Note: Descriptions are shown in the official language in which they were submitted.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
1
Genetically Modified Plants with altered Starch
This invention relates to genetically modified plants, and
in particular to genetically modified maize and wheat. The
genetically modified plants have an altered starch synthesising
ability following the introduction, by recombinant DNA
techniques, of one or more gene sequences coding for enzymes in
the starch or glycogen biosynthetic pathway into the plant.
Starch is a complex polymer of glucosyl residues. It is
the major form in which carbohydrate is stored in the tissues
and cells of most species of higher plants. It is accumulated
in the leaves of plants during the day as a result of
photosynthesis and is used to supply the needs of the plant for
energy and biosynthesis during the night. Starch is also
accumulated in non-photosynthetic cells, especially those
involved in reproduction such as in seeds, fruits and tubers.
Therefore, starch is of great importance to the productivity of
the plant and its survival.
Starch is also highly significant to man. Firstly, it
forms a major component of animal diets, supplying man and his
domestic animals with a large portion of their carbohydrate
intake. Secondly, the type of starch in a plant affects the
quality of the processed plant product. Thirdly, starch is used
industrially in the production of paper, textiles, plastics and
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
2
adhesives, as well as providing the raw material for some bio-
reactors. Starch from different species have preferred uses.
On a world scale, starch producing crops are agriculturally and
economically by far the most important, and these crops include
wheat, maize, rice and potatoes. The type of starch will affect
the quality of a processed product and the profitability of the
processed crop. In addition, the quantity and quality of starch
present in the harvested organ of a plant will affect the gross
yield and the processing efficiency. It is also known that some
starch producing crops produce a lower yield of starch when
grown under higher temperature conditions. This reduction in
yield is undesirable in terms of gross yield and processing
efficiency of the crop. Starch yield may be measured in terms
of the number of seeds harvested or the weight of the seeds
harvested.
In plants, i.e. vascular plants, the starch consists of
linear chain and branched chain glucans known as amylose and
amylopectin respectively. Starch with various amounts of
amylose and amylopectin are found in different plants.
Typically, plant starch contains 10-25o amylose, the remainder
being amylopectin, the branched chain glucan. Amylopectin
contains short chains and long chains, the short chains ranging
from 5-30 glucose units and the long chains ranging from 30-100
glucose units, or more. It is thought that the ratio of amylose
to amylopectin and the distribution of short to long chains in
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
3
the amylopectin fraction affect the physical properties of
starch, e.g. thermal stabilisation, retrogradation and
viscosity. These properties also affect the utility of starch,
as mentioned above. Starches from different plants have
different properties, which also affects their suitability for
processing under certain conditions and for certain uses. It
can be seen, therefore, that modifying the starch generated in a
plant can have particular utility in the downstream processing
or the yield of the starch in the plant storage organ. It can
also be seen that providing a plant having an improved starch
yield when grown under higher temperature conditions compared
with unmodified plants is also desirable.
Waxy corn starch lacks amylose and this starch has unique
properties. Also, most mutations in the waxy locus of maize,
which encodes starch granule bound synthase I (GBSSI), result in
plants which produce much reduced amylose. When no functioning
GBSSI is synthesised in the homozygous waxy mutant it also lacks
amylose (Echt & Schwartz, 1981).
The genetic modifications of the present invention produce
altered starch composition and properties, which properties are
ideally beneficial in terms of starch processing. The genetic
modifications surprisingly also affected starch yields under
more stringent growing conditions.
In the last few years this concept of modifying starch
properties has been postulated and put into practice in varying
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
4
degrees. In the patent literature Ir~ternational Patent
Application, Publication No. WO 94/11520 (Zeneca) described
constructs having a target gene which encodes an enzyme involved
in the starch or glycogen biosynthetic pathway under control of
a gene switch, for example, a chemical or temperature controlled
on-off mechanism. Various crops were postulated as being
suitable for use in the method but no plant transformation was
actually carried out. Some constructs were made but no examples
or results were given. International Patent Application,
Publication No. WO 94/09144 (Zeneca) was very similar to the
just described application. Only the first steps in the
transformation process were demonstrated. No results are given
for any plant, and only the transformation of tomato is
described with reference to the exemplary methodology, although
other plants are mentioned. International Patent Application,
Publication No. WO 92/11376 (Amylogene) described introducing
antisense genes for GBSSI in to potatoes to down-regulate
amylose production with the intention of producing a potato with
practically no amylose-type starch. Whilst great detail is
given of methodology, no actual results from transformed plants
are given and no plant transformations other than potato are
postulated. Only a small number of constructs are actually
produced to enable one to carry out the invention. The results
for potato were eventually published in the scientific
literature by Visser et al in 1991. Increases in the
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
amylopectin content of the starch was seen. Further scientific
papers on altering GBSSI in potato using antisense GBSSI
constructs, e.g. Visser et al (1991a) and Kuipers et al (1994),
have shown actual transformation and alteration of starch
composition.
In terms of successful transformation using non-plant
derived starch-related genes, in International Patent
Application, Publication No. WO 92/11382 (Calgene) and their
later publication (Shewmaker et al, 1994) potato was actually
transformed with E.coli glgA (Glycogen synthase) and E.coli glgC
(ADPG pyrophosphorylase). Higher specific gravity measurements
were obtained from transformed potato plants compared with two
control events, as well as altered starch characteristics.
It can be seen, therefore, that work to date has involved
introducing certain genes involved in glycogen biosynthesis
specifically into potato. The effects and their potential
usefulness for other plants and other non-plant derived starch-
related genes has only been postulated.
This invention seeks to transform cereal crops and
specifically wheat and maize with an enzyme involved in the
synthesis of microbial glycogen, namely glycogen synthase (E. C.
2.4.1.21).
This invention also seeks to identify properties of the
starch in these transformed plants which are particularly useful
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
6
and/or advantageous in the downstream processing of starch or
the plant itself.
The present invention provides transgenic wheat or maize
plants, said plants having therein a chimaeric gene comprising a
promoter, a coding sequence for glycogen synthase, which coding
sequence is derived from a microorganism, and a terminator.
As used herein, the term chimaeric gene refers to a
combination of nucleic acid sequences for each part of the
chimaeric gene, which sequences have been engineered into
relationship by recombinant DNA techniques, which sequences may
also be in their separate parts endogenous or exogenous to the
plant into which the chimaeric gene is to be introduced.
A construct and a chimaeric gene comprising nucleic acid
causing the expression of the sequences above mentioned are also
aspects of the invention.
Plant cells containing a chimaeric gene comprising a
nucleic acid sequence encoding glycogen synthase are also an
aspect of this invention, as are other plant parts, such as for
example, seed of the transformed plant containing a chimaeric
gene according to the invention. Seed of the transformed plants
grown on average at more than 20°C can exhibit a higher weight
than seed of the control plants grown on average at more than
20°C. Seed of the transformed plants can in addition or
alternatively exhibit less of a loss in yield compared with
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00100848
7
control plants when both are grown on average at a higher
temperature of more than 20°C.
The present invention also provides a method of altering
the starch in maize or wheat plants, the method comprising the
steps of stably introducing into the plant genome a nucleic acid
sequence encoding glycogen synthase under the direction of a
suitable promoter and a suitable terminator, and regenerating a
plant having an altered genome.
The present invention also provides a starch obtained from
transformed wheat or maize, said starch having an altered chain
length and/or processing property compared with control starch
from a non-transformed plant.
The present invention also provides a method of reducing
the loss of starch yield in wheat or maize grown under high
temperature conditions, the method comprising the steps of
stably introducing into the plant genome a nucleic acid sequence
encoding glycogen synthase under the direction of a suitable
promoter and a suitable terminator, and regenerating a plant
having an altered genome.
The chain length and/or branching of the starch may be
increased or decreased. Other parameters which may be altered
include the degree of retrogradation, the viscosity, the pasting
temperature, the gelling temperature, each of which may be
increased or decreased. The starch may also have modified
properties for chemical derivitisation. The yield of starch in
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
8
seed may also be less affected under more stringent growing
conditions, and in particular under growing conditions where the
temperature on average is greater than 23°C, and more preferably
on average greater than 25°C and even more preferably is on
average about 27°C, 30°C, 33°C or 36°C, or more,
or
incrementally in full degrees from 23°C upwards to 36°C. Using
the inventive method a greater than normal starch yield in seeds
can be achieved at higher temperatures during seed growth
compared with control seed grown at 20°C.
The turnover of starch in leaves is of central importance
to the growth of the plant. A change in the structure of the
starch in the granule without a complementary change in other
enzymes of starch breakdown might be expected to restrict the
export of carbon from the leaf at night. This might be expected
to cause an altered ratio of source to sink with a subsequent
effect on yield.
Preferably the promoter is capable of directing expression
in a particular tissue of the plant and/or at particular stages
of development of the plant. The promoter may be heterologous
or homologous to the plant. Preferably the promoter directs
expression to the endosperm of the plant seed. A preferred
promoter is the high molecular weight glutenin (HMWG) gene of
wheat. Other suitable promoters will be known to the skilled
man, such as the promoters of gliadin, branching enzyme, ADPG
pyrophosphorylase, starch synthase and actin, for example.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
9
Preferably the chimaeric gene also contains a sequence that
encodes a transit peptide which provides for translocation of
the glycogen synthase and/or a marker gene to the plant plastid.
Suitable transit peptides include those from the small sub-unit
of the ribulose bisphosphate carboxylase enzyme (ssu of Rubisco)
from pea, maize or sunflower, for example. Combinations of
transit peptides may also be used. Other suitable transit
peptides for transporting to the amyloplast will be known to
those skilled in the art, such as the transit peptide for the
plant plastid acyl carrier protein (ACP) or for GBSSI.
The coding sequence encoding glycogen synthase is a
sequence obtained from a microorganism, such as a unicellular
organism, for example, bacteria, which sequence has the
necessary ability to encode glycogen synthase.
Suitably the glycogen synthase is derived from a bacterial
source such as E.coli (for example, Baecker, P.A. et al, 1983 or
Kumar, A. et al 1986), Agrobacterium (Uttaro, A.D., & Ugalde,
R.A. 1994), Salmonella (Leung, P.S.C. & Preiss, J. 1987), or
Bacillus (Kiel, J.A. et al 1994). Standard methods of cloning
by hybridisation or polymerase chain reaction (PCR) techniques
may be used to isolate the sequences from such organisms: for
example, molecular cloning techniques such as those described by
Sambrook, J. et al 1989 and the PCR techniques described by
Innis, M.A., et al 1990. Other microbial sequences may be
obtained in a similar manner.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
Depending on the homology of the nucleotide sequences
encoding glycogen synthases, different conditions of
stringencies may be used in the hybridisation procedures. By
way of example and not limitation, hybridisation procedures
using such conditions of high stringency are as follows:
hybridisation to filter-bound DNA in 0.5 M NaHP04, 7% sodium
dodecyl sulfate (SDS), 1 mM EDTA at 65°C, and washing in
O.IxSSC/0.1%SDS at 68°C (Ausubel F.M. et al, eds., 1989, Current
Protocols in Molecular Biology, Vol. I, Green Publishing
Associates, Inc., and John Wiley and Sons, Inc., New York, at p.
2 . 10 . 3 ) . Other conditions of high stringency which may be used
are well known in the art. Hybridisation procedures using
conditions of moderate stringency that may be used are as
follows: hybridisation to filter-bound DNA in 0.5 M NaHP04, 7%
sodium dodecyl sulfate (SDS) , 1 mM EDTA at 65°C, and washing in
0.2xSSC/0.1% SDS at 42°C (Ausubel et al, 1989, supra). Other
conditions of moderate stringency which may be used are well-
known in the art.
The chimaeric gene may comprise one or more additional
coding sequences from the starch or glycogen biosynthetic
pathway, such as, for example, branching enzyme (EC 2.4.1.18).
The transformation techniques for the method of the
invention are advantageously direct DNA transfer techniques,
such as electroporation, microinjection or DNA bombardment (the
biolistic approach). Alternatively, plant cell transformation
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
11
using plant vectors introduced into plant pathogenic bacteria,
such as Agrobacterium-mediated transfer (Cheng, M. et al
(1997)), may be used. In any transformation method positive or
negative selectable markers may be used, at least initially, in
order to determine whether transformation has actually occurred.
Useful negative selectable markers include enzymes which confer
resistance to an antibiotic, such as gentamycin, hygromycin,
kanamycin and the like, or resistance to a herbicide, such as
asulam or basta. Alternatively, markers which provide a
compound identifiable by a colour change, such as GUS, or
luminescence, such as luciferase, may be used.
The chimaeric gene may also comprise a gene switch
mechanism which determines under what conditions or when the
coding sequence is to be expressed. The gene switch may be a
chemically induced promoter or a temperature controlled
promoter, for example.
In order that the invention may be easily understood and
readily carried into effect, reference will now be had, by way
of example, to the following diagrammatic drawings in which:
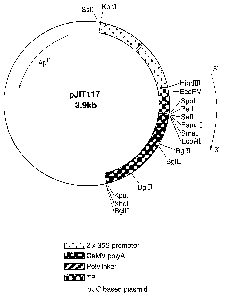
Figure 1 shows a map of the plasmid pJIT117 used in the
preparation of the plamid of Figure 2;
Figure 2 shows a map of the plasmid pBSl7R used in the
sticky-feet polymerase chain reaction;
Figure 3 shows a diagrammatic representation of the steps
in the sticky-feet polymerase chain reaction;
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
12
Figure 4 shows a map of the plasmid pBSHMVJGP used in the
preparation of the plasmid of Figure 6;
Figure 5 shows a map of the plasmid pDV02000 used in the
preparation of the plasmid of Figure 6;
Figure 6 shows a map of the plasmid pDV03000 used in the
preparation of the plasmid of Figure 7;
Figure 7 shows a map of the plasmid pDV03191 according to
one aspect of the invention and used in the transformation
process of the invention;
Figure 8 shows a standard chromatogram of glucose at 1mM
concentration;
Figure 9 shows a standard chromatogram of maltose at 1mM
concentration;
Figure 10 shows a standard chromatogram of maltotriose at
1mM concentration;
Figure 11 shows a standard chromatogram of maltohexaose at
1mM concentration;
Figure 12 shows a standard chromatogram of a mixture of
maltotriose, maltotetraose, maltopentaose, maltohexaose and
maltoheptaose each at 1mM concentration;
Figure 13 shows a chromatogram of an isoamylase digest of
wheat starch from wheat plants according to the invention;
Figure 14 shows a graph of starch branch chain lengths for
starch from the seed of a single transgenic wheat plant compared
with starch from the seed of a control wheat plant;
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
13
Figure 15 shows a graph of starch branch chain lengths for
starch from the seed of a further single transgenic wheat plant
compared with starch from the seed from a control wheat plant;
Figure 16 shows a comparison of branch chain length for a
family of starches from the seed of transgenic lines against a
family of starches from the seed of control wheat plants;
Figure 17 shows a western blot of proteins extracted from
the seed of transgenic maize plants according to the invention.
Figure 18 shows the differences in two lines of wheat in a
dry weight of seed at two different temperatures; and
Figure 19 shows the effect of temperature on the rate of
starch synthesis in two transgenic lines of wheat compared to a
control.
The invention will now be described, by way of example,
with reference to an embodiment for incorporating glgA from
E.coli strain LCB618 into wheat and maize.
EXAMPLE 1
Construction of crlgA plasmids used for particle bombardment of
wheat embryos.
Isolation of E.coli chromosomal DNA
The coding sequence for glgA was originally isolated by PCR
using chromosomal DNA from the E.coli strain LCB618 as
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
14
template. E.coli LCB618 was obtained from E.coli Genetic Stock
Center, Yale University, U.S.A.
E.coli LCB618 was grown up in 100m1 LB o/n at 37°C. Cells
were pelleted and re-suspended in 9.5m1 lOmM Tris-HCl, 1mM EDTA
(TE) pH8.0 and 0.5m1 10% w/v Sodium dodecyl sulphate (SDS) and
50,1 proteinase K 20mg/ml were added. The mixture was incubated
at 37°C for 1 hour to lyse cells. 1.8m1 of 5M NaCl followed by
l.5ml of CTAB (cetyl trimethyl ammonium bromide)/NaCl solution
(10% w/v CTAB in 0.7M NaCl) were added and the mixture incubated
at 65°C for 20 minutes. The lysate was extracted with an equal
volume of chloroform and centrifuged at 6000g to separate the
layers. The upper layer was removed to a fresh tube and DNA was
precipitated by the addition of 0.6 volumes isopropanol. The
DNA was removed from the solution with a sealed pasteur pipette,
placed into a fresh tube and washed with 70 % ethanol . The DNA
was dried in vacuo and re-suspended in TE pH8Ø The DNA was
purified on a CsCl gradient.
Sticky-~eet PCR
In order for the E.coli glycogen synthase to function in
plants the protein has to be transported into the amyloplast.
This transport can be facilitated by attachment of a plastid
transit peptide to the amino terminus of the E.coli polypeptide.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
The coding sequence for the transit peptide (TP) from the
small subunit of the ribulose bisphosphate carboxylase enzyme
(ssu of Rubisco) pea has been cloned and the TP shown to target
(3-glucuronidase (GUS) protein to chloroplasts (Guerineau et al,
1988) .
The plasmid pJIT117 (Guerineau et a1, 1988), the map of
which is shown in Figure l, has several restriction sites
downstream of the ssuTP which can be used for subcloning of
coding sequences, however, the subcloning must create a
translational fusion between the transit peptide and the coding
sequence, and the Cys-Met amino acid sequence at the junction
must be maintained.
We have previously used pJIT117 to attach the ssu transit
peptide to the coding sequence for E.coli ADPG PPase g1gC16
using restriction digestion and PCR. The TP-g1gC16 DNA, herein
known as SEQ.ID. No.l, was subsequently transferred to the
vector pBluescript (Stratagene Ltd., Cambridge, UK) to create
pBSl7R (the map for which is shown in Figure 2) and this plasmid
was useful in generating a similar construct for glgA.
The glgA coding sequence has no convenient restriction
sites at the 5' end. Therefore, to ensure that the open reading
frame was in a translational fusion with the ssu transit peptide
and to maintain the integrity of the Cys-Met cleavage site,
plasmid pBSl7R was used to substitute the glgA sequence for the
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
16
g1gC16 sequence with a technique called sticky-feet PCR
(Clackson and Winter, 1989).
This technique is explained diagrammatically with reference
to Figure 3. In this technique, PCR primers are designed to the
5' and 3' ends of the acceptor sequence of chromosomal or
genomic DNA and the sequences which are to be attached to the
acceptor from a donator plasmid. In Step A, PCR is used to
amplify the sequences which are to be inserted in the donator.
In Step B the amplified acceptor DNA fragment is annealed to the
donator plasmid which has been made single-stranded and carries
uracil residues instead of thymidine residues by using a
specific type of E.coli host. In Step C, a new strand is
synthesised, using the donator plasmid as template and the
acceptor fragment as primer, with a combination of Taq
polymerase, T7 DNA polymerase (Sequenase) and T4 DNA ligase.
The new double-stranded plasmid is a hybrid with one strand of
the uracil-containing donator and one strand incorporating the
acceptor fragment.
This hybrid plasmid is then transferred into a normal
E.coli host where the uracil-containing strand is degraded and
the acceptor strand replicated. A double-stranded plasmid
incorporating the acceptor DNA can then be recovered. As an
alternative, in Step D (not shown), the hybrid plasmid can be
used in a PCR reaction with primers which will amplify out the
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
17
acceptor DNA with the required fragments from the donator
attached.
In this particular example, glgA sticky-feet primers were
designed as follows:
SEQ. ID. No. 4 GLGASF5 (Pl)
TGGTGGAAGAGTAAAGTGCATGCAGGTTTTACATGTATGTTCA
ssu TP 3'end ~ glgA 5' end --
SEQ. ID. No. 5 GLGASF3 (P2)
TCGCTCCTGTTTATGCCCTAGATCTCTATTTCGAGCGATAGTAAAGCTCACGGT
~glgC 3 ' end ~ glgA 3 ' end-
The PCR primers are designed to the 5' and 3' ends of the
glgA cDNA sequence.
The 5' end primer (SEQ. ID. No. 4) also has sequences which
are homologous to the ssu-TP.
The 3' end primer (SEQ. ID. No. 5) also incorporates
sequences which are homologous to the 3' end of the glgC coding
sequence. These primers are used in a PCR process to amplify a
glgA fragment with extensions which will overlap onto the
sequences in pBSl7R. This is represented by Step A of Figure 3.
Plasmid pBSl7R is made into a template for sticky-feet PCR
by transferring the plasmid into the E.coli host CJ236 (Raleigh
et al, 1989). This host is deficient in the enzyme dUTPase,
(i.e. dut-) which results in deoxyuridine being incorporated into
the DNA instead of thymidine. The absence of another enzyme
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
18
uracyl N-glycosylase (ung-) means that the deoxyuridines cannot
then be removed from the DNA.
In Step B of Figure 3, the extended glgA DNA (2) is
annealed to the uracil-containing template which has been
isolated as single-stranded DNA (3), and a new strand is
synthesised as per Step C above. The new double-stranded
plasmid is a hybrid (5) with one strand of the uracil-containing
template (3) and the other strand consisting of the plasmid
backbone and the glgA fragment now with ssu-TP and a 3' glgC
fragment attached at 5' and 3' ends respectively (4).
In Step D (not shown) , the hybrid plasmid is used in a PCR
reaction with primers (SEQ. ID. No. 6) (P3)(see below) and SEQ.
ID. No. 5 (P2) which will amplify out the extended glgA.
With reference to Figure 3, the experimental details are as
follows:
The primers GLGASFS (P1) (SEQ. ID. No. 4) vs GLGASF3 (P2)
( SEQ . ID . No . 5 ) were kinased and used to ampl i fy the glgA open
reading frame with extension sequences using E.coli LCB618
genomic DNA (1) as template. The DNA (2) was purified with
GeneClean (BIO 101, Ltd.). The sticky-feet template DNA,
single-stranded uracil pBSl7R DNA (3), was isolated from 5m1
overnight cultures of the dut- ung- E.coli strain CJ236.
The sticky-feet PCR reaction was carried out in 10,1 volume
containing 20ng ss uracil pBSl7R (3); 200ng glgA DNA (2), 1~1 x
Taq polymerase buffer, 1.0,1 2mM mixture of dATP, dTTP, dCTP,
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
19
dGTP (2mM dNTPs); 2.5 units Taq polymerase. The mix was
overlaid with 30,1 mineral oil and cycled once at 94°C, 3 min;
72°C, 2 min; 40°C, 2 min. and then cooled to room temperature.
10.1 of a solution containing 2.0,1 x5 Sequenase buffer (200mM
Tris-HC1 pH 7.5; 100mM MgCl2, 250mM NaCl), 1.51 O.lmM
Dithiothreitol (DTT); 2.0,1 lOmM Adenosine 5' triphosphate
(ATP); 4 units T4 DNA lipase; 6.5 units Sequenase was then added
and the reaction incubated at room temperature for 30 minutes.
Generation of TP-glgA DNA
1Ø1 of the reaction containing the hybrid plasmid (3 + 4)
was taken and diluted to 101 with lOmM TE at pH8Ø 1.0,1 of
the diluted sample was used in a PCR reaction in order to obtain
the TP-glgA coding sequence (Step C of Figure 3). The primers
used were TPSSU5 (P3) (SEQ. ID. No. 6) vs GLGASF3 (P2) (SEQ. ID.
No. 5) .
SEQ. ID. No. 6 TPSSU5 (P3)
ACGTAGATCTATGGCTTCTATGATATCCTCTTC
The primers both have restriction sites for BglII,
therefore after purification, the amplified DNA was digested
with BglII and subcloned into the BamHI site of pDV03000 (see
below) .
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
Construction of pDV03000 vector
Transgenic wheat and maize plants are generated by particle
bombardment of embryos and it is not necessary to use binary
vectors. For expression of the glgA protein the coding sequence
is placed under the control of an endosperm-specific promoter.
One such suitable promoter is that from the High Molecular
Weight Glutenin (HMWG) gene of wheat (Bartels and Thompson,
1986). Primers (P4) and (P5) (SEQ. ID. Nos. 7 and 8
respectively) were designed so that the 430bp HMWG promoter,
(the nucleotide sequence of which is given in SEQ. ID. No. 3)
could be isolated by PCR and subcloned via EcoRI and Clal
restriction sites into pBluescript to generate the plasmid
pBSHMWGP (Figure 4).
A second set of PCR primers were designed to obtain the
nopaline synthase terminator from plasmid pDV02000, the map of
which is shown in Figure 5. This plasmid was previously
constructed in our laboratory as an intermediate vector for the
sub-cloning of coding sequences. The 5' primer, NTPRIMES (P6)
(SEQ. ID. No. 9), has a BamHI restriction site, while the 3'
primer NTP3NXS2 (P7) (SEQ. ID. No. 10), has restriction sites
for Notl, Xhol and SacII. The amplified DNA was digested with
BamHI and SacII and ligated into the pBSHMWGP plasmid to
generate pDV03000 (the map of which is shown in Figure 6).
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
21
SEQ. ID. No. 7 HMWGPR05 (P4)
GACATCGATCCCAGCTTTGAGTGGCCGTAGATTTGC
SEQ. ID. No. 8 HMWGPR03 (P5)
GACGAATTCGGATCTCTAGTTTGTGGTGCTCGGTGTTGT
SEQ. ID. No. 9 NTPRIME5 I(P6)
CAGGATCCGAATTTCACCCGATCGTTCAAACA
SEQ. ID. No. 10 NTP3NXS2 (P7)
GACCCGCGGCTCGAGGCGGCCGCCCGATCTAGTAACATAGATGACACCGC
pDV03000 vector has the HMWG promoter-nos terminator
sequences separated by unique restriction sites for EcoRI, Pstl,
Smal and BamHI.
Construction of pDV03191
TP-glgA DNA amplified from the sticky-feet PCR sample with
primers TPSSU5 vs GLGASF3 (Step D, Figure 3) was digested with
BglII, purified and ligated into the BamHI site of pDV03000.
Plasmid pDV03191 (the map of which is shown in Figure 7) was
confirmed by restriction enzyme digestion and by sequencing of
the junctions between promoter and coding sequence. E.coli XL1
Blue (Stratagene Ltd., UK) harbouring pDV03191 was deposited by
Advanced Technologies (Cambridge) Limited of 210 Cambridge
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
22
Science Park, Cambridge CB4 OWA, under the Budapest Treaty on
the International Recognition of the Deposit of Micro-organisms
for the purposes. of Patent Procedures at the National Collection
of Industrial and Marine Bacteria (NCIMB), 23 Machar Street,
Aberdeen, Scotland GB on 4 August 1998 under accession number
NCIMB 40962. The micro-organism is E.coli XL1 Blue: strain
LCB618 containing PDV03191. The DNA for E.coli glgA was
inserted as described above into pBluescript with the ssu
transit peptide, the HMWG promoter and nos terminator. The
vector is useful for altering starch properties.
Transformation of wheat
Methods for the transformation of wheat by particle
bombardment are well known in the art, for example see Vasil et
al, 1992 .
Immature embryos of wheat are used to initiate embryogenic
callus. The callus is subcultured and used for particle
bombardment with gold particles coated with plasmid DNA.
Two plasmids are used per bombardment, one plasmid carries
the construct of interest, in this case pDV03191. The second
plasmid carries the selectable marker which expresses the gene
responsible for resistance to the herbicide Basta. Plants
resistant to Basta are generally found to also have the
recombinant gene of interest present.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
23
Bombarded calli are grown on Basta selection media and
surviving calli are transferred to regeneration medium. Rooted
plants are transferred to soil and grown to maturity in a growth
room.
Primary transformant wheat plants (To) are selfed to produce
transgenic seed.
Seed are extracted for protein and the protein analysed by
western blotting for the presence of E.coli glgA polypeptide.
EXAMPLE 2
Biochemical Analysis of c~rlgA transformed maize
1. Expression of glgA protein
Soluble protein samples were prepared from individual maize
grain derived from transformed maize plants. Each grain was
pulverised in a pestle and mortar until a fine powder was
obtained. A portion of this powder (100-200mg) was placed in an
Eppendorf tube and 5001 of ice cold extraction buffer (50mM
HEPES, pH 8.0; lOmM DTT; lOmM EDTA) added. The powder was
homogenised with a micropestle to release soluble proteins.
The extract was centrifuged at 13000 rpm for 1 minute and
the supernatant decanted into a fresh Eppendorf tube and stored
on ice. The total protein content in the soluble protein sample
was assayed using The Bradford dye binding method (Bradford, M.
1976 ) .
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
24
An aliquot of the soluble protein sample containing 100mg
total protein was placed into an Eppendorf tube and excess
acetone (ca l.5ml) was added to precipitate the proteins. The
proteins were collected by centrifuging the sample at 13000 rpm
for 5 minutes. The acetone was decanted off and the samples
were air-dried until all the residual acetone had evaporated.
SDS PAGE loading buffer (4% (w/v) SDS; 12% (w/v) glycerol;
50mM Tris-HCl pH 6.8; 20 (v/v) (3-mercaptoethanol; 0.01% Serva
blue G) in an amount of 100,1 was added to the protein sample
contained in the Eppendorf tube. Samples were boiled for 1
minute before loading onto a polyacrylamide gel.
Electrophoresis was carried out according to the method of
Schagger and Von Jagow (1987). The resolving gel composition was
loo acrylamide, 3% bis-acrylamide. Gels were run at 50 V
constant for 16 hours.
Separated proteins were transferred from the acrylamide gel
onto PVDF membrane by electroblotting (Transfer buffer: 20%
methanol; 25mM Tris-HCl pH 8.3; 190mM glycine. Run in a Biorad
blotting apparatus at 50 V for 3 hours).
To detect expression of glgA the membrane was challenged
with a rabbit anti-glgA antiserum (raised glgA-GST fusion
protein purified from E.coli). Specific cross-reacting proteins
were detected using an anti-rabbit IgG-alkaline phosphatase
conjugate secondary antibody and visualised by the NBT/BCIP
reaction.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
NuPacreTM Electrophoresis
Alternatively, an aliquot of the soluble protein sample,
containing 100mg total protein was placed into an Eppendorf tube
and excess acetone (ca l.5ml) was added to precipitate the
proteins. The proteins were collected by centrifuging the
sample at 13000 rpm for 5 minutes. The acetone was decanted off
and the samples were air-dried until all the residual acetone
had evaporated.
NuPageTM loading buffer (2% (w/v) SDS; 10% (w/v) sucrose; 25
mM Tris-HC1 pH 8.5; 1% (v/v) (3-mercaptoethanol; 0.5 mM EDTA;
0.02% Serva blue 6250; 0.006% Phenol Red) 100 ~.1, was added to
the protein sample contained in the Eppendorf tube. Samples
were heated at 100°C for 1 minute before loading onto a
polyacrylamide gel. Electrophoresis was carried out on NuPageTM
precast gels according to the manufacturer's instructions
(Novex, San Diego CA) . Gels were run at 200 V constant for 60
minutes using MES SDS running buffer (20 mM MES/20 mM Tris-HCl
pH 7.3; 1% (w/v) SDS; 1 mM EDTA).
Separated proteins were transferred from the acrylamide gel
onto PVDF membrane by electroblotting (Transfer buffer: 200
methanol; 25 mM Bis-Tris/25 mM Bicine pH 8.3; 1 mM EDTA. Run in
a Novex electroblotting apparatus at 25 V for 1.5 hours).
To detect expression of glgA the membrane was challenged
with a rabbit anti-glgA antiserum (raised against glgA-GST
fusion protein purified from E. coli). Specific cross-reacting
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
26
proteins were detected using an anti-rabbit IgG-horse Radish
Peroxidase conjugate secondary antibody and visualised using
enhanced chemiluminesence (ECL) as supplied by Amersham
International.
Several transformed lines were found to express a 50 kDa
protein in their grain, which was not present in control grain
derived from non-transformed maize plants.
2. Preparation of wheat starch
Starch was extracted from grain of separate field grown
samples of two of the glgA expressing lines and a control line.
Wheat grains of each sample (3-4g) were placed in a mortar, 30m1
of 1% sodium bisulphate was added and placed on ice for 30
minutes. The grains were then gently pulverised using a pestle.
The solution was filtered through a nylon filter sieve and
collected in a centrifuge tube. The pulverised wheat grains
were re-extracted with a further 30m1 of 1% sodium bisulphate
and the filtrates were combined. The filtrate was centrifuged
at 6000 rpm for 5 minutes. After decanting off the supernatant,
the pellet of extracted starch was re-suspended in water and
centrifuged at 6000 rpm for 5 minutes. This was repeated once.
The resulting starch pellet was re-suspended in acetone,
centrifuged at 6000 rpm for 5 minutes and the supernatant
decanted away. This was repeated once and the starch left to
air dry. Once dried the starch was stored at -20°C.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
27
3. Branch chain length analysis of wheat starch
Portions of the starch samples were digested with
isoamylase and the resulting unbranched linear glucan chains
were analysed by HPLC.
75mg of isolated wheat starch was placed in a l5ml Pyrex
boiling tube and suspended in 3.Om1 of water. The sample was
placed in a boiling water bath for 6 minutes, occasionally
removed and vortex mixed. The sample was cooled to room
temperature and 250,1 of 200mM sodium acetate, pH 3.5 and 180
units of isoamylase enzyme added. The samples were made up to a
final volume of 3.8m1 with water. After mixing, the sample was
placed in a 37°C water bath for 4 hours. The samples were
occasionally vortex mixed throughout this incubation period. At
the end of the incubation the sample was placed in a boiling
water bath for 2 minutes, and then allowed to cool to 4°C. The
sample was centrifuged at 3,400 rpm for 20 minutes. The
resulting supernatant was transferred to Eppendorf tubes and
centrifuged at 13000 rpm for 15 minutes. Finally, the sample
was filtered through a 0.2mm syringe filter and stored at 4°C
until required.
Separate isoamylase digest samples were normalised to a
constant total glucan content by digesting a portion of the
sample to glucose using a,-amylase and amyloglucosidase.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
28
Two 1001 aliquots of isoamylase digested starch were
placed in two separate Eppendorf tubes (one is to be used as a
blank). To one aliquot was added: 5001 of 200mM sodium acetate
pH 4.8; 50.1 of a-amylase solution containing 10 units of a-
amylase; 1001 of amyloglucosidase solution containing 10 units
of amyloglucosidase and water to a final volume of l.Oml. To
the second (blank) aliquot was added: 500.1 of 200mM sodium
acetate pH 4.8 and 400,1 of water. The samples were left to
digest at 25°C for 16 hours.
The glucose content of the digest and blanks was assayed
spectrophotometrically using a coupled enzyme assay. An aliquot
of the total glucose digest or the blank was added to a cuvette
containing in a final volume of 990 ~1 100mM HEPES, pH 8.0; 5mM
MgCl2; 4mM NAD; 1mM ATP and 1 unit of hexokinase. The optical
density (OD) of the reaction mixture at 340nm was measured prior
to the addition of 101 containing 1 unit of glucose-6-
phosphate dehydrogenase. The OD at 340nm was monitored until
there was no further change and the difference in OD after the
addition of glucose-6-phosphate dehydrogenase compared to before
the addition of glucose-6-phosphate dehydrogenase was
determined. This figure was used to determine the total glucose
amounts in the original isoamylase digests. These samples were
diluted with water to a standard concentration of 8mM total
glucose and stored at 4°C until required for HPLC analysis.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
29
The samples were then analysed by Dionex HPLC using a
Dionex PA 100 column and PED-Integrated Amperometric detection.
The solvent flow rate was l.Om1/min and a gradient system was
developed. Solvent 1 consisted of 100mM NaOH and Solvent 2 was
100mM NaOH, 0.60M sodium acetate. The gradient profile was as
shown in Table l, with the pulsed electrochemical detection
(PED) parameters shown in Tables 2.1 and 2.2.
Table 1
Gradient Profile
Event Start Solvent 1 (~) Solvent 2 ($)
Time (min)
0 100 0
1 100 0
2 100 0
30 0 100
30.1 100 0
35 100 0
Table 2.1
Waveform Table
Time (sec) Potential (V)
0 0.1
0.5 0.1
0.51 0.6
0.59 0.6
0.6 -0.6
0.65 -0.6
CA 02365279 2001-08-28
WO 00/55331 PCTlGB00/00848
Table 2.2
Integration
Begin (sec) End (sec)
0.3 0.5
Three isoamylase digestions were performed for each sample
and three aliquots of each isoamylase digest were analysed by
the HPLC system. Separate chromatogram peaks were assigned to
specific linear glucan sizes by reference to standard mixtures
containing linear glucans of known numbers of glucose molecules
(see Figures 8-12). Peak areas were abstracted from the primary
data and averaged for the replicate chromatograms.
Figures 8 to 12 are HPLC traces of standards for various
sugars. The standards in Figures 8-12 allow the peak area for
each peak of the inventive sample of Figure 13 to be converted
to a quantitative representation of the number of glucan chains
in each peak, and the position (on the x-axis) of each peak to
the number of glucose residues in each chain, i.e. the chain
length. In Figures 14 and 15 this conversion has been done for
wheat starch extracted from a single transgenic line and its
paired control. In Figure 16, a family of starches from
transgenic lines are compared with a family of controls. Figure
16 clearly shows that the transgenic starches have a different
chain length distribution from the control starches. The starch
has therefore been altered, which alteration affects its
processing capabilities.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
31
EXAMPLE 3
Maize plants transformed with glcrA recombinant ctene
In the transformation step, immature maize embryos are used
instead of wheat and are subject to particle bombardment with
gold particles coated with plasmid DNA. Methods for the
transformation of maize are well known in the art, for example
see Gordon-Kamm et al (1990) and Fromm et al (1990). After
rooted primary transformant plants (To) are transferred to soil
and grown to maturity, maize plants are back-crossed to produce
transgenic seed which can be extracted and analysed according to
Example 2. Further back-crossing is performed to introgres the
transgene into elite varieties and selfing of transgenic plants
is performed to obtain plants and seed which are homozygous for
the transgene. Seed from these generations can also be
extracted and analysed according to method 2.
Seed from a number of back-crossed primary transformants
were shown to be expressing the glgA protein. The plants grown
up from the remaining seeds were subsequently selfed and progeny
seed were extracted for protein and western blotting according
to Example 2. Figure 17 shows the presence of glgA polypeptide
in seed from two of these second generation lines 2-AM4-5'-2 and
2-AM4-6'-1.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
32
EXAMPLE 4
Viscometry measurements of transgenic wheat seed extracts.
Flour was extracted from T2 and T3 progeny seed of primary
transformant wheat line 72.11B which was shown to be expressing
the glgA polypeptide by western blots. 4g of ground sample (140
moisture) was mixed with 25m1 water or with 24.5m1 water + 0.5m1
10% AgN03 solution. The presence of silver nitrate will inhibit
any amylase activity in the slurry and allows the true viscosity
developed by the flour to be assessed.
The slurry was subject to rapid viscometric analysis (RVA)
using standard profile 1 (Table 3). Results of the RVA are
tabulated in Table 4 and Table 5 below.
Standard 1: Idle temperature . 50 ~ 1°C
End Test (HH:MM:SS) . 00:13:00
Table 3
Time Type Value
(HH:MM:SS)
00:00:00 Speed 960 rpm
00:01:00 Speed 160 rpm
00:01:00 Temp. 50C
00:04:45 Temp. 95C
00:07:15 Temp. 95C
00:11:00 Temp. 50C
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
33
Table 4
RVA STD without AaN03
Pasting Peak BKD Final
temperature viscosity viscosity
CYMMIT control 87.2 191 61 222
72.11B/62 87.2 181 57 208
72.11B/39/4 88.1 182 53 223
72.11B/49/11 86.3 184 53 230
I72.11B/41/22 88.1 I 185 I 52 I 226
I
Table 5
RVA Modified with AaN03
Pasting Peak BKD Final Peak Final
temp. visc. visc. AgN03 visc.
- AgN03
Peak -
standard FV std
CYMMIT ctrl 86.4 251 98 267 60 45
72.11B/62 87.2 251 99 259 70 51
72.11B/39/4 87.3 238 86 265 56 42
72.11B/49/11 87.2 234 80 267 50 37
72.11B/41/22 86.5 244 87 273 59 47
The RvA method is described in Edwards et al (1999).
EXAMPLE 5
Differential scanning calorimetry of glgA transgenic wheat seed
extracts
Wheat kernels were cleaned and water was added to the
sample (90mg). The sample was allowed to condition in the
analysis chamber at ambient temperature for 24 hours before
cycling using the following conditions:
Stabilisation: lh 25min at 25°C
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
34
Raise temperature to 110°C at 1.2°C/minute
Cool to 25°C at 1.2°C/minute.
The DSC results are shown in Table 6. The DSC method is
described in the book of Frazier et al (1997).
Table 6
Peak
1 Peak
2
(amylopectin)
(amylose-lipid
complex)
Onset Temp. Enthalpy Onset Temp. Enthalpy
Point peak Point Peak
CYMMIT ctrl. 52 60 6.9 80.2 92.5 1.9
72.11B/62 52 59 6.7 82 93 1.4
72.11B/39/4 52 60 6.8 80 93 1.9
72.11B/49/11 52.3 59.6 6.4 80 93 1.8
~72.11B/41/22 51.7 59.4 6.8 I 80.2 92 1.8
~ ~ I
Example 6
Growth of plants and plant seed under high temperature
conditions
Seeds were planted in 6-inch pots in M2 compost (5-6 seeds
per pot). They were grown to anthesis in a greenhouse at 15-
25°C under a l6hr photoperiod in daylight supplemented with
sodium light (photosynthetically active radiation - 160~.mol.m-z
S-1~ ~ Plants were watered regularly (every day in summer) and
were fed weekly with Phostrogen (Phostrogen, Corwne, Clwyd, UK)
at a concentration of 1.7 gl-1. On the day of anthesis plants
were tagged. 5 days post anthesis (p. a.) plants were repotted
to 1 plant per pot. Plants from each line were moved at 5 days
p.a. into controlled environment cabinets set at either 20°C or
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
27°C for a 16 hr day length. Plants were grown in cabinets
until seed maturity (approx. 70 days p.a.). Mature seeds were
harvested and then weighed and average seed weight calculated.
The results are given in Table 7 below.
Table 7
27C 20C
Line Dry Weight Dry Weight Change in ~ loss
wt in wt
Control 0.0360.0028 0.0450.0097 0.0087 19
(Cimmyt 101)
72.11b 0.0370.0045 0.0400.0068 0.0034 8.4
79.42a 0.0380.0024 0.0430.009 0.0045 10.6
T~Vhere ~ loss in weight = ((wt at 20°C - wt at 27°C)/wt at
20°C) * 100
As plants were grown under identical conditions with
temperatures from 5 days p.a. being the only variable, it is
statistically relevant to compare changes in seed weight within
lines and not just with controls. It is clear that lines 72.11b
and 79.42a are markedly less sensitive to temperatures above
25°C than controls, losing 10.60 and 8.40 less dry weight
respectively, than the cimmyt 101 control This reduction in
seed weight loss is advantageous in countries with hotter
climates and increases the starch yield, in seeds at least, of
transformed plants compared with control plants.
The results were similar when the experiment was repeated
(see Table 8).
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
36
Table 8
27C 20C
Line Seed weight (g) Seed weight(g) Change
in change
weight
(g)
Control 0.032 0.0002 0.039 0.0004 0.007 17.9
Cimmyt 101
72.11b 0.033 0.0015 0.038 0.0003 0.005 13.2
79.42a 0.028 0.0009 0.031 0.0004 0.003 9.7
Example 7
Measurement of rate of starch synthesis
Starch synthesis
The rate of starch synthesis was measured by following
incorporation of [U-14C] sucrose into starch. Sixty wheat
endosperm were placed in a manometer flask containing a centre
well and sidearm. The centre well contained 10% KOH and a piece
of fluted filter paper to aid absorbtion of CO2. The endosperm
were placed in incubation medium (lOmM Mes-NaOH, 319mM sorbitol,
60mM KCl, 6mM MgCl2, pH5.6). After equilibration for 30 minutes
20mM (U-14C] sucrose 37KBq (final concentration) was added from
the sidearm and the flasks incubated for a further 3 hours. At
the end of the incubation the medium and KOH paper were removed
from the flask and the radioactivity determined.
The endosperm were removed from the flask and placed into a
screw top eppendorf. The tissue was washed 5 times with 1 ml
incubation medium (as above) per wash and then frozen in liquid
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
37
nitrogen. TCA (lml) was added to a final concentration of 10%
and the tissue allowed to thaw. The endosperm were ground using
a micropestle and then centrifuged for 5 minutes at 10,0008.
The supernatant was removed and the pellet washed in lml
methanol:KC1 (75%:1% v:w) by re-suspension and centrifuged
(10,0008, 5 minutes) (MSE microcentaur) five times. The
radioactivity was determined in all of the washes from the above
steps.
0.5m1 50mM acetic acid-NaOH buffer (pH4.8) was added to the
tissue and boiled for 30 minutes. After cooling, starch present
in the sample was digested by adding 40 units amyloglucosidase
and 40 units a-amylase and incubating at 37°C for 12-16 hours.
Digests were centrifuged for 10 minutes at 10,0008. The
supernatant (degraded starch) was then added to 4m1
scintillation fluid (Ecoscint A) and the radioactive counts were
determined using a liquid scintillation counter (Tri-carb 300C).
Controls contained a) boiled tissue and b) tissue stopped at
zero time. The amount of 14C label present in the degraded
starch was calculated by subtracting the values for the control
samples from the digests. The nmoles of hexose incorporated
into starch was then derived from the amount of 14C label
incorporated into starch. The rate of starch synthesis may
therefore be derived from the time of incubation. The results
are shown in Figure 19.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
38
References:
Baecker, P.A., Preston, A., Furlong, C.E. and Preiss J. (1983)
Biosynthesis of bacterial glycogen. Primary Structure of E.coli
ADPG glucose synthetase as deduced from the nucleotide sequence
of the glgC gene. J. Biol. Chem. 258 (8), 5084-5088.
Bartels, D. and Thompson, R.D. (1986). Synthesis of messenger-
RNAs coding for abundant endosperm proteins during wheat-grain
development. Plant Sci., 46 (2) 117-125.
Bradford, M. (1976) A rapid and sensitive method for the
quantitation of microgram quantities of protein utilizing the
principle of protein dye binding. Anal. Biochem. 72, (1-2), 248-
254.
Cheng, M., Fry, J.E., Pan, S.Z., Zhou H.P., Hironaka C.M.,
Duncan D.R., Conner, T.W., and Wan, Y.C. (1997) Genetic
transformation of wheat mediated by Agrobacterium tumefaciens.
Plant Physiology, 115 (3), 971-980.
Clackson, T. and Winter, G. (1989). "Sticky-Feet"-directed
mutagenesis and its application to swapping antibody domains.
Nucl. Acids Res., 17, 10163-10170.
Echt, C.S. and Schwarz, D. (1981) Evidence for the inclusion of
controlling elements within the structural gene at the waxy
locus in maize. Genetics, 99, 275-284.
Edwards, E., Fulton, D.C., Hylton, C.M., Jobling, S.A., Gidley,
M., Rossner, U., Martin, C. and Smith, A.M. (1999). A combined
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
39
reduction in activity of starch synthases II and III of potato
has novel effects on the starch of tubers. Plant J. 17:251-161.
Frazier, P.J., Donal, A.M. and Richmond, P. Starch: Structure
and Functionality (1997). Royal Society of Chemistry,
Cambridge, UK.
Fromm, M.E., Morrish, F., Armstrong, C., Williams, R., Thomas,
J. and Klein, T.M. (1990) Inheritance and expression of chimeric
genes in the progeny of transgenic maize plants.
Bio/Technology, 8 (9), 833-839.
Geurineau, F., Woolston, S., Brooks, L. and Mullineaux, P.
(1988). An expression cassette for targeting foreign proteins
into chloroplasts. Nucl. Acids Res., 16 (23), 11380.
Gordon-Kamm, W.J., Spencer, T.M., Mangans, M.L., Adams, R.T.,
Dames, R.J. , Start, W.G. , O'Brien, J.V. , Chambers, S.A. , Adams,
W.J. et a1. (1990) Transformation of maize cells and
regeneration of fertile transgenic plants. Plant Cell, 2 (7),
603-618.
Innis, M.A., Gelfand, D.H., Sninsky, J.J. and White, T.J.
(1990). PCR Protocols. A Guide to Methods and Applications.
Published Academic Press.
Kiel, J.A., Boels, J.M., Beldman, G. and Venema, G. (1994)
Glycogen in Bacillus subtilis: molecular characterisation of an
operon encoding enzymes involved in glycogen biosynthesis and
degradation. Mol. Microbiol., 11(1), 203-218.
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
Kuipers, A.G.J; Jacobsen, E; Visser, R.G.F._, (1994). Formation
and deposition of amylose in the potato tuber starch granule are
affected by the reduction of granule-bound starch synthase gene
expression. Plant Cell, 6 (1), 43-52.
Kumar, A., Larsen, C.E., Preiss, J. (1986) Biosynthesis of
bacterial glycogen primary structure of E. coli ADP-glucose a-
1,4-glucan, 4-glucosyltransferase as deduced from the nucleotide
sequence of the glgA gene. J. Biol. Chem., 261 (34), 16256-
16259.
Leung, P., and Preiss J. (1987) Cloning ADP glucose
pyrophosphorylase glgC with glycogen synthase glgA structural
genes from Salmonella-typhimurium. J. Bacteriol., 169 (9), 4349-
4354.
Raleigh, E.A., Lech, K., and Brent, R. (1989) Current Protocols
in Molecular Biology, Eds. Ausubel F.M. et al. Publishing
Associates and Wiley Interscience, New York, Unit 1.4
Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular
Cloning: A Laboratory Manual. Publd. Cold Spring Harbor, U.S.A.
Schagger and Von Jagow (1987). Tricine-SDS-Polyacrylamide gel
electrophoresis for the separation of proteins in the range from
1-100 kDA. Analy. Biochem., 166(2), 368-379.
Shewmaker, C.K; Boyer, C.D; Wiesenborn, D.P; Thompson, D.B;
Boersig, M.R; Oakes, J.V. (1994). Expression of Escherichia
coli glycogen synthase in the tubers of transgenic potatoes
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
41
(Solanum tuberosum) results in a highly branched starch. Pl.
Physiol, 104(4), 1159-1166.
Uttaro, A.D. and Ugalde, R.A. (1994) A chromosomal cluster of
genes encoding ADP-glucose synthetase, glycogen synthase and
phosphoglucomutase in Agrobacterium tumefaciens. Gene, 150 (1),
117-122.
Vasil, V., Castillo, A.M., Fromm, M.E. and Vasil, I.K. (1992).
Herbicide-resistant transgenic wheat plants obtained by
microprojectile bombardment of regenerable embryogenic callus.
Bio/Technology, 10(6), 667-674
Visser, G.F.; Stolte, A; Jacobsen, E, (1991) Expression of a
chimaeric granule bound starch synthase-GUS gene in transgenic
potato plants. P1. Mol. Biol, 17 (4), 691-699.
Visser, R.G.F.; Somhorst, I.; Kuipers, G.J.; Ruys, N.J.;
Feenstra, W.J.; Jacobsen, (1991a). Inhibition of the expression
of the gene for granule bound starch synthase in potato by
antisense constructs. Mol. Gen Genet., 225 (2), 289-296.
Materials Abbreviations
LB - Luria broth
TF - Tris-HC1, 1mM EDTA
SDS - sodium dodecyl sulphate
CTAB - cetyl trimethyl ammonium bromide
dATP - 2' - deoxy adenosine 5' triphosphate
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
42
dTTP - 2' - deoxy thymidine 5' triphosphate
dCTP - 2' - deoxy cytosine 5' triphosphate
dGTP - 2' - deoxy guanosine
DTT - dithiothreitol
ATP - adenosine 5' triphosphate
HEPES N-[2-hydroxyethyl] piperazine-N'-[2-ethanesulfonic
acid]
NBT - nitroblue tetrazolium
BCIP - 5-bromo-4-chloro-3-indolyl phosphate
GST - glutathione S transferase
NAD - nicotinamide adenine dinucleotide
IgG - immunoglobulin G
Mes - 2-[N-morpholino]ethane sulfonic acid
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
43
~~ ~~.4PEST TREATY ON THE INTERNATIONAL
RECOGNII yi:~ (11~ THE DEPOSIT OF MICROORGANISMS
FOR THE PIJi'.!'OSES OF PATENT PROCEDURE
Advanced Technologies (Cambridge) Ltd
INTERNATIONAL FORM
210 Cambridge Science Park, RECEIPT IN THE CASE OF AN ORIGINAL DEPOSIT
Cambridge. issued pursuant to Rule 7.1 by the
CB4 4WA INTERNATIONAL DEPOSITARY AUTHORITY
identified at the bottom of this page
NAME AND ADDRESS
OF DEPOSITOR
I. IDENTIFICATION OF THE MICROORGANISM
Identification reference given by the Accession number given by the
DEPOSITOR: INTERNATIONAL DEPOSITARY AUTHORITY:
Escherichia coli NCIMB 40962
(XLl Blue MRF' pDV03191 )
Il. SCIENTIFIC DESCRIPTION AND/OR PROPOSED TAXONOMIC DESIGNATION
The microorganism identified under I above was accompanied by:
a scientific description
a proposed taxonomic designation
(Mark with a cross where applicable)
IIL RECEIPT AND ACCEPTANCE
This International Depositary Authority accepts the microorganism identified
under I above, which was received by it on
4 August 1998 (date of the original deposit)I
IV. RECEIPT OF REQUEST FOR CONVERSION
The microorganism identified under 1 above was received by this International
Depositary Authority on
(date of the original deposit) and a request to convert the original deposit
to a deposit under the Budapest Treaty was received by it
on (date of receipt of request for conversion)
V. INTERNATIONAL DEPOSITARY AUTHORITY
Name: NCIMB Ltd., Signatures) of persons) having the power to represent the
International Depositary Authority or of authorised
ofFcial(s): '
~p~.~c~ ~ir.~-Cl
Address:23 St Machar Drive.
Aberdeen, Date: 19 August 1998
AB24 3RY.
Scotland.
Where Kule 6/4(a) appnes. such pate is the aa~e on wmcn me mama m imarnauonai
veposnary Ru~nonry way avyumcu.
Form BP/4 (sole pagc)
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
44
BUi:.aPFST TREATY ON THE li'v'r'ERNATIONAL
RECOGNITION ~:F THE DEPOSIT OF MICROORGANISMS
FOR THE PURi';.a>ES OF PATENT PROCEDURE
Advanced Technologies (Cambridge) Ltd.,
INTERNATIONAL FORM
210 Cambridge Science Park, VIABILITY STATEMENT
Cambridge. issued pursuant to Rule 10.2 by the
CB4 4WA INTERNATIONAL DEPOSITARY AUTHORITY
identified on the following page
NAME AND ADDRESS OF THE PARTY
TO WHOM THE VIABILITY STATEMENT
IS ISSUED
I. DEPOSTfOR II. IDENTIFICATION OF THE MICROORGANISM
Name: Accession number given by the
AS ABOVE INTERNATIONAL DEPOSITARY AUTHORITY:
Address: NCIMB 40962
Date of the deposit or of the transferl
4 August 1998
III. VIABILITY STATEMENT
The viability of the
microorganism identified
under II above was
tested on 8 August
1998 2. On that date,
the said
microorganism was:
3
viable
3
no longer viable
Indicate the date of the original deposit or, where a new deposit or a
transfer has been made, the most recent relevant date
(date of the new deposit or date of the uansfer).
In the cases referred to in Rule 10.2(a)(ii) and (iii), refer to the most
recent viability test.
Mark with a cross the applicable box.
Form BP/9 (first page)
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
IV. CONDITIONS UNDER WHICH
THE VIABILi i ~' ': ~' ST
HAS BEEN PERFORMEb4
y. INTERNATIONAL DEPOSITARY
AUTHORITY
Name: NCIMB Ltd., Signatures) of persons) having
the power
to represent the International
Depositary
Address: 23 St Machar Drive,Authority or of authorised official(s):
Aberdeen,
A24 3RY,
Scotland. Date: 19 August 1998
Fill in if the information has been requested and if the results of the test
were negative.
Form BP/9 (second and last page)
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
1
SEQUENCE LISTING
<W~.> Auvanced Technologies (Cambridge) Limited
<120> Genetically Modified Plants with altered Starch
<130> RD-ATC-20
<140>
<141>
<160> 10
<170> PatentIn Ver. 2.1
<210> 1
<211> 1467
<212> DNA
<213> ArtificialSequence
<220>
<221> CDS
<222> (1)..(171)
<223> Pea
ssu transit
peptide
<220>
<221> CDS
<222> (172)..(1467)
<223> E. coligCl6
gl
<220>
<223> Description Artifici..l Sequence:
of Pea
ssu
TP
linked to E. g1gC16 CDS
coli
<400> 1
atg get tct atatcctct tcagetgtg actacagtc agccgtget 48
atg
Met Ala Ser IleSerSer SerAlaVal ThrThrVal SerArgAla
Met
1 5 10 15
tct acg gtg tcggccgcg gtggetcca ttcggcggc ctcaaatcc 96
caa
Ser Thr Val SerAlaAla ValAlaPro PheGlyGly LeuLysSer
Gln
20 25 30
atg act gga ccagttaag aaggtcaac actgacatt acttccatt 144
ttc
Met Thr Gly ProValLys LysValAsn ThrAspIle ThrSerIle
Phe
35 40 45
aca agc aat ggaagagta aagtgcatg cttagttta gagaagaac 192
ggt
Thr Ser Asn GlyArgVal LysCysMet LeuSerLeu GluLysAsn
Gly
50 55 60
gat cac tta ttggcgcgc cagctgcca ttgaaatct gttgccctg 240
atg
Asp His Leu LeuAlaArg GlnLeuPro LeuLysSer ValAlaLeu
Met
65 70 75 80
ata ctg gcg ggacgtggt acccgcctg aaggattta accaataag 288
gga
Ile Leu Ala GlyArgGly ThrArgLeu LysAspLeu ThrAsnLys
Gly
85 90 95
cga gca aaa ccg gcc gta cac ttc ggc ggt aag ttc cgc att atc gac 336
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
2
Arg Ala Lys Pro Ala Val His Phe Gly Gly Lys Phe Arg Ile i-ie Asp
100 105 110.
ttt gcg ctg tct aac tgc atc aac tcc ggg atc cgt cgt atg ggc gtg 384
Phe Ala Leu Ser Asn Cys Ile Asn Ser Gly Ile Arg Arg Met Gly Val
115 120 125
atc acc cag tac cag tcc cac act ctg gtg cag cac att cag cgc ggc 432
Ile Thr Gln Tyr Gln Ser His Thr Leu Val Gln His Ile Gln Arg Gly
130 135 140
tgg tca ttc ttc aat gaa gaa atg aac gag ttt gtc gat ctg ctg cca 480
Trp Ser Phe Phe Asn Glu Glu Met Asn Glu Phe Val Asp Leu Leu Pro
145 150 155 160
gca cag cag aga atg aaa ggg gaa aac tgg tat cgc ggc acc gca gat 528
Ala Gln Gln Arg Met Lys Gly Glu Asn Trp Tyr Arg Gly Thr Ala Asp
165 170 175
gcg gtc acc caa aac ctc gac att atc cgt cgt tat aaa gcg gaa tac 576
Ala Val Thr Gln Asn Leu Asp Ile Ile Arg Arg Tyr Lys Ala Glu Tyr
180 185 190
gtg gtg atc ctg gcg ggc gac cat atc tac aag caa gac tac tcg cgt 624
Val Val Ile Leu Ala Gly Asp His Ile Tyr Lys Gln Asp Tyr Ser Arg
195 200 205
atg ctt atc gat cac gtc gaa aaa ggt gta cgt tgt acc gtt gtt tgt 672
Met Leu Ile Asp His Val Glu Lys Gly Val Arg Cys Thr Val Val Cys
210 215 220
atg cca gta ccg att gaa gaa gcc tcc gca ttt ggc gtt atg gcg gtt 720
Met Pro Val Pro Ile Glu Glu Ala Ser Ala Phe Gly Val Met Ala Val
225 230 235 240
gat gag aac gat aaa act atc gaa ttc gtg gaa aaa cct get aac ccg 768
Asp Glu Asn Asp Lys Thr Ile Glu Phe Val Glu Lys Pro Ala Asn Pro
245 250 255
ccg tca atg ccg aac gat ccg agc aaa tct ctg gcg agt atg ggt atc 816
Pro Ser Met Pro Asn Asp Pro Ser Lys Ser Leu Ala Ser Met Gly Ile
260 265 270
tac gtc ttt gac gcc gac tat ctg tat gaa ctg ctg gaa gaa gac gat 864
Tyr Val Phe Asp Ala Asp Tyr Leu Tyr Glu Leu Leu Glu Glu Asp Asp
275 280 285
cgc gat gag aac tcc agc cac gac ttt ggc aaa gat ttg att ccc aag 912
Arg Asp Glu Asn Ser Ser His Asp Phe Gly Lys Asp Leu Ile Pro Lys
290 295 300
atc acc gaa gcc ggt ctg gcc tat gcg cac ccg ttc ccg ctc tct tgc 960
Ile Thr Glu Ala Gly.Leu Ala Tyr Ala His Pro Phe Pro Leu Ser Cys
305 310 315 320
gta caa tcc gac ccg gat gcc gag ccg tac tgg cgc gat gtg ggt acg 1008
Val Gln Ser Asp Pro Asp Ala Glu Pro Tyr Trp Arg Asp Val Gly Thr
325 330 335
ctg gaa get tac tgg aaa gcg aac ctc gat ctg gcc tct gtg gtg ccg 1056
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
3
Leu Glu Ala Tyr Trp Lys Ala Asn Leu Asp Leu A~a Ser Val Val Pro
340 345 . 350
aaa ctg gat atg tac gat cgc aat tgg cca att cgc acc tac aat gaa 1104
Lys Leu Asp Met Tyr Asp Arg Asn Trp Pro Ile Arg Thr Tyr Asn Glu
355 360 365
tca tta ccg cca gcg aaa ttc gtg cag gat cgc tcc ggt agc cac ggg 1152
Ser Leu Pro Pro Ala Lys Phe Val Gln Asp Arg Ser Gly Ser His Gly
370 375 380
atg acc ctt aac tca ctg gtt tcc gac ggt tgt gtg atc tcc ggt tcg 1200
Met Thr Leu Asn Ser Leu Val Ser Asp Gly Cys Val Ile Ser Gly Ser
385 390 395 400
gtg gtg gtg cag tcc gtt ctg ttc tcg cgc gtt cgc gtg aat tca ttc 1248
Val Val Val Gln Ser Val Leu Phe Ser Arg Val Arg Val Asn Ser Phe
405 410 415
tgc aac att gat tcc gcc gta ttg tta ccg gaa gta tgg gta ggt cgc 1296
Cys Asn Ile Asp Ser Ala Val Leu Leu Pro Glu Val Trp Val Gly Arg
420 425 430
tcg tgc cgt ctg cgc cgc tgc gtc atc gat cgt get tgt gtt att ccg 1344
Ser Cys Arg Leu Arg Arg Cys Val Ile Asp Arg Ala Cys Val Ile Pro
435 440 445
gaa ggc atg gtg att ggt gaa aac gca gag gaa gat gca cgt cgt ttc 1392
Glu Gly Met Val Ile Gly Glu Asn Ala Glu Glu Asp Ala Arg Arg Phe
450 455 460
tat cgt tca gaa gaa ggc atc gtg ctg gta acg cgc gaa atg cta cgg 1440
Tyr Arg Ser Glu Glu Gly Ile Val Leu Val Thr Arg Glu Met Leu Arg
465 470 475 480
aag tta ggg cat aaa cag gag cga taa 1467
Lys Leu Gly His Lys Gln Glu Arg
485
<210> 2
<211> 488
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Pea ssu TP
linked to E. coli g1gC16 CDS
<400> 2
Met Ala Ser Met Ile Ser Ser Ser Ala Val Thr Thr Val Ser Arg Ala
1 5 10 15
Ser Thr Val Gln Ser Ala Ala Val Ala Pro Phe Gly Gly Leu Lys Ser
20 25 30
Met Thr Gly Phe Pro Val Lys Lys Val Asn Thr Asp Ile Thr Ser Ile
35 40 45
Thr Ser Asn Gly Gly Arg Val Lys Cys Met Leu Ser Leu Glu Lys Asn
50 55 60
Asp His Leu Met Leu Ala Arg Gln Leu Pro Leu Lys Ser Val Ala Leu
65 70 75 80
Ile Leu Ala Gly Gly Arg Gly Thr Arg Leu Lys Asp Leu Thr Asn Lys
85 90 95
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
4
Arg Ala Lys Pro Ala Val His Phe Gly Glr Lys Phe Arg Ile Ile Asp
100 105 110
Phe Ala Leu Ser Asn Cys Ile Asn Ser Gly Ile Arg A~5 Met Gly Val
115 120 125
Ile Thr Gln Tyr Gln Ser His Thr Leu Val Gln His Ile Gln Arg Gly
130 135 140
Trp Ser Phe Phe Asn Glu Glu Met Asn Glu Phe Val Asp Leu Leu Pro
145 150 155 160
Ala Gln Gln Arg Met Lys Gly Glu Asn Trp Tyr Arg Gly Thr Ala Asp
165 170 175
Ala Val Thr Gln Asn Leu Asp Ile Ile Arg Arg Tyr Lys Ala Glu Tyr
180 185 190
Val Val Ile Leu Ala Gly Asp His Ile Tyr Lys Gln Asp Tyr Ser Arg
195 200 205
Met Leu Ile Asp His Val Glu Lys Gly Val Arg Cys Thr Val Val Cys
210 215 220
Met Pro Val Pro Ile Glu Glu Ala Ser Ala Phe Gly Val Met Ala Val
225 230 235 240
Asp Glu Asn Asp Lys Thr Ile Glu Phe Val Glu Lys Pro Ala Asn Pro
245 250 255
Pro Ser Met Pro Asn Asp Pro Ser Lys Ser Leu Ala Ser Met Gly Ile
260 265 270
Tyr Val Phe Asp Ala Asp Tyr Leu Tyr Glu Leu Leu Glu Glu Asp Asp
275 280 285
Arg Asp Glu Asn Ser Ser His Asp Phe Gly Lys Asp Leu Ile Pro Lys
290 295 300
Ile Thr Glu Ala Gly Leu Ala Tyr Ala His Pro Phe Pro Leu Ser Cys
305 310 315 320
Val Gln Ser Asp Pro Asp Ala Glu Pro Tyr Trp Arg Asp Val Gly Thr
325 330 335
Leu Glu Ala Tyr Trp Lys Ala Asn Leu Asp Leu Ala Ser Val Val Pro
340 345 350
Lys Leu Asp Met Tyr Asp Arg Asn Trp Pro Ile Arg Thr Tyr Asn Glu
355 360 365
Ser Leu Pro Pro Ala Lys Phe Val Gln Asp Arg Ser Gly Ser His Gly
370 375 380
Met Thr Leu Asn Ser Leu Val Ser Asp Gly Cys Val Ile Ser Gly Ser
385 390 395 400
Val Val Val Gln Ser Val Leu Phe Ser Arg Val Arg Val Asn Ser Phe
405 410 415
Cys Asn Ile Asp Ser Ala Val Leu Leu Pro Glu Val Trp Val Gly Arg
420 425 430
Ser Cys Arg Leu Arg Arg Cys Val Ile Asp Arg Ala Cys Val Ile Pro
435 440 445
Glu Gly Met Val Ile Gly Glu Asn Ala Glu Glu Asp Ala Arg Arg Phe
450 455 460
Tyr Arg Ser Glu Glu Gly Ile Val Leu Val Thr Arg Glu Met Leu Arg
465 470 475 480
Lys Leu Gly His Lys Gln Glu Arg
485
<210> 3
<211> 421
<212> DNA
<213> Triticum aestivum
<220>
<221> promoter
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
<222> (1) . . (421)
<223> High Molecular Weight Giuc~nir_.Promoter Genomic
DNA
<400> 3
cccagctttg agtggccgta gatttgcaaa agcaatggct aacagacaca tattctgcca 60
aaccccaaga aggataatca cttttcttag ataaaaaaga acagaccaat atacaaacat 120
ccacacttct gcaaacaata catcagaact aggattacgc cgattacgtg gctttagcag 180
actgtccaaa aatctgtttt gcaaagctcc aattgctcct tgcttatcca gcttcttttg 240
tgttggcaaa ctgcgctttt ccaaccgatt ttgttcttct cgcgctttct tcttagccta 300
aacaaacctc accgtgcacg cagccatggt cctgaacctt cacctcgtcc ctataaaagc 360
ctagccaacc ttcacaatct tatcatcacc cacaacaccg agcaccacaa actagagatc 420
c 421
<210> 4
<211> 43
<212> DNA
<213> Artificial Sequence
<220>
<221> primer bind
<222> (1) . . (19)
<223> Primer to 3' end of ssu transit peptide
<220>
<221> primer bind
<222> (20) . . (43)
<223> Primer to 5' end of glgA CDS
<220>
<223> Description of Artificial Sequence: Oligo primer
<400> 4
tggtggaaga gtaaagtgca tgcaggtttt acatgtatgt tca 43
<210> 5
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<221> primer bind
<222> (1) . . (19)
<223> Primer to 3' end of glgC CDS
<220>
<221> primer bind
<222> (26) . . (54)
<223> Primer to 3' end of glgA CDS
<220>
<223> Description of Artificial Sequence: Oligo primer
<400> 5
tcgctcctgt ttatgcccta gatctctatt tcgagcgata gtaaagctca cggt 54
<210> 6
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
6
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Oligo primer
<220>
<221> primer bind
<222> (11)..(33)
<223> Primer to 5' end of ssu transit peptide
<400> 6
acgtagatct atggcttcta tgatatcctc ttc 33
<210> 7
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Oligo primer
<220>
<221> primer bind
<222> (10) . . (36)
<223> Primer to 5' end of HMWG promoter
<400> 7
gacatcgatc ccagctttga gtggccgtag atttgc 36
<210> 8
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Oligo primer
<220>
<221> primer bind
<222> (10) . . (39)
<223> primer to 3' end of HMWG promoter
<400> 8
gacgaattcg gatctctagt ttgtggtgct cggtgttgt 39
<210> 9
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<221> primer bind
<222> (9) . . (32)
<223> Primer to 5' end of nopaline synthase terminator
CA 02365279 2001-08-28
WO 00/55331 PCT/GB00/00848
7
<220>
<223> Description of Artificial Sequence: Oligo primer
<400> 9
caggatccga atttcacccg atcgttcaaa ca 32
<210> 10
<211> 50
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Oligo primer
<220>
<221> primer bind
<222> (23)..(50)
<223> Primer to 3' end of nopaline synthase terminator
<400> 10
gacccgcggc tcgaggcggc cgcccgatct agtaacatag atgacaccgc 50