Note: Descriptions are shown in the official language in which they were submitted.
CA 02279490 1999-08-03
WO 98/33925 PCT/US98/01860
~'rlufIZNAo'" Amidotra_nsferase - A Novel Essential Translational Component
CROSS REFERENCE TO RELATED APPLICATIONS
This application, is related to U.S. Provisional Application Serial No.
60/037,275;
filed February 3, 1997, which is herein incorporated by reference in its
entirety.
FIELD OF THE INVENTION
The present invention is in the field of inhibitors of protein translation,
particularly translation of proteins within microorganisms and organelles.
This
invention relates to newly identified polynucleotides and polypeptides, and
their
production and uses, as well as their variants, agonists and antagonists, and
their uses.
In particular, in these and in other regards, the invention relates to novel
polynucleotides and polypeptides of the Glu-tRNA~'° Amidotransferase
family,
hereinafter referred to as "Glu-tRNA°'° AdT" or "AdT". The
present invention further
provides methods and compositions for use in identifying and using protein
translation inhibitors as antibacterial, antifungal or herbicidal agents.
BACKGROUND O:F THE INVENTION
Prior to their :incorporation into protein, amino acids are chemically linked
to
small RNA molecules called transfer RNA (tRNA). For each of the 20 different
amino acids, a specific enzyme catalyzes its linkage to the 3' end of its
specific tRNA
molecule. While the general mechanism of protein biosynthesis (the translation
CA 02279490 1999-08-03
WO 98133925 PCT/US98/01860
-2-
process) is conserved throughout the living kingdom there exist two different
pathways for the formation of GIntRNA~'". While the two pathways for
GIntRNAG'"
formation are evolutionarily conserved, the reason for existence of
the~different
pathways is as yet not known. In gram-negative eubacteria and in the cytoplasm
of
eukaryotic cells the enzyme glutaminyl-tRNA synthetase (GInRS) acylates
glutamine
directly to the cognate tRNA to provide GIntRNA~'". Interestingly, GInRS is
not
detectable in several biological systems. In certain organisms and organelles
including the archae, gram-positive eubacteria, mitochondria and chloroplasts
a
different pathway of GIntRNA~'" formation, a transamidation pathway is
operative
(Curnow et al. ( 1996) Nature 3 82: 5 89-590; Curnow et al. ( 1997) Proc.
Natl. Acad.
Sci. USA 94(22):11819-11826; Schon et al. (1988) Biochimie 70(3):391-394;
Wilcox
& Nirenberg ( 1968) Proc. Natl. Acad. Sci. USA 61 ( 1 ):229-236; Schon et al.
( 1988)
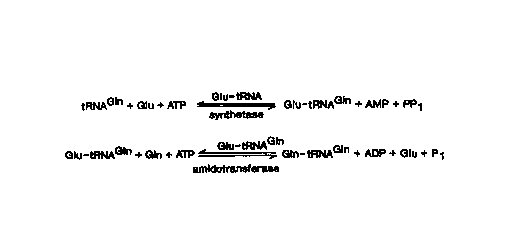
Nature 331:187-190. This pathway (depicted in Figure 1 ) is initiated by
misacylation
of tRNAG'" by glutamyl-tRNA synthetase (GIuRS) farming GIutRNA~'". The
incorrectly charged tRNA is then converted to GIntRNAG'" by GIutRNA~'"
amidotransferase (AdT). AdT catalyzes the amidation of glutamate to glutamine
only
when the glutamate is covalently attached to tRNAGIn. It has been shown that
the
partially purified GIutRNA~'" amidotransferase activity from Bacillus
megaterium in
the presence of ATP, Mg++, and an amide-nitrogen donor (glutamine) will carry
out
the amidation of GIutRNA~'" to GIntRNA°'" (Wilcox & Nirenberg, 1968).
CA 02279490 1999-08-03
WO 98/33925 PCT/US98I01860
-3-
Subsequent work demonstrated, in vitro, that the amidation proceeds through
the
activated intermediate (phospho-GIutRNA°'") (Wilcox ( 1969) Cold Spring
Harb.
Symp. Quant. Biol. 34:521-528; Wilcox (1969) Eur J. Biochem 11(3):405-412).
Since
the initial aminoacylation product, GIutRNA~'", would be toxic to the cell due
to the
fact that it would result in faulty protein translation, it must be converted
to the
correctly charged tRNA.
It appears that this pathway is the primary source of GIntRNA~'" within these
cells and may act as a regulatory mechanism for glutamine metabolism.
Evolutionarily, it has been suggested that glutamine was the last amino acid
formed.
Therefore it may be postulated that cells which employ the transamidation
pathway
utilized the gene encoding GIuRS to generate the AdT. Likewise, in the cells
in
which the direct glutaminylation pathway operates, the enzyme GInRS may have
evolved from a GIuR~S gene duplication (Rogers & Soll (1995) J. Mol. Evol. 40
(5)
p476-81 ). This is reasonable since both enzymes are required to specifically
recognize and bind tItNAGIn and free glutamine. However, database searches
and, in
particular, a detailed analysis of the Mycoplasma genome (Fraser et al. (1995)
Science
270(5235):397-403), the only gram-positive organism sequenced and published to
date, have shown no significant homologies to GIuRS and GInRS in the currently
available sequence information. Thus, the amidotransferase may not have
significant
homology to the ami.noacyl-tRNA synthetases. Despite the unquestioned
CA 02279490 1999-08-03
wo ~39zs rcT,US~srois~w
-4-
evolutionary and biochemical significance in understanding this system, there
have
been very few investigations of this enzyme to date (Wilcox & Nirenberg, 1968;
Wilcox, 1969; Strauch et al. (1988) .I. Bacteriol. 170:916-920; and Jahn
(1990) J. Biol.
Che»t. 265(14):8059-64).
SUMMARY OF THE INVENTION
The present invention is based, in part, on the isolation and characterization
of
a heterotrimeric protein designated AdT that is involved in generating
GIntRNA°'"
from GIutRNA°'". This invention further provides polypeptides that have
been
identified as novel AdT polypeptides by homology between the amino acid
sequence
of GIutRNA~'" AdT and a known amino acid sequence.
This invention further provides polynucleotides that encode AdT
polypeptides. In particular, this invention provides the polynucleotide
sequence
encoding GIutRNAG'" AdT comprising the sequence set out in Figure 3 (SEQ ID
NO:1 ), or a variant thereof, such as naturally occurring allelic variants of
AdT and
polypeptides encoded thereby. Thus, this invention provides polynucleotides
that
hybridize to AdT polynucleotide sequences, particularly under stringent
conditions.
This invention provides GIutRNA~'" AdT protein from B. subtilis comprising
the amino acid sequences encoded by the nucleotide sequence of Figure 3 (SEQ
ID
NOS:1, 3, 5 and 7), as well as biologically, diagnostically, prophylactically,
clinically
CA 02279490 1999-08-03
WO 9&33925 PCT/US98/i11860
-5-
or therapeutically usefc~l variants thereof, and compositions comprising the
same.
Particularly preferred variants include AdT polypeptides encoded by naturally
occurring alleles of the AdT gene. Methods for producing the aforementioned
AdT
polypeptides are also provided by this invention.
The invention also provides isolated nucleic acid molecules encoding AdT,
particularly B. subtilis AdT, including mRNAs, cDNAs, and genomic DNAs,
including biologically, diagnostically, prophylactically, clinically or
therapeutically
useful variants thereof; and compositions comprising the same.
In accordance 'with yet another aspect of the invention, there are provided
inhibitors to such AdT polypeptides, useful as antibacterial agents,
antifungal agents
and herbicides. Thus, the present invention provides compositions and methods
for
use in identifying agonists and antagonists of the AdT protein.
This invention provides compositions and methods for (i) assessing AdT
expression, (ii) treating disease, for example, diseases associated with
excessive or
deficient amounts of available AdT, (iii) assaying genetic variation, and (iv)
and
administering an AdT' polypeptide or polynucleotide to a cell or to a
multicellular
organism to raise an immunological response. In certain preferred embodiments
of
this aspect of the invesntion there are provided antibodies against AdT
polypeptides.
This invention also provides compositions and methods for protecting plants,
especially crop plants. For example, this invention provides antagonists of
AdT
CA 02279490 1999-08-03
WO 98133925 PCT/ITS98H11860
-6-
which are useful as herbicides, as well as the herbicidal compositions which
include
such inhibitors of AdT. This invention also provides non-inhibited mutants of
AdT
and functional derivatives thereof which are resistant to inhibition from
certain
herbicides, especially herbicides containing inhibitors of AdT. The
polynucleotides
coding for the non-inhibited AdT can be placed in plants by various
transformation
methods so as to render the plants tolerant or resistant to certain herbicides
containing
inhibitors of AdT. Therefore, methods of treating weeds utilizing the
application of
AdT inhibitors to transgenic plants containing the non-inhibited mutants of
AdT are
also encompassed by this invention.
Various changes and modifications within the spirit and scope of the disclosed
invention will become readily apparent to those skilled in the art from
reading the
following description and from reading the other parts of the present
disclosure.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1 shows the transamidation pathway for the formation of GlntI~NAc~"
Figure 2 shows the gene arrangement of the AdT gene.
Figure 3 shows the nucleic acid sequence of the AdT protein from B. subtilis.
DESCRIPTION OF THE INVENTION
I. General Description
CA 02279490 1999-08-03
wo 9zs rcrms9srois6o
The present invention is based, in part, on the identification and
characterization of a heterotrimeric protein that is responsible for
generating GIntRNA
within a cell, herein after the AdT protein. The present invention
specifically
provides the amino aciid sequences of each of the three subunits of an AdT
protein
isolated from B. subtilis, as well as nucleotide sequences that encode the AdT
protein.
The AdT protein and nucleic acid molecules can serve as targets in methods for
identifying agents for use in inhibiting protein synthesis, particularly
antimicrobial,
antifungal and herbicide agents.
II. Specific Embodiments
A. AdT Protein
Prior to the present invention the art had taught that there was an enzyme
involved in converting GIutRNA°'" to GIntRNA~'" . However the isolation
and
characterization of the protein responsible for generating GIntRNA remained
unknown. The present invention provides, in part, the amino acid sequences of
the
three subunits of the .8. subtilis AdT protein. Quite unexpectedly, this AdT
protein
was found to be a het:erotrimeric protein.
In one emboduiment, the present invention provides the ability to produce a
previously unlaiown protein using the cloned nucleic acid molecules herein
described
or by synthesizing a protein having the amino acid sequence herein disclosed.
CA 02279490 1999-08-03
wo ~3~s rcr~rs9srois6o
_8_
As used herein, the AdT protein refers to a protein that has the amino acid
sequence B. subtilis AdT encoded by the polynucleotide of Fig. 1, allelic
variants
thereof and conservative substitutions thereof that have AdT activity. The AdT
protein is comprised of 3 subunits: the A (SEQ )D N0:4), B (SEQ ID N0:6) and C
(SEQ )D N0:8) subunits, referred to herein collectively as aAdT, bAdT and cAdT
subunits, respectively. For the s2lce of convenience, the collective subunits
will be
referred to as the AdT protein or the AdT protein of the present invention. A
skilled
artisan can readily recognize within the context whether a single subunit or
the
collective protein is being referred to.
The polypeptides of the invention include the polypeptides encoded by SEQ
1D NO:1 (Figure 3) as well as polypeptides and fragments, particularly those
which
have the biological activity of AdT and also those which have at least 70%
sequence
identity to the polypeptides encoded by SEQ ID NO:1 or the relevant portion,
preferably at least 80% identity to the polypeptides encoded by SEQ ID NO:1,
and
more preferably at least 90% similarity (more preferably at least 90%
identity) to the
polypeptides encoded by SEQ m NO:1 and still more preferably at least 95%
similarity (still more preferably at least 95% identity) to the polypeptides
encoded by
SEQ B7 NO:1 and also include portions of such polypeptides. '
The AdT proteins of the present invention include the specificall~r identified
and characterized variant herein described as well as allelic variants,
conservative
CA 02279490 1999-08-03
WO 98/33925 PCTIUS'9~01860
-9-
substitution variants and homologues that can be isolated/generated and
characterized
without undue experimentation following the methods outlined below. For the
sake
of convenience, all AdT proteins will be collectively referred to as the AdT
proteins
or the AdT proteins of the present invention.
The term "AdT proteins" includes all naturally occurring allelic variants of
the
B. subtilis AdT protein that possess normal AdT activity. In general, allelic
variants
of the AdT protein will have a slightly different amino acid sequence than
that
specifically encoded by SEQ ID NO:1 but will be able to convert GIutRNA to
GIntRNA. Allelic variants, though possessing a slightly different amino acid
sequence than those reciited above, will posses the ability to generate
GIntRNA.
Typically, allelic variants of the AdT protein will contain conservative amino
acid
substitutions from the A.dT sequences herein described or will contain a
substitution
of an amino acid from a. corresponding position in an AdT homologue (an AdT
protein isolated from an organism other than B. subtilis).
The AdT proteins of the present invention are preferably in isolated form. As
used herein, a protein is said to be isolated when physical, mechanical or
chemical
methods are employed to remove the AdT protein from cellular constituents that
are
normally associated with the protein. A skilled artisan can readily employ
standard
purification methods to. obtain an isolated AdT protein. One purification
scheme is
CA 02279490 1999-08-03
WO 98r33925 PCT/US98/01860
-10-
outlined in Example 1. The nature and degree of isolation will depend on the
intended use.
The cloning of an AdT encoding nucleic acid molecule makes it possible to
generate defined fragments of the AdT proteins of the present invention. As
discussed
S below, fragments of the AdT proteins of the present invention are
particularly useful
in generating subunit specific antibodies, in identifying agents that bind to
a AdT
protein and in isolating homologues of the B. subtilis AdT protein.
Fragments of the AdT proteins can be generated using standard peptide
synthesis technology and the amino acid sequences disclosed herein.
Alternatively,
recombinant methods can be used to generate nucleic acid molecules that encode
a
fragment of the AdT protein.
Fragments of the AdT protein subunits that contain particularly interesting
structures can be identified using art-known methods such as immunogenicity,
Chou-Fasman, Gamier-Robson, Kyte-Doolittle, Eisenberg, Karplus-Schultz or
Jameson-Wolf analysis. Fragments containing such residues are particularly
useful in
generating subunit specific anti-AdT antibodies.
As described below, members of the AdT family of proteins can be used for,
but are not limited to: 1 ) a target to identify agents that block or
stimulate AdT
activity, 2) a target or bait to identify and isolate binding partners that
bind an AdT
CA 02279490 1999-08-03
WO 98/33925 PCTIUS98f01860
-11-
protein, 3)identifying agents that block or stimulate the activity of an AdT
protein and
4) an assay target to identify AdT mediated activity or disease.
B. Anti-AdT Antibodies
The present invention further provides antibodies that selectively bind one or
more of the AdT proteins of the present invention, or to a specific subunit of
an AdT
protein of the present invention. The most preferred antibodies will bind to
either an
entire heterotrimeric AdT protein but not to an isolated subunit or will bind
to an
isolated subunit but not to the assembled trimeric protein. Anti- AdT
antibodies that
are particularly contemplated include monoclonal and poiyclonal antibodies as
well as
fragments containing. the antigen binding domain and/or one or more complement
determining regions of these antibodies.
Antibodies ace generally prepared by immunizing a suitable mammalian host
using an AdT protein, or fragment, in isolated or immunoconjugated form
(Harlow,
Antibodies, Cold Spn.~ing Harbor Press, NY ( 1989)). Regions of the AdT
protein that
show immunogenic structure can readily be identified using art-known methods.
Other important regions and domains can readily be identified using protein
analytical
and comparative methods known in the art.
Fragments containing these residues are particularly suited in generating
specific classes of anti-AdT antibodies.
CA 02279490 1999-08-03
wo ~3ns rcT~s9srois6o
-12-
Methods for preparing a protein for use as an immunogen and for preparing
immunogenic conjugates of a protein with a carrier such as BSA, KLH, or other
carrier proteins are well known in the art. In some circumstances, direct
conjugation
using, for example, carbodiimide reagents may be used; in other instances
linking
reagents such as those supplied by Pierce Chemical Co., Rockford, IL, may be
effective.
Administration of an AdT immunogen is conducted generally by injection
over a suitable time period and with use of a suitable adjuvant, as is
generally
understood in the art. During the immunization schedule, titers of antibodies
can be
taken to determine adequacy of antibody formation.
While the polyclonal antisera produced in this way may be satisfactory for
some applications, for pharmaceutical compositions, monoclonal antibody
preparations are preferred. Immortalized cell lines which secrete a desired
monoclonal antibody may be prepared using the standard method of Kohler and
1 S Milstein or modifications which effect immortalization of lymphocytes or
spleen
cells, as is generally known. The immortalized cell lines secreting the
desired
antibodies are screened by immunoassay in which the antigen is the AdT protein
or
peptide fragment. When the appropriate immortalized cell culture secreting the
desired antibody is identified, the cells can be cultured either in vitro or
by production
in ascites fluid.
CA 02279490 1999-08-03
WO 98/33925 PCT/US98/01860
-13-
The desired monoclonal antibodies are then recovered from the culture
supernatant or from the ascites supernatant. Fragments of the monoclonals or
the
polyclonal antisera which contain the immunologically significant portion can
be used
as antagonists, as well as the intact antibodies. Use of immunologically
reactive
fragments, such as the Fab, Fab', of F(ab')2 fragments is often preferable,
especially in
a therapeutic context, as these fragments are generally less immunogenic than
the
whole immunoglobulin.
The antibodies; or fragments may also be produced, using current technology,
by recombinant mean;. Regions that bind specifically to the desired regions of
the
transporter can also bc; produced in the context of chimeric or CDR grafted
antibodies
of multiple species origin.
As described t>elow, anti-AdT antibodies are useful as modulators of AdT
activity, are useful in immunoassays for detecting AdT expression/activity and
for
purifying homologues of the B. subtilis AdT protein.
C. AdT Encodung Nucleic Acid Molecules
As described ;above, the present invention is based, in part, on isolating
nucleic
acid molecules from .8. subtilis that encode the three subunits of the AdT
protein.
Accordingly, the pre.oent invention further provides nucleic acid molecules
that
encode the AdT protE;in, as herein defined, preferably in isolated form. For
CA 02279490 1999-08-03
WO 98J33925 PCT1US98101860
-14-
convenience, all AdT encoding nucleic acid molecules will be referred to as
AdT
encoding nucleic acid molecules, the AdT genes, or AdT. The nucleotide
sequence of
the B. subtilis nucleic acid molecule that encodes each of the subunits of AdT
is
provided in SEQ ID NO:1. The start and stop codons for each of subunits A (SEQ
ID N0:4), B (SEQ ID N0:6) and C (SEQ ID N0:8) are designated in the nucleotide
sequence for AdT provided in Figure 3.
Further preferred embodiments of the invention are polynucleotides that are at
least 70% sequence identical over their entire length to a polynucleotide
encoding
AdT polypeptides having an amino acid sequence encoded by SEQ ID NO:I, and
polynucleotides which are complementary to such polynucleotides.
Alternatively,
most highly preferred are polynucleotides that comprise a region that is at
least 80%
identical over their entire length to a polynucleotide encoding AdT
polypeptide and
polynucleotides complementary thereto. In this regard, polynucleotides at
least 90%
identical over their entire length to the same are particularly preferred, and
among
these particularly preferred polynucleotides, those with at least 95% are
especially
preferred. Furthermore, those with at least 97% are highly preferred among
those
with at least 95%, and among these those with at least 98% and at least 99%
are
particularly highly preferred, with at least 99% being the more preferred.
CA 02279490 1999-08-03
wo ~3gzs rcr~s9grois6o
-15-
The invention :further relates to variants of the herein above described
polynucleotides which. encode for variants of the polypeptides having the
deduced
amino acid sequences of SEQ ID NO:1.
Variants that are fragments of the polypeptides of the invention may be
employed for producing the corresponding full-length polypeptide by peptide
synthesis; therefore, these variants may be employed as intermediates for
producing
the full-length polypptides. Variants that are fragments of the
polynucleotides of the
invention may be used to synthesize full-length polynucleotides of the
invention.
Variants that are fragments of the polynucleotides of the invention may be
used to
synthesize full-length polynucleotides of the invention. Such methods are
widely
available, such as those disclosed in WO 97/26340 and WO 97/3$716.
A fragment is a variant polypeptide having an amino acid sequence that
entirely is the same as. part but not all of the amino acid sequence of the
aforementioned polypeptides. AdT polypeptides fragments may be "free-
standing,"
or comprised within a. larger polypeptide of which they form a part or region,
most
preferably as a single continuous region, a single larger polypeptide.
Further particularly preferred embodiments are polynucleotides encoding AdT
variants, which have the amino acid sequence of the AdT polypeptides encoded
by
SEQ ID NO:1 in which several, a few, 10 to 1 S, 5 to 10, 1 to 5, 1 to 3, 2, 1
or no
amino acid residues are substituted, deleted or added, in any combination.
Especially
CA 02279490 1999-08-03
WO 98133925 PCT/US98/01860
-16-
preferred among these are silent substitutions, additions and deletions, which
do not
alter the properties and activities of AdT.
As used herein, a "nucleic acid molecule" is defined as an RNA or DNA
molecule that encodes a peptide as defined above, or is complementary to a
nucleic
acid sequence encoding such peptides. Particularly preferred nucleic acid
molecules
will have a nucleotide sequence identical to or complementary to the B.
subtilis DNA
sequences herein disclosed. Specifically contemplated are genomic DNA,
polycistronic mRNA and antisense molecules, as well as nucleic acids based on
an
alternative backbone or including alternative bases, whether derived from
natural
sources or synthesized. Such nucleic acid molecules, however, are defined
further as
being novel and unobvious over any prior art nucleic acid molecules encoding
non-AdT proteins isolated from organisms other than B. subtilis.
As used herein, a nucleic acid molecule is said to be "isolated" when the
nucleic acid molecule is substantially separated from contaminant nucleic acid
molecules that encode polypeptides other than AdT. A skilled artisan can
readily
employ nucleic acid isolation procedures to obtain an isolated AdT encoding
nucleic
acid molecule.
The present invention further provides fragments of the AdT encoding nucleic
acid molecules of the present invention. As used herein, a fragment of an AdT
encoding nucleic acid molecule r efers to a small portion of the entire
protein encoding
CA 02279490 1999-08-03
wo ~39ZS rcTrtrs9srois6o
-17-
sequence. The size of the fragment will be determined by its intended use. For
example, if the fi~agment is chosen so as to encode an active portion of the
AdT
protein, such an active domain or effector binding site, then the fi-agment
will need to
be .large enough to encode the functional regions) of the AdT protein. If the
fragment
is to be used as a nuclleic acid probe or I'CR primer, then the fragment
leitgttf is
chosen so as to obtain a relatively small number of false positives during
probing/priming. Fragments of the B. subtilis AdT nucleic acid molecule that
are
particularly useful as selective hybridization probes or PCR can be readily
determined
using art-known methods.
Fragments of the AdT encoding nucleic acid molecules of the present
invention (i.e., synthetic oligonucleotides) that are used as probes or
specific primers
for the polymerase chain reaction (PCR), or to synthesize gene sequences
encoding
AdT proteins, can easily be synthesized by chemical techniques, for example,
the
phosphotriester method of Matteucci, et al., J Am Chem Soc ( 1981 ) 103 :3185-
3191 or
1 S using automated synthesis methods. ~ In addition, larger DNA segments can
readily be
prepared by well known methods, such as synthesis of a group of
oligonucleotides
that define various modular segments of the AdT gene, followed by ligation of
oligonucleotides to guild the complete modified AdT gene.
The AdT encoding nucleic acid molecules of the present invention may firrther
be modified so as to contain a detectable label for diagnostic and probe
purposes. As
CA 02279490 1999-08-03
WO 98/33925 PCT/US98/~01860
-18-
described above, such probes can be used to identify nucleic acid molecules
encoding
other allelic variants or homologues of the AdT proteins and as described
below, such
probes can be used to diagnose the presence of a AdT protein as a means for
diagnosing a pathological condition caused by AdT mediated translation. A
variety of
such labels are known in the art and can readily be employed with the AdT
encoding
molecules herein described. Suitable labels include, but are not limited to,
biotin,
radiolabeled nucleotides, biotin, and the like. A skilled artisan can employ
any of the
art-known labels to obtain a labeled AdT encoding nucleic acid molecule.
D. Isolation of Other AdT Encoding Nucleic Acid Molecules
The identification of the AdT protein from B. subtilis and the corresponding
nucleic acid molecules, has made possible the identification of and isolation
of AdT
proteins from organisms other than B. subtilis, hereinafter referred to
collectively as
AdT homologues. The preferred source of the AdT homologues are pathogenic
microorganisms such as bacteria and fungi, as well as plants in which it is
desirable to
control growth. The most preferred sources are gram positive bacteria,
pathogenic
fizngi and plant organelles such as chloroplasts.
Essentially, a skilled artisan can readily use the amino acid sequence of the
B.
subtilis AdT protein to generate antibody probes to screen expression
libraries
prepared from cells. Typically, polyclonal antiserum from mammals such as
rabbits
CA 02279490 1999-08-03
WO 98133925 PCT/US98l01860
-19-
immunized with the purified protein (as described below) or monoclonal
antibodies
can be used to probe an expression library, prepared from a target organism,
to obtain
the appropriate coding; sequence for AdT protein homologue. The cloned cDNA
sequence can be expressed as a fusion protein, expressed directly using its
own
control sequences, or expressed by constructing an expression cassette using
control
sequences appropriatE; to the particular host used for expression of the
enzyme.
Alternatively, a portion of the AdT encoding sequence herein described can be
synthesized and used as a probe to retrieve DNA encoding a member of the AdT
family of proteins from organisms other than B. subtilis. Oligomers containing
approximately 18-20 nucleotides (encoding about a 6-7 amino acid stretch) are
prepared and used to screen genomic DNA or cDNA libraries to obtain
hybridization
under stringent conditions or conditions of sufficient stringency to eliminate
an undue
level of false positives. This method can be used to identify and isolate
altered and
variants of the AdT encoding sequences.
Additionally, pairs of oligonucleotide primers can be prepared for use in a
polymerase chain reaction (PCR) to selectively amplify/clone an AdT-encoding
nucleic acid molecule, or fragment thereof. A PCR denature/anneal/extend cycle
for
using such PCR primers is well lrnown in the art and can readily be adapted
for use in
isolating other AdT Encoding nucleic acid molecules. Regions of the B.
subtilis AdT
gene that are particularly well suited for use as a probe or as primers can be
readily
CA 02279490 1999-08-03
wo ~3ns rcrms~ois6o
-20-
identified. In general, the preferred primers will flank one or more of the
subunit
encoding regions of the B. subtilis AdT gene.
Homologues of the herein disclosed AdT proteins will share homology. In
general, nucleic acid molecules that encode AdT homologues will hybridize to
the
B. subtilis sequences under high stringency. Such sequences will typically
contain at
least 70% homology, preferably at least 80%, most preferably at least 90%
homology
to the B. subtilis sequences.
E. Recombinant DNA Molecules Containing an AdT Encoding Nucleic Acid
Molecule
The present invention further provides recombinant DNA molecules that
contain one or more of the AdT encoding sequences herein described, or a
fragment of
the herein-described nucleic acid molecules. As used herein, an recombinant
DNA
molecule is a DNA molecule that has been subjected to molecular manipulation
in
vitro. Methods for generating recombinant DNA molecules are well known in the
art,
for example, see Sambrook et al., Molecular Cloning (1989). In the preferred
recombinant DNA molecules, an AdT encoding DNA sequence that encodes an AdT
protein, or AdT subunit is operably linked to one or more expression control
sequences and/or vector sequences. The recombinant DNA molecule can encode
CA 02279490 1999-08-03
wo ins rc~r~s9srois6o
-21-
either a single subunit: of the AdT protein, or can encode an operon that
contains all
three of the AdT subunits.
The choice of vector and/or expression control sequences to which one of the
AdT encoding sequences of the present invention is operably linked depends
directly,
as is well known in tl-.~e art, on the functional properties desired, e.g.,
protein
expression, and the host cell to be transformed. A vector contemplated by the
present
invention is at least capable of directing the replication or insertion into
the host
chromosome, and preferably also expression, of an AdT encoding sequence
included
in the recombinant DNA molecule.
Expression control elements that are used for regulating the expression of an
operably linked protein encoding sequence are known in the art and include,
but are
not limited to, induci ble promoters, constitutive promoters, secretion
signals,
enhancers, transcription terminators and other regulatory elements.
Preferably, an
inducible promoter that is readily controlled, such as being responsive to a
nutrient in
the host cell's medium, is used.
In one embodiment, the vector containing an AdT encoding nucleic acid
molecule will include a prokaryotic replicon, i.e., a DNA sequence having the
ability
to direct autonomous replication and maintenance of the recombinant DNA
molecule
intrachromosomally in a prokaryotic host cell, such as a bacterial host cell,
transformed therewith. Such replicons are well known in the art. In addition,
vectors
CA 02279490 1999-08-03
WO 98J33925 PCT/US98rol860
-22-
that include a prokaryotic replicon may also include a gene whose expression
confers
a detectable marker such as a drug resistance. Typical bacterial drug
resistance genes
are those that confer resistance to ampicillin or tetracycline.
Vectors that include a prokaryotic replicon can further include a prokaryotic
or
viral promoter capable of directing the expression (transcription and
translation) of the
AdT encoding sequence in a bacterial host cell, such as E. coli. A promoter is
an
expression control element formed by a DNA sequence that permits binding of
RNA
polymerase and transcription to occur. Promoter sequences compatible with
bacterial
hosts are typically provided in plasmid vectors containing convenient
restriction sites
Z O for insertion of a DNA segment of the present invention. Typical of such
vector
plasmids are pUCB, pUC9, pBR322 and pBR329 available from Biorad Laboratories
(Richmond, CA), pPL and pKK223 available from Pharmacia, Piscataway, NJ.
Expression vectors compatible with eukaryotic cells, preferably those
compatible with vertebrate cells, can also be used to variant recombinant DNA
molecules that contain an AdT encoding sequence. Eukaryotic cell expression
vectors
are well known in the art and are available from several commercial sources.
Typically, such vectors are provided containing convenient restriction sites
for
insertion of the desired DNA segment. Typical of such vectors are PSVL and
pKSV-10 (Pharmacia), pBPV-1/pML2d (International Biotechnologies, Inc.), pTDTl
CA 02279490 1999-08-03
WO 98133925 PCT/US98I01860
-23-
(ATCC, #31255), the vector pCDMB described herein, and the like eukaryotic
expression vectors.
Eukaryotic cell expression vectors used to construct the recombinant DNA
molecules of the presc;nt invention may further include a selectable marker
that is
effective in an eukaryotic cell, preferably a drug resistance selection
marker. A
preferred drug resistance marker is the gene whose expression results in
neomycin
resistance, i.e., the neomycin phosphotransferase (neo) gene. Southern et al.,
JMoI
Anal Genet ( 1982) 1::327-341. Alternatively, the selectable marker can be
present on
a separate plasmid, and the two vectors are introduced by cotransfection of
the host
cell, and selected by culturing in the presence of the appropriate drug for
the
selectable marker.
F. Host Cells Containing an Exogenously Supplied AdT Encoding Nucleic
Acid Molecule
The present invention further provides host cells transformed with a nucleic
acid molecule that encodes an AdT protein of the present invention, either the
entire
heterotrimeric protein or one or more subunits. The host cell can be either
prokaryotic
or eukaryotic. Eukaryotic cells useful for expression of an AdT protein are
not
limited, so long as the cell line is compatible with cell culture methods and
compatible with the propagation of the expression vector and expression of an
AdT
CA 02279490 1999-08-03
WO 98/33925 PCT/US98/01860
-24-
gene. Preferred eukaryotic host cells include, but are not limited to, yeast,
insect and
mammalian cells, preferably vertebrate cells such as those from a mouse, rat,
monkey
or human fibroblastic cell line, the most preferred being cells that do not
naturally
express an AdT protein.
Any prokaryotic host can be used to express an AdT-encoding recombinant
DNA molecule. The preferred prokaryotic host is E. coli.
Transformation of appropriate cell hosts with an recombinant DNA molecule
of the present invention is accomplished by well known methods that typically
depend
on the type of vector used and host system employed. With regard to
transformation
of prokaryotic host cells, electroporation and salt treatment methods are
typically
employed, see, for example, Cohen et al., Proc Acad Sci USA (1972) 69:2110;
and
Maniatis et al., Molecular Cloning, A Laboratory Manual, Cold Spring Harbor
Laboratory, Cold Spring Harbor, NY (1982). With regard to transformation of
vertebrate cells with vectors containing recombinant DNAs, electroporation,
cationic
I S lipid or salt treatment methods are typically employed, see, for example,
Graham et
al., Yirol (1973) 52:456; Wigler et al., Proc Natl Acad Sci USA (1979) 76:1373-
76.
Successfully transformed cells, i.e., cells that contain an recombinant DNA
molecule of the present invention, can be identified by well known techniques.
For
example, cells resulting from the introduction of an recombinant DNA of the
present
invention can be cloned to produce single colonies. Cells from those colonies
can be
CA 02279490 1999-08-03
WO 98/33925 PCT/US98~101860
-25-
harvested, lysed and tlheir DNA content examined for the presence of the
recombinant
DNA using a method such as that described by Southern, JMoI Biol (1975)
98:503, or
Berent et al., Biotech (1985) 3:208 or the proteins produced from the cell
assayed via
an immunological method.
G. Production of an AdT Protein Using an recombinant DNA Molecule
The present invention further provides methods for producing an AdT protein
that uses one of the AdT encoding nucleic acid molecules herein described. In
general
terms, the production of a recombinant AdT protein typically involves the
following
steps.
First, a nucleic; acid molecule is obtained that encodes an AdT protein, such
as
the nucleic acid molecule depicted in Figure 3 (SEQ ID NO:1 ) or an AdT
subunit.
The AdT encoding nucleic acid molecule is then preferably placed in an
operable
linkage with suitable control sequences, as described above, to generate an
expression
unit containing the AdT encoding sequence. The expression unit is used to
transform
a suitable host and thc; transformed host is cultured under conditions that
allow the
production of the Ad"C protein. Optionally the AdT protein is isolated from
the
medium or from the cells; recovery and purification of the protein may not be
necessary in some instances where some impurities may be tolerated.
CA 02279490 1999-08-03
WO 98133925 PCTIUS98/01860
-26-
Each of the foregoing steps can be done in a variety of ways. For example, the
desired coding sequences may be obtained from genomic fragments and used
directly
in an appropriate host. The construction of expression vectors that are
operable in a
variety of hosts is accomplished using an appropriate combination of replicons
and
control sequences. The control sequences, expression vectors, and
transformation
methods are dependent on the type of host cell used to express the gene and
were
discussed in detail earlier. Suitable restriction sites can, if not normally
available, be
added to the ends of the coding sequence so as to provide an excisable gene to
insert
into these vectors. A skilled artisan can readily adapt any hostlexpression
system
known in the art for use with AdT encoding sequences to produce an AdT
protein.
H. Identification of Agents that Bind to an AdT Protein
Another embodiment of the present invention provides methods for identifying
agents that are agonists or antagonists of the AdT proteins herein described.
Specifically, agonists and antagonists of an AdT protein can be identified by
the
ability of the agent to bind to an AdT protein and/or the ability to inhibit
AdT activity.
Activity assays for AdT activity and binding assays using an AdT protein are
suitable
for use in high through-put screening methods.
In detail, in one embodiment, an AdT protein is mixed with an agent. After
mixing under conditions that allow association of AdT protein with the agent,
the
CA 02279490 1999-08-03
wo ~3~s pcrivs9srois6o
-27-
mixture is analyzed to determine if the agent bound the AdT protein. Agonists
and
antagonists are identifiued as being able to bind to an AdT protein.
Alternatively or
consecutively, as described below, AdT activity can be directly assessed as a
means
for identifying agonists and antagonists of AdT activity.
The AdT protein used in the above assay can be: an isolated and fully
characterized protein, a single subunit of an AdT protein, a partially
purified protein, a
cell that has been altered to express an AdT protein or a fraction of a cell
that has been
altered to express an AdT protein. Further, the AdT protein can be the entire
AdT
protein, a specific fragment of the AdT protein or a single subunit of the AdT
protein.
It will be apparent to one of ordinary skill in the art that so long as the
AdT protein
can be assayed for agc;nt binding, e.g., by a shift in molecular weight or
activity, as
described in the Exalr.~ples, the present assay can be used. The AdT protein
is
particularly well suited for high through-put screening methods.
The source of the AdT protein will be based on the intended use of the
modulating agent. For example, microbial AdT protein is used to identify AdT
inhibitors that have bactericidal activity whereas chloroplast derived AdT
protein is
used to identify AdT inhibitors that have herbicide activity.
The method used to identify whether an agent binds to an AdT protein will be
based primarily on the nature of the AdT protein used. For example, a gel
retardation
assay can be used to determine whether an agent binds to a soluble fragment of
an
CA 02279490 1999-08-03
w0 98/33925 PCT/US98/01860
-28-
AdT protein. Alternatively, immunodetection and biochip technologies can be
adopted for use with an AdT protein. A skilled artisan can readily employ
numerous
art-known techniques for determining whether a particular agent binds to an
AdT
protein.
Agents can be further tested for the ability to modulate the activity of an
AdT
protein using a cell-free assay system or a cellular assay system. Example 1
provides
one such methods that can be used to assay for AdT activity.
As used herein, an agent is said to antagonize AdT activity when the agent
reduces AdT activity. The preferred antagonist will selectively antagonize
AdT, not
affecting any other cellular proteins, particularly other proteins involved in
translation.
Further, the preferred antagonist will reduce AdT activity by more than 50%,
more
preferably by more than 90%, most preferably eliminating all AdT activity.
As used herein, an agent is said to agonize AdT activity when the agent
increases AdT activity. The preferred agonist will increase AdT activity by
more than
50%, more preferably by more than 90%, most preferably more than doubling the
level of AdT activity.
The preferred antagonists and agonists will be selective for a specific
species,
genus, family, order or kingdom of organisms. Agents can be screened using one
AdT protein, or a combination of AdT proteins, to aid in identifying agents
for target
specificity. For example, several different microbial AdT proteins can be used
to
CA 02279490 1999-08-03
wo ~39ZS rc~r~s~ois6o
-29-
identify general antimicrobial agents whereas chloroplast derived AdT proteins
can be
used to identify herbicide agents.
Agents that are assayed in the above method can be randomly selected or
rationally selected or designed. As used herein, an agent is said to be
randomly
S selected when the agent is chosen randomly without considering the specific
sequences of the AdT 'protein. An example of randomly selected agents is the
use a
chemical library or a peptide combinatorial library, or a growth broth of an
organism
or plant extract.
As used herein, an agent is said to be rationally selected or designed when
the
agent is chosen on a nonrandom basis that takes into account the sequence of
the
target site andlor its conformation in connection with the agent's action.
Agents can
be rationally selected or rationally designed by utilizing the peptide
sequences that
make up the AdT protein. For example, a rationally selected peptide agent can
be a
peptide whose amino acid sequence is identical to a fragment of an AdT
protein.
The agents of the present invention can be, as examples, peptides, small
molecules, and vitamin derivatives, as well as carbohydrates. A skilled
artisan can
readily recognize that there is no limit as to the structural nature of the
agents of the
present invention. One class of agents of the present invention are peptide
agents
whose amino acid sequences are chosen based on the amino acid sequence of the
AdT
CA 02279490 1999-08-03
WO 98r339Z5 PCT/US98/01860
-30-
protein. Small peptide agents can serve as competitive inhibitors of AdT
protein
assembly.
The peptide agents of the invention can be prepared using standard solid phase
(or solution phase) peptide synthesis methods, as is known in the art. In
addition) the
DNA encoding these peptides may be synthesized using commercially available
oligonucleotide synthesis instrumentation and produced recombinantly using
standard
recombinant production systems. The production using solid phase peptide
synthesis
is necessitated if non-gene-encoded amino acids are to be included.
Another class of agents of the present invention are antibodies immunoreactive
with critical positions of the AdT protein. As described above, antibodies are
obtained by immunization of suitable mammalian subjects with peptides,
containing
as antigenic regions, those portions of the AdT protein intended to be
targeted by the
antibodies. Critical regions include the domains identified in Figure 2. Such
agents
can be used in competitive binding studies to identify second generation
inhibitory
agents.
K. Uses of Agents th$t Bind to an AdT Protein
As provided in the Background section, the AdT proteins are involved in
protein translation, particularly protein translation in gram positive
microorganisms,
fungi and cellular organelles, particularly chloroplasts. Agents that bind an
AdT
CA 02279490 1999-08-03
WO 98133925 PCT/US98/01860
-31-
protein and act as an agonist or antagonist can be used to modulate
translation in these
organism'and serves :~s a basis for an antibacterial, antifungal or herbicide
agents. In
detail, protein translation that requires AdT can be modulated by
administering to an
organism an agent that binds to an AdT protein and acts as an agonist or
antagonist of
AdT activity.
As used herein, an organism can be any organism, so long as it is desirable to
modulate protein translation in the organism, for example to control the
growth of an
infectious agent in a mammalian subject or to act as an herbicide agent. The
invention
is particularly useful in the treatment of human subjects for controlling
microbial
growth.
As used herein, protein translation that requires AdT refers to protein
translation that would not occur without the presence of an active AdT
protein. As
used herein, an agent is said to modulate AdT meditated protein translation
when the
agent reduces the del;ree of protein translation.
The use of the AdT modulating agents will be based primarily on the target
AdT protein used to :identify the agent as well as the activity/selectivity of
the agent.
For example, an AdT inhibitory agent, that is used as an antimicrobial agent,
is
preferably isolated using one or more microbial AdT proteins. Herbicide agent
will
be preferably identified using chloroplast AdT protein as a target.
CA 02279490 1999-08-03
WO 98133925 PCT/US98/01860
-32-
L. Administration of Agonists and Antagonists of an AdT Protein
The administration of agonists and antagonists of the AdT protein will be
dependent on their intended purpose. For example, to control microbial growth
in a
mammalian subject, an AdT inhibitory agent can be administered via parenteral,
subcutaneous, intravenous, intramuscular, intraperitoneal, transdermal, or
buccal
routes. Alternatively, or concurrently, administration may be by the oral
route. The
dosage administered will be dependent upon the age, health, and weight of the
recipient, kind of concurrent treatment, if any, frequency of treatment, and
the nature
of the effect desired. For example, to treat microbial infection, an agent
that
modulates AdT activity is administered systemically or locally to the
individual being
treated. As described below, there are many methods that can readily be
adapted to
administer such agents.
The present invention further provides compositions containing an antagonist
or agonist of an AdT protein that is identified by the methods herein
described. The
determination of optimal ranges of effective amounts of each component is
within the
skill of the art and is based on the intended use.
In addition to the AdT modulating agent, the compositions of the present
invention may contain other ingredients, such as suitable pharmaceutically
acceptable
Garners comprising excipients and auxiliaries which facilitate processing of
the active
compounds into preparations which can be used pharmaceutically for delivery to
the
CA 02279490 1999-08-03
wo 98/33925 - PCT/US98101860
-33-
site of action. Suitabh; formulations for parenteral administration include
aqueous
solutions of the active compounds in water-soluble variant, for example, water-
soluble
salts. In addition, suspensions of the active compounds and as appropriate,
oily
injection suspensions may be administered. Suitable lipophilic solvents or
vehicles
include fatty oils, for example, sesame oil, or synthetic fatty acid esters,
for example,
ethyl oleate or triglyce.rides. Aqueous inj ection suspensions may contain
substances
which increase the viscosity of the suspension and include, for example,
sodium
carboxymethyl cellulose, sorbitol, and/or dintran. Optionally, the suspension
may
also contain stabilizers. Liposomes can also be used to encapsulate the agent
for
delivery into the cell.
The pharmaceutical formulation for systemic administration according to the
invention may be formulated for enteral, parenteral or topical administration.
Indeed,
all three types of formulations may be used simultaneously to achieve systemic
administration of the active ingredient.
Suitable formulations for oral administration include hard or soft gelatin
capsules, pills, tablets, including coated tablets, elixirs, suspensions,
syrups or
inhalations and controlled release variants thereof.
M. Combination Therapy
CA 02279490 1999-08-03
WO 98/33925 PCT/US98l01860
-34-
The agents of the present invention that modulate AdT activity can be
provided alone, or in combination with another agents that modulate protein
synthesis
microbial, fungal or plant growth. For example, an agent of the present
invention that
reduces microbial AdT activity can be administered in combination with other
antimicrobial agents. As used herein, two agents are said to be administered
in
combination when the two agents are administered simultaneously or are
administered
independently in a fashion such that the agents will act at the same time.
N. Methods for Identifying the Presence of an AdT protein or gene
The present invention further provides methods for identifying cells or
organisms expressing an AdT protein or an AdT gene. Such methods can be used
to
diagnose the presence of an organism that expresses an AdT protein. The
methods of
the present invention are particularly useful in the determining the presence
of
pathogenic microorganisms. Specifically, the presence of an AdT protein can be
identified by determining whether an AdT protein, or nucleic acid encoding an
AdT
protein, is expressed. The expression of an AdT protein can be used as a means
for
diagnosing the presence of an organism that relies on AdT mediated
translation.
A variety of immunological and molecular genetic techniques can be used to
determine if an AdT protein is expressed/produced in a particular cell or
sample. In
general, an extract containing nucleic acid molecules or an extract containing
proteins
CA 02279490 1999-08-03
WO 98/33925 PCT/US98I01860
-35-
is prepared. The extras;t is then assayed to determine whether an AdT protein,
or an
AdT encoding nucleic acid molecule, is produced in the cell.
For example, to perform a diagnostic test based on nucleic acid molecules, a
suitable nucleic acid s~unple is obtained and prepared using conventional
techniques.
S DNA can be prepared, for example, simply by boiling a sample in SDS. The
extracted nucleic acid can then be subjected to amplification, for example by
using the
polymerase chain reaction (PCR) according to standard procedures, to
selectively
amplify an AdT encoding nucleic acid molecule or fragment thereof. The size or
presence of a specific amplified fragment (typically following restriction
endonuclease digestion) is then determined using gel electrophoresis or the
nucleotide
sequence of the fragment is determined (for example, see Weber and May Am J
Hum
Genet (1989) 44:388-.339; Davies, J. et al. Nature (I994) 371:130-136)). The
resulting size of the fragment or sequence is then compared to the known AdT
proteins encoding sequences, for example via hybridization probe. Using this
method, the presence of an AdT protein can be identified.
To perform a .diagnostic test based on proteins, a suitable protein sample is
obtained and prepared using conventional techniques. Protein samples can be
prepared, for example;, simply by mixing a sample with SDS followed by salt
precipitation of a protein fraction. The extracted protein can then be
analyzed to
determine the presence of an AdT protein using known methods. For example, the
CA 02279490 1999-08-03
WO 98133925 PCT/US98/01860
-36-
presence of specific sized or charged variants of a protein can be identified
using
mobility in an electric filed. Alternatively, antibodies can be used for
detection
purposes. A skilled artisan can readily adapt known protein analytical methods
to
determine if a sample contains an AdT protein.
Alternatively, AdT expression can also be used in methods to identify agents
that decrease the level of expression of the AdT gene. For example, cells or
tissues
expressing an AdT protein can be contacted with a test agent to determine the
effects
of the agent on AdT expression. Agents that activate AdT expression can be
used as
an agonist of AdT activity whereas agents that decrease AdT expression can be
used
as an antagonist of AdT activity.
O. Preparation and Use of Herbicides
As discussed herein, the transamidation pathway is operative in chloroplasts.
The ability to identify AdT inhibitors which specifically inhibit plastid
isoforms of
AdT can be useful in designing herbicides that are not toxic or harmful to
humans and
animals. Thus, the ability to develop herbicides that inhibit only chloroplast
isoforms
of enzymes such as Adt but do not inhibit cytosolic (i.e., the fluid portion
of the
cytoplasm exclusive of organelles) AdT or human AdT, would provide a new form
of
highly effective herbicide that is also less toxic to humans. However, Adt
inhibitors
which are not limited to the chloroplasts may also find utility in use as an
herbicide.
CA 02279490 1999-08-03
WO 98/33925 PCTIUS98/01860
-37-
An identified compound which inhibits function of the wild-type AdT enzyme
is utilized as an active ingredient in an herbicide. The active ingredient is
normally
applied in the form of compositions together with one or more agriculturally
acceptable carriers, and can be applied to the crop area or plant to be
treated,
S simultaneously or in succession, with further compounds. These additional
compounds can include fertilizers, other herbicides, fungicides, bactericides,
nematicides, or mixtures of several of these preparations, together with
further
carriers, surfactants or application-promoting adjuvants. The herbicide may be
applied as a seed coating, a ground spray, incorporated into the soil, or
applied
directly to the plant. :Preferably, the active ingredient of the present
invention or an
agrochemical composition which contains at least one of the active ingredients
of the
present invention are applied as a leaf preparation. Methods of herbicide
preparation
and application are well known to one skilled in the art.
Resistant mutants to the AdT-inhibiting compound can be identified by
mutagenizing cells or organisms and growing the mutagenized populations in the
presence of a concentration of the inhibitor sufficient to inhibit growth of
the wild-
type cells or organisms, and selecting cells or organisms from the populations
that are
able to grow more rapidly than wild-type cells or organisms. Mutagenesis can
be
accomplished by any one of the means well known to one skilled in the art,
including:
chemical mutatgenesis (e.g., ethyl rnethanesulfonate); ultraviolet radiation;
X-ray
CA 02279490 1999-08-03
WO 98/33925 PCT/US98/018G0
-38-
exposure; and gamma radiation (see, e.g., Watson et al., Recombinant DNA,
Second
Edition ( 1992) Chapter 11:191-211; Freifelder, Molecular Biology ( 1987)
Chapter
11:293-313). The mutant individuals which have the ability to tolerate or
resist the
normally toxic levels of the inhibitor are genetically purified, the gene
encoding the
mutant AdT is isolated, and the DNA sequence of the mutant gene is determined
and
translated into a predicted amino acid sequence. The amino acids which differ
between the wild-type AdT enzyme and the mutant AdT enzyme are assumed to be
responsible for the inhibitor-resistant phenotype of the newly-identified
mutant.
The coding DNA sequence for the mutant AdT can be introduced into the
plant cell in a number of different ways that are well known to those of skill
in the art.
Examples of such methods include micro inj ection, electroporation,
Agrobacterium-
mediated transformation, direct gene transfer, and micro projectile
bombardment.
Techniques for producing herbicide resistance in plants by incorporating DNA
encoding and expressing enzymes resistant to herbicides are well known (see,
e.g.,
LT.S. Patent No. 5,145,777; U.S. Patent No. 5,290,926), including techniques
for
adding a chloroplast transit sequence upstream from an herbicide gene so that
the
protein product is transported into the cholorplasts (Comai et aL, Nature (
1985)
313:741-744; U.S. Patent No. 4,940,835; U.S. Patent No. 5,188,642). In the
same
manner, the gene coding for a mutant AdT may be substituted for one of the
other
herbicide resistance genes of the references. Since AdT performs its function
in the
CA 02279490 1999-08-03
WO 98!33925 PCTIUS98/01860
-39-
chloroplast, it may be particularly relevant to use a plastid transit sequence
to ensure
expression'in the chloroplast or other plastid as is known in the art.
Following introduction of the mutant AdT gene into plant cells and the
regeneration of transfolzned plants from such cells, conventional methods of
plant
husbandry and plant breeding can be used to maintain and increase the
transformed
plants. The transformed plants can also be used in conventional hybridization
schemes to produce new plant types which also carry the novel mutant AdT gene
(see,
e.g., Fehr and Hadley, Hybridization of Crop Plants (1980); Jensen, Plant
Breeding
Methodology (1988); Allard, Principles of Plant Breeding (1960).
Plants which e:Kpress a gene which is tolerant or resistant to an inhibitor of
AdT can be grown in soil and the herbicide containing the AdT inhibitor can be
applied to inhibit weed growth. Since the weed plants will not be carrying the
mutant
AdT gene, the weeds will be susceptible to the herbicide containing the AdT
inhibitor.
The following examples are intended to illustrate, but not to limit, aspects
of
the present invention.
'l~he invention. will be further described by reference to the following
detailed
examples. These examples are provided for purposes of illustration only, and
are not
intended to be limiting unless otherwise specified.
CA 02279490 1999-08-03
WO 98133925 PCT/US98~1860
-40-
F~xnerimental Procedures
Preparation and purification of recombinant Bacillus subtilis
GIntRNA°~°
amidotransferase. E. coli BL21 (DE3) harboring pABC were incubated overnight
in
3 mL LB medium (10 g bactotryptone, 5 g yeast extract, 10 g NaCI) with 50
(g/mL
ampicillin at 37°C. The culture was scaled up to 1 L and again allowed
to incubate at
37°C overnight. Cells were harvested via centrifugation (4000 x g for 5
minutes at
4°C) and resuspended in 20 mL Buffer A (25 mM Hepes~KOH, pH 7.5, 25 mM
KCI,
mM MgCl2 and 1 mM DTT). This step and all subsequent steps were performed
at 4°C unless otherwise specified. The cells were lysed by sonication
(4 x 15
10 seconds) and centrifuged at 100,000 x g for one hour. The enzyme was then
purified
to homogeneity, as determined by SDS~polyacrylamide gel electrophoresis, via a
series of chromatographic steps using a Pharmacia FPLC system. The supernatant
was first applied to a Q~sepharose (HR 16/10) column (strong anion exchange)
and the
activity was eluted by a linear gradient from 0 to 1 M NaCI in Buffer A. The
active
fractions from this column were applied to a Superdex-200 (HR 26/100) column
(gel
filtration) and the activity was eluted isocratically in Buffer A. The
fractions from
this column which contained activity were pooled and applied onto a MonoQ (HR
10/10) column and the activity as eluted with a linear gradient from 150 to
300 mM
NaCl in Buffer A. Active fractions from this column were pooled and dialyzed
against Buffer A + 200 mM NaCI in 50% glycerol for 12 hours and stored at
70°C.
CA 02279490 1999-08-03
wo ~~zs pc~rws9sroisso
-41-
In vivo expressed Bacillus subtilis tItNA~'° isolation and
purification. A
3 mL culture of E. coli :DHSa/pGPl-2/pBTT (encoding tRNAGIn) in LB medium
(10 g bactotryptone, S i; yeast extract, 10 g NaCI) with 50 (g/mL ampicillin
and
(g/mL kanamycin was incubated at 37°C overnight. The culture was scaled
up to
5 1 L and overnight incubation was repeated. Cells were harvested via
centrifugation at
4000 x g for 5 minutes at 4°C and resuspended in 10 mL lysis buffer (20
mM
Tris~HCl, pH 7.4 and 20 mM MgCl2). Total nucleic acids were isolated by two
sequential extractions with equal volumes of water saturated phenol followed
by
isopropanol precipitation of the aqueous phase. The nucleic acid pellet was
collected
10 via centrifugation at 11),000 x g for 15 minutes at 4°C. The pellet
was resuspended in
5 mL of 200 mM Tris~OAc, pH 9.0 and incubated at 37°C for 1 hour to
ensure
complete deacylation of the tRNA. The nucleic acids were recovered by ethanol
precipitation followed by centrifugation at 10,000 x g for 15 minutes at
4°C. The
pellet was resuspended in 100 mM NaCI, incubated overnight at 4°C and
ethanol
precipitated. The tRhTAGIn was purified by a two-step anion exchange
chromatography protocol. The nucleic acids were resuspended in S mL of Buffer
1
(140 mM NaOAc, pH 4.5) and 1 gm DE52 resin/100 OD260 was added. The resin
was washed with 200 mL Buffer A and 150 mL Buffer 2 (140 mM NaOAc, pH 4.5 +
300 mM NaCI) and tlhe tRNA was eluted with 100 mL Buffer 3 (140 mM NaOAc,
pH 4.5 + 1 M NaCI). the nucleic acids were recovered by ethanol precipitation
CA 02279490 1999-08-03
WO 98133925 PGT/US98J01860
-42-
followed by centrifugation at 10,000 x g for 15 minutes at 4°C and
resuspended in
mM Tris~HCl, pH 7.4, 1 mM MgCl2 and 1 mM DTT and applied onto a Pharmacia
MonoQ (HR 10/10) column. The tIRNA was eluted with a gradient of 450 to 750 mM
NaCI in 10 mM Tris~HCl, pH 7.4, 1 mM MgCl2 and 1 mM DTT. Fractions
5 containing the tIRNAGIn, based on ability to be aminoacylated with both Glu
and Gln,
were pooled and used as substrates in the amidotransferase assays.
Aminoacylation reactions. The procedure for the formation of radiolabelled
GIntRNA~'" was adapted from Jahn, D. et al. ( 1990). Unless otherwise noted,
these
reactions were conducted at 37°C in a buffer consisting of 10 mM ATP,
50 mM
10 Hepes~KOH pH 7.0, 25 mM KCI, 15 mM MgCl2, and 5 mM DTT. The concentration
of tRNAGIn, recovered from E. coli DHSa harboring the plasmids pGPl2 and pBTT
(see above), and L14C(U)-glutamate (300 mCi/rriMol) was IO (M. GIuRS was
isolated from B. subtilis and then partially purified by DEAF-sepharose
chromatography. The reactions were allowed to progress for various lengths of
time
depending upon the assay. Aliquots from this mixture were then added to the
amidotransferase assay mixtures either directly or following water saturated
phenol
extraction, ethanol precipitation, and resuspension in the aminoacylation
buffer.
Amidotransferase reactions. The procedure for the formation of
radiolabelled GIntRNA~'" from GlntltNA°'" was adapted from Jahn, D. et
al. ( 1990).
Unless otherwise noted, these reactions were conducted at 37°C in a
buffer consisting
CA 02279490 1999-08-03
wo ~s rcT~rs9srois6o
-43-
of 1 mM ATP, 5 mM Hepes~KOH pH 7.0, 2.5 mM KCI, 1.5 mM MgCl2, and 0.5 mM
dithiothreitol (DTT). '.Che concentration of L14C(Ln-GIutRNAc'° was 1
(M and the
concentration of Lglutamine was 1 mM. Aliquots (0 to 20 (L) from fractions
obtained
during purification of the enzyme were added and the mixture was incubated for
various lengths of timE: depending upon the assay followed by quenching with
10 (L,
3 M NaOAc, pH 5Ø The mixture was extracted with an equal volume of
water-saturated phenol and the aqueous and organic phases were separated by
centrifilgation at 15,000 x g at room temperature for 60 seconds. The aqueous
phase
was removed, 3x volumes of ethanol were added and the tRNA was precipitated at
70°C for 15 minutes. The precipitated tRNA was recovered by
centrifugation at
15,000 x g at 4°C for 15 minutes. The pellet obtained was resuspended
in 50 (L 0.01
N KOH and deaminoacylated at 65°C for 10 minutes. The base was
neutralized with
1.3 (L, 0.1 N HCl (to pH ( 6 to 7) and the solution was dried completely-under
vacuum. The dried pellet was resuspended in 3 (L double-distilled H20 and
spotted
onto a TLC place (ce:llulose, Aldrich). The front was allowed to migrate 3.5
to 5
hours in one of two solvent systems (A. 20:1:5 isopropyl alcohol:formic
acid:water or
B. 2:1:6:6 ammonia:water:chloroform:methanol). The plate was dried at
85°C,
exposed to an activated phosphoroimaging plate {(12 hours) and the image was
analyzed using MacBas v2Ø In this way, the conversion of Glu to Gln was
measured.
CA 02279490 1999-08-03
WO 9$r33925 PG"T/US98~1860
_ø4._
Example 1
characterization of DNA Fras~nent Encoding AdT
Three genes; one transcript of correct size hybridizing with three probes is
provided in Figure 3 (SEQ ID NO:1 ). Open reading frames for each of the
subunits is
provided.
Example 2
(characterization of B.subtilis AdT Protein
The amino acid sequence of B. subtilis AdT is encoded by the nucleotide
sequence of Figure 3 (SEQ ID NO:1 ).
The molecular weights of the three subunits is computed to be: 53.039 Kd, A
subunit; 53.314 Kd, B subunit; and 10.859 Kd, C subunit. The amino acid
sequences
of each of the subunits A, B and C are provided in SEQ ID NOs: 4, 6 and 8,
respectively.
The sizes of the subunits were confirmed via polyacrylamide gel
electrophoresis.
Example 3
Preparation of polyclonal antiserum containing anti-AdT antibodies
A polyclonal antiserum containing anti-AdT antibodies was obtained by
administered to rabbits recombinant AdT (trimeric protein) to rabbits
following
CA 02279490 1999-08-03
wo ~s rcrrtrs~ois6o
-as-
lmown methods. Incubation of the antiserum with a B. subtilis extract
containing AdT
protein completely inhibited AdT activity.
Using polyacrylamide gel electrophoresis, the antiserum was shown to contain
antibodies immunoreactive to the assembled AdT protein. The polycional
antisera
was significantly less imlnunoreactive to non-assembled, individual subunits.
Example 4
P rQductir,n and u~ rif cation of AdT
Table I. Activi
ell extracts
from E. coli
BL21(DE3)
harboring
various vectors.
Glutamine Recovered
Vector pMole Relative Activity
pABC 2.03 t 0.28 1.000
pA 0.02 t 0.02 0.010
pg 0.03 t 0.01 0.015
pC 0.03 t 0.02 0.017
CA 02279490 1999-08-03
WO 98/33925 PGT/US98~1860
-46-
Table II.
Purification
of Bacillus
subtilis
GIntRNAGIn
amidotransferase
over
expressed
in Escherichia
coli BL21(DE3).
PurificationTotal Total Total Specific
Step volumeprotein activity'activity Yield -fold
PurificationmL mg units unitslmg
x
103
S-100 15 190 13.5 '70 100 1
Q~sepharoseFF12 40 8.1 200 60 3
Superdex-20010 4.6 13.4 3000 99 40
MonoQ 3.5 1.0 8.3 9000 61 130
a One unit
is defined
as 1 pMole
glutamine
produced
per minute
at 37C under
the
assay conditions
described
in materials
and methods.
Example 5
Identi ins inhibitors of AdT activity
Purified amidotransferase is used in an assay to identify inhibitors of AdT
activity. The assay used to identify inhibitors of AdT activity comprises:
(a) incubating a first sample of AdT and its substrate;
(b) measuring an uninhibited reactivity of the AdT from step (a);
(c) incubating a first sample of AdT and its substrate in the presence of a
second sample comprising an inhibitor compound;
(d) measuring an inhibited reactivity of the AdT from step (c); and,
CA 02279490 1999-08-03
wo ~392s rc~r~s9sromo
-47-
(e) comparing the inhibited reactivity to the uninhibited reactivity of the
AdT.
Inhibitors of Ad'T identified using this process are utilized as
antibacterial,
antifungal and herbicidal agents.
Example 6
identification of inhibitor-resistant AdT mutants
Purified amidot~~ansferase and an identified inhibitor of AdT is used in an
assay to identify inhibitor-resistant AdT mutants. The assay used to identify
inhibitor-resistant AdT mutants comprises
(a) incubating a first sample of AdT and its substrate in the presence of a
second sample comprising an AdT inhibitor;
(b) measuring an unmutated reactivity of the AdT from step (a);
{c) incubating a first sample of a mutated AdT and its substrate in the
presence of a second s~unple comprising an AdT inhibitor;
(d) measuring a mutated reactivity of the mutated AdT from step (c); and,
(e) comparing the mutated reactivity to the unmutated reactivity of the
AdT.
Inhibitor-resisxant AdT mutants identified using this process are utilized in
the
production of cells and organisms resistant to AdT inhibitors.
CA 02279490 1999-08-03
WO 98/33925 PCT/US98/01860
-48-
Example 7
Diagnostic assays
Nucleic acids for diagnosis are obtained from cells or tissues. Genomic DNA
may be used directly for dectection or may be amplified enzymatically by using
PCR
or other amplification techniques prior to analysis. The genomic DNA can be
compared to the polynucleotide coding for amidotransferase as provided in SEQ
ID
NO:1. Deletions and insertions can be detected by a change in size of the
amplified
product in comparison to SEQ ID NO:1. Point mutations can be identified by
hybridizing amplified DNA to labeled AdT polynucleotide sequences. Perfectly
matched sequences can be distinguished from mismatched duplexes by RNASE
digestion or by differences in melting temperatures. DNA sequence differences
may
also be detected by alterations in the electrophoretic mobility of the DNA
fragments
in gels, with or without denaturing agents, or by direct DNA sequencing.
Sequence
changes at specific locations also may be revealed by nuclease protection
assays, such
as RNase and S 1 protection or a chemical cleavage method.
Cells carrying mutations or polymorphisms in the gene of the invention may
also be detected at the DNA level by a variety of techniques. For example,
RTPCR
can be used to detect mutations. It is particularly preferred to used RTPCR in
conduction with automated detection systems, such as, for example, GeneScan.
RNA
or cDNA may also be used for the same purpose, PCR or RTPCR. As an example,
PCR primers complementary to the nucleic acid encoding AdT can be used to
identify
CA 02279490 1999-08-03
WO 98/33925 PCT/US98~01860
-49-
and analyze mutations. 'these primers may be used for amplifying AdT DNA
isolated
from a sample derived from an organism. The invention also provides these
primers
with 1, 2, 3 or 4 nucleotides removed from the S' and/or the 3' end. The
primers may
be used to amplify the gene isolated from an infected individual such that the
gene
may then be subject to various techniques for elucidation of the DNA sequence.
In
this way, mutations in the AdT DNA sequence may be detected and used to
diagnose
infection and to serotypc; or classify the infectious agent.
Increased or decreased expression of AdT polynucleotide can be measured
using any of the methods well known in the art for the quantitation of
polynucleotides,
such as, for example, amplification, PCR, RTPCR, RNase protection, Northern
blotting and other hybridization methods.
In addition, a diagnostic assay in accordance with the intention for detecting
over-or under- expression of AdT protein compared to normal control tissue
samples.
Assay techniques that c,an be sued to determine levels of AdT protein, in a
sample
derived from a host are well-known to those of skill in the art. Such assay
methods
include radioimmunoassay, competitive-binding assays, Western Blot analysis
and
ELISA assays.
CA 02279490 1999-08-03
WO 98J33925 PCT/US98/01860
-50-
Example 8
Production of transformed plants
A mutated AdT-encoding DNA sequence that confers resistance to AdT
inhibitors is isolated from an inhibitor-resistant AdT mutant using
mutagenesis and
isolation techniques well known to one of skill in the art. The coding
sequence for the
mutant AdT gene is then introduced into a plant cell and whole transformed
plants are
regenerated from the transformed plant cell using a number of different
techniques
well known to those of skill in the art. The transformed plants are used in
conventional plant breeding schemes to produce new varieties of plants which
also
carry the mutant AdT gene. Crop plants carrying the mutant AdT gene are grown
in
production and an herbicide comprising an AdT inhibitor is applied to the crop
to
control weeds.
Although the present invention has been described in detail with reference to
examples above, it is understood that various modifications can be made
without
departing from the spirit of the invention. All cited references referred to
in the
application are hereby incorporated by reference.
CA 02279490 1999-08-03
WO 9$133925 PCT/US98/01860
-51
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: Soll, Dieter
(ii) TITLE OF INVENTION: GLU-TRAM AMIDOTRANSFERASE - A NOVEL
ESSENTIAL TRANSLATIONAL COMPONENT
(iii) NUMBER OF SEQUENCES: 8
(ivl CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: MORGAN, LEWIS & BOCKIUS LLP
(B) STREET: 1800 M Street, N.PJ.
(C) CITY: Washington
(D) STATE: D.C.
(E) COUNTRY: USA
(F) ZIP: 20036-5869
(el COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
~;B) COMPUTER: T_BI~! PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: PatentIn Release #1.0, Version #1.30
(vi; CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER: US Unassigned
(B) FILING CATS: 03-FEB-1998
(C) CLASSIFICATION:
(viii PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER: US 60/037,275
(B) FILING GATE: 03-FEB-1997
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: Ad,ler, Reid G.
(B) REGISTRATION NUMBER: 30,988
(C) REFERENC:E/DOCKET NUMBER: 044574-5029-WO
(ix'; TELECOMMUNICP.TION INFORMATION:
(A) TELEPHONE: 202-967-7000
(B) TELEFAX: 202-467-7176
(2) INFORMATION FOR SE:Q ID N0:1:
i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3995 base pairs
(B) TYPE: nucleic acid
(C) STRANDEI)NESS: single
( D) TOPOLOG7.': linear
(ii) MOLECULE TYPE: CDNA
(ix) FEATURE:
(A) NAME/KE'.t: CDS
(B) LOCATION: join(1..54, 58..390, 399..1866, 1870..3303, 3310
..3321, 3325..3348, 3352..3929, 3433..3471, 3975
..34(30, 3489..3495)
CA 02279490 1999-08-03
WO 98/33925 PGT/US98/01860
-52-
(xi)SEQUENCE SEQ ID
DESCRIPTION: NO:
l:
GAATTCGATCCT GTCTCAAGGCGTTTT GTTGCT TTAAAGGGCTTG TTT 48
GluPheAspPro ValSerArgArgPhe ValAla LeuLysGlyLeu Phe
1 5 10 15
TTGATATGATCA GTATTATATGACTTA ACGGAG AAATATGTGGAG GTG 96
LeuIle Ser ValLeuTyrAspLeu ThrGlu LysTyrValGlu Val
20 25 30
GATCATATGTCA CGAATTTCAATAGAA GAAGTA AAGCACGTTGCG CAC 144
AspHisMetSer ArgIleSerIleGlu GluVal LysHisValAla His
35 40 45
CTTGCAAGACTT GCGATTACTGAAGAA GAAGCA AAAATGTTCACT GAA 192
LeuA'._aArgLeu AlaIleThrGluGlu GluAla LysMetPheThr Glu
50 55 60
CAGC:'CGACAGT ATCATTTCATTTGCC GAGGAG CTTAATGAGGTT AAC 240
GlnLeuAspSer IleIleSerPheAla GluGlu LeuAsnGluVal Asn
"., 70 75
ACAGr:.AATGTG GAGCCTACAACTCAC GTGCTG AAAATGAAAAAT GTC 288
ThrAshAsnVal GluProThrThrHis ValLeu LysMetLysAsn Val
80 85 90 95
ATGAG.=~GAAGAT GAAGCGGGTAAAGGT CTTCCG GTTGAGGATGTC ATG 336
MetArgGluAsp GluAlaGlyLysGly LeuPro ValGluAspVal Met
100 105 110
AAAAA'.'GCGCCT GACCATAAAGACGGC TATATT CGTGTGCCATCA ATT 389
LysAsnAlaPro AspHisLysAspGly TyrIle ArgValProSer Ile
115 120 125
CTGGASTAAAGG AGGGACACAAGAATG TCATTA TTTGATCATAAA ATC 432
LeuAsp Arg ArgAspThrArgMet SerLeu PheAspHisLys Ile
130 135 140
ACAG~,TTAAAA CAGCTCATACATAAA AAAGAG ATTAAGATTTCT GAT 480
ThrG1;:LeuLys GlnLeuIleHisLys LysGlu IleLysIleSer Asp
145 150 155
CTGGT1'GATGAA TCTTATAAACGCATC CAAGCG GTTGATGATAAG GTA 528
LeuValAspGlu SerTyrLysArgIle GlnAla ValAspAspLys Val
160 165 170
CAAGCCTTTTTG GCATTAGATGAAGAA AGAGCG CGCGCATACGCG AAG 576
GlnAlaPheLeu AlaLeuAspGluGlu ArgAla ArgAlaTyrAla Lys
175 180 185 190
GAGCT'.'GATGAG GCGGTTGACGGCCGT TCTGAG CACGGTCTTCTT TTC 624
GluLeuAspGlu AlaValAspGlyArg SerGlu HisGlyLeuLeu Phe
195 200 205
GGTATGCCGATC GGCGTAAAAGATAAT ATCGTA ACAAAAGGGCTG CGC 672
GlyMetProIle GlyValLysAspAsn IleVal ThrLysGlyLeu Arg
210 215 220
CA 02279490 1999-08-03
WO PCT/US98~1860
98/33925
-53-
ACA TGC AGC AAA CTCGAA AACTTTGATCCG GAT 720
ACA TCC ATT ATT
TAC
ThrT::rCysSerSer LysIleLeuGlu AsnPheAspPro Asp
Ile
T_Ir
225 230 235
GCTACTGTCGTTCAG CGC:CTTCAAGAC GCTGAAGCGGTC GGA 768
ACA
ATC
AlaTh~ValValGln ArgLeuGlnAsp AlaGluAlaVal ThrIleGly
240 245 250
AAACTGAACATGGAC GAi~TTCGCCATG GGGTCATCTACA TCA 816
GAA
AAC
LysLeuAsnMetAsp GluPheAlaMet GlySerSerThr Glu Ser
Asn
255 260 265 270
GCTTACAAGCTGACG AA;4AACCCTTGG AACCTGGATACA GTTCCCGGC 864
AlaTyrLysLeuThr LysAsnProTrp AsnLeuAspThr ValProGly
275 28 0 285
GGTTCAAGCGGCGGA TC'TGCAGCTGCG GTTGCTGCGGGA GAAGTTCCG 912
GlySerSerGlyGly SerAlaAlaAla ValAlaAlaGly GluValPro
290 295 300
TTTTCTCTTGGATCT GACACAGGCGGC TCCATCCGTCAG CCGGCATCT 960
PheSe_-LeuGlySer AspThrGlyGly SerIleArgGln ProAlaSer
305 310 315
TTC':GCGGCGTTGTC GGATTAAAACCT ACATACGGACGT GTATCTCGT 1008
PheCysGlyValVal GlyLeuLysPro ThrTyrGlyArg ValSerArg
320 325 330
TACGGCCTGGTCGCA TTTGCGTCTTCA TTGGACCAAATC GGACCGATT 1056
TyrGlyLeuValAla PheAlaSerSer LeuAspGlnIle GlyProIle
335 390 345 350
ACACGTACGGTTGAG GP.TAACGCGTTT TTACTTCAAGCG ATTTCCGGC 1104
ThrArgThrValGlu As;pAsnAlaPhe LeuLeuGlnAla IleSerGly
355 360 365
GTAGACAAAATGGAC TC:TACGAGTGCA AATGTGGACGTG CCTGATTTT 1152
ValAspLysMetAsp SeerThrSerAla AsnValAspVal ProAspPhe
370 375 380
CTTTC'_"TCATTAACT GGCGACATCAAA GGACTGAAAATC GCCGTTCCG 1200
LeuSerSerLeuThr GlyAspIleLys GlyLeuLysIle AlaValPro
385 390 395
AAAGAATACCTTGGT GAAGGTGTCGGC AAAGAAGCGAGA GAATCTGTC 1248
LysGluTyrLeuGly G:LuGlyValGly LysGluAlaArg GluSerVal
400 ~ 405 910
TTGGCAGCGCTG G'PCCTTGAA CTCGGCGCTACA TGGGAAGAA 1296
AAA GGT
LeuAlaAlaLeu ValLeu Gly Thr TrpGluGlu
Lys Glu Ala
Gly
Leu
415 420 425 430
GTG CTTCCG AGTAAA TACCTG 1344
TCT CAC TAC CTG
GCG
CTT
GCG
ACA
TAT
ValSerLeuPro SerLys TyrLeu
His Tyr Leu
Ala
Leu
Ala
Thr
Tyr
435 440 445
TCA TCTGAA GGCATC 1392
TCT GCG CGC
TCA
GCG
AAC
CTT
GCA
CGC
TTT
GAC
Ser SerGlu Ala GlyIle
Ser Ala Asn Arg
Ser Leu
Ala
Arg
Phe
Asp
450 455 46;
CA 02279490 1999-08-03
WO PCT/US981~01860
98133925
-54-
TACGGCTACCGC ACAGACAACGCG GATAACCTG ATCGACCTTTACAAG 1440
TyrGlyTyrArg ThrAspAsnAla AspAsnLeu IleAspLeuTyrLys
465 470 475
CAAACGCGCGCT GAAGGTTTCGGA AATGAAGTC AAACGCCGCATCATG 1488
GlnThrArgAla GluGlyPheGly AsnGluVal LysArgArgIleMet
480 485 490
CTCGGAACGTTT GCTTTAAGCTCA GGCTACTAC GATGCGTACTACAAA 1536
LeuGlyThrPhe AlaLeuSerSer GlyTyrTyr AspAlaTyrTyrLys
495 500 505 510
AAAGCGCAAAAA GTGCGTACGTTG ATTAAGAAG GATTTCGAGGACGTA 1584
LysAlaGlnLys ValArgThrLeu IleLysLys AspPheGluAspVal
515 520 525
TTTGAAAAATAT GATGTTATTGTT GGACCGACT ACACCGACACCTGCG 1632
PheGl:,LysTyr AspValIleVal GlyProThr ThrProThrProAla
530 535 540
TTTAAAATCGGT GAAAACACGAAG GATCCGCTC ACAATGTACGCAAAC 1680
PheL_.rsIleGly GluAsnThrLys AspProLeu ThrMetTv_rAlaAsn
545 550 555
GATA'_CTTAACG ATTCCGGTCAAC CTTGCGGCG TACCGGGAATCAGGT 1728
AspIleLeuThr IleProValAsn LeuAlaAla TyrArgGluSerGly
560 565 570
GCCATGCGGTTA GCAGACGGACTT CCGCTCGGC CTGCAAATCATCGGA 1776
AlaMetArgLeu AlaAspGlyLeu ProLeuGly LeuGlnIleIleGly
575 580 585 590
AAACACTTTGAT GAAGCACTGTAT ACCGCGTTG CTCATGCATTTGAAC 1824
LysHisPheAsp GluAlaLeuTyr ThrAlaLeu LeuMetHisLeuAsn
595 600 605
AAGCAACAGACC ATCATAAAGCAA AACCTGAAC TGTAAGGGG 1866
LysGlnGlnThr IleIleLysGln AsnLeuAsn CysLysGly
610 615 620
TGAAAAGAATTG AACTTTGAAACG GTAATCGGA CTTGAAGTCCACGTT 1914
ys GluLeu AsnPheGluThr ValIleGly LeuGluVaiHisVal
625 630 635
GAGTTAAAAACA AAATCAAAAATT TTCTCAAGC TCTCCAACGCCATTC 1962
GluLeuLysThr LysSerLysIle PheSerSer SerProThrProPhe
640 645 650
GGCGCGGAGGCG AATACGCAGACA AGCGTTATT GACCTCGGATATCCG 2010
GlyAlaGluAla AsnThrGlnThr SerValIle AspLeuGlyTyrPro
655 660 665
GGCGTCCTGCCT GTTCTGAACAAA GAAGCCGTT GAATTCGCAATGAAA 2058
GlyValLeuPro ValLeuAsnLys GluAlaVal GluPheAlaMetLys
670 675 680
GCCGCTATGGCG CTCAACTGTGAG ATCGCAACG GATACGAAGTTTGAC 2106
AlaAlaMetAla LeuAsnCysGlu IleAlaThr AspThrLysPheAsp
665 690 695
CA 02279490 1999-08-03
WO -55- PGTIUS98/01860
98J33925
CGCAAAAACTAT TTCTA'TCCTGAC AACCCGAAAGCG TATCAGATTTCT 2154
ArgLysAsnTyr PheTy.rProAsp AsnProLysAla TyrGlnIleSer
700 705 710 715
CAATTTGATAAG CCAATCGGCGAA AACGGCTGGATC GAAATTGAAGTC 2202
GlnPheAspLys ProIleGlyGlu AsnGlyTrpIle GluIleGluVal
720 725 730
GGCGGCAAAACA AAACGCATCGGC ATCACGCGCCTT CATCTTGAAGAG 2250
GlyGlyLysThr LysArgIleGly IleThrArgLeu HisLeuGluGlu
735 ' 740 745
GATGCCGGAAAA CTGACGCATACG GGCGACGGCTAT TCTCTTGTTGAC 2298
AspAlaGlyLys LeuThrHisThr GlyAspGlyTyr SerLeuValAsp
750 755 760
TTCAACCGTCAA GGAACGCCGCTT GTTGAGTNCGTA TCAGAGCCGGAC 2346
PheAsnArgGln GlyThrProLeu ValGluXaaVal SerGluProAsp
765 770 775
ATCCGCACGCCG GAAGAANCGTAC GCATATCTTGAA AAGCTGAAATCC 2394
IleArgThrPro GluGluXaaTyr AlaTyrLeuGlu LysLeuLysSer
780 785 790 795
ATCA~CCAATAT ACAGGCGTTTCT GACTGTAAAATG GAAGAAGGCTCA 2442
IleIleGlnTyr ThrGlyValSer AspCysLysMet GluGluGlySer
800 805 810
CTTC:;C'~GTGAC GCCAATATCTCT CTTCGTCCGATC GGCCAAGAGGAA 2490
LeuArc~ysAsp AlaAsnIleSer LeuArgProIle GlyGlnGluGlu
8 15 820 825
TTCGGCACAAAA ACAGAATTGAAA AACTTGAACTCC TTTGCGTTTGTT 2538
PheGlyThrLys ThrGluLeuLys AsnLeuAsnSer PheAlaPheVal
830 835 840
CAAAAAGGCCTT GAGCA.TGAAGAA AAACGCCAGGAG CAGGTTCTTCTT 2586
GlnLysGlyLeu GluHisGluGlu LysArgGlnGlu GlnValLeuLeu
84~ 850 855
TCCGG~TTCTTC ATCCA.GCAAGAA ACTCGCCGTTAT GATGAAGCAACG 2634
SerGlyPhePhe IleGlnGlnGlu ThrArgArgTyr AspGluAlaThr
860 86.5 870 875
AAGAAAACCATT CTTAT'GCGTGTC AAAGAGGGATCT GACGACTACCGT 2682
LysLysThrIle LeuMeetArgVal LysGluGlySer AspAspTyrArg
880 885 890
TACTTCCCAGAG CCAGATCTAGTC GAGCTCTACATT GATGATGAATGG 2730
TyrPheProGlu ProA~~pLeuVal GluLeuTyrIle AspAspGluTrp
895 900 905
AAGGAACGCGTA AAAGC:AAGCATT CCTGAGCTTCCG GATGAGCGCCGC 2778
LysGluArgVal LysAl.aSerIle ProGluLeuPro AspGluArgArg
910 915 920
AAGCGTTATATC GAAGAGCTTGGC TTCGCTGCATAT GACGCAATGGTT 2826
LysArgTyrIle GluGluLeuGly PheAlaAlaTyr AspAlaMetVal
925 930 935
CA 02279490 1999-08-03
wo rcr~s~ois~w
~s
-5 6-
CTGACGCTGACAAAA GAAATG GCTGAT TTCTTCGAA GAAACCGTTCAA 2874
LeuThrLeuThrLv_sGluMet AlaAsp_PhePheGlu GluThrValGln
940 945 950 955
AAAGGCGCTGAAGCT AAACAA GCGTCT AACTGGCTG ATGGGTGAAGTG 2922
LysGiyAlaGluAla LysGln AlaSer AsnTrpLeu MetGlyGluVal
960 965 970
TCAGCTTACCTAAAC GCAGAA CAAAAA GAGCTTGCC GATGTTGCCCTG 2970
SerAlaTyrLeuAsn AlaGlu GlnLys GluLeuAla AspValAlaLeu
975 980 985
ACACCTGAAGGCCTT GCCGGC ATGATC AAATTGATT GAAAAAGGAACC 3018
ThrProGluGlyLeu AlaGly MetIle LysLeuIle GluLysGlyThr
990 995 1000
ATTTC'"TCTAAGATC GCGAAG AAAGTG TTTAAAGAA TTGATTGAAAAA 3066
IleSerSerLysIle AlaLys LysVal PheLysGlu LeuIleGluLys
1005 1010 1015
GGCGGCGACGCTGAG AAGATT GTGAAA GAGAAAGGC CTTGTTCAGATT 3114
GlyG1_rAspAlaGlu LysIle ValLys GluLysGly LeuValGlnIle
1020 1025 1030 1035
TCTGA~GAAGGCGTG CTTCTG AAGCTT GTCACTGAG GCGCTTGACAAC 3162
SerAspGluGlyVal LeuLeu LysLeu ValThrGlu AlaLeuAspAsn
1040 1045 1050
AATCC'_"CAATCA ATCGAA TTT AAA GGA GACCGCGCGATC 3210
GAC AAC AAA
AsnPrcGlnSer IieGluAspPhe Lys Gly AspArgAlaIle
Asn Lys
1055 1060 1065
GGCTTCCTAGTC GGACAGATTATG AAA TCC GGACAAGCCAAC 3258
GCG AAA
GlyPheLeuVal GlyGlnIleMet Lys Ser GlyGlnAlaAsn
Ala Lys
1070 1075 1080
CCGCCGATGGTC AACAAAATTCTG CTT GAA AAAAAACGC 3303
GAA ATT
ProProMetVal AsnLysIleLeu Leu Glu LysLysArg
Glu Ile
1085 1090 1095
TAATAA AGC TTT 3348
P.AA AGC TTA
CCT TGG
TAG TCA
AGG AAT
CTG
CTT
Lys Ser Arg Phe
Ser Leu Leu
Pro Leu Trp
Ser
Asn
1100 1105 1 110
TGAGATAAAGAC AAGATGAGGGCC CGA CTT ACTTCTTTGTCG 3396
AGC TCA
AspLysAsp LysMetArgAla Arg Leu ThrSerLeuSer
Ser Ser
1115 1120 1125
TTGGTTCCGGCC AAATTGGACAGC ATG TTA TCGGCTTGCGCG 3449
CCT TAA
LeuValProAla LysLeuAspSer Met Leu SerAlaCysAla
Pro
1130 113 5 1140
GTTTATCCTGAG TCAATTCTTCCT CGA GAT TGACACGGTGAT 3492
TAA AAG
ValTyrProGlu SerIleLeuPro Arg Asp HisGlyAsp
Lys
1145 1150
ATC 3495
Ile
1155
CA 02279490 1999-08-03
wo 9813392s - 5 7 - rc°r~trs9sl~ols6o
(2) INFORMATION FOR Sf~Q ID N0:2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENG'.~H: 1'_55 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE T'tPE: protein
(xi) SEQUENCE Df'sSCRIPTION: SEQ ID N0:2:
Glu Phe Asp Pro Va1 Sf=_r Arg Arg Phe Val Ala Leu Lys Gly Leu Phe
1 5 10 15
Leu Iie Ser Val Leu Tyr Asp Leu Thr Glu Lys Tyr Val Glu Val Asp
20 25 30
His Met Ser Arg Iie Ser Ile Glu Glu Val Lys His Val Ala His Leu
35 40 45
Ala Arg Leu Ala Ile Tlzr Glu Glu Glu Ala Lys Met Phe Thr Glu Gln
~0 55 60
Leu As_c Ser Ile lle S.=_r Phe Ala Glu Glu Leu Asn Glu Val Asn Thr
65 '70 75 80
Asp Asn Val Glu Pro Tinr Thr His Val Leu Lys Met Lys Asn Val Met
85 90 95
Arg Glu Asp Glu Ala Gly Lys Gly Leu Pro Val Glu Asp Val Met Lys
100 105 110
Asn Ala Pro Asp His Lys Asp Gly Tyr Ile Arg Val Pro Ser Ile Leu
115 120 125
Asp Arg Arg Asp Thr Arg Met Ser Leu Phe Asp His Lys Ile Thr Glu
130 135 140
Leu Lys Gln Leu Ile His Lys Lys Glu Ile Lys Ile Ser Asp Leu Val
145 150 155 160
Asp Glu Ser Tyr Lys Arg Ile Gln Ala Val Asp Asp Lys Val Gln Ala
165 170 175
Phe Leu Ala Leu Asp Glu Glu Arg Ala Arg Ala Tyr Ala Lys Glu Leu
180 185 190
Asp Glu Ala Val Asp Gly Arg Ser Glu His Gly Leu Leu Phe Gly Met
195 200 205
Pro Ile Gly Val Lys A.sp Asn Ile Val Thr Lys Gly Leu Arg Thr Thr
210 215 220
Cys Ser Ser Lys Ile L~eu Glu Asn Phe Asp Pro Ile Tyr Asp Ala Thr
225 230 235 290
Val Val Gln Arg Leu Gln Asp Ala Glu Ala Val Thr Ile Gly Lys Leu
245 250 255
Asn Met Asp Glu Phe P.la Met Gly Ser Ser Thr Glu Asn Ser Ala Tyr
260 265 270
CA 02279490 1999-08-03
WO 98133925 -58- PCT/US48/01860
Lys Leu Thr Lys Asn Pro Trp Asn Leu Asp Thr Val Pro Gly Gly Ser
275 280 285
Ser Gly Gly Ser Ala Ala Ala Val Ala Ala Gly Glu Val Pro Phe Ser
290 295 300
Leu Gly Ser Asp Thr Gly Gly Ser Ile Arg Gln Pro Ala Ser Phe Cys
305 310 315 320
Gly Val Val Gly Leu Lys Pro Thr Tyr Gly Arg Val Ser Arg Tyr Gly
325 330 335
Leu Val Ala Phe Ala Ser Ser Leu Asp Gln Ile Gly Pro Ile Thr Arg
340 345 350
Thr Val Glu Asp Asn Ala Phe Leu Leu Gln Ala Ile Ser Gly Val Asp
355 360 365
Lys Met Asp Ser Thr Ser Ala Asn Val Asp Val Pro Asp Phe Leu Ser
37C 375 380
Ser Le:: Thr Gly Asp Ile Lys Gly Leu Lys Ile Ala Val Pro Lys Glu
385 390 395 400
Tyr Leu Gly Glu Gly Val Gly Lys Glu Ala Arg Glu Ser Val Leu Ala
405 410 415
Ala Leu Lys Val Leu Glu Gly Leu Gly Ala Thr Trp Glu Glu Val Ser
920 425 430
Leu Pro His Ser Lys Tyr Ala Leu Ala Thr Tyr Tyr Leu Leu Ser Ser
935 490 445
Ser Glu Ala Ser Ala Asn Leu Ala Arg Phe Asp Gly Ile Arg Tyr Gly
450 455 460
Tyr Arg Thr Asp Asn Ala Asp Asn Leu Ile Asp Leu Tyr Lys Gln Thr
465 470 475 480
Arg Ala Glu Gly Phe Gly Asn Glu Val Lys Arg Arg Ile Met Leu Gly
485 490 495
Thr Phe Ala Leu Ser Ser Gly Tyr Tyr Asp Ala Tyr Tyr Lys Lys Ala
500 505 510
Gln Lys Val Arg Thr Leu Ile Lys Lys Asp Phe Glu Asp Val Phe Glu
515 520 525
Lys Tyr Asp Val Ile Val Gly Pro Thr Thr Pro Thr Pro Ala Phe Lys
530 535 590
Ile Gly Glu Asn Thr Lys Asp Pro Leu Thr Met Tyr Ala Asn Asp Ile
545 550 555 560
Leu Thr Ile Pro Val Asn Leu Ala Ala Tyr Arg Glu Ser Gly Ala Met
565 570 575
Arg Leu Ala Asp Gly Leu Pro Leu Gly Leu Gln Ile Ile Gly Lys His
580 585 590
CA 02279490 1999-08-03
wo 92s -59- rcr~rs9srois6o
Phe Asp Glu Ala Leu Tyr Thr Ala Leu Leu Met His Leu Asn Lys Gln
595 600 605
Gln Thr ile Ile Lys G:Ln Asn Leu Asn Cys Lys Gly Lys Glu Leu Asn
610 615 620
Phe Glu Thr Val Ile G:Ly Leu Glu Val His Val Glu Leu Lys Thr Lys
625 6:30 635 640
Ser Lys Ile Phe Ser Se=r Ser Pro Thr Pro Phe Gly Ala Glu Ala Asn
645 650 655
Thr Gln Thr Ser Val I:Le Asp Leu Gly Tyr Pro Gly Val Leu Pro Val
660 665 670
Leu Asn Lys Glu Ala Val Glu Phe Ala Met Lys Ala Ala Met Ala Leu
675 680 6s5
Asn Cys Glu Ile Ala Thr Asp Thr Lys Phe Asp Arg Lys Asn Tyr Phe
690 695 700
Tyr Pro Asp Asn Pro L~,rs Ala Tyr Gln Ile Ser Gln Phe Asp Lys Pro
705 7:L0 715 720
Ile G1_: Glu Asn Gly T;_p Ile Glu Ile Glu Val Gly Gly Lys Thr Lys
725 730 735
Arg Ile Gly Ile Thr A:rg Leu His Leu Glu Glu Asp Ala Gly Lys Leu
740 745 750
Thr His Thr Gly Asp G.Ly Tyr Ser Leu Val Asp Phe Asn Arg Gln Gly
755 760 765
Thr Pro Leu Val Glu Xaa Val Ser Glu Pro Asp Ile Arg Thr Pro Glu
770 775 ?80
Glu Xaa Tyr Ala Tyr L~su Glu Lys Leu Lys 5er Ile Ile Gln Tyr Thr
785 790 795 800
Gly Val Ser Asp Cys Lys Met Glu Glu Gly Ser Leu Arg Cys Asp Ala
ao5 slo s15
Asn Ile Ser Leu Arg Pro Ile Gly Gln Glu Glu Phe Gly Thr Lys Thr
820 825 830
Glu Leu Lys Asn Leu Asn Ser Phe Ala Phe Val Gln Lys Gly Leu Glu
835 840 B45
His Glu Glu Lys Arg Gln Glu Gln Val Leu Leu Ser Gly Phe Phe Ile
850 855 860
Gln Gln Glu Thr Arg Arg Tyr Asp Glu Ala Thr Lys Lys Thr Ile Leu
865 B70 875 880
Met Arg Val Lys Glu Gly Ser Asp Asp Tyr Arg Tyr Phe Pro Glu Pro
885 890 895
Asp Leu Val Glu Leu Tyr Ile Asp Asp Glu Trp Lys Glu Arg Val Lys
900 905 910
CA 02279490 1999-08-03
WO 98/33925 -60- PGT/ITS98/01860
Ala Se. Ile Pro Glu Leu Pro Asp Glu Arg Arg Lys Arg Tyr Ile Glu
915 920 925
Glu Leu Gly Phe Ala Ala Tyr Asp Ala Met Val Leu Thr Leu Thr Lys
930 935 940
Glu Met Ala Asp Phe Phe Glu Glu Thr Val Gln Lys Gly Ala Glu Ala
945 950 955 960
Lys Gin Ala Ser Asn Trp Leu Met Gly Glu Val Ser Ala Tyr Leu Asn
965 970 975
Ala Glu Gln Lys Glu Leu Ala Asp Val Ala Leu Thr Pro Glu Gly Leu
980 985 990
Ala Gl.,r Met Ile Lys Leu Ile Glu Lys Gly Thr Ile Ser Ser Lys Ile
995 1000 1005
Ala Ls Lys Val Phe Lys Glu Leu Ile Glu Lys Gly Gly Asp Ala Glu
lCiC 1015 1020
Lys I_e Val Lys Glu Lys Gly Leu Val Gln Ile Ser Asp Glu Gly Val
1025 1030 1035 1040
Leu Lw.: Lys Leu Val Thr Giu Ala Leu Asp Asn Asn Pro Gln Ser Ile
1045 1050 1055
Glu Asp Phe Lys Asn Gly Lys Asp Arg Ala Ile Gly Phe Leu Val G1y
1060 1065 1070
Gln i~_e Met Lys Ala Ser Lys Gly Gln Ala Asn Pro Pro Met Val Asn
1075 1080 1085
Lys I_e Leu Leu Glu Glu Ile Lys Lys Arg Lys Ser Ser Pro Arg Leu
1090 1095 1100
Leu P!-:e Leu Trp Ser Asn Asp Lys Asp Lys Met Arg Ala Arg Ser Leu
1105 1110 1115 1120
Ser T::= Ser Leu Ser Leu Val Pro Ala Lys Leu Asp Ser Met Pro Leu
1125 1130 1135
Ser A_a Cys Ala Val Tyr Pro Glu Ser Ile Leu Pro Arg Asp Lys His
1140 1145 1150
Gly Asp Ile
1155
(2) IN:ORMATION FOR SEQ ID N0:3:
') SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1461 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
iii) MOLECULE TYPE: cDNA
(ix) FEATURE:
(A) NAME/KEY: CDS
CA 02279490 1999-08-03
WO 98r33925 -61- PCT/US'981~01860
(B) LOCATIOPd: 1..1458
(xi)SEQUENCE ID
DESCRIPTION: N0:3:
SEQ
ATGTCA TTATTTGATCATAAA ATCACAGAATTA AAACAGCTCATA CAT 48
MetSer LeuPheAspHisLys IleThrGluLeu LysGlnLeuIie His
1 5 10 15
AAAAAA GAGATTAAGA'PTTCT GATCTGGTTGAT GAATCTTATAAA CGC 96
LysLys GluIleLysI:LeSer AspLeuValAsp GluSerTyrLys Arg
20 25 30
ATCCAA GCGGTTGATGATAAG GTACAAGCCTTT TTGGCATTAGAT GAA I94
IleGln AlaValAspAspLys ValGlnAlaPhe LeuAlaLeuAsp Glu
35 90 45
GAAAGA GCGCGCGCATi~CGCG AAGGAGCTTGAT GAGGCGGTTGAC GGC 192
GluArg AlaArgAiaT~rAla LysGluLeuAsp GluAlaValAsp Gly
0 55 60
CGTTCT GAGCACGGTC'TTCTTTTCGGTATG CCGATCGGCGTAAAA GAT 240
ArgSer GluHisGlv_LuauLeuPheG1yMet ProIleGlyValLys Asp
65 '70 75 80
AATATC GTAACAAAAGGG CTGCGCACAACA TGCTCCAGCAAAATT CTC 288
AsnIle ValThrLysG.lyLeuArgThrThr CysSerSerLysIle Leu
85 90 95
GAAAAC TTTGATCCGA'TTTACGATGCTACT GTCGTTCAGCGCCTT CAA 336
GluAsn PheAspProIle TyrAspAlaThr ValValGlnArgLeu Gln
100 105 110
GACGCT GAAGCGGTCACA ATCGGAAAACTG AACATGGACGAATTC GCC 384
AspAla GluAlaValT:hrIleGlyLysLeu AsnMetAspGluPhe Ala
115 120 125
ATGGGG TCATCTACAG.AAAACTCAGCTTAC AAGCTGACGAAAAAC CCT 432
MetGly SerSerThrGlu AsnSerAlaTyr LysLeuThrLysAsn Pro
i~u 135 140
TGGAAC CTGGATACAGTT CCCGGCGGTTCA AGCGGCGGATCTGCA GCT 480
TrpAsn LeuAspThrVal ProGlyGlySer SerGlyGlySerAla Ala
145 150 155 160
GCGGTT GCTGCGGGAGAA GTTCCGTTTTCT CTTGGATCTGACACA GGC 528
AlaVal AlaAl~aGlyGlu ValProPheSer LeuGlySerAspThr Gly
165 170 175
GGCTCC ATCCGTCAGCCG GCATCTTTCTGC GGCGTTGTCGGATTA AAA 576
GlySer IleArgGlnPro AlaSerPheCys GlyValValGlyLeu Lys
180 185 190
CCTACA TACGGACGTGTA TCTCGTTACGGC CTGGTCGCATTTGCG TCT 624
ProThr TyrGlyArgVal SerArgTyrGly LeuValAlaPheAla Ser
195 200 205
TCATTG GACCAAATCGGA CCGATTACACGT ACGGTTGAGGATAAC GCG 672
SerLeu AspGlnIleG~lyProIleThrArg ThrValGluAspAsn Ala
2i0 215 220
CA 02279490 1999-08-03
wo 9sr33ns -62- rcrrtrs9sroisso
TTTTA CTTCAA ATTTCCGGCGTA GACAAAATGGAC TCTACGAGT 720
GCG
PheLeuLeuGlnAla IleSerGlyVal AspLysMetAsp SerThrSer
225 230 235 240
GCAAATGTGGACGTG CCTGATTTTCTT TCTTCATTAACT GGCGACATC 768
AlaAsnValAspVal ProAspPheLeu SerSerLeuThr GlyAspIle
245 250 255
AAAGGACTGAAAATC GCCGTTCCGAAA GAATACCTTGGT GAAGGTGTC 816
LysGlyLeuLysIle AlaValProLys GluTyrLeuGly GluGlyVal
260 265 270
GGCAAAGAAGCGAGA GAATCTGTCTTG GCAGCGCTGAAA GTCCTTGAA 864
GlyLysGluAlaArg GluSerValLeu AlaAlaLeuLys ValLeuGlu
275 280 285
GGTCTCGGCGCTACA TGGGAAGAAGTG TCTCTTCCGCAC AGTAAATAC 912
GlyLeuGlyAlaThr TrpGluGluVal SerLeuProHis SerLysTyr
290 295 300
GCGCTTGCGACATAT TACCTGCTGTCA TCTTCTGAAGCG TCAGCGAAC 960
AlaLeuAlaThrTyr TyrLeuLeuSer SerSerGluAla SerAlaAsn
305 310 315 320
CTTGCACGCTTTGAC GGCATCCGCTAC GGCTACCGCACA GACAACGCG 1008
LeuAlaArgPheAsp GlyIleArgTyr GlyTyrArgThr AspAsnAla
325 330 335
GATAACCTGATCGAC CTTTACAAGCAA ACGCGCGCTGAA GGTTTCGGA 1056
AspAsnLeuIleAsp LeuTyrLysGln ThrArgAlaGlu GlyPheGly
340 345 350
AATGAAGTCAAACGC CGCATCATGCTC GGAACGTTTGCT TTAAGCTCA 1104
AsnGluValLysArg ArgIleMetLeu GlyThrPheAla LeuSerSer
355 360 365
GGCTACTACGATGCG TACTACAAAAAA GCGCAAAAAGTG CGTACGTTG 1152
GlyTyrTyrAspAla TyrTyrLysLys AlaGlnLysVal ArgThrLeu
370 375 380
ATTAAGAAGGATTTC GAGGACGTATTT GAAAAATATGAT GTTATTGTT 1200
IleLysLysAspPhe GluAspValPhe GluLysTyrAsp ValIleVal
385 390 395 400
GGACCGACTACACCG ACACCTGCGTTT AAAATCGGTGAA AACACGAAG 1248
GlyProThrThrPro ThrProAlaPhe LysIleGlyGlu AsnThrLys
405 410 915
GATCCGCTCACAATG TACGCAAACGAT ATCTTAACGATT CCGGTCAAC 1296
AspProLeuThrMet TyrAlaAsnAsp IleLeuThrIle ProValAsn
420 425 430
CTTGCGGCGTACCGG GAATCAGGTGCC ATGCGGTTAGCA GACGGACTT 1344
LeuAlaAlaTyrArg GluSerGlyAla MetArgLeuAla AspGlyLeu
435 440 445
CCGCTCGGCCTGCAA ATCATCGGAAAA CACTTTGATGAA GCACTGTAT 1392
ProLeuGlyLeuGln IleIleGlyLys HisPheAspGlu AlaLeuTyr
450 955 960
CA 02279490 1999-08-03
wo ~~ns -63- rc~r~s~ois6o
ACC GCG TTG CTC ATG CAT TTG AAC AAG CAA CAG ACC ATC ATA AAG CAA 1440
Thr Ala Leu Leu Met Hi_s Leu Asn Lys Gln Gln Thr Ile Ile Lys Gln
465 4''0 475 480
AAC CTG AAC TGT AAG GGG TGA 1461
Asn Leu Asn Cys Lys Gly
485
(2) INFORMATION FOR SE~Q ID N0:4:
:.'i) SEQUENCE CFiARACTERISTICS:
(A) LENGTH: 486 amino acids
(B) TYPE;: amino acid
(D) TOPOhOGY: linear
(ii) MOLECULE T'.CPE: protein
(xi) SEQUENCE DF~SCRIPTION: SEQ ID N0:4:
Met Ser Leu Phe Asp His Lys Ile Thr Glu Leu Lys Gln Leu Ile His
1 5 10 15
Lys Lys Glu Ile Lys I:Le Ser Asp Leu Val Asp Glu Ser Tyr Lys Arg
20 25 30
Ile Gin Ala Val Asp Asp Lys Val Gln Ala Phe Leu Aia Leu Asp Glu
35 40 45
Glu Arg Ala Arg Ala Tyr Ala Lys Glu Leu Asp Glu Ala Val Asp Gly
50 55 60
Arg Ser Glu His Gly Leu Leu Phe Gly Met Pro Ile Gly Val Lys Asp
65 '70 75 80
Asn Ile Val Thr Lys G.Ly Leu Arg Thr Thr Cys Ser Ser Lys Ile Leu
85 90 95
Glu Asn Phe Asp Pro I.le Tyr Asp Ala Thr Val Val Gln Arg Leu Gln
100 105 110
Asp Ala Glu Ala Val T:hr Ile Gly Lys Leu Asn Met Asp Glu Phe Ala
115 120 125
Met Gly Ser Ser Thr Glu Asn Ser Ala Tyr Lys Leu Thr Lys Asn Pro
130 135 140
Trp Asn Leu Asp Thr Val Pro Gly Gly Ser Ser Gly Gly Ser Ala Ala
145 150 155 160
Ala Val Ala Ala Gly Glu Val Pro Phe Ser Leu Gly Ser Asp Thr Gly
165 170 175
Gly Ser Ile Arg Gln Pro Ala Ser Phe Cys Gly Val Val Gly Leu Lys
180 185 190
Pro Thr Tyr Gly Arg Val Ser Arg Tyr Gly Leu Val Ala Phe Ala Ser
195 200 205
Ser Leu Asp Gln Ile Gly Pro Ile Thr Arg Thr Val Glu Asp Asn Ala
210 215 220
CA 02279490 1999-08-03
wo ~33ns -64- rc~r~rs9srois6o
Phe Leu Leu Gln Ala Ile Ser Gly Val Asp Lys Met Asp Ser Thr Ser
225 230 235 240
Ala Asn Val Asp Val Pro Asp Phe Leu Ser Ser Leu Thr Gly Asp Ile
245 250 255
Lys Gly Leu Lys Ile Ala Val Pro Lys Glu Tyr Leu Gly Glu Gly Val
260 265 270
Gly Lys Glu Ala Arg Glu Ser Val Leu Ala Ala Leu Lys Val Leu Glu
275 280 285
Gly Leu Gly Ala Thr Trp Glu Glu Val Ser Leu Pro His Ser Lys Tyr
290 295 300
Ala Leu Ala Thr Tyr Tyr Leu Leu Ser Ser Ser Glu Ala Ser Ala Asn
305 310 315 320
Leu Ala Arg Phe Asp Gly Ile Arg Tyr Gly Tyr Arg Thr Asp Asn Ala
325 330 335
Asp Asn Leu Ile Asp Leu Tyr Lys Gln Thr Arg Ala Glu Gly Phe Gly
340 345 350
Asn Glu Val Lys Arg Arg Ile Met Leu Gly Thr Phe Ala Leu Ser Ser
355 360 365
Gly Tyr Tyr Asp Ala Tyr Tyr Lys Lys Ala Gln Lys Val Arg Thr Leu
370 375 380
Ile Lys Lys Asp Phe Glu Asp Val Phe Glu Lys Tyr Asp Val Ile Val
385 390 395 900
Gly Pro Thr Thr Pro Thr Pro Ala Phe Lys Ile Gly Glu Asn Thr Lys
405 410 415
Asp Pro Leu Thr Met Tyr Ala Asn Asp Ile Leu Thr Ile Pro Val Asn
420 425 430
Leu Ala Ala Tyr Arg Glu Ser Gly Ala Met Arg Leu Ala Asp Gly Leu
435 440 495
Pro Leu Gly Leu Gln Ile Ile Gly Lys His Phe Asp Glu Ala Leu Tyr
450 455 460
Thr Ala Leu Leu Met His Leu Asn Lys Gln Gln Thr Ile Ile Lys Gln
465 470 475 480
Asn Leu Asn Cys Lys Gly
485
(2) INFORMATION FOR SEQ ID N0:5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1431 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
CA 02279490 1999-08-03
wo 9as rcr~s~ois6o
-65-
(ix) FEATURE:
(A) NAME/fCEY : CDS
(B) LOCATION:: 1..1428
(xi)SEQUENCE
DESCRIPTION:
SEQ
ID
N0:5:
TTGAAC TTTGAAACGGTA ATCGGACTTGAA GTCCACGTTGAG TTAAAA 48
LeuAsn PheGluThrVal IleGly.LeuGlu ValHisValGlu LeuLys
1 5 10 15
ACAAAA TCAAAAATTTTC TCAAGCTCTCCA ACGCCATTCGGC GCGGAG 96
ThrLys SerLysIlePhf:SerSerSerPro ThrProPheGly AlaGlu
20 25 30
GCGAAT ACGCAGACAAG(:GTTATTGACCTC GGATATCCGGGC GTCCTG 144
AlaAsn ThrGlnThrSer ValIleAspLeu GlyTyrProGly ValLeu
35 40 95
CCTGTT CTGAACAAAGAA GCCGTTGAATTC GCAATGAAAGCC GCTATG 192
ProVa'_LeuAsnLysGlu AlaValGluPhe AlaMetLysAla AlaMet
~i: 55 60
GCGCT~ AACTGTGAGAT(.GCAACGGATACG AAGTTTGACCGC AAAAAC 240
AlaLeu AsnCysGluIle AlaThrAspThr LysPheAspArg LysAsn
65 70 75 80
TATTTC TATCCTGACAAC CCGAAAGCGTAT CAGATTTCTCAA TTTGAT 288
TyrPhe TyrProAspAsn ProLysAlaTyr GlnIleSerGln PheAsp
85 90 95
AAGCCA ATCGGCGAAAAC GGCTGGATCGAA ATTGAAGTCGGC GGCAAA 336
LysPro IleGlyGluAsn GlyTrpIleGlu IleGluValGly GlyLys
100 105 110
ACAAAA CGCATCGGCATC ACGCGCCTTCAT CTTGAAGAGGAT GCCGGA 384
ThrLys ArgIleGlyIl~sThrArgLeuHis LeuGluGluAsp AlaGly
115 120 125
AAACTG ACGCATACGGGC GACGGCTATTCT CTTGTTGACTTC AACCGT 432
LysLeu ThrHisThrGl~yAspGlyTyrSer LeuValAspPhe AsnArg
130 135 140
CAAGGA ACGCCGCTTGT'TGAGTNCGTATCA GAGCCGGACATC CGCACG 480
GlnGly ThrProLeuVa.1GluXaaValSer GluProAspIle ArgThr
145 150 155 160
CCGGAA GAANCGTACGC;4TATCTTGAAAAG CTGAAATCCATC ATCCAA 528
ProGlu GluXaaTyrAla TyrLeuGluLys LeuLysSerIle IleGln
165 170 175
TATACA GGCGTTTCTGAC TGTAAAATGGAA GAAGGCTCACTT CGCTGT 576
TyrThr GlyValSerAsp CysLysMetGlu GluGlySerLeu ArgCys
180 185 190
GACGCC AATATCTCTCT'TCGTCCGATCGGC CAAGAGGAATTC GGCACA 624
AspAla AsnIleSerLeu ArgProIleGly GlnGluGluPhe GlyThr
195 200 205
CA 02279490 1999-08-03
wo -66- PCT/US98/101860
92s
AAA ACAGAATTGAAA AACTTGAACTCC TTTGCGTTTGTT CAAAAAGGC 672
Lys ThrGluLeuLys AsnLeuAsnSer PheAlaPheVal GlnLysGly
210 215 220
CTT GAGCATGAAGAA AAACGCCAGGAG CAGGTTCTTCTT TCCGGCTTC 720
Leu GluHisGluGlu LysArgGlnGlu GlnValLeuLeu SerGlyPhe
225 230 235 240
TTC ATCCAGCAAGAA ACTCGCCGTTAT GATGAAGCAACG AAGAAAACC 768
Phe IleGlnGlnGlu ThrArgArgTyr AspGluAlaThr LysLysThr
~
245 250 255
ATT CTTATGCGTGTC AAAGAGGGATCT GACGACTACCGT TACTTCCCA 816
Ile LeuMetArgVal LysGluGlySer AspAspTyrArg TyrPhePro
260 265 270
GAG CCAGATCTAGTC GAGCTCTACATT GATGATGAATGG AAGGAACGC 864
Glu ProAspLeuVal GluLeuTyrIle AspAspGluTrp LysGluArg
275 280 285
GTA AAAGCAAGCATT CCTGAGCTTCCG GATGAGCGCCGC AAGCGTTAT 912
Val LysAlaSerIle ProGluLeuPro AspGluArgArg LysArgTyr
290 295 300
ATC GAAGAGCTTGGC TTCGCTGCATAT GACGCAATGGTT CTGACGCTG 960
Ile GluGluLeuGly PheAlaAlaTyr AspAlaMetVal LeuThrLeu
305 310 315 320
ACA AAAGAAATGGCT GATTTCTTCGAA GAAACCGTTCAA AAAGGCGCT 1008
Thr LysGluMetAla AspPhePheGlu GluThrValGln LysGlyAla
325 330 335
GAA GCTAAACAAGCG TCTAACTGGCTG ATGGGTGAAGTG TCAGCTTAC 1056
Glu AlaLysGlnAla SerAsnTrpLeu MetGlyGluVal SerAlaTyr
340 345 350
CTA AACGCAGAACAA AAAGAGCTTGCC GATGTTGCCCTG ACACCTGAA 1104
Leu AsnAlaGluGln LysGluLeuAla AspValAlaLeu ThrProGlu
355 360 365
GGC CTTGCCGGCATG ATCAAATTGATT GAAAAAGGAACC ATTTCTTCT 1152
Gly LeuAlaGlyMet IleLysLeuIle GluLysGlyThr IleSerSer
370 375 380
AAG ATCGCGAAGAAA GTGTTTAAAGAA TTGATTGAAAAA GGCGGCGAC 1200
Lys IleAlaLysLys ValPheLysGlu LeuIleGluLys GlyGlyAsp
385 390 395 400
GCT GAGAAGATTGTG AAAGAGAAAGGC CTTGTTCAGATT TCTGACGAA 1248
Ala GluLysIleVal LysGluLysGly LeuValGlnIle SerAspGlu
405 410 415
GGC GTGCTTCTGAAG CTTGTCACTGAG GCGCTTGACAAC AATCCTCAA 1296
Gly ValLeuLeuLys LeuValThrGlu AlaLeuAspAsn AsnProGln
420 425 430
TCA ATCGAAGACTTT AAA GGA CGCGCGATC GGCTTCCTA 1344
AAC AAA
GAC
Ser IleGluAspPhe LysAsnGlyLys AspArgAlaIle GlyPheLeu
435 440 445
CA 02279490 1999-08-03
WO PCT/US98/01860
98133925
-67-
GTCGGA CAG ATG GCG TCC GGA GCC AAC CCG CCG ATG 1392
ATT AA;A AAA CAA
ValGly Gln Met Ala Ser Gly Ala Asn Pro Pro Met
Ile Lys Lys Gln
450 455 460
GTCAAC AAA CTG GAA GAA AAA CGC TAA 1431
ATT CT'T ATT AAA
ValAsn Lys Leu Glu Glu Lys Arg
Ile Leu Ile Lys
465 470 975
(2) INFORMATION FOR SEQ ID N0:6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 476 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:6:
Leu Asn Phe Glu Thr Val Ile Gly Leu Glu Val His Val Glu Leu Lys
1 5 10 15
Thr Lys Ser Lys Iie Phe Ser Ser Ser Pro Thr Pro Phe Gly Ala Glu
20 25 30
Ala Asn Thr Gln Thr Ser Val Ile Asp Leu Gly Tyr Pro Gly Val Leu
35 40 45
Pro Val Leu Asn Lys Glu Ala Val Glu Phe Ala Met Lys Ala Ala Met
50 55 60
Ala Leu Asn Cys Glu Ile Ala Thr Asp Thr Lys Phe Asp Arg Lys Asn
65 70 75 80
Tyr Phe Tyr Pro Asp Asn Pro Lys Ala Tyr Gln Ile Ser Gln Phe Asp
85 90 95
Lys Pro Ile Gly Glu Asn Gly Trp Ile Glu Ile Glu Val Gly Gly Lys
100 105 110
Thr Lys Arg Ile Gly Ile Thr Arg Leu His Leu Glu Glu Asp Ala Gly
115 120 125
Lys Leu Thr His Thr Gly Asp Gly Tyr Ser Leu Val Asp Phe Asn Arg
130 135 140
Gln Gly Thr Pro Leu Va.l Glu Xaa Val Ser Glu Pro Asp Ile Arg Thr
145 1°_~0 155 160
Pro Glu Glu Xaa Tyr Al.a Tyr Leu Glu Lys Leu Lys Ser Ile Ile Gln
165 170 175
Tyr Thr Gly Val Ser A~~p Cys Lys Met Glu Glu Gly Ser Leu Arg Cys
180 185 190
Asp Ala Asn Ile Ser Leu Arg Pro Ile Gly Gln Glu Glu Phe Gly Thr
195 200 205
Lys Thr Glu Leu Lys A:;n Leu Asn Ser Phe Ala Phe Val Gln Lys Gly
210 215 220
CA 02279490 1999-08-03
wo ~392s -68- rcTiUS9srois6o
Leu Glu His Glu Glu Lys Arg Gln Glu Gln Val Leu Leu Ser Gly Phe
225 230 235 240
Phe Ile Gln Gln Glu Thr Arg Arg Tyr Asp Glu Ala Thr Lys Lys Thr
245 250 255
Ile Leu Met Arg Val Lys Glu Gly Ser Asp Asp Tyr Arg Tyr Phe Pro
260 265 270
Glu Pro Asp Leu Val Glu Leu Tyr Ile Asp Asp Glu Trp Lys Glu Arg
275 280 285
Val Lys Ala Ser Ile Pro Glu Leu Pro Asp Glu Arg Arg Lys Arg Tyr
290 295 300
Ile Glu Glu Leu Gly Phe Ala Ala Tyr Asp Ala Met Val Leu Thr Leu
305 310 315 320
Thr Lys Glu Met Ala Asp Phe Phe Glu Glu Thr Val Gln Lys Gly Ala
325 330 335
Glu Aia Lys Gln Ala Ser Asn Trp Leu Met Gly Glu Val Ser Ala Tyr
340 345 350
Leu Asn Ala Glu Gln Lys Glu Leu Ala Asp Val Ala Leu Thr Pro Glu
355 360 365
Gly Leu Ala Gly Met Ile Lys Leu Ile Glu Lys Gly Thr Ile Ser Ser
370 375 380
Lys Ile Ala Lys Lys Val Phe Lys Glu Leu Ile Glu Lys Gly Gly Asp
385 390 395 400
Ala Glu Lys Ile Val Lys Glu Lys Gly Leu Val Gln Ile Ser Asp Glu
405 410 415
Gly Val Leu Leu Lys Leu Val Thr Glu Ala Leu Asp Asn Asn Pro Gln
420 425 930
Ser Ile Glu Asp Phe Lys Asn Gly Lys Asp Arg Ala Ile Gly Phe Leu
435 440 445
Val Gly Gln Ile Met Lys Ala Ser Lys Gly Gln Ala Asn Pro Pro Met
450 455 460
Val Asn Lys Ile Leu Leu Glu Glu Ile Lys Lys Arg
465 470 475
(2) INFORMATION FOR SEQ ID NO:7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 291 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
(ix) FEATURE:
CA 02279490 1999-08-03
wo zs rcTivs9srois6o
(A) NAME/KE'.f: CDS
(B) LOCATION: 1..288
-69-
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:7:
ATG TCA CGA ATT TCA A7.'A GAA GAA GTA AAG CAC GTT GCG CAC CTT GCA 48
Met Ser Arg Ile Ser Ile Glu Glu Val Lys His Val Ala His Leu Ala
1 5 10 I5
AGA CTTGCG GAA GAA GCA TTC ACTGAACAGCTC 96
ATT GAA AAA
ACT ATG
Arg LeuAlaIle ThrGl_uGluGlu AlaLysMetPhe ThrGluGlnLeu
20 25 30
GAC AGTATCATT TCATTTGCCGAG GAGCTTAATGAG GTTAACACAGAC 144
Asp SerIleIle SerPheAlaGlu GluLeuAsnGlu ValAsnThrAsp
35 40 45
AAT GTGGAGCCT ACAAC:TCACGTG CTGAAAATGAAA AATGTCATGAGA 192
Asn ValGluPro ThrThrHisVal LeuLysMetLys AsnValMetArg
50 55 60
GAA GATGAAGCG GGTAF,AGGTCTT CCGGTTGAGGAT GTCATGAAAAAT 240
Glu AspGluAla GlyLysGlyLeu ProValGluAsp ValMetLysAsn
65 TO 75 g0
GCG CCTGACCAT AAAGP~CGGCTAT ATTCGTGTGCCA TCAATTCTGGAC 288
Ala ProAspHis LysA~;pGlyTyr IleArgValPro SerIleLeuAsp
85 90 95
TAA 291
(2) INFORMATION FOR SE;Q ID N0:8:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 96 amino acids
(B) TYPE: amino acid
(D) TOPOhOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DE'~SCRIPTION: SEQ ID N0:8:
Met Ser Arg ile Ser Il.e Glu Glu Val Lys His Val Ala His Leu Ala
1 5 10 15
Arg Leu Ala Ile Thr Gl.u Glu Glu Ala Lys Met Phe Thr Glu Gln Leu
20 25 30
Asp Ser Ile Ile Ser Phe Ala Glu Glu Leu Asn Glu Val Asn Thr Asp
35 40 45
Asn Val Glu Pro Thr Th.r His Val Leu Lys Met Lys Asn Val Met Arg
50 55 60
Glu Asp Glu Ala Gly Lys Gly Leu Pro Val Glu Asp Val Met Lys Asn
65 70 75 80
Ala Pro Asp His Lys Asp Gly Tyr Ile Arg Val Pro Ser Ile Leu Asp
85 90 95