Note: Descriptions are shown in the official language in which they were submitted.
CA 02280894 1999-08-10
- WO 98/36085 PCT/US98/030b8
Production of Mature Proteins in Plants
FiE;ld of the Invention
The present invention relates to the production of mature proteins in plant
cells, and in
particular, to the production of proteins in mature secreted form.
background of the Invention
A major commercial focus of biotechnology is the recombinant production of
proteins,
including both industrial enzymes and proteins that have important therapeutic
uses.
Therapeutic proteins are convmonly produced recombinantly by microbial
expression
to systems, such as in E, coli and the yeast system S. cerevisiae. To date,
the cost of recombinant
proteins produced in a microbial host has limited the availability of a
variety of therapeutically
important proteins, such as human serum albumin (HSA) and a,l-antitrypsin
(AAT), to the extent
thavt the proteins are in short supply.
Some therapeutic proteins appear to rely on glycosylation for optimal activity
or stability,
and. the general inability of microbial systems to glycosylate or properly
glycosylate mammalian
proteins has also limited the usefulness of these recombinant expression
systems. In some cases,
proper protein folding cannot take place, because of the need for mammalian-
specifc foldases or
other folding conditions,
To some extent, protein expression in cultured mammalian cells, or in
transgenic animals
may overcome the limitations of microbial expression systems. However, the
cost per weight ratio
of the protein is still high in mammalian expression systems, and the risk of
protein contamination
by :mammalian viruses may be a significant regulatory problem. Protein
production by transgenic
animals also carries the risk of genetic variation from one generation to
another. The attendant risk
is variation in the recombinant protein produced, for example, variation in
protein processing to
yield a nature active protein with different N-terminal residue.
It would therefore be desirable to produce selected therapeutic and industrial
proteins in a
prol:ein expression system that largely overcomes problems associated with
microbial and
marnmalian-cell systems. In particular, production of the proteins should
allow large volume
production at low cost, and yield properly processed and glycosylated
proteins. Tee production
3o system should also have a relatively stable genotype from generation to
generation. These aims are
achieved, in the present invention, for the therapeutic proteins AAT, HSA, and
antithrombin III
(ATZIl7, and the industrial enzyme subtilisin BPN'.
SUBSTITUTE SHEET {RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
~um~ ~1-antitry~sin
Human at-antitrypsin (AAT) is a monomer with a molecular weight of about 52Kd.
Normal AAT contains 394 residues, with three compiex oligosaccharide units
exposed to the surface
of the
molecule, linked to asparagines 46, 83, and 247 (Carrell, P., et al., Nature
(1982) 298:329).
AAT is the major plasma proteinase inhibitor whose primary function is to
control the
proteolytic activity of trygsin, elastase, and chymotrypsin in plasma. In
particular, the protein is a
potent inhibitor of neutrophil elastase, and a deficiency of AAT has been
observed in a number of
patients with chronic emphysema of the lungs. A proportion of individuals with
serum deficiency
of AAT may progress to cirrhosis and Iiver failure (e.g., Wu, Y., et al.,
BioEssays _I~(4):163
(1991).
Because of the key role of AAT as an elastase inhibitor, and because of the
prevalence of
genetic diseases resulting in deficient serum levels of AAT, there has been an
active interest in
recombinant synthesis of AAT, for human therapeutic use. To date, this
approach has not been
satisfactory for AAT produced by recombinant methods, for the reasons
discussed above.
Human Antithrombin III
Antithrombin III (ATIII) is the major inhibitor of thrombin and factor Xa, and
to a lesser
extent, other serine proteases generated during the coagulation process, e.g.,
factors IXa, XIa, and
XIIa. The inhibitory effect of ATIII is accelerated dramatically by heparin.
In patients with a
history of deep vein thrombosis and pulmonary embolism, the prevalence of
ATIII deficiency is 2-
39~.
ATIII protein has been useful in treating hereditary ATIII deficiency and has
wide clinical
applications for the prevention of thrombosis in high risk situations, such as
surgery and delivery,
and for treating acute thrombotic episodes, when used in combination with
heparin.
ATIII is a glycoprotein with a molecular weight of 58,200, having 432 amino
acids and
containing three disulfide linkages and four asparagine-linked biantennary
carbohydrate chains.
Because of the key role of ATIII as an-anti-thrombotic agent, and because of
the broad clinical
potential in anti-thrombosis therapy, there has been an active interest in
recombinant synthesis of
3o ATIII, for human therapeutic use. To date, this approach has not been
satisfactory for ATIII
produced by microbial or mammalian recombinant methods, for the reasons
discussed above.
Human Serum Albumin
Serum albumin is the main protein component of plasma. Its main function is
regulation of
colloidal osmotic pressure in the bloodstream. Serum albumin binds numerous
ions and small
molecules, including Ca2+, Na+, K+, fatty acids, hormones, bilirubin and
certain drugs.
2
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
Human serum albumin (FISA) is expressed as a 609 amino acid prepro-protein
which is
further processed by removal of an amino-terminal peptide and an additional
six amino acid residues
to form the mature protein. The mature protein found in human serum is a
monomeric, _
unglycosylated protein 585 amino acids in length (66 kDal), with a globular
structure maintained by
i7 disulfide bonds. The pattern of disulfide links forms a structural unit of
one small and two large
disulfide-linked double loops (Geisow, M.J. et al. (197?) Biochem. J. 163:477-
484) which forms a
high-affinity bilirubin binding site.
HSA is used to expand blood volume and raise low blood protein levels in cases
of shock,
trauma, and post-surgical recovery. HSA is often administered in emergency
situations to stabilize
blood pressure.
Because of the key role of HSA as an osmotic stabilizing agent, and because of
its broad
clinical potential in, e.g., plasma replacement therapy, there has been an
active interest in
recombinant synthesis of HSA for human therapeutic use. This approach has not
been satisfactory
for HSA produced by microbial or mammalian recombinant methods, for the
reasons discussed
I5 above.
Subtilisin BPN'
Subtilisin BPN' (BPN') is an important industrial enzyme, particularly for use
as a
detergent enzyme. Several groups have reported amino acid substitution
modifications of the
enzyme that are effective in enhancing the activity, pH optimum, stability
and/or therapeutic use of
2o the enzyme.
BPN' is expressed in as a 381 amino acid preproenzyme, including 35 amino acid
sequence
required for secretion and a 77 amino acid moiety which serves as a chaperon
to facilitate folding.
Studies indicate that the pro moiety acts in trans outside of cells.
To date, large-scale production of BPN' is predominantly by microbial
fermentation, which
25 has relatively high costs associated with it. In addition, the enzyme tends
to auto-degrade at optimal
fermentation growth-medium conditions.
Summary of the Invention
In one aspect, the invention includes a method of producing, in monocot plant
cells, a
30 mature heterologous protein selected from the group consisting of (i)
mature, glycosylated ai
antitrypsin (AAT) having the same N-terminal amino acid sequence as mature AAT
produced in
humans and a glycosylation pattern which increases serum halflife
substantially over that of non
glycosylated mature AAT; (ii) mature, glycosylated antithrombin III (ATIII)
having the same N
terminai amino acid sequence as mature ATIII produced in humans; (iii) mature
human serum
35 albumin (HSA) having the same N-terminal amino acid sequence as mature HSA
produced in
3
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
humans and having the folding pattern of native mature HSA as evidenced by its
bilirubin-binding
characteristics; and (iv) mature, active subtilisin BPN' (BPN'), glycosylated
or non-glycosylated,
having the same N-terminal amino acid sequence as BPN' produced in Bacillus.
The method includes obtaining monocot cells transformed with a chimeric gene
having (i) a
monocot transcriptional regulatory region, inducible by addition or removal of
a small molecule, or
during seed maturation, (ii) a first DNA sequence encoding the heterologous
protein, and (iii) a
second DNA sequence encoding a signal peptide. The second DNA sequence is
operably linked to
the transcriptional regulatory region and to the first DNA sequence. The first
DNA sequence is in
translation-frame with the second DNA sequence, and the two sequences encode a
fusion protein.
The transformed cells are cultivated under conditions effective to induce the
transcriptional
regulatory region, thereby promoting expression of the fusion protein and
secretion of the mature
heterologous protein from the transformed cells. The mature heterologous
protein produced by the
transformed cells is then isolated.
In one embodiment of the method, the first DNA sequence encodes pro-subtilisin
BPN'
(proBPN'), the cultivating includes cultivating the transformed cells at a pH
between 5 and 6, and
the isolating step includes incubating the proBPN' to under condition
effective to allow its
autoconversion to active mature BPN'. In another embodiment, the first DNA
sequence encodes
mature BPN', and the cells are transformed with a second chimeric gene
containing (i) a transcript-
ional regulatory region inducible by addition or removal of a small molecule,
(ii) a third DNA
sequence encoding the pro-peptide moiety of BPN', and (iii) a fourth DNA
sequence encoding a
signal polypeptide. The fourth DNA sequence is operabiy linked to the
transcriptional regulatory
region and to the third DNA sequence, and the signal polypeptide is in
translation-frame with the
pro-peptide moiety and is effective to facilitate secretion of expressed pro-
peptide moiety from the
transformed cells. The cultivating step includes cultivating the transformed
cells at a pH between 5
and 6, and the isolating step includes incubating the mature BPN' and the pro-
moiety under
conditions effective to allow the conversion of BPN' by the pro- moiety to
active mature BPN'.
In another embodiment of the method, the signal peptide is the lZAmy3D signal
peptide
(SEQ )D NO:1) or the RAmylA signal peptide (SEQ ID N0:4). The coding sequence
of the signal
peptide may be a codon-optimized sequence, such as the codon-optimized RAmy3D
sequence
identified as SEQ ID N0:3. The first DNA sequence may also be codon-optimized.
Exemplary
codon-optimized signal peptide-heterologous protein fusion protein coding
sequences include 3D-
AAT (SEQ ID N0:18), 3D-ATIII (SEQ ID NO:I9), and 3D-HSA (SEQ ID N0:20). The
first
DNA sequence may further contain codon substitutions which eliminate one or
more potential
glycosylation sites present in the native amino acid sequence of the
heterologous protein, such as the
colon-optimized sequence encoding 3D-proBPN' (SEQ ID N0:21).
4
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98103068
- In other embodiments of the method, the transcriptional regulatory region
may be a
promoter derived from a rice or barley ~,-amylase gene, .including RAmyIA,
ltAmylB, l2Amy2A,
RAmy3A, RAmy3B, RAmy3C, RAmy3D, RAmy3E, pM/C, gKAmy141, gKAmy155, Amy32b, or
HV 18. The chimeric gene may further include, between the transcriptional
regulatory region and the
fusion protein coding sequence, the 5' untranslated region (5' UTR) of an
inducible monocot gene
such as one of the rice or barley ~,-amylase genes described above. One
preferred 5' UTR is that
from the RAmylA gene, which is effective to enhance the stability of the gene
transcript. The
chimeric gene may further include, downstream of the coding sequence, the 3'
untranslated region
(3' UTR) from an inducible monocot gene, such as one of the rice or barley a-
amylase genes
to mentioned above. One preferred 3' UTR is from the RAmyIA gene.
Where the method is employed in protein production in a monocot cell culture,
preferred
promoters are the RAmy3D and RAmy3E gene promoters, which are upregulated by
sugar
depletion in cell culture. Where the gene is employed in grotein production in
germinating seeds, a
preferred promoter is the RAmyIA gene promoter, which is upregulated by
gibbereIlic acid during
seed germination. Where gene is upregulated during seed maturation, a
preferred promoter is the
barley endosperm-specific B1-hordein promoter.
The invention also includes a mature heterologous protein produced by the
above method.
The protein has a glycosylation pattern characteristic of the monocot plant in
which the protein is
produced. The glycosyated protein is selected from the group consisting of (i)
mature glycosylated
o,1-antitrypsin (AAT) having the same N-terminal amino acid sequence as mature
AAT produced in
humans and having a glycosylation pattern which increases serum halflife
substantially over that of
non-glycosylated mature AAT; (ii) mature glycosylated antithrombin III (ATIII)
having the same N-
terminal amino acid sequence as mature ATIII produced in humans; and (iii}
mature glycosylated
subtilisin BPN' {BPN') having the same N-terminal amino acid sequence as BPN'
produced in
Bacillus.
The invention also includes plant cells and seeds capable of producing the
mature
heterologous proteins according to the above method. '
These and other objects and features of the invention will be more fully
understood when
the following detailed description of the invention is read in conjunction
with the accompanying
3o drawings.
Brief Description of the Figures
Fig. 1 shows, in the lower row, the amino acid sequence of a RAmy3D signal
sequence
portion employed in the invention, identified as SEQ ID NO:1; in the middle
row, the
corresponding native coding sequence, identified as SEQ ID N0:2; and in the
upper row, a
corresponding codon-optimized sequence, identified as SEQ ID N0:3;
5
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCTNS98/03068
Fig. 2 illustrates the components of a chimeric gene constructed in accordance
with an
embodiment of the invention;
Figs. 3A and 3B illustrate the construction of an exemplary transformation
vector for use in
transforming a monocot plant, for production of a mature protein in cell
culture in accordance with
one embodiment of the invention (native mature AAT coding sequence under
control of the
RAmy3D promoter and signal sequence);
Fig. 4 illustrates factors in the metabolic regulation of AAT production in
rice cell culture;
Fig. 5 shows immunodetection of AAT using antibody raised against the C-
terminal regioa
of AAT;
l0 Fig. 6 shows Western blot analysis of AAT produced by transformed rice cell
lines 18F,
11B, and 27F;
Fig. 7 shows the time course of elastase:AAT complex formation in human and
rice-
produced forms of AAT;
Fig. 8 shows an N-terminal sequence for mature ~,I-antitrypsin (AAT) produced
in
accordance with the invention, identified herein as SEQ ID N0:22;
Fig. 9 shows a Western blot of ATIII produced in accordance with the
invention;
Fig. 10 shows a Western blot of Flant-produced BPN', comparing expression from
codon-
optimized and native coding sequences;
Fig. 11 compares the specific activity of BPN' codon-optimized (AP106} vs.
BPN' native
(AP101) expression in rice callus cell culture; and
Fig. 12 shows a western blot of HSA produced in germinating seeds in
accordance with the
invention.
Brief Description of the Sequences
SEQ ID NO:1 is the amino acid sequence of the RAmy3D signal peptide;
SEQ ID N0:2 is the native sequence encoding the RAmy3D signal peptide;
SEQ iD N0:3 is a codon-optimized sequence encoding the RAmy3D signal peptide;
SEQ ID N0:4 is the amino acid sequence of the RAmylA signal peptide;
SEQ ID N0:5 is the 5' UTR derived from the RAmylA gene;
SEQ ID N0:6 is the 3' UTR derived from the RAmyIA gene;
SEQ ID N0:7 is the amino acid sequence of mature ~,1-antitrypsin (AAT);
SEQ ID N0:8 is the native DNA coding sequence of mature AAT;
SEQ ID N0:9 is the amino acid sequence of mature antithrombin III (ATII>7;
SEQ ID NO:IO is the native DNA coding sequence of mature ATIII;
SEQ ID NO:11 is the amino acid sequence of mature human serum albumin (HSA);
6
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
SEQ ID N0:12 is the native DNA coding sequence of mature IiSA;
SEQ ID N0:13 is the amino acid sequence of native proBPN';
SEQ ID N0:14 is the native DNA coding sequence of proBPN';
SEQ ID NO:IS is the amino acid sequence of the "pro" moiety of
BPN';
SEQ ID N0:16 is the amino acid sequence of native mature BPN';
SEQ ID N0:17 is the amino acid sequence of a mature BPN' variant
in which alI potential
N-glycosylation sites are removed according to Table 2;
SEQ ID N0:18 is a colon-optimized sequence encoding the RAmy3D
signal
sequence/mature ai-antitrypsin fusion protein;
SEQ ID N0:19 is a sequence encoding the RAmy3D signal sequence/mature
antithrombin
III fusion protein, with a colon-optimized RAmy3D coding sequence
fused to the native mature
ATIII coding sequence;
SEQ ID N0:20 is a sequence encoding the RAmy3D signal sequence/mature
human serum
albumin fusion protein, with a colon-optimized lRAmy3D coding
sequence fused to the native
mature HSA coding sequence;
SEQ ID N0:21 is a colon-optimized sequence encoding the RAmy3D
signal
sequence/prosubtilisin BPN' fusion protein;
SEQ ID N0:22 is the N-terminal sequence of mature ~,,-antitrypsin
produced in accordance
with the invention;
2o SEQ ID N0:23 is an oligonucleotide used to prepare the intermediate
p3DProSig construct
of Example l;
SEQ ID N0:24 is the complement of SEQ ID N0:23;
SEQ iD N0:25 is an oligonucleotide used to prepare the intermediate
p3DProSigENDlink
construct of Example 1;
SEQ ID N0:26 is the complement of SEQ ID N0:25;
SEQ ID N0:27 is one of six oligonucleotides used to prepare the
intermediate p lAProSig
construct of Example 1;
SEQ ID NO:28 is one of six oligonucleotides used to prepare the
intermediate plAProSig
construct of Example 1;
3o SEQ ID N0:29 is one of six oligonucleotides used to prepare the
intermediate plAProSig
construct of Example 1;
SEQ ID N0:30 is one of six oligonucleotides used to prepare the
intermediate plAProSig
construct of Example l;
SEQ ID N0:31 is one of six oligonucleotides used to prepare the
intermediate plAProSig
construct of Example 1;
7
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
SEQ ID N0:32 is one of six oligonucleotides used to prepare the intermediate
plAProSig
construct of Example 1; ,
SEQ ID N0:33 is the N-terminal primer used to PCR-amplify the AAT coding
sequence
according to Example 1; and
SEQ ID N0:34 is the C-terminal primer used to PCR-amplify the AAT coding
sequence
according to Example 1.
Detailed Description of the Invention
I. Definitions:
The terms below have the following meaning, unless indicated otherwise in the
specif cation.
"Cell culture" refers to cells and cell clusters, typically callus cells,
growing on or
suspended in a suitable growth medium.
"Germination" refers to the breaking of dormancy in a seed and the resumption
of metabolic
activity in the seed, including the production of enzymes effective to break
down starches in the
seed endosperm.
"Inducible" means a promoter that is upregulated by the presence or absence of
a small
molecules. It includes both indirect and direct inducement.
"Inducible during germination" refers to promoters which are substantially
silent but not
totally silent prior to germination but are turned on substantially (greater
than 25 % ) during
germination and development in the seed. Examples of promoters that are
inducible during
germination are presented below.
"Small molecules", in the context of promoter induction, are typically small
organic or
bioorganic molecules less than about 1 kDal. Examples of such small molecules
include sugars,
sugar-derivatives (including phosphate derivatives), and plant hormones (such
as, gibberellic or
absissic acid).
"Specifically regulatable" refers to the ability ~of a small molecule to
preferentially affect
transcription from one promoter or group of promoters (e.g., the a-amylase
gene farnity), as
opposed to non-specific effects, such as, enhancement or reduction of global
transcription within a
cell by a small molecule.
"Seed maturation" or "grain development" refers to the period starting with
fertilization in
which metabolizable reserves, e.g., sugars, oligosaccharides, starch,
phenolics, amino acids, and
proteins, are deposited, with and without vacuole targeting, to various
tissues in the seed (grain),
e.g., endosperm, tests, aleurone layer, and scutellar epithelium, leading to
grain enlargement, grain
filling, and ending with .grain desiccation.
8
SUBSTITUTE SHEET (RULE 2fij
CA 02280894 1999-08-10
WO 98/36085 PCT/CTS98/03068
"Inducible during seed maturation" refers to promoters which are turned on
substantially
(greater than 25~Y) during seed maturation.
"Heterologous DNA" or "foreign DNA" refers to DNA which has been introduced
into
plant cells from another source, or which is from a plant source, including
the same plant source,
but which is under the control of a promoter or terminator that does not
normally regulate
expression of the heterologous DNA.
"Heterologous protein" is a protein, including a polypeptide, encoded by a
heteroiogous
DNA. A "transcription regulatory region" or "promoter" refers to nucleic acid
sequences that
influence and/or promote initiation of transcription. Promoters are typically
considered to include
to regulatory regions, such as enhancer or inducer elements.
A "chimeric gene," in the context of the present invention, typically
comprises a promoter
sequence operably linked to DNA sequence that encodes a heterologous gene
product, e.g., a
selectable marker gene or a fusion protein gene. A chimeric gene may also
contain further
transcription regulatory elements, such as transcription termination signals,
as well as translation
regulatory signals, such as, termination codons.
"Operably linked" refers to components of a chimeric gene or an expression
cassette that
function as a unit to express a~ heterologous protein. For example, a promoter
operably linked to a
heterologous DNA, which encodes a protein, promotes the production of
functional mRNA
corresponding to the heterologous DNA.
2o A "product" encoded by a DNA molecule includes, for example, RNA molecules
and
polypeptides.
"Removal" in the context of a metabolite includes both physical removal as by
washing and
the depletion of the metabolite through the absorption and metabolizing of the
metabolite by the
cells.
"Substantially isolated" is used in several contexts and typically refers to
the at least partial
purification of a protein or polypeptide away from unrelated or contaminating
components.
Methods and procedures for the isolation or purification of proteins or
polypeptides are known in,
the art.
"Stably transformed" as used herein refers to a cereal cell or plant that has
foreign nucleic
acid stably integrated into its genome which is transmitted through multiple
generations.
"al-antitrypsin or "AAT" refers to the protease inhibitor which has an amino
acid sequence
substantially identical or homologous to AAT protein identified by SEQ ID
N0:7.
"Antithrombin III" or "ATIII" refers to the heparin-activated inhibitor of
thrombin and
factor Xa, and which has an amino acid sequence substantially identical or
homologous to AT1II
protein identified by SEQ ID N0:9.
9
SU9ST1TUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
- "Human serum albumin" or "HSA" refers to a protein which has an amino acid
sequence
substantially identical or homologous to the mature HSA protein identified by
SEQ ID NO:11.
"Subtilisin" or "subtilisin BPN"' or "BPN"' refers to the protease enzyme
produced
naturally by B. amyloliquefaciens, and having the sequence of SEQ ID N0:16, or
a sequence
homologous therewith.
"proBPN"' refers to a form of BPN' having an approximately 78 amino-acid "pro"
moiety
that functions as a chaperon polypeptide to assist in folding and activation
of the BPN', and having
the sequence in SEQ ID N0:13, or a sequence homologous therewith.
"Codon optimization" refers to changes in the coding sequence of a gene to
replace native
to codons with those corresponding to optimal codons in the host plant.
A DNA sequence is "derived from" a gene, such as a rice or barley a,-amylase
gene, if it
corresponds in sequence to a segment or region of that gene. Segments of genes
which may be
derived from a gene include the promoter region, the S' untranslated region,
and the 3' untranslated
region of the gene.
II. Transformed plant cell
The plants used in the process of the present invention are derived from
monocots,
particularly the members of the taxonomic family known as the Gramineae. This
family includes all
members of the grass family of which the edible varieties are known as
cereals. The cereals include
a wide variety of species such as wheat (Trincum sps.), rice (Oryza sps.)
barley (Hordeum sps.)
oats, (Avena sps.) rye (Secale sps.), corn (Zea sps.) and millet (Pennisettum
sps.). In the present
invention, preferred family members are rice and barley.
Plant cells or tissues derived from the members of the family are transformed
with
expression constructs (i.e., plasmid DNA into which the gene of interest has
been inserted) using a
variety of standard techniques (e.g., electroporation, protoplast fusion or
microparticle
bombardment}. The expression construct includes a transcription regulatory
region (promoter)
whose transcription is specifically upregulated by the piesence of absence of
a small molecule, such
as the reduction or depletion of sugar, e.g., sucrose, in culture medium, or
in plant tissues, e.g.,
germinating seeds. In the gresent invention, particle bombardment is the
preferred transformation
procedure.
The construct also includes a gene encoding a mature heterologous protein in a
form
suitable for secretion from plant cells. The gene encoding the recombinant
heterologous protein is
placed under the control of a metabolically regulated promoter. Metabolically
regulated promoters
are those in which mRNA synthesis or transcription, is repressed or
upregulated by a small
3s metabolite or hormone molecule, such as the rice RAmy3D and I:ZAmy3E
promoters, which are
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WD 98/36085 PCTlUS98/03068
upregulated by sugar-depletion in. cell culture. For protein production in
germinating seeds from
regenerated transgenic plants, a preferred promoter is the_Ramy lA promoter,
which is up-regulated
by gibberellic acid during seed germination. The expression construct also
utilizes additional_
regulatory DNA sequences e.g., preferred codons, termination sequences, to
promote efficient
translation of AAT, as will be described.
A. Plant Expression Vector
Expression vectors for use in the present invention comprise a chimeric gene
(or expression
cassette), designed for operation in plants, with companion sequences upstream
and downstream
to from the expression cassette. The companion sequences will be of plasmid or
viral origin and
provide necessary characteristics to the vector to permit the vectors to move
DNA from bacteria to
the desired plant host. Suitable transformation vectors are described in
related application PCT WO
95/14099, published May 25, 1995, which is incorporated by reference herein.
Suitable
components of the expression vector, including an inducible promoter, coding
sequence for a signal
peptide, coding sequence for a mature heterologous protein, and suitable
termination sequences are
discussed below. One exemplary vector is the p3D(AAT)v1.0 vector illustrated
in Figs 3A and 3B.
Al. Promoters
The transcription regulatory or promoter region is chosen to be regulated in a
manner
allowing for induction under selected cultivation conditions, e.g., sugar
depletion in culture or
water uptake followed by gibberellic acid production in germinating seeds.
Suitable promoters, and
their method of selection are detailed in above-cited PCT application WO
95/14099. Examples of
such promoters include those that transcribe the cereal o,-amylase genes and
sucrose synthase genes,
and are repressed or induced by small molecules, Iike sugars, sugar depletion
or phytohormones
such as gibberellic acid or absissic acid. Representative promoters include
the promoters from the
rice ~,-amylase RAmyIA, RAmyIB, RAmy2A, RAmy3A, RAmy3B, RAmy3C, RAmy3D, and
RAmy3E genes, and from the pM/C, gKt1my141, gKAmy155, Amy32b, and HVl8 barley
a-
amylase genes. These promoters are described, for example, in ADVANCr~S IN
PLANT
$IpTECFiIQOLOGY Ryu, D.D.Y., et al, Eds., Elsevier, Amsterdam, 1994, p.37, and
references cited
therein. Other suitable promoters include the sucrose synthase and sucrose-6-
phosphate-synthetase
(SPS) promoters from rice and barley.
Other suitable promoters include promoters which are regulated in a manner
allowing for
induction under seed-maturation conditions. Examples of such promoters include
those associated
with the following monocot storage proteins: rice glutelins, oryzins, and
prolamines, barley
hordeins, wheat gliadins and glutelins, maize zeros and glutelins, oat
glutelins, and sorghum
11
SU~STITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
- WO 98/36085 PCT/US98/03068
kafirins, millet pennisetins, and rye secaiins. _
A preferred promoter for expression in germinating seeds is the rice a-amylase
RAmyIA
promoter, which is upregulated by gibberellic acid. Preferred promoters for
expression in cell
culture are the rice a-amylase RAmy3D and RAmy3E promoters which are strongly
upregulated by
sugar depletion in the culture. These promoters are also active during seed
germination. A
preferred promoter for expression in maturing seeds is the barley endosperm-
specific Bl-hordein
promoter (Brandt, A., et al., (1985) Carlsberg Res. Commun. 50:333-345}.
The chimeric gene may further include, between the promoter and coding
sequences, the 5'
untransiated region (5' UTR) of an inducible monocot gene, such as the 5' UTR
derived from one
to of the rice or barley a-amylase genes mentioned above. One preferred 5' UTR
is that derived from
the RAmylA gene, which is effective to enhance the stability of the gene
transcript. This 5' UTR
has the sequence given by SEQ ID NO:S herein.
A2. Signal Sequgnces
In addition to encoding the protein of interest, the chimeric gene encodes a
signal sequence
(or signal peptide) that allows processing and translocation of the protein,
as appropriate. Suitable
signal sequences are described in above-referenced PCT application WO
95/14099. One preferred
signal sequence is identified as SEQ ID NO:1 and is derived from the RAmy3D
promoter. Another
preferred signal sequence is identified as SEQ ID N0:4 and is derived from the
RAmyIA promoter.
The plant signal sequence is placed in frame with a heterologous nucleic acid
encoding a mature
protein, forming a construct which encodes a fusion protein having an N-
terminal region
correspanding to the signal peptide and, immediately adjacent to the C-
terminal amino acid of the
signal peptide, the N-terminal amino acid of the mature heterologous protein.
The expressed fusion
protein is subsequently secreted and processed by signal peptidase cleavage
precisely at the junction
of the signal peptide and the mature protein, to yield the mature heterologous
protein.
In another embodiment of the invention, the coding sequence in the fusion
protein gene, in
at least the coding region for the signal sequence, may tie colon-optimized
for optimal expression in
plant cells, e.g., rice cells, as described below. The upper row in Fig. 1
shows one codon-
optimized coding sequence for the RAmy3D signal sequence, identified herein as
SEQ ID N0:3.
A3. Naturally-Occurring Heterologous Protein Coding~g"q ~ n~P~
(i) ~1-Anti sin: Mature human AAT is composed of 394 amino acids, having the
sequence identified herein as SEQ iD N0:7. The protein has N-glycosyiation
sites at asparagines
46, 83 and 247. The corresponding native DNA coding sequence is identified
herein as SEQ ID
N0:8.
12
SU9ST1TUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
- (ii) Antithrombin III: Mature human ATIII is composed of 432 amino acids,
having the
sequence identified herein as SEQ ID N0:9. The protein has N-glycosylation
sites at the four
asparagine residues 96, 135, 155, and 192. The corresponding native DNA coding
sequence is
identified herein as SEQ ID NO:10.
(iii) Human serum albumin: Mature HSA as found in human serum is composed of
585
amino acids, having the sequence identified herein as SEQ ID N0:11. The
protein has no N-linked
glycosylation sites. The corresponding native DNA coding sequence is
identified herein as SEQ ID
N0:12.
(iv) Subtilisin BPN': Native proBPN' as produced in B. amyloliquefaciens is
composed of
l0 352 amino acids, having the sequence identified herein as SEQ 117 N0:13,
The corresponding native
DNA coding sequence is identifed herein as SEQ ID NO: i4. The proBPN'
polypeptide contains a
77 amino acid "pro" moiety which is identified herein as SEQ ID N0:15. The
remainder of the
polypeptide, which forms the mature active BPN', is a 275 amino acid sequence
identified herein by
SEQ ID NO: i6. Native BPN' as produced in Bacillus is not glycosylated.
A4. Codon-Optimized Coding Sequences
In accordance with one aspect of the invention, it has been discovered that a
severalfold
enhancement of expression level can be achieved in plant cell culture by
modifying the native
coding sequence of a heterologous gene by contain predominantly or
exciusively, highest-frequency
codons found in the plant cell host.
The method will be illustrated for expression of a heterologous gene in rice
plant cells, it
being recognized that the method is generally applicable to any monocot. As a
first step, a
representative set of known coding gene sequence from rice is assembled. The
sequences are then
analyzed for codon frequency for each amino acid, and the most frequent codon
is selected for each
amino acid. This approach differs from earlier reported codon matching
methods, in which more
than one frequent codon is selected for at least some of the amino acids. The
optimal codons
selected in this manner for rice and barley are shown in-Table 1.
Table 1
Amino Acid Rice Preferred Codon Barley Preferred
Codon
I
AIa A GCC
Arg R CGC
Asn N AAC
13
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
~ ,
Amino Acid Rice Preferred Barley Preferred
Codon Codon
Asp D GAC
Cys C UGC
GIn Q CAG
Glu E GAG
Gly G GGC
His H CAC
Ile I AUC
Leu L CUC
Lys K AAG
Phe F UUC
Pro P CCG CCC
Ser S AGC UCC
Thr T ACC
TYr Y UAC
Val V GUC GUG
stop UAA UGA
As indicated above, the fusion protein coding sequence in the chimeric gene is
constructed
such that the final (C-terminal) codon in the signal sequence is immediately
followed by the codon
for the N-terminal amino acid in the mature form of the heterologous protein.
Exemplary fusion
protein genes, in accordance with the present invention, are identified herein
as follows:
SEQ ID N0:18, corresponding to codon-optimized coding sequences of the fusion
protein
consisting of RAmy3D signal sequence/mature a,-antitrypsin;
SEQ ID NO:I9, corresponding to the fusion protein coding sequence consisting
of the
l0 codon-optimized RAmy3D signal sequence and the native mature antithrombin
III sequence;
SEQ ID N0:20, corresponding to the fusion protein coding sequence consisting
of the
codon-optimized RAmy3D signal sequence and the native mature human serum
albumin sequence;
SEQ ID N0:2I, corresponding to codon-optimized coding sequence of the fusion
protein
RAmy3D signal sequence/prosubtilisin BPN'. In this instance, prosubtilisin is
considered the
"mature" protein, in that secreted prosubtilisin can autocatatyze to active,
mature subtilisin.
In a preferred embodiment, the BPN' coding sequence is further modified to
eliminate
I4
SU~STlTUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98136085 PCT/US98/03068
potential N-glycosylation sites, as native BPN' is not glycosylated. Table 2
illustrates preferred -
codon substitutions, which eliminate all potential N-glycosylation sites in
subtilisin BPN'. SEQ ID
N0:17 corresponds to a mature BPN' amino acid sequence containing the
substitutions presented in
Table 2.
Table z
N Glycosylation Location (Asn) (in Amino Acid
Sites mature Substitution
protein)
Asn Asn Ser 61 Thr Asn Ser
Asn Asn Ser 76 Thr Asn Ser
Asn Met Ser 123 Thr Met Ser
Asn Gly Thr 2I8 Ser Gly Thrt
Asn Trp Thr 240 Thr Trp Thr
'improved thcrmostability; Bryan, et al., Proteiru: Structure, Function, and
Genetics 1:326 (1986).
A5. Transcription and Translation Terminators
The chimeric gene may also include, downstream of the coding sequence, the 3'
untranslated region (3' UTR) from an inducible monocot gene, such as one of
the rice or barley a-
amylase genes mentioned above. One preferred 3' UTR is that derived from the
RAmylA gene,
whose sequence is given by SEQ ID N0:6. This sequence includes non-coding
sequence 5' to the
polyadenylation site, the polyadenylation site, and the transcription
termination sequence. The
transcriptional termination region may be selected, particularly for stability
of the mRNA to
enhance expression. Polyadenylation tails (Alber and Kawasaki, 1982, Mol. and
Appl. Genet.
_1:419-434) are also commonly added to the expression cassette to optimize
high levels of
2Q transcription and proper transcription termination, respectively.
Polyadenylation sequences include
but are not limited to the Agrobacterium octopine synthetase signal (Gielen,
et al., EMBO J. ~:835-
846 (I984) or the nopaline synthase of the same species (Depicker, et al.,
Mol. Appl. Genet. _1:561-
573 (1982).
Since the ultimate expression of the heterologous protein will be in a
eukaryotic cell (in this
case, a member of the grass family), it is desirable to determine whether any
portion of the cloned
gene contains sequences which will be processed out as introns by the host's
splicing machinery. If
so, site-directed mutagenesis of the "intron" region may be conducted to
prevent losing a portion of
the genetic message as a false intron code (Reed and Maniatis, Cell 41:95-105
(1985).
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/IJS98/03068
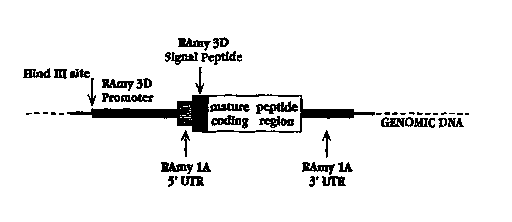
Fig. 2 shows the elements of one preferred chimeric gene constructed in
accordance with
the invention, and intended particularly for use in protein expression in a
rice cell suspension
culture. The gene includes, in a 5' to 3' direction, the promoter from the
RAmy3D gene, which is
inducible in cell culture with sugar depletion, the 5' UTR from the RAmylA
gene, which confers
enhanced stability on the gene transcript, the RAmy3D signal sequence coding
region, as identified
above, the coding region of a heterologous protein to be produced, and a 3'
UTR region from the
RAmyIA gene.
BI. Plant Transformation
For transformation of plants, the chimeric gene is placed in a suitable
expression vector
designed for operation in plants. The vector includes suitable elements of
plasmid or viral origin
that provide necessary characteristics to the vector to permit the vectors to
move DNA from bacteria
to the desired plant host. Suitable transformation vectors are described in
related application PCT
WO 95/14099, published May 25, 1995, which is incorporated by reference
herein. Suitable
components of the expression vector, inciuding the chimeric gene described
above, are discussed
below. One exemplary vector is the p3Dv1.0 vector described in Example 1.
A. Transformation Vector
Vectors containing a chimeric gene of the present invention may also include
selectable
markers for use in plant cells (such as the nptIl kanamycin resistance gene,
for selection in
kanamycin-containing or the phosphinothricin acetyltransferase gene, for
selection in medium
containing phosphinothricin (PPT).
The vectors may also include sequences that allow their selection and
propagation in a
secondary host, such as sequences containing an origin of replication and a
selectable marker such
as antibiotic or herbicide resistance genes, e.g., FiPH (Hagio et al., Planr
Cell Reports ,~:329
(1995); van der Elzer, Plant Mol. Biol. x:299-302 (/985). Typical secondary
hosts include bacteria
and yeast. In one embodiment, the secondary host is ~scherichia coli, the
origin of replication is a
colEl-type, and the selectable marker is a gene encoding ampicillin
resistance. Such sequences are
well known in the art and are commercialiy available as well (e.g., Clontech,
Palo Alto, CA;
3o Stratagene, La Jolla, CA).
The vectors of the present invention may also be modified to intermediate
plant
transformation plasmids that contain a region of homology to an Agrobacterium
tumefaciens vector,
a T-DNA border region from Agrobacterium tumefaciens, and chimeric genes or
expression
cassettes (described above). Further, the vectors of the invention may
comprise a disarmed plant
tumor inducing plasmid of Agrobacterium tumefaciens.
16
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98!36085 PCT/US98/03068
The vector described in Example I, and having a promoter from the RAmy3D gene,
is
suitable for use in a method of mature protein productionin cell culture,
where the RAmy3D
promoter is induced by sugar depletion in cell culture medium. Other promoters
may be selected
for other applications, as indicated above. For example, for mature protein
expression in
germinating seeds, the coding sequence may be placed under the control of the
rice a-amylase
RAmylA promoter, which is inducible by gibberellic acid during seed
germination.
B. Transformation of plp ant cell
Various methods for direct or vectored transformation of plant cells, e.g.,
plant protoplast
cells, have been described, e.g., in above-cited PCT application WO 95/14099.
As noted in that
reference, promoters directing expression of selectable markers used for plant
transformation (e. g.,
nptlI) should operate effectively in plant hosts. One such promoter is the nos
promoter from native
Ti plasmids (Herrera-Estrella, et al., Nature 303:209-213 (i983). Others
include the 35S and 19S
promoters of cauliflower mosaic virus {Odell, et al., Nature ~,~I :810-812
{I985) and the Z'
promoter (Velten, et al., EMBO J. x:2723-2730 {1984).
In one preferred embodiment, the embryo and endosperm of mature seeds are
removed to
exposed scutulum tissue cells. The cells may be transformed by DNA bombardment
or injection, or
by vectored transformation, e.g., by Agrobacteriu»z infection after bombarding
the scuteller cells
with microparticles to make them susceptible to Agrobacterium infection
(Bidney et al., Plant Mol.
Biol. 18:301-313, 1992).
One preferred transformation follows the methods detailed generally in
Sivamani, E. et al.,
Plant Cell Reports x:465 (1996); Zhang, S., et al., Plant Cell Reports 15:465
(1996); and Li, L.,
et al., Plant Cell Reports 12:250 (1993). Briefly, rice seeds are sterilized
by standard methods, and
callus induction from the seeds is carried out on MB media with 2,4D. During a
first incubation
period, callus tissue forms around the embryo of the seed. By the end of the
incubation period,
(e.g., 14 days at 28~C) the calli are about 0.25 to 0.5 cm in diameter. Callus
mass is then detached
from the seed, and placed on fresh NB media, and incubated again for about 14
days at 28~C. After
the second incubation period, satellite calli developed around the original
"mother" callus mass.
These satellite calli were slightly smaller, more compact and defined than the
original tissue. It was
these calli were transferred to fresh media. The "mother " calti was not
transferred. The goal was to
select only the strongest, most vigorous growing tissue for further culture.
Calli to be bombarded are selected from 14-day-old subcultures. The size,
shape, color and
density are all important in selecting calli in the optimal physiological
condition for transformation.
The calli should be between .8 and 1.1 mm in diameter. The calli should appear
as spherical
masses with a rough exterior.
17
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
Transformation is by particle bombardment, as detailed in the references cited
above. After
the transformation steps, the cells are typically grown under conditions that
permit expression of the
selectable marker gene. In a preferred embodiment, the selectable marker gene
is HPH. It is
preferred to culture the transformed cells under multiple rounds of selection
to produce a uniformly
stable transformed cell line.
IV. Cell Culture Production of Mature Heterolog us Protein
Transgenic cells, typically callus cells, are cultured under conditions that
favor plant cell
growth, until the cells reach a desired cell density, then under conditions
that favor expression of
l0 the mature protein under the control of the given promoter. Preferred
culture conditions are
described below and in Example 2. Purification of the mature protein secreted
into the medium is
by standard techniques known by those of skill in the art.
Production of mature AAT: In a preferred embodiment, the culture medium
contains a
phosphate buffer, e. g., the 20 mM phosphate buffer, pH 6.8 described in
Example 2, to reduce
AAT degradation catalyzed by metals. Alternatively, or in addition, a metal
chelating agent, such
as EDTA, may be added to the medium.
Following the cell culture method 'described in Example 2, cell culture media
was partially
purified and the fraction containing AAT was analyzed by Western blot, as
shown in Fig. 4. The
first two lanes ("phosphate") show AAT bands both in the presence and absence
of elastase ("+E"
and "-E"), where the higher molecular weight bands in the presence of elastase
correspond roughly
to a 58-59 kdal AAT/elastase complex. Also as seen in the figure, expression
was high in the
absence of sucrose, but nearly undetectable in the presence of sucrose.
To ascertain the degree of glycosylation (as determined by apparent molecular
weight by
SDS-PAGE) the protein produced in culture was fractionated by SDS-PAGE and
immunodetected
with a labeled antibody raised against the C-terminal portion of AAT, as shown
in Fig. S. Lane 4
contains human AAT, and its migration position corresponds to about 52 kdal.
In lane 3 is the
plant-produced AAT, having an apparent molecular weight of about 49-50 kdal,
indicating an extent
of glycosylation of up to 60-$0~'a of the glycosylation found in human AAT
(non-glycosylated AAT
has a molecular weight of 45 kdal).
Similar results are shown in the Western blots in Fig. 6. Lanes I-3 in this
figure
correspond to decreasing amount (I5, 10, and 5 ng) of human AAT; lane 4, to 10
~l supernatant
from a non-expressing plant cell line; lanes 5 and 6, to 10 ~l supernatant
from AAT-expressing
plant cell lines I1B and 27F, respectively, and lane 7, to 10 p,l supernatant
from cell line 27F plus
250 ng trypsin. The upward mobility shift in lane 7 is indicative of
association between trypsin and
the plant-produced AAT.
18
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/LTS98/03068
The ability of plant-produced AAT to bind to elastase is demonstrated in Fig.
7, which-
shows the shift in molecular weight over a 30 minute binding interval for the
52 kdal human AAT
(lanes 1-4) and the 49-50 kdal plant-produced AAT.
To demonstrate that the mature protein is produced in secreted form, with the
desired N-
terminus, a chimeric gene constructed as above, and having the coding sequence
for mature al-
antitrypsin was expressed and secreted in cell culture as described in Example
2. The isolated
_ protein was then sequenced at its N-terminal region, yielding the N-terminal
sequence shown fn Fig.
8. This sequence, which is identified herein as SEQ ID N0:22, has the same N-
terminal residues
as native mature a,-antitrypsin.
Production of mature ATIII: In a preferred embodiment, the culture medium
contains a
MES buffer, pH 6.8. Western blot analysis of the ATIII protein produced, shown
in lanes 4 and 6
in Fig. 9, shows a band corresponding to ATIII (lane 1) in cell lines 42 and
46, when grown in the
absence (but not in the presence) of sucrose.
Production of mature BPN': In one embodiment of the invention, in which BPN'
is secreted
I5 as the proBPN' form of the enzyme, the chaperon "pro" moiety of the enzyme
facilitates enzyme
folding and is cleaved from the enzyme, leaving the active mature form of
BPN'. In another
embodiment, the mature enzyme is co-expressed and co-secreted with the "pro"
chaperon moiety,
with conversion of the enzyme to active form occurring in presence of the free
chaperon (Eder et
al., Biochem. (1993) 32:18-26; Eder et al, (1993) J. Mol. Biol. 223:293-304).
In yet another
embodiment of the invention, the BPN' is secreted in inactive form at a pH
that may be in the 6-8
range, with subsequent activation of the inactive form, e.g., after enzyme
isolation, by exposure to
the "pro" chaperon moiety, e. g., immobilized to a solid support.
In both of these embodiments, the culture medium is maintained at a pH of
between 5 and
6, preferably about 5.5 during the period of active expression and secretion
of BPN', to keep the
BPN', which is normally active at alkaline pH, at a pH below optimal activity.
Codon optimization to the host plant's most frequent codons yielded a
severalfold
enhancement in the level of expressed heterologous protein in cell culture as
shown in Fig. 11. The
extent of enhancement is seen from the Western blot analysis shown in Fig. 10
for two cells lines
and further substantiated in Fig. 11. Lane 2 (second from left) in Fig. 10
shows a Western blot of
BPN' obtained in culture from cells transformed with a native proBPN' coding
sequence. Two
bands observed correspond to a lower molecular weight protein whose
approximately 35 kdal
molecular weight corresponds to that of proBPN'. The upper band corresponds to
a somewhat
higher molecular weight species, possibly glycosylated.
The first lane in the figure shows BPN' polypeptides produced in culture by
plant cells
transformed with the codon-optimized proBPN' sequence identified by SEQ ID
N0:21. For
19
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
- WO 98/36085 PCT/US98/03068
comparative purposes, the same volume of culture medium, adjusted for cell
density, was applied in
both lanes 1 and 2. As seen, the amount of BPN' enzyme produced with a colon-
optimized
sequence was severalfold higher than for subtilisin BPN' produced with the
native coding sequence..
Further, a dark band or bands corresponding to mature peptide (molecular
weight 27.5 kdal) was
observed. However, it should be noted that directly above the band at 35kD is
a more pronounced
band which may be pro mature product yet to be cleaved into active form.
Fig. 11 compares the specific activity of BPN' colon-optimized (AP106) versus
BPN'
native (AP101) expression in rice callus cell culture, assayed using the
chromogenic peptide
substrate suc-Ala-Ala-Pro-Phe-pNA as described by DelMar, E.G. et al. (1979;
Anal. Biochem.
l0 99:316-320). As shown if Fig. I1, several of the cell lines transformed
with colon-optimized
chimeric genes produced levels of BPN', as evidenced by measured specific
activity in culture
medium, that were 2-5 times the highest levels observed for plant cells
transformed with native
proBPN' sequence.
In accordance with another aspect of the invention, it has been found that the
transformed
plant cell culture is able to express and secrete BPN' at a cell culture pH,
pH 5.5, which largely
inhibits self-degradation of mature, active BPN'. To assay for optimal pH
conditions, the assay
disclosed in DeIMar, et al. (supra) is used to test the media derived from
BPN' transformed cell
lines under various pH conditions. Transformed rice callus cells are cultured
in a MES medium
under similar conditions as disclosed in Example 2, but where the pH of the
medium is maintained
at a selected pH between 5 and 8Ø At each pH, the total amount of expressed
and secreted BPN'
is determined by Western blot analysis. BPN' activity can be tested in the
assay described by
DelMar (supra).
V. Production of Mature Heterolo n"s Protein in ~Prminating~g~s
In this embodiment, monocot cells transformed as above are used to regenerate
plants, seeds
from the plants are harvested and then germinated, and the mature protein is
isolated from the
germinated seeds.
Plant regeneration from cultured protoplasts or callus tissue is carried by
standard methods,
e.g., as described in Evans et al., HALtt~BOOK OF PLaNT CELL L Es Vol. 1:
(MacMiltan
3o Publishing Co. New York, 1983); and Vasil LR. (ed.), E L CuL~ryRn ArrD
~oMA~rrc CELL
~$NETICS OF PLANTS, Acad. Press, Orlando, Vol. I, 1984, and Vol. III, 1986,
and as described in
the above-cited PCT application.
A. Seed Germination Condition
The transgenic seeds obtained from the regenerated plants are harvested, and
prepared for
germination by an initial steeping step, in which the seeds immersed in or
sprayed with water to
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98103068
increase the moisture content of the seed to between 35-45%. This initiates
germination. Steeping
typically takes place in a steep tank which is typically ftted with a conical
end to allow the seed to
flow freely out. The addition of compressed air to oxygenate the steeping
process is an option.
The temperature is controlled at approximately 22~C depending on the seed.
After steeping, the seeds are transferred to a germination compartment which
contains air
saturated with water and is under controlled temperature and air flows. The
typical temperatures
are between I2-25oC and germination is permitted to continue for from 3 to 7
days.
Where the heterologous protein coding gene is operably linked to a inducibie
promoter
requiring a metabolite such as sugar or plant hormone, e.g., 2 to 100 p,M
gibberellic acid, this
l0 metabolite is added, removed or depleted from the steeping water medium
and/or is added to the
water saturated air used during germination. The seed absorbs the aqueous
medium and begins to
germinate, expressing the heterologous protein. The medium may then be
withdrawn and the
malting begun, by maintaining the seeds in a moist temperature controlled
aerated environment. In
this way, the seeds may begin growth prior to expression, so that the
expressed product is less
likely to be partially degraded or denatured during the grocess.
More specifically, the temperature during the imbibition or steeping phase
will be
maintained in the range of about IS-25oC, while the temperature during the
germination will usually
be about 20~C. The time for the imbibition will usually be from about 1 to 4
days, while the
gernunation time will usually be an additional 1 to 10 days, more usually 3 to
7 days. Usually, the
2o time for the malting does not exceed about ten days. The period for the
malting can be reduced by
using giant hormones during the imbibition, particularly gibberellic acid.
To achieve maximum production of recombinant protein from malting, the malting
procedure may be modified to accommodate de-hulled and de-embryonated seeds,
as described in
above-cited PCT application WO 95114099. In the absence of sugars from the
endosperm, there is
expected to be a S to 10 fold increase in ltAmy3D promoter activity and thus
expression of
heterologous protein. Alternatively when embryoless half seeds are incubated
in 10 mM CaCl2 and
5 p,M gibberellic acid, there is a SO fold increase in RAmyIA promoter
activity.
Prgduction of mature HSA: Following the germination conditions as outlined
above and
further detailed in Example 3, supernatant was analyzed by Western blot.
Western blot analysis
shows production of HSA in germinating rice seeds, with seed samples taken 24,
72, and 120 hours
after induction with gibberellin. HSA production was highest approximately 24
hours post-
induction (lanes 3 and 4, Fig. 12). Bilirubin binding, a measure of correct
folding of plant-
produced HSA, is assayed according to the method presented in Example 3.
VI. Production of Mature Heterologous Protein in Maturin Seeds
21
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
In this embodiment, monocot cells transformed as above are used to regenerate
punts, anti
seeds from the plants are allowed to mature, typically in the field, with
consequent production of
heterologous protein in the seeds.
Following seed maturation, the seeds and their heterologous proteins may be
used directly,
that is, without protein isolation, where for example, the heterologous
protein is intended to confer
a benefit on the seed as a whole, for example, to enrich the seed in the
selected protein.
Alternatively, the seeds may be fractionated by standard methods to obtain the
heterologous
protein in enriched or purified form. In one general approach, the seed is
first milled, then
suspended in a suitable extraction medium, e.g., an aqueous or an organic
solvent, to extract the
l0 protein or metabolite of interest. If desired the heterologous protein can
be further fractionated and
purified, using standard purification methods.
The following examples are provided by way of illustration only and not by way
of
limitation. Those of skill will readily recognize a variety of noncritical
parameters which could be
changed or modified to yield essentially similar results.
General Methods
Generally, the nomenclature and laboratory procedures with respect to standard
recombinant
DNA technology can be found in Sambrook, et al., MOLECULAR Ct,orrn~ro - A
LABORATORY
~r , Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 1989 and in
S.B.
Gelvin and R.A. Schilperoot, PLANT MOLECULAR BIOLOGY, 1988. Other general
references are
provided throughout this document. The procedures therein are known in the art
and are provided
for the convenience of the reader.
xam le 1
Construction of a Transforming Vector Containing
a Colon-Optimized n,-antitrYpsin Setluence
A. Hv~romvcin Resistance Gene Insertion:
The 3 kb BamHI fragment containing the 35S promoter-Hph-NOS was removed from
the
plasmid pMON410 (Monsanto, St. Louis, MO) and placed into an site-directed
mutagenized BgllT
site in the pUCl8 at 1463 to form the plasmid pUCHl8+.
B. Terminator Insertion:
pOSgIABKS is a 5 kb BamHI-Kpnl fragment from lambda clone ~,OSglA (Huang, N.,
et
al., (1990) Nuc. Acids Res. 18:7007) cloned into pBluescript KS- (Stratagene,
San Diego, CA).
22
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
Plasmid pOSgIABKS was digested with Mspl and blunted with T4 DNA polymerase
followed by
SpeI digestion. The 350 by terminator fragment was subcloned into pUCI9 (New
England
BioLabs, Beverly, MA), which had been digested with BamHI, blunted with T4 DNA
polymerase
and digested with Xbal, to form pUCl9/terminator.
C. RAmy 3D Promoter Insertion:
. A 1.1 kb NheI-PstI fragment derived from plASI.S (Huang, N. et al. (1993)
Plant Mol.
Biol. 23:737-747), was cloned into the vector pGEMSzf jmuitiple cloning site
(MCS) (Promega,
Madison, WI): ApaI, AatII, SphI, Ncol, SstII, EcoRV, SpeI, NotI, PstI, SaII,
NdeI, SacI, MIuI,
l0 NsiI~ at the SpeI and Pstl sites to form pGEMSzf (3DlNheI PstI). pGEMSzf
(3DlNheI Pst~ was
then digested with PstI and SacI, and two non kinased 30mers having the
complementary sequences
5' GCTTG ACCTG TAACT CGGGC CAGGC GAGCT 3' (SEQ ID N0:23) and 5' CGCCT
AGCCC GAGTT ACAGG TCAAG CAGCT 3' (SEQ ID N0:24) were Iigated in to form
p3DProSig. The promoter fragment prepared by digesting p3DProSig with NcoI,
blunting with T4
DNA polymerase, and digesting with SstI was subcloned into pUCl9/terminator
which had been
digested with EcoRI, blunted with T4 DNA polymerase and digested with SstI, to
form
p3DProSigEND.
D. Multiple Cloning Site Insertion:
p3DProSigEND was digested with SstI and SmaI followed by the ligation of a new
synthetic
linker fragment constructed with the non-kinased complementary
oligonucleotides 5' AGCTC
CATGG CCGTG GCTCG AGTCT AGACG CGTCC CC 3' (SEQ ID N0:25} and 5' GGGGA
CGCGT CTAGA CTCGA GCCAC GGCCA TGG 3' (SEQ ID N0:26) to form
p3DProSigENDlink.
E. p3DProSigENDlink Flankine Site Modification:
p3DProSigENDlink was digested with SalI and blunted with T4 DNA polymerase
followed
by EcoRV digestion. The blunt fragment was then inserted into pBluescript KS+
(Stratagene) in
the EcoRV site so that the HindIII site is proximal to the promoter and the
EcoRI is proximal to the
terminator sequence. The HindIII EcoRI fragment was then moved into the
polylinker of
pUCHl8+ to farm the p3Dv1.0 expression vector.
F. RAmyIA Promoter insertion:
A 1.9 kb NheI PstI fragment derived from subclone pOSG2CA2.3 from lambda clone
~,OSg2 (Huang et al. (1990) Plant Mol. Biol. I4:655-668), was cloned into the
vector pGEMSzf at
23
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
the SpeI and PstI sites to form pGEMSzf (lAINheI Pstl). pGEMSzf (lAlNheI Pstn
was digested
with Pstl and SacI and two non-kinased 35mers and four- kinased 32mers were
Iigated in, with the
complementary sequences as follows: 5' GCATG CAGGT GCTGA ACACC ATGGT GAACA
AACAC 3' (SEQ ID N0:27); 5' TTCTT GTCCC TTTCG GTCCT CATCG TCCTC CT 3' (SEQ
ID N0:28); 5' TGGCC TCTCC TCCAA CTTGA CAGCC GGGAG CT 3' (SEQ ID 0:29); 5'
TTCAC CATGG TGTTC AGCAC CTGCA TGCTG CA 3' (SEQ ID N0:30); 5' CGATG AGGAC
CGAAA GGGAC AAGAA GTGTT TG 3' (SEQ ID N0:31); 5' CCCGG CTGTC AAGTT
GGAGG AGAGG CCAAG GAGGA 3' (SEQ ID N0:32) to form plAProSig. The HindIII-SacI
0.8
kb promoter fragment was subcloned from pIAProSig into the p3Dv1.0 vector
digested with
l0 HindIII-SacI to yield the plAvl.O expression vector.
G. construction of p3D-AAT Plasmid
Two PCR grimers were used to amplify a fragment encoding AAT according to the
sequence disclosed as Genbank Accession No. K01396: N-terminal primer 5' GAGGA
TCCCC
AGGGA GATGC TGCCC AGAR 3' (SEQ ID N0:33) and C-terminal primer 5' CGCGC TCGAG
TTATT TTTGG GTGGG ATTCA CCAC 3' (SEQ ID N0:34). The N-terminal primer
amplifies to
a blunt site for in-frame insertion with the end of the p3D signal peptide and
the C-terminal primer
contains a XhoI site for cloning the fragment into the vector as shown in
Figs. 3A and 3B.
Alternatively, the sequence encoding mature AAT (SEQ ID N0:8) or colon-
optimized AAT may be
2o chemically synthesized using techniques known in the art, incorporating a
XhoI restriction site 3' of
the termination colon for insertion into the expression vector as described
above.
J I 2
P~uction of mature ."-antitrypsin in cell culture
After selection of transgenic callus, callus cells were suspended in liquid
culture containing
AA2 media ('Thompson, J.A., et al., Plant Science 47:123 (1986), at 3%
sucrose, pH 5.8.
Thereafter, the cells were shifted to phosphate-buffered media (20 mM
phosphate buffer, pH 6.8)
using 10 mL mufti-well tissue culture plates and shaken at 120 rpm in the dark
for 48 hours. The
supernatant was then removed and stored at -80~C prior to western blot
analysis.
Supernatants were concentrated using Centricon-IO filters (Amicon cat. #4207)
and washed
with induction media to remove substances interfering with electrophoretic
migration. Samples
were concentrated approximately 10 fold, and mature AAT was purified by SDS
PAGE
electrophoresis. The purified protein was extracted from the electrophoresis
medium, and
sequenced at its N-terminus, giving the sequence shown in Fig. 8, identified
herein as SEQ 7~
N0:22.
24
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
Example 3
HSA Induction in Germinating Seeds
After selection of transgenie plants which tested positive for the presence of
a codon-
optimized HSA gene driven by the GA3-responsive RAmylA promoter, seeds were
harvested and
imbibed for 24 hours with 100 rpm orbital shaking in the dark at ZS~C. GA3 was
added to a final
concentration of Sp.M and incubated for an additional 24-120 hours. Total
soluble protein was
isolated by double grinding each seed in 120 ~cI grinding buffer and
centrifuging at 23,000 x g for 1
minute at 4oC. The clear supernatant was carefully removed from the pellet and
transferred to a
fresh tube.
Biliruhin bindin assax
Bilirubin binding to its high-affinity site on mature HSA is assayed using the
method
described by Jacobsen, J. et al. (1974; Clin. Chem. 20:783) and Reed, R.G. et
al. (1975;
Biochemistry 14:4578-4583). Briefly, the concentration of free bilirubin in
equilibrium with
protein-bound biiirubin is determined by the rate of peroxide-peroxidase
catalyzed oxidation of free
bilirubin. Stock solutions of bilirubin (Nutritional Biochemicals Corp.) are
prepared fresh daily in
5 mM NaOH containing 1mM EDTA and the concentration determined using a molar
absorptivity
of 47,500 M'1 cm 1 at 440 nm. An aliquot containing between S and 30 nmol
bilirubin is added to a
1 cm cuvette containing I ml PBS and approximately 30 nmol HSA at 37~C. An
absorbance
spectrum between 500 and 350 nm is recorded. Aliquots of horseradish
peroxidase (Sigma), 0.05
mg/ml in PBS, and 0.05% ethyl hydrogen peroxide (Ferrosan; Malmo Sweden) are
added and the
change in absorbance at ~,max is recorded for 3-5 minutes. The concentrations
of free and bound
billirubin calculated from the oxidation rate observed using varying
concentrations of total bilirubin
are used to construct a Scatchard plot from which the association constant for
a single binding site is
determined.
Although the invention has been described with reference to particular
embodiments, it will
be appreciated that a variety of changes and modifications can be made without
departing from the
invention.
35
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
SEQUENCE LISTING
(1 ) GENERAL INFORMATTON
(i) APPLICANT: Applied Phytologics, Inc. -
(ii) TITLE OF THE INVENTION: Production of Mature
Proteins
in Plants
(iii) NUMBER OF SEQUENCES: 34
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: Dehlinger & Associates -
(B) STREET: P.O. Box 60850
(C) CITY: Palo Alto
(D) STATE: CA
(E) COUNTRY: USA
(F} ZIP: 94306
(v) COMPUTER
READABLE
FORM:
(A) MEDIUM TYPE: Diskette
(B) COMPUTER: IBM Compatible
(C) OPERATING SYSTEM: DOS
(D) SOFTWARE: FastSEQ for Windows Version 2.0
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER: PCT/US98/03068
(B) FTLING DATE: 13-FEB--1998
(C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA:
(A) APPLICATTON NUMBER: 60/038,169
(B) FILING DATE: 13-FEB-1997
(A) APPLICATION NUMBER: 60/037,991
(B) FILING DATE: 13-FEB-1997
(A) APPLICATION NUMBER: 60/038,170
(B) FILING DATE: 13-FEB-1997
(A) APPLICATION NUMBER: 60/038,168
(B) FILING DATE. 13-FEB-1997 -
(viii) ATTORNEY/AGENT INFORMATION: -
(A) NAME: Petithory, Joanne R
(B) REGISTRATION NUMBER: P42,995
(C) REFERENCE/DOCKET NUMBER: 0665-0007.41
(ix) TELECOMMUNICATION
INFORMATION:
(A) TELEPHONE: 650-324-0880
(B) TELEFAX: 650-324-0960
(2) INFORMATION FOR SEQ ID NO:1:
(7.) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 25 amino acids -
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE
TYPE:
peptide
(vii) IMMEDIATE SOURCE:
(B) CLONE: 3D signal peptide sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:1:
26
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
Met Lys Asn Thr Ser Ser Leu Cys Leu Leu Leu Leu Val Val Leu Cys
1 5 10 15
Ser Leu Thr Cys Asn Ser Gly Gln Ala
20 25
(2) INFORMATION FOR SEQ ID N0:2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 75 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY. linear
(vii) IMMEDIATE SOURCE:
(B) CLONE: native 3D signal peptide DNA sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:2:
ATGAAGAACA CCAGCAGCTT GTGTTTGCTG CTCCTCGTGG TGCTCTGCAG CTTGACCTGT60
AACTCGGGCC AGGCG 75
(2) INFORMATION FOR SEQ ID N0:3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 75 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(vii) IMMEDIATE SOURCE:
(B) CLONE: codon-optimized 3D signal peptide DNA sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:3:
ATGAAGAACA CCTCCTCCCT CTGCCTCCTG CTGCTCGTGG TCCTCTGCTC CCTGACCTGC60
AACAGCGGCC AGGCC 75
(2) INFORMATION FOR SEQ ID N0:4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 25 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY. linear
(ii) MOLECULE TYPE: peptide
(vii) IMMEDIATE SOURCE:
{B) CLONE: RAmylA signal peptide
(xi} SEQUENCE DESCRIPTION: SEQ ID N0:4:
Met Val Asn Lys His Phe Leu Ser Leu Ser Val Leu Ile Val
Leu Leu
1 5 10 15
Gly Leu Ser Ser Asn Leu Thr Ala Gly
20 25
(2) INFORMATION FOR SEQ ID NO: S:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 51 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(vii) IMMEDIATE SOURCE:
(B) CLONE: RAmy lA 5' untranslated region {UTR)
(xi) SEQUENCE DESCRIPTTON: SEQ ID N0:5:
27
SUBSTITUTE SHEET {MULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/L1S98/03068
ATCAATCATC CATCTCCGAA GTGTGTCTGC AGCATGCAGG TGCTGAACAC C 51
(2) INFORMATION FOR SEQ ID N0:6: -
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 321 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(vii) IMMEDTATE SOURCE:
(B) CLONE: RAmy 1A 3' untranslated region (UTR)
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:6:
GCGCACGATG ACGAGACTCT CAGTTTAGCA GATTTAACCT GCGATTTTTA CCCTGACCGG 60
TATACGTATA TACGTGCCGG CAACGAGCTG TATCCGATCC GAATTACGGA TGCAATTGTC 120
CACGAAGTAC TTCCTCCGTA AATAAAGTAG GATCAGGGAC ATACATTTGT ATGGTTTTAC 180
GAATAATGCT ATGCAATAAA ATTTGCACTG CTTAATGCTT ATGCATTTTT GCTTGGTTCG 240
ATTGTACTGG TGAATTATTG TTACTGTTCT TTTTACTTCT CGAGTGGCAG TATTGTTCTT 300
CTACGAAAAT TTGATGCGTA G 32l
(2) INFORMATION FOR SEQ ID N0:7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 394 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear -
(ii) MOLECULE TYPE: protein
(vii) IMMEDIATE SOURCE:
(B) CLONE: mature AAT amino acid sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:7:
Glu AspProGln GlyAsp AlaAlaGln LysThr AspThrSer HisHis
1 5 10 15
Asp GlnAspHis ProThr PheAsnLys IleThr ProAsnLeu AlaGlu
20 25 30
Phe AlaPheSex LeuTyr ArgGlnLeu AlaHis GlnSerAsn SerThr
35 40 45
Asn IlePhePhe SerPro ValSerIle AlaThr AlaPileAla MetLeu
50 55 60
Ser LeuGlyThr LysAla AspThrHis AspGlu IleLeuGlu GlyLeu
65 70 75 80
Asn PheAsnLeu ThrGlu IleProGlu AlaGln IleHisGlu GlyPhe
85 90 95
Gln GluLeuLeu ArgThr LeuAsnGln ProAsp SerGlnLeu GlnLeu
100 105 110
Thr ThrGlyAsn GlyLeu PheLeuSer.GluGly LeuLysLeu ValAsp
11.5 12 12
0 5
Lys PheLeuGlu AspVal LysLysLeu TyrHis 5erGluAla PheThr
130 135 140
Val AsnPheGly AspThr GluGluAla LysLys GlnIleAsn AspTyr
145 150 155 160
Val GluLysGly ThrGln G1yLysIle ValAsp LeuValLys GluLeu
165 170 175
Asp ArgAspThr ValPhe AlaLeuVal AsnTyr IlePhePhe LysGly
180 185 190
Lys TrpGluArg ProPhe GluValLys AspThr GluGluGlu AspPhe
195 200 205
His ValAspGln ValThr ThrValLys ValPro MetMetLys ArgLeu
210 215 220
Gly MetPheAsn IleGln HisCysLys LysLeu SerSerTrp ValLeu
225 230 235 240
Leu MetLysTyr LeuGly AsnAlaThr AlaIle PhePheLeu ProAsp
245 250 X55
28
SU9ST1TUTE SHEET (RULE 26j
CA 02280894 1999-08-10
WO 98f36085 PCT/US98/U3068
G1u Gly Lys Leu Gln His Leu Glu Asn Glu Leu Thr His Asp
Ile Ile
260 265 - .. 270
Thr Lys Phe Leu Glu Asn Glu Asp Arg Arg Ser Ala Ser Leu
His Leu
275 280 285 _
Pro Lys Leu Ser Ile Thr Gly Thr Tyr Asp Leu Lys Ser Val
Leu Gly
290 295 300
Gln Leu Gly Ile Thr Lys Val Phe Ser Asn Gly Ala Asp Leu
Ser Gly
305 310 315 320
Val Thr Glu Glu Ala Pro Leu Lys Leu Ser Lys Ala Val His
Lys Ala
325 330 335
Val Leu Thr Ile Asp Glu Lys Gly Thr Glu Ala Ala Gly Ala
Met Phe
340 345 350
Leu Glu Ala Ile Pro Met Ser Ile Pro Pro Glu Val Lys Phe
Asn Lys
355 360 365
Pro Phe VaI Phe Leu Met Ile Glu Gln Asn Thr Lys Ser Pro
Leu Phe
370 375 380
Met Gly Lys Val Val Asn Pro Thr Gln Lys
385 390
(2) INFORMATION FOR SEQ ID N0:8:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1185 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(vii) IMMEDIATE SOURCE:
(B) CLONE. native coding sequence of mature AAT
(xi) SEQUENCE DESCRIPTION: SEQ ID~N0:8:
GAGGATCCCC AGGGAGATGC TGCCCAGAAG ACAGATACAT CCCACCATGA 60
TCAGGATCAC
CCAACCTTCA ACAAGATCAC CCCCAACCTG GCTGAGTTCG CCTTCAGCCT 120
ATACCGCCAG
CTGGCACACC AGTCCAACAG CACCAATATC TTCTTCTCCC CAGTGAGCAT 180
CGCTACAGCC
TTTGCAATGC TCTCCCTGGG GACCAAGGCT GACACTCACG ATGAAATCCT 240
GGAGGGCCTG
AATTTCAACC TCACGGAGAT TCCGGAGGCT CAGATCCATG AAGGCTTCCA 300
GGAACTCCTC
CGTACCCTCA ACCAGCCAGA CAGCCAGCTC CAGCTGACCA CCGGCAATGG 360
CCTGTTCCTC
AGCGAGGGCC TGAAGCTAGT GGATAAGTTT TTGGAGGATG TTAAAAAGTT 420
GTACCACTCA
GAAGCCTTCA CTGTCAACTT CGGGGACACC GAAGAGGCCA AGAAACAGAT 480
CAACGATTAC
GTGGAGAAGG GTACTCAAGG GAAAATTGTG GATTTGGTCA AGGAGCTTGA 540
CAGAGACACA
GTTTTTGCTC TGGTGAATTA CATCTTCTTT AAAGGCAAAT GGGAGAGACC 600
CTTTGAAGTC
AAGGACACCG AGGAAGAGGA CTTCCACGTG GACCAGGTGA CCACCGTGAA 660
GGTGCCTATG
ATGAAGCGTT TAGGCATGTT TAACATCCAG CACTGTAAGA AGCTGTCCAG 720
CTGGGTGCTG
CTGATGAAAT ACCTGGGCAA TGCCACCGCC ATCTTCTTCC TGCCTGATGA 780
GGGGAAACTA
CAGCACCTGG AAAATGAACT CACCCACGAT ATCATCACCA AGTTCCTGGA 840
AAATGAAGAC
AGAAGGTCTG CCAGCTTACA TTTACCCAAA CTGTCCATTA CTGGAACCTA 900
TGATCTGAAG
AGCGTCCTGG GTCAACTGGG CATCACTAAG GTCTTCAGCA ATGGGGCTGA 960
CCTCTCCGGG
GTCACAGAGG AGGCACCCCT GAAGCTCTCC AAGGCCGTGC ATAAGGCTGT 1020
GCTGACCATC
GACGAGAAAG GGACTGAAGC TGCTGGGGCC ATGTTTTTAG AGGCCATACC 1080
CATGTCTATC
CCCCCCGAGG TCAAGTTCAA CAAACCCTTT GTCTTCTTAA TGATTGAACA 1140
AAATACCAAG
TCTCCCCTCT TCATGGGAAA AGTGGTGAAT CCCACCCAAA AATAA 1185
(2) INFORMATION FOR SEQ ID N0:9:
(i) SEQUENCE CHARACTERISTTCS:
(A) LENGTH: 432 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(vii) IMMEDIATE SOURCE:
(B) CLONE: mature ATIII as sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:9:
His Gly Sex Pro Va1 Asp Ile Cys Thr Ala Lys Pro Arg Asp Ile Pro
29
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PC'd'/CTS98/03068
1 5 10 15
Met AsnPro MetCys TleTyrArg SerPro GluLysLys AlaThr Glu
20 25 30 _
Asp GluGly SerGlu GlnLysIle ProGlu AlaThrAsn ArgArg Val
35 40 45
Trp GluLeu SerLys AlaAsnSer ArgPhe AlaThrThr PheTyr Gln
50 55 60
His LeuAla AspSer LysAsnAsp AsnAsp AsnIlePhe LeuSer Pro
65 70 75 g0
Leu SerIle SerThr AlaPheAla MetThr LysLeuGly AlaCys Asn
85 9p g5
Asp ThrLeu GlnGln LeuMetGlu ValPhe LysPheAsp ThrIle Ser
100 105 110
Glu LysThr SerAsp GlnIleHis PhePhe PheAlaLys LeuAsn Cys
115 120 125
Arg LeuTyr ArgLys AlaAsnLys SerSer LysLeuVal SerA1a Asn
130 135 140
Arg LeuPhe GlyAsp LysSerLeu ThrPhe AsnGluThr TyrGln Asp
145 150 155 160
Ile SerGlu LeuVal TyrGlyAla LysLeu GlnProLeu AspPhe Lys
165 170 175
Glu AsnAla GluGln SerArgAla AlaIle AsnLysTrp ValSer Asn
180 185 190
Lys ThrGlu GlyArg IleThrAsp ValIle ProSerGlu AlaIle Asn
195 200 205
Glu LeuThr ValLeu ValLeuVal AsnThr IleTyrPhe LysGly Leu
210 215 220
Trp LysSer LysPhe SerProGlu AsnThr ArgLysGlu LeuPhe Tyr
225 230 235 240
_
Lys AlaAsp Gly'GluSerCysSer AlaSer MetMetTyr GlnGlu Gly
245 250 255
Lys PheArg T~rArg ArgValAla GluGly ThrGlnVal LeuGlu Leu
260 265 270
Pro PheLys GlyAsp AspIleThr MetVal LeuIleLeu ProLys Pro
275 280 2g5
Glu LysSer LeuAla LysValGlu LysGlu LeuThrPro GluVal Leu
290 295 300
Gln GluTrp LeuAsp GluLeuGlu GluMet MetLeuVal ValHis Met
305 310 315 320
Pro ArgPhe ArgIle GluAspG1y PheSer LeuLysG1u GlnLeu Gln
325 330 335
Asp MetGly LeuVal AspLeuPhe SerPro G1uLysSer LysLeu Pro
340 345 350
Gly IleVal AlaGlu GlyArgAsp AspLeu TyrValSer AspA1a Phe
355 360 365
His LysAla PheLeu GluValAsn GluGlu GlySerGlu AlaAla Ala
370 375 380
Ser ThrAla ValVal TleAlaGly ArgSer LeuAsnPro AsnArg Val
3B5 390 . 395 400
Thr PheLys AlaAsn ArgProPhe LeuVal PheIleArg GluVal Pro
405 410 415
Leu AsriThr IleIle PheMetGly ArgVal AlaAsnPro CysVal Lys
420 425 430
(2) INFORMATION FORSEQ ID N0:10:
(i) QUENCE CS:
SE CHARACTERISTI
(A) LENGTH: 1299base irs
pa
(B) TYPE: c id
nuclei ac
(C) STRANDEDNESS : ngle
si
(D) TOPOLOGY: near -
li
(vii) IMMEDIATE SOURCE:
(B) CLONE: native ATIII DNA sequence -
(xi) SEQUENCE DESCRIPTION: SEQ TD N0:10:
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
W O 98136085 PCT/US98/03068
CACGGAAGCC CTGTGGACAT CTGCACAGCC.AAGCCGCGGG ACATTCCCAT GAATCCCATG60
TGCATTTACC GCTCCCCGGA GAAGAAGGCA ACTGAGGATG AGGGCTCAGA ACAGAAGATC120
CCGGAGGCCA CCAACCGGCG TGTCTGGGAA CTGTCCAAGG CCAATTCCCG CTTTGCTACC180
ACTTTCTATC AGCACCTGGC AGATTCCAAG AATGACAATG ATAACATTTT CCTGTCACCC240
CTGAGTATCT CCACGGCTTT TGCTATGACC AAGCTGGGTG CCTGTAATGA CACCCTCCAG300
CAACTGATGG AGGTATTTAA GTTTGACACC ATATCTGAGA AAACATCTGA TCAGATCCAC360
TTCTTCTTTG CCAAACTGAA CTGCCGACTC TATCGAAAAG CCAACAAATC CTCCAAGTTA420
GTATCAGCCA ATCGCCTTTT TGGAGACAAA TCCCTTACCT TCAATGAGAC CTACCAGGAC480
ATCAGTGAGT TGGTATATGG AGCCAAGCTC CAGCCCCTGG ACTTCAAGGA AAATGCAGAG540
CAATCCAGAG CGGCCATCAA CAAATGGGTG TCCAATAAGA CCGAAGGCCG 600
AATCACCGAT
GTCATTCCCT CGGAAGCCAT CAATGAGCTC ACTGTTCTGG TGCTGGTTAA CACCATTTAC660
TTCAAGGGCC TGTGGAAGTC AAAGTTCAGC CCTGAGAACA CAAGGAAGGA ACTGTTCTAC720
AAGGCTGATG GAGAGTCGTG TTCAGCATCT ATGATGTACC AGGAAGGCAA GTTCCGTTAT780
CGGCGCGTGG CTGAAGGCAC CCAGGTGCTT GAGTTGCCCT TCAAAGGTGA TGACATCACC840
ATGGTCCTCA TCTTGCCCAA GCCTGAGAAG AGCCTGGCCA AGGTGGAGAA 900
GGAACTCACC
CCAGAGGTGC TGCAGGAGTG GCTGGATGAA TTGGAGGAGA TGATGCTGGT GGTTCACATG960
CCCCGCTTCC GCATTGAGGA CGGCTTCAGT TTGAAGGAGC AGCTGCAAGA CATGGGCCTT1020
GTCGATCTGT TCAGCCCTGA AAAGTCCAAA CTCCCAGGTA TTGTTGCAGA AGGCCGAGAT1080
GACCTCTATG TCTCAGATGC ATTCCATAAG GCATTTCTTG AGGTAAATGA AGAAGGCAGT1140
GAAGCAGCTG CAAGTACCGC TGTTGTGATT GCTGGCCGTT CGCTAAACCC 1200
CAACAGGGTG
ACTTTCAAGG CCAACAGGCC CTTCCTGGTT TTTATAAGAG AAGTTCCTCT GAACACTATT1260
ATCTTCATGG GCAGAGTAGC CAACCCTTGT GTTAAGTAA 1299
30
(2) TNFORMATION FOR SEQ ID N0:11:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: S85 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(vii) IMMEDIATE SOURCE:
(B) CLONE: mature HSA amino acid sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:11:
Asp Ala HisLys SerGlu ValAlaHis ArgPheLys AspLeu GlyGlu
1 5 10 15
Glu Asn PheLys AlaLeu ValLeuIle AlaPheAla GlnTyr LeuGln
20 25 30
Gln Cys ProPhe GluAsp HisValLys LeuValAsn GluVal ThrGlu
35 40 45
Phe Ala LysThr CysVal AlaAspGlu SerAlaGlu AsnCys AspLys
50 55 60
Ser Leu HisThr LeuPhe GlyAspLys LeuCysThr ValAla ThrLeu
65 70 75 80
Arg Glu ThrTyr GlyGlu MetAlaAsp CysCysAla LysGln GluPro
85 90 95
Glu Arg AsnGlu CysPhe LeuGlnHis LysAspAsp AsnPro AsnLeu
100 lOS' I10
Pro Arg LeuVal ArgPro GluValAsp ValMetCys ThrAla PheHis
115 120 125
Asp Asn GluGlu ThrPhe LeuLysLys TyrLeuTyr GluIle AlaArg
130 135 140
Arg His ProTyr PheTyr AlaProGlu LeuLeuPhe PheAla LysArg
145 150 155 160
Tyr Lys AlaAla PheThr GluCysCys GlnAlaAla AspLys AlaAla
165 170 175
Cys Leu LeuPro LysLeu AspGluLeu ArgAspGlu GlyLys AlaSer
180 185 190
Ser Ala LysGln ArgLeu LysCysAla SerLeuGln LysPhe GlyGlu
195 200 205
Arg Ala PheLys AlaTrp AlaValAla ArgLeuSer GlnArg PhePro
210 215 220
Lys Ala GluPhe AlaGlu ValSerLys LeuValThr AspLeu ThrLys
225 230 235 240
Val His ThrGlu CysCys HisGlyAsp LeuLeuGlu CysAla AspAsp
31
SUBSTfTUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
245 250 255
Arg AlaAspLeu AlaLys TyrIle CysGluAsn Gln-Asp SerIleSer
260 265 270
Ser LysLeuLys GluCys CysGlu LysProLeu LeuGlu Lys-SerHis
275 280 285
Cys IleAlaGlu ValGlu AsnAsp GluMetPro AlaAsp LeuProSer
290 295 300
Leu AlaAlaAsp PheVal GluSer LysAspVal CysLys AsnTyrAla
305 310 315 - .- 320
Glu AlaLysAsp ValPhe LeuGly MetPheLeu TyrGlu TyrAlaArg
325 330 - 335
Arg HisProAsp TyrSer ValVal LeuLeuLeu ArgLeu AlaLysThr
340 345 350
Tyr GluThrThr LeuGlu LysCys CysAlaAla AlaAsp Pro-HisGlu
355 360 365
Cys TyrAlaLys ValPhe AspGlu PheLysPro LeuVal GluGluPro
370 375 380
Gln AsnLeuIle LysGln AsnCys G1uLeuPhe LysGln LeuGlyGlu
385 390 395 400
Tyr LysPheGln AsnAla LeuLeu ValArgTyr ThrLys LysValPro
405 410 415
G1n ValSerThr ProThr LeuVal GluValSer ArgAsn LeuGlyLys
420 425 430
Val GlySerLys CysCys LysHis ProGluAla LysArg MetProCys
435 440 445
Ala GluAspTyr LeuSer ValVal LeuAsnGln LeuCys ValLeuHis
450 455 460
Glu LysThrPro ValSer AspArg ValThrLys CysCys ThrGluSer
465 470 475 480
Leu -ValAsnArg ArgPro CysPhe SerAlaLeu GluVal AspGluThr
485 490 495
Tyr ValProLys GluPhe AsnAla GluThrPhe ThrPhe HisAlaAsp
500 505 510.
Ile CysThrLeu SerGlu LysG1u ArgGlnIle LysLys GlnThrAla
515 520 525
Leu ValGluLeu ValLys HisLys ProLysAla ThrLys GluGlnLeu
530 535 540
Lys AlaValMet AspAsp PheAla AlaPheVal GluLys CysCysLys
545 550 555 560
Ala AspAspLys GluThr CysPhe AlaGluGlu GlyLys LysLeuVal
565 570 575
Ala AlaSerGln AlaAla LeuGly Leu
580 585 - . -
(2) INFORMATION FOR SEQ ID N0:12: -
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1865 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(vii) IMMEDIATE SOURCE:
(B) CLONE: native coding sequence of mature HSA
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:12:
AGATGCACAC AAGAGTGAGG TTGCTCATCG GTTTAAAGAT TTGGGAGAAG AAAATTTCAA 60
AGCCTTGGTG TTGATTGCCT TTGCTCAGTA TCTTCAGCAG TGTCCATTTG AAGATCATGT 120
AAAATTAGTG AGTCAGCTGA180
AATGAAGTAA
CTGAATTTGC
AAAAACATGT
GTAGCTGATG
AAATTGTGAC AAATCACTTC ATACCCTTTT TGGAGACAAA TTATGCACAGTTGCAACTCT240
TCGTGAAACC TATGGTGAAA TGGCTGACTG CTGTGCAAAA CAAGAACCTGAGAGAAATGA300
ATGCTTCTTG CAACACAAAG ATGACAACCC AAACCTCCCC CGATTGGTGAGACCAGAGGT360
TGATGTGATG TGCACTGCTT TTCATGACAA TGAAGAGACA TTTTTGAAAAAATACTTATA420
TGAAATTGCC AGAAGACATC CTTACTTTTA TGCCCCGGAA CTCCTTTTCTTTGCTAAAAG480
GTATAAAGCT GCTTTTACAG AATGTTGCCA AGCTGCTGAT AAAGCTGCCTGCCTGTTGCC540
AAAGCTCGAT GAACTTCGGG ATGAAGGGAA GGCTTCGTCT GCCAAACAGAGACTCAAATG600
32
SU~STiTUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCTlUS98/03068
TGCCAGTCTC CAAAA.ATTTG GAGAAAGAGC TTTCAAAGCA TGGGCAGTGG CTCGCCTGAG 660
CCAGAGATTT CCCAAAGCTG AGTTTGCAGA AGTTTCCAAG TTAGTGACAG ATCTTACCAA 720
AGTCCACACG GAATGCTGCC ATGGAGATCT GCTTGAATGT GCTGATGACA GGGCGGACCT 780
TGCCAAGTAT ATCTGTGAAA ATCAGGATTC GATCTCCAGT AAACTGAAGG AATGCTGTGA 840
AAAACCTCTG TTGGAAAA.AT CCCACTGCAT TGCCGAAGTG GAAAATGATG AGATGCCTGC 900
TGACTTGCCT TCATTAGCTG CTGATTTTGT TGAAAGTAAG GATGTTTGCA AAAACTATGC 960
TGAGGCAAAG GATGTCTTCC TGGGCATGTT TTTGTATGAA TATGCAAGAA GGCATCCTGA 1020
TTACTCTGTC GTGCTGCTGC TGAGACTTGC CAAGACATAT GAAACCACTC TAGAGAAGTG 1080
CTGTGCCGCT GCAGATCCTC ATGAATGCTA TGCCAAAGTG TTCGATGAAT TTAAACCTCT 1140
TGTGGAAGAG CCTCAGAATT TAATCAAACA AAACTGTGAG CTTTTTAAGC AGCTTGGAGA 1200
GTACAAATTC CAGAATGCGC TATTAGTTCG TTACACCAAG AAAGTACCCC AAGTGTCAAC 1260
' TCCAACTCTT GTAGAGGTCT CAAGAAACCT AGGAAAAGTG GGCAGCAAAT GTTGTAAACA 1320
TCCTGAAGCA AAAAGAATGC CCTGTGCAGA AGACTATCTA TCCGTGGTCC TGAACCAGTT 1380
ATGTGTGTTG CATGAGAAAA CGCCAGTAAG TGACAGAGTC ACAAAATGCT GCACAGAGTC 1440
CTTGGTGAAC AGGCGACCAT GCTTTTCAGC TCTGGAAGTC GATGAAACAT ACGTTCCCAA 1500
AGAGTTTAAT GCTGAAACAT TCACCTTCCA TGCAGATATA TGCACACTTT CTGAGAAGGA 1560
GAGACAAATC AAGAAACAAA CTGCACTTGT TGAGCTTGTG AAACACAAGC CCAAGGCAAC 1620
AAAAGAGCAA CTGAAAGCTG TTATGGATGA TTTCGCAGCT TTTGTAGAGA AGTGCTGCAA 1680
GGCTGACGAT AAGGAGACCT GCTTTGCCGA GGAGGGTAAA AAACTTGTTG CTGCAAGTCA 1740
AGCTGCCTTA GGCTTATAAC ATCTACATTT AAAAGCATCT CAGCCTACCA TGAGAATAAG 1800
AGAAAGAAAA TGAAGATCAA AAGCTTATTC ATCTGTTTTC TTTTTCGTTG GTGTAAAGCC 1860
AACAC
1865
(2) INFORMATION FOR SEQ ID N0:13:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 352 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(vii) IMMEDIATE SOURCE:
(B) CLONE: native proBPN' amino acid sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:13:
Ala Gly LysSer AsnGlyGlu LysLys TyrIIe ValGlyPhe LysGln
1 5 10 15
Thr Met SerThr MetSerAla AlaLys LysLys AspValIle SerGlu
20 25 30
Lys Gly GlyLys ValGlnLys GlnPhe LysTyr ValAspAla AlaSer
35 40 45
Ala Thr LeuAsn GluLysAla ValLys GluLeu LysLysAsp ProSer
55 60
45 Val Ala TyrVal GluGluAsp HisVal AlaHis AlaTyrAla GlnSer
65 70 75 80
Val Pro TyrGly ValSerGln IleLys AlaPro AlaLeuHis SerGln
85 90 95
Gly Tyr ThrGIy SerAsnVal LysVal AlaVal IleAspSer GlyIle
50 100 105 110
Asp Ser SerHis ProAspLeu LysVal AlaGly GlyAlaSer MetVal
115 120 125
Pro Ser GluThr AsnProPhe GlnAsp AsnAsn SerHisGly ThrHis
130 135 140
!55Val Ala GlyThr ValAlaAla LeuAsn AsnSer IleGlyVal LeuGly
145 150 155 160
Val Ala ProSer AlaSerLeu TyrAla ValLys ValLeuGly AlaAsp
165 170 175
Gly Ser GlyGln TyrSerTrp IleIle AsnGly IleGluTrp AlaIle
180 185 190
Ala Asn AsnMet AspValIle AsnMet SerLeu GlyGlyPro SerGly
195 200 205
Ser Ala AlaLeu LysAlaAla ValAsp LysAla ValAlaSer GlyVal
210 215 220
Val Val ValAla AlaAlaGly AsnGlu GlyThr SerGlySer SerSer
225 230 235 240
Thr Val GlyTyr ProGlyLys TyrPro SerVal IIeAlaVal GlyAla
33
SUBSTITUTE SHEET (RULE 26j
CA 02280894 1999-08-10
WO 98/36085 PCT/U898/03068
245 250 -255
Val Asp Ser Ser Asn Gln Arg Ala Ser Phe Ser Ser Val Gly
Pro Glu
260 265 - - 270
Leu Asp Val Met Ala Pro Gly Val Ser Ile Gln Ser Thr Leu
Pro Gly
275 280 285
Asn Lys Tyr Gly Ala Tyr Asn Gly Thr Ser Met Ala Ser Pro
His Val
290 295 300
Ala Gly Ala Ala Ala Leu Ile Leu Ser Lys His Pro Asn Trp
Thr Asn
305 310 315 320
Thr Gln Val Arg Ser Ser Leu Glu Asn Thr Thr Thr Lys Leu
Gly Asp
325 330 335
Ser Phe Tyr Tyr Gly Lys Gly Leu Ile Asn Val Gln Ala Ala
Ala Gln
340 345 -- 350
(2) INFORMATION FOR SEQ ID N0:14:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1056 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(vii} IMMEDIATE SOURCE:
(B) CLONE: native proBPN~ coding sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:14:
GCAGGGAAAT CAAACGGGGA AAAGAAATAT ATTGTCGGGT TTAAACAGAC 60
AATGAGCACG
ATGAGCGCCG CTAAGAAGAA AGATGTCATT TCTGAAAAAG GCGGGAAAGT 120
GCAAAAGCAA
TTCAAATATG TAGACGCAGC TTCAGCTACA TTAAACGAAA AAGCTGTAAA 180
AGAATTGAAA
AAAGACCCGA GCGTCGCTTA CGTTGAAGAA GATCACGTAG CACATGCGTA 240
CGCGCAGTCC
GTGCCTTACG GCGTATCACA AATTAAAGCC CCTGCTCTGC ACTCTCAAGG 300
CTACACTGGA
TCAAATGTTA AAGTAGCGGT TATCGACAGC GGTATCGATT CTTCTCATCC 360
TGATTTAAAG
GTAGCAGGCG GAGCCAGCAT GGTTCCTTCT GAAACAAATC CTTTCCAAGA 420
CAACAACTCT
CACGGAACTC ACGTTGCCGG CACAGTTGCG GCTCTTAATA ACTCAATCGG 480
TGTATTAGGC
GTTGCGCCAA GCGCATCACT TTACGCTGTA AAAGTTCTCG GTGCTGACGG 540
TTCCGGCCAA
TACAGCTGGA TCATTAACGG AATCGAGTGG GCGATCGCAA ACAATATGGA 600
CGTTATTAAC
ATGAGCCTCG GCGGACCTTC TGGTTCTGCT GCTTTAAAAG CGGCAGTTGA 660
TAAAGCCGTT
GCATCCGGCG TCGTAGTCGT TGCGGCAGCC GGTAACGAAG GCACTTCCGG 720
CAGCTCAAGC
ACAGTGGGCT ACCCTGGTAA ATACCCTTCT GTCATTGCAG TAGGCGCTGT 780
TGACAGCAGC
AACCAAAGAG CATCTTTCTC AAGCGTAGGA CCTGAGCTTG ATGTCATGGC 840
ACCTGGCGTA
TCTATCCAAA GCACGCTTCC TGGAAACAAA TACGGGGCGT ACAACGGTAC 900
GTCAATGGCA
TCTCCGCACG TTGCCGGAGC GGCTGCTTTG ATTCTTTCTA AGCACCCGAA 960
CTGGACAAAC
ACTCAAGTCC GCAGCAGTTT AGAAAACACC ACTACAAAAC TTGGTGATTC 1020
TTTC
TACTAT
_ 1056
_
GGAAAAGGGC TGATCAACGT ACAGGCGGCA GCTCAG =- -
(2) INFORMATION FOR SEQ ID N0:15: _
(i) SEQUENCE CHARACTERISTICS:
(A} LENGTH: 77 amino acids .
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
(vii) IMMEDIATE SOURCE:
(B) CLONE: subtilisin BPN~ pro-peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:15:
Ala Gly Lys Ser Asn Gly Glu Lys Lys Tyr Ile Val Gly Phe Lys Gln
1 5 ZO 15
Thr Met Ser Thr Met Ser Ala Ala Lys Lys Lys Asp Val Ile Sex Glu
20 25 30
Lys Gly Gly Lys Val Gln Lys Gln Phe Lys Tyr Val Asp Ala Ala Ser
35 40 45
Ala Thr Leu Asn Glu Lys Ala Val Lys Glu Leu Lys Lys Asp Pro Ser
50 55 50
34-i
SUBSTITUTE SHEET (RULE 2fi)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
Val Ala Tyr Val Glu Glu Asp His 'Val Ala~His Ala Tyr
65 70 75
(2) INFORMATION FOR SEQ ID N0:16:
(i) SEQUENCE CHARACTERISTICS:
- (A) LENGTH: 275 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
' (ii) MOLECULE TYPE: protein
(vii) IMMEDIATE SOURCE:
(B) CLONE: native mature BPN~ amino acid sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:16:
Ala Gln SerValPro TyrGly ValSerGln IleLys AlaProAla Leu
1 5 10 15
His Ser GlnGlyTyr ThrGly SerAsnVal LysVal AlaValIle Asp
20 25 30
Ser Gly IleAspSer SerHis ProAspLeu LysVal AlaGlyGly Ala
35 40 45
Ser Met ValProSer GluThr AsnProPhe GlnAsp AsnAsnSer His
50 55 60
Gly Thr HisValAla GlyThr ValAlaAla LeuAsn AsnSerIle Gly
65 70 75 80
Val Leu GlyValAla ProSer AlaSerLeu TyrAla ValLysVal Leu
85 90 95
Gly Ala AspGlySer GlyGln TyrSerTrp IleIle AsnGlyIle Glu
100 105 110
Trp Ala IleAlaAsn AsnMet AspValIle AsnMet SerLeuGly Gly
115 120 125
Pro Ser GlySerAla AlaLeu LysAlaAla ValAsp LysAlaVal A1a
130 135 140
Ser Gly ValValVal ValAla AlaAlaGly AsnGlu GlyThrSer Gly
145 150 155 160
Ser Ser SerThrVal GlyTyr ProGlyLys TyrPro SerValIle Ala
165 170 175
Val Gly AlaValAsp SerSer AsnGlnArg AlaSer PheSerSer Val
180 185 190
Gly Pro GluLeuAsp ValMet AlaProGly ValSer IleGlnSer Thr
195 200 205
Leu Pro GlyAsnLys TyrGly AlaTyrAsn GlyThr SerMetAla Ser
210 215 220
Pro His ValAlaGly AlaAla AlaLeuIle LeuSer LysHisPro Asn
225 230 235 240
Trp Thr AsnThrGln ValArg SerSerLeu GluAsn ThrThrThr Lys
245 250 255
Leu Gly Asp.SerPhe TyrTyr GlyLys~Gly LeuIle AsnValGln Ala
260 265 270
Ala Ala Gln
275
!55 (2) INFORMATION FOR SEQ ID N0:17:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 275 amino acids
(B) TYPE: amino acid
GO (D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(vii) IMMEDIATE SOURCE:
(B) CLONE: amino acid sequence of mature BPN~ variant
34-ii
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/LTS98/03068
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:17:
Ala Gln Ser Val Pro Tyr Gly Val Ser Gln Ile Lys Ala Pro Ala Leu
1 5 10 15
His SerGln GlyTyrThr GlySer AsnVal LysValAla ValIle Asp
20 25 30
Ser GlyIle AspSerSer HisPro AspLeu LysValAla GlyGly Ala
35 40 45 _
Ser MetVal ProSerGlu ThrAsn ProPhe GlnAspThr AsnSer His
50 55 60 -
Gly ThrHis ValAlaGly ThrVal AlaAla LeuThrAsn SexIle Gly
65 70 75 80
Val LeuGly ValAlaPro SerAla SerLeu TyrAlaVal LysVal Leu
- 85 90 95
Gly AlaAsp GlySerGly GlnTyr SerTrp IleIleAsn GlyIle Glu
100 105 110
Trp AlaIle AlaAsnAsn MetAsp ValIle ThrMetSer LeuGly Gly
115 120 125
Pro SerGly SerAlaAla LeuLys AlaAla ValAsplaysAlaVal Ala
130 135 140
Ser GlyVal ValValVal A1aAla AlaGly AsnGluGly ThrSer G1y
145 150 155 160
Ser SerSer ThrValGly TyrPro GlyLys TyrProSer ValIle Ala
165 170 175
Val GlyAla ValAspSer SerAsn GlnArg AlaSerPhe SerSer Val
180 185 190
Gly ProGlu LeuAspVal MetAla ProGly ValSerLle GlnSer Thr
195 200 205
Leu ProGly AsnLysTyr GlyAla TyrSer GlyThrSer MetAla Ser
210 215 ~ 220
Pro HisVal AlaGlyAla AlaAla LeuIle LeuSerLys HisPro Thr
225 230 235 240
Trp ThrAsn ThrGlnVal ArgSer SerLeu GluAsnThr ThrThr Lys
245 250 255
Leu GlyAsp SerPheTyr TyrGly LysGly LeuIleAsn ValGln Ala
260 265 270
Ala AlaGln
275
(2) INFORMATION FOR SEQ ID N0:18: -
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1260 base pairs
(S) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(vii) IMMEDIATE SOURCE: -
(B) CLONE: codon-optimized~3D signal peptide-AAT DNA sequence
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:18:
ATGAAGAACA CCTCCTCCCT CTGCCTCCTG CTGCTCGTGG TCCTCTGCTC CCTGACCTGC 60
AACAGCGGCC AGGCCGAGGACCCGCAGGGCGACGCCGCCCAGAAGACCGACACCAGCCAC120
CACGACCAGG ACCACCCGACGTTCAACAAGATCACCCCGAATTTGGCCGAATTCGCCTTC180
AGCCTGTACC GCCAGCTCGCGCACCAGTCCAACTCCACCAACATCTTCTTCAGCCCGGTG240
AGCATCGCCA CCGCCTTCGCCATGCTGTCCCTGGGTACCAAGGCGGACACCCACGACGAG300
ATCCTCGAAG GGCTGAACTTCAACCTGACGGAGATCCCGGAGGCGCAGATCCACGAGGGC360
TTCCAGGAGC TGCTCAGGACGCTCAACCAGCCGGACTCCCAGCTCCAGCTCACCACCGGC420
AACGGGCTCT TCCTGTCCGAGGGCCTCAAGCTCGTCGATAAGTTCCTGGAGGACGTGAAG480
AAGCTCTACC ACTCCGAGGCGTTCACCGTCAACTTCGGGGACACCGAGGAGGCCAAGAAG540
CAGATCAACG ACTACGTCGAGAAGGGGACCCAGGGCAAGATCGTGGACCTGGTCAAGGAA-600
TTGGACAGGG ACACCGTCTTCGCGCTCGTCAACTACATCTTCTTCAAGGGCAAGTGGGAG660
CGCCCGTTCG AGGTGAAGGACACCGAGGAGGAGGACTTCCACGTCGACCAGGTCACCACC720
GTCAAGGTCC CGATGATGAAGAGGCTCGGCATGTTCAACATCCAGCACTGCAAGAAGCTC780
TCCAGCTGGG TGCTCCTCATGAAGTACCTGGGGAACGCCACCGCCATCTTCTTCCTGCCG840
34-iii -
SLD~STITUTE SHEET (RULE 26j
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
GACGAGGGCA AGCTCCAGCA CCTGGAGAAC GAGCTGACGC ACGACATCAT CACGAAGTTC 900
CTGGAGAACG AGGACAGGCG CTCCGCTAGC CTCCACCTCC CGAAGCTGAG CATCACCGGC 960
ACGTACGACC TGAAGAGCGT GCTGGGCCAG CTGGGCATCA CGAAGGTCTT CAGCAACGGC 1020
GCGGACCTCT CCGGCGTGAC GGAGGAGGCC CCCCTGAAGC TCTCCAAGGC CGTGCACAAG 1080
GCGGTGCTCA CGATCGACGA GAAGGGGACG GAAGCTGCCG GGGCCATGTT CCTGGAGGCC 1140
ATCCCCATGT CCATCCCGCC CGAGGTCAAG TTCAACAAGC CCTTCGTCTT CCTGATGATC 1200
GAGCAGAACA CGAAGAGCCC CCTCTTCATG GGGAAGGTCG TCAACCCCAC GCAGAAGTGA 1260
(2) INFORMATION FOR SEQ ID N0:19:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1382 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single .
(D) TOPOLOGY: linear
(vii) IMMEDIATE SOURCE:
(B) CLONE: codon-optimized 3D signal peptide-ATIII DNA sequen
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:19:
ATGAAGAACA CCTCCTCCCT CTGCCTCCTG CTGCTCGTGG TCCTCTGCTC CCTGACCTGC 60
AACAGCGGCC AGGCCCACGG AAGCCCTGTG GACATCTGCA CAGCCAAGCC GCGGGACATT 120
CCCATGAATC CCATGTGCAT TTACCGCTCC CCGGAGAAGA AGGCAACTGA GGATGAGGGC 180
TCAGAACAGA AGATCCCGGAGGCCACCAACCGGCGTGTCTGGGAACTGTC CAAGGCCAAT240
TCCCGCTTTG CTACCACTTTCTATCAGCACCTGGCAGATTCCAAGAATGA CAATGATAAC300
ATTTTCCTGT CACCCCTGAGTATCTCCACGGCTTTTGCTATGACCAAGCT GGGTGCCTGT360
AATGACACCC TCCAGCAACTGATGGAGGTATTTAAGTTTGACACCATATC TGAGAAAACA420
TCTGATCAGA TCCACTTCTTCTTTGCCAAACTGAACTGCCGACTCTATCG AAAAGCCAAC480
AAATCCTCCA AGTTAGTATCAGCCAATCGCCTTTTTGGAGACAAATCCCT TACCTTCAAT540
GAGACCTACC AGGACATCAGTGAGTTGGTATATGGAGCCAAGCTCCAGCC CCTGGACTTC600
AAGGAAAATG CAGAGCAATCCAGAGCGGCCATCAACAAATGGGTGTCCAA TAAGACCGAA660
GGCCGAATCA CCGATGTCATTCCCTCGGAAGCCATCAATGAGCTCACTGT TCTGGTGCTG720
GTTAACACCA TTTACTTCAAGGGCCTGTGGAAGTCAAAGTTCAGCCCTGA GAACACAAGG780
AAGGAACTGT TCTACAAGGCTGATGGAGAGTCGTGTTCAGCATCTATGAT GTACCAGGAA840
GGCAAGTTCC GTTATCGGCGCGTGGCTGAAGGCACCCAGGTGCTTGAGTT GCCCTTCAAA900
GGTGATGACA TCACCATGGTCCTCATCTTGCCCAAGCCTGAGAAGAGCCT GGCCAAGGTG960
GAGAAGGAAC TCACCCCAGAGGTGCTGCAGGAGTGGCTGGATGAATTGGA GGAGATGATG1020
CTGGTGGTTC ACATGCCCCGCTTCCGCATTGAGGACGGCTTCAGTTTGAA GGAGCAGCTG1080
CAAGACATGG GCCTTGTCGATCTGTTCAGCCCTGAAAAGTCCAAACTCCC AGGTATTGTT1140
GCAGAAGGCC GAGATGACCTCTATGTCTCAGATGCP-TTCCATAAGGCATT TCTTGAGGTA1200
AATGAAGAAG GCAGTGAAGCAGCTGCAAGTACCGCTGTTGTGATTGCTGG CCGTTCGCTA1260
AACCCCAACA GGGTGACTTTCAAGGCCAACAGGCCCTTCCTGGTTTTTAT AAGAGAAGTT1320
CCTCTGAACA CTATTATCTTCATGGGCAGAGTAGCCAACCCTTGTGTTAA GTAACTCGAG1380
CC 1382
(2) INFORMATION FOR SEQ ID N0:20:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1940 base pairs.
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(vii) IMMEDIATE SOURCE:
(B) CLONE: codon-optimized 3D signal peptide-HSA DNA sequence
(xi) SEQUENCE DESCRTPTTON: SEQ ID N0:20:
ATGAAGAACA CCTCCTCCCT CTGCCTCCTG CTGCTCGTGG TCCTCTGCTC CCTGACCTGC 60
AACAGCGGCC AGGCCAGATG CACACAAGAG TGAGGTTGCT CATCGGTTTA AAGATTTGGG 120
AGAAGAAAAT TTCAAAGCCT TGGTGTTGAT TGCCTTTGCT CAGTATCTTC AGCAGTGTCC 180
ATTTGAAGAT CATGTAAAAT TAGTGAATGA AGTAACTGAA TTTGCAAAAA CATGTGTAGC 240
TGATGAGTCA GCTGAAAATT GTGACAAATC ACTTCATACC CTTTTTGGAG ACAAATTATG 300
CACAGTTGCA ACTCTTCGTG AAACCTATGG TGAAATGGCT GACTGCTGTG CAAAACAAGA 360
ACCTGAGAGA AATGAATGCT TCTTGCAACA CAAAGATGAC AACCCAAACC TCCCCCGATT 420
GGTGAGACCA GAGGTTGATG TGATGTGCAC TGCTTTTCAT GACAATGAAG AGACATTTTT 480
34-iv
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/CTS98103068
GAAAA.AATAC TTATATGAAA TTGCCAGAAG ACATCCTTAC TTTTATGCCC CGGAACTCCT540
TTTCTTTGCT AAAAGGTATA AAGCTGCTTT TACAGAATGT TGCCAAGCTG CTGATAAAGC600
TGCCTGCCTG TTGCCAAAGC TCGATGAACT TCGGGATGAA GGGAAGGCTT CGTCTGCCAA660
ACAGAGACTC AAATGTGCCA GTCTCCAAAA ATTTGGAGAA AGAGCTTTCA AAGCATGGGC720
AGTGGCTCGC CTGAGCCAGA GATTTCCCAA AGCTGAGTTTGCAGAAGTTT CCAAGTTAGT780
GACAGATCTT ACCAAAGTCC ACACGGAATG CTGCCATGGA GATCTGCTTG AATGTGCTGA840
TGACAGGGCG GACCTTGCCA AGTATATCTG TGAAAATCAG GATTCGATCT CCAGTAAACT900
GAAGGAATGC TGTGAAAAAC CTCTGTTGGA AAAATCCCAC TGCATTGCCG AAGTGGAAAA960
TGATGAGATG CCTGCTGACT TGCCTTCATT AGCTGCTGAT TTTGTTGAAA GTAAGGATGT1020
TTGCAAAAAC TATGCTGAGG CAAAGGATGT CTTCCTGGGC ATGTTTTTGT 1080
ATGAATATGC
AAGAAGGCAT CCTGATTACT CTGTCGTGCT GCTGCTGAGA CTTGCCAAGA CATATGAAAC1140
CACTCTAGAG AAGTGCTGTG CCGCTGCAGA TCCTCATGAA TGCTATGCCA AAGTGTTCGA1200
TGAATTTAAA CCTCTTGTGG AAGAGCCTCA GAATTTAATC AAACAAAACT GTGAGCTTTT1260
TAAGCAGCTT GGAGAGTACA AATTCCAGAA TGCGCTATTA GTTCGTTACA CCAAGAAAGT1320
ACCCCAAGTG TCAACTCCAA CTCTTGTAGA GGTCTCAAGA AACCTAGGAA 1380
AAGTGGGCAG
CAAATGTTGT AAACATCCTG AAGCAAAAAG AATGCCCTGT GCAGAAGACT ATCTATCCGT1440
GGTCCTGAAC CAGTTATGTG TGTTGCATGA GAAAACGCCA GTAAGTGACA GAGTCACAAA1500
ATGCTGCACA GAGTCCTTGG TGAACAGGCG ACCATGCTTT TCAGCTCTGG AAGTCGATGA1560
AACATACGTT CCCAAAGAGT TTAATGCTGA AACATTCACC TTCCATGCAG ATATATGCAC1620
ACTTTCTGAG AAGGAGAGAC AAATCAAGAA ACAAACTGCA CTTGTTGAGC 1680
TTGTGAAACA
CAAGCCCAAG GCAACAAAAG AGCAACTGAA AGCTGTTATG GATGATTTCG CAGCTTTTGT1740
AGAGAAGTGC TGCAAGGCTG ACGATAAGGA GACCTGCTTT GCCGAGGAGG GTAAAAAACT1800
TGTTGCTGCA AGTCAAGCTG CCTTAGGCTT ATAACATCTA CATTTA&AAG CATCTCAGCC1860
TACCATGAGA ATAAGAGAAA GAAAATGAAG ATCAAAAGCT TATTCATCTG TTTTCTTTTT1920
CGTTGGTGTA AAGCCAACAC 1940
(2) TNFORMATION FOR SEQ ID N0:21:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1140 base pairs
(B) TYPE: nucleic acid
(C} STRANDEDNESS: single
(D) TOPOLOGY: linear
(vii} IMMEDIATE SOURCE:
(B) CLONE: codon-optimized 3D signal peptide-BPN~ DNA sequene
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:21:
ATGAAGAACA CCTCCTCCCTCTGCCTCCTGCTGCTCGTGGTCCTCTGCTC CCTGACCTGC60
AACAGCGGCC AGGCCGCTGGCAAGAGCAACGGGGAGAAGAAGTACATCGT CGGCTTCAAG120
CAGACCATGA GCACCATGAGCGCCGCCAAGAAGAAGGACGTCATCAGCGA GAAGGGCGGC180
AAGGTACAGA AGCAGTTCAAGTACGTGGACGCCGCCAGCGCCACCCTCAA CGAGAAGGCC240
GTCAAGGAGC TGAAGAAGGACCCGAGCGTCGCCTACGTCGAGGAGGACCA CGTCGCCCAC300
GCATATGCAC AGAGCGTCCCGTACGGCGTCAGCCAGATCAAGGCCCCGGC CCTCCACAGC360
CAGGGCTACA CCGGCAGCAACGTCAAGGTCGCCGTCATCGACAGCGGCAT CGACAGCAGC420
CACCCGGACC TCAAGGTCGCCGGCGGAGCTAGCATGGTCCCGAGCGAGAC CAACCCGTTC480
CAGGACACCA ACAGCCATGGCACCCACGTCGCCGGCACCGTCGCCGCCCT CACCAACAGC540
ATCGGCGTCC TCGGCGTCGCCCCGAGCGCCAGCCTCTACGCCGTCAAGGT ACTCGGCGCC600
GACGGCAGCG GCCAGTACAGCTGGATCATCAACGGCATCGAGTGGGCCAT CGCCAACAAC660
ATGGACGTCA TCACCATGAGCCTCGGCGGCCCGAGCGGCAGCGCCGCCCT CAAGGCCGCC720
GTCGACAAGG CCGTCGCCAGCGGCGTCGTCGTCGTCGCCGCCGCCGGCAA CGAGGGCACC780
AGCGGCAGCA GCAGCACCGTCGGCTACCCGGGCAAGTACCCGAGCGTCAT CGCCGTCGGC840
GCCGTGGACA GCAGCAACCAGCGCGCGAGCTTCAGCAGCGTCGGCCCGGA GCTGGACGTC900
ATGGCCCCGG GCGTCAGCATCCAGAGCACCCTCCCGGGCAACAAGTACGG CGCCTACAGC960
GGCACCAGCA TGGCCAGCCCGCACGTCGCCGGCGCCGCTGCACTCATCCT CAGCAAGCAC1020
CCGACCTGGA CCAACACCCAGGTCCGCAGCAGCCTGGAGAACACCACCAC CAAGCTCGGC1080
GACAGCTTCT ACTACGGCAAGGGCCTCATCAACGTCCAGGCCGCCGCCCA GTGACTCGAG1140
(2) INFORMATION FOR SEQ ID N0:22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii} MOLECULE TYPE: peptide
34-v
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98!36085 PCT/US98/03068
(vii) IMMEDIATE SOURCE:
(B) CLONE: N-terminus o~ mature AAT
(xi) SEQUENCE DESCRIPTION. SEQ ID N0:22:
Glu Asp Pro Gln Gly Asp Ala Ala Gln Lys Thr Asp Thr
1 5 10
(2) INFORMATION FOR SEQ ID N0:23:
(i) SEQUENCE CHARACTERISTICS:
' (A) LENGTH: 30 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ TD N0:23:
GCTTGACCTG TAACTCGGGC CAGGCGAGCT 30
(2) INFORMATION FOR SEQ ID N0:24:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUEI~TCE DESCRIPTION: SEQ ID N0:24:
CGCCTAGCCC GAGTTACAGG TCAAGCAGCT 30
(2) INFORMATION FOR SEQ ID N0:25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 37 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:25:
AGCTCCATGG CCGTGGCTCG AGTCTAGACG CGTCCCC 37
(2) INFORMATION FOR SEQ ID N0:26:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:26:
GGGGACGCGT CTAGACTCGA GCCACGGCCA TGG 33
(2) INFORMATION FOR SEQ ID N0:27:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 35 base pairs
X55 (B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
34-vi
SUBSTITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
WO 98/36085 PCT/US98/03068
(xi) SEQUENCE DESCRIPTION: SEQ-ID N0:27:
GCATGCAGGT GCTGAACACC ATGGTGAACA AACAC 35
(2) INFORMATION FOR SEQ ID N0:28:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH. 32 base pairs
(B) TYPE. nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTTON: SEQ ID N0:28:
TTCTTGTCCC TTTCGGTCCT CATCGTCCTC CT _ 32
(2) INFORMATION FOR SEQ ID N0:29:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid __.
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:29:
TGGCCTCTCC TCCAACTTGA CAGCCGGGAG CT 32
(2) INFORMATION FOR SEQ TD N0:30:
(i) SEQUENCE CHARACTERTSTICS:
(A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single -
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:30:
TTCACCATGG TGTTCAGCAC CTGCATGCTG CA 32
(2) INFORMATION FOR SEQ ID N0:31:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single .
(D) TOPOLOGY: linear
(xi} SEQUENCE DESCRIPTION: SEQ ID N0:31:
CGATGAGGAC CGAAAGGGAC AAGAAGTGTT TG 32
(2) INFORMATION FOR SEQ ID N0:32:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 35 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear -
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:32:
34-vii
SU~STITUTE SHEET (RULE 26)
CA 02280894 1999-08-10
W4 98136485 PCT/CJS98/03068
CCCGGCTGTC AAGTTGGAGG AGAGGCCAAG GAGGA 35
(2) INFORMATION FOR SEQ ID N0:33:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:33:
GAGGATCCCC AGGGAGATGC TGCCCAGAA 2g
(2) INFORMATION FOR SEQ ID N0:34:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 34 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:34:
CGCGCTCGAG TTATTTTTGG GTGGGATTCA CCAC 34
34-viii