Note: Descriptions are shown in the official language in which they were submitted.
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
PLANT ACYLTRANSFERASES
TECHNICAL FIELD
The invention relates to alteration of oil content in plants.
BACKGROUND
Triacylglycerols produced in plant tissues (e.g., canola seeds) are a
significant
source of fatty acids in the human diet. Diets rich in animal fat appear to
promote
maladies such as heart disease in humans. The ability to modulate
triacylglycerol
synthesis in plants could allow for production of fatty acid compositions that
are more
beneficial for humans or more efficiently synthesized by the plant. Cloning
and
to characterization of plant genes encoding enzymes involved in
triacylglycerol
synthesis and metabolism represents a major advance toward controlling
triacylglycerol synthesis in plants.
SUMMARY
A new plant acyltransferase gene from Brassica napus has been cloned and
characterized. The protein encoded by this gene is involved in triacylglycerol
synthesis. Partial DNA sequences of the B. napus gene confirm that it is
related to an
Arabidopsis thaliana putative acyltransferase gene. Antisense expression of
the B.
napus sequence decreases total oil content.
In one aspect, the invention features an isolated nucleic acid having at least
80% (e.g., 85%, 90%, or 99%) sequence identity to the nucleotide sequence of
SEQ
ID NO:3, or to a fragment of the nucleotide sequence of SEQ ID NO:3, the
fragment
being at least 15 nucleotides in length. An isolated nucleic acid having at
least 80%
(e.g., 85%, 90%, or 99%) sequence identity to the nucleotide sequence of SEQ
ID
NO:4, or to a fragment of the nucleotide sequence of SEQ ID NO:4, the fragment
being at least 15 nucleotides in length is also featured.
In another aspect, the invention features an isolated nucleic acid that
includes
a first and second region, the first region having at least 80% sequence
identity to the
nucleotide sequence of SEQ ID NO:3 and the second region having at least 80%
sequence identity to the nucleotide sequence of SEQ ID NO:4. The nucleic acid
can
3o encode a diacylglycerol acyltransferase polypeptide. The first and second
regions can
-1-
CA 02372632 2008-02-25
be separated by approximately 600 nucleotides. The nucleic acid can be the
insert of
pMB 143. An expression vector including the nucleic acid operably linked to an
expression control element also is featured. The nucleic acid can be operably
linked in
antisense orientation.
In another aspect, the invention features a transgenic plant and progeny
thereof
that include an exogenous nucleic acid encoding a diacylglycerol
acyltransferase
polypeptide operably linked to a regulatory element, and seed produced by such
plants. The nucleic acid can include a first and second region, the first
region having at
least 80% sequence identity to the nucleotide sequence of SEQ ID NO:3, the
second region having at least 80% sequence identity to the nucleotide sequence
of SEQ
ID NO:4. The transgenic plant can produce seeds that exhibit a statistically
significantly
altered oil content as compared to seeds produced by a corresponding plant
lacking the
nucleic acid encoding the diacylglycerol acyltransferase polypeptide. The
plant can be a
soybean plant or a Brassica plant.
The invention also features a nucleic acid that includes the nucleotide
sequence of SEQ ID NO:3 or SEQ ID NO:4 or the nucleotide sequence exactly
complementary to SEQ ID NO:3 or SEQ ID NO:4. The nucleic acid also can include
the RNA equivalent of the nucleotide sequence of SEQ ID NO:3 or SEQ ID NO:4,
or
an RNA equivalent that is exactly complementary to the nucleotide sequence of
SEQ ID
NO:3 or SEQ ID NO:4. The nucleotide sequences of SEQ ID NO:3 and SEQ ID NO:4
are partial sequences of the new B. napus gene.
In another aspect, the invention features a nucleic acid that (1) hybridizes
under
stringent conditions to a DNA molecule consisting of the nucleotide sequence
of SEQ
ID NO:3 or SEQ ID NO:4, and (2) encodes a plant acyltransferase. In addition,
the invention includes a nucleic acid (1) having a nucleotide sequence which
is at least
80% (e.g., at least 82, 85, 90, 92, 95, 98, or 99%) identical to the
nucleotide sequence of
SEQ ID NO:3 or SEQ ID NO:4, and (2) encoding a plant acyltransferase. As used
herein, the term "stringent conditions" means hybridization at 42 C in the
presence of
50% formamide; a first wash at 65 C with 2X SSC containing 1% SDS; followed by
a
second wash at 65 C with 0.1 X SSC.
In accordance with an aspect of the present invention, there is provided an
isolated nucleic acid having at least 95% sequence identity to the nucleotide
sequence
of SEQ ID NO:3.
In accordance with another aspect of the present invention, there is provided
a
transgenic plant cell comprising an exogenous nucleic acid encoding a
diacylglycerol
acyltransferase polypeptide operably linked to a regulatory element, said
nucleic acid
comprising a first and second region, said first region having at least 95%
sequence
identity to the nucleotide sequence of SEQ ID NO:3.
-2-
CA 02372632 2009-01-16
In accordance with an aspect of the present invention, there is provided an
isolated
nucleic acid having at least 95% sequence identity to the nucleotide sequence
of SEQ ID
NO:3, wherein said nucleic acid encodes a diacylglycerol acyltransferase
polypeptide.
In accordance with another aspect of the present invention, there is provided
a
transgenic plant cell comprising an exogenous nucleic acid encoding a
diacylglycerol
acyltransferase polypeptide operably linked to a regulatory element, said
nucleic acid
comprising a first and second region, said first region having at least 95%
sequence
identity to the nucleotide sequence of SEQ ID NO:3 and said second region
having at least
80% sequence identity to the nucleotide sequence of SEQ ID NO:4.
The nucleotide sequence fragments described below can be used to hybridize
against cDNA or genomic DNA libraries from a variety of sources to clone genes
related to the B. napus acyltransferase gene. In addition, the sequence
fragments can be
used to design additional primers for further sequencing of the B. napus gene
or for PCR
amplification of portions of the B. napus gene.
Unless otherwise defined, all technical and scientific terms used herein have
the
same meaning as commonly understood by one of ordinary skill in the art to
which
this invention belongs. Although methods and materials similar or equivalent
to those
described herein can be used to practice the invention, suitable methods and
materials
are described below. In case of conflict, the present specification, including
definitions,
will control. In addition, the materials, methods, and examples are
illustrative only and
not intended to be limiting.
Other features and advantages of the invention will be apparent from the
following detailed description, and from the claims.
DESCRIPTION OF DRAWINGS
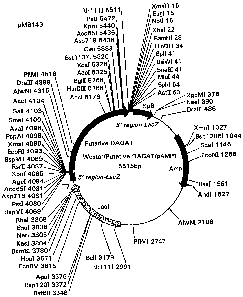
Figure 1 is a schematic of a construct containing the acyltransferase.
Restriction sites are marked.
Figure 2 is a schematic of a construct containing the acyltransferase in
antisense orientation. Restriction sites are marked.
3
CA 02372632 2009-01-16
DETAILED DESCRIPTION
Triacylglycerols (TAGs) are synthesized by the sequential transfer of acyl
chains
to the glycerol backbone by a series of enzymes in the Kennedy pathway
(glycerol-3-
phosphate acyltransferase, lysophosphatidic acid acyltransferase, and
diacylglycerol
acyltransferase). See, Topfer et al., Science, 1995, 268:681-686.
Diacylglycerol can be
used to form TAGs or membrane glycerolipids, and is the substrate for
diacylglycerol
acyltransferase (DAGAT, E.C. 2.3.1.20). DAGAT transfers a third acyl chain to
diacylglycerol, forming a TAG, and is the only enzyme unique to TAG synthesis
in the
Kennedy pathway. Thus, the reaction catalyzed by
3a
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
DAGAT represents a key branchpoint in TAG synthesis. As described herein, a
1.4
kB gene that encodes DAGAT has been identified from Brassica and can be used
to
alter total oil content in plants.
DAGAT Nucleic Acid Molecules
The invention features isolated nucleic acids having at least 80% sequence
identity, e.g., 85%, 90%, 95%, or 99% sequence identity, to the nucleic acid
of SEQ
ID NO:3 or SEQ ID NO:4, or fragments of the nucleic acid of SEQ ID NO:3 or 4
that
are at least about 15 nucleotides (nt) in length (e.g., at least 18, 20, 22,
24, 26, 28, or
30 nt). In one embodiment, the nucleic acid includes a first region having at
least
80% sequence identity to the nucleotide sequence of SEQ ID NO:3 and a second
region having at least 80% sequence identity to the nucleotide sequence of SEQ
ID
NO:4. The first and second regions can be separated by about 590 to about 700
nucleotides, e.g., about 600 nucleotides.
Generally, percent sequence identity is calculated by determining the number
of matched positions in aligned nucleic acid sequences, dividing the number of
matched positions by the total number of aligned nucleotides, and multiplying
by 100.
A matched position refers to a position in which identical nucleotides occur
at the
same position in aligned nucleic acid sequences. The total number of aligned
nucleotides refers to the number of nucleotides from SEQ ID NO:3 or NO:4 that
are
being aligned. Nucleic acid sequences can be aligned by the Clustal algorithm
of
MEGALIGN (DNASTAR, Madison, WI, 1997) sequence alignment software. In
this method, sequences are grouped into clusters by examining the distances
between
all pairs. Clusters are aligned as pairs, then as groups. A gap penalty of 100
and a
gap length penalty of 2 are used in the alignments.
Isolated nucleic acid molecules of the invention can be produced by standard
techniques. As used herein, "isolated" refers to a sequence corresponding to
part or
all of a gene encoding a DAGAT polypeptide, but free of sequences that
normally
flank one or both sides of the wild-type gene in a plant genome. As used
herein,
"polypeptide" refers to a chain of least eight amino acids. An isolated
nucleic acid
can be, for example, a DNA molecule, provided one of the nucleic acid
sequences
normally found immediately flanking that DNA molecule in a naturally-occurring
-4-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
genome is removed or absent. Thus, an isolated nucleic acid includes, without
limitation, a DNA molecule that exists as a separate molecule (e.g., a cDNA or
genomic DNA fragment produced by PCR or restriction endonuclease treatment)
independent of other sequences as well as recombinant DNA that is incorporated
into
a vector, an autonomously replicating plasmid, a virus (e.g., a retrovirus,
adenovirus,
or herpes virus), or into the genomic DNA of a prokaryote or eukaryote. In
addition,
an isolated nucleic acid can include a recombinant DNA molecule that is part
of a
hybrid or fusion nucleic acid. A nucleic acid existing among hundreds to
millions of
other nucleic acids within, for example, cDNA libraries or genomic libraries,
or gel
1o slices containing a genomic DNA restriction digest, is not to be considered
an isolated
nucleic acid.
Isolated nucleic acid molecules are at least about 15 nucleotides in length.
For
example, the nucleic acid molecule can be about 20 to 35, 40-50, 50-100, or
greater
than 150 nucleotides in length, e.g., 200-300, 300-500, 500-1000, or 1100-1500
nucleotides in length. Such fragments, whether encoding a polypeptide or not,
can be
used as probes, primers, and diagnostic reagents. In some embodiments, the
isolated
nucleic acid molecules encode a full-length DAGAT polypeptide. Nucleic acid
molecules can be DNA or RNA, linear or circular, and in sense or antisense
orientation. The nucleic acid molecules also can be complementary to the
nucleotide
sequences of SEQ ID NO:3 or SEQ ID NO:4. A nucleic acid encoding a DAGAT
polypeptide may or may not contain introns within the coding sequence.
Polymerase chain reaction (PCR) techniques can be used to produce nucleic
acid molecules of the invention. PCR refers to a procedure or technique in
which
target nucleic acids are amplified. Sequence information from the ends of the
region
of interest or beyond typically is employed to design oligonucleotide primers
that are
identical in sequence to opposite strands of the template to be amplified. PCR
can be
used to amplify specific sequences from DNA as well as RNA, including
sequences
from total genomic DNA or total cellular RNA. Primers are typically 14 to 40
nucleotides in length, but can range from 10 nucleotides to hundreds of
nucleotides in
length. General PCR techniques are described, for example in PCR Primer: A
Laboratory Manual, Ed. by Dieffenbach, C. and Dveksler, G., Cold Spring Harbor
Laboratory Press, 1995.
-5-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
Nucleic acids encoding DAGAT polypeptides also can be produced by
chemical synthesis, either as a single nucleic acid molecule or as a series of
oligonucleotides. For example, one or more pairs of long oligonucleotides
(e.g., >100
nucleotides) can be synthesized that contain the desired sequence, with each
pair
containing a short segment of complementarity (e.g., about 15 nucleotides)
such that a
duplex is formed when the oligonucleotide pair is annealed. DNA polymerase is
used
to extend the oligonucleotides, resulting in a double-stranded nucleic acid
molecule
per oligonucleotide pair, which then can be ligated into a vector.
It should be appreciated that many different nucleic acids will encode a
1o polypeptide having a particular DAGAT amino acid sequence. The degeneracy
of the
genetic code is well known in the art, i.e., many amino acids are encoded by
more
than one nucleotide codon. It should also be appreciated that certain amino
acid
substitutions can be made within polypeptide sequences without affecting the
function
of the polypeptide. Conservative amino acid substitutions or substitutions of
similar
amino acids often are tolerated without affecting polypeptide function.
Similar amino
acids can be those that are similar in size and/or charge properties.
Similarity between
amino acids has been assessed in the art. For example, Dayhoff et al. (1978)
in Atlas
of Protein Sequence and Structure, Vol. 5, Suppl. 3, pp. 345-352, provides
frequency
tables for amino acid substitutions that can be employed as a measure of amino
acid
similarity.
Transgenic Plants
The invention features transgenic plants that have altered total oil content,
i.e.,
increased or decreased oil content. Suitable plant species include, for
example,
Brassica spp. such as B. napus, B. campestris, B. juncea, and B. rapa (canola-
type
and high erucic acid rapeseed), soybean, sunflower, castor bean, safflower,
crambe,
palm, coconut, corn, cottonseed, olive, peanut, flax, and sesame. Canola, soy,
sunflower, and safflower plants having increased oil content are particularly
useful.
Table I provides relative percent oil and protein content on a dry weight
basis (unless
indicated otherwise) of suitable oilseed plants.
The present invention describes a novel method of making transgenic plants
that produce seeds with a statistically significant alteration in oil content.
As used
-6-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
herein, "statistically significant" refers to a p-value of less than 0.05,
e.g., a p-value of
less than 0.025 or a p-value of less than 0.01, using an appropriate measure
of
statistical significance, e.g., a one-tailed two sample t-test. By using this
method,
plants can be produced that exhibit an altered oil content in their seeds. The
altered
oil content is statistically significant relative to the oil content of
unmodified seeds.
Plants produced by the method of the present invention can produce seeds
having an
increase in oil of from about I% to about 25% over the oil content in seeds
produced
by unmodified control plants. For example, the increase in oil content for
plants
described herein can be from about 2% to about 20%, from about 4% to about
15%,
from about 5% to about 10%, or from about 10% to about 20%, relative to
unmodified plants.
TABLE 1
Relative Percent Oil and Protein Content of Oilseed Plants
Plant % Oil % Protein Key
Soybean -20 -40 C
(Glycine max)
Rapeseed 40-44 38-41 (oil C; D
(Brassica na us free meal)
Sunflower 40 D
(Helianthus annus)
Castor bean 50 A
(Ricinus communis)
Safflower 36.8-47.7 15.4-22.5 D
(Carthamus tinctorius)
Crambe 30-35 -28 B
(Crambe ab ssinica
Palm 20 C; per fresh fruit bunch (-20%
(Elaeis guineensis) moisture);
>50 Dried kernels
Coconut 34 3.5 D; coconut flesh (50% moisture);
(Cocos nucifera) 69 dried kernels
Maize 3.1-5.7 6-12 C; D
Zea mays)
Cottonseed 25-30 25-30 D; kernel
(Gossypium hirsutum)
Olive 19.6 1.6 fruit (52.4% moisture)
(Olea europaea)
Peanut 36-56 25-30 C; (unknown moisture)
(Arachis by o aea
Flax 35-45 D; per fruit capsule (--10
(Linum usitatissimum) seeds/fruit)
-7-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
Sesame 53.3-57.5 25-30 D; (5-7% moisture)
(Sesamum indicum)
(A) Brigham RD, 1993, Castor: Return of an old crop, p 380-3. In New Crops,
Janick, J & Simon, JE, eds. Wiley, NY.
(B) Grombacher et al., Cooperative Extension, Institute of Agriculture and
Natural Resources, University of Nebraska-Lincoln, Crambe production,
Publication
G93-1126A, G1126 (Field Crops), F-17 (Misc. Crops); see also
pubs@unlvm.unl.edu
(C) In Principles of Cultivar Development, 1987, Fehr, WR, ed., Macmillan
Publishing Co., NY.
(D) In 5'h Edition Bailey's Industrial Oil & Fat Products, Vol. 2, Edible Oil
& Fat Products: Oils and Oil Seeds, 1996, Hui, YH, ed., Wiley, NY.
A plant described herein may be used as a parent to develop a plant line, or
may itself be a member of a plant line, i.e., it is one of a group of plants
that display
little or no genetic variation between individuals for total oil content. Such
lines can
be created by several generations of self-pollination and selection, or
vegetative
propagation from a single parent using tissue or cell culture techniques known
in the
art. Additional means of breeding plant lines from a parent plant are known in
the art.
In general, plants of the invention can be obtained by introducing at least
one
exogenous nucleic acid encoding a DAGAT polypeptide into plant cells. As used
herein, the term "exogenous" refers to a nucleic acid that is introduced into
the plant.
Exogenous nucleic acids include those that naturally occur in the plant and
have been
introduced to provide one or more additional copies, as well as nucleic acids
that do
not naturally occur in the plant. Typically, a nucleic acid construct
containing a
nucleic acid encoding a DAGAT polypeptide is introduced into a plant cell.
Seeds
produced by a transgenic plant can be grown and selfed (or outcrossed and
selfed) to
obtain plants homozygous for the construct. Seeds can be analyzed to identify
those
homozygotes having the desired expression of the construct. Transgenic plants
can be
entered into a breeding program, e.g., to increase seed, to introgress the
novel
construct into other lines or species, or for further selection of other
desirable traits.
Alternatively, transgenic plants can be obtained by vegetative propagation of
a
transformed plant cell, for those species amenable to such techniques.
Progeny of a transgenic plant are included within the scope of the invention,
provided that such progeny exhibit altered oil content. Progeny of an instant
plant
-8-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
include, for example, seeds formed on F1, F2, F3, and subsequent generation
plants, or
seeds formed on BC1, BC2, BC3, and subsequent generation plants.
Transgenic techniques for use in the invention include, without limitation,
Agrobacterium-mediated transformation, electroporation, and particle gun
transformation. Illustrative examples of transformation techniques are
described in
WO 99/43202 and U.S. Patent 5,204,253 (particle gun) and U.S. Patent 5,188,958
(Agrobacterium). Transformation methods utilizing the Ti and Ri plasmids of
Agrobacterium spp. typically use binary type vectors. Walkerpeach, C. et al.,
in Plant
Molecular Biology Manual, S. Gelvin and R. Schilperoort, eds., Kluwer
Dordrecht,
1o C1:1-19 (1994). If cell or tissue cultures are used as the recipient tissue
for
transformation, plants can be regenerated from transformed cultures by
techniques
known to those skilled in the art. In addition, various plant species can be
transformed using the pollen tube pathway technique.
Nucleic acid constructs suitable for producing transgenic plants of the
invention include a nucleic acid encoding a DAGAT polypeptide operably linked
to a
regulatory element such as a promoter. Standard molecular biology techniques
can be
used to generate nucleic acid constructs. To increase oil content in plants,
the nucleic
acid encoding a DAGAT polypeptide is operably linked to the regulatory element
in
sense orientation.
Suitable promoters can be constitutive or inducible, and can be seed-specific.
As used herein, "constitutive promoter" refers to a promoter that facilitates
the
expression of a nucleic acid molecule without significant tissue- or temporal-
specificity. An inducible promoter may be considered to be a "constitutive
promoter", provided that once induced, expression of the nucleic acid molecule
is
relatively constant or uniform without significant tissue- or temporal-
specificity.
Suitable promoters are known (e.g., Weising et al., Ann. Rev. Genetics 22:421-
478
(1988)). The following are representative examples of promoters suitable for
use
herein: regulatory sequences from fatty acid desaturase genes (e.g.,
Brassicafad2D or
fad2F, see WO 00/07430); alcohol dehydrogenase promoter from corn; light
inducible promoters such as the ribulose bisphosphate carboxylase (Rubisco)
small
subunit gene promoters from a variety of species; major chlorophyll a/b
binding
-9-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
protein gene promoters; the 19S promoter of cauliflower mosaic virus (CaMV);
as
well as synthetic or other natural promoters that are either inducible or
constitutive.
In one embodiment, regulatory sequences are seed-specific, i.e., the
particular gene
product is preferentially expressed in developing seeds and expressed at low
levels or
not at all in the remaining tissues of the plant. Non-limiting examples of
seed-specific
promoters include napin, phaseolin, oleosin, and cruciferin promoters.
Additional regulatory elements may be useful in the nucleic acid constructs of
the present invention, including, but not limited to, polyadenylation
sequences,
enhancers, introns, and the like. Such elements may not be necessary for
expression
of a DAGAT polypeptide, although they may increase expression by affecting
transcription, stability of the mRNA, translational efficiency, or the like.
Such
elements can be included in a nucleic acid construct as desired to obtain
optimal
expression of the acyltransferase nucleic acid in the host cell(s). Sufficient
expression, however, may sometimes be obtained without such additional
elements.
A reference describing specific regulatory elements is Weising et al., Ann.
Rev.
Genetics 22:421-478 (1988).
In some situations, a decreased oil content may be desired. A feature of the
invention is that DAGAT activity can be reduced by gene silencing, antisense,
ribozymes, cosuppression, or mutagenesis techniques, resulting in a decrease
in oil
content. Gene silencing techniques, such as that described in WO 98/36083 are
useful. Antisense RNA has been used to inhibit plant target genes in a tissue-
specific
manner. See, for example, U.S. Patent Nos. 5,453,566, 5,356,799, and
5,530,192.
Antisense nucleic acid constructs include a partial or a full-length coding
sequence
operably linked to at least one suitable regulatory sequence in antisense
orientation.
Expression of DAGAT also can be inhibited by ribozyme molecules designed
to cleave DAGAT mRNA transcripts. While various ribozymes that cleave mRNA at
site-specific recognition sequences can be used to destroy DAGAT mRNAs,
hammerhead ribozymes are particularly useful. Hammerhead ribozymes cleave
mRNAs at locations dictated by flanking regions that form complementary base
pairs
with the target mRNA. The sole requirement is that the target RNA contain a 5'-
UG-
3' nucleotide sequence. The construction and production of hammerhead
ribozymes is
-10-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
well known in the art. See, for example, U.S. Patent No. 5,254,678. Hammerhead
ribozyme sequences can be embedded in a stable RNA such as a transfer RNA
(tRNA) to increase cleavage efficiency in vivo. Perriman, R. et al., Proc.
Natl. Acad.
Sci. USA, 92(13):6175-6179 (1995); de Feyter, R. and Gaudron, J., Methods in
Molecular Biology, Vol. 74, Chapter 43, "Expressing Ribozymes in Plants",
Edited by
Turner, P.C, Humana Press Inc., Totowa, NJ (1997). RNA endoribonucleases such
as
the one that occurs naturally in Tetrahymena thermophila, and which have been
described extensively by Cech and collaborators also are useful. See, for
example,
U.S. Patent No. 4,987,071.
The phenomenon of co-suppression also has been used to inhibit plant target
genes in a tissue-specific manner. Co-suppression of an endogenous gene using
a
full-length cDNA sequence as well as a partial cDNA sequence are known. See,
for
example, WO 94/11516, and U.S. Patent Nos. 5,451,514 and 5,283,124. Co-
suppression of DAGAT activity in plants can be achieved by expressing, in the
sense
orientation, the entire or partial coding sequence of a DAGAT gene.
Mutagenesis can also be used to reduce acytltransferase activity in plants.
Mutagenic agents can be used to induce random genetic mutations within a
population
of seeds or regenerable plant tissue. Suitable mutagenic agents include, for
example,
ethyl methyl sulfonate, methyl N-nitrosoguanidine, ethidium bromide,
diepoxybutane,
x-rays, UV rays, and other mutagens known in the art. The treated population,
or a
subsequent generation of that population, is screened for reduced oil content
or
reduced DAGAT activity that results from the mutation. Mutations can be in any
portion of a gene, including the coding region, introns, and regulatory
elements, that
render the resulting gene product non-functional or with reduced activity.
Suitable
types of mutations include, for example, insertions or deletions of
nucleotides, and
transitions or transversions in the wild-type coding sequence. Such mutations
can
lead to deletion or insertion of amino acids, and conservative or non-
conservative
amino acid substitutions in the corresponding gene product.
Characterization of Oils
Techniques that are routinely practiced in the art can. be used to extract,
process, and analyze the oils produced by plants of the instant invention.
Typically,
-ll-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
plant seeds are cooked, pressed, and extracted to produce crude oil, which is
then
degummed, refined, bleached, and deodorized. Generally, techniques for
crushing
seed are known in the art. For example, soybean seeds can be tempered by
spraying
them with water to raise the moisture content to, e.g., 8.5%, and flaked using
a
smooth roller with a gap setting of 0.23 to 0.27 mm. Depending on the type of
seed,
water may not be added prior to crushing. Application of heat deactivates
enzymes,
facilitates further cell rupturing, coalesces the oil droplets, and
agglomerates protein
particles, all of which facilitate the extraction process.
The majority of the seed oil can be released by passage through a screw press.
1o Cakes expelled from the screw press are then solvent extracted, e.g., with
hexane,
using a heat traced column. Alternatively, crude oil produced by the pressing
operation can be passed through a settling tank with a slotted wire drainage
top to
remove the solids that are expressed with the oil during the pressing
operation. The
clarified oil can be passed through a plate and frame filter to remove any
remaining
fine solid particles. If desired, the oil recovered from the extraction
process can be
combined with the clarified oil to produce a blended crude oil.
Once the solvent is stripped from the crude oil, the pressed and extracted
portions are combined and subjected to normal oil processing procedures (i.e.,
degumming, caustic refining, bleaching, and deodorization). Degumming can be
performed by addition of concentrated phosphoric acid to the crude oil to
convert
non-hydratable phosphatides to a hydratable form, and to chelate minor metals
that
are present. Gum is separated from the oil by centrifugation. The oil can be
refined
by addition of a sufficient amount of a sodium hydroxide solution to titrate
all of the
fatty acids and removing the soaps thus formed.
Deodorization can be performed by heating the oil to 500 F (260 C) under
vacuum, and slowly introducing steam into the oil at a rate of about 0.1
ml/minute/100 ml of oil. After about 30 minutes of sparging, the oil is
allowed to
cool under vacuum. The oil is typically transferred to a glass container and
flushed
with argon before being stored under refrigeration. If the amount of oil is
limited, the
oil can be placed under vacuum, e.g., in a Parr reactor and heated to 500 F
for the
-12-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
same length of time that it would have been deodorized. This treatment
improves the
color of the oil and removes a majority of the volatile substances.
Oil content can be measured by NMR using AOCS Method AM 2-93 and
AOCS Recommended Practice AK 4-95 or by near infra-red reflectance
spectroscopy
(NIR) using AOCS Method AK 3-94 and AOCS Procedure AM 1-92. Oil
composition can be analyzed by extracting fatty acids from bulk seed samples
(e.g., at
least 10 seeds). Fatty acid TAGs in the seed are hydrolyzed and converted to
fatty
acid methyl esters. Percentages of fatty acids typically are designated on a
weight
basis and refer to the percentage of the fatty acid methyl ester in comparison
with the
lo total fatty acid methyl esters in the sample being analyzed. Seeds having
an altered
fatty acid composition may be identified by techniques known to the skilled
artisan,
e.g., gas-liquid chromatography (GLC) analysis of a bulked seed sample, a
single seed
or a single half-seed. Half-seed analysis is well known in the art to be
useful because
the viability of the embryo is maintained and thus those seeds having what
appears to
be a desired fatty acid profile may be planted to form the next generation.
However,
bulk seed analysis typically yields a more accurate representation of the
fatty acid
profile of a given genotype. Fatty acid composition can also be determined on
larger
samples, e.g., oil obtained by pilot plant or commercial scale refining,
bleaching and
deodorizing of endogenous oil in the seeds.
The following examples are to be construed as merely illustrative of how one
skilled in the art can make and use the DAGAT gene fragments, and does not
limit the
scope of the invention described in the claims.
EXAMPLES
Example 1 - Cloning of a Brassica Acyl Transferase: A mouse acyl
CoA:diacylglycerol acyltransferase (DAGAT) gene was recently identified (Case
et
al., Proc Natl Acad Sci USA 95:13018-13023, 1998). The deduced mouse DAGAT
amino acid sequence was used to search for similar plant sequences in GenBank
and
dbEST databases. The mouse DAGAT protein sequence aligned with significant
sequence identity and similarity to a putative Arabidopsis thaliana acyl-
CoA:cholesterol acyltransferase (ACAT; GenBank Accession No. 3135276, locus
ATA0003058). However, upon aligning the A. thaliana ACAT with the mouse
-13-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
DAGAT and mouse ACAT (Case et al., JBiol Chem 273:26755-26764, 1998)
sequences, it was found that the A. thaliana protein had greater homology to
mouse
DAGAT than to mouse ACAT. Thus, the type of acyltransferase encoded by the A.
thaliana gene was unclear.
The genomic DNA sequence encoding the A. thaliana protein (GenBank
Accession No. 3135276) was used to design two PCR primers for amplifying
candidate DAGAT or ACAT genomic sequences from different sources. The 5'
DAGAT-1 primer had the sequence
caucaucaucauACTGCCATGGACAGGTGTGATTCTGCTTTTT TATCA (SEQ ID
NO:1), and the 3' DAGAT02 primer had the sequence
cuacuacuacuaCTAGAGACAGGGCAATGTAGAAAGTATGTA (SEQ ID NO:2).
Lowercase sequences were used for cloning into the pAMPI vector (Gibco, BRL).
PCR amplification using DAGAT- I and DAGAT-2 primers was carried out as
follows. Each l001i1 PCR reaction mixture contained 50ng of genomic DNA, 200 M
of each dNTP, 1X buffer B (Gibco BRL), I M DAGAT-1 primer, 1 M DAGAT-2
primer, 3mM magnesium sulfate, and 2 l Elongase enzyme (Gibco BRL). The
reaction mixture was denatured at 94 C for 3 minutes, followed by 30 cycles of
denaturation at 94 C for 1 minute, annealing at 50 C for 2 minutes, and
extension at
72 C for 3 minutes. A final extension incubation was performed at 72 C for 10
minutes after cycling. Based on the sequence of A. thaliana genomic DNA in
GenBank Accession No. 3135276, a 1369 bp fragment was expected to be amplified
from Brassica.
An approximately 1.4 kb fragment was amplified from genomic DNA isolated
from Brassica napus variety Westar under PCR conditions described above. The
amplified DNA fragment was cloned into the pAMPI vector (Gibco BRL) and
partially sequenced from the 5' end using a T7 universal primer and from the
3' end
using a Sp6 universal primer. Figure 1 contains a restriction map of the pAMPI
vector containing the putative DAGAT fragment, which has been designated
pMB 143. The partial sequence of the 5' end using the T7 primer was
-14-
CA 02372632 2001-10-31
WO 00/66749 PCT/USOO/12316
ATGGACAGGTGTGATTCTGCTTTVVFATCAGGTGTCACGTTGATGCTCCTC
ACTTGCATTGTGTGGCTGAAGTTGGTTTCTTACGCTCATACTAACTATGAC
ATAAGAACCCTAGCTAATTCATCTGATAAGGTAAAAGAAGTGATATAATA
TTGGTCACTTGCATTGTGTTTTACTATTTTGACCAGACACTGTTGAAAACT
GTAGGCCAATCCTGAAGTCTCCTACTATGTTAGCTTGAAGAGCTTGGCGTA
TTTCATGCTTGCTCCCACATTGTGTTATCAGGTAATCTGATGCGTCTTCTGC
TAATTGTATCATACATTATCTTTCACTTGCAAAAGTTTCTTGTCTAAAACCT
TGCGTCTTCGCTTTACCCAGCCGAGCTATCCACGTTCT (SEQ ID NO:3).
The partial sequence of the 3' end using the Sp6 primer was
1o ATCAATCTTGTCTTACTCAAAAATCATATTATGTTTACGTTANTAACCAAA
ATTCATGTACGCACTGTCTACCTTTGTCAGTATTGGAGAATGTGGAATATG
GTATGGTTCTCTTCTTGAACATCCCCTTCTTTTTTTATACAAAGCA GATTAA
GAAAAGCTTATTGAGATCTTGT VV1TFCTAATAGCCTGTTCATAAATGGAT
GGTTCGACATGTATACTTTCCGTGCCTTCGCAGAAATATACCGAAAGTGA
GTGTAGTTAATTGCGATGATCGATAT 'I-=CTGTGCTTCATAAATTTAAC
CCTCCACTCATCCAGGTACCCGCTATTATCCTTGCTTTCTTAGTCT
CTGCAGTCTTTCATGAGGTATAATACATACTTTCTACATTGNCCCTGTCTC
(SEQ ID NO:4).
The sequence of the B. napus clone obtained using the T7 primer had an
overall identity of 76.5% with the corresponding region of the A. thaliana
gene,
suggesting that the 1.4 kb cloned B. napus fragment encoded a protein that may
be
related to the A. thaliana DAGAT or ACAT.
Plasmid pMB 143 will be deposited with the American Type Culture
Collection (ATCC), 10801 University Blvd., Manassas, VA, 20110-2209.
Example 2 - Reduction of Oil Content in Brassica Plants Expressing
Antisense DAGAT: The putative DAGAT genomic DNA cloned in pAMP1 was
excised with a Smal/SnaBi digestion. The putative DAGAT fragment was purified
and inserted at the blunted, EcoRl site of pMB 110 to generate pMB 171. This
construct contains cruciferin promoter and cruciferin termination sequences.
The
antisense orientation of the putative DAGAT insert in pMB 171 was confirmed
with
restriction analysis. The cruciferin promoter/putative DAGAT/cruciferin
termination
cassette was released from pMB 171 by partially digesting with Xbal and
completely
digesting with Xhol, and cloned into the XhoI/XbaI sites of pMOG800 to
generate
pMB 170. Figure 2 provides a diagram of pMB 170.
The construct pMB 170 was used to transform Agrobacterium LBA4404. The
resulting Agrobacterium transformants were each co-cultivated separately with
B.
-15-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
napus variety Westar hypocotyls and cultured consecutively on incubation,
selection
(containing kanamycin) and regeneration media until green shoots were
produced.
Regenerated plantlets were transferred to the greenhouse and grown to
maturity.
Each Ti plant (N=56) was selfed and the resulting T2 seeds were harvested from
each
individual Ti plant.
Oil, protein, chlorophyll, glucosinolate, oleic acid (18:1), linoleic acid
(18:2),
and a-linolenic acid (18:3) content were determined in the T2 seed samples.
Oil
content was measured by NIR using a Foss NIR Systems model 6500 Feed and
Forage Analyzer (Foss North America, Eden Prairie, MN) calibrated according to
1o manufacturer's recommendations. Canola seed samples, which represented wide
ranges of the sample constituents listed above, were collected for
calibration. Lab
analysis results were determined using accepted methodology (i.e., oil, AOCS
Method
Ak 3-94; moisture, AOCS Method Ai 2-75; fatty acid, AOCS Method CE 1e-91 and
AOCS Method CE 2-66; chlorophyll, AOCS Method CC 13D-55; protein, AOCS
Method BA 4e-93; and glucosinolates, AOCS Method Ak 1-92). Instrument response
also was measured for each sample. A calibration equation was calculated for
each
constituent by means of chemometrics. These equations are combined into one
computer file and are used for prediction of the constituents contained in
unknown
canola samples.
T2 seed samples containing unknown levels of the above constituents were
prepared by removing foreign material from the sample. Cleaned whole seed was
placed into the instrument sample cell and the cell was placed into the
instrument
sample assembly. Analysis was carried out according to manufacturer
instructions
and was based on AOCS Procedure Am 1-92. The results are predicted and
reported
as % constituent (% oil and protein are reported based on dry weight).
Conversion
from `dry weight' basis to `as is' basis for oil and protein can be calculated
using the
following formula:
constituent (as is) = constituent (dry wt.) x [1-(% moisture/100)].
The average dried oil content of the T2 seeds (39.62 1.95%, n=56) was not
significantly different from that of the control (Westar, 39.92 1.63, n=5).
There were
ten Ti plants, however, that produced T2 seeds having a lower total oil
content. In
-16-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
these T2 seeds, the average oil content was 36.63 0.64%. Table 2 provides an
analysis of eight of these 10 T2 seeds.
Approximately 10-20 T2 seeds from each of the ten selected plants were
planted, and a nickel size portion of the leaf tissue was taken from plants
about 2.5
weeks post-germination. Tissue samples were dried in a food dehydrator at 135
C for
8-16 hours. DNA was isolated using the Qiagen Dneasy96 Plant Kit, and
resuspended in 150 1 of buffer. PCR amplification was performed in a 20 I
volume
containing 1X PCR buffer containing 1.5mM MgC12 (Qiagen PCR Core Kit), 0.2mM
dNTP, 0.5 units Taq polymerase (Qiagen), 0.5 M cruciferen primer (5'-CTT TAT
1o GGA TGA GCT TGA TTG AG-3', SEQ ID NO:5), and 0.5 M acyltransferase primer
(5'-CCG CTC TAG AGG GAT CCA AGC-3', SEQ ID NO:6), 0.4% sucrose, and
0.008% cresol. Amplification conditions included 30 cycles of denaturation at
94 C
for 30 seconds, annealing at 55 C for 30 seconds, and extension at 72 C for 1
minute.
PCR products were analyzed 1.2% agarose gel electrophoresis. As indicated in
Table
1, the selected plants tested positive for the transgene.
TABLE 2
Analysis of T2 Seeds With Lower Oil Content
SAMPLE OIL PROT CHLOR GLUC C18:1 C 18:2 C 18:3 (# pos.
plants/Total
# plants
tested)
Y350071 35.69 26.07 78.12 3.66 70.57 10.03 7.74 15/18
Y350070 36.35 24.87 78.57 2.60 72.02 8.18 7.34 11/18
Y350095 36.73 26.55 88.44 1.38 73.12 7.80 7.62 7/9
Y350079 36.79 27.96 22.75 2.96 66.67 14.97 7.53 17/18
Y350052 37.02 26.02 74.34 3.44 67.80 14.09 7.62 8/18
Y350077 37.04 25.70 70.80 2.91 71.30 9.16 8.07 15/18
Y350089 37.13 25.07 59.60 2.28 71.37 9.93 7.26 8/15
Y350078 37.66 25.64 43.00 2.35 74.75 7.48 6.94 11/18
Average 36.80 25.99 64.45 2.70 70.95 10.23 7.51
Std Dev. 0.58 0.97 21.79 0.72 2.65 2.81 0.34
-17-
CA 02372632 2001-10-31
WO 00/66749 PCT/US00/12316
OTHER EMBODIMENTS
It is to be understood that while the invention has been described in
conjunction with the detailed description thereof, the foregoing description
is intended
to illustrate and not limit the scope of the invention, which is defined by
the scope of
the appended claims. Other aspects, advantages, and modifications are within
the
scope of the following claims.
-18-
CA 02372632 2009-01-16
SEQUENCE LISTING
<110> CARGILL, INCORPORATED
<120> PLANT ACYLTRANSFERASES
<130> 8978-69 JHW
<150> US 60/132,417
<151> 1999-05-04
<160> 6
<170> Patentln Ver. 2.0
<210> 1
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetically generated primer
<400> 1
actgccatgg acaggtgtga ttctgctttt ttatca 36
<210> 2
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetically generated primer
<400> 2
ctagagacag ggcaatgtag aaagtatgta 30
<210> 3
<211> 396
<212> DNA
<213> Brassica napus
<400> 3
atggacaggt gtgattctgc ttttttatca ggtgtcacgt tgatgctcct cacttgcatt 60
gtgtggctga agttggtttc ttacgctcat actaactatg acataagaac cctagctaat 120
tcatctgata aggtaaaaga agtgatataa tattggtcac ttgcattgtg ttttactatt 180
ttgaccagac actgttgaaa actgtaggcc aatcctgaag tctcctacta tgttagcttg 240
aagagcttgg cgtatttcat gcttgctccc acattgtgtt atcaggtaat ctgatgcgtc 300
ttctgctaat tgtatcatac attatctttc acttgcaaaa gtttcttgtc taaaaccttg 360
cgtcttcgct ttacccagcc gagctatcca cgttct 396
19
CA 02372632 2009-01-16
<210> 4
<211> 410
<212> DNA
<213> Brassica napus
<220>
<221> misc feature
<222> (1) _.(410)
<223> n = A,T,C or G
<400> 4
atcaatcttg tcttactcaa aaatcatatt atgtttacgt tantaaccaa aattcatgta 60
cgcactgtct acctttgtca gtattggaga atgtggaata tggtatggtt ctcttcttga 120
acatcccctt ctttttttat acaaagcaga ttaagaaaag cttattgaga tcttgttttt 180
tctaatagcc tgttcataaa tggatggttc gacatgtata ctttccgtgc cttcgcagaa 240
atataccgaa agtgagtgta gttaattgcg atgatcgata tttttttctg tgcttcataa 300
atttaaccct ccactcattt ttttccaggt acccgctatt atccttgctt tcttagtctc 360
tgcagtcttt catgaggtat aatacatact ttctacattg nccctgtctc 410
<210> 5
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetically generated primer
<400> 5
ctttatggat gagcttgatt gag 23
<210> 6
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetically generated primer
<400> 6
ccgctctaga gggatccaag c 21