Note: Descriptions are shown in the official language in which they were submitted.
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
1
POLYSATURATED FATTY ACID (PUFA) ELONGASE FROM CAENORHABDITIS ELEGANS
The present invention relates to polyunsaturated fatty acid (PUFA) elongases.
More
specifically, the invention relates to a DNA sequence from C. elegans encoding
a PUFA
elongase.
Unsaturated fatty acids are essential components required for normal cellular
function, being
involved in a diverse number of roles ranging from membrane fluidity to acting
as signal
molecules (Gill, L, Valivety, R. (1997). Trends ~iotechnol. 15, 401-409;
Broun, P., et al
(1999) Ann. Rev. Nutr. 19, 197-216). In particular, the class of fatty acids
known as the
polyunsaturated fatty acids (PUFAs) has attracted considerable interest as
pharmaceutical
and nutraceutical compounds (Broun supra; Horrobin, D. F. (1990) Reviews in
Contemp
Pharmacotherpy 1, 1-45).
The synthesis of PUFAs i.e. fatty acids of 18 carbons or more in length and
containing two
or more double bonds, is thought to be catalyzed in a variety of organisms by
a specific fatty
acid elongase enzyme. This elongase is responsible for the addition of 2
carbon units to an
18 carbon PUFA, resulting in a 20 carbon fatty acid. An example of this
reaction is the
elongation of Y-linolenic acid (GLA; 18:306'~~12) to di-homo-y-linolenic acid
(DHGLA;
20:348°"°'4) in which the tri-unsaturated 18 carbon fatty acid
is elongated by the addition of a
two carbon unit to yield the tri-unsaturated 20 carbon fatty acid. Since there
is considerable
interest in the production of long chain PUFAs of more than 18 carbons in
chain length, for
example arachidonic acid and eicosapentanoic acid, the identification of this
enzyme is of
both academic and commercial interest.
At present, there are no examples of identified cloned genes encoding PUFA
elongases,
though a number of genes encoding enzymes likely to be involved in other
aspects of lipid
synthesis have been identified. For example, an Arabidopsis gene (FAE1) has
been shown
to be required for the synthesis of very long chain monounsaturated fatty
acids (such as
erucic acid; 20:14") (James, D. W. et al, (1995) Plant Cell7, 309-319).
However, it is clear
that this enzyme does not recognize di- and tri-unsaturated 18 carbon fatty
acids, for
example, linoleic acid, 18:209~'z or a,-linolenic acid, 18:39''2,15
respectively, as substrates,
SUBSTITUTE SHEET (RULE 26)
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
2
and is therefore not involved in the synthesis of long chain PUFAs (Millar &
Kunst (1997),
Plant Journal 12, 121-131). This in itself is not surprising; since, of the
plant kingdom, only
a very few lower plant species, such as the moss Physcomicotrella patens
(Girke et al.,
(1998), Plant J, 15: 39-48); are capable of synthesising long chain PLJFAs,
and therefore
Arabidopsis would not be expected to contain any such enzymes (Napier et al.
(1997),
Biochem J, 328: 717-720; Napier et al., (1999) Trends in Plant Sci 4, 2-5).
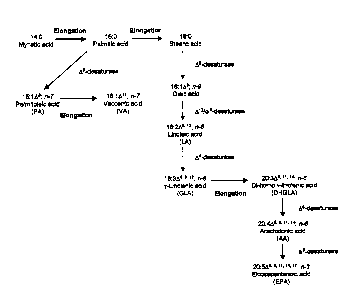
A schematic diagram representing a generalized pathway for the product of
PLTFAs is
shown in Figure 1. Biochemical characterisation of mammalian elongation
systems (most
notably from liver microsomes) has indicated that a mammalian elongase
consists of four
subunits, made up of a condensing enzyme, a (3-ketoreductase, a dehydrase and
an enoyl
reductase (reviewed in Cinti, D. L., et al (1992) Prog. Lipid Res. 31, 1-51).
The
Arabidopsis FAEI gene product encodes a polypeptide of 56kDa, which shows very
limited homology to condensing enzymes such as chalcone synthase and stillbene
synthase
(James, D. W. supra). Although FAEI is normally only expressed in seed
tissues, ectopic
expression in non-seed tissue (or heterologously in yeast) revealed that FAEI
could direct
the synthesis of erucic acid (Millar, A. A., Kunst, L. (1997) Plant J. 12, 121-
131).
Three fatty acid elongase activities have been characterised from the yeast S.
cerevisiae.
Again, this organism does not synthesis PUFAs, and therefore does not contain
genes
encoding a PUFA elongase. One gene ELO1, was identified on the basis of a
screen to
isolate mutants defective in elongation of 14 carbon (i.e. medium) chain
saturated fatty
acids (Toke & Martin (1996) J Biol Chem 271, 18413-18422). Complementation of
elol
mutants restored viability, and the ELOl gene product was shown to encode a
polypeptide
which was responsible for the specific elongation of 14:0 fatty acids to 16:0
fatty acids.
Two related genes were also detected in the genome of S. cerevisiae, and their
function
determined by disruption. These two genes, subsequently named EL02 and EL03,
were
shown to be involved in the elongation of the very long chain saturated fatty
acids found in
sphingolipid molecules (Oh et al (1997), J. Biol Chem 272, 17376-17384). In
particular,
EL02 was required for elongation of fatty acids up to 24 carbons, and EL03 was
required
for elongation of the 24 carbon fatty acid to 26 carbons. However, neither
gene was
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
3
essential for viability. Examination of the these three fatty acid elongases
revealed the
presence of a conserved "histidine box" motif (Shanklin et al., (1994),
Biochemistry, 33,
12787-12794) (His-X-X-His-His, where X is any amino acid) towards the centre
of the
polypeptide sequences. Importantly, there was no detectable homology between
the yeast
elongases (EL01,2,3) and the plant very long chain mono-unsaturated fatty acid
elongase
(FAE1) (Oh et al, supra).
In order to identify genes encoding PUFA elongases, it is necessary to study
systems in
which the synthesis of PUFAs is well documented; a good example of this is the
model
animal system C. elegans, a small free-living worm (Tanaka et al., (1996),
Lipids 31,
1173-1178). C. elegans, like most other animals, and in contrast to higher
plants,
synthesises PUFAs such as arachidonic acid (AA; ZO:4 05~g~31,14) as precursors
to a class of
molecules known as the eicosanoids, which in turn serve as precursors for
compounds such
as prostaglandins and leucotrienes (Horrobin, (1990), Reviews in Contemp
Pharmacotherpy,
1:1-45). The presence of AA and other long chain polyunsaturated fatty acids
in C. elegans
is well documented (Tanaka et al., (1996), Lipids 31, 1173-1178). The complete
sequence
of the nematode's genome is now publicly available (The C elegans consortium,
1998,
Science 282, 2012-2018: Database at
http:llwww.sanger.ac.uklProjectslC elganslblast server,shtml).
An object of the invention is to provide an isolated PUFA elongase.
Using the above-mentioned C. elegans genomic sequence, together with suitable
search
strings, the inventors identified eight related putative open reading frames
(ORFs) encoding
for PUFA elongases. A number of different search criteria were applied to
identify a
number of (ORFs) which were likely to encode polypeptides with fatty acid
elongase
activities. These ORFs were then subject to functional characterisation by
heterologous
expression in yeast, allowing the identification of a PUFA elongase.
Accordingly, a first aspect of the invention provides an isolated polypeptide
comprising a
functional long chain polyunsaturated fatty acid (PUFA) elongase i.e. the
polypeptide has
the function of extending the chain length of an 18 carbon PUFA to 20 carbons
in length.
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
4
This polypeptide can be used to elevate PUFA levels in animals, thereby
providing a ready
source of PLTFAs.
The polypeptide may be from a eukaryote.
The polypeptide may comprise at least a portion of the amino acid shown in SEQ
>D. 15, or
variants thereof.
For the purposes of the present application, the term "variant" in relation to
a certain
sequence means a protein or polypeptide which is derived from the sequence
through the
insertion or deletion of one or more amino acid residues or the substitution
of one or more
ammo acid residues with amino acid residues having similar properties, e.g.
the replacement
of a polar amino acid residue with another polar amino acid residue, or the
replacement of a
non-polar amino acid residue with another non-polar amino acid residue. In all
cases,
variants must have an elongase function as defined herein.
A second aspect of the invention provides a polypeptide having at least 60 %
homology to a
polypeptide according to a first aspect of the invention. The polypeptide may
have at least
$0%, or as much as 90% or more homology to a polypeptide according to a first
aspect of
the invention.
The polypeptide according to either aspect of the invention may include a
sequence motif
responsible for Endoplasmic Reticulum (ER) - retention. This allows the
polypeptide to be
specifically located or targeted to the ER of a cell.
The polypeptide may also be able to elongate palmitoleic acid (PA; 16:1~~) to
vacceric acid
(VA; 18:14"). Thus, the polypeptide is also capable of elongation of a ~9-
monounsaturated
16C fatty acid.
Preferably, the polypeptide is from an animal, more preferably, the animal is
an invertebrate
such as a worm. Where the animal is a worm, it is preferably C. elegans.
Alternatively, the
animal is a vertebrate, preferably a mammal such as a human, rat or mouse.
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
A third aspect of the invention provides an isolated DNA sequence, preferably
a cDNA
sequence, encoding a polypeptide according to a first or second aspect of the
invention.
This DNA sequence may be used to engineer transgenic organisms.
Preferably, the DNA sequence comprises the sequence shown in SEQ 1D NO: 7 or
variants
of that sequence due, for example, to base substitutions, deletions, and/or
additions.
A fourth aspect of the invention provides an engineered organism, such as a
transgenic
animal, engineered to express a polypeptide according to a first or second
aspect of the
invention. The engineered organism may be engineered to express elevated
levels of the
polypeptide, thereby providing a supply of polypeptide at a reduced cost as a
reduced
number of organisms need be used.
Preferably, the engineered organism is a mammal such as a rat, mouse or
monkey.
A fifth aspect of the invention provides an engineered organism containing a
synthetic
pathway for the production of a polypeptide according to a first or second
aspect of the
invention. This has the advantage of allowing greater control over the
production of PUFAs
by the pathway by an organism.
The pathway may include D'-fatty acid desaturase, and/or 46-fatty acid
desaturase.
The engineered organism according to a fourth or fifth aspect of the invention
may be a
lower eukaryote, such as yeast. Alternatively, the transgenic organism may be
a fish.
A sixth aspect of the invention provides a transgenic plant engineered to
express a
polypeptide according to a first aspect of the invention.
A seventh aspect of the invention provides a transgenic plant containing a DNA
sequence
according to a third aspect of the invention.
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
6
An eighth aspect of the invention provides a method of producing a PUFA
comprising
carrying out an elongase reaction catalysed by a polypeptide according to a
first or second
aspect of the invention.
The PUFA may be di-homo-gamma-linoleic acid (20:308'"''4), arachidonic acid
(20:4058°"''4), eicosapentanoic acid (20:5~5~8~"~'4~"), docosatrienoic
acid (22:3~3~'6~'9),
docosatetraenoic acid (22:4~'~'°~'3~'6), docosapentaenoic acid
(22:50'''°~~3,16,19) or
docosahexaenoic acid (22:54''''°,13.16,19)_
The PLJFA may be a 24 carbon fatty acid with at least 4 double bonds.
A ninth aspect of the invention provides a PUFA produced by a method according
to an
eighth aspect of the invention.
The PLTFA may be used in foodstuffs, dietary supplements or pharmaceutical
compositions.
A tenth aspect of the invention provides a foodstuff comprising a PUFA
according to a fifth
aspect of the invention. The foodstuff can be fed to an animal.
An eleventh aspect of the invention provides a dietary supplement comprising a
PUFA
according to a fifth aspect of the invention. The dietary supplement can be
supplied to an
animal to augment its PUFA levels.
An twelfth aspect of the invention provides a pharmaceutical composition
comprising a
polypeptide according to a first or second aspect of the invention or a PLTFA
according to a
ninth aspect of the invention.
Preferably, the pharmaceutical composition comprises a pharmaceutically-
acceptable
diluent, earner, excipient or extender. This allows the composition to be
supplied in a form
which best suits the pharmaceutical application in question. Fox example, a
topical
application would preferably be a cream or lotion, whereas if the composition
was to be
ingested a different form would be more suitable.
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
7
A thirteenth aspect of the invention provides a method of treatment of an
animal, such as a
mammal, or a plant, comprising supplying to the animal or plant a DNA sequence
according
to a third aspect of the invention, a foodstuff according to a tenth aspect of
the invention, a
dietary supplement according to an eleventh aspect of the invention, a
pharmaceutical
composition according to a twelfth aspect of the invention or a PUFA according
to a ninth
aspect of the invention.
Preferably, the mammal is a human.
The invention will now be further described, by way of example only, with
reference to SEQ
>D1 to 16, and Figures 2 to 11, in which;
SEQ 1D 1 to 8 show the putative ORFs encoding PUFA elongases A to H
respectively; and
SEQ ID9 to 16 show the deduced amino acid sequences of the putative ORFs of
SEQ ID
NO: 1 to 8 respectively; and
Figures 2 to 9 show hydrophobicity plots for each of PUFA elongases A to H
respectively.
Figure 10 shows an amino acid sequence line-up comparing the C'. elegans ORF
F56H11.4
(Z68749) with related sequences.
Figure 11 shows chromatograms of fatty acid methyl esters from transformed
yeast.
Introduction to general strategy
Initially the C. elegans databases were searched for any sequences which
showed low levels
of homology to yeast ELO genes (EL02 and EL03) using the TBLASTN programme. A
similar search was carried out using short (20 to 50 amino acid) stretches of
ELO genes
which were conserved amongst the three ELO polypeptide sequences. C. elegans
sequences
which were identified by this method were then used themselves as search
probes, to
identify any related C. elegans genes which the initial search with the yeast
sequences failed
to identify. This was necessary because the level of homology between the
yeast ELO genes
CA 02365096 2001-09-14
WO OOJ55330 PCTIGB00/01035
8
and ~ worm genes is always low (see BLAST scores later). To allow for a more
sensitive
search of worm sequences, a novel approach was adopted to circumvent the major
drawback
with searches using the BLAST programmes, namely that the search string (i.e.
the input
search motif) must be longer than 1 S characters for the algorithm to work.
Thus, if it was
desired to search for a short motif (like a histidine box), then the BLAST
programme would
not be capable of doing this. A complete list of all the predicted ORFs
present in the C.
elegans genome exists as a database called Wormpep, which is freely available
from the
Sanger WWW site (http://www.sanger.ac.uk/Projects/C_eleganslwebace front
end.shtml).
The latest version of Wormpep was down loaded to the hard disc of a Pentium
PC, and
re-formatted as a Microsoft Word6 document, resulting in a document of about
3,500 pages.
This was then searched using the "Search & Replace" function of Word6, which
also allows
for the introduction of "wildcard" characters into the search motif. So, for
example, it is
possible to search both for the short text string HPGG, which would identify
any predicted
worm ORF present in the Wormpep 3,500 page document containing this motif, or
alternatively search with HPGX (where X is a wild card character). Clearly,
such (manual)
searches of a 3,500 page document are extremely time-consuming and demanding,
also
requiring visual inspection of each and every identified ORF. For example,
searching with a
motif such as HXXHH identifies in excess of 300 different ORFs. However, by
using a
number of different short search strings (as outlined below), and combining
these with other
methods for identifying putative elongase enzymes, a number of candidate ORFs
have been
identified.
Database search using the FAE1 polypeptide sequence
As a negative control, to demonstrate that the FAEl gene sequence was unlikely
to provide a
useful search sequence in the identification of C.elegans sequences encoding
for PUFA
elongases, the GenBank databases
(http://www.ncbi.nlm.nih.gov/Web/Search/index.html)
were searched using the Arabidopis FAE1 polypeptide sequence to identify
related genes or
expressed sequence transcripts (ESTs). GenBank is the NIH genetic sequence
database, an
annotated collection of all publicly available DNA sequences (Nucleic Acid
Research (1998)
26, 1-7). There are approximately 2,162,000,000 bases in 3,044,000 sequence
records as of
December 1998. The search was carried out using the BLAST2 (Basic Local
Alignment
Search Tool) algorithm (Altschul et al., (1990) ,I Mol Biol 215,403,410)
Although a number
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
9
of plant ORFs and ESTs were reported as being related, no animal sequences
were identified
by this search, confirming the observation that FAE1 was unlikely to be a
suitable candidate
as a search template for PUFA elongases.
Database search using yeast ELO sequences
Using the three yeast fatty acid elongase sequences (ELO 1, 2, 3) as probes, a
number of
putative ORFs in the DNA of C. elegans-derived cosmid sequences which form the
C.
elegans genomic sequence database were identified. Moreover, an extensive and
time-consuming search of a downloaded copy of the WormPep database
(ftp://ftp.sanger.ac.uk./pub/databases/wormpep) using manual search strings in
MSWord 6,
identified a number of C. elegans ORFs which contained presumptive histidine
boxes.
Wormpep contains predicted proteins from the Caenorhabditis elegans genome
sequence
project, which is carried out jointly by the Sanger Centre in Cambridge, UK
and Genome
Sequencing Center in St. Louis, USA. The current Wormpep database, Wormpep 16,
contains 16,332 protein sequences (7,120,115 residues). Search strings used
included
[HXXHH], [HXXXHH], [QXXHH] and [YHH]. Comparison of the data from the two
different searches indicated a small (<10) number of putative ORFs as
candidate elongases.
The histidine box motifs are shown in bold in SEQ >D 9 to 16.
Hydrophobicity plot analysis
Since the fatty acid elongase reaction is predicted to be carried out on the
cytosolic face of
the endomembrane system (Toke & Martin (1996), supra; Oh et al (1997), supra),
the
putative C. elegans ORFs were examined for potential membrane spanning
domains, via
Kyte & Doolittle hydrophobicity plots (J. Mol Biol, (1982), 157, 105-.132).
This revealed a
number of ORFs with possible membrane-spanning domains, and also indicated a
degree of
similarity in the secondary-structure of a number of identified ORFs.
Screening for ER-retention signal sequences
The inventors postulated that since fatty acid elongases are expected to be
endoplasmic
reticulum (ER) membrane proteins, they might be expected to have peptide
signals which
are responsible for "ER-retention". In the case of ER membrane proteins, this
signal often
takes the form of a C-terminal motif [K-K-XZ_,-Stop], or similar variants
thereof (Jackson et
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
al., (1990), EMBO J., 9, 3153-3162). Further sequence analysis of the C.
elegans putative
elongases revealed that 4 ORFs (F41H10.7, F41H10.8, F56H11.4, Y53F4B.c) had
C-terminal motifs that exactly matched this search pattern, and that a further
2 ORFs
(F11E6.5, C40H1.4) had related sequences. These sequence motifs are underlined
in SEQ
m 9 to 13, 15 and 16.
Chromosome mapping
Since the inventors had previously observed that C.elegans genes involved in
the synthesis
of PUFA may exist in tandem (for example the 05 and 06 desaturases required
for AA and
GLA synthesis, respectively, are < 1 kB apart on chromosome N (Michaelson et
al., (1998),
FEBS Letts 439, 215-218), the positions of the putative C. elegans elongase
ORFs were
determined using the Sanger Centre's WebAce C. elegans server
(http://www.sanger.ac.uk/Projects/C elegans/webace-front end.shtml).. This
indicated that
two pairs of putative elongases were in close proximity to each other on the
C. elegans
chromosome N.
F41H10.7 and F41H10.8 were identified as being approximately 10 Kb apart on
chromosome N, and F56H11.3 and F56H11.4 were identified as being approximately
2 Kb
apart on chromosome N.
Putative C. elegans fatty acid elongases
The positions of the putative OIRF's in the C. elegans genome are shown below
i.e.
chromosome number, and map position in centiMorgans, together with the GenBank
database accession numbers.
The designations used employ the same method as used on the Sanger Centre's C
elegans
database, i.e. ORF C40H1.4 is predicted coding sequence 4 on cosmid C40H1.
Elon ase Cosmid Saner ID GenBank Acc Chromosome
Code
A C40H 1.4 Z 19154 III
CA 02365096 2001-09-14
WO 00155330 PCT/GB00/01035
11
B D2024.3 U41011 N, 7.68
C F11E6.5 281058 IV, 18.8
D F41H10.7* U61954 N, 29.8
E F41H10.8* U61954 N, 29.8
F F56H11.3# 268749 IV, 2.5
G F56H11.4~ 268749 IV, 2.5
H Y53F4B.c 292860 II
* ors indicates genes in tandem
Comparison of C. elegans putative elongase ORFs with yeast genes:
Each of the three yeast ELO polypeptides were compared against all of the worm
putative
elongase translated ORF sequences, and then ranked in order of similarity (as
measured by
the BLAST score) (Altschul et al (1990), supra)
The results are shown below, with the ORF sequences ranked from most similar
to least
similar, and the BLAST scores are shown in brackets:
Yeast ELO1 (14 to 16 carbon fatty acid elongase)
G (262) > E (241) > D (225) > C (219) > A (216) > F (215) > H (197) > B (172)
Yeast EL02 (24 carbon sphingolipid elongase)
E(231)>C(226)>G(189)>A(181)>F(166)>D(150)>H(141)>B(140)
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
12
Yeast EL03 (24 to 26 sphingolipid elongase)
D(171)>G(163)>F(154)>A(152)>E(150)>C(131)>B(132)>H(128)
It is clear from the numeric values of the BLAST scores that the sequences are
related, but
the levels of homology are low. For comparison, the BLAST score for homology
between
two related worm proteins, the 05 and the 46 desaturase is in excess of 500.
Analysis of potential sphingolipid ancestry
Previously, the inventors had noted the similarities between the fatty acid D6
desaturase and
sphingolipid desaturases in plants, and that the two distinct enzymes could
have arisen from
one ancestral gene. Moreover, it was considered likely that the sphingolipid
desaturase
predated the fatty acid desaturase, and may in fact have been the ancesteral
progenitor.
Therefore it is plausible that the next step in the arachidonic acid
biosynthetic pathway has
also evolved from the sphingolipid metabolic pathway. It is therefore
considered highly
significant that some of the C. elegans ORF putative elongases have similarity
to
sphingolipid enzymes. For this reason, these ORFs are considered to be very
clear
candidates for PUFA elongases. It has previously been considered that the C.
elegans 05
and D6 fatty acid desaturases have evolved from 1 ancestral gene (Michaelson
et al., ( 1998),
FEBS Letts 439, 215-218). It is also significant that one pair of C. elegans
putative
elongase ORFs (F & G) genetically maps close to the 05/06 fatty acid
desaturase genes,
with both gene pairs being located at the top end of chromosome IV.
Cosmid San eg r ID GenBank Acc Chromosome Encoded Peptide
Code
W08D2.4 270271 IV, 3.06 06 fatty acid desaturase
T13F2.1 281122 IV, 3.06 45 fatty acid desaturase
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
13
Cloning of Desaturase and Elongase Genes in Yeast Expression Vectors
Putative elongases sequences F56H11.4 and F41H10.8 were cloned by PCR into the
pYES2 vector (Invitrogen). A C. elegans mixed stage cDNA library was used as a
PCR
template. F56H11.4 was amplified using primers:
56h114.for 5'-GCGGGTACCATGGCTCAGCATCCGCTC-3' and;
56h1 l4.rev 5'-GCGGGATCCTTAGTTGTTCTTCTTCTT-3'.
F41H10.8 was amplified using primers:
41h108.for 5'-GCGGGTACCATGCCACAGGGAGAAGTC-3' and;
41h108.rev 5'-GCGGGATCCTTATTCAATTTTTCTTTT-3'.
Amplified sequences were then restricted using Kpnl and BamHI (underlined in
the
forward and reverse primers, respectively), purified using the Qiagen PCR
purification kit,
and ligated into a KpnI/BamHI cut pYes2 vector.
An ORF encoding the Mortierella alpina 45-fatty acid desaturase (Michaelson,
L. V., et al
(1998) J. Biol. Chem. 273, 19055-19059) was amplified using primers:
MadS.for 5'-GCGAATTCACCATGGGTACGGACCAAGGA-3' and;
MadS.rev 5'-GCGGAGCTCCTACTCTTCCTTGGGACG-3',
and restricted using EcoRI and SacI, gel purified as described and ligated
into a EcoRIlSacI
cut pESC-TRP vector (Stratagene) to generate pESC/~5.
An ORF encoding the borage O6-fatty acid desaturase (Sayanova, O., et al
(1997) Proc.
Natl. Acad. Sci USA 94, 4211-4216) was restricted from pGEM3 using BamHI and
XhoI
and ligated into a BamHIlXhoI cut pESC-TRP vector to generate pESC/0''.
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
14
A double construct was also generated by ligating the BamHIlXhoI borage 46
insert into the
pESC/OS construct described previously, generating pESC/(05,06).
Functional Characterisation in Yeast
Elongases and desaturase constructs were introduced in Saccharomyces
cerevisiae
W303-lA using a lithium acetate based method (Elble, R. (1992) Biotechniques
13, 18-20)
and expression of the transgenes was induced by addition of galactose to 2%
(w/v) as
described in Napier et al (Napier, J. A., et al (1998) Biochem J330, 611-614;
Michaelson
L. V., supra; Michaelson, L. V., (1998) FEBS Letts 439, 215-218). Yeast
transformants
containing pYES2-derived constructs were grown on synthetic minimal media (SD,
the
composition of which is defined in Sherman, F ( 1991 ) Methods in Enzymology
194, 3-21 );
synthetic minimal medium minus uracil; pESC-derived constructs were grown on
SD
minimal medium minus tryptophan. Co-transformed yeast (containing both pYES2
and
pESC derivatives) were grown on SD minimal medium minus uracil and tryptophan.
Prior
to induction, cultures were grown in the presence of 2% raffinose and
supplemented with
0.5 mM of the appropriate fatty acid substrate in the presence of 1 % tergitol-
(NP40)
(Sigma). All cultures were then grown for a further 48-h unless indicated.
Fatty Acid Analysis
To identify the elongation reaction responsible for the synthesis of di-homo-y-
linolenic acid
(DHGLA; 20:308"~'4) from GLA, this latter fatty acid was supplied as the
(exogenous)
substrate.
Lipids were extracted from transformed and control yeast by homogenisation in
MeOH-CHC13 using a modification of the method of Bligh and Dyer (Dickenson &
Lester
(1999) Biochim Biophys Acta 1426, 347-357). The resulting CHC13 phase was
evaporated
to dryness under nitrogen gas and the samples were transmethylated with 1M HCl
in
methanol at 80 °C for 1 hour. Fatty acid methyl esters (FAMES) were
extracted in hexane
and purified using a small column packed with Florisil. Analysis of FAMES was
conducted using a Hewlett Packard 5880A Series Gas Chromatograph equipped with
a
25M x 0.32mm RSL-SOOBP bonded capilliary column and a flame ionisation
detector.
Fatty acids were identified by comparison of retention times with FAME
standards (Sigma)
WO 00/$5330 CA 02365096 2001-09-14 pCT/GB00/01035
separated on the same GC. Quantitation was carried out using peak height area
integrals
expressed as a total of all integrals (Bligh, E.G. & Dyer, W.J. (1959) Can. J.
Biochem.
Physiol. 37, 911-917).
Total fatty acids extracted from yeast cultures were analysed by gas
chromatography (GC)
of methyl ester derivatives. Lipids were extracted, transmethylated and the
fatty acid
methyl esters (FAMEs) analysed as described by Sayanova et al.
Figure 11 shows chromatograms of fatty acid methyl esters from yeast
transformed with the
control (empty) plasmid pYES2 (Fig. 11A) or with ORF F56H11.4 in pYES2 (Fig.
11B).
Exogenous substrate in the form of GLA was supplied to the cultures. Two novel
peaks
are observed in (B); these peaks (annotated as 20:3 and 18:1 *) were
identified (against
known standards) as DHGLA and vaccenic acid, respectively. Detection was by
flame
ionisation.
One cDNA ORF tested in this manner displayed a high level of elongase activity
on the
GLA substrate, converting 44% to DHGLA. The identity of this elongation
product was
confirmed as DHGLA by comparison with a known standard (the standards used
were
known standards for either DHGLA, AA, EPA or VA from Sigma Chemicals, Ltd.),
using
GCMS analysis using a Kratos MS80RFA (Napier, J. A., supra; Michaelson, L. V.,
supra;
Michaelson, L. V., supra). The deduced amino acid sequence of the functional
elongase
clone identified it as being encoded by the C. elegans gene F56H11.4, and
comparison with
the yeast ELO genes showed low homology confined to a few short amino acid
motifs (see
Fig. 10). Some similarity with a mouse gene Cig30 (Tvrdik, P., (1997) J. Biol.
Chem. 272,
31738-31746), which has been implicated in the recruitment of brown adipose
tissue in
liver tissue, was also observed, as well as a potential human homologue
encoded by a gene
located on chromosome 4q25, BAC 207d4. The most closely related C. elegans
ORFs,
F41H10.8 (U61954) and F56H11.3 (Z68749) are also shown, as is part of a
related human
gene present on chromosome N (present on BAC clone B207d4; AC004050). The
GenBank accession numbers are given for all sequences.
The range of fatty acids synthesised by C. elegans can potentially require a
number of
different elongation reactions (Tanaka, T., (1996) Lipids 31, 1173-1178). The
substrate-specificity of the F56H11.4 PUFA elongase was therefore determined
using a
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
16
range of exogenously supplied fatty acids. This revealed that GLA is the major
substrate,
with a number of other fatty acids being elongated at a lower efficiency (see
Table 1 ).
Although most of these substrates are polyunsaturated fatty acids, it was
unexpectedly
observed that palmitoleoic. acid (PA; 16:1 09) was also elongated by F56H11.4
to yield
vaccenic acid (VA; 18:1 0"). The biosynthetic pathway for VA is unclear, but
the data
indicate that it may be synthesised by elongation of 0~-monounsaturated 16C
fatty acid.
The C. elegans PUFA elongase ORF F56H11.4 maps to the top of chromosome IV (at
4.32
cM) with a related sequence (F56H11.3; 51 % similarity) located 1,824bp
downstream.
Another C. elegans gene (F41H10.8) was also observed, which is present on
chromosome
IV, and which shows a slightly higher level (53%) of similarity to the PUFA
elongase than
F56H11.3 (see Fig. 10). However, when a PCR product encoding ORF F41H10.8 was
expressed in yeast in a manner identical to that used for F56H11.4, the former
failed to
direct the elongation of any fatty acids, despite the provision of a range of
substrates (see
Table II).
In order to reconstitute the PUFA biosynthetic pathway in a heterologous
system, the
PLTFA elongase F56H11.4 was expressed in yeast in conjunction with either the
06- or
OS-fatty acid desaturases previously isolated and characterised by the
inventor (Napier, J.
A., supra; Michaelson, L. V., supra). Expression of the OG-fatty acid
desaturase and
F56H11.4 was carried out in the presence of two different substrates (LA or
ALA) while
the 4'-fatty acid desaturase and the elongase were expressed in the presence
of GLA only.
This demonstrated that was possible to combine a desaturase and an elongase in
yeast to
generate significant amounts of a final "product" (see Table III). In the case
of the
elongase and the 4G-fatty acid desaturase, the reactions proved highly
efficient with the
production of 4.5% of DHGLA from the LA substrate. This resulted from 25%
desaturation of the LA substrate to GLA, which was then elongated to DHGLA at
a similar
level of efficiency (18%). This is lower than the % conversion observed for
GLA when
supplied exogenously (see Table I), indicating that the in vivo production of
substrates for
elongation may be rate-limiting.
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
17
If ALA was used as a substrate, 27% of this was initially O6-desaturated to
yield
octadecatetraenoic acid (OTA; 18:4 06.9,'z,'s) but only 8% of was subsequently
elongated to
yield eicosatetraenoic acid (20:4 ~8~"~'4~"). Thus, the conversion efficiency
of ALA to the
final 20-carbon tetraenoic PUFA was only about 2.2%.
Since DHGLA is an n-6 fatty acid, whilst the OTA-derived eicostetraenoic acid
is an n-3
type, this demonstrates that the elongase is capable of accepting both forms
of essential
fatty acid, albeit with different efficiencies. Verification was also provided
that the 20C
PUFAs synthesised in the yeast expression system were generated by the O6-
desaturation of
18C substrates which were subsequently elongated, as the O6-desaturase showed
no activity
on 20:2 or 20:3 substrates (see Table IIII).
The combination of the D5-desaturase and the elongase also demonstrated that
these two
enzymes could work in tandem, although the efficiency of this overall
conversion was
lower (3.3% AA from GLA) which was due to the previously observed low activity
of the
OS-desaturase enzyme itself (Michaelson, L. V., supra; Michaelson, L. V.,
supra). Thus,
although nearly 45% of the GLA substrate was elongated to DHGLA, only 7.5% of
this
was then desaturated to AA (see Table III).
Finally, the production of either AA or eicosapentanoic acid (EPA;
20:5~5~8~"~'4~") in yeast
from dienoic or trienoic 18 carbon substrates was achieved via expression of
all three
enzymes (the two desaturases and the F56H11.4 PUFA elongase) simultaneously.
As
shown in Table IV, small but significant amounts of AA were produced when the
yeast was
supplied with the 18C dienoic fatty acid LA.
GC-Mass Spectroscopy (MS) Analysis
Peak identification and confirmation were carried out by GC-MS using a Kratos
MS80RFA
using known standards (Sigma). The identity of this 20C PLTFA was verified by
GCMS,
indicating that the conversion efficiency from LA was 0.65%. When ALA was used
as a
substrate, 12.5% of the (46-desaturated and elongated) eicosatetraenoic n-3
fatty acid was
OS-desaturated, resulting in a total conversion of 0.3% of the ALA substrate
to EPA (the
identity of EPA was confirmed by GCMS).
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
18
Expression of C. elegans elongase in plants
In order to express C. elegans elongase in plants, the following protocol is
an example of a
process which can be used to create the transgenic plants. C. elegans ORF
sequence can be
subcloned into a plant expression vector pJD330, which comprises a viral 35S
promoter,
and a Nos terminator. The resulting cassette or promoter/coding
sequence/terminator can
then be subcloned into the plant binary transformation vector pain 19, and the
resulting
plasmid introduced into Agrobacterium tumefaciens. This Agrobacterium strain
can then be
used to transform Arabidopsis by the vacuum-infiltration of inflorescences,
and the seeds
harvested and plated onto selective media containing kanamycin. Since pain 19
confers
resistance to this antibotic, only transformed plant material will grow.
Resistant lines can
therefore be identified and self fertilized to produce homozygous material.
Leaf material
can then be analyzed for expression of C. elegans elongase.
Fatty acid methyl ester analysis can be carried out as previously described.
CA 02365096 2001-09-14
WO 00/55330 19 PCT/GB00/01035
.. .r n ..a
, n~, ~~ ..
~ ~ V, ~ C
N ,n , , . , N . .
, , , , .
r
oanon o
+a +~ ~ +~
+ Ya+~~~,
~~~nn
N V1 r, ~O . ~ , ,
, . , , , , O
rr y~ CO
r N "', n
.
C .-,
, ~ ~O +1 +1
e0
0~
A
q~
Y, n, ,. v..,
O,v W
N
p .~-v ~ ..i
~ M
O O O
~
~ 'H .-n '~
.w
p v, a,
a aA
O ~
~ .-i , v
. Y n , v , M n
~
~
~
r. h
C
+1
'H
+1
. ~
b
M
~
'
n . n . , n . v
n , .
f
~ Y
~
~
~
+ y'~.
y'~e
ae
r.
.. -. .. n
, , , v1
N h ... M ~ v ,
, , t~ ,
W
.~'!0 0
o
-
~i ~i +1 +1
'~
.. o
ao .r . . . .
'' 'o ~c v . .
r ~ . .
n ~
, ,
n
I ii
~
+ '~ g fl +
n
~0 0~
N M VI ~ t.i , , ~ ,
, v n W 1 , ~
M
+1 1t '1i
n
f Y ~ n , . , , . .
, . , v .
.
yl rr
~)
~ ~
M M
O O
+ 0~ T
01
n
,O , , , .
:g.~~a , , , ,
, , ,
m~~~
+1 -H 'i~
. +1 ~
.
, , , , ,
~ ,
~ h
. , , , , ,
Y
~r ' p~ ~ p,
ooO~o'o~po~~ ~AN~ i~
SUBSTITUTE SHEET (RULE 26)
CA 02365096 2001-09-14
WO 00/55330 2p PCT/GB00/01035
Ov N N
oo
C M M N
~
+I +I +I
+I
' (~ N
~ M
M N '~ M
Vi
NMV1.-l ~~ i~~~~N ~i ~
a
~.
w
O M ~ N
N
i-I +I ii
+I
+ O I~ M
~ M
M ~ ~ N
~n
N M ~t ~ ~ ~ ~ ~ i ~
~ ~ N ~ O
N O~ M
C O '-' .-.
~
O O
d' 00 .d.
'~'~
-H
M vi vi
~: ~
N -r ~ d' ~ ~ i v ~
W O~ ~ ~ ~ ~
d
a
d N N 'n
O CO O
M """
~ O O
+ 00 ~O
-. vi O
~
N
. mt ~ ~ ~ ~
~ ~
N M N
O O '~ O
O
-H +I i1
~ 1-1
' ~ ~ +~ rr
rr
V V' M ~D
0 ~ O
N N v1 ~ ~ ~ ~ ~ ~ i
-~ M ~ ~ i
O
_
x
_ ,~ _
w
O N N
M
o +~+~ +~
+ os ~r
'~ '~
M N " ~
'"~ Q~ O
N vi
s
C M ~ i ~ ~ ~ ~
N ~
OW O ~D
O .-1 O
'."'
.-:
-f-I -fi
i-i
+I
O 'n N
~ .--~
~ h
N M
d
a
U v0 N ~D
d'
O N
i-I +i +i
+i
+ ~ V7 M
~ N
00 M ~ ~!'
M ~O -~ O
N
-~
'
n ~ ~ . ~ ~
M ~ ~ ~
N ~D
v~
O O '~
O
-H -H
+I
M 00
~ Qv
O~ O
'~ 'd'
.-~ i!1 i ~ ~ ~ ~
v1 N ~ ~ i
O~ l~
M
O O "'
.-.
+I ii
O ii
+ O Ov
~ u1
~ O N
~
.-~ v~
~t N
b
N
d b
y # ~ o
b aa d a
d d
' ~' ooaoxoa W ' A
o~o' - ..
o o d
H o v~ ,~ .-, ,-, .-.a c~ N A s t~ ~
,-. ,.aN w a w
.-.
CA 02365096 2001-09-14
WO 00/55330 2 j PCT/GB00/01035
00 N V1 h
M M ~ N M
-fl -!-i ~ -H
i-I
' 00 ~ ~ O N
Qv V1 ~. M 01
N N ~ ~!1 ~ ~ ~ ~ ~i ~ ~
...r ~ ~ ~ ~ ~ ~ i
~r d
~ a
_
d' M 00 N M 00 N
Ov
w +i -+~ o +I +~ -: o
o
-~ O~ ~G '~'~ N 'H -H
l'~ ~I
h ~t ~ N ~ M
N N ~ ~n .-.,~ ~' ~ ~ C~
N ~ ~ O~ ~
M l~
Oy N et
O O O
o ii +I -ti
r; .--~ V: pp
O
N C~ ~ v~ ~n
~O ~ ~
~O ~ I~ 00 M ~t N
M
~ C O G O ~ ~ O O
O
.o + ,~. ii ~i M M -H i-I
-H ii ~-I
C1 d r, M d; N N ,n d: '~t 'ct
h ~O C;
...y'i O ~O t'~ ~ M ~~ ~ OG
V1 N W .-n~ ~ Wn
~i;
rr
C '-'
O N
'n O
-H ii ~ ii
+I
' t~ ~O ~ N
wt
M et' ~ ~
Y1
N N Wi .-r ~ ~ ~ ~ ~~ ~ i
i M ~ ~ ~
a
~D N ~h N
~; .-. M r; N 'n
t~ M N
~ O O ~ O N O
,i~ C
-1-l~ Ov 'f'~ -H -H
~ N ~ O -i-I
00 00 ~ ~ Os M ~ I~
cJ ~ v1 t~
.-r .r O et ~ t~ M n ~i .-i
.-i t~ N ~ .--n ~ ~ oO
~
v1 I~ N
r; n1 ~; ~; 'n
r; -
O O
O ' N I~ ~
N ~ ~ .-~ ..~ 00
~
N et 'd' u'W,i i ~ W1iv n i
v ~ i i i i
d
M 00 .-r
.-; N N ~t M
r.;
N +i +f '~ ~ O
+I
C ~' t~ O ~ ~ M
N ~: ~ N o0 O
,ri
N d' vi rt ~ i mt' ~
.-, v ~ ~
b
N
d
V ~ G c~
' ~ ~
a o -~ cv, o d d cv M ~r W d d
.-. *. d ~ d d
x aH
b ~~~~~~d aH o o ors o d
H v ~ ~ ~~~~~-.,~ ~ac~oNA N ~Nw a c~oa~
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
22
SEQ ID1
C40H1.4
atggagcttgccgagttctggaatgatctcaacaccttcaccatctacggaccgaatcac
acagatatgaccacaaaatacaaatattcatatcacttcccaggtgaacaggtggcggat
ccgcagtattggacgattttattccagaaatattggtatcattcgatcacaatatcagtt
ctttatttcattttaattaaggtgattcaaaagtttatggagaatcgaaaaccattcact
ttgaaatacccattgattctttggaatggagctcttgcagcattcagtataattgccaca
ttgcggttctctattgatcctctacgatcactatatgctgaaggattctacaaaactctg
tgctattcgtgtaatccaactgatgtggctgcattttggagctttgcattcgctctttcc
aagattgttgaacttggagacactatgttcattattttgagaaaacggccattgatcttt
ttacactactatcatcatgcagcagtgttaatctacactgtccattctggtgccgagcat
actgcagctggtcgtttctacatcctaatgaactacttcgcacattctctcatgtatact
tactacacagtttctgccatgggatacagattaccgaaatgggtatcaatgactgtcaca
actgttcaaacaactcaaatgttagctggagtcggaataacttggatggtgtacaaagtg
aaaactgaatacaagcttccttgtcaacaatccgtagccaatttgtatctcgcattcgtc
atctatgtcacatttgccattcttttcattcaattcttcgtcaaggcatacattatcaag
tcgtcgaagaagtcgaaatcggtgaagaacgaataa
SEQ ID2
D2024.3
atggcaaaatacgactacaatccgaagtatgggttagaaaattacagcatattccttccc
tttgagacatcttttgatgcatttcgatcgacaacatggatgcaaaatcactggtatcaa
tcaattacagcatctgtcgtgtatgtagccgtcatttttacaggaaagaaggtggttctc
atctacaaaaaatcacgagttattacttttgagtctagccttcagaatgcaattaagaat
cgaaaccgaaaatcacttaatagttctcaaatgtttcagattatggaaaagtacaagccc
ttccaactggacacaccactcttcgtctggaattcatttttagccattttctcaattctc
gggttcctccgaatgacacctgaatttgtatggagttggtcagcagaaggaaactcattc
aaatattcaatttgtcattcatcttatgctcaaggagtcactggtttctggactgaacaa
ttcgcaatgagcaaacttttcgagctcatcgacacaatcttcatcgttcttcgtaaacgt
ccactcatcttccttcactggtatcatcatgtaactgttatgatctacacatggcacgcg
tacaaggatcacactgcatcaggacggtggttcatttggatgaattatggagttcatgct
cttatgtattcctactatgctcttcgttctctgaaattccgtcttccaaaacaaatggca
atggttgttactactctccaacttgctcaaatggttatgggagtaatcatcggagtcact
gtctaccgtatcaagtcatcgggtgaatactgccaacagacatgggacaatttgggatta
tgctttggagtttatttcacatatttccttcttttcgccaacttcttctaccatgcatat
gttaagaaaaacaaccgtacagtaaattatgaaaataattcaaaaaatttccccgatctc
gttttaatttacctgagaaaaaaggtttcaagaaaatcgaaaaatcggcaatgttcagaa
aataattataaaattcaattttcatcaaattttgttaatgttgatggaaaaaaacataag
aaaacatatgaacttattcttccaagaagaaaaatgaccacaattttaacttttctattt
ggaaaaaatcgaattttttcgaaatatcagaaaaatcgaaaaaacatttcgattcctgtt
gatttcgaaattctggagccaaaagaagatatcaatgctaacatcgctgagccatccatc
acaacgaggtccgccgccgcacgaagaaaagttcaaaaagctgattag
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
23
SEQ ID3
F11E6.5
atggcagcagcacaaacaagtccagcagccacgctcgtcgatgttttgacaaaaccatgg
agtctggatcagactgattcttacatgtctacatttgtaccattatcctataaaatcatg
attggttatctcgtcaccatctacttcgggcaaaaattaatggctcacagaaaaccattc
gatctccaaaatacacttgctctctggaacttcgggttttcactgttctcgggaatcgcc
gcctataagcttattccagaactattcggagttttcatgaaggacgggtttgtcgcttcc
tactgtcaaaacgagaactactacaccgatgcatcaactggattctggggctgggccttt
gtgatgtcgaaagctccagaactaggggatactatgttcttggtccttcgtaaaaaacca
gttatcttcatgcactggtatcatcatgccctcacatttgtctacgcagtagtcacatac
tctgagcatcaggcatgggctcgttggtctttggctctcaaccttgccgtccacactgtt
atgtatttctacttcgccgttcgcgccttgaacatccaaactccacgcccagtggcaaag
ttcatcactactattcaaattgtccaatttgtcatctcatgctacatttttgggcatttg
gtattcattaagtctgctgattctgttcctggttgcgctgttagctggaatgtgctatcg
atcggaggactcatgtacatcagttatttgttcctttttgccaagttcttctacaaggcc
tacattcaaaaacgctcaccaaccaaaaccagcaagcaggagtag
SEQ ID4
F41H10.7
atgtcatcggacgatcgtggcactagaaccttcaagatgatggatcaaattcttggaaca
aacttcacttatgaaggtgccaaagaagttgctcgaggccttgaaggtttctcagcaaag
cttgccgtcggatatattgccactatttttggactgaaatattatatgaaagaccgaaaa
gccttcgatctcagtactccattaaacatttggaatggtattctttcgacattcagctta
ttgggattcttattcacttttcctactttgttatcagttatcagaaaggatggatttagt
cacacctattcccatgtctctgagctttacactgacagtacctctggatattggatcttc
ctttgggttatctcaaagattccggaacttttggatacagtattcattgttcttcgcaag
agaccacttattttcatgcactggtaccatcacgcattgaccggttactatgctcttgtc
tgctaccatgaggatgctgtccatatggtttgggttgtatggatgaattatattattcat
gcattcatgtatggatactatcttctgaaatctctgaaagttccaattccaccatcagtt
gctcaagcaatcaccacatctcaaatggttcaattcgcagttgccattttcgcacaagtt
catgtttcctataaacactatgttgagggagttgaaggattagcctactcgttcagagga
acagctatcggatttttcatgcttactacctacttctatctatggattcaattctacaaa
gagcactatcttaagaatggaggcaaaaagtacaatttggcaaaggatcaggcaaaaact
caaacaaagaaggctaactaa
SEQ ID5
F41H10.8 (ce477)
atgccacagg gagaagtctc attctttgag gtgctgacaa ctgctccatt
cagtcatgag ctctcaaaaa agcatattgc acagactcag tatgctgctt
tctggatctc aatggcatat gttgtcgtta tttttgggct caaggctgtc
atgacaaacc gaaaaccatt tgatctcacg ggaccactga atctctggaa
tgcgggtctt gctattttct caactctcgg atcacttgcc actacatttg
gacttctcca cgagttcttc agccgtggat ttttcgaatc ttacattcac
atcggagact tttataatgg actttctgga atgttcacat ggcttttcgt
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
24
tctctcaaaa gttgctgaat tcggagatac actttttatt attcttcgta
aaaagccatt gatgttcctt cattggtatc atcatgtgct tacaatgaat
tatgctttta tgtcatttga agctaatttg ggatttaata cttggattac
atggatgaat ttctcagttc actcaattat gtatggatat tatatgcttc
gttcttttgg tgtcaaggtt ccagcatgga ttgccaagaa tattacaaca
atgcaaattc ttcaattcgt tattactcat ttcattcttt tccacgttgg
atatttggca gttactggac aatctgttga ctcaactcca ggatattatt
ggttctgcct tctcatggaa atctcttatg tcgttctgtt cggaaacttc
tactatcaat catacatcaa gggaggtggc aagaagttta atgcagagaa
gaagactgaa aagaaaattg aataa
SEQ ID6
F56H11.3
atgtatttgaattatttcgcgacggaaatcttccatcgtagtgcggtttgtgaaacagaa
gcttgtcgctcgtcaaaaataatgattgctgacgtgttcaaatggaaattcgatgcaaac
gaattgtggagtcttttaacgaatcaggatgaagttttcccgcatattagagcacggcga
ttcattcaagaacattttggtctattcgtccagatggcaattgcatatgtcattttggtg
ttctcaatcaaaaggttcatgagggatcgtgaaccatttcaactcaccacagctcttcgt
ctctggaacttcttcctctccgtcttctcaatttatggttcctggacaatgtttccattt
atggttcaacaaataagactttatggtctctacggatgtggatgcgaagcactttcaaac
cttccgagtcaagcagaatattggcttttcctgacgatcttgtccaaagctgtggagttt
gttgatacatttttcttggttctccggaaaaaaccactcatcttcctacactggtatcat
catatggcaacatttgtcttcttctgcagtaattacccgactccatcgtcacaatcacgc
gtcggagttatcgtcaacctgttcgtgcatgccttcatgtacccatactatttcacccga
tcaatgaacatcaaagttcctgcgaaaatttcaatggctgttacagttcttcaattgact
caattcatgtgctttatctatggatgtactctcatgtactactcgttggccactaatcag
gcacgatacccctcaaatacacctgcgacactccaatgtttgtcctacactctacatttg
ctttga
SEQ ID7
F56H11.4 (Ce 166)
atg gctcagcatc cgctcgttca acggcttctc gatgtcaaat tcgacacgaa
acgatttgtg gctattgcta ctcatgggcc aaagaatttc cctgacgcaga
aggtcgcaa gttctttgct gatcactttg atgttactat tcaggcttcaa
tcctgtacat
ggtcgttgtg ttcggaacaa aatggttcat gcgtaatcgt caaccattcc
aattgactat tccactcaac atctggaatttcatcctcgc cgcattttcc
atcgcaggag ctgtcaaaat gaccccagag ttctttggaa ccattgccaa
caaaggaatt gtcgatcctactgc aaagtgtttg atttcacgaa aggagagaat
ggatactgggt gtggctctt catggcttcc aaacttttcg aacttgttga
caccatcttc ttggttctccgtaaacgtcc actcatgttc cttcactggt
atcaccatat tctcaccatg atctacgcctggtactctca tccattgacc
ccaggattca acagatacgg aatttatctt aactttgtcg
tccacgcctt catgtactct tactacttcc ttcgctcgat gaagattcgc gtgc
caggattcatcgccca agctatcaca tctcttcaaa tcgttcaatt catcatctc
CA 02365096 2001-09-14
WO 00155330 PCTlGB00I01035
t tgcgccgttcttgctcatct tggttatctc atgcacttca ccaat
gccaactgt gatttcgagc catcagtatt caagctcgca
gttttcatgg acacaacata cttggctctt ttcgtcaact tcttcctcca
atcatatgtt
ctccgcggag gaaaagacaa gtacaaggca gtgccaaaga agaagaacaa ctaa
SEQ ID8
Y53F4B.c
atgtcggccg aagtgtccga acgattcaaa gtttggacag gaaacaatga
gaccatcatc tattccccat tcgagtacga ttccacgttg ctcatcgagt
catgtcggtg tacttatcag ctgcttatat tattgcgaca aatttattac
agagatatat ggagtcacgg aaacctaaaa cttttactag catggaacgg
ttttttggca gtgttcagta ttatgggtac atggagattt ggaatcgaat
tctacgatgc tgttttcaga agaggcttca tcgattcgat ctgcctggct
gtaaatccac gttcaccgtc cgcattctgg gcatgcatgt tcgctctatc
gaaaatcgcc gagtttgggg acacgatgtt cttggtgctg aggaaacggc
cggttatatt ccttcactgg tatcatcacg ctgttgttct gatcctttct
tggcatgctg caatcgaact cacagctcca ggacgctggt ttatttttat
gaactatttg gtgcattcaa taatgtatac atactacgca ataacatcaa
tcggctatcg tcttcccaaa atcgtttcaa tgactgttac attccttcaa
actcttcaaa tgctcattgg tgtcagcatt tcttgcattg tgctttattt
gaagcttaat ggagagatgt gccaacaatc ctacgacaat ctggcgttga
gcttcggaat ctacgcctca ttcctggtgc tattctccag tttcttcaac
aatgcatatt tggtaaaaaa ggacaagaaa cccgatgtga agaaggatta
a
SEQ ID9
A
1 MELAEFWNDL NTFTIYGPNH TDMTTKYKYS YHFPGEQVAD PQYWTILFQK
51 YWYHSITISV LYFILIKVIQ KFMENRKPFT LKYPLILWNG ALAAFSIIAT
101 LRFSIDPLRS LYAEGFYKTL CYSCNPTDVA AFWSFAFALS KIVELGDTMF
151 IILRKRPLIF LHYYHHAAVL IYTVHSGAEH TAAGRFYILM NYFAHSLMYT
201 YYTVSAMGYR LPKWVSMTVT TVQTTQMLAG VGITWMVYKV KTEYKLPCQQ
251 SVANLYLAFV IYVTFAILFI QFFVKAYIIK SSKKSKSVKN E*
SEQ ID10
B
1 MAKYDYNPKY GLENYSIFLP FETSFDAFRS TTWMQNHWYQ SITASVVYVA
51 VIFTGKKVVL IYKKSRVITF ESSLQNAIKN RNRKSLNSSQ MFQIMEKYKP
101 FQLDTPLFVW NSFLAIFSIL GFLRMTPEFV WSWSAEGNSF KYSICHSSYA
151 QGVTGFWTEQ FAMSKLFELI DTIFIVLRKR PLIFLHWYHH VTVMIYTWHA
201 YKDHTASGRW FIWMNYGVHA LMYSYYALRS LKFRLPKQMA MWTTLQLAQ
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00J01035
26
251 MVMGVIIGVT VYRTKSSGEY CQQTWDNLGL CFGVYFTYFL LFANFFYHAY
301 VKKNNRTVNY ENNSKNFPDL VLIYLRKKVS RKSKNRQCSE NNYKIQFSSN
351 FVNVDGKKHK KTYELILPRR KMTTILTFLF GKNRIFSKYQ KNRKNISIPV
401 DFEILEPKED INANIAEPSI TTRSAAARRK VQKAD*
SEQ ID11
C
1 MAAAQTSPAA TLVDVLTKPW SLDQTDSYMS TFVPLSYKIM IGYLVTIYFG
51 QKLMAHRKPF DLQNTLALWN FGFSLFSGIA AYKLIPELFG VFMKDGFVAS
101 YCQNENYYTD ASTGFWGWAF VMSKAPELGD TMFLVLRKKP VIFMHWYHHA
151 LTFWAWTY SEHQAWARWS LALNLAVHTV MYFYFAVRAL NIQTPRPVAK
201 FITTIQIVQF VISCYIFGHL VFIKSADSVP GCAVSWNVLS IGGLMYISYL
251 FLFAKFFYKA YIQKRSPTKT SK E*
SEQ ID12
D
1 MSSDDRGTRT FKMMDQILGT NFTYEGAKEV ARGLEGFSAK LAVGYIATIF
51 GLKYYMKDRK AFDLSTPLNI WNGILSTFSL LGFLFTFPTL LSVIRKDGFS
101 HTYSHVSELY TDSTSGYWIF LWVISKIPEL LDTVFIVLRK RPLIFMHWYH
151 HALTGYYALV CYHEDAVHMV WWWMNYIIH AFMYGYYLLK SLKVPIPPSV
201 AQAITTSQMV QFAVAIFAQV HVSYKHYVEG VEGLAYSFRG TAIGFFMLTT
251 YFYLWIQFYK EHYLKNGGKK YNLAKDQAKT QTKKAN*
SEQ ID13
E
MPQGEVSFFE VLTTAPFSHE LSKKHIAQTQ YAAFWISMAY VWIFGLKAV
MTNRKPFDLT GPLNLWNAGL AIFSTLGSLA TTFGLLHEFF SRGFFESYIH
IGDFYNGLSG MFTWLFVLSK VAEFGDTLFI ILRKKPLMFL HWYHHVLTMN
YAFMSFEANL GFNTWITWMN FSVHSIMYGY YMLRSFGVKV PAWIAKNITT
MQILQFVITH FILFHVGYLA VTGQSVDSTP GYYWFCLLME ISYWLFGNF
YYQSYIKGGG KKFNAEKKTE KKIE*
SEQ ID14
F
1 MYLNYFATEI FHRSAVCETE ACRSSKIMIA DVFKWKFDAN ELWSLLTNQD
51 EVFPHIRARR FIQEHFGLFV QMAIAYVILV FSIKRFMRDR EPFQLTTALR
101 LWNFFLSVFS IYGSWTMFPF MVQQIRLYGL YGCGCEALSN LPSQAEYWLF
CA 02365096 2001-09-14
WO 00/55330 PCT/GB00/01035
27
151 LTILSKAVEF VDTFFLVLRK KPLIFLHWYH HMATFVFFCS NYPTPSSQSR
201 VGVIVNLFVH AFMYPYYFTR SMNIKVPAKI SMAVTVLQLT QFMCFIYGCT
251 LMYYSLATNQ ARYPSNTPAT LQCLSYTLHL L*
SEQ ID15
G
MAQHPLVQRL LDVKFDTKRF VAIATHGPKN FPDAEGRKFF ADHFDVTIQA
SILYMVVVFG TKWFMRNRQP FQLTIPLNIW NFILAAFSIA GAVKMTPEFF
GTIANKGIVA SYCKVFDFTK GENGYWVWLF MASKLFELVD TIFLVLRKRP
LMFLHV~IYHHI LTMIYAWYSH PLTPGFNRYG IYLNFVVHAF MYSYYFLRSM
KIRVPGFIAQ AITSLQIVQF IISCAVLAHL GYLMHFTNAN CDFEPSVFKL
AVFMDTTYLA LFVNFFLQSY VLRGGKDKYK AVPKKKNN*
SEQ ID16
H
MSAEVSERFKVWTGNNETIIYSPFEYDSTLLIESCRCTYQLLILLRQI
YYRDIWSHGNLKACDXLLLAWNGFLAVFSIMGTWRFGIEFYDAVFRXG
FIXSICLAVNPRSPSAFWACMFALSKIAEFGDTMFLVLRKRPVIFLHWYHH
AVVLILSWHAAIELTAPGRWFIFMNYLVHSIMYTYYAITSIGYRXPKIVSMT
VTFLQTLQMLIGVSISCIVLYLKLNGEMCQQSYDNLALSFGIYASFLVLSSFF
NNAYLVKKDKKPDVKKD*
CA 02365096 2001-09-14
ELONGASE.APP.txt
SEQUENCE LISTING
<110> the university of Bristol
<120> POLYUNSATURATED FATTY ACID (PUFA) ELONGASE FROM CAENORHABDITIS ELEGANS
<130> >1470003
<140> PCT/GB00/01035
<141> March 20, 2000
<160> 22
<170> Patentln Ver. 2.1
<210> 1
<211> 27
<212> DNA
<213> C. elegans
<400> 1
gcgggtacca tggctcagca tccgctc 27
<210> 2
<211> 27
<212> DNA
<213> C. elegans
<400> 2
gcgggatcct tagttgttct tcttctt 27
<210> 3
<211> 27
<212> DNA
<213> C. elegans
<400> 3
gcgggtacca tgccacaggg agaagtc 27
<210> 4
<211> 27
<212> DNA
<213> c. elegans
<400> 4
gcgggatcct tattcaattt ttctttt 27
<210> 5
<211> 29
<212> DNA
<213> C. elegans
<400> 5
gcgaattcac catgggtacg gaccaagga 2g
<210> 6
<211> 27
<212> DNA
Page 1
CA 02365096 2001-09-14
ELONGASE.APP.tXt
<213> C. ~legans
<400> 6
gcggagctcc tactcttcct tgggacg 27
<210> 7
<211> 876
<212> DNA
<213> c. elegans
<400> 7
atggagcttg ccgagttctg gaatgatctc aacaccttca ccatctacgg accgaatcac 60
acagatatga ccacaaaata caaatattca tatcacttcc caggtgaaca ggtggcggat 120
ccgcagtatt ggacgatttt attccagaaa tattggtatc attcgatcac aatatcagtt 180
ctttatttca ttttaattaa ggtgattcaa aagtttatgg agaatcgaaa accattcact 240
ttgaaatacc cattgattct ttggaatgga gctcttgcag cattcagtat aattgccaca 300
ttgcggttct ctattgatcc tctacgatca ctatatgctg aaggattcta caaaactctg 360
tgctattcgt gtaatccaac tgatgtggct gcattttgga gctttgcatt cgctctttcc 420
aagattgttg aacttggaga cactatgttc attattttga gaaaacggcc attgatcttt 480
ttacactact atcatcatgc agcagtgtta atctacactg tccattctgg tgccgagcat 540
actgcagctg gtcgtttcta catcctaatg aactacttcg cacattctct catgtatact 600
tactacacag tttctgccat gggatacaga ttaccgaaat gggtatcaat gactgtcaca 660
actgttcaaa caactcaaat gttagctgga gtcggaataa cttggatggt gtacaaagtg 720
aaaactgaat acaagcttcc ttgtcaacaa tccgtagcca atttgtatct cgcattcgtc 780
atctatgtca catttgccat tcttttcatt caattcttcg tcaaggcata cattatcaag 840
tcgtcgaaga agtcgaaatc ggtgaagaac gaataa
876
<210> 8
<211> 1308
<212> DNA
<213> c. elegans
<400> 8
atggcaaaat acgactacaa tccgaagtat gggttagaaa attacagcat attccttccc 60
tttgagacat cttttgatgc atttcgatcg acaacatgga tgcaaaatca ctggtatcaa 120
tcaattacag catctgtcgt gtatgtagcc gtcattttta caggaaagaa ggtggttctc 180
atctacaaaa aatcacgagt tattactttt gagtctagcc ttcagaatgc aattaagaat 240
cgaaaccgaa aatcacttaa tagttctcaa atgtttcaga ttatggaaaa gtacaagccc 300
ttccaactgg acacaccact cttcgtctgg aattcatttt tagccatttt ctcaattctc 360
gggttcctcc gaatgacacc tgaatttgta tggagttggt cagcagaagg aaactcattc 420
aaatattcaa tttgtcattc atcttatgct caaggagtca ctggtttctg gactgaacaa 480
ttcgcaatga gcaaactttt cgagctcatc gacacaatct tcatcgttct tcgtaaacgt 540
ccactcatct tccttcactg gtatcatcat gtaactgtta tgatctacac atggcacgcg 600
tacaaggatc acactgcatc aggacggtgg ttcatttgga tgaattatgg agttcatgct 660
cttatgtatt cctactatgc tcttcgttct ctgaaattcc gtcttccaaa acaaatggca 720
atggttgtta ctactctcca acttgctcaa atggttatgg gagtaatcat cggagtcact 780
gtctaccgta tcaagtcatc gggtgaatac tgccaacaga catgggacaa tttgggatta 840
tgctttggag tttatttcac atatttcctt cttttcgcca acttcttcta ccatgcatat 900
gttaagaaaa acaaccgtac agtaaattat gaaaataatt caaaaaattt ccccgatctc 960
gttttaattt acctgagaaa aaaggtttca agaaaatcga aaaatcggca atgttcagaa 1020
aataattata aaattcaatt ttcatcaaat tttgttaatg ttgatggaaa aaaacataag 1080
aaaacatatg aacttattct tccaagaaga aaaatgacca caattttaac ttttctattt 1140
ggaaaaaatc gaattttttc gaaatatcag aaaaatcgaa aaaacatttc gattcctgtt 1200
gatttcgaaa ttctggagcc aaaagaagat atcaatgcta acatcgctga gccatccatc 1260
acaacgaggt ccgccgccgc acgaagaaaa gttcaaaaag ctgattag 1308
<210> 9
<211> 825
<212> DNA
<213> c. elegans
Page 2
CA 02365096 2001-09-14
ELONGASE.APP.txt
<400> 9
atggcagcag cacaaacaag tccagcagcc acgctcgtcg atgttttgac aaaaccatgg 60
agtctggatc agactgattc ttacatgtct acatttgtac cattatccta taaaatcatg 120
attggttatc tcgtcaccat ctacttcggg caaaaattaa tggctcacag aaaaccattc 180
gatctccaaa atacacttgc tctctggaac ttcgggtttt cactgttctc gggaatcgcc 240
gcctataagc ttattccaga actattcgga gttttcatga aggacgggtt tgtcgcttcc 300
tactgtcaaa acgagaacta ctacaccgat gcatcaactg gattctgggg ctgggccttt 360
gtgatgtcga aagctccaga actaggggat actatgttct tggtccttcg taaaaaacca 420
gttatcttca tgcactggta tcatcatgcc ctcacatttg tctacgcagt agtcacatac 480
tctgagcatc aggcatgggc tcgttggtct ttggctctca accttgccgt ccacactgtt 540
atgtatttct acttcgccgt tcgcgccttg aacatccaaa ctccacgccc agtggcaaag 600
ttcatcacta ctattcaaat tgtccaattt gtcatctcat gctacatttt tgggcatttg 660
gtattcatta agtctgctga ttctgttcct ggttgcgctg ttagctggaa tgtgctatcg 720
atcggaggac tcatgtacat cagttatttg ttcctttttg ccaagttctt ctacaaggcc 780
tacattcaaa aacgctcacc aaccaaaacc agcaagcagg agtag g25
<210> 10
<211> 861
<212> DNA
<213> C. elegans
<400> 10
atgtcatcgg acgatcgtgg cactagaacc ttcaagatga tggatcaaat tcttggaaca 60
aacttcactt atgaaggtgc caaagaagtt gctcgaggcc ttgaaggttt ctcagcaaag 120
cttgccgtcg gatatattgc cactattttt ggactgaaat attatatgaa agaccgaaaa 180
gccttcgatc tcagtactcc attaaacatt tggaatggta ttctttcgac attcagctta 240
ttgggattct tattcacttt tcctactttg ttatcagtta tcagaaagga tggatttagt 300
cacacctatt cccatgtctc tgagctttac actgacagta cctctggata ttggatcttc 360
ctttgggtta tctcaaagat tccggaactt ttggatacag tattcattgt tcttcgcaag 420
agaccactta ttttcatgca ctggtaccat cacgcattga ccggttacta tgctcttgtc 480
tgctaccatg aggatgctgt ccatatggtt tgggttgtat ggatgaatta tattattcat 540
gcattcatgt atggatacta tcttctgaaa tctctgaaag ttccaattcc accatcagtt 600
gctcaagcaa tcaccacatc tcaaatggtt caattcgcag ttgccatttt cgcacaagtt 660
catgtttcct ataaacacta tgttgaggga gttgaaggat tagcctactc gttcagagga 720
acagctatcg gatttttcat gcttactacc tacttctatc tatggattca attctacaaa 780
gagcactatc ttaagaatgg aggcaaaaag tacaatttgg caaaggatca ggcaaaaact 840
caaacaaaga aggctaacta a 861
<210> 11
<211> 825
<212> DNA
<213> C. elegans
<400> 11
atgccacagg gagaagtctc attctttgag gtgctgacaa ctgctccatt cagtcatgag 60
ctctcaaaaa agcatattgc acagactcag tatgctgctt tctggatctc aatggcatat 120
gttgtcgtta tttttgggct caaggctgtc atgacaaacc gaaaaccatt tgatctcacg 180
ggaccactga atctctggaa tgcgggtctt gctattttct caactctcgg atcacttgcc 240
actacatttg gacttctcca cgagttcttc agccgtggat ttttcgaatc ttacattcac 300
atcggagact tttataatgg actttctgga atgttcacat ggcttttcgt tctctcaaaa 360
gttgctgaat tcggagatac actttttatt attcttcgta aaaagccatt gatgttcctt 420
cattggtatc atcatgtgct tacaatgaat tatgctttta tgtcatttga agctaatttg 480
ggatttaata cttggattac atggatgaat ttctcagttc actcaattat gtatggatat 540
tatatgcttc gttcttttgg tgtcaaggtt ccagcatgga ttgccaagaa tattacaaca 600
atgcaaattc ttcaattcgt tattactcat ttcattcttt tccacgttgg atatttggca 660
gttactggac aatctgttga ctcaactcca ggatattatt ggttctgcct tctcatggaa 720
atctcttatg tcgttctgtt cggaaacttc tactatcaat catacatcaa gggaggtggc 780
aagaagttta atgcagagaa gaagactgaa aagaaaattg aataa 825
Page 3
CA 02365096 2001-09-14
ELONGASE.APP.tXt
<210> 12
<211> 846
<212> DNA
<213> C. elegans
<400> 12
atgtatttga attatttcgc gacggaaatc ttccatcgta gtgcggtttg tgaaacagaa 60
gcttgtcgct cgtcaaaaat aatgattgct gacgtgttca aatggaaatt cgatgcaaac 120
gaattgtgga gtcttttaac gaatcaggat gaagttttcc cgcatattag agcacggcga 180
ttcattcaag aacattttgg tctattcgtc cagatggcaa ttgcatatgt cattttggtg 240
ttctcaatca aaaggttcat gagggatcgt gaaccatttc aactcaccac agctcttcgt 300
ctctggaact tcttcctctc cgtcttctca atttatggtt cctggacaat gtttccattt 360
atggttcaac aaataagact ttatggtctc tacggatgtg gatgcgaagc actttcaaac 420
cttccgagtc aagcagaata ttggcttttc ctgacgatct tgtccaaagc tgtggagttt 480
gttgatacat ttttcttggt tctccggaaa aaaccactca tcttcctaca ctggtatcat 540
catatggcaa catttgtctt cttctgcagt aattacccga ctccatcgtc acaatcacgc 600
gtcggagtta tcgtcaacct gttcgtgcat gccttcatgt acccatacta tttcacccga 660
tcaatgaaca tcaaagttcc tgcgaaaatt tcaatggctg ttacagttct tcaattgact 720
caattcatgt gctttatcta tggatgtact ctcatgtact actcgttggc cactaatcag 780
gcacgatacc cctcaaatac acctgcgaca ctccaatgtt tgtcctacac tctacatttg 840
ctttga 846
<210> 13
<211> 866
<212> DNA
<213> C. elegans
<400> 13
atggctcagc atccgctcgt tcaacggctt ctcgatgtca aattcgacac gaaacgattt 60
gtggctattg ctactcatgg gccaaagaat ttccctgacg cagaaggtcg caagttcttt 120
gctgatcact ttgatgttac tattcaggct tcaatcctgt acatggtcgt tgtgttcgga 180
acaaaatggt tcatgcgtaa tcgtcaacca ttccaattga ctattccact caacatctgg 240
aatttcatcc tcgccgcatt ttccatcgca ggagctgtca aaatgacccc agagttcttt 300
ggaaccattg ccaacaaagg aattgtcgat cctactgcaa agtgtttgat ttcacgaaag 360
gagagaatgg atactgggtg tggctcttca tggcttccaa acttttcgaa cttgttgaca 420
ccatcttctt ggttctccgt aaacgtccac tcatgttcct tcactggtat caccatattc 480
tcaccatgat ctacgcctgg tactctcatc cattgacccc aggattcaac agatacggaa 540
tttatcttaa ctttgtcgtc cacgccttca tgtactctta ctacttcctt cgctcgatga 600
agattcgcgt gccaggattc atcgcccaag ctatcacatc tcttcaaatc gttcaattca 660
tcatctcttg cgccgttctt gctcatcttg gttatctcat gcacttcacc aatgccaact 720
gtgatttcga gccatcagta ttcaagctcg cagttttcat ggacacaaca tacttggctc 780
ttttcgtcaa cttcttcctc caatcatatg ttctccgcgg aggaaaagac aagtacaagg 840
cagtgccaaa gaagaagaac aactaa 866
<210> 14
<211> 801
<212> DNA
<213> C. elegans
<400> 14
atgtcggccg aagtgtccga acgattcaaa gtttggacag gaaacaatga gaccatcatc 60
tattccccat tcgagtacga ttccacgttg ctcatcgagt catgtcggtg tacttatcag 120
ctgcttatat tattgcgaca aatttattac agagatatat ggagtcacgg aaacctaaaa 180
cttttactag catggaacgg ttttttggca gtgttcagta ttatgggtac atggagattt 240
ggaatcgaat tctacgatgc tgttttcaga agaggcttca tcgattcgat ctgcctggct 300
gtaaatccac gttcaccgtc cgcattctgg gcatgcatgt tcgctctatc gaaaatcgcc 360
gagtttgggg acacgatgtt cttggtgctg aggaaacggc cggttatatt ccttcactgg 420
tatcatcacg ctgttgttct gatcctttct tggcatgctg caatcgaact cacagctcca 480
ggacgctggt ttatttttat gaactatttg gtgcattcaa taatgtatac atactacgca 540
ataacatcaa tcggctatcg tcttcccaaa atcgtttcaa tgactgttac attccttcaa 600
actcttcaaa tgctcattgg tgtcagcatt tcttgcattg tgctttattt gaagcttaat 660
Page 4
CA 02365096 2001-09-14
ELONGASE.APP.tXt
ggagagatgt gccaacaatc ctacgacaat ctggcgttga gcttcggaat ctacgcctca 720
ttcctggtgc tattctccag tttcttcaac aatgcatatt tggtaaaaaa ggacaagaaa 780
cccgatgtga agaaggatta a
801
<210>
15
<211>
291
<212>
PRT
<213> elegans
C.
<400>
15
MetGlu LeuAla GluPheTrpAsn AspLeuAsn ThrPheThr IleTyr
1 5 10 15
GlyPro AsnHis ThrAspMetThr ThrLysTyr LysTyrSer TyrHis
20 25 30
PhePro GlyGlu GlnValAlaAsp ProGlnTyr TrpThrIle LeuPhe
35 40 45
GlnLys TyrTrp TyrHisSerIle ThrIleSer ValLeuTyr PheIle
50 55 60
LeuIle LysVal IleGlnLysPhe MetGluAsn ArgLysPro PheThr
65 70 75 80
LeuLys TyrPro LeuIleLeuTrp AsnGlyAla LeuAlaAla PheSer
85 90 95
IleIle AlaThr LeuArgPheSer IleAspPro LeuArgSer LeuTyr
100 105 110
AlaGlu GlyPhe TyrLysThrLeu CysTyrSer CysAsnPro ThrAsp
115 120 125
ValAla AlaPhe TrpSerPheAla PheAlaLeu SerLysIle ValGlu
130 135 140
LeuGly AspThr MetPheIleIle LeuArgLys ArgProLeu IlePhe
145 150 155 160
LeuHis TyrTyr HisHisAlaAla ValLeuIle TyrThrVal HisSer
165 170 175
GlyAla GluHis ThrAlaAlaGly ArgPheTyr IleLeuMet AsnTyr
180 185 190
PheAla HisSer LeuMetTyrThr TyrTyrThr ValSerAla MetGly
195 200 205
TyrArg LeuPro LysTrpValSer MetThrVal ThrThrVal GlnThr
210 215 220
ThrGln MetLeu AlaGlyValGly IleThrTrp MetValTyr LysVal
225 230 235 240
LysThr GluTyr LysLeuProCys GlnGlnSer ValAlaAsn LeuTyr
245 250 255
LeuAla PheVal IleTyrValThr PheAlaIle LeuPheIle GlnPhe
260 265 270
PheVal LysAla TyrIleIleLys SerSerLys LysSerLys SerVal
Page 5
CA 02365096 2001-09-14
ELONGASE.APP.tXt
' 2?5 280 285
Lys Asn Glu
290
<210>
16
<211>
435
<212>
PRT
<213> elegan s
c.
<400>
16
MetAla LysTyrAsp TyrAsnPro LysTyrGly LeuGluAsn TyrSer
1 5 10 15
IlePhe LeuProPhe GluThrSer PheAspAla PheArgSer ThrThr
20 25 30
TrpMet GlnAsnHis TrpTyrGln SerIleThr AlaSerVal ValTyr
35 40 45
ValAla ValIlePhe ThrGlyLys LysValVal LeuIleTyr LysLys
50 55 60
SerArg ValIleThr PheGluSer SerLeuGln AsnAlaIle LysAsn
65 70 75 80
ArgAsn ArgLysSer LeuAsnSer SerGlnMet PheGlnIle MetGlu
85 90 95
LysTyr LysProPhe GlnLeuAsp ThrProLeu PheValTrp AsnSer
100 105 110
PheLeu AlaIlePhe SerIleLeu GlyPheLeu ArgMetThr ProGlu
115 120 125
PheVal TrpSerTrp SerAlaGlu GlyAsnSer PheLysTyr SerIle
130 135 140
cysHis SerSerTyr AlaGlnGly ValThrGly PheTrpThr GluGln
145 150 155 160
PheAla MetSerLys LeuPheGlu LeuIleAsp ThrIlePhe IleVal
165 170 175
LeuArg LysArgPro LeuIlePhe LeuHisTrp TyrHisHis ValThr
180 185 190
ValMet IleTyrThr TrpHisAla TyrLysAsp HisThrAla SerGly
195 200 205
ArgTrp PheIleTrp MetAsnTyr GlyValHis AlaLeuMet TyrSer
21 0 215 220
TyrTyr AlaLeuArg SerLeuLys PheArgLeu ProLysGln MetAla
225 230 235 240
MetVal ValThrThr LeuGlnLeu AlaGlnMet ValMetGly ValIle
245 250 255
IleGly ValThrVal TyrArgIle LysSerSer GlyGluTyr cysGln
260 265 270
Page 6
CA 02365096 2001-09-14
S
ELONGASE.APP.tXt
Gln Thr Trp Asp Asn Leu Gly Leu Cys Phe Gly Val Tyr Phe Thr Tyr
275 280 285
Phe Leu Leu Phe Ala Asn Phe Phe Tyr His Ala Tyr Val Lys Lys Asn
290 295 300
Asn Arg Thr Val Asn Tyr Glu Asn Asn Ser Lys Asn Phe Pro Asp Leu
305 310 315 320
Val Leu Ile Tyr Leu Arg Lys Lys Val Ser Arg Lys Ser Lys Asn Arg
325 330 335
Gln Cys Ser Glu Asn Asn Tyr Lys Ile Gln Phe Ser Ser Asn Phe Val
340 345 350
Asn Val Asp Gly Lys Lys His Lys Lys Thr Tyr Glu Leu Ile Leu Pro
355 360 365
Arg Arg Lys Met Thr Thr Ile Leu Thr Phe Leu Phe Gly Lys Asn Arg
370 375 380
Ile Phe Ser Lys Tyr Gln Ly5 Asn Arg Lys Asn Ile Ser Ile Pro Val
385 390 395 400
Asp Phe Glu Ile Leu Glu Pro Lys Glu Asp Ile Asn Ala Asn Ile Ala
405 410 415
Glu Pro Ser Ile Thr Thr Arg Ser Ala Ala Ala Arg Arg Lys Val Gln
420 425 430
Lys Ala Asp
435
<210> 17
<211> 274
<212> PRT
<213> C. elegans
<400> 17
Met Ala Ala Ala Gln Thr Ser Pro Ala Ala Thr Leu Val Asp Val Leu
1 5 10 15
Thr Lys Pro Trp Ser Leu Asp Gln Thr Asp Ser Tyr Met Ser Thr Phe
20 25 30
Val Pro Leu Ser Tyr Lys Ile Met Ile Gly Tyr Leu Val Thr Ile Tyr
35 40 45
Phe Gly Gln Lys Leu Met Ala His Arg Lys Pro Phe Asp Leu Gln Asn
50 55 60
Thr Leu Ala Leu Trp Asn Phe Gly Phe Ser Leu Phe Ser Gly Ile Ala
65 70 75 80
Ala Tyr Lys Leu Ile Pro Glu Leu Phe Gly Val Phe Met Lys Asp Gly
85 90 95
Phe Val Ala Ser Tyr Cys Gln Asn Glu Asn Tyr Tyr Thr Asp Ala Ser
100 105 110
Page 7
CA 02365096 2001-09-14
ELONGASE.APP.txt
Thr Gly Phe Trp Gly Trp Ala Phe Val Met Ser Lys Ala Pro Glu Leu
115 120 125
Gly Asp Thr Met Phe Leu Val Leu Arg Lys Lys Pro Val Ile Phe Met
130 135 140
His Trp Tyr His His Ala Leu Thr Phe Val Tyr Ala Val Val Thr Tyr
145 150 155 160
Ser Glu His Gln Ala Trp Ala Arg Trp Ser Leu Ala Leu Asn Leu Ala
165 170 175
Val His Thr Val Met Tyr Phe Tyr Phe Ala Val Arg Ala Leu Asn Ile
180 185 190
Gln Thr Pro Arg Pro Val Ala Lys Phe Ile Thr Thr Ile Gln Ile Val
195 200 205
Gln Phe Val Ile Ser Cys Tyr Ile Phe Gly His Leu Val Phe Ile Lys
210 215 220
Ser Ala Asp Ser Val Pro Gly Cys Ala Val Ser Trp Asn Val Leu Ser
225 230 235 240
Ile Gly Gly Leu Met Tyr Ile Ser Tyr Leu Phe Leu Phe Ala Lys Phe
245 250 255
Phe Tyr Lys Ala Tyr Ile Gln Lys Arg Ser Pro Thr Lys Thr Ser Lys
260 265 270
Gln Glu
<210> 18
<211> 286
<212> PRT
<213> C. elegans
<400> 18
Met Ser Ser Asp Asp Arg Gly Thr Arg Thr Phe Lys Met Met Asp Gln
1 5 10 15
Ile Leu Gly Thr Asn Phe Thr Tyr Glu Gly Ala Lys Glu Val Ala Arg
20 25 30
Gly Leu Glu Gly Phe Ser Ala Lys Leu Ala Val Gly Tyr Ile Ala Thr
35 40 45
Ile Phe Gly Leu Lys Tyr Tyr Met Lys Asp Arg Lys Ala Phe Asp Leu
50 55 60
Ser Thr Pro Leu Asn Ile Trp Asn Gly Ile Leu Ser Thr Phe Ser Leu
65 70 75 80
Leu Gly Phe Leu Phe Thr Phe Pro Thr Leu Leu Ser Val Ile Arg Lys
85 90 95
Asp Gly Phe Ser His Thr Tyr Ser His Val Ser Glu Leu Tyr Thr Asp
100 105 110
Ser Thr Ser Gly Tyr Trp Ile Phe Leu Trp Val Ile Ser Lys Ile Pro
Page 8
CA 02365096 2001-09-14
ELONGASE.APP.tXt
' lI5 120 125
Glu Leu Leu Asp Thr Val Phe Ile Val Leu Arg Lys Arg Pro Leu Ile
130 135 140
Phe Met His Trp Tyr His His Ala Leu Thr Gly Tyr Tyr Ala Leu Val
145 150 155 160
Cys Tyr His Glu Asp Ala Val His Met Val Trp Val Val Trp Met Asn
165 170 175
Tyr Ile Ile His Ala Phe Met Tyr Gly Tyr Tyr Leu Leu Lys Ser Leu
180 185 190
Lys Val Pro Ile Pro Pro Ser Val Ala Gln Ala Ile Thr Thr Ser Gln
195 200 205
Met Val Gln Phe Ala Val Ala Ile Phe Ala Gln Val His Val Ser Tyr
210 215 220
Lys His Tyr Val Glu Gly Val Glu Gly Leu Ala Tyr Ser Phe Arg Gly
225 230 235 240
Thr Ala Ile Gly Phe Phe Met Leu Thr Thr Tyr Phe Tyr Leu Trp Ile
245 250 255
Gln Phe Tyr Lys Glu His Tyr Leu Lys Asn Gly Gly Lys Lys Tyr Asn
260 265 270
Leu Ala Lys Asp Gln Ala Lys Thr Gln Thr Lys Lys Ala Asn
275 280 285
<210> 19
<211> 274
<212> PRT
<213> C. elegans
<400> 19
Met Pro Gln Gly Glu Val Ser Phe Phe Glu Val Leu Thr Thr Ala Pro
1 5 10 15
Phe Ser His Glu Leu Ser Lys Lys His Ile Ala Gln Thr Gln Tyr Ala
20 25 30
Ala Phe Trp Ile Ser Met Ala Tyr Val Val Val Ile Phe Gly Leu Lys
35 40 45
Ala Val Met Thr Asn Arg Lys Pro Phe Asp Leu Thr Gly Pro Leu Asn
50 55 60
Leu Trp Asn Ala Gly Leu Ala Ile Phe Ser Thr Leu Gly Ser Leu Ala
65 70 75 80
Thr Thr Phe Gly Leu Leu His Glu Phe Phe Ser Arg Gly Phe Phe Glu
85 90 95
Ser Tyr Ile His Ile Gly Asp Phe Tyr Asn Gly Leu Ser Gly Met Phe
100 105 110
Thr Trp Leu Phe Val Leu Ser Lys Val Ala Glu Phe Gly Asp Thr Leu
115 120 125
Page 9
CA 02365096 2001-09-14
ELONGASE.APP.tXt
Phe Ile Ile Leu Arg Lys Lys Pro Leu Met Phe Leu His Trp Tyr His
130 135 140
His Val Leu Thr Met Asn Tyr Ala Phe Met Ser Phe Glu Ala Asn Leu
145 150 155 160
Gly Phe Asn Thr Trp Ile Thr Trp Met Asn Phe Ser Val His Ser Ile
165 170 175
Met Tyr Gly Tyr Tyr Met Leu Arg Ser Phe Gly Val Lys Val Pro Ala
180 185 190
Trp Ile Ala Lys Asn Ile Thr Thr Met Gln Ile Leu Gln Phe Val Ile
195 200 205
Thr His Phe Ile Leu Phe His Val Gly Tyr Leu Ala Val Thr Gly Gln
210 215 220
Ser Val Asp Ser Thr Pro Gly Tyr Tyr Trp Phe Cys Leu Leu Met Glu
225 230 235 240
Ile Ser Tyr Val Val Leu Phe Gly Asn Phe Tyr Tyr Gln Ser Tyr Ile
245 250 255
Lys Gly Gly Gly Lys Lys Phe Asn Ala Glu Lys Lys Thr Glu Lys Lys
260 265 270
Ile Glu
<210> 20
<211> 281
<212> PRT
<213> C. elegans
<400> 20
Met Tyr Leu Asn Tyr Phe Ala Thr Glu Ile Phe His Arg Ser Ala Val
1 5 10 15
Cys Glu Thr Glu Ala Cys Arg Ser Ser Lys Ile Met Ile Ala Asp Val
20 25 30
Phe Lys Trp Lys Phe Asp Ala Asn Glu Leu Trp Ser Leu Leu Thr Asn
35 40 45
Gln Asp Glu Val Phe Pro His Ile Arg Ala Arg Arg Phe Ile Gln Glu
50 55 60
His Phe Gly Leu Phe Val Gln Met Ala Ile Ala Tyr Val Ile Leu Val
65 70 75 80
Phe Ser Ile Lys Arg Phe Met Arg Asp Arg Glu Pro Phe Gln Leu Thr
85 90 95
Thr Ala Leu Arg Leu Trp Asn Phe Phe Leu Ser Val Phe Ser Ile Tyr
100 105 110
Gly Ser Trp Thr Met Phe Pro Phe Met Val Gln Gln Ile Arg Leu Tyr
115 120 125
Page 10
CA 02365096 2001-09-14
ELONGASE.APP.tXt
Gly Leu Tyr Gly Cys Gly Cys Glu Ala Leu Ser Asn Leu Pro Ser Gln
130 135 140
Ala Glu Tyr Trp Leu Phe Leu Thr Ile Leu Ser Lys Ala Val Glu Phe
145 150 155 160
Val Asp Thr Phe Phe Leu Val Leu Arg Lys Lys Pro Leu Ile Phe Leu
165 170 175
His Trp Tyr His His Met Ala Thr Phe Val Phe Phe Cys Ser Asn Tyr
180 185 190
Pro Thr Pro Ser Ser Gln Ser Arg Val Gly Val Ile Val Asn Leu Phe
195 200 205
Val His Ala Phe Met Tyr Pro Tyr Tyr Phe Thr Arg Ser Met Asn Ile
210 215 220
Lys Val Pro Ala Lys Ile Ser Met Ala Val Thr Val Leu Gln Leu Thr
225 230 235 240
Gln Phe Met Cys Phe Ile Tyr Gly Cys Thr Leu Met Tyr Tyr Ser Leu
245 250 255
Ala Thr Asn Gln Ala Arg Tyr Pro Ser Asn Thr Pro Ala Thr Leu Gln
260 265 270
Cys Leu Ser Tyr Thr Leu His Leu Leu
275 280
<210> 21
<211> 288
<212> PRT
<213> C. elegans
<400> 21
Met Ala Gln His Pro Leu Val Gln Arg Leu Leu Asp Val Lys Phe Asp
1 5 10 15
Thr Lys Arg Phe Val Ala Ile Ala Thr His Gly Pro Lys Asn Phe Pro
20 25 30
Asp Ala Glu Gly Arg Lys Phe Phe Ala Asp His Phe Asp Val Thr Ile
35 40 45
Gln Ala Ser Ile Leu Tyr Met Val Val Val Phe Gly Thr Lys Trp Phe
50 55 60
Met Arg Asn Arg Gln Pro Phe Gln Leu Thr Ile Pro Leu Asn Ile Trp
65 70 75 80
Asn Phe Ile Leu Ala Ala Phe Ser Ile Ala Gly Ala Val Lys Met Thr
85 90 95
Pro Glu Phe Phe Gly Thr Ile Ala Asn Lys Gly Ile Val Ala Ser Tyr
100 105 110
Cys Lys Val Phe Asp Phe Thr Lys Gly Glu Asn Gly Tyr Trp Val Trp
115 120 125
Leu Phe Met Ala Ser Lys Leu Phe Glu Leu Val Asp Thr Ile Phe Leu
Page 11
CA 02365096 2001-09-14
ELONGASE.APP.tXt
130 ' 135 140
Val Leu Arg Lys Arg Pro Leu Met Phe Leu His Trp Tyr His His Ile
145 150 155 160
Leu Thr Met Ile Tyr Ala Trp Tyr Ser His Pro Leu Thr Pro Gly Phe
165 170 175
Asn Arg Tyr Gly Ile Tyr Leu Asn Phe Val Val His Ala Phe Met Tyr
180 185 190
Ser Tyr Tyr Phe Leu Arg Ser Met Lys Ile Arg Val Pro Gly Phe Ile
195 200 205
Ala Gln Ala Ile Thr Ser Leu Gln Ile Val Gln Phe Ile Ile Ser Cys
210 215 220
Ala Val Leu Ala His Leu Gly Tyr Leu Met His Phe Thr Asn Ala Asn
225 230 235 240
Cys Asp Phe Glu Pro Ser Val Phe Lys Leu Ala Val Phe Met Asp Thr
245 250 255
Thr Tyr Leu Ala Leu Phe Val Asn Phe Phe Leu Gln Ser Tyr Val Leu
260 265 270
Arg Gly Gly Lys Asp Lys Tyr Lys Ala Val Pro Lys Lys Lys Asn Asn
275 280 285
<210> 22
<211> 269
<212> PRT
<213> C. elegans
<400> 22
Met Ser Ala Glu Val Ser Glu Arg Phe Lys Val Trp Thr Gly Asn Asn
1 5 10 15
Glu Thr Ile Ile Tyr Ser Pro Phe Glu Tyr Asp Ser Thr Leu Leu Ile
20 25 30
Glu Ser Cys Arg Cys Thr Tyr Gln Leu Leu Ile Leu Leu Arg Gln Ile
35 40 45
Tyr Tyr Arg Asp Ile Trp Ser His Gly Asn Leu Lys Ala Cys Asp Xaa
50 55 60
Leu Leu Leu Ala Trp Asn Gly Phe Leu Ala Val Phe Ser Ile Met Gly
65 70 75 80
Thr Trp Arg Phe Gly Ile Glu Phe Tyr Asp Ala Val Phe Arg Xaa Gly
85 90 95
Phe Ile Xaa Ser Ile Cys Leu Ala Val Asn Pro Arg Ser Pro Ser Ala
100 105 110
Phe Trp Ala Cys Met Phe Ala Leu Ser Lys Ile Ala Glu Phe Gly Asp
115 120 125
Page 12
CA 02365096 2001-09-14
ELONGASE.APP.tXt
Thr Met PheLeu ValLeuArg LysArgProVal IlePheLeu HisTrp
130 135 140
Tyr His HisAla ValValLeu IleLeuSerTrp HisAlaAla IleGlu
145 150 155 160
Leu Thr AlaPro GlyArgTrp PheIlePheMet AsnTyrLeu ValHis
165 170 175
Ser Ile MetTyr ThrTyrTyr AlaIleThrSer IleGlyTyr ArgXaa
180 185 190
Pro Lys IleVal SerMetThr ValThrPheLeu GlnThrLeu GlnMet
195 200 205
Leu Ile GlyVal SerIleSer CysIleValLeu TyrLeuLys LeuAsn
210 215 220
Gly Glu MetCys GlnGlnSer TyrAspAsnLeu AlaLeuSer PheGly
225 230 235 240
Ile Tyr AlaSer PheLeuVal LeuSerSerPhe PheAsnAsn AlaTyr
245 250 255
Leu Val LysLys AspLysLys ProAspValLys LysAsp
260 265
Page 13
CA 02365096 2001-09-14
gcgggtacca~ggctcagcatccgctco
PRIMER1.GBS.tXt
Page 1
CA 02365096 2001-09-14
PRIMER2.GBS.tXt
gcgggatcct~agttgttcttcttctto
Page 1
CA 02365096 2001-09-14
PRIMER3.GBS.tXt
gcgggtaccatgccacagggagaagtco
Page 1
CA 02365096 2001-09-14
PRIMER4.GBS.tXt
gcgggatccttattcaatttttctttto
Page 1
CA 02365096 2001-09-14
PRIMERS.GBS.tXt
gcgaattcaccatgggtacggaccaagga~
Page 1
CA 02365096 2001-09-14
PRIMER6.GBS.tXt
gcggagctcctactcttccttgggacgo
Page 1
CA 02365096 2001-09-14
SEQID1.GBS.tXt
atggagcttgccgagttctggaatgatctcaacaccttcaccatctacggaccgaatcac
acagatatgaccacaaaatacaaatattcatatcacttcccaggtgaacaggtggcggat
ccgcagtattggacgattttattccagaaatattggtatcattcgatcacaatatcagtt
ctttatttcattttaattaaggtgattcaaaagtttatggagaatcgaaaaccattcact
ttgaaatacccattgattctttggaatggagctcttgcagcattcagtataattgccaca
ttgcggttctctattgatcctctacgatcactatatgctgaaggattctacaaaactctg
tgctattcgtgtaatccaactgatgtggctgcattttggagctttgcattcgctctttcc
aagattgttgaacttggagacactatgttcattattttgagaaaacggccattgatcttt
ttacactactatcatcatgcagcagtgttaatctacactgtccattctggtgccgagcat
actgcagctggtcgtttctacatcctaatgaactacttcgcacattctctcatgtatact
tactacacagtttctgccatgggatacagattaccgaaatgggtatcaatgactgtcaca
actgttcaaacaactcaaatgttagctggagtcggaataacttggatggtgtacaaagtg
aaaactgaatacaagcttccttgtcaacaatccgtagccaatttgtatctcgcattcgtc
atctatgtcacatttgccattcttttcattcaattcttcgtcaaggcatacattatcaag
tcgtcgaagaagtcgaaatcggtgaagaacgaataao
Page 1
CA 02365096 2001-09-14
SEQID2.GBS.tXt
atggcaaaatacgactacaatccgaagtatgggttagaaaattacagcatattccttccc
tttgagacatcttttgatgcatttcgatcgacaacatggatgcaaaatcactggtatcaa
tcaattacagcatctgtcgtgtatgtagccgtcatttttacaggaaagaaggtggttctc
atctacaaaaaatcacgagttattacttttgagtctagccttcagaatgcaattaagaat
cgaaaccgaaaatcacttaatagttctcaaatgtttcagattatggaaaagtacaagccc
ttccaactggacacaccactcttcgtctggaattcatttttagccattttctcaattctc
gggttcctccgaatgacacctgaatttgtatggagttggtcagcagaaggaaactcattc
aaatattcaatttgtcattcatcttatgctcaaggagtcactggtttctggactgaacaa
ttcgcaatgagcaaacttttcgagctcatcgacacaatcttcatcgttcttcgtaaacgt
ccactcatcttccttcactggtatcatcatgtaactgttatgatctacacatggcacgcg
tacaaggatcacactgcatcaggacggtggttcatttggatgaattatggagttcatgct
cttatgtattcctactatgctcttcgttctctgaaattccgtcttccaaaacaaatggca
atggttgttactactctccaacttgctcaaatggttatgggagtaatcatcggagtcact
gtctaccgtatcaagtcatcgggtgaatactgccaacagacatgggacaatttgggatta
tgctttggagtttatttcacatatttccttcttttcgccaacttcttctaccatgcatat
gttaagaaaaacaaccgtacagtaaattatgaaaataattcaaaaaatttccccgatctc
gttttaatttacctgagaaaaaaggtttcaagaaaatcgaaaaatcggcaatgttcagaa
aataattataaaattcaattttcatcaaattttgttaatgttgatggaaaaaaacataag
aaaacatatgaacttattcttccaagaagaaaaatgaccacaattttaacttttctattt
ggaaaaaatcgaattttttcgaaatatcagaaaaatcgaaaaaacatttcgattcctgtt
gatttcgaaattctggagccaaaagaagatatcaatgctaacatcgctgagccatccatc
acaacgaggtccgccgccgcacgaagaaaagttcaaaaagctgattago
Page 1
CA 02365096 2001-09-14
SEQID3.GBS.txt
atggcagcagcacaaacaagtccagcagccacgctcgtcgatgttttgacaaaaccatgg
agtctggatcagactgattcttacatgtctacatttgtaccattatcctataaaatcatg
attggttatctcgtcaccatctacttcgggcaaaaattaatggctcacagaaaaccattc
gatctccaaaatacacttgctctctggaacttcgggttttcactgttctcgggaatcgcc
gcctataagcttattccagaactattcggagttttcatgaaggacgggtttgtcgcttcc
tactgtcaaaacgagaactactacaccgatgcatcaactggattctggggctgggccttt
gtgatgtcgaaagctccagaactaggggatactatgttcttggtccttcgtaaaaaacca
gttatcttcatgcactggtatcatcatgccctcacatttgtctacgcagtagtcacatac
tctgagcatcaggcatgggctcgttggtctttggctctcaaccttgccgtccacactgtt
atgtatttctacttcgccgttcgcgccttgaacatccaaactccacgcccagtggcaaag
ttcatcactactattcaaattgtccaatttgtcatctcatgctacatttttgggcatttg
gtattcattaagtctgctgattctgttcctggttgcgctgttagctggaatgtgctatcg
atcggaggactcatgtacatcagttatttgttcctttttgccaagttcttctacaaggcc
tacattcaaaaacgctcaccaaccaaaaccagcaagcaggagtago
Page 1
CA 02365096 2001-09-14
atgtcatcgg'acgatc t SEQzo4.GBS.txt
g ggcactagaaccttcaagatgatggatcaaattcttggaaca
aacttcacttatgaaggtgccaaagaagttgctcgaggccttgaaggtttctcagcaaag
cttgccgtcggatatattgccactatttttggactgaaatattatatgaaagaccgaaaa
gccttcgatctcagtactccattaaacatttggaatggtattctttcgacattcagctta
ttgggattcttattcacttttcctactttgttatcagttatcagaaaggatggatttagt
cacacctattcccatgtctctgagctttacactgacagtacctctggatattggatcttc
ctttgggttatctcaaagattccggaacttttggatacagtattcattgttcttcgcaag
agaccacttattttcatgcactggtaccatcacgcattgaccggttactatgctcttgtc
tgctaccatgaggatgctgtccatatggtttgggttgtatggatgaattatattattcat
gcattcatgtatggatactatcttctgaaatctctgaaagttccaattccaccatcagtt
gctcaagcaatcaccacatctcaaatggttcaattcgcagttgccattttcgcacaagtt
catgtttcctataaacactatgttgagggagttgaaggattagcctactcgttcagagga
acagctatcggatttttcatgcttactacctacttctatctatggattcaattctacaaa
gagcactatcttaagaatggaggcaaaaagtacaatttggcaaaggatcaggcaaaaact
caaacaaagaaggctaactaao
Page 1
CA 02365096 2001-09-14
SEQIDS.GBS.txt
atgccacagggagaagtctcattctttgaggtgctgacaactgctccattcagtcatgag
ctctcaaaaaagcatattgcacagactcagtatgctgctttctggatctcaatggcatat
gttgtcgttatttttgggctcaaggctgtcatgacaaaccgaaaaccatttgatctcacg
ggaccactgaatctctggaatgcgggtcttgctattttctcaactctcggatcacttgcc
actacatttggacttctccacgagttcttcagccgtggatttttcgaatcttacattcac
atcggagacttttataatggactttctggaatgttcacatggcttttcgttctctcaaaa
gttgctgaattcggagatacactttttattattcttcgtaaaaagccattgatgttcctt
cattggtatcatcatgtgcttacaatgaattatgcttttatgtcatttgaagctaatttg
ggatttaatacttggattacatggatgaatttctcagttcactcaattatgtatggatat
tatatgcttcgttcttttggtgtcaaggttccagcatggattgccaagaatattacaaca
atgcaaattcttcaattcgttattactcatttcattcttttccacgttggatatttggca
gttactggacaatctgttgactcaactccaggatattattggttctgccttctcatggaa
atctcttatgtcgttctgttcggaaacttctactatcaatcatacatcaagggaggtggc
aagaagtttaatgcagagaagaagactgaaaagaaaattgaataaa
Page 1
CA 02365096 2001-09-14
SEQID6.GBS.txt
atgtatttgaattatttcgcgacggaaatcttccatcgtagtgcggtttgtgaaacagaa
gcttgtcgctcgtcaaaaataatgattgctgacgtgttcaaatggaaattcgatgcaaac
gaattgtggagtcttttaacgaatcaggatgaagttttcccgcatattagagcacggcga
ttcattcaagaacattttggtctattcgtccagatggcaattgcatatgtcattttggtg
ttctcaatcaaaaggttcatgagggatcgtgaaccatttcaactcaccacagctcttcgt
ctctggaacttcttcctctccgtcttctcaatttatggttcctggacaatgtttccattt
atggttcaacaaataagactttatggtctctacggatgtggatgcgaagcactttcaaac
cttccgagtcaagcagaatattggcttttcctgacgatcttgtccaaagctgtggagttt
gttgatacatttttcttggttctccggaaaaaaccactcatcttcctacactggtatcat
catatggcaacatttgtcttcttctgcagtaattacccgactccatcgtcacaatcacgc
gtcggagttatcgtcaacctgttcgtgcatgccttcatgtacccatactatttcacccga
tcaatgaacatcaaagttcctgcgaaaatttcaatggctgttacagttcttcaattgact
caattcatgtgctttatctatggatgtactctcatgtactactcgttggccactaatcag
gcacgatacccctcaaatacacctgcgacactccaatgtttgtcctacactctacatttg
ctttgao
Page 1
CA 02365096 2001-09-14
SEQID7.GBS.txt
atggctcagcatccgctcgttcaacggcttctcgatgtcaaattcgacacgaaacgattt
gtggctattgctactcatgggccaaagaatttccctgacgcagaaggtcgcaagttcttt
gctgatcactttgatgttactattcaggcttcaatcctgtacatggtcgttgtgttcgga
acaaaatggttcatgcgtaatcgtcaaccattccaattgactattccactcaacatctgg
aatttcatcctcgccgcattttccatcgcaggagctgtcaaaatgaccccagagttcttt
ggaaccattgccaacaaaggaattgtcgatcctactgcaaagtgtttgatttcacgaaag
gagagaatggatactgggtgtggctcttcatggcttccaaacttttcgaacttgttgaca
ccatcttcttggttctccgtaaacgtccactcatgttccttcactggtatcaccatattc
tcaccatgatctacgcctggtactctcatccattgaccccaggattcaacagatacggaa
tttatcttaactttgtcgtccacgccttcatgtactcttactacttccttcgctcgatga
agattcgcgtgccaggattcatcgcccaagctatcacatctcttcaaatcgttcaattca
tcatctcttgcgccgttcttgctcatcttggttatctcatgcacttcaccaatgccaact
gtgatttcgagccatcagtattcaagctcgcagttttcatggacacaacatacttggctc
ttttcgtcaacttcttcctccaatcatatgttctccgcggaggaaaagacaagtacaagg
cagtgccaaagaagaagaacaactaa~
Page 1
CA 02365096 2001-09-14
SEQID8.GBS.tXt
atgtcggccgaagtgtccgaacgattcaaagtttggacaggaaacaatgagaccatcatc
tattccccattcgagtacgattccacgttgctcatcgagtcatgtcggtgtacttatcag
ctgcttatattattgcgacaaatttattacagagatatatggagtcacggaaacctaaaa
cttttactagcatggaacggttttttggcagtgttcagtattatgggtacatggagattt
ggaatcgaattctacgatgctgttttcagaagaggcttcatcgattcgatctgcctggct
gtaaatccacgttcaccgtccgcattctgggcatgcatgttcgctctatcgaaaatcgcc
gagtttggggacacgatgttcttggtgctgaggaaacggccggttatattccttcactgg
tatcatcacgctgttgttctgatcctttcttggcatgctgcaatcgaactcacagctcca
ggacgctggtttatttttatgaactatttggtgcattcaataatgtatacatactacgca
ataacatcaatcggctatcgtcttcccaaaatcgtttcaatgactgttacattccttcaa
actcttcaaatgctcattggtgtcagcatttcttgcattgtgctttatttgaagcttaat
ggagagatgtgccaacaatcctacgacaatctggcgttgagcttcggaatctacgcctca
ttcctggtgctattctccagtttcttcaacaatgcatatttggtaaaaaaggacaagaaa
cccgatgtgaagaaggattaao
Page 1
CA 02365096 2001-09-14
SEQID9.GBS.txt
MELAEFWNDL~ITFTIYGPNHTDMTTKYKYSYHFPGEQVADPQYWTILFQKYWYHSITISV
LYFILIKVIQKFMENRKPFTLKYPLILWNGALAAFSIIATLRFSIDPLRSLYAEGFYKTL
CYSCNPTDVAAFWSFAFALSKIVELGDTMFIILRKRPLIFLHYYHHAAVLIYTVHSGAEH
TAAGRFYILMNYFAHSLMYTYYTVSAMGYRLPKWVSMTVTTVQTTQMLAGVGITWMVYKV
KTEYKLPCQQSVANLYLAFVIYVTFAILFIQFFVKAYIIKSSKKSKSVKNEO
Page 1
CA 02365096 2001-09-14
SEQID10.GBS.tXt
MAKYDYNPKItGLENYSIFLPFETSFDAFRSTTWMQNHWYQSITASVVYVAVIFTGKKWL
IYKKSRVITFESSLQNAIKNRNRKSLNSSQMFQIMEKYKPFQLDTPLFVWNSFLAIFSIL
GFLRMTPEFVWSWSAEGNSFKYSICHSSYAQGVTGFWTEQFAMSKLFELIDTIFIVLRKR
PLIFLHWYHHVTVMIYTWHAYKDHTASGRWFIWMNYGVHALMYSYYALRSLKFRLPKQMA
MVVTTLQLAQMVMGVIIGVTVYRIKSSGEYCQQTWDNLGLCFGVYFTYFLLFANFFYHAY
VKKNNRTVNYENNSKNFPDLVLIYLRKKVSRKSKNRQCSENNYKIQFSSNFVNVDGKKHK
KTYELILPRRKMTTILTFLFGKNRIFSKYQKNRKNISIPVDFEILEPKEDINANIAEPSI
TTRSAAARRKVQKADD
Page 1
CA 02365096 2001-09-14
SEQID11.GBS.tXt
MAAAQTSPAA'fLVDVLTKPWSLDQTDSYMSTFVPLSYKIMIGYLVTIYFGQKLMAHRKPF
DLQNTLALWNFGFSLFSGIAAYKLIPELFGVFMKDGFVASYCQNENYYTDASTGFWGWAF
VMSKAPELGDTMFLVLRKKPVIFMHWYHHALTFVYAVVTYSEHQAWARWSLALNLAVHTV
MYFYFAVRALNIQTPRPVAKFITTIQIVQFVISCYIFGHLVFIKSADSVPGCAVSWNVLS
IGGLMYISYLFLFAKFFYKAYIQKRSPTKTSKQE~
Page 1
CA 02365096 2001-09-14
SEQID12.GBS.txt
MSSDDRGTRl'~FKMMDQILGTNFTYEGAKEVARGLEGFSAKLAVGYIATIFGLKYYMKDRK
AFDLSTPLNIWNGILSTFSLLGFLFTFPTLLSVIRKDGFSHTYSHVSELYTDSTSGYWIF
LWVISKIPELLDTVFIVLRKRPLIFMHWYHHALTGYYALVCYHEDAVHMVWVVWMNYIIH
AFMYGYYLLKSLKVPIPPSVAQAITTSQMVQFAVAIFAQVHVSYKHYVEGVEGLAYSFRG
TAIGFFMLTTYFYLWIQFYKEHYLKNGGKKYNLAKDQAKTQTKKAN~
Page 1
CA 02365096 2001-09-14
SEQID13.GBS.tXt
MPQGEVSFFE~VLTTAPFSHELSKKHIAQTQYAAFWISMAYVVVIFGLKAVMTNRKPFDLT
GPLNLWNAGLAIFSTLGSLATTFGLLHEFFSRGFFESYIHIGDFYNGLSGMFTWLFVLSK
VAEFGDTLFIILRKKPLMFLHWYHHVLTMNYAFMSFEANLGFNTWITWMNFSVHSIMYGY
YMLRSFGVKVPAWIAKNITTMQILQFVITHFILFHVGYLAVTGQSVDSTPGYYWFCLLME
ISYWLFGNFYYQSYIKGGGKKFNAEKKTEKKIEO
Page 1
CA 02365096 2001-09-14
SEQID14.GBS.txt
MYLNYFATEI~FHRSAVCETEACRSSKIMIADVFKWKFDANELWSLLTNQDEVFPHIRARR
FIQEHFGLFVQMAIAYVILVFSIKRFMRDREPFQLTTALRLWNFFLSVFSIYGSWTMFPF
MVQQIRLYGLYGCGCEALSNLPSQAEYWLFLTILSKAVEFVDTFFLVLRKKPLIFLHWYH
HMATFVFFCSNYPTPSSQSRVGVIVNLFVHAFMYPYYFTRSMNIKVPAKISMAVTVLQLT
QFMCFIYGCTLMYYSLATNQARYPSNTPATLQCLSYTLHLLD
Page 1
CA 02365096 2001-09-14
SEQID15.GBS.tXt
MAQHPLVQRL'~LDVKFDTKRFVAIATHGPKNFPDAEGRKFFADHFDVTIQASILYMVWFG
TKWFMRNRQPFQLTIPLNIWNFILAAFSIAGAVKMTPEFFGTIANKGIVASYCKVFDFTK
GENGYWVWLFMASKLFELVDTIFLVLRKRPLMFLHWYHHILTMIYAWYSHPLTPGFNRYG
IYLNFVVHAFMYSYYFLRSMKIRVPGFIAQAITSLQIVQFIISCAVLAHLGYLMHFTNAN
CDFEPSVFKLAVFMDTTYLALFVNFFLQSYVLRGGKDKYKAVPKKKNNO
Page 1
CA 02365096 2001-09-14
SEQID16.GBS.txt
MSAEVSERFICVWTGNNETIIYSPFEYDSTLLIESCRCTYQLLILLRQIYYRDIWSHGNLK
ACDXLLLAWNGFLAVFSIMGTWRFGIEFYDAVFRXGFIXSICLAVNPRSPSAFWACMFAL
SKIAEFGDTMFLVLRKRPVIFLHWYHHAWLILSWHAAIELTAPGRWFIFMNYLVHSIMY
TYYAITSIGYRXPKIVSMTVTFLQTLQMLIGVSISCIVLYLKLNGEMCQQSYDNLALSFG
IYASFLVLSSFFNNAYLVKKDKKPDVKKD~
Page 1