Note: Descriptions are shown in the official language in which they were submitted.
CA 02255113 1998-12-16
CONDUCTIVE METAL-CONTAINING NUCLEIC ACIDS
FIELD OF THE INVENTION
The invention is in the field of conductive polymers, particularly conductive
nucleic
acids, such as DNA, as well as methods for producing and using such compounds.
BACKGROUND OF THE INVENTION
Polymeric molecular conductors are known. For example, some naturally
occurring
proteins facilitate electron transfer in such fundamental biological processes
as
photosynthesis and respiration. Electron transfer in such systems is generally
understood to
occur as the result of quantum mechanical 'tunnelling' of electrons along
pathways, molecular
orbitals, that connect one atom to the next in the polymer.
It has been proposed that the stacked aromatic bases of DNA may act as a'7r-
way' for
the transfer of electrons (Dandliker et al., 1997; Hall et al., 1996; Arkin et
al., 1996). This
proposal is based on a theory that the stacked arrangement of bases on
complementary strands
juxtaposes the shared electrons in the n orbitals of the aromatic nitrogen
bases, facilitating
quantum mechanical tunnelling along the stack of base pairs. A number of
experiments have
supported the view that this effect exists, while other experiments have
provided contrary
evidence that the effect is limited or non-existent.
For example, experiments have been reported to demonstrate that photoinduced
electron transfer may occur between two metallointercalators tethered at
either end of a 15-
base pair DNA duplex (Murphy et al., 1993). On the other hand, kinetic
analysis of distance-
dependent electron transfer in a DNA hairpin has been used to show that DNA is
a poor
conductor, only somewhat more effective than proteins as a conductor of
electrons (Lewis et
al., 1997; Taubes, 1997).
-1-
CA 02255113 2008-03-31
United States Patent Nos. 5,591,578; 5,705,348; 5,770,369; 5,780,234 and
5,824,473
issued to Meade et al. on, respectively, 7 January 1997, 6 January 1998, 23
June 1998, 14
July 1998 and 20 October 1998 disclose nucleic acids that are covalently
modified with
electron transfer moieties along the nucleic acid backbone. Meade et al. teach
that such
modifications are necessary for nucleic acids to efficiently mediate electron
transfer.
The theory of 7r-orbital-mediated conductance along a nucleic acid duplex
suggests
that, as a precondition, such conductance requires a stable duplex with
stacked base pairs.
The effect on duplex stability of the binding of metal ions to nucleic acids,
particularly DNA,
has been studied extensively for nearly 40 years. In general, cations that
bind primarily to the
phosphate backbone will stabilize the duplex conformation, whereas those that
bind to the
bases will tend to denature the duplex. These effects are readily demonstrated
with thermal
denaturation profiles (Tm measurements). Experiments of this sort show that
most
monovalent cations, such as Na+, which tend to interact with the phosphate
backbone,
stabilize the duplex. This effect is reflected in the finding that there is
approximately a 12 C
increase in Tm for each 10-fold increase in monovalent cation concentration
(Marmur and
Doty 1962). An exception to this general principle is Ag+, which binds tightly
to nitrogen
bases, destabilizes the duplex, and therefore decreases the duplex Tm (Guay
and Beauchamp
1979). Similarly, multivalent ions, particularly polyamines, which interact
with the phosphate
backbone are very effective duplex stabilizers.
For divalent metal cations, a series can be written in increasing order of DNA
destabilization: Mg2+, Co2+, Ni2+, Mn2+, Zn2+, Cd2+, Cu2+ (Eichorn 1962;
Eichorn and Shin
1968). At one end of the spectrum, Mg2+ increases the Tm at all
concentrations; at the other
end of the spectrum, sufficiently high concentrations of Cu2+ will lead to
complete
denaturation of the duplex at room temperature (Eichom and Shin 1968). This
series also
correlates with the ability of the divalent cations to bind to the bases
(Hodgson 1977;
Swaminathan and Sundaralingham 1979).
-2-
CA 02255113 1998-12-16
Cations are also involved in promoting several other structural transitions
and
dismutations in nucleic acids. It has previously been reported that Zn2+ and
some other
divalent metal ions bind to duplex DNA at pHs above 8 and cause a
conformational change
(Lee et al., 1993). Preliminary characterization of the resulting structure
incorporating zinc
showed that it retained two antiparallel strands but that it was distinct from
normal 'B' DNA:
it did not bind ethidium bromide, it appeared to lose the imino protons of
both A-T and G-C
base pairs upon addition of a stoichiometric amount of ZnZ+, and it contained
at least 5%
fewer base pairs per turn than'B' DNA.
SUMMARY OF THE INVENTION
The invention provides an electrical conductor comprising an electron source
electrically coupled to a conductive metal-containing nucleic acid duplex (CM-
CNA). An
electron sink may also be electrically coupled to the CM-CNA. The CM-CNA
comprises a
first strand of nucleic acid and a second strand of nucleic acid. The first
and the second
nucleic acid strands include a plurality of nitrogen-containing aromatic bases
covalently
linked by a backbone (the backbone may be made up of phosphodiester bonds, as
in DNA or
RNA, or altelnative structures as discussed below). The nitrogen-containing
aromatic bases
of the first nucleic acid strand are joined by hydrogen bonding to the
nitrogen-containing
aromatic bases of the second nucleic acid strand. The nitrogen-containing
bases on the first
and the second nucleic acid strands form hydrogen-bonded base pairs in stacked
arrangement
along the length of the CM-CNA. At least some, and preferably each, of the
hydrogen-
bonded base pairs comprises an interchelated divalent metal cation coordinated
to a nitrogen
atom in one of the aromatic nitrogen-containing bases.
The electron source electrically coupled to the CM-CNA may be an electron
donor
molecule capable of donating an electron to the conductive metal-containing
nucleic acid
duplex.Similarly, the electron sink may be an electron acceptor molecule
capable of accepting
an electron from the CM-CNA. The electron donor molecule may be a fluorescent
molecule,
such as fluorescein. Similarly, the electron acceptor molecule may be a
fluorescent molecule,
-3-
CA 02255113 1998-12-16
such as rhodamine. It will be appreciated that some molecules may act both as
electron
donors and electron acceptors in various embodiments of the invention.
The CM-CNA may be made of deoxyribonucleic acid strands, which together
produce
metal-containing DNA ("M-DNA"). The nitrogen-containing aromatic bases in the
nucleic
acid may be the naturally occurring bases: adenine, thymine, guanine and
cytosine.
In various embodiments, the divalent metal cation used to make CM-CNA may be
Zn2+, Co2+, or Niz+. Some divalent metal cations will not produce CM-CNA, and
the present
invention provides simple assays to determine whether a particular divalent
metal cation will
work to produce CM-CNA.
The divalent metal cations may be substituted for the imine protons of
aromatic
nitrogen-containing bases in the CM-CNA. In one embodiment, the divalent metal
cations
may be substituted for the N3 imine proton of thymine, or the imine protons of
the Nl
nitrogen atom of guanine.
The invention provides a method for making conductive metal-containing nucleic
acid
duplexes. A nucleic acid duplex is subjected to basic conditions in the
presence of a divalent
metal cation under conditions effective to form a conductive metal-containing
nucleic acid
duplex. Electron sources and sinks may be electrically coupled to the
conductive metal-
containing nucleic acid duplex, which may take the form of various embodiments
discussed
above.
The invention provides a method for detecting the formation of conductive
metal-
containing nucleic acid duplexes from first and second nucleic acid strands.
The nucleic acid
strands are mixed under conditions which allow complementary stands to
hybridize and
subjected to basic conditions in the presence of a divalent metal cation under
conditions
effective to form a conductive metal-containing nucleic acid duplex if the
first and second
strands are complementary. An electron source is provided electrically coupled
to the
-4-
CA 02255113 1998-12-16
conductive metal-containing nucleic acid duplex. Conductance of electrons
between the
electron source and the conductive metal-containing nucleic acid duplex is
then tested to
determine whether a CM-CNA has formed. The CM-CNA may take the form of various
embodiments discussed above.
CM-CNAs of the invention may be used to carry electrons. They may also be used
to
raise antibodies in an animal, producing antibodies to CM-CNA. This latter use
takes
advantage of the finding that in some embodiments and under certain conditions
CM-CNAs
may be nuclease resistant.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1: shows the release of protons on formation of M-DNA. Upon addition of
NiC121 protons are released and KOH was added (left axis) to maintain the pH
at 8.5. After
each addition 10 l was removed to assess the formation of M-DNA by the
ethidium
fluorescence assay (Lee et al, 1993) (right axis). The experiment was
performed in a 10 mL
volume, with 1.1 mM in base pairs of calf thymus DNA. The DNA was dialyzed
against
water and sheared by passing through a 30 gauge needle five times. Arrow (a)
indicates the
putative point at which M-DNA formation began. This lag phase is proportional
to the DNA
concentration (data not shown) and may be due to the initial binding of the
metal ion to the
outside of the helix. Arrow (b) indicates the point at which 1.1 mM of H+ had
been released,
beyond which precipitation of the M-DNA was observed.
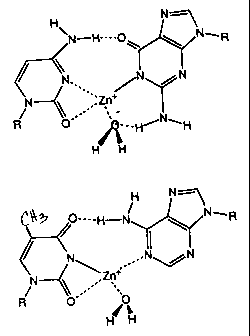
Figure 2: shows a putative structure of M-DNA showing G-C and A-T base pairs.
Putative hydrogen bonds and interactions between Zn2+ and its coordinating
groups are shown
as dotted lines.
Figure 3: shows the fluorescence of fluorescein-labelled oligonucleotides
during the
formation of M-DNA (see Table 1 for the sequences of the 20-mer and 54-mer)
(a) Effect of
Znz+ on the 20-mer duplex. (i) F1-20-mer duplex without Zn2+; (ii) F 1-20mer
duplex with
Zn2+; (iii) F1-20-mer-Rh duplex in the absence of Znz+; (iv) F1-20mer-Rh
duplex in the
-5-
CA 02255113 1998-12-16
presence of ZnZ+; (v) addition of EDTA after the formation of M-DNA. (b)
Effect of Zn2+ on
the 54-mer duplex. (i) F1-54-mer-Rh with D-site binding protein (lug/ml) (the
site is located
at the centre of the 54-mer duplex) in the presence of ZnZ+; (ii) addition of
proteinase K
(50ug/ml) after 3,000 seconds; (iii) Fl-54mer-Rh duplex wit Zn2+. The
experiments were
performed in 20mM NaBO3 buffer, pH 9.0 at 20 C with 10mM NaCl and 1 mM ZnZ+
as
appropriate. Fluorescence intensities are normalized with respect to the
fluorescence intensity
of the F1-20-mer-duplex either in the absence or presence of Zn2+.
Figure 4: shows the nuclease resistance of M-DNA. The amount of duplex DNA
remaining as a function of time was assessed by the ethidium fluorescence
assay (under
conditions where M-DNA rapidly reverts to B-DNA, pH 8, 0.1 mM EDTA, so that
ethidium
can bind the DNA). The digestion was performed at 37 C in 10mM Tris-HCI, pH
7.4, 5 mM
MgC12, 1 mM NiC121 1 mg/ml gelatin, and 0.2 g/ml DNase I. The NiZ+ form of M-
DNA was
preformed for the assay at pH 9 before adding it to the digestion buffer; B-
DNA was added
directly to the digestion buffer. The graph shows that the M-DNA is resistant
to nuclease
digestion while B-DNA is digested in about 10 minutes. The results also
demonstrate that the
Ni2+ form of M-DNA is stable at physiological pH, a characteristic which
facilitates the use of
Niz+-M-DNA to mediate physiological responses in vivo, such as DNA
immunization (in
which the DNA 'vaccine' expresses an antigenic protein) or antisense
applications (in which
injected M-DNA inhibits the expression of a complementary gene).
Figure 5: shows that M-DNA is immunogenic. Balb/C mice were immunized
interperitoneally three times at ten day intervals with 10 g of nickel-
containing M-DNA,
with and without methylated bovine serum albumin (Me-BSA). The first injection
was with
complete Freunds adjuvant and subsequent injections were with incomplete
Freunds
adjuvant. Three days after the final injection, blood was obtained by tail
bleeding and the
serum was tested for the present of antiM-DNA antibodies using nickel M-DNA
coated
polyvinylchloride plates in a SPIRA assay, using methods known in the art
(Braun and Lee,
1988).
-6-
CA 02255113 1998-12-16
Figure 6: is a schematic illustration showing as aspect of the invention that
comprises
a method of sequence analysis, in which one sequence that is susceptible to
cutting by a
restriction endonuclease may be distinguished from another sequence that is
not susceptible
on the basis of the presence or absence of conductance in an M-DNA duplex
formed between
an electron donor, shown as "F" for fluorescein in the figure, and an electron
acceptor, shown
as "R" for rhodamine in the figure.
Figure 7: is a schematic illustration showing a nucleic acid coupled to
electrode 10
and subjected to conditions suitable to form M-DNA, while measurements are
taken of the
conductance of the CM-CNA by cyclic voltammetry, both in the absence and
presence of a
DNA-binding drug.
DETAILED DESCRIPTION OF THE INVENTION
The invention provides a CM-CNA duplex comprising an electron source
electrically
coupled to a nucleic acid duplex in which at least some of the stacked,
aromatic nitrogen-
containing base pairs chelate divalent metal cations. In such an embodiment,
the metal-
containing nucleic acid duplex acts as an electron acceptor, receiving
electrons from the
electron donor. In one embodiment of this aspect of the invention, the imine
protons of a
DNA duplex may be replaced by Znz+, Coz+ or Ni2+. The metal-containing DNA
duplex may,
for example be electronically coupled to molecular electron donors or electron
sinks, such as
fluorescein or rhodamine respectively, by covalent attachment.
In one aspect, the present invention provides a method for converting a
nucleic acid
duplex to CM-CNA. A nucleic acid duplex is treated with sufficient base in the
presence of
an adequate concentration of an appropriate divalent metal ion to result in
the chelation of the
divalent metal ion by the aromatic nitrogen bases of the nucleic acid. Such
treatment is
carried out for a sufficient period of time to produce a modified duplex
comprising the
divalent metal cation coordinated to nitrogen atoms in the aromatic nitrogen-
containing bases
of base pairs.
-7-
CA 02255113 2008-03-31
In one embodiment, conditions for converting DNA, such as a B-DNA, to M-DNA
comprise subjecting the DNA to a solution at pH 8.5 or greater, with
approximate
concentrations of divalent metal ions as follows: 0.1 mM Zn2+ or 0.2mM Co2+ or
0.2mM
Ni2+. The conditions necessary to form M-DNA will vary depending on the metal
ion or ions
used and the nature of the nucleic acid. Those skilled in this art will
appreciate that routine
experiments may be carried out to determine appropriate conditions, varying
parameters such
as pH, nucleic acid concentration, metal ion concentration and the ratio of
the metal ion
concentration to the nucleic acid concentration. In some embodiments, a pH
equal to or
greater than 8, or greater than 8.5, may be required, and a suitable nucleic
acid to metal ion
ratio maybe about 1:1.5 to about 1:2Ø
The CM-CNA may be electrically coupled to an electron source and an electron
sink.
For example, molecular electron donors and acceptors may act respectively as
electron
sources and electron sinks. In alternative embodiments, the electron donors
and acceptors
may be in solution, interacting transiently with the CM-CNA, or they may be in
the form of a
solid support, such as an electrode.
Solid phase supports to which the CM-CNA is attached may serve as electron
sources,
sinks or both. For example, immobilized arrays of CM-CNA may be prepared in
accordance
with the teaching of United States Patent No. 5,556,752, issued 17 September
1996 to
Lockhart et al. (the "'752 Patent"). Such immobilized arrays may then be used,
as described
therein and modified as necessary, to detect hybridization. In accordance with
the present
invention, the step of hybridizing the target nucleic acid to the immobilized
probe may be
followed, or accompanied, by the step of converting resulting duplexes to CM-
CNA under
basic conditions in the presence of a divalent metal cation, as described
herein. Electron
donors and acceptors may be provided in such a system, as described in the'752
Patent, so
that the conductivity of the resulting CM-CNA duplex is detectable at the
surface of the
immobilization substrate, as is also described in the '752 Patent. It will be
appreciated that
such a system involves the use of CM-CNA as a conductor, which is one aspect
of the present
invention.
-8-
CA 02255113 2008-03-31
The formation of CM-CNA may be used to assay a variety of nucleic acid
interactions. For example, the amplification of a target sequence using the
polymerase chain
reaction (PCR) may be assayed using methods of the present invention. In one
aspect of such
an assay, one PCR primer is provided with an electron donor moiety and the
other PCR
primer is provided with an electron acceptor moiety. In accordance with this
aspect of the
present invention, following PCR amplification cycles, the reaction mixture
may be subjected
to basic conditions in the presence of a divalent metal cation to promote the
formation of
CM-CNA. If the PCR amplification has been successful, CM-CNA will have formed
and will
be detectable as disclosed herein as a result of characteristic conductance
between the
electron donor and the electron acceptor. Unsuccessful amplification will
leave the electron
transfer moieties on the primers electrically uncoupled. In some embodiments,
this approach
may have the advantage of allowing detection of amplification without the need
to separate
the PCR primers from the PCR reaction mixture following amplification cycles.
In
accordance with this aspect of the invention, a kit may be provided comprising
PCR primers
having electron donors and electron acceptors, together with instructions for
subjecting an
amplification reaction mixture to basic conditions in the presence of a
divalent metal cation to
form CM-CNA. Such kits with appropriate instructions may also be provided for
the other
aspects of the invention disclosed herein
Ligation of nucleic acids may also be assayed using methods of the present
invention,
wherein successful ligation is detectable by the formation of CM-CNA. In such
a system, one
of the nucleic acid duplexes to be ligated may be provided with an electron
donor moiety,
while the other nucleic acid duplex to be ligated is provided with an electron
acceptor moiety.
Ligation and subsequent formation of CM-CNA electrically couples the electron
transfer
moieties, producing a signal under appropriate conditions that is indicative
of successful
ligation. A kit may be provided for such a reaction, comprising an electron
donor label and an
electron acceptor label, together with instructions for coupling the electron
donor and electron
acceptor to nucleic acids that are to be ligated, and subjecting the ligation
reaction mixture to
basic conditions in the presence of a divalent metal cation to form CM-CNA.
In one embodiment of the invention, conditions are adapted to convert a B-DNA
duplex to M-DNA. In one aspect of the invention, M-DNA is formed at pHs at or
above 8 in
-9-
CA 02255113 1998-12-16
In one embodiment of the invention, conditions are adapted to convert a B-DNA
duplex to M-DNA. In one aspect of the invention, M-DNA is formed at pHs at or
above 8 in
the presence of sufficient amounts (preferably, in some embodiments, about 0.1
mM,
provided the nucleic acid concentration is less than about 0.1 mM) of Znz+,
NiZ+ or Co2+. In
such an embodiment, Mg2+ or Ca2+ may not serve to produce M-DNA (Lee et al.,
1993). A
wide variety of bacterial and synthetic DNAs may dismutate to M-DNA under
these
conditions. In some embodiments, the process of M-DNA formation may be
reversible by
lowering the pH and/or addition of EDTA. In some embodiments, Ni-M-DNA
requires
EDTA to be converted to 'B' DNA at pHs greater than about 7. In some
embodiments,
poly[d(AT)] may not be convertible to M-DNA under such conditions. Unlike B-
DNA,
ethidium will not bind to some embodiments of M-DNA, and this property forms
the basis of
a rapid and sensitive "ethidium fluorescence assay" that may be used to
monitor M-DNA
formation (Lee et al., 1993).
M-DNA may be readily interconverted with B-DNA; therefore, useful techniques
for
manipulation of DNA such as cutting and splicing, and for the self-assembly of
a variety of
structures (such as two and three-way junctions) may be used with M-DNA
forming
sequences, as are well known in the art (Lilley and Clegg, 1993; Seeman and
Kallenbach,
1994). In addition, the binding of sequence-specific proteins to CM-CNAs may
be
manipulated in some embodiments to interfere with conductance of the M-DNA so
as to
mimic electric switches and resistors.
One aspect of the invention provides methods for detecting particular
sequences in
genomic analysis, such as methods for detecting particular mutations. In one
such aspect of
the invention, as shown in Fig. 6, the nucleic acid to be analysed (shown as
wild type or
mutant) is amplified by PCR using primers, one of the primers being labelled
with an electron
donor and the other being labeled with an electron acceptor, such as
fluorescein (shown as
"F") and rhodamine (shown as "R") respectively. Following amplification, the
nucleic acid is
treated with a restriction enzyme which may or may not cut the amplified
sequence,
-10-
CA 02255113 1998-12-16
depending upon the nature of the amplified sequence. For example, the
restriction enzyme
may only cut one allele of a gene, leaving other alleles or non-wild type
sequences uncleaved
(as shown in Fig. 6, where X and Y represent a mutant base pair). Following
treatment with
the restriction enzyme the amplified sequence may be subjected to conditions
suitable for the
formation of a CM-CNA, for example amplified DNA duplexes may be converted to
M-
DNA. The fluorescence of the sample may then be measured. If the amplified
duplex spans
the region between the primers, as in the mutant gene of Figure 6, then the
fluorescence of the
amplified nucleic acid will be quenched by electron transfer along the CM-CNA.
If, on the
other hand, as in the wild type gene of Figure 6, the amplified duplex has
been cut by the
restriction enzyme, the fluorescence of the electron donor will not be
quenched. A sample in
which half of the sequences formed CM-CNA and half did not, such as may be the
case in an
analysis of a sample from an individual with an autosomal recessive mutation,
the degree of
fluorescence may be intermediate. Sequence analysis in accordance with this
aspect of the
invention may be carried out in an automated fashion. For example, in one
aspect this
approach may simultaneously use multiple reaction wells to carry out such
reactions, each
well containing different reagents, such as different primers or different
restriction enzymes,
to yield an abundance of information about a particular sample in a relatively
short time
without the necessity of electrophoresis or other more time-consuming steps.
In an alternative aspect, the invention provides a sensor for monitoring the
presence of
nucleic acid binding moieties in a sample. In one embodiment of this aspect of
the invention,
as shown in Figure 7, a nucleic acid duplex capable of forming a CM-CNA is
attached
between an electron sink and an electron donor, such as ferrocene, the nucleic
acid is exposed
to a sample under conditions that favour the formation of a CM-CNA duplex, the
binding of a
moiety to the nucleic acid is detected by a change in the conductance of the
nucleic acid. For
example, DNA binding molecules may convert M-DNA back to B-DNA under such
conditions, and thereby prevent or reduce the CM-CNA mediated quenching of a
fluorescent
electron donor that is coupled to the nucleic acid. In one embodiment, the
nucleic acid may be
coupled to an electrode (for example as described in Braun et al., Nature,
391: 775-778, 1998,
incorporated herein by reference), such as gold electrode 10 of Figure 7, and
the electrode
-11-
CA 02255113 2008-03-31
may then be used to measure the conductance of the CM-CNA while the electrode
is exposed
to a sample. In some embodiments, for example, such conductance measurements
may utilize
cyclic voltammetry (shown as CV in Figure 7). Such, assays involving the
detection of
variations in the conductance of a CM-CNA may be used in various embodiments
of the
invention to detect interactions between nucleic acids and a wide variety of
other moieties,
such as small molecules, triplex-forming oligonucleotides and DNA-binding
proteins.
In some embodiments, the conductance of CM-CNAs may be enhanced by
modification of the nucleic acid with electron transfer moieties, such as is
taught in the
following U.S. Patent Nos. 5,591,578; 5,705,348; 5,770,369; 5,780,234 and
5,824,473 issued
to Meade et al. on respectively 7 January 1997, 6 January 1998, 23 June 1998,
14 July 198
and 20 October 1998.
In the context of the present invention, 'conductive' means capable of
conducting
electrons. An electron source in accordance with the present invention may be
any compound
or substance capable of providing electrons, such as an atomic or molecular
conductor.
Similarly, an electron sink (or acceptor) may be any compound or composition
capable of
accepting electrons. A nucleic acid duplex comprises hybridized strands of
nucleic acid
molecules. A strand of nucleic acid comprises at least two nucleotides
covalently linked by a
backbone. The backbone may be made up of polymeric phosphodiester bonds, as in
DNA or
RNA. Alternatively, other backbone structures may be effective to
appropriately align the
aromatic nitrogen-containing bases in a stacked arrangement capable of
chelating metal ions
and conducting electrons. For example, phosphoramide, phosphorothioate,
phosphorodithioate, O-methylphosphoroamidite or peptide nucleic acid linkages
may be
effective to form such a backbone. Similarly, other components of the backbone
may vary in
accordance with the invention, encompassing deoxyribose moieties, ribose
moieties, or
combinations thereof. If RNA is used, those skilled in this art will
appreciate that conditions
must be adapted to account for the fact that RNA is labile in basic solution,
so that conversion
of RNA to CM-CNA may require modified reaction conditions which avoid
hydrolysis of the
RNA. In one aspect of the invention, the nitrogen-containing aromatic bases
are preferably
-12-
CA 02255113 2008-03-31
those that occur in native DNA and RNA: adenine, thymine, cytosine guanine or
uracil.
However, those skilled in this art will understand that alternative nitrogen-
containing
aromatic bases may be utilized, preferably they are capable of interchelating
a divalent metal
ion, coordinated to a nitrogen atom in the aromatic nitrogen-containing base
and stacking, to
produce a conductive metal-containing nucleic acid duplex. In accordance with
these
variations in the structure of the molecules of the invention, alternative
divalent metal ions
may be utilized, again depending upon the ability of such ions to participate
with the other
substituents of the molecules of the invention in the formation of a
conductive metal-
containing nucleic acid duplex. The present application sets out assays for
the creation of
such a duplex, so that others may routinely identify functional substitutions
and variations in
the structure of the molecules of the invention. Accordingly, although various
embodiments
of the invention are exemplified herein, other adaptations and modifications
may be made
within the scope of the invention. The following examples are merely
illustrative of
alternative embodiments of the invention and are not comprehensive nor
limiting in their
scope.
EXAMPLE 1
Conductance of CM-CNA
The conductance of CM-CNA was investigated by preparing duplexes of 20 base
pairs
of DNA with fluorescein (the electron donor) and rhodamine (the electron
acceptor) at
opposite ends of the duplex. Methods for such attachment are disclosed in
Kessler, 1995, and
Haugland, referenced below. Fluorescein and rhodamine fluoresce at different
wavelengths,
so that it is possible to distinguish fluorescence of the electron donor from
fluorescence of the
electron acceptor. Under conditions which favour B-DNA (pH less than about 8.0
in the
presence of EDTA) the fluorescence of the fluorescein electron donor is
partially quenched
and the fluorescence of the rhodamine electron acceptor is partially enhanced.
This appears to
be an example of through space energy transfer (Forster resonance energy
transfer or FRET)
which has been well-documented in a number of different laboratories (Cheung,
H.C. 1991
and Clegg, R.M., 1992). FRET quenching is understood to be due to dipole-
dipole
interactions along a molecule (not electron conductance) and is highly
distance dependent
-13-
CA 02255113 1998-12-16
(the effect decreasing with interatom distance in sixth order relationship:
1/r6); the value of
25% quenching measured for the 20 base pair duplex is in accordance with the
expected
FRET behaviour for this length of helix (Clegg, 1992). As shown in Figure 3a,
the
fluorescence intensity is relatively stable at pH 9 although at long times
there is some loss
due to photobleaching.
On addition of Zn2+ (1 mM) to the DNA (pH 9), the fluorescence is quenched up
to
95% over a period of 1 hr. This rate of increasing quenching mirrors the known
rate of
formation of M-DNA under these conditions (Lee et al., 1993). Upon reformation
of B-DNA
by addition of an excess of EDTA (2 mM) after 4,000 sec., the quenching is
rapidly reversed.
These results are summarized in Table 1.
As a control, the 20-mer duplex (without metal ions) with a fluorescein label
shows
only a small decrease in intensity due to photobleaching, similar to the
effect noted above
with respect to ordinary B-DNA (Figure 3a). Similarly a mixture of two
duplexes, one
labelled with fluorescein and the other labelled with rhodamine, show minimal
quenching
either as B-DNA or M-DNA (see Table 1).
To measure the fluorescence life time of the fluorescein when it is attached
to the 20-
mer oligonucleotide having fluorescein at one end and rhodamine at the other
end, the
fluorescein is irradiated with a picosecond pulse of laser light and the
fluorescence decay of
the excited fluorescein is then followed for several nanoseconds. Normally (as
is the case
with B-DNA) the t%2 for decay is about 3 nanoseconds. Upon conversion to the
ZnZ+ form of
M-DNA as described above, the t%2 drops to about 0.3 nanoseconds. This
extremely fast
decay is consistent with electron conductivity by the M-DNA helix.
Electron transfer in the Zn2+ isomer of M-DNA was investigated in a longer
helix of
54 base pairs (this 54mer has an estimated length of over 150A). This 54mer
also contained
the recognition site for the D-site binding protein (Roesler et al., 1992) in
the middle of the
sequence. As shown in Figure 3b, there is no quenching in the absence of metal
ions in the
-14-
CA 02255113 1998-12-16
54mer, this may be because the fluorophores are well separated so that there
is no FRET.
However, upon addition of Znz+ under appropriate conditions to form M-DNA (1mM
Zn2+,
pH 9), the fluorescent intensity rapidly drops to 25% of the initial value,
demonstrating
efficient conductance over the length of the 54mer.
In the presence of the D-site binding protein, under conditions appropriate to
form M-
DNA, the fluorescence intensity of the 54mer only drops slowly. However, as
judged from
the ethidium fluorescence assay (Lee et al., 1993), the majority of the 54mer
DNA is in the
form of M-DNA (which does not bind ethidium). This demonstrates that the D-
site DNA-
binding protein is interrupting the flow of electrons along the 54mer M-DNA
duplex. As a
control, the D-site binding protein has no effect on the quenching of the 20-
mer (which as no
D-site binding sequence, see Table 1). On addition of protease at 3000 seconds
to the D-site
binding protein:54mer M-DNA complex, the protein is cleaved and the
fluorescence intensity
begins to drop, eventually reaching the minimum value of 25% of the initial
fluorescence
value. This experiment is a simple example of a bioreactive electronic switch
comprising
CM-CNA and a DNA-binding protein capable of disrupting the conductive duplex.
Such a
switch is also analogous to an electronic memory element, having two
interchangeable states,
conductive and non-conductive.
-15-
CA 02255113 1998-12-16
Table 1. Normalized Fluorescence of the Fluorescein-labelled oligonucleotides
Oligonucleotide Treatment Fluorescence
F1-20-mer duplex none 1
F1-20-mer duplex +Zn2+ 0.98
F1-20-mer duplex +Zn2+ at pH 8.0 0.92
F1-20-mer single strand none 0.87
F1-20-mer duplex + none 0.97
Rh-20-mer duplex
F1-20-mer-Rh duplex none 0.73
F1-20-mer-Rh duplex +Zn2+ 0.05
F1-20-mer-Rh duplex +Znz+ + EDTA 0.87
F1-20-mer-Rh duplex +ZnZ+ at pH 8.0 0.92
F1-20-mer-Rh duplex +Coz+ 0.05
F1-20-mer-Rh duplex +CoZ+ +EDTA 0.7
F1-20-mer-Rh duplex +Ni2+ 0.06
F1-20-mer-Rh duplex +Ni2+ +EDTA 0.7
F1-20-mer-Rh duplex +Mg2+ 0.83
F1-20-mer-Rh duplex +D-site binding protein + Zn2+ 0.06
F1-54-mer-Rh +D-site binding protein + Zn2+ 1
F1-54-mer-Rh +Zn2+ 0.21
Conversion to M-DNA was performed in 20mM NaBO3 buffer, pH 9Ø Fluorescence
assays were carried out in 20 mM Tris pH 8Ø Other conditions were as
follows: 10mM NaCI
at 20 C and 1 mM Zn2+ or 0.2 mM CoZ+ or 0.2 mM Ni2+ or 2 mM EDTA as
appropriate.
Excitation was at 490 nm with emission measured at 520 nm. Fluorescence
intensities are
normalized with respect to the fluorescence intensity of the F1-20-mer-duplex
either in the
absence or presence of ZnZ+ and were measured after 3,000 sec.
Sequences and nomenclature: The oligonucleotides were labelled 5' with
fluorescein
(Fl) or rhodamine (Rh) using standard attachment methods and constructs, for
example as
used in DNA sequencing. The fluorescein 20-mer was as follows: SEQ ID 1: F 1-
5'-d(GTC
ACG ATG GCC CAG TAG TT). The rhodamine 20-mer was as follows: SEQ ID 2: Rh-5'-
3 5 d(AAC TAC TGG GCC ATC GTG AC). The same unlabelled sequence was used to
produce
the F1-20-mer-duplex. The F1-54-mer was as follows: SEQ ID 3: Fl-5'-d(GCT ATG
ATC
CAA AGG CCG GCC CCT TAC GTC AGA GGC GAG CCT CCA GGT CCA GCT) (The
D-site is underlined). The Rh-54mer was as follows: SEQ ID 4: Rh-5'-d(AGC TGG
ACC
-16-
CA 02255113 1998-12-16
TGG AGG CTC GCC TCT GAC GTA AGG GGC CGG CCT TTG GAT CAT AGC). The
same unlabelled sequence was used to produce the F1-54-mer duplex.
This Example demonstrates a method for converting a nucleic acid duplex to a
conductive metal-containing nucleic acid duplex, in this case M-DNA. The
excited electron
on the fluorescein is rapidly transmitted down the M-DNA helix to the
rhodamine;
demonstrating rapid and efficient electron transfer along the M-DNA. The CoZ+
and Ni2+
isomers of M-DNA show quenching of the fluorescein by up to 95% even in the
absence of
the rhodamine acceptor (Table 1). This indicates that the M-DNA can itself act
as an electron
acceptor.
EXAMPLE 2
Physical Properties of M-DNA
The mobility of linear or covalently closed circular forms of M-DNA in agarose
gels
is only slightly less than that of B-DNA (indicating that treatment in
accordance with the
invention to produce M-DNA need not cause condensation or aggregation of the
DNA).
NMR studies show that the imino protons of T(pKa 9.9) and G (pKa 9.4) may not
be present
in M-DNA, illustrating that the imine protons may be replaced by the divalent
metal cation in
M-DNA. The release of protons during the formation of M-DNA may be indicative
of this
phenomenon. As shown in Figure 1, M-DNA begins to form at about 0.7 mM NiC12
(as
judged from the ethidium fluorescence assay); there is a concomitant release
of protons so
that KOH may be added to maintain the pH at 8.5. At 1.8 mM NiC121 M-DNA
formation is
virtually complete and the complex starts to precipitate. This suggests that
one proton is
released per Ni2+ atom per base pair during the formation of M-DNA. The Zn2+
and Co2+
isomers of M-DNA also release protons during formation, and precipitation of
the complex
may occur at a lower concentration of divalent metal ion than with Ni2+. These
results are
consistent with the metal ion being coordinated to the N3 position of T and N1
of G in the
base pairs.
-17-
CA 02255113 1998-12-16
Based on these observations, a putative structure for M-DNA can be modelled as
shown in Figure 2. This model reflects experimental results relating to one
aspect of the
present invention, and does not limit the invention to any such putative
structure. The model
may nevertheless be helpful to others in practising routine variations of the
invention. In this
putative structure, the A-T and G-C base pairs are isomorphous, which is a
common feature
of stable helical nucleic acid structures (Paleck, 1991). Compared to a Watson-
Crick base
pair, insertion of the metal ion with an imino N-metal bond of 2A (Swaminathan
and
Sundralingham, 1979; DeMeester, 1973; McGall and Taylor, 1973) requires a 20 -
30
rotation of the bases which opens up the minor groove. One hydrogen bond is
retained in both
base pairs, which may facilitate rapid reformation of normal B-DNA without
denaturation of
the helix on removal of the metal ion. The coordination geometry of the metal
ion may be
distorted square planar with the solvent providing the fourth ligand in some
embodiments.
The UV-Vis spectrum of the Co2+ and Ni2+ isomers of M-DNA have peaks in the
visible with
E of 20 and 60 mol-1 cm-1 respectively; an observation which is consistent
with this
geometry (Lever, 1988). In this putative model of an M-DNA duplex, the metal
ion is buried
within the helix and d-n bonding may occur with the aromatic bases above and
below the
metal ion. The putative model helix could be considered as a distorted member
of the B-type
helix family in agreement with the unremarkable CD spectrum (Lee et al,
1993.). On average
the model metal-metal distance is 4A.
EXAMPLE 3
M-DNA Is Nuclease Resistant
The nuclease resistance of M-DNA was established by assaying the amount of
duplex
M-DNA remaining as a function of time in the presence of DNase I, as shown in
Figure 4.
The amount of DNA was assessed by the ethidium fluorescence assay (under
conditions
where M-DNA rapidly reverts to B-DNA, i.e. pH 8 in the presence of EDTA, so
that
ethidium can bind the DNA for the purpose of the assay). The digestion was
performed at
37 C in 10mM Tris-HCI, pH 7.4, 5 mM MgC121 1 mM NiC121 1 mg/ml gelatin, and
0.2 g/ml
DNase I. The Niz+ form of M-DNA was preformed for the assay at pH 9, before
adding it to
the digestion buffer; B-DNA was added directly to the digestion buffer. The
graph shows that
-18-
CA 02255113 2008-03-31
M-DNA is resistant to nuclease digestion while B-DNA is digested in about 10
minutes. The
results also demonstrate that the Ni2+ form of M-DNA is stable at
physiological pH, a
characteristic which may facilitate the use of Ni-M-DNA to mediate
physiological responses
in vivo, such as DNA immunization (in which the DNA vaccine expresses an
antigenic
protein) or antisense applications (in which injected M-DNA inhibits the
expression of a
gene).
EXAMPLE 4
M-DNA Is Immunogenic
B-DNA is generally not immunogenic. However, synthetic or modified nucleic
acids
that are nuclease resistant may be capable of producing an antibody response
under certain
conditions (Braun and Lee, 1988).
To test the immunogenicity of M-DNA, Balb/C mice were immunized
interperitoneally three times at ten day intervals with 10 g of nickel-
containing M-DNA,
with and without methylated bovine serum albumin (Me-BSA). The first injection
was with
complete Freunds adjuvant and subsequent injections were with incomplete
Freunds
adjuvant. Three days after the final injection, blood was obtained by tail
bleeding and the
serum was tested for the present of antiM-DNA antibodies using nickel M-DNA
coated
polyvinylchloride plates in a SPIRA assay, using methods known in the art
(Braun and Lee,
1988).
The results, shown in Figure 5, demonstrate that the mice immunized with M-DNA
(with and without Me-BSA) show antibody titres to M-DNA up to about 1:1000
dilution. The
control sera from an unimmunized mouse contains no antibodies to M-DNA. The
ability of
M-DNA to elicit an immune response is consistent with the finding that M-DNA
may be
nuclease resistant (see Braun and Lee, 1988). Accordingly, in some
embodiments, M-DNA
may be useful for immunizing a host, for example in methods as disclosed in
U.S. Patent
Nos. 5,679,647; 5,804,566 or 5,830,877 issued to Carson et al. on,
respectively 21 October
1997, 8 September 1998 and 3 November 1998.
- 19-
CA 02255113 1998-12-16
REFERENCES
1. Dandliker, P. J., Holmlin, R. E. & Barton, J. K. Science 275,1465-1468
(1997).
2. Hall, D. B., Holmlin, R. E. & Barton, J. K. Nature 382, 731-735 (1996).
3. Arkin, M. R., Stemp, E. D. A., Holmlin, R. E., Barton, J. K., Hormann, A.,
Olson, E.
J. C. & Barbara, P. F. Science 273,475-479 (1996).
4. Murphy, C. J., Arkin, M. R., Jenkins, Y., Ghatlia, N. D., Bossmann, S. H.,
Turro, N.J.
& Barton, J. K. Science 262,1025-1029 (1993).
5. Lewis, F. D., Wu, T., Zhang, Y., Letsinger, R. L., Greenfield, S. R., &
Wasielewski,
M. R.Science 277,673-676 (1997).
6. Taubes, G. Science 275,1420-1421 (1997).
7. Lee, J. S., Latimer, L. J. P. & Reid, R. S. Biochem. Cell Biol. 71, 162-168
(1993).
8. Palecek, E. CRC Crit. Rev. Biochem. Mol. Biol. 26,151-226 (1991).
9. Yagil, G. CRC Crit. Rev. Biochem. Mol. Biol. 26, 475-559 (1991).
10. Swaminathan, V. & Sundralingham, M. CRC Crit. Rev. Biochem. Mol. Biol. 14,
245-
336 (1979).
11. DeMeester, P., Goodgame, D. M. L., Skapski, A. C. & Warnke, Z. Biochem.
Biophys.
Acta 324,301-303 (1973).
12. McGall, M. J. & Taylor, M. R.Biochem. Biophys. Acta 390,137-139 (1973).
13. Lever, A. B. P. "Inorganic Electronic Spectroscopy" (Elsevier, Amsterdam)
(1988).
14. Cheung, H. C. in "Topics in Fluorescence Spectroscopy" pp 128-171, ed.
Lakowicz,
J. R. (Plenum, New York) (1991).
15. Clegg, R. M. Methods in Enzymology 211,353-371 (1992).
16. Roesler, W. J., McFie, P. J. & Dauvin, C. J. Biol. Chem. 267, 21235-21243
(1992).
17. Lilley, D. M. J. & Clegg, R. M. Ann. Rev. Biophys. Biomol. Str. 22, 299-
328 (1993).
18. Seeman, N. C. & Kallenbach, N. R.Ann. Rev. Biophys. Biomol. Str. 23, 53-86
(1994).
19. Brunger, A. 1. X-PLOR Manual, version 3.1 (rare University Press New Haven
USA
(1993).
20. Braun, R.P. and Lee, J.S. J. Immunol. v.141, 2084-2089 (1988).
-20-
CA 02255113 1998-12-16
21. Kessler, C. in Nonisotopic Probing, Blotting and Sequencing, L.J. Kricka,
Ed.,
Academic Press (1995) pp. 3-40.
22. Haugland, R.P. Handbook of Fluorescent Probes and Reserch Chemicals, 6th
Ed.,
p.157.
-21-
CA 02255113 1999-02-10
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: The University of Saskatchewan
(ii) TITLE OF INVENTION: CONDUCTIVE METAL-CONTAINING NUCLEIC
ACIDS
(iii) NUMBER OF SEQUENCES: 4
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: Smart & Biggar
(B) STREET: 2200 - 650 WEST GEORGIA STREET
(C) CITY: Vancouver
(D) STATE: British Columbia
(E) COUNTRY: Canada
(F) ZIP: V6B 4N8
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: 3.5" Diskette, 1.44 Mb
(B) COMPUTER: IBM PC Compatible
(C) OPERATING SYSTEM: Windows 95
(D) SOFTWARE: Notepad
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER: 2,255,113
(B) FILING DATE: 16 DECEMBER 1998
(vii) PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER: 2,218,443
(B) FILING DATE: 16 DECEMBER 1997
(A) APPLICATION NUMBER: 2,229,386
(B) FILING DATE: 11 FEBRUARY 1998
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: Brian G. Kingwell
(C) REFERENCE/DOCKET NUMBER: 81527-5
(ix) TELECOMMUNICATION INFORMATION:
(A) TELEPHONE: (604) 682-7295
(B) TELEFAX: (604) 682-0274
(2) INFORMATION FOR SEQUENCE IDENTIFICATION NUMBER: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 1:
GTCACGATGG CCCAGTAGTT 20
(2) INFORMATION FOR SEQUENCE IDENTIFICATION NUMBER: 2:
(i) SEQUENCE CHARACTERISTICS:
- 22 -
CA 02255113 1999-02-10
(A) LENGTH: 20
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2:
AACTACTGGG CCATCGTGAC 20
(2) INFORMATION FOR SEQUENCE IDENTIFICATION NUMBER: 3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 3:
GCTATGATCC AAAGGCCGGC CCCTTACGTC AGAGGCGAGC CTCCAGGTCC AGCT 54
(2) INFORMATION FOR SEQUENCE IDENTIFICATION NUMBER: 4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 4:
AGCTGGACCT GGAGGCTCGC CTCTGACGTA AGGGGCCGGC CTTTGGATCA TAGC 54
- 23 -