhibits the cis clcavaL~e. (The cleavable
sequence may also havc an invcrsed orientation, and cis cleavage will thus
SUBSTITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96/27026 ~1~ 02380
not be possible). Howcvcr, rclcascd ribozymc may approach thc nucleic
acid sequence in a correct orientation, and cleave it, thereby releasing more
ribozymes to the medium. In order to prevent spontaneous trans cleavage
of non-released ribozymes, it is possible to ensure spatial separation
5 similarly as indicatcd abovc.
Detection of the presence of activated ribozymes in the catalytic
system may take various forms depending on the type of catalytic activity
of the ribozyme. Where, for example, the activity is cleavage or spliced out,
a label may be linked to the part to be cleaved or spliced out and detection
10 of such freed label is then an indication of the presence of the catalytically
active initiation ribozymc in the mcdium.
In some cases the activation of the ribozyme brings to a change
in the distance between two regions of the ribozyme, such as where two
distant regions are brought together by ligation or rearrangement, by splicing
15 out of an interfering region, or wherein two initially adjacent regions are
separated for example by opening of a closecl circle. In such case it is
possible to attach a fluorescellt marker on one region of the ribozyme and
a moiety, such as Rodamine which quenches the light emission from the
fluroescent marker on the other region of the ribozyme. Rodamine has a
O quenching effect on the light emission of a fluorescent label when the two
are adjacent and no such effect when the two are separated. By monitoring
the change in light emission of the fluorescent label, it is possible to
determine whether the two regions are adjacent (for example in the case of
a closed-circle inactive ribozyme) or separatecl (when the ribozyme has been
25 opened and activatecl).
The label may also be carried on a substrate which is not
associated with the ribozyme and on which the catalytically active ribozyme
may exert its catalytic activity. For cxample, the labcl may be carried on
a nucleic acid sequence, which is cleaved or spliced-out as a result of a
SUBSTITUTE SHEET (RULE 26)
-
CA 02213622 1997-08-22
W O 96~7026 ~liU'_-/02380
3 _
ribozyme's activity, whereby the label is released to the medium. The
detection will in such a case be based on the presence of a free label in the
medium.
The "ping-pong" cross activation, whether by cross-cleavage,
S cross-ligation, cross-splicing, cross rearrangement or alternating cycles of
various catalytic actions, substantially amplifies the reaction resulting in a
signal that indicating in a short time whether the initiation catalytically
active ribozyme was present in the medium. While prior art amplification-
detection methods such as PCR or LCR require several hours to be
10 complete, the amplification-detcction method of the invcntion is completed
in a much shorter time period.
Where the catalytically active initiation ribozyme serves as a
reporter ribozyme for the presence of other biomolecules, a detection system
is required in which a catalytically active initiatioll ribozyme is gencrated
15 only in the presence of the assayed biomolecule. This may be performed in
one of the following embodiments, referred to herein as the ~ctivation
embodiment", the "h-anscri~ti~n c~m~ 7zcnt the ''ase~7Zbly en~bodi--
ment ' and the "completion ~mbodu7lent".
According to the activation embodiment, the initiation (reporter)
~0 ribozyme is ~ prio~i inactivc. This inactivity may bc a rcsult of thc absenceof magnesium ions, which are required for the ribzoyme's catalytic activity,
from the medium; it may be a result of the presence of an inhibitory moiety
in the medium; it may be a result of the presence in the medium of an
oligonucleotide which hybridizes to sequence which is to be cleaved to
25 either activate or free the ribozyme into the system, which cleavage is not
possible as long as thc scqucncc is doublc-strandcd; ctc. Accordillg to this
embodiment, the ribozyme is linked to a recognition biomolecule which is
capable of specifically recognizing and binding to the biomolecule which is
to be assayed in the sample. For example, whcre the assayed biomoleucle
SU8STITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96127026 ~ 96/02380
' - 24 -
is an oligonucleotide sequence, the recognition biomolecule is the comple-
mentary sequence; where the assayed biomolecule is an enzyme, the
recognition biomolecule may be a substrate; where the assayed biomolecule
is an antigen, the recognition biomolecule may be an antibody which
S specifically interacts with the antigen; etc.
The ribozyme linked to the recognition molecule is then allowed
to interact with the assayed biomolecules, and unbound ribozymes are then
separated and washed away. Such separation can be carried out, on the
basis of size difference between the complex of bound ribozymes and
10 assayed biomolecules and that of free ribozymes; by, a priori, immobilizing
the assayed biomolecules and then washing away free molecules of
ribozymes; etc. After said separation, conditions are changed so as to
activate the ribozyme, for example, by addition of lacking magnesium ions;
by modifying or removing thc inhibitory moicty to stop its inhibitory
15 activity; by melting the double-strandcd non-cleavable sequence to a single
stranded cleavable sequence; etc. Only if the assayed biomolecule is
present, ribozymes which are bound thereto are retained, and only these
retained ribozymes are activated by an appropriate change of conditions.
The transcription embodiment of the invention can be utilized
20 where the assayed biomolecule is a nucleic acid sequence. The detection
phase of this embodiment can be carried out generally as described in Israel
Patent Applications Nos. ln~894 and 1118~7 (and their counterpart PCT
Applications Nos. WO 94/29481 and ) with the "triggering
oligon~lcleiti~" being said initiation ribozyme. The detection system of
25 this embodiment comprises two oligonucleotide molecules, the first
comprising a sequence complementary to the ~'- part of the assayed nucleic
acid sequence and the second comprising a sequence complementary to the
3'- part of the assayed nuclcic acid sequence. The first oligonucleotide
molecule comprises upstrcam from the sequence complementary to the
SUBSTITUTE S~EET (RULE 26)
. CA 02213622 1997-08-22
W O 96/27026 P~~ 5'~02380
assayed biomolecule, a functiollal promoter, a sequcnce that cocles for an
initiation ribozyme sequence and a sequence that is capable of being cleaved
by said detecting ribozymc (also esscntially a DNA sequence). The second
oligonucleotide molecule comprises, downstream from the complementary
S 3'- part of the assayed sequence, a triggering oligonucleotide template, the
transcriptional product of which is capable of triggering transcription of
sequences of initiation ribozymes as will be explained in detail hereinafter.
If the assayed biomolecule is not present in the test sample, then
the triggering oligonucleotide sequence is not transcribed, since only
presence of the assayed biomolecule brings togcther the two molecules
required to produce thc appropriate template of said triggering oligonucleo-
tide sequence: namely the first oligonucleotide molecule carrying the
functional promotor and the second oligonucleotide molecule carrying the
triggering oligonucleotide template. If the assayed biomolecules are present,
and in the presence of transcription system, the triggering sequence is
produced and in turn is able to bring about production of transcripts
cont~ining initiation ribozyme linked to a sequence capable of being cleaved
thereby. After self-clcavagc, these transcripts release to the medium
catalytically active initiation ribozyme.
According to the assembly embodiment of the invention, also
appropriate in cases where the assayed biomolccule is a nucleic acid
sequence, the detection systcm compriscs a third oligonucleotide, comprising
a sequence complementary to the ~'- portion of the assayed nucleic acid
sequence, and a fourther oligonucleotide comprising a sequence complemen-
tary to the remaining, 3'- portion of the assayed nucleic acid sequence.
Each of these oligonucleotidcs comprises also one portion (for example,
half) of a ribozyme and both parts together constitute a full, functionally
active ribozyme. In accordallce with this embodiment, the function of the
assayed nucleic acid sequencc is to bring these two oligonucleotides
SUBSTITUTE SHEET (RULE 26)
=
CA 02213622 1997-08-22
W O 96/27026 ~ 96102380
. - ~6 -
together, thus yielding a functionally active initiation ribozymc. Thus, in the
presence of an assayed nucleic acid sequence in the sample, the functional
initiation ribozyme will be generated which could then be detected in the
catalytic system of the invention.
In accordance with the completion embodiment of the invention,
the detection system comprises a seventh oli~onuclcotide and the assayed
sequence, complexes with the seventh oligonucleotide to yield a catalytically
active initiation ribozyme. For example, the assayed sequence may form
part of the catalytic core of the ribozyme. Thus, in accordance with this
embodiment, the ribozyme is a pr iori incomplete, and only in the presence
of the assayed sequence it becomcs a complete, catalytically active ribozyme
which may then be detectcd in the catalytic system.
The assayed sequcnce may complcte the ribozyme by hybridizing
at its 3'-end to a sequencc on Onc sidc of thc ribozymc's missillg portion,
and by hybridizing at its ~'-end to a sequence on the other side of the
ribozyme's missing portion, thus bridging the missing portion and creating
a functional initiation ribozymc.
The assaycd scquellcc may also bc able to complete the
ribozyme's missing portion by ~ everse e~on splicing wherein the assayed
sequence is inserted into the ribozyme through suitable cleavage and ligation
reactions. Said reverse exon splicing may be carried out by other
ribozymes present in the medium.
ln order to decrease the noise level of the method of the
invention and decrease false positive results, it is possible to combine two
2~ or more embodiments of the invclltion to doubly ensure that no catalytically active initiation ribozymes are produced in the absence of assayed
biomolecules. For examplc, it is possible to combine the assembly and
activation embodiments of thc invcntion, whcrcby catalytically active
ribozymes will be generatcd only as a result of two accumulative conditions:
SUBSTITUTE SHEET tRULE 26)
. CA 02213622 1997-08-22
W O 96~7026 PCTrUS96/02380 - ~7 -
assembly of a full ribozyme from its two parts in a magnesium-less mixture,
and after washing away free incomplete ribozymes, activating of the full
ribozyme by addition of magnesium ions.
The ribozymes used in most embodiments of the detection system
S of the invention are in most cases universal, i.e. the same ribozyme can be
used to detect different assayed biomolecules since the specificity is acquired
by the attached recognition biomolecule (in the activation embodiment), or
by the first and second oligonucleotide molecules (in the transcription
embodiment) or by third and fourth oligonucleotide sequences (in the
10 assembly embodimcllt). Thc fifth oligolluclcotidc in thc case of the
completion embodiment of the invention has to be tailor made for each
specific nucleic acid to be assayed, since the sequence recognizing the
assayed sequence is part of the ribozyme itself.
The present invention also providcs reagents required for carrying
15 out the above method as well as a kit comprising said reagents.
In the following thc invclltion will bc dcscribe~l with rcfcrcncc to
~;omc noll-limitill~, (3rawin~~,s and cxamplc~i:
In the drawings various symbols are used which in the context of
the present invention have the following meanings:
Straight line ( ) - DNA strand
Wavy line (~ RNA strand
A,B,C, etc. ................ - sequences in the coding strand of a
DNA
A',B',C', etc. ............. - sequences in the complementary
non-coding DNA strand
a,b,c, etc. ................ - RNA scqucnces
a',b'c', etc. - RNA scqucnces complementàry to
a,b,c
S~JBSTITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W 096/27026 ~ ,./02380
- 28 -
immobilization on a solid support
* detectable label
inhibitory moiety.
S BRIEF DESCRIPI'ION OF THE DRAWINGS
Fig. 1 shows an embodiment of the catalytic system of the invcntion
comprising two species of composite molecules activated by cross-cleavage;
Fig. 2(a) and 2(b) show a catalytic system in accordance with an
embodiment of the invcntioll comprisillg OllCSpCCiCS of composite molecule
10 activated by cross-cleavage or cross-splicing: wherein the ribozyme is in
the form of a closed circle (Fig. 2(a)); wherein the ribozyme requires
splicing for becoming active (Fig. (b));
Fig. 3 shows various manners in which composite molecules can be
separated from one anothcr: by immobilization to distinct sites of the
15 reaction vessel (3A); by linkage to largc beads (3B); by linkage to charged
moieties (3C); and by placing each species of composite molecules at
opposite sides of a porous membrane;
Fig. 4 shows an example of the detcction system according to the
activation embodiment of the invention wherein the ribozyme is activated
20 by addition of magnesium ions;
Fig. 5 shows another example of the detection system according to
the activation embodiment of the invcntion wherein the ribozyme is
activated by modification of an inhibitory moiety;
Fig. 6 shows a detection system in accordancc with the transcription
25 embodiment of the invention;
Fig. 7(a) and 7(1)) show a detection system in accordance with the
assembly embodiment of the invelltion; where the rccognitioll sequence of
the ribozyme hybridize to the assaycd nucleic acid sequence (Fig. 7(a)); or
SUBSTITUTE SH EET (RULE 26)
. CA 02213622 1997-08-22
W O 96/27026 ~~ -t02380
_ ~9 _
where the openecl stemp-ll of the ribozyme hybridizes with the assayed
sequence (Fig. (b));
Fig. 8 shows an example of the catalytic system of the invention
comprising two species of ribozymes activated by cross-ligation;
Fig. 9 shows yet another example of the detection system according
to the activation embodiment of the invention wherein the ribozyme is
activated by rendering the cleavable sequence single stranded.
Fig. 10 shows the cleavage results of a dcteclion system comprising
the stem-II open ribozyme of Fig. 7(b);
Fig. 11 shows the cleavage results of a catalytic system comprising
the closed-circle compositc molecule of Fig. (b); and
Fig. 12 shows the cleavage results of ribozyme in the presence of
untreated blood and denaturing agents.
DESCRIPI'ION OF SPECIFIC EMBODIMENTS
Catalytic system
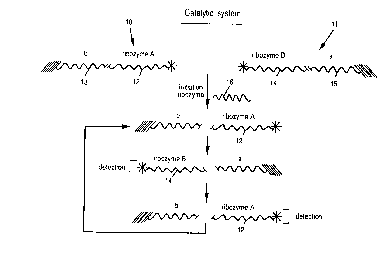
Refercncc is first ma~le to Fig. 1 showing one manncr of
constructing the catalytic system of the invention. The catalytic system
comprises two species of composite molecules 10 and 11. Composite
0 molecule 10 comprises one type of labelled ribozyme which will be denoted
ribozyme A (12) linkcd to an RNA sequence denoted b (13). Composite
molecule 11 comprises another type of labelle(J ribozyme which will be
denoted ribozymc B (l 1) an~l an RNA sequence a (l5). Ribozyme B in
molecule 11 is capablc of clcavinL~ sequellce b in molecule 10 and
ribozyme A in molecule 10 is capable of cleavin~ sequence a in mole-
cule 11. Initially molcculcs 10 ancJ ll are not able to interact since each is
~ immobilizcd to ~ distinct sitc of thc rcaction vessel. Initiation ribozyme 16
is also capablc of clcaving scqucnce b in molccule 10.
SUBSTITUTE SH EET (RULE 26)
CA 02213622 1997-08-22
WO 96127026 ~,-lIU' _~ 102380
--30--
If initiation ribozyme is present in thc rcaction mixturc then
sequence b is cleaved releasing free ribozyme A (12) to the reaction
mixture. Free ribozyme A (12) is able to diffuse within the reaction vessel
to cleave molecule 11, thus releasing to the reaction mixture free
S ribozyme B (14). Free ribozyme B (14) is again able to migrate through the
reaction vessel to cleave molecule 10 to release again free ribozyme A (12)
and the cycle is repeated again and again in a positive-feedback m~nnP.r.
Since both ribozyme A and B are labelled, detection of either or both in the
supernatant indicates thc presellce of initiation ribozyme 16 in the reaction
10 mixture.
The following is an example of molecule 10 comprising a
ribozyme 12 of the type hammerhead linked to sequence 13 which is then
labelled at 3'-end by biotin: (Capital letters: "-O-methylated; small
letters: RNA)
S'CCA cugauga gGCC GAAA GGCc gaa acGUguc CGU AAA-
The following is an example of molecule 11 comprising
another ribozyme l~ of the type hammerhead which is capable of cleaving
O seqeuence 13 present in molccule 10. Ribozyme I 1 is linked to sequence
15 capable of being cleavecl by ribozyme 12 of molecule which is then
labelled at its 3'-end by biotim (Capital letters: '-O-methylated; small
letters: RNA)
5 5'-GAG ACG cugauga gGCC GAAA GGCc gaa acAC guc UGG AAA
Although ribozymes A and B are referred to as different
ribozymes and sequences a and b are refcrred to as (:lifferent sequences, both
the ribozymes and sequences may be actually idelltical. In such a case self-
SUBSTITUTE SHEET(RULE 26)
. CA 02213622 1997-08-22
W 096127026 l~ 3~02380 - 31 -
cleavage in each molecule 10 or 11 is avoided by linking the ribozyme to
its attached sequence in such a proximity which does not enable cleavage in
cis, while free ribozymes are able to cleave composite molecules in trans.
This can be done by linking the ribozyme imme~liately adjacent to its
S potentially cleavable sequence. This is because the ribozyme should be
spaced from its potcntially clcavable scqucncc by xcvcral nuclctoidcs for
efficient cleavage to take place. Therefore, when the ribozyme in the
composite molecule is linked directly with no spacing to the cleavable
sequence, there is no possibility of cis cleaving and the sequences can be
cleaved only in trans while cnablillg only cleavage in trans.
Reference is now made to Fib. (a) which shows another
alternative where only one species of composite molecules is present in the
reaction vessel. The catalytic system comprises a single species of
molecules 17', each comprising a ribozyme C' (18') and a cleavable
sequence c' (19'). The molecule 17' is in the form of a closed circle and
thus ribozyme 18' is initially inactive.
If catalytically active iniation ribozyme 16N is present in the
media, it is able to cleave the s;cquence c' and open the ribozyme to become
active. Open ribozyme 18' can in turn open, by cleavage, additional
"O composite molecules 17' turning them active. Detection can be carried out
by using a fluorescent label (F) and Rodamine (Rd). When the two are
adjacent as in the closed molecule the light emission of the fluorescent label
is quenched and when they are separated, as in the open molecule sequence,
the light emission of the fluorescent label becomes stronger.
Reference is now made to Fig. (b) which shows yet another
alternative for the catalytic system comprising only a single species of
composite molecule. Ribozyme 17" has in its core region an extra
nucleotide sequcnce 18" which renders the ribozyme inactive. At the
SUBSTITUTE SH EET (RULE 26)
CA 02213622 1997-08-22
W 096/27026 PCTrUS96/02380
' - 3~ -
tçrmin~l of the extra sequence is a blocking group 19 which does not allow
spontaneous ligation of the open end.
Initiation ribozyme 16, which has the catalytic activity of
splicing is capable of both cleaving out extra sequence 18 and blocking
5 group 19 and then of ligating the free ends to give a functional ribozyme.
Functional ribozyme is then capable of splicing other ribozymes in the
reaction medium cau~inp an amplification of the reaction. Detection
according to one option (Option 1) is carried out essentially as described in
Fig. 2(a), but in this case, initially the Rodamine (Rd) at the fluorescent
10 group (F) are separated and only upon activation of the ribozyme they
become adjacent, so that active ribozyme is detected by quenching of light
emission. According to the second mode of detection (Option '~) the spliced
out group comprising free extra sequence 18 and blocking group 19
carries a detectable label.
The advantages of the mode of Fig. (b) resides in a very low
"noise" level since in order for an inactive ribozyme to become spontane--
ously active (not in the presence of an initiation ribozyme) two spontaneous
occurrences must happen spontaneous cleavage (at a probability of 10~6/min
in 10 mM MgCI at physiological pH and at a temperature of 37~C) and
~0 spontaneous ligation (at a probability of 10 7/min) giving a very low
probability for spontaneous activation (10 l3/min).
The label attached to either or both ribozymes A, B or C
(Fig. 1) may be any detectable label known in the art such as a radioisotope,
a fluorescent label, an enzyme which in the presence of a substrate is
'~5 capable of producing a color rcaction, etc.
Fig. 3 shows various manners in which the two composite
molecules 10 and 11 of the first embodiment are positioned so as to avoid
mutual interaction but allow interaction between free ribozymes 12 and 14
and composite molecules. It should be understood that the same principles
SUBSTITUTE SH EET (RULE 26)
- CA 02213622 1997-08-22
W O 96127026 ~liU~96/02380
--33--
apply also to the other manner for constructing the catalytic system of the
invention, i.e. where only one species of composite molecule is present,
,res~ te~l in Fig. 2.
In Fig. 3(A) molecules 10 and 11 are immobilized onto distinct
and separate sites of the reaction vessel 18 while ribozymes 12 and 14
diffuse freely in the reaction mixture.
Fig. 3(B) shows molecules 10 and 11 which are immobilized
onto beads 19 the size of which prevents interaction between said molecules.
However, free ribozyme 12 and 14 are able to diffuse freely in the reaction
mixture and interact with the composite immobilized molecules.
Fig. 3(C) shows another example of separating composite
molecules wherein composite molecules 10 and ll are attached to charged
moieties bearing the samc charge 37. The electrical rejection between their
attached moieties eliminates the possibility of intcraction between
molecules 10 and 11. However, free ribozyme 12 and 14 which are
essçn1i~1ly uncharged are able to interact with the composite molecule.
Fig. 3(D) shows yet another example of separating the
composite molecules 10 and 11 by placing them at opposite ends of a porous
membran~ 34 which ~crves as a sievc, blocking passage of large
''O molcculcs lO and ll whilc allowing passagc of thc ~mallcr frcc
ribozymes 12 and 11.
Another manner for ensuring that the two composite
molecules 10 and 11 cJo not interact is by the use of blocker molecules
which are complementary to a specific sequence rendering it double
stranded. According to this manner, composite molecule 10 comprises a
blocker molecule which renders the cleavable sequence b and part of the
catalytic region of ribozyme A double stranded. In the partially double
stranded composite molccule lO, the ribozyme is not active due to the fact
that its catalytic region is clouble strancled. Composite molecule 11 is
SUBSTITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96/27026 ~-l/U~ 02380
- 34 -
blocked in a similar manner. If initiation ribozyme is present in the reaction
mixture, it displaces a part of the blocker molecule present on the composite
molecule 10, and then the initiation molecule is able to cleave sequence b.
Once sequence b is cleaved ribozyme A is also rendered active since the
S partially displaced blocker molecule completely falls off composite
molecule 10 turning its catalytic region to become single stranded and
active. Active ribozyme A then displaces the blocker molecule of composite
molecule 11 in a similar manner as described above cleaving the cleavable
sequence a, turning Ribozyme B single stranded and active. Ribozyme B
10 then activates composite molecule 10 in a similar manner to the activation
of the initiation ribozyme described above, and cross activation of the two
composite molecules can then proceed.
Reference is now made to Fig. 8 which shows another
alternative for constructing the catalytic system of the invention. The
15 catalytic system comprises two species of ribozymes 80 and 81 which are
active when fully assembled but are inactive when separated to their parts
80a, 80b and 81a, 81b, respcctively. Full ribozyme 80 is capable of ligating
ribozyme parts 81:~ and 81b to produce a full and active ribozyme 81. Full
ribozyme 81 is capable of ligating ribozyme parts 80a and 80b to produce
20 a full and active ribozyme 80, so that cross-activation proceeds by cross-
ligation.
Initiation ribozyme 86 or 86' is capable of creating a full and
active ribozyme 80 cither by clcaving a full but immobilized ribozyme from
a location wherein it is spatially separated from ribozyme parts 81a and 81b,
25 for example, in one of the mamlers specified in Fig. 3 (Fig. 8 top left) or by
being ablc to ligate parts 80a and 80b to form the full and active
ribozyme 80 (Fig. 8 top right).
SUBSTITUTE SH EET (RULE 26)
. CA 02213622 1997-08-22
WO g6n7026 ~ 96/02380
- 35 -
DETECTION SYSTEM
Where the method of the invention is to be used to aid in the
detection of biomolecules other than ribozymes, the invention includes also
a detection system capablc of producing catalytically active initiation
ribozyme only in the presence of the assayed molecule.
Fig. 4 shows one example of the activation embodiment of the
invention. In this examplc, the assayc~J biomolcculc is an immobilizcd
nucleic acid sequence A (41) for example a DNA sequence. Immobilization
can be carried out in accordancc with any method known in the art, for
example, with the aid of a cross-lillkill~ agcllt or by trapping the a~sayed
nucleic acid molecule betwccn two porous membranes which permit passage
of smaller molecules. Where the assayed biomolecule is a protein it can be
immobilized onto bcads carryillg appropriatc trapping agents, such as
suitable immobilized antibodies, directed against regions which are not
required for detection, etc. Altematively, the assayed biomolecule can be
immobilized onto a nitroccllulose shcct ancl another protein, such as
albumin, should then be applied onto the nitrocellulose sheet in order to
saturate all the shects' vacant locations and avoicl, in the next step, non-
specific absorptioll.
The detection system also comprises first complex molecule 42
comprising ribozyme 43, linked to cleavable sequence c' (44) capable of
being cleaved by active ribozymc, and further comprises sequence a' (45)
complementary to the assayc~l sequence A (41). Ribozyme 43 does not
self-cleave since molecule 42 is kept in magnesium-less reaction mixture
which eliminates the ribozymes catalytic activity. This can be done, for
example, by keeping complex molecule 42 in a magnesium-less EDTA--
~ containing reaction mixture.
SUBSTITUTE SHEET (RULE ~6)
CA 02213622 1997-08-22
W 096127026 ~l~ 02380 - 36 -
An example of ribozyme ~13 linkcd to the cleavage sequence
c' (44) of the type hammerhead (Capital letter~ 0-methylated; small
letters: RNA; over- and undcrlined: DNA)
5 5'--GCAACAGTGGAGGAAAGCC UACguc UGG UACGU CCA cugauga
gGCC GAAA GGCc gaa acGUAGU AAA
Molecules 41 and 42 are allowed to hybridize to give
immobilized hybrid 46. Frec molcculcs 42 are washed away, and to the
10 immobilized hybrid 46 are added magnesium ions in a concentration
sufficient to activate ribozymes. In the presence of such a concentration of
magnesium ion ribozymc 13 is ablc to cleave scquence c', thus releasing
itself to the reaction mixturc while leaving immobilizcd cleaved hybrid 47.
Free and catalytically activc ribozyme 43 can serve as the initiation
15 ribozyme in the catalytic system.
Fig. ~ shows another example of the activation embodiment of
the invention. Assayed biomolecule 51, which comprises nucleic acid
sequence A, for example a DNA sequence is immobilized as described
above. The detection system comprises a second complex molecule 52
20 comprising ribozyme 53 linked to sequence c' (54) which can be cleaved
by catalytically active ribozyme and sequence a' (55) complementary to
sequence A in the assayed biomolecule. The complex molecule also
comprises an inhibitory moiety 58 which, while present in its unmodified
form, inhibits the catalytic activity of ribozyme 53. An example of an
5 inhibitory molecule is a nucleic acid sequencc complementary to part of the
ribozyme. In the prcscllcc of such a scqucllcc, thc ribozymc folcls to an
inactive three-dimensional form.
Molecules 51 and 52 are allowed to hybridize to give
immobilized hybrid 56, and free molecules 52 are washed away. To
30 separate hybrid 56 are addcd modifying substanccs which are ablc to intcract
SUBSTITUTE SH EET (RULE 26)
- CA 02213622 1997-08-22
W 096t27026 1~11~'_-'02380
- 37 -
with the inhibitory moiety 58 and modify it to an un-inhibiting form. For
example, where the inhibitory moiety is a nucleic acid sequence which
causes folding of the ribozyme, the modifying substance may be a sequence
complementary to the inhibitory moiety, which hybridizes and blocks the
S inhibitory moiety, thus allowing the ribozyme to rcfold to its active form.
Alternatively, the modifying substances may be substances able to remove
or cleave the inhibitory moiety, thus termin~ting its inhibitory action.
Active ribozyme is then able to cleave sequence c, thus releasing itself
from immobilized cleaved hybrid 57. Catalytically active free ribozyme 53
then serves as the initiation ribozyme in the catalytic system.
Reference is now made to Fig. 9 which shows another example
of the activation embodiment of the invention. Molecule 91 comprises the
sequence of the initiation ribozyme 90, attached to sequence c, cleavable by
the ribozyme, and to sequence a and b which are capable of hybridizing
with the assayed biomolecule bcing, for example the assayed DNA sequence
of molecule 9~. Molecule 91 further comprises blocker DNA sequence 92
which comprises sequence B and C, capable of hybridizing to sequences b
and c of molecule 91, respectively, to give a double-stranded structure.
Blocker DNA sequence 92 is linked via linker sequence 93. The
ribozyme 90 is not capable of cleaving sequence c since its sequence region
is double stranded (through hybridization to blocker sequence 92).
The assayed molccule 94 is then introduced to the reaction
mixture. If the assayed biomolccule is complementary to a and b of
molecule 91, then blocker sequence 92 is displaced by the assayed mole-
~ 25 cule 94 to give hybrid molecule 95. In hybrid molecule 95, the cleavable
sequence c is single stranded enabling the ribozyme to cleave it and thus to
be freed to the reaction mixture as a catalytically active ribozyme 96 which
serves as the initiation ribozyme in the catalytic system.
SUBSTITUTE SH EET (RULE 26)
CA 02213622 1997-08-22
W 096/27026 1~-lIV~96/02380
' - 38 -
According to this embodiment conditiolls such as temperature,
the length of the part of the recognition biomolecule b which is double
stranded, etc. must be chosen with care so that the assayed molecule is able
to displace blocker molecule 92 e~senti~lly only if the assayed biomolecule
sequences A and B are perfectly matched to recognition sequences a and b.
Reference is now made to Fig. 6 which shows the transcription
embodiment of the detection system of the invention appropriate where the
biomolecule is a nucleic acid sequence. According to this specific
embodiment a correct DNA template, which cventually brings to the
10 transcription of initiation ribozyme, is assemblcd from its parts only in the presence of the assayed nuclcic acid sequcnce. The detection system
comprises a first oligonuclcotide molccule 61, being essentially DNA,
comprising from 3' ,~'; a double-strallded promotor, a sequence R coding
for the complementary sequcnce of the initiatioll ribozymc, a sequcnce C
15 coding for a sequence cleavablc by a catalytically active ribozyme, and a
sequence Dl complementary to the ~ ' -part of the assayed nucleic acid
scquence. The detection systcm furthcr comprises second oligonucleotide
molcculc 62, bcing cssclltially DNA comprisillg from 3'~': a scqucncc D2
complementary to thc 3'-part of thc assayecl nuclcic acid scquence and a
20 triggering oligonuclcotide tcmplate (TRIG). if assayed nucleic acid se-
quence 63 is present, ancl under appropriate hybridization conditions,
sequence Dl of molecule 61 and sequence D2 of molecule 62 hybridize with
sequences Dl' and D2', respectively, of the assayed nucleic acid sequence
to give hybrid 6~.
In the presence of transcription system non-template strand
oligonucleotide 65 is produced, comprising from 3'~': triggering
oligonucleotide sequence tri~, dt' and dl' scquences, sequences c' r',
complementary with those of thc clcavable nuclcic acid sequenccs and the
initiation ribozyme, respectivcly.
SUBSTITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96n7026 ~liU~_-/02380
- 39 -
To the reaction mixture are added molecules of back promotor
construct 66 comprising a single-stranded DNA promotor linked to a
sequence capable of hybridizing with the oligonucleotide triggering sequence
TRIG. Under appropriate conditions the back promotor construct 66
S hybridizes with molecule 65 to give hybrid 67. In the presence of a DNA
polymerase the single-stranded promotor is completed to give a functional
double-stranded promotor in hybrid 68.
Hybrid 68 can serve, in the presence of transcription rcagents,
as a template for the production of final oligonucleotide transcript 69
10 comprising from at its S'-end: an initiation ribozyme sequence r and a
cleavable sequence c capable of being cleaved by said ribozyme.
The ribozyme r cleaves cleavable sequence c thus releasing
itself to the surrounclillg mcdium in thc form of frcc ribozyme 70. Free
ribozyme 70 can serve as the catalytically active initiation ribozyme in the
15 catalytic system.
One mode of thc asscmbly emboclimellt of the invention is
shown in Fig. 7(a). Thc dctcctioll systcm comprises assayed
biomolecule 71, comprisillg a nuclcic acid scqllellce A~ A~. In addition, the
dctection systcm compri~ics a part of a ribozymc 72, comprisillg oligonu--
O cleotide sequcnce a'~ complemcntary to sequellce Al ancl another part of a
ribozyme 73 comprising oligonuclcoti~le sequence a2' complementary to
sequence A2. The parts of ribozyme 72 and 73 together constitute a
complete ribozyme if the two parts are asscmbled.
If assayed moleculc 71 is present in the medium, it can
25 hybridize to a, of ribozyme part - 72 and a2 of ribozyme - part 73 bringing
the two parts togethcr to form functional ribozymc-assayed sequence hybrid
76, which may scrvc as an initiatioll ribozymc in a catalytic system, for
example, by clcaving molcculc 77, which clcavage may be rcquircd to
initiate the amplification cascade in thc catalytic system.
SUBSTITUTE SH EET (RULE 26)
CA 02213622 1997-08-22
W 096r27026 . ~ /02380
- 40 -
Another mode of the assembly mode of the invcntion is shown
in Fig. 7(b).
Ribozyme 79 of the hammer-head type has been constructed
in which Stem-II has been shortened to have only one complementary
nucleotide (represented by one line in the Fig. 2b) and the rem~inin~ portion
of the stem have been opened to give arms a and b. Ribozyme 79 is
capable of hybridizing with sequence 70 to form Stems I and III and then
perform the catalytic activity, for example, cleavage of sequence 70.
However, ~ prioli ribozyme 79 is incapable of cleaving the cleavable
10 sequence 70 since its Stem-II is open and inactive. Arms a and b of the
opened core have been constructed to be complementary to sequences A
and B of the assayed sequcnce, for example, DNA sequence 80.
In the presence of assayed DNA sequence 80, the arms a and b
of the stem-II ribozyme 79 are hybridized to the assayed sequence to
15 produce a fully double-stranded Stem-II and thus the ribozyme becomes
catalytically activc and can scrvc as an initiatioll ribozyme in a catalytic
system where, for example, cleavable sequence 70 is part of an enzyme in
the catalytic system which requires cleavage for its activation.
20 EXAMPLES
Example 1: Detection of ~n assayed nucleic acid sequence using a
ribozyme with ~n open sten~
Ribozyme HH8 was dissected into two parts at the loop of
25 stemp-II. Each of the two ribozyme halves of HH8 (HH8-3 and HH8 5)
has a different additional 17 bases tail sequence complementary to the
LAMTAR0 DNA target molecule. In the presence of the target, the two
halves are brought together and form an active ribozyme. In the LAMTAR0
target the two sequences complemelltary to the ribozyme halves are
30 continuous. In the other LAMTAR moleculcs (LAMTAR1 through
SUBSTITUTE SHEET(RULE 26)
. CA 022l3622 l997-08-22
W O 96/27026 r~l/~ 02380
. - 41 ~
LAMTAR4) the two sequences are separated by 1 to 4 non-complementary
bases respectively. The ribozyme substrate is SB8-''4 which contains the
sequence recognized by HH8.
(a) Method:
1. Sequences:
Oligodeoxyribonucleotides were synthesized on an Applied
Biosystem 381A DNA synthesizer according to the manufacturer's recom-
mended protocol. The Ampliscribe kit (Epicenter Tcchnologies) was used
ln for all RNA synthesis 1~32 Pl UTP 13nnnCi/mmoll was purchased from
Rotem Industries Ltd., Isracl.
DNA targets:
LAMTARO:
S'GCTCCGAGTCCACCTGCACGCCGACCAGTGCCGTÇTTCGGGA 3'
LAMTARl:
S'GCTCCGAGTCCACCTGCACGCI'CGACCAGTGCCGTGTTCGGGA 3'
LAMTAR~:
S'GCTCCGAGTCCACCTGCACGCI-l'CGACCAGTGCCGTGTTCGGGA 3'
LAMTAR3:
~0 5'GCTCCGAGTCCACCTGCACGCl'ATCGACCAGTGCCGTGTTCGGGA 3'
LAMTAR~:
5'GCTCCGAGTCCACCTGCACGCrA'l'ACGACCAGTGCCGTGTTCGGGA 3'
UnderJined-complelllenlar~ to ril)oz~me half;
I~l)l(l-non-complemenlar)~ addilional sequence.
s
RNA transcripts:
SB-''~ (substrale for HH8):
S'GGUCACAAUGUCGGUCGAGUUCCA 3'
HH8-3 (ribozyme hal~:
5'GGCGACCCUGAUGAGGCCGCGUGCAGGUGGACUCG 3'
HH8-5(ribozynle hal~:
5'GGAACACGGCACUGGUCGGGCCGAAACAUUAA 3'
Underlined-complelllenlar~ to largel.
2. Preparation of RNA:
DNA oligonucleotides were syllthesized according to (1) above.
The oligonucleotides wcrc purified by electrophoresis O~ % polyacryl-
amide 7M urea gcl, UV-shadowccl and clutcd ovcrnight at room tcmperature
SUBSTITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96/27026 PCTrUS96102380
-- 4
in O.SM Tris-CI (pH-7.0), ().l% SDS and ().l mM EDTA. Elutcd DNA
was precipitated with 0.1 volumes of 3M sodium acetate and 3 volumes of
ethanol, resuspended in 1 mM Tris-CI (pH-7.0) and 0.1 mM EDTA and
stored at -20~C until use. Purified oligonucleotides were annealed to
5 complementary non-template T7 RNA polymerase promoter oligonucleotide
(TAA TAC GAC TCA CTA TAG G) in '~0 mM each by incubating at 95~C
for 15 seconds and at 7()"C, 6()oc, 55~C, 5()~C, 45~C, 40~C and 37~C for
S min at each temperature. Transcription reaction mixture (S0 ml) contained
2 mg of annealed DNA, 1X reaction buffer, 10 mM Dithiothreitol, '' mM
ATP, CTP and GTP, 1 mM UTP, ~ mCi [a3~ P~ UTP and 1.1 mM MgCI2.
The mixture was incubated for 1 hr at 37~C and then for S minutes at 80~C
to deactivate the enzyme. The RNA was precipitated as described.
Transcripts were purified by elcctrophoresis on 15% polyacrylamide 7 M
urea gel. RNA was located by autoracliography and eluted as described.
15 Eluted RNA was prccipitatcd as dcscribed, resuspcnclecl in n.1X TE and
counted in scintillation flour (Luma LSC).
3. Cleavage reaction:
Reactions ~10 ml) were normally carried out in the presence
'~O of O.S pmol ribozyme, sn mM Tris-CI (ph-7.5), 1 mM EDTA (pH-7.5),
0.05% SDS and 30 mM MgCI2. The reactions were preincubated at 95~C
for 1 min in order to eliminatc alternative RNA conformations which may
have formed during storage at -''()~C. The reactions were incubated at 37~C
for 1 hour and stopped by adding a dye solution containing 10 M urea and
S 10 mM EDTA and put on ice. The samples were denatured at 80~C for 5
min and run on 15% polyacrylamide 7 M urea gel.
SUBSTITUTE SHEET(RU~E 26)
- CA 02213622 1997-08-22
W O 96/27026 PCTAUSg6/02380
' - 43 -
(b) Results:
The results of the polyacrylamide gel are shown in Fig. 10.
Ribozyme without a target did not produce appreciable cleavage. When the
target was added, enhanced cleavage resulted. The enhancement was
5 greatest in LAMTAR4 Ribozymes tested.
Example 2 Catalytic system comprised a closed-circle composite
molecule
The SLS-prccursor ribozyme (Rz) is a circular Rz with 11 bp
long stemp-II. The two recognition arms are connected in tandem with an
extra cleavage base separating them. Therefore this robozime has no
activity, but serves as a tcmplate for active ribozymes. Once it has been
cleavecJ, the "open" ribozymc bccomcs activc (as shown schcmatically in
15 Fig. ~(b)). ln order to initiate the catalytic system an initiation ribozyme
must be present. Spontaneous cleavage of RNA occurs at a rate of 1 event
per 1 o6 molecules per minute in 30 mM MgCI2 at 37~C. A circular
ribozyme spontaneously clcavcd at the cleavage sitc bccomcs active and is
capable of serving as the tri~gcr.
o
(a) Methods
1. Sequences:
Oligodeoxyribollucleotides were synthesized on an Applied
Biosystem 381A DNA synthesizcr according to the manufacturer's recom-
25 mended protocol. The Ampliscribe kit (Epicenter Tcchnologies) was used
- for all RNA synthesis 1~32 Pl UTP [3000Ci/mmol~ was purchased from
Rotem Industries Ltcl., Israel.
The RNA precursor (SLS's) sequellces were: 5' GGU CAG
CAG UCG AA ~Recognition arm I] X ~Recognition arm III] CUG AUG
30 AGA CUG CUG ACC A 3'
SUBSTITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96/27026 PCTnUS96102380
' - 44 -
SLS code Recognition arm 1 Recognition arm III Cleavage Site
107 CGCG AAUU ~U
108 CUAG AAUU A~
113 UAUA AAUU A/U
115 UGCA AAUU A/U
208 CUAG ACGU A/U
213 UAUA ACGU U
215 UGCA ACGU ~U
313 UAUA AGCU A/U
2. Preparation of RNA
PrcparationofRNA was conductcd as describcd in Example
1 above.
3. Spontaneous cleavage reaction:
Reactions (1() ,1l1) were normally carried out in the presence of
S pmol precursor ribozymc (SLS-transcripts), S0 mM Tris-CI (pH-7.5), 1
mM EDTA(pH-7.~),().n5~~SDSalld300 mM MgCI2. The reactions were
20 preincubated at 9~~C for 1 min in order to elimin~te altcrnative RNA
conformations which may havc formcd furin~ storage at -~noc. The
reactions were incubatcd at 37~Cforl hour or ovcrnight and stopped by
adding a dye solution containing 1û M urea and 10 mM EDTA and put on
ice. The samples were denatured at 80~C for 5 min and run on 15%
''5 polyacrylamide 7 M urea ~cl.
(b) Results '
Thc rcsults of thc polyacrylamidc gcl arc shown in Fig. 11.
A panel of 8 different circular ribozymcs was examined for cascade activity.
The cascade was initiated, hy spolltaneous cleavagc as described above. As
SUBSTITUTE SHEET (RULE 26)
. CA 02213622 1997-08-22
W 096/Z7026 ~ 96/02380
- 4~ -
shown in Fig. 1, one of the ribozymes (~313) demollstrated a functioning
catalytic cascade resulting in amplification in a positive feedback manner.
Example III Ribozyme inhibition by blood and various materials
S used for DNA preparation
All amplification techniques require a sample preparation step
to release nucleic acids and eliminate inhibition of the reactions by blood
components. Several materials like SDS, phenol an~J guanidinium are used
10 in these preparations. An amplification re~ction without the need of a
sample prep step is to be performed. Ribozyme activity in the presence of
blood, with and without nucleic acid releasing agents, was tested. Both
RNA-only and modified ribozymes were examined.
15 (a) Method
1. Oligonucleotides:
Oligodeoxyribonucleotides were purchased from the unit for
molecular biology of the H<-clcl,lcs~ Hospital, Mount Scopus, Jerusalem. The
modified ribozyme was synthcsized by RPI, Bouklcr, Co. The Ampliscribe
20 T7 transcription kit (Epicenter Technologies) was used for all RNA
synthesis. ~y32p~ ATP L6()(K)ci/mmol] an~l Lcx32P~ UTP L3()00Ci/mmol] were
purchased from Rotem Industries Ltd, Israel. T4 Polynucleotide kinase was
purchased from NEB, Beverly, Ma.
2. Ribozymes:
DS-~-RzA3-6: 5' GCAACAGTGGAGGAAAGCCUACgucUGGUACGUCCA
c~lga-lgagGCCGAAAGGCcgaaacGUAGUAAA 3'
., ribonucleoti~lcs;
~ U~ ,asc '2'-0-Mctll) ] modificalioll;
Undcrlincd u~",c,~as-;- dco~ ribonuc]colidcs.
HH8(RNA transcripl):
5'GGCGACCCUGAUGAGGCCGAAAGGCCGAAACAUUAA 3'.
SUBSTITUTE SHEET (RULE 26)
CA 02213622 1997-08-22
W O 96/27026 PCTnUS96/02380
--46--
3. Labeling of modified ribozyme:
50 pmoles of the ribozyme, 10 units of T4 Polynucleotide
kinase and '~0 ,L~Ci[y32P] ATP were incubated in lX reaction buffer in a total
volume of 10 ,ul at '~5~C for 1 hr. The reaction was terminated by
5 incubation at 60~C for 10 minutes.
4. Preparation of RNA:
HH8 template DNA oligonucleotide was purified by electro-
phoresis on 15% polyacrylamide 7 M urea gel, UV-shadowcd and eluted
overnight at room temperature in 0.5 M Tris-CI (pH-7.5), 0.1% SDS and
0.1 mM EDTA. Eluted DNA was precipitated with 0.1 volume of 3 M
sodium acetate and 3 volumcs of ethanol, resuspenclcd in 0.1 XTE (1 mM
Tris-CI pH-7.0 and 0.1 mM EDTA) and stored at - 0~C until use. Purified
oligonucleotide was annealed to complementary non-template T7 RNA
15 polymerase promoter oligonucleotide (TAATACGACTCACTATAGG) in
'~0 ,uM each by incubatillg at 9~~C for 15 SCCOlldS all~l at 7n~C, 60~C, 55~C,
50~C, 45~C, 40~C and 37~C for ~ min at each temperature. Transcription
reaction mixture (50 ,ul) contained '' ,uM of annealed DNA, lX reaction
buffer, 10 mM Dithiothreitol, mM ATP, CTP and GTP, 1 mM UTP, 25
'~0 mCi [a32 P] UTP and 1.1 mM MgCI2. The mixture was incubated for 1 hr
at 37~C and then for 5 minutes at 8()~C to deactivatc the enzyme. The RNA
was precipitated as described. Transcripts were purified by electrophoresis
on 15% polyacrylamide 7 M urca gel. RNA was located by autoradiography
and eluted as described. Eluted RNA was precipitated as described,
2~ resuspended in 0.1X TE and counted in scintillation flour (Luma LSC).
5. Cleavage reaction:
Reactions (1(),UI) wcrc normally carricd out in the presence of
0.5 pmol modified ribozymc or HH8, 50 mM Tris-CI (pH-7.5), 1 mD
SUBSTITUTE SH EET (RULE 26)
. CA 02213622 1997-08-22
W 096/27026 1~1~ '02380
--47--
EDTA (pH-7.5), 0.05% SDS and 30 mM MgCl~ l of whole blood or
O.~,ul of whole blood and the additional component were added (See Fig.
1). The reactions were incubated at 37~C for 1 hour and stopped by adding
a dye solution cont~inin~ 10 M urea and 10 mM EDTA and put on ice. The
5 samples were denatured at 80~C for S min and run on 1~% polyacrylamide
7 M urea gel.
(b) Results
The results are shown in Fig. 1'', wherein 0.5 pmol or
10 ribozymes were present in each sample an(i 1 ,ul blood was mixed with
either 10% SDS 4 M guanidine isocyanate or phenol chloroform 1:1, 1 ,~cl
of each trealed blood sample or the agent alone was added to the reaction
test. The results of ribozyme activity is not inhibited by either 1% SDS,
0.35 M guanidine or by ~% phcnc)l~)chloroform whethcr blood is present or
15 not. Untreated blood dcgradcs thc RNA part of thc ribozymcs. These
results show one of thc advalltagcs of the rihozyme based detection method
of the invention in that denaturating agents used in the preparation of the
catalytic sample do not hinder catalytic activity of the ribizoyme.
SUBSTITUTE SHEET(RULE Z6)