Note: Descriptions are shown in the official language in which they were submitted.
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
1
NUCLEIC ACID ENZYME FOR RNA CLEAVAGE
TECHNICAL FIELD
The invention relates to a novel ribozyme
construction for the specific recognition and cleavage of RNA,
and biotechnological as well as therapeutic uses thereof.
BACKGROUND ART
Though enzymatic activity has long been considered
the exclusive domain of proteins, discoveries in molecular
biology over the past couple of decades have led to the
realization that ribonucleic acid (RNA) can also function as an
enzyme. RNA enzymes are often referred to as ribozymes.
Ribozyme substrates are generally confined to RNA
molecules, and enzymatic activities of ribozymes include the
cleavage and/or ligation of RNA molecules. The cleavage
activity may be intramolecular, known as cis-acting or
intermolecular, known as trans-acting. There are at least five
classes of ribozymes known, including Group I introns, Group II
introns, hammerhead, hairpin, and delta ribozymes. The last
three are derived from plant satellites and viroids.
Since 1982, several unexpected diseases caused by
RNA-based pathogenic agents have emerged. These include the
lethal Acquired Immune Deficiency Syndrome (AIDS) and delta
hepatitis, a particularly virulent form of fulminant hepatitis
caused by a viroid-like RNA agent. These blood-borne diseases
are spread at the RNA level, manifest themselves in cells of
patients, and are by now present within the bloodstream of
millions of individuals. Conventional biotechnology, with its
reliance on recombinant DNA methods and DNA-level intervention
schemes, has been slow to provide valid approaches to combat
these diseases.
Two forms of delta ribozymes, namely genomic and
antigenomic, are derived, and referred to by, the polarity of
the hepatitis delta virus (HDV) genome from which the ribozyme
is generated. Like hammerhead and hairpin ribozymes, the delta
ribozymes cleave a phosphodiester bond of their RNA substrates
and give rise to reaction products containing a 5'-hydroxyl and
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
2
a 2',3'-cyclic phosphate termini. They are metalloenzymes and
a low concentration (cl mM) of magnesium (Mg2+) or calcium
(Ca2+) is required for delta ribozyme cleavage. Both genomic
strand and antigenomic strand forms exhibit self-cleavage
activity, and it has been suggested that they are involved in
the process of viral replication (Lazinski, D. W., and Taylor,
J. M. (1995) RNA 1, 225-233).
Delta ribozymes derived from the genome of HDV are of
interest in the development of a gene regulation system in
which the designed ribozymes would down-regulate the expression
of a target gene. The facts that delta ribozymes are derived
from HDV and that this pathogen naturally replicates in animal
systems, suggest that this catalytic RNA could be used to
control gene expression in human cells. Like other ribozymes,
the designed ribozyme should specifically cleave its target
substrates while leaving other cellular RNA molecules intact.
Trans-acting ribozymes carry out intermolecular
cleavage activity. Some trans-acting delta ribozymes have been
developed by removing a single-stranded junction which connects
the catalytic portion to the substrate portion in cis-acting
delta ribozymes. This results in two separate molecules, one
possessing the substrate sequence and the other the catalytic
property (Been, M.D. and Wichhan, G.S. (1997) Eur. J. Biochem.,
247, 741-753). Interactions between such delta ribozymes and
the substrate occur through the formation of a helix, referred
as the P1 stem. However, the example of the trans-acting
ribozyme disclosed by Been et al. (supra) was not useful for
cleaving long substrate molecules, such as those having
therapeutic applications.
In United States Patent No. 5,225,337, issued on
July 6, 1993 in the names of Hugh D. Robertson et al., there
are disclosed ribozymes derived from a specific domain present
in the HDV RNA for specifically cleaving targeted RNA sequences
and uses thereof for the treatment of disease conditions which
involve RNA expression, such as AIDS. These ribozymes consist
of at least 18 consecutive nucleotides from the conserved
region of HDV isolates between residues 611 and 771 on the
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
3
genomic strand and between residues 845 and 980 on the
complementary antigenomic strand. These ribozymes are proposed
to fold into an axe-head model secondary structure (Branch,
A. D., and Robertson, H. D. (1991) Proc. Natl Acad. Sci. USA
88, 10163-10167). The ribozymes developed according to this
model structure require the substrate to be bound to the
ribozyme through the formation of two helices, one located on
either side of the cleavage site. Further, such ribozymes
apparently require a 12-15 nucleotide recognition sequence in
the substrate in order to exhibit the desired activity. Such a
long recognition sequence is not practical in the development
of therapeutic or diagnostic applications.
In United States Patent No. 5,625,047, issued on
April 29, 1997 in the names of Michael D. Been et al., there
are disclosed enzymatic RNA molecules proposed to fold into a
pseudoknot model secondary structure (discussed below). The
method disclosed for the development of efficient ribozymes
requires a short recognition sequence of only 7 to 8
nucleotides in the substrate, a preference for a guanosine base
immediately 3' to the cleavage site, a preference for U, C or A
immediately 5' to the cleavage site, and the availability of a
2'-hydroxyl group for cleavage to occur. Thus, the specificity
of recognition of these ribozymes is limited to 6 or 7 base
pairing nucleotides with the substrate and a preference of the
first nucleotide located 5' to the cleavage site. Neither
tertiary interaction(s) between the base paired nucleotides and
another region of the ribozyme, nor single-stranded nucleotides
are involved to define the specificity of recognition of these
ribozymes. Because the recognition features are limited, these
ribozymes have a limited specificity, and thus, are not
practical for further clinical or biotechnical applications.
A pseudoknot-like structure for delta ribozymes has
been proposed by Perrotta and Been (Perrotta, A. T., and Been,
M. D. (1991) Nature 350, 434-436). This model structure
consists of two stems (P1 and P2), two stem-loops (P3 and P4)
and three single-stranded regions (J1/2, J1/4 and J4/2). An
additional stem, named P1.1, has been formed by two GC base
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
4
pairs between nucleotides from the J1/4 junction and the P3
loop (Ferre-D'Amare, A.R., Zhou, K. and Doudna, J.A. (1998)
Nature, 350, 434-436).
It would be highly desirable to be provided with a
novel delta ribozyme for the cleavage of both small and large
RNA substrates for which the specificity of recognition is well
defined. Such specificity would yield optimal conditions for
further therapeutical and biotechnological developments of
delta ribozymes.
SUMMARY OF THE INVENTION
One aim of the present invention is to provide a
novel delta ribozyme for the cleavage of RNA substrates for
which the specificity is defined by a domain composed of at
least 7 nucleotides. It is also an aim to provide a method for
the development of such ribozymes.
In one aspect, the invention provides a method for
cleaving a nucleic acid substrate with a nucleic acid enzyme at
a cleavage site comprising mixing the substrate with the
enzyme, wherein the substrate includes a 7 nucleotide sequence
with at least 6 nucleotides 3' to the cleavage site and at
least 1 nucleotide 5' to the cleavage site of formula:
5'-H'4GNNHNN-3'
wherein each
N is a nucleotide which may be the same or different,
H is a nucleotide selected from the group consisting of A,
U, C, and T, and
4 is the site of cleavage, and
H' is a ribonucleotide selected from the group consisting
of A, U, and C,
wherein
(i) the first nucleotide 3' to the cleavage site is
capable of forming a wobble pair with the enzyme,
(ii) the second, third, fifth, and sixth nucleotides 3' to
the cleavage site are capable of forming conventional Watson-
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
Crick base pairs with the enzyme,
(iii) the fourth nucleotide 3' to the cleavage site is
capable of forming a triplet with the enzyme comprising a non-
conventional Watson-Crick base pair and a conventional Watson-
5 Crick base pair, and
(iv) the ribonucleotide directly 5' to the cleavage site
does not form a base pair with the enzyme; and
the enzyme comprises a substrate binding portion which is
capable of base pairing to the 6 nucleotides 3' to the cleavage
site of the substrate and which binding portion comprises the
sequence:
3'-UNNXNN-5'
wherein each
N is a nucleotide which may be the same or different, and
X is a nucleotide selected from the group consisting of T,
U, A, and G,
whereby binding of the substrate to the enzyme effects cleavage
of the substrate at the cleavage site.
In another aspect, the invention provides a nucleic
acid enzyme capable of recognizing and cleaving a nucleic acid
substrate at a cleavage site comprising a substrate binding
portion which is capable of base pairing to the 6 nucleotides
3' to the cleavage site of the substrate and which binding
portion comprises the sequence:
3'-UNNXNN-5'
wherein each
N is a nucleotide which may be the same or different, and
X is a nucleotide selected from the group consisting of T,
U, A, G, and
the substrate includes a 7 nucleotide sequence with at least 6
nucleotides 3' to the cleavage site and at least 1 nucleotide
5' to the cleavage site of formula:
CA 02330570 2009-06-08
87036-1
6a
5'-H'vGNNHNN-3'
wherein each
N is a nucleotide which may be the same or
different,
H is a nucleotide selected from the group
consisting of A, U, C, and T,
V is the site of cleavage, and
H' is a ribonucleotide selected from the group
consisting of A, U, and C,
wherein
(i) the first nucleotide 3' to the cleavage site
is capable of forming a wobble pair with the enzyme,
(ii) the second, third, fifth, and sixth
nucleotides 3' to the cleavage site are capable of forming
conventional Watson-Crick base pairs with the enzyme,
(iii) the fourth nucleotide 3' to the cleavage
site is capable of forming a triplet with the enzyme
comprising a non-conventional Watson-Crick base pair and a
conventional Watson-Crick base pair, and
(iv) the first ribonucleotide directly 5' to the
cleavage site does not form a base pair with the enzyme.
According to one aspect of the present invention,
there is provided a nucleic acid enzyme capable of
recognizing and cleaving a nucleic acid substrate at a
cleavage site, said nucleic acid enzyme which when bound to
the substrate comprises:
CA 02330570 2009-06-08
87036-1
6b
(i) a first nucleotide sequence
' -GiG2G3UaC5C6Ai3Ci4CisUCi6Ci7 UCGCG15G14 U13NiN2N3N4N5N6N7 Gi7Gi6G7C8A9Ui0
G11C12S1B1Y-3' ; and
(ii) a second nucleotide sequence
5 5 ' -B2KS2G12C11A10U9G$G'CUAAGG6GSA4C3C2C1- 3 ' ;
wherein a first non-variable nucleotide forms a conventional
Watson-Crick base pair with a second non-variable nucleotide
wherein the first and second non-variable nucleotides have
the same superscript, except the two G' form a homopurine
base pair;
S1 and S2 are each independently selected from the
group consisting of G and C such that S' and S2 form a
conventional Watson-Crick base pair;
B1 and B2 are each independently selected from the
group consisting of G, C, U and T;
K is selected from the group consisting of
G, U and T;
Y is selected from the group consisting of
C, U and T;
N1NzN3N4N5N6N' forms a substrate binding region;
Nl, N2, N3, N4, N5 and N6 are each a nucleotide
which may independently be the same or different;
N' i s U;
N' is capable of forming a wobble pair with the
substrate;
Nl, N2, N3, NS and N6 are capable of forming
conventional Watson-Crick base pairs with the substrate; and
CA 02330570 2009-06-08
87036-1
6c
N4 is capable of forming a conventional Watson-
Crick base pair with the substrate and capable of forming a
triplet by means of a non-conventional Watson-Crick base
pair with a nucleotide in the nucleic acid enzyme,
wherein
(a) Y and B2 form a conventional Watson-Crick base
pair;
(b) B1 and K form a conventional Watson-Crick base
pair; or
(c) Bi, K, Y and B2 together form a loop, and
wherein the enzyme is adapted to bind to the substrate such
that the enzyme is incapable of interacting with nucleotide
residues in the substrate at positions -1 and -2 directly 5'
to the cleavage site and the enzyme is capable of forming a
GU wobble pair with the nucleotide residue (G) in the
substrate directly 3' to the cleavage site.
BRIEF DESCRIPTION OF THE DRAWINGS
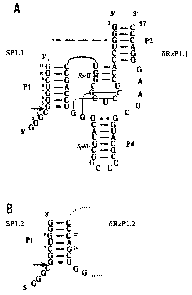
Figure 1 illustrates the secondary structure and
nucleotide sequences of two trans-acting antigenomic delta
ribozymes of the invention and complementary substrates;
panel A is the secondary structure of the complex formed
between 5RzP1.1 and a substrate Spl.l; panel B is the
P1 region of the complex formed between 6RzP1.2 and a
substrate Spl.2; the rest of the structure is identical to
bRzPl.l as in panel A;
Figure 2 illustrates the secondary structure of a
ribozyme in accordance with the invention, with an
ultrastable L4 loop; in the inset is the sequence of a
14-nucleotide long substrate;
CA 02330570 2009-06-08
87036-1
6d
Figure 3 illustrates the secondary structure of a
ribozyme in accordance with the invention; the inset shows
the ultrastable L4 loop;
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
7
Figure 4 shows a two-dimensional representation of a
catalytic trimolecular complex (RzA: RzB:S) of the invention;
Figure 5 shows results from Example 3, namely
comparative analyses of the cleavage reactions catalyzed by
delta ribozymes;
Figure 6 shows a two-dimensional representation of a
catalytic trimolecular complex (RzA: RzB:S); the influence of
2'-OH groups individually at positions 9 to 15 on RzB by
replacing the ribonucleotide at these positions with the
corresponding deoxy-ribonucleotide is shown; the symbol -
represents a two-fold diminution of activity compared to an
unmodified RzB while the symbol = represents an unchanged
catalytic activity; symbols + and ++ respectively represent an
increased activity of 1.5- and 2- fold; horizontal bars
represent base pairs; wobble and homopurine base pairs are
respectively represented by one and two ovals; the arrow
indicates the site of catalytic cleavage;
Figure 7 shows in Panel A the structural and
functional features of virion DNA, including the viral direct
repeat (DR) sequences (boxed), and the protein (=) and RNA
(AAA) species found at the 5' ends of the minus and plus DNA
strands, respectively; the dashed line indicates the presence
of the single stranded gap; the RNA products are depicted by
wavy lines; the target area is located in pre-S2 and S regions,
and is indicated by the scissors symbol; panel B illustrates
the secondary struction of an engineered ribozyme of the the
invention, such that the substrate binding region is 5'GGGAUAU-
3', complementary to HBV mRNA substrates; the recognition site
on the mRNA is located on the pres-S2 and S mRNA (2.1 kb, as
shown in Panel A); the arrow indicates the cleavage site.
DESCRIPTION OF THE PREFERRED EMBODIMENTS OF THE INVENTION
The subject invention provides for a method of
designing selective nucleic acid enzymes, such that a nucleic
acid substrate is cleaved at a specified cleavage site by the
nucleic acid enzyme. This method includes the selection of
certain substrate sequences and, within the enzymes, certain
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
8
substrate binding sequences, such that efficient cleavage at a
specified site in the nucleic acid substrate can take place.
The subject invention also provides for nucleic acid enzymes
designed using such method.
For the purpose of the present invention the
following abbreviations are defined: "A" is a nucleotide
comprising adenine including both the ribo- and deoxyribo-
forms; "G" is a nucleotide comprising guanine including both
the ribo- and deoxyribo-forms; "C" is a nucleotide comprising
cytidine including both the ribo- and deoxyribo-forms; "U" is
a nucleotide comprising uracil; "T" is a nucleotide comprising
thymine; "R" is a nucleotide comprising purine, which purine
is selected from the group consisting of A and G; and "Y" is a
nucleotide comprising pyrimidine, which pyrimidine is selected
from the group consisting of U, C, and T.
Selection of Substrate Sequence
Substrate nucleic acid includes any nucleic acid
sequence which can act as a substrate for a nucleic acid enzyme
of the invention. As such it includes ribonucleotides,
deoxyribonucleotides, or mixtures of both. Nucleotides may
also include synthetic or modified nucleotides.
The nucleic acid enzymes of the invention can be used
to target a large number of nucleic acid substrates so long as
certain conditions of the recognition mechanism are met. The
nucleic acid substrate must include a 7 nucleotide sequence
with at least 6 nucleotides 3' to the cleavage site and at
least 1 nucleotide 5' to the cleavage site of formula:
5'-H'yGNNHNN-3'
wherein each
N is a nucleotide which may be the same or different,
H is a nucleotide selected from the group consisting of A,
U, C, and T, and
y is the site of cleavage, and
H' is a ribonucleotide selected from the group consisting
of A, U, and C.
CA 02330570 2000-10-27
WO 99/55856 - PCT/CA99/00391
9
The first nucleotide 3' to the cleavage site is
capable of forming a wobble pair with the enzyme. The wobble
base pair (G-U) at the cleavage site is required to maintain a
high level of cleavage. Conventional Watson-Crick base pairs
such as A-U and G-C, as well as mismatches at this position
decrease the cleavage activity.
The second, third, fifth, and sixth nucleotides 3' to
the cleavage site are capable of forming conventional Watson-
Crick base pairs with the enzyme.
The fourth nucleotide 3' to the cleavage site is
capable of forming a conventional Watson-Crick base pair with
the substrate binding region of the enzyme. Additionally, such
base pair interacts with a nucleotide elsewhere in the ribozyme
(i.e. the nucleic acid enzyme) to form a triplet by means of a
non-conventional Watson-Crick base pair. Non-conventional
Watson-Crick base pairs include Hoogsteen pairs and reversed-
Hoogsteen pairs. The position requires an A, U, or C.
The ribonucleotide directly 5' to the cleavage site
does not form a base pair with the ribozyme.
Preferably, the substrate molecule does not contain
two consecutive pyrimidine nucleotides directly 5' to the
cleavage site.
In another preferred aspect, the substrate comprises
the sequence 5'-H',~GNNHNNN-3', more preferably the sequence 5'-
NNRH'4.GNNHNNN-3', wherein R is G or A.
In one embodiment, the substrate preferably comprises
the sequence 5'-RRRH'yGNNHNNN-3'. More preferably, such
sequence is selected from the group consisting of
5'-GGGCyGNNI1NNN-3', 5'-GGGCyGNNCNNN-3', 5'-GGGUyGNNUNNN-3',
5'-GGGUyGNNCNNNN-3', and 5'-AAAC4,GNNUNNN-3'.
In another embodiment, the substrate preferably
comprises the sequence 5'-YHRH'yGNNHNNN-3', wherein Y is C, U,
or T. It is preferred that the four nucleotides directly 5' to
the cleavage site are chosen such that Y is C or U, preferably
C; H is one of U, C, or A, preferably U or C, more preferably
U; R is preferably A; and H is A, C, or U, preferably A or C,
more preferably A.
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
It is preferable that the four nucleotides directly
5' to the cleavage site do not form a hairpin structure.
Selection of Ribozyme Sequence
By ribozymes, it is meant a nucleic acid enzyme, in
5 other words any nucleic acid sequence having enzymatic
activity, i.e. the ability to catalyze a reaction. As such it
includes nucleic acid sequences made up of ribonucleotides,
deoxyribonucleotides, or mixtures of both. Nucleotides may
also include synthetic or modified nucleotides.
10 The selection of the sequence of the substrate
binding region of the ribozyme, should be done such that the
binding region comprises the sequence 3'-UNNXNN-5',
wherein each N is a nucleotide which may be the same or
different, and X is a nucleotide selected from the group
consisting of T, U, A, and G.
The invention preferably provides for a nucleic acid
enzyme with a secondary structure which comprises three or more
distinct double-stranded regions, or stem-regions. This
includes regions of base-pairing which may or may not be capped
by a single-stranded loop, to form a stem-loop region.
Preferably, the nucleic acid ribozyme includes two or more
distinct single-stranded regions, one of which includes a
substrate binding region which will base pair to the substrate.
More preferably there are two single stranded regions.
The invention preferably contemplates the use of
nucleic acid enzymes derived from hepatitis delta virus, known
as delta ribozymes.
Generation of Ribozyme and Substrate
Trans-acting delta ribozymes of the invention were
generated based on the pseudoknot-like structure proposed by
Perrotta and Been, by removing the single-stranded region
(region J1/2) located at the junction between the P1 and P2
stems. In addition, the P2 stem was elongated, by introducing,
for instance, three G-C base pairs, and by shortening the P4
stem.
Figure 1 illustrates an example of two ribozymes,
6RzP1.1 and 5RzP1.2, in accordance with one aspect of the
CA 02330570 2008-09-26
87036-1
11
invention. The base paired regions of the pseudoknot-like
structure are numbered according to Perrotta and Been
(Perrotta, A. T., and Been, M. D. (1991) Nature 350, 434-436).
The dashed line represents the J1/2 single-stranded region
joining the substrate and ribozyme molecules present in the
cis-form. This single-stranded area was eliminated to produce
a trans-acting ribozyme of the invention. The arrow indicates
the cleavage site. The homopurine basepair at the top of the
P4 stem is represented by two dots (G= =G), while the wobble
base pair is represented by a single dot (G=U). The two small
dotted lines illustrate the P1.1 stem formed by two GC base
pairs.
In another aspect, the invention provides for a
ribozyme with an elongated P2 stem and shortened P4 stem, which
further comprises a modification of the L4 loop. Figures 2 and
3 show ribozymes in accordance with this embodiment. S and Rz
represent substrate and ribozyme respectively.
In one aspect, the invention provides for a
bimolecular ribozyme. This may be achieved by removal of the
L4 loop. Figure 4 shows a ribozyme in accordance with this
embodiment.
Applications
Ribozyme activity can be optimized by chemically
synthesizing ribozymes with modifications that prevent their
degradation by serum ribonucleases (see e.g., Eckstein et al.,
International Publication No. WO 92/07065; Perreault et al.,
Nature 1990, 344:565; Pieken et al., Science 1991, 253:314; and
Chowrira et al., 1993 J. Biol. Chem. 268, 19458, which describe
various chemical modifications that can be made to the sugar
moieties of enzymatic RNA molecules modifications which
enhance their efficacy in cells, and removal of helix-
containing bases to shorten RNA synthesis times and reduce
chemical requirements.
In one aspect, the invention provides a substrate
molecule which is a target RNA, such as a viral RNA, or an RNA
crucial to the life cycle of a pathogen, or an RNA manifested
CA 02330570 2008-09-26
87036-1
12
as a result of an inherited disease, based on the substrate
specificity described herein.
Ribozymes are added directly, or can be complexed
with cationic lipids, packaged within liposomes, or otherwise
delivered to target cells. The RNA or RNA complexes can be
locally administered to relevant tissues ex vivo, or in vivo
through injection, aerosol inhalation, infusion pump or stent,
with or without their incorporation in biopolymers.
Sullivan et al., (WO 94/02595) describes
general methods for delivery of enzymatic RNA molecules.
Rybozymes may be administered to cells by a variety
of methods known to those familiar to the art,
including, but not restricted to, encapsulation in liposomes,
by iontophoresis, or by incorporation into other vehicles, such
as hydrogels, cyclodextrins, biodegradable nanocapsules, and
bioadhesive microspheres. For some indications, ribozymes may
be directly delivered ex vivo to cells or tissues with or
without the aforementioned vehicles. Alternatively, the
RNA/vehicle combination is locally delivered by direct
injection or by use of a catheter, infusion pump or stent.
Other routes of delivery include, but are not limited to,
intravascular, intramuscular, subcutaneous or joint injection,
aerosol inhalation, oral (tablet or pill form), topical,
systemic, ocular, intraperitoneal and/or intrathecal delivery.
More detailed descriptions of ribozyme delivery and
administration are provided in Sullivan, et al., ("Method and
Reagent for Treatment of Arthritic Conditions" U.S.S.N.
08/152,487, filed November 12, 1993).
Another means of accumulating high concentrations of
a ribozyme(s) within cells is to incorporate the ribozyme-
encoding sequences into a DNA expression vector. Transcription
of the ribozyme sequences are driven from a promoter for
eukaryotic RNA polymerase I(po1 I), RNA polymerase II (pol
TI), or RNA polymerase III (pol III). Transcripts from pol II
or po7. III promoters will be expressed at high levels in all
cells; the levels of a given pol II promoter in a given cell
CA 02330570 2008-09-26
87036-1
13
type will depend on the nature of the gene regulatory sequences
(enhancers, silencers, etc.) present nearby. Prokaryotic RNA
polymerase promoters are also used, providing that the
prokaryotic RNA polymerase enzyme is expressed in the
appropriate cells (Elroy-Stein, 0. and Moss, B., 1990, Proc.
Natl. Acad. Sci. U S A, 87, 6743-7; Gao, X. and Huang;, L.,
1993, Nucleic Acids Res., 21, 2867-72).
Several investigators have demonstrated that
ribozymes expressed from such promoters can function in
].0 mammalian cells (e.g. Kashani-Sabet, M., et al., 1992,
Antisense Res. Dev., 2, 3-15; Ojwang, J. 0., et al., 1992,
Proc. Natl. Acad. Sci. U S A, 89, 10802-6).
The above ribozyme transcription units can be
incorporated into a variety of vectors for introduction into
mammalian cells, including but not restricted to, plasmid DNA
vectors, viral DNA vectors (such as adenovirus or
adeno-associated vectors), or viral RNA vectors (such as
retroviral, Semliki forest virus, hepatitis delta virus, and
sindbis virus vectors).
Thus, ribozymes of the present invention that cleave
target inRNA and thereby inhibit and/or reduce target activity
have many potential therapeutic uses, and there are reasonable
modes of delivering the ribozymes in a number of the possible
indications.
By "inhibit" is meant that the activity or level of
target RNA is reduced below that observed in the absence of the
ribozyme, and preferably is below that level observed in the
presence of an inactive RNA molecule able to bind to the same
site on the RNA, but unable to cleave that RNA.
By "vectors" is meant any nucleic acid and/or
viral-based construct used to deliver a desired nucleic acid.
Examples
Example 1: Preparation of Ribozymes, Substrates, and Plasmids.
Construction of plasmids carrying ribozymes of the
invention. The antigenomic ribozyme sequence of the hepatitis
delta virus described by Makino et al (Makino, S. et al. (1987)
CA 02330570 2008-09-26
87036-1
14
Nature 329, 343-346) was used as the basis for
generating trans-acting delta ribozymes of the
invention. Briefly, the construction was performed as follows.
Two pairs of complementary and overlapping oligonucleotides,
representing the entire length of the ribozyme (57 nt), were
synthesized and subjected to an annealing process prior to
cloning into pUC19. The annealed oligonucleotides were ligated
to HindIIl and SmaI co-digested p[7C19 to give rise to a plasmid
harboring the delta ribozyme (referred to as p6RzPl.1). The
l0 minigene was designed so as to have unique Sphl and Smal
restriction sites. The sequence of the T7 RNA promoter was
included at the 5' end of the ribozyme so as to permit in vitro
transcription. Variations based on this "wild type', ribozyme
are constructed by replacing the Sphl-SmaT fragment of pSRzPl.l
by an oligonucleotide duplex containing the desired sequence.
The sequences of engineered ribozymes were confirmed by DNA
sequencing. Plasmids containing wild type and mutant ribozymes
were then prepared using Qiagen tip-100 (Qiagen Inc.), digested
with Smal, purified by phenol and chloroform extraction and
precipitated for further use as templates for in vitro
transcription reactions.
Synthesis of Ribozymes and Substrates. Ribozyme:
in vitro transcription reactions contained 5 g linearized
recombinant plasmid DNA as template, 27 units RNAGuard (RNase
inhibitor (Pharmacia), 4 mM of each rNTP (Pharmacia), 80 mM
HEPES-KOH pH 7.5, 24 mM MgC12, 2 mM spermidine, 40 mM DTT, 0.01
unit Pyrophosphatase (Boehringer Mannheim) and 25 gg purified
T7 RNA polymerase in a final volume of 50 u.L, and were
incubated at 37 C for 4 hr. Substrates: Deoxyoligonucleotides
(500 pmoles) containing the substrate and the T7 promoter
sequence were denatured by heating at 95 C for 5 min in a 20 AL
mixture containing 10 mM Tris-HC1 pH 7.5, 10 mM MgC12, 50 mM
KC12, and allowed to cool slowly to 37 C. The in vitro
transcription reactions were carried out using the resulting
partial duplex formed as template under the same conditions as
described for the production of the ribozyrne.
After incubation, the reaction mixtures were
*Trade-mark
CA 02330570 2008-09-26
87036-1
is
fractionated by denaturing 20% polyacrylamide gel
electrophoresis (PAGE, 19:1 ratio of acrylamide to
bisacrylamide) containing 45 mM Tris-borate pH 7.5, 7 M urea
and 1 mM EDTA. The reaction products were visualized by W
shadowing. The bands corresponding to the correct sizes of
either ribozymes or substrates were cut out, and the
transcripts eluted overnight at 4 C in a.solution containing
0.1t SDS and 0.5 M ammonium acetate. The transcripts were then
precipitated by the addition of 0.1 vol 3 M sodium acetate pH
l0 5.2 and 2.2 vol ethanol. Transcript yield was determined by
spectrophotometry.
Synthesis and PcltYfication of RNA and RNA/DNA Mixed
Polymex: RNA and RNA-DNA mixed polymers were sythesized on an
automated oligonucleotide synthesizer, and deprotected
according to previously described procedures (Perreault, J.P.,
and Altman, S. (1992) U. Mol. Biol. 226, 339-409).
These polymers were purified by 20% PAGE.
Major bands were excised and eluted as described
above.
End-Iabel2ing of RNA with [y-32P]ATP. Purified
transcripts (10 pmoles) were dephosphorylated in a 20 AL
reaction mixture containing 200 mM Tris-HC1 pH 8.0, 10 units
RNA guard, and 0.2 unit calf intestine alkaline phosphatase
(Pharmacia). The mixture was incubated at 37 C for 30 min, and
then extracted twice with a same volume of phenol:chloroform
(1:1). Dephosphorylated transcripts (1 pmole) were end-
labelled in a mixture containing 1.6 pmole (y-32P]ATP, 10 mM
Tris-HC1 pH 7.5, 10 mM MgC12, 50 mM KC1 and 3 units T4
polynucleotide kinase (Pharmacia) at 37 C for 30 min. Excess
[y-32P]ATP was removed by applying the reaction mixture onto a
spin column packed with a G-50 Sephadex*gel matrix (Pharmacia).
The concentration of labelled transcripts was adjusted to 0.o1
pmol per mL by the addition of water.
Example 2: Kinetics
Cleavage reactions. To initiate a cleavage reaction,
various concentrations of ribozymes were mixed with trace
*Trade-mark
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
16
amounts of substrate (final concentration <i nM) in a 18 L
reaction mixture containing 50 mM Tris-HC1 pH 7.5, and
subjected to denaturation by heating at 95 C for 2 min. The
mixtures were quickly placed on ice for 2 min and equilibrated
to 37 C for 5 min prior to the initiation of the reaction.
Unless stated otherwise, cleavage was initiated by the addition
of MgC12 to 10 mM final concentration. The cleavage reactions
were incubated at 37 C, and followed for 3.5 hours or until the
endpoint of cleavage was reached. The reaction mixtures were
periodically sampled (2-3 L), and these samples were quenched
by the addition of 5 L stop solution containing 95% formamide,
10 mM EDTA, 0.05% bromophenol blue and 0.05% xylene cyanol.
The resulting samples were analyzed by a 20% PAGE as described
above. Both the substrate and the reaction product bands were
detected using a Molecular Dynamic radioanalytic scanner after
exposition of the gels to a phosphoimaging screen.
Kinetic analysis. Measurement of pseudo-first-order
rate constant (kcat, KM and kcat/KM) were performed under single
turnover conditions. Briefly, trace amounts of end-labelled
substrate (<1 nM) were cleaved by various ribozyme
concentrations (5 to 500 nM). The fraction cleaved was
determined, and the rate of cleavage (kob$) obtained from
fitting the data to the equation At = Aoo(1-e-kt) where At is the
percentage of cleavage at time t, Aoo is the maximum percent
cleavage (or the end point of cleavage), and k is the rate
constant (kobs)= Each rate constant was calculated from at
least two measurements. The values of kobs obtained were then
plotted as a function of ribozyme concentrations for
determination of the other kinetic parameters: kcat, Ky and
kcat/Km= Values obtained from independent experiments varied
less than 15%. The requirement for Mg2+ by both ribozymes was
studied by incubating the reaction mixtures with various
concentrations of MgC12 (1 to 500 mM) in the presence of an
excess of ribozyme (500 nM) over substrate (< inM). The
concentrations of Mg2+ at the half maximal velocity were
determined for both ribozymes. Determination of equilibrium
dissociation constants (Kd).. For mismatched substrates which
CA 02330570 2000-10-27
WO 99/55856 - PCT/CA99/00391
17
could not be cleaved by the ribozyme, the equilibrium
dissociation constants were determined. Eleven different
ribozyme concentrations, ranging from 5 to 600 nM, were
individually mixed with trace amounts of end-labelled
substrates (< 1nM) in a 9 L solution containing 50 mM Tris-HC1
pH 7.5, heated at 95 C for 2 min and cooled to 37 C for 5 min
prior to the addition of MgC12 to a final concentration of 10
mM, in a manner similar to that of a regular cleavage reaction.
The samples were incubated at 37 C for 1.5 h, at which 2 L of
sample loading solution (50% glycerol, 0.025% of each
bromophenol blue and xylene cyanol) was added, and the
resulting mixtures were electrophoresed through a nondenaturing
polyacrylamide gel (20% acrylamide with a 19:1 ratio of
acrylamide to bisacrylamide, 45 mM Tris-borate buffer pH 7.5
and 10 mM MgC12). Polyacrylamide gels were pre-run at 20 W for
1 h prior to sample loading, and the migration was carried out
at 15 W for 4.5 h at room temperature. Quantification of bound
and free substrates was performed following an exposure of the
gels to a phosphoimaging screen as described earlier.
Example 3: Determination of Ribozyme and Substrate Sequence
Specificity
A number of ribozymes and substrates were made, some
of which are in accordance with the invention and others of
which are comparative examples. Analysis of the kinetic
parameters of cleavage reactions carried out using said
ribozymes and substrates led to the characterizations of the
method for selecting the ribozyme and substrate sequences. A
summary of the kinetic data is given below.
i) Selection of a substrate comprising the sequence 5'-
H,4G1UNHIITN-3' or 5'RRRH'4,GNNFIIITNN-3' and a ribozyme comprising
the sequence 3' -U1ITNXNN-5' .
Two forms of trans-acting delta ribozymes, bRzPl.l
and bRzP1.2 were used with their corresponding substrates
(11 nt) SP1.1 and SP1.2 for the kinetic studies (see Table 1).
The sequences of bRzP1.1 , bRzP1.2, SP1.1 and SP2.2 are given
in Fig. 1. bRzP1.2 differs from bRzP1.1 in that bRzP1.2 has
CA 02330570 2000-10-27
WO 99/55856 - PCT/CA99/00391
18
two nucleotides, at positions 22 and 24 of 6RzP1.1,
interchanged (5'-CCCAGCU-3').
TABLE 1
Kinetic parameters 6RzP.1 6RzP.2
kcat(min-1) 0.34 0.02 0.13 0.01
KM'(nM) 17.9 5.6 16.7 6.4
kcat/KM'(min-1=M-1) 1.89 x 107 0.81 x 107
KMg(mM) 2.2 1.0 2.1 0.8
Table 1. Kinetic parameters of wild type ribozyme
(6RzP1.1) and mutant ribozyme (6RzP1.2). Under single
turnover conditions, trace amounts of end-labelled
substrate (<1 nM) were cleaved by various concentrations
of ribozyme (5 to 600 nM). Reactions carried out under
these conditions displayed monophasic kinetics. The
values were calculated from at least two independent
experiments, and standard variations were less than 15%.
In order to compare the specificity of the delta
ribozyme with various substrates, bRzPl.l was used under single
turnover conditions as described above. The cleavage reactions
were performed with a trace amount of each substrate (<1 nM)
and 500 nM SRzPl.l. Under these conditions, the observed rates
reflect the rates of cleavage without interference from either
product dissociation or inhibition. For each substrate both
the observed cleavage rate constants (kobs) and the extent of
cleavage were calculated and compared to those of the wild type
substrate, as shown in Table 2.
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
19
d) 0 01 1!1 N 01 W CO GD OD N
r"'I v~(V 0~ I %D i i N N U1 01 00 'W OD rl
=~ ~~y ~ 4a U r I N r~ rl O r~ rl rl N 41 fD 41 'b 41 w
f11 ri N
4'930
Oj ~ N
=~= 44 rl t0 N M [- O 10 Ol -0 Pn
(D Oro ~ O 1 I 0 rl O N I I I 1 0 0 0 0
( D r-i O O O O O O O O
id 4-) 'd 4=1
pl b-ri fd O% m
rl LJ r1 V N Lll r1 0 Ln
0-ri O(!1 d.. O eM N O O O = ' . N til =
r=1 O
0 3 a~-~
r-i
dl 4-1 U m 1~d =~ +1 +1 +I +1 +1 +1 ,+1 +i +1 +1
~
id 0 0 p 4+ m r- ao r A.4 A A N N
G O " '", ao 0o ro ro o cd rt! r0 rt r-i c=i
dl ~A m V -0 N cy ~-i lfl M L~ Li ti L} O N O f+1
a1 v Q) >=1
b 4 (D
Id -ri .L 1 4J
N l~ %0 %O L, tf1 N ri
=N O N O O C! 0 0 0 0 O O 0 0
M Ql 1) .C~ -.
r-{ ro y rom r1 0 o 0 0 0 0 0 0 0
to r.1 O 0 13{ +1 +1 +1 +1 +1 +1 +1 +1 +1 +1
4-1 .~V Q v N 01 t~ %0 r=1 t0 rl %D r-1
td o A,q ov N r A.Q A.R r+ r- ri ri
cl1 'aO r . 0 o 0 0 o 0 0 0
a a91i
ad .N -r+ tn
E ~ =r1 fQ m 1J ~p
11 a u0i v o-
ONp)
m v~ -H ~ cn c~i U u u~ u ~(i U QI I ~I ul
4}-i N (a y ~ p ~ ~c~ ~~ry 0
C l ~I
0 ~ ~ ul ~l 9 c8 i ~ 1 Ch C9 ~
4J N r-I In
~ ri "' fd U U
w ao
0 ro -~
i 0 r
~~Hb
.~..~
43 r-i ri
Cl rl = 4) ri
= ri ~
0 toi a 3 ~ 4J
'O
N N
~~~-~ ~
u41 rd
N () d)
=1 NJ~
N 11 0~) i=~ H
~ao v ~'i ori
r- o ~ ~ u~ ~n ~n ~n r r oo ao m rn H H
~ rn oo CJ C7 U' C7 Ch CJ U U L7 C7
N~ m Rn cn ~n v~ U) cn cn mU) U) cn cn U) vo
H+1 p m
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
0
4J
O ~
4.3 ~ N
0 p fx rt
,u ~ O 3I r-I
,t-- =~i .~ U ~ O
qw,-) w I~ E
Gl t0 O H
m >+ w -.i i
(D Q -~ 4J 4.)
H 0 94 ~ ~ o
r0 ro 4J 0 .O r
~ A W U +~- rn
0 E a w
4, .-i
m 1J (d G)
00 -li W 4.- lu
4.) +J ro
- rt 0 43 99
wv b1~ I
~ ~ y 0 o ro
rw O rt w a~ -
ry ~1 -ri U
at 13
~ ~ U .~," ~ M
O W 44 .N N ~-
m AG
~ rt ~
~ aD =ri w
m N ~ ~d ~ ~-~i
r~1 U ~ .S~ 1=1 r i
U 'Cf G) r~
~ H q
~o = w a~ u
~ a =
I 4.) A ro M v
4-1 ro 1! 43 w ~
~ C H U v 3
cE -r-I 4a p
a .~ w
~ ~ ~ ~ b ~
U .C .4 G) O rl
3 .ci .ci H
-f1 .u 1J
Wtn `
+.i m n
0 v~ '.! ~4 C7
r-i U V~-1 ~'dN <
10 r-I C,"
rr: ~ Gl -.i
~
4) =H N 1.> a.)
11 'J 11 bl 10 pro
~ U Q I>d IA 4)
~ O U 1 ~
0i
~ D rt ar. p, b1
O ~ g 4J 1./ -t~".1
r-1 W 44 W
~ U O 0 ~
aJ W .O G) =.i 'Cf
.~-1~ sJ tn 3 N
m
~d 0 ~ w ro
m U u G0) Or-i
O.u 41 N N 0
Ri 'C~ '0 a ~ U
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
21
Further trans-acting delta ribozyme variants were
produced using plasmid pSRzPl.l. The variants have either A23
or C24 mutated to one of the other three possible bases. The
six resulting delta ribozyme variants are named for the altered
s nucleotide (bRzPl-A23C, -A23G, -A23U, -C24A, -C24G, and -C24U;
Table 3). Complementary or compensatory substrates (Table 3)
were generated in which either position 7 or 8 of the wild type
substrate (SP1.1) was altered in order to restore the Watson-
Crick base pair formation of the P1 stem between the substrates
1o and the ribozyme variants.
TABLE 3
Transcripts Sequence
Substrates
SP1.1 1GGGCGGGUCGG11
15 SG7A GGGCGGAUCGG
SG7C GGGCGGCUCGG
SG7U GGGCGGUUCGG
SU8A GGGCGGGACGG
SUBC GGGCGGGCCGG
20 SU8G GGGCGGGGCGG
SU8G-9mers 1GCGGGGCGG9
Ribozymes
bRzPl.l 20CCGACCU26
6RzPl-A23C CCGCCCU
25 SRzPl-A23G CCGGCCU
6RzPl-A23U CCGUCCU
6RzP1-C24A CCGAACU
bRzPl-C24G CCGAGCU
bRzPl-C24U CCGAUCU
30 The extent of cleavage of the SRzPl-C24N ribozyme
variants were compared with that of the wild type ribozyme
SRzPl.l for each of 4 substrates (A), and correspondingly, the
extent of cleavage of the bRzPl-C24N ribozyme variants were
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
22
compared with that of SRzP1.1 for each of the other 4
substrates (B). The results are shown in Figure S. The base
pair formed between the ribozyme and the substrate is indicated
by the capital and lower case letters, respectively, on each
s bar of the histogram. The values are an average calculated
from at least two independent experiments.
Complementary pairs of substrates and ribozymes were
used for kinetic studies to obtain the experimental data
required for the calculation of apparent Km (K,t,' ) and apparent
so k2 values and the results are shown in Table 4.
CA 02330570 2000-10-27
WO 99/55856 - PCT/CA99/00391
23
g~ .P a~~ o o M~~ I 4'
,0
u
U1 v U v td
~ ~ ~ ~ ~ 3 rt ~ d
0 0 0 4J
a ~a +1 z P~ +1 +1 +1
x -=i tn 'LS rd
.i o o , ~ -~ O 'o 4-)
o c o o U,,,_, p O O
v Li' = rz
.ii -r-I (A -ri
v 1] 4..1 E~ 1J 3-1
Ci' O O .rJ CS
r N lfl v O
~ o a k
vw~ ~ 0 r= .'~
Ln %o 0% m N m n ty)-~ 3-0 m
+i +i +1 +1 +1 +1 +i 0 =d 4-3 ~ry ~+
N Ln a r m ao m rA v [!] U 00 -rl
a a r H v ~o r U) v ~-I v f0 rh dJ
,O .L." v U] 4-)
~i 41 ~-I i4 ri
~vy rv.~ f~s b 4-+
O N v .~-1 r V v
+i +i +i +1 +i +1 +i fd 4.) 1 J v A
~ M o a O O A
rl 1] r1 v
a a c ~ b~
p U1 id W
N n un
H bi ^ +i +1 +i O d A T1 ~'i bl O
N A m `~ ' a =r 4~i I
, u~ -r-1 v
N 1!1 -i N N u1 ~ U tn '+ A
b ^ O 4J
43 rO O ~
C-1 b ra -ri b r-i
O ca x 5 r-, 0
o A> ~~ a~i o y
w ~.a a ~ ~ a .u i v ~
L; o m v'db -I-) b ~
' 1
a! v ao ~
4J r= = -~
~-1 ei ri N ~ rl N ~ V v 4..) M v v ~i v
W7W7 ~ N v ~ -~ u~i
r-i W r-I fEQ-I ~ (d
o 0 0 0 0 0 a ri v 'J .IJ ~-I b1 O
+i +1 ,'
.d 4 i td v v
A +1 ~
A M O O ~-1 N N 0 ~ ~ U J-~ r"'I ~ .~-I- UI IA
O ~ O O O 0 M cd O M (O -m p M
-H H aro,
= v 4J rn~'fr
cn M r~ v w a 10 01 -ri -rl v-rl RS U2 p
~ ~ M 5 H r-+ .~ S a
, ,
o a a a a a a a ~~ v A v~ N 4J ~4
N N N N N N N (D ,j-) (d A -ri rO
{4' ~O W W ~O W W ~O 'H U ~ ~ H ru ~i H
~
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
24
ii) Selection of a substrate comprising the sequence 5'-
H'4GNNHNN-3' or 5'-YHRH4GNNHNNN-3'
A collection of 13 substrates including all single
mutants for positions -4 to -1 compared to the original
substrate were synthesized. Positions -4 to -1 refer to the
four nucleotides directly 5' to the cleavage site, position -1
being right next to the cleavage site and position -4 being the
furthest from the cleavage site, as shown in Figure 2. For
each mutant, trace amounts of 5'-32P-labeled substrates (<1 nM)
were incubated in the presence of an excess of ribozyme (200
nM), and the maximal cleavage percentages (i.e. end-point)
(pre-steady state conditions) determined as a comparative
parameter. The Applicant observed that the base requirement
varies for each position. At position -1, the base preference
was A > C > U >> G, where a guanosine at this position rendered
the substrate uncleavable. At position -2, an A improved the
cleavage efficiency compared to the original G, while a
substrate with a U was poorly cleaved and a C gave an
uncleavable substrate. In contrast at position -3, C, U and A
gave substrates that have a two fold improved cleavage compared
to the wildtype G. Finally at position -4, the presence of a
pyrimidine (i.e. C or U) improved the maximal percentage of
cleavage by at least two fold compared to a purine (i.e. G or
A).
In order to assess accurately the base requirement at each
position, kinetic analysis were performed under pre-steady-
state conditions. Pseudo first-order cleavage rate constants
(k2 and Kn,') were measured with an excess of ribozyme (5 to 600
nM) and trace amounts of end-labeled substrate (<0.1 nM).
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
TABLE 5
Position Identity KM' k2 k2/3CM' Specificity
(nb!) (min-1) (n1M-linin-1) index
-1 C 31.52 0.22 6.66 x 10-3 1.00
U 33.2 0.11 3.34 x 10-3 0.50
A 14.27 0.27 1.79 x 10-2 2.68
G na na na na
-2 G 31.52 0.22 6.66 x 10-3 1.00
A 28.7 0.33 1.15 x 10-2 1.73
C na na na na
U 94 0.08 8.19 x 10-4 0.12
5 -3 G 31.52 0.22 6.66 x 10-3 1.00
A 9.93 0.20 1.99 x 10-2 3.02
C 11.3 0.24 2.10 x 10-2 3.15
U 8.76 0.20 2.32 x 10-2 3.48
-4 G 31.52 0.22 6.66 x 10-3 1.00
A 27.14 0.12 4.45 x 10-3 0.67
C 11.81 0.27 1.86 x 10-2 2.79
U 16.42 0.23 1.40 x 10-2 2.10
Table 5.Kinetic analysis of the collection of single mutated
substrates. Pseudo first-order cleavage rate constants (k2 and
Kml) were measured using an excess of ribozyme (5 to 600 nM)
10 and trace amounts of end-labelled substrate (< 0.1 nM).
Apparent second-order rate constants (k2/Km') were calculated
and their relative specificity determined as compared to the
original substrate. The values were calculated from at least
two independent experiments, and errors were less than 25%.
15 Sequence for position -4 to -1 are indicated for each
substrate.
Then, apparent second-order rate constants (k2/Km') were
calculated and a specificity index determined, fixing
arbitrarily as 1.00 the values of the original substrate (i.e.
20 _4GGGC_1). At position -1, the presence of a uridine resulted in
a similar relative specificity (0.50) while the presence of an
adenine increased the relative specificity to 2.68. This
increase appears mainly as a result of a FtR,' decrease of 2
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
26
fold. For position -2, the presence of a purine (i.e. G or A)
gave similar relative specificity (1.73, compared to 1.00,
respectively). In contrast, the presence of a uridine resulted
in a poorly cleaved substrate, while when a cytosine was
present, the substrate was uncleavable. In the case of the
uridine at position -2, the specificity was evaluated to be
reduced from 8 fold to 0.12 compared to the original substrate
(i.e. 1.00). The decrease in specificity appears to result
from a 3 fold increase of the K,,,' and a 3 fold decrease of the
k2 value. These results show a clear preference for purine in
position -2, and a pyrimidine should be avoided in that
position.
For position -3, when the guanosine of the original
substrate was replaced by any other base (i.e. A, C, or U), the
Km' was lowered by 3 fold while the k2 remained almost
identical, resulting in an specificity increase ranging from
3.02 to 3.48. Finally for position -4, a purine (G and A)
yield a substrate with about the same specificity (i.e. 0.67
and 1.00). However, the presence of a pyrimidine in position
-4 improved the specificity by at least two fold with 2.79 and
2.10 for a C and a U, respectively. Specifically, the presence
of a C or a U the Km' was lowered, while the k2 remained almost
identical. Thus, it appears clear that the base requirement
from position -4 to -1 of the substrate, contributes
significantly and differently to the ability of the substrate
to be cleaved.
Based on the observation that mutations in position
-3 were those that most strongly increased the relative
specificity, the Applicant investigated whether or not a larger
amount of Mg2+ in the cleavage reaction would affect the kinetic
parameters of these substrates. Under single turnover
conditions, in which the ribozyme and substrate concentrations
were kept at 200 nM and 1 nM, respectively, the Applicant found
that the ribozyme cleaved these substrates at Mg2+
concentrations as low as 1 mM, which is the estimated
physiological concentration of Mg2+ (Ananovoranich, S. and
Perreault, J.P. (1998) J. Biol. Chem., 273, 13182-13188, and
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
27
Trut, T.W. (1994) Mol. Cell. Biochem., 140, 1-22). A maximum
kob8 for each substrate was observed when the concentration of
Mg2+ was 10 mM. The requirement for magnesium at half-maximal
velocity (KMg) was similar for these mutated substrates and the
original substrate, varying between 1.5 to 2.2 mM. Similar
experiments were also performed with several other substrates
from the collection and identical results were obtained,
suggesting that the differences of the kinetic parameters for
various substrates were not related to different affinity for
the magnesium.
Notably, the cleavage assays performed with the
initial collection of substrates (i.e. single mutants)
indicated that the presence of a pyrimidine in the position -2
either reduces the cleavage activity or yields an uncleavable
substrate. Specifically, a uridine decreases the relative
specificity by 8 fold while a cytosine inhibits the cleavage
.completely (see Table 6). One plausible explanation of such
results is that when a C is present at position -1 and followed
by a pyrimidine (i.e. C or U) at position -2, both nucleotides
of the substrate may interact with nucleotides located on the
ribozyme resulting in inactive substrate/ribozyme complex. It
seems reasonable to suggest that base-pairing may be formed
with the ribozyme guanosines at position 27 and 28 of the J1/4
junction, which new base pairs will compete with formation of
the P1.1 stem (Fig. 2). In this case, a cytosine in position
-2 will form two consecutive GC base pairs. Similarly, a
uridine in position -2 allows formation of a GC follow by a GU,
which will be less stable than two GC's, yielding a reduced
activity compared to the absence of activity. In order to learn
more about the nucleotide preference in position -2, taking
into account the neigboring positions, a second collection of
substrates with more than one mutation were synthesized.
First, the Applicant verified whether a cytosine at
position -2 after non-cytosine at position -1 has a detrimental
effect. Based on the previous results, a substrate with an
adenine in position -1 and a cytosine in position -2,
S-A_1C_2, was synthesized and further tested for cleavage
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
28
efficiency. A moderate extent of cleavage of 14% was observed
at 200 nM ribozyme, which is less than the substrates including
either the sequence C_1G_2 or A_1G_2. In comparison to the
substrate with the sequence A_1G_2 the S-A_1C_2 substrate showed
a virtually identical apparent KM (KM') while the cleavage
constant (k2) was reduced by approximately 4 fold, yielding a
4-fold reduction of the relative specificity (i.e. from 2.68 to
0.60; Table 6). These results suggest that the presence of a
cytosine at position -2 reduces significantly the cleavage of a
substrate. Moreover, if this cytosine is followed by a second
cytosine in position -1, the result is an uncleavable RNA
molecule (see above).
TABLE 6
Mutant icmi kZ K2/ICM' Specificity
(nM) (min'1) (nM`lmin"1) index
SC_1G_2 31.5 0.22 6.98 x 10-3 1
SA_1 14.3 0.27 1.89 x 10-2 2.68
SA_1C_2 15.4 0.06 3.9 x 10-3 0.6
SA_1C_2C_3 15.2 0.039 2.57 x 10-3 0.4
SA_1A_2C_3C_4 16.5 0.25 1.52 x 10-2 2.28
Table 6.ICinetic analysis of the collection of multiple mutated
substrates. Pseudo first-order cleavage rate constants (k2 and
Km') were measured using an excess of ribozyme (5 to 600 nM)
and trace amounts of end-labelled substrate (<0.1 nM).
Apparent second-order rate constants (k2/Km') were calculated
and their relative specificity determined as compared to the
original experiments, and errors were less than 25t. Sequence
for position -4 to -1 are indicated for each substrate.
Secondly, the Applicant verified whether a cytosine
at position -2 followed by a cytosine at position -3 gives a
cleavable substrate. In other words, two consecutive
cytosines, regardless of their positions, will yield
uncleavable substrates. Therefore, the Applicant synthesized
the substrate S-A_1C_2C_3 and verified its ability to be
cleaved. The S-A_1C_2C_3 put together was cleaved with kinetic
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
29
parameters almost identical to the the substrate S-A_1C_2
substrate except that the k2 was slightly reduced to 0.039 min-1
compared to 0.062 min'1, resulting in a small reduction of the
relative specificity (i.e. from 0.60 to 0.40; Table 6). These
results show that the presence of a cytosine at position -3
following a cytosine at position -2 reduced slightly the
cleavage activity, and did not significantly modify the ability
of a substrate to be cleaved. Thus, a cytosine at position -3
does not have the same influence as that at position -2.
Thirdly, the Applicant asked whether two consecutive
cytosines at positions -4 and -3 give a similar effect yielding
uncleavable (or less cleaved) substrate. A substrate
containing cytosines at positions -3 and -4 and adenines in
position -1 and -2 was synthesized. Adenines were included in
position -1 and -2 because this residue appears to give a
readily cleaved substrate as compared to the single mutation
collection (see above). The S-A_lA_2C-3C_4 mutant has a maximum
cleavage of 61%. Moreover, the Applicant determined a KM' of
16.5 nM and a k2 value increased to 0.25 min-1, resulting in a
substrate with a relative specificity of 2.28 as compared to
the original substrate (Table 6). Thus, the presence of two
consecutive cytosines at position -3 and -4 has no detrimental
effect.
Finally, the Applicant asked whether it is possible
to compensate for the detrimental effect of the presence of two
consecutive cytosines at positions -1 and -2, by including the
one at position -2 in a hairpin structure. A longer RNA
substrate (i.e. 18-mer compared to 14mer) including a hairpin
at 5'-end, which involved the C_Z in the last base pair of the
helix was chemically synthesized and then tested. This
substrate was poorly cleaved. Only trace amounts of products
were detected (i.e. maximum percentage cleavage of <2.0 t), and
as a consequence, no more extensive characterization was
possible. If the sequence was drawn in order to avoid the
formation of the 5'-end hairpin (i.e. C_2 remains single strand;
S-hp-), no cleavage at all was observed. These two results
showed that the presence of a base-paired cytosine at position
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
-2 gave minimal activity as compared to this cytosine in single
strand. However, the improvement was very limited.
Example 4: L4 loop Modifications
A modified form of 6RzP1.1 described above was made
5 by replacing the L4 loop sequence GCUU which is relatively
unstable, with the ultrastable L4 loop (WCG) (shown on the
right in Figure 3). The kinetic parameters (kcat and KM) and
dissociation constant (Kd) were virtually identical.
Example 5: Bimolecular ribozyme
10 A modified form of SRzPl.l described above was made by
dividing the L4 loop into two resulting in two fragments,
namely, RzA and RzB (as shown in Figure 4). The RzA consists
of 37 nucleotides encompassing a substrate recognition site (P1
stem), P3 stem and portions of P2 and P4 stems. The RzB
15 consists of 20 nucleotides which is able to base pair to RzA to
form a bimolecular ribozyme complex. RzA and RzB were
synthesized as described in Example 1. Because both RzA and
RzB are relatively small, they can be chemically synthesized.
Therefore, this bimolecular delta ribozyme allows the
20 introduction of any chemically modified nucleoside.
Example 6: Deoxyribonucleotide modifications
Example 5 describes a bimolecular ribozyme. Modified
versions of the ribozyme described in Example 5 were made by
replacing one ribonucleotide in RzB with a deoxyribonucletide
25 individually at positions 9 to 15. This resulted in 7
different RzB's each containing one deoxyribonucleic acid.
The influence of 2'-OH groups in RzB on the catalytic
activity of RzA:RzB complex was analyzed. 0.066 uM of a mix of
cold and end-labeled RNA substrates were incubated in presence
30 of 0.066 uM of RzA and 0.2 uM of various RzB RNA/DNA mixed
polymers. The incubation was performed in 50 mM Tris-HC1 pH
8.0 and 50 mM MgC12 at 37 C. An aliquot was removed after one
hour and the reaction stopped by the addition of an excess of
stop solution (xc, bb, formamide). Reaction mixtures were
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
31
fractioned on 20% polyacrylamide gel electrophoresis and were
exposed on x-ray films. Fully deoxyribonucleotide RzB
molecules are not able to support a cleavage activity.
Individual deoxy substitution mutants were subjected to
catalytic cleavage. All of the reconstituted complexes were
active to different extents. S and P respectively represent
substrate and product species. As an example, dGg stands for
GGCGCAUGgCUAAGGGACCC where uppercase and lowercase letters
respectively represent ribo- and deoxyribonucleotides. The
results are shown in Figure 6 and Table 7.
Table 7 shows the quantification of time course
experiments performed. Rate and extent of cleavage values were
obtained from fitting the experimental data to the equation
At = Aalpha (1-e-kt) were At is the percentage of cleavage at
time t, Aalpha is the maximum cleavage and k is the reaction
rate. Data analysis was performed with GraFit Version 3.01
from Erithacus Software.
TABLE 7
Species Rate (min-1) Extent M
RzB 5.7 x 10-2 27.01
dG9 3.3 x 10-2 9.80
dClO 2.4 x 10-2 30.42
dUll 4.6 x 10-2 45.87
dA12 4.0 x 10-2 26.79
dAl3 1.8 x 10-2 27.46
dG14 8.0 x 10-2 61.44
dGlS 7.8 x 10-2 54.15
Table 7.Rate and extent of substrate cleavage using 2'-OH
modified ribozymes.
CA 02330570 2008-09-26
87036-1
32
Figure 6 illustrates the sequence of the ribozymes of
this Example and shows the efficiency of cleavage of the
substrate molecules as a function of the position of the
deoxyribonucleic acid.
S Example 7: Cleavage of EDAg mRNA.
Plasmids encoding the HDAg mRNA and delta ribozymes.
The pKSAgS plasmid carries the S-HDAg mRNA in pBluescript KS+
(Stratagene). Briefly, the S-HDAg mRNA insert (positions 900
to 1679 of the vHDV.5 variant (according to Lafontaine, D.,
mercure, S. and Perreault, J.-P. (1997) Nucleic Acids Res., 25,
123-125) were retrieved by PCR amplification using pSVL(AgS)
(Chao, M_, Hsieh, S.X. and Taylor, J. (1990) J. Virol., 64,
5066-5069) as template. The oligonucleotides used in this PCR
had restriction sites situated at their 51 ends so as to
facilitate subsequent cloning: HDV1679.66:
5'CCGGATCCCTCGGGCTCGGGCG 3' (underlined is the Bam Hl
restriction site) and HDV900.914: 5'CCAAGCTTCGAAGAGGAAAGAAG 3'
(underlined is the Hind III restriction site). Plasmid DNA
(pSVL(AgS), 50 ng), 0.4 mM of each oligonucleotide, 200 mM
dNTPs, 1.25 mM MgCl21 10 mM Tris-HC1 pH 8.3, 50 mM KC1, and 1 U
Taq DNA polymerase were mixed together in a final volume of 100
L. The Applicant performed one low stringent PCR cycle (94 C
for 5 min, 53 C for 30 s, 72 C for 1 min), followed by 35
cycles at higher stringency (94 C for 1 min, 62 C for 30 s,
72 C for 1 min). The mixture was fractionated by electro-
phoresis in a 1t agarose gel in 1X TBE buffer (90 mM Tris-
borate, 2 mM EDTA pH 8.0), the expected band excised and eluted
using the QIAquick*gel extraction kit (Qiagen), and finally
digested and ligated into pBluescript KS+. The strategy used
for the construction of plasmids carrying ribozymes with
modified substrate recognition domains is described above. All
constructs were verified by DNA sequencing.
RNA Synthesis. In vitro transcriptf on; HDAg mRNA was
transcribed from Hind III-linearized pKSAgS, while ribozymes
were transcribed from Sma I-linearized ribozyme encoding
plasmids as described in Example 1. Small substrates (11-nt)
*Trade-mark
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
33
were synthesized as described in Example 1.
Oligonucleot.ide probing. DNA oligonucleotides
complementary to the potential target sites were purchased from
Gibco-BRL and 5'-end labelled using T4 polynucleotide kinase
(Pharmacia) in the presence of 10 Ci [y-32P]ATP. Labelled
oligonucletiodes (- 2 500 cpm; - 0.05 nM) and unlabelled mRNA
(2.4 to 1 200 nM) were hybridized together for 10 min at 25 C
in a solution containing 50 mM Tris-HC1 pH 7.5 and 10 mM MgC12
in a final volume of 15 l. Loading solution (2 L of 1X TBE,
10 mM MgC121 40% glycerol, 0.25% bromophenol blue and 0.25t
xylene cyanol) was added, and the resulting solutions
fractionated on native 5% PAGE gels (30:1 ratio of acrylamide
to bisacrylamide, 50 mM Tris-borate pH 8.3, 10 mM MgC12 and 5%
glycerol) at 4 C in the presence of recirculating 50 mM Tris-
borate pH 8.3 and 10 mM MgCl2 buffer. The dried gels were
analyzed with the aid of a PhosphorImager (Molecular Dynamics).
RNase H probing was performed using the same oligonucleotides.
In these experiments randomly labelled S-HDAg mRNA (-10 000
cpm; -10 nM) and unlabelled oligonucleotides (1 M) were
annealed as described for gel shift assays for 10 min, then
0.2 U of E. coli RNase H (Pharmacia) was added and the reaction
incubated at 37 C for 20 min. The reactions were stopped by
the addition of stop-solution (3 L of 97% formamide, 10 mM
EDTA, 0.25% bromophenol blue and 0.25% xylene cyanol),
fractionated on 5% denaturing PAGE gels, and analyzed by
autoradiography.
In vitro cleavage assays and kinetic analyses.
Cleavage assays were performed at 37 C under single turnover
conditions with either randomly labelled mRNA (- 10 nM) or 5'-
end labelled small substrates (<inM), and an excess of ribozyme
(2,5 M) in 10 L final volume containing 50 mM Tris-HC1 pH 8.0
and 10 mM MgC12. A pre-incubation of 5 min at 37 C preceeded
the addition of the Tris-magnesium buffer which initiates the
reaction. After an incubation of 1 to 3 hrs at 37 C, stop-
solution (5 1) was added and the mixture quickly stored at
-20 C until its fractionation on 5% denaturing PAGE gels and
subsequent autoradiography. Cleavage sites of the active
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
34
ribozymes were verified by primer extension assays as described
previously (Cote, F. and Perreault, J.-P. (1997) J. Mol. Biol.,
273, 533-543). Briefly, oligonucleotides were synthesized to
have complementary sequence to positions downstream (- 100
positions) from the cleavage site according to the mRNA. For
example, for the cleavage site of Rz-12, the oligonucleotide
primer, 5'CTTTGATGTTCCCCAGCCAGG-3' (21mer), was used in the
reverse transcriptase reaction containing the ribozyme cleavage
reaction mixture.
Active ribozymes (Rz-1, -11 and -12) were
characterized under single turnover conditions essentially as
described in Example 1.
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
;
41
~
0
N
a
m
bl rl l0 M \D sN fn 10 W ri
N M OD rl n -o N l- OD w
l11 V~ d~ M N N r1 10 %0
~ . . . . . . . .
r~
U U1 O M O N M O O 1A
Ry N M V~ d~ UI U1 \O rl ri
m
41
u
m
~
44
0
m
N
~
N
0
a0 O
U
Eõ4 ~ ~ ~ C(ry7 U U Q
~ U U U p [7 D 'U.~ CU9
.-1
ri N M d~ t0 I`01 ~-I N
N 1 I t I I I I ~ ~
O N N N N N N N ~ ~
~ a a a c~ a a a a a
04
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
36
v
M.0
.~ a"
4J b
'vi y v sr ~n rn N H,d
4J =
fu m=ri
C7 A', (ry ~ry C7 U C7 U U U r-1 4JH
4) bb
a~
u u c~ ~a ~n a a cn u~~ a~ b~
L) ~ cD cD 0 0 aov~
A p
CUU'J U ~ 6 9 9 U ~ u C7
~-I N N
~4.) ~4
tO w>i
u u~ v ~i 6 8 9 u cUi~ c~3 ~ -~ O.A
u CU7 0 u u '.7 U U fd -rl u
~ ~
u u 0 L) E) to
c, ED 0 a ~.~rorn
o4J 4-) -r-I
>
f-1 rtf U ,~
C7 C7 cUi aU
u u Ei ~ ~ c~i = (o b' o
d m aNi
~~~u ~:2 1 ~
u U `J U
I U 1d UO L."
C7 g C9 U I I U U 0
N N
-) g6~u E~ ~ ~aH~
a
ma~
rO ~~~ U u ~ ~ ~~ ai -~ rci
L)~ 08 ~o
~ 8 N p ~ ~
u~ -ri -.i
~
~ u cn ~~~~~a u u a~ m o0 ~ a~i
m~ c~ u~ d~ a u~ a c~ u u~
~ 41o~
En
,~ = cu v
rl N 00 V~ O ~D N CO V~ O l0 N CI N~i N
rl ~D ~-1 e~ N r+1 r1 eM sN t!1 tD ~O [~ C^ rõ~ 0 4-1 Q
oda~iul A
H En -H H
CA 02330570 2000-10-27
WO 99/55856 PCT/CA99/00391
37
Of the nine ribozymes examined, three, namely Rzl,
Rzll, and Rz12, specifically cleaved a derivative HDV mRNA.
The most active ribozyme under steady-state conditions,
displaying multiple turnovers, was Rz-12. As can be observed
from Table 8, the sequence of the substrate for this ribozyme
(positions 87-97) is 5'CAGUyGGGUGG-3'. This accords with the
sequence preferences shown in Table 5.
Example 8: Cleavage Assay of a ribozyme of the invention
against 552 nt-HBV RNA substrate.
500 nM of a delta ribozyme as shown in Figure 7 was
incubated with 1 nM randomly-labelled 552 nt-HBV (human
hepatitis B virus) mRNA at 37 C in the presence of 50 mM Tris-
HC1 pH 7.5 and 10 mM MgC12. A single exponential equation was
used to fit data to kobs = 0.031 min-1 with 28% cleavage. This
demonstrates that a ribozyme of the invention cleaves mRNA from
the human hepatitis B virus.
CA 02330570 2001-04-27
1
SEQUENCE LISTING
<110> Perreault, Jean-Pierre
Ananvoranich, Sirinart
Lafontaine, Daniel
Universite de Sherbrooke
<120> Nucleic Acid Enzyme for RNA Cleavage
<130> 77473-5
<140> PCT/CA99/00391
<141> 1999-04-29
<150> CA 2,230,203
<151> 1998-04-29
<160> 54
<170> PatentIn Ver. 2.0
<210> 1
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: nucleotide
sequence comprised of ribonucleotides or a
combination of ribonucleotides and
deoxyribonucleotides
<220>
<221> miscfeature
<222> (1) _.. (11)
<223> n is a, c, g, t, u, a nucleotide, unknown, or other
<400> 1
nnrhgnnhnn n 11
<210> 2
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: nucleotide
sequence comprised of ribonucleotides or a
combination of ribonucleotides and
deoxyribonucleotides
<220>
<221> miscfeature
<222> (6) _.. (11)
<223> n is a, c, g, t, u, a nucleotide, unknown, or other
CA 02330570 2001-04-27
2
<400> 2
rrrhgnnhnn n 11
<210> 3
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: nucleotide
sequence comprised of ribonucleotides or a
combination of ribonucleotides and
deoxyribonucleotides
<220>
<221> miscfeature
<222> (6) _.. (11)
<223> n is a, c, g, t, u, a nucleotide, unknown, or other
<400> 3
gggcgnnunn n 11
<210> 4
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: nucleotide
sequence comprised of ribonucleotides or a
combination of ribonucleotides and
deoxyribonucleotides
<220>
<221> miscfeature
<222> (6) _.. (11)
<223> n is a, c, g, t, u, a nucleotide, unknown, or other
<400> 4
gggcgnnhnn n 11
<210> 5
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: nucleotide
sequence comprised of ribonucleotides or a
combination of ribonucleotides and
deoxyribonucleotides
<220>
<221> miscfeature
<222> (6) _.. (11)
<223> n is a, c, g, t, u, a nucleotide, unknown, or other
CA 02330570 2001-04-27
3
<400> 5
gggugnnunn n 11
<210> 6
<211> 12
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: nucleotide
sequence comprised of ribonucleotides or a
combination of ribonucleotides and
deoxyribonucleotides
<220>
<221> miscfeature
<222> (6) _.. (12)
<223> n is a, c, g, t, u, a nucleotide, unknown, or other
<400> 6
gggugnncnn nn 12
<210> 7
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: nucleotide
sequence comprised of ribonucleotides or a
combination of ribonucleotides and
deoxyribonucleotides
<220>
<221> miscfeature
<222> (6) _.. (11)
<223> n is a, c, g, t, u, a nucleotide, unknown, or other
<400> 7
aaacgnnunn n 11
<210> 8
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: nucleotide
sequence comprised of ribonucleotides or a
combination of ribonucleotides and
deoxyribonucleotides
<220>
<221> miscfeature
<222> (6) _.. (11)
<223> n is a, c, g, t, u, a nucleotide, unknown, or other
CA 02330570 2001-04-27
4
<400> 8
yhrhgnnhnn n 11
<210> 9
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 9
gggcgggucg 9 11
<210> 10
<211> 10
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 10
gggcgggucg 10
<210> 11
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 11
gggcaggucg g 11
<210> 12
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 12
gggccggucg g 11
<210> 13
<211> 11
<212> RNA
<213> Artificial Sequence
CA 02330570 2001-04-27
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 13
gggcgagucg g 11
<210> 14
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 14
gggcgugucg g 11
<210> 15
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 15
gggcggaucg g 11
<210> 16
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 16
gggcgguucg g 11
<210> 17
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 17
11
gggcgggccg 9
CA 02330570 2001-04-27
6
<210> 18
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 18
gggcggggcg g 11
<210> 19
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 19
gggcggguag g 11
<210> 20
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 20
gggcggguug g 11
<210> 21
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 21
gggcgggucu g 11
<210> 22
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
CA 02330570 2001-04-27
7
<400> 22
11
gggcgggucg U
<210> 23
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 23
gggcggcucg g 11
<210> 24
<211> 11
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 24
gggcgggacg g 11
<210> 25
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 25
ggagggcggg ucgg 14
<210> 26
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 26
ggaggggggg ucgg 14
<210> 27
<211> 14
<212> RNA
<213> Artificial Sequence
CA 02330570 2001-04-27
8
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 27
ggacggcggg ucgg 14
<210> 28
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 28
ggaggccggg ucgg 14
<210> 29
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 29
ggaaggcggg ucgg 14
<210> 30
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 30
ggagagcggg ucgg 14
<210> 31
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 31
ggaggacggg ucgg 14
CA 02330570 2001-04-27
9
<210> 32
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 32
ggagggaggg ucgg 14
<210> 33
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 33
ggauggcggg ucgg 14
<210> 34
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 34
ggagugcggg ucgg 14
<210> 35
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 35
ggaggucggg ucgg 14
<210> 36
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
CA 02330570 2001-04-27
<400> 36
ggaggguggg ucgg 14
<210> 37
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 37
ggagggaggg ucgg 14
<210> 38
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 38
ggaggcaggg ucgg 14
<210> 39
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 39
ggagccaggg ucgg 14
<210> 40
<211> 14
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 40
ggaccaaggg ucgg 14
<210> 41
<211> 22
<212> DNA
<213> Artificial Sequence
CA 02330570 2001-04-27
11
<220>
<223> Description of Artificial Sequence: DNA PCR
primer
<400> 41
ccggatccct cgggctcggg cg 22
<210> 42
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: DNA PCR
primer
<400> 42
ccaagcttcg aagaggaaag aag 23
<210> 43
<211> 813
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 43
caccgcggug gcggccgcuc uagaacuagu ggaucccucg ggcucgggcg gcgaguccag 60
cagucuccuc uuuacagaaa auguaagagu acugaggacu gccgccucua gccgagauga 120
gccgguccga gucgaggaag aaccgcggag ggagagaaga gauccucgag cagugggugg 180
ccggaagaaa gaaguuagag gaacucgaga gagaccuccg gaagacaaag aagaaacuca 240
agaagauaga ggacgaaaau cccuggcugg ggaacaucaa aggaauucuc ggaaagaagg 300
auaaggaugg agagggggcu ccccccgcga agagggcccg aacggaccag auggagguag 360
acuccggacc ucggaagagg ccucucaggg gaggauucac cgacaaggag aggcaggauc 420
ccgacgaagg aaggcccucg agaacaagaa gaagcagcua ucggcgggag gcaagaaccu 480
cagcaaggag gaagaagagg aacucaggag guugaccgag gaagacgaga gaagggaaag 540
aagaguagcc ggcccgccgg uugggggugu gaacccccuc gaagguggau cgaggggagc 600
gcccgggggc ggcuucgucc ccaaucugca gggagucccg gagucccccu ucucucggac 660
cggggagggg cuggacauca ggggaaacca gggauuucca uaggauauac ucuucccagc 720
cgauccgccc uuuucucccc agaguugucg accccaguga auaaagcggg uuuccacuca 780
cagguuugcg ucucgcgucc uucuuuccuc uuc 813
<210> 44
<211> 57
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 44
ggguccaccu ccucgcgguc cgaccugggc augcggcuuc gcauggcuaa gggaccc 57
CA 02330570 2001-04-27
12
<210> 45
<211> 57
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 45
ggguccaccu ccucgcgguc ccagcugggc augcggcuuc gcauggcuaa gggaccc 57
<210> 46
<211> 57
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 46
ggguccaccu ccucgcgguc cgaccugggc augccuucgg gcauggcuaa gggaccc 57
<210> 47
<211> 57
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 47
ggguccaccu ccucgcgguc cgcccugggc augcggcuuc gcauggcuaa gggaccc 57
<210> 48
<211> 57
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 48
ggguccaccu ccucgcgguc cggccugggc augcggcuuc gcauggcuaa gggaccc 57
<210> 49
<211> 57
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
CA 02330570 2001-04-27
13
<400> 49
ggguccaccu ccucgcgguc cguccugggc augcggcuuc gcauggcuaa gggaccc 57
<210> 50
<211> 57
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 50
ggguccaccu ccucgcgguc cgaacugggc augcggcuuc gcauggcuaa gggaccc 57
<210> 51
<211> 57
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 51
ggguccaccu ccucgcgguc cgagcugggc augcggcuuc gcauggcuaa gggaccc 57
<210> 52
<211> 57
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 52
ggguccaccu ccucgcgguc cgaucugggc augcggcuuc gcauggcuaa gggaccc 57
<210> 53
<211> 37
<212> RNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: synthetic RNA
sequence
<400> 53
ggguccaccu ccucgcgguc cgaccugggc augcggc 37
<210> 54
<211> 18
<212> RNA
<213> Artificial Sequence
CA 02330570 2001-04-27
14
<220>
<223> Description of Artificial Sequence: synthetic
sequence which is comprised of ribonucleotides or
a combination of both ribonucleotides and
deoxyribonucleotides
<400> 54
ggcauggcua agggaccc 18