Note: Descriptions are shown in the official language in which they were submitted.
CA 02236113 1998-04-29
,~.. ~''~ vc ! ~~~s'' ~~
~,.~~~~1 V W I" ~ Y V/ 1'1
1
Specification
Negative Strand RNA Viral Vector Having Autonomous
Replication Capability
Field of the Invention
The present invention relates to a viral vector for the
gene therapy. More specifically, this invention relates to a
negative strand RNA viral vector.
Backcrrour~d of the Invention
As to the gene therapy for humans and animals,
therapeutic effectiveness and safety are very important
factors. Especially, therapy performed by using "viral
vector" obtained by the viral gene recombination needs to be
very cautiously carried out, when such undeniable
possibilities exist as that gene may be inserted to
unspecified sites of chromosomal DNA, that the recombinant
virus and pathogenic virus may be released to the natural
environment, and that the expression level of gene
transfected into cells cannot be controlled, or the like,
even though its therapeutic effectiveness is recognized.
These days, a great number of gene therapies using
recombinant viruses are performed, and many clinical
protocols of gene therapy are proposed. Characteristics of
these recombinant viral vectors largely depend on those of
viruses from which said vectors are derived.
The basic principle of viral vector is a method for
transferring the desired gene into targeted cells by
utilizing the viral infectivity. By "infectivity" in this
CA 02236113 1998-04-29
2
specification is meant the "capability of a virus to transfer
its nucleic acid, etc. into cells through its retaining
adhesiveness to cells and fusion capability to membrane".
With the surface of recombinant viral vectors genetically
manipulated to insert a desired gene are associated the
nucleocapsid and envelope proteins, etc. which are derived
from the virus and confer the infectivity on the recombinant
virus. These proteins enable the transfer of the enclosed
recombinant gene into cells. Such recombinant viral vectors
can be used for the purpose of not only gene therapy, but
also production of cells expressing a desired gene as well as
transgenic animals.
Viral vectors are classified into three classes
comprising the retroviral vector, DNA viral vector and RNA
viral vector.
These days, the vectors most frequently used in gene
therapy are retroviral vectors derived from retroviruses.
Retroviruses replicate through the following processes.
First, upon viral infection established, they generate
complementary DNAs (cDNAs) using their own reverse
transcriptase as at least part of catalysts and their own RNA
templates. After several steps, said cDNAs are incorporated
into host chromosomal DNAs, becoming the proviruses.
Proviruses are transcribed by the DNA-dependent RNA
polymerase derived from the host, generating viral RNAs,
which is packaged by the gene products encoded by their own
genes, becoming viral particles.
CA 02236113 1998-04-29
3
In general, retroviral vectors used in gene therapy, etc.
are capable of carrying out processes up to provirus
generation. However, they are deficient viruses deprived of
genes necessary for their packaging so that they do not form
viral particles from provirus. Retroviruses are exemplified
by, for example, mouse leukemia virus, feline leukemia virus,
baboon type C oncovirus, human immunodeficiency virus, adult
T cell leukemia virus, etc. Furthermore, recombinant
retroviral vectors hitherto reported include those derived
from mouse leukemia virus [see Virology, 65, 1202 (1991),
Biotechniques, 9, 980 (1989), Nucleic Acids Research, 18,
3587 (1990), Molecular and Cellular Biology, 7, 887 (1987),
Proceedings of National Academy of Sciences of United States
of America, 90, 3539 (1993), Proceedings of National Academy
of Sciences of United States of America, 86, 3519 (1989),
etc.] and those derived from human immunodeficiency virus
(see Journal of Clinical Investigation, 88, 1043 (1991)],
etc.
Retroviral vectors are produced aiming at efficiently
integrating a specific DNA into chromosomal DNA. However,
since the insertion position of the desired gene is
unpredictable, there is undeniable possibilities such as the
damage of normal genes, activation of oncogenes, and
excessive or suppressive expression of desired gene, due to
inactivation by insertion. In order to solve these problems,
a transient expression system using DNA viral vectors which
can be used as extrachromosomal genes has been developed.
CA 02236113 1998-04-29
4
DNA viral vectors are derived from DNA viruses, having
DNA as genetic information within viral particles.
Replication of said DNA is carried out by repeating the
process of generating complementary DNA strand using DNA-
dependent DNA replicase derived from host as at least one of
catalysts with its own DNA as template. The actual gene
therapy using adenoviral vector, a DNA viral vector usable as
extrachromosomal gene, is exemplified by the article in
[Nature Genetics, 3, 1-2 (1993)]. However, since, in the
case of DNA viral vectors, the occurrence of their
undesirable recombination with chromosomal DNA within nucleus
is also highly possible, they should be very carefully
applied as vectors for gene therapy.
Recently, RNA viral vectors based on RNA viruses have
been developed as conceivably more safer vectors than DNA
viral vectors described above. RNA viruses replicate by
repeating the processes for generating complementary strands
using their own RNA-dependent RNA replicase as the catalyst
with their own RNA as template.
The genome RNA of positive strand RNA viruses have dual
functions as the messenger RNA (hereafter simply called
mRNA), which generate proteins, depending on the
translational functions of host cells, necessary for the
replication and viral particle formation. In other words,
the genome RNA itself of positive strand RNA viruses has a
disseminative capability. In the present specification, by
"disseminative capability" is meant "the capability to form
CA 02236113 1998-04-29
infectious particles or their equivalent complexes and
disseminate them to other cells following the transfer of
nucleic acid into host cells by infection or artificial
techniques and the intracellular replication of said nucleic
acid". Sindbis virus classified to positive strand RNA
viruses and Sendai virus classified to negative strand RNA
viruses have both infectivity and disseminative capability.
Adeno-satellite virus classified in Parboviruses is
infectious but not disseminative (mixed infection with
adenovirus is required for the formation of viral
particles.). Furthermore, the positive strand RNA derived
from Sindbis virus which is artificially transcribed in vitro
is disseminative (forming infectious viral particles when
transfected into cells), but neither positive nor negative
RNA strands of Sendai virus artificially transcribed in vitro
is disseminative (generating no infectious viral particles
when transfected into cells).
In view of the advantage that the genome RNA functions as
mRNA at the same time, the development of RNA viral vectors
derived from positive strand RNA viruses preceded [see
Bio/Technology, 11, 916-920 (1993), Nucleic Acids Research,
23, 1495-1501 (1995), Human Gene Therapy, 6, 1161-1167
(1995), Methods in Cell Biology, 43, 43-53 (1994), Methods in
Cell Biology, 43, 55-78 (1994)]. For example, RNA viral
vectors derived from Semliki forest virus (SFV) and Sindbis
virus are basically of the RNA structure wherein the
structural gene regions related to the viral structure are
CA 02236113 1998-04-29
6
deleted, and a group of genes encoding proteins necessary for
viral transcription and replication are retained with a
desired foreign gene being linked downstream of the
transcription promotor. Direct transfer of such recombinant
RNA or cDNA which can transcribe said RNA [Nucleic Acids
Research, 23, 1495-1501 (1995)] into cells by microinjection,
etc. allows autonomous replication of RNA vectors containing
the foreign gene, and the transcription of the foreign gene
inserted downstream of the transcription promotor, resulting
in the expression of the desired products from the foreign
gene within cells. Furthermore, the present inventors
succeeded in forming an infectious but not disseminative
complex by the co-presence of cDNA unit (helper) for
expressing the viral structural gene and that for expressing
said RNA vector in the packaging cells. However,
recombination between RNA derived from helper and vector RNA
often occurred during packaging, resulting in the emergence
of infectious particles. Then, it was elucidated that spike
proteins present in the icosohedral capsid characteristic of
positive strand RNA viruses catalyzed this recombination.
Therefore, the introduction of variation into spike proteins
has been attempted to solve these problems [Bio/Technology,
11, 916-920 (1993)].
Positive strand RNA viral vectors are expected to be
useful as RNA vectors with autonomous replicating capability,
but their use as vectors for gene therapy poses the following
problems.
CA 02236113 1998-04-29
7
1. Since they are of the icosohedral structure, the size
of foreign gene allowed to be inserted is limited to 3,700
nucleotides at most.
2. Until nucleic acids are released from the packaged
complex into the cell and replicated, as many as five
processes are required, including cellular adhesion,
endocytosis, membrane fusion, decapsidation and translation
of replication enzymes.
3. A possible formation of disseminative viral particles
even in a minute quantity during packaging cannot be denied.
Especially, even with attenuated viral particles, the inside
RNA itself has disseminative potency and may belatedly be
amplified, making it difficult to check.
4. Since these vectors are derived from viruses
transmitted to animals by insects such as mosquitoes, when
animals and humans to which such vector genes are transferred
are mix-infected with wild type viruses, disseminative
recombinants may be formed, possibly further creating a risk
of said recombinants being scattered to the natural
environment by insects.
Such problems described above are conceived to be
basically overcome if RNA viral vectors derived from negative
strand RNA viruses are constructed. That is, since negative
strand RNA viruses do not have the capsid of icosohedral
structure, and also since the envelope size of particles is
known to vary depending on the inside RNA content, they are
supposed to be much less restricted by the size of foreign
CA 02236113 1998-04-29
g
genes to be inserted when used as RNA viral vectors.
Furthermore, since a group of proteins required for
transcription and replication are packaged into particles,
only two processes are required, including cellular adhesion
and membrane fusion, until nucleic acids are released from
packaged complex and replicated. In addition, viral RNA
alone is not disseminative, and disseminative particles can
be easily identified, because they readily fuse with cell
membrane and proliferate within cells. Therefore, the
presence of disseminative particles can be easily detected.
Furthermore, negative strand RNA viruses are not transmitted
by insects.
In spite of many advantages of negative strand RNA
viruses which may be used as the source of industrially
useful viral vectors, no negative strand RNA vectors
applicable for gene therapy has become available until now.
This is probably due to tremendous difficulties in re-
constituting viral particles via viral cDNA. Since the gene
manipulation on the DNA level is required to insert foreign
genes into vectors, so far as viral particles are not
reconstructed from viral cDNA with a foreign gene inserted,
it is difficult to use negative strand RNA viruses as a
vector. "Reconstruction of viral particles" refers to the
formation of original virus or recombinant virus in vitro or
intracellularly from artificially prepared viral genome
nucleic acids.
As described above, it has been clearly demonstrated
CA 02236113 1998-04-29
9
that, even if RNA of negative strand RNA viruses (vRNA; viral
RNA) or its complementary strand RNA (cRNA; complementary
RNA) alone is transferred into cells, no negative strand RNA
virus can be generated. This is a definitely different point
from the case of positive strand RNA viruses. Although, in
Tokkai H4-211377, "methods for preparing cDNA corresponding
to negative strand RNA viral genome and infectious negative
strand RNA virus" are described, the entire experiments of
said document described in "EMBO. J., 9, 379-384 (1990)" were
later proved to be not reproducible, so that the authors
themselves had to withdraw all the article contents [ref.
EMHO. J., 10, 3558 (1991)]. Therefore, it is obvious that
techniques described in Tokkai H4-211377 do not correspond to
the related art of the present invention.
With regard to the reconstitution system for negative
strand RNA viruses, there are reports on influenza virus
[Annu. Rev. Microbiol., 47, 765-790 (1993); Curr. Opin.
Genet. Dev., 2, 77-81 (1992)]. Influenza virus is an eight-
segmented negative strand RNA virus. According to these
literatures, a foreign gene was first inserted to a cDNA
corresponding to one of said segments, and the RNA
transcribed from the cDNA corresponding to all eight segments
including the one inserted with said foreign gene was
assembled with the virus-derived NP protein to form an RNP.
Then, the virus-reconstitution system was established by
providing cells with these RNPs and RNA-dependent RNA
polymerase. In addition, as with negative single stranded
CA 02236113 1998-04-29
1
RNA viruses, virus-reconstitution from cDNA was reported with
rabies virus belonging to rhabdoviruses [J. Virol., 68, 713-
719 (1994)].
However, it has been difficult to use these virus
reconstitution techniques as such for constructing vectors
for gene therapy because of the following problems.
1. Reconstituted viruses were identified only by the
expression of marker gene, RT-PCR, etc. No re-constitution
system usable for the production of vector viruses in a
satisfactory yield has been established.
2. Differing from the case of positive strand RNA
viruses, in order to form complexes with infectivity but
deficient in disseminative potency as vectors for gene
therapy, it is necessary to enclose factors required for
primary transcription and replication within the complex. No
technique for amplifying these complexes in a large scale has
been established.
3. For the purpose of intracellularly providing factors
necessary for viral reconstitution, cells to which cDNAs are
introduced are mix-infected with helper viruses such as wild
type viruses and recombinant vaccinia virus, etc. It is not
easy to separate these natural type viruses added.
Furthermore, as one problem with regard to RNA viral
vectors in general, it is conceivably necessary to beforehand
provide inhibitors for replication of RNA viral vectors which
have no effects on host's replication and transcription,
providing for the case where RNA replicated and transcribed
CA 02236113 1998-04-29
11
in large amounts exerts undesirable effects on treated humans
and animals. However, no such inhibitors have been
developed.
Disclosure of the Invention
Problems to be solved by the present invention are to
develop negative strand RNA viral vectors for practical use,
methods for efficiently preparing said vectors, and
inhibitors for the replication of said vectors:
Present inventors first attempted to reconstitute Sendai
virus from nucleic acids of said virus which is a typical
negative strand RNA virus, and conceived to be industrially
most useful as a vector from the viewpoints of safety and
convenience. First, in order to apply to the reconstitution
test, various investigations were performed using cDNA
derived from Sendai virus DI (defective interfering)
particles [see EMBO J., 10, 3079-3085 (1991)] or cDNA of
Sendai virus minigenome as experimental materials. As a
result, they found efficient conditions regarding weight
ratios among materials to be transferred into host cells,
including cDNA, cDNAs concerning the transcription and
replication, and the recombinant vaccinia virus to provide a
unit for expressing the T7RNA polymerase. Furthermore, the
present inventors obtained full-length cDNAs corresponding to
both positive and negative strands, constructed plasmids for
inducing the intracellular biosynthesis of either positive or
negative strand RNA of Sendai virus, and transferred said
plasmids into host cells wherein cDNAs concerning the
CA 02236113 1998-04-29
12
transcription and replication were expressed. As a result,
they first succeeded in re-constructing Sendai virus
particles from cDNAs derived thereof.
In addition, the present inventors found that Sendai
virus could be reconstructed without using recombinant
vaccinia virus as T7-RNA polymerase expression unit. That
is, when the full-length RNA of Sendai virus transcribed in
vitro was transferred into cells, and cDNAs encoding enzymes
for initial transcription and replication were transcribed
under the control of T7 promotor, viral particles were re-
constructed. This indicates that, if cells which express
group of all enzymes required for initial transcription and
replication are constituted, the recombinant Sendai virus,
eventually complexes described above can be formed entirely
without using helper viruses such as vaccinia virus. Since
cells which express group of all enzymes required for initial
transcription and replication were already described [J.
Virology, 68, 8413-8417 (1994)], those skilled in the art may
form such cells with reference to said article. Cells
described in said reference are the one derived from the 293
cell line which carries three of Sendai virus genes, namely
NP, P/C and L, on its chromosome, expressing proteins encoded
by the three genes, NP, P/C and L.
From numerous examples of viral vectors, if viral
particles can be efficiently reconstructed from nucleic
acids, it is obvious that those skilled in the art are able
to readily exchange a desired viral gene, insert a foreign
CA 02236113 1998-04-29
13
gene, or inactivate or delete a desired gene. For example,
an article on the use of DI particles [J. Virol., 68, 8413-
8417 (1994)] clearly indicates that, when RNA deficient in at
least a part of structural genes of Sendai virus, but normal
of genes for replication enzymes is transferred into cells,
the succeeding autonomous replication may be able to proceed,
if group of enzymes necessary for the initial transcription
and replication are provided in the cells. Therefore, once
an RNA molecule containing a foreign gene transcribed from
"specific viral cDNA deficient in at least a part of
structural genes but normal in genes for the replication
enzyme group" can be enclosed in the viral structure
comprising the initial transcription and replication enzymes,
complexes which are infectious to and autonomously
replicating but deficient in the disseminative potency, and
functional as the foreign gene vector can be formed. Such
complexes are extremely useful as a vector for gene therapy.
That is, in the present invention, with a negative strand RNA
virus, it becomes possible to prepare complexes which are
infectious as well as autonomously replicative but is
deficient in the disseminative potency, for example,
complexes comprising the initial transcription and
replication enzymes.
The present inventors further developed a method for
amplifying the same complex by transfecting said complex to
cells which express the structural proteins corresponding to
genes in RNA of the complex which have been deleted or
CA 02236113 1998-04-29
14
inactivated. Further, taking avian eggs into consideration
as the most suitable medium for proliferating Sendai virus in
order to amplify the said complex, the inventors found that
transgenic avians, their eggs and cells which carry at least
one or more genes out of M, F and HN genes of Sendai virus on
chromosome are suitable for amplifying complexes. Methods
for preparing transgenic avians have been reported [Poultry
Sci., 65, 1445-1458 (1986); Bio/Technology, 12, 60-63
(1994)], and those skilled in the art can appropriately
produce transgenic birds carrying at least one or more genes
out of M, F and HN genes on their chromosomes. Preferably,
proteins encoded by genes related to the deficiency in
disseminative capability of RNA contained in the complex
among M, F and HN genes, are
expressed in transgenic birds.
The present inventors also developed a method for
preparing the complex described above. In the following,
cases related to Sendai virus are exemplified. Genome of
Sendai virus Z is a single stranded RNA comprising 15384
nucleotides [Virology, 108, 318-324 (1981)]. Its entire base
sequence has been determined from cDNA clones prepared by
using reverse transcriptase [Nucleic Acids Research, 11,
7313-7330 (1983); Nucleic Acids Research, 12, 7965-7972
(1984); Nucleic Acids Research, 14, 1545-1563 (1986)]. Since
its genome RNA is a negative strand, a group of enzymes for
transcription and replication in the viral particles perform
both transcription and replication with the genome RNA as
CA 02236113 1998-04-29
template. At least six proteins including NP, P/C, M, F, HN
and L are known as proteins encoded by the genome RNA. It
has been elucidated that, of these proteins, NP, P/C and L
are factors essential and sufficient for replication [Journal
of Virology, 68, 8413-8417 (1994)], and M, F and HN are
components necessary for constructing the viral structure.
Based on these facts, when a specific RNA virus from which
RNA is derived is Sendai virus, it is possible to reconstruct
an infectious complex by transferring both 1) cDNA
transcribable to DNA, and 2) a gene encoding the RNA
polymerase necessary for transcribing said cDNA within cells
or an RNA molecule itself transcribed from said cDNA in vitro
into cells wherein all the genes for the autonomous
replication, NP, P/C and L, and a group of genes, out of M, F
and HN genes, for the deficiency of RNA dissemination are
expressed. In this case, all genes for the autonomous
replication, NP, P/C and L, and genes, out of M, F and HN
genes, for the deficiency of RNA disseminative capability may
be transiently expressed by transfecting cells with the
plasmids coding for the respective genes. However, genes
related to the deficiency of RNA disseminative capability at
least are preferably incorporated into
chromosomes to be stably expressed.
The present inventors further developed a method for
producing said complex thus re-constituted in large
quantities, wherein said complex is replicated by
transfecting it to cells having no genes related to the
CA 02236113 1998-04-29
I6
autonomous replication but expressing genes, from among M, F
and HN genes, related to the deficiency in the RNA
disseminative potency. In this case, as cells having no
genes related to the autonomous replication but expressing a
group of genes, from among M, F and HN genes, related to the
deficiency in the RNA disseminative capability, transgenic
avian eggs expressing said group of genes are preferable for
the production of complex on a large scale.
Furthermore, the present inventors produced cells for
propagating the complex containing said RNA and proteins.
More specifically, said cells are those with genes
corresponding to a group of genes related to the deficiency
in infectious particle-forming capability of the RNA retained
by said complex, and capable of intracellularly producing
proteins encoded by said genes. In the case wherein the
specific RNA virus from which RNA is derived is Sendai virus,
cells which have at least more than one genes from among M, F
and HN genes on their chromosomes or animals having such
cells are used. In addition, M, F and HN genes are not
necessarily of wild type. Any of those with functions
equivalent to those of the wild type will be usable. That
is, any gene may be used where said gene has complementarity
to the wild type for deficient virus when functionally
introduced into cells. Preferable cells to be used are host
cells for Sendai virus. It is preferable that proteins
encoded by genes corresponding to those related to the
deficiency in infectious particle-forming capability, from
CA 02236113 1998-04-29
17
among M, F and HN genes in the vector viral RNA, are
intracellularly produced.
Hitherto only the enhancement of expression efficiency
has been emphasized with conventional RNA virus vectors, and
little efforts have been made for developing compounds to
suppress the RNA replication to prevent unfavorable results
due to excessive expression. In this respect, the present
inventors developed an inhibitor for the negative strand
virus vector which specifically inhibits the RNA-dependent
RNA replication and RNA-dependent RNA transcription without
affecting the transcription and translation of cell-derived
RNAs leading only to the inhibition of RNA-dependent RNA
replication.
That is, the present invention comprises the followings.
1. A complex comprising an RNA molecule derived from a
specific disseminative negative strand RNA virus and viral
structural components containing no nucleic acids, having the
infectivity and autonomous RNA replicating capability, but
deficient in the disseminative capability.
2. The complex of description 1, wherein said specific
RNA virus is a negative strand RNA virus having non-segmented
genome.
3. The complex of description 2, wherein said specific
RNA virus is Sendai virus.
4. An RNA molecule comprising Sendai viral RNA or Sendai
viral cRNA, wherein said RNA molecule is defective in that at
least more than one gene coding for the M, F and HN proteins
CA 02236113 1998-04-29
Ig
are deleted or inactivated.
5. A complex comprising the RNA of description 4 and
viral structural components containing no nucleic acids
derived from Sendai virus, having the infectivity and
autonomous RNA replicating capability, but deficient in the
disseminative capability.
6. A DNA molecule comprising a template DNA transcribable
to the RNA molecule of description 4 in vitro or
intracellularly.
7. The complex of any one of descriptions 1-3 or 5,
wherein the RNA molecule contained in said complex comprises
a foreign gene.
8. The complex of descriptions 3 or 5, wherein the RNA
molecule contained in said complex comprises a foreign gene.
9. The RNA molecule of description 4 comprising a foreign
gene.
10. The DNA molecule of description 6 comprising a
foreign gene.
11. An inhibitor for RNA replication contained in the
complex of any one of descriptions 1-3, 5, 7 or 8 comprising
an inhibitory drug for the RNA-dependent RNA replication.
12. A host whereto the complex of any one of descriptions
1-3, 5, 7 or 8 can disseminate.
13. The host of description 12 comprising a group of
genes related to the infectivity of the complex of any one of
descriptions 1-3, 5, 7 or 8 on its chromosomes, and capable
of replicating the same copies of said complex when infected
CA 02236113 1998-04-29
19
with it.
14. The host of descriptions 12 or 13, wherein said host
is animals, or cells, tissues, or eggs derived from it.
15. The host of description 14 wherein said animal is
mamma 1 ian .
16. The host of description 14 wherein said animal is
avian.
17. A host comprising a group of genes related to the
infectivity of the complex of any one of descriptions 3, 5 or
8 on its chromosomes, and capable of replicating the same
copies of said complex when infected with it.
18. A host comprising at least more than one gene of the
M, F and HN genes of Sendai virus or genes having functions
equivalent to them on its chromosomes.
19. A host comprising the M gene of Sendai virus or its
functionally equivalent gene on its chromosomes.
20. A host comprising the M, NP, P/C and L genes of
Sendai virus on its chromosomes (wherein each gene may be
substituted with its functionally equivalent gene,
respectively).
21. A host comprising the M, F and HN genes of Sendai
virus on its chromosomes (wherein each gene may be
substituted with its functionally equivalent gene,
respectively).
22. A host comprising the M, F, HN, NP, P/C and L genes
of Sendai virus on its chromosomes (wherein each gene may be
substituted with its functionally equivalent gene,
CA 02236113 1998-04-29
respectively).
23. The host of any one of descriptions 17-22, wherein
said host is animal, or cell, tissue or egg derived from it.
24. The host of description 23, wherein said animal is
mammal ian .
25. The host of description 23, wherein said animal is
avian.
26. A kit consisting of the following three components,
a. the RNA molecule contained in the complex of any one
of descriptions 1-3, 5, 7 or 8, or cRNA of said RNA, or a
unit capable of biosynthesizing said RNA or said cRNA,
b. a group of enzymes required for replicating said RNA
or said cRNA, or a unit capable of biosynthesizing said group
of enzymes, and
c. a group of proteins related to the infectivity of said
complex, or a unit for biosynthesizing said group of
proteins.
27. A kit consisting of the following three components,
a. the RNA molecule contained in the complex of any one
of descriptions 1-3, 5, 7 or 8, or cRNA of said RNA, or a
unit capable of biosynthesizing said RNA or said cRNA,
b. a group of enzymes required for replicating said RNA
or said cRNA, or a unit capable of biosynthesizing said group
of enzymes, and
c. the host of any one of descriptions 12-25.
28. A kit consisting of the following two components,
a. the complex of any one of descriptions 1-3, 5, 7 or 8,
CA 02236113 1998-04-29
21
and
b. the host of any one of descriptions 12-25.
29. A kit consisting of the following three components,
a. the RNA molecule contained in the complex of any one
of descriptions 3, 5 or 8, or cRNA of said RNA, or a unit
capable of biosynthesizing said RNA or said cRNA,
b. all NP, P/C and L proteins of Sendai virus, or a unit
for biosynthesizing said group of proteins, and
c. a group of proteins related to the infectivity of said
complex, or a unit for biosynthesizing said group of
proteins.
30. A kit consisting of the following three components,
a. the RNA molecule contained in the complex of any one
of descriptions 3, 5 or 8, cRNA of said RNA, or a unit
capable of biosynthesizing said RNA or said cRNA,
b. all NP, P/C and L proteins of Sendai virus, or a unit
capable of biosynthesizing said group of proteins, and
c. the host of any one of descriptions 17-25.
31. A kit consisting of the following two components,
a. the complex of any one of descriptions 3, 5 or 8, and
b. the host of any one of descriptions 17-25.
32. A method for producing the complex of any one of
descriptions 1-3, 5, 7 or 8 by introducing three components
of descriptions 26a, 26b and 26c into a host.
33. A method for producing the complex of any one of
descriptions 1-3, 5, 7 or 8 by introducing both components
of descriptions 27a and 27b into the host of description 27c.
CA 02236113 1998-04-29
22
34. A method for amplifying and producing the complex of
description 28a by transfecting said complex to the host of
description 28b.
35. A method for producing the complex of any one of
descriptions 3, 5 or 8 by introducing the three components of
descriptions 29a, 29b and 29c into a host.
36. A method for producing the complex of any one of
descriptions 3, 5 or 8 by introducing both components of
descriptions 30a and 30b into the host of description 30c.
37. A method for amplifying and producing the complex of
description 31a by transfecting said complex into the host of
description 31b.
38. The RNA molecule of description 9 wherein a gene
corresponding to the M gene is deleted or inactivated.
39. The RNA molecule of description 9 wherein all the
genes corresponding to the M, F and HN genes are deleted or
inactivated.
40. A kit consisting of the following three components,
a. the RNA molecule of description 38,
b. the host of description 20, and
c. the host of description 19.
41. A method for producing a complex by introducing the
RNA molecule of description 40a into the host of description
40b, and amplifying and producing said complex by
transfecting it into the host of description 40c.
42. A complex produced by the method of description 41.
43. A kit consisting of the following three components,
CA 02236113 1998-04-29
23
a. the RNA molecule of description 39,
b. the host of description 22, and
c. the host of description 21.
44. A method for producing a complex by introducing the
RNA molecule of description 43a into the host of description
43b, and amplifying and producing said complex by
transfecting it into the host of description 43c.
45. A complex produced by the method of description 44.
46. An inhibitor for RNA replication contained in the
complex of either descriptions 42 or 45 comprising an
inhibitory drug of the RNA-dependent RNA replication.
47. A method for preparing the foreign proteins, wherein
said method comprises the process of introducing the complex
of description 7 to a host and the process of recovering the
expressed foreign proteins.
48. A method for preparing the foreign proteins of
description 47, wherein the host is a cell expressing a group
of genes, from among those related to the disseminative
capability, which are deficient in the RNA molecule contained
in the complex of description 7.
49. A culture medium or chorio-allantoic fluid containing
the expressed foreign proteins, wherein said culture medium
or chorio-allantoic fluid is obtained by inoculating the
complex of description 7 into a host and recovering it.
Any negative strand RNA viruses with disseminative
capability may be used as materials in the present invention.
Although incomplete viruses such as defective interfering
CA 02236113 1998-04-29
24
particles (DI particles) and synthetic oligonucleotide may
also be used as partial materials, in general, they must have
the base sequence equivalent to that of the virus with
disseminative capability. Negative strand RNA viruses of the
present invention include, for example, Sendai virus,
Newcastle disease virus, mumps virus, measles virus,
respiratory syncytial virus, rinderpest virus of cattle and
canine distemper virus of Paramyxoviridae, influenza virus of
Orthomyxoviridae, vesicular stomatitis virus and rabies virus
of Rhabdoviridae.
As the negative strand viral material, recombinant
negative strand viruses derived from any viruses described
above and retaining the disseminative capability may be used.
For example, the recombinant negative strand virus may be the
one with the gene for the immunogenicity inactivated or a
partial region of gene altered to enhance the efficiency of
RNA transcription and replication.
RNAs contained in the RNA-protein complex of the present
invention can be obtained by transcribing modified cDNAs
derived from any viruses or recombinant viruses described
above in vitro or intracellularly. In RNAs thus obtained, at
least one gene related to the disseminative capability of the
original virus must be deleted or inactivated, but the gene
related to the autonomous replication should not. In
addition, RNA molecules with artificial sequences, which are
obtained by transcribing, in vitro or intracellularly, DNA
formed by inserting the genes for the autonomous replication
CA 02236113 1998-04-29
into cDNA having both terminus structures of the virus genome
such as DI molecule, may be also used.
As described above, in the case of Sendai virus, "the
genes related to autonomous replication" refer to any one of
the NP, P/C and L genes, and "the gene related to the
disseminative capability" refers to any one of the M, F and
HN genes. Therefore, the RNA molecule of Sendai virus Z
strain deficient only in the M gene, for example, is suitable
as a component contained in the "complex" of the present
invention. Also, the RNA molecule having all the M, F and HN
genes deleted or inactivated are also preferable as the
component contained in the "complex" of the present
invention. On the other hand, it is necessary for the genes
encoding the NP, P/C and L proteins to be expressed from RNA.
However, the sequences of these genes are not necessarily the
same as those of virus, and may be modified by introducing
variations, or replacing by the corresponding gene derived
from other viruses, so far as the transcription and
replication activity of the resulting RNA is similar to or
higher than that of the natural one.
"Virus structural component free of nucleic acid" of the
present invention includes, for example, virus with only its
RNA removed. As such structural component is used the one
which complements the infectivity and autonomous replicating
capability at the early stage, but not the disseminative
capability. In the case of Sendai virus, the complex
composed of its RNA with only the M gene deleted, and Sendai
CA 02236113 1998-04-29
26
virus having only its RNA deleted have the infectivity and
autonomous replicating capability, but no disseminative
capability. Complex may contain other components so long as
it is provided with no disseminative capability. For
example, complex may contain adhering molecule, ligand,
receptors, etc. on its envelope surface for facilitating the
adherence to specific cells.
The RNA molecule contained in the complex can have an
inserted foreign gene at its appropriate site. In order to
express a desired protein, the foreign gene encoding said
protein is inserted. In the case of Sendai viral RNA, a
sequence of bases of 6 multiplication in number is preferably
inserted between sequences R1 (5'-AGGGTCAAAGT-3') and R2 (5~-
GTAAGAA.AAA-3') [Journal of Virology, Vol. 67, No. 8 (1993),
p.4822-4830]. Levels of expression of the foreign gene
inserted into RNA can be regulated by virtue of the site of
gene insertion and the base sequence flanking the foreign
gene. For example, in the case of Sendai viral RNA, it is
known that there are increasing levels of expression of the
inserted gene with decreasing distance of said gene from the
NP gene. Preferred host cells for the introduction of the
complex to express desired proteins are those expressing
genes deleted in the RNA molecule composed of said complex.
For this, transgenic avian eggs expressing said genes are
most preferable for preparing proteins in large quantities.
For example, proteins thus expressed can be recovered from
the culture medium when host cells are cultured cells, and
CA 02236113 1998-04-29
27
chorio-allantoic fluid when host cells are chicken eggs,
using standard techniques. In Examples 5 and 6 is used a
disseminative complex in place of non-disseminative complex
of the present invention. However, it will be clear to those
skilled in the art that similar results are obtained with the
complex of the present invention as with the disseminative
complex in these examples when "cells expressing genes
deleted from among genes for disseminative capability in the
RNA molecule contained in the complex" are used as host
cells.
Furthermore, the present inventor has confirmed that, for
the efficient reconstitution of Sendai virus particles, cDNA
to be introduced into cells is preferably in the circular
form rather than in the linear form, and, for viral particle
formation at a high efficiency, the transcription of the
positive strand RNA is preferred to that of the negative
strand RNA within cells. Although these conditions may not
necessarily be applicable to the reconstitution of all other
negative strand RNA viruses, it is possible to search for
appropriate conditions for the reconstitution of other
negative strand RNA viruses based on the disclosure of the
present invention and conventional technology, indicating a
possibility for establishing techniques to produce basic
materials of desired negative strand viral vectors, that is,
the viral reconstitution systems.
As the "RNA replication inhibitor" of the present
invention, any drugs to inhibit RNA-dependent RNA replication
CA 02236113 1998-04-29
28
may be applied, and, for example, Ribavirin, TJ13025, etc.
are preferably used. Such replication inhibitors are
effective, for example, when health deterioration is noticed
with the cellular amplification of recombinant RNA, or when
the control of intracellular expression of foreign genes
derived from recombinant RNA is required.
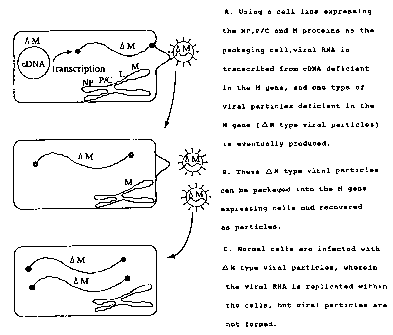
As an embodiment of the present invention, processes for
reconstituting the complex of the present invention from
cDNA with the M gene deleted of Sendai virus (steps A-B), and
those for amplifying said complex (steps B-C) are shown in
Fig. 1.
Brief description of the drawings
Figure 1 is a schematic representation of a process for
generating complexes of the present invention from cDNA
deficient in the M gene of Sendai virus (steps A-~B) and
further amplifying said complexes (steps BBC).
Figure 2 is a schematic representation of the
construction of a pUCl8/T7(+)HVJRz.DNA.
Figure 3 is a schematic representation of the
construction of a pUCl8/T7(-)HVJRz.DNA.
Figure 4 is a graphical representation showing the
relationship between the time after the infection of SeVgp120
into CV-1 cells and levels of HAU and gp120 expression.
Best mode for carrying out the Invention
In the following, the present invention will be
concretely described with reference to Examples, but not be
limited to them.
CA 02236113 1998-04-29
29
Example 1. Preparation of Sendai virus transcription units
pUCl8/T7(-)HVJRz.DNA and pUCl8/T7(+)HVJR2.DNA
Plasmid pUCl8/T7(-)HVJRz.DNA was constructed by inserting
a DNA molecule comprising T7 RNA polymerase promotor, Sendai
virus cDNA designed to be transcribed to the negative strand
RNA and the ribozyme gene in this order into pUCl8 vector.
Also, plasmid pUCl8/T7(+)HVJRz.DNA was constructed by
inserting a DNA molecule comprising T7 RNA polymerase
promotor, Sendai virus cDNA designed to be transcribed to the
positive strand RNA and the ribozyme gene in this order into
pUCl8 vector. Constructions of pUCl8/T7(-)HVJRz.DNA and
pUCl8/T7(+)HVJRz.DNA are shown in Figs. 1 and 2,
respectively.
Example 2.Reconstitution experiment of Sendai virus from
cDNA
LLC-MK2 cells (2 x 106) trypsinized in a usual manner
were placed in a 60-mm diameter plastic dish, and incubated
in MEM medium (MEM supplemented with 10$ FBS) (2 ml) in a 5~
COZ atmosphere at 37°C for 24 h. After removing the medium
and washing with PBS (1 ml), a suspension of recombinant
vaccinia virus vTF7-3 expressing T7 polymerase in PHS (0.1
ml) was added to the cells at the multiplicity of infection
(moi) of 2. The dish was gently agitated every 15 min to
thoroughly spread the viral solution for 1 h infection.
After removing the viral solution and washing with PBS (1
ml), a medium containing cDNA, which was prepared as follows,
was added to the dish.
CA 02236113 1998-04-29
Nucleic acids shown in Tables 1 and 2 (containing
plasmids expressing factors required for the replication of
Sendai virus, pGEM-L, pGEM-P/C and pGEM-NP) were placed in a
1.5-ml sampling tube, and adjusted to a total volume of 0.1
ml with HBS (Hepes buffered saline; 20 mM Hepes pH 7.4
containing 150 mM NaCl). In those tables, (-) and (+)cDNAs
represent plasmids pUCl8/T7(-)HVJRz.DNA and pUCl8/T7(+)HVJRz.
DNA, respectively, and /C and /L indicate that cDNA is
introduced into cells in the circular form and linear form
after the treatment with restriction enzyme MluI,
respectively.
On the other hand, in a polystyrene tube were placed HBS
(0.07 ml), DOTAP (Boehringer Mannheim) (0.03 ml). To this
tube was added the nucleic acid solution described above, and
the mixture was left standing as such for 10 min. Then, to
this mixture was added the cell culture medium described
above (2 ml, MEM supplemented with 10~ FBS) followed by the
vaccinia virus inhibitors, rifampicin and cytosine
arabinoside C (C/Ara/C), to the final concentrations of 0.1
mg/ml and 0.04 mg/ml, respectively, resulting in the
preparation of the medium containing cDNA described above.
The dish described above was incubated in a 5~ COz
atmosphere at 37°C for 40 h. The cells in the dish were
harvested using a rubber policeman, transferred to an
Eppendorf tube, sedimented by centrifuging at 6,000 rpm for 5
min, and re-suspended in PBS (1 ml). Aliquots of this cell
suspension, as such or after diluted, were inoculated to 10-
CA 02236113 1998-04-29
31
days old developing embryonated chicken eggs. That is, the
cell suspension was diluted with PBS to the cell numbers
shown in Table 1, and eggs inoculated with its 0.5-ml
aliquots were incubated at 35°C for 72 h, then at 4°C
overnight. Chorio-allantoic fluid was recovered as virus
solution from these eggs using a syringe with a needle.
Hemagglutinin unit (HAU) and plaque forming unit (PFU) of
the recovered virus solution were assayed as follows.
HAU was determined as follows. Chicken blood was
centrifuged at 400 x g for 10 min and the supernatant was
discarded. Precipitates thus obtained were suspended in 100
volumes of PBS, and centrifuged at 400 x g for 10 min to
discard the supernatant. This procedure was repeated twice
to prepare an 0.1~ blood cell solution. Two-fold serial
dilutions of virus solutions were prepared, and 0.05 ml each
dilution to be assayed was dispensed into each well of 96-
well titer plate. The blood cell solution (0.05 ml each) was
further added to each well, gently swirled to ensure a
thorough mixing, and left at 4°C for 40 min. The highest
virus dilution to cause the hemagglutination observable with
the naked eye was taken as HAU.
PFU was assayed as follows. CV-1 cells were grown to a
monolayer on a 6-well culture plate. After the culture
medium was discarded, a virus solution 10-fold serially
diluted (0.1 ml each) was dispensed into each well of the
culture plate to infect the cells at 37°C for 1 h. During
the infection, a mixture of 2 x MEM free of serum and melted
CA 02236113 1998-04-29
32
2~ agar (55°C) was prepared, and trypsin was added to the
mixture to a final concentration of 0.0075 mg/ml. After 1 h
infection and removal of the virus solution, the culture
medium mixed with agar (3 ml each) was added to each well of
the culture plate, and incubated under a 5~ COZ atmosphere at
37°C for 3 days. Phenol red (0.1~) (0.2 ml) was added to
each well, incubated at 37°C for 3 h, and then removed.
Unstained plaques were counted to estimate the virus titer as
PFU/ml.
Table 1 shows Sendai virus template cDNAs transfected
into LLC-2 cells, amounts of cDNA factors, pGEM-L, pGEM-P/C,
and pGEM-NP, required for the RNA replication, incubation
time, cell numbers inoculated to chicken eggs, HAU and PFU
values.
CA 02236113 1998-04-29
33
Table 1
Template Total pGEM pGEM pGEM IncubationAmount HAU PFU
cDNA amount-L -P/C -NP time (h) of cells
(N9) (Ng) (N9) (N9.)
(+)cDNA/C10 4 2 4 40 1.00x105 512 2x109
(+)cDNA/C10 4 2 4 40 1.00x105 256 9x108
(+)cDNA/C10 4 2 4 40 1.00x106 256 9x108
(+)cDNA/L10 4 2 4 40 1.00x105 ~2 ~10
(+)cDNA/L10 4 2 4 40 1.00x105 ~2 ~10
(+)cDNA/L10 4 2 4 40 1.00x106 ~2 ~10
(-)cDNA/L10 4 2 4 40 1.00x10' ~2 ~10
(-)cDNA/L10 4 2 4 40 1.00x105 ~2 ~10
(-)cDNA/L10 4 2 4 40 1.00x106 ~2 X10
(-)cDNA/C10 4 2 4 40 1.00x10' ~2 ~10
(-)cDNA/C10 4 2 4 40 1.00x105 ~2 ~10
(-)cDNA/C10 4 2 4 40 1.00x106 4 8x10'
Samples showing both HAU and PFU were sedimented by
ultra-centrifugation, re-suspended, purified by a sucrose
density gradient centrifugation from 20~ to 60~, and
fractionated by 12.5 SDS-PAGE. Each protein contained in
these samples was the same in size as that of Sendai virus.
These results demonstrated that Sendai virus can be
reconstituted by introducing cDNAs into cells, and that virus
CA 02236113 1998-04-29
34
particles are more efficiently reconstituted by introducing
cDNAs transcribing positive strand RNAs as compared with
those transcribing negative strand RNAs, and further by
introducing cDNAs in the circular form rather in the linear
form.
Example 3. Survey of RNA replication factors required for
Sendai virus reconstitution
Experiments were performed to examine whether all three
plasmids expressing the L, P/C and NP proteins were required
for the reconstitution of Sendai virus. Experimental methods
were similar to those described in Example 2 except that any
combinations of two out of pGEM-L, pGEM-P/C and pGEM-NP
plasmids or only one out of them, instead of all these three
combined as in Example 2, were introduced together with a
template cDNA into cells.
Table 2 shows Sendai virus template cDNAs introduced into
LLC-MK2 cells, amounts of the cDNA factors required for RNA
replication including pGEM-L, pGEM-P/C and pGEM-NP,
incubation time, number of cells inoculated into chicken
eggs, and values of HAU and PFU.
CA 02236113 1998-04-29
Table 2
Template Total pGEM pGEM pGEM Incubation Number of HAU PFU
cDNA amount -L -P/C -NP time cells
( y~g ) ( h ) inoculated
(+)cDNA/C10 4 2 4 40 1.00x105 256 6x108
(+)cDNA/C10 4 2 4 40 1.00x106 512 4x109
(+)cDNA/C 10 0 2 4 40 1.00x106 [2 [10
(+)cDNA/C 10 0 2 4 40 1.00x106 [2 [10
(+)cDNA/C 10 4 0 4 40 1.00x106 [2 [10
(+)cDNA/C 10 4 0 4 40 1.00x106 [2 [10
(+)cDNA/C 10 4 2 0 40 1.00x106 [2 [10
(+)cDNA/C 10 4 2 0 40 1.00x106 [2 [10
(+)cDNA/C 10 0 0 4 40 1.00x10° [2 [10
(+)cDNA 10 0 0 4 40 1.00x106 [2 [10
(+)cDNA/C 10 0 2 0 40 1.00x106 [2 [10
(+)cDNA/c 10 0 2 0 40 1.00x106
[2 [10
(+)eDNA/C 10 4 0 0 40 1.00x106 [2 [10
CA 02236113 1998-04-29
36
As shown in Table 2, no virus reconstitution was observed
by introducing any combinations of two out of these three
factors into cells, confirming the necessity of all three
proteins L, P/C and NP for the virus reconstitution.
Example 4. Reconstitution experiment of Sendai virus in vitro
from transcribed RNAs
Since the reconstitution of Sendai virus from the
functional cDNA clones was described in Example 2, it was
further examined whether transcription products of said cDNAs
in vitro, that is, vRNA and cRNA, can support similar
reconstitution.
After the Sendai virus transcription units, pUCl8/T7(-
)HVJRz.DNA and pUCl8/T7(+)HVJRz.DNA, were linearized with
restriction enzyme MluI, using these DNAs as templates, RNA
synthesis was performed in vitro with a purified T7
polymerase preparation (EPICENTRE TECHNOLOGIES: Ampliscribe
T7 Transcription Kit). The method for synthesizing in vitro
RNAs essentially followed the protocols provided with the
kit. Using RNA products thus obtained in place of cDNAs in
Example 2, similar experiments were performed, and the
virus production was estimated by HA test. Results are shown
in Table 3.
CA 02236113 1998-08-20
- 37 -
Table 3
TemplateTotal pGEM- pGEM- pGEM- Incubation Number HAU PFU
of
cDNA amountL P/C NP time (h) cells
(ug) (ug) (ug) (fig) inoculated
in vitro10 4 2 4 40 1.00x106 512 2x109
(-) RNA
in vitro10 4 2 4 40 1.00x106 512 ND
(-) RNA
in vitro10 4 2 4 40 1.00X106 2 5X103
(+) RNA
in vitro10 4 2 4 40 1.00x106 <2 ND
(+) RNA
These results indicate that virus can be
reconstituted by introducing either negative or positive sense
strand RNAs into cells.
Example 5. Expression of foreign genes inserted into Sendai
viral vectors in host cells
I. Preparation of Sendai virus vector "pSeVgp120"
inserted with a foreign gene (HIV-1 gp120)
Using a set of primers comprising primer a (5'-
TGCGGCCGCCGTACGGTGGCAATGAGTGAAGGAGAAGT-3' (SEQ ID NO:1) and
primer d (5'-TTGCGGCCGCGATGAACTTTCACCCTAAGTTTTTVTTACTACGGCG-
TACGTCATCTTTTTTCTCTCTGC-3' (SEQ ID N0:2), the HIV-1gp120 gene
was amplified on "pN1432" by the standard PCR techniques. PCR
products were sub;ected to TA cloning,
76432-11
CA 02236113 1998-04-29
38
digested with NotI, and then inserted into the NotI site of
"pSeVl8"'. Then, E. coli cells were transformed with this
recombinant plasmid. DNAs were extracted from each colony of
E. coli by the "Miniprep" method, digested with DraIII, and
then electrophoresed. Positive clones (designated "clone 9"
hereafter) were selected by confirming to contain DNA
fragments of the size expected from the insertion. After DNA
fragments were confirmed to have the authentic nucleotide
sequence, DNAs were purified by a cesium chloride density
gradient centrifugation. pSeVl8+ inserted with the gp120
gene is designated "pSeVgp120" hereafter.
2. Reconstitution of Sendai virus containing pSeVgp120
(SeVgp120) and analysis of gp120 expression
Except for the further transfection of pSeVgp120 into
LLCMK2 cells, in addition to pGEM-NP, pGEM-P/C and pGEM-L,
chorio-allantoic fluid was recovered from embryonated chicken
eggs and assayed for the viral HAU by exactly as described in
Example 2. The recovered virus was also examined for the
expression of gp120 by ELISA as follows.
Samples (100 pl each) were dispensed into each well of a
96-well plate which had been coated with monoclonal antibody
against HIV-1, and incubated at 37°C for 60 min. After
washing with PBS, HRP-linked anti-HIV-1 antibody (100 pl
each) was added to each well, and incubated at 37°C for 60
min. After washing with PBS, tetramethylbenzidine was added
to each well, and amounts of reaction product converted by
the action of HRP under acidic conditions were determined by
CA 02236113 1998-04-29
39
following the optical density at 450 nm to estimate the
expression amount of gp120. Results are shown in the left-
hand column in Table 4.
The virus solution thus obtained was inoculated to CV-1
cells, and similarly examined as follows. CV-1 cells were
dispensed to a culture plate at 5 x 105 cells/plate, grown,
and then the culture medium was discarded. After washing
with PBS(-), the viral solution was added to the cells at the
multiplicity of infection of 10, and incubated at room
temperature for 1 h. After the virus solution was discarded,
washed with PBS(-), a plain MEM medium (MEM medium
supplemented with antibiotics AraC and Rif, and trypsin) was
added to the cells, and incubated at 37°C for 48 h. After
the reaction, the medium was recovered and assayed for HAU
(by a similar method as described in Example 2) and examined
for the expression of gp120 (by ELISA). Results are shown in
the center column of Table 4. In addition, the supernatant
of CV-1 cell culture medium was inoculated to embryonated
chicken eggs again, and the virus solution thus obtained was
assayed for HAU and also examined for the gp120 expression
(by ELISA). Results are shown in the right hand column of
Table 4.
CA 02236113 1998-04-29
4D
Table4
( ~t g~ml )
Chorio-allantoic CV-1 medium (F1) Chorio-allantoic
fluid (F1) gp120 (HAU) fluid (F2)
gp120 (HAU)
gp120 (HAU)
0.10 ( 4) 3.46 (128)
0.15 (32) 1.81 (128) 1.56, 1.21
(512, 512)
0.05 (32) 2.20 (128)
As shown in Table 4, markedly high concentrations of
gp120 were detected in CV-1 cells in culture (center column
of the Table), and also in the chorio-allantoic fluids from
embryonated chicken eggs inoculated again with the virus
(right-hand column of the Table). In the left-hand and
center columns of the Table are shown the mean values of
three clones.
Furthermore, the expression of gp120 was analyzed by
Western blotting. After the culture medium of CV-1 cells
infected with SeVgp120 was centrifuged at 20,000 rpm for 1 h
to sediment virus, the supernatant was treated with either
TCA (10$, v/v) for 15 min on ice or 70~ ethanol at -20°C, and
centrifuged at 15,000 rpm for l5 min. Proteins thus
precipitated were mixed to react with an "SDS-PAGE sample
buffer" (Daiichi Chemicals) at 90°C for 3 min, and then
subjected to electrophoresis on 10°s SDS-polyacrylamide gel
CA 02236113 1998-04-29
41
(SDS-PAGE). Proteins thus fractionated were transferred to
PVDF membranes (Daiichi Chemicals), reacted with monoclonal
antibody 902 at room temperature for 1 h, and then washed
with T-TBS. The membranes were reacted with anti-mIgG
(Amersham) at room temperature for 1 h, and washed with T-
TBS. The membranes were then reacted with HRP-linked protein
A (Amersham) at room temperature for 1 h, washed with T-TBS,
and 4-chloro-1-naphthol (4CNPlus) (Daiichi Chemicals) was
added to detect gp120. As a result, protein bands were
visualized at positions corresponding to the expected
molecular weight of gp120.
In addition, effects of postinfection time of CV-1 cells
transfected with SeVgp120 on the HAU value and gp120
expression amount were analyzed. CV-1 cells (5 x 106)
dispensed to 10-cm plate were infected with SeVgp120 at the
multiplicity of infection of 10, and the culture medium (1 ml
each) was postinfectionally recovered at 30, 43, 53 and 70 h,
mixed with an equal volume of the fresh medium, and subjected
to HAU assay, gp120 expression examination (by ELISA) and
Western blotting. Results are shown in Figure 4. As clearly
shown in Fig. 3, the production of gp120 tends to increase
with the increasing HA titer of Sendai virus.
Example 6. Analyses of SeVgp120 propagation and gp120
expression level in various types of cells
Using similar methods as those in Example 5 except for
the use of various types of cells, HAU and gp120 expression
levels (by ELISA) were assayed. Results are shown in Table
CA 02236113 1998-04-29
42
5.
Table 5
Cell type Time (postinfection)HAU rgp120 (pg/ml)
CV-1 96 32 2.5
LLCMK2 48 16 0.5
CHO 55 4 0.46
NIH3T3 48 4 0.25
MT4 24 16 0.8
MOLT4/ 24 16 1.2
In the left-hand column of the Table are shown the
postinfectional times of various types of cells transfected
with SeVgp120. As a result, SeVgp120 propagation and gp120
expression were detected in all types of cells tested.
Example 7. Studies on the expression of luciferase gene
inserted into the Sendai viral vector in host cells.
In order to isolate the luciferase gene for inserting to
vectors, the luciferase gene bounded by the engineered NotI
sites on both termini was constructed by the standard PCR
using a set of primers [5'-AAGCGGCCGCCAAAGTTCACGATGGAAGAC-3')
(30mer) (SEQ ID NO: 3)] and [5'-TGCGGCCGCGATGAACTTTCACCC-
TAAGTTTTTCTTACTACGGATTATTACAATTTGGACTTTCCGCCC-3' (69mer) (SEQ
ID NO: 4) with "pHvluciRT4" as a template. The PCR product
was cloned into the Notl window of pSeVl8+ to obtain Sendai
virus vector to which the luciferase gene was inserted:
Then, this recombinant vector was transfected into LLCMK2
CA 02236113 1998-04-29
43
cells, and inoculated into embryonated chicken eggs. Chorio-
allantoic membranes of developing eggs were excised out,
twice washed with cold PBS(-), and, after the addition of a
lysis buffer (Picagene WAKO) (25 girl) and thorough mixing,
centrifuged at 15,000 rpm for 2 min. To the supernatant (5
girl each) was added the substrate (IATRON) (50 pl), and the
mixture was dispensed into each well of a 96-well plate.
Fluorescent intensity was measured with a luminometer
(Luminous CT-9000D, DIA-IATRON), and the enzyme activity was
expressed as counts per second (CPS). As a result, an
extremely high luciferase activity was detected with CV-1
cells at 24-h postinfection (Table 6). In this case, Sendai
virus which did not carry the luciferase gene was used as
control (represented by "SeV" in the table). Results
obtained from two clones are shown in the table.
Table 6
Fluorescence intensity (counts/10 sec)
Chorio-allantoic CV-1 (24h postinfection)
membrane
Luc/SeV 669187
2891560 8707815
SeV 69 48
23 49
CA 02236113 1998-04-29
44
industrial applicability
In the present invention, a system has been established
allowing the efficient rescue of viral particles from cDNAs
of negative strand viruses, and also a method has been
developed enabling the production and amplification of
"complexes comprised of RNAs derived from disseminative
specific negative strand RNA virus and viral structural
components containing no nucleic acids so as to have the
infectivity and autonomous RNA replicating capability but no
disseminative potency". Since said complexes can replicate
only within infected cells, these techniques are especially
useful in the fields of gene therapy, etc. wherein
therapeutical safety is highly appreciated.
CA 02236113 1998-04-29
Sequence Listing
SEQUENCE IDENTIFICATION NUMBER: 1
LENGTH: 38
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: other nucleic acid (synthetic DNA)
SEQUENCE
TGCGGCCGCC GTACGGTGGC AATGAGTGAA GGAGAAGT 3g
SEQUENCE IDENTIFICATION NUMBER: 2
LENGTH: 69
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: other nucleic acid (synthetic DNA)
SEQUENCE
TTGCGGCCGC GATGAACTIT CACCCTAAGT TTITVTTACT ACGGCGTACG TCATCTTTTT 60
69
SEQUENCE IDENTIFICATION NUMBER: 3
LENGTH: 30
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: other nucleic acid (synthetic DNA)
SEQUENCE
CA 02236113 1998-08-20
- 46 -
AAGCGGCCGC CAAAGTTCAC GATGGAAGAC 30
SEQUENCE IDENTIFICATION NUMBER: 4
LENGTH: 69
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: other nucleic acid (synthetic DNA)
SEQUENCE
TGCGGCCGCG ATGAACTTTC ACCCTAAGTT TTTCTTACTA CGGATTATTA CAATTTGGAC 60
1 0 TTTCCGCCC 69
76432-11