Note: Descriptions are shown in the official language in which they were submitted.
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 1 -
USE OF PEX IN THE TREATMENT OF METABOLIC BONE DISEASES
BACKGROUND OF THE INVENTION
(a) Field of the Invention
The invention relates to the use of PEX in the
treatment of metabolic bone diseases, such as osteoma-
lacia and osteoporosis.
(b) Description of Prior Art
Mutations in the PEX gene are responsible for X-
linked hypophosphatemic rickets (HYP) . To gain insight
into the role of PEX in normal physiology we have
cloned the human full-length cDNA and studied its tis-
sue expression, subcellular localization, and peptidase
activity. We show that the cDNA encodes a 749 amino
acid protein structurally related to a family of neu-
tral endopeptidases that include neprilysin (NEP) as
prototype. By Northern blot analysis, the size of the
full-length PEX transcript is 6.5 kb. PEX expression,
as determined by semi-quantitative PCR, is high in bone
and in tumor tissue associated with the paraneoplastic
syndrome of renal phosphate wasting. PEX is glycosyl-
ated in the presence of canine microsomal membranes and
partitions exclusively in the detergent phase from Tri-
ton X-114 extractions of transiently transfected COS
cells. Immunofluorescence studies in A293 cells
expressing PEX tagged with a c-myc epitope show a pre-
dominant cell-surface location for the protein with its
C-terminal domain in the extracellular compartment,
substantiating the assumption that PEX, like other mem-
bers of the neutral endopeptidase family, is a type II
integral membrane glycoprotein. Cell membranes from
cultured COS cells transiently expressing PEX effi-
ciently degrade exogenously added PTH-derived peptides,
demonstrating for the first time that recombinant PEX
can function as an endopeptidase. PEX peptidase activ-
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 2 -
ity may provide a convenient target for pharmacological
intervention in states of altered phosphate homeostasis
and in metabolic bone diseases.
X-linked hypophosphatemic rickets (HYP) is the
most common inherited disorder of renal phosphate wast-
ing
characterized by severe hypophosphatemia, renal
phosphate wasting, reduced serum concentrations of
1,25-dihydroxyvitamin D levels, and defective bone min-
eralization. Until recently, much of our understanding
of HYP has been facilitated by the availability of two
murine homologues, the Hyp and Gy mice, which exhibit
many of the phenotypic features of HYP. Through
positional cloning, however, a gene which spans the
deleted region Xp22.1 in HYP patients, or is mutated in
non-deletion patients with the disorder, was identified
(designated PEX) and its partial cDNA sequence reported
(The HYP Consortium (1995) Nature Genetics 11, 130-
136) . The predicted human PEX gene product, as well as
its murine homologue (Du, L. et al. (1996) Genomics 36,
22-28), exhibit homology to a family of neutral endo-
peptidases involved in either activation or degradation
of a number of peptide hormones. It has been postulated
that PEX metabolizes a peptide hormone that modulates
renal tubular phosphate handling. Such an activity
could involve either the processing of a phosphate-
reabsorbing hormone precursor to its active form or the
inactivation of a circulating phosphaturic factor.
These speculations notwithstanding, the physiologic
function of the PEX gene product and the mechanisms
that lead to the renal and skeletal abnormalities of
HYP remain to be defined.
Oncogenous hypophosphatemic osteomalacia (OHO)
is a rare acquired disorder of phosphate homeostasis
with biochemical and physical abnormalities similar to
HYP. This syndrome is associated with a variety of his-
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 3 -
tologically distinct, usually benign, mesenchymal
tumors whose excision promptly reverses the metabolic
abnormalities and results in cure of the bone disease.
It is generally thought that a factor(s) produced by
these tumors promotes phosphaturia and inhibits the
renal conversion of 25-hydroxyvitamin D to 1,25-dihy-
droxyvitamin D. The nature of the phosphaturic sub-
stance remains unknown and is likely distinct from both
parathyroid hormone (PTH) and calcitonin, two polypep-
tide hormones known to inhibit the renal tubular reab-
sorption of phosphorus. Because of the striking simi-
larity in the clinical presentation of patients with
OHO and HYP, it is postulated that the factor causing
phosphaturia in OHO is the active form of the PEX sub-
strate. The identification and characterization of the
putative PEX substrate, referred to as phosphatonin,
however, will require first a better understanding of
PEX function.
To date, there is still a need to understand how
local factors produced in the bone regulate bone forma-
tion and bone resorption. Derangement of these factors
leads to metabolic bone diseases. Pharmacological
manipulation of such factors may serve as a novel
approach to the treatment of these disorders.
It would be highly desirable to be provided with
a tool in the treatment of metabolic bone diseases,
such as osteomalacia and osteoporosis.
SU'N.MARY OF THE INVENTION
One aim of the present invention is to provide a
tool in the treatment of metabolic bone diseases, such
as osteomalacia and osteoporosis.
Another aim of the present invention is to pro-
vide the use of PEX in the treatment of metabolic bone
diseases, such as osteomalacia and osteoporosis.
CA 02343713 2006-11-24
WO 00/18954 PCT/CA99/00895
- 4 -
Another aim of the present invention is to pro-
vide a method of diagnostic of metabolic bone diseases,
such as osteomalacia and osteoporosis.
Toward this objective, we have cloned a cDNA
encoding the full-length human PEX protein, and deter- _
mined the tissue distribution of PEX transcripts. In
addition, we have examined the subcellular localization
of recombinant PEX protein and demonstrated its pepti-
dase activity.
In accordance with the present invention there
is provided a method for the diagnosis of metabolic
bone diseases in a patient, which comprises the step of
determining the level of PTHrP in a biological sample
of a patient wherein an alteration of PTHrP levels from
that of a normal individual is indicative of metabolic
bone diseases and/or metabolic bone diseases predispo-
sition.
In accordance with the present invention there
is provided a method for the treatment of metabolic
bone diseases, which comprises administering to a
patient a compound for the modulation of PEX enzymatic
activity.
In accordance with the present invention there
is provided the use of a PEX enzymatic activity
modulator for treating osteomalacia, osteoporosis,
osteopetrosis, Paget's disease or x-linked
hypophosphatemia rickets (HYP), wherein said
modulator modulates endogenous levels of PTH and/or
PTHrP to regulate bone formation.
In accordance with the present invention there
is provided the method of screening for a PEX
CA 02343713 2006-11-24
4a
enzymatic activity modulator for use as a candidate
compound for treating osteomalacia, osteoporosis,
osteopetrosis, Paget's disease or x-linked
hypophosphatemia rickets (HYP), the method
comprising: (i) contacting PEX with a substrate for
PEX and a test compound; and (ii) determining whether
degradation of the substrate is modulated in the
presence of the test compound; wherein when
degradation of the substrate is modulated said
compound is identified as having a therapeutic
potential
In accordance with the present invention there
is provided a method for the treatment of metabolic
bone diseases, which comprises modulating PTH and
PTHrP levels that regulate osteoblast activity in a
patient to modulate bone breakdown and bone
formation.
In accordance with the present invention there
is provided the use of modulation of PTH and PTHrP
levels that regulate osteoblast activity for the
treatment of metabolic bone diseases.
CA 02343713 2005-04-28
- 5 -
In accordance with the present invention there is
provided a non-human transgenic mammal to study the role
of PEX in bone development and homeostasis, whose germ
cells and somatic cells contain a PEX gene construct for
expression of PEX in osteoblast consisting essentially of
a recombinant PEX gene sequence under the control of a
proximal promoter of a pro-al(I) collagen gene, the PEX
gene construct being introduced into the mammal, or an
ancestor of the mammal, at an embryonic stage.
The non-human mammal is preferably a mouse and the
proximal promoter is preferably murine pro-al(I) collagen
gene, more preferably a 2.3 kb fragment thereof.
For the purpose of the present invention the
following terms are defined below.
The expression "metabolic bone diseases" includes
osteomalacia, osteoporosis, osteopetrosis, Paget's
disease, X-linked hypophosphatemic rickets (HYP) and
other metabolic bone diseases resulting from altered
activity of PEX and/or altered levels of PTH and/or
PTHrP.
BRIEF DESCRIPTION OF THE DRAWINGS
Fig. 1 illustrates PEX mRNA expression in OHO
tumors;
Fig. 2A illustrates human PEX cDNA cloned from OHO
tumors (SEQ ID NOS:1-2);
Fig. 2B illustrates human PEX and human NEP protein
alignment (SEQ ID NOS:3-4);
Fig. 2C illustrates the TMpred output for PEX;
Fig. 3 illustrates PEX expression in human tissues;
Fig. 4 illustrates a Northerm blot analysis of PEX
mRNA;
Fig. 5 illustrates in vitro translation of human PEX
cRNA;
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 6 -
Figs. 6A-6B illustrate TRITONT"' X-114 extraction
and immunofluorescent localization of PEX;
Figs. 7A-7C illustrate HPLC analysis of the
hydrolysis of [D-Ala2, Leu5] enkephalin;
Figs. 8A-8C illustrate the hydrolysis of PTH-
derived
peptides by PEX endopeptidase activity; and
Fig. 9 illustrates Schematic representation of
phosphate handling in the proximal renal tubule in nor-
mal, OHO, and HYP states.
DETAILED DESCRIPTION OF THE INVENTION
PEX is a Cell Membrane-Associated Protein
Previous studies have established that NEP, ECE-1
and Kell blood group glycoprotein are integral membrane
proteins. We have used extraction with the detergent
TRITONTM X-114 and immunochemical localization to examine
whether PEX is also a membrane-associated protein. For
identification of PEX, we generated a construct in which
the carboxyl terminus sequences of PEX are modified by a
human c-myc tag. The epitope tag was inserted immediately
upstream of the potential prenylation motif so that any
lipid modification of the PEX protein may proceed uninter-
rupted.
TRITONTM X-114 is a detergent that forms an aque-
ous solution at 4 C but separates into hydrophobic and
aqueous phases when the temperature is raised to 30-37 C.
This property has been used as an indicator of the hydro-
phobic nature of proteins, with integral membrane proteins
partitioning exclusively in the detergent phase while
highly hydrophilic proteins associate with the aqueous
phase. TRITONT'" X-114 extracts from COS-7 cells tran-
siently expressing PEX tagged with the c-myc epitope
showed that PEX partitions nearly exclusively into the
detergent phase. This finding indicates that PEX is a
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 7 -
membrane-associated protein and is consistent with the
prediction from sequence analysis that it is an integral
membrane protein.
To determine the subcellular localization of PEX,
the distribution of recombinant protein expressed in sta=
bly transfected A293 cells was examined using immunofluo-
rescence. When cells were fixed and permeabilized, myc-
tagged PEX immunostaining was detected primarily on the
cell surface, but in a number of cells staining was also
observed intracellularly, although no signal was observed
in the nucleus. If permeabilization was omitted, staining
was localized exclusively to the plasma membrane, while
untransfected cells or cells transfected with vector alone
showed no immunofluorescent staining. Since the myc-tag
was inserted in the carboxyl end of PEX, these findings
further corroborate the sequence-based prediction that PEX
is a Type II integral membrane protein with its large C-
terminal hydrophilic domain containing the active enzy-
matic site in the extracellular compartment.
Recombinant PEX protein has peptidase activity
The subcellular localization and sequence similar-
ity between PEX and NEP strongly suggest that PEX func-
tions as a membrane-bound metallopeptidase. However, no
peptidase activity has yet been ascribed to PEX. As shown,
when [D-Ala2, Leu5] enkephalin, used to assay for NEP
activity, was incubated with cell membrane preparations
from vector-transfected COS cells or COS cells expressing
equivalent amounts of recombinant human NEP or PEX pro-
teins, as determined by Western blot analysis, production
of Tyr-D-Ala-Gly from the substrate was evident only in
NEP-expressing membrane preparations. While the PEX
sequence preserves two of the residues critical for cata-
lytic activity of NEP (equivalent to E646 and H711), it
lacks a residue equivalent to R102 shown to be crucial for
the dipeptidylcarboxypeptidase activity of NEP. Therefore,
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 8 -
unlike NEP, PEX has no dipeptidylcarboxypeptidase activ-
ity.
To test for peptidase activity of recombinant PEX,
cell membrane preparations from vector-transfected COS
cells or COS cells expressing recombinant PEX protein wer.e-
incubated with human parathyroid hormone PTH (1-34) and
PTH (1-38) . As shown, PEX activity was able to degrade
both peptides in a very characteristic pattern. Therefore,
PEX functions as an endopeptidase, and more specifically
we have shown for the first time that it degrades PTH. PTH
is the first and only known substrate of PEX.
These observations make two important points:
PEX is a membrane bound protein with its active
enzymatic site in the extracellular compartment. The cells
with the highest level of PEX expression are the osteo-
blasts (bone forming cells). These cells are also the site
of action of circulating PTH at the level of the bone. PTH
stimulates these cells to produce factors (nature unknown)
which in turn stimulate other bone cells, specifically the
osteoclasts, to break down bone. Since PEX likely inacti-
vates PTH in contact with osteoblasts, it would result in
decreased stimulation of osteoclasts and therefore less
bone breakdown.
Alternatively, osteoblasts produce parathyroid
hormone-related peptide, PTHrP, which is important in the
development of normal bone density. PTHrP shares many of
the structural features of PTH and may therefore also
serve as substrate for PEX. Our previous studies using
PTHrP heterozygous-null mice generated by gene targeting
have shown that decreased levels of PTHrP in the skeletal
microenvironment lead to a premature form of osteoporosis.
PEX in osteoblasts may therefore modulate local PTHrP
levels and thus bone formation. Inhibition of PEX enzy-
matic activity may allow higher local concentrations of
PTHrP and therefore better bone formation.
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 9 -
By examining PTH breakdown fragments, we can now
design peptide and non-peptide activators and inhibitors
of PEX enzymatic activity.
By modulating PTH and PTHrP levels that regulate
osteoblast activity, PEX may play a critical role in thp,
pathogenesis of osteomalacia and osteoporosis. By pharma-
cological modulation of PEX activity, it will be possible
to modulate bone breakdown and bone formation. This would
be a totally novel approach to the treatment of these
metabolic bone diseases.
Experimental procedure
Tumor Tissues
Patient I was a 55 year-olid woman who presented
with a two-year history of progressively increasing
bone pain and difficulty in walking. X-rays of the lum-
bosacral spine showed diffuse osteopenia. Biochemical
investigation showed the serum calcium level to be nor-
mal while serum phosphorus was low (0.41 to 0.57
mmol/L; normal, 0.8-1.6 mmol/L). Alkaline phosphatase
was 232 U/L (normal, 30-105 U/L) and tubular reabsorp-
tion of phosphate while the patient was hypophospha-
temic was decreased to 63% (normal, >80%). A search for
a tumor was negative and the patient was treated with
1,25-dihydroxyvitaminD3 and oral phosphate. Five years
later a right hand mass was discovered and was surgi-
cally removed. On histopathological examination, it was
a fibrous hemangioma. Postoperatively, the patient
noted increasing strength in her lower extremities and
marked decrease in her pain. The serum phosphorus nor-
malized (0.96 mmol/L) and the t'ubular reabsorption of
phosphate improved but did not completely normalize
(71-76%). No recurrence of the tumor has been found ten
years later.
Patient II was a 21 year old man with classic
features of OHO. Resection of a benign extraskeletal
CA 02343713 2001-03-23
Ki - -==-'"' "UENCHEN 05 : 8- 9- 0: 14:56 ;514 288 8389 +49 89 : CA 009900895
OvQ-OfI-nOOC..VVV V.JT114 u1t/1LLl VV1411 lllll. JlY LVV VJVJ ll=J.
~l G JJJJ
- 10 -
cho ndror.tia from the plantar surface of the foot resulted
in complete reversal of the biochemical and clinical
abnormalities associated with the syndrome.
Tumor tissue obtained from these two patients at
surgery was frozen immediately in liquid nitrogen and
stored at -700C. -
PEX Etcpress3att d-n O80 alaaociatod Twnars
RNA was extracted from tumor tissue using the
RNeasy''m Total RNA kit (Qiagen, Chat,sworth, CA) and
reverse transcribed using oligo(dT) primer and Super-
script zI (BRL) reverse transcriptase for 1 hour at
42 C in a=final reaction volume of 30 }il. The resulting
cDNA was then amplified using human PEX-specific oligo-
nucleotide primers PEX-1 (5'-GGAGGAATTGGTTGAGGGCG -3'
SEQ ID N0:5) and PF'X-2 (5'-GTAGACCACCAAGGATCCAG -3' SEQ
ID NO:6), designed from the published cDNA sequence
(1298 and 1807 are the nucleotide positions of the 5'
end of the sense and antisense primers, respectively)
(The HYP Consortium (1995) Nature Genetics 11, 130-
136). Following amplification (35 cycles), an aliquot
of the PCR reaction was fractionated on an 1% agarose
gel and visualized following staining with ethidium
bromide.
Cloniag af FttI1-Length PEX cmUl
Cloning of the 5' end of pEx cDNA was accotn-
plished by anchored PCR. Total cellul.ar RNA was
extracted f rom tumor II and mRNA was prepared. 1. 5 Ug
of mRNA was reverse transcribed into cDNA using 100 ng
of a PEX-specific antisense oligomer (PEX-2) and 200
units of Superscript YI (BRL) reverse transcriptase for
1 hour at 420C in a final reaction volume of 30 ul. The
resulting cDNA was size fractionated on a 1ik agarose
gel and fragments =corresponding to >600 bp were puri-
fied and resus?oended in H20. The 3' end of the first
strand cDNA was homopol.yrner tailed with dGTP using 1 ul
AMENDED SHEET
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 11 -
of Terminal deoxynucleotidyl transferase (TdT) at 37 C
for 30 minutes in a volume of 50 ul. Following heat
inactivation of the enzyme, the RNA template was
removed by incubation with RNase H and the tailed cDNA
5. was purified by phenol-chloroform extraction followed
by ammonium acetate precipitation. The purified tailed
cDNA was resuspended in H20 and an aliquot was used for
anchored PCR analysis along with 200 ng of an internal
PEX specific antisense primer (PEX-3, 5'-
CGTGCCCAGAACTAGGGTGCCACC-3' (SEQ ID NO:7); nucleotide
98 of the published human cDNA sequence is the 5' end
of the primer) and 200 ng of oligodC as the sense
primer. Forty cycles of PCR were performed using 0.5 p1
of Taq polymerase (Promega Biotec, Madison, WI) in a
reaction volume of 50 }zl. Cycling parameters were: 1
minute of denaturation at 94 C, 2 minutes of annealing
at 55 C and 2 minutes of extension at 72 C. The PCR
products were fractionated on a 1% agarose gel and a
band of 700 bp was isolated, purified, and ligated into
pPCRII vector (Invitrogen). Following transformation
into INVaF' bacteria, clones containing the appropriate
size insert were sequenced.
To clone the 3' end of PEX cDNA, an aliquot of
an amplified unidirectional cDNA library in pCDNA3 vec-
tor (Invitrogen) generated from mRNA obtained from
tumor I was grown overnight in LB medium and plasmid
DNA extracted. DNA (0.5 pg) was subjected to PCR using
a PEX-specific sense oligomer (PEX-1) and an antisense
oligomer corresponding to the SP6 RNA polymerase bind-
ing site sequences present in the pCDNA3 vector.
Thirty-five cycles of amplification were performed in a
50 ul reaction volume with each cycle consisting of 1
min denaturation at 94 C, 1 min annealing at 55 C and 1
min extension at 72 C. Amplified products were frac-
tionated on a 1% agarose gel and a 1.2 kb fragment cor-
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 12 -
responding to the 3' end of PEX cDNA was subcloned and
sequenced.
For expression studies, an EcoRV (in the
polylinker of pPCRII) /AccI (in the PEX sequence) frag-
ment containing the 5' end of PEX cDNA was ligated into
the pPCRII vector containing the 3' end of PEX cDNA
following digestion with AccI and EcoRV. The resulting
plasmid was restricted with KpnI and NotI excising the
full length PEX cDNA that was then inserted into pCDNA3
vector digested at the KpnI/NotI sites in the
polylinker region, resulting in plasmid pPEX. The full-
length PEX cDNA was sequenced using an Applied Biosys-
tems 373A automated sequencer.
Tissue Expression of PEX mRNA
PEX expression was examined in normal human tis-
sues and in the Saos-2 human osteoblastic osteosarcoma
cell line, by RT-PCR using oligonucleotides PEX-4 (5'-
CTGGAT-CCTTGGTGGTCTAC-3' SEQ ID NO:8) and PEX-5 (5'-
CACTGTGCAACTGTCTCAG-3' SEQ ID NO:9) as sense and
antisense primers (2398 and 2895 are the nucleotide
positions of the 5' end of these primers designed from
the full-length human PEX cDNA) . Semiquantitative PCR
analysis for PEX expression in human tissues was
performed as previously described, following
normalization for GAPDH message in all samples
containing PEX transcripts.
Northern-blot Analysis
Total RNA was obtained from Tumor I and human
Saos-2 osteosarcoma cells using the RNeasy Total RNA
kit (Qiagen) and oligo(dT)-purified poly(A)+ RNA was
isolated from Saos-2 total RNAusing standard proce-
dures. Twenty micrograms of Tumor I total RNA and 20 pg
of Saos-2 poly(A)+ RNA were fractionated on 1% denatur-
ing agarose gel, and transferred to nylon membrane
(Hybond N+, Amersham). Hybridization was performed with
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 13 -
32P-labeled full-length human PEX cDNA (3.1 kb) in 7 mM
Tris-HC1, 50% formamide, 10% dextran sulfate, 4 X SSC,
2 x Denhardt's solution and heat-denatured salmon sperm
DNA (100 ug/ml). The blot was washed in 0.1 X SSC, 0.1%
SDS for 20 min at 50 C, and subjected to autoradiogra-
phy
for 4 days.
In Vitro Transcription, Translation, and Analysis of
Products
Plasmid pPEX was linearized with NotI and sense
RNA strand was transcribed using T7 RNA polymerase.
Translation reactions in rabbit reticulocyte lysate
were performed in the presence of [3H]leucine according
to the manufacturer's recommendations (Promega) with or
without canine pancreas microsomal membranes. Products
were analyzed by SDS-polyacrylamide gel electrophoresis
(SDS-PAGE; 80). Autoradiography was performed after
treating the gel with EN3HANCE (Dupont NEN), as previ-
ously described.
Generation of myc-tagged PEX, Transfection in COS-7
Cells, and Triton X-114 Extraction
Plasmid pPEX-myc was generated by PCR amplifica-
tion of PEX cDNA using oligonucleotide PEXMycl as the
sense primer (5'-TTGGATGTCAACGCCTCG -3' SEQ ID NO:10,
519 is the nucleotide position of the 5' end of this
primer designed from the cloned human PEX cDNA) and
PEXMyc2 as the antisense (5'-CTACCACAATCTACAGTTGTT-
CAGGTCCTCTTCGCTAATCAGCTTTTGTTCCATAGAGTCCATGCCTCTG-3'
SEQ ID NO:11) primer. The latter encodes the human c-
myc tag sequences (underlined) and PEX sequences
corresponding to the carboxyl terminal of the mature
protein (742RGMDSMEQKLISEEDLNNCRLW*). Following PCR,
the amplified fragment was ligated to the pPCR II
vector, excised by digestion with KpnI/NotI and
inserted into the corresponding sites in the polylinker
region of pCDNA3. The in-frame fusion protein was
verified by DNA sequencing.
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 14 -
COS-7 cells maintained in Dulbecco's modified
Eagle's medium (DMEM, 4,500 mg/L glucose with L-gluta-
mine; JRH Biosciences, Lenexa, KS) supplemented with
10% fetal calf serum (FCS; GIBCO) and antibiotics
(pen/strep) were plated at a density of 3 x 105
cells/well in 6-well cluster plates 24 h prior to
transfection. Cells were washed with twice with PBS and
incubated with 2}zg of pPEX-myc plasmid DNA in 1 ml of
DMEM containing 0.1% BSA, and DEAE-dextran (Pharmacia
LKB) for 3.5 h at 37 C. Following incubation, the
transfection medium was aspirated, the cells were
shocked with 10% DMSO in PBS for 2 min, and then cul-
tured in DMEM with 10% calf serum at 37 C for 48 h.
Triton X-114 extraction were performed on cultured
cells expressing myc-tagged PEX as described. The sam-
ples were then analyzed by immunoblotting using the
9E10 anti-myc monoclonal antibody.
Stable Transfection of A293 Cells and Immunofluores-
cence
A293 cells maintained in DMEM with 10% FCS were
transfected with the pPEX-myc plasmid by electropora-
tion and selection initiated using G418 (600 mg/ml for
14 days and then decreased to 400 mg/ml). Populations
of stably transfected cells were recovered at the end
of the selection period. For myc-tagged PEX indirect
immunofluorescence, stably transfected cells plated on
gelatin-coated coverslips were washed twice with PBS,
fixed in 4% parafolmaldehyde and in some experiments
permeabilized with 0.5% Triton X-100. Cells were
blocked with 10% FCS in DMEM for 30 min, washed and
incubated for 1 hr at 37 C with the 9E10 anti-myc mono-
clonal antibody (1:500 dilution). Cells were subse-
quently washed and incubated in turn with fluorescein-
conjugated sheep anti-mouse secondary antibody (1:250
dilution). Coverslips were rinsed extensively with PBS,
mounted in medium (glycerol:Tris; 1:1) containing 2.5%
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 15 -
1,4-diazabicyclo-(2,2,2) octane (Sigma) and examined
with fluorescent microscopy using appropriate filters.
Assay for membrane-bound endopeptidase activity
COS-7 cells transiently transfected with pCDNA3
vector alone, with vector containing human NEP cDNA
(generous gift of P. Crine, Universite de Montreal), or
with pPEX plasmid, were washed and scraped in PBS. Fol-
lowing brief centrifugation, the cell pellets were
resuspended in 50 mM Tris-HC1, pH 7.4 and disrupted by
sonication. Homogenates were fractionated by sequential
centrifugation at 1,000 x g for 10 min and then at
100,000 x g for 60 min. The final precipitate was
washed with 50 mM Tris-HC1, pH 7.4, resuspended in the
same buffer, and assayed for endopeptidase activity.
The protein concentration in membrane fractions was
determined by the method of Bradford with bovine serum
albumin as standard.
[D-Ala2,Leu5] enkephalin (500 M) was incubated
with COS cell membrane preparations (-60 pg of protein)
in 100 mM Tris-HC1, pH 7.0, at 37 C for 30 min (final
volume 30 ul). The reaction was terminated by the addi-
tion of 100 ul 0.1% TFA (v/v). Production of Tyr-D-Ala-
Gly was monitored using reversed-phase HPLC (Bondpak C-
18 reverse phase column, Waters) with a U.V. detector
set at 214 nm. A linear solvent gradient of 0% B to 40%
B in 60 min was used with a flow rate of 1.5 ml/min
(mobile phase A=0.1% TFA (v/v); mobile phase B=80% ace-
tonitrile/0.1% TFA). Tyr-D-Ala-Gly was identified by
co-chromatography with marker synthetic peptide. For
assessing PEX endopeptidase activity, 10 g of PTH [1-
38] and PTH [1-34] peptides (Peninsula Laboratories;
Belmont, CA) were added to the membrane preparations.
For HPLC analysis of hydrolysis products, a linear sol-
vent gradient of 0% to 50% solution B was used at a
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 16 -
rate of 1.5 ml/min. MALDI-TOF mass spectrometry was
performed on specific peptide fragments.
RESULTS
Cloning of Human PEX cDNA
At the initiation of these studies, PEX expres- _
sion had been reported in minute amounts only in leuko-
cytes and fetal brain. We postulated that in states of
hypophosphatemia PEX expression may be increased and
therefore opted to use the OHO tumor as a tissue source
that may express considerably more PEX. Tissues
obtained from two tumors associated with OHO were used
to obtain total RNA and analysis for PEX mRNA expres-
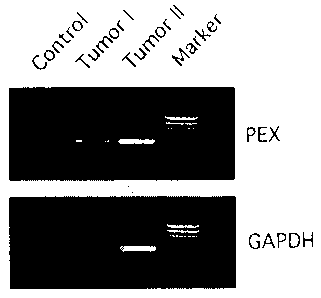
sion was assessed by RT-PCR. As shown in Fig.1, PEX
transcripts were readily amplified from both tumor sam-
ples demonstrating the expected 509 bp fragment pre-
dicted from the published partial human PEX sequence
(The HYP Consortium (1995) Nature Genetics 11, 130-
136) . Total RNA extracted from two tumors associated
with OHO was reverse transcribed and amplified by PCR
(35 cycles) using human PEX-specific primers, PEX-1 and
PEX-2, designed from the published human sequence. The
expected 509 bp amplified fragment was obtained from
both tumor samples. Control, no cDNA added to the
amplification reaction, i.e. negative control; Marker,
0174 DNA digested with HaeIII restriction endonuclease.
The cloning of the 3' end of PEX transcript was
performed by rapid amplification of the 3' end of the
cDNA (3' RACE), while the 5' of the cDNA was amplified
by anchored PCR, as described in Experimental Proce-
dures. Fig. 2A shows the nucleotide and predicted amino
acid sequence of the full-length human PEX cDNA cloned
from tumor tissues. Nucleotide and deduced amino acid
sequence of tumor-derived human PEX cDNA (Fig. 2A). The
numbering begins at the 5' end nucleotide as determined
by anchored PCR. Amino acids are given below each codon
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 17 -
using the single letter code. The putative start codon
is indicated as /1 along with the deduced amino acid
translation. Two stop codons preceding the predicted
initiation ATG are in bold type. Asterisk (*) indicates
an in-frame stop codon, while a large asterisk (*) _
denotes the putative prenylation site. A potential
polyadenylation signal in the 3' untranslated region is
underlined. Nine potential N-glycosylation sites are
boxed. The sequence has been assigned GenBank accession
No. (U82970).
The composite cDNA reveals a single open reading
frame encoding a protein of 749 amino acids which dis-
plays homology (34.2% identity, 70% similarity) to
human neprilysin (NEP; EC 3.4.24.11), and other members
of the membrane-bound metalloendopeptidase family
encompassing endothelin-converting enzyme-1 (ECE-1; 66%
similarity) and the Kell antigen (60% similarity), sug-
gesting that PEX is a novel member of this family of
neutral endopeptidases, as previously suggested (The
HYP Consortium (1995) Nature Genetics 11, 130-136).
Like the other members, PEX is a likely a glycoprotein
with eight potential N-glycosylation sites and 10 cys-
teine residues that may be important for the proper
folding and hence native conformation of the protein.
The ATG codon at position 604 was assigned as
the initiator methionine since it is preceded by two
in-frame TGA termination codons 36 and 63 basepairs
upstream and conforms favorably to the Kozak consensus
for vertebrate initiation of translation. The cloned
cDNA identifies the first 3 and the last 108 amino
acids of the predicted PEX gene product in addition to
the published partial sequence. These additional amino
acids comprise residues such as E642 and H710 that are
shared by NEP, and may be critical for the formation of
the active site of the protein and hence its enzymatic
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 18 -
activity. Three amino acid residues predicted from our
cDNA clone differ from the published partial human PEX
sequence, D363A (GAC to GCC), R403W (AGG to TGG), and
A641G (GCG to GGA) . To confirm that these alterations
did not arise because of PCR errors, PEX sequences were _
amplified from Saos-2 human osteosarcoma cells (see
below) and sequenced. In addition, the same alterations
were subsequently described in the murine PEX cDNA,
suggesting possible cloning artifacts in the published
partial human PEX sequence. Our cloned sequences also
encompass 603 nucleotides of the 5' untranslated
region, and 276 nucleotides of the 3' untranslated
region, including the canonical polyadenylation signal
AATAAA, 19 nt upstream of the poly(A) tract. The human
and the published mouse PEX cDNA sequences share exten-
sive homology within the protein coding region (96%
identity) as well as in the 5' and 3' non coding
regions.
TMpred analysis of the human PEX sequence pre-
dicts that the protein has no apparent N-terminal sig-
nal sequence but has a single membrane-spanning helical
domain comprising amino acid residues 21-39 (Fig. 2C).
TMpred analysis of the PEX sequence showing a single
membrane-spanning domain encompassing amino acid resi-
dues 21-39 (arrowhead). Numbers on the horizontal axis
refer to the amino acid sequence. Amino acid homology
between PEX and human NEP cDNA (Fig. 2B). Sequence com-
parison was performed using the LALIGN program.
This predicts its transmembrane topology to be
that of a type II integral membrane protein with a 20-
residue N-terminal cytoplasmic tail and a C-terminal of
700 amino acid residues containing the catalytic domain
in the extracellular compartment. Unexpectantly, a CXXX
box motif comprising amino acid residues 746CRLW was
also identified at the carboxyl terminus of PEX. This
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 19 -
motif may serve as a site for prenylation, a post-
translational lipid modification involved in a number
of processes including facilitating membrane attach-
ment, targeting of proteins to specific subcellular
membrane compartments, promoting protein-protein inter-
actions
and regulating protein function.
Tissue Expression of PEX mRNA
We next examined PEX expression in a number of
fetal and adult tissues and compared the level of
expression to OHO tumor RNA using semi-quantitative RT-
PCR (Fig. 3). Quantitative RT-PCR amplification of the
PEX transcripts from total RNA prepared from human tis-
sues and OHO-associated tumor. Relative expression lev-
els for the PEX transcript were measured by quantifying
PEX product in reversed-transcribed RNA samples that
have been previously normalized for GAPDH levels. The
specific primers used were as follows: for PEX, the
forward primer was PEX-4 and the reverse primer PEX-5;
for GAPDH, the primers were as previously described.
PCR products were electrophoresed on a 1.5% agarose gel
and stained with ethidium bromide. Control, negative
control; Marker, 0174 DNA digested with HaeIII restric-
tion endonuclease. Below, shown are the relative levels
of PEX transcripts in various human tissues compared to
those in the tumor.
PEX transcripts were expressed in human fetal
calvarium and to a lesser degree in fetal kidney and
skeletal muscle while no expression was apparent in
fetal liver. PEX expression was also observed in the
human osteoblastic osteosarcoma cell line, Saos-2. In
adult tissues, PEX mRNA was identified in kidney, but
not in liver, or endomyocardium. Recent studies have
also reported PEX expression in human fetal bone,
skeletal muscle, and liver as well as fetal and adult
ovary and lung (Beck, L. et al. (1997) J. Clin. Invest.
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99100895
- 20 -
99, 1200-1209; Grieff, M. et al. (1997) Biochem. Bio-
phys. Res. Commun. 231, 635-639). Analysis following
normalization for GAPDH message in all tissues contain-
ing PEX transcript disclosed that bone PEX expression
5. is 2-10 fold higher than in other normal tissues exam-
ined.
In comparison, OHO tumor PEX expression was twice
the levels observed in fetal calvarium, consistent with
its relative "overabundance" in these tissues.
Northern Blot Analysis
To determine the size of the full-length PEX
transcript, we isolated total RNA from tumor I (quan-
tity of available tissue was insufficient for poly(A)+
RNA extraction) and poly(A)+ RNA from human Saos-2
osteosarcoma cells. This cell line was used since it is
readily available and successful amplification of PEX
sequences has been performed by RT-PCR (see above).
Aliquots (20 pg of each) were examined by Northern-blot
analysis using the cloned human PEX cDNA as probe. A
single transcript of approximately 6.5 kb was readily
detected only in the Saos-2-derived poly(A)+ sample and
contrasts with the predicted size of the cloned
sequence of 3.1 kb (Fig. 4). Approximately 20 pg of
poly(A+)RNA prepared from Saos-2 cells and 20 pg of
total RNA prepared from tumor I tissue were resolved on
1% agarose gel containing formaldehyde and then trans-
ferred to a nylon membrane. Following hybridization
with radiolabeled PEX cDNA, the blot was washed and the
signal detected by autoradiography. A transcript of
-6.5 kb was observed only in the lane containing Saos-2
poly(A+)RNA. There is suggestion of an additional band
corresponding to a transcript of '3.8 kb. Arrows indi-
cate the position of the 28S (approx. 4.8 kb) and 18S
(approx. 1.8 kb) ribosomal RNA.
This finding would therefore predict a -4 kb 5'
untranslated region for PEX cDNA, consistent with pub-
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 21 -
lished data from Northern blot analysis of PEX expres-
sion in mouse calvaria (Du, L. et al. (1996) Genomics
36, 22-28). A less well defined band was also detected
in the Saos-2 sample corresponding to a potential tran-
5. script of -3.8 kb, although the nature of this tran-
script
remains unclear. Northern analysis of total RNA
samples from tumor I and Saos-2 cells (results not
shown) did not reveal any signal for PEX, consistent
with the relatively low expression levels of the PEX
transcript, previously described (The HYP Consortium
(1995) Nature Genetics 11, 130-136; Beck, L. et al.
(1997) J. Clin. Invest. 99, 1200-1209; Grieff, M. et
al. (1997) Biochem. Biophys. Res. Commun. 231, 635-
639) . This finding contrasts sharply with PEX expres-
sion levels demonstrated in murine calvaria and cul-
tured osteoblasts (Du, L. et al. (1996) Genomics 36,
22-28) and may reflect tissue and species differences.
in vitro translation of PEX cRNA
In vitro translation studies using full-length
human PEX cRNA were performed in the rabbit reticulo-
cyte lysate cell-free system. In the absence of micro-
somal membranes, PEX cRNA was translated into an -86 kD
protein, as predicted from the cloned cDNA sequence
(Fig. 5) Plasmid pPEX was linearized and sense RNA
strand transcribed using T7 RNA polymerase. Translation
of PEX cRNA was performed using rabbit reticulocyte
lysate in the absence (minus) and presence (plus) of
canine pancreas rough microsomes. Products were elec-
trophoresed in a SDS-polyacrylamide gel (10%) and visu-
alized by autoradiography. Arrowhead in lane 2 indi-
cates full-length human PEX protein. The addition of
microsomal membranes results in the appearance of
higher molecular weight forms that likely represent
glycosylated products.
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 22 -
Following addition of canine microsomal mem-
branes to the translation mixture, products of higher
molecular weight (-100 kD) became apparent, consistent
with N-glycosylation of PEX at the eight potential gly-
cosylation sites deduced from the predicted sequence.
PEX is a Cell Membrane-Associated Protein
Previous studies have established that NEP, ECE-
1 and Kell blood group glycoprotein are integral mem-
brane proteins. We have used extraction with the deter-
gent Triton X-114 and immunofluorescent localization to
examine whether PEX is also a membrane-associated pro-
tein. For identification of PEX, we generated a con-
struct in which the carboxyl terminus sequences of PEX
are modified by a human c-myc tag. The epitope tag was
inserted immediately upstream of the putative prenyla-
tion motif so that any potential lipid modification of
the PEX protein may proceed uninterrupted.
Triton X-114 is a detergent that forms an aque-
ous solution at 4 C but separates into hydrophobic and
aqueous phases when the temperature is raised to 30-
37 C. This property has been used as an indicator of
the hydrophobic nature of proteins, with integral mem-
brane proteins partitioning exclusively in the deter-
gent phase while highly hydrophilic proteins associate
with the aqueous phase. Triton X-114 extracts from COS-
7 cells transiently expressing PEX tagged with the c-
myc epitope showed that PEX partitions nearly exclu-
sively into the detergent phase (Fig. 6A) . Extraction
and partitioning of PEX expressed in COS-7 cells with
Triton X-114 (Fig. 6A) Plasmid pPEX-myc was tran-
siently transfected in COS-7 cells and 48 h later cells
were extracted with Triton X-114. Whole cell extracts,
as well as detergent and aqueous phases, were analyzed
by SDS-PAGE and immunoblotted with an anti-myc mono-
clonal antibody. Right margin indicates Mr x 10-3.
CA 02343713 2001-03-23
WO 00/18954 - 23 PCT/CA99/00895
-
This finding indicates that PEX is a membrane-
associated protein and is consistent with the predic-
tion from sequence analysis that it is an integral mem-
brane protein.
To determine the subcellular localization of
PEX, the distribution of recombinant protein expressed
in stably transfected A293 cells was examined using
immunofluorescence. When cells were fixed and permeabi-
lized, myc-tagged PEX immunostaining was detected pri-
marily on the cell surface, but in a number of cells
staining was also observed intracellularly, although no
signal was observed in the nucleus (Fig. 6B). If perme-
abilization was omitted, staining was localized exclu-
sively to the plasma membrane (Fig. 6C), while untrans-
fected cells or cells transfected with vector alone
showed no immunofluorescent staining. Localization of
PEX using indirect immunofluorescence in stably trans-
fected A293 cells with (Fig. 6B) and without (Fig. 6C)
permeabilization with Triton X-100, respectively.
Staining was carried out using the 9E10 anti-myc mono-
clonal antibody, followed by fluorescein-labeled secon-
dary (sheep anti-mouse) antibody. Arrowheads indicate
intracellular (B) and plasma membrane staining (C).
Since the myc-tag was inserted in the carboxyl
end of PEX, these findings further corroborate the
sequence-based prediction that PEX is a type II inte-
gral membrane protein with its large C-terminal hydro-
philic domain in the extracellular compartment.
Recombinant PEX protein has endopeptidase activity
The subcellular localization and sequence simi-
larity between PEX and NEP strongly suggest that PEX
functions as a membrane-bound metallopeptidase. How-
ever, no peptidase activity has been ascribed to PEX.
As shown in Fig. 7A, when [D-Ala2, Leu5] enkephalin,
used to assay for NEP activity, was incubated with cell
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 24 -
membrane preparations from vector-transfected COS cells
or COS cells expressing equivalent amounts of recombi-
nant human NEP or PEX proteins, as determined by West-
ern blot analysis, production of Tyr-D-Ala-Gly from the
5. substrate was evident only in NEP-expressing membrane
preparations. Cell membrane preparations from vector
transfected COS-7 cells (Fig. 7A) or from cells tran-
siently expressing human NEP (Fig. 7B) or, human PEX
cDNAs (Fig. 7C) were incubated in the presence of [D-
Alaz,Leu5]enkephalin (500 M) and hydrolysis products
were resolved by HPLC as described in the Experimental
Procedures section. Tyr-D-Ala-Gly was identified by
chromatography of synthetic marker peptide.
While the PEX sequence preserves two of the
residues critical for catalytic activity of NEP
(equivalent to E646 and H711), it lacks a residue
equivalent to R102 shown to be crucial for the dipepti-
dylcarboxypeptidase activity of NEP. Therefore, unlike
NEP, PEX has no dipeptidylcarboxypeptidase activity,
but likely functions as an endopeptidase.
To examine recombinant human PEX for endopepti-
dase activity, cell membrane preparations from COS
cells transiently expressing the protein were incubated
with human PTH [1-38] or PTH [1-34] and the cleavage
products were analyzed by reverse-phase high pressure
liquid chromatography (HPLC), as shown in Fig. 8. Human
PTH [1-38] was incubated with cell membrane prepara-
tions from vector transfected COS-7 cells (Fig. 8A) or
from cells transiently expressing human PEX and
hydrolysis products were resolved by HPLC (Fig. 8B).
Chromatographic profile of products arising from the
hydrolysis of PTH [1-34] when incubated with cell mem-
branes from COS-7 cells transiently expressing PEX
(Fig. 8C). The novel product with a molecular weight of
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 25 -
630 likely corresponds to the terminal pentapeptide
DVHNF of human PTH [1-34].
A parallel preparation from vector transfected
COS cells did not appreciably cleave PTH [1-38]. How-
ever, in the presence of PEX, both PTH peptides were
hydrolyzed in a highly reproducible pattern resulting
in the formation of several peaks that absorb at 214
nm. Mass spectrometry of the peptide materials recov-
ered from two product peaks gave m/z values of 861 and
630, respectively. While the former product was present
in hydrolysates from both PTH [1-38] and PTH [1-34],
the latter product was identified only in the PTH [1-
34] hydrolysate and likely corresponds to the carboxyl
terminal pentapeptide DVHNF of human PTH [1-34]. These
findings provide the first direct evidence that recom-
binant PEX possesses endopeptidase activity and suggest
that its substrate specificity may not be restricted to
the putative phosphatonin but may include other circu-
lating hormones or perhaps bone-derived auto-
crine/paracrine regulatory factors that regulate renal
phosphate handling.
DISCUSSION
To gain insight into the role of PEX in normal
physiology we have cloned the human full-length cDNA
and studied its expression, subcellular localization,
and peptidase activity. The cloned human PEX cDNA
encodes a protein whose deduced amino acid sequence is
identical to the published partial (The HYP Consortium
(1995) Nature Genetics 11, 130-136) and to the full-
length sequences reported more recently (Beck, L. et
al. (1997) J. Clin. Invest. 99, 1200-1209; Grieff, M.
et al. (1997) Biochem. Biophys. Res. Commun. 231, 635-
639; Guo, R. and Quarles, L. D. (1997) J. Bone Miner.
Res. 12, 1009-1017) . Its deduced topology is that of a
type II integral membrane glycoprotein and in the pres-
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 26 -
ent study we have provided experimental evidence to
support this prediction. We have shown that PEX is gly-
cosylated in the presence of canine microsomal mem-
branes and partitions exclusively in the detergent
phase following extraction with Triton X-114, consis-
tent
with the prediction from sequence analysis that it
is an integral membrane glycoprotein. Nevertheless, the
observed hydrophobic nature of PEX, need not be attrib-
uted solely to it being an integral membrane protein.
Lipophilic modification is known to cause cell membrane
association, presumably through hydrophobic interaction
of the modifying group with the lipid bilayer. Signaled
by the C-terminal tetrapeptide CRLW motif, post-trans-
lational attachment of isoprenoids via a thioether
linkage to the cysteine residue would be sufficient to
promote effective membrane association. Further studies
will be necessary to determine if such lipid modifica-
tion of PEX does indeed take place. Of interest, how-
ever, is the observation that a nonsense mutation
within this motif (R747Stop) has been reported to co-
segregate with HYP and is likely to be associated with
an inactive PEX gene product. Finally, the localiza-
tion of PEX expressed in A293 cells is also consistent
with the protein being membrane-associated and corrobo-
rates the sequence-based prediction that PEX is a type
II integral membrane protein with its large C-terminal
hydrophilic domain in the extracellular compartment.
While protein expression was detected mostly on the
cell surface, in some cells the signal was also local-
ized intracellularly. This localization of the
expressed protein would indicate that a portion of PEX
activity is located in a membrane-bound compartment,
possibly the Golgi membranes. The Golgi localization
described for ECE-1 activity in cultured endothelial
cells is proposed to promote the efficient conversion
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 27 -
of big endothelin-1 because of the co-localization and
concentration of enzyme and substrate through the con-
stitutive secretory pathway. It is possible then, that
in parallel fashion, the PEX enzyme mediates both
intracellular and cell-surface conversions of its puta- _
tive substrate.
The finding that wild-type PEX transcripts are
expressed in relative overabundance in OHO tumors poses
a question in trying to understand the pathophysiology
of these disorders. That is, how do we reconcile the
apparently disparate observations that overexpression
of PEX in OHO and loss of function in HYP patients,
both lead to similar derangement in phosphate homeosta-
sis? One of the physiological functions of PEX may well
be the inactivation of a factor that normally promotes
renal phosphate excretion (Fig. 9). The diagrams indi-
cate events proposed to occur at the level of the
proximal renal tubule. A putative circulating phospha-
turic hormone (PHa) interacts with its renal receptor
(PR) and inhibits phosphate reabsorption across the
renal brush border membrane (-I) by decreasing NaPi
activity. Downward arrows indicate the degree of phos-
phate excretion. PEX expressed predominantly in extra-
renal tissues modulates the levels of circulating PHa
by converting it to its inactive form (PHi).
In patients with OHO, the hyperphosphaturia that
characterizes the syndrome would be the consequence of
unregulated and excessive elaboration of the phosphatu-
ric factor by the tumor. The modestly elevated PEX lev-
els that we have documented in these tumors may arise
either in response to the severe hypophosphatemia or to
the abnormally high levels of the active phosphaturic
factor. Yet, the increased PEX expression may not be
sufficient to accommodate the increased substrate load,
resulting in abnormally high circulating levels of the
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 28 -
active phosphaturic hormone. The inactivation of PEX
observed in HYP patients would similarly cause
decreased turnover of this humoral phosphaturic factor
and thereby lead to renal phosphate wasting.
This model is also consistent with the observa- -
tion that the Hyp phenotype is neither corrected nor
transferred following cross transplantation of kidneys
in normal and Hyp mice. Thus, when Hyp mice are
engrafted with a normal kidney, phosphaturia ensues
since circulating levels of the phosphaturic agent are
excessive. On the other hand, engraftment of mutant
kidneys in normal mice will not affect renal tubular
phosphate handling of the recipients since circulating
levels of the phosphaturic substance will be normally
regulated by the enzymatic activity of extrarenal wild-
type PEX. Indeed, analysis of the tissue distribution
of PEX mRNA by RT-PCR has confirmed its expression in
extrarenal tissues and particularly bone. Our present
findings and those of others (Du, L. et al. (1996)
Genomics 36, 22-28; Beck, L. et al. (1997) J. Clin.
Invest. 99, 1200-1209; Grieff, M. et al. (1997) Bio-
chem. Biophys. Res. Commun. 231, 635-639; Guo, R. and
Quarles, L. D. (1997) J. Bone Miner. Res. 12, 1009-
1017) showing high levels of PEX expression in cells of
the osteoblast lineage would be consistent with the
intrinsic osteoblast defect postulated to exist in HYP
patients and in Hyp mice.
Finally, although the deduced structure of PEX
clearly suggests that it is a metalloprotease, no pep-
tidase activity had been ascribed to the protein. The
preservation of the catalytic glutamate and histidine
residues (equivalent to E646 and H711 of NEP; Fig. 2B)
would argue for such an activity. In addition, the wide
range of PEX mutations in HYP patients that align with
regions required for protease activity in NEP suggests
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 29 -
that PEX also functions as a protease. Here, for the
first time, we provide experimental evidence that
recombinant PEX indeed functions as an endopeptidase.
Unlike NEP, however, the protein does not possess
dipeptidylcarboxypeptidase activity since it lacks a
residue equivalent to R102 of NEP. Our unexpected obser-
vation that PEX effectively degrades PTH raises the
question of whether circulating PTH is the putative
phosphatonin. Although extracts from some OHO tumors
have been reported to stimulate renal adenylate cyclase
and this activity was inhibited by PTH antagonists,
most studies have excluded PTH and PTH-related peptide
(PTHrP) activity in OHO-associated tumors. Moreover,
calcium homeostasis is generally preserved in patients
with HYP. It is more likely, therefore, that the enzyme
is rather promiscuous in its substrate specificity. PEX
may indeed modulate PTH bioavailability and bioactiv-
ity, particularly at the level of the osteoblast, as
well as the hormonal and paracrine/autocrine effects of
factors produced by osteoblasts involved in regulating
phosphate reabsorption and osteoblast maturation and
mineralization. Although additional work will be
required to clarify many of these issues, the avail-
ability of full-length human PEX cDNA now provides us
with the opportunity to study the biology of PEX, iden-
tify its substrate(s), elucidate its role in pathologi-
cal states characterized by dysregulated phosphate
homeostasis, and determine its suitability as target
for therapeutic intervention in the treatment of meta-
bolic bone diseases.
While the invention has been described in con-
nection with specific embodiments thereof, it will be
understood that it is capable of further modifications
and this application is intended to cover any varia-
tions, uses, or adaptations of the invention following,
CA 02343713 2001-03-23
WO 00/18954 PCT/CA99/00895
- 30 -
in general, the principles of the invention and
including such departures from the present disclosure
as come within known or customary practice within the
art to which the invention pertains and as may be
applied to the essential features hereinbefore set
forth, and as follows in the scope of the appended
claims.
CA 02343713 2001-03-23
- 30a -
SEQUENCE LISTING
<110> McGILL UNIVERSITY
KARAPLIS, Andrew C.
GOLTZMAN, David
LIPMAN, Mark L.
HENDERSON, Janet E.
<120> USE OF PEX IN THE TREATMENT OF METABOLIC
BONE DISEASES
<130> 1770-214PCT FC/
<140> PCT/CA99/00895
<141> 1999-09-27
<150> CA 2,245,903
<151> 1998-09-28
<160> 11
<170> FastSEQ for Windows Version 3.0
<210> 1
<211> 3130
<212> DNA
<213> Unknown
<220>
<221> CDS
<222> (604) ... (2848)
<223> Human PEX
<400> 1
gatccactag taacggccgc cagtgtggtg gaattcaagg gactcacaca ctgaaagaat 60
atctttgatg aagacaattc aggcaagcag aatgattctt gcaacagaat tacatgatta 120
attgagatct tgaagtgggt ccggtgaatc ctggccacct aacttatcat gatttggggg 180
agtttcacga gaatccagtt ttgataaaac aattgttttt ttcctcccca agtgactata 240
catttaaata gctaaaacat ctgttcagca acatagtaaa acatatatac tcggaacgct 300
tgagagaaga gcctgccaaa cagggacttt gctgagggag agcaccaaga taaagcaaca 360
ctgtttgttt tgtctagtca ggggggaaag ccaaggcaac caatattttg gtttttataa 420
ttttcatttg tgaagaatta tttgagaaag ggttggcgag gggagatttc ctgacggcag 480
tttcttaagc tgtccattag tagaagagca agagagcctt ggatgtcaac gcctcgctct 540
tgagaccagc caccaaacca cgaaaagtga ctttcttctc gtgtgctctc tacggccctt 600
ctg atg gaa gca gaa aca ggg agc agc gtg gag act gga aag aag gcc 648
Met Glu Ala Glu Thr Gly Ser Ser Val Glu Thr Gly Lys Lys Ala
1 5 10 15
aac aga ggc act cga att gcc ctg gtc gtg ttt gtc ggt ggc acc cta 696
Asn Arg Gly Thr Arg Ile Ala Leu Val Val Phe Val Gly Gly Thr Leu
20 25 30
gtt ctg ggc acg atc ctc ttt cta gtg agt caa ggt ctc tta agt ctc 744
Val Leu Gly Thr Ile Leu Phe Leu Val Ser Gln Gly Leu Leu Ser Leu
35 40 45
caa gct aaa cag gag tac tgc ctg aag cca gaa tgc atc gaa gcg gct 792
Gln Ala Lys Gln Glu Tyr Cys Leu Lys Pro Glu Cys Ile Glu Ala Ala
50 55 60
CA 02343713 2001-03-23
- 30b -
gct gcc atc tta agt aaa gta aat ctg tct gtg gat cct tgt gat aat 840
Ala Ala Ile Leu Ser Lys Val Asn Leu Ser Val Asp Pro Cys Asp Asn
65 70 75
ttc ttc cgg ttc gct tgt gat ggc tgg ata agc aat aat cca att ccc 888
Phe Phe Arg Phe Ala Cys Asp Gly Trp Ile Ser Asn Asn Pro Ile Pro
80 85 90 95
gaa gat atg cca agc tat ggg gtt tat cct tgg ctg aga cat aat gtt 936
Glu Asp Met Pro Ser Tyr Gly Val Tyr Pro Trp Leu Arg His Asn Val
100 105 110
gac ctc aag ttg aag gaa ctt ttg gag aaa tca atc agt aga agg cgg 984
Asp Leu Lys Leu Lys Glu Leu Leu Glu Lys Ser Ile Ser Arg Arg Arg
115 120 125
gac acc gaa gcc ata cag aaa gcc aaa atc ctt tat tca tcc tgc atg 1032
Asp Thr Glu Ala Ile Gln Lys Ala Lys Ile Leu Tyr Ser Ser Cys Met
130 135 140
aat gag aaa gcg att gaa aaa gca gat ggc aag cca ctg cta cac atc 1080
Asn Glu Lys Ala Ile Glu Lys Ala Asp Gly Lys Pro Leu Leu His Ile
145 150 155
cta cgg cat tca cct ttc cgc tgg ccc gtg ctt gaa tct aat att ggc 1128
Leu Arg His Ser Pro Phe Arg Trp Pro Val Leu Glu Ser Asn Ile Gly
160 165 170 175
cct gaa ggg gtt tgg tca gag aga aag ttc agc ctt ctg cag aca ctt 1176
Pro Glu Gly Val Trp Ser Glu Arg Lys Phe Ser Leu Leu Gln Thr Leu
180 185 190
gca acg ttt cgt ggt caa tac agc aat tct gtg ttc atc cgt ttg tat 1224
Ala Thr Phe Arg Gly Gln Tyr Ser Asn Ser Val Phe Ile Arg Leu Tyr
195 200 205
gtg tcc cct gat gac aaa gca tcc aat gaa cat atc ttg aag ctg gac 1272
Val Ser Pro Asp Asp Lys Ala Ser Asn Glu His Ile Leu Lys Leu Asp
210 215 220
caa gca aca ctc tcc ctg gcc gtg agg gaa gac tac ctt gat aac agt 1320
Gln Ala Thr Leu Ser Leu Ala Val Arg Glu Asp Tyr Leu Asp Asn Ser
225 230 235
aca gaa gcc aag tct tat cgg gat gcc ctt tac aag ttc atg gtg gat 1368
Thr Glu Ala Lys Ser Tyr Arg Asp Ala Leu Tyr Lys Phe Met Val Asp
240 245 250 255
act gcc gtg ctt tta gga gct aac agt tcc aga gca gag cat gac atg 1416
Thr Ala Val Leu Leu Gly Ala Asn Ser Ser Arg Ala Glu His Asp Met
260 265 270
aag tca gtg ctc aga ttg gaa att aag ata gct gag ata atg att cca 1464
Lys Ser Val Leu Arg Leu Glu Ile Lys Ile Ala Glu Ile Met Ile Pro
275 280 285
CA 02343713 2001-03-23
- 30c -
cat gaa aac cga acc agc gag gcc atg tac aac aaa atg aac att tct 1512
His Glu Asn Arg Thr Ser Glu Ala Met Tyr Asn Lys Met Asn Ile Ser
290 295 300
gaa ctg agt gct atg att ccc cag ttc gac tgg ctg ggc tac atc aag 1560
Glu Leu Ser Ala Met Ile Pro Gln Phe Asp Trp Leu Gly Tyr Ile Lys
305 310 315
aag gtc att gac acc aga ctc tac ccc cat ctg aaa gac atc agc ccc 1608
Lys Val Ile Asp Thr Arg Leu Tyr Pro His Leu Lys Asp Ile Ser Pro
320 325 330 335
tcc gag aat gtg gtg gtc cgc gtc ccg cag tac ttt aaa gat ttg ttt 1656
Ser Glu Asn Val Val Val Arg Val Pro Gln Tyr Phe Lys Asp Leu Phe
340 345 350
agg ata tta ggg tct gag aga aag aag acc att gac aac tat ttg gtg 1704
Arg Ile Leu Gly Ser Glu Arg Lys Lys Thr Ile Asp Asn Tyr Leu Val
355 360 365
tgg aga atg gtt tat tcc aga att cca aac ctt agc agg cgc ttt cag 1752
Trp Arg Met Val Tyr Ser Arg Ile Pro Asn Leu Ser Arg Arg Phe Gln
370 375 380
tat aga tgg ctg gaa ttc tca agg gta atc cag ggg acc aca act ttg 1800
Tyr Arg Trp Leu Glu Phe Ser Arg Val Ile Gln Gly Thr Thr Thr Leu
385 390 395
ctg cct caa agg gac aaa tgt gta aac ttt att gaa agt gcc ctc cct 1848
Leu Pro Gln Arg Asp Lys Cys Val Asn Phe Ile Glu Ser Ala Leu Pro
400 405 410 415
tat gtt gtt gga aag atg ttt gta gat gtg tac ttc cag gaa gat aag 1896
Tyr Val Val Gly Lys Met Phe Val Asp Val Tyr Phe Gln Glu Asp Lys
420 425 430
aag gaa atg atg gag gaa ttg gtt gag ggc gtt cgc tgg gcc ttt att 1944
Lys Glu Met Met Glu Glu Leu Val Glu Gly Val Arg Trp Ala Phe Ile
435 440 445
gac atg cta gag aaa gaa aat gag tgg atg gat gca gga acg aaa agg 1992
Asp Met Leu Glu Lys Glu Asn Glu Trp Met Asp Ala Gly Thr Lys Arg
450 455 460
aaa gcc aaa gaa aag gcg aga gct gtt ttg gca aaa gtt ggc tat cca 2040
Lys Ala Lys Glu Lys Ala Arg Ala Val Leu Ala Lys Val Gly Tyr Pro
465 470 475
gag ttt ata atg aat gat act cat gtt aat gaa gac ctc aaa gct atc 2088
Glu Phe Ile Met Asn Asp Thr His Val Asn Glu Asp Leu Lys Ala Ile
480 485 490 495
aag ttt tca gaa gcc gac tac ttt ggc aac gtc cta caa act cgc aag 2136
Lys Phe Ser Glu Ala Asp Tyr Phe Gly Asn Val Leu Gln Thr Arg Lys
500 505 510
CA 02343713 2001-03-23
- 30d -
tat tta gca cag tct gat ttc ttc tgg cta aga aaa gcc gtt cca aaa 2184
Tyr Leu Ala Gln Ser Asp Phe Phe Trp Leu Arg Lys Ala Val Pro Lys
515 520 525
aca gag tgg ttt aca aat ccg acg act gtc aat gcc ttc tac agt gca 2232
Thr Glu Trp Phe Thr Asn Pro Thr Thr Val Asn Ala Phe Tyr Ser Ala
530 535 540
tcc acc aac cag atc cga ttt cca gca gga gag ctc cag aag cct ttc 2280
Ser Thr Asn Gln Ile Arg Phe Pro Ala Gly Glu Leu Gln Lys Pro Phe
545 550 555
ttt tgg gga aca gaa tat cct cga tct ctg agt tat ggt gct ata gga 2328
Phe Trp Gly Thr Glu Tyr Pro Arg Ser Leu Ser Tyr Gly Ala Ile Gly
560 565 570 575
gta att gtc gga cat gaa ttt aca cat gga ttt gat aat aat ggt aga 2376
Val Ile Val Gly His Glu Phe Thr His Gly Phe Asp Asn Asn Gly Arg
580 585 590
aaa tat gat aaa aat gga aac ctg gat cct tgg tgg tct act gaa tca 2424
Lys Tyr Asp Lys Asn Gly Asn Leu Asp Pro Trp Trp Ser Thr Glu Ser
595 600 605
gaa gaa aag ttt aag gaa aaa aca aaa tgc atg att aac cag tat agc 2472
Glu Glu Lys Phe Lys Glu Lys Thr Lys Cys Met Ile Asn Gln Tyr Ser
610 615 620
aac tat tat tgg aag aaa gct ggc tta aat gtc aag ggg aag agg acc 2520
Asn Tyr Tyr Trp Lys Lys Ala Gly Leu Asn Val Lys Gly Lys Arg Thr
625 630 635
ctg gga gaa aat att gct gat aat gga ggc ctg cgg gaa gct ttt agg 2568
Leu Gly Glu Asn Ile Ala Asp Asn Gly Gly Leu Arg Glu Ala Phe Arg
640 645 650 655
gct tac agg aaa tgg ata aat gac aga agg cag gga ctt gag gag cct 2616
Ala Tyr Arg Lys Trp Ile Asn Asp Arg Arg Gln Gly Leu Glu Glu Pro
660 665 670
ctt cta cca ggc atc aca ttc acc aac aac cag ctc ttc ttc ctg agt 2664
Leu Leu Pro Gly Ile Thr Phe Thr Asn Asn Gln Leu Phe Phe Leu Ser
675 680 685
tat gct cat gtg agg tgc aat tcc tac aga cca gaa gct gcc cga gaa 2712
Tyr Ala His Val Arg Cys Asn Ser Tyr Arg Pro Glu Ala Ala Arg Glu
690 695 700
caa gtc caa att ggt gct cac agt ccc cct cag ttt agg gtc aat ggt 2760
Gln Val Gln Ile Gly Ala His Ser Pro Pro Gln Phe Arg Val Asn Gly
705 710 715
gca att agt aac ttt gaa gaa ttc cag aaa gct ttt aac tgt cca ccc 2808
Ala Ile Ser Asn Phe Glu Glu Phe Gln Lys Ala Phe Asn Cys Pro Pro
720 725 730 735
CA 02343713 2001-03-23
- 30e -
aat tcc acg atg aac aga ggc atg gac tcc tgc cga ctc t ggtagctggg 2858
Asn Ser Thr Met Asn Arg Gly Met Asp Ser Cys Arg Leu
740 745
acgctggttt atggcatcct gagacagttg cacagtgcca gcggaggctg cactgagcct 2918
tcatcgccca ttgctttagg cctggaggag ctttcatttt tagtgcattt tcattatttg 2978
ggtaggtgac ctgcttggat ctagacagca tctgttcaaa gttgtagggc ttataaagtg 3038
gaatataaga agaactaagt atgtttcttt agaaaatcaa accaacaaaa ataaatccct 3098
aggctacttt tgttaaaaaa aaaaaaaaaa aa 3130
<210> 2
<211> 749
<212> PRT
<213> Unknown
<220>
<223> Human PEX
<400> 2
Met Glu Ala Glu Thr Gly Ser Ser Val Glu Thr Gly Lys Lys Ala Asn
1 5 10 15
Arg Gly Thr Arg Ile Ala Leu Val Val Phe Val Gly Gly Thr Leu Val
20 25 30
Leu Gly Thr Ile Leu Phe Leu Val Ser Gln Gly Leu Leu Ser Leu Gln
35 40 45
Ala Lys Gln Glu Tyr Cys Leu Lys Pro Glu Cys Ile Glu Ala Ala Ala
50 55 60
Ala Ile Leu Ser Lys Val Asn Leu Ser Val Asp Pro Cys Asp Asn Phe
65 70 75 80
Phe Arg Phe Ala Cys Asp Gly Trp Ile Ser Asn Asn Pro Ile Pro Glu
85 90 95
Asp Met Pro Ser Tyr Gly Val Tyr Pro Trp Leu Arg His Asn Val Asp
100 105 110
Leu Lys Leu Lys Glu Leu Leu Glu Lys Ser Ile Ser Arg Arg Arg Asp
115 120 125
Thr Glu Ala Ile Gln Lys Ala Lys Ile Leu Tyr Ser Ser Cys Met Asn
130 135 140
Glu Lys Ala Ile Glu Lys Ala Asp Gly Lys Pro Leu Leu His Ile Leu
145 150 155 160
Arg His Ser Pro Phe Arg Trp Pro Val Leu Glu Ser Asn Ile Gly Pro
165 170 175
Glu Gly Val Trp Ser Glu Arg Lys Phe Ser Leu Leu Gln Thr Leu Ala
180 185 190
Thr Phe Arg Gly Gln Tyr Ser Asn Ser Val Phe Ile Arg Leu Tyr Val
195 200 205
Ser Pro Asp Asp Lys Ala Ser Asn Glu His Ile Leu Lys Leu Asp Gln
210 215 220
Ala Thr Leu Ser Leu Ala Val Arg Glu Asp Tyr Leu Asp Asn Ser Thr
225 230 235 240
Glu Ala Lys Ser Tyr Arg Asp Ala Leu Tyr Lys Phe Met Val Asp Thr
245 250 255
Ala Val Leu Leu Gly Ala Asn Ser Ser Arg Ala Glu His Asp Met Lys
260 265 270
Ser Val Leu Arg Leu Glu Ile Lys Ile Ala Glu Ile Met Ile Pro His
275 280 285
CA 02343713 2001-03-23
- 30f -
Glu Asn Arg Thr Ser Glu Ala Met Tyr Asn Lys Met Asn Ile Ser Glu
290 295 300
Leu Ser Ala Met Ile Pro Gln Phe Asp Trp Leu Gly Tyr Ile Lys Lys
305 310 315 320
Val Ile Asp Thr Arg Leu Tyr Pro His Leu Lys Asp Ile Ser Pro Ser
325 330 335
Glu Asn Val Val Val Arg Val Pro Gln Tyr Phe Lys Asp Leu Phe Arg
340 345 350
Ile Leu Gly Ser Glu Arg Lys Lys Thr Ile Asp Asn Tyr Leu Val Trp
355 360 365
Arg Met Val Tyr Ser Arg Ile Pro Asn Leu Ser Arg Arg Phe Gln Tyr
370 375 380
Arg Trp Leu Glu Phe Ser Arg Val Ile Gln Gly Thr Thr Thr Leu Leu
385 390 395 400
Pro Gln Arg Asp Lys Cys Val Asn Phe Ile Glu Ser Ala Leu Pro Tyr
405 410 415
Val Val Gly Lys Met Phe Val Asp Val Tyr Phe Gln Glu Asp Lys Lys
420 425 430
Glu Met Met Glu Glu Leu Val Glu Gly Val Arg Trp Ala Phe Ile Asp
435 440 445
Met Leu Glu Lys Glu Asn Glu Trp Met Asp Ala Gly Thr Lys Arg Lys
450 455 460
Ala Lys Glu Lys Ala Arg Ala Val Leu Ala Lys Val Gly Tyr Pro Glu
465 470 475 480
Phe Ile Met Asn Asp Thr His Val Asn Glu Asp Leu Lys Ala Ile Lys
485 490 495
Phe Ser Glu Ala Asp Tyr Phe Gly Asn Val Leu Gln Thr Arg Lys Tyr
500 505 510
Leu Ala Gln Ser Asp Phe Phe Trp Leu Arg Lys Ala Val Pro Lys Thr
515 520 525
Glu Trp Phe Thr Asn Pro Thr Thr Val Asn Ala Phe Tyr Ser Ala Ser
530 535 540
Thr Asn Gln Ile Arg Phe Pro Ala Gly Glu Leu Gln Lys Pro Phe Phe
545 550 555 560
Trp Gly Thr Glu Tyr Pro Arg Ser Leu Ser Tyr Gly Ala Ile Gly Val
565 570 575
Ile Val Gly His Glu Phe Thr His Gly Phe Asp Asn Asn Gly Arg Lys
580 585 590
Tyr Asp Lys Asn Gly Asn Leu Asp Pro Trp Trp Ser Thr Glu Ser Glu
595 600 605
Glu Lys Phe Lys Glu Lys Thr Lys Cys Met Ile Asn Gln Tyr Ser Asn
610 615 620
Tyr Tyr Trp Lys Lys Ala Gly Leu Asn Val Lys Gly Lys Arg Thr Leu
625 630 635 640
Gly Glu Asn Ile Ala Asp Asn Gly Gly Leu Arg Glu Ala Phe Arg Ala
645 650 655
Tyr Arg Lys Trp Ile Asn Asp Arg Arg Gln Gly Leu Glu Glu Pro Leu
660 665 670
Leu Pro Gly Ile Thr Phe Thr Asn Asn Gln Leu Phe Phe Leu Ser Tyr
675 680 685
Ala His Val Arg Cys Asn Ser Tyr Arg Pro Glu Ala Ala Arg Glu Gln
690 695 700
Val Gln Ile Gly Ala His Ser Pro Pro Gln Phe Arg Val Asn Gly Ala
705 710 715 720
Ile Ser Asn Phe Glu Glu Phe Gln Lys Ala Phe Asn Cys Pro Pro Asn
725 730 735
CA 02343713 2001-03-23
- 30g -
Ser Thr Met Asn Arg Gly Met Asp Ser Cys Arg Leu Trp
740 745
<210> 3
<211> 749
<212> PRT
<213> Unknown
<220>
<223> Human PEX
<400> 3
Met Glu Ala Glu Thr Gly Ser Ser Val Glu Thr Gly Lys Lys Ala Asn
1 5 10 15
Arg Gly Thr Arg Ile Ala Leu Val Val Phe Val Gly Gly Thr Leu Val
20 25 30
Leu Gly Thr Ile Leu Phe Leu Val Ser Gln Gly Leu Leu Ser Leu Gln
35 40 45
Ala Lys Gln Glu Tyr Cys Leu Lys Pro Glu Cys Ile Glu Ala Ala Ala
50 55 60
Ala Ile Leu Ser Lys Val Asn Leu Ser Val Asp Pro Cys Asp Asn Phe
65 70 75 80
Phe Arg Phe Ala Cys Asp Gly Trp Ile Ser Asn Asn Pro Ile Pro Glu
85 90 95
Asp Met Pro Ser Tyr Gly Val Tyr Pro Trp Leu Arg His Asn Val Asp
100 105 110
Leu Lys Leu Lys Glu Leu Leu Glu Lys Ser Ile Ser Arg Arg Arg Asp
115 120 125
Thr Glu Ala Ile Gln Lys Ala Lys Ile Leu Tyr Ser Ser Cys Met Asn
130 135 140
Glu Lys Ala Ile Glu Lys Ala Asp Ala Lys Pro Leu Leu His Ile Leu
145 150 155 160
Arg His Ser Pro Phe Arg Trp Pro Val Leu Glu Ser Asn Ile Gly Pro
165 170 175
Glu Gly Val Trp Ser Glu Arg Lys Phe Ser Leu Leu Gln Thr Leu Ala
180 185 190
Thr Phe Arg Gly Gln Tyr Ser Asn Ser Val Phe Ile Arg Leu Tyr Val
195 200 205
Ser Pro Asp Asp Lys Ala Ser Asn Glu His Ile Leu Lys Leu Asp Gln
210 215 220
Ala Thr Leu Ser Leu Ala Val Arg Glu Asp Tyr Leu Asp Asn Ser Thr
225 230 235 240
Glu Ala Lys Ser Tyr Arg Asp Ala Leu Tyr Lys Phe Met Val Asp Thr
245 250 255
Ala Val Leu Leu Gly Ala Asn Ser Ser Arg Ala Glu His Asp Met Lys
260 265 270
Ser Val Leu Arg Leu Glu Ile Lys Ile Ala Glu Ile Met Ile Pro His
275 280 285
Glu Asn Arg Thr Ser Glu Ala Met Tyr Asn Lys Met Asn Ile Ser Glu
290 295 300
Leu Ser Ala Met Ile Pro Gln Phe Asp Trp Leu Gly Tyr Ile Lys Lys
305 310 315 320
Val Ile Asp Thr Arg Leu Tyr Pro His Leu Lys Asp Ile Ser Pro Ser
325 330 335
Glu Asn Val Val Val Arg Val Pro Gln Tyr Phe Lys Asp Leu Phe Arg
340 345 350
CA 02343713 2001-03-23
- 30h -
Ile Leu Gly Ser Glu Arg Lys Lys Thr Ile Ala Asn Tyr Leu Val Trp
355 360 365
Arg Met Val Tyr Ser Arg Ile Pro Asn Leu Ser Arg Arg Phe Gln Tyr
370 375 380
Arg Trp Leu Glu Phe Ser Arg Val Ile Gln Gly Thr Thr Thr Leu Leu
385 390 395 400
Pro Gln Trp Asp Lys Cys Val Asn Phe Ile Glu Ser Ala Leu Pro Tyr
405 410 415
Val Val Gly Lys Met Phe Val Asp Val Tyr Phe Gln Glu Asp Lys Lys
420 425 430
Glu Met Met Glu Glu Leu Val Glu Gly Val Arg Trp Ala Phe Ile Asp
435 440 445
Met Leu Glu Lys Glu Asn Glu Trp Met Asp Ala Gly Thr Lys Arg Lys
450 455 460
Ala Lys Glu Lys Ala Arg Ala Val Leu Ala Lys Val Gly Tyr Pro Glu
465 470 475 480
Phe Ile Met Asn Asp Thr His Val Asn Glu Asp Leu Lys Ala Ile Lys
485 490 495
Phe Ser Glu Ala Asp Tyr Phe Gly Asn Val Leu Gln Thr Arg Lys Tyr
500 505 510
Leu Ala Gln Ser Asp Phe Phe Trp Leu Arg Lys Ala Val Pro Lys Thr
515 520 525
Glu Trp Phe Thr Asn Pro Thr Thr Val Asn Ala Phe Tyr Ser Ala Ser
530 535 540
Thr Asn Gln Ile Arg Phe Pro Ala Gly Glu Leu Gln Lys Pro Phe Phe
545 550 555 560
Trp Gly Thr Glu Tyr Pro Arg Ser Leu Ser Tyr Gly Ala Ile Gly Val
565 570 575
Ile Val Gly His Glu Phe Thr His Gly Phe Asp Asn Asn Gly Arg Lys
580 585 590
Tyr Asp Lys Asn Gly Asn Leu Asp Pro Trp Trp Ser Thr Glu Ser Glu
595 600 605
Glu Lys Phe Lys Glu Lys Thr Lys Cys Met Ile Asn Gln Tyr Ser Asn
610 615 620
Tyr Tyr Trp Lys Lys Ala Gly Leu Asn Val Lys Gly Lys Arg Thr Leu
625 630 635 640
Gly Glu Asn Ile Ala Asp Asn Gly Gly Leu Arg Glu Ala Phe Arg Ala
645 650 655
Tyr Arg Lys Trp Ile Asn Asp Arg Arg Gln Gly Leu Glu Glu Pro Leu
660 665 670
Leu Pro Gly Ile Thr Phe Thr Asn Asn Gln Leu Phe Phe Leu Ser Tyr
675 680 685
Ala His Val Arg Cys Asn Ser Tyr Arg Pro Glu Ala Ala Arg Glu Gln
690 695 700
Val Gln Ile Gly Ala His Ser Pro Pro Gln Phe Arg Val Asn Gly Ala
705 710 715 720
Ile Ser Asn Phe Glu Glu Phe Gln Lys Ala Phe Asn Cys Pro Pro Asn
725 730 735
Ser Thr Met Asn Arg Gly Met Asp Ser Cys Arg Leu Trp
740 745
<210> 4
<211> 747
<212> PRT
<213> Unknown
<220>
CA 02343713 2001-03-23
- 30i -
<223> Human PEX
<400> 4
Met Glu Ser Gln Met Asp Ile Thr Asp Ile Asn Thr Pro Lys Pro Lys
1 5 10 15
Lys Lys Gln Arg Trp Thr Pro Leu Glu Ile Ser Leu Ser Val Leu Val
20 25 30
Leu Leu Leu Thr Ile Ile Ala Val Thr Met Ile Ala Leu Tyr Ala Thr
35 40 45
Tyr Asp Asp Gly Ile Cys Lys Ser Ser Asp Cys Ile Lys Ser Ala Ala
50 55 60
Arg Leu Ile Gln Asn Met Asp Ala Thr Thr Glu Pro Cys Thr Asp Phe
65 70 75 80
Phe Lys Tyr Ala Cys Gly Gly Trp Leu Lys Arg Asn Val Ile Pro Glu
85 90 95
Thr Ser Ser Arg Tyr Gly Asn Phe Asp Ile Leu Arg Asp Glu Leu Glu
100 105 110
Val Val Leu Lys Asp Val Leu Gln Glu Pro Lys Thr Glu Asp Ile Val
115 120 125
Ala Val Gln Lys Ala Lys Ala Leu Tyr Arg Ser Cys Ile Asn Glu Ser
130 135 140
Ala Ile Asp Ser Arg Gly Gly Glu Pro Leu Leu Lys Leu Leu Pro Asp
145 150 1.55 160
Ile Tyr Gly Trp Pro Val Ala Thr Glu Asn Trp Glu Gln Lys Tyr Gly
165 170 175
Ala Ser Trp Thr Ala Glu Lys Ala Ile Ala Gln Leu Asn Ser Lys Tyr
180 185 190
Gly Lys Lys Val Leu Ile Asn Leu Phe Val Gly Thr Asp Asp Lys Asn
195 200 205
Ser Val Asn His Val Ile His Ile Asp Gln Pro Arg Leu Gly Leu Pro
210 215 220
Ser Arg Asp Tyr Tyr Glu Cys Thr Gly Ile Tyr Lys Glu Ala Cys Thr
225 230 235 240
Ala Tyr Val Asp Phe Met Ile Ser Val Ala Arg Leu Ile Arg Gln Glu
245 250 255
Glu Arg Leu Pro Ile Asp Glu Asn Gln Leu Ala Leu Glu Met Asn Lys
260 265 270
Val Met Glu Leu Glu Lys Glu Ile Ala Asn Ala Thr Ala Lys Pro Glu
275 280 285
Asp Arg Asn Asp Pro Met Leu Leu Tyr Asn Lys Met Thr Leu Ala Gln
290 295 300
Ile Gln Asn Asn Phe Ser Leu Glu Ile Asn Gly Lys Pro Phe Ser Trp
305 310 315 320
Leu Asn Phe Thr Asn Glu Ile Met Ser Thr Val Asn Ile Ser Ile Thr
325 330 335
Asn Glu Glu Asp Val Val Val Tyr Ala Pro Glu Tyr Leu Thr Lys Leu
340 345 350
Lys Pro Ile Leu Thr Lys Tyr Ser Ala Arg Asp Leu Gln Asn Leu Met
355 360 365
Ser Trp Arg Phe Ile Met Asp Leu Val Ser Ser Leu Ser Arg Thr Tyr
370 375 380
Lys Glu Ser Arg Asn Ala Phe Arg Lys Ala Leu Tyr Gly Thr Thr Ser
385 390 395 400
Glu Thr Ala Thr Trp Arg Arg Cys Ala Asn Tyr Val Asn Gly Asn Met
405 410 415
Glu Asn Ala Val Gly Arg Leu Tyr Val Glu Ala Ala Phe Ala Gly Glu
420 425 430
CA 02343713 2001-03-23
- 30j -
Ser Lys His Val Val Glu Asp Leu Ile Ala Gln Ile Arg Glu Val Phe
435 440 445
Ile Gln Thr Leu Asp Asp Leu Thr Trp Met Asp Ala Glu Thr Lys Lys
450 455 460
Arg Ala Glu Glu Lys Ala Leu Ala Ile Lys Glu Arg Ile Gly Tyr Pro
465 470 475 480
Asp Asp Ile Val Ser Asn Asp Asn Lys Leu Asn Asn Glu Tyr Leu Glu
485 490 495
Leu Asn Tyr Lys Glu Asp Glu Tyr Phe Glu Asn Ile Ile Gln Asn Leu
500 505 510
Lys Phe Ser Gln Ser Lys Gln Leu Lys Lys Leu Arg Glu Lys Val Asp
515 520 525
Lys Asp Glu Trp Ile Ser Gly Ala Ala Val Val Asn Ala Phe Tyr Ser
530 535 540
Ser Gly Arg Asn Gln Ile Val Phe Pro Ala Gly Ile Leu Gln Pro Pro
545 550 555 560
Phe Phe Ser Ala Gln Gln Ser Asn Ser Leu Asn Tyr Gly Gly Ile Gly
565 570 575
Met Val Ile Gly His Glu Ile Thr His Gly Phe Asp Asp Asn Gly Arg
580 585 590
Asn Phe Asn Lys Asp Gly Asp Leu Val Asp Trp Trp Thr Gln Gln Ser
595 600 605
Ala Ser Asn Phe Lys Glu Gln Ser Gln Cys Met Val Tyr Gln Tyr Gly
610 615 620
Asn Phe Ser Trp Asp Leu Ala Gly Gly Gln His Leu Asn Gly Ile Asn
625 630 635 640
Thr Leu Gly Glu Asn Ile Ala Asp Asn Gly Gly Leu Gly Gln Ala Tyr
645 650 655
Arg Ala Tyr Gln Asn Tyr Ile Lys Lys Asn Gly Glu Glu Lys Leu Leu
660 665 670
Pro Gly Leu Asp Leu Asn His Lys Gln Leu Phe Phe Leu Asn Phe Ala
675 680 685
Gln Val Trp Cys Gly Thr Tyr Arg Pro Glu Tyr Ala Val Asn Ser Ile
690 695 700
Lys Thr Asp Val His Ser Pro Gly Asn Phe Arg Ile Ile Gly Thr Leu
705 710 715 720
Gln Asn Ser Ala Glu Phe Ser Glu Ala Phe His Cys Arg Lys Asn Ser
725 730 735
Tyr Met Asn Pro Glu Lys Lys Cys Arg Val Trp
740 745
<210> 5
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Human PEX-specific primer
<400> 5
ggaggaattg gttgagggcg 20
<210> 6
<211> 20
<212> DNA
<213> Artificial Sequence
CA 02343713 2001-03-23
- 30k -
<220>
<223> Human PEX-specific primer
<400> 6
gtagaccacc aaggatccag 20
<210> 7
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Human PEX-specific primer
<400> 7
cgtgcccaga actagggtgc cacc 24
<210> 8
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide PEX-4 used as primer
<400> 8
ctggatcctt ggtggtctac 20
<210> 9
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide PEX-5 used as primer
<400> 9
cactgtgcaa ctgtctcag 19
<210> 10
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide PEXMycl used as primer
<400> 10
ttggatgtca acgcctcg 18
<210> 11
<211> 70
<212> DNA
CA 02343713 2001-03-23
- 301 -
<213> Artificial Sequence
<220>
<223> Oligonucleotide PEXMyc2 used as a primer
<400> 11
ctaccacaat ctacagttgt tcaggtcctc ttcgctaatc agcttttgtt ccatagagtc 60
catgcctctg 70