Note: Descriptions are shown in the official language in which they were submitted.
CA 02335646 2001-O1-19
"u WO 00/05388 PCT/EP99/04892
Genes of the DEAD box protein family, their expression products and use
Description
The present invention relates to the preparation of novel nucleic acids from
ciliates coding for expression products, preferably for RNA helicases from
the family of DEAD box proteins, and the use thereof.
The modulation of the RNA structure has an essential function in cellular
processes, such as, for example, in pre-mRNA splicing, in RNA transport or
in protein translation, as the cellular RNA is present in the cell in
different
secondary and tertiary structures and, in addition, a large number of RNA-
binding proteins provides for further structuring of the RNA. Proteins from
the family of the so-called DEAD protein family, inter alia, are involved in
these modulation processes. The members of this protein superfamily,
which characteristically contain a number of homologous protein
sequences, so-called "protein boxes", are named after the highly
conserved tetrapeptide Asp-Glu-Ala-Asp, in the single-letter code DEAD,
as a motif. This protein superfamily includes in particular RNA helicases.
The characteristic protein sequences of DEAD proteins are highly
conserved in evolution (cf. Figure 1 ).
The DEAD superfamily is divided into various subfamilies, which according
to their sequence motif are called DEAD, DEAH or DExH subfamily. All
family members have an ATP-binding and RNA-binding function and also
an ATP hydrolysis and for the most part an RNA helicase function (Fig. 2).
A conserved region comprising approx. 300 amino acids and flanked by
nonconserved amino acid sequences of varying length is characteristic of
the various members of the DEAD box protein family
CA 02335646 2001-O1-19
w WO 00/05388 2 PCT/EP99/04892
(Schmid S.R., Lindner P., Mol Cell Biol 1991 11: 3463-3471 ).
The so-called homology boxes (synonymously conserved motifs), one of
which is the "DEAD box", are located within the conserved region. The
homology boxes confer not only structural but also functional similarity on
the members of the DEAD box family. Considering the homology boxes
(see Figure 2), DEAD box proteins are putative ATP-dependent RNA
helicases which take part in a plurality of cellular processes and are
connected with secondary structure modification of RNA molecules
(Fuller-Pace F.V. Trends Cell Biol. 1994 4:271-274, Pause A.,
Sonenberg N., Curr Opin Struct Biol 1993 3:953-959). Helicase-dependent
processes have been described likewise for: translation initiation, nuclear
and mitochondrial RNA splicing, mRNA transport, ribosome assembly and
spliceosome assembly, mRNA stabilization and mRNA degradation (lost I.,
Dreyfus M., Nature 1994 372:193-196).
The functions of RNA helicases corresponding to the DEAD homology
boxes in cellular processes, preferably in the context of protein
biosynthesis, allow specifically these enzymes to be employed with regard
to pharmaceutical, agricultural or biotechnological and analytical
applications.
Important pharmaceutical applications are the development of substances
which inhibit specifically bacterial, parasitical and viral helicases or
helicases originating from pathogenic fungi, but which have no inhibitory
effect on human helicases. Since helicases are for the most part essential
enzymes, it is possible to achieve destruction of the pathogen (bacterium,
fungus, parasite/protozoon, virus) by specific inhibition of these enzymes.
According to Missel et al., switching off the gene for a DEAD box protein
leads to decreased growth in particular protozoa (Trypanosoma,
Leishmania, Crithidia) (Missel A., Souza A.E., Norskau G, Goringer H.U.,
Mol Cell Biol 1997 17:4895-903). In the malaria pathogen Plasmodium
falsiparum helicases control protein translation, mitosis and DNA repair
(Thelu J, Burnod J, Bracchi V, Ambroise-Thomas P, DNA Cell
CA 02335646 2001-O1-19
WO 00/05388 3 PCT/EP99/04892
Biol 1994 13: 1109-1115). Helicases are essential for the initiation of
translation, in the spliceosome, in the cell cycle and assembly of ribosomes
in yeast. Thus for example, the DEAD box protein ROK1 is essential for the
viability of yeast, for pre-rRNA processing and for mitotic growth. Switching
off ROK1 blocks 18S rRNA synthesis (Venema J., Bousquet-Antonelli C.,
Gelugne J.P., Caizegues-Ferrer M., Tollenrey, Mol Cell Biol 1997 17: 3398-
3407).
Approx. 80% of all "positive-stranded" RNA viruses whose genomes have
been sequenced code for at least one putative helicase. Examples are NS3
of hepatitis C virus, helicases of human coronavirus and adeno-associated
virus, and vaccinia virus helicase (Kadare G., Haenni A.L., J Virol 1997:
2583-2590). Possible roles for viral helicases are (i) proof reading during
replication, (ii) transcription initiation by unwinding the RNA and preventing
loop formation behind RNA polymerase, (iii) translation initiation. Vaccinia
virus helicase is essential for the life cycle of the virus and is nucleic
acid-
specific.
There are indications that at least some helicases can be very specifically
activated. Thus, for example, DpbA from E. coli (Fuller-Pace F.V., Nicol
S.M., Reid A.D., Land D.P., EMBO J 1993 12:3619-3626) and SIt22 from
yeast (Xu D., Nouraini S., Field D., Tang S.J., Friesen J.D., Nature 1996
381: 709-716) need specific RNA ligands for activation.
In addition, DEAD box proteins are described in association with diseases.
The amplification of a specific gene in cancer cells (N-myc) is linked to the
fact that a DEAD box protein is coamplified with N-myc, which points to a
role of this protein in the degeneration of cancer cells (George R.E.,
Kenyon R.M., McGuckin A.G., Malcolm A.J., Pearson A.D., Lunec J.,
Oncogene 1996 12: 1583-7). Mutations of an RNA helicase are connected
with Werner's syndrome - premature aging - (Yu C., Oshima J., Wijsman
E.M., Nakura J. et al., Am J Hum Genet 1997 60: 330-341 ) and with
xeroderma pigmentosum (Kobayashi T., Kuraoka I., Sailo M., Nakatsu Y.
Tanaka A. et al., Huma Mut 1997 9: 322-331 ). Furthermore, a possible
CA 02335646 2001-O1-19
WO 00/05388 4 PCT/EP99/04892
connection between DEAD box proteins and connective tissue diseases
has been postulated (Valdez B.C., Henning D., Perlaky L., Busch R.K.,
Busch H., Biochem Biophis Res Commun 1997 234: 335-340). Additionally
a connection is known between defective DNA repair and a mutation in the
helicase domain of the XNP/ATR-X gene (Villard L., Lossi AM, Cardoso C,
Proud V, Chiaroni P, Colleaux L, Schwartz C, Fontes M Genomics 1997
43: 149-155).
In addition to helicases from humans or from various pathogens, helicases
from plants are also of the greatest interest. Some DEAD box proteins from
plants have become known (Lorkovic Z.J., Hermann R.G., Oelmialler R.,
Mol Cell Biol 1997 17: 2257-2265 and Aubourg et al., Gene 1997 199(1-2):
241-253). Although these proteins are structurally similar to DEAD box
proteins from organisms other than plants, they form nevertheless a
subgroup (Fig. 3). Fig. 3 shows the phylogenetic relationship of various
eIFA4, one of the best characterized members of the DEAD box protein
family, from various organisms. The plant proteins are much more closely
related to each other than to eIF4A from animal eukaryotes. An application
which is of interest to agricultural production is the stimulation of the
activity
of plant-specific RNA helicases in order to increase protein expression of
economically relevant proteins. In this case it is possible either to
stimulate
helicases intrinsic to plants (for example by overexpression) or to express
helicases plant-like heterologously in useful plants.
It is therefore the object of the present invention to provide novel nucleic
acids coding preferably for RNA helicases.
The object is achieved by providing nucleic acids which code for RNA
helicases and which are obtained preferably from ciliates, particularly
preferably Tetrahymena thermophila. The nucleic acids of the invention
code for expression products originating from the family of DEAD box
proteins and thus also for RNA helicases.
CA 02335646 2001-O1-19
WO 00/05388 5 PCT/EP99/04892
Expression products, preferably proteins from the DEAD protein
superfamily, in accordance with this invention are those having conserved
motifs among which one conserved motif comprises the amino acid
sequence DEAD. Preferably the proteins comprise an RNA helicase activity
and ATPase activity.
The present invention therefore relates to nucleic acids coding for RNA
helicases having a nucleic acid sequence depicted in SEQ ID No. 13 or
SEQ ID No. 15 or a functional variant thereof, and parts thereof having at
least 8 nucleotides, preferably having at least 15 or 20 nucleotides, in
particular having at least 100 nucleotides, especially having at least 300
nucleotides (called "nucleic acids of the invention" in the following).
The nucleic acid of the invention having the nucleic acid sequence depicted
in SEQ ID No. 13 ("Hc1" in the following) codes for an amino acid
sequence depicted in SEQ ID No. 14.
The nucleic acid of the invention having the nucleic acid sequence depicted
in SEQ ID No. 15 ("Hc2" in the following) codes for an amino acid
sequence depicted in SEO ID No. 16.
Expression of the nucleic acids of the invention in E. coli led to an
expression product which shows similar enzymatic activities to those of an
RNA helicase. Further experiments according to the present invention
confirmed that the nucleic acid is a nucleic acid coding for an RNA
helicase, in particular due to the presence of the characteristic homology
boxes such as in SEQ ID No. 14 and SEQ ID No. 16, which are
represented in Figurel.
In a preferred embodiment the nucleic acid of the invention is a DNA or
RNA, preferably a double-stranded DNA, and in particular a DNA having a
nucleic acid sequence coding for RNA helicases.
CA 02335646 2001-O1-19
WO 00/05388 6 PCT/EP99/04892
The term "functional variant" means according to the present invention a
nucleic acid which is functionally related to RNA helicases having the
described homology boxes.
In a wider sense, the term "variants" means according to the present
invention nucleic acids having a homology, in particular a sequence identity
of approx. 60%, preferably of approx. 75%, in particular of approx. 90% and
especially of approx. 95%.
The parts of the nucleic acid of the invention may be used, for example, for
preparing individual epitopes, as probes for identifying further functional
variants or as antisense nucleic acids. For example, a nucleic acid of at
least approx. 8 nucleotides is suitable as an antisense nucleic acid, a
nucleic acid of at least approx. 15 nucleotides is suitable as a primer in the
PCR method, a nucleic acid of at least approx. 20 nucleotides is suitable
for identifying further variants and a nucleic acid of at least approx. 100
nucleotides is suitable as a probe.
In particular it is possible to use the nucleic acids of the invention in
order
to construct complementary and/or antisense nucleic acids which hybridize
with Hc1 or Hc2 themselves or with related nucleic acids. Introducing the
complementary and/or antisense nucleic acid into the target cell prevents
expression of related RNA helicases or related expression products.
Antisense nucleic acids obtainable from the nucleic acids of the invention
may therefore be used for the specific regulation of gene expression. In this
case either the target cell may be transfected according to known methods
with the anti-gene which is then transcribed in the cell, or in vitro
synthesized antisense RNA or DNA is introduced into the target cell by
microinjection. It is known that antisense RNA complementary to the coding
region of the target mRNA can inhibit gene expression.
CA 02335646 2001-O1-19
WO 00/05388 7 PCT/EP99/04892
The duplex strand formed by mRNA and antisense RNA is susceptible to
fast degradation by RNAses.
Inhibition of transcription and translation by the antisense technique
discussed has also been successfully carried out in plant cells (van der Krol
A.R. et al. Nature 1988 333: 866).
In a further preferred embodiment the nucleic acid of the invention
comprises one or more noncoding sequences and/or a poly (A) sequence.
The noncoding sequences are, for example, intron sequences or regulatory
sequences, such as promoter sequences or enhancer sequences, for the
controlled expression of expression products, preferably of RNA helicases.
In a further embodiment the nucleic acid of the invention is therefore
included in a vector, preferably in an expression vector or a vector effective
for gene therapy.
The expression vectors may be, for example, prokaryotic or eukaryotic
expression vectors. Examples of prokaryotic expression vectors are for
expression in E. coli, for example, the T7 expression vector pGMlO or
pGEX-4T-1 GST (Pharmacia Biotech), which codes for an N-terminal Met-
Ala-His6 tag, which facilitates advantageous purification of the expressed
protein via a Ni2+-NTA column. Suitable eukaryotic expression vectors for
expression in Saccharomyces cerevisiae are, for example, the vectors
p426Met25 or p426GAL 1 (Mumberg et al. (1994) Nucl. Acids Res., 22,
5767), while suitable vectors for expression in insect cells are, for example,
baculovirus vectors as disclosed in EP-B1-0127839 or EP-B1-0549721,
and suitable vectors for expression in mammalian cells are, for example,
SV40 vectors which are generally available.
In general the expression vectors also include regulatory sequences
suitable for the host cell such as, for example, the trp promoter for
expression in E. coli
CA 02335646 2001-O1-19
WO 00/05388 8 PCT/EP99/04892
(see e.g. EP-B1-0154133) in E. coli, the ADH-2 promoter for expression in
yeasts (Russet et al. (1983), J. Biol. Chem. 258, 2674), the baculovirus
polyhedrin promoter for expression in insect cells (see e.g. EP-B1-
0127839) or the early SV40 promoter or LTR promoters, for example of
MMTV (mouse mammary tumour virus; Lee et al. (1981 ) Nature, 214, 228).
The recombinant proteins obtained in this way are purified using suitable
methods (e.g. affinity chromatography, HPLC, FPLC) and dissolved
(guanidine, urea). Characterization of the proteins and determination of
enzyme activity are carried out with the aid of established assays (RNA
binding, ATPase activity, helicase activity).
Examples of vectors effective for gene therapy are virus vectors, preferably
adenovirus vectors, in particular replication-deficient adenovirus vectors, or
adeno-associated virus vectors, for example an adeno-associated virus
vector consisting exclusively of two inserted terminal repeats (ITR).
Examples of suitable adenovirus vectors are described in McGrory, W.J. et
al. (1988) Virol. 163, 614; Gluzman, Y. et al. (1982) in "Eukaryotic Viral
Vectors" (Gluzman, Y. ed.) 187, Cold Spring Harbor Press, Cold Spring
Habor, New York; Chroboczek, J. et al. (1992) Virol. 186, 280; Karlsson, S.
et al. (1986) EMBO J.. 5, 2377 or W095/00655.
Examples of suitable adeno-associated virus vectors are described in
Muzyczka, N. (1992) Curr. Top. Microbiol. Immunol. 158, 97; W095/23867;
Samulski, R.J. (1989) J. Virol, 63, 3822; W095/23867; Chiorini, J.A. et al.
(1995) Human Gene Therapy 6, 1531 or Kotin, R.M. (1994) Human Gene
Therapy 5, 793.
It is also possible to obtain vectors effective for gene therapy by
complexing the nucleic acid of the invention with liposomes. Suitable for
this purpose are lipid mixtures as described in Felgner, P.L. et al. (1987)
Proc. Natl. Acad. Sci, USA 84,
CA 02335646 2001-O1-19
WO 00/05388 9 PCT/EP99/04892
7413; Behr, J.P. et al. (1989) Proc. Natl. Acad. Sci. USA 86, 6982; Felgner,
J.H. et al. (1994) J. Biol. Chem. 269, 2550 or Gao, X. & Huang, L. (1991 )
Biochim. Biophys. Acta 1189, 195. The liposomes are prepared by binding
the DNA ionically on the surface of the liposomes, in a ratio such that a
positive net charge remains and that the DNA is completely complexed by
the liposomes.
The nucleic acids of the invention can, for example, be synthesized
chemically, e.g. by the phosphotriester method (see, e.g., Uhlman, E. &
Peyman, A. (1990) Chemical Reviews, 90, 543, No. 4), either on the basis
of the sequence disclosed in SEQ ID No. 13 and SEQ ID No. 15 or on the
basis of the peptide sequence disclosed in SEQ ID No. 14 and SEQ ID No.
16 and making use of the genetic code.
A further possibility of obtaining the nucleic acids of the invention
themselves and variants is that of using a suitable probe (see, e.g.,
Sambrook, J. et al. (1989) Molecular Cloning. A laboratory manual. 2nd
Edition, Cold Spring Harbor, New York) to isolate them from a suitable
gene library. Examples of suitable probes are single-stranded DNA
fragments which have a length of from approx. 100 to 1000 nucleotides,
preferably of a length of from approx. 200 to 500 nucleotides, in particular
of a length of from approx. 300 to 400 nucleotides, and whose sequence
can be derived from the nucleic acid sequence depicted in Figures 4 and 6.
The invention further relates to the use of the nucleic acids of the invention
for specifically affecting protein biosynthesis.
Experiments show that increased helicase activity leads to enhanced
unwinding of the target RNA. This makes the target RNA available for
binding partners such as proteins, in particular enzymes or enzyme
complexes for ribosomal translation or for degradation by the
degradosome. Depending on the target RNA, this leads to a) increased
translation and therefore increased protein biosynthesis, b) faster
degradation by the degradosome and thus decreased protein biosynthesis.
Inhibition of the helicase activity may
CA 02335646 2001-O1-19
WO 00/05388 10 PCT/EP99/04892
inhibit degradation by the degradosome and lead to a decreased protein
biosynthesis. This is based on the finding that important biosynthetic
processes can be specifically regulated by selective inhibition or activation
of helicases.
For this purpose, the nucleic acids are expressed in a recombinant manner
in suitable target organisms as described.
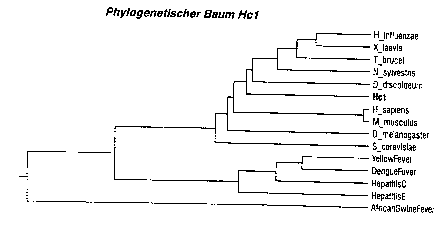
The nucleic acids of the invention, preferably Hc1, are an outstanding
model for various eukaryotic RNA helicases, preferably from humans and
parasites (Fig. 3A). The genetic relationship of Hc1 with the relevant
eukaryotic helicases is close enough in order to draw conclusions from
experiments with Hc1 about structure and function of other eukaryotic
helicases (e.g. human). Figure 3A shows the genetic relationship of some
helicases from various organisms in comparison with Hc1. Particularly
surprising are the great structural similarity between Hc1 and mammalian
helicases from humans and mice and the great structural difference
between Hc1 and known viral helicases.
On the basis of phylogenetic studies depicted in Figure 3B, the nucleic
acids of the invention, preferably Hc2, prove to be an outstanding model for
various eukaryotic RNA helicases, preferably from plants. Figure 3B shows
the genetic relationship of some RNA helicases from various organisms in
comparison with Hc2. Particularly surprising is the great structural
similarity
between Hc2 and RNA helicases from plants. The recombinant expression
of Hc2 makes it possible to use this new enzyme as a model for
investigating in particular helicases from plants and the structure and
function thereof and for developing suitable inhibitors or activators of these
important enzymes. Thus it has been postulated for PRH75 from spinach
that this enzyme needs a very specific RNA ligand in order to be active
(Lorkovic Z.J., Herrmann R.G., Oelmuller R., Mol Cell Biol 17(4): 2257-
2265 (1997)).
CA 02335646 2001-O1-19
WO 00/05388 11 PCT/EP99/04892
The invention therefore further relates to the specific heterologous
expression - by means of overexpression according to known methods - of
the nucleic acids of the invention, preferably Hc2, in suitable useful plants
for the potential increase in the biosynthesis of relevant proteins.
The nucleic acids of the invention, preferably Hc2, may be introduced into
plants by recombinant DNA techniques. A method which may be used is
the introduction of the foreign gene with the aid of Agrobacterium
tumefaciens. This involves introducing the foreign gene into the genome of
the bacterium in a known manner. Infecting the target plant leads to stable
integration of the genes of the bacterium including the foreign gene into the
genome of the plant (Chilton M.D. et al., Cell 1977 11:263, Barton K.A. et
al., Cell 1983 32: 1033). It is preferred to employ this method for the
transformation of dicotyledons. Known methods such as calcium phosphate
precipitation, PEG treatment, electroporation or a combination of these
methods may be employed for the transformation of monocotyledons
(Potrykus I. et al., Mol Gen Genet 1985 199: 183; Lorz H. et al., Mol Gen
Genet 1985 199: 178; Fromm M et al. Nature 1986 319:791; Uchimiya H. et
al., Mol Gen Genet 1986 204: 204). It is also possible to introduce the
foreign DNA into plant cells with the aid of the so-called gene gun (Klein
T.M. et al. Nature 1987 327: 70).
The invention further relates to the use of the nucleic acids as selection
markers in molecular biology. Conventionally, antibiotics are used as
selection markers. Molecular-biologically modified organisms carry a gene
which confers resistance to an antibiotic. The organisms are grown in
antibiotic-containing medium such that only the carriers of the resistance
gene are able to develop. Analogously, helicase genes may be used as
"resistance genes". It has been shown (Mullner et al, patent application
DPA 19545126.0) that overexpression of a helicase gene in murine cells
confers on these cells tolerance of an otherwise toxic substance,
leflunomide. It is thus possible to employ the nucleic acids of the invention
as selection markers in molecular biology, with the nucleic acids of the
invention having to be introduced into cells using a suitable vector as
described, and to select using a suitable substance, such as leflunomide),
CA 02335646 2001-O1-19
WO 00/05388 12 PCT/EP99/04892
of which the cells are tolerant due to overexpression of the helicase.
The present invention furthermore also relates to the expression products,
preferably polypeptides and polypeptide fragments (encoded by Hc1 and
Hc2) themselves, having amino acid sequences depicted in SEQ ID No. 14
and SEQ ID No. 16 or a functional variant thereof, and parts thereof having
at least six amino acids, preferably having at least 12 amino acids, in
particular having at least 65 amino acids and especially having 257 amino
acids Hc1 and 255 amino acids Hc2 (subsequently called "polypeptides of
the invention"). For example, a polypeptide which is approx. 6-12 amino
acids in length, preferably approx. 8 amino acids in length, can contain an
epitope which, after having been coupled to a carrier, is used for preparing
specific polyclonal or monoclonal antibodies (in this regard, see, e.g.,
US 5,656,435). Polypeptides whose length is at least approx. 65 amino
acids can also be used directly, without any carrier, for preparing polyclonal
or monoclonal antibodies.
In accordance with the present invention, the term "functional variant"
means polypeptides which are functionally related to the peptides of the
invention, i.e. which have an RNA helicase activity. Variants also mean
allelic variants or polypeptides which are derived from other human cells or
tissue.
In the broader sense, it also means polypeptides which possess a
sequence homology, in particular a sequence identity, with the poly-
peptides having the amino acid sequences depicted in Figures 5 and 7 of
approx. 70%, preferably of approx. 80%, in particular of approx. 90%,
especially of approx. 95%. They furthermore also include deletion of the
polypeptide in the range of approx. 1 - 60, preferably of approx. 1 - 30, in
particular of approx. 1 - 15, especially of approx. 1 - 5 amino acids. In
addition, they also include fusion proteins which comprise the above-
described polypeptides of the invention,with the
CA 02335646 2001-O1-19
WO 00/05388 13 PCT/EP99/04892
fusion proteins already themselves possessing the function of an RNA
helicase or only being able to acquire the specific function after the fusion
moiety has been eliminated. Especially, they include fusion proteins having
a moiety of, in particular, nonhuman sequences of approx. 1 - 200,
preferably approx. 1 - 150, in particular of approx. 1 - 100, especially of
approx. 1 - 50 amino acids. Examples of nonhuman peptide sequences are
prokaryotic peptide sequences, for example from E. coli galactosidase or a
so-called histidine tag, e.g. a Met-Ala-His6 tag. A fusion protein containing
a so-called histidine tag is particularly advantageously suitable for
purifying
the expressed protein via metal ion-containing columns, for example via a
Ni2+-NTA column. "NTA" stands for the chelating agent nitrilotriacetic acid
(Qiagen GmbH, Hilden).
The parts of the polypeptide of the invention represent, for example,
epitopes which can be specifically recognized by antibodies.
By comparing with known helicases, it was found that the polypeptides of
the invention is a member of the so-called DEAD superprotein family.
Figure 1 shows the conserved motifs which are characteristic of this class
of RNA helicases. All these motifs are highly conserved within the family
and are also found in the polypeptides of the invention.
The polypeptide of the invention is prepared, for example, by expressing
the nucleic acid of the invention in a suitable expression system, as already
described above, using methods which are generally known to the skilled
worker. The present invention therefore also relates to a method for
preparing a polypeptide of the invention, with a nucleic acid of the invention
being expressed in a suitable host cell and being isolated, where
appropriate.
In particular, said parts of the polypeptide can also be synthesized by
classical peptide synthesis (Merrifield technique).They are
CA 02335646 2001-O1-19
WO 00/05388 14 PCT/EP99/04892
particularly suitable for obtaining antisera which can be used for screening
suitable gene expression libraries in order to gain access to other functional
variants of the polypeptide of the invention.
The present invention furthermore also relates to antibodies which react
specifically with the polypeptide of the invention, with the abovementioned
parts of the polypeptide either themselves being immunogenic or being
able to be made immunogenic, or to have their immunogenicity increased,
by being coupled to suitable carriers such as, for example, bovine serum
albumin.
The antibodies are either polyclonal or monoclonal. Their production, which
is also an aspect of the present invention, is carried out, for example,
according to generally known methods, by immunizing a mammal, for
example a rabbit, with the polypeptide of the invention or said parts thereof,
where appropriate in the presence of, for example, Freund's adjuvant
and/or aluminum hydroxide gels (see, e.g., Diamond, B.A. et al. (1981 ) The
New England Journal of Medicine, 1344). The polyclonal antibodies which
have been generated in the animal due to an immunological reaction can
then be readily isolated from the blood using generally known methods and
purified, for example, via column chromatography.
Monoclonal antibodies can, for example, be prepared using the known
method of Winter & Milstein (Winter, G. & Milstein, C. (1991 ) Nature, 349,
293).
The present invention furthermore also relates to a pharmaceutical which
comprises a nucleic acid of the invention or a polypeptide of the invention
and, where appropriate, suitable additives or excipients, and to a process
for producing a pharmaceutical for treating cancer, autoimmune diseases,
in particular multiple sclerosis or rheumatoid arthritis, Alzheimer's disease,
allergies, in particular neurodermatitis, type I allergies or type IV
allergies,
arthrosis, atherosclerosis, osteoporosis, acute and chronic infectious
diseases and/or diabetes, and/or for affecting the cell metabolism, in
particular in association with immunosuppression, especially in association
CA 02335646 2001-O1-19
WO 00/05388 15 PCT/EP99/04892
with transplants and/or genetic diseases, in particular Werner's syndrome,
Bloom's syndrome, xeroderma pigmentosa and connective tissue diseases,
in which pharmaceutical a nucleic acid of the invention, for example a so-
called antisense nucleic acid, or a polypeptide of the invention is formulated
together with pharmaceutically acceptable additives and/or excipients.
A pharmaceutical which comprises the nucleic acid of the invention in
naked form or in the form of one of the above-described vectors which are
effective for gene therapy, or in the form in which it is complexed with
liposomes, is especially suitable for gene-therapeutic application in
humans.
Examples of suitable additives and/or excipients are a physiological sodium
chloride solution, stabilizers, proteinase inhibitors, nuclease inhibitors,
etc.
The present invention furthermore also relates to a diagnostic agent which
comprises the nucleic acids of the invention, the polypeptides of the
invention or antibodies of the invention and, where appropriate, suitable
additives and/or excipients, and to a process for preparing a diagnostic
agent for diagnosing cancer, autoimmune diseases, in particular multiple
sclerosis or rheumatoid arthritis, Alzheimer's disease, allergies, in
particular
neurodermatitis, type I allergies or type IV allergies, arthrosis,
atherosclerosis, osteoporosis, acute and chronic infectious diseases and/or
diabetes, and/or for analyzing the cell metabolism, in particular the immune
status, especially in association with transplants and/or for analyzing
genetic diseases, in particular Werner's syndrome, Bloom's syndrome,
xeroderma pigmentosa and connective tissue diseases, in which diagnostic
agent suitable additives and/or excipients are added to a nucleic acid of the
invention, a polypeptide of the invention or antibodies of the invention. For
example, according to the present invention, the nucleic acid of the
invention can be used to prepare a diagnostic agent which is based on the
CA 02335646 2001-O1-19
WO 00/05388 16 PCT/EP99/04892
polymerase chain reaction (PCR diagnostics, e.g. as described in EP-
0200362) or on a Northern blot. These tests are based on the specific
hybridization of the nucleic acid of the invention with the complementary
opposite strand, usually of the corresponding mRNA. In this connection, the
nucleic acid of the invention can also be modified, as described, for
example, in EP 0063879. Preference is given to labeling a DNA fragment of
the invention with suitable reagents, for example radioactively with
a-P32-dATP or nonradioactively with biotin, using generally known
methods, and incubating it with isolated RNA which has preferably been
bound to suitable membranes composed, for example, of cellulose or
nylon. It is furthermore advantageous to fractionate the isolated RNA
according to size, e.g. by means of agarose gel electrophoresis, before
hybridization and binding to a membrane. In this way, when the quantity of
RNA examined from each tissue sample is the same, it is then possible to
determine the quantity of mRNA which has been labeled specifically by the
probe.
Another diagnostic agent comprises the polypeptide of the invention or the
immunogenic parts thereof which have been described in more detail
above. The polypeptide, or the parts thereof, which are preferably bound to
a solid phase, for example composed of nitrocellulose or nylon, can, for
example, be brought into contact in vitro with the body fluid, e.g. blood, to
be investigated in order thereby to be able to react, for example, with
autoimmune antibody. The antibody-peptide complex can then, for
example, be detected using labeled anti-human IgG or anti-human IgM
antibodies. The label is, for example, an enzyme, such as peroxidase,
which catalyzes a color reaction. The presence of autoimmune antibodies,
and the quantity of these antibodies which are present, can thus be
detected readily and rapidly via the color reaction.
Another diagnostic agent comprises the antibodies of the invention
themselves. These antibodies can be used, for example, to readily and
rapidly investigate a human tissue sample to determine whether the poly-
peptide in question is present. In this case, the antibodies of the invention
are, for example, labeled with an enzyme as already described above. The
specific
CA 02335646 2001-O1-19
WO 00/05388 17 PCT/EP99/04892
antibody-peptide complex can thereby be detected readily and just as
rapidly via an enzymatic color reaction.
The present invention also relates to an assay for identifying functional
interactors, such as, for example, inhibitors or stimulators, comprising a
nucleic acid of the invention, a polypeptide of the invention or the
antibodies of the invention, and, where appropriate, suitable additives
and/or excipients.
An example of a suitable assay for identifying functional interactors is the
so-called two-hybrid system (Fields, S. & Sternglanz, R. (1984) Trends in
Genetics, 10, 286). In this assay, a cell, for example a yeast cell, is
transformed or transfected with one or more expression vectors which
express a fusion protein comprising the polypeptide of the invention and a
DNA-binding domain from a known protein, for example from Gal4 or LexA
from E. coli, and/or which express a fusion protein comprising an unknown
polypeptide and a transcription-activating domain, for example from Gal4,
herpesvirus VP16 or B42. In addition, the cell contains a reporter gene, for
example the E. coli LacZ gene, "green fluorescence protein" or the yeast
amino acid biosynthesis genes His3 or Leu2, which is controlled by
regulatory sequences such as, for example, the IexA promoter/operator or
by a so-called upstream activation sequence (UAS) which is present in the
yeast. The unknown polypeptide is, for example, encoded by a DNA
fragment which is derived from a gene library, for example from a human
gene library. Normally, the described expression vectors are used to
prepare a cDNA gene library directly in yeast so that the assay can be
performed immediately thereafter.
For example, the nucleic acid of the invention is cloned in a yeast
expression vector in functional unity onto the nucleic acid encoding the
IexA DNA-binding domain, such that the transformed yeast expresses a
fusion protein composed of the polypeptide of the invention and the LexA
DNA-binding domain. In another yeast expression vector, cDNA fragments
from a
CA 02335646 2001-O1-19
WO 00/05388 18 PCT/EP99/04892
cDNA gene library are cloned in functional unity onto the nucleic acid
encoding the Gal4 transcription-activating domain, such that the
transformed yeast expresses a fusion protein composed of an unknown
polypeptide and the Gal4 transcription-activating domain. The yeast which
is transformed with the two expression vectors, and which is, for example,
Leu2-, additionally contains a nucleic acid which encodes Leu2 and which
is controlled by the LexA promoter/operator. If a functional interaction takes
place between the polypeptide of the invention and the unknown
polypeptide, the Gal4 transcription-activating domain then binds, via the
LexA DNA-binding domain, to the LexA promoter/operator, resulting in the
latter being activated and the Leu2 gene being expressed. This then
enables the Leu2- yeast to grow on minimal medium which does not
contain any leucine.
When the LacZ reporter gene or the green fluorescence protein reporter
gene is used instead of an amino acid biosynthesis gene, activation of
transcription can be detected by blue- or green-fluorescent colonies being
formed. However, the blue color or fluorescence color can also be readily
quantified in a spectrometer, for example at 585 nM in the case of a blue
color.
In this way, it is possible to screen gene expression libraries readily and
rapidly for polypeptides which interact with the polypeptides of the
invention. The new polypeptides which have been found can then be
isolated and subjected to further characterization.
Another possibility of applying the two-hybrid system is that of using other
substances, such as, for example, chemical compounds, to influence the
interaction between the polypeptide of the invention and a known or
unknown polypeptide. In this way, it is also readily possible to find novel,
valuable, chemically synthesizable active compounds which can be
employed as therapeutic agents. The present invention is therefore not only
directed toward a process for finding polypeptide-like interactors but also
extends to a process for finding substances which are able to interact with
the above-described
CA 02335646 2001-O1-19
WO 00/05388 19 PCT/EP99/04892
protein-protein complex. In accordance with the present invention, such
peptide-like interactors, and also chemical interactors, are therefore
designated functional interactors which may have an inhibitory or
stimulatory effect.
Description of the figures and the important sequences
Figure 1 shows diagrammatically the conserved regions (homology boxes)
of the proteins of the DEAD protein superfamily. The numbers between the
regions indicate the distances in amino acids between the homo boxes.
Figure 2 diagrammatically describes the conserved regions and known
functions thereof of the expressed proteins according to Fuller Pace F.V.
(1994), supra.
Figures 3A and 3B describe the phylogenetic trees of Hc1 and Hc2 and
establishes the evolutionary connections. These figures were prepared
using the program: Lasergene (Modul MegAlign 3.1.7) by DNASTAR Inc.,
with the aid of the Clustal algorithm (Higgins D.G., Sharp P.M., CABIOS
(1989), Vol. 5, no. 2, 151-153).
SEQ ID No. 13 shows the nucleic acid sequence of Hc1.
SEQ ID No. 14 shows the amino acid sequence corresponding to SEQ ID
No. 13.
SEQ ID No. 15 shows the nucleic acid sequence of Hc2.
SEQ ID No. 16 shows the amino acid sequence corresponding to SEQ ID
No. 15.
CA 02335646 2001-O1-19
WO 00/05388 20 PCT/EP99/04892
The following examples serve to further illustrate the invention without
restricting said invention to the products and embodiments described in the
examples.
Examples
The practical work which led to the present invention is mainly based on
established known methods in microbiology, molecular biology and
recombinant DNA technology.
Example 1: Cultivation of Tetrahymena thermophila
Tetrahymena thermophila, strain B18681V, was inoculated in PPYS
medium (10 g/I proteose peptone No. 3 DIFCO, 1 g/I yeast extract DIFCO,
10 mg sodium citrate, 24. 3 mg FeCl3) in a 500 ml flask (100,000 cells/ml
PPYS) and incubated at 25°C and 100-150 rpm for 2-3 days, up to a
cell
density of approx. 1 million/ml.
Example 2: Isolation of mRNA
Total RNA was isolated from 200 ml of shaker culture of Tetrahymena
thermophila, strain B18681V, according to Chomczynski & Sacchi, (1987).
For this purpose approx. 2 million cells were lysed in the presence of
guanidine thiocyanate/sarkosyl/beta-mercaptoethanol. After adding sodium
acetate and chloroform/isoamyl alcohol/phenol (25:24:1 ), the mixture is
mixed well, incubated on ice for 15 min and centrifuged thereafter at
10,000 x g, 4°C for 20 min. 1 volume of isopropanol is added to the
aqueous phase which is then left standing at -20°G for at least 30 min.
The
RNA pellet is obtained by centrifugation (10 min, 10,000 x g, 4°C
). The
pellet is then washed twice, dried and resuspended in DEPC water. After
incubating at 55-60°C for 10 - 15 min, it is possible to store the RNA
at -
80°C. mRNA is purified from total RNA via oligo(dT)-Sepharose (Clontech
mRNA Separator Kit #K1040-2).
CA 02335646 2001-O1-19
WO 00/05388 21 PCT/EP99/04892
Example 3: Preparation of cDNA
mRNA was transcribed into cDNA according to CLONTECH (CapFinderT""
PCR cDNA Library Construction Kit #K1051-1 ).
Example 4: Amplification of specific gene fragments
Specific gene fragments were amplified with the aid of the polymerase
chain reaction (PCR). A standard PCR mixture contains 10 mM Tris-HCI,
pH 8.3, 50 mM KCI, 1.5 mM MgCl2, 0.001 % gelatin, 75 NM dNTP, 0.3 ng of
each primer, 0.5,u1 of cDNA, 0.5 U of Taq polymerase.
The primers
(2) 5'-GTTCTACCnATTCTGTG-3' and
(3) 5'-ACnGGTTCnGGTAAGAC-3' were used for amplifying the
fragment Hc1, the primers
(4) 5'-ATAGAATTCCCnACnAGAGAAnTnGCT-3' and
(8) 5'-ATAGGATCCGTTCTACCnATTCTGTG-3' were used for amplifying
the fragment Hc2, with n being any nucleotide. The PCR program was
5 min 95°C, 95°C/37s - 50°C/37s - 72°C/37s - 30
cycles.
Example 5: Cloning and sequencing of the fragments
After PCR the PCR product is fractionated on a 1 % agarose gel. The
specific fragments are excised and purified using OIAGEN Gel Extraction
Kit. The purified fragments are directly employed for cloning (Invitrogen
Original TA Cloning Kit #K2000-01 ). Positive clones are grown in shaker
culture, and the plasmid DNA is purified using QIAGEN Maxi-Prep Kit.
Sequencing is performed using the AbiPrism Model 377 automated
sequencer.
Recombinant expression
The gene fragments Hc1 and Hc2 are cloned into a suitable vector,
preferably pGEX-4T-1 GST fusion vector (Pharmacia Biotech). For this
purpose Hc1 and Hc2 are prepared from Tetrahymena thermophila cDNA
by PCR using suitable primers. A standard PCR mixture contains 10 mM
Tris-HCI, pH 8.3, 50 mM KCI, 1.5 mM MgCl2, 0.001 % gelatine, 75 ,uM
dNTP, 0.3 ng of each primer, 0.5 ,ul of cDNA, 0.5 U of Taq polymerase. The
primers
CA 02335646 2001-O1-19
WO 00/05388 22 PCT/EP99/04892
(2A) 5' ATAAGAATGCGGCCGCTGTTCTACCGATTCTGTGAATATA 3';
(3A) 5' CGCGGATCCTC ACT GGT TCG GGT AAG ACT GCT ACT TTC
TCT 3' were used for amplifying the fragment Hcl, the primers
(7A) 5' TATAGAATTCCCCACTAGAGAACTCGCTATGCAAATCGAA 3'
(8A) 5' ATAAGAATGCGGCCGCGTTCTACCGATTCTGTGGACATAG 3'
were used for amplifying the fragment Hc2. The primer (2A) contains a Notl
cleavage site, the primer (3A) contains a BamHl cleavage site, the primer
(7A) contains an EcoRl cleavage site and the primer (8A) contains a Notl
cleavage site. The PCR program was 5 min 95°C, 95°C/37s -
50°C/37s -
72°C/37s - 30 cycles.
The fragments to be cloned are purified via a 1 % agarose gel (QIAgen Gel
Extraction Kit) and the ends to be cloned are prepared by digestion with
Notl and BamHl (Hc1) or EcoRl and Notl (Hc2). The vector pGEX-4T-1 is
prepared likewise by digestion with Notl and BamHl (Hc1 ) or EcoRl and
Notl (Hc2). Vector and insert are ligated at 16°C overnight, the
ligation
mixtures are employed for transformation of competent TOP10F'
(Invitrogen) E. coli cells. Positive clones are picked and used for protein
expression. The construct pGEX-Hc1 or pGEX-Hc2 permits translation of a
fusion protein consisting of 257 amino acids (28.3 kDa) of Hc1 and
glutathione S-transferase (24 kDa) or of 255 amino acids (28.1 kDa) of Hc2
and glutathione S-transferase (24 kDa). The fusion protein contains all
homology boxes (DEAD, SAT,...) which characterize the members of the
protein family. For recombinant protein expression an overnight culture is
induced with IPTG and the fusion protein is purified from the supernatant in
a batch process using glutathione Sepharose 4B or via a glutathione
Sepharose 4B column. The glutathione S-transferase is cleaved with
thrombin. For this purpose, for example, 100 Ng of GST fusion protein are
incubated with one unit of thrombin proteinase in 1 x PBS at 22°C for
16 h.
The gene product of Hc1 or Hc2 is removed via gel filtration using, for
example, a Superdex 200 HR 10/30 column (Pharmacia Biotech). The
BioRad gel filtration chromatography standard (Ref. 151-1901 ) can be used
as a standard.
CA 02335646 2001-O1-19
WO 00/05388 23 PCT/EP99/04892
Example 6: ATPase activity
The activity is determined as described in the literature, for example
according to Jaramillo et al., Mol Cell Biol 1991 11:5992; Rozen et al., Mol
Cell Biol 1990 10: 1134, Ladomery M. et al. Nucl. Acid Res. 1997, 25:965-
973 or Dong F. et al. Proc. Natl. Acad. Sci. USA 1996, 93: 14456-14461 or
patent PCT/US97/01614. A concrete example is described in the following.
The reaction mixture for measuring the ATPase activity contains 150 mM
NaCI, 5 mM KCI, 1.5 mM MgCl2, 20 mM Hepes/KOH, pH 7, 1 mM
dithiothreitol, 1 mM PMSF, lO,uM ATP and 0.2,u1 of 32P-ATP in a total
volume of 50,u1. The reaction mixture is heated to 37°C and Hc1 or Hc2
is
added. After 30 min at 37°C the reaction is stopped by adding 400,u1 of
7%
activated carbon in 50 mM HCI and 5 mM H3P04. The samples are mixed
and centrifuged at 13,000 rpm for 15 min. The released radioactivity in the
supernatant is measured in a scintillation counter.
Example 7: helicase activity
The helicase activity of Hc1 or Hc2 can be monitored by means of
dissociation of double-stranded RNA. The substrate can be any RNA
oligomer which is labeled on one strand, for example with 32P. The
reaction mixture contains in a 10 NI mixture 32P-labeled helicase substrate,
Hc1 or Hc2 at various concentrations, 2 mM ATP, 5 mM dithiothreitol and
50 Ng of bovine serum albumin in 20 mM Tris-HCI. The reaction is carried
out at 37°C for 30 min and stopped by heating.
The reaction mixture is applied to a 16 cm x 18 cm 12% non-denaturing
polyacrylamide gel and fractionated at a constant current of 25 mA. The gel
is dried in vacuo and exposed to film (e.g. Kodak RPXRP-5 film, -70°C
).
Example 8: Antisense
The RNA opposite strands of the DNA fragments Hc1, Hc2 or of DNA
sequences homologous to Hc1 or Hc2 or of part sequences of Hc1, Hc2 or
homologs can be used as antisense strands. Normally a plasmid is
constructed which carries the desired antisense sequence and
CA 02335646 2001-O1-19
WO 00/05388 24 PCT/EP99/04892
selection markers, for example neomycin, a promoter, which controls the
expression of antisense RNA, and RNA-stabilizing sequences. The
transfected sequences are transcribed in the cell and the transcript is
hybridized with the target DNA. On the other hand it is also possible to
introduce in vitro synthesized sequences into the cell by microinjection.
It is also conceivable to employ oligonucleotides. These may either be
prepared synthetically or generated by restriction digestion of Hc1, Hc2 or
homologs. The oligonucleotides must be highly pure. This is achieved by
2 - 5 lyophilizations. Pure oligonucleotides are taken up, for example, in
HEPES-buffered saline, pH 7.4.
Example 9: Gene probe for detecting novel members of the DEAD box
protein family
The fragments Hc1 and Hc2 are used in order to isolate novel DEAD box
proteins from suitable organisms. For this purpose the specific gene
fragments are amplified with the aid of the polymerase chain reaction
(PCR) and simultaneously labeled with digoxigenin, according to
Boehringer Mannheim PCR DIG Probe Synthesis Kit #1636 090. Plasmid
DNA of the cloned fragments Hc1 or Hc2 is used as a template. A PCR
mixture contains ExpandT"" High Fidelity buffer (Boehringer Mannheim
#1636 090), 20 NM dATP, 200 NM dGTP, 200,uM dCTP, 130 NM dTTP,
70 NM DIG-11-dUTP, 0.3 ng of each primer, 100 pg plasmid DNA, 2.6 U
Taq polymerase.
The primers
(2) 5'-GTTCTACCnATTCTGTG-3' and
(3) 5'-ACnGGTTCnGGTAAGAC-3' were used for amplifying the
fragment Hc1, the primers
(4) 5'-ATAGAATTCCCnACnAGAGAAnTnGCT-3' and
(8) 5'-ATAGGATCCGTTCTACCnATTCTGTG-3' were used for amplifying
the fragment Hc2.
The PCR program was 5 min 95°C, 95°C/37s - 50°C/37s -
72°C/37s - 30
cycles.
CA 02335646 2001-O1-19
WO 00/05388 25 PCT/EP99/04892
The PCR reaction mixture containing the labeled fragments is directly
employed for hybridization studies. For this purpose the PCR product is
denatured at 95°C for 10 min and then the hybridization solution DIG
Easy
Hyb (Boehringer Mannheim Ref. 1603558) is added (conc. 2 NI/ml). This
hybridization solution is used for screening cDNA libraries of suitable
organisms at low stringency (hybridization temperature 30 - 50°C).
Example 10: Antibodies
Protein expressed in a recombinant manner is purified by a suitable
method and used for the purpose of generating polyclonal antibodies in a
suitable organism, for example a rat or a rabbit. For this purpose, the fusion
protein is purified, for example, initially via glutathione Sepharose and then
via SDS-PAGE. The band containing the fusion protein is excised from the
gel, ground up and injected for example into a rabbit or a rat. The obtained
antiserum is passed through an IgG column, and the antibodies are eluted
at low pH in 1 M Tris-HCI pH 8. The antibodies are dialyzed against 25 mM
HEPES pH 7.9, 12 mM MgCl2, 0.5 mM EDTA, 2 mM dithiothreitol, 17%
glycerol, 100 mM KCI.
Example 11: Use of the antibodies for isolating novel DEAD box proteins
The antibodies obtained as described in the previous section are used for
the purpose of isolating novel DEAD box proteins from suitable organisms.
For this purpose, the antibodies are covalently coupled to a suitable matrix,
for example Sephadex G50 (Pharmacia). The Sephadex is used to pack,
for example, BioRad columns 1.5 x 10 cm or 2.5 x 10 cm and the columns
are equilibrated with 20 ml of buffer A (0.05 M Tris-HCI, 0.15 M NaCI,
0.005 M EDTA, 0.1 % NP40, pH 8.0). Subsequently, 3.0 g of protein A-
Sepharose beads (Pharmacia CL-4B) are applied to the column. Leave
overnight at 4°C. The antibody solution is applied to the column and
allowed to drip through with a flow rate of -100 ml/h at 4°C. Then the
column is washed several times: with 250 ml of buffer A (plus 0.5% NP40),
then with 125 ml of 0.1 M borate buffer, pH 9.0, then with 125 ml of borate
buffer, pH 8.0, then with 125 ml of 0.2 M triethanolamine, pH 8.2. The Fc
region of the antibody is coupled to protein A - Sepharose via crosslinking.
Then the column is washed again, specifically, once with buffer B (0.15 M
CA 02335646 2001-O1-19
WO 00/05388 26 PCT/EP99/04892
Tris-HCI, 0.15 M NaCI, 1 mM EDTA pH 8.0, 10% glycerol, 10% NP-40),
once with buffer C (0.05 M Tris-HCI, 0.5 M LiCI, 1 mM EDTA, pH 8.0, 10%
glycerol), and once with buffer D (0.01 M Pipes, 5 mM NaCI, 1 mM EDTA,
pH 8.0, 10% glycerol). The non-crosslinked antibody is eluted with citrate
buffer. The column is stored in borate buffer, pH 8.0 with 0.02% Na N3.
In order to isolate novel DEAD box proteins, cell lysate from suitable
organisms is applied to the antibody column. The column is washed
several times and the bound proteins are eluted with a suitable buffer, for
example glycine, pH 3, in Tris-HC1, pH 8.
Example 12: Overexpression in useful plants
The gene fragments Hc1 and Hc2 can be heterologously expressed in
useful plants. The gene transfer can be mediated, for example, by
Agrobacterium tumefaciens. A typical A. tumefaciens vector (Ti plasmid)
contains a replication origin (ori Agro) which permits replication in
Agrobacterium, a replication origin on E. coli which ensures functional
replication in E. coli, a plurality of resistance genes, for example against
kanamycin and spectinomycin, insertion sites for introducing the foreign
gene and directed T-DNA flanking sequences which ensure recognition of
beginning and end of the foreign gene in the gene transfer. A. tumefaciens
is transformed with the Ti plasmid.
To infect the useful plant, leaf disks are punched out of said plant and put
into a shallow dish (Petri dish). Subsequently, a solution of recombinant
agrobacteria is added and after a few minutes the leaf disks are transferred
onto a medium with feeder cells (e.g. filter paper). Injured cells on the
edges of the leaf disks release factors which lead to infection of the plant
cells by the agrobacterium. After 2-3 days, the leaf disks are transferred
onto a sprout-stimulating medium containing an antibiotic which destroys
the agrobacteria (e.g. cefotaxime) and cultivated for 2-3 weeks.
CA 02335646 2001-O1-19
WO 00/05388 27 PCT/EP99/04892
The sprouts are transferred onto a root-inducing medium and after a further
2-3 weeks planted in soil.
Example 13: Diagnostic probes
The gene fragments Hc1 or Hc2 or homologous gene fragments or parts
thereof (at least 20 base pairs long) which comprise the homology boxes
characteristic of RNA helicases can be employed as diagnostic probes. For
this purpose, the DNA fragments are immobilized on a suitable matrix (e.g.
nylon membrane, chip). mRNA of a patient is purified and transcribed into
cDNA via reverse transcription by, for example, MMLV reverse
transcriptase, 2 h at 37°C. Simultaneously, the cDNA is labeled, for
example with 32P or digoxigenin. The cDNA is diluted in a suitable
hybridization buffer, for example DIG EasyHyb (Boehringer Mannheim
Ref. 1603558) and hybridized with the immobilized DNA under stringent
conditions.
Example 14: Selection markers
The gene fragments Hc1 and Hc2 may be employed as selection markers
in molecular biology. It has been shown that overexpression of an RNA
helicase in mouse cells confers tolerance of the substance leflunomide on
said cells (Mullner patent). In order to use Hc1 or Hc2 as a selection
marker, an expression vector is constructed which contains Hc1 or Hc2 and
a gene to be expressed. The gene to be expressed can be located beside
or within the helicase gene. The vector is used to transform suitable host
cells (e.g. cloning into a pGEX vector and introduction into E. coli). When
the gene to be expressed is located beside the He gene, transformants
become tolerant of leflunomide in the case of successful introduction of the
vector. The success of the ligation has to be checked, for example via
blue/white screening. When the gene to be expressed is located within the
He gene, the He gene is destroyed in the case of successful ligation and
the transformants lose their tolerance of leflunomide in the case of
successful introduction of the vector.
CA 02335646 2001-O1-19
WO 00/05388 1 PCT/EP99/04892
SEQUENCE LISTING
<110> Aventis Research 6 Technologies Deutschland GnsbH i Co KG
<120> Neue Gene dez DEAD Box Protein-Familie, deren
Expressionsprodukte and VerHendung
<150> DE 198 20 608.9
<151> 1998-07-22
<160> 16
<210> 1
<211> 16
<212> DNA
<223> Artiticiel sequence
<220>
<223> cDNA
<400> 1
G~_'TCTACCAT TCTGTG 16
<210> 2
<211> 15
<212> DNA
<213> Artificiel sequence
<220>
<223> cDNA
<400> 2
ACGGTTCGGT AAGAC 15
<210> 3
<211> 23
<212> DNA
<213> Artificiel sequence
<220>
<223> cDNA
<400> 3
ATAGAATTCC CACAGAGAAT GCT 23
<210> 4
<211> 25
<212> DNA
<213> Artificiel sequence
<220>
<223> cDNA
CA 02335646 2001-O1-19
WO 00/05388 2 PCT/EP99/04892
<400> 4 z _ _ . ..___
ATAGGATCCG TTCTACCATT CTGTG - 25
<210>5
<211>90
<212>DNA
<213>Artificiel sequence
<220>
<223>cDNA
<400> 5
ATAAGAATGC GGCCGCTGTT CTACCGATTC TGTGAATATA 40
<210>6
<211>4I
<212>DNA
<213>Rrtificiel sequence
<220>
<223>cDNA
<400> 6
CGCGGATCCT CACTGGTTCG GGTAAGACTG CTACTTTCTC T 91
<210> 7
<211> 91
<212> DNA
<213> Artificiel sequence
<220>
<223> cDNA
<900> 7
CGCGGATCC~_' CACTGGTTCG GGTAAGACTG CTACTTTCTC T 41
<210> 8
<211> 40
<212> DNA
<213> Attificiel sequence
<220>
<223> cDNA
<900> B
ATAAGAATGC GGCCGCGTTC TACCGATTCT GTGGACA':AG 40
CA 02335646 2001-O1-19
WO 00/05388 3 PCT/EP99/04892
J
<210> 9
<211> 16
<212> DNA
<213> Artificiel sequence .
<220>
<223> cDNA
<400> 9
GTTCTACCAT TCTGTG 16
<210> 10
<211> 15
<212> DNA
<213> Artificiel sequence
<220>
<223> cDNA
<900> 10
ACGGTTCGGT AAGAC 15
<210> 1I
<211> 23
<212> DNA
<213> Artificiel sequence
<220>
<223> cDNA
<400> 11
ATAGAATTCC CACAGAGAAT GCT 23
<2i0> 12
<211> 25
<212> DNA
<213> Artificiel sequence
<220>
<223> cDNA
<400> 12
ATAGGATCCG '_"TCTACCATT CTGTG 25
<210> 13
<211> 771
<212> DNA
<213> Tetrahymena thermophila
<220>
<223> cDNA
. ~ CA 02335646 2001-O1-19
WO 00/05388 4 PCT/EP99/04892
<400> I3
CCCACTAGAG AACTCGCTAT GCAAATCGAA AGAGAGTCCG AAAGATTTGG TAAATCC_TCT60
AAGCTTARAT GTGCTTGTAT CTATGGTGGT GCTGACAAAT ACTCCTAAAG AGCACTTCTC12G
CAATAAGGTG TAGATGTAGT TATTGCTACT CCTGGTAGAC TTATTGACTT TTTAGAAAGT180
GAAACTACTA CTTTACGTAG AGTTACTTAT CTCGTATTAG ATGAAGCAGA TAGAATGTTA240
GATATGGGTT TTGAAATTTA AATTAGAAAA ATCTTGGGTT AAATTAGACC TGATCGTTAA300
ACATTGATGT TTTCTGCTAC CTGGCCTAAG AATGTTTAGA ATCTTGCTTA AGATTATTGC360
AAGAATACTC CCGTTTATGT TCAAATCGGA AAACATGAAT TAGCTATTAA CGAAAGAATT420
AAATAAATTG TTTATGTTAC AGATCAATCA AAGAAAATCA ATCAACTTAT CAAGCAATTA490
GATTGTTTGA CTTAGAAAGA TAAAGTATTG ATTTTCGCTT AAACAAAGAA GGGATGTGAA540
AGCATGAGTC GTATTTTGAA TAAAGAAGGA TTTAAGTGTC TTGCTATCCA TGGTGACAAA600
GCCTAAAAAG ACAGAGACTA TGTTATGAAC AAGTTCAAAA GCGGAGAATG CAGAATCCTT660
ATTGCTACAG ACGTAGCCAG TAGAGGTTTG GATGTTAAGG ATGTCTCCCA CGTATTTAAT720
TACGATTTCC CAAAGGTTAT GGAAGACTAT GTCCACAGAA TCGGTAGAAC G 771
<210>14
<211>257
<212>Protein
<213>Tetrahymena thermophila
<220>
<223>PRT
<400> 19
Pro Thr Arg Glu Leu Ala Met Gln :ie Glu Arg Glu Ser Giu Arg Phe
I 5 10 15
Gly Lys Ser Ser Lys Leu Lys Cys Ala Cys Ile Tyr Gly Gly Ala Asp
20 25 30
Lys Tyr Ser Gln Arg Ala Leu Leu Gln Gln Gly Val Asp Val Val Ile
35 40 95
Ala Thr Pro Gly Arg Leu Ile Asp Phe Leu Glu Se. Glu Th: Thr Thr
50 55 60
Leu Arg Arg Val Thr Tyr Leu Va_ Leu Asp Glu Ala Pap n=g Met Leu
65 70 75 80
Asp Met Gly Phe G_u Ile Gln Ile Arg Lys :le Leu Gly Gln Ile Arg
65 90 95
Pro Asp A=g Gln :'hr Leu Met Phe Ser Ala T:m Trp Pro Lys Asp Val
100 105 1.0
CA 02335646 2001-O1-19
WO 00/05388 5 PCT/EP99/04892
Gln Asn Leu Ala Gln Asp Tyt Cys Lys Asn Thr Pro Val Tyr Val Gln v'vI
115 120 125
Ile Gly Lys His Glu Leu Ala Ile Asn Glu Arg Ile Lys Gln Ile Val
I30 135 140 .
Tyr Vai Thr Asp Gln Ser Lys Lys Ile Asn Gln Leu Iie Lys Gln Leu
145 150 155 160
Asp Cys Leu Thr Gln Lys Asp Lys Val Leu '_le Phe Ala Gln Thr Lys
165 170 175
Lys Gly Cys Glu Ser Met Ser Arg Ile Leu Asn Lys Glu Gly Phe Lys
180 185 190
Cys Leu Ala Ile His Gly Asp Lys Ala Gln Lys Asp Arg Asp Tyr Val
195 200 205
Met Asn Lys Phe Lys Ser Gly Glu Cys Arg Iie Leu Ile Ala Thr Asp
210 215 220
Val Ala Ser Arg Gly Leu Asp Val Lys Asp Val Ser His Val Phe Asn
225 230 235 240
Tyr Asp Phe Pro Lys Val Met Glu Asp Tyr Val His Arg Ile Gly Arg
245 250 255
Thr
<210> 15
<211> 765
<212> DNA
<213> Tetrahymena thermophila
<220>
<223> cDNA
<400> 15
CCCACCAGAG AATTAGCCCA 60
ATAAACTATC ACCGTTATTA
TGTACTTAGG TGAATTCTTG
AAGGTCTCCG CCTATGCTTGCACTGGTGGT ACTGATCCCA AGGAAGATAG 120
AAAGAGATTA
AGAGAAGGTG TCCAAGTCGTTGTTGGTACC CCTGGTAGAG TTTTGGATTT 1B0
AATC':AAAAG
AAGACTTTAG TCACCGATCAC~_'TAAAATTA TTCATTTTGG ACGAAGCCGA 240
TGAAATGTTA
GGAAGAGGTT TCAAGGATCAAATTAACAAA ATCTTCTAAA ACTTACCCCA 300
CGATATCTAG
GTTGCTC':TT TCTCTGCTAC:ATGGCT.~.CC GAAAT'::.T'_"G AAATTACCAA360
GTAATTTATG
AGAGACCCCG CTA:.TATCCTTGTCAAGRAT GATGACTTGA CTTTGGACGG 420
TF.TTAAATAA
TTCTACATCG CCTTAGATAAGGAAGAATGG AAGTTTGACA CCTTAG':C~~A 4BO
ATTATACAAT
AACATCGAAA TCGCTTAAGCTATTATCTAT TGCAACACCA AGAAGAGAGT 540
CGATGAP.TTA
AGAGACAAGC TTATTGAAAAGAATATGACC GTCTCTGCTA TGCACGGTGA 600
AATGGACCAA
TAnAACAGAG ATCTTATTAT_GAAGGAATTC AGAACCGG'_A CCTCCAGAGT 660
TCTTATCACT
CA 02335646 2001-O1-19
WO 00/05388 6 PCT/EP99/04892
ACTGATTTGC TCTCCAGAGG TATTGATATC CATCAAGTCA ACTTGGTTAT CAACTACGAG 720
TTACCCCTTA AGAAGGAATG TTATATTCAC AGAATCGGTA GAACA 765
<210>16
<211>255
<212>Protein
<213>Tetrahymeaa thermophila
<220>
<223>PRT
<400> 16
Pro Thz Azg Glu Leu Ala Gln Gln Thr Ile Thr Val Ile Met Tyr Leu
1 5 10 I5
Gly Glu Phe Leu Lys Val Ser Ala Tyz Ala Cys Thr Gly Gly Thr Asp
20 25 30
Pro Lys Glu Asp Arg Lys Arg Leu Arg Glu Gly Val Gln Val Val Val
35 90 45
Gly Thr Pro Gly Arg Val Leu Rsp Leu Iie Gln Lys Lys Thr Leu Val
50 55 60
Thr Asp His Leu Lys Leu Phe I1e Leu Asp Glu Ala Asp Glu Met Leu
65 70 75 BO
Gly Arg Gly Phe Lys Rsp Gln Ile Asn Lys Ile Phe Gln Asn Leu Pro
85 90 g5
H_s Asp Ile Gln Val Ala Leu Phe Ser Ala Th_ Met Ala Pro Glu Ile
100 105 110
Leu Glu Ile Thr Lys Gln Phe Met Arg Asp Pro A:a Thr Ile Leu Val
115 I20 I25
Lys Asn Asp Asp Leu Thr Leu Asp Gly Ile Lys G1n Phe Tyr '_le Ala
130 I35 140
Leu Asp Lys Glu Glu Trp Lys Phe Asp Thr Leu Val G'_u Leu Tyr Asn
145 150 155 160
Asn Ile Giu Ile Ala Gin Ala Ile ile Tyz Cys Asn Thr Lys Lys Arg
165 170 175
Val Asp Glu Leu Arg Asp Lys Leu 11e Glu Lys Asn Met Thr Val Ser
180 185 190
A1 a Met His Gly Glu Met Asp Gl.~. Gl.~. As.~. A_g Psp Leu I:e Met Lys
195 200 2C5
Glu Phe Arg Thr Gly Thr Sez Arg Vai Leu _'le Thr Thr Asp Leu Leu
210 2i5 220
Ser Gly Gly Ile Asp Ile His Gln Val Asn Leu Val :le Asn Tyr Asp
225 230 235 290
Leu Pro Leu Lys Lys Glu Cys Tyr Ile His Arg Ile Gly Azg Thr
245 250 255