Note: Descriptions are shown in the official language in which they were submitted.
CA 02246892 1998-09-09
CRYPTIC REGULATORY ELEMI!:NTS IN PLANTS
Field of Invention
This invention relates to cryptic regulator~~ elements within plants.
Background and Prior Art
Bacteria from the genus Agrobacterium have the ability to transfer
specific segments of DNA (T-DNA) to plant cells, where they stably integrate
into the nuclear chromosomes. Analyses of plants harbouring the T-DNA have
revealed that this genetic element may be integrated at numerous locations,
and
can occasionally be found within genes. One strategy which may be exploited
to identify integration events within genes is to transform plant cells with
specially designed T-DNA vectors which contain a reporter gene, devoid of cis-
acting transcriptional and translational expression signals (i.e.
promoterless),
located at the end of the T-DNA. Upon integration, the initiation codon of the
promoterless gene (reporter gene) will be juxtaposed to plant sequences. The
consequence of T-DNA insertion adjacent to, andl downstream of, gene
promoter elements may be the activation of reporter gene expression. The
resulting hybrid genes, referred to as T-DNA-mediated gene fusions, consist of
unselect~i plant promoters residing at their natural location within the
chromosome, and the coding sequence of a mark~:r gene located on the inserted
T-DNA (Fobert et al., 1991, Plant Mol. Biol. 17, 837-851).
It has generally been assumed that activation of promoterless or
enhancerless marker genes result from T-DNA irvsertions within or immediately
adjacent to genes. The recent isolation of severa:~ T-DNA insertional mutants
(Koncz et al., 1992, Plant Mol. Biol. 20, 963-976; reviewed in Feldmann,
1991, Plant J. 1, 71-82; Van Lijsebettens et al., 1991, Plant Sci. 80, 27-37;
Walden et al., 1991, Plant J. 1: 281-288; Yanof<,~ky et al., 1990, Nature 346,
CA 02246892 1998-09-09
-2-
35-39), shows that this is the case for at least some insertions. However,
other
possibilities exist. One of these is that integration of the T-DNA activates
silent regulatory sequences that are not associated with genes. Lindsey et al.
(1993, Transgenic Res. 2, 33-47) referred to such sequences as "pseudo-
promoters" and suggested that they may be responsible for activating marker
genes in some transgenic lines.
Inactive regulatory sequences that are bur ied in the genome but with the
capability of being functional when positioned adjacent to genes have been
described in a variety of organisms, where they rave been called "cryptic
promoters" (Al-Shawi et al., 1991, Mol. Cedl. Biol. 11, 4207-4216; Fourel et
al., 1992, Mol. Cell. Biol. 12, 5336-5344.; Irniger et al., 1992, Nucleic
Acids
Res. 20, 4733-4739; Takahashi et al. , 1991, Jpn J. Cancer Res. 82, 1239-
1244). Cryptic promoters can be found in the int:rons of genes, such as those
encoding for yeast actin (Irniger et al., 1992, Nu~~leic Acids Res. 20, 4733-
4739), and a mammalian melanoma-associated amtigen (Takahashi et al., 1991,
Jpn J. Cancer Res. 82, 1239-1244). It has been suggested that the cryptic
promoter of the yeast actin gene may be a relict of a promoter that was at one
time active but lost function once the coding regi~~n was assimilated into the
exon-intron structure of the present-day gene (Irndger et al., 1992, Nucleic
Acids Res. 20, 4733-4739). A cryptic promoter leas also been found in an
untranslated region of the second exon of the woodchuck N-myc proto-
oncogene (Fourel et al., 1992, Mol. Cell. Biol. 12, 5336-5340. This cryptic
promoter is responsible for activation of a N-myc:2, a functional processed
gene
which arose from retropositon of N-myc transcript (Fourel et al. , 1992, Mol.
Cell. Biol. 12, 5336-534.4). These types of regulatory sequences have not yet
been isolated from plants.
Other regulatory elements are located within the 5' and 3' untranslated
regions (UTR) of genes. These regulatory elements can modulate gene
expression in plants through a number of mechanisms including translation,
CA 02246892 1998-09-09
-3-
transcription and RNA stability. For example, some regulatory elements are
known to enhance the translational efficiency of rnRNA, resulting in an
increased accumulation of recombinant protein by many folds. Some of those
regulatory elements contain translational enhancer sequences or structures,
such
as the Omega sequence of the 5' leader of the tobacco mosaic virus (Gallie and
Walbot, 1992, Nucleic Acid res. 20, 4631-4638), the 5' alpha-beta leader of
the
potato virus X (Tomashevskaya et al, 1993, J. Gen. Virol. 74, 2717-2724), and
the 5' leader of the photosystem I gene psaDb of 1~'icotiana sylvestris
(Yamamoto
et al., 1995, J. Biol. Chem 270, 12466-12470). Other 5' regulatory elements
affect gene expression by quantitative enhancement of transcription, as with
the
UTR of the thylakoid protein genes PsaF, PetH and PetE from pea (Bone et al.,
199, Plant J. 6, 513-523), or by repression of trans~;,ription, as for the 5'
UTR of
the pollen-specific LAT59 gene from tomato (Curie and McCor~rnick, 1997, Plant
Cell 9, 2025-2036). Some 3' regulatory regions contain sequences that act as
mRNA instability determinants, such as the DST element in the Small Auxin-Up
RNA (SAUR) genes of soybean and Arabidopisis (Newman et al., 1993, Plant
Cell 5, 701-714). Other translational enhancers are also well documented in
the
literature (e.g. Helliwell and Gray 1995, Plant Mol. Bio. vol 29, pp. 621-626;
Dickey L.F. al. 1998, Plant Cell vol 10, 475-484; Dunker B.P. et al. 1997
Mol. Gen. Genet. vol 254, pp. 291-296). However, there have been no reports
of these types of cryptic regulatory elements, nor have any cryptic regulatory
elements of this kind been isolated from plants.
The present invention discloses transgenir plants generated by tagging
with a promoterless GUS ((3-glucuronidase) T-DPJA vector and the isolation
and characterization of cryptic regulatory elements identified using this
protocol. Cloning and characterization of these insertion sites uncovered
unique
cryptic regulatory elements not conserved among related species. In one of the
plants of interest, GUS expression was spatially and developmentally regulated
with in seed tissue. The isolated regulatory element specific to this tissue
has
not been previously isolated or characterized in any manner. In another plant,
CA 02246892 1998-09-09
-4-
10
a novel constitutive regulatory element was identified that is expressed in
tissues throughout the plant and across a broad range of plant species.
Furthermore, novel non-translated 5' sequences have been identified that
function as post transcriptional regulatory elements.
S~xnmary of Invention
This invention relates to cryptic regulator: elements within plants.
Several transgenic tobacco plants, including T218 and T1275, were
identified using the method of this invention that contain novel regulatory
elements. These regulatory elements were found not to be active in the native
plant.
Plant T218 contains a 4.65 kb EcoRI fragment containing the 2.15 kb
promoterless GUS-nos gene and 2.5 kb of 5' flanking DNA. Deletion of the
region approximately between 2.5 and 1.0 kb of 'the 5' flanking region did not
alter GUS expression, as compared to the entire 1.65 kb GUS fusion. A
further deletion to 0.5 kb of the 5' flanking site resulted in complete loss
of
GUS activity. Thus the region between 1.0 and ().5 of the 5' flanking region
of
the tobacco DNA contains the elements essential to gene activation. This
region is contained within a XbaI - SnaBI restriction site fragment of the
flanking tobacco DNA. Expression of a gene operatively associated with the
regulatory region was only observed in seed tissues, more specifically seed-
coat
tissue.
A second transgenic tobacco plant, T1275, contained a 4.38 kb
EcoRIIXbaI fragment containing the 2.15 kb promoterless GUS-nos gene and
2.23 kb of 5' flanking tobacco DNA (2225 bp). Expression of the cloned
fragment in transgenic tobacco, N. tabacum c.v. Petit Havana, SRI and
CA 02246892 1998-09-09
-5-
transgenic B. napus c.v. Westar was observed in leaf, stem, root, developing
seed and flower. By transient expression analysi;~, GUS activity was also
observed in leaf tissue of soybean, alfalfa, Arabidopsis, tobacco, B. napus,
pea
and suspension cultured cells of oat, corn, wheat and barley. The
transcription
start site for the GUS gene in transgenic tobacco was located in the plant DNA
upstream of the insertion site. A set of deletions within the plant DNA
revealed the presence of a core promoter element located within a 62 by region
from the transcriptional start site, the occurrence of at least one negative
regulatory element located within an XbaI-SspI fragment, a transcriptional
enhancer located within the BstYI DraI fragment, and a at least one post
transcriptional regulatory element located within a NdeI-SmaI fragment.
This invention therefore provides for isolated nucleic acids that
comprise cryptic regulatory elements within plants. This invention also is
directed to cryptic regulatory elements that comp rise at least one of: a
promoter, a core promoter element, a negative regulatory element, a
transcriptional enhancer, a translational enhancer and a post transcriptional
regulatory element.
Furthermore, this invention relates to a cryptic regulatory element
comprising a nucleic acid that is substantially homologous to the nucleotide
sequence of SEQ ID NO:1. This invention also relates to a nucleic acid
comprising at least 19 contiguous nucleotides of nucleotides 1 to 993 of SEQ
ID NO:1, or, comprising a nucleotide sequence consisting of at least 19
contiguous nucleotides of nucleotides 1 to 467 of SEQ ID NO:1. This
invention also relates to a vector comprising the nucleic acids as defined
above
This invention is also directed to a cryptic. regulatory element
comprising a nucleic acid fragment bounded by F,'coRI-SmaI restriction sites
defined by the restriction map of Figure 2 (B). Furthermore, this invention
relates to a cryptic regulatory element comprising; an XbaI - SmaI fragment,
of
CA 02246892 1998-09-09
-6-
the restriction map of Figure 2 (B) of about 2 kb. Also considered within the
scope of the present invention is a cryptic regulatory element comprising an
XbaI and SnaBI fragment as defined by the restrintion map of Figure 2 (B),
wherein the fragment is of about 500 bp. This imrention also is directed to a
cryptic regulatory element comprising an XbaI and SnaBI fragment, as defined
by the restriction map of Figure 2 (B), wherein the fragment is of about 1.5
kb,
or a cryptic regulatory element comprising a Hiru~III and SnaBI fragment,
defined by the restriction map of Figure 2 (B), wherein the fragment is of
about
1.9 kb. Furthermore, this invention also embraces a cryptic regulatory element
comprising an EcoRI and SnaBI fragment defined'. by the restriction map of
Figure 2, wherein the fragment is of about 2 kb.
This invention also embraces a regulatory element characterized in that
it is substantially homologous with the sequence ~iefmed by SEQ ID N0:2.
This invention is also directed to a cryptic regulatory element that comprises
at
least an 18 by contiguous sequence of SEQ ID N~0:2. Furthermore, this
regulatory element functions in diverse plant species when introduced on a
cloning vector. This invention also relates to a c:himeric gene construct
comprising a DNA of interest for which constitutive expression is desired, and
a constitutive regulatory element, comprising at least an 18 by contiguous
sequence of SEQ ID NO: 2.
This invention also embraces cryptic regulatory elements comprising an
XbaI - SmaI fragment, an Xbal - Ndel fragment, an SphI - SmaI fragment, a
PstI - SmaI fragment, an SspI - SmaI fragment, a BstYI - SmaI fragment, a
DraI - SmaI fragment, a NdeI-SmaI fragment, a XbaI-BstYI fragment, or a
BstYI-DraI fragment as defined by Figure 13(C).
This invention also includes a plant cell which has been
transformed with a chimeric gene construct, or a cloning vector comprising a
cryptic plant regulatory element. Furthermore, this invention embraces
CA 02246892 1998-09-09
-'j_
transgenic plants containing chimeric gene constructs, or cloning vectors
comprising cryptic plant regulatory elements.
This invention further relates to any trans;;enic plant containing a
cryptic regulatory element, having a DNA sequence substantially homologous
to SEQ ID NO: 1, or SEQ ID N0:2, operatively linked to a DNA region that is
transcribed into RNA.
Also included in the present invention a method of conferring
expression of a gene in a plant, comprising operatively linking an exogenous
DNA of interest, for which expression is desired with a cryptic regulatory
element as defined above, to produce a chimeric ,gene construct, and
introducing the chimeric gene construct into a plant capable of expressing the
chimeric gene construct. This invention also embraces a method of modulating
expression of a gene in a plant, comprising operatively linking an exogenous
DNA of interest, for which expression is desired with a promoter of interest
and the cryptic regulatory element as defined above and introducing the
chimeric construct in to a plant.
This invention also relates to the above method wherein the plant-
derived cryptic regulatory element is a seed-coat specific or constitutive
regulatory element. Furthermore, this invention embraces the above method
wherein the seed-coat specific regulatory element comprises a nucleic acid
that
is substantially homologous with the sequence of SEQ ID NO:1, or constitutive
regulatory element comprises a nucleic acid that :is substantially homologous
with the sequence of SEQ ID N0:2. This invention also relates to the above
method wherein the nucleic acid comprises at least a 19 by contiguous sequence
of SEQ ID NO:1, or the nucleic acid comprises apt least an 18 by contiguous
sequence of SEQ ID N0:2.
.* ,. .,
CA 02246892 1998-09-09
_g_
According to the present invention there is also provided a seed coat-
specific cryptic regulatory element contained within a DNA sequence, or
analogue thereof, as shown in SEQ ID NO: 1. Furthermore, there is provided
a constitutive regulatory element contained within a DNA sequence, or
analogue thereof, as shown in SEQ ID NO: 2.
This invention also relates to a cloning vector containing a seed coat-
specific cryptic regulatory element, which is contained within a DNA sequence,
or analogue thereof, as shown in SEQ ID NO: 1 and a gene encoding a protein.
This invention also relates to a cloning vector containing a constitutive
cryptic
regulatory element, which is contained within a I)NA sequence, or analogue
thereof, as shown in SEQ ID NO: 2 and a gene encoding a protein.
This invention also includes a plant cell which has been transformed
with a cloning vector as described above, and to a transgeluc plant containing
a
cloning vector as described above, operatively linked to a gene encoding a
protein.
Brief Description of the Drawings
Figure 1 depicts the fluorogenic analyses of GUS expression in the plant
T218. Each bar represents the average t one standard deviation of three
samples. Nine different tissues were analyzed: leaf (L), stem (S), root (R),
anther (A), petal (P), ovary (O), sepal (Se), seeds 10 days post anthesis (S1)
and seeds 20 days post-anthesis (S2). For all measurements of GUS activity,
the fraction attributed to intrinsic fluorescence, as determined by analysis
of
untransformed tissues, is shaded black on the graph. Absence of a black area
at the bottom of a histogram indicates that the relative contribution of the
background fluorescence is too small to be apparent.
CA 02246892 1998-09-09
-9-
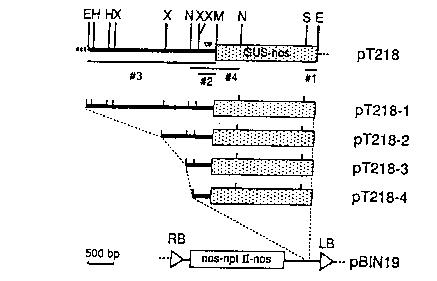
Figure 2 shows the cloning of the GUS fusion in plant T218 (pT218)
and construction of transformation vectors. Plant DNA is indicated by the
solid line and the promoterless GUS-nos gene is ;indicated by the open box.
The transcriptional start site and presumptive TATA box are located by the
closed and open arrow heads respectively. Figure 2 (A) shows DNA probes
#1, 2, 3, and RNA probe #4 (all listed under the pT218 restriction map). The
EcoRI fragment in pT218 was subcloned in the pBINl9 polylinker to create
pT218-1. Fragments truncated at the XbaI, SnaE~I and XbaI sites were also
subcloned to create pT218-2, pT218-3 and pT2lf;-4. Figure 2 (B) shows the
restriction map of the plant DNA upstream from the GUS insertion site.
Abbreviations for the endonuclease restriction sites are as follows: EcoRI
(E),
HindIII (H), XbaI (X), SnaBI (N), SmaI (M), Sstl (S).
Figure 3 shows the expression pattern of promoter fusions during seed
development. GUS activity in developing seeds ( 4-20 days postanthesis (dpa))
of (Fig. 3a) plant T218 (~-~) and (Fig. 3b) plants transformed with vectors
pT218-1 (O-O), pT218-2 (0-0), pT218-3 (0-~ and pT218-4 (0-O) which are
illustrated in Figure 2. The 2 day delay in the pesak of GUS activity during
seed development, seen with the pT218-2 transformant, likely reflects
greenhouse variation conditions.
Figure 4 shows GUS activity in 12 dpa seeds of independent
transformants produced with vectors pT218-1 (O), pT218-2 (~), pT218-3 (~
and pT218-4 (0). The solid markers indicate the plants shown in Figure 3 (b)
and the arrows indicate the average values for plants transformed with pT218-1
or pT218-2.
Figure 5 shows the mapping of the T218 GUS fusion termini and
expression of the region surrounding the insertion site in untransformed
plants.
Figure 5 (A) shows the mapping of the GUS mR:(~A termini in plant T218.
The antisense RNA probe from subclone #4 (Figure 2) was used for
CA 02246892 1998-09-09
-10-
hybridization with total RNA of tissues from untransformed plants (10 ~cg) and
from plant T218 (30 ~,g). Arrowheads indicate the anticipated position of
protected fragments if transcripts were initiated at the same sites as the
T218
GUS fusion. Figure 5 (B) shows the results of au RNase protection assay using
the antisense (relative to the orientation of the GtlS coding region) RNA
probe
from subclone a (see Figure 7) against 30 wg total RNA of tissues from
untransformed plants. The abbreviations used ar~~ as follows: P, untreated
RNA probe; -, control assay using the probe and tRNA only; L, leaves from
untransformed plants; 8, 10, 12, seeds from untr;~nsformed plants at 8, 10,
and
12 dpa, respectively; T10, seeds of plant T218 at 10 dpa; +, control
hybridization against unlabelled in vitro-synthesi~:ed sense RNA from subclone
c (panel a) or subclone a (panel b). The two hybridizing bands near the top of
the gel are end-labelled DNA fragment of 3313 and 1049 bp, included in all
assays to monitor losses during processing. Molecular weight markers are in
number of bases.
Figure 6 provides the nucleotide sequence of pT218 (top line) (SEQ ID
NO: 1) and pIS-1 (bottom line). Sequence identity is indicated by dashed
lines.
The T-DNA insertion site is indicated by a vertical line after by 993. This
site
on pT218 is immediately followed by a 12 by filler DNA, which is followed by
the T-DNA. The first nine amino acids of the GUS gene and the GUS
initiation codon (*) are shown. The major and n-.iinor transeriptional start
site is
indicated by a large and small arrow, respectively. The presumptive TATA
box is identified and is in boldface. Additional putative TATA and CART
boxes are marked with boxes. The location of direct (1-5) and indirect (6-8)
repeats are indicated by arrows.
Figure 7 shows the base composition of rf:gion surrounding the T218
insertion site cloned from untransformed plants. The site of T-DNA insertion
in plant T218 is indicated by the vertical arrow. The position of the 2
genomic
CA 02246892 1998-09-09
-11-
clones pIS-1 and pIS-2, and of the various RNA ~~robes (a-e) used in RNase
protection assays are indicated beneath the graph ,
Figure 8 shows the Southern blot analyse; of the insertion site in
Nicotiana species. DNA from N. tomentosiformis (N torn), N. sylvestris (N
syl), and N. tabacum (N tab) were digested with HindIII (H), XbaI (X) and
EcoRI (E) and hybridized using probe #2 (Figure 2). Lambda HindIII markers
(kb) are indicated.
Figure 9 shows the AT content of 5' non-coding regions of plant genes.
A program was written in PASCAL to scan Genl3ank release 75.0 and to
calculate the AT contents of the S' non-coding (solid bars) and the coding
regions (hatched bars) of all plant genes identified as "Magnoliophyta"
(flowering plants). The region -200 to -1 and + l to +200 were compared.
Shorter sequences were also accepted if they wens at least 190 by long. The
horizontal axis shows the ratio of the AT content (%). The vertical axis shows
the number of the sequences having the specified AT content ratios.
Figure 10 shows the constitutive expression of GUS in all tissues of
plant T1275, including leaf segments (a), stem cross-sections (b), roots (c),
flower cross-sections (d), ovary cross-sections (e;I, immature embryos (fj,
mature embryos (g), and seed cross-sections (h).
Figure 11 shows GUS specific activity within a variety of tissues
throughout the plant T1275, including leaf (L), stem (S), root (R), anther
(A),
petal (P), ovary (O), sepal (Se), seeds 10 days post anthesis (S1), and seeds,
20
days post anthesis (S2).
Figure 12 shows the restriction map of the cryptic regulatory element of
pT1275. Figure 12 (A) shows the plant DNA fused with GUS. Figure 12 (B)
CA 02246892 1998-09-09
-12-
shows the restriction map of the plant DNA. The arrow indicates the GUS
mRNA start site within the cryptic regulatory region.
Figure 13 shows deletion constructs of thE; T1275 regulatory element.
Figure 13 (A) shows the 5' endpoints of each construct as indicated by the
restriction endonuclease site, relative to the full length T1275 regulatory
element, the arrow indicates the transcriptional start site. Plant DNA is
indicated by the solid line, the promoterless GUS-nos gene is indicated by the
open box and the shaded box indicates the region coding for the amino terminal
peptide fused to GUS . The XbaI fragment in pT 1275 was subcloned to create
pT1275-GUS-nos. Deletion constructs truncated at the SphI, PstI, SspI,
BstYI, and DraI sites were also subcloned to create -1639-GUS-nos, -1304-
GUS-nos, -684-GUS-nos, -394-GUS-nos, and --197-GUS-nos, respectively.
Figure 13 (B) shows further deletion constructs of -62-GUS-nos, -12-GUS-nos,
-62(-tsr)-GUS-nos and +30-GUS-nos, relative to -197-GUS-nos (see Figure 13
(A)). Figure 13 (C) shows the restriction map of the plant DNA of pT1275
upstream from the GUS insertion site.
Figure 14 shows the GUS specific activity, mRNA, and protein levels in
leaves of individual, regenerated, greenhouse-grown transgenic plants
containing T1275-GUS-nos (T plants), or 35S-Gl:lS-nos (S plants). Figure 14
(A) shows the levels of GUS expression in leave; from randomly selected
plants containing either T1275-GUS-nos (left-hand side) or 35S-GUS-nos
(right-hand side). Figure 14 (B) shows the level of accumulated GUS mRNA
measured by RNase protection assay and densitometry of autoradiograms in
leaves from the same randomly selected plants containing either T1275-GUS-
nos (left-hand side) or 35S-GUS-nos (right-hand side). Figure 14 (C) shows a
Western blot of GUS fusion protein obtained from T1275-GUS-nos and 35S-
GUS-nos plants. Leaf extracts were equally loaded onto gels and GUS was
detected using anti-GUS antibodies. The molecular weight markers are
CA 02246892 1998-09-09
-13-
indicated on the right-hand side of the gel; untrar~sformed control (SR1) and
GUS produced in E. coli (Ec).
Figure 15 shows deletion and insertion constructs of the 5' untranslated
leader region of T1275 regulatory element and construction of transformation
vectors. The constructs are presented relative to T1275-GUS-nos or 35S-GUS-
nos. The arrow indicates the transcriptional start site. Plant DNA is
indicated
by the solid line labeled T1275, the 35S regulatory region by the solid line
labelled CaMV35S, the NdeI - SmaI region by a 'filled in box, the shaded box
coding for the amino terminal peptide, and the promoterless GUS-nos gene is
indicated by an open box. The deletion construct removing the NdeI - SmaI
fragment of T1275-GUS-nos is identified as T1275-N-GUS-nos. The NdeI -
SmaI fragment from T1275-GUS-nos was also introduced into 35S-GUS-nos to
produce 35S+N-Gus-nos.
Figure 16 shows the region surrounding tlae insertion site in
untransformed plants, positions of various probe, used for RNase protection
assays, and results of the RNase protection assay. Figure 16 (A) shows a
restriction map of the insertion site and various probes used for the assay
(IP:
insertion point of GUS in transformed plants; *: that T1275 probe ended at the
BstYl site, not the IP; **: probe 7 included 600bp of the T1275 plant sequence
and 400 by of the GUS gene). Figure 16 (B) shows results of an RNase
protection
assay of RNA isolated from leaf (L), stem (St), root (R.), flower bud (F) and
developing
seed (Se) tissues of tobacco transformed with T1275-CiUS-nos (10 pg RNA) and
untransformed tobacco (30 pg RNA). Undigested prohe (P), tRNA negative control
(-)
lanes and markers are indicated. RNase protection assays shown used a probe to
detect
sense transcripts between about -446 and +596 of T12'75-GUS-nos or between
about
446 to +169 of untransformed tobacco. The protected fragment in transformed
plants is
about 596 by (upper arrowhead) and, if present, accummlated transcripts
initiated at this
site in untransformed plants are predicted to protect a iFragment of about 169
by (lower
arrowhead). Upper band in RNA-containing lanes was added to samples to
indicate loss
of sample during assay.
CA 02246892 1998-09-09
-14-
Figure 17 shows the levels of mRNA , as well as the ratio between GUS
specific activity and mRNA levels in leaves of individual, regenerated,
greenhouse-grown transgenic plants containing 71275-GUS-nos, or 35S-GUS-
nos constructs, with or without the Ndel Smal fragment (see Figure 15).
Figure 17 (A) shows the level of accumulated GI1S mRNA measured by RlVase
protection assay and densitometry of autoradiogr;~ms in leaves from the same
randomly selected plants containing either 71275-GUS-nos, 71275-N-GUS-
nos. Figure 17 (B) shows the level of accumulat~:d GUS mRIVA measured by
RlVase protection for 35S-GUS-nos or 35S+N-GUS-nos. Figure 17 (C) shows
the ratio between GUS specific activity and mRNA levels in leaves of
individual, regenerated, greenhouse-grown traps~;enic plants containing T 1275-
GUS-nos, 71275-N-GUS-nos, 35S-GUS-nos, or 35S+N-GUS-nos constructs.
Detailed Description of the Preferred Embodiments
This invention relates to cryptic regulatory elements identified in plants.
More specifically, this invention relates to cryptic: promoters, negative
regulatory elements, transcriptional enhancer elements and other post
transcriptional regulatory elements identified in plants.
T-DNA tagging with a promoterless (3-glcGCUronidase (GUS) gene
generated several transgenic Nicotiana tabacum plants that expressed GUS
activity. Examples, which are not to be considered limiting in any manner, of
transgenic plants displaying expression of the promoterless reporter gene,
include a plant that expressed GUS only in devel~~ping seed coats, 7218, and
another plant that expressed GUS in all organs, 71275 (see co-pending patent
applications US serial No. 08/593121 and PCT/C".A97/00064, both of which are
incorporated by reference).
CA 02246892 1998-09-09
-15-
Cloning and deletion analysis of the GUS fusions in both of these plants
revealed that the regulatory regions were located in the plant DNA proximal to
the GUS gene:
~ In T218, a cryptic regulatory region was identified between an EcoRI-
SmaI fragment, and further deletion analyses localized a cryptic
regulatory element to an approximately 0.5 kb region between a XbaI
and a SnaBI restriction endonuclease site ~~f the 5' flanking tobacco
DNA (see Figure 2). This region spans from nucleotide 1 to nucleotide
467 of SEQ ID NO: 1.
~ In T1275, a regulatory region was identif.ed within an XbaI - SmaI
fragment, which comprises several cryptic; regulatory elements which
were localized to several regions throughout the upstream region and
include a minimal promoter region between DraI and NdeI sites (see
Figure 13), negative regulatory elements between XbaI and BstYI, a
transcriptional enhancer between BstYI and DraI, and a translational
enhancer regulatory element between the .NdeI-SmaI sites.
However, it is to be understood that other portions of the isolated disclosed
regulatory elements within T218 and T1275 may also exhibit activities in
directing organ specificity, tissue specificity, or a combination thereof, or
temporal activity, or developmental activity, or a combination thereof, or
other
regulatory attributes including, negative regulatory elements, enhancer
sequences, or post transcriptional regulatory elements, including sequences
that
affect stability of the transcription or initiation complexes or stability of
the
transcript.
Thus, the present invention includes cryptic regulatory elements
obtained from plants that are capable of conferring, or enhancing expression
upon gene of interest linked in operative association therewith. Furthermore,
CA 02246892 1998-09-09
-16-
the present invention includes cryptic regulatory elements obtained from
plants
capable of mediating the translational efficiency of a transcript produced
from a
gene of interest linked in operative association therewith. It is to be
understood
that the cryptic regulatory elements of the present invention may also be used
in
combination with other regulatory elements, either cryptic or otherwise, such
as
promoters, enhancers, or fragments thereof, and the like.
The term cryptic regulatory element refer, to regulatory elements that
are inactive in the control of expression at their native location. These
inactive
regulatory sequences are buried in the genome including intergenic regions or
regions of genes that are not involved in the regulation of XXX but are
capable
of being functional when positioned adjacent to a gene.
By "regulatory element" or "regulatory region", it is meant a portion of
nucleic acid typically, but not always, upstream of a gene, and may be
comprised of either DNA or RNA, or both DNA and RNA. The regulatory
elements of the present invention includes those ~Nhich are capable of
mediating
organ specificity, or controlling developmental o~~ temporal gene activation.
Furthermore, "regulatory element" includes pronnoter elements, core promoter
elements, elements that are inducible in response to an external stimulus,
elements that are activated constitutively, or elements that decrease or
increase
promoter activity such as negative regulatory elements or transcriptional
enhancers, respectively. "Regulatory elements" ass used herein, also includes
elements that are active following transcription initiation or transcription,
for
example, regulatory elements that modulate gene expression such as
translational and transcriptional enhancers, translational and transcriptional
repressors, and mRNA stability or instability determinants. In the context of
this disclosure, the term "regulatory element" also refers to a sequence of
DNA, usually, but not always, upstream (5') to the coding sequence of a
structural gene, which includes sequences which control the expression of the
coding region by providing the recognition for RIVA polymerase and/or other
CA 02246892 1998-09-09
-17-
factors required for transcription to start at a particular site. An example
of a
regulatory element that provides for the recognition for RNA polymerase or
other transcriptional factors to ensure initiation at a particular site is a
promoter
element. A promoter element comprises a core yromoter element, responsible
for the initiation of transcription, as well as other regulatory elements (as
listed
above) that modify gene expression. It is to be understood that nucleotide
sequences, located within introns, or 3' of the coding region sequence may
also
contribute to the regulation of expression of a coding region of interest. A
regulatory element may also include those elements located downstream (3') to
the site of transcription initiation, or within trans~~ribed regions, or.
both. In the
context of the present invention a post-transcriptional regulatory element may
include elements that are active following transcription initiation, for
example
translational and transcriptional enhancers, translational and transcriptional
repressors, and mRNA stability determinants.
An example of a cryptic regulatory element of the present invention,
which is not to be considered limiting in any mariner, is an organ-specific,
and
temporally-specific element obtained from plant '.0218. Such an element is a
seed-specific regulatory element. More preferably, the element is a seed-coat
specific regulatory element as described herein, or an analogue thereof, or a
nucleic acid fragment localized between EcoRI - SmaI sites, as defined in
restriction map of Figure 2 (B) or a fragment thereof. The seed coat-specific
regulatory element may also be defined by a nucleic acid comprising
substantial
homology (similarity) with the nucleotide sequence comprising nucleotides 1-
467, or 1-993, of SEQ ID NO:l. For example, which is not to be considered
limiting in any manner, the nucleic acid may exhibit 80% similarity to the
nucleotide sequence comprising nucleotides 1-46"7, or 1-993, of SEQ ID NO:1.
Furthermore, the seed-coat specific nucleotide sequence may be defined as
comprising at least a 19 by fragment of nucleotides 1-467, or 1-993 as defined
within SEQ ID NO:1.
CA 02246892 1998-09-09
-18-
Another example of a cryptic regulatory element of an aspect of the
present invention includes, but is not limited to, a constitutive regulatory
element obtained from the plant T1275, as described herein and analogues or
fragments thereof, or a nucleic acid fragment loc,~lized between XbaI - SmaI,
as
identified by the restriction map of Figure 12 (B) or a fragment thereof.
Furthermore, the constitutive regulatory element may be defined as a nucleic
acid fragment localized between XbaI - SmaI as i~dentifred by the restriction
map of Figure 13 (C) or a fragment thereof. The: constitutive cryptic
regulatory element may also be defined by a nucleotide sequence comprising at
least an 18 by fragment of the regulatory region defined in SEQ ID N0:2, or
by a nucleic acid comprising from about 80% similarity to the nucleotide
sequence of SEQ ID N0:2.
Another cryptic regulatory element of the present invention includes,
but is not limited to, a post-transcriptional or trar~slational enhancer
regulatory
element localized between NdeI - SmaI (see Figure 15), or the post-
transcriptional or translational enhancer regulatory element may comprise the
nucleotide sequence as defined by nucleotides 2086-2224 of SEQ ID N0:2 or
an analog thereof, or the element may comprise 80 % similarity to the
nucleotide sequence of nucleotides 2086-2224 of SEQ ID N0:2.
Furthermore, other regulatory elements of the present invention include
negative regulatory elements (for example located within an XbaI-BstYI
fragment as defined by Figure 13 (C)), a transcriptional enhancer localized
within the BstYI-DraI fragment of Figure 13 (C) , a core promoter element
located within the DraI-NdeI fragment of Figure 13 (C), or a regulatory
element or post-transcriptional element downstres~m of the transcriptional
start
site.
An "analogue" of the above identified cryptic regulatory elements
includes any substitution, deletion, or additions t~~ the sequence of a
regulatory
element provided that said analogue maintains at least one regulatory property
CA 02246892 1998-09-09
-19-
associated with the activity of the regulatory element. Such properties
include
directing organ specificity, tissue specificity, or a combination thereof, or
temporal activity, or developmental activity, or a combination thereof, or
other
regulatory attributes including, negative regulatory elements, enhancer
sequences, or sequences that affect stability of the transcription or
translation
complexes or stability of the transcript.
There are several types of regulatory elements, including those that are
developmentally regulated, inducible and constiW tive. A regulatory element
that is developmentally regulated, or controls the differential expression of
a
gene under its control, is activated within certain organs or tissues of an
organ
at specific times during the development of that organ or tissue. However,
some regulatory elements that are developmentally regulated may preferentially
be active within certain organs or tissues at specific developmental stages,
they
may also be active in a developmentally regulated manner, or at a basal level
in
other organs or tissues within the plant as well.
An inducible regulatory element is one th,~t is capable of directly or
indirectly activating transcription of one or more DNA sequences or genes in
response to an inducer. In the absence of an inducer the DNA sequences or
genes will not be transcribed. Typically the protein factor, that binds
specifically to an inducible regulatory element to activate transcription, is
present in an inactive form which is then directly or indirectly converted to
the
active form by the inducer. The inducer can be a chemical agent such as a
protein, metabolite, growth regulator, herbicide or phenolic compound or a
physiological stress imposed directly by heat, cold, salt, or toxic elements
or
indirectly through the action of a pathogen or disease agent such as a virus.
A
plant cell containing an inducible regulatory element may be exposed to an
inducer by externally applying the inducer to the cell or plant such as by
spraying, watering, heating or similar methods.
CA 02246892 1998-09-09
-20-
A constitutive regulatory element directs the expression of a gene
throughout the various parts of a plant and continuously throughout plant
development. Examples of known constitutive regulatory elements include
promoters associated with the CaMV 35S transcript. (Odell et al., 1985,
Nature, 313: 810-812), the rice actin 1 (Zhang et al, 1991, Plant Cell, 3:
1155-
1165) and triosephosphate isomerase 1 (Xu et al, 1994, Plant Physiol. 106:
459-467) genes, the maize ubiquitin 1 gene (Cornejo et al, 1993, Plant Mol.
Biol. 29: 637-646), the Arabidopsis ubiquitin 1 and 6 genes (Holtorf et al,
1995, Plant Mol. Biol. 29: 637-646), and the tobacco translational initiation
factor 4A gene (lVlandel et al, 1995 Plant Mol. Biol. 29: 995-1004).
The term "constitutive" as used herein does not necessarily
indicate that a gene under control of the constitutive regulatory element is
expressed at the same level in all cell types, but that the gene is expressed
in a
wide range of cell types even though variation in abundance is often observed.
The present invention is further directed to a chimeric gene
construct containing a DNA of interest operatively linked to a regulatory
element of the present invention. Any exogenous gene can be used and
manipulated according to the present invention tc~ result in the expression of
said exogenous gene.
The chimeric gene construct of the: present invention can further
comprise a 3' untranslated region. A 3' untranslated region refers to that
portion of a gene comprising a DNA segment that contains a polyadenylation
signal and any other regulatory signals capable of effecting mRNA processing
or gene expression. The polyadenylation signal is usually characterized by
effecting the addition of polyadenylic acid tracks to the 3' end of the mRNA
precursor. Polyadenylation signals are commonly recognized by the presence
of homology to the canonical form S' AATAAA-3' although variations are not
uncommon.
CA 02246892 1998-09-09
-21-
Examples of suitable 3' regions are; the 3' transcribed non-
translated regions containing a polyadenylation signal of Agrobacterium tumor
inducing (Ti) plasmid genes, such as the nopaline: synthase (Nos gene) and
plant
genes such as the soybean storage protein genes and the small subunit of the
ribulose-1, 5-bisphosphate carboxylase (ssRUBISCO) gene. The 3' untranslated
region from the structural gene of the present construct can therefore be used
to
construct chimeric genes for expression in plants .
The chimeric gene construct of the: present invention can also
include further enhancers, either translation or transcription enhancers, as
may
be required. These enhancer regions are well known to persons skilled in the
art, and can include the ATG initiation codon and adjacent sequences. The
initiation codon must be in phase with the reading; frame of the coding
sequence
to ensure translation of the entire sequence. The translation control signals
and
initiation codons can be from a variety of origins, both natural and
synthetic.
Translational initiation regions may be provided :From the source of the
transcriptional initiation region, or from the strucaural gene. The sequence
can
also be derived from the regulatory element selected to express the gene, and
can be specifically modified so as to increase translation of the mRNA.
To aid in identification of txansfonmed plant cells, the constructs
of this invention may be further manipulated to include plant selectable
markers. Useful selectable markers include enzymes which provide for
resistance to an antibiotic such as gentamycin, hygromycin, kanamycin, and the
like. Similarly, enzymes providing for production of a compound identifiable
by colour change such as GUS ((3-glucuronidase), or luminescence, such as
luciferase are useful.
Also considered part of this invention are transgenic plants
containing the chimeric gene construct comprising a regulatory element of the
present invention. Methods of regenerating whole plants from plant cells are
CA 02246892 1998-09-09
-22-
known in the art. In general, transformed plant cells are cultured in an
appropriate medium, which may contain selectivf; agents such as antibiotics,
where selectable markers are used to facilitate identification of transformed
plant cells. Once callus forms, shoot formation c:an be encouraged by
employing the appropriate plant hormones in acc~crdance with known methods
and the shoots transferred to rooting medium for regeneration of plants. The
plants may then be used to establish repetitive generations, either from seeds
or
using vegetative propagation techniques.
The constructs of the present invention can be introduced into
plant cells using Ti plasmids, Ri plasmids, plant virus vectors, direct DNA
transformation, micro-injection, electroporation, etc. For reviews of such
techniques see for example Weissbach and Weissbach, Methods for Plant
Molecular Biology, Academy Press, New York 'VIII, pp. 421-463 (1988);
Geierson and Corey, Plant Molecular Biology, 2d Ed. (1988); and Miki and
Iyer, Fundamentals of Gene Transfer in Plants. :fn Plant Metabolism, 2d Ed.
DT. Dermis, DH Turpin, DD Lefebrve, DB Lay:aell (eds), Addison Wesly,
Langmans Ltd. London, pp. 561-579 (1997). The present invention further
includes a suitable vector comprising the chimeric gene construct.
The DNA sequences of the present invention thus include the DNA
sequences of SEQ ID NO: 1 and 2, the regulatory regions and fragments
thereof, as well as analogues of, or nucleic acid ~;equences comprising about
80% similarity with the nucleic acids as defined in SEQ ID NO's: 1 and 2.
Analogues (as defined above), include those DN~~ sequences which hybridize
under stringent hybridization conditions (see Maliiatis et al. , in Molecular
Cloning (A Laboratory Manual), Cold Spring Harbor Laboratory, 1982, p.
387-389) to the DNA sequence of SEQ ID NO: 1 or 2, provided that said
sequences maintain at least one regulatory property of the activity of the
regulatory element as defined herein.
CA 02246892 1998-09-09
-23-
An example of one such stringent hybridi;~ation conditions may be
hybridization in 4XSSC at 65 °C, followed by washing in O.1XSSC at 65
°C for
an hour. Alternatively an exemplary stringent hybridization condition could be
in 50% formamide, 4XSSC at 42°C. Analogues also include those DNA
sequences which hybridize to the sequences of SEQ ID NO: 1 or 2 under
relaxed hybridization conditions, provided that said sequences maintain at
least
one regulatory property of the activity of the regulatory element. Examples of
such non-hybridization conditions includes hybridization in 4XSSC at
50°C or
with 30-40 % formamide at 42 ° C .
There are several lines of evidence that suggest that the seed coat-
specific expression of GUS activity in the plant T'218 is regulated by a
cryptic
regulatory element. The region surrounding the regulatory element and
transcriptional start site for the GUS gene are not transcribed in
untransformed
plants. Transcription was only observed in plant T218 when T-DNA was
inserted in cis. DNA sequence analysis did not uncover a long open reading
frame within the 3.3 kb region cloned. Moreover, the region is very AT rich
and predicted to be noncoding (data not shown) by the Fickett algorithm
(Fickett, 1982, Nucleic Acids Res. 10, 5303-5318) as implemented in DNASIS
7.0 (Hitachi). Southern blots revealed that the insertion site is within the
N.
tomentosiformis genome and is not conserved among related species as would
be expected for a region with an important gene.
Furthermore, Northern analysis demonstrate that the transcript,
associated with the regulatory region and corresponding to the native plant
sequence, does not accumulate in developing seeds or leaves of untransformed
plants. This indicates that in native plants, the regulatory region as defined
as
pT218, is silent.
Similarly, results indicate that the constih~tive expression of GUS
activity in the plant T1275 is regulated by a cryptic regulatory element.
RNase
CA 02246892 1998-09-09
-24-
protection assays performed on the region spanning the regulatory element and
downstream region did not reveal a transcript for the sense strand (see Figure
16, Table 2). RNase protection assays were performed using RNA from organs
of untransformed tobacco and probes that spanned the T 1275 sequence from
about -2055 by to +1200 by relative to the transcriptional start site. In all
tissues
tested (leaf, stem, root, flower bud, petal, ovary and developing seed)
protected
fragments were not detected, in the sense orientation relative to the GUS
coding
region, with all probes (Figure 16; see also PCT C.'A97/00064, which is
incorporated by reference). Furthermore, GenB~ink searches revealed no
significant sequence similarity with the T1275 sequence. An amino acid
identity
of about 66% with two open reading frames on the; antisense strand of the
genomic sequence of T1275 (between about -141 E. and -1308, nucleotides 636-
746 of SEQ ID N0:2; and between about -541 and -395, nucleotides 1513-1659
of SEQ ID N0:2 relative to the transcriptional start) and an open reading
frame of
a partial Arabidopsis expressed sequence (GenBank Accession No. W43439) was
identified. The sequence which lies downstream of sequences at the T-DNA
insertion point in untransformed tobacco shows no significant similarity in
GenBank searches. These data suggest that this region is silent in
untransformed
plants and that the insertion of the T-DNA activatf;d a cryptic promoter.
Southern analysis indicates that the 2.2 kb regulatory region of T1275
does not hybridize with DNA isolated from soybean, potato, sunflower,
Arabidopsis, B. napus, B. oleracea, corn, wheat or black spruce. However,
transient assays indicate that this regulatory region can direct expression of
the
GUS coding region in all plant species tested including canola, tobacco,
soybean,
alfalfa, pea, arabidopsis, corn, wheat and barley (Table 3), indicating that
this
regulatory element is useful for directing gene expression in both dicot and
monocot plants.
The transcriptional start site was delimited by RNase protection assay to a
single position about 220 by upstream of the translational initiation codon of
the
CA 02246892 1998-09-09
-25-
GUS coding region in the T-DNA. The sequence around the transcriptional start
site exhibits similarity with sequences favored at the transcriptional start
site
compiled from available dicot plant genes (T/A TIC A+, A C/A CIA A/C/T A A
A/T). Sequence similarity is not detected about 30 by upstream of the
transcriptional start site with the TATA-box consensus compiled from available
dicot plant genes (C T A T A A/T A T/A A).
Deletions in the upstream region indicate that negative regulatory
elements and enhancer sequences exist within the full length regulatory
region.
For example, deletion of the 5' region to BstYI (-394 relative to the
transcriptional start site) resulted in a 3 to 8 fold increase in expression
of the
gene associated therewith (see Table 6), indicating the occurrence of at least
one negative regulatory element within the XbaI-.BstYI portion of the full
length
regulatory element. Other negative regulatory elements also exist within the
XbaI- BstYI fragment as removal of an XbaI-PstI fragment also resulted in
increased activity (-1305-GUS-nos; Table 6). An enhancer is also localized
within the BstYI-DraI fragment as removal of this region results in a 4 fold
loss
in activity of the remaining regulatory region (-1!~7-GUS-nos; Table 6).
5' deletions of the promoter (see Figures l~s(A) and (B) and analysis by
transient expression using biolistics showed that the promoter was active
within a
fragment 62 by from the transcriptional start site indicating that the core
promoter
has a basal level of expression (see Table 5). Deleaion of a fragment
containing
the transcriptional start site (see -62(-tsr)/GUS/no~; in Figure 13 (B); Table
5) did
not eliminate expression, however deletions to -12 by and further (i.e. +30)
did
eliminate expression indicating that the region defined by -(62-12) by
(nucleotides 1992-2042 of SEQ ID N0:2) contained the core promoter. DNA
sequence searches did not reveal conventional core promoter motifs found in
plant genes such as the TATA box.
CA 02246892 1998-09-09
-26-
A number of the 5' promoter deletion clonea (Figure 13 (A)) were
transferred into tobacco by Agrobacterium-mediated transformation using the
vector pRD400. Analysis of GUS specific activity in leaves of transgenic
plants
(see Table 6) confirmed the transient expression data down to the -197
fragment
(nucleotides 1857- of SEQ ID N0:2). Histochemical analysis of organs sampled
from the transgenic plants indicated GUS expression in leaf, seeds and flowers
as
a minimum as not all the organs were analyzed.
A comparison of GUS specific activities in the leaves of transgenic
tobacco SR1 transformed with the T1275-GUS-nos gene and the 355-GUS-nos
genes revealed a similar range of values (Figure. 14(A)). Furthermore, the GUS
protein levels detected by Western blotting were similar between plants
transformed with either gene when the GUS specific activities were similar
(Figure. 14(C)). Analysis of GUS mRNA levels by RNase protection however
revealed that the levels of mRNA were about 60 fold (mean of 13 measurements)
lower in plants transformed with the T1275-GUS-nos gene (Figure 14(B)
suggesting the existence of a post-transcriptional regulatory element in the
mRNA leader sequence.
Further analysis confirmed the presence of a regulatory sequence within
the NdeI-SmaI fragment of the mRNA leader sequence that had a significant
impact on the level of GUS specific activity expressed in leaves. Deletion of
the
NdeI-SmaI fragment from the T1275-GUS-nos gene (Figure 15) resulted in a 46-
fold reduction in the amount of GUS specific activity that could be detected
in
leaves of transgenic tobacco cv Delgold (see Tablf; 7). Addition of the same
fragment to a 355-GUS-nos gene (Figure 15) construct increased the amount of
GUS specific activity by 4-fold (see Table 7). The; data is consistent with
the
presence of a post-transcriptional regulatory element in this fragment.
The Ndel Smal regulatory elements situated downstream of the
transcriptional start site functions both at a transcriptional, and post-
CA 02246892 1998-09-09
-27-
transcriptional level. The levels of rnRNA examined from transgenic tobacco
plants transformed with either T1275-GUS-nos, T1275-N-GUS-nos, 35S-GUS-
nos, or 35S+N-GUS-nos, are higher in transgenic ~alants comprising the Ndel
Smal fragment under the control of the T1275 promoter but lower in those under
control of the 35S promoter, than in plants comprising constructs that lack
this
region (Figure 17 (A)). This indicates that this region functions by either
modulating transcriptional rates, or the stability of the transcript, or both.
The Ndel Smal region also functions post-~.ranscriptionally. The ratio of
GUS specific activity to relative RNA level in individual transgenic tobacco
plants that lack the Ndel Smal fragment is lower, and when averaged indicates
an
eight fold reduction in GUS activity per RNA, than in plants comprising this
region (Figure 17 (B)). Similarly, an increase, by an average of six fold, in
GUS
specific activity is observed when the Ndel Smal region is added within the
35S
untranslated region (Figure 17 (B)). The GUS specific activity:relative RNA
levels are similar in constructs containing the Nde~! Smal fragment (T1275-GUS-
nos and 35S+N-GUS-nosy. These results indicate that the Ndel Smal fragment
modulates gene expression post-transcriptionally. Further experiments suggest
that this region is a novel translational enhancer. 'translation of
transcripts in
vitro demonstrate an increase in translational efflc iency of RNA containing
the
Ndel to Smal fragment (see Table 8). Furthermorf;, the levels of protein
produced
using mRNAs comprising the Ndel-Smal fragment are greater than those
produced using the known translational enhancer of Alfalfa Mosaic Virus RNA4
(refJ. These results indicate that this region functions post-
transcriptionally, as a
translational enhancer.
As this is the first report of cryptic regulatory elements in plants, it is
impossible to estimate the degree to which cryptic regulatory elements may
contribute to the high frequencies of promoterles;~ marker gene activation in
plants. It is interesting to note that transcription~~l GUS fusions in
Arabidopsis
occur at much greater frequencies (54%) than translational fusions (1.6% ,
CA 02246892 1998-09-09
-28-
Kertbundit et al., 1991, Proc. Natl. Acad. Sci. L'SA 88, 5212-5216). The
possibility that cryptic promoters may account for some fusions was recognized
by Lindsey et al. (1993, Transgenic Res. 2, 33-f7).
The cryptic regulatory elements of the present invention may be used to
control the expression of any given gene within a plant. Furthermore, the
cryptic regulatory elements as described herein may be used in conjunction
with
other regulatory elements, such as tissue specific , inducible or constitutive
promoters, enhancers, or fragments thereof, and the like. For example, the
regulatory region or a fragment thereof as defined herein may be used to
regulate gene expression of a gene of interest spatially and developmentally
within developing seed coats. Some examples of such uses, which are not to be
considered limiting, include:
1. Modification of storage reserves in seed coats, such as starch by
the expression of yeast invertase to mobilize the starch or
expression of the antisense transcript of ADP-glucose
pyrophosphorylase to inhibit starch biosynthesis.
2. Modification of seed color contributed by condensed tannins in
the seed coats by expression of antisense transcripts of the
phenylalanine ammonia lyase or chalcone synthase genes.
3. Modification of fibre content in seed-derived meal by expression
of antisense transcripts of the caffeic acid-o-methyl transferase or
cinnamoyl alcohol dehydrogenase genes.
4. Inhibition of seed coat maturation by expression of ribonuclease
genes to allow for increased seed ;size, and to reduce the relative
biomass of seed coats, and to aid in dehulling of seeds.
CA 02246892 1998-09-09
-29-
5. Expression of genes in seed coats ~~oding for insecticidal proteins
such as a-amylase inhibitor or protease inhibitor.
6. Partitioning of seed metabolites such as glucosinolates into seed
coats for nematode resistance.
7. Nucleotide fragments of the regulatory region of at least 19 by as
a probe in order to identify analogous regions within other plants
etc.
Similarly, a constitutive regulatory element may also be used to drive
the expression within all organs or tissues, or both of a plant of a gene of
interest, and such uses are well established in the literature. For example,
fragments of specific elements within the 35S Ca:MV promoter have been
duplicated or combil~ed with other promoter fragments to produce chimeric
promoters with desired properties (e.g. U.S. 5,4~~1,288, 5,424,200,
5,322,938, 5,196,525, 5,164,316). As indicated above, a constitutive
regulatory element or a fragment thereof, as defined herein, may also be used
along with other promoter, enhancer elements, or fragments thereof ,
translational enhancer elements or fragments thereof in order to control gene
expression. Furthermore, oligonucleotides of 18 bps or longer are useful as
probes or PCR primers in identifying or amplifying related DNA or RNA
sequences in other tissues or organisms.
Thus this invention is directed to regulatory elements and gene
combinations comprising these cryptic regulatory elements. Further this
invention is directed to such regulatory elements and gene combinations in a
cloning vector, wherein the gene is under the control of the regulatory
element
and is capable of being expressed in a plant cell transformed with the vector.
This invention further relates to transformed plant cells and transgenic
plants
regenerated from such plant cells. The regulatory element, and regulatory
CA 02246892 1998-09-09
-30-
element-gene combination of the present invention can be used to transform any
plant cell for the production of any transgenic plant. The present invention
is
not limited to any plant species.
While this invention is described in detail with particular reference to
preferred embodiments thereof, said embodiments are offered to illustrate but
not limit the invention.
EXAMPLES
Transfer of binary constructs to Agrobact~~rium and leaf disc
transformation of Nicotiana tabacum SRl were performed as described by
Fobert et al. (1991, Plant Mol. Biol. 17, 837-851'.). Plant tissue was
maintained on 100 ~,g/ml kanamycin sulfate (Sigma) throughout in vitro
culture.
Nine-hundred and forty transgenic plants were produced. Several
hundred independent transformants were screened for GUS activity in
developing seeds using the fluorogenic assay. One of these, T218, was chosen
for detailed study because of its unique pattern of GUS expression.
Furthermore, following the screening of transfonnants in a range of plant
organs, T1275 was selected which exhibited high level, constitutive expression
of GUS.
Characterization of a Seed Coat-Specific GUS Fusion - T218
Fluorogenic and histological GUS assays were performed according to
Jefferson (Plant Mol. Biol. Rep. , 1987, 5, 387-405), as modified by Fobert et
al. (Plant Mol. Biol. , 1991, 17, 837-851 ). For i nitial screening, leaves
were
harvested from in vitro grown plantlets. Later flowers corresponding to
developmental stages 4 and 5 of Koltunow et al. (Plant Cell, 1990, 2,
1201-1224) and beige seeds, approximately 12-lei dpa (Chen et al., 1988,
CA 02246892 1998-09-09
-31-
EMBO J. 7, 297-302), were collected from plants grown in the greenhouse.
For detailed, quantitative analysis of GUS activity, leaf, stem and root
tissues
were collected from kanamycin resistant F 1 progeny of the different
transgenic
lines grown in vitro. Floral tissues were harvested at developmental stages
8-10 (Koltunow et al., 1990, Plant Cell 2, 1201-1224) from the original
transgenic plants. Flowers of these plants were also tagged and developing
seeds were collected from capsules at 10 and 20 dpa. In all cases, tissue was
weighed, immediately frozen in liquid nitrogen, and stored at -80°C.
Tissues analyzed by histological assay were at the same developmental
stages as those listed above. Different hand-cut ~;ections were analyzed for
each
organ. For each plant, histological assays were performed on at least two
different occasions to ensure reproducibility. Except for floral organs, all
tissues were assayed in phosphate buffer according to Jefferson (1987, Plant
Mol. Biol. Rep. 5, 387-405), with 1 mM X-Gluc (Sigma) as substrate. Flowers
were assayed in the same buffer containing 20 % (v/v) methanol (Kosugi et al.
,
1990, Plant Sci. 70, 133-140).
Tissue-specific patterns of GUS expression were only found in seeds.
For instance, GUS activity in plant T218 (Figure 1) was localized in seeds
from
9 to 17 days postanthesis (dpa). GUS activity was not detected in seeds at
other stages of development or in any other tissuf~ analyzed which included
leaf,
stem, root, anther, ovary, petal and sepal (Figure: 1). Histological staining
with
X-Gluc revealed that GUS expression in seeds at 14 dpa was localized in seed
coats but was absent from the embryo, endosperm, vegetative organs and floral
organs (results not shown).
The seed coat-specificity of GUS expression was confirmed with the
more sensitive fluorogenic assay of seeds derived from reciprocal crosses with
untransformed plants. The seed coat differentiates from maternal tissues
called
the integuments which do not participate in double fertilization (Esau, 1977,
CA 02246892 1998-09-09
-32-
Anatomy of Seed Plants. New York: John Wiley and Sons). If GUS activity is
strictly regulated, it must originate from GUS fu~~ions transmitted to seeds
maternally and not by pollen. As shown in Tablf: 1, this is indeed the case.
As
a control, GUS fusions expressed in embryo and endosperm, which are the
products of double fertilization, should be transmitted through both gametes.
This is illustrated in Table 1 for GUS expression driven by the napin promoter
(BngNAPI, Baszczynki and Fallis, 1990, Plant Nlol. Biol. 14, 633-635) which
is active in both embryo and endosperm (data not: shown).
Table 1. GUS activity in seeds at 14 days post anthesis.
Cross (JUS Acs ivit,~!
nmol~e MUlminlmg Protein
T218 T218 1.09 t 0.39
T218 WTa 3.02 t 0.19
WT T218 0.04 t 0.005
WT WT 0.04 t 0.005
NAP-5b NAP-5 14.6 t 7.9
NAP-5 WT 3.42 t 1.60
WT NAP-5 2.91 t 1.97
a WT, untransformed plants
b Transgenic tobacco plants with the GUS gene fused to the
napin, BngNAPl, promoter (Baszczynski and Fallis, 1990, Plant
Mol. Biol. 14, 633-635).
Cloning and Analysis of the Seed Coat-Specific' GUS Fusion
Genomic DNA was isolated from freeze-dried leaves using the protocol
of Sanders et al. (1987, Nucleic Acid Res. 15, 1543-1558). Ten micrograms of
T218 DNA was digested for several hours with ~?coRI using the appropriate
manufacturer-supplied buffer supplemented with 2.5 mM spermidine. After
electrophoresis through a 0.8% TAE agarose gel, the DNA size fraction around
CA 02246892 1998-09-09
-33-
4-6 kb was isolated, purified using the GeneClean kit (BIO 101 Inc., LaJolla,
CA), ligated to phosphatase-treated EcoRI-digested Lambda GEM-2 arms
(Promega) and packaged in vitro as suggested by the supplier. Approximately
125,000 plaques were transferred to nylon filters (Nytran, Schleicher and
Schuell) and screened by plaque hybridization (Rutledge et al. , 1991, Mol.
Gen. Genet. 229, 31-40), using the 3' (termination signal) of the nos gene as
probe (probe #l, Figure 2). This sequence, contained in a 260 by SstIlEcoRI
restriction fragment from pPRF-101 (Fobert et a~'., 1991, Plant Mol. Biol. 17,
837-851), was labelled with [a-32P]-dCTP (NEN) using random priming
(Stratagene). After plaque purification, phage D;VA was isolated (Sambrook et
al., 1989, A Laboratory Manual. New York: Cold Spring Harbor Laboratory
Press), mapped and subcloned into pGEM-4Z (Promega).
The GUS fusion in plant T218 was isolated as a 4.7 kb EcoRI fragment
containing the 2.2kb promoterless GUS-nos gene at the T-DNA border of
pPRF120 and 2.5 kb of 5' flanking tobacco DNA, (pT218, Figure 2), using the
nos 3' fragment as probe (probe #1, Figure 2). ~Co confirm the ability of the
flanking DNA to activate the GUS coding region, the entire 4.7 kb fragment
was inserted into the binary transformation vector pBIN 19 (Bevan, 1984, Nucl.
Acid Res. 12, 8711-8721), as shown in Figure 2. Several transgenic plants
were produced by Agrobacterium-mediated transi:ormation of leaf discs. Plants
were transformed with a derivative which contained the 5' end of the GUS gene
distal to the left border repeat. This orientation is the same as that of the
GUS
gene in the binary vector pB1101 (Jefferson, 198',x, Plant Mol. Biol. Rep. 5,
387-405). Southern blots indicated that each plant contained 1-4 T-DNA
insertions at unique sites. The spatial patterns of GUS activity were
identical to
that of plant T218. Histologically, GUS staining was restricted to the seed
coats of 14 dpa seeds and was absent in embryos and 20 dpa seeds (results not
shown). Fluorogenic assays of GUS activity in dLeveloping seeds showed that
expression was restricted to seeds between 10 and 17 dpa, reaching a maximum
at 12 dpa (Figure 3 (a) and 3 (b)). The 4.7 kb fragment therefore contained
all
CA 02246892 1998-09-09
-34-
of the elements required for the tissue-specific and developmental regulation
of
GUS expression.
To locate regions within the flanking plant DNA responsible for seed
coat-specificity, truncated derivatives of the GUS fusion were generated
(Figure
2) and introduced into tobacco plants. Deletion of the region approximately
between 2.5 and 1.0 kb, 5' of the insertion site (pT218-2, Figure 2) did not
alter expression compared with the entire 4.7 kb GUS fusion (Figures 3b and
4). Further deletion of the DNA, to the SnaBI rE;striction site approximately
0.5 kb, 5' of the insertion site (pT218-3, Figure :Z), resulted in the
complete
loss of GUS activity in developing seeds (Figure~~ 3b and 4). This suggests
that
the region approximately between 1.0 and 0.5 kb, 5' of the insertion site
contains elements essential to gene activation. GUS activity in seeds remained
absent with more extensive deletion of plant DN~~ (pT218-4, Figures 2, 3b and
4) and was not found in other organs including leaf, stem, root, anther,
petal,
ovary or sepal from plants transformed with any of the vectors (data not
shown).
The transcriptional start site for the GUS gene in plant T218 was
determined by RNase protection assays with RNr~ probe #4 (Figure 2) which
spans the T-DNA/plant DNA junction. For RNase protection assays, various
restriction fragments from pIS-1, pIS-2 and pT218 were subcloned into the
transcription vector pGEM-4Z as shown in Figures 7 and 2, respectively. A
440bp HindIII fragment of the tobacco acetohydroxyacid synthase SURA gene
was used to detect SURA and SURF mRNA. DrfA templates were linearized
and transcribed in vitro with either T7 or SP6 po lymerases to generate strand-
specific RNA probes using the Promega transcription kit and [a 32P]CTP as
labelled nucleotide. RNA probes were further processed as described in
Ouellet et al. (1992, Plant J. 2, 321-330). RNase protection assays were
performed as described in Ouellet et al., (1992, i°lant J. 2, 321-330),
using 10-
30 ~.g of total RNA per assay. Probe digestion vas done at 30°C for 15
min
CA 02246892 1998-09-09
-35-
using 30 ~,g mln RNase A (Boehringer Mannheim) and 100 units ml-' RNase
T1 (Boehringer Mannheim). Figure 5 shows that two termini were mapped in
the plant DNA. The major 5' terminus is situated at an adenine residue, 122
by upstream of the T-DNA insertion site (Figure 6). The sequence at this
transcriptional start site is similar to the consensus sequence for plant
genes
(C/TTC IATCA; Joshi, 1987 Nucleic Acids Res. 15, 6643-6653). A TATA
box consensus sequence is present 37 by upstrearn of this start site (Figure
6).
The second, minor terminus mapped 254 by from the insertion site in an area
where no obvious consensus motifs could be identified (Figure 6).
The tobacco DNA upstream of the insertion site is very AT-rich
( > 75 % , see Figure 7). A search for promoter-like motifs and scaffold
attachment regions (SAR), which are often associated with promoters (Breyne
et al., 1992, Plant Cell 4, 463-471; Gasser and I,aemmli, 1986, Cell 46, 521-
530), identified several putative regulatory elements in the first 1.0 kb of
tobacco DNA flanking the promoterless GUS gene (data not shown).
However, the functional significance of these sequences remains to be
determined.
Cloning and Analysis of the Insertion Site from Untransformed Plants
A lambda DASH genomic library was prepared from DNA of
untransformed N. tabacum SRl plants by Stratag~~ne for cloning of the
insertion
site corresponding to the gene fusion in plant T2i18. The screening of 500,000
plaques with probe #2 (Figure 2) yielded a single lambda clone. The EcoRI
and XbaI fragments were subcloned in pGEM-4Z. to generate pIS-1 and pIS-2.
Figure 7 shows these two overlapping subclones, pIS-1 (3.0 kb) and pIS-2 (1.1
kb), which contain tobacco DNA spanning the insertion site (marked with a
vertical arrow). DNA sequence analysis (using dideoxy nucleotides in both
directions) revealed that the clones, pT218 and p:fS-1, were identical over a
length of more than 2.5 kb, from the insertion site to their 5' ends, except
for a
CA 02246892 1998-09-09
-36-
12 by filler DNA insert of unknown origin at the T-DNA border (Figure 6 and
data not shown). The presence of filler DNA is ;~ common feature of T-
DNA/plant DNA junctions (Gheysen et al., 1991, Gene 94, 155-163). Gross
rearrangements that sometimes accompany T-DNA insertions (Gheysen et al.,
1990, Gene 94, 155-163; and 1991, Genes Dev. :5, 287-297) were not found
(Figure 6) and therefore could not account for thf: promoter activity
associated
with this region. The region of pIS-1 and pIS-2, 3' of the insertion site is
also
very AT-rich (Figure 7).
To determine whether there was a gene associated with the pT218
promoter, more than 3.3 kb of sequence contained with pIS-1 and pIS-2 was
analyzed for the presence of long open reading frames (ORFs). However, none
were detected in this region (data not shown). T~~ determine whether the
region
surrounding the insertion site was transcribed in ~antransformed plants,
Northern blots were performed with RNA from leaf, stem, root, flower and
seeds at 4, 8, 12, 14, 16, 20 and 24 dpa. Total F~IVA from leaves was isolated
as described in Ouellet et al., (1992, Plant J. 2, :321-330). To isolate total
RNA from developing seeds, 0.5 g of frozen tissue was pulverized by grinding
with dry ice using a mortar and pestle. The powder was homogenized in a 50
ml conical tube containing 5 ml of buffer (1 M Tris HCI, pH 9.0, 1 % SDS)
using a Polytron homogenizer. After two extractions with equal volumes of
phenol:chloroform:isoamyl alcohol (25:24:1), nucleic acids were collected by
ethanol precipitation and resuspended in water. 'Che RNA was precipitated
overnight in 2M LiCI at 0°C, collected by centri~Fugation, washed in
70%
ethanol and resuspended in water. Northern blot hybridization was performed
as described in Gottlob-McHugh et al. (1992, Plcznt Physiol. 100, 820-825).
Probe #3 (Figure 2) which spans the entire region of pT218 5' of the insertion
did not detect hybridizing RNA bands (data not shown). To extend the
sensitivity of RNA detection and to include the rc;gion 3' of the insertion
site
within the analysis, RNase protection assays were. performed with 10 different
RNA probes that spanned both strands of pIS-1 and pIS-2 (Figure 7). Even
CA 02246892 1998-09-09
-37-
after lengthy exposures, protected fragments could not be detected with RNA
from 8, 10, 12 dpa seeds or leaves of untransforrned plants (see Figure 5 for
examples with two of the probes tested). The spE;ciflc conditions used allowed
the resolution of protected RNA fragments as small as 10 bases (data not
shown). Failure to detect protected fragments was not due to problems of RNA
quality, as control experiments using the same samples detected
acetohydroxyacid synthase (AHAS) SURA and SL'RB mRNA which are
expressed at relatively low abundance (data not s)hown). Conditions used in
the
present work were estimated to be sensitive enough to detect low-abundance
messages representing 0.001-0.01 % of total mRTTA levels (Ouellet et al.,
1992,
Plant J. 2, 321-330). Therefore, the region flanl;ing the site of T-DNA
insertion does not appear to be transcribed in untransformed plants.
Genomic Origins of the Insertion Site
Southern blots were performed to determine if the insertion site is
conserved among Nicotiana species. Genomic DNA (5 ~,g) was isolated,
digested and separated by agarose gel electrophoresis as described above.
After capillary transfer on to nylon filters, DNA was hybridized, and probes
were labeled, essentially as described in Rutledge: et al. (1991, Mol. Gen.
Genet. 229, 31-40). High-stringency washes were in 0.2 x SSC at 65
°C while
low-stringency washes were in 2 x SSC at room ~:emperature. In Figure 8,
DNA of the allotetraploid species N. tabacum and the presumptive progenitor
diploid species N. tomentosiformis and N. sylvestris (Okamuro and Goldberg,
1985, Mol. Gen. Genet. , 198, 290-298) were hybridized with probe #2 (Figure
2). Single hybridizing fragments of identical size; were detected in N.
tabacum
and N. tomentosiformis DNA digested with Hind:(II, XbaI and EcoRI, but not in
N. sylvestris. Hybridizations with pIS-2 (Figure 8) which spans the same
region but includes DNA 3' of the insertion site :Melded the same results.
They
did not reveal hybridizing bands, even under conditions of reduced stringency,
in additional Nicotiana species including N. rushca, N. glutinosa, N.
CA 02246892 1998-09-09
-38-
megalosiphon and N. debneyi (data not shown). Probe #3 (Figure 2) revealed
the presence of moderately repetitive DNA specific to the 1V. tomentosiformis
genome (data not shown). These results suggest that the region flanking the
insertion site is unique to the N. tomentosiformis genome and is not conserved
among related species as might be expected for rf:gions that encode essential
genes.
Characterization of a Constitutive GUS fusion -T1275
From the transgenic plants produced (see above), one of these, T1275,
was chosen for detail~l study because of its high level and constitutive
expression of GUS (see also US patent application 08/593,121 and
PCT/CA97100064, both of which are incorporated by reference).
Fluorogenic and histological GUS assays 'were performed as outlined
above. For initial screening, leaves were harvested from in vitro grown
plantlets. Later nine different tissues: leaf (L), ~~tem (S), root (R), anther
(A),
petal (P), ovary (O), sepal (Se), seeds 10 days past anthesis (S1) and seeds
20
days post-anthesis (S2), were collected from plants grown in the greenhouse
and analyzed.
GUS activity in plant T1275 was found in all tissues. Figure 10 shows
the constitutive expression of GUS by histochemical staining with X-Gluc of
T1275, including leaf (a), stem (b), root (c), flower {d), ovary (e), embryos
(f
and g), and seed (h).
Constitutive GUS expression was confirmed with the more sensitive
fluorogenic assay of plant tissue from transformed plant T1275. These results
are shown in Figure 11. GUS expression was evident in all tissue types
including leaf (L), stem (S), root (R), anther (A).. pistil (P), ovary (O),
sepal
(Se), seeds at 10 dpa (S1) and 20 dpa (S2). Furthermore, the level of GUS
CA 02246892 1998-09-09
-39-
expression in leaves was comparable to the level of expression in transformed
plants containing the constitutive promoter CaMV 35S in a GUS - nos fusion.
As reported by Fobert et al. ( 1991, Plant Molecu lar Biology, 17 : 837-851 )
GUS activity in transformed plants containing pBa121 (Clontech), which
contains a CaMV 35S - GUS - nos chimeric gene, was as high as 18,770 t
2450 (pmole MU per minute per mg protein).
Cloning and Analysis of the Constitutive Promoter - GUS Fusion
Genomic DNA was isolated from leaves according to Hattori et al.
(1987, Anal. Biochem. 165, 70-74). Ten ~.g of T'1275 total DNA was digested
with EcoRI and XbaI according to the manufacturer's instructions. The
digested DNA was size-fractionated on a 0.7 % agarose gel. The DNA
fragments of about 4 to 6 kb were isolated from the gel using the Elu-Quick
kit
(Schleicher and Schuell) and ligated to lambdaGF:M-2 arms previously digested
with EcoRI and XbaI and phosphatase-treated. About 40,000 plaques were
transferred to a nylon membrax>e (Hybond, Amersham) and screened with the
sap-labelled 2kb GUS insert isolated form pBI121, essentially as described in
Rutledge et al. (1991, Mol. Gen Genet. 229, 31-40). The positive clones were
isolated. The XbaI-EcoRI fragment (see restriction map Figure 12) was
isolated from the lambda phage and cloned into pTZl9R previously digested
with XbaI and EcoRI and treated with intestinal <;alf phosphatase.
The plant DNA sequence within the clone:, SEQ ID N0:2, has not been
previously reported in sequence data bases. It is not observed among diverse
species as Southern blots did not reveal bands hybridizing with the fragment
in
soybean, potato, sunflower, Arabidopsis, B. napus, B. oleracea, corn, wheat or
black spruce (data not shown). In tobacco, Southern blots did not reveal
evidence for gross rearrangements at or upstream of the T-DNA insertion site
(data not shown).
CA 02246892 1998-09-09
-40-
The T1275 Regulatory Element is Cryptic
The 4.2kb fragment containitng about 2.2k:b of the T 1275 promoter
fused to the GUS gene and the nos 3' was isolated) by digesting pTZ-T1275 with
S HindIII and EcoRI. The isolated fragment was li,gated into the pRD400 vector
(Datla et al. , 1992, Gene, 211:383-384) previously digested with HindIII and
EcoRI and treated with calf intestinal phosphatase. Transfer of the binary
vector to Agrobacterium tumefaciens and leaf disc. transformation of N.
tabacum SRl were performed as described above. GUS activity was examined
in several organs of many independent transgenic lines. GUS rnRNA was also
examined in the same organ by RNase protection assay (Melton et al, 1984,
Nucleic Acids Res. 121: 7035 - 7056) using a probe that mapped the mRNA 5'
end in both untransformed and transgenic tissues. RNA was isolated from
frozen-ground tissues using the TRIZOL Reagent: (Life Technologies) as
described by the manufacturer. For each assay 10 - 30 ug of total RNA was
hybridized to RNA probes described in Figure lfi (A). Assays were performed
using the RPAII kit (Ambion CA) as described b;y the manufacturer. The
protected fragments were separated on a 5 % Long Ranger acrylamide (J.J.
Baker, N.J.) denaturing gel which was dried and exposed to Kodak X-RP film.
RNase protection assays performed with 1Z1VA from leaves, stem, root,
developing seeds and flowers of transgenic tobacco revealed a single protected
fragment in all organs indicating a single transcription start site that was
the
same in each organ, whereas RNA from untransformed tobacco tissues did not
reveal a protected fragment (Figure 16 (B)). The insertion site, including
1200
by downstream, was cloned from untransformed tobacco as a PCR fragment
and sequenced. A composite restriction map of the insertion site was
assembled as shown in Figure 16 (A). RNA probes were prepared that spanned
the entire region as shown in Figure 16 (A). Rr'ase protection assays did not
reveal transcripts from the sense strand as summarized in Table 2. These data
suggest that the insertion site is transcriptionally silent in untransformed
CA 02246892 1998-09-09
-41-
tobacco and is activated by T-DNA insertion. The region upstream of the
insertion site is therefore another example of a pl;~nt cryptic regulatory
element.
Table 2.
Summary of the RNase Protection Assays of the insertion site in
untransformed tobacco. Sce Figure 16 (.~) for probe positions.
Probe Rnase Protection Assay result
Looking for "sense" RNAs (relative to the T1275 promoter)
C8-EcoRI many bands, all in tRNA (negative control)
A10-HindIII no bands
2-21-HindIII no bands
1-4 SmaI many bands, all in tIRNA
7-EcoRI faint bands, all in tRNA
Constitutive Activity of the T1275 Regulatory Element
For analysis of transient expression of Gt7S activity mediated by
biolistics (Sandford et al, 1983, Methods Enzymol, 217: 483-509), the Xbal -
EcoRl fragment was subcloned in pUCl9 and G~JS activity was detected by
staining with X-Gluc as described above. Leaf tissue of greenhouse-grown
plants or cell suspension cultures were examined for the number of blue spots
that stained. As shown in Table 3, the T1275 - GUS nos gene was active in
each of the diverse species examined and can direct expression of a gene of
interest in all plant species tested. Leaf tissue of canola, tobacco, soybean,
CA 02246892 1998-09-09
-42-
alfalfa, pea and arabidopsis, and cell suspensions of oat, corn, wheat and
barley
exhibited GUS-positive blue spots after transient lbombardment-mediated assays
and histochemical GUS activity staining. This suggests that the T1275
regulatory element may be useful for directing gene expression in both dicot
and monocot plants.
CA 02246892 1998-09-09
-43-
TABLE 3
Transient Expression of GUS Activity in Tissues of Diverse Plant Species
Ti~ue Source Species GUS Activity
~
Leaf Soybean + + +
Alfalfa + +
Arabidopsis +
Leaf disc Tobacco + +
B. napes +
Pea +
Cell Cultures Oat +
Corn +
Wheat +
Barley + +
* Numbers of blue spots: 1 - 10 (+), 10 - 100 (-i- +), 100 - 400 (+ + +)
For analysis of GUS expression in different organs, lines derived from
progeny of the above lines were examined in detail. Table 4 shows the GUS
specific activities in one of these plants. It is exyressed in leaf, stem,
root,
developing seeds and the floral organs, sepals, pedals, anthers, pistils and
ovaries at varying levels, confirming constitutive expression. Introduction of
the same vector into B. napes also revealed expression of GUS activity in
these
organs (data not shown) indicating that constitutive expression was not
specific
to tobacco. Examination of GUS mRNA in the tobacco organs showed that the
transcription start sites was the same in each (Fig;ure 16 (B)) and the level
of
mRNA was similar except in flower buds where it was lower (Table 4).
CA 02246892 1998-09-09
-44-
TABLE 4
GUS Specific Activity and Relative RNA Levels in the
Organs of Progeny of Transgenic Line T64
Organ Relative GUS GUS Specific
&NA Activity
Levels in T64 (picomollMUlmin/mg
protein)
Progeny (grey
scale units) Transformed Untransformed
Tobacco T64 Tobacco
Leaf 1774 988.32 3.02
Stem 1820 826.48 7.58
Root 1636 4078.45 22.18
14 day post 1790 253.21 10.03
anthesis
Seeds
Flower - 715 2.59 ND*
buds
Petals ND * 28. 24 1. 29
Anthers ND * 4. 64 0. 35
Pistils ND* 9.76 1.72
Sepals ND* 110.02 2.48
Ovary ND* 4.42 2.71
* Not Done
Identification of Regulatory Elements within the Full Length T1275
Regulat~ry Element
An array of deletions of the full length regulatory region of T1275 were
prepared, as identified in Figures 13 (A) and (B), for further analysis of the
cryptic regulatory element.
5' deletions of the promoter (see Figures 13(A) and (B) and analysis by
transient expression using biolistics showed that the promoter was active
within
CA 02246892 1998-09-09
-45-
a fragment 62bp from the transcriptional start site indicating that the core
promoter has a basal level of expression (see Tablie 5).
Table 5.
Transient GUS activity detected in soybean leaves by staining with X-gluc
after particle bombardment. Vectors illustrated in Figures. 13 (A) and (B).
Genes (iUS staining
1. T1275-GUS-nos
2. -1639-GUS-nos v
3. -1304-GUS-nos v
4. -684-GUS-nos v
5. -394-GUS-nos v
6. -197-GUS-nos
7. -62-GUS-nos +
8. -62(-tsr)-GUS-nos
9. -12-GUS-nos -
10.
+30-GUS-nos
-
Deletion of a fragment containing the trar~scriptional start site (see -62(-
tsr)/GUS/nos in Figure 14(B), Table 5) did not eliminate expression, however
deletions to -12 by and further (ie+30) did eliminate expression indicating
that
the region defined by -(62-12) (nucleotides 1992-2042 of SEQ ID N0:2) by
contained the core promoter. DNA sequence se~~rches did not reveal
conventional core promoter motifs within this region as are typically found in
plant genes, such as the TATA box.
A number of the 5' promoter deletion clones (Figure 13(A)) were
transferred into tobacco by Agrobacterium-mediated transformation using the
vector pRD400. Analysis of GUS specific activity in leaves of transgenic
CA 02246892 1998-09-09
-46-
plants (see Table 6) confirmed the transient expression data down to the -197
fragment (nucleotide 1857 of SEQ ID N0:2).
Table 6.
GUS specific activities in leaves of greenhouse-grown transgenic tobacco,
SRl, transformed with the T1275-GUS-nos gene fusion and 5' deletion
clones (see Figure 13 A). MeanfSE(n)
Genes GLIS specific activities
pmoles MU/min/mg protein
1. T1275-GUS-nos a!83~171 (27)
2. -1639-GUS-nos '~87~188 (26)
3. -1403-GUS-nos E~32~217 (10)
4. -683-GUS-nos in progress
5. -394-GUS-nos 16271340 (13)
6. -197-GUS-nos 47574 (27)
Histochemical analysis of organs sampled from tlae transgenic plants indicated
GUS expression in leaf, seeds and flowers as a minimum as not all the organs
were analyzed.
Activity of the T1275 Regulatory Element
Analysis of leaves of randomly-selected, l;reenhouse-grown plants
regenerated from culture revealed a wide range of GUS specific activities
(Figure 14 (A); T plants). Plants transformed with pBI 121 (CLONETECH)
which contains the 35S GUS-nos gene yielded comparable specific activity
levels (Figure 14 (A); S plants). Furthermore, the GUS protein levels detected
by Western blotting were similar between plants transformed with either gene
when the GUS specific activities were similar (Fig;ure. 14(C)).
CA 02246892 1998-09-09
-47-
Generally, the level of GUS mRNA in the: leaves as determined by
RNase protection (Figure 14 (B)) correlated with the GUS specific activities,
however, the level of GUS mRNA was about 60 i:old (mean of 13
measurements) lower in plants transformed with the T1275-GUS-nos gene
(Figure 14(B)) when compared with plants transformed with 35S-GUS-nos.
Since the levels of protein and the activity of extractable protein were
similar in plants transformed with T1275-GUS-nos or 35S-GUS-nos, yet the
mRNA levels were dramatically different, these results suggested the existence
of
a regulatory element downstream of the transcriptional start site in the
sequence
of T1275-derived transcript.
Post-Transcriptional Regulatory Elements witlxin T1275
An experiment was performed to determine the presence of a post-
transcriptional regulatory element within the T 12 7 5 leader sequence. A
portion
of the sequence downstream from the transcriptional initiation site was
deleted in
order to examine whether this region may have an effect on translational
efficiency (determined by GUS extractable activit:~), mRNA stability or
transcription.
Deletion of the Ndel-Smalfragment from the T1275-GUS-nos gene
(Figure 15; T1275-N-GUS-nos; including nucleotides 2086-2224 of SEQ ID
N0:2) resulted in a 46-fold reduction in the amount of GUS specific activity
that
could be detected in leaves of transgenic tobacco c:v Delgold (see Table 7).
Addition of the same fragment to a 35S-GUS-nos gene (Figure 15; 35S+N-GUS-
nos) construct increased the amount of GUS specific activity by 4-fold (see
Table
7).
CA 02246892 1998-09-09
-48-
Table 7.
GUS specific activity in leaves of greenhouse-grown transgenic tobacco cv
Delgold transformed with vectors designed to assess the presence of cryptic
regulatory sequences within the transcribed sequence derived from the
T1275 GUS gene fusion (see Figure 15). Mean~SE(n).
Gene GUS specific activity
pmoles MU/min/mg protein
1. T1275-GUS-nos 557+183 (21)
2. T1275-N-GUS-nos 1f.+3 (22)
3. P35S-GUS-nos 1818+692 (15)
4. P35S+N-GUS-nos 699~~+3148 (23)
'The data presented in Table 7 are consistent with the presence of a post-
transcriptional regulatory sequence in the NdeI-SmaI fragment.
The Ndel Smal fragment functions as a transcriptional enhancer or mRNA
stability determinant
The levels of mRNA were determined in leaves obtained from plants
transformed with either T1275-GUS-nos, T1275-rJ-GUS-nos, 35S-GUS-nos, or
35S+N-GUS-nos (Figure 17 (A)). Relative RNA :levels were determined by
ribonuclease protection assay (Ambion RPAII Kit ) in the presence of a 32P-CTP
labeled in vitro transcribed probe and autoradiographic quantification using
Kodak Digital Science 1D Image Analysis Software. Hybridization conditions
used during RNase protection assay were overnigrit at 42-45 degrees in 80%
formamide, 100 mM sodium citrate pH 6.4, 300 m~M sodium acetate pH 6.4, 1
mM EDTA.
The levels of mRNA examined from transi;enic tobacco plants
transformed with either T1275-GUS-nos, T1275-rJ-GUS-nos, 35S-GUS-nos, or
CA 02246892 1998-09-09
-49-
35S+N-GUS-nos, were higher in transgenic plants comprising the Ndel Smal
fragment under the control of the T1275 promoter but lower in those under the
control of the 35S promoter, than in plants comprising constructs that lack
this
region (Figure 17 (A)). This indicates that this region functions by either
modulating transcriptional rates, or the stability of the transcript, or both.
The Ndel Smal fragment functions as a translational enhancer
Analysis were performed in order to determine whether the Ndel Smal
region functions post-transcriptionally. The GUS specific activity:relative
RNA
level was determined from the GUS specific activity measurements, and relative
RNA levels in greenhouse grown transgenic plants (figure 17 (B)). The ratio of
GUS specific activity to relative RNA level in individual transgenic tobacco
plants comprising the Ndel Smal fragment is higher than in plants that do not
comprise this region (Figure 17 {B)). Similar results are obtained when the
data are averaged, indicating an eight fold reduction in GUS activity per
RNA. Similarly, an increase, by an average of siz fold, in GUS specific
activity is observed when the lVdel Smal region is added within the 35S
untranslated region (Figure 17 (B)). The GUS s~~ecific activity:relative RNA
levels are similar in constructs containing the Nde.! Smal fragment (T1275-GUS-
nos and 35S+N-GUS-nosy. These results indicate that the Ndel Smal fragment
modulates gene expression post-transcriptionally.
Further experiments, involving in vitro translation, suggest that this region
is a novel translational enhancer. For these experiments, fragments, from
approximately 3' of the transcriptional start site to the end of the
terminator, were
excised from the constructs depicted in Figure 1 S using appropriate
restriction
endonucleases and ligated to pGEM4Z at an approximately similar distance from
the transcriptional start site used by the prokaryotic T7 RNA polymerase.
Another construct containing the AMV enhancer in the 5' UTR of a GUS-nos
fusion was similarly prepared. This AMV-GUS-nos construct was created by
restriction endonuclease digestion of an AMV-GUS-nos fusion, with BgIII and
EcoRl, from pBI525 (Dada et al., 1993, Plant Science 94: 139-149) and ligation
CA 02246892 1998-09-09
-$
with pGEM4Z (Promega) digested with BamHl arid EcoRl. Transcripts were
prepared in vitro in the presence of m'G(5')ppp(5')G Cap Analog (Ambion).
Transcripts were translated in vitro in Wheat Germ Extract (Promega) in the
presence of 35S-Methionine and fold enhancement calculated from TCA
precipitable cpms.
Translation of transcripts in vitro demonstrate an increase in translational
efficiency of RNA containing the Ndel to Smal fragment (see Table 8).
Table 8
In vitro translation of mRNA obtained from transgenic tobacco plants
transformed with vectors with or without a Nd~I Smal fragment obtained
from the T1275 GUS gene fusion (see Figure 15) using wheat germ eztract.
in vitro translation
in vitro transcript fold enhancement
T1275-GUS-nos 3.7
T1275-N-GUS-nos 1.0
AMV-GUS-nos 1.9
The levels of protein produced using mRNAs comprising the Ndel Smal
fragment are also greater than those produced using the known translational
enhancer of Alfalfa Mosaic Virus RNA4 (Jobling S.A. and Gehrke L. 1987,
Nature, vol 325 pp. 622-625; Datla R.S.S. et al 1993 Plant Sci. vol 94, pp.
139-
149). These results indicate that this region functions post-
transcriptionally, as a
translational enhancer.
All scientific publications and patent documents are incorporated herein
by reference.
CA 02246892 1998-09-09
-$1-
The present invention has been described with regard to preferred
embodiments. However, it will be obvious to persons skilled in the art that a
number of variations and modifications can be made without departing from the
scope of the invention as described in the following claims.
CA 02246892 1999-02-19
-52-
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT:
(A) NAME: Brian Miki
(A) NAME: Theresa. Ouellet
(A) NAME: Jiro Hattori
(A) NAME: Elizabeth Foster
(A) NAME: Helene Labbe
(A) NAME: Teresa Martin-Heller
(A) NAME: Lihing Tian
(A) NAME: Daniel Brown
(ii) TITLE OF INVENTION: Cryptic Regulatory Elements in Plants
(iii) NUMBER OF SEQUENCES: 2
(iv) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: PatentIn Release #1.0, Version #1.30 (EPO)
(v) CURRENT APPLICATION DATA
(A) APPLICATION NUMBER: 2,246,892
(B) FILING DATE: September, 09.1998
(2) INFORMATION FOR SEQ ID NO: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1070 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(vii) IMMEDIATE SOURCE:
(B) CLONE: pT218
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 1:
TCTAGACTTG TCTTTTCTTT ACATAATCCT CTTCTTCTTT TTTTTGTTAG TTTCTTCTGT 60
TTTATCCAAA AAACGAATTA TTGATTAAGA AATACACCAG ACAAGTTTTT TACTTCTTTT 120
TCTTTTTTTT TTTGTGGTAA AAAATTACAC CTGGACAAGT TTATCACGAA AATGAAAATT 180
CA 02246892 1999-02-19
-53-
GCTATTTAAG GGATGTAGTT CCGGACTATT GTGTTAACAA AATAAATAAA240
TGGAAGATAA
TAAAAAGTTT ATACAGTTAG ATCTCTCTAT CTTATTTATA ACAATACTTT300
AACAGTCATC
ACTATAACCG TCAAATTTAT TTTGAAACAA TTATGTTACT ATAACAGTAT360
AATTTTCATG
TTTATTATAG CAACCAAA.AA ATATCGAAAC GTTATAGAGC GATTTGATTG420
AGATACGATT
TATCATTATC CACATATTTT CGTAAGCCCA CTACGTACGA TGAAAGTAAA480
ATTACTCCTC
CCAATTTAAA GTTGCAAAAA TCCAATAGAT CTTCAACTGG CGTTATGTTA540
TTCAATACTT
GGTAATGACT CCTTTTTAAC TTTTCATCTT TTTCTTTCAT TAAAAGAAAG600
TAATTTGAAG
TTTCTAGAAG AGAAGTGTTT TAACACTTCT TTATCTGTGT TTCTAGAAGA660
AGCTCTACTA
AAAATAGAAA ATGTGTCCAC CTCAAAAACA GGCAAATCTC CACCTATTTA720
ACTAAAGGTG
TTTTATTTTG GATTAATTAA GATATAGTAA TAAACGGAGT TTTGAGTTGA780
AGATCAGTTA
TACAGTGAAT TTTAAGATGT GTACCGATTT ACATTTATGT TTCGCACATA840
AACTTTATTT
TAAGAAGTCC GATTTGGAAA TACTAGATTT GCAATTCATG TGGTTGAAGA900
TGTCAATCAG
ATTTAAGTTA TATACAATGA TGATATAAAG CTATTAGTGC AAATTAATCG960
AATTTTTATA
ATTACTAAAA ATTATTATTC TATTAATTTA CCTCCCCAAC CCGTCGACCG1020
TGCTATCGTG
CGGTACCCGG TGGTCAGTCC CTTATGTTAC AACCCCAACC 1070
GTCCTGTAGA
(2) INFORMATION FOR SEQ ID NO: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2224 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(vii) IMMEDIATE SOURCE:
(B) CLONE: pT1275
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
NO: 2:
TCTAGACTTACAGAAAGTCTCTAACACGTGAGGGAATGATCCCTTTCCTTACCTCCCTGT 60
AGAGATATTGGCTTTTCAACAACTAGTACATAAATATGCGACTTTGACCGTGTATCCCCA 120
GTCAAAAGGGAACTTCACCCTCCTAGTTCTTTATTTCCAACATACATGGGGAGTAATGCT 180
AAATTTACATAGAAGAATAATAAAATGAACTGTAACTAATGATGTACTGTTCCAAAGAGA 240
CA 02246892 1999-02-19
54-
TGAGGACGTCAACATATTTATTCCTTCAGCCCTTTTCAGAATAATACCATAAGTAGAAGA 300
AATGGCACATAAAATGAAGTCCTCGGCAAGTCAAATGTAAATCTGAACCCACCCAGCTAA 360
CCCAGTGAACTCAACTTTCCTGGATAGATCAGCACTCCTTCATGACATTGCATGCCTTCT 420
CTTTAAAGAGCCGCTTGATCTCTGAAAACCAAATGAATCTCCACAGAGAGATTTCGAGCT 480
CCATGAGACGCCTTTTGGTTCTTGATTTACTAAACCTATAAAAATGAAAGGAAGTAGGAC 540
AACTGCATTTTGCCGCTTAAGATGCTTCGGCGCTTTGTGAATTTTAAGTCATGAGAAAGT 600
ACAATGTTGGAATCTCACATTAGAACAATGTATTTGTAATAACCTAGGAAAGCAAAGCTA 660
GAAGGGAGGTGCAGCTAAATCTTCTTCTACCTTGTTATCCTTGCATTTCTTGAGGAGGAG 720
GAACTGTCCTCGCAGGTGCAAAATCTGCAGTCGCCCAAAAGGATATTCAGAAGTATATTA 780
CAACATGTTTAATGGTTAACCAAGTGAAAGATCAAAATAGTCATTAGAACAAAATGCGTG 840
CTCAGAGCGTATCTACTAGTTCATCAACCCAGTACACATCTCTGAATTTCATCTCTTGCC 900
GTTGAACTAAGTCAATTGGTCAAAGACGCATAACATGAGAGACACTCATAAAACGGCTGA 960
ATAACATGCAGAAGACGTCATGCGCCTTAGGTCTCATTATGCATGAGATTATTAGTTATA 1020
TGCTCCTTCAGTTTGACTAGAAATGAAAAATCAGTTAAGCCTGTAACGAAATGATAACCT 1080
GCTTCAAGAAGATTAGACTATTTTTCATAAAATATGCAGTGCCGTGAAATAGATACTTAA 1140
TCTTAGGCAGGAAAAATCTTCTATTGGGCCATAATAAGAACTACCAATTAGAAAGGAGGT 1200
AGAAAGCTCCGATACTGTTATGAAGGCCATTCTAAGTGCTGATGTGAATTTCCCAATACA 1260
AAATGACAACAAAAACAAAAGCCTCAATCCTAAGCTAGTTGGGGTCGCTATATAAATCCT 1320
CGACATCCATTTAACTCCACTTGGACTCCTTTCTTTCCAATATTTTAATATTGTTAGATT 1380
AATCATAAAATTGCTTAGCTTTCTACTGGCACTTAACCTACTGCAACCCTCCTCTTCTGG 1440
GATTCCAACACAAACAACTAAGAGGAATTTGAAAAAAAGAAAGCAAATGTGAGAAGAGAC 1500
AAAATGTACAATGATACCTCTTCTTGCAGCAAAGGAGGCAGGTTCTCTGCTGAGACAAGG 1560
TTCTCTATTTCCTGCAAGACCTTCGTATCTTTTATTCGAGACCATGTATGTGGAGGTAAC 1620
GCCAGCAATAGTGCTGTCAGCACATCGTTGCTTGCAGGGGATCTTCTGCAAGCATCTCTA 1680
TTTCCTGAAGGTCTAACCTCGAAGATTTAAGATTTAATTACGTTTATAATTACAAAATTG 1740
ATTCTAGTATCTTTAATTTAATGCTTATACATTATTAATTAATTTAGTACTTTCAATTTG 1800
TTTTCAGAAATTATTTTACTATTTTTTATAAAATAAAAGGGAGAAAATGGCTATTTAAAT 1860
ACTAGCCTATTTTATTTCAATTTTAGCTTAAAATCAGCCCCAATTAGCCCCAATTTCAAA 1920
CA 02246892 1999-02-19
-55-
TTCAAATGGTCCAGCCCAATTCCTAAATAACCCACCCCTAACCCGCCCGGTTTCCCCTTT 1980
TGATCCAGGCCGTTGATCATTTTGATCAACGCCCAGAATTTCCCCTTTTCCTTTTTTAAT 2040
TCCCAAACACCCCTAACTCTATCCCATTTCTCACCAACCGCCACATATGAATCCTCTTAT 2100
CTCTCAAACTCTCTCGAACCTTCCCCTAACCCTAGCAGCCTCTCATCATCCTCACCTCAA 2160
AACCCACCGGAATACATGGCTTCTCAAGCCGTGGAAACCTTATACTCACCTCCCTTTGCT 2220
CTTA
2224