Note: Descriptions are shown in the official language in which they were submitted.
CA 02268036 1999-04-12
wo gyms rc~rms~nszi4
1
COMPOUNDS AND METHODS FOR DIAGNOSIS OF TUBERCULOSIS
' TECHNICAL FIELD
The present invention relates generally to the detection of Mycobacterium
tuberculosis infection. The invention is more particularly related to
polypeptides comprising
a Mycobacterium tuberculosis antigen, or a portion or other variant thereof,
and the use of
such polypeptides for the serodiagnosis of Mycobacterium tuberculosis
infection.
BACKGROUND OF THE INVENTION
Tuberculosis is a chronic, infectious disease, that is generally caused by
infection with Mycobacterium tuberculosis. It is a major disease in developing
countries, as
well as an increasing problem in developed areas of the world, with about 8
million new
cases and 3 million deaths each year. Although the infection may be
asymptomatic for a
considerable period of time, the disease is most commonly manifested as an
acute
inflammation of the lungs, resulting in fever and a nonproductive cough. If
left untreated,
serious complications and death typically result.
Although tuberculosis can generally be controlled using extended antibiotic
therapy, such treatment is not sufficient to prevent the spread of the
disease. Infected
individuals may be asymptomatic, but contagious, for some time. In addition,
although
compliance with the treatment regimen is critical, patient behavior is
difficult to monitor.
Some patients do not complete the course of treatment, which can lead to
ineffective
treatment and the development of drug resistance.
Inhibiting the spread of tuberculosis will require effective vaccination and
accurate, early diagnosis of the disease. Currently, vaccination with live
bacteria is the most
efficient method for inducing protective immunity. The most common
Mycobacterium for
this purpose is Bacillus Calmette-Guerin (BCG), an avirulent strain of
Mycobacterium bovis.
However, the safety and efficacy of BCG is a source of controversy and some
countries, such
as the United States, do not vaccinate the general public. Diagnosis is
commonly achieved
using a skin test, which involves intradermal exposure to tuberculin PPD
(protein-purified
derivative). Antigen-specific T cell responses result in measurable incubation
at the injection
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
2
site by 48-72 hours after injection, which indicates exposure to Mycobacterial
antigens.
Sensitivity and specificity have, however, been a problem with this test, and
individuals
vaccinated with BCG cannot be distinguished from infected individuals.
While macrophages have been shown to act as the principal effectors of
M. tuberculosis immunity, T cells are the predominant inducers of such
immunity. The
essential role of T cells in protection against M. tuberculosis infection is
illustrated by the
frequent occurrence of M. tuberculosis in AIDS patients, due to the depletion
of CD4 T cells
associated with human immunodeficiency virus (HIV) infection. Mycobacterium-
reactive
CD4 T cells have been shown to be potent producers of gamma-interferon (IFN-
y), which, in
turn, has been shown to trigger the anti-mycobacterial effects of macrophages
in mice. While
the role of IFN-y in humans is less clear, studies have shown that 1,25-
dihydroxy-vitamin D3,
either alone or in combination with IFN-y or tumor necrosis factor-alpha,
activates human
macrophages to inhibit M. tuberculosis infection. Furthermore, it is known
that IFN-y
stimulates human macrophages to make 1,25-dihydroxy-vitamin D3. Similarly, IL-
12 has
been shown to play a role in stimulating resistance to M. tuberculosis
infection. For a review
of the immunology of M. tuberculosis infection see Chan and Kaufmann, in
Tuberculosis:
Pathogenesis, Protection and Control, Bloom (ed.), ASM Press, Washington, DC,
1994.
Accordingly, there is a need in the art for improved diagnostic methods for
detecting tuberculosis. The present invention fulfills this need and further
provides other
related advantages.
SUMMARY OF THE INVENTION
Briefly stated, the present invention provides compositions and methods for
diagnosing tuberculosis. In one aspect, polypeptides are provided comprising
an antigenic
portion of a soluble M. tuberculosis antigen, or a variant of such an antigen
that differs only
in conservative substitutions and/or modifications. In one embodiment of this
aspect, the
soluble antigen has one of the following N-terminal sequences:
(a) Asp-Pro-Val-Asp-Ala-Val-Ile-Asn-Thr-Thr-Cys-Asn-Tyr-Gly-Gln- ,
Val-Val-Ala-Ala-Leu (SEQ ID NO: 115);
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/1$214
3
(b) Ala-Val-Glu-Ser-Gly-Met-Leu-Ala-Leu-Gly-Thr-Pro-Ala-Pro-Ser
(SEQ ID NO: 116);
(c) Ala-Ala-Met-Lys-Pro-Arg-Thr-Gly-Asp-Gly-Pro-Leu-Glu-Ala-Ala-
Lys-Glu-Gly-Arg (SEQ ID NO: 117);
(d) Tyr-Tyr-Trp-Cys-Pro-Gly-Gln-Pro-Phe-Asp-Pro-Ala-Trp-Gly-Pro
(SEQ ID NO: 118);
(e) Asp-Ile-Gly-Ser-Glu-Ser-Thr-Glu-Asp-Gln-Gln-Xaa-Ala-Val (SEQ ID
NO: 119);
(fj Ala-Glu-Glu-Ser-Ile-Ser-Thr-Xaa-Glu-Xaa-Ile-Val-Pro (SEQ ID
NO: 120);
(g) Asp-Pro-Glu-Pro-Ala-Pro-Pro-Val-Pro-Thr-Thr-Ala-Ala-Ser-Pro-Pro-
Ser (SEQ ID NO: 121 );
(h) Ala-Pro-Lys-Thr-Tyr-Xaa-Glu-Glu-Leu-Lys-Gly-Thr-Asp-Thr-Gly
(SEQ ID NO: 122);
(i) Asp-Pro-Ala-Ser-Ala-Pro-Asp-Val-Pro-Thr-Ala-Ala-Gln-Leu-Thr-Ser-
Leu-Leu-Asn-Ser-Leu-Ala-Asp-Pro-Asn-Val-Ser-Phe-Ala-Asn (SEQ
ID NO: 123);
(j) Xaa-Asp-Ser-Glu-Lys-Ser-Ala-Thr-Ile-Lys-Val-Thr-Asp-Ala-Ser;
(SEQ ID NO: 129)
(k) Ala-Gly-Asp-Thr-Xaa-Ile-Tyr-Ile-Val-Gly-Asn-Leu-Thr-Ala-Asp;
(SEQ ID NO: 130) or
(I) Ala-Pro-Glu-Ser-Gly-Ala-Gly-Leu-Gly-Gly-Thr-Val-Gln-Ala-Gly;
(SEQ ID NO: 131)
wherein Xaa may be any amino acid.
In a related aspect, polypeptides are provided comprising an immunogenic
portion of an M. tuberculosis antigen, or a variant of such an antigen that
differs only in
conservative substitutions and/or modifications, the antigen having one of the
following N-
terminal sequences:
CA 02268036 1999-04-12
WO 98116645 PCT/US97/18214
4
(m) Xaa-Tyr-Ile-Ala-Tyr-Xaa-Thr-Thr-Ala-Gly-Ile-Val-Pro-Gly-Lys-Ile-
Asn-Val-His-Leu-Val; (SEQ ID NO: 132) or
(n) Asp-Pro-Pro-Asp-Pro-His-Gln-Xaa-Asp-Met-Thr-Lys-Gly-Tyr-Tyr-
Pro-Gly-Gly-Arg-Arg-Xaa-Phe; (SEQ ID NO: 124)
wherein Xaa may be any amino acid.
In another embodiment, the soluble M. tuberculosis antigen comprises an
amino acid sequence encoded by a DNA sequence selected from the group
consisting of the
sequences recited in SEQ ID NOS: 1, 2, 4-10, 13-25, 52, 94 and 96, the
complements of said
sequences, and DNA sequences that hybridize to a sequence recited in SEQ ID
NOS: 1, 2,
4-10, 13-25, 52, 94 and 96 or a complement thereof under moderately stringent
conditions.
In a related aspect, the polypeptides comprise an antigenic portion of a
M. tuberculosis antigen, or a variant of such an antigen that differs only in
conservative
substitutions and/or modifications, wherein the antigen comprises an amino
acid sequence
encoded by a DNA sequence selected from the group consisting of the sequences
recited in
SEQ ID NOS: 26-51, 133, 134, 158-178 and 196, the complements of said
sequences, and
DNA sequences that hybridize to a sequence recited in SEQ ID NOS: 26-51, 133,
134, 158-
178 and 196 or a complement thereof under moderately stringent conditions.
In related aspects, DNA sequences encoding the above polypeptides,
recombinant expression vectors comprising these DNA sequences and host cells
transformed
or transfected with such expression vectors are also provided.
In another aspect, the present invention provides fusion proteins comprising a
first and a second inventive polypeptide or, alternatively, an inventive
polypeptide and a
known M. tuberculosis antigen.
In further aspects of the subject invention, methods and diagnostic kits are
provided for detecting tuberculosis in a patient. The methods comprise: (a)
contacting a
biological sample with at least one of the above polypeptides; and (b)
detecting in the sample
the presence of antibodies that bind to the polypeptide or polypeptides,
thereby detecting
M. tuberculosis infection in the biological sample. Suitable biological
samples include whole
blood, sputum, serum, plasma, saliva, cerebrospinal fluid and urine. The
diagnostic kits
comprise one or more of the above polypeptides in combination with a detection
reagent.
CA 02268036 1999-04-12
WO 98/16645 PCTIUS97/18Z14
The present invention also provides methods for detecting M. tuberculosis
infection comprising: (a) obtaining a biological sample from a patient; (b)
contacting the
" sample with at least one oligonucleotide primer in a polymerase chain
reaction, the
oligonucleotide primer being specific for a DNA sequence encoding the above
polypeptides;
5 and (c) detecting in the sample a DNA sequence that amplifies in the
presence of the first and
second oligonucleotide primers. In one embodiment, the oligonucleotide primer
comprises at
least about 10 contiguous nucleotides of such a DNA sequence.
In a further aspect, the present invention provides a method for detecting
M. tuberculosis infection in a patient comprising: (a) obtaining a biological
sample from the
patient; (b) contacting the sample with an oligonucleotide probe specific for
a DNA sequence
encoding the above polypeptides; and (c) detecting in the sample a DNA
sequence that
hybridizes to the oligonucleotide probe. In one embodiment, the
oligonucleotide probe
comprises at least about 15 contiguous nucleotides of such a DNA sequence.
In yet another aspect, the present invention provides antibodies, both
polyclonal and monoclonal, that bind to the polypeptides described above, as
well as methods
for their use in the detection of M. tuberculosis infection.
These and other aspects of the present invention will become apparent upon
reference to the following detailed description and attached drawings. All
references
disclosed herein are hereby incorporated by reference in their entirety as if
each was
incorporated individually.
BRIEF DESCRIPTION OF THE DRAWINGS AND SEQUENCE IDENTIFIERS
Figure 1 A and B illustrate the stimulation of proliferation and interferon-y
production in T cells derived from a first and a second M. tuberculosis-immune
donor,
respectively, by the 14 Kd, 20 Kd and 26 Kd antigens described in Example 1.
Figures 2A-D illustrate the reactivity of antisera raised against secretory M.
tuberculosis proteins, the known M. tuberculosis antigen 85b and the inventive
antigens
Tb38-1 and TbH-9, respectively, with M. tuberculosis lysate (lane 2), M.
tuberculosis
secretory proteins (lane 3), recombinant Tb38-1 (lane 4), recombinant TbH-9
(lane 5) and
- 30 recombinant 85b (lane 5).
CA 02268036 1999-04-12
WO 98J16645 PCT/US97/18214
6
Figure 3A illustrates the stimulation of proliferation in a TbH-9-specific T
cell
clone by secretory M. tuberculosis proteins, recombinant TbH-9 and a control
antigen,
TbRal 1.
Figure 3B illustrates the stimulation of interferon-y production in a TbH-9-
specific T cell clone by secretory M. tuberculosis proteins, PPD and
recombinant TbH-9.
Figure 4 illustrates the reactivity of two representative polypeptides with
sera
from M. tuberculosis-infected and uninfected individuals, as compared to the
reactivity of
bacterial lysate.
Figure 5 shows the reactivity of four representative polypeptides with sera
from M. tuberculosis-infected and uninfected individuals, as compared to the
reactivity of the
38 kD antigen.
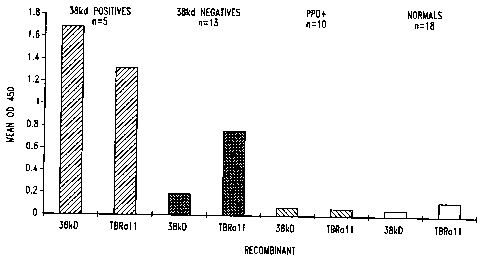
Figure 6 shows the reactivity of recombinant 38 kD and TbRal 1 antigens with
sera from M. tuberculosis patients, PPD positive donors and normal donors.
Figure 7 shows the reactivity of the antigen TbRa2A with 38 kD negative sera.
Figure 8 shows the reactivity of the antigen of SEQ ID NO: 60 with sera from
M. tuberculosis patients and normal donors.
Figure 9 illustrates the reactivity of the recombinant antigen TbH-29 (SEQ ID
NO: 137) with sera from M. tuberculosis patients, PPD positive donors and
normal donors as
determined by indirect ELISA.
Figure 10 illustrates the reactivity of the recombinant antigen TbH-33 (SEQ
ID NO: 140) with sera from M. tuberculosis patients and from normal donors,
and with a pool
of sera from M. tuberculosis patients, as determined both by direct and
indirect ELISA
Figure 11 illustrates the reactivity of increasing concentrations of the
recombinant antigen TbH-33 (SEQ ID NO: 140) with sera from M. tuberculosis
patients and
from normal donors as determined by ELISA.
SEQ. ID NO. 1 is the DNA sequence of TbRal .
SEQ. ID NO. 2 is the DNA sequence of TbRalO.
SEQ. ID NO. 3 is the DNA sequence of TbRal 1.
SEQ. ID NO. 4 is the DNA sequence of TbRal2. -
CA 02268036 1999-04-12
WO 98I1b645 PCT/US97/18214
7
SEQ. ID NO. 5 is the DNA sequence of TbRal3.
SEQ. ID NO. 6 is the DNA sequence of TbRal6.
' SEQ. ID NO. 7 is the DNA sequence of TbRa17.
SEQ. ID NO. 8 is the DNA sequence of TbRal8.
SEQ. ID NO. 9 is the DNA sequence of TbRal9.
SEQ. ID NO. 10 is the DNA sequence of TbRa24.
SEQ. ID NO. 11 is the DNA sequence of TbRa26.
SEQ. ID NO. 12 is the DNA sequence of TbRa28.
SEQ. ID NO. 13 is the DNA sequence of TbRa29.
SEQ. ID NO. 14 is the DNA sequence of TbRa2A.
SEQ. ID NO. 15 is the DNA sequence of TbRa3.
SEQ. ID NO. 16 is the DNA sequence of TbRa32.
SEQ. ID NO. 17 is the DNA sequence of TbRa35.
SEQ. ID NO. 18 is the DNA sequence of TbRa36.
SEQ. ID NO. 19 is the DNA sequence of TbRa4.
SEQ. ID NO. 20 is the DNA sequence of TbRa9.
SEQ. ID NO. 21 is the DNA sequence of TbRaB.
SEQ. ID NO. 22 is the DNA sequence of TbRaC.
SEQ. ID NO. 23 is the DNA sequence of TbRaD.
SEQ. ID NO. 24 is the DNA sequence of YYWCPG.
SEQ. ID NO. 25 is the DNA sequence of AAMK.
SEQ. ID NO. 26 is the DNA sequence of TbL-23.
SEQ. ID NO. 27 is the DNA sequence of TbL-24.
SEQ. ID NO. 28 is the DNA sequence of TbL-25.
SEQ. ID NO. 29 is the DNA sequence of TbL-28.
SEQ. ID NO. 30 is the DNA sequence of TbL-29.
SEQ. ID NO. 31 is the DNA sequence of TbH-5.
SEQ. ID NO. 32 is the DNA sequence of TbH-8.
SEQ. ID NO. 33 is the DNA sequence of TbH-9.
SEQ. ID NO. 34 is the DNA sequence of TbM-1.
CA 02268036 1999-04-12
WO 98/16645 PCT/ITS97/18Z14
8
SEQ. ID NO. 35 is the DNA sequence of TbM-3.
SEQ. ID NO. 36 is the DNA sequence of TbM-6.
SEQ. ID NO. 37 is the DNA sequence of TbM-7. '
SEQ. ID NO. 38 is the DNA sequence of TbM-9.
SEQ. ID NO. 39 is the DNA sequence of TbM-12.
SEQ. ID NO. 40 is the DNA sequence of TbM-13.
SEQ. ID NO. 41 is the DNA sequence of TbM-14.
SEQ. ID NO. 42 is the DNA sequence of TbM-15.
SEQ. ID NO. 43 is the DNA sequence of TbH-4.
SEQ. ID NO. 44 is the DNA sequence of TbH-4-FWD.
SEQ. ID NO. 45 is the DNA sequence of TbH-12.
SEQ. ID NO. 46 is the DNA sequence of Tb38-1.
SEQ. ID NO. 47 is the DNA sequence of Tb38-4.
SEQ. ID NO. 48 is the DNA sequence of TbL-17.
SEQ. ID NO. 49 is the DNA sequence of TbL-20.
SEQ. ID NO. 50 is the DNA sequence of TbL-21.
SEQ. ID NO. 51 is the DNA sequence of TbH-16.
SEQ. ID NO. 52 is the DNA sequence of DPEP.
SEQ. ID NO. 53 is the deduced amino acid sequence of DPEP.
SEQ. ID NO. 54 is the protein sequence of DPV N-terminal Antigen.
SEQ. ID NO. 55 is the protein sequence of AVGS N-terminal Antigen.
SEQ. ID NO. 56 is the protein sequence of AAMK N-terminal Antigen.
SEQ. ID NO. 57 is the protein sequence of YYWC N-terminal Antigen.
SEQ. ID NO. 58 is the protein sequence of DIGS N-terminal Antigen.
SEQ. ID NO. 59 is the protein sequence of AEES N-terminal Antigen.
SEQ. ID NO. 60 is the protein sequence of DPEP N-terminal Antigen.
SEQ. ID NO. 61 is the protein sequence of APKT N-terminal Antigen.
SEQ. ID NO. 62 is the protein sequence of DPAS N-terminal Antigen.
SEQ. ID NO. 63 is the deduced amino acid sequence of TbM-1 Peptide.
SEQ. ID NO. 64 is the deduced amino acid sequence of TbRal . -
CA 02268036 1999-04-12
WO 98/16645 PCT/US97I18214
9
SEQ. ID NO. 65 is the deduced amino acid sequence of TbRalO.
SEQ. ID NO. 66 is the deduced amino acid sequence of TbRal 1.
SEQ. ID NO. 67 is the deduced amino acid sequence of TbRal2.
SEQ. ID NO. 68 is the deduced amino acid sequence of TbRal3.
SEQ. ID NO. 69 is the deduced amino acid sequence of TbRal6.
SEQ. ID NO. 70 is the deduced amino acid sequence of TbRal7.
SEQ. ID NO. 71 is the deduced amino acid sequence of TbRal 8.
SEQ. ID NO. 72 is the deduced amino acid sequence of TbRal9.
SEQ. ID NO. 73 is the deduced amino acid sequence of TbRa24.
SEQ. ID NO. 74 is the deduced amino acid sequence of TbRa26.
SEQ. ID NO. 75 is the deduced amino acid sequence of TbRa28.
SEQ. ID NO. 76 is the deduced amino acid sequence of TbRa29.
SEQ. ID NO. 77 is the deduced amino acid sequence of TbRa2A.
SEQ. ID NO. 78 is the deduced amino acid sequence of TbRa3.
SEQ. ID NO. 79 is the deduced amino acid sequence of TbRa32.
SEQ. ID NO. 80 is the deduced amino acid sequence of TbRa35.
SEQ. ID NO. 81 is the deduced amino acid sequence of TbRa36.
SEQ. ID NO. 82 is the deduced amino acid sequence of TbRa4.
SEQ. ID NO. 83 is the deduced amino acid sequence of TbRa9.
SEQ. ID NO. 84 is the deduced amino acid sequence of TbRaB.
SEQ. ID NO. 85 is the deduced amino acid sequence of TbRaC.
SEQ. ID NO. 86 is the deduced amino acid sequence of TbRaD.
SEQ. ID NO. 87 is the deduced amino acid sequence of YYWCPG.
SEQ. ID NO. 88 is the deduced amino acid sequence of TbAAMK.
SEQ. ID NO. 89 is the deduced amino acid sequence of Tb38-1.
SEQ. ID NO. 90 is the deduced amino acid sequence of TbH-4.
SEQ. ID NO. 91 is the deduced amino acid sequence of TbH-8.
SEQ. ID NO. 92 is the deduced amino acid sequence of TbH-9.
SEQ. ID NO. 93 is the deduced amino acid sequence of TbH-12.
- 30 SEQ. ID NO. 94 is the DNA sequence of DPAS.
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
SEQ. ID NO. 95 is the deduced amino acid sequence of DPAS.
SEQ. ID NO. 96 is the DNA sequence of DPV.
SEQ. ID NO. 97 is the deduced amino acid sequence of DPV.
SEQ. ID NO. 98 is the DNA sequence of ESAT-6.
5 SEQ. ID NO. 99 is the deduced amino acid sequence of ESAT-6.
SEQ. ID NO. 100 is the DNA sequence of TbH-8-2.
SEQ. ID NO. 101 is the DNA sequence of TbH-9FL.
SEQ. ID NO. 102 is the deduced amino acid sequence of TbH-9FL.
SEQ. ID NO. 103 is the DNA sequence of TbH-9-1.
10 SEQ. ID NO. 104 is the deduced amino acid sequence of TbH-9-1.
SEQ. ID NO. 105 is the DNA sequence of TbH-9-4.
SEQ. ID NO. 106 is the deduced amino acid sequence of TbH-9-4.
SEQ. ID NO. 107 is the DNA sequence of Tb38-1F2 IN.
SEQ. ID NO. 108 is the DNA sequence of Tb38-1F2 RP.
SEQ. ID NO. 109 is the deduced amino acid sequence of Tb37-FL.
SEQ. ID NO. 110 is the deduced amino acid sequence of Tb38-IN.
SEQ. ID NO. 111 is the DNA sequence of Tb38-1F3.
SEQ. ID NO. 112 is the deduced amino acid sequence of Tb38-1F3.
SEQ. ID NO. 113 is the DNA sequence of Tb38-1F5.
SEQ. ID NO. 114 is the DNA sequence of Tb38-1F6.
SEQ. ID NO. 115 is the deduced N-terminal amino acid sequence of DPV.
SEQ. ID NO. 116 is the deduced N-terminal amino acid sequence of AVGS.
SEQ. ID NO. 117 is the deduced N-terminal amino acid sequence of AAMK.
SEQ. ID NO. 118 is the deduced N-terminal amino acid sequence of YYWC.
SEQ. ID NO. 119 is the deduced N-terminal amino acid sequence of DIGS.
SEQ. ID NO. 120 is the deduced N-terminal amino acid sequence of AAES.
SEQ. ID NO. 121 is the deduced N-terminal amino acid sequence of DPEP.
SEQ. ID NO. 122 is the deduced N-terminal amino acid sequence of APKT.
SEQ. ID NO. 123 is the deduced N-terminal amino acid sequence of DPAS.
SEQ. ID NO. 124 is the protein sequence of DPPD N-terminal Antigen. _
CA 02268036 1999-04-12
WO 98/16645 PCT/US97118214
11
SEQ ID NO. 125-128 are the protein sequences of four DPPD cyanogen bromide
fragments.
SEQ ID NO. 129 is the N-terminal protein sequence of XDS antigen.
SEQ ID NO. 130 is the N-terminal protein sequence of AGD antigen.
SEQ ID NO. 131 is the N-terminal protein sequence of APE antigen.
SEQ ID NO. 132 is the N-terminal protein sequence of XYI antigen.
SEQ ID NO. 133 is the DNA sequence of TbH-29.
SEQ ID NO. 134 is the DNA sequence of TbH-30.
SEQ ID NO. 135 is the DNA sequence of TbH-32.
SEQ ID NO. 136 is the DNA sequence of TbH-33.
SEQ ID NO. 137 is the predicted amino acid sequence of TbH-29.
SEQ ID NO. 138 is the predicted amino acid sequence of TbH-30.
SEQ ID NO. 139 is the predicted amino acid sequence of TbH-32.
SEQ ID NO. 140 is the predicted amino acid sequence of TbH-33.
SEQ ID NO: 141-146 are PCR primers used in the preparation of a fusion protein
containing TbRa3, 38 kD and Tb38-1.
SEQ ID NO: 147 is the DNA sequence of the fusion protein containing TbRa3, 38
kD
and Tb38-1.
SEQ ID NO: 148 is the amino acid sequence of the fusion protein containing
TbRa3,
38 kD and Tb38-1.
SEQ ID NO: 149 is the DNA sequence of the M. tuberculosis antigen 38 kD.
SEQ ID NO: 150 is the amino acid sequence of the M. tuberculosis antigen 38
kD.
SEQ ID NO: 1 S 1 is the DNA sequence of XP 14.
SEQ ID NO: 152 is the DNA sequence of XP24.
SEQ ID NO: 153 is the DNA sequence of XP31.
SEQ ID NO: 154 is the 5' DNA sequence of XP32.
SEQ ID NO: 155 is the 3' DNA sequence of XP32.
SEQ ID NO: 156 is the predicted amino acid sequence of XP14.
SEQ ID NO: 157 is the predicted amino acid sequence encoded by the reverse
complement of XP 14.
CA 02268036 1999-04-12
WO 98/16645 PCT/ITS97/18214
12
SEQ ID NO: 158 is the DNA sequence of XP27.
SEQ ID NO: 159 is the DNA sequence of XP36.
SEQ ID NO: 160 is the 5' DNA sequence of XP4. '
SEQ ID NO: 161 is the 5' DNA sequence of XPS.
SEQ ID NO: 162 is the 5' DNA sequence of XP 17.
SEQ ID NO: 163 is the 5' DNA sequence of XP30.
SEQ ID NO: 164 is the 5' DNA sequence
of XP2.
SEQ ID NO: 165 is the 3' DNA sequence
of XP2.
SEQ ID NO: 166 is the 5' DNA sequence
of XP3.
SEQ ID NO: 167 is the 3' DNA sequence
of XP3.
SEQ ID NO: 168 is the 5' DNA sequence of XP6.
SEQ ID NO: 169 is the 3' DNA sequence of XP6.
SEQ ID NO: 170 is the S' DNA sequence of XP 18:
SEQ ID NO: 171 is the 3' DNA sequence of XP18.
SEQ ID NO: 172 is the 5' DNA sequence of XP19.
SEQ ID NO: 173 is the 3' DNA sequence of XP 19.
SEQ ID NO: 174 is the 5' DNA sequence of XP22.
SEQ ID NO: 175 is the 3' DNA sequence of XP22.
SEQ ID NO: 176 is the 5' DNA sequence of XP25.
SEQ ID NO: 177 is the 3' DNA sequence of XP25.
SEQ ID NO: 178 is the full-length DNA sequence of TbH4-XP 1.
SEQ ID NO: 179 is the predicted amino acid sequence of TbH4-XP1.
SEQ ID NO: 180 is the predicted amino acid sequence encoded by the reverse
complement of TbH4-XP 1.
SEQ ID NO: 181 is a first predicted amino acid sequence encoded by XP36.
SEQ ID NO: 182 is a second predicted amino acid sequence encoded by XP36.
SEQ ID NO: 183 is the predicted amino acid sequence encoded by the reverse
complement of XP36.
SEQ ID NO: 184 is the DNA sequence of RDIF2.
SEQ ID NO: 185 is the DNA sequence of RDIFS. -
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
13
SEQ ID NO: 186 is the DNA sequence of RDIFB.
SEQ ID NO: 187 is the DNA sequence of RDIF 10.
SEQ ID NO: 188 is the DNA sequence of RDIF 11.
SEQ ID NO: 189 is the predicted amino acid sequence of RDIF2.
S SEQ ID NO: 190 is the predicted amino acid sequence of RDIFS.
SEQ iD NO: 191 is the predicted amino acid sequence of RDIFB.
SEQ ID NO: 192 is the predicted amino acid sequence of RDIF
10.
SEQ ID NO: 193 is the predicted amino acid sequence of RDIF
11.
SEQ ID NO: 194 is the 5' DNA sequence of RDIF 12.
SEQ ID NO: 195 is the 3' DNA sequence of RDIF12.
SEQ ID NO: 196 is the DNA sequence of RDIF7.
SEQ ID NO: 197 is the predicted amino acid sequence of RDIF7.
SEQ ID NO: 198 is the DNA sequence of DIF2-1.
SEQ ID NO: 199 is the predicted amino acid sequence of DIF2-1.
SEQ ID NO: 200-207 are PCR primers used in the preparation
of a fusion protein
containing TbRa3,
38 kD, Tb38-l
and DPEP (hereinafter
referred to as
TbF-2).
SEQ ID NO: 208 is the DNA sequence of the fusion protein
TbF-2.
SEQ ID NO: 209 is the amino acid sequence of the fusion protein
TbF-2.
DETAILED DESCRIPTION OF THE INVENTION
As noted above, the present invention is generally directed to compositions
and methods for diagnosing tuberculosis. The compositions of the subject
invention include
polypeptides that comprise at least one antigenic portion of a M. tuberculosis
antigen, or a
variant of such an antigen that differs only in conservative substitutions
and/or modifications.
Polypeptides within the scope of the present invention include, but are not
limited to, soluble
M. tuberculosis antigens. A "soluble M. tuberculosis antigen" is a protein of
M. tuberculosis
origin that is present in M. tuberculosis culture filtrate. As used herein,
the term
"polypeptide" encompasses amino acid chains of any length, including full
length proteins
(i.e., antigens), wherein the amino acid residues are linked by covalent
peptide bonds. Thus,
CA 02268036 1999-04-12
wo 9sn6sas rc~rrt~s9~nsma
14
a polypeptide comprising an antigenic portion of one of the above antigens may
consist
entirely of the antigenic portion, or may contain additional sequences. The
additional
sequences may be derived from the native M. tuberculosis antigen or may be
heterologous,
and such sequences may (but need not) be antigenic.
An "antigenic portion" of an antigen (which may or may not be soluble) is a
portion that is capable of reacting with sera obtained from an M. tuberculosis-
infected
individual (i.e., generates an absorbance reading with sera from infected
individuals that is at
least three standard deviations above the absorbance obtained with sera from
uninfected
individuals, in a representative ELISA assay described herein). An "M.
tuberculosis-infected
individual" is a human who has been infected with M. tuberculoses (e.g., has
an intradermal
skin test response to PPD that is at least 0.5 cm in diameter). Infected
individuals may
display symptoms of tuberculosis or may be free of disease symptoms.
Polypeptides
comprising at least an antigenic portion of one or more M. tuberculosis
antigens as described
herein may generally be used, alone or in combination, to detect tuberculosis
in a patient.
The compositions and methods of this invention also encompass variants of
the above polypeptides. A "variant," as used herein, is a polypeptide that
differs from the
native antigen only in conservative substitutions and/or modifications, such
that the antigenic
properties of the polypeptide are retained. Such variants may generally be
identified by
modifying one of the above polypeptide sequences, and evaluating the antigenic
properties of
the modified polypeptide using, for example, the representative procedures
described herein:
A "conservative substitution" is one in which an amino acid is substituted for
another amino acid that has similar properties, such that one skilled in the
art of peptide
chemistry would expect the secondary structure and hydropathic nature of the
polypeptide to
be substantially unchanged. In general, the following groups of amino acids
represent
conservative changes: (1) ala, pro, gly, glu, asp, gln, asn, ser, thr; (2)
cys, ser, tyr, thr; (3) val,
ile, leu, met, ala, phe; (4) lys, arg, his; and {5) phe, tyr, trp, his.
Variants may also (or alternatively) be modified by, for example, the deletion
or addition of amino acids that have minimal influence on the antigenic
properties, secondary
structure and hydropathic nature of the polypeptide. For example, a
polypeptide may be
conjugated to a signal (or leader) sequence at the N-terminal end of the
protein which co- _
CA 02268036 1999-04-12
WO 98/16645 PCTlUS97/18214
translationally or post-translationally directs transfer of the protein. The
polypeptide may
also be conjugated to a linker or other sequence for ease of synthesis,
purification or
" identification of the polypeptide (e.g., poly-His), or to enhance binding of
the polypeptide to a
solid support. For example, a polypeptide may be conjugated to an
immunoglobulin Fc
5 region.
In a related aspect, combination polypeptides are disclosed. A "combination
polypeptide" is a polypeptide comprising at least one of the above antigenic
portions and one
or more additional antigenic M. tuberculosis sequences, which are joined via a
peptide
linkage into a single amino acid chain. The sequences may be joined directly
(i.e., with no
10 intervening amino acids) or may be joined by way of a linker sequence
(e.g., Gly-Cys-Gly)
that does not significantly diminish the antigenic properties of the component
polypeptides.
In general, M. tuberculosis antigens, and DNA sequences encoding such
antigens, may be prepared using any of a variety of procedures. For example,
soluble
antigens may be isolated from M. tuberculosis culture filtrate by procedures
known to those
15 of ordinary skill in the art, including anion-exchange and reverse phase
chromatography.
Purified antigens may then be evaluated for a desired property, such as the
ability to react
with sera obtained from an M. tuberculosis-infected individual. Such screens
may be
performed using the representative methods described herein. Antigens may then
be partially
sequenced using, for example, traditional Edman chemistry. See Edman and Berg,
Eur. J.
Biochem. 80:116-132, 1967.
Antigens may also be produced recombinantly using a DNA sequence that
encodes the antigen, which has been inserted into an expression vector and
expressed in an
appropriate host. DNA molecules encoding soluble antigens may be isolated by
screening an
appropriate M. tuberculosis expression library with anti-sera (e.g., rabbit)
raised specifically
against soluble M. tuberculosis antigens. DNA sequences encoding antigens that
may or may
not be soluble may be identified by screening an appropriate M. tuberculosis
genomic or
cDNA expression library with sera obtained from patients infected with M.
tuberculosis.
Y
Such screens may generally be performed using techniques well known in the
art, such as
those described in Sambrook et al., Molecular Cloning: A Laboratory Manual,
Cold Spring
Harbor Laboratories, Cold Spring Harbor, NY, 1989.
CA 02268036 1999-04-12
WO 98116645 PCT/US97/182I4
16
DNA sequences encoding soluble antigens may also be obtained by screening
an appropriate M. tuberculosis cDNA or genomic DNA library for DNA sequences
that
hybridize to degenerate oligonucleotides derived from partial amino acid
sequences of
isolated soluble antigens. Degenerate oligonucleotide sequences for use in
such a screen may
be designed and synthesized, and the screen may be performed, as described
(for example) in
Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor
Laboratories, Cold Spring Harbor, NY (and references cited therein).
Polymerase chain
reaction (PCR) may also be employed, using the above oligonucleotides in
methods well
known in the art, to isolate a nucleic acid probe from a cDNA or genomic
library. The library
screen may then be performed using the isolated probe.
Regardless of the method of preparation, the antigens described herein are
"antigenic." More specifically, the antigens have the ability to react with
sera obtained from
an M. tuberculosis-infected individual. Reactivity may be evaluated using, for
example, the
representative ELISA assays described herein, where an absorbance reading with
sera from
infected individuals that is at least three standard deviations above the
absorbance obtained
with sera from uninfected individuals is considered positive.
Antigenic portions of M. tuberculosis antigens may be prepared and identified
using well known techniques, such as those summarized in Paul, Fundamental
Immunology,
3d ed., Raven Press, 1993, pp. 243-247 and references cited therein. Such
techniques include
screening polypeptide portions of the native antigen for antigenic properties.
The
representative ELISAs described herein may generally be employed in these
screens. An
antigenic portion of a polypeptide is a portion that, within such
representative assays,
generates a signal in such assays that is substantially similar to that
generated by the full
length antigen. In other words, an antigenic portion of a M. tuberculosis
antigen generates at
least about 20%, and preferably about 100%, of the signal induced by the full
length antigen
in a model ELISA as described herein.
Portions and other variants of M. tuberculosis antigens may be generated by
synthetic or recombinant means. Synthetic polypeptides having fewer than about
100 amino
acids, and generally fewer than about 50 amino acids, may be generated using
techniques
well known in the art. For example, such polypeptides may be synthesized using
any of the _
CA 02268036 1999-04-12
WO 98/16645 PGT/U897/18214
17
commercially available solid-phase techniques, such as the Mernfield solid-
phase synthesis
method, where amino acids are sequentially added to a growing amino acid
chain. See
' Merrifield, J. Am. Chem. Soc. 85:2149-2146, 1963. Equipment for automated
synthesis of
polypeptides is commercially available from suppliers such as Applied
BioSystems, Inc.,
Foster City, CA, and may be operated according to the manufacturer's
instructions. Variants
of a native antigen may generally be prepared using standard mutagenesis
techniques, such as
oligonucleotide-directed site-specific mutagenesis. Sections of the DNA
sequence may also
be removed using standard techniques to permit preparation of truncated
polypeptides.
Recombinant polypeptides containing portions and/or variants of a native
antigen may be readily prepared from a DNA sequence encoding the polypeptide
using a
variety of techniques well known to those of ordinary skill in the art. For
example,
supernatants from suitable host/vector systems which secrete recombinant
protein into culture
media may be first concentrated using a commercially available filter.
Following
concentration, the concentrate may be applied to a suitable purification
matrix such as an
affinity matrix or an ion exchange resin. Finally, one or more reverse phase
HPLC steps can
be employed to further purify a recombinant protein.
Any of a variety of expression vectors known to those of ordinary skill in the
art may be employed to express recombinant polypeptides as described herein.
Expression
may be achieved in any appropriate host cell that has been transformed or
transfected with an
expression vector containing a DNA molecule that encodes a recombinant
polypeptide.
Suitable host cells include prokaryotes, yeast and higher eukaryotic cells.
Preferably, the host
cells employed are E. coli, yeast or a mammalian cell line, such as COS or
CHO. The DNA
sequences expressed in this manner may encode naturally occurring antigens,
portions of
naturally occurnng antigens, or other variants thereof.
In general, regardless of the method of preparation, the polypeptides
disclosed
herein are prepared in substantially pure form. Preferably, the polypeptides
are at least about
80% pure, more preferably at least about 90% pure and most preferably at least
about 99%
pure. For use in the methods described herein, however, such substantially
pure polypeptides
may be combined.
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
18
In certain specific embodiments, the subject invention discloses polypeptides
comprising at least an antigenic portion of a soluble M. tuberculosis antigen
(or a variant of
such an antigen), where the antigen has one of the following N-terminal
sequences:
(a) Asp-Pro-Val-Asp-Ala-Val-Ile-Asn-Thr-Thr-Cys-Asn-Tyr-Gly-Gln-
Val-Val-Ala-Ala-Leu (SEQ ID NO: 115);
(b) Ala-Val-Glu-Ser-Gly-Met-Leu-Ala-Leu-Gly-Thr-Pro-Ala-Pro-Ser
(SEQ ID NO: 116);
(c) Ala-Ala-Met-Lys-Pro-Arg-Thr-Gly-Asp-Gly-Pro-Leu-Glu-Ala-Ala-
Lys-Glu-Gly-Arg (SEQ ID NO: 117);
(d) Tyr-Tyr-Trp-Cys-Pro-Gly-Gln-Pro-Phe-Asp-Pro-Ala-Trp-Gly-Pro
(SEQ ID NO: 118);
(e) Asp-Ile-Gly-Ser-Glu-Ser-Thr-Glu-Asp-Gln-Gln-Xaa-Ala-Val (SEQ ID
NO: 119);
(fj Ala-Glu-Glu-Ser-Ile-Ser-Thr-Xaa-Glu-Xaa-Ile-Val-Pro (SEQ ID
NO: 120);
(g) Asp-Pro-Glu-Pro-Ala-Pro-Pro-Val-Pro-Thr-Thr-Ala-Ala-Ser-Pro-Pro-
Ser (SEQ ID NO: 121);
(h) Ala-Pro-Lys-Thr-Tyr-Xaa-Glu-Glu-Leu-Lys-Gly-Thr-Asp-Thr-Gly
(SEQ ID NO: 122);
(i) Asp-Pro-Ala-Ser-Ala-Pro-Asp-Val-Pro-Thr-Ala-Ala-Gln-Gln-Thr-Ser-
Leu-Leu-Asn-Ser-Leu-Ala-Asp-Pro-Asn-Val-Ser-Phe-Ala-Asn (SEQ
ID NO: 123);
(j) Xaa-Asp-Ser-Glu-Lys-Ser-Ala-Thr-Ile-Lys-Val-Thr-Asp-Ala-Ser;
(SEQ ID NO: 129)
(k) Ala-Gly-Asp-Thr-Xaa-Ile-Tyr-Ile-Val-Gly-Asn-Leu-Thr-Ala-Asp;
(SEQ ID NO: 130) or
(1) Ala-Pro-Glu-Ser-Gly-Ala-Gly-Leu-Gly-Gly-Thr-Val-Gln-Ala-Gly;
(SEQ ID NO: 131)
wherein Xaa may be any amino acid, preferably a cysteine residue. A DNA
sequence
encoding the antigen identified as (g) above is provided in SEQ ID NO: 52, the
deduced
CA 02268036 1999-04-12
WO 98/16645 PCT/US97I18214
19
amino acid sequence of which is provided in SEQ ID NO: 53. A DNA sequence
encoding
the antigen identified as (a) above is provided in SEQ ID NO: 96; its deduced
amino acid
' sequence is provided in SEQ ID NO: 97. A DNA sequence corresponding to
antigen (d)
above is provided in SEQ ID NO: 24, a DNA sequence corresponding to antigen
(c) is
provided in SEQ ID NO: 25 and a DNA sequence corresponding to antigen (I) is
disclosed in
SEQ ID NO: 94 and its deduced amino acid sequence is provided in SEQ ID NO:
95.
In a further specific embodiment, the subject invention discloses polypeptides
comprising at least an immunogenic portion of an M. tuberculosis antigen
having one of the
following N-terminal sequences, or a variant thereof that differs only in
conservative
substitutions and/or modifications:
(m) Xaa-Tyr-Ile-Ala-Tyr-Xaa-Thr-Thr-Ala-Gly-Ile-Val-Pro-Gly-Lys-Ile-
Asn-Val-His-Leu-Val; (SEQ ID NO: 132) or
(n) Asp-Pro-Pro-Asp-Pro-His-Gln-Xaa-Asp-Met-Thr-Lys-Gly-Tyr-Tyr-
Pro-Gly-Gly-Arg-Arg-Xaa-Phe; (SEQ ID NO: 124)
wherein Xaa may be any amino acid, preferably a cysteine residue.
In other specific embodiments, the subject invention discloses polypeptides
comprising at least an antigenic portion of a soluble M. tuberculosis antigen
(or a variant of
such an antigen) that comprises one or more of the amino acid sequences
encoded by (a) the
DNA sequences of SEQ ID NOS: 1, 2, 4-10, 13-25, 52, 94 and 96, (b) the
complements of
such DNA sequences, or {c) DNA sequences substantially homologous to a
sequence in (a) or
(b).
In further specific embodiments, the subject invention discloses polypeptides
comprising at least an antigenic portion of a M. tuberculosis antigen (or a
variant of such an
antigen), which may or may not be soluble, that comprises one or more of the
amino acid
sequences encoded by (a) the DNA sequences of SEQ ID NOS: 26-51, 133, 134, 158-
178 and
196, (b) the complements of such DNA sequences or (c) DNA sequences
substantially
homologous to a sequence in (a) or (b).
In the specific embodiments discussed above, the M. tuberculosis antigens
include variants that are encoded DNA sequences which are substantially
homologous to one
CA 02268036 1999-04-12
WO 98/16645 PCT/I1S9~/182I4
or more of DNA sequences specifically recited herein. "Substantial homology,"
as used
herein, refers to DNA sequences that are capable of hybridizing under
moderately stringent
conditions. Suitable moderately stringent conditions include prewashing in a
solution of SX
SSC, 0.5% SDS, 1.0 mM EDTA (pH 8.0); hybridizing at 50°C-65°C,
SX SSC, overnight or,
S in the event of cross-species homology, at 45°C with O.SX SSC;
followed by washing twice
at 65°C for 20 minutes with each of 2X, O.SX and 0.2X SSC containing
O.I% SDS). Such
hybridizing DNA sequences are also within the scope of this invention, as are
nucleotide
sequences that, due to code degeneracy, encode an immunogenic polypeptide that
is encoded
by a hybridizing DNA sequence.
10 In a related aspect, the present invention provides fusion proteins
comprising a
f rst and a second inventive polypeptide or, alternatively, a polypeptide of
the present
invention and a known M. tuberculosis antigen, such as the 38 kD antigen
described above or
ESAT-6 (SEQ ID NOS: 98 and 99), together with variants of such fusion
proteins. The
fusion proteins of the present invention may also include a linker peptide
between the first
1 S and second polypeptides.
A DNA sequence encoding a fusion protein of the present invention is
constructed using known recombinant DNA techniques to assemble separate DNA
sequences
encoding the first and second polypeptides into an appropriate expression
vector. The 3' end
of a DNA sequence encoding the first polypeptide is ligated, with or without a
peptide linker,
20 to the 5' end of a DNA sequence encoding the second polypeptide so that the
reading frames
of the sequences are in phase to permit mRNA translation of the two DNA
sequences into a
single fusion protein that retains the biological activity of both the first
and the second
polypeptides.
A peptide linker sequence may be employed to separate the first and the
second polypeptides by a distance sufficient to ensure that each polypeptide
folds into its
secondary and tertiary structures. Such a peptide linker sequence is
incorporated into the
fusion protein using standard techniques well known in the art. Suitable
peptide linker
sequences may be chosen based on the following factors: (1) their ability to
adopt a flexible
extended conformation; (2) their inability to adopt a secondary structure that
could interact
with functional epitopes on the first and second polypeptides; and (3) the
lack of hydrophobic ,
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
21
or charged residues that might react with the polypeptide functional epitopes.
Preferred
peptide linker sequences contain Gly, Asn and Ser residues. Other near neutral
amino acids,
' such as Thr and Ala may also be used in the linker sequence. Amino acid
sequences which
may be usefully employed as linkers include those disclosed in Maratea et al.,
Gene 40:39-46,
1985; Murphy et al., Proc. Natl. Acad. Sci. USA 83:8258-8562, 1986; U.S.
Patent
No. 4,935,233 and U.S. Patent No. 4,751,180. The linker sequence may be from 1
to about
50 amino acids in length. Peptide linker sequences are not required when the
first and second
polypeptides have non-essential N-terminal amino acid regions that can be used
to separate
the functional domains and prevent steric hindrance.
In another aspect, the present invention provides methods for using the
polypeptides described above to diagnose tuberculosis. In this aspect, methods
are provided
for detecting M. tuberculosis infection in a biological sample, using one or
more of the above
polypeptides, alone or in combination. In embodiments in which multiple
polypeptides are
employed, polypeptides other than those specifically described herein, such as
the 38 kD
antigen described in Andersen and Hansen, Infect. Immun. 57:2481-2488, 1989,
may be
included. As used herein, a "biological sample" is any antibody-containing
sample obtained
from a patient. Preferably, the sample is whole blood, sputum, serum, plasma,
saliva,
cerebrospinal fluid or urine. More preferably, the sample is a blood, serum or
plasma sample
obtained from a patient or a blood supply. The polypeptide(s) are used in an
assay, as
described below, to determine the presence or absence of antibodies to the
polypeptide(s) in
the sample, relative to a predetermined cut-off value. The presence of such
antibodies
indicates previous sensitization to mycobacterial antigens which may be
indicative of
tuberculosis.
In embodiments in which more than one polypeptide is employed, the
polypeptides used are preferably complementary (i.e., one component
polypeptide will tend
to detect infection in samples where the infection would not be detected by
another
component polypeptide). Complementary polypeptides may generally be identified
by using
each polypeptide individually to evaluate serum samples obtained from a series
of patients
known to be infected with M. tuberculosis. After determining which samples
test positive (as
described below) with each polypeptide, combinations of two or more
polypeptides may be
CA 02268036 1999-04-12
WO 98/16645 PGT/US99/18214
22
formulated that are capable of detecting infection in most, or all, of the
samples tested. Such
polypeptides are complementary. For example, approximately 25-30% of sera from
tuberculosis-infected individuals are negative for antibodies to any single
protein, such as the
38 kD antigen mentioned above. Complementary polypeptides may, therefore, be
used in
S combination with the 38 kD antigen to improve sensitivity of a diagnostic
test.
There are a variety of assay formats known to those of ordinary skill in the
art
for using one or more polypeptides to detect antibodies in a sample. See,
e.g., Harlow and
Lane, Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, 1988,
which is
incorporated herein by reference. In a preferred embodiment, the assay
involves the use of
polypeptide immobilized on a solid support to bind to and remove the antibody
from the
sample. The bound antibody may then be detected using a detection reagent that
contains a
reporter group. Suitable detection reagents include antibodies that bind to
the
antibody/polypeptide complex and free polypeptide labeled with a reporter
group (e.g., in a
semi-competitive assay). Alternatively, a competitive assay may be utilized,
in which an
antibody that binds to the polypeptide is labeled with a reporter group and
allowed to bind to
the immobilized antigen after incubation of the antigen with the sample. The
extent to which
components of the sample inhibit the binding of the labeled antibody to the
polypeptide is
indicative of the reactivity of the sample with the immobilized polypeptide.
The solid support may be any solid material known to those of ordinary skill
in the art to which the antigen may be attached. For example, the solid
support may be a test
well in a microtiter plate or a nitrocellulose or other suitable membrane.
Alternatively, the
support may be a bead or disc, such as glass, fiberglass, latex or a plastic
material such as
polystyrene or polyvinylchloride. The support may also be a magnetic particle
or a fiber
optic sensor, such as those disclosed, for example, in U.S. Patent No.
5,359,681.
The polypeptides may be bound to the solid support using a variety of
techniques known to those of ordinary skill in the art, which are amply
described in the patent
and scientific literature. In the context of the present invention, the term
"bound" refers to
both noncovalent association, such as adsorption, and covalent attachment
(which may be a
direct linkage between the antigen and functional groups on the support or may
be a linkage
by way of a cross-linking agent). Binding by adsorption to a well in a
microtiter plate or to a _
CA 02268036 1999-04-12
WO 98/16645 PCT/US97118214
23
membrane is preferred. In such cases, adsorption may be achieved by contacting
the
polypeptide, in a suitable buffer, with the solid support for a suitable
amount of time. The
- contact time varies with temperature, but is typically between about 1 hour
and 1 day. In
general, contacting a well of a plastic microtiter plate (such as polystyrene
or
polyvinylchloride) with an amount of polypeptide ranging from about 10 ng to
about 1 pg,
and preferably about 100 ng, is sufficient to bind an adequate amount of
antigen.
Covalent attachment of polypeptide to a solid support may generally be
achieved by first reacting the support with a bifunctional reagent that will
react with both the
support and a functional group, such as a hydroxyl or amino group, on the
polypeptide. For
example, the polypeptide may be bound to supports having an appropriate
polymer coating
using benzoquinone or by condensation of an aldehyde group on the support with
an amine
and an active hydrogen on the polypeptide (see, e.g., Pierce Immunotechnology
Catalog and
Handbook, 1991, at A12-A13).
In certain embodiments, the assay is an enzyme linked immunosorbent assay
(ELISA). This assay may be performed by first contacting a polypeptide antigen
that has
been immobilized on a solid support, commonly the well of a microtiter plate,
with the
sample, such that antibodies to the polypeptide within the sample are allowed
to bind to the
immobilized polypeptide. Unbound sample is then removed from the immobilized
polypeptide and a detection reagent capable of binding to the immobilized
antibody-
polypeptide complex is added. The amount of detection reagent that remains
bound to the
solid support is then determined using a method appropriate for the specific
detection reagent.
More specifically, once the polypeptide is immobilized on the support as
described above, the remaining protein binding sites on the support are
typically blocked.
Any suitable blocking agent known to those of ordinary skill in the art, such
as bovine serum
albumin or Tween 20TM (Sigma Chemical Co., St. Louis, MO) may be employed. The
immobilized polypeptide is then incubated with the sample, and antibody is
allowed to bind
to the antigen. The sample may be diluted with a suitable diluent, such as
phosphate-buffered
saline (PBS) prior to incubation. In general, an appropriate contact time
(i.e., incubation
time) is that period of time that is sufficient to detect the presence of
antibody within a
M. tuberculosis-infected sample. Preferably, the contact time is sufficient to
achieve a level
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
24
of binding that is at least 95% of that achieved at equilibrium between bound
and unbound
antibody. Those of ordinary skill in the art will recognize that the time
necessary to achieve
equilibrium may be readily determined by assaying the level of binding that
occurs over a
period of time. At room temperature, an incubation time of about 30 minutes is
generally
sufficient.
Unbound sample may then be removed by washing the solid support with an
appropriate buffer, such as PBS containing 0.1% Tween 20TM. Detection reagent
may then be
added to the solid support. An appropriate detection reagent is any compound
that binds to
the immobilized antibody-polypeptide complex and that can be detected by any
of a variety
of means known to those in the art. Preferably, the detection reagent contains
a binding agent
(such as, for example, Protein A, Protein G, immunoglobulin, lectin or free
antigen)
conjugated to a reporter group. Preferred reporter groups include enzymes
(such as
horseradish peroxidase), substrates, cofactors, inhibitors, dyes,
radionuclides, luminescent
groups, fluorescent groups and biotin. The conjugation of binding agent to
reporter group
may be achieved using standard methods known to those of ordinary skill in the
art.
Common binding agents may also be purchased conjugated to a variety of
reporter groups
from many commercial sources (e.g., Zymed Laboratories, San Francisco, CA, and
Pierce,
Rockford, IL).
The detection reagent is then incubated with the immobilized antibody-
polypeptide complex for an amount of time sufficient to detect the bound
antibody. An
appropriate amount of time may generally be determined from the manufacturer's
instructions
or by assaying the level of binding that occurs over a period of time. Unbound
detection
reagent is then removed and bound detection reagent is detected using the
reporter group.
The method employed for detecting the reporter group depends upon the nature
of the
reporter group. For radioactive groups, scintillation counting or
autoradiographic methods
are generally appropriate. Spectroscopic methods may be used to detect dyes,
luminescent
groups and fluorescent groups. Biotin may be detected using avidin, coupled to
a different
reporter group (commonly a radioactive or fluorescent group or an enzyme).
Enzyme
reporter groups may generally be detected by the addition of substrate
(generally for a
specific period of time), followed by spectroscopic or other analysis of the
reaction products. ,
CA 02268036 1999-04-12
WO 98/16645 PCT/US9?/1$214
To determine the presence or absence of anti-M. tuberculosis antibodies in the
sample, the signal detected from the reporter group that remains bound to the
solid support is
' generally compared to a signal that corresponds to a predetermined cut-off
value. In one
preferred embodiment, the cut-off value is the average mean signal obtained
when the
5 immobilized antigen is incubated with samples from an uninfected patient. In
general, a
sample generating a signal that is three standard deviations above the
predetermined cut-off
value is considered positive for tuberculosis. In an alternate preferred
embodiment, the cut
off value is determined using a Receiver Operator Curve, according to the
method of Sackett
et al., Clinical Epidemiology: A Basic Science for Clinical Medicine, Little
Brown and Co.,
10 1985, pp. 106-107. Briefly, in this embodiment, the cut-off value may be
determined from a
plot of pairs of true positive rates (i.e., sensitivity) and false positive
rates (100%-specificity)
that correspond to each possible cut-off value for the diagnostic test result.
The cut-off value
on the plot that is the closest to the upper left-hand corner (i.e., the value
that encloses the
largest area) is the most accurate cut-off value, and a sample generating a
signal that is higher
15 than the cut-off value determined by this method may be considered
positive. Alternatively,
the cut-off value may be shifted to the left along the plot, to minimize the
false positive rate,
or to the right, to minimize the false negative rate. In general, a sample
generating a signal
that is higher than the cut-off value determined by this method is considered
positive for
tuberculosis.
20 In a related embodiment, the assay is performed in a rapid flow-through or
strip test format, wherein the antigen is immobilized on a membrane, such as
nitrocellulose.
In the flow-through test, antibodies within the sample bind to the immobilized
polypeptide as
the sample passes through the membrane. A detection reagent (e.g., protein A-
colloidal gold)
then binds to the antibody-polypeptide complex as the solution containing the
detection
25 reagent flows through the membrane. The detection of bound detection
reagent may then be
performed as described above. In the strip test format, one end of the
membrane to which
polypeptide is bound is immersed in a solution containing the sample. The
sample migrates
along the membrane through a region containing detection reagent and to the
area of
immobilized polypeptide. Concentration of detection reagent at the polypeptide
indicates the
presence of anti-M. tuberculosis antibodies in the sample. Typically, the
concentration of
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/I8214
26
detection reagent at that site generates a pattern, such as a line, that can
be read visually. The
absence of such a pattern indicates a negative result. In general, the amount
of polypeptide
immobilized on the membrane is selected to generate a visually discernible
pattern when the
biological sample contains a level of antibodies that would be sufficient to
generate a positive
signal in an ELISA, as discussed above. Preferably, the amount of polypeptide
immobilized
on the membrane ranges from about 25 ng to about 1 p.g, and more preferably
from about
50 ng to about 500 ng. Such tests can typically be performed with a very small
amount (e.g.,
one drop) of patient serum or blood.
Of course, numerous other assay protocols exist that are suitable for use with
the polypeptides of the present invention. The above descriptions are intended
to be
exemplary only.
In yet another aspect, the present invention provides antibodies to the
inventive polypeptides. Antibodies may be prepared by any of a variety of
techniques known
to those of ordinary skill in the art. See, e.g., Harlow and Lane, Antibodies:
A Laboratory
Manual, Cold Spring Harbor Laboratory, 1988. In one such technique, an
immunogen
comprising the antigenic polypeptide is initially injected into any of a wide
variety of
mammals (e.g., mice, rats, rabbits, sheep and goats). In this step, the
polypeptides of this
invention may serve as the immunogen without modification. Alternatively,
particularly for
relatively short polypeptides, a superior immune response may be elicited if
the polypeptide
is joined to a carrier protein, such as bovine serum albumin or keyhole limpet
hemocyanin.
The immunogen is injected into the animal host, preferably according to a
predetermined
schedule incorporating one or more booster immunizations, and the animals are
bled
periodically. Polyclonal antibodies specific for the polypeptide may then be
purified from
such antisera by, for example, affinity chromatography using the polypeptide
coupled to a
suitable solid support.
Monoclonal antibodies specific for the antigenic poiypeptide of interest may
be prepared, for example, using the technique of Kohler and Milstein, Eur. J.
Immunol.
6:511-519, 1976, and improvements thereto. Briefly, these methods involve the
preparation
of immortal cell lines capable of producing antibodies having the desired
specificity (i.e.,
reactivity with the polypeptide of interest): Such cell lines may be produced,
for example, ,
CA 02268036 1999-04-12
wo Xmas rc-r~s9~nszia
27
from spleen cells obtained from an animal immunized as described above. The
spleen cells
are then immortalized by, for example, fusion with a myeloma cell fusion
partner, preferably
one that is syngeneic with the immunized animal. A variety of fusion
techniques may be
employed. For example, the spleen cells and myeloma cells may be combined with
a
nonionic detergent for a few minutes and then plated at low density on a
selective medium
that supports the growth of hybrid cells, but not myeloma cells. A preferred
selection
technique uses HAT (hypoxanthine, aminopterin, thymidine) selection. After a
sufficient
time, usually about 1 to 2 weeks, colonies of hybrids are observed. Single
colonies are
selected and tested for binding activity against the polypeptide. Hybridomas
having high
reactivity and specificity are preferred.
Monoclonal antibodies may be isolated from the supernatants of growing
hybridoma colonies. In addition, various techniques may be employed to enhance
the yield,
such as injection of the hybridoma cell line into the peritoneal cavity of a
suitable vertebrate
host, such as a mouse. Monoclonal antibodies may then be harvested from the
ascites fluid or
the blood. Contaminants may be removed from the antibodies by conventional
techniques,
such as chromatography, gel filtration, precipitation, and extraction. The
polypeptides of this
invention may be used in the purification process in, for example, an affinity
chromatography
step.
Antibodies may be used in diagnostic tests to detect the presence of
M. tuberculosis antigens using assays similar to those detailed above and
other techniques
well known to those of skill in the art, thereby providing a method for
detecting
M. tuberculosis infection in a patient.
Diagnostic reagents of the present invention may also comprise DNA
sequences encoding one or more of the above polypeptides, or one or more
portions thereof.
For example, at least two oligonucleotide primers may ~be employed in a
polymerase chain
reaction (PCR) based assay to amplify M. tuberculosis-specific cDNA derived
from a
biological sample, wherein at least one of the oligonucleotide primers is
specific for a DNA
molecule encoding a polypeptide of the present invention. The presence of the
amplified
cDNA is then detected using techniques well known in the art, such as gel
electrophoresis.
Similarly, oligonucleotide probes specific for a DNA molecule encoding a
polypeptide of the
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
28
present invention may be used in a hybridization assay to detect the presence
of an inventive
polypeptide in a biological sample.
As used herein, the term "oligonucleotide primer/probe specific for a DNA '
molecule" means an oligonucleotide sequence that has at least about 80%,
preferably at least
S about 90% and more preferably at least about 95%, identity to the DNA
molecule in question.
Oligonucleotide primers and/or probes which may be usefully employed in the
inventive
diagnostic methods preferably have at least about 10-40 nucleotides. In a
preferred
embodiment, the oligonucleotide primers comprise at least about 10 contiguous
nucleotides
of a DNA molecule encoding one of the polypeptides disclosed herein.
Preferably,
oligonucleotide probes for use in the inventive diagnostic methods comprise at
least about 15
contiguous oligonucleotides of a DNA molecule encoding one of the polypeptides
disclosed
herein. Techniques for both PCR based assays and hybridization assays are well
known in
the art (see, for example, Mullis et al. Ibid; Ehrlich, Ibid). Primers or
probes may thus be
used to detect M. tuberculosis-specific sequences in biological samples. DNA
probes or
primers comprising oligonucleotide sequences described above may be used
alone, in
combination with each other, or with previously identified sequences, such as
the 38 kD
antigen discussed above.
The following Examples are offered by way of illustration and not by way of
limitation.
EXAMPLES
EXAMPLE 1
PURIFICATION AND CHARACTERIZATION OF POLYPEPTIDES
FROM M. TUBERCULOSIS CULTURE FILTRATE
This example illustrates the preparation of M. tuberculosis soluble
polypeptides from culture filtrate. Unless otherwise noted, all percentages in
the following
example are weight per volume.
CA 02268036 1999-04-12
wo Xmas rc~rn~rs9~nszia
29
M. tuberculosis (either H37Ra, ATCC No. 25177, or H37Rv, ATCC
No. 25618) was cultured in sterile GAS media at 37°C for fourteen days.
The media was
then vacuum filtered (leaving the bulk of the cells) through a 0.45 p filter
into a sterile 2.5 L
bottle. The media was then filtered through a 0.2 p. filter into a sterile 4 L
bottle. NaN3 was
then added to the culture filtrate to a concentration of 0.04%. The bottles
were then placed in
a 4°C cold room.
The culture filtrate was concentrated by placing the filtrate in a 12 L
reservoir
that had been autoclaved and feeding the filtrate into a 400 ml Amicon stir
cell which had
been rinsed with ethanol and contained a 10,000 kDa MWCO membrane. The
pressure was
maintained at 60 psi using nitrogen gas. This procedure reduced the 12 L
volume to
approximately 50 ml.
The culture filtrate was then dialyzed into 0.1 % ammonium bicarbonate using
a 8,000 kDa MWCO cellulose ester membrane, with two changes of ammonium
bicarbonate
solution. Protein concentration was then determined by a commercially
available BCA assay
(Pierce, Rockford, IL).
The dialyzed culture filtrate was then lyophilized, and the polypeptides
resuspended in distilled water. The polypeptides were then dialyzed against
0.01 mM 1,3
bis[tris(hydroxymethyl)-methylamino)propane, pH 7.5 (Bis-Tris propane buffer),
the initial
conditions for anion exchange chromatography. Fractionation was performed
using gel
profusion chromatography on a POROS 146 II Q/M anion exchange column 4.6 mm x
100 mm (Perseptive BioSystems, Framingham, MA) equilibrated in 0.01 mM Bis-
Tris
propane buffer pH 7.5. Polypeptides were eluted with a linear 0-0.5 M NaCI
gradient in the
above buffer system. The column eluent was monitored at a wavelength of 220
nm.
The pools of polypeptides eluting from the ion exchange column were
dialyzed against distilled water and lyophilized. The resulting material was
dissolved in 0.1%
trifluoroacetic acid (TFA) pH 1.9 in water, and the polypeptides were purified
on a Delta-Pak
C18 column (Waters, Milford, MA) 300 Angstrom pore size, 5 micron particle
size (3.9 x
150 mm). The polypeptides were eluted from the column with a linear gradient
from 0-60%
dilution buffer (0.1% TFA in acetonitrile). The flow rate was 0.75 ml/minute
and the HPLC
eluent was monitored at 214 nm. Fractions containing the eluted polypeptides
were collected
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
to maximize the purity of the individual samples. Approximately 200 purified
polypeptides
were obtained.
The purified polypeptides were then screened for the ability to induce T-cell
'
proliferation in PBMC preparations. The PBMCs from donors known to be PPD skin
test
5 positive and whose T cells were shown to proliferate in response to PPD and
crude soluble
proteins from MTB were cultured in medium comprising RPMI 1640 supplemented
with
10% pooled human serum and SO p,g/ml gentamicin. Purified polypeptides were
added in
duplicate at concentrations of 0.5 to 10 ~,g/mL. After six days of culture in
96-well round-
bottom plates in a volume of 200 ~1, 50 ~,l of medium was removed from each
well for
10 determination of iFN-y levels, as described below. The plates were then
pulsed with
1 ~,Ci/well of tritiated thymidine for a further 18 hours, harvested and
tritium uptake
determined using a gas scintillation counter. Fractions that resulted in
proliferation in both
replicates three fold greater than the proliferation observed in cells
cultured in medium alone
were considered positive.
15 IFN-y was measured using an enzyme-linked immunosorbent assay (ELISA).
ELISA plates were coated with a mouse monoclonal antibody directed to human
IFN-y
(Chemicon) in PBS for four hours at room temperature. Wells were then blocked
with PBS
containing 5% (W/V) non-fat dried milk for 1 hour at room temperature. The
plates were
then washed six times in PBS/0.2% TWEEN-20 and samples diluted 1:2 in culture
medium
20 in the ELISA plates were incubated overnight at room temperature. The
plates were again
washed and a polyclonal rabbit anti-human IFN-y serum diluted 1:3000 in
PBS/10% normal
goat serum was added to each well. The plates were then incubated for two
hours at room
temperature, washed and horseradish peroxidase-coupled anti-rabbit IgG
(Jackson Labs.) was
added at a 1:2000 dilution in PBS/5% non-fat dried milk. After a further two
hour incubation
25 at room temperature, the plates were washed and TMB substrate added. The
reaction was
stopped after 20 min with 1 N sulfuric acid. Optical density was determined at
450 nm using
570 nm as a reference wavelength. Fractions that resulted in both replicates
giving an OD
two fold greater than the mean OD from cells cultured in medium alone, plus 3
standard
deviations, were considered positive.
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18Z14
31
For sequencing, the polypeptides were individually dried onto
BiobreneT"' (Perkin Elmer/Applied BioSystems Division, Foster City, CA)
treated glass fiber
- filters. The filters with polypeptide were loaded onto a Perkin
Elmer/Applied BioSystems
Division Procise 492 protein sequencer. The polypeptides were sequenced from
the amino
' S terminal and using traditional Edman chemistry. The amino acid sequence
was determined
for each polypeptide by comparing the retention time of the PTH amino acid
derivative to the
appropriate PTH derivative standards.
Using the procedure described above, antigens having the following
N-terminal sequences were isolated:
(a) Asp-Pro-Val-Asp-Ala-Val-Ile-Asn-Thr-Thr-Xaa-Asn-Tyr-Gly-Gln
Val-Val-Ala-Ala-Leu (SEQ ID NO: 54);
(b) Ala-Val-Glu-Ser-Gly-Met-Leu-Ala-Leu-Gly-Thr-Pro-Ala-Pro-Ser
(SEQ ID NO: 55);
(c) Ala-Ala-Met-Lys-Pro-Arg-Thr-Gly-Asp-Gly-Pro-Leu-Glu-Ala-Ala-
Lys-Glu-Gly-Arg (SEQ ID NO: 56);
(d) Tyr-Tyr-Trp-Cys-Pro-Gly-Gln-Pro-Phe-Asp-Pro-Ala-Trp-Gly-Pro
(SEQ ID NO: 57);
(e) Asp-Ile-Gly-Ser-Glu-Ser-Thr-Glu-Asp-Gln-Gln-Xaa-Ala-Val (SEQ ID
NO: 58);
(f) Ala-Glu-Glu-Ser-Ile-Ser-Thr-Xaa-Glu-Xaa-Ile-Val-Pro (SEQ ID
NO: 59);
(g) Asp-Pro-Glu-Pro-Ala-Pro-Pro-Val-Pro-Thr-Ala-Ala-Ala-Ala-Pro-Pro-
Ala (SEQ ID NO: 60); and
(h) Ala-Pro-Lys-Thr-Tyr-Xaa-Glu-Glu-Leu-Lys-Gly-Thr-Asp-Thr-Gly
(SEQ ID NO: 61);
wherein Xaa may be any amino acid.
An additional antigen was isolated employing a rnicrobore HPLC purification
step in addition to the procedure described above. Specifically, 20 p,l of a
fraction comprising
a mixture of antigens from the chromatographic purification step previously
described, was
purified on an Aquapore C18 column (Perkin Elmer/Applied Biosystems Division,
Foster
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
32
City, CA) with a 7 micron pore size, column size 1 mm x 100 mm, in a Perkin
Elmer/Applied
Biosystems Division Model 172 HPLC. Fractions were eluted from the column with
a linear
gradient of 1%/minute of acetonitrile (containing O.OS% TFA) in water (O.OS%
TFA) at a
flow rate of 80 p,l/minute. The eluent was monitored at 2S0 nm. The original
fraction was
S separated into 4 major peaks plus other smaller components and a polypeptide
was obtained
which was shown to have a molecular weight of 12.OS4 Kd (by mass spectrometry)
and the
following N-terminal sequence:
(i) Asp-Pro-Ala-Ser-Ala-Pro-Asp-Val-Pro-Thr-Ala-Ala-Gln-Gln-Thr-Ser-
Leu-Leu-Asn-Asn-Leu-Ala-Asp-Pro-Asp-Val-Ser-Phe-Ala-Asp (SEQ
ID NO: 62).
This polypeptide was shown to induce proliferation and IFN-y production in
PBMC
preparations using the assays described above.
Additional soluble antigens were isolated from M. tuberculosis culture
filtrate
as follows. M. tuberculosis culture filtrate was prepared as described above.
Following
1 S dialysis against Bis-Tris propane buffer, at pH S.S, fractionation was
performed using anion
exchange chromatography on a Poros QE column 4.6 x 100 mm (Perseptive
Biosystems)
equilibrated in Bis-Tris propane buffer pH S.S. Polypeptides were eluted with
a linear 0-1.S
M NaCI gradient in the above buffer system at a flow rate of 10 ml/min. The
column eluent
was monitored at a wavelength of 214 nm.
The fractions eluting from the ion exchange column were pooled and
subjected to reverse phase chromatography using a Poros R2 column 4.6 x 100 mm
(Perseptive Biosystems). Polypeptides were eluted from the column with a
linear gradient
from 0-100% acetonitrile (0.1 % TFA) at a flow rate of S ml/min. The eluent
was monitored
at 214 nm.
Fractions containing the eluted polypeptides were lyophilized and resuspended
in 80 ~1 of aqueous 0.1% TFA and further subjected to reverse phase
chromatography on a
Vydac C4 column 4.6 x 1 SO mm (Western Analytical, Temecula, CA) with a linear
gradient
of 0-100% acetonitrile (0.1% TFA) at a flow rate of 2 ml/min. Fluent was
monitored at 214
nm.
CA 02268036 1999-04-12
WO 98116645 PCT/US97118214
33
The fraction with biological activity was separated into one major peak plus
other smaller components. Western blot of this peak onto PVDF membrane
revealed three
- major bands of molecular weights 14 Kd, 20 Kd and 26 Kd. These polypeptides
were
determined to have the following N-terminal sequences, respectively:
' S (j) Xaa-Asp-Ser-Glu-Lys-Ser-Ala-Thr-Ile-Lys-Val-Thr-Asp-Ala-Ser;
(SEQ ID NO: 129)
(k) Ala-Gly-Asp-Thr-Xaa-Ile-Tyr-Ile-Val-Gly-Asn-Leu-Thr-Ala-Asp;
(SEQ ID NO: 130) and
{l) Ala-Pro-Glu-Ser-Gly-Ala-Gly-Leu-Gly-Gly-Thr-Val-Gln-Ala-Gly;
(SEQ ID NO: 131 ), wherein Xaa may be any amino acid.
Using the assays described above, these polypeptides were shown to induce
proliferation and
IFN-y production in PBMC preparations. Figs. 1 A and B show the results of
such assays
using PBMC preparations from a first and a second donor, respectively.
DNA sequences that encode the antigens designated as (a), (c), (d) and (g)
above were obtained by screening a M. tuberculosis genomic library using 32P
end labeled
degenerate oligonucleotides corresponding to the N-terminal sequence and
containing
M. tuberculosis codon bias. The screen performed using a probe ~rresponding to
antigen (a)
above identified a clone having the sequence provided in SEQ ID NO: 96. The
polypeptide
encoded by SEQ ID NO: 96 is provided in SEQ ID NO: 97. The screen performed
using a
probe corresponding to antigen (g) above identified a clone having the
sequence provided in
SEQ ID NO: 52. The polypeptide encoded by SEQ ID NO: 52 is provided in SEQ ID
NO: 53. The screen performed using a probe corresponding to antigen {d) above
identified a
clone having the sequence provided in SEQ ID NO: 24, and the screen performed
with a
probe corresponding to antigen (c) identified a clone having the sequence
provided in SEQ ID
NO: 25.
The above amino acid sequences were compared to known amino acid
sequences in the gene bank using the DNA STAR system. The database searched
contains
some 173,000 proteins and is a combination of the Swiss, PIR databases along
with translated
protein sequences (Version 87). No significant homologies to the amino acid
sequences for
antigens (a)-(h) and (1) were detected.
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
34
The amino acid sequence for antigen (i) was found to be homologous to a
sequence from M. leprae. The full length M. leprae sequence was amplified from
genomic
DNA using the sequence obtained from GENBANK. This sequence was then used to
screen '
an M. tuberculosis library and a full length copy of the M. tuberculosis
homologue was
obtained (SEQ ID NO: 94).
The amino acid sequence for antigen (j) was found to be homologous to a
known M. tuberculosis protein translated from a DNA sequence. To the best of
the
inventors' knowledge, this protein has not been previously shown to possess T-
cell
stimulatory activity. The amino acid sequence for antigen (k) was found to be
related to a
sequence from M. leprae.
In the proliferation and IFN-y assays described above, using three PPD
positive donors, the results for representative antigens provided above are
presented in Table
1:
TABLE 1
RESULTS OF PBMC PROLIFERATION AND IFN-y ASSAYS
Sequence Proliferation IFN-y
(a) +
(c) +++ +++
(d) ++ ++
(g) +++ +++
(h) +++ +++
In Table 1, responses that gave a stimulation index (SI) of between 2 and 4
(compared to cells cultured in medium alone) were scored as +, as SI of 4-8 or
2-4 at a
concentration of 1 ~g or less was scored as ++ and an SI of greater than 8 was
scored as +++.
The antigen of sequence (i) was found to have a high SI (+++) for one donor
and lower SI
(++ ~d +) for the two other donors in both proliferation and IFN-y assays.
These results
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
indicate that these antigens are capable of inducing proliferation and/or
interferon-y
production.
EXAMPLE 2
S USE OF PATIENT SERA TO ISOLATE M. TUBERCULOSIS ANTIGENS
This example illustrates the isolation of antigens from M. tuberculosis lysate
by screening with serum from M. tuberculosis-infected individuals.
Dessicated M. tuberculosis H37Ra (Difco Laboratories) was added to a 2%
10 NP40 solution, and alternately homogenized and sonicated three times. The
resulting
suspension was centrifuged at 13,000 rpm in microfuge tubes and the
supernatant put through
a 0.2 micron syringe filter. The filtrate was bound to Macro Prep DEAE beads
(BioRad,
Hercules, CA). The beads were extensively washed with 20 mM Tris pH 7.5 and
bound
proteins eluted with 1 M NaCI. The NaCI elute was dialyzed overnight against
10 mM Tris,
15 pH 7.5. Dialyzed solution was treated with DNase and RNase at 0.05 mg/ml
for 30 min. at
room temperature and then with a-D=mannosidase, 0.5 U/mg at pH 4.5 for 3-4
hours at room
temperature. After returning to pH 7.5, the material was fractionated via FPLC
over a Bio
Scale-Q-20 column (BioRad). Fractions were combined into nine pools,
concentrated in a
Centriprep 10 (Amicon, Beverley, MA) and screened by Western blot for
serological activity
20 using a serum pool from M. tuberculosis-infected patients which was not
immunoreactive
with other antigens of the present invention.
The most reactive fraction was run in SDS-PAGE and transferred to PVDF. A
band at approximately 85 Kd was cut out yielding the sequence:
(m) Xaa-Tyr-Ile-Ala-Tyr-Xaa-Thr-Thr-Ala-Gly-Ile-Val-Pro-Gly-Lys-Ile-
25 Asn-Val-His-Leu-Val; (SEQ ID NO: 132), wherein Xaa may be any
amino acid.
Comparison of this sequence with those in the gene bank as described above,
revealed no significant homologies to known sequences.
A DNA sequence that encodes the antigen designated as (m) above was
30 obtained by screening a genomic M. tuberculosis Erdman strain library using
labeled
CA 02268036 1999-04-12
WO 98J16645 PGTIITS97/18214
36
degenerate oligonucleotides corresponding to the N-terminal sequence of SEQ ID
N0:137. A
clone was identified having the DNA sequence provided in SEQ ID NO: 198. This
sequence
was found to encode the amino acid sequence provided in SEQ ID NO: 199.
Comparison of
these sequences with those in the genebank revealed some similarity to
sequences previously
identified in M. tuberculosis and M. bovis.
EXAMPLE 3
PREPARATION OF DNA SEQUENCES ENCODING M TUBERCULOSIS ANTIGENS
This example illustrates the preparation of DNA sequences encoding
M. tuberculosis antigens by screening a M. tuberculosis expression library
with sera obtained
from patients infected with M. tuberculosis, or with anti-sera raised against
M. tuberculosis
antigens.
1 S A. PREPARATION OF M. TUBERCULOSIS SOLUBLE ANTIGENS USING RABBIT ANTI-SERA
RAISED AGAINST M. TUBERCULOSIS SUPERNATANT
Genomic DNA was isolated from the M. tuberculosis strain H37Ra. The DNA
was randomly sheared and used to construct an expression library using the
Lambda ZAP
expression system (Stratagene, La Jolla, CA). Rabbit anti-sera was generated
against
secretory proteins of the M. tuberculosis strains H37Ra, H37Rv and Erdman by
immunizing a
rabbit with concentrated supernatant of the M. tuberculosis cultures.
Specifically, the rabbit
was first immunized subcutaneously with 200 p,g of protein antigen in a total
volume of 2 ml
containing 100 p,g muramyl dipeptide (Calbiochem, La Jolla, CA) and 1 ml of
incomplete
Freund's adjuvant. Four weeks later the rabbit was boosted subcutaneously with
100 ~,g
antigen in incomplete Freund's adjuvant. Finally, the rabbit was immunized
intravenously ,
four weeks later with 50 ~.g protein antigen. The anti-sera were used to
screen the expression
library as described in Sambrook et al., Molecular Cloning: A Laboratory
Manual, Cold
Spring Harbor Laboratories, Cold Spring Harbor, NY, 1989. Bacteriophage
plaques
expressing immunoreactive antigens were purified. Phagemid from the plaques
was rescued
and the nucleotide sequences of the M. tuberculosis clones deduced.
CA 02268036 1999-04-12
WO 98116645 PCT/US97/18214
37
Thirty two clones were purified. Of these, 25 represent sequences that have
not been previously identified in M. tuberculosis. Proteins were induced by
IPTG and
' purified by gel elution, as described in Skeiky et al., J. Exp. Med.
181:1527-1537, 1995.
Representative partial sequences of DNA molecules identified in this screen
are provided in
' 5 SEQ ID NOS: 1-25. The corresponding predicted amino acid sequences are
shown in SEQ
ID NOS: 64-88.
On comparison of these sequences with known sequences in the gene bank
using the databases described above, it was found that the clones referred to
hereinafter as
TbRA2A, TbRAl6, TbRAl8, and TbRA29 (SEQ ID NOS: 77, 69, 71, 76) show some
homology to sequences previously identified in Mycobacterium leprae but not in
M. tuberculosis. TbRAI l, TbRA26, TbRA28 and TbDPEP (SEQ ID NOS: 66, 74, 75,
53)
have been previously identified in M. tuberculosis. No significant homologies
were found to
TbRA 1, TbRA3, TbRA4, TbRA9, TbRA 10, TbRA 13, TbRA 17, TbRA 19, TbRA29,
TbRA32, TbRA36 and the overlapping clones TbRA35 and TbRAl2 (SEQ ID NOS: 64,
78,
82, 83, 65, 68, 76, 72, 76, 79, 81, 80, 67, respectively). The clone TbRa24 is
overlapping
with clone TbRa29.
B. USE OF SERA FROM PATIENTS HAVING PULMONARY OR PLEURAL TUBERCULOSIS TO
IDENTIFY DNA SEQUENCES ENCODING M. TUBERCULOSIS ANTIGENS
The genomic DNA library described above, and an additional H37Rv library,
were screened using pools of sera obtained from patients with active
tuberculosis. To prepare
the H37Rv library, M. tuberculosis strain H37Rv genomic DNA was isolated,
subjected to
partial Sau3A digestion and used to construct an expression library using the
Lambda Zap
expression system (Stratagene, La Jolla, Ca). Three different pools of sera,
each containing
sera obtained from three individuals with active pulmonary or pleural disease,
were used in
the expression screening. The pools were designated TbL, TbM and TbH,
referring to
relative reactivity with H37Ra lysate (i.e., TbL = low reactivity, TbM =
medium reactivity
Y. and TbH = high reactivity) in both ELISA and immunoblot format. A fourth
pool of sera
from seven patients with active pulmonary tuberculosis was also employed. All
of the sera
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/182I4
38
lacked increased reactivity with the recombinant 38 kD M. tuberculosis H37Ra
phosphate-
binding protein.
All pools were pre-adsorbed with E. coli lysate and used to screen the H37Ra
and H37Rv expression libraries, as described in Sambrook et al., Molecular
Cloning: A
Laboratory Manual, Cold Spring Harbor Laboratories, Cold Spring Harbor, NY,
1989.
Bacteriophage plaques expressing immunoreactive antigens were purified.
Phagemid from
the plaques was rescued and the nucleotide sequences of the M. tuberculosis
clones deduced.
Thirty two clones were purified. Of these, 31 represented sequences that had
not been previously identified in human M. tuberculosis. Representative
sequences of the
DNA molecules identified are provided in SEQ ID NOS:: 26-51 and 100. Of these,
TbH-8-2
(SEQ. ID NO. 100) is a partial clone of TbH-8, and TbH-4 (SEQ. ID NO. 43) and
TbH-4-
FWD (SEQ. ID NO. 44) are non-contiguous sequences from the same clone. Amino
acid
sequences for the antigens hereinafter identified as Tb38-1, TbH-4, TbH-8, TbH-
9, and
TbH-12 are shown in SEQ ID NOS.: 89-93. Comparison of these sequences with
known
sequences in the gene bank using the databases identified above revealed no
significant
homologies to TbH-4, TbH-8, TbH-9 and TbM-3, although weak homologies were
found to
TbH-9. TbH-12 was found to be homologous to a 34 kD antigenic protein
previously
identified in M. paratuberculosis (Acc. No. 528515). Tb38-1 was found to be
located 34
base pairs upstream of the open reading frame for the antigen ESAT-6
previously identified
in M. bovis (Acc. No. U34848) and in M. tuberculosis (Sorensen et al., Infec.
Immun.
63:1710-1717, 1995).
Probes derived from Tb38-1 and TbH-9, both isolated from an H37Ra library,
were used to identify clones in an H37Rv library. Tb38-1 hybridized to Tb38-
1F2, Tb38-
1F3, Tb38-1F5 and Tb38-1F6 (SEQ. ID NOS: 107, 108, 111, 113, and 114). (SEQ ID
NOS:
107 and 108 are non-contiguous sequences from clone Tb38-IF2.) Two open
reading frames
were deduced in Tb38-IF2; one corresponds to Tb37FL (SEQ. ID. NO. 109), the
second, a
partial sequence, may be the homologue of Tb38-1 and is called Tb38-IN (SEQ.
ID NO. 110).
The deduced amino acid sequence of Tb38-1F3 is presented in SEQ. ID. NO. 112.
A TbH-9
probe identified three clones in the H37Rv library: TbH-9-FL (SEQ. ID NO. 1 O1
), which
may be the homologue of TbH-9 (R37Ra), TbH-9-1 (SEQ. ID NO. 103), and TbH-8-2
(SEQ.
CA 02268036 1999-04-12
wo ms~as rcr~s9~nszi4
39
ID NO. 105) is a partial clone of TbH-8. The deduced amino acid sequences for
these three
clones are presented in SEQ ID NOS: 102, 104 and 106.
Further screening of the M. tuberculosis genomic DNA library, as described
above, resulted in the recovery of ten additional reactive clones,
representing seven different
genes. One of these genes was identified as the 38 Kd antigen discussed above,
one was
determined to be identical to the l4Kd alpha crystallin heat shock protein
previously shown
to be present in M. tuberculosis, and a third was determined to be identical
to the antigen
TbH-8 described above. The determined DNA sequences for the remaining five
clones
(hereinafter referred to as TbH-29, TbH-30, TbH-32 and TbH-33) are provided in
SEQ ID
NO: 133-136, respectively, with the corresponding predicted amino acid
sequences being
provided in SEQ ID NO: 137-140, respectively. The DNA and amino acid sequences
for
these antigens were compared with those in the gene bank as described above.
No
homologies were found to the 5' end of TbH-29 (which contains the reactive
open reading
frame), although the 3' end of TbH-29 was found to be identical to the M.
tuberculosis
cosmid Y227. TbH-32 and TbH-33 were found to be identical to the previously
identified
M. tuberculosis insertion element IS6110 and to the M. tuberculosis cosmid
Y50,
respectively. No significant homologies to TbH-30 were found.
Positive phagemid from this additional screening were used to infect E. coli
XL-1 Blue MRF', as described in Sambrook et al., supra. Induction of
recombinant protein
was accomplished by the addition of IPTG. Induced and uninduced lysates were
run in
duplicate on SDS-PAGE and transferred to nitrocellulose filters. Filters were
reacted with
human M. tuberculosis sera (1:200 dilution) reactive with TbH and a rabbit
sera (1:200 or
1:250 dilution) reactive with the N-terminal 4 Kd portion of lacZ. Sera
incubations were
performed for 2 hours at room temperature. Bound antibody was detected by
addition of'ZSI-
labeled Protein A and subsequent exposure to film for variable times ranging
from 16 hours
to 11 days. The results of the immunoblots are summarized in Table 2.
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
TABLE 2
Human M. tb Anti-lacZ
Antisen Sera Sera
5
TbH-29 45 Kd 45 Kd
TbH-30 No reactivity 29 Kd
TbH-32 12 Kd 12 Kd
TbH-33 16 Kd 16 Kd
Positive reaction of the recombinant human M. tuberculosis antigens with both
the human M. tuberculosis sera and anti-lacZ sera indicate that reactivity of
the human M.
tuberculosis sera is directed towards the fusion protein. Antigens reactive
with the anti.-lacZ
sera but not with the human M. tuberculosis sera may be the result of the
human M.
tuberculosis sera recognizing conformational epitopes, or the antigen-antibody
binding
kinetics may be such that the 2 hour sera exposure in the immunoblot is not
sufficient.
Studies were undertaken to determine whether the antigens TbH-9 and Tb38-1
represent cellular proteins or are secreted into M. tuberculosis culture
media. In the first
study, rabbit sera were raised against A) secretory proteins of M.
tuberculosis, B) the known
secretory recombinant M. tuberculosis antigen 85b, C) recombinant Tb38-l and
D)
recombinant TbH-9, using protocols substantially as described in Example 3A.
Total M.
tuberculosis lysate, concentrated supernatant of M. tuberculosis cultures and
the recombinant
antigens 85b, TbH-9 and Tb38-1 were resolved on denaturing gels, immobilized
on
nitrocellulose membranes and duplicate blots were probed using the rabbit sera
described
above.
The results of this analysis using control sera (panel I) and antisera (panel
II) '
against secretory proteins, recombinant 85b, recombinant Tb38-1 and
recombinant TbH-9 are
shown in Figures 2A-D, respectively, wherein the lane designations are as
follows: 1 )
molecular weight protein standards; 2) 5 p,g of M. tuberculosis lysate; 3) 5
p,g secretory
proteins; 4) SO ng recombinant Tb38-1; 5) 50 ng recombinant TbH-9; and 6) 50
ng
recombinant 85b. The recombinant antigens were engineered with six terminal
histidine
CA 02268036 1999-04-12
WO 98/16645 PGT/US97/18214
41
residues and would therefore be expected to migrate with a mobility
approximately 1 kD
larger that the native protein. In Figure 2D, recombinant TbH-9 is lacking
approximately 10
kD of the full-length 42 kD antigen, hence the significant difference in the
size of the
immunoreactive native TbH-9 antigen in the lysate lane (indicated by an
arrow). These
results demonstrate that Tb38-1 and TbH-9 are intracellular antigens and are
not actively
secreted by M. tuberculosis.
The finding that TbH-9 is an intracellular antigen was confirmed by
determining the reactivity of TbH-9-specific human T cell clones to
recombinant TbH-9,
secretory M. tuberculosis proteins and PPD. A TbH-9-specific T cell clone
(designated
131 TbH-9) was generated from PBMC of a healthy PPD-positive donor. The
proliferadve
response of 131 TbH-9 to secretory proteins, recombinant TbH-9 and a control
M.
tuberculosis antigen, TbRa11, was determined by measuring uptake of tritiated
thymidine, as
described in Example 1. As shown in Figure 3A, the clone I3ITbH-9 responds
specifically
to TbH-9, showing that TbH-9 is not a significant component of M. tuberculosis
secretory
proteins. Figure 3B shows the production of IFN-y by a second TbH-9-specific T
cell clone
(designated PPD 800-10) prepared from PBMC from a healthy PPD-positive donor,
following stimulation of the T cell clone with secretory proteins, PPD or
recombinant TbH-9.
These results further confirm that TbH-9 is not secreted by M. tuberculosis.
C. USE OF SERA FROM PATIENTS HAVING EXTRAPULMONARY TUBERCULOSIS TO IDENTIFY
DNA SEQUENCES ENCODING M. TUBERCULOSIS ANTIGENS
Genomic DNA was isolated from M. tuberculosis Erdman strain, randomly
sheared and used to construct an expression library employing the Lambda ZAP
expression
system (Stratagene, La Jolla, CA). The resulting library was screened using
pools of sera
' obtained from individuals with extrapulmonary tuberculosis, as described
above in Example
3B, with the secondary antibody being goat anti-human IgG + A + M (H+L)
conjugated with
alkaline phosphatase.
- Eighteen clones were purified. Of these, 4 clones (hereinafter referred to
as
XP14, XP24, XP31 and XP32) were found to bear some similarity to known
sequences. The
determined DNA sequences for XP14, XP24 and XP31 are provided in SEQ ID NOS:
151-
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
42
153, respectively, with the S' and 3' DNA sequences for XP32 being provided in
SEQ ID
NOS: 154 and 155, respectively. The predicted amino acid sequence for XP14 is
provided in
SEQ ID NO: 156. The reverse complement of XP14 was found to encode the amino
acid '
sequence provided in SEQ ID NO: 157.
Comparison of the sequences for the remaining 14 clones (hereinafter referred -
to as XP 1-XP6, XP 17-XP 19, XP22, XP25, XP27, XP30 and XP36) with those in
the
genebank as described above, revealed no homologies with the exception of the
3' ends of
XP2 and XP6 which were found to bear some homology to known M. tuberculosis
cosmids.
The DNA sequences for XP27 and XP36 are shown in SEQ ID NOS: 158 and 159,
respectively, with the 5' sequences for XP4, XPS, XP17 and XP30 being shown in
SEQ ID
NOS: 160-163, respectively, and the S' and 3' sequences for XP2, XP3, XP6,
XP18, XP19,
XP22 and XP25 being shown in SEQ ID NOS: 164 and 165; 166 and 167; 168 and
169; 170
and 171; 172 and 173; 174 and 175; and 176 and 177, respectively. XP 1 was
found to
overlap with the DNA sequences for TbH4, disclosed above. The full-length DNA
sequence
for TbH4-XPI is provided in SEQ ID NO: 178. This DNA sequence was found to
contain an
open reading frame encoding the amino acid sequence shown in SEQ ID NO: 179.
The
reverse complement of TbH4-XP 1 was found to contain an open reading frame
encoding the
amino acid sequence shown in SEQ ID NO: 180. The DNA sequence for XP36 was
found to
contain two open reading frames encoding the amino acid sequence shown in SEQ
ID NOS:
181 and 182, with the reverse complement containing an open reading frame
encoding the
amino acid sequence shown in SEQ ID NO: 183.
Recombinant XP1 protein was prepared as described above in Example 3B,
with a metal ion affinity chromatography column being employed for
purification.
Recombinant XP 1 was found to stimulate cell proliferation and IFN-y
production in T cells
isolated from an M. tuberculosis-immune donors.
D. PREPARATION OF M. TUBERCULOSIS SOLUBLE ANTIGENS USING RABBIT ANTI-SERA
RAISED AGAINST M. TUBERCULOSIS FRACTIONATED PROTEINS
M. tuberculosis lysate was prepared as described above in Example 2. The
resulting material was fractionated by HPLC and the fractions screened by
Western blot for -
CA 02268036 1999-04-12
WO 98/16645 PCT/tTS97t18214
43
serological activity with a serum pool from M. tuberculosis-infected patients
which showed
little or no immunoreactivity with other antigens of the present invention.
Rabbit anti-sera
- was generated against the most reactive fraction using the method described
in Example 3A .
The anti-sera was used to screen an M. tuberculosis Erdman strain genomic DNA
expression
' S library prepared as described above. Bacteriophage plaques expressing
immunoreactive
antigens were purified. Phagemid from the plaques was rescued and the
nucleotide sequences
of the M. tuberculosis clones determined.
Ten different clones were purified. Of these, one was found to be TbRa35,
described above, and one was found to be the previously identified M.
tuberculosis antigen,
HSP60. Of the remaining eight clones, six (hereinafter referred to as RDIF2,
RDIFS, RDIFB,
RDIF 10, RDIF 11 and RDIF 12) were found to bear some similarity to previously
identified
M. tuberculosis sequences. The determined DNA sequences for RDIF2, RDIFS,
RDIFB,
RDIF10 and RDIF11 are provided in SEQ ID NOS: 184-188, respectively, with the
corresponding predicted amino acid sequences being provided in SEQ ID NOS: 189-
193,
respectively. The 5' and 3' DNA sequences for RDIF12 are provided in SEQ ID
NOS: 194
and 195, respectively. No significant homologies were found to the antigen
RDIF-7. The
determined DNA and predicted amino acid sequences for RDIF7 are provided in
SEQ ID
NOS: 196 and 197, respectively. One additional clone, referred to as RDIF6 was
isolated,
however, this was found to be identical to RDIFS.
Recombinant RDIF6, RDIFB, RDIF 10 and RDIF 11 were prepared as
described above. These antigens were found to stimulate cell proliferation and
IFN-y
production in T cells isolated from M. tuberculosis-immune donors.
EXAMPLE 4
PURIFICATION AIVD CHARACTERIZATION OF A POLYPEPTIDE FROM TUBERCULIN PURIFIED
PROTEIN DERIVATIVE
An M. tuberculosis polypeptide was isolated from tuberculin purified protein
derivative (PPD) as follows.
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
44
PPD was prepared as published with some modification (Seibert, F. et al.,
Tuberculin purified protein derivative. Preparation and analyses of a large
quantity for
standard. The American Review of Tuberculosis 44:9-25, 1941). M. tuberculosis
Rv strain
was grown for 6 weeks in synthetic medium in roller bottles at 37°C.
Bottles containing the
bacterial growth were then heated to 100°C in water vapor for 3 hours.
Cultures were sterile '
filtered using a 0.22 p, filter and the liquid phase was concentrated 20 times
using a 3 kD cut-
off membrane. Proteins were precipitated once with 50% ammonium sulfate
solution and
eight times with 25% ammonium sulfate solution. The resulting proteins (PPD)
were
fractionated by reverse phase liquid chromatography (RP-HPLC) using a C 18
column (7.8 x
300 mM; Waters, Milford, MA) in a Biocad HPLC system (Perceptive Biosystems,
Framingham, MA). Fractions were eluted from the column with a linear gradient
from 0-
100% buffer (0.1 % TFA in acetonitrile). The flow rate was 10 ml/minute and
eluent was
monitored at 214 nm and 280 nm.
Six fractions were collected, dried, suspended in PBS and tested individually
in M. tuberculosis-infected guinea pigs for induction of delayed type
hypersensitivity (DTH)
reaction. One fraction was found to induce a strong DTH reaction and was
subsequently
fractionated further by RP-HPLC on a microbore Vydac C18 column (Cat. No.
218TP5115)
in a Perkin Elmer/Applied Biosystems Division Model 172 HPLC. Fractions were
eluted
with a linear gradient from S-100% buffer (0.05% TFA in acetonitrile) with a
flow rate of 80
p.l/minute. Eluent was monitored at 215 nm. Eight fractions were collected and
tested for
induction of DTH in M. tuberculosis-infected guinea pigs. One fraction was
found to induce
strong DTH of about 16 mm induration. The other fractions did not induce
detectable DTH.
The positive fraction was submitted to SDS-PAGE gel electrophoresis and found
to contain a
single protein band of approximately 12 kD molecular weight.
This polypeptide, herein after referred to as DPPD, was sequenced from the
amino terminal using a Perkin Elmer/Applied Biosystems Division Procise 492
protein
sequencer as described above and found to have the N-terminal sequence shown
in SEQ ID
NO:: 124. Comparison of this sequence with known sequences in the gene bank as
described
above revealed no known homologies. Four cyanogen bromide fragments of DPPD
were
isolated and found to have the sequences shown in SEQ ID NOS: 125-128.
CA 02268036 1999-04-12
wo mr~sas rc~r~smnma
EXAMPLE 5
SYNTHESIS OF SYNTHETIC POLYPEPTIDES
' S Polypeptides may be synthesized on a Millipore 9050 peptide synthesizer
using FMOC chemistry with HPTU (O-Benzotriazole-N,N,N',N'-tetramethyluronium
hexafluorophosphate) activation. A Gly-Cys-Gly sequence may be attached to the
amino
terminus of the peptide to provide a method of conjugation or labeling of the
peptide.
Cleavage of the peptides from the solid support may be carried out using the
following
10 cleavage mixture: trifluoroacetic
acid:ethanedithiolahioanisole:water:phenol (40:1:2:2:3).
After cleaving for 2 hours, the peptides may be precipitated in cold methyl-t-
butyl-ether. The
peptide pellets may then be dissolved in water containing 0.1 %
trifluoroacetic acid (TFA) and
lyophilized prior to purification by C18 reverse phase HPLC. A gradient of 0-
60%
acetonitrile (containing 0.1 % TFA) in water (containing 0.1 % TFA) may be
used to elute the
15 peptides. Following lyophilization of the pure fractions, the peptides may
be characterized
using electrospray mass spectrometry and by amino acid analysis.
This procedure was used to synthesize a TbM-1 peptide that contains one and
a half repeats of a TbM-1 sequence. The TbM-1 peptide has the sequence
GCGDRSGGNLDQIRLRRDRSGGNL (SEQ ID NO: 63).
EXAMPLE 6
USE OF REPRESENTATIVE ANTIGENS FOR SERODIAGNOSIS OF TUBERCULOSIS
This Example illustrates the diagnostic properties of several representative
antigens.
Assays were performed in 96-well plates were coated with 200 ng antigen
4
diluted to 50 p,L in carbonate coating buffer, pH 9.6. The wells were coated
overnight at 4°C
(or 2 hours at 37°C). The plate contents were then removed and the
wells were blocked for 2
hours with 200 p,L of PBS/1% BSA. After the blocking step, the wells were
washed five
CA 02268036 1999-04-12
WO 98/16645 PGT/US9~/18214
46
times with PBS/0.1 % Tween 20T"". 50 p,L sera, diluted 1:100 in PBS/0. I %
Tween 20T""l0.1
BSA, was then added to each well and incubated for 30 minutes at room
temperature. The
plates were then washed again five times with PBS/0.1% Tween 20T"".
The enzyme conjugate (horseradish peroxidase - Protein A, Zymed, San
Francisco, CA) was then diluted 1:10,000 in PBS/0.1% Tween 20T""/0.1% BSA, and
50 p,L of
the diluted conjugate was added to each well and incubated for 30 minutes at
room
temperature. Following incubation, the wells were washed five times with
PBS/0.1 % Tween
20T"". 100 p,L of tetramethylbenzidine peroxidase (TMB) substrate (Kirkegaard
and Perry
Laboratories, Gaithersburg, MD) was added, undiluted, and incubated for about
15 minutes.
The reaction was stopped with the addition of 100 p.L of 1 N HzS04 to each
well, and the
plates were read at 450 nm.
Figure 4 shows the ELISA reactivity of two recombinant antigens isolated
using method A in Example 3 (TbRa3 and TbRa9) with sera from M. tuberculosis
positive
and negative patients. The reactivity of these antigens is compared to that of
bacterial lysate
isolated from M. tuberculosis strain H37Ra (Difco, Detroit, MI). In both
cases, the
recombinant antigens differentiated positive from negative sera. Based on cut-
off values
obtained from receiver-operator curves, TbRa3 detected 56 out of 87 positive
sera, and
TbRa9 detected 111 out of 165 positive sera.
Figure 5 illustrates the ELISA reactivity of representative antigens isolated
using method B of Example 3. The reactivity of the recombinant antigens TbH4,
TbH 12,
Tb38-1 and the peptide TbM-1 (as described in Example 4) is compared to that
of the 38 kD
antigen described by Andersen and Hansen, Infect. Immun. 57:2481-2488, 1989.
Again, all
of the polypeptides tested differentiated positive from negative sera. Based
on cut-off values
obtained from receiver-operator curves, TbH4 detected 67 out of 126 positive
sera, TbH 12
detected 50 out of 125 positive sera, 38-1 detected 61 out of 101 positive
sera and the TbM-1
peptide detected 25 out of 30 positive sera.
The reactivity of four antigens (TbRa3, TbRa9, TbH4 and TbHl2) with sera
from a group of M. tuberculosis infected patients with differing reactivity in
the acid fast stain
of sputum (Smithwick and David, Tubercle 52:226, 1971 ) was also examined, and
compared
CA 02268036 1999-04-12
WO 98116645 PCT/I1S97118214
47
to the reactivity of M. tuberculosis lysate and the 38 kD antigen. The results
are presented in
Table 3, below:
TABLE 3
REACTIVITY OF ANTIGENS WITH SERA FROM M. TUBERCULOSIS PATIENTS
Acid ELISA
Fast Values
Patient Sputum Lysate
381cD
TbRa9
TbHl2
TbH4
TbRa3
Tb01 B93I-2 ++++ 1.853 0.634 0.998 1.022 1.030 1.314
TbO1B93I-19 ++++ 2.657 2.322 0.608 0.837 1.857 2.335
TbO1B93I-8 +++ 2.703 0.527 0.492 0.281 0.501 2.002
TbO1B93I-10 +++ 1.665 1.301 0.685 0.216 0.448 0.458
TbO1B93I-11 +++ 2.817 0.697 0.509 0.301 0.173 2.608
TbO1B93I-15 +++ 1.28 0.283 0.808 0.218 1.537 0.811
TbO1B93I-16 +++ 2.908 >3 0.899 0.441 0.593 1.080
TbO1B93I-25 +++ 0.395 0.131 0.335 0.211 0.107 0.948
TbO1B93I-87 +++ 2.653 2.432 2.282 0.977 1.221 0.857
TbO1B93I-89 +++ 1.912 2.370 2.436 0.876 0.520 0.952
TbO1B94I-108 +++ 1.639 0.341 0.797 0.368 0.654 0.798
Tb01 B94I-201+++ 1.721 0.419 0.661 0.137 0.064 0.692
TbO1B93I-88 ++ 1.939 1.269 2.519 1.381 0.214 0.530
TbO1B93I-92 ++ 2.355 2.329 2.78 0.685 0.997 2.527
TbO1B94I-109 ++ 0.993 0.620 0.574 0.441 0.5 2.558
TbO1B94I-210 ++ 2.777 >3 0.393 0.367 1.004 1.315
TbO1B94I-224 ++ 2.913 0.476 0.251 1.297 1.990 0.256
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
48
Acid ELISA
Fast Values
Patient Sputum Lysate
38kD
TbRa9
TbHl2
TbH4
TbRa3
TbO1B93I-9 + 2.649 0.278 0.210 0.140 0.181 1.586
TbO1B93I-14 + >3 1.538 0.282 0.291 0.549 2.880
Tb01 B93I-21 + 2.645 0.739 2.499 0.783 0.536 1.770
Tb01 B93I-22 + 0.714 0.451 2.082 0.285 0.269 1.159
TbO1B93I-31 + 0.956 0.490 1.019 0.812 0.176 1.293
TbO1B93I-32 2.261 0.786 0.668 0.273 0.535 0.405
TbOlB93I-52 - 0.658 0.114 0.434 0.330 0.273 1.140
TbO1B93I-99 - 2.118 0.584 1.62 0.119 0.977 0.729
TbO1B94I-130 1.349 0.224 0.86 0.282 0.383 2.146
Tb01 B94I-131 0.685 0.324 1.173 0.059 0.118 1.431
AT4-0070 Normal 0.072 0.043 0.092 0.071 0.040 0.039
AT4-0105 Normal 0.397 0.121 0.118 0.103 0.078 0.390
3/15/94-1 Normal 0.227 0.064 0.098 0.026 0.001 0.228
4/15/93-2 Normal 0.114 0.240 0.071 0.034 0.041 0.264
5/26/94-4 Normal 0.089 0.259 0.096 0.046 0.008 0.053
5/26/94-3 Normal 0.139 0.093 0.085 0.019 0.067 0.01
Based on cut-off values obtained from receiver-operator curves, TbRa3
detected 23 out of 27 positive sera, TbRa9 detected 22 out of 27, TbH4
detected 18 out of 27
and TbH 12 detected 15 out of 27. If used in combination, these four antigens
would have a
theoretical sensitivity of 27 out of 27, indicating that these antigens should
complement each
other in the serological detection of M. tuberculosis infection. In addition,
several of the
recombinant antigens detected positive sera that were not detected using the
38 kD antigen,
indicating that these antigens may be complementary to the 38 kD antigen. '
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
49
The reactivity of the recombinant antigen TbRall with sera from
M. tuberculosis patients shown to be negative for the 38 kD antigen, as well
as with sera from
- PPD positive and normal donors, was determined by ELISA as described above.
The results
are shown in Figure 6 which indicates that TbRal 1, while being negative with
sera from PPD
positive and normal donors, detected sera that were negative with the 38 kD
antigen. Of the
thirteen 38 kD negative sera tested, nine were positive with TbRal l,
indicating that this
antigen may be reacting with a sub-group of 38 kD antigen negative sera. In
contrast, in a
group of 38 kD positive sera where TbRal 1 was reactive, the mean OD 450 for
TbRal l was
lower than that for the 38 kD antigen. The data indicate an inverse
relationship between the
presence of TbRal 1 activity and 38 kD positivity.
The antigen TbRa2A was tested in an indirect ELISA using initially 50 ~,1 of
serum at 1:100 dilution for 30 minutes at room temperature followed by washing
in PBS
Tween and incubating for 30 minutes with biotinylated Protein A (Zymed, San
Francisco,
CA) at a 1:10,000 dilution. Following washing, 50 p,l of streptavidin-
horseradish peroxidase
(Zymed) at 1:10,000 dilution was added and the mixture incubated for 30
minutes. After
washing, the assay was developed with TMB substrate as described above. The
reactivity of
TbRa2A with sera from M. tuberculosis patients and normal donors in shown in
Table 4. The
mean value for reactivity of TbRa2A with sera from M. tuberculosis patients
was 0.444 with
a standard deviation of 0.309. The mean for reactivity with sera from normal
donors was
0.109 with a standard deviation of 0.029. Testing of 38 kD negative sera
(Figure 7) also
indicated that the TbRa2A antigen was capable of detecting sera in this
category.
TABLE 4
REACTIVITY OF TBRA2A WITH SERA FROM M. TUBERCULOSIS PATIENTS AND FROM NORMAL
DotJORs
Serum ID Status OD 450
Tb85 TB 0.680
Tb86 TB 0.450
Tb87 TB 0.263
Tb88 TB 0.275
- ~ Tb89 TB 0.403
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
Tb91 TB 0.393
Tb92 TB 0.401
Tb93 TB 0.232
Tb94 TB 0.333
Tb95 TB 0.435
Tb96 TB 0.284
Tb97 TB 0.320
Tb99 TB 0.328
Tb 100 TB 0.817
Tb 1 O TB 0.607
l
Tb 102 TB 0.191
Tb 103 TB 0.228
Tb107 TB 0.324
TbI09 TB 1.572
Tb112 TB 0.338
DL4-0176 Normal 0.036
AT4-0043 Normal 0.126
AT4-0044 Normal 0.130
AT4-0052 Normal 0.135
AT4-0053 Normal 0.133
AT4-0062 Normal 0.128
AT4-0070 Normal 0.088
AT4-0091 Normal 0.108
AT4-0100 Normal 0.106
AT4-0105 Normal 0.108
AT4-0109 Normal 0.105
The reactivity of the recombinant antigen (g) (SEQ ID NO: 60) with sera from
M. tuberculosis patients and normal donors was determined by ELISA as
described above.
Figure 8 shows the results of the titration of antigen (g) with four M.
tuberculosis positive
5 sera that were all reactive with the 38 kD antigen and with four donor sera.
All four positive
sera were reactive with antigen (g).
The reactivity of the recombinant antigen TbH-29 (SEQ ID NO: I37) with
sera from M. tuberculosis patients, PPD positive donors and normal donors was
determined
by indirect ELISA as described above. The results are shown in Figure 9. TbH-
29 detected
10 30 out of 60 M. tuberculosis sera, 2 out of 8 PPD positive sera and 2 out
of 27 normal sera.
Figure 10 shows the results of ELISA tests (both direct and indirect) of the
antigen TbH-33 (SEQ ID NO: 140) with sera from M. tuberculosis patients and
from normal
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
51
donors and with a pool of sera from M. tuberculosis patients. The mean OD 450
was
demonstrated to be higher with sera from M. tuberculosis patients than from
normal donors,
with the mean OD 450 being significantly higher in the indirect ELISA than in
the direct
ELISA. Figure 11 is a titration curve for the reactivity of recombinant TbH-33
with sera
S from M. tuberculosis patients and from normal donors showing an increase in
OD 450 with
increasing concentration of antigen.
The reactivity of the recombinant antigens RDIF6, RDIF8 and RDIF10 (SEQ
ID NOS: 184-187, respectively) with sera from M. tuberculosis patients and
normal donors
was determined by ELISA as described above. RDIF6 detected & out of 32 M.
tuberculosis
sera and 0 out of 15 normal sera; RDIF8 detected 14 out of 32 M. tuberculosis
sera and 0 out
of 15 normal sera; and RDIF 10 detected 4 out of 27 M. tuberculosis sera and 1
out of 15
normal sera. In addition, RDIF10 was found to detect 0 out of 5 sera from PPD-
positive
donors.
EXAMPLE 7
PREPARATION AND CHARACTERIZATION OF M. TUBERCULOSIS FUSION PROTEINS
A fusion protein containing TbRa3, the 38 kD antigen and Tb38-1 was
prepared as follows.
Each of the DNA constructs TbRa3, 38 kD and Tb38-1 were modified by PCR
in order to facilitate their fusion and the subsequent expression of the
fusion protein TbRa3-
38 kD-Tb38-1. TbRa3, 38 kD and Tb38-1 DNA was used to perform PCR using the
primers
PDM-64 and PDM-65 (SEQ ID NO: 141 and 142), PDM-57 and PDM-58 (SEQ ID NO: 143
and 144), and PDM-69 and PDM-60 (SEQ ID NO: 145-146), respectively. In each
case, the
DNA amplification was performed using 10 p,l 1 OX Pfu buffer, 2 ~l 10 mM
dNTPs, 2 ~l each
of the PCR primers at 10 ~.M concentration, 81.5 ~,1 water, 1.5 ~.l Pfu DNA
polymerase
(Stratagene, La Jolla, CA) and 1 ~,l DNA at either 70 ng/~tl {for TbRa3) or 50
ng/~,l (for 38
kD and Tb38-1). For TbRa3, denaturation at 94°C was performed for 2
min, followed by 40
cycles of 96°C for 15 sec and 72°C for 1 min, and lastly by
72°C for 4 min. For 38 kD,
denaturation at 96°C was performed for 2 min, followed by 40 cycles of
96°C for 30 sec,
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
52
68°C for 15 sec and 72°C for 3 min, and finally by 72°C
for 4 min. For Tb38-1 denaturation
at 94°C for 2 min was followed by 10 cycles of 96°C for 15 sec,
68°C for 15 sec and 72°C for
1.5 min, 30 cycles of 96°C for 15 sec, 64°C for 15 sec and
72°C for 1.5, and finally by 72°C '
for 4 min.
The TbRa3 PCR fragment was digested with NdeI and EcoRI and cloned
directly into pT7~L2 IL 1 vector using NdeI and EcoRI sites. The 38 kD PCR
fragment was
digested with Sse8387I, treated with T4 DNA polymerase to make blunt ends and
then
digested with EcoRI for direct cloning into the pT7~L2Ra3-I vector which was
digested with
StuI and EcoRI. The 38-1 PCR fragment was digested with Eco47III and EcoRI and
directly
subcloned into pT7~L2Ra3/38kD-17 digested with the same enzymes. The whole
fusion was
then transferred to pET28b using NdeI and EcoRI sites. The fusion construct
was confirmed
by DNA sequencing.
The expression construct was transformed to BLR pLys S E. coli (Novagen,
Madison, WI) and grown overnight in LB broth with kanamycin (30 ~g/ml) and
chloramphenicol (34 ~,g/ml). This culture (12 ml) was used to inoculate 500 ml
2XYT with
the same antibiotics and the culture was induced with IPTG at an OD560 of 0.44
to a final
concentration of 1.2 mM. Four hours post-induction, the bacteria were
harvested and
sonicated in 20 mM Tris (8.0), 100 mM NaCI, 0.1 % DOC, 20 p,g/ml Leupeptin, 20
mM
PMSF followed by centrifugation at 26,000 X g. The resulting pellet was
resuspended in 8 M
urea, 20 mM Tris (8.0), 100 mM NaCI and bound to Pro-bond nickel resin
(Invitrogen,
Carlsbad, CA). The column was washed several times with the above buffer then
eluted with
an imidazole gradient (50 mM, 100 mM, 500 mM imidazole was added to 8 M urea,
20 mM
Tris (8.0), 100 mM NaCI). The eluates containing the protein of interest were
then dialzyed
against 10 mM Tris (8.0).
The DNA and amino acid sequences for the resulting fusion protein
(hereinafter referred to as TbRa3-38 kD-Tb38-1) are provided in SEQ ID NO: 147
and 148,
respectively.
A fusion protein containing the two antigens TbH-9 and Tb38-1 (hereinafter
referred to as TbH9-Tb38-1) without a hinge sequence, was prepared using a
similar
CA 02268036 1999-04-12
WO 98/16645 PCTIUS97/18Z14
53
procedure to that described above. The DNA sequence for the TbH9-Tb38-1 fusion
protein is
provided in SEQ ID NO: 151.
' A fusion protein containing TbRa3, the antigen 38kD, Tb38-1 and DPEP was
prepared as follows.
Each of the DNA constructs TbRa3, 38 kD and Tb38-1 were modified by PCR
and cloned into vectors essentially as described above, with the primers PDM-
69 (SEQ ID
N0:145 and PDM-83 (SEQ ID NO: 200) being used for amplification of the Tb38-lA
fragment. Tb38-lA differs from Tb38-1 by a DraI site at the 3' end of the
coding region that
keeps the final amino acid intact while creating a blunt restriction site that
is in frame. The
TbRa3/38kD/Tb38-lA fusion was then transferred to pET28b using NdeI and EcoRl
sites.
DPEP DNA was used to perform PCR using the primers PDM-84 and PDM-
85 (SEQ ID NO: 201 and 202, respectively) and 1 ~1 DNA at 50 ng/p,l.
Denaturation at 94 °C
was performed for 2 min, followed by 10 cycles of 96 °C for 15 sec, 68
°C for 15 sec and 72
°C for 1.5 min; 30 cycles of 96 °C for 15 sec, 64 °C for
15 sec and 72 °C for 1.5 min; and
finally by 72 °C for 4 min. The DPEP PCR fragment was digested with
EcoRI and Eco72I
and clones directly into the pET28Ra3/38kD/38-lA construct which was digested
with DraI
and EcoRI. The fusion construct was confirmed to be correct by DNA sequencing.
Recombinant protein was prepared as described above. The DNA and amino acid
sequences
for the resulting fusion protein (hereinafter referred to as TbF-2) are
provided in SEQ ID NO:
203 and 204, respectively.
EXAMPLE 8
USE OF M. TUBERCULOSIS FUSION PROTEINS FOR
SERODIAGNOSIS OF TUBERCULOSIS
The effectiveness of the fusion protein TbRa3-38 kD-Tb38-1, prepared as
described above, in the serodiagnosis of tuberculosis infection was examined
by ELISA.
The ELISA protocol was as described above in Example 6, with the fusion
protein being coated at 200 ng/well. A panel of sera was chosen from a group
of tuberculosis
patients previously shown, either by ELISA or by western blot analysis, to
react with each of
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
54
the three antigens individually or in combination. Such a panel enabled the
dissection of the
serological reactivity of the fusion protein to determine if all three
epitopes functioned with
the fusion protein. As shown in Table 5, all four sera that reacted with TbRa3
only were '
detectable with the fusion protein. Three sera that reacted only with Tb38-1
were also
detectable, as were two sear that reacted with 38 kD alone. The remaining 15
sera were all
positive with the fusion protein based on a cut-off in the assay of mean
negatives +3 standard
deviations. This data demonstrates the functional activity of all three
epitopes in the fusion
protein.
1 O TABLE 5
REACTIVITY OF TRI-PEPTIDE FUSION PROTEIN WITH SERA FROM M. TUBERCULOSIS
PATIENTS
Serum ID Status ELISA Fusion Fusion
and/or recombinantRecombinant
Western OD 450 Status
Blot
Reactivity
with
Individual
proteins
38kd Tb38-1
TbRa3
O 1 B 93I-40TB - - + 0.413 +
O1B93I-41 TB - + + 0.392 +
O1B93I-29 TB + _ + 2.217 +
O1B93I-109TB + t + 0.522 +
O1B93I-132TB + + + 0.937 +
5004 TB ~ + + 1.098 +
15004 TB + + + 2.077 +
39004 TB + + + 1.675 +
68004 TB + + + 2.388 +
99004 TB - + t 0.607 +
107004 TB - + ~ 0.667 +
92004 TB + ~ ~ 1.070 +
97004 TB + - t 1.152 + ,
118004 TB + - ~ 2.694 +
173004 TB + + + 3.258 +
175004 TB + - + 2.514 +
274004 TB - - + 3.220 + '
276004 TB - + - 2.991 +
282004 TB + - - 0.824 +
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
289004 TB - - + 0.848 +
308004 TB - + - 3.338 +
- 314004 TB - + - 1.362 +
317004 TB + - - 0.763 +
312004 TB - - + 1.079 +
D 176 PPD - - - 0.145 -
D 162 PPD - - - 0.073 -
D161 PPD - - - 0.097 -
D27 PPD - - - 0.082 -
A6-124 NORMAL - - - 0.053 -
A6-125 NORMAL - - - 0.087 -
A6-126 NORMAL - - - 0.346 +
A6-127 NORMAL - - - 0.064 -
A6-128 NORMAL - - - 0.034 -
A6-129 NORMAL - - - 0.037 -
A6-130 NORMAL - - - 0.057 -
A6-131 NORMAL - - - 0.054 -
A6-132 NORMAL - - 0.022 -
A6-133 NORMAL - - 0.147 -
A6-134 NORMAL - - - 0.101 -
A6-135 NORMAL - - 0.066 -
A6-136 NORMAL - - 0.054 -
A6-137 NORMAL - - - 0.065 -
A6-138 NORMAL - - - 0.041 -
A6-139 NORMAL - - - 0.103 -
A6-140 NORMAL - - - 0.212 -
A6-141 NORMAL - - - 0.056 -
A6-142 NORMAL - - - 0.051 -
The reactivity of the fusion protein TbF-2 with sera from M. tuberculosis-
infected patients was examined by ELISA using the protocol described above.
The results of
. these studies (Table 6) demonstrate that all four antigens function
independently in the fusion
5 protein.
CA 02268036 1999-04-12
WO 98/16645 PCT/US97/18214
56
TABLE 6
REACTIVITY OF TBF-2 FUSION PROTEIN WITH TB AND NORMAL SERA
Serum Status TbF StatusTbF-2 StatusELISA
ID OD450 OD450 Reactivity
38 TbRa3 Tb38-1DPEP
kD
B931-40 TB 0.57 + 0.321 + _ + _ +
B931-41 TB 0.601 + 0.396 + + + + _
B931-109TB 0.494 + 0.404 + + + +_ _
B931-132TB 1.502 + 1.292 + + + + t
5004 TB 1.806 + 1.666 + + _
15004 TB 2.862 + 2.468 + + + + -
39004 TB 2.443 + 1.722 + + + + _
68004 TB 2.871 + 2.575 + + + + _
99004 TB 0.691 + 0.971 + _ + _
107004 TB 0.875 + 0.732 + + _
92004 TB 1.632 + 1.394 + + f _
97004 TB 1.491 + 1.979 + + _ +
118004 TB 3.182 + 3.045 + + f _ _
173004 TB 3.644 + 3.578 + + + + _
175004 TB 3.332 + 2.916 + + + - _
274004 TB 3.696 + 3.716 + + - +
276004 TB 3.243 + 2.56 + - _ + _
282004 TB 1.249 + 1.234 + + _ _ _
289004 TB 1.373 + 1,17 + _ + _ _
308004 TB 3.708 + 3.355 + _ + _
314004 TB 1.663 + 1.399 + - - + _
317004 TB 1.163 + 0.92 + + _ _ _
312004 TB 1.709 + 1.453 + - + _ _
380004 TB 0.238 - 0.461 + _ +
451004 TB 0.18 - 0.2 - - - +_
478004 TB 0.188 - 0.469 + - _ +_
410004 TB 0.384 + 2.392 + _ _ +
411004 TB 0.306 + 0.874 + + _ +
421004 TB 0.357 + 1.456 + + +
528004 TB 0.047 0.196 - - +
A6-87 Normal 0.094 0.063 - _ _
A6-88 Normal 0.214 - 0.19 - - _ _
A6-89 Normal 0.248 - 0.125 - - _ _
A6-90 Normal 0.179 - 0.206 - - - _
A6-91 Normal 0.135 - 0.151 - - - _ _
A6-92 Normal 0.064 - 0.097 - - - _
A6-93 Normal 0.072 - 0.098 - - - _
A6-94 Normal 0.072 - 0.064 - - _
A6-95 Normal 0.125 - 0.159 - - - _
A6-96 Normal 0.121 0.12 - - _ _
Cut-off ~ 0.284 0.266
~ ~
CA 02268036 1999-04-12
WO 98/16645 PGT/US97/18214
57
One of skill in the art will appreciate that the order of the individual
antigens
within the fusion protein may be changed and that comparable activity would be
expected
- provided each of the epitopes is still functionally available. In addition,
truncated forms of
the proteins containing active epitopes may be used in the construction of
fusion proteins.
S
From the foregoing, it will be appreciated that, although specific embodiments
of the invention have been described herein for the purpose of illustration,
various
modifications may be made without deviating from the spirit and scope of the
invention.
CA 02268036 1999-10-12
58
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT:
(A) NAME: Corixa Corporation
(B) STREET: 1124 Columbia Street
(C) CITY: Seattle
(D) STATE: Washington
(E) COUNTRY: USA
(F) POSTAL CODE (ZIP): 98104
(G) TELEPHONE: (206) 754-5711
(H) TELEFAX: (206) 754-5715
(I) TELEX:
(ii) TITLE OF INVENTION: Compounds and Methods for
Diagnosis of Tuberculosis
(iii) NUMBER OF SEQUENCES: 209
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: Osler, Hoskin & l3arcourt
(B) STREET: 50 O'Connor Street
(C) CITY: Ottawa
(D) STATE: Ontario
(E) COUNTRY: Canada
(F) ZIP: K1P 6L2
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: PatentIn Release #1.0, Version #1.30
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER: 2,268,036
(B) FILING DATE: 7-OCT-1997
(C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER: US 08/729,622
(B) FILING DATE: 11-OCT-1996
(vii) PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER: US 08/818,111
(B) FILING DATE: 13-MAR-1997
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: Aitken, David W.
(B) REFERENCE/DOCKET NUMBER: 13590
(2) INFORMATION FOR SEQ ID N0:1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 766 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:1:
CGAGGCACCG GTAGTTTGAA CCAAACGCAC AATCGACGGG CAAACGAACG GAAGAACACA 60
CA 02268036 1999-10-12
59
ACCATGAAGATGGTGAAATCGATCGCCGCAGGTCTGACCGCCGCGGCTGCAATCGGCGCC 120
GCTGCGGCCGGTGTGACTTCGATCATGGCTGGCGGCCCGGTCGTATACCAGATGCAGCCG 180
GTCGTCTTCGGCGCGCCACTGCCGTTGGACCCGGCATCCGCCCCTGACGTCCCGACCGCC 240
GCCCAGTTGACCAGCCTGCTCAACAGCCTCGCCGATCCCAACGTGTCGTTTGCGAACAAG 300
GGCAGTCTGGTCGAGGGCGGCATCGGGGGCACCGAGGCGCGCATCGCCGACCACAAGCTG 360
AAGAAGGCCGCCGAGCACGGGGATCTGCCGCTGTCGTTCAGCGTGACGAACATCCAGCCG 420
GCGGCCGCCGGTTCGGCCACCGCCGACGTTTCCGTCTCGGGTCCGAAGCTCTCGTCGCCG 480
GTCACGCAGAACGTCACGTTCGTGAATCAAGGCGGCTGGATGCTGTCACGCGCATCGGCG 540
ATGGAGTTGCTGCAGGCCGCAGGGNAACTGATTGGCGGGCCGGNTTCAGCCCGCTGTTCA 600
GCTACGCCGCCCGCCTGGTGACGCGTCCATGTCGAACACTCGCGCGTGTAGCACGGTGCG 660
GTNTGCGCAGGGNCGCACGCACCGCCCGGTGCAAGCCG'TCCTCGAGATAGGTGGTGNCTC 720
GNCACCAGNGANCACCCCCNNNTCGNCNNTTCTCGNTG1VTGNATGA 766
(2) INFORMATION
FOR
SEQ
ID N0:2:
(i) S EQUENCE RACTERISTICS:
CHA
(A) LENGTH:752 base
pairs
(B) TYPE: ucleic
n acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi) TION:
SEQUENCE SEQ ID
DESCRIP N0:2:
ATGCATCACCATCACCATCACGATGAAGTCACGGTAGAGACGACCTCCGT CTTCCGCGCA60
GACTTCCTCAGCGAGCTGGACGCTCCTGCGCAAGCGGGTACGGAGAGCGC GGTCTCCGGG120
GTGGAAGGGCTCCCGCCGGGCTCGGCGTTGCTGGTAGTC;AAACGAGGCCC CAACGCCGGG180
TCCCGGTTCCTACTCGACCAAGCCATCACGTCGGCTGGTCGGCATCCCGA CAGCGACATA240
TTTCTCGACGACGTGACCGTGAGCCGTCGCCATGCTGAF,TTCCGGTTGGA AAACAACGAA300
TTCAATGTCGTCGATGTCGGGAGTCTCAACGGCACCTACGTCAACCGCGA GCCCGTGGAT360
TCGGCGGTGCTGGCGAACGGCGACGAGGTCCAGATCGGCAAGCTCCGGTT GGTGTTCTTG420
ACCGGACCCAAGCAAGGCGAGGATGACGGGAGTACCGGGGGCCCGTGAGC GCACCCGATA480
GCCCCGCGCTGGCCGGGATGTCGATCGGGGCGGTCCTCCGACCTGCTACG ACCGGATTTT540
CCCTGATGTCCACCATCTCCAAGATTCGATTCTTGGGAGGCTTGAGGGTC NGGGTGACCC600
CCCCGCGGGCCTCATTCNGGGGTNTCGGCNGGTTTCACCCCNTACCNACT GCCNCCCGGN660
TTGCNAATTCNTTCTTCNCTGCCCNNAAAGGGACCNTTANCTTGCCGCTN GAAANGGTNA720
TCCNGGGCCCNTCCTNGAANCCCCNTCCCCCT 752
(2) INFORMATION FOR SEQ ID N0:3:
CA 02268036 1999-10-12
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:813 base airs
p
(B) TYPE: ucleic
n acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:3:
CATATGCATCACCATCACCATCACACTTCTAACCGCCC.AGCGCGTCGGGG GCGTCGAGCA60
CCACGCGACACCGGGCCCGATCGATCTGCTAGCTTGAG'TCTGGTCAGGCA TCGTCGTCAG120
CAGCGCGATGCCCTATGTTTGTCGTCGACTCAGATATCGCGGCAATCCAA TCTCCCGCCT180
GCGGCCGGCGGTGCTGCAAACTACTCCCGGAGGAATTTCGACGTGCGCAT CAAGATCTTC240
ATGCTGGTCACGGCTGTCGTTTTGCTCTGTTGTTCGGG'rGTGGCCACGGC CGCGCCCAAG300
ACCTACTGCGAGGAGTTGAAAGGCACCGATACCGGCCA(JGCGTGCCAGAT TCAAATGTCC360
GACCCGGCCTACAACATCAACATCAGCCTGCCCAGTTA(:TACCCCGACCA GAAGTCGCTG420
GAAAATTACATCGCCCAGACGCGCGACAAGTTCCTCAGCGCGGCCACATC GTCCACTCCA480
CGCGAAGCCCCCTACGAATTGAATATCACCTCGGCCACATACCAGTCCGC GATACCGCCG540
CGTGGTACGCAGGCCGTGGTGCTCAMGGTCTACCACAA<:GCCGGCGGCAC GCACCCAACG600
ACCACGTACAAGGCCTTCGATTGGGACCAGGCCTATCGC:AAGCCAATCAC CTATGACACG660
CTGTGGCAGGCTGACACCGATCCGCTGCCAGTCGTCTTC:CCCATTGTTGC AAGGTGAACT720
GAGCAACGCAGACCGGGACAACWGGTATCGATAGCCGCC:NAATGCCGGCT TGGAACCCNG780
TGAAATTATCACAACTTCGCAGTCACNAAANAA 813
(2) INFORMATION
FOR
SEQ
ID N0:4:
(i) S EQUENCE CHARACTERISTICS:
(A) LENGTH: 447 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:4:
CGGTATGAACACGGCCGCGT CCGATAACTT CCAGCTGTCCCAGGGTGGGC AGGGATTCGC60
CATTCCGATCGGGCAGGCGA TGGCGATCGC GGGCCAGATCCGATCGGGTG GGGGGTCACC120
CACCGTTCATATCGGGCCTA CCGCCTTCCT CGGCTTGGGTGTTGTCGACA ACAACGGCAA180
CGGCGCACGAGTCCAACGCG TGGTCGGGAG CGCTCCGGCGGCAAGTCTCG GCATCTCCAC240
CGGCGACGTGATCACCGCGG TCGACGGCGC TCCGATCAACTCGGCCACCG CGATGGCGGA300
CGCGCTTAACGGGCATCATC CCGGTGACGT CATCTCGGTGAACTGGCAAA CCAAGTCGGG360
CGGCACGCGTACAGGGAACG TGACATTGGC CGAGGGACCCCCGGCCTGAT TTCGTCGYGG420
ATACCACCCGCCGGCCGGCC AATTGGA 447
CA 02268036 1999-10-12
61
(2) INFORMATION
FOR
SEQ
ID N0:5:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 604 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:5:
GTCCCACTGCGGTCGCCGAG TATGTCGCCC AGCAAATGTCTGGCAGCCGC CCAACGGAAT60
CCGGTGATCCGACGTCGCAG GTTGTCGAAC CCGCCGCCGCGGAAGTATCG GTCCATGCCT120
AGCCCGGCGACGGCGAGCGC CGGAATGGCG CGAGTGAGGAGGCGGGCAAT TTGGCGGGGC180
CCGGCGACGGNGAGCGCCGG AATGGCGCGA GTGAGGAGGTGGNCAGTCAT GCCCAGNGTG240
ATCCAATCAACCTGNATTCG GNCTGNGGGN CCATTTGACAATCGAGGTAG TGAGCGCAAA300
TGAATGATGGAAAACGGGNG GNGACGTCCG NTGTTCTGGTGGTGNTAGGT GNCTGNCTGG360
NGTNGNGGNTATCAGGATGT TCTTCGNCGA AANCTGAT(~NCGAGGAACAG GGTGTNCCCG420
NNANNCCNANGGNGTCCNAN CCCNNNNTCC TCGNCGANATCANANAGNCG NTTGATGNGA480
NAAAAGGGTGGANCAGNNNN AANTNGNGGN CCNAANAAPdCNNNANNGNNG NNAGNTNGNT540
NNNTNTTNNCANNNNNNNTG NNGNNGNNCN NNNCAANCPdNNTNNNNGNAA NNGGNTTNTT600
NAAT 604
(2) INFORMATION
FOR
SEQ
ID N0:6:
(i) S EQUENCE CHARACTERISTICS:
(A) LENGTH: 633
base pairs
(B) TYPE: nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:6:
TTGCANGTCGAACCACCTCA CTAAAGGGAACAAAAGCTNGAGCTCCACCG CGGTGGCGGC60
CGCTCTAGAACTAGTGKATM YYYCKGGCTGCAGSAATYCGGYACGAGCAT TAGGACAGTC120
TAACGGTCCTGTTACGGTGA TCGAATGACCGACGACATCCTGCTGATCGA CACCGACGAA180
CGGGTGCGAACCCTCACCCT CAACCGGCCGCAGTCCCGYAACGCGCTCTC GGCGGCGCTA240
CGGGATCGGTTTTTCGCGGY GTTGGYCGACGCCGAGGYCGACGACGACAT CGACGTCGTC300
ATCCTCACCGGYGCCGATCC GGTGTTCTGCGCCGGACTGGACCTCAAGGT AGCTGGCCGG360
GCAGACCGCGCTGCCGGACA TCTCACCGCGGTGGGCGGCCATGACCAAGC CGGTGATCGG420
CGCGATCAACGGCGCCGCGG TCACCGGCGGGCTCGAACTGGCGCTGTACT GCGACATCCT480
GATCGCCTCCGAGCACGCCC GCTTCGNCGACACCCACGCCCGGGTGGGGC TGCTGCCCAC540
CTGGGGACTCAGTGTGTGCT TGCCGCAAAAGGTCGGCATCGGNCTGGGCC GGTGGATGAG600
CA 02268036 1999-10-12
62
CCTGACCGGC GACTACCTGT CCGTGACCGA CGC 633
(2) INFORMATION
FOR SEQ
ID N0:7:
(i) S EQUENCE S:
CHARACTERISTIC
(A) LENGTH:1362 basepairs
(B) TYPE:ucleic
n acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY: linear
(xi) S EQUENCE CRIPTION:
DES SEQ ID
N0:7:
CGACGACGACGGCGCCGGAGAGCGGGCGCGAACGGCGA'rCGACGCGGCCCTGGCCAGAGT 60
CGGCACCACCCAGGAGGGAGTCGAATCATGAAATTTGTCAACCATATTGAGCCCGTCGCG 120
CCCCGCCGAGCCGGCGGCGCGGTCGCCGAGGTCTATGCCGAGGCCCGCCGCGAGTTCGGC 180
CGGCTGCCCGAGCCGCTCGCCATGCTGTCCCCGGACGAGGGACTGCTCACCGCCGGCTGG 240
GCGACGTTGCGCGAGACACTGCTGGTGGGCCAGGTGCCGCGTGGCCGCAAGGAAGCCGTC 300
GCCGCCGCCGTCGCGGCCAGCCTGCGCTGCCCCTGGTGCGTCGACGCACACACCACCATG 360
CTGTACGCGGCAGGCCAAACCGACACCGCCGCGGCGATCTTGGCCGGCACAGCACCTGCC 420
GCCGGTGACCCGAACGCGCCGTATGTGGCGTGGGCGGCAGGAACCGGGACACCGGCGGGA 480
CCGCCGGCACCGTTCGGCCCGGATGTCGCCGCCGAATAC;CTGGGCACCGCGGTGCAATTC 540
CACTTCATCGCACGCCTGGTCCTGGTGCTGCTGGACGAAACCTTCCTGCCGGGGGGCCCG 600
CGCGCCCAACAGCTCATGCGCCGCGCCGGTGGACTGGTCiTTCGCCCGCAAGGTGCGCGCG 660
GAGCATCGGCCGGGCCGCTCCACCCGCCGGCTCGAGCCGCGAACGCTGCCCGACGATCTG 720
GCATGGGCAACACCGTCCGAGCCCATAGCAACCGCGTTC;GCCGCGCTCAGCCACCACCTG 780
GACACCGCGCCGCACCTGCCGCCACCGACTCGTCAGGTCiGTCAGGCGGGTCGTGGGGTCG 840
TGGCACGGCGAGCCAATGCCGATGAGCAGTCGCTGGACGAACGAGCACACCGCCGAGCTG 900
CCCGCCGACCTGCACGCGCCCACCCGTCTTGCCCTGCTGACCGGCCTGGCCCCGCATCAG 960
GTGACCGACGACGACGTCGCCGCGGCCCGATCCCTGCTC'GACACCGATGCGGCGCTGGTT 1020
GGCGCCCTGGCCTGGGCCGCCTTCACCGCCGCGCGGCGCATCGGCACCTGGATCGGCGCC 1080
GCCGCCGAGGGCCAGGTGTCGCGGCAAAACCCGACTGGGTGAGTGTGCGCGCCCTGTCGG 1140
TAGGGTGTCATCGCTGGCCCGAGGGATCTCGCGGCGGCGAACGGAGGTGGCGACACAGGT 1200
GGAAGCTGCGCCCACTGGCTTGCGCCCCAACGCCGTCGTGGGCGTTCGGTTGGCCGCACT 1260
GGCCGATCAGGTCGGCGCCGGCCCTTGGCCGAAGGTCCAGCTCAACGTGCCGTCACCGAA 1320
GGACCGGACGGTCACCGGGGGTCACCCTGCGCGCCCAAGGAA 1362
(2) INFORMATION FOR SEQ ID N0:8:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1458 base pairs
CA 02268036 1999-10-12
63
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi) S EQUENCE CRIPTION:EQ ID
DES S N0:8:
GCGACGACCCCGATATGCCGGGCACCGTAGCGAAAGCCGTCGCCGACGCACTCGGGCGCG60
GTATCGCTCCCGTTGAGGACATTCAGGACTGCGTGGAGGCCCGGCTGGGGGAAGCCGGTC120
TGGATGACGTGGCCCGTGTTTACATCATCTACCGGCAGCGGCGCGCCGAGCTGCGGACGG180
CTAAGGCCTTGCTCGGCGTGCGGGACGAGTTAAAGCTGAGCTTGGCGGCCGTGACGGTAC240
TGCGCGAGCGCTATCTGCTGCACGACGAGCAGGGCCGGCCGGCCGAGTCGACCGGCGAGC300
TGATGGACCGATCGGCGCGCTGTGTCGCGGCGGCCGAGGACCAGTATGAGCCGGGCTCGT360
CGAGGCGGTGGGCCGAGCGGTTCGCCACGCTATTACGC.AACCTGGAATTCCTGCCGAATT420
CGCCCACGTTGATGAACTCTGGCACCGACCTGGGACTGCTCGCCGGCTGTTTTGTTCTGC480
CGATTGAGGATTCGCTGCAATCGATCTTTGCGACGCTGcJGACAGGCCGCCGAGCTGCAGC540
GGGCTGGAGGCGGCACCGGATATGCGTTCAGCCACCTGCGACCCGCCGGGGATCGGGTGG600
CCTCCACGGGCGGCACGGCCAGCGGACCGGTGTCGTTTCTACGGCTGTATGACAGTGCCG660
CGGGTGTGGTCTCCATGGGCGGTCGCCGGCGTGGCGCC'.CGTATGGCTGTGCTTGATGTGT720
CGCACCCGGATATCTGTGATTTCGTCACCGCCAAGGCCGAATCCCCCAGCGAGCTCCCGC780
ATTTCAACCTATCGGTTGGTGTGACCGACGCGTTCCTGC;GGGCCGTCGAACGCAACGGCC840
TACACCGGCTGGTCAATCCGCGAACCGGCAAGATCGTCGCGCGGATGCCCGCCGCCGAGC900
TGTTCGACGCCATCTGCAAAGCCGCGCACGCCGGTGGCGATCCCGGGCTGGTGTTTCTCG960
ACACGATCAATAGGGCAAACCCGGTGCCGGGGAGAGGCC;GCATCGAGGCGACCAACCCGT1020
GCGGGGAGGTCCCACTGCTGCCTTACGAGTCATGTAATC;TCGGCTCGATCAACCTCGCCC1080
GGATGCTCGCCGACGGTCGCGTCGACTGGGACCGGCTCGAGGAGGTCGCCGGTGTGGCGG1140
TGCGGTTCCTTGATGACGTCATCGATGTCAGCCGCTACC',CCTTCCCCGAACTGGGTGAGG1200
CGGCCCGCGCCACCCGCAAGATCGGGCTGGGAGTCATGGGTTTGGCGGAACTGCTTGCCG1260
CACTGGGTATTCCGTACGACAGTGAAGAAGCCGTGCGGTTAGCCACCCGGCTCATGCGTC1320
GCATACAGCAGGCGGCGCACACGGCATCGCGGAGGCTGGCCGAAGAGCGGGGCGCATTCC1380
CGGCGTTCACCGATAGCCGGTTCGCGCGGTCGGGCCCGAGGCGCAACGCACAGGTCACCT1440
CCGTCGCTCCGACGGGCA 1458
(2) INFORMATION FOR SEQ ID N0:9:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 862 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
CA 02268036 1999-10-12
64
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:9:
ACGGTGTAATCGTGCTGGATCTGGAACCGCGTGGCCCGCTACCTACCGAGATCTACTGGC 60
GGCGCAGGGGGCTGGCCCTGGGCATCGCGGTCGTCGTAGTCGGGATCGCGGTGGCCATCG 120
TCATCGCCTTCGTCGACAGCAGCGCCGGTGCCAAACCGGTCAGCGCCGACAAGCCGGCCT 180
CCGCCCAGAGCCATCCGGGCTCGCCGGCACCCCAAGCACCCCAGCCGGCCGGGCAAACCG 240
AAGGTAACGCCGCCGCGGCCCCGCCGCAGGGCCAAAACCCCGAGACACCCACGCCCACCG 300
CCGCGGTGCAGCCGCCGCCGGTGCTCAAGGAAGGGGACGATTGCCCCGATTCGACGCTGG 360
CCGTCAAAGGTTTGACCAACGCGCCGCAGTACTACGTCGGCGACCAGCCGAAGTTCACCA 420
TGGTGGTCACCAACATCGGCCTGGTGTCCTGTAAACGCGACGTTGGGGCCGCGGTGTTGG 480
CCGCCTACGTTTACTCGCTGGACAACAAGCGGTTGTGG'rCCAACCTGGACTGCGCGCCCT 540
CGAATGAGACGCTGGTCAAGACGTTTTCCCCCGGTGAGCAGGTAACGACCGCGGTGACCT 600
GGACCGGGATGGGATCGGCGCCGCGCTGCCCATTGCCGCGGCCGGCGATCGGGCCGGGCA 660
CCTACAATCTCGTGGTACAACTGGGCAATCTGCGCTCGCTGCCGGTTCCGTTCATCCTGA 720
ATCAGCCGCCGCCGCCGCCCGGGCCGGTACCCGCTCCGGGTCCAGCGCAGGCGCCTCCGC 780
CGGAGTCTCCCGCGCAAGGCGGATAATTATTGATCGCTGATGGTCGATTCCGCCAGCTGT 840
GACAACCCCTCGCCTCGTGCCG 862
(2) INFORMATION
FOR SEQ
ID N0:10:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 622 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE :
DESCRIPTION:
SEQ ID
N0:1.0
TTGATCAGCACCGGCAAGGC GTCACATGCC TCCCTGGGTGTGCAGGTGAC CAATGACAAA60
GACACCCCGGGCGCCAAGAT CGTCGAAGTA GTGGCCGGT'GGTGCTGCCGC GAACGCTGGA120
GTGCCGAAGGGCGTCGTTGT CACCAAGGTC GACGACCGCCCGATCAACAG CGCGGACGCG180
TTGGTTGCCGCCGTGCGGTC CAAAGCGCCG GGCGCCACGGTGGCGCTAAC CTTTCAGGAT240
CCCTCGGGCGGTAGCCGCAC AGTGCAAGTC ACCCTCGGCAAGGCGGAGCA GTGATGAAGG300
TCGCCGCGCAGTGTTCAAAG CTCGGATATA CGGTGGCACCCATGGAACAG CGTGCGGAGT360
TGGTGGTTGGCCGGGCACTT GTCGTCGTCG TTGACGATCGCACGGCGCAC GGCGATGAAG420
ACCACAGCGGGCCGCTTGTC ACCGAGCTGC TCACCGAGGCCGGGTTTGTT GTCGACGGCG480
TGGTGGCGGTGTCGGCCGAC GAGGTCGAGA TCCGAAATGCGCTGAACACA GCGGTGATCG540
GCGGGGTGGACCTGGTGGTG TCGGTCGGCG GGACCGGNGTGACGNCTCGC GATGTCACCC600
CA 02268036 1999-10-12
CGGAAGCCAC CCGNGACATT CT 622
(2) INFORMATION
FOR SEQ
ID N0:11:
(i) S EQUENCE RACTERISTICS:
CHA
(A) LENGTH:1200 basepairs
(B) TYPE:ucleic
n acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY: linear
(xi) S EQUENCE EQ ID :
DESCRIPTION: NO:11
S
GGCGCAGCGGTAAGCCTGTTGGCCGCCGGCACACTGGTGTTGACAGCATG CGGCGGTGGC60
ACCAACAGCTCGTCGTCAGGCGCAGGCGGAACGTCTGGGTCGGTGCACTG CGGCGGCAAG120
AAGGAGCTCCACTCCAGCGGCTCGACCGCACAAGAAAA'rGCCATGGAGCA GTTCGTCTAT180
GCCTACGTGCGATCGTGCCCGGGCTACACGTTGGACTACAACGCCAACGG GTCCGGTGCC240
GGGGTGACCCAGTTTCTCAACAACGAAACCGATTTCGCCGGCTCGGATGT CCCGTTGAAT300
CCGTCGACCGGTCAACCTGACCGGTCGGCGGAGCGGTGCGGTTCCCCGGC ATGGGACCTG360
CCGACGGTGTTCGGCCCGATCGCGATCACCTACAATATCAAGGGCGTGAG CACGCTGAAT420
CTTGACGGACCCACTACCGCCAAGATTTTCAACGGCAC(:ATCACCGTGTG GAATGATCCA480
CAGATCCAAGCCCTCAACTCCGGCACCGACCTGCCGCCAACACCGATTAG CGTTATCTTC540
CGCAGCGACAAGTCCGGTACGTCGGACAACTTCCAGAAATACCTCGACGG TGTATCCAAC600
GGGGCGTGGGGCAAAGGCGCCAGCGAAACGTTCAGCGGC~GGCGTCGGCGT CGGCGCCAGC660
GGGAACAACGGAACGTCGGCCCTACTGCAGACGACCGAC:GGGTCGATCAC CTACAACGAG720
TGGTCGTTTGCGGTGGGTAAGCAGTTGAACATGGCCCAGATCATCACGTC GGCGGGTCCG780
GATCCAGTGGCGATCACCACCGAGTCGGTCGGTAAGACAATCGCCGGGGC CAAGATCATG840
GGACAAGGCAACGACCTGGTATTGGACACGTCGTCGTTC:TACAGACCCAC CCAGCCTGGC900
TCTTACCCGATCGTGCTGGCGACCTATGAGATCGTCTGC:TCGAAATACCC GGATGCGACG960
ACCGGTACTGCGGTAAGGGCGTTTATGCAAGCCGCGATTGGTCCAGGCCA AGAAGGCCTG1020
GACCAATACGGCTCCATTCCGTTGCCCAAATCGTTCCAAGCAAAATTGGC GGCCGCGGTG1080
AATGCTATTTCTTGACCTAGTGAAGGGAATTCGACGGTGAGCGATGCCGT TCCGCAGGTA1140
GGGTCGCAATTTGGGCCGTATCAGCTATTGCGGCTGCTGGGCCGAGGCGG GATGGGCGAG1200
(2) INFORMATION FOR SEQ ID N0:12:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1155 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:12:
CA 02268036 1999-10-12
66
GCAAGCAGCTGCAGGTCGTGCTGTTCGACGAACTGGGCATGCCGAAGACCAAACGCACCA 60
AGACCGGCTACACCACGGATGCCGACGCGCTGCAGTCGTTGTTCGACAAGACCGGGCATC 120
CGTTTCTGCAACATCTGCTCGCCCACCGCGACGTCACCCGGCTCAAGGTCACCGTCGACG 180
GGTTGCTCCAAGCGGTGGCCGCCGACGGCCGCATCCAC;ACCACGTTCAACCAGACGATCG 240
CCGCGACCGGCCGGCTCTCCTCGACCGAACCCAACCTGCAGAACATCCCGATCCGCACCG 300
ACGCGGGCCGGCGGATCCGGGACGCGTTCGTGGTCGGGGACGGTTACGCCGAGTTGATGA 360
CGGCCGACTACAGCCAGATCGAGATGCGGATCATGGGGCACCTGTCCGGGGACGAGGGCC 420
TCATCGAGGCGTTCAACACCGGGGAGGACCTGTATTCG'rTCGTCGCGTCCCGGGTGTTCG 480
GTGTGCCCATCGACGAGGTCACCGGCGAGTTGCGGCGCCGGGTCAAGGCGATGTCCTACG 540
GGCTGGTTTACGGGTTGAGCGCCTACGGCCTGTCGCAGCAGTTGAAAATCTCCACCGAGG 600
AAGCCAACGAGCAGATGGACGCGTATTTCGCCCGATTCGGCGGGGTGCGCGACTACCTGC 660
GCGCCGTAGTCGAGCGGGCCCGCAAGGACGGCTACACC'.PCGACGGTGCTGGGCCGTCGCC 720
GCTACCTGCCCGAGCTGGACAGCAGCAACCGTCAAGTG(:GGGAGGCCGCCGAGCGGGCGG 780
CGCTGAACGCGCCGATCCAGGGCAGCGCGGCCGACATCATCAAGGTGGCCATGATCCAGG 840
TCGACAAGGCGCTCAACGAGGCACAGCTGGCGTCGCGCRTGCTGCTGCAGGTCCACGACG 900
AGCTGCTGTTCGAAATCGCCCCCGGTGAACGCGAGCGGGTCGAGGCCCTGGTGCGCGACA 960
AGATGGGCGGCGCTTACCCGCTCGACGTCCCGCTGGAGGTGTCGGTGGGCTACGGCCGCA 1020
GCTGGGACGCGGCGGCGCACTGAGTGCCGAGCGTGCATC;TGGGGCGGGAATTCGGCGATT 1080
TTTCCGCCCTGAGTTCACGCTCGGCGCAATCGGGACCGRGTTTGTCCAGCGTGTACCCGT 1140
CGAGTAGCCTCGTCA 1155
(2) INFORMATION
FOR SEQ
ID N0:13:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 1771 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:13:
GAGCGCCGTCTGGTGTTTGA ACGGTTTTAC CGGTCGGCATCGGCACGGGC GTTGCCGGGT60
TCGGGCCTCGGGTTGGCGAT CGTCAAACAG GTGGTGCTC;AACCACGGCGG ATTGCTGCGC120
ATCGAAGACACCGACCCAGG CGGCCAGCCC CCTGGAACGTCGATTTACGT GCTGCTCCCC180
GGCCGTCGGATGCCGATTCC GCAGCTTCCC GGTGCGACGGCTGGCGCTCG GAGCACGGAC240
ATCGAGAACTCTCGGGGTTC GGCGAACGTT ATCTCAGTGGAATCTCAGTC CACGCGCGCA300
ACCTAGTTGTGCAGTTACTG TTGAAAGCCA CACCCATGCCAGTCCACGCA TGGCCAAGTT360
GGCCCGAGTAGTGGGCCTAG TACAGGAAGA GCAACCTAGCGACATGACGA ATCACCCACG420
CA 02268036 1999-10-12
67
GTATTCGCCA CCGCCGCAGCAGCCGGGAACCCCAGGTTATGCTCAGGGGC AGCAGCAAAC480
GTACAGCCAG CAGTTCGACTGGCGTTACCCACCGTCCCCGCCCCCGCAGC CAACCCAGTA540
CCGTCAACCC TACGAGGCGTTGGGTGGTACCCGGCCGGGTCTGATACCTG GCGTGATTCC600
GACCATGACG CCCCCTCCTGGGATGGTTCGCCAACGCCCTCGTGCAGGCA TGTTGGCCAT660
CGGCGCGGTG ACGATAGCGGTGGTGTCCGCCGGCATCGGCGGCGCGGCCG CATCCCTGGT720
CGGGTTCAAC CGGGCACCCGCCGGCCCCAGCGGCGGCC~~AGTGGCTGCCA GCGCGGCGCC780
AAGCATCCCC GCAGCAAACATGCCGCCGGGGTCGGTCG:~ACAGGTGGCGG CCAAGGTGGT840
GCCCAGTGTC GTCATGTTGGAAACCGATCTGGGCCGCCAGTCGGAGGAGG GCTCCGGCAT900
CATTCTGTCT GCCGAGGGGCTGATCTTGACCAACAACCACGTGATCGCGG CGGCCGCCAA960
GCCTCCCCTG GGCAGTCCGCCGCCGAAAACGACGGTAACCTTCTCTGACG GGCGGACCGC1020
ACCCTTCACG GTGGTGGGGGCTGACCCCACCAGTGATA'.PCGCCGTCGTCC GTGTTCAGGG1080
CGTCTCCGGG CTCACCCCGATCTCCCTGGGTTCCTCCTC:GGACCTGAGGG TCGGTCAGCC1140
GGTGCTGGCG ATCGGGTCGCCGCTCGGTTTGGAGGGCA(:CGTGACCACGG GGATCGTCAG1200
CGCTCTCAAC CGTCCAGTGTCGACGACCGGCGAGGCCGGCAACCAGAACA CCGTGCTGGA1260
CGCCATTCAG ACCGACGCCGCGATCAACCCCGGTAACTC:CGGGGGCGCGC TGGTGAACAT1320
GAACGCTCAA CTCGTCGGAGTCAACTCGGCCATTGCCAC;GCTGGGCGCGG ACTCAGCCGA1380
TGCGCAGAGC GGCTCGATCGGTCTCGGTTTTGCGATTCC;AGTCGACCAGG CCAAGCGCAT1440
CGCCGACGAG TTGATCAGCACCGGCAAGGCGTCACATGC;CTCCCTGGGTG TGCAGGTGAC1500
CAATGACAAA GACACCCCGGGCGCCAAGATCGTCGAAGTAGTGGCCGGTG GTGCTGCCGC1560
GAACGCTGGA GTGCCGAAGGGCGTCGTTGTCACCAAGGTCGACGACCGCC CGATCAACAG1620
CGCGGACGCG TTGGTTGCCGCCGTGCGGTCCAAAGCGCC'GGGCGCCACGG TGGCGCTAAC1680
CTTTCAGGAT CCCTCGGGCGGTAGCCGCACAGTGCAAGT'CACCCTCGGCA AGGCGGAGCA1740
GTGATGAAGG TCGCCGCGCAGTGTTCAAAGC 1771
(2) INFORMATION Q ID N0:14:
FOR SE
(i) SEQUENCE CHARACTERISTICS :
(A) LENGTH: 1058 baseairs
p
(B) TYPE: nu cleic
acid
(C) STRANDED NESS: e
singl
(D) TOPOLOGY : linear
(xi) SEQUENCE DESCRIPTION: Q ID N0:14:
SE
CTCCACCGCG GTGGCGGCCG CCGGGCTGCA GGAATTCGGC60
CTCTAGAACT AGTGGATCCC
ACGAGGATCC GACGTCGCAG CCGCCGCCGCGGAAGTATCG GTCCATGCCT120
GTTGTCGAAC
AGCCCGGCGA CGGCGAGCGC GGCGGGCAAT TTGGCGGGGC180
CGGAATGGCG CGAGTGAGG:~1
CA 02268036 1999-10-12
68
CCGGCGACGGCGAGCGCCGGAATGGCGCGAGTGAGGAG'GCGGGCAGTCATGCCCAGCGTG 240
ATCCAATCAACCTGCATTCGGCCTGCGGGCCCATTTGACAATCGAGGTAGTGAGCGCAAA 300
TGAATGATGGAAAACGGGCGGTGACGTCCGCTGTTCTGGTGGTGCTAGGTGCCTGCCTGG 360
CGTTGTGGCTATCAGGATGTTCTTCGCCGAAACCTGATGCCGAGGAACAGGGTGTTCCCG 420
TGAGCCCGACGGCGTCCGACCCCGCGCTCCTCGCCGAGATCAGGCAGTCGCTTGATGCGA 480
CAAAAGGGTTGACCAGCGTGCACGTAGCGGTCCGAACAACCGGGAAAGTCGACAGCTTGC 540
TGGGTATTACCAGTGCCGATGTCGACGTCCGGGCCAATCCGCTCGCGGCAAAGGGCGTAT 600
GCACCTACAACGACGAGCAGGGTGTCCCGTTTCGGGTACAAGGCGACAACATCTCGGTGA 660
AACTGTTCGACGACTGGAGCAATCTCGGCTCGATTTCTGAACTGTCAACTTCACGCGTGC 720
TCGATCCTGCCGCTGGGGTGACGCAGCTGCTGTCCGGTGTCACGAACCTCCAAGCGCAAG 780
GTACCGAAGTGATAGACGGAATTTCGACCACCAAAATCACCGGGACCATCCCCGCGAGCT 840
CTGTCAAGATGCTTGATCCTGGCGCCAAGAGTGCAAGGCCGGCGACCGTGTGGATTGCCC 900
AGGACGGCTCGCACCACCTCGTCCGAGCGAGCATCGACCTCGGATCCGGGTCGATTCAGC 960
TCACGCAGTCGAAATGGAACGAACCCGTCAACGTCGAC'.CAGGCCGAAGTTGCGTCGACGC 1020
GTTGNTCGAAACGCCCTTGTGAACGGTGTCAACGGNAC 1058
(2) INFORMATION
FOR SEQ
ID N0:15:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 542 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:1.5:
GAATTCGGCACGAGAGGTGA TCGACATCAT CGGGACCAGCCCCACATCCT GGGAACAGGC60
GGCGGCGGAGGCGGTCCAGC GGGCGCGGGA TAGCGTCGP,TGACATCCGCG TCGCTCGGGT120
CATTGAGCAGGACATGGCCG TGGACAGCGC CGGCAAGATCACCTACCGCA TCAAGCTCGA180
AGTGTCGTTCAAGATGAGGC CGGCGCAACC GCGCTAGCA.CGGGCCGGCGA GCAAGACGCA240
AAATCGCACGGTTTGCGGTT GATTCGTGCG ATTTTGTGTCTGCTCGCCGA GGCCTACCAG300
GCGCGGCCCAGGTCCGCGTG CTGCCGTATC CAGGCGTGCATCGCGATTCC GGCGGCCACG360
CCGGAGTTAATGCTTCGCGT CGACCCGAAC TGGGCGATCCGCCGGNGAGC TGATCGATGA420
CCGTGGCCAGCCCGTCGATG CCCGAGTTGC CCGAGGAAACGTGCTGCCAG GCCGGTAGGA480
AGCGTCCGTAGGCGGCGGTG CTGACCGGCT CTGCCTGCGCCCTCAGTGCG GCCAGCGAGC540
GG 542
(2) INFORMATION FOR SEQ ID N0:16:
CA 02268036 1999-10-12
69
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:913 base
pairs
(B) TYPE: ucleic
n acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi) SEQUENCE :
DESCRIPTION:
SEQ ID
N0:16
CGGTGCCGCCCGCGCCTCCGTTGCCCCCATTGCCGCCGTCGCCGATCAGC TGCGCATCGC60
CACCATCACCGCCTTTGCCGCCGGCACCGCCGGTGGCGCCGGGGCCGCCG ATGCCACCGC120
TTGACCCTGGCCGCCGGCGCCGCCATTGCCATACAGCACCCCGCCGGGGG CACCGTTACC180
GCCGTCGCCACCGTCGCCGCCGCTGCCGTTTCAGGCCGGGGAGGCCGAAT GAACCGCCGC240
CAAGCCCGCCGCCGGCACCGTTGCCGCCTTTTCCGCCCGCCCCGCCGGCG CCGCCAATTG300
CCGAACAGCCAMGCACCGTTGCCGCCAGCCCCGCCGCCcJTTAACGGCGCT GCCGGGCGCC360
GCCGCCGGACCCGCCATTACCGCCGTTCCCGTTCGGTGCCCCGCCGTTAC CGGCGCCGCC420
GTTTGCCGCCAATATTCGGCGGGCACCGCCAGACCCGCCGGGGCCACCAT TGCCGCCGGG480
CACCGAAACAACAGCCCAACGGTGCCGCCGGCCCCGCCGTTTGCCGCCAT CACCGGCCAT540
TCACCGCCAGCACCGCCGTTAATGTTTATGAACCCGGTACCGCCAGCGCG GCCCCTATTG600
CCGGGCGCCGGAGNGCGTGCCCGCCGGCGCCGCCAACGC:CCAAAAGCCCG GGGTTGCCAC660
CGGCCCCGCCGGACCCACCGGTCCCGCCGATCCCCCCG7.'TGCCGCCGGTG CCGCCGCCAT720
TGGTGCTGCTGAAGCCGTTAGCGCCGGTTCCGCSGGTTC;CGGCGGTGGCG CCNTGGCCGC780
CGGCCCCGCCGTTGCCGTACAGCCACCCCCCGGTGGCGC;CGTTGCCGCCA TTGCCGCCAT840
TGCCGCCGTTGCCGCCATTGCCGCCGTTCCCGCCGCCAC;CGCCGGNTTGG CCGCCGGCGC900
CGCCGGCGGCCGC 913
(2) INFORMATION
FOR SEQ
ID N0:17:
(i) S EQUENCE CHARACTERISTICS:
(A) LENGTH: 1872 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:17:
GACTACGTTGGTGTAGAAAA ATCCTGCCGC CCGGACCCTTAAGGCTGGGA CAATTTCTGA60
TAGCTACCCCGACACAGGAG GTTACGGGAT GAGCAATTCGCGCCGCCGCT CACTCAGGTG120
GTCATGGTTGCTGAGCGTGC TGGCTGCCGT CGGGCTGGGCCTGGCCACGG CGCCGGCCCA180
GGCGGCCCCGCCGGCCTTGT CGCAGGACCG GTTCGCCGACTTCCCCGCGC TGCCCCTCGA240
CCCGTCCGCGATGGTCGCCC AAGTGGCGCC ACAGGTGGTCAACATCAACA CCAAACTGGG300
CTACAACAACGCCGTGGGCG CCGGGACCGG CATCGTCATCGATCCCAACG GTGTCGTGCT360
GACCAACAACCACGTGATCG CGGGCGCCAC CGACATCAA'PGCGTTCAGCG TCGGCTCCGG420
CA 02268036 1999-10-12
CCAAACCTACGGCGTCGATGTGGTCGGGTATGACCGCACCCAGGATGTCGCGGTGCTGCA480
GCTGCGCGGTGCCGGTGGCCTGCCGTCGGCGGCGATCGGTGGCGGCGTCGCGGTTGGTGA540
GCCCGTCGTCGCGATGGGCAACAGCGGTGGGCAGGGCGGAACGCCCCGTGCGGTGCCTGG600
CAGGGTGGTCGCGCTCGGCCAAACCGTGCAGGCGTCGGATTCGCTGACCGGTGCCGAAGA660
GACATTGAACGGGTTGATCCAGTTCGATGCCGCAATCCAGCCCGGTGATTCGGGCGGGCC720
CGTCGTCAACGGCCTAGGACAGGTGGTCGGTATGAACACGGCCGCGTCCGATAACTTCCA780
GCTGTCCCAGGGTGGGCAGGGATTCGCCATTCCGATCGGGCAGGCGATGGCGATCGCGGG840
CCAAATCCGATCGGGTGGGGGGTCACCCACCGTTCATA'rCGGGCCTACCGCCTTCCTCGG900
CTTGGGTGTTGTCGACAACAACGGCAACGGCGCACGAG'PCCAACGCGTGGTCGGAAGCGC960
TCCGGCGGCAAGTCTCGGCATCTCCACCGGCGACGTGA'CCACCGCGGTCGACGGCGCTCC1020
GATCAACTCGGCCACCGCGATGGCGGACGCGCTTAACGGGCATCATCCCGGTGACGTCAT1080
CTCGGTGAACTGGCAAACCAAGTCGGGCGGCACGCGTA(:AGGGAACGTGACATTGGCCGA1140
GGGACCCCCGGCCTGATTTGTCGCGGATACCACCCGCCGGCCGGCCAATTGGATTGGCGC1200
CAGCCGTGATTGCCGCGTGAGCCCCCGAGTTCCGTCTCC:CGTGCGCGTGGCATTGTGGAA1260
GCAATGAACGAGGCAGAACACAGCGTTGAGCACCCTCCC:GTGCAGGGCAGTTACGTCGAA1320
GGCGGTGTGGTCGAGCATCCGGATGCCAAGGACTTCGGC:AGCGCCGCCGCCCTGCCCGCC1380
GATCCGACCTGGTTTAAGCACGCCGTCTTCTACGAGGTGCTGGTCCGGGCGTTCTTCGAC1440
GCCAGCGCGGACGGTTCCGNCGATCTGCGTGGACTCATC:GATCGCCTCGACTACCTGCAG1500
TGGCTTGGCATCGACTGCATCTGTTGCCGCCGTTCCTACGACTCACCGCTGCGCGACGGC1560
GGTTACGACATTCGCGACTTCTACAAGGTGCTGCCCGAP.TTCGGCACCGTCGACGATTTC1620
GTCGCCCTGGTCGACACCGCTCACCGGCGAGGTATCCGCATCATCACCGACCTGGTGATG1680
AATCACACCTCGGAGTCGCACCCCTGGTTTCAGGAGTCCCGCCGCGACCCAGACGGACCG1740
TACGGTGACTATTACGTGTGGAGCGACACCAGCGAGCGCTACACCGACGCCCGGATCATC1800
TTCGTCGACACCGAAGAGTCGAACTGGTCATTCGATCCTGTCCGCCGACAGTTNCTACTG1860
GCACCGATTCTT 1872
(2) INFORMATION FOR
SEQ ID
N0:18:
(i) SE QUENCE :
CHARACTERISTICS
(A) LENGTH: 1482 baseairs
p
( B) TYPE:
nucleic
acid
( C) STRANDEDNESS:
single
( D) TOPOLOGY: linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:1'B:
CTTCGCCGAA 60
ACCTGATGCC
GAGGAACAGG
GTGTTCCCG'P
GAGCCCGACG
GCGTCCGACC
CA 02268036 1999-10-12
71
CCGCGCTCCT CGCCGAGATCAGGCAGTCGCTTGATGCGACAAAAGGGTTGACCAGCGTGC120
ACGTAGCGGT CCGAACAACCGGGAAAGTCGACAGCTTGCTGGGTATTACCAGTGCCGATG180
TCGACGTCCG GGCCAATCCGCTCGCGGCAAAGGGCGTATGCACCTACAACGACGAGCAGG240
GTGTCCCGTT TCGGGTACAAGGCGACAACATCTCGGTGAAACTGTTCGACGACTGGAGCA300
ATCTCGGCTC GATTTCTGAACTGTCAACTTCACGCGTGCTCGATCCTGCCGCTGGGGTGA360
CGCAGCTGCT GTCCGGTGTCACGAACCTCCAAGCGCAAGGTACCGAAGTGATAGACGGAA420
TTTCGACCAC CAAAATCACCGGGACCATCCCCGCGAGC'rCTGTCAAGATGCTTGATCCTG480
GCGCCAAGAG TGCAAGGCCGGCGACCGTGTGGATTGCCCAGGACGGCTCGCACCACCTCG540
TCCGAGCGAG CATCGACCTCGGATCCGGGTCGATTCAGCTCACGCAGTCGAAATGGAACG600
AACCCGTCAA CGTCGACTAGGCCGAAGTTGCGTCGACGCGTTGCTCGAAACGCCCTTGTG660
AACGGTGTCA ACGGCACCCGAAAACTGACCCCCTGACGGCATCTGAAAATTGACCCCCTA720
GACCGGGCGG TTGGTGGTTATTCTTCGGTGGTTCCGGC'.PGGTGGGACGCGGCCGAGGTCG780
CGGTCTTTGA GCCGGTAGCTGTCGCCTTTGAGGGCGACGACTTCAGCATGGTGGACGAGG840
CGGTCGATCA TGGCGGCAGCAACGACGTCGTCGCCGCCGAAAACCTCGCCCCACCGGCCG900
AAGGCCTTAT TGGACGTGACGATCAAGCTGGCCCGCTCATACCGGGAGGACACCAGCTGG960
AAGAAGAGGT TGGCGGCCTCGGGCTCAAACGGAATGTAF1CCGACTTCGTCAACCACCAGG1020
AGCGGATAGC GGCCAAACCGGGTGAGTTCGGCGTAGATC~CGCCCGGCGTGGTGAGCCTCG1080
GCGAACCGTG CTACCCATTCGGCGGCGGTGGCGAACAGC:ACCCGATGACCGGCCTGACAC1140
GCGCGTATCG CCAGGCCGACCGCAAGATGAGTCTTCCCGGTGCCAGGCGGGGCCCAAAAA1200
CACGACGTTA TCGCGGGCGGTGATGAAATCCAGGGTGCC:CAGATGTGCGATGGTGTCGCG1260
TTTGAGGCCA CGAGCATGCTCAAAGTCGAACTCTTCCAP.,CGACTTCCGAACCGGGAAGCG1320
GGCGGCGCGG ATGCGGCCCTCACCACCATGGGACTCCCGGGCTGACACTTCCCGCTGCAG1380
GCAGGCGGCC AGGTATTCTTCGTGGCTCCAGTTCTCGGCGCGGGCGCGATCGGCCAGCCG1440
GGACACTGAC TCACGCAGGGTGGGAGCTTTCAATGCTCTTGT 1982
(2) INFORMATION Q ID N0:19:
FOR SE
(i) SEQUENCE CHARACTERISTICS :
(A) LENGTH: 876 base irs
pa
(B) TYPE: nucleic
acid
(C) STRANDEDNESS: e
singl
(D) TOPOLOGY : linear
(xi) SEQUENCE DESCRIPTION:
SEQ ID N0:19:
GAATTCGGCA CGAGCCGGCG GACCAGATGG 60
ATAGCTTCTG GGCCGCGGCC CTCGAGGGTT
CGTGCTCGGG GCCACCGCCG GAGGGCCTGC 120
GGCGCACCAC CCTGACCGG'r AACACGCCGA
CGGTCACTCG TTGCTGCTGG 180
ACGCCACCAA CCCGGCGGTG
GTTGCCTACG ACCCGGCCTT
CA 02268036 1999-10-12
72
CGCCTACGAA TCGNGGAAAGCGGACTGGCCAGGATGTGCGGGGAGAACCC 240
ATCGGCTACA
GGAGAACATCTTCTTCTACATCACCGTCTACAACGAGCCGTACGTGCAGCCGCCGGAGCC 300
GGAGAACTTCGATCCCGAGGGCGTGCTGGGGGGTATCTACCGNTATCACGCGGCCACCGA 360
GCAACGCACCAACAAGGNGCAGATCCTGGCCTCCGGGG'rAGCGATGCCCGCGGCGCTGCG 420
GGCAGCACAGATGCTGGCCGCCGAGTGGGATGTCGCCGCCGACGTGTGGTCGGTGACCAG 480
TTGGGGCGAGCTAAACCGCGACGGGGTGGTCATCGAGACCGAGAAGCTCCGCCACCCCGA 540
TCGGCCGGCGGGCGTGCCCTACGTGACGAGAGCGCTGGAGAATGCTCGGGGCCCGGTGAT 600
CGCGGTGTCGGACTGGATGCGCGCGGTCCCCGAGCAGA'.CCCGACCGTGGGTGCCGGGCAC 660
ATACCTCACGTTGGGCACCGACGGGTTCGGTTTTTCCGACACTCGGCCCGCCGGTCGTCG 720
TTACTTCAACACCGACGCCGAATCCCAGGTTGGTCGCGGTTTTGGGAGGGGTTGGCCGGG 780
TCGACGGGTGAATATCGACCCATTCGGTGCCGGTCGTGC~GCCGCCCGCCCAGTTACCCGG 840
ATTCGACGAAGGTGGGGGGTTGCGCCCGANTAAGTT 876
(2) INFORMATION
FOR SEQ
ID N0:20:
(i) S EQUENCE S:
CHARACTERISTIC
(A) LENGTH:1021 basepairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY:
linear
(xi) S EQUENCE :
DESCRIPTION:
SEQ ID
N0:2'0
ATCCCCCCGGGCTGCAGGAATTCGGCACGAGAGACAAAP.TTCCACGCGTT AATGCAGGAA60
CAGATTCATAACGAATTCACAGCGGCACAACAATATGTC'GCGATCGCGGT TTATTTCGAC120
AGCGAAGACCTGCCGCAGTTGGCGAAGCATTTTTACAGCCAAGCGGTCGA GGAACGAAAC180
CATGCAATGATGCTCGTGCAACACCTGCTCGACCGCGACCTTCGTGTCGA AATTCCCGGC240
GTAGACACGGTGCGAAACCAGTTCGACAGACCCCGCGAGGCACTGGCGCT GGCGCTCGAT300
CAGGAACGCACAGTCACCGACCAGGTCGGTCGGCTGACAGCGGTGGCCCG CGACGAGGGC360
GATTTCCTCGGCGAGCAGTTCATGCAGTGGTTCTTGCAGGAACAGATCGA AGAGGTGGCC420
TTGATGGCAACCCTGGTGCGGGTTGCCGATCGGGCCGGGGCCAACCTGTT CGAGCTAGAG480
AACTTCGTCGCACGTGAAGTGGATGTGGCGCCGGCCGCATCAGGCGCCCC GCACGCTGCC540
GGGGGCCGCCTCTAGATCCCTGGGGGGGATCAGCGAGTGGTCCCGTTCGC CCGCCCGTCT600
TCCAGCCAGGCCTTGGTGCGGCCGGGGTGGTGAGTACC~~TCCAGGCCAC CCCGACCTCC660
CGGNAAAAGTCGATGTCCTCGTACTCATCGACGTTCCAGGAGTACACCGC CCGGCCCTGA720
GCTGCCGAGCGGTCAACGAGTTGCGGATATTCCTTTAACGCAGGCAGTGA GGGTCCCACG780
GCGGTTGGCCCGACCGCCGTGGCCGCACTGCTGGTCAGG'PATCGGGGGGT CTTGGCGAGC840
CA 02268036 1999-10-12
73
AACAACGTCG GCAGGAGGGG TGGAGCCCGC CGGATCCGCA GACCGGGGGG GCGAAAACGA 900
CATCAACACC GCACGGGATC GATCTGCGGA GGGGGGTGCG GGAATACCGA ACCGGTGTAG 960
GAGCGCCAGC AGTTGTTTTT CCACCAGCGA AGCGTTTTCG GGTCATCGGN GGCNNTTAAG 1020
T
1021
(2) INFORMATION FOR SEQ ID N0:21:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 321 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:21:
CGTGCCGACG AACGGAAGAA CACAACCATG AAGATGGTcJA AATCGATCGC CGCAGGTCTG 60
ACCGCCGCGG CTGCAATCGG CGCCGCTGCG GCCGGTGTGA CTTCGATCAT GGCTGGCGGN 120
CCGGTCGTAT ACCAGATGCA GCCGGTCGTC TTCGGCGCGC CACTGCCGTT GGACCCGGNA 180
TCCGCCCCTG ANGTCCCGAC CGCCGCCCAG TGGACCAGNC TGCTCAACAG NCTCGNCGAT 240
CCCAACGTGT CGTTTGNGAA CAAGGGNAGT CTGGTCGAGG GNGGNATCGG NGGNANCGAG 300
GGNGNGNATC GNCGANCACA A 321
(2) INFORMATION
FOR
SEQ
ID N0:22:
(i) S EQUENCE CHARACTERISTICS:
(A) LENGTH: 373 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) EQUENCE DESCRIPTION: SEQ
S ID N0:2:2:
TCTTATCGGTTCCGGTTGGC GACGGGTTTT GGGNGCGGGTGGTTAACCCG CTCGGCCAGC60
CGATCGACGGGCGCGGAGAC GTCGACTCCG ATACTCGGCGCGCGCTGGAG CTCCAGGCGC120
CCTCGGTGGTGNACCGGCAA GGCGTGAAGG AGCCGTTGNAGACCGGGATC AAGGCGATTG180
ACGCGATGACCCCGATCGGC CGCGGGCAGC GCCAGCTGA.TCATCGGGGAC CGCAAGACCG240
GCAAAAACCGCCGTCTGTGT CGGACACCAT CCTCAAACCAGCGGGAAGAA CTGGGAGTCC300
GGTGGATCCCAAGAAGCAGG TGCGCTTGTG TATACGTTGGCCATCGGGCA AGAAGGGGAA360
CTTACCATCGCCG 373
(2) INFORMATION FOR SEQ ID N0:23:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 352 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
CA 02268036 1999-10-12
74
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:23:
GTGACGCCGTGATGGGATTCCTGGGCGGGGCCGGTCCGCTGGCGGTGGTGGATCAGCAAC 60
TGGTTACCCGGGTGCCGCAAGGCTGGTCGTTTGCTCAGGCAGCCGCTGTGCCGGTGGTGT 120
TCTTGACGGCCTGGTACGGGTTGGCCGATTTAGCCGAG.ATCAAGGCGGGCGAATCGGTGC 180
TGATCCATGCCGGTACCGGCGGTGTGGGCATGGCGGCTGTGCAGCTGGCTCGCCAGTGGG 240
GCGTGGAGGTTTTCGTCACCGCCAGCCGTGGNAAGTGGGACACGCTGCGCGCCATNGNGT 300
TTGACGACGANCCATATCGGNGATTCCCNCACATNCGA;~GTTCCGANGGAGA 352
(2) INFORMATION FOR SEQ ID N0:24:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 726 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID :
NO::?4
GAAATCCGCG TTCATTCCGT TCGACCAGCG GCTGGCGATAATCGACGAAG TGATCAAGCC60
GCGGTTCGCG GCGCTCATGG GTCACAGCGA GTAATCAGC:AAGTTCTCTGG TATATCGCAC120
CTAGCGTCCA GTTGCTTGCC AGATCGCTTT CGTACCGTC:ATCGCATGTAC CGGTTCGCGT180
GCCGCACGCT CATGCTGGCG GCGTGCATCC TGGCCACGGGTGTGGCGGGT CTCGGGGTCG240
GCGCGCAGTC CGCAGCCCAA ACCGCGCCGG TGCCCGACTACTACTGGTGC CCGGGGCAGC300
CTTTCGACCC CGCATGGGGG CCCAACTGGG ATCCCTACF~CCTGCCATGAC GACTTCCACC360
GCGACAC~CGA CGGCCCCGAC CACAGCCGCG ACCCATCCTC GAAGGTCCCG420
ACTACCCCGG
TGCTTGACGA TCCCGGTGCT GCGCCGCCGC CCCCGGCTGCCGGTGGCGGC GCATAGCGCT980
CGTTGACCGG GCCGCATCAG CGAATACGCG TATAAACCCGGGCGTGCCCC CGGCAAGCTA540
CGACCCCCGG CGGGGCAGAT TTACGCTCCC GTGCCGATGGATCGCGCCGT CCGATGACAG600
AAAATAGGCG ACGGTTTTGG CAACCGCTTG GAGGACGCTTGAAGGGAACC TGTCATGAAC660
GGCGACAGCG CCTCCACCAT CGACATCGAC AAGGTTGTTACCCGCACACC CGTTCGCCGG720
ATCGTG 726
(2) INFORMATION FOR SEQ ID N0:25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 580 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID
N0:2.5:
CGCGACGACG ACGAACGTCG GGCCCACCAC CGCCTATGCc.;TTGATGCAGG CGACCGGGAT60
CA 02268036 1999-10-12
GGTCGCCGAC CATATCCAAGCATGCTGGGT GCCCACTGAGCGACCTTTTG ACCAGCCGGG120
CTGCCCGATG GCGGCCCGGTGAAGTCATTG CGCCGGGGCTTGTGCACCTG ATGAACCCGA180
ATAGGGAACA ATAGGGGGGTGATTTGGCAG TTCAATGTCGGGTATGGCTG GAAATCCAAT240
GGCGGGGCAT GCTCGGCGCCGACCAGGCTC GCGCAGGCGGGCCAGCCCGA ATCTGGAGGG300
AGCACTCAAT GGCGGCGATGAAGCCCCGGA CCGGCGACGGTCCTTTGGAA GCAACTAAGG360
AGGGGCGCGG CATTGTGATGCGAGTACCAC TTGAGGGTGGCGGTCGCCTG GTCGTCGAGC420
TGACACCCGA CGAAGCCGCCGCACTGGGTG ACGAACTC.~1AAGGCGTTACT AGCTAAGACC480
AGCCCAACGG CGAATGGTCGGCGTTACGCG CACACCTTCCGGTAGATGTC CAGTGTCTGC540
TCGGCGATGT ATGCCCAGGAGAACTCTTGG ATACAGCGCT 580
(2) INFORMATION
FOR SEQ ID N0:26:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 160 base pairs
(B) TYPE: nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION:
SEQ ID N0:26:
AACGGAGGCG CCGGGGGTTTTGGCGGGGCC GGGGCGGTC;GGCGGCAACGG CGGGGCCGGC60
GGTACCGCCG GGTTGTTCGGTGTCGGCGGG GCCGGTGGGGCCGGAGGCAA CGGCATCGCC120
GGTGTCACGG GTACGTCGGCCAGCACACCG GGTGGATCC;G 160
(2) INFORMATION FOR SEQ ID N0:27:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 272 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:27:
GACACCGATA CGATGGTGAT GTACGCCAAC GTTGTCGACA CGCTCGAGGC GTTCACGATC 60
CAGCGCACAC CCGACGGCGT GACCATCGGC GATGCGGCCC CGTTCGCGGA GGCGGCTGCC 120
AAGGCGATGG GAATCGACAA GCTGCGGGTA ATTCATACCG GAATGGACCC CGTCGTCGCT 180
GAACGCGAAC AGTGGGACGA CGGCAACAAC ACGTTGGCGT TGGCGCCCGG TGTCGTTGTC 240
GCCTACGAGC GCAACGTACA GACCAACGCC CG 272
(2) INFORMATION FOR SEQ ID N0:28:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 317 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
CA 02268036 1999-10-12
76
(D) TOPOLOGY:
linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:28:
GCAGCCGGTGGTTCTCGGACTATCTGCGCACGGTGACGCAGCGCGACGTGCGCGAGCTGA 60
AGCGGATCGAGCAGACGGATCGCCTGCCGCGGTTCATGCGCTACCTGGCCGCTATCACCG 120
CGCAGGAGCTGAACGTGGCCGAAGCGGCGCGGGTCATCGGGGTCGACGCGGGGACGATCC 180
GTTCGGATCTGGCGTGGTTCGAGACGGTCTATCTGGTACATCGCCTGCCCGCCTGGTCGC 240
GGAATCTGACCGCGAAGATCAAGAAGCGGTCAAAGATCCACGTCGTCGACAGTGGCTTCG 300
CGGCCTGGTTGCGCGGG 317
(2) INFORMATION FOR SEQ ID N0:29:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 182 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:'<?9:
GATCGTGGAG CTGTCGATGA ACAGCGTTGC CGGACGCGC:G GCGGCCAGCA CGTCGGTGTA 60
GCAGCGCCGG ACCACCTCGC CGGTGGGCAG CATGGTGATG ACCACGTCGG CCTCGGCCAC 120
CGCTTCGGGC GCGCTACGAA ACACCGCGAC ACCGTGCGC:G GCGGCGCCGG ACGCCGCCGT 180
GG 182
(2) INFORMATION FOR SEQ ID N0:30:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 308 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:30:
GATCGCGAAG TTTGGTGAGC AGGTGGTCGA CGCGAAAGTC TGGGCGCCTG CGAAGCGGGT 60
CGGCGTTCAC GAGGCGAAGA CACGCCTGTC CGAGCTGCTG CGGCTCGTCT ACGGCGGGCA 120
GAGGTTGAGA TTGCCCGCCG CGGCGAGCCG GTAGCAAAGC TTGTGCCGCT GCATCCTCAT 180
GAGACTCGGC GGTTAGGCAT TGACCATGGC GTGTACCGCG TGCCCGACGA TTTGGACGCT 240
CCGTTGTCAG ACGACGTGCT CGAACGCTTT CACCGGTGAA GCGCTACCTC ATCGACACCC 300
ACGTTTGG 308
(2) INFORMATION FOR SEQ ID N0:31:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 267 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
CA 02268036 1999-10-12
77
(D) TOPOLOGY:
linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:31:
CCGACGACGAGCAACTCACGTGGATGATGGTCGGCAGCGG CATTGAGGACGGAGAGAATC 60
CGGCCGAAGCTGCCGCGCGGCAAGTGCTCATAGTGACCGG CCGTAGAGGGCTCCCCCGAT 120
GGCACCGGACTATTCTGGTGTGCCGCTGGCCGGTAAGAGC GGGTAAAAGAATGTGAGGGG 180
ACACGATGAGCAATCACACCTACCGAGTGATCGAGATCGT CGGGACCTCGCCCGACGGCG 290
TCGACGCGGCAATCCAGGGCGGTCTGG 267
(2) INFORMATION
FOR SEQ
ID N0:32:
(i) S EQUENCE RACTERISTICS:
CHA
(A) LENGTH:1539 basepairs
(B) TYPE: ucleic
n acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY: linear
(xi) S EQUENCE EQ ID :
DESCRIPTION: N0:32
S
CTCGTGCCGAAAGAATGTGAGGGGACACGATGAGCAATC;ACACCTACCGA GTGATCGAGA60
TCGTCGGGACCTCGCCCGACGGCGTCGACGCGGCAATCC;AGGGCGGTCTG GCCCGAGCTG120
CGCAGACCATGCGCGCGCTGGACTGGTTCGAAGTACAGTCAATTCGAGGC CACCTGGTCG180
ACGGAGCGGTCGCGCACTTCCAGGTGACTATGAAAGTCGGCTTCCGCTGG AGGATTCCTG240
AACCTTCAAGCGCGGCCGATAACTGAGGTGCATCATTAF,GCGACTTTTCC AGAACATCCT300
GACGCGCTCGAAACGCGGTTCAGCCGACGGTGGCTCCGCCGAGGCGCTGC CTCCAAAATC360
CCTGCGACAATTCGTCGGCGGCGCCTACAAGGAAGTCGGTGCTGAATTCG TCGGGTATCT420
GGTCGACCTGTGTGGGCTGCAGCCGGACGAAGCGGTGCTCGACGTCGGCT GCGGCTCGGG480
GCGGATGGCGTTGCCGCTCACCGGCTATCTGAACAGCGA.GGGACGCTACG CCGGCTTCGA540
TATCTCGCAGAAAGCCATCGCGTGGTGCCAGGAGCACATCACCTCGGCGC ACCCCAACTT600
CCAGTTCGAGGTCTCCGACATCTACAACTCGCTGTACAACCCGAAAGGGA AATACCAGTC660
ACTAGACTTTCGCTTTCCATATCCGGATGCGTCGTTCGATGTGGTGTTTC TTACCTCGGT720
GTTCACCCACATGTTTCCGCCGGACGTGGAGCACTATCTGGACGAGATCT CCCGCGTGCT780
GAAGCCCGGCGGACGATGCCTGTGCACGTACTTCTTGCTCAATGACGAGT CGTTAGCCCA840
CATCGCGGAAGGAAAGAGTGCGCACAACTTCCAGCATGAGGGACCGGGTT ATCGGACAAT900
CCACAAGAAGCGGCCCGAAGAAGCAATCGGCTTGCCGGAGACCTTCGTCA GGGATGTCTA960
TGGCAAGTTCGGCCTCGCCGTGCACGAACCATTGCACTACGGCTCATGGA GTGGCCGGGA1020
ACCACGCCTAAGCTTCCAGGACATCGTCATCGCGACCAAAACCGCGAGCT AGGTCGGCAT1080
CCGGGAAGCATCGCGACACCGTGGCGCCGAGCGCCGCTGCCGGCAGGCCG ATTAGGCGGG1140
CAGATTAGCCCGCCGCGGCTCCCGGCTCCGAGTACGGCGCCCCGAATGGC GTCACCGGCT1200
CA 02268036 1999-10-12
78
GGTAACCACGCTTGCGCGCCTGGGCGGCGGCCTGCCGGATCAGGTGGTAG ATGCCGACAA1260
AGCCTGCGTGATCGGTCATCACCAACGGTGACAGCAGCCGGTTGTGCACC AGCGCGAACG1320
CCACCCCGGTCTCCGGGTCTGTCCAGCCGATCGAGCCGCCCAAGCCCACA TGACCAAACC1380
CCGGCATCACGTTGCCGATCGGCATACCGTGATAGCCAAGATGAAAATTT AAGGGCACCA1440
ATAGATTTCGATCCGGCAGAACTTGCCGTCGGTTGCGGGTCAGGCCCGTG ACCAGCTCCC1500
GCGACAAGAACCGTATGCCGTCGATCTCGCCTCGTGCCG 1539
(2) INFORMATION
FOR SEQ
ID N0:33:
(i) SEQUENCE RACTERISTICS:
CHA
(A) LENGTH:851 base
pairs
(B) TYPE:ucleic
n acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi) SEQUENCE :
DESCRIPTION:
SEQ ID
N0::33
CTGCAGGGTGGCGTGGATGAGCGTCACCGCGGGGCAGGCCGAGCTGACCG CCGCCCAGGT60
CCGGGTTGCTGCGGCGGCCTACGAGACGGCGTATGGGC'~GACGGTGCCCC CGCCGGTGAT120
CGCCGAGAACCGTGCTGAACTGATGATTCTGATAGCGA(:CAACCTCTTGG GGCAAAACAC180
CCCGGCGATCGCGGTCAACGAGGCCGAATACGGCGAGATGTGGGCCCAAG ACGCCGCCGC240
GATGTTTGGCTACGCCGCGGCGACGGCGACGGCGACGGC:GACGTTGCTGC CGTTCGAGGA300
GGCGCCGGAGATGACCAGCGCGGGTGGGCTCCTCGAGCAGGCCGCCGCGG TCGAGGAGGC360
CTCCGACACCGCCGCGGCGAACCAGTTGATGAACAATG7.'GCCCCAGGCGC TGAAACAGTT420
GGCCCAGCCCACGCAGGGCACCACGCCTTCTTCCAAGCTGGGTGGCCTGT GGAAGACGGT480
CTCGCCGCATCGGTCGCCGATCAGCAACATGGTGTCGATGGCCAACAACC ACATGTCGAT540
GACCAACTCGGGTGTGTCGATGACCAACACCTTGAGCTC',GATGTTGAAGG GCTTTGCTCC600
GGCGGCGGCCGCCCAGGCCGTGCAAACCGCGGCGCAAAACGGGGTCCGGG CGATGAGCTC660
GCTGGGCAGCTCGCTGGGTTCTTCGGGTCTGGGCGGTGGGGTGGCCGCCA ACTTGGGTCG720
GGCGGCCTCGGTACGGTATGGTCACCGGGATGGCGGAAAATATGCANAGT CTGGTCGGCG780
GAACGGTGGTCCGGCGTAAGGTTTACCCCCGTTTTCTGGATGCGGTGAAC TTCGTCAACG840
GAAACAGTTAC 851
(2) INFORMATION FOR SEQ ID N0:34:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 254 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:34:
CA 02268036 1999-10-12
79
GATCGATCGGGCGGAAATTTGGACCAGATTCGCCTCCGGCGATAACCCAATCAATCGAAC 60
CTAGATTTATTCCGTCCAGGGGCCCGAGTAATGGCTCGCAGGAGAGGAACCTTACTGCTG 120
CGGGCACCTGTCGTAGGTCCTCGATACGGCGGAAGGCGTCGACATTTTCCACCGACACCC 180
CCATCCAAACGTTCGAGGGCCACTCCAGCTTGTGAGCGAGGCGACGCAGTCGCAGGCTGC 240
GCTTGGTCAAGATC 254
(2) INFORMATION
FOR SEQ
ID N0:35:
(i) S EQUENCE RACTERISTICS:
CHA
(A) LENGTH:1227 basepairs
(B) TYPE:ucleic
n acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY: linear
(xi) S EQUENCE EQ ID :
DESCRIPTION: N0::35
S
GATCCTGACCGAAGCGGCCGCCGCCAAGGCGAAGTCGC'CGTTGGACCAGG AGGGACGGGA60
CGATCTGGCGCTGCGGATCGCGGTTCAGCCGGGGGGGT(sCGCTGGATTGC GCTATAACCT120
TTTCTTCGACGACCGGACGCTGGATGGTGACCAAACCG(:GGAGTTCGGTG GTGTCAGGTT180
GATCGTGGACCGGATGAGCGCGCCGTATGTGGAAGGCGC:GTCGATCGATT TCGTCGACAC240
TATTGAGAAGCAAGGTTCACCATCGACAATCCCAACGCC;ACCGGCTCCTG CGCGTGCGGG300
GATTCGTTCAACTGATAAAACGCTAGTACGACCCCGCGGTGCGCAACACG TACGAGCACA360
CCAAGACCTGACCGCGCTGGAAAAGCAACTGAGCGATGC;CTTGCACCTGA CCGCGTGGCG420
GGCCGCCGGCGGCAGGTGTCACCTGCATGGTGAACAGCACCTGGGCCTGA TATTGCGACC480
AGTACACGATTTTGTCGATCGAGGTCACTTCGACCTGGGAGAACTGCTTG CGGAACGCGT540
CGCTGCTCAGCTTGGCCAAGGCCTGATCGGAGCGCTTGT'CGCGCACGCCG TCGTGGATAC600
CGCACAGCGCATTGCGAACGATGGTGTCCACATCGCGGTTCTCCAGCGCG TTGAGGTATC660
CCTGAATCGCGGTTTTGGCCGGTCCCTCCGAGAATGTGC'CTGCCGTGTTG GCTCCGTTGG720
TGCGGACCCCGTATATGATCGCCGCCGTCATAGCCGACACCAGCGCGAGG GCTACCACAA780
TGCCGATCAGCAGCCGCTTGTGCCGTCGCTTCGGGTAGGACACCTGCGGC GGCACGCCGG840
GATATGCGGCGGGCGGCAGCGCCGCGTCGTCTGCCGGTCCCGGGGCGAAG GCCGGTTCGG900
CGGCGCCGAGGTCGTGGGGGTAGTCCAGGGCTTGGGGTTCGTGGGATGAG GGCTCGGGGT960
ACGGCGCCGGTCCGTTGGTGCCGACACCGGGGTTCGGCGAGTGGGGACCG GGCATTGTGG1020
TTCTCCTAGGGTGGTGGACGGGACCAGCTGCTAGGGCGACAACCGCCCGT CGCGTCAGCC1080
GGCAGCATCGGCAATCAGGTGAGCTCCCTAGGCAGGCTAGCGCAACAGCT GCCGTCAGCT1140
CTCAACGCGACGGGGCGGGCCGCGGCGCCGATAATGTTGAAAGACTAGGC AACCTTAGGA1200
ACGAAGGACGGAGATTTTGTGACGATC 1227
CA 02268036 1999-10-12
(2) INFORMATION FOR SEQ ID N0:36:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 181 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:36:
GCGGTGTCGG CGGATCCGGC GGGTGGTTGA ACGGCAACGG CGGGGCCGGC GGGGCCGGCG 60
GGACCGGCGC TAACGGTGGT GCCGGCGGCA ACGCCTGG'TT GTTCGGGGCC GGCGGGTCCG 120
GCGGNGCCGG CACCAATGGT GGNGTCGGCG GGTCCGGCGG ATTTGTCTAC GGCAACGGCG 180
G 181
(2) INFORMATION FOR SEQ ID N0:37:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 290 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:37:
GCGGTGTCGGCGGATCCGGCGGGTGGTTGAACGGCAACGGCGGTGTCGGCGGCCGGGGCG 60
GCGACGGCGTCTTTGCCGGTGCCGGCGGCCAGGGCGGCC;TCGGTGGGCAGGGCGGCAATG 120
GCGGCGGCTCCACCGGCGGCAACGGCGGTCTTGGCGGCGCGGGCGGTGGCGGAGGCAACG 180
CCCCGGACGGCGGCTTCGGTGGCAACGGCGGTAAGGGTGGCCAGGGCGGNATTGGCGGCG 240
GCACTCAGAGCGCGACCGGCCTCGGNGGTGACGGCGGTGACGGCGGTGAC 290
(2) INFORMATION FOR SEQ ID N0:38:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 34 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:38:
GATCCAGTGG CATGGNGGGT GTCAGTGGAA GCAT 34
(2) INFORMATION FOR SEQ ID N0:39:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 155 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:39:
CA 02268036 1999-10-12
81
GATCGCTGCT CGTCCCCCCC TTGCCGCCGA CGCCACCGGT CCCACCGTTA CCGAACAAGC 60
TGGCGTGGTC GCCAGCACCC CCGGCACCGC CGACGCCGGA GTCGAACAAT GGCACCGTCG 120
TATCCCCACC ATTGCCGCCG GNCCCACCGG CACCG 155
(2) INFORMATION FOR SEQ ID N0:40:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 53 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:~40:
ATGGCGTTCA CGGGGCGCCG GGGACCGGGC AGCCCGGNGG GGCCGGGGGG TGG 53
(2) INFORMATION FOR SEQ ID N0:41:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 132 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:~Il:
GATCCACCGC GGGTGCAGAC GGTGCCCGCG GCGCCACCC:C GACCAGCGGC GGCAACGGCG 60
GCACCGGCGG CAACGGCGCG AACGCCACCG TCGTCGGNGG GGCCGGCGGG GCCGGCGGCA 120
AGGGCGGCAA CG 132
(2) INFORMATION FOR SEQ ID N0:42:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 132 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:42:
GATCGGCGGC CGGNACGGNC GGGGACGGCG GCAAGGGCGG NAACGGGGGC GCCGNAGCCA 60
CCNGCCAAGA ATCCTCCGNG TCCNCCAATG GCGCGAATGG CGGACAGGGC GGCAACGGCG 120
GCANCGGCGG CA 132
(2) INFORMATION FOR SEQ ID N0:43:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 702 base pairs
(By TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:43:
CA 02268036 1999-10-12
82
CGGCACGAGGATCGGTACCCCGCGGCATCGGCAGCTGCCGATTCGCCGGGTTTCCCCACC 60
CGAGGAAAGCCGCTACCAGATGGCGCTGCCGAAGTAGGGCGATCCGTTCGCGATGCCGGC 120
ATGAACGGGCGGCATCAAATTAGTGCAGGAACCTTTCAGTTTAGCGACGATAATGGCTAT 180
AGCACTAAGGAGGATGATCCGATATGACGCAGTCGCAGACCGTGACGGTGGATCAGCAAG 240
AGATTTTGAACAGGGCCAACGAGGTGGAGGCCCCGATGGCGGACCCACCGACTGATGTCC 300
CCATCACACCGTGCGAACTCACGGNGGNTAAAAACGCCGCCCAACAGNTGGTNTTGTCCG 360
CCGACAACATGCGGGAATACCTGGCGGCCGGTGCCAAAGAGCGGCAGCGTCTGGCGACCT 420
CGCTGCGCAACGCGGCCAAGGNGTATGGCGAGGTTGATGAGGAGGCTGCGACCGCGCTGG 480
ACAACGACGGCGAAGGAACTGTGCAGGCAGAATCGGCCGGGGCCGTCGGAGGGGACAGTT 540
CGGCCGAACTAACCGATACGCCGAGGGTGGCCACGGCCGGTGAACCCAACTTCATGGATC 600
TCAAAGAAGCGGCAAGGAAGCTCGAAACGGGCGACCAAGGCGCATCGCTCGCGCACTGNG 660
GGGATGGGTGGAACACTTNCACCCTGACGCTGCAAGGCGACG 702
(2) INFORMATION FOR SEQ ID N0:44:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 298 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi} SEQUENCE DESCRIPTION: SEQ ID N0:44:
GAAGCCGCAG CGCTGTCGGG CGACGTGGCG GTCAAAGCGG CATCGCTCGG TGGCGGTGGA 60
GGCGGCGGGG TGCCGTCGGC GCCGTTGGGA TCCGCGATC',G GGGGCGCCGA ATCGGTGCGG 120
CCCGCTGGCG CTGGTGACAT TGCCGGCTTA GGCCAGGGP,A GGGCCGGCGG CGGCGCCGCG 180
CTGGGCGGCG GTGGCATGGG AATGCCGATG GGTGCCGCGC ATCAGGGACA AGGGGGCGCC 240
AAGTCCAAGG GTTCTCAGCA GGAAGACGAG GCGCTCTACA CCGAGGATCC TCGTGCCG 298
(2) INFORMATION FOR SEQ ID N0:45:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1058 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi} SEQUENCE DESCRIPTION: SEQ ID N0:45:
CGGCACGAGG ATCGAATCGC GTCGCCGGGA GCACAGCGTC GCACTGCACC AGTGGAGGAG 60
CCATGACCTA CTCGCCGGGT AACCCCGGAT ACCCGCAAGC GCAGCCCGCA GGCTCCTACG 120
GAGGCGTCAC ACCCTCGTTC GCCCACGCCG ATGAGGGTGC GAGCAAGCTA CCGATGTACC 180
TGAACATCGC GGTGGCAGTG CTCGGTCTGG CTGCGTACT'r CGCCAGCTTC GGCCCAATGT 240
CA 02268036 1999-10-12
83
TCACCCTCAGTACCGAACTCGGGGGGGGTGATGGCGCAGTGTCCGGTGACACTGGGCTGC 300
CGGTCGGGGTGGCTCTGCTGGCTGCGCTGCTTGCCGGGGTGGTTCTGGTGCCTAAGGCCA 360
AGAGCCATGTGACGGTAGTTGCGGTGCTCGGGGTACTCGGCGTATTTCTGATGGTCTCGG 420
CGACGTTTAACAAGCCCAGCGCCTATTCGACCGGTTGGGCATTGTGGGTTGTGTTGGCTT 480
TCATCGTGTTCCAGGCGGTTGCGGCAGTCCTGGCGCTCTTGGTGGAGACCGGCGCTATCA 540
CCGCGCCGGCGCCGCGGCCCAAGTTCGACCCGTATGGACAGTACGGGCGGTACGGGCAGT 600
ACGGGCAGTACGGGGTGCAGCCGGGTGGGTACTACGGTCAGCAGGGTGCTCAGCAGGCCG 660
CGGGACTGCAGTCGCCCGGCCCGCAGCAGTCTCCGCAGCCTCCCGGATATGGGTCGCAGT 720
ACGGCGGCTATTCGTCCAGTCCGAGCCAATCGGGCAGTGGATACACTGCTCAGCCCCCGG 780
CCCAGCCGCCGGCGCAGTCCGGGTCGCAACAATCGCACCAGGGCCCATCCACGCCACCTA 840
CCGGCTTTCCGAGCTTCAGCCCACCACCACCGGTCAGTGCCGGGACGGGGTCGCAGGCTG 900
GTTCGGCTCCAGTCAACTATTCAAACCCCAGCGGGGGCGAGCAGTCGTCGTCCCCCGGGG 960
GGGCGCCGGTCTAACCGGGCGTTCCCGCGTCCGGTCGCGCGTGTGCGCGAAGAGTGAACA 1020
GGGTGTCAGCAAGCGCGGACGATCCTCGTGCCGAATTC 1058
(2) INFORMATION FOR SEQ ID N0:46:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 327 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:46:
CGGCACGAGA GACCGATGCC GCTACCCTCG CGCAGGAGGC AGGTAATTTC GAGCGGATCT 60
CCGGCGACCT GAAAACCCAG ATCGACCAGG TGGAGTCGAC GGCAGGTTCG TTGCAGGGCC 120
AGTGGCGCGG CGCGGCGGGG ACGGCCGCCC AGGCCGCGGT GGTGCGCTTC CAAGAAGCAG 180
CCAATAAGCA GAAGCAGGAA CTCGACGAGA TCTCGACGAA TATTCGTCAG GCCGGCGTCC 240
AATACTCGAG GGCCGACGAG GAGCAGCAGC AGGCGCTGT'C CTCGCAAATG GGCTTCTGAC 300
CCGCTAATAC GAAAAGAAAC GGAGCAA 327
(2) INFORMATION FOR SEQ ID N0:47:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 170 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:47:
CGGTCGCGAT GATGGCGTTG TCGAACGTGA CCGATTCTG'T ACCGCCGTCG TTGAGATCAA 60
CA 02268036 1999-10-12
84
CCAACAACGT GTTGGCGTCG GCAAATGTGC CGNACCCGTG GATCTCGGTG ATCTTGTTCT 120
TCTTCATCAG GAAGTGCACA CCGGCCACCC TGCCCTCGGN TACCTTTCGG 170
(2) INFORMATION FOR SEQ ID N0:48:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 127 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:48:
GATCCGGCGG CACGGGGGGT GCCGGCGGCA GCACCGCTGG CGCTGGCGGC AACGGCGGGG 60
CCGGGGGTGG CGGCGGAACC GGTGGGTTGC TCTTCGGCAA CGGCGGTGCC GGCGGGCACG 120
GGGCCGT 127
(2) INFORMATION FOR SEQ ID N0:49:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 81 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:49:
CGGCGGCAAG GGCGGCACCG CCGGCAACGG GAGCGGCGC;G GCCGGCGGCA ACGGCGGCAA 60
CGGCGGCTCC GGCCTCAACG G 81
(2) INFORMATION FOR SEQ ID N0:50:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 149 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:50:
GATCAGGGCT GGCCGGCTCC GGCCAGAAGG GCGGTAACGG AGGAGCTGCC GGATTGTTTG 60
GCAACGGCGG GGCCGGNGGT GCCGGCGCGT CCAACCAAGC CGGTAACGGC GGNGCCGGCG 120
GAAACGGTGG TGCCGGTGGG CTGATCTGG 149
(2) INFORMATION FOR SEQ ID N0:51:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 355 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:51:
CA 02268036 1999-10-12
CGGCACGAGATCACACCTACCGAGTGATCGAGATCGTCGGGACCTCGCCCGACGGTGTCG 60
ACGCGGNAATCCAGGGCGGTCTGGCCCGAGCTGCGCAGACCATGCGCGCGCTGGACTGGT 120
TCGAAGTACAGTCAATTCGAGGCCACCTGGTCGACGGAGCGGTCGCGCACTTCCAGGTGA 180
CTATGAAAGTCGGCTTCCGCCTGGAGGATTCCTGAACCTTCAAGCGCGGCCGATAACTGA 240
GGTGCATCATTAAGCGACTTTTCCAGAACATCCTGACGCGCTCGAAACGCGGTTCAGCCG 300
ACGGTGGCTCCGCCGAGGCGCTGCCTCCAAAATCCCTGCGACAATTCGTCGGCGG 355
(2) INFORMATION
FOR
SEQ
ID N0:52:
(i) S EQUENCE S:
CHARACTERISTIC
(A) LENGTH:999 base airs
p
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS: le ;
sing
(D) TOPOLOGY:
linear
(xi) EQUENCE :
S DESCRIPTION:
SEQ ID
NO:.'>2
ATGCATCACCATCACCATCACATGCATCAGGTGGACCCC:AACTTGACACG TCGCAAGGGA60
CGATTGGCGGCACTGGCTATCGCGGCGATGGCCAGCGCC:AGCCTGGTGAC CGTTGCGGTG120
CCCGCGACCGCCAACGCCGATCCGGAGCCAGCGCCCCCGGTACCCACAAC GGCCGCCTCG180
CCGCCGTCGACCGCTGCAGCGCCACCCGCACCGGCGACACCTGTTGCCCC CCCACCACCG240
GCCGCCGCCAACACGCCGAATGCCCAGCCGGGCGATCCC;AACGCAGCACC TCCGCCGGCC300
GACCCGAACGCACCGCCGCCACCTGTCATTGCCCCAAAC;GCACCCCAACC TGTCCGGATC360
GACAACCCGGTTGGAGGATTCAGCTTCGCGCTGCCTGCT'GGCTGGGTGGA GTCTGACGCC420
GCCCACTTCGACTACGGTTCAGCACTCCTCAGCAAAACCACCGGGGACCC GCCATTTCCC480
GGACAGCCGCCGCCGGTGGCCAATGACACCCGTATCGTGCTCGGCCGGCT AGACCAAAAG540
CTTTACGCCAGCGCCGAAGCCACCGACTCCAAGGCCGCGGCCCGGTTGGG CTCGGACATG600
GGTGAGTTCTATATGCCCTACCCGGGCACCCGGATCAACCAGGAAACCGT CTCGCTCGAC660
GCCAACGGGGTGTCTGGAAGCGCGTCGTATTACGAAGTCAAGTTCAGCGA TCCGAGTAAG720
CCGAACGGCCAGATCTGGACGGGCGTAATCGGCTCGCCCGCGGCGAACGC ACCGGACGCC780
GGGCCCCCTCAGCGCTGGTTTGTGGTATGGCTCGGGACCGCCAACAACCC GGTGGACAAG840
GGCGCGGCCAAGGCGCTGGCCGAATCGATCCGGCCTTTGGTCGCCCCGCC GCCGGCGCCG900
GCACCGGCTCCTGCAGAGCCCGCTCCGGCGCCGGCGCCGGCCGGGGAAGT CGCTCCTACC960
CCGACGACACCGACACCGCAGCGGACCTTACCGGCCTGA ggg
(2) INFORMATION FOR SEQ ID N0:53:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 332 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
CA 02268036 1999-10-12
86
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:53:
Met His His His His His His Met His Gln Val Asp Pro Asn Leu Thr
1 5 10 15
Arg Arg Lys Gly Arg Leu Ala Ala Leu ,~11a Ile Ala Ala Met Ala Ser
20 25 30
Ala Ser Leu Val Thr Val Ala Val Pro Ala Thr Ala Asn Ala Asp Pro
35 90 45
Glu Pro Ala Pro Pro Val Pro Thr Thr i~la Ala Ser Pro Pro Ser Thr
50 55 60
Ala Ala Ala Pro Pro Ala Pro Ala Thr 1?ro Val Ala Pro Pro Pro Pro
65 70 75 80
Ala Ala Ala Asn Thr Pro Asn Ala Gln F?ro Gly Asp Pro Asn Ala Ala
85 90 95
Pro Pro Pro Ala Asp Pro Asn Ala Pro F?ro Pro Pro Val Ile Ala Pro
100 105 110
Asn Ala Pro Gln Pro Val Arg Ile Asp Asn Pro Val Gly Gly Phe Ser
115 120 125
Phe Ala Leu Pro Ala Gly Trp Val Glu Ser Asp Ala Ala His Phe Asp
130 135 140
Tyr Gly Ser Ala Leu Leu Ser Lys Thr Thr Gly Asp Pro Pro Phe Pro
145 150 155 160
Gly Gln Pro Pro Pro Val Ala Asn Asp T'hr Arg Ile Val Leu Gly Arg
165 170 175
Leu Asp Gln Lys Leu Tyr Ala Ser Ala Glu Ala Thr Asp Ser Lys Ala
180 185 190
Ala Ala Arg Leu Gly Ser Asp Met Gly Glu Phe Tyr Met Pro Tyr Pro
195 200 205
Gly Thr Arg Ile Asn Gln Glu Thr Val Ser Leu Asp Ala Asn Gly Val
210 215 220
Ser Gly Ser Ala Ser Tyr Tyr Glu Val Lys Phe Ser Asp Pro Ser Lys
225 230 235 240
Pro Asn Gly Gln Ile Trp Thr Gly Val Ile Gly Ser Pro Ala Ala Asn
245 250 255
Ala Pro Asp Ala Gly Pro Pro Gln Arg Trp Phe Val Val Trp Leu Gly
260 265 270
Thr Ala Asn Asn Pro Val Asp Lys Gly A.la Ala Lys Ala Leu Ala Glu
275 280 285
Ser Ile Arg Pro Leu Val Ala Pro Pro P:ro Ala Pro Ala Pro Ala Pro
290 295 300
Ala Glu Pro Ala Pro Ala Pro Ala Pro Ala Gly Glu Val Ala Pro Thr
305 310 315 320
CA 02268036 1999-10-12
87
Pro Thr Thr Pro Thr Pro Gln Arg Thr Leu Pro Ala
325 330
(2) INFORMATION FOR SEQ ID N0:59:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:54:
Asp Pro Val Asp Ala Val Ile Asn Thr 'Phr Xaa Asn Tyr Gly Gln Val
1 5 :LO 15
Val Ala Ala Leu
(2) INFORMATION FOR SEQ ID N0:55:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:55:
Ala Val Glu Ser Gly Met Leu Ala Leu Gly Thr Pro Ala Pro Ser
1 5 10 15
(2) INFORMATION FOR SEQ ID N0:56:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:56:
Ala Ala Met Lys Pro Arg Thr Gly Asp Gly Pro Leu Glu Ala Ala Lys
1 5 10 15
Glu Gly Arg
(2) INFORMATION FOR SEQ ID N0:57:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:5'7:
Tyr Tyr Trp Cys Pro Gly Gln Pro Phe Aap Pro Ala Trp Gly Pro
1 5 10 15
CA 02268036 1999-10-12
88
(2) INFORMATION FOR SEQ ID N0:58:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 14 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:58:
Asp Ile Gly Ser Glu Ser Thr Glu Asp Gln Gln Xaa Ala Val
1 5 10
(2) INFORMATION FOR SEQ ID N0:59:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:.'i9:
Ala Glu Glu Ser Ile Ser Thr Xaa Glu Xaa Ile Val Pro
1 5 10
(2) INFORMATION FOR SEQ ID N0:60:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 17 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:E~O:
Asp Pro Glu Pro Ala Pro Pro Val Pro Thr Ala Ala Ala Ala Pro Pro
1 5 10 15
Ala
(2) INFORMATION FOR SEQ ID N0:61:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:61:
Ala Pro Lys Thr Tyr Xaa Glu Glu Leu Lys Gly Thr Asp Thr Gly
1 5 10 15
(2) INFORMATION FOR SEQ ID N0:62:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 amino acids
(B) TYPE: amino acid
CA 02268036 1999-10-12
89
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:62:
Asp Pro Ala Ser Ala Pro Asp Val Pro Thr Ala Ala Gln Gln Thr Ser
1 5 10 15
Leu Leu Asn Asn Leu Ala Asp Pro Asp Val Ser Phe Ala Asp
20 25 30
(2) INFORMATION FOR SEQ ID N0:63:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:i53:
Gly Cys Gly Asp Arg Ser Gly Gly Asn Leu Asp Gln Ile Arg Leu Arg
1 5 10 15
Arg Asp Arg Ser Gly Gly Asn Leu
(2) INFORMATION FOR SEQ ID N0:64:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 187 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:64:
Thr Gly Ser Leu Asn Gln Thr His Asn Arg Arg Ala Asn Glu Arg Lys
1 5 10 15
Asn Thr Thr Met Lys Met Val Lys Ser Ile Ala Ala Gly Leu Thr Ala
20 25 30
Ala Ala Ala Ile Gly Ala Ala Ala Ala Gly Val Thr Ser Ile Met Ala
35 40 45
Gly Gly Pro Val Val Tyr Gln Met Gln Pro Val Val Phe Gly Ala Pro
50 55 60
Leu Pro Leu Asp Pro Ala Ser Ala Pro Asp Val Pro Thr Ala Ala Gln
65 70 75 gp
Leu Thr Ser Leu Leu Asn Ser Leu Ala Asp Pro Asn Val Ser Phe Ala
85 90 95
Asn Lys Gly Ser Leu Val Glu Gly Gly Ile Gly Gly Thr Glu Ala Arg
100 105 110
Ile Ala Asp His Lys Leu Lys Lys Ala Ala Glu His Gly Asp Leu Pro
115 120 125
Leu Ser Phe Ser Val Thr Asn Ile Gln P:ro Ala Ala Ala Gly Ser Ala
CA 02268036 1999-10-12
130 135 140
Thr Ala Asp Val Ser Val Ser Gly Pro Lys Leu Ser Ser Pro Val Thr
195 150 155 160
Gln Asn Val Thr Phe Val Asn Gln Gly Gly Trp Met Leu Ser Arg Ala
165 170 175
Ser Ala Met Glu Leu Leu Gln Ala Ala Gly Xaa
180 185
(2) INFORMATION FOR SEQ ID N0:65:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 148 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:65:
Asp Glu Val Thr Val Glu Thr Thr Ser Val Phe Arg Ala Asp Phe Leu
1 5 :LO 15
Ser Glu Leu Asp Ala Pro Ala Gln Ala Gly Thr Glu Ser Ala Val Ser
20 25 30
Gly Val Glu Gly Leu Pro Pro Gly Ser Ala Leu Leu Val Val Lys Arg
35 40 45
Gly Pro Asn Ala Gly Ser Arg Phe Leu Leu Asp Gln Ala Ile Thr Ser
50 55 60
Ala Gly Arg His Pro Asp Ser Asp Ile Phe Leu Asp Asp Val Thr Val
65 70 75 80
Ser Arg Arg His Ala Glu Phe Arg Leu Glu Asn Asn Glu Phe Asn Val
85 90 95
Val Asp Val Gly Ser Leu Asn Gly Thr T'yr Val Asn Arg Glu Pro Val
100 105 110
Asp Ser Ala Val Leu Ala Asn Gly Asp Glu Val Gln Ile Gly Lys Leu
115 120 125
Arg Leu Val Phe Leu Thr Gly Pro Lys Gln Gly Glu Asp Asp Gly Ser
130 135 140
Thr Gly Gly Pro
145
(2) INFORMATION FOR SEQ ID N0:66:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 230 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:66:
Thr Ser Asn Arg Pro Ala Arg Arg Gly A:rg Arg Ala Pro Arg Asp Thr
CA 02268036 1999-10-12
91
1 5 10 15
Gly Pro Asp Arg Ser Ala Ser Leu Ser Leu Val Arg His Arg Arg Gln
20 25 30
Gln Arg Asp Ala Leu Cys Leu Ser Ser Thr Gln Ile Ser Arg Gln Ser
35 40 45
Asn Leu Pro Pro Ala Ala Gly Gly Ala .Ala Asn Tyr Ser Arg Arg Asn
50 55 60
Phe Asp Val Arg Ile Lys Ile Phe Met Leu Val Thr Ala Val Val Leu
65 70 75 80
Leu Cys Cys Ser Gly Val Ala Thr Ala ;~la Pro Lys Thr Tyr Cys Glu
85 90 95
Glu Leu Lys Gly Thr Asp Thr Gly Gln Ala Cys Gln Ile Gln Met Ser
100 105 110
Asp Pro Ala Tyr Asn Ile Asn Ile Ser Leu Pro Ser Tyr Tyr Pro Asp
115 120 125
Gln Lys Ser Leu Glu Asn Tyr Ile Ala Gln Thr Arg Asp Lys Phe Leu
130 135 140
Ser Ala Ala Thr Ser Ser Thr Pro Arg Glu Ala Pro Tyr Glu Leu Asn
145 150 155 160
Ile Thr Ser Ala Thr Tyr Gln Ser Ala 7:1e Pro Pro Arg Gly Thr Gln
165 7.70 175
Ala Val Val Leu Xaa Val Tyr His Asn Ala Gly Gly Thr His Pro Thr
180 185 190
Thr Thr Tyr Lys Ala Phe Asp Trp Asp Gln Ala Tyr Arg Lys Pro Ile
195 200 205
Thr Tyr Asp Thr Leu Trp Gln Ala Asp Thr Asp Pro Leu Pro Val Val
210 215 220
Phe Pro Ile Val Ala Arg
225 230
(2) INFORMATION FOR SEQ ID N0:67:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 132 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:67:
Thr Ala Ala Ser Asp Asn Phe Gln Leu Ser Gln Gly Gly Gln Gly Phe
1 5 10 15
Ala Ile Pro Ile Gly Gln Ala Met Ala I.le Ala Gly Gln Ile Arg Ser
20 25 30
Gly Gly Gly Ser Pro Thr Val His Ile G:ly Pro Thr Ala Phe Leu Gly
35 40 45
CA 02268036 1999-10-12
92
Leu Gly Val Val Asp Asn Asn Gly Asn Gly Ala Arg Val Gln Arg Val
50 55 60
Val Gly Ser Ala Pro Ala Ala Ser Leu Gly Ile Ser Thr Gly Asp Val
65 70 75 80
Ile Thr Ala Val Asp Gly Ala Pro Ile Asn Ser Ala Thr Ala Met Ala
85 90 95
Asp Ala Leu Asn Gly His His Pro Gly Asp Val Ile Ser Val Asn Trp
100 105 110
Gln Thr Lys Ser Gly Gly Thr Arg Thr Gly Asn Val Thr Leu Ala Glu
115 120 125
Gly Pro Pro Ala
130
(2) INFORMATION FOR SEQ ID N0:68:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 100 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:Ei8:
Val Pro Leu Arg Ser Pro Ser Met Ser Pro Ser Lys Cys Leu Ala Ala
1 5 1.0 15
Ala Gln Arg Asn Pro Val Ile Arg Arg Arg Arg Leu Ser Asn Pro Pro
20 25 30
Pro Arg Lys Tyr Arg Ser Met Pro Ser Fro Ala Thr Ala Ser Ala Gly
35 40 45
Met Ala Arg Val Arg Arg Arg Ala Ile Trp Arg Gly Pro Ala Thr Xaa
50 55 60
Ser Ala Gly Met Ala Arg Val Arg Arg Trp Xaa Val Met Pro Xaa Val
65 70 75 80
Ile Gln Ser Thr Xaa Ile Arg Xaa Xaa Gly Pro Phe Asp Asn Arg Gly
85 90 95
Ser Glu Arg Lys
100
(2) INFORMATION FOR SEQ ID N0:69:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 163 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:69:
Met Thr Asp Asp Ile Leu Leu Ile Asp Thr Asp Glu Arg Val Arg Thr
1 5 10 15
CA 02268036 1999-10-12
93
Leu Thr Leu Asn Arg Pro Gln Ser Arg Asn Ala Leu Ser Ala Ala Leu
20 25 30
Arg Asp Arg Phe Phe Ala Xaa Leu Xaa Asp Ala Glu Xaa Asp Asp Asp
35 40 45
Ile Asp Val Val Ile Leu Thr Gly Ala Asp Pro Val Phe Cys Ala Gly
50 55 60
Leu Asp Leu Lys Val Ala Gly Arg Ala Asp Arg Ala Ala Gly His Leu
65 70 75 gp
Thr Ala Val Gly Gly His Asp Gln Ala Gly Asp Arg Arg Asp Gln Arg
85 90 95
Arg Arg Gly His Arg Arg Ala Arg Thr Gly Ala Val Leu Arg His Pro
100 105 110
Asp Arg Leu Arg Ala Arg Pro Leu Arg Arg His Pro Arg Pro Gly Gly
115 120 125
Ala Ala Ala His Leu Gly Thr Gln Cys Val Leu Ala Ala Lys Gly Arg
130 135 190
His Arg Xaa Gly Pro Val Asp Glu Pro Asp Arg Arg Leu Pro Val Arg
145 150 155 160
Asp Arg Arg
(2) INFORMATION FOR SEQ ID N0:70:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 344 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:70:
Met Lys Phe Val Asn His Ile Glu Pro V'al Ala Pro Arg Arg Ala Gly
1 5 10 15
Gly Ala Val Ala Glu Val Tyr Ala Glu Ala Arg Arg Glu Phe Gly Arg
20 25 30
Leu Pro Glu Pro Leu Ala Met Leu Ser Pro Asp Glu Gly Leu Leu Thr
35 40 45
Ala Gly Trp Ala Thr Leu Arg Glu Thr Leu Leu Val Gly Gln Val Pro
50 55 60
Arg Gly Arg Lys Glu Ala Val Ala Ala Ala Val Ala Ala Ser Leu Arg
65 70 75 80
Cys Pro Trp Cys Val Asp Ala His Thr Thr Met Leu Tyr Ala Ala Gly
85 90 95
Gln Thr Asp Thr Ala Ala Ala Ile Leu Ala Gly Thr Ala Pro Ala Ala
100 105 110
Gly Asp Pro Asn Ala Pro Tyr Val Ala T:rp Ala Ala Gly Thr Gly Thr
115 120 125
CA 02268036 1999-10-12
94
Pro Ala Gly Pro Pro Ala Pro Phe Gly Pro Asp Val Ala Ala Glu Tyr
130 135 140
Leu Gly Thr Ala Val Gln Phe His Phe Ile Ala Arg Leu Val Leu Val
145 150 155 160
Leu Leu Asp Glu Thr Phe.Leu Pro Gly Gly Pro Arg Ala Gln Gln Leu
165 170 175
Met Arg Arg Ala Gly Gly Leu Val Phe Ala Arg Lys Val Arg Ala Glu
180 185 190
His Arg Pro Gly Arg Ser Thr Arg Arg :Leu Glu Pro Arg Thr Leu Pro
195 200 205
Asp Asp Leu Ala Trp Ala Thr Pro Ser Glu Pro Ile Ala Thr Ala Phe
210 215 220
Ala Ala Leu Ser His His Leu Asp Thr Ala Pro His Leu Pro Pro Pro
225 230 235 240
Thr Arg Gln Val Val Arg Arg Val Val Gly Ser Trp His Gly Glu Pro
245 :?50 255
Met Pro Met Ser Ser Arg Trp Thr Asn Glu His Thr Ala Glu Leu Pro
260 265 270
Ala Asp Leu His Ala Pro Thr Arg Leu Ala Leu Leu Thr Gly Leu Ala
275 280 285
Pro His Gln Val Thr Asp Asp Asp Val Ala Ala Ala Arg Ser Leu Leu
290 295 300
Asp Thr Asp Ala Ala Leu Val Gly Ala Leu Ala Trp Ala Ala Phe Thr
305 310 315 320
Ala Ala Arg Arg Ile Gly Thr Trp Ile Gly Ala Ala Ala Glu Gly Gln
325 330 335
Val Ser Arg Gln Asn Pro Thr Gly
340
(2) INFORMATION FOR SEQ ID N0:71:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 485 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:71:
Asp Asp Pro Asp Met Pro Gly Thr Val Ala Lys Ala Val Ala Asp Ala
1 5 10 15
Leu Gly Arg Gly Ile Ala Pro Val Glu Asp Ile Gln Asp Cys Val Glu
20 25 30
Ala Arg Leu Gly Glu Ala Gly Leu Asp Asp Val Ala Arg Val Tyr Ile
35 40 45
Ile Tyr Arg Gln Arg Arg Ala Glu Leu A:rg Thr Ala Lys Ala Leu Leu
CA 02268036 1999-10-12
50 55 60
Gly Val Arg Asp Glu Leu Lys Leu Ser Leu Ala Ala Val Thr Val Leu
65 70 75 g0
Arg Glu Arg Tyr Leu Leu His Asp Glu Gln Gly Arg Pro Ala Glu Ser
85 90 95
Thr Gly Glu Leu Met Asp Arg Ser Ala Arg Cys Val Ala Ala Ala Glu
100 105 110
Asp Gln Tyr Glu Pro Gly Ser Ser Arg Arg Trp Ala Glu Arg Phe Ala
115 120 125
Thr Leu Leu Arg Asn Leu Glu Phe Leu 1?ro Asn Ser Pro Thr Leu Met
130 135 140
Asn Ser Gly Thr Asp Leu Gly Leu Leu Ala Gly Cys Phe Val Leu Pro
145 150 155 160
Ile Glu Asp Ser Leu Gln Ser Ile Phe Ala Thr Leu Gly Gln Ala Ala
165 170 175
Glu Leu Gln Arg Ala Gly Gly Gly Thr Czly Tyr Ala Phe Ser His Leu
180 185 190
Arg Pro Ala Gly Asp Arg Val Ala Ser Thr Gly Gly Thr Ala Ser Gly
195 200 205
Pro Val Ser Phe Leu Arg Leu Tyr Asp Ser Ala Ala Gly Val Val Ser
210 215 220
Met Gly Gly Arg Arg Arg Gly Ala Cys Nlet Ala Val Leu Asp Val Ser
225 230 235 240
His Pro Asp Ile Cys Asp Phe Val Thr F,la Lys Ala Glu Ser Pro Ser
245 250 255
Glu Leu Pro His Phe Asn Leu Ser Val Gly Val Thr Asp Ala Phe Leu
260 265 270
Arg Ala Val Glu Arg Asn Gly Leu His A.rg Leu Val Asn Pro Arg Thr
275 280 285
Gly Lys Ile Val Ala Arg Met Pro Ala Ala Glu Leu Phe Asp Ala Ile
290 295 300
Cys Lys Ala Ala His Ala Gly Gly Asp Pro Gly Leu Val Phe Leu Asp
305 310 315 320
Thr Ile Asn Arg Ala Asn Pro Val Pro Gly Arg Gly Arg Ile Glu Ala
325 330 335
Thr Asn Pro Cys Gly Glu Val Pro Leu Leu Pro Tyr Glu Ser Cys Asn
390 345 350
Leu Gly Ser Ile Asn Leu Ala Arg Met Leu Ala Asp Gly Arg Val Asp
355 360 365
Trp Asp Arg Leu Glu Glu Val Ala Gly Val Ala Val Arg Phe Leu Asp
370 375 380
Asp Val Ile Asp Val Ser Arg Tyr Pro Plze Pro Glu Leu Gly Glu Ala
385 390 395 400
CA 02268036 1999-10-12
96
Ala Arg Ala Thr Arg Lys Ile Gly Leu Gly Val Met Gly Leu Ala Glu
405 410 415
Leu Leu Ala Ala Leu Gly Ile Pro Tyr .Asp Ser Glu Glu Ala Val Arg
420 425 430
Leu Ala Thr Arg Leu Met Arg Arg Ile Gln Gln Ala Ala His Thr Ala
435 440 445
Ser Arg Arg Leu Ala Glu Glu Arg Gly Ala Phe Pro Ala Phe Thr Asp
450 455 460
Ser Arg Phe Ala Arg Ser Gly Pro Arg Arg Asn Ala Gln Val Thr Ser
465 470 475 480
Val Ala Pro Thr Gly
485
(2) INFORMATION FOR SEQ ID N0:72:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 267 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:72:
Gly Val Ile Val Leu Asp Leu Glu Pro Arg Gly Pro Leu Pro Thr Glu
1 5 7.0 15
Ile Tyr Trp Arg Arg Arg Gly Leu Ala Leu Gly Ile Ala Val Val Val
20 25 30
Val Gly Ile Ala Val Ala Ile Val Ile Ala Phe Val Asp Ser Ser Ala
35 40 45
Gly Ala Lys Pro Val Ser Ala Asp Lys Fro Ala Ser Ala Gln Ser His
50 55 60
Pro Gly Ser Pro Ala Pro Gln Ala Pro Gln Pro Ala Gly Gln Thr Glu
65 70 75 80
Gly Asn Ala Ala Ala Ala Pro Pro Gln Gly Gln Asn Pro Glu Thr Pro
85 90 95
Thr Pro Thr Ala Ala Val Gln Pro Pro Pro Val Leu Lys Glu Gly Asp
100 105 110
Asp Cys Pro Asp Ser Thr Leu Ala Val Lys Gly Leu Thr Asn Ala Pro
115 120 125
Gln Tyr Tyr Val Gly Asp Gln Pro Lys Phe Thr Met Val Val Thr Asn
130 135 140
Ile Gly Leu Val Ser Cys Lys Arg Asp Va l Gly Ala Ala Val Leu Ala
145 150 155 160
Ala Tyr Val Tyr Ser Leu Asp Asn Lys A:rg Leu Trp Ser Asn Leu Asp
165 1'70 175
Cys Ala Pro Ser Asn Glu Thr Leu Val Lys Thr Phe Ser Pro Gly Glu
CA 02268036 1999-10-12
97
180 185 190
Gln Val Thr Thr Ala Val Thr Trp Thr Gly Met Gly Ser Ala Pro Arg
195 200 205
Cys Pro Leu Pro Arg Pro Ala Ile Gly Pro Gly Thr Tyr Asn Leu Val
210 215 220
Val Gln Leu Gly Asn Leu Arg Ser Leu :Pro Val Pro Phe Ile Leu Asn
225 230 235 240
Gln Pro Pro Pro Pro Pro Gly Pro Val Pro Ala Pro Gly Pro Ala Gln
245 :?50 255
Ala Pro Pro Pro Glu Ser Pro Ala Gln Gly Gly
260 265
(2) INFORMATION FOR SEQ ID N0:73:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 97 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:73:
Leu Ile Ser Thr Gly Lys Ala Ser His Ala Ser Leu Gly Val Gln Val
1 5 1.0 15
Thr Asn Asp Lys Asp Thr Pro Gly Ala Lys Ile Val Glu Val Val Ala
20 25 30
Gly Gly Ala Ala Ala Asn Ala Gly Val Fro Lys Gly Val Val Val Thr
35 40 45
Lys Val Asp Asp Arg Pro Ile Asn Ser Ala Asp Ala Leu Val Ala Ala
50 55 60
Val Arg Ser Lys Ala Pro Gly Ala Thr Val Ala Leu Thr Phe Gln Asp
65 70 75 80
Pro Ser Gly Gly Ser Arg Thr Val Gln Val Thr Leu Gly Lys Ala Glu
85 90 95
Gln
(2) INFORMATION FOR SEQ ID N0:74:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 364 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:74:
Gly Ala Ala Val Ser Leu Leu Ala Ala G:ly Thr Leu Val Leu Thr Ala
1 5 10 15
Cys Gly Gly Gly Thr Asn Ser Ser Ser Ser Gly Ala Gly Gly Thr Ser
20 25 30
CA 02268036 1999-10-12
98
Gly Ser Val His Cys Gly Gly Lys Lys Glu Leu His Ser Ser Gly Ser
35 40 45
Thr Ala Gln Glu Asn Ala Met Glu Gln Phe Val Tyr Ala Tyr Val Arg
50 55 60
Ser Cys Pro Gly Tyr Thr Leu Asp Tyr Asn Ala Asn Gly Ser Gly Ala
65 70 75 80
Gly Val Thr Gln Phe Leu Asn Asn Glu 'Phr Asp Phe Ala Gly Ser Asp
85 90 95
Val Pro Leu Asn Pro Ser Thr Gly Gln Pro Asp Arg Ser Ala Glu Arg
100 105 110
Cys Gly Ser Pro Ala Trp Asp Leu Pro '.Phr Val Phe Gly Pro Ile Ala
115 120 125
Ile Thr Tyr Asn Ile Lys Gly Val Ser '.Chr Leu Asn Leu Asp Gly Pro
130 135 190
Thr Thr Ala Lys Ile Phe Asn Gly Thr ile Thr Val Trp Asn Asp Pro
145 150 155 160
Gln Ile Gln Ala Leu Asn Ser Gly Thr Asp Leu Pro Pro Thr Pro Ile
165 7.70 175
Ser Val Ile Phe Arg Ser Asp Lys Ser Gly Thr Ser Asp Asn Phe Gln
180 185 190
Lys Tyr Leu Asp Gly Val Ser Asn Gly Ala Trp Gly Lys Gly Ala Ser
195 200 205
Glu Thr Phe Ser Gly Gly Val Gly Val Gly Ala Ser Gly Asn Asn Gly
210 215 220
Thr Ser Ala Leu Leu Gln Thr Thr Asp Gly Ser Ile Thr Tyr Asn Glu
225 230 235 240
Trp Ser Phe Ala Val Gly Lys Gln Leu Asn Met Ala Gln Ile Ile Thr
245 250 255
Ser Ala Gly Pro Asp Pro Val Ala Ile Thr Thr Glu Ser Val Gly Lys
260 265 270
Thr Ile Ala Gly Ala Lys Ile Met Gly Gln Gly Asn Asp Leu Val Leu
275 280 285
Asp Thr Ser Ser Phe Tyr Arg Pro Thr Gln Pro Gly Ser Tyr Pro Ile
290 295 300
Val Leu Ala Thr Tyr Glu Ile Val Cys Ser Lys Tyr Pro Asp Ala Thr
305 310 315 320
Thr Gly Thr Ala Val Arg Ala Phe Met Gln Ala Ala Ile Gly Pro Gly
325 330 335
Gln Glu Gly Leu Asp Gln Tyr Gly Ser Ile Pro Leu Pro Lys Ser Phe
340 345 350
Gln Ala Lys Leu Ala Ala Ala Val Asn Ala Ile Ser
355 360
CA 02268036 1999-10-12
99
(2) INFORMATION FOR SEQ ID N0:75:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 309 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:'75:
Gln Ala Ala Ala Gly Arg Ala Val Arg ~Arg Thr Gly His Ala Glu Asp
1 5 10 15
Gln Thr His Gln Asp Arg Leu His His Gly Cys Arg Arg Ala Ala Val
20 25 30
Val Val Arg Gln Asp Arg Ala Ser Val Ser Ala Thr Ser Ala Arg Pro
35 40 45
Pro Arg Arg His Pro Ala Gln Gly His Arg Arg Arg Val Ala Pro Ser
50 55 60
Gly Gly Arg Arg Arg Pro His Pro His His Val Gln Pro Asp Asp Arg
65 70 75 80
Arg Asp Arg Pro Ala Leu Leu Asp Arg Thr Gln Pro Ala Glu His Pro
85 90 95
Asp Pro His Arg Arg Gly Pro Ala Asp E'ro Gly Arg Val Arg Gly Arg
100 105 110
Gly Arg Leu Arg Arg Val Asp Asp Gly Arg Leu Gln Pro Asp Arg Asp
115 120 125
Ala Asp His Gly Ala Pro Val Arg Gly Arg Gly Pro His Arg Gly Val
130 135 140
Gln His Arg Gly Gly Pro Val Phe Val Arg Arg Val Pro Gly Val Arg
145 150 155 160
Cys Ala His Arg Arg Gly His Arg Arg V'al Ala Ala Pro Gly Gln Gly
165 170 175
Asp Val Leu Arg Ala Gly Leu Arg Val Glu Arg Leu Arg Pro Val Ala
180 185 190
Ala Val Glu Asn Leu His Arg Gly Ser Gln Arg Ala Asp Gly Arg Val
195 200 205
Phe Arg Pro Ile Arg Arg Gly Ala Arg Leu Pro Ala Arg Arg Ser Arg
210 215 220
Ala Gly Pro Gln Gly Arg Leu His Leu Asp Gly Ala Gly Pro Ser Pro
225 230 235 240
Leu Pro Ala Arg Ala Gly Gln Gln Gln Pro Ser Ser Ala Gly Gly Arg
245 2'50 255
Arg Ala Gly Gly Ala Glu Arg Ala Asp Pro Gly Gln Arg Gly Arg His
260 265 270
His Gln Gly Gly His Asp Pro Gly Arg G.ln Gly Ala Gln Arg Gly Thr
275 280 285
CA 02268036 1999-10-12
100
Ala Gly Val Ala His Ala Ala Ala Gly Pro Arg Arg Ala Ala Val Arg
290 295 300
Asn Arg Pro Arg Arg
305
(2) INFORMATION FOR SEQ ID N0:76:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 580 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:'76:
Ser Ala Val Trp Cys Leu Asn Gly Phe '.Chr Gly Arg His Arg His Gly
1 5 .LO 15
Arg Cys Arg Val Arg Ala Ser Gly Trp Arg Ser Ser Asn Arg Trp Cys
20 25 30
Ser Thr Thr Ala Asp Cys Cys Ala Ser hys Thr Pro Thr Gln Ala Ala
35 40 45
Ser Pro Leu Glu Arg Arg Phe Thr Cys C:ys Ser Pro Ala Val Gly Cys
50 55 60
Arg Phe Arg Ser Phe Pro Val Arg Arg Leu Ala Leu Gly Ala Arg Thr
65 70 75 g0
Ser Arg Thr Leu Gly Val Arg Arg Thr Leu Ser Gln Trp Asn Leu Ser
85 90 95
Pro Arg Ala Gln Pro Ser Cys Ala Val Thr Val Glu Ser His Thr His
100 105 110
Ala Ser Pro Arg Met Ala Lys Leu Ala Arg Val Val Gly Leu Val Gln
115 120 125
Glu Glu Gln Pro Ser Asp Met Thr Asn His Pro Arg Tyr Ser Pro Pro
130 135 140
Pro Gln Gln Pro Gly Thr Pro Gly Tyr Ala Gln Gly Gln Gln Gln Thr
145 150 155 160
Tyr Ser Gln Gln Phe Asp Trp Arg Tyr Pro Pro Ser Pro Pro Pro Gln
165 170 175
Pro Thr Gln Tyr Arg Gln Pro Tyr Glu Ala Leu Gly Gly Thr Arg Pro
180 185 190
Gly Leu Ile Pro Gly Val Ile Pro Thr Met Thr Pro Pro Pro Gly Met
195 200 205
Val Arg Gln Arg Pro Arg Ala Gly Met Leu Ala Ile Gly Ala Val Thr
210 215 220
Ile Ala Val Val Ser Ala Gly Ile Gly G.ly Ala Ala Ala Ser Leu Val
225 230 235 240
Gly Phe Asn Arg Ala Pro Ala Gly Pro Se r Gly Gly Pro Val Ala Ala
CA 02268036 1999-10-12
101
245 250 255
Ser Ala Ala Pro Ser Ile Pro Ala Ala Asn Met Pro Pro Gly Ser Val
260 265 270
Glu Gln Val Ala Ala Lys Val Val Pro Ser Val Val Met Leu Glu Thr
275 280 285
Asp Leu Gly Arg Gln Ser Glu Glu Gly ;Ser Gly Ile Ile Leu Ser Ala
290 295 300
Glu Gly Leu Ile Leu Thr Asn Asn His 'Jal Ile Ala Ala Ala Ala Lys
305 310 315 320
Pro Pro Leu Gly Ser Pro Pro Pro Lys 'Chr Thr Val Thr Phe Ser Asp
325 330 335
Gly Arg Thr Ala Pro Phe Thr Val Val C~ly Ala Asp Pro Thr, Ser Asp
340 345 350
Ile Ala Val Val Arg Val Gln Gly Val Ser Gly Leu Thr Pro Ile Ser
355 360 365
Leu Gly Ser Ser Ser Asp Leu Arg Val C~ly Gln Pro Val Leu Ala Ile
370 375 380
Gly Ser Pro Leu Gly Leu Glu Gly Thr Val Thr Thr Gly Ile Val Ser
385 390 395 900
Ala Leu Asn Arg Pro Val Ser Thr Thr C~ly Glu Ala Gly Asn Gln Asn
405 410 915
Thr Val Leu Asp Ala Ile Gln Thr Asp Ala Ala Ile Asn Pro Gly Asn
420 425 430
Ser Gly Gly Ala Leu Val Asn Met Asn Ala Gln Leu Val Gly Val Asn
435 440 445
Ser Ala Ile Ala Thr Leu Gly Ala Asp Ser Ala Asp Ala Gln Ser Gly
450 455 460
Ser Ile Gly Leu Gly Phe Ala Ile Pro Val Asp Gln Ala Lys Arg Ile
465 470 475 480
Ala Asp Glu Leu Ile Ser Thr Gly Lys Ala Ser His Ala Ser Leu Gly
485 490 495
Val Gln Val Thr Asn Asp Lys Asp Thr Pro Gly Ala Lys Ile Val Glu
500 505 510
Val Val Ala Gly Gly Ala Ala Ala Asn Ala Gly Val Pro Lys Gly Val
515 520 525
Val Val Thr Lys Val Asp Asp Arg Pro Ile Asn Ser Ala Asp Ala Leu
530 535 540
Val Ala Ala Val Arg Ser Lys Ala Pro Gly Ala Thr Val Ala Leu Thr
545 550 555 560
Phe Gln Asp Pro Ser Gly Gly Ser Arg Thr Val Gln Val Thr Leu Gly
565 5'70 575
Lys Ala Glu Gln
580
CA 02268036 1999-10-12
102
(2) INFORMATION FOR SEQ ID N0:77:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 233 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:'77:
Met Asn Asp Gly Lys Arg Ala Val Thr Ser Ala Val Leu Val Val Leu
1 5 :LO 15
Gly Ala Cys Leu Ala Leu Trp Leu Ser Gly Cys Ser Ser Pro Lys Pro
20 25 30
Asp Ala Glu Glu Gln Gly Val Pro Val :ier Pro Thr Ala Ser Asp Pro
35 40 45
Ala Leu Leu Ala Glu Ile Arg Gln Ser Leu Asp Ala Thr Lys Gly Leu
50 55 60
Thr Ser Val His Val Ala Val Arg Thr 9'hr Gly Lys Val Asp Ser Leu
65 70 75 g0
Leu Gly Ile Thr Ser Ala Asp Val Asp Val Arg Ala Asn Pro Leu Ala
85 g0 95
Ala Lys Gly Val Cys Thr Tyr Asn Asp Glu Gln Gly Val Pro Phe Arg
100 105 110
Val Gln Gly Asp Asn Ile Ser Val Lys L~eu Phe Asp Asp Trp Ser Asn
115 120 125
Leu Gly Ser Ile Ser Glu Leu Ser Thr Ser Arg Val Leu Asp Pro Ala
130 135 140
Ala Gly Val Thr Gln Leu Leu Ser Gly Val Thr Asn Leu Gln Ala Gln
145 150 155 160
Gly Thr Glu Val Ile Asp Gly Ile Ser Thr Thr Lys Ile Thr Gly Thr
165 170 175
Ile Pro Ala Ser Ser Val Lys Met Leu Asp Pro Gly Ala Lys Ser Ala
180 185 190
Arg Pro Ala Thr Val Trp Ile Ala Gln Asp Gly Ser His His Leu Val
195 200 205
Arg Ala Ser Ile Asp Leu Gly Ser Gly Ser Ile Gln Leu Thr Gln Ser
210 215 220
Lys Trp Asn Glu Pro Val Asn Val Asp
225 230
(2) INFORMATION FOR SEQ ID N0:78:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 66 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
CA 02268036 1999-10-12
103
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:78:
Val Ile Asp Ile Ile Gly Thr Ser Pro Thr Ser Trp Glu Gln Ala Ala
1 5 10 15
Ala Glu Ala Val Gln Arg Ala Arg Asp Ser Val Asp Asp Ile Arg Val
20 25 30
Ala Arg Val Ile Glu Gln Asp Met Ala Val Asp Ser Ala Gly Lys Ile
35 40 45
Thr Tyr Arg Ile Lys Leu Glu Val Ser Phe Lys Met Arg Pro Ala Gln
50 55 60
Pro Arg
(2) INFORMATION FOR SEQ ID N0:79:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 69 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:79:
Val Pro Pro Ala Pro Pro Leu Pro Pro Leu Pro Pro Ser Pro Ile Ser
1 5 7.0 15
Cys Ala Ser Pro Pro Ser Pro Pro Leu E'ro Pro Ala Pro Pro Val Ala
20 25 30
Pro Gly Pro Pro Met Pro Pro Leu Asp Pro Trp Pro Pro Ala Pro Pro
35 40 45
Leu Pro Tyr Ser Thr Pro Pro Gly Ala E~ro Leu Pro Pro Ser Pro Pro
50 55 60
Ser Pro Pro Leu Pro
(2) INFORMATION FOR SEQ ID N0:80:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 355 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:80:
Met Ser Asn Ser Arg Arg Arg Ser Leu Arg Trp Ser Trp Leu Leu Ser
1 5 10 15
Val Leu Ala Ala Val Gly Leu Gly Leu Ala Thr Ala Pro Ala Gln Ala
20 25 30
Ala Pro Pro Ala Leu Ser Gln Asp Arg Phe Ala Asp Phe Pro Ala Leu
35 40 45
CA 02268036 1999-10-12
104
Pro Leu Asp Pro Ser Ala Met Val Ala Gln Val Ala Pro Gln Val Val
50 55 60
Asn Ile Asn Thr Lys Leu Gly Tyr Asn ,Asn Ala Val Gly Ala Gly Thr
65 70 75 80
Gly Ile Val Ile Asp Pro Asn Gly Val 'Val Leu Thr Asn Asn His Val
85 90 95
Ile Ala Gly Ala Thr Asp Ile Asn Ala Phe Ser Val Gly Ser Gly Gln
100 105 110
Thr Tyr Gly Val Asp Val Val Gly Tyr Asp Arg Thr Gln Asp Val Ala
115 120 125
Val Leu Gln Leu Arg Gly Ala Gly Gly Leu Pro Ser Ala Ala Ile Gly
130 135 140
Gly Gly Val Ala Val Gly Glu Pro Val Val Ala Met Gly Asn Ser Gly
145 150 155 160
Gly Gln Gly Gly Thr Pro Arg Ala Val Pro Gly Arg Val Val Ala Leu
165 7.70 175
Gly Gln Thr Val Gln Ala Ser Asp Ser Leu Thr Gly Ala Glu Glu Thr
180 185 190
Leu Asn Gly Leu Ile Gln Phe Asp Ala Ala Ile Gln Pro Gly Asp Ser
195 200 205
Gly Gly Pro Val Val Asn Gly Leu Gly Gln Val Val Gly Met Asn Thr
210 215 220
Ala Ala Ser Asp Asn Phe Gln Leu Ser Gln Gly Gly Gln Gly Phe Ala
225 230 235 240
Ile Pro Ile Gly Gln Ala Met Ala Ile A.la Gly Gln Ile Arg Ser Gly
245 250 255
Gly Gly Ser Pro Thr Val His Ile Gly Pro Thr Ala Phe Leu Gly Leu
260 265 270
Gly Val Val Asp Asn Asn Gly Asn Gly Ala Arg Val Gln Arg Val Val
275 280 285
Gly Ser Ala Pro Ala Ala Ser Leu Gly Ile Ser Thr Gly Asp Val Ile
290 295 300
Thr Ala Val Asp Gly Ala Pro Ile Asn Ser Ala Thr Ala Met Ala Asp
305 310 315 320
Ala Leu Asn Gly His His Pro Gly Asp Val Ile Ser Val Asn Trp Gln
325 330 335
Thr Lys Ser Gly Gly Thr Arg Thr Gly A,sn Val Thr Leu Ala Glu Gly
340 345 350
Pro Pro Ala
355
(2) INFORMATION FOR SEQ ID N0:81:
CA 02268036 1999-10-12
105
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 205 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:81:
Ser Pro Lys Pro Asp Ala Glu Glu Gln Gly Val Pro Val Ser Pro Thr
1 5 10 15
Ala Ser Asp Pro Ala Leu Leu Ala Glu Ile Arg Gln Ser Leu Asp Ala
20 25 30
Thr Lys Gly Leu Thr Ser Val His Val Ala Val Arg Thr Thr Gly Lys
35 40 45
Val Asp Ser Leu Leu Gly Ile Thr Ser Ala Asp Val Asp Val Arg Ala
50 55 60
Asn Pro Leu Ala Ala Lys Gly Val Cys 'Phr Tyr Asn Asp Glu Gln Gly
65 70 75 80
Val Pro Phe Arg Val Gln Gly Asp Asn Ile Ser Val Lys Leu Phe Asp
85 90 95
Asp Trp Ser Asn Leu Gly Ser Ile Ser Glu Leu Ser Thr Ser Arg Val
100 105 110
Leu Asp Pro Ala Ala Gly Val Thr Gln Leu Leu Ser Gly Val Thr Asn
115 120 125
Leu Gln Ala Gln Gly Thr Glu Val Ile Asp Gly Ile Ser Thr Thr Lys
130 135 140
Ile Thr Gly Thr Ile Pro Ala Ser Ser Val Lys Met Leu Asp Pro Gly
145 150 155 160
Ala Lys Ser Ala Arg Pro Ala Thr Val Trp Ile Ala Gln Asp Gly Ser
165 170 175
His His Leu Val Arg Ala Ser Ile Asp Leu Gly Ser Gly Ser Ile Gln
180 185 190
Leu Thr Gln Ser Lys Trp Asn Glu Pro Val Asn Val Asp
195 200 205
(2) INFORMATION FOR SEQ ID N0:82:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 286 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:82:
Gly Asp Ser Phe Trp Ala Ala Ala Asp Gln Met Ala Arg Gly Phe Val
1 5 10 15
Leu Gly Ala Thr Ala Gly Arg Thr Thr L~~u Thr Gly Glu Gly Leu Gln
20 25 30
CA 02268036 1999-10-12
106
His Ala Asp Gly His Ser Leu Leu Leu Asp Ala Thr Asn Pro Ala Val
35 40 45
Val Ala Tyr Asp Pro Ala Phe Ala Tyr Glu Ile Gly Tyr Ile Xaa Glu
50 55 60
Ser Gly Leu Ala Arg Met Cys Gly Glu .Asn Pro Glu Asn Ile Phe Phe
65 70 75 80
Tyr Ile Thr Val Tyr Asn Glu Pro Tyr Val Gln Pro Pro Glu Pro Glu
85 90 95
Asn Phe Asp Pro Glu Gly Val Leu Gly Gly Ile Tyr Arg Tyr His Ala
100 105 110
Ala Thr Glu Gln Arg Thr Asn Lys Xaa c,ln Ile Leu Ala Ser Gly Val
115 120 125
Ala Met Pro Ala Ala Leu Arg Ala Ala Gln Met Leu Ala Ala Glu Trp
130 135 140
Asp Val Ala Ala Asp Val Trp Ser Val '.Phr Ser Trp Gly Glu Leu Asn
145 150 155 160
Arg Asp Gly Val Val Ile Glu Thr Glu hys Leu Arg His Pro Asp Arg
165 170 175
Pro Ala Gly Val Pro Tyr Val Thr Arg Ala Leu Glu Asn Ala Arg Gly
180 185 190
Pro Val Ile Ala Val Ser Asp Trp Met Arg Ala Val Pro Glu Gln Ile
195 200 205
Arg Pro Trp Val Pro Gly Thr Tyr Leu Thr Leu Gly Thr Asp Gly Phe
210 215 220
Gly Phe Ser Asp Thr Arg Pro Ala Gly Arg Arg Tyr Phe Asn Thr Asp
225 230 235 240
Ala Glu Ser Gln Val Gly Arg Gly Phe Gly Arg Gly Trp Pro Gly Arg
245 250 255
Arg Val Asn Ile Asp Pro Phe Gly Ala Gly Arg Gly Pro Pro Ala Gln
260 265 270
Leu Pro Gly Phe Asp Glu Gly Gly Gly Leu Arg Pro Xaa Lys
275 280 285
(2) INFORMATION FOR SEQ ID N0:83:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 173 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:83:
Thr Lys Phe His Ala Leu Met Gln Glu Gln Ile His Asn Glu Phe Thr
1 5 10 15
Ala Ala Gln Gln Tyr Val Ala Ile Ala Val Tyr Phe Asp Ser Glu Asp
20 25 30
CA 02268036 1999-10-12
107
Leu Pro Gln Leu Ala Lys His Phe Tyr Ser Gln Ala Val Glu Glu Arg
35 40 45
Asn His Ala Met Met Leu Val Gln His Leu Leu Asp Arg Asp Leu Arg
50 55 60
Val Glu Ile Pro Gly Val Asp Thr Val .Arg Asn Gln Phe Asp Arg Pro
65 70 75 80
Arg Glu Ala Leu Ala Leu Ala Leu Asp Gln Glu Arg Thr Val Thr Asp
85 90 95
Gln Val Gly Arg Leu Thr Ala Val Ala ,~lrg Asp Glu Gly Asp Phe Leu
100 105 110
Gly Glu Gln Phe Met Gln Trp Phe Leu Gln Glu Gln Ile Glu Glu Val
115 120 125
Ala Leu Met Ala Thr Leu Val Arg Val Ala Asp Arg Ala Gly Ala Asn
130 135 140
Leu Phe Glu Leu Glu Asn Phe Val Ala Arg Glu Val Asp Val Ala Pro
145 150 155 160
Ala Ala Ser Gly Ala Pro His Ala Ala (~ly Gly Arg Leu
165 170
(2) INFORMATION FOR SEQ ID N0:84:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 107 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:84:
Arg Ala Asp Glu Arg Lys Asn Thr Thr Niet Lys Met Val Lys Ser Ile
1 5 L0 15
Ala Ala Gly Leu Thr Ala Ala Ala Ala Ile Gly Ala Ala Ala Ala Gly
20 25 30
Val Thr Ser Ile Met Ala Gly Gly Pro V'al Val Tyr Gln Met Gln Pro
35 40 45
Val Val Phe Gly Ala Pro Leu Pro Leu Asp Pro Xaa Ser Ala Pro Xaa
50 55 60
Val Pro Thr Ala Ala Gln Trp Thr Xaa Leu Leu Asn Xaa Leu Xaa Asp
65 70 75 80
Pro Asn Val Ser Phe Xaa Asn Lys Gly Ser Leu Val Glu Gly Gly Ile
85 90 95
Gly Gly Xaa Glu Gly Xaa Xaa Arg Arg Xaa Gln
100 105
(2) INFORMATION FOR SEQ ID N0:85:
(i) SEQUENCE CHARACTERISTICS:
CA 02268036 1999-10-12
108
(A) LENGTH: 125 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:85:
Val Leu Ser Val Pro Val Gly Asp Gly Phe Trp Xaa Arg Val Val Asn
1 5 10 15
Pro Leu Gly Gln Pro Ile Asp Gly Arg Gly Asp Val Asp Ser Asp Thr
20 25 30
Arg Arg Ala Leu Glu Leu Gln Ala Pro ;Ser Val Val Xaa Arg Gln Gly
35 40 45
Val Lys Glu Pro Leu Xaa Thr Gly Ile Lys Ala Ile Asp Ala Met Thr
50 55 60
Pro Ile Gly Arg Gly Gln Arg Gln Leu :Ile Ile Gly Asp Arg Lys Thr
65 70 75 80
Gly Lys Asn Arg Arg Leu Cys Arg Thr 1?ro Ser Ser Asn Gln Arg Glu
85 90 95
Glu Leu Gly Val Arg Trp Ile Pro Arg Ser Arg Cys Ala Cys Val Tyr
100 105 110
Val Gly His Arg Ala Arg Arg Gly Thr Tyr His Arg Arg
115 120 125
(2) INFORMATION FOR SEQ ID N0:86:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 117 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:86:
Cys Asp Ala Val Met Gly Phe Leu Gly Gly Ala Gly Pro Leu Ala Val
1 5 1.0 15
Val Asp Gln Gln Leu Val Thr Arg Val Fro Gln Gly Trp Ser Phe Ala
20 25 30
Gln Ala Ala Ala Val Pro Val Val Phe Leu Thr Ala Trp Tyr Gly Leu
35 40 45
Ala Asp Leu Ala Glu Ile Lys Ala Gly Glu Ser Val Leu Ile His Ala
50 55 60
Gly Thr Gly Gly Val Gly Met Ala Ala Val Gln Leu Ala Arg Gln Trp
65 70 75 80
Gly Val Glu Val Phe Val Thr Ala Ser Arg Gly Lys Trp Asp Thr Leu
85 90 95
Arg Ala Xaa Xaa Phe Asp Asp Xaa Pro Tyr Arg Xaa Phe Pro His Xaa
100 105 110
Arg Ser Ser Xaa Gly
CA 02268036 1999-10-12
109
115
(2) INFORMATION FOR SEQ ID N0:87:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 103 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:87:
Met Tyr Arg Phe Ala Cys Arg Thr Leu Met Leu Ala Ala Cys Ile Leu
1 5 10 15
Ala Thr Gly Val Ala Gly Leu Gly Val Gly Ala Gln Ser Ala Ala Gln
20 25 30
Thr Ala Pro Val Pro Asp Tyr Tyr Trp Cys Pro Gly Gln Pro Phe Asp
35 40 45
Pro Ala Trp Gly Pro Asn Trp Asp Pro '.Cyr Thr Cys His Asp Asp Phe
50 55 60
His Arg Asp Ser Asp Gly Pro Asp His :ier Arg Asp Tyr Pro Gly Pro
65 70 75 80
Ile Leu Glu Gly Pro Val Leu Asp Asp 1?ro Gly Ala Ala Pro Pro Pro
85 90 95
Pro Ala Ala Gly Gly Gly Ala
100
(2) INFORMATION FOR SEQ ID N0:88:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 88 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:88:
Val Gln Cys Arg Val Trp Leu Glu Ile Gln Trp Arg Gly Met Leu Gly
1 5 10 15
Ala Asp Gln Ala Arg Ala Gly Gly Pro Ala Arg Ile Trp Arg Glu His
20 25 30
Ser Met Ala Ala Met Lys Pro Arg Thr Gly Asp Gly Pro Leu Glu Ala
35 40 45
Thr Lys Glu Gly Arg Gly Ile Val Met Arg Val Pro Leu Glu Gly Gly
50 55 60
Gly Arg Leu Val Val Glu Leu Thr Pro Asp Glu Ala Ala Ala Leu Gly
65 70 75 80
Asp Glu Leu Lys Gly Val Thr Ser
CA 02268036 1999-10-12
110
(2) INFORMATION FOR SEQ ID N0:89:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 95 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:89:
Thr Asp Ala Ala Thr Leu Ala Gln Glu ;~la Gly Asn Phe Glu Arg Ile
1 5 10 15
Ser Gly Asp Leu Lys Thr Gln Ile Asp Gln Val Glu Ser Thr Ala Gly
20 25 30
Ser Leu Gln Gly Gln Trp Arg Gly Ala Ala Gly Thr Ala Ala Gln Ala
35 90 45
Ala Val Val Arg Phe Gln Glu Ala Ala Asn Lys Gln Lys Gln Glu Leu
50 55 60
Asp Glu Ile Ser Thr Asn Ile Arg Gln Ala Gly Val Gln Tyr Ser Arg
65 70 75 80
Ala Asp Glu Glu Gln Gln Gln Ala Leu Ser Ser Gln Met Gly Phe
85 90 95
(2) INFORMATION FOR SEQ ID N0:90:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 166 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:90:
Met Thr Gln Ser Gln Thr Val Thr Val Asp Gln Gln Glu Ile Leu Asn
1 5 7.0 15
Arg Ala Asn Glu Val Glu Ala Pro Met Ala Asp Pro Pro Thr Asp Val
20 25 30
Pro Ile Thr Pro Cys Glu Leu Thr Xaa Xaa Lys Asn Ala Ala Gln Gln
35 40 45
Xaa Val Leu Ser Ala Asp Asn Met Arg Glu Tyr Leu Ala Ala Gly Ala
50 55 60
Lys Glu Arg Gln Arg Leu Ala Thr Ser L~eu Arg Asn Ala Ala Lys Xaa
65 70 75 80
Tyr Gly Glu Val Asp Glu Glu Ala Ala Thr Ala Leu Asp Asn Asp Gly
85 90 95
Glu Gly Thr Val Gln Ala Glu Ser Ala Gly Ala Val Gly Gly Asp Ser
100 105 110
Ser Ala Glu Leu Thr Asp Thr Pro Arg Val Ala Thr Ala Gly Glu Pro
115 120 125
Asn Phe Met Asp Leu Lys Glu Ala Ala Arg Lys Leu Glu Thr Gly Asp
CA 02268036 1999-10-12
111
130 135 140
Gln Gly Ala Ser Leu Ala His Xaa Gly Asp Gly Trp Asn Thr Xaa Thr
145 150 155 160
Leu Thr Leu Gln Gly Asp
165
(2) INFORMATION FOR SEQ ID N0:91:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 5 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:91:
Arg Ala Glu Arg Met
1 5
(2) INFORMATION FOR SEQ ID N0:92:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 263 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:92:
Val Ala Trp Met Ser Val Thr Ala Gly G'ln Ala Glu Leu Thr Ala Ala
1 5 10 15
Gln Val Arg Val Ala Ala Ala Ala Tyr Glu Thr Ala Tyr Gly Leu Thr
20 25 30
Val Pro Pro Pro Val Ile Ala Glu Asn Arg Ala Glu Leu Met Ile Leu
35 40 45
Ile Ala Thr Asn Leu Leu Gly Gln Asn Thr Pro Ala Ile Ala Val Asn
50 55 60
Glu Ala Glu Tyr Gly Glu Met Trp Ala Gln Asp Ala Ala Ala Met Phe
65 70 75 80
Gly Tyr Ala Ala Ala Thr Ala Thr Ala Thr Ala Thr Leu Leu Pro Phe
85 90 95
Glu Glu Ala Pro Glu Met Thr Ser Ala Gly Gly Leu Leu Glu Gln Ala
100 105 110
Ala Ala Val Glu Glu Ala Ser Asp Thr Ala Ala Ala Asn Gln Leu Met
115 120 125
Asn Asn Val Pro Gln Ala Leu Lys Gln Leu Ala Gln Pro Thr Gln Gly
130 135 140
Thr Thr Pro Ser Ser Lys Leu Gly Gly Leu Trp Lys Thr Val Ser Pro
145 150 155 160
His Arg Ser Pro Ile Ser Asn Met Val Ser Met Ala Asn Asn His Met
CA 02268036 1999-10-12
112
165 170 175
Ser Met Thr Asn Ser Gly Val Ser Met 'Phr Asn Thr Leu Ser Ser Met
180 185 190
Leu Lys Gly Phe Ala Pro Ala Ala Ala Ala Gln Ala Val Gln Thr Ala
195 200 205
Ala Gln Asn Gly Val Arg Ala Met Ser ;ier Leu Gly Ser Ser Leu Gly
210 215 220
Ser Ser Gly Leu Gly Gly Gly Val Ala Ala Asn Leu Gly Arg Ala Ala
225 230 235 240
Ser Val Arg Tyr Gly His Arg Asp Gly C~ly Lys Tyr Ala Xaa Ser Gly
245 250 255
Arg Arg Asn Gly Gly Pro Ala
260
(2) INFORMATION FOR SEQ ID N0:93:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 303 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:93:
Met Thr Tyr Ser Pro Gly Asn Pro Gly Tyr Pro Gln Ala Gln Pro Ala
1 5 10 15
Gly Ser Tyr Gly Gly Val Thr Pro Ser F'he Ala His Ala Asp Glu Gly
20 25 30
Ala Ser Lys Leu Pro Met Tyr Leu Asn Ile Ala Val Ala Val Leu Gly
35 40 45
Leu Ala Ala Tyr Phe Ala Ser Phe Gly Pro Met Phe Thr Leu Ser Thr
50 55 60
Glu Leu Gly Gly Gly Asp Gly Ala Val Ser Gly Asp Thr Gly Leu Pro
65 70 75 80
Val Gly Val Ala Leu Leu Ala Ala Leu Leu Ala Gly Val Val Leu Val
85 90 95
Pro Lys Ala Lys Ser His Val Thr Val Val Ala Val Leu Gly Val Leu
100 105 110
Gly Val Phe Leu Met Val Ser Ala Thr Phe Asn Lys Pro Ser Ala Tyr
115 120 125
Ser Thr Gly Trp Ala Leu Trp Val Val Leu Ala Phe Ile Val Phe Gln
130 135 140
Ala Val Ala Ala Val Leu Ala Leu Leu Val Glu Thr Gly Ala Ile Thr
145 150 155 160
Ala Pro Ala Pro Arg Pro Lys Phe Asp Pro Tyr Gly Gln Tyr Gly Arg
165 170 175
CA 02268036 1999-10-12
113
Tyr Gly Gln Tyr Gly Gln Tyr Gly Val Gln Pro Gly Gly Tyr Tyr Gly
180 185 190
Gln Gln Gly Ala Gln Gln Ala Ala Gly Leu Gln Ser Pro Gly Pro Gln
195 200 205
Gln Ser Pro Gln Pro Pro Gly Tyr Gly Ser Gln Tyr Gly Gly Tyr Ser
210 215 220
Ser Ser Pro Ser Gln Ser Gly Ser Gly Tyr Thr Ala Gln Pro Pro Ala
225 230 235 240
Gln Pro Pro Ala Gln Ser Gly Ser Gln C~ln Ser His Gln Gly Pro Ser
245 250 255
Thr Pro Pro Thr Gly Phe Pro Ser Phe :>er Pro Pro Pro Pro Val Ser
260 265 270
Ala Gly Thr Gly Ser Gln Ala Gly Ser Ala Pro Val Asn Tyr Ser Asn
275 280 285
Pro Ser Gly Gly Glu Gln Ser Ser Ser Pro Gly Gly Ala Pro Val
290 295 300
(2) INFORMATION FOR SEQ ID N0:94:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:507 base pairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY:
linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:94:
ATGAAGATGGTGAAATCGATCGCCGCAGGT CTGACCGCCGCGGCTGCAAT CGGCGCCGCT60
GCGGCCGGTGTGACTTCGATCATGGCTGGC GGCCCGGTCGTATACCAGAT GCAGCCGGTC120
GTCTTCGGCGCGCCACTGCCGTTGGACCCG GCATCCGCCCCTGACGTCCC GACCGCCGCC180
CAGTTGACCAGCCTGCTCAACAGCCTCGCC GATCCCAACGTGTCGTTTGC GAACAAGGGC240
AGTCTGGTCGAGGGCGGCATCGGGGGCACC GAGGCGCGCATCGCCGACCA CAAGCTGAAG300
AAGGCCGCCGAGCACGGGGATCTGCCGCTG TCGTTCAGCGTGACGAACAT CCAGCCGGCG360
GCCGCCGGTTCGGCCACCGCCGACGTTTCC GTCTCGGGTCCGAAGCTCTC GTCGCCGGTC420
ACGCAGAACGTCACGTTCGTGAATCAAGGC GGCTGGATGCTGTCACGCGC ATCGGCGATG480
GAGTTGCTGCAGGCCGCAGGGAACTGA 507
(2) INFORMATION FOR SEQ ID N0:95:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 168 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:95:
CA 02268036 1999-10-12
114
Met Lys Met Val Lys Ser Ile Ala Ala Gly Leu Thr Ala Ala Ala Ala
1 5 :LO 15
Ile Gly Ala Ala Ala Ala Gly Val Thr Ser Ile Met Ala Gly Gly Pro
20 25 30
Val Val Tyr Gln Met Gln Pro Val Val I?he Gly Ala Pro Leu Pro Leu
35 40 45
Asp Pro Ala Ser Ala Pro Asp Val Pro Thr Ala Ala Gln Leu Thr Ser
50 55 60
Leu Leu Asn Ser Leu Ala Asp Pro Asn Val Ser Phe Ala Asn Lys Gly
65 70 75 80
Ser Leu Val Glu Gly Gly Ile Gly Gly 7.'hr Glu Ala Arg Ile Ala Asp
85 90 95
His Lys Leu Lys Lys Ala Ala Glu His Gly Asp Leu Pro Leu Ser Phe
100 105 110
Ser Val Thr Asn Ile Gln Pro Ala Ala Ala Gly Ser Ala Thr Ala Asp
115 120 125
Val Ser Val Ser Gly Pro Lys Leu Ser ~~er Pro Val Thr Gln Asn Val
130 135 140
Thr Phe Val Asn Gln Gly Gly Trp Met Leu Ser Arg Ala Ser Ala Met
145 150 155 160
Glu Leu Leu Gln Ala Ala Gly Asn
165
(2) INFORMATION FOR SEQ ID N0:96:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 500 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:96:
CGTGGCAATGTCGTTGACCG TCGGGGCCGG GGTCGCCTCCGCAGATCCCG TGGACGCGGT60
CATTAACACCACCTGCAATT ACGGGCAGGT AGTAGCTGCGCTCAACGCGA CGGATCCGGG120
GGCTGCCGCACAGTTCAACG CCTCACCGGT GGCGCAGTCCTATTTGCGCA ATTTCCTCGC180
CGCACCGCCACCTCAGCGCG CTGCCATGGC CGCGCAATTGCAAGCTGTGC CGGGGGCGGC240
ACAGTACATCGGCCTTGTCG AGTCGGTTGC CGGCTCCTGCAACAACTATT AAGCCCATGC300
GGGCCCCATCCCGCGACCCG GCATCGTCGC CGGGGCTAGGCCAGATTGCC CCGCTCCTCA360
ACGGGCCGCATCCCGCGACC CGGCATCGTC GCCGGGGCT.AGGCCAGATTG CCCCGCTCCT420
CAACGGGCCGCATCTCGTGC CGAATTCCTG CAGCCCGGGGGATCCACTAG TTCTAGAGCG480
GCCGCCACCGCGGTGGAGCT 500
(2) INFORMATION FOR SEQ ID N0:97:
CA 02268036 1999-10-12
115
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 96 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:97:
Val Ala Met Ser Leu Thr Val Gly Ala Gly Val Ala Ser Ala Asp Pro
1 5 10 15
Val Asp Ala Val Ile Asn Thr Thr Cys Asn Tyr Gly Gln Val Val Ala
20 25 30
Ala Leu Asn Ala Thr Asp Pro Gly Ala Ala Ala Gln Phe Asn Ala Ser
35 40 45
Pro Val Ala Gln Ser Tyr Leu Arg Asn Phe Leu Ala Ala Pro Pro Pro
50 55 60
Gln Arg Ala Ala Met Ala Ala Gln Leu C7ln Ala Val Pro Gly Ala Ala
65 70 75 80
Gln Tyr Ile Gly Leu Val Glu Ser Val Ala Gly Ser Cys Asn Asn Tyr
85 90 95
(2) INFORMATION FOR SEQ ID N0:98:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 154 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:98:
ATGACAGAGC AGCAGTGGAA TTTCGCGGGT ATCGAGGCCG CGGCAAGCGC AATCCAGGGA 60
AATGTCACGT CCATTCATTC CCTCCTTGAC GAGGGGAAGC AGTCCCTGAC CAAGCTCGCA 120
GCGGCCTGGG GCGGTAGCGG TTCGGAAGCG TACC 154
(2) INFORMATION FOR SEQ ID N0:99:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 51 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:99:
Met Thr Glu Gln Gln Trp Asn Phe Ala Gly Ile Glu Ala Ala Ala Ser
1 5 10 15
Ala Ile Gln Gly Asn Val Thr Ser Ile His Ser Leu Leu Asp Glu Gly
20 25 30
Lys Gln Ser Leu Thr Lys Leu Ala Ala Ala Trp Gly Gly Ser Gly Ser
35 40 45
CA 02268036 1999-10-12
116
Glu Ala Tyr
(2) INFORMATION FOR SEQ ID NO:100:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 282 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:100:
CGGTCGCGCA CTTCCAGGTG ACTATGAAAG TCGGCTTCC;G NCTGGAGGAT TCCTGAACCT 60
TCAAGCGCGG CCGATAACTG AGGTGCATCA TTAAGCGAC:T TTTCCAGAAC ATCCTGACGC 120
GCTCGAAACG CGGCACAGCC GACGGTGGCT CCGNCGAGC~C GCTGNCTCCA AAATCCCTGA 180
GACAATTCGN CGGGGGCGCC TACAAGGAAG TCGGTGCTGA ATTCGNCGNG TATCTGGTCG 240
ACCTGTGTGG TCTGNAGCCG GACGAAGCGG TGCTCGACGT CG 282
(2) INFORMATION
FOR SEQ
ID N0:101:
(i) S EQUENCE S:
CHARACTERISTIC
(A) LENGTH:3058 basepairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY:
linear
(xi) SEQUENCE EQ ID N0:101:
DESCRIPTION:
S
GATCGTACCCGTGCGAGTGCTCGGGCCGTTTGAGGATGGAGTGCACGTGTCTTTCGTGAT 60
GGCATACCCAGAGATGTTGGCGGCGGCGGCTGACACCCTGCAGAGCATCGGTGCTACCAC 120
TGTGGCTAGCAATGCCGCTGCGGCGGCCCCGACGACTGGGGTGGTGCCCCCCGCTGCCGA 180
TGAGGTGTCGGCGCTGACTGCGGCGCACTTCGCCGCACATGCGGCGATGTATCAGTCCGT 240
GAGCGCTCGGGCTGCTGCGATTCATGACCAGTTCGTGGCCACCCTTGCCAGCAGCGCCAG 300
CTCGTATGCGGCCACTGAAGTCGCCAATGCGGCGGCGGCCAGCTAAGCCAGGAACAGTCG 360
GCACGAGAAACCACGAGAAATAGGGACACGTAATGGTGGATTTCGGGGCGTTACCACCGG 420
AGATCAACTCCGCGAGGATGTACGCCGGCCCGGGTTCGGCCTCGCTGGTGGCCGCGGCTC 480
AGATGTGGGACAGCGTGGCGAGTGACCTGTTTTCGGCCGCGTCGGCGTTTCAGTCGGTGG 590
TCTGGGGTCTGACGGTGGGGTCGTGGATAGGTTCGTCGGCGGGTCTGATGGTGGCGGCGG 600
CCTCGCCGTATGTGGCGTGGATGAGCGTCACCGCGGGGCAGGCCGAGCTGACCGCCGCCC 660
AGGTCCGGGTTGCTGCGGCGGCCTACGAGACGGCGTATGGGCTGACGGTGCCCCCGCCGG 720
TGATCGCCGAGAACCGTGCTGAACTGATGATTCTGATAGCGACCAACCTCTTGGGGCAAA 780
ACACCCCGGCGATCGCGGTCAACGAGGCCGAATACGGCGAGATGTGGGCCCAAGACGCCG 890
CCGCGATGTTTGGCTACGCCGCGGCGACGGCGACGGCGACGGCGACGTTGCTGCCGTTCG 900
CA 02268036 1999-10-12
117
AGGAGGCGCCGGAGATGACCAGCGCGGGTGGGCTCCTCGAGCAGGCCGCCGCGGTCGAGG 960
AGGCCTCCGACACCGCCGCGGCGAACCAGTTGATGAACAATGTGCCCCAGGCGCTGCAAC 1020
AGCTGGCCCAGCCCACGCAGGGCACCACGCCTTCTTCCAAGCTGGGTGGCCTGTGGAAGA 1080
CGGTCTCGCCGCATCGGTCGCCGATCAGCAACATGGTGTCGATGGCCAACAACCACATGT 1140
CGATGACCAACTCGGGTGTGTCGATGACCAACACCTTGAGCTCGATGTTGAAGGGCTTTG 1200
CTCCGGCGGCGGCCGCCCAGGCCGTGCAAACCGCGGCGC;AAAACGGGGTCCGGGCGATGA 1260
GCTCGCTGGGCAGCTCGCTGGGTTCTTCGGGTCTGGGCGGTGGGGTGGCCGCCAACTTGG 1320
GTCGGGCGGCCTCGGTCGGTTCGTTGTCGGTGCCGCAGGCCTGGGCCGCGGCCAACCAGG 1380
CAGTCACCCCGGCGGCGCGGGCGCTGCCGCTGACCAGCC;TGACCAGCGCCGCGGAAAGAG 1440
GGCCCGGGCAGATGCTGGGCGGGCTGCCGGTGGGGCAGATGGGCGCCAGGGCCGGTGGTG 1500
GGCTCAGTGGTGTGCTGCGTGTTCCGCCGCGACCCTATGTGATGCCGCATTCTCCGGCGG 1560
CCGGCTAGGAGAGGGGGCGCAGACTGTCGTTATTTGACC;AGTGATCGGCGGTCTCGGTGT 1620
TTCCGCGGCCGGCTATGACAACAGTCAATGTGCATGACAAGTTACAGGTATTAGGTCCAG 1680
GTTCAACAAGGAGACAGGCAACATGGCCTCACGTTTTAT'GACGGATCCGCACGCGATGCG 1740
GGACATGGCGGGCCGTTTTGAGGTGCACGCCCAGACGGT'GGAGGACGAGGCTCGCCGGAT 1800
GTGGGCGTCCGCGCAAAACATTTCCGGTGCGGGCTGGAGTGGCATGGCCGAGGCGACCTC 1860
GCTAGACACCATGGCCCAGATGAATCAGGCGTTTCGCAA,CATCGTGAACATGCTGCACGG 1920
GGTGCGTGACGGGCTGGTTCGCGACGCCAACAACTACGAGCAGCAAGAGCAGGCCTCCCA 1980
GCAGATCCTCAGCAGCTAACGTCAGCCGCTGCAGCACAATACTTTTACAAGCGAAGGAGA 2040
ACAGGTTCGATGACCATCAACTATCAATTCGGGGATGTCGACGCTCACGGCGCCATGATC 2100
CGCGCTCAGGCCGGGTTGCTGGAGGCCGAGCATCAGGCCATCATTCGTGATGTGTTGACC 2160
GCGAGTGACTTTTGGGGCGGCGCCGGTTCGGCGGCCTGCCAGGGGTTCATTACCCAGTTG 2220
GGCCGTAACTTCCAGGTGATCTACGAGCAGGCCAACGCCCACGGGCAGAAGGTGCAGGCT 2280
GCCGGCAACAACATGGCGCAAACCGACAGCGCCGTCGGCTCCAGCTGGGCCTGACACCAG 2340
GCCAAGGCCAGGGACGTGGTGTACGAGTGAAGTTCCTCGCGTGATCCTTCGGGTGGCAGT 2400
CTAAGTGGTCAGTGCTGGGGTGTTGGTGGTTTGCTGCTTGGCGGGTTCTTCGGTGCTGGT 2960
CAGTGCTGCTCGGGCTCGGGTGAGGACCTCGAGGCCCAGGTAGCGCCGTCCTTCGATCCA 2520
TTCGTCGTGTTGTTCGGCGAGGACGGCTCCGACGAGGCGGATGATCGAGGCGCGGTCGGG 2580
GAAGATGCCCACGACGTCGGTTCGGCGTCGTACCTCTCGGTTGAGGCGTTCCTGGGGGTT 2640
GTTGGACCAGATTTGGCGCCAGATCTGCTTGGGGAAGGCGGTGAACGCCAGCAGGTCGGT 2700
GCGGGCGGTGTCGAGGTGCTCGGCCACCGCGGGGAGTTT~GTCGGTCAGAGCGTCGAGTAC 2760
CCGATCATATTGGGCAACAACTGATTCGGCGTCGGGCTGGTCGTAGATGGAGTGCAGCAG 2820
CA 02268036 1999-10-12
118
GGTGCGCACCCACGGCCAGGAGGGCTTCGGGGTGGCTGCCATCAGATTGGCTGCGTAGTG2880
GGTTCTGCAGCGCTGCCAGGCCGCTGCGGGCAGGGTGGCGCCGATCGCGGCCACCAGGCC2940
GGCGTGGGCGTCGCTGGTGACCAGCGCGACCCCGGACAGGCCGCGGGCGACCAGGTCGCG3000
GAAGAACGCCAGCCAGCCGGCCCCGTCCTCGGCGGAGGTGACCTGGATGCCCAGGATC 3058
(2) INFORMATION FOR SEQ ID N0:102:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 391 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:7.02:
Met Val Asp Phe Gly Ala Leu Pro Pro C~lu Ile Asn Ser Ala Arg Met
1 5 1.0 15
Tyr Ala Gly Pro Gly Ser Ala Ser Leu Val Ala Ala Ala Gln Met Trp
20 25 30
Asp Ser Val Ala Ser Asp Leu Phe Ser P,la Ala Ser Ala Phe Gln Ser
35 40 45
Val Val Trp Gly Leu Thr Val Gly Ser T'rp Ile Gly Ser Ser Ala Gly
50 55 60
Leu Met Val Ala Ala Ala Ser Pro Tyr Val Ala Trp Met Ser Val Thr
65 70 75 80
Ala Gly Gln Ala Glu Leu Thr Ala Ala Gln Val Arg Val Ala Ala Ala
85 90 95
Ala Tyr Glu Thr Ala Tyr Gly Leu Thr Val Pro Pro Pro Val Ile Ala
100 105 110
Glu Asn Arg Ala Glu Leu Met Ile Leu Ile Ala Thr Asn Leu Leu Gly
115 120 125
Gln Asn Thr Pro Ala Ile Ala Val Asn Glu Ala Glu Tyr Gly Glu Met
130 135 140
Trp Ala Gln Asp Ala Ala Ala Met Phe Gly Tyr Ala Ala Ala Thr Ala
145 150 155 160
Thr Ala Thr Ala Thr Leu Leu Pro Phe Glu Glu Ala Pro Glu Met Thr
165 170 175
Ser Ala Gly Gly Leu Leu Glu Gln Ala Ala Ala Val Glu Glu Ala Ser
180 185 190
Asp Thr Ala Ala Ala Asn Gln Leu Met Asn Asn Val Pro Gln Ala Leu
195 200 205
Gln Gln Leu Ala Gln Pro Thr Gln Gly T:hr Thr Pro Ser Ser Lys Leu
210 215 220
Gly Gly Leu Trp Lys Thr Val Ser Pro H.is Arg Ser Pro Ile Ser Asn
225 230 235 240
CA 02268036 1999-10-12
119
Met Val Ser Met Ala Asn Asn His Met S er Met Thr Asn Ser Gly Val
245 250 255
Ser Met Thr Asn Thr Leu Ser Ser Met Leu Lys Gly Phe Ala Pro Ala
260 265 270
Ala Ala Ala Gln Ala Val Gln Thr Ala Ala Gln Asn Gly Val Arg Ala
275 280 285
Met Ser Ser Leu Gly Ser Ser Leu Gly Ser Ser Gly Leu Gly Gly Gly
290 295 300
Val Ala Ala Asn Leu Gly Arg Ala Ala Ser Val Gly Ser Leu Ser Val
305 310 315 320
Pro Gln Ala Trp Ala Ala Ala Asn Gln Ala Val Thr Pro Ala Ala Arg
325 330 335
Ala Leu Pro Leu Thr Ser Leu Thr Ser Ala Ala Glu Arg Gly Pro Gly
340 345 350
Gln Met Leu Gly Gly Leu Pro Val Gly Gln Met Gly Ala Arg Ala Gly
355 360 365
Gly Gly Leu Ser Gly Val Leu Arg Val Pro Pro Arg Pro Tyr Val Met
370 375 380
Pro His Ser Pro Ala Ala Gly
385 390
(2) INFORMATION FOR SEQ ID N0:103:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:1725 base
pairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY:
linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:103:
GACGTCAGCACCCGCCGTGCAGGGCTGGAGCGTGGTCGGTTTTGATCTGCGGTCAAGGTG 60
ACGTCCCTCGGCGTGTCGCCGGCGTGGATGCAGACTCGA.TGCCGCTCTTTAGTGCAACTA 120
ATTTCGTTGAAGTGCCTGCGAGGTATAGGACTTCACGATTGGTTAATGTAGCGTTCACCC 180
CGTGTTGGGGTCGATTTGGCCGGACCAGTCGTCACCAACGCTTGGCGTGCGCGCCAGGCG 240
GGCGATCAGATCGCTTGACTACCAATCAATCTTGAGCTCCCGGGCCGATGCTCGGGCTAA 300
ATGAGGAGGAGCACGCGTGTCTTTCACTGCGCAACCGGAGATGTTGGCGGCCGCGGCTGG 360
CGAACTTCGTTCCCTGGGGGCAACGCTGAAGGCTAGCAATGCCGCCGCAGCCGTGCCGAC 420
GACTGGGGTGGTGCCCCCGGCTGCCGACGAGGTGTCGCTGCTGCTTGCCACACAATTCCG 480
TACGCATGCGGCGACGTATCAGACGGCCAGCGCCAAGGCCGCGGTGATCCATGAGCAGTT 540
TGTGACCACGCTGGCCACCAGCGCTAGTTCATATGCGGACACCGAGGCCGCCAACGCTGT 600
GGTCACCGGCTAGCTGACCTGACGGTATTCGAGCGGAAGGATTATCGAAGTGGTGGATTT 660
CA 02268036 1999-10-12
120
CGGGGCGTTACCACCGGAGATCAACTCCGCGAGGATGTACGCCGGCCCGGGTTCGGCCTC 720
GCTGGTGGCCGCCGCGAAGATGTGGGACAGCGTGGCGAGTGACCTGTTTTCGGCCGCGTC 780
GGCGTTTCAGTCGGTGGTCTGGGGTCTGACGGTGGGGTCGTGGATAGGTTCGTCGGCGGG 840
TCTGATGGCGGCGGCGGCCTCGCCGTATGTGGCGTGGA'.PGAGCGTCACCGCGGGGCAGGC 900
CCAGCTGACCGCCGCCCAGGTCCGGGTTGCTGCGGCGG(:CTACGAGACAGCGTATAGGCT 960
GACGGTGCCCCCGCCGGTGATCGCCGAGAACCGTACCGAF1CTGATGACGCTGACCGCGAC 1020
CAACCTCTTGGGGCAAAACACGCCGGCGATCGAGGCCAATCAGGCCGCATACAGCCAGAT 1080
GTGGGGCCAAGACGCGGAGGCGATGTATGGCTACGCCGC:CACGGCGGCGACGGCGACCGA 1140
GGCGTTGCTGCCGTTCGAGGACGCCCCACTGATCACCAACCCCGGCGGGCTCCTTGAGCA 1200
GGCCGTCGCGGTCGAGGAGGCCATCGACACCGCCGCGGC:GAACCAGTTGATGAACAATGT 1260
GCCCCAAGCGCTGCAACAGCTGGCCCAGCCAGCGCAGGGCGTCGTACCTTCTTCCAAGCT 1320
GGGTGGGCTGTGGACGGCGGTCTCGCCGCATCTGTCGCC;GCTCAGCAACGTCAGTTCGAT 1380
AGCCAACAACCACATGTCGATGATGGGCACGGGTGTGTC;GATGACCAACACCTTGCACTC 1440
GATGTTGAAGGGCTTAGCTCCGGCGGCGGCTCAGGCCGTGGAAACCGCGGCGGAAAACGG 1500
GGTCTGGGCGATGAGCTCGCTGGGCAGCCAGCTGGGTTC'.GTCGCTGGGTTCTTCGGGTCT 1560
GGGCGCTGGGGTGGCCGCCAACTTGGGTCGGGCGGCCTC:GGTCGGTTCGTTGTCGGTGCC 1620
GCCAGCATGGGCCGCGGCCAACCAGGCGGTCACCCCGGC'.GGCGCGGGCGCTGCCGCTGAC 1680
CAGCCTGACCAGCGCCGCCCAAACCGCCCCCGGACACAT'GCTGGG 1725
(2} INFORMATION FOR SEQ ID N0:104:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 359 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:109:
Val Val Asp Phe Gly Ala Leu Pro Pro Glu Ile Asn Ser Ala Arg Met
1 5 10 15
Tyr Ala Gly Pro Gly Ser Ala Ser Leu Val Ala Ala Ala Lys Met Trp
20 25 30
Asp Ser Val Ala Ser Asp Leu Phe Ser Ala Ala Ser Ala Phe Gln Ser
35 40 45
Val Val Trp Gly Leu Thr Val Gly Ser Trp Ile Gly Ser Ser Ala Gly
50 55 60
Leu Met Ala Ala Ala Ala Ser Pro Tyr V.al Ala Trp Met Ser Val Thr
65 70 75 80
Ala Gly Gln Ala Gln Leu Thr Ala Ala G.ln Val Arg Val Ala Ala Ala
85 90 95
CA 02268036 1999-10-12
121
Ala Tyr Glu Thr Ala Tyr Arg Leu Thr Val Pro Pro Pro Val Ile Ala
100 105 110
Glu Asn Arg Thr Glu Leu Met Thr Leu '.Chr Ala Thr Asn Leu Leu Gly
115 120 125
Gln Asn Thr Pro Ala Ile Glu Ala Asn Gln Ala Ala Tyr Ser Gln Met
130 135 140
Trp Gly Gln Asp Ala Glu Ala Met Tyr CJly Tyr Ala Ala Thr Ala Ala
145 150 155 160
Thr Ala Thr Glu Ala Leu Leu Pro Phe Glu Asp Ala Pro Leu Ile Thr
165 7.70 175
Asn Pro Gly Gly Leu Leu Glu Gln Ala Val Ala Val Glu Glu Ala Ile
180 185 190
Asp Thr Ala Ala Ala Asn Gln Leu Met Asn Asn Val Pro Gln Ala Leu
195 200 205
Gln Gln Leu Ala Gln Pro Ala Gln Gly Val Val Pro Ser Ser Lys Leu
210 215 220
Gly Gly Leu Trp Thr Ala Val Ser Pro His Leu Ser Pro Leu Ser Asn
225 230 235 240
Val Ser Ser Ile Ala Asn Asn His Met ~;er Met Met Gly Thr Gly Val
245 2'50 255
Ser Met Thr Asn Thr Leu His Ser Met heu Lys Gly Leu Ala Pro Ala
260 265 270
Ala Ala Gln Ala Val Glu Thr Ala Ala Glu Asn Gly Val Trp Ala Met
275 280 285
Ser Ser Leu Gly Ser Gln Leu Gly Ser Ser Leu Gly Ser Ser Gly Leu
290 295 300
Gly Ala Gly Val Ala Ala Asn Leu Gly Arg Ala Ala Ser Val Gly Ser
305 310 315 320
Leu Ser Val Pro Pro Ala Trp Ala Ala A.la Asn Gln Ala Val Thr Pro
325 330 335
Ala Ala Arg Ala Leu Pro Leu Thr Ser Leu Thr Ser Ala Ala Gln Thr
340 345 350
Ala Pro Gly His Met Leu Gly
355
(2) INFORMATION FOR SEQ ID N0:105:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3027 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:105:
AGTTCAGTCG AGAATGATAC TGACGGGCTG TATCCACGA'P GGCTGAGACA ACCGAACCAC 60
CA 02268036 1999-10-12
122
CGTCGGACGCGGGGACATCGCAAGCCGACGCGATGGCG'rTGGCCGCCGAAGCCGAAGCCG 120
CCGAAGCCGAAGCGCTGGCCGCCGCGGCGCGGGCCCGTGCCCGTGCCGCCCGGTTGAAGC 180
GTGAGGCGCTGGCGATGGCCCCAGCCGAGGACGAGAACGTCCCCGAGGATATGCAGACTG 240
GGAAGACGCCGAAGACTATGACGACTATGACGACTATGAGGCCGCAGACCAGGAGGCCGC 300
ACGGTCGGCATCCTGGCGACGGCGGTTGCGGGTGCGGT'.PACCAAGACTGTCCACGATTGC 360
CATGGCGGCCGCAGTCGTCATCATCTGCGGCTTCACCGGGCTCAGCGGATACATTGTGTG 420
GCAACACCATGAGGCCACCGAACGCCAGCAGCGCGCCG(:GGCGTTCGCCGCCGGAGCCAA 480
GCAAGGTGTCATCAACATGACCTCGCTGGACTTCAACAAGGCCAAAGAAGACGTCGCGCG 540
TGTGATCGACAGCTCCACCGGCGAATTCAGGGATGACTTCCAGCAGCGGGCAGCCGATTT 600
CACCAAGGTTGTCGAACAGTCCAAAGTGGTCACCGAAGGCACGGTGAACGCGACAGCCGT 660
CGAATCCATGAACGAGCATTCCGCCGTGGTGCTCGTCGC:GGCGACTTCACGGGTCACCAA 720
TTCCGCTGGGGCGAAAGACGAACCACGTGCGTGGCGGC7.'CAAAGTGACCGTGACCGAAGA 780
GGGGGGACAGTACAAGATGTCGAAAGTTGAGTTCGTACC:GTGACCGATGACGTACGCGAC 840
GTCAACACCGAAACCACTGACGCCACCGAAGTCGCTGACJATCGACTCAGCCGCAGGCGAA 900
GCCGGTGATTCGGCGACCGAGGCATTTGACACCGACTCTGCAACGGAATCTACCGCGCAG 960
AAGGGTCAGCGGCACCGTGACCTGTGGCGAATGCAGGTTACCTTGAAACCCGTTCCGGTG 1020
ATTCTCATCCTGCTCATGTTGATCTCTGGGGGCGCGACGGGATGGCTATACCTTGAGCAA 1080
TACGACCCGATCAGCAGACGGACTCCGGCGCCGCCCGTGCTGCCGTCGCCGCGGCGTCTG 1140
ACGGGACAATCGCGCTGTTGTGTATTCACCCGACACGTC:GACCAAGACTTCGCTACCGCC 1200
AGGTCGCACCTCGCCGGCGATTTCCTGTCCTATACGACC'AGTTCACGCAGCAGATCGTGG 1260
CTCCGGCGGCCAAACAGAAGTCACTGAAAACCACCGCCP.,AGGTGGTGCGCGCGGCCGTGT 1320
CGGAGCTACATCCGGATTCGGCCGTCGTTCTGGTTTTTGTCGACCAGAGCACTACCAGTA 1380
AGGACAGCCCCAATCCGTCGATGGCGGCCAGCAGCGTGA.TGGTGACCCTAGCCAAGGTCG 1440
ACGGCAATTGGCTGATCACCAAGTTCACCCCGGTTTAGGTTGCCGTAGGCGGTCGCCAAG 1500
TCTGACGGGGGCGCGGGTGGCTGCTCGTGCGAGATACCGGCCGTTCTCCGGACAATCACG 1560
GCCCGACCTCAAACAGATCTCGGCCGCTGTCTAATCGGCCGGGTTATTTAAGATTAGTTG 1620
CCACTGTATTTACCTGATGTTCAGATTGTTCAGCTGGATTTAGCTTCGCGGCAGGGCGGC 1680
TGGTGCACTTTGCATCTGGGGTTGTGACTACTTGAGAGAATTTGACCTGTTGCCGACGTT 1740
GTTTGCTGTCCATCATTGGTGCTAGTTATGGCCGAGCGGAAGGATTATCGAAGTGGTGGA 1800
CTTCGGGGCGTTACCACCGGAGATCAACTCCGCGAGGATGTACGCCGGCCCGGGTTCGGC 1860
CTCGCTGGTGGCCGCCGCGAAGATGTGGGACAGCGTGGCGAGTGACCTGTTTTCGGCCGC 1920
GTCGGCGTTTCAGTCGGTGGTCTGGGGTCTGACGACGGGATCGTGGATAGGTTCGTCGGC 1980
CA 02268036 1999-10-12
123
GGGTCTGATGGTGGCGGCGGCCTCGCCGTATGTGGCGTC~GATGAGCGTCACCGCGGGGCA 2040
GGCCGAGCTGACCGCCGCCCAGGTCCGGGTTGCTGCGG(:GGCCTACGAGACGGCGTATGG 2100
GCTGACGGTGCCCCCGCCGGTGATCGCCGAGAACCGTGC:TGAACTGATGATTCTGATAGC 2160
GACCAACCTCTTGGGGCAAAACACCCCGGCGATCGCGGTCAACGAGGCCGAATACGGGGA 2220
GATGTGGGCCCAAGACGCCGCCGCGATGTTTGGCTACGC:CGCCACGGCGGCGACGGCGAC 2280
CGAGGCGTTGCTGCCGTTCGAGGACGCCCCACTGATCAC:CAACCCCGGCGGGCTCCTTGA 2340
GCAGGCCGTCGCGGTCGAGGAGGCCATCGACACCGCCGC:GGCGAACCAGTTGATGAACAA 2400
TGTGCCCCAAGCGCTGCAACAACTGGCCCAGCCCACGAAAAGCATCTGGCCGTTCGACCA 2460
ACTGAGTGAACTCTGGAAAGCCATCTCGCCGCATCTGTC;GCCGCTCAGCAACATCGTGTC 2520
GATGCTCAACAACCACGTGTCGATGACCAACTCGGGTGTGTCGATGGCCAGCACCTTGCA 2580
CTCAATGTTGAAGGGCTTTGCTCCGGCGGCGGCTCAGGC;CGTGGAAACCGCGGCGCAAAA 2640
CGGGGTCCAGGCGATGAGCTCGCTGGGCAGCCAGCTGGGTTCGTCGCTGGGTTCTTCGGG 2700
TCTGGGCGCTGGGGTGGCCGCCAACTTGGGTCGGGCGGC:CTCGGTCGGTTCGTTGTCGGT 2760
GCCGCAGGCCTGGGCCGCGGCCAACCAGGCGGTCACCCC:GGCGGCGCGGGCGCTGCCGCT 2820
GACCAGCCTGACCAGCGCCGCCCAAACCGCCCCCGGACACATGCTGGGCGGGCTACCGCT 2880
GGGGCAACTGACCAATAGCGGCGGCGGGTTCGGCGGGGTTAGCAATGCGTTGCGGATGCC 2940
GCCGCGGGCGTACGTAATGCCCCGTGTGCCCGCCGCCGGGTAACGCCGATCCGCACGCAA 3000
TGCGGGCCCTCTATGCGGGCAGCGATC 3027
(2) INFORMATION FOR SEQ ID N0:106:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 396 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:106:
Val Val Asp Phe Gly Ala Leu Pro Pro Glu Ile Asn Ser Ala Arg Met
1 5 10 15
Tyr Ala Gly Pro Gly Ser Ala Ser Leu Val Ala Ala Ala Lys Met Trp
20 25 30
Asp Ser Val Ala Ser Asp Leu Phe Ser Ala Ala Ser Ala Phe Gln Ser
35 40 45
Val Val Trp Gly Leu Thr Thr Gly Ser Trp Ile Gly Ser Ser Ala Gly
50 55 60
Leu Met Val Ala Ala Ala Ser Pro Tyr Val Ala Trp Met Ser Val Thr
65 70 75 80
Ala Gly Gln Ala Glu Leu Thr Ala Ala G.ln Val Arg Val Ala Ala Ala
CA 02268036 1999-10-12
124
85 90 95
Ala Tyr Glu Thr Ala Tyr Gly Leu Thr Val Pro Pro Pro Val Ile Ala
100 105 110
Glu Asn Arg Ala Glu Leu Met Ile Leu :Lle Ala Thr Asn Leu Leu Gly
115 120 125
Gln Asn Thr Pro Ala Ile Ala Val Asn (ilu Ala Glu Tyr Gly Glu Met
130 135 140
Trp Ala Gln Asp Ala Ala Ala Met Phe Gly Tyr Ala Ala Thr Ala Ala
145 150 155 160
Thr Ala Thr Glu Ala Leu Leu Pro Phe Glu Asp Ala Pro Leu Ile Thr
165 T_70 175
Asn Pro Gly Gly Leu Leu Glu Gln Ala Val Ala Val Glu Glu Ala Ile
180 185 190
Asp Thr Ala Ala Ala Asn Gln Leu Met Asn Asn Val Pro Gln Ala Leu
195 200 205
Gln Gln Leu Ala Gln Pro Thr Lys Ser l:le Trp Pro Phe Asp Gln Leu
210 215 220
Ser Glu Leu Trp Lys Ala Ile Ser Pro His Leu Ser Pro Leu Ser Asn
225 230 235 240
Ile Val Ser Met Leu Asn Asn His Val :cer Met Thr Asn Ser Gly Val
245 250 255
Ser Met Ala Ser Thr Leu His Ser Met Leu Lys Gly Phe Ala Pro Ala
260 265 270
Ala Ala Gln Ala Val Glu Thr Ala Ala Gln Asn Gly Val Gln Ala Met
275 280 285
Ser Ser Leu Gly Ser Gln Leu Gly Ser Ser Leu Gly Ser Ser Gly Leu
290 295 300
Gly Ala Gly Val Ala Ala Asn Leu Gly Arg Ala Ala Ser Val Gly Ser
305 310 315 320
Leu Ser Val Pro Gln Ala Trp Ala Ala A.la Asn Gln Ala Val Thr Pro
325 330 335
Ala Ala Arg Ala Leu Pro Leu Thr Ser Leu Thr Ser Ala Ala Gln Thr
340 345 350
Ala Pro Gly His Met Leu Gly Gly Leu Pro Leu Gly Gln Leu Thr Asn
355 360 365
Ser Gly Gly Gly Phe Gly Gly Val Ser Asn Ala Leu Arg Met Pro Pro
370 375 380
Arg Ala Tyr Val Met Pro Arg Val Pro Ala Ala Gly
385 390 395
(2) INFORMATION FOR SEQ ID N0:107:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1616 base pairs
CA 02268036 1999-10-12
125
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY:
linear
(xi) S EQUENCE EQ ID 7:
DESCRIPTION: N0::10
S
CATCGGAGGGAGTGATCACCATGCTGTGGCACGCAATG(:CACCGGAGTAAATACCGCACG 60
GCTGATGGCCGGCGCGGGTCCGGCTCCAATGCTTGCGGCGGCCGCGGGATGGCAGACGCT 120
TTCGGCGGCTCTGGACGCTCAGGCCGTCGAGTTGACCG(:GCGCCTGAACTCTCTGGGAGA 180
AGCCTGGACTGGAGGTGGCAGCGACAAGGCGCTTGCGG(:TGCAACGCCGATGGTGGTCTG 240
GCTACAAACCGCGTCAACACAGGCCAAGACCCGTGCGATGCAGGCGACGGCGCAAGCCGC 300
GGCATACACCCAGGCCATGGCCACGACGCCGTCGCTGCC:GGAGATCGCCGCCAACCACAT 360
CACCCAGGCCGTCCTTACGGCCACCAACTTCTTCGGTA7.'CAACACGATCCCGATCGCGTT 420
GACCGAGATGGATTATTTCATCCGTATGTGGAACCAGGC:AGCCCTGGCAATGGAGGTCTA 480
CCAGGCCGAGACCGCGGTTAACACGCTTTTCGAGAAGC7.'CGAGCCGATGGCGTCGATCCT 540
TGATCCCGGCGCGAGCCAGAGCACGACGAACCCGATCT7.'CGGAATGCCCTCCCCTGGCAG 600
CTCAACACCGGTTGGCCAGTTGCCGCCGGCGGCTACCCAGACCCTCGGCCAACTGGGTGA 660
GATGAGCGGCCCGATGCAGCAGCTGACCCAGCCGCTGCAGCAGGTGACGTCGTTGTTCAG 720
CCAGGTGGGCGGCACCGGCGGCGGCAACCCAGCCGACGAGGAAGCCGCGCAGATGGGCCT 780
GCTCGGCACCAGTCCGCTGTCGAACCATCCGCTGGCTGGTGGATCAGGCCCCAGCGCGGG 840
CGCGGGCCTGCTGCGCGCGGAGTCGCTACCTGGCGCAGGTGGGTCGTTGACCCGCACGCC 900
GCTGATGTCTCAGCTGATCGAAAAGCCGGTTGCCCCCTC'GGTGATGCCGGCGGCTGCTGC 960
CGGATCGTCGGCGACGGGTGGCGCCGCTCCGGTGGGTGCGGGAGCGATGGGCCAGGGTGC 1020
GCAATCCGGCGGCTCCACCAGGCCGGGTCTGGTCGCGCCGGCACCGCTCGCGCAGGAGCG 1080
TGAAGAAGACGACGAGGACGACTGGGACGAAGAGGACGACTGGTGAGCTCCCGTAATGAC 1140
AACAGACTTCCCGGCCACCCGGGCCGGAAGACTTGCCAA.CATTTTGGCGAGGAAGGTAAA 1200
GAGAGAAAGTAGTCCAGCATGGCAGAGATGAAGACCGATGCCGCTACCCTCGCGCAGGAG 1260
GCAGGTAATTTCGAGCGGATCTCCGGCGACCTGAAAACCCAGATCGACCAGGTGGAGTCG 1320
ACGGCAGGTTCGTTGCAGGGCCAGTGGCGCGGCGCGGCGGGGACGGCCGCCCAGGCCGCG 1380
GTGGTGCGCTTCCAAGAAGCAGCCAATAAGCAGAAGCAGGAACTCGACGAGATCTCGACG 1440
AATATTCGTCAGGCCGGCGTCCAATACTCGAGGGCCGACGAGGAGCAGCAGCAGGCGCTG 1500
TCCTCGCAAATGGGCTTCTGACCCGCTAATACGAAAAGAAACGGAGCAAAAACATGACAG 1560
AGCAGCAGTGGAATTTCGCGGGTATCGAGGCCGCGGCAAGCGCAATCCAGGGAAAT 1616
(2) INFORMATION FOR SEQ ID N0:108:
(i) SEQUENCE CHARACTERISTICS:
CA 02268036 1999-10-12
126
(A) LENGTH:432 base pairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY:
linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:108:
CTAGTGGATGGGACCATGGCCATTTTCTGC AGTCTCAC'.CGCCTTCTGTGTTGACATTTTG 60
GCACGCCGGCGGAAACGAAGCACTGGGGTC GAAGAACGGCTGCGCTGCCATATCGTCCGG 120
AGCTTCCATACCTTCGTGCGGCCGGAAGAG CTTGTCGTAGTCGGCCGCCATGACAACCTC 180
TCAGAGTGCGCTCAAACGTATAAACACGAG AAAGGGCGAGACCGACGGAAGGTCGAACTC 240
GCCCGATCCCGTGTTTCGCTATTCTACGCG AACTCGGCGTTGCCCTATGCGAACATCCCA 300
GTGACGTTGCCTTCGGTCGAAGCCATTGCC TGACCGGC7.'TCGCTGATCGTCCGCGCCAGG 360
TTCTGCAGCGCGTTGTTCAGCTCGGTAGCC GTGGCGTCC;CATTTTTGCTGGACACCCTGG 420
TACGCCTCCGAA 432
(2) INFORMATION FOR SEQ ID N0:109:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 368 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:109:
Met Leu Trp His Ala Met Pro Pro Glu Xaa Asn Thr Ala Arg Leu Met
1 5 10 15
Ala Gly Ala Gly Pro Ala Pro Met Leu Ala Ala Ala Ala Gly Trp Gln
20 25 30
Thr Leu Ser Ala Ala Leu Asp Ala Gln A.la Val Glu Leu Thr Ala Arg
35 40 45
Leu Asn Ser Leu Gly Glu Ala Trp Thr Gly Gly Gly Ser Asp Lys Ala
50 55 60
Leu Ala Ala Ala Thr Pro Met Val Val Trp Leu Gln Thr Ala Ser Thr
65 70 75 80
Gln Ala Lys Thr Arg Ala Met Gln Ala Thr Ala Gln Ala Ala Ala Tyr
85 90 95
Thr Gln Ala Met Ala Thr Thr Pro Ser Leu Pro Glu Ile Ala Ala Asn
100 105 110
His Ile Thr Gln Ala Val Leu Thr Ala Thr Asn Phe Phe Gly Ile Asn
115 120 125
Thr Ile Pro Ile Ala Leu Thr Glu Met Asp Tyr Phe Ile Arg Met Trp
130 135 140
Asn Gln Ala Ala Leu Ala Met Glu Val T:yr Gln Ala Glu Thr Ala Val
145 150 155 160
CA 02268036 1999-10-12
127
Asn Thr Leu Phe Glu Lys Leu Glu Pro Met Ala Ser Ile Leu Asp Pro
165 170 175
Gly Ala Ser Gln Ser Thr Thr Asn Pro :Ile Phe Gly Met Pro Ser Pro
180 185 190
Gly Ser Ser Thr Pro Val Gly Gln Leu 1?ro Pro Ala Ala Thr Gln Thr
195 200 205
Leu Gly Gln Leu Gly Glu Met Ser Gly I?ro Met Gln Gln Leu Thr Gln
210 215 220
Pro Leu Gln Gln Val Thr Ser Leu Phe Ser Gln Val Gly Gly Thr Gly
225 230 235 240
Gly Gly Asn Pro Ala Asp Glu Glu Ala Ala Gln Met Gly Leu Leu Gly
245 250 255
Thr Ser Pro Leu Ser Asn His Pro Leu Ala Gly Gly Ser Gly Pro Ser
260 265 270
Ala Gly Ala Gly Leu Leu Arg Ala Glu Ser Leu Pro Gly Ala Gly Gly
275 280 285
Ser Leu Thr Arg Thr Pro Leu Met Ser C:ln Leu Ile Glu Lys Pro Val
290 295 300
Ala Pro Ser Val Met Pro Ala Ala Ala Ala Gly Ser Ser Ala Thr Gly
305 310 315 320
Gly Ala Ala Pro Val Gly Ala Gly Ala Nlet Gly Gln Gly Ala Gln Ser
325 ?.30 335
Gly Gly Ser Thr Arg Pro Gly Leu Val P,la Pro Ala Pro Leu Ala Gln
340 345 350
Glu Arg Glu Glu Asp Asp Glu Asp Asp Trp Asp Glu Glu Asp Asp Trp
355 360 365
(2) INFORMATION FOR SEQ ID NO:110:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 100 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:110:
Met Ala Glu Met Lys Thr Asp Ala Ala Thr Leu Ala Gln Glu Ala Gly
1 5 10 15
Asn Phe Glu Arg Ile Ser Gly Asp Leu Lys Thr Gln Ile Asp Gln Val
20 25 30
Glu Ser Thr Ala Gly Ser Leu Gln Gly Gln Trp Arg Gly Ala Ala Gly
35 40 45
Thr Ala Ala Gln Ala Ala Val Val Arg Phe Gln Glu Ala Ala~Asn Lys
50 55 60
Gln Lys Gln Glu Leu Asp Glu Ile Ser T:hr Asn Ile Arg Gln Ala Gly
65 70 75 80
CA 02268036 1999-10-12
128
Val Gln Tyr Ser Arg Ala Asp Glu Glu (~ln Gln Gln Ala Leu Ser Ser
85 90 95
Gln Met Gly Phe
100
(2) INFORMATION FOR SEQ ID N0:111:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 396 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:7.11:
GATCTCCGGCGACCTGAAAACCCAGATCGACCAGGTGGAGTCGACGGCAGGTTCGTTGCA 60
GGGCCAGTGGCGCGGCGCGGCGGGGACGGCCGCCCAGGC;CGCGGTGGTGCGCTTCCAAGA 120
AGCAGCCAATAAGCAGAAGCAGGAACTCGACGAGATCTC;GACGAATATTCGTCAGGCCGG 180
CGTCCAATACTCGAGGGCCGACGAGGAGCAGCAGCAGGC:GCTGTCCTCGCAAATGGGCTT 240
CTGACCCGCTAATACGAAAAGAAACGGAGCAAAAACATGACAGAGCAGCAGTGGAATTTC 300
GCGGGTATCGAGGCCGCGGCAAGCGCAATCCAGGGAAATGTCACGTCCATTCATTCCCTC 360
CTTGACGAGGGGAAGCAGTCCCTGACCAAGCTCGCA 396
(2) INFORMATION FOR SEQ ID N0:112:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 80 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:112:
Ile Ser Gly Asp Leu Lys Thr Gln Ile A.sp Gln Val Glu Ser Thr Ala
1 5 10 15
Gly Ser Leu Gln Gly Gln Trp Arg Gly Ala Ala Gly Thr Ala Ala Gln
20 25 30
Ala Ala Val Val Arg Phe Gln Glu Ala Ala Asn Lys Gln Lys Gln Glu
35 40 45
Leu Asp Glu Ile Ser Thr Asn Ile Arg Gln Ala Gly Val Gln Tyr Ser
50 55 60
Arg Ala Asp Glu Glu Gln Gln Gln Ala Leu Ser Ser Gln Met Gly Phe
65 70 75 80
(2) INFORMATION FOR SEQ ID N0:113:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 387 base pairs
(B) TYPE: nucleic acid
CA 02268036 1999-10-12
129
(C) STRANDEDNESS:
single
(D) TOPOLOGY:
linear
(xi) EQUENCE
S DESCRIPTION:
SEQ ID
NO::L13:
GTGGATCCCGATCCCGTGTTTCGCTATTCTACGCGAAC'.CCGGCGTTGCCCTATGCGAACA 60
TCCCAGTGACGTTGCCTTCGGTCGAAGCCATTGCCTGAC:CGGCTTCGCTGATCGTCCGCG 120
CCAGGTTCTGCAGCGCGTTGTTCAGCTCGGTAGCCGTGGCGTCCCATTTTTGCTGGACAC 180
CCTGGTACGCCTCCGAACCGCTACCGCCCCAGGCCGCTGCGAGCTTGGTCAGGGACTGCT 240
TCCCCTCGTCAAGGAGGGAATGAATGGACGTGACATTTC:CCTGGATTGCGCTTGCCGCGG 300
CCTCGATACCCGCGAAATTCCACTGCTGCTCTGTCATG7.'TTTTGCTCCGTTTCTTTTCGT 360
ATTAGCGGGTCAGAAGCCCATTTGCGA 3g7
(2) INFORMATION FOR SEQ ID N0:114:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 272 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:1.14:
CGGCACGAGGATCTCGGTTG GCCCAACGGC GCTGGCGAGGGCTCCGTTCC GGGGGCGAGC60
TGCGCGCCGGATGCTTCCTC TGCCCGCAGC CGCGCCTGGATGGATGGACC AGTTGCTACC120
TTCCCGACGTTTCGTTCGGT GTCTGTGCGA TAGCGGTGACCCCGGCGCGC ACGTCGGGAG180
TGTTGGGGGGCAGGCCGGGT CGGTGGTTCG GCCGGGGAC'.GCAGACGGTCT GGACGGAACG240
GGCGGGGGTTCGCCGATTGG CATCTTTGCC CA 272
(2) INFORMATION FOR SEQ ID N0:115:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:115:
Asp Pro Val Asp Ala Val Ile Asn Thr Thr Cys Asn Tyr Gly Gln Val
1 5 10 15
Val Ala Ala Leu
(2) INFORMATION FOR SEQ ID N0:116:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
CA 02268036 1999-10-12
130
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0::116:
Ala Val Glu Ser Gly Met Leu Ala Leu Gly Thr Pro Ala Pro Ser
1 5 :LO 15
(2) INFORMATION FOR SEQ ID N0:117:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:7_17:
Ala Ala Met Lys Pro Arg Thr Gly Asp Gly Pro Leu Glu Ala Ala Lys
1 5 7.0 15
Glu Gly Arg
(2) INFORMATION FOR SEQ ID N0:118:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:118:
Tyr Tyr Trp Cys Pro Gly Gln Pro Phe P,sp Pro Ala Trp Gly Pro
1 5 10 15
(2) INFORMATION FOR SEQ ID N0:119:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 14 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:119:
Asp Ile Gly Ser Glu Ser Thr Glu Asp Gln Gln Xaa Ala Val
1 5 10
(2) INFORMATION FOR SEQ ID N0:120:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:120:
Ala Glu Glu Ser Ile Ser Thr Xaa Glu Xaa Ile Val Pro
1 5 10
CA 02268036 1999-10-12
131
(2) INFORMATION FOR SEQ ID N0:121:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 17 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:121:
Asp Pro Glu Pro Ala Pro Pro Val Pro Thr Thr Ala Ala Ser Pro Pro
1 5 7_0 15
Ser
(2) INFORMATION FOR SEQ ID N0:122:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:1.22:
Ala Pro Lys Thr Tyr Xaa Glu Glu Leu hys Gly Thr Asp Thr Gly
1 5 10 15
(2) INFORMATION FOR SEQ ID N0:123:
(i} SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:123:
Asp Pro Ala Ser Ala Pro Asp Val Pro Thr Ala Ala Gln Leu Thr Ser
1 5 10 15
Leu Leu Asn Ser Leu Ala Asp Pro Asn Val Ser Phe Ala Asn
20 25 30
(2) INFORMATION FOR SEQ ID N0:124:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:124:
Asp Pro Pro Asp Pro His Gln Xaa Asp Met Thr Lys Gly Tyr Tyr Pro
1 5 10 15
Gly Gly Arg Arg Xaa Phe
CA 02268036 1999-10-12
132
(2) INFORMATION FOR SEQ ID N0:125:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 7 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:125:
Asp Pro Gly Tyr Thr Pro Gly
1 5
(2) INFORMATION FOR SEQ ID N0:126:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 10 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(ix) FEATURE:
(A) NAME/KEY: Modified-site
(B) LOCATION: 2
(D) OTHER INFORMATION: /product== "OTHER"
/note= "x:aa = Pro or Thr"
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:126:
Xaa Xaa Gly Phe Thr Gly Pro Gln Phe Tyr
1 5 10
(2) INFORMATION FOR SEQ ID N0:127:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 9 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(ix) FEATURE:
(A) NAME/KEY: Modified-site
(B) LOCATION: 3
(D) OTHER INFORMATION: /product= "OTHER"
/note= "Xaa = Gln or Leu"
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:127:
Xaa Pro Xaa Val Thr Ala Tyr Ala Gly
1 5
(2) INFORMATION FOR SEQ ID N0:128:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 9 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
CA 02268036 1999-10-12
133
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0::128:
Xaa Xaa Xaa Glu Lys Pro Phe Leu Arg
1 5
(2) INFORMATION FOR SEQ ID N0:129:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:129:
Xaa Asp Ser Glu Lys Ser Ala Thr Ile Lys Val Thr Asp Ala Ser
1 5 7.0 15
(2) INFORMATION FOR SEQ ID N0:130:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:130:
Ala Gly Asp Thr Xaa Ile Tyr Ile Val Gly Asn Leu Thr Ala Asp
1 5 10 15
(2) INFORMATION FOR SEQ ID N0:131:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:131:
Ala Pro Glu Ser Gly Ala Gly Leu Gly Gly Thr Val Gln Ala Gly
1 5 10 15
(2) INFORMATION FOR SEQ ID N0:132:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:132:
Xaa Tyr Ile Ala Tyr Xaa Thr Thr Ala Gly Ile Val Pro Gly Lys Ile
1 5 10 15
Asn Val His Leu Val
CA 02268036 1999-10-12
134
(2) INFORMATION
FOR
SEQ
ID N0:133:
(i) S EQUENCE RACTERISTICS:
CHA
(A) LENGTH:882 base
pairs
(B) TYPE:ucleic
n acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID NO:133:
GCAACGCTGTCGTGGCCTTTGCGGTGATCGGTTTCGCCTCGCTGGCGGTGGCGGTGGCGG 60
TCACCATCCGACCGACCGCGGCCTCAAAACCGGTAGAGGGACACCAAAACGCCCAGCCAG 120
GGAAGTTCATGCCGTTGTTGCCGACGCAACAGCAGGCGC:CGGTCCCGCCGCCTCCGCCCG 180
ATGATCCCACCGCTGGATTCCAGGGCGGCACCATTCCGGCTGTACAGAACGTGGTGCCGC 240
GGCCGGGTACCTCACCCGGGGTGGGTGGGACGCCGGCTTCGCCTGCGCCGGAAGCGCCGG 300
CCGTGCCCGGTGTTGTGCCTGCCCCGGTGCCAATCCCGC~TCCCGATCATCATTCCCCCGT 360
TCCCGGGTTGGCAGCCTGGAATGCCGACCATCCCCACCGCACCGCCGACGACGCCGGTGA 420
CCACGTCGGCGACGACGCCGCCGACCACGCCGCCGACCP~CGCCGGTGACCACGCCGCCAA 480
CGACGCCGCCGACCACGCCGGTGACCACGCCGCCAACGACGCCGCCGACCACGCCGGTGA 540
CCACGCCACCAACGACCGTCGCCCCGACGACCGTCGCCC'.CGACGACGGTCGCTCCGACCA 600
CCGTCGCCCCGACCACGGTCGCTCCAGCCACCGCCACGC'CGACGACCGTCGCTCCGCAGC 660
CGACGCAGCAGCCCACGCAACAACCAACCCAACAGATGC'CAACCCAGCAGCAGACCGTGG 720
CCCCGCAGACGGTGGCGCCGGCTCCGCAGCCGCCGTCCGGTGGCCGCAACGGCAGCGGCG 780
GGGGCGACTTATTCGGCGGGTTCTGATCACGGTCGCGGCTTCACTACGGTCGGAGGACAT 840
GGCCGGTGATGCGGTGACGGTGGTGCTGCCCTGTCTCAA.CGA 882
(2) INFORMATION FOR SEQ ID N0:134:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 815 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:134:
CCATCAACCA ACCGCTCGCG CCGCCCGCGC CGCCGGATCC GCCGTCGCCG CCACGCCCGC 60
CGGTGCCTCC GGTGCCCCCG TTGCCGCCGT CGCCGCCGTC GCCGCCGACC GGCTGGGTGC 120
CTAGGGCGCT GTTACCGCCC TGGTTGGCGG GGACGCCGCC GGCACCACCG GTACCGCCGA 180
TGGCGCCGTT GCCGCCGGCG GCACCGTTGC CACCGTTGCC ACCGTTGCCA CCGTTGCCGA 240
CCAGCCACCC GCCGCGACCA CCGGCACCGC CGGCGCCGCC CGCACCGCCG GCGTGCCCGT 300
CA 02268036 1999-10-12
135
TCGTGCCCGTACCGCCGGCACCGCCGTTGCCGCCGTCACCGCCGACGGAACTACCGGCGG 360
ACGCGGCCTGCCCGCCGGCGCCGCCCGCACCGCCATTGGCACCGCCGTCACCGCCGGCTG 420
GGAGTGCCGCGATTAGGGCACTGACCGGCGCAACCAGCtaCAAGTACTCTCGGTCACCGAG 480
CACTTCCAGACGACACCACAGCACGGGGTTGTCGGCGGACTGGGTGAAATGGCAGCCGAT 540
AGCGGCTAGCTGTCGGCTGCGGTCAACCTCGATCATGA'CGTCGAGGTGACCGTGACCGCG 600
CCCCCCGAAGGAGGCGCTGAACTCGGCGTTGAGCCGATCGGCGATCGGTTGGGGCAGTGC 660
CCAGGCCAATACGGGGATACCGGGTGTCNAAGCCGCCGCGAGCGCAGCTTCGGTTGCGCG 720
ACNGTGGTCGGGGTGGCCTGTTACGCCGTTGTCNTCGAACACGAGTAGCAGGTCTGCTCC 780
GGCGAGGGCATCCACCACGCGTTGCGTCAGCTCGT 815
(2) INFORMATION
FOR SEQ
ID N0:135:
(i) SEQUENCE S:
CHARACTERISTIC
(A) LENGTH:1152 basepairs
(B) TYPE:ucleic
n acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY: linear
(xi) SEQUENCE EQ ID
DESCRIPTION: N0:7.35:
S
ACCAGCCGCCGGCTGAGGTCTCAGATCAGAGAGTCTCCC~GACTCACCGGGGCGGTTCAGC 60
CTTCTCCCAGAACAACTGCTGAAGATCCTCGCCCGCGAAP.CAGGCGCTGATTTGACGCTC 120
TATGACCGGTTGAACGACGAGATCATCCGGCAGATTGATATGGCACCGCTGGGCTAACAG 180
GTGCGCAAGATGGTGCAGCTGTATGTCTCGGACTCCGTGTCGCGGATCAGCTTTGCCGAC 240
GGCCGGGTGATCGTGTGGAGCGAGGAGCTCGGCGAGAGC:CAGTATCCGATCGAGACGCTG 300
GACGGCATCACGCTGTTTGGGCGGCCGACGATGACAACGCCCTTCATCGTTGAGATGCTC 360
AAGCGTGAGCGCGACATCCAGCTCTTCACGACCGACGGCCACTACCAGGGCCGGATCTCA 420
ACACCCGACGTGTCATACGCGCCGCGGCTCCGTCAGCAA.GTTCACCGCACCGACGATCCT 480
GCGTTCTGCCTGTCGTTAAGCAAGCGGATCGTGTCGAGGAAGATCCTGAATCAGCAGGCC 540
TTGATTCGGGCACACACGTCGGGGCAAGACGTTGCTGAGAGCATCCGCACGATGAAGCAC 600
TCGCTGGCCTGGGTCGATCGATCGGGCTCCCTGGCGGAGTTGAACGGGTTCGAGGGAAAT 660
GCCGCAAAGGCATACTTCACCGCGCTGGGGCATCTCGTCCCGCAGGAGTTCGCATTCCAG 720
GGCCGCTCGACTCGGCCGCCGTTGGACGCCTTCAACTCGATGGTCAGCCTCGGCTATTCG 780
CTGCTGTACAAGAACATCATAGGGGCGATCGAGCGTCACAGCCTGAACGCGTATATCGGT 840
TTCCTACACCAGGATTCACGAGGGCACGCAACGTCTCGTGCCGAATTCGGCACGAGCTCC 900
GCTGAAACCGCTGGCCGGCTGCTCAGTGCCCGTACGTAATCCGCTGCGCCCAGGCCGGCC 960
CGCCGGCCGAATACCAGCAGATCGGACAGCGAATTGCCGCCCAGCCGGTTGGAGCCGTGC 1020
ATACCGCCGGCACACTCACCGGCAGCGAACAGGCCTGGCACCGTGGCGGCGCCGGTGTCC 1080
CA 02268036 1999-10-12
136
GCGTCTACTT CGACACCGCC CATCACGTAG TGACACGTCG GCCCGACTTC CATTGCCTGC 1140
GTTCGGCACG AG 1152
(2) INFORMATION FOR SEQ ID N0:136:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:655 base pairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY:
linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:136:
CTCGTGCCGATTCGGCAGGGTGTACTTGCC GGTGGTGTANGCCGCATGAGTGCCGACGAC 60
CAGCAATGCGGCAACAGCACGGATCCCGGT CAACGACGC;CACCCGGTCCACGTGGGCGAT 120
CCGCTCGAGTCCGCCCTGGGCGGCTCTTTC CTTGGGCACiGGTCATCCGACGTGTTTCCGC 180
CGTGGTTTGCCGCCATTATGCCGGCGCGCC GCGTCGGGC;GGCCGGTATGGCCGAANGTCG 240
ATCAGCACACCCGAGATACGGGTCTGTGCA AGCTTTTTGAGCGTCGCGCGGGGCAGCTTC 300
GCCGGCAATTCTACTAGCGAGAAGTCTGGC CCGATACGGATCTGACCGAAGTCGCTGCGG 360
TGCAGCCCACCCTCATTGGCGATGGCGCCG ACGATGGCGCCTGGACCGATCTTGTGCCGC 420
TTGCCGACGGCGACGCGGTAGGTGGTCAAG TCCGGTCTACGCTTGGGCCTTTGCGGACGG 480
TCCCGACGCTGGTCGCGGTTGCGCCGCGAA AGCGGCGGGTCGGGTGCCATCAGGAATGCC 540
TCACCGCCGCGGCACTGCACGGCCAGTGCC GCGGCGATGTCAGCCATCGGGACATCATGC 600
TCGCGTTCATACTCCTCGACCAGTCGGCGG AACAGCTCGATTCCCGGACCGCCCA 655
(2) INFORMATION FOR SEQ ID N0:137:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 267 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:137:
Asn Ala Val Val Ala Phe Ala Val Ile Gly Phe Ala Ser Leu Ala Val
1 5 10 15
Ala Val Ala Val Thr Ile Arg Pro Thr Ala Ala Ser Lys Pro Val Glu
20 25 30
Gly His Gln Asn Ala Gln Pro Gly Lys Phe Met Pro Leu Leu Pro Thr
35 40 45
Gln Gln Gln Ala Pro Val Pro Pro Pro Pro Pro Asp Asp Pro Thr Ala
50 55 60
Gly Phe Gln Gly Gly Thr Ile Pro Ala V~sl Gln Asn Val Val Pro Arg
65 70 75 80
CA 02268036 1999-10-12
137
Pro Gly Thr Ser Pro Gly Val Gly Gly 'Phr Pro Ala Ser Pro Ala Pro
85 90 95
Glu Ala Pro Ala Val Pro Gly Val Val Pro Ala Pro Val Pro Ile Pro
100 105 110
Val Pro Ile Ile Ile Pro Pro Phe Pro oily Trp Gln Pro Gly Met Pro
115 120 125
Thr Ile Pro Thr Ala Pro Pro Thr Thr 1?ro Val Thr Thr Ser Ala Thr
130 135 140
Thr Pro Pro Thr Thr Pro Pro Thr Thr I?ro Val Thr Thr Pro Pro Thr
145 150 155 160
Thr Pro Pro Thr Thr Pro Val Thr Thr I?ro Pro Thr Thr Pro Pro Thr
165 170 175
Thr Pro Val Thr Thr Pro Pro Thr Thr Val Ala Pro Thr Thr Val Ala
180 185 190
Pro Thr Thr Val Ala Pro Thr Thr Val Ala Pro Thr Thr Val Ala Pro
195 200 205
Ala Thr Ala Thr Pro Thr Thr Val Ala Pro Gln Pro Thr Gln Gln Pro
210 215 220
Thr Gln Gln Pro Thr Gln Gln Met Pro Thr Gln Gln Gln Thr Val Ala
225 230 235 240
Pro Gln Thr Val Ala Pro Ala Pro Gln Pro Pro Ser Gly Gly Arg Asn
245 250 255
Gly Ser Gly Gly Gly Asp Leu Phe Gly Gly Phe
260 265
(2) INFORMATION FOR SEQ ID N0:138:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 174 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:138:
Ile Asn Gln Pro Leu Ala Pro Pro Ala Pro Pro Asp Pro Pro Ser Pro
1 5 10 15
Pro Arg Pro Pro Val Pro Pro Val Pro Pro Leu Pro Pro Ser Pro Pro
20 25 30
Ser Pro Pro Thr Gly Trp Val Pro Arg Ala Leu Leu Pro Pro Trp Leu
35 40 45
Ala Gly Thr Pro Pro Ala Pro Pro Val Pro Pro Met Ala Pro Leu Pro
50 55 60
Pro Ala Ala Pro Leu Pro Pro Leu Pro Pro Leu Pro Pro Leu Pro Thr
65 70 75 80
Ser His Pro Pro Arg Pro Pro Ala Pro Pro Ala Pro Pro Ala Pro Pro
85 90 95
CA 02268036 1999-10-12
138
Ala Cys Pro Phe Val Pro Val Pro Pro Ala Pro Pro Leu Pro Pro Ser
100 105 110
Pro Pro Thr Glu Leu Pro Ala Asp Ala Ala Cys Pro Pro Ala Pro Pro
115 120 125
Ala Pro Pro Leu Ala Pro Pro Ser Pro I?ro Ala Gly Ser Ala Ala Ile
130 135 140
Arg Ala Leu Thr Gly Ala Thr Ser Ala Ser Thr Leu Gly His Arg Ala
145 150 155 160
Leu Pro Asp Asp Thr Thr Ala Arg Gly C:ys Arg Arg Thr Gly
165 7.70
(2) INFORMATION FOR SEQ ID N0:139:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 35 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:139:
Gln Pro Pro Ala Glu Val Ser Asp Gln F,rg Val Ser Gly Leu Thr Gly
1 5 10 15
Ala Val Gln Pro Ser Pro Arg Thr Thr Ala Glu Asp Pro Arg Pro Arg
20 25 30
Asn Arg Arg
(2) INFORMATION FOR SEQ ID N0:140:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 104 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:140:
Arg Ala Asp Ser Ala Gly Cys Thr Cys Arg Trp Cys Xaa Pro His Glu
1 5 10 15
Cys Arg Arg Pro Ala Met Arg Gln Gln His Gly Ser Arg Ser Thr Thr
20 25 30
Pro Pro Gly Pro Arg Gly Arg Ser Ala Arg Val Arg Pro Gly Arg Leu
35 40 45
Phe Pro Trp Ala Gly Ser Ser Asp Val P:he Pro Pro Trp Phe Ala Ala
50 55 60
Ile Met Pro Ala Arg Arg Val Gly Arg P.ro Val Trp Pro Xaa Val Asp
65 70 75 80
Gln His Thr Arg Asp Thr Gly Leu Cys L:ys Leu Phe Glu Arg Arg Ala
85 90 95
CA 02268036 1999-10-12
139
Gly Gln Leu Arg Arg Gln Phe Tyr
100
(2) INFORMATION FOR SEQ ID N0:141:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 53 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:191:
GGATCCATAT GGGCCATCAT CATCATCATC ACGTGATCGA CATCATCGGG ACC 53
(2) INFORMATION FOR SEQ ID N0:142:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 42 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:7.42:
CCTGAATTCA GGCCTCGGTT GCGCCGGCCT CATCTTGAAC GA 42
(2) INFORMATION FOR SEQ ID N0:143:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 31 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:143:
GGATCCTGCA GGCTCGAAAC CACCGAGCGG T 31
(2) INFORMATION FOR SEQ ID N0:144:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 31 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:144:
CTCTGAATTC AGCGCTGGAA ATCGTCGCGA T 31
(2) INFORMATION FOR SEQ ID N0:145:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
CA 02268036 1999-10-12
140
(xi) SEQUENCE DESCRIPTION: SEQ ID N0::145:
GGATCCAGCG CTGAGATGAA GACCGATGCC GCT 33
(2) INFORMATION FOR SEQ ID N0:146:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:146:
GAGAGAATTC TCAGAAGCCC ATTTGCGAGG ACA 33
(2) INFORMATION FOR SEQ ID N0:147:
(i)SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1993base rs
pai
(B) TYPE: nucleicacid
(C) STRANDEDNESS:single
(D) TOPOLOGY: ear
lin
(ix)FEATURE:
(A) NAME/KEY:
CDS
(B) LOCATION: ..1276
152
(xi)SEQUENCE DESCRIPTION: ID 0:147:
SEQ N
TGTTCTTCGA CCCACCC-~A ACAGCTG TTC TCCTCGCCGA 60
CGGCAGGCTG
GTGGAGGAAG
GG
AGCATGCGGA CGGGGGACGTC AAG GACGCCAAGC 120
AACCGCCCGA
TACGTCGCCG
GACTGT
GCGGAAATTG GTGAAA CGTTTG CAT 172
AAGAGCACAG ATT ACG
AAAGGTATGG
C
ValLys Ile Leu HisThr
Arg
1 5
CTGTTGGCC GTG TTG ACC GCG CTGCTG CTAGCAGCG GCGGGC 220
GCT CCG
LeuLeuAla Val Leu Thr Ala LeuLeu LeuAlaAla AlaGly
Ala Pro
10 15 20
TGTGGCTCG AAA CCA CCG GGT CCTGAA ACGGGCGCC GGCGCC 268
AGC TCG
CysGlySer Lys Pro Pro Gly ProGlu ThrGlyAla GlyAla
Ser Ser
25 30 35
GGTACTGTC GCG ACT ACC GCG TCGCCG GTGACGTTG GCGGAG 316
CCC TCG
GlyThrVal Ala Thr Thr Ala SerPro ValThrLeu AlaGlu
Pro Ser
40 45 50 55
ACCGGTAGC ACG CTG CTC CCG TTCAAC CTGTGGGGT CCGGCC 364
TAC CTG
ThrGlySer Thr Leu Leu Pro PheAsn LeuTrpGly ProAla
Tyr Leu
60 65 70
TTTCACGAG AGG TAT CCG GTC ATCACC GCTCAGGGC ACCGGT 412
AAC ACG
PheHisGlu Arg Tyr Pro Val IleThr AlaGlnGly ThrGly
Asn Thr
75 80 85
TCTGGTGCC GGG ATC GCG GCC GCCGGG ACGGTCAAC ATTGGG 460
CAG GCC
SerGlyAla Gly Ile Ala Ala AlaGly ThrValAsn IleGly
Gln Ala
CA 02268036 1999-10-12
141
90 95 100
GCCTCCGACGCC TATCTGTCG GAAGGTGAT ATGGCCGCG CACAAGGGG 508
AlaSerAspAla TyrLeuSer GluGlyAsp MetAlaAla HisLysGly
105 110 115
CTGATGAACATC GCGCTAGCC ATCTCCGCT CAGCAGGTC AACTACAAC 556
LeuMetAsnIle AlaLeuAla IleSerAla G7_nGlnVal AsnTyrAsn
120 125 130 135
CTGCCCGGAGTG AGCGAGCAC CTCAAGCTG AACGGAAAA GTCCTGGCG 604
LeuProGlyVal SerGluHis LeuLysLeu A:>nGlyLys ValLeuAla
140 145 150
GCCATGTACCAG GGCACCATC AAAACCTGG GACGACCCG CAGATCGCT 652
AlaMetTyrGln GlyThrIle LysThrTrp AspAspPro GlnIleAla
155 160 165
GCGCTCAACCCC GGCGTGAAC CTGCCCGGC AC;CGCGGTA GTTCCGCTG 700
AlaLeuAsnPro GlyValAsn LeuProGly ThrAlaVal ValProLeu
170 175 180
CACCGCTCCGAC GGGTCCGGT GACACCTTC TTGTTCACC CAGTACCTG 748
HisArgSerAsp GlySerGly AspThrPhe Le~uPheThr GlnTyrLeu
185 190 195
TCCAAGCAAGAT CCCGAGGGC TGGGGCAAG TCGCCCGGC TTCGGCACC 796
SerLysGlnAsp ProGluGly TrpGlyLys SeerProGly PheGlyThr
200 205 210 215
ACCGTCGACTTC CCGGCGGTG CCGGGTGCG CTGGGTGAG AACGGCAAC 844
ThrValAspPhe ProAlaVal ProGlyAla LeuGlyGlu AsnGlyAsn
220 225 230
GGCGGCATGGTG ACCGGTTGC GCCGAGACA CCGGGCTGC GTGGCCTAT 892
GlyGlyMetVal ThrGlyCys AlaGluThr ProGlyCys ValAlaTyr
235 240 245
ATCGGCATCAGC TTCCTCGAC CAGGCCAGT CAACGGGGA CTCGGCGAG 940
IleGlyIleSer PheLeuAsp GlnAlaSer GlnArgGly LeuGlyGlu
250 255 260
GCCCAACTAGGC AATAGCTCT GGCAATTTC TTGTTGCCC GACGCGCAA 988
AlaGlnLeuGly AsnSerSer GlyAsnPhe LeuLeuPro AspAlaGln
265 270 275
AGCATTCAGGCC GCGGCGGCT GGCTTCGCA TCGAAAACC CCGGCGAAC 1036
SerIleGlnAla AlaAlaAla GlyPheAla SerLysThr ProAlaAsn
280 285 290 295
CAGGCGATTTCG ATGATCGAC GGGCCCGCC CCGGACGGC TACCCGATC 1084
GlnAlaIleSer MetIleAsp GlyProAla ProAspGly TyrProIle
300 305 310
ATCAACTACGAG TACGCCATC GTCAACAAC CGGCAAAAG GACGCCGCC 1132
IleAsnTyrGlu TyrAlaIle ValAsnAsn ArgGlnLys AspAlaAla
315 320 325
ACCGCGCAGACC TTGCAGGCA TTTCTGCAC TGGGCGATC ACCGACGGC 1180
ThrAlaGlnThr LeuGlnAla PheLeuHis TrpAlaIle ThrAspGly
330 335 340
AACAAGGCCTCG TTCCTCGAC CAGGTTCAT TTCCAGCCG CTGCCGCCC 1228
AsnLysAlaSer PheLeuAsp GlnValHis Ph~~GlnPro LeuProPro
CA 02268036 1999-10-12
142
345 350 355
GCG GTG GTG AAG TTG TCT GAC GCG TTG ATC GCG ACG ATT TCC AGC 1273
Ala Val Val Lys Leu Ser Asp Ala Leu Ile A:La Thr Ile Ser Ser
360 365 3'70
TAGCCTCGTTGACCACCACGCGACAGCAACCTCCGTCGC~GCCATCGGGCTGCTTTGCGGA 1333
GCATGCTGGCCCGTGCCGGTGAAGTCGGCCGCGCTGGCC:CGGCCATCCGGTGGTTGGGTG 1393
GGATAGGTGCGGTGATCCCGCTGCTTGCGCTGGTCTTGGTGCTGGTGGTGCTGGTCATCG 1453
AGGCGATGGGTGCGATCAGGCTCAACGGGTTGCATTTC~PTCACCGCCACCGAATGGAATC 1513
CAGGCAACACCTACGGCGAAACCGTTGTCACCGACGCGTCGCCCATCCGGTCGGCGCCTA 1573
CTACGGGGCGTTGCCGCTGATCGTCGGGACGCTGGCGAC:CTCGGCAATCGCCCTGATCAT 1633
CGCGGTGCCGGTCTCTGTAGGAGCGGCGCTGGTGATCGTGGAACGGCTGCCGAAACGGTT 1693
GGCCGAGGCTGTGGGAATAGTCCTGGAATTGCTCGCCGC~AATCCCCAGCGTGGTCGTCGG 1753
TTTGTGGGGGGCAATGACGTTCGGGCCGTTCATCGCTCATCACATCGCTCCGGTGATCGC 1813
TCACAACGCTCCCGATGTGCCGGTGCTGAACTACTTGCGCGGCGACCCGGGCAACGGGGA 1873
GGGCATGTTGGTGTCCGGTCTGGTGTTGGCGGTGATGG7.'CGTTCCCATTATCGCCACCAC 1933
CACTCATGACCTGTTCCGGCAGGTGCCGGTGTTGCCCCGGGAGGGCGCGATCGGGAATTC 1993
(2) INFORMATION FOR SEQ ID N0:148:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 374 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NC>:148:
Val Lys Ile Arg Leu His Thr Leu Leu Ala Val Leu Thr Ala Ala Pro
1 5 10 15
Leu Leu Leu Ala Ala Ala Gly Cys Gly Ser Lys Pro Pro Ser Gly Ser
20 25 30
Pro Glu Thr Gly Ala Gly Ala Gly Thr Val Ala Thr Thr Pro Ala Ser
35 40 45
Ser Pro Val Thr Leu Ala Glu Thr Gly Ser Thr Leu Leu Tyr Pro Leu
50 55 60
Phe Asn Leu Trp Gly Pro Ala Phe His Glu Arg Tyr Pro Asn VaI Thr
65 70 75 80
Ile Thr Ala Gln Gly Thr Gly Ser Gly Ala Gly Ile Ala Gln Ala Ala
85 90 95
Ala Gly Thr Val Asn Ile Gly Ala Ser Asp Ala Tyr Leu Ser Glu Gly
100 105 110
Asp Met Ala Ala His Lys Gly Leu Met Asn Ile Ala Leu Ala Ile Ser
115 120 125
CA 02268036 1999-10-12
143
Ala Gln Gln Val Asn Tyr Asn Leu Pro Gly Val Ser Glu His Leu Lys
130 135 140
Leu Asn Gly Lys Val Leu Ala Ala Met Tyr Gln Gly Thr Ile Lys Thr
145 150 1.'i5 160
Trp Asp Asp Pro Gln Ile Ala Ala Leu Asn P~_.o Gly Val Asn Leu Pro
165 170 175
Gly Thr Ala Val Val Pro Leu His Arg Ser Asp Gly Ser Gly Asp Thr
180 185 190
Phe Leu Phe Thr Gln Tyr Leu Ser Lys Gln A:>p Pro Glu Gly Trp Gly
195 200 205
Lys Ser Pro Gly Phe Gly Thr Thr Val Asp Phe Pro Ala Val Pro Gly
210 215 220
Ala Leu Gly Glu Asn Gly Asn Gly Gly Met Val Thr Gly Cys Ala Glu
225 230 235 240
Thr Pro Gly Cys Val Ala Tyr Ile Gly Ile Se:r Phe Leu Asp Gln Ala
245 250 255
Ser Gln Arg Gly Leu Gly Glu Ala Gln Leu Gl.y Asn Ser Ser Gly Asn
260 265 270
Phe Leu Leu Pro Asp Ala Gln Ser Ile Gln Al.a Ala Ala Ala Gly Phe
275 280 285
Ala Ser Lys Thr Pro Ala Asn Gln Ala Ile Ser Met Ile Asp Gly Pro
290 295 300
Ala Pro Asp Gly Tyr Pro Ile Ile Asn Tyr Gl.u Tyr Ala Ile Val Asn
305 310 31.5 320
Asn Arg Gln Lys Asp Ala Ala Thr Ala Gln Thr Leu Gln,Ala Phe Leu
325 330 335
His Trp Ala Ile Thr Asp Gly Asn Lys Ala Seer Phe Leu Asp Gln Val
340 345 350
His Phe Gln Pro Leu Pro Pro Ala Val Val Lys Leu Ser Asp Ala Leu
355 360 365
Ile Ala Thr Ile Ser Ser
370
(2) INFORMATION FOR SEQ ID N0:149:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1993 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:149:
TGTTCTTCGA CGGCAGGCTG GTGGAGGAAG GGCCCACCGA ACAGCTGTTC TCCTCGCCGA 60
AGCATGCGGA AACCGCCCGA TACGTCGCCG GACTGTCGGG GGACGTCAAG GACGCCAAGC 120
GCGGAAATTG AAGAGCACAG AAAGGTATGG CGTGAAAATT CGTTTGCATA CGCTGTTGGC 180
CA 02268036 1999-10-12
144
CGTGTTGACCGCTGCGCCGCTGCTGCTAGCAGCGGCGGGCTGTGGCTCGA 240
AACCACCGAG
CGGTTCGCCTGAAACGGGCGCCGGCGCCGGTACTGTCGCGACTACCCCCGCGTCGTCGCC 300
GGTGACGTTGGCGGAGACCGGTAGCACGCTGCTCTACCCGCTGTTCAACCTGTGGGGTCC 360
GGCCTTTCACGAGAGGTATCCGAACGTCACGATCACCGCTCAGGGCACCGGTTCTGGTGC 420
CGGGATCGCGCAGGCCGCCGCCGGGACGGTCAACATTG(~GGCCTCCGACGCCTATCTGTC 480
GGAAGGTGATATGGCCGCGCACAAGGGGCTGATGAACA'.CCGCGCTAGCCATCTCCGCTCA 540
GCAGGTCAACTACAACCTGCCCGGAGTGAGCGAGCACC'.CCAAGCTGAACGGAAAAGTCCT 600
GGCGGCCATGTACCAGGGCACCATCAAAACCTGGGACGACCCGCAGATCGCTGCGCTCAA 660
CCCCGGCGTGAACCTGCCCGGCACCGCGGTAGTTCCGCTGCACCGCTCCGACGGGTCCGG 720
TGACACCTTCTTGTTCACCCAGTACCTGTCCAAGCAAGATCCCGAGGGCTGGGGCAAGTC 780
GCCCGGCTTCGGCACCACCGTCGACTTCCCGGCGGTGCC:GGGTGCGCTGGGTGAGAACGG 840
CAACGGCGGCATGGTGACCGGTTGCGCCGAGACACCGGCiCTGCGTGGCCTATATCGGCAT 900
CAGCTTCCTCGACCAGGCCAGTCAACGGGGACTCGGCGAGGCCCAACTAGGCAATAGCTC 960
TGGCAATTTCTTGTTGCCCGACGCGCAAAGCATTCAGGC:CGCGGCGGCTGGCTTCGCATC 1020
GAAAACCCCGGCGAACCAGGCGATTTCGATGATCGACGGGCCCGCCCCGGACGGCTACCC 1080
GATCATCAACTACGAGTACGCCATCGTCAACAACCGGCAAAAGGACGCCGCCACCGCGCA 1140
GACCTTGCAGGCATTTCTGCACTGGGCGATCACCGACGGCAACAAGGCCTCGTTCCTCGA 1200
CCAGGTTCATTTCCAGCCGCTGCCGCCCGCGGTGGTGAAGTTGTCTGACGCGTTGATCGC 1260
GACGATTTCCAGCTAGCCTCGTTGACCACCACGCGACAC~CAACCTCCGTCGGGCCATCGG 1320
GCTGCTTTGCGGAGCATGCTGGCCCGTGCCGGTGAAGTC:GGCCGCGCTGGCCCGGCCATC 1380
CGGTGGTTGGGTGGGATAGGTGCGGTGATCCCGCTGCTTGCGCTGGTCTTGGTGCTGGTG 1440
GTGCTGGTCATCGAGGCGATGGGTGCGATCAGGCTCAAC:GGGTTGCATTTCTTCACCGCC 1500
ACCGAATGGAATCCAGGCAACACCTACGGCGAAACCGTTGTCACCGACGCGTCGCCCATC 1560
CGGTCGGCGCCTACTACGGGGCGTTGCCGCTGATCGTCGGGACGCTGGCGACCTCGGCAA 1620
TCGCCCTGATCATCGCGGTGCCGGTCTCTGTAGGAGCGGCGCTGGTGATCGTGGAACGGC 1680
TGCCGAAACGGTTGGCCGAGGCTGTGGGAATAGTCCTGGAATTGCTCGCCGGAATCCCCA 1740
GCGTGGTCGTCGGTTTGTGGGGGGCAATGACGTTCGGGCCGTTCATCGCTCATCACATCG 1800
CTCCGGTGATCGCTCACAACGCTCCCGATGTGCCGGTGCTGAACTACTTGCGCGGCGACC 1860
CGGGCAACGGGGAGGGCATGTTGGTGTCCGGTCTGGTGTTGGCGGTGATGGTCGTTCCCA 1920
TTATCGCCACCACCACTCATGACCTGTTCCGGCAGGTGCCGGTGTTGCCCCGGGAGGGCG 1980
CGATCGGGAATTC 1993
CA 02268036 1999-10-12
145
(2) INFORMATION FOR SEQ ID N0:150:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 374 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:7.50:
Met Lys Ile Arg Leu His Thr Leu Leu Ala Val Leu Thr Ala Ala Pro
1 5 7.0 15
Leu Leu Leu Ala Ala Ala Gly Cys Gly Ser Lys Pro Pro Ser Gly Ser
20 25 30
Pro Glu Thr Gly Ala Gly Ala Gly Thr Val Ala Thr Thr Pro Ala Ser
35 40 45
Ser Pro Val Thr Leu Ala Glu Thr Gly ~cer Thr Leu Leu Tyr Pro Leu
50 55 60
Phe Asn Leu Trp Gly Pro Ala Phe His Glu Arg Tyr Pro Asn Val Thr
65 70 75 80
Ile Thr Ala Gln Gly Thr Gly Ser Gly Ala Gly Ile Ala Gln Ala Ala
85 90 95
Ala Gly Thr Val Asn Ile Gly Ala Ser P,sp Ala Tyr Leu Ser Glu Gly
100 105 110
Asp Met Ala Ala His Lys Gly Leu Met Asn Ile Ala Leu Ala Ile Ser
115 120 125
Ala Gln Gln Val Asn Tyr Asn Leu Pro Gly Val Ser Glu His Leu Lys
130 135 140
Leu Asn Gly Lys Val Leu Ala Ala Met Tyr Gln Gly Thr Ile Lys Thr
145 150 155 160
Trp Asp Asp Pro Gln Ile Ala Ala Leu Asn Pro Gly Val Asn Leu Pro
165 170 175
Gly Thr Ala Val Val Pro Leu His Arg Ser Asp Gly Ser Gly Asp Thr
180 185 190
Phe Leu Phe Thr Gln Tyr Leu Ser Lys G1n Asp Pro Glu Gly Trp Gly
195 200 205
Lys Ser Pro Gly Phe Gly Thr Thr Val Asp Phe Pro Ala Val Pro Gly
210 215 220
Ala Leu Gly Glu Asn Gly Asn Gly Gly Met Val Thr Gly Cys Ala Glu
225 230 235 240
Thr Pro Gly Cys Val Ala Tyr Ile Gly Ile Ser Phe Leu Asp Gln Ala
245 250 255
Ser Gln Arg Gly Leu Gly Glu Ala Gln Leu Gly Asn Ser Ser Gly Asn
260 265 270
Phe Leu Leu Pro Asp Ala Gln Ser Ile Gln Ala Ala Ala Ala Gly Phe
275 280 285
CA 02268036 1999-10-12
146
Ala Ser Lys Thr Pro Ala Asn Gln Ala Ile Ser Met Ile Asp Gly Pro
290 295 300
Ala Pro Asp Gly Tyr Pro Ile Ile Asn Tyr Glu Tyr Ala Ile Val Asn
305 310 315 320
Asn Arg Gln Lys Asp Ala Ala Thr Ala Gln Thr Leu Gln Ala Phe Leu
325 330 335
His Trp Ala Ile Thr Asp Gly Asn Lys Ala Ser Phe Leu Asp Gln Val
340 345 350
His Phe Gln Pro Leu Pro Pro Ala Val Val Lys Leu Ser Asp Ala Leu
355 360 365
Ile Ala Thr Ile Ser Ser
370
(2) INFORMATION
FOR
SEQ
ID N0:151:
(i) SEQUENCE S:
CHARACTERISTIC
(A) LENGTH:1777 basepairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY:
linear
(xi) EQ ID
SEQUENCE N0:7.51:
DESCRIPTION:
S
GGTCTTGACCACCACCTGGGTGTCGAAGTCGGTGCCCGC~ATTGAAGTCCAGGTACTCGTG 60
GGTGGGGCGGGCGAAACAATAGCGACAAGCATGCGAGCAGCCGCGGTAGCCGTTGACGGT 120
GTAGCGAAACGGCAACGCGGCCGCGTTGGGCACCTTGTTCAGCGCTGATTTGCACAACAC 180
CTCGTGGAAGGTGATGCCGTCGAATTGTGGCGCGCGAAC:GCTGCGGACCAGGCCGATCCG 240
CTGCAACCCGGCAGCGCCCGTCGTCAACGGGCATCCCGTTCACCGCGACGGCTTGCCGGG 300
CCCAACGCATACCATTATTCGAACAACCGTTCTATACTT'TGTCAACGCTGGCCGCTACCG 360
AGCGCCGCACAGGATGTGATATGCCATCTCTGCCCGCAC:AGACAGGAGCCAGGCCTTATG 420
ACAGCATTCGGCGTCGAGCCCTACGGGCAGCCGAAGTAC:CTAGAAATCGCCGGGAAGCGC 480
ATGGCGTATATCGACGAAGGCAAGGGTGACGCCATCGTC:TTTCAGCACGGCAACCCCACG 540
TCGTCTTACTTGTGGCGCAACATCATGCCGCACTTGGAAGGGCTGGGCCGGCTGGTGGCC 600
TGCGATCTGATCGGGATGGGCGCGTCGGACAAGCTCAGCCCATCGGGACCCGACCGCTAT 660
AGCTATGGCGAGCAACGAGACTTTTTGTTCGCGCTCTGGGATGCGCTCGACCTCGGCGAC 720
CACGTGGTACTGGTGCTGCACGACTGGGGCTCGGCGCTC'GGCTTCGACTGGGCTAACCAG 780
CATCGCGACCGAGTGCAGGGGATCGCGTTCATGGAAGCGATCGTCACCCCGATGACGTGG 890
GCGGACTGGCCGCCGGCCGTGCGGGGTGTGTTCCAGGGTTTCCGATCGCCTCAAGGCGAG 900
CCAATGGCGTTGGAGCACAACATCTTTGTCGAACGGGTGCTGCCCGGGGCGATCCTGCGA 960
CAGCTCAGCGACGAGGAAATGAACCACTATCGGCGGCCATTCGTGAACGGCGGCGAGGAC 1020
CGTCGCCCCACGTTGTCGTGGCCACGAAACCTTCCAATCGACGGTGAGCCCGCCGAGGTC 1080
CA 02268036 1999-10-12
147
GTCGCGTTGGTCAACGAGTACCGGAGCTGGCTCGAGGAAACCGACATGCCGAAACTGTTC 1140
ATCAACGCCGAGCCCGGCGCGATCATCACCGGCCGCAT(:CGTGACTATGTCAGGAGCTGG 1200
CCCAACCAGACCGAAATCACAGTGCCCGGCGTGCATTT(:GTTCAGGAGGACAGCGATGGC 1260
GTCGTATCGTGGGCGGGCGCTCGGCAGCATCGGCGACCTGGGAGCGCTCTCATTTCACGA 1320
GACCAAGAATGTGATTTCCGGCGAAGGCGGCGCCCTGCTTGTCAACTCATAAGACTTCCT 1380
GCTCCGGGCAGAGATTCTCAGGGAAAAGGGCACCAATCGCAGCCGCTTCCTTCGCAACGA 1440
GGTCGACAAATATACGTGGCAGGACAAAGGTCTTCCTATTTGCCCAGCGAATTAGTCGCT 1500
GCCTTTCTATGGGCTCAGTTCGAGGAAGCCGAGCGGATC:ACGCGTATCCGATTGGACCTA 1560
TGGAACCGGTATCATGAAAGCTTCGAATCATTGGAACACzCGGGGGCTCCTGCGCCGTCCG 1620
ATCATCCCACAGGGCTGCTCTCACAACGCCCACATGTAC:TACGTGTTACTAGCGCCCAGC 1680
GCCGATCGGGAGGAGGTGCTGGCGCGTCTGACGAGCGAAGGTATAGGCGCGGTCTTTCAT 1740
TACGTGCCGCTTCACGATTCGCCGGCCGGGCGTCGCT 1777
(2) INFORMATION FOR SEQ ID N0:152:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 324 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID
N0:7.52:
GAGATTGAAT CGTACCGGTC TCCTTAGCGG TGAATGCCCA TATCACGCAC60
CTCCGTCCC;G
GGCCATGTTC TGGCTGTCGA CCTTCGCCCC GTTGGTAAAC CCAGGGTTTG120
ATGCCCGGAC
ATCAGTAATT CCGGGGGACG GTTGCGGGAA ATGTGCGTGA GCCGCGGCGC180
GGCGGCCAC~G
CGCCGTCGCC CAGGCGACCG CTGGATGCTC CGGCGACGTA GCCAGCGTTT240
AGCCCCGGTG
GGCGCGTGTC GTCCACAGTG GTACTCCGGT CGCGGTGCCT GGGTGAAGAC300
GACGACGCGG
CGTGACCGAC GCCGCCGATT CAGA 324
(2) INFORMATION FOR SEQ ID N0:153:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1338 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID
N0:153:
GCGGTACCGC CGCGTTGCGC TGGCACGGGA CTGAACCACT TCGCCTCGCG60
CCTGTACGF,C
AACGATTGAC GAACCGCTCG TGCGGCGGCT AAGGTGTGGG GTGATGTCGT120
GTGGGTGCT'C
CGATGACCGG CGCGGCACCC GGCCACTACG CGTCGAAGA.C GTCCTCGCCG CCCGCAGCGA 180
CA 02268036 1999-10-12
148
GCACGACTTCCAGCCCGACTCGATCGGCGTGCTGACCCGTCCTGTCGCTATGGCTGCCTG 240
GGAAGCTCGCGTTCGGAAGCGATTTGCGTTCCTCACTGACCTCGACGCCGACGAGCAGCG 300
GTGGGCCGCCTGCGACGAACGGCACCGCCGCGAAGTGGAGAACGCGCTGGCGGTGCTGCG 360
GTCCTGATCA ACCTGCCGGC GATCGTGCCG TTCCGCTGGC ACGGTTGCGG CTGGACGCGG 420
CTGAATCGACTAGATGAGAGCAGTTGGGCACGAATCCGGCTGTGGTGGTGAGCAAGACAC 480
GAGTACTGTCATCACTATTGGATGCACTGGATGACCGG(:CTGATTCAGCAGGACCAATGG 540
AACTGCCCGGGGCAAAACGTCTCGGAGATGATCGGCGTC:CCCTCGGAACCCTGCGGTGCT 600
GGCGTCATTCGGACATCGGTCCGGCTCGCGGGATCGTGGTGACGCCAGCGCTGAAGGAGT 660
GGAGCGCGGCGGTGCACGCGCTGCTGGACGGCCGGCAGACGGTGCTGCTGCGTAAGGGCG 720
GGATCGGCGAGAAGCGCTTCGAGGTGGCGGCCCACGAGTTCTTGTTGTTCCCGACGGTCG 780
CGCACAGCCACGCCGAGCGGGTTCGCCCCGAGCACCGCCiACCTGCTGGGCCCGGCGGCCG 840
CCGACAGCACCGACGAGTGTGTGCTACTGCGGGCCGCACiCGAAAGTTGTTGCCGCACTGC 900
CGGTTAACCGGCCAGAGGGT.CTGGACGCCATCGAGGATC;TGCACATCTGGACCGCCGAGT 960
CGGTGCGCGCCGACCGGCTCGACTTTCGGCCCAAGCACAP.ACTGGCCGTCTTGGTGGTCT 1020
CGGCGATCCCGCTGGCCGAGCCGGTCCGGCTGGCGCGTAGGCCCGAGTACGGCGGTTGCA 1080
CCAGCTGGGTGCAGCTGCCGGTGACGCCGACGTTGGCGGCGCCGGTGCACGACGAGGCCG 1140
CGCTGGCCGAGGTCGCCGCCCGGGTCCGCGAGGCCGTGGGTTGACTGGGCGGCATCGCTT 1200
GGGTCTGAGCTGTACGCCCAGTCGGCGCTGCGAGTGATC;TGCTGTCGGTTCGGTCCCTGC 1260
TGGCGTCAATTGACGGCGCGGGCAACAGCAGCATTGGCC~GCGCCATCCTCCGCGCGGCCG 1320
GCGCCCACCGCTACAACC 1338
(2) INFORMATION FOR SEQ ID N0:154:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 321 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) EQUENCE DESCRIPTION: SEQ
S ID N0:1.54:
CCGGCGGCACCGGCGGCACC GGCGGTACCG GCGGCAACGGCGCTGACGCCGCTGCTGTGG 60
TGGGCTTCGGCGCGAACGGC GACCCTGGCT TCGCTGGCGGCAAAGGCGGTAACGGCGGAA 120
TAGGTGGGGCCGCGGTGACA GGCGGGGTCG CCGGCGACCzGCGGCACCGGCGGCAAAGGTG 180
GCACCGGCGGTGCCGGCGGC GCCGGCAACG ACGCCGGCP,GCACCGGCAATCCCGGCGGTA 240
AGGGCGGCGACGGCGGGATC GGCGGTGCCG GCGGGGCCGGCGGCGCGGCCGGCACCGGCA 300
ACGGCGGCCATGCCGGCAAC C 321
CA 02268036 1999-10-12
149
(2) INFORMATION FOR SEQ ID N0:155:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 492 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:_L55:
GAAGACCCGGCCCCGCCATATCGATCGGCTCGCCGACTACTTTCGCCGAACGTGCACGCG 60
GCGGCGTCGGGCTGATCATCACCGGTGGCTACGCGCCCAACCGCACCGGATGGCTGCTGC 120
CGTTCGCCTCCGAACTCGTCACTTCGGCGCAAGCCCGAC:GGCACCGCCGAATCACCAGGG 180
CGGTCCACGATTCGGGTGCAAAGATCCTGCTGCAAATCC:TGCACGCCGGACGCTACGCCT 240
ACCACCCACTTGCGGTCAGCGCCTCGCCGATCAAGGCGC:CGATCACCCCGTTTCGTCCGC 300
GAGCACTATCGGCTCGCGGGGTCGAAGCGACCATCGCGGATTTCGCCCGCTGCGCGCAGT 360
TGGCCCGCGATGCCGGCTACGACGGCGTCGAAATCATGC~GCAGCGAAGGGTATCTGCTCA 420
ATCAGTTCCTGGCGCCGCGCACCAACAAGCGCACCGACTCGTGGGGCGGCACACCGGCCA 480
ACCGTCGCCGGT 492
(2) INFORMATION FOR SEQ ID N0:156:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 536 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:1.56:
Phe Ala Gln His Leu Val Glu Gly Asp Ala Val Glu Leu Trp Arg Ala
1 5 10 15
Asn Ala Ala Asp Gln Ala Asp Pro Leu Gln Pro Gly Ser Ala Arg Arg
20 25 30
Gln Arg Ala Ser Arg Ser Pro Arg Arg L~eu Ala Gly Pro Asn Ala Tyr
35 40 45
His Tyr Ser Asn Asn Arg Ser Ile Leu C'ys Gln Arg Trp Pro Leu Pro
50 55 60
Ser Ala Ala Gln Asp Val Ile Cys His Leu Cys Pro His Arg Gln Glu
65 70 75 80
Pro Gly Leu Met Thr Ala Phe Gly Val Glu Pro Tyr Gly Gln Pro Lys
85 90 95
Tyr Leu Glu Ile Ala Gly Lys Arg Met Ala Tyr Ile Asp Glu Gly Lys
100 105 110
Gly Asp Ala Ile Val Phe Gln His Gly Asn Pro Thr Ser Ser Tyr Leu
115 120 125
CA 02268036 1999-10-12
150
Trp Arg Asn Ile Met Pro His Leu Glu Gly Leu Gly Arg Leu Val Ala
130 135 140
Cys Asp Leu Ile Gly Met Gly Ala Ser Asp Lys Leu Ser Pro Ser Gly
145 150 155 160
Pro Asp Arg Tyr Ser Tyr Gly Glu Gln Arg Asp Phe Leu Phe Ala Leu
165 170 175
Trp Asp Ala Leu Asp Leu Gly Asp His Val Val Leu Val Leu His Asp
180 185 190
Trp Gly Ser Ala Leu Gly Phe Asp Trp Ala Asn Gln His Arg Asp Arg
195 200 205
Val Gln Gly Ile Ala Phe Met Glu Ala l:le Val Thr Pro Met Thr Trp
210 215 220
Ala Asp Trp Pro Pro Ala Val Arg Gly Val Phe Gln Gly Phe Arg Ser
225 230 235 240
Pro Gln Gly Glu Pro Met Ala Leu Glu His Asn Ile Phe Val Glu Arg
245 250 255
Val Leu Pro Gly Ala Ile Leu Arg Gln Leu Ser Asp Glu Glu Met Asn
260 265 270
His Tyr Arg Arg Pro Phe Val Asn Gly Gly Glu Asp Arg Arg Pro Thr
275 280 285
Leu Ser Trp Pro Arg Asn Leu Pro Ile Asp Gly Glu Pro Ala Glu Val
290 295 300
Val Ala Leu Val Asn Glu Tyr Arg Ser Trp Leu Glu Glu Thr Asp Met
305 310 315 320
Pro Lys Leu Phe Ile Asn Ala Glu Pro Gly Ala Ile Ile Thr Gly Arg
325 330 335
Ile Arg Asp Tyr Val Arg Ser Trp Pro Asn Gln Thr Glu Ile Thr Val
340 345 350
Pro Gly Val His Phe Val Gln Glu Asp Ser Asp Gly Val Val Ser Trp
355 360 365
Ala Gly Ala Arg Gln His Arg Arg Pro Gly Ser Ala Leu Ile Ser Arg
370 375 380
Asp Gln Glu Cys Asp Phe Arg Arg Arg Arg Arg Pro Ala Cys Gln Leu
385 390 395 400
Ile Arg Leu Pro Ala Pro Gly Arg Asp Ser Gln Gly Lys Gly His Gln
405 410 415
Ser Gln Pro Leu Pro Ser Gln Arg Gly Arg Gln Ile Tyr Val Ala Gly
420 425 430
Gln Arg Ser Ser Tyr Leu Pro Ser Glu Leu Val Ala Ala Phe Leu Trp
435 440 445
Ala Gln Phe Glu Glu Ala Glu Arg Ile Thr Arg Ile Arg Leu Asp Leu
450 455 460
Trp Asn Arg Tyr His Glu Ser Phe Glu Ser Leu Glu Gln Arg Gly Leu
CA 02268036 1999-10-12
151
465 470 475 480
Leu Arg Arg Pro Ile Ile Pro Gln Gly (:ys Ser His Asn Ala His Met
985 490 495
Tyr Tyr Val Leu Leu Ala Pro Ser Ala Asp Arg Glu Glu Val Leu Ala
500 505 510
Arg Leu Thr Ser Glu Gly Ile Gly Ala Val Phe His Tyr Val Pro Leu
515 520 525
His Asp Ser Pro Ala Gly Arg Arg
530 535
(2) INFORMATION FOR SEQ ID N0:157:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 284 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:1.57:
Asn Glu Ser Ala Pro Arg Ser Pro Met Leu Pro Ser Ala Arg Pro Arg
1 5 7.0 15
Tyr Asp Ala Ile Ala Val Leu Leu Asn C~lu Met His Ala Gly His Cys
20 25 30
Asp Phe Gly Leu Val Gly Pro Ala Pro Asp Ile Val Thr Asp Ala Ala
35 40 45
Gly Asp Asp Arg Ala Gly Leu Gly Val Asp Glu Gln Phe Arg His Val
50 55 60
Gly Phe Leu Glu Pro Ala Pro Val Leu Val Asp Gln Arg Asp Asp Leu
65 70 75 80
Gly Gly Leu Thr Val Asp Trp Lys Val Ser Trp Pro Arg Gln Arg Gly
85 90 95
Ala Thr Val Leu Ala Ala Val His Glu Trp Pro Pro Ile Val Val His
100 105 110
Phe Leu Val Ala Glu Leu Ser Gln Asp Arg Pro Gly Gln His Pro Phe
115 120 125
Asp Lys Asp Val Val Leu Gln Arg His Trp Leu Ala Leu Arg Arg Ser
130 135 140
Glu Thr Leu Glu His Thr Pro His Gly A.rg Arg Pro Val Arg Pro Arg
145 150 155 160
His Arg Gly Asp Asp Arg Phe His Glu A.rg Asp Pro Leu His Ser Val
165 170 175
Ala Met Leu Val Ser Pro Val Glu Ala Glu Arg Arg Ala Pro Val Val
180 185 190
Gln His Gln Tyr His Val Val Ala Glu Val Glu Arg Ile Pro Glu Arg
195 200 205
CA 02268036 1999-10-12
152
Glu Gln Lys Val Ser Leu Leu Ala Ile Ala Ile Ala Val Gly Ser Arg
210 215 220
Trp Ala Glu Leu Val Arg Arg Ala His I?ro Asp Gln Ile Ala Gly His
225 230 235 240
Gln Pro Ala Gln Pro Phe Gln Val Arg His Asp Val Ala Pro Gln Val
245 :?50 255
Arg Arg Arg Gly Val Ala Val Leu Lys Asp Asp Gly Val Thr Leu Ala
260 265 270
Phe Val Asp Ile Arg His Ala Leu Pro Gly Asp Phe
275 280
(2) INFORMATION FOR SEQ ID N0:158:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 264 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:158:
ATGAACATGTCGTCGGTGGTGGGTCGCAAGGCCTTTGCGCGATTCGCCGGCTACTCCTCC 60
GCCATGCACGCGATCGCCGGTTTCTCCGATGCGTTGCGC;CAAGAGCTGCGGGGTAGCGGA 120
ATCGCCGTCTCGGTGATCCACCCGGCGCTGACCCAGACACCGCTGTTGGCCAACGTCGAC 180
CCCGCCGACATGCCGCCGCCGTTTCGCAGCCTCACGCCC;ATTCCCGTTCACTGGGTCGCG 240
GCAGCGGTGCTTGACGGTGTGGCG 264
(2) INFORMATION FOR SEQ ID N0:159:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:1171 basepairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY:
linear
(xi) EQUENCE
S DESCRIPTION:
SEQ ID
N0:1.59:
TAGTCGGCGACGATGACGTCGCGGTCCAGGCCGACCGCTTCAAGCACCAGCGCGACCACG 60
AAGCCGGTGCGATCCTTACCCGCGAAGCAGTGGGTGAGC:ACCGGGCGTCCGGCGGCAAGC 120
AGTGTGACGACACGATGTAGCGCGCGCTGTGCTCCATTGCGCGTTGGGAATTGGCGATAC 180
TCGTCGGTCATGTAGCGGGTGGCCGCGTCATTTATCGACTGGCTGGATTCGCCGGACTCG 240
CCGTTGGACCCGTCATTGGTTAGCAGCCTCTTGAATGCGGTTTCGTGCGGCGCTGAGTCG 300
TCGGCGTCATCATCGGCGAGGTCGGGGAACGGCAGCAGGTGGACGTCGATGCCGTCCGGA 360
ACCCGTCCTGGACCGCGGCGGGCAACCTCCCGGGACGACCGCAGGTCGGCAACGTCGGTG 420
ATCCCCAGCCGGCGCAGCGTTGCCCCTCGTGCCGAATTCGGCACGAGGCTGGCGAGCCAC 480
CGGGCATCACCAAGCAACGCTTGCCCAGTACGGATCGTCACTTCCGCATCCGGCAGACCA 540
CA 02268036 1999-10-12
153
ATCTCCTCGCCGCCCATCGTCAGATCCCGCTCGTGCGT'.CGACAAGAACGGCCGCAGATGT 600
GCCAGCGGGTATCGGAGATTGAACCGCGCACGCAGTTC'.PTCAATCGCTGCGCGCTGCCGC 660
ACTATTGGCACTTTCCGGCGGTCGCGGTATTCAGCAAG(:ATGCGAGTCTCGACGAACTCG 720
CCCCACGTAACCCACGGCGTAGCTCCCGGCGTGACGCGGAGGATCGGCGGGTGATCTTTG 780
CCGCCACGCTCGTAGCCGTTGATCCACCGCTTCGCGGTGCCGGCGGGGAGGCCGATCAGC 840
TTATCGACCTCGGCGTATGCCGACGGCAAGCTGGGCGCGTTCGTCGAGGTCAAGAACTCC 900
ACCATCGGCACCGGCACCAAGGTGCCGCACCTGACCTAC:GTCGGCGACGCCGACATCGGC 960
GAGTACAGCAACATCGGCGCCTCCAGCGTGTTCGTCAAC:TACGACGGTACGTCCAAACGG 1020
CGCACCACCGTCGGTTCGCACGTACGGACCGGGTCCGAC:ACCATGTTCGTGGCCCCAGTA 1080
ACCATCGGCGACGGCGCGTATACCGGGGCCGGCACAGTGGTGCGGGAGGATGTCCCGCCG 1140
GGGGCGCTGGCAGTGTCGGCGGGTCCGCAAC 1171
(2) INFORMATION FOR SEQ ID N0:160:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 227 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:7.60:
GCAAAGGCGG CACCGGCGGG GCCGGCATGA ACAGCCTCGA CCCGCTGCTA GCCGCCCAAG 60
ACGGCGGCCA AGGCGGCACC GGCGGCACCG GCGGCAACC~C CGGCGCCGGC GGCACCAGCT 120
TCACCCAAGG CGCCGACGGC AACGCCGGCA ACGGCGGTGA CGGCGGGGTC GGCGGCAACG 180
GCGGAAACGG CGGAAACGGC GCAGACAACA CCACCACCGC CGCCGCC 227
(2) INFORMATION FOR SEQ ID N0:161:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 304 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:161:
CCTCGCCACC ATGGGCGGGC AGGGCGGTAG CGGTGGCGC'C GGCTCTACCC CAGGCGCCAA 60
GGGCGCCCAC GGCTTCACTC CAACCAGCGG CGGCGACGGC GGCGACGGCG GCAACGGCGG 120
CAACTCCCAA GTGGTCGGCG GCAACGGCGG CGACGGCGGC AATGGCGGCA ACGGCGGCAG 180
CGCCGGCACG GGCGGCAACG GCGGCCGCGG CGGCGACGGC GCGTTTGGTG GCATGAGTGC 240
CAACGCCACC AACCCTGGTG AAAACGGGCC AAACGGTAA.C CCCGGCGGCA ACGGTGGCGC 300
CGGC 304
CA 02268036 1999-10-12
154
(2) INFORMATION
FOR
SEQ
ID N0:162:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:1439 base
pairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY:
linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:162:
GTGGGACGCTGCCGAGGCTGTATAACAAGGACAACATCGACCAGCGCCGGCTCGGTGAGC 60
TGATCGACCTATTTAACAGTGCGCGCTTCAGCCGGCAGGGCGAGCACCGCGCCCGGGATC 120
TGATGGGTGAGGTCTACGAATACTTCCTCGGCAATTTCGCTCGCGCGGAAGGGAAGCGGG 180
GTGGCGAGTTCTTTACCCCGCCCAGCGTGGTCAAGGTGATCGTGGAGGTGCTGGAGCCGT 240
CGAGTGGGCGGGTGTATGACCCGTGCTGCGGTTCCGGAC~GCATGTTTGTGCAGACCGAGA 300
AGTTCATCTACGAACACGACGGCGATCCGAAGGATGTC7.'CGATCTATGGCCAGGAAAGCA 360
TTGAGGAGACCTGGCGGATGGC;GAAGATGAACCTCGCCATCCACGGCATCGACAACAAGG 420
GGCTCGGCGCCCGATGGAGTGATACCTTCGCCCGCGACC:AGCACCCGGACGTGCAGATGG 480
ACTACGTGATGGCCAATCCGCCGTTCAACATCAAAGACTGGGCCCGCAACGAGGAAGACC 540
CACGCTGGCGCTTCGGTGTTCCGCCCGCCAATAACGCCAACTACGCATGGATTCAGCACA 600
TCCTGTACAACTTGGCGCCGGGAGGTCGGGCGGGCGTGGTGATGGCCAACGGGTCGATGT 660
CGTCGAACTCCAACGGCAAGGGGGATATTCGCGCGCAAATCGTGGAGGCGGATTTGGTTT 720
CCTGCATGGTCGCGTTACCCACCCAGCTGTTCCGCAGCACCGGAATCCCGGTGTGCCTGT 780
GGTTTTTCGCCAAAAACAAGGCGGCAGGTAAGCAAGGGTCTATCAACCGGTGCGGGCAGG 840
TGCTGTTCATCGACGCTCGTGAACTGGGCGACCTAGTGGACCGGGCCGAGCGGGCGCTGA 900
CCAACGAGGAGATCGTCCGCATCGGGGATACCTTCCACGCGAGCACGACCACCGGCAACG 960
CCGGCTCCGGTGGTGCCGGCGGTAATGGGGGCACTGGCC:TCAACGGCGCGGGCGGTGCTG 1020
GCGGGGCCGGCGGCAACGCGGGTGTCGCCGGCGTGTCCTTCGGCAACGCTGTGGGCGGCG 1080
ACGGCGGCAACGGCGGCAACGGCGGCCACGGCGGCGACGGCACGACGGGCGGCGCCGGCG 1140
GCAAGGGCGGCAACGGCAGCAGCGGTGCCGCCAGCGGCTCAGGCGTCGTCAACGTCACCG 1200
CCGGCCACGGCGGCAACGGCGGCAATGGCGGCAACGGCGGCAACGGCTCCGCGGGCGCCG 1260
GCGGCCAGGGCGGTGCCGGCGGCAGCGCCGGCAACGGCGGCCACGGCGGCGGTGCCACCG 1320
GCGGCGCCAGCGGCAAGGGCGGCAACGGCACCAGCGGTGCCGCCAGCGGCTCAGGCGTCA 1380
TCAACGTCACCGCCGGCCACGGCGGCAACGGCGGCAATGGCCGCAACGGCGGCAACGGC 1439
(2) INFORMATION FOR SEQ ID N0:163:
(i) SEQUENCE CHARACTERISTICS:
CA 02268036 1999-10-12
155
(A) LENGTH: 329 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:7_63:
GGGCCGGCGGGGCCGGATTT TCTCGTGCCT TGATTGTCGCTGGGGATAACGGCGGTGATG60
GTGGTAACGGCGGGATGGGC GGGGCTGGCG GGGCTGGC<7GCCCCGGCGGGGCCGGCGGCC120
TGATCAGCCTGCTGGGCGGC CAAGGCGCCG GCGGGGCCC~GCGGGACCGGCGGGGCCGGCG180
GTGTTGGCGGTGACGGCGGG GCCGGCGGCC CCGGCAACC:AGGCCTTCAACGCAGGTGCCG240
GCGGGGCCGGCGGCCTGATC AGCCTGCTGG GCGGCCAACTGCGCCGGCGGGGCCGGCGGGA300
CCGGCGGGGCCGGCGGTGTT GGCGGTGAC 329
(2) INFORMATION FOR SEQ ID N0:164:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 80 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO::L64:
GCAACGGTGG CAACGGCGGC ACCAGCACGA CCGTGGGGAT GGCCGGAGGT AACTGTGGTG 60
CCGCCGGGCT GATCGGCAAC 80
(2) INFORMATION FOR SEQ ID N0:165:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 392 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO::L65:
GGGCTGTGTC GCACTCACAC CGCCGCATTC GGCGACGTTG GCCGCCCAAT ATCCAGCTCA 60
AGGCCTACTA CTTACCGTCG GAGGACCGCC GCATCAAGGT GCGGGTCAGC GCCCAAGGAA 120
TCAAGGTCAT CGACCGCGAC GGGCATCGAG GCCGTCGT(:G CGCGGCTCGG GCAGGATCCG 180
CCCCGGCGCA CTTCGCGCGC CAAGCGGGCT CATCGCTC(:G AACGGCGGCG ATCCTGTGAG 240
CACAACTGAT GGCGCGCAAC GAGATTCGTC CAATTGTCAA GCCGTGTTCG ACCGCAGGGA 300
CCGGTTATAC GTATGTCAAC CTATGTCACT CGCAAGAACC GGCATAACGA TCCCGTGATC 360
CGCCGACAGC CCACGAGTGC AAGACCGTTA CA 392
(2) INFORMATION FOR SEQ ID N0:166:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 535 base pairs
CA 02268036 1999-10-12
156
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY:
linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:166:
ACCGGCGCCACCGGCGGCACCGGGTTCGCCGGTGGCGCCGGCGGGGCCGGCGGGCAGGGC 60
GGTATCAGCGGTGCCGGCGGCACCAACGGCTCTGGTGGCGCTGGCGGCACCGGCGGACAA 120
GGCGGCGCCGGGGGCGCTGGCGGGGCCGGCGCCGATAACCCCACCGGCATCGGCGGCGCC 180
GGCGGCACCGGCGGCACCGGCGGAGCGGCCGGAGCCGGCGGGGCCGGTGGCGCCATCGGT 240
ACCGGCGGCACCGGCGGCGCGGTGGGCAGCGTCGGTAACGCCGGGATCGGCGGTACCGGC 300
GGTACGGGTGGTGTCGGTGGTGCTGGTGGTGCAGGTGCGGCTGCGGCCGCTGGCAGCAGC 360
GCTACCGGTGGCGCCGGGTTCGCCGGCGGCGCCGGCGGi~GAAGGCGGACCGGGCGGCAAC 420
AGCGGTGTGGGCGGCACCAACGGCTCCGGCGGCGCCGG(:GGTGCAGGCGGCAAGGGCGGC 480
ACCGGAGGTGCCGGCGGGTCCGGCGCGGACAACCCCACCGGTGCTGGTTTCGCCG 535
(2) INFORMATION
FOR
SEQ
ID N0:167:
(i) SEQUENCE RACTERISTICS:
CHA
(A) LENGTH:690 base
pairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:7.67:
CCGACGTCGCCGGGGCGATACGGGGGTCACCGACTACTACATCATCCGCA CCGAGAATCG60
GCCGCTGCTGCAACCGCTGCGGGCGGTGCCGGTCATCGGAGATCCGCTGG CCGACCTGAT120
CCAGCCGAACCTGAAGGTGATCGTCAACCTGGGCTACGGCGACCCGAACT ACGGCTACTC180
GACGAGCTACGCCGATGTGCGAACGCCGTTCGGGCTGTGGCCGAACGTGC CGCCTCAGGT240
CATCGCCGATGCCCTGGCCGCCGGAACACAAGAAGGCAT'CCTTGACTTCA CGGCCGACCT300
GCAGGCGCTGTCCGCGCAACCGCTCACGCTCCCGCAGATCCAGCTGCCGC AACCCGCCGA360
TCTGGTGGCCGCGGTGGCCGCCGCACCGACGCCGGCCGA.GGTGGTGAACA CGCTCGCCAG420
GATCATCTCAACCAACTACGCCGTCCTGCTGCCCACCGTGGACATCGCCC TCGCCTGGTC480
ACCACCCTGCCGCTGTACACCACCCAACTGTTCGTCAGGCAACTCGCTGC GGGCAATCTG540
ATCAACGCGATCGGCTATCCCCTGGCGGCCACCGTAGGTTTAGGCACGAT CGATAGCGGG600
CGGCGTGGAATTGCTCACCCTCCTCGCGGCGGCCTCGGACACCGTTCGAA ACATCGAGGG660
CCTCGTCACCTAACGGATTCCCGACGGCAT 690
(2) INFORMATION FOR SEQ ID N0:168:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 407 base pairs
CA 02268036 1999-10-12
157
(B) TYPE: nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:168:
ACGGTGACGGCGGTACTGGC GGCGGCCACGGCGGCAACGGCGGGAATCCCGGGTGGCTCT 60
TGGGCACAGCCGGGGGTGGC GGCAACGGTGGCGCCGGCAGCACCGGTACTGCAGGTGGCG 120
GCTCTGGGGGCACCGGCGGC GACGGCGGGACCGGCGGGCGTGGCGGCCTGTTAATGGGCG 180
CCGGCGCCGGCGGGCACGGT GGCACTGGCGGCGCGGGCGGTGCCGGTGTCGACGGTGGCG 240
GCGCCGGCGGGGCCGGCGGG GCCGGCGGCAACGGCGGCGCCGGGGGTCAAGCCGCCCTGC 300
TGTTCGGGCGCGGCGGCACC GGCGGAGCCGGCGGCTACGGCGGCGATGGCGGTGGCGGCG 360
GTGACGGCTTCGACGGCACG ATGGCCGGCCTGGGTGGTACCGGTGGC 407
(2) INFORMATION FOR SEQ ID N0:169:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 468 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID 9:
N0:16
GATCGGTCAG CGCATCGCCC TCGGCGGCAA GGTCTCACCG AAGAACATCG60
GCGATTCCGC
TGCACGCGGC GGCGCGGACC AGCCCGCTGC GTCGAACGCC TCCAGCAGGC120
GCTGCGGCC~C
ACAGCCAGTC CTTGGCGGCC TGCGAGGCGA GTCACCGGTG TAGATCGCCG180
ACACGTCGGT
GGATGCCCGC CTCCGCCAAC GCATTCCGGC GTCTTTGTGA TGCTCGACGA240
ACGCCCGCGC
TCACCGCGAT GTCTGCGGCC ACCACGGGCC GGTGGCCCCG CTGGCCAGTA300
GCCCGGCGAA
GCGCCGCGAC GTCGGCGGCC AGGTCGTCGG GCGCAGCGCT CCGGCGCGAC360
GGATGTGCC'G
GCCCGAAAAA CGACCCCTCA CCCAGCTGGG ATATCCCTTG CCGTCCTGGG420
TCCCGCTGGC
CGATATTGGA CGCGCATGCC CCGACCGCGT CACCACCG 468
ACAGGCCGGC
(2) INFORMATION FOR SEQ ID N0:170:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 219 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID
N0:170:
GGTGGTAACG GCGGCCAGGG TGGCATCGGC AGAGAGGCGC CGACGGCGCC60
GGCGCCGGCG
GGCCCCAATG CTAACGGCGC AAACGGCGAG GCGGTGGTAA CGGTGGCGAC120
AACGGCGGTA
GGCGGCGCCG GCGGCAATGG CGGCGCGGGC AGGCGGCCGG GTACACCGAC180
GGCAACGCGC
CA 02268036 1999-10-12
158
GGCGCCACGG GCACCGGCGG CGACGGCGGC AACGGCGGC 219
(2) INFORMATION FOR SEQ ID N0:171:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 494 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:171:
TAGCTCCGGC GAGGGCGGCA AGGGCGGCGA CGGTGGCCAC GGCGGTGACG GCGTCGGCGG 60
CAACAGTTCC GTCACCCAAG GCGGCAGCGG CGGTGGCGGC GGCGCCGGCG GCGCCGGCGG 120
CAGCGGCTTT TTCGGCGGCA AGGGCGGCTT CGGCGGCGi~C GGCGGTCAGG GCGGCCCCAA 180
CGGCGGCGGT ACCGTCGGCA CCGTGGCCGG TGGCGGCGGC AACGGCGGTG TCGGCGGCCG 240
GGGCGGCGAC GGCGTCTTTG CCGGTGCCGG CGGCCAGG(~C GGCCTCGGTG GGCAGGGCGG 300
CAATGGCGGC GGCTCCACCG GCGGCAACGG CGGCCTTGGC GGCGCGGGCG GTGGCGGAGG 360
CAACGCCCCG GCTCGTGCCG AATCCGGGCT GACCATGGAC AGCGCGGCCA AGTTCGCTGC 420
CATCGCATCA GGCGCGTACT GCCCCGAACA CCTGGAACAT CACCCGAGTT AGCGGGGCGC 480
ATTTCCTGAT CACC 494
(2) INFORMATION FOR SEQ ID N0:172:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 220 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:172:
GGGCCGGTGG TGCCGCGGGC CAGCTCTTCA GCGCCGGAGG CGCGGCGGGT GCCGTTGGGG 60
TTGGCGGCAC CGGCGGCCAG GGTGGGGCTG GCGGTGCCGG AGCGGCCGGC GCCGACGCCC 120
CCGCCAGCAC AGGTCTAACC GGTGGTACCG GGTTCGCTGG CGGGGCCGGC GGCGTCGGCG 180
GCCAGAGCGG CAACGCCATT GCCGGCGGCA TCAACGGCTC 220
(2) INFORMATION FOR SEQ ID N0:173:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 388 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:173:
ATGGCGGCAA CGGGGGCCCC GGCGGTGCTG GCGGGGCCGG CGACTACAAT TTCCAACGGC 60
GGGCAGGGTG GTGCCGGCGG CCAAGGCGGC CAAGGCGGCC TGGGCGGGGC AAGCACCACC 120
ACGCCCGCGC
CA 02268036 1999-10-12
159
TGATCGGCCTAGCCGCACCCGGGAAAGCCGATCCAACA.GGCGACGATGCCGCCTTCCTTG 180
CCGCGTTGGACCAGGCCGGCATCACCTACGCTGACCCAGGCCACGCCATAACGGCCGCCA 240
AGGCGATGTGTGGGCTGTGTGCTAACGGCGTAACAGGTCTACAGCTGGTCGCGGACCTGC 300
GGGACTACAATCCCGGGCTGACCATGGACAGCGCGGCCAAGTTCGCTGCCATCGCATCAG 360
GCGCGTACTGCCCCGAACACCTGGAACA 388
(2) INFORMATION
FOR
SEQ
ID N0:174:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 400 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0::174:
GCAAAGGCGGCACCGGCGGG GCCGGCATGA ACAGCCTCGACCCGCTGCTA GCCGCCCAAG60
ACGGCGGCCAAGGCGGCACC GGCGGCACCG GCGGCAACGCCGGCGCCGGC GGCACCAGCT120
TCACCCAAGGCGCCGACGGC AACGCCGGCA ACGGCGGTGACGGCGGGGTC GGCGGCAACG180
GCGGAAACGGCGGAAACGGC GCAGACAACA CCACCACCCiCCGCCGCCGGC ACCACAGGCG240
GCGACGGCGGGGCCGGCGGG GCCGGCGGAA CCGGCGGAACCGGCGGAGCC GCCGGCACCG300
GCACCGGCGGCCAACAAGGC AACGGCGGCA ACGGCGGCACCGGCGGCAAA GGCGGCACCG360
GCGGCGACGGTGCACTCTCA GGCAGCACCG GTGGTGCCGG 400
(2) INFORMATION
FOR
SEQ
ID N0:175:
(i) S EQUENCE CHARACTERISTICS:
(A) LENGTH: 538 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi)
SEQUENCE
DESCRIPTION:
SEQ
ID N0:175:
GGCAACGGCGGCAACGGCGG CATCGCCGGC ATTGGGCGGCAACGGCGTTCCGGGACGGGC 60
AGCGGCAACGGCGGCCAACG GCGGCAGCGG CGGCAACGGCGGCAACGCCGGCATGGGCGG 120
CAACAGCGGCACCGGCAGCG GCGACGGCGG TGCCGGCGGGAACGGCGGCGCGGCGGGCAC 180
GGGCGGCACCGGCGGCGACG GCGGCCTCAC CGGTACTGGCGGCACCGGCGGCAGCGGTGG 240
CACCGGCGGTGACGGCGGTA ACGGCGGCAA CGGAGCAGATAACACCGCAAACATGACTGC 300
GCAGGCGGGCGGTGACGGTG GCAACGGCGG CGACGGTGGCTTCGGCGGCGGGGCCGGGGC 360
CGGCGGCGGTGGCTTGACCG CTGGCGCCAA CGGCACCGGCGGGCAAGGCGGCGCCGGCGG 420
CGATGGCGGCAACGGGGCCA TCGGCGGCCA CGGCCCACTCACTGACGACCCCGGCGGCAA 480
CGGGGGCACCGGCGGCAACG GCGGCACCGG CGGCACCGGCGGCGCGGGCATCGGCAGC 538
CA 02268036 1999-10-12
160
(2) INFORMATION
FOR SEQ
ID N0:176:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 239 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:176:
GGGCCGGTGGTGCCGCGGGC CAGCTCTTCA GCGCCGGAGG 60
CGCGGCGGGT GCCGTTGGGG
TTGGCGGCACCGGCGGCCAG GGTGGGGCTG GCGGTGCCGGAGCGGCCGGC GCCGACGCCC120
CCGCCAGCACAGGTCTAACC GGTGGTACCG GGTTCGCTGGCGGGGCCGGC GGCGTCGGCG180
GCCACGGCGGCAACGCCATT GCCGGCGGCA TCAACGGC'PCCGGTGGTGCC GGCGGCACC239
(2) INFORMATION
FOR SEQ
ID N0:177:
(i) S EQUENCE CHARACTERISTICS:
(A) LENGTH: 985 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:177:
AGCAGCGCTACCGGTGGCGC CGGGTTCGCC GGCGGCGCC;GGCGGAGAAGG CGGAGCGGGC60
GGCAACAGCGGTGTGGGCGG CACCAACGGC TCCGGCGGC;GCCGGCGGTGC AGGCGGCAAG120
GGCGGCACCGGAGGTGCCGG CGGGTCCGGC GCGGACAAC:CCCACCGGTGC TGGTTTCGCC180
GGTGGCGCCGGCGGCACAGG TGGCGCGGCC GGCGCCGGC'.GGGGCCGGCGG GGCGACCGGT240
ACCGGCGGCACCGGCGGCGT TGTCGGCGCC ACCGGTAGT'GCAGGCATCGG CGGGGCCGGC300
GGCCGCGGCGGTGACGGCGG CGATGGGGCC AGCGGTCTCGGCCTGGGCCT CTCCGGCTTT360
GACGGCGGCCAAGGCGGCCA AGGCGGGGCC GGCGGCAGCGCCGGCGCCGG CGGCATCAAC420
GGGGCCGGCGGGGCCGGCGG CAACGGCGGC GACGGCGGGGACGGCGCAAC CGGTGCCGCA480
GGTCTCGGCGACAACGGCGG GGTCGGCGGT GACGGTGGGGCCGGTGGCGC CGCCGGCAAC540
GGCGGCAACGCGGGCGTCGG CCTGACAGCC AAGGCCGGCGACGGCGGCGC CGCGGGCAAT600
GGCGGCAACGGGGGCGCCGG CGGTGCTGGC GGGGCCGGCGACAACAATTT CAACGGCGGC660
CAGGGTGGTGCCGGCGGCCA AGGCGGCCAA GGCGGCTTGGGCGGGGCAAG CACCACCTGA720
TCGGCCTAGCCGCACCCGGG AAAGCCGATC CAACAGGCG.~CGATGCCGCC TTCCTTGCCG780
CGTTGGACCAGGCCGGCATC ACCTACGCTG ACCCAGGCC:~1CGCCATAACG GCCGCCAAGG840
CGATGTGTGGGCTGTGTGCT AACGGCGTAA CAGGTCTACAGCTGGTCGCG GACCTGCGGG900
AATACAATCCCGGGCTGACC ATGGACAGCG CGGCCAAGT'PCGCTGCCATC GCATCAGGCG960
CGTACTGCCCCGAACACCTG GAACA g85
CA 02268036 1999-10-12
161
(2) INFORMATION
FOR SEQ
ID N0:178:
(i) S EQUENCE
CHARACTERISTICS:
(A) LENGTH:2138 basepairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi) S EQUENCE CRIPTION:EQ ID 8:
DES S N0:17
CGGCACGAGGATCGGTACCCCGCGGCATCGGCAGCTGCCGATTCGCCGGGTTTCCCCACC 60
CGAGGAAAGCCGCTACCAGATGGCGCTGCCGAAGTAGGGCGATCCGTTCGCGATGCCGGC 120
ATGAACGGGCGGCATCAAATTAGTGCAGGAACCTTTCAGTTTAGCGACGATAATGGCTAT 180
AGCACTAAGGAGGATGATCCGATATGACGCAGTCGCAGACCGTGACGGTGGATCAGCAAG 240
AGATTTTGAACAGGGCCAACGAGGTGGAGGCCCCGATGGCGGACCCACCGACTGATGTCC 300
CCATCACACCGTGCGAACTCACGGCGGCTAAAAACGCCGCCCAACAGCTGGTATTGTCCG 360
CCGACAACATGCGGGAATACCTGGCGGCCGGTGCCAAAGAGCGGCAGCGTCTGGCGACCT 420
CGCTGCGCAACGCGGCCAAGGCGTATGGCGAGGTTGATGAGGAGGCTGCGACCGCGCTGG 480
ACAACGACGGCGAAGGAACTGTGCAGGCAGAATCGGCCGGGGCCGTCGGAGGGGACAGTT 540
CGGCCGAACTAACCGATACGCCGAGGGTGGCCACGGCCCiGTGAACCCAACTTCATGGATC 600
TCAAAGAAGCGGCAAGGAAGCTCGAAACGGGCGACCAAC~GCGCATCGCTCGCGCACTTTG 660
CGGATGGGTGGAACACTTTCAACCTGACGCTGCAAGGCC~ACGTCAAGCGGTTCCGGGGGT 720
TTGACAACTGGGAAGGCGATGCGGCTACCGCTTGCGAGC~CTTCGCTCGATCAACAACGGC 780
AATGGATACTCCACATGGCCAAATTGAGCGCTGCGATGGCCAAGCAGGCTCAATATGTCG 840
CGCAGCTGCACGTGTGGGCTAGGCGGGAACATCCGACTTATGAAGACATAGTCGGGCTCG 900
AACGGCTTTACGCGGAAAACCCTTCGGCCCGCGACCAAP,TTCTCCCGGTGTACGCGGAGT 960
ATCAGCAGAGGTCGGAGAAGGTGCTGACCGAATACAACAACAAGGCAGCCCTGGAACCGG 1020
TAAACCCGCCGAAGCCTCCCCCCGCCATCAAGATCGACCCGCCCCCGCCTCCGCAAGAGC 1080
AGGGATTGATCCCTGGCTTCCTGATGCCGCCGTCTGACGGCTCCGGTGTGACTCCCGGTA 1140
CCGGGATGCCAGCCGCACCGATGGTTCCGCCTACCGGATCGCCGGGTGGTGGCCTCCCGG 1200
CTGACACGGCGGCGCAGCTGACGTCGGCTGGGCGGGAAGCCGCAGCGCTGTCGGGCGACG 1260
TGGCGGTCAAAGCGGCATCGCTCGGTGGCGGTGGAGGCGGCGGGGTGCCGTCGGCGCCGT 1320
TGGGATCCGCGATCGGGGGCGCCGAATCGGTGCGGCCCGCTGGCGCTGGTGACATTGCCG 1380
GCTTAGGCCAGGGAAGGGCCGGCGGCGGCGCCGCGCTGGGCGGCGGTGGCATGGGAATGC 1940
CGATGGGTGCCGCGCATCAGGGACAAGGGGGCGCCAAGTCCAAGGGTTCTCAGCAGGAAG 1500
ACGAGGCGCTCTACACCGAGGATCGGGCATGGACCGAGGCCGTCATTGGTAACCGTCGGC 1560
CA 02268036 1999-10-12
162
GCCAGGACAGTAAGGAGTCGAAGTGAGCATGGACGAAT'TGGACCCGCATGTCGCCCGGGC1620
GTTGACGCTGGCGGCGCGGTTTCAGTCGGCCCTAGACGGGACGCTCAATCAGATGAACAA1680
CGGATCCTTCCGCGCCACCGACGAAGCCGAGACCGTCGAAGTGACGATCAATGGGCACCA1740
GTGGCTCACCGGCCTGCGCATCGAAGATGGTTTGCTGAAGAAGCTGGGTGCCGAGGCGGT1800
GGCTCAGCGGGTCAACGAGGCGCTGCACAATGCGCAGGCCGCGGCGTCCGCGTATAACGA1860
CGCGGCGGGCGAGCAGCTGACCGCTGCGTTATCGGCCATGTCCCGCGCGATGAACGAAGG1920
AA.TGGCCTAAGCCCATTGTTGCGGTGGTAGCGACTACGCACCGAATGAGCGCCGCAATGC1980
GGTCATTCAGCGCGCCCGACACGGCGTGAGTACGCATTGTCAATGTTTTGACATGGATCG2040
GCCGGGTTCGGAGGGCGCCATAGTCCTGGTCGCCAATA'PTGCCGCAGCTAGCTGGTCTTA2100
GGTTCGGTTACGCTGGTTAATTATGACGTCCGTTACCA 2138
(2) INFORMATION FOR SEQ ID N0:179:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 460 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:179:
Met Thr Gln Ser Gln Thr Val Thr Val Asp Gln Gln Glu Ile Leu Asn
1 5 7.0 15
Arg Ala Asn Glu Val Glu Ala Pro Met Ala Asp Pro Pro Thr Asp Val
20 25 30
Pro Ile Thr Pro Cys Glu Leu Thr Ala Ala Lys Asn Ala Ala Gln Gln
35 40 45
Leu Val Leu Ser Ala Asp Asn Met Arg Glu Tyr Leu Ala Ala Gly Ala
50 55 60
Lys Glu Arg Gln Arg Leu Ala Thr Ser Leu Arg Asn Ala Ala Lys Ala
65 70 75 g0
Tyr Gly Glu Val Asp Glu Glu Ala Ala Thr Ala Leu Asp Asn Asp Gly
85 90 95
Glu Gly Thr Val Gln Ala Glu Ser Ala Gly Ala Val Gly Gly Asp Ser
100 105 110
Ser Ala Glu Leu Thr Asp Thr Pro Arg Val Ala Thr Ala Gly Glu Pro
115 120 125
Asn Phe Met Asp Leu Lys Glu Ala Ala Arg Lys Leu Glu Thr Gly Asp
130 135 140
Gln Gly Ala Ser Leu Ala His Phe Ala Asp Gly Trp Asn Thr Phe Asn
145 150 155 160
Leu Thr Leu Gln Gly Asp Val Lys Arg Phe Arg Gly Phe Asp Asn Trp
165 1'70 175
CA 02268036 1999-10-12
163
Glu Gly Asp Ala Ala Thr Ala Cys Glu Ala Ser Leu Asp Gln Gln Arg
180 185 190
Gln Trp Ile Leu His Met Ala Lys Leu Ser Ala Ala Met Ala Lys Gln
195 200 205
Ala Gln Tyr Val Ala Gln Leu His Val Trp Ala Arg Arg Glu His Pro
210 215 220
Thr Tyr Glu Asp Ile Val Gly Leu Glu Arg Leu Tyr Ala Glu Asn Pro
225 230 235 240
Ser Ala Arg Asp Gln Ile Leu Pro Val Tyr Ala Glu Tyr Gln Gln Arg
245 250 255
Ser Glu Lys Val Leu Thr Glu Tyr Asn Asn Lys Ala Ala Leu Glu Pro
260 265 270
Val Asn Pro Pro Lys Pro Pro Pro Ala Ile Lys Ile Asp Pro Pro Pro
275 280 285
Pro Pro Gln Glu Gln Gly Leu Ile Pro Gly Phe Leu Met Pro Pro Ser
290 295 300
Asp Gly Ser Gly Val Thr Pro Gly Thr c;ly Met Pro Ala Ala Pro Met
305 310 315 320
Val Pro Pro Thr Gly Ser Pro Gly Gly (ily Leu Pro Ala Asp Thr Ala
325 :330 335
Ala Gln Leu Thr Ser Ala Gly Arg Glu Ala Ala Ala Leu Ser Gly Asp
340 345 350
Val Ala Val Lys Ala Ala Ser Leu Gly CJly Gly Gly Gly Gly Gly Val
355 360 365
Pro Ser Ala Pro Leu Gly Ser Ala Ile Gly Gly Ala Glu Ser Val Arg
370 375 380
Pro Ala Gly Ala Gly Asp Ile Ala Gly Leu Gly Gln Gly Arg Ala Gly
385 390 395 400
Gly Gly Ala Ala Leu Gly Gly Gly Gly Met Gly Met Pro Met Gly Ala
405 910 415
Ala His Gln Gly Gln Gly Gly Ala Lys Ser Lys Gly Ser Gln Gln Glu
420 425 930
Asp Glu Ala Leu Tyr Thr Glu Asp Arg Ala Trp Thr Glu Ala Val Ile
435 440 945
Gly Asn Arg Arg Arg Gln Asp Ser Lys Glu Ser Lys
450 455 460
(2) INFORMATION FOR SEQ ID N0:180:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 277 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:180:
CA 02268036 1999-10-12
164
Ala Gly Asn Val Thr Ser Ala Ser Gly Pro His Arg Phe Gly Ala Pro
1 5 10 15
Asp Arg Gly Ser Gln Arg Arg Arg Arg His Pro Ala Ala Ser Thr Ala
20 25 30
Thr Glu Arg Cys Arg Phe Asp Arg His Val Ala Arg Gln Arg Cys Gly
35 40 45
Phe Pro Pro Ser Arg Arg Gln Leu Arg Arg Arg Val Ser Arg Glu Ala
50 55 60
Thr Thr Arg Arg Ser Gly Arg Arg Asn His Arg Cys Gly Trp His Pro
65 70 75 80
Gly Thr Gly Ser His Thr Gly Ala Val Arg Arg Arg His Gln Glu Ala
85 90 95
Arg Asp Gln Ser Leu Leu Leu Arg Arg .~lrg Gly Arg Val Asp Leu Asp
100 105 110
Gly Gly Gly Arg Leu Arg Arg Val Tyr Arg Phe Gln Gly Cys Leu Val
115 120 125
Val Val Phe Gly Gln His Leu Leu Arg Pro Leu Leu Ile Leu Arg Val
130 135 140
His Arg Glu Asn Leu Val Ala Gly Arg Arg Val Phe Arg Val Lys Pro
195 150 155 160
Phe Glu Pro Asp Tyr Val Phe Ile Ser Arg Met Phe Pro Pro Ser Pro
165 170 175
His Val Gln Leu Arg Asp Ile Leu Ser Leu Leu Gly His Arg Ser Ala
180 185 190
Gln Phe Gly His Val Glu Tyr Pro Leu Pro Leu Leu Ile Glu Arg Ser
195 200 205
Leu Ala Ser Gly Ser Arg Ile Ala Phe F'ro Val Val Lys Pro Pro Glu
210 215 220
Pro Leu Asp Val Ala Leu Gln Arg Gln V'al Glu Ser Val Pro Pro Ile
225 230 235 240
Arg Lys Val Arg Glu Arg Cys Ala Leu Val Ala Arg Phe Glu Leu Pro
245 250 255
Cys Arg Phe Phe Glu Ile His Glu Val Gly Phe Thr Gly Arg Gly His
260 265 270
Pro Arg Arg Ile Gly
275
(2) INFORMATION FOR SEQ ID N0:181:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 192 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
CA 02268036 1999-10-12
165
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:181:
Arg Val Ala Ala Ser Phe Ile Asp Trp Leu Asp Ser Pro Asp Ser Pro
1 5 10 15
Leu Asp Pro Ser Leu Val Ser Ser Leu Leu Asn Ala Val Ser Cys Gly
20 25 30
Ala Glu Ser Ser Ala Ser Ser Ser Ala Arg Ser Gly Asn Gly Ser Arg
35 40 45
Trp Thr Ser Met Pro Ser Gly Thr Arg Pro Gly Pro Arg Arg Ala Thr
50 55 60
Ser Arg Asp Asp Arg Arg Ser Ala Thr Ser Val Ile Pro Ser Arg Arg
65 70 75 80
Ser Val Ala Pro Arg Ala Glu Phe Gly 'Thr Arg Leu Ala Ser His Arg
85 90 95
Ala Ser Pro Ser Asn Ala Cys Pro Val ;erg Ile Val Thr Ser Ala Ser
100 105 110
Gly Arg Pro Ile Ser Ser Pro Pro Ile 'Jal Arg Ser Arg Ser Cys Val
115 120 125
Asp Lys Asn Gly Arg Arg Cys Ala Ser Gly Tyr Arg Arg Leu Asn Arg
130 135 140
Ala Arg Ser Ser Ser Ile Ala Ala Arg (:ys Arg Thr Ile Gly Thr Phe
145 150 155 160
Arg Arg Ser Arg Tyr Ser Ala Ser Met Arg Val Ser Thr Asn Ser Pro
165 7.70 175
His Val Thr His Gly Val Ala Pro Gly Val Thr Arg Arg Ile Gly Gly
180 185 190
(2) INFORMATION FOR SEQ ID N0:182:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 196 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:182:
Gln Glu Arg Pro Gln Met Cys Gln Arg Val Ser Glu Ile Glu Pro Arg
1 5 10 15
Thr Gln Phe Phe Asn Arg Cys Ala Leu Pro His Tyr Trp His Phe Pro
20 25 30
Ala Val Ala Val Phe Ser Lys His Ala Ser Leu Asp Glu Leu Ala Pro
35 40 45
Arg Asn Pro Arg Arg Ser Ser Arg Arg Asp Ala Glu Asp Arg Arg Val
50 55 60
Ile Phe Ala Ala Thr Leu Val Ala Val A;sp Pro Pro Leu Arg Gly Ala
65 70 75 80
CA 02268036 1999-10-12
166
Gly Gly Glu Ala Asp Gln Leu Ile Asp Leu Gly Val Cys Arg Arg Gln
85 90 95
Ala Gly Arg Val Arg Arg Gly Gln Glu Leu His His Arg His Arg His
100 105 110
Gln Gly Ala Ala Pro Asp Leu Arg Arg Arg Arg Arg His Arg Arg Val
115 120 125
Gln Gln His Arg Arg Leu Gln Arg Val Arg Gln Leu Arg Arg Tyr Val
130 135 140
Gln Thr Ala His His Arg Arg Phe Ala Arg Thr Asp Arg Val Arg His
145 150 155 160
His Val Arg Gly Pro Ser Asn His Arg .Arg Arg Arg Val Tyr Arg Gly
165 170 175
Arg His Ser Gly Ala Gly Gly Cys Pro Ala Gly Gly Ala Gly Ser Val
180 185 190
Gly Gly Ser Ala
195
(2) INFORMATION FOR SEQ ID N0:183:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 311 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:183:
Val Arg Cys Gly Thr Leu Val Pro Val Pro Met Val Glu Phe Leu Thr
1 5 7.0 15
Ser Thr Asn Ala Pro Ser Leu Pro Ser Ala Tyr Ala Glu Val Asp Lys
20 25 30
Leu Ile Gly Leu Pro Ala Gly Thr Ala Lys Arg Trp Ile Asn Gly Tyr
35 40 45
Glu Arg Gly Gly Lys Asp His Pro Pro Ile Leu Arg Val Thr Pro Gly
50 55 60
Ala Thr Pro Trp Val Thr Trp Gly Glu Fhe Val Glu Thr Arg Met Leu
65 70 75 80
Ala Glu Tyr Arg Asp Arg Arg Lys Val Pro Ile Val Arg Gln Arg Ala
85 90 95
Ala Ile Glu Glu Leu Arg Ala Arg Phe Asn Leu Arg Tyr Pro Leu Ala
100 105 110
His Leu Arg Pro Phe Leu Ser Thr His Glu Arg Asp Leu Thr Met Gly
115 120 125
Gly Glu Glu Ile Gly Leu Pro Asp Ala Glu Val Thr Ile Arg Thr Gly
130 135 140
Gln Ala Leu Leu Gly Asp Ala Arg Trp L~~u Ala Ser Leu Val Pro Asn
145 150 155 160
CA 02268036 1999-10-12
167
Ser Ala Arg Gly Ala Thr Leu Arg Arg Leu Gly Ile Thr Asp Val Ala
165 170 175
Asp Leu Arg Ser Ser Arg Glu Val Ala Arg Arg Gly Pro Gly Arg Val
180 185 190
Pro Asp Gly Ile Asp Val His Leu Leu Pro Phe Pro Asp Leu Ala Asp
195 200 205
Asp Asp Ala Asp Asp Ser Ala Pro His Glu Thr Ala Phe Lys Arg Leu
210 215 220
Leu Thr Asn Asp Gly Ser Asn Gly Glu Ser Gly Glu Ser Ser Gln Ser
225 230 235 240
Ile Asn Asp Ala Ala Thr Arg Tyr Met 'rhr Asp Glu Tyr Arg Gln Phe
245 250 255
Pro Thr Arg Asn Gly Ala Gln Arg Ala :Leu His Arg Val Val Thr Leu
260 265 270
Leu Ala Ala Gly Arg Pro Val Leu Thr 1-Iis Cys Phe Ala Gly Lys Asp
275 280 285
Arg Thr Gly Phe Val Val Ala Leu Val Leu Glu Ala Val Gly Leu Asp
290 295 300
Arg Asp Val Ile Val Ala Asp
305 310
(2) INFORMATION FOR SEQ ID N0:184:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:2072 basepairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY:
linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0:1.84:
CTCGTGCCGATTCGGCACGAGCTGAGCAGCCCAAGGGGC:CGTTCGGCGAAGTCATCGAGG 60
CATTCGCCGACGGGCTGGCCGGCAAGGGTAAGCAAATCAACACCACGCTGAACAGCCTGT 120
CGCAGGCGTTGAACGCCTTGAATGAGGGCCGCGGCGACTTCTTCGCGGTGGTACGCAGCC 180
TGGCGCTATTCGTCAACGCGCTACATCAGGACGACCAACAGTTCGTCGCGTTGAACAAGA 240
ACCTTGCGGAGTTCACCGACAGGTTGACCCACTCCGATGCGGACCTGTCGAACGCCATCC 300
AGCAATTCGACAGCTTGCTCGCCGTCGCGCGCCCGTTCTTCGCCAAGAACCGCGAGGTGC 360
TGACGCATGACGTCAATAATCTCGCGACCGTGACCACCACGTTGCTGCAGCCCGATCCGT 420
TGGATGGGTTGGAGACCGTCCTGCACATCTTCCCGACGCTGGCGGCGAACATTAACCAGC 480
TTTACCATCCGACACACGGTGGCGTGGTGTCGCTTTCCGCGTTCACGAATTTCGCCAACC 540
CGATGGAGTTCATCTGCAGCTCGATTCAGGCGGGTAGCCGGCTCGGTTATCAAGAGTCGG 600
CCGAACTCTGTGCGCAGTATCTGGCGCCAGTCCTCGATGCGATCAAGTTCAACTACTTTC 660
CA 02268036 1999-10-12
168
CGTTCGGCCT GAACGTGGCC AGCACCGCCT CGACACTC:CCTAAAGAGATCGCGTACTCCG720
AGCCCCGCTT GCAGCCGCCC AACGGGTACA AGGACACC:ACGGTGCCCGGCATCTGGGTGC780
CGGATACGCC GTTGTCACAC CGCAACACGC AGCCCGGTTGGGTGGTGGCACCCGGGATGC840
AAGGGGTTCA GGTGGGACCG ATCACGCAGG GTTTGCTGACGCCGGAGTCCCTGGCCGAAC900
TCATGGGTGG TCCCGATATC GCCCCTCCGT CGTCAGGGCTGCAAACCCCGCCCGGACCCC960
CGAATGCGTA CGACGAGTAC CCCGTGCTGC CGCCGATCGGTTTACAGGCCCCACAGGTGC1020
CGATACCACC GCCGCCTCCT GGGCCCGACG TAATCCCGGGTCCGGTGCCACCGGTCTTGG1080
CGGCGATCGT GTTCCCAAGA GATCGCCCGG CAGCGTCGGAAAACTTCGACTACATGGGCC1140
TCTTGTTGCT GTCGCCGGGC CTGGCGACCT TCCTGTTCGGGGTGTCATCTAGCCCCGCCC1200
GTGGAACGAT GGCCGATCGG CACGTGTTGA TACCGGCGATCACCGGCCTGGCGTTGATCG1260
CGGCATTCGT CGCACATTCG TGGTACCGCA CAGAACATCCGCTCATAGACATGCGCTTGT1320
TCCAGAACCG AGCGGTCGCG CAGGCCAACA TGACGATG;~CGGTGCTCTCCCTCGGGCTGT1380
TTGGCTCCTT CTTGCTGCTC CCGAGCTACC TCCAGCAAGTGTTGCACCAATCACCGATGC1440
AATCGGGGGT GCATATCATC CCACAGGGCC TCGGTGCCATGCTGGCGATGCCGATCGCCG1500
GAGCGATGAT GGACCGACGG GGACCGGCCA AGATCGTG(:TGGTTGGGATCATGCTGATCG1560
CTGCGGGGTT GGGCACCTTC GCCTTTGGTG TCGCGCGGCAAGCGGACTACTTACCCATTC1620
TGCCGACCGG GCTGGCAATC ATGGGCATGG GCATGGGCTGCTCCATGATGCCACTGTCCG1680
GGGCGGCAGT GCAGACCCTG GCCCCACATC AGATCGCTC:GCGGTTCGACGCTGATCAGCG1740
TCAACCAGCA GGTGGGCGGT TCGATAGGGA CCGCACTGATGTCGGTGCTGCTCACCTACC1800
AGTTCAATCA CAGCGAAATC ATCGCTACTG CAAAGAAAGTCGCACTGACCCCAGAGAGTG1860
GCGCCGGGCG GGGGGCGGCG GTTGACCCTT CCTCGCTACCGCGCCAAACCAACTTCGCGG1920
CCCAACTGCT GCATGACCTT TCGCACGCCT ACGCGGTGGTATTCGTGATAGCGACCGCGC1980
TAGTGGTCTC GACGCTGATC CCCGCGGCAT TCCTGCCGAAACAGCAGGCT 2040
AGTCATCGAA
GAGCACCGTT GCTATCCGCA TGACGTCTGC TT 2072
(2) INFORMATION FOR SEQ ID N0:185:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1923 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID :
N0:185
TCACCCCGGA GAAGTCGTTC GTCGACGACC TGGACATCG.A 60
CTCGCTGTCG ATGGTCGAGA
TCGCCGTGCA GACCGAGGAC AAGTACGGCG TCAAGATCCC 120
CGACGAGGAC CTCGCCGGTC
TGCGTACCGT CGGTGACGTT GTCGCCTACA TCCAGAAGC'T 180
CGAGGAAGAA AACCCGGAGG
CA 02268036 1999-10-12
169
CGGCTCAGGCGTTGCGCGCGAAGATTGAGT 240
CGGAGAAC;CC
CGATGCGGCA
CGAGCAGATC
GGTGCGTTTCACCCACATCGCAAGCTCGAG 300
ACGCCCGTCG
TCCTCTTGCA
CGCTCAGCCA
GGTTGGCGTGTCGCCGCCTTCCAGCAAGTGTTCCCACCAC CCTCGCGAAA360
ACGAAGGGAC
GGTGACTGATCCGCGGACCACATAGTCGATGCCACCGTGG CGCCGGGTCC420
CTGACAATTG
GAGTTGGCGGGGGCCGAATTGCGGCATTGCGTCGAAGGCCAGCGGATCCCGGCGCCCGCC480
CGGCGTGGCTGGTGTTTTGGGCCGCCGGATGGCCACGACGAGAACGACGATGGCGGCGAT540
GAACAGCGCCACGGCAATCACGACCAGCAGATTTCCCACGCATACCCTCTCGTACCGCTG600
CGCCGCGGTTGGTCGATCGGTCGCATATCGATGGCGCCGTTTAACGTAACAGCTTTCGCG660
GGACCGGGGGTCACAACGGGCGAGTTGTCCGGCCGGGAACCCGGCAGGTCTCGGCCGCGG720
TCACCCCAGCTCACTGGTGCACCATCCGGGTGTCGGTG,~GCGTGCAACTCAAACACACTC780
AACGGCAACGGTTTCTCAGGTCACCAGCTCAACCTCGACCCGCAATCGCTCGTACGTTTC840
GACCGCGCGCAGGTCGCGAGTCAGCAGCTTTGCGCCGGCAGCTTTCGCCGTGAAGCCGAC900
CAGGGCATCGTAGGTTGCGCCACCGGTGACATCGTGCTCGGCGAGGTGGTCGGTCAAGCC960
GCGATATGAGCAGGCATCCAGTGCCAGGTAGTTGCTGGAGGTGATGTCCGCCAAGTAGGC1020
GTGGACGGCAACAGGGGCAATACGATGCGGCGGTGGTAGCCGGGTCAAGACCGAATAGGT1080
TTCCACAGCCGCGTGCGCGATCAGATGGACGCCACGGTTGAGCGCGCGCACGGCGGCCTC1140
GTGCCCTTCGTGCCAGGTCGCGAATCCGGCAACCAGCAC:GCTGGTGTCTGGTGCGATCAC1200
CGCCGTGTGCGATCGAGCGTTTCCCGAACGATTTCGTCGGTCAACGGGGGCAGGGGACGT1260
TCTGGCCGTGCGACGAGAACCGAGCCTTCCCGAACGAGTTCGACACCGGTCGGGGCCGGC1320
TCAATCTCGATGCGCCCATCGCGCTCGGTGATCTCCACC:TGGTCGTTCCCGCGCAAGCCA1380
AGGCGCTCGCGAATCCGCTTGGGAATCACCAGACGTCCTGCGACATCGATGGTTGTTCGC1440
ATGGTAGGAAATTTACCATCGCACGTTCCATAGGCGTGTCCTGCGCGGGATGTCGGGACG1500
ATCCGCTAGCGTATCGAACGATTGTTTCGGAAATGGCTGAGGGAGCGTGCGGTGCGGGTG1560
ATGGGTGTCGATCCCGGGTTGACCCGATGCGGGCTGTCGCTCATCGAGAGTGGGCGTGGT1620
CGGCAGCTCACCGCGCTGGATGTCGACGTGGTGCGCACACCGTCGGATGCGGCCTTGGCG1680
CAGCGCCTGTTGGCCATCAGCGATGCCGTCGAGCACTGGCTGGACACCCATCATCCGGAG1740
GTGGTGGCTA GTTCTCTCAGCTCAACGTGACCACGGTGATGGGCACCGCG1800
TCGAACGGGT
CAGGCCGGCG GCCAAACGTGGTGTCGACGT 1860
GCGTGATCGC GCATTTCCAT
CCTGGCGGCG
ACCCCCAGCG GGCAACGGT'rCCGCAGACAA 1920
AGGTCAAGGC GGCTCAGGTC
GGCGGTCACT
ACC
1923
(2) INFORMATION FOR SEQ ID N0:186:
CA 02268036 1999-10-12
170
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:1055 basepairs
(B) TYPE: ucleic
n acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi) SEQUENCE CRIPTION:EQ ID 6:
DES S N0:18
CTGGCGTGCCAGTGTCACCGGCGATATGACGTCGGCATTCAATTTCGCGGCCCCGCCGGA 60
CCCGTCGCCACCCAATCTGGACCACCCGGTCCGTCAATTGCCGAAGGTCGCCAAGTGCGT 120
GCCCAATGTGGTGCTGGGTTTCTTGAACGAAGGCCTGCCGTATCGGGTGCCCTACCCCCA 180
AACAACGCCAGTCCAGGAATCCGGTCCCGCGCGGCCGATTCCCAGCGGCATCTGCTAGCC 240
GGGGATGGTTCAGACGTAACGGTTGGCTAGGTCGAAACCCGCGCCAGGGCCGCTGGACGG 300
GCTCATGGCAGCGAAATTAGAAAACCCGGGATATTGTCCGCGGATTGTCATACGATGCTG 360
AGTGCTTGGTGGTTCGTGTTTAGCCATTGAGTGTGGATGTGTTGAGACCCTGGCCTGGAA 420
GGGGACAACGTGCTTTTGCCTCTTGGTCCGCCTTTGCCGCCCGACGCGGTGGTGGCGAAA 480
CGGGCTGAGTCGGGAATGCTCGGCGGGTTGTCGGTTCCGCTCAGCTGGGGAGTGGCTGTG 540
CCACCCGATGATTATGACCACTGGGCGCCTGCGCCGGA(iGACGGCGCCGATGTCGATGTC 600
CAGGCGGCCGAAGGGGCGGACGCAGAGGCCGCGGCCATGGACGAGTGGGATGAGTGGCAG 660
GCGTGGAACGAGTGGGTGGCGGAGAACGCTGAACCCCGC:TTTGAGGTGCCACGGAGTAGC 720
AGCAGCGTGATTCCGCATTCTCCGGCGGCCGGCTAGGAGAGGGGGCGCAGACTGTCGTTA 780
TTTGACCAGTGATCGGCGGTCTCGGTGTTCCCGCGGCCGGCTATGACAACAGTCAATGTG 840
CATGACAAGTTACAGGTATTAGGTCCAGGTTCAACAAGC~AGACAGGCAACATGGCAACAC 900
GTTTTATGACGGATCCGCACGCGATGCGGGACATGGCGGGCCGTTTTGAGGTGCACGCCC 960
AGACGGTGGAGGACGAGGCTCGCCGGATGTGGGCGTCCGCGCAAAACATCTCGGGNGCGG 1020
GCTGGAGTGGCATGGCCGAGGCGACCTCGCTAGAC 1055
(2) INFORMATION FOR SEQ ID N0:187:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 359 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:187:
CCGCCTCGTT GTTGGCATAC TCCGCCGCGG CCGCCTCGAC CGCACTGGCC GTGGCGTGTG 60
TCCGGGCTGA CCACCGGGAT CGCCGAACCA TCCGAGATC,A CCTCGCAATG ATCCACCTCG 120
CGCAGCTGGT CACCCAGCCA CCGGGCGGTG TGCGACAGCG CCTGCATCAC CTTGGTATAG 180
CCGTCGCGCC CCAGCCGCAG GAAGTTGTAG TACTGGCCCA CCACCTGGTT ACCGGGACGG 240
GAGAAGTTCA GGGTGAAGGT CGGCATGTCG CCGCCGAGG'P AGTTGACCCG GAAAACCAGA 300
CA 02268036 1999-10-12
171
TCCTCCGGCA GGTGCTCGGG CCCGCGCCAC ACGACAARCCCGACGCCGGG ATAGGTCAG359
(2) INFORMATION FOR SEQ ID N0:188:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 350 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: $EQ ID
N0:188:
AACGGGCCCG TGGGCACCGC TCCTCTAAGG GCTCTCGTTGGTCGCATGAA GTGCTGGAAG60
GATGCATCTT GGCAGATTCC CGCCAGAGCA AAACAGCCGCTAGTCCTAGT CCGAGTCGCC120
CGCAAAGTTC CTCGAATAAC TCCGTACCCG GAGCGCCAAACCGGGTCTCC TTCGCTAAGC180
TGCGCGAACC ACTTGAGGTT CCGGGACTCC TTGACGTCCAGACCGATTCG TTCGAGTGGC240
TGATCGGTTC GCCGCGCTGG CGCGAATCCG CCGCCGAGCGGGGTGATGTC AACCCAGTGG300
GTGGCCTGGA AGAGGTGCTC TACGAGCTGT CTCCGATCGAGGACTTCTCC 350
(2) INFORMATION FOR SEQ ID N0:189:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 679 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:1.89:
Glu Gln Pro Lys Gly Pro Phe Gly Glu Val Ile Glu Ala Phe Ala Asp
1 5 1.0 15
Gly Leu Ala Gly Lys Gly Lys Gln Ile Asn Thr Thr Leu Asn Ser Leu
20 25 30
Ser Gln Ala Leu Asn Ala Leu Asn Glu G'ly Arg Gly Asp Phe Phe Ala
35 40 45
Val Val Arg Ser Leu Ala Leu Phe Val A.sn Ala Leu His Gln Asp Asp
50 55 60
Gln Gln Phe Val Ala Leu Asn Lys Asn Leu Ala Glu Phe Thr Asp Arg
65 70 75 80
Leu Thr His Ser Asp Ala Asp Leu Ser Asn Ala Ile Gln Gln Phe Asp
85 90 95
Ser Leu Leu Ala Val Ala Arg Pro Phe Phe Ala Lys Asn Arg Glu Val
100 105 110
Leu Thr His Asp Val Asn Asn Leu Ala T:hr Val Thr Thr Thr Leu Leu
115 120 125
Gln Pro Asp Pro Leu Asp Gly Leu Glu Tlzr Val Leu His Ile Phe Pro
130 135 140
CA 02268036 1999-10-12
172
Thr Leu Ala Ala Asn Ile Asn Gln Leu Tyr His Pro Thr His Gly Gly
145 150 155 160
Val Val Ser Leu Ser Ala Phe Thr Asn Phe Ala Asn Pro Met Glu Phe
165 170 175
Ile Cys Ser Ser Ile Gln Ala Gly Ser Arg Leu Gly Tyr Gln Glu Ser
180 185 190
Ala Glu Leu Cys Ala Gln Tyr Leu Ala Pro Val Leu Asp Ala Ile Lys
195 200 205
Phe Asn Tyr Phe Pro Phe Gly Leu Asn Val Ala Ser Thr Ala Ser Thr
210 215 220
Leu Pro Lys Glu Ile Ala Tyr Ser Glu Pro Arg Leu Gln Pro Pro Asn
225 230 235 240
Gly Tyr Lys Asp Thr Thr Val Pro Gly Ile Trp Val Pro Asp Thr Pro
245 250 255
Leu Ser His Arg Asn Thr Gln Pro Gly 'Trp Val Val Ala Pro Gly Met
260 265 270
Gln Gly Val Gln Val Gly Pro Ile Thr Gln Gly Leu Leu Thr Pro Glu
275 280 285
Ser Leu Ala Glu Leu Met Gly Gly Pro Asp Ile Ala Pro Pro Ser Ser
290 295 300
Gly Leu Gln Thr Pro Pro Gly Pro Pro Asn Ala Tyr Asp Glu Tyr Pro
305 310 315 320
Val Leu Pro Pro Ile Gly Leu Gln Ala Pro Gln Val Pro Ile Pro Pro
325 330 335
Pro Pro Pro Gly Pro Asp Val Ile Pro CJly Pro Val Pro Pro Val Leu
340 395 350
Ala Ala Ile Val Phe Pro Arg Asp Arg Pro Ala Ala Ser Glu Asn Phe
355 360 365
Asp Tyr Met Gly Leu Leu Leu Leu Ser F'ro Gly Leu Ala Thr Phe Leu
370 375 380
Phe Gly Val Ser Ser Ser Pro Ala Arg Gly Thr Met Ala Asp Arg His
385 390 395 400
Val Leu Ile Pro Ala Ile Thr Gly Leu Ala Leu Ile Ala Ala Phe Val
405 410 415
Ala His Ser Trp Tyr Arg Thr Glu His Pro Leu Ile Asp Met Arg Leu
420 425 430
Phe Gln Asn Arg Ala Val Ala Gln Ala Asn Met Thr Met Thr Val Leu
435 440 445
Ser Leu Gly Leu Phe Gly Ser Phe Leu Leu Leu Pro Ser Tyr Leu Gln
450 455 460
Gln Val Leu His Gln Ser Pro Met Gln S~~r Gly Val His Ile Ile Pro
465 470 475 480
Gln Gly Leu Gly Ala Met Leu Ala Met P:ro Ile Ala Gly Ala Met Met
CA 02268036 1999-10-12
173
485 490 495
Asp Arg Arg Gly Pro Ala Lys Ile Val Leu Val Gly Ile Met Leu Ile
500 505 510
Ala Ala Gly Leu Gly Thr Phe Ala Phe Gly Val Ala Arg Gln Ala Asp
515 520 525
Tyr Leu Pro Ile Leu Pro Thr Gly Leu Ala Ile Met Gly Met Gly Met
530 535 540
Gly Cys Ser Met Met Pro Leu Ser Gly Ala Ala Val Gln Thr Leu Ala
545 550 555 560
Pro His Gln Ile Ala Arg Gly Ser Thr Leu Ile Ser Val Asn Gln Gln
565 570 575
Val Gly Gly Ser Ile Gly Thr Ala Leu Met Ser Val Leu Leu Thr Tyr
580 585 590
Gln Phe Asn His Ser Glu Ile Ile Ala 'Phr Ala Lys Lys Val Ala Leu
595 600 605
Thr Pro Glu Ser Gly Ala Gly Arg Gly Ala Ala Val Asp Pro Ser Ser
610 615 620
Leu Pro Arg Gln Thr Asn Phe Ala Ala Gln Leu Leu His Asp Leu Ser
625 630 635 640
His Ala Tyr Ala Val Val Phe Val Ile Ala Thr Ala Leu Val Val Ser
645 Ei50 655
Thr Leu Ile Pro Ala Ala Phe Leu Pro hys Gln Gln Ala Ser His Arg
660 665 670
Arg Ala Pro Leu Leu Ser Ala
675
(2) INFORMATION FOR SEQ ID N0:190:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 120 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:190:
Thr Pro Glu Lys Ser Phe Val Asp Asp Leu Asp Ile Asp Ser Leu Ser
1 5 10 15
Met Val Glu Ile Ala Val Gln Thr Glu Asp Lys Tyr Gly Val Lys Ile
20 25 30
Pro Asp Glu Asp Leu Ala Gly Leu Arg Thr Val Gly Asp Val Val Ala
35 40 45
Tyr Ile Gln Lys Leu Glu Glu Glu Asn Pro Glu Ala Ala Gln Ala Leu
50 55 60
Arg Ala Lys Ile Glu Ser Glu Asn Pro Asp Ala Ala Arg Ala Asp Arg
65 70 75 80
CA 02268036 1999-10-12
174
Cys Val Ser Pro Thr Ser Gln Ala Arg Asp Ala Arg Arg Pro Leu Ala
85 90 95
Arg Ser Ala Arg Leu Ala Cys Arg Arg Leu Pro Ala Ser Val Pro Thr
100 105 110
Thr Arg Arg Asp Pro Arg Glu Arg
115 120
(2) INFORMATION FOR SEQ ID N0:191:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 89 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:191:
Leu Ala Cys Gln Cys His Arg Arg Tyr .Asp Val Gly Ile Gln Phe Arg
1 5 10 15
Gly Pro Ala Gly Pro Val Ala Thr Gln Ser Gly Pro Pro Gly Pro Ser
20 25 30
Ile Ala Glu Gly Arg Gln Val Arg Ala ciln Cys Gly Ala Gly Phe Leu
35 40 45
Glu Arg Arg Pro Ala Val Ser Gly Ala Leu Pro Pro Asn Asn Ala Ser
50 55 60
Pro Gly Ile Arg Ser Arg Ala Ala Asp Ser Gln Arg His Leu Leu Ala
65 70 75 80
Gly Asp Gly Ser Asp Val Thr Val Gly
(2) INFORMATION FOR SEQ ID N0:192:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 119 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:192:
Ala Ser Leu Leu Ala Tyr Ser Ala Ala A.la Ala Ser Thr Ala Leu Ala
1 5 10 15
Val Ala Cys Val Arg Ala Asp His Arg Asp Arg Arg Thr Ile Arg Asp
20 25 30
His Leu Ala Met Ile His Leu Ala Gln Leu Val Thr Gln Pro Pro Gly
35 40 45
Gly Val Arg Gln Arg Leu His His Leu Gly Ile Ala Val Ala Pro Gln
50 55 60
Pro Gln Glu Val Val Val Leu Ala His His Leu Val Thr Gly Thr Gly
65 70 75 80
CA 02268036 1999-10-12
175
Glu Val Gln Gly Glu Gly Arg His Val Ala Ala Glu Val Val Asp Pro
85 90 95
Glu Asn Gln Ile Leu Arg Gln Val Leu Gly Pro Ala Pro His Asp Lys
100 105 110
Pro Asp Ala Gly Ile Gly Gln
115
(2) INFORMATION FOR SEQ ID N0:193:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 116 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:193:
Arg Ala Arg Gly His Arg Ser Ser Lys Gly Ser Arg Trp Ser His Glu
1 5 10 15
Val Leu Glu Gly Cys Ile Leu Ala Asp S er Arg Gln Ser Lys Thr Ala
20 25 30
Ala Ser Pro Ser Pro Ser Arg Pro Gln Ser Ser Ser Asn Asn Ser Val
35 40 45
Pro Gly Ala Pro Asn Arg Val Ser Phe Ala Lys Leu Arg Glu Pro Leu
50 55 60
Glu Val Pro Gly Leu Leu Asp Val Gln Thr Asp Ser Phe Glu Trp Leu
65 70 75 80
Ile Gly Ser Pro Arg Trp Arg Glu Ser Ala Ala Glu Arg Gly Asp Val
85 '-30 95
Asn Pro Val Gly Gly Leu Glu Glu Val Leu Tyr Glu Leu Ser Pro Ile
100 105 110
Glu Asp Phe Ser
115
(2) INFORMATION FOR SEQ ID N0:194:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 811 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:194:
TGCTACGCAGCAATCGCTTTGGTGACAGATGTGGATGCCGGCGTCGCTGCTGGCGATGGC 60
GTGAAAGCCGCCGACGTGTTCGCCGCATTCGGGGAGAAC.ATCGAACTGCTCAAAAGGCTG 120
GTGCGGGCCGCCATCGATCGGGTCGCCGACGAGCGCACG'TGCACGCACTGTCAACACCAC 180
GCCGGTGTTCCGTTGCCGTTCGAGCTGCCATGAGGGTGC'rGCTGACCGGCGCGGCCGGCT 240
TCATCGGGTCGCGCGTGGATGCGGCGTTACGGGCTGCGGGTCACGACGTGGTGGGCGTCG 300
CA 02268036 1999-10-12
176
ACGCGCTGCTGCCCGCCGCGCACGGGCCAA GCCACCGGGCTGCCAGCGGG 360
ACCCGGTGCT
TCGACGTGCGCGACGCCAGCGCGCTGGCCCCGTTGTTG'~GCCGGTGTCGATCTGGTGTGTC 420
ACCAGGCCGCCATGGTGGGTGCCGGCGTCAACGCCGCCGACGCACCCGCCTATGGCGGCC 480
ACAACGATTTCGCCACCACGGTGCTGCTGGCGCAGATGTTCGCCGCCGGGGTCCGCCGTT 540
TGGTGCTGGCGTCGTCGATGGTGGTTTACGGGCAGGGGCGCTATGACTGTCCCCAGCATG 600
GACCGGTCGACCCGCTGCCGCGGCGGCGAGCCGACCTGGACAATGGGGTCTTCGAGCACC 660
GTTGCCCGGGGTGCGGCGAGCCAGTCATCTGGCAATTGGTCGACGAAGATGCCCCGTTGC 720
GCCCGCGCAGCCTGTACGCGGCAGCAAGACCGCGCAGGAGCACTACGCGCTGGCGTGGTC 780
GGAAACGAATGGCGGTTCCGTGGTGGCGTTG 811
(2) INFORMATION
FOR SEQ
ID N0:195:
(i) S EQUENCE S:
CHARACTERISTIC
(A) LENGTH:966 base airs
p
(B) TYPE:ucleic
n acid
(C) STRANDEDNESS: le
sing
(D) TOPOLOGY: linear
(xi) S EQUENCE CRIPTION: 5:
DES SEQ ID
N0:19
GTCCCGCGATGTGGCCGAGCATGACTTTCGGCAACACCC~GCGTAGTAGTCGAAGATATCG 60
GACTTTGTGGTCCCGGTGGCGGGATAGAGCACCTGTCGC~CGTTGGTCAGCGTCACCCGTT 120
GCTCGGACGCCGAACCCATGCTTTCAACGTAGCCTGTCGGTCACACAAGTCGCGAGCGTA 180
ACGTCACGGTCAAATATCGCGTGGAATTTCGCCGTGACCJTTCCGCTCGCGGACAATCAAG 240
GCATACTCACTTACATGCGAGCCATTTGGACGGGTTCGATCGCCTTCGGGCTGGTGAACG 300
TGCCGGTCAAGGTGTACAGCGCTACCGCAGACCACGACF~TCAGGTTCCACCAGGTGCACG 360
CCAAGGACAACGGACGCATCCGGTACAAGCGCGTCTGCGAGGCGTGTGGCGAGGTGGTCG 420
ACTACCGCGATCTTGCCCGGGCCTACGAGTCCGGCGACGGCCAAATGGTGGCGATCACCG 480
ACGACGACATCGCCAGCTTGCCTGAAGAACGCAGCCGGGAGATCGAGGTGTTGGAGTTCG 540
TCCCCGCCGCCGACGTGGACCCGATGATGTTCGACCGCAGCTACTTTTTGGAGCCTGATT 600
CGAAGTCGTCGAAATCGTATGTGCTGCTGGCTAAGACACTCGCCGAGACCGACCGGATGG 660
CGATCGTGGATCGCCCCACCGGCCGTGAATGCAGGAAAAATAAGAGCCGCTATCCACAAT 720
TCGGCGTCGAGCTCGGCTACCACAAACGGTAGAACGATCGAGACATTCCCGAGCTGAAGT 780
GCGGCGCTATAGAAGCCGCTCTGCGCGATTATCAAACGC.AAAATACGCTTACTCATGCCA 840
TCGGCGCTGCTCACCCGATGCGACGTTTTTGCCACGCTCCACCGCCTGCCGCGCGACCTC 900
AAGTGGGCATGCATCCCACCCGTTCCCGGAAACCGGTTCCGGCGGGTCGGCTCATCGCTT 960
CATCCT 966
CA 02268036 1999-10-12
177
(2) INFORMATION
FOR SEQ
ID N0:196:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:2367 basepairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY:
linear
(xi) S EQUENCE 6:
DESCRIPTION:
SEQ ID
N0:19
CCGCACCGCCGGCAATACCGCCAGCGCCAC GTTTGCGCCGTTGCCCCCGT 60
CGTTACCGCC
TGCCGCCCGTCCCGCCGGCCCCGCCGATGGAGTTCTCATCGCCAAAAGTACTGGCGTTGC 120
CACCGGAGCCGCCGTTGCCGCCGTCACCGCCAGCCCCGCCGACTCCACCGGCCCCACCGA 180
CTCCGCCGCTGCCACCGTTGCCGCCGTTGCCGATCAACATGCCGCTGGCGCCACCCTTGC 240
CACCCACGCCACCGGCTCCGCCCACCCCGCCGACACCA;~GCGAGCTGCCGCCGGAGCCAC 300
CATCACCACCTACGCCACCGACCGCCCAGACACCAGCGACCGGGTCTTCGTGAAACGTCG 360
CGGTGCCACCACCGCCGCCGTTACCGCCAACCCCACCGGCAACGCCGGCGCCGCCATCCC 420
CGCCGGCCCCGGCGTTGCCGCCGTTGCCGCCGTTGCCGAACAACAACCCGCCGGCGCCGC 480
CGTTGCCGCCCGCGCCGCCGGTCCCGCCGGCGCCGCCGACGCCAAGGCCGCTGCCGCCCT 540
TGCCGCCATCACCACCCTTGCCGCCGACCACATCGGGT~PCTGCCTCGGGGTCTGGGCTGT 600
CAAACCTCGCGATGCCAGCGTTGCCGCCGCTTCCCCCGC~GCCCCCCCGTGGCGCCGTCAC 660
CACCGATACCACCCGCGCCACCGGCGCCACCGTTGCCGC:CATCACCGAATAGCAACCCGC 720
CGGCGCCACCATTGCCGCCAGCTCCCCCTGCGCCACCG7.'CGGCGCCGGAGGCGGCACTGG 780
CAGCCCCGTTACCACCGAAACCGCCGCTACCACCGGTAGAGGTGGCAGTGGCGATGTGTA 840
CGAAAGCGCCGCCTCCGGCGCCGCCGCTACCACCCCCAC',TGCCGGCGGCTACACCGTCGG 900
ACCCGTTGCCACCATCACCGCCAAAGGCGCTCGCAATGT'CGCCCTGCGCGACTCCGCCGT 960
CGCCGCCGTTGCCGCCGCCGCCACCGGCAGCGGCGGTACCGCCGTCACCACCGGCACCGC 1020
CGGTGGCCTTGCCCGAGCCTGCCGTCGCGGTGGCACCGTCGCCGCCGGTGCCACCGGTCG 1080
GCGTGCCGGCAGTGCCATGGCCGCCCGTGCCGCCGTCGCCGCCGGTTTGATCACCGATGC 1140
CGGACACATCTGCCGGGCTGTCCCCGGTGCTGGCCGCGGGGCCGGGCGTGGGATTGACCC 1200
CGTTTGCCCCGGCGAGGCCGGCGCCGCCGGTACCACCGGCGCCGCCATGGCCGAACAGCC 1260
CGGCGTTGCCGCCGTTACCGCCCGCACCCCCGATGCCTGCGGCCACGCTGGTGCCGCCGA 1320
CACCGCCGTTGCCGCCGTTGCCCCACAACCACCCCCCGTTCCCACCGGCACCGCCGGCCG 1380
CGCCGGTACCACCGGCCCCGCCGTTGCCGCCGTTGCCGA'TCAACCCGGCCGCGCCTCCGC 1440
TGCCGCCGGTTTGACCGAACCCGCCAGCCGCGCCGTTGCC AACAGCAACC 1500
ACCGTTGCCA
CGCCGGCCGC CCGGGTGCCGTCCCGTCGGC 1560
GCCAGGCTGC GCCGTTTCCG
ATCAACGGGC
GCCCCAAAAG GGCGCATTCACCGCACCCAG TCAACAGCGG 1620
CGCCTCGGTG CAGACTCCGC
CA 02268036 1999-10-12
178
CTTCAGTGCTGGCATACCGACCCGCGGCCGCAGTCAAC'GCCTGCACAAACTGCTCGTGAA1680
ACGCTGCCACCTGTACGCTGAGCGCCTGATACTGCCGA.GCATGGGCCCCGAACAACCCCG1790
CAATCGCCGCCGACACTTCATCGGCAGCCGCAGCCACCACTTCCGTCGTCGGGATCGCCG1800
CGGCCGCATTAGCCGCGCTCACCTGCGAACCAATAGTCGATAAATCCP.AAGCCGCAGTTG1860
CCAGCAGCTGCGGCGTCGCGATCACCAAGGACACCTCGCACCTCCGGATACCCCATATCG1920
CCGCACCGTGTCCCCAGCGGCCACGTGACCTTTGGTCGCTGGCTGGCGGCCCTGACTATG1980
GCCGCGACGGCCCTCGTTCTGATTCGCCCCGGCGCGCAGCTTGTTGCGCGAGTTGAAGAC2040
GGGAGGACAGGCCGAGCTTGGTGTAGACGTGGGTCAAG'TGGGAATGCACGGTCCGCGGCG2100
AGATGAATAGGCGGACGCCGATCTCCTTGTTGCTGAGTCCCTCACCGACCAGTAGAGCCA2160
CCTCAAGCTCTGTCGGTGTCAACGCGCCCCAGCCACTTGTCGGGCGTTTCCGTGCACCGC2220
GGCCTCGTTGCGCGTACGCGATCGCCTCATCGATCGATi'~ACGCAGTTCCTTCGGCCCAGG2280
CATCGTCGAACTCGCTGTCACCCATGGATTTTCGAAGGGTGGCTAGCGACGAGTTACAGC2340
CCGCCTGGTAGATCCCGAAGCGGACCG 2367
(2) INFORMATION FOR SEQ ID N0:197:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 376 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:1.97:
Gln Pro Ala Gly Ala Thr Ile Ala Ala w~er Ser Pro Cys Ala Thr Val
1 5 1.0 15
Gly Ala Gly Gly Gly Thr Gly Ser Pro V'al Thr Thr Glu Thr Ala Ala
20 25 30
Thr Thr Gly Arg Gly Gly Ser Gly Asp Val Tyr Glu Ser Ala Ala Ser
35 40 45
Gly Ala Ala Ala Thr Thr Pro Thr Ala Gly Gly Tyr Thr Val Gly Pro
50 55 60
Val Ala Thr Ile Thr Ala Lys Gly Ala Arg Asn Val Ala Leu Arg Asp
65 70 75 80
Ser Ala Val Ala Ala Val Ala Ala Ala Ala Thr Gly Ser Gly Gly Thr
85 90 95
Ala Val Thr Thr Gly Thr Ala Gly Gly Leu Ala Arg Ala Cys Arg Arg
100 105 110
Gly Gly Thr Val Ala Ala Gly Ala Thr G.ly Arg Arg Ala Gly Ser Ala
115 120 125
Met Ala Ala Arg Ala Ala Val Ala Ala G:Ly Leu Ile Thr Asp Ala Gly
130 135 140
CA 02268036 1999-10-12
179
His Ile Cys Arg Ala Val Pro Gly Ala Gly Arg Gly Ala Gly Arg Gly
145 150 155 160
Ile Asp Pro Val Cys Pro Gly Glu Ala Gly Ala Ala Gly Thr Thr Gly
165 170 175
Ala Ala Met Ala Glu Gln Pro Gly Val Ala Ala Val Thr Ala Arg Thr
180 185 190
Pro Asp Ala Cys Gly His Ala Gly Ala Ala Asp Thr Ala Val Ala Ala
195 200 205
Val Ala Pro Gln Pro Pro Pro Val Pro Thr Gly Thr Ala Gly Arg Ala
210 215 220
Gly Thr Thr Gly Pro Ala Val Ala Ala Val Ala Asp Gln Pro Gly Arg
225 230 235 240
Ala Ser Ala Ala Ala Gly Leu Thr Glu Pro Ala Ser Arg Ala Val Ala
245 250 255
Thr Val Ala Lys Gln Gln Pro Ala Gly Arg Ala Arg Leu Pro Gly Cys
260 265 270
Arg Pro Val Gly Ala Val Ser Asp Gln Arg Ala Pro Gln Lys Arg Leu
275 280 285
Gly Gly Arg Ile His Arg Thr Gln Gln '.Chr Pro Leu Asn Ser Gly Phe
290 295 300
Ser Ala Gly Ile Pro Thr Arg Gly Arg ;ier Gln Arg Leu His Lys Leu
305 310 315 320
Leu Val Lys Arg Cys His Leu Tyr Ala Glu Arg Leu Ile Leu Pro Ser
325 330 335
Met Gly Pro Glu Gln Pro Arg Asn Arg Arg Arg His Phe Ile Gly Ser
340 345 350
Arg Ser His His Phe Arg Arg Arg Asp Arg Arg Gly Arg Ile Ser Arg
355 360 365
Ala His Leu Arg Thr Asn Ser Arg
370 375
(2) INFORMATION FOR SEQ ID N0:198:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2852 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID
N0:198:
GGCCAAAACG CCCCGGCGAT CGCGGCCACC GAGGCCGCC'rACGACCAGATGTGGGCCCAG 60
GACGTGGCGG CGATGTTTGG CTACCATGCC GGGGCTTCGGCGGCCGTCTCGGCGTTGACA 120
CCGTTCGGCC AGGCGCTGCC GACCGTGGCG GGCGGCGGTGCGCTGGTCAGCGCGGCCGCG 180
GCTCAGGTGA CCACGCGGGT CTTCCGCAAC CTGGGCTTG(3CGAACGTCCGCGAGGGCAAC 240
CA 02268036 1999-10-12
180
GTCCGCAACGGTAATGTCCGGAACTTCAAT 300
CTCGGCTC;GG
CCAACATCGG
CAACGGCAAC
ATCGGCAGCGGCAACATCGGCAGCTCCAAC 360
ATCGGGTTTG
GCAACGTGGG
TCCTGGGTTG
ACCGCAGCGCTGAACAACATCGGTTTCGGCAACACCGGCA GCAACAACATCGGGTTTGGC420
AACACCGGCAGCAACAACATCGGGTTCGGCAATACCGG'AG ACGGCAACCGAGGTATCGGG480
CTCACGGGTAGCGGTTTGTTGGGGTTCGGCGGCCTGAA.CT CGGGCACCGGCAACATCGGT540
CTGTTCAACTCGGGCACCGGAAACGTCGGCATCGGCAACT CGGGTACCGGGAACTGGGGC600
ATTGGCAACTCGGGCAACAGCTACAACACCGGTTTTGGCA ACTCCGGCGACGCCAACACG660
GGCTTCTTCAACTCCGGAATAGCCAACACCGGCGTCGGCA ACGCCGGCAACTACAACACC720
GGTAGCTACAACCCGGGCAACAGCAATACCGGCGGCTTCA ACATGGGCCAGTACAACACG780
GGCTACCTGAACAGCGGCAACTACAACACCGGCTTGGC.AA ACTCCGGCAATGTCAACACC890
GGCGCCTTCATTACTGGCAACTTCAACAACGGCTTCTTGT GGCGCGGCGACCACCAAGGC900
CTGATTTTCGGGAGCCCCGGCTTCTTCAACTCGACCAG'rG CGCCGTCGTCGGGATTCTTC960
AACAGCGGTGCCGGTAGCGCGTCCGGCTTCCTGAACTCCG GTGCCAACAATTCTGGCTTC1020
TTCAACTCTTCGTCGGGGGCCATCGGTAACTCCGGCCTGG CAAACGCGGGCGTGCTGGTA1080
TCGGGCGTGATCAACTCGGGCAACACCGTATCGGGTTTGT TCAACATGAGCCTGGTGGCC1140
ATCACAACGCCGGCCTTGATCTCGGGCTTCTTCAACAC(:G GAAGCAACATGTCGGGATTT1200
TTCGGTGGCCCACCGGTCTTCAATCTCGGCCTGGCAAAC:C GGGGCGTCGTGAACATTCTC1260
GGCAACGCCAACATCGGCAATTACAACATTCTCGGCAGC:G GAAACGTCGGTGACTTCAAC1320
ATCCTTGGCAGCGGCAACCTCGGCAGCCAAAACATCTTGG GCAGCGGCAACGTCGGCAGC1380
TTCAATATCGGCAGTGGAAACATCGGAGTATTCAATGTC;G GTTCCGGAAGCCTGGGAAAC1440
TACAACATCGGATCCGGAAACCTCGGGATCTACAACATC;G GTTTTGGAAACGTCGGCGAC1500
TACAACGTCGGCTTCGGGAACGCGGGCGACTTCAACCAAG GCTTTGCCAACACCGGCAAC1560
AACAACATCGGGTTCGCCAACACCGGCAACAACAACATC'G GCATCGGGCTGTCCGGCGAC1620
AACCAGCAGGGCTTCAATATTGCTAGCGGCTGGAACTCGG GCACCGGCAACAGCGGCCTG1680
TTCAATTCGGGCACCAATAACGTTGGCATCTTCAACGCGG GCACCGGAAACGTCGGCATC1740
GCAAACTCGGGCACCGGGAACTGGGGTATCGGGAACCCGG GTACCGACAATACCGGCATC1800
CTCAATGCTGGCAGCTACAACACGGGCATCCTCAACGCCG GCGACTTCAACACGGGCTTC1860
TACAACACGGGCAGCTACAACACCGGCGGCTTCAACGTCG GTAACACCAACACCGGCAAC1920
TTCAACGTGG TACCGGCAGCTATAACCCGG GTGACACCAACACCGGCTTC1980
GTGACACCAA
TTCAATCCCG TTCGACACGG GCGACTTCAA 2040
GCAACGTCAA CAATGGCTTC
TACCGGCGCT
TTGGTGGCGG 2100
GCGATAACCA
GGGCCAGATT
GCCATCGATC
TCTCGGTCAC
CACTCCATTC
ATCCCCATAA 2160
ACGAGCAGAT
GGTCATTGAC
GTACACAACG
TAATGACCTT
CGGCGGCAAC
CA 02268036 1999-10-12
181
ATGATCACGGTCACCGAGGCCTCGACCGTTTTCCCCCP,AACCTTCTATCTGAGCGGTTTG2220
TTCTTCTTCGGCCCGGTCAATCTCAGCGCATCCACGCT'GACCGTTCCGACGATCACCCTC2280
ACCATCGGCGGACCGACGGTGACCGTCCCCATCAGCATTGTCGGTGCTCTGGAGAGCCGC2340
ACGATTACCTTCCTCAAGATCGATCCGGCGCCGGGCATCGGAAATTCGACCACCAACCCC2900
TCGTCCGGCTTCTTCAACTCGGGCACCGGTGGCACATCTGGCTTCCAAAACGTCGGCGGC2460
GGCAGTTCAGGCGTCTGGAACAGTGGTTTGAGCAGCGCGATAGGGAATTCGGGTTTCCAG2520
AACCTCGGCTCGCTGCAGTCAGGCTGGGCGAACCTGGGCAACTCCGTATCGGGCTTTTTC2580
AACACCAGTACGGTGAACCTCTCCACGCCGGCCAATGTCTCGGGCCTGAACAACATCGGC2640
ACCAACCTGTCCGGCGTGTTCCGCGGTCCGACCGGGACGATTTTCAACGCGGGCCTTGCC2700
AACCTGGGCCAGTTGAACATCGGCAGCGCCTCGTGCCGAATTCGGCACGAGTTAGATACG2760
GTTTCAACAATCATATCCGCGTTTTGCGGCAGTGCATCAGACGAATCGAACCCGGGAAGC2820
GTAAGCGAATAAACCGAATGGCGGCCTGTCAT 2852
(2) INFORMATION FOR SEQ ID N0:199:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 943 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:1.99:
Gly Gln Asn Ala Pro Ala Ile Ala Ala Thr Glu Ala Ala Tyr Asp Gln
1 5 L0 15
Met Trp Ala Gln Asp Val Ala Ala Met Phe Gly Tyr His Ala Gly Ala
20 25 30
Ser Ala Ala Val Ser Ala Leu Thr Pro Phe Gly Gln Ala Leu Pro Thr
35 90 45
Val Ala Gly Gly Gly Ala Leu Val Ser A.la Ala Ala Ala Gln Val Thr
50 55 60
Thr Arg Val Phe Arg Asn Leu Gly Leu Ala Asn Val Arg Glu Gly Asn
65 70 75 80
Val Arg Asn Gly Asn Val Arg Asn Phe Asn Leu Gly Ser Ala Asn Ile
85 90 95
Gly Asn Gly Asn Ile Gly Ser Gly Asn Ile Gly Ser Ser Asn Ile Gly
100 105 110
Phe Gly Asn Val Gly Pro Gly Leu Thr Ala Ala Leu Asn Asn Ile Gly
115 120 125
Phe Gly Asn Thr Gly Ser Asn Asn Ile G.ly Phe Gly Asn Thr Gly Ser
130 135 140
Asn Asn Ile Gly Phe Gly Asn Thr Gly A;sp Gly Asn Arg Gly Ile Gly
CA 02268036 1999-10-12
182
145 150 155 160
Leu Thr Gly Ser Gly Leu Leu Gly Phe Gly Gly Leu Asn Ser Gly Thr
165 170 175
Gly Asn Ile Gly Leu Phe Asn Ser Gly Thr Gly Asn Val Gly Ile Gly
180 185 190
Asn Ser Gly Thr Gly Asn Trp Gly Ile Gly Asn Ser Gly Asn Ser Tyr
195 200 205
Asn Thr Gly Phe Gly Asn Ser Gly Asp Ala Asn Thr Gly Phe Phe Asn
210 215 220
Ser Gly Ile Ala Asn Thr Gly Val Gly Asn Ala Gly Asn Tyr Asn Thr
225 230 235 240
Gly Ser Tyr Asn Pro Gly Asn Ser Asn Thr Gly Gly Phe Asn Met Gly
245 250 255
Gln Tyr Asn Thr Gly Tyr Leu Asn Ser Gly Asn Tyr Asn Thr Gly Leu
260 265 270
Ala Asn Ser Gly Asn Val Asn Thr Gly Ala Phe Ile Thr Gly Asn Phe
275 280 285
Asn Asn Gly Phe Leu Trp Arg Gly Asp l3is Gln Gly Leu Ile Phe Gly
290 295 300
Ser Pro Gly Phe Phe Asn Ser Thr Ser Ala Pro Ser Ser Gly Phe Phe
305 310 315 320
Asn Ser Gly Ala Gly Ser Ala Ser Gly F?he Leu Asn Ser Gly Ala Asn
325 330 335
Asn Ser Gly Phe Phe Asn Ser Ser Ser Gly Ala Ile Gly Asn Ser Gly
340 345 350
Leu Ala Asn Ala Gly Val Leu Val Ser C~ly Val Ile Asn Ser Gly Asn
355 360 365
Thr Val Ser Gly Leu Phe Asn Met Ser Leu Val Ala Ile Thr Thr Pro
370 375 380
Ala Leu Ile Ser Gly Phe Phe Asn Thr Gly Ser Asn Met Ser Gly Phe
385 390 395 400
Phe Gly Gly Pro Pro Val Phe Asn Leu Gly Leu Ala Asn Arg Gly Val
405 410 415
Val Asn Ile Leu Gly Asn Ala Asn Ile Gly Asn Tyr Asn Ile Leu Gly
420 425 430
Ser Gly Asn Val Gly Asp Phe Asn Ile Leu Gly Ser Gly Asn Leu Gly
435 440 445
Ser Gln Asn Ile Leu Gly Ser Gly Asn Val Gly Ser Phe Asn Ile Gly
450 455 460
Ser Gly Asn Ile Gly Val Phe Asn Val Gly Ser Gly Ser Leu Gly Asn
465 470 475 480
Tyr Asn Ile Gly Ser Gly Asn Leu Gly Ile Tyr Asn Ile Gly Phe Gly
485 490 495
CA 02268036 1999-10-12
183
Asn Val Gly Asp Tyr Asn Val Gly Phe Gly Asn Ala Gly Asp Phe Asn
500 505 510
Gln Gly Phe Ala Asn Thr Gly Asn Asn Asn Ile Gly Phe Ala Asn Thr
515 520 525
Gly Asn Asn Asn Ile Gly Ile Gly Leu Ser Gly Asp Asn Gln Gln Gly
530 535 540
Phe Asn Ile Ala Ser Gly Trp Asn Ser Gly Thr Gly Asn Ser Gly Leu
545 550 555 560
Phe Asn Ser Gly Thr Asn Asn Val Gly Ile Phe Asn Ala Gly Thr Gly
565 570 575
Asn Val Gly Ile Ala Asn Ser Gly Thr Gly Asn Trp Gly Ile Gly Asn
580 585 590
Pro Gly Thr Asp Asn Thr Gly Ile Leu Asn Ala Gly Ser Tyr Asn Thr
595 600 605
Gly Ile Leu Asn Ala Gly Asp Phe Asn 'rhr Gly Phe Tyr Asn Thr Gly
610 615 620
Ser Tyr Asn Thr Gly Gly Phe Asn Val Gly Asn Thr Asn Thr Gly Asn
625 630 635 640
Phe Asn Val Gly Asp Thr Asn Thr Gly Ser Tyr Asn Pro Gly Asp Thr
695 (i50 655
Asn Thr Gly Phe Phe Asn Pro Gly Asn Val Asn Thr Gly Ala Phe Asp
660 665 670
Thr Gly Asp Phe Asn Asn Gly Phe Leu Val Ala Gly Asp Asn Gln Gly
675 680 685
Gln Ile Ala Ile Asp Leu Ser Val Thr Thr Pro Phe Ile Pro Ile Asn
690 695 700
Glu Gln Met Val Ile Asp'Val His Asn Val Met Thr Phe Gly Gly Asn
705 710 715 720
Met Ile Thr Val Thr Glu Ala Ser Thr Val Phe Pro Gln Thr Phe Tyr
725 730 735
Leu Ser Gly Leu Phe Phe Phe Gly Pro Val Asn Leu Ser Ala Ser Thr
740 745 750
Leu Thr Val Pro Thr Ile Thr Leu Thr Ile Gly Gly Pro Thr Val Thr
755 760 765
Val Pro Ile Ser Ile Val Gly Ala Leu Glu Ser Arg Thr Ile Thr Phe
770 775 780
Leu Lys Ile Asp Pro Ala Pro Gly Ile Gly Asn Ser Thr Thr Asn Pro
785 790 795 800
Ser Ser Gly Phe Phe Asn Ser Gly Thr Gly Gly Thr Ser Gly Phe Gln
805 810 815
Asn Val Gly Gly Gly Ser Ser Gly Val T:rp Asn Ser Gly Leu Ser Ser
820 825 830
CA 02268036 1999-10-12
184
Ala Ile Gly Asn Ser Gly Phe Gln Asn Leu Gly Ser Leu Gln Ser Gly
835 840 845
Trp Ala Asn Leu Gly Asn Ser Val Ser Gly Phe Phe Asn Thr Ser Thr
850 855 860
Val Asn Leu Ser Thr Pro Ala Asn Val Ser Gly Leu Asn Asn Ile Gly
865 870 875 880
Thr Asn Leu Ser Gly Val Phe Arg Gly Pro Thr Gly Thr Ile Phe Asn
885 890 895
Ala Gly Leu Ala Asn Leu Gly Gln Leu Asn Ile Gly Ser Ala Ser Cys
900 905 910
Arg Ile Arg His Glu Leu Asp Thr Val Ser Thr Ile Ile Ser Ala Phe
915 920 925
Cys Gly Ser Ala Ser Asp Glu Ser Asn Pro Gly Ser Val Ser Glu
930 935 940
(2) INFORMATION FOR SEQ ID N0:200:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 53 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:200:
GGATCCATAT GGGCCATCAT CATCATCATC ACGTGATCGA CATCATCGGG ACC 53
(2) INFORMATION FOR SEQ ID N0:201:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 42 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:201:
CCTGAATTCA GGCCTCGGTT GCGCCGGCCT CATCTTGAAC GA 42
(2) INFORMATION FOR SEQ ID N0:202:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 31 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:202:
GGATCCTGCA GGCTCGAAAC CACCGAGCGG T 31
(2) INFORMATION FOR SEQ ID N0:203:
(i) SEQUENCE CHARACTERISTICS:
CA 02268036 1999-10-12
185
(A) LENGTH: 31 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:203:
CTCTGAATTC AGCGCTGGAA ATCGTCGCGA T 31
(2) INFORMATION FOR SEQ ID N0:.204:
(ij SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:204:
GGATCCAGCG CTGAGATGAA GACCGATGCC GCT 33
(2) INFORMATION FOR SEQ ID N0:205:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 38 base pairs
(B) TYPE: nucleic acid
(Cj STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:205:
GGATATCTGC AGAATTCAGG TTTAAAGCCC ATTTGCGA 38
(2) INFORMATION FOR SEQ ID N0:206:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:206:
CCGCATGCGA GCCACGTGCC CACAACGGCC 30
(2) INFORMATION FOR SEQ ID N0:207:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 37 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:207:
CTTCATGGAA TTCTCAGGCC GGTAAGGTCC GCTGCGG 37
(2) INFORMATION FOR SEQ ID N0:208:
CA 02268036 1999-10-12
186
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:7676 basepairs
(B) TYPE:
nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
(xi) S EQUENCE CRIPTION: 8:
DES SEQ ID
N0:20
TGGCGAATGGGACGCGCCCTGTAGCGGCGCATTAAGCGCGGCGGGTGTGGTGGTTACGCG 60
CAGCGTGACCGCTACACTTGCCAGCGCCCTAGCGCCCGCTCCTTTCGCTTTCTTCCCTTC 120
CTTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCTC'TAAATCGGGGGCTCCCTTTAGG 180
GTTCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAA,?~ACTTGATTAGGGTGATGGTTC 240
ACGTAGTGGGCCATCGCCCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTT 300
CTTTAATAGTGGACTCTTGTTCCAAACTGGAACAACAC'PCAACCCTATCTCGGTCTATTC 360
TTTTGATTTATAAGGGATTTTGCCGATTTCGGCCTATTGGTTAAAAAATGAGCTGATTTA 420
ACAAAAATTTAACGCGAATTTTAACAAAATATTAACGT'CTACAATTTCAGGTGGCACTTT 480
TCGGGGAAATGTGCGCGGAACCCCTATTTGTTTATTTT'.CCTAAATACATTCAAATATGTA 540
TCCGCTCATGAATTAATTCTTAGAAAAACTCATCGAGCATCAAATGAAACTGCAATTTAT 600
TCATATCAGGATTATCAATACCATATTTTTGAAAAAGCC:GTTTCTGTAATGAAGGAGAAA 660
ACTCACCGAGGCAGTTCCATAGGATGGCAAGATCCTGG7.'ATCGGTCTGCGATTCCGACTC 720
GTCCAACATCAATACAACCTATTAATTTCCCCTCGTCAAAAATAAGGTTATCAAGTGAGA 780
AATCACCATGAGTGACGACTGAATCCGGTGAGAATGGCAAAAGTTTATGCATTTCTTTCC 890
AGACTTGTTCAACAGGCCAGCCATTACGCTCGTCATCAAP.ATCACTCGCATCAACCAAAC 900
CGTTATTCATTCGTGATTGCGCCTGAGCGAGACGAAATACGCGATCGCTGTTAAAAGGAC 960
AATTACAAACAGGAATCGAATGCAACCGGCGCAGGAACACTGCCAGCGCATCAACAATAT 1020
TTTCACCTGAATCAGGATATTCTTCTAATACCTGGAATGCTGTTTTCCCGGGGATCGCAG 1080
TGGTGAGTAACCATGCATCATCAGGAGTACGGATAAAAT'GCTTGATGGTCGGAAGAGGCA 1140
TAAATTCCGTCAGCCAGTTTAGTCTGACCATCTCATCTGTAACATCATTGGCAACGCTAC 1200
CTTTGCCATGTTTCAGAAACAACTCTGGCGCATCGGGCTTCCCATACAATCGATAGATTG 1260
TCGCACCTGATTGCCCGACATTATCGCGAGCCCATTTATACCCATATAAATCAGCATCCA 1320
TGTTGGAATTTAATCGCGGCCTAGAGCAAGACGTTTCCCGTTGAATATGGCTCATAACAC 1380
CCCTTGTATTACTGTTTATGTAAGCAGACAGTTTTATTGTTCATGACCAAAATCCCTTAA 1440
CGTGAGTTTTCGTTCCACTGAGCGTCAGACCCCGTAGAAAAGATCAAAGGATCTTCTTGA 1500
GATCCTTTTTTTCTGCGCGTAATCTGCTGCTTGCAAACAAAAAAACCACCGCTACCAGCG 1560
GTGGTTTGTTTGCCGGATCAAGAGCTACCAACTCTTTTTCCGAAGGTAACTGGCTTCAGC 1620
AGAGCGCAGATACCAAATACTGTCCTTCTAGTGTAGCCG'T CCACTTCAAG 1680
AGTTAGGCCA
CA 02268036 1999-10-12
187
AACTCTGTAGCACCGCCTACATACCTCGCTCTGCTAATCC GGCTGCTGCC1740
TGTTACCAGT
AGTGGCGATAAGTCGTGTCTTACCGGGTTGGACTCAAGACGATAGTTACCGGATAAGGCG1800
CAGCGGTCGGGCTGAACGGGGGGTTCGTGCACACAGCCCA AACGACCTAC1860
GCTTGGAGCG
ACCGAACTGAGATACCTACAGCGTGAGCTATGAGAAAGCGCCACGCTTCCCGAAGGGAGA1920
AAGGCGGACAGGTATCCGGTAAGCGGCAGGGTCGGAAC:AGGAGAGCGCACGAGGGAGCTT1980
CCAGGGGGAAACGCCTGGTATCTTTATAGTCCTGTCGGGTTTCGCCACCTCTGACTTGAG2040
CGTCGATTTTTGTGATGCTCGTCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCG2100
GCCTTTTTACGGTTCCTGGCCTTTTGCTGGCCTTTTGC'TCACATGTTCTTTCCTGCGTTA2160
TCCCCTGATTCTGTGGATAACCGTATTACCGCCTTTGAGTGAGCTGATACCGCTCGCCGC2220
AGCCGAACGACCGAGCGCAGCGAGTCAGTGAGCGAGGAAGCGGAAGAGCGCCTGATGCGG2280
TATTTTCTCCTTACGCATCTGTGCGGTATTTCACACCGCATATATGGTGCACTCTCAGTA2340
CAATCTGCTCTGATGCCGCATAGTTAAGCCAGTATACACTCCGCTATCGCTACGTGACTG2400
GGTCATGGCTGCGCCCCGACACCCGCCAACACCCGCTGACGCGCCCTGACGGGCTTGTCT2460
GCTCCCGGCATCCGCTTACAGACAAGCTGTGACCGTCTCCGGGAGCTGCATGTGTCAGAG2520
GTTTTCACCGTCATCACCGAAACGCGCGAGGCAGCTGCGGTAAAGCTCATCAGCGTGGTC2580
GTGAAGCGATTCACAGATGTCTGCCTGTTCATCCGCGTC:CAGCTCGTTGAGTTTCTCCAG2640
AAGCGTTAATGTCTGGCTTCTGATAAAGCGGGCCATGT7.'AAGGGCGGTTTTTTCCTGTTT2700
GGTCACTGATGCCTCCGTGTAAGGGGGATTTCTGTTCA7.'GGGGGTAATGATACCGATGAA2760
ACGAGAGAGGATGCTCACGATACGGGTTACTGATGATGAACATGCCCGGTTACTGGAACG2820
TTGTGAGGGTAAACAACTGGCGGTATGGATGCGGCGGGACCAGAGAAAAATCACTCAGGG2880
TCAATGCCAGCGCTTCGTTAATACAGATGTAGGTGTTCC:ACAGGGTAGCCAGCAGCATCC2940
TGCGATGCAGATCCGGAACATAATGGTGCAGGGCGCTGP,CTTCCGCGTTTCCAGACTTTA3000
CGAAACACGGAAACCGAAGACCATTCATGTTGTTGCTCF,GGTCGCAGACGTTTTGCAGCA3060
GCAGTCGCTTCACGTTCGCTCGCGTATCGGTGATTCATTCTGCTAACCAGTAAGGCAACC3120
CCGCCAGCCTAGCCGGGTCCTCAACGACAGGAGCACGATCATGCGCACCCGTGGGGCCGC3180
CATGCCGGCGATAATGGCCTGCTTCTCGCCGAAACGTTTGGTGGCGGGACCAGTGACGAA3240
GGCTTGAGCGAGGGCGTGCAAGATTCCGAATACCGCAAGCGACAGGCCGATCATCGTCGC3300
GCTCCAGCGAAAGCGGTCCTCGCCGAAAATGACCCAGAGCGCTGCCGGCACCTGTCCTAC3360
GAGTTGCATGATAAAGAAGACAGTCATAAGTGCGGCGACGATAGTCATGCCCCGCGCCCA3420
CCGGAAGGAGCTGACTGGGTTGAAGGCTCTCAAGGGCATCGGTCGAGATCCCGGTGCCTA3480
ATGAGTGAGCTAACTTACAT GCGCTCACTGCCCGCTTTCC 3540
TAATTGCGTT AGTCGGGAAA
CCTGTCGTGCCAGCTGCATT GGGAGAGGCG 3600
AATGAATCGG GTTTGCGTAT
CCAACGCGCG
CA 02268036 1999-10-12
188
TGGGCGCCAGGGTGGTTTTTCTTTTCACCAGTGAGACGGGCAACAGCTGATTGCCCTTCA3660
CCGCCTGGCCCTGAGAGAGTTGCAGCAAGCGGTCCACGCTGGTTTGCCCCAGCAGGCGAA3720
AATCCTGTTTGATGGTGGTTAACGGCGGGATATAACATGAGCTGTCTTCGGTATCGTCGT3780
ATCCCACTACCGAGATATCCGCACCAACGCGCAGCCCGGACTCGGTAATGGCGCGCATTG3840
CGCCCAGCGCCATCTGATCGTTGGCAACCAGCATCGCAGTGGGAACGATGCCCTCATTCA3900
GCATTTGCATGGTTTGTTGAAAACCGGACATGGCACTCCAGTCGCCTTCCCGTTCCGCTA3960
TCGGCTGAATTTGATTGCGAGTGAGATATTTATGCCAGCCAGCCAGACGCAGACGCGCCG4020
AGACAGAACTTAATGGGCCCGCTAACAGCGCGATTTGCTGGTGACCCAATGCGACCAGAT4080
GCTCCACGCCCAGTCGCGTACCGTCTTCATGGGAGAAAATAATACTGTTGATGGGTGTCT4140
GGTCAGAGACATCAAGAAATAACGCCGGAACATTAGTGCAGGCAGCTTCCACAGCAATGG4200
CATCCTGGTCATCCAGCGGATAGTTAATGATCAGCCCACTGACGCGTTGCGCGAGAAGAT4260
TGTGCACCGCCGCTTTACAGGCTTCGACGCCGCTTCGT'PCTACCATCGACACCACCACGC4320
TGGCACCCAGTTGATCGGCGCGAGATTTAATCGCCGCGi~CAATTTGCGACGGCGCGTGCA4380
GGGCCAGACTGGAGGTGGCAACGCCAATCAGCAACGAC'CGTTTGCCCGCCAGTTGTTGTG4440
CCACGCGGTTGGGAATGTAATTCAGCTCCGCCATCGCCGCTTCCACTTTTTCCCGCGTTT4500
TCGCAGAAACGTGGCTGGCCTGGTTCACCACGCGGGAAACGGTCTGATAAGAGACACCGG4560
CATACTCTGCGACATCGTATAACGTTACTGGTTTCACA'.CTCACCACCCTGAATTGACTCT4620
CTTCCGGGCGCTATCATGCCATACCGCGAAAGGTTTTGC:GCCATTCGATGGTGTCCGGGA4680
TCTCGACGCTCTCCCTTATGCGACTCCTGCATTAGGAAC7CAGCCCAGTAGTAGGTTGAGG4740
CCGTTGAGCACCGCCGCCGCAAGGAATGGTGCATGCAAGGAGATGGCGCCCAACAGTCCC4800
CCGGCCACGGGGCCTGCCACCATACCCACGCCGAAACAAGCGCTCATGAGCCCGAAGTGG4860
CGAGCCCGATCTTCCCCATCGGTGATGTCGGCGATATAGGCGCCAGCAACCGCACCTGTG4920
GCGCCGGTGATGCCGGCCACGATGCGTCCGGCGTAGAGGATCGAGATCTCGATCCCGCGA4980
AATTAATACGACTCACTATAGGGGAATTGTGAGCGGATP~ACAATTCCCCTCTAGAAATAA5040
TTTTGTTTAACTTTAAGAAGGAGATATACATATGGGCCATCATCATCATCATCACGTGAT5100
CGACATCATCGGGACCAGCCCCACATCCTGGGAACAGGCGGCGGCGGAGGCGGTCCAGCG5160
GGCGCGGGATAGCGTCGATGACATCCGCGTCGCTCGGGT'CATTGAGCAGGACATGGCCGT5220
GGACAGCGCCGGCAAGATCACCTACCGCATCAAGCTCGAAGTGTCGTTCAAGATGAGGCC5280
GGCGCAACCGAGGGGCTCGAAACCACCGAGCGGTTCGCCTGAAACGGGCGCCGGCGCCGG5340
TACTGTCGCGACTACCCCCGCGTCGTCGCCGGTGACGTTGGCGGAGACCGGTAGCACGCT5400
GCTCTACCCGCTGTTCAACCTGTGGGGTCCGGCCTTTCACGAGAGGTATCCGAACGTCAC5460
GATCACCGCTCAGGGCACCGGTTCTGGTGCCGGGATCGCG CCGGGACGGT5520
CAGGCCGCCG
CA 02268036 1999-10-12
189
CAACATTGGGGCCTCCGACGCCTATCTGTCGGAAGGTGATATGGCCGCGCACAAGGGGCT5580
GATGAACATCGCGCTAGCCATCTCCGCTCAGCAGGTCAACTACAACCTGCCCGGAGTGAG5640
CGAGCACCTCAAGCTGAACGGAAAAGTCCTGGCGGCCATGTACCAGGGCACCATCAAAAC5700
CTGGGACGACCCGCAGATCGCTGCGCTCAACCCCGGCGTGAACCTGCCCGGCACCGCGGT5760
AGTTCCGCTGCACCGCTCCGACGGGTCCGGTGACACCTTCTTGTTCACCCAGTACCTGTC5820
CAAGCAAGATCCCGAGGGCTGGGGCAAGTCGCCCGGCTTCGGCACCACCGTCGACTTCCC5880
GGCGGTGCCGGGTGCGCTGGGTGAGAACGGCAACGGCGGCATGGTGACCGGTTGCGCCGA5940
GACACCGGGCTGCGTGGCCTATATCGGCATCAGCTTCCTCGACCAGGCCAGTCAACGGGG6000
ACTCGGCGAGGCCCAACTAGGCAATAGCTCTGGCAATT'TCTTGTTGCCCGACGCGCAAAG6060
CATTCAGGCCGCGGCGGCTGGCTTCGCATCGAAAACCCCGGCGAACCAGGCGATTTCGAT6120
GATCGACGGGCCCGCCCCGGACGGCTACCCGATCATCA;~CTACGAGTACGCCATCGTCAA6180
CAACCGGCAAAAGGACGCCGCCACCGCGCAGACCTTGCAGGCATTTCTGCACTGGGCGAT6290
CACCGACGGCAACAAGGCCTCGTTCCTCGACCAGGTTCi~TTTCCAGCCGCTGCCGCCCGC6300
GGTGGTGAAGTTGTCTGACGCGTTGATCGCGACGATTTCCAGCGCTGAGATGAAGACCGA6360
TGCCGCTACCCTCGCGCAGGAGGCAGGTAATTTCGAGC(~GATCTCCGGCGACCTGAAAAC6420
CCAGATCGACCAGGTGGAGTCGACGGCAGGTTCGTTGCAGGGCCAGTGGCGCGGCGCGGC6480
GGGGACGGCCGCCCAGGCCGCGGTGGTGCGCTTCCAAGAAGCAGCCAATAAGCAGAAGCA6540
GGAACTCGACGAGATCTCGACGAATATTCGTCAGGCCGGCGTCCAATACTCGAGGGCCGA6600
CGAGGAGCAGCAGCAGGCGCTGTCCTCGCAAATGGGCTTTGTGCCCACAACGGCCGCCTC6660
GCCGCCGTCGACCGCTGCAGCGCCACCCGCACCGGCGAC:ACCTGTTGCCCCCCCACCACC6720
GGCCGCCGCCAACACGCCGAATGCCCAGCCGGGCGATCC;CAACGCAGCACCTCCGCCGGC6780
CGACCCGAACGCACCGCCGCCACCTGTCATTGCCCCAAACGCACCCCAACCTGTCCGGAT6840
CGACAACCCGGTTGGAGGATTCAGCTTCGCGCTGCCTGC;TGGCTGGGTGGAGTCTGACGC6900
CGCCCACTTCGACTACGGTTCAGCACTCCTCAGCAAAAC;CACCGGGGACCCGCCATTTCC6960
CGGACAGCCGCCGCCGGTGGCCAATGACACCCGTATCGTGCTCGGCCGGCTAGACCAAAA7020
GCTTTACGCCAGCGCCGAAGCCACCGACTCCAAGGCCGC'.GGCCCGGTTGGGCTCGGACAT7080
GGGTGAGTTCTATATGCCCTACCCGGGCACCCGGATCAP,CCAGGAAACCGTCTCGCTTGA7140
CGCCAACGGGGTGTCTGGAAGCGCGTCGTATTACGAAGTCAAGTTCAGCGATCCGAGTAA7200
GCCGAACGGCCAGATCTGGACGGGCGTAATCGGCTCGCCCGCGGCGAACGCACCGGACGC7260
CGGGCCCCCTCAGCGCTGGTTTGTGGTATGGCTCGGGACCGCCAACAACCCGGTGGACAA7320
GGGCGCGGCCAAGGCGCTGGCCGAATCGATCCGGCCTTTGGTCGCCCCGCCGCCGGCGCC7380
GGCACCGGCTCCTGCAGAGCCCGCTCCGGCGCCGGCGCCGGCCGGGGAAGTCGCTCCTAC7440
CA 02268036 1999-10-12
190
CCCGACGACACCGACACCGCAGCGGACCTTACCGGCCTGAGAATTCTGCAGATATCCATC7500
ACACTGGCGGCCGCTCGAGCACCACCACCACCACCACTGAGATCCGGCTGCTAACAAAGC7560
CCGAAAGGAAGCTGAGTTGGCTGCTGCCACCGCTGAGC.AATAACTAGCATAACCCCTTGG7620
GGCCTCTAAACGGGTCTTGAGGGGTTTTTTGCTGAAAGGAGGAACTATATCCGGAT 7676
(2) INFORMATION FOR SEQ ID N0:209:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 802 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:209:
Met Gly His His His His His His Val :Lle Asp Ile Ile Gly Thr Ser
1 5 :LO 15
Pro Thr Ser Trp Glu Gln Ala Ala Ala (~lu Ala Val Gln Arg Ala Arg
20 25 30
Asp Ser Val Asp Asp Ile Arg Val Ala Arg Val Ile Glu Gln Asp Met
35 40 45
Ala Val Asp Ser Ala Gly Lys Ile Thr Tyr Arg Ile Lys Leu Glu Val
50 55 60
Ser Phe Lys Met Arg Pro Ala Gln Pro Arg Gly Ser Lys Pro Pro Ser
65 70 75 80
Gly Ser Pro Glu Thr Gly Ala Gly Ala Gly Thr Val Ala Thr Thr Pro
85 90 95
Ala Ser Ser Pro Val Thr Leu Ala Glu Thr Gly Ser Thr Leu Leu Tyr
100 105 110
Pro Leu Phe Asn Leu Trp Gly Pro Ala F~he His Glu Arg Tyr Pro Asn
115 120 125
Val Thr Ile Thr Ala Gln Gly Thr Gly S'er Gly Ala Gly Ile Ala Gln
130 135 140
Ala Ala Ala Gly Thr Val Asn Ile Gly A.la Ser Asp Ala Tyr Leu Ser
145 150 155 160
Glu Gly Asp Met Ala Ala His Lys Gly Leu Met Asn Ile Ala Leu Ala
165 170 175
Ile Ser Ala Gln Gln Val Asn Tyr Asn Leu Pro Gly Val Ser Glu His
180 185 190
Leu Lys Leu Asn Gly Lys Val Leu Ala Ala Met Tyr Gln Gly Thr Ile
195 200 205
Lys Thr Trp Asp Asp Pro Gln Ile Ala Ala Leu Asn Pro Gly Val Asn
210 215 220
Leu Pro Gly Thr Ala Val Val Pro Leu His Arg Ser Asp Gly Ser Gly
225 230 235 240
CA 02268036 1999-10-12
191
Asp 'Thr Phe Leu Phe Thr Gln Tyr Leu Ser Lys Gln Asp Pro Glu Gly
245 250 255
Trp Gly Lys Ser Pro Gly Phe Gly Thr Thr Val Asp Phe Pro Ala Val
260 265 270
Pro Gly Ala Leu Gly Glu Asn Gly Asn Gly Gly Met Val Thr Gly Cys
275 280 285
Ala Glu Thr Pro Gly Cys Val Ala Tyr Ile Gly Ile Ser Phe Leu Asp
290 295 300
Gln Ala Ser Gln Arg Gly Leu Gly Glu ;~11a Gln Leu Gly Asn Ser Ser
305 310 315 320
Gly Asn Phe Leu Leu Pro Asp Ala Gln Ser Ile Gln Ala Ala Ala Ala
325 :330 335
Gly Phe Ala Ser Lys Thr Pro Ala Asn Gln Ala Ile Ser Met Ile Asp
340 345 350
Gly Pro Ala Pro Asp Gly Tyr Pro Ile :Cle Asn Tyr Glu Tyr Ala Ile
355 360 365
Val Asn Asn Arg Gln Lys Asp Ala Ala Thr Ala Gln Thr Leu Gln Ala
370 375 380
Phe Leu His Trp Ala Ile Thr Asp Gly Asn Lys Ala Ser Phe Leu Asp
385 390 395 400
Gln Val His Phe Gln Pro Leu Pro Pro Ala Val Val Lys Leu Ser Asp
405 X110 415
Ala Leu Ile Ala Thr Ile Ser Ser Ala C~lu Met Lys Thr Asp Ala Ala
420 425 930
Thr Leu Ala Gln Glu Ala Gly Asn Phe Glu Arg Ile Ser Gly Asp Leu
435 440 445
Lys Thr Gln Ile Asp Gln Val Glu Ser Thr Ala Gly Ser Leu Gln Gly
450 455 460
Gln Trp Arg Gly Ala Ala Gly Thr Ala Ala Gln Ala Ala Val Val Arg
465 470 475 480
Phe Gln Glu Ala Ala Asn Lys Gln Lys Gln Glu Leu Asp Glu Ile Ser
485 490 495
Thr Asn Ile Arg Gln Ala Gly Val Gln T'yr Ser Arg Ala Asp Glu Glu
500 505 510
Gln Gln Gln Ala Leu Ser Ser Gln Met Gly Phe Val Pro Thr Thr Ala
515 520 525
Ala Ser Pro Pro Ser Thr Ala Ala Ala Pro Pro Ala Pro Ala Thr Pro
530 535 540
Val Ala Pro Pro Pro Pro Ala Ala Ala Asn Thr Pro Asn Ala Gln Pro
545 550 555 560
Gly Asp Pro Asn Ala Ala Pro Pro Pro Ala Asp Pro Asn Ala Pro Pro
565 570 575
Pro Pro Val Ile Ala Pro Asn Ala Pro Gln Pro Val Arg Ile Asp Asn
CA 02268036 1999-10-12
192
580 585 590
Pro Val Gly Gly Phe Ser Phe Ala Leu Pro Ala Gly Trp Val Glu Ser
595 600 605
Asp Ala Ala His Phe Asp Tyr Gly Ser Ala Leu Leu Ser Lys Thr Thr
610 615 620
Gly Asp Pro Pro Phe Pro Gly Gln Pro Pro Pro Val Ala Asn Asp Thr
625 630 635 640
Arg Ile Val Leu Gly Arg Leu Asp Gln Lys Leu Tyr Ala Ser Ala Glu
645 650 655
Ala Thr Asp Ser Lys Ala Ala Ala Arg :Leu Gly Ser Asp Met Gly Glu
660 665 670
Phe Tyr Met Pro Tyr Pro Gly Thr Arg :Ile Asn Gln Glu Thr Val Ser
675 680 685
Leu Asp Ala Asn Gly Val Ser Gly Ser i~la Ser Tyr Tyr Glu Val Lys
690 695 700
Phe Ser Asp Pro Ser Lys Pro Asn Gly Gln Ile Trp Thr Gly Val Ile
705 710 715 720
Gly Ser Pro Ala Ala Asn Ala Pro Asp Ala Gly Pro Pro Gln Arg Trp
725 '130 735
Phe Val Val Trp Leu Gly Thr Ala Asn Asn Pro Val Asp Lys Gly Ala
740 745 750
Ala Lys Ala Leu Ala Glu Ser Ile Arg F?ro Leu Val Ala Pro Pro Pro
755 760 765
Ala Pro Ala Pro Ala Pro Ala Glu Pro Ala Pro Ala Pro Ala Pro Ala
770 775 780
Gly Glu Val Ala Pro Thr Pro Thr Thr Pro Thr Pro Gln Arg Thr Leu
785 790 795 800
Pro Ala