Note: Descriptions are shown in the official language in which they were submitted.
CA 02244735 2006-10-24
WO 9727480 PCP/IB97/00071
Description
Technical Field
The present invention relates to an in-vitro method of assessing the
chemotherapy of patients who are HIV positive,
for use by physicians treating such patients based on
the phenotypic drug sensitivity of human HIV strains for inhibitors of
one or more enzymes of the pol gene of HIV, as well as a method for
simultaneously determining the phenotypic drug sensitivity of two or
more of the enzymes of the pol gene of HIV to inhibitors thereof.
Background Art
To date, several chemotherapeutic regimens have been developed
for treating HIV infected patients. Certain of these regimens have been
approved for clinical use, and others are the subject of on-going clinical
trials. It can be assumed that the number of approved chemotherapeutic
regimens will increase steadily in the near future. Increasingly,
combination therapy or multiple drug treatment regimens are being used
because of the development of drug-resistant HIV variants daring
therapy. Although these chemotherapeutic regimens have been shown to
exert an effect on virological (viral load), immunological and clinical
parameters of HIV disease, practical experience teaches that these effects
are transient. In particular, one finds that the HIV strains infecting an
individual patient after a. while start to display reduced sensitivity to the
drug or drug combination with which said patient is being treated. The
loss of efficacy of the chemotherapy can vary from patient to patient,
from drug to drug, or from drug combination to drug combination. It is
well established that the loss of efficacy to a particular type of
chemotherapy can be associated with a genotypic pattern of amino acid
changes in the genome of the HIV strains infecting the patient. This
probably renders these HIV strains less susceptible to the chemotherapy.
CA 02244735 1998-07-23
WO 97/27480 PCT/IB97/00071
2
As an HIV infected patient is exposed to several chemotherapeutic
regimens over extended periods of time, more complex patterns of
amino acid changes in the genome of infecting HIV strains occur which
for the present defeat a rational approach to the further treatment of the
infected patient. As implied in the previous explanation, one can
routinely determine the genotypic changes occurring in HIV strains
exposed to different chemotherapeutic regimens involving single or
multiple anti-HIV drugs, but thus far it has proven very difficult to
derive from these data information enabling a physician in charge of
prescribing the chemotherapy whether or not it is sensible to initiate or
continue a particular chemotherapeutic regimen. In other words, the
genotypic information which is available on a limited scale, cannot
routinely be translated into phenotypic information enabling the
responsible physician to make the crucial decision as to which
chemotherapy a patient should preferably follow. The problem also
exists for drug-naive patients who become infected by drug-resistant
HIV strains.
Viral load monitoring is becoming a routine aspect of HIV care.
However, viral load number alone cannot be used as a basis for deciding
which drugs to use alone or in combination.
Combination therapy is becoming increasingly the
chemotherapeutic regimen of choice. When a person using a
combination of drugs begins to experience drug failure, it is impossible
to know with certainty which of the drugs in the combination is no
longer active. One cannot simply replace all of the drugs, because of the
limited number of drugs currently available. Furthermore, if one
replaces an entire chemotherapeutic regimen, one may discard one or
more drugs which are active for that particular patient. Furthermore, it is
possible for viruses which display resistance to a particular inhibitor to
also display varying degrees of cross-resistance to other inhibitors.
Ideally, therefore, every time a person has a viral load test and a
viral load increase is detected, a drug sensitivity/resistance test should
CA 02244735 1998-07-23
WO 97/27480 PCT/1B97/00071
3
also be carried out. Until effective curative therapy is developed,
management of HIV disease will require such testing.
Currently there does exist a phenotyping method which is based
on virus isolation from plasma in the presence of donor peripheral blood
mononuclear cells (PBMCs), and subsequent phenotyping in said cells
(Japour, A.J., et al. (1993) Antimicrobial Agents and Chemotherapy;
Vol. 37, No. 5, p1095-1101). This co-cultivation method, which is
advocated by the AIDS Clinical Trial Group (ACTG) - particularly for
phenotyping AZT (synonymous herein with zidovudine/Retrovir
(Retrovir is a Trade Mark)) resistance, is time-consuming, costly and too
complex to be used on a routine basis.
A phenotypic recombinant virus assay for assessment of drug
susceptibility of HIV Type 1 isolates to reverse transcriptase (RT)
inhibitors has been developed by Kellam, P. and Larder, B.A.
(Antimicrobial Agents and Chemotherapy (1994) Vol. 38, No. 1, p23-
30). This procedure allows the generation of viable virus by
homologous recombination of a PCR-derived pool of RT coding
sequences into an RT-deleted, noninfectious proviral clone,
pHIVORTB stEII. Analysis of two patients during the course of
zidovudine therapy showed that this approach produced viruses which
accurately exhibited the same genotype and phenotype as that of the
original infected PBL DNA. However, the procedure involves isolation
of the patient virus by co-cultivation of patient plasma or patient PBMCs
with donor PBMCs. Such prior cultivation of virus may distort the
original virus composition. Furthermore, this method, although allowing
one to determine the sensitivity of the isolates to various inhibitors, does
not provide the physician with information as to whether to continue
with the existing chemotherapeutic regimen or to alter the therapy.
Also when one enzyme only of the pol gene is being studied, the
method does not readily lend itself to routine phenotypic assessment of
combination therapy which conventionally involves the use of one
protease and 2 RT inhibitors.
#t J4'iYJA \ c Y i 11) li ad
CA 02244735 2006-10-24
4
The nested PCR (polymerase chain reaction) procedure used in the
recombinant virus assay can lead to a situation where the recombinant
virus does not truly reflect the situation with the HIV strains infecting
the patient under investigation. This problem resides in DNA sequence
homology and the minimum amount of homology required for
homologous recombination in mammalian cells (C. Rubnitz, J. and
Subramini, S. (1984) Molecular and Cellular Biology Vol. 4, No. 11,
p2253-2258). Accordingly, any phenotypic assay based on the
recombinant virus approach should endeavour to ensure that as much as
possible of the patient material is amplified and that there is maximum
recombination.
Thus, the RNA extraction and nested PCR procedures employed
should ensure that the viral genetic material is amplified such that the
amplified material maximally reflects the viral genetic diversity in the
patient being investigated.
In current clinical practice there is therefore a hard-felt need (a) to
determine rapidly and on a routine basis the phenotypic drug sensitivity
of HIV strains infecting a particular patient, (b) to process the thus
obtained data into easily understood information, and (c) to initiate,
continue or adjust on the basis of said information the chemotherapy
prescribed for said particular patients.
Disclosure of the Invention
According to a first aspect of the invention there is provided an in-vitro
method of assessing HIV chemotherapy of patients who are HIV
positive, which comprises transfecting a cell line susceptible to infection
by HIV with a sequence from the pol gene of HIV, obtained by isolating
viral RNA from a sample of a biological material from a patient and
reverse transcribing the desired region of said pol gene, and a HIV-DNA
construct from which said sequence has been deleted, culturing said
transfected cells so as to create a stock of chimeric viruses,
assessing the
phenotypic sensitivity of said chimeric viruses to an inhibitor of said
CA 02244735 1998-07-23
enzyme encoded by the pol gene of HIV and assigning a value thereto,
constructing a data set comprising said value for chimeric virus
sensitivity and the corresponding value for a chimeric wild-type strain of
HIV, repeating the sensitivity assessment for at least two further
5 inhibitors and thereby constructing at least three such data sets in total,
representing said data sets in two dimensional or three dimensional
graphical form such that the difference between the chimeric and wild-
type sensitivities in the case of each data set provides a visual measure of
the resistance of the chimeric stock to treatment by the inhibitor in
question, and selecting the optimum inhibitor(s) on the basis of the
graphical representation of the resistances so measured.
The method according to the invention yields phenotypic
information on individual HIV infected patients on a large scale,
economically and rapidly. The method is applicable to all currently
available chemotherapeutic regimens and it is expected to be equally
applicable to future chemotherapeutic regimens.
The method according to the invention provides the physician
with phenotypic data on patient HIV strains which can be immediately
used to determine whether a particular chemotherapeutic regimen should
be initiated, continued or adjusted.
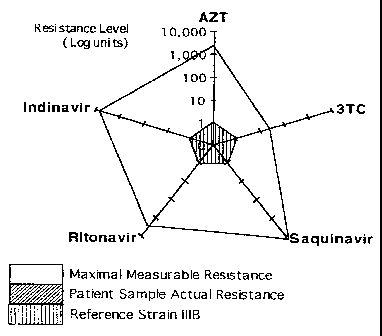
Preferably, the data sets are represented on a polygonal or quasi-
circular graph comprising:
(a) a plurality of normalised axes extending radially from an
origin, each axis corresponding to one data set or inhibitor
or combination thereof;
(b) the axes being normalised such that the sensitivity values
for wild-type HIV for the various inhibitors are equal on
each axis, the data points for wild-type HIV being
optionally represented and connected to form a regular
polygon whose vertices lie on the axes and whose center is
defined by the origin;
AMENDED SHEET
CA 02244735 1998-07-23
WO 97/27480 PCT/1B97/00071
6
(c) on each axis a data point representing the sensitivity value
of the chimeric HIV stock against the inhibitor
corresponding to said axis is plotted, the chimeric data
points being optionally connected to form a regular or
irregular polygon the shape of which represents the
resistance of the chimeric stock to a range of inhibitors.
A polygonal or quasi-circular graph provides the advantage that
the patient's resistance to a number of drugs is characterised in terms of
the degree of divergence between the polygon representing the patient's
chimeric HIV stock and the polygon representing the wild-type strain.
The areas of the polygons will generally diverge more in some areas than
in others, indicating in each case a greater or lesser degree of resistance
to the inhibitor whose axis passes through the area in question.
Thus, the method according to the invention takes a chimeric HIV
stock and provides a map of the resistance of this stock across a range of
inhibitors. In this way the map or graph provides a technical
characterisation of an aspect of the chimeric stock which is not obtained
by conventional measurements.
In a preferred embodiment, the normalised axes are equiangular
from one another.
Further, preferably, each axis has a logarithmic scale.
Further, preferably, eccentric data points in the chimeric polygon,
if represented, identify inhibitors whose usefulness can be assumed to be
of little or no benefit to the patient, while data points lying within, on or
close outside the wild-type polygon identify inhibitors whose usefulness
can be assumed to be of substantial benefit to the patient.
When worst case values are represented along with the chimeric
and wild-type HIV, a usually irregular polygon encloses the chimeric
and wild-type polygons. The meaning of the term "eccentric" as used
above denotes a data point lying relatively close to the worst-case border
1,:' 11,40A
CA 02244735 1998-07-23
WO 97/27480 PCT/1B97/00071
7
and relatively far from the wild-type polygon. Similarly the term "close
outside the wild-type polygon" refers to relative closeness to the wild-
type polygon when compared to the distance from the worst-case border.
The method as hereinabove defined is limited in the sense that the
measurable resistance against an inhibitor is dependent on the particular
range of concentrations of the inhibitor used. Also one must endeavour
to reduce the effects of biological variability. Accordingly, it is
desirable to obtain a value for maximum or worst-case measurable
resistance where it is assumed that a given inhibitor has no effect. This
concentration, e.g. 100 M, is generally the maximum concentration that
can practically be tested, but may also be derived from e.g.
pharmacological, toxicological or pharmacokinetic studies. The
comparison of the resistance level of the patient under investigation and
the maximum measurable resistance determines what is the significant
level of resistance for the patient under investigation. The maximum
measurable resistance and the actual resistance can be suitably shown on
a bar graph as hereinafter described.
In a still further preferred embodiment of the invention each of
said three or more data sets further comprises a value for worst-case
measurable resistance for the inhibitor in question, said worst case
values being represented on said graphical representations such that the
data point for the chimeric stock can be compared both to wild-type and
to worst-case HIV, thereby providing an assessment of the relative value
of the inhibitor in a particular case.
Experiments with in excess of 150 patient samples have revealed a
close correlation between resistance development and therapy history as
hereinafter further illustrated in the Examples. A close correlation has
been found with the data generated in accordance with the invention
relative to classical virus isolation and phenotyping techniques.
The method in accordance with the invention can thus be used for
an individualised and more rational management of HIV chemotherapy.
Thus, use of the method according to the invention in combination with
CA 02244735 2006-10-24
WO 97127480 PCTlIB97/00071
8
the proper administration of anti-HIV drugs should ultimately lead to
better treatment and survival of patients infected with the HIV virus.
The method according to the invention has particular application
where an individual patient has been receiving many different drugs and
his mutation pattern is not readily interpreted by attending physicians.
According to a further aspect of the invention there is provided a
method of assessing HIV chemotherapy of patients who'are HIV
positive, which comprises the steps of:
(a) periodically assessing the phenotypic sensitivity of a
patient's HIV strains by a method hereinabove described;
(b) issuing instructions to maintain the chemotherapy with the selected
inhibitor
while the patient's HIV strains remain susceptible to the
selected chemotherapy; and
(c) to select a different inhibitor if and when the susceptibility
to the original inhibitor decreases.
*
We have coined the term "Antivirogram" for the clinical
management device disclosed herein and this term will be used
hereinafter in the specification. This device provides the physician with
a clear representation of the relative changes and susceptibilities for
different inhibitors which are or which may be used in the clinical
management of individual HIV-infected patients.
By HIV herein is generally meant HIV-1. However, the invention
is also applicable to HIV-2.
* Trade-mark
CA 02244735 2006-10-24
9
Preferably, the phenotypic sensitivity of said chimeric viruses to
inhibitors of at least two enzymes encoded by the pol gene of HIV is
simultaneously assessed.
In a further aspect of the invention there is provided a method of
determining the phenotypic drug sensitivity of individual HIV strains in
a patient to inhibitors of at least two enzymes encoded by the pol gene of
HIV, which comprises transfecting a cell line susceptible to infection by
HIV with a sequence from the pol gene of HIV, obtained by isolating
viral RNA from a sample of a biological material from a patient and
reverse transcribing the desired region of said pol gene, and a HIV-DNA
construct from which said sequence has been deleted, culturing said
transfected cells so as to create a stock of chimeric viruses
and assessing
the phenotypic sensitivity of said chimeric viruses to inhibitors of said
enzymes encoded by the pol gene of HIV.
The desired sequence from the pol gene is isolated from a sample
of a biological material obtained from the patient whose phenotypic drug
sensitivity is being determined. A wide variety of biological materials
can be used for the isolation of the desired sequence.
Thus, the biological material can be selected from plasma, serum
or a cell-free body fluid selected from semen and vaginal fluid. Plasma
is particularly preferred and is particularly advantageous relative to the
use of PBMCs as used in the prior art described above.
Alternatively, the biological material can be whole blood to which
an RNA stabiliser has been added.
In a still further embodiment, the biological material can be a solid
tissue material selected from brain tissue or lymph nodal tissue, or other
tissue obtained by biopsy.
As hereinafter demonstrated, when a biological material such as
plasma is used in the isolation of the desired sequence, a minimal
CA 02244735 2006-10-24
WO 9797480 PCT/1B97/00071
volume of plasma can be used, typically about 100-250 1, more
particularly of the order of 2O0 1.
Further, preferably the two enzymes selected will be selected from
HIV RT, protease and integrase.
5 Viral RNA is conveniently isolated
by methods known per se, for example the method of Boom,
R. et al. (Journal of Clinical Microbiology (1990) Vol. 28, No. 3, p.495-
503).
In the case of plasma, serum and cell-free body fluids, one can
10 also use the QlAamp viral RNA kit marketed by the Qiagen group of
companies.
Preferably, the cell line susceptible to infection by HIV is a CD4+
T-cell line.
Further, preferably, the CD4+ T-cell line is the MT4 cell line or
the HeLa CD4} cell line.
Reverse transcription can be carried out with a commercial kit
such as the GeneAmp Reverse Transcriptase Kit marketed by Perkin
Elmer.
The desired region of the patient pol gene is preferably reverse
transcribed using a specific downstream primer.
In the case where the sequence to be reverse transcribed is that
coding for reverse transcriptase or reverse transcriptase and protease, the
downstream primer is preferably OUT3 : 5'-CAT TGC TCT CCA ATT
ACT GTG ATA TTT CTC ATG-3' (SEQ ID NO: 1).
In a particularly preferred embodiment a patient's HIV RT gene
and HIV protease gene are reverse transcribed using the HIV-1 specific
OUT 3 primer and a genetically engineered reverse transcriptase lacking
* Trade-mark
WO 97ZZ7480 CA 02244735 2006-10-24
PCT/IB97/00071
11
RNase H activity, such that the total RNA to be transcribed is converted
to cDNA without being degraded. Such a genetically engineered reverse
transcriptase, the Expand* reverse
transcriptase, can be obtained from Boehringer Mannheim GmbH.
*
Expand reverse transcriptase is a RNA. directed DNA polymerase.
The enzyme is a genetically engineered version of the Moloney Murine
Leukaemia Virus reverse transcriptase (M-MuLV-RT). Point mutation
within the RNase H sequence reduces the RNase H activity to below the
detectable level. Using this genetically engineered reverse transcriptase
enables one to obtain higher amounts of full length cDNA transcripts.
Following reverse transcription the transcribed DNA is amplified
using the technique of PCR.
Preferably, the product of reverse transcription is amplified using
a nested PCR technique.
Preferably, in the case where the region of interest is the RT
region, a nested PCR technique is used using inner and outer primers as
described by Kellam, P. and Larder, B.A. (1994 supra). When the
region of interest is that spanning the RT and protease genes, the specific
primers used are preferably a combination of OUT 3/IN 3 (downstream)
and RVP 5 (upstream).
The primer RVP 5 (Maschera, B., et al. Journal of Virology, 69,
5431-5436) has the sequence 5'-GGGAAGATCTGGCC
TTCCTACAAGGG-3' (SEQ ID NO: 2).
A schematic representation of the amplification is set forth in Fig.
3 and is described in greater detail in Example 2.
The amplification of the protease cDNA actually involves a hemi-
nested PCR procedure as will be apparent from Fig. 3.
* Trade-mark
CA 02244735 2006-10-24
WO 97127480 PCT/1B97/00071
12
The nested PCR technique has the advantage over the known
simple PCR techniques in that it enables one to obtain the most specific
PCR product.
However, to obtain an even higher fidelity and yield during PCR,
one can make use of a mixture of thermostable polymerases (Barnes,
W.M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2216-2220). Such a
polymerase mixture is available from Boehringer Mannheim GmbH,
namely the Expand (Expand is a Trade Marky high fidelity PCR system.
Using this system we have obtained increased sensitivity, namely a
sensitivity which is ten times or greater than that obtained with a
conventional PCR procedure using Taq polymerase alone.
When the region of the pol gene is that embracing the RT and
protease genes, preferably the HIV-DNA construct is one from which
the RT and protease genes are deleted and is the plasmid pGEMT3-
APRT as deposited at the Belgian Coordinated Collections of
Microorganisms-BCCM LMBP-Collection on November 8, 1996 under
the number LMBP3590.
However, several approaches can be adopted to generate a
plasmid containing the HIV-1 provirus carrying a deletion for the
protease as well as for the RT gene. One possibility is the introduction
of the desired deletion by means of oligonucleotide-mediated
inutagenesis. However, the procedure adopted hereinafter in Example 2
involves the generation of the desired construct by making use of
specific restriction enzymes and subcloning procedures, as hereinafter
described. Although the final results depend on the available restriction
sites a major advantage of this procedure is that one can obtain
conclusive results rapidly.
To ensure the most efficient outcome for the transfection, the
PCR-product, being transfected, should ideally be purified by anion
exchange spin columns in a manner known per se. A suitable kit is the
QlAquick' PCR Purification Kit marketed by the Qiagen group of
companies.
* Trade-mark
CA 02244735 1998-07-23
WO 97/27480 PCT/M97/00071
13
Transfection can be achieved by electroporation or, alternatively,
by the use of lipids, especially cathionic lipids, DEAE dextran, CaHPO4,
etc.
In the case of lipid transfection one can avail of a PERFECT
(PERFECT is a Trade Mark) transfection kit marketed by Invitrogen
B.V. of Leek, the Netherlands.
Thus, for transfection an HIV-DNA construct from which the
gene or genes of choice from the pol gene has/have been deleted is used
in conjunction with the product obtained following amplification.
The construct can be the plasmid pHIVART (obtainable from the
Medical Research Council (MRC)) if it is the RT gene only that is
deleted. When the RT and protease genes are both deleted a suitable
HIV DNA construct is the plasmid pGEMT3-zPRT described herein and
which is a high copy vector. Such plasmids are linearised prior to
transfection according to methods known per se.
A particular advantage of using a construct coding for more than
one pol gene enzyme, for example a OPRT construct, is that one is more
likely to include more of the original patient material in the construct
than if a single gene is used, so that the amplified material reflects to a
greater extent the viral genetic diversity in the particular patient being
investigated.
It will be appreciated that it is preferable that the specific primers
selected for the nested PCR are located outside the body sequences of
the target enzymes to be amplified and investigated. It will furthermore
be appreciated that a combination of RT and protease is likely to provide
better results for studying RT than RT alone, because forty more amino
acids are patient borne relative to the situation with RT alone. For
studying the protease, one should be aware that the first nine amino acids
of the protease are still derived from the construct's (pGEMT3-OPRT )
wild-type backbone.
CA 02244735 1998-07-23
14
When the transfection of the cells is achieved through
electroporation, the parameters selected are optimized to achieve good
cell growth and virus production. The electroporation can conveniently
be conducted at approximately 250 F and 300V. Preferably, the
electroporation is conducted in the presence of about 10 g of linearised
plasmid e.g. pHIVORTBstEII and about 5 g of amplified PCR product
e.g. RT PCR product. Upon successful intracellular homologous
recombination, new chimeric HIV is formed within 5 to 10 days. With
known techniques typical cultivation times are 12-14 days before
chimeric HIV is formed. Culture supernatant aliquots are stored at -
70 C or lower temperatures.
It is readily seen that one can use the above methods for isolating
and amplifying other HIV genes, e.g. the integrase gene, or more than- -
one other HIV gene, e.g. both the RT and the integrase gene, and
transfecting a CD4+ T-cell with the respective integrase or RT/integrase
PCR products in conjunction with an appropriate linearised HIV-DNA
construct from which the relevant gene (or genes) is deleted.
The newly formed chimeric viruses are titrated and then analysed
for their phenotypic sensitivity (i.e. susceptibility) to the different pol
gene enzyme inhibitors, preferably in an automated cell-based assay.
Preferably, the phenotypic drug sensitivity of the chimeric viruses
and of the wild HIV strain, which is suitably a recombinant wild HIV
strain, to one or more RT, protease or integrase inhibitor(s) is expressed
as an inhibitory concentration (IC value).
The susceptibilites of the chimeric viruses and of the wild type
HIV strain to one or more RT inhibitors and/or one or more protease
inhibitors and/or one or more integrase inhibitors can be expressed as for
example 50% or 90% inhibitory concentrations (IC50 or IC90 values).
Preferably, RT inhibitors are selected from nucleoside RT
inhibitors such as AZT, ddl (didanosine/Videx (Videx is a Trade Mark),
ddC (zalcitabine), 3TC (larnivudine), d4T (stavudine), non-nucleoside
AMENDED SHEET
CA 02244735 2006-10-24
WO 97/27480 PCT/11197100071
RT inhibitors such as delavirdine (U 9051125 (BMAP)/Rescriptor ),
loviride (alpha-APA), nevirapine (B 1-RG-
587/Viramune* and tivirapine (8-Cl-
TIBO(R86183)), protease inhibitors such as saquinavir, indinavir and
5 ritonavir and integrase inhibitors such as caffeic acid phenylethyl ester
(CAPE).
Suitable RT and/or protease inhibitors and/or integrase inhibitors
are selected from nucleoside RT inhibitors such as AZT, ddl, ddC, 3TC,
d4T, 1592U89 and the like, non-nucleoside RT inhibitors such as
10 loviride, nevirapine, delaviridine, ateviridine, and tivirapine (8-Cl TIBO)
and the like, protease inhibitors such as saquinavir, indinavir and
ritonavir and the like, and integrase inhibitors such as caffeic acid
phenylethyl ester (CAPE) and HIV integrase inhibitors of the type
described in WO 95/08540 and GB 2,271,566.
15 The method according to the invention may compromise the step of
comparing the phenotypic drug sensitivity of patient HIV strains with
one or more RT inhibitors and/or one or more protease inhibitors, and/or
one or more integrase inhibitors to that of a wild type HIV strain. For an
easy-to-understand representation of the relative changes in
susceptibility to the different drug compounds (or combinations) tested,
an Antivirogram graph, is constructed.
The graph should be interpreted as follows : eccentric data points
in the Antivirogram* identify chemotherapeutic regimens unlikely to
benefit the HIV infected patient any further, whereas data points within
or on the reference polygon, or only slightly beyond the reference
polygon, identify chemotherapeutic regimens likely to benefit the HIV
infected patient.
The methods according to the invention in combination with the
administration of the correct anti-HIV drugs should ultimately lead to
better treatment, improved quality of life and improved survival of HIV
infected patients ; i.e. ineffective treatment (due to the presence of or
* Trade-mark
CA 02244735 2006-10-24
WO 97/27480 PCT/1B97/00071
16
emergence of resistant HIV strains) can be prevented or halted, and
effective chemotherapy can be initiated in good time.
Brief Description of the Drawings
Fig. 1 is a schematic representation of the construction of the
plasmid pGEMT3-APRT;
Fig. 2 is a further and complementary schematic representation
of the construction of the plasmid pGEMT3-APRT;
Fig. 3 is a schematic representation of that part of the HIV-
HXB2D sequence containing protease and RT genes;
Fig. 4A-H is a complete sequence for that part of the HIV-
HXB2D sequence containing protease and RT genes;
Fig. 5 is an Antivirogram for a patient harbouring 3TC resistant
HIV strains as described in Example 5;
Fig. 6 is an Antivirogram for a drug-naive patient harbouring
wild type like HIV strains as described in Example 6;
Fig. 7A is a bar graph showing relative change in drug
susceptibility for the patient of Example 7;
Fig. 7B is an Antivirogram for the patient the subject of
Example 7;
Fig. 8A is a bar graph showing relative change in drug
susceptibility for the patient of Example 8;
* Trade-mark
CA 02244735 2006-10-24
WO 97/Z7480 PCT/11397/00071
17
*
Fig. 8B is an Antivirogram for the patient the subject of
Example 8;
Fig. 9A is a bar graph showing relative change in drug
susceptibility for the patient of Example 9;
Fig. 9B is an Antivirogram for the patient the subject of
Example 9;
Fig. 1OA is a bar graph showing relative change in drug
susceptibility for the patient of Example 10;
Fig. lOB is an Antivirogramfor the patient the subject of
Example 10;
Fig. 11A is a bar graph showing relative change in drug
susceptibility for the patient of Example 11;
Fig.11B is an Antivirogram for the patient the subject of
Example 11;
Fig. 12A is a bar graph showing relative change in drug
susceptibility for the patient of Example 12; and
Fig. 12B is an Antivirogram for the patient the subject of
Example 12.
The invention will be further illustrated by the following
.20 Examples.
* Trade-mark
CA 02244735 2006-10-24
WO 97127480 PCT/IB97/00071
18
Modes for Carrying Out the Invention
Example 1
Protocol
1. Extraction and amplification of viral RNA.
RNA was isolated from 100 l of plasma according to the method
described by Boom, R. et al. (1990, supra), and was reverse transcribed
with the GeneAmp reverse transcriptase kit (Perkin Elmer) as described
by the manufacturer and using an HIV-1 specific downstream primer
(OUT3 : 5'-CAT TG C TCT CCA ATT ACT GTG ATA 'ITI' CTC ATG-
3'; SEQ ID NO: 1). PCR on reverse transcribed RNA was performed
with inner and outer primers as described by Kellam, P. and Larder, B.A.
(1994, supra). After chloroform extraction and centrifugation on
Centricon 100 columns or centrifugation on anion-exchange spin
columns (Quiagen), the isolated PCR product was ready for use in the
transfection reactions.
2. Production and isolation of plasmid.
Production of pHIV RT (MCR) plasmid was performed in E. coll.
Plasmid DNA was isolated from overnight cultures making use of Qiagen
columns as described by the manufacturer. Yield of the isolated plasmid
was determined spectrophotometrically by A260/280 measurement
(optical density measurement at X. = 260 and 280 nm). About 250 g of
ultrapure plasmid DNA was obtained from 500 ml of bacterial culture.
The identity of the isolated plasmid was confirmed by restriction
analysis. Subsequently, the isolated plasmid DNA was linearised with
BstEII and purified again by a classical phenol/chloroform extraction.
3. Transfection of cells.
MT4 cells were subcultured at a density of 250,000 cells/ml before
transfection (exponential growth phase). Cells were pelleted and
* Trade-mark
CA 02244735 1998-07-23
WO 97/27480. PCT/IB97/00071
19
resuspended in phosphate buffered saline (PBS) at a concentration of 3.1
10E6 cells/ml. A 0.8 ml portion (2.5 10E6 cells/ml) was used for each
transfection. Transfection was performed with the Bio-Rad Gene pulser
making use of 0.4 cm electrode cuvettes. Cells were electroporated in the
= 5 presence of 1O g of linearised pHIVORT plasmid and approximately 5 g
of RT PCR product at 250 F and 300 V, followed by a 30-min
incubation at room temperature. Subsequently, 10 ml of fresh culture
medium was added to the cell suspension and incubation was performed
at 37 C in a humidified atmosphere of 5%C02.
4. Culture and follow-up of transfected cells.
During 7 to 10 days following the transfection, cells were
monitored for the appearance of cytopathogenic effect (CPE). In the
absence thereof, cells were subcultured in different flasks. Subsequently,
culture supernatants of transfected cells were used to create a stock of
recombinant virus and stored in 1.5 ml aliquots at -70 C.
5. Analysis of recombinant virus from patient viral RNA.
After titration of the new viruses, the stocks were used for antiviral
experiments in the presence of serial dilutions of different HIV inhibitors.
Titres of the harvested supernatants were determined by limited serial
dilution titration of virus in MT4 cells.
Viruses with a useful titre were used in antiviral experiments. For
this purpose, 96-well microtitre plates were filled with 100 l of complete
culture medium. Subsequently, stock solutions of compounds were
added in 25 l volumes to series of duplicate wells. HIV- and mock-
infected cell samples were included for each drug (or drug combination).
Exponentially growing MT4 cells were then transferred to the microtitre
plates at a density of 150,000 cells/ml. The cell cultures were then
incubated at 37 C in a humidified atmosphere of 5% CO2. Five days
after infection, the viability of the mock- and HIV-infected cells was
examined spectrophotometrically by the MTT method (Pauwels, R. et al.
- J. Virol. Meth. (1988), 20 : 309-321) as described in Section 6 below.
CA 02244735 2006-10-24
WO 97127480 PCT/M97/00071
6. MTT assay.
To each well of the microtiter plates, 20 l of a solution of MTT
(7.5 mg/ml in PBS) was added. The plates were further incubated at
37 C for 1 h. Then, 150 l of medium was removed without disturbing
5 the MT4 cell clusters containing the formazan crystals. Solubilization of
the formazan crystals was achieved by adding l00 15% Triton X-100 * in
acidified isopropanol (5 ml concentrated HCI per litre solvent).
Complete dissolution of the formazan crystals was obtained after the
plates had been placed on a plate shaker for 10 min. Finally the
10 absorbances were read at two wavelengths (540 and 690 rim). From
these optical density (OD) data, 50% inhibitory (IC50) and 50% cytotoxic
(CC50) concentrations were derived.
Example 2
Construction of a pHIVARTBstEll-variant with deletion of the HIV-1
15 protease and reverse transcriptase gene.
The protocol described in Example 1 was repeated, except that the
sequence of the HIV pot gene of interest was that coding for RT and
protease and the construct prepared was pGEMT3-APRT as described
below. Other modifications relative to the procedure set out in Example
20 1 are set out below.
For amplification of viral RNA, reverse transcription from RNA to
DNA was again carried out with the OUT3 primer. However, for the
nested PCR procedure the primers used are as shown in Fig. 3. Thus, it
will be observed that the nested PCR procedure uses as outer primers
RVP5 and OUT3 and as inner primers RVP5 and IN3. Thus, this nested
procedure is, in effect a hemi-nested PCR procedure.
Production and isolation of pGEMT3-APRT.
The final pGEMT3-DPRT construct is a derivative of pGEM9-
Zf(-) (Promega).
* Trade-mark
CA 02244735 1998-07-23
WO 97/27480 PCT/097100071
21
In short, the pGEMT3-APRT construct is built up by introducing
the desired insert HIV-HXB2 (a protease and reverse transcriptase-
deleted proviral HIV-1 clone, including flanking human sequences) into
the Xbal restriction site of the vector pGEM9-Zf(-). The proviral genome
has been deleted from the AhdI site within the protease gene
(surrounding amino acid 9) to the BstEII site of the pHIVARTBstEIl
construct (MRC Repository reference : ADB23 1). At the junction of the
AProRT deletion Smal and BstEII sites are located which can be used for
linearisation of the proviral construct prior to transfection. The
construction of pGEMT3-APRT is schematically represented in Figs 1
and 2. The yield of pGEMT3-APRT was about lmg out of 500m1
bacterial culture.
As indicated above, the plasmid pGEMT3-APRT was deposited at
the Belgian Coordinated Collections of Microorganisms-BCCM LMBP-
Collection on November 8, 1996 under the number LMBP3590.
It was not expected that the introduction of the proviral genome
into another vector (pGEM9-Zf(-) instead of pIB 120) would cause major
problems. pIB 120, a derivative of pEMBL8(-) (according to information
provided by Kodak Scientific Imaging Systems), and pGEM9-ZF(-) are
similar vectors. Nevertheless the proviral vector piB 120HIV may be
unstable in recA+ E. coli host cells (Maschera, B., et al. J. Virol. (1995)
69, 5431-5436. Therefore the stability of the pGEMT3-APRT construct
should be verified after every new preparation of plasmid.
HIV-HXB2 se uence:
The region of interest within the HIV-HXB2D sequence
(nucleotide 1800 to 4400) is represented in Fig. 3 (schematically) and
Fig. 4 (complete sequence). The location of several genes, restriction
sites, primers and deletions (APro, ART, AProRT) are also indicated.
The sequence of HIV-1 (isolate HXB2, reference genome, 9718bp)
was obtained from the National Center for Biotechnology Information
CA 02244735 1998-07-23
WO 97/27480 PCT/IB97/00071
22
(NCBI), National Library of Medicine, National Institutes of Health via
the ENTREZ Document Retrieval System.
Genbank name: HIVHXB2CG
Genbank Accesion No: 03455
NCBI Seq.ID No: 327742
Regions of recombination:
In combination with RT-PCR fragments generated by RVP5 and
OUT3/IN3 primers, the pGEMT3-OPRT vector can be used to transfect
MT4 cells as described in Example 1, Section 3. The region for
recombination at the 5'-end of OProRT contains 188 nucleotides. The
region for recombination at the 3'-end of AProRT is similar to the one
described earlier (Kellam, P. and Larder, B.A. (1994) supra) and contains
130 nucleotides.
The length of these regions for recombination is not unimportant.
Previous data (Bandyopadhyay, P.K. et al. Proc. Natl. Acad. Sci. U.S.A.,
(1984) 81, 3476-3480; Rubnitz, J. and Subramani, S. (1984) supra)
demonstrate that a 10-fold reduction in recombination frequency may
occur when sequence homology is reduced from 214 to 163 base pairs.
Furthermore, sufficient recombination events should occur within the
electroporated cells to ensure that the generated viral phenotype is a
reliable reflection of the quasi-species present in the treated HIV-
positive patient. Optimisation of recombinant events can first be
achieved by adjusting the ratio of linearised proviral vector to RT-PCR
fragment that is used for electroporation of the target cells. The standard
method therefore,with typical results of outcome, has previously been
described by Kellam, P. and Larder, B.A. (1994 supra ). As a
consequence, it was decided to increase the initial input of about 2 g of
PCR product (with 1O g of vector) to about 5 g or more. The result
thereof was reflected in a faster appearance of visible virus growth
(cytopathogenic effect) in the culture of transfected cells.
CA 02244735 1998-07-23
WO 97/27480 PCTIIB97/00071
23
Another option for optimisation of recombination events would be
the design of primers resulting in longer recombination sequences.
Nevertheless, the real input in the transfection reaction always
depends on the yield after PCR. Some samples have a high yield and as a
result there will be a higher input of amplified material in the
transfection reaction (with better results on efficiency of recombination).
However, despite a lower recombination efficiency, samples having a
low yield can also be transfected and will result in viable virus with a
reliable reflection of the virus population.
Example 3
Alternative primers for RT-PCR of the ProRT sequence:
New primers (A-D) have been designed relative to those used in
Example 2 and should result in longer recombination sequences at both 5'
and 3' end of the ProRT region. Two primers were designed at both the
5' and 3' end of the respective region to allow nested PCR. As indicated
in Figs. 3 and 4 the direct repeat present at the 5' end of the ProRT region
was taken into account when designing the respective primers. The new
primers are as follows:
A PRTO-5 : 5'-GCCCCTAGGA-AAAAGGGCTG-TTGG (SEQ ID NO: 3)
B PRTI-5 : 5'-TGAAAGATTG-TACTGAGAGA-CAGG (SEQ ID NO: 4)
C PRTI-3 : 5'-GATATTTCTC-ATGTTCATCT-TGGG (SEQ ID NO: 5)
D PRTO-3 : 5'-AGGTGGCAGG-TTAAAATCAC-TAGC (SEQ ID NO: 6)
Example 4
Construction of an alternative AProRT vector:
As mentioned above, construction of an alternative ProRT deleted
vector can be achieved by oligonucleotide-mediated mutagenesis.
However, it is also possible to enlarge the ProRT deletion from the
current 3'-end to the next Kpnl site in the RT gene (40 base pairs further
WO97/27480 CA 02244735 2006-10-24 PCT/IB97100071
24
downstream). Ligation of a Klenow-treated KpnI site to a Klenow-
treated BstEII site will restore the initial BstEII recognition sequence.
As such, this alternative vector behaves similarly to the pGMT3-EPRT
vector described in Example 2, but has a slightly larger RT deletion-
Example 5
An HIV infected patient who received AZT from December 1989
until an undocumented later date, and switched to a combined
chemotherapy of AZT + 3TC (1:1) from February 1994 until October
1995 donated plasma whose susceptibility to a number of RT inhibitors
was determined according to the above described protocol of Example 1.
Recombinant wild type HIV strain recIHB was used in said protocol as a
reference HIV virus. Table 1 shows the IC50 values (p.M) measured and
the ratio of said values. The Antivirogram is shown in Fig. 5.
Table I
Anti-HIV-1 activity
IC50 (M)
Drug Ex p. I Ex p. 2 Mean (1) recIIIB ref (2) ratio (1)/(2)
loviride 0.1 0.12 0.11 0.05 2
tivirapine 0.019 0.018 0.019 0.01 1.5
AZT 0.001 0.002 0.002 0.004 0.4
3TC 31.6 100 65.8 0.56 118
d4T 0.07 0.49 0.06 0.12 0.5
ddI 2.0 0.8 1.4 2.83 0.5
ddC 0.2 0.2 0.2 0.38 0.5
AZT+3TC 0.001 0.001 0.001 0.002 0.5
(1:l
From these data, one can determine that monotherapy with 3TC is
unlikely to benefit this particular patient. Combined therapy of AZT +
3TC (the current therapy), however, is still likely to exert a positive
effect.
* Trade-mark
WO 97/27480 CA 02244735 2006-10-24 PCr/1B97/00071
Example 6
A drug-naive HIV infected patient donated plasma whose
susceptibility to a number of RT inhibitors was determined according to
the above described protocol of Example 1. Recombinant wild type HIV
5 strain reclIIB was used in said protocol as a reference HIV virus. Table 2
shows the IC50 values ( M) measured and the ratio of said values. An
Antivirogram was prepared and shown in Fig. 6.
Table 2
Anti-HIV-1 activity
IC M
Drug Ex p. 1 Ex p. 2 Mean (1) recIIIB ref (2) ratio (1)/(2)
3TC 1.81 2.02 1.91 3.08 1
ddl 3.07 4.47 3.77 8.58 0.4
ddC 1.45 1.47 1.46 2.21 1
AZT 0.04 0.05 0.05 0.06 1
d4T 1.31 0.97 1.14 1.74 1
AZT+3TC 0.05 0.04 0.05 0.02 3
(1:1)
DDC+D4T 0.62 0.44 0.53 0.77 1
(1:1)
3TC+d4T 0.42 0.44 0.43 1.11 0.4
1:1
From these data one can determine that the patient is infected with
HIV strains closely resembling the wild type HIV. None of the drug
regimens is to be excluded, so chemotherapy can be initiated with a drug
such as AZT having a positive track record.
Example 7
A drug-naive HIV-infected patient donated plasma whose
susceptibility to a number of RT inhibitors was determined according to
the protocol set out in Example 1. Recombinant wild type HIV strain
* Trade-mark
CA 02244735 2006-10-24
wo 97/27480 PCT/M97/00071
26
recIIIB was used as a reference HIV virus. Table 3 shows the IC50 values
(gM) measured and the ratio of said values. A bar graph showing
relative change in drug susceptibility is shown in Fig. 7A. An
Antivirogratn*was also prepared and is shown in Fig. 7B.
Table 3
Anti-HIV-1 activity
IC50 ( M)
Drug Ex p. 1 Ex p. 2 Mean (1) recIIIB ref (2) ratio (1)/(2)
AZT 0.052 0.050 0.051 0.023 2
3TC 2.173 ND 2.173 1.381 2
ddl 0.475 0.429 0.452 0.648 0.7
ddC 1.042 1.389 1.216 1.616 0.8
d4T 1.142 1.657 1.399 1.368 1
loviride 0.035 0.025 0.030 0.024 1
tivirapine 0.042 0.049 0.046 0.021 2
From these data one can determine that the patient is infected with
a HIV strain closely resembling the wild type HIV. None of the drug
regimens is to be excluded, so chemotherapy can be initiated with a drug
such as AZT, 3TC or others having a positive track record.
Example
An HIV-infected patient with a therapy history including AZT,
3TC and loviride donated plasma whose susceptibility to a-,number of RT
inhibitors was determined according to the protocol set out in Example 1.
Recombinant wild type HIV strain recIIIB was used as a reference HIV
virus. Table 4 shows the IC50 values ( M) measured and the ratio of said
values. A bar graph showing relative change in drug susceptability is
shown in Fig. 8A. An Antivirogram was also prepared and is shown in
Fig. 8B.
* Trade-mark
CA 02244735 2006-10-24
WO 97/27480 PCT/IB97/00071
27
Table 4
Anti-HIV-1 activity
IC,50 M)
Drug Ex p. 1 Ex p. 2 Mean 1 reciIB ref (2) ratio (1)/(2)
AZT 18.264 22.251 20.257 0.084 241
3TC > 100.000 > 100.000 > 100.000 6.304 > 16
ddl 26.861 15.435 21.148 1.586 13
ddC 9.290 8.506 8.898 1.931 5
d4T 7.500 7.097 7.298 5.465 1
loviride > 100.000 > 100.000 > 100.000 0.037 > 2717
tivirapine 1.626 1.604 1.615 0.021 78
From this data one can determine that the patient is infected with a
HIV strain displaying a decreased susceptibility towards most of the
nucleoside and non-nucleoside antiretroviral drugs examined. Therapy
can still be initiated with D4T or DDC. The possibility of including
protease-inhibitors into the therapy can be considered.
Example 9
An HIV-infected patient with a therapy history including multiple
nucleoside analogue RT-inhibitors donated plasma whose susceptibility
to a number of RT inhibitors was determined according to the protocol
set out in Example 1. Recombinant wild type HIV strain recIIIB was
used as a reference HIV virus. Table 5 shows the IC50 values (p M)
measured and the ratio of said values. A bar graph showing relative
change in drug susceptibility is shown in Fig. 9A. An Antivirogram was
also prepared and is shown in Fig. 9B.
* Trade-mark
CA 02244735 2006-10-24
WO 97/27480 PCT/1B97/00071
28
Table 5
Anti-HIV-1 activity
IC hiM
Drug Exp. 1 Exp. 2 Mean (1) recIIIB ref ratio (1)/(2)
(2)
AZT > 100.000 ND > 100.000 0.291 > 344
3TC > 100.000 > 100.000 > 100.000 16.670 > 6
ddl > 100.000 > 100.000 > 100.000 4.757 > 21
ddC 60.079 73.049 66.564 3.444 19
d4T > 100.000 > 100.000 > 100.000 12.030 > 8
loviride 0.064 0.058 0.061 0.065 0.9
tivira ine 0.052 0.043 0.048 0.042 1
From this data one can determine that the patient is infected with a
HIV strain displaying a decreased susceptibility towards all nucleoside
analogue antiretroviral drugs. Non-nucleoside antiretroviral drugs should
not be excluded from therapy. Here also, the possibility of including
protease inhibitors into the therapy can be considered.
Example 10
A drug-naive HIV-infected patient donated plasma whose
susceptibility to a number of RT inhibitors and protease inhibitors was
determined according to the protocol set out in Example 1. Recombinant
wild type HIV strain reciiB was used as a reference HIV virus. Table 6
shows the IC50 values ( M) measured and the ratio of said values. A bar
graph showing relative change in drug susceptibility is shown in Fig.
10A. An Antivirograrn was also prepared and is shown in Fig. lOB.
* Trade-mark
WO 97/27480 CA 02244735 2006-10-24 PCT/IB97/00071
29
Table 6
Anti-HIV-1 activity
IC M
Drug Ex p. 1 Ex p. 2 Mean (1) recIIIB ref (2) ratio(l)/(2)
AZT 0.019 0.019 0.019 0.041 0.5
3TC 1.525 1.718 1.622 4.608 0.4
saquinavir 0.003 0.003 0.003 0.006 0.4
ritonavir 0.022 0.017 0.019 0.033 0.6
indinavir 0.013 0.013 0.013 0.016 0.8
From these data one can determine that the patient is infected with
HIV strains closely resembling the wild type HIV. None of the drug
regimens is to be excluded, so that chemotherapy can be initiated with a
drug such as AZT, 3TC or others having a positive track record.
Example 11
An HIV infected patient with a therapy history including RT and
protease inhibitors donated plasma whose susceptibility to a number of
RT inhibitors and protease inhibitors was determined according to the
protocol set out in Example 1. Recombinant wild type HIV strain
recilb was used as a reference HIV virus. Table 7 shows the IC50
values (p M) measured and the ratio of said values. A bar graph showing
relative change in drug susceptibility is shown in Fig. I IA. An
Antivirogram*was also prepared and is shown in Fig. 1 lB.
* Trade-mark
CA 02244735 2006-10-24
WO 97/27480 PCT/M97/00071
Table 7
Anti-HIV-1 activity
IC50 M
Drug Exp. 1 Exp. 2 Mean (1) reciiB ref ratio (1)/(2)
(2)
AZT ND 0.015 0.015 0.047 0.3
3TC > 100.000 93.962 96.981 5.178 19
saquinavir 0.014 0.015 0.014 0.012 1
ritonavir 1.198 1.739 1.468 0.062 24
indinavir 0.229 0.416 0.323 0.027 12
From these data one can determine that the patient is infected with
a HIV strain displaying a decreased susceptibilty towards the RT-
5 inhibitor 3TC and protease inhibitors indinavir and ritonavir.
Accordingly, chemotherapy can be adjusted with drugs such as AZT or
saquinavir having a positive track record.
Example 12
An HIV infected patient with a therapy history including RT and
10 protease inhibitors donated plasma whose susceptibility to a number of
RT inhibitors and protease inhibitors was determined according to the
protocol set out in Example 1. Table 8 shows the IC50 values (gM)
measured and the ratio of said values. A bar graph showing relative
change in drug susceptibility is shown in Fig. 12A. An Antivirogram*
15 was also prepared and is shown in Fig. 12B.
*Trade-mark
WO 97/27480 CA 02244735 2006-10-24 PCTIM97/00071
31
Table 8
Anti-HIV-1 activity
IC50 (M)
Drug Exp. 1 Exp. 2 Mean (1) recI1IB ref ratio (1)/(2)
(2)
AZT 3.833 3.355 3.594 0.041 88
3TC > 100.000 > 100.000 > 100.000 4.608 22
saquinavir 0.350 0.352 0.351 0.006 56
ritonavir 1.610 1.530 1.570 0.033 47
indinavir 0.124 ND 0.124 0.016 8
From these data one can determine that the patient is infected with
a HIV strain displaying a decreased susceptibility towards RT-inhibitors
3TC and AZT and protease inhibitors indinavir, ritonavir and saquinavir.
Example 13
Comparison of Phenotyping relative to Genotyping
Plasma samples were obtained from HIV-infected individuals who
had been receiving non-nucleoside RT inhibitor (NNRTI) long-term
monotherapy. HIV-RNA was extracted, reverse-transcribed and
amplified as described in Example 1. Starting from outer PCR material
of positive samples, the first 785 nucleotides of the RT gene were
amplified and this material was further used for genotyping.
Briefly, the 785 nucleotide fragment was subjected to cycle-
sequencing reactions using the ThermoSequenase*
fluorescent labelled primer cycle sequencing kit with 7-
deaza-dGTP from Amersham (cat# RPN2438). Four sequencing
primers, chosen to allow for sequence determination in both directions
from nucleotide 27 to nucleotide 681 of the RT gene, were used for each
sample. The reactions were analysed on an ALF *
automatic sequencer (Pharmacia). The generated sequences were
exported to a Power Macintosh and further analysed with the
* Trade-mark
CA 02244735 2006-10-24
WO 97/27480 PCT/1B97/00071
32
GeneWorks 2.5 software (Oxford Molecular Group Inc.). Resulting
amino acid sequences were compared with the corresponding sequence
of the laboratory HIV-1 clone HXB2D and resistance-associated
mutations identified in patient material. The results are shown in Table
9 where the one-letter amino acid code is used.
Table
RESISTANCE ASSOCIATED MUTATIONS FOLD RESISTANCE TO
P M D K A K K V E YJ M G T K AZT 3TC NNRTI NNRTI
41 67 70 98 101 103 108 138 181 184 190 215 219 1 2
I N 1 1 5 437
2 S 1 0.4 10 53
3 N 0.4 1 3 87
4 0.3 0.1 1 0.4
5 1 0.4 3
6 N 1 0.3 10 245
7 S 1 0.04 11 57
8 N I I 3 81
9 C 2 1 >1432 12
N 0.4 0.1 5 172
11 L N R N V F Q 94 >8 321 669
12 L Y 28 2 1 3
13 E KIN A G/A Not 1 1> 1 455
Oct 14 N 1 1 9 349
N 2 1 >2424 449
16 N V 1 >8 21 115
17 N, 1 1 2 102
18 S 1 1 9 181
19 N I 1 78 260
G 0.4 1 2 25
21 N 1 0.4 4 68
22 I 1 0.4 3
23 N 0.2 0.2 9
24 Q 1 I 59
P = Patient
* Trade-mark
WO 97/27480 CA 02244735 2006-10-24 PC1'/11B97/00071
33
The top row of Table 9 shows the aminoacids (AA) found in the
wild type sequence and their position. Amino acids changes at these
positions are shown for each patient in the following rows. Only the
positions at which changes were observed in patient material are shown.
The right part of Table 9 presents the fold resistance to different RT
inhibitors as determined by the method according to the invention for
each of the patients' samples. NNRTI I is the non-nucleoside RT
inhibitor that was administered to the patients. NNRTI 2 is another non-
nucleoside RT inhibitor for which cross-resistance with the first one was
observed to some extent.
The genotyping results regarding nucleoside analog RT inhbitors
resistance are as follows :
- M41 L, D67N, K70R, T215F/Y and K219Q/E are AZT
resistance-associated mutations (Larder, B. and Kemp, S. (1989)
Science 246, 1155-1158; Kellam, P. et al. (1992) PNAS 89,
1934-1938). Their presence, individually or in different
combinations, in the genome of HIV isolated from patient
material correlates with the phenotypic resistance as determined
by the Antivirogram generated (patients 11 and 12).
- The same applies to resistance to 3TC associated with the
M184V mutation (Tisdale, M. et al. (1993) PNAS 90, 5653-
5656) which is observed only in the patients which show
phenotypic resistance to the drug (patients 11 and 16).
The genotyping results regarding NNRTIs resistance are as follows :
- Three patients (3, 4 and 12) have no NNRTI resistance-
associated mutation and are phenotypically sensitive to the drug.
- Most of the patients who show phenotypic resistance to the
NNRTIs have a NNRTI resistance-associated mutation at
position 103 (K 103N/S).
* Trade-mark
CA 02244735 1998-07-23
WO 97/27480. PCT/IB97/00071
34
- One patient (9) has the Y181C NNRTI resistance-associated
mutation and shows a high phenotypic resistance (>1432 fold) to
NNRTI 1.
- Patient 13 has several NNRTI resistance-associated mutations
(K 101 E, K 103N partially and G 190A partially). This patient also
shows a high phenotypic resistance (>1466 fold) to NNRTI 1.
The E138A mutation observed in this sample is not associated so
far with resistance. However, another mutation at this same
position, i.e. E138K, has been demonstrated to play an important
role in resistance to the TSAO compounds (Balzarini, J. et al.
(1993) PNAS 90, 6952-9656). The role of the E138A mutation
still needs to be assessed.
- Patient 20 has the A98G NNRTI resistance-associated mutation
and shows phenotypic resistance to the tested NNRTIs.
- Patient 22 has the V1081 NNRTI resistance-associated mutation
but does not show any phenotypic resistance to the tested
NNRTIs.
- Patient 24 shows no NNRTI resistance-associated mutation (the
K101Q mutation is found in several HIV-1 wild type genomes)
but is phenotypically resistant to the tested NNRTIs.
WO 97/7,7480 CA 02244735 2006-10-24 PCT/IB97/00071
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT:
(A) NAME: VIRCO N.V.
(B) STREET: Drie Eikenstraat 661
(C) CITY: Edegem
(E) COUNTRY: Belgium
(F) POSTAL CODE (ZIP): B-2650
(A) NAME: DE BETHUNE, Marie-Pierre
(B) STREET: Tweeleeuwenstraat 15
(C) CITY: Everburg
(E) COUNTRY: Belgium
(F) POSTAL CODE (ZIP): B-3078
(A) NAME: HERTOGS, Kurt
(B) STREET: Sint Vincentiusstraat 53
(C) CITY: Antwerpen
(E) COUNTRY: Belgium
(F) POSTAL CODE (ZIP): B-2018
(A) NAME: PAUWELS, Rudi
(B) STREET: Damstraat 166
(C) CITY: Weerde
(E) COUNTRY: Belgium
(F) POSTAL CODE (ZIP): B-1982
(ii) TITLE OF. INVENTION: Method of assessing the chemotherapy of
patients who are HIV positive based on the phenotypic drug
sensitivity of human HIV strains
(iii) NUMBER OF SEQUENCES: 6
(iv) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: Patentln Release #1.0, Version #1.30 (EPO)
(vi) PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER: EP 96200175.6
(B) FILING DATE: 26-JAN-1996
(2) INFORMATION FOR SEQ ID NO: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
(iii) HYPOTHETICAL: NO
CA 02244735 1998-07-23
WO 97/27480 PCT/M97/00071
36
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Human immunodeficiency virus type 1
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 1:
CATTGCTCTC CAATTACTGT GATATTTCTC ATG 33
(2) INFORMATION FOR SEQ ID NO: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 26 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
(iii) HYPOTHETICAL: NO
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Human immunodeficiency virus type 1
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2:
GGGAAGATCT GGCCTTCCTA CAAGGG 26
(2) INFORMATION FOR SEQ ID NO: 3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
(iii) HYPOTHETICAL: NO
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Human immunodeficiency virus type 1
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 3:
GCCCCTAGGA AAAAGGGCTG TTGG 24
CA 02244735 1998-07-23
WO 97/27480 PCT/M97/00071
37
(2) INFORMATION FOR SEQ ID NO: 4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
(iii) HYPOTHETICAL: NO
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Human immunodeficiency virus type 1
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 4:
TGAAAGATTG TACTGAGAGA CAGG 24
(2) INFORMATION FOR SEQ ID NO: 5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
(iii) HYPOTHETICAL: NO
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Human immunodeficiency virus type 1
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 5:
GATATTTCTC ATGTTCATCT TGGG 24
(2) INFORMATION FOR SEQ ID NO: 6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
(iii) HYPOTHETICAL: NO
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Human immunodeficiency virus type 1
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 6:
AGGTGGCAGG TTAAAATCAC TAGC 24
CA 02244735 1998-07-23
WO 97/27480 PCTlIB97/00071
38
BELGIAN COORDINATED COLLECTIONS OF MICROORGANISMS - BCCM
LMBP-COLLECTION
= Page I of Form BCCM/LMBP/BP/4/96-07 Receipt in the case of an original
deposit
Budapest Treaty on the International Recognition of the Deposit of
Microorganisms for
the Purposes of Patent Procedure
Receipt in the case of an original deposit issued pursuant to Rule 7.1 by the
International Depositary Authority BCCM/LMBP identified at the bottom of next
page
International Form BCCM/LMBP/BP/4/96-07
3E
To : Name of the depositor : VIRGO nv.
C
Address : Drie Eikenstraat, 661
2650 Edegem
W I. Identification of the microorganism:
11 Identification reference given by the depositor:
pGEMT3APRT
C
1.2 Accession number given by the International Depositary Authority:
C
LMBP3590
SUBSTITUTE SHEET (RULE 26)
CA 02244735 1998-07-23
WO 97/27480 PCT/IB97/00071
39
BELGIAN COORDINATED COLLECTIONS OF MICROORGANISMS - BCCM
LMBP-COLLECTION
Page 2 of Form BCCM/LMBP/BP/4/96-07 Receipt in the case of an original deposit
11. Scientific description and/or proposed taxonomic designation
The microorganism identified under I above was accompanied by:
(mark with a cross the applicable box(es)}:
a scientific description
^ a proposed taxonomic designation
III. Receipt and acceptance
This International Depositary Authority accepts the microorganism identified
under I
above, which was received by it on (date of original deposit) : November 08,
1996
IV. International Depositary Authority
Belgian Coordinated Collections of Microorganisms (BCCM)
Laboratorium voor Moleculaire Biologie - Plasmidencollectie (LMBP)
Universiteit Gent
K.L. Ledeganckstraat 35
B-9000 Gent, Belgium
Signature(s) of person(s) having the power to represent the International
Depositary
Authority or of authorized official(s):
Date . November 19, 1996 Lic Martine Vanhoucke
? 1 M3jt i
SUBSTITUTE SHEET (RULE 26)