Note: Descriptions are shown in the official language in which they were submitted.
.
CA 02366129 2001-12-21
-1-
IATORS OF EXTRACEL,I:ULAR MATRIX REMODELING
BACKGROUND OF THE INVENTION
Implantation and Placentation in the Human
Formation and Organisation of the Human Placenta.
The first step in human implantation involves the attachment of embryonic
trophoblasts to the surface epithelium of the endometrium (1,13). After this
initial interaction, the trophoblasts proliferate, invade the underlying
stroma,
and differentiate into chorionic villi which are composed of two layers: the
inner
cell layer which is comprised of mitoticatly active cytotrophoblasts and the
outer syncytial trophoblast which is a terminally differentiated
multinucleated
cell formed by the fusion of post-mitotic cytotrophoblasts. Subsequently, the
cytotrophoblasts at the tips of the villi proliferate and rupture the outer
syncytial
trophoblast to form cellular columns that extend into the decidua (14). These
extravillous cytrotrophoblast columns are believed to anchor the placenta to
the decidua. Cytotrophoblasts dissociate from the extravillous columns and
invade deeply into the maternal decidua. Some of these cytotrophoblasts will
penetrate the basal lamina of the uterine arteries and replace the endothelial
and smooth muscle cells of these blood vessels, thereby ensuring a
continuous blood supply to the develmping fetus (15). The remainder of the
invasive cytotrophoblasts undergo differentiation and fusion to form placental
bed giant cells, large multinucleated cells that lie in intimate contact with
the
thick ECM surrounding decidual cells (15,16). Unlike tumor cells, the invasion
of trophoblasts into the underlying maternal tissues is highly regulated.
Cyclic Remodeling of the Human Endometrium
The epithelial and stromal cells of the human endometrium undergo cyclic
proliferation, differentiation and shedding in response to the gonadal
steroids,
17(i-estradiol (E2) and progesterone (P4) (10-13). After menstruation, the
CA 02366129 2001-12-21
-2-
endometrium regenerates under the influence of E2 to produce a dense
cellular stroma containing narrow tubular glands. Following ovulation, under
the influence of P4, glandular secretion and the decidualisation become the
predominant features of the endometrium. The decidua is believed to fulfill
paracrine, nutritional, immunoregulatory and embryoregulatory functions
throughout pregnancy (6). in the absence of pregnancy, decidual cells
surrounding the spiral arterioles are well positioned to mediate the vascular
disruption and rapid tissue degradation associated with menstruation.
Celiular Mechanisms underlying Im_piantation and Placentation
Remodeling of the ECM of is a key event underlying the morphological and
functional maturation of the human endometrium. For example,
decidualisation involves the transformation of the interstitial type ECM
surrounding the stromal cells, which is rich in collagen types I, III, V and
VI, to
a basement membrane-like ECM in which in the predominant structural
components are laminin, fibronectin, heparan sulfate proteoglycan and
collagen type V. Alterations in the composition of the ECM mediate the
terminal differentiation of endometrial stromal cells into decidua and
regulates
trophoblast invasion into the underlying maternal tissues.
Matrix metalloproteinase (MMPs) play a key role in maintaining the balance
between the deposition and degradation of the endometrial ECM. MMP-2, -3,
-7, -9 and -11 are spatiotemporally expressed in the endometrium during the
menstrual cycle(19-22). In addition, human trophoblastic cells have also been
shown to produce several MMPs, including MMP-1, -2, -3, -7, -9, -11, and -14
in vitro and in vivo (22-28). However, MMP-2 and MMP-9 are believed to be
the key regulators of extravillous cytotrophoblast invasion. For example,
MMP-9 function-perturbing antibodies are capable of reducing the invasive
capacity of these cells in vitro (24). The production of MMP-9 is down-
regulated during the third trimester of pregnancy, paralleling the decline in
trophoblast invasiveness associated with gestational age (25). Reduced
MMP-9 activity has also been detected in cytotrophoblasts isolated from
CA 02366129 2001-12-21
-3-
placentae obtained from women with preeclampsia, a disease in which
trophoblast invasion is compromised (27). In addition to promoting
trophoblast invasion, MMP-2 and -9 which are also secreted by the syncytial
trophoblast play roles) in the formation of the chorionic villous(28).
TIMPs are the major endogenous regulators of MMP proteolytic activity (29-
30). TIMP-1, -2, and -3 are produced by human trophoblastic cells and
decidual cells, they have a role in autocrine and paracrine regulation of MMP
activity during human implantation (14, 24, 31). TIMP-1, -2 and -3 have been
shown to be capable of reducing trophoblast invasion in vitro (14, 24).
ADAMS
The ADAMs (A Disintegrin And Metalloproteinase) are a gene family of
transmembrane proteins that contain a disintegrin and a metalloprotease
domain(33). Thus, the ADAMs have the potential to act as adhesion
molecules and/or proteinases. Two generic functions have been proposed for
the ADAM proteases:
(1) local activation of signalling pathways by the shedding of cell
surface cytokines and growth factors; and
(2) cell migration/invasion by the degradation of the ECM(34, 35).
Although 5 ADAM subtypes (ADAMs-9, -10, 12, 17, and -28) have been
shown to act as metalloproteases in vitro, only ADAMS-9, -10, and -17 are
known to be catalytically active in vivo. In particular ADAM-9 is responsible
for
the shedding of HB-EGF from cultured cells(36), ADAM-10 acts as a
sheddase in the Nofch signalling pathway(37-40) and ADAM-17 is involved in
multiple ectodomain-shedding events, most notably the release of
TNFa (41-42).
ADAM-10 and snake venom metalloproteases (SVMPs), the closest relatives
of ADAMs, have been shown to cleave purified ECM components in vitro(43-
CA 02366129 2001-12-21
44). ADAM-9 has been shown to promote the migration of fibroblasts in vitro
whereas(46) and ADAM-13 expression has been detected in cranial neural-
crest cells, a highly migratory population of cells in the Xenopus embryo(47).
However, there is currently no direct evidence linking ADAM protease activity
with the adhesive and migratory behaviour of specific cell populations in
vivo.
Recent cloning studies have identified new members of the ADAM family,
known as ADAMTS, in C elegans, Drosophila and mammals(6-7). In contrast
to the ADAMS, ADAMTS are secreted proteins which do not contain the
cysteine rich, EGF-like, transmembrane and cytoplasmic domains
characteristic of the other members of the ADAM gene family. To date, 12
members of the ADAMTS subfamily have been identified in the human:
ADAMTS-1,-2,-3,-4,-5,-6,-7,-8,-9,-10,-12 and von ~Ilebrand factor-cleaving
protease (48-54).
Structural and Functional Organisation of ADAMTS Subtypes
ADAMTS are characterised by four structural and functional subunits: an
amino terminal prodomain, a catalytic domain, a disintegrin-like domain, and
an ECM binding domain (which is composed of a central thrombospondin
(TSP) type 1 motif, a spacer region and a variable number of TSP-like motifs)
at the carboxy terminal end of the protein (6-7, 48-54). Overall, the
predicted
mature forms of the ADAMTS proteins exhibit 20-4.0% similarity to one
another.
Prnrinmain
The prodomain of the distinct ADAMTS subtypes vary in length but exhibit
short stretches of homology that correspond to consensus sequences. In
particular, the prodomains of the ADAMTS subtypes contain a Cys-switch
motif, found in MMPs and SVMPs, that is involved in maintaining the en2yme
in its latent form(55). A conserved furin activation site has also been
identified
in the prodomains of the ADAMTS. This putative cleavage site is located
CA 02366129 2001-12-21
-5-
immediately prior to the amino terminal sequence of the mature protein
suggesting that unlike MMPs, the prodomains of the ADAMTS subtypes are
proteolytically cleaved by these endopeptidases in the Golgi apparatus and
the proteins secreted in their active form(54, 57-58).
Catalytic Domain
The catalytic domain of ADAMTS contains consensus sequences also found
in the proteolytic domains of MMPs, SVMPs, or ADAMs(59). To date, the
specific substrates) of many ADAMTS subtypes have not been identified.
However, ADAMTS-4 and -8 have been shown to degrade aggrecan, a large
chondroitin sulphate found in cartilage(60-61). ADAMTS-4 is also capable of
degrading versican and the brain specific ECM protein, BEHAB(62-63).
Procollagens-I and -II have been identified as substrates for ADAMTS-1, -2
and -3(64-65). Finally, the metalloprotease domain of ADAMTS-12 has been
shown to be proteolytically active using the a2-macroglobulin complex
formation assay(54, 66).
Disintegrin Domain
The disintegrin domain is located immediately after the catalytic domain of
the
ADAMTS. This region shows limited homology to the disintegrin domains of
ADAMs and SVMPs(48-54). To date, the biological functions) of this distinct
ADAMTS domain is not known.
ECM Binding Domain
The ECM Binding Domain contains an internal thrombospondin type 1 (TSP-
1 ) motif that has two conserved regions, one of which is involved in the
binding sulphated glycosaminoglycan chains of heparin, heparan sulfate and
chondroitin sulphate and the other in the binding of the thrombospondin
receptor, CD36 (67-69). A spacer region, of variable length, separates this
internal TSP motif from the TSP-1 repeats located at the carboxyl end of the
protein. The spacer region exhibits the least sequence homology between the
distinct members of the ADAMTS subfamily. The number of TSP-1 motifs at
CA 02366129 2001-12-21
-6-
the carboxy terminal of the protein is also highly variable among the ADAMTS
subtypes (48-54). The biological significance of these structural variations
is
not known.
The TSP-1 motifs and the spacer region are required for ADAMTS-ECM
interactions. The deletion of the central or the 2 carboxyl terminal TSP
motifs
of ADAMTS-1 have been shown to inhibit binding to ECM(57). The truncation
of the spacer region also significantly reduces the ability of the mature
protein
to interact with the ECM. In contrast, deletion mutants corresponding to
either
the carboxy terminal TSP-1 or spacer region of ADAMTS-1 were capable of
forming tight interactions with the ECM.
Truncated forms of ADAMTS-4 lacking the spacer region and/or TSP-1 motifs
did not exhibit any protease activity in vitro (61 ). Similarly, peptides
corresponding to different regions of the TSP-1 motif and/or spacer region of
ADAMTS-4 reduced the cleavage of its substrate, aggrecan, in a dose-
dependent manner.
Tissue Distribution of the ADAMTS subtypes
ADAMTS subtypes have been detected in human adult and fetal tissues (56-
59; Tables 1 and 2, respectively). In addition, ADAMTS-1,-2, -3 and -8 mRNA
levels have been shown to be highly regulated during the early stages of
embryonic development of the mouse (70; Table 3).
The Cell Biology of the ADAMTS
Recent gene knockout studies have highlighted the important biological roles
that members of the ADAMTS gene family play in embryonic development
and tissue morphogenesis (71-72).
ADAMTS-1 gene knockout mice exhibit growth retardation and aberrant
development of the kidneys, adrenal glands and urogenital tract.
Abnormalities in the ovaries and uterine tissues of female mice null mutant
for
CA 02366129 2001-12-21
-7-
ADAMTS-1 were also observed. In particular, the ovaries of these mice
contained fewer mature follicles(73).
Mice null mutant for ADAMTS-2 did not exhibit any abnormal phenotype at
birth. However, as these mice matured, their skin became fragile. At the
structural level, the skin of these mice mimicked the defects described for
the
connective tissue disorder known as dermatosparaxis in animals and Ehlers-
Danlos syndrome in humans(74-75). Furthermore, male mice null mutant for
ADAMTS-2 were infertile.
Aberrant expression of ADAMTS subtypes has also been associated with the
pathogenesis of disease. For example, ADAMTS-12 mRNA levels were found
to be significantly higher in colorectal, renal and pancreatic carcinomas than
matched normal tissues (54). Similarly, increased ADAMTS-4 expression and
proteolytic activity have been detected in gliomas of the CNS (65). ADAMTS-4
and -5 have also been shown to play key roles in the degradation of cartilage
during the progression of arthritic diseases(76). Finally, it has been
proposed
that ADAMTS-4 is involved in the degradation of the ECM in the brains of
patients diagnosed with Alzheimer's disease (77).
Regulation of ADAMTS Expression
The factors capable of regulating ADAMTS expression remain poorly
characterised. TGF-X31 but not TGF-a, It_-a, IL-1 Vii, aFGF or EGF increased
ADAMTS-12 mRNA levels in human fetal fibroblasts (54). Lipoplysaccharides
were shown to increase ADAMTS-1 mRNA levels in renal and cardiac tissues
of adult mice whereas P4 appears to be a key regulator of this ADAMTS
subtype in the rat ovary (57, 78).
CA 02366129 2001-12-21
-$-
Regulation of ADAMTS Activity
Proteolytic Modification
Recent studies show that ADAMTS-1, -4 and -12 undergo a second
proteolytic cleavage, resulting in the formation of an amino terminal fragment
containing the metalloproteinase, disintegrin-like and the central TSP-1
domains of the mature proteins (54, 58, 60). The truncated forms of these
proteins have modified adhesion properties. For example, the amino terminal
fragment of ADAMTS-1 binds to the ECM with a lower affinity and is less
effective at regulating the proliferation of endothelial cells than the mature
form(58). Carboxyl terminal fragments of ADAMTS-4 and -12, are also
capable of binding to the ECM(54, 60).
Endogenous Regulatory Factors: TIMP-3, but not TIMPs-1,-2, or -4, has been
shown to be a potent inhibitor of recombinant ADMTS-4 and ADAMTS-5 in
vitro (79). To date, the molecular mechanisms) by which TIMP-3 inhibit these
two ADAMTS subtypes are not known. TIMP-3 is capable of binding to the
ECM(80).
Papilin is an ECM glycoprotein, identified in Drosophila, C elegans and
mammals, which inhibits the proteolytic activity of ADAMTS-1 in vitro. Papilin
shares a set of protein domains (the papilin cassette) with the ADAMTS (81 ).
In particular, the papilin cassette contains a complete TSP type 1 repeat, a
spacer region, and six incomplete TSP type 1 domains. In binding assays,
papilin was capable of interacting with ADAMTS-1 and the enzyme-substrate
complex, but does not compete with procollagen for the catalytic site of this
proteinase.
Alterations in the expression levels of papilin have a profound effect on the
development of the Drosphila embryo(81 ). For example, overexpression of
papilin resulted in the aberrant development of the Malphigian tubules,
muscle, trachea, CNS, and death of these embryos. Similarly, inhibition of
papilin expression in adult C elegans significantly reduced brood sizes and
CA 02366129 2001-12-21
_g_
often caused the death of the parental animal(81 ). The small number of
offspring, which survived were infertile. The mechanisms by which papilin
modulates these morphogenetic events are not known. Furthermore, the
biological roles) of papilin in mammalian tissue morphogenesis has not been
determined.
SUMMARY OF THE INVENTION
This invention includes novel peptides that mediate cellular invasion.
Cellular
invasion can be present in both disease and non-disease related conditions
as in the case of cancer and in the case of pregnancy. This invention also
includes the use of these peptides to modulate cellular invasion and
pharmaceutically acceptable formulations comprising the peptide, which may
be administered for the treatment of both disease and non-disease states
involing cellular invasiveness. These peptides are further defined as the
lacking a metalloproteinase domain and/or a disintigrin domain, but including
a Thrombospondin Type-1 domain.
Other aspects and features of the present invention will become apparent to
those ordinarily skilled in the art upon review of the following description
of
specific embodiments of the invention in conjunction with the accompanying
fig a res.
BRIEF DESCRIPTION OF THE DRAWINGS
LIST OF FIGURES
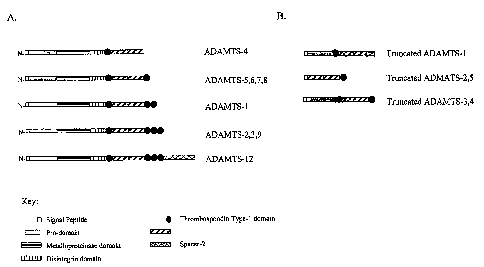
Figure 1 shows schematic diagrams of the structures of the distinct
ADAMTS (panel A) and trun-ADAMTS subtypes (panel B).
Figure 2 is an autoradiogram of Southern blots containing a RT-PCR
products generated using total RNA extracted from first
trimester placenta (lane A) or deciduas parietalis (lane B) and
CA 02366129 2001-12-21
-10-
oligonucleotides specific for ADAMTS-1, -2, -3, -5, -7, -9 or
GAPDH as primers.
Figure 3 is an autoradiogram of Southern blots containing a RT-PCR
products generated using total RNA extracted from primary
cultures of highly extravillous cytotrophoblasts (lane A) or
poorly invasive JEG-3 choriocarcinoma cells (lane B) and
oligonucleotides specific for ADAMTS-1, -2, -4, -5, -6, -10, -12
or GAPDH as primers.
Figure 4 is a nucleotide sequence and predicted amino acid sequence
of trun-ADAMTS-1. The conserved TSP-1 motifs are
underlined. Putative glycosylation sites are marked with an
asterisk.
Figure 5 is a nucleotide sequence and predicted amino acid sequence
of trun-ADAMTS-2. The conserved TSP-1 motifs are
underlined. Putative glycosylation sites are marked with an
asterisk.
Figure 6 is a nucleotide sequence and predicted amino acid sequence
of trun-ADAMTS-3. The conserved TSP-1 motifs are
underlined. Putative glycosylation sites are marked with an
asterisk.
Figure 7 is a nucleotide sequence and predicted amino acid sequence
of trun-ADAMTS-4. The conserved TSP-1 motifs are
underlined. Putative glycosylation sites are marked with an
asterisk.
Figure 8 is a nucleotide sequence and predicted amino acid sequence
of trun-ADAMTS-5. The conserved TSP-1 motifs are
underlined. Putative glycosylation sites are marked with an
asterisk.
Figure 9 is a partial nucleotide sequence of trun-ADAMTS-6 isolated from
human decidual tissues.
CA 02366129 2001-12-21
-11-
Figure 10 is a partial nucleotide sequence of trun-ADAMTS-7 isolated
from human decidual tissues.
Figure 11 shows a photomicrograph of an ethidium bromide stained gel
containing RT-PCR products generated from a pallet of
human tissues using primers specific for trun-ADAMTS-5 (b).
The template cDNAs were synthesised from poly A+ isolated
from the human brain (1 ), colon (2) kidney (3), liver (4),
pancreas (5), skeletal muscle (6), testis (7), heart (8),
peripheral blood leukocytes (9), ovary (10), mammary gland
(11 ), placenta (12), prostate (13), and skin (14).
Figure 12 is an autoradiogram of an SDS-polyacrylamide gel containing
radiolabeled protein products generated from pc-AT5 (lane A)
or an empty pcDNA3 vector (lane B) using an in vitro
transcription/translation kit.
Figure 13 is an autoradiogram of a Western blot containing total protein
lysates prepared from the ECM, cells or conditioned medium
of COS-7 cells transfected with pc-AT5 and probed with a
monoclonal antibody directed against the V5-epitope of the
recombinant protein (lanes A-C, respectively). Protein lystaes
extracted from the ECM and cells of COS-7 cells transfected
with an empty pcDNA3 vector (lanes D and E, respectively)
served as controls for these studies. The Amersham ECL
system was used to detect antibody bound to antigen.
Figure 14 is a bar graph representing the results obtained from EVT
invasion assay. EVT invasion into Matrigel containing ECM
deposited by untransfected COS-7 cells was considered to be
100%. The ECM deposited by COS-7 cells transfected by pc
AT5 reduced trophoblast invasion by 55% whereas those
deposited by cells transfected with pc-ATScon or trun
ADAMTS-1 had no significant effect. Values are the means of
four independent experiments. Error bars represent s.e.m.
CA 02366129 2001-12-21
-12-
LIST OF TABLES
Table 1 shows the tissue distribution of the distinct ADAMTS subtypes
in adult human tissues.
Table 2 shows the tissue distribution of the distinct ADAMTS subtypes
in human fetal tissues.
Table 3 shows the expression of the distinct ADAMTS subtypes during
the mouse embryonic development.
Table 4 shows the nucleotide sequences of the degenerate primers
and PCR conditions used for the identification of distinct
ADAMTS subtypes in human tissues and cells.
Table 5 shows the ADAMTS subtypes present in first trimester
placenta and decidual tissues.
Table 6 shows the primer sequences and PCR conditions for the
semiquantitative analysis of the distinct ADAMTS subtypes.
Table 7 shows the nucleotide and amino acid sequence homology
between the trun-ADAMTS, papilin and ADAMTS subtypes.
Table 8 shows the distribution of the distinct trun-ADAMTS subtypes
in the human genome.
Table 9 shows the primer sequences and PCR conditions for the
generation of a cDNA corresponding to the coding region of
trun-ADAMTS-5.
Table 10 shows the I.M.A.G.E. clones corresponding to the TSP-1 and
spacer regions of selected ADAMTS subtypes.
DETAILED DESCRIPTION
The following description describes a first embodiment of the present
invention.
CA 02366129 2001-12-21
-13-
Identification of the ADAMTS subtypes present in first trimester placenta
and decidual tissues.
Degenerate oligonucleotides corresponding to two conserved regions,
identified in the carboxyl terminal regions of all of the known ADAMTS
subtypes, were prepared and used as primers in a reverse transcription-
polymerase chain reaction (RT-PCR). The primers sequences and PCR
conditions used are listed in Table 4. Template cDNAs were synthesised
from total RNA extracted from first trimester human placenta and decidua
parietalis. The resultant RT-PCR products (400-650 bp) were subcloned into
the RT-PCR II vector (Invitrogen) and subjected to DNA sequence analysis.
Multiple ADAMTS subtypes were detected in the total RNA extracts prepared
from the human placental and decidual tissues (Table 5). Surprisingly, a
similar repertoire of ADAMTS subtypes was detected in these two
reproductive tissues. However, semiquantitative RT-PCR using primers
specific for each of these ADAMTS subtypes or the housekeeping gene,
GAPDH (Table6), followed by Southern blotting demonstrated that the levels
of mRNA transcripts encoding these proteases were significantly higher in the
total RNA extracts prepared from the human placenta (Fig. 2).
Identification of the ADAMTS subtypes present in invasive and non-
invasive human trophoblastic cells: We examined the repertoire of
ADAMTS present in primary cultures of highly invasive extravillous
cytotrophoblasts (EVTs) and the poorly invasive JEG-3 choriocarcinoma cell
line by degenerate and semi-quantitative RT-PCR using the conditions and
primers described above.
ADAMTS-1 mRNA transcripts were readily detectable in invasive and non-
invasive trophoblastic cells. In contrast, to the extravillous
cytotrophoblasts
which only ADAMTS-6 and -9 mRNA transcripts were detected in JEG-3
cells. The differential expression of distinct ADAMTS subtypes is associated
CA 02366129 2001-12-21
-14-
with the differentiation of human trophoblastic cells along the invasive and
non-invasive pathways. (Fig. 3)
Identification of Novel, Truncated ADAMTS Subtypes in Human Decidua.
In addition to the known ADAMTS subtypes, the partial cDNA sequences of 7
novel members of the ADAMTS gene family were detected in total RNA
extracted from the decidua. To date, full length cDNA clones corresponding to
5 of these novel ADAMTS subtypes have been isolated from a human uterus
5'-stretch cDNA library using a CIonCapture cDNA selection kit (Clontech;
Figs 4-7). Sequence homology and the identification of putative open reading
frames and potentially important consensus of these clones were performed
using computer software provided by the NCBI (Blast and ORF Finder) or the
Swiss Institute of Bioinformatics (Expert Protein Analysis System; ExPASy).
DNA and amino acid sequence analysis demonstrated that each of these
novel ADAMTS subtypes exhibited significant homology to a known ADAMTS
subtypes and the ECM protein, papilin (Figs 4-10). Further analysis
demonstrated that these full length cDNAs contained the central and/or
carboxyl-terminal TSP-1 motifs characteristic of the ADAMTS but did not have
a disintegrin-like or a proteolytic domain (Fig. 1). These cDNAs represent a
unique ADAMTS subfamily which we have chosen to name trun-ADAMTs (for
the purpose of this application). Other features common of the known
ADAMTS and the trun-ADAMTS subtypes are 1 ) a putative signal peptide at
the amino terminus 2) the lack of a transmembrane domain, and 3) the
presence of at least one putative glycosylation site in the spacer region.
The Unigene Resource System (NCBI) was used to demonstrate that the
trun-ADAMTS subtypes are distinct gene products whose functions have yet
to be determined (Table 7). Importantly, there was no correlation between the
chromosomal locations of the trun-ADAMTS and ADAMTS subtypes. These
observations indicate that the trun-ADAMTS cDNAs do not simply represent
CA 02366129 2001-12-21
-15-
alternatively spliced forms of the mRNA transcripts encoding the functional
ADAMTS proteases.
To date, our studies have focused on one of the novel trun-ADAMTS
subtypes identified in decidua, trun-ADAMTS-5. This trun-ADAMTS subtypes
exhibits significant homology to ADAMTS-10, one of the predominant
ADAMTS subtypes present in EVTs.
Tissue Distribution of dun-ADAMTS-5. PCR was performed using primers
specific for trun-ADAMTS-5 (Table 8) and template cDNAs generated from a
pallet of adult tissues. In addition to decidua, significant levels of trun-
ADAMTS-5 were only detected in the prostate (Fig 12).
Characterisation of the trun-ADAMTS-5 protein species and its
expression in a stably transfected COS-7 cell line. A cDNA corresponding
to the coding region of trun-ADAMTS-5 was generated from the full length
clone by PCR. The resultant PCR product was ligated into the pcDNA3.1
mammalian expression vector at the HindIIlIXho1 sites (pc-AT5). Proteins
expressed by pCDNA3.1 vectors are tagged at the carboxyl-terminus with a
peptide containing a V5 epitope derived from paramyxovirus. pcDNA3.1
vectors containing the trun-ADAMTS cDNA, inserted "out-of-frame" at the
Kpn 1/BamH 1 sites (pc-ATScon) and the Lac Z gene (pc-LacZ) were also
prepared.
Aliquots (1 ug) of pc-AT5 or an empty pcDNA3.1 vector were transcribed and
translated using a coupled reticulocyte TNT T7 kit (Promega) in the presence
of [35Sj methionine (Amersham) according to the manufacturer's instructions.
The translational protein products were analysed by SDS-PAGE
electrophoresis followed by overnight autoradiography. A trun-ADAMTS-5
protein species of approximately 60 kDa was generated using this in vitro
system.
CA 02366129 2001-12-21
-16-
COS-7 cells were transfected with pc-ATS, pc-LacZ, or an empty pcDNA3
vector using Fugene6 (Roche Diagnostics) and cultured for a further 48h. The
transfection efficiency of these COS-7 cell cultures was determined to be >
80% using ~i-galactosidase histochemistry. The culture medium from COS-7
cells transfected with pc-ATS, or an empty pcDNA3 was collected prior to the
cells being detached from the culture dishes using PBS containing EDTA
(5mM). The cells were then pelleted, washed with ice cold PBS and treated
with RIPA buffer. The ECM deposited by these transfected COS-7 cells was
harvested from the culture dish using Laemmli sample buffer.
Western blots were prepared using aliquots of these cellular lysates, culture
media or ECM fractions and probed with a mouse monoclonal antibody
directed against the HA epitope. A single recombinant trun-ADAMTS protein
species (approximately 75 kDa) was detected in the ECM fraction but not the
culture medium obtained from the COS-7 cells transfected with psecTag-T5.
These observations demonstrate that trun-ADAMTS-5 is a secreted protein
that associates with the ECM. In addition, the higher Mr of the trun-ADAMTS-
5 protein species produced by COS-7 cells shows that the mature form of this
protein is likely glycosylated.
To generate stably transfected cell lines, COS-7 cells were transfected with
pc-ATS, or pc-ATScon. Forty eight h after transfection, the cells were
trypsinised and subcultured in medium containing the antibiotic, Neomycin
(G418; 500 ug/ml). Approximately 30 of the cellular foci that formed in these
cultures were selected and expanded. The expression of a HA-tagged sense
or non-sense fusion proteins in the expanded cellular clones was confirmed
by Western blot analysis as described above.
Trun-ADAMTS-5 reduces th~ invasive capacity of EVTs in vitro.
Untransfected COS-7 cells or our 2 stably transfected COS-7 cell lines were
plated on Matrigel-coated Transwell inserts containing polycarbonate filters
with 8 um pores. After 72 h, the COS-7 cells were detached using PBS
CA 02366129 2001-12-21
-17-
containing EDTA (5 mM). EVTs (2 x 105 cells) were then plated on the filters
coated with ECM deposited by the COS-7 cells. After a further 72h of culture,
the EVT cultures were axed and immunostained with a monoclonal antibody
directed against cytokeratins 8 and 18 using a protocol previously described
(Appendix ). The filters were dissected from the inserts with a scalpel blade
and mounted on microscope slides such that the underside of the filters faced
upward. Trophoblast invasion was determined by counting the numbers of
immunostained cells that had penetrated the ECM and appeared on the
underside of the filters.
The ECM deposited by the COS-7 cells expressing trun-ADAMTS-5
significantly reduced the number of EVTs that appeared on the underside of
the filters (Fig. 13). In contrast, there were no significant differences in
the
number of EVTs that penetrated the ECM deposited by untransfected COS-7
cells or those transfected with pc-ATScon.
OTHER MATERIALS AND METHODS
Tissues
Tissue collected from over 50 primary ovarian tumor specimens are collected.
Women recruited for these studies fulfill the following criteria: 1 ) have
been
diagnosed with a primary epithelial ovarian carcinoma 2) have never received
chemotherapy and/or radiation therapy and 3) have no familial ovarian cancer.
Patients with benign, borderline epithelial neoplasms or primary peritoneal
carcinoma are also be excluded from this study.
Samples of the primary ovarian tumor mass and ascites fluid obtained at the
time
of surgery. The ratio of tumor:normal tissue confirmed to be >80% before
inclusion. Normal ovarian tissues obtained during the surgical resection of
the
ovary serve as controls for these studies. The tissues are harvested for total
RNA
extraction or placed in Tissue-Tek OCT embedding media (VWR, Mississauga,
ON) and snap-frozen for subsequent cryosectioning and histochemical analysis.
Ascites cells will be retrieved directly onto microscope slides (cytospin)
using a
CA 02366129 2001-12-21
-18-
Cytofuge-2 for histochemical analysis or collected for total RNA extraction by
centrifugation.
Tumor histology is done to classify cells according to the WHO system by two
pathologists on two independent occasions. Tumors are graded as well- (grade
1 ), moderately- (grade 2) or poorly (grade 3) differentiated. Clinical
disease stage
is established according to the criteria outlined by FIGO.
Cells
Primary cultures of OSE cells isolated from normal or malignant ovarian
tissues. In
addition, cultures of OVCAR-3 (well-differentiated) or SKOV-3 (poorly
differentiated) ovarian carcinoma cell lines could be used.
Identification of the ADAMTS and trun-ADAMTS subtyp~s present in normal
OSE, ovarian tumor masses, and ascitic cells
A reverse-transcription polymerase chain reaction (RT-PCR) strategy is
employed.
RT-PCR will be performed using degenerate oligonucleotides corresponding to
two conserved regions present in all of the known ADAMTS or trun-ADAMTS
subtypes as primers. Template cDNAs are synthesised from the total RNA
extracts prepared from normal OSE, or grade-1 and grade-3 ovarian tumor
masses and the corresponding ascitic cells. The resultant PCR products are
subcloned and subjected to DNA sequence analysis.
Cellular Distribution of the ADAMTS and trun-ADAMTS in ovarian tumor
masses and corr~sponding ascitic cells obtained at different stages of the
disease states
In situ hybridisation histochemistry is performed using a Rapid In Situ
Hybridisation kit (Roche Diagnostics, PQ). The cytospin cellular preparations
and
frozen sections prepared from normal ovary or ovarian tumor masses, obtained
at
different stages of the disease state, is probed with oligonucleotides
specific for
each of the ADAMTS and trun-ADAMTS subtypes identified in these two tissue
types.
CA 02366129 2001-12-21
-19-
Regulated expression of ADAMTS and trun-ADAMTS during the neopiastic
progression of OSE cells in vivo and in vitro
Northern blots containing total RNA extracted from ovarian tumor masses and
corresponding acites cells, obtained at different stages of the disease state,
cultures of normal OSE cells or the ovarian carcinoma cells are prepared. The
blots are probed with radiolabled cDNAs specific for the ADAMTS or trun-
ADAMTS subtypes identified in ovarian tumors. To standardise the amount of
RNA in each lane, the blots are reprobed with a cDNA specific for the 28S rRNA
subunit.
Specific roles for the ADAMTS and trun ADAMTS subtypes in the
proliferation and metastatic behaviour of normal OSE and ovarian
carcinoma cells
To examine the proliferation and invasive capacity of normal OSE, OVCAR-3 or
SKOV-3 cells grown on ECM deposited by COS-7 cells, are stably transfected
with each of the trun-ADAMTS subtypes. The effects of sense or antisense
oligonucelotides, specific for each ADAMTS or trun-ADAMTS subtype, on the
proliferation and migratory behaviour of normal OSE or ovarian carcinoma cells
is
be determined. These cells are cultured on collagen type 1, fibrin or
Matrigel, 3
representative biomatrices that are likely to be encountered by ovarian cancer
cells in vivo. Compensatory changes in the repertoire of ADAMTS subtypes
expressed by the normal or carcinoma cells following treatment with the
antisense oligonucelotides is determined by Northern blot analysis.
CA 02366129 2001-12-21
-20-
REFERENCES
1. Serle E, Aplin JD, Li T-C, Warren MA, Graham RA, Seif MW, Cooke
ID (1994). Fertil Steril 62:989-996.
2. Karamardian LM, Grimes DA (1992). Am J Obstet Gyneocol
167:1391-1398.
3. Khong TY, DeWolf F, Robertson WB, Brosens I (1996) Br J Obstet
Gynaecol 93:1049-1059.
4. Redline RW, Patterson P (1995) Hum Pathol 26:594-600.
5. Krebs C, Macara LM, Leiser R, Bowman AW, Greer IA, Kingdom JCP
(1996) Am J Obstet Gynecol 175: 1534-1542.
6. Tang BL (2001 ) Int J Biochem Cell Biol 33:33-44.
7. Kaushal GP, Shah VS (2000) J Clin Invest 105:1335-1337.
8. Hertig AT, Rock J, Adams EC (1956). Am J Anat 98:435-493.
9. Pijnenborg R, Dixon G, Robertson WB, Brosens I (1980) Placenta 1:3-
19.
10. Aplin JD (1991) J Cell Sci 99:681-692.
11. Pijnenborg R, Bland JM, Robertson WB, Brosens I (1980) Placenta
4:387-414.
12. Pijnenborg R, Bland JM, Robertson WB, Dixon G, Brosens 1 (1981 )
Placenta 2:303-316. 13. AI-Lamki RS, Skepper JN, Burton GJ (1999)
Hum reprod 14:496-504.
14. Graham CH, Lala PK (1991 ) J Cell Physiol 148:228-234.
15. Graham CH, McCrae KR, Lala PK (1993) Trophoblast Res 7:237-250.
CA 02366129 2001-12-21
-21-
16. Noyes RW, Hertig AT, Rock J (1950). Fertil Steril 1:3-25.
17. Aplin JD (2000) Sem Cell Dev Biol 11:115-125.
18. Aplin JD, Charlton Ak, Ayad S (1988) Cell Tissue Res 253:231-
240.
19. Tabibzadeh S, Babaknia A (1995). Mol Hum Reprod 10:1579-
1602.
20. RodgersWH, Matrisian LM, Giudice LC, Dsupin B, Cannon P,
Svitek C (1994) J Clin Invest 94:946-953.
21. Irwin JC, Kirk D, Gwatkin RBL (1996) J Clin Invest 97:438-447.
22. Fata JE, Ho AT, Leco KJ, Moorehead RA, Khoka R (2000) Cell Mol
Life Sci 57:77-95.
23. Fisher SJ, Cui TY, Zhang L, Hartman L, Grahl K, Guo-Yang K, Tarpey
J, Damsky, CH (1989) J Cell Biol 109:891-902.
24. Librach CL, Werb Z, Fitzgerald ML, Chiu K, Corwin NM, Esteves RA,
Grobelny D, Galardy R, Damsky Ch, Fisher SJ (1991) J Cell Biol
113:437-449.
25. Polette M, Nawrocki B, Pintiaux A, Massenat, C, Maquoi E, Volders L,
Schnapps. JP, Birembaul P, Foidart JM. (1994) Lab Invest 71:838-
846.
26. Nawrocki B, Polette M, Marchand V, Maquoi E, Berochia A,
Tournier JM, Foidart JM, Birembault P. (1996) Placenta
17:565-572.
27. Graham CH, McCrae KR (1996) Am J Obstet Gynecol 175:555-562.
28. Sawicki G, Radomski MW, Winten-Lowen B, Krzymien A,
Guilbert LJ (2000) Biol Reprod 63:1390-1395.
29. Denhardt DT, Feng B, Edwards DR, Cocuzzi ET, Malyankar (1993)
Pharmacol Ther 59: 329-341.
CA 02366129 2001-12-21
-22-
30. Gomez DE, Alonso DF, Yoshigi H, Thorgeirsson UP (1997) Eur J Cell
Biol 74:111-122.
31. Riley SC, Leask R, benison FC, Wisely K, Calder AA, Howe DC
(1999) J Endocrinol 162:351359.
32. Nothnick WB (2000) Biol Reprod 63:905-912.
33. Wolfsberg TG, Primakoff P, Myles DG, white JM (1995)
J Cell Biol 131:275-278.
34. Black RA, White JM (1998) Curr opin Cell Biol 10:654-
659.
35. Sclondorff J, Blobel CP (1999) J Cell Sci 112:36063-36017.
36. Izumi Y, Hirata M, Hasuwa H, Iwamoto R, Umata T, Miyakato K
(1998) EMBO J 17: 7260-7272.
37. Artavanis-Tsakonas S, Rand MD, Lake RJ
(19990 Science 284:770-776.
38. Blobel CP (1997) Cell 90:589-592.
39. Pan D, Rubin GM (1997) Cell 90:271-280.
40. Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P. (1999) Science
283:91-94.
41. Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL,
Wolfson MF (1997) Nature 385:729-733.
42. Moss ML, Jin SL, Milla ME, Burkhart W, Carter HL, Chen WJ
(1997) Nature 385:733736.
43. Jeon OH, Kim DS (1999) Eur J Biochem 263:526-533.
44. Millichip MI, Dallas DJ, Wu E, Dale S, McKie N (1998) Biochem
Biophys Res Commun 245:594-598.
45. Kamiguti AS, Hay CR, Zuzel M (1996) Biochem J 320:635-641.
46. Nath D, Slocombe PM, Webster A, Stephens PE, Docherty AJ,
Murphy G (2000) J Cell Sci 113:2319-2328.
CA 02366129 2001-12-21
-23-
47. Alfandari D, Wolfsberg TG, White JM, DeSimone DW (1997) Dev
Biol 182:314-330.
48. Kuno K, Kanada N, Nakashima E, Fujiki F, Ichimura F,
Matsushima K (1997) J Biol Chem 272:556-562.
49. Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, Link JR,
Ellis DM, Tortorella MD, Pratta MA, Hollis JM, Wynn R, Duke JL,
George HJ, Hillman MC, Murphy K, Wiswall BH, Copeland RA,
Decicco CP, Bruckner R, Nagase H, Ito Y, Newton RC, Magdola RL,
Trazskos JM, Hollis GF, Amer EC, Burn TC (1999) J Biol Chem
274:23442-23450.
50. HurskainenTL, Hirohata S, Seldin MF, Apte SS (1999) J Biol Chem
274:25555-25563.
51. Vazquez F, Hastings G, Ortega MA, Lane TF, Oikemus S,
Lombardo M, Iruela-Arispe ML (1999) J Biol Chem 274:23349-
23357.
52. Tang BL, Hong W (1999) FEES Lett 445:223-225.
53. Clark ME, Kelner GS, Turbeville LA, Boyer A, Arden KC, Maki RA
(2000) Genomics 67:343350.
54. Cal S, Arguelles JM, Fernandez PL, Lopes-Otin C (2001) J Biol
Chem 276:17932-17940.
55. Fujikawa K, Suzuki H, McMullen B, Chung D (2001 ) Blood
98:1662-1666.
56. Rawlings ND, Barrett AJ (1995) Methods Enzymol 248:183-228.
57. Kuno K, Terashima Y, Matsushima K (1999). J Biol Chem 274:
18821-18826.
58. Rodriguez-Mazaneque JC, Miichanowski AB, Dufour EK, Leduc R,
Iruela-Arispe ML (2000) J Biol Chem 275:33471-33479.
59. Stocker W, Grams F, Baumann U, Reinemer P, Gomis-Ruth FX,
Mackay DB, Bode W (1995) Protein Sci 4:823-840.
CA 02366129 2001-12-21
-24-
60. Tortorella M, Pratta M, Rui-Qin L, Abbaszade 1, Ross H, Burn T,
Arner E (2000) J Biol Chem 275:25791-25797.
61. Tortorella M, Malfait A, Deccico C, Arner E, Nagase H (2001 )
Osteoarthritis Cartilage 6;539-552.
62. Nakamura H, Fujii Y, Inoki I, Sugimoto K, Tanzawa K, Matsuki H,
Miura R, Yamaguchi Y, Okada Y (2000) J Biol Chem 275:38885-
38890.
63. Sandy JD, Westling J, Kenagy RD, Iruela-Arispe ML, Verscharen C,
Rodriguez-Mazaneque JC, Zimmermann DR, Lemire JM, Fischer
JW, Wight TN, Clowes AW (2001 ) J Biol Chem 276:13372-13378.
64. Prockop DJ, Sieron AL, Li SW (1998) Matrix Biol 16:399-408.
65. Matthews RT, Gary SC, Zerillo C, Pratta M, Solomon K, Amer EC,
Hockfield S (2000) J Biol Chem 275:22695-22703.
66. Zimmermann DR, Lemire JM, Fischer JW, Wight TN, Clowes
AW (2001 ) J Biol Chem 276:13372-13378.
67. Bornstein P, Sage EH (1994) Methods Enzymol 248:183-283.
68. Gantt SM, Clavijo P, Bai X, Esko JD, Sinnis P (1997) J Biol
Chem 272:19205=19213.
69. Magnetto S, Bruno-Bossio G, Voland C, Lecerf J, Lawler J,
delmas P, Siverstein R, Clezardin P (1998) Cell Biochem Funct
16:211-221.
70. Fernandes RJ, Hirohata S, Engle JM, Colige A, Cohn DH, Eyre DR,
Apte SS (2001 ) J Biol Chem 276:31502-31509.
71. Shindo T, Kurihara H, Kuno K, Yokoyama H, Wada T, Kurihara Y, Imai
T, Wang Y, Ogata M, Nishimatsu H, Moriyama N, Oh-hashi Y, Morita H,
Ishikawa T, Nagai R, Yazaki Y, Matsushiman K (2000) J Clin Invest
105:1345-1352.
72. Li SW, Arita M, Fertala A, Bao Y, Kopen GC, Langsjo TK, Hyttinen
MM, Helminen HJ, Prockop DJ (2001) Biochem J 355:271-278.
CA 02366129 2001-12-21
-25-
73. Espey LL, Yoshioka S, Russell DL, Fuji S, Richards
JS (2000) Biol
Reprod 62:1090-1095.
74. Colige A, Sieron AL, Li SW, Scharze U, Petty E, Wertelecki
W, ~Icox
W, Karkow D, Cohn DH, Reardon W (1997) Am J Hum Genet
65:308-
317
75. Giunta C, Seperti-Furga A, Spranger S, Cole WG, Steinmann
B (1999) J
Bone Jt Surg Am Vol 81:225-238.
76. Bayliss MT, Hutton S, Hayward J, Maciewicz RA (2001
)
Osteoaerthritis Cartilage 6:553-560 77. Satoh K, Suzuki
N, Yokota H
(2000) Neurosci Lett 289:177-180.
78. Robker RL, Russell DL, Espey LL, Lydon JP, O'Malley
BW,
Richards, JS (2000) Proc Natl Acad Sci USA 97:4689-4694.
79. Kashiwagi M, Tortorella M, Nagase H, Brew K (2001 )
J Biol Chem
276:12501-12504.
80. Yu WH, Yu S, Meng Q, Brew K, Woessner, JF, Jr (2000)
J Biol Chem
275:31226-31232
81. Kramerova IA, Kawaguchi N, Fessler LI, Nelson RE, Chen
Y, Kramerov
AA, Kusche-Gullberg M, Kramer JM, Ackley BD, Sieron
AL, Prockop DJ,
Fessler JH (2000) Development 127:54755485.
82. Irwin JC, Kirk D, King RJ Quigley MM, Gwatkin RB (1989)
Fertil Steril
52:761-768.
83. Ilic D, Genbacev O, Jin F, Caceres E, Almeida EAC, Bellingard-
Dubouchaud V, Schaefer EM, Damsky CH, Fisher SJ (2001 ) Am J
Pathol 158:93-108.
CA 02366129 2001-12-21
H
Id
. . . ~. . . .~ . ~ . .
.
a
m
c . . + . . . + . +
~ +
,
Y
+ . . . . . .;. . . . .
N
+ + ~ + ~ ~ + ~ +
J ~ +
. . + . . . + + . ,
J ~ +
+ ~ -E + + ~ . ~. . .
~'a
c
.~. . . . . .~.
m
m ~~ + + + ~ ~ ,+t, ~ + ~ .
Z
N M ~ tI~ t0 f~ 00 Q7 ~ N
Q
a
CA 02366129 2001-12-21
H
7
t
f-
C
m .
Q +
N
r
m
m
Y
N
W
C
' ~ r
C + W
Y m
C9
O
. . . .
a o
+ ' l
~ " + + +
v
- N M 00
a as a
CA 02366129 2001-12-21
T 4: N I N S D TO
rnFNTIFY ADAMTS SUBTYPES IN HUMAN TRUES AND CELLS
PRIMER SE UENCES PCR CONDITIONS
FORWARD PRIMER: 1 min at 94C
GATTCTGYMGNAGNACNTGYGGNGG 2 min at SOC
REVERSE PRIMER: GATTCRCANCTNGTNCCRTC 3 min at 72C
Cycle Repeated 35 times
where:Y=CorT, M=Aor C, R=Aorta, N=A, T,C,o
G
TABLE 5~ IDENTIF] Cf ATION OF THE ADAMTS SUBES PRESENT IN FIRST
T T T -PCR
Number of KnownADAMTS Subtypes
Identified Clones/tissueAnal sed
out of 40
ADAMTS SubtypePlacenta Decidua
ADAMTS-1 4 5
ADAMTS-2 10 6
ADAMTS-4 8 4
ADAM'TS-5 5 2
ADAMTS-7 6 1
ADAMTS-9 3 1
Total = Total =19
36
CA 02366129 2001-12-21
Table 6: Primer sequences and PCR conditions for the semiquantitative analysis
of
ADAMTS subtype mRNA levels.
Primer Sequence Estimated PCR Conditions
PCR
Product
Size
ADAMTS-1 Forward: S'- CGAGTGTGCAAAGGAAGTGA-3' Denawring:94'C30s
Reverse: S'- CTACCCCCATAATCCCACCT-3'399bp Annealing:65'C30s
Extension:72'C60s
35 c
cles
ADAMTS-2 Forward: S'- CCTATGACTGGCTGCTGGAT-3' Denaturing:94'C30s
Reverse: S'-CTCCCAAAGTGCTGGGATAA-3'310 by Annealing:6S'C30s
Extension:72'C60s
35 c
cles
ADAMTS-4 Forward: S'-AATCCAGGGTGGTGGTGATA-3' Denawring:94'C30s
Reverse: S'-TACTCAGGAGGCTGAGGCAT-3'349 by Annealing:60'C30s
Extension:72'C60s
35 c
cles
ADAMTS-5 Forward: S'-GGGCATGGTAACTGTTTGCT-3' Denaturing:94'C30s
Reverse: S'-CCTCTTCCCTGTGCAGTAGC-3'~4 by Annealing:6S'C30s
Extension:72'CbOs
3S c
cles
ADAMTS-6 Forward: S'-TGACAGTCCAGCACCTTCAG-3' Denaturing:94'C30s
Reverse: S'-CTACGTGCTTGCATTCTCCA-3'340 by Annealing:60'C30s
Extension:72'C60s
35 c
cles
ADAMTS-7 Forward: S'-CCATGTGGTGTACAAGCGTC-3' Denaturing:94'C30s
Reverse: S'-GGTCCTCCTCCTCATCTTCC-3'3gg by Annealing:SS'C30s
Extension:72'C60s
35 c
cles
ADAMTS-8 Forward: S'-AAGAAGAGGAGGCAGAAGGC-3' Denawring:94'C30s
Reverse: S'-TCTGTCTGGTGAGCAGGATG-3'380 by Annealing:6S'C30s
Extension:72'C60s
35 c
cles
ADAMTS-9 Forward: S'- CCCAGCCTGGACACATTACT-3' Denawring:94'C30s
Reverse: S'- CCCAGCCTGGACACATTACT-3'428 by Annealing:6S'C30s
Extension:72'C60s
3S ccles
ADAMTS-10 Forward: S'-CAATGTCCTCATTGACGCTG-3'_ _ 94'C30s
Denawring:
Reverse: S'CTGGGAAGCACCGTTAACAT-3'360bp Annealing:65'C30s
Extension:72'C60s
3S cles
ADAMTS-12 Forward:5'-GTGCAGCGAGGAGTACATCA-3' Denawring:94'C30s
Reverse: S'-GCGTTTTCTTTCTCCAGTGC-3'488 by Annealing:6S'C30s
Extension:72'C60s
35 c
cles
GAPDH Forward: S'-TGTTCGTCATGGGTGTGAACCA-3' Denaturing:94'C30s
I
Reverse: S'-TGGCAGGTTfTTTTCTAGACGGCA-3'378 by Annealing:SS'C30s
Extension:72'C90s
27 c
cles
CA 02366129 2001-12-21
3~
AB N
3 CHORIOCARCINOMA CELLS USING DEGENRRATE RT-PAR
Number of KnownADAMTS Subtypes
Identified Clones/cella se uenced
out of 40
ADAMTS SubtypeEVTs JEG3 cells
ADAMTS-I 7 14
ADAMTS-2 0 12
ADAMTS-4 10 1
ADAMTS-5 0 8
ADAMTS-6 6 0
ADAMTS-10 14 0
ADAMTS-12 3 0
Total = Total =35
40
TABLE 8: IDENTIFICATION OF THE trun-ADAMTS SUBTYPES PRESENT IN FIRST
TRIMESTER PLACENTA AND DECIDUAL TISSUES USING DEGENERATE RT-PCR
Number of trun- ADAMTS subtypes out of 40 clones/
Tissue analysed
trun ADAMTS Decidua Placenta
1 3 1
2 2 1
3 4 0
4 3 2
8 0
6 1 0
7 1 0
Total =21 Total=4
CA 02366129 2001-12-21
TABLE 9: AMINO ACID SEQUENCE HOMOLOGY (%) BETWEEN THE trun-ADAMTS,
the TSP-1 MOTIFS AND SPACER REGIONS ADAMTS AND PAPILIN
TRUN-ADAMTS SUBTYPES
1 2 3 ~ 4 __ ~~ 5
ADAMTS
_
-1 - 51 - 16
-2 - - 16 - -
-3 - 23 - - -
-4 - - 11 46 -
-5 - - - - 12
-6 - - 50 13 -
-7 11 - - 16
_g _ . _ _
-- -
-9 15 - _ _
- -.
-10 _ 20 _ - 45
-
-12 43 23 - 0 .
Papilin 33 ~ 26 ~ 38 30 28
j
TABLE 10: DISTRIBUTION OF THE trun-ADAMTS SUBTYPES IN THE HUMAN GENOME
trun ADAMTS Chromosome mRNA Sequence Unigene
1 9 hypothetical protein HS149184
(C9 of 8 gene)
2 15 hypothetical protein HS170345
FLJ13170
3 9 hypothetical protein HS342601
KIAA0605
4 1 hypothetical protein HS96657
DKFZP434K17721
9 hypothetical protein HS128138
670426809
6 5 Image clones HS18705
2543501 RP11-90E5
366669
7 15 Image clone BAC Gone
2359701
CA 02366129 2001-12-21
TABLE 11: Primer Sequences and PCR Conditions Used To Determine The Tissue
Distribution of trun-ADAMTS-5
Trun-ADAMTS-5
Forward Primer: tggggcccatggagtgaatgc
Reverse Primer: tggaccccatctccgttgcag
Yields PCR product of 636bp
94C for 30 sec
68C for 1 min
72 C for 1 min
TABLE 12: LM.A.G.E. CLONES CORRESPONDING TO THE TSP-1 and SPACER
REGIONS OF SELECTED ADAMTS SUBTYTPES
ADAMTS SUBTYPE _ LM.A.G.E. CLONE #
__ _,__-
ADAMTS-1 2288437
.. _.._ - _... __
742630
ADAMTS-10 253523
ADAMTS-12 2421547