Note: Descriptions are shown in the official language in which they were submitted.
CA 02342970 2001-04-12
TITLE: FUSION PROTEINS
Field of the Invention
The present invention relates to conjugate or fusion type proteins comprising,
for example,
C3 (see below). Although, in the following, fusion-type proteins of the
present invention,
will be particularly discussed in relation to the use to facilitate generation
of axons, it is to be
understood that the fusion proteins may be exploited in other contexts.
The present invention in partcular pertains to the field of mammalian nervous
system repair
(e.g. repair of a central nervous system (CNS) lesion site or a peripheral
nervous system
(PNS) lesion site), axon regeneration and axon sprouting.
The Rho family GTPases regulate axon growth and regeneration. Inactivation of
Rho with
Clostridium botulinum C3 exotransferase (hereinafter simply referred to as C3)
can stimulate
regeneration and sprouting of injured axons; C3 is a toxin obtainable from
Closteridium
botulinum (see Tigyi, et al. (1996)Journal ofNeurochemistry.66:537-548, Jin
and Strittmatter
(1997) J. Neurosci.17:6256-6263). Compounds of the C3 family from Closteridium
botulinum inactive Rho by ADP-ribosylation.
The present invention in particular relates to a means of delivery of C3
protein (e.g. C3 itself
or other active analogs such as C3-like transferases - see below) or other Rho
antagonists to
repair damage in the nervous system. The means of delivery may take the form
of chimeric
(i.e. conjugate) C3-like Rho antagonists. These conjugate antagonists provide
a significant
improvement over C3 compounds (alone) because they are 3 to 4 orders of
magnitude more
potent with respect to the stimulation of axon growth on inhibitory substrates
than
recombinant C3 alone. Examples of these Rho antagonists have been made as
recombinant
proteins created to facilitate penetration of the cell membrane (i.e. to
enhance cell uptake of
the antagonists), improve dose-response when applied to neurons to stimulate
growth, on
growth inhibitory substrates, and to inactivate Rho. Examples of these
conjugate Rho
antagonists are described below in relation to the designations C3APL, C3APS,
C3-TL, C3-
2
CA 02342970 2001-04-12
TS, C3-RTS, C3BASIC1 (a random, basic charge sequence added to the C-terminal
of C3),
C3BASIC2 (a random, basic charge sequence added to th C-terminal of C3) and
C3BASIC3
(the reverse TAT sequence added to the C-terminal of C3).
Background
Traumatic injury of the spinal cord results in permanent functional
impairment. Most of the
deficits associated with spinal cord injury result from the loss of axons that
are damaged in the
central nervous system (CNS). Similarly, other diseases of the CNS are
associated with axonal
loss and retraction, such as stroke, HIV dementia, prion diseases, Parkinson's
disease,
Alzheimer's disease, multiple sclerosis and glaucoma. Common to all of these
diseases is the loss
of axonal connections with their targets, and the ability to stimulate growth
of axons from the
affected or diseased neuronal population would improve recovery of lost
neurological functions.
For example, following a white matter stroke, axons are damaged and lost, even
though the
neuronal cell bodies are alive. Treatments that are effective in eliciting
sprouting from injured
axons are equally effective in treating some types of stroke (Boston life
sciences, Sept. 6, 2000
Press release ). Similarly, although the the following discussion will
generally relate to delivery
of Rho antagonists, etc. to a traumatically damaged nervous system, this
invention may also be
applied to damage from unknown causes, such as during multiple sclerosis, HIV
dementia,
Parkinson's disease, Alzheimer's disease, prion diseases or other diseases of
the CNS were axons
are damaged in the CNS environment.
It has been proposed to use various Rho antagonists as agents to stimulate
regeneration of (cut)
axons, i.e. nerve lesions; please see, for example, Canadian Patent
application nos. 2,304,981
(McKerracher et al) and 2,300,878 (Strittmatter). These patent application
documents propose
the use of known Rho antagonists such as for example C3, chimeric C3 proteins,
etc. (see blow)
as well as substances selected from among known
trans-4-amino(alkyl)-1-pyridylcarbamoylcyclohexane compounds (also see below)
or Rho kinase
inhibitors for use in the regeneration of axons. C3 inactivates Rho by ADP-
ribosylation and is
fairly non-toxic to cells (Dillon and Feig (1995)Methods in Enzymology: Small
GTPases and
their regulators Part. B.256:174-184).
3
CA 02342970 2001-04-12
While the following discussion will generally relate or be directed at repair
in the CNS, the
techniques described herein may be extented to use in PNS repair. Treatment
with Rho
antagonists could be used to enhance the rate of axon growth in the PNS.
As mentioned above, traumatic injury of the spinal cord results in permanent
functional
impairment. Axon regeneration does not occur in the adult mammalian CNS
because substrate-
bound growth inhibitory proteins block axon growth. Many compounds, such as
trophic factors,
enhance neuronal differentiation and stimulate axon growth in tissue culture.
However, most
factors that enhance growth and differentiation are not able to promote axon
regenerative growth
on inhibitory substrates. To demonstrate that a compound known to stimulate
axon growth in
tissue culture most accurately reflects the potential for therapeutic use in
axon regeneration in
the CNS, it is important for the cell culture studies to include the
demonstration that a compound
can permit axon growth on growth inhibitory substrates. An example of trophic
and
differentiation factors that stimulate growth on permissive substrates in
tissue culture, are
neurotrophins such as nerve growth factor (NGF) and brain-derived growth
factor. NGF,
however, does not promote growth on inhibitory substrates (Lehmann, et al.
(1999) 19: 7537-
7547) and it has not been effective in promoting axon regeneration in vivo.
BDNF is not effective
to promote regeneration in vivo either (Mansour-Robaey, et al. (1994) 91: 1632-
1636)
Targeting intracellular signalling mechanisms involving Rho and the Rho kinase
promotes axon
regeneration has been proposed (see, for example, the above mentioned Canadian
Patent
application nos. 2,304,981 (McKerracher et al)). For demonstration that
inactivation of Rho
promotes axon regeneration on growth inhibitory substrates, recombinant C3, a
protein that
inactivates by ADP ribosylation of the effector domain was used. While such a
C3 protein can
effectively promote regeneration, it has been noted that such a C3 protein
does not easily
penetrate into cells, and high doses must therefore be applied for it to be
effective.
The high dose of recombinant C3 needed to promote functional recovery presents
a practical
4
CA 02342970 2001-04-12
constraint or limitation on the use of C3 in vivo to promote regeneration
(Lehmann, et al. (1999)
19: 7537-7547). In tissue culture studies, it has, for example, been
determined that the minimum
amount of C3 that can be used to induce growth on inhibitory substrates is 25
ug/ml. If the cells
area not titurated, even this dose is ineffective (Figure 1 ). In the context
of the present invention
it has been determined, for example, that at least 40 ug/20 g mouse needs to
be applied to
injured mouse spinal cord or rat optic nerve. Calculating doses that would be
required to treat an
adult human on an equivalent dose per weight scale up used for our rat and
mice experiments,
it would be necessary to apply 120 mg/kg of C3 (i.e. alone) to the injured
human spinal cord.
This large of amount of recombinant protein creates significant problems for
manufacturing, due
to the large scale protein purification and cost. It also limits the dose
ranging that can be tested
because of the large amount of protein needed for minimal effective doses.
Another related limitation with respect to the use of C3 to promote repair in
the injured CNS
is that it does not easily penetrate the plasma membrane of living cells. In
tissue culture studies
when C3 is applied to test biological effects it has been microinjected
directly into the cell
(Ridley and Hall (1992) 70: 389-399), or applied by trituration ofthe cells to
break the plasma
membrane (Lehmann, et al. (1999) 19: 7537-7547, Jin and Strittmatter (1997)
17: 6256-6263).
In the case of axon injury in vivo, the C3 protein is likely able to enter the
cell because injured
axons readily take up substances from their environment.
Summary of the invention
The term "Rho antagonists" as used herein includes, but is not restricted to,
(known ) C3,
including C3 chimeric proteins, and like Rho antogonists .
The term "nerve injury site" refers to a site of traumatic nerve injury or
nerve injury caused by
disease. The nerve injury site may be a single nerve (eg sciatic nerve) or a
nerve tract comprised
of many nerves (eg. damaged region of the spinal cord). The nerve injury site
may be in the
central nervous system of peripheral nervous system in any region needing
repair. The nerve
injury site may form as a result of damage caused by stroke. The nerve injury
site may be in the
5
CA 02342970 2001-04-12
brain as a result of surgery, brain tumour removal or therapy following a
cancerous lesion. The
nerve injury site may result from Parkinson's disease, Alzheimer's disease,
Amyotrophic lateral
sclerosis, diabetes or any other type of neurodegenerative disease.
The term "pharmaceutically acceptable carrier" or "adjuvant" and
"physiologically acceptable
vehicle" and the like are to be understood as referring to an acceptable
carrier or adjuvant that
may be administered to a patient, together with a compound of this invention,
and which does
not destroy the pharmacological activity thereof.
It is to be understood herein, that if a "range" or "group of substances" is
mentioned with respect
to a particular characteristic (e.g. amino acid groups, temperature, pressure,
time and the like) of
the present invention, the present invention relates to and explicitly
incorporates herein each and
every specific member and combination of sub-ranges or sub-groups therein
whatsoever. Thus,
any specified range or group is to be understood as a shorthand way of
referring to each and every
member of a range or group individually as well as each and every possible sub-
ranges or sub-
groups encompassed therein; and similarly with respect to ariy sub-ranges or
sub-groups therein.
Thus, for example,
- with respect to a sequence comprising up to 50 base units it is to be
understood as
specifically incorporating herein each and every individual unit, as well as
sub-range of
units;
- and similarly with respect to other parameters such as low pressures,
concentrations,
elements, etc...
It is also to be understood herein that "g" or "gm" is a reference to the gram
weight unit; that "C"
is a reference to the Celsius temperature unit; and "psig" is a reference to
"pounds per square inch
guage".
CA 02342970 2001-04-12
In accordance with the present invention a conjugate or fusion protein
comprising a
therapeutically active agent is provided whereby the active agent may be
delivered across a cell
wall membrane, the conjugate or fusion protein comprising a transport
subdomain(s) or
moiety(ies) in addition to an active agent moiety(ies). More particularly, as
discussed herein,
in acccordance with the present invention a conjugate or fusion protein is
provided wherein the
therapeutically active agent is one able to faciliate (for facilitating) axon
growth (e.g.
regeneration) i.e. a conjugate or fusion proteinin the form of a conjugate Rho
antagonist.
The present invention in acccordance with an aspect thereof provides a drug
delivery construct
or conjugate [e.g. able to (for) suppressing) the inhibition of neuronal axon
growth at a central
nervous system (CNS) lesion site or a peripheral nervous system (PNS) lesion
site] comprising
at least one transport agent region and an active agent region not naturally
associated with the
active agent region, wherein the transport agent region is able to facilitate
(i.e. facilitates) the
uptake of the active agent region into a mammalian (i.e. human or animal)
tissue or cell, and
wherein the active agent region is an active therapeutic agent region able
(i.e. has the capacity
or capability) to facilitate axon growth (e.g. regeneration) , including a
derivative or homologue
thereof (i.e. pharmaceutically acceptable chemical equivalents thereof -
pharmaceutically
acceptable derivative or homologue).
In accordance with the present invention the active agent region may be an ADP-
ribosyl
transferase C3 region. In accordance with the present invention the ADP-
ribosyl transferase C3
may be selected from the group consisting of ADP-ribosyl transferase derived
from Closteridum
botulinum and a recombinat ADP-ribosyl transferase.
In accordance with another aspect the present invention provides a drug
conjugate consisting of
a transport polypeptide moiety (e.g. rich in base pairs e.g. 50 base pairs or
more ) covalently
linked to an active cargo moiety (e.g. by a labile bond (i.e. a bond readily
cleavable or subject
to chemical change in the interior target cell environment)) wherein the
transport polypeptide
moiety is able to or has the capability to facilitates) the uptake of the
active cargo moiety into
7
CA 02342970 2001-04-12
a mammalian (e.g. human or animal) tissue or cell (for example, a transport
subdomain of HIV
Tat protein, a transport homeoprotein (e.g. the homeodomain of antennopedia),
or a variation
derivative or homolog thereof, (i.e. pharmaceutically acceptable chemical
equivalents thereof))
[by a receptor independent processses] and wherein the active cargo moiety is
an active
therapeutic moiety able (i.e. has the capacity or capability) to facilitate
(i.e. for facilitating) axon
growth (e.g. regeneration).
In acccordance with the present nvention the transport polypeptide moiety may
be selected from
the group consisting of a transport subdomain of HIV Tat protein, the
homeodomain of
antennopedia, and a functional derivative and analog thereof [i. e. by the
adddition of polyamine,
or any random sequence enriched in basic aminoacids] - [i.e. pharmaceutically
acceptable
chemical equivalents thereof] and wherein the active cargo moiety is selected
from the group
consisting of C3 protein able (i.e. has the capacity or capability) to
facilitate (i.e. for facilitating)
axon growth (e.g. regeneration).
In acccordance with the present invention the C3 protein may be ADP-ribosyl
transferase C3.
In accordance with the present invention the ADP-ribosyl transferase C3 imay
be selected from
the group consisting of ADP-ribosyl transferase derived from Closteridum
botulinum and a
recombinat ADP-ribosyl transferase. In accordance with the present invention
the transport
polypeptide moiety may include an active contiguous amine acid sequence as
described herein
In accordance with an additional aspect the present invention provides a
fusion protein [e.g. able
to (for) suppressing) the inhibition of neuronal axon growth at a central
nervous system (CNS)
lesion site or a peripheral nervous system (PNS) lesion site] consisting of a
carboxy terminal
active cargo moiety and an amino terminal transport moiety, wherein the
terminal transport
moiety is selected from the group consisting of a transport subdomain of HIV
Tat protein, a
transport homeoprotein (e.g. the homeodomain of antennopedia), and a
functional derivatives and
analogs thereof (i.e. pharmaceutically acceptable chemical equivalents
thereof) and wherein the
active cargo moiety consists of a C3 protein.
8
CA 02342970 2001-04-12
In accordance with the presetn invention the C3 protein may be ADP-ribosyl
transferase C3.
In accordance with the presetn invention the ADP-ribosyl transferase C3 is
selected from the
group consisting of ADP-ribosyl transferase derived from C.'losteridum
botulinum and a
recombinat ADP-ribosyl transferase.
The present invention in particular provides a fusion protein (e.g. able to
(for) suppressing the
inhibition of neuronal axon growth at a central nervous system (C'.NS) lesion
site or a peripheral
nervous system (PNS) lesion site) consisting of a carboxy terminal active
cargo moiety and an
amino terminal transport moiety, wherein the terminal transport moiety
consists of the
homeodomain of antennopedia and the active cargo moiety consists of a C3
protein (i.e. as
described herein).
The present invention also in particular provides a fusion protein (e.g. able
to (for) suppressing
the inhibition of neuronal axon growth at a central nervous system (CNS)
lesion site or a
peripheral nervous system (PNS) lesion site) consisting of a carboxy terminal
active cargo moiety
and an amino terminal transport moiety, wherein the terminal transport moiety
consists of a
transport subdomain of HIV Tat protein and the active cargo moiety consists of
a C3 protein (i.e.
as descried herein).
The present invention in a furthe aspect provides for the use of a member
selected from the group
consisting of a drug delivery construct as described herein, a drug conjugate
as described herein
and a fusion protein as described herein (e.g. including pharmaceutically
acceptable chemical
equivalents thereof) for suppressing the inhibition of neuronal axon growth.
The present invention a pharmaceutical composition (e.g. for suppressing the
inhibition of
neuronal axon growth), the pharmaceutical composition comprising a
pharmaceutically
acceptable diluent or carrier and an effective amount of an active member
selected from the
group consisting of a drug delivery construct as described herein, a drug
conjugate as described
9
CA 02342970 2001-04-12
herein, and a fusion protein as described herein (e.g. including
pharmaceutically acceptable
chemical equivalents thereof).
The present invention further provides for the use of a member selected from
the group
consisting of a drug delivery construct as described herein, a drug conjugate
as described herein,
and a fusion protein as described herein (e.g. including pharmaceutically
acceptable chemical
equivalents thereof) for the manufacture of a pharmaceutical composition (e.g.
for suppressing
the inhibition of neuronal axon growth).
The present invention also relates to a methd for preparing a conjugate or
fusion protein as
defined above comprising
- cultivating a host cell under conditions hich provide for the expression of
the
conjugate or fusion protein within the cell ; and
- recovering the conjugate or fusion protein by affnity purification under non-
denaturing conditions.
The present invention in particular provides a fusion protein selected from
the group consisting
of C3APL, C3APS, C3-TL, C3-TS C3-RTS, C3BASIC2 and C3BASIC3 and
pharmaceutically
acceptable chemical equivalents thereof.
The present invention in another aspect provides a method of suppressing the
inhibition of
neuronal axon growth comprising administering (e.g.delivering) a member
selected from the
group consisting of a drug delivery construct as described herein, a drug
conjugate as described
herein and a fusion protein as described herein (e.g. including
pharmaceutically acceptable
chemical equivalents thereof) (e.g. directly) to a central nervous system
(CNS) lesion site or a
peripheral nervous system (PNS) lesion site (of a patient), in an ~unount
effective to counteract
said inhibition.
CA 02342970 2001-04-12
The present invention, for example, provides recombinant Rho antagonists
comprising C3
enzymes with basic stretches of amino acids added to the C3 coding sequence to
facilitate the
uptake thereof into tissue or cells for the repair and/or promotion of repair
in the CNS, even in
the lack of traumatic axon damage.
The invention in particular provides C3 proteins, which may have additional
amino acids added
to the carboxy terminal end of the C3 proteins. Examples of such proteins
includes:
C3APL: (C3 antennapedia -long) created by annealing sequences from the
antennapedia
transcription factor to the 5' end of the sequence encoding C3 cDNA. The long
antennapedia
sequence of 60 amino acids containing the homeodomain of antennapedia, was
used;
C3APS: A short 11 amino acid sequence of antennapedia that has transmembrane
transport
properties was fused to the carboxy terminal of C3 to create C3APS;
C3-TL: C3 tat-long created by fusing amino acids 27 to 72 of the carboxy
terminal of C3 protein.
C3-TS: C3 tat-short created by fusing the amino acids YGRKRRQRRR to the C3
protein; and
C3-T RS: C3 tat-short created by fusing the amino acids RRQRRKKR to the C3
protein.
It has been found that conjugate or fusion protein antagonists of the present
invention are
effective to stimulate repair in the C'NS after spinal cord injury. It is
obvious that the cell
permeability would now allow treatment of victims of stoke and
neurodegenerative disease with
the new antagonists because Rho signalling pathway is important in repair
after stroke (Hitomi,
et al. (2000) 67: 1929-39.). Treatment with Rho antagonists in the adhesive
delivery system could
be used to enhance the rate of axon growth in the PNS. Also, evidence in the
literature now links
Rho signalling with formation of Alzheimer's disease tangles through its
ability to activate PKN
which then phosphorylates tau and neurofilaments (Morissette, et al. (2000)
278: H1769-74.,
Kawamata, et al. ( 1998) 18 : 7402-10., Amano, et al. ( 1996) 271: 648-50.,
Watanabe, et al. ( 1996)
271: 645-8.). Therefore, Rho antagonists are expected to be useful in the
treatment of
11
CA 02342970 2001-04-12
Alzheimer's disease. The new chimeric C3 drugs should are able to diffuse
readily and therefore
can promote repair for diseases that are neurodegenerative. Examples include,
but are not limited
to stroke, traumatic brain injury, Parkinson's disease, Alzheimer's disease
and ALS. Moreover,
it is now well established that Rho signalling antagonists are effective in
the treatment of other
diseases. These include, but are not limited to eye diseases such as glaucoma
(Honjo, et al. (2001)
42: 137-44., Rao, et al. (2001) 42: 1029-1037.), cancer cell migration and
metastasis (Sahai, et
al. (1999) 9: 136-45., Takamura, et al. (2001) 33: 577-81., Imamura, et al.
(2000) 91: 811-6.).
The effect of the Rho signalling pathway on smooth muscle relaxation are well
established. This
has led to the identification of Rho signalling antagonists as effective in
treatment of
hypertension (Chitaley, et al. (2001) 3: 139-144., Somlyo (1997) 389: 908-911,
Uehata, et al.
(1997) 389: 990-994) , asthma (Nakahara, et al. (2000) 389: 103-6., Ishizaki,
et al. (2000) 57:
976-83), and vascular disease (Miyata, et al. (2000) 20: 2351-8., Robertson,
et al. (2000) 131: 5-
9.)as well as penile erectile dysfunction (Chitaley, et al. (2001) 7: 119-22.)
Rho GTPases include members of the Rho, Rac and Cdc42 family of proteins. Our
invention
concerns Rho family members of the Rho class. Rho proteins consist of
different variants
encoded by different genes. For example, PC 12 cells express RhoA, RhoB and
RhoC
(Lehmann et al 1999 IBID); PC12 cells: Pheochromocytom cell ligne (Greene A
and Tischler,
A S PNAS 73 :2424 ( 1976). To inactivate Rho proteins inside cells, Rho
antagonists of the
C3 family type are effective because they inactivate all forms of Rho (eg.
RhoA, Rho B etc).
In contrast, gene therapy techniques, such as introduction of a domainant
negative RhoA
family member into a diseased cell, will only inactivate that specific RhoA
family member.
Recombinant C3 proteins, or C3 proteins that retain the ribosylation activity
are also effective
in our delivery system and are covered by this invention. In addition, Rho
kinase is a
well-known target for active Rho, and inactivating Rho kinase has the same
effect as
inactiving Rho, at least in terms of neurite or axon growth (Kimura and
Schubert
(1992)Journal of Cell Biology.116:777-783, Keino-Masu, et al.
(1996)Ce11.87:175-185,
Matsui, et al. (1996)EMBO J.15:2208-2216, Matsui, et al. (1998)J. Cell
Bio1.140:647-657,
Ishizaki (1997)FEBS Lett.404:118-124), the biological activity that concerns
this invention
12
CA 02342970 2001-04-12
The C3 polypeptides of the present invention include biologically active
fragments and
analogs of C3; fragments encompass amino acid sequences having truncations of
one or
more amino acids , wherein the truncation may originate from the amino
terminus, carboxy
terminus, or from the interior of the protein. Analogs of the invention
involve an insertion or
a substitution of one or more amino acids.. Fragments and analogs will have
the biological
property of C3 that is capable of inactivation Rho GTPases. Also encompassed
by the
invention are chimeric polypeptides comprising C3 amino acid sequences fused
to
heterologous amino acid sequences. Said heterologous sequences encompass those
which,
when formed into a chimera with C3 retain one or more biological or
immunological
properties of C3. A host cell transformed or transfected with nucleic acids
encoding C3
protein or c3 chimeric protein are also encompassed by the invention. Any host
cell which
produces a polypeptide having at least one of the biological properties of a
C3 may be used.
Specific examples include bacterial, yeast, plant, insect or mammalian cells.
In addition, C3
protein may be produced in transgenic animals. Transformed or transfected host
cells and
transgenic animals are obtained using materials and methods that are routinely
available to
one skilled in the art. Host cells may contain nucleic acid sequences having
the full-length
gene for C3 protein including a leader sequence and a C-terminal membrane
anchor sequence
(see below) or, alternatively, may contain nucleic acid sequences lacking one
or both of the
leader sequence and the C-terminal membrane anchor sequence. In addition,
nucleic acid
fragments, variants and analogs which encode a polypeptide capable of
retaining the
biological activity of C3 may also be resident in host expression systems.
The Rho antogaonist that is a recombinant proteins can be made according to
methods present
in the art. The proteins of the present invention may be prepared from
bacterial cell extracts,
or through the use of recombinant techniques. In general, C3 proteins
according to the
invention can be produced by transformation (transfection, transduction, or
infection) of a
host cell with all or part of a C3-encoding DNA fragment in a suitable
expression vehicle.
Suitable expression vehicles include: plasmids, viral particles, and phage.
For insect cells,
baculovirus expression vectors are suitable. The entire expression vehicle, or
a part thereof,
can be integrated into the host cell genome. In some circumstances, it is
desirable to employ
an inducible expression vector.
13
CA 02342970 2001-04-12
Those skilled in the field of molecular biology will understand that any of a
wide variety of
expression systems can be used to provide the recombinant protein. The precise
host cell used
is not critical to the invention. The C3 protein can be produced in a
prokaryotic host (e.g., E.
coli or B. subtilis) or in a eukaryotic host (e.g., Saccharomyces or Pichia;
mammalian cells,
e.g., COS, NIH 3T3, CHO, BHK, 293, or HeLa cells; or insect cells).
Proteins and polypeptides can also be produced by plant cells. For plant cells
viral expression
vectors (e.g., cauliflower mosaic virus and tobacco mosaic virus) and plasmid
expression
vectors (e.g., Ti plasmid) are suitable. Such cells are available from a wide
range of sources
(e.g., the American Type Culture Collection, Rockland, Md.). The methods of
transformation
or transfection and the choice of expression vehicle will depend on the host
system selected.
The host cells harbouring the expression vehicle can be cultured in
conventional nutrient
media adapted as need for activation of a chosen gene, repression of a chosen
gene, selection
of transformants, or amplification of a chosen gene. One expression system is
the mouse 3T3
fibroblast host cell transfected with a pMAMneo expression vector (Clontech,
Palo Alto,
Calif.). pMAMneo provides an RSV-LTR enhancer linked to a dexamethasone-
inducible
MMTV-LTR promotor, an S V40 origin of replication which allows replication in
mammalian
systems, a selectable neomycin gene, and SV40 splicing and polyadenylation
sites. DNA
encoding a C3 protein would be inserted into the pMAMneo vector in an
orientation
designed to allow expression. The recombinant C3 protein would be isolated as
described
below. Other preferable host cells that can be used in conjunction with the
pMAMneo
expression vehicle include COS cells and CHO cells (ATCC Accession Nos. CRL
1650 and
CCL 61, respectively).
C3 polypeptides can be produced as fusion proteins. For example, expression
vectors can be
used to create lacZ fusion proteins. The pGEX vectors can be used to express
foreign
polypeptides as fusion proteins with glutathione S-transferase (GST). In
general, such fusion
proteins are soluble and can be easily purified from lysed cells by adsorption
to
14
CA 02342970 2001-04-12
glutathione-agarose beads followed by elution in the presence of free
glutathione. The pGEX
vectors are designed to include thrombin or factor Xa protease cleavage sites
so that the
cloned target gene product can be released from the GST moiety. Another
stategy to make
fusion proteins is to use the His tag system.
In an insect cell expression system, Autographa californica nuclear
polyhedrosis virus
AcNPV), which grows in Spodoptera frugiperda cells, is used as a vector to
express foreign
genes. A C3 coding sequence can be cloned individually into non-essential
regions (for
example the polyhedrin gene) of the virus and placed under control of an AcNPV
promoter,
e.g., the polyhedrin promoter. Successful insertion of a gene encoding a C3
polypeptide or
protein will result in inactivation of the polyhedrin gene and production of
non-occluded
recombinant virus (i.e., virus lacking the proteinaceous coat encoded by the
polyhedrin gene).
These recombinant viruses are then used to infect spodoptera frugiperda cells
in which the
inserted gene is expressed (see, Lehmann et al for an example of making
recombinant MAG
protein).
In mammalian host cells, a number of viral-based expression systems can be
utilised. In cases
where an adenovirus is used as an expression vector, the C3 nucleic acid
sequence can be
ligated to an adenovirus transcription/translation control complex, e.g., the
late promoter and
tripartite leader sequence. This chimeric gene can then be inserted into the
adenovirus
genome by in vitro or in vivo recombination. Insertion into a non-essential
region of the viral
genome (e.g., region E1 or E3) will result in a recombinant virus that is
viable and capable of
expressing a C3 gene product in infected hosts.
Specific initiation signals may also be required for efficient translation of
inserted nucleic
acid sequences. These signals include the ATG initiation codon and adjacent
sequences. In
cases where an entire native C3 gene or cDNA, including its own initiation
codon and
adjacent sequences, is inserted into the appropriate expression vector, no
additional
translational control signals may be needed. In other cases, exogenous
translational control
CA 02342970 2001-04-12
signals, including, perhaps, the ATG initiation codon, must be provided.
Furthermore, the
initiation codon must be in phase with the reading frame of the desired coding
sequence to
ensure translation of the entire insert. These exogenous translational control
signals and
initiation codons can be of a variety of origins, both natural and synthetic.
The efficiency of
expression may be enhanced by the inclusion of appropriate transcription
enhancer elements,
transcription terminators.
In addition, a host cell may be chosen which modulates the expression of the
inserted
sequences, or modifies and processes the gene product in a specific, desired
fashion. Such
modifications (e.g., glycosylation) and processing (e.g., cleavage) of protein
products may be
important for the function of the protein. Different host cells have
characteristic and specific
mechanisms for the post-translational processing and modification of proteins
and gene
products. Appropriate cell lines or host systems can be chosen to ensure the
correct
modification and processing of the foreign protein expressed. To this end,
eukaryotic host
cells that possess the cellular machinery for proper processing of the primary
transcript,
glycosylation, and phosphorylation of the gene product can be used. Such
mammalian host
cells include, but are not limited to, CHO, VERO, BHK, HeLa, COS, MDCK, 293,
3T3,
WI38, and in particular, choroid plexus cell lines.
Alternatively, a C3 protein can be produced by a stably-transfected mammalian
cell line. A
number of vectors suitable for stable transfection of mammalian cells are
available to the
public; methods for constructing such cell lines are also publicly available.
In one example,
cDNA encoding the C3 protein can be cloned into an expression vector that
includes the
dihydrofolate reductase (DHFR) gene. Integration of the plasmid and,
therefore, the C3
protein-encoding gene into the host cell chromosome is selected for by
including 0.01-300
~M methotrexate in the cell culture medium (as described in Ausubel et al.,
supra). This
dominant selection can be accomplished in most cell types.
Recombinant protein expression can be increased by DHFR-mediated amplification
of the
16
CA 02342970 2001-04-12
transfected gene. Methods for selecting cell lines bearing gene amplifications
are known in
the art; such methods generally involve extended culture in medium containing
gradually
increasing levels of methotrexate. DHFR-containing expression vectors commonly
used for
this purpose include pCVSEII-DHFR and pAdD26SV(A). Any of the host cells
described
above or, preferably, a DHFR-deficient CHO cell ligne (e.g., CHO DHFR cells,
ATCC
Accession No. CRL 9096) are among the host cells preferred for DHFR selection
of a
stably-transfected cell line or DHFR-mediated gene amplification.
A number of other selection systems can be used, including but not limited to
the herpes
simplex virus thymidine kinase, hypoxanthine-guanine
phosphoribosyltransferase, and
adenine phosphoribosyltransferase genes can be employed in tk, hgprt, or aprt
cells,
respectively. In addition, gpt, which confers resistance to mycophenolic acid
; neo, which
confers resistance to the aminoglycoside G-418; and hygro, which confers
resistance to
hygromycin. can be used.
Alternatively, any fusion protein can be readily purified by utilising an
antibody specific for
the fusion protein being expressed. For example, a system described in
Janknecht et al.
(1981) Proc. Natl. Acad. Sci. USA 88, 8972, allows for the ready purification
of
non-denatured fusion proteins expressed in human cell lines. In this system,
the gene of
interest is subcloned into a vaccinia recombination plasmid such that the
gene's open reading
frame is translationally fused to an amino-terminal tag consisting of six
histidine residues.
Extracts from cells infected with recombinant vaccinia virus are loaded onto
Ni2+
nitriloacetic acid-agarose columns, and histidine-tagged proteins are
selectively eluted with
imidazole-containing buffers.
Alternatively, C3 or a portion thereof, can be fizsed to an immunoglobulin Fc
domain. Such
a fusion protein can be readily purified using a protein A column.
17
CA 02342970 2001-04-12
To test Rho antagonists for activity, a tissue culture bioassay system was
used. This bioassay
is used to define activity of Rho antagonists that will be effective in
promoting axon
regeneration in spinal cord injury, stroke or neurodegenerative disease.
Neurons do not grow neurites on inhibitory myelin substrates. When neurons are
placed on
inhibitory substrates in tissue culture, they remain rounded. When an
effective Rho antagonist
is added, the neurons are able to grow neurites on myelin substrates. The time
that it takes for
neurons to growth neurites upon the addition of a Rho antagonist is the same
as if neurons
had been plated on growth permissive substrate such as laminin or polylysine,
typically 1 to 2
days in cell culture. The results can be scored visually. If needed, a
quantitative assessment of
neurite growth can be performed. This involved measuring the neurite length in
a) control
cultures where neurons are plated on myelin substrates and left untreated b)
in positive
control cultures, such as neurons plated on polylysine c) or treating cultures
with different
concentrations of the test antagonist.
To test C3 in tissue culture, it has been found that the best concentration is
25-50 ug/ml.
Thus, high concentrations of this Rho antagonist are needed as compared to the
growth
factors used to stimulate neurite outgrowth. Growth factors, such as nerve
growth factor
(NGF) are used at concentrations of 1- 100 ng/ml in tissue culture. However,
growth factors
are not able to overcome growth inhibition by myelin. Our tissue culture
experiments are all
performed in the presence of the growth factor BDNF for retinal ganglion
cells, or NGF for
PC12 cells. When growth factors have been tested in vivo, typically the
highest
concentrations possible are used, in the ug/ml range. Also they are often
added to the CNS
with the use of pumps for prolonged delivery (eg. Ramer et al, IBID). For in
vivo experiments
the highest concentrations possible was used when working with C3 stored as a
frozen 1
mg/ml solution.
The Rho antagonist C3 is stable at 37 C for at least 24 hours. The stability
of C3 was tested
in tissue culture with the following experiment. The C3 was diluted in tissue
culture medium,
18
CA 02342970 2001-04-12
left in the incubator at 37C for 24 hours, then added to the bioassay system
described above,
using retinal ganglion cells as the test cell type. These cells were able to
extend neurites on
inhibitory substrates when treated with C3 stored for 24 hours at 37C.
Therefore , the
minimun stability is 24 hours. This is in keeping with the stability
projection based on amino
acid composition (see sequence data, below).
A compound can be confirmed as a Rho antagonist in one of the following ways:
a) Cells are cultured on a growth inhibitory substrate as above, and exposed
to the candidate
Rho antagonist;
b) Cells of step a) are homogenized and a pull-down assay is perlbrmed. This
assay is based
on the capability of GST-Rhotektin to bind to GTP-bound Rho. Recombinant GST-
Rhotektin
or GST rhotektin binding domain (GST-RBD) is added to the cell homogenate made
from
cells cultured as ina). It has been found that inhibitory substrates activate
Rho, and that this
activated Rho is pulled down by(GST-RBD). Rho antagonists will block
activation of Rho,
and therefore, an effective Rho antagonist will block the detection of Rho
when cell are
cultured as described by a) above;
c) An alternate method for this pull-down assay would be to use the GTPase
activating
protein, Rho-GAP as bait in the assay to pull down activated Rho, as described
(Diekmann
and Hall, 1995. In Methods in Enzymology Vol. 256 part B 207-215).
Another method to confirm that a compound is a Rho antagonist is as follows:
When added to living cells antagonists that inactivate Rho by ADP-ribosylation
of the
effector domain can be identified by detecting a molecular weight shift in Rho
(Lehmann et
al, 1999 Ibid). The molecular weight shift can be detected after treatment of
cells with Rho
antagonist by homogenizing the cells, separating the proteins in the cellular
homogenate by
SDS polyacrylamide gel electrophoresis. The proteins are transferred to
nitrocellulose paper,
then Rho is detected with Rho-specific antibodies by a Western blotting
technique.
19
CA 02342970 2001-04-12
Another method to confirm that compound is a Rho-kinase antagonist is as
follows:
a) Recombinant Rho kinase tagged with myc epitope tag, or a GST tag is
expressed in Hela
cells or another suitable cell type by transfection.
b) The kinase is purified from cell homogenates by immunoprecipation using
antibodies
directed against the myc tag or the GST tag.
c) The recovered immunoprecipitates from b) are incubated with [32P] ATP and
histone type
2 as a substrate in the presence or absence of the Rho kinase. In the absence
of Rho kinase
activity the Rho kinase antigens is able to block the phosphorylation activity
of Rho kinase
(i.e. phosphorylation of hislore), and as such identified the compound as a
Rho kinase
antagonist.
Turning now to the trasport side of the conjugates of the present invention ,
known methods
are available to add transport sequences that allow proteins to penetrate into
the cell;
examples include membrane translocating sequence (Rojas (1998) 16: 370-375),
Tat-
mediated protein delivery (Vives (1997) 272: 16010-16017), polyargine
sequences (blender
et al. 2000, PNAS 24: 13003-13008) and antennapedia (Derossi (1996) 271: 18188-
18193).
Examples of known tranport agents, moities, subdomains and the like are also
shown for
example in Canadian patent document no. 2,301,157 (conjugates containing
homeodomain of
antennopedia) as well as in U.S. patent 5,652,122, 5,670,617, 5,674,980,
5,747,641, and
5,804604 (conjugates containing amino acids of Tat HIV protein (hereinafter
Tat HIV protein
is sometimes simply referred to as Tat); the entire contents of each of these
patent documents
is incorporated herein by reference.
Several receptor -mediated transport strategies have been used to try and
improve function of
ADP ribosylases: these methods include fusing C2 and C3 sequences ENRfu(Wilde,
et al.
(2001) 276: 9537-9542.) and use of receptor-mediated transport with the
diptheria toxin
receptorENRfu(Aullo, et al. (1993) 12: 921-31.). These methods have not been
demonstrated
to dramatically increase the potency of C3. Moreover, these proteins require
receptor-
CA 02342970 2001-04-12
mediated transport. This means that the cells must express the receptor, and
must express
sufficient quantities of the receptor to significantly improve transport. In
the case of dipthera
toxin, not all cells express the appropriate receptor, limiting its potential
use. The clinical
importance for any of these has not been tested or shown.
One strategy which may be used in accordance with the present invention is to
exploit the
antennapedia homeodomain that is able to transport proteins across the plasma
membrane by
a receptor-independent mechanism (Derossi (1996) 271: 18188-18193); an
alternate strategy
is to exploit tat-mediated delivery (Vives (1997) 272: 16010-16017, Fawell
(1994) 91: 664-
668, Frankel (1988) SS: 1189-1193).
The Anntenapedia strategy has been used for protein translocation into neurons
(Derossi
(1996) 271: 18188-18193). Anntenapedia has, for examle, been used to transport
biotin-
labelled peptides in order to demonstrate the efficacy of the technique; see
U. S. Patent no.
6,080,724 (the entire contents of this patent re incorporated herein by
reference).
Antennapedia enhances growth and branching of neurons in vitro (Block-Gallego
(1993) 120:
485-492). Homeoproteins are transcription factors that regulate development of
body
organization, and antennapedia is a Drosophila homeoprotein. Tat on the other
hand is a
regulatory protein from human immunodeficiency virus (HIV) that is expressed
in the long
terminal repeat. It is a highly basic protein that is found in the nucleus and
can transport
reported genes into cell. Moreover, tat-linked proteins can penetrate cells
after intraperotoneal
injection, and it can even cross the blood brain barrier to enter cells within
the brain
(Schwarze, et al. (1999) 285: 1569-72).
In the context of axon growth on inhibitory substrates, axon regeneration
after injury, or axon
regeneration in the brain or spinal cord, no method using these transport
sequences has been
devised. In particular, it should be noted that the ability of antennapedia to
enhance growth
was tested with neurons placed on laminin-coated coverslips. Laminin supports
axon growth
and overrides growth inhibition (David, et al. (1995) 42: 594-602) thus, it is
not a suitable
21
CA 02342970 2001-04-12
substrate to test the potential for regeneration. There is an enormous wealth
of literature over
the last 20 years on substances that promote axon growth under such favourable
tissue culture
conditions, but none of these has lead to clinical advances in the treatment
of spinal cord
injury. The effect of antennapedia was shown to act as similar to a growth
factors. Growth
factors do not over come growth inhibition by CNS growth inhibitory substrates
(Lehmann, et
al. (1999) 19: 7537-7547, Cai, et al. (1999) 22: 89-101). Growth factors
applied in vivo do
not support regeneration, only sprouting (Schnell, et al. (1994) 367: 170-
173).
The transport sequence may be added to the n-terminal sequence of the C3
protein.
Aternatively, the transport sequence may be added on the C-terminal end of the
C3 protein;
because the C-terminal is already quite basic, this should enhance further the
transport
properties
The new chimeric C3 may be used to treat spinal cord injury to promote
functional repair. We
have demonstrated that both C3APL and C3APS can overcome growth inhibition on
complex
inhibitory substrates that include myelin and mixed condroitin sulfate
proteoglycans. Further,
we demonstrate that C3APL can promote functional recovery after application to
injured
spinal cord in adult mice. The new chimeric protein may be used to promote
axon
regeneration and reduce scarring after CNS injury. Scarring is a barrier to
nerve regeneration.
The advantage of the new chimeric C3 is that ability to treat the injured
axons after a
significant delay between the injury and the treatment. Also, the new
recombinant protein
may be useful in the treatment of chronic injury. The chimeric C:3 can also be
used to treat
neurodegenerative diseases such as Alzheimer's disease and Parkinson's disease
where
penetration of the Rho antagonist to the affected neuronal population is
required for effective
treatment. The chimeric C3 will also be of benefit for the treatment of stroke
and traumatic
brain injury. Moreover, much evidence suggests efficacy in the treatment of
cancer cell
migration. Rho antagonists are also useful in the treatment of disease
involving smooth
muscle, such as vascular disease, hypertention, asthma, and penile
dysfunction.
22
CA 02342970 2001-04-12
For treatment of spinal cord injury, the conjugate Rho antagonists of the
present invention
may be used in conjunction cell transplantation. Many different cell
transplants have been
extensively tested for their potential to promote regeneration and repair,
including , but a not
S restricted to, Schwann cells, fibroblasts modified to express growth
factors, fetal spinal cord
transplants, macrophages, embryonic or adult stem cells, and olfactory
ensheathing glia.
C3APL and C3APshort may be used in conjunction with neurotrophins, apoptosis
inhibitors,
or other agents that prevent cell death. They may be used in conjunction with
cell adhesion
molecules such as L1, laminin, and artifical growth matrices that promote axon
growth. The
chimeric C3 constructs of the present invention may also be used in
conjunction with the use
of antibodies that block growth inhibitory protein substrates to promote axon
growth.
Examples of such antibody methods are the use of IN-1 or related antibodies
(Schnell and
Schwab (1990) 343: 269-272)or through the use of therapeutic vaccine
approaches (Huang
( 1999) 24: 63 9-647).
BRIEF DESCRIPTION OF THE FIGURES
In drawings which llustrate example embodiments of the present invention:
Figure 1 illustrates the Dose response of normal C3 with and without
trituration;
Figure 2 illustrates ADP ribosyation by C3AL and C3APS, but not C3 after
passively adding
the comppounds to PC12 cells;
Figure 3A illustrates that C3APL penetrates cells;
Figure 3B illustrates a lower level of cell peneration by C3 as compared to
figure 3A;
Figure 4 illustrates the effectiveness of C3APL and C3APS at low doses;
Figure 5 illustrates the effectiveness of C3APL and C3APS at low doses;
23
CA 02342970 2001-04-12
Figure 6 illustrates the effeciveness of CAPL to stimulate axon regeneration
of primary
neurons;
Figure 7 illustrates the effectiveness of C3APL to promore functional recovery
ater spinal
cors injury;
Figure 8 illustrates effectiveness of TAT transport sequences to enhance
growth as C3-TAT
chimeras; and
Figures 9A and 9B illustrate axon regeneration after spinal cord injury and
treatment with
C3APL.
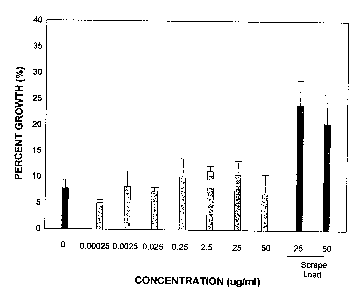
Referring to figure 1, PC12 cells were plated on inhibitory myelin substrates.
Unmodified C3
added to the tissue culture medium at concentration from 0.00025 - 50 ug/ml
did not
significantly improve neurite outgrowth over the untreated control (0). C3 was
only effective
n stimulating neurite outgrowth for cells plated on myelin substrates after
scrape -loading.
This figure demonstrates the limited or no penetration in cells when passively
added to the
tissue culture medium. Please see example 4 below for techniques.
Referring to figure 2 this figure provides a demonstration of the new C3
ribosylated Rho
C3APL and C3APS ADP ribosylate Rho. Western blot of showing RhoA in untreated
cells
(lane 1), and cells treated with C3 APL (lane 2) or C3APS (lane 3). When Rho
is ADP
ribosylated by C3 it undergoes a molecular weight shift, as observed for lanes
2 and 3. Please
see example 4 below for techniques.
Referring to figure 3 this figure shows intracellular activty after treatment
with C3APL.
Detection that the new fusion C3 penetrates into the cell wash.
Immunocytochemistry
with anti-C3 antibody of PC12 cells plated on myelin and treated with C3 (A)
or C3APL (B).
Cells in A (figure 3A) are not immuoreactive because C3 has not penetrated
into the cells.
Cells in B (figure B) are immunoreactive and they are able to extend neurites
on myelin
substrates. Please see example 4 below for techniques.
24
CA 02342970 2001-04-12
Turnng to figure 4 it shows C3-antennopedia proteins promote growth on
ihibitory
substrates. The dose response experiment shows that C3APL and C3APS promote
more
neurite growth per cell than control PC 12 cells plated on myelin. PC 12 cells
were plated on
myelin and either scrape loaded with unmodified C3 (C3 50) left untreated (0)
or treated with
various concentrations of C3APL. Compared to C3 used at 25 ug/ml, C3APS is
effective at
stimulating more cells to grow neurites at 0.0025 ug/ml, a dose 10,000 X less.
Please see
example 4 below for techniques.
Figure 5 shows a dose response experiment showing that C3APL and C3APS elicit
long
neurites to grow when cells are plated on inhibitory substrates. PC 12 cells
were plated on
myelin and either scrape loaded with unmodified C3 (C3 50) left untreated (0)
or treated with
various concentrations of C3APL. Compared to C3 used at 25 ug/ml, C3APS is
effective at
stimulating more cells to longer neurite growth at 0.0025 ug/ml, a dose 10,000
X less.
Please see example 4 below for techiniques.
As may be seen in figure 6 shows primary neurons growing on inhibitory
substrates after
treatment with C3APL. Rat retinal ganglion cells were plated on myelin
substrates and treated
with different concentrations of C3APL. Concentrations of 0.025 and above
promoted
significantly longer neurites. This dose is 1000X lower than that of C3 need
to promote
growth on myelin.
Referring to figure 7 this figure shows behavioral recovery after treatment of
adult mice with
C3APL. Dose-response experiment. Mice received a dorsal hemisection of the
spinal cord
and were left untreated (transection), were treated with fibrin alone (fibrin)
or were treated
with fibrin plus C3 at the indicated concentrations given in ug/mouse. Each
point represents
one animal. The BBB score was assessed 24 hours after treatment. Animals
treated with C3
exhibed a significant improvement in behavioural recovery than untreated
animals. The
effective dose of 0.5 ug is 100X less than unmodified C3 used. Please see
example 6.
CA 02342970 2001-04-12
Referrring to figure 8 tis figure shows promotion of axon growth by TAT-C3
chimeric
proteins. The dose response experiment shows that C3-TS and C3-TL promote more
neurite
growth per cell than control PC12 cells plated on myelin. PC12 cells were
plated on myelin
and either scrape loaded with unmodified C3 (scrape load) left untreated
(myelin) or treated
S with various concentrations of C3-TS (grey bars) or C3-TL (black bars).
Compared to C3
used at 25 ug/ml, C3-TL is effective at stimulating more cells to grow
neurites at 0.0025
ug/ml, a dose 10,000 X less than C3.
Refering to figure 9A and 9B these figures show axon regeneration in injured
spinal cord, i.e.
anatomical regeneration after treatment with C3APL. Section of the spinal cord
after
anterograde labeling with WGA HRP. A) Sprouting of cut axons in to the dorsal
white
matter. Arrows show regenerating axons distal to the lesion. B) Same section 3
mm from the
lesion site. Arrows show regenerating axons
DETAILED DESCRIPTION
Method for making the C'3APL and C'3AP-short proteins
C3APL is the name given to the protein made by ligating a cDNA encoding C3
(Dillon and Feig (1995) 256: 174-184) with cDNA encoding the antennapedia
homeodomain
(Bloch-Gallego (1993) 120: 485-492). The stop codon at the 3' end of the DNA
was replaced
with an EcoRl site by polymerase chain reaction using the primers 5'GAA TTC
TTT AGG
ATT GAT AGC TGT GCC 3' (SEQ ID NO: 1) and 5'GGT GGC GAC CAT CCT CCA
AAA 3' (SEQ ID NO: 2). The PCR product was sub-cloned into a pSTBlue-1 vector
(Novagen, city), then cloned into a pGEX-4T vector using Baml-~ I and Not I
restriction site.
This vector was called pGEX-4T/C3. The antennapedia sequence used to add to
the 3' end of
C3 in pGEX-4T/C3 was created by .PCR from the pET-3a vector (Bloch-Gallego
(1993)
120: 485-492, Derossi (1994) 269: 10444-10450), subcloned into a pSTBlue-1
blunt vector,
26
CA 02342970 2001-04-12
then cloned into the pGEX-4T/C3, using the restriction sites EcoR I and Sal I,
creating
pGEX-4T/C3 APL
A shorter version of the Antennapedia (pGEX-4T/C3APS) was also made. This
chimeric
sequence was made by ligating oligonucleotides encoding the short antennapedia
peptide
(Maiael (1999) 126: 3183-3190) into the pGEX-4T/C3 vector cut with EcoR I and
Sal I. To
confirm the sequence of C3APL, the coding sequence from both strands was
sequenced. The
recombinant C3APL and C3APS cDNAs were separately transformed into bacteria ,
and after
the recombinant proteins were produced, a bacterial homogenate obtained by
sonication, and
the homogenate cleared by centrifugation. Glutathione-agrose beads (Sigma)
were added to
the cleared lysate and placed on a rotating plate for 2-3 hours, then washed
extensively. To
remove the glutathione S transferase sequence from the recombinant protein,
20U of
Thrombin was added, the beads were left on a rotator overnight at 4°C.
After cleavage with
thrombin, the beads were loaded into an empty 20m1 column, and the proteins
eluted with
PBS. Aliquots containing recombinant protein were pooled and 100~1s p-
aminobenzamidine
agrose beads (Sigma) were added and left mixing for 45 minutes at 4°C
to remove thrombin,
then recombinant protein was isolated from the beads by centrifugation. Purity
of the sample
was determined by SDS-PAGE, and bioactivity bioassay with PC 12 cells. (See
Lehmann et al
IBID)
Other possible methods for making bioactive chimeric proteins
The Rho antagonist is a recombinant proteins can be made according to methods
present in
the art. The proteins of the present invention may be prepared from bacterial
cell extracts, or
through the use of recombinant techniques by transformation, transfection, or
infection of a
host cell with all or part of a C3-encoding DNA fragment with an antennapedia-
derived
transport sequence in a suitable expression vehicle. Those skilled in the
field of molecular
biology will understand that any of a wide variety of expression systems can
be used to
provide the recombinant protein. The precise host cell used is not critical to
the invention.
27
CA 02342970 2001-04-12
Any fusion protein can be readily purified by utilising either affinity
purification techniques
or more traditional column chromatography. Aff'mity techniques include GST, an
antibody
specific for the fusion protein being expressed, and histidine-tagged proteins
are selectively
eluted with imidazole-containing buffers. Alternatively, recombinant protein
can be fused to
an immunoglobulin Fc domain. Such a fusion protein can be readily purified
using a protein
A column. It is envisioned that small molecule mimetics of the above described
antagonists
are also encompassed by the invention.
Testing the bioactivity of C3APL, C3APS, C3TL and C3-TS
To test the eflricacy of C3APL , C3APS, C3-TL and C3-TS a number of
experiments were
performed with PC 12 cells, a neural cell line, grown on growth inhibitory
substrates. PC 12
cells were plated on myelin substrates as described (Lehmann et al , IBID).
C3, C3APL ,
C3APS, C3-TL or C3-TS were added at different concentrations without
trituration (please
refere to figures 4, 5 and 8for concentrations used). C3 added passively to
the culture
medium in this way was not able to promote neurite growth in the growth
inhibitory
substrates because cells must be triturated for C3 to enter the cells and be
active (Fig. 1).
Both C3APL and C3APLS were able to ADP ribosylates Rho to cause a shift in the
molecular weight of RhoA (Figure 2). Both C3APL and C3APLS were able to
promote
neurite growth and enter neurons after being added passively to the culture
medium (Fig. 3,
Figure 4 and 5). Dose response experiment where concentrations of 0.25ng/ml,
2.5 ng/ml, 25
ng/ml, 250 ng/ml and 2.5 p.g/ml and 25 pg/ml were tested and showed that C3APL
and
C3APLS helped more neurons differentiate neurites at does 10,000 fold less
than C3 (Fig. 4)
Dose response experiments where concentrations of 0.25ng/ml, 2.5 ng/ml, 25
ng/ml, 250
ng/ml and 2.5 ~g/ml and 25 ~g/ml were tested and showed that (:3APL was able
to promote
long neurite growth when added at a minimum concentration of 0.0025 ug/ml
(Fig. 5). These
concentrations of 2.5 ng/ml and 25 ng/ml for C3APL and C3APLS, represent
10,000 and
1,000 times less than the dose needed with C3, respectively. Moreover, at the
highest
concentration tested, 50 ug/ml, these two new Rho antagonists did not exhibit
toxic effect on
PC12 cells, and were able to stimulate neurite outgrowth on growth inhibitory
substrates.
28
CA 02342970 2001-04-12
C3-TL and C3-TS also were tested at concentrations of 0.25ng/ml, 2.5 ng/ml, 25
ng/ml, 250
ng/ml and 2.5 p.g/ml and 25 ~,g/ml and were found to be able to promote
neurite growth on
myelin substrates at doses significantly less than C3 (Figure 8).
To verify the ability of C3APL and C3APS to promote growth from primary
neurons, primary
retinal cultures were prepared, and the neurons were plated on myelin
substrates as described
with respect to example 5. In the absence of treatment with C3APL orC3APS, the
cells
remained round and are not able to grow neurites. When treated with C3APL or
C3APS,
retinal neurons were able to extend long neurites on inhibitory myelin
substrates (Figure 6).
Next, was tested the ability of C3APL and C3APS to promote growth on a
different type of
growth inhibitory substrate relevant to the type of growth inhibitory proteins
found at glial
scars. Chamber slides were coated with a mixture of chondroitin sulfate
proteoglycans
(Chemicon), and then plated retinal neurons. The neurons were not able to
extend neuritis on
the proteoglycan substrates, but when treated with C3APL or C3APS, they
extended long
neurites (not shown). These studies demonstrate that C3APL and C3APS can be
used to
promote neurite growth on myelin and on proteoglycans, the major classes of
inhibitory
substrates that prevent repair after injury in the CNS.
Testing ability of C3APL to promote regeneration and functional recovery after
spinal cord
injury
To test if C3APL could promote repair after spinal cord injury, fully adult
mice were
used.(as described with resepcet to example 6). A dorsal hemisection was made
at T8, and
mice were treated with different amounts (figure 7) of C3APL in a fibrin glue
as described
(McKerracher, US patent pending (delivery patent)). In previous known
experiments with
C3, it was found that 40-50 ug was needed to promote anatomical regeneration
in optic nerve
(Lehmann et all IBID). We tested different doses (see figure 7) of C3APL
ranging form 1 ug
to 50 ug and assessed animals for behavioural recovery according the BBB scale
(Basso
(1995) 12: 1-21)
The day following surgery and application of C3APL behavioural testing began
The animals
were placed in an open field environment that consisted of a rubber mat
approximately 4' X
29
CA 02342970 2001-04-12
3' in size. The animals were left to move randomly, the movement of the
animals were
videotaped. For each test two observers scored the animals for ability to move
ankle, knee
and hip joints in the early phase of recovery. Previously C3 treatment of mice
was seen to
lead to functional recovery observable 24 hours after treatment. In mice
treated with C3APL,
functional recovery could be observed as early as 24 hours after spinal cord
injury (Fig. 7).
Untreated mice exhibit a function recovery score according to the BBB scale
averaging 0,
whereas mice treated with C3 are able to walk and have a BBB score averaging 8
(Fig.7). At
higher concentrations of 50 ug the mice died within 24 hours. However, of the
mice that
survive, they exhibited good long-term functional recovery. These results
demonstrate that
C3APL effectively promotes functional recovery early after spinal cord injury,
and that it is
ei~ective at much lower doses than C3. However, at high concentrations, C3APL
appears to
exhibit toxicity, and therefore careful doing will be required for clinical
use.
Qualitative observations of the video tapes showed thatoOnly animals that
received C3APL
reached the late phase of recovery after 30 days of treatment. Untreated
control animals did
not typically pass beyond the early phase of recovery. These results indicate
that the
application of C3APL improved long-term functional recovery after spinal cord
injury
compared to no treatement, injury alone, or fibrin adhesive alone.
EXAMPLE 1. DNA and protein sequence details of C3APL
Nucleotide sequence of C3APL. The start site, is in the GST sequence of the
plasmid (not
shown). The vector with the GST sequence is commercially available and thus
the entire
GST sequence including the start was not sequenced. It was desired to
determine only the
sequence 3 ' to the thrombin cleavage site which releases C3 conjugate from
the GST
sequence. The GST sequence is cleaved with thrombin.
Both strands were sequenced to verify that there were no errors in the
sequence.
Bold is the stop codon.
CA 02342970 2001-04-12
Nucleotide sequence of coding sequence for the protein C3APL (SEQ ID NO: 3)
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC ,AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA A'TG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC C(:T ATT AGT GCT TTT C.AG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC GTG ATG GAA TCC CGC AAA CGC GCA AGG
CAG ACA TAC ACC CGG TAC CAG ACT CTA GAG CTA CTAG AAG GAG TTT CAC
TTC AAT CGC TAC TTG ACC CGT CGG CGA AGG ATC GAG ATC GCC CAC GCC
CTG TGC CTC ACG GAG CGC CAG ATA AAG ATT TGG TTC CAG AAT CGG CGC
ATG AAG TGG AAG AAG GAG AAC TGA 3'
Amino acid sequence of C3APL (SEQ ID NO: 4)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGV1NGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFKVA
31
CA 02342970 2001-04-12
KGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLS SDGKQIIITATMMGTAINPKEFV
MESRKRARQTYTRYQTLELEKEFHFNRYLTRRRRIEIAHALCLTERQIKIWFQNRRMK
WKKEN
Physical characteristics of C3APL
Molecular Weight 34098.03 Daltons
295 Amino Acids
48 Strongly Basic(+) Amino Acids (K,R)
28 Strongly Acidic(-) Amino Acids (D,E)
89 Hydrophobic Amino Acids (A,I,L,F,W,V)
94 Polar Amino Acids (N,C,Q,S,T,Y)
9.847 Isolectric Point
20.524 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 79.48
EXAMPLE 2. DNA and protein sequence details of C3APSt
Nucleotide sequence of C3APS (SEQ ID NO: 5). The start site, is in the GST
sequence of the
plasmid, not shown here.
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
32
CA 02342970 2001-04-12
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT C.AG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC CGC CAG ATC AAG ATT TGG TTC CAG AAT
CGT CGC ATG AAG TGG AAG AAG GTC GAC TCG AGC GGC CGC ATC GTG ACT
GAC TGA 3'
Amino acid sequence for C3APS (SEQ ID NO: 6)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFKVA
KGSKAGYIDPI SAFQGQLEMLLPKHSTYHIDDMRLS SDGKQIIITATMMGTAINPKEFR
QIKIWFQNRRMKWKKVD S S GRIVTD
Physical charteristics of C3APS
Molecular Weight 29088.22 Daltons
257 Amino Acids
38 Strongly Basic(+) Amino Acids (K,R)
23 Strongly Acidic(-) Amino Acids (D,E)
79 Hydrophobic Amino Acids (A,I,L,F,W,V)
83 Polar Amino Acids (N,C,Q,S,T,Y)
33
CA 02342970 2001-04-12
9.745 Isolectric Point
15.211 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 78.34
EXAMPLE 3. METHOD FOR M,4KING THE C3APL AND C3APS PRO:T'EINS
C3APL is the name given to the protein encoded by cDNA made by ligating the
functional domain of C3 transferase and the homeobox region of the
transcription factor
called anntenopedia (Block-Gallego (1993) 120: 485-492) in the following way.
A cDNA
encoding C3 (Dillon and Feig (1995) 256: 174-184) the plasmid vector pGEX-2T
was used
for the C3 portion of the chimeric protein. The stop codon at the 3' end of
the DNA was
replaced with an EcoRl site by polymerase chain reaction using the primers
5'GAA TTC
TTT AGG ATT GAT AGC TGT GCC 3' (SEQ ID NO: 1) and S'GGT GGC GAC CAT CCT
CCA AAA 3' (SEQ ID NO: 2). The PCR product was sub-cloned into a pSTBlue-1
vector
(Novagen, city), then cloned into a pGEX-4T vector using BamH I and Not I
restriction site.
This vector was called pGEX-4T/C3. The pGEX-4T vector has a 5' glutathione S
transferase
(GST), sequence for use in aWnity purification. The anntenopedia sequence used
to add to
the 3' end of C3 in pGEX-4T/C3 was created by PCR from the pET-3 a vector
(Bloch-
Gallego (1993) 120: 485-492, Derossi (1994) 269: 10444-10450). The primers
used were
5'GAA TCC CGC AAA CGC GCA AGG CAG 3' (SEQ ID NO: 7) and 5'TCA GTT CTC
CTT CTT CCA CTT CAT GCG 3' (SEQ ID NO: 8). The PCR product obtained from the
reaction was subcloned into a pSTBlue-1 blunt vector, then cloned into the
pGEX-4T/C3,
using the restriction sites EcoR I and Sal I, creating pGEX-4T/C3APL.
A shorter version of the Antennapedia (pGEX-4T/C3AP-short) was also made. This
chimeric sequence was made by ligating oligonucleotides encoding the short
anntenapedia
pepide (Maizel (1999) 126: 3183-3190) into the pGEX-4T/C3 vector cut with EcoR
I and Sal
I. For pGEX-4T/C3AP-short the sequences of the oligos made were 5'AAT TCC GCC
AGA
TCA AGA TTT GGT TCC AGA ATC GTC GCA TGA AGT GGA AGA AGG 3' (SEQ ID
NO: 9)
34
CA 02342970 2001-04-12
and 5'GGC GGT CTA GTT CTA AAC CAA GCT CTT AGC AGC GTA GTT CAC CTT
CTT CCA GCT 3' (SEQ ID NO: 10). The two strands were annealed together by
mixing
equal amounts of the oligonucleotides, heating at 72 °C for 5 minutes
and then leaving them
at room temperature for 15 minutes. 'The oligonucleotides were ligated into
the pGEX4T/C3
vector. To confirm the sequences of C3APL, the coding sequence from both
strands of
pGEX-4T/C3APL were sequenced.
To prepare recombinant C3APL and C3APS proteins, the plasmids containing the
corresponding cDNAs (pGEX-4T/C3APL and pGEX-4T/C3AP-short) were transformed
into
bacteria , strain XL-1 blue competent E. coli. The bacteria were grow were
grown in L-broth
(lOglL Bacto-Tryptone, 5glL Yeast Extract, lOglL NaCI) with ampicillin at 50
ug/ml(BMC-
Roche), in a shaking incubator for 1 hr at 37 °C and 300 rpm.
Isopropyl ~3-
thiogalactopyranoside (IPTG), (Gibco) was added to a final concentration of
0.5 mM to
induce the production of recombinant protein and the culture was grown for a
further 6 hours
at 37° C and 250 rpm. Bacteria pellets were obtained by centrifugation
in 250 ml centrifuge
bottles at 7000rpm for 6 minutes at 4 °C. Each pellet was re-suspended
in 10 ml of Buffer A
(50mM Tris, pH 7.5, 50mM NaCI, 5mM MgClz, 1mM DTT) plus 1mM PMSF. All re-
suspended pellets were pooled and transferred to a 100 ml plastic beaker on
ice. The
remaining buffer A with PMSF was added to the pooled sample. The bacteria
sample was
sonicated 6 x 20 seconds using a Branson Sonifier 450 probe sonicator. Both
the bacteria and
probe were cooled on ice 1 minute between sonications. The sonicate was
centrifuged in a
Sorvall SS-34 rotor at 16,000 rpm for 12 minutes at 4°C to clarify the
supernatant. The
supernatant was transferred into fresh SS-34 tubes and re-spun at 12,000 rpm
for 12 minutes
at 4°C. Up to 20 ml of Glutathione-agrose beads (Sigma) were added to
the cleared lysate
and placed on a rotating plate for 2-3 hours. The beads were washed 4 times
with buffer B,
(buffer A, NaCI is 150mM, no PSMF) then 2 times with buffer C (buffer B +
2.5mM CaClz).
The final wash was poured out till the beads created a thick slurry. To remove
the glutathione
S transferase sequence from the recombinant protein, 20U of Thrombin (Bovine,
Plasminogen-free, Calbiochem) was added, the beads were left on a rotator
overnight at 4°C.
After cleavage with thrombin the beads were loaded into an empty 20m1 column.
Approximately 20 aliquots of 1 ml were collected by elution with PBS. Samples
of each
aliquote of 0.5u1 were spotted on nitrocellulose and stained with Amido Black
to determine
CA 02342970 2001-04-12
the protein peak. Aliquots containing C3 were pooled and 100p.1s p-
aminobenzamidine agrose
beads (Sigma) were added and left mixing for 45 minutes at 4°C. This
last step removed the
thrombin from the recombinant protein sample. The recombinant protein was
centrifuged to
remove the beads and then concentrated using a centriprep-10 concentrator
(Amicon). The
concentrated recombinant protein was desalted with a PD-10 column (Pharmacia,
containing
Sephadex G-25M) and 10 O.SmI aliquots were collected. A dot-blot was done on
these
samples to determine the protein peak, and the appropriate aliquots pooled,
filter-sterilized,
and stored at - 80°C. A protein assay (Biorad) was used to determine
the concentration of
recombinant protein. Purity of the sample was determined by SDS-PAGE, and
bioactivity
bioassay with PC12 cells.
EXAMPLE ~: TESTING OF EFFEC.4CY OF C3APL AND C3APS IN TISSUE CULTURE
To test the ability of C3APL and C:3APS to overcome growth inhibition, PC12
cells were
plated on myelin, a growth inhibitory substrate. The myelin was purified from
bovine brain
(Norton and Poduslo (1973) 21: 749-757). In some other experiments chondroitin
sulfate
proteoglycan (CSPG) substrates were made from a purchased protein composition
(Chemicon). Before coating coverslips or wells of a 96 well plate, they were
coated with
poly-L-lysine (0.025pg/ml) (Sigma, St. Louis, MO, washed with water and
allowed to dry.
Myelin stored as a lmg/ml solution at: -80 C was thawed at 37C, and vortexed.
The myelin
was plated at 8 ug/well of a 8 well chamber Lab-Tek slides (Nuc, Naperville,
IL). The
myelin solution was left to dry overnight in a sterile tissue culture hood.
The next morning
the substrate was washed gently with phosphate buffered saline, and then cells
in media were
added to the substrate. PC-12 cells (L,ehmann et al., 1999) were grown in DMEM
with 10%
horse serum (HS) and 5% fetal bovine serum (FBS). Two days prior to use the PC-
12 cells
were differentiated by 50 ng/ml of nerve growth factor (NGF). After the cells
were primed,
Sml of trypsin was added to the culture dish detach the cells, the cells were
pelleted and re-
suspended in 2m1 of DMEM with 1% HS and 50 ng/ml of nerve growth factor.
Approximately, 5000 to 7000 cells were then plated on 8 well chamber Lab-Tek
slides (Nuc,
Naperville, IL) coated myelin. The cells were placed on the test substrates at
37°C for 3-4
hours to allow the cells to settle. The original media was carefully removed
by aspiration,
36
CA 02342970 2001-04-12
taking care not to disrupt the cells and replaced with DMEM with 1% HS,
50ng/ml ofNGF
and varying amounts of the C3, C3AfL, or C3APS, depending on the dose desired.
After two
days, the cells were fixed (4% paraformaldehyde and 0.5% glutaraldehyde). For
control
experiments with unmodified C3, NGF primed PC 12 cells were trypsinized to
detach them
form the culture dish, the cells were washed once with scrape loading buffer
(in mM: 114
KCL, 15 NaCI, 5.5 MgClz, and 10 Tris-HCL) and then the cells were scraped with
a rubber
policeman into 0.5 ml of scraping buffer in the presence of 25 or 50 pg/ml C3
transferase.
The cells were pelleted and resuspended in 2m1 of DMEM, 1% HS and 50 ng/ml
nerve
growth factor before plating. At least four experiments were analyzed for each
treatment.
For each well, twelve images were collected with a 20X objective using a Zeiss
Axiovert
microscope. For each image, the numbers of cells with and without neurites
were counted
and the lengths of the neurites were determined. Since myelin is phase dense,
cells plated on
myelin substrates were immuno-stained with anti-~3III tubulin antibody before
analysis.
Quantitative analysis of neurite outgrowth was with the aid of Northern
Eclipse software
(Empix Imaging, Mississauga, Ontario, Canada). Data analysis and statistics
were with
Microsoft Excel.
To examine ADP ribosylation by C3, C3APL, and C3APS, the compounds were added
to
PC 12 cell cultures, as described above. The cells were harvested by
centrifugation, cell
homogenates prepared and the proteins separated by SDS polyacrylamidegel
electrophoresis.
The Proteins were then transfered to nitrocellulose and the Western blots
probed with anti-
Rho Antibody (LJ~I).
EXAMPLE 5: TESTING ABILITY OF C3APL AND C3APS TO O vERRIDE INHIBITION
OFMULTIPLE GROWTH INHIBITORY PROTEINS
Myelin substrates were made as described in example 4 and plated on tissue
culture chamber
slides. P 1 to P3 rat pups were decapitated, the heads washed in ethanol and
the eye removed
and placed in a petri dish with Hanks buffered saline solution (HBSS, from
Gibco). A Noel
was cut in the cornea, the lens removed, and the retina squeezed out.
Typically, four retinas
37
CA 02342970 2001-04-12
per preparation were used. The retinas were removed to a 15 ml tube and the
volume brought
to 7 ml. A further 7 mls of dissociation enzymes and papain were added. The
dissociation
enzyme solution was made as follows: 30 mg DL cystein was added to a 15 ml
tube (Sigma
DK cyctein hydrochloride), and 70 ml HBSS, 280 ul of lOmg.ml bovine serum
albumin were
added and the solution mixed and pH adjusted to 7 with 0.3 N NaOH. The
dissociate solution
was filter-sterilized and kept frozen in 7 ml aliquots, and before use 12.5
units papain per ml
(Worthington) was added. After adding the dissociation solution to the retina,
the tube was
incubated for 30 min. on a rocking tray at 37C. The retinas were then gently
triturated,
centrifuged and washed with HB S S . 'Che HB S S was replaced with growth
medium (DMEM
(Gibco), 10 % fetal bovine serum, and 50 ng/ml brain derived neurotrophic
factor (BDNF)
vitamins, penicillin-streptomycin, in the presence or absence of C3APL or
C3APS. Cells
were plated on test substrates of myelin or CSPG in chamber slides prepared as
described in
example 4 , above. A quantitative analysis was completed as described for
example 4 above.
Neurons were visualized by fluorescent microscopy with anti-(3III tublin
antibody, which
detects growing retinal ganglion cells (RGCs).
EXAMPLE 6. TREATMENT OFIN,IURED MOUSE SPINAL CORD WITH C3APL AND
MEASUREMENT OF RECOVERY OF MOTOR FUNCTION IN TREATED MICE.
Adult Balb-c mice were anaesthetized with 0.6 ml/kg hypnorm, 2.5 mg/kg
diazepam and 35
mg/kg ketamine. This does gives about 30 minutes of anesthetic, which is
sufficient for the entire
operation. A segment of the thorac spinal column was exposed by removing the
vertebrae and
spinus processus with microrongeurs (Fine Science Tools). A spinal cord lesion
was then made
dorsally, extending past the central canal with fine scissors, and the lesion
was recut with a fine
knife. This lesion renders all of the control animals parapelegic. The
paravertebral muscle were
closed with reabsorbable sutures, and the skin was closed with 2.0 silk
sutures. After surgery, the
38
CA 02342970 2001-04-12
bladder was manually voided every 8-10 hours until the animals regained
control, typically 2-3
days. Food was placed in the cage for easy access, and sponge-water used for
easy accessibility
of water after surgery. Also, animals received subcutaneous injection
Buprenorphine (0.05 a 0.1
mg/kg) every 8-12 hours for the first 3 days. Any animals that lost 15-20% of
body weight were
killed.
Rho antagonists were delivered locally to the site of the lesion by a fibrin-
based tissue
adhesive delivery system (McKerracher, US patent submitted). Recombinant C3APL
was
mixed with fibrinogen and thrombin in the presence of CaCl2, Fibrinogen is
cleaved by
thrombin, and the resulting fibrin monomers polymerize into a three-
dimensional matrix. We
added C3APL as part of a fibrin adhesive, which polymerized within about 10
seconds after
being placed in the injured spinal cord. We tested C3APL applied to the spinal
cord lesion
site after the lesion was made. For control we injected fibrin adhesive alone,
or transected the
cord without further treatment. For behavioural testing, the BB.B scoring
method was used to
examine locomotion in an opern field environment (Basso (1995) 12: 1-21). The
environment was a rubber mat mat approximately 4' X 3' in size, and animals
were placed on
the mat and videotaped for about 4 minutes. Care was taken not to stimulate
the peroneal
region or touch the animals excessively during the taping session. The video
tapes were
digitized and observed by two observers to assign BBB scores. The BBB score,
modified for
mice, was as follows:
Score Description
1 No observable hindlimb (HL) movement.
2 Slight movement of one or two joints.
3 Extensive movement of one joint and /or slight movement of one other joint..
4 Extensive movement of two joints.
5 Slight movement of all three joints of the HL,.
6 Slight movement of two joints and extensive movement of the third.
7 Extensive movement of two joints and slight movement of the third.
8 Extensive movement of all three joints of the HL walking with no weight
support.
9 Extensive movement of all three joints, walking with weight support.
10 Frequent to consistent dorsal stepping with weight support.
39
CA 02342970 2001-04-12
11 Frequent plantar stepping with weight support.
12 Consistent plantar stepping with weight support, no coordination.
13 Consistent plantar stepping with consistent weight support, occasional FL-
HL
coordination.
14 Consistent plantar stepping with consistent weight support, frequent FL-HL,
coordination.
Consistent plantar stepping with consistent weight support, consistent FL-HL
coordination; Predominant paw position during locomotion is rotated internally
or externally, or
consistent FL-HL, coordination with occasional dorsal stepping.
10 16 Consistent plantar stepping with consistent weight support, consistent
FL-HL
coordination; predominant paw position is parallel to the body; frequent to
consistent toe drag,
or curled toes, trunk instability.
17 Consistent plantar stepping with consistent weight support, consistent FL-
HI,
coordination; predominant paw position is parallel to the body, no toe drag,
some trunk
15 instability.
18 Consistent plantar stepping with consistent weight support, consistent FL-
HL
coordination; predominant paw position is parallel to the body, no toe drag
and consistent
stability in the locomotion.
EXAMPLE . 7 TREATMENT OF IN.IURED MOUSE SPINAL (,ORD WITH C3APL AND
ASSESSMENT OFANATOMICAL RECOVERY.
Mice that received a spinal cord injury and treated as controls or with C3APL,
as described
for example 6 were assessed for morphological changes to the scar and for axon
regeneration.
To study axon regeneration, the corticospinal axons were identified by
anterograde labeling.
For anterograde labeling studies, the animals were anaesthetized as above, and
the cranium
over the motor cortex was removed. With the fine glass micropippetter (about
100 um in
diameter) 1e cerbral cortex was injected with 2-4 u1 of horse radishe
peroxidase conjugated to
wheat germ agglutinin (2%), a marquer that is taken up by nerve cells and
transported
antergradely into the axon that extends into the spinal cord. After injection
of the anterograde
tracer, the cranium was replaced, and the skin closed with 5-0 silk sutures.
The animals were
CA 02342970 2001-04-12
sacrificed with chloral hydrate (4.9 mg/10 g) after 48 hours, and perfused
with 4%
paraformaldehyde in phosphate bufTer as a fixative. The spinal cord was
removed,
cryoprotected with sucrose and cryostat sections placed on slides for
histological
examination.
EXAMPLE 8. DNA and protein sequence details of C3-TL
The TAT coding sequence was obtained by polymerase chain reaction of the
plasmid
SVCMV-TAT(obtained form Dr. Eric Cohen, Universite de Montreal) that contains
the entire
Tat coding sequence. To isolate the transport sequence of the Tat protein PCR
was used. The
first primer (5'GAATCCAAGCACCAGGAAGTCAGCC 3'- (SEQ ID NO: 11)) and the
second primer (5' ACC AGCCACCACCTTCTGATA 3' - (seq id no: 12)) used
corresponded
to amino acids 27 to 72 of the HIV TAT protein. Upon verification and
purification, the PCR
product was sub cloned into a pSTBlue-1 blunt vector. This transport segment
of the TAT
protein was then cloned into pGEX-4T/C3, using the restriction sites EcoR I
and Sac I. The
new C3-tat fusion protein was called C3-TL. Recombinant protein was made as
described in
Example 3.
DNA sequence of C3-TL (seq id no: 13)
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA A.~1T AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT AT'G TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT AT'T AGT ACT TCA TTA A'TG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AAG CAT CCA GGA AGT CAG CCT AAA ACT
41
CA 02342970 2001-04-12
GCT TGT ACC AAT TGC TAT TGT AAA AAG TGT TGC T'TT CAT TGC CAA GTT
TGT TTC ATA ACA AAA GCC TTA GGC ATC TCC TAT GGC AGG AAG CGG AGA
CAG CGA CGA AGA GCT CAT CAG AAC AGT CAG ACT CAT CAA GCT TCT CTA
TCA AAG CAG TAA 3'
The protein sequence of C3-TL (seq id no: 14)
GS SRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIV S
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAK.AKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFKVA
KGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLS SDGKQIIITATMMGTAINPKEF~~.
HPGSQPI~TACThICYCKKCCFHCQVCFITI~AL(BISYGRKR1~Q QNSQTHQASLS
KQ.
Molecular Weight 32721.40 Daltons
291 Amino Acids
43 Strongly Basic(+) Amino Acids (K,R)
21 Strongly Acidic(-) Amino Acids (D,E)
82 Hydrophobic Amino Acids (A,I,L,F,W,V)
104 Polar Amino Acids (N,C,Q,S,T,Y)
9.688 Isolectric Point
22.655 Charge at PH 7.0
Total number of bases translated is 876
% A = 37.44 [328]
G = 17.58 [154]
T = 28.31 [248]
C = 16.67 [ 146]
42
CA 02342970 2001-04-12
EXAMPLE 9. DNA AND PROTEIN SEQ UENCE DETAILS OF C3-TS
A shorter tat construct was also made called C3-TS. To make the shorter C3 tat
fusinon
protein the following oligonucleotrides were 5'AAT TCT ATG GTC LTA AAA AAC GTC
GTC AAC GTC GTC GTG 3' (SEQ ID NO: 15)
and 5' GAT ACC AGC ATT TTT TLC AGC ALT TLC AGC AGC ACA GCT 3' (SEQ ID
NO: 16. The two oligonucleotide strands were annealed together by combining
equal
amounts of the oligonucleotides, heating at 72 °C for S minutes and
then letting the
oligonucleotide solution cool at room temperature for 15 minutes. . The
oligonucleotides
were ligated into the pGEX4T/C3 vector. The constructs was sequenced. All
plasmids were
transformed into XL-1 blue competent cells. Recombinant protein was made as
described in
Example 3.
Nucleotide sequence of C3-TS (SEQ ID NO: 17)
5' GGA TCC TCT ALA GTC GAC CTG CAL GCA TLC AAT' GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAL GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAL TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA LTA TCA TAT ACT AAA AGC GCT ALT GAA ATA
AAT GGA AAG CTA ALA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT ALA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
43
CA 02342970 2001-04-12
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT C AG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC TAT GGT GCT AAA AAA CGT CGT CAA CGT
CGT CGT GTC GAC TCG AGC GGC CCG CAT CGT GAC TGA 3'
The protein sequence of C3-TS (SEQ ID NO: 18)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGV1NGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAK,AKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFKVA
KGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPKEFY
GAKKRRQRRRVDSSGPHRD
Molecular Weight 26866.62 Daltons
23 8 Amino Acids
36 Strongly Basic(+) Amino Acids (K,R)
21 Strongly Acidic(-) Amino Acids (D,E)
71 Hydrophobic Amino Acids (A,I,L,F,W,V)
78 Polar Amino Acids (N,C,Q,S,T,Y)
9.802 Isolectric Point
15.212 Charge at PH 7.0
Total number of bases translated is 717
A = 38.91 [279]
% G = 17.43 [125]
44
CA 02342970 2001-04-12
T = 28.45 [204]
C = 15.20 [ 109]
EXAMPLE 10.
The following example illustrates how a coding sequence can be modified
without affecting
the efficacy of the transplated protein. The example shows modifications to
C3basic3 that
would not affect the activity. Sequences may include the entire GST sequence,
as shown here
that includes the start site, which would not be removed enzymatically. Also,
the transport
sequence shown in this example has changes in amino acid composition
surrounding the
active sequence dues to a difference in the cloning strategy, and the his tag
has been omitted.
However, the active region is : R R K Q R R K R R . This sequence is contained
in the
C3BASIC3, and is the active transport sequence in the sequence below. Also
note that the C-
terminal region of the protein after this active region differs from C3BASIC1.
That is because
the cloning strategy was changed, the restriction sites differ, and therefore
non-essential
amino acids 3 terminal to the transport sequence are transplated and included
in the protein.
Nucleic acid sequence: (SEQ ID NO: 19)
1413 base pairs
single strand
liniear sequence
5' ATG TCC CCT ATA CTA GGT TAT TGG AAA ATT AAG GGC CTT GTG CAA CCC
ACT CGA CTT CTT TTG GAA TAT CTT GAA GAA AAA T.AT GAA GAG CAT TTG
TAT GAG CGC GAT GAA GGT GAT AAA TGG CGA AAC ,AAA AAG TTT GAA TTG
GGT TTG GAG TTT CCC AAT CTT CCT TAT TAT ATT GAT GGT GAT GTT AAA
TTA ACA CAG TCT ATG GCC ATC ATA CGT TAT ATA GCT GAC AAG CAC AAC
ATG TTG GGT GGT TGT CCA AAA GAG CGT GCA GAG ATT TCA ATG CTT GAA
GGA GCG GTT '-I"TG GAT ATT ACiA TAC GGT GTT TCG AGA ATT GCA TAT AGT
AAA GAC TTT GAA ACT CTC AAA GTT GAT TTT CTT AGC AAG CTA CCT GAA
CA 02342970 2001-04-12
ATG CTG AAA ATG TTC GAA GAT CGT TTA TGT CAT AAA ACA TAT TTA AAT
GGT GAT CAT GTA ACC CAT CCT GAC TTC ATG TTG TAT GAC GCT CTT GAT
GTT GTT TTA TAC ATG GAC CCA ATG TGC CTG GAT GCG TTC CCA AAA TTA
GTT TGT TTT AAA AAA CGT ATT GAA GCT ATC CCA C.AA ATT GAT AAG TAC
TTG AAA TCC AGC AAG TAT ATA GCA TGG CCT TTG CAG GGC TGG CAA GCC
ACG TTT GGT GGT GGC GAC CAT CCT CCA AAA TCG GAT CTG GTT CCG CGT
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT AC'.T TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC ,AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT AT'G TTA TTT ANA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT ANA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT C.AG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AGA AGG AAA CAA AGA AGA A AA AGA
AGA CTG CAG GCG GCC GCA TCG TGA 3'
Amino acid sequence (SEQ ID NO: 20)
479 amino acids
linear, single strand
MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRNKKF'ELGLEFPNLPY
YIDGDVKLTQSMAIIRYIADKHNMLGGCPKERAEISMLEGAVLDIRYGVSRIAYSKDF
ETLKVDFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALDVVLYMDPMCL
DAFPKLVCFKKRIEAIPQIDKYLKSSKYIAWPLQGWQATFGGGDHPPKSDLVPRGSSR
46
CA 02342970 2001-04-12
VDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVSYTKS
ASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFXGDDPAYLGTEFQ
NTLLNSNGTINKTAFEKAKAKFLNXDRLEYGYISTSLMNVSQFAGRPIITKFKVAKGS
KAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLS SDGKQIIITATMMGTAINPKEFRRKQ
RRKRRLQAAAS.
Molecular Weight 53813.02 Daltons
470 Amino Acids
68 Strongly Basic(+) Amino Acids (K,R)
55 Strongly Acidic(-) Amino Acids (D,E)
149 Hydrophobic Amino Acids (A,I,L,F,W,V)
121 Polar Amino Acids (N,C,Q,S,T,Y)
9.137 Isolectric Point
14.106 Charge at PH 7.0
Total number of bases translated is 1413
A = 34.61 [489]
G = 19.75 [279]
% T = 29.51 [417]
C = 15.99 [226]
Ambiguous = 0.14 [2]
A+T = 64.12 [906]
% C+G = 35.74 [505]
Davis,Botstein,Roth Melting Temp C. 79.20
EXAMPLE 1 1. ADDITIONAL CHIMERIC C3 PROTEINS THAT WOULD BE EFFECTIVE TO
STIMULATE REPAIR IN THE CNS.
47
CA 02342970 2001-04-12
The following sequences could be added to the amino terminal or carboxy
terminal of C3 or a
truncated C3 that retains its enzymatic activity.
1) Sequences of polyarginine as decribed (blender, et al. (2000) 97: 13003-
8.). These
could be from 6 to 9 or more arginines.
2) Sequences of poly-lSyine
3) Sequences of polyhistidine
4) Sequences of arginine and lysine mixed.
5) Basic stretchs of amino acids containing non-basic amino acids stretch
where the
sequence added retains transport characteristics.
6) Sequences of 5- 15 amino acids containing at least 50 % basic amino acids
7) Sequences longer than 15 -30 amino acids containing at least 30 % basic
amino
acids.
8) Sequences longer than 50 amino acids containing at least 18 % basic amino
acids.
9) Any of the above where the amino acids are chemically modified, such as by
addition
of cyclohexl side chains, other side chains, different alkyl spacers.
EXAMPLE 12. ADDITIONAL C'HIMERIC' C3 PROTEIN'S THAT WOULD BE EFFECTIVE TD
STIMULATE
REPAIR IN THI? CNS.
ENBbu
C3basicl : C3 fused to a randomly designed basic tail
C3basic2: C 3 fused to a randomly designed basic tail
C3 basic3: C3 fused to the reverse Tat sequence
EXAMPLE 12. ADDITIONAL C'HIMERIC' C3 PROTEIN' THAT WOULD BE EFFECTIVE TO
STIMULATE
REP 91R IN THE CNS.
We have designed the following DNA encoding a chimeric C3 with membrane
transport
properties. The protein is designated C3BASIC1. This sequence was designed
with C3 fused
to a random basic sequence. The construct was made to encode the peptide given
below.
K R R R R R P K K R R R A K R R (SEQ ID NO: 21)
48
CA 02342970 2001-04-12
The construct was made by synthesizing the two oligonucleotides given below,
annealing
them together, and ligating them into the pGEX-4T/C3 vector with an added
histidine tag.
5' AAG AGA AGG CGA AGA AGA CCT AAG AAG AGA CGA AGG GCG AAG
AGG AGA 3' (SEQ ID NO: 22)
5' TTC TCT TCC GCT TCT TCT GGA TTC TTC TCT GCT TCC CGC TTC
TCC TCT 3' (SEQ ID NO 23)
DNA sequence C3BASIC1 (SEQ ID NO: 24)
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG 'TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT ANA GGC CiAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG 'TTT TTA AAT ANA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AAG AGA AGG CC~A AGA AGA CCT AAG
AAG AGA CGA AGG GCG AAG AGG AGA CAC CAC CAC CAC CAC CAC GTC
GAC TCG AGC GGC CGC ATC GTG ACT GAC TGA 3'
49
CA 02342970 2001-04-12
Protein sequence (SEQ ID NO: 25)
GS SRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIV S
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFXGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAKAKFLNXDRLEYGYISTSLMNVSQFAGRPIITKFKVA
KGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPKEFK
RRRRR1PKKRRRAKRRI~E IHVDSSGRIVTD.
Molecular Weight 29897.03 Daltons
263 Amino Acids
44 Strongly Basic(+) Amino Acids (K,R)
23 Strongly Acidic(-) Amino Acids (D,E)
75 Hydrophobic Amino Acids (_A,I,L,F,W,V)
79 Polar Amino Acids (N,C,Q,S,T,Y)
10.024 Isolectric Point
22.209 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 78.56
EXAMPLE 13. ADDITIONAL CHIMERI(_' C3 PROTEIN THAT WO ULD BE EFFECTIVE TO
STIMULATE
REPAIR IN THE CNS.
We have designed the following DNA encoding a chimeric C3 with membrane
transport
properties. The protein is designated C3BASIC2. This sequence was designed
with C3 fused
to a random basic sequence. The construct was made to encode the peptide given
below.
K R R R R K K R R Q R R R (SEQ ID NO: 26)
The construct was made by synthesizing the two oligonucleotides given below,
annealing
them together, and ligating them into the pGEX4T/C3 vector with an added
histidine tag.
CA 02342970 2001-04-12
5' AAG CGT CGA CGT AGA AAG AAA CGT AGA CAG CGT AGA CGT 3' (SEQ
ID NO: 27)
5' TTC GCA GCT GCA TCT TTC TTT GCA TCT GTC GCA TCT GCA 3'
(SEQ ID NO: 28)
DNA sequence C3BASIC2 (SEQ ID NO: 29)
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG 'TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA G'TA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT ANA GGC CTAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT ANA GAT
AGA CTT CJAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA A'l'A ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AAG CGT CGA CGT AGA AAG AAA CGT
AGA CAG CGT AGA CGT CAC'. CAC CAC CAC CAC CAC GTC GAC TCG AGC
GGC CGC ATC GTG ACT GAC TGA 3'
Protein sequence (SEQ ID NO: 30)
GS SRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNI~TPENIMLFXGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAKAKFLNXDRLEYGYISTSLMNVSQFAGRPIITKFKVA
KGSKAGYIDPISAFQGQLEMLLPEZHSTYHIDDMRLSSDGKQIIITATMMGTATNPKEFK
RRRRKKRRQR1E~RI-~-l(I-1HVD S SGRIVTD.
Molecular Weight 29572.61 Daltons
51
CA 02342970 2001-04-12
260 Amino Acids
42 Strongly Basic(+) Amino Acids (K,R)
23 Strongly Acidic(-) Amino Acids (D,E)
74 Hydrophobic Amino Acids (A,1,L,F,W,V)
80 Polar Amino Acids (N,C,Q,S,T,Y)
9.956 Isolectric Point
20.210 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 78.45
EXAMPLE 14. ADDITIONAL C'HIMERIC' C3 PROTEIN THAT WOULD BE EFFECTIVE TO
STIMULATE
REPAIR IN THE CNS.
We have designed the following DNA encoding a chimeric C3 with membrane
transport
properties. The protein is designated C3BASIC3. This sequence was designed
with C3 fused
to a the reverse Tat sequence. The construct was made to encode the peptide
given below
R R K Q R R K R R (SEQ ID NO: 31)
The construct was made by synthesizing the two oligonucleotides given below,
annealing
them together, and ligating them into the pGEX4T/C3 vector with an added
histidine tag, then
subcloning root pGEX-4T/C3.
5' AGA AGG AAA CAA AGA AGA AAA AGA AGA 3' (SEQ ID NO: 32)
5' TCT TCC TTT GTT TCT TCT TTT TCT TCT 3' (SEQ ID NO: 33)
DNA sequence C3BASIC3 (SEQ ID NO: 34)
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
52
CA 02342970 2001-04-12
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT AT'G TTA TTT ANA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT ANA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA (JTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA G.AC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA AT'A ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TT'C AGA AGG AAA CAA AGA AGA AAA AGA
AGA CAC CAC CAC CAC CAC CAC GTC GAC TCG AGC (1GC CGC ATC GTG ACT
GAC TGA 3'
Protein sequence (SEQ ID NO: 35)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFP SNLIKQVELLDKSFNKMKTPENIMLFXGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAKAKFLNXDRLEYGYISTSLMNVSQFAGRPIITKFKVA
KGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPKEFR
RKQRRI VD S S GRIV TD.
Molecular Weight 29441.47 Daltons
260 Amino Acids
39 Strongly Basic(+) Amino Acids (K,R)
23 Strongly Acidic(-) Amino Acids (D,E)
76 Hydrophobic Amino Acids (A,I,L,F,W,V)
80 Polar Amino Acids (N,C,Q,S,T,Y)
9.833 Isolectric Point
17.211 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 78.29
Example 15: Modifications of sequences. Any of sequences given in examples 1,
2, 8, 9, 10,
11, 12 and 13 could be modified to retain C3 enzymatic activity( and effective
transport
53
CA 02342970 2001-04-12
sequences). For example amino acids encoded from DNA at the 3' end of the
sequence that
represents the translation of the restriction sites used in cloning can be
removed without
affecting activity. Some of the amino terminal amino acids may also be removed
without
affecting activity. The C3 portion of teh protein could be truncated to
include just the amino
acids needed for activity. The transport sequences could be modified to add or
remove one
or more amino unessential acids.
54
CA 02342970 2001-04-12
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: LISA MCKERRACHER
(ii) TITLE OF INVENTION: FUSION PROTEINS
(iii) NUMBER OF SEQUENCES: 35
(iv) CORRESPONDENCE ADDRESS:
(A) ADRESSEE: BROULLETTE KOSIE
(B) STREET: 1100 RENE-LESVEQUE BLVD WEST
(C) PROV/STATE: QUEBEC
(D) COUNTRY: CANADA
(E) POSTAL/ZIP CODE: H3B SC9
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE:ASCII (TExT)
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE:
(C) CLASSIFICATION:
(vii) ATTORNEY/AGENT INFORMATION:
(A) NAME: BROULLETTE KOSIE
CA 02342970 2001-04-12
(B) REGISTRATION NO.:
(C) REFERENCE/DOCKET NO.: 06447-004-CA-O1
(D) TEL. NO.: (514) 397 8500
(E) FAX NO.: (514) 397 8515
(2) INFORMATION FOR SEQ ID NO: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE: S' cDNA primer - C3 sequence
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IlVIIVtEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
56
CA 02342970 2001-04-12
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES 1N SEQ ID NO:
(~) SEQUENCE DESCRIPTION: SEQ ID NO: l
5'GAA TTC TTT AGG ATT GAT AGC TGT GCC 3'
(2) INFORMATION FOR SEQ ID NO: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE: 3' cDNA primer - C3 sequence
57
CA 02342970 2001-04-12
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RES1DUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2:
5'GGT GGC GAC CAT CCT CCA AAA 3'
(2) INFORMATION FOR SEQ ID NO: 3:
(i) SEQUENCE CHARACTERISTICS:
58
CA 02342970 2001-04-12
(A) LENGTH: 888 BASE PAIRS
(B) TYPE: cDNA
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
IO
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
IS
(ix) FEATURE:
(A) NAME/KEY: C3APL
20 (B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
25 (B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
30 (G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
59
CA 02342970 2001-04-12
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES 1N SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 3: (Nucleotide sequence of protein
C3 APL)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT (iCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG 'TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC CiAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG 'TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC GTG ATG GAA TCC C'GC AAA CGC GCA AGG
CAG ACA TAC ACC CGG TAC CAG ACT CTA GAG CTA GAG AAG GAG TTT CAC
TTC AAT CGC TAC TTG ACC CGT CGG CGA AGG ATC CiAG ATC GCC CAC GCC
CTG TGC CTC ACG GAG CGC CAG ATA AAG ATT TGG TTC CAG AAT CGG CGC
ATG AAG TGG AAG AAG GAG AAC TGA
(2) INFORMATION FOR SEQ ID NO: 4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 295 AMINO ACIDS
(B) TYPE: PROTEIN
CA 02342970 2001-04-12
(C) STRANDEDNESS SINGLE:
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES 1N SEQ ID NO:
61
CA 02342970 2001-04-12
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 4: (amino acid sequence of C3APL)
GS SRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSE
KEAIVSYTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMI~TPE
NIMLFRGDDPAYLGTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGY
ISTSLMNV SQFAGRPIITQFKVAKGSKAGYIDPISAFQGQLEMLLPRHST
YHIDDMRLS SDGKQIIITATMMGTAINPKEF~' '~I~:~ ~;P~; P.;~'~. ~
~~1:"~"i'P'~~'Q'i'1.:1:~.
LEKE YL IAHALCLTERQ Q ~ N
(2) INFORMATION FOR SEQ ID NO: 5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 774 BASE PAIRS
(B) TYPE: cDNA
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) Il~~IMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
62
CA 02342970 2001-04-12
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RES1DLTES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 5: (Nucleotide sequence of C3APS)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG 'TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC CiAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG 'TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA (rTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT C'.AG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
63
CA 02342970 2001-04-12
GCT ATC AAT CCT AAA GAA TTC CGC CAG ATC AAG ATT TGG TTC CAG AAT
CGT CGC ATG AAG TGG AAG A.AG GTC GAC TCG AGC GGC CGC ATC GTG ACT
GAC TGA
(2) INFORMATION FOR SEQ ID NO: 6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 257 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
64
CA 02342970 2001-04-12
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 6:(amino acid sequence of C3APS)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSE
KEAIVSYTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNI~TPE
NIMLFRGDDPAYLGTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGY
ISTSLMNVSQFAGRPIITQFKVAKGSKAGYIDPISAFQGQLEMLLPRHST
YHIDDMRLS SDGKQIIITATMMGTAINPKEFRQIKIWFQNWRMKWKKVDS
SGRIVTD
(2) INFORMATION FOR SEQ ID NO: 7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 BASE PAIRS
(B) TYPE:
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: 5' cDNA primer - antennapedia sequence
(v) FRAGMENT TYPE:
CA 02342970 2001-04-12
(V1) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 7: (EX 3)
5'GAA TCC CGC AAA CGC GCA AGG CAG 3'
66
CA 02342970 2001-04-12
(2) INFORMATION FOR SEQ ID NO: 8:
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 27 BASE PAIRS
(B) TYPE:NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: 3' cDNA primer
(v) FRAGMENT TYPE: antennapedia sequence
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
67
CA 02342970 2001-04-12
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 8: (EX 3)
5'TCA GTT CTC CTT CTT CCA CTT CAT GCG 3'
(2) INFORMATION FOR SEQ ID NO: 9:
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 54 BASR PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY:LINEAR
(ii) MOLECULE TYPE: OLIGONEUCLEOTIDE STRAND 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
68
CA 02342970 2001-04-12
(B) LOCATION:
(D) OTHER INFORMATION
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 9: (EX 3)
5'AAT TCC GCC AGA TCA AGA TTT GGT TCC AGA ATC GTC GCA TGA AGT GGA
AGA AGG 3'
(2) INFORMATION FOR SEQ ID NO: 10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54 BSE PAIRS
(B) TYPE:NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: OLIGONUCLEOTIDE STRAND 2
69
CA 02342970 2001-04-12
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES 1N SEQ ID NO:
(xi) SEQUENCE DESCRIPTION' SEQ ID NO: 10: (EX 3)
5'GGC GGT CTA GTT CTA AAC CAA GCT CTT AGC AGC: GTA GTT CAC CTT CTT
CCA GCT 3'
CA 02342970 2001-04-12
(2) INFORMATION FOR SEQ ID NO: 11:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 26 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: TAT 5' PRIMER
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
71
CA 02342970 2001-04-12
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 11:
5'GAATCCAAGCATCCAGGAAGTCAGCC3'
(2) INFORMATION FOR SEQ ID NO: 12:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:TAT 3' PRIMER
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
72
CA 02342970 2001-04-12
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 12: (ex 8)
5' ACC AGC CAC CAC CTT CTG ATA 3'
(2) INFORMATION FOR SEQ ID NO: 13:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 876 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3-TL SEQUENCE
73
CA 02342970 2001-04-12
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 13: (DNA sequence of C3-TL)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
74
CA 02342970 2001-04-12
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG 'CAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
S AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG 'CTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA G.AC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AAG CAT CCA GGA AGT CAG CCT AAA ACT
GCT TGT ACC AAT TGC TAT TGT AAA AAG TGT TGC TTT CAT TGC CAA GTT
1 S TGT TTC ATA ACA AAA GCC TTA GGC ATC TCC TAT GGC AGG AAG CGG AGA
CAG CGA CGA AGA GCT CAT C.AG AAC AGT CAG ACT CAT CAA GCT TCT CTA
TCA AAG CAG TAA
(2) INFORMATION FOR SEQ ID NO: 14
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 291 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3-TL PROTEIN SEQUENCE
(v) FRAGMENT TYPE:
CA 02342970 2001-04-12
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
1 S (B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: I4: (The protein sequence of C3-TL)
GS SRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFP SNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFKVA
KGSKAGYIDPI SAFQGQLEMLLPRHSTYHIDDMRLS SDGKQIIITATMMGTAINPKEF~.
IMP(~SQPKTACTI~TCYCKKCCF~C~Q'7CFITKAi,CBISYG Q QI~SQTHQASLS
76
CA 02342970 2001-04-12
(2) INFORMATION FOR SEQ ID NO: 15:
S (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: TS OL1GONUCLEOTIDE STRAND1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
77
CA 02342970 2001-04-12
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(~) SEQUENCE DESCRIPTION: SEQ ID NO: 15: (EX 9)
5'AAT TCT ATG GTC GTA AAA AAC GTC GTC AAC GTC GTC GTG 3'
(2) INFORMATION FOR SEQ ID NO: 16:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: TS OLIGONUCLEOTIDE STRAfVD 2
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
78
CA 02342970 2001-04-12
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 16: (EX 9)
5' GAT ACC AGC ATT TTT TGC AGC AGT TGC AGC AGC ACA GCT 3'
(2) INFORMATION FOR SEQ ID NO: 17:
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 756 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
79
CA 02342970 2001-04-12
(ii) MOLECULE TYPE: C3-TS cDNA
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 17: (Nucleotide sequence of C3-TS)
CA 02342970 2001-04-12
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT CJCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT AC'T TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC .AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC TAT GGT GCT AAA AAA CGT CGT CAA CGT
CGT CGT GTC GAC TCG AGC GGC CCG CAT CGT GAC TGA
(2) INFORMATION FOR SEQ ID NO: 18
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 251 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRAIVDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3-TS PROTEIN
(v) FRAGMENT TYPE:
81
CA 02342970 2001-04-12
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 18: (The protein sequence of C3-TS)
GS SRVDLQACNAYS1NQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFKVA
KGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPKEFY
GAKKRRQRRRVDS SGPHRD
82
CA 02342970 2001-04-12
(2) INFORMATION FOR SEQ ID NO: 19:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:
(B) TYPE:
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
83
CA 02342970 2001-04-12
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 19: (EX 10)
ATGTCCCCTATACTAGGTTATTGGAAAATTAAGGGCCTTGTGCAACCCACTCGAC
TTCTTTTGGAATATCTTGAAGAAAAATATGAAGAGCATTTGTATGAGCGCGATGA
AGGTGATAAATGGCGAAACAAAAAGTTTGAATTGGGTTTGGAGTTTCCCAATCTT
CCTTATTATATTGATGGTGATGTTAAATTAACACAGTCTATGGCCATCATACGTTA
TATAGCTGACAAGCACAACATGTTGGGTGGTTGTCCAAAAGAGCGTGCAGAGAT
TTCAATGCTTGAAGGAGCGGTTTTGGATATTAGATACGGTGTTTCGAGAATTGCA
TATAGTAAAGACTTTGAAACTCTCAAAGTTGATTTTCTTAGCAAGCTACCTGAAA
TGCTGAAAATGTTCGAAGATCGTTTATGTCATAAAACATATTTAAATGGTGATCA
TGTAACCCATCCTGACTTCATGTTGTATGACGCTCTTGATGTTGTTTTATACATGG
ACCCAATGTGCCTGGATGCGTTCCCAAAATTAGTTTGT'rTTAAAAAACGTATTGA
AGCTATCCCACAAATTGATAAGTACTTGAAATCCAGCAAGTATATAGCATGGCCT
TTGCAGGGCTGGCAAGCCACGTTTGGTGGTGGCGACCATCCTCCAAAATCGGATC
TGGTTCCGCGTGGATCCTCTAGAGTCGACCTGCAGGCATGCAATGCTTATTCCAT
TAATCAAAAGGCTTATTCAAATACTTACCAGGAGTTTACTAATATTGATCAAGCA
AAAGCTTGGGGTAATGCTCAGTATAAAAAGTATGGACTAAGCAAATCAGAAAAA
GAAGCTATAGTATCATATACTAAAAGCGCTAGTGAAAT'AAATGGAAAGCTAAGA
CAAAATAAGGGAGTTATCAATGGATTTCCTTCAAATTTAATAAAACAAGTTGAAC
TTTTAGATAAATCTTTTAATAAAATGAAGACCCCTGAA.~1ATATTATGTTATTTAN
AGGCGACGACCCTGCTTATTTAGGAACAGAATTTCAAA.ACACTCTTCTTAATTCA
AATGGTACAATTAATAAAACGGCTTTTGAAAAGGCTAAAGCTAAGTTTTTAAATA
NAGATAGACTTGAATATGGATATATTAGTACTTCATTAATGAATGTTTCTCAATTT
GCAGGAAGACCAATTATTACAAAATTTAAAGTAGCAAAAGGCTCAAAGGCAGGA
TATATTGACCCTATTAGTGCTTTTCAGGGACAACTTGAAATGTTGCTTCCTAGACA
84
CA 02342970 2001-04-12
TAGTACTTATCATATAGACGATATGAGATTGTCTTCTGATGGTAAACAAATAATA
ATTACAGCAACAATGATGGGCACAGCTATCAATCCTAAAGAATTCAGAAGGAAA
CAAAGAAGAAAAAGAAGACTGCAGGCGGCCGCATCGTGA
S
(2) INFORMATION FOR SEQ ID NO: 20: (EX 10)
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:
(B) TYPE:
(C) STRANDEDNESS:
(D) TOPOLOGY:
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
CA 02342970 2001-04-12
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES 1N SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 20: (EX 10)
MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRIVI~KF'ELGLEFPNLPY
YIDGDVKLTQSMAIIRYIADKI~INMI,GGCPKERAEISMLEGAVLDIRYGVSRIAYSKDF
ETLKVDFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALD
VVLYMDPMCLDAFPKLVCFKKRIEAIPQIDKYLKS SKYIAWPLQGWQATFGGGDHPP
KSDLVPRGSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKS
EKEAIVSYTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKM
KTPENIMLFXGDDPAYLGTEFQNTLLNSNGTINKTAFEKAKAKFLNXDRLEYGYISTS
LMNV SQFAGRPIITKFKVAKGSKAGYIDPI SAFQGQLEML:LPRHSTYHIDDMRLS SDG
KQIIITATMMGTAINPKEFRRKQRRKRRLQAAAS
(2) INFORMATION FOR SEQ ID NO: 21:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:16 AMINO ACIDS
(B) TYPE: AMINO ACID
86
CA 02342970 2001-04-12
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC1 TRANSPORT SEQUENCE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES 1N SEQ ID NO:
87
CA 02342970 2001-04-12
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 21: (C3basicl)
K R R R R R P K K R R R A K R R
(2) INFORMATION FOR SEQ ID NO: 22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:48 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: OLIGONUCLEOTIDE STRAND 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
88
CA 02342970 2001-04-12
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 22: ( EX 12)
AAG AGA AGG CGA AGA AGA CCT AAG AAG AGA CGA AGG GCG AAG
AGG AGA
(2) INFORMATION FOR SEQ ID NO: 23:
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 48 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY:LINEAR
(ii) MOLECULE TYPE: C3BASIC1 OLIGONUCLEOTIDE STRAND 2
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
89
CA 02342970 2001-04-12
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 23: (EX 12)
TTC TCT TCC GCT TCT TCT GGA TTC TTC TCT GCT TCC CGC TTC
TCC TCT
90
CA 02342970 2001-04-12
(2) INFORMATION FOR SEQ ID NO: 24:
(i) SEQUENCE .CHARACTERISTICS:
(A) LENGTH: 792 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
91
CA 02342970 2001-04-12
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 24: (EX 12 DNA sequence)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT AC',T TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG 'CAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT ANA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT ANA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA T'TC AAG AGA AGG CGA AGA AGA CCT AAG
AAG AGA CGA AGG GCG AAG AGG AGA CAC CAC CAC CAC CAC CAC GTC
GAC TCG AGC GGC CGC ATC G'rG ACT GAC TGA
(2) INFORMATION FOR SEQ ID NO: 25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 263 AMINO ACIDS
(B) TYPE: AMINO ACID
92
CA 02342970 2001-04-12
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BAS IC 1 PROTEIN SEQUENCE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMIV1EDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
93
CA 02342970 2001-04-12
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 25: (EX 12 Protein sequence)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPS
NLIKQVELLDKSFNKMKTPENIMLFXGDDPAYLGTEFQNTLLNSNGTINKTAFEKAK
AKFLNXDRLEYGYISTSLMNVSQ
FAGRPIITKFKVAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLS SDGKQIIITAT
MMGTAINPKEFI<:RRRRR.I'KK
RRRAKRR3VDSSGRIVTD.
(2) INFORMATION FOR SEQ ID NO: 26:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 AMINO ACIDS
(B) TYPE:AMINO ACLD
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC2 TRANSPORT SEQUENCE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
94
CA 02342970 2001-04-12
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 26: (EX 13)
K R R R R K K R R Q R R R
(2) INFORMATION FOR SEQ ID NO: 27:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
CA 02342970 2001-04-12
(ii) MOLECULE TYPE: C3BASIC2 OLIGONEUCLOTDE STRAND 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 27: (EX 13)
96
CA 02342970 2001-04-12
AAG CGT CGA CGT AGA AAG AAA CGT AGA CAG CGT AGA CGT
(2) INFORMATION FOR SEQ ID NO: 28
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 36 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC 2 OLIGONUCLEOTIDE STRAND 2
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
97
CA 02342970 2001-04-12
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 28: (EX 13)
TTC GCA GCT GCA TCT TTC TTT GCA TCT GTC GCA TCT GCA
(2) INFORMATION FOR SEQ ID NO: 29:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 783 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC2 cDNA
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
98
CA 02342970 2001-04-12
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
S (D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 29: (EX 13 DNA sequence)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG 'TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT ANA GGC CTAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG 'TTT TTA AAT ANA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA A.TG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
99
CA 02342970 2001-04-12
AAG GCA GGA TAT ATT GAC CC'T ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA AT A ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA G AA TTC AAG CGT CGA CG'T AGA AAG AAA CGT
AGA CAG CGT AGA CGT CAC CAC CAC CAC CAC CAC GTC GAC TCG AGC
GGC CGC ATC GTG ACT GAC TGA
(2) INFORMATION FOR SEQ ID NO: 30:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 260 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC2 PROTE11~T
(v) FRAGMENT TYPE:
(vi) ORIG1NAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAMIE/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
100
CA 02342970 2001-04-12
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES 1N SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 30: (EX 13 Protein sequence
GS SRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIV S
YTKSASEINGKLRQNKGVINGFPS
NLIKQVELLDKSFNKMKTPENIMLFXGDDPAYLGTEFQNTLLNSNGTINKTAFEKAK
AKFLNXDRLEYGYISTSLMNVSQ
FAGRPIITKFKVAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITAT
MMGTAINPKEFk;RRRRKKRR
QRl VDSSGRIVTD.
(2) INFORMATION FOR SEQ ID NO: 31:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 9 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
101
CA 02342970 2001-04-12
(ii) MOLECULE TYPE: C3BASIC3 TRANSPORT PEPTIDE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER Il'JFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES 1N SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 31:
102
CA 02342970 2001-04-12
RRKQRRKRR
(2) INFORMATION FOR SEQ ID NO: 32
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC3 OLIGONUCLEOTIDE STRAND 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
103
CA 02342970 2001-04-12
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 32: (EX 14)
AGA AGG AAA CAA AGA AGA AAA AGA AGA
(2) INFORMATION FOR SEQ ID NO: 33:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC3 OLIGONUCLEOTIDE STRAND 2
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
104
CA 02342970 2001-04-12
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 33: (EX 14)
TCT TCC TTT GTT TCT TCT TTT TCT TCT
(2) INFORMATION FOR SEQ ID NO: 34:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 771 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STR.ANDEDNESS: SINGLE
105
CA 02342970 2001-04-12
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: cDNA C3BASIC3
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
106
CA 02342970 2001-04-12
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 34: (EX 14 DNA sequence)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG 'TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT ANA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG 'CTT TTA AAT ANA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA AT'A ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AGA AGG AAA CAA AGA AGA AAA AGA
AGA CAC CAC CAC CAC CAC CAC G TC GAC TCG AGC GGC CGC ATC GTG ACT
GAC TGA
(2) INFORMATION FOR SEQ ID NO: 35:
107
CA 02342970 2001-04-12
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 256 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC3
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
108
CA 02342970 2001-04-12
(B) LOCATION:
(D) OTHER INFORMATION:
(X) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(~) SEQUENCE DESCRIPTION: SEQ ID NO: 35: (EX 14)
GS SRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFXGDDPAYLG
TEFQNTLLNSNGTINKTAFEKAKAKFLNXDRLEYGYISTSLMNVSQFAGRPIITKFKVA
109
CA 02342970 2001-04-12
KGSKAGYIDPI SAFQGQLEMLLPRHSTYHIDDMRL,S SDGKQIIITATMMGTAINPKEFR
RKQRRI VDSSGRIVTD.
110
CA 02342970 2001-04-12
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: LISA MCKERRACHER
(ii) TITLE OF INVENTION: FUSION PROTEINS
(iii) NUMBER OF SEQUENCES: 35
(iv) CORRESPONDENCE ADDRESS:
(A) ADRESSEE: BROULLETTE KOSIE
(B) STREET: 1100 RENE-LESVEQUE BLVD WEST
(C) PROV/STATE: QUEBEC
(D) COUNTRY: CANADA
(E) POSTAL/ZIP CODE: H3B SC9
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE:ASCII (TExT)
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE:
(C) CLASSIFICATION:
(vii) ATTORNEY/AGENT INFORMATION:
(A) NAME: BROULLETTE KOSIE
(B) REGISTRATION NO.:
(C) REFERENCE/DOCKET NO.: 06447-004-CA-O1
(D) TEL. NO.: (514) 397 8500
(E) FAX NO.: (514) 397 8515
(2) INFORMATION FOR SEQ ID NO: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 BASE PAIRS
(B) TYPE: NUCLEIC ACID
CA 02342970 2001-04-12
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE: 5' cDNA primer - C3 sequence
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 1:
5'GAA TTC TTT AGG ATT GAT AGC TGT GCC 3'
(2) INFORMATION FOR SEQ ID NO: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 BASE PAIRS
2
CA 02342970 2001-04-12
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE: 3' cDNA primer - C3 sequence
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2:
5'GGT GGC GAC CAT CCT CCA AAA 3'
(2) INFORMATION FOR SEQ ID NO: 3:
(i) SEQUENCE CHARACTERISTICS:
3
CA 02342970 2001-04-12
(A) LENGTH: 888 BASE PAIRS
(B) TYPE: cDNA
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY: C3APL
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES 1N SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 3: (Nucleotide sequence of protein
C3APL)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT CAA
AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA GCA AAA
GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA TCA GAA
AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA AAT GGA
AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA AAT TTA ATA
AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG AAG ACC CCT GAA
4
CA 02342970 2001-04-12
AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT TTA GGA ACA GAA TTT
CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT AAT AAA ACG GCT TTT GAA
AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT AGA CT'T GAA TAT GGA TAT ATT
AGT ACT TCA TTA ATG AAT GTC TCT CAA TTT GCA GGA AGA CCA ATT ATT ACA
CAA TTT AAA GTA GCA AAA GGC TCA AAG GCA GGA TAT ATT GAC CCT ATT AGT
GCT TTT CAG GGA CAA CTT GAA ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT
ATA GAC GAT ATG AGA TTG TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA
ACA ATG ATG GGC ACA GCT ATC AAT CCT AAA GAA TTC GTG ATG GAA TCC CGC
AAA CGC GCA AGG CAG ACA TAC ACC CGG TAC CAG AC",T CTA GAG CTA GAG
AAG GAG TTT CAC TTC AAT CGC TAC TTG ACC CGT CGCJ CGA AGG ATC GAG ATC
GCC CAC GCC CTG TGC CTC ACG GAG CGC CAG ATA AAG ATT TGG TTC CAG AAT
CGG CGC ATG AAG TGG AAG AAG GAG AAC TGA
(2) INFORMATION FOR SEQ ID NO: 4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 295 AMINO ACIDS
(B) TYPE: PROTEIN
(C) STRANDEDNESS SINGLE:
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
CA 02342970 2001-04-12
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 4: (amino acid sequence of C3APL)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSE
KEAIVSYTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPE
NIMLFRGDDPAYLGTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGY
ISTSLMNVSQFAGRPIITQFKVAKGSKAGYIDPISAFQGQLEMLLPRHST
YHIDDMRLSSDGKQIIITATMMGTAINPKEFVMESRKRARQ'CYTRYQTLE
LEKEFHFNRYLTRRRRIEIAHALCLTERQIKIWFQNRRMICWKKEN
(2) INFORMATION FOR SEQ ID NO: 5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 774 BASE PAIRS
(B) TYPE: cDNA
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IM1V1EDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
6
CA 02342970 2001-04-12
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 5: (Nucleotide sequence of C3APS)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT CAA
AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA GCA AAA
GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA TCA GAA
AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA AAT GGA
AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA AAT TTA ATA
AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG AAG ACC CCT GAA
AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT TTA GGA ACA GAA TTT
CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT AAT AAA ACG GCT TTT GAA
AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT AGA CTT GAA TAT GGA TAT ATT
AGT ACT TCA TTA ATG AAT GTC TCT CAA TTT GCA GGA AGA CCA ATT ATT ACA
CAA TTT AAA GTA GCA AAA GGC TCA AAG GCA GGA TAT ATT GAC CCT ATT AGT
GCT TTT CAG GGA CAA CTT GAA ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT
ATA GAC GAT ATG AGA TTG TCT TCT GAT GGT AAA CA,A ATA ATA ATT ACA GCA
ACA ATG ATG GGC ACA GCT ATC AAT CCT AAA GAA TTC CGC CAG ATC AAG ATT
TGG TTC CAG AAT CGT CGC ATG AAG TGG AAG AAG GTC GAC TCG AGC GGC
CGC ATC GTG ACT GAC TGA
(2) INFORMATION FOR SEQ ID NO: 6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 257 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
7
CA 02342970 2001-04-12
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 6:(amino acid sequence of C3APS)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSE
KEAIVSYTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNI~TPE
NIMLFRGDDPAYLGTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGY
ISTSLMNVSQFAGRPIITQFKVAKGSKAGYIDPISAFQGQLEI~~,LPRHST
YHIDDMRLS SDGKQIIITATMMGTAINPKEFRQ1KIWFQNRRMKWKKVDS
SGRIVTD
(2) INFORMATION FOR SEQ ID NO: 7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 BASE PAIRS
(B) TYPE:
(C) STRANDEDNESS: SINGLE
8
CA 02342970 2001-04-12
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: 5' cDNA primer - antennapedia sequence
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 7: (EX 3)
5'GAA TCC CGC AAA CGC GCA AGG CAG 3'
(2) INFORMATION FOR SEQ ID NO: 8:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 BASE PAIRS
(B) TYPE:NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
9
CA 02342970 2001-04-12
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: 3' cDNA primer
(v) FRAGMENT TYPE: antennapedia sequence
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 8: (EX 3)
5'TCA GTT CTC CTT CTT CCA CTT CAT GCG 3'
(2) INFORMATION FOR SEQ ID NO: 9:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54 BASR PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
CA 02342970 2001-04-12
(D) TOPOLOGY:LINEAR
(ii) MOLECULE TYPE: OLIGONEUCLEOTIDE STRAND 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANTSM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 9: (EX 3)
5'AAT TCC GCC AGA TCA AGA TTT GGT TCC AGA ATC GTC GCA TGA AGT GGA
AGA AGG 3'
(2) INFORMATION FOR SEQ ID NO: 10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54 BSE PAIRS
(B) TYPE:NUCLEIC ACID
11
CA 02342970 2001-04-12
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: OLIGONUCLEOTIDE STRAND 2
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 10: (EX 3)
5'GGC GGT CTA GTT CTA AAC CAA GCT CTT AGC AGC GTA GTT CAC CTT CTT
CCA GCT 3'
(2) INFORMATION FOR SEQ ID NO: 11:
(i) SEQUENCE CHARACTERISTICS:
12
CA 02342970 2001-04-12
(A) LENGTH: 26 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: TAT 5' PRIMER
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 11:
5'GAATCCAAGCATCCAGGAAGTCAGCC 3'
(2) INFORMATION FOR SEQ ID NO: 12:
(i) SEQUENCE CHARACTERISTICS:
13
CA 02342970 2001-04-12
(A) LENGTH: 21 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:TAT 3' PRIMER
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) INIIVIEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 12: (ex 8)
5' ACC AGC CAC CAC CTT CTG ATA 3'
(2) INFORMATION FOR SEQ ID NO: 13:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 876 BASE PAIRS
14
CA 02342970 2001-04-12
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3-TL SEQUENCE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGE S
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 13: (DNA sequence of C3-TL)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT CAA
AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA GCA AAA
GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA TCA GAA
AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA AAT GGA
AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TT'T CCT TCA AAT TTA ATA
AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG AAG ACC CCT GAA
AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT TTA GGA ACA GAA TTT
CA 02342970 2001-04-12
CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT AAT AAA ACG GCT TTT GAA
AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT AGA CTT GAA TAT GGA TAT ATT
AGT ACT TCA TTA ATG AAT GTC TCT CAA TTT GCA GGA AGA CCA ATT ATT ACA
CAA TTT AAA GTA GCA AAA GGC TCA AAG GCA GGA TAT ATT GAC CCT ATT AGT
GCT TTT CAG GGA CAA CTT GAA ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT
ATA GAC GAT ATG AGA TTG TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA
ACA ATG ATG GGC ACA GCT ATC AAT CCT AAA GAA TTC AAG CAT CCA GGA
AGT CAG CCT AAA ACT GCT TGT ACC AAT TGC TAT TGT AAA AAG TGT TGC TTT
CAT TGC CAA GTT TGT TTC ATA ACA AAA GCC TTA GGC ATC TCC TAT GGC AGG
AAG CGG AGA CAG CGA CGA AGA GCT CAT CAG AAC AGT CAG ACT CAT CAA
GCT TCT CTA TCA AAG CAG TAA
(2) INFORMATION FOR SEQ ID NO: 14:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 291 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3-TL PROTEIN SEQUENCE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
16
CA 02342970 2001-04-12
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 14: (The protein sequence of C3-TL)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVSYT
KSASE1NGKLRQNKGVINGFPSNLIKQVELLDKSFNKM1{TPENIMLFRGDDPAYLGTEFQ
NTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFKVAKGSKA
GYIDPI SAFQGQLEMLLPRH STYHIDDMRLS SDGKQIIITATMMGTAINPKEFKHPGSQPK
TACTNCYCKKCCFHCQVCFITKALGISYGRKRRQRRRAHQNSQTHQASLSKQ
(2) INFORMATION FOR SEQ ID NO: 15:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: TS OLIGONUCLEOTIDE STRAND 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
17
CA 02342970 2001-04-12
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 15: (EX 9)
5'AAT TCT ATG GTC GTA AAA AAC GTC GTC AAC GTC GTC GTG 3'
(2) INFORMATION FOR SEQ ID NO: 16:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: TS OLIGONUCLEOTIDE STRAND 2
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
18
CA 02342970 2001-04-12
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 16: (EX 9)
5' GAT ACC AGC ATT TTT TGC AGC AGT TGC AGC AGC ACA GCT 3'
(2) INFORMATION FOR SEQ ID NO: 17:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 756 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3-TS cDNA
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
19
CA 02342970 2001-04-12
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 17: (Nucleotide sequence of C3-TS)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT CAA
AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA GCA AAA
GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA TCA GAA
AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA AAT GGA
AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA AAT TTA ATA
AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG AAG ACC CCT GAA
AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT TTA GGA ACA GAA TTT
CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT AAT AAA ACG GCT TTT GAA
AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT AGA CTT GAA TAT GGA TAT ATT
AGT ACT TCA TTA ATG AAT GTC TCT CAA TTT GCA GGA AGA CCA ATT ATT ACA
CAA TTT AAA GTA GCA AAA GGC TCA AAG GCA GGA TAT ATT GAC CCT ATT AGT
GCT TTT CAG GGA CAA CTT GAA ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT
ATA GAC GAT ATG AGA TTG TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA
ACA ATG ATG GGC ACA GCT ATC AAT CCT AAA GAA TTC TAT GGT GCT AAA AAA
CGT CGT CAA CGT CGT CGT GTC GAC TCG AGC GGC CCG CAT CGT GAC TGA
(2) INFORMATION FOR SEQ ID NO: 18:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 251 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3-TS PROTEIN
(v) FRAGMENT TYPE:
CA 02342970 2001-04-12
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 18: (The protein sequence of C3-TS)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVSYT
KSASEINGKLRQNKGV1NGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYLGTEFQ
NTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFKVAKGSKA
GYIDPISAFQGQLEMLLPRHSTYHIDDMRLS SDGKQIIITATMMGTAINPKEFYGAKKRRQ
RRRVDS SGPHRD
(2) INFORMATION FOR SEQ ID NO: 19:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:
(B) TYPE:
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
21
CA 02342970 2001-04-12
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) 1MNIEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 19: (EX 10)
ATGTCCCCTATACTAGGTTATTGGAAAATTAAGGGCCTTGTGCAACCCACTCGACTT
CTTTTGGAATATCTTGAAGAAAAATATGAAGAGCATTTGTATGAGCGCGATGAAGGT
GATAAATGGCGAAACA,AAAAGTTTGAATTGGGTTTGGAGTTTCCCAATCTTCCTTAT
TATATTGATGGTGATGTTAAATTAACACAGTCTATGGCCATCATACGTTATATAGCT
GACAAGCACAACATGTTGGGTGGTTGTCCAAAAGAGCGTGCAGAGATTTCAATGCTT
GAAGGAGCGGTTTTGGATATTAGATACGGTGTTTCGAGAATTGCATATAGTAAAGAC
TTTGAAACTCTCAAAGTTGATTTTCTTAGCAAGCTACCTGAAATGCTGAAAATGTTC
GAAGATCGTTTATGTCATAAAACATATTTAAATGGTGATCATGTAACCCATCCTGAC
TTCATGTTGTATGACGCTCTTGATGTTGTTTTATACATGGACCCAATGTGCCTGGATG
CGTTCCCAAAATTAGTTTGTTTTAAAAAACGTATTGAAGCTATCCCACAAATTGATA
AGTACTTGAAATCCAGCAAGTATATAGCATGGCCTTTGCAGGGCTGGCAAGCCACGT
22
CA 02342970 2001-04-12
TTGGTGGTGGCGACCATCCTCCAAAATCGGATCTGGTTCCGCGTGGATCCTCTAGAG
TCGACCTGCAGGCATGCAATGCTTATTCCATTAATCAAAAGGCTTATTCAAATACTT
ACCAGGAGTTTACTAATATTGATCAAGCAAAAGCTTGGGGTAATGCTCAGTATAAAA
AGTATGGACTAAGCAAATCAGAAAAAGAAGCTATAGTATCATATACTAAAAGCGCT
AGTGAAATAAATGGAAAGCTAAGACAAAATAAGGGAGTTATCAATGGATTTCCTTC
AAATTTAATAAAACAAGTTGAACTTTTAGATAAATCTTTTAATAAAATGAAGACCCC
TGAAAATATTATGTTATTTANAGCiCGACGACCCTGCTTATTTAGGAACAGAATTTCA
AAACACTCTTCTTAATTCAAATGCTTACAATTAATAAAACGGCTTTTGAAAAGGCTAA
AGCTAAGTTTTTAAATANAGATAGACTTGAATATGGATATATTAGTACTTCATTAAT
GAATGTTTCTCAATTTGCAGGAAGACCAATTATTACAAAATTTAAAGTAGCAAAAGG
CTCAAAGGCAGGATATATTGACCCTATTAGTGCTTTTCAGGGACAACTTGAAATGTT
GCTTCCTAGACATAGTACTTATCATATAGACGATATGAGATTGTCTTCTGATGGTAA
ACAAATAATAATTACAGCAACAATGATGGGCACAGCTATCAATCCTAAAGAATTCA
GAAGGAAACAAAGAAGA,AAAAGAAGACTGCAGGCGGCCGCATCGTGA
(2) INFORMATION FOR SEQ ID NO: 20: (EX 10)
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:
(B) TYPE:
(C) STRANDEDNESS:
(D) TOPOLOGY:
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
23
CA 02342970 2001-04-12
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 20: (EX 10)
MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRNKKFELGLEFPNLPYYID
GDVKLTQSMAIIRYIADKHNML,GGCPKERAEISMLEGAVLDIRYGVSRIAYSKDFETLKV
DFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALD
VVLYMDPMCLDAFPKLVCFKKRIEAIPQIDKYLKS SKYIAWPLQGWQATFGGGDHPPKS
DLVPRGSSRVDLQACNAYS1NQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEA
IVSYTKSASEINGKLRQNKGV1NGFPSNLIKQVELLDKSFNKM
KTPENIMLFXGDDPAYLGTEFQNTLLNSNGTINKTAFEKAKAKFLNXDRLEYGYISTSLM
NVSQFAGRPIITKFKVAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIIT
ATMMGTAINPKEFRRKQRRKRRLQAAAS
(2) INFORMATION FOR SEQ ID NO: 21:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:16 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC1 TRANSPORT SEQUENCE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
24
CA 02342970 2001-04-12
(vii) IIVllVIEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ B7 NO: 21: (C3basicl)
K R R R R R P K K R R R A K R R
(2) INFORMATION FOR SEQ ID NO: 22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:48 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: OLIGONUCLEOTIDE STRAND 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
CA 02342970 2001-04-12
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 22: ( EX 12)
AAG AGA AGG CGA AGA AGA CCT AAG AAG AGA CGA AGG GCG AAG
AGG AGA
(2) INFORMATION FOR SEQ B7 NO: 23:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 48 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY:LINEAR
(ii) MOLECULE TYPE: C3BASIC 1 OLIGONUCLEOTIDE STRAND 2
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
26
CA 02342970 2001-04-12
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 23: (EX 12)
TTC TCT TCC GCT TCT TCT GGA TTC TTC TCT GCT TCC CGC TTC TCC
TCT
(2) INFORMATION FOR SEQ ID NO: 24:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 792 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
27
CA 02342970 2001-04-12
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 24: (EX 12 DNA sequence)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT CAA
AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA GCA AAA
GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA TCA GAA
AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA AAT GGA
AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA AAT TTA ATA
AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG AAG ACC CCT GAA
AAT ATT ATG TTA TTT ANA GGC GAC GAC CCT GCT TAT' TTA GGA ACA GAA TTT
CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT AA'C AAA ACG GCT TTT GAA
AAG GCT AAA GCT AAG TTT TTA AAT ANA GAT AGA CTT GAA TAT GGA TAT ATT
AGT ACT TCA TTA ATG AAT GTT TCT CAA TTT GCA GGA AGA CCA ATT ATT ACA
AAA TTT AAA GTA GCA AAA GGC TCA AAG GCA GGA TAT ATT GAC CCT ATT AGT
GCT TTT CAG GGA CAA CTT GAA ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT
ATA GAC GAT ATG AGA TTG TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA
ACA ATG ATG GGC ACA GCT ATC AAT CCT AAA GAA TTC AAG AGA AGG CGA
AGA AGA CCT AAG AAG AGA CGA AGG GCG AAG AGG AGA CAC CAC CAC
CAC CAC CAC GTC GAC TCG AGC GGC CGC ATC GTG ACT GAC TGA
28
CA 02342970 2001-04-12
(2) INFORMATION FOR SEQ ID NO: 25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 263 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC 1 PROTEIN SEQUENCE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 25: (EX 12 Protein sequence)
GS SRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIV SYT
29
CA 02342970 2001-04-12
KSASEINGKLRQNKGVINGFPS
NLIKQVELLDKSFNI~TPENIMLFXGDDPAYLGTEFQNTLLNSNGTINKTAFEKAKAKF
LNXDRLEYGYISTSLMNVSQ
FAGRPIITKFKVAKGSKAGYIDPI SAFQGQLEMLLPRHSTYHIDDMRLS SDGKQIIITATM
MGTAINPKEFKRRRRRPKK
RRRAI VD S SGRIVTD.
(2) INFORMATION FOR SEQ ID NO: 26:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 AMINO ACIDS
(B) TYPE:AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC2 TRANSPORT SEQUENCE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IIVINIEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
CA 02342970 2001-04-12
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 26: (EX 13)
K R R R R K K R R Q R R R
(2) INFORMATION FOR SEQ ID NO: 27:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC2 OLIGONEUCLOTDE STRAND 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
31
CA 02342970 2001-04-12
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 27: (EX I3)
AAG CGT CGA CGT AGA AAG AAA CGT AGA CAG CGT AGA CGT
(2) INFORMATION FOR SEQ ID NO: 28:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 36 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC 2 OLIGONUCLEOTIDE STRAND 2
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
32
CA 02342970 2001-04-12
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 28: (EX I3)
TTC GCA GCT GCA TCT TTC TTT GCA TCT GTC UCA TCT GCA
(2) INFORMATION FOR SEQ ID NO: 29:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 783 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC2 cDNA
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
33
CA 02342970 2001-04-12
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 29: (EX 13 DNA sequence)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT CAA
AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA GCA AAA
GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA TCA GAA
AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA AAT GGA
AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA AAT TTA ATA
AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG AAG ACC CCT GAA
AAT ATT ATG TTA TTT ANA GGC GAC GAC CCT GCT TAT TTA GGA ACA GAA TTT
CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT AA'T AAA ACG GCT TTT GAA
AAG GCT AAA GCT AAG TTT TTA AAT ANA GAT AGA CTT GAA TAT GGA TAT ATT
AGT ACT TCA TTA ATG AAT GTT TCT CAA TTT GCA GGA AGA CCA ATT ATT ACA
AAA TTT AAA GTA GCA AAA GGC TCA AAG GCA GGA TAT ATT GAC CCT ATT AGT
GCT TTT CAG GGA CAA CTT GAA ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT
ATA GAC GAT ATG AGA TTG TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA
ACA ATG ATG GGC ACA GCT ATC AAT CCT AAA G AA TTC AAG CGT CGA CGT
AGA AAG AAA CGT AGA CAG CGT AGA CGT CAC CAC CAC CAC CAC CAC
GTC GAC TCG AGC GGC CGC ATC GTG ACT GAC TGA
(2) INFORMATION FOR SEQ ID NO: 30:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 260 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: L1NEAR
(ii) MOLECULE TYPE: C3BASIC2 PROTEIN
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
34
CA 02342970 2001-04-12
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 30: (EX 13 Protein sequence
GS SRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVSYT
KSASEINGKLRQNKGVINGFPS
NLIKQVELLDKSFNKMKTPENIMLFXGDDPAYLGTEFQNTLLNSNGTINKTAFEKAKAKF'
LNXDRLEYGYI S T S LMNV S Q
FAGRPIITKFKVAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRL,SSDGKQIIITATM
MGTA1NPKEFKRRRRKKRR
Q VDSSGRIVTD.
(2) INFORMATION FOR SEQ ID NO: 31:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 9 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC3 TRANSPORT PEPTIDE
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
CA 02342970 2001-04-12
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 31:
RRKQRRKRR
(2) INFORMATION FOR SEQ ID NO: 32:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC3 OLIGONUCLEOTIDE STRAND 1
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
36
CA 02342970 2001-04-12
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 32: (EX 14)
AGA AGG AAA CAA AGA AGA AAA AGA AGA
(2) INFORMATION FOR SEQ ID NO: 33:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC3 OLIGONUCLEOTIDE STRAND 2
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
37
CA 02342970 2001-04-12
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 33: (EX 14)
TCT TCC TTT GTT TCT TCT TTT TCT TCT
(2) INFORMATION FOR SEQ ID NO: 34:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 771 BASE PAIRS
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: cDNA C3BASIC3
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
38
CA 02342970 2001-04-12
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 34: (EX 14 DNA sequence)
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GC',T TAT TCC ATT AAT CAA
AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA GCA AAA
GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA TCA GAA
AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA AAT GGA
AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA AAT TTA ATA
AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG AAG ACC CCT GAA
AAT ATT ATG TTA TTT ANA GGC GAC GAC CCT GCT TAT TTA GGA ACA GAA TTT
CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT AA'T AAA ACG GCT TTT GAA
AAG GCT AAA GCT AAG TTT TTA AAT ANA GAT AGA CTT GAA TAT GGA TAT ATT
AGT ACT TCA TTA ATG AAT GTT TCT CAA TTT GCA GGA AGA CCA ATT ATT ACA
AAA TTT AAA GTA GCA AAA GGC TCA AAG GCA GGA TAT ATT GAC CCT ATT AGT
GCT TTT CAG GGA CAA CTT GAA ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT
ATA GAC GAT ATG AGA TTG TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA
ACA ATG ATG GGC ACA GCT ATC AAT CCT AAA GAA TTC AGA AGG AAA CAA
AGA AGA AAA AGA AGA CAC CAC CAC CAC CAC CAC G TC GAC TCG AGC GGC
CGC ATC GTG ACT GAC TGA
39
CA 02342970 2001-04-12
(2) INFORMATION FOR SEQ ID NO: 35:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 256 AMINO ACIDS
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: C3BASIC3
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID NO:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 35: (EX 14)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVSYT
KSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNI~TPENIMLFXGDDPAYLGTEFQ
NTLLNSNGTINKTAFEKAKAKFLNXDRLEYGYISTSLMNVSQFAGRPIITKFKVAKGSKA
CA 02342970 2001-04-12
GYIDPISAFQGQLEML,LPRHSTYHIDDMRLS SDGKQIIITATMMGTAINPKEFRRKQRRKR
~VDSSGRIVTD.
41