Note: Descriptions are shown in the official language in which they were submitted.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
1
DESCRIPTION
METHODS AND COMPOSITIONS FOR THE IDENTIFICATION OF GROWTH
FACTOR MIMETICS, GROWTH FACTORS AND INHIBITORS
FIELD OF THE INVENTION
'The present invention relates to the field of growth factors, inhibitors and
their
receptors.
1 o BACKGROUND OF THE INVENTION
The following is a discussion of the relevant art, none of which is admitted
to be prior
art to the appended claims.
Conventional means, i.e., protein purification and expression screening have
lead to
the identification of a limited number of growth factors and cytokines.
In hematopoiesis, for example, cytokines are required for development of all
mature,
highly specialized blood cells. All blood cells are believed to derive from a
common pool of
pluripotent stem cells which are able to undergo self renewal or give rise to
progenitor cells.
Cytokines control the complex process involving continuous coordinated
differentiation and
proliferation of stem and progenitor cells. Many cytokines are locally
produced by bone
2 0 marrow stromal cells. Others are produced in other areas of the body. Some
cytokines have
broad specificity, acting on pluripotent stem cells leading to their
differentiation, self renewal,
and proliferation. Others act late in hematopoiesis on cells of particular
lineages. Many
cytokines also affect the activity of mature cells, playing a role in the
immune response to
extrinsic antigens. Many of the known cytokines and their major roles in
hematopoiesis are
described in Table 1 (Metcalf, D., and Nicola, N.A. 1995. In The Hemopoietic
colony-
Stimulating Factors. Cambridge University press, New York; Canard, R.E., and
Gearing,
A.J.H. 1994. In The Cytokine Facts Book. Academic Press Inc., San Diego, CA;
Hamblin,
A.S. 1993. In Cytokines and Cytokine Receptors. Oxford University Press Inc.,
New York).
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
TABLE 1
IL-3 Broad specificity, acts on pluripotent
stem cells for
differentiation, self renewal, and
proliferation.
Myeloid progenitors develop into early
erythrocytes,
neutrophils, eosinophils, basophils,
macrophages,
and megakaryocytes.
GM-CSF Broad specificity, acts on pluripotent
stem cells for
differentiation, self renewal, and
proliferation.
Gives rise to neutrophils, macrophages,
and
eosinophils.
G-CSF Acts late in hematopoiesis to promote
development
of primarily neutrophils and their
precursors.
M-CSF Acts late in hematopoiesis to promote
macrophage
development
Erythropoietin Produced by the kidney. Responsible
for terminal
erythrocyte development and regulation
of red cell
development. Stimulates erythrocytes
and
megakaryocytes to develop in presence
of IL-3 or
GM-CSF.
IL-1 Primes stem cells to become responsive
to CSFs.
Induces other cells to produce GM-CSF
IL-2 Promotes T cell division and activation
of NK and B
cells.
IL-4 Promotes mast cell production.
IL-5 Promotes eosinophil differentiation.
IL-6 Induces B cell differentiation.
IL-7 Produced by bone marrow and thymic
stromal cells.
Is important in the proliferation
and differentiation
of pro-B cells and early thymocytes.
IL-8 Activated neutrophils.
IL-9 Stimulates T cell proliferation. Differentiation
factor for erythrocytes.
IL-10 Inhibits cytokine production.
IL-11 Differentiation factor for megakaryocytes.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
3
SCF (Kit-ligand) Promotes colony formation from early
hematopoietic progenitor cells by
synergising with
other growth factors. Promotes mast
cell
proliferation. Promotes survival of
hematopoietic
stem cells, and other cell types.
Plays roles in cell
adhesion and migration.
TNF- a, ~i Enhances B and T cell proliferation,
activates
granulocytes and macrophages, induces
cytokine
secretion.
gamma-interferon Enhances B cell proliferation and
differentiation,
activates macrophages, increases secretion
of other
cytokines.
LIF Supports survival of hematopoietic
stem cells.
TPO Stimulators platelet production.
flk2/flt3 ligand Stimulates proliferation of early
hematopoietic cell
precursors.
Besides natural growth factors and cytokines, agonist antibodies have been
discovered. Agonist antibodies are antibodies that mimic the natural ligand of
a receptor.
Agonist antibodies have been identified in hematopoiesis and related growth
and
differentiation pathways, using conventional monoclonal antibody technology.
Agonist
antibodies have been identified against Hepatoma transmembrane kinase (Htk), a
tyrosine
kinase in CD34+ human bone marrow cells and a human hepatocellular carcinoma
cell line
(Bennett, B.D., Wang, Z., Kuang, W.J., Wang, A., Groopman, J.E., Goeddel,
D.V., Scadden,
D.T., J. Biol. Chem. 269:14211-14218, 1994). Agonist antibodies have also been
raised to
1o flt3/flk2 (Bennett et al. U.S. Patent No. 5,635,388).
An antibody that mimics erythropoietin has been identified through screening
of monoclonal
antibodies generated against the erythropoietin receptor. This antibody
promotes receptor
response via dimerization (Schneider et al., Blood 89:473,1997). Other
antibodies that mimic
ligands have also been identified (Kahan et al. Proc. Natl. Acad. Sci., USA
75:4209, 1978).
In some disease states antibodies that bind to receptors can mimic the effect
of a
natural ligand. In hyperthyroid disease (Graves disease) there is the abnormal
production of
antibodies that bind to TSH receptors and activate these receptors (Kosugi,
S., Ban, T., Kohn,
L.D., Mol. Endocrinol. 7:114-130, 1993; Lundgate, M.E,. Vassart, G.,
Baillieres Clin.
Endocrinol. Metab. 9:95-113, 1995).
CA 02316755 2000-06-27
WO 99138008 PCTlUS99/01331
4
In contrast to antibodies that promote proliferation or differentiation,
inhibitory
antibodies have been identified that bind to cell surface receptors and
efficiently interfere with
ligand binding. These antibodies act by competing with the native molecule for
the same
binding site, or by blocking the native binding site by binding to a site in
close proximity.
There are many examples in the literature of these types of inhibitory
antibodies, including
antibodies against the c-fms receptor (Sudo, T., Nishikawa, S., Ogawa, M.,
Kataoka, H.,
Ohno, N., Izawa, A., Hayashi, S., Nishikawa, S., Oncogene 11: 2469-2476,
1995), and
against the c-kit receptor (Kodama, H., Nose, M., Niida, S., Nishikawa, S.,
Nishikawa, S.,
Exp. Hematol. 22: 979-984, 1994).
Alternatively, some inhibitory antibodies may act by mimicking a native
inhibitory
molecule. Cytokines with inhibitory effects on hematopoiesis have been
identified
(Quesenberry, P.J. 1995. Hemopoietic stem cells, progenitor cells, and
cytokines. In Williams
Hematology, Fifth Edition. Eds. Beutler, E., Lichtman, M.A., Coller, B.S.,
Kipps, T.J.
McGraw-Hill United States). These inhibitory molecules act by suppressing cell
division
during S phase, modulating surface cytokine receptor expression, or by
suppressing release
of cytokines from cells. Transforming growth factor-B (TGF-B) inhibits early
stem cells
while stimulating more mature cells. Other cytokines with inhibitory effects
include H-
subunit ferritin, Prostaglandin E1 and E2, Inhibin, and Lactoferrin
(Quesenberry 1995).
Inhibitors of hematopoiesis in the chemokine family include macrophage-
inflammatory
protein-la, macrophage-inflammatory protein-2a, platelet factor-4, interleukin-
8, interferon
inducible protein-10, as well as a few small peptides (Quesenberry, 1995).
Action of the
inhibitor may require dimerization of a receptor, or a conformational change
of a receptor
triggered by binding. An inhibitory antibody may mimic an inhibitory ligand
similarly by
promoting dimerization or change in receptor conformation.
SUMMARY OF THE INVENTION
The present invention utilizes an immune system approach for the
identification of
new growth factor mimetics, growth factor receptors, native growth factors,
growth factor
receptor antagonists, and inhibitors involved in differentiation pathways,
developmental
3 0 pathways, cell survival and functional activation or inhibition of cells.
Such pathways
include, but are not limited to, hematopoiesis, nervous system development and
regeneration
CA 02316755 2000-06-27
WO 99/38008 PCT/US99101331
and organ/tissue development and regeneration. In a preferred embodiment the
invention
concerns the identification of growth factors, mimetics and inhibitors that
affect growth and
differentiation of hematopoietic cells including pluripotent stem cells,
multipotent progenitor
cells and unipotent cells at various stages within each hematopoietic lineage
including
5 granulocytes, monocytes, macrophages, eosinophils, megakaryocytes, mast
cells, erythroid
cells, T-lymphocytes, B-lymphocytes and dendritic cells.
The broad range screening and identification methods of the current invention
allow
for the selection and screening through a large repertoire of binding
molecules to cell surface
molecules to find those that have proliferative, differentiative,
developmental, cell survival,
functional activation or functional inhibitory effects. In preferred aspects
the method uses
combinatorial libraries and phagemids to identify first generation mimetic
molecules (agonist
antibodies) or inhibitor molecules (inhibitory antibodies) (e.g. antibodies
that mimic natural
inhibitors or antagonist antibodies) that behave as agonists or inhibitors in
proliferation,
differentiation, survival or activation of various cell lineages. First
generation agorusts and
inhibitors can be used directly as therapeutic agents (e.g., for agonists:
amplification of
clinically relevant cell types; for ex vivo proliferation and differentiation
for gene therapy
purposes; e.g., for inhibitors: inhibition of the proliferation of normal
cells only so that
cancerous cells will continue to divide and be more susceptible to
chemotherapeutic agents
or inhibition of the proliferation or differentiation of a population of cells
that is involved in
2 0 a disease or disorders, such as cells involved in allergic reactions), as
reagents in diagnostics,
and in basic and clinical research (e.g., antibodies for cell sorting of cells
such as
hematopoietic cells and for the identification of cells, such as hematopoietic
cells). First
generation agonists and inhibitors can also be used to clone the receptors,
and ultimately, the
native factors they mimic. Native factors that result from these studies could
be used
2 5 clinically in many disease states. For example, beneficiaries of new
hematopoietic growth
factors and mimetics include, but are not limited to, all patients, including
HIV infected
patients that suffer from disease or treatment related immunosuppression,
including
chemotherapy, bone marrow transplants, and myeloproliferative disorders, and
will augment
the armamentarium of this class of therapeutic agents.
CA 02316755 2000-06-27
WO 99/38008 PCT/(JS99/01331
6
One aspect of the invention involves the immunization of an animal with human
or
marine stem/progenitor cells so as to generate immune repertoire libraries of
antibodies raised
to epitopes displayed on the surface of these cells. As mixtures of primary
cells are utilized,
immunogens will be expressing thousands of potential epitopes which represent
portions of
cell surface receptors involved in a variety of different stages of
development, differentiation,
survival, activation and inactivation for a variety of cell types. The
epitopes of interest are
those cell surface receptors that link to a signal transduction cascade that
lead to enhancement
or inhibition of proliferation, differentiation, development, survival or
activation. For
example, to raise antibodies that effect cells of hematopoietic origin, bone
marrow cells would
be one choice of cells to be injected.
The present methods of discovery of new growth factors, inhibitors and related
molecules is applicable to other differentiation/developmental pathways in
addition to
hematopoiesis. For example, an animal (rabbit, chicken, or other animal) can
be immunized
with marine or human embryonal carcinoma cells, such as marine P 19 cells
(McBurney,
M.W., Rogers, B.J., Developmental Biology 89:503-508, 1982) or human NTera-
2cl.DI cells
(Andrews, P.W., Damjanov, L, Simon, D., Banting, G.S., Carlin, C., Dracopoli,
N.C., Fogh,
J., Laboratory Investigation 50:147-162, 1984). These cells are
pluripotential, but
differentiate poorly under normal culture conditions. They can be induced to
differentiate into
neuronal and glial cells (generally in presence of inducing agents such as
retinoic acid), or
2 0 cardiac muscle and skeletal muscle (for example, in the presence of DMSO).
Immunization
with these cells will allow for the identification of agonist antibodies,
inhibitory antibodies
and growth factors that are involved in differentiation into these and other
pathways.
Alternatively, an animal could be immunized with human or mouse pluripotent
teratocarcinoma cells. In this case, agonist or inhibitory antibodies could be
identified that
2 5 would promote or inhibit development of endodermal derived tissues and
organs, such as
lung, liver, pancreas, stomach, esophagus, pharynx, intestines, or salivary
glands.
Immunization is also possible with marine pluripotent embryonic stem cells.
Embryonic stem cells (ES cells) are derived from early mammalian cells that
are totipotent
and capable of in vitro proliferation. They can differentiate to all three
embryonic germ layers
30 and their derivatives. (Evans, M.; Kaufman, M., Nature 292: 154, 1981;
Martin, G., Proc.
CA 02316755 2000-06-27
WO 99/38808 PCTNS99/01331
7
Natl. Acad. Sci. U.S.A.,78: 7634, 1981). In addition, human ES cell lines
could be used as
immunogens (Thomson, J.A., Itskovitz-Eldor, J., Shapiro, S.S., Waknitz, M.A.,
Swwiergiel,
J.J., Marshal, V.S., Jones, J.M., Science 282: 1145-1147, 1998; Bongso, A.,
C.Y. Fong, S.C.
Ng, and S. Ratnam, Hum. Reprod. 9: 2110, 1994). In this case, agonist or
inhibitory
antibodies could be identified that promote or inhibit differentiation into a
variety of
embryonic structures.
Following immunization, combinatorial antibody fragment libraries are
constructed
from an immunized animal. The present invention also encompasses these
combinatorial
libraries directed to surface components on the various cells used for
immunization. These
l0 libraries will yield a higher frequency of specific antibodies to antigens
of interest as
compared with naive libraries or synthetic libraries. Libraries generated from
an animal that
has undergone 1-3 booster immunizations will also result in antibody fragments
with higher
affinities, due to affinity maturation.
Alternatively, in a less preferred embodiment existing antibody libraries,
such as
synthetic antibody phage display libraries, may be screened. However, as these
are not
targeted libraries, agonist or inhibitory antibodies of interest may not be
present. A number
of synthetic antibody phage display libraries contain a broad assortment of
binding
specificities. Libraries have been created using random oligonucleotide
synthesis (Barbas,
C.F. III, Bain, J.D., Hoekstra, D.M., Lerner, R.A., Proc. Natl. Acad. Sci U S
A 89: 4457-
4461, 1992; Soderlind, E., Vergeles, M., Borrebaeck, C.A., Gene 160: 269-272,
1995). In
addition, large phage display libraries of human single chain Fv (scFv)
antibody fragments
were constructed by combining germline VH genes with synthetic heavy chain
CDR3 regions
and various light chain sequences (de Kruif, J., Boel, E., Logtenberg, T., J.
Mol. Biol. 248:
97-105, 1995; Akamatsu, Y., Cole, M.S., Tso, J.Y., Tsurushita, N., Jlmmunol.
151: 4651-
2 5 4659, 1993). Other synthetic libraries have also been reported (Fuchs, P.,
Dubel, S., Breitling,
F., Braunagel, M., Klewinghaus, L, Little, M., Cell Biophys. 21: 81-91, 1992;
Hoogenboom,
H.R., and Winter, G., JMoI. Biol. 227: 381-388, 1992).
In the present invention, expression libraries from an immunized animal are
preferably
constructed as either Fab fragment libraries or single chain variable region
libraries (scFv).
3 o Fab fragment libraries, that maintain the native antigen recognition site,
are useful to ensure
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
8
that affinity is maintained. Single chain libraries are useful because the
entire binding domain
will be contained on one polypeptide.
Antibody or antibody fragments can be presented to target cells for screening
purposes
in a variety of ways. First, they can be surface displayed on bacteriophage,
phagemids,
prokaryotic cells such as E. coli, eukaryotic cells such as mammalian cells or
yeast or
displayed on ribosomes. The surface displayed antibodies are screened by
contacting the
entity that carries the surface displayed antibody (bacteriophage, phagemid,
bacterial cell or
mammalian cell) with the target cells so that binding to target cells occurs,
removing unbound
entities, amplifying any entities that remain bound for further testing in
bioassays.
Alternatively, co-cultivation can be used for screening purposes. The antibody
producing
cells, bacterial cells producing bacteriophage or phagemids, bacterial cells
producing
antibodies, eukaryotic cells producing antibodies) are grown in the presence
of target cells
(co-cultivation). Antibodies are presented on the surface of cells,
bacteriophages or
phagemids or are secreted into the medium. A third, means of presentation to
target cells is
by utilizing secreted antibodies.
Antibody display has been done on the surface of bacteriophage (Huse, W.D.,
Sastry,
L., Iverson, S.A., Kang, A.S., Alting-Mees, M., Burton, D.R., Benkovic, S.J.,
and Lerner,
R.A., Science 246:1275-1281,1989; McCafferty, J., Griffiths, A.D., Winter, G.,
Chiswell,
D.J., Nature 348:552-554, 1990; Chang, C.N., Landolfi, N.F., and Queen, C.,
J., Immunol.
2 0 147:3610-3614, 1991); phagemids (Barbas III, C.F., Kang, C.F., Lemer,
R.A., and Benkovic,
S.J., Proc. Natl. Acad. Sci., USA, 88: 7978-7982, 1991) and on the surface of
prokaryotic
cells, such as E. coli (Fuchs, P., Breidling, F., Belscheaus, T., Little, M.,
Biotechnology 9:
1369-1372, 1991; Francisco, J.A. et al., Proc. Natl. Acad Sci., USA, 90: 10444-
10448, 1993;
Chen, G., Cloud, J., Georgiou, G., Iverson, B.L., Biotechnol. Prog. 12: 572-
574, 1996).
2 5 Antibodies have been expressed intracellularly and secreted in eukaryotic
cells. Antibodies
have also been expressed on the surface of eukaryotic cells (yeast). Mammalian
CHO cells
and COS cells have been well utilized for antibody secretion (Trill, J.J.,
Shatzman, A.R.,
Ganguly, S., Curr. Opin. Biotechnol. 6: 553-560, 1995; Fouser, L.A., Swanberg,
S.L., Lin,
B.Y., Benedict, M., Kelleher, K., Gumming, D.A., Riedel, G.E., Biotechnology
(N ~ 10:
3 0 1121-1127, 1992). The GPI anchor is well utilized for anchoring a variety
of proteins to the
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
9
cell surface. In eukaryotic cells, antibody fragments could be displayed on
the surface of the
plasma membrane using a GPI anchor. Fusion proteins linked to a GPI anchor
have been used
extensively for the expression of heterologous proteins on the cell surface
(Scallon, B.J.,
Kado-Fong, H., Nettleton, M.Y., Kochan, J.P., Biotechnology 10: 550-556,
1992). In yeast,
antibody expression has been done intracellularly (Carlson, J.R. and Weissman,
LL., Mol.
Cell. Biol., 8:2647-2650, 1988; Bowdish, K. S., Tang, Y. Hicks, J.B., and
Hilvert, D., J. Biol.
Chem. 266: 11901-11908, 1991) and on the surface (Boder, E.T. and Wittrup,
K.D., Nat.
Biotechnol., 15: 553-557, 1997)). A variety of stable vectors and efficient
promoters and
secretion signals are available in yeast for engineering the secretion of any
protein of interest.
to These have been reviewed in Moir, D.T., Davidow, L.S., Methods Enzymol.
194: 491-507,
1991. Cell surface expression of heterologous proteins on the surface of yeast
was reviewed
recently (Georgiou, G., Stathopoulos, C., Daugherty, P.S., Nayak, A.R.,
Iverson, B.L.,
Curtiss, R. IIL, Nat. Biotechnol. 15: 29-34, 1997). Cloning fragments
downstream from the
leader sequence Mating Factor alpha has been used successfully for secretion
of heterologous
proteins (Swart, A.C., Swart, P., Roux, S.P., van der Merwe, K.J., Pretorius,
LS., Steyn A.J.,
Endocr. Res. 21: 289-295, 1995). In addition, numerous heterologous proteins
have been
produced at greater than gram per liter levels in the methyiotropic yeast
Pichia pastoris using
the methanol oxidase promoter (Sreekrishna, K., Brankamp, R.G., Kropp, K.E.,
Blankenship,
D.T., Tsay, J.T., Smith, P.L., Wierschke, J.D., Subramaniam, A., Birkenberger,
L.A., Gene
2 0 190: 55-62, 1997). Those of ordinary skill in the art based on these and
other known
techniques can readily achieve surface display and secretion of antibody
molecules and
fragments.
Those who practice the art appreciate that other means for presenting
antibodies or
antibody fragments to target cells that are not surface display or secreted
are possible and are
2 5 suitable for use in the present invention. These methods include, but are
not limited to,
prokaryotic ribosome display (thanes, J. and Pluckthun, A., Proc. Natl. Acad.
Sci. U.S.A., 94:
4937-4942, 1997) and eukaryotic ribosome display (Translocus Therapeutics,
Cambridge,
United Kingdom).
Preferably, the libraries are constructed in phagemid vectors which allows for
rapid
3 0 screening. Phagemids require superinfection with helper phage.
Superinfection will provide
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
the remaining phage components needed for packaging plasmids into phagemid
particles.
Each phagemid will contain approximately one or more antibody binding sites,
displayed on
the surface as a coat protein fusion, per phagemid particle. The remaining
coat proteins will
be contributed by helper phage and will therefore be wildtype and allow for
efficient
5 reinfection of phagemids into E. coli for amplification. The functional
domain of the gene III
coat protein is preferred as the fusion partner for display of the antibody
fragment. However,
it is also possible to utilize any full length or functional domain coat
protein of the phage,
such as gene VIII, that allows for phage surface display (Kang, A.S., Barbas
III, C.F., Janda,
K.D., Benkovic, S.J., and Lerner, R.A. Proc. Natl. Acad. Sci. USA, 88:4363-
4366, 1991).
10 The initial screening of the surface displayed antibody molecules or
fragments is
preferably by panning {DeKruif, J., Terstappen, L., Boel, E., Logtenberg, T.,
Proc. Natl. Acad.
Sci. USA 92: 3938-3942, 1995). Panning of monomeric antigen binding sites has
the
advantage of sorting clones based on amity as well as specificity, and will
therefore skew
the population towards isolation of high affinity binders. Typically there are
several rounds
of selection and amplification so as to enrich for binding molecules. Panning
is conducted
utilizing target cells expressing the cell surface receptor toward which the
antibody is directed
or a closely related receptor. Alternatively, cells can be separated by
fluorescence activated
cell sorter (FACS) sorting or magnetic sorting. Phage that adhere to a
specific population of
cells can be grown out by eluting and infection of bacterial cells.
2 0 Another alternative for initial screening detects binding of antibody
molecules or
fragments surface displayed on phage to target cells expressing the cell
surface receptor
toward which the antibody is directed or a closely related receptor and which
results in the
bound phage become internalized by endocytosis of the receptor. These phage
can be
identified, for example, by eluting surface bound phage and lysing cells
followed by
2 5 electroporation to recover internalized phage DNA.
Another possible method of identification of specifically bound antibodies
involves
the use of radiolabelled cell lysates or membrane preparations which are
electrophoresed in
one or two dimensional gels and transferred to an immobilized solid support
membrane.
Phage are hybridized and eluted from specific spots of interest. Spots can be
chosen based
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
11
on comparing the pattern of membrane proteins exhibited by targets cells which
have the cell
surface molecules of interest and unrelated cells which should not.
Surface displayed antibodies that specifically bind to target cells (those
that remain
on the surface of the target cell or those that are internalized) are then
screened against various
target cells and cell lines for the ability to promote or inhibit
proliferation, differentiation, cell
survival, or activation utilizing various bioassays or receptor assays.
Dimerization is often a
prerequisite for activation of many receptors including hematopoietic
receptors (The
Hematopoietic Colony-Stimulating Factors, D.M. Metcalf, N.A. Nicola (1995)
Cambridge
University Press, New York). Thus, subsequent to panning, antibody fragments
identified as
binding cell surface molecules on target cells are tested in bioassays or
receptor assays as
dimers. Inhibitory antibodies that act as antagonists may not need to be
dimers, especially if
they merely block the receptor from binding of an agonist, however if the
inhibitory antibody
mimics a native inhibitory molecule it may be required to be a dimer to be
functional.
The present invention offers several advantages over other methods for
identification
of growth related molecules. Prior approaches to the isolation of growth
factors utilized
purification of active factors from conditioned medium by separative
biochemistry. These
methods required the factors to be present in significant quantities. Other
methods use direct
expression screening of cDNA pools, using cell lines as bioassays. In this
case, the level of
transcription is a factor in successful identification of the factor, as is
proper folding of the
2 0 polypeptide chain in the expression screen. Also, it is increasingly
becoming obvious that
many of the regulators were designed to function most efficiently when acting
in combination,
these approaches are limited in that they can only identify factors that act
singly to affect
proliferation and/or differentiation.
In distinction, the present invention selects directly for binding molecules
and then
2 5 screens through the entire repertoire of binding molecules to find those
with proliferative or
differentiative effects. A large immune repertoire as a combinatorial library,
allows for
random association of heavy and light chain binding regions, increasing
candidate agonists
and inhibitors even further. Also, the claimed methods are not subject to
limitations of
traditional monoclonal antibody technology (limited by the number of B cell
fusions to
3 0 myeloma cells per immunized animal) and will enable screening through a
large immune
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
12
repertoire. In addition, the screening methods are not limited to the type of
regulator or to its
origin or site of production, as it is known that both local control through
cell-cell contact, and
humoral regulatory molecules are involved in proliferation, development,
differentiation, cell
survival, functional activation and functional inhibition. Furthermore, the
present screening
methods do not presuppose any biochemical function, extensive sequence
similarity with
known genes, or site of origin. The claimed identification and screening
methods offer
several practical advantages over conventional screening methods including:
rapidity,
cost-effectiveness, and simplicity. The current methods also offer
simultaneous multi-sample
analysis, which renders more Ab fragments available for screening.
The present invention also encompasses methods for screening antibodies
(agonist or
inhibitory) by co-culturing cells that express the antibodies in the presence
of cells (target cell)
that express receptors that the antibodies bind.
Once agonist or inhibitory antibodies are identified they can be synthesized
using
standard recombinant techniques known to those who practice the art. These
antibodies and
the methods of making them, are also encompassed by the claimed invention. The
present
invention further encompasses methods for the use of such antibodies to
identify the receptors
to which the antibodies bind and to identify the native growth factors or
inhibitory factors that
the antibodies may mimic.
Furthermore, the present invention encompasses use of agonist antibodies to
treat a
2 0 patient with a deficiency in a particular cell type by stimulating the
proliferation or
differentiation of the cell type or its precursors and the use of inhibitory
antibodies to inhibit
a particular cell type, and the use of antibodies to identify particular cell
types.
Also, encompassed by the present invention is use of agonist and inhibitory
antibodies
to identify and isolate specific populations of cells.
2 5 In a first aspect, the invention features a method to identify agonist or
inhibitory
antibodies to receptors involved in cellular proliferation, differentiation,
survival or activation
comprising the steps of immunizing an animal with stem/progenitor cells having
surface
molecules comprising the receptors, so as to generate a plurality of immune
cells expressing
one or more antibodies to the surface molecules, creating a library from the
plurality of
3 0 immune cells comprising nucleic acid sequences encoding the antibodies,
cloning the nucleic
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
13
acid sequences from the library into surface display vectors so that the
antibodies are surface
displayed, and screening the surface displayed antibodies using target cells
to identify agonist
or inhibitory antibodies to the receptors.
By "receptor involved in cellular proliferation, differentiation, survival or
activation"
is meant a receptor linked to a signal transduction cascade that leads to
enhancement or
inhibition of proliferation, differentiation, development, survival or
activation of the cell that
displays the receptor. Proliferation is active division and progression
through the various
stages of the cell cycle as detected by changes in the rate of protein
synthesis, chromosome
replication, cell size or cell number. Differentiation involves changes in
cell morphology,
1 o behavior or function that lead to the production of different types of
cells with specialized
functions, as a result of exposure to extrinsic factors, or changes in gene
expression by which
cells mature and become less pluripotent. Activation is the process by which
cells leave Go
and enter G,, but do not synthesize DNA or divide until a second signal is
received.
Activation involves the expression of a specific set of activation genes and
activation antigens.
For example, activation antigens for B cells include CD23, surface IgM, and
CD40. Survival
is meant that cells do not undergo apoptosis or programmed cell death or
necrosis. Although
the present invention encompasses antibodies directed to yet undiscovered
receptors, the
identity of the receptors is not necessary as one of ordinary skill in the art
would be able to
detect binding of the antibodies to the receptors by the resultant behavior
exhibited by the cell,
2 0 i.e., proliferation, differentiation, activation, or survival.
In preferred embodiments the stem/progenitor cells are selected from the group
consisting of unsorted human bone marrow cells, human peripheral blood cells
originating
from human bone marrow, sorted human bone marrow cells, unsorted marine bone
marrow
cells, sorted marine bone marrow cells, fetal liver cells, yolk sac cells,
cells derived from the
2 5 marine AGM region, human or marine embryonal carcinoma cells or lines,
human or mouse
pluripotential teratocarcinoma cells or lines, marine pluripotent embryonic
cells, human
embryonic stem (ES) cell lines, cells of neural origin, cells involved in
organ or tissue
regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
mononuclear cells, human bone marrow CD34+ cells, FDCP-mix marine
hematopoietic stem
30 cell line, B6SUTA marine hematopoietic stem cell line, P19 teratocarcinoma
cells, and
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
14
NTera-2 pluripotent embryonal carcinoma cells, the antibodies are scFv or Fab
fragments, the
surface display vectors are phagemids, the library is a combinatorial library.
In another embodiment, the invention features a method to identify agonist
antibodies
to growth factor receptors comprising the steps of immunizing an animal with
stem/progenitor
cells having surface molecules comprising the growth factor receptors, so as
to generate a
plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from the plurality of immune cells comprising nucleic acid sequences
encoding the
antibodies, cloning the nucleic acid sequences from the library into surface
display vectors
so that the antibodies are surface displayed, and screening the surface
displayed antibodies
using target cells to identify agonist antibodies to growth factor receptors.
By "agonist antibody" is meant entire antibodies or fragments thereof,
including Fab
fragments and scFv fragments, that bind to a cell surface receptor that is
linked to a signal
transduction cascade that leads to the proliferation, differentiation,
activation, survival or
preservation of the viability of the cell that displays the cell surface
receptor.
By "growth factor receptor" is meant a molecule that binds to a growth factor
or
another ligand and participates in a signal transduction cascade that leads to
the proliferation,
differentiation, survival or activation of a cell which has the growth factor
receptor presented
on its plasma membrane (i.e., cell surface).
By "immunizing an animal" is meant injecting one or more animals by any route
of
2 0 administration, e.g., intraperitoneally, intravenously, subcutaneously, or
intramuscularly with
cells, e.g., stem/progenitor cells, so that an immune response is generated by
the animal to
molecules on the surface of the cell. Primary immunizing can be in the
presence of adjuvant.
Primary immunization is often followed by boosts with the same antigen to
produce a greater
response and increased affinity of antibodies produced. Typically, 1 to 3
booster
2 5 immunizations are given at intervals of 2-8 weeks, usually at 3-4 weekly
intervals. Typically,
more than a single animal is immunized.
By "stem/progenitor cells" is meant to include cells that are pluripotent and
cells that
are multipotent and also end stage or terminally differentiated cells derived
from pluripotent
or multipotent cells that have cell surface receptors involved in cellular
proliferation,
3 o differentiation, activation or survival. Pluripotent cells (stem cells)
are able to differentiate
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/OI331
into all cell lineages. For example, human embryonic stem (ES) cells are able
to differentiate
to all three embryonic germ layers and their derivatives. The least mature
hematopoietic stem
cells are able to form mature hematopoietic cells of all lineages and
repopulate the
hematopoietic tissues of an animal. Multipotent cells (progenitor cells) are
restricted to
5 differentiating to a limited number of lineages (typically two or three).
Hematopoietic
stem/progenitor cells are present in bone marrow and in circulating blood.
Stem/progenitor
cells include, but are not limited to, unsorted human bone marrow cells, human
peripheral
blood cells originating from human bone marrow, sorted human bone marrow
cells, unsorted
marine bone marrow cells, sorted marine bone marrow cells, fetal liver cells,
yolk sac cells,
10 cells derived from the marine AGM region, human or marine embryonal
carcinoma cells or
lines, human or mouse pluripotential teratocarcinoma cells or lines, marine
pluripotent
embryonic cells, human embryonic stem (ES) cell lines, cells of neural origin,
cells involved
in organ or tissue regeneration, human bone marrow cells that have undergone
RBC lysis,
human bone marrow mononuclear cells, human bone marrow CD34+ cells, FDCP-mix
marine
15 hematopoietic stem cell line, B6SUTA marine hematopoietic stem cell line,
P19
teratocarcinoma cells, and NTera-2 pluripotent embryonal carcinoma cells.
Cells for
immunization can be obtained from any of these sources.
By "surface molecules" is meant plasma membrane (cell membrane) protein
molecules where at least a portion of the molecule is on the extracellular
face of the plasma
2 0 membrane (cell membrane).
By "plurality of immune cells expressing one or more antibodies" is meant a
collection of antibody producing cells from blood or lymphoid organs, such as
bone marrow,
spleen and lymph nodes from an immunized animal.
By "library" is meant a collection of nucleic acid molecules representing the
immune
2 5 repertoire of an organism, which in the present invention would include
molecules encoding
for antibodies and fragments directed toward the growth factor receptors of
the relevant
stem/progenitor cell. Libraries are typically presented as nucleic acid
molecules cloned in a
vector, e.g. surface display vector.
By "surface display vector" is meant a nucleic acid vector that carries at
least the
3 0 functional domain of a surface display protein, i.e., the leader sequence
that directs export
CA 02316755 2000-06-27
WO 99138008 PCT/US99/OI331
16
from the cells, and a cloning site to insert antibody sequences, which results
in an antibody
fusion gene. The surface display vector when in a host cell allows for the
production of a
fusion protein consisting of a surface display protein or functional domain
thereof and an
antibody or fragment thereof so that the antibody or fragment is surface
displayed. That is the
antibody or fragment is presented on the surface as a functional binding
polypeptide, linked
to the domain of the surface display protein. Surface display can be on the
surface of a
bacteriophage, phagemid, bacterial cell, or mammalian cells. Examples of
surface display
vectors include, but are not limited to, pCOMB3, SurfLAP, pCANTABSE and
pEXmide 3.
Those who practice the art are familiar with these and other surface display
vectors and can
readily construct other similar vectors. Antibodies that are surface displayed
are able to bind
antigen presented from the extracellular side of the plasma (cell) membrane in
which they are
located.
By "screening" is meant using methods for identifying antibodies that are
agonist
antibodies or are inhibitory antibodies. For agonist antibodies, screening
involves identifying
antibodies that bind to receptors on target cells, so as to trigger or
activate a receptor that
participates in a signal transduction cascade that leads to the proliferation,
differentiation,
survival or activation of that cell. Screening may involve only the
determination of the ability
of the antibody to bind to the receptor on a target cell (including both
antibodies that remain
on the cell surface after binding and those that are internalized after
binding) and/or assays
2 0 that determine cellular responses associated with binding of the antibody
to the receptor; these
include bioassays that detect differentiation, proliferation, cell survival or
activation (such as
changes in transcription of downstream effector genes) and biochemical assays
that detect
chemical processes such as phosphorylation. For inhibitory antibodies
screening involves
identifying antibodies that bind to receptors on cells, such that the binding
of the antibody to
2 5 the receptor results in the inhibition of growth, proliferation,
differentiation, activation or
survival of that cell. Screening may involve only the determination of the
ability of the
antibody to bind to the receptor on a cell (including both antibodies that
remain on the cell
surface after binding and those that are internalized after binding) and/or
assays that determine
cellular responses associated with binding of the antibody to the receptor;
these include
3 0 bioassays that detect differentiation, proliferation, cell survival or
activation (such as changes
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
17
in transcription of downstream effector genes) and biochemical assays that
detect chemical
processes such as phosphorylation.
By "target cells" is meant cells that display receptors to which agonist or
inhibitory
antibodies bind. The growth factor receptor on the target cell is the same as
or highly related
to the receptor present on the stem/progenitor cell. For hematopoietic cells,
these may
include, but are not limited to, primary hematopoietic cells, several factor
dependent marine
cell lines and human leukemia cell lines. Target cells could be the same
primary cell type, a
population of cell types, or cell line used as an immunogen or another cell
type or cell line.
Target cells are chosen based on the immunogen used. For example, immunizing
broadly
l0 with human primary bone marrow aspirates treated with red blood cell lysing
reagents and
centrifugation to remove red blood cells and platelets should result in
antibodies with
specificities against a wide variety of cell surface receptors. Target cells
in this case could
include the same immunogen where a large variety of antibodies would be
isolated, or target
cells could include a sorted population of cells, such as CD34+ cells to
identify antibodies
against a specific cell lineage. Another target option with the broad
immunogen would be to
utilize the marine hematopoietic cell line FDCP-mix. In this case, a more
uniform population
of cells, capable of in vitro growth has its advantages, however, agonist
antibodies identified
using a marine target would have to be further characterized for its ability
to promote effects
on human cells. Alternatively, with an immunogen composed of a sorted
population of cells,
2 o such as CD34+ cells, one would likely use the CD34+ cells as the target,
or the FDCP-mix cell
line. Thus, the appropriate choice of immunogen and target cell are key to
identifying
relevant antibodies. Those of ordinary skill in the art would readily be able
to determine the
appropriate target cell based on the nature of the antibody sought, i.e., an
antibody directed
to a receptor that only participates in the growth of precursor cells would
require the target
2 5 cells to be early lineage cells.
In a preferred embodiment, the invention features a method to identify agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
stem/progenitor cells having surface molecules comprising the growth factor
receptors, so as
to generate a plurality of immune cells expressing one or more antibodies to
the surface
3 o molecules, creating a library from the plurality of cells, comprising
nucleic acid sequences
CA 02316755 2000-06-27
WO 99/3$008 PC"T/US99/01331
18
encoding the antibodies, cloning the nucleic acid sequences from the library
into viral display
vectors so that the antibodies are displayed on the surface of virus, and
screening the
antibodies displayed on the surface of the virus using target cells to
identify agonist antibodies
to the growth factor receptors.
By "viral display vectors" is meant vectors that allow for display of an
antibody or
fragment thereof on the surface of a virus. Preferably the viral vectors are
bacteriophage (e.g.,
fl, M13, fd, ~,, T3, T4, T7) or phagemids vectors. Bacteriophage are viruses
able to infect
bacterial cells, replicate and package their genome and upon release from a
cell are able to
continue this cycle by infecting other cells. Phagemids are virus that are
missing essential
genes required for the above described life cycle, which are provided by
separate helper
phage. Those of ordinary skill in the art are familiar with viruses,
bacteriophage and
phagemid vectors which are suitable for use in the claimed invention. Examples
of viral
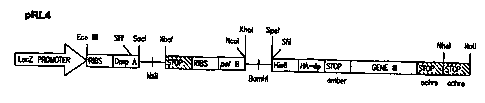
display vectors include pRL4 (Prolifaron, LLC, San Diego, CA), pCOMB3 (Burton,
D.R. and
Barbas, C.F. III. Advances in Immunology 57:191-280, 1994), SurfZAP Vector
(Stratagene,
La Jolla, CA), pCANTABSE (Pharmacia, Piscataway, NJ), pEXmide 3 (Soderlind,
E.,
Lagerkvist, A.C., Duenas, M., Malmborg, A.C., Ayala, M., Danielsson, L.,
Borrebaeck, C.A.
Biotechnology 11:503-507,1993).
In further preferred embodiments the viral vectors are selected from the group
consisting of bacteriophage and phagemid vectors.
2 o In another preferred embodiment, the invention features a method to
identify agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
stem/progenitor cells having surface molecules comprising the growth factor
receptors, so as
to generate a plurality of immune cells expressing one or more antibodies to
the surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
2 5 sequences encoding scFv fragments of the antibodies, cloning the nucleic
acid sequences from
the library into phagemid vectors so that the scFv fragments are displayed on
the surface of
phagemid, and screening using target cells, the scFv fragments displayed on
the surface of the
phagemid to identify those that are agonist antibodies for the growth factor
receptors.
By "scFv fragment" is meant single chain antibody fragment whereby the
complete
3 o antigen binding domain is contained within a single polypeptide.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
19
In another preferred embodiment, the invention features, a method to identify
agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
stem/progenitor cells having surface molecules comprising the growth factor
receptors, so as
to generate a plurality of immune cells expressing one or more antibodies to
the surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding scFv fragments of the antibodies, cloning the nucleic acid
sequences from
the library into surface display vectors so that the scFv fragments are
surface displayed, and
screening using target cells the surfaced displayed scFv fragments to identify
those that are
agonist antibodies for the growth factor receptors.
In a further preferred embodiment the surface display vector is a
bacteriophage vector
which allows for expression of the antibody on the surface of bacteriophage.
In another preferred embodiment, the invention features a method to identify
agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
stem/progenitor cells having surface molecules comprising the growth factor
receptors, so as
to generate a plurality of immune cells expressing one or more antibodies to
the surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding scFv fragments of the antibodies, cloning the nucleic acid
sequences from
the library into phagemid vectors so that the scFv fragments are displayed on
the surface of
phagemid, panning the scFv fragments displayed on phagemids for binding to
cell surface
2 0 molecules on target cells, and screening in a functional assay the scFv
fragments that bind the
cell surface molecules to identify those that are agonist antibodies for the
growth factor
receptors.
By "panning" is meant exposing a surface display library (bacteriophage,
phagemid
or cell) to antigens of interest, i.e., cell surface molecules which include
receptors to which
2 5 the antibodies or antibody fragments are directed, which are displayed on
target cells to enrich
for antibodies that specifically bind to target cells. For example, the target
cells can be
immobilized, e.g., on a plastic surface, or fixed, or captured by
centrifugation. After specific
antibodies or fragments displayed on bacteriophage, phagemid or whole cells
bind to target
cells the remainder of the surface display library are removed by washing.
Binding and
3 0 washing are followed by elution (e.g., by low pH) of bacteriophage,
phagemid, cells which
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
display specific binding antibodies. The panning process is typically repeated
for several
rounds. If bacteriophage or phagemid are utilized then, after each round
eluted phagemid or
bacteriophage may be amplified in host cells. Alternatively, fluorescence
activated cell
sorting (FACS), or magnetic cell sorting can be used for high throughput
screening to identify
5 relevant binding antibodies. Methods for FACS sorting of surface displayed
molecules are
described in: Georgiou, G., Stathopoulos, C. Daugherly, P.S., Nayak, A.R.,
Iverson, B.L., and
Curtiss R. III. Nature Biotechnology 15: 29-34, 1996. These methods can be
easily adapted
to screen surface displayed antibody fragments by those of ordinary skill in
the art. Panning
for scSv fragments can be carried out as dimers, due to one cloning strategy
employed, but
10 could also be carried out as monomers. Alternatively, bacteriophage or
phagemid that
recognize receptors can also be identified using an internalization approach
in which the
bacteriophage or phagemid that remain bound to the cell surface are removed
and then cells
are lysed to recover internalized bacteriophage or phagemids. Another
alternative, is to isolate
membrane proteins from target cells and bind antibodies to specific fractions
of these
15 membrane proteins.
By "binding to cell surface molecules "is meant binding of the antigen binding
domain
of an antibody fragment to the antigenic determinant on the receptor to which
it was raised
or a closely related antigenic determinant, such as a determinant on a
receptor of the same or
a related family or a related receptor in another species.
2 0 By "screening in a functional assay" is meant examination of antibodies
(agonist or
inhibitory), that have been surface expressed, either individually or in pools
to determine their
effect on target cells (i.e., binding of antibodies to growth factor receptors
displayed on target
cells that effects proliferation, differentiation, development, survival or
activation). Only
antibody fragments that have been determined to bind to target cells by
panning, are
2 5 subsequently screened in the functional assays. In functional assays for
agonist antibody
fragments it is scFv dimers that are screened. Screening of the scFv dimer
molecules can
occur as surface displayed molecules, soluble scFv molecules or as secreted
molecules. In
functional assays for inhibitory antibody fragments the antibody fragments are
screened
independently as monomers and as dimers and as either surface displayed
molecules or
3 0 soluble molecules. If the scFv fragment acts as an antagonist, binding to
a growth factor
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
21
receptor and blocking its activation, it may do so as a monomer. However, if
the scFv
fragment acts as a mimetic of a native inhibitory molecule, it may need to be
a dimer as in the
case of agonist antibodies. Screening in functional assays involves either
bioassays (such as
examination of colony formation, measurement of DNA synthesis by either 3H
thymidine
incorporation or BrdU incorporation, assaying for changes in gene
transcription, or
measurement of cellular enzymes which can be used to screen for effects on
proliferation) or
biochemical assays (such as receptor phosphorylation). Those of ordinary skill
in the art are
familiar with bioassays and biochemical assays suitable for detecting cell
proliferation,
differentiation, survival and activation.
In another preferred embodiment, the invention features a method to identify
agonist
antibodies to growth factor receptors comprising the steps of an immunizing
animal with
stem/progenitor cells having surface molecules comprising the growth factor
receptors, so as
to generate a plurality of immune cells expressing one or more antibodies to
the surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding Fab fragments of the antibodies, cloning the nucleic acid
sequences from
the library into phagemid vectors so that the Fab fragments are displayed on
the surface of
phagemid, panning the Fab fragments displayed on phagemids for binding to cell
surface
molecules on target cells, dimerizing the Fab fragments that bind the cell
surface molecules,
and screening the dimerized Fab fragments to identify those that are agonist
antibodies for the
2 0 growth factor receptors.
By "Fab fragment" is meant a fragment of an antibody where heavy chain
variable
region and the light chain variable regions are contained on separate
polypeptides. The
polypeptides are covalently bound to each other by at least one disulfide
bridge. Constant
regions present may be native or from different species, also known as hybrid
or chimeric Fab.
Panning is carried out using target cells as previously described, although
Fab
fragments are generally monomers.
By "dimerizing" is meant causing the association of two antibody binding
domains
contained on antibody fragments so that binding can occur to more than one
growth factor
receptor. One way that dimerization can be achieved is by linking individual
antibody
3 0 fragments to dimerization domains such as jun (Kostelny, S.A., Cole, M.S.,
and Tso, J.Y. J.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/0133t
22
Immunol. 148:1547-1553, 1992; deKruif, 3. and Logtenberg, T. J. Biol. Chem.
271:7630-
7634, 1996), which allow the two protein fragments to stably associate through
the interaction
of these domains.
"Screening in functional assays" is carried out using target cells as
previously
described for scFv fragments. The monomeric Fab molecules that bind to target
cells are
diinerized and screened as dimers. These can be surface displayed, but are
preferably
screened as soluble molecules.
In another preferred embodiment, the invention features a method to identify
agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
stem/progenitor cells having surface molecules comprising the growth factor
receptors, so as
to generate a plurality of immune cells expressing one or more antibodies to
the surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding Fab fragments of the antibodies, cloning the nucleic acid
sequences from
the library into surface display vectors so that the Fab fragments are surface
displayed,
panning the surface displayed Fab fragments for binding to cell surface
molecules on target
cells, dimerizing the Fab fragments that bind the cell surface molecules, and
screening in a
functional assay the dimerized Fab fragments to identify those that are
agonist antibodies for
the growth factor receptors.
In a fiirther preferred embodiment the surface display vector is a
bacteriophage vector
2 0 which allows for expression of the antibody on the surface of
bacteriophage.
In other preferred embodiments of the methods to identify agonist antibodies,
the
animal is a rabbit or a chicken; the stem/progenitor cells are selected from
the group
consisting unsorted human bone marrow cells, human peripheral blood cells
originating from
human bone marrow, sorted human bone marrow cells, unsorted marine bone marrow
cells,
2 5 sorted marine bone marrow cells, fetal liver cells, yolk sac cells, cells
derived from the marine
AGM region, human or marine embryonal carcinoma cells or lines, human or mouse
pluripotential teratocarcinoma cells or lines, marine pluripotent embryonic
cells, human
embryonic stem (ES) cell lines, cells of neural origin, cells involved in
organ or tissue
regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
3 0 mononuclear cells, human bone marrow CD34+ cells, FDCP-mix marine
hematopoietic stem
CA 02316755 2000-06-27
WO 99/38008 PGT/US99/01331
23
cell line, B6SUTA marine hematopoietic stem cell line, P19 teratocarcinoma
cells, and
NTera-2 pluripotent embryonal carcinoma cells.
The choice of preferred animals is twofold: Rabbits are routinely used
successfully
for production of antibodies and are generally the first choice, provided that
species
differences with regard to the immunogen will permit generation of an immune
response.
However, chickens provide a more evolutionarily distinct environment in which
to elicit
antibodies. This is an important consideration because rabbits and mice do not
elicit an
immune response to some human antigens. However, those skilled in the art are
familiar with
other animals which are suitable for use.
l0 Cells useful in the present invention include primary cells isolated from
an animal,
cells cultured in vitro from primary cells and established cell lines that can
be grown
continually in culture.
By "unsorted human bone marrow cells" is meant samples of human bone marrow
aspirates that have undergone minimal manipulation, for example aspirates
treated to lyse red
blood cells and to remove platelets by centrifugation, such that all other
lineages are present.
By "human peripheral blood cells originating from human bone marrow" is meant
cells circulating in the peripheral blood such as mononuclear cells or CD34+
cells that were
released into the blood stream and had their origin in bone marrow.
By "sorted human bone marrow cells" is meant human bone marrow cells that have
2 0 undergone a separation process whereby cell surface markers or cell
determinants are used to
separate mixtures of cells into different populations. Sorting can be via FACS
(Fluorescent
Activated Cell Sorter) or magnetic separation using antibody cocktails,
microbeads and
magnets. Sorting can be positive selection for the cells of interest (e.g.,
CD34+ cells, which
are selected with a-CD 34 antibodies to isolate cells expressing the CD34
antigen) or negative
selection whereby undesirable cells are removed from the population (e.g., use
of a CD38
antibody to remove CD38+ cells from a given population).
Unsorted and sorted marine bone marrow cells are as described for human cells
except
that the relevant marine surface antigen applies.
By "fetal liver cells" is meant cells that have migrated from the blood
islands of the
3 0 yolk sac to the fetal liver which develop into hematopoietic cells
including embryonic
CA 02316755 2000-06-27
WO 99/38008 PCTNS99/01331
24
erythroid cells, macrophages and stem/progenitor cells. They are obtained as
described in
Jordan, C.T., McKearn, J.P., Lemischka, LR. Cell 61:953-963, 1990. In
addition, one could
use a specific antibody, AA4.1, to enrich for a subpopulation of fetal liver
tissue that includes
multipotential stem/progenitor cells (Jordan, C.T., McKearn, J.P., Lemischka,
LR. Cell
61:953-963, 1990). Such cells have as surface markers CD34 and AC 133.
By "yolk sac cells" is meant cells which are the origin of the first
hematopoietic cells
in a developing organism. Cells within the extraembryonic tissue in yolk sac
migrate from
blood islands of the yolk sac to the fetal liver where they build up to a
large population of
hematopoietic cells including embryonic erythroid cells, macrophages, stem
cells, and
progenitor cells.
By "human or marine embryonal carcinoma cells or lines" is meant pluripotent
cells
that can be derived from teratocarcinomas or by direct culture of normal
embryos that can be
cultured in vitro, are pluripotent, and can be induced to differentiate in
presence of various
inducing agents.
By "human or mouse pluripotential teratocarcinoma cells or lines" is meant
transplantable cancer cells derived from teratoma, a disorganized mass of
cells containing
many varieties of differentiated tissue, mixed with undifferentiated stem
cells that continue
to divide and generate more of the differentiate tissues. Teratocarcinoma
cells can be grown
in culture as permanent cell lines and in a suitable medium they will continue
to proliferate
2 0 indefinitely without differentiating. However, these cells are
multipotential and can be
induced to undergo multilineage differentiation. If the medium is changed by
adding an
inducer of differentiation, such as retinoic acid, or if the cells are allowed
to aggregate, the
cells can be triggered to differentiate into a variety of apparently normal
specialized cell types.
By "marine pluripotent embryonic cells" is meant cells derived from marine
blastocysts that when injected into blastocysts, the cells can colonize the
germline and
reconstitute a mouse.
By "human embryonic stem (ES) cell lines" is meant cell lines derived from
early
mammalian embryo that are totipotent and capable of in vitro proliferation
(Thomson, J.A.,
Itskovitz-Eldor, J., Sllapiro, S.S., Waknitz, M.A., Swwiergiel, J.J., Marshal,
V.S., Jones, J.M.,
CA 02316755 2001-08-03
CA 02316755 2000-06-27
WO 99(38008 PGT/US99/01331
Science 282: 1145-1147, 1998; Bongso, A., C.Y. Fong, S.C. Ng, and S. Ratnam,
Hum.
Reprod. 9: 2110, 1994).
By "cell derived from the marine AGM" is meant cells derived from an area
comprising dorsal aorta, gonads and mesonephros in mammalian embryos that
contains a high
5 level of hematopoietic stem/progenitor cells during marine embryogenesis
(Medvinsky, A.L.,
Samoylina, N.L., Muller, A.M., Dzierzak, E.A., Nalure 364:64-67, 1993;
Medvinsky, A.L.,
Gan, 0.1., Semenova, M.L., Samoylina, N.L., Blood 87:557-566, 1996).
By "cells involved in organ or tissue regeneration" is meant cells that divide
by
duplication such as kidney cells, endothelial cells that form the lining of
blood vessels, and
10 liver cells (Michalopoulous, G.K., and DeFrances, M.C. Science 276:60-66,
1997), as well
as other cell populations that are renewed by means of stem cells, such as
epidermis, bone,
skeletal muscle, and nervous tissue (McICay, R., Science 276:66-71, 1997).
By "hematopoietic cells" is meant blood forming cells.
By "cells of neural origin" is meant cells that originate from ectoderm and
develop
15 into neural tube or neural crest to ultimately form the central nervous
system and the
peripheral nervous system.
By "human bone marrow cells that have undergone RBC lysis"is meant whole bone
marrow aspirates that were depleted of the majority of red blood cells (RBCs)
and platelets
through RBC lysis and washes. This population is expected to include all other
hematopoietic
2 0 lineages.
By "human bone mononuclear cells" is meant cells in circulating blood and bone
marrow that contain a single nucleus such as monocytes, lymphocytes, and NK
cells. These
cells can be isolated from the remaining red blood cells, platelets, and
granulocytes on
Histopaque ~''~' 1077 solution of polysucrose and sodium diatrizoate adjusted
to a density of 1.077
25 g/m (SIGMA Diagnostics, St. Louis, MO) or a similar product.
By "human bone marrow CD34' cells' is meant a collection of stem/progenitor
cells
derived from human bone marrow that express the CD34 surface marker on the
cell surface.
Typically, these cells are isolated using magnetic or FAC sorting using
antibodies specific for
the CD34 molecule.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
26
By "FDCP-mix marine hematopoietic stem cell line" is meant marine cell lines
that
were cloned and isolated from long-term marine bone-marrow cultures infected
with a
recombinant of the Molony marine leukemia virus and the src oncogene of the
Rous sarcoma
virus (Spooncer, E., Heyworth, C.M., Dunn, A., Dexter, T. M. Differentiation
31: 111-118,
1986). These cell lines have many of the characteristics of hematopoietic stem
cells.
By "B6SUTA marine hematopoietic stem cell line" is meant a factor-dependent
hematopoietic cell line established from nonadherent cell populations removed
from
continuous mouse bone marrow cultures (Greenberger, J.S., Sakakeeny, M.A.,
Humphries,
R.K., Eaves, C.J. and Eckner, R.J., Proc. Natl. Acad Sci. LISA 80:2931-2935,
1983).
By "P 19 teratocarcinoma cells" is meant a cell line initiated by McBurney, et
al,
(McBurney, M.W. and Rogers, B.J., Developmental Biology 89: 503-508, 1982)
that is a
teratocarcinoma cell line derived from an embryonal carcinoma induced in a
C3H/He strain
mouse.
By "NTera-2 pluripotential embryonal carcinoma cells" is meant a pluripotent
human
embryonal carcinoma cell line which was derived from a single cell clone of
NTera-2 cells
which were established from a nude mouse xenograft tumor of Tera-2 cells
(Andrews, P.W.,
Damjanov, L, Simon, D., Banting, G.S., Carlin, C., Dracopoli, N.C. Fogh, J.,
Laboratory
Investigation 50:147-162, 1984). The Tera-2 cells were isolated from a lung
metastasis from
a human male with primary embryonal carcinoma of the testis.
2 0 In another preferred embodiment, the invention features a method to
identify agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
human bone marrow cells having surface molecules comprising the growth factor
receptors,
harvesting primary and/or secondary lymphoid organs from the animals and
isolating RNA
from the organs, creating a library from the RNA, comprising nucleic acid
sequences
2 5 encoding scFv fragments of the antibodies, cloning the nucleic acid
sequences from the library
into phagemid vectors so that the scFv fragments are displayed on the surface
of phagemid,
panning the scFv fragments displayed on phagemids for binding to cell surface
molecules on
target cells, and screening in a functional assay the scFv fragments that bind
the cell surface
molecules to identify those that are agonist antibodies for the growth factor
receptors.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
27
By "harvesting" is meant sacrificing the animals and collecting primary and/or
secondary lymphoid organs such as blood, spleen and bone marrow.
By "primary and secondary lymphoid organs" is meant to include but is not
limited
to organs such as blood, bone marrow, spleen, and lymph nodes.
By "isolating RNA" is meant lysing cells in the presence of agents that
inhibit RNase
activity, such as phenol and guanidine thiocyanate (Molecular Research Center,
Inc.,
Cincinnati, OH) and separating RNA from DNA and proteins by centrifugation or
other
methods known to those who practice the art.
In further preferred embodiments, the stem/progenitor cells are selected from
the
1 o group consisting of unsorted human bone marrow cells, human peripheral
blood cells
originating from human bone marrow, sorted human bone marrow cells, unsorted
marine bone
marrow cells, sorted marine bone marrow cells, fetal liver cells, yolk sac
cells, cells derived
from the marine AGM region, human or marine embryonal carcinoma cells or
lines, human
or mouse pluripotential teratocarcinoma cells or lines, marine pluripotent
embryonic cells,
human embryonic stem (ES) cell lines, cells of neural origin, cells involved
in organ or tissue
regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
mononuclear cells, human bone marrow CD34'' cells, FDCP-mix marine
hematopoietic stem
cell line, B6SUTA marine hematopoietic stem cell line, P19 teratocarcinoma
cells, and
NTera-2 pluripotent embryonal carcinoma cells.
2 0 In another preferred embodiment, the invention features a method to
identify agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
human bone marrow cells having surface molecules comprising the growth factor
receptors,
harvesting primary or secondary lymphoid organs from the animals and isolating
RNA from
the organs, creating a library from the RNA, comprising nucleic acid sequences
encoding Fab
2 5 fragments of the antibodies, cloning the nucleic acid sequences from the
library into phagemid
vectors so that the Fab fragments are displayed on the surface of phagemid,
panning the Fab
fragments displayed on phagemids for binding to cell surface molecules on
target cells,
dimerizing the Fab fragments that bind the cell surface molecules, and
screening in a
functional assay the dimerized Fab fragments to identify those that are
agonist antibodies for
3 0 the growth factor receptors.
CA 02316755 2000-06-27
WO 99/38008 PCTNS99/01331
28
In further preferred embodiments, the stem/progenitor cells are selected from
the
group consisting of unsorted human bone marrow cells, human peripheral blood
cells
originating from human bone marrow, sorted human bone marrow cells, unsorted
marine bone
marrow cells, sorted marine bone marrow cells, fetal liver cells, yolk sac
cells, cells derived
from the marine AGM region, human or marine embryonal carcinoma cells or
lines, human
or mouse pluripotential teratocarcinoma cells or lines, marine pluripotent
embryonic cells,
human embryonic stem (ES) cell lines, cells of neural origin, cells involved
in organ or tissue
regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
mononuclear cells, human bone marrow CD34+ cells, FDCP-mix marine
hematopoietic stem
cell line, B6SUTA marine hematopoietic stem cell line, P19 teratocarcinoma
cells, and
NTera-2 pluripotent embryonal carcinoma cells.
The invention also features a method to make agonist antibodies to growth
factor
receptors comprising the steps of immunizing an animal with stemlprogenitor
cells having
surface molecules comprising the growth factor receptors, so as to generate a
plurality of
immune cells expressing one or more antibodies to the surface molecules,
creating a library
from the plurality of immune cells, comprising nucleic acid sequences encoding
the
antibodies, cloning the nucleic acid sequences from the library into surface
display vectors
so that the antibodies are surface displayed, screening using target cells the
antibodies that are
surface displayed to identify antibodies that are agonist antibodies for the
growth factor
2 0 receptors cells, and synthesizing the agonist antibodies.
By "synthesizing the agonist antibodies" is meant introducing the nucleic acid
encoding the agonist antibody into a cell for the synthesis of the antibody
and preferably its
secretion into extracellular medium such that it can be isolated. High level
expression of
heterologous proteins has been achieved in many different systems including E.
coli,
2 5 Saccharomyces cerevisiae, Pichia pastoris, mammalian cells, plants, and
insect cells.
Engineered antibody fragments have been expressed in many of these organisms
including
bacteria (Pluckthun, A., Nature 347: 497-498, 1990), yeast (Carlson, J.R. and
Weissman, LL.,
Mol. Cell. Biol. 8:2647-2650, 1988), plants (Hiatt, A., Cafferkey, R. Bowdish,
K., Nature
342: 76-78, 1989), and mammalian cells. The incorporation of a molecular tag
such as HIS6
3 0 allows rapid and efficient purification through nickel-chetate
chromatography from the
CA 02316755 2000-06-27
WO 99/38008 PCTNS99101331
29
medium or from cell lysates (Kroiher, M., Raffioni, S., Steele, R.E., Biochim.
Biophys. Acta.
1250: 29-34, 1995; Burks, E.A. and Iverson, B.L., Biotechnol. Prog. 1 l: 112-
114, 1995).
Those who practice the art are familiar with these and other methods for
synthesis of
heterologous proteins and could readily adapt those methods to the synthesis
of antibodies.
In another preferred embodiment, the invention features a method to make
agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
stem/progenitor cells having surface molecules comprising the growth factor
receptors, so as
to generate a plurality of immune cells expressing one or more antibodies to
the surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding the antibodies, cloning the nucleic acid sequences from the
library into
viral display vectors so that the antibodies are displayed on the surface of
virus, screening
using target cells the antibodies displayed on the surface of the virus for
antibodies that are
agonist antibodies to the growth factor receptors and synthesizing the agonist
antibodies.
In a further preferred embodiment, the viral display vector is a bacteriophage
vector
or a phagemid vector.
In another preferred embodiment, the invention features a method to make
agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
stem/progenitor cells having surface molecules comprising the growth factor
receptors, so as
to generate a plurality of immune cells expressing one or more antibodies to
the surface
2 0 molecules, creating a library from the plurality of immune cells,
comprising nucleic acid
sequences encoding scFv fragments of the antibodies, cloning the nucleic acid
sequences from
the library into phagemid so that the scFv fragments are displayed on the
surface of phagemid,
panning the scFv fragments displayed on phagemids for binding to cell surface
molecules on
target cells, 'screening in a functional assay the scFv fragments that bind
the cell surface
2 5 molecules to identify those that are agonist antibodies for the growth
factor receptors and
synthesizing the agonist antibodies.
In another preferred embodiment, the invention features a method to make
agonist
antibodies to growth factor receptors comprising the steps of immunizing an
animal with
stem/progenitor cells having surface molecules comprising the growth factor
receptors, so as
3 0 to generate a plurality of immune cells expressing one or more antibodies
to the surface
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding Fab fragments of the antibodies, cloning the nucleic acid
sequences from
the library into phagemid vectors so that the Fab fragments are displayed on
the surface of
phagemid, panning the Fab fragments displayed on phagemids for binding to cell
surface
5 molecules on target cells, dimerizing the Fab fragments that bind the cell
surface molecules,
and screening in a functional assay the dimerized Fab fragments to identify
those that are
agonist antibodies for the growth factor receptors and synthesizing the
agonist antibodies.
In further preferred embodiments, in the methods to identify and to make
agonist
antibodies to growth factor receptors the library is a combinatorial library;
the target cells are
10 selected from the group consisting of unsorted human bone marrow cells,
human peripheral
blood cells originating from human bone marrow, sorted human bone marrow
cells, unsorted
marine bone marrow cells, sorted marine bone marrow cells, fetal liver cells,
yolk sac cells,
cells derived from the marine AGM region, human or marine embryonal carcinoma
cells or
lines, human or mouse pluripotential teratocarcinoma cells or lines, marine
pluripotent
15 embryonic cells, human embryonic stem (ES) cell lines, cells of neural
origin, cells involved
in organ or tissue regeneration, human bone marrow cells that have undergone
RBC lysis,
human bone marrow mononuclear cells, human bone marrow CD34+ cells, FDCP-mix
marine
hematopoietic stem cell line, B6SUTA marine hematopoietic stem cell line, P19
teratocarcinoma cells, and NTera-2 pluripotent embryonal carcinoma cells; the
screening of
2 0 the scFv fragments is by bioassay for proliferation, differentiation or
activation of target cells;
the screening of the scFv fragments is by assaying for changes in
transcription of dovvnstream
genes or phosphorylation of the receptor; the screening of the dimerized Fab
fragments is by
bioassay for proliferation, differentiation or activation of target cells; the
screening of the
dimerized Fab fragments is by assaying for changes in transcription of
downstream genes or
2 5 phosphorylation of the receptor.
By "combinatorial library" is meant a collection of nucleic acid molecules
representing the immune repertoire of an organism where rearranged heavy and
light chain
V genes are combined at random creating many artificial V gene combinations.
This
significantly expands the number of antibody molecules that can be screened.
Combinatorial
3 0 libraries can be constructed as described in "Phage Display of Peptides
and Proteins" Eds:
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
31
Brian K. Kay, Jill Winter, and John McCafferty; Chapter 6: "Construction and
Screening of
Antibody Display Libraries" by John McCafferty and Kevin S. Johnson. Academic
Press, San
Diego, CA 1996. The combinatorial libraries of the present invention represent
a collection
of nucleic acid sequences encoding the immune repertoire of an organism which
contains
nucleic acid sequences encoding antibodies or fragments thereof directed to
surface molecules
on the particular cell type utilized for immunization. The rearrangement of
heavy and light
chain V genes serves to simply expand the number of antibody molecules
directed to these
surface molecules, as artificial V gene combinations are produced, including
those directed
to cell surface molecules. This organized collection of nucleic acids has
increased use for
l0 isolating antibody molecules or fragments thereof that specifically bind to
receptors linked
to a signal transduction cascade that leads to enhancement or inhibition of
cellular
proliferation, differentiation, activation or survival, e.g. growth factor
receptors, displayed on
the cell surface of the cell type utilized for immunization.
By "bioassay for proliferation or differentiation" is meant to also include
cell survival
and activation. For example such assays include, but are not limited to
measuring Brdu
incorporation, tritiated thymidine incorporation, changes in cellular enzyme
levels such as
mitochondrial dehydrogenase, visible colony formation, pH change, Ca~
concentration
change and changes in gene transcription.
By "assaying for changes in the transcription of downstream genes" is meant
assaying
2 0 for changes in transcription of any gene that is regulated as part of a
signal transduction
cascade, e.g., nuclear transcription factors such as c-myc, c jun, NF-x(3 and
c-fos. For.
example, exposing starved (serum and growth factor deprived) target cells to
candidate
antibodies for a brief period of time (e.g., 2 hrs) and isolating RNA from the
cells. Changes
in transcription can be detected (quantified) by methods that include RT-PCR
using primers
2 5 specific for one or more downstream genes, followed by agarose gel
electrophoresis or by
hybridizing RNA to silicon wafers having bound probes for specific genes. By
utilizing such
chips many genes can be assayed for simultaneously.
By "assaying for phosphorylation of the receptor" is meant the
immunoprecipitation
of the receptor with the antibody and then providing 'y-ATP and examining
phosphorylated
3 0 products by SDS-PAGE or high-throughput screens for phosphorylation of the
receptor.
CA 02316755 2000-06-27
WO 99138008 PCT/US99101331
32
The invention also features a method to identify growth factor receptors
comprising
the steps of generating agonist antibody to the receptors, and using the
agonist antibody to
identify the receptors.
Once an agonist antibody is identified, it can be used to identify the
receptor to which
it is binding. Those of ordinary skill in the art are familiar with techniques
such as
immunoprecipitation and/or immunoaffinity purification (Springer, T.A. (1997),
"Isolation
of Proteins Using Antibodies," In Current Protocols in Immunology (J.E.
Coligan, A.M.
Kruisbeek, D.H. Margulies, E.M. Shevach, W. Strober, eds.) pp. 821-829, John
Wiley &
Sons, New York) which are useful for the identification of the receptor.
l0 In another preferred embodiment, the invention features a method to
identify growth
factor receptors comprising the steps of immunizing an animal with
stem/progenitor cells
having surface molecules comprising growth factor receptors, so as to generate
a plurality of
immune cells expressing one or more antibodies to the surface molecules,
creating a library
from the plurality of immune cells, comprising nucleic acid sequences encoding
scFv
fragments of the antibodies, cloning the nucleic acid sequences from the
library into surface
display vectors so that the scFv fragments are surface displayed, screening
using target cells
the surface displayed scFv fragments to identify those that are agonist
antibodies for the
growth factor receptors and using the agonist antibodies to identify the
receptors.
In a further preferred embodiment the surface display vector is a
bacteriophage vector
2 0 and surface display is on the surface of bacteriophage.
In another preferred embodiment, the invention features a method to identify
growth
factor receptors comprising the steps of immunizing an animal with
stem/progenitor cells
having surface molecules comprising the growth factor receptors, so as to
generate a plurality
of immune cells expressing one or more antibodies to the surface molecules,
creating a library
2 5 from the plurality of immune cells, comprising nucleic acid sequences
encoding scFv
fragments of the antibodies, cloning the nucleic acid sequences from the
library into
phagemid vectors so that the scFv fragments are displayed on the surface of
phagemid,
panning the scFv fragments displayed on phagemids for binding to cell surface
molecules on
target cells, screening in a functional assay said scFv fragments that bind
the cell surface
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
33
molecules to identify those that are agonist antibodies for the growth factor
receptors, and
using the agonist antibody to identify the receptors.
In another preferred embodiment, the invention features a method to identify
growth
factor receptors comprising the steps of immunizing an animal with
stem/progenitor cells
having surface molecules comprising the growth factor receptors, so as to
generate a plurality
of immune cells expressing one or more antibodies to the surface molecules,
creating a library
from the plurality of immune cells, comprising nucleic acid sequences encoding
scFv
fragments of the antibodies, cloning the nucleic acid sequences from the
library into surface
display vectors so that the scFv fragments are surface displayed, panning the
surface displayed
scFv fragments for binding to cell surface molecules on target cells,
screening in a functional
assay said scFv fragments that bind the cell surface molecules to identify
those that are
agonist antibodies for the growth factor receptors, and using the agonist
antibody to identify
the receptors.
In another preferred embodiment, the invention features a method to identify
growth
factor receptors comprising the steps of immunizing an animal with
stem/progenitor cells
having surface molecules comprising the growth factor receptors, so as to
generate a plurality
of immune cells expressing one or more antibodies to the surface molecules,
creating a library
from the plurality of immune cells, comprising nucleic acid sequences encoding
Fab
fragments of the antibodies, cloning the nucleic acid sequences from the
library into cell
2 0 surface display vectors so that the Fab fragments are surface displayed,
panning the surface
displayed Fab fragments for binding to cell surface molecules on target cells,
dimerizing the
Fab fragments that bind the cell surface molecules, screening in a functional
assay the
dimerized Fab fragments to identify those that are agonist antibodies for the
growth factor
receptors and using the agonist antibody to identify the receptors.
2 5 In a further preferred embodiments the surface display vector is a
bacteriophage vector
and surface display is on the surface of bacteriophage.
In another preferred embodiment, the invention features a method to identify
growth
factor receptors comprising the steps of immunizing an animal with
stem/progenitor cells
having surface molecules comprising the growth factor receptors, so as to
generate a plurality
3 0 of immune cells expressing one or more antibodies to the surface
molecules, creating a library
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
34
from the plurality of immune cells, comprising nucleic acid sequences encoding
Fab
fragments of the antibodies, cloning the nucleic acid sequences from the
library into phagemid
vectors so that the Fab fragments are displayed on the surface of phagemid,
panning the Fab
fragments displayed on phagemids for binding to cell surface molecules on
target cells,
dimerizing the Fab fragments that bind the cell surface molecules, screening
in a functional
assay the dimerized Fab fragments to identify those that are agonist
antibodies for the growth
factor receptors and using the agonist antibody to identify the receptors.
In a further preferred embodiment the growth factor receptor is a
hernatopoietic
growth factor receptor.
1 o By "hematopoietic growth factor receptor" is meant a receptor expressed on
cells of
hematopoietic origin that participates in the proliferation, differentiation,
cell survival or
functional activation of stem/progenitor or mature blood cells.
The invention fiu~ther features a method to identify growth factors comprising
the
steps of generating an agonist antibody to growth factor receptor, using the
agonist antibody
to identify the receptor, and using the receptor to identify the growth
factor.
There are a number of ways in which the native growth factor could be
identified by
using the growth factor receptor. For example, it is possible to transform
cells with the cloned
receptor under an inducible promoter for expression, followed by screening
cDNA pools for
effects in bioassay. For example, there is the use of the Cytosensor
Microphysiometer System
2 0 (Molecular Devices, Sunnyvale, CA), a biosensor to monitor receptor-
mediated responses
without previous knowledge of the signal transduction pathway. Alternatively,
it is possible
to make chimeric receptors to set up a well defined bioassay, such as
increases in specific
reporter gene expression. For example, the extracellular domain of the new
receptor could be
fused to the intracellular domain of a well characterized receptor, especially
one in which
2 5 downstream signaling events have been defined. Then, cDNA pools could be
screened in
bioassay for reporter gene expression. Those of ordinary skill in the art are
familiar with these
and other useful techniques.
In another preferred embodiment, the invention features a method to identify
growth
factors comprising the steps of immunizing an animal with stem/progenitor
cells having
3 0 surface molecules comprising the growth factor receptors, so as to
generate a plurality of
CA 02316755 2000-06-27
WO 99/38008 PGT/US99/01331
immune cells expressing one or more antibodies to the surface molecules,
creating a library
from the plurality of immune cells, comprising nucleic acid sequences encoding
scFv
fragments of the antibodies, cloning the nucleic acid sequences from the
library into surface
display vectors so that the scFv fragments are surface displayed, screening
the surface
5 displayed scFv fragments to identify those that are agonist antibodies for
the growth factor
receptors using the agonist antibody to identify the receptor, and using the
receptors to
identify the growth factor.
In a fiutller preferred embodiment, the surface display vector is a
bacteriophage vector
and surface display is on the surface of bacteriophage.
10 In another preferred embodiment, the invention features a method to
identify growth
factors comprising the steps of immunizing an animal with stem/progenitor
cells having
surface molecules comprising the growth factor receptors,. so as to generate a
plurality of
immune cells expressing one or more antibodies to the surface molecules
creating a library
from the plurality of immune cells, comprising nucleic acid sequences encoding
scFv
15 fragments of the antibodies, cloning the nucleic acid sequences from the
library into phagemid
vectors so that the scFv fragments are displayed on the surface of phagemid,
panning the scFv
fragments displayed on phagemids for binding to cell surface molecules on
target cells, and
screening in a functional assay the scFv fragments that bind the cell surface
molecules to
identify those that are agonist antibodies for the growth factor receptors,
using the agonist
2 0 antibodies to identify the receptor, and using the receptors to identify
the growth factor.
In another preferred embodiment, the invention features a method to identify
growth
factors comprising the steps of immunizing an animal with stern/progenitor
cells having
surface molecules comprising the growth factor receptors, so as to generate a
plurality of
immune cells expressing one or more antibodies to the surface molecules,
creating a library
2 5 from the plurality of immune cells, comprising nucleic acid sequences
encoding Fab
fragments of the antibodies, cloning the nucleic acid sequences from the
library into surface
display vectors so that the Fab fragments are surface displayed, panning the
surface displayed
Fab fragments for binding to cell surface molecules on target cells,
dimerizing the Fab
fragments that bind the cell surface molecules, screening in a functional
assay the dimerized
3 0 Fab fragments to identify those that are agonist antibodies for the growth
factor receptors
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
36
using the agonist antibody to identify the receptor, and using the receptors
to identify the
growth factor.
In a further preferred embodiment, the surface display vector is a
bacteriophage vector
and surface display is on the surface of bacteriophage.
In another preferred embodiment, the invention features a method to identify
growth
factors comprising the steps of immunizing an animal with stem/progenitor
cells having
surface molecules comprising the growth factor receptors, so as to generate a
plurality of
immune cells expressing one or more antibodies to the surface molecules,
creating a library
from the plurality of immune cells, comprising nucleic acid sequences encoding
Fab
fragments of the antibodies, cloning the nucleic acid sequences from the
library into phagemid
vectors so that the Fab fragments are displayed on the surface of phagemid,
panning the Fab
fragments displayed on phagemids for binding to cell surface molecules on
target cells,
dimerizing the Fab fragments that bind the cell surface molecules, screening
in a functional
assay the dimerized Fab fragments to identify those that are agonist
antibodies for the growth
factor receptors, using the agonist antibodies to identify the receptor, and
using the receptors
to identify the growth factor.
In further preferred embodiments the growth factor is a hematopoietic growth
factor,
the growth factor effects proliferation, differentiation, activation or
survival of the following
cell types: unsorted human bone marrow cells, human peripheral blood cells
originating from
2 0 human bone marrow, sorted human bone marrow cells, unsorted marine bone
marrow cells,
sorted marine bone marrow cells, fetal liver cells, yolk sac cells, cells
derived from the marine
AGM region, human or marine embryonal carcinoma cells or lines, human or mouse
pluripotential teratocarcinoma cells or lines, marine pluripotent embryonic
cells, human
embryonic stem (ES) cell lines, cells of neural origin, cells involved in
organ or tissue
2 5 regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
mononuclear cells, human bone marrow CD34+ cells, FDCP-mix marine
hematopoietic stem
cell line, B6SUTA marine hematopoietic stem cell line, P19 teratocarcinoma
cells, and
NTera-2 pluripotent embryonal carcinoma cells.
The invention also features a method for screening for agonist antibodies
comprising
3 0 the steps of growing cells expressing antibody fragments in the presence
of target cells
CA 02316755 2000-06-27
WO 99/38005 PCT/US99/01331
37
expressing receptors to which the antibodies are directed, and screening the
antibody
fragments to identify those that are agonist antibodies.
By "cells expressing antibody fragments" is meant prokaryotic or eukaryotic
cells that
carry cloned copies of antibody fragments linked to promoters and signal
sequences specific
to the host cell to enable cell surface antibody display, that may include
transmembrane
dimerization domains (Lemmon, M.A., Treutlein, H.R., Adams, P.D., Brunger,
A.T.,
Engelman, D.M. Nat. Struct. Biol. 1:157-163, 1994); or to enable secretion of
monomers and
dimers of the antibody fragments.
By "in the presence of target cells" is meant coculturing cells expressing
antibodies
or fragments and cells expressing a receptor to which the antibody binds.
Screening involves other bioassays or biochemical assays to determine if the
antibody
fragment stimulates proliferation, differentiation, activation or survival of
the target cell.
In preferred embodiments the cells expressing antibody fragments are bacterial
cells,
mammalian cells or yeast.
By "bacterial cell" is meant any prokaryotic cell, usually E. toll of any
genotype or
bacterial cells producing cell surface displayed molecules
By "mammalian cell" is meant any cell derived from mammalian origin, usually
clonal with ability to grow in vitro.
By "yeast" is meant yeast cells, typically Saccharomyces cerevisiae,
2 0 Schitzosacchamyces pombe, or Pichia pastoris that allows for the
expression of plasmids or
integrated DNA engineered to produce antibody molecules, which are surface
displayed or
secreted.
In another embodiment, the invention features agonist antibodies to growth
factor
receptors produced by immunizing an animal with stem/progenitor cells.
In preferred embodiments the stem/progenitor cells are selected from the group
unsorted human bone marrow cells, human peripheral blood cells originating
from human
bone marrow, sorted human bone marrow cells, unsorted marine bone marrow
cells, sorted
marine bone marrow cells, fetal liver cells, yolk sac cells, cells derived
from the marine AGM
region, human or marine embryonal carcinoma cells or lines, human or mouse
pluripotential
3 0 teratocarcinoma cells or Lines, marine pluripotent embryonic cells, human
embryonic stem
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
38
(ES) cell lines, cells of neural origin, cells involved in organ or tissue
regeneration, human
bone marrow cells that have undergone RBC lysis, human bone marrow mononuclear
cells,
human bone marrow CD34+ cells, FDCP-mix marine hematopoietic stem cell line,
B6SUTA
marine hematopoietic stem cell line, P19 teratocarcinoma cells, and NTera-2
pluripotent
embryonal carcinoma cells.
In another preferred embodiment, the invention features agonist antibodies to
growth
factor receptors produced by the methods of the present invention.
The invention also features combinatorial libraries produced from immune cells
generated by immunizing an animal with stem/progenitor cells. In another
embodiment the
l0 invention features combinatorial libraries encoding antibody molecules or
fragments thereof
comprising nucleic acid sequences from immune cells of animals immunized with
stem/progenitor cells. In a further embodiment, the invention features
combinatorial antibody
fragment libraries directed to surface molecules on stem/progenitor cells. In
a still further
embodiment the invention features a combinatorial library produced by
immunizing an animal
with stem/progenitor cells having surface molecules comprising receptors
involved in cell
proliferation, differentiation, survival or activation so as to generate a
plurality of immune
cells expressing one or more antibodies or antibody fragments thereof of the
surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding the antibodies or antibody fragments thereof, cloning the
nucleic acid
2 0 sequences from the library into surface display vectors so that the
antibodies are surface
displayed. In a preferred embodiment the receptor is a growth factor receptor.
In still another embodiment, the invention features a method of producing a
combinatorial antibody library encoding antibodies or fragments thereof to
receptors involved
in cell proliferation, differentiation, survival or activation comprising the
steps of immunizing
2 5 an animal with stem/progenitor cells having surface molecules comprising
the receptors, so
as to generate a plurality of immune cells expressing one or more antibodies
or antibody
fragments to the surface molecules, obtaining nucleic acid sequences encoding
variable and
constant regions of the antibody fragments, from the plurality of immune
cells, randomly
combining nucleic acid sequences encoding variable regions of the antibody
fragments to
3 0 produce a combinatorial library of nucleic acid sequences encoding the
antibody fragments,
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
39
and cloning the nucleic acid sequences of the library into surface display
vectors so that the
antibody fragments are surface displayed. In a preferred embodiment the
receptor is a growth
factor receptor.
By "randomly combining" is meant combining nucleic acid sequences encoding
rearranged heavy and light chain variable genes to create many artificial
variable gene
combinations, e.g., by using a PCR overlap reaction.
In preferred embodiments, the stem/progenitor cells are selected from the
group
consisting of unsorted human bone marrow cells, human peripheral blood cells
originating
from human bone marrow, sorted human bone marrow cells, unsorted marine bone
marrow
l0 cells, sorted marine bone marrow cells, fetal liver cells, yolk sac cells,
cells derived from the
marine AGM region, human or marine embryonal carcinoma cells or lines, human
or mouse
pluripotential teratocarcinoma cells or lines, marine pluripotent embryonic
cells, human
embryonic stem (ES) cell lines, cells of neural origin, cells involved in
organ or tissue
regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
mononuclear cells, human bone marrow CD34+ cells, FDCP-mix marine
hematopoietic stem
cell line, B6SUTA marine hematopoietic stem cell line, P19 teratocarcinoma
cells, and
NTera-2 pluripotent embryonal carcinoma cells.; the animal is a rabbit or a
chicken; the
antibody or fragments thereof is a scFv fragment; the antibody or fragments
thereof is a Fab
fragment; the antibodies or fragments thereof encoded by the combinatorial
library are surface
2 0 displayed on phagemeids; the method of producing further includes the step
of screening
serum obtained from the immunized animal for binding to the stem/progenitor
cell, prior to
obtaining nucleic acid sequences encoding the antibody fragments.
The invention also features various equivalent aspects directed toward the
identification, synthesis and use of antibodies that are inhibitory to the
proliferation,
2 5 differentiation, activation or survival of cells. These aspects are
similar to those concerning
agonist antibodies and one of ordinary skill in the art would be able to adapt
these so as to
apply to inhibitory antibodies.
Furthermore, the invention features a method to identify inhibitory antibodies
involved in cellular proliferation, differentiation or activation comprising
the steps of
3 o immunizing an animal with stem/progenitor cells having receptors involved
in cellular
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
proliferation, differentiation or activation so as to generate a plurality of
immune cells
expressing one or more antibodies to the surface molecules, creating a library
from the
plurality of immune cells, comprising nucleic acid sequences encoding the
antibodies, cloning
the nucleic acid sequences from the library into surface display vectors so
that the antibodies
5 are surface displayed, and screening the surface displayed antibodies for
those that inhibit
proliferation, differentiation or activation of target cells, so as to
identify inhibitory antibodies.
By "inhibitory antibody" is meant entire antibodies or fragments thereof,
including
Fab fragments and scFv fragments, etc., that bind to a cell surface receptor
that is linked to
a signal transduction cascade that is involved in cellular proliferation,
differentiation,
10 activation, inhibition or survival and inhibits the proliferation,
differentiation, activation or
survival of a cell. Inhibitory antibodies could inhibit by blocking the action
of a receptor that
promotes proliferation, differentiation, activation or survival of the cell or
by mimicking an
inhibitory molecule that binds to a receptor that sends a negative growth
signal.
By "proliferation" is meant active division and progression through the
various stages
15 of the cell cycle as detected by changes in the rate of protein synthesis,
chromosome
replication, cell size or cell number.
By "differentiation" is meant changes in cell morphology, behavior or function
that
lead to the production of different types of cells with specialized functions,
as a result of
exposure to extrinsic factors, or changes in gene expression. Differentiation
refers to the
2 0 process by which cells mature and become less pluripotent, as that term is
used in the art.
By "activation" is meant the process by which cells leave Go and enter G,, but
do not
synthesize DNA or divide until a second signal is received. Activation is
associated with the
expression of a specific set of activation genes and activation antigens.
By "survival" is meant that cells do not undergo apoptosis or programmed cell
death
2 5 or necrosis.
Receptors involved in proliferation, differentiation, activation or survival
could
represent the same receptors that bind agonist antibodies if the inhibitory
antibody is acting
as an antagonist or blocking antibody. The receptor may be different from the
growth factor
receptor that agonist antibodies bind if the inhibitory antibody is mimicking
an inhibitory
3 0 factor.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
41
In another preferred embodiment, the invention features a method to identify
inhibitory antibodies for cellular proliferation, differentiation or
activation comprising the
steps of immunizing an animal with stem/progenitor cells having surface
molecules
comprising receptors involved in cellular proliferation, differentiation or
activation, so as to
generate a plurality of immune cells expressing one or more antibodies to the
surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding the antibodies, cloning the nucleic acid sequences from the
library into
viral display vectors so that the antibodies are displayed on the surface of
virus, and screening
the antibodies displayed on the surface of the virus for those that inhibit
proliferation,
1 o differentiation or activation of target cells so as to identify inhibitory
antibodies.
In a further preferred embodiment the viral vector is selected from the group
consisting of bacteriophage and phagemid vectors.
In another preferred embodiment, the invention features a method to identify
inhibitory antibodies for cellular proliferation, differentiation or
activation comprising the
steps of immunizing an animal with stem/progenitor cells having surface
molecules
comprising receptors involved in cellular proliferation differentiation or
activation, so as to
generate a plurality of immune cells expressing one or more antibodies to the
surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding scFv fragments of the antibodies, cloning the nucleic acid
sequences from
2 o the library into phagemid vectors so that the scFv fragments are displayed
on the surface of
phagemid, and screening the scFv fragments displayed on the surface of the
phagemid for
those that inhibit proliferation, differentiation or activation of target
cells so as to identify
inhibitory antibodies.
In another preferred embodiment, the invention features a method to identify
2 5 inhibitory antibodies for cellular proliferation, differentiation or
activation comprising the
steps of immunizing an animal with stem/progenitor cells having surface
molecules
comprising receptors involved in cellular proliferation, differentiation or
activation; so as to
generate a plurality of immune cells expressing one or more antibodies to the
surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
3 0 sequences encoding scFv fragments of the antibodies, cloning the nucleic
acid sequences from
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
42
the library into surface display vectors so that the scFv fragments are
surface displayed, and
screening the 'surface displayed scFv fragments for those that inhibit
proliferation,
differentiation or activation of target cells so as to identify inhibitory
antibodies.
In a further preferred embodiment the surface display vector is a
bacteriophage vector
and surface display is on the surface of bacteriophage.
In another preferred embodiment, the invention features a method to identity
inhibitory antibodies for cellular proliferation, differentiation or
activation comprising the
steps of immunizing an animal with stem/ progenitor cells having surface
molecules
comprising receptors involved in cellular proliferation, differentiation or
activation, so as to
generate a plurality of immune cells expressing one or more antibodies to the
surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
sequences encoding scFv fragments of the antibodies, cloning the nucleic acid
sequences from
the library into phagemid vectors so that the scFv fragments are displayed on
the surface of
phagemid, panning the scFv fragments displayed on phagemids for binding to
cell surface
molecules on target cells, and screening in a functional assay the scFv
fragments that bind the
cell surface molecules for those that inhibit proliferation, differentiation
or activation of target
cells so as to identify inhibitory antibodies.
Panning and screening are carried out as previously described.
In another preferred embodiment, the invention features a method to identity
2 0 inhibitory antibodies for cellular proliferation, differentiation or
activation comprising the
steps of immunizing an animal with stem/ progenitor cells having surface
molecules
comprising receptors involved in cellular proliferation, differentiation or
activation, so as to
generate a plurality of immune cells expressing one or more antibodies to the
surface
molecules, creating a library from the plurality of immune cells, comprising
nucleic acid
2 5 sequences encoding scFv fragments of the antibodies, cloning the nucleic
acid sequences from
the library into surface display vectors so that the scFv fragments are
surface displayed,
panning the surface displayed scFv fragments for binding to cell surface
molecules on target
cells, and screening in a functional assay the scFv fragments that bind the
cell surface
molecules for those that inhibit proliferation, differentiation or activation
of target cells so as
3 0 to identify inhibitory antibodies.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
43
In a further preferred embodiment the surface display vector is a
bacteriophage vector
and surface display is on the surface of bacteriophage.
In another preferred embodiment the invention features a method to identify
inhibitory
antibodies for cellular proliferation, differentiation or activation
comprising the steps of
immunizing an animal with stem/progenitor cells having surface molecules
comprising
receptors involved in cellular proliferation, differentiation or activation,
so as to generate a
plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from the plurality of immune cells, comprising nucleic acid
sequences encoding Fab
fragments of the antibodies, cloning the nucleic acid sequences from the
library into phagemid
vectors so that the Fab fragments are displayed on the surface of phagemid,
panning the Fab
fragments displayed on phagemids for binding to cell surface molecules on
target cells,
dimerizing Fab fragments the bind cell surface molecules, and screening in a
functional assay
as both monomer and dimerized the Fab fragments that bind the cell surface
molecules for
those that inhibit proliferation, differentiation or activation of target
cells so as to identify
inhibitory antibodies.
Panning and screening are carried out as with scFv fragments.
By "dimerizing Fab fragments" is meant that a representative portion of the
Fab
fragments that bind to cell surface molecules are dimerized. The remainder of
the Fab
fragments are left as monomers. Both monomers and dimers are tested in the
functional
2 0 assays.
By "screening in a functional assay as both monomer and dimerized Fab
fragments"
is meant that monomers and dimers are independently screened (see discussion
on scFV
fragments). In addition the Fab fragments (monomers and dimers) may be either
surface
displayed or soluble molecules.
2 5 In another preferred embodiment, the invention features a method to
identify
inhibitory antibodies for cellular proliferation, differentiation or
activation comprising the
steps of immunizing an animal with stem/progenitor cells having surface
molecules
comprising receptors involved in cellular proliferation, differentiation or
activation, so as to
generate a plurality of immune cells expressing one or more antibodies to the
surface
3 0 molecules, creating a library from the plurality of immune cells,
comprising nucleic acid
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
44
sequences encoding Fab fragments of the antibodies, cloning the nucleic acid
sequences from
the library into surface display vectors so that the Fab fragments are surface
displayed,
panning the surface displayed Fab fragments for binding to cell surface
molecules on target
cells, dimerizing Fab fragments that bind said cell surface molecules, and
screening in a
functional assay both monomers and dimerized Fab fragments bind to cell
surface molecules
on target cells for those that inhibit proliferation, differentiation or
activation of target cells
so as to identify inhibitory antibodies.
In a further preferred embodiment the surface display vector is a
bacteriophage vector
and surface display is on the surface of bacteriophage.
In still further preferred embodiments of the methods to identify inhibitory
antibodies
the animal is a rabbit or a chicken; the stem/progenitor cells are selected
from the group
consisting of unsorted human bone marrow cells, human peripheral blood cells
originating
from human bone marrow, sorted human bone marrow cells, unsorted marine bone
marrow
cells, sorted marine bone marrow cells, fetal liver cells, yolk sac cells,
cells derived from the
marine AGM region, human or marine embryonal carcinoma cells or lines, human
or mouse
pluripotential teratocarcinoma cells or lines, marine pluripotent embryonic
cells, human
embryonic stem (ES) cell lines, cells of neural origin, cells involved in
organ or tissue
regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
mononuclear cells, human bone marrow CD34+ cells, FDCP-mix marine
hematopoietic stem
2 0 cell line, B6SUTA marine hematopoietic stem cell line, P 19
teratocarcinoma cells, and
NTera-2 pluripotent embryonal carcinoma cells.
In another preferred embodiment the invention features a method to identify
inhibitory
antibodies for cellular proliferation, differentiation or activation
comprising the steps of
immunizing an animal with stem/progenitor cells having surface molecules
comprising
2 5 receptors involved in cellular proliferation differentiation or
activation, harvesting primary
or secondary lymphoid organs from the animals and isolating RNA from the
organs, creating
a library from the RNA, comprising nucleic acid sequences encoding scFv
fragments of the
antibodies, cloning the nucleic acid sequences from the library into phagemid
vectors so that
the scFv fragments are displayed on the surface of phagemid, panning the scFv
fragments
3 0 displayed on phagemids for binding to cell surface molecules on target
cells, and screening
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
in a functional assay the scFv fragments that bind the cell surface molecules
for those that
inhibit proliferation, differentiation or activation of target cells so as to
identify inhibitory
antibodies.
In a further preferred embodiment the stem/progenitor cells are selected from
the
5 group consisting of unsorted human bone marrow cells, human peripheral blood
cells
originating from human bone marrow, sorted human bone marrow cells, unsorted
marine bone
marrow cells, sorted marine bone marrow cells, fetal liver cells, yolk sac
cells, cells derived
from the marine AGM region, human or marine embryonal carcinoma cells or
lines, human
or mouse pluripotential teratocarcinoma cells or lines, marine pluripotent
embryonic cells,
10 human embryonic stem (ES) cell lines, cells of neural origin, cells
involved in organ or tissue
regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
mononuclear cells, human bone marrow CD34+ cells, FDCP-mix marine
hematopoietic stem
cell line, B6SUTA marine hematopoietic stem cell line, P19 teratocarcinoma
cells, and
NTera-2 pluripotent embryonal carcinoma cells.
15 In another preferred embodiment the invention features a method to identify
inhibitory
antibodies for cellular proliferation, differentiation or activation
comprising the steps of
immunizing an animal with stem/progenitor cells having surface molecules
comprising
receptors involved in cellular proliferation differentiation or activation,
harvesting primary
or secondary lymphoid organs from the animals and isolating RNA from the
organs, creating
2 0 a library from the RNA, comprising nucleic acid sequences encoding Fab
fragments of the
antibodies, cloning the nucleic acid sequences from the library into phagemid
vectors so that
the Fab fragments are displayed on the surface of phagemid, panning the Fab
fragments
displayed on phagemids for binding to cell surface molecules on target cells,
dimerizing Fab
fragments that bind said cell surface molecules, and screening in a functional
assay both
2 5 monomers and dimerized Fab fragments that bind said cell surface molecules
for those that
inhibit proliferation, differentiation or activation of target cells so as to
identify inhibitory
antibodies.
In a further preferred embodiment the stem/progenitor cells are selected from
the
group consisting of unsorted human bone marrow cells, human peripheral blood
cells
3 0 originating from human bone marrow, sorted human bone marrow cells,
unsorted marine bone
CA 02316755 2000-06-27
WO 99/38008 PGT/US99/01331
46
marrow cells, sorted marine bone marrow cells, fetal liver cells, yolk sac
cells, cells derived
from the marine AGM region, human or marine embryonal carcinoma cells or
lines, human
or mouse pluripotential teratocarcinoma cells or lines, marine pluripotent
embryonic cells,
human embryonic stem (ES) cell lines, cells of neural origin, cells involved
in organ or tissue
regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
mononuclear cells, human bone marrow CD34+ cells, FDCP-mix marine
hematopoietic stem
cell line, B6SUTA marine hematopoietic stem cell line, P 19 teratocarcinoma
cells, and
NTera-2 pluripotent embryonal carcinoma cells.
The invention also features a method to make inhibitory antibodies for
cellular
proliferation, differentiation or activation comprising the steps of
immunizing an animal with
stem/progenitor cells having surface molecules comprising receptors involved
in cellular
proliferation, differentiation or activation so as to generate a plurality of
immune cells
expressing one or more antibodies to the surface molecules, creating a library
from the
plurality of immune cells, comprising nucleic acid sequences encoding the
antibodies, cloning
the nucleic acid sequences from the library into surface display vectors so
that the antibodies
are surface displayed, screening the surface displayed antibodies for those
that inhibit
proliferation, differentiation or activation of target cells so as to identify
inhibitory antibodies,
and synthesizing the inhibitory antibodies.
Synthesis of inhibitory antibodies is carried out as previously described for
agonist
2 0 antibodies.
In another preferred embodiment, the invention features a method to make
inhibitory
antibodies for cellular proliferation, differentiation or activation
comprising the steps of
immunizing an animal with stem/progenitor cells having surface molecules
comprising
receptors involved in cellular proliferation, differentiation or activation so
as to generate a
2 5 plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from the plurality of immune cells, comprising nucleic acid
sequences encoding the
antibodies, cloning the nucleic acid sequences from the library into viral
display vectors so
that the antibodies are displayed on the surface of virus, screening the
antibodies displayed
on the surface of the virus for those that inhibit growth, proliferation,
differentiation or
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
47
activation of target cells so as to identify inhibitory antibodies, and
synthesizing the inhibitory
antibodies.
In a further preferred embodiment the viral vector is a bacteriophage vector
or a
phagemid vector.
In another preferred embodiment the invention features a method to make
inhibitory
antibodies for cellular proliferation, differentiation or activation
comprising the steps of
immunizing an animal with stem/progenitor cells having surface molecules
comprising
receptors involved in cellular proliferation, differentiation or activation,
so as to generate a
plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from the plurality of immune cells, comprising nucleic acid
sequences encoding scFv
fragments of the antibodies, cloning the nucleic acid sequences from the
library into phagemid
so that the scFv fragments are displayed on the surface of phagemid, panning
the scFv
fragments displayed on phagemids for binding to cell surface molecules on
target cells,
screening by functional assay the scFv fragments that bind the cell surface
molecules for those
that inhibit proliferation, differentiation or activation of target cells so
as to identify inhibitory
antibodies, and synthesizing the inhibitory antibodies.
in another preferred embodiment the invention features a method to make
inhibitory
antibodies for cellular proliferation, differentiation or activation
comprising the steps of
immunizing an animal with stem or progenitor cells having surface molecules
comprising
2 0 receptors involved in cellular proliferation, differentiation or
activation, so as to generate a
plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from said plurality of immune cells, comprising nucleic acid
sequences encoding Fab
fragments of the antibodies, cloning said nucleic acid sequences from the
library into
phagemid vectors so that the Fab fragments are displayed on the surface of
phagemid, panning
2 5 the Fab fragments displayed on phagemids for binding to cell surface
molecules on target
cells, dimerizing Fab fragments that bind the cell surface molecules, and
screening by
functional assays both monomers and dimerized Fab fragments that bind the cell
surface
molecules for those that inhibit proliferation, differentiation or activation
of target cells so as
to identify inhibitory antibodies, and synthesizing the inhibitory antibodies.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
48
In further preferred embodiments in the methods to identify inhibitory
antibodies of
claims the library is a combinatorial library; the target cells are selected
from the group
consisting of unsorted human bone marrow cells, human peripheral blood cells
originating
from human bone marrow, sorted human bone marrow cells, unsorted marine bone
marrow
cells, sorted marine bone marrow cells, fetal liver cells, yolk sac cells,
cells derived from the
marine AGM region, human or marine embryonal carcinoma cells or lines, human
or mouse
pluripotential teratocarcinoma cells or lines, marine pluripotent embryonic
cells, human
embryonic stem (ES) cell lines, cells of neural origin, cells involved in
organ or tissue
regeneration, human bone marrow cells that have undergone RBC lysis, human
bone marrow
mononuclear cells, human bone marrow CD34' cells, FDCP-mix marine
hematopoietic stem
cell line, B6SUTA marine hematopoietic stem cell line, P19 teratocarcinoma
cells, and
NTera-2 pluripotent embryonal carcinoma cells; the screening by functional
assay of the scFv
fragments is by bioassay for the inhibition of proliferation, differentiation
or activation of
target cells; the screening by functional assay of the dimerized Fab fragments
is by bioassay
for the inhibition of proliferation, differentiation or activation of target
cells.
In another embodiment the invention features a method to identify receptors
involved
in cellular proliferation, differentiation or activation comprising the steps
of generating an
inhibitory. antibody to the receptors, and using the inhibitory antibody to
identify the receptor.
Identification of the receptor using the inhibitory antibody to the receptor
is carried
2 0 out as previously described for agonist antibodies.
In another preferred embodiment, the invention features a method to identify
receptors
involved in cellular proliferation, differentiation or activation comprising
the steps of
immunizing an animal with stem/progenitor cells having surface molecules
comprising
receptors involved in cellular proliferation, differentiation or activation,
so as to generate a
2 5 plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from the plurality of immune cells, comprising nucleic acid
sequences encoding scFv
fragments of the antibodies, cloning the nucleic acid sequences from the
library into surface
display vectors so that the scFv fragments are surface displayed, screening
the surface
displayed scFv fragments for those that inhibit proliferation, differentiation
or activation of
CA 02316755 2000-06-27
WO 99138008 PC1'/US99/01331
49
target cells so as to identify inhibitory antibodies, and using the inhibitory
antibodies to
identify the receptors.
In a further preferred embodiment, the surface display vector is a
bacteriophage vector
and surface display is on the surface of a bacteriophage.
In another preferred embodiment the invention features a method to identify
receptors
involved in cellular proliferation, differentiation or activation comprising
the steps of
immunizing an animal with stem/progenitor cells having surface molecules
receptors involved
in cellular proliferation, differentiation or activation, so as to generate a
plurality of immune
cells expressing one or more antibodies to the surface molecules, creating a
library from the
1 o plurality of immune cells, comprising nucleic acid sequences encoding scFv
fragments of the
antibodies, cloning the nucleic acid sequences from the library into phagemid
vectors so that
the scFv fragments are displayed on the surface of phagemid, panning the scFv
fragments
displayed on phagemids for binding to cell surface molecules on target cells,
screening in a
functional assay the scFv fragments that bind said cell surface molecules for
those that inhibit
proliferation, differentiation, or activation of target cells sb as to
identify inhibitory antibodies,
and using the inhibitory antibody to identify the receptors.
In another preferred embodiment, the invention features a method to identify
receptors
involved in cellular proliferation, differentiation or activation comprising
the steps of
immunizing an animal with stem/progenitor cells having surface molecules
comprising
2 0 receptors involved in cellular proliferation differentiation or
activation, so as to generate a
plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from the plurality of immune cells, comprising nucleic acid
sequences encoding Fab
fragments of the antibodies, cloning the nucleic acid sequences from the
library into surface
display vectors so that the Fab fragments are displayed on the surface of
cells, panning the
2 5 Fab fragments displayed on the surface of cells for binding to cell
surface molecules on target
cells, dimerizing Fab fragments that bind the cell surface molecules,
screening in a functional
assay both monomer and dimerized Fab fragments for those that inhibit
proliferation,
differentiation or activation of target cells so as to identify inhibitory
antibodies, and using
the inhibitory antibody to identify said receptors.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
In a further preferred embodiment, the surface display vector is a
bacteriophage vector
and surface display is on the surface of a bacteriophage.
In another preferred embodiment, the invention features a method to identify
receptors
involved in cellular proliferation, differentiation or activation comprising
the steps of
5 immunizing an animal with stem/progenitor cells having surface molecules
comprising
receptors involved in cellular proliferation differentiation or activation, so
as to generate a
plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from the plurality of immune cells, comprising nucleic acid
sequences encoding Fab
fragments of the antibodies, cloning the nucleic acid sequences from the
library into phagemid
1 o vectors so that the Fab fragments are displayed on the surface of
phagemid, panning the Fab
fragments displayed on phagemids for binding to cell surface molecules on
target cells,
dimerizing Fab fragments that bind said cell surface molecules, screening in a
functional assay
both monomer and dimerized Fab fragments for those that inhibit proliferation,
differentiation
or activation of target cells so as to identify inhibitory antibodies, and
using the inhibitory
15 antibody to identify the receptors.
In a further preferred embodiment of the methods to identify the receptor
involved in
cellular proliferation, differentiation or activation, the receptor is a
hematopoietic receptor.
The invention further features a method to identify inhibitory factors to
cellular
proliferation, differentiation or activation comprising the steps of
generating inhibitory
2 0 antibodies to receptors involved in cellular proliferation,
differentiation or activation, using
the inhibitory antibody to identify the receptor, and using the receptors to
identify the
inhibitory factor.
The receptor is used to identify the inhibitory factor as previously described
for
identification of growth factors.
2 5 In another preferred embodiment, the invention features a method to
identify
inhibitory factors to cellular proliferation, differentiation or activation
comprising the steps
of immunizing an animal with stem/progenitor cells having surface molecules
comprising
receptors involved in cellular proliferation, differentiation or activation,
so as to generate a
plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
3 0 a library from the plurality of immune cells, comprising nucleic acid
sequences encoding scFv
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
51
fragments of the antibodies, cloning the nucleic acid sequences from the
library into surface
display vectors so that the scFv fragments are surface displayed on the
surface of cells,
screening the surface displayed scFv fragments for those that inhibit
proliferation,
differentiation or activation of target cells so as to identify inhibitory
antibodies, using the
inhibitory antibody to identify the receptor, and using the receptors to
identify the inhibitory
factor.
In a further preferred embodiment, the surface display vector is a
bacteriophage vector
and surface display is on the surface of a bacteriophage.
In another preferred embodiment, the invention features a method to identify
1 o inhibitory factors to cellular proliferation, differentiation or
activation comprising the steps
of immunizing an animal with stem/progenitor cells having surface molecules
comprising
receptors involved in cellular proliferation, differentiation or activation,
so as to generate a
plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from the plurality of immune cells, comprising nucleic acid
sequences encoding scFv
fragments of the antibodies, cloning the nucleic acid sequences from the
library into phagemid
vectors so that said scFv fragments are displayed on the surface of phagemid,
panning the
scFv fragments displayed on phagemids for binding to cell surface molecules on
target cells,
and screening in a functional assay the scFv fragments that bind the cell
surface molecules for
those that inhibit proliferation, differentiation or activation of target
cells so as to identify
2 0 inhibitory antibodies, using the inhibitory antibodies to identify the
receptor, and using the
receptors to identify the inhibitory factor.
In another preferred embodiment, the invention features a method to identify
inhibitory factors comprising the steps of immunizing an animal with
stem/progenitor cells
having surface molecules comprising receptors involved in cellular
proliferation,
2 5 differentiation or activation, so as to generate a plurality of immune
cells expressing one or
more antibodies to the surface molecules, creating a library from the
plurality of immune
cells, comprising nucleic acid sequences encoding Fab fragments of the
antibodies, cloning
the nucleic acid sequences from the library into surface display vectors so
that the Fab
fragments are surface displayed, panning the surface displayed Fab fragments
for binding to
3 0 cell surface molecules on target cells, dimerizing Fab fragments that bind
the cell surface
CA 02316755 2000-06-27
WO 99/38008 PCTIUS99/01331
52
molecules, screening in a functional assay both monomer and dimerized Fab
fragments for
those that inhibit proliferation, differentiation or activation of target
cells so as to identify
inhibitory antibodies, using the inhibitory antibody to identify the receptor,
and using the
receptors to identify the inhibitory factor.
In a further preferred embodiment, the surface display vector is a
bacteriophage vector
and surface display is on the surface of a bacteriophage.
In another preferred embodiment, the invention features a method to identify
inhibitory factors to cellular proliferation, differentiation or activation
comprising the steps
of immunizing an animal with stem/progenitor cells having surface molecules
comprising
receptors involved in cellular proliferation, differentiation or activation,
so as to generate a
plurality of immune cells expressing one or more antibodies to the surface
molecules, creating
a library from the plurality of immune cells, comprising nucleic acid
sequences encoding Fab
fragments of the antibodies, cloning the nucleic acid sequences from the
library into phagemid
vectors so that Fab fragments are displayed on the surface of phagemid,
panning the Fab
fragments displayed on phagemids for binding to cell surface molecules on
target cells,
dimerizing Fab fragments that bind the cell surface molecules, screening in a
functional assay
both monomer and the dimerized Fab fragments for those that inhibit
proliferation,
differentiation or activation of target cells so as to identify inhibitory
antibodies, using the
inhibitory antibodies to identify the receptor, and using the receptors to
identify the inhibitory
2 0 factor.
In a further preferred embodiment of the methods to identify inhibitory
factors;
inhibitory factor is a hematopoietic factor.
In another embodiment, the invention features a method for screening for
inhibitory
antibodies for cellular proliferation, differentiation, activation or survival
comprising the steps
2 5 of growing cells expressing antibody fragments in the presence of target
cells expressing
receptors to which said antibodies are directed, and screening the antibody
fragments for those
that inhibit proliferation, differentiation, activation or survival of target
cells so as to identify
inhibitory antibodies.
In further preferred embodiments the cells expressing antibody fragments are
selected
3 0 from the group consisting of bacterial cells, mammalian cells and yeast.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
53
Also, the invention features inhibitory antibodies to receptors involved in
proliferation, differentiation, activation or survival made by immunizing an
animal with
stem/progenitor cells.
In preferred embodiments the stem/progenitor cells selected from the group
consisting
of unsorted human bone marrow cells, human peripheral blood cells originating
from human
bone marrow, sorted human bone marrow cells, unsorted marine bone marrow
cells, sorted
marine bone marrow cells, fetal liver cells, yolk sac cells, cells derived
from the marine AGM
region, human or marine embryonal carcinoma cells or lines, human or mouse
pluripotential
teratocarcinoma cells or lines, marine pluripotent embryonic cells, human
embryonic stem
(ES) cell lines, cells of neural origin, cells involved in organ or tissue
regeneration, human
bone marrow cells that have undergone RBC lysis, human bone marrow mononuclear
cells,
human bone marrow CD34+ cells, FDCP-mix marine hematopoietic stem cell line,
B6SUTA
marine hematopoietic stem cell line, P19 teratocarcinoma cells, and NTera-2
pluripotent
embryonal carcinoma cells.
In further preferred embodiments the invention features inhibitory antibodies
to
receptors involved in proliferation, differentiation, activation or survival
made by any of the
methods of making inhibitory antibodies.
In addition, the invention features a method for treating a patient having a
disease or
disorder characterized by a deficiency in a cell population comprising the
step of
2 0 administering to the patient a therapeutically effective amount of an
agonist antibody
produced by immunizing an animal with stem/progenitor cells that stimulates
the proliferation
or differentiation of cell of the population.
By "disease or disorder characterized by a deficiency in a cell population" is
meant
for example diseases, disorders or treatment related to suppression of
hematopoiesis where
2 5 less than the normal number of cells of a given lineage or lineages are
present in a patient.
For example, patients undergoing chemotherapy show subnormal levels of
neutrophils and
platelets. Anemias are examples of subnormal levels of red blood cells.
By "administering" is meant provision of an antibody.
By "therapeutically effective amount" is meant an amount which at least
partially
3 o alleviates or abrogates some of the symptoms associated with the disease
or condition.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
54
In a preferred embodiment, the cell population is an hematopoietic cell
population.
In another preferred embodiment, the invention features a method for treating
a patient
having a disease or disorder characterized by a deficiency in a cell
population comprising the
step of contacting ex vivo patient's cells with an agonist antibody produced
by immunizing
an animal with stem/progenitor cells that stimulates the proliferation or
dii~erentiation of the
cells of the population.
By "ex vivo" is meant outside the body. Those of ordinary skill in the art are
familiar
with techniques for the isolation of cells from a patient, conditions for the
maintenance of
cells outside a patient and the reintroduction of cells into a patient.
In a preferred embodiment, the cell population is an hematopoietic cell
population.
In another embodiment, the invention features a method for treating a patient
having
a disease or disorder characterized by an increase in a cell population
comprising the step of
administering to the patient a therapeutically effective amount of an
inhibitory antibody
produced by immunizing an animal with stem/progenitor cells that inhibits the
proliferation
or differentiation of the cell population.
By "disease or disorder characterized by an increase in a cell population" is
meant
diseases or disorders where greater than the normal number of cells of a given
lineage or
lineages are present in a patient. For example, chronic myeloid leukemia is a
disease
characterized by myeloid proliferation due the presence of a chromosomal
translocation in the
2 0 hematopoietic stem cells. Thrombocytopenia is a disease characterized by
an increase in the
numbers of megakaryocytes.
In a preferred embodiment, the cell population is an hematopoietic cell
population.
In a further embodiment, the invention features a method for treating a
patient having
a disease or disorder characterized by cells exhibiting abnormal growth
comprising the steps
2 5 of administering to the patient an amount of an inhibitory antibody that
inhibits the
proliferation or differentiation of normal cells and that was produced by
immunizing an
animal with stem/progenitor cells, and killing the cells exhibiting abnormal
growth.
By "cells exhibiting abnormal growth" is meant cells that do not exhibit
growth
control, such as cancer cells.
CA 02316755 2000-06-27
WO 99/38008 PCTNS99/01331
By "killing" is meant subjecting the cells that exhibit abnormal growth to an
agent that
results in cell death, such as radiation or a chemotherapeutic agents. Those
of ordinary skill
in the art are familiar with the administration of such agents to patients.
Killing can be done
in vivo or ex vivo.
5 In a preferred embodiment, the cells are hematopoietic cells.
In a further embodiment, the invention features a method for treating a
patient having
a disease or disorder characterized by cells exhibiting abnormal growth
comprising the steps
of administering to the patient an amount of an antibody that specifically
binds to the cells and
is linked to a toxin or radiolabel and that was produced by immunizing an
animal with
1 o stem/progenitor cells, such that the antibody binds the cells exhibiting
abnormal growth and
results in their death.
The antibodies of the present invention can be coupled to a toxin protein to
form a
fusion immunotoxin that can be used to treat malignancies or other disease
states. Toxic
protein that can be linked to antibody binding domains include diphtheria
toxin, Pseudomonas
15 exotoxin A, among others (see Hertler, A.A., Frankel, A.E., J. Clin. Oncol.
7:1932-42, 1989).
Those of skill in the art are familiar with the construction of such fusion
immunotoxins.
The invention also provides a method for identifying cells comprising the step
of
contacting cells with an antibody specific for the cell type produced by
immunizing an animal
with stem/progenitor cells under conditions were the antibody specifically
binds the cells, and
2 0 detecting binding of the antibody.
By "specifically binds" is meant binds to an antigenic determinant is specific
for a
particular cell type.
In preferred embodiments, the cells are hematopoietic cells; the antibody is
linked to
a radiolabel.
2 5 Those of skill in the art are familiar with the use of antibodies as
imaging agents (see
Breitz, H.B., Tyler, A., Bjorn, M.J., Lesley, T., Weiden, P.L., Clin. Nucl.
Med. 22:b15-620,
1997 and Li Destri, G., Greco, S., Rinzivillo, C., Racalbuto, A., Curreri, R.,
Di Cataldo, A.,
Surg. Today 28:1233-1236, 1998).
In another embodiment, the invention features a method for isolating a
population of
3 0 specific cells comprising the step of contacting a sample potentially
containing the specific
CA 02316755 2001-08-03
CA 02316755 2000~06-27
WO 99/38008 PCT/US99/01331
56
cells with an antibody that specifically binds to the population of cells
produced by
immunizing an animal with stem/progenitor cells under conditions werc the
antibody
specifically binds the population of cells, and using the bound antibody to
isolate the
population of cells.
By "using the bound antibody to isolate" is meant using the identified
antibodies in
cell isolation strategies such as fluorescence-activated cell sorting (FACS),
or magnetic
sorting procedures. In FACS sorting, the antibody is often conjugated to
fluorescent
molecules. In magnetic sorting procedures, the antibody is linked directly or
indirectly to
magnetic microbeads.
In a preferred embodiment, the cells are hematopoietic cells.
In a further embodiment, the invention features a method for amplifying a
population
of cells comprising the step of contacting the population of cells with an
agonist antibody that
stimulates proliferation or differentiation of the cells and that was produced
by immunizing
an animal with stem/progenitor cells.
Hy "amplifying" is meant causing the proliferation of a cell or cells.
In a preferred embodiment, the cells are hematopoietic cells.
In another embodiment the invention provides a method of modulating activity
of a
receptor involved in cellular proliferation, differentiation, survival or
activation which is
present on the surface of a stem/progenitor cell by binding to the receptor an
antibody or
fragment thereof produced by immunizing an animal with the stem/progenitor
cells.
By "modulating is meant stimulating, inhibiting, or blocking the activation of
the
receptor.
In a further preferred embodiment, the receptor is a growth factor receptor.
Other~features and advantages of the invention will be apparent from the
following
description of the preferred embodiments thereof, and from the claims.
BRIEF DESCRIPTION OF THE FIGURES
3 o Fig. 1 is a diagrammatic representation of a vector used for phage
display.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
57
Figs. 2A and B are diagrammatic representations of the primers utilized in a
scFv
cloning scheme.
Figs. 3A, 3B, 3C and 3D are diagrammatic representations of the primers used
in a
Fab cloning scheme.
Fig. 4 is a diagrammatic representation of a ScFv cloning scheme.
Figs. SA and SB are diagrammatic representations of vectors used in a Fab
cloning
scheme.
DESCRIPTION OF THE PREFERRED EMBODIMENTS
The following examples are provided for further illustrating various aspects
and
embodiments of the present invention and are in no way intended to be limiting
in scope.
Those in the art would appreciate that the examples concerning hematopoietic
cells are
applicable to the screening of other factors that participate in the
proliferation, differentiation,
activation and survival of cells of other systems. In addition, those of
ordinary skill in the art
appreciate and would be readily able to adapt these methods to identify
inhibitory antibodies,
receptors and inhibitory factors.
Example 1: Screening for agonist antibodies for hematopoietic growth factor
receptors
Immunization
2 0 Chickens and rabbits are immunized with each of the following separately:
human
bone marrow cells (sorted and unsorted ) (Poietic Technologies, Germantown,
MD), marine
bone marrow cells(sorted and unsorted) (Rabbit & Rodent Diagnostic Associates,
San Diego,
CA), fetal liver cells (obtained as described in the Jordan, C.T., McKearn,
J.P., Lemischka,
LR. Cell 61:953-963, 1990). In addition, one could use a specific antibody,
AA4.1, to enrich
for the stem cells, yolk sac cells (Rabbit & Rodent Diagnostic Associates, San
Diego, CA),
mononuclear cell preparations (Poietic Technologies, Germantown, MD).
Immunization can
also be with marine hematopoietic cell lines such as FDCP-Mix (Spooncer, E.,
Heyworth,
C.M., Dunn. A., Dexter, T.M. Differentiation 31:111-118, 1986) and B6SUTA
(Greenberger, J.S., Sakakeeny, M.A., Humphries, R.K., Eaves, C.J. Eckner, R.J.
Proc. Natl.
Acad. Sci. USA 80: 2931-2935, 1983). Also, hematopoietic stem/progenitor cells
isolated
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
58
from circulating blood, as previously described. In addition, immunization can
be with other
cells such as P 19 cells, NTera-2 cells and human ES cell lines. Sorting of
bone marrow cells
is carried out via magnetic sorting using antibodies (e.g.. a-CD 34) attached
to magnetic
beads. The cells are precoated with monoclonal antibodies that recognize cell
surface
molecules. The monoclonal antibodies are attached to magnetic beads through
secondary
antibodies conjugated to the beads. The cells are then removed with a magnet.
Magnetic
sorting can be positive selection where the cells of interest are bound by the
antibody (e.g.,
stem cell populations bound to antibody) (Miltenyi Biotec, Auburn, CA) or
negative sorting
where undesired cells are captured by the magnet (Stem Cell Technologies,
Vancouver BC,
Canada).
In fluorescence activated sorting, cells tagged with antibodies conjugated to
fluorescent molecules are sorted electronically on a flow cytometer such as
Becton-Dickinson
FACS IV or FACS vantage cytometer or an equivalent sorting machine. The
fluorescent
molecule conjugated antibodies recognize specific cell surface antigens. The
antibodies are
conjugated to fluorescent markers such as fluorescein isothiocyanate (FITC) or
Phycoerythrin
(PE) (Becton-Dickinson, San Jose, CA; Biosource International, Camarillo,
CA.).
Immunization can be by any acceptable route including but not limited to
intraperitoneal, subcutaneous, intramuscular, intravenous. Immunizing, can be
in the presence
or absence of adjuvant. Generally 1-10 million cells are injected at primary
immunization.
2 0 Animals immunized broadly, such as with bone marrow preparations where
RBCs are lysed
and platelets are removed have all of the remaining lineages present,
including
stem/progenitor cells. By using these types of bone marrow preparations, the
response to the
antigen should be very broad with antibodies generated against a wide variety
of surface
receptors. Animals immunized with a sorted population, e.g. CD34+cells, will
generate less
2 5 diversity in the immune response, but may facilitate identification of an
antibody with effects
specific to stem cells or to one lineage.
Following primary immunization, animals are boosted, one or more times, with
the
same immunogens to generate secondary immune responses. Libraries generated
from
secondary immune responses should result in antibody fragments with higher
affinities, due
3 0 to affinity maturation.
CA 02316755 2000-06-27
WO 99/38008 PGTNS99/01331
59
At preimmunization and following booster immunizations, blood is collected and
serum is prepared for evaluation of serum antibodies. Indirect cellular ELISAs
with
preimmune and immune serum against target cells are used to screen for
antibodies against
uncharacterized cellular antigens. In these whole cell ELISAs, target cells (1
x 10' cells/ml)
are dispensed into microtiter plates (1 x 106 cells/well), and pelleted gently
(1500rpm/4°C/5').
Culture supernatant are removed by aspiration. Cells are washed briefly in
wash buffer
(PBS/1% BSA/0.1% NaN3) and pelleted. Cells are resuspended in 100 ul solutions
containing
serum dilutions at 1/50, 1/100, 1/500, 1/1000, 1/5000. Cells and antibodies
are incubated 1
hour at 4°C. Samples are again pelleted gently. Supernatant is removed
by aspiration and
cells are washed twice in wash buffer. Pellets are resuspended in 100 ul of
2° antibody-
enzyme conjugate (secondary goat anti-rabbit or goat anti-chicken antibody
congugated to
alkaline phosphatase or horse radish peroxidase). Plates are incubated at
4°C/lhr and cells are
then washed twice in wash buffer. Pellets are resuspended in developer
reagent, 100 uUwell.
Developer is PNPP (Sigma, St. Louis, MO) for alkaline phosphatase secondary
antibody or
ABTS (Sigma, St. Louis, MO} for horseradish peroxidase antibody. The enzyme
reaction is
allowed to proceed and read visually or on microtiter plate reader at
appropriate wavelength.
Only immunizations that produce a positive serum ELISA test are carried to the
next stage
of the method (library construction).
2 0 Library construction
Within one week after final boost, following a positive serum ELISA test,
animals are
harvested and primary and secondary lymphoid organs (spleen, bone marrow, and
blood) are
rescued. Peripheral blood lymphocytes are isolated from blood on a Ficoll or
Percol cushion
(Sigma, St. Louis, MO). RNA is isolated from spleen, bone marrow, and
peripheral blood
2 5 lymphocytes by a phenol/guanidine thiocyanate procedure (Tri Reagent,
Molecular Research
Center, Cincinnati, OH). RNA is reverse transcribed to cDNA using oligo(dT},5
primers,
deoxynucleotides, and AMV Reverse Transcriptase using a first strand cDNA
synthesis kit
(Boehringer Mannheim, Indianapolis, IN).
Fab fragment and/or single chain variable region (scFv) expression libraries
are
3 0 constructed from each immunized animal. The surface display vector for
each library is pRL4
(see Figure 1), which enables display of chimeric expression products on the
surface of
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
packaged phagemid particles. pRL4 is a modified version of pComb3H (Barbas,
C.F. III and
Burton, D. R. 1994. Monoclonal Antibodies from Combinatorial Libraries. Cold
Spring
Harbor Laboratory Course Manual, Cold Spring Harbor, N.Y.; Burton, D.R.;
Barbas, C.F. III.
Advances in Immunology 57:191-280, 1994; Lang, LM., Chuang, T.L., Barbas, C.F.
3rd,
5 Schleef, R.R. J. Biol. Chem. 271: 30126-30135, 1996) It was constructed by
PCR
amplification with primers NPCAMB-F 1 (CACCATGGCGCATACCCGTACGACGTT
CCGGACTACGCTTCTTAGGAGGGTGGTGGCTCT, SEQ. ID. NO. 1) and NPC3AMB-B
(GCTTACAATTTCCCAGATCTGCG, SEQ. ID. NO. 2) and template plasmid pComb3H
(Barbas, C.F. III and Burton, D.R, Cold Spring Laboratory Course Manual,
Monoclonal
10 Antibodies from Combinatorial Libraries" 1994). This amplifies a fragment
containing
hemagglutinin decapeptide (HA-dp) (Field, J., Nikawa, J.L, Broek, D.,
MacDonald, B.,
Rodgers, L., Wilson, LA., Lerner, R.A., Wigler, M. Mol. Cell. BioL 8:2159-
2165, 1988), an
amber stop colon (TAG) and gene III. The resulting PCR product was used as the
template
in a second PCR reaction with primers NPC3AMB-F2 (GAGGAGGAGGAGGA
15 GGAGACTAGTGGCCAGGCCGGCCAGCACCATCACCATCACCATGGCGCATACC
CGT, SEQ. ID. NO. 3) and NPC3AMB-B {SEQ. ID. NO. 2), adding a HIS6 sequence
and SfiI
and SpeI restriction sites to the 5' end. The final PCR product was cloned
into vector pComb
3H and confirmed by sequence analysis, and functional expression. In pRL4,
antibody
fragments are cloned into the SfiI restriction site.
20 In pRL4, the single chain antibody fragments are cloned downstream of the
E. coli
lacZ promoter, ribosome binding site, and omp A leader sequence. These
elements allow
induction of expression by IPTG, and secretion to the periplasm via the Omp A
leader
sequence.
In the final hybrid Fab expression construct in pRL4, the light and heavy
chains are
2 5 cloned as a single SfiI fragment. In this way, the light chain fragments
are cloned downstream
of the E. coli lacZ promoter, ribosome binding site, and omp A leader
sequence. These
elements allow induction of expression by IPTG, and secretion out of the cell
via the omp A
leader sequence. The light chain fi~agments are followed by sequences provided
by the PCR
primers and include a stop colon, a second ribosome binding site, and the E.
coli pel B leader
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
61
sequence. Hybrid heavy chain genes are fused in frame with filamentous phage
gene III (gIII)
sequences (amino acids 230-406). An amber stop codon is present at the fusion
junction. In
a sup E bacterial host such as ER2357 (New England Biolabs, Beverly, MA), the
amber
mutation is suppressed. Upon promoter induction, a single polycistronic
message is
transcribed and translated as two polypeptides, a light chain and a heavy
chain-gene III fusion
protein. Following synthesis the polypeptides are transported to the bacterial
periplasmic
space as directed by the leader sequences. In the periplasmic space the heavy
chain-pIII
fusion proteins are inserted into the membrane, and the light and heavy chains
are associated
covalently through disulfide bonds, forming the antigen binding sites. The
human constant
region CH l and C~, sequences include the cysteines that form the disulfide
bond between
heavy and light chains. Upon superinfection with helper phage, these fragments
are exported
out of the cell on the surface of phage as Fab-pIII fusions. In a non-sup E
host, such as
TOP10F' (Invitrogen, Carlsbad, CA), the amber stop codon is recognized
yielding soluble Fab
fragments: Other features of pRL4 include two molecular tags, HA and His6. The
HA tag
is recognized by HA.11 antibody (Babco, Berkeley, CA). The His6 tag allows
affinity
purification of antibody fragments by Nickel-chelate chromatography (Quiagen,
Santa Carita,
CA).
ScFv fray, m~ tints
2 0 Single chain variable regions libraries (scFv)are constructed from each
immunized
animal. Single chain libraries are useful because the entire binding domain is
contained on
one polypeptide. The light chain variable region is separated from heavy chain
variable
region by a linker region. The use of short linkers (< 11 amino acids) favors
a dimeric
complex where VH of one ScFv associates with VL of another ScFv molecule and
visa versa,
2 5 these molecules are termed diabodies (Kortt, A.A., Malky, R.L., Caldwell,
J.B., Gruen, L.C.,
Ivanci, N., Lawerence, M.G. et al. Eur. J. Biochem. 221:151-157, 1994). This
is because
folding of monomeric ScFv is impaired with linkers < 11 amino acids (Alfthan,
K., Takkinen,
K., Sizman, D., Soderlund, H., and Teeri, T.T. Protein-Eng. 8:725-731, 1995).
Longer linkers
(> 11 amino acids) favors folding of monomeric ScFv into a single antigen
binding domain,
3 0 thus precluding dimer formation. In the present example scFv fragments are
constructed with
short linkers, 7 amino acids in length, or long linkers, 18 amino acids in
length.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
62
The design of pRL4 allows for dimerization of scFv antigen binding domains on
the
phage surface and in soluble form as detailed below. When the plasmid is
transformed into
a supE bacterial host such as ER2537 (F' Sup E, New England Biolabs, Beverly,
MA), the
amber mutation is suppressed approximately fifty percent of the time. In this
way half of the
expressed scFvs are fused with the filamentous phage gene III protein (amino
acids 230-406)
and the other half will be terminated just prior to gene III to produce
soluble scFv. Both the
scFv-pIII fusion and soluble scFv products have the Omp A signal sequence and
will be
transported to the periplasm where they will be able to form dimeric scFv
complexes, termed
diabodies (Kortt, A.A., Malby, R.L., Caldwell, J.B., Gruen, L.C., Ivanci, N.,
Lawrence, M.C.
et al. Eur. J Biochem. 221: 151-157, 1994). Diabodies are expected to fold
such that the VH
of one scFv will pair with the V,, of a second scFv-pIII resulting in divalent
antibody
fragments. Upon superinfection with helper phage, these diabodies are exported
out of the
cell on the surface of phage as pIII-antibody fragments. In a non-sup E host,
such as TOP 1 OF'
(InVitrogen, Carlsbad, CA), the amber stop codon is recognized yielding
soluble scFv
diabodies.
Figure 2A & B is a diagrammatic representation of the primers utilized in a
scFv
cloning scheme. All immunoglobulin variable regions are composed of
hypervariable
regions, also known as complementarity determining regions (CDRs) proposed to
form the
antigen binding site, and less variable regions, known as framework regions
(FR). Oligos are
2 0 designed such that they will anneal to and amplify all framework regions
of published
antibodies of a given organism. Antibody sequences from many organisms have
been
compiled in a single source: Kabat, E.A., Wu, T.T., Perry, H.M., Gottesman,
K.S., and
Foeller, C. 1991. Sequences of Proteins of Immunological Interest. Vol. 1-3.
U.S. Department
of Health and Human Services, Public Health Service, National Institutes of
Health, NIH
Publication No. 91-3242. Sets of primers for amplification of the entire
immunoglobulin
repertoire of various animals have been developed and used successfully as
described (Barbas,
C.F. III, and Burton, D.R. 1994. Monoclonal Antibodies from Combinatorial
Libraries. Cold
Spring Harbor Laboratory Course Manual). These primers include restriction
sites for
cloning, and a linker sequence between the light and heavy chain variable
regions that is used
3 0 in PCR overlap extension, and to provide the amino acids to link the light
and heavy chain
CA 02316755 2000-06-27
WO 99/38008 PCTNS99/01331
63
variable regions into a single polypeptide. Linker sequences can be short (for
example coding
for 7 amino acids) or long (for example coding for 18 amino acids). Rabbit
variable light
chain "forward" primers bind to a sequence within framework 1 (FRl) of a
specific light chain
family or families. Degeneracies are incorporated into the primers such that a
single primer
will amplify other closely related FR1 variable region sequences. Restriction
sites are
incorporated in the 5' end of each primer to facilitate cloning. Similarly,
there are "reverse"
primers for rabbit variable light chain sequences each annealing with a
specific family or
families of rabbit light chain FR4 regions. A restriction site and a linker
sequence is
incorporated in the 3' end of each of the "reverse" primers. For a single
chain short library,
l0 the sequence is 5'-GGAAGAAGAGGAACC -3' (SEQ. ID. NO. 34). For the rabbit
heavy
chain variable region fragments, the sequences binding to FR regions are
designed similarly,
except primers are designed to anneal to published rabbit heavy chain
sequences (Kabat, E.A.,
et al. 1991 ). A restriction site and linker sequence are incorporated in the
5' end of each of
these primers for use during the second round of PCR amplification. The linker
sequence
incorporated in the heavy chain variable region primers is the reverse
complement of the light
chain linker sequence 5'-GGTGGTTCGTCTAGATCTTCC-3'(SEQ. ID. NO. 35). Briefly,
variable heavy (VH) and light (V~) chains are amplified separately, then
combined for an
additional round of PCR amplification by overlap extension through the linker
region to
generate scFv products. This cloning step also permits random association of
heavy and light
2 0 chains. The entire products are amplified using oligos that bind to the
extreme S' and 3' ends
of the resulting annealed products. The final PCR products, each containing
one VH region
and one V~ region separated by a short linker(? amino acids), is cloned into
pRL4 (Figure 1).
In order to construct scFv long libraries, the method is the same except that
the sequence of
the linker incorporated into the light chain reverse primers is extended by 33
nucleotides,
2 5 coding for an additional 11 amino acids. In this case, the sequence
CCCACCACCGCCC
GAGCCACCGCCACCAGAGGA (SEQ. ID. NO. 36) is included just 3' of the nucleotides
that encode the 7 amino acid short linker. The heavy chain primers for scFv
long libraries are
identical to those used in scFv short libraries. One of ordinary skill in the
art can extrapolate
from these designs and design other primers to amplify large immunoglobulin
repertoire
3 0 libraries from many different species.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
64
Heavy and light chain variable region genes are amplified separately by PCR
using
cDNA as the template. Primers used will vary depending on the animal
immunized.
Construction of scFv libraries from rabbit is as follows (see Fig. 2): light
chain variable region
genes are amplified from first strand cDNA using forward Kappa primers RSCVK-1
(GGGCCCAGGCGGCCGAGCTCGTGMTGACCCAGACTCCA, SEQ. ID. N0.4),
RSCVK-2 (GGGCCC AGGCGGCCGAGCTCGATMTGACCCAGACTCCA, SEQ. ID. NO.
5), RSCVK-3 (GGGCCCAGGCGGCCGAGCTCGTGATGACCCAGACTGAA, SEQ. ID.
NO. 6), and forward lambda primer RSClambdal RSCL-1 (GGGCCCAGGC
GGCCGAGCTCGTGCTGACTCAGTCGCCCTC, SEQ. ID. NO. 7) (GibcoBRL,
Gaithersburg, MD) (see Fig. 2A). These primers bind to framework 1 (FRl )
sequence of the
different light chain families (Kabat, E.A. et al. 1991 ) and add restriction
enzyme sites 5' of
the variable sequences for cloning purposes. Reverse primers for Kappa light
chain
amplification are RKB9J0-B (GGAAGATCTAGAGGAACCACCTAGGATCTCCAGC
TCGGTCCC, SEQ. ID. NO. 8), RKB9J10-B (GGAAGATCTAGAGGAACCACCTT
TGATTTCCACATTGGTGCC, SEQ. ID. NO. 9) RKB42J0-B (GGAAGATCTAGAGG
AACCACCTTTGACSACCACCTCGGTCCC, SEQ. ID. NO. 10), and reverse primer for
lambda light chain amplification is Rjlambda0-B (GGAAGATCTAGAGGAACCAC
CGCCTGTGACGGTCAGCTGGGTCCC, SEQ. ID. NO. 11) (GibcoBRL, Gaithersburg,
MD) (see Fig. 2A). These primers bind to FR4 sequences of the kappa and lambda
light chain
2 0 families, and include an extension sequence that is used to generate the
linker sequence by
PCR overlap extension. In the light chain reactions, each Kappa forward primer
is paired with
each Kappa reverse primer in nine separate reactions. The tenth light chain
reaction uses the
two lambda primers. Heavy chain variable region genes are amplified using
forward primers
RSCVHO1 (GGTGGTTCCTCTAGATCTTCCCAGTCGGTGGAGGAGTCCRGG, SEQ. ID.
NO. 12), RSCVH02 (GGTGGTTCC TCTAGATCTTCCCAGTCGGTGAAGGAGTCCGAG,
SEQ. ID. NO. 13), RSCVH03 (GGTGGTTCCTCTAGATCTTCCCAGTCGYTGGAG
GAGTCCGGG, SEQ. ID. NO. 14), and RSCVH04 (GGTGGTTCCTCTAGATC
TTCCCAGSAGCAGCTGRTGGAGTCCGG, SEQ. ID. NO. 15) (GibcoBRL, Gaithersburg,
MD) {see Fig. 2B). These oligonucleotides bind to variable region framework 1
sequence
3 0 and include a 5' extension used to generate linker sequence by overlap PCR
extension. The
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
reverse primer for heavy chain amplification is RSCG-B (CCTGGCCGGCCTGGC
CACTAGTGACTGAYGGAGCCTTAGGTTGCCC, SEQ. ID. NO. 16) (GibcoBRL,
Gaithersburg, MD) (see Fig. 2B). RSCG-B binds to rabbit constant FR4/CHl
sequence and
carries an extension of restriction sites to facilitate cloning. In the heavy
chain PCR reactions
5 each forward primer is paired with the reverse primer for four separate
heavy chain reactions.
Following heavy and light chain amplification and gel purification of
fragments, all
light chain and heavy chain fragments are combined and an additional round of
PCR
amplification allows for overlap extension. In this final overlap PCR
reaction, light chain
variable region fragments and heavy chain variable region fragments are
annealed, and
1 o amplified. This PCR reaction uses primers RSC-F (GAGGAGGAGGAG
GAGGAGGCGGGGCCCAGGCGGCCGAGCTC, SEQ. ID. NO. 17) (GibcoBRL,
Gaithersburg, MD) (see Fig. 2A) and RSC-B {GAGGAGGAGGAGGAGGAGCCT
GGCCGGCCTGGCCACTAGTG, SEQ. ID. NO. 18) (Gibco/BRL, Gaithersburg, MD) (see
Fig. 2B) which bind to restriction sites engineered onto light and heavy chain
variable region
15 sequences. This cloning step also permits random association of heavy and
light chains. The
PCR products are restricted with Sfi I. The final Sfi I restricted fragments,
each containing
one heavy chain variable region and one light chain variable region separated
by a linker, is
cloned into the Sfi I site of pRL4 (see Fig. 4). Ligation is transformed into
E. coli ER2537
genotype: F'proA+B+lacIq/O(lacZ)M15/ fhuA2(tonA)~(lac-proAB) supE thi-1
~(hsdMS-
2 0 mcrB)5 and ampicillin resistant (AmpR) colonies are selected on a fraction
of all the
transformants. The remainder of the transformants are amplified in liquid
culture. Colony
counts are used to determine colony size. To confirm that antibody fragments
have been
successfully cloned in plasmids, random analysis of individual clones by
restriction digest to
check for appropriately sized inserts is performed. A single library of scFv
fragments will
2 5 contain greater than 10' individual members.
In the final single chain expression construct in pRL4, the single chain
antibody
fragments are cloned downstream of the E. coli lacZ promoter, ribosome binding
site, and
omp A leader sequence. These elements allow induction of expression by IPTG,
and secretion
out of the cell via the omp A leader sequence when expressed in the suppressor
strain ER2537.
3 0 The single chain fragments are fused in frame with filamentous phage gene
III (gIII)
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
66
sequences (amino acids 230-406). The gIII protein product, pIII, is a minor
coat protein
necessary for infectivity. Upon promoter induction by IPTG, the single chain
antibody-pIII
fusion is synthesized and transported to the bacterial periplasmic space. In
the periplasmic
space, the scFv-gene III fusion proteins are inserted into the membrane. Upon
superinfection
with helper phage, these fragments are exported out of the cell on the surface
of phage as pIII-
antibody fragments. Other possible proteins to be used for fusion on the
surface of phagemids
include filamentous coat protein pVIII and other coat proteins.
For chicken scFv libraries, the method is essentially the same, but the
primers used are
specific to chicken light and heavy chain sequences (Kabat, E.A. et al., 1991
). The primer
extension sequences and the linker sequences are the same as the rabbit
primers to facilitate
cloning into pRL4. Based on the exemplified primers and the sequences
disclosed in Kabat,
E.A. et al., those of ordinary skill in the art could readily design primers
for any species and
construct scFv libraries.
Fab fragments
Fab fragment libraries, that maintain the native antigen recognition site, are
useful to
ensure that affinity is maintained.
Primers are designed based on published FR and constant region sequences
{Kabat,
E.A., et al. 1991). In the current scheme primers are used for hybrid antibody
fragment
2 0 construction (rabbit variable/human constant), however, primers can be
designed similarly for
constant regions from any species. For example, Fab fragment libraries derived
from rabbit
could have both variable and constant regions from rabbit. In this case, the
variable region
forward primers would remain the same, but reverse primers would contain
constant region
sequence from either rabbit kappa light chain, rabbit lambda light chain or
rabbit heavy chain
2 5 sequence. Hybrid Fab libraries can also be constructed. These libraries
are composed of
variable regions from immunized animals and constant regions (C~ and CH1) from
human
IgGI. For hybrid Fab, each Fab fragment will be composed of variable regions
from the
immunized animals and constant regions (CL and CHl) from human IgGl. Sets of
primers for
amplification of the immunoglobulin repertoire of rabbits and chickens, and
for generation
3 0 of chimeric Fabs, have been developed (Barbas, C.F. III, and Burton, D.R.
1994. Monoclonal
Antibodies from Combinatorial Libraries. Cold Spring Harbor Laboratory Course
Manual,
CA 02316755 2000-06-27
WO 99/38008 PGT/US99/01331
67
and November 1997 course on Monoclonal Antibodies from Combinatorial
Libraries, Cold
Spring Harbor Laboratory). "Forward" primers are paired with "reverse" primers
to PCR
amplify the rabbit variable kappa sequences, and one forward primer is paired
with one
reverse primer for lambda light chain variable region sequence amplification.
The human
constant kappa sequence is amplified from a phage display plasmid carrying a
human
antibody Fab (Persson, M.A., Caothien, R.H., and Burton, D.A. Proc. Natl.
Acad. Sci. U.S.A.
88:2432-2436, 1991 ) using a "forward" primer that anneals to published human
kappa
constant region sequence (Kabat, E.A., et al. 1991), and a reverse primer that
anneals to pelB
leader sequence in the phage display plasmid. An overlap PCR reaction where
all light chain
fragments are pooled with the human constant kappa fragment generates hybrid
light chain
fragments. A similar approach is used for the heavy chain to generate hybrid
heavy chain
fragments. In the final and third round of PCR reactions, the hybrid light
chains are pooled
with the hybrid heavy chains and overlap PCR amplification is performed with
external
primers to generate fragments, each carrying both hybrid light chain and
hybrid heavy chain
sequences.
Heavy and light chain variable region genes are amplified by PCR using cDNA as
the
template. For rabbit Fab fragment libraries, construction is as follows: light
chain variable
region genes are amplified using the forward primers RSCVKI (SEQ. ID. NO. 4),
RSCVK2
(SEQ. ID. NO. S), RSCVK3 {SEQ. ID. NO. 6) (Operon Technologies, Alameda, CA)
(see
Fig. 3A). These primers bind to framework I (FRl) sequence of the different
Kappa light
chain families (Kabat, E.A. et al., 1991 ) and incorporate restriction enzyme
sites S' of the
variable sequences for cloning purposes. Design and utilization of rabbit
primers for the
amplification of mRNAs encoding rabbit kappa light and gamma heavy chains for
the
construction of an antibody library from this species is described in: Lang,
LM., Barbas, C.F.
2 5 3rd, Schleef, R.R. Gene 172: 295-298, 1996. The lambda light chains are
amplified with the
forward primer RSCL-1 (SEQ. ID. NO. 7). Reverse primers for Kappa light chain
amplification are RHHybKI-B (AGATGGTGCAGCCACAGTTCGTTTGATTT
CCACATTGGTGCC, SEQ. ID. NO. 19), RHHybK2-B (AGATGGTGCAGCCAC
AGTTCGTAGGATCTCCAGCTCGGTCCC, SEQ. ID. NO. 20), RHHybK3-B
{AGATGGTGCAGCCACAGTTGCTTTGACSACCACCTCGGTCCC, SEQ. ID. NO. 21)
CA 02316755 2000-06-27
WO 99138008 PCT/US99/01331
68
(Operon Technologies, Alameda, CA) (see Fig. 3A). The reverse primer for the
lambda light
chains is RHHybL-B (AGATGGTGCAGCCACAGTTCGGCCTGTGACGGT
CAGCTGGGTCCC, SEQ. ID. NO. 22). These primers bind to FR4 sequence of the
light
chain families (Kabat, E.A. et al., 1991 ), and include an extension of human
constant region
sequence. The extension enables human kappa chain constant region sequences to
be added
in a second round of PCR reactions. In the light chain reactions, each kappa
forward primer
is paired with each kappa reverse primer in nine separate PCR reactions. The
tenth light chain
PCR reaction uses the lambda forward and reverse primers. The human constant
kappa
sequences can be amplified from any human kappa chain clone. In this case, the
forward
primer, HKC-F (CGAACTGT GGCTGCACCATCTGTC, SEQ. ID. NO. 23) (see Fig. 3B)
binds to human constant kappa sequence as published in Kabat, E.A. et al.,
1991. The reverse
primer, lead B (GGCCATGGCTGGTTGGGCAGC, SEQ. ID. NO. 24), binds to plasmid
sequences of a human kappa chain cloned into pComb3H. Alternatively, a primer
may be
designed to incorporate the sequences (e.g. the stop codon, the ribosome
binding site, and the
pelB leader sequence) necessary for expression in E.coli. Following gel
purification of the
fragments, a second PCR reaction is performed where all variable region PCR
fragments are
combined with the human constant kappa PCR fragments and primers RSC-F (SEQ.
ID. NO.
17) (see Fig. 3A) and lead B (SEQ. ID. NO. 24). This reaction enables fusions
of light chain
variable regions to human constant kappa sequences.
Heavy chain variable region genes are amplified using the forward primers
RhyVHI
(GCTGCCCAACCAGCCATGGCCCAGTCGGTGGAGGAGTCCRGG, SEQ. ID. NO. 25),
RhyVH2 (GCTGCCCAACCAGCCATGGCCCAGTCG GTGAAGGAGTCCGAG, SEQ. ID.
NO. 26), RhyVH3 (GCTGCCCAACCAGC CATGGCCCAGTCGYTGGAGGAGTCCGGG,
SEQ. ID. NO. 27), and RhyVH4 (GCTGCCCAACCAGCCATGGCCCAGSAGCAG
2 5 CTGRTGGAGTCCGG, SEQ. ID. NO. 28) as described in Lang, LM., Barbas, C.F.
3'~,
Schleef, R.R, Gene 172: 295-8, 1996 (see Fig. 3C). These primers include 5'
sequences that
encode the pel B leader sequence. The rabbit heavy chain reverse primer,
RHHyIgGCHl-B
(CGATGGGCCCTTGGTGGAGGCTGARGAGAYGGTGACCAGGGTGCC, SEQ. ID. NO.
29) binds to rabbit constant FR4 sequence and has an extension of human
constant CH1
3 0 sequence. Therefore, four separate PCR reactions are performed with each
forward primer
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
69
and the reverse primer. The human constant IgG sequence can be amplified by
PCR using
a human antibody clone in pCOMB3H as the template and a forward primer that
anneals to
human constant IgG sequence, HIgGCHl-F (GCCTCCACCAAGGGCCCATCGGTC, SEQ.
ID. NO. 30) and a reverse primer, dp-seq (AGAAGCGTAGTCCGGAACGTC, SEQ. ID.
NO. 31) that anneals to plasmid sequences (Fig. 3). Alternatively, the human
sequence can
be amplified from any human antibody cDNA library using primers that anneal to
constant
region sequences as published in Kabat, E.A. et al., 1991 and incorporate a
restriction site on
the reverse primer far cloning purposes. Following gel purification of the
fragments, a second
set of heavy chain PCR reactions is performed where all heavy chain variable
region
amplified fragments are combined with the human heavy chain constant region
fragment. An
overlap PCR extension reaction using primers lead VH (GCTGCCCAACCAGCCATGGCC,
SEQ. ID. NO. 32) and dp-ex (GAGGAG GAGGAGGAGGAGAGAAGCGTAGTC
CGGAACGTC, SEQ. ID. NO. 33) (see Fig. 3D) enables fusions of heavy chain
variable
regions to human constant region sequences. In the final overlap PCR reaction,
hybrid light
chains and hybrid heavy chains are annealed, and amplified using primers RSC-F
(SEQ. ID.
NO. 17) and dp-ex (SEQ. ID. NO. 33). The resulting fragment is restricted with
appropriate
enzymes, gel-purified and cloned as a single Sfrl fragment containing both
hybrid light and
heavy chains into Sfil digested pRL4. Figure 5 is a diagrammatic
representation of a Fab
cloning scheme illustrating the final hybrid Fab fragments cloned into pRL4.
Ligations are
2 0 transformed into E. coli ER2537 and ampicillin resistant (AmpR) colonies
are selected on a
fraction of all of the transformation. The remainder is amplified in liquid
culture. Colony
counts are used to determine library size.
For chicken Fab fragment libraries, the method is essentially the same, but
the primers
used are specific to chicken light and heavy chain sequences (Kabat, E.A. et
al., 1991 ). The
2 5 primer extension sequences are the same as the rabbit primers to
facilitate cloning into pRL4
and to generate the hybrid Fab fusions.
After libraries are constructed they are screened using target cells.
Tar eg t cells
3 0 Target cells are selected based on the immunogen that was used, the
desired product,
and the goal of the screen. For very broad range screens, it is important that
the majority of
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
hematopoietic cell lineages are present in the immunizing population of cells.
For example,
immunizing with a population consisting of mononuclear cells, expected to
include stem cells,
progenitor cells, lymphocytes, monocytes, and NK cells would be fairly broad
and has the
potential to generate agonist antibodies or inhibitory antibodies to stem
cells, progenitor cells
5 of different lineages, lymphocytes, monocytes, or NK cells. An even broader
screen would
use whole bone marrow aspirates that were depleted of the majority of red
blood cells (RBCs)
and platelets through RBC lysis and washes. This population is expected to
include all other
hematopoietic lineages. Once these libraries are constructed, target cells for
screening
depends on the goal of the screen. To identify relevant antibodies for
multiple lineages at
10 once, the target cell population could be the same as the immunogen, a
diverse population of
cells, and likewise very broad. These studies will uncover interesting
agonists and inhibitors
against a wide variety of cells at once. For high throughput screening on a
diverse population
of cells it is necessary to confirm that each member of the antibody library
is exposed to each
cell lineage, at various stages of maturation. An alternative to this is to
limit the target
15 population to a single lineage to identify antibodies relevant to a single
lineage. For example
neutrophils can be isolated from cells of other lineages using light
scattering signals in flow
cytometry and a combination of the monoclonal antibodies, CD 11 b, CD 15, and
CD 16 as
described in: Terstappen, L.W., Safford, M., Loken, M.R. Leukemia 4: 657-
663,1990. Many
other hematopoietic lineages can isolated for use as a target cell to identify
relevant antibodies
2 0 against a specific lineage.
Alternatively, one can identify relevant antibodies that play a role human
hematopoietic stem cell renewal or differentiation. Studies suggest that the
CD34' CD38-
fraction of human bone marrow cells contains stem cells capable of long term
engraft~nent and
multilineage differentiation (reviewed in "Williams Hematology" Fifth Edition
Editors: Ernest
25 Beutler, Marshall A. Lichtman, Barry S. Coller, Thomas J. Kipps McGraw-
Hill, U.S.A. in
chapter "Hemopoietic stem cell, progenitor cells, and cytokines" by Peter J.
Quesenberry).
Because stem cells represent a very small fraction of human bone marrow cells
(less than 1
in 1 X 105 bone marrow cells), ligands specific to the surface of primitive
human stem cells
could be identified from antibody libraries where human CD34+ sorted primary
human cells
3 o were used as immunogen. In this case, the target cells could be the CD34+
population and
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
71
antibodies that recognize CD34+CD38- cells could be identified by FACS
sorting, magnetic
sorting, or by panning on such target cells. Another target in this case could
be marine
hematopoietic stem cell lines such as FDCP-Mix or a marine multipotent
hematopoietic stem
cell line (Spooncer, E., Heyworth, C.M., Dunn. A., Dexter, T.M. 1986.
Differentiation
31:111-118). The advantage of using a cell line as a target is its
availability, clonal properties,
and ease to work with.
Therefore, target cells are selected based upon criteria that include the
immunogen that
was used, the desired end product, the range of the screen desired,
availability, ease to work
with.
If the initial target cell is a marine cell, eventually testing will be
carried out in the
appropriate human cell line.
For hematopoiesis preferred primary cells and cell lines are lysed whole
blood, human
bone marrow mononuclear cells, FDCP-mix, and CD34+ sorted bone marrow cells.
For P19
cells the preferred target are p19 cells and for NTERA cells the preferred
target cells are
NTERA cells.
Pannine
Phage (bacteriophage or phagemid) displayed antibody libraries are selected
for phage
carrying antibody fragments that bind to various target cells, and remain on
the cell surface
2 0 or are internalized, including mouse cell lines, human cell lines, and
sorted and unsorted
primary bone marrow samples, using a method termed panning, as well as using
FACS
sorting and magnetic cell sorting. Methods for whole cell panning have been
described
previously (Siegel, D.L., Chang, T.Y., Russell, S.L., and Bunya, V.Y. 1997. J.
Immunol.
Methods 206.:73-85).
2 5 Prior to any selection strategy, initial libraries are electroporated into
host cells
(ER2537). Library cultures are grown to log phase and superinfected with
helper phage, such
as VCSM13, a commercially available helper phage (Stratagene, La Jolla, CA).
Superinfection provides the remaining phage components needed for packaging
plasmids into
phagemid particles. Following overnight growth, phagemids in the culture
supernate are
3 0 precipitated with polyethylene glycol (PEG). PEG supernatants are used in
panning (cell
CA 02316755 2001-08-03
CA 01316755 1000-06-27
WO 99138008 PGTNS99I01331
72
surface, internalization and membrane), FACS sorting, and magnetic sorting to
purify binding
antibodies from non-binders.
In panning, antibody-phage libraries are incubated with target cells, and the
non-
adherent phage arc removed with multiple washes. A typical panning protocol is
as follows:
1. Block phage particles with PBS + 1 %BSA or 10% FBS + 4% milk powder +
NaN~ (except when internalized antibodies are assayed).
2. Add target cells to blocked phages (approximately 5 X 106 cells).
3. Mix and rotate slowly at 4° C or 37 ° C.
4. Wash cells twice with 1 ml ice cold PBS/1%BSA/NaN~ or room temperature
PBSII%BSA/l~laN~.
5. Specific antibody-phage bound to cells can be eluted by low pH, for example
with 76 mM citric acid ph 2.5 in PBS for 5 to 10 minutes at room temperature.
6. Neutralize eluted phage with 1M Tris~ T" HCI pH 7.4.
7. After neutralization, antibody-phage can be used to infect ER253? bacteria
and
amplify during overnight growth for the next round of panning.
Generally, 3-4 rounds of panning are performed on each library. Phage ELISAs
using
commercially available secondary antibody {sheep anti-M13 antibody-HRP) or
soluble
antibody ELISAs using a commercially available HA..11 antibody (Babco,
Berkeley, CA) that
recognizes the HA tag incorporated into each antibody from PRL4 sequences, can
be
2o performed on whole cells following each round of panning to allow
estimation of the
enrichment of binding antibodies over non-binders. Following the last round of
panning, the
antibody-phage can be picked as single colonies from agar plates, grown as
monoclonal
antibody-phage and screened by ELISA on whole cells for identification of
specific binders.
Specifically~the antibody-phage are infected into ToplOF' bacteria and plated
for single
2 5 colonies. Single colonies are picked form agar plates, grown and induced
with IPTG. Soluble
antibody is screened by ELISA on whole cells for identification of specific
binders. In
addition to live cells, screening can be done against intact, mildly fixed
target cells.
Where the target cell can be defined by cell surface markers, relevant
antibody-phage
can be identified by FACS or magnetic sorting. In the case where CD34' cells
were used as
3 0 immunogen for example, and the desired antibody fragments bind to either
CD34'CD3 ~ cells
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
73
or CD34+CD38+ cells, the phage-antibody library is added to the CD34+
population of cells
and incubated to allow antibody-phage binding to the cell surface. Following
binding, cells
are washed, pelleted and resuspended in fluorescein isothiocyanate (FITC)-
conjugated CD38
antibody solution. After incubation, and washing, cell sorting is performed.
FACS sorting
has been used successfully to identify phage antibodies specific for subsets
of blood
leukocytes {DeKruif, J., Terstappen, L., Boel, E., Logtenberg, T. Proc. Natl.
Acad Sci. U.S.A.
92:3938-3942, 1995). Phage are eluted from the two populations CD34+ CD38-
cells or
CD34+CD38+ cells at low pH, neutralized and used to infect bacterial cells.
Phage or soluble
antibody can then be prepared from individual colonies for further analysis.
In the case of
antibodies directed to the CD34+ CD38- cells, phages interacting with
structures present on
all or a majority of cells in the heterogeneous population would be absorbed
out by the CD38+
cells, effectively resulting in the enrichment of phages specific for the CD38-
cells. The
considerable shear forces exerted on the cells and attached phage during
fluorescence-
activated cell sorting would result in selection for relatively high-affinity
antibodies.
Alternatively, adsorber cells can be used to get rid of phage that bind non-
specifically
to cell surfaces or that bind to common antigens not specific to the
stem/progenitor cell, such
as receptors involved in cellular housekeeping functions. Adsorber cells for
CD34+
stem/progenitor cells, are the more differentiated CD34- cells, which can be
doped in with the
CD34+ cells.
The phage-antibody library is added to a 1:5 mixture of CD34+:CD34-cell
population
(for example, mononuclear.cells, which are primarily CD34' can be added to
presorted CD34+
cells) and incubated to allow antibody-phage binding to the cell surface.
Following binding,
cells are washed, pelleted and resuspended in fluorescein isothiocyanate
(FITC)-conjugated
CD34 antibody solution and phycoerythrin (PE)-conjugated CD38 antibody
solution. After
2 5 incubation, and washing, cell sorting is performed to collect the two
separate populations of
cells; CD34+CD38+and CD34+CD38'. Phage are eluted from the two populations at
low pH,
neutralized and used to infect bacterial cells. Phage or soluble antibody can
then be prepared
from individual colonies for further analysis. In this way, phages interacting
with structures
present on all or a majority of cells in the heterogeneous population would be
adsorbed out
CA 02316755 2000-06-27
WO 99/38008 PCTNS99/01331
74
by the presence of CD34' mononuclear cells, effectively resulting in the
enrichment of phages
specific for the CD34+ cells.
As an alternative to FACS sorting, one can also use magnetic sorting to
identify
antibodies/phage-antibodies specific for CD34+ CD38' cells and CD34+CD38+
cells. Magnetic
sorting may yield a higher background of non-specific antibodies, but may be
less rigorous
than FACS sorting.
If the target cell cannot be readily identified by cell surface markers, such
as with
NTera-2 cells, relevant antibody-phage can be identified using the same
techniques, of sorting
by FACS or magnetic sorting in the presence or absence of excess adsorber
cells. For
selections in the presence of excess adsorber cell, cell populations of
interest can be
biotinylated, an excess of irrelevant adsorber cells is added, such as a
fibroblast cell line,
phage-antibodies to the mixture are bound, and the cells of interest with
phage-antibodies
attached are pulled out using a streptavadin-PE conjugate.
For any particular species of target cell type an adsorber cell is selected
that is further
differentiated than the target cell or of a distinct or unrelated cell type.
Typically, the adsorber
cell lacks specific surface molecules present on the target cell, but would
have surface
molecules common to many cells. A preferred adsorber cell is a fibroblast.
Phage displaying antibodies that recognize receptors can also be identified
using an
internalization protocol. For example, mammalian cells are washed with ice
cold buffer (for
example, PBS with 5% FBS), then exposed to phagemids displaying monomeric or
dimeric
antibodies for 15 minutes to one hour incubation on ice. Cells are then washed
with ice cold
buffer, put into fresh culture medium, and incubated at 37 ° C to
permit internalization. Cell
are then lysed to recover internalized phagemids. Phagemids can be recovered
by direct
infection of phagemid particles into bacterial cells, or alternatively,
phagemids can be
2 5 recovered by electroporation of bacterial cells, selecting for antibiotic
resistance encoded by
the phagemid.
Alternatively, phage displaying antibodies that recognize receptors can be
screened
by membrane panning. Proteins are isolated from cells of interest, for example
a stem cell
line, or NTera-2 cell cell line as well as from an unrelated cell type, for
example a fibroblast
3 0 cell line. Proteins are separated by electrophoresis on one or two
dimensional polyacrylamide
CA 02316755 2000-06-27
WO 99/38008 PCTNS99/01331
gels. Following electrophoresis, proteins are transferred to a membrane
support such as
nitrocellulose or nylon. Proteins can be stained prior to transfer,
alternatively, identical gels
are run so that one can be stained with either silver or coomassie blue to
detect protein bands
or spots, while the other is used for membrane panning. Specific regions of
the membrane
5 may be chosen for panning based on comparisons of the cell line of interest
with an unrelated
cell type, alternatively larger areas of membrane containing many spots may be
chosen.
Patterns can be compared and regions chosen that are dissimilar between the
two cell lines.
These regions are cut out of the membrane as a solid support for panning.
Phage are applied
to the regions and allowed to bind. The membrane is washed and the phage are
eluted from
1 o the membrane and infected into bacterial cells and amplified.
From the above screening methods, one can build a diverse collection of
antibody
binding sites displayed on the surface of phage that bind non-covalently to
random cell
surface targets on hematopoietic cells or other cells. After FACS sorting,
magnetic sorting,
or panning, clones can be analyzed individually.
Dimerization
Following panning to isolate high affinity antibody binders, bioassays for
functional
screens of agonist antibodies are carried out. Dimerization is often a
prerequisite for
activation of many receptors and thus bioassays focus on agonist antibodies
that stimulate
2 0 receptors via promotion of dimerization. As previously described, single
chain multivalency
is approached in linker design. Fab fragment multivalency can be approached in
a number
of ways. A number of recent reports in the literature have shown success in
dimeric antibody
fragment formation which is applicable to phage display (DeKruif, J., and
Logtenberg, T.
1996. J. Biol. Chem. 271:7630-7634, Pack, P., and Pluckthun, A. 1992.
Biochemistry
31:1579-1584, and Holliger, P., and Winter, G. 1993. Current Opin. Biotech.
4:446-449).
Divalent Fabs can be created in at least two ways. In one approach
dimerization is achieved
by addition of a dimerization domain to pRL4, forming pRL8 (see Fig. SB).
There are a
number of dimerization domains (lexA, Zn fingers, fos, jun etc.) that can be
utilized in these
vectors to obtain multivalency of Fab fragments. Dimerization domains are
selected from,
3 o but not limited to, the following: jun (DeKruif, J. and Logtenberg, T. J.
Biol. Chem. 271:7630-
7634, 1996; Kostelny, S.A., Cole, M.S., and Tso, J.Y. J.Immunol. 148:1547-
1553, 1992) the
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
76
LexA dimerization region (Kim, B. and Little, J.W. Science 255:203-206, 1992),
the yeast
GCN4 dimerization domain (van Heeckeren, W.J., Sellers, J.W., Struhl, K.
Nucleic Acids Res.
20:3721-3724,1992), Gin invertase from the bacteriophage Mu (Spaeny-Dekking,
L.,
Schlicher, E., Franken, K., van de Putte, P., Goosen, N. J. Bacteriol. 34:1779-
1786, 1995),
E. coli NTRC protein dimerization domain (Klose, K.E., North, A.K., Stedman,
K.M., Kustu,
S. J. Mol. Biol. 241:233-245, 1994), and HSV-1 ICP4 dimerization domain
(Gallinari, P.,
Wiebauer, K., Nardi, M.C., Jiricny, J. J. Virol. 68:3809-3820, 1994). Also, a
high temperature
dimer domain from thermus organisms can be utilized (MacBeath, G., Kast, P.,
Hilvert, D.,
Biochemistry 37:100062-73, 1998 and MacBeath, G., Kast, P., Hilvert, D.,
Science 279:1958-
61, 1998). These are functional domains that when incorporated into a molecule
allow for
dimerization to occur. Those of ordinary skill in the art are familiar with
these and other
dimerization domains and their use to dimerize proteins. Following the panning
or sorting
steps of Fab libraries, the library of panned molecules are restricted with
Sac I and Spe I and
cloned into pRL8. Subcloning to pRL8 vector individually or en masse following
FACS
sorting or panning allows expression of dimeric soluble binding Fabs for
analysis in
bioassays. In pRL8, the antibody fragments are transported to the periplasmic
space and form
dimers there. The advantage of this approach is that it permits panning of
monomeric Fab
fragments, favoring high affinity Fabs.
Another approach uses a secondary antibody. pRL4 has the hemagglutinin
2 o decapeptide tag recognized by the commercially available HA.11 antibody
(Babco, Berkeley,
CA). Fabs identified in FACS sorting or panning to be tested in bioassay are
preincubated
with HA.11 which will promote dimerization, prior to addition to bioassays.
Bioassays
2 5 Once binding scFv's are identified by panning, the individual clones, each
expressing
a unique dimerized scFv on the phage surface, are tested for proliferation,
differentiation,
activation or survival effects on target cells. In addition, soluble dimerized
scFv's are
examined in bioassays. A simple transformation of the selected phage into
ToplOF' will allow
bacterially produced soluble scFv's to be secreted into the periplasm. Lysates
of individual E.
3 0 coli transformants can be tested for agonist effects. Fab antibodies are
transferred to pRL8
CA 02316755 2001-08-03
CA 02316755 2000-06-27
WO 99!38008 PC'TNS99/01331
77
(sec Fig. 5B) for dimerization and expressed as soluble dimerized Fab
fragments in bacterial
host ToplOF'.
A number of bioassays can be used in high-throughput screening. Those of
ordinary
skill in the art are familiar with these and other suitable bioassays. Several
non-radioactive
assays have been developed in which either DNA synthesis or enzyme activity
can be
analyzed. For example, an MTT cell proliferation assay(catalogue number 64000)
(Promega
Corporation, Madison, VJI) that is based on an assay described by Mosmann
(Mossman, T.
1983. J. Immu>sol. Methods 65:55-57) can be used. This protocol is fast and
easy, and yields
results within a single day. In the assay, MTT (3-[4,5-dimethylthiazol-2-yl]-
2,5-diphenyl-
l0 tetrazolium bromide), a tetrazolium salt, is converted into a blue formazan
product by
rnitochondrial dehydrogenase activity in living cells. The dehydrogenase
content, and
therefore the amount of colored product produced, is proportional to cell
number. The colored
product is detectable in an ELISA plate reader at 570nm. Assays are performed
in triplicate,
en masse in 96 well microtiter plates. Briefly, primary hematopoictic cells or
growth factor
dependent cell lines are plated in 100 pl aliquots in culture medium in 96-
well plates.
Following addition of various concentrations of antibodies or control growth
factors, cells are
incubated for 48-72 hours at 37'C and 5% CO~ in a fully humidified atmosphere.
MTT is
added to each well, and proliferation monitored via ELISA plate reader.
For example, in proliferation assays using TF-1 cells, bacterial cells
containing
phagmids expressing antibodies are grown overnight at 3TC in 96 well deep well
plates in
1 ml of a media that is a mixture of mammalian cell media and bacterial media
(in the ease
of TF-1 cells: RPMI 2.7/SD 0.3/Carb 100ug/ml). TF-1 cells are a human bone
marrow
erythroleukemia cell line that responds to multiple cytokines (Kitamura, T.,
Tange, T.,
Terasawa, T:, Chiba, S., Kuwaki, T., Miyagawa, K., Piao, Y.F., Miyazono, K.,
Urabe, A.,
Takaku, F., Cell Physiol. 140:323-334, 1989; Kitamura, T., Tojo, A., Kuwaki,
T., Chiba, S.,
Miyazono, K., Urabe, A., Takaku, F., Blood 73:375-380, 1989; Kitamura, T.,
Takaku, F.,
Miyajima, A., Int. Immunol. 3:571-577, 1991) On the following day, the
overnight cultures
are subcultured 1/10 to fresh trays, and placed at 3TC for 2 hours. Following
induction with
IPTG at 37~C for 4 hours, the plates are centrifuged al 2000 rpm/15' at room
temperature. 50
ul each culture supen-tate are filtered in 96 well filter trays (Millipore)
'"' to sterile 96 well assay
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
78
plates. Mammalian cells are prewashed to remove growth factor and resuspended
at a
concentration of 1 X 105 cells/ml. 50 ul cells are added to each well. Assay
plates are
incubated in 3TC/5% COz incubator for 72 hours. At 72 hours, the trays are
developed by
adding 40 ul media/MTS/PMS per well. MTS is an improved more soluble version
of MTT.
Both assays are based on the cellular conversion of tetrazolium salt. A MTS
proliferation
assay kit (catalogue number 65421 ) can be purchased from Promega, Inc.
(Madison, WI).
Plates are kept at 3TC/COz incubator and read at OD,~ at 1 hour, 4 hours, 8
hours with
microplate reader.
The activities of cytokines are often synergistic. Synergy could be manifested
through
1 o the binding of ligands to two different receptors which then sends the
correct signal, or via a
priming effect whereby interaction of ligand/receptor primes the cell to
respond to a second
cytokine. Furthermore, cytokines that act early in lineage development are
more often
synergistic than cytokines that act at later stages in a developmental
pathway. Therefore,
suboptimal concentrations of growth factors can be used in these bioassays to
examine
synergism. Conditions for suboptimal concentrations are determined for each
assay. This is
done by adding serial dilutions of growth factors, individually and as a
mixture, to the assays
and determining the levels below which a single factor does not promote a
response compared
to the mixture, and the level below which the mixture does not promote a
response in the
bioassay. Bone marrow stromal cells can also be added in bioassays to provide
other
2 0 necessary factors that may play a role in a synergistic response.
In addition, cell proliferation can be examined by monitoring DNA synthesis. A
non-
radioactive, colorimetric assay that examines 5-bromo-2'-deoxy-uridine (BrdU)
incorporation
(Boehringer Mannheim, Indianapolis, IN) can be performed in microtiter plate
format. Here,
cells are cultured in 96-well plates and incubated with BrdU and sub-optimal
concentrations
2 5 of cytokines. The amount of BrdU is determined after labeling with a
peroxidase labeled anti-
BrdU antibody. Final results are analyzed by ELISA plate reader at 405nm.
A radioactive mitogenesis assay that measures the rate of DNA synthesis as an
indication of proliferation (Raines and Ross, Methods of Enzymol. 109: 749-
773, 1985) can
also be used. In these assays, changes in rate of incorporation of [3H]-
thymidine in target cells
3 0 is examined. Again, these assays permit concurrent and rapid screening of
many antibody
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
79
fragments. They have been widely used as a convenient method of assessing the
stimulatory
and inhibitory effects on the growth of many different cells. Cells are
cultured in suspension
until they reach exponential growth rate. Cells are then washed free of the
medium in which
they were cultured, and replated in fresh medium. Cells are aiiquoted into 96
well plates in
a total volume of 100 ul at a concentration of about 1-2 x 105 cells/ml.
Dilutions of phage
supernatant, soluble dimerized Fab or ScFv antibodies are added and cells are
incubated for
18-48 hours in a gassed C02 incubator at a temp of 37°C. Following
incubation,
[3H]thymidine (937kBq) is added to each well and incubated for a further 4
hours. The cells
are then removed from the incubator and counted directly in a bench top
microplate
scintillation counter such as Packard Top Count NXT Instrument (Packard,
Meriden, CT).
Alternatively cells can be serially transferred to GF/C filters on a Millipore
cell harvester
(Millipore, Bedford, MA) or similar apparatus. Radioactivity associated with
acid-insoluble
material retained on the filter is then determined. Dilutions of commercially
available growth
factors are applied to positive control wells. Negative controls would include
supernatants
from cells carrying non-insert containing plasmids or irrelevant antibodies
treated similarly.
The relative growth promoting activities of the standard and the diluents of
the phage
supernatants under test are compared to quantify the growth promoting activity
in the sample.
Activation can be tested for by assaying second messengers or by
transcriptional
readout assays.
2 o Survival can be assayed, for example, by monitoring apoptosis using assays
such as
tunel assays or by other methods known to those who practice the art.
Other useful assays to analyze cellular signal transduction, the activity of
kinases and
phosphatases and ultimately cellular activities as a result of agonist
activity include
measurement of the generation of second messengers, e.g. cAMP, Ca++,
diacylglycerol
(DAG), and isositol 1,4,5-triphosphate (IP3). Measurement of spikes in
intracellular calcium
concentration, intracellular pH and membrane potential in high throughput
screening assays
can be performed using instruments such as the FLIPR Fluormetric Imaging Plate
Reader
System (Molecular Devices, Sunnyvale, CA). A number of fluorescent probes are
available
for examination of second messenger concentrations (Molecular Probes, Eugene
OR).
3 0 Measurement of concentrations of second messengers can also be done on the
single cell level
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
(DeBernardi, M.A. and Brooker, G. Proc. Natl. Acad. Sci USA 93:4577-4582,
1996). In
addition, assays that examine other signaling events such as phosphorylation,
apoptosis or
levels of RNA or protein of specific genes would be useful. For example. most
cytokines
have been shown to activate the enzyme PI 3-K (reviewed in Silvennoinen, O.,
Ihle, J.N.
5 Signaling by the Hematopoietic Cytokine Receptors, R.G. Landes company,
Austin, TX
1996). Furthermore, the Jak family of tyrosine kinases have been shown to be
central
mediators for cytokine receptor signaling (Ihle, J.N., Witthuhn, B.A., Quelle,
F. W. Annu. Rev.
Immunol. 13:369-398, 1995). In addition, several other tyrosine kinases, e.g.,
members of the
Src family, are activated in response to certain cytokine stimulations. In the
case of RNA or
10 proteins, c-Jun, c-Fos and Nfic(3 are rapidly and transiently upregulated
upon cytokine
stimulation, while c-Myc induction is slower. These proteins are required for
G1 transition
and proliferation (reviewed in Silvennoinen, O., Ihle, J.N. Signaling by
Hematopoietic
Cytokine Receptors, R.G. Landes Company, Austin, TX 1996). High throughput
screens that
detect increases in these transcripts could be utilized.
15 In transcriptional read out assays, changes in the transcription of
specific genes are
observed following exposure of cells to a growth factor or growth factor
mimetic (agonist or
inhibitory antibody). For example, in a myc read-out assay, cells such as IL-3
dependent
FDCP-mix cell line is starved of IL-3 growth factor for 8 hours, followed by
exposure to
growth factor mimetics, or native growth factors for 2 hours at 37°C.
At this time, the cells
2 0 are harvested, RNA is isolated, and reverse transcriptase-polymerase chain
reactions (RT-
PCR) are performed with primers specific for the myc gene. The RT-PCR
reactions are
electrophoresed in horizontal agarose gels for quantitation of PCR product. In
this case
expression of a single gene is being monitored.
Alternatively assay for changes in expression of genes can be monitored using
CHIP
2 5 technology, agonist antibodies could be identified under conditions of
high probe sensitivity
and a dynamic range. In this way, up to 10,000 could be analyzed for changes
in expression.
Desired genes that could be monitored could include c-myc, c-jun, NF-K~i,
among others.
These genes are downstream of various signal transduction pathways and their
expression
should change upon a mitogenic response. In one type of commercially available
CHIP
3 0 {Affymetrix, Santa Clara, CA), oligonucleotides from desired test genes
can be printed out
CA 02316755 2000-06-27
WO 99138008 PCT/US99/01331
81
onto glass surface. Target cells are exposed to test agonist antibodies. RNA
is isolated from
the cells exposed to test agonist antibodies, copied to cDNA, and in vitro
transcribed in the
presence of biotin. Hybridization of in vitro transcribed, biotinylated mRNA
is used as probe
in the arrays. Chips are then scanned to determine genes that show increases
in transcription
upon exposure to test agonist antibodies. In another version of CHIP
technology (Incyte, Palo
Alto, CA), the amount of DNA is not normalized on the glass, therefore, one
would set up a
competitive hybridization. RNA is isolated from the cells before and after
exposure to
agonist. cDNA is made from each sample whereby one cDNA reaction has one label
incorporated, for example, Cy-3, and the other cDNA population has a different
label
incorporated, for example Cy-5. Signals are detected and compared on a dual
laser scan to
collect images.
Visual assay
All scFvs or Fabs that show a proliferative response in the above assays are
also tested
in traditional methylcellulose colony forming assays (Stem Cell Technologies,
Vancover BC,
Canada). In these assays, colony growth, and morphological changes are scored
via light
microscope.
Visual examination for proliferation or differentiation effects in semi-solid
agar
cultures or methylcellulose can be performed using unsorted or sorted primary
hematopoietic
cells, and stem cell lines (Stem Cell Technologies, Vancouver BC, Canada)
{Eaves, C.J.,
Assays of hemopoietic progenitor cells. Williams Hematology 5 (eds. E.
Beutler, M.A.
Lichtman, B.S. Coller & T.J. Kipps), McGraw-Hill, Inc., pp L22-L26, 1995).
Hematopoietic colony assays whereby a culture medium that maximizes the growth
and differentiation of hematopoietic cells is utilized. Addition of
methlycellulose allows
2 5 clonal progeny of a single progenitor cell to stay together and
facilitates the recognition and
enumeration of distinct colonies. All necessary components are added to a
basic
methylcellulose medium (such as Iscove's MDM, BSA, (3-mercaptoethanol, L-
glutamine)
except colony-stimulating factor supplements. Test antibodies (phage
supernatants, soluble
antibodies) are added to see if they can substitute for growth factors.
Hematopoietic cells in
3 0 methylcellulose culture are incubated for 10-12 days following the
addition of antibodies in
a 37°C humidified atmosphere of S% COZ in air. After 10-12 days of
incubation, colonies are
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
82
counted using an inverted microscope. After another 8-10 days, colonies are
counted again.
Comparisons are made between media containing antibodies and controls with and
without
growth factors. In addition, colonies can be picked from methylcellulose and
individual cells
examined cytologically by staining with Wright's stain (see Atlas of
Hematological Cytology,
F.G.J. Hayhoe and R.J. Flemans, Wiley-InterScience 1970).
Another example of a visual assay is Pichia co-cultivation with bone marrow
cells.
For example, after one or more rounds of panning CD34+ or human fetal liver
libraries by
either FACS, or magnetic selection, the antibody genes are cloned from the
panned library
into a Pichia expression vector so the antibody genes are under control of the
pGAP promoter
and secreted from cells using the a-factor leader sequence (InVitrogen,
Carlsbad, CA) (Das,
R.C., Shultz, J.L., Lehman, D.J., Mol. Gen. Genet. 218:240-8, 1989). Pichi
atransformants
are selected on YPD + Zeocin. Colonies are replica plated to "Iscoves plates"
(Iscoves media
plus agar). Cells are scraped off plates in 5 ml Iscoves media, centrifuged,
resuspended in 0.6
ml Iscoves media and counted. Methylcellulose medium that does not contain
added growth
factors (catalogue #4230 Stem Cell Technologies, Vancouver BC, Canada) is
thawed. 5000
Pichia cells and 50,000 human bone marrow CD34+ cells are plated in the medium
per 3 cm
dish. Plates are incubated in a humidified atmosphere at 37'C 5% C02. Scoring
is done at
5-12 days. If a bone marrow colony is noted, the nearby yeast is plucked from
the
methylcellulose, and retested in a similar way. The antibody genes can be
fingerprinted from
2 0 Pichia by a whole cell PCR and restriction digest with EcoRII. The
antibody released into the
medium can be purified and used in FACS sorting to examine the population of
cells the
antibody binds to, in the presence of other known fluorescein conjugated
antibodies. Here,
the primary antibody is secreted from yeast. The secondary antibody is HA.11-
FITC
conjugate (Babco, Berkeley, CA).
Stem Cell Assays
Candidates identified in stem cell specific assays are further analyzed for
their ability
to stimulate renewal in serum free medium (Stem Cell Technologies, Vancouver
BC,
Canada), and the ability of cells cultured in the presence of agonists to
generate long term
3 0 culture initiating cells (LTC-IC) (Stem Cell Technologies, Vancover BC,
Canada). Agonists
that appear to expand LTC-IC in vitro are subjected to DNA analysis, including
restriction
CA 02316755 2000-06-27
WO 99/38008 PC'TNS99/01331
83
enzyme footprinting and DNA sequencing, in order to identify unique clones.
Unique agonists
are further assayed to determine whether cells cultured in the presence of the
agonist support
long term engraftment in irradiated mice (Hodgson, G.S. and Bradley, T.R.
Nature 281:381-
382, 1979; Harrison, D.E. Blood 55:77-81, 1980) or in NOD/SCID mice
(Conneally, E.,
Cashman, J., Petzer, A., Eares, C. Proc. Natl. Acad. Sci. U.SA. 94: 9836-9841,
1997).
Agonists that prove interesting by these criteria are subject to sequence
analysis, binding
studies, and used to identify the receptor through immunoprecipitation,
protein sequence
analysis, and database searches.
Rece~,tor effects
Even if the receptor is unknown, if the receptor encodes a kinase or the
receptor causes
kinase activity, receptor phosphorylation can be examined by incubating
receptor-containing
membrane extracts with [Y-32PJATP in the presence of the test compound
(antibody). Then
the extracts are examined for receptor phosphorylation by gel electrophoresis
and
1 S autoradiography. These and other techniques for examining receptor
phosphorylation are
known to those who practice the art.
Synthesis of antibodies
Once antibody fragments are identified in bioassays they are selected for high
level
2 o expression as soluble antibody fragments. Soluble ScFv fragments or Fab
fragments can be
isolated in bacterial hosts such as ToplOF' (Invitrogen, Carlsbad, CA).
Single colonies of cells are grown up and the soluble antibody fragments are
purified
by methods such as nickel-chelate chromatography, using the His6 sequence
engineered in
the vectors.
2 5 Those of ordinary skill in the art using known techniques would be able to
synthesize
antibodies in other organisms such as yeast, mammalian, insect, and plants
(Carlson, J.R. and
Weissman, LL., Mol. Cell. Biol., 8:2647-2650, 1988; Trill, J.J., Shatzman,
A.R., Ganguly, S.
Curr. Opin. Biotechnol 6:553-560, 1995; Hiatt, A., Cafferkey, R. Bowdish, K.
Nature 342:
76-78, 1989).
CA 02316755 2001-08-03
CA 02316755 2000-06-27
WO 99/38008 PCTNS99/0133t
84
Example 2: Identification of the growth factor receptor
The first generation agonist molecules are grouped into categories depending
on cell
type that appears to be proliferating and/or differentiating. The receptor can
be identified by
using a number of approaches including immunoprecipitation and affinity
chromatography
and chemical crosslinking followed by protein sequencing and database
searches.
In immunoprecipitation, cells can be radiolabelled, lysed in the presence of
detergent
Tl~.t
such as Triton X-100, and the binding proteins precipitated by the antibody,
and analyzed by
SDS-PAGE followed by autoradiography. Therefore this will allow detection of
the receptor,
characterization of its molecular size, and identification of other subunits
or associated
to proteins. Additionally, proteins can be electrophoretically transferred to
a sheet of
polyvinylidene difluoride (PVDF) and stained with Comassie blue. The separated
bands can
be excised, and analyzed in an automated protein sequencer, followed by data
base searches
to determine whether the receptor is new, or previously identified.
Additionally, the proteins
can be cleaved with proteolytic enzymes to determine intcmal amino acid
sequences.
In conventional immunoaffinity purification, the antibody is covalently
attached to a
TA1
solid-phase matrix. Typically, antibodies are coupled to Sepharose. A cell
lysate, prepared
in the presence of detergent such as Triton X-100 for lysis and solubilization
of integral
membrane proteins is prepared. Lysates are applied to the columns, the column
is washed,
and the binding proteins are eluted by brief exposure to high-pH, or low-pH
buffer. Each is
followed by partial amino acid sequencing, and database searches.
Alternatively, specific
elution can be done with soluble scFv. Often a precolumn is used to reduce non-
specific
binding. If multiple species are present, one can determine which is the
ligand-binding
subunit for the agonist by ccll surface radioreceptor cross-linking of
radiolabeled scFv to
intact cells.
An expression cloning approach could also be used. A cDNA library is
constructed
from cells responding to the antibody. The library is cloned into a mammalian
expression
vector and transfccted into mammalian cells. Cells expressing the receptor are
identified by
affnity chromatography or "routine panning" where a single species of antibody-
phage are
placed in contact with a pool of cells, each expressing a different eDNA
clone.
CA 02316755 2000-06-27
WO 99/38008 PGT/US99/01331
Once the receptor is identified it can be cloned using standard molecular
biology
techniques. Transient transfection assays with cDNAs encoding the receptor in
mammalian
cells can be carried out to demonstrate that it is the receptor the antibody
recognizes. For
example COS cells can be utilized for receptor expression (Neil, J.D.,
Sellers, J.C., Musgrove,
5 L.C., Duck, L.W. Mol. Cell. Endrocrinol. 127:143-154, 1997).
Identification of the natural ligand
The native ligand of a newly identified receptor can be identified by
expressing the
receptor heterologously in mammalian cells or in yeast, preferably in cells
that normally do
1 o not express the receptor. Pools of secreted factors from expressed cDNA
libraries can be
added to receptor expressing cells and analyzed in the same bioassays used to
identify the
agonist (see Lee, F. et al. Proc. Natl. Acad. Sci. 82:4360, 1985). In addition
to transient
expression systems, permanent expression systems in retroviral vectors can
also be used in
these assays (Rayner, J.R. and Gonda, T.J. Mol. Cell. Biol., 14:880, 1994). A
positive
15 response is linked back to an individual clone expressing the specific
ligand cDNA. In
addition, competition experiments can be performed with the ligand and the
agonist antibody
where one is labeled with'ZSI or fluorescent compounds.
'Those of ordinary skill in the art are familiar with these techniques and
others useful
for identifying the native ligand once the receptor is known.
Screenin~~ for inhibitory antibodies
Inhibitory antibodies are screened using the same assays as utilized to screen
for
agonist antibodies, panning and bioassays. In bioassays however, additional
growth factors,
such as IL-3, are included in the culture to enable the cells to grow, and
inhibition of
2 5 proliferation, differentiation, survival or activation is observed.
Example 3: Phag_e display libraries to hematopoeitic cells and other cell
types
Animals (rabbits) were immunized individually with the following cells or cell
lines:
human bone marrow mononuclear cells, human bone marrow CD34+ cells, human bone
3 0 marrow cells that have undergone RBC lysis, human fetal liver cells, human
NTera-2 cell line,
marine P 19 cell line, and mouse FDCP-mix cell line. Rabbits were immunized
with whole
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
86
cells, according to the protocol in Example 1, 3-5 times in 3 week intervals
until a strong
positive response was detected.
Prior to immunization, approximately S ml of blood was obtained from each
animal.
The blood was processed to serum immediately by allowing the blood to clot,
centrifuging
and pelleting of cells, and transferring serum to fresh tubes. Serum from each
animal was
stored at -20°C. After primary immunizations and two boosts, blood was
again collected
from each animal and processed to serum. Whole cell ELISAs were performed
comparing
preimmune serum to post-immune serum. Dilutions of serum were made, e.g.,
1/50, 1/100,
11500, 1/1000, 1/5000 in 5% milk in PBS. Cells of the same type used for the
immunization(1
X 106) were applied to plates, serum dilutions were added and the cells were
incubated. The
secondary antibody was an enzyme-conjugated antibody to the host animal IgG
that is being
tested, for example goat anti-rabbit IgG-alkaline phosphatase conjugate or
peroxidase
conjugate. Results were read visually or plates were centrifuged, supernatants
removed and
placed in fresh wells for ELISA plate reading. Readings are graphed to
determine the half
maximum serum response, or that dilution that gives a response that is equal
to one half of the
maximum response.
When a positive response was detected, the animals were sacrificed and spleen,
bone
marrow and peripheral blood lymphocytes were collected. RNA was isolated from
these
organs and libraries were constructed as in Example 1 to produce antibody
fragment libraries
displayed on the surface of phagemids. Ligations were electroporated into
ER2537. A small
aliquot was plated to determine efficiency of cloning and potential diversity
of libraries as in
Example 1. Results are shown in Table 2.
Table 2
Name/Type of Antigen Serum ResponsePotential Diversity
Ab
B 1 /scFv shorthBM mononuclear 1 /300 5.2 x 10
cells
B2/scFv long hBM mononuclear 1/300 1.3 x 108
B3/chimeric hBM mononuclear 1/300 1.6 x 10
Fab
F l /scFv shorthBM CD34+ 1 /400 3.0 x 10$
F2/scFv long hBM CD34+ 1 /400 1.5 x 1 Og
F3/chimeric hBM CD34+ 1/400 5.2 x 108
Fab
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
87
Name/Type of Antigen Serum ResponsePotential Diversity
A6
G1/scFv short hBM CD34+ 1/1000 2.3 x 10~
G2/scFv long hBM CD34+ 1/1000 1.9 x 10
J1/scFv short hBM RBC lysis 1/250 3.5 x 10~
J2/scFv long hBM RBC lysis 1/250 4.1 x 10
K1/scFv short hBM RBC lysis 1/1000 5.3 x 10~
K2/scFv long hBM RBC lysis 1/1000 4.9 x 10
T 1 /scFv shorth fetal liver 1 / 1000 1. 8 x 1 Oy
T2/scFv long h fetal liver 1/1000 1.9 x l0y
U l/scFv shorth fetal liver 1 /750 5 x 10
U2/scFv long h fetal liver 1/750 1.6 x 10y
PQ1/scFv shortn NTera-2 1/500 nd
PQ2/scFv long h NTera-2 1/500 nd
MN 1 /scFv m P 19 1 /500 1.0 x 10
short
MN2/scFv long m P 19 1 /500 2.5 X 1 Oy
BBCC1/ scFv m FDCP-mix 1/1000 nd
short
BBCC2/ scFv m FDCP-mix 1/1000 nd
long
h = human
BM a bone marrow
m = marine
nd = actual
value not
determined
yet.
Those of ordinary skill in the art will recognize that using the above
described
protocols other animals, such as chickens, and could be immunized with other
cell types and
lines such as yolk sac cells, cells derived from the marine AGM region, marine
pluripotent
embryonic cells, human embryonic stem {ES) cell lines, cells of neural origin,
cells involved
in organ or tissue regeneration, in order to generate additional libraries of
phage displayed
antibody fragments to a variety of receptors on a number of cell types.
Pharmaceutical Formulations And Routes Of Administration
The agents described herein can be administered to a human patient per se, or
in
pharmaceutical compositions where it is mixed with suitable carriers or
excipient(s).
Techniques for formulation and administration of the compounds of the instant
application
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
88
may be found in "Remington's Pharmaceutical Sciences," Mack Publishing Co.,
Easton, PA,
latest edition.
Routes Of Administration
Suitable routes of administration may, for example, include oral, rectal,
transmucosal,
or intestinal administration; parenteral delivery, including intramuscular,
subcutaneous,
intramedullary injections, as well as intrathecal, direct intraventricular,
intravenous,
intraperitoneal, intranasal, or intraocular injections.
Alternately, one may administer the agents in a local rather than systemic
manner.
l0 Furthermore, one may administer the agents in a targeted drug delivery
system, for
example, in a liposome coated with tumor-specific antibody. The liposomes will
be targeted
to and taken up selectively by the tumor.
Composition/Formulation
The pharmaceutical compositions of the present invention may be manufactured
in a
manner that is itself known, e.g., by means of conventional mixing,
dissolving, granulating,
dragee-making, levigating, emulsifying, encapsulating, entrapping or
lyophilizing processes.
Pharmaceutical compositions for use in accordance with the present invention
thus
may be formulated in conventional manner using one or more physiologically
acceptable
2 0 Garners comprising excipients and auxiliaries which facilitate processing
of the active
molecules into preparations which can be used pharmaceutically. Proper
formulation is
dependent upon the route of administration chosen.
For injection, the agents of the invention may be formulated in aqueous
solutions,
preferably in physiologically compatible buffers such as Hanks's solution,
Ringer's solution,
2 5 or physiological saline buffer. For transmucosal administration,
penetrants appropriate to the
barrier to be permeated are used in the formulation. Such penetrants are
generally known in
the art.
For oral administration, the agents can be formulated readily by combining the
active
molecules with pharmaceutically acceptable carriers well known in the art.
Such Garners
3 0 enable the compounds of the invention to be formulated as tablets, pills,
dragees, capsules,
liquids, gels, syrups, slurries, suspensions and the like, for oral ingestion
by a patient to be
CA 02316755 2000-06-27
WO 99/38008 PCTNS99/01331
89
treated. Pharmaceutical preparations for oral use can be obtained solid
excipient, optionally
grinding a resulting mixture, and processing the mixture of granules, after
adding suitable
auxiliaries, if desired, to obtain tablets or dragee cores. Suitable
excipients are, in particular,
fillers such as sugars, including lactose, sucrose, mannitol, or sorbitol;
cellulose preparations
such as, for example, maize starch, wheat starch, rice starch, potato starch,
gelatin, gum
tragacanth, methyl cellulose, hydroxypropyhnethyl-cellulose, sodium
carboxymethylcellulose,
and/or polyvinylpyrrolidone (PVP). If desired, disintegrating agents may be
added, such as
the cross-linked polyvinyl pyrrolidone, agar, or alginic acid or a salt
thereof such as sodium
alginate.
Dragee cores are provided with suitable coatings. For this purpose,
concentrated sugar
solutions may be used, which may optionally contain gum arabic, talc,
polyvinyl pyrrolidone,
carbopol gel, polyethylene glycol, and/or titanium dioxide, lacquer solutions,
and suitable
organic solvents or solvent mixtures. Dyestuffs or pigments may be added to
the tablets or
dragee coatings for identification or to characterize different combinations
of active
compound doses.
Pharmaceutical preparations which can be used orally include push-fit capsules
made
of gelatin, as well as soft, sealed capsules made of gelatin and a
plasticizes, such as glycerol
or sorbitol. The push-fit capsules can contain the active ingredients in
admixture with filler
such as lactose, binders such as starches, and/or lubricants such as talc or
magnesium stearate
2 0 and, optionally, stabilizers. In soft capsules, the active compounds may
be dissolved or
suspended in suitable liquids, such as fatty oils, liquid paraffin, or liquid
polyethylene glycols.
In addition, stabilizers may be added. All formulations for oral
administration should be in
dosages suitable for such administration.
For buccal administration, the agents may take the form of tablets or lozenges
2 5 formulated in conventional manner.
For administration by inhalation, the agents for use according to the present
invention
are conveniently delivered in the form of an aerosol spray presentation from
pressurized packs
or a nebuliser, with the use of a suitable propellant, e.g.,
dichlorodifluoromethane,
trichlorofluoromethane, dichlorotetrafluoroethane, carbon dioxide or other
suitable gas. In
3 0 the case of a pressurized aerosol the dosage unit may be determined by
providing a valve to
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
deliver a metered amount. Capsules and cartridges of e.g. gelatin for use in
an inhaler or
insufflator may be formulated containing a powder mix of the compound and a
suitable
powder base such as lactose or starch.
The agents may be formulated for parenteral administration by injection, e.g.,
by bolus
5 injection or continuous infusion. Formulations for injection may be
presented in unit dosage
form, e.g., in ampoules or in mufti-dose containers, with an added
preservative. The
compositions may take such forms as suspensions, solutions or emulsions in
oily or aqueous
vehicles, and may contain formulatory agents such as suspending, stabilizing
and/or
dispersing agents.
l0 Pharmaceutical formulations for parenteral administration include aqueous
solutions
of the active molecules in water-soluble form. Additionally, suspensions of
the active
molecules may be prepared as appropriate oily injection suspensions. Suitable
lipophilic
solvents or vehicles include fatty oils such as sesame oil, or synthetic fatty
acid esters, such
as ethyl oleate or triglycerides, or liposomes. Aqueous injection suspensions
may contain
15 substances which increase the viscosity of the suspension, such as sodium
carboxymethyl
cellulose, sorbitol, or dextran. Optionally, the suspension may also contain
suitable stabilizers
or agents which increase the solubility of the compounds to allow for the
preparation of highly
concentrated solutions.
Alternatively, the active ingredient may be in powder form for constitution
with a
20 suitable vehicle, e.g., sterile pyrogen-free water, before use.
The agents may also be formulated in rectal compositions such as suppositories
or
retention enemas, e.g., containing conventional suppository bases such as
cocoa butter or
other glycerides.
In addition to the formulations described previously, the agents may also be
2 5 formulated as a depot preparation. Such long acting formulations may be
administered by
implantation (for example subcutaneously or intramuscularly) or by
intramuscular injection.
Thus, for example, the agents may be formulated with suitable polymeric or
hydrophobic
materials (for example as an emulsion in an acceptable oil) or ion exchange
resins, or as
sparingly soluble derivatives, for example, as a sparingly soluble salt.
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
91
A pharmaceutical carrier for the hydrophobic agents of the invention is a
cosolvent
system comprising benzyl alcohol, a nonpolar surfactant, a water-miscible
organic polymer,
and an aqueous phase. The cosolvent system may be the VPD co-solvent system.
VPD is a
solution of 3% w/v benzyl alcohol, 8% w/v of the nonpolar surfactant
polysorbate 80, and
65% w/v polyethylene glycol 300, made up to volume in absolute ethanol. The
VPD co-
solvent system (VPD:SV~ consists of VPD diluted 1:1 with a 5% dextrose in
water solution.
This co-solvent system dissolves hydrophobic compounds well, and itself
produces low
toxicity upon systemic administration. Naturally, the proportions of a co-
solvent system may
be varied considerably without destroying its solubility and toxicity
characteristics.
Furthermore, the identity of the co-solvent components may be varied: for
example, other
low-toxicity nonpolar surfactants may be used instead of polysorbate 80; the
fraction size of
polyethylene glycol may be varied; other biocompatible polymers may replace
polyethylene
glycol, e.g. polyvinyl pyrrolidone; and other sugars or polysaccharides may
substitute for
dextrose.
Alternatively, other delivery systems for hydrophobic pharmaceutical compounds
may
be employed. Liposomes and emulsions are well known examples of delivery
vehicles or
carriers for hydrophobic drugs. Certain organic solvents such as
dimethylsulfoxide also may
be employed, although usually at the cost of greater toxicity. Additionally,
the compounds
may be delivered using a sustained-release system, such as semipermeable
matrices of solid
2 0 hydrophobic polymers containing the therapeutic agent. Various of
sustained-release
materials have been established and are well known by those skilled in the
art. Sustained-
release capsules may, depending on their chemical nature, release the
compounds for a few
weeks up to over 100 days. Depending on the chemical nature and the biological
stability of
the therapeutic reagent, additional strategies for protein stabilization may
be employed.
2 5 The pharmaceutical compositions also may comprise suitable solid or gel
phase
carriers or excipients. Examples of such carriers or excipients include but
are not limited to
calcium carbonate, calcium phosphate, various sugars, starches, cellulose
derivatives, gelatin,
and polymers such as polyethylene glycols.
Many of the agents of the invention may be provided as salts with
pharmaceutically
3 o compatible counterions. Pharmaceutically compatible salts may be formed
with many acids,
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
92
including but not limited to hydrochloric, sulfuric, acetic, lactic, tartaric,
malic, succinic, etc.
Salts tend to be more soluble in aqueous or other protonic solvents that are
the corresponding
free base forms.
Effective Dosase
Pharmaceutical compositions suitable for use in the present invention include
compositions wherein the active ingredients are contained in an amount
effective to achieve
its intended purpose. More specifically, a therapeutically effective amount
means an amount
of agent effective to prevent, alleviate or ameliorate symptoms of disease or
prolong the
survival of the subject being treated. Determination of a therapeutically
effective amount is
well within the capability of those skilled in the art, especially in light of
the detailed
disclosure provided herein.
For any agent used in the methods of the invention, the therapeutically
effective dose
can be estimated initially from cell culture assays. For example, a dose can
be formulated in
animal models to achieve a circulating concentration range that includes the
ICso as
determined in cell culture (e.g., the concentration of the test molecule which
promotes or
inhibits cellular proliferation or differentiation). Such information can be
used to more
accurately determine useful doses in humans.
Toxicity and therapeutic efficacy of the agents described herein can be
determined by
standard pharmaceutical procedures in cell cultures or experimental animals,
e.g., for
determining the LDso (the dose lethal to 50% of the population) and the EDso
(the dose
therapeutically effective in 50% of the population). The dose ratio between
toxic and
therapeutic effects is the therapeutic index and it can be expressed as the
ratio between LDso
and EDso. Molecules which exhibit high therapeutic indices are preferred. The
data obtained
2 5 from these cell culture assays and animal studies can be used in
formulating a range of dosage
for use in human. The dosage of such molecules lies preferably within a range
of circulating
concentrations that include the EDSO with little or no toxicity. The dosage
may vary within
this range depending upon the dosage form employed and the mute of
administration utilized.
The exact formulation, route of administration and dosage can be chosen by the
individual
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
93
physician in view of the patient's condition. (See e.g., Fingl et al., 1975,
in "The
Pharmacological Basis of Therapeutics", Ch. 1 p. l ).
Dosage amount and interval may be adjusted individually to provide plasma
levels of
the active moiety which are sufficient to promote or inhibit cellular
proliferation or
differentiation or minimal effective concentration (MEC). The MEC will vary
for each agent,
but can be estimated from in vitro data using described assays. Dosages
necessary to achieve
the MEC will depend on individual characteristics and route of administration.
However,
HPLC assays or bioassays can be used to determine plasma concentrations.
Dosage intervals can also be determined using MEC value. Compounds should be
l0 administered using a regimen which maintains plasma levels above the MEC
for 10-90% of
the time, preferably between 30-90% and most preferably between 50-90%.
In cases of local administration or selective uptake, the effective local
concentration
of the agent may not be related to plasma concentration.
The amount of agent administered will, of course, be dependent on the subject
being
treated, on the subject's weight, the severity of the affliction, the manner
of administration and
the judgment of the prescribing physician.
Theranv
Antibodies and factors encompassed by the claimed invention are useful for the
2 0 amplification of a variety of clinically relevant cell types. Treatment
can be in vivo or ex vivo.
For example, agonist antibodies are useful to treat patients suffering from a
deficiency in a
hematopoietic cell population caused by disease, disorder or treatment related
to for example
suppression of hematopoiesis where less than the normal number of cells of a
given lineage
or lineages are present in a patient. The following represent only some
examples of the
2 5 conditions that can be treated with the antibodies and factors of the
claimed invention, those
who practice the art would be able to identify other diseases and conditions
that would benefit
from such treatment. For example, HIV-infected patients, patients undergoing
chemotherapy,
bone marrow transplant patients, and patients suffering from
myeloproliferative disorders
show subnormal levels of specific hematopoietic lineages. Neutropenia is a
decrease in the
3 0 number of circulating, terminally differentiated neutrophils. Neutropenia
appears secondary
to or in association with a number of pathophysiologic conditions, including
viral and severe
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
94
bacterial infections; exposure to drugs, such as certain antibiotics and
anticonvulsants;
autoimmune processes, such as those seen with systemic lupus erythematosus;
marrow
infiltrative processes, such as leukemia, metastatic tumor, and myelofibrosis;
and inborn
errors of metabolism, such as propionic acidemia and isovaleric acidemia. Some
chronic
primary neutropenic disorders such as cyclic neutropenia are transmitted
genetically, while
others occur in association with disorders of immune function, such as that
produced by
infection with Epstein-Barr or human immunodeficiency virus. Other conditions
may be
associated with chronic idiopathic neutropenia including Shwachman syndrome, X-
linked
agammaglobulinemia, dysgammaglobulinemia, and depressed cellular immunity.
Acquired
agranulocytosis is a blood disorder characterized by a reduction in the number
of granular
leukocytes in the circulating blood due to an impairment in granulocyte
production in bone
marrow. Patients undergoing renal dialysis often suffer from treatment related
anemia with
subnormal levels of red blood cells. In aplastic anemia, bone marrow
suppression can cause
pancytopenia or may affect only the red blood cells, the white cells, or the
platelets. Agonist
antibodies and factors will augment the armamentarium of therapeutic agents
for these and
other diseases and disorders characterized by deficiencies in specific cell
populations, such
as hematopoietic cells.
Antibodies and factors encompassed by the claimed invention are also useful
for the
inhibition of clinically relevant cell types. For example, abnormal growth of
hematopoietic
2 0 cell populations include diseases where greater than the normal number of
cells of a given
lineage or lineages are present in a patient. Chronic myeloid leukemia is a
disease
characterized by myeloid proliferation due the presence of a chromosomal
translocation in the
hematopoietic stem cells. Thrombocytopenia is a disease characterized by an
increase in the
numbers of rriegakaryocytes. Polycythemia vera is a form of myeloproliferative
syndrome
2 5 in which the granulocyte, monocyte, and platelet counts as well as the red
cell count are
usually elevated. This disorder is caused by a neoplastic clone of stem cells
which proliferate
excessively. In allergic diseases such as rhinitis, asthma, and exzema, there
is excessive
production of basophils, mast cells and eosinophils. Inhibitors of eosinophil,
mast cell and
basophil production identified by the claimed invention can serve as
therapeutics for severe
3 0 allergy sufferers. In addition, inhibitors can be used clinically in
chemotherapy to inhibit the
CA 02316755 2000-06-27
WO 99/38008 PCT/US99/01331
proliferation of normal cells, while cancerous cells will continue through the
cell cycle and
therefore be more susceptible to chemotherapeutic agents.
The molecules encompassed by the claimed invention can also be used for ex
vivo
proliferation and differentiation of cells. This is useful for gene therapy
purposes, for example
5 for traditional viral vector approaches, and for autologous bone marrow
transplants. Gene
transfer into hematopoietic progenitor cells is a major goal of gene therapy
for malignant and
non-malignant disease. Most gene therapy vectors in use require mitosis for
integration of
viral genome into cellular DNA. Integration is preferred since a single stem
cell will divide
to form many progeny cells. Agonist antibodies, factors, or combinations of
the above
l0 administered before or during vector exposure can promote proliferation of
stem cells ex vivo.
In addition, antibodies of the present invention can be radiolabeled for
radioimmunotherapy or conjugared to toxins to deliver such toxins to specific
cell types and
result in the killing of those cells.
15 Diagnosis and Purification
The molecules encompassed by the claims can be used in diagnostics where the
antibody fragments specific to a cell lineage are conjugated to fluorescent
markers or used as
primary antibodies with secondary antibodies that are conjugated to
fluorescent markers, and
used in flow cytometry analysis to identify clinically relevant cell types,
such as in the blood
2 0 or bone marrow of a patient. In addition, the molecules can be used in
cell isolation strategies
such as fluorescence-activated cell sorting {FACS), or magnetic sorting
procedures. In
fluorescence-activated cell sorting, cells tagged with fluorescent molecules
are sorted
electronically on a flow cytometer such as a Becton-Dickinson (San Jose,
California) FACS
IV cytometer or equivalent instrument. The fluorescent molecules are
antibodies that
2 5 recognize specific cell surface antigens. The antibodies are conjugated to
fluorescent markers
such as fluorescein isothiocyanate (FITC) or Phycoerythrin (PE). In magnetic
sorting
procedures, the antibody is linked directly or indirectly to magnetic
microbeads. Cells are
precoated with agonist or inhibitory antibodies that recognize cell surface
molecules, e.g.,
receptors involved in proliferation, differentiation, activation or survival.
The antibodies are
30 attached to magnetic beads conjugated with a secondary immunoglobulin that
binds to the
agonist or inhibitory antibody, such as to the HA molecular tag engineered
into each antibody.
CA 02316755 2000-06-27
WO 99138008 PGT/US99/01331
96
The cells are then removed with a magnet. Magnetic sorting can be positive
selection where
cells of interest are bound by the antibody and hence the magnet, or negative
selection where
undesired cells are isolated onto the magnet.
Alternatively, radiolabeled antibodies can be used for diagnostic purposes.
Other embodiments are within the claims.
CA 02316755 2000-10-26
97
SEQUENCE LISTING
<110> Bowdish, Katherine Susan
<120> METHODS AND COMPOSITIONS FOR THE
IDENTIFICATION OF GROWTH FACTOR MIMETICS, GROWTH FACTORS AND
INHIBITORS
<130> 24750-2702
<140> 09/235,499
<141> 1999-O1-22
<150> US 60/072,253
<151> 1998-O1-23
<160> 36
<170> FastSEQ for Windows Version 4.0
<210> 1
<211> 60
2 0 <212> DNA
<213> SYNTHETIC
<400> 1
caccatggcg catacccgta cgacgttccg gactacgctt cttaggaggg tggtggctct 60
<210> 2
<211> 23
<212> DNA
<213> SYNTHETIC
<400> 2
3 0 gcttacaatt tcccagatct gcg 23
<210> 3
<211> 70
<212> DNA
<213> SYNTHETIC
<400> 3
gaggaggagg aggaggagac tagtggccag gccggccagc accatcacca tcaccatggc 60
gcatacccgt 70
40 <210> 4
<211> 38
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 24
<223> The letter "m" stands for a or c
<400> 4
gggcccaggc ggccgagctc gtgmtgaccc agactcca 38
<210> 5
<211> 38
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 24
<223> The letter "m" stands for a or c
<400> 5
gggcccaggc ggccgagctc gatmtgaccc agactcca 38
CA 02316755 2000-10-26
98
<210> 6
<211> 38
<212> DNA
<213> SYNTHETIC
<400> 6
gggcccaggc ggccgagctcgtgatgacccagactgaa 38
<210> 7
<211> 40
<212> DNA
<213> SYNTHETIC
<400> 7
gggcccaggc ggccgagctcgtgctgactcagtcgccctc 40
<210> 8
<211> 42
<212> DNA
2 <213> SYNTHETIC
O
<400> 8
ggaagatcta gaggaaccacctaggatctccagctcggtccc 42
<210> 9
<211> 42
<212> DNA
<213> SYNTHETIC
<400> 9
ggaagatcta gaggaaccacctttgatttccacattggtgcc 42
30
<210> 10
<211> 42
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 28
<223> The letter stands c or g
"s" for
<400> 10
4 ggaagatcta gaggaaccacctttgacsaccacctcggtccc 42
0
<210> 11
<211> 45
<212> DNA
<213> SYNTHETIC
<400> 11
ggaagatcta gaggaaccaccgcctgtgacggtcagctgggtccc 45
<210> 12
50 <211> 42
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 40
<223> The letter stands a or g.
"r" for
<400> 12
ggtggttcct ctagatcttcccagtcggtggaggagtccrgg 42
60 <210> 13
<211> 42
<212> DNA
CA 02316755 2000-10-26
99
<213> SYNTHETIC
<400> 13
ggtggttcct ctagatcttcccagtcggtgaaggagtccgag 42
<210> 14
<211> 42
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 28
<223> The letter stands or t.
"y" for c
<400> 14
ggtggttcct ctagatcttcccagtcgytggaggagtccggg 42
<210> 15
<211> 44
2 <212> DNA
O
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 25 , 34
<223> The letter stands or g.
"s" for c
The letter "r" stands or g.
for a
<400> 15
ggtggttcct ctagatcttcccagsagcagctgrtggagtccgg 44
30 <210> 16
<211> 46
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 29
<223> The letter stands or t.
"y" for c
<400> 16
cctggccggc ctggccactagtgactgayggagccttaggttgccc 46
40
<210> 17
<211> 41
<212> DNA
<213> SYNTHETIC
<400> 17
gaggaggagg aggaggaggcggggcccaggcggccgagctc 41
<210> 18
<211> 41
50 <212> DNA
<213> SYNTHETIC
<400> 18
gaggaggagg aggaggagcctggccggcctggccactagtg 41
<210> 19
<211> 42
<212> DNA
<213> SYNTHETIC
<400> 19
6 agatggtgca gccacagttcgtttgatttccacattggtgcc 42
0
CA 02316755 2000-10-26
100
<210> 20
<211> 42
<212> DNA
<213> SYNTHETIC
<400> 20
agatggtgca gccacagttcgtaggatctccagctcggtccc 42
<210> 21
<211> 42
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 28
<223> The letter stands or g.
"s" for c
<400> 21
agatggtgca gccacagttgctttgacsaccacctcggtccc 42
<210> 22
<211> 45
<212> DNA
<213> SYNTHETIC
<400> 22
agatggtgca gccacagttcggcctgtgacggtcagctgggtccc 45
<210> 23
<211> 24
3 <212> DNA
O
<213> SYNTHETIC
<400> 23
cgaactgtgg ctgcaccatctgtc 24
<210> 24
<211> 21
<212> DNA
<213> SYNTHETIC
<400> 24
4 ggccatggct ggttgggcagc 21
0
<210> 25
<211> 42
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 40
<223> The letter stands
"r" for a
or g.
50 <400> 25
gctgcccaac cagccatggcccagtcggtggaggagtccrgg 42
<210> 26
<211> 42
<212> DNA
<213> SYNTHETIC
<400> 26
gctgcccaac cagccatggcccagtcggtgaaggagtccgag 42
60 <210> 27
<211> 42
<212> DNA
CA 02316755 2000-10-26
101
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 28
<223> The letter "y" stands for c or t.
<400> 27
gctgcccaac cagccatggc ccagtcgytg gaggagtccg gg 42
<210> 28
<211> 44
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 25, 34
<223> The letter "s" stands for c or g.
The letter "r" stands for a or g.
<400> 28
gctgcccaac cagccatggc ccagsagcag ctgrtggagt ccgg 44
<210> 29
<211> 45
<212> DNA
<213> SYNTHETIC
<220>
<221> misc_feature
<222> 25, 30
3 0 <223> The "r" stands for a or g.
The letter "y" stands for c or t.
<400> 29
cgatgggccc ttggtggagg ctgargagay ggtgaccagg gtgcc 45
<210> 30
<211> 24
<212> DNA
<213> SYNTHETIC
<400> 30
4 0 gcctccacca agggcccatc ggtc 24
<210> 31
<211> 21
<212> DNA
<213> SYNTHETIC
<400> 31
agaagcgtag tccggaacgt c 21
<210> 32
50 <211> 21
<212> DNA
<213> SYNTHETIC
<400> 32
gctgcccaac cagccatggc c 21
<210> 33
<211> 39
<212> DNA
<213> SYNTHETIC
60 <400> 33
gaggaggagg aggaggagag aagcgtagtc cggaacgtc 39
CA 02316755 2000-10-26
102
<210> 34
<211> 15
<212> DNA
<213> SYNTHETIC
<400> 34
ggaagaagag gaacc 15
<210> 35
<211> 21
<212> DNA
<213> SYNTHETIC
<400> 35
ggtggttcgt ctagatcttc c 21
<210> 36
<211> 33
<212> DNA
2 0 <213> SYNTHETIC
<400> 36
cccaccaccg cccgagccac cgccaccaga gga 33