Note: Descriptions are shown in the official language in which they were submitted.
CA 02338185 2001-02-05
Thermostable in vitro complex with polymerase activity
Description
The invention concerns a thermostable in vitro complex for the template-
dependent
elongation of nucleic acids, a thermostable in vitro complex and DNA sequences
and
vectors coding therefor. The invention additionally concerns the use of the
inventive
complexes in methods for the template-dependent elongation of nucleic acids
such as
PCR reactions, reverse transcription, DNA labelling or DNA sequencing in which
an
in vitro template-dependent DNA strand synthesis occurs. Finally the invention
also
concerns kits or reagent kits for carrying out the inventive methods.
DNA polymerases belong to a group of enzymes which use single-stranded DNA as
a template for the synthesis of a complementary DNA strand. These enzymes play
a
major role in nucleic acid metabolism including the processes of DNA
replication,
repair and recombination. DNA polymerases have been identified in all cellular
organisms from bacterial to human cells, in many viruses as well as in
bacteriophages
(Kornberg, A. & Baker, T.A. (1991) DNA Replication WH Freeman, New York,
NY). The archaebacteria and eubacteria are usually combined to form the
prokaryote
group which are organisms without a real cell nucleus in contrast to the
eukaryotes
which are organisms with a real cell nucleus. A common feature of many
polymerases from the diverse organisms is often a similarity of the amino acid
sequence and a similarity of structure (Wang, J., Sattar, A.K.M.A.; Wang,
C.C.,
Karam, J.D., Konigsberg, W.H. & Steitz, T.A. (1997) Crystal Structure of pol
a,
family replication DNA polymerase from bacteriophage RB69.Cell 89, 1087-1099).
Organisms such as humans have numerous DNA-dependent polymerases which are,
however, not all responsible for DNA replication but some also carry out DNA
repair. Replicative DNA polymerases are usually composed in vivo of protein
complexes with several units which replicate the chromosomes of the cellular
organisms and viruses. A general property of these replicating polymerases is
in
general a high processivity which means their ability to polymerise thousands
of
nucleotides without dissociating from the DNA template (Kornberg, A. & Baker,
T.A. (1991) DNA Replication, WH Freeman, New York, NY).
CA 02338185 2001-02-05
-2-
Highly processive replication mechanisms are known in the prior art which are
on
the one hand cellular mechanisms and, on the other hand, the replication
mechanisms
occurring in the bacteriophages T4 and T7.
The replication apparatus comprises numerous components. These include among
others, a) proteins having polymerase activity, b) proteins which are involved
in the
formation of a clamp structure, one of the functions of the clamp structure
being to
bind a polymerase activity to its template, to stabilize the binding and thus
to
correspondingly change the dissociation constant, c) proteins which load the
clamp
onto the template, d) proteins which stabilize the template and optionally e)
proteins
which guide the polymerase onto the template.
Proteins mentioned under b) form structures which are either open or closed,
for
example circular or semi-circular structures. Such structures can be formed by
one
or several species of proteins. One of the said protein species may have a
polymerase
activity.
The proteins responsible for the formation of these structures are referred to
in the
following as "sliding clamp proteins" or "clamp proteins" provided they have
no
polymerase activity.
Reference is made to the replication apparatus of the bacteriophages T4 or T7
as an
example of a processive replication apparatus which does not have a closed
circular
shape.
Reference is made to the replication apparatus of the bacterium E. coli as an
example
of a processive replication apparatus which has a closed circular shape
(Stukenberg,
P.T., Studwell-Vaughan, P.S. & O'Donnel, M. (1991) Mechanisms of the (3 clamp
of DNA polymerase III holoenzyme. J. Biol. Chem. 266, 11328-11334; Kuriyan, J.
& O'Donnel, M. (1993) Sliding clamps of DNA polymerases. J. Mol. Biol. 234,
915-925).
CA 02338185 2001-02-05
-3-
It is known that the replication apparatus in archaebacteria is similar to the
eukaryotic replication apparatus although the genome organisation in
eukaryotes and
archaebacteria is completely different and the cellular structure of the
eubacteria is
similar to that of the archaea (Edgell, D.R. and Doolittle, W.F. (1997),
Archaea and
the origins) of DNA replication proteins. Cell 89, 995-998).
The sliding clamp is frequently bound to an elongation protein via one or
several
other proteins, in other words it is coupled to the elongation protein. Such a
coupling protein is referred to herein in the following as a coupling protein
or
coupling subunit in which the coupling may take place via a plurality of
coupling
proteins.
An elongation protein should be understood herein as a protein or complex
having
polymerase activity which has one or several of the following properties: use
of
RNA as a template to synthesize DNA and/or RNA, use of DNA as a template to
synthesize DNA and/or RNA, synthesis of RNA, synthesis of DNA, synthesis of
nucleic acids from DNA and RNA, exonuclease activity in the S'-3' direction
and
exonuclease activity in the 3'-S' direction, strand displacement activity,
thermostability and processivity or non-processivity.
The three-dimensional structure of various sliding clamp proteins has already
been
determined:
- that of the eukaryotic proliferating cell nuclear antigen (PCNA) (Krishna,
T.S.R.,
Kong, X.-P., Gary, S., Burgers, P.M. & Kuriyan, J. (1994) Crystal structure of
the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233-1243;
Gulbis, J.M., Kelman, Z., Hurwitz, J., O'Donnel, M. & Kuriyan, J. (1996)
Structure of the C-terminal region of p21 WAF 1/CIP 1 complexed with human
PCNA. Cell 87, 297-306),
- that of the ~3 subunit of the polymerase III of the eubacterium Escherichia
coli
(Kong, X.-P., Onrust, R., O'Donnel, M. & Kuriyan.; J. ( 1992) Three
dimensional
structure of the (3 subunit of Escherichia coli DNA polymerase III holoenzyme;
a sliding DNA clamp. Cell 69, 425-437)
- and that of the gene-4S protein of the bacter-iophage T4 protein (Kelman,
Zvi,
Hurwitz, J. O'Donnel, Mike (1998) Structure, 6, 121-12S).
CA 02338185 2001-02-05
-4-
The overall structure of these sliding clamps is very similar; the pictures of
the
circular total protein structure of PCNA, of the ~i subunit and gp45 rings are
superimposable when laid on top of one another (Kelman, Z. & O'Donnel, M.
(1995) Structural and functional similarities of prokaryotic and eukaryotic
sliding
clamps. Nucleic Acids Res. 23, 3613-3620). Each ring has comparable dimensions
and a central opening which is large enough to encircle duplex DNA i.e. a DNA
double strand composed of the two complementary DNA strands.
The sliding clamp cannot position itself in vivo around the DNA but must be
clamped around the DNA. In prokaryotes and eukaryotes such a protein complex
is
composed of numerous subunits which are referred to as the y complex in the
eubacterium Escherichia coli and as the replication factor C (RF-C) in humans
(Kelman, Z. & O'Donnell, M. ( 1994) DNA replication - enzymology and
mechanisms. Curr. Opin. Gent. Dev. 4, 185-195). The; protein complex
recognises
the 3'-end of the primer in the primer template duplex and positions the
sliding
clamp around the DNA in the presence of ATP.
In the case of the bacteriophage T7 the same object i.e. a processive DNA
synthesis,
is achieved by means of a protein complex with a different structure. The
phage
expresses its own catalytic polymerase, T7 polymerase., the gene product of
gene 5
which binds to a protein from the host Escherichia coli i.e. thioredoxin and
thus
enables a highly processive DNA replication as a replicase (Proc. Natl. Acad.
Sci.
USA 1992, Oct. 15; 80(20):9774-9778 Genetic analysis of the interaction
between
bacteriophage T7 DNA polymerase and Escherichia coli thiorexin, Himawan JS,
Richardson CC). In this case there is also clamp formation but this clamp does
not
have the same structure as for example in the case of the eukaryotic PCNA.
It is often necessary, such as in the case of the human polymerase 8, that
coupling
proteins have to create a connection between the catalytically active part of
the
polymerase and the processivity factor (sliding clamp). In the case of humans
this is
the small subunit of the 8 polymerase (Zhang, S.-J., Zeng, X.-R., Zhang, P.,
Toomey, N.L., Chuang, R.Y., Chang, L.-S., and :Lee, M.Y.W.T. (1994). A
conserved region in the amino terminus of DNA polymerase 8 is involved in
poliferating cell nuclear antigen binding. J. Biol. Chem. 270, 7988-7992).
However,
CA 02338185 2001-02-05
-5-
in the case of T7 polymerase the processivity factor binds directly to the
catalytic
unit of the polymerase.
DNA polymerases are characterized among others by two properties, their
elongation rate i.e. the number of nucleotides which they can incorporate per
second
into a growing DNA strand and their dissociation constant. If the polymerase
dissociates again from the strand after each step of incorporating one of the
nucleotides into the growing chain (i.e. one elongation step occurs per
binding
event), then the processivity has the value 1 and the polymerase is not
processive.
This synthesis is referred to as distributive. If the polymerase remains
connected to
the strand for repeated nucleic acid incorporations, then the replication
modus is
referred to as processive and can reach a value of several thousand (see also:
Methods in Enzymology Volume 262, DNA replication, edited by J.L. Campbell,
Academic Press 1995, pp. 270-280).
Processivity is a desirable property for most in vitro applications such as
PCR or
sequencing processes but the thermostable enzymes that have been used up to
now
in these reactions only possess processivity to a slight extent whereas the
temperature-sensitive T7 polymerase associated with thioredoxin has a
processivity
of several thousand nucleotides. In comparison the thermostable DNA
polymerases
from Thermr~.r thermophilzrs or Thermos aqzraticr~s only have a processivity
of about
50 nucleotides (Biochim. Biophys. Acta 1995 Nov. 7; 1264(2):243-248
Inactivation
of the 5'-3' exonuclease of Thermos aquaticus DNA polymerase. Merkens LS,
Bryan SK, Moses RE).
The US patents 4,683,195, 4,800,195 and 4,683,202 describe the application of
such
thermostable DNA polymerases in the polymerase chain reaction (PCR). In PCR
DNA is newly synthesized using primers, templates (also referred to as
matrices),
nucleotides, a DNA polymerase, an appropriate buffer and suitable reaction
conditions. The starting point is usually a double-stranded DNA sequence of
which a
certain target region is to be amplified. Two primers are used for this which
are
complementary to flanking regions of the target sequence, each being on a
partial
strand of the DNA double strand. However, in order to hybridize the primers,
the
DNA double strands are firstly denatured and in particular thermally melted.
After
CA 02338185 2001-02-05
-6-
hybridization of the primers, they are elongated by means of the polymerase.
Subsequently denaturation is again carried out in order to separate the newly
formed
DNA strands from the template strands whereupon, in addition to the original
template strands, the nucleic acid strands formed in the first step are also
available as
a template for a further elongation cycle, these are each again hybridized
with
primers and a new elongation takes place. This procedure is carried out in
cycles
each with thermal denaturation as intermediate steps. A thermostable
polymerase
which survives the cyclic thermal melting of the DNA strands is preferably
used for
the PCR. Thus Taq DNA polymerase is often used (US patent 4,965,188). However,
the processivity of Taq DNA polymerase is relatively low compared to T7
polymerase as described above.
DNA polymerases are also used in DNA sequence determination (Sanger et al.,
Proc. Natl. Acad. Sci., USA 74:5463-5467 (1997)). A T7 DNA polymerase is
frequently used for sequencing according to Sanger (Tabor, S. and Richardson,
C.C.
Proc. Natl. Acad Sci., USA 86:4076-4080 (1989)). Subsequently the cycle
sequencing method was developed (hurray, V. (1989) ,Nucleic Acids Re.s. 17,
8889)
which does not require a single-stranded template and allows initiation of the
sequence reaction with relatively small amounts of template. The templates
that can
be used for this are for example the Taq polymerase mentioned above (US patent
5,075,216) or the polymerase from Thermotoga neapolitana (WO 96/10640) or
other thermostable volvmerases. Recent methods couvle the exponential
amplification and sequencing of a DNA fragment in one step so that it is
possible to
directly sequence genomic DNA. One of the methods, the so-called DEXAS method
(Nucleic Acids Res. 1997 May 15;25(10):2032-2034 Direct DNA sequence
determination from total genomic DNA. Kilger C, Paabo S, Biol. Chem. 1997 Feb;
378(2):99-105 Direct exponential amplification and sequencing (DEXAS) of
genomic DNA. Kilger C, Paabo S and DE 19653439.9 and DE 19653494.1), uses a
polymerase with a reduced ability to discriminate against dideoxynucleotides
(ddNTPs), compared to deoxynucleotides (dNTPs) as well as a reaction buffer,
two
primers which are preferably not present in equimolar amounts and the above-
mentioned nucleotides in order to then obtain a complete, sequence-specific
DNA
ladder of a fragment in several cycles which is flanked by the primers. A
fi~rther
development of this method comprises the use of a polymerase mixture in which
one
of the two polymerases discriminates between ddNTPs and dNTPs whereas the
CA 02338185 2001-02-05
_7-
second has a reduced discrimination ability (Nucleic Acidr Res. 1997 May 15;
26( 10):2032-2034 Direct DNA sequence determination from total genomic DNA.
Kilger C, Paabo S).
DNA polymerases are also used for the reverse transcription of RNA into DNA.
In
this case RNA serves as a template and the polymerase synthesizes a
complementary
DNA strand. The thermostable DNA polymerase from the organism Thermos
thermusphilus (Tth) (US patent 5,322,770) is for example used in this case.
The polymerase may also have a proof reading activity i.e. a 3'-5' exonuclease
activity. This property is particularly desirable when the product to be
synthesized
should be produced with a low rate of nucleotide incorporation errors. The
polymerases from the organism Pyrococcus wosei are an example of this.
Most of the above-mentioned enzymes that are usually used in PCR reactions are
not
actually replication enzymes in vivo but are mostly enzymes which are assumed
to be
involved in DNA repair which is why their processivity is relatively small.
Hence an object of the present invention was t:o combine several of the
aforementioned properties of polymerases in particular that of high
processivity and
thermostability for use in in vitro reactions.
This object was achieved according to the invention by providing a
thermostable in
vitro complex comprising a thermostable sliding clamp protein and a
thermostable
elongation protein having polymerase activity. The inventive complex can thus
be
used for the template-dependent elongation of nucleic acid(s).
This complex can be used in in vitro reactions such as in PCR reactions and
has a
high processivity in these reactions. An additional advantage is that the
complex has
a low error rate in the nucleotide incorporation i.e. an increased accuracy of
base
incorporation. Hence this complex can be used advantageously for the
elongation,
amplification, reverse transcription, DNA labelling and sequencing of nucleic
acids.
CA 02338185 2001-02-05
_ g _
The said applications each represent particularly preferred embodiments of the
invention.
If the inventive complex is used to amplify nucleic acids it was surprisingly
found
that the amplification product produced in this process has a particularly low
rate of
erroneous base incorporation.
The use of such a complex, for example in standard PCR reactions, ensures a
simple
handling and a high processivity as shown for example in Fig. 26.
In one embodiment it is intended that the sliding clamp protein is linked to
the
elongation protein in the in vitro complex according to t:he invention.
In a preferred embodiment of the inventive thermostable in vitro complex the
sliding
clamp protein and the elongation protein are linked by a coupling protein.
The coupling between the sliding clamp protein and the elongation protein
having
polymerase activity can be achieved by covalent and also by non-covalent
binding. In
a preferred alternative embodiment a direct coupling between the sliding clamp
protein and elongation protein is envisaged.
In this case the sliding clamp protein andlor the elongation protein can be
derived
from archaebacteria.
In a preferred embodiment of the inventive complex it is a prokaryotic in
vitro
complex.
In a preferred embodiment the inventive prokaryotic complex can be an
archaebacterial in vitro complex.
In a preferred alternative embodiment the inventive prokaryotic complex can be
a
eubacterial in vitro complex.
CA 02338185 2001-02-05
-9-
In an alternative embodiment of the complex according to the invention it is a
eukaryotic in vitro complex.
In this connection a prokaryotic in vitro complex is one in which the sliding
clamp
protein is of prokaryotic origin irrespective of the origin of the elongation
protein.
Correspondingly a eubacterial complex is one in which the sliding clamp
protein is of
eubacterial origin irrespective of the origin of the elongation protein.
Correspondingly an archaebacterial complex is one in which the sliding clamp
protein is of archaebacterial origin irrespective of the origin of the
elongation
protein. Furthermore a eukaryotic complex is correspondingly one in which the
sliding clamp protein is of eukaryotic origin irrespective of the origin of
the
elongation product.
The present invention also concerns those thermostable irr vitro complexes in
which
the proteins of which the complexes are composed are partly derived from
archaebacteria, eukaryotes and eubacteria. In this respect any permutations of
the irr
vitro complexes with regard to their protein components or their respective
origin
are a subject matter of the present invention.
In this connection origin in the above sense is intended to denote any source
in which
the gene, the gene information or the protein is based.
This is independent of the actual manner in which the sliding clamp protein or
elongation protein is obtained such as by chemical synthesis, genetic
engineering
methods or isolation from natural sources.
The invention in particular concerns a thermostable prokaryotic in vitro
complex for
the elongation and especially for the template-dependent elongation of nucleic
acids
which comprises a thermostable sliding clamp protein (fig. 20) which wholly or
partially encircles the complementary nucleic acid strands, and a thermostable
protein having polymerase activity (fig. 21 and fig. 22), this protein or this
protein
complex being coupled to or associated with the sliding clamp protein.
CA 02338185 2001-02-05
-10-
In the scope of the present invention the term elongation protein having
polymerise
activity also encompasses protein complexes having polymerise activity or
subunits
of such complexes which carry the polymerise activity.
Thermostable in the sense of the present invention means that the irr vitro
complex
incorporates nucleotides into growing nucleic acid strands with high
processivity at
the low as well as at the high temperatures which occur in PCR or other
reactions
e.g. DNA sequencing.
PCR usually for example comprises the steps of denaturation (70°C to
98°C),
annealing (40°C to 78°C) and DNA strand synthesis (60°C
to 76°C). Hence this
complex must be fiznctional at least between ca. 60°(: and ca.
70°C, in particular
preferably between 60°C and 76°C and particularly preferably
between 40°C and
98°C. There should be no signs of irreversible denaturation of the
complex or of
individual components during the entire reaction which could prevent or
inhibit the
elongation reaction.
The sliding clamp
The following details are intended to illustrate the fiznction and possible
forms of the
sliding clamp and of the sliding clamp protein.
The function of the sliding clamp protein is to bind the elongation protein to
the
DNA. As already mentioned above the sliding clamp protein itself surrounds the
single-stranded or double-stranded nucleic acid wholly or partially or by
association
with the protein having polymerise activity or as the case may be with the
protein
complex having polymerise activity or a subunit thereof and thus forms a clamp
around the nucleic acid. In any case the processivity is significantly
increased by at
least one and a half fold by this clamp formation (example 5 and fig. 23).
This means that the processivity of the inventive in vitro complex is at least
one and
a half fold of an elongation protein alone or a protein complex having
polymerise
activity or a subunit thereof without a sliding clamp (example 5 and fig. 23).
CA 02338185 2001-02-05
According to the invention homologues or functional analogues of the
proliferating
cell nuclear antigen protein complex coded in the human genome or homologues
of
the likewise circular (3-clamp protein complex from E coli which are derived
from
thermostable organisms or are thermostable or, if they are non-thermostable,
can be
made thermostable, or are derived from non-thermostable organisms and are
thermostable or can subsequently be made thermostable by modifying the amino
acid
sequence, can be used for example as sliding clamps (Eijsink VG, van der Zee
JR,
van den Burg B, Vriend G, Venema G, FEBS Lett 1991 Apr 22; 282(1):13-16,
Improving the thermostability of the neutral protease of Bacillus
stearothermophilus
by replacing a buried asparagine by leucine, Bertus Van den Burg, Gert Vriend,
Oene R. Veltman, Gerard Venema and Vincent G.H. Eijsink Engineering An enzyme
to resist boiling PNAS 1998 95:2056-2060). Homologous sequences are understood
herein in the following as sequences which are characterized by having a
sequence
that is similar to one or several other sequences and namely to such an extent
that it
cannot be assumed to be a coincidental similarity. The degree of sequence
similarity
is expressed in percent and is referred to as homology. Sometimes the term
sequence
identity is also used. A homologue is a nucleic acid or amino acid sequence
which is
a homologous sequence to a reference sequence.
The sliding clamp can be composed of several components. The sliding clamp
identified in the human genome is composed of three PCNA-protein components
(SEQ ID NO:11 ) (homotrimer) and the sliding clamp identified in the E coli
genome
is composed of two components (SEQ ID N0:35) (homodimer).
A sliding clamp in the sense of the present invention is understood in
particular as
any protein that has the functional property of increasing polymerase
processivity
(example 5 and fig. 23) and/or which reduces the error rate. For this purpose
the
sliding clamp can have a circular three-dimensional structure or can form a
circular
three-dimensional structure by coupling to another protein by which means it
is able
to wholly or partially encircle single and double-stranded nucleic acids.
CA 02338185 2001-02-05
- 12 -
A sliding clamp in the sense of the present invention is in particular
understood as a
protein which
1. has a sequence identity of at least 20 %, preferably of at least 25 % and
more
preferably of at least 30 °ro to the human PCNA amino acid sequence
(eukaryotes) (SEQ ID NO:11 ) over a length of at least 100 amino acids in a
sequence alignment or which
2. has a sequence identity of at least 20 %, preferably of at least 25 % and
more
preferably of at least 30 % to the bacterial (3-clamp sequence from E. coli
(eubacteria) (SEQ ID N0:35) over a length of at least 100 amino acids in a
sequence alignment or which
3. has a sequence identity of at least 20 %, preferably of at least 25 % and
more
preferably of at least 30 % to the amino acid sequence of the PCNA homologue
from Archaeoglobus.fulgiclus (archaebacteria) (SEQ ID N0:12) over a length of
at least 100 amino acids in a sequence alignment.
All sequence alignments disclosed herein were generated using the BLAST
algorithm
according to Altschul, S.F., Gish, W. Miller, W., Myers, E.W., and Lipman,
D.J., J.
Mol. Biol. 215, 403-410 (1990).
The sliding clamp according to the invention can have one or several of the
aforementioned features.
In the sense of the present invention sliding clamps or sliding clamp proteins
are also
to be understood as proteins which contain one or both of the following
consensus
sequences (of region 1 and region 2) and deviate at not more than four
positions
from region 1 (SEQ ID N0:39) or at not more than four positions from region 2
(SEQ ID N0:40) (fig. 4):
Region 1
(SEQ ID N0:39):
[GAVLIMPFW]-D-X-X-X-[GAVL11VIPFW]-X-X-[GAVLIMPFW]-X-[GAVLIMPFW]-
X-[GAVLIMPFW]-X-X-X-X-F-X-X-Y-X-X-D
and/or
CA 02338185 2001-02-05
-13-
Region 2
(SEQ ID N0:40):
[GAVLIMPFW]-X(3)-L-A-P-[KRHDE]-[GAVLIMPFW]-E
The amino acids are denoted according to the standard fUPAC - single letter -
nomenclature and shown in accordance with the Prosite pattern description
standard.
The following amino acid groups are frequently pooled together:
G,A,V,L,I,M,P,F or W (amino acids with non-polar side chains)
S,T,N,Q,Y or C (amino acid with uncharged polar side chains)
K,R,H,D or E (amino acid with charged and polar side chains)
In addition X denotes any desired amino acid or insertion or deletion in the
sequences or sequence protocols.
Furthermore a hidden Markov model was generated from the multiple alignment of
human PCNA homologues shown in fig. 12. Hence a sliding clamp in the sense of
the present invention is also especially to be understood as any protein which
has a
score of more than 20 preferably 25 and most preferably 30 with the hidden
Markov
model (referred to as HMM in the following) generated in this manner (fig. 12)
whereby a score is the output value of a HMM analysis. The hidden Markov model
and the corresponding scores were calculated using the hmmfs programme
(version
1.8.4, July 1997) from the l~-IMMER package (~:R protein and DNA hidden
Markov Model (version 1.8) by Sean Eddy, Dept. of Genetics, Washington
University School of Medicine, St. Louis, USA).
Markov models with a hidden profile (profile HMMs) can also be referred to in
a
short form as hidden Markov models and are abbreviated herein as HMM are
statistical models of the consensus of the primary structure of a sequence
family. The
profiles use position-specific scores for amino acids (or nucleotides) and
position-
specific scores for the opening or extension of an insertion or deletion.
Methods for
setting up profiles from multiple alignments were introduced by Taylor (
1986),
Gribskov et al. ( I 987), Barton ( 1990) and Heinikof~ ( 1996).
HMMs provide a completely probabilistic description of profiles i.e. the
teachings of
Bayes determine how the entire probability (assessment) parameters should be
set
CA 02338185 2001-02-05
- 14-
(cf. Krogh et al. 1994, Eddy 1996 and Eddy 1998). The pivotal idea is that a
HMM
is a finite model which describes a probability distribution over an unlimited
number
of possible sequences. The HMM is composed of a number of states which
correspond to the columns of a multiple alignment as it is usually shown. Each
state
emits symbols (remainders) corresponding to the (state-specific) symbol
emission
probabilities and the states are linked together by state transition
probabilities. A
series of states is generated starting from an initial state by transition
from one state
to the other according to the state transition probabilities until an end
state is
reached. Each state then emits symbols corresponding to the emission
probability
distribution of this state which generates an observable sequence of symbols.
The attribute hidden is derived from the fact that the underlying state
sequence
cannot be observed; it is the symbol sequence that is observed. An estimation
of the
transition and emission probabilities (the training of the model) is achieved
by
dynamic programming algorithms which are implemented in the HMMER package.
With an existing HN11VI and a given sequence it is possible to calculate the
probability
that the HMM can generate the sequence in question. The HMMER package
provides a numerical quantity (the score or output value) which is
proportional to
this probability i.e. the information content of the sequence is stated in
bits and
measured according to the HMM.
Reference is made to the following literature references in connection with
HMM:
Barton, G.J. (1990):
Protein multiple alignment and flexible pattern matching.
Methods enzymol. 183: 403-427
Eddy, S.R. (1996):
Hidden Markov models
Curr. Opin. Strct. Biol. 6: 361-365
CA 02338185 2001-02-05
-15-
Eddy, S.R. (1998):
Profile hidden Markov models
Bioinformatics. 14: 75 5-763
Gribskov, M. McLachlan, A.D. and Eisenberg D. (1987):
Profile analysis: Detection of distantly related proteins
Proc. Natl. Acad. Sci. USA 84: 4355-5358
Heinikoff, S. ( 1996):
Scores for sequence searches and alignment
Curr. Opin. Strct. Biol. 6: 353-360
Krogh, A., Brown, M., Mian, I. S., Sjolander, K. and Haussler, D. ( 1994):
Hidden Markov models in computational biology: Applications to protein
modelling.
J. Mol. Biol. 235: 1501-1531
Taylor, W.R. (1986):
Identification of protein sequence homology by consensus template alignment
J. Mol. Biol. 188: 233-258
A HMM was generated from the multiple alignment of E. coli (3-clamp homologues
shown in fig. 13. Hence a sliding clamp in the sense of the present invention
is also in
particular to be understood as any protein which has a score of more than 25,
preferably 30 and most preferably 35 in the HMM generated in this manner (fig.
13).
The sliding clamps can be composed of several components which are bound
firmly
together by a characteristic binding such that a stable circular molecular
complex is
formed which cannot be readily dissociated from the nucleic acid. This enables
a firm
but non-covalent binding to the nucleic acid which does not hinder free
movement
on the nucleic acid. Moreover, the sliding clamp proteins that increase
processivity
have characteristic local molecular properties in the region of interaction
with the
CA 02338185 2001-02-05
-16-
DNA which facilitate free movability and which can be facilitated by water
molecules
intercalated in this region.
A further preferred embodiment of the present invention is in particular a
thermostable prokaryotic in vitro complex in which the sliding clamp protein
is one
of the following: AF 0335 from Archaeoglobus ficlgid2cs (SEQ ID N0:12) (fig
24),
MJ0247 from Methanococctrs jarn~aschii (SEQ ID N0:13), PHLA008 from
Pyrococcrrs horikoschii (SEQ ID N0:14), MTH1312 from Methanobacterium
Thermoazetotrophicus (SEQ ID N0:15) as well as AE000761 7 from Aquifex
aeolicus (SEQ ID N0:36).
In particular those thermostable in vitro complexes are a subject matter of
this
application in which the sliding clamp protein contains an amino acid sequence
which
is selected from the group comprising SEQ ID NO: 1 I, 12, 13, 14, 15 and 36.
The sliding clamp loader
A further preferred embodiment is one in which the inventive complex includes
a
sliding clamp loader. A sliding clamp loader is understood herein as a
protein,
protein complex or subunit of a protein which comprises a homologue of the
replication factor C protein complex identified in humans.
In humans this protein complex is composed of five subunits and is coded by
five
separate genes in humans. The four small subunits each coded by one gene
(referred
to herein as sliding clamp loader 1 ) form a protein complex in humans. The
protein
of the large subunit is coded by one gene in humans (referred to herein as
sliding
clamp loader 2). The sequences of the four small subunits are shown as SEQ ID
NO:1, 32, 33, 34, the sequence of the large subunit is shown as SEQ ID N0:6.
According to the invention homologues or fiznctional analogues of any of the
above-
mentioned sequences SEQ ID NO:1, 32, 33, 34 can be used singly or in any
combination as the sliding clamp loader 1. In this connection the homologues
can be
of prokaryotic as well as eubacterial or archaebacterial or eukaryotic origin.
CA 02338185 2001-02-05
- 1 7 -
According to the invention homologues or functional analogues of the above-
mentioned sequence SEQ ID N0:6 can be used singly or in any combination as the
sliding clamp loader 2. In this connection the homologues can be of
prokaryotic as
well as eubacterial or archaebacterial or eukaryotic origin.
A protein can also be understood as a sliding clamp loader 1 in the sense of
the
present invention which has an at least 20 %, preferably at least 25 % and
even more
preferably at least 30 % sequence identity to the human (eukaroytic) amino
acid
sequence (SEQ ID NO:1, 32, 33, 34) over a length of at least 100 amino acids
in a
sequence alignment.
A protein can also be understood as a sliding clamp loader 2 in the sense of
the
present invention which has an at least 20 %, preferably at least 25 % and
even more
preferably at least 30 % sequence identity to the human (eukaryotic) amino
acid
sequence (SEQ ID N0:6) over a length of at least 150 amino acids in a sequence
alignment.
Sliding clamp loader homologues in the sense of the above definition are for
example
the genes from archaebacteria listed in fig. 1. These genes correspond to the
sequences SEQ ID NO: 2, 3, 4 and 5 for the sliding clamp loader 1 and to the
sequences SEQ ID NO: 7, 8, 9 and 10 for the sliding clamp loader 2.
A protein can also be understood as a sliding clamp loader 1 in the sense of
the
present invention which contains a sequence in accordance with the following
consensus sequence and which differs at no more than four positions from this
sequence (see also fig. 6 for the alignment):
SEQ ID N0:41:
C-N-Y-X-S-[KRHDE]-I-I-X-[GAVLIMPFW]-[GAVL,:fMPFW]-Q-S-R-C-X-X-F-
R-F-X-P-[GAVLIIVVIPFW]
A protein can also be understood as a sliding clamp loader 2 in the sense of
the
present invention which contains a sequence in accordance with the following
CA 02338185 2001-02-05
-18-
consensus sequence and which differs at no more than four positions from this
sequence (see also fig. 7 for the alignment):
SEQ ID N0:42:
K-X-X-L-L-X-G-P-P-G-X-G-K-T-[STNQYC]-X-[GAVLIMPFWJ-X-X-
[GAVLIMPFW]
In addition a HMM was generated from the multiple alignment of sequences of
the
sliding clamp loader 1 shown in fig. 14 comprising the human sequence and some
homologous sequences thereto from archaebacteria. Consequently a sliding clamp
loader 1 is also understood in the sense of the present invention as a protein
which
has a score of more than 25, preferably more than 30 and most preferably more
than
35 in the HMM generated in this manner (see also fig. 14 for the alignment).
In addition a HMM was generated from the multiple alignment of sequences of
the
sliding clamp loader 2 shown in fig. 15 comprising the human sequence and some
homologous sequences thereto from archaebacteria. C',onsequently a sliding
clamp
loader 2 is also understood in the sense of the present invention as a protein
which
has a score of more than 15, preferably more than 20 and most preferably more
than
25 in the HMM generated in this manner (see also fig. 15 for the alignment).
The inventive in vitro complex may also contain a protein homologous to the
eubacterium Escherichia coli y-complex or parts thereof as the sliding clamp
loader
1 or sliding clamp loader 2.
Consequently a sliding clamp loader in the sense of the present invention can
be a
sliding clamp loader 1 alone, a sliding clamp loader 2 alone or a combination
of one
or several sliding clamp loaders 1 or sliding clamp loaders 2 each as defined
above.
Furthermore in one embodiment the inventive thermostable in vitro complex for
the
elongation of nucleic acids additionally contains a compound which releases
energy
when cleaved such as ATP, GTP, CTP, TTP, dATP, dGTP, dCTP or dTTP.
CA 02338185 2001-02-05
- 19-
Without wanting to be limited thereto in the following, a sliding clamp loader
appears to assemble the components of the sliding clamp around the
uninterrupted
DNA strand and to remove these again when the reaction is completed. In this
connection the sliding clamp can have a ring-shaped three-dimensional
structure or
can form a ring-shaped three-dimensional structure by coupling to another
protein by
which means it is able to wholly or partially encircle single-stranded or
double-
stranded nucleic acids. The sliding clamp loader can be advantageous when the
template nucleic acid molecule is present in a closed ring shape.
In particular those thermostable in vitro complexes are a subject matter of
this
application in which the sliding clamp loader 1 comprises an amino acid
sequence
which is selected from the group comprising SEQ ID NO: 2, 3, 4 and 5.
In particular those thermostable in vitro complexes are a subject matter of
this
application in which the sliding clamp loader 2 comprises an amino acid
sequence
which is selected from the group comprising SEQ ID NO: 7, 8, 9 and 10.
The coupling protein
The function of the coupling protein is to connect an elongation protein with
one or
several sliding clamp proteins or with a sliding clamp protein complex. Hence
a
coupling protein in the sense of the present invention is to be understood in
particular as any protein which has the function described above. Those
coupling
proteins are preferred which are of archaebacterial origin.
Coupling proteins in the sense of the present invention are for example
homologues
or functional analogues of singly or in any combination of each of the
sequences
listed in fig. 1 which are homologues to the human sequence of the small
subunit of
polymerase 8 which is referred to herein as the coupling subunit (sequence
name:
DPD2 I-fUMAN, shown in the sequence protocol as SEQ >D NO:16). In the sense
of the present invention a coupling protein can be a protein which comprises a
sequence that is selected from groups comprising the sequences SEQ ID NO: 17,
18,
19, 20 and 21.
CA 02338185 2001-02-05
-20-
A coupling protein in the sense of the present invention is in particular to
be
understood as a protein which has a sequence identity to the human
(eukaryotic)
amino acid sequence (SEQ ID N0:16) of at least 18 °io, preferably of at
least 22
and even more preferably of at least 26 % over a length of at least 150 amino
acids
in a sequence alignment.
A coupling protein in the sense of the present invention is in particular to
be
understood as a protein which has a sequence identity to the amino acid
sequence
(SEQ ID N0:19) from Pyrococcus horikoshii of at least 20 %, preferably a
sequence identity of at least 25 % and even more preferably a sequence
identity of
more than 30 % over a length of at least 150 amino acids in a sequence
alignment.
A coupling protein in the sense of the present invention is also to be
understood as
any protein which contains the following consensus sequence and deviates from
this
sequence at not more than four positions. The generation of the consensus
sequence
is shown in fig. 5 which is disclosed herein as SEQ ID N0:43.
SEQ ID N0:43:
[FLJ-[GAVLIMPFWJ-X-X-[GAVLIMPFW]-X-G-X( 13)-[GAVLIMPFWJ-X-[YRJ-
[GAVLIMPFWJ-X-[GAVLIMPFWJ-A-G-[DNJ-[GAVLIMPFWJ-[GAVLIMPFWJ-
[DSJ
In addition a HMM was generated from the multiple alignment of homologues to
the
human coupling subunit or coupling protein shown in fig. 16. Hence a coupling
subunit in the sense of the present invention is to be understood as any
protein which
has a score of more than 10, preferably of more than 15 and most preferably of
more
than 20 in the HMM generated in this manner.
In particular those thermostable in vitro complexes are a subject matter of
this
application in which the coupling protein comprises an amino acid sequence
selected
from the group comprising SEQ ID NO: 17, 18, 19, 20 and 21.
CA 02338185 2001-02-05
-21 -
Elongation protein
An elongation protein can be used within the scope of the present invention
which
has the features defined above. In addition it is possible to use forms of
elongation
proteins which are described in the following and of which at least some are
already
known in the prior art.
It is known that some elongation proteins require the presence of a coupling
protein
in order to have any polymerase activity at all. It is also known that some
elongation
proteins can bind directly to sliding clamp proteins whereas other elongation
proteins
require the presence of a coupling protein in order to bind to the sliding
clamp
proteins. Furthermore elongation proteins can combine both of the above-
mentioned
properties i.e. bind to a sliding clamp protein via a coupling protein or
directly i.e.
without a coupling protein.
Preferred elongation proteins for the inventive in vitro complex can be
selected from
the group which comprises the organisms Carboxydothermrrs hydrogenoformans,
Thermrrs aqrraticZrs, Thernnrs caldophihrs, lhernarrs chliarophilus, Thermrrs
filiformis, Thernnrs fZcnnss, Thermos oshimai, Tlrermus tuber, Thermrrs
scotodrrctrrs,
Thermos silvarrus, Thermrrs species ZO~, Thermrrs species sp. 17, Thermos
thermrrsphilus, Therotoga maritima, Therotoga rreapolitaraa, Thermosipho
africanrrs, Afraerocellum thermophilum, Bacillus caldotenax or Bacillus
stearothermophilrrs.
The elongation proteins listed in fig. 1 from Archaebacteria (SEQ ID NO: 23,
24,
25, 26) which are homologous or fianctionally analogous to the human
elongation
protein (SEQ ID N0:22) are for example also suitable as elongation proteins.
In particular those thermostable irr vitro complexes are a subject matter of
the
invention in which the elongation protein comprises an amino acid sequence
which is
selected from the group comprising SEQ ID NO: 23, 24, 25, 26, 27, 28, 29, 30
and
31.
CA 02338185 2001-02-05
-22-
An elongation protein in the sense of the present invention is in particular
to be
understood as a protein which has a sequence identity of at least 20 %,
preferably of
at least 25 % and most preferably of at least 30 % to the human (eukaryotic)
amino
acid sequence (SEQ ID N0:22) over a length of at least 200 amino acids in a
sequence alignment.
An elongation protein in the sense of the present invention is also in
particular
understood as a protein which contains the following consensus sequence (SEQ
ID
N0:44) and does not differ at more than four positions from this sequence.
Fig. 8
shows the alignment which forms the basis for the consensus sequence.
SEQ ID N0.44:
D-[GAVLIMPFW]-[GAVLIMPFW]-X-X-Y-N-X-X-X-F-D-X-P-Y-
[GAVLIMPFW]-X-X-R-A
In addition a HMM was generated from the multiple alignment of homologues to
the
human elongation protein (SEQ ID N0:22) shown in fig. 17. Hence an elongation
protein in the sense of the present invention is to be understood as any
protein which
has a score of more than 20, preferably of more than 25 and most preferably of
more
than 30 in the HMM generated in this manner.
Hence an elongation protein in the sense of the present invention is in
particular to
be understood as a protein which has a sequence identity of at least 25 %,
preferably
of at least 30 % and most preferably of at least 35 %, to the archaebacterial
amino
acid sequence (SEQ ID N0:27) over a length of at least 400 amino acids in a
sequence alignment. For example the proteins derived from Archaebacteria
having
SEQ ID N0:27, 28, 29. 30 or 31 are suitable.
Hence an elongation protein in the sense of the present invention is also in
particular
understood as any protein which contains the following consensus sequence
referred
to herein as SEQ 117 N0:45 and which differs from this sequence at not more
than
four positions. (Fig. 9 shows the alignment which forms the basis for the
consensus
sequence).
CA 02338185 2001-02-05
- 23 -
SEQ ID N0:45:
A-[GAVLIMPFW]-R-T-A-[GAVLIMPFW]-A-[GAVLIMPFW]-[GAVLIMPFW]-
T-E-G-[GAVLIMPFW]-V-X-A-P-[GAVLIMPFW]-E-G-I-A-X-V-[KRHDE]
In addition a HMM was generated from the multiple alignment of homologues to
the
archaebacterial elongation protein (SEQ ID N0:27) shown in fig. 18. Hence an
elongation protein in the sense of the present invention is to be understood
as any
protein which has a score of more than 35, preferably of more than 40 and even
more preferably of more than 45 in the HMM generated in this manner (fig. I
8).
The elongation protein can also be of eubacterial origin.
Hence an elongation protein in the sense of the present invention is in
particular also
to be understood as a protein which has a sequence identity of at least 25 %,
preferably of at least 30 % and most preferably of at least 35 % to the
eubacterial
amino acid sequence (SEQ ID N0:37) over a length of at least 300 amino acids
in a
sequence alignment.
Hence an elongation protein in the sense of the present invention is also in
particular
understood as any protein which contains the following consensus sequence and
which differs from this sequence at not more than eight positions. (Fig.10).
SEQ ID N0:46:
[GAVLIMPFW]-P-V-G-[GAVLIMPFW]-G-R-G-S-X-[GAVLIMPFW]-G-S-
[GAVLIMPFW]-V-A-X-A-[GAVLIMPFW]-X-I-T-D-[GAVLIMPFW]-D-P-
[GAVLIMPFW]-X-X-X-[GAVLIMPFW]-L-F-E-R-F-I,-N-P-E-R-[GAVLIMPFW]-
S-M-P-D
In addition a HMM was generated from the multiple alignment of homologues to
the
eubacterial elongation protein (SEQ ID N0:37) shown in fig. 19. Hence an
elongation protein in the sense of the present invention is to be understood
as any
protein which has a score of more than 20, preferably of more than 25 and most
preferably of more than 30 in the HMM generated in this manner.
CA 02338185 2001-02-05
-24-
In particular those thermostable irr vitro complexes are a subject matter of
this
application in which the elongation protein comprises an amino acid sequence
which
is selected from the group comprising SEQ ID N0:38.
Use of the in vitro complex according to the invention:
Previously DNA polymerases such as e.g. DNA polymerase I from Pyrococcus
_frrriosrrs (US No. 5,545,552) or Pyrococcrrs species (EP-A-0 547 359) were
used as
elongation proteins for standard PCR reactions without a coupling protein and
without a sliding clamp. A characteristic of these enzymes is that they are
thermostable and frequently possess a 3'-5' exonuclease activity (proof
reading
activity). Only recently was a heterodimer with polymerase activity discovered
in
Pyrococcrrs.firriosrrs (Uemori, T., Sato, Y., Kato, L, Doi, H,, and Ishino, Y.
(1997).
A novel DNA polymerase in the hyperthermophilic archaeon,
Pyrococcrrs,frrriosrrs:
gene cloning, expression and characterization. Genes to Cells 2, 499-512).
It is also possible to optimize the properties of these proteins by deletions
or
mutations or by attaching amino acids. These modified proteins are also a
subject
matter of the present invention provided they form the inventive irr vitro
complex for
the elongation of nucleic acids and fulfil the functions which are specified
above in
more detail.
Fig. 3 illustrates by way of example an embodiment of the inventive
thermostable in
vitro complex in which the sliding clamp binds to the elongation protein by
means of
a coupling protein.
Furthermore the inventive in vitro complex or the inventive reaction mixture
containing this complex can contain a nucleic acid which is for example the
nucleic
acid to be elongated, to be sequenced, to be amplified or to be reversely
transcribed
and/or a primer in which case this primer is preferably hybridized to a
nucleic acid.
Primers are usually oligonucleotides that are complementary to a target
sequence
which enables them to bind to it and in opposite orientation with their 3'-
ends facing
CA 02338185 2001-02-05
-25-
one another, they enclose the nucleic acid section to be elongated, to be
sequenced,
to be amplified or to be reversely transcribed. They serve as the starting
point of
enzyme activity and usually provide a free 3'OH end for the polymerise to
incorporate a nucleotide.
During the use of the inventive thermostable irmitro complex for the
amplification,
elongation, reverse transcription and/or sequencing, the inventive complex may
be
present in a reaction mixture preferably in a suitable buffer. Suitable
buffers are those
that are used for PCR, sequencing, nucleic acid labelling and other ifs vitro
nucleic
acid elongation reactions by means of polymerise. Suitable buffers are
described for
example in Methods in Molecular Biology, vol. 15 Humana Press Totowa, New
Jersey, 1993, publ. Bruce A. White.
When the inventive thermostable in vitro complex is used for elongation,
amplification, reverse transcription and/or sequencing, a nucleotide or a
mixture of
nucleotides can be present or used or included in addition to the inventive
complex.
Deoxynucleotides can be selected from dGTP, dATP, dTTP and dCTP but are not
limited to these. In addition derivatives of deoxynucleotides can also be used
which
are defined as those deoxynucleotides which are able to be incorporated by a
thermostable polymerise into growing nucleic acid molecules. Such derivatives
include the thionucleotides 7-deaza-2'-dGTP, 7-deaza-2'-dATP, digoxigenin-dUTP
(Boehringer Mannheim)-dATP as well as deoxyinosine triphosphate which can also
be used as a substitute deoxynucleotide for dATP, dGTP, dTTP or dCTP but are
not
limited to these. It is also possible to use labelled deoxynucleotides. It is
also
possible to use pyrenes and pyrene derivatives. In this connection all known
and/or
suitable labels for the purpose of the invention can be present or used.
Dideoxynucleotides can be selected from ddGTP, ddATP, ddTTP and ddCTP, but
are not limited to these. Alternatively it is also possible to use derivatives
of dideoxy-
nucleotides which are defined as those dideoxynucleotides that are able to be
incorporated by a polymerise into growing nucleic acid molecules that are
synthesized in the reaction. Such derivatives can be radioactive
dideoxynucleotides
(ddATP, ddGTP, ddTTP and ddCTP) or dideoxynucleotides (ddATP, ddGTP,
ddTTP and ddCTP) that are labelled with e.g. FITC, CyS, Cy5.5, Cy7, Texas-red
or
CA 02338185 2001-02-05
-26-
other dyes but are not limited to these. As part of a sequencing using the
inventive
thermostable in vitro complex it is also possible to use labelled
deoxynucleotides
together with unlabelled dideoxynucleotides.
Ribonucleotides can be selected from GTP, ATP, TTP and CTP but are not limited
to these. In addition derivatives of ribonucleotides can also be used
according to the
invention which are defined as those ribonucleotides which are able to be
incorporated by a polymerase into growing nucleic acid molecules which are
synthesized in the reaction. Such derivatives can be radioactive
ribonucleotides
(ATP, GTP, TTP and CTP) or ribonucleotides (ATP, CiTP, TTP and CTP) that are
labelled with e.g. FITC, CyS, Cy5.5, Cy7 and Texas-red or others but are not
limited
to these.
During the use of the protein complex for amplification, elongation or
sequencing it
may prove to be advantageous when a pyrophosphatase is present in the reaction
or
in the reaction mixture.
Reaction mixture containing the thermostable in vitro complex according to the
invention: '
A further subject matter of the present invention is a reaction mixture which
contains
the ifi vitro complex according to the invention. Provision can also be made
for the
reaction mixture to additionally contain one or several elongation proteins
which
have at least one or several of the above-mentioned properties or activities.
Such an
additional elongation protein is advantageously a thermostable polymerase.
Such a
reaction mixture allows an increased processivity compared with using the
known
thermostable polymerases.
Identification of the genes, cloning the genes, expression of these and
purification of the proteins of inventive in vitro complexes:
The complexes, which are preferably completely or partially composed of
recombinant proteins, can usually be prepared by the following steps:
Preparation of
CA 02338185 2001-02-05
-27-
the nucleic acid fragment which codes for the desired protein, ligation into
an
expression vector, transformation into a host, expression and purification of
the
protein (fig. 25). In accordance with the present invention the genes and
especially
those from the Archaebacteria may contain inteins which can firstly be removed
(Proc, Natl, Acac~ Sci. USA 1992 Jun. 15, 89(12):5577-5581, Intervening
sequences in an Archaea DNA polymerase gene, Perler FB, Comb DG, Jack WE,
Moran LS, Qiang B, Kucera RB, Benner J, Slatko BE, Nwankwo DO, Hempstead
SK, et al).
Further genes and/or proteins which are suitable for the inventive complex can
for
example be identified by homology searches in databases that contain genomes
from
prokaryotes. Suitable programs for this include for example the programs
BLAST,
BLASTP and FASTA but are not limited to these (Altschul, Stephen F., Thomas L.
Madden, Alejandro A. Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller and
David J. Lipman ( 1997), Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs, Nucleic Acids Res. :25:3389-3402. W.R.
Pearson
& D.J. Lipman PNAS (1988) 85:2444-2448).
They can also be identified by using DNA probes in order to screen for
suitable
genes in for example total genomic banks from prokaryotes or eukaryotes. The
experimental methods required for this can be found in Maniatis et al.
(Molecular
Cloning (2°d edition, 3 volume set): A laboratory Manual, Cold Spring
Harbor
Laboratory Press, N.Y. (1989)) or in Ausubel et al (Current Protocols in
Molecular
Biology, John Wiley and Sons ( 1988)).
The purified nucleic acid of the genes for the proteins of which the inventive
complexes are composed can for example be provided by isolating it from a
genomic
bank of the relevant organism or by synthetic DNA preparation each being
combined
if desired with an amplification by means of PCR with the aid of primers that
are
specific for the desired gene section. Standard methods are described in
Maniatis et
al. (Molecatlar Cloning: A laboratory Manual, Cold Spring Harbor Laboratory
Press, N.Y. (1989)).
CA 02338185 2001-02-05
-28-
The genes of the proteins of the inventive in vitro complexes can be cloned
using
numerous methods and thus made available for protein expression in the host
organism by means of an expression vector. Standard methods are described in
Maniatis et al. (Molecular C'loning.~ A laboratory th'arnral, Cold Spring
Harbor
Laboratory Press, N.Y. (1989)). In this connection the genes of the complexes
can
for example be firstly cloned into a high copy vector e.g. pUCl8, past or
pBR322
and only afterwards be recloned into a prokaryotic expression vector e.g.
pTrc99,
pQE30, pQE31 or pQE32, or alternatively directly cloned into a prokaryotic or
viral
expression vector. Vectors in this connection are understood as nucleic acids
which
are able to transport another nucleic acid molecule into or between different
organisms or genetic backgrounds. As a rule they are able to autonomously
replicate
and/or express (expression vectors) the operatively linked nucleic acid
molecule.
Operatively linked as used herein means that the transported nucleic acid
molecule is
linked with the vector in such a manner that it is under the transcriptional
and
translational control of expression control sequences of the vector and can be
expressed in a host cell. Bacterial and viral expression systems, their
preferred
applications and a selection of vector systems are described for example in
Gene
Expression Technology, (Meth. Enzymol. vol 185, Goeddel Ed., Academic Press,
N.Y. (1990)). Suitable vectors for the present invention should enable the
proteins to
be expressed at different strengths due to the fact that they have one or all
of the
following properties: ( 1 ) promoters or transcription initiation sites either
directly
adjacent to the start of the protein or as a fusion protein, (2) operators can
be used
to switch the gene expression on or off, (3) ribosomal binding sites for an
improved
translation and (4) termination sites for the transcription or translation
which lead to
an improved stability of the transcript.
Expression vectors that are compatible with eukaryotic cells and preferably
with
vertebrate cells can also be used. Some known vectors are pSVL and pKSV-10
(Pharmacia), pBPV-1/pML2d (International Biotechnologies, Inc.), pAcHLT-ABC
(Pharmingen) and pTDTI (ATCC 31255). It is also possible to use a retroviral
expression vector.
Hence further subject matters of the present invention are DNA sequences which
code for the inventive thermostable iyi vitro complexes and appropriate
vectors
preferably expression vectors.
CA 02338185 2001-02-05
-29-
The vectors according to the invention contain at least one gene for the
sliding clamp
protein and at least one gene for the coupling protein or at least one gene
for a
sliding clamp loader 1 and/or 2 or at least one gene for an elongation
protein. In one
embodiment the vectors simultaneously contain several of the various genes
mentioned above, for example the genes for an elongation protein and a sliding
clamp protein.
Within the scope of the invention it is preferred that the vector contains
additional
suitable restriction cleavage sites and optionally polylinkers for the
insertion of
further DNA sequences in addition to the DNA sequences that are already
contained
therein. It is particularly preferred that the spatial arrangement of the DNA
sequence
already present and the additional insertion site leads to the formation of a
fusion
protein after expression.
It is additionally preferred that the inventive vector contains promoter
and/or
operator regions and it is particularly preferred that such promoter and/or
operator
regions are inducible or repressible. This considerably simplifies the control
of
expression in host cells and can be made to be particularly efficient.
Such promoter/operator regions can also occur several times in one expression
vector which may enable several DNA sequences to be expressed independently
using only one expression vector.
A further subject matter of the present invention is a host cell containing
one or
several inventive vectors) wherein expression to form proteins can take place
in this
host cell under suitable conditions. Suitable conditions include for example
the
presence of a repressor, inducer or a derepressor.
There are standard protocols for transformation, phage infection and cell
culture in
Maniatis et al. (Molecular Cloning: A laboratory tl~lanual, Cold Spring Harbor
Laboratory Press, N.Y.). Among the numerous available E. coli strains that are
suitable for transformation, the following are preferred JM101 (ATCC No.
33876),
XI,I (Stratagene), RRI (ATCC No. 31343) M15[prep4] (QIAGEN) and BL21
(Pharmacia). Protein expression can for example also occur with the aid of the
E.
CA 02338185 2001-02-05
-30-
coli strand INVaF' (Invitrogen). The transformants are cultured under suitable
growth conditions for the host strain. Thus most E. coli strains are for
example
cultured in LB medium at 30°C to 42°C until the logarithmic or
stationary growth
phase is reached. The proteins can be purified from a transformed culture, and
this
can either be from a cell pellet after centrifugation or from the culture
liquid. If the
proteins are purified from the cell pellet, the cells are resuspended in a
suitable buffer
and disrupted by means of ultrasonic treatment, enzymatic treatment or
freezing and
thawing. If they are purified from the culture suspension either with or
without a
fusion protein, the supernatant is separated from the cells by means of known
procedures such as centrifugation.
The proteins of the inventive complexes can be separated and purified either
from
the supernatant of the culture solution or from the cell extract by known
separation
or purification procedures. These methods are for example those which are
based on
solubilities such as salt precipitations and solvent precipitations, methods
which
utilize the differences in molecular weights such as dialysis,
ultrafiltration, gel
filtration and SDS polyacrylamide gel electrophoresis, methods which utilize
differences in charge such as ion exchange chromatography, methods which
utilize
differences in hydrophobicity such as reverse phase HPLC (High Performance
Liquid Chromatography), methods which utilize particular affinities such as
affinity
chromatography (example 6, 7 fig. 24 and fig. 25) and methods which utilize
differences in the isoelectric point such as isoelectric focussing. It is also
conceivable
that cell extracts can be made either from the organism which carries the gene
of the
accessory complex which fulfils the inventive object or from the recombinant
host
organism e.g. E. coli. If these extracts are used it is possible to omit other
purification steps.
The methods described above can be used in many combinations in order to
prepare
proteins of the in vitro complex.
Elongation of nucleic acids
The inventive thermostable in vitro complexes can be used to elongate nucleic
acids
e.g, for the polymerase chain reaction (example 3, 4 and fig. 21, fig. 22),
DNA
CA 02338185 2001-02-05
-31 -
sequencing, to label nucleic acids and for other reactions which comprise the
in vitro
synthesis of nucleic acids.
Hence a further subject matter of the present invention is a method for the
template-
dependent elongation of nucleic acids in which the nucleic acid is denatured
if
necessary, is provided with at least one primer under hybridization
conditions, the
primer being sufficiently complementary to a flanking region of a desired
nucleic acid
sequence of the template strand and a primer elongation is carried out in the
presence of nucleotides with the aid of a polymerase, the inventive
thermostable in
vitro complex being used as the polymerase.
Methods are known to a person skilled in the art for the template-dependent
elongation of nucleic acids in which the elongation is initiated with a primer
that has
been hybridized to the template nucleic acid and provides a free 3'-OH end for
the
elongation. In particular a PCR polymerase chain reaction is carried out for
the
amplification.
Reverse transcription
The thermostable irt vitro complex according to the invention can also be used
for
reverse transcription in which either the inventive complex itself has reverse
transcriptase activity or a suitable enzyme is additionally added which has
reverse
transcriptase activity irrespective of whether the thermostable in vitro
complex itself
has a reverse transcriptase activity.
An inventive thermostable in vib°o complex whose elongation protein
itself has a
reverse transcriptase activity is also used for the reverse transcription of
RNA into
DNA which is preferred according to the invention. This reverse transcriptase
activity may be the only polymerase activity of the elongation protein but it
may also
be present in addition to an existing 5'-3'-DNA polymerase activity. An
embodiment
of the in vitro complex that is preferred according to the invention contains
the
elongation protein from the organism Carboxydothermus hydrogenoformans as
disclosed in EP-A 0 834 569.
CA 02338185 2001-02-05
-32-
Sequencing
A further preferred use of the inventive in vitro complex is to sequence
nucleic acids
according to the method of Sanger. Starting with at least one primer which is
sufficiently complementary to a part of the nucleic acid to be sequenced, a
template-
dependent elongation is carried out. When sequencing RNA it is necessary to
carry
out a reverse transcription. In the scope of this preferred embodiment the
respective
derivatives described above are also regarded to be suitable as
deoxynucleotides or
dideoxynucleotides. In particular it is preferable for the inventive method of
nucleic
acid elongation that the generated nucleic acids are labelled. For this it is
possible to
use labelled primers and/or labelled deoxynucleotides and/or labelled
dideoxynucleotides and/or labelled ribonucleotides or appropriate derivatives
of each
of these like those that have for example already been described above.
Labelling of nucleic acids
A further subject matter of the present invention is a method for labelling
nucleic
acids e.g. by inserting individual breaks in the phosphodiester bonds of the
nucleic
acid chain and replacing a nucleotide at the sites of the breaks by a labelled
nucleotide with the aid of a polymerase, in which an inventive thermostable in
vitro
complex is used as the polymerase.
A preferred method is the method that is generally referred to as nick
translation
which enables a simple labelling of nucleic acids. All labelled
ribonucleotides or
deoxyribonucleotides or derivatives thereof that have already been described
above
are suitable for this provided the polymerase accepts them as a substrate.
Labelling in the above sense is also a labelling which occurs as part of a PCR
reaction whereby in this case labelled nucleotides or derivatives thereof are
incorporated into the nucleic acid sequence.
CA 02338185 2001-02-05
- 33 -
The present invention additionally concerns a kit for the elongation and/or
amplification and/or reverse transcription and/or sequencing of nucleic acids
wherein
this kit can contain one or several containers.
The kit itself comprises
a) an inventive thermostable ira vitro complex or
b) a thermostable in vitro complex and optionally, separately therefrom, an
elongation protein having polymerase activity and optionally one or several
components which are selected from the group comprising primers, buffer
substances, nucleotides, ATP, other cofactors and pyrophosphatase.
In this case it is possible that the thermostable irJ vitro complex according
to the
invention is present in the said kit in the form of its individual components
i.e. as a
thermostable sliding clamp protein and thermostable elongation protein that
are
separated or combined in one container and which does not form as such until a
later
time.
In particular a subject matter of the present invention is a kit for the
elongation,
amplification, reverse transcription, labelling and sequencing of nucleic
acids
additionally containing deoxynucleotides or derivatives thereof.
The kit according to the invention can optionally also contain ribonucleotides
or
derivatives thereof especially when an elongation protein is used which
accepts
ribonucleotides as a substrate.
A preferred embodiment of the inventive kit is a kit for sequencing nucleic
acids
containing dideoxynucleotides or derivatives thereof for chain termination in
addition
to deoxynucleotides and derivatives thereof.
Furthermore a subject matter of the present invention is in particular a kit
for the
reverse transcription of nucleic acids in which either the inventive complex
itself has
reverse transcriptase activity or a suitable enzyme is additionally present
which has
reverse transcriptase activity and deoxynucleotides or derivatives thereof may
be
present in the reaction mixture.
CA 02338185 2001-02-05
-34-
In a further particularly preferred embodiment the kit contains primers and/or
deoxynucleotides and/or dideoxynucleotides and/or ribonucleotides and/or their
respective derivatives in a labelled form.
Especially when sequencing nucleic acids it is necessary to insert a label.
Suitable
labels have already been described above in the form of examples. Suitable
labelling
agents are included in a preferred embodiment of the inventive kit.
Within the scope of the present invention it is additionally possible that the
kit (also
referred to as reagent kit herein) is used to label nucleic acids. In this
case the
reagent kit contains the components a) or b) and labelled nucleotides wherein
buffer
substances, ATP or other cofactors and/or pyrophosphatase may also be present.
A kit is especially preferred which contains a suitable buffer as described
above. It is
also preferred that the kit according to the invention additionally contains a
pyrophosphatase, ATP and/or other cofactors.
A further subject matter of the present invention is the use of a thermostable
sliding
clamp protein in in vitro methods for the elongation, amplification, labelling
and
sequencing or reverse transcription of nucleic acids.
The thermostable irt vitro complex according to the invention can be used for
the
purposes of sequencing, amplification, reverse transcription and such like
using short
as well as long nucleic acid fragments to achieve an overall reduction in the
error
rate (incorporation of incorrect nucleotides) compared to the use of
thermostable
elongation proteins alone.
In a preferred alternative the thermostable in vitro complex is used for the
sequencing, amplification and reverse transcription of long nucleic acid
fragments.
In a fizrther preferred alternative the thermostable in vitro complex is used
for the
sequencing, amplification and reverse transcription of those nucleic acid
fragments
which form a secondary structure.
CA 02338185 2001-02-05
-35-
The following examples in conjunction with the figures are intended to further
elucidate the invention and illustrate the advantages:
Figures:
Sequence names are often used in the following which refer to the protein or
nucleic
acid sequences in the gene bank and the EMBL database. They show the
following:
Fig. l tabulation of protein sequence names of the inventive in vitro complex
and
values from paired alignments and multiple alignments between Archaebacteria
and
between Archaebacteria and the corresponding human genes;
Fig. 2 a table similar to that of fig. 1 but limited to the sliding clamp
pratein and
elongation protein of E. coli and A. aeolicu.s;
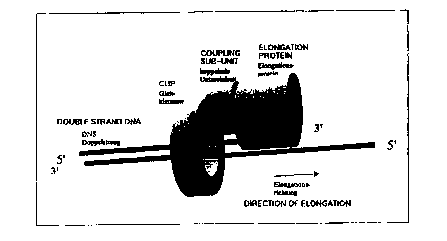
Fig. 3 a schematic representation of the inventive thermostable in vitro
complex;
Fig. 4 alignments of two conserved regions of the sliding clamp protein;
Fig. 5 an alignment of a conserved region of the coupling protein;
Fig. 6 an alignment of a conserved region of the sliding clamp loader;
Fig. 7 an alignment of a conserved region of the sliding clamp loader 2;
Fig. 8 an alignment of a conserved region of the elongation protein 1;
Fig. 9 an alignment of a conserved region of the elongation protein 2;
Fig. 10 an alignment of a conserved region of the elongation protein from
eubacteria;
CA 02338185 2001-02-05
-36-
Fig. 1 I an alignment of a conserved region of the sliding clamp from
eubacteria;
Fig. 12 to 19 multiple alignments of various protein sequences for the
generation of
Hidden Markov models (H1V1NI);
Fig. 20 a chromatographic analysis of a recombinant sliding clamp
(Archaeoglobus
fulgidus PCNA (AF 0335));
Fig. 21 and 22 the result of a PCR using an elongation protein as a component
of a
thermostable in vitro complex;
Fig. 23 comparison of the activity of an elongation protein with and without a
sliding clamp protein;
Fig. 24 the result of the purification of a sliding clamp protein;
Fig. 25 the expression and purification of Archaeoglobus J'rrlgidrrs DNA
polymerise;
Fig. 26 the results of using an inventive ira vitro complex in the PCR;
Fig. 27 the results of a Y2H experiment;
Fig. 28 the PCR amplification result of the human collagen gene using the
inventive
thermostable ifr vitro complex;
Fig. 29 a tabulated overview of those genes that correspond to the protein
sequence
numbers stated in fig. 1 and which can be obtained from the respective
databases;
and
CA 02338185 2001-02-05
-37-
Fig. 30 a tabulation of the background information for the various databases
in the
English language which can be used to obtain the nucleic acid sequence data
and
amino acid sequence data that are stated herein.
Detailed description of the figures
Fig. 1:
Figure 1 is a tabulation of protein sequence names of the inventive
thermostable in
vitro complex and values from paired alignments and multiple alignments
between
Archaebacteria and between Archaebacteria and the corresponding human genes.
In
this connection the annotation ~~~" denotes the percentage identity (%) to the
corresponding human gene calculated from the paired alignment (see figures)
using
BLAST 2Ø4 [Feb-24-1998] and the annotation ~~Z'~ denotes the percentage
identity
to the corresponding gene of Archaeoglobus , fiilgidus, calculated from the
paired
alignment (see figures) using BLASTP 2Ø4 [Feb-24-1998] and the annotation
~~3"
denotes the percentage identity to the corresponding human gene calculated
from the
paired alignment using FASTA 3.1t02 [March,1998]~. 'The methods are described
in
more detail by: Altschul, Stephen F. Thomas L. Madden, Alejandro A. SchaiTer,
Jinghui Zhang, Zheng Zhang, Webb Miller and David J. Lipman (1997), "Gapped
BLAST and PSI-BLAST: a new generation of protein database search programs,
Nucleic Acids Res. 25:3389-3402 and W.R. Pearson & D.J. Lipman Proc. Natl.
Acad. Sci. (1988) 85:2444-2448 Fig. 1 shows the sequence names from the
databases and also the SEQ ID numbers for the sliding clamp loader 1, the
sliding
clamp loader 2, the sliding clamp, the coupling protein and the elongation
protein 1,
the values in parentheses each representing the percentage identity per number
of
amino acids. In the case of the elongation protein 2 the values refer to the
percentage
sequence identity relative to the Archaeoglobus fulgidu.s sequence. In this
case the
sequence names from the databases and their SEQ ID NO are also shown.
Fig. 2:
Fig. 2 shows protein sequences, paired alignments and multiple alignments from
eubacteria of the replication apparatus. The annotation' denotes the
percentage
CA 02338185 2001-02-05
-38-
identity to the corresponding gene from E. coli calculated from the paired
alignment
(see attachment) using BLASTP 2Ø4 [Feb-24-1998]#. The method is described in
more detail in: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller and David J. Lipman (1997), gapped
BLAST and PSI-BLAST: a new generation of protein database search programs,
Nucleic Acids Res. 25:3389-3402.
Fig. 3:
Fig. 3 shows a sketch of a possible form of the inventive thermostable ira
vitro
complex in which the sliding clamp binds to the elongation protein by means of
a
coupling protein.
Fig. 4:
Fig. 4 shows alignments of two conserved regions of the sliding clamp protein
as
well as the consensus sequences derived therefrom. The following genes are
shown:
human PCNA (from SEQ ID NO:11) and the corresponding sequences from
Archaeoglobus firlgidus (from SEQ ID N0:12), from tl~TethanococcTrs janashii
(from
SEQ ID N0:13), from Pyrococcus horikoschii (from SEQ ID N0:14) and from
Methanococcus thermoautothrophicus (from SEQ ID NO:15).
Fig. 5:
Fig. 5 shows an alignment of a conserved region of the coupling protein and
the
consensus sequences derived therefrom The following genes are shown: PfuORF2,
DPD2 HUMAN, AF 1790 and MJ0702. The SEQ ID numbers can be taken from
fig. 1.
Fig. 6:
Fig. 6 shows an alignment of a conserved region of the coupling subunit and
the
consensus sequences derived therefrom. The following genes are shown
CA 02338185 2001-02-05
-39-
AC11 HUMAN, AF2060, MTH0241, PHBN012 and MJ1422. The SEQ ID
numbers can be taken from fig. 1.
Fig. 7:
Fig. 7 shows an alignment of a conserved region of the sliding clamp loader 2
and
the consensus sequences derived therefrom. The following genes are shown
AC I 5 HUMAN, MJ0884, AF I I 95, MTH0240 and MTH0240. The SEQ ID
numbers can be taken from fig. 1.
Fig. 8:
Fig. 8 shows an alignment of a conserved region of the elongation protein 1
and the
consensus sequences derived therefrom. The following genes are shown:
DPOD HUMAN, MJ0885, MTH 1208, PHBT047 and DPOL ARCFU. The SEQ
ID numbers can be taken from fig. 1.
Fig. 9:
Fig. 9 shows an alignment of a conserved region of the elongation protein 2
and the
consensus sequences derived therefrom. The following genes are shown: AF1722,
MJ1630, PfuORF3, MTH1536 and PHBN021. The SF?Q ID numbers can be taken
from fig. 1.
Fig. 10:
Fig. 10 shows an alignment of a conserved region of the elongation protein
from
eubacteria and the consensus sequences derived therefrom. The following genes
are
shown: DP3A ECOLI:DNAPoI III, alpha subunit, Escherichia coli, BB0579: DNA
Pol III, alpha subunit, Borrelia bTrrgdorferi, DP3A HELPY: DNA Pol III, alpha
subunit, Helicobacter pylori AA50: Aquifex aeolicus, section 50 and
DP3A SALTY: DNA Pol III, alpha subunit, Salmonella typhimurium).
CA 02338185 2001-02-05
-40-
Fig. 11:
Fig. 11 shows an alignment of a conserved region of the sliding clamp from
eubacteria and the consensus sequences derived therefrom. The following genes
are
shown: AAPOL3B, DP3B ECOI,I, S.TYPHIM, DP3B PROMI, DP3B PSEPU
and DP3B_STRCO (AAPOL3B: Aqz~ifex aeolicns section 93: DP3B ECOLI: DNA
Pol III, beta chain, ~scherichia coli S.TYPHIM: DNA Pol III, beta chain,
Salmonella typhimtrrizem P3B PROMI: DNA Pol III, beta chain Proteres mirabilis
DP3B PSEPU: DNA Pol III, beta chain, Psercdomonas putidcr DP3B STRCO:
DNA Pol III, beta chain, Streptomyces coelicolor).
Fig. 12:
Fig. 12 shows a multiple alignment of the sliding clamp protein sequences for
generating the HMM.
Fig. 13:
Fig. 13 shows a multiple alignment of the eubacterial sliding clamp protein
sequences
to generate the HMM (AAPOL3B: Aquifex aeolicus section 93: DP3B ECOLI:
DNA Pol III, beta chain, F,scherichia coli S. TYPHIM: DNA Pol III, beta chain,
Salomonella typhimitrium P3B PROMI: DNA Pol III, beta chain, Protezrs
mirabili.s
DP3B PSEPU: DNA Pol III, beta chain, Pseudomonas putida DP3B STRCO:
DNA Pol III, beta chain, Streptomyces coelicolor.
Fig. 14:
Fig. 14 shows a multiple alignment of the sliding clamp loader 1 protein
sequences
for generating the HMM.
CA 02338185 2001-02-05
-41 -
Fig. 15:
Fig. 15 shows a multiple alignment of the sliding clamp loader 2 protein
sequences
for generating the HMM.
Fig. 16:
Fig. 16 shows a multiple alignment of the protein sequences of the coupling
proteins
for generating the HMM.
Fig. 17:
Fig. 17 shows a multiple alignment of the sequences of the elongation proteins
1 for
generating the hidden Markov model.
Fig. 18:
Fig. 18 shows a multiple alignment of the sequences of the elongation proteins
2 for
generating the hidden Markov model.
Fig. 19:
Fig. 19 shows a multiple alignment of the sequences of the eubacterial
elongation
proteins for generating the I-PVIM. The following sequences are shown:
DP3A ECOLI: DNA Pol III, alpha subunit, Esche~°ichia coli, BB0579:
DNA Pol
III, alpha subunit, Borrelia burgdorferi, DP3A HELPY: DNA Pol III, alpha
subunit, Helicobacter pylori AA50: Aguifex aeoliczrs, section 50 and
DP3A SALTY: DNA Pol III, alpha subunit, Salmonella typhiniuriz~m.
CA 02338185 2001-02-05
- 42 -
Fig. Z0: (example 2)
Fig. 20 shows a chromatographic analysis of recombinant Archaeoglohus
firlgidrrs
PCNA (AF0335):
Recombinant Archaeoglohr~.s.fulgidrrs PCNA (AF 0335) is present as a trimer
under
native conditions. Fig. 20A shows proteins with a His tag (histidine tag) in
fractions
15 (lane 1), 17 (lane 2), 19 (lane 3), 21 (lane 4), 23 (lane 5), 25 (lane 6)
of the
chromatography carried out without urea. Fig. 20B shows proteins with a His
tag in
fractions 10 (lane 1), 11 (lane 2), 12 (lane3), 13 (lane 4), 14 (lane S), 15
(lane 6), 16
(lane 7), 17 (lane 8) of the denaturing chromatography carried out in the
presence of
urea.
Fig. 21: (example 3)
Fig. 21 shows the result of a PCR using an elongation protein as a component
of an
inventive thermostable in vitro complex:
I pl (lane 4), 2.5 pl (lane 5) and 5 pl (lane 6) Pyrococcu.s horilroshii DNA
polymerase (PH1947; crude extract see fig. 25) were each used individually in
standard PCR reactions for an activity comparison with 1 unit Taq polymerase
(lane
2) and 1 pl Archaeoglohrrs.firlgidus DNA polymerase (AF 0497) crude extract
(see.
fig. 25) (lane 3); lane 1 shows a DNA size marker (New England Biolabs;
mixture of
1 kb DNA ladder and 100 by DNA ladder).
Fig. 22: (example 4)
Fig. 22 shows the result of a PCR using an elongation protein as a component
of an
inventive thermostable in vitro complex.
Samples of the PCR using Archaeoglobrrs firlgidus DNA polymerase AF 0497 were
taken after various numbers of cycles (Z) and separated on a 1 % agarose gel
in 1 x
TAE bufl"er (40 mM Tris acetate; 20 mM sodium acetate; 10 mM EDTA; pH 7.2) at
v/cm. Lane 2: 16Z; lane 3: 21Z; lane 4: 26Z; lane 5: 28Z; lane 6: 30Z; lane 7:
32Z; lane 8: 34Z; lane 9: 36Z; lane 10: 38Z; lane 11: 40Z; lane 1 shows a DNA
size
marker (New England Biolabs; mixture of 1 kb DNA ladder and 100 by DNA
ladder). The upper section shows reaction products of Taq polymerase; the
lower
CA 02338185 2001-02-05
- 43 -
section shows reaction products of Archaeoglobrrs firlgidrrs DNA polymerase AF
0497.
Fig. 23: (example 5)
Fig. 23 shows a comparison of the activity of an elongation protein with and
without
a sliding clamp protein. Sample I represents an enzyme-free mixture. Samples 2-
12
contained additionally 3 ~l each of a 1:1000 dilution of a fraction of
recombinant
Archaeoglobrrs fulgidus DNA polymerase (initial concentration 7.5 ~g/pl).
Samples
3-7 and 8-12 additionally contained 0.5; l; 2; 4 and 8 pl of a fraction of
recombinant
sliding clamp protein from Archaeoglobus _ fzrlgidus 1?CNA. The intensities
were
evaluated using AIDA: intensity of background lane l: 46.4; lane 2= 258.5;
lane 3=
164.4; lane 4= 122.8; lane 5= 162.1; lane 6= 297.4; lane' 7= 359.5
Fig. 24: (example 6)
Fig. 24 shows the result of purifying a sliding clamp protein: lane 1 shows a
molecular weight standard (BIO RAD cat. No. 161-0317). For lane 2 500 ~l
bacteria were sedimented directly before induction, they were treated and
applied
according to the manufacturer's instructions for operating NuPage gels (NOVEX;
fig. 25). Lane 3 shows the same amount of bacteria 16 hours after the elution.
Lanes
4 and 5 each show 8 ~1 of the two eluates of the Ni-NTA agarose column after
dialysis. Highly purified fractions of the sliding clamp protein of the
organism
Archaeoglobzrs .fulgidus were obtained by purification over Ni-NTA agarose
(Qiagen) (see lanes 4 and 5).
Fig. 25: (example 7)
Fig. 25 shows the expression and purification of A~°chaeoglobzrs
fulgidus DNA
polymerase (see also example 7):
Lane 1 shows a molecular weight standard (BIO RAD cat. No. 161-0317). For lane
2 500 pl bacteria were sedimented directly before induction, they were treated
and
applied according to the manufacturer's instructions for operating NuPage
gels.
Lane 3 shows the same amount of bacteria 16 hours after the elution. Lanes 4
and 5
CA 02338185 2001-02-05
-44-
each show 8 pl of the two eluates of the Ni-NTA agarose column after dialysis.
Lane
6 shows 8 ml of a dialysed crude extract. Lanes 4 and 5 show highly purified
fractions of the Archaeoglobrr.s . frrlgidTrs DNA polymerase which are
obtained by
purification over Ni-NTA agarose.
Fig. 26: (example 8)
Fig. 26 shows the results of using an inventive ire vitro complex in the PCR.
Lane 1
shows a PCR reaction without using a sliding clamp whereas lane 2 shows the
result
of a PCR reaction using a sliding clamp protein.
Fig. 27 (example 9)
Fig. 27 shows the results of a yeast two hybrid experiment referred to herein
as the
Y2H experiment in which cells that carry the empty pGAD424 vector (Clontech,
Palo Alto, USA) are placed in row A such that the transcription activation
domain is
expressed and cells which carry the pGAD424 vector from which the Sacharomyces
cereve.siae gene CDC48 is expressed as a fusion protein with the transcription
activation domain are placed in row B and row C contains cells which carry the
pGAD424 vector from which the sliding clamp gene fram Archaeoglobus firlgidus
is
expressed as a fusion protein with the transcription activation domain, row D
contains no cells and row E contains cells which carry the pGAD424 vector from
which the elongation protein gene from Archaeoglobus fi~lgidus is expressed as
a
fusion protein with the transcription activation domain.
Column 1 is provided with cells which carry the empty pGBT9 vector (Clontech,
Palo Alto, USA) such that the DNA binding domain is expressed, column 2 is
provided with cells which carry the pGBT9 vector from which the Saccharomyces
cerevisiae gene UFD3 is expressed as a fusion protein with the DNA binding
domain, column 3 is provided with cells which carry the pGBT9 vector from
which
the sliding clamp protein from Archaeoglobus fulgidus is expressed as a fusion
protein with the DNA binding domain, column 4 is provided with cells which
carry
the pGBT9 vector from which the coupling protein from Archaeoglobus fi~lgidus
is
expressed as a fusion protein with a DNA binding domain and column 5 is
provided
CA 02338185 2001-02-05
- 45 -
with cells which carry the pGBT9 vector from which the elongation protein from
Archaeoglobrr.s.firlgidns is expressed as a fusion protein with the DNA
binding
domain.
Fig. 28: (example 10)
Fig. 28 shows the PCR amplification result of the human collagen gene using
the
inventive thermostable in vitro complex. The expected amplificate has a size
of about
I kb in both cases. Lane 1 shows a molecular weight marker, lane 2 shows the
result
of the amplification using an inventive elongation protein without a sliding
clamp and
lane 3 shows the result of the amplification using the inventive thermostable
in vitro
complex.
Examples:
The invention is described in more detail by the following examples but is not
limited
thereto.
Example 1:
DNA is purified from the organism Archaeoglobzts fulgid~r.s (DSM No. 4304) by
known methods. The organisms were cultured by the DSM ("Deutsche Sammlung
fiir Mikroorganismen"). In order to clone the appropriate genes (sliding clamp
loader
1 and 2, sliding clamp, elongation proteins 1 and 2, coupling subunit) into
the
expression vectors pTrc99 and pQE30, primers were developed for each gene
which
span the complete open reading frame including the stop codon. The primer
sequences only additionally contain the start codon for cloning into pTRC99.
The
corresponding primers for cloning into pQE30 contain the nucleotides which
immediately follow the start codon as the first gene-specific sequences.
Restriction
ends are added to the primers which facilitate the directed cloning into the
expression vector. PCR reactions (about 35 cycles) are carried out using about
200
ng total genomic DNA at the appropriate annealing temperatures and the
resulting
products are purified. After purification the products are treated with
restriction
CA 02338185 2001-02-05
-46-
enzymes and purified over an agarose gel in order to prepare them for
ligation. The
expression vector is linearized by means of restriction enzymes, purified and
diluted
in such a manner that it is ready for ligation with the arnplificates of the
genes of the
inventive irt vitro complex from the above PCR. The ligation is set up and
after
incubation an aliquot is transformed into one of the E. coli strains
INValphaF'
(Invitrogen) or XL,1 blue or M15 [prep4). At least 3 positive colonies are
picked
from each gene, plasmid DNA is prepared and the inserts are checked for
completeness and correctness by means of DNA sequencing. Correct clones are
selected and again placed on agar plates for isolatian (ampicillin; ampicillin
and
kanamycin). Colonies are picked and overnight cultures are prepared. An
aliquot
(500 pl) of the overnight culture is added to a one to five litre culture of
LB
(ampicillin: 80 mg/1 or additionally 25 pg/ml kanamycin for M15[prep4)
strains. The
cultures grow up to an OD~~" of 0.8 at 37°C. IPTG (125 mg/1) is added
for
induction. These cultures now grow for a further 4-21 hours. The cultures are
centrifuged and starting from the recombinant proteins expressed by the vector
pQE30, are extracted and purified according to protocol 8 and 11 from the
QIAexpressionist (third edition, QIAGEN). In alternative purification
protocols, the
pellets are taken up in a buffer (buffer A: 50 mM Tris-HCI pH 7.9, 50 mM
dextrose,
I mM EDTA). After centrifugation the cells are taken up again but buffer A now
additionally contains lysozyme (4 mg/ml). After incubation ( 15 min) the same
volume of buffer B is added (B: 10 mM Tris-HCl (p:H 7.9), 50 mM KCI, 1 mM
EDTA, 1 mM PMSF, 0.5 % Tween 20, 0.5 % Nonidet P40) and lysed by incubating
at 75°C for one hour. After centrifugation the supernatant is removed
and the
overexpressed proteins are precipitated by means of (NH.,)~SOa. The pellets
are
pooled after centrifugation and the proteins are resuspended in buffer A. The
resuspended proteins are dialysed against storage buffer (50 mM Tris-HCI, (pH
7.9),
50 mM KCI, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 50 % glycerol) and
subsequently stored at +4°C to -70°C.
Reactions are set up as follows in order to test the activity of the proteins.
Aliquots
of the proteins are combined in different configurations and molarities;
sliding clamp
loader I, 2 with the sliding clamp, coupling subunit and elongation protein 1,
or
sliding clamp loader 1, 2 with the sliding clamp with and without the coupling
subunit
and elongation proteins 2 or sliding clamp and elongation protein 1 or 2 and
finally
only elongation protein 1 or 2; the above-mentioned storage buffer served as
the
CA 02338185 2001-02-05
-47-
buffer. DNA polymerization activity is measured by the incorporation of
(methyl 3H)
TTP into trichloric acid-insoluble material or by incorporation of digoxigenin
dUTP
into unlabelled DNA double-strand regions with free internal 3' ends (lshino,
Y.,
Iwasaki, H., Fukui, H., Mineno, J., Kato, L, & Schinigawa, H. (1992) Biochemie
74,
131-136). In order to determine the processivity the above-mentioned protein
mixtures are used in primer elongation experiments. An M13 single-stranded
template is added to 10 mM Tris-HCI (pH 9.4) and heated (92°C) and
cooled (room
temperature) together with a universal primer (New England Biolabs) (5'-FITC
labelled). Serial dilutions of the thus generated template primer mixtures are
added to
a reaction composed of nucleotides (about 200 pM to 1 mM), reaction buffer
(final
concentration: 50 mM KCI, 10 mM Tris-HCl (pH 8.3), 1.5-5 mM MgCh, ATP (0
mM - 200 mM) and protein-stabilizing agents and incubated for 10 minutes at
37°C,
52°C, 62°C, 68°C, 74°C and 78°C. An aliquot
is loaded onto an automated
sequencer for analysis (e.g. Alf, Pharmacia Biotech). Alternatively the
increase of
processivity can be analysed qualitatively according to Maga G., Jonssom Z.O.,
Stucki M., Spadari S. and Hiibscher U. (J. Mol. Biol. 1999; 285: 259-267, Dual
Mode of Interaction of DNA polymerase and Proliferating Cell Nuclear Antigen
in
Primer Binding and DNA synthesis) by detecting a stimulation of the
incorporation of
nucleotides into double-stranded regions with free internal 3' ends under
suitable
reaction conditions (see fig. 23). The above-mentioned protein mixtures also
serve to
measure fidelity and exonuclease activity using the methods described in
Kohler et al.
(Proc. Natl. Acad. Sci. USA 88:7958-7962 (1991) or Chase et al. (J. Biol.
Chem.,
249:4545-4552 (1972). The protein mixtures are also used in the PCR (fig. 26,
Methods in Molecular Biology, vol. 15 Humana Press Totowa, New Jersey, 1993,
edited by Bruce A. White).
Example 2: Trimerization of PCNA
In the following experiments it is shown that recombinant
A~chaeoglobus_fulgidus
PCNA protein (AF 0335) is present as a trimer under native conditions and can
thus
adopt a suitable structure for clamp formation.
For the experiment shown in fig. 20A, 350 pl Ni-NTA agarose eluate of PCNA
(see
fig. 24) and 150 pl of a crude DNA polymerase fraction (see fig. 25) were made
up
CA 02338185 2001-02-05
-48-
to 1 ml with storage buffer without glycerol (see fig. 25) and the proteins
were
separated according to their molecular weight in the same buffer on a Superdex
200
HR (Pharmacia) FPLC column according to the manufacturer's instructions. I ml
fractions were collected during the entire run. For the experiment shown in
fig. ZOB,
the same amounts of the same protein fractions were made up to 1 ml in storage
buffer without glycerol containing 8 M urea, denatured for 10 minutes at
95°C and
subsequently the proteins were separated according to molecular weight in the
same
buffer as shown for fig. 20A. 8 gl of each fraction was subsequently separated
on a
NuPage Bis-Tris gel (NOVEX; see fig. 25) and blotted by means of a blot module
(NOVEX) onto a nitrocellulose membrane according to the manufacturer's
instructions. Proteins with a His tag were detected according to the
manufacturer's
instructions using the RGS His antibody (QIAGEN) and the DIG luminescent
detection kit for nucleic acids (Boehringer Mannheim). Fig. 20 A shows
proteins
with a His tag in the fractions 15 (lane 1), 17 (lane 2), 19 (lane 3), 21
(lane 4), 23
(lane 5), 25 (lane 6) in the chromatography which was carried out without
urea. Fig.
20 B shows proteins with a His tag in the fractions 10 (lane 1 ), 11 (lane 2),
12 (lane
3), 13 (lane 4), 14 (lane 5), 15 (lane 6), 16 (lane 7), 17 (lane 8) in the
chromatography which was carried out in the presence of urea. Archaeoglobrrs
firlgidrrs DNA polymerase (AF0497) has a calculated molecular weight of Mr =
90
kDa. Archaeoglobus.firlgidzrs PCNA (AF0335) has a calculated molecular weight
of
Mr = 27 kDa. IfArchaeoglobrrs,fulgidrrs PCNA (AF 335) is present as a
homotrimer
like the homologous protein from eukaryotes, the native factor therefore has a
theoretical molecular weight of Mr = 81 kDa. The results shown in fig. 20A
confirm
this assumption according to which native PCNA has a similar molecular weight
to
the DNA polymerase for the recombinant protein: both proteins elute under
native
conditions in the same fractions from the gel filtration column (fig. 20A,
lanes I-3).
Most of the PCNA elutes somewhat later than the DNA polymerase peak and
correlates with the somewhat smaller size of the postulated trimer (81 kDa
compared
to 90 kDa). The data shown in fig. 20B prove that this observation is based on
an
oligomerization of PCNA since under denaturing conditions which do not allow
protein/protein interactions, PCNA elutes considerably later from the column
than
the DNA polymerase (fig. 20B, lanes 1-7) which corresponds to the lower
molecular
weight of the monomer.
CA 02338185 2001-02-05
-49-
Example 3: Isolation, preparation and use of an elongation protein (Pyrocoecus
horikoshii DNA polymerase (PH 1947)) to form the inventive in vitro complex
The elongation protein from Pyrococcus horikoshii (Pyrococcus horikoshii-DNA
polymer (PH1947)) was used in PCR reactions and leads to an efficient
amplification
of a specific DNA product.
1 (lane 4), 2.5 (lane 5) and 5 pl (lane 6) Pyrococcu.s horikoshii DNA
polymerase
(PH1947; crude extract see fig. 25) were used individually in standard PCR
reactions
to compare the activity with 1 unit Taq polymerase (lane 2) and 1 pl
Archavoglobrrs
.frrlgidus DNA polymerase (AF497) crude extract (see fig. 25) (lane 3); lane 1
shows
a DNA size marker (New England Biolabs; mixture of 1 kb DNA molecular weight
size marker and 100 by DNA molecular weight size marker). In addition to the
enzyme, each reaction contained 5 ng template DNA (421 by Rsa I fragment with
adapters cloned into PCR 2.1; (Kaiser C., v. Stein (J., Laux G., Hoffmann M.,
Electrophoresis 1999; 20: 261-268, Functional Genomics in Cancer Research:
Identification of target genes of the Epstein-Barr virus nuclear antigen 2 by
subtractive cDNA cloning and high throughput differential screening using high-
density agarose gel); 0.2 mM each of dATP, dTTP, dC',TP and dGTP; 1.5 pM each
of the specific adapter primers (Kaiser et al., (1999)); 50 mM KCI; 2 mM MgCh,
10
mM Tris HCl (pH 8.3 for Taq reactions; pH 7.5 for Archaeoglobrr.s firlgidus
and
Pyrococcrrs horikoshii polymerase reactions) in a total volume of 50 pl per
reaction.
The samples were subjected to 40 cycles comprising 30 s at 95°C; 30 s
at 55°C and
120 s at 68°C. Subsequently 5 ul of the reactions was removed and
separated at 10
V/cm on a 1 % agarose gel in 1 x TAE buffer (40 mM Tris acetate; 20 mM sodium
acetate; 10 mM EDTA; pH 7.2).
Example 4: Use of an elongation protein
The Archaeoglobus fulgidus DNA polymerase AF 0497 can generate PCR products
as efficiently as Taq polymerase. 1 unit Taq polymerase and 1 pl Archaeoglobus
firlgidus DNA polymerase AF 0497 crude extract (see fig. 25) were used
individually
in standard PCR reactions to compare the activity. In addition to the enzyme
each
reaction contained 20 ng M 13 MP 18 ssDNA; 0.2 mM each of dATP, dTTP, dCTP
CA 02338185 2001-02-05
-50-
and dGTP; 1.5 ~M DNA of each of the primers with the following nucleotide
sequences:
GGATTGACCGTAATGGGATAGGTTACGTT (SEQ ID NO: 47) or
AGCGGATAACAATTTCACACAGGAAACAG (SEQ ID NO: 48)
in 50 mM KCI; 2 mM MgCh; 10 mM Tris-HCl (pH 8.3 for Taq reactions, pH 7.5 for
Archaeoglobns fi~lgidr~s polymerase reactions) in a total volume of 50 pl per
reaction. The samples ran through various numbers of cycles comprising 30 s at
95°C; 30 s at 59°C and 60 s at 68°C. After different
numbers of cycles (Z), 5 pl of
the mixtures was removed and separated at 10 V/cm on a 1 % agarose gel in 2 x
TAE buffer (40 mM Tr~is acetate, 20 mM sodium acetate; 10 mM EDTA; pH 7.2)
(see fig. 22).
Example 5: Preparation and use of an inventive thermostable in vitro complex
The following components of an inventive irr vitro complex were present among
others in a final volume of 50 pl: 10 mM Tris-HCI, pH 7.5; 50 mM KCI; 2 mM
MgCh; 10 pg BSA (can also be omitted); 0.5 mM digoxigenin dUTP (DIG-dUTP,
Boehringer Mannheim); 40 pM; 0.5 pg poly dA/40 rrNl oligodT (20mer) hybrid.
Sample I without an elongation protein. Samples 2-12 each additionally
contained
3 ~l of a 1:1000 dilution of a fraction of recombinant
Archaeoglobus,firlgidz~s DNA
polymerase (elongation protein). Samples 3-7 as well as 8-12 additionally
contained
0. S; I ; 2; 4 and 8 girl of a fraction of recombinant sliding clamp protein
from
Archaeoglobzrs fulgidns.
The samples were incubated for 30 minutes at 68°C and nucleic
acids were
subsequently precipitated with 3 parts by volume ethanol / 3 M sodium acetate
pH
5.2 (30/1). The precipitate was resuspended in 20 pl 100 mM Tris-HCl (pH 7.9)
and
lrl aliquots were added dropwise to individual wells of a 96-well silent
screen
plate containing nylon 66 Biodyne B 0.45 pm pore (Nalge Nunc) and the nucleic
acids were fixed to the membranes for 10 minutes at 70°C. The
incorporated
digoxigenin dUTP (Boehringer Mannheim) was detected by means of the DIG
luminescent detection kit for nucleic acids (Boehringer Mannheim). In order to
detect the chemiluminescence, an X-ray film was subsequently placed on the
membrane for 30 s. PCNA considerably stimulated the incorporation of DIG-dUTP
CA 02338185 2001-02-05
-51 -
by the DNA polymerase (compare lanes 3-7 with lane 2). The PCNA fraction used
has no endogenous polymerase activity (lanes 8-12).
Example 6: Amplification, cloning, expression and purification of a sliding
clamp protein from Archaeoglobus fulgidus
After amplification of the inventive sliding clamp protein gene from
Archaeoglobrrs
frrlgidrr.s, the gene was cloned into the expression vectors pTrc99 and pQE30.
The
expression, purification and gel-electrophoretic separation of the
Archaeoglobirs
.firlgidrrs sliding clamp protein (PCNA (AF 0335)) was carried out as shown
for the
elongation protein (the DNA polymerase AF0497) in fig. 25.
Example 7: Preparation of an elongation protein
Expression and purification of the Archaeoglobu.s firlgidrr.s DNA polymerase:
pQE30 plasmid DNA (QIAGEN) with a gene inserted in the correct reading frame
for the elongation protein, the Archac.~oglobrr.s firlgiclrrs DNA polymerase
AF 0497,
was transformed in competent E. coli M 15 [prep4] according to the
instructions
from the QIAexprssionist (third edition: QIAGEN). Transfer to 1 1 culture
medium
and induction of protein expression were also carried out according to the
schemes
given in these instructions. After 16 hours induction time, the bacteria were
sedimented for 10 minutes at 5000 g. The QIAexpressionist procedure (third
edition:
QIAGEN; protocol 8, protocol 11 ) was used to obtain highly purified
fractions. Only
the elution of the bound proteins was carried out with 2 x 2 ml elution buffer
and not
as described in these instructions with 4 x 0.5 ml elution buffer. In order to
obtain
crude extracts of recombinant proteins, the bacterial sediments were
alternatively
washed with 100 ml buffer A (50 mM Tris-HCI, pH 7.9; 50 mM glucose; 1 mM
EDTA) and, after centrifuging again, they were resuspended in 50 ml buffer A
containing 4 mg/ml lysozyme. After 15 minutes at room temperature, 50 ml lysis
buffer (10 mM Tris-HCl pH 7.9; 50 mM KCI; 1 mM EDTA; 0.5 % Tween 20; 0.5
IGPAL) was added and E. coli proteins were denatured by incubating for 60 min
at
75°C in a water bath. After subsequent centrifugation for 15 minutes at
27,000 g, the
supernatant was precipitated by slow addition with permanent stirring of 40 mg
crystalline ammonium sulphate per ml extract. Precipitated proteins were
sedimented
CA 02338185 2001-02-05
-52-
for 10 minutes at 27,000 g and the sediment was resuspended in 20 ml bui~'er
A.
These crude extracts as well as the elution fractions from the Ni NTA agarose
were
each dialysed twice for 8 hours against at least 50 parts per volume storage
buffer
(buffer ( I 0 mM Tris-HCl pH 7.9; 50 mM KCI; 1 mM EDTA; 50 % glycerol; 1 mM
dithiothreitol; 0.5 mM phenylmethylsulfonyl fluoride) and stored at -
20°C. In order
to analyse the protein composition of the fractions obtained, aliduots thereof
were
separated electrophoretically on NuPage'''f 10 % Bis-Tris gels (NOVEX)
according
to the manufacturer's instructions and the proteins obtained were stained with
Coomassie brilliant blue. Lane 1 shows a molecular weight standard (BIO RAD
cat.
No. 161-0317). For lane 2, 500 pl bacteria were sedimented directly before
induction and treated and applied according to the instructions for rurming
NuPage
gels. Lane 3 shows the same amount of bacteria 16 hours after the elution.
Lanes 4
and S each show 8 pl of the two eluates of the Ni-NTA agarose column after
dialysis. Lane 6 shows 8 ml of a dialysed crude extract. Highly purified
fractions of
the Archaeoglobus,firlgidus DNA polymerase are obtained by purification over
Ni-
NTA agarose (lanes 4 and 5). However, this enzyme is already the dominant
polypeptide in the crude extract.
Example 8: Use of an inventive in vitro complex in the PCR
The use of Archaeoglohn.s fiilgidus PCNA (AF 0335) in PCR reactions containing
Archaeoglobrss fulgidus DNA polymerase (AF 0497) led to a more efficient
amplification of a specific DNA product compared to a PCR reaction without
PCNA. Evaluation of the amplified amounts of DNA according to AIDA (Advanced
Image Data Analyzer, software version AIDA 2.1, Raytest Company) shows a
background of 94, a value of 104.9 for lane 1 and a value of 228.4 for lane 2.
If the
background is subtracted from the values for lanes 1 and 2, it follows that
lane 2
shows the result of a 12.3-fold processive stimulation by PCNA in the
reaction.
0.3 pl Archaeoglobus fulgid~rs DNA polymerase (7.5 ug/pl protein concentration
of
AF497; crude extract see fig. 25) was used individually in PCR reactions to
analyse
the stimulation of the PCR activity by 0 pl and 0.01 pl Archaeoglobus
_fulgidzis
PCNA (Ni-NTA eluate; fig. 24). In addition to the enzyme and 0.05 pl (2.8 pg)
of
the sliding clamp protein from Archaeoglobus frrlgidr~s (homologous to
proliferating
CA 02338185 2001-02-05
-53-
cell nuclear antigen PCNA), each reaction contained a non-purified PCNA gene
fragment amplified by means of PCR (PCR reaction for the specific
amplification of
the PCNA fragment: 0.5 pl Archaeoglob~rs , firlgidze.s polymerase; 0.2 mM each
of
dATP, dTTP, dCTP and dGTP; 1.5 pM of each of the specific primers (5'-ACG
CGC GGA TCC ATA GAC GTC ATA ATG AC(: GG-3' (SEQ ID N0:49);
5'-TAC GGG GTA CCC GAG CCA AAA TTG GGT AAA G-3' (SEQ ID N0:50);
50 mM KCI; 2 mM MgCh; 10 mM Tris-HCI (pH 7.5) as well as 50 ng of a pQE30
plasmid which carries the coding sequences of Archaeoglobr~s _fi~lgidt~s PCNA
inserted into the BamHI and Kpn I restriction sites in a total volume of 50 pl
per
reaction with a cycle number of 40 comprising 30 s at 95°C; 30 s at
61°C; 240 s at
68°C; 0.2 mM each of dATP, dTTP, dCTP and dGTP; 1.5 pM of each of the
specific primers (S'-ACG CGC GGA TCC ATA GAC GTC ATA ATG ACC GG-3'
(SEQ ID NO: 51 ) and 5'-TAC GGG GTA CCC GAG CCA AAA TTG GGT AAA
G-3' (SEQ ID N0:52); 50 mM KCI; 2 mM MgCh; 10 mM Tris HCl pH 7.5 in a
total volume of 50 pl per reaction. The samples ran through 40 cycles
comprising 30
s at 95°C; 120 s at 61°C; 240 s at 68°C. Subsequently 5
pl of the mixtures was
removed and separated at 10 V/cm on a 1 % agarose gel in I x TAE buffer (40 mM
Tris acetate; 20 mM sodium acetate; 10 mM EDTA; pH 7.2).
Example 9: Y2H experiments
Interactions of proteins of the inventive complex from
.4rchaeoglobus.frelgidus were
demonstrated with a Y2H system. The coding regions of genes from
Archaeoglobtrs
fi~lgidus, whose gene products were used in the inventive thermostable in
vitro
complex were amplified by means of PCR, cloned into the vectors pGBT9
(vertical
columns of fig. 27) and pGAD424 (horizontal rows of fig. 27) and expressed as
hybrid proteins by gap repair in yeast PJ69-4a (for pGAD424) and PJ69-4alpha
(for
pGBT9). A positive control was also amplified by means of PCR, cloned into the
vectors pGBT9 (see also vertical columns of fig. 27) and pGAD424 (see also
horizontal rows of fig. 27) and expressed as hybrid proteins by gap repair in
the
yeast strain PJ69-4a (for pGAD424) and PJ69-4alpha (for pGBT9). Diploid cells
which contained both vectors were generated by pairing according to the raster
shown in fig. 27. Expression of three independent reporters (HIS3, ADE2 and
MEL 1 ) was measured. Fig. 27 shows the growth on medium without histidine and
adenine. The cells which grow in this experiment are those which carry both
vectors
CA 02338185 2001-02-05
-54-
and in which additionally the expression products of both these vectors bind
to one
another. Transcription of the reporter genes is initiated as a result of the
binding of
the expression products such that histidine and adenine auxotrophy is
abolished and
these cells are able to grow in the said medium.
All the clones that were positive here were also positive with respect to the
expression of the MEL 1 gene. The yeast two hybrid (Y2H) system was used
according to the instructions of the Clontech Company (yeast protocols
handbook,
PT3024-1). Fig. 27 shows binding between the proteins of the positive control,
the
elongation protein and the sliding clamp protein, the sliding clamp protein
and the
sliding clamp protein and the coupling protein and the sliding clamp protein.
Example 10: Use of an inventive thermostable in vitro complex
The inventive thermostable ire vitro complex can be used very well for the
amplification of genomic DNA fragments. An effcient amplification occurs even
when using small amounts of a template or of an elongation protein. In example
10 a
section of the human collagen gene was amplified using the inventive
thermostable in
vitro complex and also using an elongation protein alone. The following were
used in
a total volume of 50 pl: 0.5 pl nucleotide mix (25 niM initial solution
comprising
each nucleotide A, C, G and T), 0.2 pl of each primer ( 100 pmol/pl initial
solution
"collagen forward" 5'-TAA AGG GTC ACC GTG GCT TC-3' (SEQ ID N0:53),
100 pmol/pl initial solution of "collagen reverse" 5'-CGA ACC ACA TTG GCA
TCA TC-3' (SEQ ID NO: 54), 0.8 pl DNA (200 ng/pl human genomic DNA from
Boehringer Mannheim), 5 pl 10 x PCR buffer (pH 7.5) (100 mM Tris-HCI, pH 7.5,
500 mM KCI, 15 mM MgCh), 1 pl elongation protein (AF1722, 7.5 pg/pl protein
concentration) and 8 pl sliding clamp protein (AF 0335, protein concentration
0.3
pglml) and subjected to the following cycle in a PCR machine: initially 5 min
at 95°
then 30 times 30 s at 95°C, 30 s at 59°C and finally 6 min at
68°C. Fig. 28 clearly
shows the advantage of using the inventive thermostable in vitro complex.
The features of the invention disclosed in the aforementioned description and
in the
claims can be important individually as well as in any combination for the
realization
of the invention in its various embodiments.
CA 02338185 2001-02-05
- 1 -
- SEQUENCE PROTOCOL
(1) GENERAL INFORMATION:
(i) APPLICANT:
(A) NAME: LION bioscience AG
(B) STREET: Im Neuenheimer Feld 517
(C) CITY: Heidelberg
(E) COUNTRY: DE
(F) POSTCODE: 69120
(ii) TITLE OF THE INVENTION: Ac~~essory complexes with
polymerase activity
(iii) NUMBER OF SEQUENCES: 54
(iv) COMPUTER-READABLE FORM:
(A) DATA CARRIER: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: PatentIn Release #1.0, Version #1.30 (EPO)
(2) INFORMATION FOR SEQ ID NO: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 340 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: both
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Homo Sapiens
(xi)SEQUENCE
DESCRIPTION:
SEQ
ID
NO:
1:
MetGluThr SerAla LeuLysGln GlnGluGln ProAlaAla ThrLys
1 5 10 15
IleArgAsn LeuPro TrpValGlu LysTyrArg ProGlnThr LeuAsn
20 25 30
AspLeuIle SerHis GlnAspIle LeuSerThr IleGlnLys PheIle
35 40 45
AsnGluAsp ArgLeu ProHisLeu LeuLeuTyr GlyProPro GlyThr
50 55 60
GlyLysThr SerThr IleLeuAla CysAlaLys GlnLeuTyr LysAsp
65 70 75 80
LysGluPhe GlySer MetValLeu GluLeuAsn AlaSerAsp AspArg
85 90 95
GlyIleAsp IleIle ArgGlyPro IleLeuSer PheAlaSer ThrArg
100 105 110
CA 02338185 2001-02-05
' 2 -
Thr Ile Phe Lys Lys Gly Phe Lys Leu Val Ile Leu Asp Glu Ala Asp
115 120 125
Ala Met Thr Gln Rsp Ala Gln Asn Ala Leu Arg Arg Val Ile Glu Lys
130 135 140
Phe Thr Glu Asn Thr Arg Phe Cys Leu Ile Cys Asn Tyr Leu Ser Lys
145 150 155 160
Ile Ile Pro Ala Leu Gln Ser Arg Cys Thr Arg Phe Arg Phe Gly Pro
165 170 175
Leu Thr Pro Glu Leu Met Val Pro Arg Leu Glu His. Val Val Glu Glu
180 185 190
Glu Lys Val Asp Ile Ser Glu Asp Gly Met Lys Ala Leu Val Thr Leu
195 200 205
Ser Ser Gly Asp Met Arg Arg Ala Leu Asn Ile Leu Gln Ser Thr Asn
210 215 220
Met Ala Phe Gly Lys Val Thr Glu Glu Thr Val Tyr Thr Cys Thr Gly
225 230 235 290
His Pro Leu Lys Ser Asp Ile Ala Asn Ile Leu Asp Trp Met Leu Asn
295 250 255
Gln Asp Phe Thr Thr Ala Tyr Arg Asn Ile Thr Glu Leu Lys Thr Leu
260 265 270
Lys Gly Leu Ala Leu His Asp Ile Leu Thr Glu Ile His Leu Phe Val
275 280 285
His Arg Val Asp Phe Pro Ser Ser Val Arg Ile His Leu Leu Thr Lys
290 295 300
Met Ala Asp Ile Glu Tyr Arg Leu Ser Val Gly Thr Asn Glu Lys Ile
305 310 315 320
Gln Leu Ser Ser Leu Ile Ala Ala Phe Gln Val Thr Arg Asp Leu Ile
325 330 335
Val Ala Glu Ala
340
(2) INFORMATION FOR SEQ ID NO: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 319 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeoglobus fulgidus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2:
CA 02338185 2001-02-05
- 3 -
a Met Glu Asn Phe Glu Ile Trp Val Glu Lys Tyr Arg Pro Arg Thr Leu
1 5 10 15
Asp Glu Val Val Gly Gln Asp Glu Val Ile Gln Arg Leu Lys Gly Tyr
20 25 30
Val Glu Arg Lys Asn Ile Pro His Leu Leu Phe Ser Gly Pro Pro Gly
35 40 95
Thr Gly Lys Thr Ala Thr Ala Ile Ala Leu Ala Arg Asp Leu Phe Gly
50 55 60
Glu Asn Trp Arg Asp Asn Phe Ile Glu Met Asn Ala Ser Asp Glu Arg
65 70 75 80
Gly Ile Asp Val Val Arg His Lys Ile Lys Glu Phe Ala Arg Thr Ala
85 90 95
Pro Ile Gly Gly Ala Pro Phe Lys Ile Ile Phe Leu Asp Glu Ala Asp
100 105 110
Ala Leu Thr Ala Asp Ala Gln Ala Ala Leu Arg Arg Thr Met Glu Met
115 120 125
Tyr Ser Lys Ser Cys Arg Phe Ile Leu Ser Cys Asn Tyr Val Ser Arg
130 135 140
Ile Ile Glu Pro Ile Gln Ser Arg Cys Ala Val Phe Arg Phe Lys Pro
145 150 155 160
Val Pro Lys Glu Ala Met Lys Lys Arg Leu Leu Glu Ile Cys Glu Lys
165 170 175
Glu Gly Val Lys Ile Thr Glu Asp Gly Leu Glu Ala Leu Ile Tyr Ile
180 185 190
Ser Gly Gly Asp Phe Arg Lys Ala Ile Asn Ala Leu Gln Gly Ala Ala
195 200 205
Ala Ile Gly Glu Val Val Asp Ala Asp Thr Ile Tyr Gln Ile Thr Ala
210 215 220
Thr Ala Arg Pro Glu Glu Met Thr Glu Leu Ile Gln Thr Ala Leu Lys
225 230 235 290
Gly Asn Phe Met Glu Ala Arg Glu Leu Leu Asp Arg Leu Met Val Glu
245 250 255
Tyr Gly Met Ser Gly Glu Asp Ile Val Ala Gln Leu Phe Arg Glu Ile
260 265 270
Ile Ser Met Pro Ile Lys Asp Ser Leu Lys Val Gln Leu Ile Asp Lys
275 280 285
Leu Gly Glu Val Asp Phe Arg Leu Thr Glu Gly Ala Asn Glu Arg Ile
290 295 300
Gln Leu Asp Ala Tyr Leu Ala Tyr Leu Ser Thr Leu Ala Lys Lys
305 310 315
CA 02338185 2001-02-05
- 4 -
(2) INFORMATION FOR SEQ ID NO: 3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1847 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanococcus jannaschii
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 3:
Met Val Ile Ile Met Glu Lys Pro Trp Val Glu Lys Tyr Arg Pro Lys
1 5 10 15
Thr Leu Asp Asp Ile Val Gly Gln Asp Glu Ile Val. Lys Arg Leu Lys
20 25 30
Lys Tyr Val Glu Lys Lys Ser Met Pro His Leu Leu Phe Ser Gly Pro
35 40 45
Pro Gly Val Gly Lys Cys Leu Thr Gly Asp Thr Lys Val Ile Val Asn
50 55 60
Gly Glu Ile Arg Glu Ile Gly Glu Val Ile Glu Glu. Ile Ser Asn Gly
65 70 75 80
Lys Phe Gly Val Thr Leu Thr Asn Asn Leu Lys Val Leu Gly Ile Asp
85 90 95
GluAspGly LysIleArg GluPheAsp ValGlnTyr ValTyrLys Asp
100 105 110
LysThrAsn ThrLeuIle LysIleLys ThrLysMet GlyArgGlu Leu
115 120 125
LysValThr ThrTyrHis ProLeuLeu IleAsnHis LysAsnGly Glu
130 135 140
IleLysTrp GluLysAla GluAsnLeu LysValGly AspLysLeu Ala
145 150 155 160
Thr Pro Arg Tyr Ile Leu Phe Asn Glu Ser Asp Tyr Asn Glu Glu Leu
165 170 175
Ala Glu Trp Leu Gly Tyr Phe Ile Gly Asp Gly His Ala Asp Lys Glu
180 185 190
Ser Asn Lys Ile Thr Phe Thr Asn Gly Asp Glu Lys Leu Arg Lys Arg
195 200 205
Phe Ala Glu Leu Thr Glu Lys Leu Phe Lys Asp Ala Lys Ile Lys Glu
210 215 220
Arg Ile His Lys Asp Arg Thr Pro Asp Ile Tyr Val Asn Ser Lys Glu
225 230 235 240
CA 02338185 2001-02-05
- 5 -
Ala ValGlu PheIleAsp LysLeuGly LeuArgGly LysLysA1a Asp
245 250 255
Lys ValArg IleProLys GluIleMet ArgSerAsp AlaLeuArg Ala
260 265 270
Phe LeuArg AlaTyrPhe AspCysAsp GlyGlyIle GluLysHis Ser
275 280 285
Ile ValLeu SerThrAla SerLysGlu MetAlaGlu AspLeuVal Tyr
290 295 300
Ala LeuLeu ArgPheGly IleIleAla LysLeuArg GluLysVal Asn
305 310 315 320
Lys AsnAsn AsnLysVal TyrTyrHis IleValIle SerAsnSer Ser
325 330 335
Asn LeuArg ThrPheLeu AspAsnIle GlyPheSer GlnGluArg Lys
340 345 350
Leu Lys Lys Leu Leu Glu Ile Ile Lys Asp Glu Asn Pro Asn Leu Asp
355 360 365
ValIleThr IleAspLys GluLysIle ArgTyrIle ArgAspArg Leu
370 375 38C!
LysValLys LeuThrArg AspIleGlu LysAspAsn TrpSerTyr Asn
385 390 395 400
LysCysArg LysIleThr GlnGluLeu LeuLysGlu IleTyrTyr Arg
405 410 415
LeuGluGlu LeuLysGlu IleGluLys AlaLeuGlu.GluAsnIle Leu
420 425 430
IleAspTrp AspGluVal AlaGluArg RrgLysGlu IleAlaGlu Lys
435 490 445
ThrGlyIle ArgSerAsp ArgIleLeu GluTyrIle RrgGlyLys Arg
450 955 460
Lys Pro Ser Leu Lys Asn Tyr Ile Lys Ile Ala Asn Thr Leu Gly Lys
465 470 475 480
Asn Ile Glu Lys Ile Ile Asp Ala Met Arg Ile Phe Ala Lys Lys Tyr
485 490 495
Ser Ser Tyr Ala Glu Ile Gly Lys Met Leu Asn Met Trp Asn Ser Ser
500 505 510
Ile Lys Ile Tyr Leu Glu Ser Rsn Thr Gln Glu Ile Glu Lys Leu Glu
515 520 525
Glu Ile Arg Lys Thr Glu Leu Lys Leu Val Lys Glu Ile Leu Asn Asp
530 535 540
Glu Lys Leu Ile Asp Ser Ile Gly Tyr Val Leu Phe Leu Ala Ser Asn
545 550 555 560
Glu Ile Tyr Trp Asp Glu Ile Val Glu Ile Glu Gln Leu Asn Gly Glu
565 570 575
CA 02338185 2001-02-05
- 6 -
Phe Thr Ile Tyr Asp Leu His Val Pro Arg Tyr His Asn Phe Ile Gly
580 585 590
Gly Asn Leu Pro Thr Ile Leu His Asn Thr Thr Ala Ala Leu Cys Leu
595 600 605
Ala Arg Asp Leu Phe Gly Glu Asn Trp Arg Asp Asn Phe Leu Glu Leu
610 615 620
Asn Ala Ser Val Ser Lys Asp Thr Pro Ile Leu Val Lys Ile Asp Gly
625 630 635 640
Lys Val Lys Arg Thr Thr Phe Glu Glu Leu Asp Lys Ile Tyr Phe Glu
645 650 655
Thr Asn Asp Glu Asn Glu Met Tyr Lys Lys Val Asp Asn Leu Glu Val
660 665 670
Leu Thr Val Asp Glu Asn Phe Arg Val Arg Trp Arg Lys Val Ser Thr
675 680 685
Ile Ile Arg His Lys Val Asp Lys Ile Leu Arg Ile Lys Phe Glu Gly
690 695 700
Gly Tyr Ile Glu Leu Thr Gly Asn His Ser Ile Met Met Leu Asp Glu
705 710 715 720
Asn Gly Leu Val Ala Lys Lys Ala Ser Asp Ile Lys Val Gly Asp Cys
725 730 735
Phe Leu Ser Phe Val Ala Asn Ile Glu Gly Glu Lys Rsp Arg Leu Asp
740 745 750
Leu Lys Glu Phe Glu Pro Lys Asp Ile Thr Ser Arg Val Lys Ile Ile
755 760 765
Asn Asp Phe Asp Ile Asp Glu Asp Thr Ala Trp Met Leu Gly Leu Tyr
770 775 780
Val Ala Glu Gly Ala Val Gly Phe Lys Gly Lys Thr Ser Gly Gln Val
785 790 795 800
Ile Tyr Thr Leu Gly Ser His Glu His Asp Leu Ile Asn Lys Leu Asn
805 810 815
Asp Ile Val Asp Lys Lys Gly Phe Ser Lys Tyr Glu Asn Phe Thr Gly
820 825 830
Ser Gly Phe Asp Arg Lys Arg Leu Ser Ala Lys Gln Ile Arg Ile Leu
835 840 845
Asn Thr Gln Leu Ala Arg Phe Val Glu Glu Asn Phe Tyr Asp Gly Asn
850 855 860
Gly Arg Arg Ala Arg Asn Lys Arg Ile Pro Asp Ile Ile Phe Glu Leu
865 870 875 880
Lys Glu Asn Leu Arg Val Glu Phe Leu Lys Gly Leu Ala Asp Gly Asp
885 890 895
Ser Ser Gly Asn Trp Arg Glu Val Val Arg Ile Ser Ser Lys Ser Asp
900 905 910
CA 02338185 2001-02-05
_ 7 _
Asn Leu LeuIle AspThr ValTrpLeuAla ArgIle SerGlyIleGlu
915 920 925
Ser Ser IlePhe GluAsn GluAlaArgLeu IleTrp LysGlyGlyMet
930 935 940
Lys Trp LysLys SerAsn LeuLeuProAla GluPro IleIleLysMet
945 950 955 960
Ile Lys LysLeu GluAsn LysIleAsnGly AsnTrp ArgTyrIleLeu
965 970 975
Arg His GlnLeu TyrGlu GlyLysLysArg ValSer LysAspLysIle
980 985 990
Lys Gln IleLeu GluMet ValAsnValGlu LysLeu SerAspLysGlu
995 1000 1005
Lys Glu ValTyr AspLeu LeuLysLysLeu SerLys ThrGluLeuTyr
1010 1015 1020
Ala Leu ValVal LysGlu IleGluIleIle AspTyr AsnAspPheVal
1025 1030 1035 1040
Tyr Asp ValSer ValPro AsnAsnGluMet PhePhe AlaGlyAsnVal
1045 1050 1055
Pro Ile Leu Leu His Asn Ser Asp Glu Arg Gly Ile Asp Val Ile Arg
1060 1065 1070
ThrLysVal LysAspPheAla Arg Lys ProIleGly ValPro
Thr Asp
1075 1080 1085
PheLysIle IlePheLeuAsp Glu Asp AlaLeuThr AspAla
Ser Ala
1090 1095 1100
GlnAsnAla LeuArgArgThr Met Lys TyrSerAsp CysArg
Glu Val
1105 1110 1115 1120
PheIleLeu SerCysLeuThr Gly Ala LysIleThr ProAsp
Asp Leu
1125 1130 1135
Glu Arg Glu Ile Lys Ile Glu Asp Phe Ile Lys Met Phe Glu Glu Arg
1140 1145 1150
Lys Leu Lys His Val Leu Asn Arg Asn Gly Glu Asp Leu Val Leu Ala
1155 1160 1165
Gly Val Lys Phe Asn Ser Lys Ile Val Asn His Lys Val Tyr Arg Leu
11?0 1175 1180
Val Leu Glu Ser Gly Arg Glu Ile Glu Ala Thr Gly Asp His Lys Phe
1185 1190 1195 1200
Leu Thr Arg Asp Gly Trp Lys Glu Val Tyr Glu Leu Lys Glu Asp Asp
1205 1210 1215
Glu Val Leu Val Tyr Pro Ala Leu Glu Gly Val Gly Phe Glu Val Asp
1220 1225 1230
Glu Arg Arg Ile Ile Gly Leu Asn Glu Phe Tyr Glu Phe Leu Thr Asn
1235 1240 1245
CA 02338185 2001-02-05
_ Tyr GluIleLys LeuGlyTyr LysPro LeuGlyLys AlaLysSer Tyr
1250 1255 1260
Lys GluLeuIle ThrArgAsp LysGlu LysIleLeu SerArgVal Leu
1265 1270 1275 1280
Glu LeuSerAsp LysTyrSer LysSer GluIleArg ArgLysIle Glu
1285 1290 1295
Glu GluPheGly IleLysIle SerLeu ThrThrIle LysAsnLeu Ile
1300 1305 1310
Asn GlyLysIle AspGlyPhe AlaLeu LysTyrVal ArgLysIle Lys
1315 1320 1325
Glu LeuGlyTrp AspGluIle ThrTyr AspAspGlu LysAlaGly Ile
1330 1335 1340
Phe AlaArgLeu LeuGlyPhe IleIle GlyAspGly HisLeuSer Lys
1345 1350 1355 1360
Ser LysGluGly ArgIleLeu IleThr AlaThrIle AsnGluLeu Glu
1365 1370 1375
Gly IleLysLys AspLeuGlu LysLeu GlyIleLys AlaSerAsn Ile
1380 1385 1390
Ile GluLysAsp IleGluHis LysLeu AspGlyArg GluIleLys Gly
1395 1400 1405
Lys ThrSerPhe IleTyrIle AsnAsn LysAlaPhe TyrLeuLeu Leu
1410 1915 1420
Asn PheTrpG1y ValGluIle GlyAsn LysThrIle AsnGlyTyr Asn
1425 1430 1435 1440
Ile ProLysTrp IleLysTyr GlyAsn LysPheVal LysArgGlu Phe
1445 1450 1455
Leu ArgGlyLeu PheGlyAla AspGly ThrLysPro TyrIleLys Lys
1460 1465 1470
Tyr AsnIleAsn GlyIleLys LeuGly IleArgVal GluAsnIle Ser
1475 1480 1485
Lys AspLysThr LeuGluPhe PheGlu GluValLys LysMetLeu Glu
1490 1495 1500
Glu PheGluVal GluSerTyr IleLys ValSerLys IleAspAsn Lys
1505 1510 1515 1520
Asn Leu Thr Glu Leu Ile Val Lys Ala Asn Asn Lys Asn Tyr Leu Lys
1525 1530 1535
Tyr Ser Arg Ile Tyr TyrGlu Asp Phe Ala
Leu Ser Ala Lys Asn Arg
1540 1545 1550
Leu Gly Glu Tyr Arg LysGlu Tyr Asp Ile
Val Leu Ile Ala Lys Ile
1555 1560 1565
Leu Glu Ile Ala Asn LeuLys Ala Gly Glu
Lys Glu Ala Glu Asp Lys
1570 1575 1580
CA 02338185 2001-02-05
g _
_ Ser Leu Arg Glu Leu Ala Arg Lys Tyr Asn Val Pro Val Asp Phe Ile
1585 1590 1595 1600
Ile Rsn Gln Leu Lys Gly Lys Asp Ile Gly Leu Pro Arg Asn Phe Met
1605 1610 1615
Thr Phe Glu Glu Phe Leu Lys Glu Lys Val Val Asp Gly Lys Tyr Val
1620 1625 1630
Ser Glu Arg Ile Ile Lys Lys Glu Cys Ile Gly Tyr Arg Asp Val Tyr
1635 1640 1645
Asp Ile Thr Cys His Lys Asp Pro Ser Phe Ile Ala Asn Gly Phe Val
1650 1655 1660
Ser His Asn Cys Asn Tyr Pro Ser Lys Ile Ile Pro Pro Ile Gln Ser
1665 1670 1675 1680
Arg Cys Ala Val Phe Arg Phe Ser Pro Leu Lys Lys Glu Asp Ile Ala
1685 1690 1695
Lys Lys Leu Lys Glu Ile Ala Glu Lys Glu Gly Leu Asn Leu Thr Glu
1700 1705 1710
Ser Gly Leu Glu Ala Ile Ile Tyr Val Ser Glu Gly Asp Met Arg Lys
1715 1720 1725
Ala Ile Asn Val Leu Gln Thr Ala Ala Ala Leu Ser Asp Val Ile Asp
1730 1735 1740
Asp Glu Ile Val Tyr Lys Val Ser Ser Arg Ala Arg Pro Glu Glu Val
1745 1750 1755 1760
Lys Lys Met Met Glu Leu Ala Leu Asp Gly Lys Phe Met Glu Ala Arg
1765 1770 1775
Asp Leu Leu Tyr Lys Leu Met Val Glu Trp Gly Met Ser Gly Glu Asp
1780 1785 1790
Ile Leu Asn Gln Met Phe Arg Glu Ile Asn Ser Leu Asp Ile Asp Glu
1795 1800 1805
Arg Lys Lys Val Glu Leu Ala Asp Ala Ile Gly Glu Thr Asp Phe Arg
1810 1815 1820
Ile Val Glu Gly Ala Asn Glu Arg Ile Gln Leu Ser Ala Leu Leu Ala
1825 1830 1835 1840
Lys Met Ala Leu Met Gly Arg
1845
(2) INFORMATION FOR SEQ ID NO: 4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 855 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Pyrococcus horikoshii
CA 02338185 2001-02-05
- 10 -
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 4:
Met His Asn Met Glu Glu Val Arg Glu Val Lys Val Leu Glu Lys Pro
1 5 10 15
Trp Val Glu Lys Tyr Arg Pro Gln Arg Leu Asp Glu Ile Val Gly Gln
20 25 30
Glu His Ile Val Lys Arg Leu Lys His Tyr Val Lys Thr Gly Ser Met
35 40 45
Pro His Leu Leu Phe Ala Gly Pro Pro Gly Val Gl:y Lys Cys Leu Thr
50 55 60
Gly Asp Thr Lys Val Ile Ala Asn Gly Gln Leu Phe Glu Leu Arg Glu
65 70 75 80
Leu Val Glu Lys Ile Ser Gly Gly Lys Phe Gly Pro Thr Pro Val Lys
85 90 95
Gly Leu Lys Val Ile Gly Ile Asp Glu Asp Gly Lys Leu Arg Glu Phe
100 105 110
Glu Val Gln Tyr Val Tyr Lys Asp Lys Thr Glu Arq_ Leu Ile Arg Ile
115 120 125
Arg Thr Arg Leu Gly Arg Glu Leu Lys Val Thr Pro Tyr His Pro Leu
130 135 140
Leu Val Asn Arg Arg Asn Gly Glu Ile Lys Trp Val. Lys Ala Glu Glu
145 150 155 160
Leu Lys Pro Gly Asp Lys Leu Ala Val Pro Arg Phe Leu Pro Ile Val
165 170 175
Thr Gly Glu Asp Pro Leu Ala Glu Trp Leu Gly Tyr Phe Leu Gly Gly
180 185 190
Gly Tyr Ala Asp Ser Lys Glu Asn Leu Ile Met Phe Thr Asn Glu Asp
195 200 205
Pro Leu Leu Arg Gln Arg Phe Met Glu Leu Thr Glu Lys Leu Phe Ser
210 215 220
Asp Ala Arg Ile Arg Glu Ile Thr His Glu Asn Gly Thr Ser Lys Val
225 230 235 240
Tyr Val Asn Ser Lys Lys Ala Leu Lys Leu Val Asn Ser Leu Gly Asn
245 250 255
Ala His Ile Pro Lys Glu Cys Trp Arg Gly Ile Arg Ser Phe Leu Arg
260 265 270
Ala Tyr Phe Asp Cys Asn Gly Gly Val Lys Gly Asn Ala Ile Val Leu
275 280 285
Ala Thr Ala Ser Lys Glu Met Ser Gln Glu Ile Ala Tyr Ala Leu Ala
290 295 300
CA 02338185 2001-02-05
- 11 -
- Gly Phe Gly Ile Ile Ser Arg Ile Gln Glu Tyr Arg Val Ile Ile Ser
305 310 315 320
Gly Ser Asp Asn Val Lys Lys Phe Leu Asn Glu Ile Gly Phe Ile Asn
325 330 335
Arg Asn Lys Leu Glu Lys Ala Leu Lys Leu Val Lys Lys Asp Asp Pro
340 345 350
Gly His Asp Gly Leu Glu Ile Asn Tyr Glu Leu Ile Ser Tyr Val Lys
355 360 365
Asp Arg Leu Arg Leu Ser Phe Phe Asn Asp Lys Arg Ser Trp Ser Tyr
370 375 380
Arg Glu Ala Lys Glu Ile Ser Trp Glu Leu Met Lys Glu Ile Tyr Tyr
385 390 395 400
Arg Leu Asp Glu Leu Glu Lys Leu Lys Glu Ser Leu Ser Arg Gly Ile
405 410 415
Leu Ile Asp Trp Asn Glu Val Ala Lys Arg Ile Glu Glu Val Ala Glu
420 425 430
Glu Thr Gly Ile Arg Ala Asp Glu Leu Leu Glu Tyr Ile Glu Gly Lys
435 440 445
Arg Lys Leu Ser Phe Lys Asp Tyr Ile Lys Ile Ala Lys Val Leu Gly
450 455 460
Ile Asp Val Glu His Thr Ile Glu Ala Met Arg Val Phe Ala Arg Lys
465 470 475 480
Tyr Ser Ser Tyr Ala Glu Ile Gly Arg Arg Leu Gly Thr Trp Asn Ser
485 490 495
Ser Val Lys Thr Ile Leu Glu Ser Asn Ala Val Asn Val Glu Ile Leu
500 505 510
Glu Arg Ile Arg Lys Ile Glu Leu Glu Leu Ile Glu Glu Ile Leu Ser
515 520 525
Asp Glu Lys Leu Lys Glu Gly Ile Ala Tyr Leu Ile Phe Leu Ser Gln
530 535 540
Asn Glu Leu Tyr Trp Asp Glu Ile Thr Lys Val Glu Glu Leu Arg Gly
545 550 555 560
Glu Phe Ile Ile Tyr Asp Leu His Val Pro Gly Tyr His Asn Phe Ile
565 570 575
Ala Gly Asn Met Pro Thr Val Val His Asn Thr Thr Ala Ala Leu Ala
580 585 590
Leu Ser Arg Glu Leu Phe Gly Glu Asn Trp Arg His Asn Phe Leu Glu
595 600 605
Leu Asn Ala Ser Asp Glu Arg Gly Ile Asn Val Ile Arg Glu Lys Val
610 615 620
Lys Glu Phe Ala Arg Thr Lys Pro Ile Gly Gly Ala Ser Phe Lys Ile
625 630 635 640
CA 02338185 2001-02-05
- 12 -
Ile Phe Leu Asp Glu Ala Asp Ala Leu Thr Gln Asp Ala Gln Gln Ala
645 650 655
Leu Arg Arg Thr Met Glu Met Phe Ser Ser Asn Val Arg Phe Ile Leu
660 665 670
Ser Cys Asn Tyr Ser Ser Lys Ile Ile Glu Pro Ile Gln Ser Arg Cys
675 680 685
Ala Ile Phe Arg Phe Arg Pro Leu Arg Asp Glu Asp Ile Ala Lys Arg
690 695 700
Leu Arg Tyr Ile Ala Glu Asn Glu Gly Leu Glu Leu Thr Glu Glu Gly
705 710 715 720
Leu Gln Ala Ile Leu Tyr Ile Ala Glu Gly Asp Met Arg Arg Ala Ile
725 730 735
Asn Ile Leu Gln Ala Ala Ala Ala Leu Asp Lys Lys Ile Thr Asp Glu
740 795 750
Asn Val Phe Met Val Ala Ser Arg Ala Arg Pro Glu Asp Ile Arg Glu
755 760 765
Met Met Leu Leu Ala Leu Lys Gly Asn Phe Leu Lys Ala Arg Glu Lys
770 775 780
Leu Arg Glu Ile Leu Leu Lys Gln Gly Leu Ser Gly Glu Asp Val Leu
785 790 795 800
Ile Gln Met His Lys Glu Val Phe Asn Leu Pro Ile Asp Glu Pro Thr
805 810 815
Lys Val Tyr Leu Ala Asp Lys Ile Gly Glu Tyr Asn Phe Arg Leu Val
820 825 830
Glu Gly Ala Asn Glu Met Ile Gln Leu Glu Ala Leu Leu Ala Gln Phe
835 840 845
Thr Leu Val Gly Lys Lys Lys
850 855
(2) INFORMATION FOR SEQ ID NO: 5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 321 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanobacterium thermoautotrophicum
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 5:
Met Ile Ile Met Asn Gly Pro Trp Val Glu Lys Tyr Arg Pro Gln Lys
1 5 10 15
CA 02338185 2001-02-05
- 13 -
Leu Asp Asp Ile Val Gly Gln Glu His Ile Ile Pro Arg Leu Lys Arg
20 25 30
Tyr Val Glu Glu Lys Ser Met Pro Asn Leu Met Phe Thr Gly Pro Ala
35 40 45
Gly Val Gly Lys Thr Thr Ala Ala Leu Ala Leu Ala Arg Glu Ile Leu
50 55 60
Gly Glu Tyr Trp Arg Gln Asn Phe Leu Glu Leu Asn Ala Ser Asp Ala
65 70 75 80
Arg Gly Ile Asp Thr Val Arg Thr Ser Ile Lys Asn Phe Cys Arg Leu
85 90 95
Lys Pro Val Gly Ala Pro Phe Arg Ile Ile Phe Leu Asp Glu Val Asp
100 105 110
Asn Met Thr Lys Asp Ala Gln His Ala Leu Arg Arg Glu Met Glu Met
115 120 125
Tyr Thr Lys Thr Ser Ser Phe Ile Leu Ser Cys Asn Tyr Ser Ser Lys
130 135 140
Ile Ile Asp Pro Ile Gln Ser Arg Cys Ala Ile Phe Arg Phe Leu Pro
145 150 155 160
Leu Lys Gly His Gln Ile Ile Lys Arg Leu Glu Tyr Ile Ala Glu Lys
165 170 175
Glu Rsn Leu Glu Tyr Glu Ala His Ala Leu Glu Thr Ile Val Tyr Phe
180 185 190
Ala Glu Gly Asp Leu Arg Lys Ala Ile Asn Leu Leu Gln Ser Ala Ala
195 200 205
Ser Leu Gly Glu Lys Ile Thr Glu Ser Ser Ile Tyr Asp Val Val Ser
210 215 220
Arg Ala Arg Pro Lys Asp Val Arg Lys Met Ile Lys Thr Ile Leu Asp
225 230 235 240
Gly Lys Phe Met Glu Ala Arg Asp Met Leu Arg Glu Ile Met Val Leu
245 250 255
Gln Gly Ile Ser Gly Glu Asp Met Val Thr Gln Ile Tyr Gln Glu Leu
260 265 270
Ser Arg Leu Ala Met Glu Gly Glu Val Asp Gly Asp Arg Tyr Val Gly
275 280 285
Leu Ile Asp Ala Ile Gly Glu Tyr Asp Phe Arg Ile Arg Glu Gly Ala
290 295 300
Asn Pro Arg Ile Gln Leu Glu Ala Leu Leu Ala Arg Phe Leu Glu His
305 310 315 320
Ala
CA 02338185 2001-02-05
- 14 -
(2) INFORMATION FOR SEQ ID N0: 6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1148 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Homo Sapiens
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 6:
Met Asp Ile Arg Lys Phe Phe Gly Val Ile Pro Ser Gly Lys Lys Leu
1 5 10 15
Val Ser Glu Thr Val Lys Lys Asn Glu Lys Thr Lys Ser Asp Glu Glu
20 25 30
Thr Leu Lys Ala Lys Lys Gly Ile Lys Glu Ile Lys Val Asn Ser Ser
35 40 45
Arg Lys Glu Asp Asp Phe Lys Gln Lys Gln Pro Ser Lys Lys Lys Arg
50 55 60
Ile Ile Tyr Asp Ser Asp Ser Glu Ser Glu Glu Thr Leu Gln Val Lys
65 70 75 80
Asn Ala Lys Lys Pro Pro Glu Lys Leu Pro Val Ser Ser Lys Pro Gly
85 90 95
Lys Ile Ser Arg Gln Asp Pro Val Thr Tyr Ile Ser Glu Thr Asp Glu
100 105 110
Glu Asp Asp Phe Met Cys Lys Lys Ala Ala Ser Lys Ser Lys Glu Asn
115 120 125
Gly Arg Ser Thr Asn Ser His Leu Gly Thr Ser Asn Met Lys Lys Asn
130 135 140
Glu Glu Asn Thr Lys Thr Lys Asn Lys Pro Leu Ser Pro Ile Lys Leu
145 150 155 160
Thr Pro Thr Ser Val Leu Asp Tyr Phe Gly Thr Gly Ser Val Gln Arg
165 170 175
Ser Asn Lys Lys Met Val Ala Ser Lys Arg Lys Glu Leu Ser Gln Asn
180 185 190
Thr Asp Glu Ser Gly Leu Asn Asp Glu Ala Ile Ala Lys Gln Leu Gln
195 200 205
Leu Asp Glu Asp Ala Glu Leu Glu Arg Gln Leu His Glu Asp Glu Glu
210 215 220
Phe Ala Arg Thr Leu Ala Met Leu Asp Glu Glu Pro Lys Thr Lys Lys
225 230 235 240
CA 02338185 2001-02-05
- 15 -
Ala Arg Lys Asp Thr Glu Ala Gly Glu Thr Phe Ser Ser Val Gln Ala
245 250 255
Asn Leu Ser
Lys Ala Glu
Lys His Lys
Tyr Pro His
Lys Val Lys
Thr
260 265 270
Ala Gln Val Ser Asp
Glu Rrg Lys
Ser Tyr Ser
Pro Arg Lys
Gln Ser
275 280 285
Lys Tyr Glu Ser Ser Glu Ser Gln Gln His Se.r Lys Ser
Lys Ser Ala
290 295 300
Asp Lys Ile Gly Glu Ser Ser Pro Lys Ala Ser Ser Lys
Val Leu Ala
305 310 315 320
Ile Met Lys Arg Lys Glu Ser Ser Tyr Lys Glu Ile Glu
Lys Pro Val
325 330 335
Ala Ser Lys Arg Lys Asn Ala Ile Lys Leu Lys Gly Glu
Glu Thr Lys
340 345 350
Thr Pro Lys Lys Thr Ser Ser Pro Ala Lys Lys Glu Ser
Lys Val Ser
355 360 365
Pro Glu Asp Ser Glu Lys Arg Thr Asn Tyr Gln Ala Tyr
Lys Arg Ser
370
375
380
Tyr Leu Rsn Arg Glu Pro Lys Ala Leu Gly Ser Lys Glu
Gly Ile Pro
385 390 395 400
Lys Gly A1a Glu Asn Leu Glu Gly Leu Ile Phe Val Ile
Cys Thr Gly
405 410 415
Val Leu Glu Ser Ile Arg Asp Glu Ala Lys Ser Leu Ile
Glu Glu Arg
420 425 430
Tyr Gly Gly Lys Val Gly Asn Val Ser Lys Lys Thr Asn
Thr Tyr Leu
435 440 445
Val Met Gly Arg Asp Gly Gln Ser Lys Ser Asp Lys Ala
Ser Ala Ala
450 455 460
Leu Gly Thr Lys Ile Asp Glu Asp Gly Leu Leu Asn Leu
Ile Ile Arg
465 470 475 480
Thr Met Pro Gly Lys Ser Lys Tyr Glu Ile Ala Val Glu
Lys Thr Glu
485 490 495
Met Lys Lys Glu Ser Leu Glu Arg Thr Pro Gln Lys Asn
Lys Val Gln
500 505 510
Gly Lys Arg Pro Ser Lys Lys Glu Ser Glu Ser
Lys Ile Ser Lys Lys
515 520 525
Ser Arg Pro
Thr Ser Lys
Arg Asp Ser
Leu Ala Lys
Thr Ile Lys
Lys
530 535 540
Glu Thr Asp
Val Phe Trp
Lys Ser Leu
Asp Phe Lys
Glu Gln Val
Ala
545 550 555 560
Glu Glu Thr
Ser Gly Asp
Ser Lys Ala
Arg Asn Leu
Ala Asp Asp
Ser
565 570 575
CA 02338185 2001-02-05
- 16 -
Ser Glu Asn Lys Val Glu Asn Leu Leu Trp Val Asp Lys Tyr Lys Pro
580 585 590
Thr Ser Leu Lys Thr Ile Ile Gly Gln Gln Gly Asp Gln Ser Cys Ala
595 ~ 600 605
Asn Lys Leu Leu Arg Trp Leu Arg Asn Trp Gln Lys Ser Ser Ser Glu
610 615 620
Asp Lys Lys His Ala Ala Lys Phe Gly Lys Phe Ser Gly Lys Asp Asp
625 630 635 640
Gly Ser Ser Phe Lys Ala Ala Leu Leu Ser Gly Pro Pro Gly Val Gly
645 650 655
Lys Thr Thr Thr Ala Ser Leu Va.l Cys Gln Glu Leu Gly Tyr Ser Tyr
660 665 670
Val Glu Leu Asn Ala Ser Asp Thr Arg Ser Lys Ser Ser Leu Lys Ala
675 680 685
Ile Val Ala Glu Ser Leu Asn Asn Thr Ser Ile Lys Gly Phe Tyr Ser
690 695 700
Asn Gly Ala Ala Ser Ser Val Ser Thr Lys His Ala Leu Ile Met Asp
705 710 715 720
Glu Val Asp Gly Met Ala Gly Asn Glu Asp Arg Gly Gly Ile Gln Glu
725 730 735
Leu Ile Gly Leu Ile Lys His Thr Lys Ile Pro Ile Ile Cys Met Cys
740 745 750
Asn Asp Arg Asn His Pro Lys Ile Arg Ser Leu Val His Tyr Cys Phe
755 760 765
Asp Leu Arg Phe Gln Arg Pro Arg Val Glu Gln Ile Lys Gly Ala Met
770 775 780
Met Ser Ile Ala Phe Lys Glu Gly Leu Lys Ile Pro Pro Pro Ala Met
785 790 795 800
Asn Glu Ile Ile Leu Gly Ala Asn Gln Asp Ile Arg Gln Val Leu His
805 810 815
Asn Leu Ser Met Trp Cys Ala Arg Ser Lys Ala Leu Thr Tyr Asp Gln
820 825 830
Ala Lys Ala Asp Ser His Arg Ala Lys Lys Asp Ile Lys Met Gly Pro
835 840 845
Phe Asp Val Ala Arg Lys Val Phe Ala Ala Gly Glu Glu Thr Ala His
850 855 860
Met Ser Leu Val Asp Lys Ser Asp Leu Phe Phe His Asp Tyr Ser Ile
865 870 875 880
Ala Pro Leu Phe Val Gln Glu Asn Tyr Ile His Val Lys Pro Val Ala
885 890 895
Ala Gly Gly Asp Met Lys Lys His Leu Met Leu Leu Ser Arg Ala Ala
900 905 910
CA 02338185 2001-02-05
- 17 -
Asp Ser Ile Cys Asp Gly Asp Leu Val Asp Ser Gln Ile Arg Ser Lys
915 920 925
Gln Asn Trp Ser Leu Leu Pro Ala Gln Ala Ile Tyr Ala Ser Val Leu
930 935 940
Pro Gly Glu Leu Met Arg Gly Tyr Met Thr Gln Phe Pro Thr Phe Pro
945 950 955 960
Ser Trp Leu Gly Lys His Ser Ser Thr Gly Lys His Asp Arg Ile Val
965 970 975
Gln Asp Leu Ala Leu His Met Ser Leu Arg Thr Tyr Ser Ser Lys Arg
980 985 990
Thr Val Asn Met Asp Tyr Leu Ser Leu Leu Arg Asp Ala Leu Val Gln
995 1000 1005
Pro Leu Thr Ser Gln Gly Val Asp Gly Val Gln Asp Val Val Ala Leu
1010 1015 1020
Met Asp Thr Tyr Tyr Leu Met Lys Glu Asp Phe Glu Asn Ile Met Glu
1025 1030 1035 1040
Ile Ser Ser Trp Gly Gly Lys Pro Ser Pro Phe Ser Lys Leu Asp Pro
1045 1050 1055
Lys Val Lys Ala Ala Phe Thr Arg Ala Tyr Asn Lys Glu Ala His Leu
1060 1065 1070
Thr Pro Tyr Ser Leu Gln Ala Ile Lys Ala Ser Arg His Ser Thr Ser
1075 1080 1085
Pro Ser Leu Asp Ser Glu Tyr Asn Glu Glu Leu Asn Glu Asp Asp Ser
1090 1095 1100
Gln Ser Asp Glu Lys Asp Gln Asp Ala Ile Glu Thr Asp Ala Met Ile
1105 1110 1115 1120
Lys Lys Lys Thr Lys Ser Ser Lys Pro Ser Lys Pro Glu Lys Asp Lys
1125 1130 1135
Glu Pro Arg Lys Gly Lys Gly Lys Ser Ser Lys Lys
1140 1145
(2) INFORMATION FOR SEQ ID NO: 7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 479 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeoglobus fulgidus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 7:
CA 02338185 2001-02-05
- 18 -
Met Leu Trp Val Glu Lys Tyr Arg Pro Lys Thr Leu Glu Glu Val Val
1 5 10 15
Ala Asp Lys Ser Ile Ile Thr Arg Val Ile Lys Trp Ala Lys Ser Trp
20 25 30
Lys Arg Gly Ser Lys Pro Leu Leu Leu Ala Gly Pro Pro Gly Val Gly
35 40 45
Lys Thr Ser Leu Ala Leu Ala Leu Ala Asn Thr Met Gly Trp Glu Ala
50 55 60
Val Glu Leu Asn Ala Ser Asp Gln Arg Ser Trp Arg Val Ile Glu Arg
65 70 75 80
Ile Val Gly Glu Gly Ala Phe Asn Glu Thr Ile Ser Asp Glu Gly Glu
85 90 95
Phe Leu Ser Ser Arg Ile Gly Lys Leu Lys Leu Ile Ile Leu Asp Glu
100 105 110
Val Asp Asn Ile His Lys Lys Glu Asp Val Gly Gly Glu Ala Ala Leu
115 120 125
Ile Arg Leu Ile Lys Arg Lys Pro Ala Gln Pro Leu Ile Leu Ile Ala
130 135 140
Asn Asp Pro Tyr Lys Leu Ser Pro Glu Leu Arg Asn Leu Cys Glu Met
145 150 155 160
Ile Asn Phe Lys Arg Leu Thr Lys Gln Gln Val Ala Arg Val Leu Glu
165 170 175
Arg Ile Ala Leu Lys Glu Gly Ile Lys Val Asp Lys Ser Val Leu Leu
180 185 190
Lys Ile Ala Glu Asn Ala Gly Gly Asp Leu Arg Ala Ala Ile Asn Asp
195 200 205
Phe Gln Ala Leu Ala Glu Gly Lys Glu Glu Leu Lys Pro Glu Asp Val
210 215 220
Phe Leu Thr Lys Arg Thr Gln Glu Lys Asp Ile Phe Arg Val Met Gln
225 230 235 240
Met Ile Phe Lys Thr Lys Asn Pro Ala Val Tyr Asn Glu Ala Met Leu
245 250 255
Leu Asp Glu Ser Pro Glu Asp Val Ile His Trp Val Asp Glu Asn Leu
260 265 270
Pro Leu Glu Tyr Ser Gly Val Glu Leu Val Asn Ala Tyr Glu Ala Leu
275 280 285
Ser Arg Ala Asp Ile Phe Leu Gly Arg Val Arg Arg Arg Gln Phe Tyr
290 295 300
Arg Leu Trp Lys Tyr Ala Ser Tyr Leu Met Thr Val Gly Val Gln Gln
305 310 315 320
Met Lys Glu Glu Pro Lys Lys Gly Phe Thr Arg Tyr Arg Arg Pro Ala
325 330 335
CA 02338185 2001-02-05
- 19 -
Val Trp Gln Met Leu Phe Gln Leu Arg Gln Lys Arg Glu Met Thr Arg
340 345 350
Lys Ile Leu Glu Lys Ile Gly Lys Tyr Ser His Leu Ser Met Arg Lys
355 360 365
Ala Arg Thr Glu Met Phe Pro Val Ile Lys Leu Leu Leu Lys Glu Leu
370 375 380
Asp Val Asp Lys Ala Ala Thr Ile Ala Ala Phe Tyi: Glu Phe Thr Lys
385 390 395 400
Glu Glu Leu Glu Phe Leu Val Gly Glu Lys Gly Asp Glu Ile Trp Lys
405 410 415
Tyr Val Glu Lys His Gly Met His Arg Ile Glu Asp Glu Thr Phe Leu
420 425 430
Glu Ser Phe Val Lys Ala Glu Lys Glu Glu Lys Glu Glu Ser Val Glu
435 440 445
Glu Val Ala Glu Glu Lys Pro Glu Glu Glu Arg Glu Glu Pro Arg Ala
450 455 460
Arg Lys Lys Ala Gly Lys Asn Leu Thr Leu Asp Ser Phe Phe Ser
465 470 475
(2) INFORMATION FOR SEQ ID NO: 8:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 516 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanococcus jannaschii
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 8:
Met Leu Ser Trp Val Glu Lys Tyr Arg Pro Lys Ser Leu Lys Asp Val
1 5 10 15
Ala Gly His Glu Lys Val Lys Glu Lys Leu Lys Thr Trp Ile Glu Ser
20 25 30
Tyr Leu Lys Gly Glu Thr Pro Lys Pro Ile Leu Leu Val Gly Pro Pro
35 40 45
Gly Cys Gly Lys Thr Thr Leu Ala Tyr Ala Leu Ala Asn Asp Tyr Gly
50 55 60
Phe Glu Val Ile Glu Leu Asn Ala Ser Asp Lys Arg Asn Ser Ser Ala
65 70 75 80
Ile Lys Lys Val Val Gly His Ala Ala Thr Ser Ser Ser Ile Phe Gly
85 90 95
CA 02338185 2001-02-05
_ - 20 -
Lys Lys Phe Leu Ile Val Leu Asp Glu Val Asp Gly Ile Ser Gly Lys
100 105 110
GluAsp AlaGlyGly ValSerGlu LeuIleLys Val.IleLys LysAla
115 120 125
LysAsn ProIleIle LeuThrAla AsnAspAla TyrAlaPro SerIle
130 135 140
ArgSer LeuLeuPro TyrValGlu ValIleGln LeuAsnPro ValHis
145 150 155 160
ThrAsn SerValTyr LysValLeu LysLysIle Ala.GluLys GluGly
165 170 175
LeuAsp ValAspAsp LysThrLeu LysMetIle AlaGlnHis SerAla
180 185 190
GlyAsp LeuArgSer AlaIleAsn AspLeuGlu AlaLeuAla LeuSer
195 200 205
Gly LeuSer TyrGluAla AlaGlnLysLeu ProAspArg LysArg
Asp
210 215 220
GluAla AsnIle PheAspAla LeuArgValIle LeuLysThr ThrHis
225 230 235 240
TyrGly IleAla ThrThrAla LeuMetAsnVal AspGluThr ProAsp
245 250 255
ValVal IleGlu TrpIleAla GluAsnValPro LysGluTyr GluLys
260 265 270
ProGlu GluVal AlaArgAla PheGluTyrLeu SerLysAla AspArg
275 280 285
TyrLeu GlyArg ValMetArg ArgGlnAsnTyr SerPheTrp LysTyr
290 295 300
AlaThr ThrLeu MetThrAla GlyValAlaLeu SerLysAsp GluLys
305 310 315 320
TyrArg LysTrp ThrProTyr SerTyrProLys IlePheArg LeuLeu
325 330 335
ThrLys ThrLys AlaGluArg GluIleLeuAsn LysIleLeu LysLys
340 345 350
IleGly GluLys ThrHisThr SerSerLysArg AlaArgPhe AspLeu
355 360 365
GlnMet LeuLys LeuLeuAla LysGluAsnPro SerValAla AlaAsp
370 375 380
LeuVal AspTyr PheGluIle LysGluAspGlu LeuLysVal LeuVal
385 390 395 400
GlyAsp LysLeu AlaSerGlu IleLeuLysIle LeuLysGlu LysLys
405 410 415
LysLeu GluArg LysLysLys LysGluLysGlu LysLeuGlu LysGlu
420 425 430
CA 02338185 2001-02-05
- 21 -
Lys Lys Lys Glu Glu Lys Ala Lys Glu Lys Gln Ser Asn Leu Ile Ile
435 440 445
Gln Pro Lys Glu Ile Lys Glu Glu Val Lys Ala Glu Val Glu Lys Lys
450 455 q6p
Glu Glu Val Lys Glu Lys Ile Val Glu Lys Pro Lya Ala Glu Glu Val
465 470 475 480
Lys Glu Lys Ser Lys Thr Glu Glu Lys Glu Thr Lys Lys Asp Lys Lys
485 490 495
Lys Gly Lys Lys Lys Lys Glu Asp Lys Gly Lys Gln Leu Thr Leu Asp
500 505 510
Ala Phe Phe Lys
515
(2) INFORMATION FOR SEQ ID NO: 9:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 468 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Pyrococcus horikoshii
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 9:
Met Pro Asp Val Pro Trp Ile Glu Lys Tyr Arg Pro Arg Lys Leu Ser
1 5 10 15
Glu Ile Val Asn Gln Glu Gln Ala Leu Glu Lys Val Arg Ala Trp Ile
20 25 30
Glu Ser Trp Leu His Gly Asn Pro Pro Lys Lys Lys Rla Leu Leu Leu
35 40 45
Ala Gly Pro Pro Gly Ser Gly Lys Thr Thr Thr Val Tyr Ala Leu Ala
50 55 60
His Glu Tyr Asn Phe Glu Val Ile Glu Leu Asn Ala Ser Asp Glu Arg
65 70 75 80
Thr Tyr Asn Lys Ile Ala Arg Tyr Val Gln Ala Ala Tyr Thr Met Asp
85 90 95
Ile Met Gly Lys Arg Arg Lys Ile Ile Phe Leu Asp Glu Ala Asp Asn
100 105 110
Ile Glu Pro Ser Gly Ala Pro Glu Ile Ala Lys Leu Ile Asp Lys Ala
115 120 125
Arg Asn Pro Ile Ile Met Ala Ala Asn His Tyr Trp Glu Val Pro Lys
130 135 140
CA 02338185 2001-02-05
- 22 -
Glu Ile Arg Asp Arg Ala Glu Leu Val Glu Tyr Lys Arg Leu Asn Gln
145 150 155 160
Arg Asp Val Ile Ser Ala Leu Val Arg Ile Leu Lys Arg Glu Gly Ile
165 170 175
Thr Val Pro Lys Glu Ile Leu Thr Glu Ile Ala Lys Arg Ser Ser Gly
180 185 190
Asp Leu Arg Ala Ala Ile Asn Asp Leu Gln Thr Ile Val Ala Gly Gly
195 200 205
Tyr Glu Asp Ala Lys Tyr Val Leu Ala Tyr Arg Asp Val Glu Lys Thr
210 215 220
Val Phe Gln Ser Leu Gly Met Val Phe Ser Ser Asp Asn Ala Lys Arg
225 230 235 240
Ala Lys Leu Ala Leu Met Asn Leu Asp Met Ser Pro Asp Glu Phe Leu
245 250 255
Leu Trp Val Asp Glu Asn Ile Pro His Met Tyr Leu Lys Pro Glu Glu
260 265 270
Met Ala Arg Ala Tyr Glu Ala Ile Ser Arg Ala Asp Ile Tyr Leu Gly
275 280 285
Arg Ala Gln Arg Thr Gly Asn Tyr Ser Leu Trp Lys Tyr Ala Ile Asp
290 295 30C)
Met Met Thr Ala Gly Val Ala Val Ala Gly Thr Lye, Lys Lys Gly Phe
305 310 315 320
Ala Lys Phe Tyr Pro Pro Asn Thr Leu Lys Met Leu. Ala Glu Ser Lys
325 330 335
Glu Glu Arg Ser Ile Arg Asp Ser Ile Ile Lys Lys Ile Met Lys Glu
340 345 350
Met His Met Ser Lys Leu Glu Ala Leu Glu Thr Met Lys Ile Leu Arg
355 360 365
Thr Ile Phe Glu Asn Asn Leu Asp Leu Ala Ala His Phe Thr Val Phe
370 375 380
Leu Glu Leu Thr Glu Lys Glu Val Glu Phe Leu Ala Gly Lys Glu Lys
385 390 395 400
Ala Gly Thr Ile Trp Gly Lys Thr Leu Ser Ile Arg Arg Arg Ile Lys
405 410 415
Glu Thr Glu Lys Ile Glu Glu Lys Ala Val Glu Glu Lys Val Glu Glu
420 425 430
Glu Glu Ala Glu Glu Glu Glu Glu Glu Glu Arg Lys Glu Glu Glu Lys
435 440 445
Pro Lys Ala Glu Lys Lys Lys Gly Lys Gln Val Thr Leu Phe Asp Phe
450 455 460
Ile Lys Lys Asn
465
CA 02338185 2001-02-05
- 23 -
(2) INFORMATION FOR SEQ ID NO: 10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 479 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanobacterium thermoautotrrophicum
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 10:
Met Ser Trp Thr Glu Lys Tyr Arg Pro Gly Ser Phe Asp Glu Val Val
1 5 10 15
Gly Asn Gln Lys Val Ile Ala Glu Ile Lys Glu Trp Ile Lys Ala Trp
20 25 30
Lys Ala Gly Lys Pro Gln Lys Pro Leu Leu Leu Val Gly Pro Pro Gly
35 40 45
Thr Gly Lys Thr Thr Leu Ala His Ile Ile Gly Lys Glu Phe Ser Asp
50 55 60
Thr Leu Glu Leu Asn Ala Ser Asp Arg Arg Ser Gln Asp Ala Leu Met
65 70 75 80
Arg Ser Ala Gly Glu Ala Ser Ala Thr Arg Ser Leu Phe Asn His Asp
85 90 95
Leu Lys Leu Ile Ile Leu Asp Glu Val Asp Gly Ile His Gly Asn Glu
100 105 110
Asp Arg Gly Gly Val Gln Ala Ile Asn Arg Ile Ile Lys Glu Ser Arg
115 120 125
His Pro Met Val Leu Thr Ala Asn Asp Pro Tyr Ser Lys Arg Leu Gln
130 135 140
Ser Ile Lys Pro Arg Cys Arg Val Leu Asn Leu Arg Lys Val His Thr
145 150 155 160
Ser Ser Ile Ala Ala Ala Leu Arg Arg Ile Cys Arg Ala Glu Gly Ile
165 170 175
Glu Cys Pro Asp Asp Val Leu Arg Glu Leu Ala Lys Arg Ser Arg Gly
180 185 190
Asp Leu Arg Ser Ala Ile Asn Asp Leu Glu Ala Met Ala Glu Gly Glu
195 200 205
Glu Arg Ile Gly Glu Glu Leu Leu Lys Met Gly Glu Lys Asp Ala Thr
210 215 220
Ser Asn Leu Phe Asp Ala Val Arg Ala Val Leu Lys Ser Arg Asp Val
225 230 235 240
CA 02338185 2001-02-05
- 24 -
Ser Lys Val Arg Glu Ala Met Arg Val Asp Asp Asp Pro Thr Leu Val
245 250 255
Leu Glu Phe Ile Ala Glu Asn Val Pro Arg Glu Tyr Glu Lys Pro Asn
260 265 270
Glu Ile Ser Arg Ala Tyr Asp Met Leu Ser Arg Ala Asp Ile Phe Phe
275 280 285
Gly Arg Ala Val Arg Thr Arg Asn Tyr Thr Tyr Trp Arg Tyr Ala Ser
290 295 300
Glu Leu Met Gly Pro Gly Val Ala Leu Ala Lys Asp Lys Thr Tyr Arg
305 310 315 320
Lys Phe Val Arg Tyr Thr Gly Ser Ser Ser Phe Arg Ile Leu Gly Lys
325 330 335
Thr Arg Lys Gln Arg Ser Leu Arg Asp Ser Val Ala Ala Lys Met Ala
340 345 350
Gly Lys Met His Ile Ser Pro Lys Val Ala Ile Ser Met Phe Pro Tyr
355 36C 365
Met Glu Ile Leu Phe Glu Asn Asp Glu Met Ala Tyr Asp Ile Ser Glu
370 375 380
Phe Leu Glu Leu Arg Asp Glu Glu Ile Lys Leu Phe Arg Lys Arg Lys
385 390 395 400
Ile Lys Ala Pro Lys Arg Lys Lys Thr Pro Arg Lys Ala Glu Ile Lys
405 410 415
Val Gly Pro Leu Tyr Ser Gln Lys Lys Asp Lys Gly Ala Asp Lys Ser
420 425 430
Ile Asn Asp Lys Ala Thr Asp Lys Ser Ala Lys Thr Pro Ile Lys Ser
435 440 445
Ser Lys Lys Asp Asp Arg Pro Arg Asp Glu Ser Ser Ser Ser Ser Asp
450 455 460
Asp Lys Lys Pro Lys Glu Lys Gln Thr Ser Leu Phe Gln Phe Ser
465 470 475
(2) INFORMATION FOR SEQ ID NO: 11:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 261 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(Vi) INITIAL ORIGIN:
(A) ORGANISM: Homo sapiens
CA 02338185 2001-02-05
- 25 -
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 11:
Met Phe Glu Ala Arg Leu Val Gln Gly Ser Ile Leu Lys Lys Val Leu
1 5 10 15
Glu Ala Leu Lys Asp Leu Ile Asn Glu Ala Cys Trp Asp Ile Ser Ser
20 25 30
Ser Gly Val Asn Leu Gln Ser Met Asp Ser Ser His Val Ser Leu Val
35 40 45
Gln Leu Thr Leu Arg Ser Glu Gly Phe Asp Thr Tyr Arg Cys Asp Arg
50 55 60
Asn Leu Ala Met Gly Val Asn Leu Thr Ser Met Ser Lys Ile Leu Lys
65 70 75 80
Cys Ala Gly Asn Glu Asp Ile Ile Thr Leu Arg Ala Glu Asp Asn Ala
85 90 95
Asp Thr Leu Ala Leu Val Phe Glu Ala Pro Asn Gln Glu Lys Val Ser
100 105 110
Asp Tyr Glu Met Lys Leu Met Asp Leu Asp Val Glu Gln Leu Gly Ile
115 120 125
Pro Glu Gln Glu Tyr Ser Cys Val Val Lys Met Pro Ser Gly Glu Phe
130 135 140
Ala Arg Ile Cys Arg Asp Leu Ser His Ile Gly Asp Ala Val Val Ile
145 150 155 160
Ser Cys Ala Lys Asp Gly Val Lys Phe Ser Ala Ser Gly Glu Leu Gly
165 170 175
Asn Gly Asn Ile Lys Leu Ser Gln Thr Ser Asn Val Asp Lys Glu Glu
180 185 190
Glu Ala Val Thr Ile Glu Met Asn Glu Pro Val Gln Leu Thr Phe Ala
195 200 205
Leu Arg Tyr Leu Asn Phe Phe Thr Lys Ala Thr Pro Leu Ser Ser Thr
210 215 220
Val Thr Leu Ser Met Ser Ala Asp Val Pro Leu Val Val Glu Tyr Lys
225 230 235 240
Ile Ala Asp Met Gly His Leu Lys Tyr Tyr Leu Ala Pro Lys Ile Glu
245 250 255
Asp Glu Glu Gly Ser
260
(2) INFORMATION FOR SEQ ID NO: 12:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 245 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
CA 02338185 2001-02-05
- 26 -
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeoglobus fulgidus
(xi)SEQUENCE DESCRIPTION: 12:
SEQ
ID
NO:
Met IleAsp ValIleMet ThrGlyGlu LeuLeuLysThr ValThrArg
1 5 10 15
Ala IleVal AlaLeuVal SerGluAla ArgIleHisPhe LeuGluLys
20 25 30
Gly LeuHis SerArgAla ValAspPro AlaAsnValAla MetValIle
35 90 45
Val AspIle ProLysAsp SerPheGlu ValTyrAsnIle AspGluGlu
50 55 60
Lys ThrIle GlyValAsp MetAspArg IlePheAspIle SerLysSer
65 70 75 80
Ile Ser Thr Lys Asp Leu Val Glu Leu Ile Val Glu Asp Glu Ser Thr
85 90 95
Leu Lys Val Lys Phe Gly Ser Val Glu Tyr Lys Val Ala Leu Ile Asp
100 105 110
Pro Ser Ala Ile Arg Lys Glu Pro Arg Ile Pro Glu Leu Glu Leu Pro
115 120 125
Ala Lys Ile Val Met Asp Ala Gly Glu Phe Lys Lys Ala Ile Ala Ala
130 135 140
Ala Asp Lys Ile Ser Asp Gln Val Ile Phe Arg Ser Asp Lys Glu Gly
145 150 155 160
Phe Arg Ile Glu Ala Lys Gly Asp Val Asp Ser Ile Val Phe His Met
165 170 175
Thr Glu Thr Glu Leu Ile Glu Phe Asn Gly Gly Glu Ala Arg Ser Met
180 185 190
Phe Ser Val Asp Tyr Leu Lys Glu Phe Cys Lys Val Ala Gly Ser Gly
195 200 205
Asp Leu Leu Thr Ile His Leu Gly Thr Asn Tyr Pro Val Arg Leu Val
210 215 220
Phe Glu Leu Val Gly Gly Arg Ala Lys Val Glu Tyr Ile Leu Ala Pro
225 230 235 240
Arg Ile Glu Ser Glu
245
(2) INFORMATION FOR SEQ ID NO: 13:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 247 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
CA 02338185 2001-02-05
- 27 -
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanococcus jannaschii
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 13:
Met Phe Arg Gly Val Met Glu Ser Ala Lys Glu Phe Lys Lys Val Val
1 5 10 15
Asp Thr Ile Ser Thr Leu Leu Asp Glu Ile Cys Phe Glu Val Asp Glu
20 25 30
Glu Gly Ile Lys Ala Ser Ala Met Asp Pro Ser His Val Ala Leu Val
35 40 45
Ser Leu Glu Ile Pro Arg Leu Ala Phe Glu Glu Tyr Glu Ala Asp Ser
50 55 60
His Asp Ile Gly Ile Asp Leu Glu Ala Phe Lys Lys Val Met Asn Arg
65 70 75 80
Ala Lys Ala Lys Asp Arg Leu Ile Leu Glu Leu Asp Glu Glu Lys Asn
85 90 95
Lys Leu Asn Val Ile Phe Glu Asn Thr Gly Lys Arg Lys Phe Ser Leu
100 105 110
Rla Leu Leu Asp Ile Ser Ala Ser Ser Val Lys Val Pro Glu Ile Glu
115 120 125
Tyr Pro Asn Val Ile Met Ile Lys Gly Asp Ala Phe Lys Glu Ala Leu
130 135 140
Lys Asp Ala Asp Leu Phe Ser Asp Tyr Val Ile Leu Lys Ala Asp Glu
145 150 155 160
Asp Lys Phe Val Ile His Ala Lys Gly Asp Leu Asn Glu Asn Glu Ala
165 170 175
Ile Phe Glu Lys Asp Ser Ser Ala Ile Ile Ser Leu Glu Val Lys Glu
180 185 190
Glu Ala Lys Ser Ala Phe Asn Leu Asp Tyr Leu Met Asp Met Val Lys
195 200 205
Gly Val Ser Ser Gly Asp Ile Ile Lys Ile Tyr Leu Gly Asn Asp Met
210 215 220
Pro Leu Lys Leu Glu Tyr Ser Ile Ala Gly Val Asn Leu Thr Phe Leu
225 230 235 240
Leu Ala Pro Arg Ile Glu Gly
245
(2) INFORMATION FOR SEQ ID N0: 14:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 299 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
CA 02338185 2001-02-05
- 28 -
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Pyrococcus horikoshii
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 14:
Met Pro Phe Glu Ile Val Phe Glu Gly Ala Lys Glu Phe Ala Gln Leu
1 5 10 15
Ile Glu Thr Ala Ser Arg Leu Ile Asp Glu Ala Ala. Phe Lys Val Thr
20 25 30
Glu Glu Gly Ile Ser Met Arg Ala Met Asp Pro Ser Arg Val Val Leu
35 40 45
Ile Asp Leu Asn Leu Pro Ser Ser Ile Phe Ser Lys Tyr Glu Val Asp
50 55 60
Gly Glu Glu Thr Ile Gly Val Asn Met Asp His Leu Lys Lys Val Leu
65 70 75 80
Lys Arg Gly Lys Ala Lys Asp Thr Leu Ile Leu Arg Lys Gly Glu Glu
85 90 95
Asn Phe Leu Glu Ile Ser Leu Gln Gly Thr Ala Thr Arg Thr Phe Arg
100 105 110
Leu Pro Leu Ile Asp Val Glu Glu Ile Glu Val Glu Leu Pro Asp Leu
115 120 125
Pro Tyr Thr Ala Lys Val Val Val Leu Gly Glu Val Leu Lys Glu Ala
130 135 140
Val Lys Asp Ala Ser Leu Val Ser Asp Ser Ile Lys Phe Met Ala Lys
145 150 155 160
Glu Asn Glu Phe Ile Met Arg Ala Glu Gly Glu Thr Gln Glu Val Glu
165 170 175
Val Lys Leu Thr Leu Glu Asp Glu Gly Leu Leu Asp Ile Glu Val Gln
180 185 190
Glu Glu Thr Lys Ser Ala Tyr Gly Val Ser Tyr Leu Ala Asp Met Val
195 200 205
Lys Gly Ile Gly Lys Ala Asp Glu Val Thr Met Arg Phe Gly Asn Glu
210 215 220
Met Pro Met Gln Met Glu Tyr Tyr Ile Arg Asp Glu Gly Arg Leu Thr
225 230 235 240
Phe Leu Leu Ala Pro Arg Val Glu Glu
245
(2) INFORMATION FOR SEQ ID NO: 15:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 244 amino acids
CA 02338185 2001-02-05
- 29 -
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanobacterium thermoautotrophicum
(xi)SEQUENCE 15:
DESCRIPTION:
SEQ
ID
NO:
MetPheLys AlaGluLeu AsnAsp ProAsnIleLeu ArgThrSer Phe
1 5 10 15
AspAlaIle SerSerIle ValAsp GluValGlnIle GlnLeuSer Ala
20 25 30
GluGlyLeu ArgLeuAsp AlaLeu AspArgSerHis IleThrTyr Val
35 40 95
HisLeuGlu LeuLysAla GluLeu PheAspGluTyr ValCysAsp Glu
50 55 60
Pro Glu Arg Ile Asn Val Asp Thr Glu Glu Leu Met Lys Val Leu Lys
65 70 75 80
Arg Ala Lys Ala Asn Asp Arg Val Ile Leu Ser Thr Asp Glu Gly Asn
85 90 95
Leu Ile Ile Gln Phe Glu Gly Glu Ala Val Arg Thr Phe Lys Ile Arg
100 105 110
Leu Ile Asp Ile Glu Tyr Glu Thr Pro Ser Pro Pro Glu Ile Glu Tyr
115 120 125
Glu Asn Glu Phe Glu Val Pro Phe Gln Leu Leu Lys Asp Ser Ile Ala
130 135 140
Asp Ile Rsp Ile Phe Ser Asp Lys Ile Thr Phe Arg Val Asp Glu Asp
145 150 155 160
Arg Phe Ile Ala Ser Ala Glu Gly Glu Phe Gly Asp Ala Gln Ile Glu
165 170 175
Tyr Leu His Gly Glu Arg Ile Asp Lys Pro Ala Arg Ser Ile Tyr Ser
180 185 190
Leu Asp Lys Ile Lys Glu Met Leu Lys Ala Asp Lys Phe Ser Glu Thr
195 200 205
Ala Ile Ile Asn Leu Gly Asp Asp Met Pro Leu Lys Leu Thr Leu Lys
210 215 220
Met Ala Ser Lys Glu Gly Glu Leu Ser Phe Leu Leu Ala Pro Arg Ile
225 230 235 240
Glu Ala Glu Glu
(2) INFORMATION FOR SEQ ID NO: 16:
CA 02338185 2001-02-05
- 30 -
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 469 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Homo Sapiens
(xi)SEQUENCE 16:
DESCRIPTION:
SEQ
ID
N0:
MetPheSer GluGlnAla AlaGlnArg AlaHisThr LeuLeuSer Pro
1 5 10 15
ProSerAla AsnAsnAla ThrPheAla ArgValPro ValAlaThr Tyr
20 25 30
ThrAsnSer SerGlnPro PheArgLeu GlyGluArg SerPheSer Arg
35 40 45
GlnTyrAla HisIleTyr AlaThrArg LeuIleGln MetArgPro Phe
50 55 60
LeuGluAsn ArgAlaGln GlnHisTrp GlySerGly ValGlyVal Lys
65 70 75 80
LysLeuCys GluLeuGln ProGluGlu LysCysCys ValValGly Thr
85 90 95
LeuPheLys AlaMetPro LeuGlnPro SerIleLeu ArgGluVal Ser
100 105 110
GluGluHis AsnLeuLeu ProGlnPro ProArgSer LysTyrIle His
115 120 125
ProAspAsp GluLeuVal LeuGluAsp GluLeuGln ArgIleLys Leu
130 135 140
Lys Gly Thr Ile Asp Val Ser Lys Leu Val Thr Gly Thr Val Leu Ala
145 150 155 160
Val Phe Gly Ser Val Arg Asp Asp Gly Lys Phe Leu Val Glu Asp Tyr
165 170 175
Cys Phe Ala Asp Leu Ala Pro Gln Lys Pro Ala Pro Pro Leu Asp Thr
180 185 190
Asp Arg Phe Val Leu Leu Val Ser Gly Leu Gly Leu Gly Gly Gly Gly
195 200 205
Gly Glu Ser Leu Leu Gly Thr Gln Leu Leu Val Asp Val Val Thr Gly
210 215 220
Gln Leu Gly Asp Glu Gly Glu Gln Cys Ser Ala Ala His Val Ser Arg
225 230 235 240
Val Ile Leu Ala Gly Asn Leu Leu Ser His Ser Thr Gln Ser Arg Asp
245 250 255
CA 02338185 2001-02-05
- 31 -
Ser Ile Asn Lys Ala Lys Tyr Leu Thr Lys Lys Thr Gln Ala Ala Ser
260 265 270
Val Glu Ala Val Lys Met Leu Asp Glu Ile Leu Leu Gln Leu Ser Ala
275 280 285
Ser Val Pro Val Asp Val Met Pro Gly Glu Phe Asp Pro Thr Asn Tyr
290 295 300
Thr Leu Pro Gln Gln Pro Leu His Pro Cys Met Phe Pro Leu Ala Thr
305 310 315 320
Ala Tyr Ser Thr Leu Gln Leu Val Thr Asn Pro Tyr Gln Ala Thr Ile
325 330 335
Asp Gly Val Arg Phe Leu Gly Thr Ser Gly Gln Asn Val Ser Asp Ile
340 345 350
Phe Arg Tyr Ser Ser Met Glu Asp His Leu Glu Ile Leu Glu Trp Thr
355 360 365
Leu Arg Val Arg His Ile Ser Pro Thr Ala Pro Asp Thr Leu Gly Cys
370 375 380
TyrPro PheTyr LysThrAsp ProPheIlePhe ProGluCys ProHis
385 390 395 400
ValTyr PheCys GlyAsnThr ProSerPheGly SerLysIle IleArg
405 410 915
GlyPro GluAsp GlnThrVal LeuLeuValThr ValProAsp PheSer
420 425 430
AlaThr GlnThr AlaCysLeu ValAsnLeuArg SerLeuAla CysGln
435 440 445
Pro Ile Ser Phe Ser Gly Phe Gly Ala Glu Asp Asp Asp Leu Gly Gly
450 455 460
Leu Gly Leu Gly Pro
465
(2) INFORMATION FOR SEQ ID NO: 17:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 488 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeoglobus fulgidus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 17:
Met Val Ile Lys Asn Ile Asp Ala Ala Thr Val Ala Lys Lys Phe Leu
1 5 10 15
CA 02338185 2001-02-05
- 32 -
Val Arg GlyTyrAsn IleAspPro LysAlaAla GluLeuIle CysLys
20 25 30
Ser Gly LeuPheSer AspGluLeu ValAspLys IleCysArg IleAla
35 40 45
Asn Gly GlyPheIle IleGluLys SerValVal GluGluPhe LeuArg
SO 55 60
Asn Leu SerAsnLeu LysProAla ThrLeuThr ProArgPro GluGlu
65 70 75 80
Arg Lys ValGluGlu ValLysAla SerCysIle AlaLeuLys ValIle
85 90 95
Lys Asp IleThrGly LysSerSer CysGlnGly AsnValGlu AspPhe
100 105 110
Leu Met TyrPheAsn SerArgLeu GluLysLeu SerArgIle IleArg
115 120 125
Ser Arg ValAsnThr ThrProIle AlaHisAla GlyLysVal ArgGly
130 135 140
Asn Val SerValVal GlyMetVal AsnGluVal TyrGluArg GlyAsp
145 150 155 160
Lys Cys TyrIleArg LeuGluAsp ThrThrGly ThrIleThr CysVal
165 170 175
Ala Thr GlyLysAsn AlaGluVal AlaArgGlu LeuLeuGly AspGlu
180 185 190
Val Ile GlyValThr GlyLeuLeu LysGlySer SerLeuTyr AlaAsn
195 200 205
Arg Ile ValPhePro AspValPro IleAsnGly AsnGlyGlu LysLys
210 215 220
Arg Asp Phe Tyr Ile Val Phe Leu Ser Asp Thr His Phe Gly Ser Lys
225 230 235 240
Glu Phe Leu Glu Lys Glu Trp Glu Met Phe Val Arg Trp Leu Lys Gly
245 250 255
Glu Val Gly Gly Lys Lys Ser Gln Asn Leu Ala Glu Lys Val Lys Tyr
260 265 270
Ile Val Ile Ala Gly Asp Ile Val Asp Gly Ile Gly Val Tyr Pro Gly
275 280 285
Gln Glu Asp Asp Leu Ala Ile Ser Asp Ile Tyr Gly Gln Tyr Glu Phe
290 295 300
Ala Ala Ser His Leu Rsp Glu Ile Pro Lys Glu Ile Lys Ile Ile Val
305 310 315 320
Ser Pro Gly Asn His Asp Ala Val Arg Gln Ala Glu Pro Gln Pro Ala
325 330 335
Phe Glu Gly Glu Ile Arg Ser Leu Phe Pro Lys Asn Val Glu His Val
340 345 350
CA 02338185 2001-02-05
- 33 -
Gly Asn Pro Ala Tyr Val Asp Ile Glu Gly Val Lys Val Leu Ile Tyr
355 360 365
His Gly Arg Ser Ile Asp Asp Ile Ile Ser Lys Ile Pro Arg Leu Ser
370 375 380
Tyr Asp Glu Pro Gln Lys Val Met Glu Glu Leu Leu Lys Arg Arg His
385 390 395 400
Leu Ser Pro Ile Tyr Gly Gly Arg Thr Pro Leu Ala Pro Glu Arg Glu
405 410 915
Asp Tyr Leu Val Ile Glu Asp Val Pro Asp Ile Leu His Cys Gly His
420 425 430
Ile His Thr Tyr Gly Thr Gly Phe Tyr Arg Gly Val Phe Met Val Asn
435 440 445
Ser Ser Thr Trp Gln Ala Gln Thr Glu Phe Gln Lys Lys Val Asn Leu
450 455 960
Asn Pro Met Pro Gly Asn Val Ala Val Tyr Arg Pro Gly Gly Glu Val
465 470 475 480
Ile Arg Leu Arg Phe Tyr Gly Glu
485
(2) INFORMATION FOR SEQ ID NO: 18:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 594 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanococcus jannaschii
(xi)SEQUENCE 18:
DESCRIPTION:
SEQ
ID
NO:
MetGluIle IleAsnLys PheLeuAsp LeuGluAla LeuLeuSer Pro
1 5 10 15
ThrValTyr GluLysLeu LysAsnPhe AspGluGlu LysLeuLys Arg
20 25 30
LeuIleGln LysIleArg GluPheLys LysTyrAsn AsnAlaPhe Ile
35 40 45
LeuLeuAsp GluLysPhe LeuAspIle PheLeuGln LysRspLeu Asp
50 55 60
GluIleIle AsnGluTyr LysAspPhe AspPheIle PheTyrTyr Thr
65 70 75 80
GlyGluGlu GluLysGlu LysProLys GluValLys LysGluIle Lys
85 90 95
CA 02338185 2001-02-05
- 34 -
LysGlu ThrGluGlu LysIleGlu LysGluLys IleGluPhe ValLys
100 105 110
LysGlu GluLysGlu GlnPheIle LysLysSer AspGluAsp ValGlu
115 120 125
GluLys LeuLysGln LeuIleSer LysGluGlu LysLysGlu AspPhe
130 135 140
AspAla GluArgAla LysArgTyr GluHisIle ThrLysIle LysGlu
145 150 155 160
SerVal AsnSerArg IleLysTrp IleAlaLys AspIleAsp AlaVal
165 170 175
IleGlu IleTyrGlu AspSerAsp ValSerGly LysSerThr CysThr
180 185 190
GlyThr IleGluAsp PheValLys TyrPheArg AspArgPhe GluArg
195 200 205
LeuLys ValPheIle GluArgLys AlaGlnArg LysGlyTyr ProLeu
210 215 220
LysAsp IleLysLys MetLysGly GlnLysAsp IlePheVal ValGly
225 230 235 240
IleVal SerAspVal AspSerThr ArgAsnGly AsnLeuIle ValArg
245 250 255
IleGlu AspThrGlu AspGluAla ThrLeuIle LeuProLys GluLys
260 265 270
IleGlu AlaGlyLys IleProAsp AspIleLeu LeuAspGlu ValIle
275 280 285
Gly Ala Ile Gly Thr Val Ser Lys Ser Gly Ser Ser Ile Tyr Val Asp
290 295 300
Glu Ile Ile Arg Pro Ala Leu Pro Pro Lys Glu Pro Lys Arg Ile Asp
305 310 315 320
Glu Glu Ile Tyr Met Ala Phe Leu Ser Asp Ile His Val Gly Ser Lys
325 330 335
Glu Phe Leu His Lys Glu Phe Glu Lys Phe Ile Arg Phe Leu Asn Gly
340 345 350
Asp Val Asp Asn Glu Leu Glu Glu Lys Val Val Ser Arg Leu Lys Tyr
355 360 365
Ile Cys Ile Ala Gly Asp Leu Val Asp Gly Val Gly Val Tyr Pro Gly
370 375 380
Gln Glu Glu Asp Leu Tyr Glu Val Asp Ile Ile Glu Gln Tyr Arg Glu
385 390 395 400
Ile Ala Met Tyr Leu Asp Gln Ile Pro Glu His Ile Ser Ile Ile Ile
405 410 415
Ser Pro Gly Asn His Asp Ala Val Arg Pro Ala Glu Pro Gln Pro Lys
420 425 430
CA 02338185 2001-02-05
- 35 -
- Leu Pro Glu Lys Ile Thr Lys Leu Phe Asn Arg Asp Asn Ile Tyr Phe
435 940 495
Val Gly Asn Pro Cys Thr Leu Asn Ile His Gly Phe Asp Thr Leu Leu
450 455 460
Tyr His Gly Arg Ser Phe Asp Asp Leu Val Gly Gln Ile Arg Ala Ala
465 470 475 480
Ser Tyr Glu Asn Pro Val Thr Ile Met Lys Glu Leu Ile Lys Arg Arg
485 490 495
Leu Leu Cys Pro Thr Tyr Gly Gly Arg Cys Pro Ile Ala Pro Glu His
500 505 510
Lys Asp Tyr Leu Val Ile Asp Arg Rsp Ile Asp Ile Leu His Thr Gly
515 520 525
His Ile His Ile Asn Gly Tyr Gly Ile Tyr Arg Gly Val Val Met Val
530 535 540
Asn Ser Gly Thr Phe Gln Glu Gln Thr Asp Phe Gln Lys Arg Met Gly
545 550 555 560
Ile Ser Pro Thr Pro Ala Ile Val Pro Ile Ile Asn Met Ala Lys Val
565 570 575
Gly Glu Lys Gly His Tyr Leu Glu Trp Asp Arg Gly Val Leu Glu Val
580 585 590
Arg Tyr
(2) INFORMATION FOR SEQ ID NO: 19:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 622 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Pyrococcus horikoshii
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 19:
Met Asp Glu Phe Val Lys Gly Leu Met Lys Asn Gly Tyr Leu Ile Thr
1 5 10 15
Pro Ser Ala Tyr Tyr Leu Leu Val Gly His Phe Asn Glu Gly Lys Phe
20 25 30
Ser Leu Ile Glu Leu Ile Lys Phe Ala Lys Ser Arg Glu Thr Phe Ile
35 40 45
Ile Asp Asp Glu Ile Ala Asn Glu Phe Leu Lys Ser Ile Gly Ala Glu
50 55 60
CA 02338185 2001-02-05
- - 36 -
Val Glu Leu Pro Gln Glu Ile Lys Glu Gly Tyr Ile Ser Thr Gly Glu
65 70 75 80
Gly Ser Gln Lys Val Pro Asp His Glu Glu Leu Glu Lys Ile Thr Asn
85 90 95
Glu Ser Ser Val Glu Ser Ser Ile Ser Thr Gly Glu Thr Pro Lys Thr
100 105 110
Glu Glu Leu Gln Pro Thr Leu Asp Ile Leu Glu Glu Glu Ile Gly Asp
115 120 125
Ile Glu Gly Gly Glu Ser Ser Ile Ser Thr Gly Asp Glu Val Pro Glu
130 135 140
Val Glu Asn Asn Asn Gly Gly Thr Val Val Val Phe Asp Lys Tyr Gly
145 150 155 160
Tyr Pro Phe Thr Tyr Val Pro Glu Glu Ile Glu Glu Glu Leu Glu Glu
165 170 175
Tyr Pro Lys Tyr Glu Asp Val Thr Ile Glu Ile Asn Pro Asn Leu Glu
180 185 190
Val Val Pro Ile Glu Lys Asp Tyr Glu Ile Lys Phe Asp Val Arg Arg
195 20U 205
Val Lys Leu Lys Pro Pro Lys Val Lys Ser Gly Ser Gly Lys Glu Gly
210 215 220
Glu Ile Ile Val Glu Ala Tyr Ala Ser Leu Phe Arg Ser Arg Leu Arg
225 230 235 240
Lys Leu Arg Arg Ile Leu Arg Glu Asn Pro Glu Val Ser Asn Val Ile
245 250 255
Asp Ile Lys Lys Leu Lys Tyr Val Lys Gly Asp Glu Glu Val Thr Ile
260 265 270
Ile Gly Leu Val Asn Ser Lys Lys Glu Thr Ser Lys Gly Leu Ile Phe
275 280 285
Glu Val Glu Asp Gln Thr Asp Arg Val Lys Val Phe Leu Pro Lys Asp
290 295 300
Ser Glu Asp Tyr Arg Glu Ala Leu Lys Val Leu Pro Asp Ala Val Val
305 310 315 320
Ala Phe Lys Gly Val Tyr Ser Lys Arg Gly Ile Phe Phe Ala Asn Arg
325 330 335
Phe Tyr Leu Pro Asp Val Pro Leu Tyr Arg Lys Gln Lys Pro Pro Leu
340 345 350
Glu Glu Lys Val Tyr Ala Val Leu Thr Ser Asp Ile His Val Gly Ser
355 360 365
Lys Glu Phe Cys Glu Lys Ala Phe Ile Lys Phe Leu Glu Trp Leu Asn
370 375 380
Gly Tyr Val Glu Ser Lys Glu Glu Glu Glu Ile Val Ser Arg Ile Arg
385 390 395 400
CA 02338185 2001-02-05
37
Tyr Leu Ile Ile Ala Gly Asp Val Val Asp Gly Ills Gly Ile Tyr Pro
405 410 415
Gly Gln Tyr Ser Asp Leu Ile Ile Pro Asp Ile Phe Asp Gln Tyr Glu
420 425 430
Ala Leu Ala Asn Leu Leu Ser Asn Val Pro Lys Hip; Ile Thr Ile Phe
435 490 445
Ile Gly Pro Gly Asn His Asp Ala Ala Arg Pro Ala Ile Pro Gln Pro
450 455 460
Glu Phe Tyr Glu Glu Tyr Ala Lys Pro Leu Tyr Lys Leu Lys Asn Thr
465 470 475 480
Val Ile Ile Ser Asn Pro Ala Val Ile Arg Leu His Gly Arg Asp Phe
485 490 495
Leu Ile Ala His Gly Arg Gly Ile Glu Asp Val Val Ser Phe Val Pro
500 505 510
Gly Leu Thr His His Lys Pro Gly Leu Pro Met Val Glu Leu Leu Lys
515 520 525
Met Arg His Leu Ala Pro Thr Phe Gly Gly Lys Val Pro Ile Ala Pro
530 535 540
Asp Pro Glu Asp Leu Leu Val Ile Glu Glu Val Pro Asp Leu Val Gln
545 550 555 560
Met Gly His Val His Val Tyr Asp Thr Ala Val Tyr Arg Gly Val Gln
565 570 575
Leu Val Asn Ser Ala Thr Trp Gln Ala Gln Thr Glu Phe Gln Lys Met
580 585 590
Val Asn Ile Val Pro Thr Pro Gly Leu Val Pro Ile Val Asp Val Glu
595 600 605
Ser Ala Arg Val Ile Lys Val Leu Asp Phe Ser Arg Trp Cys
610 615 620
(2) INFORMATION FOR SEQ ID NO: 20:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 482 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanobacterium thermoautotrophicum
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 20:
Met Asn Glu Ile Ile Gly Lys Phe Ala Arg Glu Gly Ile Leu Ile Glu
1 5 10 15
CA 02338185 2001-02-05
- 38 -
Asp Asn Ala Tyr Phe Arg Leu Arg Glu Met Asp Asp Pro Ala Ser Val
20 25 30
Ser Ser Glu Leu Ile Val Lys Ile Lys Ser Asn Gl;y Gly Lys Phe Thr
35 40 45
Val Leu Thr Ser Glu Met Leu Asp Glu Phe Phe Glu Ile Asp Asn Pro
50 55 60
Ala Glu Ile Lys Ala Arg Gly Pro Leu Met Val Pro Ala Glu Arg Asp
65 70 75 80
Phe Asp Phe Glu Val Ile Ser Asp Thr Ser Asn Arg Ser Tyr Thr Ser
85 90 95
Gly Glu Ile Gly Asp Met Ile Rla Tyr Phe Asn Ser Arg Tyr Ser Ser
100 105 110
Leu Lys Asn Leu Leu Ser Lys Arg Pro Glu Leu Lys Gly His Ile Pro
115 120 125
Ile Ala Asp Leu Arg Gly Gly Glu Asp Val Val Ser Ile Ile Gly Met
130 135 140
Val Asn Asp Val Arg Asn Thr Lys Asn Asn His Arg Ile Ile Glu Leu
145 150 155 160
Glu Rsp Asp Thr Gly Glu Ile Ser Val Val Val His Asn Glu Asn His
165 170 175
Lys Leu Phe Glu Lys Ser Glu Lys Ile Val Arg Asp Glu Val Val Gly
180 185 190
Val His Gly Thr Lys Lys Gly Arg Phe Val Val Ala Ser Glu Ile Phe
195 200 205
His Pro Gly Val Pro Arg Ile Gln Glu Lys Glu Met Asp Phe Ser Val
210 215 220
Ala Phe Ile Ser Asp Val His Ile Gly Ser Gln Thr Phe Leu Glu Asp
225 230 235 240
Ala Phe Met Lys Phe Val Lys Trp Ile Asn Gly Asp Phe Gly Ser Glu
245 250 255
Glu Gln Arg Ser Leu Ala Ala Asp Val Lys Tyr Leu Val Val Ala Gly
260 265 270
Asp Ile Val Asp Gly Ile Gly Ile Tyr Pro Gly Gln Glu Lys Glu Leu
275 280 285
Leu Ile Arg Asp Ile His Glu Gln Tyr Glu Glu Rla Ala Arg Leu Phe
290 295 300
Gly Asp Ile Arg Ser Asp Ile Lys Ile Val Met Ile Pro Gly Asn His
305 310 315 320
Asp Ser Ser Arg Ile Ala Glu Pro Gln Pro Ala Ile Pro Glu Glu Tyr
325 330 335
Ala Lys Ser Leu Tyr Ser Ile Arg Asn Ile Glu Phe Leu Ser Asn Pro
340 345 350
CA 02338185 2001-02-05
- 39 -
SerLeuVal SerLeuAsp Gly Arg ThrLeu IleTyrHisGly
Val Rrg
355 360 365
SerPheAsp AspMetAla MetSerVal AsnGly LeuSerHisGlu Arg
370 375 380
SerAspLeu IleMetGlu GluLeuLeu GluLys ArgHisLeuAla Pro
385 390 395 400
IleTyrGly GluArgThr ProLeuAla SerGlu IleGluAspHis Leu
405 410 415
ValIleAsp GluValPro HisValLeu HisThr GlyHisValHis Ile
420 425 430
AsnAlaTyr LysLysTyr LysGlyVal HisLeu IleAsnSerGly Thr
435 440
445
PheGlnSer GlnThrGlu PheGlnLys IleTyr AsnIleValPro Thr
450
455 460
CysGlyGln ValProVal LeuAsnArg GlyVal MetLysLeuLeu Glu
465 470 475 480
PheSer
(2) INFORMATION FOR SEQ ID NO: 21:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 613 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Pyrococcus furiosus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 21:
Met Asp Glu Phe Val Lys Ser Leu Leu Lys Ala Asn Tyr Leu Ile Thr
1 5 10 15
Pro Ser Ala Tyr Tyr Leu Leu Arg Glu Tyr Tyr Glu Lys Gly Glu Phe
20 25 30
Ser Ile Val Glu Leu Val Lys Phe Ala Arg Ser Arg Glu Ser Tyr Ile
35 40 45
Ile Thr Asp Ala Leu Ala Thr Glu Phe Leu Lys Val Lys Gly Leu Glu
50 55 60
Pro Ile Leu Pro Val Glu Thr Lys Gly Gly Phe Val Ser Thr Gly Glu
65 70 75 80
Ser Gln Lys Glu Gln Ser Tyr Glu Glu Ser Phe Gly Thr Lys Glu Glu
85 90 95
CA 02338185 2001-02-05
- 40 -
Ile Ser Gln Glu Ile Lys Glu Gly Glu Ser Phe Ile Ser Thr Gly Ser
100 105 110
Glu Pro Leu Glu Glu Glu Leu Asn Ser Ile Gly Ile Glu Glu Ile Gly
115 120 125
Ala Asn Glu Glu Leu Val Ser Asn Gly Asn Asp Asn Gly Gly Glu Ala
130 135 140
Ile Val Phe Asp Lys Tyr Gly Tyr Pro Met Val Ty:r Ala Pro Glu Glu
145 150 155 160
Ile Glu Val Glu Glu Lys Glu Tyr Ser Lys Tyr Glu Asp Leu Thr Ile
165 170 175
Pro Met Asn Pro Asp Phe Asn Tyr Val Glu Ile Lys Glu Asp Tyr Asp
180 185 190
Val Val Phe Asp Val Arg Rsn Val Lys Leu Lys Pro Pro Lys Val Lys
195 200 205
Asn Gly Asn Gly Lys Glu Gly Glu Ile Ile Val Glu Ala Tyr Ala Ser
210
215
220
Leu Phe Arg Ser Arg Leu Lys Lys Leu Arg Lys Ile Leu Arg Glu Asn
225 230 235 240
Pro Glu Leu Asp Asn Val Val Asp Ile Gly Lys Leu Lys Tyr Val Lys
245 250 255
Glu Asp Glu Thr Val Thr Ile Ile Gly Leu Val Asn Ser Lys Arg Glu
260 265 270
Val Asn Lys Gly Leu Ile Phe Glu Ile Glu Asp Leu Thr Gly Lys Val
275 280 285
Lys Val Phe Leu Pro Lys Asp Ser Glu Asp Tyr Arg Glu Ala Phe Lys
290 295 300
Val Leu Pro Asp Ala Val Val Ala Phe Lys Gly Val Tyr Ser Lys Arg
305 310 315 320
Gly Ile Leu Tyr Ala Asn Lys Phe Tyr Leu Pro Asp Val Pro Leu Tyr
325 330 335
Arg Arg Gln Lys Pro Pro Leu Glu Glu Lys Val Tyr Ala Ile Leu Ile
340 345 350
Ser Asp Ile His Val Gly Ser Lys Glu Phe Cys Glu Asn Ala Phe Ile
355 360 365
Lys Phe Leu Glu Trp Leu Asn Gly Asn Val Glu Thr Lys Glu Glu Glu
370 375 380
Glu Ile Val Ser Arg Val Lys Tyr Leu Ile Ile Ala Gly Asp Val Val
385 390 395 400
Asp Gly Val Gly Val Tyr Pro Gly Gln Tyr Ala Asp Leu Thr Ile Pro
405 410 415
Asp Ile Phe Asp Gln Tyr Glu Ala Leu Ala Asn Leu Leu Ser His Val
420 425 430
CA 02338185 2001-02-05
- 41 -
ProLysHisIle ThrMetPhe IleAla ProGly HisAspAla Ala
Asn
435 440 445
ArgGlnAlaIle ProGlnPro GluPhe TyrLysGlu TyrAlaLys Pro
450 455 460
IleTyrLysLeu LysAsnAla ValIle IleSerAsn ProAlaVal Ile
465 470 475 480
ArgLeuHisGly ArgAspPhe LeuIle AlaHisGly ArgGlyIle Glu
485 490 495
AspValValGly SerValPro GlyLeu ThrHisHis LysProGly Leu
500 505 510
ProMetValGlu LeuLeuLys MetArg HisValAla ProMetPhe Gly
515 520 525
GlyLysValPro IleAlaPro AspPro GluAspLeu LeuValIle Glu
530 535 540
GluValProAsp ValValHis MetGly HisValHis ValTyrAsp Ala
545 550 555 560
ValValTyrArg GlyValGln LeuVal AsnSerAla ThrTrpGln Ala
565 570 575
GlnThrGluPhe GlnLysMet ValAsn IleValPro ThrProAla Lys
580 585 590
ValProValVal AspIleAsp ThrAla LysValVal LysValLeu Asp
595 600 605
PheSerGlyTrp Cys
610
(2) INFORMATION FOR SEQ ID NO: 22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1107 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Homo Sapiens
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 22:
MetAsp LysArgArg Pro Pro Gly Gly Val Pro Pro
Gly Gly Pro Lys
1 5 10 15
ArgAla GlyGlyLeu Trp Asp Asp Ala Pro Trp Pro
Arg Asp Asp Ser
20 25 30
GlnPhe GluAspLeu Ala Met Glu Met Glu Ala Glu
Glu Leu Glu His
35 40 45
CA 02338185 2001-02-05
- 42 -
Arg Leu Gln Glu Gln Glu Glu Glu Glu Leu Gln Ser Val Leu Glu Gly
50 55 60
Val Ala Asp Gly Gln Val Pro Pro Ser Ala Ile Asp Pro Arg Trp Leu
65 70 75 80
Arg Pro Thr Pro Pro Ala Leu Asp Pro Gln Thr Glu Pro Leu Ile Phe
85 90 95
Gln Gln Leu Glu Ile Asp His Tyr Val Gly Pro Ala Gln Pro Val Pro
100 105 110
Gly Gly Pro Pro Pro Ser Arg Gly Ser Val Pro Val Leu Arg Ala Phe
115 120 125
Gly Val Thr Asp Glu Gly Phe Ser Val Cys Cys Hip; Ile His Gly Phe
130 135 140
Ala Pro Tyr Phe Tyr Thr Pro Ala Pro Pro Gly Phe Gly Pro Glu His
145 150 155 160
Met Gly Asp Leu Gln Arg Glu Leu Asn Leu Ala Ile Ser Arg Asp Ser
165 170 175
Arg Gly Gly Arg Glu Leu Thr Gly Pro Ala Val Leu Ala Val Glu Leu
180 185 190
Cys Ser Arg Glu Ser Met Phe Gly Tyr His Gly His Gly Pro Ser Pro
195 200 205
Phe Leu Arg Ile Thr Val Ala Leu Pro Arg Leu Val Ala Pro Ala Arg
210 215 220
Arg Leu Leu Glu Gln Gly Ile Arg Val Ala Gly Leu Gly Thr Pro Ser
225 230 235 240
Phe Ala Pro Tyr Glu Ala Asn Val Asp Phe Glu Ile Arg Phe Met Val
245 250 255
Asp Thr Asp Ile Val Gly Cys Asn Trp Leu Glu Leu Pro Ala Gly Lys
260 265 270
Tyr Ala Leu Arg Leu Lys Glu Lys Ala Thr Gln Cys Gln Leu Glu Ala
275 280 285
Asp Val Leu Trp Ser Asp Val Val Ser His Pro Pro Glu Gly Pro Trp
290 295 300
Gln Arg Ile Ala Pro Leu Arg Val Leu Ser Phe Asp Ile Glu Cys Ala
305 310 315 320
Gly Arg Lys Gly Ile Phe Pro Glu Pro Glu Arg Asp Pro Val Ile Gln
325 330 335
Ile Cys Ser Leu Gly Leu Arg Trp Gly Glu Pro Glu Pro Phe Leu Arg
340 345 350
Leu Ala Leu Thr Leu Arg Pro Cys Ala Pro Ile Leu Gly Ala Lys Val
355 360 365
Gln Ser Tyr Glu Lys Glu Glu Asp Leu Leu Gln Ala Trp Ser Thr Phe
370 375 380
CA 02338185 2001-02-05
- 43 -
Ile Arg Ile Met Asp Pro Asp Val Ile Thr Gly Tyr Asn Ile Gln Asn
385 390 395 400
Phe Asp Leu Pro Tyr Leu Ile Ser Arg Ala Gln Thr_ Leu Lys Val Gln
405 410 915
Thr Phe Pro Phe Leu Gly Arg Val Ala Gly Leu Cys Ser Asn Ile Arg
420 425 430
Asp Ser Ser Phe Gln Ser Lys Gln Thr Gly Arg Arg_ Asp Thr Lys Val
435 440 445
Val Ser Met Val Gly Arg Val Gln Met Asp Met Leu Gln Val Leu Leu
450 455 460
Arg Glu Tyr Lys Leu Arg Ser His Thr Leu Asn Rla. Val Ser Phe His
465 470 4?5 480
Phe Leu Gly Glu Gln Lys Glu Asp Val Gln His Ser Ile Ile Thr Asp
485 490 495
Leu Gln Asn Gly Asn Asp Gln Thr Arg Arg Arg Leu Ala Val Tyr Cys
500 505 510
Leu Lys Asp Ala Tyr Leu Pro Leu Arg Leu Leu Glu Arg Leu Met Val
515 520 525
Leu Val Asn Ala Val Glu Met Ala Arg Val Thr Gly Val Pro Leu Ser
530 535 540
Tyr Leu Leu Ser Arg Gly Gln Gln Val Lys Val Val Ser Gln Leu Leu
545 550 555 560
Arg Gln Ala Met His Glu Gly Leu Leu Met Pro Val Val Lys Ser Glu
565 570 575
Gly Gly Glu Asp Tyr Thr Gly Ala Thr Val Ile Glu Pro Leu Lys Gly
580 585 590
Tyr Tyr Asp Val Pro Ile Ala Thr Leu Asp Phe Ser Ser Leu Tyr Pro
595 600 605
Ser Ile Met Met Ala His Asn Leu Cys Tyr Thr Thr Leu Leu Arg Pro
610 615 620
Gly Thr Ala Gln Lys Leu Gly Leu Thr Glu Asp Gln Phe Ile Arg Thr
625 630 635 640
Pro Thr Gly Asp Glu Phe Val Lys Thr Ser Val Arg Lys Gly Leu Leu
645 650 655
Pro Gln Ile Leu Glu Asn Leu Leu Ser Ala Arg Lys Arg Ala Lys Ala
660 665 670
Glu Leu Ala Lys Glu Thr Asp Pro Leu Arg Arg Gln Val Leu Asp Gly
675 680 685
Arg Gln Leu Ala Leu Lys Val Ser Ala Asn Ser Val Tyr Gly Phe Thr
690 695 700
Gly Ala Gln Val Gly Lys Leu Pro Cys Leu Glu Ile Ser Gln Ser Val
705 710 715 720
CA 02338185 2001-02-05
_ 44 _
Thr Gly Phe Gly Arg Gln Met Ile Glu Lys Thr Lys Gln Leu Val Glu
725 730 735
Ser Lys Tyr Thr Val Glu Asn Gly Tyr Ser Thr Ser Ala Lys Val Val
740 745 750
Tyr Gly Asp Thr Asp Ser Val Met Cys Arg Phe Gl.y Val Ser Ser Val
755 760 765
Ala Glu Ala Met Ala Leu Gly Arg Glu Ala Ala Asp Trp Val Ser Gly
770 775 780
His Phe Pro Ser Pro Ile Arg Leu Glu Phe Glu Lys Val Tyr Phe Pro
785 790 795 800
Tyr Leu Leu Ile Ser Lys Lys Arg Tyr Ala Gly Leu Leu Phe Ser Ser
805 810 815
Arg Pro Asp Ala His Asp Arg Met Asp Cys Lys Gly Leu Glu Ala Val
820 825 830
Arg Arg Asp Asn Cys Pro Leu Val Ala Asn Leu Val Thr Ala Ser Leu
835 840 845
Arg Arg Leu Leu Ile Asp Arg Asp Pro Glu Gly Ala Val Ala His Ala
850 855 860
Gln Asp Val Ile Ser Asp Leu Leu Cys Asn Arg IlE= Asp Ile Ser Gln
865 870 875 880
Leu Val Ile Thr Lys Glu Leu Thr Arg Ala Ala Ser_ Asp Tyr Ala Gly
885 890 895
Lys Gln Ala His Val Glu Leu Ala Glu Arg Met Ar<_~ Lys Arg Asp Pro
900 905 910
Gly Ser Ala Pro Ser Leu Gly Asp Arg Val Pro Tyt: Val Ile Ile Ser
915 920 925
Ala Ala Lys Gly Val Ala Ala Tyr Met Lys Ser Glu Asp Pro Leu Phe
930 935 940
Val Leu Glu His Ser Leu Pro Ile Asp Thr Gln Tyr Tyr Leu Glu Gln
945 950 955 960
Gln Leu Ala Lys Pro Leu Leu Arg Ile Phe Glu Pro Ile Leu Gly Glu
965 970 975
Gly Arg Ala Glu Ala Val Leu Leu Arg Gly Asp His Thr Arg Cys Lys
980 985 990
Thr Val Leu Thr Gly Lys Val Gly Gly Leu Leu Ala Phe Ala Lys Arg
995 1000 1005
Arg Asn Cys Cys Ile Gly Cys Arg Thr Val Leu Ser His Gln Gly Ala
1010 1015 1020
Val Cys Glu Phe Cys Gln Pro Arg Glu Ser Glu Leu Tyr Gln Lys Glu
1025 1030 1035 1040
Val Ser His Leu Asn Ala Leu Glu Glu Arg Phe Ser Arg Leu Trp Thr
1045 1050 1055
CA 02338185 2001-02-05
- 45 -
Gln Cys Gln Arg Cys Gln Gly Ser Leu His Glu Asp Val Ile Cys Thr
1060 1065 1070
Ser Arg Asp Cys Pro Ile Phe Tyr Met Arg Lys Lys Val Arg Lys Asp
1075 1080 1085
Leu Glu Asp Gln Glu Gln Leu Leu Arg Arg Phe Gly Pro Pro Gly Pro
1090 1095 1100
Glu Ala Trp
1105
(2) INFORMATION FOR SEQ ID NO: 23:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 781 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeoglobus fulgidus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 23:
Met Glu Arg Val Glu Gly Trp Leu Ile Asp Ala Asp Tyr Glu Thr Ile
1 5 10 15
Gly Gly Lys Ala Val Val Arg Leu Trp Cys Lys Asp Asp Gln Gly Ile
20 25 30
Phe Val Ala Tyr Asp Tyr Asn Phe Asp Pro Tyr Phe Tyr Val Ile Gly
35 40 95
Val Asp Glu Asp Ile Leu Lys Asn Ala Ala Thr Ser Thr Arg Arg Glu
50 55 60
Val Ile Lys Leu Lys Ser Phe Glu Lys Ala Gln Leu Lys Thr Leu Gly
65 70 75 80
Arg Glu Val Glu Gly Tyr Ile Val Tyr Ala His His Pro Gln His Val
85 90 95
Pro Lys Leu Arg Asp Tyr Leu Ser Gln Phe Gly Asp Val Arg Glu Ala
100 105 110
Asp Ile Pro Phe Ala Tyr Arg Tyr Leu Ile Asp Lys Asp Leu Ala Cys
115 120 125
Met Asp Gly Ile Ala Ile Glu Gly Glu Lys Gln Gly Gly Val Ile Arg
130 135 140
Ser Tyr Lys Ile Glu Lys Val Glu Arg Ile Pro Arg Met Glu Phe Pro
145 150 155 160
Glu Leu Lys Met Leu Val Phe Asp Cys Glu Met Leu Ser Ser Phe Gly
165 170 175
CA 02338185 2001-02-05
- 46 -
Met Pro Glu Pro Glu Lys Asp Pro Ile Ile Val Ile Ser Val Lys Thr
180 185 190
Asn Asp Asp Asp Glu Ile Ile Leu Thr Gly Asp Glu Arg Lys Ile Ile
195 200 205
Ser Asp Phe Val Lys Leu Ile Lys Ser Tyr Asp Pro Asp Ile Ile Val
210 215 220
Gly Tyr Asn Gln Asp Ala Phe Asp Trp Pro Tyr Leu Arg Lys Arg Ala
225 230 235 240
Glu Arg Trp Asn Ile Pro Leu Asp Val Gly Arg Asp Gly Ser Asn Val
245 250 255
Val Phe Arg Gly Gly Arg Pro Lys Ile Thr Gly Arg_ Leu Asn Val Asp
260 265 270
Leu Tyr Asp Ile Ala Met Arg Ile Ser Asp Ile Lys Ile Lys Lys Leu
275 280 285
Glu Asn Val Ala Glu Phe Leu Gly Thr Lys Ile Glu Ile Ala Asp Ile
290 295 300
Glu Ala Lys Asp Ile Tyr Arg Tyr Trp Ser Arg Gly Glu Lys Glu Lys
305 310 315 320
Val Leu Asn Tyr Ala Arg Gln Asp Ala Ile Asn Thr Tyr Leu Ile Ala
325 330 335
Lys Glu Leu Leu Pro Met His Tyr Glu Leu Ser Lys Met Ile Arg Leu
340 345 350
Pro Val Asp Asp Val Thr Arg Met Gly Arg Gly Lys Gln Val Asp Trp
355 360 365
Leu Leu Leu Ser Glu Ala Lys Lys Ile Gly Glu Ile Ala Pro Asn Pro
370 375 380
Pro Glu His Ala Glu Ser Tyr Glu Gly Ala Phe Val Leu Glu Pro Glu
385 390 395 400
Arg Gly Leu His Glu Asn Val Ala Cys Leu Asp Phe Ala Ser Met Tyr
405 410 415
Pro Ser Ile Met Ile Ala Phe Asn Ile Ser Pro Asp Thr Tyr Gly Cys
420 425 430
Arg Asp Asp Cys Tyr Glu Ala Pro Glu Val Gly His Lys Phe Arg Lys
435 440 445
Ser Pro Asp Gly Phe Phe Lys Arg Ile Leu Arg Met Leu Ile Glu Lys
450 455 460
Arg Arg Glu Leu Lys Val Glu Leu Lys Asn Leu Ser Pro Glu Ser Ser
465 470 475 480
Glu Tyr Lys Leu Leu Asp Ile Lys Gln Gln Thr Leu Lys Val Leu Thr
485 490 495
Asn Ser Phe Tyr Gly Tyr Met Gly Trp Asn Leu Ala Arg Trp Tyr Cys
500 505 510
CA 02338185 2001-02-05
_ 47 _
His Pro Cys Ala Glu Ala Thr Thr Ala Trp Gly Arg His Phe Ile Arg
515 520 525
Thr Ser Ala Lys Ile Ala Glu Ser Met Gly Phe Lys Val Leu Tyr Gly
530 535 54()
Asp Thr Asp Ser Ile Phe Val Thr Lys Ala Gly Met. Thr Lys Glu Asp
545 550 555 560
Val Asp Arg Leu Ile Asp Lys Leu His Glu Glu Leu Pro Ile Gln Ile
565 570 575
Glu Val Asp Glu Tyr Tyr Ser Ala Ile Phe Phe Val. Glu Lys Lys Arg
580 585 590
Tyr Ala Gly Leu Thr Glu Asp Gly Arg Leu Val Val. Lys Gly Leu Glu
595 600 605
Val Arg Arg Gly Asp Trp Cys Glu Leu Rla Lys Lys Val Gln Arg Glu
610 615 62G
Val Ile Glu Val Ile Leu Lys Glu Lys Asn Pro Glu Lys Ala Leu Ser
625 630 635 640
Leu Val Lys Asp Val Ile Leu Arg Ile Lys Glu Gly Lys Val Ser Leu
645 650 655
Glu Glu Val Val Ile Tyr Lys Gly Leu Thr Lys Lys Pro Ser Lys Tyr
660 665 670
Glu Ser Met Gln Ala His Val Lys Ala Ala Leu Lys Ala Arg Glu Met
675 680 685
Gly Ile Ile Tyr Pro Val Ser Ser Lys Ile Gly Tyr Val Ile Val Lys
690 695 700
Gly Ser Gly Asn Ile Gly Asp Arg Ala Tyr Pro Ile Asp Leu Ile Glu
705 710 715 720
Asp Phe Asp Gly Glu Asn Leu Arg Ile Lys Thr Lys Ser Gly Ile Glu
725 730 735
Ile Lys Lys Leu Asp Lys Asp Tyr Tyr Ile Asp Asn Gln Ile Ile Pro
740 745 750
Ser Val Leu Arg Ile Leu Glu Arg Phe Gly Tyr Thr Glu Ala Ser Leu
755 760 765
Lys Gly Ser Ser Gln Met Ser Leu Asp Ser Phe Phe Ser
770 775 780
(2) INFORMATION FOR SEQ ID NO: 24:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1634 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanococcus jannaschii
CA 02338185 2001-02-05
_ 48 _
(xi)SEQUENCE
DESCRIPTION:
SEQ
ID
N0:
24:
Met LysIle Ile Asn
Gly Asp Asp
Met Ala
Ser Leu
Met
Gly
Lys
Ile
1 5 10 15
Thr AlaVal Ile : Ile
Tyr Tyi Leu
Lys Tyr
Thr Leu
Ile
Glu
Asp
Lys
20 25 30
Asn Lys Asp Arg PheLys Pro Phe ValGlu
Ser Asp Tyr Tyr
Ile
Leu
35 40 45
Leu GluLys Val Glu GluAsp Ile LysIle LysGlu
His Asn Glu.
Lys
50 55 60
PheLeu LysAsn Asp Leu LysPhe ValGluAsnIle GluVal
Leu Leu
65 70 75 80
ValLysLys IleIle Leu Arg GluLys GluValIleLys IleIle
Lys
85 90 95
AlaThrHis ProGln Lys Val LysLeu ArgLysIleLys GluCys
Pro
100 105 110
GluIleVal LysGlu Ile Tyr HisAsp IleProPheAla LysArg
Glu
115 120 125
TyrLeuIle AspAsn Glu Ile ProMet ThrTyrTrpAsp PheGlu
Ile
130 135 140
AsnLysLys ProVal Ser Ile IlePro LysLeuLysSer ValAla
Glu
145 150 155 160
PheAspMet GluVal Tyr Asn AspThr GluProAsnPro GluArg
Arg
165 170 175
AspProIle LeuMet Ala Ser TrpAsp GluRsnGlyGly LysVal
Phe
180 185 190
IleThrTyr LysGlu Phe Asn ProAsn IleGluValVal LysAsn
His
195 200 205
GluLysGlu LeuIle Lys Lys IleGlu ThrLeuLysGlu TyrAsp
Ile
210 215 220
ValIleTyr ThrTyr Asn Gly Phe AspPheProTyr LeuLys
Asp Asn
225 230 235 240
AlaArgAla LysIle Tyr Gly Ile LeuGlyLys AspGly
Ile Asp Asn
245 250 255
GluGluLeu LysIle Lys Arg GluTyrArgSer TyrIle
Gly Gly
Met
260 265 270
ProGlyArg ValHis Ile Asp IleSerArgArg Leu
Leu Tyr . Leu
Pro
275 280 285
LysLeuThr Lys Thr Leu Tyr Leu Phe
Tyr Glu Asp Asn Gly
Val Val
290 295 300
CA 02338185 2001-02-05
- 49 -
- Ile Glu Lys Leu Lys Ile His Thr Lys Ile Val
Pro Asp Tyr Trp Ala
305 310 315 320
Asn Asn Asp Lys Thr Leu Glu Tyr Ser Leu Gln
Ile Asp Ala Lys Tyr
325 330 335
Thr Tyr Lys Ile Gly Lys Phe Phe Pro Leu Glu
Tyr Val Met Phe Ser
340 345 350
Arg Ile Val Asn Gln Thr Phe Glu Ile Thr Arg
Pro Met Ser Ser Gly
355 360 365
Gln Met Val Glu Tyr Leu Met Lys Arg Ala Phe~ Asn
Leu Lys Glu Met
370 375
380
Ile Val Pro Asn Lys Pro Glu Glu Glu Tyr Arg Val
Asp Arg Arg Leu
385 390 395 400
Thr Thr Tyr Glu Gly Gly Val Lys Glu Pro Glu Met
Tyr Lys Gly Phe
405 410 415
Glu Asp Ile Ile Ser Met Phe Arg Cys His Pro Thr
Asp Lys Gly Lys
420 425 430
Val Val Val Lys Gly Lys Ile Val Asn Ile Glu Lys
Gly Asp Val Glu
435 440 q45
Gly Asn Tyr Val Leu Gly Asp Gly Trp Gln Lys Lys
Ile Val Lys Val
450 455 460
Trp Lys Tyr Glu Tyr Glu Glu Leu Ile Asn Val Leu
Gly Asn Gly Lys
465 470 475 480
Cys Thr Pro Asn His Lys Pro Leu Arg Tyr Lys His
Ile Ile Lys Lys
485 490 495
Lys Ile Asn Lys Asn Asp Leu Val Arg Asp Ile Lys
Tyr Tyr Ala Ser
500 505 510
Leu Leu Thr Lys Phe Lys Glu Gly Lys Leu Ile Lys
Gly Leu Cys Asp
515 520 525
Phe Glu Thr Ile Gly Asn Glu Lys Tyr Ile Asn Asp
Tyr Asp Met Glu
530 535 540
Asp Phe Ile Leu Lys Ser Leu Ile Gly Ile Leu Glu
Glu Leu Ala Gly
545 550 555 560
His Leu Leu Gly
Arg Lys
Arg
Asp
Ile
Glu
Tyr
Phe
Asp
Ser
Ser
Arg
565 570 575
Lys Arg Ile Glu
Ser Asp
His
Gln
Tyr
Arg
Val
Glu
Ile
Thr
Val
Asn
580 585 590
Glu Lys Asp Leu
Phe Ile Glu Phe
Lys Ile Lys
Tyr Ile Phe
Lys Lys
595 600 605
Asn Tyr Glu Ile
Leu Tyr Val Thr
Arg Arg Lys
Lys Gly Thr
Lys Ala
610 615 620
Leu Gly Cys
Ala Lys Lys
Asp Ile Tyr
Leu Lys Ile
Glu Glu Ile
Leu
625 630 635 640
CA 02338185 2001-02-05
- 50 -
Lys Asn Lys Glu Lys Tyr Leu Pro Asn Ala Ile Leu Arg Gly Phe Phe
645 650 655
Glu Gly Asp Gly Tyr Val Asn Thr Val Arg Arg Ala Val Val Val Asn
660 665 670
Gln Gly Thr Asn Asn Tyr Asp Lys Ile Lys Phe I1~~ Ala Ser Leu Leu
675 680 685
Asp Arg Leu Gly Ile Lys Tyr Ser Phe Tyr Thr Ty.r Ser Tyr Glu Glu
690 695 7p0
Arg Gly Lys Lys Leu Lys Arg Tyr Val Ile Glu IlE= Phe Ser Lys Gly
705 710 715 720
Asp Leu Ile Lys Phe Ser Ile Leu Ile Ser Phe Ile Ser Arg Arg Lys
725 730 735
Asn Asn Leu Leu Asn Glu Ile Ile Arg Gln Lys Thr Leu Tyr Lys Ile
740 745 750
Gly Asp Tyr Gly Phe Tyr Asp Leu Asp Asp Val Cys Val Ser Leu Glu
755 760 765
Ser Tyr Lys Gly Glu Val Tyr Asp Leu Thr Leu Glu Gly Arg Pro Tyr
770 775 780
Tyr Phe Ala Asn Gly Ile Leu Thr His Asn Ser Leu Tyr Pro Ser Ile
785 790 795 800
Ile Ile Ser Tyr Asn Ile Ser Pro Asp Thr Leu Rsp Cys Glu Cys Cys
805 810 815
Lys Asp Val Ser Glu Lys Ile Leu Gly His Trp Phe Cys Lys Lys Lys
820 825 830
Glu Gly Leu Ile Pro Lys Thr Leu Arg Asn Leu Ile Glu Arg Arg Ile
835 890 845
Asn Ile Lys Arg Arg Met Lys Lys Met Ala Glu Ile Gly Glu Ile Asn
850 855 860
Glu Glu Tyr Asn Leu Leu Asp Tyr Glu Gln Lys Ser Leu Lys Ile Leu
865 870 875 880
Ala Asn Ser Ile Leu Pro Asp Glu Tyr Leu Thr Ile Ile Glu Glu Asp
885 890 895
Gly Ile Lys Val Val Lys Ile Gly Glu Tyr Ile Asp Asp Leu Met Arg
900 905 910
Lys His Lys Asp Lys Ile Lys Phe Ser Gly Ile Ser Glu Ile Leu Glu
915 920 925
Thr Lys Asn Leu Lys Thr Phe Ser Phe Asp Lys Ile Thr Lys Lys Cys
930 935 940
Glu Ile Lys Lys Val Lys Ala Leu Ile Arg His Pro Tyr Phe Gly Lys
945 950 955 960
Ala Tyr Lys Ile Lys Leu Arg Ser Gly Arg Thr Ile Lys Val Thr Arg
965 970 975
CA 02338185 2001-02-05
- 51 -
Gly His Ser Leu Phe Lys Tyr Glu Asn Gly Lys I1e Val Glu Val Lys
980 985 990
Gly Asp Asp Val Arg Phe Gly Asp Leu Ile Val Val Pro Lys Lys Leu
995 1000 1005
Thr Cys Val Asp Lys Glu Val Val Ile Asn Ile Pro Lys Arg Leu Ile
1010 1015
1020
Asn Ala Asp Glu Glu Glu Ile Lys Asp Leu Val Ile Thr Lys His Lys
1025 1030
1035 1040
Asp Lys Ala Phe Phe Val Lys Leu Lys Lys Thr Leu Glu Asp Ile Glu
1045 1050 1055
Asn Asn Lys Leu Lys Val Ile Phe Asp Asp Cys Ile Leu Tyr Leu Lys
1060 1065
1070
Glu Leu Gly Leu Ile Asp Tyr Asn Ile Ile Lys Lys Ile Asn Lys Val
1075 1080
1085
Asp Ile Lys Ile Leu Asp Glu Glu Lys Phe Lys A1<~ Tyr Lys Lys Tyr
1090 1095 1100
Phe Asp Thr Val Ile Glu His Gly Asn Phe Lys Lys Gly Arg Cys Asn
1105 1110 1115
1120
Ile Gln Tyr Ile Lys Ile Lys Asp Tyr Ile Ala Asn Ile Pro Asp Lys
1125 1130
1135
Glu Phe Glu Asp Cys Glu Ile Gly Ala Tyr Ser Gly Lys Ile Asn Ala
1140 1145
1150
Leu Leu Lys Leu Asp Glu Lys Leu Ala Lys Phe Leu Gly Phe Phe Val
1155 1160
1165
Thr Arg Gly Arg Leu Lys Lys Gln Lys Leu Lys Gly Glu Thr Val Tyr
1170 1175
1180
Glu Ile Ser Val Tyr Lys Ser Leu Pro Glu Tyr Gln Lys Glu Ile Ala
1185 1190 1195
1200
Glu Thr Phe Lys Glu Val Phe Gly Ala Gly Ser Met Val Lys Asp Lys
1205 1210
1215
Val Thr Met Asp Asn Lys Ile Val Tyr Leu Val Leu Lys Tyr Ile Phe
1220 1225
1230
Lys Cys Gly Asp Lys Asp Lys Lys His Ile Pro Glu Glu Leu Phe Leu
1235 1240
1245
Ala Ser Glu Ser Val Ile Lys Ser Phe Leu Asp Gly Phe Leu Lys Ala
1250 1255
1260
Lys Lys Asn Ser His Lys Gly Thr Ser Thr Phe Met Ala Lys Asp Glu
1265 1270 1275
1280
Lys Tyr Leu Asn Gln Leu Met Ile Leu Phe Asn Leu Val Gly Ile Pro
1285 1290
1295
Thr Arg Phe Thr Pro Val Lys Asn Lys Gly Tyr Lys :Leu Thr Leu Asn
1300 1305
1310
CA 02338185 2001-02-05
- 52 -
Pro Lys Tyr Gly Thr Val Lys Asp Leu Met Leu Asp Glu Val Lys Glu
1315 1320 1325
Ile Glu Ala Phe Glu Tyr Ser Gly Tyr Val Tyr Asp Leu Ser Val Glu
1330 1335 1340
Asp Asn Glu Asn Phe Leu Val Asn Asn Ile Tyr Ala His Asn Ser Val
1395 1350 1355
1360
Tyr Gly Tyr Leu Ala Phe Pro Arg Ala Arg Phe Tyr Ser Arg Glu Cys
1365 1370
1375
Ala Glu Ile Val Thr Tyr Leu Gly Arg Lys Tyr Ile Leu Glu Thr Val
1380 1385 1390
Lys Glu Ala Glu Lys Phe Gly Phe Lys Val Leu Tyr Ile Asp Thr Asp
1395 1400
1405
Gly Phe Tyr Ala Ile Trp Lys Glu Lys Ile Ser Lys Glu Glu Leu Ile
1410 1415 1420
Lys Lys Ala Met Glu Phe Val Glu Tyr Ile Asn Ser Lys Leu Pro Gly
1425 1430 1435
1440
Thr Met Glu Leu Glu Phe Glu Gly Tyr Phe Lys Arg Gly Ile Phe Val
1445 1450 1455
Thr Lys Lys Arg Tyr Ala Leu Ile Asp Glu Asn Gly Arg Val Thr Val
1460 1465 1470
Lys Gly Leu Glu Phe Val Arg Arg Asp Trp Ser Asn Ile Ala Lys Ile
1475 1480
1485
Thr Gln Arg Arg Val Leu Glu Ala Leu Leu Val Glu Gly Ser Ile Glu
1490 1495
1500
Lys Ala Lys Lys Ile Ile Gln Asp Val Ile Lys Asp Leu Arg Glu Lys
1505 1510 1515
1520
Lys Ile Lys Lys Glu Asp Leu Ile Ile Tyr Thr Gln Leu Thr Lys Asp
1525 1530 1535
Pro Lys Glu Tyr Lys Thr Thr Ala Pro His Val Glu Ile Ala Lys Lys
1540 1545
1550
Leu Met Arg Glu Gly Lys Arg Ile Lys Val Gly Asp Ile Ile Gly Tyr
1555 1560
1565
Ile Ile Val Lys Gly Thr Lys Ser Ile Ser Glu Arg Ala Lys Leu Pro
1570 1575 1580
Glu Glu Val Asp Ile Asp Asp Ile Asp Val Asn Tyr Tyr Ile Asp Asn
1585 1590 1595
1600
Gln Ile Leu Pro Pro Val Leu Arg Ile Met Glu Ala 'Val Gly Val Ser
1605 1610 1615
Lys Asn Glu Leu Lys Lys Glu Gly Ala Gln Leu Thr Leu Asp Lys Phe
1620 1625
1630
Phe Lys
CA 02338185 2001-02-05
- 53 -
(2) INFORMATION FOR SEQ ID NO: 25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1235 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Pyrococcus horikoshii
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 25:
Met Ile Leu Asp Ala Asp Tyr Ile Thr Glu Asp Gly Lys Pro Ile Ile
1 5 10 15
Arg Ile Phe Lys Lys Glu Asn Gly Glu Phe Lys Val Glu Tyr Asp Arg
20 25 30
Asn Phe Arg Pro Tyr Ile Tyr Ala Leu Leu Arg Asp Asp Ser Ala Ile
35 40 45
Asp Glu Ile Lys Lys Ile Thr Ala Gln Arg His Gly Lys Val Val Arg
50 55 60
Ile Val Glu Thr Glu Lys Ile Gln Arg Lys Phe Leu Gly Arg Pro Ile
65 70 75 80
Glu Val Trp Lys Leu Tyr Leu Glu His Pro Gln Asp Val Pro Ala Ile
85 90 95
Arg Asp Lys Ile Arg Glu His Pro Ala Val Val Asp Ile Phe Glu Tyr
100 105 110
Asp Ile Pro Phe Ala Lys Arg Tyr Leu Ile Asp Lys Gly Leu Thr Pro
115 120 125
Met Glu Gly Asn Glu Lys Leu Thr Phe Leu Ala Val Asp Ile Glu Thr
130 135 140
Leu Tyr His Glu Gly Glu Glu Phe Gly Lys Gly Pro Val Ile Met Ile
145 150 155 160
Ser Tyr Ala Asp Glu Glu Gly Ala Lys Val Ile Thr Trp Lys Lys Ile
165 170 175
Asp Leu Pro Tyr Val Glu Val Val Ser Ser Glu Arg Glu Met Ile Lys
180 185 190
Arg Leu Ile Arg Val Ile Lys Glu Lys Asp Pro Asp Val Ile Ile Thr
195 200 205
Tyr Asn Gly Asp Asn Phe Asp Phe Pro Tyr Leu Leu Lys Arg Ala Glu
210 215 220
Lys Leu Gly Ile Lys Leu Leu Leu Gly Arg Asp Asn Ser Glu Pro Lys
225 230 235 240
CA 02338185 2001-02-05
- 54 -
MetGln LysMet Gly LeuAla GluIle:Lys Gly
Asp Val Arg
Ser Ile
245 250 255
HisPhe AspLeu PheProVal IleArgArg ThrIleAsn Leu Thr
Pro
260 265 270
TyrThr LeuGlu AlaValTyr GluAlaIle PheGlyLys ProLysGlu
275 280 285
LysVal TyrAla AspGluIle AlaLysAla TrpGluThr GlyGluGly
290 295 300
LeuGlu ArgVal AlaLysTyr SerMetGlu AspAlaLys ValThrTyr
305 310 315 320
GluLeu GlyArg GluPhePhe ProMetGlu AlaGlnLeu AlaArgLeu
325 330 335
ValGly GlnPro ValTrpAsp ValSerArg SerSerThr GlyAsnLeu
340 345 350
ValGlu TrpPhe LeuLeuArg LysAlaTyr GluArgAsn GluLeuAla
355 360 365
ProAsn LysPro AspGluLys GluTyrGlu ArgArgLeu ArgGluSer
370 375 380
TyrGlu GlyGly TyrValLys GluProGlu LysGlyLeu TrpGluGly
385 390 395 400
IleVal SerLeu AspPheArg SerLeuTyr ProSerIle IleIleThr
405 410 415
HisAsn ValSer ProAspThr LeuRsnArg GluGlyCys GluGluTyr
420 425 430
AspVal AlaPro LysValGly HisArgPhe CysLysAsp PheProGly
435 440 445
PheIle ProSer LeuLeuGly GlnLeuLeu GluGluArg GlnLysIle
450 455 460
LysLys ArgMet LysGluSer LysAspPro ValGluLys LysLeuLeu
465 470 475 4g0
AspTyr ArgGln ArgAlaIle LysIleLeu AlaAsnSer IleLeuPro
485 490 495
AspGlu TrpLeu ProIleVal GluAsnGlu LysValArg PheValLys
500 505 510
IleGly AspPhe IleAspArg GluIleGlu GluAsnAla GluArgVal
515 520 525
LysArg Gly GluThrGlu IleLeuGlu Lys LeuLysAla
Asp Val Asp
530 535 540
LeuSer PheAsn GluThr LysLysSer LeuLys LysValLys
Arg Glu
545 550 555 560
AlaLeu Ile Tyr Lys Ser IleLysLeu
Arg Ser Val
His Gly Tyr
Arg
565 570 575
CA 02338185 2001-02-05
- 55 -
Lys Ser Gly Arg Arg Ile Lys Ile Thr Ser Gly His Ser Leu Phe Ser
580 585 590
Val Lys Asn Gly Lys Leu Val Lys Val Arg Gly Asp Glu Leu Lys Pro
595 600 605
Gly Asp Leu Val Val Val Pro Gly Arg Leu Lys Leu Pro Glu Ser Lys
610 615 620
Gln Val Leu Asn Leu Val Glu Leu Leu Leu Lys Leu Pro Glu Glu Glu
625 630 635 640
Thr Ser Asn Ile Val Met Met Ile Pro Val Lys Gly Arg Lys Asn Phe
645 650 655
Phe Lys Gly Met Leu Lys Thr Leu Tyr Trp Ile Phe Gly Glu Gly Glu
660 665 670
Arg Pro Arg Thr Ala Gly Arg Tyr Leu Lys His Leu Glu Arg Leu Gly
675 680 685
Tyr Val Lys Leu Lys Arg Arg Gly Cys Glu Val Leu Asp Trp Glu Ser
690 695
700
Leu Lys Arg Tyr Arg Lys Leu Tyr Glu Thr Leu Ile Lys Asn Leu Lys
705 710 715 720
Tyr Asn Gly Asn Ser Arg Ala Tyr Met Val Glu Phe Asn Ser Leu Arg
725 730 735
Asp Val Val Ser Leu Met Pro Ile Glu Glu Leu Lys Glu Trp Ile Ile
740 745 750
Gly Glu Pro Arg Gly Pro Lys Ile Gly Thr Phe Ile Asp Val Asp Asp
755 760 765
Ser Phe Ala Lys Leu Leu Gly Tyr Tyr Ile Ser Ser Gly Asp Val Glu
770 775 780
Lys Asp Arg Val Lys Phe His Ser Lys Asp Gln Asn Val Leu Glu Asp
785 790 795 800
Ile Ala Lys Leu Ala Glu Lys Leu Phe Gly Lys Val Arg Arg Gly Arg
805 810 815
Gly Tyr Ile Glu Val Ser Gly Lys Ile Ser His Ala Ile Phe Arg Val
820 825 830
Leu Ala Glu Gly Lys Arg Ile Pro Glu Phe Ile Phe Thr Ser Pro Met
835 840 845
Asp Ile Lys Val Ala Phe Leu Lys Gly Leu Asn Gly Asn Ala Glu Glu
850 855 860
Leu Thr Phe Ser Thr Lys Ser Glu Leu Leu Val Asn Gln Leu Ile Leu
865 870 875 880
Leu Leu Asn Ser Ile Gly Val Ser Asp Ile Lys Ile Glu His Glu Lys
885 890 895
Gly Val Tyr Arg Val Tyr Ile Asn Lys Lys Glu Ser Ser Asn Gly Asp
900 905 910
CA 02338185 2001-02-05
- 56 -
Ile Val Leu Asp Ser Val Glu Ser Ile Glu Val Glu Lys Tyr Glu Gly
915 920 925
Tyr Val TyrAsp LeuSerVal GluAspAsn GluASIlPheLeuValGly
930 935 9qp
Phe Gly LeuLeu TyrAlaHis AsnSerTyr TyrGly TyrTyrGlyTyr
945 950 955 960
Ala Lys AlaArg TrpTyrCys LysGluCys AlaGlu SerValThrAla
965 970 975
Trp Gly ArgGln TyrIleAsp LeuValArg ArgGlu LeuGluAlaArg
980 985 990
Gly Phe LysVal LeuTyrIle AspThrAsp GlyLeu TyrAlaThrIle
995 1000 1005
Pro Gly ValLys AspTrpGlu GluValLys ArgArg AlaLeuGluPhe
1010 1015 1020
Val Asp TyrIle AsnSerLys LeuProGly ValLeu GluLeuGluTyr
1025 1030 1035 1040
Glu Gly PheTyr AlaArgGly PhePheVal ThrLys LysLysTyrAla
1045 1050 1055
Leu Ile AspGlu GluGlyLys IleValThr ArgGly LeuGluIleVal
1060 1065 1070
Arg Arg AspTrp SerGluIle AlaLysGlu ThrGln AlaArgValLeu
1075 1080 1085
Glu Ala IleLeu LysHisGly AsnValGlu GluAla ValLysIleVal
1090 1095 1100
Lys Asp ValThr GluLysLeu ThrAsnTyr GluVal ProProGluLys
1105 1110 1115 1120
Leu Val IleTyr GluGlnIle ThrArgPro IleAsn GluTyrLysAla
1125 1130 1135
Ile Gly ProHis ValAlaVal RlaLysArg LeuMet AlaArgGlyIle
1140 1145 1150
Lys Val LysPro GlyMetVal IleGlyTyr IleVal LeuArgGlyAsp
1155 1160 1165
Gly Pro IleSer LysArgAla IleSerIle GluGlu PheAspProArg
1170 1175 1180
Lys His LysTyr AspAlaGlu TyrTyrIle GluAsn ValLeuPro
Gln
1185 1190 1195 1200
Ala Val GluArg IleLeuLys PheGly Lys Glu Leu
Ala Tyr Arg Asp
1205 1210 1215
Arg Trp Lys Lys GlyLeu Ile Val
Gln Thr Gln Gly Lys
Val Ala
Trp
1220 1225 1230
Lys Lys
Ser
1235
CA 02338185 2001-02-05
- 57 -
(2) INFORMATION FOR SEQ ID NO: 26:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 586 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanobacterium thermoautotrophicum
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 26:
Met Glu Asp Tyr Arg Met Val Leu Leu Asp Ile Asp Tyr Val Thr Val
1 5 10 15
Asp Glu Val Pro Val Ile Arg Leu Phe Gly Lys Asp Lys Ser Gly Gly
20 25 30
Asn Glu Pro Ile Ile Ala His Asp Arg Ser Phe Arg~ Pro Tyr Ile Tyr
35 40 45
Ala Ile Pro Thr Asp Leu Asp Glu Cys Leu Arg Glu Leu Glu Glu Leu
50 55 60
Glu Leu Glu Lys Leu Glu Val Lys Glu Met Arg Asp Leu Gly Arg Pro
65 70 75 80
Thr Glu Val Ile Arg Ile Glu Phe Arg His Pro Gln Asp Val Pro Lys
85 90 95
Ile Arg Asp Arg Ile Arg Asp Leu Glu Ser Val Arg Asp Ile Arg Glu
100 105 110
His Asp Ile Pro Phe Tyr Arg Arg Tyr Leu Ile Asp Lys Ser Ile Val
115 120 125
Pro Met Glu Glu Leu Glu Phe Gln Gly Val Glu Val Asp Ser Ala Pro
130 135 140
Ser Val Thr Thr Asp Val Arg Thr Val Glu Val Thr Gly Arg Val Gln
145 150 155
160
Ser Thr Gly Ser Gly Ala His Gly Leu Asp Ile Leu Ser Phe Asp Ile
165 170 175
Glu Val Arg Asn Pro His Gly Met Pro Asp Pro Glu Lys Asp Glu Ile
180 185 190
Val Met Ile Gly Val Ala Gly Asn Met Gly Tyr Glu Ser Val Ile Ser
195 200 205
Thr Ala Gly Asp His Leu Asp Phe Val Glu Val Val Glu Asp Glu Arg
210 215 220
Glu Leu Leu Glu Arg Phe Ala Glu Ile Val Ile Asp Lys Lys Pro Asp
225 230 235
240
CA 02338185 2001-02-05
- 58 -
Ile Leu Val Gly Tyr Rsn Ser Asp Asn Phe Asp Pha_ Pro Tyr Ile Thr
245 250 255
Arg Arg Ala Ala Ile Leu Gly Ala Glu Leu Asp Leu Gly Trp Asp Gly
260 265 270
Ser Lys Ile Arg Thr Met Arg Arg Gly Phe Ala Asn Ala Thr Ala Ile
275 280 285
Lys Gly Thr Val His Val Asp Leu Tyr Pro Val Met. Arg Arg Tyr Met
290 295 300
Asn Leu Asp Arg Tyr Thr Leu Glu Arg Val Tyr Gln Glu Leu Phe Gly
305 310 315 320
Glu Glu Lys Ile Asp Leu Pro Gly Asp Arg Leu Trp Glu Tyr Trp Asp
325 330 335
Arg Asp Glu Leu Arg Asp Glu Leu Phe Arg Tyr Ser Leu Asp Asp Val
340 345 350
Val Ala Thr His Arg Ile Ala Glu Lys Ile Leu Pro Leu Asn Leu Glu
355 360 365
Leu Thr Arg Leu Val Gly Gln Pro Leu Phe Asp Ile Ser Arg Met Ala
370 375 380
Thr Gly Gln Gln Ala Glu Trp Phe Leu Val Arg Lys Ala Tyr Gln Tyr
385 390 395 400
Gly Glu Leu Val Pro Asn Lys Pro Ser Gln Ser Asp Phe Ser Ser Arg
405 410 415
Arg Gly Arg Arg Ala Val Gly Gly Tyr Val Lys Glu Pro Glu Lys Gly
420 425 430
Leu His Glu Asn Ile Val Gln Phe Asp Phe Arg Ser Leu Tyr Pro Ser
435 440 445
Ile Ile Ile Ser Lys Asn Ile Ser Pro Asp Thr Leu Thr Asp Asp Glu
450 455 460
Glu Ser Glu Cys Tyr Val Ala Pro Glu Tyr Gly Tyr Arg Phe Arg Lys
465 470 475 480
Ser Pro Arg Gly Phe Val Pro Ser Val Ile Gly Glu Ile Leu Ser Glu
485 490 495
Arg Val Arg Ile Lys Glu Glu Met Lys Gly Ser Asp Asp Pro Met Glu
500 505 510
Arg Lys Ile Leu Asn Val Gln Gln Glu Ala Leu Lys Arg Leu Ala Asn
515 520 525
Thr Met Tyr Gly Val Tyr Gly Tyr Ser Arg Phe Arg Trp Tyr Ser Met
530 535 540
Glu Cys Ala Glu Ala Ile Thr Ala Trp Gly Rrg Asp Tyr Ile Lys Lys
545 550 555 560
Thr Ile Lys Thr Ala Glu Glu Phe Gly Phe His Thr Val Tyr Ala Asp
565 570 575
CA 02338185 2001-02-05
- 59 -
Thr Asp Gly Phe Tyr Ala Thr Tyr Arg Gly
580 585
(2) INFORMATION FOR SEQ ID NO: 27:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1143 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeoglobus fulgidus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 27:
Met Asp Ala Thr Leu Asp Arg Phe Phe Pro Leu Phe Glu Ser Glu Ser
1 5 10 15
Asn Glu Asp Phe Trp Arg Ile Glu Glu Ile Arg Arg~ Tyr His Glu Ser
20 25 30
Leu Met Val Glu Leu Asp Arg Ile Tyr Arg Ile Ala Glu Ala Ala Arg
35 40 45
Lys Lys Gly Leu Asp Pro Glu Leu Ser Val Glu Ile Pro Ile Ala Lys
50 55 60
Asn Met Ala Glu Arg Val Glu Lys Leu Met Asn Leu Gln Gly Leu Ala
65 70 75 80
Lys Arg Ile Met Glu Leu Glu Glu Gly Gly Leu Ser Arg Glu Leu Ile
85 90 95
Cys Phe Lys Val Ala Asp Glu Ile Val Glu Gly Lys Phe Gly Glu Met
100 105 110
Pro Lys Glu Glu Ala Ile Asp Lys Ala Val Arg Thr Ala Val Ala Ile
115 120 125
Met Thr Glu Gly Val Val Ala Ala Pro Ile Glu Gly Ile Ala Arg Val
130 135 140
Arg Ile Asp Arg Glu Asn Phe Leu Arg Val Tyr Tyr Ala Gly Pro Ile
145 150 155 160
Arg Ser Ala Gly Gly Thr Ala Gln Val Ile Ser Val Leu Val Ala Asp
165 170 175
Tyr Val Arg Arg Lys Ala Glu Ile Gly Arg Tyr Val Pro Thr Glu Glu
180 185 190
Glu Ile Leu Rrg Tyr Cys Glu Glu Ile Pro Leu Tyr Lys Lys Val Ala
195 200 205
Asn Leu Gln Tyr Leu Pro Ser Asp Glu Glu Ile Arg Leu Ile Val Ser
210 215 220
CA 02338185 2001-02-05
- 60 -
Asn Cys Pro Ile Cys Ile Asp Gly Glu Pro Thr Glu Ser Ala Glu Val
225 230 235 240
Ser Gly Tyr Arg Asn Leu Pro Arg Val Glu Thr Asn Arg Val Arg Gly
245 250 255
Gly Met Ala Leu Val Ile Ala Glu Gly Ile Ala Leu Lys Ala Pro Lys
260 265 270
Leu Lys Lys Met Val Asp Glu Val Gly Ile Glu Gly Trp Glu Trp Leu
275 280 285
Asp Ala Leu Ile Lys Gly Gly Gly Asp Ser Gly Set: Glu Glu Glu Lys
290 295 300
Ala Val Ile Lys Pro Lys Asp Lys Tyr Leu Ser Asp Ile Val Ala Gly
305 310 315 320
Arg Pro Val Leu Ser His Pro Ser Arg Lys Gly Gly Phe Arg Leu Arg
325 330 335
Tyr Gly Arg Ala Arg Asn Ser Gly Phe Ala Thr Val Gly Val Asn Pro
340 345 350
Ala Thr Met Tyr Leu Leu Glu Phe Val Ala Val Gly Thr Gln Leu Lys
355 360 365
Val Glu Arg Pro Gly Lys Ala Gly Gly Val Val Pro Val Ser Thr Ile
370 375 380
Glu Gly Pro Thr Val Arg Leu Lys Asn Gly Asp Val Val Lys Ile Asn
385 390 395 400
Thr Leu Ser Glu Ala Lys Ala Leu Lys Gly Glu Val Ala Ala Ile Leu
405 410 415
Asp Leu Gly Glu Ile Leu Ile Asn Tyr Gly Asp Phe Leu Glu Asn Asn
420 425 430
His Pro Leu Ile Pro Ala Ser Tyr Thr Tyr Glu Trp Trp Ile Gln Glu
435 440 445
Ala Glu Lys Ala Gly Leu Arg Gly Asp Tyr Arg Lys Ile Ser Glu Glu
450 455 460
Glu Ala Leu Lys Leu Cys Asp Glu Phe His Val Pro Leu His Pro Asp
465 470 475 480
Tyr Thr Tyr Leu Trp His Asp Ile Ser Val Glu Asp Tyr Arg Tyr Leu
485 490 495
Arg Asn Phe Val Ser Asp Asn Gly Lys Ile Glu Gly Lys His Gly Lys
500 505 510
Ser Val Leu Leu Leu Pro Tyr Asp Ser Arg Val Lys Glu Ile Leu Glu
515 520 525
Ala Leu Leu Leu Glu His Lys Val Arg Glu Ser Phe Ile Val Ile Glu
530 535 540
Thr Trp Arg Ala Phe Ile Arg Cys Leu Gly Leu Asp Glu Lys Leu Ser
545 550 555 560
CA 02338185 2001-02-05
- 61 -
Lys Val Ser Glu Val Ser Gly Lys Asp Val Leu Glu Ile Val Asn Gly
565 570 575
Ile Ser Gly Ile Lys Val Arg Pro Lys Ala Leu Ser Arg Ile Gly Ala
580 585 590
Arg Met Gly Arg Pro Glu Lys Ala Lys Glu Arg Lys Met Ser Pro Pro
595 600 605
Pro His Ile Leu Phe Pro Val Gly Met Ala Gly Gly Asn Thr Arg Asp
610 615
620
Ile Lys Asn Ala Ile Asn Tyr Thr Lys Ser Tyr Asn Ala Lys Lys Gly
625 630 635 640
Glu Ile Glu Val Glu Ile Ala Ile Arg Lys Cys Pro Gln Cys Gly Lys
645 650 655
Glu Thr Phe Trp Leu Lys Cys Asp Val Cys Gly Glu. Leu Thr Glu Gln
660 665 670
Leu Tyr Tyr Cys Pro Ser Cys Arg Met Lys Asn Thr Ser Ser Val Cys
675 680 685
Glu Ser Cys Gly Arg Glu Cys Glu Gly Tyr Met Lys Arg Lys Val Asp
690 695 700
Leu Arg Glu Leu Tyr Glu Glu Ala Ile Ala Asn Leu Gly Glu Tyr Asp
705 710 715 720
Ser Phe Asp Thr Ile Lys Gly Val Lys Gly Met Thr Ser Lys Thr Lys
725 730 735
Ile Pro Glu Arg Leu Glu Lys Gly Ile Leu Arg Val Lys His Gly Val
740 745 750
Phe Val Phe Lys Asp Gly Thr Ala Arg Phe Asp Ala Thr Asp Leu Pro
755 760 765
Ile Thr His Phe Lys Pro Ala Glu Ile Gly Val Ser Val Glu Lys Leu
770 775 780
Arg Glu Leu Gly Tyr Glu Arg Asp Tyr Lys Gly Ala Glu Leu Lys Asn
785 790 795 800
Glu Asn Gln Ile Val Glu Leu Lys Pro Gln Asp Val Ile Leu Pro Lys
805 810 815
Ser Gly Ala Glu Tyr Leu Leu Arg Val Ala Asn Phe Ile Asp Asp Leu
820 825 830
Leu Val Lys Phe Tyr Lys Met Glu Pro Phe Tyr Asn Ala Lys Ser Val
835 840 845
Glu Asp Leu Ile Gly His Leu Val Ile Gly Leu Ala Pro His Thr Ser
850 855 860
Ala Gly Val Leu Gly Arg Ile Ile Gly Phe Ser Asp Val Leu Ala Gly
865 870 875 880
Tyr Ala His Pro Tyr Phe His Rla Ala Lys Arg Arg .Asn Cys Asp Gly
885 890 895
CA 02338185 2001-02-05
- 62 -
Asp Glu Asp Cys Phe Met Leu Leu Leu Asp Gly Leu Leu Asn Phe Ser
900 905 910
Arg Lys Phe Leu Pro Asp Lys Arg Gly Gly Gln Met. Asp Ala Pro Leu
915 920 925
Val Leu Thr Ala Ile Val Asp Pro Arg Glu Val Asp Lys Glu Val His
930 935
940
Asn Met Asp Ile Val Glu Arg Tyr Pro Leu Glu Phe Tyr Glu Ala Thr
945 950 955 960
Met Arg Phe Ala Ser Pro Lys Glu Met Glu Asp Tyr Val Glu Lys Val
965 970 975
Lys Asp Arg Leu Lys Asp Glu Ser Arg Phe Cys Gly Leu Phe Phe Thr
980 985 990
His Asp Thr Glu Asn Ile Ala Ala Gly Val Lys Glu Ser Ala Tyr Lys
995 1000 1005
Ser Leu Lys Thr Met Gln Asp Lys Val Tyr Arg Gln Met Glu Leu Ala
1010 1015 1020
Arg Met Ile Val Ala Val Asp Glu His Asp Val Ala Glu Arg Val Ile
1025 1030 1035
1040
Asn Val His Phe Leu Pro Asp Ile Ile Gly Asn Leu Arg Ala Phe Ser
1045 1050 1055
Arg Gln Glu Phe Arg Cys Thr Arg Cys Asn Thr Lys Tyr Arg Arg Ile
1060 1065 1070
Pro Leu Val Gly Lys Cys Leu Lys Cys Gly Asn Lys Leu Thr Leu Thr
1075 1080 1085
Val His Ser Ser Ser Ile Met Lys Tyr Leu Glu Leu Ser Lys Phe Leu
1090 1095 110()
Cys Glu Asn Phe Asn Val Ser Ser Tyr Thr Lys Gln Arg Leu Met Leu
1105 1110 1115 1120
Leu Glu Gln Glu Ile Lys Ser Met Phe Glu Asn Gly Thr Glu Lys Gln
1125 1130 1135
Val Ser Ile Ser Asp Phe Val
1140
(2) INFORMATION FOR SEQ ID NO: 28:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1139 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanococcus jannaschii
CA 02338185 2001-02-05
- 63 -
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 28:
Met Ile Val Met Val His Val Ala Cys Ser Glu Asn Met Lys Lys Tyr
1 5 10 15
Phe Glu Asn Ile Val Asp Glu Val Lys Lys Ile Tyr Rrg Ile Ala Glu
20 25 30
Glu Cys Arg Lys Lys Gly Phe Asp Pro Thr Asp Gl.u Val Glu Ile Pro
35 40 45
Leu Ala Ala Asp Met Ala Asp Arg Val Glu Gly Leu Val Gly Pro Lys
50 55 60
Gly Val Ala Glu Arg Ile Arg Glu Leu Val Lys Glu Leu Gly Lys Glu
65 70 75 80
Pro Ala Ala Leu Glu Ile Ala Lys Glu Ile Val Glu Gly Lys Phe Gly
85 90 95
Asn Phe Asp Lys Glu Lys Lys Ala Glu Gln Ala Val Arg Thr Ala Leu
100 105
110
Ala Val Leu Thr Glu Gly Ile Val Ala Ala Pro Leu Glu Gly Ile Ala
115 120
125
Asp Val Lys Ile Lys Lys Asn Pro Asp Gly Thr Glu Tyr Leu Ala Ile
130 135
140
Tyr Tyr Ala Gly Pro Ile Arg Ser Ala Gly Gly Thr Ala Gln Ala Leu
145 150
155 160
Ser Val Leu Val Gly Asp Phe Val Arg Lys Ala Met Gly Leu Asp Arg
165 170
175
Tyr Lys Pro Thr Glu Asp Glu Ile Glu Arg Tyr Val Glu Glu Val Glu
180 185
190
Leu Tyr Gln Ser Glu Val Gly Ser Phe Gln Tyr Asn Pro Thr Ala Asp
195 200
205
Glu Ile Arg Thr Ala Ile Arg Asn Ile Pro Ile Glu Ile Thr Gly Glu
210 215 220
Ala Thr Asp Asp Val Glu Val Ser Gly His Arg Asp Leu Pro Arg Val
225 230
235 240
Glu Thr Asn Gln Leu Arg Gly Gly Ala Leu Leu Val Leu Val Glu Gly
245 250
255
Val Leu Leu Lys Ala Pro Lys Ile Leu Arg His Val .asp Lys Leu Gly
260 265
270
Ile Glu Gly Trp Asp Trp Leu Lys Asp Leu Met Ser Lys Lys Glu Glu
275 280 285
Lys Glu Glu Glu Lys Asp Glu Lys Val Asp Asp Glu Glu Ile Asp Glu
290 295 300
Glu Glu Glu Glu Ile Ser Gly Tyr Trp Arg Asp Val hys Ile Glu Ala
305 310 315
320
CA 02338185 2001-02-05
- 64 -
° Asn Lys Lys Phe Ile Ser Glu Val Ile Ala Gly Arg Pro Val Phe Ala
325 330 335
His Pro Ser Lys Val Gly Gly Phe Arg Leu Arg Tyr Gly Arg Ser Arg
340 345 350
Asn Thr Gly Phe Ala Thr Gln Gly Phe His Pro Ala Leu Met Tyr Leu
355 360 365
Val Asp Glu Phe Met Ala Val Gly Thr Gln Leu Lys Thr Glu Arg Pro
370 375
380
Gly Lys Ala Thr Cys Val Val Pro Val Asp Ser Ile Glu Pro Pro Ile
385 390 395 400
Val Lys Leu Lys Asn Gly Asp Val Ile Arg Val Asp Thr Ile Glu Lys
405 410 415
Ala Met Asp Val Arg Asn Arg Val Glu Glu Ile Leu Phe Leu Gly Asp
420 425 430
Val Leu Val Asn Tyr Gly Asp Phe Leu Glu Asn Asn His Pro Leu Leu
435 440 445
Pro Ser Cys Trp Cys Glu Glu Trp Tyr Glu Lys Ile Leu Ile Ala Asn
450 455 460
Asn Ile Glu Tyr Asp Lys Asp Phe Ile Lys Asn Pro Lys Pro Glu Glu
465 470 475 480
Ala Val Lys Phe Ala Leu Glu Thr Lys Thr Pro Leu His Pro Arg Phe
485 490 495
Thr Tyr His Trp His Asp Val Ser Lys Glu Asp Ile Ile Leu Leu Arg
500 505 510
Asn Trp Leu Leu Lys Gly Lys Glu Asp Ser Leu Glu Gly Lys Lys Val
515 520 525
Trp Ile Val Asp Leu Glu Ile Glu Glu Asp Lys Lys Ala Lys Arg Ile
530 535 540
Leu Glu Leu Ile Gly Cys Cys His Leu Val Arg Asn Lys Lys Val Ile
545 550 555 560
Ile Glu Glu Tyr Tyr Pro Leu Leu Tyr Ser Leu Gly Phe Asp Val Glu
565 570 575
Asn Lys Lys Asp Leu Val Glu Asn Ile Glu Lys Ile Leu Glu Ser Ala
580 585 590
Lys Asn Ser Met His Leu Ile Asn Leu Leu Ala Pro Phe Glu Val Arg
595 600 605
Arg Asn Thr Tyr Val Tyr Val Gly Ala Arg Met Gly .Arg Pro Glu Lys
610 615 620
Ala Ala Pro Arg Lys Met Lys Pro Pro Val Asn Gly Leu Phe Pro Ile
625 630 635 640
Gly Asn Ala Gly Gly Gln Val Arg Leu Ile Asn Lys ;41a Val Glu Glu
645 650 655
CA 02338185 2001-02-05
- 65 -
Asn Asn Thr Asp Asp Val Asp Val Ser Tyr Thr Arg Cys Pro Asn Cys
660 665 670
Gly Lys Ile Ser Leu Tyr Arg Val Cys Pro Phe Cys Gly Thr Lys Val
675 680 685
Glu Leu Asp Asn Phe Gly Arg Ile Lys Ala Pro Leu Lys Asp Tyr Trp
690 695
700
Tyr Ala Ala Leu Lys Arg Leu Gly Ile Asn Lys Pro Gly Asp Val Lys
705 710 715 720
Cys Ile Lys Gly Met Thr Ser Lys Gln Lys Ile Val Glu Pro Leu Glu
725 730 735
Lys Ala Ile Leu Arg Ala Ile Asn Glu Val Tyr Val Phe Lys Asp Gly
740 745 750
Thr Thr Arg Phe Asp Cys Thr Asp Val Pro Val Thr His Phe Lys Pro
755 760 765
Asn Glu Ile Asn Val Thr Val Glu Lys Leu Arg Glu Leu Gly Tyr Asp
770 775 780
Lys Asp Ile Tyr Gly Asn Glu Leu Val Asp Gly Glu Gln Val Val Glu
785 790 795 800
Leu Lys Pro Gln Asp Val Ile Ile Pro Glu Ser Cys Ala Glu Tyr Phe
805 810 815
Val Lys Val Ala Asn Phe Ile Asp Asp Leu Leu Glu Lys Phe Tyr Lys
820 825 830
Val Glu Arg Phe Tyr Asn Val Lys Lys Lys Glu Asp Leu Ile Gly His
835 840 845
Leu Val Ile Gly Met Ala Pro His Thr Ser Ala Gly Met Val Gly Arg
850 855 860
Ile Ile Gly Tyr Thr Lys Ala Asn Val Gly Tyr Ala His Pro Tyr Phe
865 870 875 880
His Ala Ala Lys Arg Arg Asn Cys Asp Gly Asp Glu Asp Ser Phe Phe
885 890 895
Leu Leu Leu Asp Ala Phe Leu Asn Phe Ser Lys Lys Phe Leu Pro Asp
900 905 910
Lys Arg Gly Gly Gln Met Asp Ala Pro Leu Val Leu Thr Thr Ile Leu
915 920 925
Asp Pro Lys Glu Val Asp Gly Glu Val His Asn Met Asp Thr Met Trp
930 935 940
Ser Tyr Pro Leu Glu Phe Tyr Glu Lys Thr Leu Glu Met Pro Ser Pro
945 950 955 960
Lys Glu Val Lys Glu Phe Met Glu Thr Val Glu Asp Arg Leu Gly Lys
965 970 975
Pro Glu Gln Tyr Glu Gly Ile Gly Tyr Thr His Glu 'Phr Ser Arg Ile
980 985 990
CA 02338185 2001-02-05
- 66 -
AspLeuGly ProLysVal CysAlaTyr LysThr LeuGlySerMet Leu
995 1000 1005
GluLysThr ThrSerGln LeuSerVal AlaLys LysIleArgAla Thr
1010 1015 1020
AspGluArg AspValAla GluLysVal IleGln Sei:HisPheIle Pro
1025 1030 1035
1040
AspLeuIle GlyAsnLeu ArgAlaPhe SerArg GlnAlaValArg Cys
1045 1050 1055
LysCysGly AlaLysTyr ArgArgIle ProLeu LysGlyLysCys Pro
1060 1065 1070
LysCysGly SerAsnLeu IleLeuThr ValSer LysGlyAlaVal Glu
1075 1080 1085
LysTyrMet AspValAla GluLysMet AlaGlu GluTyrAsnVal Asn
1090 1095 1100
AspTyrIle LysGlnArg LeuLysIle IleLys GluGlyIleAsn Ser
1105 1110 1115 1120
IlePheGlu AsnGluLys SerArgGln ValLys LeuSerAspPhe Phe
1125 1130 1135
LysIleGly
(2) INFORMATION FOR SEQ ID N0: 29:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1434 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Pyrococcus horikoshii
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 29:
MetValLeuMet GluLeuPro LysGlu MetGluGlu TyrPheSer Met
1 5 10 15
LeuGlnArgGlu IleAspLys AlaTyr GluIleAla LysLysAla Arg
20 25 30
AlaGlnGlyLys AspProSer LeuAsp ValGluIle ProGlnAla Ser
35 40 45
AspMetAlaGly ArgValGlu SerLeu ValGlyPro :ProGlyVal Ala
50 55 60
GluArgIleArg GluLeuVal LysGlu TyrGlyLys GluIleAla Ala
65 70 75 80
CA 02338185 2001-02-05
- 67 -
Leu Lys Ile Val Asp Glu Ile Ile Asp Gly Lys Phe Gly Asp Leu Gly
85 90 95
Ser Lys Glu Lys Tyr Ala Glu Gln Ala Val Arg Thr Ala Leu Ala Ile
100 105 110
Leu Thr Glu Gly Val Val Ser Ala Pro Ile Glu Gly Ile Ala Ser Val
115 120 125
Lys Ile Lys Arg Asn Thr Trp Ser Asp Asn Ser Glu Tyr Leu Ala Leu
130 135 140
Tyr Tyr Ala Gly Pro Ile Arg Ser Ser Gly Gly Thr Ala Gln Ala Leu
145 150 155 160
Ser Val Leu Val Gly Asp Tyr Val Arg Arg Lys Leu Gly Leu Asp Arg
165 170 175
Phe Lys Pro Ser Glu Lys His Ile Glu Arg Met Val Glu Glu Val Asp
180 185 190
Leu Tyr His Arg Thr Val Ser Arg Leu Gln Tyr His Pro Ser Pro Glu
195 200 205
Glu Val Arg Leu Ala Met Arg Asn Ile Pro Ile Glu Ile Thr Gly Glu
210 215 220
Ala Thr Asp Glu Val Glu Val Ser His Arg Asp Ile Pro Gly Val Glu
225 230 235 240
Thr Asn Gln Leu Arg Gly Gly Ala Ile Leu Val Leu Ala Glu Gly Val
245 250 255
Leu Gln Lys Ala Lys Lys Leu Val Lys Tyr Ile Asp Lys Met Gly Ile
260 265 270
Glu Gly Trp Glu Trp Leu Lys Glu Phe Val Glu Ala Lys Glu Lys Gly
275 280 285
Glu Glu Ile Glu Glu Glu Gly Ser Ala Glu Ser Thr Val Glu Glu Thr
290 295 300
Lys Val Glu Val Asp Met Gly Phe Tyr Tyr Ser Leu Tyr Gln Lys Phe
305 310 315 320
Lys Ser Glu Ile Ala Pro Asn Asp Lys Tyr Ala Lys Glu Ile Ile Gly
325 330 335
Gly Arg Pro Leu Phe Ser Asp Pro Ser Arg Asn Gly Gly Phe Arg Leu
340 345 350
Arg Tyr Gly Arg Ser Arg Val Ser Gly Phe Ala Thr 'rrp Gly Ile Asn
355 360 365
Pro Ala Thr Met Ile Leu Val Asp Glu Phe Leu Ala :Cle Gly Thr Gln
370 375 380
Leu Lys Thr Glu Arg Pro Gly Lys Gly Ala Val Val Thr Pro Val Thr
385 390 395 400
Thr Ile Glu Gly Pro Ile Val Lys Leu Lys Asp Gly Ser Val Val Lys
405 410 415
CA 02338185 2001-02-05
- 68 -
Val Asp Asp Tyr Lys Leu Ala Leu Lys Ile Arg Asp Glu Val Glu Glu
420 425 430
Ile Leu Tyr Leu Gly Asp Ala Val Ile Ala Phe Gly Asp Phe Val Glu
435 440 445
Asn Asn Gln Thr Leu Leu Pro Ala Asn Tyr Cys Glu Glu Trp Trp Ile
450 455 460
Leu Glu Phe Thr Lys Ala Leu Asn Glu Ile Tyr Glu Val Glu Leu Lys
465 470 475 480
Pro Phe Glu Val Asn Ser Ser Glu Asp Leu Glu Glu Ala Ala Asp Tyr
485 490 495
Leu Glu Val Asp Ile Glu Phe Leu Lys Glu Leu Leu Lys Asp Pro Leu
500 505 510
Arg Thr Lys Pro Pro Val Glu Leu Ala Ile His Phe Ser Glu Ile Leu
515 520 525
Gly Ile Pro Leu His Pro Tyr Tyr Thr Leu Tyr Trp Asn Ser Val Lys
530 535 540
Pro Glu Gln Val Glu Lys Leu Trp Arg Val Leu Lys Glu His Ala His
545 550 555 560
Ile Asp Trp Asp Asn Phe Arg Gly Ile Lys Phe Ala Arg Arg Ile Val
565 570 575
Ile Pro Leu Glu Lys Leu Arg Asp Ser Lys Arg Ala Leu Glu Leu Leu
580 585 590
Gly Leu Pro His Lys Val Glu Gly Lys Asn Val Ile Val Asp Tyr Pro
595 600 605
Trp Ala Ala Ala Leu Leu Thr Pro Leu Gly Asn Leu Glu Trp Glu Phe
610 615 620
Arg Ala Lys Pro Leu His Thr Thr Ile Asp Ile Ile Asn Glu Asn Asn
625 630 635 640
Glu Ile Lys Leu Arg Asp Arg Gly Ile Ser Trp Ile Gly Ala Arg Met
645 650 655
Gly Arg Pro Glu Lys Ala Lys Glu Arg Lys Met Lys Pro Pro Val Gln
660 665 670
Val Leu Phe Pro Ile Gly Leu Ala Gly Gly Ser Ser Arg Asp Ile Lys
675 680 685
Lys Ala Ala Glu Glu Gly Lys Val Ala Glu Val Glu Ile Ala Leu Phe
690 695 700
Lys Cys Pro Lys Cys Gly His Val Gly Pro Glu His Ile Cys Pro Asn
705 710 715 720
Cys Gly Thr Arg Lys Glu Leu Ile Trp Val Cys Pro Arg Cys Asn Ala
725 730 735
Glu Tyr Pro Glu Ser Gln Ala Ser Gly Tyr Asn Tyr Thr Cys Pro Lys
740 745 750
CA 02338185 2001-02-05
- 69 -
Cys Asn Val Lys Leu Lys Pro Tyr Ala Lys Arg Lys Ile Lys Pro Ser
755 760 765
Glu Leu Leu Lys Arg Ala Met Asp Asn Val Lys Val Tyr Gly Ile Asp
770 775 780
Lys Leu Lys Gly Val Met Gly Met Thr Ser Gly Trp Lys Met Pro Glu
785 790 795 800
Pro Leu Glu Lys Gly Leu Leu Arg Ala Lys Asn Asp Val Tyr Val Phe
805 810 815
Lys Asp Gly Thr Ile Arg Phe Asp Ala Thr Asp Ala Pro Ile Thr His
820 825 830
Phe Arg Pro Arg Glu Ile Gly Val Ser Val Glu Lys Leu Arg Glu Leu
835 840 845
Gly Tyr Thr His Asp Phe Glu Gly Asn Pro Leu Val Ser Glu Asp Gln
850 855 860
Ile Val Glu Leu Lys Pro Gln Asp Ile Ile Leu Sei: Lys Glu Ala Gly
865 870 875 880
Lys Tyr Leu Leu Lys Val Ala Lys Phe Val Asp Asp Leu Leu Glu Lys
885 890 895
Phe Tyr Gly Leu Pro Arg Phe Tyr Asn Ala Glu Lys Met Glu Asp Leu
900 905 910
Ile Gly His Leu Val Ile Gly Leu Ala Pro His Thr Ser Ala Gly Ile
915 920 925
Val Gly Arg Ile Ile Gly Phe Val Asp Ala Leu Val Gly Tyr Ala His
930 935 940
Pro Tyr Phe His Ala Ala Lys Arg Arg Asn Cys Phe Pro Gly Asp Thr
945 950 955 960
Arg Ile Leu Val Gln Ile Asn Gly Thr Pro Gln Arg Val Thr Leu Lys
965 970 975
Glu Leu Tyr Glu Leu Phe Asp Glu Glu His Tyr Glu Ser Met Val Tyr
980 985 990
Val Arg Lys Lys Pro Lys Val Asp Ile Lys Val Tyr Ser Phe Asn Pro
995 1000 1005
Glu Glu Gly Lys Val Val Leu Thr Asp Ile Glu Glu Val Ile Lys Ala
1010 1015 1020
Pro Ala Thr Asp His Leu Ile Arg Phe Glu Leu Glu Leu Gly Ser Ser
1025 1030 1035 1040
Phe Glu Thr Thr Val Asp His Pro Val Leu Val Tyr Glu Asn Gly Lys
1045 1050 1055
Phe Val Glu Lys Arg Ala Phe Glu Val Arg Glu Gly Asn Ile Ile Ile
1060 1065 1070
Ile Ile Asp Glu Ser Thr Leu Glu Pro Leu Lys Val Ala Val Lys Lys
1075 1080 1085
CA 02338185 2001-02-05
_ _ 70 _
Ile Glu Phe Ile Glu Pro Pro Glu Asp Phe Val Phe Ser Leu Asn Ala
1090 1095
17.00
Lys Lys Tyr His Thr Val Ile Ile Asn Glu Asn Il.e Val Thr His Gln
1105 1110 1115 1120
Cys Asp Gly Asp Glu Asp Ala Val Met Leu Leu Leu Asp Ala Leu Leu
1125 1130
1135
Asn Phe Ser Arg Tyr Tyr Leu Pro Glu Lys Arg Gly Gly Lys Met Asp
1140 1145
1150
Ala Pro Leu Val Ile Thr Thr Arg Leu Asp Pro Arg Glu Val Asp Ser
1155 11.60 1165
Glu Val His Asn Met Asp Ile Val Arg Tyr Tyr Pro Leu Glu Phe Tyr
1170 1175
1180
Glu Ala Thr Tyr Glu Leu Lys Ser Pro Lys Glu Leu Val Gly Val Ile
1185 1190 1195 1200
Glu Arg Val Glu Asp Arg Leu Gly Lys Pro Glu Met Tyr Tyr Gly Leu
1205 1210
1215
Lys Phe Thr His Asp Thr Asp Asp Ile Ala Leu Gly Pro Lys Met Ser
1220 1225 1230
Leu Tyr Lys Gln Leu Gly Asp Met Glu Glu Lys Val. Lys Arg Gln Leu
1235 1240
1245
Asp Val Ala Arg Arg Ile Arg Ala Val Asp Glu His Lys Val Ala Glu
1250 1255
12E~0
Thr Ile Leu Asn Ser His Leu Ile Pro Asp Leu Arg Gly Asn Leu Arg
1265 1270 1275 1280
Ser Phe Thr Arg Gln Glu Phe Arg Cys Val Lys Cys Asn Thr Lys Phe
1285 1290
1295
Arg Arg Pro Pro Leu Asp Gly Lys Cys Pro Ile Cys Gly Gly Lys Ile
1300 1305 1310
Val Leu Thr Val Ser Lys Gly Ala Ile Glu Lys Tyr Leu Gly Thr Ala
1315 1320
1325
Lys Met Leu Val Thr Glu Tyr Lys Val Lys Asn Tyr Thr Arg Gln Rrg
1330 1335
1340
Ile Cys Leu Thr Glu Arg Asp Ile Asp Ser Leu Phe Glu Thr Val Phe
1345 1350
1355 1360
Pro Glu Thr Gln Leu Thr Leu Leu Val Asn Pro Asn Asp Ile Cys Gln
1365 1370
1375
Arg Ile Ile Met Glu Arg Thr Gly Gly Ser Lys Lys Ser Gly Leu Leu
1380 1385
1390
Glu Asn Phe Ala Asn Gly Tyr Asn Lys Gly Lys Lys Glu Glu Met Pro
1395 1400
1405
Lys Lys Gln Arg Lys Lys Glu Gln Glu Lys Ser Lys Lys Arg Lys Val
1410 1415 1420
CA 02338185 2001-02-05
_ 71
a- Ile Ser Leu Asp Asp Phe Phe Ser Arg Lys
1425 1430
(2) INFORMATION FOR SEQ ID N0: 30:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1092 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Methanobacterium thermoautot:rophicum
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 30:
Met Met Asp Tyr Phe Asn Glu Leu Glu Arg Glu Thr Glu Arg Leu Tyr
1 5 10 15
Glu Ile Ala Rrg Lys Ala Arg Ala Arg Gly Leu Asp Val Ser Thr Thr
20 25 30
Pro Glu Ile Pro Leu Ala Lys Asp Leu Ala Glu Arg Val Glu Gly Leu
35 40 45
Val Gly Pro Glu Gly Ile Ala Arg Arg Ile Lys Glu Leu Glu Gly Asp
50 55 60
Arg Gly Arg Glu Glu Val Ala Phe Gln Ile Ala Ala Glu Ile Ala Ser
65 70 75 80
Gln Ala Val Pro Asp Asp Asp Pro Glu Glu Arg Glu Lys Leu Ala Asp
85 90 95
Gln Ala Leu Arg Thr Ala Leu Ala Ile Leu Thr Glu Gly Val Val Ala
100 105 110
Ala Pro Leu Glu Gly Ile Ala Arg Val Arg Ile Lys Glu Asn Phe Asp
115 120 125
Lys Ser Arg Tyr Leu Ala Val Tyr Phe Ala Gly Pro Ile Arg Ser Ala
130 135 140
Gly Gly Thr Ala Ala Ala Leu Ser Val Leu Ile Ala Asp Tyr Ile Arg
145 150 155 160
Leu Ala Val Gly Leu Asp Arg Tyr Lys Pro Val Glu Arg Glu Ile Glu
165 170 175
Arg Tyr Val Glu Glu Val Glu Leu Tyr Glu Ser Glu Val Thr Asn Leu
180 185 190
Gln Tyr Ser Pro Lys Pro Asp Glu Val Arg Leu Ala Ala Ser Lys Ile
195 200 205
Pro Val Glu Val Thr Gly Glu Pro Thr Asp Lys Val Glu Val Ser His
210 215 220
CA 02338185 2001-02-05
- 72 -
Arg Asp Leu Glu Arg Val Glu Thr Asn Asn Ile Arg_ Gly Gly Ala Leu
225 230 235 240
Leu Ala Met Val Glu Gly Val Ile Gln Lys Ala Pro Lys Val Leu Lys
245 250 255
Tyr Ala Lys Gln Leu Lys Leu Glu Gly Trp Asp Trp Leu Glu Lys Phe
260 265 270
Ser Lys Ala Pro Lys Lys Gly Glu Gly Glu Glu Lys, Val Val Val Lys
275 280 285
Ala Asp Ser Lys Tyr Val Glu Asp Ile Ile Gly Gly Arg Pro Val Leu
290 295 300
Ala Tyr Pro Ser Glu Lys Gly Ala Phe Arg Leu Arg Tyr Gly Arg Ala
305 310 315 320
Arg Asn Thr Gly Leu Ala Ala Met Gly Val His Pro Ala Thr Met Glu
325 330 335
Leu Leu Gln Phe Leu Ala Val Gly Thr Gln Met Lys Ile Glu Arg Pro
340 345 350
Gly Lys Gly Asn Cys Val Val Pro Val Asp Thr Ile Asp Gly Pro Val
355 360 365
Val Lys Leu Arg Asn Gly Asp Val Ile Arg Ile Glu Asp Ala Glu Thr
370 375 380
Ala Ser Arg Val Arg Ser Glu Val Glu Glu Ile Leu Phe Leu Gly Asp
385 390 395 400
Met Leu Val Ala Phe Gly Glu Phe Leu Arg Asn Asn His Val Leu Met
405 910 415
Pro Ala Gly Trp Cys Glu Glu Trp Trp Ile Gln Thr Ile Leu Ser Ser
420 425 430
Pro Lys Tyr Pro Gly Asp Asp Pro Leu Asn Leu Ser Tyr Tyr Arg Thr
435 440 445
Arg Trp Asn Glu Leu Glu Val Ser Ala Gly Asp Ala Phe Arg Ile Ser
450 455 460
Glu Glu Tyr Asp Val Pro Leu His Pro Arg Tyr Thr Tyr Phe Tyr His
465 470 475 480
Asp Val Thr Val Arg Glu Leu Asn Met Leu Arg Glu Trp Leu Asn Thr
485 490 495
Ser Gln Leu Glu Asp Glu Leu Val Leu Glu Leu Arg Pro Glu Lys Arg
500 505 510
Ile Leu Glu Ile Leu Gly Val Pro His Arg Val Lys Asp Ser Arg Val
515 520 525
Val Ile Gly His Asp Asp Ala His Ala Leu Ile Lys Thr Leu Arg Lys
530 535 540
Pro Leu Glu Asp Ser Ser Asp Thr Val Glu Ala Leu Asn Arg Val Ser
545 550 555 560
CA 02338185 2001-02-05
- - 73 -
Pro Val Arg Ile Met Lys Lys Ala Pro Thr Tyr Ile Gly Thr Arg Val
565 570 575
Gly Arg Pro Glu Lys Thr Lys Glu Arg Lys Met Arg Pro Ala Pro His
580 585 590
Val Leu Phe Pro Ile Gly Lys Tyr Gly Gly Ser Arc_~ Arg Asn Ile Pro
595 600 605
Asp Ala Ala Lys Lys Gly Ser Ile Thr Val Glu Ile Gly Arg Ala Thr
610 615 620
Cys Pro Ser Cys Arg Val Ser Ser Met Gln Ser Ile Cys Pro Ser Cys
625 630 635 640
Gly Ser Arg Thr Val Ile Gly Glu Pro Gly Lys Arg Asn Ile Asn Leu
645 650 655
Ala Ala Leu Leu Lys Arg Ala Ala Glu Asn Val Ser Val Arg Lys Leu
660 665 670
Asp Glu Ile Lys Gly Val Glu Gly Met Ile Ser Ala Glu Lys Phe Pro
675 680 685
Glu Pro Leu Glu Lys Gly Ile Leu Arg Ala Lys Asn Asp Val Tyr Thr
690 695 700
Phe Lys Asp Ala Thr Ile Arg His Asp Ser Thr Asp Leu Pro Leu Thr
705 710 715 720
His Phe Thr Pro Arg Glu Val Gly Val Ser Val Glu Arg Leu Arg Glu
725 730 735
Leu Gly Tyr Thr Arg Asp Cys Tyr Gly Asp Glu Leu Glu Asp Glu Asp
740 745 750
Gln Ile Leu Glu Leu Arg Val Gln Asp Val Val Ile Ser Glu Asp Cys
755 760 765
Ala Asp Tyr Leu Val Arg Val Ala Asn Phe Val Asp Asp Leu Leu Glu
770 775 780
Rrg Phe Tyr Asp Leu Glu Arg Phe Tyr Asn Val Lys Thr Arg Glu Asp
785 790 795 800
Leu Val Gly His Leu Ile Ala Gly Leu Ala Pro His Thr Ser Ala Ala
805 810 815
Val Leu Gly Arg Ile Ile Gly Phe Thr Gly Ala Ser Ala Cys Tyr Ala
820 825 830
His Pro Tyr Phe His Ser Ala Lys Arg Arg Asn Cys Asp Ser Asp Glu
835 840 845
Asp Ser Val Met Leu Leu Leu Asp Ala Leu Leu Asn Phe Ser Lys Ser
850 855 860
Tyr Leu Pro Ser Ser Arg Gly Gly Ser Met Asp Ala Pro Leu Val Leu
865 870 875 880
Ser Thr Arg Ile Asp Pro Glu Glu Ile Asp Asp Glu Ser His Asn Ile
885 890 895
CA 02338185 2001-02-05
_ 79 _
Asp Thr Met Asp Met Ile Pro Leu Glu Val Tyr Glu Arg Ser Phe Asp
900 905 910
His Pro Arg Pro Ser Glu Val Leu Asp Val Ile Asp Asn Val Glu Lys
915 920 925
Arg Leu Gly Lys Pro Glu Gln Tyr Thr Gly Leu Met Phe Ser His Asn
930 935 940
Thr Ser Arg Ile Asp Glu Gly Pro Lys Val Cys Leu Tyr Lys Leu Leu
945 950 955 960
Pro Thr Met Lys Glu Lys Val Glu Ser Gln Ile Thr Leu Ala Glu Lys
965 970 975
Ile Arg Ala Val Asp Gln Arg Ser Val Val Glu Gly Val Leu Met Ser
980 985 990
His Phe Leu Pro Asp Met Met Gly Asn Ile Arg Ala Phe Ser Arg Gln
995 1000 1005
Lys Val Arg Cys Thr Lys Cys Asn Arg Lys Tyr Arg Arg Ile Pro Leu
1010 1015 1020
Ser Gly Glu Cys Arg Cys Gly Gly Asn Leu Val Leu Thr Val Ser Lys
1025 1030 1035 1040
Gly Ser Val Ile Lys Tyr Leu Glu Ile Ser Lys Glu Leu Ala Ser Arg
1045 1050 1055
Tyr Pro Ile Asp Pro Tyr Leu Met Gln Arg Ile Glu Ile Leu Glu Tyr
1060 1065 1070
Gly Val Asn Ser Leu Phe Glu Ser Asp Arg Ser Lys Gln Ser Ser Leu
1075 1080 1085
Asp Val Phe Leu
1090
(2) INFORMATION FOR SEQ ID NO: 31:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1263 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Pyrococcus furiosus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 31:
Met Glu Leu Pro Lys Glu Ile Glu Glu Tyr Phe Glu Met Leu Gln Arg
1 5 10 15
Glu Ile Asp Lys Ala Tyr Glu Ile Ala Lys Lys Ala Arg Ser Gln Gly
20 25 30
CA 02338185 2001-02-05
- 75 -
Lys Pro Thr Asp GluIle Thr
Asp Ser Val Pro Asp
Gln Met
Ala Ala
35 40 45
Gly ValGluSer Leu GlyProProGly AlaGln ArgIle
Arg Val Val
50 55 60
ArgGlu LeuLeuLys GluTyr AspLysGluIle ValAlaLeu Ile
Lys
65 70 75 80
ValAsp GluIleIle GluGly LysPheGlyAsp PheGlySer LysGlu
85 90 95
LysTyr AlaGluGln AlaVal ArgThrAlaLeu AlaIleLeu ThrGlu
100 105 110
GlyIle ValSerAla ProLeu GluGlyIleAla AspValLys IleLys
115 120 125
ArgAsn ThrTrpAla AspAsn SerGluTyrLeu AlaLeuTyr TyrAla
130 135 140
GlyPro IleArgSer SerGly GlyThrAlaGln AlaLeuSer ValLeu
145 150 155 160
ValGly AspTyrVal RrgArg LysLeuGlyLeu AspArgPhe LysPro
165 170 175
SerGly LysHisIle GluArg MetValGluGlu ValAspLeu TyrHis
180 185 190
ArgAla ValSerArg LeuGln TyrHisProSer ProAspGlu ValArg
195 200 205
LeuAla MetArgAsn IlePro IleGluIleThr GlyGluAla ThrAsp
210 215 220
AspVal GluValSer HisArg AspValGluGly ValGluThr AsnGln
225 230 235 240
LeuArg GlyGlyAla IleLeu ValLeuAlaGlu GlyValLeu GlnLys
245 250 255
AlaLys LysLeuVal LysTyr IleAspLysMet GlyIleAsp GlyTrp
260 265 270
GluTrp LeuLysGlu PheVal GluAlaLysGlu LysGlyGlu GluIle
275 280 285
GluGlu SerGluSer LysAla GluGluSerLys ValGluThr ArgVal
290 295 300
GluVal GluLysGly PheTyr LysLeuTyr GluLysPhe ArgAla
Tyr
305 310 315 320
GluIle ProSer GluLys LysGlu IleIleGly Gly
Ala Tyr Arg
Ala
325 330 335
ProLeu Phe ProSer Phe Leu Arg
Ala Glu Arg Tyr
Gly Asn
Gly
Gly
340 345 350
Gly Ser Pro
Arg :Ile Ala
Ser Asn
Arg
Val
Ser
Gly
Phe
Ala
Thr
Trp
355 360 :365
CA 02338185 2001-02-05
- 76 -
Thr Met Val Leu Val Asp Glu Phe Leu Ala Ile Gly Thr Gln Met Lys
370 375 380
Thr Glu Arg Pro Gly Lys Gly Ala Val Val Thr Pro Ala Thr Thr Ala
385 390 395 400
Glu Gly Pro Ile Val Lys Leu Lys Asp Gly Ser Val Val Arg Val Asp
405 410 415
Asp Tyr Asn Leu Ala Leu Lys Ile Arg Asp Glu Val Glu Glu Ile Leu
420 425 430
Tyr Leu Gly Asp Ala Ile Ile Ala Phe Gly Asp Phe Val Glu Asn Asn
435 440 445
Gln Thr Leu Leu Pro Ala Asn Tyr Val Glu Glu Trp Trp Ile Gln Glu
450 455 460
Phe Val Lys Ala Val Asn Glu Ala Tyr Glu Val Glu Leu Arg Pro Phe
465 470 475 480
Glu Glu Asn Pro Arg Glu Ser Val Glu Glu Ala Ala Glu Tyr Leu Glu
485 490 495
Val Asp Pro Glu Phe Leu Ala Lys Met Leu Tyr Asp Pro Leu Arg Val
500 505 510
Lys Pro Pro Val Glu Leu Ala Ile His Phe Ser Glu Ile Leu Glu Ile
515 520 525
Pro Leu His Pro Tyr Tyr Thr Leu Tyr Trp Asn Thr Val Asn Pro Lys
530 535 540
Asp Val Glu Arg Leu Trp Gly Val Leu Lys Asp Lys Ala Thr Ile Glu
545 550 555 560
Trp Gly Thr Phe Arg Gly Ile Lys Phe Ala Lys Lys Ile Glu Ile Ser
565 570 575
Leu Asp Asp Leu Gly Ser Leu Lys Arg Thr Leu Glu Leu Leu Gly Leu
580 585 590
Pro His Thr Val Arg Glu Gly Ile Val Val Val Asp Tyr Pro Trp Ser
595 600 605
Ala Ala Leu Leu Thr Pro Leu Gly Asn Leu Glu Trp Glu Phe Lys Ala
610 615 620
Lys Pro Phe Tyr Thr Val Ile Asp Ile Ile Asn Glu Asn Asn Gln Ile
625 630 635 640
Lys Leu Arg Asp Arg Gly Ile Ser Trp Ile Gly Ala Arg Met Gly Arg
645 650 655
Pro Glu Lys Ala Lys Glu Arg Lys Met Lys Pro Pro Val Gln Val Leu
660 665 670
Phe Pro Ile Gly Leu Ala Gly Gly Ser Ser Arg Asp Ile Lys Lys Ala
675 680 685
Ala Glu Glu Gly Lys Ile Ala Glu Val Glu Ile Ala Phe Phe Lys Cys
690 695 700
CA 02338185 2001-02-05
_ 77 _
ProLys CysGly HisValGly ProGluThr Pro GluCysGly
Leu
Cy~s
705 710 715 720
IleArg LysGlu LeuIleTrp ThrCysPro LysCy~oGly GluTyr
Ala
725 730 735
ThrAsn SerGln AlaGluGly TyrSerTyr SerCysPro LysCysAsn
740 745 750
ValLys LeuLys ProPheThr LysArgLys IleLysPro SerGluLeu
755 760 765
LeuAsn ArgAla MetGluAsn ValLysVal TyrGlyVal AspLysLeu
770 775 780
LysGly ValMet GlyMetThr SerGlyTrp LysIleAla GluProLeu
785 790 795 800
GluLys GlyLeu LeuArgAla LysAsnGlu ValTyrVal PheLysAsp
805 810 815
GlyThr IleArg PheAspAla ThrAspAla ProIleThr HisPheArg
820 825 830
ProArg GluIle GlyValSer ValGluLys LeuArgGlu LeuGlyTyr
835 840 845
ThrHis AspPhe GluGlyLys ProLeuVal SerGluAsp GlnIleVal
850 855 860
GluLeu LysPro GlnAspVal IleLeuSer LysGluAla GlyLysTyr
865 870 875 880
LeuLeu ArgVal AlaArgPhe ValAspAsp LeuLeuGlu LysPheTyr
885 890 895
GlyLeu ProArg PheTyrAsn AlaGluLys MetGluAsp LeuIleGly
900 905 910
HisLeu ValIle GlyLeuAla ProHisThr SerAlaGly IleValGly
915 920 925
ArgIle IleGly PheValAsp AlaLeuVal GlyTyrAla HisProTyr
930 935 94p
PheHis AlaAla LysArgArg AsnCysAsp GlyAspGlu AspSerVal
945 950 955 960
MetLeu LeuLeu AspAlaLeu LeuAsnPhe SerArgTyr TyrLeuPro
965 970 975
GluLys ArgGly GlyLysMet AspAlaPro LeuValIle ThrThrArg
980 985 990
LeuAsp ProArg GluValAsp SerGluVal HisAsnMet AspValVal
995 1000 1005
ArgTyr TyrPro LeuGluPhe GluAla ThrTyrGlu LeuLysSer
Tyr
1010 1015 1020
ProLys GluLeu Val IleGluGly ValGlu LeuGly
Val Asp
Arg Arg
1025 1030 1035 1040
CA 02338185 2001-02-05
_ 78
Lys Pro Glu Met Tyr Tyr Gly Ile Lys Phe Thr His Asp Thr Asp Asp
1045 1050 1055
Ile Ala Leu Gly Pro Lys Met Ser Leu Tyr Lys Gln Leu Gly Asp Met
1060 1065 1070
Glu Glu Lys Val Lys Arg Gln Leu Thr Leu Ala Glu Arg Ile Arg Ala
1075 1080 1085
Val Asp Gln His Tyr Val Ala Glu Thr Ile Leu Asn. Ser His Leu Ile
1090 1095 1100
Pro Asp Leu Arg Gly Asn Leu Arg Ser Phe Thr Arg Gln Glu Phe Arg
1105 1110 1115 1120
Cys Val Lys Cys Asn Thr Lys Tyr Arg Arg Pro Pro Leu Asp Gly Lys
1125 1130 1135
Cys Pro Val Cys Gly Gly Lys Ile Val Leu Thr Val Ser Lys Gly Ala
1140 1145 1150
Ile Glu Lys Tyr Leu Gly Thr Ala Lys Met Leu Val Ala Asn Tyr Asn
1155 1160 1165
Val Lys Pro Tyr Thr Arg Gln Arg Ile Cys Leu Thr Glu Lys Asp Ile
1170 1175 1180
Asp Ser Leu Phe Glu Tyr Leu Phe Pro Glu Ala Gln Leu Thr Leu Ile
1185 1190 1195 1200
Val Asp Pro Asn Asp Ile Cys Met Lys Met Ile Lys Glu Arg Thr Gly
1205 1210 1215
Glu Thr Val Gln Gly Gly Leu Leu Glu Asn Phe Asn Ser Ser Gly Asn
1220 1225 1230
Asn Gly Lys Lys Ile Glu Lys Lys Glu Lys Lys Ala Lys Glu Lys Pro
1235 1240 1245
Lys Lys Lys Lys Val Ile Ser Leu Asp Asp Phe Phe Ser Lys Arg
1250 1255 126()
(2) INFORMATION FOR SEQ ID N0: 32:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 363 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Homo sapiens
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 32:
Met Gln Ala Phe Leu Lys Gly Thr Ser Ile Ser Thr Lys Pro Pro Leu
1 5 10 15
CA 02338185 2001-02-05
_ 79 _
Thr Lys Asp Arg Gly Val Ala Ala Ser Ala Gly Ser Ser Gly Glu Asn
20 25 30
Lys Lys AlaLys ValPro ValGlu Arg Cys
Pro Trp Lys Pro
Tyr Lys
35 40 45
Val Asp GluValAla PheGln GluValVal AlaValLeu Lys
Glu Lys
50 55 60
Ser Leu GluGly AspLeu AsnLeuLeu PheTyrGly Pro
Ala Pro Pro
65 70 75 80
Gly Thr GlyLysThr SerThrIle LeuAlaAla AlaArgGlu LeuPhe
85 90 95
Gly Pro GluLeuPhe ArgLeuArg ValLeuGlu LeuAsnAla SerAsp
100 105 110
Glu Arg GlyIleGln ValValArg GluLysVal LysAsnPhe AlaGln
115 120 125
Leu Thr ValSerGly SerArgSer AspGlyLys ProCysPro ProPhe
130 135 140
Lys Ile ValIleLeu AspGluAla AspSerMet ThrSerAla AlaGln
145 150 155 160
Ala Ala LeuArgArg ThrMetGlu LysGluSer LysThrThr ArgPhe
165 170 175
Cys Leu IleCysAsn TyrValSer ArgIleIle GluProLeu ThrSer
180 185 190
Arg Cys SerLysPhe ArgPheLys ProLeuSer AspLysIle GlnGln
195 200 205
Gln Arg LeuLeuAsp IleAlaLys LysGluAsn ValLysIle SerAsp
210 215 220
Glu Gly IleAlaTyr LeuValLys ValSerGlu GlyAspLeu ArgLys
225 230 235 240
Ala Ile ThrPheLeu GlnSerAla ThrArgLeu ThrGlyGly LysGlu
245 250 255
Ile Thr GluLysVal IleThrAsp IleAlaGly ValIlePro AlaGlu
260 265 270
Lys Ile AspGlyVal PheAlaAla CysGlnSer GlySerPhe AspLys
275 280 285
Leu Glu AlaValVal LysAspLeu IleAspGlu GlyHisAla AlaThr
290 295 300
Gln Leu Val Gln LeuHisAsp ValValVal GluAsnAsn LeuSer
Asn .
305 310 315 320
Asp Lys Gln Ser IleIleThr GluLysLeu AlaGluVal AspLys
Lys
325 330 335
Cys Leu Gly AspGlu LeuGln LeuIleSer LeuCys
Ala Asp Ala His
340 345 350
CA 02338185 2001-02-05
_ 80 _
Ala Thr Val Met Gln Gln Leu Ser Gln Asn Cys
355 360
(2) INFORMATION FOR SEQ ID NO: 33:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 329 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Homo Sapiens
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 33:
Asn Leu Val Gln Cys Gly Asp Phe Pro His Leu Leu Val Tyr Gly Pro
1 5 10 15
Ser Gly Ala Gly Lys Lys Thr Arg Ile Met Cys Ile Leu Arg Glu Leu
20 25 30
Tyr Gly Val Gly Val Glu Lys Leu Arg Ile Glu His Gln Thr Ile Thr
35 40 45
Thr Pro Ser Lys Lys Lys Ile Glu Ile Ser Thr Ile Ala Ser Asn Tyr
50 55 60
His Leu Glu Val Asn Pro Ser Asp Ala Gly Asn Ser Asp Arg Val Val
65 70 75 80
Ile Gln Glu Met Leu Lys Thr Val Ala Gln Ser Gln Gln Leu Glu Thr
85 90 95
Rsn Ser Gln Arg Asp Phe Lys Val Val Leu Leu Thr Glu Val Asp Lys
100 105 110
Leu Thr Lys Asp Ala Gln His Ala Leu Arg Arg Thr Met Glu Lys Tyr
115 120 125
Met Ser Thr Cys Arg Leu Ile Leu Cys Cys Asn Ser Thr Ser Lys Val
130 135 140
Ile Pro Pro Ile Arg Ser Arg Cys Leu Ala Val Arg Val Pro Ala Pro
145 150 155 160
Ser Ile Glu Asp Ile Cys His Val Leu Ser Thr Val Cys Lys Lys Glu
165 170 175
Gly Leu Asn Leu Pro Ser Gln Leu Ala His Arg Leu Ala Glu Lys Ser
180 185 190
Cys Arg Asn Leu Arg Lys Ala Leu Leu Met Cys Glu Ala Cys Arg Val
195 200 205
Gln Gln Tyr Pro Phe Thr Ala Asp Gln Glu Ile Pro Glu Thr Asp Trp
210 215 220
CA 02338185 2001-02-05
- 81 -
Glu Val Tyr Leu Arg Glu Thr Ala Asn Ala Ile Val Ser Gln Gln Thr
225 230 235 240
Pro Gln Arg Leu Leu Glu Val Arg Gly Arg Leu Tyr Glu Leu Leu Thr
245 250 255
His Cys Ile Pro Pro Glu Ile Ile Met Lys Gly Leu Leu Ser Glu Leu
260 265 270
Leu His Asn Cys Asp Gly Gln Leu Lys Gly Glu Val Ala Gln Met Ala
275 280 285
Ala Tyr Tyr Glu His Arg Leu Gln Leu Gly Ser Lys Ala Ile Tyr His
290 295 300
Leu Glu Ala Phe Val Ala Lys Phe Met Ala Leu Tyr Lys Lys Phe Ile
305 310 315 320
Gln Asp Gly Leu Glu Gly Met Met Phe
325
(2) INFORMATION FOR SEQ ID NO: 34:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 354 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Homo Sapiens
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 34:
Met Glu Val Glu Ala Val Cys Gly Gly Ala Gly Glu Val Glu Ala Gln
1 5 10 15
Asp Ser Asp Pro Ala Pro Ala Phe Ser Lys Ala Pro Gly Ser Ala Gly
20 25 30
His Tyr Glu Leu Pro Trp Val Glu Lys Tyr Arg Pro Val Lys Leu Asn
35 40 45
Glu Ile Val Gly Asn Glu Asp Thr Val Ser Arg Leu Glu Val Phe Ala
50 55 60
Arg Glu Gly Asn Val Pro Asn Ile Ile Ile Ala Gly Pro Pro Gly Thr
65 70 75 g0
Gly Lys Thr Thr Ser Ile Leu Cys Leu Ala Arg Ala Leu Leu Gly Pro
85 90 95
Ala Leu Lys Asp Ala Met Leu Glu Leu Asn Ala Ser Asn Asp Arg Gly
100 105 110
Ile Asp Val Val Arg Asn Lys Ile Lys Met Phe Ala Gln Gln Lys Val
115 120 125
CA 02338185 2001-02-05
- 82 -
Thr Leu Pro Lys Gly Arg His Lys Ile Ile Ile Leu Asp Glu Ala Asp
130 135 140
Ser Met Thr Asp Gly Ala Gln Gln Ala Leu Arg Arg Thr Met Glu Ile
145 150 155 160
Tyr Ser Lys Thr Thr Arg Phe Ala Leu Ala Cys Asn Ala Ser Asp Lys
165 170 175
Ile Ile Glu Pro Ile Gln Ser Arg Cys Ala Val Leu Arg Tyr Thr Lys
180 185 190
Leu Thr Asp Ala Gln Ile Leu Thr Arg Leu Met Asn Val Ile Glu Lys
195 20C 205
Glu Arg Val Pro Tyr Thr Asp Asp Gly Leu Glu Ala Ile Ile Phe Thr
210 215 220
Ala Gln Gly Asp Met Arg Gln Ala Leu Asn Asn Leu Gln Ser Thr Phe
225 230 235 240
Ser Gly Phe Gly Phe Ile Asn Ser Glu Asn Val Phe Lys Val Cys Asp
245 250 255
Glu Pro His Pro Leu Leu Val Lys Glu Met Ile Gln His Cys Val Asn
260 265 270
Ala Asn Ile Asp Glu Ala Tyr Lys Ile Leu Ala His Leu Trp His Leu
275 280 285
Gly Tyr Ser Pro Glu Asp Ile Ile Gly Asn Ile Phe Arg Val Cys Lys
290 295 300
Thr Phe Gln Met Ala Glu Tyr Leu Lys Leu Glu Phe Ile Lys Glu Ile
305 310 315 320
Gly Tyr Thr His Met Lys Ile Ala Glu Gly Val Asn Ser Leu Leu Gln
325 330 335
Met Ala Gly Leu Leu Ala Arg Leu Cys Gln Lys Thr Met Ala Pro Val
340 345 350
Ala Ser
(2) INFORMATION FOR SEQ ID NO: 35:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 366 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Escherichia coli
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 35:
CA 02338185 2001-02-05
- 83 -
Met Lys Phe Thr Val Glu Arg Glu His Leu Leu Lys Pro Leu Gln Gln
1 5 10 15
Val Ser Gly Pro Leu Gly Gly Arg Pro Thr Leu Pro Ile Leu Gly Asn
20 25 30
Leu Leu Leu Gln Val Ala Asp Gly Thr Leu Ser Leu Thr Gly Thr Asp
35 40 q5
Leu Glu Met Glu Met Val Ala Arg Val Ala Leu Val Gln Pro His Glu
50 55 60
Pro Gly Ala Thr Thr Val Pro Ala Arg Lys Phe Phe Asp Ile Cys Arg
65 70 75 80
Gly Leu Pro Glu Gly Ala Glu Ile Ala Val Gln Leu Glu Gly Glu Arg
85 90 95
Met Leu Val Arg Ser Gly Arg Ser Arg Phe Ser Leu Ser Thr Leu Pro
100 105 110
Ala Ala Asp Phe Pro Asn Leu Asp Asp Trp Gln Ser Glu Val Glu Phe
115 120 125
Thr Leu Pro Gln Ala Thr Met Lys Arg Leu Ile Glu Ala Thr Gln Phe
130 135 140
Ser Met Ala His Gln Asp Val Arg Tyr Tyr Leu Asn Gly Met Leu Phe
145 150 155 160
Glu Thr Glu Gly Glu Glu Leu Arg Thr Val Ala Thr Asp Gly His Arg
165 170 175
Leu Ala Val Cys Ser Met Pro Ile Gly Gln Ser Leu Pro Ser His Ser
180 185 190
Val Ile Val Pro Arg Lys Gly Val Ile Glu Leu Met Arg Met Leu Asp
195 200 205
Gly Gly Asp Asn Pro Leu Arg Val Gln Ile Gly Ser Asn Asn Ile Arg
210 215 220
Ala His Val Gly Asp Phe Ile Phe Thr Ser Lys Leu Val Asp Gly Arg
225 230 235 240
Phe Pro Asp Tyr Arg Arg Val Leu Pro Lys Asn Pro Asp Lys His Leu
245 250 255
Glu Ala Gly Cys Asp Leu Leu Lys Gln Ala Phe Ala Arg Ala Ala Ile
260 265 270
Leu Ser Asn Glu Lys Phe Arg Gly Val Arg Leu Tyr Val Ser Glu Asn
275 280 285
Gln Leu Lys Ile Thr Ala Asn Asn Pro Glu Gln Glu Glu Ala Glu Glu
290 295 300
Ile Leu Asp Val Thr Tyr Ser Gly Ala Glu Met Glu Ile Gly Phe Asn
305 310 315 320
Val Ser Tyr Val Leu Asp Val Leu Asn Ala Leu Lys Cys Glu Asn Val
325 330 335
CA 02338185 2001-02-05
_ 84 _
Arg Met Met Leu Thr Asp Ser Val Ser Ser Val Gln Ile Glu Asp Ala
340 345 350
Ala Ser Gln Ser Ala Ala Tyr Val Val Met Pro Met Arg Leu
355 360 365
(2) INFORMATION FOR SEQ ID NO: 36:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 363 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Aquifex Aeolicus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 36:
Met Arg Val Lys Val Asp Arg Glu Glu Leu Glu Glu Val Leu Lys Lys
1 5 10 15
Ala Arg Glu Ser Thr Glu Lys Lys Ala Ala Leu Pro Ile Leu Ala Asn
20 25 30
Phe Leu Leu Ser Ala Lys Glu Glu Asn Leu Ile Val Arg Ala Thr Asp
35 40 45
Leu Glu Asn Tyr Leu Val Val Ser Val Lys Gly Glu Val Glu Glu Glu
50 55 60
Gly Glu Val Cys Val His Ser Gln Lys Leu Tyr Asp Ile Val Lys Asn
65 70 75 80
Leu Asn Ser Ala Tyr Val Tyr Leu His Thr Glu Gly Glu Lys Leu Val
85 90 95
Ile Thr Gly Gly Lys Ser Thr Tyr Lys Leu Pro Thr Ala Pro Ala Glu
100 105 110
Asp Phe Pro Glu Phe Pro Glu Ile Val Glu Gly Gly Glu Thr Leu Ser
115 120 125
Gly Asn Leu Leu Val Asn Gly Ile Glu Lys Val Glu 'ryr Ala Ile Ala
130 135 140
Lys Glu Glu Ala Asn Ile Ala Leu Gln Gly Met Tyr :Leu Arg Gly Tyr
145 150 155 160
Glu Asp Arg Ile His Phe Val Gly Ser Asp Gly His Arg Leu Ala Leu
165 170 175
Tyr Glu Pro Leu Gly Glu Phe Ser Lys Glu Leu Leu :Cle Pro Arg Lys
180 185 190
Ser Leu Lys Val Leu Lys Lys Leu Ile Thr Gly Ile C~lu Asp Val Asn
195 200 205
CA 02338185 2001-02-05
- 85 -
Ile Glu Lys Ser Glu Asp Glu Ser Phe Ala Tyr Phe Ser Thr Pro Glu
210 215 220
Trp Lys Leu Ala Val Arg Leu Leu Glu Gly Glu Phe Pro Asp Tyr Met
225 230 235 240
Ser Val Ile Pro Glu Glu Phe Ser Ala Glu Val Leu Phe Glu Thr Glu
245 250 255
Glu Val Leu Lys Val Leu Lys Arg Leu Lys Ala Leu Ser Glu Gly Lys
260 265 270
Val Phe Pro Val Lys Ile Thr Leu Ser Glu Asn Leu Ala Ile Phe Glu
275 280 285
Phe Ala Asp Pro Glu Phe Gly Glu Ala Arg Glu Glu Ile Glu Val Glu
290 295 300
Tyr Thr Gly Glu Pro Phe Glu Ile Gly Phe Asn Gly Lys Tyr Leu Met
305 310 315 320
Glu Ala Leu Asp Ala Tyr Asp Ser Glu Arg Val Trp Phe Lys Phe Thr
325 330 335
Thr Pro Asp Thr Ala Thr Leu Leu Glu Ala Glu Asp Tyr Glu Lys Glu
340 345 350
Pro Tyr Lys Cys Ile Ile Met Pro Met Arg Val
355 360
(2) INFORMATION FOR SEQ ID NO: 37:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1160 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Escherichia coli
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 37:
Met Ser Glu Pro Arg Phe Val His Leu Arg Val His Ser Asp Tyr Ser
1 5 10 15
Met Ile Asp Gly Leu Ala Lys Thr Ala Pro Leu Val Lys Lys Ala Ala
20 25 30
Ala Leu Gly Met Pro Ala Leu Ala Ile Thr Asp Phe Thr Asn Leu Cys
35 40 45
Gly Leu Val Lys Phe Tyr Gly Ala Gly His Gly Ala Gly Ile Lys Pro
50 55 60
Ile Val Gly Ala Asp Phe Asn Val Gln Cys Asp Leu Leu Gly Asp Glu
65 70 75 80
CA 02338185 2001-02-05
- 86 -
Leu Thr His Leu Thr Val Leu Ala Ala Asn Asn Thr Gly Tyr Gln Asn
85 90 95
Leu Thr Leu Leu Ile Ser Lys Ala Tyr Gln Arg Gly Tyr Gly Ala Ala
100 105 110
Gly Pro Ile Ile Asp Arg Asp Trp Leu Ile Glu Leu Asn Glu Gly Leu
115 120 125
Ile Leu Leu Ser Gly Gly Arg Met Gly Asp Val Gly Arg Ser Leu Leu
130 135 140
Arg Gly Asn Ser Ala Leu Val Asp Glu Cys Val Ala Phe Tyr Glu Glu
145 150 155 160
His Phe Pro Asp Arg Tyr Phe Leu Glu Leu Ile Arg Thr Gly Arg Pro
165 170 175
Asp Glu Glu Ser Tyr Leu His Ala Ala Val Glu Leu Ala Glu Ala Arg
180 185 190
Gly Leu Pro Val Val Ala Thr Asn Asp Val Arg Phe Ile Asp Ser Ser
195 200 205
Asp Phe Asp Ala His Glu Ile Arg Val Ala Ile His Asp Gly Phe Thr
210 215 220
Leu Asp Asp Pro Lys Arg Pro Arg Asn Tyr Ser Pro Gln Gln Tyr Met
225 230 235 240
Arg Ser Glu Glu Glu Met Cys Glu Leu Phe Ala Asp Ile Pro Glu Ala
245 250 255
Leu Ala Asn Thr Val Glu Ile Ala Lys Arg Cys Asn Val Thr Val Arg
260 265 270
Leu Gly Glu Tyr Phe Leu Pro Gln Phe Pro Thr Gly Asp Met Ser Thr
275 280 285
Glu Asp Tyr Leu Val Lys Arg Ala Lys Glu Gly Leu Glu Glu Arg Leu
290 295 300
Ala Phe Leu Phe Pro Asp Glu Glu Glu Arg Leu Lys Arg Arg Pro Glu
305 310 315 320
Tyr Asp Glu Arg Leu Glu Thr Glu Leu Gln Val Ile Asn Gln Met Gly
325 330 335
Phe Pro Gly Tyr Phe Leu Ile Val Met Glu Phe Ile Gln Trp Ser Lys
340 345 350
Asp Asn Gly Val Pro Val Gly Pro Gly Arg Gly Ser Gly Ala Gly Ser
355 360 365
Leu Val Ala Tyr Ala Leu Lys Ile Thr Asp Leu Asp Pro Leu Glu Phe
370 375 380
Asp Leu Leu Phe Glu Arg Phe Leu Asn Pro Glu Arg 'Val Ser Met Pro
385 390 395 400
Asp Phe Asp Val Asp Phe Cys Met Glu Lys Arg Asp Gln Val Ile Glu
405 410 415
CA 02338185 2001-02-05
_ 87 _
His Val Ala Asp Met Tyr Gly Arg Asp Ala Val Ser Gln Ile Ile Thr
420 425 930
Phe Gly Thr Met Ala Ala Lys Ala Val Ile Arg Asp Val Gly Arg Val
435 940 495
Leu Gly His Pro Tyr Gly Phe Val Asp Arg Ile Ser Lys Leu Ile Pro
450 455 460
Pro Asp Pro Gly Met Thr Leu Ala Lys Ala Phe Glu Ala Glu Pro Gln
465 470 475 480
Leu Pro Glu Ile Tyr Glu Ala Asp Glu Glu Val Lys Ala Leu Ile Asp
985 490 495
Met Ala Arg Lys Leu Glu Gly Val Thr Arg Asn Ala Gly Lys His Ala
500 505 510
Gly Gly Val Val Ile Ala Pro Thr Lys Ile Thr Asp Phe Ala Pro Leu
515 520 525
Tyr Cys Asp Glu Glu Gly Lys His Pro Val Thr Gln Phe Asp Lys Ser
530 535 540
Asp Val Glu Tyr Ala Gly Leu Val Lys Phe Asp Phe Leu Gly Leu Arg
545 550 555 560
Thr Leu Thr Ile Ile Asn Trp Ala Leu Glu Met Ile Asn Lys Arg Arg
565 570 575
Ala Lys Asn Gly Glu Pro Pro Leu Asp Ile Ala Ala Ile Pro Leu Asp
580 585 590
Asp Lys Lys Ser Phe Asp Met Leu Gln Arg Ser Glu Thr Thr Ala Val
595 600 605
Phe Gln Leu Glu Ser Arg Gly Met Lys Asp Leu Ile Lys Arg Leu Gln
610 615 620
Pro Asp Cys Phe Glu Asp Met Ile Ala Leu Val Ala Leu Phe Arg Pro
625 630 635 640
Gly Pro Leu Gln Ser Gly Met Val Asp Asn Phe Ile Asp Arg Lys His
645 650 655
Gly Arg Glu Glu Ile Ser Tyr Pro Asp Val Gln Trp Gln His Glu Ser
660 665 670
Leu Lys Pro Val Leu Glu Pro Thr Tyr Gly Ile Ile Leu Tyr Gln Glu
675 680 685
Gln Val Met Gln Ile Ala Gln Val Leu Ser Gly Tyr Thr Leu Gly Gly
690 695 700
Ala Asp Met Leu Arg Arg Ala Met Gly Lys Lys Lys Pro Glu Glu Met
705 710 715 720
Ala Lys Gln Arg Ser Val Phe Ala Glu Gly Ala Glu Lys Asn Gly Ile
725 730 735
Asn Ala Glu Leu Ala Met Lys Ile Phe Asp Leu Val ~~lu Lys Phe Ala
740 745 750
CA 02338185 2001-02-05
_ 88 _
Gly Tyr Gly Phe Asn Lys Ser His Ser Ala Ala Tyr Ala Leu Val Ser
755 760 765
Tyr Gln Thr Leu Trp Leu Lys Ala His Tyr Pro Ala Glu Phe Met Ala
770 775 780
Ala Val Met Thr Ala Asp Met Asp Asn Thr Glu Lys Val Val Gly Leu
785 790 795 800
Val Asp Glu Cys Trp Arg Met Gly Leu Lys Ile Leu Pro Pro Asp Ile
805 810 815
Asn Ser Gly Leu Tyr His Phe His Val Asn Asp Asp Gly Glu Ile Val
820 825 830
Tyr Gly Ile Gly Ala Ile Lys Gly Val Gly Glu Gly Pro Ile Glu Ala
835 840 845
Ile Ile Glu Ala Arg Asn Lys Gly Gly Tyr Phe Arg Glu Leu Phe Asp
850 855 860
Leu Cys Ala Arg Thr Asp Thr Lys Lys Leu Asn Arg Arg Val Leu Glu
865 870 875 g80
Lys Leu Ile Met Ser Gly Ala Phe Asp Arg Leu Gly Pro His Arg Ala
885 890 895
Ala Leu Met Asn Ser Leu Gly Asp Ala Leu Lys Ala Ala Asp Gln His
900 905 910
Ala Lys Ala Glu Ala Ile Gly Gln Ala Asp Met Phe Gly Val Leu Ala
915 920 925
Glu Glu Pro Glu Gln Ile Glu Gln Ser Tyr Ala Ser Cys Gln Pro Trp
930 935 940
Pro Glu Gln Val Val Leu Asp Gly Glu Arg Glu Thr Leu Gly Leu Tyr
945 950 955 960
Leu Thr Gly His Pro Ile Asn Gln Tyr Leu Lys Glu Ile Glu Arg Tyr
965 970 975
Val Gly Gly Val Arg Leu Lys Asp Met His Pro Thr Glu Arg Gly Lys
980 985 990
Val Ile Thr Ala Ala Gly Leu Val Val Ala Ala Arg Val Met Val Thr
995 1000 1005
Lys Arg Gly Asn Arg Ile Gly Ile Cys Thr Leu Asp .Asp Arg Ser Gly
1010 1015 1020
Arg Leu Glu Val Met Leu Phe Thr Asp Ala Leu Asp Lys Tyr Gln Gln
1025 1030 1035 1040
Leu Leu Glu Lys Asp Arg Ile Leu Ile Val Ser Gly Gln Val Ser Phe
1045 1050 1055
Asp Asp Phe Ser Gly Gly Leu Lys Met Thr Ala Arg Glu Val Met Asp
1060 1065 1070
Ile Asp Glu Ala Arg Glu Lys Tyr Ala Arg Gly Leu Ala Ile Ser Leu
1075 1080 1085
CA 02338185 2001-02-05
_ 89 _
Thr Asp Arg Gln Ile Asp Asp Gln Leu Leu Asn Arg Leu Arg Gln Ser
1090 1095 1100
Leu Glu Pro His Arg Ser Gly Thr Ile Pro Val His Leu Tyr Tyr Gln
1105 1110 1115 1120
Arg Ala Asp Ala Arg Ala Arg Leu Arg Phe Gly Ala Thr Trp Arg Val
1125 1130 1135
Ser Pro Ser Asp Arg Leu Leu Asn Asp Leu Arg Gly Leu Ile Gly Ser
1190 1145 1150
Glu Gln Val Glu Leu Glu Phe Asp
1155 1160
(2) INFORMATION FOR SEQ ID NO: 38:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1161 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: Aquifex Aeolicus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 38:
Met Ser Lys Asp Phe Val His Leu His Leu His Thr Gln Phe Ser Leu
1 5 10 15
Leu Asp Gly Ala Ile Lys Ile Asp Glu Leu Val Lys Lys Ala Lys Glu
20 25 30
Tyr Gly Tyr Lys Ala Val Gly Met Ser Asp His Gly Asn Leu Phe Gly
35 40 45
Ser Tyr Lys Phe Tyr Lys Ala Leu Lys Ala Glu Gly Ile Lys Pro Ile
50 55 60
Ile Gly Met Glu Ala Tyr Phe Thr Thr Gly Ser Arg Phe Asp Arg Lys
65 70 75 80
Thr Lys Thr Ser Glu Asp Asn Ile Thr Asp Lys Tyr Asn His His Leu
85 90 95
Ile Leu Ile Ala Lys Asp Asp Lys Gly Leu Lys Asn Leu Met Lys Leu
100 105 110
Ser Thr Leu Ala Tyr Lys Glu Gly Phe Tyr Tyr Lys Pro Arg Ile Asp
115 120 125
Tyr Glu Leu Leu Glu Lys Tyr Gly Glu Gly Leu Ile Ala Leu Thr Ala
130 135 140
Cys Leu Lys Gly Val Pro Thr Tyr Tyr Ala Ser Ile Asn Glu Val Lys
145 150 155 160
CA 02338185 2001-02-05
- 90 -
Lys Ala Glu Glu Trp Val Lys Lys Phe Lys Asp Ile Phe Gly Asp Asp
165 170 175
Leu Tyr Leu Glu Leu Gln Ala Asn Asn Ile Pro Glu Gln Glu Val Ala
180 185 190
Asn Arg Asn Leu Ile Glu Ile Ala Lys Lys Tyr Asp Val Lys Leu Ile
195 200 205
Ala Thr Gln Asp Ala His Tyr Leu Asn Pro Glu Asp Arg Tyr Ala His
210 215 220
Thr Val Leu Met Ala Leu Gln Met Lys Lys Thr Ile His Glu Leu Ser
225 230 235 240
Ser Gly Asn Phe Lys Cys Ser Asn Glu Asp Leu His Phe Ala Pro Pro
245 250 255
Glu Tyr Met Trp Lys Lys Phe Glu Gly Lys Phe Glu Gly Trp Glu Lys
260 265 270
Ala Leu Leu Asn Thr Leu Glu Val Met Glu Lys Thr Ala Asp Ser Phe
275 280 285
Glu Ile Phe Glu Asn Ser Thr Tyr Leu Leu Pro Lys Tyr Asp Val Pro
290 295 300
Pro Asp Lys Thr Leu Glu Glu Tyr Leu Arg Glu Leu Ala Tyr Lys Gly
305 310 315 320
Leu Arg Gln Arg Ile Glu Arg Gly Gln Ala Lys Asp Thr Lys Glu Tyr
325 330 335
Trp Glu Arg Leu Glu Tyr Glu Leu Glu Val Ile Asn Lys Met Gly Phe
340 345 350
Ala Gly Tyr Phe Leu Ile Val Gln Asp Phe Ile Asn Trp Ala Lys Lys
355 360 365
Asn Asp Ile Pro Val Gly Pro Gly Arg Gly Ser Ala Gly Gly Ser Leu
370 375 380
Val Ala Tyr Ala Ile Gly Ile Thr Asp Val Asp Pro Ile Lys His Gly
385 390 395 400
Phe Leu Phe Glu Arg Phe Leu Asn Pro Glu Arg Val Ser Met Pro Asp
405 410 415
Ile Asp Val Asp Phe Cys Gln Asp Asn Arg Glu Lys 'Val Ile Glu Tyr
420 425 430
Val Arg Asn Lys Tyr Gly His Asp Asn Val Ala Gln :Lle Ile Thr Tyr
435 440 .~45
Asn Val Met Lys Ala Lys Gln Thr Leu Arg Asp Val Ala Arg Ala Met
450 455 460
Gly Leu Pro Tyr Ser Thr Ala Asp Lys Leu Ala Lys heu Ile Pro Gln
465 470 475 480
Gly Asp Val Gln Gly Thr Trp Leu Ser Leu Glu Glu Met Tyr Lys Thr
485 490 495
CA 02338185 2001-02-05
- 91 -
Pro Val Glu Glu Leu Leu Gln Lys Tyr Gly Glu His Arg Thr Asp Ile
500 505 510
Glu Asp Asn Val Lys Lys Phe Arg Gln Ile Cys Glu Glu Ser Pro Glu
515 520 525
Ile Lys Gln Leu Val Glu Thr Ala Leu Lys Leu Glu Gly Leu Thr Arg
530 535 540
His Thr Ser Leu His Ala Ala Gly Val Val Ile Ala Pro Lys Pro Leu
545 550 555 560
Ser Glu Leu Val Pro Leu Tyr Tyr Asp Lys Glu Gly Glu Val Ala Thr
565 570 575
Gln Tyr Asp Met Val Gln Leu Glu Glu Leu Gly Leu Leu Lys Met Asp
580 585 590
Phe Leu Gly Leu Lys Thr Leu Thr Glu Leu Lys Leu Met Lys Glu Leu
595 600 605
Ile Lys Glu Arg His Gly Val Asp Ile Asn Phe Leu Glu Leu Pro Leu
610 615 620
Asp Asp Pro Lys Val Tyr Lys Leu Leu Gln Glu Gly Lys Thr Thr Gly
625 630 635 640
Val Phe Gln Leu Glu Ser Arg Gly Met Lys Glu Leu Leu Lys Lys Leu
645 650 655
Lys Pro Asp Ser Phe Asp Asp Ile Val Ala Val Leu Ala Leu Tyr Arg
660 665 670
Pro Gly Pro Leu Lys Ser Gly Leu Val Asp Thr Tyr Ile Lys Arg Lys
675 680 685
His Gly Lys Glu Pro Val Glu Tyr Pro Phe Pro Glu Leu Glu Pro Val
690 695 700
Leu Lys Glu Thr Tyr Gly Val Ile Val Tyr Gln Glu Gln Val Met Lys
705 710 715 720
Met Ser Gln Ile Leu Ser Gly Phe Thr Pro Gly Glu Rla Asp Thr Leu
725 730 735
Arg Lys Ala Ile Gly Lys Lys Lys Ala Asp Leu Met Ala Gln Met Lys
740 745 750
Asp Lys Phe Ile Gln Gly Ala Val Glu Arg Gly Tyr Pro Glu Glu Lys
755 760 765
Ile Arg Lys Leu Trp Glu Asp Ile Glu Lys Phe Ala Ser Tyr Ser Phe
770 775 780
Asn Lys Ser His Ser Val Ala Tyr Gly Tyr Ile Ser Tyr Trp Thr Ala
785 790 795 800
Tyr Val Lys Ala His Tyr Pro Ala Glu Phe Phe Ala Val Lys Leu Thr
805 810 815
Thr Glu Lys Asn Asp Asn Lys Phe Leu Asn Leu Ile Lys Asp Ala Lys
820 825 830
CA 02338185 2001-02-05
- 92 -
Leu Phe Gly Phe Glu Ile Leu Pro Pro Asp Ile Asn Lys Ser Asp Val
835 840 845
Gly Phe Thr Ile Glu Gly Glu Asn Arg Ile Arg Phe Gly Leu Ala Arg
850 855 860
Ile Lys Gly Val Gly Glu Glu Thr Ala Lys Ile Ile Val Glu Ala Arg
865 870 875 880
Lys Lys Tyr Lys Gln Phe Lys Gly Leu Ala Asp Phe Ile Asn Lys Thr
885 890 895
Lys Asn Arg Lys Ile Asn Lys Lys Val Val Glu Ala Leu Val Lys Ala
900 905 910
Gly Ala Phe Asp Phe Thr Lys Lys Lys Arg Lys Glu Leu Leu Ala Lys
915 920 925
Val Ala Asn Ser Glu Lys Ala Leu Met Ala Thr Gln Asn Ser Leu Phe
930 935 990
Gly Ala Pro Lys Glu Glu Val Glu Glu Leu Asp Pro Leu Lys Leu Glu
945 950 955 960
Lys Glu Val Leu Gly Phe Tyr Ile Ser Gly His Pro Leu Asp Asn Tyr
965 970 975
Glu Lys Leu Leu Lys Asn Arg Tyr Thr Pro Ile Glu Asp Leu Glu Glu
980 985 990
Trp Asp Lys Glu Ser Glu Ala Val Leu Thr Gly Val Ile Thr Glu Leu
995 1000 1005
Lys Val Lys Lys Thr Lys Asn Gly Asp Tyr Met Ala Val Phe Asn Leu
1010 1015 1020
Val Asp Lys Thr Gly Leu Ile Glu Cys Val Val Phe Pro Gly Val Tyr
1025 1030 1035 1040
Glu Glu Ala Lys Glu Leu Ile Glu Glu Rsp Arg Val Val Val Val Lys
1045 1050 1055
Gly Phe Leu Asp Glu Asp Leu Glu Thr Glu Asn Val Lys Phe Val Val
1060 1065 1070
Lys Glu Val Phe Ser Pro Glu Glu Phe Ala Lys Glu Met Arg Asn Thr
1075 1080 1085
Leu Tyr Ile Phe Leu Lys Arg Glu Gln Ala Leu Asn Gly Val Ala Glu
1090 1095 1100
Lys Leu Lys Gly Ile Ile Glu Asn Asn Arg Thr Glu Asp Gly Tyr Asn
1105 1110 1115 1120
Leu Val Leu Thr Val Asp Leu Gly Asp Tyr Phe Val Asp Leu Ala Leu
1125 1130 1135
Pro Gln Asp Met Lys Leu Lys Ala Asp Arg Lys Val Val Glu Glu Ile
1140 1145 1150
Glu Lys Leu Gly Val Lys Val Ile Ile
1155 1160
CA 02338185 2001-02-05
- 93 -
(2) INFORMATION FOR SEQ ID NO: 39:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 64 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: both
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: synthetic
xi) SEQUENCE DESCRIPTION: SEQ ID NO: 39:
[GAVLIMPFW]-D-X-X-X-[GAVLIMPFWJ-X-X-[GAVLIMPFW]-X-[GAVLIMPFW]-X-
[GAVLIMPFW]-X-X-X-X-F-X-X-Y-X-X-D 64
(2) INFORMATION FOR SEQ ID NO: 40:
(i) SEQUENCE CHARACTERISTICS:
(R) LENGTH: 28 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: both
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: synthetic
xi) SEQUENCE DESCRIPTION: SEQ ID NO: 40:
[GAVLIMPFW]-X(3)-L-A-P-[KRHDE]-[GAVLIMPFW]-E 28
(2) INFORMATION FOR SEQ ID NO: 41:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 51 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: both
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: synthetic
xi) SEQUENCE DESCRIPTION: SEQ ID NO: 41:
C-N-Y-X-S-[KRHDE]-I-I-X-[GAVLIMPFW)-[GAVLIMPFW]-Q-S-R-C-X-X-F-R-F-X-P-
[GAVLIMPFW] 51
CA 02338185 2001-02-05
_ 94 _
(2) INFORMATION FOR SEQ ID NO: 42:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 41 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: both
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: synthetic
xi) SEQUENCE DESCRIPTION: SEQ ID NO: 42:
K-X-X-L-L-X-G-P-P-G-X-G-K-T-[STNQYC]-X-[GAVLIMPFW]-X-:~-[GAVLIMPFW] 41
(2) INFORMATION FOR SEQ ID NO: 43:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: BO amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: both
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: synthetic
xi) SEQUENCE DESCRIPTION: SEQ ID NO: 43:
[FL]-[GAVLIMPFW]-X-X-[GAVLIMPFW]-X-G-X(13)-[GAVLIMPFW]-X-[YR)-
[GAVLIMPFW]-X-[GAVLIMPF'4V]-A-G-(DN]-[GAVLIMPFW]-[GAVLIMPFW]-[DS] 80
(2) INFORMATION FOR SEQ ID NO: 44:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 44 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: both
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: synthetic
xi) SEQUENCE DESCRIPTION: SEQ ID NO: 44:
D-[GAVLIMPFWJ-[GAVLIMPFW)-X-X-Y-N-X-X-X-F-D-X-P-Y-[GAVLIMPFW)-X-X-R-A 44
(2) INFORMATION FOR SEQ ID N0: 95:
(i) SEQUENCE CHARACTERISTICS:
CA 02338185 2001-02-05
- - 95 -
(A) LENGTH: 78 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: both
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: synthetic
xi) SEQUENCE DESCRIPTION: SEQ ID N0: 45:
A-[GAVLIMPFW]-R-T-A-[GAVLIMPFW]-A-[GAVLIMPFW]-[GAVLIMPFW]-T-E-G-[GAVLIMPFW]-V-
X-A-
P-[GAVLIMPFW]-E-G-I-A-X-V-[KRHDE)-I 7g
(2) INFORMATION FOR SEQ ID NO: 46:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 118 amino acids
(B) TYPE: amino acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: both
(ii) TYPE OF MOLECULE: protein
(vi) INITIAL ORIGIN:
(A) ORGANISM: synthetic
xi) SEQUENCE DESCRIPTION: SEQ ID NO: 46:
[GAVLIMPFW)-P-V-G-[GAVLIMPFW)-G-R-G-S-X-[GAVLIMPFW]-G-S-[GAVLIMPFW]-V-A-X-A-
[GAVLIMPFW)-X-I-T-D-[GAVLIMPFW]-D-P-[GAVLIMPFWJ-X-X-X-[GRVLIMPFW)-L-F-E-R-F-L-
N-P-
E-R-[GAVLIMPFW)-S-M-P-D 118
(2) INFORMATION FOR SEQ ID NO: 47:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 base pairs
(B) TYPE: nucleic acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: DNA
(vi) INITIAL ORIGIN:
(A) ORGANISM: M13 MP18 ss DNA (phage)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 47:
GGATTGACCG TAATGGGATA GGTTACGTT 29
CA 02338185 2001-02-05
- 96 -
(2) INFORMATION FOR SEQ ID NO: 48:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 base pairs
(B) TYPE: nucleic acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: DNA
(vi) INITIAL ORIGIN:
(A) ORGANISM: M13 MP18 ss DNA (phage)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 48:
AGCGGATAAC AATTTCACAC AGGAAACAG 29
(2) INFORMATION FOR SEQ ID NO: 49:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: DNA
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeglobus fulgidus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 49:
ACGCGCGGAT CCATAGACGT CATAATGACC GG 32
(2) INFORMATION FOR SEQ ID NO: 50:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 31 base pairs
(B) TYPE: nucleic acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: DNA
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeglobus fulgidus
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 50:
CA 02338185 2001-02-05
97
TACGGGGTAC CCGAGCCAAA ATTGGGTAAA G 31
(2) INFORMATION FOR SEQ ID NO: 51:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: DNA
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeglobus fulgidus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 51:
ACGCGCGGAT CCATAGACGT CATAATGACC GG 32
(2) INFORMATION FOR SEQ ID NO: 52:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 31 base pairs
(B) TYPE: nucleic acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: DNA
(vi) INITIAL ORIGIN:
(A) ORGANISM: Archaeglobus fulgidus
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 52:
TACGGGGTAC CCGAGCCAAA ATTGGGTAAA G 31
(2) INFORMATION FOR SEQ ID NO: 53:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 base pairs
(B) TYPE: nucleic acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: DNA
(vi) INITIAL ORIGIN:
CA 02338185 2001-02-05
- 98 -
(A) ORGANISM: Human Collagen Forward
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 53:
TAAAGGGTCA CCGTGGTTC 19
(2) INFORMATION FOR SEQ ID NO: 54:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20 base pairs
(B) TYPE: nucleic acid
(C) STRAND FORM: single strand
(D) TOPOLOGY: linear
(ii) TYPE OF MOLECULE: DNA
(vi) INITIAL ORIGIN:
(A) ORGANISM: Human Collagen Reverse
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 54:
CGAACCACAT TGGCATCATC 20