Note: Descriptions are shown in the official language in which they were submitted.
CA 02236998 1998-05-07
WO 97/17433 PCTIUS96/18088
TISSUE-SPECIFIC AND TARGET RNA-SPECIFIC
RIBOZYMES
BACKGROUND OF THE INVENTION
Field of the Invention
The invention pertains to the use of tissue-specific and target RNA-specific
ribozymes for treatment of cancers and bacterial, parasitic and viral
infections. More
specifically, the invention relates to a ribozyme targeted to the RNA
polymerase 1(A)
under the control of the probasin promoter.
Background Art
A ribozyme is a catalytic RNA molecule that cleaves RNA in a sequence specific
manner. The use of ribozyme as potential gene regulators in mammalian cells
and
antiviral agents has been suggested, but subject to serious questions
regarding technical
feasibility. For example, it is not known how ribozymes can be introduced to
target cells
or how they can be directed to the same subcellular compartments as their
target RNAs.
Other questions concern the effects of target RNA secondary structure on
ribozymal
activity. The art has not been successful in answering any of these questions.
Furthermore, because ribozymes are a form of antisense technology, the
problems encountered in applying antisense technology to disease treatment are
also
encountered in the use of ribozyme technology. For example, it has been shown
that the
expression of antisense RNA in transgenic mice did not invariably lead to a
reduction in
target RNA molecules, and when reduction in target RNA molecules did occur, it
was
not predictably paralleled by a reduction in protein. Even when protein levels
were
reduced sometimes no biological effect was detected (Whitton, J. Lindsay
"Antisense
Treatment of Viral Infection" Adv. in Virus Res. Vol. 44, 1994).
The experience in the art suggests that it is also not clear whether ribozymes
work best when free, or when embedded in an unrelated large RNA molecule
(Whitton,
1994). At present, sufficient data are not available, either in vitro or in
cell culture to
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
2
allow systematic comparison of the transactivities of free ribozymes with
their embedded
equivalents. There have been some studies that focus on the potential use of
ribozyme
technology in the treatment of cancer. In these studies, ribozymes have been
directed
against both c-fos and c-ras oncogenes in cell culture and showed some
suppression of
the malignant potential of cells when transplanted into mice. Nevertheless,
these
ribozymes specifically target an oncogene.
There has been no suggestion in the literature that tissue-specific cancers or
other tissue-specific disease can be treated by delivering to that tissue a
ribozyme having
a tissue-specific promoter, and that it is targeted to an RNA that is
essential for cell
survival. The invention provides such a ribozyme capable of treating tissue-
specific
cancers and other tissue-specific diseases.
The magnitude of the prostate cancer problem requires little introduction.
Approximately 44,000 men die each year of prostate cancer and about 10,000,000
men
have precancerous conditions of the prostate. It is clear that new approaches
to therapy
are needed. Animal models for testing therapeutic approaches are just becoming
available, and will require a number of years for validation. However, the
present
invention provides important reagents to address this problem.
One of the difficulties in using gene therapy to treat prostate cancer is the
long
standing problem of target-specific delivery. However, the recently developed
probasin
promoter provides target target-specificity enabling systemic delivery of the
present
ribozyme-encoding vector (Greenburg et al. Mol. Endocrinol. 8:230-239, 1994).
The
present vector consists of a tandem array of 3 hammerhead ribozymes, the 5'
and 3' of
which are designed to autocatalytically cleave themselves from the primary
transcript.
This novel construct eliminates problems inherent with extensive residual
flanking
sequences which might otherwise be present to compromise catalytic activity
and
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
3
specificity. The present constructs couple the prostate specific probasin
promoter to
triple-ribozymes targeted at mRNAs critical for prostate cell growth.
Endogenous delivery of a ribozyme under the control of a tissue-specific or
other
promoter can be complicated by "leakiness", where low levels of transcription
occur in
extraneous tissues. This could present a considerable therapeutic problem,
depending
upon the cellular target chosen. The present ribozyme compensates for this
problem by
targeting a cellular target which is associated with high levels of product
(that is, RNA
polymerase I produces large amounts of cellular ribosomal RNA). Thus, in the
event
promoter leakiness occurs in unintended tissues, it is not likely that cell
death would
occur. This choice, therefore, provides a needed level of safety, and
targeting of pol I
would be applicable to many selected tissues using other promoters.
A common problem in gene therapy is the difficulty in delivering the ribozyme
to
the correct tissue. The present invention avoids this difficulty by targeting
the ribozyme
to non-cellular RNAs in cells to which ribozyme constructs can be efficiently
delivered.
IV liposome delivery will be effective for treatment of HBV hepatitis. IV
and/or
extracorporeal treatment will effectively delivery construct to erythrocytes
for treatment
of malarial infection. And topical (with or without iv) administration will
effectively
deliver ribozyme construct to cervical epithelium in
dysplastic/precancerous/cancerous
HPV 16 cervical lesions. This latter example is of extreme importance for
treatment of
dysplastic/carcinoma in situ lesions diagnosed via abnormal Pap smears. A
second
advantageous facet of the non-cellular target ribozymes is that even if
promoter
leakiness and/or extraneous delivery and/or expression of the ribozyme occurs
in
unintended cells, the ribozymes should not cleave any cellular RNAs.
SUMMARY OF THE INVENTION
The invention provides tissue-specific and target RNA-specific ribozymes.
These ribozymes can be used to destroy target-specific neoplasms and to treat
viral
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
4
infections, among other uses. The ribozymes of the present invention comprise
a 5'
autocatalytically cleaving ribozyme sequence, a catalytic ribozyme comprising
a target =
RNA-specific binding site and a 3' autocatalytically cleaving ribozyme.
The invention also provides nucleic acids which encode the ribozymes of the
invention. These nucleic acids can be used to express the ribozymes of the
invention at
the selected site. The nucleic acids of the invention comprise a tissue-
specific promoter
binding site upstream from a sequence encoding a 5' autocatalytically cleaving
ribozyme
sequence, a catalytic ribozyme comprising a target RNA-specific binding site
and a 3'
autocatalytically cleaving ribozyme sequence.
A method of treating a subject having a proliferative disease of a specific
tissue
by inhibiting cell proliferation in the tissue, comprising administering to
the subject the
nucleic acid of claim 5, wherein the target-specific promoter binding sequence
is specific
for the diseased tissue, whereby the ribozyme encoded by the nucleic acid is
expressed,
ribosomal RNA production in the tissue is inhibited, cell proliferation is
inhibited, and
the proliferative disease treated is provided.
A method is provided for treating a subject having prostate cancer, comprising
administering to the subject the nucleic acid of claim 7, whereby the ribozyme
encoded
by the nucleic acid is expressed in the prostate and the prostate cancer is
treated.
A method of treating an infection in a subject, comprising administering to
the
subject the nucleic acid of claim 1, wherein the encoded target RNA-specific
binding site
is specific for an RNA unique to the infectious agent, whereby the ribozyme
encoded by
the nucleic acid is expressed and the infectious agent is killed is also
provided. =
CA 02236998 1998-05-07
WO 97/17433 PCTIUS96/18088
BRIEF DESCRIPTION OF THE FIGURES
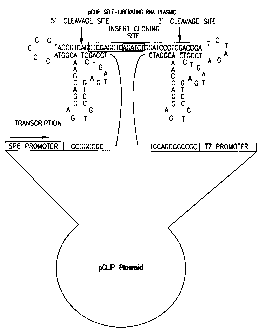
Fig. 1 shows DNA encoding the parental double ribozyme starting at Not I site.
Underlined sequences are Not I sites (GCGGCCGC) and Bgl II (AGATCT). Sequence
5 of parental double ribozyme starting at Not I site. Underlined sequences are
Not I sites
(GCGGCCGC) and Bgl II (AGATCT)
Fig. 2 shows DNA encoding the entire sequence with the internal Pol I targeted
triple ribozyme in bold. Underlined sequences are Not I (GCGGCCGC) and Bgl II
(AGATCT). This is the entire sequence with the internal Pol I targeted triple
ribozyxne
in bold. Underlined sequences are Not I (GCGGCCGC) and Bgl II (AGATCT)
Fig. 3 shows the two dimensional structure of parent double ribozyme into
which
the core ribozyme was cloned, as its reverse complement DNA.
DETAILED DESCRIPTION OF THE INVENTION
Ribozymes
The invention provides tissue-specific and target RNA-specific ribozymes.
These ribozymes can be used to destroy tissue-specific neoplasms and to treat
viral,
bacterial or parasitic infections, among other uses. The ribozymes of the
present
invention comprise a 5' autocatalytically cleaving ribozyme sequence, a
catalytic
ribozyme comprising a target RNA-specific binding site and a 3'
autocatalytically
cleaving ribozyme. One example of the present ribozyme is shown in by its DNA
coding
sequence in Fig. 2 and in SEQ ID NO: 1. The nucleotides numbered 1-164 encode
the
ribozyme, including the 5' and 3' autocatalytic ribozyme sequences. The 5'
autocatalytically cleaving ribozyme, catalytic ribozyme, and 3'
autocatalytically cleaving
= ribozyme of this exemplary ribozyme are shown separately in SEQ ID No:1.
,,. ,.
CA 02236998 2004-11-03
WO 97/17433 PCT/US96/18088
6
Alternatively, the 5' autocatalytically cleaving ribozyme can be replaced with
another stretch of transcribed RNA. In this RNA, the first 20-30 nt (or
longer) are
followed by a sequence which represents the reverse complement of the
initial,20-30 nt.
This way, the construct would presumably still be capped at the 5' end the way
pol II
transcripts are, but the initial nucleotides should not alter the specificity
of the
nucleotides on the 5' side of the targeted middle ribozyme. Based upon the
fact that the
transcript would be capped, it should be exported efficiently to the
cytoplasm. For
present triple ribozyme construct having both 5' and 3' autocatalytic
ribozymes, it is
expected that there will be some diffusion mediated transport to the cytoplasm
of the
internal targeted ribozyme, although this altemative 5' end should increase
the
cytoplasmic proportion.
The invention provides ribozymes that have the unique characteristic of being
both target RNA-specific in their catalytic action, and subject to tissue-
specific
expression. In the example shown in Fig. I and SEQ ID NO: 1, the target RNA
specificity is conferred by an RNA binding site that specifically binds a
sequence that is
unique to RNA polymerase I(A) (ribosomal RNA polymerase). It will be
understood
that an RNA sequence unique fbr any RNA can be the target of the present
target RNA-
specific ribozyme. The determination of unique sequences is routine given the
availability of numerous computer databases (GenBank) and computer programs
(Genetics Computer Group, PCGENE and BLAST) which can search for and find any
matches between nucleic acid sequences. A unique DNA sequence located on one
of
the databases will have a corresponding unique RNA sequence.
One example of the catalytic sequence of the present ribozymes is also shown
as
its DNA coding sequence in Fig. I and SEQ ID NO:1. Other catalytic sequences
include those known in the art. A number of sequence variation have defined
permissible nucleotide alteration in "stem" regions (Fedor and Uhlenbeck Proc.
Nat.
Acad. Sci. 87:1668-1672, 1990). Those skilled in the art will understand that
any
catalytic sequence, even those not yet discovered, can be used to construct a
ribozyme
* trademark
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
7
of the invention when it is routinely combined with the autocatalytically
cleaving
ribozymes and RNA binding site as described herein.
= One example of the 5' and 3' autocatalytically cleaving ribozymes that are
expressed with the catalytic ribozyme of the invention are shown in Fig. 1 and
SEQ ID
NO: 1, and also in Figs. 2 and 3. As further described below, these ribozymes
are
important for the expression of the catalytic ribozyme, because they cleave
off of the
ribozyme transcript as soon as they are transcribed to produce a catalytic
ribozyme
having minimal extraneous 5' or 3' sequences. Thus, the target-specific
binding site and
the catalytic sequence that comprise the catalytic ribozyme are in the correct
configuration to bind and cleave the target RNA. The extraneous sequences in
the
exemplified construct are the result of the cloning procedure. It is
understood that with
the selection of an alternative cloning scheme some or all of these extraneous
nucleotides can be elinzinated.
Ribozyme Encoding Nucleic Acids
The invention also provides nucleic acids which encode the ribozymes of the
invention. These nucleic acids can be used to express the ribozymes of the
invention at
the selected site. The site can be tissue-specific in the case of treating
tissue-specific
cancers, or it can be target-specific in the case of ribozymes that prevent
replication of
infectious agents to treat infection (e.g. hepatitis, herpes, malaria,
tuberculosis, etc.).
The nucleic acids of the invention comprise a tissue-specific promoter binding
site upstream from a sequence encoding a 5' autocatalytically cleaving
ribozyme
sequence, a catalytic ribozyme comprising a target RNA-specific binding site
and a 3'
autocatalytically cleaving ribozyme sequence.
The tissue-specific promoter binding site in the ribozyme-producing construct
results in tissue-specific expression of the ribozyme in tissue(s) that
actively transcribe
RNA from the selected promoter. Thus, only the target RNA in tissue that
utilizes the
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
8
promoter will be cleaved by the ribozyme. The exemplary ribozyme shown in Fig.
I and
SEQ ID NO:1 uses the binding site for the probasin promoter, a promoter-
specific for
prostate epithelium (Greenburg et al., 1994)). This tissue-specific promoter
binding site
has the sequence shown in (Greenburg et al., 1994).
As expected, other tissue-specific promoters can be used in the present
nucleic
acid constructs. Examples of these promoters include the binding sites for
prostate-
specific antigen (prostate), albumin (liver), fatty acid binding protein
(ilium), whey acidic
protein (breast), smooth muscle action (smooth muscle), etc. It will also be
clear that
target-specific promoters not yet identified can be used to target expression
of the
present ribozymes to the selected tissue(s). Once a target-specific promoter
is identified
its binding sequence can be routinely determined by routine methods such as
sequence
analysis. The promoter is defined by deletion analysis, mutagenesis,
footprinting, gel
shifts and transfection analyses (Sambrook et al., Molecular Cloning: A
Laboratory
Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, New York,
1989).
In the ribozyme-encoding nucleic acid of the invention, the nucleic acid
encoding
the 5' autocatalytically cleaving ribozyme can have the sequence of
nucleotides 1-54
shown in SEQ ID NO:1. In the ribozyme-encoding nucleic acid of the invention,
the
nucleic acid encoding the 3' autocatalytically cleaving ribozyme can have the
sequence
of nucleotides 111-164 shown in SEQ ID NO:1.
It is understood that other 5' and 3' autocatalytically cleaving ribozymes may
be
developed that can be encoded by the present nucleic acids. These ribozymes
can be
developed according to the methods of Taira et al. (Nuc. Acids Res.
19(19):5125-5130,
1991).
The present nucleic acid encodes a catalytic ribozyme that contains two
separable functional regions: a highly conserved catalytic sequence which
cleaves the
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
9
target RNA (also known as the "catalytic core"), and flanking regions which
include a
target RNA-specific binding site. By nucleic acid complementarity, the binding
site
directs the ribozyme core to cleave a specific site on the target RNA
molecule. The
= length of flanking sequences have implications not only for specificity, but
also for the
cleavage efficiency of the individual ribozyme molecules. In the present
catalytic
ribozyme, the flanking sequences are highly specific for the target RNA, yet
allow ready
dissociation from the target RNA once cleavage occurs. This permits cycling of
the
ribozyme (with an expected Kcat of about 1 cleavage per minute) and reduces
the amount
of ribozyme required to be effective. A range of binding/dissociation values
from 16-21
Kcal should be effective.
The complexity of human RNA is about 100 fold lower than that for human
DNA, and specificity can be achieved with as few as 12-15 base pairs. The
stability of
the RNA-RNA duplex is effected by several factors, such as GC content,
temperature,
pH, ionic concentration, and structure. The nearest neighbor rules can provide
a useful
estimate of the stability of the duplex (Castanotto et al. "Antisense
Catalatic RNAs as
Therapeutic Agents" Advances in Pharmacol. 25:289-317, 1994).
As described above, the encoded RNA binding site is unique, so the encoding
nucleic acid sequence will be the corresponding unique DNA sequence. The RNA
binding site can comprise a sequence that binds to an RNA sequence unique to
ribosomal RNA polymerase I(A) subunit. The ribosomal RNA polymerase binding
site
encoding DNA can have the sequence shown in Fig. 3. This is a sequence from
the
RNA polymerase I(A) subunit.
The catalytic ribozyme of the invention also includes a catalytic sequence,
which
cleaves the target RNA near the middle of the site to which the target RNA-
specific
binding site binds. In the hammerhead type of ribozyme, the catalytic sequence
is
generally highly conserved. The conserved catalytic core residues are 5'
CUGANGA 3'
and 5' GAAA 3' linked by an evolutionarily conserved stem-loop structure.
CA 02236998 2004-11-03
= _ _.
WO 97/17433 PCT/I3S96/18088
The most conserved and probably most efficiently cleaved sequence on the
target
RNA is 5' GUC 3'. However, XUN (N = A, U or C) can also be cleaved
efficiently.
Such cleavage sites are ubiquitous in most RNAs allowing essentially all RNA's
to be
targeted (Whitton, J. Lindsay "Antisense Treatment of Viral Infection" Adv. in
Virus
5 Res. Vol. 44, 1994).
With regard to the selection of the appropriate sites on target RNA, it is
known
that target site secondary structure can have an effect on cleavage in vitro
(Whitton,
1994). Thus, the selected target molecule's sequence can be routinely screened
for
10 potential secondary structure, using the program RNAFOLD*(from the PCGENE
group
of programs or available on the Internet). Thus, reasonable predictions of
target
accessibility can be made. Computer assisted RNA folding (Castanotto et al.,
1994),
along with computational analysis for 3-dimensional modeling of RNA (Major et
al.,
Science 253:1255-1260, 1991 and Castanotto et al., 1994) is certainly
effective in
guiding the choice of cleavage sites.
The internal ribozyme can be targeted to noncellular RNAs necessary for growth
of parasites, virus life cycles, etc., and expression can be driven with
tissue-specific or
virus-specific promoters. Three important examples which are specifically
presented in
the applicaxion are:
A) Use of the albumin promoter with a Hepatitis B virus target (chosen to
cleave the viral RNA pregenome, S protein, and polymerase/reverse
transcriptase
transcripts using the same ribozyme target site);
B) Use of generic promoters active in erythrocytes, using a ribozyme targeted
to
highly conserved regions of the EMP-1 protein family from P. faiciparum, which
are
necessary for cytoadherence and antigenic variation in malaria; and
C) Use of the HPV promoter, with a ribozyme targeted to a specific site near
the translational start site of the E6 protein, a site known to be critical
for expression of
both the E6 and E7 proteins which are intimately involved in cervical
carcinogenesis.
* trademark
CA 02236998 1998-05-07
WO 97/17433 PCTIUS96/18088
11
One example of the nucleic acid of the invention has the nucleotides in the
sequence shown in the Sequence Listing as SEQ ID NO: 1. This exemplary nucleic
acid
includes a probasin promoter, upstream from a sequence that encodes the 5'
autocatalytically cleaving ribozyme having the sequence shown in SEQ ID NO: 1,
the
ribosomal RNA binding site encoding DNA having the sequence shown in the
Sequence
Listing as SEQ ID NO:1 and the 3' autocatalytically cleaving ribozyme having
the
sequence shown in SEQ ID NO: 1.
Alternatively, silent base substitutions in the promoter binding site and
ribozyme
encoding sequence can be made that express the same ribozyme in the same
tissue.
Thus, a nucleic acid having substantially the nucleotide sequence shown in SEQ
ID
NO: 1, which encodes the ribozyme shown in SEQ ID NO: 1, is provided. The
nucleic
acid can vary based on the characteristics/definition of the promoter chosen,
and will
have 80%-99% sequence identity with SEQ ID NO: 1, more preferably, it will
have
90%-99% sequence identity with SEQ ID NO: 1. Other modifications could include
for
example, changes (or deletion) of nucleotides inserted for cloning purposes
(Fig. 2),
which include -1 to -8, +69 to +76. In Figure 3, the box includes extraneous
nucleotides
that are a function of cloning choices and, thus, can be modified. The
unpaired bases
can be any base, determined only by the cloning scheme chosen. If one of the
bases of a
pair is changed, the other must be changed in a complementary fashion.
Furthermore,
the ribozyme-coding sequence can be altered in ways that modify the ribozyme
sequence, but do not effect the ribozyme's target RNA-specificity or negate
its cleavage
activity. For example, changes in the stem loop regions of the 5', 3', and
internal
ribozyme (Fig. 2) could be incorporated into other constructs while
maintaining catalytic
activity (Fedor and Uhlenbeck, 1990).
Synthesis of the Ribozyme Producing Construct
Typically, the RNA binding and core sequences are synthesized as reverse
complementary oligonucleotides and are cloned into a vector that will allow
production
of the relevant RNA containing the ribozyme. The present ribozymes are
prepared by
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
12
synthesis of an oligonucleotide (5' GGA AGA TCT TTC AAA GAC TGA TGA CTC
CGT GAG GAC GAA ACG AGG ATC AGA TCT TCC 3') and its reverse
complement. The Bgl II site used in cloning is underlined. Following
appropriate
restriction digestion, in this particular case Bgl II, the double-stranded DNA
oligonucleotide is cloned into the multiple cloning site within the parent
vector (Figure
1).
Functional Testing
Once sequenced, these ribozymes are functionally tested. The test can involve
transcription of the ribozyme using one of the two possible bacterial
promoters, in this
case SP-6 or T-7, (in the presence of trace amounts of radioactivity) followed
by
evaluating the autocatalytic cleavage of the ribozyme by electrophoresis. Data
from
these tests are provided in the Examples.
Additional testing procedures encompass incubation of in vitro transcribed
ribozymes with in vitro synthesized target RNA transcript or with cytoplasmic
RNA
preparations. Following incubations, RNAs are examined by standard Northern
blot
analyses to verify specific degradation of target RNA transcripts.
The triple-ribozyme that has been constructed can be further tested by
subcloning it behind one of the tissue-specific promoters that will drive
expression of the
vector in a tissue-specific manner in the target. Data from these tests are
provided in the
Examples.
Finally, the triple-ribozyme experimental approach is further validated by
doing
in vivo studies in nzice. Two such studies have been performed as described in
the
Examples. The first case used a control vector more easily monitored than the
pol I
ribozyme consisting of the probasin promoter driving expression of algal green
fluorescent protein (pBGFT). This test vector is used to ascertain the
effectiveness of
our in vivo delivery system. A second experiment has also been carried out in
which the
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
13
pol I ribozyme is introduced. In both cases where either the triple ribozyme
or the green
fluorescent protein was introduced, the animals were euthanized at various
time post
operatively, autopsied, and various tissue were examined for activity of the
vector by
immunohistochemistry.
Delivery
The nucleic acids of the invention can be in a vector for delivering the
nucleic
acid to the site for expression of the ribozyme. The vector can be one of the
commercially available preparations, such as the pGM plasmid (Promega) Vector
delivery can be by liposome, using commercially available liposome
preparations or
newly developed liposomes having the features of the present liposomes. Other
delivery
methods can be adopted routinely tested in methods taught herein. An example
of a
delivery method using liposomes is further described in the Examples.
The modes of administration of the liposome will vary predictably according to
the disease being treated and the tissue being targeted. For lung (e.g.,
tuberculosis,
cancer) and liver (e.g., hepatitis and cancer) which are both sinks for
liposomes,
intravenous administration is reasonable. For many other localized pathologic
conditions including cancers, infections (e.g., hepatitis, cystitis,
proctitis, cervicitis, etc.)
as well as precancerous conditions, catheterization of an artery upstream from
the organ
is a preferred mode of delivery, because it avoids significant clearance of
the liposome
by the lung and liver. For lesions at a number of other sites (e.g., skin
cancer, human
papilloma virus infection, herpes (oral or genital) and precancerous cervical
dysplasia),
topical delivery is expected to be effective and may be preferred, because of
its
convenience.
Leukemias and other conditions such as malaria, may also be more readily
treated by ex vivo administration of the ribozyme.
CA 02236998 2004-11-03
.. , _.
WO 97/17433 PCT/US96/18088
14
The liposomes may be administered topically, parenterally (e.g.,
intravenously),
by intramuscular injection, by intraperitoneal injection, transdermally,
excorporeally or
the like, although IV or topical administration is typically preferred. The
exact amount
of the liposomes required will vary from subject to subject, depending on the
species,
age, weight and general condition of the subject, the severity of the disease
that is being
treated, the particular compound used, its mode of administration, and the
like. Thus, it
is not possible to specify an exact amount. However, an appropriate amount may
be
determined by one of ordinary skill in the art using only routine
experimentation given
the teachings herein. Generally, dosage will approximate that which is typical
given in
the Examples.
Parenteral administration, if used, is generally characterized by injection.
Injectables can be prepared in conventional forms, either as liquid solutions
or
suspensions, solid forms suitable for solution or suspension in liquid prior
to injection, or
as emulsions. A more recently revised approach for parenteral administration
involves
use of a slow release or sustained release system, such that a constant level
of dosage is
maintained. See, e.g., U.S. Patent No. 3,710,795.'
Topical administration can be by creams, gels, suppositories and the like. Ex
vivo (excorporeal) delivery can be as typically used in other contexts.
Transgenic Animals
The invention provides a transgenic non-human animal, containing, in a germ or
somatic cell, a nucleic acid comprising a target-specific promoter binding
site upstream
from a sequence encoding a 5' autocatalytically cleaving ribozyme sequence, a
cataTytic
ribozyme comprising a target RNA-specific binding site and a 3'
autocatalytically
cleaving ribozyme sequence, wherein the animal expresses a ribozyme comprising
a 5'
autocatalytically cleaving ribozyme sequence, a catalytic ribozyrne comprising
a target
RNA-specific binding site and a 3' autocatalytically cleaving ribozyme
sequence.
CA 02236998 1998-05-07
WO 97/17433 PCTlUS96/18088
The nucleic acid can be the nucleic acid shown in Fig.1 and SEQ ID NO: 1.
Alternatively, silent base substitutions in the promoter binding site and
ribozyme
encoding sequence can be made that express the same ribozyme in the same
tissue. For
example, these substitutions can be as described above.
5
The transgenic non-human animal of the invention is useful, because the animal
does not express a phenotype associated with the target RNA (e.g., with the
protein it
encodes). As used herein the term "phenotype" includes morphology, biochemical
profiles (e.g., changes in amounts of RNA or protein expressed, etc.) and
other
10 parameters that are affected by the knockout. For example, cell death of
otherwise
healthy cells can be a measure of altered phenotype resulting from ribozyme
expression.
Transformed Host Cells
The present ribozymes can be expressed in a transformed cell line. The
15 transformed cell can be used to validate both the specificity of the
ribozyme's expression
and the specificity and cleavage activity against the target RNA. An example
of such a
screening function is described in the Examples.
Screening Methods
The transgenic animals and transformed host cells of the invention can be used
in
a method of screening a compound for its ability to cause the animal or host
cell to
express a phenotype associated with the target RNA. The method requires
administering the compound to the animal/cell and assessing the compounds
ability to
cause expression of the phenotype. If the phenotype is restored, the compound
is
considered to be effective. For example an L-dopa functional knockout
transgenic
animal can be made and used to screen for drugs that restore an L-dopa
associated
phenotype.
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
16
Treating Proliferative Diseases
A method of treating a subject having a proliferative disease of a specific
tissue is
provided. The treatment is carried out by inhibiting cell proliferation in the
specific
tissue, and this is accomplished by administering to the subject a nucleic
acid encoding a
ribozyme that is targeted to an RNA that is essential to cell survival or
replication, and
containing a target-specific promoter binding sequence that is specific for
the diseased
tissue. The ribozyme encoded by the nucleic acid is expressed in the diseased
tissue,
production of an essential RNA in the tissue is inhibited, cell proliferation
is inhibited in
the tissue, cell death ensues and the proliferative disease treated.
The proliferative diseases that can be treated by the present method include
almost all cancers for which a target-specific promoter exists, including,
prostate, breast,
colon, pancreatic, lung and liver.
For example, the invention provides a method of treating a subject having
prostate cancer, comprising administering to the subject the nucleic acid
shown in SEQ
ID NO: 1, whereby the ribozyme encoded by the nucleic acid is expressed in the
prostate
and the prostate cancer is treated.
Treating Viral Infection
A method of treating a viral infection in a subject, comprising administering
to
the subject a nucleic acid of the invention, wherein the encoded target RNA-
specific
binding site is specific for an RNA unique to the infectious agent, whereby
the ribozyme
encoded by the nucleic acid is expressed and the infectious agent is killed.
Transcription can be driven using a virus specific promoter or a tissue-
specific promoter
which will selectively express the targeted ribozyme in virus-infected tissue,
i.e., using
the liver- specific albumin promoter for expression of a targeted ribozyme
directed
against hepatitis B virus.
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
17
In the context of determining anti-viral efficacy, ribozyme expressing cell
lines
can be compared with their ribozyme negative counterparts for their ability to
support
viral infection/replication/yield. In a manner similar to that described
above, ribozyme
expressing cell lines can be obtained and assayed; and in all cases the
abilities of the
ribozyme to prevent infection can be determined.
The present invention will be illustrated in further detail in the following
non-
limiting examples.
EXAMPLES
ANALYSIS OF RIBOZYME GENE T'HERAPY IN PROSTATE CANCER
This example presents the vectors and a novel strategy to utilizing prostate
targeted expression of a hammerhead ribozyme to kill normal and neoplastic
prostate
epithelium. The ribozyme is a highly innovative, triple-ribozyme targeted to
destroy
cells by attacking essential RNA(s). The 5' and 3' ribozymes have been
designed to
undergo autocatalytic cleavage during transcription, freeing the internal
ribozyme (at
high levels) within the cells.
Intracellular expression of a hammerhead ribozyme, targeted towards an
essential cellular RNA (such as RNA polymerase 1(A)), results in the death of
the cell.
If the ribozyme is targeted to a specific tissue in a constrained manner, then
only cells in
that tissue will be affected by expression of the ribozyme. Targeting
selectively to
prostate can be achieved via the rat probasin promoter (pb) (or the prostate
specific
antigen promoter). Because tissue-specific prostate targeting exists using the
probasin
promotor, delivery of vectors systemically or by direct introduction into the
prostate will
result in death of transfected prostate cancer cells with some collateral
damage also
being observed in the remaining normal prostate epithelium. Because of the
rather
unique specificity of the probasin promoter, no additional collateral damage
is expected
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
18
to be observed elsewhere in the body. This is also expected to be true for the
prostate-
specific antigen.
Examples of targets include the I(A) subunit of RNA polymerase I and II. Other
internal targeted ribozymes are tested for in vitro and in vivo activity by
the methods
described.
Synthesis
The primary double ribozyme vector depicted in Figure 1 was constructed. The
two flanking ribozymes (bases 1 to 54 and 66 to 120) are capable of self-
cleavage. A
third ribozyme (Figure 2)(bases 64 to 105) targeted to pol I mRNA is cloned
between
the flanking ribozymes. This internal ribozyme has 19 bases within two regions
(TTCAAAGA-catalytic core-ACGAGGATCAG) that are anti-sense to the pol I
message and interact by base pairing in regions with minimal secondary
structures to
effect cleavage.
The internal pol I ribozyme was prepared by synthesis of an oligonucleotide
(5'
GGA AGA TCT TTC AAA GAC TGA TGA CTC CGT GAG GAC GAA ACG AGG
ATC AGA TCT TCC 3')(only the ribozyme sense strand is shown in its reverse
complement). The Bgl II site used in cloning is underlined. Following
appropriate
restriction digestion with Bgl II the double-stranded oligonucleotide was
cloned into the
Bgl II site within the parent vector (Figure 1).
Delivery
The prostate specific promoter, when coupled to the triple ribozyme construct,
will be delivered to the prostate systemically via liposomes. Various routes
of
introduction into the blood vascular system (some bypassing the lung and
liver) are
evaluated as described. Orthotopic routes can also be utilized. The liposome
vehicle is
expected to be efficient enough to deliver the molecule to prostate cancer
cells, because
of a high degree of vascularization). Thus, curing or at least reducing tumor
burden by
this gene therapy approach is reasonably expected.
i 1
CA 02236998 2004-11-03
WO 97/17433 PCT/US96/18088
19
The following liposome proparations were used in these studies: (a)
lipofectamine reagent (GIBCO BRL, Gaithersburg,lvID) is a polycationic lipid
composed of a positively charged lipid, DOSPA, and the neutral lipid, DOPE, in
a 3:1
molar ration; (b) the cationic lipid, DDAB, used in combination with DOPE at
2:1 or
0.6:1.0 ratios (Brunette et al. Mol. Cell. Biol. 14:2411-2418, 1994); and (c)
DIVIRIE in
combination with DOPE in a 1:1 molar ratio (Felgner et al. Methods (Orlando)
5:67-75,
1995), obtained from VICAL Corp. (San Diego, CA). Liposome reagents were
stored
at 4 C prior to transfection.
Testing
Once sequenced, these ribozymes are functionally tested: The test mechanism
involves transcription of the triple ribozyme using one of the two possible
bacterial
promoters, in this case SP-6 or T-7 present in the pCRII vector (Invitrogen,
San Diego,
Ca), (in the presence of trace amounts of radioactivity) followed by
evaluating the
. autocatalytic cleavage of the ribozyme by electrophoresis. This was carried
out with the
pol I ribozyme and cleavage was observed, i.e., first a 113 bp fragment was
produced
that included the iirternal targeting ribozyme and the 3' ribozyme, followed
by the
appearance of a 74 bp fragment containing the internal pol I ribozyme.
The ribozyme was subsequently tested by transient transfection it into C3H10T
1/2 mouse fibroblast cells. This demonstrated that pol I RNA was degraded when
triple
ribozyme expression was induced. Thus, two phases of the activity of the
molecule have
been examined and demonstrated to occur. Autocatalytic cleavage of the cis
ribozymes
and functional degradation of pol I mRNA in trans occur as expected.
A subsequent step is to introduce the triple-ribozyme into a vector under the
control of a tissue-specific promoter such as probasin or prostate specific
antigen that
will target expression in a tissue-specific manner. This was done by taking
the Not I
fragment containing the entire 3', 5' and intenlal ribozymes (Fig. 2) and
subcloning it
into a vector containing the probasin promoter region (-426 to +28 (Greenburg
et al.,
* trademark
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
1994)). This promoter has been demonstrated to target gene expression to the
prostate.
Vectors are tested in vivo in transgenic mice expressing the triple ribozyme
anti-
pol I construct. Transgenic mice are generated by standard pronuclear
injection as
5 described in Hogan (Manipulating the mouse embryo: a laboratory manual, Cold
Spring
Harbor, NY 1986). Prior to injection, constructs are separated from vector DNA
by
restriction digestion (H'ind III and Sac II) of the plasmid, followed by
sucrose gradient
fractionation. Isolated constructs are dialyzed against 10mm tris ph 8.0,
0.1mm EDTA
before injection. In these mice, once probasin expression becomes apparent at
the 3-5
10 week stage of post natal development destruction of prostate epithelium is
expected to
occur. The efficiency of generating transgenic mice is 10-15% with twenty-six
mice
delivered, i.e. 2-3 transgenic mice are predicted. Another way to verify the
present
ribozymes' functionality is to introduce the vector into tissue culture, e.g.,
human PC3
prostate cancer cells, and observe cell death in response to activation of the
ribozyme.
15 C3H10T 1/2 mouse fibroblasts have been confirmed as sensitive to the pol I
triple
ribozyme.
Further validation of this gene therapy approach is obtained through in vivo
studies. Two such studies have been performed. In the first case, a control
vector,
20 more easily monitored than the pol I ribozyme, which consists of the
probasin promoter
driving expression of the algal green fluorescent protein was used to
ascertain the
effectiveness of the in vivo liposomal delivery system. The procedure involves
mice
that are anesthetized and prepared by a surgical procedure to expose the
descending
aorta. A 30 gauge catheter is placed in the descending aorta followed by
introduction of
300,ug vector/kilogram body weight (vector: liposome ratio of 10gg/40,u1). The
results
verify that expression of the green fluorescent protein occurs in dorsolateral
prostate
epithelium but not in the lung (a normal target of liposomes). Further studies
determine
if expression is observed in liver and other tissue including the kidney,
adrenal gland and
the brain. A second experiment has also been carried out in which the po1 I
ribozyme
was introduced in vivo using the same method described above. In both cases
where
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
21
either the triple ribozyme or the green fluorescent protein was introduced,
animals were
euthanized at various time post operatively, autopsied, and tissues were
examined for
activity of the vector by immunohistochemistry. There was evidence of some
apoptotic
cell death in prostate epithelium at seven days following administration.
In vivo studies are conducted in transgenic mice bearing prostate tumors.
Tumors are induced by probasin directed expression of genes such as EcoRl (a
restriction enzyme), cfos (a proto-oncogene), or a modified version of lamin
(a nuclear
matrix molecule). Administration of the ribozyme is as described above.
Target Choice
Various molecules have been chosen for targeting. The first is RNA polymerase
1(A). This represents an excellent target, because it is an abundant RNA. If
there is a
leakiness (low levels of transcription) of the probasin promoter in other than
prostate
cells, they are predicted to survive the presence of limited levels of the Pol
I ribozyme.
Other potential targets will be examined including phosphofructokinase
(ribozyme
targets nt 178, 121, or 162 depending on tissue involved), RNA pol II subunit
14.4 kd
(ribozyme targets nt 83 or 884), mRNA pol II 140 kd subunit (ribozyme targets
nt 204),
and RNA pol 1123 kd subunit (ribozyme targets nt 143).
Another potential target of interest is the 70 kD subunit of replication
protein A.
It is needed for formation of DNA replication centers/foci, so it should
disrupt DNA
replication without danger of introducing errors. The sequence reference is
Kim, et al.,
Mol. Cell. Biol. 12:3050-3059, 1992.
RI[BOZYME GENE THERAPY IN PARASITIC INFECTION
The methods described above are applicable to the present context, except
where specified. For example, The administration mode will be different for
parasitic
infection than for prostate cancer and will depend upon tissue site.
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
22
For malaria (Plasmodiumfalciparum) the EMP-1 proteins, which are necessary
for cytoadherence and present a problem because they cause rapid antigenic
variation,
are targeted. Specifically, highly conserved GTCs in exon II are targeted.
Pertinent
EMBL accession numbers are L42246, L42244, L42245, L42247, L40600-L40609,
L42636. See Smith, J. et al. Cel182:101-110, 1995, and Su, X. et al. Cell
82:89-100,
1995. A promoter active in red blood cells can be used and treatment also
could be
extracorporeal.
RIBOZYME GENE THERAPY IN BACTERIAL INFECTION
The methods described above are applicable to the present context, except
where specified. For example, The administration mode will be different for
bacterial
infection than for prostate cancer, and will depend on the targeted tissue.
For Mycobacterium tuberculosis, a transcribed fragment which is essential for
cell entry will be targeted. The EMBL accession number is X70901. See Arruda,
S. et
al. Science 261:1454-1457, 1993. The target will be near the N-terminus of the
open
reading frame coding for the 52 kD protein, from the 1535 bp fragment.
RIBOZYME GENE THERAPY IN VIRAL INFECTION
The methods described above are applicable to the present context, except
where specified. For example, the administration mode will be different for
viral
infection than for prostate cancer.
Human papillomavirus type 16 E6 and E7 proteins are translated from a
bicistronic mRNA. Antisense oligonucleotides to the translational start site
of E6 inhibit
synthesis of both E6 and E7. The target will be a GTT, cleaving at nt 108
(Tan, et al., J. Gen. Virol. 75:2663-2670, 1994). Original numbering is from
Seedort, et al., Virology
CA 02236998 1998-05-07
WO 97/17433 PCT/US96/18088
23
145:181-185, 1985. IV and topical administration should be an effective
combination.
Topical administration should be effective for dysplastic/precancerous
lesions.
Hepatitis B virus is a partially single-stranded DNA virus, and it is now
thought
that integration of the viral genome is not the critical incident. Rather, the
viral genome
is made into an extended RNA intermediate, which is then reverse transcribed
into
DNA. A target with multiple advantage has been chosen. It will cut the viral
RNA
pregenome in the first place, and its located in both the S (envelope) and
polymerase/reverse transcriptase domains. The cut is at a GTC, after nt 438 in
the HBV
subtype ayw. The EMBL accession number is X02496, and the original sequence
reference is Galibert, et al., Nature 281, 646-650, 1979. Expression could be
driven by
the albuming promoter and IV administration should be highly effective.
While the foregoing invention has been described in some detail for purposes
of
clarity and understanding, it will be appreciated by one skilled in the art
from a reading
of this disclosure that various changes in form and detail can be made without
departing
from the true scope of the invention and appended claims.
Throughout this application various publications are referenced within
parentheses. The disclosures of these publications in their entireties are
hereby
incorporated by reference into this application in order to more fully
describe the state of
the art to which this invention pertains.
CA 02236998 1998-05-29
24
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: MEDICAL UNIVERSITY OF SOUTH CAROLINA;
171 Ashley Avenue
Charleston, South Carolina
29464; and
PENN STATE RESEARCH FOUNDATION
(ii) TITLE OF INVENTION:
TISSUE-SPECIFIC AND TARGET RNA-SPECIFIC RIBOZYMES
(iii) NUMBER OF SEQUENCES: 1
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: GOWLING, STRATHY & HENDERSON
(B) STREET: Suite 2600, 160 Elgin Street
(C) CITY: Ottawa
(D) PROVINCE: Ontario
(E) COUNTRY: Canada
(F) POSTAL CODE: K1P 1C3
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: MS-DOS 6.20
(D) SOFTWARE: PatentIn Release #1.0, Version #1.30
(vi) CURRENT APPLICATION DATA:
INTERNATIONAL APPLICATION NUMBER: PCT/US96/18088
CANADIAN APPLICATION NUMBER: Not Yet assigned
FILING DATE: NOVEMBER 8, 1996
(viii) PATENT AGENT INFORMATION:
(A) NAME: Eli J. McKhool
(B) REFERENCE NUMBER: 08-879558CA
CA 02236998 1998-05-29
5 (2) INFORMATION FOR SEQ ID NO:1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 184 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
10 (D) TOPOLOGY: linear
(ii) MOLECULE TYPE: oligonucleotide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:1:
GCGGCCGCTC GAGCTCTGAT GAGTCCGTGA GGACGAAACG GTACCCGGTA CCGTCAGCTC 60
GAGCTCAGAT CTTTCAAAGA CTGATGACTC GCTGAGGACG AAACGAGGAT CAGATCTGGA 120
15 TCCGTCGACG GATCTAGATC CGTCCTGATG AGTCGTGAGG ACGAAACGGA TCTGCAGCGG 180
CCGC 184