Note: Descriptions are shown in the official language in which they were submitted.
CA 02325593 2000-12-06
990200 BT / AL
1
Process for the fermentative preparation of L-amino acids
using coryneform bacteria
The invention relates to a process for the fermentative
preparation of L-amino acids, in particular L-threonine,
using coryneform bacteria in which the glyA gene is
attenuated.
Prior art
L-Amino acids are used in animal nutrition, in human
medicine and in the pharmaceuticals industry.
It is known that amino acids are prepared by fermentation
from strains of coryneform bacteria, in particular
Corynebacterium glutamicum. Because of their great
importance, work is constantly being undertaken to improve
the preparation processes. Improvements to the processes
can relate to fermentation measures, such as e. g. stirring
and supply of oxygen, or the composition of the nutrient -
media, such as e. g. the sugar concentration during the
fermentation, or the working up to the product form by
e. g. ion exchange chromatography, or the intrinsic output
properties of the microorganism itself.
Methods of mutagenesis, selection and mutant selection are
used to improve the output properties of these
microorganisms. Strains which are resistant to
antimetabolites, such as e. g. the threonine analogue a-
amino-(3-hydroxyvaleric acid (AHV), or are auxotrophic for
metabolites of regulatory importance and produce L-amino
acids such as e. g. threonine are obtained in this manner.
Methods of the recombinant DNA technique have also been
employed for some years for improving the strain of
Corynebacterium glutamicum strains which produce L-amino
acids, by amplifying individual amino acid biosynthesis
genes and investigating the effect on the L-amino acid
--'-CA 02325593 2000-12-06
990200 BT / AI,
2
production. Review articles in this context are to be
found, inter alia, in Kinoshita ("Glutamic Acid Bacteria",
in: Biology of Industrial Microorganisms, Demain and
Solomon (Eds.), Benjamin Cummings, London, UK, 1985, 115-
142), Hilliger (BioTec 2, 40-44 (1991)), Eggeling (Amino
Acids 6, 261-272 (1994)), Jetten and Sinskey (Critical
Reviews in Biotechnology 15, 73-103 (1995)) and Sahm et al.
(Annuals of the New York Academy of Science 782, 25-39
(1996) ) .
Object of the invention
The inventors had the object of providing new principles
for improved processes for the fermentative preparation of
L-amino acids with coryneform bacteria.
Description of the invention
L-Amino acids are used in human medicine and in the
pharmaceuticals industry, in the foodstuffs industry and
especially in animal nutrition. There is therefore a
general interest in providing new improved processes for
the preparation of amino acids.
Where L-amino acid is mentioned below, this means L-
threonine or L-isoleucine.
The invention provides a process for the fermentative
preparation of L-amino acids using coryneform bacteria in
which at least the nucleotide sequence which codes for the
glyA gene product (glyA gene) is attenuated, in particular
expressed at a low level, the desired product is
concentrated in the medium or in the cells and the L-amino
acid is isolated.
The strains employed preferably already produce L-amino
acids before attenuation of the glyA gene.
CA 02325593 2000-12-06 - w - --
990200 BT / AL
3
Preferred embodiments are to be found in the claims.
The term "attenuation" in this connection describes the
reduction or elimination of the intracellular activity of
one or more enzymes (proteins) in a microorganism which are
coded by the corresponding DNA (here the glyA gene), for
example by using a weak promoter or using a gene or allele
which codes for a corresponding enzyme with a low activity
or inactivates the corresponding gene or enzyme (protein),
and optionally combining these measures.
The microorganisms to which the present invention relates
can prepare amino acids from glucose, sucrose, lactose,
fructose, maltose, molasses, starch, cellulose or from
glycerol and ethanol. They can be representatives of
coryneform bacteria, in particular of the genus
Corynebacterium. Of the genus Corynebacterium, there may be
mentioned in particular the species Corynebacterium
glutamicum, which is known among experts for its ability to
produce L-amino acids.
Suitable strains of the genus Corynebacterium, in
particular of the species Corynebacterium glutamicum, are
in particular the known wild-type strains
Corynebacterium glutamicum ATCC13032
Corynebacterium acetoglutamicum ATCC15806
Corynebacterium acetoacidophilum ATCC13870
Corynebacterium melassecola ATCC17965
Corynebacterium thermoaminogenes FERM BP-1539
Brevibacterium flavum ATCC14067
Brevibacterium lactofermentum ATCC13869 and
Brevibacterium divaricatum ATCC14020
and L-amino acid-producing mutants or strains prepared
therefrom
such as, for example, the L-threonine-producing strains
Corynebacterium glutamicum ATCC21649
CA 02325593 2000-12-06
990200 BT / AL
9
Brevibacteriurn flavurn BB69
Brevibacterium flavum DSM5399
Brevibacterium lactofermentum FERM-BP 269
Brevibacterium lactofermentum TBB-10 and
such as, for example, the L-isoleucine-producing strains
Corynebacterium glutamicum ATCC 19309
Corynebacterium glutamicum ATCC 14310
Corynebacterium glutamicum ATCC 14311
Corynebacterium glutamicum ATCC 15168
Corynebacterium ammoniagenes ATCC 6871.
It has been found that coryneform bacteria produce L-amino
acids in an improved manner after attenuation of the glyA
gene.
The glyA gene codes for the enzyme serine
hydroxymethyltransferase (EC 2.1.2.1). The nucleotide
sequence of the glyA gene has been described in Japanese
Laid-Open Specification JP-A-08107788. The glyA gene -
described in the text reference mentioned can be used
according to the invention. Alleles of the glyA gene which
result from the degeneracy of the genetic code or due to
sense mutations of neutral function can furthermore be
used.
To achieve an attenuation, either the expression of the
glyA gene or the catalytic properties of the gene product
can be reduced or eliminated. The two measures are
optionally combined.
The gene expression can be reduced by suitable culturing or
by genetic modification (mutation) of the signal structures
of gene expression. Signal structures of gene expression
are, for example, repressor genes, activator genes,
operators, promoters, attenuators, ribosome binding sites,
the start codon and terminators. The expert can find
information on this e. g. in the patent application WO
CA 02325593 2000-12-06
990200 BT / AL
96/15246, in Boyd and Murphy (Journal of Bacteriology 170:
5949 (1988)), in Voskuil. and Chambliss (Nucleic Acids
Research 26: 3548 (1998), in Jensen and Hammer
(Biotechnology and Bioengineering 58: 191 (1998)), in Patek
5 et al. (Microbiology 142: 1297 (1996)) and in known
textbooks of genetics and molecular biology, such as e. g.
the textbook by Knippers ("Molekulare Genetik" [Molecular
Genetics], 6th edition, Georg Thieme Verlag, Stuttgart,
Germany, 1995) or that by Winnacker ("Gene and Klone"
[Genes and Clones], VCH Verlagsgesellschaft, Weinheim,
Germany, 1990).
Mutations which lead to a change or reduction in the
catalytic properties of enzyme proteins are known from the
prior art; examples which may be mentioned are the works by
Qiu and Goodman (Journal of Biological Chemistry 272: 8611-
8617 (1997)), Sugimoto et al. (Bioscience Biotechnology and
Biochemistry 61: 1760-1762 (1997)) and Mockel ("Die
Threonindehydratase aus Corynebacterium glutamicum:
Aufhebung der allosterischen Regulation and Struktur des
Enzyms" [Threonine dehydratase from Corynebacterium
glutamicum: Cancelling the allosteric regulation and
structure of the enzyme], Reports from the Julich Research
Centre,Jul-2906, ISSN09442952, Julich, Germany, 1994).
Comprehensive descriptions can be found in known textbooks
of genetics and molecular biology, such as e. g. that by
Hagemann ("Allgemeine Genetik" [General Genetics], Gustav
Fischer Verlag, Stuttgart, 1986).
Possible mutations are transitions, transversions,
insertions and deletions. Depending on the effect of the
amino acid exchange on the enzyme activity, missense
mutations or nonsense mutations are referred to. Insertions
or deletions of at least one base pair in a gene lead to
frame shift mutations, as a consequence of which incorrect
amino acids are incorporated or translation is interrupted
prematurely. Deletions of several colons typically lead to
w ~ CA 02325593 2000-12-06w
990200 BT / AL
6
a complete loss of the enzyme activity. Instructions on
generation of such mutations are prior art and can be found
in known textbooks of genetics and molecular biology, such
as e. g. the textbook by Knippers ("Molekulare Genetik"
[Molecular Genetics], 6th edition, Georg Thieme Verlag,
Stuttgart, Germany, 1995), that by Winnacker ("Gene and
Klone" [Genes and Clones], VCH Verlagsgesellschaft,
Weinheim, Germany, 1990) or that by Hagemann ("Allgemeine
Genetik" [General Genetics], Gustav Fischer Verlag,
Stuttgart, 1986).
By way of example, the glyA gene was attenuated by removal
of the natural promoter and insertion of a regulatable
control element lying upstream. The lacI-tac system was
used as the control element. To be able to achieve
incorporation of the lacI-tac system upstream of the
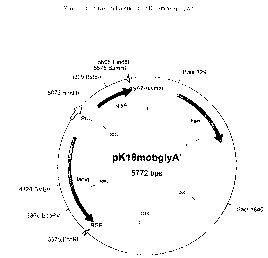
chromosomal glyA gene, the integration plasmid pKl8mobglyA'
(figure 1) was prepared. The plasmid pKl8mobglyA' contains
the tac promoter (Amann et al., Gene 25: 167-178 (1983); De
Boer et al., Proceedings of the National Academy of
Sciences of the United States of America USA 80: 21-25
(1983)) and directly downstream of the tac promoter a 5'-
terminal sequence of the glyA gene shown in SEQ ID No 1.
The plasmid furthermore contains the lacI gene which codes
for the Lac inhibitor (Farabaugh, Nature 274: 765-769
(1978); Stark et al., Gene 51: 255-267 (1987)). The
sequence of the lacI-tac-5'glyA unit is shown in SEQ ID No
2. Plasmid pKl8mobglyA' is capable of replication in
Escherichia coli but not in Corynebacterium glutamicum.
After transformation and homologous recombination by means
of a "cross-over" event which effects integration, an
intact copy of the glyA gene, expression of which can be
controlled or regulated by the lacI-tac control element
lying upstream, and an inactive copy of the glyA gene
truncated on the 3'-terminus, including the natural
promoter, are obtained. The sequence of the lacI-tac-glyA
unit is shown in SEQ ID No 3. SEQ ID No 9 shows the known
CA 02325593 2000-12-06 -- -
990200 BT / AL
7
amino acid sequence of the glyA gene product. By addition
of suitable concentrations of the lactose analogue
isopropyl thiogalactoside (Fiirste et al., Gene 48: 119-131
(1986)), the expression of the glyA gene can be controlled
or the cell content of serine hydroxymethyltransferase can
be attenuated or adjusted.
Further instructions and explanations on integration
mutagenesis are to be found, for example, in Schwarzer and
Piihler (Bio/Technology 9,84-87 (1991)), Peters-Wendisch et
al. (Microbiology 144, 915-927 (1998)) or Fitzpatrick et
al. (Applied Microbiology Biotechnology 42, 575-580
(1994)).
An example of an amino acid-producing strain of coryneform
bacteria with an attenuated glyA gene is the threonine
producer Corynebacterium glutamicum DM368-2::pK18mobglyA'.
In addition, it may be advantageous for the production of
amino acids to amplify one or more enzymes of the -
particular biosynthesis pathway, of glycolysis, of
anaplerosis, of the citric acid cycle or of amino acid
2C export, in addition to attenuation of the glyA gene.
Thus, for example, for the preparation of L-threonine
~ at the same time the hom gene which codes for homoserine
dehydrogenase (Peoples et al., Molecular Microbiology 2,
63-72 (1988)) or the homdr allele which codes for a "feed
back resistant" homoserine dehydrogenase (Archer et al.,
Gene 107, 53-59 (1991)) and/or
~ at the same time the gap gene which codes for
glyceraldehyde 3-phosphate dehydrogenase (Eikmanns et
al., Journal of Bacteriology 174: 6076-6086 (1992)), or
~ at the same time the pyc gene which codes for pyruvate
carboxylase (Peters-Wendisch et al., Microbiology 144:
915-927 (1998)), or
CA 02325593 2000-12-06
990200 BT / AL
8
~ at the same time the mqo gene which codes for
malate:quinone oxidoreductase (Molenaar et al., European
Journal of Biochemistry 254, 395-403 (1998)), or
~ at the same time the thrE gene which codes for threonine
export (DE 199 41 478.5; DSM 12840)
can be over-expressed.
For the production of amino acids it may furthermore be
advantageous to attenuate, in addition to the glyA gene,
~ the pck gene which codes for phosphoenol pyruvate
carboxykinase (DE 199 50 409.1; DSM 13047) and/or
~ the poxB gene which codes for pyruvate oxidase
(DE 199 51 975.7; DSM 13114)
at the same time.
Finally, in addition to attenuation of the glyA gene it may
be advantageous for the production of amino acids to
eliminate undesirable side reactions (Nakayama: "Breeding
of Amino Acid Producing Microorganisms", in: Overproduction
of Microbial Products, Krumphanzl, Sikyta, Vanek (eds.),
Academic Press, London, UK, 1982).
The culture medium to be used must meet the requirements of
the particular strains in a suitable manner. Descriptions
of culture media for various microorganisms are contained
in the handbook "Manual of Methods for General
Bacteriology" of the American Society for Bacteriology
(Washington D.C., USA, 1981). Sugars and carbohydrates,
such as e. g. glucose, sucrose, lactose, fructose, maltose,
molasses, starch and cellulose, oils and fats, such as
e. g. Soya oil, sunflower oil, groundnut oil and coconut
fat, fatty acids, such as e. g. palmitic acid, stearic acid
and linoleic acid, alcohols, such as e. g. glycerol and
ethanol, and organic acids, such as e. g. acetic acid, can
CA 02325593 2000-12-06
990200 BT / AL
9
be used as the source of carbon. These substance can be
used individually or as a mixture. Organic nitrogen-
containing compounds, such as peptones, yeast extract, meat
extract, malt extract, corn steep liquor, soya bean flour
and urea, or inorganic compounds, such as ammonium
sulphate, ammonium chloride, ammonium phosphate, ammonium
carbonate and ammonium nitrate, can be used as the source
of nitrogen. The sources of nitrogen can be used
individually or as a mixture. Phosphoric acid, potassium
dihydrogen phosphate or dipotassium hydrogen phosphate or
the corresponding sodium-containing salts can be used as
the source of phosphorus. The culture medium must
furthermore comprise salts of metals, such as e. g.
magnesium sulfate or iron sulfate, which are necessary for
growth. Finally, essential growth substances, such as amino
acids and vitamins, can be employed in addition to the
abovementioned substances. Suitable precursors can moreover
be added to the culture medium. The starting substances
mentioned can be added to the culture in the form of a
single batch, or can be fed in during the culture in a
suitable manner.
Basic compounds, such as sodium hydroxide, potassium
hydroxide, ammonia or aqueous ammonia, or acid compounds,
such as phosphoric acid or sulfuric acid, can be employed
in a suitable manner to control the pH. Antifoams, such as
e. g. fatty acid polyglycol esters, can be employed to
control the development of foam. Suitable substances having
a selective action, such as e. g. antibiotics, can be added
to the medium to maintain the stability of plasmids. To
maintain aerobic conditions, oxygen or oxygen-containing
gas mixtures, such as e. g. air, are introduced into the
culture. The temperature of the culture is usually 20°C to
45°C, and preferably 25°C to 40°C. Culturing is continued
until a maximum of the desired product has formed. This
3'> target is usually reached within 10 hours to 160 hours.
' CA 02325593 2000-12-06
990200 BT / AL
Methods for the determination of L-amino acids are known
from the prior art. The analysis can thus be carried out as
described by Spackman et al. (Analytical Chemistry, 30,
(1958), 1190) by anion exchange chromatography with
5 subsequent ninhydrin derivatization, or it can be carried
out by reversed phase HPLC, for example as described by
Lindroth et al. (Analytical Chemistry (1979) 51: 1167-
1174).
The following microorganism has been deposited at the
10 Deutsche Sammlung fur Mikrorganismen and Zellkulturen (DSMZ
- German Collection of Microorganisms and Cell Cultures,
Braunschweig, Germany) in accordance with the Budapest
Treaty:
~ Escherichia coli strain DHSa.mcr/pKl8mobglyA' as
DSM 13170
CA 02325593 2000-12-06
990200 BT / AL
11
Examples
The present invention is explained in more detail in the
following with the aid of embodiment examples.
The isolation of plasmid DNA from Escherichia coli and all
techniques of restriction, Klenow and alkaline phosphatase
treatment were carried out by the method of Sambrook et al.
(Molecular cloning. A laboratory manual (1989) Cold Spring
Harbour Laboratory Press). The transformation of
Escherichia coli was carried out by the method of Chung et
al. (Proceedings of the National Academy of Sciences of the
United States of America USA (1989) 86: 2172-2175), unless
described otherwise.
Example 1
Cloning and sequencing of the glyA gene from
Corynebacterium glutamicum ATCC13032
The glyA gene was cloned in the E. coli cloning vector
pUCl8 (Norrander et al., Gene (1983) 26: 101-106, Roche
Diagnostics, Mannheim, Germany). The cloning was carried
out in two steps. The gene from Corynebacterium glutamicum
ATCC13032 was first amplified by a polymerase chain
reaction (PCR) by means of the following oligonucleotide
primers derived from Japanese Laid-Open Specification JP-A-
08107788.
glyAl-forward:
5'-GCT TGC AGC GTT TTG CTC TGC C-3'
glyAl-reverse:
5'-ACC CGT AAC CTC TTC CAC ATA GG-3'
CA 02325593 2000-12-06
990200 BT / AL
ZL
The PCR reaction was carried out in 30 cycles in the
presence of 200 ~M deoxynucleotide triphosphates (dATP,
dCTP, dGTP, dTTP), in each case 1 uM of the corresponding
oligonucleotide, 100 ng chromosomal DNA from
Corynebacterium glutamicum ATCC13032, 1/10 volume 10-fold
reaction buffer and 2.6 units of a heat-stable Taq-/Pwo-DNA
polymerase mixture (Expand High Fidelity PCR System from
Roche Diagnostics, Mannheim, Germany) in a Thermocycler
(PTC-100, MJ Research, Inc., Watertown, USA) under the
following conditions: 94°C for 30 seconds, 64°C for 1
minute and 68°C for 3 minutes.
The amplified fragment about 1.7 kb in size was then
subsequently ligated with the aid of the SureClone Ligation
Kit (Amersham Pharmacia Biotech, Uppsala, Sweden) into the
SmaI cleavage site of the vector pUCl8 in accordance with
the manufacturer's instructions. The E. coli strain DHSamcr
(Grant et al., Proceedings of the National Academy of
Sciences of the United States of America USA (1990) 87:
4645-4649) was transformed with the entire ligation batch.
Transformants were identified with the aid of their
carbenicillin resistance on LB-agar plates containing
50 ug/mL carbenicillin. The plasmids were prepared from 7
of the transformants and checked for the presence of the
1.7 kb PCR fragment as an insert by restriction analysis.
The recombinant plasmid formed in this way is called
pUCl8glyA in the following.
The nucleotide sequence of the 1.7 kb PCR fragment in
plasmid pUCl8glyA was determined by the dideoxy chain
termination method of Sanger et al. (Proceedings of the
National Academy of Sciences of the United States of
America USA (1977) 74: 5463-5467). For this, the complete
insert of pUCl8glyA was sequenced with the aid of the
following primers.
Universal primer: 5'-GTA AAA CGA CGG CCA GT-3'
CA 02325593 2000-12-06
990200 BT / AL
13
Reverse primer: 5'-GGA AAC AGC TAT GAC CAT G-3'
The nucleotide sequences obtained were analysed with the
Lasergene program package (Biocomputing Software for
Windows, DNASTAR, Madison, USA). The result of the analysis
was identification of an open reading frame of 1302 by in
length. The corresponding gene was called the glyA gene.
The associated gene product comprises 434 amino acids and
is reproduced as SEQ ID No 4.
Example 2
Construction of a vector for reduced expression of glyA
A DNA fragment 1418 by in size which contains the glyA gene
without its own promoter region was cut out of the plasmid
pUCl8glyA described under example 1 with the restriction
enzymes EcoRI and TfiI. The 5' and 3' ends of this fragment
were treated with Klenow enzyme. The resulting DNA fragment
was ligated in the vector pVWEx2, previously linearized
with BamHI, treated with Klenow enzyme and dephosphorylated
(Wendisch, "Physiologische and NMR-spektroskopische
Untersuchungen zur in vivo-Aktivitat zentraler
Stoffwechselwege im Wildstamm and in rekombinanten Stammen
von Corynebacterium glutamicum" [Physiological and NMR-
spectroscopic analyses of the in vivo activity of central
metabolic pathways in the wild-type strain and in
recombinant strains of Corynebacterium glutamicum], Reports
from the Jiilich Research Centre, Jiil-3397, ISSN09442952,
Jiilich, Germany, 1997), such that the glyA gene lies in the
same orientation directly after the tac promoter of the
vector which can be induced with isopropyl (3-D-
thiogalactoside (IPTG). The E. coli strain DHSamcr (Grant
et al., Proceedings of the National Academy of Sciences of
the United States of America USA (1990) 87: 4645-4699) was
transformed with the entire ligation batch. Transformants
CA 02325593 2000-12-06
990200 BT / AI.
14
were identified with the aid of their tetracycline
resistance on LB-agar plates containing 15 ug/mL
tetracycline. The plasmids were prepared from 12
transformants and checked for the presence of the 1418 by
fragment as an insert in the correct orientation with
respect to the tac promoter by restriction analysis. The
recombinant plasmid formed in this manner is called
pVWEx2glyA in the following.
A DNA fragment which contains lacI, the gene for the
repressor of the tac promoter, the tac promoter and the
first 438 by of the cloned glyA gene of corynebacterium
glutamicum was then amplified from the plasmid pVWEx2glyA
by a polymerase chain reaction (PCR) by means of the
following oligonucleotide primers.
glyA2-forward (with the attached EcoRI recognition sequence
identified by underlining):
5'-CCG GAA TTC TCA CTG CCC GCT TTC CAG TC-3'
glyA2-reverse (with the attached BamHI recognition sequence
identified by underlining):
5'-CGG GAT CCC AGC TTT CCG GAG AAG TTC AAC-3'
The PCR reaction was carried out in 30 cycles in the
presence of 200 ~.M deoxynucleotide triphosphates (dATP,
dCTP, dGTP, dTTP), in each case 1 uM of the corresponding
oligonucleotide, 100 ng plasmid DNA of pVWEx2glyA, 1/10
volume of 10-fold reaction buffer and 2.6 units of a heat-
stable Taq-/Pwo-DNA polymerase mixture (Expand High
Fidelity PCR System from Roche Diagnostics, Mannheim,
Germany) in a Thermocycler (PTC-100, MJ Research, Inc.,
Watertown, USA) under the following conditions: 99°C for 30
seconds, 58°C for 30 seconds and 72°C for 2 minutes.
The amplified fragment about 2.0 kb in size was
subsequently digested with EcoRI and BamHI, isolated with
CA 02325593 2000-12-06
990200 BT / AL
the aid of the NucleoSpin Extract 2 in 1 Kit from Macherey-
Nagel (Diiren, Germany) in accordance with the
manufacturer's instructions and then ligated in the vector
pKl8mob, which had also been cleaved with EcoRI and BamHI
and dephosphorylated (Schafer et al., Gene (1994) 145: 69-
73). The E. coli strain DHSamcr (Grant et al., Proceedings
of the National Academy of Sciences of the United States of
America USA (1990) 87: 4645-4649) was transformed with the
entire ligation batch. Transformants were identified with
10 the aid of their kanamycin resistance on LB-agar plates
containing 50 ug/mL kanamycin. The plasmids were prepared
from 12 of the transformants and checked for the presence
of the 2.0 kb PCR fragment as an insert by restriction
analysis. The recombinant plasmid formed in this way is
15 called pKlBmobglyA' in the following (see figure 1).
Example 3
Construction of the strain Corynebacterium glutamicum
ATCC13032::pK18mobglyA' with reduced, regulatable glyA
expression
By means of electroporation (Haynes et al., FEMS
Microbiology Letters (1989) 61: 329-334), the blank vector
pZl (Menkel et al., Applied and Environmental Microbiology
(1989) 64: 549-554) and the plasmid pKl8mobglyA' described
2~, in example 2 were introduced into the wild-type strain
Corynebacterium glutamicum ATCC13032 (Abe et al., Journal
of General and Applied Microbiology (1967) 13: 279-301).
After transformation with pZl, the transformants were
identified with the aid of their kanamycin resistance on
LBWS-agar places containing 15 ~,g/mL kanamycin (Liebl et
al., fEMS Microbiology Letters (1989) 65: 299-304). The
plasmids were prepared from 3 of the transformants and
checked for the presence of the pZl blank vector by
CA 02325593 2000-12-06
990200 BT / AL
16
restriction analysis. The control strain Corynebacterium
glutamicum ATCC13032/pIl was formed in this manner.
After transformation with pKlBmobglyA' the plasmid had to
integrate into the chromosome of Corynebacterium glutamicum
ATCC13032 via homologous recombination of the cloned 5' end
of glyA. The kanamycin-resistant clones obtained were
identified on LBWS-agar places containing 15 ~g/mL
kanamycin and 1 mM isopropyl (3-D-thiogalactoside (IPTG)
(Liebl et al., FEMS Microbiology Letters (1989) 65: 299-
304). Correct integration of pKl8mobglyA' in the chromosome
was checked in 2 resulting integration mutants by a
polymerase chain reaction (PCR) by means of the following
oligonucleotide primers.
Reverse primer (RSP):
5'-GGA AAC AGC TAT GAC CAT G-3'
glyA2-reverse:
5'-CGG GAT CCC AGC TTT CCG GAG AAG TTC AAC-3'
The PCR reaction was carried out in 30 cycles in the
presence of 200 uM deoxynucleotide triphosphates (dATP,
dCTP, dGTP, dTTP), in each case 1 uM of the corresponding
oligonucleotide, 100 ng chromosomal DNA from
Corynebacterium glutamicum ATCC13032::pK18mobglyA', 1/10
volume 10-fold reaction buffer and 2.6 units of a heat-
stable Taq-/Pwo-DNA polymerase mixture (Expand High
Fidelity PCR System from Roche Diagnostics, Mannheim,
Germany) in a Thermocycler (PTC-100, MJ Research, Inc.,
Watertown, USA) under the following conditions: 94°C for 30
seconds, 48°C for 30 seconds and 72°C for 2 minutes. The
strain Corynebacterium glutamicum ATCC13032::pK18mobglyA',
in which the glyA gene is present under the control of the
tac promoter which can be induced with isopropyl (3-D-
thiogalactoside (IPTG), was formed in this manner.
CA 02325593 2000-12-06
990200 BT / AL
17
Example 9
Determination of the serine hydroxymethyltransferase
activity coded by the glyA gene in the strain
Corynebacterium glutamicum ATCC13032::pK18mobglyA'
To obtain the crude extracts for determination of the
serine hydroxymethyltransferase activity coded by glyA, the
strains C. glutamicum ATCC13032/pZl and C. glutamicum
ATCC13032::pK18mobglyA' described in example 3 were
precultured in 100 mL Brain Heart Infusion-Medium (Difco
Laboratories, Detroit, USA) with 25 ~g kanamycin/mL and
100 ~M isopropyl ~-D-thiogalactoside (IPTG) for 14 hours at
30°C. The cells were then washed once with 0.9o(w/v) sodium
chloride solution and 100 mL portions of CgXII medium were
inoculated with this suspension such that the OD6oo (optical
1~~ density at 600 nm) was 0.5. The medium was identical to the
medium described by Keilhauer et al. (Journal of
Bacteriology (1993) 175: 5593-5603), but additionally
comprised 25 ~g kanamycin/mL and 0, 10 or 100 ~M isopropyl
~-D-thiogalactoside (IPTG). The composition of the medium
described by Keilhauer et al. is shown in table 1.
CA 02325593 2000-12-06
990200 BT / AL
1 f3
Table 1
Composition of medium CGXII
Component Concentration
(NH4 ) 2SOa 20
g /L
Urea 5 g/L
KHzPOq 1 g/L
K2HP0q 1 g/L
MgSOq x 7 HZO 0.25 g/L
3-Morpholinopropanesulfonic acid 42 g/L
CaClz 10 mg/L
FeSOq x 7 H20 10 mg/L
MnS09 x HZO 10 mg/L
ZnS04 x 7H20 1 mg/L
CuSOq 0.2 mg/L
NiCl2 x 6 Hz0 0.02 mg/L
Biotin 0.2 mg/L
Glucose 40 g/L
Protocatechuic acid 30 mg/L
Culturing of the two strains was carried out at 30°C. After
S 10 hours, the cells were washed once with 50 mM 4-(2-
hydroxyethyl)-1-piperazinethanesulfonic acid/sodium
hydroxide buffer (pH 7.0) gewaschen, centrifuged off (10
minutes at 5000 revolutions per minute with a Minifuge RF
from Heraeus, Osterode, Germany) and resuspended in 200 mM
4-(2-hydroxyethyl)-1-piperazinethanesulfonic acid/sodium
hydroxide buffer (pH 7.0) such that the final volume was
5 mL. 50 ~L 2 mM pyridoxal 5-phosphate solution and 50 ~.L
100 mM dithiothreitol solution were added to this cell
suspension and the cells were broken down. The cells were
1_'- broken down at 0°C by an ultrasonic disintegrator (Branson
Sonifier W-250, Branson Sonic Power Co, Danbury, USA;
ultrasonic exposure time 6 minutes, pulse length 1000,
CA 02325593 2000-12-06
990200 BT / AL
19
ultrasonic intensity 2.5). After the ultrasonic treatment,
the cell debris was separated off by centrifugation (30
minutes at 4°C and 13000 revolutions per minute in a
coolable Sigma 202 MK centrifuge from Sigma-Aldrich,
S Deisenhofen, Germany). The supernatant was employed
directly as the cell-free crude extract for determination
of the enzyme activity.
The protein determination in the cell-free crude extracts
was carried out photometrically by the method of Bensadoun
and Weinstein (Analytical Biochemistry (1976) 70: 241-250).
The protein content was determined here via a calibration
curve plotted with bovine serum albumin as the standard.
To determine the activity of the serine
hydroxymethyltransferase in the cell-free crude extracts, a
discontinuous enzyme test in which the glycine form from
the substrate threonine was quantified was used. The
reaction batches were incubated in the following
composition (modified according to Scrimgeour and
Huennekens, Methods in Enzymology (1962) Vol. V: 838-843,
Academic Press) for 15 minutens at 37°C inkubiert: 20 mM
threonine, 200 ~M pyridoxal 5-phosphate, 900 ~M
tetrahydrofolate, 100 mM 4-(2-hydroxyethyl)-1-
piperazinethanesulfonic acid/sodium hydroxide buffer (pH
7.0) and 1.0-1.5 mg protein (from the crude extract) in a
final volume of 1 mL. The reaction was stopped by addition
of 0.25 volume 25o (w/v) trichloroacetic acid solution, the
batches were incubated for 15 minutes at 0°C and the
denatured protein was centrifuged off (15 minutes at 4°C
and 13000 revolutions per minute in a coolable Sigma 202 MK
centrifuge from Sigma-Aldrich, Deisenhofen, Germany). The
quantitative determination of the glycine formed in the
enzyme test from the supernatant was carried out by means
of reversed phase HPLC (Lindroth et al., Analytical
Chemistry (1979) 51: 1167-1174). An HPLC apparatus of the
HP1100 series (Hewlett-Packard, Waldbronn, Germany)
CA 02325593 2000-12-06
990200 BT / AL
connected to a fluorescence detector (G1321A) was used; the
system was controlled and the data evaluated with an HP-
Chem-Station (Hewlett-Packard). 1 ~L of the amino acid
solution to be analysed was mixed in an automatic precolumn
derivatization with 20 ~L ortho-phthalaldehyde/2-
mercaptoethanol ready-to-use reagent (Pierce Europe BV,
Oud-Beijerland, The Netherlands). The fluorescent thio-
substituted isoindoles formed here (Jones et al., Journal
of Chromatography (1983) 266: 471-482) were separated over
10 a combined precolumn (40x4 mm Hypersil ODS 5) and main
column (Hypersil ODS 5, both columns from CS-
Chromatographie Service GmbH, Langerwehe, Germany) with a
gradient programme with an increasingly non-polar phase
(methanol). The polar eluent was sodium acetate (0.1 molar,
15 pH 7.2); the flow rate was 0.8 mL per minute. Flourescence
detection of the derivatized amino acids took place at an
excitation wavelength of 230 nm and an emission wavelength
of 450 nm. The glycine concentrations were calculated via a
comparison with an external standard and asparagine as an -
20 additional internal standard.
The results of the enzyme test with threonine as the
substrate are listed in table 2.
Table 2:
Strain IPTG Serine
concentrationhydroxymethyltransferase
(uM) activity (nmol
glycine/minute/mg protein)
ATCC13032/pZl
0 0.9
0 0.3
ATCC1.3032::pKlBmobglyA'
10 0.7
CA 02325593 2000-12-06
990200 BT / AL
21
Example 5
Construction of the strain Brevibacterium flavum DM368-
2::pK18mobglyA' with reduced regulatable glyA expression
By means of electroporation (Haynes et al., FEMS
Microbiology Letters (1989) 61: 329-334), the blank vector
pZl (Menkel et al., Applied and Environmental Microbiology
(1989) 64: 549-554) and the plasmid pKl8mobglyA' described
in example 2 were introduced into the threonine-forming
strain Brevibacterium flavum DM368-2. The strain DM368-2 is
described in EP-B-0 385 940 and deposited as DSM5399.
After transformation with pZl, the transformants were
identified with the aid of their kanamycin resistance on
LBWS-agar plates containing 15 ~g/mL kanamycin (Liebl et
al., FEMS Microbiology Letters (1989) 65: 299-304). The
plasmids were prepared from 3 of the transformants and
checked for the presence of the pZl blank vector by
restriction analysis. The control strain Brevibacterium
flavum DM368-2/pZl was formed in this manner.
After transformation with pKl8mobglyA' the plasmid had to
integrate into the chromosome of Brevibacterium flavum
DM368-2 via homologous recombination of the cloned 5' end
of glyA. The kanamycin-resistant clones obtained were
identified on LBWS-agar plates containing 15 ~g/mL
kanamycin and 1 mM isopropyl a-D-thiogalactoside (Liebl et
al., FEMS Microbiology Letters (1989) 65: 299-304). Correct
integration of pKl8mobglyA' in the chromosome was checked
in 4 resulting integration mutants by a polymerase chain
reaction (PCR), as already described in example 3, with
3(:' 100 ng chromosomal DNA of Brevibacterium flavum DM368-
2::pK18mobglyA' as the template. The strain Brevibacterium
flavum DM368-2::pK18mobglyA', in which the glyA gene is
CA 02325593 2000-12-06
990200 BT / AL
22
present under the control of the tac promoter which can be
induced with isopropyl (3-D-thiogalactoside (IPTG), was
formed in this manner.
Example 6
Determination of the serine hydroxymethyltransferase
activity coded by glyA in the strain Brevibacterium flavum
DM368-2::pK18mobglyA'
The crude extracts for the determination of the serine
hydroxymethyltransferase activity coded by glyA in the
strains B. flavum DM368-2/pZl and B. flavum DM368-
2::pK18mobglyA' described in example 5 were obtained as
already described in example 4. The protein determination
in the cell-free crude extracts obtained and the
discontinuous enzyme test, in which the glycine formed from
the substrate threonine is quantified, was [sic] likewise
carried out as described in example 4.
The results of this enzyme test with threonine as the
substrate are listed in table 3.
Table 3:
Strain IPTG Serine hydroxymethyltransferase
concentrationactivity (nmol glycine/minute/mg
(uM) protein)
DM368-2/pZl
0 1.6
0 <0.1
DM368-2::pK18mobglyA'
10 0.8
100 i.7
CA 02325593 2000-12-06
990200 BT / AL
23
Example 7
Preparation of L-threonine with Brevibacterium flavum
To investigate their threonine formation, the strains B.
flavum DM368-2/pZl and DM368-2::pK18mobglyA' described in
example 5 were precultured in 100 mL Brain Heart Infusion
Medium (Difco Laboratories, Detroit, USA) with 25 ug
kanamycin/mL and 100 ~M isopropyl (3-D-thiogalactoside
(IPTG) for 14 hours at 30°C. The cells were then washed
once with 0.9o(w/v) sodium chloride solution and 60 mL
portions of CgXII medium were inoculated with this
suspension such that the OD6oo (optical density at 600 nm)
was 0.5. The medium was identical to the medium described
by Keilhauer et al. (Journal of Bacteriology (1993) 175:
5593-5603), but additionally comprised 25 ~g kanamycin/mL
1' and 0, 10 or 100 ~,M isopropyl ~3-D-thiogalactoside (IPTG).
Culturing of the two strains was carried out at 30°C over a
period of 72 hours. After 48 and 72 hours samples were in
each case taken and the cells were centrifuged off briefly
(5 minutes at 13000 revolutions per minute with a Biofuge
pico from Heraeus, Osterode, Germany).
The quantitative determination of the extracellular amino
acid concentrations from the culture supernatant was
carried out as already described in example 4 by means of
reversed phase HPLC (Lindroth et al., Analytical Chemistry
(1979) 51: 1167-1174). The threonine concentrations were
calculated via a comparison with an external standard and
asparagine as an additional internal standard.
The results are listed in table 4.
CA 02325593 2000-12-06 w
990200 BT / AL
29
Table 4:
Strain fPTG L-Threonine
concentration(g/1)
~M 48 hours 72 hours
DM368-2/pZl
0 1.27 1.32
10 1.32 1.44
DM368-2::pK18mobglyA'
0 1.41 1.60
CA 02325593 2000-12-06
990200 BT / AL
The following figures are attached:
Figure l: Map of the plasmid pKlBmobglyA'. The length data
are to be understood as approx. values.
The abbreviations and designations used have the following
meaning.
~ BamHI: Restriction endonuclease from Bacillus
amyloliquefaciens
~ BglII: Restriction endonuclease from Bacillus globigii
~ BstEII: Restriction endonuclease from Bacillus
10 stearothermophilus
~ EcoRI: Restriction endonuclease from Escherichia coli
~ EcoRV: Restriction endonuclease from Escherichia coli
~ HindIII: Restriction endonuclease from Haemophilus
influenzae
15 ~ SacI: Restriction endonuclease from Streptomyces
achromogenes
~ kan: Kanamycin resistance gene
~ lacIq: Gene for the repressor of the tac promoter Ptac
~ Ptac: tac promoter
20 ~ glyA': 5' part of the serine hydroxymethyltransferase
gene
~ glyA2-reverse: Primer for checking an integration
~ RSP: Reverse standard primer for checking an integration
2 ~;
CA 02325593 2001-02-21
' ' 26
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT:
(A) NAME: Degussa-Hills Aktiengesellschaft
(B) STREET: Weissfrauenstrasse 9
(C) CITY: Frankfurt am Main
(D) COUNTRY: Germany
(E) POSTAL CODE (ZIP): D-60287
(i) APPLICANT:
(A) NAME: Forschungszentrum Jiilich GmbH
(B) CITY: Jiilich
(C) COUNTRY: Germany
(D) POSTAL CODE (ZIP): D-52425
(ii) TITLE OF INVENTION: PROCESS FOR THE FERMENTATIVE PREPARATION
OF L-AMINO ACIDS USING CORYNEFORM BACTERIA
(iii) NUMBER OF SEQUENCES: 4
(iv) CORRESPONDENCE ADDRESS:
(A) NAME: Marks & Clerk
(B) STREET: 280 Slater Street, Suite 1800
(C) CITY: Ottawa
(D) STATE: Ontario
(E) COUNTRY: Canada
(F) POSTAL CODE (ZIP): K1P 1C2
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Diskette
(B) COMPUTER: IBM PC
(C) OPERATING SYSTEM: MS DOS
(D) SOFTWARE: PatentIn Ver. 2.1
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER: 2,325,593
(B) FILING DATE: 2000-12-06
(C) CLASSIFICATION: Unknown
(vii) PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER: 199 59 329.9
(B) FILING DATE: 1999-12-09
(C) CLASSIFICATION: Unknown
(viii) PATENT AGENT INFORMATION:
(A) NAME: Richard J. Mitchell
(B) REGISTRATION NUMBER:
(C) REFERENCE/DOCKET NUMBER: 10036-6
(ix) TELECOMMUNICATION INFORMATION:
(A) TELEPHONE: 613-236-9561
(B) TELEFAX: 613-230-8821
(2) INFORMATION FOR SEQ ID NO.: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 438
(B) TYPE: nucleic acid
CA 02325593 2001-02-21
27
(C) STRANDEDNESS:
(D) TOPOLOGY:
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Corynebacterium glutamicum ATCC13032
(ix) FEATURE:
(A) NAME/KEY: N_region
(B) LOCATION: (1)..(438)
(C) OTHER INFORMATION: 5'glyA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO.: 1:
ATGACCGATG CCCACCAAGC GGACGATGTC CGTTACCAGC CACTGAACGA GCTTGATCCT 60
GAGGTGGCTG CTGCCATCGC TGGGGAACTT GCCCGTCAAC GCGATACATT AGAGATGATC 120
GCGTCTGAGA ACTTCGTTCC CCGTTCTGTT TTGCAGGCGC AGGGTTCTGT TCTTACCAAT 180
AAGTATGCCG AGGGTTACCC TGGCCGCCGT TACTACGGTG GTTGCGAACA AGTTGACATC 240
ATTGAGGATC TTGCACGTGA TCGTGCGAAG GCTCTCTTCG GTGCAGAGTT CGCCAATGTT 300
CAGCCTCACT CTGGCGCACA GGCTAATGCT GCTGTGCTGA TGACTTTGGC TGAGCCAGGC 360
GACAAGATCA TGGGTCTGTC TTTGGCTCAT GGTGGTCACT TGACCCACGG AATGAAGTTG 420
AACTTCTCCG GAAAGCTG 438
(2) INFORMATION FOR SEQ ID NO.: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2000
(B) TYPE: nucleic acid
(C) STRANDEDNESS:
(D) TOPOLOGY:
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Knstliche Sequenz
(ix) FEATURE:
(C) OTHER INFORMATION: Description of the synthetic sequence:
lacI-tac-5'glyA
(ix) FEATURE:
(A) NAME/KEY: gene
(B) LOCATION: Complement ((6)..(1097))
(C) OTHER INFORMATION: lacI
(ix) FEATURE:
(A) NAME/KEY: promotor
(B) LOCATION: (1391)..(1439)
(C) OTHER INFORMATION: tac
CA 02325593 2001-02-21
28
(ix) FEATURE:
(A) NAME/KEY: N_region
(B) LOCATION: (1562)..(1999)
(C) OTHER INFORMATION: 5'glyA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO.: 2:
AATTCTCACT GCCCGCTTTC CAGTCGGGAA ACCTGTCGTG CCAGCTGCAT TAATGAATCG 60
GCCAACGCGC GGGGAGAGGC GGTTTGCGTA TTGGGCGCCA GGGTGGTTTT TCTTTTCACC 120
AGTGAGACGG GCAACAGCTG ATTGCCCTTC ACCGCCTGGC CCTGAGAGAG TTGCAGCAAG 180
CGGTCCACGC TGGTTTGCCC CAGCAGGCGA AAATCCTGTT TGATGGTGGT TGACGGCGGG 240
ATATAACATG AGCTGTCTTC GGTATCGTCG TATCCCACTA CCGAGATATC CGCACCAACG 300
CGCAGCCCGG ACTCGGTAAT GGCGCGCATT GCGCCCAGCG CCATCTGATC GTTGGCAACC 360
AGCATCGCAG TGGGAACGAT GCCCTCATTC AGCATTTGCA TGGTTTGTTG AAAACCGGAC 420
ATGGCACTCC AGTCGCCTTC CCGTTCCGCT ATCGGCTGAA TTTGATTGCG AGTGAGATAT 480
TTATGCCAGC CAGCCAGACG CAGACGCGCC GAGACAGAAC TTAATGGGCC CGCTAACAGC 540
GCGATTTGCT GGTGACCCAA TGCGACCAGA TGCTCCACGC CCAGTCGCGT ACCGTCTTCA 600
TGGGAGAAAA TAATACTGTT GATGGGTGTC TGGTCAGAGA CATCAAGAAA TAACGCCGGA 660
ACATTAGTGC AGGCAGCTTC CACAGCAATG GCATCCTGGT CATCCAGCGG ATAGTTAATG 720
ATCAGCCCAC TGACGCGTTG CGCGAGAAGA TTGTGCACCG CCGCTTTACA GGCTTCGACG 780
CCGCTTCGTT CTACCATCGA CACCACCACG CTGGCACCCA GTTGATCGGC GCGAGATTTA 840
ATCGCCGCGA CAATTTGCGA CGGCGCGTGC AGGGCCAGAC TGGAGGTGGC AACGCCAATC 900
AGCAACGACT GTTTGCCCGC CAGTTGTTGT GCCACGCGGT TGGGAATGTA ATTCAGCTCC 960
GCCATCGCCG CTTCCACTTT TTCCCGCGTT TTCGCAGAAA CGTGGCTGGC CTGGTTCACC 1020
ACGCGGGAAA CGGTCTGATA AGAGACACCG GCATACTCTG CGACATCGTA TAACGTTACT 1080
GGTTTCACAT TCACCACCCT GAATTGACTC TCTTCCGGGC GCTATCATGC CATACCGCGA 1140
AAGGTTTTGC ACCATTCGAT GGTGTCAACG TAAATGCATG CCGCTTCGCC TTCGCGCGCG 1200
AATTGCAAGC TGATCCGGGC TTATCGACTG CACGGTGCAC CAATGCTTCT GGCGTCAGGC 1260
AGCCATCGGA AGCTGTGGTA TGGCTGTGCA GGTCGTAAAT CACTGCATAA TTCGTGTCGC 1320
TCAAGGCGCA CTCCCGTTCT GGATAATGTT TTTTGCGCCG ACATCATAAC GGTTCTGGCA 1380
AATATTCTGA AATGAGCTGT TGACAATTAA TCATCGGCTC GTATAATGTG TGGAATTGTG 1940
AGCGGATAAC AATTTCACAC AGGAAACAGA ATTAAAAGAT ATGACCATGA TTACGCCAAG 1500
CTTGCATGCC TGCAGGTCGA CTCTAGAGGA TCATTCGTCT TGTGAAAGGT TAGCTGACCT 1560
GATGACCGAT GCCCACCAAG CGGACGATGT CCGTTACCAG CCACTGAACG AGCTTGATCC 1620
CA 02325593 2001-02-21
' 29
TGAGGTGGCT GCTGCCATCG CTGGGGAACT TGCCCGTCAA CGCGATACAT TAGAGATGAT 1680
CGCGTCTGAG AACTTCGTTC CCCGTTCTGT TTTGCAGGCG CAGGGTTCTG TTCTTACCAA 1740
TAAGTATGCC GAGGGTTACC CTGGCCGCCG TTACTACGGT GGTTGCGAAC AAGTTGACAT 1800
CATTGAGGAT CTTGCACGTG ATCGTGCGAA GGCTCTCTTC GGTGCAGAGT TCGCCAATGT 1860
TCAGCCTCAC TCTGGCGCAC AGGCTAATGC TGCTGTGCTG ATGACTTTGG CTGAGCCAGG 1920
CGACAAGATC ATGGGTCTGT CTTTGGCTCA TGGTGGTCAC TTGACCCACG GAATGAAGTT 1980
GAACTTCTCC GGAAAGCTGG 2000
(2) INFORMATION FOR SEQ ID NO.: 3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2866
(B) TYPE: nucleic acid
(C) STRANDEDNESS:
(D) TOPOLOGY:
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Knstliche Sequenz
(ix) FEATURE:
(C) OTHER INFORMATION: Description of the synthetic sequence:
lacI-tac-glyA
(ix) FEATURE:
(A) NAME/KEY: gene
(B) LOCATION: Complement ((6)..(1097))
(C) OTHER INFORMATION: lacI
(ix) FEATURE:
(A) NAME/KEY: promotor
(B) LOCATION: (1391)..(1434)
(C) OTHER INFORMATION: tac
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: (1562)..(2863)
(C) OTHER INFORMATION: glyA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO.: 3:
AATTCTCACT GCCCGCTTTC CAGTCGGGAA ACCTGTCGTG CCAGCTGCAT TAATGAATCG 60
GCCAACGCGC GGGGAGAGGC GGTTTGCGTA TTGGGCGCCA GGGTGGTTTT TCTTTTCACC 120
AGTGAGACGG GCAACAGCTG ATTGCCCTTC ACCGCCTGGC CCTGAGAGAG TTGCAGCAAG 180
CGGTCCACGC TGGTTTGCCC CAGCAGGCGA AAATCCTGTT TGATGGTGGT TGACGGCGGG 290
ATATAACATG AGCTGTCTTC GGTATCGTCG TATCCCACTA CCGAGATATC CGCACCAACG 300
CA 02325593 2001-02-21
' ' 30
CGCAGCCCGG ACTCGGTAAT GGCGCGCATT GCGCCCAGCG CCATCTGATC GTTGGCAACC 360
AGCATCGCAG TGGGAACGAT GCCCTCATTC AGCATTTGCA TGGTTTGTTG AAAACCGGAC 420
ATGGCACTCC AGTCGCCTTC CCGTTCCGCT ATCGGCTGAA TTTGATTGCG AGTGAGATAT 480
TTATGCCAGC CAGCCAGACG CAGACGCGCC GAGACAGAAC TTAATGGGCC CGCTAACAGC 540
GCGATTTGCT GGTGACCCAA TGCGACCAGA TGCTCCACGC CCAGTCGCGT ACCGTCTTCA 600
TGGGAGAAAA TAATACTGTT GATGGGTGTC TGGTCAGAGA CATCAAGAAA TAACGCCGGA 660
ACATTAGTGC AGGCAGCTTC CACAGCAATG GCATCCTGGT CATCCAGCGG ATAGTTAATG 720
ATCAGCCCAC TGACGCGTTG CGCGAGAAGA TTGTGCACCG CCGCTTTACA GGCTTCGACG 780
CCGCTTCGTT CTACCATCGA CACCACCACG CTGGCACCCA GTTGATCGGC GCGAGATTTA 840
ATCGCCGCGA CAATTTGCGA CGGCGCGTGC AGGGCCAGAC TGGAGGTGGC AACGCCAATC 900
AGCAACGACT GTTTGCCCGC CAGTTGTTGT GCCACGCGGT TGGGAATGTA ATTCAGCTCC 960
GCCATCGCCG CTTCCACTTT TTCCCGCGTT TTCGCAGAAA CGTGGCTGGC CTGGTTCACC 1020
ACGCGGGAAA CGGTCTGATA AGAGACACCG GCATACTCTG CGACATCGTA TAACGTTACT 1080
GGTTTCACAT TCACCACCCT GAATTGACTC TCTTCCGGGC GCTATCATGC CATACCGCGA 1190
AAGGTTTTGC ACCATTCGAT GGTGTCAACG TAAATGCATG CCGCTTCGCC TTCGCGCGCG 1200
AATTGCAAGC TGATCCGGGC TTATCGACTG CACGGTGCAC CAATGCTTCT GGCGTCAGGC 1260
AGCCATCGGA AGCTGTGGTA TGGCTGTGCA GGTCGTAAAT CACTGCATAA TTCGTGTCGC 1320
TCAAGGCGCA CTCCCGTTCT GGATAATGTT TTTTGCGCCG ACATCATAAC GGTTCTGGCA 1380
AATATTCTGA AATGAGCTGT TGACAATTAA TCATCGGCTC GTATAATGTG TGGAATTGTG 1440
AGCGGATAAC AATTTCACAC AGGAAACAGA ATTAAAAGAT ATGACCATGA TTACGCCAAG 1500
CTTGCATGCC TGCAGGTCGA CTCTAGAGGA TCATTCGTCT TGTGAAAGGT TAGCTGACCT 1560
G ATG ACC GAT GCC CAC CAA GCG GAC GAT GTC CGT TAC CAG CCA CTG AAC 1609
Met Thr Asp Ala His Gln Ala Asp Asp Val Arg Tyr Gln Pro Leu Asn
1 5 10 15
GAG CTT GAT CCT GAG GTG GCT GCT GCC ATC GCT GGG GAA CTT GCC CGT 1657
Glu Leu Asp Pro Glu Val Ala Ala Ala Ile Ala Gly Glu Leu Ala Arg
20 25 30
CAA CGC GAT ACA TTA GAG ATG ATC GCG TCT GAG AAC TTC GTT CCC CGT 1705
Gln Arg Asp Thr Leu Glu Met Ile Ala Ser Glu Asn Phe Val Pro Arg
35 40 45
TCT GTT TTG CAG GCG CAG GGT TCT GTT CTT ACC AAT AAG TAT GCC GAG 1753
Ser Val Leu Gln Ala Gln Gly Ser Val Leu Thr Asn Lys Tyr Ala Glu
50 55 60
CA 02325593 2001-02-21
' ' 31
GGT TAC CCT GGC CGC CGT TAC TAC GGT GGT TGC GAA CAA GTT GAC ATC 1801
Gly Tyr Pro Gly Arg Arg Tyr Tyr Gly Gly Cys Glu Gln Val Asp Ile
65 70 75 80
ATTGAGGAT CTTGCACGT GATCGTGCG AAGGCTCTCTTC GGTGCAGAG 1849
IleGluAsp LeuAlaArg AspArgAla LysAlaLeuPhe GlyAlaGlu
85 90 95
TTCGCCAAT GTTCAGCCT CACTCTGGC GCACAGGCTAAT GCTGCTGTG 1897
PheAlaAsn ValGlnPro HisSerGly AlaGlnAlaAsn AlaAlaVal
100 105 110
CTGATGACT TTGGCTGAG CCAGGCGAC AAGATCATGGGT CTGTCTTTG 1945
LeuMetThr LeuAlaGlu ProGlyAsp LysIleMetGly LeuSerLeu
115 120 125
GCTCATGGT GGTCACTTG ACCCACGGA ATGAAGTTGAAC TTCTCCGGA 1993
AlaHisGly GlyHisLeu ThrHisGly MetLysLeuAsn PheSerGly
130 135 140
AAGCTGTAC GAGGTTGTT GCGTACGGT GTTGATCCTGAG ACCATGCGT 2041
LysLeuTyr GluValVal AlaTyrGly ValAspProGlu ThrMetArg
195 150 155 160
GTTGATATG GATCAGGTT CGTGAGATT GCTCTGAAGGAG CAGCCAAAG 2089
ValAspMet AspGlnVal ArgGluIle AlaLeuLysGlu GlnProLys
165 170 175
GTAATTATC GCTGGCTGG TCTGCATAC CCTCGCCACCTT GATTTCGAG 2137
ValIleIle AlaGlyTrp SerAlaTyr ProArgHisLeu AspPheGlu
180 185 190
GCTTTCCAG TCTATTGCT GCGGAAGTT GGCGCGAAGCTG TGGGTCGAT 2185
AlaPheGln SerIleAla AlaGluVal GlyAlaLysLeu TrpValAsp
195 200 205
ATGGCTCAC TTCGCTGGT CTTGTTGCT GCTGGTTTGCAC CCAAGCCCA 2233
MetAlaHis PheAlaGly LeuValAla AlaGlyLeuHis ProSerPro
210 215 220
GTTCCTTAC TCTGATGTT GTTTCTTCC ACTGTCCACAAG ACTTTGGGT 2281
ValProTyr SerAspVal ValSerSer ThrValHisLys ThrLeuGly
225 230 235 240
GGACCTCGT TCCGGCATC ATTCTGGCT AAGCAGGAGTAC GCGAAGAAG 2329
GlyProArg SerGlyIle IleLeuAla LysGlnGluTyr AlaLysLys
245 250 255
CTGAACTCT TCCGTATTC CCAGGTCAG CAGGGTGGTCCT TTGATGCAC 2377
LeuAsnSer SerValPhe ProGlyGln GlnGlyGlyPro LeuMetHis
260 265 270
GCAGTTGCT GCGAAGGCT ACTTCTTTG AAGATTGCTGGC ACTGAGCAG 2425
AlaValAla AlaLysAla ThrSerLeu LysIleAlaGly ThrGluGln
275 280 285
TTCCGTGAC CGTCAGGCT CGCACGTTG GAGGGTGCTCGC ATTCTTGCT 2473
PheArgAsp ArgGlnAla ArgThrLeu GluGlyAlaArg IleLeuAla
290 295 300
CA 02325593 2001-02-21
' 32
GAG CGTCTG ACTGCT TCTGATGCG AAGGCCGCT GGCGTGGATGTC TTG 2521
Glu ArgLeu ThrAla SerAspAla LysAlaAla GlyValAspVal Leu
305 310 315 320
ACC GGTGGC ACTGAT GTGCACTTG GTTTTGGCT GATCTGCGTAAC TCC 2569
Thr GlyGly ThrAsp ValHisLeu ValLeuAla AspLeuArgAsn Ser
325 330 335
CAG ATGGAT GGCCAG CAGGCGGAA GATCTGCTG CACGAGGTTGGT ATC 2617
Gln MetAsp GlyGln GlnAlaGlu AspLeuLeu HisGluValGly Ile
390 345 350
ACT GTGAAC CGTAAC GCGGTTCCT TTCGATCCT CGTCCACCAATG GTT 2665
Thr ValAsn ArgAsn AlaValPro PheAspPro ArgProProMet Val
355 360 365
ACT TCTGGT CTGCGT ATTGGTACT CCTGCGCTG GCTACCCGTGGT TTC 2713
Thr SerGly LeuArg IleGlyThr ProAlaLeu AlaThrArgGly Phe
370 375 380
GAT ATTCCT GCATTC ACTGAGGTT GCAGACATC ATTGGTACTGCT TTG 2761
Asp IlePro AlaPhe ThrGluVal AlaAspIle IleGlyThrAla Leu
385 390 395 900
GCT AATGGT AAGTCC GCAGACATT GAGTCTCTG CGTGGCCGTGTA GCA 2809
Ala AsnGly LysSer AlaAspIle GluSerLeu ArgGlyArgVal Ala
405 410 915
AAG CTTGCT GCAGAT TACCCACTG TATGAGGGC TTGGAAGACTGG ACC 2857
Lys LeuAla AlaAsp TyrProLeu TyrGluGly LeuGluAspTrp Thr
420 425 430
ATC GTCTAA 2866
Ile Val
(2) INFORMATION FOR SEQ ID NO.: 9:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 939
(B) TYPE: amino acid
(C) STRANDEDNESS:
(D) TOPOLOGY:
(ii) MOLECULE TYPE: polypeptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO.: 4:
Met Thr Asp Ala His Gln Ala Asp Asp Val Arg Tyr Gln Pro Leu Asn
1 5 10 15
Glu Leu Asp Pro Glu Val Ala Ala Ala Ile Ala Gly Glu Leu Ala Arg
20 25 30
Gln Arg Asp Thr Leu Glu Met Ile Ala Ser Glu Asn Phe Val Pro Arg
35 40 45
Ser Val Leu Gln Ala Gln Gly Ser Val Leu Thr Asn Lys Tyr Ala Glu
50 55 60
CA 02325593 2001-02-21
33
' Gly Tyr Pro Gly Arg Arg Tyr Tyr Gly Gly Cys Glu Gln Val Asp Ile
~ 65 70 75 80
~ Ile Glu Asp Leu Ala Arg Asp Arg Ala Lys Ala Leu Phe Gly Ala Glu
85 90 95
Phe Ala Asn Val Gln Pro His Ser Gly Ala Gln Ala Asn Ala Ala Val
100 105 110
Leu Met Thr Leu Ala Glu Pro Gly Asp Lys Ile Met Gly Leu Ser Leu
115 120 125
Ala His Gly Gly His Leu Thr His Gly Met Lys Leu Asn Phe Ser Gly
130 135 140
Lys Leu Tyr Glu Val Val Ala Tyr Gly Val Asp Pro Glu Thr Met Arg
145 150 155 160
Val Asp Met Asp Gln Val Arg Glu Ile Ala Leu Lys Glu Gln Pro Lys
165 170 175
Val Ile Ile Ala Gly Trp Ser Ala Tyr Pro Arg His Leu Asp Phe Glu
180 185 190
Ala Phe Gln Ser Ile Ala Ala Glu Val Gly Ala Lys Leu Trp Val Asp
195 200 205
Met Ala His Phe Ala Gly Leu Val Ala Ala Gly Leu His Pro Ser Pro
210 215 220
Val Pro Tyr Ser Asp Val Val Ser Ser Thr Val His Lys Thr Leu Gly
225 230 235 240
Gly Pro Arg Ser Gly Ile Ile Leu Ala Lys Gln Glu Tyr Ala Lys Lys
245 250 255
Leu Asn Ser Ser Val Phe Pro Gly Gln Gln Gly Gly Pro Leu Met His
260 265 270
Ala Val Ala Ala Lys Ala Thr Ser Leu Lys Ile Ala Gly Thr Glu Gln
275 280 285
Phe Arg Asp Arg Gln Ala Arg Thr Leu Glu Gly Ala Arg Ile Leu Ala
290 295 300
Glu Arg Leu Thr Ala Ser Asp Ala Lys Ala Ala Gly Val Asp Val Leu
305 310 315 320
Thr Gly Gly Thr Asp Val His Leu Val Leu Ala Asp Leu Arg Asn Ser
325 330 335
Gln Met Asp Gly Gln Gln Ala Glu Asp Leu Leu His Glu Val Gly Ile
340 345 350
Thr Val Asn Arg Asn Ala Val Pro Phe Asp Pro Arg Pro Pro Met Val
355 360 365
Thr Ser Gly Leu Arg Ile Gly Thr Pro Ala Leu Ala Thr Arg Gly Phe
370 375 380
CA 02325593 2001-02-21
' - 34
Asp Ile Pro Ala Phe Thr Glu Val Ala Asp Ile Ile Gly Thr Ala Leu
_- 385 390 395 400
Ala Asn Gly Lys Ser Ala Asp Ile Glu Ser Leu Arg Gly Arg Val Ala
905 910 915
Lys Leu Ala Ala Asp Tyr Pro Leu Tyr Glu Gly Leu Glu Asp Trp Thr
420 425 430
Ile Val