Note: Descriptions are shown in the official language in which they were submitted.
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-1-
DESCRIPTION
VOLATILE MATRICES FOR MATRIX-ASSISTED LASER
DESORPTION/IONIZATION MASS SPECTROMETRY
1.0 BACKGROUND OF THE INVENTION
1.1 Field of the Invention
This invention relates to volatile photoabsorbing matrices having a low
sublimation
temperature for use in the mass spectrometric analysis of large, nonvolatile
molecules. This
invention also relates to methods for preparing samples containing large.
nonvolatile analyte
I 0 molecules for laser desorption mass spectrometry employing such matrices.
1.2 Description of Related Art
Approximately 4,000 human disorders are attributed to genetic causes. Hundreds
of
genes responsible for various disorders have been mapped, and sequence
information is being
I S accumulated rapidly. A principal goal of the Human Genome Project is to
find all genes
associated with each disorder. The definitive diagnostic test for any specific
genetic disease (or
predisposition to disease) will be the identification of polymorphic
variations in the DNA
sequence of affected cells that result in alterations of gene function.
Furthermore, response to
specific medications may depend on the presence of polymorphisms. Developing
DNA (or
20 RNA) screening as a practical tool for medical diagnostics requires a
method that is
inexpensive, accurate, expeditious, and robust.
Genetic polymorphisms and mutations can manifest themselves in several forms,
such
as point polymorphisms or point mutations where a single base is changed to
one of the three
other bases; deletions where one or more bases are removed from a nucleic acid
sequence and
25 the bases flanking the deleted sequence are directly linked to each other;
insertions where new
- bases are inserted at a particular point in a nucleic acid sequence adding
additional length to the
overall sequence; and expansions and reductions of repeating sequence motifs.
Large insertions
and deletions, often the result of chromosomal recombination and rearrangement
events, can
lead to partial or complete loss of the activity of a gene. Of these forms of
polymorphism, in
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-2-
general the most difficult type of change to screen for and detect is the
point polymorphism
because it represents the smallest degree of molecular change.
Although a number of genetic defects can be linked to a specific single point
mutation
within a gene, e.g. sickle cell anemia, many are caused by a wide spectrum of
different
mutations throughout the gene. A typical gene that might be screened could be
anywhere from
1,000 to 100,000 bases in length, though smaller and larger genes do exist. Of
that amount of
DNA, only a fraction of the base pairs actually encode the protein. These
discontinuous protein
coding regions are called exons and the remainder of the gene is referred to
as introns. Of these
two types of regions, exons often contain the most important sequences to be
screened. Several
complex procedures have been developed for scanning genes in order to detect
polymorphisms.
These procedures are applicable to both exons and introns.
In terms of current use, most of the methods to scan or screen genes employ
slab or
capillary gel electrophoresis for the separation and detection step in the
assays. Gel
electrophoresis of nucleic acids primarily provides relative size information
based on mobility
through the gel matrix. If calibration standards are employed, gel
electrophoresis can be used
to measure absolute and relative molecular weights of large biomolecules with
some moderate
degree of accuracy; even then, the accuracy is typically only 5% to 10%. Also
the molecular
weight resolution is limited. In cases where two DNA fragments with the
identical number of
base pairs can be separated, for example, by using high concentration
polyacrylamide gels, it is
still not possible to identify which band on a gel corresponds to which DNA
fragment without
performing secondary labeling experiments. Thus, gel electrophoresis
techniques can only
determine size and cannot provide any information about changes in base
composition or
sequence without performing more complex sequencing reactions. Gel-based
techniques, for
the most part, are dependent on labeling or staining methods to visualize and
discriminate
between different nucleic acid fragments.
Many methods in use today capable of screening broadly for genetic
polymorphisms
suffer from technical complication and are labor and time intensive. Single
strand
conformational polymorphism (SSCP) (Orita et al., 1989), denaturing gradient
gel
electrophoresis (DGGE) (Abrams et al., 1990), chemical cleavage at mismatch
(CCM) (Saleeba
and Cotton, 1993), enzymatic mismatch cleavage (EMC) (Youil et al., 1995), and
cleavage
fragment length polymorphism (CFLP) procedures are currently gel-based, making
them
___._ ..__ __ __._. . ..__ T. ._.._._____~.__.....
CA 02302036 2003-02-06
77718-61(S)
-3-
cumbersome to automate and perform efficiently. Thus, there is a need for new
methods that
can provide cost effective and expeditious means for screening genetic
material in an effort to
detect genetic mutations and diagnose related medical conditions simply,
quicI:3y, accurately,
and inexpensively.
Another approach that is having some success is to employ mass spectrometry to
screen
for and detect genetic mutations as well as to sequence nucleic acids. In
order to measure the
mass of nonvolatile high molecular weight molecules, typically greater than
1000 Da, in a mass
spectrometer, the analyte molecules must frst be volatilized or converted into
gas-phase ions.
Although direct laser desorption of the neat analvte is one approach to
volatilizing the
molecule, the energy deposited into the anah~te may induce fragmentation and
lead to results
that are ambiguous or dimcult to analyze. The late 1980's saw the rise of two
new mass
spectrometric techniques which are potentially suitable for genetic screening
tests by
successfully measuring the masses of intact very large biomolecules, namely,
matti.Y-assisted
laser desorption/ionization (MALDI) time-of flight mass spectrometry (TOF MS)
(Tanaka
1 S et al., 1988; Spengler et al., 1989) and electrospray ionization (ES)
combined with a variety of
mass analyzers. The MALDI mass spectrometric technique can also be used with
methods
other than time-of flight, for example, magnetic sector, Fourier-transform ion
cyclotron
resonance, quadrupole, and quadrupole trap.
MALDI-TOF MS involves laser pulses focused on a small sample plate on which
analyte molecules (i.e. nucleic acids) are embedded in either a solid or
liquid matrix which is
typically a small, highly absorbing material. such as a small aromatic organic
moiecule. The
volatilization of intact fragile molecules benefits from the use of matrix-
assisted laser
desorption ionization because the radiative energy from the laser pulse is
coupled indirectly into
the anaiyte through the matrix molecules. Typically, the analyte molecules are
crystallized with
a large molar excess of a photoabsorbing matrix (see U.S. Patent Nos. 4.920?64
and x.118,937).
An advance in MALDI analysis of polynucleotides was the discovery of
3-hydroxypicolonic acid (3-HPA) as a suitable matrix for mixed-base
oligonucleotides
(Wu, et al., 1993).
The Laser pulses transfer energy to the matrix causing a microscopic ablation
and
concomitant ionization of the anaIyte molecules, producing a 'aseous plume of
intact, charged
nucleic acids in single-stranded form. It is thoueht that upon laser
excitation the matrix
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-4-
molecules are rapidly heated and ejected into the gas phase, carrying analyte
molecules into the
expansion plume of molecules and ions. It is thought that gas-phase ion-
molecule collisions
subsequently ionize the neutral analyte molecules in the near-surface region,
often via proton
transfer. The matrix thus functions as both an energy- and charge-transfer
species. If double-
s stranded nucleic acids are analyzed, the MALDI-TOF MS typically results in
detection of
mostly charged denatured single-stranded nucleic acids.
The ions generated by the laser pulses are accelerated to a fixed kinetic
energy by a
strong electric field and then passed through an electric field-free region in
vacuum, traveling
with a velocity corresponding to their respective mass-to-charge ratios (m/z).
Thus, the smaller
m/z ions will travel through the vacuum region faster than the larger m/z ions
thereby causing a
separation. At the end of the electric field-free region, the ions collide
with a detector that
generates a signal as each set of ions of a particular mass-to-charge ratio
strikes the detector.
Usually for a given assay, 10 to 100 mass spectra resulting from individual
laser pulses are
summed together to make a single composite mass spectrum with an improved
signal-to-noise
ratio.
The mass of an ion (such as a charged nucleic acid) is measured by using its
velocity to
determine the mass-to-charge ratio by time-of flight analysis. In other words,
the mass of the
molecule directly correlates with the time it takes to travel from the sample
plate to the detector.
The entire process takes only microseconds. In an automated apparatus, tens to
hundreds of
samples can be analyzed per minute. In addition to speed, MALDI-TOF MS has one
of the
largest mass ranges for mass spectrometric devices. The current mass range for
MALDI-TOF
MS is from 1 to 1,000,000 Da (measured recently for a protein) (Nelson et al.,
1995).
The performance of a mass spectrometer is measured by its sensitivity, mass
resolution
and mass accuracy. Sensitivity is measured by the amount of material needed;
it is generally
desirable and possible with mass spectrometry to work with sample amounts in
the femtomole
and low picomole range. Mass resolution, m/Om, is the measure of an
instrument's ability to
produce separate signals from ions of similar mass. Mass resolution is defined
as the mass, m,
of an ion signal divided by the full width of the signal, Dm, usually measured
between points of
half maximum intensity. Mass accuracy is the measure of error in designating a
mass to an ion
signal. The mass accuracy is defined as the ratio of the mass assignment error
divided by the
mass of the ion and can be represented as a percentage.
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-5-
To be able to detect any point polymorphism directly by MALDI-TOF mass
spectrometry, one would need to resolve and accurately measure the masses of
nucleic acids in
which a single base change has occurred (in comparison to the wild type
nucleic acid). A single
base change can be a mass difference of as little as 9 Da. This value
represents the difference
between the two bases with the closest mass values, A and T (A = 2'-
deoxyadenosine-5'-
phosphate - 313.19 Da; T = 2'-deoxythymidine-5'-phosphate - 304.20 Da; G =2'-
deoxyguanosine-5'-phosphate = 329.21 Da; and C = 2'-deoxycytidine-5'-phosphate
= 289.19
Da). If during the mutation process, a single A changes to T or a single T to
A, the mutant
nucleic acid containing the base transversion will either decrease or increase
by 9 Da in total
mass as compared to the wild type nucleic acid. For mass spectrometry to
directly detect these
transversions, it must therefore be able to detect a minimum mass change, Om,
of
approximately 9 Da.
For example, in order to fully resolve (which may not be necessary) a point-
mutated
(A to T or T to A) heterozygote 50-base single-stranded DNA fragment having a
mass, m, of
1 S ~ 15,000 Da from its corresponding wild type nucleic acid, the required
mass resolution is
m/Om = 15,000/9 ~ 1,700. However, the mass accuracy needs to be significantly
better than 9
Da to increase quality assurance and to prevent ambiguities where the measured
mass value is
near the half way point between the two theoretical masses. For an analyte of
15,000 Da, in
practice the mass accuracy needs to be Om ~ ~3 Da = 6 Da. In this case, the
absolute mass
accuracy required is (6/15,000)* 100 = 0.04%. Often a distinguishing level of
mass accuracy
relative to another known peak in the spectrum is sufficient to resolve
ambiguities. For
example, if there is a known mass peak 1000 Da from the mass peak in question,
the relative
position of the unknown to the known peak may be known with greater accuracy
than that
provided by an absolute, previous calibration of the mass spectrometer.
In addition, the ability to separate DNA fragments ( 1 ) differing in only one
base in
length and (2) of reasonable length (e.g., of sizes corresponding to at least
primer size, around
20 to 30 bases or so up to about 50 bases in length) is critical to achieving
even rudimentary
DNA sequencing by MALDI-MS. For laser desorption mass spectroscopy techniques
to
successfully analyze macromolecules requires that one stably laser-desorb
molecules into a
vapor phase, and separate and detect (and thereby determine the mass of) the
volatilized
molecules by mass spectroscopy. The ability to stably desorb the macromolecule
depends on
CA 02302036 2003-02-06
77718-61(5)
-6-
the availability of a suitable light absorbing matrix that will allow one to
stably laser-desorb
DNA molecules from a solid state to a gaseous state. and permit separation of
DNA molecules
having only a nucleotide or so difference in length. Putting that into
perspective, the difference
in mass between a polynucleotide having 30 versus 31 nucleotide represents
about a 3%
difference in mass (about 9610 v. 310, assuming an average m.w. of 310 for
each nucleotide).
If one applies this to a DNA molecule of 100 nucleotides in length, a modest
sequence by DNA
sequencing standards, the separation system must distinguish among DNA
molecules differing
by only I % in mass.
Thus, there is a need for the development of MS techniques and related
materials for
practicing these techniques that have enhanced resolution, accuracy, and
sensitivity. The ability
to stably desorb the molecule from a solid matrix that absorbs light at the
laser wavelength,
without radiation damage and fragmentation of the sample is particularly
important as
fragmentation can lead to complex spectra and decreased resolution and
sensitivity.
Although MALDI generates less energetic analyte ions than direct laser
desorption, thus
decreasing the thermal degradation of the analyte, the ions nevertheless
contain significant
internal energy, which may result in fragmentation. Among the few matrix
molecules that have
been found to desorb/ionize intact DNA, 3-HPA is currently the most widely
used (Wu et al.,
1993; Wu et al., 1994) ). Using a matrix mixture of~ 3-HPA with picolinic
acid,
oligonucleotides have been detected that are greater than 500 bases (up to
about 200 kDa) in
length (Tang e~ al., 1994; Liu et al., 1995). However, as the length of the
oligonucleotide
increases, the mass resolution is degraded by widening kinetic energy spreads,
prompt
fragmentation, delayed fragmentation (metastable decay), and the formation of
matrix adducts.
Thus, there is a need to develop MS materials and methods that minimize
fragmentation of the
analyte ions during the MALDI process, extend the accessible mass range for
mass
spectrometric detection. and enhance the utility of the MS techniques.
CA 02302036 2003-02-06
77718-61(S)
6a
2.0 SZT1~IARY OF THE INVENTION
According to one aspect of the present invention,
there is provided a method for determining the mass of a
large organic molecule, said method comprising:
(a) contacting said large organic molecule with a
photoabsorbing low-sublimation temperature matrix to produce
a matrix: molecule mixture; (b) desorbing and ionizing said
molecule; and (c) determining the mass of said ionized large
organic molecule by mass spectrometry.
According to another aspect of the present
invention, there is provided a method for preparing a sample
of large organic molecules for mass spectral analysis, said
method comprising: a) providing a solution comprising said
large organic molecule to be analyzed, a volatile, light
absorbing hydroxy-bearing matrix composition, and a solvent;
and b) evaporating said solvent to provide a solid matrix
material containing the molecule to be analyzed.
It is therefore a goal of the present invention to
provide compositions and methods relating to the preparation
of samples containing nonvolatile analyte molecules for mass
analysis using a photoabsorbing, low-sublimation temperature
matrix. These matrix molecules provide a means for
desorbing and ionizing nonvolatile, nonthermally-labile
organic molecules
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
_'7_
such as biomolecules and synthetic polymers. Minimizing fragmentation of the
parent analyte
ion and/or reducing adduct formation leads to increased detection sensitivity
and/or increased
resolution and/or extension of the usable mass range.
The deleterious effects associated with widening kinetic energy spreads,
fragmentation
and the formation of matrix-analyte adducts are reduced by employing a matrix
system, as
disclosed herein, having lower intermolecular binding energies associated with
increased
volatility. Lower binding energies can reduce fragmentation by minimizing the
internal energy
of the desorbed analyte, and can reduce adduct formation by lowering the
binding energy of the
analyte with its surrounding molecules. The desorption of a volatile matrix at
room
temperature but cooled to maintain low vapor pressure in the mass spectrometer
may also
require less energy. Because a vacuum is required for the mass spectrometry,
volatile,
crystalline matrices which sublimate or evaporate readily at room temperature
are typically
cooled to reduce their vapor pressures to practical levels, which is below
about 10'~ Torr in the
desorption plume. This consequently means that the analyte internal energy may
also be lower.
It is therefore an advantage of the present invention to use liquids or low
sublimation
temperature solids as matrices because such systems generally enable lower
desorption/ionization temperatures.
The present invention relates to a method for volatilization and mass
spectrometric
analysis of nonvolatile, or nonthermally labile, large organic molecules
including biomolecules
such as nucleic acids, for example, DNA and RNA; proteins and peptide nucleic
acids (PNA);
oligosaccharides, and other high molecular weight polymers.
The invention generally provides a method for determining the mass of a large
organic
molecule. The method typically includes contacting a large organic molecule,
the mass of
which one desires to determine, with a photoabsorbing, or light absorbing, low-
sublimation
temperature matrix to produce a matrix:molecule mixture. This contacting step
may be carried
out by dissolving the large organic molecule to be analyzed in a solution
containing the matrix.
The matrix:molecule mixture is then irradiated by a light source, such as a
laser, to desorb,
ionize, and produce an ionized large organic molecule. The ionized large
organic molecule is
then separated from other constituents, such as the matrix:molecule mixture or
other
matrix:molecule adducts, using mass spectrometry and the mass of the ionized
large organic
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
_g_
molecule determined. While any mass spectrometry is contemplated for use with
the present
invention, time-of flight mass spectrometry is preferred.
The matrix:molecule mixture typically comprises a physical mixture of the
matrix with
the molecule to be analyzed. It may or may not contain adducts of the matrix
with the
molecule. Although if adducts are formed, they will typically be only weakly
associated such
that they may be readily dissociated upon irradiation. desorption, and
ionization.
As used herein the term "a" encompasses embodiments wherein it refers to a
single
element as well as embodiments including one or more of such elements.
In performing the mass spectrometry, it is preferable to use a cooled sample
stage.
Generally, the sample stage is cooled to less than 273° K, typically to
from about 150° K to
200° K or to about 180 K. While it is contemplated that the sample
stage may be cooled by any
suitable means, it may typically be cryongenically cooled by liquid nitrogen.
In creating the matrix:molecule mixture, for example, by dissolving the large
organic
molecule in a solution containing the matrix, one of skill in the art will
understand that the
solution containing the matrix may generally contain one or more solvents.
Preferably the
solvents will be water and/or organic solvents, such as ethanol, methanol,
toluene. acetone, and
acetonitrile. After the matrix:molecule mixture is formed, the solvents are
substantially
evaporated, typically to dryness. In preferred embodiments, the solvents are
evaporated at
room temperature. After evaporating the solvent, the resulting solid or
crystalline molecule-
matrix mixture is cooled to a vapor pressure between about 10-' ° Torr
and about 10-5 Torr.
The matrix for use in the present invention is generally a volatile, light-
absorbing,
hydroxy-bearing matrix. As used herein, volatile matrices are those that are
volatile at room
temperature at ambient or reduced pressures. In preferred aspects, the matrix
may be a phenol,
a hydroxyquinoline, or a hydroxynaphthalene. Where the matrix is a phenol, it
will preferably
be 4-nitrophenol. Where the matrix is a hydroxyquinoline, it will preferably
be 8-
hydroxyquinoline. It is also generally preferred that the matrix have a
molecular weight of
between about 90 Da and about 400 Da. Different classes of analyte molecules
may also
require different matrix systems. The matrix should typically not react or
interact strongly with
the analyte and the analyte should be soluble in the matrix crystals.
In particular embodiments the matrix has a high sublimation rate between the
temperatures of 20°C to 200°C (or a low sublimation
temperature). The low-sublimation
CA 02302036 2000-02-25
WO 98/54751 PCTNS98/11003
-9-
temperature matrix may typically have a sublimation rate at room temperature
of at least 0.1
pm.miri' at a pressure of about 10-5 Torr or less and preferably the
sublimation rate at these
conditions is from about 0.01 ~m.miri ~ to about 0.1 mm~miri'. Also provided
are
embodiments where the matrix is a crystalline solid.
As used herein the terms "photo absorbing" or "light absorbing" refer to the
ability of
the matrix to absorb the desorption light sufficiently strong to aid in the
desorption and
ionization of the large organic molecule. Typically the matrices will absorb
light between the
wavelengths of approximately 200 nm and approximately 20,000 nm although it
will be
understood that this absorption is not continuous. It is further preferred
that the photoabsorbing
matrix have an absorption coefficient greater than about 10 L~cm-~.mol-', up
to and including
an absorption coefficient of 106 L.cm-~.mol~~, at the wavelength of the
desorbing and ionizing
radiation. The method of the invention is useful for determining the mass of
virtually any large
organic molecule. For example, the mass of a polymer may be determined using
the methods
of the invention. In preferred aspects of the invention, the polymer to be
analyzed will be a
biopolymer, such as a nucleic acid, a polypeptide, a peptide nucleic acid
(PNA), an
oligosaccharide, or a mass-modif ed derivative thereof. Where the molecule to
be analyzed is a
nucleic acid, it will be understood that it may be, for example, a DNA or an
RNA.
The analyte should typically be purified to minimize the presence of salt ions
and other
molecular contaminants. These impurities may reduce the intensity and quality
of the mass
spectrometric signal to a point where either (i) the signal is undetectable or
unreliable, or (ii) the
mass accuracy and/or resolution is below the value necessary for the
particular application, such
as to detect the type of polymorphism expected or sequence the analyte. A
preferred method to
purify the analyte is to immobolize it on a solid support and wash it remove
impurities, such as
sodium and potassium ions. The analyte may then be released from the solid
support and
contacted with the matrix.
The size of the analyte to be analyzed should also be within the range where
there is
sufficient mass resolution and accuracy. Mass accuracy and resolution
significantly degrade as
the mass of the analyte increases. Currently, the detection of single
nucleotide polymorphisms
(SNPs) above said mass value is difficult above a mass of approximately 30,000
Da for
oligonucleotides (~ 100 bases) although this range may increase with further
advances in MS-
related technology. Third, because all molecules within a sample are
visualized during mass
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-10-
spectrometric analysis (i.e. it is not possible to selectively label and
visualize certain molecules
and not others as one can with gel electrophoresis methods), samples may
preferably be
partitioned prior to analysis to remove unwanted products from the spectrum.
It is contemplated that the method of the invention will allow for the mass
determination
of any large organic molecule having a mass of greater than about 1,000 Da.
More specifically,
one may determine the mass of a molecule having a mass of greater than about
27,000 Da,
greater than about 30,000 Da, greater than about 50,000 Da, greater than about
75,000 Da,
greater than about 100,000 Da, greater than about 150.000 Da, greater than
about 175,000 Da,
greater than about 200,000 Da, greater than about 250,000 Da, or even greater
than about
315,000 Da. The organic molecule will typically have a mass of less than
5.00,000 Da,
3,000,000 Da or 1,000,000 DA. In some embodiments. the organic molecule may
have a mass
of less than 500,000 or 300,000 Daltons.
To perform the desorbing step, one will generally expose the matrix:molecule
mixture
to a source of energy to desorb the large organic molecule from the matrix.
The source of
energy used for desorption of the large organic molecule will preferably be a
laser beam. The
laser beam used to desorb and ionize the large organic molecule may be any
laser but is
preferably a pulsed laser. Typically, the desorption step will include
applying an energy of
about 20 kV followed by a pulse of energy of about 2.7 kV. Preferably, the
pulse of energy
comprises light having a wavelength of about 355 nm. The mass of the large
organic molecule
may then be determined by summing the mass spectra over a number of laser
pulses, preferably
about 200 laser pulses or about 1000 laser pulses, or any number of pulses
therebetween, such
as, for example, about 250 laser pulses, about 300 laser pulses, about 350
laser pulses, about
500 laser pulses, about 750 laser pulses, etc. Of course, it is contemplated
that one may sum
the mass spectra of less than about 200 pulses or more than about 1000 pulses,
but it will be
understood that lower numbers of pulses, especially very low numbers of pulses
such as 10 or
20 or 50 pulses, etc., may give less accurate results, and higher numbers of
pulses becomes
unnecessarily repetitive and lower the efficiency and cost-effectiveness of
the method.
In another aspect, the invention also provides a method for preparing a sample
of large
organic molecules for mass spectral analysis. This method typically includes
providing a
solution comprising a large organic molecule to be analyzed, a matrix molecule
comprising a
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-11-
volatile, light-absorbing hydroxy-bearing matrix molecule. and a solvent. and
evaporating the
solvent to provide a solid crystalline matrix containing the molecule to be
analyzed.
The present invention applies to MALDI mass spectrometry of all classes of
nonvolatile, large organic compounds, with synthetic polymers and biopolymers
preferred. The
present invention is particularly preferred for mass analysis of biopolymers
such as nucleic
acids, proteins, PNAs and oligosaccharides due to the fragile nature of these
molecules. The
method utilizes pulsed laser desorption/ionization mediated by a matrix
followed by mass
spectrometric separation and detection of the analyte molecules. The matrix
may be a
crystalline solid or a liquid at room temperature, with crystalline solids
being preferred. The
preferred matrix has a high sublimation rate in vacuum at room temperature and
absorbs the
desorption light strongly.
Therefore in accordance with the present invention, there is provided
crystalline solid,
light absorbing compounds having hydroxy functionalities, but not carboxylic
fimctionalities,
for use as a matrix in mass analysis. In preferred embodiments the matrix
compounds may be
phenols, hydroxyquinolines or hydroxynaphthalenes. The crystalline solids, 8-
hydroxyquinoline and 4-nitrophenol, which are volatile at room temperature,
are particularly
preferred as matrices in accordance with the present invention.
The less energetic, more facile desorption/ionization from these volatile
matrices
minimizes fragmentation and extends the high mass limit for generation of
intact analyte
molecules. These crystalline matrices exhibit increased sensitivity for
detection of both low (8-
hydroxyquinoline) and high (4-nitrophenol) molecular weight analytes. Analyte
molecules,
including DNA, exceeding 250 kDa molecular weight can be detected by this
method.
There is provided embodiments where the analyte is a large organic molecule of
greater
than about 1,000 Da. Also provided are embodiments where the large organic
analyte is a
polymer. In certain embodiments the polymer is a biopolymer. In further
embodiments the
biopolymer is a polynucleic acid, and in still further embodiments the
biopolymer is an
oligonucleotide. Additionally provided are embodiments where the biopolymer is
a protein,
polypeptide, or oligosaccharide.
In yet other embodiments, the sample is placed on a cooled sample stage in
order to
maintain a low vapor pressure of the sample in the vacuum chamber of the mass
spectrometer.
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-12-
The sample stage is cooled below about 273°K, more typically between
about 170 to about
190°K, and most typically to about 180°K.
3.0 BRIEF DESCRIPTION OF THE DRAWINGS
The following drawings form part of the present specification and are included
to
further demonstrate certain aspects of the present invention. The invention
may be better
understood by reference to one or more of these drawings in combination with
the detailed
description of specific embodiments presented herein.
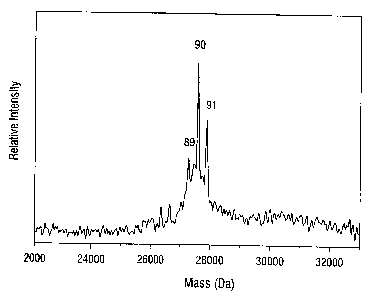
FIG. 1 is a laser desorption/ionization time-of flight mass spectrum of a
mixture of
single-stranded DNA oligomers 89, 90, and 91 nucleotides in length obtained
using 8-
hydroxyquinoline as the matrix. The laser wavelength was 355 nm.
FIG. 2 is a laser desorption/ionization time-of flight mass spectrum of a
double-
I S stranded PCR product at 315 kDa per strand (greater than approximately
1000 nucleotides in
length) using a 4-nitrophenol matrix. The laser wavelength was 355 nm.
4.0 DESCRIPTION OF ILLUSTRATIVE EMBODIMENTS
In accordance with the present invention, methods are provided for the
preparation of
samples for analysis by mass spectroscopy to minimize undesired fragmentation.
Select light
absorbing molecules, containing hydroxy functionalities (but not carboxylic
functionalities) and
having significant sublimation rates at room temperature under vacuum, are
used as matrices in
MALDI mass spectrometry. Hydroxy functionalities offer advantage over
carboxylic
functionalities due to their increased acidity in the excited state (Huppert
et al., 1981 ) and also
typically provide lower intermolecular binding energies to increase
volatility. Representative
examples of matrix compounds include, but are not limited to,
hydroxyquinolines, phenols, and
hydroxynaphthalenes.
Samples are prepared by dissolving the analyte in a solution containing the
matrix
molecule, with the bulk of the solution being one or more solvents which are
subsequently
allowed to evaporate before mass analysis begins. Typically, the anaIyte will
be present in the
solution at a concentration of about 0.05 M to about I .0 M.
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-13-
The solvent evaporation may be conducted at a temperature range of about
20°C to
about 30°C, with room temperature, about 25°C being most
preferred. The evaporation results
in the formation of a crystalline matrix, composed in part (between about 30%
by weight to
about 100% by weight) of the subject matrix molecule. Typically the matrix
molecular weight
is greater than about 90 Da, preferably between about 90 Da and about 400 Da.
Due to the high
volatility of these matrix molecules under vacuum at room temperature, the
samples must be
cooled in the mass spectrometers vacuum system to a vapor pressure between
about 10-~° Torr
and about 10-5 Ton, but not exceeding about 10-5 Torr. These matrix molecules
are termed
herein as volatile, light-absorbing, hydroxy-bearing matrix molecules. As used
herein the term
volatile refers to a molecule having a sublimation rate at room temperature of
greater than or
equal to 0.1 ~m.miri' at a pressure of about 10-5 Torr or less. and the term
light absorbing
refers to a molecule having an absorption coefficient greater than about 10
l.crri'.mol-~.
Two low-sublimation-temperature molecules in particular function effectively
as
matrices for MALDI of nonvolatile organic molecules for detection by mass
spectrometry. The
compounds, 8-hydroxyquinoline (8HQ) and 4-nitrophenol (4NP), both contain a
hydroxy
functional group. The former is especially effective for high-resolution
analysis of DNA less
than approximately 100 nucleotides (30 kDa), and the latter is especially
effective for sensitive
detection of higher mass molecules.
Compounds contemplated for analysis using the present invention include a vast
array
of large organic molecules. As used herein, the term "large organic molecule"
refers to a
compound having a molecular weight of greater than about 1000 Da. Also as used
herein, the
term "nonvolatile" refers to a molecule which, when present in its pure, neat
form and heated,
does not sublimate intact to any significant extent. Also included in the
definition of
nonvolatile compounds are compounds which, when present in their pure neat
form, cannot be
practically analyzed by mass spectrometry when conventional gas chromatography
methods are
employed in the sampling process. Representative of such organic compounds are
polynucleic
acids, polypeptides, oligosaccharides, PNAs and synthetic polymers. Polymeric
compounds are
also contemplated for analysis using the present invention. In particular
biopolymers which are
subject to fragmentation during mass analysis. Representative biopolymers
include polymers
of amino acids, nucleic acids, saccharides, carbohydrates and polypeptides.
CA 02302036 2003-02-06
77718-61 (S)
-14-
The mass spectrometry may be accomplished by one of several techniques such as
time-
of flight, magnetic sector or ion trap. Preferably, the mass spectrometry
technique for use with
the present invention will be time-of flight.
The volatility of the matrix crystals necessitates that the sample stage of
the mass
spectrometer be cooled to substantially below room temperature where the
sublimation rate is
between about 0.1 wm.miri ~ and about 0.1 mm.min~~~. A preferred approach is
to use a liquid
nitrogen cooled sample stage, accomplished by flowing liquid nitrogen through
a copper
sample holder. Thus, the sample is cooled to less than 273°K,
preferably between about 170
and 190 °K or to about 180 °K.
Wavelengths from the ultraviolet to infrared may be employed, depending on the
cooled
matrix being analyzed. Generally, one of skill in the an will understand ;hat
the appropriate
wavelength will be one where light absorption is significant for the molecule
i3eing analyzed.
The disclosed low-sublimation temperature matrices and methods for using them
to
determine the mass of a large organic molecule or prepare a large organic
molecule for mass
spectral analysis may be used in a variety of MS applications, such as MS
sequencing of
nucleic acids; MS analysis of single nucleotide polymorphisms (SNPs); and MS
analysis of
simple sequence repeats (SSRs), short tandem repeats (STlts), and
microsatellite repeats
(MRs).
For example, the methods disclosed herein may be used in nucleic acid
sequencing
methods involving obtaining nucleic acid fragments using a four base Sanger
sequencing
reaction, performing MS on the products and determining the nucleic acid
sequence from the
mass differences between the peaks. The nucleic acid fragments may be obtained
by
hybridizing a DNA primer to a DNA template and extending the primer by a DNA
polymerase
in the presence of deoxy- and dideoxy- nucleotides. T:ze DNA template may
generally contain
the DNA fragment to be sequenced and a region complementary to the primer. The
DNA
primers may also contain a biotin which allows for capture to a solid phase
and a single,
chemically cleavable internal linkage (such as a ~'-or 3'-(S)-phosphorothioate
linkage which is
cleavabIe by a silver ion catalyzed reaction). The cleavage chemistry of the
internal linkage
combined with the biotin capture are described in L'.S. Patent No. x,700,642.
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-15-
The nucleic acid fragments may be further processed prior to MS analysis.
Generally,
these processing steps involve binding the nucleic acid fragments to a
streptavidin solid
support, washing the bound fragments, and cleaving at the internal cleavage
site to release the
- nucleic acid fragment from the solid support. Typically the bound fragments
are first washed
with a denaturant, such as aqueous NaOH, to remove unbound DNA and enzyme and
then with
a series of ammonium acetate washes. Following cleavage, the cleaved extension
products may
be prepared for MS analysis by drying; mixing the solid residue with the
matrix material and
ammonium citrate solution; spotting the mixture by pipette onto a plate; and
allowing the
mixture to dry.
The methods for MS SNP analysis are very similar to the DNA sequencing methods
except that only dideoxynucleotides are employed.
These low-sublimation temperature matrices may also be used for analyzing
SSRs,
STRs, and MRs involving the determination of the number of repetitive units
contained in
amplification products by MS. The amplification products are typically
obtained by
hybridizing a DNA primer to a DNA target molecule and extending the primer by
a DNA
polymerase. Similar to the sequencing methods, the DNA primer contains a
region
complementary to the DNA target molecule adjacent to the SSR-, STR-, or MR-
containing
region. The primer may also contain biotin and internal cleavable linkages.
The following examples are included to demonstrate preferred embodiments of
the
invention. It should be appreciated by those of skill in the art that the
techniques disclosed in
the examples which follow represent techniques discovered by the inventor to
function well in
the practice of the invention, and thus can be considered to constitute
preferred modes for its
practice. However, those of skill in the art should, in light of the present
disclosure, appreciate
that many changes can be made in the specific embodiments which are disclosed
and still
obtain a like or similar result without departing from the spirit and scope of
the invention.
5.0 EXAMPLES
The following examples are included to demonstrate preferred embodiments of
the
invention. It should be appreciated by those of skill in the art that the
techniques disclosed in
the examples which follow represent techniques discovered by the inventors to
function well in
the practice of the invention, and thus can be considered to constitute
preferred modes for its
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-16-
practice. However, those of skill in the art should, in light of the present
disclosure, appreciate
that many changes can be made in the specific embodiments which are disclosed
and still
obtain a like or similar result without departing from the spirit and scope of
the invention.
5.1 Example 1 - Materials and Methods
A time-of flight mass spectrometer similar to that previously described by Wu
et al.
(1994) and Hunter et al. (1997) was used, having pulsed delayed ion
extraction. The sample
stage was floated at 20 kV, and after some delay time (approximately several
hundred
nanoseconds, dependent on mass), ions were extracted by a 2.7 kV pulse and
focused into a I-
meter flight tube. The signal output from the dual microchannel plate detector
was amplified
and digitized with 5 ns time resolution.
Laser wavelengths of either 355 or 266 nm were employed for
desorption/ionization in
the examples below. Comparable positive and negative ion signals were observed
from
oligonucleotide analytes.
1 S The temperature of the sample on a liquid nitrogen-cooled sample stage was
maintained
at approximately 180 K as measured by thermocouple wires, low enough to
maintain a matrix
vapor pressure of less than 10'5 Torr.
5.2 EXAMPLE 2 - DNA Oligomer Analysis Employing 8-Hydroxyquinoline as a
Matrix
The preparative solution for the 8HQ matrix began by using 0.2 M 8HQ in 1:1
(volume)
acetone:butanone. To reduce alkali-metal adduct ion formation, to that initial
8HQ solution
was added an equal volume of 50 mM aqueous diammonium citrate, resulting in a
25 mM final
diammonium citrate concentration and 0.1 M 8HQ concentration. 8HQ is known to
chelate
trace amounts of metal ions, especially copper, but the addition of CDTA
(trans-1,2-
diaminocyclohexane-N,N,N',N' tetraacetic acid monohydrate) effectively
suppressed copper
adducts in the mass spectrum; a small aliquot of concentrated CDTA was added
to a much
larger volume of the 8HQ solution to yield a I OmM CDTA concentration.
The oligonucleotide sample was obtained from polymerase chain reaction (PCR)
amplification of a short tandem repeat sequence at the human TH01 (tyrosine
hydroxylase
gene) locus. One of the strands was captured, denatured, washed, then released
to produce
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-17-
single-stranded products. An aliquot of aqueous solution of this THO1
oligonucleotide
(estimated 10 pmol quantity) was first evaporated in a vacuum evaporator to
remove the water,
and then one microliter of the matrix solution was added to the dried DNA.
This resulting
solution was pipetted onto a silicon substrate mounted on a copper sample
holder. After air-
drying of the solvent and resultant crystallization of the matrix, the sample
was placed on the
cryogenically-cooled sample stage in the mass spectrometer.
8HQ is an effective matrix for high resolution studies of DNA oligomers less
than
approximately 100 nucleotides in length. FIG. 1 illustrates the mass
resolution attainable for
single-stranded oligonucleotides of about 27 kDa using 355 nm pulsed laser
light for desorption
and summing mass spectra over 200 laser pulses. DNA oligomers containing 89,
90, and 91
nucleotides have a mass resolution (m/Om) of 650, 6?~, and 700, respectively
at full width at
half height. Spectra of oligonucleotides in 8HQ matrix typically have a low
background ion
signal and high signal-to-noise levels.
5.3 EXAMPLE 3 - DNA Oligomer Analysis Employing 4-Nitrophenol as a Matrix
The preparative solution for the 4NP matrix was 0.5 M 4NP in 1:1 (volume)
methanol:water containing diammonium citrate at 50 mM final concentration. One
microliter
of the matrix solution was added to dried DNA which was a double-stranded PCR
product
estimated at 10 pmol quantity derived from an unknown cDNA insert in a vector.
This
resulting solution was pipetted onto a silicon substrate mounted on a copper
sample holder.
After air-drying of the solvent and resultant crystallization of the matrix,
the sample was placed
on the cryogenically-cooled sample stage in the mass spectrometer. FIG. 2 is
the resulting
time-of flight mass spectrum using 355 nm laser light for desorption and
summing over 1000
laser pulses yielding an estimated mass of 315 kDa which corresponds to an
estimated number
of bases exceeding 1,000. The width of the peak originates in part from the
mass difference of
the two complementary DNA strands (denatured during analysis) and partly from
adduct
formation as well as fragmentation. DNA oligomers have not previously been
reported to be
detected in this size range.
CA 02302036 2000-02-25
WO 98/54751 PCTNS98/11003
-18-
All of the compositions and methods disclosed and claimed herein can be made
and
executed without undue experimentation in light of the present disclosure.
While the
compositions and methods of this invention have been described in terms of
preferred
embodiments, it will be apparent to those of skill in the art that variations
may be applied to the
compositions and methods and in the steps or in the sequence of steps of the
method described
herein without departing from the concept, spirit and scope of the invention.
More specifically,
it will be apparent that certain agents which are both chemically and
physiologically related
may be substituted for the agents described herein while the same or similar
results would be
achieved. All such similar substitutes and modifications apparent to those
skilled in the art are
deemed to be within the spirit, scope and concept of the invention as defined
by the appended
claims.
CA 02302036 2003-02-06
~77is-6~.(S)
-19-
6.0 R>a88»NC>ES
The following references provide exemplary
procedural or other details supplementary to those'set forth
herein.
US 4,920,264 to Bod~er.
US 5,118,937 to Hillenkasnp et al.
US S,I35,870 to Willisms st al.
Abrams et al , "Comprehensive Detection of Single Base Changes in Human
Genomic DNA
Using Denaturing Gradient Gel Electrophoresis and a GC Clamp," Genomics, 7:463
475, ( 1990).
Gimon et v1., "Are Proton Transfer Reactions of Excited States Involved in W
La~e~
Desorption Ionization?," Organic Mass Spectrometry, 27:827-830 (1992).
Hooter et al., "Cryogenic Frown Solution Matrices for Analysis of DNA by Time-
of Flight
Mass Spectrometry," Analytical Chemistry, 69:3608-12 ( 1997).
Hopper st al., "Laser Studies of Proton Transfer," Advances in Chemical
Physics, 47:643-679
(1981).
Liu ~ et al. "Use of a Nitrocellulose Film Subahatc in Matrix-Assisted Laser
Desorption/Ionization Mass Spectrometry for DNA Mapping and
Screening,".Analytical
Chemistry, 67:3482-3490 ( 1995).
Nelson et al.. "Detection of Human IgM at m/z ~ 1 MDa," Rapid Commr~nications
in Mass
Spectrometry, 9:625 (1995).
Orita et oL, "Detection of Polymorphistns of Human DNA by Gel Electrophoresis
as Singfe
Strand Conformation Polymorphisms." Proc. Natl. Acvd Sci. (ISA, 86:2766-
2770,1989
Ssleeba et al., "Chemical Cleavage of Mismatch to Detect Mutations," Methods
F.tnymology,
21786-295 (1993).
Speogla~ et al., "Laser Mass Analysis in Biology," Bsr. Bunseaqes Plrys
Clrem., 93(3):396-
402, (1989).
Tanaka et al., "Protein and Polymer Analyses up to m/z 100 000 by Lascr
Ionization Time-of
flight Mass Spectrometry," Rapid Common in Mass Spectrometry, 2:151-153
(1988).
Tang et al. "Detection of 500-Nucleotide DNA by Laser Desorption Msss
Spectrometry,"
Rapid Communications in Mass Spectromerry, 8;727-730 ( 1994).
Wu et aL "Matrix-Assisted Laser Desorption Time-of Flight Mass Spectrometry of
Oligonucieotides Using 3-Hydroxypicolinic Acid as an Ultraviolet-Sensitive
Matrix,"
Rapid Communications in Mass Spectrometry, 7:142.146 (1993).
CA 02302036 2000-02-25
WO 98/54751 PCT/US98/11003
-20-
Wu et al. "Time-of Flight Mass Spectrometry of Underivatized Single-Stranded
DNA
Oligomers by Matrix-Assisted Laser Desorption," Analytical Chemistry, 66:1637-
1645
( 1994).
Youil et al., "Screening for Mutations by Enzyme Mismatch Cleavage with T4
Endonuclease
VII," Proc. Natl. Acad. Sci. USA, 92:87-91 (1995).