Note: Descriptions are shown in the official language in which they were submitted.
CA 02353577 2001-06-O1
WO 00/327$9 PCT/NL99/00737
Fruit Flavour Related Genes And Use
Thereof
The invention disclosed herein relates to strawberry
(fragaria ananassa) and lemon (citrus limon) fruit derived
genes and enzymes specifically involved in the formation of
aliphatic and/ or aromatic esters and other aroma and
flavour compounds in fruits. More specifically it relates
to a process for improving natural volatile fruit flavours
by the control of one or more than one gene implicated in
that process.
Background of the invention
Like most natural products the chemical compositi-
on of strawberry aroma and flavour is quite complex [Zabe-
takis and Holden, (1997); J. Sci. Food. Agric 74: 421-434].
Over 300 compounds have been identified which may contribu-
te to flavour/aroma. Qualitatively, the major components of
strawberry flavour and aroma may be grouped into several
chemical classes including acids, aldehydes, ketones,
alcohols, esters, and lactones. Other groups include
sulphur compounds, acetals, furans, phenols and even traces
of epoxides, and hydrocarbons. Compounds produced by these
groups, whilst often present at low levels, may have a
significant impact on the overall flavour of strawberry.
In general, fruit flavour compounds develop
during ripening when the metabolism of the fruit changes to
catabolism. Esters are proposed to be qualitatively and
quantitatively the most important class of volatiles
produced and are a key component of strawberry flavour.
Seven volatile esters, ethyl hexanoate, methyl hexanoate,
ethyl butanoate, methyl butanoate, hexyl acetate, ethyl
propionate and 2-hexenyl-acetate were reported to contribu-
te largely to the aroma associated with the fruit. However,
more then a 100 types of esters have been reported during
the years to be identified in strawberry volatile analysis
[Zabetakis and Holden, (1997); J. Sci. Food. Agric 74: 421-
434] .
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
2
In lemon, the acetate esters derived from cyclic
terpene alcohols are used frequently as flavour, fragrance,
scent and aroma components. The lower fatty acid esters of
acyclic terpene alcohols (geraniol, linalool, and citronel-
lol) are the acetates (linalyl acetate, geranyl acetate)
which are the main flavour components of lemon peel oils.
These are used for obtaining citrus notes [Bauer, K and
Garbe, D, (1985). Common fragrance and flavour materials.
Preparation, properties and uses. VCH Publishers, Florida,
USA]. Formates, propionates, butyrates occur less frequent-
ly. In addition to the cyclic and acyclic terpene esters,
lemon is a rich source for the ester methyl anthranillate,
which is a methyl ester of o-aminobenzoic acid. This
compound is known to be a major constituent of citrus Iimon
fruit and flowers and can also be found in the leaves.
Primary metabolites which include sugars, fatty
acids, nucleotides and amino acids have been proposed to
serve as precursors for the production of secondary me-
tabolites such as volatile compounds. Sugars in the fruit
in addition to acting as a carbon source, act as precursors
for the flavour compound furanones [Zabetakis and Holden,
(1996); Plant Cell Tissue and Organ Culture 45:25-29].
Amino acid metabolism leads to the formation of various
aldehydes, acids, carbonyls, aliphatic and branched-chain
alcohols, some of which serve as aroma and flavour compo-
nents. The changes in amino acid content during maturation
of fruits in relation to fruit flavour formation, have been
studied in banana, tomato and strawberry. The amino acids
valine, leucine and isoleucine have been proposed to serve
as flavour precursors in banana fruit, and are metabolised
to branched chain alcohols, 2-propanol, isoamyl alcohol,
and 2-methylbutanol, respectively [Drawert and Berger,
(1981); Bioflavour '81; Schreier, P., Ed.; de Gruyter:
Berlin]. The conversion of alanine, leucine and valine to
flavour compounds has also been demonstrated in tomato [Yu
et al., (1968); Phytochemistry 6: 1457-1465]. In strawber-
ry, alanine was proposed to be the main free amino acid
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
3
metabolised to flavour compounds. This was based mainly on
the dramatic decrease in its content just before the
formation of volatile aroma and flavour compounds commenced
in the fruit [Perez et al., (1992); J.Agric. Food Chem. 40:
2232-2235]. In addition feeding alanine to strawberry
cultures resulted in the formation of several esters such
as methyl and ethyl hexanoate which are important constitu-
ents of strawberry volatiles [Drawert and Berger, (1981);
Bioflavour '81; Schreier, P., Ed.; de Gruyter: Berlin].
Like sugars and amino acids the fatty acids in strawberry
play an important role in ester formation. In mango fruit
there have been reports that fatty acids are actively
metabolised during ripening. Changes in fatty acids and
triglycerides have been associated with changes in aroma
and flavour during mango ripening [Gholop AS and Bandyopha-
day C, J Am. Oil Chem. Soc.,52: 514-516, 1975]. Therefore
fatty acid related enzymes may have a role in the produc-
tion of flavour volatiles. Thiolase is the last enzyme in
the 13-oxidation of fatty acids. It catalyses the thiolytic
cleavage involving a molecule of CoA. The products of this
reaction are acetyl-CoA and aryl-CoA derivatives containing
two carbon atoms less than the original acyl-CoA molecule
that underwent oxidation. The acyl-CoA formed in the cleava-
ge reaction may be utilised at the final stage of the
biosynthetic pathway for ester formation in fruit. Therefore
the profile of fatty acid and amino acid precursors found in
each fruit, along with the specificity of the enzymes in the
biosynthetic pathway/process leading to ester formation
could have a key role in determining the types of esters
formed.
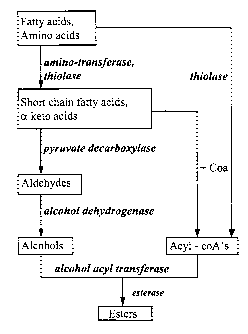
A proposed biosynthetic pathway for volatile ester
formation in fruits is illustrated in Figure 1. In fruits
the transamination and oxidative decarboxylation of amino
acids provides the precursors for volatile aroma compounds
such as aldehydes, acids, alcohols, esters and thiols. The
transamination reaction is catalysed by aminotransferases.
Aminotransferases or transaminases are enzymes promoting the
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
4
first step in the catabolism of L-amino acids by removing
the a-amino groups. As a result of this transamination
reaction, the a-amino group is transferred to the a-carbon
atom of a-ketoglutarate, leaving behind the a-keto acid
analogue of the amino acid. Aminotransferases have been
studied in various micro-organisms and some of them have
been purified [Gelfand et a1, (1997); J. Bacteriol. 130:429-
440, Lee et al., (1985); J. Gen. Microbiol. 131: 459-467].
No strawberry fruit specific aminotransferase to date has
been cloned or characterised.
Ketoacids produced by transamination can be enzyma-
tically degraded to the corresponding aldehydes or carboxy-
lic acids. The enzyme which can catalyse this reaction is
pyruvate decarboxylase. An EST encoding part of pyruvate
decarboxylase, the first to be characterised from a straw-
berry fruit has been mentioned in patent application WO
97/27295. The pyruvate decarboxylase enzyme provides precur-
sors for the biosynthesis of volatile flavour compounds.
Alcohol dehydrogenase enzymes have been implicated in the
interconversion of the aldehyde and alcohol forms of flavour
volatiles. The mechanism of ester formation has been shown
to be a coenzyme A dependent reaction. Esterification is a
result of transacylation from aryl-coenzyme A to an alcohol.
The enzyme is therefore termed as alcohol acyl transferase
(AAT) and plays a major role in the biosynthesis of volatile
esters.
It is still unclear whether esterase enzymes (Fig
1), are involved in the hydrolysis of esters or in their
synthesis [Mauricio et al., (1993) J. Agric. Food Chem. 41,
2086-2091].
Ester formation by micro-organisms has been studied
in most detail and has been cited in a number of publicati-
ons [Harada et al., (1985); Plant Cell Physiol. 26(6): 1067-
1074], Fujii et al., (1996); Yeast 12:593-598, Fujii et al.,
(1994); Applied and Enviro. Microbiology 8:2786-2792 ].
Alcohol acyl transferases (AAT's) have been identified from
both fungi [Yoshihide et al., (1978); AGRIC. Biol. Chem.
42(2):269-274], and yeast [Fujii et al., (1994); Applied and
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
Enviro. Microbiology 8:2786-2792 ] and have been the subject
of patent applications (EP 0 574 941 A2). The AAT from the
fungi Neurospora sp. which was purified to homogeneity acts
on various acyl Coenzyme A containing more than a four
5 carbon linear chain but not on acetyl coenzyme A as descri-
bed for the yeast enzyme [Yamauchi et al., (1989); Agric.
Biol. Chem. 53(6) 1551-1556] . In Cladosporium cladosporioi-
des No. 9, AAT was partially purified and was described to
form acetate esters like the yeast enzyme [Yamakawa et al.,
(1978); Agric. Biol. Chem. 42(2):269-274]. So it is clear
that the type of esters formed is dependant on the substrate
specificity of the ester forming enzyme to alcohols, and the
availability of acyl-CoAs. For example the yeast Saccha-
romyces cerevisiae (sake yeast) and brewer's yeast (bottom
fermenting yeast) AAT's show high affinity to acetyl-CoA and
the alcohols ethanol and isoamyl alcohol, giving rise to the
formation of acetate esters. These esters play an important
role in determining flavour characteristics of beer.
In plants ester formation has been studied both in
fruit and flowers. Melon, banana and strawberry AAT's have
been investigated using crude fruit extracts. The formation
of esters from aldehydes that were incubated with whole
strawberry fruit has been reported [Yamashita, et al.,
(1989); Agr. Biol. Chem. 39(12) 2303-2307]. Analysis of the
substrate specificity of the enzymes from the various
sources, revealed differences in their affinity to acyl-CoAs
and alcohols [Veda et al., (1992); Nippon Shokukin Kogyo
Gakkaishi 39(2): 183-187, Perez et al., (1996); J. AgricFood
Chem. 44: 3286-3290, Harada et al., (1985); Plant Cell
Physiol. 26(6): 1067-1074]. Maximum activity for the straw-
berry (AAT) was obtained using acetyl CoA and hexyl alcohol
as substrates, and acetyl-CoA and butyl alcohol is the
preferred substrate for the banana enzyme. A clear correla-
tion could be observed between substrate preference and
volatile esters present in both fruits. The purification
method and some properties of the strawberry AATase enzyme
have been reported. However, to date no sequence of the
peptide or of the gene has been disclosed. In Clarkia
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
6
breweri the ester benzylacetate is an important constituent
of flower scent. The purification of the protein and the
gene AAT encoding it has been reported. The flower AAT
enzyme was not reported to have affinity to aliphatic
alcohols. The enzyme has high affinity to the aromatic
alcohols such as benzyl alcohol and cinnamyl alcohol [Duda-
reva et al., (1998); The Plant Journal 14(3) 297-304]. Genes
coding for flower aromatic acyl transferases, that acylate
plant pigments, causing changes in colour tone have been the
subject of a recent patent application [EP 0810287]. Recent
ly, an aromatic amino transferase enzyme has been purified
from lactic acid bacteria and was shown to initiate the
conversion of amino acids to cheese flavour compounds [Yvon
et al., (1997); Applied and Environmental Microbiology 414
419] .
Although volatile esters are qualitatively and
quantitatively one of the most important classes of volatile
compounds in fruit flavour and aroma, there are very few
reports concerning the biochemical aspects of ester forma-
tion in fruits. An understanding of, the precursors and
characterisation of enzymes involved in the pathways leading
to the formation of flavours in fruit is essential for the
production of natural flavours. Plant derived flavour
components alone represent a world-wide market of 1.5
billion dollars. Presently, the main ways to produce plant
flavour compounds is by the synthetic route. Synthetic
organic chemicals constitute more than 80-90% (by weight and
value) of the raw materials used in flavour and fragrance
formulations. Problems often exist concerning production.
Extraction from intact plants and conventional fermentation
are currently providing alternative routes for the commerci-
al production of flavour and aroma chemicals. However, the
demand for natural flavours by the consumer has been steadi-
ly increasing, and often demand outstrips supply. In many
cases sought after flavour compounds preclude isolation.
Unlike other fruit flavours, no single chemical or class of
chemicals in particular are associated with strawberry
flavour. To date no strawberry fruit specific aminotransfe-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
7
rase, pyruvate decarboxylase, thiolase, alcohol dehydrogena-
se, or acyl-transferase enzymes at the route of this inven-
tion, involved directly or in providing precursors for ester
formation, hence flavour compounds, has been isolated and
characterized. The genes and their peptides at the route of
this invention are involved in the biosynthetic pathway for
aliphatic and/or aromatic ester production in fruit and can
therefore provide a novel method for the in-vivo and in-
vitro biotechnology production of bio-flavours, natural
flavour chemicals by recombinant means.
Summary of invention
The object of the present invention is to disclose the
genes/peptides involved in the process of volatile ester
production, particularly aliphatic esters although not
exclusively so, hence fruit flavour and aroma. This is the
first time to our knowledge, that the genes/peptides sequen-
ces from an entire pathway to the production of volatile
esters in fruit, in particularly strawberry, have been
disclosed. The nucleotide and polypeptide sequences descri-
bed in this application can serve as a tool for a vast num-
ber of applications related to ester formation and the
biotechnological engineering of natural and artificial fruit
flavours for the food industry. The invention is based on
the identification of genes which encode proteins central to
the pathway leading to volatile ester formation in fruit.
DNA sequences which encode these proteins have been cloned
and characterised. The nucleic acid/peptide sequences may be
used in expression systems, for industrial application and
/or to modify plants with the goal to produce natural and/or
synthetic flavours.
Definitions of Terms
'Nucleic acid' sequence as used herein refers to an oligo-
nucleotide, nucleotide or polynucleotide, and fragments or
portions thereof, and to DNA or RNA of genomic or synthetic
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
8
origin which may be single- or double-stranded, and repre-
sents the sense or antisense strand. Similarly, 'amino acid
sequence' as used herein refers to peptide or protein
sequence.
The term 'agonist', as used herein, refers to a
molecule which, when bound to a polypeptide causes a change
in the polypeptide which modifies the activity of the
polypeptide. Agonist may include proteins, nucleic acids,
carbohydrates, or any other molecule which binds to the
polypeptide.
The term 'antagonist' or 'inhibitor' as used
herein, refer to a molecule which when bound to a polypep-
tide, blocks or modulates the biological or immunological
activity of the polypeptide. Antagonists or inhibitors may
include proteins, nucleic acids, carbohydrates, or any other
molecules which bind to the polypeptide.
'Stringency' typically occurs in the range from
about Tm-5°C (5°C below the Tm of the probe) to about
20°C to
25°C below Tm. As will be understood by those skilled in the
art, a stringent hybridisation can be used to identify or
detect identical polynucleotide sequences or to identify or
detect similar or related polynucleotide sequences.
The term 'hybridisation' as used herein shall
include 'any process' by which a strand of nucleic acid
joins with a complementary strand through base pairing'
(Coombs J, (1994) Directory of biotechnology, Stockton
press, New York NY).
A 'deletion' is defined as a change in either
nucleotide or amino acid sequence in which one or more
nucleotides or amino acid residues, respectively, are
absent.
An 'insertion' or 'addition' is that change in
nucleotide or amino acid sequence which has resulted in the
addition of one or more nucleotides or amino acid residues,
respectively, as compared to the naturally occurring poly-
peptide ( s ) .
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
9
A 'substitution' results from the replacement of
one or more nucleotides or amino acids by different nucleo-
tides or amino acids, respectively.
As used herein, the term 'substantially purified'
refers to molecules, either nucleic or amino acid sequences,
that are removed from their natural environment, isolated or
separated, and that are at least 60~ free, preferably 75%
free, and most preferably 90o free from other components
with which they are naturally associated. The definition
'substantially homologous' means that a particular subject
sequence, for example a mutant sequence, varies from the
reference sequence by one or more substitutions, deletions,
or additions, the net effect of which does not result in an
adverse functional dissimilarity between the reference and
the subject sequence.
A 'variant' of a polypeptide as outlined in the
first and second aspects of this invention is defined as an
amino acid sequence that is different by one or more amino
acid 'substitutions'. A variant may have 'conservative'
changes, wherein a substituted amino acid has similar
structural or chemical properties eg replacement of leucine
with isoleucine. More rarely a variant may have 'non-conser-
vative' changes, eg replacement of a glycine with a tryp-
tophan. Similar minor variations may also include amino acid
deletions or insertions, or both. Guidance in determining
which and how many amino acid residues may be substituted,
inserted or deleted, without abolishing biological or
immunological activity may be found using computer program-
mes well known in the art, for example, DNAStar software.
The term 'biologically active' refers to a poly-
peptide as outlined in the first and second aspects of the
invention, having structural, regulatory, or biochemical
function of their naturally occurring counterparts. Likewi-
se, 'immunologically active' defines the capability of the
natural, recombinant or synthetic polypeptide, as outlined
in the first and second aspects of the invention, or any
oligopeptide thereof, to induce a specific immune response
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
in an appropriate animal or cells and to bind with specific
antibodies.
The term 'natural products' are those products that
are obtained directly from plants and sometimes from animal
5 sources by physical procedures. 'Nature identical compounds'
are produced synthetically but are chemically identical to
their 'natural counterparts'. Artificial flavour substances
are compounds that have not been identified in plant or
animal products for human consumption. 'Nature-identical'
10 aroma substances are with few exceptions the only synthetic
compounds used in flavours in addition to 'natural pro-
ducts'.
The definition 'host cell' refers to a cell in
which an alien process is executed by bio-interaction,
irrespective of the cell belongs to a unicellular, multi-
cellular, a differentiated organism or to an artificial
cell, cell culture or protoplast. The definition 'host cell'
in the context of this invention is to encompass the defini-
tion 'plant cell'.
'Plant cell' by definition is meant by any self-
propagating cell bounded by a semi permeable membrane and
containing one or more plastids. Such a cell requires a cell
wall if further propagation is required. 'Plant cell', as
used herein, includes without limitation, seeds, suspension
cultures, embryos, meristematic regions, callous tissues,
protoplasts, leaves, roots, shoots, gametophytes, sporophy-
tes, pollen and microspores.
With the definition 'transformed cell' or 'transg
enic cell' is meant a cell (or ancestor of said cell) into
which by means of recombinant DNA techniques, DNA encoding
the target polypeptide can be introduced.
The definition 'micro-organisms' refers to micro-
scopic organisms, such as Archaea, Bacteria, Cyanobacteria,
Microalgae, Fungi, Yeast, Viruses, Protozoa, Rotifers,
Nematodes, Micro-Crustaceans, Micro-Molluscs, Micro-Shellf-
ish, Micro-insects etc.
The definition 'plant(s)' refers to eukaryotic,
autotrophic organisms. They are characterised by direct
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
11
usage of solar energy for their primary metabolism, their
permanent cell wall and in case of multicellular individuals
their open unlimited growth. In case of heterotrophic
plants, the organisms are in an evolutionary context essen-
tially derived from autotrophic plants in their structure
and metabolism.
'Dicotyledons' (and all scientific equivalents
referring to the same group of plants) form one of the two
divisions of the flowering plants or angiospermae in which
the embryo has two or more free or fused cotyledons.
'Monocotyledons' (and all scientific equivalents
referring to the same group of plants) form one of the two
divisions of the flowering plants or angiospermae in which
the embryo has one cotyledon.
'Angiospermae' or flowering plants are seed plants
characterised by flowers as specialised organs of plant
reproduction and by carpels covering the ovaries. Gymno-
spermae are seed plants characterised by strobili as specia-
lised organs for plant repoduction and by naked sporophylls
bearing the male or female reproductive organs. 'Ornamen-
tal' plants are plants that are primarily in cultivation for
their habitus, special shape, (flower, foliage or otherwise)
colour or other characteristics which contribute to human
well being indoor as cut flowers or pot plants or outdoors
in the man made landscape. 'vegetables' are plants that are
purposely bred or selected for human consumption of foliage,
tubers, stems, fruits, flowers of parts of them and that
need an intensive cultivation regime. 'Arable crops' are
purposely bred or selected for human objectivity's (ranging
from direct or indirect consumption, feed or industrial
applications such as fibres).
The definition 'process' is the development of, a
methodology) concerning, the progress of a series of
activities in a certain context. 'Biological' process is a
process based on metabolic activity in organisms, or essen-
tially derived from that by biochemical, biophysical,
physiological, ecological or genetic means. 'Industrial'
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
12
process is an economically feasible process with high added
value, aiming at bulk production.
The definition 'homology' refers to the basic
similarity of a particular structure in different organisms,
usually as result from the descent from a common ancestor.
The definition 'promoter' is intended as a nucleo-
tide sequence sufficient to direct transcription. Also
included are those promoter elements which are sufficient to
render tissue-specific gene expression; such elements may be
located in the 5' or 3' regions of the native gene.
The definition 'operably linked' is meant that a
gene and a regulatory sequence ( s ) are connected in such a
way as to permit gene expression when the appropriate
molecules (for example, transcriptional activator proteins)
are bound to the regulatory sequence(s).
The definition 'fruit' (botanically) is the ripened
ovary of a plant and its contents. The definition 'fruit'
(agronomically linguistically) is the ripened ovary and its
contents together with any structure with which they are
combined, as in case of strawberry or apple, the receptacle.
The definition 'climacteric' is pointing at the
phase of increased respiration found at fruit ripening and
at senescence. The definition 'non-climacteric' is pointing
at no such phase being present.
The definition 'antisense' RNA is an RNA sequence
which is complementary to a sequence of bases in the corres-
ponding mRNA: complementary in the sense that each base (or
majority of bases) in the antisense strand (read in the 5'
to 3' sense) is capable of pairing with the corresponding
base (G with C, A with U), in the mRNA sequence read in the
5' to 3' sense.
The definition 'sense' RNA is an RNA sequence which
is substantially homologous to at least part of the corres-
ponding mRNA sequence.
Brief description of drawings
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
13
The present invention may be more fully understood by
reference to the following description, when read together
with the accompanying drawings:
Figure 1. The proposed biosynthetic pathway for the produc-
tion of esters in fruits.
Figure 2. Volatile ester emission during strawberry fruit
development. GC-MS chromatograms (detector response: 100 _
2x106 total ion counts) of volatiles in-vivo released by
strawberry fruits (cv. Elsanta) at different stages of
development. Developmental stages: G-green; W-white; T-
turning; P- pink; R-red; DR-dark red. The five main volatile
esters detected are marked with numbers: 1-methyl hexanoate;
2-hexyl acetate; 3-hexyl butanoate; 4-octyl acetate; 5-octyl
butyrate.A GCMS chromatogram of a mature strawberry fruit of
the cultivar Elsanta. The analysis was done using the
headspace method. Esters are marked with dots.
Figure 3. The expression pattern of strawberry alcohol acyl
transferase cDNA (SLE27, SEQ ID NO:lA) in various tissues of
the cultivar Elsanta by Northern blot analysis. The Northern
blot was hybridised with a full length cDNA fragment of
SEQID:lA (SLE27).
Figure 4. The expression pattern of a lemon alcohol acyl
transferase (CLF26, SEQ ID N0:2A) in various tissues of the
lemon cultivar Mayer by Northern blot analysis. The Northern
blot was hybridised with a full length cDNA fragment of SEQ
ID 2A (CLF26).
Figure 5. Expression analysis in various tissues of the
strawberry cultivar Elsanta by Northern blots analysis. The
Northern blots where hybridised with full length cDNA
fragments corresponding to (a) a thiolase SEQ ID 4A (SLG150)
(b) a pyruvate decarboxylase SEQ ID 5A (SLH51) and (c) an
alcohol dehydrogenase SEQ ID 6A (SLB39).
CA 02353577 2001-06-O1
WO 00/327$9 PCT/NL99/00737
14
Figure 6. Strawberry genomic Southern analysis. 10 lg DNA
was digested with restriction enzymes HindIII, EcoRI, XbaI
and XhoI, separated on an agarose gel and blotted onto a
nylon filter. The DNA was hybridised with a full length 32P-
labelled strawberry alcohol aryl transferase cDNA (SLE27)
probe, corresponding to SEQ ID lA and autoradiographed.
Figure 7. A scheme of the pRSET B (Invitrogen) vector used
for the expression of various alcohol acyl transferases in
E.coli.
Figure 8. Verification of ester formation by the SLE27
(SAAT), SEQ ID NO: 1B protein using GC-MS. GC-MS chromato-
grams (detector response: 1000 - 2x106 total ion counts) of
volatiles produced with the incubation conditions as descri-
bed below. (A) Butanol and butyl acetate standards. (B) SAAT
protein + butanol + acetyl-CoA. (C) As in B, protein absent.
(D) As in B, butanol absent. (E) As in B, acetyl-CoA absent.
(F) GFP protein + butanol + acetyl-CoA. (G) Empty pRSET B
vector elute + butanol + acetyl-CoA. Other visible peaks are
impurities from the butanol substrate.
Figure 9. Western blot loaded with protein corresponding to
four different treatments carried out on the pellet, super-
natant and elute from a Ni-NTA column after expression of
strawberry alcohol acyl transferase (SLE 27, SEQ ID N0:1A)
in E. coli. As a control for the experiment the Green
Fluorescent Protein (GFP) was partially purified in the same
way and the elute from one treatment was loaded on the gel.
The blot was hybridised with a commercial antibody (Clon-
tech) raised against an epitope fused at the N- terminus of
the recombinant protein.
Figure 10. Gene expression profiles of strawberry alcohol
acyl transferase (SLE 27, SEQ ID NO: 1A)), alcohol dehydro-
genase (SLF138, SEQ ID NO: 11A), pyruvate decarboxylase (SLH
51, SEQ ID NO: SA), alcohol dehydrogenase (SLG16, SEQ ID
N0:12A) and amino transferase (SLF96, SEQ ID NO: 3A). Ex-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
pression ratios were monitered during fruit development and
ripening in the red stage vs green fruit and red fruit v.s
turning fruit.
Figure 11. SLE27 (SAAT), SEQ ID NO: 1B (SAAT)-induced [14C]-
5 acetyl-CoA incorporation in esters. (A) Flame ionization
detector (FID)-signal of unlabelled standards of 1, hexyla-
cetate; 2, 1-hexanol; 3, octylacetate and 4, 1-octanol. (B)
and (C): Radio-GC chromatograms of labelled products formed
by the SAAT protein from 0 . 1 mM [14C] -acetyl-CoA and alcohols
10 with (B) 2 mM 1-octanol or (C) 2 mM 1-hexanol.
Figure 12.
Verification of ester formation using GC-MS (A) E. coli cell
cultures expressing the strawberry alcohol acyl transferase
(SLE27, SEQ ID NO:lA). (B) E. coli cell cultures expressing
15 the empty vector cassette. Both cultures supplemented with
1-butanol.
Figure 13.
Verification of ester formation using GC-MS A) E. coli cell
cultures expressing the melon alcohol acyl transferase (MAY
5, SEQ ID N0:23 (A). (B) E. coli cell cultures expressing
the empty vector cassette. No alcohol supplements.
Figure 14.
Verification of ester formation using GC-MS (A) E. coli cell
cultures expressing the apple alcohol acyl transferase (MAY
3, SEQ ID N0:18A). (B) E. coli cell cultures expressing the
empty vector cassette. Both cultures supplemented with 1-
butanol.
Figure 15.
Verification of ester formation using GC-MS (A) E. coli cell
cultures expressing the citrus alcohol acyl transferase
(CLF26: SEQ ID N0:2A). (B) E. coli cell cultures expressing
the empty vector cassette. Both cultures supplemented with
1-butanol.
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
16
Figure 16.
Verification of ester formation using GC-MS (A) E. coli cell
cultures expressing the banana alcohol acyl transferase (MAY
2, SEQ ID N0:17A). (B) E. coli cell cultures expressing the
empty vector cassette. No alcohol supplements.
Figure 17.
Verification of ester formation using GC-MS (A) E, coli cell
cultures expressing the apple alcohol acyl transferase
(MAY3, SEQ ID N0:18A). (B) E. coli cell cultures expressing
l0 the empty vector cassette. Both cultures supplemented with
isoamylalcohol, n-butanol, cis-2-hexen-1-of and isopropanol.
Figure 18.
Verification of ester formation using GC-MS (A) E. coli cell
cultures expressing the strawberry Vesca alcohol acyl
transferase (SUN1, SEQ ID N0:16A). (B) E. coli cell cultures
expressing the empty vector cassette. Both cultures supple-
mented with isoamylalcohol, n-butanol, cis-2-hexen-1-of and
isopropanol.
Detailed description of the invention
According to a first aspect the invention provides an
insolated DNA sequence encoding
(a) a polypeptide having an amino acid sequence as
shown in SEQ ID NO: 1B or SEQ ID NO: 2B, or
(b) a polypeptide having at least 30% homology
with the amino acid sequence SEQ ID NO: 1B or
at least 40% homology with the amino acid
sequence SEQ ID NO: 2B, or
(c) a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has alcohol acyl transferase activity and is
involved in the biosynthetic pathway for ali
phatic and /or aromatic ester production in
fruit .
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
17
Preferably, the DNA sequence encodes a polypeptide
having at least 50% homology, more preferably at least 70%
homology, with any of the amino acid sequences SEQ ID NO: 1B
or SEQ ID NO: 2B, or a fragment thereof.
With reference to the nucleic acid sequence the
invention provides an isolated DNA sequence having a nucleic
acid sequence
(a) as shown in SEQ ID NO: lA or SEQ ID NO: 2A
which encodes a polypeptide having alcohol
acyl transferase activity and being involved
in the biosynthetic pathway for aliphatic
and/or aromatic ester production in fruit, or
(b) complementary to SEQ ID NO: lA or SEQ ID NO:
2A, or
(c) which has at least 25% homology with any of
the sequences of (a) or (b), or
(d) which is capable of hybridising under strin-
gent conditions to any of the sequences (a)-
(c) .
Preferably the DNA sequence has a nucleic acid
sequence
(c) which has at least 40% homology, more prefera-
bly at least 60% homology, with any of the
sequences of (a) or (b), or
(d) which is capable of hybridising under strin-
gent conditions to said sequence (c).
According to a second aspect the invention provides
a purified and isolated polypeptide,
(a) having an amino acid sequence as shown in SEQ
ID NO: 1B or SEQ ID NO: 2B, or
(b) having at least 30% homology with the amino
acid sequence SEQ ID NO: 1B or at least 40%
homology with the amino acid sequence SEQ ID
NO: 2B, or
(c) being a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has alcohol acyl transferase activity and is
involved in the biosynthetic pathway for ali-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
18
phatic and/or aromatic ester production in
fruit.
Preferably, the polypeptide has at least 50%
homology, more preferably at least 70% homology, with any of
the amino acid sequences SEQ ID NO: 1B or SEQ TD NO: 2B, or
a fragment thereof.
It may be advantageous to produce nucleotide
sequences according to the first aspect of the invention as
defined above or derivatives thereof possessing a substanti-
ally different codon usage. It is known by those skilled in
the art that as a result of degeneracy of the genetic code,
a multitude of gene sequences, some bearing minimal homology
to the nucleotide sequences of any known and any naturally
occurring genes may be produced. The invention contemplates
each and every possible variation of the nucleotide sequen-
ces that could be made by selecting combinations based on
possible codon choices. These combinations are made in ac-
cordance with the standard triplet genetic code as applied
to the nucleotide sequence of naturally occurring gene
sequences, and all such variations are to be considered as
being specifically disclosed. In addition the nucleotide
sequences of the invention may be used in molecular biology
techniques that have not been developed, providing the new
techniques rely on properties of nucleotide sequences that
are currently known, including but are not limited to such
properties such as the triplet genetic code and specific
base pair interactions.
Altered nucleic acid sequences of this invention
include deletions, insertions, substitutions of different
nucleotides resulting in the polynucleotides that encode the
same or are functionally equivalent. Deliberate amino acid
substitution may be made on the basis of similarity in
polarity, charge, solubility, hydrophobicity, and/or the
amphipathetic nature of the residues as long as the biolo-
gical activity of the polypeptide is retained. Included in
the scope of the present invention are alleles of the
polypeptides according to the second aspect of the invention
as defined above. As used herein, an 'allele' or 'allelic
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
19
sequence' is an alternative form of the polypeptides descri-
bed above. Alleles result from a mutation, eg a change in
the nucleic acid sequence, and generally produce altered
mRNA or polypeptide whose structure or function may or may
not be altered. Any given polypeptide may have none, or more
allelic forms. Common allelic changes that give rise to
alleles are generally ascribed to natural deletions, additi-
ons or substitutions of amino acids. Each of these types of
changes may occur alone, or in combination with the others,
one or more times in a given sequence.
It is envisaged that the polynucleotide sequences
of the present invention can be used as probes for the
isolation of similar sequences from the strawberry genome
and lemon genome. By using as a probe a cDNA of the gene
sequences of the invention it would be possible by those
skilled in the art to obtain comparable gene sequences from
various tissues, particularly fruit tissues, from climacte-
ric and non climacteric plants. One aspect of the invention
is to provide for hybridisation or PCR probes which are
capable of detecting polynucleotide sequences, including
genomic sequence(s), encoding the polypeptides of the
invention, or closely related molecules. The specificity of
the probe, whether it is made from a highly specific region,
eg 10 unique nucleotides in the 5' regulatory region, or a
less specific region, e.g. in the 3' region, and the strin-
gency of the hybridisation or amplification (maximal, high,
intermediate, low) will determine whether the probe identi-
fies only naturally occurring sequences) encoding the
polypeptide, allele's or related sequences.
Additional alcohol acyltransferase sequences from
melon, strawberry, vesca, banana, apple, mango and lemon
were obtained using primers. These additional sequences are
covered by the first aspect of invention. The nucleic acid
sequences are shown in SEQ ID NO: 16A to 23A, respectively.
The amino acid sequences are shown in SEQ ID N0:16B to 23B,
respectively. Apart from the applications as disclosed
herein said sequences can be used in combination for the
synthesis of a broader range of esters via recombinant means
CA 02353577 2001-06-O1
WO 00/32789 PCTML99/00737
or via transgenic approaches and/or for the synthesis of new
flavours.
Probes may also be used for the detection of
related sequences and preferably contain at least 50% of any
5 of the nucleotides from any one of the gene encoding sequen-
ces according to the present invention. For gene sequences
of the present invention, the hybridisation probes may be
derived from the nucleotide sequence, or from genomic
sequence including promoter, enhancer element and introns.
10 Hybridisation probes may be labelled by a variety of repor-
ter groups, including radionuclides such as 32P or 355, or
enzymatic labels such as alkaline phosphatase coupled to the
probe via avidin/biotin coupling systems, and the like.
Other means for producing specific hybridisation
15 probes for encoding gene sequences of the invention include
the cloning of nucleic acid Sequences into suitable vectors
for the production of mRNA probes. Such vector are known in
the art and are commercially available and may be used to
synthesise RNA probes in-vitro by means of the addition of
20 the appropriate RNA polymerase as T7 or SP6 RNA polymerase
and the appropriately radioactively labeled nucleotides.
According to a third aspect the invention provides
a recombinant expression vector comprising a coding sequence
which is operably linked to a promoter sequence capable of
directing expression of said coding sequence in a host cell
for said vector, and a transcription termination sequence,
in which the coding sequence is a DNA sequence according to
the invention.
In a further embodiment the invention provides a
replicative cloning vector comprising an isolated DNA
sequence of the invention and a replicon operative in a host
cell for said vector.
Methods which are well known to those skilled in
the art can be used to construct expression vectors contai
ning the gene coding sequence of the invention, and approp
riate transcriptional and translational controls. These
methods include in-vitro recombinant techniques. Such
techniques are described in Sambrook et al., 1989. Molecular
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
21
cloning a laboratory manual, cold spring Harbour press,
Plain view, NY and Ausubel FM et al., (1989) Current proto-
cols in molecular biology, John Wiley and Sons, New York,
NY.
In order to express a biologically active polypep-
tide, the nucleotide sequence encoding the polypeptide, or
the functional equivalents thereof, fragments of the
polypeptide, is inserted into the appropriate expression
vector (i.e. a vector that contains the necessary elements
for the transcription or translation of the inserted coding
sequence). Specific initiation signals may also be required
for efficient translation of the polypeptides of the inven-
tion. These signals include the ATG initiation codon and
adjacent sequences. In cases where the polypeptides, their
initiation codons and upstream sequences are inserted into
the appropriate expression vector, no additional translatio-
nal control systems including the ATG initiation codon must
be provided. Furthermore, the initiation codon must be in
the correct reading frame to ensure transcription of the
entire insert. Exogenous transcriptional elements and
initation codons can be of various origins, both natural and
synthetic. The efficiency of expression may be enhanced by
the inclusion of enhancers appropriate to the cell system in
use (Scharf D et al (1994) Results Prob Cell Differ 20: 125-
62; Bittner et a1. (1987) Methods in Enzymol 153: 516-544).
In addition a host strain may be chosen for its
ability to modulate the expression of the inserted sequences
or to process the expressed protein in the desired fashion.
Such modifications of the polypeptide include, but are not
limited to, acylation, carboxylation, glycosylation, phos-
phorylation and lipidation. Past translation processing
which cleaves a 'prepro' form of the protein may also be
important for correct insertion, folding and/or function.
Different host cells which have the correct cellular machi-
nery and characteristic mechanisms for such post-translatio-
nal activities maybe chosen to ensure correct modification
and processing of the introduced, foreign protein.
CA 02353577 2001-06-O1
WO 00/32789 PCT/Ni.99/00737
22
The nucleotide sequences of the present invention
can be engineered in order to alter the coding sequence for
a variety of reasons, including but not limited to, altera-
tions which modify the cloning, processing, and/or expres-
s sion of the gene product. For example mutations may be
introduced using techniques which are well known in the
state of the art, e.g. site directed mutagenesis to insert
new restriction sites, to alter glycosylation patterns, to
change codon usage, to produce splice variants etc. Codons
may be selected to increase the rate at which expression of
the peptide occurs in a particular procaroytic or eukaroytic
expression host in accordance with the frequency with which
particular codons are utilised by the host (hurray E et al.
(1989) Nuc Acids Res 17: 477-508). Other reasons for sub-
stantially altering the nucleotide sequences) of the
invention and their derivatives, without altering the
encoded amino acid sequences include the production of RNA
transcripts having more desirable properties, such as a
greater half life, than transcripts produced from naturally
occurring sequences.
In developing the expression cassette, the various
fragments comprising the regulatory regions and open reading
frame may be subjected to different processing conditions,
such as ligation, restriction enzyme digestion, resection,
in-vitro mutagenesis, primer repair, use of linkers and
adapters and the like. Thus, nucleotide transitions, trans-
versions, insertions, deletions and the like, may be perfor-
med on the DNA which is employed in the regulatory regions
and/or open reading frame. The expression cassette may be
wholly or partially derived from natural sources endogenous
to the host cell. Furthermore, the various DNA constructs
(DNA sequences, vectors, plasmids, expression cassettes) of
the invention are isolated and/or purified, or synthesised
and thus are not naturally occurring. The invention further
contemplates the use of yet undescribed biological and non
biological based expression systems and novel hosts)
systems that can be can be utilised to contain and express
the gene coding sequences) of the invention.
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
23
'Antisense' or 'partial sense' or other techniques
may also be used to reduce the expression of genes in the
pathway leading to the production of flavour volatile
esters. Full length 'sense' techniques may be designed to
increase or reduce the expression of genes in the pathway
leading to the production of flavour volatiles. The 'anti-
sense' or 'partial sense' molecules may be designed to block
translation of mRNA by preventing the transcript from
binding to the ribosomes. Ribozymes are enzymatic RNA
molecules capable of catalysing the specific cleavage of
RNA. The mechanism of ribozyme action involves sequence-
specific hybridisation of the ribozyme molecule to comple-
mentary target RNA, followed by endonucleolytic cleavage.
Within the scope of the invention are engineered hammerhead
motif ribozyme molecules that can specifically and effi-
ciently catalyse endonucleolytic cleavage of gene sequences
of the invention. Specific ribozyme cleavage sites within
any potential RNA target are initially identified by scan-
ning the target molecule for ribozyme cleavage sites which
include the following sequences, GUA, GUU and GUC. Once
identified, short RNA sequences of between 15 and 20 ribonu-
cleotides corresponding to the region of the target gene
containing the cleavage site may be evaluated for secondary
structural features which may render the oligonucleotide
inoperable. The suitability of candidate targets may also be
evaluated by testing accessibility to hybridisation with
complementary oligonucleotides using ribonuclease protection
assays.
'Antisense' molecules and ribozymes of the inven
tion may be prepared by any method known in the art for the
synthesis of RNA molecules. These include techniques for
chemically synthesizing oligonucleotides such as solid phase
phosphoramidite chemical synthesis. Alternatively, RNA
molecules may be generated by in-vitro and in-vivo trans
cription of DNA sequences of the invention. Such DNA sequen-
ces may be incorporated into a wide variety of vectors with
suitable RNA palymerase promoters such as T7 and SP6.
Alternatively, antisense cDNA constructs that synthesise
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
24
antisense RNA constitutively or inducibly can be introduced
into cell lines, cells or tissues. RNA molecules may be
modified to increase intracellular stability and half-life.
Possible modifications include, but are not limited to, the
addition of flanking sequences, at the 5' and/or 3' end of
the molecule or the use of phosphorothioate or 2' O-methyl
rather than phosphodiesterase linkages within the backbone
of the molecule.
A variety of vector/host expression systems can be
utilised to contain and express the gene coding sequences of
the invention. These include and are not limited to micro
organisms such as bacteria (e. g. E coli, B subtilis, Strep
tomyces, Pseudomonads) transformed with recombinant bacteri
ophage, plasmid or cosmid DNA expression systems, yeast (e.g
S. cerevisiae, Kluyveromyces lactis, Pichia pastoris,
Hansenula polymorpha, Schizosacch. Pombe, Yarrowia) trans-
formed with yeast expression vectors; filamentous fungi
(aspergillus nidulans, aspergillus orizae, aspergillus
niger) transformed with filamentous fungi expression vec-
tors, insect cell systems transfected with virus expression
vectors (eg baculovirus, adenovirus, herpes or vaccinia
viruses); plant cell systems transfected with virus expres-
sion vectors (e. g. cauliflower mosaic virus, CaMV, tobacco
mosaic virus, TMV) or transformed with bacterial expression
vectors (e.g Ti or Pbr322 plasmid); or mammalian cell
systems (chinese hamster ovary (CHO), baby hamster kidney
(BHK), Hybridoma's, including cell lines of mouse, monkey,
human and the like.
In the case of plant expression vectors, the
expression of a sequence (s) of the invention may be driven
by a number of previously defined promoters, including
inducible and developmentally regulated promoters. The
invention further contemplates the use of the individual
promoters of the polynucleotide sequences) of the present
invention for this purpose. In particular an alcohol acyl
transferase promoters) pertaining to SEQ ID IA and SEQ ID
2A or any promoters particularly responsive to ripening
events may be used to drive the tissue specific expression
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
of the target genes. In addition, viral promoters such as
the 358 and the 198 promoters of CaMV (Brisson et al., 1984;
Nature, 310: 511 -514) may be used alone or in combination
with the omega leader sequence from TMV (Takamatsu et al.
5 (1987); EMBO J 6:307-311). Promoters or enhancers derived
from the genomes of plant cells, tissue specific promoters
i.e fruit specific promoters, Fbp7 (Columbo et a1. 1997;
Plant Cell 9; 703-715), 2A11 promoter (Pear et al., 1989,
Plant Molecular Biology, 13:639-651), small subunit of
10 Rubisco (Corruzzi et al., 1984; EMBO J 3:16; Brogue et al.,
1984 Science 224:838-843) or timing specific promoters such
as ripening specific promoters (the E8 promoter, Diekman and
Fisher, 1988, EMBO J, 7:3315-3320) may be used. Suitable
terminator sequences include that of the agrobacterium
15 tumefaciens nopaline synthase gene (Nos 3' end), the tobacco
ribulose bisphosphate carboxylase small subunit termination
region; and other 3' regions known to those skilled in the
art. These constructs can be introduced into plant cells by
direct DNA transformation, or pathogen mediated transfec-
20 tion. For reviews of such techniques, see Hobbs S or Murry
LE, in McGraw Hill yearbook of Science and technology
(1992), Mc Graw Hill NY, PP 191-196 or Weissbach and Weiss-
bach (1988) Methods for Plant Molecular Biology, Academic
Press, New York, NY, pp421-463, Fillatti et al., Biotech-
25 nology, 5: 726-730. Manipulation of DNA sequences in plant
cells may be carried out using the Cre/lox site specific
recombination system as outlined in patent application W091-
09957.
According to a fourth aspect the invention provides
a method for regulating aliphatic and/or aromatic ester
formation in fruit, comprising inserting into the genome of
a fruit-producing plant one or more copies of one or more
DNA sequences of the invention.
The activity of genes according to the present
invention involved in the biosynthetic pathway leading to
volatile ester formation, hence flavour, may be either
increased or reduced depending on the characteristics
desired for the modified plant part. The gene sequence may
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
26
be in the same 'sense' or 'antisense' orientation as the
endogenous target gene. Methods well known to those skilled
in the art can be used to construct recombinant vectors
which will express 'sense' or 'antisense' polynucleotides
encoding the gene sequences of the present invention. Such
technology is now well known in the art. In the case of the
'plant cell' officiating as 'host cell', the target 'plant
cell' may be part of a whole plant or may be an isolated
cell or part of a tissue which may be regenerated into a
whole plant. The target plant may be selected from any mono-
cotyledonous or dicotyledonous plant species. Suitable
plants include any fruit bearing plant such as strawberry,
citrus (lemon), banana, apple, pear, melon, sweet pepper,
peach or mango. Other suitable plant hosts include vegeta-
ble, ornamental (to include sunflower) and arable crops (to
include soybean, sunflower, corn, peanut, maize, wheat,
cotton, safflower, and rapeseed). For any particular plant
cell, the gene sequences, cDNA, genomic DNA, or synthetic
polynucleotide, used in the transformation vector construct,
may be derived from the same plant species, or may be
derived from other plant species (as there will be suffi-
cient homology to allow modification of related enzyme gene
function) . The procedure or method for preparing a transfor-
mant can be performed according to the conventional techni-
que used in the fields of molecular biology, biotechnology
and genetic engineering.
According to a fifth aspect the invention provides
a plant and propagating material thereof which contains in
its genome a DNA sequence of the invention or a vector as
defined above.
In a preferred embodiment the invention provides a
genetically modified strawberry or lemon plant and propaga-
ting material derived therefrom which has a genome compri-
sing an expression vector for over-expression or down-
regulation of an endogenous strawberry or lemon plant gene
counterpart of any of the DNA sequences of the invention.
In mammalian cells a number of viral based systems
may be utilised. In cases where the adenovirus is used as an
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
27
expression vector, a gene coding sequences) of the inventi-
on may be ligated into the adenovirus transcription/transla-
tion complex consisting of the late promoter and tripartite
leader sequence. Insertion in a non essential E1 or E3
region of the viral genome will result in a variable virus
capable of expressing the gene sequences) in infected host
cells (Logan and Shark (1984) Proc Natl Acad. Sc.i 81:3655-
59) Tn addition transcription enhancers, such as the Rous
sarcoma virus (RSV) enhancer, may be used to increase
expression in mammalian host cells.
Furthermore insect cells such as silkworm cells or
larvae themselves may be used as a host. In one such system,
Autographa californica nuclear polyhedrosis virus (AcNPV) is
used as a vector to express foreign genes in Spodoptera
frugiperda cells or in Trichoplusia larvae. The gene coding
sequences) of the invention may be cloned into the nones-
sential region of the virus, such as the polyhedrin gene,
and placed under control of a polyhedrin promoter. Succes-
sful insertion of the sequences) will render the polyhedrin
gene inactive and produce recombinant virus lacking coat
protein coat. The recombinant viruses are then used to
infect S frugiperda cells or Trichoplusia larvae in which
the gene sequence or sequences are expressed (Smith et a1.
(1993) J Virol 46:584; Engelhard et al. (1994) Proc. Natl
acad Sci, 91: 3224-7).
The control elements or regulatory sequences of the
described systems vary in their strength and specificities
and are those nontranslated regions of the vector, enhan-
cers, promoters and 3' untranslated regions, which interact
with host cellular proteins to carry out transcription and
translation. Dependent on the vector system and host utili-
sed, any number of suitable transcription and translation
elements, including constitutive and inducible promoters,
may be used. For example, when cloning in bacterial systems,
inducible promoters such as the hybrid lacZ promoter of the
Bluescript ° phagemid (Strategene, La Jolla) or pSPORT
(Gibco BRL) and ptrp-lac hybrids and the like may be used.
Other conventionally used promoters are trc and trc promo-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
28
ters. The baculovirus polyhedrin promoter may be used for
insect cells. As a promoter for filamentous organisms, for
example, amalyse, trp C, and the like may be used. As a
promoter for yeast for example glyceraldehyde-3-phosphate
dehydrogenase may be used. As a promoter for animal cell
hosts, viral promoters such as the SV40 early promoter, SV40
late promoter and the like may be used. In the yeast Saccha-
romyces cerevisiae, a number of vectors containing constitu-
tive or inducible promoters such as the alpha factor,
alcohol oxidase and PGH may be used. For review, see Grant
et al., 1987 Methods in Enzymology 153: 516-544.
For long term, high yield production of recombi-
nant proteins, stable expression is preferred. For example
cell lines which stably express the polypeptides of the
invention may be transformed using expression vectors which
contain viral origins of replication or endogenous expressi-
on elements and a selectable marker gene. Following the
introduction of the vector, cells may be allowed to grow for
one to two days in an enriched media before they are swit-
ched to selective media. The purpose of the selectable
marker is to confer resistance to selection and its presence
allows growth and recovery of cells which successfully
express the introduced sequences. Resistant clumps of stable
transformed cells can be proliferated using tissue culture
techniques appropriate to the cell type.
Although the presence/absence of the marker gene
expression suggests that the gene of interest is also
present, its presence and expression must be confirmed. For
example, if the gene sequences) of the invention are
inserted within a marker gene sequence, recombinant cells
can be identified by the absence of gene function. Alterna-
tively a marker gene can be placed in tandem with the gene
sequence of interest, under the control of a single pro-
moter. Expression of the marker gene in response to induc-
tion or selection usually indicates expression of the tandem
gene as well.
Any number of selection systems may be used to
recover the transformed cell lines. Anti-metabolite, antibi-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
29
otic or herbicide resistance can be used as the basis of
selection; for example, dhfr which confers resistance to
methotrextate (Wiger M. et al. (1980) Proc Natl Acad Sci 77:
3567-70; npt, which confers resistance to the aminoglycosi-
des neomycin and G-418 (Colbere-Garapin F. et al. (1981) J
Mo1 Bio1 150: 1-14) and als and pat, which confer resistance
to chlorsulfuron and phosphinotricin acyl transferase,
respectively (hurry supra). Additional selectable genes have
been transcribed, for example, trp8, which allows cells
utilise indole in place of tryptophan, or His D, which
allows cells to utilise histinol in place of histidine
(Hartman SC and RC Mulligan (1988) Proc Nat1 Acad Sci 85:
8047-51). Alternatively one could use visible markers such
as anthocyanidins,l3-glucuronidase and its substrates, GUS,
GFP and variants, and luciferase and its substrate, lucife-
rin, which are widely used to identify transformants, but
also to quantify the amount of stable protein expression
attributable to a specific vector system (Rhodes CA et al.
(1995) Methods Mo1 Biol 55: 121-131.
Host cells transformed with a nucleotide sequen-
ces) of the invention, may be cultured under conditions
suitable for expression and recovery of the encoded protein
from cell culture. The protein produced by recombinant cells
may be secreted or contained intracellularly depending on
the sequence and/or the vector used. As will be understood
by those skilled in the art, expression vectors containing
polynucleotides sequences of the invention, can be designed
with signal sequences for direct secretion of the protein
product, through a prokaroytic or eukaryotic cell membrane.
Other recombinant constructions may join the gene sequences
of the invention, to a nucleotide sequence encoding a
polypeptide domain, which will facilitate purification of
soluble proteins (Kroll DJ et a1. (1993). DNA Cell Biol 12:
441-53).
According to a sixth aspect the invention provides
a method for producing aromatic and/or aliphatic esters in
microorganisms, plant cells or plants comprising inserting
into the genome of the microorganism or plant one or more
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
copies of DNA sequences of the invention, and feeding an
alcohol and an acyl-CoA to the microorganism or plant
(cell) .
Alternatively, host cells which contain the gene
5 sequences or polypeptides thereof according to the invention
may be identified by a variety of procedures known to those
skilled in the art. These procedures include, but are not
limited to DNA-DNA, DNA-RNA hybridisation, amplification
using probes (portions or fragments of polynucleotides),
10 protein bioassay or immunoassay techniques which include
membrane, solution or chip based technologies for the
detection and /or quantification of the nucleic acid or
protein.
Nucleic acid amplification based assays involve the
15 use of oligonucleotides or oligomers based on the gene
sequences of the invention to detect transformants contai-
ning DNA or RNA encoding the polypeptides . As used herein
'oligonucleotides' or 'oligomers' refer to nucleic acid
sequence of at least l0 nucleotides and as many as about 60
20 nucleotides, preferably about 15-30 nucleotides, and more
preferably about 20-25 nucleotides which can be used as a
probe or amplimer. Polynucleotide sequences according to the
invention encoding alcohol acyl transferase polypeptides may
be used to detect volatile ester (aliphatic and aromatic)
25 formation in fruits. PCR as described in US patent Nos
4,683,195 and 4,965,188, provides additional uses for
oligonucleotides based on the alcohol acyl transferase
sequence. Such oligomers are chemically synthesised, but
they may be generated enzymatically or produced from a
30 recombinant source. Oligomers generally comprise two nucleo-
tide sequences, one with sense orientation (5'-3') and one
with antisense (3'-5'), employed under less stringent
conditions for detection and /or quantitation of closely
related DNA or RNA sequences. For example, polynucleotide
sequences encoding the alcohol acyl transferase protein may
be used in the hybridisation or PCR assay of plant tissues,
more specifically fruit tissues, to detect alcohol acyl
transferase protein presence. The form of such quantitative
CA 02353577 2001-06-O1
WO 00/32789 PC'T/NL99/00737
31
methods may include, Southern or Northern analysis, dot/slot
blot or other membrane based technologies; PCR technologies
such as DNA Chip, Taqman and ELISA Technologies. All of
these technologies are well known in the art and are the
basis of many commercially owned diagnostic kits. The
polynucleotide sequences according to the present invention
herein provide the basis for assays that detect signal
transduction events associated with volatile ester producti-
on, aliphatic and aromatic, hence flavour formation.
A variety of protocols for detecting and measuring
expression of the polypeptides of the invention, using
either polyclonal or monoclonal antibodies specific for the
protein are well known in the art. Examples include enzyme-
linked immunosorbent assay (ELISA), radioimmunoassay (RIA)
and fluorescent activated cell sorting (FRCS). These methods
are described, among other places, in Hampton R et al.
(1990), Serological methods, a laboratory manual, APS Press,
St Paul MN) and Maddox DE et aI. (1993, J Exp Med 158:1211).
A variety of labels and conjugation techniques are
known by those skilled in the art and can be used in various
nucleic acid and amino acid assays. Means for producing
labelled hybridisation or PCR probes for detecting sequences
related to polynucleotides as outlined in the first and
second aspects of the invention, include oligolabeling, nick
translation, end labelling of PCR amplification using a
labelled nucleotide. Alternatively, the gene sequence or
sequences of the invention, or any portion of it, may be
cloned into a vector for the production of the mRNA probe.
Such vectors are known in the art, are commercially availa-
ble, and may be used to synthesise RNA probes in-vitro by
addition of an appropriate RNA polymerase, such as T7, T3,
or SP6 and labelled nucleotides. A number of companies such
as Pharmacia Biotech (Piscataway NJ), promega (Madisson WI),
and US Biochemical Corp (Cleveland OH) supply commercial
kits and protocols for these procedures. Suitable reporter
molecules or labels include fluorescent, chemiluminescent,
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
32
or chromogenic agents as well as substrates, cofactors,
inhibitors, magnetic particles and the like.
According to another aspect of the invention,
natural, modified or recombinant polynucleotide sequences
according to the invention may be ligated to a heterologous
sequence to encode a fusion protein. A fusion protein may be
engineered with one or more additional domains added to
facilitate protein purification (i.e, a cleavage site
located between the sequence of the polypeptide and the
heterologous protein sequence) so that the polypeptide of
the invention may be purified away from the heterologous
moiety. The invention further contemplates the creation of
fusion proteins, chosen from the group of sequences of the
invention, degenerate variants thereof, functional equiva-
lents thereof, pertaining to 'fruit' of strawberry and
lemon, or gene sequences with substantial homology to the
sequences of the invention from other 'fruit' tissues
concerned with the pathway for the formation of aliphatic or
aromatic esters, hence flavour. Such purification facilita-
ting domains include, but are not limited to, metal chela-
ting peptides such as histidine-tryptophan modules that
allow purification on immobilised metals, protein A domains
that allow purification on immobilised immunoglobin, and the
domain utilised in the FLAGS extension/affinity purification
system (Immunex Corp, Seattle, WA). The introduction of a
cleavable linker such as factor XA, thrombin or enterokinase
(Invitrogen, San Diego CA) between the purification domain
and the polypeptide is useful to facilitate purification.
One such expression vector provides for the expression of a
fusion protein comprising gene sequences) which encode
polypeptides of the invention, and contains nucleic acid
encoding six histidine residues followed by thioredoxin and
the enterokinase cleavage site. The histidine residues
facilitate purification on IMIAC (immobilised metal ion
affinity chromatography as described in Porath et al.,
(1992) Protein Expression and Purification 3:263-281) while
the enterokinase cleavage site provides a means for purify-
ing the polypeptide from the fusion protein.
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
33
According to a seventh aspect the invention provi-
des an isolated DNA sequence encoding
(a) a polypetide having an amino acid sequence as
shown in SEQ ID NO: 3B, or
(b) a polypeptide having at least 80% homology
with the amino acid sequence SEQ ID NO: 3B, or
(c) a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has aminotransferase activity and is involved
in the biosynthetic pathway for aliphatic
and/or aromatic ester production in fruit.
In particular the invention provides an isolated
DNA sequence having a nucleic acid sequence
(a) as shown in SEQ ID NO: 3A which encodes a
polypeptide having aminotransferase activity
and being involved in the biosynthetic pathway
for aliphatic and/or aromatic ester production
in fruit, or
(b) complementary to SEQ ID NO: 3A, or
(c) which has at least 70 % homology with any of
the sequences of (a) or (b), or
(d) which is capable of hybridising under strin-
gent conditions to any of the sequences (a) -
(c) .
Further the invention provides a purified and
isolated polypeptide,
(a) having an amino acid sequence as shown in SEQ
ID NO: 3B, or
(b) having at least 80% homology with the amino
acid sequence SEQ ID NO: 3B, or
(c) being a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has aminotransferase activity and is involved
in the biosynthetic pathway for aliphatic
and/or aromatic ester production in fruit.
According to an eighth aspect the invention provi-
des an isolated DNA sequence encoding
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
34
(a) a polypeptide having an amino acid sequence as
shown in SEQ ID NO: 4B, or
(b) a polypeptide having at least 90% homology
with the amino acid sequence SEQ ID NO: 4B, or
(c) a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has thiolase activity and is involved in the
biosynthetic pathway for aliphatic and/or
aromatic ester production in fruit.
In particular the invention provies an isolated DNA
sequence having a nucleic acid sequence
(a) as shown in SEQ ID NO: 4A which encodes a
polypeptide having thiolase activity and being
involved in the biosynthetic pathway for ali-
phatic and/or aromatic ester production in
fruit, or
(b) complementary to SEQ ID NO: 4A, or
(c) which has at least 75 % homology with any of
the sequences of (a) or (b), or
(d) which is capable of hybridising under strin-
gent conditions to any of the sequences (a) -
(c) .
Further the invention provides a purified and
isolated polypeptide,
(a) having an amino acid sequence as shown in SEQ
ID NO: 4B, or
(b) having at least 90% homology with the amino
acid sequence SEQ ID NO: 4B, or
(c) being a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has thiolase activity and is involved in the
biosynthetic pathway for aliphatic and/or
aromatic ester production in fruit.
According to a ninth aspect the invention provides
an isolated DNA sequence encoding
(a) a polypetide having an amino acid sequence as
shown in SEQ ID NO: 5B, or
CA 02353577 2001-06-O1
WO 00/32789 PCT/N1.99/00737
(b) a polypeptide having at least 90% homology
with the amino acid sequence SEQ ID NO: 5B, or
(c) a fragment of polypeptide (a) or (b) ,
which polypeptide or fragment of polypeptide
5 has pyruvate decarboxylase activity and is
involved in the biosynthetic pathway for ali
phatic and/or aromatic ester production in
fruit.
In particular the invention provides an isolated
10 DNA sequence having a nucleic acid sequence
(a) as shown in SEQ ID NO: 5A which encodes a
polypeptide having pyruvate decarboxylase
activity and being involved in the biosynthe-
tic pathway for aliphatic and/or aromatic
15 ester production in fruit, or
(b) complementary to SEQ ID NO: 5A, or
(c) which has at least 75 % homology with any of
the sequences of (a) or (b), or
(d) which is capable of hybridising under strin
20 gent conditions to any of the sequences (a)
(c) .
Further the invention provides a purified and
isolated polypeptide,
(a) having an amino acid sequence as shown in SEQ
25 ID NO: 5B, or
(b) having at least 90% homology with the amino
acid sequence SEQ ID NO: 5B, or
(c) being a fragment of polypeptide (a) or (b), which
polypeptide or fragment of polypeptide has
30 pyruvate decarboxylase activity and is invol
ved in the biosynthetic pathway for aliphatic
and/or aromatic ester production in fruit.
According to a tenth aspect the invention provides
an isolated DNA sequence encoding
35 (a) a polypetide having an amino acid sequence as
shown in SEQ ID NO: 6B, or
(b) a polypeptide having at least 75% homology
with the amino acid sequence SEQ ID NO: 6B, or
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
36
(c) a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has alcohol dehydrogenase activity and is
involved in the biosynthetic pathway for ali-
phatic and/or aromatic ester production in
fruit.
In particular the invention provides an isolated
DNA sequence having a nucleic acid sequence
(a) as shown in SEQ ID NO: 6A which encodes a
polypeptide having alcohol dehydrogenase acti
vity and being involved in the biosynthetic
pathway for aliphatic and/or aromatic ester
production in fruit, or
(b) complementary to SEQ ID NO: 6A, or
(c) which has at least 65 % homology with any of
the sequences of (a) or (b), or
(d) which is capable of hybridising under strin-
gent conditions to any of the sequences (a) -
(C) .
Further the invention provides a purified and
isolated polypeptide,
(a) having an amino acid sequence as shown in SEQ
ID NO: 6B, or
(b) having at least 75% homology with the amino
acid sequence SEQ ID NO: 6B, or
(c) being a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has alcohol dehydrogenase activity and is
involved in the biosynthetic pathway for ali-
phatic and/or aromatic ester production in
fruit .
According to an eleventh aspect the invention
provides an isolated DNA sequence encoding
(a) a polypeptide having an amino acid sequence
selected from the group consisting of sequen
ces SEQ ID NO: 7B, 8B, 9B and lOB, or
(b) a polypeptide having at least
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
37
i) 55% homology with the amino acid sequence
of a 326 as fragment from the C termi-
nal end of the coding sequence of SEQ
ID NO: 7B, or
ii) 75% homology with the amino acid se-
quence of a 278 as fragment from the C
terminal end of the coding sequence of
of SEQ ID NO: 8B, or
iii) 65% homology with the amino acid se-
quence of a 284 as fragment from the C
terminal end of the coding sequence of
SEQ TD NO: 9B, or
iv) 80% homology with the amino acid se-
quence of a 188 as fragment from the C
terminal end of the coding sequence of
SEQ ID NO: lOB, or
(c) a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has alcohol dehydrogenase activity and is
involved in the biosynthetic pathway for ali-
phatic and/or aromatic ester production in
fruit.
In particular the invention provides an isolated
DNA sequence having a nucleic acid sequence
(a) selected from the group consisting of sequen-
ces SEQ ID NO: 7A, 8A, 9A and 10A, which enco-
des a polypeptide having alcohol dehydrogenase
activity and being involved in the biosynthe-
tic pathway for aliphatic and/or aromatic
ester production in fruit, or
(b) complementary to SEQ TD N0: 7A, 8A, 9A or 10A,
or
(c) which has at least 55% homology, preferably
at
least 65% homology, with any of the sequences
of (a) or (b), or
(d) which is capable of hybridising under strin-
gent conditions to any of the sequences (a)
-
(c) .
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
38
Further the invention provides a purified and
isolated polypeptide,
(a) having an amino acid sequence selected from
the group consisting of sequences SEQ ID NO:
7B, 8B, 9B and lOB, or
(b) having at least
i) 55~ homology with the amino acid se-
quence of a 326 as fragment from the C
terminal end of the coding sequence of
of SEQ ID NO: 7B
ii) 75% homology with the amino acid se-
quence of a 278 as fragment from the C
terminal end of the coding sequence of
SEQ ID NO: 8B, or
iii) 65o homology with the amino acid se-
quence of a 284 as fragment from the C
terminal end of the coding sequence of
SEQ ID NO: 9B, or
iv) 80% homology with the amino acid se
quence of a 188 as fragment from the C
terminal end of the coding sequence of
SEQ ID NO: lOB, or
(c) being a fragment of polypeptide (a) or (b},
which polypeptide or fragment of polypeptide
has alcohol dehydrogenase activity and is
involved in the biosynthetic pathway for ali-
phatic and/or aromatic ester production in
fruit.
According to a twelfth aspect the invention provi-
des an isolated DNA sequence encoding
(a) a polypeptide having an amino acid sequence as
shown in SEQ ID NO: 11B, or
(b) a polypeptide having at least 75~ homology
with the amino acid sequence of a 181 as frag
ment from the C terminal end of the coding
sequence of SEQ ID N0: 11B, or
(c) a fragment of polypeptide (a) or (b),
CA 02353577 2001-06-O1
WO 00/327$9 PCT/NL99/00737
39
which polypeptide or fragment of polypeptide
has alcohol dehydrogenase activity and is
involved in the biosynthetic pathway for ali
phatic and/or aromatic ester production in
fruit.
In particular the invention provides an isolated
DNA sequence having a nucleic acid sequence
(a) as shown in SEQ ID NO: 11A which encodes a
polypeptide having alcohol dehydrogenase acti
vity and being involved in the biosynthetic
pathway for aliphatic and/or aromatic ester
production in fruit, or
(b) complementary to SEQ ID NO: 11A, or
(c) which has at least 48 ~ homology with any of
the sequences of (a) or (b), or
(d) which is capable of hybridising under strin-
gent conditions to any of the sequences (a)
(c) .
Further the invention provides a purified and
isolated polypeptide,
(a) having an amino acid sequence as shown in SEQ
ID NO: 11B, or
(b) having at least 75~ homolagy with the amino
acid sequence of a 181 as fragment from the 3'
end of SEQ ID NO: 11B, or
(c) being a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has alcohol dehydrogenase activity and is
involved in the biosynthetic pathway for ali-
phatic and/or aromatic ester production in
fruit.
According to a thirteenth aspect the invention
provides an isolated DNA sequence encoding
(a) a polypeptide having an amino acid sequence
selected from the group consisting of sequen
ces SEQ ID NO: 12B and 13B, or
(b) a polypeptide having at least
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
i) 55o homology with the amino acid sequence
of a 176 as fragment from the C termi-
nal end of the coding sequence of SEQ
ID NO: 12B, or
5 ii) 35% homology with the amino acid se-
quence of a 284 as fragment from the C
terminal end of the coding sequence of
SEQ ID NO: 13B, or
(c) a fragment of polypeptide (a) or (b),
10 which polypeptide or fragment of polypeptide
has alcohol dehydrogenase activity and is
involved in the biosynthetic pathway for ali-
phatic and/or aromatic ester production in
fruit.
15 In particular the invention provides an isolated
DNA sequence having a nucleic acid sequence
(a) selected from the group consisting of sequen-
ces SEQ ID NO: 12A and 13A, which encodes a
polypeptide having alcohol dehydrogenase acti-
20 vity and being involved in the biosynthetic
pathway for aliphatic and/or aromatic ester
production in fruit, or
(b) complementary to SEQ ID NO: 12A or 13A, or
(c) which has at least 20o homology, preferably at
25 least 30% homology, with any of the sequences
of (a) or (b) , or
(d) which is capable of hybridising under strin-
gent conditions to any of the sequences (a)
(c) .
30 Further the invention provides a purified and
isolated polypeptide,
(a) having an amino acid sequence selected from
the group consisting of sequences SEQ ID NO:
12B and 13B, or
35 (b) having at least
i) 55~ homology with the amino acid sequence of a
176 as fragment from the C terminal end of
the coding sequence of SEQ ID NO: 12B, or
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
41
ii) 35% homology with the amino acid sequence
of a 284 as fragment from the C terminal
end of the coding sequence of SEQ ID NO:
13B, or
(c) being a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide
has alcohol dehydrogenase activity and is
involved in the biosynthetic pathway for ali
phatic and/or aromatic ester production in
fruit.
According to a further aspect the invention provi
des an isolated DNA sequence encoding
(a) a polypeptide having an amino acid sequence as
shown in SEQ ID N0:15B, or
(b) a polypeptide having at least 41% homology,
preferably at least 50% and more preferably at
least 80% homology, with the amino acid sequence
SEQ ID N0:15B, or
(c) a fragment of polypeptide (a) or (b), which
polypeptide or fragment of polypeptide has esterase
activity and is involved in the biosynthesis
pathway for aliphatic and/or aromatic ester produc
tion in fruit.
In particular the invention provides an isolated
DNA sequence having a nucleic acid sequence
(a) as shown in SEQ ID N0:15A which encodes a
polypeptide having esterase activity and being
involved in the biosynthetic pathway for aliphatic
and/or aromatic ester production in fruit, or
(b) complementary to SEQ ID N0:15A, or
(c) which has at least 53% homology,preferably at
least 70% homology, with any of the sequences of
(a) or (b) .
Further the invention provides a purified and
isolated polypeptide,
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
42
(a) having an amino acid sequence as shown in SEQ
ID N0:15B, or
(b) having at least 41°s homology, preferably at
least 50% and more preferably at least SOo homolo
gy, with the amino acid sequence ID N0:15B, or
(c) being a fragment of polypeptide (a) or (b),
which polypeptide or fragment of polypeptide has
esterase activity and is involved in the biosynthe
tic pathway for aliphatic and/or aromatic ester
production in fruit.
Additonally, the invention provides nucleotide
sequences which are suitable as primers for obtaining
additional alcohol acyltransferase sequences from various
fruit by cloning. These nucleotide sequences (motifs)
encode:
the amino acid sequence as set forth in SEQ ID
N0:14B, or
- Leu X X X Tyr Pro X X Gly Arg, or
- Pro Ser Arg Val X X Val Thr X Phe Leu X Lys X
Leu Ile
wherein X refers to any amino acid residue.
According to a fourteenth aspect the invention
further provides a method for regulating aliphatic and/or
aromatic ester formation in fruit comprising inserting into
the genome of a fruit-producing plant one or more copies of
one or more DNA sequences as defined above encoding various
enzymes involved in the biosynthetic pathway for aliphatic
and/or aromatic ester formation in fruit.
The levels of protein may be increased for example
by the incorporation of additional genes. The additional
genes maybe designed to give either the same or different
spatial or temporal patterns of expression in the fruit as
the target gene.
Further the invention provides a genetically
modified strawberry or lemon plant and propagating material
derived therefrom which has a genome comprising an expres-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
43
sion vector for overexpression or downregulation of an
endogenous strawberry or lemon plant gene counterpart of any
of the DNA sequences as defined above encoding various
enzymes involved in the biosynthetic pathway for aliphatic
and/or aromatic ester formation in fruit.
Further the invention provides a method for produ-
cing aromatic and/or aliphatic esters in microorganisms,
plant cells or plants comprising inserting into the genome
of the microorganism or plant one or more copies of DNA
sequences of this invention encoding a polypeptide having
alcohol acyl transferase activity, and one or more copies of
DNA sequences of this invention encoding a polypeptide
having alcohol dehydrogenase activity, and feeding aldehydes
and acyl-CoA to the microorganism or plant (cell).
In a further embodiment the above method further
comprises inserting one or more copies of DNA sequences of
this invention encoding a polypeptide having pyruvate
decarboxylase activity, and feeding alpha-keto acids and
acyl-CoA to the microorganism or plant (cell).
In a still further embodiment the above method
further comprises inserting one or more copies of DNA
sequences of this invention encoding a polypeptide having
aminotransferase activity, and feeding amino acids and acyl-
CoA to the microorganism or plant (cell).
In a still further embodiment the above method
further comprises inserting one or more copies of DNA
sequences of this invention encoding a polypeptide having
thiolase activity, and feeding amino acids and fatty acids
to the microorganism or plant (cell).
The DNA sequences of this invention encoding a
polypeptide having esterase activity can be disrupted to
increase the amount of volatile esters. Molecular strategies
to accomplish this are well known in the art.
Strawberry and citrus are among the most popular
fruits for natural flavour ingredients because of their
flavour, fragrance, aroma and scent. Examples of commerci
ally important aliphatic esters are ethyl acetate (used in
artificial fruit and brandy flavours), ethyl butyrate (the
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
44
primary constituent of apple aromas), isoamyl acetate (the
main component of banana aroma) and ethyl butyrate (has a
fruity odour, reminiscent of pineapple). Flavour usage of
these esters is among the highest of all natural flavour
ingredients. The invention further contemplates the use of
the polynucleotide sequences according to the present
invention, for the industrial production of 'fruit' flavours
which are natural to match the odour fidelity of the natural
fruit. As the chemical composition of flavour is quite
complex, both the type and ratio of compounds present being
the key determinant of flavour quality, this present inven-
tion further contemplates the production of novel flavours
by the use of the polynucleotide sequences according to the
present invention, alone or in combination, to provide novel
avenues for natural flavour production in the future.
The invention further contemplates the generation
of strawberry and lemon plants which amongst other phenoty-
pic modifications may have one of the following characte-
ristics:
(a) reduced/enhanced flavour from the reduced/en-
hanced production of volatile esters,
(b) increased disease resistance due to the enhan-
ced production of volatile esters,
(c) modified plant-insect interactions, to include
increased insect resistance.
In a further aspect, the gene sequences of the
present invention are used for the manipulation of flavour
in other 'fruits' and or/ industrial processes. The genes/
peptides of the present invention, in particular SEQID lA
(strawberry SLE27) and SEQ ID 1B (lemon CLF26) may be used
in the following industrial applications; with particular
reference to the manipulation of fruit derived products,
however, not exclusively limited to these:
a) in the processed food industry as food additives to
enhance the flavour of syrups, ice-creams, frozen
desserts, yoghurts, confectionery, and like pro
ducts,
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
b) flavouring agents for oral medications and vitam-
ins,
c) providing additional flavour/aroma in beverages,
including alcoholic beverages,
5 d) enhancing or reducing fruit flavour/aroma/fragran-
ce/scent,
e) enhancing the flavour/aroma of 'natural products
(eg flower scent however, including but not exclu-
sively limited to flower scent),
10 f) enhancing the flavour/aroma of 'synthetic or arti-
ficial products',
g) as an alternative to the industrial synthesis of
nature identical flavour/aroma substances,
h) for the production of novel combinations of arti-
15 ficial flavour substances,
i) as antibacterial or as anti-fungal agents,
j) fragrance /perfumes in the cosmetics, creams, sun-
protectant products, hair conditioners, lengthening
agent and fixative in perfumes, suspension aid
for
20 aluminium salts in anti-perspirants pharmaceuti-
cals, cleaning products, personal care products
and
animal care products,
k) disinfectant additives,
1) as degreasing solvents for electronics (etc.),
25 m) insect pheromones,
n) dye carrier, solvents, insect repellent, miticide,
scabicide, plasticiser, deodorants.
An example as way of illustration of the indus-
trial imp ortance of volatile esters (aromatic and aliphatic)
30 is the
ester
methyl
anthranillate.
Methyl
anthranillate,
a
methyl ter of o-aminobenzoic acid, apart from being a
es very
important perfumery, aroma, scent ester (it imparts a
peculiar aroma known as "foxy" to grapes of the North
american species Vitis Labrusca), the same compound is known
35 to serve as a pheromone in various insects, Results of
investiga tions aimed at testing the attractiveness of methyl
anthranil ate for the the soybean beetle Anomala rufocurea,
Motschuls ky suggested the potential use of methyl anthrani-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
46
late as a lure for mass trapping, as well as for monitoring
( Imai et al . , Applied Entomology and Zoology, 32 : 1, 45 -
48, 1997) . In addition methyl anthrenilate acts as a bird
repellent. According to the invention we propose the use of
the SEQ1D: 2A for the production of methyl anthranilate, for
a number of applied applications.
The present invention also relates to antibodies
which specifically bind to the polypeptides as claimed in
the invention, compositions comprising substantially puri-
fied polypeptides, fragments thereof, agonists or alternati-
vely antagonists, and methods of producing polypeptides. In
bacterial/prokaroyte systems a number of expression vectors
may be selected depending on the used of the polypeptide(s)
according to the invention. For example, when large quanti-
ties of the polypeptide are needed for the induction of the
antibody, vectors which drive high levels of expression of
fusion proteins that are readily purified are desirable.
Such vectors include, but are not limited to the multi-
functional E.col.i cloning and expression vectors, such as
Bluescript~ (Stratagene), in which the gene sequences)
according to the invention, can be ligated into the vector
in frame with sequences for the amino-terminal Met and the
subsequent 7 residues ofl3-galactosidase so that the hybrid
protein is produced; pPIN vectors (Van Heekle and Schuster
(1989) J Biol chem 264: 503-5509); and the like. pGEX
vectors (Promega, Madison WI) may also be used to express
foreign polypeptides as fusion proteins with glutathione-S-
transferase (GST). In general such fusion proteins are
soluble and can be easily purified from lysed cells by
absorption to gluthionine-agarose beads followed by elution
in the presence of free gluthionine. Proteins made in such
systems are designed to include heparin or factor XA
protease cleavage sites so that the cloned polypeptide of
interest can be released from the GST moiety at will.
In addition to recombinant production the amino
acid sequences of the polypeptides, or any parts thereof,
may be altered by direct synthesis or by genetic means known
by those skilled in the art, and/or combined using chemical
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
47
methods with sequences from other proteins, or any part
thereof, to produce a variant polypeptide. Fragments of the
polypeptides according to the invention may be produced by
direct peptide synthesis using solid phase techniques (c.f.
Stewert et al. (1969) Solid Phase Peptide Synthesis, WH
Freeman Co, San Fancisco, Merrifield J (1963) J Am Chem Soc
85:2149-2154). In vitro protein synthesis may be performed
using manual techniques or by automation. Automated synthe-
sis may be achieved, for example, using Applied Biosystems
431A Peptide Synthesizers (Perkin Elmer, Forster City CA) in
accordance with the instructions provided by the manufactu-
rer. Various fragments of each of the polypepides may be
chemically synthesised separately and combined using chemi-
cal methods to produce full length molecules.
The invention further contemplates a diagnostic
assay using antibodies raised to each of the polypeptides of
the present invention, with or without modification. For
example, although not exclusively limited to, alcohol acyl
transferase antibodies raised to the polypeptides as outli-
ned in the 6-8th embodiments are useful for the detection of
flavour forming esters in various fruits. Diagnostic assays
for alcohol acyl transferase include methods utilising the
antibody and a label to detect the alcohol acyl transferase
protein in various fruit extracts. Frequently the polypepti-
des and antibodies are labelled by joining them, either
covalently or non covalently, with a reporter molecule. A
variety of reporter molecules are known and several of them
have been previously described (cf. reporter genes).
A variety of protocols for measuring the polypep
tide, using either polyclonal or monoclonal antibodies
specific for the respective protein are known in the art.
Polynucleotide sequences) of the invention, SEQ ID lA
strawberry (SLE27), SEQ ID 2A, lemon alcohol acyl transfe
rase proteins (CLF26), or any part thereof, may be used for
diagnostic purposes, to detect and quantify gene expression
in various tissues (eg leaves, stems and in particular
fruits (climacteric and non climacteric)), hence volatile
ester (aliphatic and aromatic) formation, hence flavour at
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
48
various stages of plant maturation. The diagnostic assay is
useful to distinguish between absence, presence and excess
expression of the alcohol acyl transferase proteins and to
monitor expression levels during development.
The following examples are offered by way of
illustration and not by limitation.
Example 1: Method for the detection of volatiles
The composition of the headspace of the strawberries was
analysed using Solid Phase Micro-extraction for trapping and
concentrating the emitted volatiles. The fibres used were
100 um Polydimethyl siloxane (PDMS) supplied by Supelco.
Analysis was performed on a Fisons GC 8000 system, coupled
to the Fisons MD800 mass spectrometer. The gas chromatograph
was temperature programmed, with a plateau at 80 °C for 2
min., followed by a ramp of 8°C/min to 250 °C for 5 min.
The injector port was operated at 250°C for thermal desorp-
tion during 60 sec. of the SPME fibers, using the deactiva-
ted standard 0.7 ml split/splitless injection port liner
with zero split. The column used was a HP-5 with a length of
50 m and internal diameter of 0.32 mm and a film thickness
of 1.03 ~Cm, and operated with helium at a pressure of 38kPa.
Eluted components were identified by their mass spectrum,
using the Fisons Masslab software and the NIST library. The
identity of the components was verified where possible by
using known substances as reference (most of them obtained
from Aldrich).
Headspace analyses were performed both on the plant
and in the laboratory using detached fruits. The fruits were
put onto new clean aluminium foil and covered with a glass
funnel closed with aluminium foil. The SPME fiber was
inserted in the funnel through the foil closing, and the
fiber was exposed to the air in the funnel during 30 min.
Figure 2 shows a typical chromatogram obtained after head-
space measurement of red ripe fruit of strawberry grown in
the greenhouse. The importance of esters to the overall
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
49
volatile emission from the ripe strawberry fruit can be
clearly observed.
Example 2: Construction of a strawberry red fruit stage and
lemon young fruit peel cDNA library, mass excision and
random sequencing
2.1 Messenger RNA isolation and cDNA libraries construction
Total RNA was isolated from strawberry fruit red stage of
development using the method described by Manning K. [Analy-
tical Biochemistry 195, 45-50 (1991)]. The cultivar used was
Fragaria X ananassa Duch. cv. Elsanta. Plant material for
the isolation of RNA from lemon (citrus Limon cv. Meyer) was
the green peel of a small fruit (1.2 x 2 cm) of the culti-
var. Isolation of RNA from lemon was done using the same
method as described for strawberry. The cDNA libraries for
both strawberry and lemon where produced as a custom service
by (Stratagene) in the lambda zap vector. Messenger RNA was
isolated from total RNA using the polyA+ isolation kit
(Pharmacia).
2.2 Mass excision and random sequencing
The ExAssist~"/SOLRT"" system (Stratagene) was used for mass
excision of the pBluescript SK(-) phagemid. The excision was
done according to the manufacturer's instructions using 20 x
103 pfu from the non amplified library for each excision.
High quality plasmid DNA from randomly picked colonies was
extracted using the QIAGEN BioROBOT 9600. Colonies were
grown overnight in 3m1 Luria Broth medium (lOg/1 tryptone, 5
g/liter yeast extract, 5 g/liter NaCl) supplemented with 100
mg/litre Ampicillin, centrifuged at 3000 RPM for 10 min. and
the pellet was used directly for plasmid DNA isolation by
the robot. Each DNA isolation round consisted of 96 cultu-
res.
Insert size was estimated by agarose gel electrop-
horesis after restriction enzyme digestion of the pBlue-
Script (SK-) vector with EcoRI and XhoI. Inserts with length
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
above 500 by were used for sequencing. Plasmid DNA from the
selected samples were used for polymerise chain reaction
(PCR) sequencing reactions using the ABI PRISMT"" Dye Termina-
tor Cycle Sequencing Ready Reaction Kit and the MJ Research
5 PTC-200 DNA EngineT"" thermal cycler. The T3 and T7 universal
primers were used for sequencing from the 5' and 3'ends
respectively. PCR program was according to the Dye Termina-
tor manufacture's protocol (ABI PRISM). The ABI 373, 370A
and 310 sequencers (Applied Bio-systems) were used. Sequen-
10 ces were edited manually to remove vector and non reliable
sequences and submitted to the Blast homology search program
using BlastX and BlastN (Altschul et al. J. Mol. Biol. 215,
403 - 410, 1990) provided by the National Centre for Bio-
technological Information on the world wide web. The straw-
15 berry and lemon sequencing data received from the Blast
search was transferred using a self designed macro program
to a database made using the software Access (Microsoft).
Sequences with a Blastx score above 80 were considered as
having a significant homology (Pearson, Curr. Opinion.
20 Struct. Biol, l, 321 - 326, 1991) and they were used for
data interpretation in the Access and Excel programmes
(Microsoft). The cDNA clones: SLE27 (SEQ ID NO: lA), CLF26
(SEQ ID NO: 2A), SLF96 (SEQ ID NO: 3A), SLG150 (SEQ ID NO:
4A), SLH51 (SEQ ID NO: 5A), SLB39 (SEQ ID NO: 6A), SLF193
25 (SEQ ID NO: 7A), SLF122 (SEQ ID NO: 8A), SLD194 (SEQ ID NO:
9A), SLF17 (SEQ ID NO: l0A), SLF138 (SEQ ID NO: 11A), SLG16
(SEQ ID NO: 12A), SLG144 (SEQ ID N0:13A) were selected for
further analysis.
Example 3: Characterisation of selected cDNAs
30 3.1 Sequencing the entire length cDNAs
Plasmid DNA from the selected cDNAs was used for PCR sequen-
cing reactions using the ABI PRISMT"" Dye Terminator Cycle
Sequencing Ready Reaction Kit and the MJ Research PTC-200
DNA Enginer"" thermal cycler. Each individual selected cDNA
35 was sequenced completely on both strands. DNA and protein
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
51
alignment 'contigs' were performed using the GeneWorks
software (Intell:iGenetics, Oxford) .
3.2. RNA isolation from different tissues and organs of
strawberry and lemon.
RNA isolation from both strawberry and lemon tissues was
done as described for the preparation of total RNA for the
construction of the cDNA libraries [Manning K. Analytical
Biochemistry 195, 45-50 (1991)]. For both species green
house grown plant material was used. From strawberry the
different tissues/organs used for RNA isolations were; leaf,
root, petiole, flower, green fruit, white fruit, turning
fruit, red fruit, red fruit without seeds, seeds and overri-
pe fruit. For lemon the different tissues/organs used were;
root, leaf, flower bud, albedo, green fruit peel (1.2 x 2
cm), green fruit peel (2 x 3 cm), green fruit peel (3.5 x 6
cm), green fruit peel (5 x 8 cm), yellow fruit peel (2 weeks
fruit detached) .
3.3 8xpression analysis using Northern blots
Ten ~Cg of each RNA sample was denatured by glyoxal (1.5M)
prior to electrophoresis on a 1.4% agarose gel in l5mM
sodium phosphate buffer pH = 6.5 (without Ethidium bromide).
Equal loading of RNA was verified by visualising equal
amount of RNA from each sample on a 1% TAE buffer [lx .
0.04M Tris-acetate / O.OO1M ethylenediaminetetra-acetate
(EDTA)] agarose gel prior to loading. Blotting was done
using Hybond N+membrane (Amersham) in 25 mM Sodium phosphate
buffer pH = 6.5 overnight.
After fixation (2h 800c) blots were hybridised as
described by Angenent et al. 1992 (Plant Cell 4, 983-993).
The hybridisation probes were made by random labelling
oligonucleotide priming (Feinberg and Vogelstein, Anal.
Biochem. 137, 266-267, 1984) of the entire cDNAs. The
hybridised membranes were autoradiographed at -70°C with
intensifying screens. In Figure 3 the expression pattern of
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
52
strawberry alcohol acyl transferase (SLE27, SEQ ID N0: lA)
is demonstrated by a Northern blot loaded with total RNA
extracted from 10 different plant tissues and/or organs from
strawberry. The expression pattern of strawberry alcohol
acyl transferase is in perfect match with the accumulation
of volatile esters in the strawberry plant. The gene SLE27,
(SEQ ID NO: lA) is expressed at it lowest level in the white
fruit, gradually increasing as fruit develops reaching its
highest levels in the turning stage and the red stage when
the volatile ester compounds can be detected at their
maximum. In other strawberry plant tissues/organs which do
not produce those volatile fruity esters the expression of
the gene can not be detected even when the film is exposed
to a longer period after hybridisation.
The expression of the lemon alcohol acyl transfe-
rase (CLF26 SEQ ID NO; 2A) seems to be correlated with the
accumulation of volatile monoterpene compounds in the lemon
tree tissues/organs (Figure 4). These compounds can be
converted by this enzyme to their ester derivatives as a
result of alcohol acyl transferase enzyme activity. The gene
is more highly expressed in the flower bud. Analysis by Gas
chromatograph (GCMS) shows high concentrations of terpene
compounds produced in the flower bud such as limonene.
Expression analysis of montoterpene cyclases (genes directly
responsible for the formation of monoterpenes) isolated from
lemon thus shows a similar pattern of expression to the
lemon alcohol acyl transferase (CLF26, SEQ ID N0:2A).
Expression of the gene was also strongly detected in the
early stages of fruit development and in the flower bud
whilst no expression could be detected in the root and
albedo tissues. This data provides evidence for a link
between the presence of terpene compounds mainly monoterpe-
nes and the lemon alcohol acyl transferase (CLF26, SEQ ID
N0:2A).
The expression of other three genes part of the
metabolic pathway described in this invention for the
formation of volatile ester compounds, in particular alipha-
tic esters in fruits is shown in Figure 5. Thiolase (SLG150,
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
53
SEQ ID NO: 4A) alcohol dehydrogenase (SLB39, SEQ ID NO: 6A)
and pyruvate decarboxylase (SLH51, SEQ ID NO: 5A) gene
expression was monitored at different organs or tissues of
the strawberry plant. The data shown provides evidence that
thiolase (SLG150, SEQ ID NO: 4A), alcohol dehydrogenase
(SLB39, SEQ TD NO: 6A) and pyruvate decarboxylase (SLH51,
SEQ ID NO: 5A) are involved in ripening processes such as
the production of strawberry flavour and aroma compounds.
Clones SLF193 (SEQ ID NO: 7A), SLF122 (SEQ ID NO: 8A),
SLD194 (SEQ ID NO: 9A), SLF17 (SEQ TD NO: l0A), SLF138 {SEQ
ID NO: 11A), SLG16 (SEQ ID NO: 12A), SLG144 (SEQ ID N0:13A)
and SLF96 (SEQ ID NO: 3A) were also tested for the expressi-
on and they were all appearing to have upregulated expressi-
on in the ripening fruit {data not shown).
3.4 Co-ordinate expression of genes in the biosynthetic
pathway leading to aromatic and aliphatic ester formation
The co-ordinate expression pattern of genes of the biosyn-
thetic pathway leading to aromatic and aliphatic ester
formation was simultaneously monitored using arrays of
cDNAs[Ermolaeva et al., 1998, Nature Genetics, Vol, (20),
19-23] . The method allows one to simultaneous quantitatively
and statistically analyse the expression profiles of genes
pertaining to a particular metabolic pathway, such as
pathway leading to aliphatic and aromatic ester formation,
flavour, fragrance, aroma and scent. The experimental and
hybridisation conditions followed were according to Schena
et al., (PNAS, 93: 10614 - 10619). The expression levels
derived from four images were statistically analysed with an
analysis of variance modelling the factors dye, origin, and
spot levels on a logarithmic scale giving the significant
ratios of expression between the spots of origin (eg. green
versus red). As way of illustration the expression profiles
of SEQ ID lA, SEQ ID 3A, SEQ ID 5A, SEQ ID 11A, SEQ ID 12A,
analysed in the green versus red and turning versus red
stages of fruit development are outlined in Figure 10. The
co-ordinated expression pattern of the genes during straw-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
54
berry fruit development is in perfect match with the accumu-
lation of volatile ester compounds aromatic and aliphatic
during strawberry fruit development. In addition alcohol
acyl transferase enzyme activity was detected by Perez et
al., (J. Agric. Food Chem. 1996, 44, 3286-3290) to be
elevated dramatically between the white and the turning
stages of strawberry fruit development. This correlates well
with a dramatic increase in expression of the genes of the
biosynthetic pathway as shown in Figure 10. A supporting
detail is the fact that volatile ester compounds can be
detected in their maximum when the fruit is completely red
and this is probably a result of gene activation at earlier
stages between the white and turning stage of development.
3.5 DNA isolatioxi from strawberry leaf tissue.
Genomic DNA was isolated from green house grown strawberry
plants (cv. Elsanta). Young folded leaves kept 2 days in
water in the dark were used fox the isolation. 1 gram of
leaf material was ground in liquid nitrogen and immediately
added to 7 ml DNA extraction buffer (100 mM Tris-Cl, 1.4 M
NaCl, 20 mM EDTA, 2% CTAB, 0.2% f3- mercaptoethanol) . After
incubation for 30 min. at 600C one volume of chloroform:iso-
amyl alcohol (24:1) was added to the tubes. The samples were
then centrifuged for 10 min. at 4000 rpm and the aqueous
phases were transferred to 4.5 ml isopropanol and incubated
for 30 min. at -20°C. Nucleic acids were spun down at 4000
rpm for 12 min. washed with 70% ethanol and air dried.
Pellets were dissolved in 3 ml TE and 0.25 volumes of NaCl
(5M) and 0.1 volume absolute ethanol were added. Samples
were centrifuged for 10 min at 4000 rpm and supernatants
were transferred to 4.5 ml isopropanol for precipitation.
Pellets were washed with 70% ethanol dried and dissolved in
0.5 ml TE buffer (lOmM Tris-C1, 1mM EDTA pH = 8.0). RNA was
selectively precipitated by the addition of 0.6 volumes of
8M lithium chloride. The RNA was pelleted by centrifugation
10 min. 4000 rpm and the DNA was precipitated again with the
addition of 0.6 volume isopropanol. Pellets were washed with
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
70 % ethanol, dried and dissolved in 0.5 ml sterile water.
The amount of DNA was determined by comparing to known
amounts of phage lambda DNA.
3.6 Southern blot analysis for strawberry alcohol acyl
5 transferase (SLE27, SEQ ID N0:1A)
Aliquots of 2 ~,g strawberry genomic DNA were digested for
16h with the restriction enzymes: HindIII, XbaI, XhoI and
EcoRI and separated on a 0.7o TAE (0.04M Tris-acetate pH =
8.0 and 1mM EDTA pH - $.0) agarose gel. The DNA was then
10 denatured in 0.4M NaOH for 30 min. and then transferred to
a Hybond N+ membrane in 0.4M NaOH. Blot hybridisation and
probe preparation was done as described previously for
Northern blots. After hybridisation the blot was washed
first under low stringency conditions (52°C with 2 times
15 half an hour 2xSSC/0.1°s SDS) and after observation of the
results by autoradiography of the film they were washed
further under stringent conditions (65°C with 2 times half
an hour O.IxSSC/0.1~ SDS) and autoradiographed again.
In order to investigate whether the different
20 esters formed in the fruit are a result of an activity of
one single enzyme or a multigene family, a Southern blot was
performed using strawberry genomic DNA and the full length
SLE27 cDNA (SEQ ID NO:lA) which served as a probe (Figure
6). Since the cultivar used for the investigation is an
25 octaploid the interpretation of the results is more diffi-
cult than that for a diploid plant. However, the results
obtained suggest the possible existence of another homolog
of strawberry alcohol acyl transferase. This additional copy
might arise from one of the parental lines with a similar
30 function to the one isolated, or may represent another
homolog with different function or substrate specificity to
SLE27 (SEQ ID NO: 1A).
3.7 Alcohol acyltransferase sequences from other fruit.
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
56
Additional alcohol acyltransferase sequences from melon,
strawberry vesca, banana, apple, lemon and mango were
obtained using degenerate primers synthesised to the motif
sequence ID number 14B using methods well known in the art
such as polymerase chain reaction (PCR) and reverse genetic
approaches such as RT-PCR. Such techniques are described in
Sambrook et al., 1989. Molecular cloning a laboratory
manual, cold spring Harbour press, Plain view, NY and
Ausubel FM et al., (1989) Current protocols in molecular
biology, John Wiley and Sons, New York, NY. The esterase
gene was cloned using PCR techniques using the following
degenerate primer . 5'-GG (T/A) TGGGG I (T/G) CTA(T/C)
TCTTGC-3' ) and reverse genetic approaches such as RT-PCR.
Such techniques are described in Sambrook et al., 1989.
Molecular cloning a laboratory manual, cold spring Harbour
press, Plain view, NY and Ausubel FM et al., (1989) Current
protocols in molecular biology, John Wiley and Sons, New
York, NY.
Example 4
Strawberry alcohol acyl transferase (SLE27, SEQ ID N0:1B)
protein analysis
4.1 Cloning to the expression vector pRSET B
The E.coli expression vector pRSETB (Invitrogen), was used
for expression studies of strawberry alcohol acyl transfe-
rase (SLE27, SEQ ID NO: 1A). This pRSETB vector contains the
T7 promoter which can be induced by isopropyl-beta-D-
thiogalactopyranoside (IPTG) and therefore by inserting the
desired gene downstream of this promoter, the gene can be
expressed in E. coli. In addition, DNA inserts were positi-
oned downstream and in frame with a sequence that encodes an
N-terminal fusion peptide. This sequence includes (in 5' to
3' order from the N-terminal to C-terminal), an ATG transla-
tion initiation codon, a series of six Histidine residues
that function as a metal binding domain in the translated
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
57
protein, the Anti-Xpress epitope, and the enterokinase
cleavage recognition sequence.
The original pRSETB was primarily used for the
insertion of the gene encoding the Green Fluorescent Protein
(GFP). The GFP gene was fused to the pRSETB vector using the
BamHI and HindIII restriction sites located at the multiple
cloning site (MCS) as can be seen in Figure 7. This con-
struct for the expression of GFP served as control for the
experiments together with the empty pRSETB vector.
Cloning the GFP gene to the pRSETB vector inserted
an additional SaII restriction site at the 3' of the GFP
gene and together with the BamHI site located at the 5' of
the GFP gene served as sites for the cloning of the straw-
berry alcohol acyl transferase gene (SLE27, SEQ ID N0:1A).
The BamHI and SalI sites were introduced to the 5 ' and 3'
respectively of the strawberry alcohol acyl transferase
coding sequence by the use of PCR. The 452 amino acid open
reading frame of the SLE27 (SEQ ID NO:lA) clone was ampli-
fied with the pfu DNA polymerase (Stratagene) and primers
AAP165 (5'-CGGATCCGGAGAAA.ATTGAGGTCAG} and AAP166 (5'-CGTCGA-
CCATTGCACGAGCCACATAATC) according to the manufacturers
instructions. The PCR product was cloned into PCR-script
vector (Stratagene), cut out with BamHI and SalI and further
inserted (as a translation fusion) into the corresponding
restriction sites in the pRSETB vector.
4.2 Bacterial expression and partial purification using the
His tag columns.
The pRSET B vector harbouring the strawberry alcohol aryl
transferase (SLE27, SEQ ID NO: lA) was used to transform E.
coli strain BL21 Gold DE3 (Stratagene) as described by the
manufacturer. For bacterial expression typically 1 ml of
overnight liquid culture grown at 37°C in Luria Broth (LB)
medium (lOg/1 tryptone, 5 g/liter yeast extract, 5 g/liter
i~TaCl) supplemented with 100 mg/liter Ampicillin was diluted
50 times in the same medium and grown until the ODsoo rea-
ched 0.4 (usually at 37°C except when different temperatures
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
58
for growth during induction were examined). At this stage
Isopropylthio-beta-D-galactoside (IPTG) was added to a final
concentration of 1mM in order to induce expression . After
one and a half hour of induction the cells were harvested by
centrifugation at 4000 x g for 15 min. Pellet and a sample
from the supernatant were kept for SDS gel analysis. The
cells were further processed as described by the Ni-NTA Spin
Columns manufacturers (QIAGEN) for protein purification
under native conditions.
4.3. Recombinant protein detection using Western blot
The supernatant (S), pellet (P) and elute from the Ni-NTA
columns (E) samples were diluted with 2x SDS-sample buffer
((standard Laemmli, pH 6.8 Tris /glycine/SDS/glycerol + 50mM
Dithiothreitol(DTT)]. The samples were heated for 5-l0 min
at 95°C, followed by a short spin of 30 seconds, full speed.
From the samples 101 was loaded on 10% SDS-gel (mini-gel,
lmm, 7x9cm). The gels were run with standard buffer (25mM
Tris 192mM, glycine 0.1% SDS, pH 8.3) at 100-200V, 50-30mA,
ca 3hr with 16°C water cooling. After complete run, gels
were used for Coomassie Brilliant Blue (CBB) stain or
Western blot. For CBB staining, the gel was fixed during
lhour in 40% Methanol, 10% Acetic Acid in water. Then
incubated overnight with CBB staining solution (0.05% CBB-
250 in 40% Methanol, 10% Acetic acid in water) and destained
by repeated washing in 25% Methanol, 10% Acetic acid in
water. Western blot was onto Polyvinyl di fluoride (PVDF),
(Immobilon P, Amersham) pre-wetted with Methanol. Transfer
buffer was Towbin buffer plus 0.1% SDS (25mM Tris, 192mM
Glycine, 20% 0.1% SDS, pH 8.3). Transfer was submarine
(Novex blot unit, plate electrodes, 3cm distance) at 200mA
overnight plus 2hr. at 400mA, cooled in ice-water with
cooling block. Transfer was checked by using pre-stained
markers as a reference.
After transfer the blot was washed in Tris buffer
saline TBS (20mM Tris-HC1 pH7.5 / 150mM NaCl)- Tween20
(0.02%), blocked with 2% fat tree milk in TBS-Tween20, then
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
59
washed with TBS-Tween 20. Then first Antibody was anti-
Xpress (Invitrogen) 1/1000 in 2% BSA TBS-Tween for 3hr.
incubate at room temprature, then 4x washed with TBS-Tween
20. Second Antibody is GAR-AP (Boehringer) 1/3000 in 2% BSA
TBS-Tw (TBS + 0.05% Tween - 20) for 1-2hr. incubation at
room temprature, then 4x washed with TBS-Tween 20. Final
wash was in water, then in Alkaline phosphatase-buffer
(100mM Tris pH9.2, 1mM MgCl2). The staining reaction was
with nitroblue tetrazolium/5-bromo-4-chloro-3-indolphospha-
te/alkaline phosphatase-buffer (451 nitroblue tetrazolium
30mg/ml in 70% dimethylformamide, 331 5-bromo-4-chloro-3-
indol phosphate l5mg/ml in dimethylformamide in 10m1 of
alkaline phosphatase buffer). The staining reaction was
stopped by rinsing with water. Western blotting was used in
order to determine the amount of the recombinant SLE27 (SEQ
ID NO: 1B) protein in the elute after the Ni-NTA column
purification compared to the pellet and supernatant before
loading on the column. Figure 9 shows a western analysis of
the pellet supernatant and elute after partial purification
on the Ni-NTA column. 4 samples were loaded from each
pellet, supernatant and elute originating from 4 different
growth and induction of the E. coli cells. A commercial
antibody recognising an epitope peptide fused to the N-
terminus of the SLE27 recombinant protein showed relatively
high expression of SLE27 (SEQ ID NO:1B) in the elute from
the Ni-NTA column. The molecular weight of the recombinant
SLE27 (SEQ ID NO:1B) protein (SHAT) recombinant protein was
54 kDa. The enzyme exhibited a broad pH range (pH optimum
around 8.3). As a control for the experiment served the
Green Fluorescent Protein (GFP) partially purified in the
same method as done f or SLE2 7 ( SEQ ID NO :1B ) . No band was
detectable at the level of the SLE27 (SEQ ID NO:1B) recombi-
nant protein in the GFP lane.
4.4 Method for establishing alcohol acyl transferase
activity of the strawberry SLE27 (SEQ ID NO:1B)-encoded
protein.
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
The alcohol acyl transferase activity of the strawberry cDNA
SLE27 (SEQ ID NO:lA) encoded protein was established by
analysing the volatile esters produced from acyl-CoA and
different alcohols. In a 1.8 ml glass vial with a small
5 magnetic stirring bar, 325 ~.1 of buffer (50 mM Tris/HCl pH
8.0, containing 1 mM dithiothreitol), 25 ~I of protein
extract and 25 ~l of acyl-CoA (4 mM in buffer) were subse-
quently pipetted. The enzyme reaction was started by the
addition of 25~t1 of alcohol solution (160 mM in buffer). The
10 vial was quickly closed with a snap cap and placed in a
waterbath (at 35°C) with a stirring device. After 15 min
enzyme reaction with continuous stirring, the vial was
opened, 0.29 g of solid CaCl2 was added (final concentration
5M) and the vial was quickly closed again with an open top
15 crimp cap. The high CaCl2 concentration was used to stop
further enzymatic reactions and to drive the volatiles into
the headspace. After addition of CaCl2, the mixture was
incubated again at 35°C while stirring. The volatile compo-
nents released into the headspace were trapped using solid
20 phase micro-extraction (SPME; Supelco Inc., Bellefonte, PA,
USA), exposing the fused silica fiber coated with 100 lm
polydimethylsiloxane (PDMS; Supelco Inc.) to the headspace
for 15 min. The presence of esters in the alcohol stock
solutions and the spontaneous formation of esters were
25 checked by adding 25 ~1 of buffer instead of protein extract
into the reaction vial.
The volatiles trapped by SPME were separated and
identified by gas chromatography (GC 8000 Series; Inters-
cience B.V., Breda, The Netherlands) coupled to mass spec-
30 trometry (MD-800, Fisons Instruments, Interscience) as
described by Verhoeven et al.. (1997) [Chromatographia Vol.
46, No 1,2). Compounds were thermally desorbed (2500C) from
the SPME-fiber for 1 min. Separation of compounds was on a
capillary HP-5 column (50 m x 0.32 mm, film thickness 1.05
35 ~,m; Hewlett Packard) with He (37 kPa) as a carrier gas. The
oven temperature was programmed at 2 min at 80°C, then an
increase of 8°C min-1 to 250°C, and finally 5 min at
250°C.
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
61
Mass spectra of eluting compounds were generated at 70eV
each second. The MD 800 Masslab software (Interscience) was
used to record mass spectra and to calculate peak areas.
Volatile compounds were identified by screening the NIST
library for comparable mass spectra and by comparison with
authentic reference compounds.
4.5.Substrates used by the strawberry alcohol acyl transfe-
rase-encoded enzyme (SLE 27: SEQ ID NO: 1B)
The histidine-released protein elute from E. coli expressing
the strawberry alcohol acyl transferase gene (SLE27, SEQ ID
NO:lA) was incubated with both acetyl-CoA and 1-butanol. The
chromatograms obtained by SPME of volatiles released into
the headspace indicated a clear peak of acetic acid 1- butyl
ester (Figure 8B). This ester was not detected when the
protein (Figure 8C), alcohol substrate (Figure 8D) and
acetyl CoA were absent (Figure 8E). Moreover, when the SLE27
(SEQ ID NO:1B)-encoded protein elute in the assay was
replaced by an equal amount of histidine-released elute from
lysate of E. coli expressing only the histidine-tag (i.e.
the same vector without the SLE27 (SEQ ID NO:lA gene), the
ester could not be detected either (Figure 8G). To investi-
gate which substrates can be used by the SLE27(SEQ ID
NO:1B)-encoded protein to produce volatile esters with
acetyl-CoA, the alcohol species in the assay was varied.
Table 1 shows all substrates tested and the species that can
be used by the SLE27-encoded enzyme (SEQ ID N0:1B). Extensi-
ve in vitro enzyme assays using both GC-MS and radio-GC
revealed that a comprehensive array of acyl-CoAs and alco-
hols (short and long chain with even and uneven carbon
number, aromatic, cyclic and aliphatic, branched and unbran-
ched, saturated and unsaturated) can serve as substrates for
the SLE27-encoded enzyme (SEQ ID NO:1B) enzyme in vivo
(Table 1 & 2).
The alcohol acyltransferase activity of the the SLE27
encoded enzyme (SEQ ID NO:1B: SAAT) encoded protein was
established in two ways: (1) For the activity with different
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
62
alcohols and acetyl-CoA, [14C]-acetyl-CoA was used as sub-
strate and the formation of (radio-labeled) products analy-
zed using radio-gas chromatography and quantified using
liquid-scintillation counting. Radio-GLC was performed
essentially as described previously [Bouwmeester, H.J.
(1999) Plant Physiol. 121: 173-180 (1999)] Temperature
programming was as follows : 70 °C for 10 min, 10 min-1 to 270
°C and a final time of 10 min. For quantification of the
enzyme activity, 1.4 ~g SAAT protein was diluted to 100 ~,l
with 50 mM Tris-buffer (pH 8.3, to which [1'C] -acetyl-CoA
(0.1 mM at 0.2 Ci.mol-1 in routine assays) and alcohol (20 mM
in routine assays, 2 ~.l of hexane stock) were added. After
incubation for 30 min at 30 °C, the assays were cooled on
ice for 15 min and 700 ~.l of hexane were added. Assays were
vortexed, centrifuged and a 600-~1 portion of the hexane
phase was removed for liquid scintillation counting. The
ratio of hexane soluble radioactivity (esters) to the total
radioactivity added as acetyl-CoA was used to calculate
product formation. (2) For the utility of different acyl-
CoAs, assays containing 325 ~.1 of buffer (50 mM Tris/HC1 pH
8.0, containing 1 mM dithiothreitol), 25 ~,1 of protein elute
and 25 ~1 of acyl-CoA (4 mM in buffer) were stirred in a
glass vial. The enzyme reaction was started by the addition
of 25 ~.l of alcohol (160 mM in buffer). After 15-min incuba-
tion with continuous stirring (35 °C), solid CaCl2 was added
(final concentration 5 M). Volatiles released into the vial
headspace (at 35 °C with stirring) were subsequently trapped
for 15 min by exposing a fused silica fiber coated with 100
~.m polydimethylsiloxane (PDMS) to the headspace for 30 min.
The SPME-trapped volatiles were analyzed by GC-MS, as
described previously [Verhoeven et al., (1997). Chromato-
graphia 46, 63-66 ].
Table 1. Substrate specificity of the SLE27 (SAAT), SEQ ID
NO: 1B recombinant enzyme towards different types of alco-
hols. Comparison of esterification activity with different
alcohols (20 mM), and using [1'C]acetyl-CoA (0.1 mM) as acyl
donor. Activity ( mean ~ SD, n - 2) is expressed as nmol
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
63
product formed per hour per ~Cg enzyme. a- the ester product
was not reported to occur in commercial strawberry varieties
(data from Honkanen and Hirvi. 1990). b no detectable
activity. n.d. -- not described. Ester properties are from
Arctander, (1969).
Table 1
Alcohol Carbon Activity Ester formed Ester properties
no.
methanol C1:0 0.43 0.10methyl acetateethereal, sweet
ethanol C2:0 0.41t 0.07ethyl acetate pineapple, ethereal
I-propanol C3:0 2.60t 0.2G1-propyl acetatepowerful celery
odor
2-propanol C3:0 1.30t O.iS2-propyl acetateethereal, sweet,
banana, fruity
1-butanol C4:0 2.29t 0.251-butyl acetatefruity, diffusive
2-butanol C4:0 3.11 0.022-butyl acetaten.d.
3-methyl-1-butanolCS:O 3.68 0.253-methyl-1-butylfruity, banana,
acetate sweet
(Isoamylalcohol) (isoamyl acetate)
1-hexanol C6:0 8.44t 0.371-hexyl acetateapple, cherry,
pear, floral
cis-2-hexen-1-ofC6:1 6.05t 0.55cis-2-hexenyl n.d.
acetate'
cis-3-hexen-1-ofC6:1 4.06t 0.14cis-3-hexenyl green, sharp-fruity
acetate
traps-2-hexen-1-ofC6:1 9.20t O.6Smans-2-hexenylfresh-green,
acetate sweet, fruity
trnns-3-hexen-1-ofC6:1 1.25t 0.07traps-3-hexenyln.d.
acetate
1-heptanol C7:0 14.89t 4.12heptyl acetate'sweet, apricot
like
1-octanol C8:0 16.36t 2.691-octyi acetatefruity, floral,
jasmine, herba-
ceous
1-nonanol C9:0 14.00t 0.111-nonyl acetate'floral, fruity
(mushroom-
gardenia)
i-decanol C10:0 7.79 0.101-decyl acetatefloral, orange-rose
furfurylaicoholCS:O 0.72 0.06furfuryl acetate'fruity, banana
benzylalcoholC7:0 0.68 0.04benzyl acetatesweet, floral,
fruity, fresh
2-phenylethylalcoholC8:0 1.58 0.122-phenylethyl floral, rose,
acetate honey like
under-
tone, sweet,
fruity
linalool C10:0 " linalyl acetate'floral-woody
odor with faint
citrus note
Tabel 2.
Acceptance of different types of acyl-CoAs as co-substrates
by the SLE27 (SAAT), SEQ ID NO: 1B recombinant enzyme.
Esters formed after 15 min reaction of SAAT protein with the
substrates indicated were trapped using headspace-SPME and
GC-MS. a- the ester product was not reported to occur in
commercial strawberries [data from Honkanen and Hirvi. 1990
Dev. Food Sci . Amsterdam : Elsevier Scientific Publications,
125-193.]. n.d.- not described. Ester properties are from
Arctander, S. Perfume and Flavor Chemicals (Steffen Arctan-
der's Publications, Las Vegas, NV, (1969).
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
- 64 -
Tabel 2:
AcylCoAS (Carbon no.) Alcohol Ester formed Ester properties
n-propionyl CoA (C3:0) 1-butanol 1-butyl propanoate° ethereal, banana
n-butyryl CoA (C4:0) 1-propanol 1-propyl butyrate sharp, pungent, rancid,
sweaty, sickening
n-butyryl CoA (C4:0) 1-butanol 1-butyl butyrate fruity, pineapple
2-butyryl CoA (C4:0) 1-butanol 1-butyl-2-butyrate° fruity, apple,
banana
and pineapple
n-crotonoyl CoA (C4:1) 1-butanol 1-butyl crotoate° n.d.
n-hexanoyl CoA (C6:0) 1-propanol 1-propyl hexanoate° wine-like, cheese
n-decanoyl CoA (C10:0) 1-butanol 1-butyl decanoate° Brandy (Whisky-
Cognac)-like odor
1 0 benzoyl CoA (C7:0) 1-butanol 1-butyl benzoate° mild floral-balsamic
odor
Radio-GC analysis after incubations of the SLE27 (SAAT), SEQ
ID NO: 1B recombinant enzyme with [14C]-acetyl-CoA and either
octanol or hexanol showed the production of 14C-labeled
octylacetate and 1'C-labeled hexylacetate, respectively
(Figure 11). The kinetic properties of SAAT were determined
using [14C] -acetyl-CoA and five different alcohols. The
apparent Km and Vmax for acetyl-CoA were 104.2 ~.M and 44.5
nmol product.h-l.ug-1 protein, respectively. The apparent
values of Km ( in mM) and Vmax ( in nmol product . h-1. ~,g-1
protein) for octanol were 5.7 and 26.0, for hexanol 8.9 and
6.1, for butanol 46.1 and 4.2, for traps-2-hexenol 16.8 and
10.5 and for cis-2-hexenol 17.9 and 6.0, respectively.
Activity was also detected with aromatic (benzyl- and
phenylethyl-) and cyclic (furfuryl-) alcohols though activi-
ties were low compared to 1-octanol (4 to 10%). In contrast,
no activity could be detected with the terpene alcohol
linalool.
Example 5
Verification of alcohol acyl transferase activity of the
proteins from melon, strawberry vesca, banana, apple, mango
and lemon.
The alcohol acyl transferase activity of the proteins having
the sequences SEQ ID N0:16B to 23B, derived from strawberry
vests, banana, apple, mango, lemon (3x) and melon, respecti-
vely, was established in a similar way as described for
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
- 65 -
strawberry protein encoded by SEQ ID NO:lA in Example 4.
From an overnight culture of E.coli, 0.75 ml was transferred
into a 2 ml glass crimp vial, 0.3 g CaCl2 was added and a
small magnetic stirrer was put into the vial. The vial was
closed with a crimp cap, and incubated during 15 min at 50
deg. Celsius. The headspace of the vial was sampled with a
100 micro PDMS solid phase microextraction (SPME) device,
supplied by Supelco, during this period. The SPME fibre was
transferred into the injection port of a Fisons 68600
gaschromatograph, and desorbed at 250 deg. Celsius during l0
seconds. Bottom split was closed during this period of 10
seconds, and reopened afterwards. A HP-5 column was used
with 50m length, 0.32 mm ID and 1.05 micron film thickness.
The oven was programmed for 2 min. at 80 deg. Celsius, ramp
of 8 deg Celsius /min. to 250 deg. Celsius, for 5 min. at a
head pressure of 47 kPa. A MD 800 mass spectrometer was used
for the detection and identification of volatile esters, and
the resulting mass spectra from M/z 35 to 400 were compared
with the NIST database for identification. Where possible,
retention times were compared to that of authentic reference
compounds, obtained from Sigma or Aldrich.
The chromatograms obtained are shown in Fig. 12-18, of which
Fig. 12 shows the results of using SEQ ID NO:lA for compari-
son. The data obtained by the chromatograms are summarized
in the following tables 3 to 9, respectively.
Table 3.
Shows ester formation (A) E. coli cell cultures expressing
the strawberry alcohol acyl transferase (SLE27, SEQ ID
NO:lA). (B) E. co.ti cell cultures expressing the empty
vector cassette. Cultures supplemented with 1-butanol (Fig.
12) .
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
- 66 -
Peak number Retention time Ester formed
1 6.797 butanoic acid, isopropyl
ester
2 7.027 acetic acid, butyl ester
3 9.027 propionic acid, butyl ester
4 10.048 ester, not identified
5 11.028 butanoic acid, butyl ester
8 15.368 hexanoic acid, butyl ester
7 15.068 acetic acid, benzyl ester
6 13.248 pentanoic acid, butyl ester
9 17.278 butanoic acid, phenylmethyl
ester
Table 4.
Shows ester formation (A) E. coli cell cultures expressing
the melon alcohol acyl transferase (MAY5, SEQ ID N0:23A)).
(B) E. coli cell cultures expressing the empty vector
cassette. No alcohol supplements (Fig. 13).
Peak number Retention time Ester Formed
1 6.267 acetic acid, isobutyl ester
2 7.047 acetic acid, butyl ester
3 8.557 unknown ester
4 9.037 propionic acid, butyl ester
5 9.167 acetic acid, pentyl ester
6 10.048 unknown ester
7 10.438 acetic acid, 2-ethyl butyl ester
8 11.038 butanoic acid, butyl ester
9 11.408 acetic acid, hexyl ester
10 15.078 acetic acid, benzyl ester
11 15.378 hexanoic acid, butyl ester
12 17.048 acetic acid, phenyl ethyl ester
13 17.088 formic acic, unknown ester
14 17.298 butanoic acid, phenyl ethyl
ester
Table 5.
Shows ester formation (A) E. coli cell cultures expressing
the apple alcohol acyl transferase (MAY3, SEQ ID N0:18A).
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
- 67 -
(H) E. co~i cell cultures expressing the empty vector
cassette. Both cultures supplemented with isoamylalcohol, n-
butanol, cis-2-hexen-1-of and isopropanol(Fig. 14).
Peak number Retention time Ester formed
1 11.038 butanoic acid, butyl ester
2 17.089 formic acid, unknown ester
Table 6.
Shows ester formation (A) E. coli cell cultures expressing
the citrus alcohol aryl transferase (CLF26: SEQ ID N0:2A).
(H) E. coli cell cultures expressing the empty vector
cassette. Both cultures supplemented with 1-butanol (Fig.
15) .
Peak number Retention timeEster formed
1 7.047 acetic acid, butyl ester
2 10.086 propanoic acid, 2 methyl, butyl
ester
Table 7.
Shows ester formation (A) E. coli cell cultures expressing
the banana alcohol acyl transferase (MAY2, SEQ ID N0:17A).
(B) E. coli cell cultures expressing the empty vector
cassette. No alcohol supplements (Fig. 16).
Peak number Retention time Ester formed
1 17.089 formicacid, unknown ester
Table 8.
Shows ester formation (A) E. coli cell cultures expressing
the apple alcohol acyl transferase (MAY3, SEQ ID N0:18A).
(H) E. coli cell cultures expressing the empty vector
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
- 68 -
cassette. Both cultures supplemented with isoamylalcohol, n-
butanol, cis-2-hexen-1-of and isopropanol (Fig. 17).
Peak numberRetention timeEster formed
1 11.268 acetic acid, 1,2-hexenyl ester
2 15.268 butanoic acid, 1, 2-hexenyl ester
Table 9.
Shows ester formation (A) E. coli cell cultures expressing
the strawberry vesca alcohol acyl transferase (SUN1, SEQ ID
N0:16A (B) E. coli cell cultures expressing the empty vector
cassette. Both cultures supplemented with isoamylalcohol, n-
butanol, cis-2-hexen-1-of and isopropanol (Fig. 18).
Peak numberRetention timeEster formed
1 6.977 acetic acid, butyl ester
2 8.287 acetic acid, iso-amyl ester
3 11.278 acetic acid, 2 hexenyl ester
Sequence listing
INFORMATION FOR SEQ ID NO: lA (SLE27)
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
69
SEQUENCE CHARACTERISTICS:
LENGTH z6sz
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL ACYL TR.ANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 1 A
ACCTACTTTGCCAAAATGGAGAAAATTGAGGTCAGTATAAATTCCAAACA-
CACCATCAAACCATCAACTTCCTCTACACCACTTCAGCCTTACAAGCTTACC-
CTCCTGGACCAGCTCACTCCTCCGGCGTATGTCCCCATCGTGTTCTTCTACC-
CCATTACTGACCATGACTTCAATCTTCCTCAAACCCTAGCTGACTTAAGA-
CAAGCCCTTTCGGAGACTCTCACTTTGTACTATCCACTCTCTGGAAGGGT-
CAA.AAA.CAACCTATACATCGATGATTTTGAAGAAGGTGTCCCATACCTT-
GAGGCTCGAGTGAATTGTGACATGACTGATTTTCTAAGGCTTCGGAAA-
ATCGAGTGCCTTAATGAGTTTGTTCCAATA.AAACCATTTAGTATGGAAGCA-
ATATCTGATGAGCGTTACCCCTTGCTTGGAGTTCAAGTCAACGTTTTCGAT-
TCTGGAATAGCAATCGGTGTCTCCGTCTCTCACAAGCTCATCGATGGAG-
GAACGGCAGACTGTTTTCTCAAGTCCTGGGGTGCTGTTTTTCGAGGGT-
GTCGTGAAAATATCATACATCCTAGTCTCTCTGAAGCAGCATTGCTTTTCC-
CACCGAGAGATGACTTGCCTGA.AAAGTATGTCGATCAGATGGAAGCGTTAT-
GGTTTGCCGGAAAA.A.AAGTTGCTACAAGGAGATTTGTATTTGGTGTGAAAG-
CCATATCTTCAATTCAAGATGAAGCGAAGAGCGAGTCCGTGCCCAAGCCAT-
CACGAGTTCATGCCGTCACTGGTTTTCTCTGGAAACATCTAATCGCTGCTTC-
TCGGGCACTAACATCAGGTACTACTTCAACAAGACTTTCTATAGCGGCC-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
CAGGCAGTGAACTTAAGAACACGGATGAACATGGAGACAGTGTTGGATA-
ATGCCACTGGAAACTTGTTCTGGTGGGCACAGGCCATACTAGAGCTAAGT-
CATACAACACCAGAGATCAGTGATCTTAAGCTGTGTGACTTGGTTAACTTG-
CTCAATGGATCTGTCAAACAATGTAACGGTGATTACTTTGAGACTTTCAAG-
GGTAAAGAGGGATATGGAAGAATGTGCGAGTATCTAGATTTTCAGAGGAC-
TATGAGTTCTATGGAACCAGCACCGGATATTTATTTATTCTCGAGCTGGAC-
TAATTTTTTCAACCCACTTGATTTTGGATGGGGGAGGACATCATGGATTG-
GAGTTGCAGGAAAAATTGAATCTGCAAGTTGCAAGTTCATAATATTAGTTC-
CAACACAATGCGGTTCTGGAATTGAAGCGTGGGTGAATCTAGAAGAAGA-
GAAAATGGCTATGCTAGAACAAGATCCCCATTTTCTAGCGTTAGCATCTC-
CAAAGACCTTAATTTAAAGATATTGATTAAGAAAGATTATGTGGCTCGTG-
CAATGTTTCGATTTTGCAGTGAATAAGGTTTAAATTAGTTCACCAGCCAAT-
CAATAAAATGCAAGTATGATAGACTTTGTCTACGTATGTTATCCGAATGT-
GTTTCCATATGCTTGTAACCAATATAGCTCTTTATTGTAACAAATGCTCTAT-
TAAGCTTCTAGCTATAAAGTTATTTATCTATTAAAAATAA.AACTATGGAAGT-
TTTACCAAA.AAAAAA,AAAA.A
INFORMATION FOR SEQ ID NO: 1B (SLE27)
SEQUENCE CHARACTERISTICS:
LENGTH: 452
TYPE: Peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL ACYL TRANSFERASE
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
71
SEQUENCE DISCRIPTION FOR SEQ ID NO: 1B
MEKIEVSINSKHTIKPSTSSTPLQPYKLTLLDQLTPPAYVPIVFFYPITDHDFNL-
PQTLADLRQALSETLTLYYPLSGRVKNNLY117DFEEGVPYLEARVNCDMTDFL-
RLRKIECLNEFVPIKPFSMEAISDERYPLLGVQ VNVFD SGIAIGV S V SHKLIDGG-
TADCFLKSWGAVFRGCRENIIHI'SLSEAALLFPPRDDLPEKYVDQMEALWFAG-
KKVATRRFVFGVKAISSIQDEAKSESVPKPSRVHAVTGFLWKHI_.IAASRALTS-
GTTSTRLSIAAQAVNLRTR,MNMETVLDNATGNLFWWAQAILELSHTTPEIS-
DLKLCDLVNLLNGS VKQCNGDYFETFKGKEGYGRMCEYLDFQRTMS S-
MEPAPDIYLFSSWTNFFNPLDFGWGRTSWIGVAGKIESASCKFIILVPTQCGS-
GIEAWVNLEEEKMAMLEQDPHFLALASPKTLI
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
72
INFORMATION FOR SEQ 117 NO: 2A (CLF26)
SEQUENCE CHARACTERISTICS:
LENGTH: 1613
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: CITRUS LIMON ALCOHOL ACYL TR.ANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 2A
CTTATTTAAAAGTTCATCAACAAATTGTTCTACCACTTACCATTTCTCATAG-
CTCTGCAAGTTCGGATTTGACTCTTTCTCTTTTCCTCATTCCGGCCGGTGTT-
GATAGTTACATTTTGGCACAATGAAAATTCACGTTAAGGAGTCAACAATTA-
TACGCCCTGCTCAAGAAACACCCAAGCATCGCCTACAAATATCCGACCTA-
GACATGATTGTGCCATCCAATTACGTTCCCAGTGTGTATTTCTATCGGCG-
GTCCAGTGACTGCACCGATTTTTTTGAAGTTGGTTTGCTGAAGAAGGCTCT-
GAGCGAAGTTCTTGTGCCGTTTTACCCCGTTGCCGGAAGGTTGCAGAAG-
GATGAAAATCGCA.AAATTGAGATTCTATGTAACGGAGAGGGAGTTTTGTT-
TCTGGAGGCCGAAACAAGTTGTGGTATTGATGATTTCGGTGACTTCTCA-
CAAGGCTCGAAACTCCTGACGCTTGTTCCAACTGTTGGTGATACAAAGGA-
TATATCCTCCCATCCACTCTTGATGGCACAGGTAACTTATTTCAAATGTG-
GAGGCGTTTGTGTTGGAACTAGAGTGAATCATACACTGGTAGATGGAGCTT-
CAGCGTACCATATCATCAACTCATGGGCGGAGACGACGCGTGGCGTTCC-
TATTAGCACTCAACCATTCTATGATCGGACCATACTGAGTGTTGGGGTTC-
CAACTTCTCCCAAATTCCATCACATTGAATATGACCCGCCTCCTTCCAT-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
73
GAACGCTCCTCCTACCCAAA.ATCCTGAAATCATTTCTACCGCAATCCTTAAC-
CTATCACTTGATCAAATCCACACCCTCAAAGAGAAATCTAAGACAGATCAT-
GAACCCAACGTCAAGTATAGTAGGATGGCGATCCTAGCAGCACATATCTGG-
CGTAGCATGTGTAAAGCGCGCGGATTATCTGATGATCAAGTTAGCAAGTTA-
CACTTTCCTACAGACGGACGACAGAGATTGAATCCACCACTCCCGCCTGGA-
TATTTTGGAAATGTAATTTTCACCACGTCGTTGACGGCTTCATCGGGTGA-
TATCCTAAGTGAACCATTGAATCATACTGTTGAAAGAATTCAAAAAGCAT-
TAAAGCGGATGGACGATGAGTATTTGAAATCAGCACTTGCTTACCTAAAG-
CAACAGCCTGATTTAAATGCTCTACGGAAAGGAGGCCACATTTACAAGTGC-
CCTAACCTCAATATCGTCAATTTGGCAAATATGCCAATGTATGTTGCGAATT-
TTGGATGGGGCCAGCCGATATTTGCGAGGATCGTTAACACATATTATGAAG-
GGATAGCACATATTTATCCAAGTCCGAGCAATGATGGGACCTTGTCAGTG-
GTTATAAACTCGGTAGCCGATCACATGCAGCTGTTCAAGAAGTTCTTT-
TACGAGATCTTTGATTAAGGTATGAAAGACCTAGGTATTTTATATTTTCTA-
GAAATGTCACTTTTTTTTTTTTTTTTTTTTGGGGGCGCAAATGTTGTCTTAC-
TTGGAATTTTATATATTTTAATCCATGTTTTTATGGAAGGCAGTGGTGTTG-
C
INFORMATION FOR SEQ ID ISO: 2B (CLF26)
SEQUENCE CHARACTERISTICS:
LENGTH: 434
TYPE: Peptide
STRANDNESS: Single
TOPOLOGY: Linear
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
74
OTHER INFORMATION: CITRUS LIMON ALCOHOL ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 2B
MKIFM~ESTIIRPAQETPKHRLQISDLDMIVPSNYVP SVYFYRRS SDCTDF-
FEVGLLKKALSEVLVPFYPVAGRLQKDENRKIEILCNGEGVLFLEAETSCGIDD-
FGDFSQGSKLLTLVPTVGDTKDISSHPLLMAQVTYFKCGGVCVGTRVNHTL-
VDGASAYHIINSWAETTRGVPISTQPFYDRTILSVGVPTSPKFHHIEYDPPPSM-
NAPPTQNPEIISTAILNLSLDQIHTLKEKSKTDHEPNVKYSRMAILA,~~I3IWR.SM-
CKARGLSDDQVSKLHFPTDGRQRLNPPLPPGYFGNVIFTTSLTAS SGDILSEPLN-
HTVERIQKALKRMDDEYLKSALAYLKQQPDLNALRKGGHIYKCPNLNIVN-
LANMPMYVANFGWGQPIFARIVNTYYEGIAHIYPSPSNDGTLSVVINSVADHM-
QLFKKFFYEIFD.
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
INFORMATION FOR SEQ 117 NO: 3A (SLF96)
SEQUENCE CHARACTERISTICS:
LENGTH: 1586
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY AMINOTRANSFERASE
SEQUENCES DISCRIPTION FOR SEQ 117 N0:3A
AAACCGTCGGCGTCTGTAAATGCGTCGCCGCTCCGGAGAAGACAGAGTA-
CAAGACTCAGGTGAATCGCAATGCCAACATGGCCAAGCTTCAAGCCGGT-
TATCTTTTTCCAGAGATTGCGAGGAGGAGGAATGCGCACTTGCAGAAGCAC-
CCTGATGCGAAGATAATTCCACTTGGAATTGGTGATACTACCGAGCCAAT-
TCCAGAATATATAACCTCTGCAATGGCAAAGAGAGCACTTGCCATGTCCAC-
CCTAGAGGGTTACAGTGGTTATGGACCTGAACAAGGTGAAAAGCCACTGA-
GAGTTGCAATTGCTAAAACGTTTTATGGCGACCTTGGCATAGAGGAAGAT-
GACATATTTGTTTCTGATGGGGCA.AAATGTGACATATCCCGCCTTCAGGT-
TCTTTTTGGGGCGGATAAA.ACAATAGCAGTGCAAGATCCATCGTATCCGGC-
TTATGTAGACTCAAGTGTTATTATGGGCCAGACAGGACAGTATCAGAAATC-
TGTTCAGAAGTTTGGAAACATCGAGTACATGAGGTGTACTCCCGATAATG-
GATTTTTTCCTGATCTGTCCTCTACTAAGCGAACAGATATCATATTTTTCT-
GTTCACCAAACAATCCTACTGGTTCTGCTGCAACAAGGGAGCAACTGACA-
CAACTTGTAAAGTTTGCCAAGGATAATGGTTCAATCATAGTCTATGATTCT-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
76
GCATATGCCATGTATATGTCAGATGATAATCCACGCTCCATCTTTGAAATCC-
CTGGAGCTAAAGATGTTGCACTTGAGACATCATCATTTAGTAAGTATGCCG-
GATTCACTGGAGTTCGTTTGGGGTGGACTGTGGTTCCAAAGCAGTTGCAG-
TATTCAGATGGTTTTCAAGTTGCCAAGGATTTCAACCGCATTGTTTGTACTT-
GCTTCAATGGTGCATCCACTATTATCCAAGCTGGTGGTCTGGCTTGCCTT-
CAACCAAAGGGTGTTAAGGCTATGCACGGTGTGATAAATTTCTACAAA-
GAAAATACTAAGATCATAATGGAGACGTTTAACTCTCTTGGCTTTAACGTG-
TATGGAGGGACAAACGCTCCATATGTGTGGGTCCACTTCCCTGGACAAAGC-
TCCTGGGATGTGTTTGCTGAGATCCTTGAGAAGACTCATGTGGTAACCA-
CACCTGGAAGTGGCTTTGGACCTGGTGGTGAAGGTTTCATCAGGGTAAGTG-
CCTTTGGACACAGGA,AAAATATATTAGAAGCATGTAAAAGATTCAAGCAAT-
TATACAAGTGAGGACTGCGGATCTGAATTGTAGACCAGTTTCTACTGCATG-
CTAGTTGAACCTATTTGCCTCCCATTTCCGTTCTATGCTAAATATTTTAG-
CACGTTCCAATTCCGTATTCAGTTTGTCGGCTTTAGTTTATGAATTATGGA-
GATTTTAGCTATTGTAAAAA.TGATTCGATCAGCCTTGTTTTCATGTGTTA-
CACTTAATTGTTGTAACATTTGTGAGGATCAGAAGCTTTGATTCTGTTTGC-
TAGAATAGTATAATTTTACCTAAATAAAGTGGTTGATCTTTCTTGGCCTG-
C
INFORMATION FOR SEQ ID NO: 3B (SLF96)
SEQUENCE CHARACTERISTICS:
LENGTH: 397
TYPE: Peptide
STRANDNESS: Single
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
TOPOLOGY: Linear
77
OTHER INFORMATION: STRAWBERRY A~'vITNOTRANSFERASE
SEQUENCES DISCRIPTION FOR SEQ ID N0:3B
MAKLQAGYLFPEIARRRNAHLQKHPDAKIIPLGIGDTTEPIPEYITSAMAKRA-
LAMSTLEGYSGYGPEQGEKPLRVAIAKTFYGDLGIEEDDIFVSDGAKCDISR-
LQVLFGADKTIAVQDPSYPAYVDSSVIMGQTGQYQKSVQKFGNIEYMRCTPD-
NGFFPDLSSTKRTDIIFFCSPNNPTGSAATREQLTQLVKFAKDNGSIIVYDSAYA-
MYMSDDNPRSIFEIPGAKDVALETSSFSKYAGFTGVRLGWTVVPKQLQYSDG-
FQVAKDFNRIVCTCFNGASTIIQAGGLACLQPKGVKAMHGVINFYKENTKI-
IIVVIETFNSLGFNVYGGTNAPYVWVHFPGQ S S WDVFAEILEKTHWTTPGS GFGP-
GGEGFIRVSAFGHRKNILEACKRFKQLYK
CA 02353577 2001-06-O1
WO 00/327$9 PCT/NL99/00737
78
INFORMATION FOR SEQ ID NO: 4A (SLG150)
SEQUENCE CHARACTERISTICS:
LENGTH: 1775
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY THIOLASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 4A
CGCTCCTTTGATTTCCTTGTTTCAATTATCAAGAGTATGGAGAAAGCGAT-
CAACAGGCAGAAGGTTCTCCTCGACCATCTCCGACCTTCTTCTTCTTC-
CGACGACTCTTCTCTCTCCGCGTCGGTATGTGCGGCTGGGGATAGCGCTG-
CGTATGCTAGGAATCATGTCTTTGGGGACGATGTCGTCATCGTTGCAGCTT-
TTCGCACTCCACTCTGCAAGGCTAAGCGTGGCGGCTTCAAGTATACTTATG-
CTGATGATCTCCTCGCACCTGTCCTCAAGGCCGTGGTTGAGAAAACCAATC-
TCAATCCCAAGGAAGTCGGGGATATTGTTGTCGGTACCGTCTTGGCCCCAG-
GATCTCAGAGAGCTAGCGAATGCAGGATGGCTGCTTTCTATGCTGGCTTCC-
CTGAGACTGTGCCGGTTAGAACTGTGAACAGACAATGTTCGTCTGGCCTC-
CAAGCAGTTGCTGATGTTGCTGCTGCCATTAGAGCAGGGTTTTATGATATT-
GGCATTGGTGCTGGTTTGGAATCCATGACTGCAAACCCAATGGCATGG-
GAAGGGGATGTTAATCCTAAAGTAAAGATCTTTGAACAAGCCCAGAATTGC-
CTTCTTCCTATGGGAGTCACCTCAGAA.AATGTTGCTCATCGTTTTGGTGTTT-
CAAGACAGGAGCAAGATCAGGCTGCAGTTGACTCTCATAGAAAGGCAGCT-
GCTGCTGCTGCTGCTGGTAGATTTAAAGATGAAATCATCCCTGTGGCAAC-
CA 02353577 2001-06-O1
WO 00/32?89 PCT/NL99100737
79
CAAGATTGTTGATCCAAAATCTGGTGATGAGAAACCTGTTACA.ATCTCT-
GTTGATGATGGGATTCGAAACACAACATTGGCGGACCTAGCAAAGCT-
GAAGCCTGTGTTTAAGAAAGATGGGACCACCACTGCTGGTAATTCTAGT-
CAAGTTAGTGATGGTGCTGGAGCTGTTCTCTTGATGAAGAGAAGTGTTGC-
CGACCAAAAAGGATTGCCGATTCTTGGTGTATTCAGGAATTTTGTTGCT-
GTTGGTGTGGATCCTGCCATCATGGGTGTTGGCCCAGCTGCTGCAATTC-
CAGTTGCAGTTAAGGCAGCTGGTTTAGAGCTTGATGATATTGACCTTTTT-
GAGATAAATGAGGCTTTTGCATCCCAATTTGTGTATTGCCGTAACAAGCTG-
GGACTTGATCCAGA~A.AAAATCAATGTTAACGGAGGTGCAATGGCCATCGGC-
CATCCACTTGGTGCAACAGGTGCCCGGTGTGTTGCCACTCTTTTGCATGA-
GATGAAGCGTCGTGGTAAAGACTGCCGCTATGGAGTGATCTCAATGTG-
CATAGGCACAGGGATGGGTGCAGCCGCTGTTTTTGAAAGAGGAGACCG-
GACCGATGAACTCTGCAATGCTCGCAAGGTTGAATCACTCAACTTCTTATC-
CAAGGATGTTCGGTAGTAGAGAATGGTTAGTGACAGGAGCTATTCCAATCA-
ATAATGTTTGGTGGAGTCTGAA.A.ATCATAGTAAAGCACTGGAATAACGTTG-
CTAAGTTTTTCGTTGGGTACTACCTTGTTTATTGGGATGGAATACACATG-
TAGTTGGTTTGTTCTCCCAGACCTCCCACTTGTTGGCATATTCATTTTTGTC-
CAACCTAAAAAGTTCCATTTTATAGGACTTCATCTCAATAACATTGGGTTTG-
CGCCACTAAAGCAGTGCCTAAAACTGTAATTGGGTAATTTTGGTATACCT-
GTTTGCTACTTTTCTTTTCTAAGTTAATCAAGCCCTGCCCACCTCATA-
TAAA AA.A,AAA,~~,AAAAA
INFOR.~VIATION FOR SEQ ID NO: 4B (SLG150)
SEQUENCE CHARACTERISTICS:
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
LENGTH: 4 S 8
TYPE: Peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY THIOLASE
SEQUENCE DISCRIPTION FOR SEQ m NO: 4B
MEKAINRQKVLLDHLRPSS S SDDS SLSAS VCAAGDSAAYARNHVFGDDVVI-
VAAFRTPLCKAKRGGFKYTYADDLLAPVLKAVVEKTNLNPKEVGDIVVGTV-
LAPGSQRASECRMAAFYAGFPETVPVRTVNRQCSSGLQAVADVAAA.IRAGFY-
DIGIGAGLESMTANPMAWEGDVNPKVKIFEQAQNCLLPMGVTSENVAHRFGV-
SRQEQDQAAVDSHRKAA.AA.AAAGRFKDEIIPVATKIVDPKSGDEKPVTISVD-
DGIRNTTLADLAKLKP VFKKDGTTTAGNS SQV SDGAGAVLLMKRS VADQKGL-
PILGVFRNFVAVGVDPAIMGVGPAAAIPVAVKAAGLELDDff~LFEINEA-
FASQFVYCRNKLGLDPEK:INVNGGAMAIGHPLGATGARCVATLLHEMKRRGK-
DCRYGVISMCIGTGMGAAAVFERGDRTDELCNARKVESLNFLSKDVR
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
81
INFORMATION FOR SEQ ID INTO: SA (SLH51)
SEQUENCE CHARACTERISTICS:
LENGTH: 2141
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY PYRUVATE DECARBOXYLASE
SEQUENCE DISCR.iPTION FOR SEQ ID NO: SA
ATTTTCACTCAGAGTCTCAATCTTTCATCACAAAAATTCCCATTTGATCA-
CA,AAAAAGTTTCAACCTTTAAACCTCCATGGACACCAAGATTGGCTC-
CATCGACGTCTGCAAAACCGAGAACCACGACGTCGGTTGTTTACCAAACAG-
CGCCACCTCCACCGTTCA.AAACTCAGTCCCTTCCACCTCCCTCAGCTCCGC-
CGACGCCACCCTCGGCCGCCACCTGGCACGCCGCCTCGTTCAAATCGGCGT-
CACCGACGTCTTCACCGTCCCCGGCGACTTCAACTTGACCCTTCTTGACCAC-
CTCATCGCCGAGCCCGGCCTCACCAACATTGGCTGCTGCAACGAGCT-
CAACGCCGGGTACGCCGCCGACGGCTACGCGCGGTCGCGTGGCGTCGGCG-
CGTGCGTGGTGACTTTCACTGTTGGTGGACTGAGTGTGCTGAACGCGATCG-
CCGGCGCGTATAGTGAGAATTTGCCGGTGATTTGTATTGTTGGTGGGCC-
CAACTCTAACGATTATGGGACTAACCGGATTCTTCACCATACTATTGGGTT-
GCCGGACTTCAGTCAAGAGCTCCGGTGCTTTCAGACCGTGACTTGCTTT-
CAGGCTGTGGTGAATAATCTGGAGGATGCACATGAGATGATTGATACTGCA-
ATTTCGACTGCGTTGAAAGAAAGCAAGCCTGTGTATATCAGCATTGGCTG-
CAACTTGGCTGGGATTCCTCATCCTACTTTCAGCCGTGAACCTGTTCCATTT-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
az
TCATTGTCTCCAAA.ATTGAGCAATA.~GTGGGGATTAGAGGCTGCAGTG-
GAGGCTGCTGCAGAGTTCTTGAACAAGGCAGTGAAGCCAGTTATGGTGGG-
CGGGCCCAAACTGCGCTCTGCACATGCTGGTGATGCCTTTGTTGAACTGGC-
TGATGCTTCTGGATTTGCTCTGGCTGTGATGCCATCAGCAAAGGGGCAAGT-
GCCAGAGCACCACCCCCATTTCATCGGAACGTACTGGGGTGCTGTGAGCAC-
TGCCTTTTGTGCTGAGATTGTGGAGTCTGCAGATGCATACTTGTTTGCTG-
GGCCGATTTTCAATGACTACAGCTCAGTTGGGTACTCGCTCCTTCTCAA-
GAAAGAGAAGGCGATCATTGTGCAGCCAGATCGTGTGACGATAGGGAAT-
GGCCCTACATTTGGTTGTGTTCTCATGAAGGATTTCCTCTTAGGCCTAG-
CAAAGAAGCTGAAGCATAACAACACTGCTCATGAGAACTACCGCAGGATCT-
TTGTGCCTGATGGCCACCCTCTGAAGGCTGCACCCAAAGAACCTTTGAGG-
GTTAATGTTCTGTTCAAACACATTCAGAATATGCTGTCAGCTGAAACCGCT-
GTGATTGCTGAGACAGGGGACTCATGGTTTAACTGTCAGAAGCTGAAATTG-
CCACCCGGCTGCGGGTATGAGTTCCAAATGCAATATGGATCAATTGGTTG-
GTCAGTTGGAGCAACTCTTGGGTATGCTCAGGCTGTACCTGAGAAGCGAGT-
GATTTCTTTCATTGGTGATGGGAGCTTCCAGGTGACTGCTCAAGATGTGTC-
CACAATGATTCGAAATGGACAGAGAACCATTATTTTCCTGATAAACAATG-
GTGGATACACCATTGAAGTGGAAATCCATGATGGACCATACAATGTGAT-
CAAGAACTGGAACTACACTGGACTGGTTGATGCAATCCACAATGGG-
GAAGGCAAGTGCTGGACAACCAAGGTGCGTTGCGAAGAGGAGCTGATT-
GAAGCAATAGAGACTGCAAATGGACCCAAGAAGGATAGCTTCTGCTTCATT-
GAGGTGATTGTTCACAAGGATGATACCAGCA.<IAGAGTTGCTTGAGTGGGG-
GTCTAGGGTTTCTGCTGCCAACAGCCGCCCACCTAATCCTCAGTAA.A.ACTC-
TCCTGTGTCATATGAAGGCCTTCATTCACATTCACAGATTTAGATCAAGC-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
83
CAAGCTCTTGTGCAAATTTTCCTTATGTTTTTCCTGTCAACTGGAGAATGGT-
GTCTGTCAAGTTTTTTTTACACTACAGTGATTTCTGGTTTGTCTGTATATT-
TCCTTCTGAATATTAGTATCTTCTGATTTTTCAATTGATCAAATTCTGT-
GATCCTAAATGGTTTGTGG
INFORMATION FOR SEQ ID NO: SB (SLHS 1 )
SEQUENCE CHARACTERISTICS:
LENGTH: 605
TYPE: Peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY PYRUVATE DECARBOXYLASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: SB
MDTKIGSID VCKTENHDVGCLPNSATSTVQNS VPSTSLS SADATLGRHLARRL-
VQIGVTDVFTVPGDFNLTLLDHLIAEPGLT'NIGCCNELNAGYAADGYARSR-
GVGACVVTFTVGGLSVLNAIAGAYSENLPVICIVGGPNSNDYGTNRILHHTI-
GLPDFSQELRCFQTVTCFQAVVNNLEDAHEMI)7TAISTALKESKPVYISIGCN-
LAGIPHPTFSREPVPFSLSPKLSNKWGLEAAVEAAAEFLNKAVKPVMVGGPKL-
RSAHAGDAFVELADASGFALAVMPSAKGQVPEHHPHFIGTYWGAVSTAF-
CAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVTIGNGPTFGCV-
LMKDFLLGLAKKLK:HNNTAHENYRRIF VPDGI-B'LKAAPKEPLRVNVLFK-
HIQNMLSAETAVIAETGDSWFNCQKLKLPPGCGYEFQMQYGSIGWSVGATL-
GYAQAVPEKRVISFIGDGSFQVTAQDVSTMIRNGQRTIIFLINNGGYTIEVEIH-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
84
DGPYNVIKT~VNYTGLVDAI~-INGEGKCWTTKVRCEEELIEAIETANGPKKDSF-
CFIEVIVHKDDTSKELLEWGSRVSAANSRPPNPQ
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
INFORMATION FOR SEQ ID NO: 6A (SLB39)
SEQUENCE CHARACTERISTICS:
LENGTH:141 S
TYPE: cDNA
STR.ANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ m NO: 6A
TAATCTAGCTTCTGCACCAAAACTATCAGATAATTAAGAATCTGCCACAGA-
GAAAATGGTGATGTCTATCGAGCAGGAACACCCCAAGAAGGCATCTGGAT-
GGGCTGCAAGAGATTCATCTGGTGTTCTCTCTCCCTTCAGTTTCTCCAGAAG-
GGAAACCGGAGAGAAAGACGTGACGTTCAAAGTGATGTACTGTGGGATTT-
GCCATTCGGACCTTCACATGGTCAAGAATGAATGGGGCTTCTCTACCTATC-
CTCTGGTTCCAGGGCATGAGATTGTTGGTGAAGTGACGGAAGTAGGAAG-
CAATGTACA,AA.AATTCAAAGTTGGAGACAGAGTCGGTGTTGGATGCATTGT-
GGGATCTTGCCGATCTTGTGA.A.A.ATTGTACCGACCACCTTGAGAACTACTG-
CCCCAAACAGATACTCACTTACGGTGCCAAGTACTACGACGGAACCACCAC-
CTATGGCGGTTACTCTGACATTATGGTGGCCGATGAACACTTCATAGTACG-
CATCCCAGACAACTTGCCTCTTGATGGTGCTGCGCCGCTCCTATGTGCCGG-
GATTACAACCTACAGCCCCCTGAGATATTTCGGACTTGACAAGCCCGGCAT-
GCATGTAGGTGTGGTCGGCCTAGGCGGTTTAGGCCACGTCGCCGTGAAGT-
TTGCCAAGGCTATGGGAGTGAAGGTTACAGTGATTAGTACATCCCCTAA-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
86
GAAAGAGGAGGAAGCTCGTAAACACCTAGGAGCTGACTCGTTTTTGGTTAG-
CCGTGACCAAGATCA.AATGCAGGCTGCCATTGGTACCATGGATGGGAT-
CATTGACACGGTTTCTGCACAACATCCTCTCCTGCCTTTGATTGGTTTGTT-
GAAGTCTCATGGAAAGCTTGTTATGGTTGGTGCACCAGAGAAGCCTCTT-
GAACTGCCAGTTTTTCCTTTACTCATGGGAAGAAAGATGGTAGCTGGTAG-
CGGCATTGGGGGTATGAAGGAGACACAAGAGATGATAGATTTTGCAGC-
CAAGCACAACATTACAGCAGACATCGAAGTCATACCAATCGACTACTTG-
TAACACTGCTATGGAGCGTCTAGTCAAAGCAGATGTCAGATACCGTTTTGT-
CATCGACATTGGAAACACACTGAAGGCTAGCTCTTAAATTCTGCAATCCA-
GACTGGATCAATGAAGAAACAAGAACAGAAACGGAGACTGATTTAGTGT-
CATACTCGGTGTTGGTTTTCCTTGTAGCATTTTTTGTTGTCTGCTACATGA-
ATAATGATCACATGAACAACTGCCTTCTGTGATGATTTGATAATAA.AAGA-
ATACATGAACAATGATACTGCCTTCTTTTGTAATGTTTTTTACTATATAAT-
CATTTCAAATTATTTTGCTA-
TATCTCT
INFORMATION FOR SEQ ID NO: 6B (SLB39)
SEQUENCE CHARACTERISTICS:
LENGTH: 3 3 3
TYPE: Peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 6B
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
87
MVMSIEQEHPKKASGWAA.R.DSSGVLSPFSFSRRETGEKDVTFK~'MYCGICHS-
DLHMVICNEWGFSTYPLVPGHEIVGEVTEVGSNVQKFKVGDRVGVGCIVGSCR-
SCENCTDHLENYCPKQILTYGAKYYDGTTTYGGYSDIMVADEHFIVR1PDNL-
PLDGAAPLLCAGITTYSPLRYFGLDKPGMHVGWGLGGLGHVAVKFAKAM-
GVKVTVISTSPKKEEEARKHLGADSFLVSRDQDQMQAAIGTMDGIIDTVSAQH-
PLLPLIGLLKSHGKLVMVGAPEKPLELPVFPLLMGRKMVAGSGIGGM-
KETQEMIDFAAICI-EVITADIEVIPI17YL
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
88
rNFORMATIOl~T FOR SEQ LD NO: 7A (SLF193)
SEQUENCE CHARACTERISTICS:
LENGTH: 1227
TYPE: partial cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 7A
GGAAACAGGAGCAACGGACGTAAGATTCAAAGTGTTGTACTGTGGAGTAT-
GCCATTCGGACATACACATGGCCAAAAATGATTGGGGGACTTCTACCTATC-
CTATTGTACCTGGGCATGAACTTGTTGGTGTAGTAACAGAAGTAGGATG-
CAAAGTAAAGAAATTCAAA.AGTTGGAGACAAGGTCGGTGTTGGTTGCATG-
GTCGACTCAGACCAACTTGCGAA.A.ATTGTATCCATCACCTAGAA.AATTACT-
GTCCGAATCTGATACAAACCTACGGTTCTAA.ATACTACGACGGAACCATGA-
CATACGGAGGTTACTCGAACAACATGGTGACTGATGAGCACTTCATTGT-
TCGGATCCCGGACAACTTACCTCTTGATGGCGCTGCTCCGCTTCTATCTGC-
CGGGATTACAACTTACAGCCCATGGAGATATTATGGACTTGACAAACCCGG-
TATGCATCTTGGTGTTGAATGGCCTAGGCGGTTTAGGTCACGTCCGCCGT-
TAAATTTGCCAGGGCTTTGGGGCTCAAGGTTACAGTCATTAGTACCTCCCC-
TAATTAAAGAAGGAGGCAGCTATGGAACATCTCCCGCGCTGATGCATTCCC-
TGCTTAGAACTGACCAAGATCAGATGGAGGCTGCCATGAGCACAATGGAT-
GGTATCATTGACACAGTTCCTGCAGTTCGACCTCTAGAGCCTTTGATTT-
CATTGTTGAAGACTAATGGAAAAGTTGTTACCGTTGGTATAGCAGTGCAGC-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
89
CACTCGATCTCCCAGTTTTCCCTTTGATAATAGGAAGGAAGATGGTAGCTG-
GTAGTGCCATTGGAGGTATGAAAGAGACGCAAGAGATGATTGATTTTGCTG-
CTGAACATAACATAACAGCTGACATCGAGGTCATCCCGATTGATTACCT-
GAACACCGCAATGGAACGCGTTGTCA,AAAAAGATGTCAGGTTTCGATTTGT-
CATCGACGTTGAGAACACATTGTAAGTCCGCCTAAGTTTTTCATTCAATTCT-
GTTAATAAGACTATGCATTAATATATGACTGACTCTCCATAGGATGGAGT-
TATCAGTCTTCAAATTTCTAGACATATTTTGTGATCAAATAAATGGAATGGC-
TTTGTTTTCCTTTTCCACTAAGATTAGATTTCAGTTGTATTGTTTTTAAAGA-
GATTGATGTTTTTATTAATTGTAACAGTGTTATCAGTCTAATCAT-
T
INFORMATION FOR SEQ ID NO: 7B (SLF193)
SEQUENCE CHARACTERISTICS:
LENGTH: 326
TYPE: partial peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ TD N0: 7B
ETGATDVRFKVLYCGVCHSDIH~1~IAICNDWGTSTYPIVPGHELVGVVTEVG-
CKVKKFKSWRQGRCWLHGRLRPTCENCIHHI~ENYCPNLIQTYGSKYYDGTM-
TYGGYSNNMVTDEHFIVRIPDNLPLDGAAPLLCAGITTYSPWRYYGLDKPGM-
HLGVEWPRRFRSRPPLNLPGLWGSRLQSLVPPLIKEGGSYGTSPALMHSLLRT-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
DQDQMEAAMSTMDGIIDTVPAVRPLEPLISLLKTNGKWTVGIAVQPLDLPVF-
PLIIGRKMVAGSAIGGMKETQENImFAAEHNITADIEVIPII7YLNTAMERWKK-
DVRFRFV117VENTL
INFORMATION FOR SEQ ID NO: 8A (SLF122)
SEQUENCE CHARACTERISTICS:
LENGTH: 1063
TYPE: partial cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 8A
GCAAAGTGCAAAAATTTAAAGTTGGAGACAAAGTTGGTGTTGGGTGCTTGG-
TAGGCTCATGCAAAACTTGCGACAGCTGTGCTAACGATTTGGAGAACTACT-
GCCCCAAACAGATACAGACTTACGGCGCCAAGTACCTTGACGGAACAAC-
CACATACGGCGGTTACTCTGACATCATGGTGGCGGATGAGGCCTTTGTA-
ATCCGTATTCCGGACAACCTGCCTCTTGAGGGTGCTGCTCCTCTCCTATGTG-
CCGGAATCACAACTTACAGTCCCCTGAGGTATTTCGGACTTGACAAACC-
CGGCATGCATGTCGGGGTGGTTGGCCTTGGCGGTTTAGGCCATGTCGCGGT-
GAAGTTTGCCAAGGCTTTGGGGGTTAATGTCACAGTGATCAGTACCTCCGC-
TAATAAGAAAGATGAAGCTATTAAACACCTTGGTGCTGATTCTTTCTTGGT-
CAGTCGTGACCAAGATCAGATGCAGGCTGCCATGGGAACATTGGACGGTAT-
CATCGACACAGTTTCCGCAGTCCACCCCCTCCCACCTTTGATTAGTTTATT-
GAAGGCTAATGGAAAGCTTGTTATGGTTGGAGCACCAGAGAAGCCACTT-
GAGCTACCAGTTTTTTCTTTAATAATGGGAAGGAAGACTTTAGCCGGTAG-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
91
TAATATCGGAGGTATCAAGGAGACACAAGAGATGATAGATTTGGCAGC-
CAAACACAACATAACGGCCGACATCGAGATTATCCCCATCGACTATTTGAA-
CACTGCTATGGAGCGTCTTGCTAAAGGGGATGTTAGATACCGTTTTGT-
CATCGACATCGGAAACACATTGAAGCCGGCCATTTAAATTTGCATTTCGAT-
CAGAAACTGAATCAAGCGAGGTCGAGAGGCCTACGTAACAATGCAAACAT-
GTGCTAGCTTGTTCTTGGAGTAGTCTTTAGCTTTTCTCTGATGTATTCCATC-
TGTTTTGTTCATGTCCCATCTTATTATGAGAA.AAATGTGGGTACCGTGGA-
TATTGAATAAATGAAGAGCTACTGGAACGATGGTTTCA-
C
INFORMATION FOR SEQ ID NO: 8B (SLF122)
SEQUENCE CHARACTERISTICS:
LENGTH: 278
TYPE: partial peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 8B
KVQKFKVGDKVGVGCLVGSCKTCDSCANDLENYCPKQIQTYGAKYLDGTT-
TYGGYSDIMVADEAFVIRIPDNLPLEGAAPLLCAGITTYSPLRYFGLDKPGM-
HVGWGLGGLGHVAVKFAKALGVNVTVISTSANKICDEAIKHLGADSFLVSRD-
QDQMQAAMGTLDGIIDTVSAVHPLPPLISLLKANGKLVMVGAPEKPLELPVF-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
92
SLIMGRKTLAGSNIGGIKETQEMIDL_~A.ICI-~'ITADIEIIPIDYLNTAMERLAKGD-
VRYRFVIDIGNTLKPAI
INFORMATION FOR SEQ ID NO: 9A (SLD 194)
SEQUENCE CHARACTERISTICS:
LENGTH: 1228
TYPE: partial cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ 117 NO: 9A
GCAAGAGATTCATCTGGTGTCCTCTCTCCCTTCAATTTCTCCAGAAGG-
GAAACCGGAGAGAAAGACGTTATGTTCAAAGTGTTGTACTGTGGAATTTGC-
CATTCGGACCTTCACATGGTCAAGAATGAATGGGGCTTCTCTACCTATCCT-
TTGGTCCCGGGGCATGAGATTGTTGGTGAAGTTACGGAAGTAGGGAG-
CAAAGTACAA.AAATTTAAAGTTGGAGACAGAGTCGGTGTTGGATGCGTTGT-
GGGATCTTGCCGATCTTGTGAAAATTGTACCGACCACCTTGAGAACTACTG-
CCCCAAACAGATACTCACTTACGGTGCCAAGTACTACGACGGAACCACCAC-
CTATGGCGGTTACTCTGACATTATGGTGGCCGACGAACACTTCATAGTACG-
CATCCCAGACAACTTGCCTCTTGATGGCGCTGCGCCGCTCCTATGTGCCGG-
GATTACA.ACCTACAGCCCCCTGAGATATTTCGGACTTGACAAGCCCGGCAT-
GCATGTAGGTGTGGTCGGCCTAGGCGGTTTAGGCCACGTCGCCGTGAAGT-
TTGCCAAGGCTATGGGAGTGAAGGTTACAGTGATCAGTACGTCCCCTAA-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
93
GAAAGAGGAGGAAGCTCTTAAACACCTAGGAGCTGACTCGTTTTTCGTTAG-
CCGTGACCAAGATCAAATGCAGGCTGCCATTGGTACCATGGATGGGAT-
CATTGACACAGTTTCTGCACAACATCCTCTCCTGCCTTTGATTGGTTTGTT-
GAAGTCTCATGGAAAGCTTGTTATGGTTGGTGCACCAGAGAAGCCTCTT-
GAACTTCCAGTTTTTCCTTTACTCATGGGAAGAAAGATGGGTAGCTGG-
TAACCGGCATTTGGGGGTATGAAGGAGACACAAGAGATGATAGATTTTGC-
TGCCAGGCACAACATAACAGCAGACATCGAAGTCATACAATCGACTACT-
TAAACACTGCTATGGAGCGTTTAGTCAAAGCAGATGTCAGATACCGTTTT-
GTCATCGACATTGGAAACACACTGAAGGCTAGCACTTAAATTCTGCAATC-
CAGACTGTATCAATGAAGAAACAAGAACAGAAACTGAGATTGATTTGGTGT-
CATACTCCGCCTATGGTTTTCCTTACAGCATTTTTTGTTGTTTGCTACATGA-
ATAACGATCACATGAACTGTGATGATTTGATAATAAAAGAATACATAAA-
CAAAAAAAACAAAAAA AA
INFORMATION FOR SEQ ID N0: 9B (SLD 194)
SEQUENCE CHARACTERISTICS:
LENGTH: 2e3
TYPE: partial Peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID N0: 9B
ARDSSGVLSPFNFSRRETGEKDVMFKVLYCGICHSDLHMVKNEWGFSTYPL-
VPGHEIVGEVTEVGSKVQKFKVGDRVGVGCVVGSCRSCCNCTDHLEI~TYCP-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
94
KQILTYGAKYYDGTTTYGGYSDIMVADEHFIVRIPDNLPLDGAAPLLCAGIT-
TYSPLRYFGLDKPGMHVGWGLGGLGHV AVKFAKAMGVKVTVISTSPK-
KEEEALKHLGADSFFVSRDQDQMQAAIGTWGIIDTVSAQHPLLPLIGLLKSH-
GKLVMVGAPEKPLELPVFPLLMGRKIViGS W
INFORMATION FOR SEQ ID NO: l0A (SLF17)
SEQUENCE CHARACTERISTICS:
LENGTH: 852
TYPE: partial cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID N0: l0A
GTCCCCTGAGGTATTTCGGACTTGACAAACCCGGCATGCATGTCGGGGTG-
GTTGGCCTTGGCGGTTTAGGCCATGTCGCGGTGAAGTTTGCCAAGGCTTTG-
GGGGTTGAGGTCACAGTGATCAGTACCTCCGCTAATAAGAAAGATGAAGC-
TATTAAACACCTTGGTGCTGATTCTTTCTTGGTCAGTCGTGACCAAGATCA-
GATGCAGGCTGCCATGGGAACATTGGACGGTATCATCGACACAGTTTCTG-
CAGTCCACCCCCTCCCACCTTTGATTAGTTTATTGAAGGCTAATGGAAAGC-
TTGTTATGGTTGGAGCACCAGAGAAGCCACTTGAGCTACCAGTTTTTTCTT-
TAATAATGGGAAGGAAGACTTTAGCCGGTAGTAATATCGGAGGTATCAAG-
GAGACACAAGAGATGATAGATTTGGCAGCTAAACACAACATAACGGCCGA-
CATCGAGGTCATCCCCATCGATTATTTGAACACTGCAATGGAGCGTCTTGC-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
T AAAGGGGATGTTAGATACCGGTTTGTCATCGACATCGGAAACACATT-
GAAGCCGGCCACTTAAATTTGCATTTCGATCAGAAACTGAATCAAGCGAT-
GTCGAGAGGCCTACGTAACAATGTAAACATGTGCTAGCTTGTTCTTGTAG-
TAGTCTTTAGCATTTCTCTGATGTACTCCTTCTGTTTTGTTCATGTTCCATCT-
TATAATAAGATTCTTATTATGAAAAAAATATGGTACCGTGGATATTGA-
ATAAATGAAGAACTACTGGAACAATGGTTTCACAAATTATTTGTGGTGC-
T
INFORMATION FOR SEQ ID NO: 10B (SLF17)
SEQUENCE CHARACTERISTICS:
LENGTH: 188
TYPE: partial peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID N0: 10B
PLRYFGLDKPGMHVGVVGLGGLGHVAVKFAKALGVEVTVISTSANKICDEAIK-
HLGADSFLVSRDQDQMQAAMGTLDGIIDTVSAVHPLPPLISLLKANGKLVM-
VGAPEKPLELPVFSLIMGRKTLAGSMGGIKETQEMIDLAAICHNITA-
DIEVIPIDYLNTAMERLAKGDVRYRFVIDIGNTLKPAT
INFORMATION FOR SEQ ID NO: 11A (SLF138)
SEQUENCE CHARACTERISTICS:
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
LENGTH: 663
TYPE: partial cDNA
STRANDNESS: Single
TOPOLOGY: Linear
96
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 11A
AGTTTGGTCTTGATGTGGGTGGATTAAGGGGAGGGATATTGGGACTTG-
GAGGTGTTGGACACATGGGGGTGAAGATAGCAAAGGCTATGGGACAC-
CATATCACCGTGATAAGCTCTTCTGATAAGAAGA,AAAAAGAGGCCTTGGAG-
CATATTGGTGCTGATGAGTACTTGGTGAGCTCTGATGCCACCCAAATGCAA-
GAGGCTATGGACTCACTGGATTACATTATTGACACCATTCCAGTGTTCCAC-
CCTCTTGAGCCTTACCTCTCTTTGTTGAAGCTTGATGGGAAGTTGATCTT-
GATGGGTGTTATCAACACCCCATTGCAATTTGTCTCTCCATTGGTCATGCTT-
GGGGAGGAAGACGATCACCGGGAGCTTTGTGGGGAGCATGAAGGAGATG-
GAGGAGATGCTCGAGTTCTGCAAAGAGAAAGAGCTGAAACGATGATT-
GAAGTGGTGAAGATGGACTACATCAACGAAGCTTTCGAAAGGTTGGAGAA-
GAACGACGTTAGGTACAGGTTCGTTGTGGATTGTTGCCGGCAGCAATCTT-
GATCAATAAGAAAGAAAGAAGGCATCATCGAGTGTTGTCCTATTTT-
TATCGAGTACTCTGTCTCATCTTATCTTAAACAATATAAATAAACAAA-
G
INFORMATION FOR SEQ ID NO:11B (SLF138)
SEQUENCE CHARACTERISTICS:
LENGTH: 181
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
97
TYPE: partial Peptide
STRA.NDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 11B
FGLD VGGLRGGILGLGGVGHMGVKIAKAMGHHITVIS S SDKKKICEALEHI-
GADEYLVSSDATQMQEAMDSLDYIIDTIPVFHPLEPYLSLLKLDGKLILM-
GVINTPLQFVSPLVMLGEEDDHRELCGEHEGDGGDARVLQRER.AETMIEVVK-
MDYINEAFERLEKNDVRYRFVVDCCRQQS
CA 02353577 2001-06-O1
WO 00/327$9 PCT/NL99/00737
98
INFORMATION FOR SEQ ID NO: 12A (SLG16)
SEQUENCE CHARACTERISTICS:
LENGTH: 694
TYPE: partial cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ 117 NO: 12A
GTGCATTGCTATGCCTATGAAGGCAAGATGCAAGAACATCTGCAATTATGT-
GAAGACGAGTTTAAAAAGATAATGAAGATAAATTTCATGTCTGCATGGTT-
TCTGGTAAATGCCGTTGGCAGAAGAATGCGAGATCATAAATCAGGAGGTTC-
CATCATATTGTTGACCTCGATTGTTGGAGCTGAAAGAGGGCTTTATACAG-
GAGCTGTTGCCTATGGTGCATGTTCGGCAGCACTGCAGCAGTTAGTAAG-
GTCGTCGGCATTGGAGATTGGAAAATACCAGATCAGGGTTAATGCAATCG-
CACGTGGTTTGCATTTGGAAGATGAGTTTCCTAAGTCTGTGGGAATAGAGA-
GAGCA.A.AGAAGCTGGTGAATGATGCAGTTCCGCTGGAGAGATGGCTTGAT-
GTTAAA.AATGATGTGGCTTCAAGTGTCATATATTTGGTCAGTGATGGTT-
CAAGGTACATGACGGGCACAACTATATTTGTTGATGGGGCACAGTCTC-
TCGTGAGGCCTCGAATGCGTTCTTATATGTGATTCTTGCTCCTATTATATCC-
TCCTAGCCATTATTAGCTACTTAGGTTTGTTCATACTTCATAGGTGAACT-
CATTAGCTATTCTTACATTTGTTCCTTATGAATAAAGAAGTCAAGATT-
C AAAAAA
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
99
INFORMATION FOR SEQ ID NO: 12B (SLG 16)
SEQUENCE CHARACTERISTICS:
LENGTH: 176
TYPE: partial Peptide
STR.ANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 12B
VHCYAYEGKMQEHLQLCEDEFKTCIMKINFMSAWFLVNAVGRRMRDHKSGG-
SIILLTSIVGAERGLYTGAVAYGACSAALQQLVRSSALEIGKYQIRVNAIARGL-
HLEDEFPKSVGIERAKKLVNDAVPLERWLDVKNDVASSVIYLVSDGSRYMT-
GTTIFVDGAQSLVRPRMRSYM
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
100
INFORMATION FOR SEQ ID NO: 13A (SLG144)
SEQUENCE CHARACTERISTICS:
LENGTH: 1010
TYPE: partial cDNA
STR.ANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 13A
GGAAACTACCATCAATTTTGGGTCTAAGAAGATTGCAGTTGTTACTGGAGC-
CAACAAAGGGATTGGACTTGAGATTAGCAAGCAATTAGCTGCTAAAG-
GAGTTGGGGTGGTATTAACAGCAAGAGATGTGAAGAGAGGAACAGAAGCT-
GCTGAAAATCTTAAGGCTTCTGGGTTCTCTGATGTGGTATTTCATCAGCTA-
GATGTAACAGAGCCGACTACTATTGGTTCTTTGGCAAACTTTCTTGAAACG-
CAATTTGGAAAGCTTGACATATTGGTTAACAATGCAGGAGTCGTTGGATCT-
GTATACCTCACAGCCGACTATGATCCAGTGCAAACATACGAGACAGCGAG-
GGATTGTTTGAAAACAAACTATTATGGGCTCAAGCAAGTCACAGAAGCAC-
TTGTTCCGCTGCTTCA.AAA.ATCTGAAGCTGCAAGGATAGTCAATGTCTCTTC-
CGGATTAGGACAGCTAAGAAATATTGGAAATGAGAAGGCCAAGAAGGAGC-
TAGGAGATGCAGATAACCTCAACGAGGAGAAAGTGGACAAGCTAGTTGAG-
GAATTTCTGGAGGATGTGAAACAGGATTCGATAGAATCCAAAGGCTGGCC-
TCTAAGTATATCTGCCTACATTGTCTCAAAAGCAGCTCTGAATGCTTATA-
CAAGACTCTTGGCAAAGAAGTATCCCCATATTGCCATAAACGCAGTTGGTC-
CAGGTTATACCAAAACAGACCTCAATAATAATTCCGGGATTCTCACAGTT-
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
ioi
GAAGAAGCTGCAGTAGGTCCTGTGAGGCTGGCTTTGATAGCCG..~AACTA-
GAATTTCCGGCCTCTTCTTCAACAGAAATGAAGAGTCGACCTTTGATTAG-
GTCAACGTGATCCCTGATGAACTGGACTATTTTAGATTTTCAGAATGTGC-
TTGATTTTGTTGAAGTATTTATGGGATTTGTATGTATACTTTGATGTAT-
CATTGTATTAATAGAGCACATGTTGTGAT-
C
INFORMATION FOR SEQ ID NO: 13B (SLG144)
SEQUENCE CHARACTERISTICS:
LENGTH ~ 283
TYPE: partial Peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY ALCOHOL DEHYDROGENASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 13B
ETTINFGSKKIAWTGANKGIGLEISKQLAAKGVGVVLTARDVKRGTEAAENL-
KASGFSDVVFHQLDVTEPTTIGSLANFLETQFGKLDILVNNAGVVGSVYL-
TADYDPVQTYETARDCLKTNYYGLKQ VTEALVPLLQKSEAARIVNV S SGL-
GQLRNIGNEKAKICELGDADNLNEEKVDKLVEEFLEDVKQDSIESKGWPLSI-
SAYIV SKAALNAYTRLLAKKYPHIAINAVGPGYTKTDLNNNSCrILTVEEAAV G-
PVRLALIAETRISGLFFNRNEESTFD
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
102
INFORMATION FOR SEQ ID NO: 14B (Motif 1 )
SEQUENCE CHARACTERISTICS: '
LENGTH: 12
TYPE: amino acid
STR.A.NDNESS: Single
TOPOLOGY: Linear
HYPOTHETICAL SEQUENCE: NO
SEQUENCE DISCRIPTION FOR SEQ ID N0: 14B
Trp Thr Asn Phe Phe Asn Pro Leu Asp Phe Gly Trp
W T N F F N P L D F G W
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
103
INFORMATION FOR SEQ ID NO: 1 SA (MAY 1; SEST)
SEQUENCE CHARACTERISTICS:
LENGTH: 877
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: MANGO ESTERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 15A
ATGAGGCCACAAATAGTGTTATTCGGAGATTCAATAACGGAGCAATCTTT
CGGATCAGGTGGTTGGGGTTCTTCTCTTGCTGACACTTACTCTCGCAAGGC
TGATGTATTAGTTCGTGGCTATGGTGGCTACAATACTAGATGGGCATTGTT
CTTGTTATGTCACATTTTCCCTCTGCACAATAA.AATACCTCCAGCCGTCAC
CACAATTTTCTTTGGGGCTAATGATGCAGCCCTTCTTGGGAGAACGAGTG
AAAGGCAGCATGTTCCCGTGGAAGAATACAAGAACAATCTCAGAAA.AAT
GGTTCAGCATTTGAAGGAAGTCTCCCCCACGATGCTAGTTGTGCTTATTAC
TCCACCACCAATTGATGAGGAAGGGCGTAAAGCATATGCACGATCCGTTT
ATGGTGAGAAAGCTATGAAAGAGCCTGAGAGGACAAATGAA.ATGGCTGG
AGTTTATGCTAGACATTGTGTTGAACTGGCAAAAGATCTTCCTGCCATTGA
TCTGTGGTCCAAGATGCAGGAAACAGAAGGTTGGCAGAAAA.AATTCCTCA
GTGATGGGTTGCACCTTAAGTCAGAAGGCAATGCAGTGGTTCACCAAGAA
GTTGTGAGAGTTCTAAAAGAAGCATGGTTTTCTCCTGAACAAATGCCATAT
GATTTTCCTCACCAATCAGTAATTGATGGAAAACACCCTGAGAAAGCTTT
CCAACTGCAATGCCCTGCTGAATTCTAGTCAAGACAGGCTTGGAAATTTG
TTCTCTCTTTCAATTTTTCTATTTGATGAAAAGATTTGGACTGCTTTTTCCT
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
104
AGTCATGCCAAATGAAACAGTGTTAGCCTTTTGCCTATTTTATCAGATGCT
GATATGCGCTCTGTGTCGAC
INFORMATION FOR SEQ ID NO: 15B (MAY1;SEST)
SEQUENCE CHARACTERISTICS:
LENGTH: 243
TYPE: peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: MANGO ESTERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 15B
MRPQIVLFGDSITEQSFGSGGWGSSLADTYSRKADVLVRGYGGYNTRWALFL
LCHIFPLHNKIPPAVTTIFFGANDAALLGRTSERQHVPVEEYKNNLRKMVQHL
KEVSPTMLVVLITPPPIDEEGRKAYARSVYGEKAMKEPERTNEMAGVYARHC
VELAKDLPAIDLWSKMQETEGWQKKFLSDGLHLKSEGNAVVHQEVVRVLKE
AWFSPEQMPYDFPHQSVIDGKHPEKAFQLQCPAEF
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
105
INFORMATION FOR SBQ ID NO: 16A (SUN1;VAAT)
SEQUENCE CHARACTERISTICS:
LENGTH: 14 s a
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY VESCA ALCOHOL ACYL
TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 16A
ATGGAGAAAATTGAGGTCAGTATAATTTCCAAACACACCATCAAACCATC
AACTTCCTCTTCACCACTTCAGCCTTACAAGCTTACCCTGCTCGACCAGCT
CACTCCTCCATCGTATGTCCCCATGGTATTCTTCTACCCCATTACTGGCCCT
GCAGTCTTCAATCTTCAAACCCTAGCTGACTTAAGACATGCCCTTTCCGAG
ACTCTCACTTTGTACTATCCACTCTCTGGAAGGGTCAAAAACAACCTATAC
ATCGATGATTTTGAAGAGGGTGTCCCATACCTTGAGGCTCGAGTGAACTG
TGACATGAATGATTTTCTAAGGCTTCCGAAAATCGAGTGCCTAAATGAGTT
TGTTCCAATAAAACCATTTAGTATGGAAGCAATATCTGATGAGCGTTACC
CTTTGCTCGGAGTTCAAGTTAACATTTTCAACTCCGGAATAGCAATCGGGG
TCTCCGTCTCTCACAAGCTCATCGATGGAAGAACTTCAGACTGTTTTCTCA
AGTCGTGGTGTGCTGTTTTTCGTGGTTCTCGTGACAAAATCATACATCCTA
ATCTCTCTCAAGCAGCATTGCTTTTCCCACCAAGAGATGACTTGCCTGAAA
AGTATGCCCGTCAGATGGAAGGGTTATGGTTTGTCGGAAAA,AAAGTTGCT
ACAAGGAGATTTGTATTTGGTGCGAAAGCCATATCTGTAATTCAAGATGA
AGCAAAGAGCGAGTCCGTGCCCAAGCCATCACGAGTTCAGGCTGTCACTA
GTTTTCTCTGGAAACATCTAATCGCTACTTCTCGGGCACTAACATCAGGTA
CA 02353577 2001-06-O1
WO 00/32789 PCT1NL99100737
l0E
CTACTTCAACAAGACTTTCTATAGCAACCCAGGTAGTGAACATAAGATCA
CGGAGGAACATGGAGACAGTGTGGGATAATGCCATTGGAAACTTGATATG
GTTCGCTCCGGCCATACTAGAGCTAAGTCATACAACACTAGAGATCAGTG
ATCTTAAGCTGTGTGACTTGGTTAACTTGCTCAATGGATCTGTCAAACAAT
GTAACGGTGATTACTTTGAGACTTTCATGGGTAAAGAGGGATATGGAAGC
ATGTGCGAGTATCTAGATTTTCAGAGGACTATGAGTTCTATGGAACCAGC
ACCAGAGATTTATTTATTCACGAGCTGGACTAATTTTTTCAACCAACTTGA
TTTTGGATGGGGGAGGACATCATGGATTGGAGTTGCAGGAAAAATTGAAT
CTGCATTTTGCAATCTCACAACATTAGTTCCAACACCATGCGATACTGGAA
TTGAAGCGTGGGTGAATCTAGAAGAAGAAAAAATGGCTATGCTAGAACA
AGATCCCCAGTTTCTAGCACTAGCATCTCCAAAGACGCTAATTTCAAGAT
ATTGATTAAGGAAGATTATGCGGCTCGTGCAATGTTTCCATTTTGTTGTGA
TTAAGGCTTAAATTAGTTCACCAGCCAATCAATAAGATGCAAGTATGATA
GACTCGGTCTACGTATGTTATCCG
INFORMATION FOR SEQ ID NO: 16B (SUNI; VAAT)
SEQUENCE CHARACTERISTICS:
LENGTH: 4 5 5
TYPE: peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: STRAWBERRY VESCA ALCOHOL ACYL
TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 16B
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
107
MEKIEVSIISKHTIKPSTSSSPLQPYKLTLLDQLTPPSYVPMVFFYPITGPAVFNL
QTLADLRHALSETLTLYYPLSGRVKNNLYIDDFEEGVPYLEARVNCDMNDFL
RLPKIECLNEFVPIKPFSMEAISDERYPLLGVQVNIFNSGIAIGVSVSHKLIDGRT
SDCFLKSWCAVFRGSRDKIIHPNLSQAALLFPPRDDLPEKYARQMEGLWFVG
KKVATRRFVFGAKAISVIQDEAKSESVPKPSRVQAVTSFLWKHLIATSRALTS
GTTSTRLSIATQVVNIRSRRNMETVWDNAIGNLIWFAPAILELSHTTLEISDLK
LCDLVNLLNGSVKQCNGDYFETFMGKEGYGSMCEYLDFQRTMSSMEPAPEI
YLFTSWTNFFNQLDFGWGRTSWIGVAGKIESAFCNLTTLVPTPCDTGIEAWV
NLEEEKMAMLEQDPQFLALASPKTLISRY
INFORMATION FOR SEQ ID NO: 17A (MAY2; BART)
SEQUENCE CHARACTERISTICS:
LENGTH: 1291
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: BANANA ALCOHOL ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 17A
ATGAGCTTCGCTGTGACCAGAACAAGCCGGTCTTTGGTCACTCCATGCGG
GGTCACGCCGACGGGCTCGCTCGGCCTCTCCGCCATCGACCGGGTGCCCG
GCCTCAGGCATATGGTGCGGTCGCTACACGTGTTCAGGCAAGGCCGGGAG
CCGGCCAGGATCATCAGGGAAGCACTGTCGAAGGCGCTGGTGAAGTACTA
CCCCTTCGCGGGGCGGTTCGTGGACGATCCCGAGGGCGGCGGCGAGGTTC
GTGTCGCTTGCACTGGCGAGGGCGCTTGGTTCGTCGAGGCCAAGGCGGAC
TGCAGCTTGGAGGACGTGAAGTACCTCGATCTCCCGCTCATGATCCCTGA
CA 02353577 2001-06-O1
WO 00/327$9 PCT/NL99/00737
108
GGACGCGCTCCTGCCCAAGCCCTGCCCGGGACTGAACCCCCTCGACCTCC
CTCTCATGCTGCAGGTGACAGAGTTCGTGGGCGGCGGATTCGTGGTCGGC
CTCATCTCCGTCCATACCATCGCCGACGGCCTCGGCGTCGTCCAGTTCATC
AACGCCGTCGCCGAGATCGCCCGTGGCCTGCCGAAGCCCACCGTGGAGCC
TGCATGGTCCCGGGAGGTCATACCCAACCCACCTAAGCTGCCTCCCGGTG
GCCCGCCCGTGTTCCCCTCCTTCAAGCTGCTCCACGCCACCGTCGACCTAT
CCCCTGACCACATCGATCACGTCAAGTCCCGACACTTGGAGCTCACCGGC
CAGCGCTGCTCTACCTTCGACGTCGCCATCGCCAACCTGTGGCAGTCCCGC
ACGCGCGCCATCAACCTGGACCCAGGCGTCGACGTGCACGTGTGCTTCTT
CGCCAACACTCGCCACCTGTTGCGCCAGGTCGTCCTCCTGCCCCCCGAGG
ATGGCTACTACGGCAACTGCTTCTACCCGGTGACCGCCACCGCCCCAAGC
GGCAGGATCGCATCGGCCGAGCTCATCGATGTCGTCAGCATCATCAGGGA
CGCGAAGTCGAGGCTGCCGGGCGAGTTCGCCAAGTGGGCTGCCGGGGATT
TCAAGGACGACCCTTACGAGCTCAGCTTCACGTACAACTCGCTGTTCGTGT
CGGACTGGACCCGGCTCGGCTTCCTCGACGTCGACTACGGCTGGGGCAAG
CCCCTCCACGTTATACCGTTCGCGTACTTGGACATCATGGCGGTCGGCATC
ATCGGGGCGCCGCCGGCGCCGCAAAAGGGGACTCGGGTGATGGCGCAGT
GCGTCGAGAAGGAGCACATGCAGGCGTTCCTGGAAGAGATGAAAGGCTT
CGCTTAAACCAGCAGCAGTGTAGTACTTGTCAGTATCC
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
109
INFORMATION FOR SEQ ID NO: 17B {MAY2; BAAT)
SEQUENCE CHARACTERISTICS:
LENGTH: 419
TYPE: peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: BANANA ALCOHOL ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 17B
MSFAVTRTSRSLVTPCGVTPTGSLGLSAIDRVPGLRHMVRSLHVFRQGREPAR
IIREALSKALVKYYPFAGRFVDDPEGGGEVRVACTGEGAWFVEAKADCSLED
VKYLDLPLMIPEDALLPKPCPGLNPLDLPLMLQVTEFVGGGFVVGLISVHTIA
DGLGV VQFINAVAEIARGLPKPTVEPAWSREVIPNPPKLPPGGPPVFPSFKLLH
ATVDLSPDHIDHVKSRHLELTGQRCSTFDVAIANLWQSRTRAINLDPGVDVH
VCFFANTRHLLRQVVLLPPEDGYYGNCFYPVTATAPSGRIASAELIDVVSIIRD
AKSRLPGEFAKWAAGDFKDDPYELSFTYNSLFVSDWTRLGFLDVDYGWGKP
LHVIPFAYLDIMAVGIIGAPPAPQKGTRVMAQCVEKEHMQAFLEEMKGFA
INFORMATION FOR SEQ ID NO: 18A (MAY3; AAAT)
SEQUENCE CHARACTERISTICS:
LENGTH: 148 S
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: APPLE ALCOHOL ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 18A
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
110
ATGTCATTCTCAGTACTTCAGGTGAAACGATTGCAACCGGAACTTATAACT
CCGGCAAAGTCAACGCCTCAAGAAACAAAGTTTCTCTCAGATATTGACGA
CCAAGAAAGCTTGAGAGTTCAGATTCCAATCATAATGTGTTACAAAGACA
ACCCTTCACTTAATAAAAATCGTAATCCCGTTAAGGCAATTAGGGAAGCC
TTAAGTAGAGCATTAGTGTATTACTACCCCTTAGCTGGAAGGCTTAGGGA
AGGGCCTAATAGAAAGCTCGTGGTCGATTGCAATGGTGAAGGTATCTTGT
TCGTTGAGGCTTCTGCTGATGTCACACTTGAGCAACTAGGAGACA,AAATT
CTACCCCCTTGTCCACTTTTAGAGGAGTTCTTATATAATTTTCCAGGCTCTG
ATGGAATTATTGATTGTCCTTTGCTGCTGATTCAGGTGACCTGTCTTACAT
GTGGAGGTTTCATACTTGCATTGCGCCTAAACCACACAATGTGTGATGCA
GCTGGATTGCTCTTGTTCCTGACCGCCATCGCGGAGATGGCAAGAGGCGC
ACATGCACCATCTATTCTACCAGTGTGGGAGAGAGAGCTCTTGTTCGCTCG
AGATCCACCAAGAATTACATGTGCTCGTCATGAATATGAAGACGTGATTG
GTCATTCTGATGGCTCATACGCATCCAGTAACCAGTCAAACATGGTTCAA
CGATCTTTCTACTTTGGTGCCAAGGAGATGAGAGTCCTTCGAAA.ACAGAT
TCCACCCCACCTAATTTCCACTTGCTCCACATTTGACTTGATCACAGCTTG
TTTGTGGAAATGTCGCACTCTTGCACTTAACATTAATCCAAAAGAGGCTGT
TCGAGTTTCATGCATTGTCAATGCACGAGGAAAGCACAACAATGTACGTC
TTCCCTTGGGATACTATGGCAATGCATTTGCATTTCCAGCTGCAATTTCGA
AGGCTGAACCTCTATGCAAAAATCCACTGGGATATGCTTTGGAGTTGGTG
AAGAAGGCTAAAGCTACCATGAATGAAGAATACTTAAGATCAGTGGCAG
ATCTTTTGGTACTAAGAGGGCGACCTCAATATTCATCGACAGGAAGTTATT
TAATAGTTTCTGATAATACGCGTGTAGGTTTTGGAGATGTCAATTTTGGAT
GGGGACAGCCGGTATTTGCTGGACCCGTCAAGGCCTTGGATTTGATTAGC
TTCTACGTTCAACACAAAAACAACACAGAGGATGGAATATTGGTACCAAT
CA 02353577 2001-06-O1
WO 00/32789 111 PCT/NL99/00737
GTGTTTGCCATCCTCGGCCATGGAGAGATTTCAGCAGGAACTAGAGAGGA
TTACTCAGGAACCTAAGGAGGATATATGTAACAACCTTAGATCAACTAGT
CAATGATGTAAGTGTTAAACGTAATGCACTTTCTGTAATGTAGAGTTGTGT
CTCTTGGAACTTATCNCAAGAGTTATAGCTGTTATCCAAAGGTCTGAATGT
TATTAA.AAAATAGCCAATAATAAG
INFORMATION FOR SEQ ID NO: 18B (MAY3; AAAT)
SEQUENCE CHARACTERISTICS:
LENGTH: 454
TYPE: peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: APPLE ALCOHOL ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 18B
MSFSVLQVKRLQPELITPAKSTPQETKFLSDIDDQESLRVQIPIIMCYKDNPSLN
KNRNPVKAIREALSRALVYYYPLAGRLREGPNRKLVVDCNGEGILFVEASAD
VTLEQLGDKILPPCPLLEEFLYNFPGSDGIIDCPLLLIQVTCLTCGGFILALRLN
HTMCDAAGLLLFLTAIAEMARGAHAPSILPV WERELLFARDPPRITCARHEYE
DVIGHSDGSYASSNQSNMVQRSFYFGAKEMRVLRKQIPPHLISTCSTFDLITAC
L WKCRTLALNINPKEA V RV S CI VNARGKHNNV RLPLGYYGNAFAFPAAISKA
EPLCKNPLGYALELVKKAKATMNEEYLRSVADLLVLRGRPQYSSTGSYLIVS
DNTRVGFGDVNFGWGQPVFAGPVKALDLISFYVQHKNNTEDGILVPMCLPSS
AMERFQQELERITQEPKEDICNNLRSTSQ
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
112
INFORMATION FOR SEQ ID NO: 19A {MAYO; MART)
SEQUENCE CHARACTERISTICS:
LENGTH: 1380
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: MANGO ALCOHOL ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 19A
ATGATAATCACGGTGAAGGAGTCGACGATGGTCCCGCCGTCGGCGGAGAC
GCCGAGGATATCTCTGTGGAACTCCAACGCCGATCTGGTGGTTCCCCGATT
TCATACTCCCAGCGTTTACTTCTACCGGCCCACCGGGGCCATAAACTTCTT
TGATGGTAAGTTGCTCAAGGAGGCTCTCGGCAAGGCTCTGGTGCCGTTCT
ACCCAATGGCGGGGCGGTTAAAGCGTGACGAAGATGGAAGGATTGAGAT
CGATTGTAATGCTGAAGGCGTCTTGTTTGTTGAGGCCGAAACTCCCTCTGT
TATTGATGATTTTGGTGACTTTGCGCCCACTTTAGAGCTCAAGCAGCTCAT
TCCGACAGTGGATTACTCCGGCGGGATCTCTACGTATCCCCTATTGGCGTT
ACAGGTTACTCACTTCAAATGTGGTGGAGTTTCACTTGGTGTAGGTATGCA
ACACCATGCGGCAGATGGATTTTCTGGTCTTCACTTTGTAAACACATGGTC
AGACATTGCTCGTGGTCTTGATGTTAACATCACCCTGTTCATTGACCGGAC
TCTGCTCAGAGCACAGGATCCCCCTCAGCCTACTTTCCCACACACATGGA
ATACCAGGCCGCCTCCTTCCCTGAAAACTCCTCCACCAGCAGTTTCTGAGC
CTACTGCTGTCTCCATTTTTAAGTTGACGCGGGACCAGCTCAACATCCTCA
AAGCCAAGGCCAAAGAAGATGGTAACACTATCAACTATAGCTCATATGAG
ATGCTGGCGGGTCATGTCTGGAGATCTGCATGCAAGGCACGCGGCTTATC
TGATGATCAAGAGACTAAATTGTACATAGCAACTGACGGACGTGCTAGAT
CA 02353577 2001-06-O1
WO 00/32789 113 PCT/NL99/00737
TAATCCCCCCACTTCCACCTGGTTACTTTGGGAATGTGATATTTACAGCCA
CACCAATGGCAGTAGCAGGTGATCTCCAGTCAAAGCCTATATGGTATGCT
GCTGGCCAGATTCATGATGCCTTGGTTCGAATGGACAACGACTATTTAAG
GTCAGCCCTCGATTACCTAGAGCTTCAGCCTGATTTATCAGCATTAGTTCG
TGGTGCCCATACATTTAGGTGTCCAAATCTCGGGATTACTAGTTGGGTTAG
ACTGCCAATACATGATGCAGATTTTGGTTGGGGTCCACCCACATTTATGGG
GCCTGGTGGGATTGCATATGAAGGCTTATCATTTGTATTGCCAAGCCCTAC
AAATGATGGGAGCTTATCAGTTGCCATCTCTCTACAATCTGAACACATGA
AACTGTTTCAGAAGTTCTTTTATGATATTTAA
INFORMATION FOR SEQ ID NO: 19B (MAY4; MART)
SEQUENCE CHARACTERISTICS:
LENGTH: 431
TYPE: peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: MANGO ALCOHOL ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 19B
MIITVKESTMVPPSAETPRISLWNSNADLV VPRFHTPSVYFYRPTGAINFFDGK
LLKEALGKALVPFYPMAGRLKRDEDGRIEIDCNAEGVLFVEAETPSVIDDFGD
FAPTLELKQLIPTVDYSGGISTYPLLALQVTHFKCGGVSLGVGMQHHAADGF
SGLHFVNTWSDIARGLDVNITLFIDRTLLRAQDPPQPTFPHTWNTRPPPSLKTP
PPAV SEPTAVSIFKLTRDQLNILKAKAKEDGNTINYSSYEMLAGHV WRSACK
ARGLSDDQETKLYIATDGRARLIPPLPPGYFGNVIFTATPMAVAGDLQSKPIW
YAAGQIHDALVRMDNDYLRSALDYLELQPDLSALVRGAHTFRCPNLGITSW
CA 02353577 2001-06-O1
WO 00/327$9 114 PCT/NL99/00737
VRLPIHDADFGWGPPTFMGPGGIAYEGLSFVLPSPTNDGSLSVAISLQSEHMK
LFQKFFYDI
INFORMATION FOR SEQ ID NO: 20A (CLE75; LAAT2)
SEQUENCE CHARACTERISTICS:
LENGTH: 1436
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: LEMON ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 20A
ATCCACACTAATAATTCTTTCATATGCTCGGGGATGGATCTCCAAATCACC
TGCACCGAAATCATCAAGCCTTCTTCGCCGACGCCTCAACACCAAAGTAC
CTATAAACTTTCAATTATTGATCAATTAACTCCTAATGTTTACTTTTCCATC
ATTCTCTTGTATTCAAA.A.GCTGGTGAAAGTACCGCCAAAACTTCAGATCA
CCTCAAAGAATCTCTTTCAAATACATTAACCCACTACTATCCTTTAGCTGG
GCAACTCAAATATGATCAACTTATTGTTGATTGTAACGACCAAGGTGTCCC
GTTCATCGAAGCACACGTCACCAACGACATGCGTCAGCTTCTTAAAATAC
CAAATATTGATGTTCTCGAACAACTCCTACCATTCAAACCGCATGAGGGTT
TTGATTCTGATCGTTCCAACCTAACCGTTCAGGTCAATTACTTTGGTTGTG
AAGGAATGGCGATTGGTCTGTGCTTCAGACACAAAGTTATTGATGCAACA
ACGGCTGCATTCTTTGTTAAGAACTGGGGTGTAATTGCTCGTGGTGCTGGA
GAAATTAAGGACGTGATCATTGATCATGCTTCCCTGTTTCCCGCAAGAGAT
TTATCGTGCTTAACAAAGAGTGTTGACGAAGAGTTTTTGAAGCCAGAGTC
TGAAACAAAGCGCTTTGTGTTTGATGGTGCCACTATAGCTTCTTTACAAGA
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
115
AACGTTTGCAAGTTTTGAACGACGTCCAACACGCTTTGAGGTTGTGTCAGC
AGTTATTTTGGGTGCTTTGATAACTGCAACGAGAGAATCTGATGATGAGTC
TAACGTTCCTGAACGTTTGGACACGATAATTTCAGTGAATCTACGGCAGA
GAATGAATCCACCATTCCCGGAGCATTGCATGGGGAATATAATATCCGGG
GGATTAGTGTATTGGCCACTGGAGAAAAAAGTTGATTACGGGTGTTTAGC
AAAAGAGATTCATGAATCAATAAAGAAAGTGGACGATCAATTTGCGAGG
AAGTTCTATGGGGACGCAGAGTTCTTGAACCTGCCGAGGCTTGCGGGTGC
TGAGGATGTGAAGAAGCGGGAGTTTTGGGTTACTAGTTGGTGCAAAACTC
CGCTGTATGAAGCTGATTTCGGGTGGGGGAATCCTAAGTGGGCAGGCAAC
TCCATGAGGCTTAATCAGATTACTGTTTTCTTTGACAGTAGTGATGGTGAG
GGAGTTGAAGCTTGGGTGGGGTTGCCCAGAAAAGACATGGCTCGATTTGA
AAAAGATTCTGGCATCCTTGCTTACACTTCCCCTAATCCAAGCATATTTTG
AGGGTTTATTTATTTTTTATTGCACTGTTTGTTATTTGTACTGGCTTGCTGG
GAACATATTCTGGCAAATTTCGCTGATGCAAGTATCATTCTCCATAAAAAT
GTC AAAAAA
INFORMATION FOR SEQ ID NO: 20B (CLE75; LAAT2)
SEQUENCE CHARACTERISTICS:
LENGTH: 426
TYPE: Peptide
STR,ANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: LEMON ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 20B
CA 02353577 2001-06-O1
WO 00/32789 116 PCT/NL99/00737
MDLQITCTEIIKPSSPTPQHQSTYKLSIIDQLTPNVYFSIILLYSKAGESTAKTSD
HLKESLSNTLTHYYPLAGQLKYDQLIVDCNDQGVPFIEAHVTNDMRQLLKIP
NIDVLEQLLPFKPHEGFDSDRSNLTVQVNYFGCEGMAIGLCFRHKVIDATTAA
FFVKNWGVIARGAGEIKDVIIDHASLFPARDLSCLTKSVDEEFLKPESETKRFV
FDGATIASLQETFASFERRPTRFEV V SAVILGALITATRESDDESNVPERLDTIIS
VNLRQRMNPPFPEHCMGNIISGGLVYWPLEKKVDYGCLAKEIHESIKKVDDQ
FARKFYGDAEFLNLPRLAGAEDVKKREFWVTSWCKTPLYEADFGWGNPKW
AGNSMRLNQITVFFDSSDGEGVEAWVGLPRKDMARFEKDSGILAYTSPNPSIF
INFORMATION FOR SEQ ID NO: 21A (CLF35; LAAT3)
SEQUENCE CHARACTERISTICS:
LENGTH: 1648
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: LEMON ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 21A
GCTAGGCTGGCTTTCATTTAGCTTCCATCTCTTTCTCTC'fGTCAATAACTCA
TGGCTGCAATTGAA.AACAGAGTAACACTAAAGAAGCATGAGGTTACCAA
AGTCACCCCTTTCGTCAACCCCAACTCAAAGACGACGTCGTTTACTCTCGA
TCTCACCTATTTCGACTTTTTCTGGTTCAAGAATCCTCCTGTGGAACGCCTC
TTCTTCTATGAGATGACTGACTTGACGTGGGATTTATTCAACTCGGAGATC
CTCCCAAAGCTGAAGCACTCCCTTTCCTTCACTCTCCTTCATTACCTCCCTC
TTGCTGGTCACATCATGTGGCCGCTGGATGCCGCAAAGCCTGCCGTCTACT
ACTTTCCCGACCAAAACGACGGCGTTTCATTCGCAGTTGCTGAGTGGTCTT
CA 02353577 2001-06-O1
WO 00/32789 117 PC'T/NL99/00737
CCGAGTGCCACGCAGGCTTCCATCACCTCTCCGGCAACGGAATCCGCCAA
GCAGTTGAATTTCATCCTCTTGTGCCCCAGTTGTCGCTTACGGACGATAAA
GCTGAGGTAATTGCCATCCAAATAACACTGTTTCCGAATCAAGGCTTCTCA
ATTGGTGTTTCATCTCACCATGCAATTCTTGATGGAAAAACTTCGACCTTG
TTCCTGAAATCTTGGGCTTATTTGTGCAAACAATTACAATTATGCCATCAC
CCTTGTTTGTCACCTGAACTAACCCCTCTTCTCGACCGGACTGTCATCAAA
GATCCGACAGGTCAGGACATGCTGCAACTGAATAAGTGGGTTGTCGGGTC
GGATAATTCGGATCCCCAGAAGATACGGAGCTTGAAGGTTTTACCATTCT
TAGACTCTGAGTCTCTGAACAAATTGGTCCGAGCCACATTTGAGTTGACG
CGTGAAGATATTACGAAACTCAGGCACAAGGTTAATCATCAGTTATCAAA
ATCATCAAAATCAAAGCAAGTTCGTTTATCAACTTTTGTGCTTACATTAGC
TTATGTGTTTGTTTGCATGGCTAAAGCTAAATTAGCCAAAGCCAAAACTGA
AGCTGAAGCTGCAGCAGGTAATGATGAAATTA,AAAATATTATTGTGGGAT
TCACTGCGGATTATAGGAGCCGTTTGGATCCTCCAATTCCACTTAATTATT
TTGGTAACTGCAATGGGAGACATTGTGAGACTGCAAAAGCAAGTGATTTC
GTTCAAGAAAATGGGGTTGCTTTTGTTGCAGAGATGTTAAGTGATATGGTC
AA.AGGGATCGATGCGGATGCCATTGAAGCCAATGATGATAAGGTTTCAGA
AATATTGGAAATTCTGAAAGAAGGAGCAATGATTTTTTCTGTGGCTGGCTC
GACCCAATTTGATGTTTACGGGTCGGATTTCGGGTGGGGGAGGCCCAAGA
AGGTGGAGATTGTGTCAATAGATAGGACACAAGCCATCTCTTTGGCAGAG
AGAAGAGATGGAGGAGGCGGCGTTGAGGTTGGAGTTGTTTTAGAGAAGC
AACAAATGGAGGTTTTTGAATCTGTATTTGCTGATGGACTGAAAAATGAT
CTTGTTTAATTAATGATGTATCATCTAAATTTCTCAATATATTATTGGTCAT
ATTCAAAAGAAATAAATTATTGCGGATTTTTGTGACCACCAAATAAAATA
CTCTTTTTTGAAAAAAAAAAAAAAAAAA
CA 02353577 2001-06-O1
WO 00/32789 ~ 1 a PCT/NL99/00737
INFORMATION FOR SEQ ID NO: 21B (CLF35; LAAT3)
SEQUENCE CHARACTERISTICS:
LENGTH: 491
TYPE: Peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: LEMON ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 21 B
MAAIENRVTLKKHEVTKVTPFVNPNSKTTSFTLDLTYFDFFWFKNPPVERLFF
YEMTDLTWDLFNSEILPKLKHSLSFTLLHYLPLAGHIMWPLDAAKPAVYYFP
DQNDGVSFAVAEWSSECHAGFHHLSGNGIRQAVEFHPLVPQLSLTDDKAEVI
AIQITLFPNQGFSIGVSSHHAILDGKTSTLFLKSWAYLCKQLQLCHHPCLSPEL
TPLLDRTVIKDPTGQDMLQLNKWVVGSDNSDPQKIRSLKVLPFLDSESLNKL
VRATFELTREDITI~LRHKVNHQLSKSSKSKQVRLSTFVLTLAYVFVCMAKAK
LAKAKTEAEAAAGNDEIKNIIVGFTADYRSRLDPPIPLNYFGNCNGRHCETAK
ASDFVQENGVAFVAEMLSDMVKGIDADAIEANDDKVSEILEILKEGAMIFSV
AGSTQFDVYGSDFGWGRPKKVEIVSIDRTQAISLAERRDGGGGVEVGVVLEK
QQMEVFESVFADGLKNDLV
CA 02353577 2001-06-O1
WO 00/32789 l~9 PCT/NL99/00737
INFORMATION FOR SEQ ID NO: 22A (CLB82; LAAT4)
SEQUENCE CHARACTERISTICS:
LENGTH: 1520
TYPE: cDNA
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: LEMON ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 22A
AACATGGCAGCAAGCTCACTGCATGGCAAAGAAGCTACAGTTATATATCC
TTCTGAGCCAACCCCATCTACGGTTTTGTCTCTCTCAGCTCTTGATTCTCAG
CTTTTCTTGCGTTTCACTATTGAGTATCTCTTGGTCTATAGACCTCGCCCTG
GTTTGGACCCACTTGCTACCGTGGCTCGTGTCAAGTCCGCACTCGCCAAAG
CCTTGGTTCCTTACTATCCCCTCGCGGGTCGGGTCAGAGCTAAACAAGAC
GGGTCGGGCTTATTGGAAGTCGTGTGTCTAGGCCAAGGCGCTGTGTTCAT
CGAAGCCGTCGACCGTGAAAGTACGATCACCGATTTTGAGAGTGCTCCCA
GGTATGTTACTCAGTGGAGGAAACTGCTGTCGTTATACGTGGCGGATGTTC
TCAAAGGGGCCCCACCTCTTGTCGTTCAGCTGACTTGGCTTAGAGATGGA
GCCGCAGCGCTCGGTATTGGCTTTAACCATTGTGTTTGCGATGGTATCGGC
AGCGCCGAGTTCCTCAACTTGTTTACTGAGTTATGTACGAGCCGTCATAAC
GAACTGGGTGGTGGCCATTCTCTGCCGAAACCCGTTTGGGATCGCCACCT
AATGAACTCCTCCTCATCACGTCAACAGCATGCAGATACACGTGCCAGCT
CAGTGAGTCACCTGGAATTCAACAGAGTGGCTGATCTTTGTGGTTTTGTTT
CTCGTTTTTCCAACGAAAGGCTTGTTCCCACTTCAATAACGTTCGATAAAC
GACGCTTAAACGAGCTGCGGAAGCTGGCTCTGTCCACGAGTCGACCCAGT
GAGCTGGCTTACACGTCATTTGAAGTTCTTTCAGCTCATGTGTGGAGAAGC
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
120
TGGGCTAGGTCGTTGAATCTTCCGTCGAATCAAATCTTGAAGCTTCTATTT
AGCATCAATGTACGTAACCGTGTCAAGCCGAGTCTCCCCAGTGGCTATTA
TGGCGATGCATTTGTATTAGGCTGTGCTCAAACGAGGGTTAAAGATTTGA
CAGAGAAGGACTTAGGGCATGCAGCAATGTTGGTTAA.AA.AGGCGAAAGA
GAGAGTTGATAGTGAGTATGTGAAGTCGGTCATCGACTCAGTGAGTCACA
CGAGAGCGTGTCCCGACTCAGTCGGGGTGTTGATAGTGTCGCAGTGGTCA
AGGCTAGGGTTAGAGAGAGTTGACTTTGGGATGGGGAGGCCGACTCAAGT
GGGTCCCATTTGCTGCGACAGGTATTGCCTGTTTCTACCGGTTTTCAATCA
GACGGACGCTGTTAAGGTGATGGTGGCGGTCCCCACAAGTGCAGTTGACA
AGTATGAGCATCTCGCGAAGGGCTTATGCTGGTGAGGACCACACCGCATG
ATGACCCCACCATGTAATACGTTGACTTATAAACTCAGTTTGACTTTTAAC
TTTTTTAACAAGTGATGGAATTTCAGTGATTGACTCATCACTTTGATCCTG
ATCCAATAAATAATTGAATTGAGTTC
INFORMATION FOR SEQ ID NO: 22B (CLB82; LAAT4)
SEQUENCE CHARACTERISTICS:
LENGTH: 447
TYPE: Peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: LEMON ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 22B
MAASSLHGKEATVIYPSEPTPSTVLSLSALDSQLFLRFTIEYLLVYRPRPGLDPL
ATVARVKSALAKALVPYYPLAGRVRAKQDGSGLLEVVCLGQGAVFIEAVDR
CA 02353577 2001-06-O1
WO 00/32789 121 PCT/NL99/00737
ESTITDFESAPRYVTQWRKLLSLYVADVLKGAPPLVVQLTWLRDGAAALGIG
FNHCVCDGIGSAEFLNLFTELCTSRHNELGGGHSLPKPVWDRHLMNSSSSRQ
QHADTRASSVSHLEFNRVADLCGFVSRFSNERLVPTSITFDKRRLNELRKLAL
STSRPSELAYTSFEVLSAHVWRSWARSLNLPSNQILKLLFSINVRNRVKPSLPS
GYYGDAFVLGCAQTRVKDLTEKDLGHAAMLVKICAKERVDSEYVKSVIDSV
SHTRACPDSVGVLIVSQWSRLGLERVDFGMGRPTQVGPICCDRYCLFLPVFN
QTDAVKVMVAVPTSAVDKYEHLAKGLCW
INFORMATION FOR SEQ ID NO: 23A (MAYS; MEAAT)
SEQUENCE CHARACTERISTICS:
LENGTH: 1468
TYPE: cDNA
STRANDNESS: Single
~bPOLOGY: Linear
OTHER INFORMATION: HONEY DEW MELON ALCOHOL ACYL
TRANSFERASE
SEQUENCE DISCRTPTION FOR SEQ ID NO: 23A
ATGACTTCTCTTTTCACGTACGAAA.ATGCCAACCAGAATTGATTGCACCAG
CAAATCCTACACCCTATGAATTTAAACAACTTTCTGATGTGGATGATCAAC
AAAGCTTAAGGCTTCAATTGCCATTCGTAAATATCTATCCCCATAATCCAA
GTTTGGAGGGAAGAGATCCAGTGAAGGTAATAAAGGAAGCAATTGGAAA
GGCGTTGGTGTTCTACTATCCTTTAGCAGGAAGATTGAGAGAAGGGCCAG
GTAGAAAGCTTTTTGTTGAATGTACAGGTGAAGGAATCTTGTTTATTGAAG
CGGATGCAGATGTGAGCTTAGAAGAATTTTGGGATACTCTTCCATATTCAC
CA 02353577 2001-06-O1
W O 00/32789 a_ z 2 PCT/N L99/00737
TTTCAAGCATGCAGAACAATATTATACATAACGCTTTAAATTCTGATGAAG
TCCTCAATTCTCCATTATTGCTCATTCAGGTGACACGACTCAAGTGTGGAG
GTTTCATTTTTGGTCTTTGTTTCAATCATACTATGGCAGATGGTTTTGGTAT
TGTCCAATTCATGAAGGCTACAGCGGAGATAGCTCGTGGAGCTTTTGCTC
CATCTATTTTACCAGTATGGCAAAGAGCTCTCTTAACCGCAAGAGACCCTC
CCAGAATCACTTTTCGCCACTATGAATACGACCAAGTAGTCGACATGAAG
AGCGGCCTCATTCCAGTCAATAGCAAGATCGATCAATTATTCTTCTTTAGC
CAACTTCAAATCTCCACCCTTCGCCAAACTTTGCCAGCCCACCTTCACGAT
TGCCCTTCCTTCGAGGTCCTCACTGCCTATGTTTGGCGCCTCCGTACCATA
GCCCTTCAATTTAAGCCAGAGGAGGAAGTGCGGTTTCTTTGCGTAATGAA
TCTACGCTCGAAGATCGACATACCATTAGGGTATTATGGTAATGCGGTAG
TTGTTCCTGCAGTAATCACCACCGCTGCGAAGCTTTGTGGGAACCCACTTG
GTTATGCTGTAGACTTGATTAGGAAGGCCAAGGCTAAGGCAACGATGGAG
TACATAAAGTCTACGGTGGATCTTATGGTGATTAAAGGACGACCCTATTTC
ACTGTAGTTGGATCATTTATGATGTCAGACCTAACGAGAATTGGGGTTGA
AAACGTGGACTTTGGATGGGGAAAGGCCATTTTTGGAGGACCTACAACCA
CAGGGGCCAGAATTACACGAGGTTTGGTAAGCTTTTGTGTACCTTTCATGA
ATAGAAATGGAGAAAAGGGAACTGCGTTAAGTCTATGCTTGCCTCCTCCA
GCCATGGAAAGATTTAGGGCAAATGTTCATGCCTCGTTGCAAGTGAAACA
AGTGGTTGATGCAGTTGATAGCCATATGCAAACTATTCAATCTGCTTCGAA
ATAAATAATATTGTTGAAGGTGGGTCTGAGTTGACTCGACCATATCGATG
CATGCAAGCTTGATCCGGCTGCTAACAAAGCCCGAAAGGAAGCTGAGTTG
CTGT
INFORMATION FOR SEQ ID NO: 23B (MAYS; MEAAT)
CA 02353577 2001-06-O1
WO 00/32789 PCT/NL99/00737
12s
SEQUENCE CHARACTERISTICS:
LENGTH: 456
TYPE: Peptide
STRANDNESS: Single
TOPOLOGY: Linear
OTHER INFORMATION: LEMON ACYL TRANSFERASE
SEQUENCE DISCRIPTION FOR SEQ ID NO: 23B
MDFSFHVRKCQPELIAPANPTPYEFKQLSDVDDQQSLRLQLPFVNIYPHNPSL
EGRDPVKVIKEAIGKALVFYYPLAGRLREGPGRKLFVECTGEGILFIEADADV
SLEEFWDTLPYSLSSMQNNIIHNALNSDEVLNSPLLLIQVTRLKCGGFIFGLCF
NHTMADGFGIVQFMKATAEIARGAFAPSILPV WQRALLTARDPPRITFRHYEY
DQWDMKSGLIPVNSKIDQLFFFSQLQISTLRQTLPAHLHDCPSFEVLTAYVW
RLRTIALQFKPEEEVRFLCVMNLRSKIDIPLGYYGNAVVVPAVITTAAKLCGN
PLGYAVDLIRKAKAKATMEYIKSTVDLMVIKGRPYFTWGSFMMSDLTRIGV
ENVDFGWGKAIFGGPTTTGARITRGLVSFCVPFMNRNGEKGTALSLCLPPPA
MERFRANVHASLQVKQWDAVDSHMQTIQSASK