Note: Descriptions are shown in the official language in which they were submitted.
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
MULTIPLE TAG ANALYSIS
BACKGROUND OF THE INVENTION
The present invention is generally in the field of detection of molecules,
and specifically in the field of detection of multiple different molecules in
a
single assay.
The analysis of proteins in histological sections and other cytological
preparations is routinely performed using the techniques of histochemistry,
immunohistochemistry, or immunofluorescence. By performing
immunofluorescence with antibodies labeled with different colors, it has been
possible to detect simultaneously 2, 3, or even 4 different antigens present
in
cellular material. In the future, time-resolved fluorescence may permit the
extension of immunofluorescence methods to the detection of 6 to 12 different
antibodies simultaneously. Likewise, RNA detection by fluorescence in situ
hybridization permits the detection of 2 to 4 different RNAs in cellular
material,
and it may also be extended to permit the detection of 6 to 12 different RNAs
by
time-resolved fluorescence.
There is a need for a sensitive method that will permit the cytological
detection of larger numbers of proteins or RNAs simultaneously. Theoretically,
the simultaneous measurement of the concentration of 20 to 50 different
protein
(or RNA) species should be highly informative as to the specific status of
dynamic cellular processes in normal development, in stages of disease, in
response to drug treatment or gene therapy, or as a result of environmental
exposure or other deliberate or inadvertent interventions.
The study of cells by measuring the identity and concentration of a
relatively large number of proteins simultaneously (referred to as proteomics)
is
currently a very time-consuming task. Two-dimensional (2D) gel electrophoresis
is the most powerful tool for studying the expression of multiple proteins,
but
this technique is not readily adaptable to in-situ cell analysis. Typically,
many
thousands of cells are required to perform a single 2D gel analysis. In order
to
identify different protein expression profiles in heterogeneous tissue
samples,
one would need the capability to analyze the proteins expressed in a small
number of cells. This capability is most relevant in the analysis of
histological
or cytological specimens that may harbor dysplastic or pre-malignant cells.
Such
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
cells, which may precede the development of cancer, need to be identified when
present as small foci of 10 to 50 cells, before they have a chance to give
rise to
tumors. Unfortunately, the amount of protein obtained from 10 to 50 cells is
insufficient for 2D gel analysis, and is problematic even with the use of
radioisotopes to label the protein.
Mass spectroscopy is another powerful technique for protein analysis.
However, the direct analysis of proteins present in samples containing small
numbers of cells is not possible with prior mass spectroscopy technology, due
to
insufficient sensitivity. A minimum of 10,000 cells is required for mass
spectroscopic analysis of tissue samples using prior technology.
Current methods for the analysis of microarray hybridization
experiments rely on the use of a two-color signal readout system. For example,
Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of
gene expression patterns with a complementary DNA microarray. Science
270:467-70, describe an experiment where cDNA prepared from one tissue is
labeled with the dye cy3, while cDNA from another tissue is labeled with the
dye cy5. After the labeling reactions are performed, the two labeled DNAs are
mixed, and hybridized by contacting with the surface of a glass slide
containing
a cDNA microarray on its surface. At the end of the hybridization reaction,
the
microarray surface is washed to remove unhybridized material, and the glass
slide is scanned in a confocal scanning instrument designed to record
separately
the cy3 and the cy5 fluorescence intensity, which is saved as two different
computer files. Computer software is then used to calculate the fluorescence
ratio of cy3 to cy5 at each of the specific dot-addresses on the DNA
microarray.
This experimental design works very well for performing comparisons of
mRNA expression ratios between two samples.
Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R (1999)
Quantitative analysis of complex protein mixtures using isotope-coded affinity
tags. Nature Biotechnology 17:994-999, have described an approach for the
accurate quantification and concurrent sequence identification of the
individual
proteins within complex mixtures of biological origin. The method is based on
a
class of new chemical reagents termed isotope-coded affinity tags (ICATs), and
tandem mass spectrometry. These authors extracted proteins from two different
2
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
experimental states of an organism, and labeled each of the two preparations
of
total protein with two different thiol-reactive ICAT tags of different mass.
The
two labeled protein preparations were mixed, separated by liquid
chromatography, and detected on line by mass spectrometry. For each individual
protein peak, mass spectrometry permitted protein identification, as well as
measurement of the ratio of the amounts of the two proteins.
In pioneering work, Ekins and Chu (Ekins, R.P. and Chu, F.W. (1991)
Multianalyte microspot immunoasay - Microanalytical "Compact Disk" of the
future. Clinical Chemistry 37:1955-1967; Ekins, R.P. and Chu, F.W. (1994)
Developing multianalyte assays Trends in Biotechnology 12:89-94) described
assays in which different specific antibodies are laid down on a surface as an
array of microspots, subsequently a complex sample containing a mixture of
antigens is bound to the array, and a second antibody is used for detection.
In
this multiplex assay, the signal present at each address serves as an
indicator of
the concentration of each of a multiplicity of antigens present in the sample.
The
authors suggest that these microarrays could contain as many as 10,000
antibody
microspots per square centimeter.
However, a serious limitation of all existing microarray methods,
including those disclosed above, is that they do not permit the analysis of
more
than six to eight samples in a single microarray experiment. If fluorescent
dyes
are used for detection, there are problems of spectral overlap of multiple
dyes if
more than seven dyes are used. If non-amplifiable mass tags are used, there is
insufficient sensitivity of the mass spectrometric detection step, due to the
fact
that the total number of protein molecules bound on each antibody spot of the
microarray is relatively small (typically 20,000 to 1,000,000 protein
molecules
per spot, for a simple two-sample experiment, or even less for a mufti-sample
experiment).
It is therefore an object of the present invention to provide a method that
permits the indirect detection of a large number of different analytes in a
single
sample or group of samples.
It is therefore another object of the present invention to provide a method
that permits the indirect detection of a large number of different proteins in
a
single sample or group of samples.
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
It is another object of the present invention to provide a method that
permits the indirect detection of a large number of different mRNAs in a
single
sample or group of samples.
It is another object of the present invention to provide a method of
detecting indirectly modification states of a multiple analytes in a single
sample
or group of samples.
BRIEF SUMMARY OF THE INVENTION
Disclosed is a method of detecting multiple analytes in a sample in a
single assay. The method is based on encoding target molecules with signals
followed by decoding of the encoded signal. This encoding/decoding uncouples
the detection of a target molecule from the chemical and physical properties
of
the target molecule. In basic form, the disclosed method involves association
of
one or more reporter molecules with one or more target samples, association of
one or more decoding tags with the reporter molecules, and detection of the
decoding tags. The reporter molecules associate with target molecules in the
target sample(s). Generally, the reporter molecules correspond to one or more
target molecules, and the decoding tags correspond to one or more reporter
molecules. Thus, detection of particular decoding tags indicates the presence
of
the corresponding reporter molecules. In turn, the presence of particular
reporter
molecules indicates the presence of the corresponding target molecules.
This indirect detection uncouples the detection of target molecules from
the chemical and physical properties of the target molecules by interposing
decoding tags that essentially can have any arbitrary chemical and physical
properties. In particular, decoding tags can have specific properties useful
for
detection, and decoding tags within an assay can have highly ordered or
structured relationships with each other. It is the (freely chosen) properties
of
the decoding tags, rather than the (take them as they are) properties of the
target
molecules that matters at the point of detection.
The decoding tags have the additional advantage of being uncoupled
from the target molecule-specific aspects of the reporter molecules. Unlike
detection methods where a labeled molecule is bound to an analyte followed by
detection of the label, the disclosed method is not limited by the chemical
and
physical properties of the labeled molecule. This allows more convenient
4
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
detection, more sensitive detection, and more highly multiplexed detection
schemes.
The sensitivity of the disclosed method can also be enhanced by
including a signal amplification step prior to detection. In basic form,
amplification is accomplished by amplifying reporter signals on the reporter
molecules. This results in multiple reporter tags associated with each
reporter
molecule. The decoding tags are then associated with the reporter tags and
detected. Generally, the decoding tags correspond to one or more reporter tags
(and thus correspond to the reporter molecules with which the reporter tags
are
associated), and the reporter tags correspond to the reporter molecules with
which they are associated. Thus, detection of particular decoding tags
indicates
the presence of the corresponding reporter tags. In turn, the presence of
particular reporter tags indicates the presence of the corresponding reporter
molecules. In turn, the presence of particular reporter molecules indicates
the
presence of the corresponding target molecules. The reporter molecule also can
be designed to include multiple reporter tags (essentially accomplishing a pre-
assay amplification of the signal).
In one form of the disclosed method, one or more target samples and one
or more reporter molecules are brought into contact, allowing the reporter
molecules to become associated with target molecules in the target samples.
The
reporter molecules are then amplified to produce multiple reporter tags for
each
reporter molecule. The reporter tags remain associated with the reporter
molecules. One or more decoding tags are then associated with the reporter
tags,
and the decoding tags are detected. The decoding tags correspond to reporter
molecules and the reporter molecules correspond to target molecules. This
relationship means that detecting the decoding tags indicates the presence of
reporter molecules corresponding to the detected decoding tags, and that the
presence of reporter molecules indicates the presence of target molecules
corresponding to the reporter molecules.
In another form of the disclosed method, four or more target samples and
one or more reporter molecules are brought into contact, allowing the reporter
molecules become associated with target molecules in the target samples. Each
target sample is brought into contact with a different set of reporter
molecules,
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
and the reporter molecules in each set of reporter molecules are different
from
the reporter molecules in the other sets of reporter molecules. The four or
more
of the target samples are then mixed together, one or more decoding tags are
associated with the reporter molecules, and the decoding tags are detected. A
different decoding tag corresponds to each different reporter molecule such
that
each decoding tag corresponds to only one of the target samples. This
relationship means that detecting the decoding tags indicates the presence of
target molecules corresponding to the detected decoding tags. Further,
detection
of decoding tags corresponding to different target samples indicates the
presence
of the same target molecules in the corresponding target samples.
In another form of the disclosed method, one or more target samples and
one or more reporter molecules are brought into contact, allowing the reporter
molecules to become associated with target molecules in the target samples.
One or more decoding tags are then associated with the reporter molecules. A
different decoding tag corresponds to each different reporter molecule, and
the
decoding tags are not covalently coupled to the reporter molecules. The
decoding tags are then detected by disassociating the decoding tags from the
reporter molecules. The decoding tags correspond to reporter molecules and the
reporter molecules correspond to target molecules. This relationship means
that
detecting the decoding tags indicates the presence of reporter molecules
corresponding to the detected decoding tags, and that the presence of reporter
molecules indicates the presence of target molecules corresponding to the
reporter molecules.
In another form of the disclosed method, one or more target samples and
one or more reporter carriers are brought into contact. The reporter carriers
include one or more specific binding molecules, a carrier, and a plurality of
decoding tags associated with the carrier. After the reporter carriers are
associated with reporter molecules, the decoding tags are detected. The
decoding tags correspond to reporter carriers, and the reporter carriers
correspond to target molecules. This relationship means that detecting the
decoding tags indicates the presence of reporter carriers corresponding to the
detected decoding tags, and that the presence of reporter carriers indicates
the
presence of target molecules corresponding to the reporter carriers.
6
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
In preferred embodiments, a one-to-one-to-one correspondence between
target molecules, reporter molecules, and decoding tags is used. In this way,
each different type of target molecule ends up with a different and separately
detectable decoding tag associated with it. The combination of specific
binding
molecule and reporter signal results in an effective conversion of the
different
target molecules (where the target molecules may be widely chemically
divergent) into a standardized signal (the reporter signal). These
standardized
signals (which are coded for each target molecule) are then detected using a
single set of standardized conditions (during which the coding is carried
forward). The detection of the coded signals (that is, the decoding tags)
results
in effective, convenient detection of multiple target molecules in a single
assay.
Decoding tags can be detected using any suitable technique. In general,
the properties of the decoding tags will be chosen to match, or be compatible
with, a chosen detection technique. In preferred embodiments, the disclosed
method uses rapid detection techniques that allow spatial information about
analytes to be gathered. An example is matrix-assisted laser
desorption/ionization time-of flight (MALDI-TOF) mass spectroscopy and
electrophoresis (preferably used in conjunction with microdissection). Such
techniques allow automation and rapid throughput of multiple samples and
assays.
This disclosed method enables a multiplex approach to the study of
proteins and other analytes in microscopic tissue specimens. Compared to
immunofluorescence, the disclosed method offers a greatly increased capability
for multiplexing. The method is also useful for in situ mRNA profiling. The
disclosed method permits the indirect detection of a large number of different
proteins (from 20 to 50, for example, limited only by the number of specific
antibodies available).
Medical applications of this method include the analysis of the
phenotypic status of cells (growth or quiescence) and the assessment of normal
and neoplastic cells in histologic or cytologic specimens in normal and
disease
states. For example, a pathologist may use the method to link a phenotypic
state
with the protein profile of a lesion believed to contain malignant or pre-
malignant cells.
7
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
BRIEF DESCRIPTION OF THE DRAWINGS
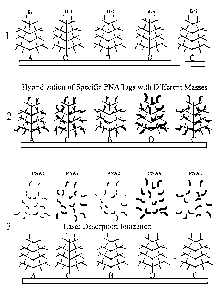
Figure 1 is a diagram illustrating and example of the disclosed method
using branched DNA for amplification. Section 1 shows different branched
DNA molecules (Brl, Br2, Br3, and Br4) associated with their cognate reporter
signals (A, B, C, and D, respectively). The reporter signals are associated
(via
specific binding molecules to which they are coupled to form reporter
molecules) to target molecules on a surface. Section 2 shows peptide nucleic
acid decoding tags (PNAI, PNA2, PNA3, and PNA4) hybridized to their
cognate reporter tags on the associated branched DNA molecules. Section 3
shows laser desorption/ionization of the decoding tags for analysis by mass
spectroscopy.
Figure 2 is a diagram illustrating an example of the disclosed method
using branched DNA for amplification. Section 1 shows different branched
DNA molecules (Brl, Br2, Br3, and Br4) associated with their cognate reporter
signals. The reporter signals are associated--via specific binding molecules
(antibodies A, B, C, and D) to which they are coupled to form reporter
molecules--to target molecules on a surface (antigens a, b, c, and d). Section
2
shows peptide nucleic acid decoding tags (PNA1, PNA2, PNA3, and PNA4)
hybridized to their cognate reporter tags on the associated branched DNA
molecules. Section 3 shows laser desorption/ionization of the decoding tags
for
analysis by mass spectroscopy.
Figure 3 is a diagram illustrating an example of the disclosed method
using branched DNA for amplification. Section 1 shows different branched
DNA molecules (Brl, Br2, Br3, and Br4) associated with their cognate reporter
signals. The reporter signals are associated--via specific binding molecules
(antigens a, b, c, and d) to which they are coupled to form reporter molecules-
-to
target molecules on a surface (antibodies A, B, C, and D). Section 2 shows
peptide nucleic acid decoding tags (PNA1, PNA2, PNA3, and PNA4) hybridized
to their cognate reporter tags on the associated branched DNA molecules.
Section 3 shows laser desorption/ionization of the decoding tags for analysis
by
mass spectroscopy.
Figure 4 is a diagram illustrating an example o~ the disclosed method
using branched DNA for amplification. Section 1 shows different branched
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
DNA molecules (Brl, Br2, Br3, and Br4) associated with their cognate reporter
signals. The reporter signals (', ", "', and "") are associated--via specific
binding
molecules to which they are coupled to form reporter molecules--to target
molecules (antigen a). The target molecules are immobilized on a surface via
interaction with a capture tags on the surface (antibody A). The different
reporter signals are associated with the same target molecules in different
target
samples. Section 2 shows peptide nucleic acid decoding tags (PNA1, PNA2,
PNA3, and PNA4) hybridized to their cognate reporter tags on the associated
branched DNA molecules. Section 3 shows laser desorption/ionization of the
decoding tags for analysis by mass spectroscopy. °
DETAILED DESCRIPTION OF THE INVENTION
Disclosed is a method using a specific signal decoding technique for
identifying and measuring the concentration of different target molecules in
an
assay. The method is especially useful for assays carried out on substrates
such
as plates, surfaces, slides, and beads. The method is particularly suited for
the
detection of macromolecular analytes present in cells or tissues. For example,
the disclosed method is useful for correlating phenotypic status with
particular
analytes in cell or tissue samples.
One form of the disclosed method is based on specific recognition of
each analyte, amplification of the recognition event, and detection of the
amplified signal. The specific recognition is accomplished with specific
binding
molecules that bind or otherwise interact specifically with an analyte of
interest.
The amplification of this interaction is mediated by a reporter signal
coupled,
tethered, or otherwise attached to the specific binding molecule. It is the
reporter signal, an oligonucleotide, that is used to produce multiple reporter
tags
that remain associated with the reporter signal (and thus also with the
specific
binding molecule and analyte). This is accomplished, for example, by
amplification of the reporter signal or hybridization of branched DNA or
oligonucleotide dendrimers to the reporter signals. Detector tags, which are
coded to include distinct, separately detectable attributes, are then
hybridized to
the reporter tags. Detection of the various detector tags results in indirect
detection of the various analytes in the sample.
9
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
In the disclosed method, multiple analytes (which may be proteins,
nucleic acid molecules, or other molecules that can be specifically
recognized)
are assayed using a signal amplification system capable of generating multiple
copies of an oligonucleotide (called a reporter tag), where the reporter
molecule
is physically anchored at or associated with a specific site on the solid
surface;
specifically, at the site where an analyte is located. An example of such a
signal
amplification system is branched DNA (Urdea, Biotechnology 12:926-928
(1994); Horn et al., Nucleic Acids Res 23:4835-4841 (1997)).
In a preferred embodiment, peptide nucleic acid (PNA) molecules
(Hanvey et al., Science 258:1481-1485 (1992)) of different sequence and
molecular weight are used as arbitrary decoding tags that bind specifically to
unique reporter tags. Laser desorption occurring on specific areas of the
cytological samples is used to generate matrix-assisted laser
desorption/ionization time-of flight (MALDI-TOF) miss spectra of the PNA
tags, which are released into the spectrometer and resolved by mass. The
intensity of each PNA decoding tag reveals the relative amount of different
DNA reporters. In other words, the PNA spectra generate scalar values that are
indirect indicators of the relative abundance of the analytes of interest at
specific
locations on the glass surface.
For protein detection, for example, suitable reporter molecules include a
specific antibody (specific binding molecules) covalently coupled to a DNA
oligonucleotide (reporter signal). Each antibody is specific for a protein
antigen
of interest. The covalently coupled oligonucleotide includes a specific DNA
sequence that serves to identify the antibody, and furthermore serves to
generate
a signal comprising amplified, surface-localized DNA. An example of a
surface-localized DNA signal of amplified DNA is the branched DNA
technology described in Urdea, Biotechnology 12:926 (1994), and Hong et al.,
Nucleic Acids Res 23:4835-4841 (1997). Another example of a surface-
localized DNA signal is the tandemly repeated DNA generated by rolling circle
reporter systems (Lizardi et al., Nature Genetics 19:225-232 (1998); U.S.
Patent
No. 5,854,033 to Lizardi; PCT Application No. WO 97/19193). In the case
where branched DNA is used for amplification, the DNA oligonucleotide that is
bound to each antibody serves to initiate the formation of a branched DNA
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
structure. Alternatively, the DNA oligonucleotide that is bound to each
antibody
can serve to initiate rolling circle replication (by serving as a primer, for
example), generating a large tandem DNA molecule that remains localized at the
site of antibody binding.
In another embodiment, referred to as multiple electrophoretic tag assay
(META), detector tags that are sequence-coded, size-coded, and fluorescently-
labeled are detected by capillary electrophoresis. Capillary electrophoresis
coupled with laser-induced fluorescence is used to generate a quantitative
profile
of the different decoding tags. The intensity of each fluorescent peak reveals
the
relative amount of each different decoding tags. In other words, the peak
sizes
of the different decoding tags are scalar values that are indirect indicators
of the
relative abundance of the analytes of interest in the sample.
Materials
Reporter Molecules
Reporter molecules are molecules that combine a specific binding
molecule with a reporter signal. Preferably, the specific binding molecule and
reporter signal are covalent coupled or tethered to each other. As used
herein,
molecules are coupled when they are covalent joined, directly or indirectly.
One
form of indirect coupling is via a linker molecule. The reporter signal can be
coupled to the specific binding molecule by any of several established
coupling
reactions. For example, Hendrickson et al., Nucleic Acids Res., 23(3):522-529
( 1995) describes a suitable method for coupling oligonucleotides to
antibodies.
As used herein, a molecule is said to be tethered to another molecule
when a loop of (or from) one of the molecules passes through a loop of (or
from)
the other molecule. The two molecules are not covalently coupled when they
are tethered. Tethering can be visualized by the analogy of a closed loop of
string passing through the hole in the handle of a mug. In general, tethering
is
designed to allow one or both of the molecules to rotate freely around the
loop.
Specific Binding Molecules
A specific binding molecule is a molecule that interacts specifically with
a particular molecule or moiety. The molecule or moiety that interacts
specifically with a specific binding molecule is referred to herein as a
target
molecule. It is to be understood that the term target molecule refers to both
11
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
separate molecules and to portions of such molecules, such as an epitope of a
protein, that interacts specifically with a specific binding molecule.
Antibodies,
either member of a receptor/ligand pair, synthetic polyamides (Dervan, P.B.
and
R.W. Burli, Sequence-specific DNA recognition by polyamides. Curr Opin Chem
Biol, 1999. 3(6): p. 688-93. Wemmer, D.E. and P.B. Dervan, Targeting the
minor groove of DNA. Curr Opin Struct Biol, 1997. 7(3): p. 355-61.), and other
molecules with specific binding affinities are examples of specific binding
molecules, useful as the affinity portion of a reporter binding molecule.
A specific binding molecule that interacts specifically with a particular
target molecule is said to be specific for that target molecule. For example,
where the specific binding molecule is an antibody that binds to a particular
antigen, the specific binding molecule is said to be specific for that
antigen. The
antigen is the target molecule. The reporter molecule containing the specific
binding molecule can also be referred to as being specific for a particular
target
molecule. Specific binding molecules preferably are antibodies, ligands,
binding
proteins, receptor proteins, haptens, aptamers, carbohydrates, synthetic
polyamides, or oligonucleotides. Preferred binding proteins are DNA binding
proteins. Preferred DNA binding proteins are zinc finger motifs, leucine
zipper
motifs, helix-turn-helix motifs. These motifs can be combined in the same
specific binding molecule.
Antibodies useful as the affinity portion of reporter binc'ting agents, can
be obtained commercially or produced using well established methods. For
example, Johnstone and Thorpe, Immunochemistry In Practice (Blackwell
Scientific Publications, Oxford, England, 1987) on pages 30-85, describe
general methods useful for producing both polyclonal and monoclonal
antibodies. The entire book describes many general techniques and principles
for the use of antibodies in assay systems.
Properties of zinc fingers, zinc finger motifs, and their interactions, are
described by Nardelli, J., T. Gibson, and P. Charnay, Zinc finger-DNA
recognition: analysis of base specificity by site- dig°ected
mutagenesis. Nucleic
Acids Res, 1992. 20(16): p. 4137-44, Jamieson, A.C., S.H. Kim, and J.A. Wells,
In vitro selection of zinc fingers with altered DNA-binding specificity.
Biochemistry, 1994. 33(19): p. 5689-95, Chandrasegaran, S. and J. Smith,
12
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
Chimeric restriction enzymes: what is next? Biol Chem, 1999. 380(7-8): p. 841-
8, and Smith, J., J.M. Berg, and S. Chandrasegaran, A detailed study of the
substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res,
1999.
27(2): p. 674-81.
One form of specific binding molecule is an oligonucleotide or
oligonucleotide derivative. Such specific binding molecules are designed for
and used to detect specific nucleic acid sequences. Thus, the target molecule
for
oligonucleotide specific binding molecules are nucleic acid sequences. The
target molecule can be a nucleotide sequence within a larger nucleic acid
molecule. An oligonucleotide specific binding molecule can be any length that
supports specific and stable hybridization between the reporter binding probe
and the target molecule. For this purpose, a length of 10 to 40 nucleotides is
preferred, with an oligonucleotide specific binding molecule 16 to 25
nucleotides long being most preferred. It is preferred that the
oligonucleotide
specific binding molecule is peptide nucleic acid. Peptide nucleic acid forms
a
stable hybrid with DNA. This allows a peptide nucleic acid specific binding
molecule to remain firmly adhered to the target sequence during subsequent
amplification and detection operations.
This useful effect can also be obtained with oligonucleotide specific
binding molecules by making use of the triple helix chemical bonding
technology described by Gasparro et al., Nucleic Acids Res. 1994 22(14):2845-
2852 (1994). Briefly, the oligonucleotide specific binding molecule is
designed
to form a triple helix when hybridized to a target sequence. This is
accomplished generally as known, preferably by selecting either a primarily
homopurine or primarily homopyrimidine target sequence. The matching
oligonucleotide sequence which constitutes the specific binding molecule will
be
complementary to the selected target sequence and thus be primarily
homopyrimidine or primarily homopurine, respectively. The specific binding
molecule (corresponding to the triple helix probe described by Gasparro et
al.)
contains a chemically linced psoralen derivative. Upon hybridization of the
reporter binding probe to a target sequence, a triple helix forms. By exposing
the triple helix to low wavelength ultraviolet radiation, the psoralen
derivative
mediates cross-linking of the probe to the target sequence.
13
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
Reporter Signals
Reporter signals are molecules or moieties that form part of a reporter
molecule and that allow amplification or detection of tie reporter molecule.
Reporter signals can have any structure that allows either amplification of
their
signal or association with a decoding tag. A single reporter signal is a
single
signal. Amplification of this single signal results in production of multiple
signals (which are referred to as reporter tags). As discussed elsewhere
herein,
this amplification is not limited to, but can include, actual amplification
(that is,
replication) of the reporter signal. Preferably, the reporter signal is
designed to
serve as a hybridization partner (to associate, for example, branched DNA or
oligonucleotide dendrimers with the probe) or as a DNA synthesis primer.
In one embodiment, the reporter signal is an oligonucleotide and includes
a sequence that serves as a hybridization partner for the tail of a branched
DNA
molecule or of an oligonucleotide dendrimer. The sequence of the probe
sequence can be arbitrarily chosen. Where multiple oligonucleotide reporter
molecules are being used in the same assay (the normal situation), it is
preferred
that the reporter signal sequence for each reporter molecule be substantially
different to limit the possibility of non-specific target detection.
In another embodiment, the oligonucleotide portion of a reporter molecule
includes a sequence, referred to as a rolling circle replication primer
sequence,
that serves as a rolling circle replication primer for an amplification target
circle
(ATC). This allows rolling circle replication of an added ATC where the
resulting tandem sequence DNA (TS-DNA) is coupled to the reporter molecule.
Because of this, the TS-DNA will be effectively immobilized at the site of the
target molecule. The sequence of the rolling circle replication primer
sequence
can be arbitrarily chosen. In a multiplex assay using multiple reporter
molecules, it is preferred that the rolling circle replication primer sequence
for
each reporter molecule be substantially different to limit the possibility of
non-
specific target detection. When the reporter signal of a reporter molecule is
used
as a rolling circle replication primer, the reporter signal can be any length
that
supports specific and stable hybridization between the reporter signal and the
primer complement portion of an amplification target circle. Generally this is
10
to 35 nucleotides long, but preferably is 16 to 20 nucleotides long.
14
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
In another embodiment, the reporter signal of a reporter molecule can
include an amplification target circle which serves as a template for rolling
circle
replication. Replication of the ATC produces TS-DNA at the site where the
reporter molecule is bound to the target molecule. For this purpose, the ATC
should be tethered to the specific binding molecule by looping the ATC around
a
tether loop. This allows the ATC to rotate freely during rolling circle
replication
while remaining coupled to the affinity portion. The tether loop can be any
material that can both form a loop and be coupled to a specific binding
molecule.
For example, linear polymers can be used for tether loops.
An amplification target circle (ATC) is a circular single-stranded DNA
molecule, generally containing from 40 to 1,000 nucleotides, preferably from
about 50 to 150 nucleotides, and most preferably from about 50 to 100
nucleotides. Portions of ATCs have specific functions making the ATC useful
for rolling circle amplification (RCA). These portions are referred to as the
primer complement portion and the reporter tag portions. The primer
complement portion and the reporter tag portion are required elements of an
amplification target circle. Those segments of the ATC that do not correspond
to a specific portion of the ATC can be arbitrarily chosen sequences. It is
preferred that ATCs do not have any sequences that are self complementary. It
is considered that this condition is met if there are no complementary regions
greater than six nucleotides long without a mismatch or ga.~vr.
An amplification target circle, when replicated, gives rise to a long DNA
molecule containing multiple repeats of sequences complementary to the
amplification target circle. This long DNA molecule is referred to herein as
tandem sequences DNA (TS-DNA). TS-DNA contains sequences
complementary to the primer complement portion and the reporter tag portions.
These sequences in the TS-DNA are referred to as primer sequences (which
match the sequence of the rolling circle replication primer) and reporter
tags.
Amplification target circles and their use are further described in U.S.
Patent No.
5;854,033.
Reporter Tags
Reporter tags are molecules or moieties that are produced during signal
amplification of reporter molecules in the disclosed method and to which
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
decoding tags can associate. Reporter tags can be any type of molecule or
moiety that can serve as a target for decoding tag association. Reporter tags
are
preferably the product of an amplification process, but need not be. For
example, multiple reporter tags can be synthesized in any desired manner and
associated en mass with a reporter molecule (thus effecting "amplification" of
the reporter molecule's signal). Preferred reporter tags are oligomers,
oligonucleotides, or nucleic acid sequences.
The oligomeric base sequences of oligomeric reporter tags can include
RNA, DNA, modified RNA or DNA, modified backbone nucleotide-like
oligomers such as peptide nucleic acid, methylphosphonate DNA, and 2'-O-
methyl RNA or DNA. Oligomeric or oligonucleotide reporter tags can have any
arbitrary sequence. The only requirement is association with decoding tags
(preferably by hybridization). In the disclosed method, multiple reporter tags
become associated with a single reporter signal (which is associated, via a
specific binding molecule, to a target molecule). The context of these
multiple
reporter tags depends upon the technique used for signal amplification. Thus,
where branched DNA is used, the branched DNA molecule includes the multiple
reporter tags on the branches. Where oligonucleotide dendrimers are used, the
reporter tags are on the dendrimer arms. Where rolling circle replication is
used,
multiple reporter tags result from the tandem repeats of complement of the
amplification target circle sequence (which includes at least one complement
of
the reporter tag sequence). In this case, the reporter tags are tandemly
repeated
in the tandem sequence DNA.
Oligonucleotide reporter tags can each be any length that supports
specific and stable hybridization between the reporter tags and the decoding
tags. For this purpose, a length of 10 to 35 nucleotides is preferred, with a
reporter tag 15 to 20 nucleotides long being most preferred.
The branched DNA for use in the disclosed method is generally known
(Urdea, Biotechnology 12:926-928 (1994), and Horn et al., Nucleic Acids Res
23:4835-4841 (1997)). As used herein, the tail of a branched DNA molecule
refers to the portion of a branched DNA molecule that is designed to interact
with the reporter signal. In general, each branched DNA molecule should have
only one tail. The branches of the branched DNA (also referred to herein as
the
16
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
arms of the branched DNA) contain reporter tag sequences. Oligonucleotide
dendrimers (or dendrimeric DNA) are also generally known (Shchepinov et al.,
Nucleic Acids Res. 25:4447-4454 (1997), and Orentas et al., J. Virol. Methods
77:153-163 (1999)). As used herein, the tail of an oligonucleotide dendrimer
refers to the portion of a dendrimer that is designed to interact with the
reporter
signal. In general, each dendrimer should have only one tail. The dendrimeric
strands of the dendrimer are referred to herein as the arms of the
oligonucleotide
dendrimer and contain reporter tag sequences.
Decoding Tags
Decoding tags are any molecule or moiety that can be associated with
reporter molecules, directly or indirectly, and which can be specifically
detected.
In particular, different decoding tags should be distinguishable upon
detection.
Decoding tags preferably are oligonucleotides, carbohydrates, synthetic
polyamides, peptide nucleic acids, antibodies, ligands, proteins, haptens,
zinc
fingers, aptamers, or mass labels.
Preferred decoding tags are molecules capable of hybridizing specifically
to an oligonucleotide reporter tag. Most preferred are peptide nucleic acid
decoding tags. Oligonucleotide or peptide nucleic acid decoding tags can have
any arbitrary sequence. The only requirement is hybridization to reporter
tags.
The decoding tags can each be any length that supports specific and stable
hybridization between the reporter tags and the decoding tags. For this
purpose,
a length of 10 to 35 nucleotides is preferred, with a reporter tag 15 to 20
nucleotides long being most preferred.
Decoding tags can be detected using any suitable detection technique.
Many molecular detection techniques are known and can be used in the
disclosed method. For example, decoding tags can be detected by nuclear
magnetic resonance, electron paramagnetic resonance, surface enhanced raman
scattering, surface plasmon resonance, fluorescence, phosphorescence,
chemiluminescence, resonance raman, microwave, mass spectrometry, or any
combination of these. Decoding tags can be separated and/or detected by mass
spectrometry, electrophoresis, or chromatography. Decoding tags can be
distinguished temporally via different fluorescent, phosphorescent, or
17
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
chemiluminescent emission lifetimes. The composition and characteristics of
decoding tags should be matched with the chosen detection method.
Decoding tags preferably are capable of being released by matrix-
assisted laser desorption-ionization (MALDI) in order to be separated and
identified (decoded) by time-of flight (TOF) mass spectroscopy, or of being
subjected to electrophoresis. A decoding tag may be any oligomeric molecule
that can hybridize to a reporter tag. For example, a decoding tag can be a DNA
oligonucleotide, an RNA oligonucleotide, or a peptide nucleic acid (PNA)
molecule. The preferred decoding tags of this invention are PNA molecules.
For MALDI-TOF detection, the decoding tags preferably are peptide
nucleic acids, where each decoding tag has a different mass to allow
separation
and separate detection in mass spectroscopy. For this purpose, it is
preferable
that each decoding tag have a similar number of nucleotide bases
complementary to the reporter tag. This allows for more consistent
hybridization characteristics while allowing the mass to vary. It is also
preferable to use combination of base composition and number of mass tags
(e.g.
the number of 8-amino-3,6-dioxaoctanoic monomers attached to the PNA
(Griffin, T.J., W. Tang, and L.M. Smith, Genetic analysis by peptide nucleic
acid affinity MALDI-TOF mass spectrometry. Nat Biotechnol, 1997. 15(12): p.
1368-72.)) to optimize the mass spectra for the set of decoding tags in a
multiple
tag analysis.
For capillary electrophoresis detection, the decoding tags preferably are
fluorescently-labeled oligonucleotides, where each decoding tag has a
different
combination of length and fluorescent label. For this purpose, it is
preferable
that each decoding tag has the same number of nucleotides complementary to
the reporter tag. It is also preferable that each decoding tag has a different
number of nucleotides not complementary to the reporter tag. This allows for
more consistent hybridization characteristics while allowing separation of the
different decoding tags during electrophoresis.
Reporter Carriers
Reporter carriers are associations of one or more specific binding
molecules, a carrier, and a plurality of decoding tags. Reporter carriers are
used
in the disclosed method to associate a large number of decoding tags with a
18
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
target molecule. The carrier can be any molecule or structure that facilitates
association of many decoding tags with a specific binding molecule. Examples
include liposomes, microparticles, nanoparticles, virons, phagmids, and
branched polymer structures.
Selenium can substitute for sulfur in methionine, resulting in the
modified amino acid selenomethionine. Selenium is approximately forty seven
mass units larger than sulfur. Mass spectrometry may be used to identify
peptides or proteins incorporating selenomethionine and methionine at a
particular ratio. Small proteins and peptides with known selenium/sulfur ratio
are preferably produced by chemical synthesis incorporating selenomethionine
and methionine at the desired ratio. Larger proteins or peptides may be by
produced from an E. coli expression system, or any other expression system
that
inserts selenomethionine and methionine at the desired ratio (Hendrickson,
W.A., J.R. Horton, and D.M. LeMaster, Selenomethionyl proteins produced for
analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct
determination of three-dimensional structure. Embo J, 1990. 9(5): p. 1665-72,
Cowie, D. and G. Cohen, Biosynthesis by Escherichia coli of active altered
proteins containing selenium instead of sulfur. Biochimica et Biophysica Acta,
1957. 26: p. 252 - 261, and Oikawa, T., et al., Metalloselenonein, the
selenium
analogue of metallothionein: synthesis and characterization of its complex
with
copper ions. Proc Natl Acad Sci U S A, 1991. 88(8): p. 3057-9.). Virion
particles, or phagemids, or various capsid proteins, or assennblies, with
these
fixed selenium/sulfur ratio proteins or peptides, following attachment of a
specific binding molecule, are examples of reporter carriers. The decoding
tags
(proteins or peptides) are preferably detected by mass spectrometry (e.g.
Matrix
Assisted Laser Desorption Ionization, MALDI, Mass Spectrometry or Secondary
Ion Mass Spectrometry, SIMS).
A general class of carriers are structures and materials designed for drug
delivery. Many such carriers are known. Liposomes are a preferred form of
carrier.
Liposomes are artificial structures primarily composed of phospholipid
bilayers. Cholesterol and fatty acids may also be included in the bilayer
construction. Liposomes may be loaded with fluorescent tags, and coated on the
19
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
outer surface with specific recognition molecules (Truneh, A., Machy, P. and
Horan, P.K., 1987, Antibody-bearing liposomes as multicolor
immunofluorescent markers for flow cytometry and imaging. J. Immunol.
Methods 100:59-71). However, the use of fluorescent liposomes in bioassays
has been limited by the constraints of detection methods for fluorescent tags.
Fluorescence-activated cell sorters typically have two or three different
excitation-emission wavelengths, and microscopes typically have three or four
excitation-emission filters. In some forms of the disclosed method, liposomes
serve as carriers for arbitrary decoding tags. By combining liposome reporter
carriers, loaded with arbitrary tags, with methods capable of separating a
very
large multiplicity of tags, it becomes possible to perform highly multiplexed
assays.
Liposomes, preferably unilamellar vesicles, ara made using established
procedures that result in the loading of the interior compartment with a very
large number (several thousand) of decoding tag molecules, where the chemical
nature of these molecules is well suited for detection by a preselected
detection
method. Preferred detection methods and corresponding arbitrary tags are as
follows: a) Mass spectrometry - oligopeptide tags; b) Electrophoresis - DNA
oligonucleotide tags; c) Liquid chromatography - DNA tags or oligopeptide
tags. Thus, one specific type of decoding tag preferably is used for each
specific
type of liposome-detector.
Each specific type of liposome reporter carrier is associated with a
specific binding molecule. The association may be direct or indirect. An
example of a direct association is a liposome containing covalently bound
antibodies on the surface of the phospholipid bilayer. An alternative,
indirect
association composition is a liposome containing covalently bound DNA
oligonucleotides of arbitrary sequence on its surface; these oligonucleotides
are
designed to recognize, by base complementarity, specific reporter molecules.
The reporter molecule may comprise an antibody-DNA covalent complex,
whereby the DNA portion of this complex can hybridize specifically with the
complementary sequence on a liposome reporter carrier. In this fashion, the
liposome reporter carrier becomes a generic reagent, which may be associated
indirectly with any desired binding molecule.
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
The use of liposome reporter carriers can be illustrated with the
following example.
1. Liposomes (preferably unilamellar vesicles with an average diameter
of 150 to 300 nanometers) are prepared using the extrusion method (Hope, M.J.,
Bally, M.B., Webb, G., and Cullis, P.R., Biochimica et Biophysica Acta, 1985,
812:55-65; MacDonald, R.C., MacDonald, R.L, Menco, B., Takeshita, K.,
Subbarao, N., and Hu, L. Biochimica et Biophysica Acta, 1991, 1061:297-303).
Other methods for liposome preparation may be used as well.
2. A solution of an oligopeptide, at a concentration 400 micromolar, is
used during the preparation of the liposomes, such that the inner volume of
the
liposomes is loaded with this specific oligopeptide, which will serve to
encode-
decode the identity of a specific analyte of interest. A liposome with an
internal
diameter of 200 nanometers will contain, on the average, 960 molecules of the
oligopeptide. Three separate preparations of liposomes are extruded, each
loaded with a different oligopeptide. The oligopeptides are chosen such that
their respective masses will be readily separable by MALDI-TOF mass
spectrometry.
3. The outer surface of the three liposome preparations is conjugated
with specific antibodies, as follows: a) the first liposome preparation is
reacted
with an antibody specific for the p53 tumor suppressor; b) the second liposome
preparation is reacted with an antibody specific for the Bcl-2 oncoprotein; c)
the
third liposome preparation is reacted with an antibody specific or the
Her2/neu
membrane receptor. Coupling reactions are performed using standard
procedures for the covalent coupling of antibodies to molecules. harboring
reactive amino groups (Hendrickson, E.R., Hatfield, T.M., Joerger, R.D.,
Majarian, W.R., and Ebersole, R.C., 1995, Nucleic Acids Research, 23:522-529;
Hermanson, G.T., Bioconjugate techniques, Academic Press, 1996, pp.528-569;
Scheffold, A., Assenmacher, M., Refiners-Schramm, L., Lauster, R., and
Radbruch, A., 2000, Nature Medicine 1:107-110). In the case of the liposomes,
the reactive amino groups are those present in the pho~phatidyl ethanolamine
moieties of the liposomes.
4. A glass slide bearing a standard formaldehyde-fixed histological
section is contacted with a mixture of all three liposome preparations,
suspended
21
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
in a buffer containing 30 mM Tris-HCI, pH 7.6, 100 mM Sodium Chloride, 1
mM EDTA, 0.1 % Bovine serum albumin, in order to allow binding of the
liposomes to the corresponding protein antigens present in the fixed tissue.
After a one hour incubation, the slides are washed twice, for 5 minutes, with
the
same buffer (30 mM Tris-HCI, pH 7.6, 100 mM Sodium Chloride, 1 mM EDTA,
0.1 % Bovine serum albumin). The slides are dried with a stream of air.
S. The slides are coated with a thin layer of matrix solution consisting of
mg/ml alpha-cyano-4-hydroxycinnamic acid, 0.1 % trifluoroacetic acid in a
50:50 mixture of acetonitrile in water. The slides are dried with a stream of
air.
10 6. The slide is placed on the surface of a modified MALDI plate, an
introduced in a Voyager DE Pro instrument (PerSeptive PE Biosystems,
Framingham, MA). The machine is run in positive-ion reflector mode, with an
ion extraction delay time of 250 ns.
7. Mass spectra are obtained from defined positions on the slide surface.
The relative amount of each of the three peaks of encoding polypeptides is
used
to decode the relative ratios of the antigens detected by the liposome-
detector
complexes.
The liposome carrier method is not limited to the detection of analytes on
histological sections. Cells obtained by sorting may also be used for analysis
according to this invention (Scheffold, A., Assenmacher, M., Refiners-Schramm,
L., Lauster, R., and Radbruch, A., 2000, Nature Medicine 1:107-110).
Target Samples
Any sample fiom any source can be used with the disclosed method. In
general, target samples should be samples that contain, or may contain, target
molecules. Examples of suitable target samples include cell samples, tissue
samples, cell extracts, components or fractions purified from another sample,
environmental samples, culture samples, tissue samples, bodily fluids, and
biopsy samples. Numerous other sources of samples are known or can be
developed and any can be used with the disclosed method. Preferred target
samples for use with the disclosed method are samples of cells and tissues.
Target samples can be complex, simple, or anywhere in between. For
example, a target sample may include a complex mixture of biological
molecules (a tissue sample, for example), a target sample may be a highly
22
CA 02371843 2001-11-06
WO 00/68434 PCT/LTS00/12391
purified protein preparation, or a single type of molecule. Target molecules
can
be any molecule or portion of a molecule that is to be detected. Thus, a
target
molecule need not be a physically separate molecule, but may be a part of a
larger molecule. Target molecules are also referred to as analytes.
Capture Arrays
A capture array (also referred to herein as an array) includes a plurality
of capture tags immobilized at identified or predetermined locations on the
array. In this context, plurality of capture tags refers to a multiple capture
tags
each having a different structure. Each predetermined location on the array
(referred to herein as an array element) has one type of capture tag (that is,
all
the capture tags at that location have the same structure). Each location will
have multiple copies of the capture tag. The spatial separation of capture
tags of
different structure in the array allows separate detection and identification
of
target molecules that become associated with the capture tags. If a decoding
tag
is detected at a given location in a capture array, it indicates that the
target
molecule corresponding to that array element was present in the target sample.
Reporter molecules and detector tags can also be immobilized in arrays.
Different modes of the disclosed method can be performed with different
components immobilized, labeled, or tagged. Arrays of reporter molecules and
decoding tags can be made and used as described below and elsewhere herein
for capture tags.
Solid-state substrates for use in capture arrays can include any solid
material to which capture tags can be coupled, directly or indirectly. This
inchtdes materials such as acrylamide, cellulose, nitrocellulose, glass,
polystyrene, polyethylene vinyl acetate, polypropylene, polymethacrylate,
polyethylene, polyethylene oxide, glass, polysilicates, polycarbonates,
teflon,
fluorocarbons, nylon, silicon rubber, polyanhydrides, polyglycolic acid,
polylactic acid, polyorthoesters, polypropylfumerate, collagen,
glycosaminoglycans, and polyamino acids. Solid-state substrates can have any
useful form including thin films or membranes, beads, bottles, dishes, fibers,
woven fibers, shaped polymers, particles and microparticles. A preferred form
for a solid-state substrate is a microtiter dish. The most preferred form of
microtiter dish is the standard 96-well type.
23
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
Planar array technology has been utilized for many years (Shalom D., S.J.
Smith, and P.O. Brown, A DNA microarray system for,analyzing complex DNA
samples using two- color fluorescent probe hybridization. Genome Res, 1996.
6(7): p. 639-45, Singh-Gasson, S., et al., Maskless fabrication of light-
directed
oligonucleotide microarrays using a digital micromirror array. Nat Biotechnol,
1999. 17(10): p. 974-8, Southern, E.M., U. Maskos, and J.K. Elder, Analyzing
and comparing nucleic acid sequences by hybridization to arrays of
oligonucleotides: evaluation using experimental models. Genomics, 1992. 13(4):
p. 1008-17, Nizetic, D., et al., Construction, arraying, and high-density
screening of large inse~°t libraries of human chromosomes X and 21:
their
potential use as reference libraries. Proc Natl Acad Sci U S A, 1991. 88(8):
p.
3233-7, Van Oss, C.J., R.J. Good, and M.K. Chaudhury, Mechanism of DNA
(Southern) and protein (Western) blotting on cellulose nitrate and other
membranes. J Chromatogr, 1987. 391(1): p. 53-65, Ramsay, G., DNA chips:
state-of the art. Nat Biotechnol, 1998. 16(1): p. 40-4, Schena, M., et al.,
Parallel
human genome analysis: microarray-based expression monitoring of 1000
genes. Proc Natl Acad Sci U S A, 1996. 93(20): p. 10614-9, Lipshutz, R.J., et
al., High density synthetic oligonucleotide arrays. Nat Genet, 1999. 21(1
Supply:
p. 20-4, Pease, A.C., et al., Light-generated oligonucleotide arrays for rapid
DNA sequence analysis. Proc Natl Acad Sci U S A, 1994. 91(11): p. 5022-6,
Maier, E., et al., Application of robotic technology to automated sequence
fingerprint analysis by oligonucleotide hybridisation. J Biotechnol, 1994.
35(2-
3): p. 191-203, Vasiliskov, A.V., et al., Fabrication ofmicroarray ofgel-
immobilized compounds on a chip by copolymerization. Biotechniques, 1999.
27(3): p. 592-4, 596-8, 600 passim, and Yershov, G., et al., DNA analysis and
diagnostics on oligonucleotide microchips. Proc Natl Acad Sci U S A, 1996.
93(10): p. 4913-8). Such arrays may be constructed upon non permeable or
permeable supports of a wide variety of support composition, for example
nylon,
cellulose, glass, polymer on glass, and many others. The array spot sizes and
density of spot packing vary over a tremendous range depending upon the
processes) and materials) used.
Methods for immobilization of oligonucleotides to solid-state substrates
are well established. Oligonucleotide capture tags can be coupled to
substrates
24
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
using established coupling methods. For example, suitable attachment methods
are described by Pease et al., Proc. Natl. Acad. Sci. USA 91(11):5022-5026
(1994), and Khrapko et al., Mol Biol (Mosk) (USSR) 25:718-730 (1991). A
method for immobilization of 3'-amine oligonucleotides on casein-coated slides
is described by Stimpson et al., Proc. Natl. Acad. Sci. USA 92:6379-6383
(1995). A preferred method of attaching oligonucleotides to solid-state
substrates is described by Guo et al., Nucleic Acids Res. 22:5456-5465 (1994).
Methods for producing arrays of oligonucleotides on solid-state
substrates are also known. Examples of such techniques are described in U.S.
Patent No. 5,871,928 to Fodor et al., U.S. Patent No. 5,654,413 to Brenner,
U.S.
Patent No. 5,429,807, and U.S. Patent No. 5,599,695 to Pease et al.
Although preferred, it is not required that a given capture array be a
single unit or structure. The set of capture tags may be distributed over any
number of solid supports. For example, at one extreme, each capture tag may be
1 S immobilized in a separate reaction tube or container.
Oligonucleotide capture tags in arrays can also be designed to have
similar hybrid stability. This would make hybridization of fragments to such
capture tags more efficient and reduce the incidence of mismatch
hybridization.
The hybrid stability of oligonucleotide capture tags can be calculated using
known formulas and principles of thermodynamics (see, for example, Santa
Lucia et al., Biochemistry 35:3555-3562 (1996); Freier et al., Proc. Natl.
Acad.
Sci. USA 83:9373-9377 (1986); Breslauer et al., Proc. Natl. Acad. Sci. USA
83:3746-3750 (1986)). The hybrid stability of the oligonucleotide capture tags
can be made more similar (a process that can be referred to as smoothing the
hybrid stabilities) by, for example, chemically modifying the capture tags
(Nguyen et al., Nucleic Acids Res. 25(15):3059-3065 (1997); Hohsisel, Nucleic
Acids Res. 24(3):430-432 (1996)). Hybrid stability can also be smoothed by
carrying out the hybridization under specialized conditions (Nguyen et al.,
Nucleic Acids Res. 27(6):1492-1498 (1999); Wood et al., Proc. Natl. Acad. Sci.
USA 82(6):1585-1588 (1985)).
Another means of smoothing hybrid stability of the oligonucleotide
capture tags is to vary the length of the capture tags. This would allow
adjustment of the hybrid stability of each capture tag so that all of the
capture
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
tags had similar hybrid stabilities (to the extent possible). Since the
addition or
deletion of a single nucleotide from a capture tag will change the hybrid
stability
of the capture tag by a fixed increment, it is understood that the hybrid
stabilities
of the capture tags in a capture array will not be equal. For this reason,
similarity of hybrid stability as used herein refers to any increase in the
similarity of the hybrid stabilities of the capture tags (or, put another way,
any
reduction in the differences in hybrid stabilities of the capture tags).
The efficiency of hybridization and ligation of oligonucleotide capture
tags to sample fragments can also be improved by grouping capture tags of
similar hybrid stability in sections or segments of a capture array that can
be
subjected to different hybridization conditions. In this way, the
hybridization
conditions can be optimized for particular classes of capture tags.
Capture Tags
A capture tag is any compound that can be used to capture or separate
compounds or complexes having the capture tag. Preferably, a capture tag is a
compound, such as a ligand or hapten, that binds to or interacts with another
compound, such as ligand-binding molecule or an antibody. It is also preferred
that such interaction between the capture tag and the capturing component be a
specific interaction, such as between a hapten and an antibody or a ligand and
a
ligand-binding molecule.
Preferred capture tags, described in the context of nucleic acid probes,
are described by Syvnen et al., Nucleic Acids Res., 14:5037 (1986). Preferred
capture tags include biotin, which can be incorporated into nucleic acids.
Capturing sample fragments on a substrate may be accomplished in several
ways. In one embodiment, capture docks are adhered or coupled to the
substrate. Capture docks are compounds or moieties that mediate adherence of a
sample fragment by binding to, or interacting with, a capture tag on the
fragment. Capture docks immobilized on a substrate allow capture of the
fragment on the substrate. Such capture provides a convenient means of
washing away reaction components that might interfere with subsequent steps.
Substrates for use in the disclosed method can include any solid material
to which components of the assay can be adhered or coupled. Examples of
substrates include, but are not limited to, materials such as acrylamide,
cellulose,
26
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
nitrocellulose, glass, polystyrene, polyethylene vinyl acetate, polypropylene,
polymethacrylate, polyethylene, polyethylene oxide, polysilicates,
polycarbonates, teflon, fluorocarbons, nylon, silicon rubber, polyanhydrides,
polyglycolic acid, polylactic acid, polyorthoesters, polypropylfumerate,
collagen, glycosaminoglycans, and polyamino acids. Substrates can have any
useful form including thin films or membranes, beads, bottles, dishes, fibers,
woven fibers, shaped polymers, particles and microparticles. Preferred forms
of
substrates are plates and beads. The most preferred form of beads are magnetic
beads.
In one embodiment, capture tags and capture docks can be
oligonucleotides. Methods for immobilizing and coupling oligonucleotides to
substrates are well established. For example, suitable attachment methods are
described by Pease et al., Proc. Natl. Acad. Sci. USA 91(11):5022-5026 (1994),
and Khrapko et al., Mol Biol (Mosk) (USSR) 25:718-730 (1991). A method for
immobilization of 3'-amine oligonucleotides on casein-coated slides is
described
by Stimpson et al., Proc. Natl. Acad. Sci. USA 92:6379-6383 (1995). A
preferred method of attaching oligonucleotides to solid-state substrates is
described by Guo et al., Nucleic Acids Res. 22:5456-5465 (1994).
In another embodiment, caputre tags and capture docks can be
anti-hybrid antibodies. Methods for immobilizing antibodies to substrates are
well established. Immobilization can be accomplished by a.~r:achment, for
example, to aminated surfaces, carboxylated surfaces or hydroxylated surfaces
using standard immobilization chemistries. Examples of attachment agents are
cyanogen bromide, succinimide, aldehydes, tosyl chloride, avidin-biotin,
photocrosslinkable agents, epoxides and maleimides. A preferred attachment
agent is glutaraldehyde. These and other attachment agents, as well as methods
for their use in attachment, are described in PYOtein immobilization:
fundamentals and applications, Richard F. Taylor, ed. (M. Dekker, New York,
1991), Johnstone and Thorpe, Immunochemistry In Practice (Blackwell
Scientific Publications, Oxford, England, 1987) pages 209-216 and 241-242, and
Immobilized Affinity Ligands, Craig T. Hermanson et al., eds. (Academic Press,
New York, 1992). Antibodies can be attached to a substrate by chemically
cross-linking a free amino group on the antibody to reactive side groups
present
27
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
within the substrate. For example, antibodies may be chemically cross-linked
to
a substrate that contains free amino or carboxyl groups using glutaraldehyde
or
carbodiimides as cross-linker agents. In this method, aqueous solutions
containing free antibodies are incubated with the solid-state substrate in the
presence of glutaraldehyde or carbodiimide. For crosslinking with
glutaraldehyde the reactants can be incubated with 2% glutaraldehyde by
volume in a buffered solution such as 0.1 M sodium cacodylate at pH 7.4. Other
standard immobilization chemistries are known by those of skill in the art.
Sorting Tags
A sorting tag is any compound that can be used to sort or separate
compounds or complexes having the sorting tag from those that do not. In
general, all capture tags can be a sorting tag. Sorting tags also include
compounds and moieties that can be detected and which can mediate the sorting
of tagged components. Such forms of sorting tags are generally not also
capture
tags. For example, a fluorescent moiety can allow sorting of components tagged
with the moiety from those that are not (or those with a different tag).
However,
such a fluorescent moiety does not necessarily have a suitable capture dock
with
which it can interact and be captured. Preferably, a sorting tag is a label,
such as
a fluorescent label, that can mediate sorting.
Method
The disclosed method is based on encoding target molecules with signals
followed by decoding of the encoded signal. This encoding/decoding uncouples
the detection of a target molecule from the chemical and physical properties
of
the target molecule. In basic form, the disclosed method involves association
of
one or more reporter molecules with one or more target samples, association of
one or more decoding tags with the reporter molecules, and detection of the
decoding tags. The reporter molecules associate with target molecules in the
target sample(s). Generally, the reporter molecules correspond to one or more
target molecules, and the decoding tags correspond to one or more reporter
molecules. Thus, detection of particular decoding tags indicates the presence
of
the corresponding reporter molecules. In turn, the presence of particular
reporter
molecules indicates the presence of the corresponding target molecules.
28
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
This indirect detection uncouples the detection of target molecules from
the chemical and physical properties of the target molecules by interposing
decoding tags that essentially can have any arbitrary chemical and physical
properties. In particular, decoding tags can have specific properties useful
for
detection, and decoding tags within an assay can have highly ordered or
structured relationships with each other. It is the (freely chosen) properties
of
the decoding tags, rather than the (take them as they are) properties of the
target
molecules that matters at the point of detection.
The decoding tags have the additional advantage of being uncoupled
from the target molecule-specific aspects of the reporter molecules. Unlike
detection methods where a labeled molecule is bound to an analyte followed by
detection of the label, the disclosed method is not limited by the chemical
and
physical properties of the labeled molecule. This allows more convenient
detection, more sensitive detection, and more highly multiplexed detection
1 S schemes.
The sensitivity of the disclosed method can also be enhanced by
including a signal amplification step prior to detection. In basic form,
amplification is accomplished by amplifying reporter signals on the reporter
molecules. This results in multiple reporter tags associated with each
reporter
molecule. The decoding tags are then associated with the reporter tags and
detected. Generally, the decoding tags correspond to one or more reporter tags
(and thus correspond to the reporter molecules with which the reporter tags
are
associated), and the reporter tags correspond to the reporter molecules with
which they are associated. Thus, detection of particular decoding tags
indicates
the presence of the corresponding reporter tags. In turn, the presence of
particular reporter tags indicates the presence of the corresponding reporter
molecules. In turn, the presence of particular reporter molecules indicates
the
presence of the corresponding target molecules. The reporter molecule also can
be designed to include multiple reporter tags (essentially accomplishing a pre-
assay amplification of the signal).
In general, the target sample is expected to contain, or is suspected of
containing, a plurality of different target molecules. The specific binding
molecule of each reporter molecule is selected to interact with one of the
target
29
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
molecules. A set of specific binding molecules can be selected to interact
with
all of the target molecules of interest in the sample. The target sample
preferably is immobilized, fixed, or adhered to a surface. Alternatively,
reporter
molecules can be associated with a sample, or the source of a sample, prior to
immobilization, fixation, or adherence. Either case allows the locations of
the
target molecules on the surface to be determined in the method by associating
decoding tags with the target molecules and detecting the location of the
decoding tags.
Any number of reporter molecules can be used when performing the
disclosed method. The disclosed method is especially useful for detection of
multiple target molecules in a single assay. This multiplexing is made
convenient in the disclosed method by the uncoupling of the chemical nature of
the target molecules and the chemical nature of the decoding tags, and
multiple
codings that can be embodied in the decoding tags. Accordingly, it is
preferred
that four or more, five or more, six or more, seven or more, eight or more,
nine
or more, ten or more, twenty or more, forty or more, eighty or more, or one
hundred or more reporter molecules be used in a single performance of the
disclosed method. The number of reporter molecules that can be used are not
limited to these ranges but can include any sub-range. For example, thirty or
more, fifty-five or more, seventy-two or more, and so on, reporter molecules
can
be used. All of the included sub-ranges are specifically contemplated.
Multiple Tag Analysis
In one form of the disclosed method, one or more target samples and one
or more reporter molecules are brought into contact, allowing the reporter
molecules to become associated with target molecules ~n the target samples.
The
reporter molecules are then amplified to produce multiple reporter tags for
each
reporter molecule. The reporter tags remain associated with the reporter
molecules. One or more decoding tags are then associated with the reporter
tags,
and the decoding tags are detected. The decoding tags correspond to reporter
molecules and the reporter molecules correspond to target molecules. This
relationship means that detecting the decoding tags indicates the presence of
reporter molecules corresponding to the detected decoding tags, and that the
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
presence of reporter molecules indicates the presence of target molecules
corresponding to the reporter molecules.
This form of the disclosed method is based on specific recognition of
analytes in a sample, amplification of the recognition event, and detection of
the
amplified signal. The specific recognition is accomplished with specific
binding
molecules that bind or otherwise interact specifically with an analyte of
interest.
The amplification of this interaction is mediated by a reporter signal
coupled,
tethered, or otherwise attached to the specific binding molecule. It is the
reporter signal, an oligonucleotide, that is used to produce multiple reporter
tags
that remain associated with the reporter signal (and thus also with the
specific
binding molecule and analyte). This is accomplished, for example, by
amplification of the reporter signal or hybridization of branched DNA or
oligonucleotide dendrimers to the reporter signals. Detector tags, which are
coded to include distinct, separately detectable attributes, are then
hybridized to
the reporter tags. Detection of the various detector tags results in indirect
detection of the various analytes in the sample.
A key to the method is the use of distinct reporter signals for each
different analyte, the subsequent association of distinct reporter tags with
the
various reporter signals, and the subsequent hybridization of distinct
decoding
tags to the various reporter tags. In this way, each different type of analyte
ends
up with a different and separately detectable decoding tag :associated with
it.
The combination of specific binding molecule and reportex signal results in an
effective conversion of the different analytes (where the analytes may be
widely
chemically divergent) into a standardized signal (the reporter signal). These
standardized signals (which are coded for each analyte) are then amplified and
detected using a single set of standardized conditions (during which the
coding
is carried forward). The detection of the coded, amplified signals (that is,
the
decoding tags) results in effective, convenient detection of multiple analytes
in a
single assay.
An example of this form of the disclosed method has the following basic
steps:
(a) bringing into contact one or more target samples and one or more
reporter molecules, where each reporter molecule includes a reporter signal
and
31
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
a specific binding molecule. The reporter signal preferably is an
oligonucleotide
coupled or tethered to the specific binding molecule. The specific binding
molecule of each reporter molecule is chosen to interact with a different
target
molecule.
(b) amplifying the reporter signal to produce multiple reporter tags for
each reporter signal, where the reporter tags remain associated with the
reporter
signal. The reporter tags produced from the reporter signal of each reporter
molecule preferably are different from the reporter tags produced from the
reporter signals of other reporter molecules.
(c) associating decoding tags to the reporter tags, where a different
decoding tag corresponds to each different reporter tag.
(d) detecting the decoding tags associated with the reporter tags. The
detected decoding tags are indicative of the location and amount of target
molecules in the target sample.
Multiple Tag Analysis of Multiple Samples
In another form of the disclosed method, four or more target samples and
one or more reporter molecules are brought into contact, allowing the reporter
molecules to become associated with target molecules in the target samples.
Each target sample is brought into contact with a different set of reporter
molecules, and the reporter molecules in each set of reporter molecules are
different from the reporter molecules in the other sets of reporter molecules.
The four or more of the target samples are then mixed together, one or more
decoding tags are associated with the reporter molecules, and the decoding
tags
are detected. A different decoding tag corresponds to each different reporter
molecule such that each decoding tag corresponds to only one of the target
samples. This relationship means that detecting the decoding tags indicates
the
presence of target molecules corresponding to the detected decoding tags.
Further, detection of decoding tags corresponding to different target samples
indicates the presence of the same target molecules in the corresponding
target
samples.
In this form of the method, it is preferred that the reporter molecules be
associated with target molecules by covalently coupling the reporter molecules
to the target molecules. For example, the reporter molecules can be associated
32
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
with target molecules by incorporating the reporter molecules during synthesis
of the target molecules, or by reacting a reactive group on the reporter
molecules
with the target molecules. A preferred way to incorporate reporter molecules
into target molecules is to include a primer portion in the reporter molecule
that
is used to prime synthesis of a nucleic acid molecule.
An example of this form of the disclosed method has the following basic
steps:
(a) bringing into contact four or more target samples and one or more
reporter molecules, where each target sample is brought into contact with a
different set of reporter molecules. The reporter molecules in each set of
reporter molecules are different from the reporter molecules in the other sets
of
reporter molecules, and the reporter molecules are associated with target
molecules in the target samples.
(b) mixing together four or more of the target samples.
(c) associating one or more decoding tags with the reporter molecules,
where a different decoding tag corresponds to each different reporter molecule
such that each decoding tag corresponds to only one of the target samples.
(d) detecting the decoding tags. The decoding tags are disassociated
from the reporter molecules during, or prior to, detection. Detection of the
decoding tags indicates the presence of target molecules corresponding to the
detected decoding tags. Detection of decoding tags corresponding to different
target samples indicates the presence of the same target molecules in the
corresponding target samples.
This form of the method can also include, prior to step (c), amplifying
the reporter molecules to produce multiple reporter tags for each reporter
molecule. The reporter tags remain associated with the reporter molecule. The
reporter tags produced from each reporter molecule are different from the
reporter tags produced from other reporter molecules, and the decoding tags
are
associated with the reporter molecules by associating the decoding tags with
the
reporter tags.
In preferred embodiments of this form of the method, each reporter
molecule can comprise a reporter signal such that each reporter molecule in
the
same set of reporter molecules has the same reporter signal. Amplification of
33
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
the reporter molecules is then accomplished by amplifying the reporter
signals.
A plurality of decoding tags can correspond to each target sample.
The reporter molecules can be associated with target molecules by
covalently coupling the reporter molecules to the target molecules. For
example,
the reporter molecules can be associated with target molecules by
incorporating
the reporter molecules during synthesis of the target molecules.
Alternatively,
the reporter molecules can be covalently coupled to the target molecules by
reacting a reactive group on the reporter molecules with the target molecules.
Each reporter molecule can comprise a primer portion and a reporter signal,
where the primer portions prime synthesis of the target molecules. The
decoding tags can then be associated with the reporter molecules by
associating
the decoding tags with the reporter signals. The reporter signals can be
oligonucleotides, and the decoding tags can be peptide nucleic acids that are
complementary to the reporter signals.
This form of the method is useful for comparing a large number of
experimental samples. In a typical case, a large batch of experimental animals
is
treated with a drug, and specific organs, such as brain, liver, kidney,
bladder,
lung, and ovary or testis are harvested for analysis. Such an experiment may
involve 24 experimental animals, where 6 different tissues are harvested at 4
time points. Using prior technology, one must then perform 24 microarray
experiments, where mRNA or protein is extracted from each experimental
organ-time point is compared to a control mRNA or protein from the same organ
or tissue. These 24 experiments consume considerable time and resources
(including the significant cost of the 24 microarrays).
One embodiment of the disclosed method allows a multiplexed, or
parallel analysis of an entire panel of mRNA samples in a single microarray
experiment. Another embodiment of the disclosed method allows a multiplexed,
or parallel analysis of an entire panel of protein samples in a single
microarray
experiment. Examples of both embodiments are described below.
In the first, an experimental animal is treated with a drug. To study
toxicity effects, messenger RNA is extracted from six different tissue
biopsies of
the experimental animal at time zero (just before drug administration), and at
34
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
another three time points (days 1, 2, 3). A total of 24 different mRNA
preparations is obtained.
The messenger RNA is labeled as follows: A first strand of cDNA is
generated using reverse transcriptase, and the RNA strand is destroyed with
alkali. After ethanol precipitation, the RNA is copied by random priming using
a
random octamer (the specific binding molecule) tethered at its 5'-end to an
arbitrary DNA zip sequence of 20 bases (the reporter signal), the zip sequence
preferably comprising 50% isoG and isoC residues (Collins ML, Irvine B, Tyner
D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen LP, Kolberg J,
Bushnell S, Urdea MS, Ho DD (1997) A branched DNA signal amplification
assay for quantification of nucleic acid targets below 100 molecules/ml.
Nucleic
Acids Res 25:2979-2984). For each of the 24 different mRNA preparations, a
unique random octamer-zip sequence is used, designed according to the rules
described by (Gerry NP, Witowski NE, Day J, Hammer RP, Barany G, Barany F
(1999) Universal DNA microarray method for multiplex detection of low
abundance point mutations. .l Mol Biol 292:251-62).
All of the 24 random-primed cDNA preparations, each harboring unique
zip sequences, are pooled together before being used for hybridization. The
pooled probes are hybridized to a cDNA microarray made on glass slides. After
overnight hybridization, the slides are washed using standard procedures, and
are then hybridized with 24 different branch DNA (bDNA) ~~r~~plifier
assemblies
(Hendricks DA, Stowe BJ, Hoo BS, Kolberg J, Irvine BD, :~euwald PD, Urdea
MS, Perrillo RP (1995) Quantitation of HBV DNA in human serum using a
branched DNA (bDNA) signal amplification assay. Am JClin Pathol 104:537-46)
tethered to isoG and isoC containing anti-zip oligos complementary to each of
the 24 random primer zip oligonucleotides. After washing excess bDNA, the
slides are hybridized with 24 PNA mass tags (these are the decoding tags),
each
capable of hybridizing to a single specific bDNA amplified tag sequence, and
each having a different molecular weight.
After washing, the microarrays are covered with a matrix solution, dried,
and placed in a mass spectrometer. The laser beam is directed sequentially to
each of the microarray dots and MALDI-TOF mass spectra are generated for all
dots.
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
The intensity of each peak, corresponding to the different PNA mass tags, is
recorded, in order to measure the expression of the mRNA corresponding to
each tag in each tissue time point (for a total of 24 data points per spot on
the
microarray).
In the above example, which involved mRNA profiling, 6 different
tissues are assayed at 4 different time points, from a total of 24
experimental
animals. Likewise, protein may be extracted from each sample. Each protein
preparation can be tagged by covalent coupling of a DNA oligonucleotide. All
24 protein preparations are then mixed together, and contacted with the
antibody
microarray. After washing unbound proteins, the bDNA oligos are amplified,
and PNA decoding tags are bound. Signal readout is performed by mass
spectrometry (electrophoresis and HPLC are less preferred options). This
experiment will yield data on the relative expression level of each protein
for
which there exists a cognate antibody on the microarray. Relative protein
expression levels are obtained for all of the 24 experimental time points.
In the second example, an experimental animal is treated with a drug.
To study toxicity effects, protein is extracted from six different tissue
biopsies of
the experimental animal at time zero (just before drug administration), and at
another three time points (days 1, 2, 3). A total of 24 different protein
preparations is obtained.
Each of the protein preparations is tagged with a unique DNA
oligonucleotide as follows: ,
The protein preparation is reacted with 2-iminothiolane to introduce
reactive sulfhydryl groups, if none is present. A DNA oligonucleotide,
containing a reactive amino group at one of its termini is reacted with a
heterobifunctional cross-linking reagent, such as SULFO-SMCC (Pierce, Inc.).
The thiol-containing proteins are incubated together with the activated
oligonucleotide, to form a covalent protein-DNA adduct. For most protein
molecules, the formation of this covalent adduct will not interfere with the
capacity of the protein to bind to its cognate antibody.
All 24 protein preparations, each harboring covalently bound unique
DNA tag sequences, are pooled together before being used for an antibody
microarray experiment. The pooled proteins are contacted with a microarray
36
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
made by spotting tens, hundreds, or thousands of specific antibodies at
different
locations on glass slides (Ekins and Chu, 1991 ). After overnight incubation,
the
slides are washed using standard immunoassay procedures, and are then
hybridized with 24 different branch DNA (bDNA) amplifier assemblies
(Hendriclcs et al., 1995).
After washing away excess bDNA, the slides are hybridized with 24
different PNA mass tags, each capable of hybridizing to a single specific bDNA
amplified tag sequence, and each having a unique molecular weight. After
washing, the microarrays are covered with a matrix solution, dried, and placed
in
a mass spectrometer. The laser beam is directed sequentially to each of the
microarray dots and MALDI-TOF mass spectra are generated for all dots. The
intensity of each peak, corresponding to the 24 different PNA mass tags, is
recorded at each dot on the microarray, in order to measure the relative
amounts
of each of the tagged proteins (each bound to its specific, cognate antibody
on
the surface of the microarray) from each of the two experimental samples.
Multiple Tag Analysis Without Amplification
The decoding tags of the disclosed method have the advantage of being
uncoupled from the target molecule-specific aspects of the reporter molecules.
Unlike detection methods where a labeled molecule is bound to an analyte
followed by detection of the label, the disclosed method is not limited by the
chemical and physical properties of the labeled molecule. This allows more
convenient detection, more sensitive detection, and more highly multiplexed
detection schemes.
In another form of the disclosed method, one or more target samples and
one or more reporter molecules are brought into contact, allowing the reporter
molecules to become associated with target molecules ~n the target samples.
One or more decoding tags are then associated with the reporter molecules. A
different decoding tag corresponds to each different reporter molecule, and
the
decoding tags are not covalently coupled to the reporter molecules. The
decoding tags are then detected by disassociating the decoding tags from the
reporter molecules. The decoding tags correspond to reporter molecules, and
the
reporter molecules correspond to target molecules. This relationship means
that
detection of the decoding tags indicates the presence of reporter molecules
37
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
corresponding to the detected decoding tags, and that the presence of reporter
molecules indicates the presence of target molecules corresponding to the
reporter molecules.
An example of this form of the disclosed method has the following basic
steps:
(a) bringing into contact one or more target samples and one or more
reporter molecules, where the reporter molecules are associated with target
molecules in the target samples.
(b) associating one or more decoding tags with the reporter molecules.
A different decoding tag corresponds to each different reporter molecule. The
decoding tags are not covalently coupled to the reporter molecules.
(c) detecting the decoding tags. The decoding tags are disassociated
from the reporter molecules during, or prior to, detection. The decoding tags
correspond to reporter molecules. The reporter molecules correspond to target
molecules. Detection of the decoding tags indicates the presence of reporter
molecules corresponding to the detected decoding tags, and the presence of
reporter molecules indicates the presence of target molecules corresponding to
the reporter molecules.
In preferred embodiments of this form of the method, each reporter
molecule can comprise one or more specific binding molecules such that the
specific binding molecule of each reporter molecule interacts with a different
target molecule.
This form of the method can also include, prior to step (c), amplifying
the reporter molecules to produce multiple reporter tags for each reporter
molecule. The reporter tags remain associated with the reporter molecule. The
reporter tags produced from each reporter molecule are different from the
reporter tags produced from other reporter molecules, and the decoding tags
are
associated with the reporter molecules by associating the decoding tags with
the
reporter tags.
As an example of this form of the method, consider a nucleic acid
construct (reporter molecule) that contains two domains, a detection domain
(specific binding molecule) and an encoding domain (reporter signal),
5'-GCATCGCATCGGATCGATCGACGGGGCAGA-3'
JS
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
The underlined sequence, 5'-GCATCGCATCGGATCGATCG-3',
represents specific binding molecule of a reporter molecule that will
specifically
hybridize to a cognate single stranded nucleic acid target molecule locus of
interest. The remaining part of this construct, 5'-ACGGGGCAGA-3',
represents the reporter signal of a reporter molecule. A PNA that is
complementary to all or part of the reporter signal sequence may act as the
decoding tag. The binding of PNA:DNA duplexes is stronger than the binding
of DNA:DNA duplexes (Chakrabarti, M.C. and F.P. Schwarz, Thermal stability
of PNAlDNA and DNAlDNA duplexes by differential scanning calorimetry.
Nucleic Acids Res, 1999. 27(24): p. 4801-6; Schwarz, F.P., S. Robinson, and
J.M. Butler, Thermodynamic comparison of PNAlDNA and DNAlDNA
hybridization reactions at ambient temperature. Nucleic Acids Res, 1999.
27(24): p. 4792-800), thus the PNA will remain bound to this construct in the
presence of a DNA strand of the same sequence. Use of isoC and isoG (Collins,
M.L., et al., A branched DNA signal amplification assay for quantification of
nucleic acid targets below 100 moleculeslml. Nucleic Acids Res, 1997. 25(15):
p. 2979-84; Switzer, C.Y., S.E. Moroney, and S.A. Benner, Enzymatic
recognition of the base pair between isocytidine and isoguanosine.
Biochemistry, 1993. 32(39): p. 10489-96; Horn, T., C.A. Chang, and M.L.
Collins, Tetrahedron Lett., 1995. 36: p. 2033-2036) in the reporter signal and
the
decoding tag will make these sequences unable to hybridize »~ith the target
sequence.
As an illustration of the use of this example method, consider the
following.
The construct is produced by mixing the single stranded nucleic acid
with the cognate decoding PNA. Within a set, such constructs are produced
individually. The DNA target molecule of interest is amplified by PCR with
modified primers. The primers are designed such that the resultant DNA strand
will contain a 3' biotin moiety, and the 5' end contains at least four
phosphorothioate linked nucleotides rather than phosphodiester linkages.
The double stranded PCR amplicons are digested by T7 gene 6 to yield
single stranded DNA (Nilciforov, T.T., et al., The use
ofphospho~°othioate
primers and exonuclecrse hyd~°olysis fo~° the
prepay°ation of single-stranded PCR
39
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
products and their detection by solid phase hybridization. PCR Methods Appl,
1994. 3(5): p. 285-91). The set of constructs is contacted with the sample
under
conditions for hybridization of the probe sequence with the cognate DNA
sample. The mixture is transferred to an avidin coated well where the PCR
amplicons are captured by the biotin-avidin interaction and the excess and
mismatched constructs are washed away. A low salt wash is performed to
release from the DNA target molecules the reporter molecules with their
associated decoding tags. The liquid phase is transferred to a well of an
autosampler for detection of the multiple PNA decoding tags.
Each PNA decoding tag sequence is detected by its unique mass
spectrum, which uniquely identifies the decoding tag. The detector is, for
example, an Electrospray Time-of Flight Mass Spectrometer. In the mass
spectrometer source the oligonucleotide is dissociated from the PNA decoding
sequence, and the PNA is detected. Alternately, the decoding sequence can be a
chromatographically or electrophoretically separable moiety. Use of any of
these techniques for multiple tag analysis enables the detection and
identification
of hundreds, or even thousands, of different decoding tags, which in turn
identifies hundreds, or even thousands, of different target molecules.
A preferred construct would utilize an improvement of a molecular
switch (Lizardi, P.M., et al., Nucleic acid process containing improved
molecular switch, 1992.United States Patent 5,118,801). Related probe
molecules labeled with a fluorophore at one terminus and a quencher at the
other
terminus have been called molecular beacons (Tyagi, S., D.P. Bratu, and F.R.
Kramer, Multicolor molecular beacons for allele discrimination. Nat
Biotechnol, 1998. 16(1): p. 49-53; Tyagi, S. and F.R. Kramer, Molecular
beacons: probes that fluoresce upon hybridization. Nat Biotechnol, 1996.
14(3):
p. 303-8; Marras, S.A., F.R. Kramer, and S. Tyagi, Multiplex detection of
single-
nucleotide variations using molecular beacons. Genet Anal, 1999. 14(5-6): p.
151-6; Vet, J.A., et al., Multiplex detection offour pathogenic retroviruses
using
molecular beacons. Proc Natl Acad Sci U S A, 1999. 96(11): p. 6394-9;
Manganelli, R., et al., Differential expression of 10 sigma factor genes in
Mycobacterium tuberculosis. Mol Microbiol, 1999. 31(2): p. 715-24) and have
been demonstrated to have better detection specificity than their linear
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
equivalent probe sequence (Bonnet, G., et al., Thermodynamic basis of the
enhanced specificity of structured DNA probes. Proc I~(atl Acad Sci U S A,
1999.
96(11): p. 6171-6). The beacons are limited in the extent of multiplexing due
to
spectral overlap of the readout fluorophores. In the current invention, the
specificity of the molecular switch is exploited, and a much higher multiplex
readout is enabled. An example of such a construct is:
5'-XMGCATCGCATCGGATCGATCGNYACGGGGCAGA-3'
where M and N indicate a (typically small) number of bases; X and Y are short
sequences that are complementary to each other, typically 5 to 8 bases; and,
detection and encoding domains as indicated above. Under the hybridization
conditions this construct will form a 'step and loop' structure (where X and Y
are hybridized to each other) which competes with the probe sequence
hybridizing to the target sequence.
Multiple Tag Analysis With Reporter Carriers
In another form of the disclosed method, one or more target samples and
one or more reporter carriers are brought into contact. The reporter carriers
include one or more specific binding molecules, a carrier, and a plurality of
decoding tags associated with the carrier. After the reporter carriers are
associated with reporter molecules, the decoding tags are detected. The
decoding tags correspond to reporter carriers, and the reporter carriers
correspond to target molecules. This relationship means that detection of the
decoding tags indicates the presence of reporter carriers corresponding to the
detected decoding tags, and that the presence of reporter carriers indicates
the
presence of target molecules corresponding to the reporter carriers.
An example of this form of the disclosed method has the following basic
steps:
(a) bringing into contact one or more target samples and one or more
reporter carriers, where each reporter carrier comprises one or more specific
binding molecules, a carrier, and a plurality of decoding tags associated with
the
carrier.
(b) detecting the decoding tags. The decoding tags correspond to
reporter carriers and the reporter carriers correspond to target molecules.
Detecting the decoding tags indicates the presence of reporter carriers
41
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
corresponding to the detected decoding tags, and the presence of reporter
carriers indicates the presence of target molecules corresponding to the
reporter
carriers.
The carrier can be a liposome. Each reporter carrier preferably includes
at least 1,000 decoding tags. The decoding tags preferably are detected by
mass
spectrometry, electrophoresis, or chromatography. The carrier can also be a
viral particle, wherein the decoding tags are viral proteins containing
selenium-
substituted methionine, where the decoding tags are detected by mass
spectrometry such that different decoding tags are distinguished by selenium-
based differences in mass.
The specific binding molecules of each reporter carrier can interact with
a different target molecule such that a different decoding tag corresponds to
each
different reporter carrier. The decoding tags can be disassociated from the
reporter carriers during, or prior to, detection.
Other Features
All forms of the disclosed method can be performed using a variety of
specific components or additional steps. For example, the reporter molecules
can be amplified using branched DNA. For this it is preferred that each
reporter
molecule comprise a reporter signal and each branched DNA comprise multiple
reporter tags and a tail which associates with the reporter signal.
The reporter molecules can also be amplified using rolling circle
amplification. For this it is preferred that each reporter molecule comprises
a
reporter signal, where the reporter signal primes rolling circle replication
of an
amplification target circle and the amplification target circle comprises one
or
more sequences complementary to the reporter signal such that replication
produces tandem sequence DNA. The tandem sequence DNA contains a
plurality of reporter tags.
The reporter molecules can also be amplified using oligonucleotide
dendrimers. The decoding tags preferably are oligonucleotides, carbohydrates,
peptide nucleic acids, antibodies, ligands, proteins, haptens, zinc fingers,
aptamers, or mass labels.
In another embodiment, the target molecules can be homing molecules.
Prior to step (a), the target samples or the source of the target samples are
42
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
exposed to the homing molecules. The result is that the presence of homing
molecules corresponding to the detected decoding tags indicates the presence
of
molecules in the target samples to which the homing molecules are directed.
For
example, the source of the tissue samples can be exposed to the homing
molecules such that the presence of homing molecules corresponding to the
detected decoding tags indicates the presence of cells or molecules in the
tissue
samples to which the homing molecules axe directed. For this embodiment, the
target molecules preferably are tumor-homing peptides with the result that the
presence of tumor-homing peptides corresponding to the detected decoding tags
indicates the presence of tumor cells in the tissue samples. In preferred
embodiments, the organisms that are the source of the tissue samples can be
exposed to the homing molecules, or the tissue samples can be exposed to the
homing molecules after the tissue samples are sectioned.
Where the target samples are cells, the cells can be sorted from other
cells. For example, the cells are sorted based on the presence, absence, or
difference in amount of a cell marker.
The disclosed method can be performed where the target samples are
organisms, and where, following step (a), derivative target samples comprising
the reporter molecules are prepared from the organisms. The disclosed method
can also be performed where the target samples are tissues, and where,
following
step (a), derivative target samples comprising the reporter nc~lecules are
prepared from the tissues. The derivative target samples preferably are tissue
sections prepared from the tissues.
In another embodiment, each decoding tag can correspond to a different
reporter tag, each reporter tag can correspond to a different reporter
molecule,
each reporter molecule can correspond to a different target molecule, or a
combination. Alternatively, each decoding tag can correspond to a single
reporter tag, each reporter tag can correspond to a single reporter molecule,
each
reporter molecule can correspond to a single target molecule, or a
combination.
Or each decoding tag can correspond to multiple reporter tags, each reporter
tag
can correspond to multiple reporter molecules, each reporter molecule can
correspond to multiple target molecules, or a combination.
43
CA 02371843 2001-11-06
WO 00/68434 PCT/LJS00/12391
The disclosed method can also include, followiilg step (b), the step of
bringing into contact the target samples and one or more capture arrays, where
different target molecules become associated with different elements of the
array
such. The result is that the array elements with which the decoding tags are
associated indicate the presence in the target samples of the target molecules
corresponding to that array element. The capture array can comprise capture
tags, where each array element comprises a different capture tag. The capture
tags preferably are oligonucleotides, antibodies, haptens, ligands, or a
combination. The capture array preferably comprises a substrate comprising
beads, plates, or slides.
The decoding tags can be distinguished temporally via different
fluorescent, phosphorescent, or chemiluminescent emission lifetimes. The
decoding tags preferably are detectable by nuclear magnetic resonance,
electron
paramagnetic resonance, surface enhanced raman scattering, surface plasmon
resonance, fluorescence, phosphorescence, chemiluminescence, resonance
raman, microwave, mass spectrometry, or a combination. The decoding tags
preferably are detected by mass spectrometry, electrophoresis, or
chromatography.
In another embodiment, the decoding tags can be peptide nucleic acids,
and the decoding tags can be detected by mass spectrometry. For this, the
different decoding tags should differ in mass. In this embodiment, each
reporter
molecule can comprise a specific binding molecule, where the specific binding
molecule of each reporter molecule interacts with a different target molecule,
and the reporter tags can be oligonucleotides, where the decoding tags are
peptide nucleic acids that are complementary to the reporter tags.
Alternatively,
each decoding tag can have the same number of nucleotide bases complementary
to the reporter tag. In this case, it is preferred that each decoding tag
comprises
a different number of 8-amino-3,6-dioxaoctanoic monomers and that the
decoding tags are detected by matrix-assisted laser desorption/ionization time-
of flight mass spectroscopy.
The decoding tags can also be fluorescently-labeled oligonucleotides,
where each decoding tag has a different combination of length and fluorescent
label. In this case, it is preferred that each reporter tag be an
oligonucleotide,
44
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
and that each decoding tag have the same number of nucleotides complementary
to the reporter tag. For this combination, it is preferred that each decoding
tag
that has the same fluorescent label has a different number of nucleotides not
complementary to the reporter tag and that the decoding tags are detected by
microdissection of the target sample and electrophoresis of the microdissected
samples.
The target molecules can be in one or more target samples, where the
target molecules include different modification states of the same target
molecules. For example, the modifications can be fragmentation, cleavage,
phosphorylation, glycosylation, methylation, alkylation, dimerization,
derivatization, depurination, conformation, or ribosylation. Where the target
molecules include different phosphorylation states of the same proteins, each
of
the reporter molecules can interact with a different protein in a different
phosphorylation state, and detection of the target molecules in the target
sample
1 S is indicative of the phosphorylation state of the proteins in the target
sample.
Where the target molecules include different glycosylation states of the same
proteins, each of the reporter molecules can interact with a different protein
in a
different glycosylation state, and detection of the target molecules in the
target
sample is indicative of the glycosylation state of the proteins in the target
sample. Where the target molecules include different poly-ADP ribosylation
states of the same proteins, each of the reporter molecules can interact with
a
different protein in a different poly-ADP ribosylation state, and detection of
the
target molecules in the target sample is indicative of the poly-ADP
ribosylation
state of the proteins in the target sample. Where the target molecules include
different fragments of the same proteins, each of the reporter molecules can
interact with a different fragment, and detection of the target molecules in
the
target sample is indicative of the fragments of the proteins in the target
sample.
Where the target molecules include different conformational states of the same
proteins, each of the reporter molecules can interact with a different protein
in a
different conformational state, and detection of the target molecules in the
target
sample is indicative of the conformational state of the proteins in the target
sample. Where at least one of the target molecules is a prion protein, the
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
conformation states can include the prion conformation of a protein and the
non-
prion conformation of the protein.
The pattern of the presence, amount, presence and amount, or absence of
decoding tags can constitute a catalog of the target molecules. Where the
target
molecules are in two or more target samples, the pattern of the presence,
amount, presence and amount, or absence of decoding tags associated with each
target sample constitutes a catalog of the target molecules in that target
sample,
the method further comprising comparing one or more catalogs with one or more
other catalogs.
The target molecules, reporter molecules, or decoding tags can be in an
array, where each target molecule, reporter molecule, or decoding tag is
immobilized at a different location in the array, and where detecting the
decoding tags is accomplished by detecting the presence, amount, presence and
amount, or absence of decoding tags in the arrays. Where the location, amount,
or location and amount of decoding tags in the arrays constitutes a pattern of
decoding tags in the arrays, the pattern of decoding tags in the arrays can be
compared with the pattern of decoding tags in arrays determined in a separate
procedure using a different one or more target molecules. The pattern of
decoding tags in the arrays can also be compared with the pattern of decoding
tags in arrays determined in a plurality of separate procedures using a
plurality
of different one or more target molecules.
Where the target molecules are associated with'cells, the reporter
molecules can be associated with the target molecules. If each reporter
molecule
comprises a sorting tag, the cells can be sorted based on the sorting tags.
The
target molecules preferably are cell surface proteins on cells such that the
reporter molecules are associated with the proteins on the cells.
Where each reporter molecule comprises a specific binding molecule, the
specific binding molecule preferably is an antibody, a ligand, a binding
protein,
a receptor protein, a hapten, aptamer, carbohydrate, or an oligonucleotide.
Where the specific binding molecules is a binding protein, it preferably is a
DNA binding protein comprising one or more zinc finger motifs, leucine zipper
motifs, helix-turn-helix motifs, or a combination.
46
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
The decoding tags preferably are detected by determining the presence,
amount, presence and amount, or absence of the decoding tags. The decoding
tags can be separated and detected by high pressure liquid chromatography. The
target sample can be immobilized, fixed, or adhered to a surface such that the
locations of the target molecules that are determined are the locations of the
target molecules on the surface.
Correspondence of Components
In preferred embodiments, a one-to-one-to-one correspondence between
target molecules, reporter molecules, and decoding tags is used. In this way,
each different type of target molecule ends up with a different and separately
detectable decoding tag associated with it. The combination of specific
binding
molecule and reporter signal results in an effective conversion of the
different
target molecules (where the target molecules may be widely chemically
divergent) into a standardized signal (the reporter signal). These
standardized
signals (which are coded for each target molecule) are then detected using a
single set of standardized conditions (during which the coding is carried
forward). The detection of the coded signals (that is, the decoding tags)
results
in effective, convenient detection of multiple target molecules in a single
assay.
Target molecules, reporter molecules, reporter tags, and decoding tags
can be mapped to each other in a variety of different ways. For example, each
decoding tag can correspond to a different reporter tag, each reporter tag can
correspond to a different reporter molecule, each reporter molecule can
correspond to a different target molecule, or there can be a combination of
these
relationships. Each decoding tag can correspond to a single reporter tag, each
reporter tag can correspond to a single reporter molecule, each reporter
molecule
can correspond to a single target molecule, or there can be a combination of
these relationships. Each decoding tag can correspond to multiple reporter
tags,
each reporter tag can correspond to multiple reporter molecules, each reporter
molecule can correspond to multiple target molecules, or there can be a
combination of these relationships. Many other combinations of relationships
are also possible.
47
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
Decoding Tag Detection
Decoding tags can be detected using any suitable technique. In general,
the properties of the decoding tags will be chosen to match, or be compatible
with, a chosen detection technique. In preferred embodiments, the disclosed
method uses rapid detection techniques that allow spatial information about
analytes to be gathered. Decoding tags preferably are detected by nuclear
magnetic resonance, electron paramagnetic resonance, surface enhanced raman
scattering, surface plasmon resonance, fluorescence, phosphorescence,
chemiluminescence, resonance raman, microwave, mass spectrometry, or any
combination of these. Decoding tags preferably are separated and/or detected
by
mass spectrometry, electrophoresis, or chromatography. Decoding tags can be
distinguished temporally via different fluorescent, phosphorescent, or
chemiluminescent emission lifetimes.
Preferred methods of detection allow the location and amount of the
decoding tags to be determined. Preferred techniques to accomplish this
include
detection by matrix-assisted laser desorption/ionization time-of flight mass
spectroscopy and detection by microdissection of the target sample and
capillary
electrophoresis of the microdissected samples.
For MALDI-TOF detection, the decoding tags preferably are peptide
nucleic acids, where each decoding tag has a different mass to allow
separation
and separate detection in mass spectroscopy. For this purpose, it is
preferable
that each decoding tag have the same number of nucleotide bases
complementary to the reporter tag. It is also preferable that each decoding
tag
have a different number of mass tags, such as 8-amino-3,6-dioxaoctanoic
monomers. This allows for more consistent hybridization characteristics while
allowing the mass to vary. Use of peptide nucleic acids in MALDI-TOF
detection is generally described by Baucom et al., Anal. Chem. 69:4894-4898
(1997), and Butler et al., Anal. Chem. 68:3283-3287 (1996).
For capillary electrophoresis detection, the decoding tags preferably are
fluorescently-labeled oligonucleotides, where each decoding tag has a
different
combination of length and fluorescent label. For this purpose, it is
preferable
that each decoding tag have the same number of nucleotides complementary to
the reporter tag. It is also preferable that each decoding tag have a
different
48
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
number of nucleotides not complementary to the reporter tag. This allows for
more consistent hybridization characteristics while allowing separation of the
different decoding tags during electrophoresis.
The disclosed method using MALDI-TOF detection can be summarized
as follows:
1. A mixture of different reporter molecules is contacted with a sample
to allow specific binding of reporter molecules to target molecules on the
surface
of the sample; then excess unbound reporter molecules are removed.
2. A nucleic acid amplification system is used to generate surface-bound
reporter tags, such that reporter tag abundance is proportional to the number
of
bound reporter molecules.
3. The surface is contacted with a mixture of decoding tags. The
decoding tags can be distinguished by molecular weight because they are of
different composition and/or contain different mass labels. The mixture of
decoding tags is allowed to bind to the reporter tags; then the excess of
decoding tags is removed.
4. The surface is processed for mass spectrometry by coating it with a
suitable matrix for laser-desorption ionization.
5. The slide is introduced in a mass spectrometer, and the laser is
directed successively to specific locations of the surface in order to obtain
desorption-ionization in specific areas, releasing the decoding tags. The
amount
of each decoding tag of different mass is measured, providing a relative
measure
of the number of bound reporter molecules of each specific class. The laser
desorption process is then repeated on many different locations on the
surface.
Amplification of Reporter Molecules
Reporter molecules can be amplified using any suitable technique. Many
amplification techniques are known and can be adapted for use in the disclosed
method. The form of amplification will generally be related to the nature of
the
reporter molecule, and in particular, to the nature of the reporter signal to
be
amplified. A major class of amplification techniques are nucleic acid
amplification techniques. Such techniques generally will be most useful when
the reporter signal is an oligonucleotide or other nucleic acid or nucleic
acid
derivative. As used herein, amplification of a reporter molecule refers to any
49
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
increase in signal (in the form of reporter tags) associated with a reporter
molecule over the signal (in the form of a reporter signal) that is part of a
reporter molecule.
Reporter signals preferably are amplified using branched DNA, rolling
circle amplification, or oligonucleotide dendrimers. The branched DNA
generally has a tail complementary to the reporter signal and also has
multiple
reporter tags. Use of branched DNA for signal amplification is generally
described by Urdea, Biotechnology 12:926-928 (1994), and Horn et al., Nucleic
Acids Res 23:4835-4841 (1997). Use of dendrimers for signal amplification is
generally described by Shchepinov et al., Nucleic Acids Res. 25:4447-4454
(1997), and Orentas et al., J. Virol. Methods 77:153-163 (1999). For rolling
circle amplification, the reporter signal primes rolling circle replication of
an
amplification target circle. Since an amplification target circle comprises
one or
more sequences complementary to the reporter tag, replication produces tandem
sequence DNA where the tandem sequence DNA contains a plurality of reporter
tags. Rolling circle replication is described in U.S. Patent No. 5,854,033.
Analysis of Modification States
Molecules can exist in a variety of states. For example, proteins can
have different phosphorylation states. The disclosed method can be used to
analyze the state of various modifications of target molecules in target
samples.
Such modifications include fragmentation, cleavage, phosphorylation,
glycosylation, methylation, allcylation, dimerization, derivatization,
depurination, conformation, or ribosylation. In basic form, this can be
accomplished by interacting different target molecules in different
modification
states with different reporter molecules specific for the different
modification
states.
An example of this is the generation of multiplexed profiles of
phosphorylated proteins. In this embodiment, the status of phosphorylated
versus non-phosphorylated forms of proteins (or between different
phosphorylation states of proteins) can be monitored through the use of a
multiplicity of sets of antibodies, with each specific for a different
phosphorylation state of a protein. The decoding tag signals obtained by
electrophoretic analysis or by MALDI-TOF spectrometry are interpreted to yield
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
ratios of the various phosphorylation states for a multiplicity of proteins,
and this
information can then be correlated with the physiological status of the cell.
This embodiment preferably is used with proteins involved in signal
transduction. Examples of proteins and antibodies suitable for this embodiment
are described by Gioeli et al., Cancer Res 59(2):279-84 (1999), and Ng et al.,
Science 283(5410):2085-2089 (1999). In a similar way, the disclosed method
can be used to multiplex profiles of the fragmentation, cleavage,
glycosylation,
methylation, alkylation, dimerization, derivatization, depurination,
conformation, or ribosylation of target molecules.
A number of peptide cleavage means are known (Means, G.E. and R.E.
Feeney, Chemical modification ofproteins. 1971, San Francisco,: Holden-Day.
x, 254; Lundblad, R.L., Chemical reagents for protein modification. 2nd ed.
1991, Boca Raton: CRC Press. 345.) and may be utilized in solution, on
surfaces, or in a matrix. Some examples include: cleavage of methionine-
containing peptide bonds with cyanogen bromide for specific cleavage at the
carboxyl side, cleavage of cysteine containing peptide bonds by conversion of
cysteine to dehydroalanine and subsequent hydrolysis, cleavage of tryptophan-
containing peptide bonds at the amino side with N-chlorosuccinimide, and
cleavage of asparagine-glycine-containing peptide bonds by hydroxylamine with
specific cleavage at the carboxyl side.
Changes in protein conformation recognized by ant5bc~dies can be
assessed using such antibodies. Prions are proteins capable of self
perpetuating
changes in conformation and function. They have been implicated in several
biological processes and diseases such as Alzheimers (Prusiner, S.B., Prions.
Proc Natl Acad Sci U S A, 1998. 95(23): p. 13363-83; Li, L. and S. Lindquist,
Creating a protein-based element of inheritance. Science, 2000. 287(5453): p.
661-4). Antibodies have been developed that recognize the different states of
prion proteins (Hardt, M., T. Baron, and M.H. Groschup, A comparative study of
imnZUnohistochemical methods_for detecting abnormal prion protein with
monoclonal and polyclonal antibodies. J Comp Pathol, 2000. 122(1): p. 43-53;
Korth, C., P. Streit, and B. Oesch, Monoclonal antibodies specific for the
native,
disease-associated isoform of the prion protein. Methods Enzymol, 1999. 309:
p. 106-22).
51
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
Sample Processing
Target samples can be processed in a variety of ways both before and
after association of reporter molecules. In the usual case, a target sample
will be
separated or purified from a source of interest. For example, cells can be
isolated from an organism or tissue, tissue can be isolated from an organism,
or
cellular contents or components can be isolated from cells. Other processing
is
possible. For example, cells, tissue, or organisms can be exposed to reporter
molecules prior to isolation of preparation of a target sample. Where the
target
samples are processed before continuing or completing the method, the
processed target sample can be referred to as a derivative target sample.
Unless
otherwise indicated, the term target sample refers to both target samples and
derivative target samples.
A target sample, or the source of a target sample, also can be "stained"
with target molecules prior to exposure to reporter molecules or prior to
sample
preparation. In this embodiment, the target molecules become associated with
some other molecules or features of the sample. For example, a tissue sample
can be stained with target molecules that bind to particular proteins of
interest.
Such target molecules are referred to herein as homing molecules. The presence
and location to these proteins can then be determined in the disclosed method
by
associating reporter molecules with the target molecules. A similar effect can
be
achieved by staining a target sample with reporter molecules themselves.
A tissue sample can be exposed to homing molecules either before or
after the tissue samples are sectioned. In a preferred embodiment the target
molecules are tumor-homing peptides. The presence of tumor-homing peptides
corresponding to the detected decoding tags indicates the presence of tumor
cells
in the tissue samples.
Where the target samples are cells, or are derived from cells, the cells can
be sorted from other cells. In particular, the cells can be sorted based on
the
presence, absence, or difference in amount of a cell marker. FACS is a useful
technique for accomplishing such cell sorting. In some embodiments, a sorting
tag can be included on the reporter molecules and sorting can be accomplished
using the sorting tags.
52
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
In some embodiments of the method, cells may be sorted prior to
preparation of the target sample or association with reporter molecules. For
example, cells in blood, such as peripheral lymphocytes and macrophages, may
be sorted into different sub-populations using a fluorescence-activated cell
sorter. Sorting may be based on the presence or absence, or on measurably
different amounts of certain cell surface markers such as proteins or
carbohydrates. After cell sorting, the cell sub-populations may be deposited
in
individual microtiter wells, or may be placed on the surface of a suitably
prepared microscope slide, where they may be fixed using conventional cytology
procedures. Sorted cells are ideally suited for analysis'in the disclosed
method
since they represent a relatively homogeneous sub-population, where the
relative
ratios of cellular components such as proteins or mRNA's are likely to be
quite
similar in every cell.
Cell sorting can also be done during the disclosed method. Thus, cells of
the immune system may be sorted according to cell surface markers such as CD
subtype. An analysis may be set up where a subset of the reporter molecules
includes an additional signal moiety whose purpose is to enable a cell sorting
step. A preferred additional signal moiety for this embodiment is a
fluorescent
dye. For example, the dye cy3 may be covalently associated with a specific
DNA reporter tag, the tag being covalently bound to an antibody specific for
the
cell surface determinant CD4. Likewise, the dye cy5 may be covalently
associated with a different specific DNA reporter tag, the tag being
covalently
bound to an antibody specific for the cell surface determinant CDB. A set of
18
other specific antibodies (directed against other cellular proteins, which may
be
either internal or surface proteins) is covalently associated with 18
different
DNA reporter tags. These tags, however, do not contain fluorescent dyes. The
set of 20 reporter molecules, which comprises 18 non-fluorescent reporters and
2
fluorescent reporters, is mixed with a population of cells from human blood.
The cells are then sorted on the basis of cy3 and cy5 fluorescence, using a
fluorescence-activated cell sorter, to obtain three different cell pools: High
CD4,
High CDB, and everything else. These pools of cells are then separately
subjected to multiple tag analysis for all 20 antibody reporter systems. In
this
analysis step, the CD4 and CD8 signals, as well as the 18 other signals, are
53
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
obtained simultaneously based on the amount of speci~'ic decoding tags
detected
by mass spectrometry. Such analysis will generate a catalog of protein
expression levels for each of the three cell pools.
Target samples and target molecules can also be immobilized on
substrates. Preferably, target molecules are associated with capture arrays
where
different target molecules become associated with different elements of the
array. In this way, the identity of the array element where decoding tags are
detected identifies the target molecule involved. Generally, association of
target
molecules to a capture array will be through association with capture tags
immobilized on the substrate. Preferred forms of capture tags are
oligonucleotides, antibodies, haptens, and ligands.
The disclosed method may be used for the detection of a panel of defined
analytes that together are indicative of the metabolic status of tissue. For
example, one may obtain a tissue biopsy form an experimental animal to assay
for the possible presence of cancer. Tissue sections are prepared, and
analyzed
with a plurality of oligonucleotide-tagged antibodies, said tagged antibodies
being specific for binding to specific tumor antigens, chemokines, cytokines,
and tumor-homing peptides (Pasqualini R, Koivunen E, Kain R, Lahdenranta J,
Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E
Aminopeptidase N is a receptor for tumor-homing peptides and a target for
inhibiting angiogenesis, Cancer Res 2000 60:722-727; Pasqualini R. Vascular
targeting with phage peptide libraries, Q JNucl Med 1999 43:159-162). A
tumor-homing peptide is an oligopeptide that has been selected on the basis of
its binding specificity for a unique type of tumor. By injecting the
experimental
animal with an equimolar mixture of a plurality of tumor-homing peptides, the
various tissues of the animal will retain the peptides depending on the
presence
or absence of tumor cells. The actual concentration of'tumor-homing peptides
bound in tumor tissue will vary according to the stage and the grade of the
tumor. By using a mixture of tumor-homing peptides, and employing the
histological analysis of the method, a profile of the relative ratios of an
entire
panel of tumor-homing peptides at any number of locations in a tissue section
can be obtained. These multiple analyte profiles derived from the retention of
injected peptides in tissue, in combination with the relative levels of
endogenous
54
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
tumor markers, growth factors, cytokines, etc., may be correlated with the
disease status of the animal.
Patterns and Catalogs of Target Molecules
Patterns of the presence, amount, presence and amount, or absence of
decoding tags constitute a catalog of the target molecules. Such catalogs can
be
used as fingerprints of target samples, and are particularly useful for
comparison
with other catalogs derived from different targets samples in the same manner.
Where the target molecules, reporter molecules, or decoding tags are in an
array,
the presence, amount, presence and amount, or absence of decoding tags in the
array constitute a pattern of decoding tags in the array. Such a pattern can
be
compared to the pattern of decoding tags in arrays determined in a separate
procedure using a different one or more target molecules. Multiple patterns
derived from multiple target samples can be compared in this way.
Illustration 1: Protein Detection Using Branched DNA Amplification
and MALDI-TOF Detection
The following illustrates an embodiment of the disclosed method used to
detect proteins where the reporter signals are amplified using branched DNA
and
where the decoding tags are detected using MALDI-TOF.
1. A branch-DNA assay is performed on the surface of a glass slide
containing a standard histological specimen of paraformald~:.~luyde-fixed
tissue or
a cytological specimen, with the objective of detecting four different
proteins on
the surface of the histological or cytological sample. Four different
antibodies
(the specific binding molecules; A, B, C, D) are used, each specific for a
specific
protein of interest. Each antibody is covalently coupled to a specific
oligonucleotide (the reporter signal; Aol, Bo2, Co3, Do4), providing each
antibody with a specific DNA tag that serves as a binding site for the branch-
DNA. Four different branch-DNA reporters are used (Aol-Brl, Bo2-Br2, Co3-
Br3, Do4-Br4) each reporter containing a tree of at least 1,000 DNA reporter
tags. After completion of the branch-DNA assay, the surface will contain
thousands of bound antibody molecules, and each antibody molecule will be
bound (via the reporter signal) by its own specific tree of branch-DNA. If the
distribution of the four proteins in tissue is non-homogenous, then the
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
distribution of surface-bound antibodies will, of course, be non-homogeneous.
Consequently, the surface distribution of the thousands of strands of branch-
DNA reporter tags Brl, Br2, Br3, and Br4 will also be non-homogeneous.
2. The surface is contacted with a mixture of four different peptide
nucleic acid (PNA) decoding tags (PNA1, PNA2, PNA3, PNA4). The PNAs are
of the same length (10 bases), but comprise four different sequences perfectly
complementary to each of the reporter tag sequences. Additionally, the PNAs
can be distinguished by molecular weight because they contain one, two, three,
or four NHZ-terminal 8-amino-3,6-dioxaoctanoic monomers (146 Daltons each)
as a mass labels for MALDI-TOF. The mixture of four PNAs is allowed to
hybridize for 15 minutes on the surface of the glass slide, and then the slide
is
washed to remove unhybridized PNA.
3. The slide is covered with a thin layer of a 10:1:1 mixture of 3-
hydroxypicolinic acid, picolinic acid, and diammonium citrate (Smirnov et al,
Analytical Biochemistry 238:19 (1996)) and then is allowed to dry in order to
embed the reporter tags, together with PNA decoding tags, within a matrix of
small molecules suitable for optimal time-delayed desorption.
4. The slide is introduced in a mass spectrometer, and the laser is
directed to specific locations on the tissue order to obtain laser desorption
in
specific areas- of approximately 500 square microns, releasing the PNA
molecules (Figure 1.3). The MALDI-TOF mass spectrum is interpreted to
document the amount of each of the four PNA decoding tags (PNA1, PNA2,
PNA3, PNA4), thus measuring indirectly the relative amounts of each protein
(A, B, C, D). Individual measurements may be performed sequentially at
different surface locations, since the laser desorption time for each spot is
rapid
(on the order of seconds).
Illustration 2: Protein Detection Using Rolling Circle Amplification and
MALDI-TOF Detection
The following illustrates an embodiment of the disclosed method used to
detect proteins where the reporter signals are amplified using rolling circle
amplification and where the decoding tags are detected using MALDI-TOF.
1. Rolling circle amplification is performed on the surface of a glass
slide containing a standard histological specimen of paraformaldehyde-fixed
56
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
tissue or a cytological specimen, with the objective of detecting four
different
proteins on the surface of the histological or cytological sample. Four
different
antibodies (the specific binding molecules; A, B, C, D) are used, each
specific
for a specific protein of interest. Each antibody is covalently coupled to a
specific oligonucleotide (the reporter signal; Aol, Bo2, Co3, Do4), providing
each antibody with a specific DNA tag that serves as a primer for rolling
circle
amplification. Four different amplification target circles are used (Aol-ATC1,
Bo2-ATC2, Co3-ATC3, Do4-ATC4) each amplification target circle containing
one or more reporter tag complements. The amplification target circles are
replicated using the reporter signals as the rolling circle replication primer
to
form tandem sequence DNA containing at least 1,000 DNA reporter tags. After
completion of rolling circle amplification, the surface will contain thousands
of
bound antibody molecules, and each antibody molecule will be coupled (via the
reporter signal) to its own specific tandem sequence DNA (TS1, TS2, TS3,
TS4). If the distribution of the four proteins in tissue is non-homogenous,
then
the distribution of surface-bound antibodies will, of course, be non-
homogeneous. Consequently, the surface distribution of the tandem sequence
DNA reporter tags TS1, TS2, TS3, and TS4 will also be non-homogeneous.
2. The surface is contacted with a mixture of four different peptide
nucleic acid (PNA) decoding tags (PNA1, PNA2, PNA3, PNA4). The PNAs are
of the same length ( 10 bases), but comprise four different sequences
perfectly
complementary to each of the reporter tag sequences. Additionally, the PNAs
can be distinguished by molecular weight because they contain one, two, three,
or four NHZ-terminal 8-amino-3,6-dioxaoctanoic monomers (146 Daltons each)
as a mass labels for MALDI-TOF. The mixture of four PNAs is allowed to
hybridize for 15 minutes on the surface of the glass slide, and then the slide
is
washed to remove unhybridized PNA.
3. The slide is covered with a thin layer of a 10:1:1 mixture of 3-
hydroxypicolinic acid, picolinic acid, and diammonium citrate (Smirnov et al,
Analytical Biochemistry 238:19 (1996)) and then is allowed to dry in order to
embed the reporter tags, together with PNA decoding tags, within a matrix of
small molecules suitable for optimal time-delayed desorption.
57
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
4. The slide is introduced in a mass spectrometer, and the laser is
directed to specific locations on the tissue order to obtain laser desorption
in
specific areas- of approximately 500 square microns, releasing the PNA
molecules (Figure 1.3). The MALDI-TOF mass spectrum is interpreted to
document the amount of each of the four PNA decoding tags (PNA1, PNA2,
PNA3, PNA4), thus measuring indirectly the relative amounts of each protein
(A, B, C, D). Individual measurements may be performed sequentially at
different surface locations, since the laser desorption time for each spot is
rapid
(on the order of seconds).
Illustration 3: Messenger RNA Detection Using Branched DNA
Amplification and Capillary Electrophoresis Detection
The following illustrates an embodiment of the disclosed method used to
detect mRNA where the reporter signals are amplified using branched DNA and
where the decoding tags are detected using capillary electrophoresis.
1. A branch-DNA assay is performed on the surface of a glass slide
containing a standard histological specimen of paraformaldehyde-fixed tissue
or
a cytological specimen, with the objective of detecting.four different mRNA on
the surface of the histological or cytological sample. Four different mRNA
probes (the specific binding molecules; A, B, C, D) are used, each specific
for a
specific mRNA of interest. Each probe is covalently coupled to a specific
oligonucleotide (the reporter signal; Aol, Bo2, Co3, Do4), providing each
mRNA probe with a specific DNA tag that serves as a binding site for the
branch-DNA. Four different branch-DNA reporters are used (Aol-Brl, Bo2-
Br2, Co3-Br3, Do4-Br4) each reporter containing a tree of at least 1,000 DNA
reporter tags. After completion of the branch-DNA assay, the surface will
contain thousands of bound probe molecules, and each probe molecule will be
bound (via the reporter signal) by its own specific tree of branch-DNA. If the
distribution of the four mRNAs in tissue is non-homogenous, then the
distribution of surface-bound probes will, of course, be non-homogeneous.
Consequently, the surface distribution of the thousands of strands of branch-
DNA reporter tags Brl, Br2, Br3, and Br4 will also be non-homogeneous
(Figure 1.1 ).
58
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
2. The surface is contacted with a mixture of four different
fluorescently-labeled DNA tags (decoding tags; DNA1, DNA2, DNA3, DNA4),
which are complementary to each of the four different reporter tag sequences.
The decoding tags are of different lengths (20, 21, 22, and 23 bases) where
only
20 bases in each are complementary to the reporter tags. The additional bases
serve to differentiate the mass of the decoding tags. The fluorescent DNA tags
are allowed to hybridize for 20 minutes on the surface of the glass slide, and
then the slide is washed to remove unhybridized decoding tags (Figure 1.2,
heavier lines indicate larger DNAs).
3. The slide is place on an Arcturus Engineering laser dissection
microscope, and different areas of interest containing anywhere from 1 cell to
1,000 cells are microdissected.
4. The microdissected cellular material, bound on the plastic cap of the
laser microdissection microscope, is dissolved in 40 ~l of 90% formamide gel
loading buffer. The material is transferred to another tube, and then loaded
into
a capillary electrophoresis instrument by contacting the tip of the capillary
with
the small 40 p l sample, and turning on the voltage.
5. The profile of fluorescent decoding tags is recorded, and the different
peak intensities are correlated with the corresponding tag in order to
generate the
relative expression profiles for the mRNA analytes that were recognized by
each
probe.
Illustration 4: Messenger RNA Detection Using Rolling Circle
Amplification and Capillary Electrophoresis Detection
The following illustrates an embodiment of the disclosed method used to
detect mRNA where the reporter signals are amplified using rolling circle
amplification and where the decoding tags are detected using capillary
electrophoresis.
1. Rolling circle amplification is performed on the surface of a glass
slide containing a standard histological specimen of paraformaldehyde-fixed
tissue or a cytological specimen, with the objective of detecting four
different
mRNA on the surface of the histological or cytological sample. Four different
mRNA probes (the specific binding molecules; A, B, C, D) are used, each
specific for a specific mRNA of interest. Each probe is covalently coupled to
a
59
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
specific oligonucleotide (the reporter signal; Aol, Bo2, Co3, Do4), providing
each mRNA probe with a specific DNA tag that serves as a primer for rolling
circle amplification. Four different amplification target circles are used
(Aol-
ATC1, Bo2-ATC2, Co3-ATC3, Do4-ATC4) each amplification target circle
containing one or more reporter tag complements. The amplification target
circles are replicated using the reporter signals as the rolling circle
replication
primer to form tandem sequence DNA containing at least 1,000 DNA reporter
tags. After completion of rolling circle amplification, the surface will
contain
thousands of bound mRNA probes, and each mRNA probe will be coupled (via
the reporter signal) to its own specific tandem sequence DNA (TS1, TS2, TS3,
TS4). If the distribution of the four mRNAs in tissue is non-homogenous, then
the distribution of surface-bound mRNA probes will, of course, be non-
homogeneous. Consequently, the surface distribution of the tandem sequence
DNA reporter tags TS 1, TS2, TS3, and TS4 will also be non-homogeneous.
2. The surface is contacted with a mixture of four different
fluorescently-labeled DNA tags (decoding tags; DNAl, DNA2, DNA3, DNA4),
which are complementary to each of the four different reporter tag sequences.
The decoding tags are of different lengths (20, 21, 22, and 23 bases) where
only
bases in each are complementary to the reporter tags. The additional bases
20 serve to differentiate the mass of the decoding tags. The fluorescent DNA
tags
are allowed to hybridize for 20 minutes on the surface of the glass slide, and
then the slide is washed to remove unhybridized decoding tags (Figure 1.2,
heavier lines indicate larger DNAs)
3. The slide is place on an Arcturus Engineering laser dissection
microscope, and different areas of interest containing anywhere from 1 cell to
1,000 cells are microdissected.
4. The microdissected cellular material, bound on the plastic cap of the
laser microdissection microscope, is dissolved in 40 ~l of 90% formamide gel
loading buffer. The material is transferred to another tube, and then loaded
into
a capillary electrophoresis instrument by contacting the tip of the capillary
with
the small 40 ~1 sample, and turning on the voltage.
5. The profile of fluorescent decoding tags is recorded, and the different
peals intensities are correlated with the corresponding tag in order to
generate the
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
relative expression profiles for the mRNA analytes that were recognized by
each
probe.
Illustration 5: Protein Detection Using Branched DNA Amplification
and Capillary Electrophoresis Detection
The following illustrates an embodiment of the disclosed method used to
detect proteins where the reporter signals are amplified using branched DNA
and
where the decoding tags are detected using capillary electrophoresis.
1. A branch-DNA assay is performed on the surface of a glass slide
containing a standard histological specimen of paraformaldehyde-fixed tissue
or
a cytological specimen, with the objective of detecting four different
proteins on
the surface of the histological or cytological sample. Four different
antibodies
(the specific binding molecules; A, B, C, D) are used, each specific for a
specific
protein of interest. Each antibody is covalently coupled to a specific
oligonucleotide (the reporter signal; Aol, Bo2, Co3, Do4), providing each
antibody with a specific DNA tag that serves as a binding site for the branch-
DNA. Four different branch-DNA reporters are used (Aol-Brl, Bo2-Br2, Co3-
Br3, Do4-Br4) each reporter containing a tree of at least 1,000 DNA reporter
tags. After completion of the branch-DNA assay, the surface will contain
thousands of bound antibody molecules, and each antibody molecule will be
bound (via the reporter signal) by its own specific tree of branch-DNA. If the
distribution of the four proteins in tissue is non-homogenous, then the
distribution of surface-bound antibodies will, of course, be non-homogeneous.
Consequently, the surface distribution of the thousands of strands of branch-
DNA reporter tags Brl, Br2, Br3, and Br4 will also be non-homogeneous.
2. The surface is contacted with a mixture of four different
fluorescently-labeled DNA tags (decoding tags; DNA1, DNA2, DNA3, DNA4),
which are complementary to each of the four different reporter tag sequences.
The decoding tags are of different lengths (20, 21, 22, and 23 bases) where
only
20 bases in each are complementary to the reporter tags. The additional bases
serve to differentiate the mass of the decoding tags. The fluorescent DNA tags
are allowed to hybridize for 20 minutes on the surface of the glass slide, and
then the slide is washed to remove unhybridized decoding tags (Figure 1.2,
heavier lines indicate larger DNAs)
61
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
3. The slide is place on an Arcturus Engineering laser dissection
microscope, and different areas of interest containing anywhere from 1 cell to
1,000 cells are microdissected.
4. The microdissected cellular material, bound on the plastic cap of the
laser microdissection microscope, is dissolved in 40 ~l of 90% formamide gel
loading buffer. The material is transferred to another tube, and then loaded
into
a capillary electrophoresis instrument by contacting the tip of the capillary
with
the small 40 ~1 sample, and turning on the voltage.
5. The profile of fluorescent decoding tags is recorded, and the different
peals intensities are correlated with the corresponding tag in order to
generate the
relative expression profiles for the protein analytes that were recognized by
each
antibody.
Illustration 6: Protein Detection Using Rolling Circle Amplification and
Capillary Electrophoresis Detection
The following illustrates an embodiment of the disclosed method used to
detect proteins where the reporter signals are amplified using rolling circle
amplification and where the decoding tags are detected using capillary
electrophoresis.
1. Rolling circle amplification is performed on,the surface of a glass
slide containing a standard histological specimen of paraformaldehyde-fixed
tissue or a cytological specimen, with the objective of detecting four
different
proteins on the surface of the histological or cytological sample. Four
different
antibodies (the specific binding molecules; A, B, C, D) are used, each
specific
for a specific protein of interest. Each antibody is covalently coupled to a
specific oligonucleotide (the reporter signal; Aol, Bo2, Co3, Do4), providing
each antibody with a specific DNA tag that serves as a primer for rolling
circle
amplification. Four different amplification target circles are used (Aol-ATC1,
Bo2-ATC2, Co3-ATC3, Do4-ATC4) each amplification target circle containing
one or more reporter tag complements. The amplification target circles are
replicated using the reporter signals as the rolling circle replication primer
to
form tandem sequence DNA containing at least 1,000 DNA reporter tags. After
completion of rolling circle amplification, the surface will contain thousands
of
bound antibody molecules, and each antibody molecule will be coupled (via the
62
CA 02371843 2001-11-06
WO 00/68434 PCT/US00/12391
reporter signal) to its own specific tandem sequence DNA (TS1, TS2, TS3,
TS4). If the distribution of the four proteins in tissue is non-homogenous,
then
the distribution of surface-bound antibodies will, of course, be non-
homogeneous. Consequently, the surface distribution of the tandem sequence
DNA reporter tags TS1, TS2, TS3, and TS4 will also be non-homogeneous.
2. The surface is contacted with a mixture of four different
fluorescently-labeled DNA tags (decoding tags; DNA1, DNA2, DNA3, DNA4),
which are complementary to each of the four different reporter tag sequences.
The decoding tags are of different lengths (20, 21, 22, and 23 bases) where
only
20 bases in each are complementary to the reporter tags. The additional bases
serve to differentiate the mass of the decoding tags. The fluorescent DNA tags
are allowed to hybridize for 20 minutes on the surface of the glass slide, and
then the slide is washed to remove unhybridized decoding tags (Figure 1.2,
heavier lines indicate larger DNAs)
3. The slide is place on an Arcturus Engineering laser dissection
microscope, and different areas of interest containing anywhere from 1 cell to
1,000 cells are microdissected.
4. The microdissected cellular material, bound on the plastic cap of the
laser microdissection microscope, is dissolved in 40 ~1 of 90% formamide gel
loading buffer. The material is transferred to another tube, and then loaded
into
a capillary electrophoresis instrument by contacting the tip ~~.~ the
capillary with
the small 40 ~l sample, and turning on the voltage.
S. The profile of fluorescent decoding tags is recorded, and the different
peak intensities are correlated with the corresponding tag in order to
generate the
relative expression profiles for the protein analytes that were recognized by
each
antibody.
63