Note: Descriptions are shown in the official language in which they were submitted.
CA 02237701 1998-07-20
Smad BINDING PROTEINS
Field of the Invention
The invention relates to a family of proteins, the SARA proteins, which
bind to receptor-regulated Smad proteins and are involved in appropriate
localization of these Smad proteins for receptor activation.
Background of the Invention
The Transforming Growth Factor-beta (TGF~) superfamily, whose
members include TGF~is, activins and bone morphogenetic proteins (BMPs),
have wide ranging effects on cells of diverse origins (Attisano and Wrana,
1998; Heldin et al., 1997; Kretzschmar and Massague, 199). Signalling by
these secreted factors is initiated upon interaction with a family of cell-
surface
transmembrane serine/threonine kinases, known as type I and type II
receptors. Ligand induces formation of a typel/typell heteromeric complex
which permits the constitutively active type II receptor to phosphorylate, and
thereby activate, the type I receptor (Wrana et al., 1994). This activated
type I
receptor then propagates the signal to a family of intracellular signalling
mediators known as Smads (Attisano and Wrana, 1998; Heldin et al., 1997;
Kretzschmar and Massague, 1998).
The first members of the Smad family identified in invertebrates were
the Drosophila MAD and the C. elegans sma genes (sma-2, sma-3 and sma-4;
Savage et al., 1996; Sekelsky et al., 1995). Currently, the family includes
additional invertebrate Smads, as well as nine vertebrate members, Smad1
through 9 (Attisano and Wrana, 1998; Heldin et al., 1997; Kretzschmar and
Massague, 1998). Smad proteins contain two conserved amino (MH1) and
carboxy (MH2) terminal regions separated by a more divergent linker region.
In general, Smad proteins can be subdivided into three groups; the receptor-
regulated Smads, which include Smad 1, 2, 3, 5 and 8, Mad, sma-2 and sma-
3; the common Smads, Smad4 and Medea, and the antagonistic Smads,
CA 02237701 1998-07-20
2
which include Smad6, 7 and 9, DAD and daf-3 (Heldin et al., 1997;
Nakayama et al., 1998; Patterson et al., 1997).
Numerous studies with vertebrate Smad proteins have provided
insights into the differential functions of these proteins in mediating
signalling.
Receptor-regulated Smads are direct substrates of specific type I receptors
and
the proteins are phosphorylated on the last two serines at the carboxy-
terminus within a highly conserved SSXS motif (Abdollah et al., 1997;
Kretzschmar et al., 1997; Liu et al., 1997b; Macias-Silva et al., 1996;
Souchelnytskyi et al., 1997). Interestingly, Smad2 and Smad3 are substrates
of TGF(i or activin receptors and mediate signalling by these ligands (Liu et
al., 1997b; Macias-Silva et al., 1996; Nakao et al., 1997a), whereas Smad1, 5
and 8 appear to be targets of BMP receptors and thereby propagate BMP
signals (Chen et al., 1997b; Hoodless et al., 1995; Kretzschmar et al., 1997;
Nishimura et al., 1998). Once phosphorylated, these Smads bind to the
common Smad, Smad4, which lacks the carboxy-terminal phosphorylation
site and is not a target for receptor phosphorylation (Lagna et al., 1996;
Zhang
et al., 1997). Heteromeric complexes of the receptor-regulated Smad and
Smad4 translocate to the nucleus where they function to regulate the
transcriptional activation of specific target genes. The antagonist Smads,
Smad6, 7 and 9 appear to function by blocking ligand-dependent signalling
by preventing access of receptor-regulated Smads to the type I receptor or
possibly by blocking formation of heteromeric complexes with Smad4
(reviewed in Heldin et al., 1997).
Analysis of the nuclear function of Smads has demonstrated that Smads
can act as transcriptional activators and that some Smads, including
Drosophila Mad, and the vertebrate Smad3 and Smad4, can bind directly to
DNA, albeit at relatively low specificity and affinity (Dennler et al., 1998;
Kim
et al., 1997; Labbe et al., 1998; Mingling et al., 1997; Zawel et al., 1998).
Localization of Smads is critical in controlling their activity and Smad
phosphorylation by the type I receptor regulates Smad activity by inducing
CA 02237701 1998-07-20
3
nuclear accumulation (Attisano and Wrana, 1998; Heldin et al., 1997;
Kretzschmar and Massague, 1998). However, little is known about how
Smad localization is controlled prior to phosphorylation and how this might
function in modulating receptor interactions with its Smad substrates.
Summa~r of the Invention
Smad proteins (Smads) transmit signals from transmembrane ser/thr
kinase receptors to the nucleus. Mammalian and non-mammalian proteins
have been identified which interact directly with Smads and are designated
the Smad Anchor for Receptor Activation or SARA proteins.
The invention provides cDNA sequences encoding this previously
undescribed family of SARA proteins which bind to receptor-regulated Smad
proteins and ensure appropriate localization of these Smad proteins for
activation by a Type I receptor of a TGF~i, activin or BMP signalling pathway.
For example, TGF~ signalling induces dissociation of Smad2 or Smad3
from SARA with concomitant formation of Smad2/Smad4 or Smad3/Smad4
complexes and nuclear translocation. In the absence of signalling, SARA
functions to recruit a particular Smad (eg. Smad2 or Smad3) to distinct
subcellular sites in the cell and interacts with the TGF(3 superfamily
receptor
complex in cooperation with the particular receptor regulated Smad.
Mutations in SARA that cause mislocalization of Smad2, and interfere with
receptor association, inhibit receptor-dependent transcriptional responses,
indicating that regulation of Smad localization is essential for TGF~3
superfamily signalling. The invention provides a novel component of the
signal transduction pathway that functions to anchor Smads to specific
subcellular sites for activation by the Type I receptor of the TGF~3, activin
or
BMP signall ing pathways.
The SARA proteins are characterised by the presence of two domains, a
double zinc finger or FYVE domain responsible for the subcellular localisation
of the SARA protein or SARA-Smad complex and a Smad-binding domain
CA 02237701 1998-07-20
4
which mediates the interaction or binding of one or more species of Smad
protein.
In accordance with one embodiment, the invention provides isolated
polynucleotides comprising nucleotide sequences encoding SARA proteins.
In accordance with a further series of embodiments, the invention
provides an isolated polynucleotide selected from the group consisting of
(a) a nucleotide sequence encoding a human SARA protein;
(b) a nucleotide sequence encoding a mammalian SARA protein;
(c) a nucleotide sequence encoding a non-mammalian SARA
1 0 protein;
(d) a nucleotide sequence encoding the human SARA amino acid
sequence of Table 2 (Sequence ID N0:2);
(e) a nucleotide sequence encoding the human SARA amino acid
sequence of Table 4 (Sequence ID N0:4;
(f) a nucleotide sequence encoding the Xenopus SARA amino acid
sequence of Table 6 (Sequence ID N0:6);
(g) a nucleotide sequence encoding the Xenopus SARA amino acid
sequence of Table 8 (Sequence ID N0:8).
In accordance with a further embodiment, the invention provides the
nucleotide sequences of Table 1 (human SARA1 or hSARA1 , Table 3 (human
SARA2), Table 5 (XSARA1 ) and Table 7 (XSARA2).
In accordance with a further embodiment, the invention provides
recombinant vectors including the polynucleotides disclosed herein and host
cells transformed with these vectors.
The invention further provides a method for producing SARA proteins,
comprising culturing such host cells to permit expression of a SARA protein-
encoding polynucleotide and production of the protein.
The invention also includes polynucleotides which are complementary
to disclosed nucleotide sequences, polynucleotides which hybridize to these
CA 02237701 1998-07-20
sequences under high stringency and degeneracy equivalents of these
sequences.
In accordance with a further embodiment, the invention provides
antisense molecules which may be used to prevent expression of a SARA
5 protein. Such antisense molecules can be synthesised by methods known to
those skilled in the art including use of in vitro transcription or chemical
synthesis. Antisense molecules of this invention may comprise modified
bases or backbone linkages, eg. by incorporating phosphothioates or similar
compounds.
The invention further includes polymorphisms and alternatively spliced
versions of the disclosed SARA genes and proteins wherein nucleotide or
amino acid substitutions or deletions do not substantially affect the
functioning of the gene or its encoded protein.
The invention also enables the identification and isolation of allelic
variants or homologues of the described SARA genes, and their
corresponding proteins, using standard hybridisation screening or PCR
techniques.
The invention provides a method for identifying allelic variants or
homologues of the described SARA genes, comprising
choosing a nucleic acid probe or primer capable of hybridizing to a
SARA gene sequence under stringent hybridisation conditions;
mixing the probe or primer with a sample of nucleic acids which may
contain a nucleic acid corresponding to the variant or homologue; and
detecting hybridisation of the probe or primer to the nucleic acid
corresponding to the variant or homologue.
In accordance with a further embodiment, the invention provides
fragments of the disclosed polynucleotides, such as polynucleotides of at
least
10, preferably 15, more preferably 20 consecutive nucleotides of the
disclosed polynucleotide sequences. These fragments are useful as probes
CA 02237701 1998-07-20
6
and PCR primers or for encoding fragments, functional domains or antigenic
determinants of SARA proteins.
In accordance with a further embodiment, the invention provides
substantially purified SARA proteins, including the proteins of Table 2
(hSARA1 ), Table 4 (hSARA2), Table 6 (XSARA1 ) and Table 8 (XSARA2).
In accordance with one embodiment, a SARA protein has a FYVE
domain, a Smad binding domain (SBD) and an amino acid sequence having
at least 50% overall identity with the amino acid sequence of hSARA1
(Sequence ID N0:2).
In accordance with a preferred embodiment, a SARA protein has a
FYVE domain having at least 65% identity of amino acid sequence with the
FYVE domain of hSARA1 and a C-terminal sequence of 550 consecutive
amino acids which have at least 50% identity with the C-terminal 550 amino
acid residues of hSARA1.
In accordance with a more preferred embodiment, a SARA protein has
a FYVE domain having at least 65% identity of amino acid sequence with the
FYVE domain of hSARA1 and wherein the portion of the SBD corresponding
to amino acid residues 721 to 740 of hSARA1 has at least 80% identity with
that portion of hSARA1.
The invention further provides a method for producing antibodies
which selectively bind to a SARA protein comprising the steps of
administering an immunogenically effective amount of a SARA
immunogen to an animal;
allowing the animal to produce antibodies to the immunogen; and
obtaining the antibodies from the animal or from a cell culture derived
therefrom.
The invention further provides substantially pure antibodies which
bind selectively to an antigenic determinant of a SARA protein. The
antibodies of the invention include polyclonal antibodies, monoclonal
antibodies and single chain antibodies.
CA 02237701 1998-07-20
7
The invention includes analogues of the disclosed protein sequences,
having conservative amino acid substitutions therein. The invention also
includes fragments of the disclosed protein sequences, such as peptides of at
least 6, preferably 10, more preferably 20 consecutive amino acids of the
disclosed protein sequences.
The invention further provides polypeptides comprising at least one
functional domain or at least an antigenic determinant of a SARA protein.
In accordance with a further embodiment, the invention provides
peptides which comprise SARA protein Smad binding domains and
polynucleotides which encode such peptides.
In accordance with a further embodiment, the invention provides a
Smad binding domain peptide selected from the group consisting of
(a) SASSQSPNPNNPAEYCSTIPPLQQAQASGALSSPPPTVMVPVGV
LKHPGAEVAQPREQRRVWFADGILPNGEVADAAKLTMNGTSS; and
(b) amino acids 589 to 672 of the XSARA sequence of Table 9.
The invention includes fragments and variants of these Smad binding
domain peptides which retain the ability to bind a Smad protein.
In accordance with a further embodiment, the invention provides
peptides which comprise SARA protein FYVE domains and polynucleotides
which encode such peptides.
In accordance with a further embodiment, the invention provides a
FYVE domain peptide selected from the group consisting of
(a) amino acids 587 to 655 of the hSARA sequence of Table 9;
(b) amino acids 510 to 578 of the XSARA sequence of Table 9; and
(c) the consensus amino acid sequence of Table 10.
The invention includes fragments and variants of these FYVE domain
peptides which retain the function of the parent peptide.
The invention further provides methods for modulating signalling by
members of the TGFa superfamily which signal through pathways which
involve a SARA protein.
CA 02237701 1998-07-20
8
Modulation of signalling by a TGF(i superfamily member through such
a pathway may be effected, for example, by increasing or reducing the
binding of the SARA protein involved in the pathway with its binding partner.
In accordance with a further embodiment, TG F(3 superfamily signalling
by a pathway involving a SARA protein described herein may be modulated
by modulating the binding of the SARA protein to a Smad binding partner, to
the subcellular localisation site (via the FYVE domain), or to the SARA
receptor.
For example, the binding of a SARA protein to a Smad binding partner
may be inhibited by a deletion mutant of the protein lacking either the SBD
domain or the FYVE domain or by the SARA protein Smad binding domain
peptides or FYVE domain peptides described herein, and effective fragments
or variants thereof.
The invention further provides methods for preventing or treating
diseases characterised by an abnormality in a TGF~i superfamily member
signalling pathway which involves a SARA protein, by modulating signalling
in the pathway, as described above.
TG F(3 signalling is important in wound healing, and excessive
signall ing is associated with scarring, with arthritis and with fibrosis in
numerous diseases, including fibrosis of the liver and kidney. TGF(3
signalling
is also involved in modulating inflammatory and immune responses and can
contribute to tumour progression.
The invention thus provides methods for modulating TGFa-dependent
cell proliferation or fibrogenesis.
The BMP signalling pathways are important in tissue morphogenesis
and in protecting tissues and restoring or regenerating tissues after tissue
damage, for example in bone, kidney, I fiver and neuronal tissue (see, for
example, Reddy, A.H. (1998), Nature Biotechnology, v. 16, pp. 247-252).
CA 02237701 1998-07-20
9
The invention further provides methods for modulating BMP-
dependent phenotypic marker expression by modulating the interactions of
SARA proteins involved in these BMP signalling pathways.
In accordance with a further embodiment, modified versions of a SARA
protein may be provided as dominant-negatives to block TGF~i, activin or
BMP signalling. These modified versions of SARA could, for example, lack
the Smad binding domain and thereby prevent recruitment of a specific Smad
or could lack the FYVE domain and thereby inhibit signalling by interfering
with appropriate subcellular localisation.
These modified versions of SARA may be provided by gene therapy,
for example using viral vectors, or naked DNA with an appropriate carrier.
Expression may be driven by inclusion in the vector of a promoter specific for
a selected target cel I type. Many examples of such specific promoters are
known to those skilled in the art.
In a further embodiment, a normal version of a SARA protein such as
hSARA1 could be provided by gene therapy to restore function in a disease
wherein SARA is mutated or non-functional.
In a further embodiment, the invention provides a pharmaceutical
composition comprising a purified SARA protein as an active ingredient.
In accordance with a further embodiment, the invention provides non-
human transgenic animals and methods for the production of non-human
transgenic animals which afford models for further study of the SARA system
and tools for screening of candidate compounds as therapeutics. For
example, knock out animals, such as mice, may be produced in which a
SARA gene is deleted from the genome. These animals or cell lines derived
therefrom may be examined for phenotypic changes and used to screen
candidate compounds for effectiveness to reverse these changes.
In a further example, transgenic animals may be produced expressing a
dominant negative mutant of a SARA protein, as described above, either
generally or in specific targeted tissues.
CA 02237701 1998-07-20
The invention provides many targets for the development of small
molecule drugs, including peptides and peptidomimetic drugs, to interfere
with the interaction of the various binding partners described herein and
thereby modulate signalling by members of the TG F[3 superfamily, including
5 TGF(3 and BMPs.
The invention further provides methods for screening candidate
compounds to identify those able to modulate signalling by a member of the
TG F(3 superfamily through a pathway involving a SARA protein.
For example, the invention provides screening methods for compounds
10 able to bind to a SARA protein which are therefore candidates for modifying
the activity of the SARA protein. Various suitable screening methods are
known to those in the art, including immobilization of a SARA protein on a
substrate and exposure of the bound SARA protein to candidate compounds,
followed by detection of compounds which have bound to the SARA protein.
The methods used to characterise the binding interactions of the SARA
proteins disclosed herein, as fully described in the examples herein, may also
be used to screen for compounds which are agonists or antagonists of the
binding of a SARA protein, to another compound of the signalling pathway,
for example, a Smad protein, a receptor, or a subcellular localisation cite.
This invention also provides methods of screening for compounds
which modulate TGF~i superfamily signalling by detecting an alteration in the
phosphorylation state of a SARA protein.
In accordance with a further embodiment, the invention provides a
method for reducing or preventing TGF(3, activin or BMP signalling by
inhibiting the activity of SARA. For example, SARA activity may be inhibited
by use of an antisense sequence to the SARA gene or by mutation of the
SARA gene.
SummarX of the Drawings
Certain embodiments of the invention are described, reference being
made to the accompanying drawings, wherein:
CA 02237701 1998-07-20
11
Figure 1 (top panel) shows interaction of full length hSARA with
bacterially expressed Smads. Full length SARA protein was produced in an in
vitro transcription/translation system in the presence of [35S]methionine and
was incubated with glutathione-sepharose beads coated with bacterially-
expressed GST fusion proteins of the indicated Smads or Smad2 subdomains.
Bound material was resolved by SDS-PAGE and visualized by
autoradiography. Migration of full length hSARA, and a translation product
that initiates from an internal methionine located upstream of the Smad
binding domain (asterisk) are indicated. The presence of approximately
equivalent amounts of GST fusion proteins was confirmed by SDS-PAGE and
coomassie staining of a protein aliquot (bottom panel).
Figure 2 shows interaction of hSARA with Smads in mammalian cells.
COS cells were transfected with Flag-tagged hSARA (Flag-SARA) either alone
or together with the indicated Myc-tagged Smad constructs. For Smad6, an
alternative version lacking the MH1 domain was used (Topper et al., 1997).
Cell lysates were subjected to an anti-Flag immunoprecipitation and
coprecipitating Smads detected by immunoblotting with anti-Myc antibodies.
The migration of anti-Flag heavy and light chains (IgG) are marked. To
confirm efficient expression of hSARA and the Smads, aliquots of total cell
lysates were immunoblotted with the anti-Flag and anti-Myc antibodies
(bottom panel). The migrations of hSARA and the Smads are indicated.
Figures 3-6 show immunoblots of lysates from COS cells transiently
transfected with various combinations of Flag or Myc-tagged hSARA, wild
type (WT) or mutant (2SA) Myc or Flag-tagged Smad2, Smad4/HA and wild
type (WT) or constitutively active (A) T(3RI/HA, cell lysates being subjected
to
immunoprecipitation with anti-Flag or anti-Myc antibodies, as indicated.
Confirmation of protein expression was performed by immunoblotting total
cell lysates prepared in parallel for the indicated tagged protein (totals,
bottom
panels).
CA 02237701 1998-07-20
12
Figure 3: Transfected cells were metabolically labelled with [32P]P04
and cell lysates subjected to immunoprecipitation with anti-Flag antibodies
for
visualization of hSARA phosphorylation (top panel) or with anti-Myc
antibodies for Smad2 phosphorylation (middle panel). Immunoprecipitates
were resolved by SDS-PAGE and visualized by autoradiography. The
migrations of hSARA AND Smad2 are indicated.
Figure 4: Lysates from transiently transfected COS cells were subjected
to immunoprecipitation with anti-Flag antibodies and Smad2 bound to SARA
was analyzed by immunoblotting with anti-Myc antibodies (IP: a-flag; blot: a-
Myc).
Figure 5: Lysates from transiently transfected COS cells were subjected
to immunoprecipitation with anti-Flag antibodies and Smad2 bound to SARA
was analyzed by immunoblotting with anti-Myc antibodies (IP: a-flag, blot: a-
Myc). Partial dissociation of SARA/Smad2 complexes induced by TGFa
signalling was enhanced by expression of Smad4.
Figure 6: Cell lysates from transiently transfected COS cells were
subjected to immunoprecipitation with anti-Flag antibodies directed towards
Smad2. Immunoprecipitates were then immunoblotted using anti-Myc or
anti-HA antibodies which recognize hSARA or Smad4, respectively.
Coprecipitating SARA (a-myc blot) and Smad4 (a-HA blot) are indicated.
Figure 7, panels A to E, shows photomicrographs of Mv1 Lu cells
transiently transfected with various combinations of Flag-Smad2, Myc-hSARA,
and constitutively active T[iRl (T[3RI*) as indicated (Tx). HSARA was
visualized with the polyclonal Myc A14 antibody and Texas-Red conjugated
goat-anti-rabbit IgG (red) and Smad2 was detected with an anti-Flag M2
monoclonal antibody followed by FITC-conjugated goat anti-mouse IgG
(green). The subcellular localization of the expressed proteins was visualized
by immunofluorescence and confocal microscopy.
CA 02237701 1998-07-20
13
Panels A, B, C, Mv1 Lu cel Is singly transfected with hSARA (A) or
Smad2 (B) are shown. Cotransfection of Smad2 with the constitutively active
T~RI (T~iRI*) results in its accumulation in the nucleus (C).
Panel D, Mv1 Lu cells were transfected with hSARA and Smad2 and the
localization of hSARA (red, panel I) and Smad2 (green, panel ii) is shown.
Colocalization of SARA and Smad2 is shown (panel iii) and appears as
yel low.
Panel E, Mv1 Lu cells were transfected with hSARA, Smad2 and
activated T~3RI (T~iRI*) and the localization of hSARA (red, panel I) and
Smad2
(green, panel ii) is shown. Colocalization of SARA and Smad2 is indicated
(panel iii). Note the shift to an orangy-red colour in the punctate spots and
an
intensification of Smad2 nuclear staining, indicative of dissociation of Smad2
from SARA and nuclear translocation.
Figure 8 is an immunoblot of COS cells transiently transfected with
various combinations of Flag-hSARA, Myc-Smad2, wild type (WT) T~RII and
either wild type or kinase-deficient (KR) versions of T~iRI. Cells were
affinity-
labelled with ['251]TGF~ and lysates immunoprecipitated with anti-Flag
antibodies. Coprecipitating receptor complexes were visualized by SDS-
PAGE and autoradiography. Equivalent receptor expression was confirmed
by visualizing aliquots of total cell lysates (bottom panel).
Figure 9 shows receptor binding to SARA in COS cells transiently
transfected with wild type T(3RI1 and kinase-deficient T(3RI and various
combinations of wild type Flag-hSARA (WT), a mutant version lacking the
Smad2 binding domain (OSBD) and Myc-Smad2. The amount of receptor
bound to SARA was determined by anti-Flag immunoprecipitation followed
by gamma counting. Data is plotted as the average of three experiments ~
S.D. Protein expression was analyzed by immunoblotting aliquots of total cell
lysates and the results from a representative experiment are shown (bottom
panel).
CA 02237701 1998-07-20
14
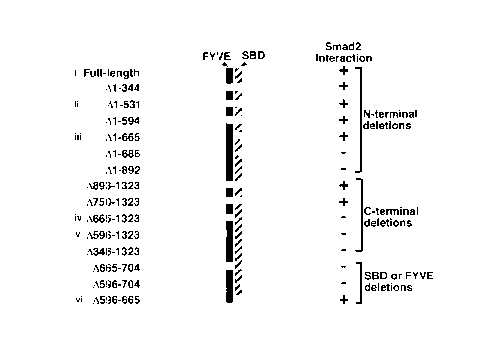
Figure 10 is a schematic representation of mutant versions of SARA.
The FYVE domain (black bar) and the Smad binding domain, SBD (striped
bar), are indicated. COS cells transiently transfected with Flag-hSARA and
Myc-Smad2 were immunoprecipitated with anti-Flag antibodies followed by
immunoblotting with anti-Myc antibodies. The presence (+) or absence (-) of
a SARA/Smad2 interaction is indicated (Smad2 interaction). Mutants used for
the subsequent localization study are marked on the left (i-vi).
Figure 11 shows an immunoblot of lysates from COS cells expressing
Flag-tagged Smad2 or Smad3 incubated with GST alone or with GST-hSARA
(665-750), which corresponds to the SBD; bound proteins were
immunoblotted using anti-Flag antibodies. The presence of Smad2 and
Smad3 bound to GST-hSARA (665-750) is indicated.
Figure 12 shows the subcellular localization of SARA mutants. Mv1 Lu
cells were transiently transfected with wild type (panel i) or mutant versions
of
Flag-hSARA (panels ii-vi, as marked on the left in Figure 10). Proteins were
visualized by immunofluorescence and confocal microscopy using a
monoclonal anti-Flag M2 monoclonal antibody followed by FITC-conjugated
goat anti-mouse IgG.
Figure 13, panels A and B, shows photomicrographs of Mv1 Lu cells
transiently transfected with Myc-hSARA (01-665) and Flag-Smad2 (panel A) or
with wild type Myc-hSARA, HA-Smad2 and hSARA (01-665) (panel B),
protein subcellular localization being visualized by immunofluorescence and
confocal microscopy. Smad2 was detected with monoclonal antibodies
followed by FITC-conjugated goat anti-mouse IgG (green) and hSARA was
visualized with the polyclonal Myc A14 antibody and Texas-Red conjugated
goat anti-rabbit IgG (red).
Figure 14 shows receptor binding to SARA in COS cells transiently
transfected with various combinations of wild type (WT) or mutant (D1-665 or
OFYVE, 595-665) Flag-hSARA together with wild type T(3RI1 and kinase-
deficient TaRI. In the case of SARA (01-665), lower DNA amounts were used
CA 02237701 1998-07-20
to obtain equivalent levels of protein expression. Cells were affinity-
labelled
with ['251]TGF~3 and lysates immunoprecipitated with anti-Flag antibodies. The
amount of receptor bound to SARA was determined by gamma counting.
Equivalent protein expression was confirmed by immunoblotting aliquots of
5 total cell lysates (lower panel).
Figure 15 shows luciferase activity of Mv1 Lu cells transfected with 3TP-
lux alone or together with the indicated amounts of wild type (WT) or mutant
(01-665 or 01-686) versions of hSARA and incubated in the presence (black
bars) or absence (open bars) of TGF~i. Luciferase activity was normalized to
~i-
10 galactosidase activity and is plotted as the mean ~S.D. of triplicates from
a
representative experiment.
Detailed Descriation of the Invention
This invention provides a family of proteins that play key roles in TGF-
15 (3, activin and bone morphogenetic protein (BMP) signal transduction
pathways. In particular, the proteins of this family interact with specific
Smad
proteins to modulate signal transduction. These proteins, termed "Smad
Anchor for Receptor Activation" (or "SARA") proteins are characterized by the
presence of two neighboring domains: (1) a double zinc finger or FYVE
domain responsible for the subcellular localization of the SARA protein or
SARA-Smad complex, and (2) a Smad binding domain ("SBD") which
mediates the interaction or binding of one or more species of Smad protein
with the particular member of the SARA family.
Cloned DNA coding sequences and corresponding amino acid
sequences for representative human and Xenopus SARA protein family
members are shown in Tables 1 and 2 (human SARA), Tables 3 and 4 (human
SARA2), Tables 5 and 6 (Xenopus SARA1) and Tables 7 and 8 (XSARA2).
Table 9 shows a comparison of the amino acid sequences of Xenopus
SARA (XSARA1) and human SARA (hSARA). Identical residues (dark grey)
and conservative changes (light grey), the FYVE domain (solid underline) and
CA 02237701 1998-07-20
16
the Smad binding domain (dashed underline) are indicated. The sequences in
XSARA1 used to design degenerate PCR primers for identifying hSARA are
shown (arrows). The amino-terminal end of the partial Xenopus cDNA
obtained in the expression screen is marked (asterisk).
SARA proteins contain a double zinc finger or FYVE domain. The
human SARA of Tables 1 and 2, identified as described in Example 2,
regulates the subcellular localization of Smad2 and Smad3 and recruits these
Smads into distinct subcellular domains. This SARA also interacts with TGF(3
receptors and TGF~i signalling induces dissociation of Smad2 or Smad3 from
the SARA protein with concomitant formation of Smad2/Smad4 complexes
and nuclear translocation. Deletion of the FYVE domain in SARA causes
mislocalization of Smad2 or Smad3, interferes with TG F(3 receptor interaction
and inhibits TGF~i-dependent transcriptional responses. Thus, this SARA
defines a component of TGF~3 signalling that fulfills an essential role in
anchoring Smad2 or Smad3 to specific subcellular domains for activation by
the receptor.
Table 10 shows alignment of the amino acid sequences of the FYVE
domains from hSARA, XSARA1, KIAA0305, FGD1, Hrs-1, Hrs-2 and EEA1.
Identical residues (dark grey) and conservative changes (light grey) are
marked. A consensus sequence (bottom) was derived from positions in which
at least 6 out of 7 residues were conserved or when proteins contained one of
only two alternate residues.
The regulation of the subcellular localization of components of
signalling pathways can be key determinants in the effective initiation and
maintenance of signalling cascades. Targeting of signal transduction proteins
to specific subcellular regions is highly regulated, often through specific
interactions with scaffolding or anchoring proteins (Faux and Scott, 1996;
Pawson and Scott, 1997). Scaffolding proteins have been defined as proteins
that bind to multiple kinases to coordinate the assembly of a cascade, while
anchoring proteins are tethered to specific subcellular regions in the cell
and
CA 02237701 1998-07-20
17
can act to bring together components of a pathway. Regulating location of
signalling components can thus coordinate the activity of a signalling
network, maintain signalling specificity or facilitate activation of a pathway
by
localizing kinases together with their downstream substrates.
As described herein, a recombinantly produced human SARA protein
bound directly and specifically to unphosphorylated Smad2 and Smad3. In
addition, receptor-dependent phosphorylation induced Smad2 to dissociate
from SARA, bind to Smad4 and translocate to the nucleus. Thus, the SARA
protein functions upstream of Smad activation to recruit Smad2 to specific
subcellular regions and also associates with the TGF~ receptor complex.
Furthermore, inducing mislocalization of Smad2 by expressing a mutant of the
SARA protein blocks TGF~-dependent transcriptional responses, indicating an
essential role for SARA-mediated localization of Smads in signalling.
Together, these results define the cloned SARA protein as a novel component
of the TG F(3 pathway that functions to anchor Smad2 to specific subcellular
sites for activation by the TGF~3 receptor kinase.
In vitro, receptor-regulated Smads are recognized by the receptor
kinases and are phosphorylated on the C-terminal SSXS motif (Abdollah et al.,
1997; Kretzschmar et al., 1997; Macias-Silva et a1.,1 996; Souchelnytskyi et
a1.,1 997). This phosphorylation is similar to receptor-dependent
phosphorylation in mammalian cells, suggesting that SARA is not absolutely
required for recognition of Smads by the receptor complex. In intact cells,
however, receptor-regulated Smads are cytosolic proteins that require
activation by transmembrane serine/threonine kinase receptors.
Consequently, access of Smads to the receptor may be poor and the specific
ocalization of SARA/Smad2 complexes may function to enhance interaction
with the TGF(3 receptor. TGF~3 receptors may have regionalized localization
and SARA may recruit Smad2 to these sites. This activity is likely to be most
critical in vivo, where ser/thr kinase receptors are often found in low
numbers
and only a small proportion need to be activated for biological responses
CA 02237701 1998-07-20
18
(Dyson and Gurdon, 1998). This may impose on the pathway a stringent
requirement for SARA to anchor Smads in these sites for receptor interaction.
The association of SARA with the TGF(3 receptor supports the notion
that it may facilitate interactions between Smad2 and the receptor kinase.
Furthermore, deletion of the FYVE domain interferes with receptor binding,
prevents the correct localization of SARA/Smad2 and blocks TGF~3 signalling
(see Example 8 below). This suggests that the localization of SARA and its
interaction with the receptor are interlinked, in agreement with the idea that
SARA acts to colocalize Smad2 with the receptor kinase. Interestingly, the
binding of the SARA protein identified in Example 2 to the receptor was
enhanced upon Smad2 expression and, on its own, SARA may interact
inefficiently with the receptor. Previously, Macias-Silva et al. (1996)
obtained
data suggesting that Smad2 interacts with the catalytic pocket of the Type I
receptor kinase domain. Therefore, it appears that wild SARA, which is not a
substrate of the kinase, may interact with regions outside of the kinase
domain, and may cooperate with Smad2 to drive association with the
receptor.
The human SARA protein identified in Example 2 did not interact with
any of the other Smads tested, indicating that it functions specifically in
Smad2 and Smad3 pathways (see Example 3). However, Smad5 localization
in 293 cells displayed a remarkably similar pattern to that of this SARA
protein
(Nishimura et al., 1998) and similar patterns were observed for endogenous
Smad1 or 5 in the kidney epithelial cell line, IMCD-3. Thus, localization of
BMP-regulated Smads (for example, Smad1, Smad5 and SmadB) may also be
regulated by a specific SARA family member.
The genes for two other SARA family member proteins were also
identified and cloned. One of these, identified in Xenopus and designated
XSARA2 (Tables 7 and 8), is related to XSARA1, while the other one, hSARA2
(Tables 3 and 4), is a human clone, related to the human SARA of Tables 1
and 2. This second human clone has been identified in EST clone KIAA0305.
CA 02237701 1998-07-20
19
A comparison of the SBD from hSARA with a similar region from the
KIAA0305 sequence indicated that the amino terminal half of the region of
the SBD was highly divergent from the amino acid sequence encoded by
KIAA0305. This suggests that the protein encoded by KIAA0305 may
mediate binding with other as yet unidentified proteins, eg. other Smads. In
contrast to the SBD, the FYVE domain of the KIAA0305 protein is more
closely related to the SARA FYVE domain (70% identity), suggesting that this
protein may be an anchor for other Smad proteins that function either in the
TGF~3 pathway or in other signalling cascades, such as the BMP signal
transduction pathway.
SAKA is not limiting in Smad activation and TGF~i signalling
It was observed that elevating Smad2 levels can saturate SARA and
yield a diffuse distribution for Smad2. Thus, the level of the SARA protein is
a
key determinant in controlling Smad2 localization. As a consequence,
endogenous Smad2 may or may not display a SARA-like distribution,
depending on the relative expression of the two proteins. Indeed, in M1 Lu
cells endogenous Smad2 displays a punctate pattern with some diffuse
staining in the cytosol (Nakao et al., 1997b). While not meaning to limit the
invention to a particular mechanism, the data are consistent with the view
that
once signalling has commenced, Smad2 will dissociate from SARA, bind to
Smad4 and translocate to the nucleus, freeing SARA to recruit additional
Smad2 from the cytosolic reservoir. This would provide a mechanism to
allow quantitative activation of Smads in the presence of high levels of TGF~3
signall ing. Furthermore, the transient nature of the SARA/Smad2 complex
insures that the SARA protein is unlikely to be a limiting component in the
activation of Smads. The observation that increasing the expression of wild
type SARA does not enhance activation of Smad2 or TGF(3-dependent
transcriptional responses is consistent with this model.
Modular Domains in SARA
CA 02237701 1998-07-20
The function of SARA in TGF~3 signalling is mediated by two
independent domains, the FYVE domain that targets SARA to specific
subcellular sites and the Smad binding domain (SBD) that mediates specific
interaction with specific Smads. In the case of the SARA protein identified in
5 Example 2, the SBD mediates interaction specifically with Smad2 and Smad3.
The Xenopus and mouse forkhead-containing DNA binding proteins, FAST1
and FAST2, also bind specifically to Smad2 and Smad3 and I ike SARA,
interact with the MH2 domains (Chen et al., 1996; Chen et al., 1997a; Labbe
et al., 1998; Liu et al., 1997a). Comparison of the SBD from this SARA with
10 the Smad Interaction Domain (SID) from these FAST proteins revealed no
regions of obvious similarity. However, since SARA acts upstream and FAST
downstream of Smad activation, these proteins may employ structurally
unrelated domains to distinguish unactivated versus activated forms of Smad2.
Thus, the SBD of this SARA protein preferentially binds unphosphorylated
15 monomeric Smad2 while the SID from FAST must bind phosphorylated
Smad2 in heteromeric complexes with Smad4. By analogy, the SBD of other
SARA family members may bind the unphosphorylated monomeric species of
other Smads that mediate signal transduction in other pathways (eg. Smads 1,
5 or 8 in the BMP signal transduction pathway).
20 In SARA, the FYVE domain functions independently of the SBD, to
mediate the subcellular targeting of the protein. FYVE domains have been
identified in other molecules that are regulated by diverse signalling
pathways, including FGD1, Hrs-1 and 2, and EEA1 (Pasteris et al., 1994;
Komada, et al., 1995; Bean, et al., 1997). It is unclear whether the FYVE
domain generally controls the localization of these other proteins. In EEA1,
however, the domain is required for the correct subcellular distribution of
the
protein (Mu et al., 1995).
Additional Roles for SARA
Controlling the localization of kinases and their substrates may allow
not only for efficient recognition and phosphorylation but may also function
CA 02237701 1998-07-20
21
to maintain specificity and suppress crosstalk between signalling pathways.
Thus, by controlling Smad localization, a SARA family member protein could
additionally function to maintain the highly specific regulation of Smad
phosphorylation by ser/thr kinase receptors that is observed in vivo and could
prevent promiscuous phosphorylation by other kinases in the cell.
Furthermore, through its interactions with a particular receptor, a SARA
protein might function to control the activity or turnover of the receptor
complex. Alternatively, SARA may also fulfill scaffolding functions to
coordinate the receptor-dependent activation of Smads with other as yet
unidentified components of a signalling pathway.
Proteins
SARA proteins may be produced by culturing a host cell transformed
with a DNA sequence encoding a selected SARA protein. The DNA
sequence is operatively linked to an expression control sequence in a
recombinant vector so that the protein may be expressed.
Host cells which may be transfected with the vectors of the invention
may be selected from the group consisting of E. coli, Pseudomonas, Bacillus
subtillus, or other bacilli, yeasts, fungi, insect cells or mammalian cells
including human cells.
For transformation of a mammalian cell for expression of a SARA
protein, the vector may be delivered to the cells by a suitable vehicle. Such
vehicles including vaccinia virus, adenovirus, retrovirus, Herpes simplex
virus
and other vector systems known to those of skill in the art.
A SARA protein may also be recombinantly expressed as a fusion
protein. For example, the SARA cDNA sequence is inserted into a vector
which contains a nucleotide sequence encoding another peptide (e.g. GST-
glutathione succinyl transferase). The fusion protein is expressed and
recovered from prokaryotic (e.g. bacterial or baculovirus) or eukaryotic
cells.
The fusion protein can then be purified by affinity chromatography based
CA 02237701 1998-07-20
22
upon the fusion vector sequence and the SARA protein obtained by
enzymatic cleavage of the fusion protein.
The protein may also be produced by conventional chemical synthetic
methods, as understood by those skilled in the art.
SARA proteins may also be isolated from cells or tissues, including
mammalian cells or tissues, in which the protein is normally expressed.
The protein may be purified by conventional purification methods
known to those in the art, such as chromatography methods, high
performance I iquid chromatography methods or precipitation.
For example, anti-SARA antibodies may be used to isolate SARA
protein which is then purified by standard methods.
Antibodies
The provision of the polynucleotide and amino acid sequences of
SARA proteins provides for the production of antibodies which bind
selectively to a SARA protein or to fragments thereof. The term "antibodies"
includes polyclonal antibodies, monoclonal antibodies, single chain
antibodies and fragments thereof such as Fab fragments.
In order to prepare polyclonal antibodies, fusion proteins containing
defined portions or all of a SARA protein can be synthesized in bacteria by
expression of the corresponding DNA sequences, as described above. Fusion
proteins are commonly used as a source of antigen for producing antibodies.
Alternatively, the protein may be isolated and purified from the recombinant
expression culture and used as source of antigen. Either the entire protein or
fragments thereof can be used as a source of antigen to produce antibodies.
The purified protein is mixed with Freund's adjuvant and injected into
rabbits or other appropriate laboratory animals. Following booster injections
at weekly intervals, the animals are then bled and the serum isolated. The
serum may be used directly or purified by various methods including affinity
chromatography to give polyclonal antibodies.
CA 02237701 1998-07-20
23
Alternatively, synthetic peptides can be made corresponding to
antigenic portions of a SARA protein and these may be used to inoculate the
animals.
In a further embodiment, monoclonal anti-SARA antibodies may be
produced by methods well known in the art. Briefly, the purified protein or
fragment thereof is injected in Freund's adjuvant into mice over a suitable
period of time, spleen cells are harvested and these are fused with a
permanently growing myeloma partner and the resultant hybridomas are
screened to identify cells producing the desired antibody. Suitable methods
for antibody preparation may be found in standard texts such as Antibody
Engineering, 2d. edition, Barreback, ED., Oxford University Press, (1995).
EXAMPLES
The examples are described for the purposes of illustration and are not
intended to limit the scope of the invention.
Methods of molecular genetics, protein and peptide biochemistry and
immunology referred to but not explicitly described in this disclosure and
examples are reported in the scientific literature and are well known to those
skilled in the art.
Example 1: Methods
Isolation of Xenopus and human SARA
To prepare a probe for library screening, the MH2 domain of Smad2
(amino acids 241-467) was subcloned into a modified pGEX4T-1 vector
containing the protein kinase A recognition site derived from pGEX2TK
(Pharmacia). This bacterial fusion protein was purified, labelled with
[3zP]yATP and used as probe to screen a .ZAP II Xenopus dorsal lip library as
described (Chen and Sudol, 1995). A screen of 1 x 106 plaques yielded four
phage which represented repeated isolates of the same clone. This partial
cDNA contained a 2.1 kb open reading frame and 1 kb of 3' untranslated
region (UTR). A full length clone was obtained by a combination of
rescreening of the same dorsal lip library using a 670 base pair EcoRl/Hpal
CA 02237701 1998-07-20
24
fragment at the 5' end of this clone and by 5' RACE (Gibco/BRL) using stage
Xenopus RNA.
To obtain a human homolog of Xenopus SARA, cDNA was synthesized
from randomly primed total RNA isolated from HepG2 cells. This cDNA was
5 subjected to polymerase chain reaction (PCR) using degenerate primers as
described previously (Attisano et al., 1992). The 5' and 3' primers, designed
to encode the zinc-finger motif, correspond to
GC(A/C/G/TnCC(A/C/G/T)AA(C!T)TG(C/TATGAA(A/C/G/T)TG(C/T) and
(A/G)CA(A/G )TA(C/T)TC(A/C/G/T)G C(A/C/G/T)G G (A/G)TT(A/G )TT,
10 respectively. A 150 base pair PCR product was sequenced and then used as
probe for screening a ,ZAP human fetal brain cDNA library (Stratagene).
Eight positive plaques were obtained, two of which contained an overlap of
approximately 1 kb and covered the entire open reading frame. The sequence
of the 5' UTR was confirmed by sequencing of an expressed sequence tag
database clone (clone ID 260739).
Construction of Plasmids
For mammalian expression constructs of SARA, the open reading frame
of hSARA was amplified by PCR and was subcloned into pCMV5 in frame
with an amino-terminal Flag or Myc tag (Hoodless et al., 1996). The deletion
mutants of pCMVS-Flag-hSara0893-1323, 0346-132, 0893-1323, and 4346-
1323 were constructed by deletion of EcoRV-Hindlll, Xbal-Hindlll, Sall-
EcoRV, and Sall-Xbal fragments, respectively. PCMV5-Flag-hSara01-594 and
01-686 were obtained by partial digestion with Asp718/Sall and for pCMVS-
Flag-hSARA 0665-1323 a Asp718/Hindlll partial digest was used. PCMV5-
Flag-hSARA0596-704 was constructed by deleting Asp718 fragment. The
other h SARA mutants were constructed by PCR using appropriate primers.
PCMV5B-Myc-Smad3 and Myc-Smad6, pGEX4T-1-Smad2/MH1 (amino acids
1-181), pGEX4T-1-Smad2/linker (amino acids 186-273), pGEX4T-1-
Smad2/MH2 (amino acids 241-467) and pGEX4T-1-h SARA (amino acids 665-
750) were constructed by PCR.
CA 02237701 1998-07-20
In Vitro Protein Interactions
In vitro transcription/translation reactions were performed using the
TNT coupled reticulocyte lysate system (Promega) following the
manufacturer's instructions using T3 RNA polymerase. Translation was
5 carried out in the presence of [35S]-methionine and labelled proteins were
incubated with purified GST fusion proteins in TNTE buffer with 10% glycerol
for 2 hours at 4°C and then washed five times with the same buffer.
Bound
protein was separated by SDS-PAGE and visualized by autoradiography.
Immunoprecipitation and Immunoblotting
10 COS-1 cells transfected with LipofectAMINE (GIBCO BRL) were lysed
with lysis buffer (Wrana et al., 1994) and subjected to immunoprecipitation
with either anti-Flag M2 (IBI, Eastern Kodak) or anti-Myc (9E10) monoclonal
antibody followed by adsorption to protein-G sepharose. Precipitates were
separated by SDS-PAGE, transferred to nitrocellulose membranes and
15 immunoblotted as described previously (Hoodless et al., 1996).
Affinity-Labell ing
LipofectAMINE transfected COS-1 cells were incubated with 200 pM
['251]TG F(3 in media containing 0.2% bovine fetal serum at 37°C for 30
minutes and receptors were cross-linked to ligand with DSS as described
20 previously (Macias-Silva et al., 1996). Cell lysates were
immunoprecipitated
with anti-Flag antibody and receptors visualized by SDS-PAGE and
autoradiography. In some cases, cross-linked ['251]TGF[i was determined by
gamma counting.
Subcellular Localization by Immunofluorescent Confocal Microscopy
25 Mv1 Lu cells, plated on gelatin-coated Permanox chamber slides
(Nunc), were transfected by the calcium phosphate-DNA precipitation
method. Fixation, permeabilisation and reaction with the primary and
secondary antibodies were described previously (Hoodless et al., 1996).
Monoclonal anti-Flag antibodies were visualized by FITC-conjugated goat
anti-mouse IgG (Jackson Laboratories) and polyclonal Myc antibody (A14,
CA 02237701 1998-07-20
26
Santa Cruz) was visualized with Texas-Red-conjugated goat anti-rabbit IgG
(Jackson Laboratories). Immunofluorescence was analyzed on a Leica
confocal microscope.
Transcriptional Response Assay
Mv1 Lu cells were transiently transfected with the reporter plasmid,
p3TP-lux (Wrana et al., 1992), CMV-agal and selected constructs using
calcium phosphate transfection. Twenty-four hours after transfection, cells
were incubated overnight with or without 50 pM TGF[3. Luciferase activity
was measured using the luciferase assay system (Promega) in a Berthold
Lumat LB 9501 luminometer and was normalized to (3-galactosidase activity.
Example 2 - Identification of SARA family members
The MH2 domain of Smad2 was fused to glutathione-S-transferase
(GST) that included a kinase recognition site for protein kinase A (PKA). The
bacterially-expressed fusion protein was labelled to high specific activity
using
PKA (Chen and Sudol, 1995), and then used to screen a ~,ZAPII expression
library prepared from the dorsal blastopore lip of Xenopus. From this screen,
four clones were identified, all of which presented a repeated isolate of a
partial cDNA clone with no similarity to sequences in the GenBank database.
To confirm that the product encoded by this clone interacted with Smad2, an
in vitro transcription/translation system was used to produce [35S]methionine-
labelled protein. Translation of the cDNA yielded a protein product of
approximately 80 kDa which corresponded in size to the longest open
reading frame (ORF) identified in the sequence. Incubation of this product
with bacterially-produced GST-Smad2(MH2) resulted in efficient binding of
the translated product to the fusion protein (data not shown). Interaction
with
full length Smad2 was also observed, whereas binding to bacterially-
expressed Smad1 or Smad4 was not.
To isolate a full length cDNA, the partial clone identified in the
interaction screen was used as a probe to rescreen the same blastopore lip
I ibrary. Since the resulting clones lacked the 5' end, 5' RACE was conducted
CA 02237701 1998-07-20
27
to obtain the entire coding sequence. Analysis of the complete cDNA
sequence (Table 5) revealed a long open reading frame that was contiguous
with that of the partial clone. The predicted protein, XSARA1, is 1235 amino
acids long with an estimated molecular mass of 135 kDa (Table 6). Analysis
of the full length cDNA sequence (Table 9) revealed a region in the middle
portion of the predicted protein that had similarity to a double zinc finger
domain (recently renamed the FYVE domain; Mu et al., 1995). The FYVE
domain has been identified in a number of unrelated signalling molecules that
include FGD1, a putative guanine exchange factor for Rho/Rac that is mutated
in faciogenital dysplasia (Pasteris et al., 1994), the HGF receptor substrate
Hrs-
1 and its homolog Hrs-2 (Bean et al., 1997; Komada and Kitamura, 1995),
EEA1, a protein involved in formation of the early endosome (Mu et al., 1995)
and the yeast proteins FAB1, VPS27 and VAC1 (Piper et al., 1995; Weisman
and Wickner, 1992;Yamamoto et al., 1995). Comparison of the FYVE
domains from the vertebrate proteins with that from SARA revealed extensive
conservation of residues throughout the domain (Table 10). Thus, SARA
contains a FYVE domain that may fulfill important functions in diverse
proteins.
To investigate the role of SARA in TGF~i signalling in mammalian cells,
a human homologue was identified. Using a carboxy-terminal portion of
XSARA1, a human library was screened and a protein was identified that was
distantly related to Xenopus SARA (34% identity) and which was also
sequenced as an EST (KIAA0305). However, no homologs closer to XSARA
were identified. Thus, degenerate oligonucleotide primers were designed
encoding amino acids in X SARA (Table 9) and HepG2 RNA was used as
template for degenerate PCR. A related sequence was identified and this
partial cDNA was used to screen a human brain cDNA library. Four
overlapping clones, encoding a long open reading frame were identified and
a search of the EST database with this sequence led to the identification of
additional overlapping cDNA clones from libraries derived from T cells,
CA 02237701 1998-07-20
28
uterus, endothelial cells and melanocytes. Analysis of the contiguous
sequence revealed a long open reading frame that had a consensus start
codon preceded by stop codons in all three reading frames (Table 1).
Comparison of the predicted protein (Table 2), from this cDNA, hSARA, with
XSARA1 (Table 9) revealed an overall identity of 62%, with a divergent 558
residue amino terminal domain (35% identity) followed by a closely related
carboxy terminus (85% identity).
Example 3 hSARA interacts specifically with Smad2 and Smad3
To characterize the interaction of hSARA with Smads, the full length
protein was translated in vitro and tested for binding to bacterially-
expressed
Smad fusion proteins. Similar to the Xenopus clone, hSARA bound
specifically to full length Smad2, but not Smad1 or Smad4 (Figure 1 ). In
addition, full length Smad3, which is highly related to Smad2, also interacted
with hSARA. To define the domains of Smad2 that bound hSARA, in bacteria
various fragments of Smad2 corresponding to the MH1 domain, linker region
and MH2 domain were expressed in bacteria. Similar to the Xenopus clone,
hSARA interacted efficiently with fusion proteins that comprised the MH2
domain, while no association was detected between hSARA and either the
MH1 or non-conserved linker domains (Figure 1). Thus, hSARA specifically
interacts with Smad2 through the MH2 domain.
To confirm that hSARA also bound to Smads in mammalian cells, a
Flag epitope tag was introduced at the amino terminus of the protein to create
Flag- SARA. Transient expression of Flag- SARA in COS cells yielded a
protein of the predicted molecular weight for SARA (Figure 2) that was not
present in untransfected cells (data not shown). To investigate the
interaction
of SARA with Smads, Flag- SARA was expressed in COS cells together with
Myc-tagged versions of Smads 1, 2, 3, 4, 6 and 7. Cell lysates were subjected
to anti-Flag immunoprecipitation followed by immunoblotting with anti-Myc
antibodies. In other immunoprecipitates of cells expressing either Smad2 or
Smad3, efficient coprecipitation of either Smad with Flag- SARA was observed
CA 02237701 1998-07-20
29
(Figure 2). In contrast, none of the other Smads coprecipitated with SARA.
Specific binding of this SARA family member to both Smad2 and Smad3 is
consistent with the observation that these two proteins possess very closely
related MH2 domains (97% identity) and are both activated by TG F(3 or
activin type I receptors (Liu et a., 1997b; Macias-Silva et al., 1996; Nakao
et
al., 1997a). Together, these results demonstrate that this SARA family
member is a specific partner for receptor-regulated Smads of the TGF~/activin
signalling pathway.
Example 4- Pho~ho~rlation of Smad2 induces dissociation from SARA
Previous findings have shown that activation of TG F[3 signalling results
in phosphorylation of Smad2 or Smad3 by type I receptors on C-terminal
serine residues (Liu et al., 1997b; Macias-Silva et al., 1996). A
constitutively
active TG F(3 type I receptor was prepared by substituting a threonine in the
GS domain with an aspartate residue (Wieser et al., 1995). This activated type
I receptor induces TG F(3 signalling in the absence of type II receptors and
ligand and regulates the phosphorylation and activation of Smad proteins in a
manner similar to ligand (Macias-Silva et al., 1996; Wieser et al., 1995). COS
cells were transfected with combinations of Smad2, hSARA or both in the
presence or absence of activated T[3RI. Cells were then metabolically labelled
with [32P]phosphate and phosphorylation of either SARA or Smad2 was
assessed in immunoprecipitates. Analysis of SARA phosphorylation revealed
that the protein was basally phosphorylated and the coexpression of the
activated type I receptor did not appreciably affect the overall
phosphorylation (Figure 3). In contrast, analysis of Smad2
immunoprecipitated from total cell lysates showed that the activated type I
receptor induced strong phosphorylation of the protein as described
previously (Macias-Silva et al., 1996). These results suggest that SARA is not
phosphorylated in response to TGF~ signalling.
The phosphorylation state of Smad2 that coprecipitated with SARA was
examined. Interestingly, unlike the strong induction of Smad2
CA 02237701 1998-07-20
phosphorylation in the total cellular pool, phosphorylation of Smad2
associated with SARA was not enhanced, but rather appeared to decrease in
the presence of TG F(3 signalling (Figure 3). This suggested that receptor-
dependent phosphorylation of Smad2 might induce dissociation from SARA.
5 To examine this directly, the interaction of SARA with wild type Smad 2 or a
mutant version lacking the C-terminal phosphorylation sites (Smad2(2SA)) was
analysed. In the absence of TGFa signalling, association of SARA with either
Smad2 or Smad2(2SA) was comparable (Figure 4). In contrast, in cells
coexpressing the activated receptor, a significant decrease in the interaction
of
10 wild type Smad2 with SARA was observed. However, SARA/Smad2(2SA)
complexes were not reduced by the activated receptor. Together, these
results suggest that SARA is not phosphorylated in response to TGF~
signall ing and that it preferentially interacts with the unphosphorylated
form
of Smad2.
15 Example 5 SARA and Smad4 -form mutually exclusive complexes with
Smad2
Phosphorylation of Smad2 induces its interaction with Smad4 (Lagna et
al., 1996; Zhang et al., 1997). SARA/Smad2 complexes in COS cells
coexpressing Smad4 were assessed. In unstimulated cells, the level of
20 SARA/Smad2 complex formation was comparable either in the presence or
absence of Smad4 (Figure 5, lanes 3 and 6). However, upon activation of
TGF~ signalling, dissociation of Smad2 from SARA was significantly enhanced
by coexpression of Smad4 (Figure 5, lanes 4 and 7). These results suggested
that Smad4 and SARA might compete to form complexes with Smad2. The
25 formation of Smad2/Smad4 and Smad2/ SARA complexes in the same
transfectants was then examined. Cell lysates were subjected to
immunoprecipitation with anti-Flag antibodies directed towards tagged Smad2
and then immunoblotted for the presence of Smad4 and SARA. Consistent
with previous findings (Lagna et al., 1996; Zhang et al., 1997), interaction
of
30 Smad4 with Smad2 was strongly stimulated by the activated type I receptor
CA 02237701 1998-07-20
31
(Figure 6, lane 3 and 4). Concomitant with the formation of Smad2/Smad4
complexes, the interaction of Smad2 with SARA was disrupted by activation
of Signalling (Figure 6, lanes 6 and 7). Thus, complexes of Smad2/ SARA and
Smad2/Smad4 are mutually exclusive, supporting the notion that Smad4 may
compete for Smad2 to enhance dissociation of SARA/Smad2 complexes.
Together these results demonstrate that during TGF~i signalling, SARA/Smad2
complexes are transient and phosphorylation of Smad2 induces dissociation
and formation of heteromeric complexes with Smad4.
Example 6- SARA r~~ulates the subcellular localization of Smad2
The studies described above suggest that SARA functions upstream in
the pathway and might control the subcellular localization of Smad2. To test
this, an investigation was done to determine whether coexpression of SARA
might alter the localization of Smad2 in the TGFa-responsive epithelial cell
I ine, Mv1 Lu, using confocal microscopy. Mv1 Lu cells were used rather than
COS since the Myc antibodies crossreacted with endogenous proteins in the
COTS and obscured nuclear staining of tagged proteins. In cells expressing
SARA alone, the protein displayed a punctate staining pattern that was present
throughout the cytosolic compartment and was excluded from the nucleus
(Figure 7A). This localization of SARA was in contrast to the diffuse staining
typically observed for Smad2 in cells overexpressing the protein (Figure 7B).
Cells transiently transfected with both SARA and Smad2 were examined. In
these cells, the distribution of SARA was indistinguishable from cells
transfected with SARA alone (Figure 7D, left panel). In contrast, the
localization of Smad2 in the presence of SARA displayed a dramatic shift to a
punctate pattern (compare Figure 7B to 7D, centre panel). Moreover, analysis
of these immunofluorescent staining patterns by confocal microscopy
revealed that SARA and Smad2 precisely colocalized in the cytosol (yellow
stain, Figure 7D, right panel). Interestingly, expression of Smad2 at much
higher levels than SARA reverted the distribution of Smad2 to that observed in
cells transfected with Smad2 alone (data not shown). This supports the notion
CA 02237701 1998-07-20
32
that elevating the amount of Smad2 can saturate SARA and yield a diffuse
distribution of Smad2 throughout the cell.
Studies were conducted to determine whether activation of TGF~3
signalling induces nuclear translocation of Smad2 in the presence of SARA.
As shown in Figure 7, the localization of SARA in the cytosolic compartment
looked similar in the presence or absence of the constitutively active TGFa
type I receptor (compare Figure 7D and E, left panels). However, TGF~i
signalling caused a significant proportion of Smad2 to translocate to the
nucleus (Figure 7E, centre panel) and this correlated with a shift to an
orangy-
red colour in the cytosolic colocalization stain (Figure 7E, right panel).
Thus
activation of TGF~i signalling induces Smad2 to dissociate from SARA and
translocate to the nucleus. Taken together with our biochemical analysis,
these results indicate that SARA functions to anchor or recruit Smad2 to
specific subcellular regions prior to activation by TGF~3 signalling.
Example 7 SARA associated with the TGH3 receptor
The positioning of SARA upstream of Smad2 activation suggested that
SARA may also physically interact with the receptor. To examine this, COS
cells were cotransfected with TGF[3 receptors in the presence of SARA and
were affinity labelled using ['251]TGF~i. SARA was then immunoprecipitated
from the cell lysates and coprecipitating receptor complexes were resolved by
SDS-PAGE and visualized by autoradiography or were quantitated using a
gamma counter. Analysis of cells expressing wild type receptors type II and
type I, revealed that receptor complexes coprecipitated with SARA (Figure 8,
lane 3). Furthermore, in the presence of kinase deficient type I receptor
there
was a small increase in binding of SARA to the receptor (Figure 8, lane 2).
This is in contrast to Smad2, which interacts only with TGF[i receptor
complexes that contain kinase deficient type I receptors (Macias-Silva et al.,
1996). These data suggest that SARA associates with the TGF~ receptor.
Studies were conducted to determine whether coexpression of Smad2
might enhance the interaction of SARA with TG F(3 receptors. In cells
CA 02237701 1998-07-20
33
expressing wild type receptor I, no difference was observed in the amount of
receptor complexes that coprecipitated with SARA either in the presence or
absence of Smad2 (Figure 8, compare lanes 3 and 5). In contrast, the
association of SARA with receptor complexes containing kinase-deficient type
I receptors was enhanced by Smad2 (Figure 8, lane 4). This finding was
consistent with the previous demonstration that kinase-deficient type I
receptors stabilize interactions of Smad2 with the receptors (Macias-Silva et
al., 1996). To investigate further the requirement for Smad2 in the
interaction
of SARA with the receptor, a mutant of SARA was tested, SARA(OSBD), that
removes a small domain that mediates its interaction with Smad2 (see below
for detailed characterization). Analysis of SARA interaction with receptor
complexes containing kinase-deficient TaRI showed that wild type SARA
interacted with the receptor and that this interaction was enhanced
approximately two-fold by Smad2 (Figure 9). The OSBD mutant of SARA
retained the capacity to associate with the receptor but wild type levels of
binding required higher expression levels of the mutant protein, suggesting
that the efficiency of interaction was reduced. Importantly, unlike wild type
SARA, binding of mutant SARA to the receptor was not enhanced by the
coexpression of Smad2. Together, these data suggest that SARA can interact
with the TGF~3 receptor independently of Smad2 binding and that Smad2
cooperates to enhance the association.
Example 8 - A modular domain in SARA mediates association with Smads
To investigate the functional importance of SARA in TGF~3 signalling,
the domains in the protein that mediate both its localization to specific
subcellular regions and its interaction with Smad2 were defined. To this end,
a series of deletion mutants of SARA were constructed and tested for their
ability to interact with Smad2 in COS cells by immunoprecipitation followed
by immunoblotting. As summarized in Figure 10, loss of the first 665 amino
acids of SARA, which included the double zinc finger/FYVE domain, did not
interfere with SARA binding to Smad2. However, further deletions (01-686)
CA 02237701 1998-07-20
34
completely abol fished the interaction of Smad2 with SARA. To map the
carboxy-terminal boundary of the Smad binding domain, a number of C-
terminal truncations were also analyzed. Deletion of all residues downstream
of position 750 did not affect Smad2 interaction with SARA, while an
additional loss of 85 amino acids (0665-1323) completely abrogated binding
to Smad2. To determine whether the region defined by this deletional
analysis was sufficient to bind Smad2, the 85 amino acids referred to as the
Smad Binding Domain (SBD) were linked to GST and the fusion protein was
expressed in bacteria (G ST-h SARA(665-750)). Incubation of lysates prepared
from cells expressing Smad2 or Smad3 with GST-SBD resulted in efficient
binding of both Smads to the fusion protein (Figure 11). These studies thus
define a novel domain in SARA that mediates interaction with Smad2 and
Smad3 and which is located downstream of the FYVE domain.
Exam le 9 - The FYVE domain controls the subcellular localization of SARA
The subcellular localization of a selection of the SARA mutants was
analysed by immunofluorescence and confocal microscopy. Analysis of
truncation mutants that removed the amino terminus upstream of the FYVE
domain (01-531) yielded wild type patterns of staining (Figure 12, compare
panels i and ii). However, a further deletion (01-665) that disrupted the FYVE
domain but did not interfere with the Smad binding domain, abolished the
wild type staining pattern (Figure 12, panel iii). Similar studies of the C-
terminal domains showed that residues downstream of the FYVE domain
(0665-1323) did not alter the localization of the mutant protein (Figure 12,
panel iv), while truncations within the FYVE domain (0596-1323) led to
diffuse localization throughout the cell (Figure 12, panel v). Of note, the
0665-1323 mutant lacked the Smad binding domain, thereby indicating that
interaction with Smad2 is not required for proper SARA localization. To
confirm that FYVE domain function was required for localization of SARA, a
mutant with a small internal deletion that removes the FYVE domain (0596-
665) was tested. Consistent with the other mutants, localization of this
CA 02237701 1998-07-20
protein was clearly disrupted (Figure 12, panel vi). Since none of these
mutants interfered with Smad binding, the F',~VE domain appears to be
required to maintain the normal localization of SARA but is not involved in
mediating interactions with Smads.
5 Examule 10 SARA mediated localization of Smad2 is necessary for TGH3
si>;nallin~
The FYVE domain and the SBD function independently to control the
subcellular distribution and Smad binding activity of SARA. Thus, the
availability of mutants of SARA that interact with Smad2 but fail to target to
10 the appropriate subcellular sites allowed the question of whether SARA-
mediated localization of Smad2 was important in TGF~3 signalling to be
addressed. Studies were done to determine whether mutants of SARA that
failed to distribute in the correct subcellular domains would mislocalize
Smad2. Since expression of the internal deletion mutant that removes the
15 FYVE domain 0596-665) was low, we analyzed the truncation mutant that
deletes the non-conserved amino-terminal region as well as the FYVE domain
(01-665) was analysed. Coexpression of the SARA mutant with Smad2
showed that it failed to recruit Smad2 to the normal SARA domains (Figure
13A). A direct examination was done to determine whether the mutant could
20 cause mislocalization of Smad2. For this, cells were cotransfected with
wild
type SARA and Smad2 either in the absence or presence of SARA(01-665). In
control transfectants performed in the absence of SARA(01-665), SARA and
Smad2 were colocalized in punctate domains as described above (data not
shown). However, in the presence of SARA(01-665), the localization of wild
25 type SARA was normal but the distribution of Smad2 was clearly disrupted
and displayed a diffuse pattern (Figure 13B). Thus, SARA(~1-665) causes
mislocalization of Smad2.
A study was done to determine whether this mutant interacted with the
TGF~i receptor. In contrast to wild type SARA, the interaction of SARA(41-
30 665) with the TGF(3 receptor was substantially reduced (Figure 14). Similar
CA 02237701 1998-07-20
36
findings were obtained with SARA lacking only the FYVE domain (0596-
665;Figure 14). Thus, disruption of SARA localization interferes with Smad
localization and the interaction of SARA with the TG F(3 receptor.
Since SARA(01-665) mislocalizes Smads and interferes with receptor
association, this mutant was tested for disruption of TG F(3 signalling. The
TG F(3-responsive reporter gene 3TP-lux was transiently transfected into
Mv1 Lu cells in the presence and absence of wild type or mutant versions of
SARA. Expression of wild type SARA had no effect on TGF~3 signalling (Figure
15). In contrast, transfection with SARA(~1-665) significantly inhibited TGF(3-
dependent signalling at the lowest concentration of DNA tested, while
transfection of higher doses completely abolished responsiveness of the cells.
SARA(01-686), which lacks a functional Smad binding domain, was also
tested. Transfection of this mutant had no effect on TGF~i signalling (Figure
15) despite efficient expression of the mutant protein (Data not shown). Thus,
the inhibitory activity of the SARA mutant is dependent on interaction with
Smad2. Since this mutant functions to disrupt the localization of Smad2,
these data suggest that anchoring of Smad2 by SARA is essential for the
initiation of TGF(3 signalling.
CA 02237701 1998-07-20
37
REFEREN CES
Abdollah, S., Macias-Silva, M., Tsukazaki, T., Hayashi, H., Attisano, L.,
and Wrana, J.L. (1997). Tf~RI phosphorylation of Smad2 on Ser 465 and 467
is required for Smad2/Smad4 complex formation and signalling. ). Biol. Chem.
272, 27678-27685.
Attisano, L. and Wrana, J.L. (1998). Mads and Smads in TGFf~
signalling. Curr. Op. Cell Biol. 10, 188-194.
Attisano, L., Wrana, J.L., Cheifetz, S., and Massague, J. (1992). Novel
activin receptors: Distinct genes and alternative mRNA splicing generate a
repertoire of serine/threonine kinase receptors. Cell 68, 97-108.
Bean, A.J., Siefert, R., Chen, Y.A., Sacks, R., and Scheller, R.H. (1997).
Hrs-2 is an ATPase implicated in calcium-regulated secretion. Nature 385,
826-829.
Chen, H.I. and Sudol, M. (1995). The WW domain of Yes-associated
protein binds a proline-rich ligand that differs from the consensus
established
for Src homology 3-binding modules. Proc Natl Acad Sci USA 82, 7819-7823.
Chen, X., Rubock, M.J., and Whitman, M. (1996). A transcriptional
partner for MAD proteins in TGF-f~ signalling. Nature 383, 691-696.
Chen, X., Weisberg, E., Fridmacher, V., Watanabe, M., Naco, G., and
Whitman, M. (1997a). Smad4 and FAST-1 in the assembly of activin-
responsive factor. Nature 389, 85-89.
Chen, Y., Bhushan, A., and Vale, W. (1997b). Smad8 mediates the
signalling of the receptor serine kinase. Proc. Natl. Acad. Sci. USA 94, 12938-
12943.
Chen, Y., Lebrun, J.-J., and Vale, W. (1996). Regulation of transforming
growth factor f~- and activin-induced transcription by mammalian Mad
proteins. Proc. Natl. Acad. Sci. USA 93, 12992-12997.
Dennler, S., Itoh, S., Vivien, D., ten Dijke, P., Huet, S., and Gauthier, J.-
M. (1998). Direct binding of Smad3 and Smad4 to critical TGFf~-inducible
elements in the promoter of human plasminogen activator inhibitor-type 1
gene. EMBO J. 17, 3091-3100.
Dyson, S. and Gurdon, J.B. (1998). The Interpretation of Position in a
Morphogen Gradient as Revealed by Occupancy of Activin Receptors. Cell
93, 557-568.
CA 02237701 1998-07-20
38
Faux, M. and Scott, J.D. (1996). Molecular glue: kinase anchoring and
scaffold proteins. Cell 85, 9-12.
Heldin, C.-H., Miyazono, K., and ten Dijke, P. (1997). TGF-(3 signalling
from cell membrane to nucleus through SMAD proteins. Nature. 390, 465-
471.
Hen is, Y.I., Moustakas, A., Lin, H.Y., and Lodish, H.F. (1994). The type
II and III transforming growth factor-f~ receptors form homo-oligomers. J.
Cell
Biol. 126, 139-154.
Hoodless, P.A., Haerry, T., Abdollah, S., Stapleton, M., O'Connor,
M.B., Attisano, L., and Wrana, J.L. (1996). MADR1, a MAD-related protein
that functions in BMP2 signalling pathways. Cell 85, 489-500.
Kim, J., Johnson, K., Chen, H.J., Carroll, S., and Laughon, A. (1997).
Drosophila Mad binds to DNA and directly mediates activation of vestigial by
decapentaplegic. Nature 388, 304-308.
Komada, M. and Kitamura, N. (1995). Growth factor-induced tyrosine
phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally
conserved putative zinc finger domain. Mol Cell Biol 15, 6213-6221.
Kretzschmar, M., Liu, F., Hata, A., Doody, J., and Massague, J. (1997).
The TGF-f~ family mediator Smad1 is phosphorylated directly and activated
functionally by the BMP receptor kinase. Genes Dev. 1 1, 984-995.
Kretzschmar, M. and Massague, J. (1998). SMADs: mediators and
regulators of TGF-~i signalling. Current Opinion in Genetics & Development
8, 103-111.
Labbe, E., Silvestri, C., Hoodless, P.A., Wrana, J.L., and Attisano, L.
(1998). Smad2 and Smad3 positively and negatively regulate TGF(3-dependent
transcription through the forkhead DNA binding protein, FAST2. Molecular
Cell in press.
Lagna, G., Hata, A., Hemmati-Brivanlou, A., and Massague, J. (1996).
Partnership between DPC4 and SMAD proteins in TGF-f~ signalling pathways.
Nature 383, 832-836.
Liu, F., Pouponnot, C., and Massague, J. (1997a). Dual role of the
Smad4/DPC4 tumor suppressor in TGF~i-inducible transcriptional complexes.
Genes & Development 11, 3157-3167.
CA 02237701 1998-07-20
39
Liu, X., Sun, Y., Constantinescu, S.N., Karam, E., Weinberg, R.A., and
Lodish, H.F. (1997b). Transforming growth factor a-induced phosphorylation
of Smad3 is required for growth inhibition and transcriptional induction in
epithelial cells. Proc. Natl. Acad. Sci. USA 94, 10669-10764.
Macias-Silva, M., Abdollah, S., Hoodless, P.A., Pirone, R., Attisano, L.,
and Wrana, J.L. (1996). MADR2 is a substrate of the TGFf~ receptor and its
phosphorylation is required for nuclear accumulation and signalling. Cell 87,
1215-1224.
Mu, F.T., Callaghan, J.M., Steele-Mortimer, O., Stenmark, H., Parton,
R.G., Campbell, P.L., McCluskey, J., Yeo, J.P., Tock, E.P., and Toh, B.H.
(1995). EEA1, an early endosome-associated protein. EEA1 is a conserved
alpha-helical peripheral membrane protein flanked by cysteine "fingers" and
contains a calmodulin-binding IQ motif. J Biol Chem 270, 13503-13511.
Nakao, A., Imamura, T., Souchelnytskyi, S., Kawabata, M., Ishisaki, A.,
Oeda, E., Tamaki, K., Hanai, J.-i., Heldin, C.-H., Miyazono, K., and ten
Dijke,
P. (1997a). TGF-f~ receptor-mediated signalling through Smad2, Smad3 and
Smad4. EMBO J. 16, 5353-5362.
Nakao, A., Roijer, E., Imamura, T., Souchelnytskyi, S., Stenman, G.,
Heldin, C.-H., and ten Dijke, P. (1997b). Identification of Smad2, a human
Mad-related protein in the transforming growth factor ~i signal ing pathway.
J.
Biol. Chem. 272, 2896-2900.
Nakayama, T., Snyder, M.A., Grewal, S.S., Tsuneizumi, K., Tabata, T.,
and Christian, J.L. (1998). Xenopus Smad8 acts downstream of BMP-4 to
modulate its activity during vertebrate embryonic patterning. Development
125, 857-867.
Nishimura, R., Kato, Y., Chen, D., Harris, S.E., Mundy, G.R., and
Yoneda, T. (1998). Smad5 and DPC4 are key molecules in mediating BMP-2-
induced osteoblastic differentiation of the pluripotent mesenchymal precursor
cell line C2C12. J. Biol. Chem. 273, 1872-1879.
Pasteris, N.G., Cadle, A., Logie, L.J., Porteous, M.E., Schwartz, C.E.,
Stevenson, R.E., Glover, T.W., Wilroy, R.S., and Gorski, J.L. (1994).
Isolation
and characterization of the faciogenital dysplasia (Aarskog-Scott syndrome)
gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell 79, 669-
678.
Patterson, G., Koweek, A., Wong, A., Liu, Y., and Ruvkun, G. (1997).
The DAF-3 Smad protein antagonizes TGF-f~-related receptor signalling in the
C. elegans dauer pathway. Genes and Dev.
CA 02237701 1998-07-20
Pawson, T. and Scott, J.D. (1997). Signaling through scaffold,
anchoring and adaptor proteins. Science 278, 2075-2080.
5 Piper, R.C., Cooper, A.A., Yang, H., and Stevens, T.H. (1995). VPS27
controls vacuolar and endocytic traffic through a prevacuolar compartment in
Saccharomyces cerevisae. J Cell Biol 131, 603-617.
Savage, C., Das, P., Finelli, A., Townsend, S., Sun, C., Baird, S., and
10 Padgett, R. (1996). The C. elegans sma-2, sma-3 and sma-4 genes define a
novel conserved family of TGF-f3 pathway components. Proc. Natl. Acad. Sci.
USA 93, 790-794.
Sekelsky, J.J., Newfeld, S.J., Raftery, L.A., Chartoff, E.H., and Gelbart,
15 W.M. (1995). Genetic characterization and cloning of Mothers against dpp, a
gene required for decapentaplegic function in Drosophila melanogaster.
Genetics 139, 1347-1358.
Souchelnytskyi, S., Tamaki, K., Engstrom, U., Wernstedt, C., ten Dijke,
20 P., and Heldin, C.-H. (1997). Phosphorylation of Ser465 and Ser467 in the C
Terminus of Smad2 Mediates Interaction with Smad4 and Is Required for
Transforming Growth Factor-(i Signaling. J. Biol. Chem. 272, 28107-28115.
Topper, J.N., Cai, )., Qiu, Y., Anderson, K.R., Xu, Y.-Y., Deeds, J.D.,
25 Feeley, R., Gimeno, C.J., Woolf, E.A., Tayber, O., Mays, G.G., Sampson,
B.A.,
Schoen, F.J., Gimbrone, M.A.J., and Falb, D. (1997). Vascular MADs: two
novel MAD-related genes selectively inducible by flow in human vascular
endothelium. Proc. Natl. Acad. Sci. USA 94, 9314-9319.
30 Weisman, L.S. and Wickner, W. (1992). Molecular characterization of
VAC1, a gene required for vacuole inheritance and vacuole protein sorting. J
Biol Chem 267, 618-623.
Wieser, R., Wrana, J.L., and Massague, J. (1995). GS domain mutations
35 that constitutively activate TfSR-I, the downstream signalling component in
the
TGF-f~ receptor complex. EMBO J. 14, 2199-2208.
Wrana, J.L., Attisano, L., Carcamo, J., Zentella, A., Doody, J., Laiho, M.,
Wang, X.-F., and Massague, J. (1992). TGF-f~ signals through a heteromeric
40 protein kinase receptor complex. Cell 71, 1003-1014.
Wrana, J.L., Attisano, L., Wieser, R., Ventura, F., and Massague, J.
(1994). Mechanism of activation of the TGF-f~ receptor. Nature 370, 341-347.
CA 02237701 1998-07-20
41
Yamamoto, A., DeWald, D.B., Boronenkov, I.V., Anderson, R.A., Emr,
S.D., and Koshland, D. (1995). Novel PI(4)P 5-kinase homologue, Fab1 p,
essential for normal vacuole function and morphology in yeast. Mol Biol Cell
6, 525-539.
Yingling, J.M., Datto, M.B., Wong, C., Frederick, J.P., Liberati, N.T.,
and Wang, X.-F. (1997). Tumour Suppressor Smad4 is a Transforming Growth
Factor (3-Inducible DNA Binding Protein. Mol. Cell. Biol. 17, 7019-7028.
Zawel, L., Dai, J.L., Buckhaults, P., Zhou, S., Kinzler, K.W., Vogelstein,
B., and Kern, S.E. (1998). Human Smad3 and Smad4 are sequence-specific
transcription activators. Mol. Cell 1, 611-617.
Zhang, Y., Musci, T., and Derynck, R. (1997). The tumor suppressor
Smad4/DPC4 as a central mediator of Smad function. Curr. Biol. 7, 270-276.
CA 02237701 1998-07-20
42
TABLE 1 - hSARA1
GCATACTGAATCAGCAGGACTGGCTGGTGGTGCAGCAGACATCATGAGTAAGCACCG
AGAAGTCTGTTCCTTATCACGTGTGTAAGGGGAA.P.AAGGTTTAAACAAGTCTCTTAA
GTGGTGTTTCCTCACCGATGGAGAATTACTTCCAAGCAGAAGCTTACAACCTGGGAC
AAGGTGTTAGATGAATTTGAACAAAACGAAGATGAAACAGTTTCTTCTACTTTATTG
GATACAA.AGTGGAATAAGATTCTAGATCCCCCTTCTCACCGGCTGTCATTTAACCCT
ACTTTGGCCAGTGTGAATGAATCTGCAGTTTCTAATGAGTCACAACCACAACTGAAA
GTCTTCTCCCTGGCTCATTCAGCTCCCCTGACCACAGAGGAAGAGGATCACTGTGCT
AATGGACAGGACTGTAATCTAAATCCAGAGATTGCCACAATGTGGATTGATGAAAAT
GCTGTTGCAGAAGACCAGTTAATTAAGAGAAACTATAGTTGGGATGATCAATGCAGT
GCTGTTGAAGTGGGAGAGAAGAA.ATGTGGAAACCTGGCTTGTCTGCCAGATGAGAAG
AATGTTCTTGTTGTAGCCGTCATGCATAACTGTGATAAAAGGACATTACAAAACGAT
TTACAGGATTGTAATAATTATAATAGTCAATCCCTTATGGATGCTTTTAGCTGTTCA
CTGGATAATGAAAACAGACAAACTGATCAATTTAGTTTTAGTATAAATGAGTCCACT
GAAAAAGATATGAATTCAGAGAAACAAATGGATCCATTGAATAGACCGAAAACAGAG
GGGAGATCTGTTAACCATCTGTGTCCTACTTCATCTGATAGTCTAGCCAGTGTCTGT
TCCCCTTCACAATTAAAGGATGACGGAAGTATAGGTAGAGACCCCTCCATGTCTGCG
ATTACAAGTTTAACGGTTGATTCAGTAATCTCATCCCAGGGAACAGATGGATGTCCT
GCTGTTAP~AAAGCAAGAGAACTATATACCAGATGAGGACCTCACTGGCAAAATCAGC
TCTCCTAGGACAGATCTAGGGAGTCCAAATTCCTTTTCCCACATGAGTGAGGGGATT
TTGATGAAAA.P.AGAGCCAGCAGAGGAGAGCACCACTGAAGAATCCCTCCGGTCTGGT
TTACCTTTGCTTCTCAAACCAGACATGCCTAATGGGTCTGGAAGGAATAATGACTGT
GAACGGTGTTCAGATTGCCTTGTGCCTAATGAAGTTAGGGCTGATGAAAATGAAGGT
TATGAACATGAAGAAACTCTTGGCACTACAGAATTCCTTAATATGACAGAGCATTTC
TCTGAATCTCAGGACATGACTAATTGGAAGTTGACTAAACTAAATGAGATGAATGAT
AGCCAAGTAAACGAAGAA.A.AGGAAAAGTTTCTACAGATTAGTCAGCCTGAGGACACT
AATGGTGATAGTGGAGGACAGTGTGTTGGATTGGCAGATGCAGGTCTAGATTTAAAA
GGAACTTGCATTAGTGAAAGTGAAGAATGTGATTTCTCCACTGTTATAGACACACCA
GCAGCAAATTATCTATCTAATGGTTGTGATTCCTATGGAATGCAAGACCCAGGTGTT
TCTTTTGTTCCAAAGACTTTACCCTCCAAAGAAGATTCAGTAACAGAAGAA.A.P.AGAA
ATAGAGGAAAGCAAGTCAGAATGCTACTCAAATATTTATGAACAGAGAGGAAATGAG
GCCACAGAAGGGAGTGGACTACTTTTAAACAGCACTGGTGACCTAATGAAGAAAA.AT
TATTTACATAATTTCTGTAGTCAAGTTCCATCAGTGCTTGGGCAATCTTCCCCCAAG
GTAGTAGCAAGCCTGCCATCTATCAGTGTTCCTTTTGGTGGTGCAAGACCCAAGCAA
CCTTCTAATCTTAAACTTCAAATTCCAAAGCCATTATCAGACCATTTACAAAATGAC
TTTCCTGCAAACAGTGGAAATAATACTAAA.A.ATAAAA.ATGATATTCTTGGGAAAGCA
AAATTAGGGGAAAACTCAGCAACCAATGTATGCAGTCCATCTTTGGGAAACATCTCT
AATGTCGATACAAATGGGGAACATTTAGAAAGTTATGAGGCTGAGATCTCCACTAGA
CCATGCCTTGCATTAGCTCCAGATAGCCCAGATAATGATCTCAGAGCTGGTCAGTTT
GGAATTTCTGCCAGAAAGCCATTCACCACGCTGGGTGAGGTGGCTCCAGTATGGGTA
CCGGATTCTCAGGCTCCAAATTGCATGAAATGTGAAGCCAGGTTTACATTCACCAAA
AGGAGGCATCACTGCAGAGCATGTGGGAAGGTTTTCTGTGCTTCCTGCTGTAGCCTG
AAATGTAA.ACTGTTATACATGGACAGAAAGGAAGCTAGAGTGTGTGTAATCTGCCAT
TCAGTGCTAATGAATGCTCAAGCCTGGGAGAACATGATGAGTGCCTCAAGCCAGAGC
CCTAACCCTAACAATCCTGCTGAATACTGTTCTACTATCCCTCCCTTGCAGCAAGCT
CAGGCCTCAGGAGCTCTGAGCTCTCCACCTCCCACTGTGATGGTACCTGTGGGAGTT
TTAA.AGCACCCTGGAGCAGAAGTGGCTCAGCCCAGAGAGCAGAGGCGAGTTTGGTTT
GCTGATGGGATCTTGCCCAATGGAGAAGTTGCTGATGCAGCCAAATTAACAATGAAT
GGAACTTCCTCTGCAGGAACCCTGGCTGTGTCACACGACCCAGTCAAGCCAGTAACT
ACCAGTCCTCTACCAGCAGAGACGGATATTTGTCTATTCTCTGGGAGTATAACTCAG
GTTGGAAGTCCTGTTGGAAGTGCAATGAATCTTATTCCTGAAGATGGCCTTCCTCCC
ATTCTCATCTCCACTGGTGTAAAAGGAGACTATGCTGTGGAAGAGAAACCATCACAG
ATTTCAGTAATGCAGCAGTTGGAGGATGGTGGCCCTGACCCACTTGTATTTGTTTTA
AATGCAAATTTGTTGTCAATGGTTAAAATTGTAAATTATGTGAACAGGAAGTGCTGG
TGTTTCACAACCAAGGGAATGCATGCAGTGGGTCAGTCTGAGATAGTCATTCTTCTA
CA 02237701 1998-07-20
43
TABLE 1 - hSARA1 Continued
CAGTGTTTACCGGATGAAAAGTGTTTGCCAAAGGATATCTTTAATCACTTTGTGCAG
CTTTATCGGGATGCTCTGGCAGGGAATGTGGTGAGCAACTTGGGACATTCCTTCTTC
AGTCAAAGTTTCCTTGGCAGTAAAGAACATGGTGGATTCTTATATGTGACATCTACC
TACCAGTCACTGCAAGACCTAGTACTCCCAACCCCACCTTACTTGTTTGGGATTCTT
ATCCAGAA.ATGGGAAACTCCTTGGGCTAAAGTATTTCCTATCCGTCTGATGTTGAGA
CTTGGAGCTGAATATCGACTTTATCCATGCCCACTATTCAGTGTCAGATTTCGGAAG
CCATTGTTTGGAGAGACGGGGCATACCATCATGAATCTTCTTGCAGACTTCAGAAAT
TACCAGTATACCTTGCCAGTAGTTCAAGGTTTGGTGGTTGATATGGAAGTTCGGAAA
ACTAGCATCAAAATTCCCAGCAACAGATACAATGAGATGATGAAAGCCATGAACAAG
TCCAATGAGCATGTCCTGGCAGGAGGTGCCTGCTTCAATGAAA.AGGCAGACTCTCAT
CTTGTGTGTGTACAGAATGATGATGGAAACTATCAGACCCAGGCTATCAGTATTCAC
AATCAGCCCAGAAAAGTGACTGGTGCCAGTTTCTTTGTGTTCAGTGGCGCTCTGAAA
TCCTCTTCTGGATACCTTGCCAAGTCCAGTATTGTGGAAGATGGTGTTATGGTCCAG
ATTACTGCAGAGAACATGGATTCCTTGAGGCAGGCACTGCGAGAGATGAAGGACTTC
ACCATCACCTGTGGGAAGGCGGACGCGGAGGAACCCCAGGAGCACATCCACATCCAG
TGGGTGGATGATGACAAGAACGTTAGCAAGGGTGTCGTAAGTCCTATAGATGGGAAG
TCCATGGAGACTATAACAAATGTGAAGATATTCCATGGATCAGAATATAAAGCAAAT
GGAAAAGTAATCAGATGGACAGAGGTGTTTTTCCTAGAAAACGATGACCAGCACAAT
TGCCTCAGTGATCCTGCAGATCACAGTAGATTGACTGAGCATGTTGCCAAAGCTTTT
TGCCTTGCTCTCTGTCCTCACCTGAAACTTCTGAAGGAAGATGGAATGACCAAACTG
GGACTACGTGTGACACTTGACTCAGATCAGGTTGGCTATCAAGCAGGGAGCAATGGC
CAGCCCCTTCCCTCGCAGTACATGAATGATCTGGATAGCGCCTTGGTGCCGGTGATC
CATGGAGGGGCCTGCCAGCTTAGTGAGGGCCCCGTTGTCATGGAACTCATCTTTTAT
ATTCTGGAAAACATCGTATAAACAGAGAAGACTTCATTTTTTTCTGTTCAGACTTGT
TGCAACAGCAGTCATACCCAAATCATTTGCACTTTAAAACTGGAAGATTAAGCTTTT
GTTAACACTATTAATGGGGTGGGGAATAGGGTGGGAGTGGGGGTTTGGGAGACGGGT
GGGAAAGGGTGGTTGGGGGGACCGATGTTCCATAATTCTAAGTCTTCTATGCATTGT
CCACCAAGAAGATCTGGGCAGCTTCTGTTCCTGCACAACAGTTATGCTATCCTTGCA
GCTAATCCCCTTCTGTTACTGTTTAGACAAGAATTCCGCTCCTCTCTCAAGATTTAC
TTATGGTCATGTGCTCAGAA.ATGCTCAAATGGGTACAACCATCACCAAGGGTGGGAT
GGGAGGGCAGAGGGGAAATAAAATATAAAGCATC
CA 02237701 1998-07-20
44
TABLE 2 - hSARA1
MWIDENAVAEDQLIKRNYSWDDQCSAVEVGEKKCGNLACLPDEKNVLVVAVMHNCDK
RTLQNDLQDCNNYNSQSLMDAFSCSLDNENRQTDQFSFSINESTEKDMNSEKQMDPL
NRPKTEGRSVNHLCPTSSDSLASVCSPSQLKDDGSIGRDPSMSAITSLTVDSVISSQ
GTDGCPAVKKQENYIPDEDLTGKISSPRTDLGSPNSFSHMSEGILMKKEPAEESTTE
ESLRSGLPLLLKPDMPNGSGRNNDCERCSDCLVPNEVRADENEGYEHEETLGTTEFL
NMTEHFSESQDMTNWKLTKLNEMNDSQVNEEKEKFLQISQPEDTNGDSGGQCVGLAD
AGLDLKGTCISESEECDFSTVIDTPAANYLSNGCDSYGMQDPGVSFVPKTLPSKEDS
VTEEKEIEESKSECYSNIYEQRGNEATEGSGLLLNSTGDLMKKNYLHNFCSQVPSVL
GQSSPKWASLPSISVPFGGARPKQPSNLKLQIPKPLSDHLQNDFPANSGNNTKNKN
DILGKAKLGENSATNVCSPSLGNISNVDTNGEHLESYEAEISTRPCLALAPDSPDND
LRAGQFGISARKPFTTLGEVAPVWVPDSQAPNCMKCEARFTFTKRRHHCRACGKVFC
ASCCSLKCKLLYMDRKEARVCVICHSVLMNAQAWENMMSASSQSPNPNNPAEYCSTI
PPLQQAQASGALSSPPPTVMVPVGVLKHPGAEVAQPREQRRVWFADGILPNGEVADA
AKLTMNGTSSAGTLAVSHDPVKPVTTSPLPAETDICLFSGSITQVGSPVGSAMNLIP
EDGLPPILISTGVKGDYAVEEKPSQISVMQQLEDGGPDPLVFVLNANLLSMVKIVNY
VNRKCWCFTTKGMHAVGQSEIVILLQCLPDEKCLPKDIFNHFVQLYRDALAGNWSN
LGHSFFSQSFLGSKEHGGFLYVTSTYQSLQDLVLPTPPYLFGILIQKWETPWAKVFP
IRLMLRLGAEYRLYPCPLFSVRFRKPLFGETGHTIMNLLADFRNYQYTLPVVQGLVV
DMEVRKTSIKIPSNRYNEMMKAMNKSNEHVLAGGACFNEKADSHLVCVQNDDGNYQT
QAISIHNQPRKVTGASFFVFSGALKSSSGYLAKSSIVEDGVMVQITAENMDSLRQAL
REMKDFTITCGKADAEEPQEHIHIQWVDDDKNVSKGVVSPIDGKSMETITNVKIFHG
SEYKANGKVIRWTEVFFLENDDQHNCLSDPADHSRLTEHVAKAFCLALCTQLKLLKG
DGMTKLGLRVTLDSDQVGYQAGSNGQHLPSQYMNDFDSDLVKMIHGGACQLSEGPVV
MELIFYILENIV
CA 02237701 1998-07-20
TABLE 3 - human SARA2
ACTCCCGGCCGGGGTAGCTCTTCACTCCTCAGCGCGACGTCGTGTCGAGTTCCCAAA
AAGCTCCGCAGGGGCTGTAGGGAGGTGATCTCATCCATTAACAGCTGTGTGTTGCCA
GTTCCCAAATCTTTATCTATCTCAGACTTCTCTCCTGCATTCCAGATTCTTATATTCAG
CTGCCTTTTGGATATCTCTCCCAGGATGTTCTCAAGGCATACAAGAATTAAATTCTG
AATAAGTCTGCAGGTAGGATGGACAGTTATTTTAAAGCAGCTGTCAGTGACTTGGAC
AAACTCCTTGATGATTTTGAACAGAACCCAGATGAACAAGATTATCTCGCAGATGTA
CAAAATGCATATGATTCTAACCACTGCTCAGTTTCTTCAGAGTTGGCTTCCTCACAGC
GAACTTCATTGCTCCCAAAAGACCAAGAGTGCGTTAATAGTTGTGCCTCATCAGAAA
CAAGCTATGGAACAAATGAGAGTTCCCTGAATGAAAAAACACTCAAGGGACTTACT
TCTATACAAAATGAAAAAAATGTAACAGGACTTGATCTTCTTTCTTCTGTGGATGGT
GGTACTTCAGATGAAATCCAGCCGTTATATATGGGACGATGTAGTAAACCTATCTGT
GATCTGATAAGTGACATGGGTAACTTAGTTCATGCAACCAATAGTGAAGAAGATATT
AAAAAATTATTGCCAGATGATTTTAAGTCTAATGCAGATTCCTTGATTGGATTGGAT
TTATCTTCAGTGTCAGATACTCCCTGTGTTTCTTCAACAGACCATGATAGTGATACTG
TCAGAGAACAACAGAATGATATCAGTTCTGAATTACAAAATAGAGAAATCGGAGGA
ATCAAAGAATTGGGTATAAAAGTAGATACAACACTTTCAGATTCCTATAATTACAGT
GGAACAGAAAATTTAAAAGATAAAAAGATCTTTAATCAGTTAGAATCAATTGTTGAT
TTTAACATGTCATCTGCTTTGACTCGACAAAGTTCCAAAATGTTTCATGCCAAAGAC
AAGCTACAACACAAGAGCCAGCCATGTGGATTACTAAAAGATGTTGGCTTAGTAAA
AGAGGAAGTAGATGTGGCAGTCATAACTGCCGCAGAATGTTTAAAAGAAGAGGGCA
AGACAAGTGCTTTGACCTGCAGCCTTCCGAAAAATGAAGATTTATGCTTAAATGATT
CAAATTCAAGAGATGAAAATTTCAAATTACCTGACTTTTCCTTTCAGGAAGATAAGA
CTGTTATAAAACAATCTGCACAAGAAGACTCAAAAAGTTTAGACCTTAAGGATAAT
GATGTAATCCAAGATTCCTCTTCAGCTTTACATGTTTCCAGTAAAGATGTGCCGTCCT
CATTGTCCTGTCTTCCTGCGTCTGGGTCTATGTGTGGATCATTAATTGAAAGTAAAGC
ACGGGGTGATTTTTTACCTCAGCATGAACATAAAGATAATATACAAGATGCAGTGAC
TATACATGAAGAAATACAGAACAGTGTTGTTCTAGGTGGGGAACCATTCAAAGAGA
ATGATCTTTTGAAACAGGAAAAATGTAAAAGCATACTCCTTCAGTCATTAATTGAAG
GGATGGAAGACAGAAAGATAGATCCTGACCAGACAGTAATCAGAGCTGAGTCTTTG
GATGGTGGTGACACCAGTTCTACAGTTGTAGAATCTCAAGAGGGGCTTTCTGGCACT
CATGTCCCAGAGTCTTCTGATTGTTGTGAAGGTTTTATTAATACTTTTTCAAGCAATG
ATATGGATGGGCAAGACTTAGATTACTTTAATATTGATGAAGGCGCAAAAAGTGGC
CCACTAATTAGTGATGCTGAACTTGATGCCTTTCTGACAGAACAGTATCTTCAGACC
ACTAACATAAAGTCTTTTGAAGAAAATGTAAATGACTCTAAATCGCAAATGAATCAG
ATAGATATGAAAGGCTTAGATGATGGAAACATCAATAATATATATTTCAATGCAGA
AGCAGGAGCTATTGGGGAAAGTCATGGTATTAATATAATTTGTGAAACAGTTGATAA
ACAAAATACAATAGAAAATGGCCTTTCTTTAGGAGAAAAAAGCACTATTCCAGTTCA
ACAAGGGTTACCTACCAGTAAGTCTGAGATTACAAATCAATTATCAGTCTCTGATAT
TAACAGTCAATCTGTTGGAGGGGCCAGACCTAAGCAATTGTTTAGCCTTCCATCAAG
AACAAGGAGTTCAAAGGACCTGAATAAGCCAGATGTTCCAGATACAATAGAAAGTG
AACCCAGCACAGCAGATACCGTTGTTCCAATCACTTGTGCTATAGATTCTACAGCTG
ATCCACAGGTTAGCTTCAACTCTAATTACATTGATATAGAAAGTAATTCTGAAGGTG
GATCTAGTTTCGTAACTGCAAATGAAGATTCTGTACCTGAAAACACTTGCAAAGAAG
GCTTGGTTTTGGGCCAGAAACAGCCTACTTGGGTTCCTGATTCAGAAGCTCCAAACT
GTATGAACTGCCAAGTCAAATTTACTTTTACCAAACGGCGACACCATTGCCGAGCAT
GTGGGAAAGTATTTTGTGGTGTCTGTTGTAATAGGAAGTGTAAACTGCAATATCTAG
CA 02237701 1998-07-20
46
TABLE 3 - human SARA2 Continued
AAAAGGAAGCAAGAGTATGTGTAGTCTGCTATGAAACTATTAGTAAAGCTCAGGCA
TTTGAAAGGATGATGAGTCCAACTGGTTCTAATCTTAAGTCTAATCATTCTGATGAA
TGTACTACTGTCCAGCCTCCTCAGGAGAACCAAACATCCAGTATACCTTCACCAGCA
ACTTTGCCAGTCTCAGCACTTAAACAACCAGGTGTTGAAGGACTATGTTCCAAAGAA
CAGAAGAGAGTATGGTTTGCAGATGGTATATTGCCCAATGGTGAAGTTGCAGATAC
AACAAAATTATCATCTGGAAGTAAAAGATGTTCTGAAGACTTTAGTCCTCTCTCACC
TGATGTGCCTATGACAGTAAACACAGTGGATCATTCCCATTCTACTACAGTGGAAAA
GCCAAACAATGAGACAGGAGATATTACAAGAAATGAGATAATTCAGAGTCCTATTT
CTCAGGTTCCATCAGTGGAAAAATTGTCTATGAACACAGGAAATGAGGGGTTACCTA
CTTCTGGTTCATTTACACTAGATGATGATGTTTTTGCAGAAACTGAAGAACCATCTA
GTCCTACTGGTGTCTTAGTTAACAGCAATTTACCTATTGCTAGTATTTCAGATTATAG
GTTACTGTGTGATATTAACAAGTATGTCTGCAATAAGATTAGTCTTCTACCTAATGAT
GAGGACAGTTTGCCCCCACTTCTGGTTGCATCTGGAGAAAAGGGATCAGTGCCTGTA
GTAGAAGAACATCCATCTCATGAGCAGATCATTTTGCTTCTTGAAGGTGAAGGCTTT
CATCCTGTTACATTTGTCCTAAATGCTAATCTACTCGTGAATGTCAAATTCATATTTT
ATTCCTCAGACAAATATTGGTACTTTTCAACCAATGGATTGCATGGCTTGGGACAGG
CAGAAATTATTATTCTATTGTTATGTTTGCCAAATGAAGATACTATTCCTAAGGACAT
CTTCAGACTATTTATCACCATATATAAGGATGCTCTAAAAGGAAAATACATAGAAAA
CTTGGACAATATTACCTTTACTGAGAGTTTTCTCAGTAGCAAGGATCACGGAGGATT
CCTGTTTATTACACCTACTTTTCAGAAACTTGATGATCTCTCATTACCAAGTAATCCT
TTTCTTTGTGGAATTCTTATCCAGAAGCTTGAGATTCCCTGGGCAAAGGTTTTTCCTA
TGCGTTTAATGTTGAGATTGGGTGCAGAATATAAAGCATATCCTGCTCCTCTAACAA
GCATCAGAGGCCGAAAACCTCTTTTTGGAGAAATAGGACACACTATTATGAACTTAC
TTGTTGACCTTCGAAATTACCAGTATACCTTGCATAATATAGATCAACTGTTGATTCA
TATGGAAATGGGAAAAAGCTGCATAAAAATACCACGGAAAAAGTACAGTGATGTAA
TGAAAGTACTAAATTCTTCCAATGAGCATGTCATTAGCATTGGAGCAAGTTTCAGTA
CAGAAGCAGATTCTCATCTAGTCTGTATACAGAATGATGGAATTTATGAAACACAGG
CCAACAGTGCCACTGGCCATCCTAGAAAAGTGACAGGTGCAAGTTTTGTGGTATTCA
ATGGAGCTCTAAAAACATCTTCAGGATTTCTTGCTAAGTCCAGCATAGTTGAAGATG
GCTTAATGGTACAAATAACTCCAGAGACCATGAATGGCTTGCGGCTAGCTTTACGAG
AACAGAAAGACTTTAAAATTACATGTGGGAAAGTTGATGCAGTAGACCTGAGAGAA
TACGTGGATATCTGCTGGGTAGATGCTGAAGAAAAAGGAAACAAAGGAGTTATCAG
TTCAGTGGATGGAATATCATTACAAGGATTTCCAAGTGAAAAAATAAAACTGGAAG
CAGATTTTGAAACCGATGAGAAGATTGTAAAATGTACCGAGGTGTTCTACTTTCTAA
AGGACCAGGATTTATCTATTTTATCAACTTCTTATCAGTTTGCAAAAGAAATAGCCA
TGGCTTGTAGTGCTGCGCTGTGCCCTCACCTGAAAACTCTAAAAAGTAATGGGATGA
ATAAAATTGGACTCAGAGTTTCCATTGACACTGATATGGTTGAATTTCAGGCAGGAT
CTGAAGGCCAACTTCTGCCTCAGCATTATCTAAATGATCTTGATAGTGCTCTGATAC
CTGTGATCCATGGTGGGACCTCCAACTCTAGTTTACCATTAGAAATAGAATTAGTGT
TTTTCATTATAGAACATCTTTTTTAGTGAAAGAATGTGCCATATTACATATTGCAACC
TAATTTGTTAAAACTAACTCCAGCACTAAAGCTGAAATGCCACAAACACTAAAAGT
ATAAATATGTCTGATTTTTGAAACACATAAGCTTTGCTCTTTAGGCAGGAATGATCTT
TTCAAATCATTAGCACAATATTTAAATATCTAAAAATTTAAGAGATCCATACTTTCTG
TAGCTTTACAATTAATTTAAGTACTAAAAAGACAAGGATTTCTTTTAAGAAATTTAT
AGCATTTACTGTGTTATTTAAATGCTAAGCCAAAGTATCTGCACTTAGGTATACCTCT
TTATGCCAATAATGATTTTAATGAAGGCTCTTTTCAGATGTAACCTTATGAAGGAAA
TATCTGCTTTGTGTATATGCCAGTTAGAATACTGGTTTCTAAAGTCTGTCAAATTGTA
TTTCAGTGGCACAAAAACCAGTTTTGAGGTCTTAGACTTATAATTCTTTGAATAAAA
CA 02237701 1998-07-20
47
TABLE 3 - human SARA2
CTGATAACTTATTTGTATAATTGGAGTGGAGACCTACCTCCATAATTAGATAAACTC
TTTTTGGATTATAATCAGAATTTTGCCTTTTTTCTTCTCAAATTATTACATATGTATGT
ATTATATATCCACATATATAGTTTTCCCTGATTAAATGGATATTAAAATAATTGCGGG
TGCTTCAGGACTTTTTGCTTCTATATTTAAGTATATTGTTTTTATAGCAAGAACATAT
TCTGAATGTTTTATAAATCTTTAATAATTTATATGTAGGTAATATTTrTGTATCACAA
TGCATTATTTTTTTTCCTCCTTTCCTTCCAAACTATACCACTGTATTTACCACTTCTAA
GAGTGACTGACGACGGGCCAGATGACCCTTGAAGTAGTCATTATGTAGCAATAAAT
GAAGCCTGAAACAGGTTTTTTTACTTCCACTTTAATCCTTAGAAATTTCTTGGCAACT
TCGCATATTTTCATTGACACTGGTGTATAAGTATAAATTTAAATGAACTAATTACTTT
TGCATATTTTAAATTCTTTATATGGTAGTTATTTTTTATAACAGGATATTAACATAAG
TTAAATCCTATGTATTTGAAATTGTTACAGAGCTTTCCTCTTTACTTCAAACAGCAAA
AAAGTGGGGGGCATATTGTAGTCCTGTCATTTAAGTTATGTAAAAAATTTAATCATT
ATTTTGATGCTTTAAACATTCTCATGTGTAATATATGTTTTTGTATCAAAAACACTCA
TATATTTCAAGAAAAAGAAATTATGTTAAATAGCCCTGTTTTAAGAAAAATATTTAT
GAAGCATCTCAACTTGAAGATCAAGTCAAAGTTATAACTCAGGATCTGAGGTCTCAA
GCTAGGAGAGACTGAGAATTTTAATCAGTTTGGGCATATAGTTTGGACTGAATCACA
TCTGTAGTACTTAGCCAAAGACAATTTGGAGGAGAATATCAGCCTTCTGGAAGTAGC
TACTTCCTGAACAATGTAAAGTGTCGCAGATATTCAATAAAATGGCAACCTGTTATA
ATTTGTGAAATTTATTGAAATGGTGTAAGATGAAAACAATTGCATATCAAACCCAAT
TTATGTTTTCTAAATATAGTGTATGTATTCTGCCATGTAAGTAATTGAACAGTCTTAA
AATAACCAAATGGTAGAGGGCTGTTCCATGATGGGACAGCTTTGGATTTGTTTTCAT
AAAATCTCTACATTCAATAAAAATTGGAATTATGTGCCTGAAGTTTGGAGGCACATT
TTGAAGT
CA 02237701 1998-07-20
48
TABLE 4 - human SARA2
MDSYFKAAVSDLDKLLDDFEQNPDEQDYLQDVQNAYDSNHCSVSSELASSQRTSLLPK
DQECVNSCASSETSYGTNESSLNEKTLKGLTSIQNEKNVTGLDLLSSVDGGTSDEIQPLY
MGRCSKPICDLISDMGNLVHATNSEEDIKKLLPDDFKSNADSLIGLDLSSVSDTPCVSSTD
HDSDTVREQQNDTSSELQNREIGGIKELGIKVDTTLSDSYNYSGTENLKDKKIFNQLESIV
DFNMSSALTRQSSKMFHAKDKLQHKSQPCGLLKDVGLVKEEVDVAVITAAECLKEEGK
TSALTCSLPKNEDLCLNDSNSRDENFKLPDFSFQEDKTVIKQSAQEDSKSLDLKDNDVIQ
DSSSALHVSSKDVPSSLSCLPASGSMCGSLIESKARGDFLPQHEHKDNIQDAVTIHEEIQN
S V VLGGEPFKENDLLKQEKCKSILLQSLIEGMEDRKIDPDQTVIRAESLDGGDTSSTV VES
QEGLSGTHVPESSDCCEGFINTFSSNDMDGQDLDYFNIDEGAKSGPLISDAELDAFLTEQ
YLQTTNIKSFEENVNDSKSQMNQIDMKGLDDGNINNIYFNAEAGAIGESHGINIICETVD
KQNTIENGLSLGEKSTIPVQQGLPTSKSEITNQLS VSDINSQS VGGARPKQLFSLPSRTRS S
KDLNKPDVPDTIESEPSTADTVVPITCAIDSTADPQVSFNSNYIDIESNSEGGSSFVTANED
SVPENTCKEGLVLGQKQPTWVPDSEAPNCMNCQVKFTFTKRRHHCRACGKVFCGVCC
NRKCKLQYLEKEARVCVVCYETISKAQAFERMMSPTGSNLKSNHSDECTTVQPPQENQ
TSSIPSPATLPVSALKQPGVEGLCSKEQKRVWFADGILPNGEVADTTKLSSGSKRCSEDF
SPLSPDVPMTVNTVDHSHSTTVEKPNNETGDITRNEIIQSPISQVPS VEKLSMNTGNEGLP
TSGSFTLDDDVFAETEEPSSPTGVLVNSNLPIASISDYRLLCDINKYVCNKISLLPNDEDSL
PPLLVASGEKGS VPV VEEHPSHEQIILLLEGEGFHPVTFVLNANLLVNVKFIFYSSDKYW
YFSTNGLHGLGQAEIIILLLCLPNEDTIPKDIFRLFITIYKDALKGKYIENLDNITFTESFLSS
KDHGGFLFITPTFQKLDDLSLPSNPFLCGILIQKLEIPWAKVFPMRLMLRLGAEYKAYPAP
LTSIRGRKPLFGEIGHTIMNLLVDLRNYQYTLHNIDQLLIHMEMGKSCIKIPRKKYSDVM
KVLNSSNEHVISIGASFSTEADSHLVCIQNDGIYETQANSATGHPRKVTGASFV VFNGAL
KTSSGFLAKSSIVEDGLMVQITPETMNGLRLALREQKDFKITCGKVDAVDLREYVDICW
VDAEEKGNKGVISSVDGISLQGFPSEKIKLEADFETDEKIVKCTEVFYFLKDQDLSILSTS
YQFAKEIAMACSAALCPHLKTLKSNGMNKIGLRVSIDTDMVEFQAGSEGQLLPQHYLN
DLDSALIPVIHGGTSNSSLPLEIELVFFIIEHLF
CA 02237701 1998-07-20
49
TABLE 5 - XSARA1
CTGTAAGTTTGACTATGTAGGAAAGCATTTCTGTTATCTATGAAGTATGTTTTAGAGT
CAGACCAATAACTAAACGGTTTTCTTTTTTTTGTTTATTTCCCCTCAGATGAGACTGT
CTCTCCAAAGCTATTAGATGCTAAGTGGAATCAAATCTTAGAACCGCATTCACATAA
AGTCGCTGATAACTCCGCCCTTGACAATGTCTGTAAATCAATCATTGCTATTGAAGC
TCATCTCAAAGTCAGGTCACCCGGCTTGTCAGCCCTTGTGAGGTCCACATATGTGAA
TGGAGAAGTAGGTATTGTGGCACCTGAAATGCCCAAAATGGTGATAGGAGACACCA
TTATGGCAGAGGATTCACTTTTTAACAACACTGGTCCCTCTGAAATTGTATGCAACC
CATCTACTGTGGAGAGTCAAAGTTTACAAGCTTTAGATGATCAATCAGTGAATATTC
ACAATGAAAAAAGTGTTCTGCTCGCTGATGGCTTTTCACCATGCAGTAGCCCCAAAA
GTATTATAAACTTTGACTGCTTGACCATGGATAACGAAATGCCTTTGCACAGTCAAA
TGAGTGTTGATGACAATGACAAAGAAACTGTAACAATTTCAGTCCTTCCAACAATCA
TACAGGATACTAGTAACGTAAGCACAGACCCAGCTATCAATAAACCTGGCACTAAA
GAACCCCATAGAGCATTAAAGGAAACCACATCAGTTATTCTGCCTGAAATAAAGCC
TTACTCCACATGTGCTGCCCTTTCGTTTGAAAATAACAATAAGGTTCCCAGTTATCAA
TTAAATAATACAGATCTACTCAGCGTTTCACCAGTGGTTGAAGCATGTAGTGAGCAG
CAGCAAAAACATACATCTTCCTTGCATGAAGAAAAACTTTTTGAAGGTGTTTCTGCA
ACGGAGTCCTTTGCAGCCACTGCTGCGGAAACTGTACTGGATAATGAGGCTCTCCGT
AGTGCTGAATTCTTTGACATTGTTGTAAAGAACTTTTCTGACTCTTGTGTGATTAATG
GCGACTTGACTAAAAGTTGTGGCCTCTCTCAAGAAAGCAATGAAAAGTTTTGTGCAA
GTAAAGAGTTTGAAGGAGGGGTAGATGCTAATGTCTTGTTGGAAAATGCATGTGTA
GCTTATAAAGAAGCAATAGATTTGCCTGAAGAAAATGGAACTAATGCACCAATGTC
TCTGTACAATGGGTGTGATTCCTATGGAATGAAAAACCCAGCCGTAGCTCAAAACCC
AAAGAATTTACCTTCAAAAGAAGATTCTGTGACAGAAGAAAAAGAAATTGAAGAAA
GCAAGTCAGAATACTATACTGGTGTTTATGAACAACAAAGAGAAGATGATGTTACA
GAGAGAGGTGGACTTCTGTTAAATGCTAAGGCTGACCAAATGAAGAACAATTTGCA
TAGTCTTTGTAATCAGGTTCCATCCATGCATGGGCAAACATCACCAAAAAAGGGCAA
GATTGTGCAATCTCTCAGTGTTCCATACGGTGGAGCACGCACTAAGCAGCCAACTCA
TCTCAAACTCCATATTCCAAAGCCATTGTCTGAAATGTTGCAGAGCGATCTCATTCCT
CCAAATGCTGGCTGCAGCTCTAAATACAAAAATGACATGTTAAACAAATCAAATCA
GGGGGATAACCTGATTTCAGAATCACTGCGTGAGGATTCTGCAGTGCGCAGCCCTGT
TACTGATGCTAATGGTGATTTCCCTGGAGAATACAGGGGACCTGGCAGCTTGTGCCT
TGCAGTGTCTCCAGACAGCCCAGACAACGATCTGCTTGCCGGGCAGTTTGGGGTACC
CATCTCTAAGCCATTTACTACTCTAGGGGAAGTGGCTCCAGTCTGGGTGCCAGATTC
CCAAGCACCAAACTGCATGAAGTGCGAGGCCAGATTTACATTTACCAAAAGGAGGC
ATCACTGCCGAGCTTGTGGAAAGGTGTTCTGTGCTGCTTGTTGCAGTCTAAAATGCA
AACTACAGTACATGGATAAAAAGGAGGCTCGTGTGTGTGTTATTTGTCATTCTGTGC
TTATGAATGCTCAAGCATGGGAGAACATGTTAAGTGCATCGGTCCAAAGCCCAAAT
CCAAATAATCCTGCTGAATACTGCTCAACTATCCCTCCGATGCAGCAGGCACAAGCT
TCAGGAGCACTGAGTTCCCCACCTCCCACTGTCATGGTGCCAGTGGGTGTGTTAAAA
CATCCAGGAACTGAAGGGTCACAGTCAAAGGAACAGCGCCGTGTTTGGTTTGCTGA
TGGAATATTACCCAACGGAGAGACTGCTGACTCAGATAATGCAAACGTAACTACAG
TGGCTGGGACACTTACTGTGTCACATACCAACAATTCCACATCTTCAGAGTCTGAGA
ACACCTCTGGATTCTGTGGAAGTATAACTCAGGTTGGCAGTGCAATGAACCTTATTC
CAGAAGATGGGCTTCCTCCTATACTAATCTCTACTGGAGTAAAAGGAGATTACGCAG
TTGAGGAACGCCCTTCCCAGATGTCTGTGATGCAGCAACTAGAGGAAGGAGGACCA
GATCCTTTGGTTTTTGTTCTAAATGCAAATCTTTTGGCCATGGTTAAGATCGTGAACT
CA 02237701 1998-07-20
TABLE 5 - XSARA1 Continued
ATGTTAACAGGAAATGCTGGTGCTTTACTACAAAGGGAATGCATGCAGTGGGCCAG
GCTGAGATCGTAATCCTGTTGCAGTGCCTGCCTGATGAGAAGTGCCTGCCGAGGGAC
CTGTTTAGCCATTTTGTTGAGCTGTATCAGGAGGCAATTGCAGGTAATGTAGTGGGG
AACCTGGGGCATTCCTTCCTCAGCCAGAGTTTCCTGGGTAGTAAGGATCATGGTGGA
TTTCTTTATGTTGCACCAACCTACCAGTCCCTCCAGGACCTGGTTCTTCCTGCAGAGC
CGTACTTGTTTGGAATCCTTATTCAAAAGTGGGAGACTCCATGGGCCAAAGTGTTCC
CCATTCGGCTTATGCTGCGTTTAGGTGCAGAATACAGATTGTACCCATGTCCACTCTT
CAGTGTTCGATACAGAAAACCTCTGTTTGGGGAAACCGGACACACCATCATTAATGT
TCTAGCCGATTTCAGAAACTATCAGTATACTCTGCCAGTGGTGCAGGGCTTGGTGGT
GGATATGGAAGTCAGAAAAACTAGCATTAAAATCCCCAGCAATAGATACAATGAGA
TGATGAAAGCAATGAACAAATCCAATGAGCATGTGTTGGCCATAGGAGCATGCTTC
AACCAGATGGCAGACTCTCACCTTGTGTGTGTGCAAAACGATGATGGCAATTACCAG
ACCCAGGCAATTAGTATCCACAAACAACCACGTAAAGTGACCGGGGCCAGCTTCTTT
GTCTTCAGTGGTGCACTAAAGTCTTCTTCCGGATACCTGGCCAAATCCAGCATAGTA
GAAGATGGGGTAATGGTTCAGATCACCGCAGAGAGCATGGATGCCCTCAGACAGTC
CCTTCGGGAGATGAAGGATTTCACCATTACATGTGGAAAAGCTGATGCAGAGGAGT
CACAGGAACATGTCTATGTCCAGTGGGTGGAGGATGACAAGAACTTTAACAAAGGA
GTTTTTAGTCCAATCGATGGCAAATCAATGGAGTCTGTGACCAGCGTCAAGATTTTT
CATGGCTCAGAATACAAAGCTAGTGGAAAAATAATTCGCTGGATAGAGGTCTTCTTT
CTGGACAATGAGGAGCAACAGAGTGGCCTGAGTGACCCTGCTGATCACAGCCGACT
CACTGAAAATGTGGCCAAAGCATTCTGTTTAGCGCTTTGCCCACACCTCAAGCTACT
GAAGGAAGATGGAATGACCAGGTTAGGTCTGCGGGTGTCACTGGACTCAGACCAGG
TTGGATACCAAGCTGGGAGCAATGGGCAACTCCTGCCTGCCCGATACACCAATGATT
TGGATGGTGCTTTGGTACCAGTGATACACGGGGGCACATGCCAGTTAAGTGAAGGG
CCTGTCAGTATGGAGCTGATATTTTATATCCTTGAGAACATCTCCTAGGAAAGACAC
ATGTGTCTCCTCACAAACTGCCATCGCCCAAACCATTTGCACTTTAACCGCAAAAGA
TTCATTTTTCTTTTCTTTTGCTAACACTAGTATTAGGTCAGGGTGCGAGAGGCAGACA
CCTGAACTCTTAAACCTTCTATGCATTTTCACAGTAAGGATCAAGCTGCAGCTGGGA
ATTTCCTGTTACTAATCCAATGTGGGACGTTAGAAGTGATCGGTGGCACTGACTATC
TAGCTGTTCAACCTTCTCTGGCTCCTCTAAGGACTCTAGTGCCAGGGGGTGAGACAT
TCAAGTTTAAAACGAAAACTCTAAATACAATCAGGAATCTCACTCTGACCTCATTTA
AATCATCACTGCGACTTTTTTTCCTGCTCGCATTCTTTATTTTGCATCTTACTCAAGTT
TACATTGTCAAGACCAGCCTAAGCCTTCAGTCCTTTCTCAATTAAACTACTCGTGCAT
GGCAAGGAGACTTTCGTTGCACAGCCTGAAATATACCAATCACTTCCCAAACCACAA
GCATGAATCCAACGTTTTCCTGACTGGTTGGCTCTGCTGTGAAAGGGACAGCAATAT
TATTTTTCTACAGTTGACAAAACTTTTGTCTATGTCTGTGTCTCTCATGGGGGATTTG
TTGCCTGATGGGCAGCCTCCGGAGAGAAGAATTCCACCCGTGTGTAATATACAGTCT
AAGTGTATGGTCTGCTATGTAACACCTGTTGCGCAGTGCAAATGCACTGACTCTCTG
GAAGGCTATAGAGTTTTAAAAACGGTTAGTCTTTTAAAAAAAAAA
CA 02237701 1998-07-20
51
TABLE 6 - XSARA1
MPKMVIGDTIMAEDSLFNNTGPSEIVCNPSTVESQSLQALDDQSVNIHNEKSVLLADGFS
PCSSPKSIINFDCLTMDNEMPLHSQMS VDDNDKETVTISVLPTIIQDTSNV STDPAINKPGT
KEPHRALKETTSVILPEIKPYSTCAALSFENNNKVPSYQLNNTDLLSVSPVVEACSEQQQ
KHTSSLHEEKLFEGVSATESFAATAAETVLDNEALRSAEFFDIVVKNFSDSCVINGDLTK
SCGLSQESNEKFCASKEFEGGVDANVLLENACVAYKEAIDLPEENGTNAPMSLYNGCDS
YGMKNPAVAQNPKNLPSKEDSVTEEKEIEESKSEYYTGVYEQQREDDVTERGGLLLNA
KADQMKNNLHSLCNQVPSMHGQTSPKKGKIVQSLSVPYGGARTKQPTHLKLHIPKPLSE
MLQSDLIPPNAGCSSKYKNDMLNKSNQGDNLISESLREDSAVRSPVTDANGDFPGEYRG
PGSLCLAVSPDSPDNDLLAGQFGVPISKPFTTLGEVAPVWVPDSQAPNCMKCEARFTFT
KRRHHCRACGKVFCAACCSLKCKLQYMDKKEARVCVICHSVLMNAQAWENMLSASV
QSPNPNNPAEYCSTIPPMQQAQASGALSSPPPTVMVPVGVLKHPGTEGSQSKEQRRVWF
ADGILPNGETADSDNANVTTVAGTLTVSHTNNSTSSESENTSGFCGSITQVGSAMNLIPE
DGLPPILISTGVKGDYAVEERPSQMSVMQQLEEGGPDPLVFVLNANLLAMVKIVNYVNR
KCWCFTTKGMHAVGQAEIVILLQCLPDEKCLPRDLFSHFVELYQEAIAGNVVGNLGHSF
LSQSFLGSKDHGGFLYVAPTYQSLQDLVLPAEPYLFGILIQKWETPWAKVFPIRLMLRLG
AEYRLYPCPLFSVRYRKPLFGETGHTIINVLADFRNYQYTLPVVQGLVVDMEVRKTSIKI
PSNRYNEMMKAMNKSNEHVLAIGACFNQMADSHLVCVQNDDGNYQTQAISIHKQPRK
VTGASFFVFSGALKSSSGYLAKSSIVEDGVMVQITAESMDALRQSLREMKDFTITCGKA
DAEESQEHV YVQWVEDDKNFNKGVFSPIDGKSMES VTS VKIFHGSEYKASGKIIRWIEV
FFLDNEEQQSGLSDPADHSRLTENVAKAFCLALCPHLKLLKEDGMTRLGLRVSLDSDQV
GYQAGSNGQLLPARYTNDLDGALVPVIHGGTCQLSEGPVSMELIFYILENIS*
CA 02237701 1998-07-10
52
TABLE 7 - XSARA2
AGTTTTATTTTCAGAAGACGTTGCATCTTTATTTTAAACATTAAGTTTCACTATGTAG
TAAAACATTACTGTTGTATATACAGTATGTTGTAGACATATAACGTAACTGTTTGCTT
TGTGCTTTCTTTCCTCCTCAGATGAAACTGTCTTTCCAAAGCTGTTAGATGCTAAGTG
GAATCAATTCTTAGAACCACATTCGCATAAAGTCACTGATAAACCAGCTCTTGACAA
TGTCTGTAAATCAATCATTGCTATTGAAGCTCATCTCAAAGTCAGGTCACCCAGCTT
GACAGCCCTTGCAAGGTCCACATATGTGAATGGAGAAGTAGGTATTGTGACTCCTGA
AATGCCTAAAATGGTGATAGGAGACACCGATATGGCAGAGGATTCACTTTTTAACAC
TGGTCCCTCTGAAATTGTATGCAACTCTATTGTGGAGAGTCAAAGTTTAGAAGTTTT
AGATGATGTACCAGTGAGTATTAACAATGAAAAAAGTGTTCTTCTTGATGATGGATT
TTCTCCGTACAGTAGCCCCAAAAGTGTTCTAAACTCTGCTTGCTTGACCATGAATAA
CGGAAAGCCCTCACACGGTCAAAAAATTGTTAATGACCAAGATAAAGAAGCTGTAA
CAATTTCAGTCCTTCCAATGATCATACAGGATACTACTAACGTAAGCACAGACCCAG
CTTTCAATAAATCTGGCACTGAAGAAGCTTATAGTGCATTAAAACAAACCACATCAG
TTATTCTGCCTGAAATAAAGCCTTATTCCATACAGGCTGCCCTTTCATGTGAAAATAT
CAACAAGATACCCAGATGTCAATTAAATAATACAGATCTACTCAGCATTTCACCAGT
GGTTGAAGCATGTAGTGAGAAGCAGCAAAATCATACAACTTCCTTGCATGAAAAAA
AACTTGCAGCTGTGTCTGCAACTGCGTTCTTTCCAGTCACTGCTGCTGAAACTGTACT
AGGTAATGAAGCTCTCCATAGTGCTGATTTTTTTGACATTGTTGTAAAGAACGTTTCT
GACTCGTGTGTGTTTAATGGTGACCTAACTAGAACTAATGGACTCTCACAAGAAAAC
AATGAAATGTTTTATGCAAGTAAAGAGTTGGAAGGAGGGGTAGATGCTAATATCTT
ATTGGAAGATGCATGCATAGCTTATAAAGAAAGAATAGATTTGTCTGAAGAAAATG
GAACTAATGCACCAATGTATCTGTACAATGGGTGTGATTCCTATGGAATGAAAAACC
CTGCTGTACGTCAAAACCCAAAGAATTTACCATCAAAAGAAGATTCTGTGACAGAA
GAAAAAGAAATTGAAGAAAGCAAGTCAGAATACTATTCTGGTGTTTATGAACAACA
GAAGGAAGATGACATAACTGAGAGAGGTGGAGTCTTGTTAAATGCCAAGGTTGACC
AAATGAAGAACAGTTTGCATAGTCTTTATAATCCGGTTCCATCCATGCATGGGCAAA
CCTCACCAAAAAAGGGCAAGATTGTGCAATCCCTCAGTGTTCCATATGGTGGAGCTC
GCCCCAAGCAGCCAACTCATCTCAAACTCAATATTCCACAGCCATTGTCTGAAATGT
TACAGTGTGATCTCATTCCGCCAAATGCTGGATGCAGCTCTAAAAACAAAAATGACA
TGTTAAACAAATCAAATCGGGGGGATAACCTGATTTCAGAATCACTACGTGAGGAA
GTGCACAGCCCTGTTACTGATACAAATGGTGAAGTCCCTCGAGAAAACAGGGGACC
TGGCAGCCTGTGCCTTGCAGTGTCTCCAGACAGCCCTGACAATGATCTGCTTGCTGG
ACAGTTTGGGGTACCCATCTCTAAGCCATTTACTACTCTAGGGGATGTGGCTCCAGT
CTGGGTGCCAGATTCCCAAGCACCAAACTGCATGAAGTGCGAGGCCAGATTTACATT
TACCAAAAGGAGGCATCACTGCCGAGCTTGTGGAAAGGTATGTAAAGAAATGTGGT
GTTTCATCAGGGCAACAGTAATCACGGCAAATTATTCATAACAAAATGTGTTCAGCA
GATTCAGTTAAAGTAGACTTATAAGTTACACAGTAACAATTCATCTGCTCAGCCTCA
TTTTGAAGTAGATAAAATATATTTTATTAGGAAACTCTGGGGAGATATAAGGGAAAG
CTTGCCTAAAAGTAGATGTTCTGTATATTATTTGGTAGTCAAAGATGATTTCATGAA
AAAAGGTTATTTGTAAAAAGTACAAAATGGGTAGAGACTAGACAATAAAAAGTAAG
GAGTAAAAAACTAGGTATGTAACGTATATTAAAATAATTTTATGATTTTAATATTTA
CTGCACATTTTCTACAGTGCAGTGATTTGTATAACCATGCAATTATCAAATGCTTAGT
GCCTTCACACAAAGTGCCTTTAATAAAAATTATTTTATAAATTATCATATTTTCTTTA
TATGTAGTCATCATCTTTTTTGTCTCATTTCTTGGAATCGTTCTACTTATGTTCTACTG
ATATGTTTTTTACCCGAGACCTATCTTGTCCTCTAAAGTAATTGGCTTGTCAACTGGC
CA 02237701 1998-07-20
53
TABLE 7 - XSARA2 Continued
TGTAGGGGGATTTTCAGAGTTATAGCTTAGTACTGTTAATGAGCCATAGGTTGAAAT
AGTGCTCTAGATTTACATGTTGTACAACAGTTATTGCAATATGTGTAGGGGGGGGG
CA 02237701 1998-07-20
54
TABLE 8 - XSARA2
MPKMVIGDTDMAEDSLFNTGPSEIVCNSIVESQSLEVLDDVPVSINNEKS VLLDDGFSPY
SSPKS VLNSACLTMNNGKPSHGQKIVNDQDKEAVTIS VLPMIIQDTTNVSTDPAFNKSGT
EEAYSALKQTTS VILPEIKPYSIQAALSCENINKIPRCQLNNTDLLSISPV VEACSEKQQNH
TTSLHEKKLAAVSATAFFPVTAAETVLGNEALHSADFFDIVVKNVSDSCVFNGDLTRTN
GLSQENNEMFYASKELEGGVDANILLEDACIAYKERIDLSEENGTNAPMYLYNGCDSYG
MKNPAVRQNPKNLPS KEDS VTEEKEIEES KSEYYSGVYEQQKEDDITERGGVLLNAKVD
QMKNSLHSLYNPVPSMHGQTSPKKGKIVQSLSVPYGGARPKQPTHLKLNIPQPLSEMLQ
CDLIPPNAGCSSKNKNDMLNKSNRGDNLISESLREEVHSPVTDTNGEVPRENRGPGSLCL
AVSPDSPDNDLLAGQFGVPISKPFTTLGDVAPVWVPDSQAPNCMKCEARFTFTKRRHHC
RACGKVCKEMWCFIRATVITANYS
CA 02237701 1998-07-20
TABLE 9
hSARA~E;N - E~CGN~AC'' NVLYYAYM CDKRT ~ NDLpD~NNYNS 77
XSAR pIKR~YSW~Dp~SAVE LM 70
A MPK V~,OTI S FNNTuGPS IV Sp~---S OA ~
NPST -----VN1 EKSVL DGFSP SSP
Q 11
' --
:
hSARAAF L ENRpTDpFSFSINEST D N LNR KT R YNH~CPTSS SLASVC S L DDGSI RDPSMS157
XSARAKp ~NDK~---~TV~ISV~PTIIp~TSNVST~ 127
FD~L M ----------------~IPLHAINP
pMS ~TKEPHR
~
----
hSARA~LTSL YDS ISS GTDG V Kp -Y1 D~T KISSPR PNSF H GILMKKEPAEE TT S S 236
XSARADE L 198
L ETSVILQPEIK~PYST~A L' FE~V~NKpN~------N -L~---V~P~J~ACSEpppKHTSLH
aSY WK~FESV
hSARALLLKPDMPNGSGRNNDCERCSDCLVPNEVR~DEN~YEHE T- T ;LN~TE~FSESpD~TK LNE~DSpV
316
XSARAi ~ 250
ITA .
ATES------------------------FAfi~
II~
FDI~YVL
INFSDSCh
iTVLDN A A
A
D SCGL --
4
~ ,
I ~
-
~
I
~
J
L
I
hSARAK - LpIS~~DTNGDS . CVGLA 386
XSARA~GG ~K T: ISES~ECFSTVIDPNY- 319
pDG ~SFV T
~
~I
rCASKEFf
1------ -----V NVL rE A'
VA Ki
N
AI
LPEENG
N MS KN
A ApN N
" .,
E ~
:l I
CCflfl
II~~
hSARASNI GNE S STG L K Y NF S YL S ~VVASL t F 475
XSARA~TGV p~EDD~V G AKA~~p N- - 398
= SL~N ~ MH i' KGK I Y~
L Y'
K ,-
~
hSARAP S'N Q DH ~-F~A.S N I G A L E SA~VCSP-~LGN1SNV -1 E LES~EAE553
XSARAT TH [~
' H' M ~
O
'
l l P A SS ",M ,N S D 478
, LI
-SLRED AVRSPVT A
:. DFPGE RGP
~
hSARA I ~T R P L A '- R 'Cf I A R 633
XSARA G - L Y S L V' i S 556
CA 02237701 1998-07-20
56
TABLE 10
hSARA 587 . P ~ S L L ;~ D I ° S V , 655
XSARA 510 ~ P K' E ~ K ' A C L ~ C K ~1 D ~ . S V ' 578
KIAA0305 737 y ~ .~'~ P ~~! 'p K C ~ C K = E V T 1
i v - i
800
FGD1 720 , P i T M ~ .R ~,p E P ~ 1 K < ,~H F " A R '" ..,; D N N R - S N - D
~, A " 485
i n v ~ v
Hrs 153 D Ea V EK ~ pI FGIEK ',"~.:'P , 'p ~ 219
Hrs-2 153AA ~ D E V G ' T VK - ~_ ~-p1 _ K K FG i EK~ , P : p r 219
EEA-I 1341TpALNfRK E~J Y,:p ° 1KG V.. ~~ 1 vE ;. AK PSSK~- -PV
._~° A F D ~ 1408
. . . .. p.W. . . . . . . .C. .C. . . F. . .. .R~~pp~-1~~ ~. .. . . .. . . . .
. .. . ~. .C.