Note: Descriptions are shown in the official language in which they were submitted.
CA 02304984 2000-03-15
r.
wo 99n2003 ~ PCTNS98n2688
TITLE
BINARY VIRAL EXPRESSION SYSTEM IN PLANTS
FIELD OF INVENTION
The present invention relates to the field of molecular biology and the
genetic transformation of plants with foreign gene fragments. More
particularly,
this invention relates to a heritable plant viral expression system useful for
expressing transgenes in plants.
BACKGROUND OF THE INVENTION
Plant transgenic work is beset with low and inconsistent levels of
10 transgene expression. Episomal vectors are expected to overcome these
problems.
In microbes, episomal (plasmid) vectors are possible because these vectors can
be
maintained by selection. Although plant viruses have been used as episomal
expression vectors, their use has been restricted to transient expression
because of
lack of selection and/or their cellular toxicity (U.S. Pat. No. 4,855,237,
WO 9534668).
Plant viruses
Viruses are infectious agents with relatively simple organization and
unique modes of replication. A given plant virus may contain either RNA or
DNA, which may be either single- or double-stranded.
20 Rice dwarf virus (RDV) and wound tumor virus (WTV) are examples of
double-stranded RNA plant viruses. Single-stranded RNA plant viruses include
tobacco mosaic virus (TMV), turnip yellow mosaic virus (TYMV), rice necrosis
virus (RNV) and brome mosaic virus (BMV). The RNA in single-stranded RNA
viruses may be either a plus (+) or a minus (-) strand. Although many plant
25 viruses have RNA genomes, organization of genetic information differs
between
groups. The genome of most monopartite plant RNA viruses is a single-stranded
molecule of (+)-sense. There are at least 11 major groups of viruses with this
type
of genome. An example of this type of virus is TMV. At least six major groups
of plant RNA viruses have a bipartite genome. In these, the genome usually
30 consists of two distinct (+)-sense single-stranded RNA molecules
encapsidated in
separate particles. Both RNAs are required for infectivity. Cowpea mosaic
virus
(CPMW) is one example of a bipartite plant virus. A third major group,
containing at least six major types of plant viruses, is tripartite, with
three
(+)-sense single-stranded RNA molecules. Each strand is separately
encapsidated,
35 and all three are required for infectivity. An example of a tripartite
plant virus is
alfalfa mosaic virus (AMV). Many plant viruses also have smaller subgenomic
mRNAs that are synthesized to amplify a specific gene product. Plant viruses
with double-stranded DNA genome include Cauliflower Mosaic virus (CaMV).
CA 02304984 2000-03-15
WO 99/22003 PCT/US98I22688
Geminiviruses
Plant viruses with single-stranded DNA genomes are represented by
geminiviruses and include African Cassava Mosaic Virus (ACMV), Tomato
Golden Mosaic Virus (TGMV), and Maize Streak Virus (MSV). Geminiviruses
5 are subdivided on the basis of whether they infect monocots or divots and
whether
their insect vector is a leafhopper or a whitefly. Subgroup I geminiviruses
are
leafhopper-transmitted that infect monocotyledonous plants (e.g., Wheat Dwarf
Virus), Subgroup II geminiviruses are leafhopper-transmitted that infect
dicotyledonous plants (e.g., Beet Curly Top Virus), and Subgroup III
10 geminiviruses are whitefly-transmitted that infect dicotyledonous plants
(e.g.,
Tomato Golden Mosaic Virus, TGMV, and African Cassava Mosaic Virus,
ACMV). Subgroup I and II geminiviruses have a single (monopartite) genome.
Subgroup III geminiviruses have a bipartite genome. For example, TGMV and
ACMV consist of two circular single-stranded DNA genomes, A and B, of ca.
15 2.8 kB each in size. DNA of genome A and DNA of genome B of a given
Subgroup III virus have little sequence similarity, except for an almost
identical
common region of about 200 bp. While both DNA of genome A and DNA of
genome B are required for infection, only DNA of genome A is necessary and
sufficient for replication and DNA of genome B encodes functions required for
20 movement of the virus through the infected plant.
In both TGMV and ACMV, DNA A contains four open reading frames
(ORFs) that are expressed in a bidirectional manner and arranged similarly.
The
ORFs are named according to their orientation relative to the common region,
i.e.,
complementary (C) versus viral (V) in ACMV and leftward (L) or rightward (R)
25 in TGMV. Thus, ORFs ALI, AL2, AL3, and AR1 of TGMV are homologous to
AC1, AC2, AC3, and AV 1, respectively, of ACMV. Three major transcripts have
been identified in ACMV DNA A and these map to the AV 1 and AC 1 ORFs,
separately and the AC21AC3 ORFs together. There is experimental evidence for
the function of these ORFs. Thus, in ACMV AC 1 encodes a replication protein
30 that is essential and sufficient for replication, AC2 is required for
transactivation
of the coat protein gene, AC3 encodes a protein that is not essential for
replication
but enhances viral DNA accumulation, and AV I is the coat protein gene. Except
for the essential viral replication protein (encoded by AC1 and AL1 in ACMV
and
TGMV, respectively), geminivirus replication relies on host replication and
35 transcription machinery. Although geminiviruses are single-stranded plant
DNA
viruses, they replicate via double-stranded DNA intermediate by 'rolling
circle
replication'.
2
CA 02304984 2000-03-15
WO 99!12003 - PCTNS98I22688
Viruses as Expression Vectors
Construction of plant viruses to introduce and express non-viral foreign
genes in plants has been demonstrated (IJ.S. Pat. No. 4,855,237, WO 9534668).
When the virus is a DNA virus, the constructions can be made to the virus
itself.
S Alternatively, the virus can first be cloned into a bacterial plasmid for
ease of
constructing the desired viral vector with the foreign DNA. If the virus is an
RNA
virus, the virus is generally cloned as a cDNA and inserted into a plasmid.
The
plasmid is then used to make all of the constructions. The RNA virus is then
produced by transcribing the viral sequence of the plasmid and translation of
the
viral genes to produce the coat proteins) which encapsidate the viral RNA.
Alternatively, the cDNA can be cloned behind a heterologous plant promoter,
introduced into a plant cell, and used to transcribe the viral RNA that can
replicate
autonomously [Sablowski et al. (1995) Proc. Natl. Acad. Sci. USA vol 92,
pp 6901-6905].
Geminiviruses have many advantages as potential plant expression
vectors. These include 1 ) replication to high copy numbers
nonsymptomatically,
2) small, well-characterized genomes, 3) assembly into nucleosomes, and
4) nuclear transcription. The DNA A component of these viruses is capable of
autonomous replication in plant cells in the absence of DNA B. Vectors in
which
the coat protein ORF has been replaced by a heterologous coding sequence have
been developed and the heterologous coding sequence expressed from the coat
protein promoter [Hayes et al. (1989) Nucleic Acids Res. vol. 17, pp. 2391-
403;
Hayes et al. (1988) Nature (London) vol. 334, pp. 179-82].
Greater than full length copies of wild type TGMV A and B genomes were
transformed into petunia [Rogers et al. (1986) Cell (Cambridge, Mass.) vol.
45,
pp. 593-600]. Replication was reported in the primary transformants and in
some
of the selfed progeny consistent with its mendelian inheritance, indicating
that the
chromosomaliy-integrated master copy, not the replicon, is inherited. This
suggests that gametophytic and/or developing seed tissues lack the ability to
support replication. The report did not demonstrate whether the virus
replicated in
non-germinating seed tissue. Prior art shows that geminiviruses are not seed-
transmitted in nature [Goodman, ( 1981 ) J. Gen. Virol. vol. 54, pp. 9-21 ].
Thus,
there was no evidence that they can replicate in gametophytic tissue or
developing
seed.
Tomato Golden Mosaic Virus (TGMV) DNA A was modified by replacing
its coat protein coding sequence with that of NPT II or GUS reporter genes or
with'
that of 35S:NPT II gene and a greater than full length copy of the modified
viruses
were transformed into tobacco [Hayes et al. (1989) Nucleic Acids Res. vol. 17,
3
CA 02304984 2000-03-15
WO 99/22003 PCT/US98/22688
pp. 2391-403; Hayes et al. (1988) Nature (London) vol. 334, pp. I7.9-82].
Leave_s
of transgenic plants showed that the high levels of the reporter enzymes was
gene
copy number-dependent. However, replication of the vector and reporter gene
expression were not reported in seed and the genetic stability of the vector
in
S transgenic plants in subsequent generations was not reported. Use of the
African ,
Cassava Mosaic Virus (ACMV) in similar fashion has not been reported and it is
not known that ACMV DNA or the replication proteins) can be stably maintained
.
in progeny plants and whether it can replicate in seed tissues.
In one report, a chimeric gene in which the constitutive plant promoter,
10 355, was fused to the TGMV sequence containing ORFs AL1, AL2, and AL3
were transformed into Nicotiana benthamiana. Different transgenic lines showed
significant non-uniformity in the levels of 35S:AL1-3 gene expression as well
as
their ability to complement viral replication [Hanley-Bowdoin et al. (1989)
Plant
Cell vol. l, pp. 1057-67]. In another report, chimeric genes in which the
15 constitutive plant promoter, 35S, was fused to the coding sequence of TGMV
replication protein AL 1 were transformed into tobacco. The expression of TGMV
replication protein in the primary transformants supported the replication of
a
mutant genome A lacking the replication protein. [Hanley-Bowdoin et al. (
1990)
Proc. Natl. Acad. Sci. U.S.A. vol. 87, pp. 1446-SOJ. However, in both reports
20 neither the genetic stability of the chimeric replication protein gene
through
subsequent generations nor its ability to support viral replication in seed
tissue
was reported. In another report, chimeric genes in which the constitutive
plant
promoter, 35S, was fused separately to the coding sequences of TGMV
replication
proteins AL 1, AL2, and AL3 were transformed into tobacco (Hayes et al. (
1989)
25 Nucleic Acids Res. vol. 17, pp. 10213-22]. The TGMV replication protein was
shown to have been expressed in progeny but the genetic stability of the
chimeric
replication protein gene through subsequent generations was not reported.
Furthermore, it was not reported whether the transgenic plants will support
replication in seed tissue.
30 In another disclosure, Rogers et al. (EP 221044) demonstrated the
expression of foreign proteins in plant tissue using a modified "A" genome of
the
TGMV gemini virus. The foreign gene was inserted in place of the gene encoding
the viral coat protein and the resulting plasmid transformed into plant
tissue.
Rogers et al. did not report tissue specific expression of the foreign protein
and are
35 silent as to the genetic stability of the transforming plasmid.
All of the reported viral vectors have a major disadvantage. They were
either not shown to be stably maintained in transgenic plants and/or not
practically
useful. Thus, despite intense efforts to develop plant viral vectors and
viruses, no
4
CA 02304984 2000-03-15
. E
WO 99122003 ' ' PCTNS98/22688
commercially useful plant virus-based recombinant vectors have been developed
that are heritable and capable of episomal replication and expression in
desired
tissues) of the transgenic host plant without the need for infection every
generation. In fact, replication of plant viruses is expected to be
detrimental to the
5 growth and development of plant cells. For example, when greater than full
length copy of TGMV genome A is introduced into plant cell one-tenth as many
transgenic plants are obtained than when genome B is used or when control
transformations are done [Rogers et al. (1986) Cell (Cambridge, Mass.) vol.
45,
pp. 593-600]. The authors suggest that this may be due to expression of a gene
in
10 TGMV A DNA. Furthermore, crude extract of plants expressing tandem copies
of
both TGMV A and TGMV B genomes are unable to infect Nicotiana
benthamiana plants. This is consistent with having a low virus titer. Thus,
transgenic plants that do regenerate could be selected for low level
expression of a
toxic viral gene product and low level of viral replication. This is also
consistent
15 with the authors' finding that relatively few cells initiate release of the
virus, a
conclusion based on their observation that most of the tissues remain viable
and
nonsymptomatic. Similarly, poor replication in transgenic plants containing
35S:replication protein in other reports suggest that plants are either
selected for
poor expression of the replication protein, presumably because of its
toxicity, or
20 that the tissue-specific expression profiles of the replication gene is
different from
that of viral replication.
Recently, it was reported that 6 of 11 transgenic tobacco plants stably
transformed with a monopartite geminivirus (Tobacco Yellow Dwarf Virus) with
a functional replication gene showed episomal replication [Needham et al.
(1998)
25 Plant Cell Rep. vol. 17, pp. 631-639].
Silencing endogenous genes and transgene is an important technology [see
Senior et al. (1998) Biotechnol. Genet. Eng. Rev. vol. 15, pp. 79-119; Thomas
et
al. (1998) Plant Growth Regul. vol. 25, p. 205]. Silencing of endogenous genes
or
transgenes by viral infection or by stably incorporated virus in transgenic
plants
30 has been reported for RNA virus [Baulcombe et al. PCT Int. Appl. (1998),
Ruiz et
al. (1998) Plant Cell vol. 10, pp. 937-946], Geminiviruses [Kjemtrup et al.
(1998)
Plant J. vol. 14, pp. 91-100, Atkinson et al. Plant J. vol. 15, pp. 593-604],
and
Cauliflower Mosaic Virus [Al-Kaff et al. Science (Washington, D.C.)
279:2113-2115 (1998)]. However, regulated virus induced silening in transgenic
35 plants, which is expected to provide regulated gene silencing, has not been
reported.
To date, no plant virus-based recombinant vectors aFe known that are
heritable and capable of episomal replication and expression of foreign
proteins in
CA 02304984 2000-03-15
WO 99/Z2003 PCT/US98122688
target tissues) of a transgenic host plant without the need for infection in
every-
generation.
SUMMARY OF THE INVENTION
In one embodiment, the present invention provides a binary transgenic
5 viral expression system for replicating and increasing expression of a
target gene
comprising a) a heritable proreplicon lacking a functional replication gene
for
autonomous episomal replication and comprising:
i) cis-acting viral elements required for viral replication;
ii) a target gene comprising at least one suitable regulatory
sequence; and
iii) flanking sequences that enable the excision of the elements of (i)
and (ii); and,
b) a heritable chimeric traps-acting replication gene comprising a
regulated plant promoter operably-linked to a viral replication protein coding
sequence.
In another embodiment, the present invention provides the binary
transgenic viral expression system described above but without a target gene,
wherein expression of the traps-acting replication gene in cells containing
the
proreplicon results in replicon replication without a target gene.
20 The expression system of the present invention is useful for the controlled
replication of viral vectors in transgenic plants. Both components of the
system
are chromosomally-integrated. One component is a chimeric traps-acting
replication gene in which the coding sequence of the geminivirus replication
proteins) is placed under the control of a tissue- or development-specific
and/or
25 inducible promoter. The second component is a proreplicon, which is unable
to
replicate by itself but does so in the presence of viral replication
protein(s). The
two components may be introduced together into a transgenic plant or brought
together by crossing transgenic plants carrying the separate components. Also
provided are methods of making the expression cassettes and methods of using
30 them to produce transformed plant cells having an altered genotype and/or
phenotype.
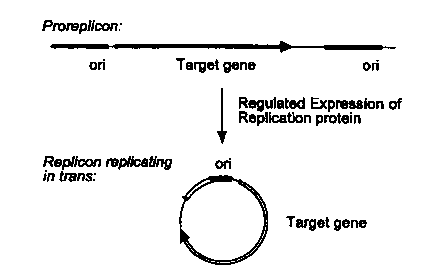
The principal aspect invention is illustrated in Figure 1. Figure 1
illustrates a scheme for transactivating replication of the proreplicon in
traps.
Regulated expression of a chromosomally integrated chimeric replication gene
35 will result in the release and replication of the replicon from a
chromosomally
integrated master copy of the proreplicon. -
The different components of the invention are heritable independently and
may be introduced together into a transgenic plant or brought together by
crossing
6
CA 02304984 2000-03-15
WO 99122003 - PCTIUS98J22688
transgenic plants carrying the separate components, such as by the method to
produce TopCross~ high oil com seed [U.S. Patent No. 5,704,160). Also
provided are methods of making the expression cassettes and methods of using
them to produce transformed plant cells having an altered genotype and/or
5 phenotype.
BRIEF DESCRIPTION OF FIGURES AND SEQUENCE DESCRIPTIONS
Figure 1 illustrates excising and activating a proreplicon via the expression
of a chimeric traps-acting replication gene.
The invention can be more fully understood from the following detailed
10 description and the accompanying sequence listing. The sequence listings
attached hereto comply with the rules governing nucleotide and/or amino acid
sequence disclosures in patent applications as set forth in 37 C.F.R. ~ 1.821-
1.825.
SEQ ID NOs:I-14 refer to primers used in the Examples.
DETAILED DESCRIPTION OF THE INVENTION
15 The present invention provides a regulated binary expression system that
uses various genetic elements of a plant virus. The expression system is
useful for
the regulated replicon replication and expression of target genes in plants
either
for producing foreign proteins or for silencing endogenous plant genes and
particularly for achieving stable expression in terminally-differentiated
cells.
20 Applicant has solved the stated problem by providing a two-component,
chromosomally-integrated viral expression system comprising a proreplicon and
a
traps-acting replication gene. The proreplicon contains the cis-acting viral
sequences required for replication and a target gene under the control of
suitable
regulatory sequences. The proreplicon is incapable of self replication in
plant
25 cells because it lacks an essential traps-activating replication gene. The
second
component of the system, a chimeric traps-acting replication gene, consists of
a
regulated promoter operably-linked to the coding region for a viral
replication
protein. Expression of the viral replication protein results in the release
and
replication of the replicon from the proreplicon.
30 Plant cells containing proreplicon replicate the replicon only in the
presence of the replication protein. Thus, regulated expression of the
chimeric
replication gene in such cells results in regulated replicon replication and
target
gene amplification. Using the present system, Applicant has demonstrated that
(i) soybean and corn seed tissue will support geminivirus replication; and
(ii) that
35 the expression system will effect the expression of target genes in
transgenic
plants. The present invention advances the art by providing plant viral
vectors
(a) which are maintained stably in the chromosome of transgenic plants; (b)
whose
replication is controlled by the regulated expression of the replication
proteins;
7
CA 02304984 2000-03-15
WO 99!22003 - PCTlUS98I22688
and (c) contain nucleic acid sequences that are either homologous to plant
endogenous genes or transgenes or that encode foreign proteins that may be
produced in the transgenic plant.
The following definitions are to be used to understand the meaning of
terms used in this disclosure.
The term "gene" refers to a nucleic acid fragment that expresses mRNA,
functional RNA, or specific protein, including regulatory sequences. The term
"native gene" refers to a gene as found in nature. The term "chimeric gene"
refers
to any gene that contains 1 ) regulatory and coding sequences that are not
found
10 together in nature, or 2) sequences encoding parts of proteins not
naturally
adjoined, or 3) parts of promoters that are not naturally adjoined.
Accordingly, a
chimeric gene may comprise regulatory sequences and coding sequences that are
derived from different sources, or comprise regulatory sequences and coding
sequences derived from the same source, but arranged in a manner different
from
1 S that found in nature. A "transgene" refers to a gene that has been
introduced into
the genome by transformation and is stably maintained. Transgenes may include,
for example, genes that are either heterologous or homologous to the genes of
a
particular plant to be transformed. Additionally, transgenes may comprise
native
genes inserted into a non-native organism, or chimeric genes. The term
20 "endogenous gene" refers to a native gene in its natural location in the
genome of
an organism.
The term "wild-type" refers to the normal gene, virus, or organism found
in nature without any known mutation.
The term "genome" refers to the complete genetic material of an organism.
25 The term "coding sequence" refers to a DNA or RNA sequence that codes
for a specific amino acid sequence and excludes the non-coding sequences. The
terms "open reading frame" and "ORF" refer to the amino acid sequence encoded
between translation initiation and termination codons of a coding sequence.
The
terms "initiation codon" and "termination codon" refer to a unit of three
adjacent
30 nucleotides ('codon') in a coding sequence that specifies initiation and
chain
termination, respectively, of protein synthesis (mRNA translation).
A "functional RNA" refers to an antisense RNA, ribozyme, or other RNA
that is not translated. It may be derived from any part of a gene, including
its open
reading frame, 5' non-coding sequence, or 3' non-coding sequence.
35 The terms "regulatory sequences" or "suitable regulatory sequences" refer
to nucleotide sequences located upstream (5' non-coding sequences), within, or
downstream (3' non-coding sequences) of a coding sequence, and which influence
the transcription, RNA processing or stability, or translation of the
associated
8
CA 02304984 2000-03-15
WO 99!22003 - PCT/US98122688
coding sequence. Regulatory sequences include enhancers, promoters,
translation
leader sequences, introns, and polyadenylation signal sequences. They include
natural and synthetic sequences as well as sequences which may be a
combination
of synthetic and natural sequences.
5 "5' non-coding sequence" refers to a nucleotide sequence located 5'
(upstream) to the coding sequence. It is present in the fully processed mRNA
upstream of the initiation codon and may affect processing of the primary
transcript to mRNA, mRNA stability or translation efficiency. (Turner et al.
(1995) Molecular Biotechnology vol. 3, p. 225).
10 "3' non-coding sequence" refers to nucleotide sequences located 3'
(downstream) to a coding sequence and include polyadenylation signal sequences
and other sequences encoding regulatory signals capable of affecting mRNA
processing or gene expression. The polyadenylation signal is usually
characterized by affecting the addition of polyadenylic acid tracts to the 3'
end of
1 S the mRNA precursor. The use of different 3' non-coding sequences is
exemplified
by ingelbrecht et al. (1989) Plant Cell vol. I, pp. b71-680.
"Promoter" refers to a nucleotide sequence, usually upstream (5') to its
coding sequence, which controls the expression of the coding sequence by
providing the recognition for RNA polymerase and other factors required for
20 proper transcription. "Promoter" includes a minimal promoter that is a
short DNA
sequence comprised of a TATA- box and other sequences that serve to specify
the
site of transcription initiation, to which regulatory elements are added for
control
of expression. "Promoter" also refers to a nucleotide sequence that includes a
minimal promoter plus regulatory elements that is capable of controlling the
25 expression of a coding sequence or functional RNA. This type of promoter
sequence consists of proximal and more distal upstream elements, the latter
elements often referred to as enhancers. Accordingly, an "enhancer" is a DNA
sequence which can stimulate promoter activity and may be an innate element of
the promoter or a heterologous element inserted to enhance the level or tissue-
30 specificity of a promoter. It is capable of operating in both orientations
(normal or
flipped), and is capable of functioning even when moved either upstream or
downstream from the promoter. Both enhancers and other upstream promoter
elements bind sequence-specific DNA-binding proteins that mediate their
effects.
Promoters may be derived in their entirety from a native gene, or be composed
of
35 different elements derived from different promoters found in nature, or
even be
comprised of synthetic DNA segments. A promoter may also contain DNA
sequences that are involved in the binding of protein factors.which control
the
9
CA 02304984 2000-03-15
WO 99122003 PCTII3S98122688
effectiveness of transcription initiation in response to .physiological .or
developmental conditions.
"Constitutive expression" refers to expression using a constitutive
promoter. "Conditional" or "regulated expression" refer to expression using a
5 regulated promoter, respectively. "Constitutive promoter" refers to
promoters that
direct gene expression in all tissues and at all times. Examples of
constitutive
promoters include CaMV 35S promoter and nopaline synthase promoter.
"Regulated promoter" refers to promoters that direct gene expression not
constitutively but in a temporally and/or spatially regulated manner and
include
both tissue-specific and inducible promoters. It includes natural and
synthetic
sequences as well as sequences which may be a combination of synthetic and
natural sequences. Different promoters may direct the expression of a gene in
different tissues or cell types, or at different stages of development, or in
response
to different environmental conditions. New promoters of various types useful
in
plant cells are constantly being discovered; numerous examples may be found in
the compilation by Okamuro et al. ( 1989) Biochemistry of Plants vol. 15,
pp. 1-82. Since in most cases the exact boundaries of regulatory sequences
have
not been completely defined, DNA fragments of different lengths may have
identical promoter activity.
20 "Tissue-specific promoter" refers to regulated promoters that are not
expressed in all plant cells but only in one or more cell types in specific
organs
(such as leaves or seeds), specific tissues (such as embryo or cotyledon), or
specific cell types (such as leaf parenchyma or seed storage cells}. These
also
include promoters that are temporally regulated (such as in early or late
embryogenesis), during fruit ripening in developing seeds or fruit, in fully
differentiated leaf, or at the onset of senescence.
"Inducible promoter" refers to those regulated promoters that can be
turned on in one or more cell types by an external stimulus (such as a
chemical,
light, hormone, stress, or pathogen).
30 The term "operably-linked" refers to the association of nucleic acid
sequences on a single nucleic acid fragment so that the function of one is
affected
by the other. For example, a promoter is operably-linked with a coding
sequence
or functional RNA when it is capable of affecting the expression of that
coding
sequence or functional RNA (i.e., that the coding sequence or functional RNA
is
35 under the transcriptional control of the promoter). Coding sequences can be
operably-linked to regulatory sequences in sense or antisense orientation.
CA 02304984 2000-03-15
WO 99122003 - PCT/US98lZ2688
The term "expression" refers to the transcription and stable accumulation_
of sense (mRNA) or functional RNA. Expression may also refer to the production
of protein.
"Altered levels" refers to the level of expression in transgenic organisms
that differs from that of normal or untransformed organisms.
"Overexpression" refers to the level of expression in transgenic organisms
that exceeds levels of expression in normal or untransformed organisms.
"Antisense inhibition" refers to the production of antisense RNA
transcripts capable of suppressing the expression of protein from an
endogenous
gene or transgene.
The terms "co-suppression" and "transwitch" refer to the production of
sense RNA transcripts capable of suppressing the expression of identical or
substantially similar transgene or endogenous genes (U.S. Patent No.
5,231,020).
The term "gene silencing" refers to inhibition or down-regulation of an of
an endogenous gene or a transgene that is substanitally similar to the target
gene.
The term "homologous to" refers to the similarity between the nucleotide
sequence of two nucleic acid molecules or between the amino acid sequences of
two protein molecules. Estimates of such homology are provided by either
DNA-DNA or DNA-RNA hybridization under conditions of stringency as is well
understood by those skilled in the art [as described in Hames and Higgins
(eds.)
Nucleic Acid Hybridisation, IR.L Press, Oxford, U.K.J; or by the comparison of
sequence similarity between two nucleic acids or proteins. Homologous genes
will
be "substantially similar" to each other.
"Substantially similar" refers to nucleic acid fragments wherein changes in
one or more nucleotide bases does not affect the ability of the nucleic acid
fragment to mediate alteration of gene expression by antisense or co-
suppression
technology. "Substantially similar" also refers to modifications of the
nucleic acid
fragments of the instant invention such as deletion or insertion of one or
more
nucleotide bases that do not substantially affect the functional properties of
the
resulting transcript.
For example, substantially similar sequences may be defined by their
ability to hybridize, under stringent conditions (O.1X SSC, 0.1% SDS,
65°C), with
specifically identified sequences. Preferred substantially similar nucleic
acid
fragments of the instant invention are those nucleic acid fragments whose DNA
sequences are at least 80% identical to specifically identified sequences,
either
over the entire length of the sequence or over a portion of the sequence. More
preferred nucleic acid fragments are at least 90% identical to specifically
identified sequences, either over the entire length of the sequence or over a
portion
11
CA 02304984 2000-03-15
WO 99J22003 PGT/US98/22688
of the sequence. Most preferred are nucleic acid fragments that are at least
95%_
identical to specifically identified sequences, either over the entire length
of the
sequence or over a portion of the sequence.
The term "binary transgenic viral expression system" describes the
expression system comprised of the proreplicon and the chirneric traps-acting
replication gene, functioning together to effect the expression of a target
gene in a
plant. Both elements of the system will be chromosomally-integrated and
heritable. Stimulation of the regulated promoter driving the traps-acting gene
will
cause the expression of viral replication proteins, which will in turn excise
the
replicon from the proreplicon and initiate replicon replication.
"Binary transgenic viral replication system" refers to a replication system
comprised of two chomosomally-integrated elements. The first element is a
proreplicon which lacks a target gene encoding a foreign protein. The second
element is comprised of a regulated promoter operably-linked to a chimeric
trans-
acting replication gene. The proreplicon and a chimeric traps-acting gene,
functioning together, will effect replication of the proreplicon in a plant in
a
regulated manner. Such a system is useful where replication of the virus is
desired
in a regulated manner but where no foreign gene expression is sought. For
example, the regulated expression of virus may be useful in confernng
resistance
to a plant to viral infection.
The term "target gene" refers to a gene on the replicon that expresses the
desired target sequence which is either a functional RNA or a mRNA encoding a
protein. The target gene is not essential for replicon replication.
Additionally,
target genes may comprise native non-viral genes inserted into a non-native
organism, or chimeric genes and will be under the control of suitable
regulatory
sequences. Thus, the regulatory sequences in the target gene may come from any
source, including the virus. Target genes may include coding sequences that
are
either heterologous or homologous to the genes of a particular plant to be
transformed. However, target genes do not include native viral genes. Proteins
encoded by target genes are known as "foreign proteins".
The terms "in cis" and "in traps" refer to the presence of DNA elements,
such as the viral origin of replication and the replication proteins) gene, on
the
same DNA molecule or different DNA molecules, respectively.
The terms "cis-acting sequence" and "cis-acting element" refer to DNA or
RNA sequence, whose function require them to be on the same molecule. An
example of a cis-acting sequence on the replicon is the viral replication
origin.
The terms "traps-acting sequence" and "traps-actingelement" refer to
DNA or RNA sequence, whose function does not require them to be on the same
12
CA 02304984 2000-03-15
WO 99/22003 PCT/US98/22688
molecule. Examples of traps-acting sequence is the replication gene (ACI or AL
1
in ACMV or TGMV geminiviruses, respectively), that can function in replication
without being on the replicon.
The term "cis-acting viral sequences" refers to viral sequences that are
S necessary for viral replication (such as the replication origin) and that
are in cis
orientation.
The terms "episome" and "replicon" refers to a DNA or RNA vector that
undergoes episomal replication in plant cells. It contains cis-acting viral
sequences, such as the replication origin, necessary for replication. It may
or may
not contain traps-acting viral sequences necessary for replication, such as
the viral
replication genes (for example, the AC 1 and AL 1 genes in ACMV and TGMV
geminiviruses, respectively). It may or may not contain a target gene for
expression in the host plant.
The term "replication-defective repIicon" refers to a replicon contains cis-
1 S acting viral sequences, such as the replication origin, necessary for
replication but
defective in an essential replication gene. Consequently, a replication-
defective
replicon can replicate episomalIy only when provided with the essential
replication protein in traps.
The term "episomal replication" and "replicon replication" refer to
replication of DNA or RNA viruses or virus-derived replicons that are not
chromosomally-integrated. It requires the presence of viral replication
protein(s),
is independent of chromosomal replication, and results in the production of
multiple copies of virus or replicons per host genome copy. The term
"autonomous episomal replication" refers to replication of a replicon that
contains
all cis- and traps-acting sequences, such as the replication gene, required
for
replication. Episomal replication does not require the presence of a
replication
gene provided in traps.
The term "replication origin" refers to a cis-acting replication sequence
essential for viral or episomal replication.
The term "proreplicon" refers to a replication-defective replicon that is
integrated into a bacterial plasmid or host plant chromosome. It is comprised
of
cis-acting viral sequences required for replication, and flanking sequences
that
enable the release of the replicon from it. In addition, the proreplicon may
contain
a target gene. In the case of RNA viruses, the flanking sequences include
regulatory sequences that allow generation of RNA transcripts that can
replicate in
the presence of replication protein. These regulatory sequences can be for
constitutive or regulated expression. Proreplicon lacks a gene encoding a
replication protein essential for replication. Therefore, it is unable to
undergo
13
CA 02304984 2000-03-15
~~:-' :.
wo 99nZ003 PCT/US98nZ688
autonomous episomal replication but can undergo episomal replication in the
presence of the replication protein provide in traps. Thus, its replication
requires
both release from the integration and the presence of the essential
replication gene
in traps. The release from integration can be triggered in different ways. For
5 example, in the case of a geminivirus, the proreplicon can be present as a
partial or
complete tandem duplication, such that a full-length replicon sequence is
flanked
by virus sequences and such that the duplicated viral sequence includes the
viral
replication origin. Thus, in this case, the proreplicon serves as a master
copy from
which replicons can be excised by replicational release in the presence of
10 replication proteins) [Bisaro, David. Recombination in geminiviruses:
Mechanisms for maintaining genome size and generating genomic diversity.
Homologous Recomb. Gene Silencing Plants (1994), 219-70. Editor(s):
Paszkowski, Jerzy. Publisher: Kluwer, Dordrecht, Germany]. In the case of RNA
virus (for example, Brome Mosaic Virus) proreplicons, the amplicon sequences
15 flanking the inactive replicon, which include regulatory sequences, allow
generation of the replicon as RNA transcripts that can replicate in traps in
the
presence of replication protein. These regulatory sequences can be for
constitutive
or regulated expression.
The terms "viral replication protein" and "replicase" refer to the viral
20 protein essential for viral replication. It can be provided in traps to the
replicon to
support its replication. Examples include, viral replication proteins encoded
by
AC1 and AL1 genes in ACMV and TGMV geminiviruses, respectively. Some
viruses have only one replication protein, others may have more than one.
Viral
replication proteins may also include replication proteins of single-stranded
RNA
25 viruses, such as the RNA-dependent RNA polymerases, when they can support
viral replication in traps, for example Brome Mosaic Virus {BMV).
The term "replication gene" refers to a gene encoding a viral replication
protein. In addition to the ORF of the replication protein the replication
gene may
also contain other overlapping or non-overlapping ORF(s) as found in viral
30 sequences in nature. These additional ORFs (while not essential for
replication)
may enhance replication and/or viral DNA accumulation. Examples of such
additional ORFs are AC3 and AL3 in ACMV and TGMV geminiviruses,
respectively.
The term "chimeric traps-acting replication gene" refers to a replication
35 gene in which the coding sequence of a replication protein is under the
control of a
regulated plant promoter other than that in the native viral replication gene.
-
The term "chromosomally-integrated" refers to the integration of a foreign
gene or DNA construct into the host DNA by covalent bonds.
14
CA 02304984 2000-03-15
WO 99122003 - PCTlUS98/22688
The term "transformation" refers to the transfer of a.foreign gene into the
genome of a host organism. Examples of methods of plant transformation include
Agrobacterium-mediated transformation (De Blaere et al. (1987) Meth. Enzymol.
vol. 143, p. 277) and particle-accelerated or "gene gun" transformation
technology
(Klein et al. (1987) Nature (London) vol. 327, pp. 70-73; U.S. Pat.
No. 4,945,050). The terms "transformed", "transformant", and "transgenic"
refer
to plants or calli that have been through the transformation process and
contain a
foreign gene integrated into their chromosome. The term "untransformed" refers
to normal plants that have not been through the transformation process.
10 The term "transiently transformed" refers to cells in which transgenes and
foreign DNA have been introduced (by such methods as agrobacterium-mediated
transformation or biolistic bombardment), but not selected for stable
maintenance.
The term "stably transformed" refers to cells that have been selected and
regenerated on a selection media following transformation.
15 The terms "genetically stable" and "heritable" refer to chromosomally-
integrated genetic elements that are stably maintained in the plant and stably
inherited by progeny through successive generations.
The terms "primary transformant" and "TO generation" refer to transgenic
plants that are of the same genetic generation as the tissue which was
initially
20 transformed, (i.e., not having gone through meiosis and fertilization since
transformation).
The terms "secondary transformants" and the "T1, T2, T3, etc.
generations" refer to transgenic plants derived from primary transformants
through one or more meiotic and fertilization cycles. Secondary transformants
25 may be derived by self fertilization of primary or secondary transformants
or
crosses of primary or secondary transformants with other transformed or
untransformed plants.
The term "derived from" refers to the obtaining of genetic material from a
specific, or identified, source.
30 The invention provides a two-component, viral expression system for the
production of transgenic plants. Both components are chromosomally-integrated
and, thus, stably maintained by themselves. One component is the proreplicon
that is unable to replicate by itself. The second component is a chimeric
trans-
acting replication gene in which the coding sequence of a viral replication is
35 operably-linked to a regulated promoter. Expression of the viral
replication
protein-under appropriate stimulus will result in the release of replicon from
the
proreplicon and its episomal replication in a cell autonomous manner. Thus,
replicon replication can be targeted to specific plant cells by controlling
the
CA 02304984 2000-03-15
r
WO 99/22003 PCT/US98/22688
expression of replication proteins) to those cells. Plants will be most
sensitive t~
cellular toxicity and/or the detrimental effect of viral replication and/or
replication
proteins) in early stages of plant growth and differentiation that involve
cell
division and differentiation. Thus, controlling the expression of the
replication
5 protein and replicon replication entirely or largely to non-dividing,
terminally-
differentiated cells will reduce the detrimental effect of replicon
replication on
plant growth and development. Examples of such terminally-differentiated cells
include, but are not limited to, the storage cells of seed and root tissues
and mature
leaf cells. Furthermore, the chromosomally-integrated proreplicon serves as a
10 master copy for replicons not only in different generations but also in the
same
generation if cell divisions occur after the onset of episomal replication.
This
strategy will also solve the problem of episomal instability through cell
divisions,
since episomes are unstable in the absence of selection. Furthermore, replicon
replication is expected to achieve high level expression of target genes
through
15 gene amplification that is heritable and cell autonomous. The target gene
expression can involve either the production of foreign proteins or the
silencing of
endogenous nuclear gene as well as transgenes by antisense inhibition or co-
suppression.
In accordance with the subject invention, novel recombinant virus
20 constructs (including transfer vectors and methods for making them and
using
them) are described. When used to transform a plant cell, the vectors provide
a
transgenic plant capable of regulated, high level expression though gene
amplification. This regulated expression could be in response to a particular
stimulus, such as the development stage, wounding of the plant (for example,
by
25 insect attack or pathogen), an environmental stress (such as heat or high
salinity),
or chemicals that induce specific promoters. Plants in which particular
tissues
and/or plant parts have a new or altered phenotype may be produced by the
subject method.
The constructs include vectors, expression cassettes and binary plasmids
30 depending upon the intended use of a particular construct. Two basic DNA
constructs are required which may be combined in a variety of ways for
transforming a plant cell and obtaining a transgenic plant. For agrobacterium-
mediated transformation, the proreplicon and chimeric replication gene may be
combined in one binary plasmid or the two may be introduced into a cell on
35 separate binary plasmids by either co-transformation or sequential
transformations. Alternatively, the two constructs may be combined by crossing
two transgenic lines containing one or the other construct.
16
CA 02304984 2000-03-15
WO 99122003 - PCTIUS98/22688
The termination region used in the target gene- in the replicon as well as in
the chimeric replication protein gene will be chosen primarily for
convenience,
since the termination regions appear to be relatively interchangeable. The
termination region which is used may be native with the transcriptional
initiation
5 region, may be native with the DNA sequence of interest, or may be derived
from
another source. The termination region may be naturally occurring, or wholly
or
partially synthetic. Convenient termination regions are available from the
Ti-plasmid of A. tumefaciens, such as the octopine synthase and nopaline
synthase
termination regions or from the genes for ~i-phaseolin, the chemically
inducible
lant gene, pIN (Hershey et al. (1991) Plant Mol. Biol. vol. 17(4), pp. 679-90;
U.S. 5,364,780).
The Proreplicon Construct
One basic construct is the proreplicon. In the case of geminiviruses, the
proreplicon is preferably present as a partial or complete tandem dimer in T-
DNA,
such that a single replicon is flanked by cis-acting viral sequences necessary
for
viral replication, including the replication origin (Figure 1). These dimers
can
serve as master copy from which replicons can be excised by replicative
release
(Bisaro, David. Recombination in geminiviruses:Mechanisms for maintaining
genome size and generating genomic diversity. Homologous Recomb. Gene
20 Silencing Plants (1994), 219-70. Editor(s): Paszkowski, Jerzy. Publisher:
Kluwer,
Dordrecht, Germany) in the presence of the replication protein. The preferable
source of proreplicon sequences is from ACMV and TGMV in which the essential
replication gene (for example, AC1) is rendered non-functional by mutation
(i.e.,
addition, rearrangement, or a partial or complete deletion of nucleotide
25 sequences). The mutation can be in the non-coding sequence, such as the
promoter, or it can be in the coding sequence of the replication protein so as
to
result either in one or more altered amino acids in the replication protein or
in a
frame shift. Preferably, the entire replication gene is deleted from the
proreplicon
such that there is no homology between the transactivating replication gene
and
30 the replicon in order to reduce virus-induced homology-based silencing of
the
transactivating replication gene during replicon replication. In addition, the
proreplicon preferentially has most or all of the coat protein gene which was
deleted and replaced by a restriction site for cloning target gene.
Proreplicons
may also contain a target genes in the replicon sequence. The coding sequence
in
35 these target genes are operably-linked to regulatory sequences that are of
viral
and/or plant origin. One or more introns may be also be present in the
cassette. ''
Other sequences, including those encoding transit peptides, secretory leader
sequences, or introns, may also be present in the proreplicon and replicon as
17
CA 02304984 2000-03-15
WO 99122003 PCTIUS98/22688
desired. How to obtain and use these sequences are well known to those skilled
in
the art. The target gene can encode a polypeptide of interest (for example, an
enzyme), or a functional RNA, whose sequence results in antisense inhibition
or
co-suppression. The nucleotide sequences of this invention may be synthetic,
5 naturally derived, or combinations thereof. Depending upon the nature of the
nucleotide sequence of interest, it may be desirable to synthesize the
sequence
with plant-preferred codons.
It is contemplated that modified proreplicons may be made that have only
the minimal origin of replication (ori) sequence. This will allow maximal room
10 for cloning target genes as well as remove all or almost all homology
between the
proreplicon and the replicase gene to reduce gene silencing of the chimeric
replication gene by the chromosomal proreplicon or the episomally replicating
replicon. The source of the minimal on sequence of bipartite geminiviruses can
be either DNA from the A genome or DNA from the B genome.
15 Target genes can encode functional RNAs to silence homologous
endogenous genes or transgenes or may encode foreign proteins. Foreign
proteins
will typically encode non-viral proteins and proteins that may be foreign to
plant
hosts. Such foreign proteins will include, for example, enzymes for primary or
secondary metabolism in plants, proteins that confer disease or herbicide
20 resistance, commercially useful non-plant enzymes, and proteins with
desired
properties useful in animal feed or human food. Additionally foreign proteins
encoded by the target genes will include seed storage proteins with improved
nutritional properties, such as the high sulfur 10 kD corn seed protein or
high
sulfur zein proteins.
25 The Chimeric. Traps-acting Replication Gene Construct
The other basic construct is a chimeric traps-acting replication gene
consisting of a regulated plant promoter operably-linked to the coding
sequence of
a replication protein. For ACMV and TGMV geminiviruses, the replication
proteins are encoded by the AC1 and AL1 ORFs, respectively. Also included are
30 the replication proteins of single-stranded RNA viruses, such as the
RNA-dependent RNA polymerise encode by the first ORF of potato virus X
(PVX) [Angell et al. (1997) The EMBOJournal, vol. 16, pp. 3675-3684].
Regulated expression of the viral replication proteins) is possible by
placing the coding sequence of the replication protein under the control of
35 promoters that are tissue-specific, developmental-specific, or inducible.
Several tissue-specific regulated genes and/or promoters have been '
reported in plants. These include genes encoding the seed storage proteins
(such
as napin, cruciferin, .beta.-conglycinin, phaseolin), zein or oil bodies
proteins
18
CA 02304984 2000-03-15
wo ~nzoo3 pc~rms9sn26ss
(such as oleosin), or genes involved in fatty acid biosynthesis, including
acyl ,
carrier protein, stearoyl-ACP desaturase, and fatty acid desaturases (fad 2-1
), and
other genes expressed during embryo development, such as Bce4, which is
expressed at high levels in Brassica seed coat cells is the EA9 gene [see, for
S example, EP 255378 and Kridi et al. (1991) Seed Science Research vol. l,
pp. 209-219]. Particularly useful for seed-specific expression is the pea
vicilin
promoter [Czako et al. ( 1992) MoL Gen. Genet. vol. 235( 1 ), pp. 33-40] that
has
been shown to be seed-specific by the use of diphtheria toxin. Another useful
promoter for expression in mature leaves are those that are switched on at the
10 onset of senescence, such as the SAG promoter from Arabidopsis [Gan et al.
(1995) Science (Washington, D. C.) vol. 270(5244), pp. 1986-8].
A class of fruit-specific promoters expressed at or during anthesis through
fruit development, at least until the beginning of ripening, is discussed in
US
4,943,674, the disclosure of which is hereby incorporated by reference. cDNA
1 S clones that are preferentially expressed in cotton fiber have been
isolated [John et
al. (1992) Proc. Natl. Acad. Sci. U.S.A, vol. 89(13), pp. 5769-73). cDNA
clones
from tomato displaying differential expression during fruit development have
been isolated and characterized [Mansson et al. ( 1985) Mol. Gen. Genet. vol.
200,
pp. 356-361; Slater et al. (1985) Plant Mol. Biol. vol. 5, pp. 137-147]. The
20 promoter for polygalacturonase gene is active in fruit ripening. The
polygalacturonase gene is described in U.S. Pat. No. 4,535,060 (issued August
13,
1985), U.S. Pat. No. 4,769,061 (issued September 6, 1988), U.S. Pat.
No. 4,801,590 (issued January 31, 1989) and U.S. Pat. No. 5,107,065 (issued
April 21, 1992), which disclosures are incorporated herein by reference.
25 Mature plastid mRNA for psbA (one of the components of photosystem II)
reaches its highest level late in fruit development, in contrast to plastid
mRNAS
for other components of photosystem I and II which decline to nondetectable
levels in chromoplasts after the onset of ripening [Piechulla et al. (1986)
Plant
Mol. Biol. vol. 7, pp. 367-376]. Recently, cDNA clones representing genes
30 apparently involved in tomato pollen [McCormick et aL, Tomato Biotechnology
( 1987) Alan R. Liss, Inc., New York) and pistil (Gasser et al. ( 1989) Plant
Cell
vol. 1, pp. I S-24] interactions have also been isolated and characterized.
Other examples of tissue-specific promoters include those that direct
expression in leaf cells following damage to the leaf (for example, from
chewing
35 insects); in tubers, (for example, patatin gene promoter); and in fiber
cells (an
example of a developmentally regulated fiber cell protein is E6 [John et al.
(1992)
Gene expression in cotton (Gossypium hirsutum L. ) fiber: cloning of the
mRNAs.
Proc. Natl. Acad Sci. U.SA. vol. 89(13), pp. 5769-73]). The E6 gene is most
19
CA 02304984 2000-03-15
WO 9922003 PCTNS98/22688
active in fiber, although low levels of transcripts are found in leaf, ovule
and -
flower.
The tissue-specificity of some "tissue-specific" promoters may not be
absolute and may be tested by one skilled in the art using the diphtheria
toxin
5 sequence. One can also achieve tissue-specific expression with "leaky"
expression by a combination of different tissue-specific promoters (Beals et
al.
(1997) Plant Cell vol. 9, pp. 1527-1545). Other tissue-specific promoters can
be
isolated by one skilled in the art (see U.S. 5,589,379). Several inducible
promoters ("gene switches") have been reported. Many are described in the
10 review by Gatz (1996) Current Opinion in Biotechnology vol. 7, pp. 168-172;
Gatz (1997) Annu. Rev. Plant Physiol. Plant Mol. Biol. vol. 48, pp. 89-108].
These include tetracycline repressor system, Lac repressor system, copper
inducible systems, salicylate inducible systems, such as the PRIa system,
glucocorticoid [Aoyama et al. (1997) N HPlant.7ournal vol. 11, pp. 605-612]
and
15 ecdysome inducible systems. Also, included are the benzene sulphonamide
(U.S. 5,364,780} and alcohol (WO 97106269 and WO 97106268~inducible
systems and glutathione S-transferase promoters. Other studies have focused on
genes inducibly regulated in response to environmental stress or stimuli such
as
increased salinity, drought, pathogen, and wounding. For example, genes
20 encoding serine proteinase inhibitors, which are expressed in response to
wounding in tomato (Graham et al. (1985) J. Biol. Chem. vol. 260, pp. 6555-
6560;
Graham et al. (1985) J. Biol. Chem. vol. 260, pp. 6561-6554} and on mRNAS
correlated with ethylene synthesis in ripening fruit and leaves after wounding
(Smith et al. (1986) Planta vol. 168, pp. 94-100). Accumulation of a
25 metallocarboxypeptidase inhibitor protein has been reported in leaves of
wounded
potato plants [Graham et al. (1981) Biochem Biophys Res Comm vol. 101,
pp. 1164-1170]. Other plant genes have been reported to be induced methyl
jasmonate, elicitors, heat-shock, anerobic stress, or herbicide safeners.
A variety of techniques are available and known to those skilled in the art
30 for introduction of constructs into a plant cell host. These techniques
include
transformation with DNA employing A. tumefaciens or A. rhiaogenes as the
transforming agent, electroporation, particle acceleration, etc. [See for
example
EP 295959 and EP 138341.] It is particularly preferred to use the binary type
vectors of Ti and Ri plasmids of Agrobacterium spp. Ti-derived vectors
transform
35 a wide variety of higher plants, including monocotyledonous and
dicotyledonous
plants, such as soybean, cotton, rape, tobacco, and rice [Pacciotti et al.
(1985)
BiolTechnology vol. 3, pp. 241; Byrne et al. (1987) Plant Cell, Tissue and
Organ
Culture vol. 8, p. 3; Sukhapinda et al. (1987) Plant Mol. Biol. vol. 8, pp.
209-216;
20
CA 02304984 2000-03-15
~,s .
WO 99122003 - PCTNS98I22688
Lori et al. (1985) Mol. Gen. Genet. vol. 199, p. 178; Potrykus (1985) Mol.
Gen'
Genet. vol. 199, p. 183; Park et al. (1995) J. Plant Biol. vol. 38(4), pp. 365-
71;
Hiei et al. (1994) Plant J. vol. 6, pp. 271-282]. The use of T-DNA for
transformation of plant cells has received extensive study and is amply
described
in EP 120516, Hoekema, In: The Binar~Plant Vector System, Offset-drukkerij
Kanters B.V., Alblasserdam, 1985, Chapter V, Knauf, et al., Genetic Analysis
of
Host Range Expression by Agrobacterium, In: Molecular Genetics of the Bacteria-
Plant Interaction, Puhler, A. ed., Springer-Verlag, New York, 1983, p. 245,
and
An et al. (1985) EMBO J. vol. 4, pp. 277-284. For introduction into plants,
the
chimeric genes of the invention can be inserted into binary vectors as
described in
the examples.
A variety of techniques are available and known to those skilled in the art
for introduction of constructs into a plant cell host. These techniques
include
transformation with DNA employing A. tumefaciens or A. rhizogenes as the
I S transforming agent, electroporation, particle acceleration, etc. [See for
example,
EP 295959 and EP 138341]. It is particularly preferred to use the binary type
vectors of Ti and Ri plasmids of Agrobacterium spp. Ti-derived vectors
transform
a wide variety of higher plants, including monocotyledonous and dicotyledonous
plants, such as soybean, cotton, rape, tobacco, and rice [Pacciotti et al.
{1985)
BiolTechnology vol. 3, p. 241; Byrne et al. (1987) Plant Cell, Tissue and
Organ
Culture vol. 8, p. 3; Sukhapinda et al. (1987) Plant Mol. Biol. vol. 8, pp.
209-216;
Lorz et al. (1985) Mol. Gen. Genet. vol. 199, p. 178; Potrykus (1985) Mol.
Gen.
Genet. vol. 199, p. 183; Park et al., J. Plant Biol. (1995), vol. 38(4), pp.
365-71;
Hiei et al. (1994) Plant J. vol. 6, pp. 271-282]. The use of T-DNA to
transform
plant cells has received extensive study and is amply described [EP 120516;
Hoekema, In: The Binary Plant Vector System. Offset-drukkerij Kanters B.V.;
Alblasserdam, ( 1985), Chapter V, Knauf, et al., Genetic Analysis of Host
Range
Expression by Agrobacterium; In: Molecular Genetics of the Bacteria-Plant
Interaction, Puhler, A. ed., Springer-Verlag, New York, 1983, p. 245; and An
et
al. (1985) EMBOJ. vol. 4, pp. 277-284]. For introduction into plants, the
chimeric genes of the invention can be inserted into binary vectors as
described in
the examples.
Transgenic plant cells are then placed in an appropriate selective medium
for selection of transgenic cells which are then grown to callus. From callus
shoots are grown and plantlets generated from the shoot by growing in rooting
medium. The various constructs normally will be joined to a marker for
selection
in plant cells. Conveniently, the marker may be resistance to a biocide,
particularly an antibiotic (such as kanamycin, 6418, bleomycin, hygromycin,
21
CA 02304984 2000-03-15
..
WO 99/22003 - PCTNS98/22688
chloramphenicol, herbicide, or the like). The particular marker used will
allow_far
selection of transformed cells as compared to cells lacking the DNA which has
been introduced. Components of DNA constructs including transcription
cassettes
of this invention may be prepared from sequences which are native (endogenous)
5 or foreign (exogenous) to the host. By "foreign" it is meant that the
sequence is
not found in the wild-type host into which the construct is introduced.
Heterologous constructs will contain at least one region which is not native
to the
gene from which the transcription initiation region is derived.
To confirm the presence of the transgenes in transgenic cells and plants, a
10 Southern blot analysis can be performed using methods known to those
skilled in
the art. Replicons can be detected and quantitated by Southern blot, since
they
can be readily distinguished from proreplicon sequences by the use of
appropriate
restriction enzymes. Expression products of the transgenes can be detected in
any
of a variety of ways, depending upon the nature of the product, and include
15 Western blot and enzyme assay. One particularly useful way to quantitate
protein
expression and to detect replication in different plant tissues is to use a
reporter
gene, such as GLTS. Once transgenic plants have been obtained, they may be
grown to produce plant tissues or parts having the desired phenotype. The
plant
tissue or plant parts, may be harvested, and/or the seed collected. The seed
may
20 serve as a source for growing additional plants with tissues or parts
having the
desired characteristics.
The present viral expression system has been used to demonstrate that
(i) soybean and corn seed tissue will support geminivirus replication; and
(ii} that
the expression system will effect expression of foreign genes in tobacco.
25 More specifically, Applicant has used a transient assay using biolistic
bombardment to show that developing soybean and corn seeds can support
replication of ACMV-A-derived vector in which the coat protein gene was
deleted. Applicant has tested episomal replication of the mutant ACMV-A DNAs
in mature tobacco leaf, developing soybean seed embryo (50-150 mg in weight),
30 developing corn seed, and corn suspension cell cultures. For this, a
greater than
full length copy of mutant ACMV-A DNA is introduced into the tissues by
biolistic bombardment and the transiently transformed tissues analyzed by
Southern blot for replication. In one mutant, called CP mutant, a 784 by
deletion
is made in the coat protein gene from Bam HI site at position 142 to Pml I
site at
35 position 926 (with refemce to Ssp I in the stem-loop structure of the on
being
position 1 }. In another mutant, called CP+REp mutant, there is a deletion in
the
replication gene AC 1 that was made by deleting 651 by Bgl iI fragment
covering
AC l, AC2, and AC3 ORFs in addition to the coat protein deletion stated above.
22
CA 02304984 2000-03-15
~'.
WO 99122003 PCTNS98/22688
While the CP mutant can replicate episomally-by itself, CP+REP mutant
cannot. However, CP+REP mutant can replicate episomally in all of the tissues
when it is co-bombarded either with CP mutant or with a chimeric replication
gene under the control of constitutively expressed 35S promoter or seed-
specific
phaseolin promoter. Since all of these tissues except the suspension culture
are
past the cell division stage, this shows that non-dividing, fully or partially
differentiated cells are capable of supporting geminivirus replication.
In another embodiment of the expression system, the viral coat protein
gene was replaced with a 1.2 kB non-viral gene. Applicant introduced more than
full length CP mutant with a 1.2 kB target gene insertion in place of the coat
protein into tobacco leaf discs on a binary vector by agrobacterium-mediated
transformation. Southern analysis of regenerated shoots showed replication of
the
modified ACMV-A genome in uprooted shoots but not in rooted shoots. Thus,
the CP mutant can tolerate at least a 1.2 kb non-viral DNA insertion. Taken
together with the ability of the CP+REP mutant to replicate in traps, it is
reasonable to expect that at least 1.85 kb of non-viral DNA can be inserted.
Applicant also obtained genetically stable transgenic tobacco plants
containing a
chromosomally-integrated more than full length of CP mutant. Retransformation
of leaf discs of these plants with a chimeric replicase gene resulted in
excision and
replication of the replicon. Applicant also introduced into tobacco via
agrobacterium-mediated transformation native or chimeric ACMV A sequences
expressing the leftward open reading frames containing ORFs AC1, AC2, and
AC3 under the control of its native promoter or other promoters that are
expressed
during plant regeneration. However, Southern analysis of transgenic plants
showed that these chimeric genes are lost or deleted. Thus, unlike in the case
of
TGMV, tobacco plants transformed with wild type ACMV DNA A or with greater
than full length copy of the coat protein replacement vector showed that the
viral
sequences were deleted in leaves of rooted transgenic plants. This observation
is
consistent with the toxicity of replication proteins) expression, since viral
sequences with partially deleted replication protein can be stably maintained.
Furthermore, when chimeric genes for expressing the replication protein (s)
are
introduced into tobacco plants, they were found to be similarly deleted in
leaves of
rooted transgenic plants. Thus, ACMV replication proteins) is detrimental to ,
plant regeneration. Applicant has shown that replicase expression is
detrimental
to plants during plant transformation/regeneration.
Applicant has shown 1 ) that a wide range of non-dividing tissues (tobacco ~'
mature leaf, corn and soybean embryos and corn suspension eulture) are capable
of supporting geminivirus replication following introduction of the
geminivirus by
23
CA 02304984 2000-03-15
WO 99/Z2003 - PCT/US98/22688
biolistic bombardment and 2) that unrooted stably-transformed shoots also
allor~
replication of geminivirus. The successful use of plant viral vectors will the
rely
on tightly regulating replication protein expression (and replicon
replication) in
non-dividing terminally-differentiated cells.
5 EXAMPLES
The present invention is further defined in the following Examples. These
Examples, while indicating preferred embodiments of the invention, are given
by
way of illustration only. From the above discussion and these Examples, one
skilled in the art can ascertain the essential characteristics of this
invention, and
10 without departing from the spirit and scope thereof, can make various
changes and
modifications of the invention to adapt it to various usages and conditions.
GENERAL METHODS
Standard recombinant DNA and molecular cloning techniques used in the
Examples are well known in the art and are described by Sambrook, J., Fritsch,
15 E.F. and Maniatis, T. Molecular Cloning: A Laborutory Manual; Cold Spring
Harbor Laboratory Press: Cold Spring Harbor, (1989} (Maniatis) and by T. J.
Silhavy, M. L. Bennan, and L. W. Enquist, Experiments with Gene Fusions, Cold
Spring Harbor Laboratory, Cold Spring Harbor, N.Y. (1984) and by Ausubel, F.
M. et al., Current Protocols in Molecular Biology, pub. by Greene Publishing
20 Assoc. and Wiley-Interscience (1987).
Restriction enzyme digestions, phosphorylations, ligations and
transformations were done as described in Sambrook et al., supra. Restriction
enzymes were obtained from New England Biolabs (Boston, MA), GIBCOBRL
(Gaithersburg, MD), or Promega (Madison, WI). Thermostable DNA polymerase
25 was obtained from Perkin Elmer (Branchburg, NJ). Growth media was obtained
from GIBCOBRL (Gaithersburg, MD).
The Agrobacterium tumefaciens strain LBA4404 was obtained from Dr. R.
Schilperoot, Leiden [Hoekema et al. (1983) Nature vol. 303, pp. 179-180].
Transformation Protocols
30 Biolistic transformations were done essentially as described in U.S. Pat.
No. 4,945,050, hereby incorporated by reference. Gold particles (1 mm in
diameter) are coated with DNA using the following technique. Ten ug of plasmid
DNAs are added to 50 mL of a suspension of gold particles (60 mg per mL).
Calcium chloride (50 uL of a 2.5 M solution) and spermidine free base (20 mL
of
35 a 1.0 M solution) are added to the particles. The suspension is vortexed
during the
addition of these solutions. After 10 min, the tubes are briefly centrifuged
(5 sec "
at 15,000 rpm) and the supernatant removed. The particles are resuspended in
200 mL of absolute ethanol, centrifuged again and the supernatant removed. The
24
CA 02304984 2000-03-15
WO 99/Z2003 - PCTIUS98/22688
ethanol rinse is performed again and the particles resuspended in a final
volume~f
30 uL of ethanol. An aliquot (5 mL) of the DNA-coated gold particles can be
placed in the center of a flying disc (Bio-Rad Labs, 861 Ridgeview Dr, Medina,
OH). The particles are then accelerated into the corn tissue with a PDS-
1000/He
5 (Bio-Rad Labs, Medina, OH), using a helium pressure of 1000 psi, a gap
distance
of 0.5 cm and a flying distance of 1.0 cm.
Where Agrobacterium transformations were done the proceedure was
accomplished were done essentially as described Park et al. (1955) J. Plant
Biol.
vol. 38(4), pp. 365-71.
10 Transformation And Expression Protocol
Transgenic plants with different constructs will be selected and
regenerated into plants in tissue culture by methods known to one skilled in
the art
and referred to above. The ability of a chimeric, traps-acting replication
gene to
replicate the replicon from the proreplicon in plant chromosome in traps will
be
15 tested following one of the following methods:
1. crossing plants where one parent contains the chimeric replication
gene and the other the proreplicon,
2. co-transformating tobacco leaf discs with two types of agrobacteria,
one containing a binary vector with a plant-selectable marker (such as
20 phosphinothricin resistance) and proreplicon and another containing a
binary
vector with a different plant-selectable marker (such as kanamycin resistance)
and
the chimeric replication gene. Transformed leaf discs will be selected and
regenerated in the presence of both selection agents, or
3. transforming tobacco with one kind of agrobacteria containing a
25 binary vector with a plant-selectable marker, obtaining regenerated
transgenic
plant and re-transforming with another kind of agrobacteria containing a
binary
vector with a different plant-selectable marker. Thus, the first binary vector
could
contain either the proreplicon or the chimeric replication gene. The binary
vector
for retransformation will contain the complementary component.
30 Replication in transgenic plant tissue will be monitored by Southern
analysis of genomic DNA (either undigested or following digestion with
restriction enzymes) that will distinguish the replicon from chromosomal
chimeric
replication protein gene and proreplicon by size. Alternatively, replication
can be
detected by reporter gene expression. For example, increased expression of the
35 GUS reporter genes on the replicon will be assayed by GUS enzyme assay or
staining and increased expression of the 10 kD corn storage protein will be
detected by western blot using antibodies specific for the 10~kD protein.
25
CA 02304984 2000-03-15
WO 9912003 - PCTNS98/22688
EXAMPLE 1
Tomato Golden Mosaic Virus (TGMV) Constructs
Partial dimer of TGMV with wild-type replication protein (plasmid pBE651 )
Plasmid pCSTA [Von Amim et al. (1992) Virology vol. 186(1),
5 pp. 286-93] was obtained from John Stanley (John Innes Center, Norwich,
United
Kingdom). It consists of a single complete TGMV-A DNA cloned at its unique
Eco RI site into Eco RI site of a plasmid pUC 19 derivative. 1922 by between
its
Nco I and Sal I sites containing the 3' ends of replication protein AL1 ORF
and
the coat protein gene were deleted by restriction digestion, fill-in, and self
ligation
10 to result in plasmid pGV650. Then, the Eco RI insert fragment from plasmid
pCSTA (containing the complete TGMV-A genome) was cloned in Eco RI site in
pGV650 to yield plasmid pGV651. Thus, plasmid pGV651 consists of an intact
TGMV-A genome and a tandem duplication of a 570 by TGMV-A sequence that
includes 206 by TGMV on sequence and 370 by 5' ALl ORF sequence adjacent
15 to the replication origin (ori), uch that an intact TGMV-A genome can be
made
from pGV651 in plant cells either by replicative release or homologous
recombination. Plasmid pGV65 i can replicate in a plant cell when introduced
by
biolistic bombardment. The Hind III fragment of pGV651 carrying the TGMV
dimer with wild-type AL I ORF was cloned into pBinl9 [Frisch et al., (1995)
20 Plant Mol. Biol. vol. 27(2), pp. 405-409] such that the AC 1 gene is
transcribed
away from the Nos:NPT II gene. The resultant binary plasmid pBE651 was
introduced into tobacco plants via Agrobacterium tumefaciens strain LBA4404.
Partial dimer of TGMV with wild-tyue replication~rotein and GUS reporter
~plasmid pBE671y
25 The unique BstBl site in plasmid pGV651 was converted to Not I
following BstB I digestion, fill-in reaction, and ligation to Nit I linkers
(New
England Biolabs catalog number 1125) to result in pGV652. Then, a 1401 by
Not I-Sac I fragment (containing the coat protein gene in plasmid pGV652) was
replaced with a 2545 by Not I-Sac I fragment from plasmid pGV662 (see below)
30 containing the GUS [Jefferson et al., The use of the Escherichia coli
b-glucuronidase gene as a gene fusion marker for studies of gene expression in
higher plants. Biochem. Soc. Traps. (1987), vol. 15(1), pp. 17-18] to yield
plasmid pGV671. In plasmid pGV671, GUS expression is under the control of
the coat protein promoter. Plasmid pGV671 can replicate when introduced into
35 plant cells by biolistic bombardment. The Hind III fragment carrying the
dimer
will be cloned into a binary vector pBinl9 to result in pBE671 and transferred
to
plaats as known to one skilled in the art. -
26
... ,.
CA 02304984 2000-03-15
WO 99/22003 - PCTIUS98I22688
TGMV nrorenlicon without GUS reporter ene (plasmid pGV654). .
Plasmid pGV654 was made from plasmid pGV652 (see description above)
by deleting a 895 by region between Bam HI -Eco RI including 709 by of the 3'
end of AL 1 ORF and the 5' regions of AL2 and AL3 ORFs following restriction
5 digestion, fill-in, and ligation. When introduced into plant cells by
biolistic
bombardment the proreplicon in plasmid pGV654 was unable to replicate without
the gene encoding the replication protein in traps.
TGMV nrorenlicons with GUS reporter eene lplasmids nGV662 and~GV6721
Three different TGMV proreplicons with GUS reporter gene will be
10 introduced into plant cells. They differ in the kind of mutation in the
replication
gene. Two (pGV662 and pGV672) were made by deletions of different sizes and
one is being made by introducing a frameshift mutation.
The 696 by Not I-Nco I fragment in plasmid pGV654 (containing most of
the coat protein gene) was replaced with a 1875 by Not I-Nco I fragment
15 containing the GUS ORF (the Nco I site includes the initiation codon) to
yield
plasmid pGV661. Since the Nco I site is downstream of the coat protein
initiation
codon, a modified coat protein promoter with a Nco I in the 5' untranslated
region
was made by nucleic acid amplification using primers [(See U.S. 4,683,195;
4,683,202; 4,965,188 to polymerase chain reaction (PCR),
20 5'-CGTCCGGATCCAATTCTCCCCATACAAGAGTATCT-3' [SEQ ID NO: 1 ]
and 5'-GTCGACCCATGGTTAAAGATCCACGAAACGCATGT [SEQ ID
N0:2] on plasmid pCSTA by 40 cycles of 60 °C, 1' annealing; 90
°C, 2'. A
663 by Bam HI+Nco I fragment derived from the PCR product and containing the
ori, and coat protein promoter without any coat protein coding sequence was
used
25 to replace the corresponding 698 by Bgl II-Nco I fragment of plasmid pGV661
to
result in plasmid pGV662.
Another replication gene deletion was made by the deletion of 772 by
between Xmn I site in AL1 ORF and Swa I site in AL3 ORF. For this, plasmid
pGV671 was digested with Swa I and partial Xmn I and then religated. Plasmids
30 pGV662 and pGV672 differ from their wild-type counterpart, plasmid pGV671,
in
having deletions in AL1, AL2 and AL3 ORFs of 709 by and 433 bp, respectively.
The Hind III fragment of plasmid pGV662 was isolated and cloned into the
Hind III site of pBinl9 and pGV674 to yield binary plasmids pBE662 and
pBE675, respectively. The Hind III fragment of pGV672 was cloned into
35 pGV674 to yield pBE672. Binary vector pGV674 was made by replacing the
Bsu 36 I-Cla I fragment in pBinl9 (carrying the nopaline synthase promoter:npt
II:3'nopaline synthase chimeric gene that confers resistance to kanamycin in
plants) with a Bsu 36 I-Cla I (fragment carrying the nopaline synthase
27
CA 02304984 2000-03-15
WO 99/22003 PCTNS98/22688
promoter:bar:3'nopaline synthase chimeric gene that confers resistance to _
phosphonithricin herbicide). pBE662 binary plasmid was introduced into plants
via Agrobacterium tumefaciens LBA4404-mediated transformation known to one
skilled in the art. [Walden, Vector Systems for Agrobacterium-mediated
5 Transformation In Method Plants Biochem. (1997), I06 (Molecular Biology),
85-102.]
EXAMPLE 2
Chimeric traps-acting TGMV replication enes
35S:TGMV renlication~(plasmid pGV653)
10 Plasmid pGV653 was derived from plasmid p35S-GFP plant vector
(Promega Corp., 7113 Benhart Dr., Raleigh, NC) by replacing the first 462 by
of
the upstream region (between Hind III and Acc I) of 35S promoter with Not I
and
then the Bam HI-Sac I fragment containing the green fluoroscent protein ORF
(between the 35S promoter and 3' region of nopaline synthase gene) with that
of
15 TGMV sequence containing AL 1, AL2, and AL3 ORFs. For the latter, a Bam HI
site was introduced at position 29 [Hamilton et al., EMBD J. 3:2197-205
(1984)]
in the S' untranslated region ( 16 by upstream of the AL 1 ORF) of TGMV AL 1
gene and the 2976 by of Bam HI-Bam HI-Sac I TGMV sequence was used to
replace the Bam HI-Sac I region to form pGV653.
20 Chemically-inducible promoter In 2-2:TGMV replication ene (plasmid~BE665~
The Mfe I and BstB 1 sites in plasmid pGV651 were converted to Spe I
following fill-in and ligation of Spe I linker (New England Biolab, catalog
no. 1087). The 1603 by Spe I fragment (containing the TGMV AL1, AL2, and
AL3 ORFs) was isolated and cloned between the inducible promoter 2-2 and the
25 3' region of 2-1 gene in plasmid pGV664 to yield plasmid pGV665. Plasmid
pGV664 was derived from pGV659 (see description below) by replacing its
Not I-Spe I fragment (containing 2-2 promoter and ACMV replication protein
gene), with that of a 2-2 promoter in which the Nco I site was modified to Spe
I
site. The latter was made by PCR such that the Nco I site at its 3' end was
30 replaced by the Spe I site, the Spe I site at the S' end of the promoter
was
destroyed, and the Bam HI site at the 5' end was changed to Bgl II. The 2561
by
BgIII-Asp718 fragment from pGVb65 containing the 2-lIN promoter:T-Rep
ORF:3' 2-2 chimeric gene was isolated from pGV665 and cloned into binary
vector pBinl9 cut with BamHI-Asp718 to result in pBE665. The chimeric gene
35 was introduced into tobacco plants viaAgrobacterium tumefaciens LBA4404.
Several shoots from each transformation of Nicotiana benthamiana with
both BA662 and BA665 rooted and regenerated into plants. All plants lacked any
observable abnormal phenotype. Applicant obtained one plant positive for the
28
CA 02304984 2000-03-15
;s
WO 99122003 ~ PCTlUS98/22688
presence of the transgene (determined by Southern blot) from each of.the BA662
and BA665 transformations. These plants were either selfed or crossed with
each
other both ways.
Seeds from the reciprocal crosses were planted on soil flats and allowed to
germinate in the growth chambers. 48 and 72 progeny seedlings from BA662 X
BA665 and BA665 X BA662 crosses, respectively, were transplanted on a gridded
flat. The phenotypes of the plants were relatively normal except that the
seedlings
varied in sizes and some of the leaves appeared crinkled as also observed on
seed
germination plates. Four leaf discs were punched out from a leaf in each plant
and
10 a pair of discs were placed in one well of two different 96 well plates
containing
agar with Murashige and Skoog salts, 30 g of sucrose, and no hormones. The
wells of one plate were flooded with 1 mL of the 30 mg/L 2-CBSU for 30 min and
then the liquid was removed without rinsing. One disc from each well was
tested
for GUS staining after 24 and 72 hrs. Out of 48 treated BA662 X BA665
15 seedlings, 3 showed GUS staining after 24 hrs and 10 more after 72. Out of
72 of
treated BA665 X BA662 seedlings, 8 showed GUS staining after 24 hrs and 23
after 72 hrs. Only discs from the strongest GUS-expressing seedlings showed
trace GUS activity without safener treatment. This 'leaky' induction may be
attributed to wounding and growth on agar. The degree of staining varied not
20 only between different GUS-positive discs but also across a disc. After 72
hours
there were not only many more individuals that stained positive compared to
24 hrs but the ones that stained after 24 hrs stained also more heavily after
72 hrs.
This indicated that the virus was replicating.
Proreplicon replication was confirmed by Southern analysis confirmed that
25 all GUS-positive plants tested were positive for both IN:Rep and
proreplicon. The
genomic DNA was isolated from leaves of the six highest GUS-expressing plants
(three each from BA662 X BA665 and BA665 X BA662 progeny) either without
or with 2-CBSU treatment. For the former, genomic DNA was isolated from
freshly harvested leaves. For the latter, leaves were cut in strips, placed on
agar,
30 treated with 30 mg/L 2-CBSU for 30 min, and placed on agar for 3 days after
removing the safener. No replicon was detected in untreated leaves and a large
increase in replicon copy number was detected in treated leaves.
Seed-specific vicilin promoter:TGMV replication sene
Plasmid pGV656 (containing a 3086 by Hind III fragment containing a
35 chimeric corn 10 kD storage protein ORF under the control of seed-specific
vicilin
promoter) was made. The chimeric gene consists of 1) an operably-linked
2308 by Hind III-Nco I vicilin promoter isolated from plasmid pGA971 [Czako et
al. (1992) Mol. Gen. Genet. vol. 235(1), pp. 33-40] and 2) the 276 by
29
CA 02304984 2000-03-15
WO 99/22003 - PGTNS98I22688
Sac I-Hind III fragment containing a 3' untranslated region of nopaline
synthase
promoter. The 394 by Spe I-Xba I fragment containing most of the 10 kD ORF
was replaced by the 1604 by Spe I fragment containing AC1-3 ORFs. (This
fragment was made by adding Spe I linker to Mfe I and Bst B 1 digested TGMV
5 clone, described above). The 56 by between the Nco I and Spe I sites was
deleted
by PCR. The 4236 by Hind III fragment (containing the chimeric vicilin
promoter:TGMV replication protein ORF:3' nopaline synthase gene) was cloned
into the Hind III site of pBinl9 binary vector. The resultant binary plasmid
pBE679 was introduced into tobacco plants via Agrobacterium tumefaciens
10 LBA4404-mediated transformation known to one skilled in the art. The
transgenic plants are in pots and will be crossed with transgenic plants
containing
the proreplicon BA662 (see above}.
Senecence-associated promoter SAG~TG1VIV replication eene ~(plasmid~BE667~
The 1.6 kB Spe I fragment containing the TGMV AL1, AL2, and AL3
15 ORFs (described above) was cloned in the Xba I site of plasmid pGV666,
downstream of a senescence associated (SAG) promoter. pGV666 was derived
from plasmid pSG516 [Gap et al. (1995) Science (Washington, DC)
vol. 270(5244), pp. 1986-8] by replacing its Bst B1-Sac I fragment (containing
3'
end of the promoter and isopentenyl transferase ORF) with a BstB I-Sac I
20 fragment (containing SAG promoter modified by PCR} to replace the Nco I
site
with Xba I/Sac I sites. The 3801 by Spe I-Sac I fragment (containing the SAG
promoter:TGMV replication gene) was isolated from pGV667 and cloned into
Xba I-Sac I digested vector pBI l0i (Clonetech Inc., 6500 Donlon Rd, Somis,
CA)
to yield binary plasmid pBE667 such that the SAG promoter:ACMV replication
25 gene is operably-linked to the 3' untranslated region of nopaline synthase
gene.
The chimeric gene in pBE667 was introduced into plants via r4grobacterium
tumefaciens LBA4404-mediated transformation known to one skilled in the art.
EXAMPLE 3
African Cassava Mosaic Virus (ACMV) Constructs
30 Partial dimer of ACMV with wild-type replication eg ne (plasmid pGV596)
Plasmid pCLV012 (ATCC 45039), which contains DNA A of African
Cassava Mosaic Virus [West Kenyan isolate 844, Stanley et al. (1983) Nature
vol. 301, pp. 260-262] was linearized with Pml I and ligated to Bam HI linker
(New England Biolab cat. no. 1071 ). Following Bam HI digestion, the 2 kB
35 fragment containing the on and AC1-3 ORFs was isolated and cloned into
Bam HI site of pSK to yield pGV592 and pGV592R. These plasmids differ in
insert orientation: the coat protein promoter is proximal to the Sal site in
the
vector in pGV592 and distal in pGV592R. BstB 1 site in AC1 gene in pGV592R
CA 02304984 2000-03-15
WO 99/22003 - PCT/US98/22688
was modified to Sac II, following restriction digestion, fill-in, and Sac II
linker
ligation. The 358 by Not I-Sac II fragment was isolated and cloned between the
Not i-Sac II cleaved pGV592 to yield pGV596a. Deletion of 140 by between
Sna B 1 and Not I sites in pGV596a containing the 3' end of the residual ACMV
5 coat protein gene following Sna BI digestion, Not I linker (New England
Biolabs,
catalog no. 1125) ligation, Not I digestion, and self ligation yielded pGV605.
Both plasmids pGV596a and pGV605 can replicate when introduced singly into
plant cells by biolistic bombardment.
ACMV uroreplicon (plasmid pGV596D1
10 Deletion of a 651 by region between the Bgl II sites that includes the 3'
end of AC1 gene in pGV596a yielded ACMV proreplicon (plasmid pGV596D).
ACMV nroreplicons with revorter gene (nlasmids pGV614D and pGV616D)
Plasmid pGV605 was modified by converting its Sac I site to Asp718 and
by destroying the Bam HI site near Sal I to form plasmid pGV611. Chimeric
15 10 kD corn storage protein gene under the control of either a seed-specific
or
constitutively-expressed promoter was used as a reporter for gene expression.
The
seed-specific chimeric gene was cloned as a 1164 by Bam HI-Bgl II fragment in
the Bam HI site of pGV611 to yield plasmid pGV614. It consists of an operably-
linked 383 by of seed-specific phaseolin promoter (from -295 to +82 by with
20 respect to the transcription start site), 450 by containing the I 0 kD ORF,
and
331 by containing 279 by of the 3' untranslated region of nopaline synthase
promoter. For the constitutive reporter gene, plasmid pGV611 was first
modified
to delete the sites between Not I and Bam HI by Bam HI digestion, fill-in,
addition of Not I linker (New England Biolabs, catalog no. 1125), and
religation
25 to yield plasmid pGV61 IN. Then, the constitutive chimeric gene was cloned
as a
1194 by Not I fragment in the Not I site of pGV611N to yield plasmid pGV616.
It consists of an operably-linked 411 by constitutively expressed 35S promoter
(from -400 to +11 by with respect to the transcription start site), 460 by
containing
the 10 kD ORF, and 323 by containing 279 by of 3' untranslated region of
30 nopaline synthase promoter. Deletion of a 651 by region between the Bgl II
sites
that includes the 3' end of replication gene AC 1 in pGV614 and pGV616 yielded
ACMV proreplicons with the reporter gene, pGV614D and pGV616D,
respectively. Plasmids pGV614D and pGV616D were linearized with Sal I
enzyme and cloned into the Sal I site of binary plasmid pZBL 1 and introduced
35 into tobacco plants via Agrobacterium tumefaciens LBA4404.
31
CA 02304984 2000-03-15
WO 99/22003 PCTNS98/22688
Chimeric ACMV Replication eenes~ Seed-specific phaseolinpromoterACMV
replication ene
Plasmid pCLV012 (ATGC 45039) containing ACMV DNA was modified
by creating an Nco I site at the initiation codon of AC 1 ORF by PCR and by
S modifying Pml I site at the 3' end of the coat protein gene to Xba I site.
The
1685 by Nco I-Nco I-Xba I fragment containing AC1-3 ORFs was used to make
seed-specific chimeric replication genes. The chimeric phaseolin:replication
gene
consists of an operably-linked 384 by Bam HI-Nco I fragment captaining 374 by
of seed-specific phaseolin promoter (from -292 to +82 by with respect to the
10 transcription start site), 1685 by containing AC1-3 ORFs (see above), and
323 by
containing 279 by of the 3' untranslated region of nopaline synthase promoter.
The phaseolin:ACMV AC 1 ORF fusion (without the 3' nopaline synthase
promoter) will be isolated as a 2120 by Bam HI-Sac I fragment and cloned into
the Bam HI-Sac I sites of binary vector pBI101 (Clonetech Co., 6500 Donlon Rd,
15 Somis, CA) to yield binary plasmid pBE625 such that the phaseolin
promoter:ACMV AC l ORF is operably-linked to the 3' untranslated region of
nopaline synthase gene. The chimeric gene in pBE625 will be introduced into
plants via Agrobacterium tumefaciens LBA4404-mediated transformation known
to one skilled in the atrt.
20 Seed-specific vicilin promoter:ACMV replication sene
Plasmid pGV656 containing a 3086 by Hind III fragment containing a
chimeric corn 10 kD storage protein ORF under the control of seed-specific
vicilin
promoter was made. The chimeric gene consists of 1 ) an operably-linked 2308
by
Hind III-Nco I vicilin promoter isolated from plasmid pGA971 [Czako et al.
25 (1992) Mol. Gen. Genet. vol. 235(1), pp. 33-~40] and 2) a 276 by Sac I-Hind
III
fragment containing the 3' untranslated legion of nopaline synthase promoter.
The
450 by Nco I-Xba I fragment containing 10 kD ORF will be replaced by the
1685 by Nco I-Nco I-Xba I fragment containing ACl-3 ORFs (see description
above). The 4321 by Hind III fragment containing the chimeric vicilin
30 promoter:ACMV replication protein ORF:3' nopaline synthase gene will be
cloned into the Hind III site of pBinl9 binary vector and introduced into
tobacco
plants via Agrobacterium tumefaciens LBA4404-mediated transformation known
to one skilled in the art.
Chemically-inducible promoter In 2-2:ACMV replication ene plasmidpBE659)
35 Plasmid pCLV012 (ATCC 45039) containing ACMV DNA was modified
by creating an Nco I site at the initiation codon of ACl ORF by PCR and by
converting Spa B 1 site at the 3' end of the coat protein gene.[Stanley et al.
(1983)
Nature vol. 301, pp. 260-262 (1983)] to multiple cloning sites (Not I, Xba I,
Spe I,
32
CA 02304984 2000-03-15
WO 99!22003 - PCTIUS98I22688
and Bam HI sites). The Nco I-Nco I-Bam HI fragment (containing the ACMV _
ORFs AC1, AC2, and AC3) was isolated and cloned between the Nco I and Bgl II
sites in H. Hershey's pIN2-1-2 vector (supra) yielding plasmid pGV659. Plasmid
PGV659 consists of a 452 by Bam HI-Nco I fragment (sequence shown in
Figure 4 of U.S. Patent No. 5,364,780) containing corn gene 2-2 promoter
[Hershey et al., Isolation and characterization of cDNA clones for RNA species
induced by substituted benzenesulfonamides in corn. Plant Mol. Biol. (1991),
17(4), 679-90)], followed by 960 by of Nco I-Nco I-Bam HI fragment (see
above),
and a 496 by Bgl II-Asp718 I fragment containing the 3' non-coding region from
corn gene 2-1 [Hershey et al., Isolation and characterization of cDNA clones
for
RNA species induced by substituted benzenesulfonamides in corn. Plant Mol.
Biol. (1991), 17(4), 679-90; U.S. Patent No. 5,364,780]. The 2538 by
Bam HI-Asp 718I fragment in pGV659 containing operably-linked chemically-
inducible in 2-2 promoter, AC1-3 ORFs, and 247 by of the 3' untranslated
region
of corn gene 2-1 will be isolated and cloned into Bam HI-Asp718 sites of
pBinl9
binary vector and the resultant binary vector will be introduced into plants
via
Agrobacterium tumefaciens LBA4404-mediated transformation known to one
skilled in the art.
Senecence-associated promoter SAG:ACMV Ren~plasmid pBE640)
A 1737 by Nco I-Nco I-Sac I fragment containing ACMV AC1-3 ORFs
was cloned into Nco I-Sac I-digested plasmid pSG516 [Gan et al. (1995) Science
(Washington, D. C.), vol. 270(5244), pp. 1986-8] to yield plasmid pGV640. A
3924 by Spe I-Sac I fragment from pGV640 (containing the SAG promoter
operably-linked to the ACMV Rep ORF) will be isolated and cloned into
Xba I-Sac I digested pB 11 O 1 to yield binary plasmid pBE676 such that the
SAG
promoter:ACMV Rep is operably-linked to the 3' untranslated region of nopaline
synthase gene. The chimeric gene in pBE676 will be introduced into plants via
via Agrobacterium tumefaciens LBA4404-mediated transformation known to one
skilled in the art.
EXAMPLE 4
Construction of Modified Proreplicons
Modified proreplicons with minimal on will be made as follows:
The I01 by minimal TGMV-A on sequence [positions 53 to 153, Orozco
et al. (1998) Virology vol. 242, pp. 346-356] will be isolated as a PCR
product
using PCR primers P1 and P2 on pGV662 template DNA. Primer P1 will have a
Not I restriction site adjacent to position 53 of the on and primer P2 will
have a
Sal I site adjacent to on position 153. Following Not I - Sal Idigestion, the
PCR
product will be cloned into Not I - Sal I digested pGV662. The resultant
plasmid,
33
CA 02304984 2000-03-15
<..
wo ~n2oo3 rcrius9snZ6ss
pGV662A, will have the on and the mutant AL 1 in pGV662 replaced. with the 1 O
1
by minimal ori.
A I92 by sequence containing the polyadenylation sequence of CaMV
[positions 7440-7638, Gen bank accession #s V00140 J02046] will be isolated as
5 a PCR product B using PCR primers P3 and P4. Primer P3 will have a Xba I
restriction site adjacent to position 7440 and primer P4 will have a Bam HI I
site
adjacent to position 7638. A 262 by sequence containing the polyadenylation
sequence of nopaline synthase gene (positions 2068-2344, Gen Bank
ACCESSION J01541 V00087) will be isolated as a PCR product C using PCR
10 primers PS and P6. Primer PS will have a Not I restriction site adjacent to
position 1068 and primer P6 will have a Bgl II site adjacent to position 2344.
Bam HI-digested PCR product B and Bgl II-digested PCR product C will be
ligated and the Bam HI- and Bgl II-resistant ligation product will be
subjected to
PCR using primers P3 and P6. The resultant 454 by PCR product comprised of
15 inverted (head-to-head) polyadenylation sequences will be digested with Xba
I
and Not I and cloned between the Xba I and Not I sites in the 3' untranslated
region following the GUS ORF in plasmid pGV662A, such that in the resultant
plasmid, pGV662B, the GUS transcript will be polyadenylated using the CaMV
polyadenylation signal sequence and the transcript from the ALI promoter in
the
20 on will be polyadenylated using the nos polyadenylation signal sequence.
A 275 by of TGMV A sequence [positions 53-228, Orozco et al. ( 1998)
Virology vol. 242, pp. 346-356] containing the minimal TGMV-A on sequence
and the coat protein promoter will be isolated as PCR fragment E using PCR
primers P7 and P8 on plasmid pGV662 template DNA. Primer P7 will have a Sac
25 I restriction site adjacent to position 53 and Primer P8 will have a Xho I -
Nco I
sites adjacent to position 228. Following digestion with Sac I and Nco I the
27~
by sequence containing the minimal on and coat protein promoter will be cloned
in Sac I-Nco I digested pGV662B to resulat in plasmid pGV662C.
The 101 by minimal TGMV-A on sequence [positions 53 to 153, Orozco
30 et al. (1998) Virology vol. 242, pp. 346-356 (1998)] will also be isolated
as a PCR
product F using PCR primers P9 and P10 on pGV662 template DNA. Primer P9
will have a Sac I restriction site adjacent to position 53 of the on and
Primer P10
will have a Bgl II site adjacent to on position 153. The phaseolin promoter
will
be isolated as a 323 by PCR product G using primers P11 and P12 on a
previously
35 described plasmid pGV614 template DNA. Primer PI 1 has a Bam HI restriction
site adjacent to position -295 (Bcl I site) and Primer P 12 has a Nco I site
at to
position +20 (Sca I site) with respect to the transcription start site of
phaseolin
promoter [Bustos et. al. ( 1991 ) EMBO J. vol. 10, pp. 1469]. Bgl II-digested
PCR
34
CA 02304984 2000-03-15
WO 99!?,ZOD3 ' ' PCTII1S9$n368S
product F and Sam HI-digested PCR product G will be ligated and the Bam HI=
and Bgl II-resistant ligation product will be subject to PCR using primers P9
and
P12. The resultant 424 by PCR fragment, containing the minimal on and
phaseolin promoter, will be cloned in pGV662 following Sac I Nco I digestion
to
result in pGV662D.
pGV662C and pGV652D will be modified pGVb62, in which the GUS
target gene will be under the control of the cast protein promoter or
phaseolin
pratnoter, respectively. The GUS sequence will be replaced by other target
genes.
These modified plasmids will be cloned as a Hind III fragment into a binary
plasmid and used to transform transgenie plants as described above.
The sequences of PCR primers (with the introduced site underlined) used
above are given below:
P1: 5'-GCTGCGQCCGCTCCAAAAGTTATATGAATTGGTAGTAAGGT-3' [SEQ 1D NO: 3]
P2: 5'-CGAGTCdACGCGCGGCCATCCGGTAAT-3' [SEQ ID N0:4]
P3: 5'-GCAGGATCCACTGGATTTTGGTTTTAGGA-3' [SEQ ID NO: S]
P4: 5'-GCATCTAGAAAATCACCAGTCTCTCTCTACA -3' [SEQ ID NO: 6]
P5: 5'-GCTGCGGCCGCTGGAGTAAAGAAGGAGTG -3' [SEQ ID NO: 7]
P6: 5'-GCCAGATCTAGTAACATAGATGACACCG-3' [SEQ ID NO:8]
P7; 5'-CGT~,fi~TCCAAAAGTTATATGAATTGGTAGTAAGGT-3' [SEQ ID NO: 9]
P8: 5'-CCTCGAGCCATGGTTTGAATTAAAGATCCACGAAA-3' [SEQ 1D NO: 10]
P9: 5'-CGTGAGCTCTCCAAAAGTTATATGAATTGGTAGTAAGGTAAGGT-3' [SEQ ID NO:I 1 ]
PIO: 5'~GGTAGATCTGCGCGGCCATCCGGTAAT-3' [SEQ 1D NO: 12]
P11: 5'-CACGGA~',~AGATCGCCGCGTC~3' [SEQ ID NO:13J
P12: 5'~C("I'CGA(3CCATGGACTCTGGATGGATGGATGATG-3' [SEQ ID N0:14 J
CA 02304984 2000-03-15
WO 99122003 ~ PCTIUS98/22688
SEQUENCE LISTING
<110> E. I. DU DE NEMOURS AND COMPANY
PONT
<120> BINARY VIRALXPRESSION SYSTEM IN PLANTS
E
<130> CL-1127-A
<140>
<141>
<150> 60/063,504
<151> OCTOBER 29, 97
19
<160> 14
<170> Microsoft Version 7.OA
Word
<210> 1
<211> 35
<212> DNA
<213> Unknown
<220>
<223> Description Unknown Organism: primers
of
<400> 1
cgtccg gatc caattctccccatacaagag tatct 35
<210> 2
<211> 35
<212> DNA
<213> Unknown
<220>
<223> Description Unknown Organism: primers
of
<400> 2
gtcgac ccat ggttaaagatccacgaaacg catgt 35
<210> 3
<211> 41
<212> DNA
<213> Unknown
<220>
<223> Description Unknown Organism: primers
of
<400> 3
gctgcggccg ctccaaaagt tatatgaatt ggtagtaagg 41
t
<210> 9
<211> 27
<212> DNA
<213> Unknown
<220>
<223> Description Unknown Organism: primers
of
<400> 9
cgagtcgacg cgcggccatc cggtaat 2-;
<210> 5
<211> 29
1
CA 02304984 2000-03-15
WO 99IZ2003 PCTNS98I22688
<212> DNA
<213> Unknown
<220>
<223> Description of Unknown Organism: primers
' <400> 5
gcaggatcca ctggattttg gttttagga 29
<210> 6
<211> 31
<212> DNA
<213> Unknown
<220>
<223> Description of Unknown Organism: primers
<400> 6
gcatctagaa aatcaccagt ctctctctac a 31
<210> 7
<211> 29
<212> DNA
<213> Unknown
<220>
<223> Description of Unknown Organism: primers
<400> 7
gctgcggccg ctggagtaaa gaaggagtg 2g
<210> 8
<211> 28
<212> DNA
<213> Unknown
<220>
<223> Description of Unknown Organism: primers
<400> 8
gccagatcta gtaacataga tgacaccg 2g
<210> 9
<211> 38
<212> DNA
<213> Unknown
<220>
<223> Description of Unknown Organism: primers
<400> 9
cgtgagctct ccaaaagtta tatgaattgg tagtaagg 3g
<210> 10
<211> 35
<212> DNA
<213> Unknown
<220> >
<223> Description of Unknown Organism: primers
<900> 10
cctcgagcca tggtttgaat taaagatcca cgaaa 35
<210> 11
<211> 49
2
CA 02304984 2000-03-15
',
WO 99/22003 PCT/US98/22688
<212> DNA
<213> Unknown
<220>
<223> Description of Unknown Organism: primers
<400> 11
cgtgagctct ccaaaagtta tatgaattgg tagtaaggta aggt 49
<210> 12
<211> 27
<212> DNA
<213> Unknown
<220>
<223> Description of Unknown Organism: primers
<400> 12
ggtagatctg cgcggccatc cggtaat 27
<210> 13
<211> 22
<212> DNA
<213> Unknown
<220>
<223> Description of Unknown Organism: primers
<900> 13
cacggatcca gatcgccgcg tc 22
<210> 14
<211> 34
<212> DNA
<213> Unknown
<220>
<223> Description of Unknown Organism: primers
<400> 19
cctcgagcca tggactctgg atggatggat gatg 34
3