Note: Descriptions are shown in the official language in which they were submitted.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
PHYTOMICS: A GENOMIC-BASED APPROACH TO HERBAL COMPOSITIONS
FIELD OF THE INVENTION
This invention relates to herbal compositions. More specifically, this
invention
provides tools and methodologies for improving the selection, testing, quality
control and
manufacture of herbal compositions, and to help guide the development of new
herbal
compositions and identify novel uses of existing herbal compositions.
BACKGROUND OF THE INVENTION
All publications and patent applications herein are incorporated by reference
to the
same extent as if each individual publication or patent application was
specifically and
individually indicated to.be'in~orporated by reference.
Herbal medicine has been in use for centuries by people of Asia and Europe. In
the
United States (US), herbs have become commercially valuable in the dietary
supplement
industry as well'as in holistic.riiedicine. Approximately one third of the US
population has
tried some form of alternative medicine at least once (Eisenberg et al., 1993,
N. Engl. J. Med.
32:246-252).
Botanicals, includiri~ herbsyhave also become a focal point for the
identification of
new active agents to treat diseases. Active compounds, derived from plant
extracts, are of
continuing interest to the'pharinaceutical industry. For example, taxol is an
antineoplastic
drug obtained from the bark of'the western yew tree. It is estimated that
approximately 50
percent of the thousands 'af driigs commonly used and prescribed today are
either derived
from a plant source or contaim~chemical~imitations of a plant compound
(Mindell, E.R., 1992,
Ecc~l Mindell's Herb Bible;'A~Fireside Book).
Currently, a number of medicinal formulations, food supplements, dietary
supplements and thelike contain herbal components or extracts from herbs.
Herbal
medicines have been u' sed 'for trieat'ing various diseases of humans and
animals in many
different countries for a very loiig.period of time (see, e.g., LA. Ross,
1999, Medicinal Plants
of the l~orld, Chemical 'Cdhstituehts, Ti°aditio~cal and Modern
Medicinal Uses,
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-2-
Humana Press; D. Moloriy,.,1.998;; The American Association of Oriental
Medicine's Complete
. ~s.. .,
Guide to Chinese Herbal Medicine, Berkley Books; Kessler et al., 1996, The
Doctor's
Complete Guide to Healing,Medicines, Berkley I~ealth/Reference Books);
Mindell, supra).
Herbal Medicines.. There are many branches of herbal medicine around the
world,
such as Ayurveda, Unani;, Sida and Traditional Chinese Medicine (TCM). While
modern
Western medicine typicatly,consists~of administering a single. chemical entity
capable of
intervening. specific biocheinioal pathways, each formula of TCM typically
contains hundreds
of chemical entities from several herbs which are designed to interact with
multiple targets in
the body in a coordinated'rrianner. ; ~~lthough empirical practice contributed
in a significant
way to the herbal composition and,.pre.~cription of these ancient herbal
medicines, they are also
supported, to a varying degree;,by,a'set of theories which all are distinct
from that of modern
Western medicine iri terms.t~f~aii"atoxriy,.pharmacology, pathology, diagnosis
treatment, etc.
Among the different herbal~iii~edicirle fields; TCM has developed a more
complete set of
theories over several'centuriesvvhich have been well documented and practiced
by local
physicians caring for a huge population (>.1.3 billion people) in greater
China and in East Asia
including~Korea and Japan:' ', . ~ ' ''
Westermmedicin~.ge~ierally uses 'purified compounds, either natural or
synthetic,
mostly directed towards va single physiological target. However, the
compositions used in
TCM are usually composed of multiple herbs and compounds which are aimed at
multiple
targets in the body based on viiique and holistic concepts. TCM mainly used
processed crude
natural products, with'yarious :combiilations and formulations, to treat
different conformations
resulting iri fewer side effects: The 'great potential of TCM has yet to be
realized for the
majority of the world°s pebple, ;'~"~ '. ~,~,
The herbs iii a typical ~'TCM prescription are assigned roles as the principal
herb and the
secondary herbs, including.~assis'tant, adjuvant and guiding herbs. The
principal herb produces
the leading effects in'treatimg~t~i~'cause'or the main symptom of a disease.
An assistant herb
helps to strengthen the effect of the principal herb and produces leading
effects in the treatment
of the accompanying symptofiis.~ There are three types of adjuvant herbs: 1)
those that enhance
the therapeutic effects of the~priricipal and assistant herbs or treat
tertiary symptoms, 2) those
that reduce or eliminate the toxicity arid other side effects of the principal
and the assistant
herbs and 3) those which act dmcoinplementary target tissues not specifically
affected by the
principal herb. A guiding herb directs 'the effect of other herbs to the
affected site and/or
coordinates and mediates the~effects~ of the other herbs in the prescription
or formulation. In
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-3-
contrast to most of the herbal medicines or supplements that consist of one or
more parts of a
single plant, the intended~effects of TCM,are directed at multiple tissues.
For example, a well,-known TCM recipe, "Ephedra Decoction" used for treating
asthma
is composed of ephedra, cinnamon twig, bitter apricot kernel and licorice.
Ephedra, is the
principal herb, which expels, cold, induces diaphoresis and facilitates the
flow of the Lung Qi
to relieve asthma, the main symptoms: , Cinnamon twig, as the assistant herb,
enhances
ephedra's induction of diaphoresis and warms the Channels to ensure the flow
of Yang Qi for
reducing headache and pantalgia.. Bitter. apricot kernel, as the adjuvant
herb, facilitates the
adverse flow of the Lung Qi and strengthens the asthma relief by ephedra.
Licorice as the
guiding herb moderates th_e' effects of both ephedra and cinnamon to ensure a
homeostasis of
the vital Qi. While each of the four,herbs clearly exhibits its respective
activity, they
,,
complement as well, as suppleri~ent: each other when they are combined. In
practice, the
principal herb can be prescribed~with one~or more secondary herbs, depending
on the
symptoms at ~a patient°s ~presetitation .(Prescriptions of Traditional
Chinese Medicine, Chapter
One, ppl0-1~6, E. Zhang, editor.in Chief, Publishing House, Shanghai
University of Traditional
Chinese Medicine,1998)., ' . ' ".. '"~ '. ,
The main theories,'of TAM that guide the treatment of sickness with herbal
medicine
and other means, such as acupW cture, are 1) the theory of Yin and Yang, 2)
the theory of Five
Elements, 3) the theoiy of Viscera and Bowel's, 4) the theory of Qi, Blood and
Body Fluid, and
5) the theory of Channels and Collaterals.
In TCM, the first~important aspect of making the proper diagnosis is to
ascertain
whether the disease is Yiri'or'Yarig: .;For example; those patients who have a
fever, are thirsty,
constipated or~have a rapid~pulse condition are of Yang character. Those
individuals who have
an aversion to cold, are 'not tliirsty, and diarrhea and a slow pulse
conclition are of Yin
character. The propeity, ~flavorrand~function ofherbs can also be classified
according to Ying
and Yang theory. For exaxriple, Herbs'df cold and cool nature belong to Ying,
while herbs
which are warm and hot~in, nature belong to Yang. Herbs with sour, bitter and
salty flavor
belong to Ying, while ~herlis~vc~ith pungent, sweet and bland flavor belong to
Yang. Herbs with
astringent and subsiding function belong to Yin, while herbs with dispersing,
ascending and
floating function belong to Yang. In TCM, the principles of treatment are
based on the
predominance or weakness' of Yiri and Yang. Herbs are prescribed according to
their property
of Ying and Yang and their function for restoring the imbalance of the Ying
and Yang. In so
doing, the benefit of treatriierit ~is achieved.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-4-
According to the theory ~of'Five Elements there are five basic substances that
constitute
the material world (i.e., wood, fire; earth, metal and water). Tn TC1VI, this
theory has been used
to explain the physiology and~patliology ~of the human body and to guide
clinical diagnosis and
treatment. Herbal physicians, have applied the laws of generation,
restriction, subjugation and
reverse restriction of the five element's to worl~,out many effective and
specific treatment
regimens, such as reinforcing, earth to generate metal (strengthening the
function of the spleen
to benefit the lung), replenishing water to nourish wood (nourishing the
essence of the kidney
to benefit the liver), supporting. earth ~to restrict the wood (supplementing
the function of the
'.
spleen to treat the hyperactivity of the liver), and strengthening water to
control fire
(replenishing the essence of the,kidney to treat hyperactivity of the heart).
Specifically, the
property of some herbs is assig~ied.to each of the five Elements for the
purposes of guiding the
-~ .
prescription of a TCM rec'ip~.~, ' . v ' °
In TCM; the internal''organs of the human body are divided into three groups:
five
;.., ° .
Viscera (the Heaxt, the Livef',the Spleen, the Lung and the Kidney), Six
Bowels (the Gall
Bladder, the Stomach, the,'Iarge Intestine, the Small Intestine, the Urinary
Bladder, and the
Triple Warmer), the Extraoidinar~ Organs (the Brain, the Medulla, the Bone,
the Blood
. ;r ,
Vessel, the Gall Bladder, a'nfalie Uterus). In TCM, the Viscera or the Bowel
are not only
anatomic units, but are also.'concepts of physiology and pathology about
interactions between
different organs. For examp'l'e; the.heart also refers to some of the mental
functions and
influence functions of blood; hair, tongue and skin. Ying-Yang and the Five
Elements
influence the interactions ainoiig'' th'e'se Viscera, Bowels and Organs. The
complexity of
interplay of the theories is.u'sed'eto°'e~plain the pathology of
diseases to which herbs are
prescribed, as discussed~lielow.
The ~prescriptiori of herbal medicine in TCM starts with the diagnosis, which
consists of
. four main items: interrogation;rimspection, auscintation and olfaction,
pulse taking and
palpation. During the interrogation phase, much information is gathered,
including the
characteristics ~of the main .syinptorris: For instance, if the main symptom
is characterized by
dull pain of epigastric regioii;'vwhich may be relieved by warming and
pressing, this suggests
the insufficiency of the Spleen-Yang: Soreness and weakness of the loins and
la~ees,
intolerance of coldness with' cold 'extremities manifests a weakness of the
Kidney-Yang.
During inspection, observationsware made for vitality; skin color and the
general appearance
and the condition of the tongue: ~ Forexample, a pale complexion corresponds
internally to the
bung of autumn; whose Qi'is diy. This may occur when Yang Qi is lacking and
the circulation
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-5-
of Qi and blood is impeded, or when the coldness in the channels and
collaterals causes them
to contract. ~ .
hi TCM, it is from Qi; ~bloodland body fluid that come energy needed by the
Viscera
and Bowels, Channels and Collaterals,ltissues and other organs for carrying-
out their
physiological functions; and'on which the formation and metabolism of Qi,
blood and body
fluid depend. Prescriptions of TC1VI consider the herbal effects on Qi and
blood for treatments.
TCM holds that Channels, Collaterals and their subsidiary parts are
distributed over the
entire body. It is through them that herbs exert influence on pathological
taxgets and achieve
the improvement of sickness. For example, ephedra acts on the Channels of the
Lung and
Urinary Bladder so as to induce sweat for relieving asthma and promoting
diuresis. As noted
. _.. ,.,a,~:.. ..
above, clinical applications~of acupuncture are also guided by the theory of
Channels and
Collaterals. ' r
In summary, while the nature or property of each herb in TCM may be assigned
as Yin
or Yang, and to one of the Five Elerilents, they act through Channels and
Collaterals and are
mediated via Qi, Blood and Fluid tovyield therapeutic effects on taxgets, such
as Viscera and
Bowels. Pathogenic factors may be~disguised as decoy through the very same
systems of
Channels and Collaterals ~o adversely affect the functions of Viscera and
Bowels and thus
cause sickness.
From the foregoing discussion, it is clear that the TCM terminology is as much
of a
philosophical concept asvan anatomical one. For example, the Heart represents
a host of
tissues, organs or systems iri the.body that contribute to a function
described in TCM. Thus,
the concept of the Heart requires a multiple dimension data set to describe
each concept of
TCM. Once this is accomplished; a molecular holistic medicine can be
developed.
U.S. IZegulatory.Proce'ss. :In the US, dietary supplements (such as botanical
products,
vitamins and minerals, ariiirio''aoids arid tissue extracts) are regulated
under the Dietary
Supplement Health and Education ~Act~of,1994 (the DSHE Act). This Act removed
the
ingredients of dietary supplements from regulation as food additives under the
Federal Food,
Drug, and Cosmetic Act: Iri vaddition, the DSHE Act requires that The Food and
Drug
Administration (FDA) bearalie burden of proof that a marketed dietary
supplement presents a
serious or unreasonable risk' under the conditions of use on the label or as
commonly
consumed. Thus, there .are cuireritly no federal regulations that establish
specific criteria for
purity, identification and inariufacturing procedixrcs for dietary
supplements. In addition, few
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-6-
published papers on herbal quality have resulted from the establishment of the
Office of
Alternative Medicine by Congress~in'1992 (Angell et al., 1998, N. Engl.
J.~Med. 339:839-841).
At the present time, the~FDA must approve each one of the chemical entities in
a drug
composition or cocktail,~and then clinical trials must be undertaken so as to
obtain separate
FDA approval for marketing,tlie drug. This process is extremely tedious and
costly. A
molecular holistic medicine may require a less arduous evaluation since the
previous use of a
particular herbal composition as a,botanical drug permits clinical trials with
multiple chemicals
at the outset (i.e., clinical trials using the herbal composition or specific
components of the
herbal composition). Recently, the.FDA has approved the testing of some herbal
medicines in
clinical trials as botanical drugs (FDA Guidance on Botanical Drugs, April,
1997). While
these events represent a positive development for health care in general, it
also raises important
issues regarding the formulation manufacturing and quality control of herbal
medicines and
dietary supplements, iricludiiig.the',traditional Chinese medicines.
., .
The multitude ofrelevarit biological responses induced by the multiple
chemicals in
herbs are not currently available 'and will be increasingly important to
support marketing
approval by the FDA. ' ~ '', ;
Herbal-based industries are coming under increasing pressure to upgrade their
current
practices (see, e.g., Angeil etal.; supra). The need to apply scientific
testing to the preparation
and administration of herbal rriediciries, and food supplements has been
highlighted by several
recent reports of toxicity resulting froril ingesting herb-based formulations.
For example, one
patient who took an herbal-based dietary supplement experienced digitalis
toxicity (Slifinan et
al., 1998, N. Engl. J. ~Med.'339:806=811). It was subsequently determined that
the herb
ingredient labeled as plantainvin the supplement was actually contaminated
with Digitalis
la~cata, an herb known to coritaiii at;least~60 cardiac glycosides. In another
instance, an herbal
preparation was found to li'e the cause of chronic lead intoxication in a
patient (Beigel et al.,
1998, N. Engl. J. Med.,33.9:827-830). This is ribt a completely unexpected
occurrence since
contamination of tradiiional'Asian herbalremedies by lead and other heavy
metals is well
documented (Woolf et.al'.,:1994, Ann. Intern. IVIed. 121:729-735).
.':
Characterization of BotariicaTs. It.is well known that the genetic identity
(e.g.,
genera, species, cultivar, variety, clone), age of herbal growth, harvest
time, the specific plant
part utilized, processing nrierhod,~geographical origin, soil type, weather
patterns, type and rate
of fertilizer, and other grov~thvfactors have a great impact on the particular
chemical
composition of any particular herb "harvested" from any particular area.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
_7_
Increasing numbers of various types of tests have been instituted to assure
the
consistent quality of herbs 'used irl~medicine and as dietary supplements;
including inspections
at the macro- and microscopic levels as' well as a variety of chemical
analyses. Recently, high
performance liquid chromatography (HPLC) profile of marker molecules in an
herbal extract
has become one reference standard. However, there are problems with this
approach,
including that some of the liioactive molecules may not adsorb UV or the
visible lights for
HPLC detection, and the 'amount of a chemical is not necessarily proportional
to its biological
potency. For these reasons, herbal manufacturers resort to a practice of
mixing raw herbs from
different sources to minimize chemical variations.
Mass spectrometry.,(MS) is ~an analytical method for determining the relative
masses
and relative abundances of components of a beam of ionized molecules or
molecular fragments
produced from a sample'im a~ high vacuum. ~MS, unlike HPLC, is not optical
density-
~',,
dependent. In practice it 'is~used iri conjunction with HPLC or capillary
electrophoresis (CE):
the HPLC separates the 'chemicals and. the MS then can be used to~ identify
what they are.
Commercial systems 'are available which integrate MS and HPLC for biological
uses. Mass
spectrometry is limited to sarriples that'are gaseous or volatile at low
pressure, or that can be so
rendered by derivatization: .' ' '
These steps are no ~loriger adequate. Recent publications report a greater
variation in
the quality of herbs by specific 'suppliers, and the difficulty of providing
biological equivalence
of herbal extracts.. Furthermore, the correlation between safety and efficacy
and chemicals in
an herb is not well defined in, most cases. Recently, in response to
complaints from consumer
groups and .regulatory agencies (Federal Register, February 6, 1997, Volume
62, No. 25,
Docket No. 96M-0417 cGIVIT'' in Manufacturing, Packing or Holding Dietary
Supplements,
Proposed Rules), some herbal rnariufacturers have begun to implement Good
Manufacturing
Practice (GMP) which requires ~stririgerit controls at all levels.
Chemical and spectTOSCOpic.methods~have been used to characterize the
components of
herbal medicines and food supplements: For'example, three new hederagenin-
based acetylated
saponins were isolated ~rom.the fruits of Gliricidia sepium using these two
methods (Kojima et
al., 1998, Phytochemistrv:48('S)$85-888). The botanical sources of Chinese
herbal drugs in a
number of commercial samples were inferred by comparing the contents of some
characteristic
constituents which were 'amalyzed with high-performance chromatography (HPLC)
or capillary
electrophoresis (CE) (Shuerin-Jyi Sheu, .1997, Journal of Food and Drug-
Analysis 5(4):285-
294). For example, the ratio .of ephedrine/pseudoephedrine was used as a
marker to
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
_g_
differentiate EphedYa.ihte~media from other species; total alkaloid contents
were used to
distinguish between species of Phellodend~on; and the contents of ginsenosides
were used to
differentiate between species of Panax. However, these methods do not provide
a direct
. :.. . . ,
measurement of the effect of the various herbs on the molecular, physiological
or
morphological responses following human treatment with the herbs.
Using gas chromatography-mass spectrometry and atomic-absorption methods, the
California Department of Health Sciences, Food and Drug Branch, recently
tested Asian
medicines obtained from herbal stores for contaminants (R. J. Ko, 1998, N. En
l~. J. Med.
339:847). Of the 260 products they tested, at least 83 (32 percent) contained
undeclared
pharmaceuticals or heavy metals, and 23 had more than one adulterant. Using
high-
performance liquid chromatography, gas chromatography, and mass spectrometry,
a
commercially available combination of eight herbs (PC-SPES), was found to
contain
estrogenic organic compoyds~(DiPaola et al., 1998, N. En,gl. J. Med. 339:785-
791). The
researchers concluded that PC _SPES has potent estrogenic activity and that
prostate cancer
patients that took PC-SPES; could confound the results,of standard therapies
and may
experience clinically significant adverse effects. Gas chromatography data was
also collected
for different samples of the traditional Chinese medicine'wei ling xian' and
correlated to the
antiinflammatory activity of he~sainples (Wei et al., Study of chemical
pattern recognition as
applied to quality assessment of the traditional Chinese medicine "wei ling
xian," Yao Hsueh
Pao 26(10): 772-772 (1991 )), yTliis study did not provide relevant HBR Array
data, such as
time course, dose dependent responses control samples to substantiate the
differential power of
the biomarkers, nor it utilize a.reiterative type. of data construction
process to establish a
comprehensive database for'characterizing effects of the herbal composition.
Changes in protein levels have also been used to characterize the effects of
herbal
compositions or specific components of herbs. fiFor example, the production of
granulocyte
colony-stimulating factor (G~CSF) from peripheral blood mononuclear cells was
found to vary
depending on which specific Chinese herb was added to the culture (Yamashiki
et al., 1992, J_.
Clin. Lab. Immunol. 37(2):83'-90). . Expression of interleukin-1 alpha
receptors was markedly
up regulated in cultured humamepiderinal keratinocytes treated with Sho-saiko-
to, the most
commonly.used herbal~medicirle irl Japan (Matsumoto et al., 1997, Jpn. J.
Pharmacol.
73(4):333-336). The expressionof FC gamma 11/111'receptors and complement
receptor 3 of
macrophages were increased'liy treatment with Toki-shakuyakusan (TSS) (J. C.
Cyong, 1997,
Nippon Yakurigaku Zasshi 110(Suppl.: l):87-92). Tetrandrine, an alkaloid
isolated from a
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-9-
natural Chinese herbal medicine; inhibited signal-induced NF-kappa B
activation in rat
alveolar macrophages (Cheri et a1:,~1997, Biochem. Biophys. Res. Coxrimun.
231(1):99-102).
The herbs Sairei-to, alismatis rhizoma'(Japanese name "Takusha") and hoelen
(Japanese name
"Bukuryou") inhibited the sy.~thesis and expression of endothelin-1 in rats
with anti-glomerular
basement membrane nephritis (Hattori et al., 1997, Nippon Jinzo Gakkai Shi
39(2):121-128).
The increase or'decrease in ~mRNA levels has also been used as an indicator of
the
effect of various herbs and herbal components. Intraperitoneal injection of
Qingyangshen
(QYS), a traditional Chinese.medicine with antiepileptic properties, and
diphenylhydantoin
sodium reduced alpha- and,beta-tublin'mRNAs and hippocampal c-fos mRNA
induction
during kainic acid-induced chronic, seizures in rats (Guo et al., 1993, J.
Tradit. Chin. Med.
13(4):281-286; Guo et al.,.~19.95, J. Tradit. Chin. Med. 15(4):292-296; Guo et
al., 1996, J_.
Tradit. Chin. Med. 16(1):48=51): fireatmerit of cultured human umbilical vein
endothelial cells
(HUVECs) with the sapomiri astragaloside IV, a component purified from
Ast~agalus
. ;a.. ,
membranaceus, decreased plasiiliriogemactivator inhibitor type I (PAI-1)
specific mRNA
expression and increased tissue=type plasminogen activator (t-PA) specific
mRNA (Zhang et
al., 1997, J. Vasc. Res. ,34(4):273-280). One component isolated from the root
of Panax
ginseng was found to be a potent inducer'of interleukin-8 (IL-8) production by
human
monocytes and by the huriian riionocytic cell line THP-1, with this induction
being
accompanied by increased' IL-8 mRNA expression (Sonoda et al., 1998,
Immunopharmacolog,~
38:287-294). . '
Recent advances in nucleic acid microarray technology enable massive parallel
mining
of information on gene expression: This process has been used to study cell
cycles,
biochemical pathways, genoirie-wide expression in yeast, cell growth, cellular
differentiation,
cellular responses to a single~chemical,compound, and genetic diseases,
including the onset
and progression of the diseases ~(M.:Schena. et al., 1998, TIBTECH 16:301).
Because cells
respond to the micro=enviibriment,~changes by changing the expression level of
specific genes,
the identities of genes expressed in a cell determine what the cell is derived
of and what
biochemical arid regulatory systems are involved, among other things (Brown et
al. 1999,
Naturea~enet.; 21 (1) supplerrierit:33): Thus,~cellular gene expression
profiles portray the
origin, the present~differentiatiomof the cell, and the cellular responses to
external stimulants.
No researchers to datea if arty, Have attempted to apply these new
technologies to study the
molecular effects of whole~herbal treatments and supplements.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-10-
Some researchers have. attempted to characterize the effects of the major
active
constituents isolated from selected herbs. For example, treatment of HLTVECs
with
notoginsenoside Rl (NRl)purifiedfrom Panax notoginsehg, resulted in a dose-
and time-
dependent increase in TPA synthesis (Zhang et al., 1994, Arteriosclerosis and
Thromobosis
14(7):1040-1046). Treatment witli~NRl did not change urokinase-type
plasminogen activator
and PAI-1 antigen synthesis rior did it effect the deposition of PAI-1 in the
extracellular
matrix. TPA mRNA increased as~mucli as twofold when HUVECs were treated with
NR1,
,.
whereas expression of PAL-1-specific mRNA was not significantly affected by
NR1. Since
most studies on P. i~otoginsehg have involved its mixture with other herbs,
the researchers
noted that it was difficult to~assess how their results relate to the
situation ih vivo when is used
therapeutically in humans ~(Id:, at 1045, second column, first paragraph). In
addition, since the
researchers only studied brie rnaj'or coriiponent~of the herb, it is not
possible to ascertain the
molecular effect of the'wliole lierb or the interactions among components of
the herb from this
study. ,,~ . . . ,
Dobashi et al. (1995,,.lsTeuroscience Letters 197:235-238) studied the effect
of two of
the main components of'saiko'agerits~'a Chinese herbal drug used to treat
nephrotic syndrome,
bronchial asthma and.chronic rheuxriatoid arthritis. Administration of SS-d
increased plasma
adrenocorticotropin (ACTH) levels; proopiomelanocortin mRNA levels in the
anterior
pituitary and the CRF~mRNA, level in the rat hypothalamus in a dose dependent
rn'anner. In
contrast, treatment with SS-a~failed to 'affect the levels of these molecular
markers. While this
study indicates that administration of SS-d may have an important role in
saiko agents-induced
CRF release and CRF gene. expression in rat hypothalamus, it fails to address
the molecular
effect of the herbal medtcatiomas a whole.
I~ojima et al. (1998;"Biol. Pharm. Bull. 4:426-428) describe the utilization
of
differential display of mRNA to isolate and identify genes transcriptionally
regulated in mouse
liver by sho-saiko-to, an herbal medicine used for treating various
inflammatory diseases in
. :. : .~ -.'' . ,w , ,
Japan. These researchers lirn'itedvheir study to the use of mRNA differential
display
techniques in investigating ~ttie'~molecular mechanisms of herbal medicine. It
also failed to
address effects in multiple'orgams of treated animals and did not provide any
guidance for
quality control, new use, and standardization of effects. In addition, the
study failed to analyze
the individual components o~,f the' herb and compare the individual results
with the results
obtained using the whole herlialniiiXture.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-11-
Ma Ji et al. (199; Chinese Medical Journal 111(1):17-23) investigated the
therapeutic
effect of the herb Astragali, yiiembrahaceus on sodium and water retention in
rats experiencing
aortocaval fistula-caused eXperimental congestive heart failure. Chronic heart
failure rats with
and without AstYaglia treatment were conipared for changes in various
morphological
characteristics (e.g., body weight, serum sodium concentration); physiological
characteristics
(e.g., mean arterial pressure, heart rate, hematocrit and plasma osmolality);
mRNA expression
levels (e.g., hypothalamic. arginine vasopressin (AVP),. AVP Vla receptor,
renal AVP VZ
receptor, aquaporin-2 (AXP2)) and protein excretion (e.g., plasma atrial
monophosphate
peptide (ANP) and urinary cyclic guanidino monophosphate (cGMP)). The
researchers found
that treatment with Astraglia~ improved cardiac and renal functions, partially
corrected
abnormal mRNA expressions of.the AVP system and AQP2, and improved the renal
reaction
to ANP. This study did'not ~a~dress using the collected data to guide the
development of new
formulations or for elucidatingvthe synergistic or other interactions among
various herbs in a
formula, or validate the differential power of the effects for quality control
purposes.
As shown by the abovevreview of relevant scientific articles, molecular-based
technology has not been .used to explore and validate cellular and molecular
responses in
biological 'systems that are treated' or challenged with multiple chemical's
at the same time,
such as herbal medicines arid TCIVI. ~Furthermore~ these recent advances have
not been
integrated with other technologies and methods to produce a process for the
systematic
exploration of biological~effects:of,herbal medicines and TCM.
~' ~" ~' SUMMARY OF THE INVENTION
This invention pro~ides'therools and methodologies for creating, maintaining,
improving and utilizing I3erbal BioResponse Arrays (HBR Arrays), wherein the
HBR Arrays
constitute data sets associ'ate'd°with particulax'herbal compositions.
The HBR Arrays of the
present invention may include'~information on'the plant-related parameters of
the herbal
constituents, marker inforriiati.on collected following the exposure of a
biosystem to the herbal
composition, and biological resporise~imformation collected following the
exposure of a
biosystem to the herbal composition..
The present inventiori'provides the tools and methodologies necessary for
establishing
standardized HBR Array's fox particular herbal compositions, wherein the
standardized HBR
Arrays are used as~benchmaxks'by which to evaluate batches of similar or
different herbal
compositions. The present invention further provides the tools and
methodologies necessary
to update and maintain the standardized.HBR Arrays. Particular embodiments of
the present
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-12-
invention involve iterative~processes whereby data for additional batches of
the herbal
composition, additional plant=related data, additional marker information,
and/or additional
BioResponse information isaperiodically added to the standardized HBR Arrays.
Thus, the
present invention provides the.tools and methodologies for creating,
maintaining, updating and
using HBR Arrays on an ongoing basis.
The present invention.provides the tools and methodologies necessary to guide
the
standardization of herbal compositions; to determine which specific components
of herbal
compositions are responsible for particular biological activities; to predict
the biological
activities of herbal compositions; for the development of improved herbal
therapeutics; for
adjusting or modifying an herbal composition; for measuring the relatedness of
different herbal
compositions; fox identifying~specihc molecules in the batch herbal
composition which retain
the desired biological actiyity;~for determining which herbal components of a
known herbal
composition can be eliminated from 'the known herbal composition while
maintaining or
improving the desired bio~lo~gical activity of the known herbal composition;
for identifying new
uses and previously unkno'wii biological activities for the batch herbal
composition; and for
using the predicted biologica~~ activity of the batch herbal composition to
aid in the design of
therapeutics which include herbal components and synthetic chemical drugs,
including the
design of therapeutics us'irigtlie methods of combinatorial chemistry.
More specifically, the present invention provides methods of establishing
standardized
I3erbal BioResponse Arrays; (~iBR .Arrays) for herbal compositions, wherein
the methods
comprise: . - , , .
a) selecting a characterized herbal.composition;
b) exposing a biosystem ~to~~abatch'of the characterized herbal composition
and collecting data
on two or more markers; wherein one of the markers is a change in gene
expression
determined through the iiseeo'f a'nucleic acid microarray, produced by the
steps comprising:
i) producing a, celfbankirig system;
ii) profiling tlie~geiie'~expressiori pattern of cells from the cell banking
system
before and after exposure to the herbal composition;
iii) selecting as markers those genes whose expression levels are changed by
' ~eXposure'to~tlie herbal composition;
c) storing the marker~data of~step b)' as~ a standardized HBR array.
The present invention further provides such methods which further comprise
exposing a
biosysteri~ to one or inore1iatclies of-the lierbal~composition, collecting
the data on one or more
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-13-
BioResponses, and adding the collected BioResponse data to the standardized
HBR Array for
that herbal composition. ,
The present inventioii~provides methods of evaluating herbal compositions,
wherein the
methods comprise exposing albiosystem to a batch of the herbal composition and
collecting
.,.
data on two or more markers; and comparing the collected marker data with a
standardized
j
i HBR Array for the same orb a substantially same herbal composition as that
of the batch herbal
compositions.
The present invention,provides, a system for predicting the biological
activity of an
herbal composition comprising: - ry . .
1). a biosystem comprising one or more different types of cells, tissues,
organs or
i~a vitro assays; 1 ' t, : . . .
2). a batch herbal :coiriposition;
3). two or more ,molecular markers;
4). a means for.'eXposing~the biosystem to the batch herbal composition and
measuring the differential respbrises ~of the molecular markers;
S). a computervprocessor, including,memory, for analyzing and storing the
differential response measureirients of the molecular markers so as to create
an Herbal
BioResponse Array (HBR ;!tray) data set for the batch herbal composition;
6). a computer processor; including memory, for comparing the HBR Array of the
batch herbal composition to one br more previously-stored HBR Arrays so as to
predict the
biological activity of the batch herbal composition, wherein the biological
activities of the
herbal compositions used to ~gerierate the one or more previously-stored HBR
Arrays are
known. ' , ...
BRIEF DESCRIPTI~N OF'I'HE DRAWINGS
Fi ure 1. ,Figure 1 'provides°,a schematic of the basic method steps
for constructing a
Standardized Herbal BioResporise'Array (HBR Array) for any selected herbal
composition.
The figure is shown iri its xriost:basic form for ease of understanding. As
discussed herein,
each of the pathways of thew'sche~natic~ can be done iteratively. Furthermore,
any information
contained in one box can be 'used to guide decisions regarding gathering
information for any
other box. In this way, nuriieroits feedback loops are also possible
throughout the scheme.
Figure 2. Figure Z~provides ~.~ schematic of the basic method steps for
constructing a an
Herbal BioResponse Array (HB~t~~Array) for any batch herbal composition and
for comparing
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
=14=
this batch IilZB Array to a elected subset of inforination from the
Standardized HBR Array.
The figure is shown in its'most.basic form for ease of understanding. As
discussed herein,
each of the.pathways of the schematic can be done iteratively. Furthermore,
any information
contained in one box can be.used.to guide decisions regarding gathering
information for any
other box. In this way, numerous feedback loops are possible throughout the
scheme.
Fi ug re 3. Figure 3 provides a schematic of the basic method steps for
establishing and
using a major data set. The figure-is shown in its most basic form for ease of
understanding.
As discussed herein, each of the pathways of the schematic can be done
iteratively.
Furthermore, any information contained in one box can be used to guide
decisions regarding
... ' . 5~ . .
gathering information for anyother box. In this way, numerous feedback loops
are possible
throughout the scheme.
Fi ure 4. Western,blbt for various herbal compositions.
A. No herbal composition. ..
B. Huang Qiiig Tang A..(HQT A) (0.2 mg/ml).
C. HQT A (4 mg/ml). '
D. HQT B (0.2 mg/ml).
E. HQT B (4 mg/rril). ,
F. Scute (0.2 mg7inl), w'
G. Scute (4 mg%ml). .. .
Figure 5. HPLC for Paeonie lactiflora pallus.
Figure 6. HPLC for Zizipha fructus.
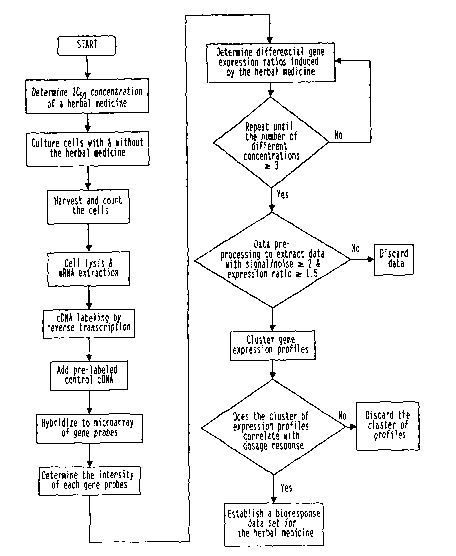
Figure 7. Figure' 7 prpvides a schematic- for establishing a bio-response data
set for an
herbal composition. The data set is based on differentially expressed gene
induced by the
herbal medicirie for moreahan three different concentrations in a mammalian
cell culture.
~ ' . .
Figure 8. Figure 8' provides a schematic for establishing a characteristic
expression
profile database or HBR Array for an herbal medicine or a complex herbal
preparation.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-15-
Fi urg- a 9. Figure' .9'~ provides a schematic for identifying an unl~nown
herbal
composition. The expression, 'profiles induced by the unknown herbal medicine
are aligned
with the expression profile database and statistical method is employed to
score the possible
identities of herbal medicines archived in the database.
,,. .,: ,., . ' .
,.: ;.
Fi ure 1 . Figure 10'~provides a schematic for extracting signature genes for
an herbal
composition or ~a complex herbal preparation.
Fi rugr a 11. ~ Figure 11''provides a'schematic for extracting signature genes
for individual
chemical constituents.in axi herbal medicine or a complex herbal preparation.
:, , , ..
Fi urg a 12. Clustered display of gene expression data from cells treated with
three types
of single-element herbal eXtr'acts (Cordyceps Sinansis Mycelium(CSM), ST024,
ST117) with
high and low concentrations, (indicated with H and L, respectively).
(A) Cluster analysis Wasperformed by the program "Cluster" (Eisen et al.,
1999) with
492 selected genes (see text).
(B) Enlarged image bf genes;up-regulated by ST117 treatment but down-regulated
by
other herbal extract treatments:. The clone ID and putative gene name are
indicated.
(C) The clustering' algorithm~separated CSM, ST024 and ST117 into 3 distinct
clusters.
The distance between each cluster as displayed by the hierarchical dendrogram
can be viewed
as the difference between~'the expression profiles of the three herbal
extracts treated cells.
Fi,~ure-13. ~ , . : '
(A) Pseudo=color'encotied display of clustering results as calculated based on
the
selected 492 genes. The bb~es girl (A) indicate the positions of the three
clusters of genes
described above. ' -~ ' . . .
(B) Enlarged''image of genes down-regulated by the CSM but up-regulated by the
others. ~ ~ ~ ' . '
(C)Genes up=regulated liy all kinds of herbal treatments.
(D) Genes down-regulated by CSM and up-regulated by the others. The IMAGE
clone
A~.
ID and putativevgene name' are.iridicated.
. .
Figure 14. Clustered display of expression data from 2 batches of multi-
element herbal
preparations of the Huaiig~'Chin .Tang (PHY906-303503 (#11) and PHY906-284003
(#12))
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-16-
treated cells mtn lugn and low concentrations (indicated with H and L,
respectively). The data
were averaged based on,'three' repeated experiments on three different dates.
Cluster analysis
was performed based on the, selected 500 genes (see text). (B) The clustering
algorithm
separated #11-L, #11H ~andp (#12-H and #12-L) into 3 distinct clusters.
Distance between
clusters or resemblance coefficient isindicated by the hierarchical clustering
dendrogram.
Figure.l5. Enlarged image~of (A) averaged and (B) individual gene expression
levels
measured by three independexit experiments. Boxl encloses genes that were down
regulated in
#11-L treated cells but up regulated in others, Box2 encloses the genes that
were up regulated
by all the herbal treatments. Box3 enclosed thel genes that showed no response
by #11-L
treatment but were dov~n regulated by the others. Box4 encloses the genes
highly down
regulated by low concentration herbal'treatments but show mild response at
high concentration
.;;.. ..
herbal treatments.~.The clorie.ID arid putative gene name are indicated beside
each gene.
Fi ure 1 . Classification of gene expression profiles in the cells treated by
herbal
.. ,. . .~, ,
medicines. Hierarchical clustering of (A) the data sets normalized with the
expression data of
the untreated control cells and (B) data sets standardized to have zero-mean
and unit-variance.
(C) The result of a non-hierarchical;clusteririg by the self organizing maps
algorithm.
Figure 17.Candidate class predictors for the classification of herbal
medicines based
on the gene expression .profiles induced by the medicines. 50 class predictors
with their
expression profiles for discriminating.#11 and #12 herbal preparations are
shown in this figure.
The IMAGE clone ~ID aridvputative~gene name are indicated beside each gene.
Fi ure 18. The gene expression profiles induced by a batch of a complex herbal
preparation of five different concentrations.' A 6x4 clustering of expression
profiles is shown
in (A), and the details of the ~gerie expression profiles for the selected
clusters are shown in (B).
Figure 19. Figure 19 illustrates how the expression profiles in Figure 18 are
categorized into three different.groups for subsequent hamming distance
calculation.
Fi ure 20. Figure~20 shows the analysis.results of gene expression profiles
induced by
five batches of a complex herbal preparation. The numbers in the table are
hamming distance.
The smaller the distance,~tlie more similar are 'the expression profiles.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-17-
Fi re 21. Shown im. (A) .is. ~ a table of integrated peak intensities of 4
chemical
constituents in HPLC analyses.of five batches of a complex herbal preparation.
Two additional
parameters, BG+B and BG/B are introduced to the table and a 6 parameter radial
plot is shown
in (B) to illustrate that:one' batch is more similar to a second batch #18
than to the other batches
by the'HPLC analysis. ~ ~ ' .
Fi re 22. A.display of the signature genes induced by a complex herbal
preparation,
the Huang Chin Tang, in Jurkat T cells.
Figure 23. Figure 23 illustrates the principle of identifying signature genes
induced by
individual chemical constituents in a mix of herbal medicines. The signature
genes are those
whose expression levels correlate with the amount of chemical constituents in
the herbal
medicine and that the correlation.coefficient is larger than 0.99 or smaller
than -0.99. (A)
shows that the R value between the, gene and Glycyrrhizin was 0.998, and (B)
shows that the
IS gene whose, expression levels:iiicrease with the decrease of Wogonin has an
R value of-0.997.
' ;
Figure 24. The signature genes induced by the chemical constituent Albiflorin
in a
complex herbal preparatiori,~ Huax~g Chin Tang, in Jurkat T cell. (A) show the
genes that were
positively correlated witli~,Albiflorin, and (B) shows the genes that were
negatively correlated
with Albiflorin.
Figure 25. Correlationvof gene expression profiles to a control group. (A) is
the gene
expression profile of a controlvgroup, and (B) is the gene expression profile
of a sample group.
(C) shows the number of genes with a differential expression ratio having
greater than 2-fold
increase with concentration of herbal treaterient.
' < ..
Fi ,gore 26. ClustersA:of.,expression profiles clustered by a non-hierarchical
analysis
program, wherein the,program is~based on a self organizing map (SOM)
principle. The X-axis
represents the herbal concentration frbm Iow to high and the Y-axis is the
gene-expression
.,, a:.:
ratio.
Figure 27. Figure 27 shows the induced and repressed genes commonly found in
two
batches of Huang Chin Tang. ,
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-18-
Fi r~ a 28. SOlVf chis'teririg'results for two batches of Huang Chin Tang. (A)
shows the
f ~ ,
SOM clustering results for.the'expression profiles of two batches of Huang
Chin Tang. (B)
shows that ten genes have similarly responded to the two batches, and (C)
shows how the
weighing factor decreases as~ cluster I and cluster j become more different.
Fi urg a 29. Calculation of S score between pairs of herbal preparations in
cluster
analysis. (A) is a tabulation,of the scores, and (B) is demonstrates how 5
batches of similar
herbal preparations are,related. ~ .~
DETAILED DESCRIPTION OF THE INVENTION
Unless defined otherwise, all technical and scientific terms used herein have
the same
meaning as commonly understood by one of ordinary skill in the art to which
this invention
.,,
belongs. Although any.inethods arid materials similar or equivalent to those
described herein
can be used in the practice or testing of the present invention, the preferred
methods and
materials are described. ,
Overview 0f the Invention
As set forth above, the present invention is directed to tools and methods
useful for
.: .>;
predicting the biological response of ari herbal composition. More
particularly, this invention
provides methods of creating Herbal BioResponse Array (HBR Array) databases as
well as
methods for using such databases to improve the design of effective herbal-
based therapeutics.
The goal of the present invention is~ the overall design, creation,
improvement and use of HBR
Arrays. for the preparation,'.testing and administration of herbal
compositions, and guide
development'of new herbal. compositidns and novel uses of existing herbal
compositions.
~p:E,, . , .
Phytomics. As used herein, depending on the context in which it is used,
"phytomics" refers ~to using' bioinformatics and statistical approaches to
address the qualitative
and quantitative aspects of the components of herbal compositions or to the
actual data bases
which are developed for addressing such aspects. ,
Herlbal BioResponse:~Array. As used herein, an HBR Array constitutes a data
set of
two or more observations' or'nieasiirements associated with an herbal
composition. The HBR
Array may include qualitative 'and quantitative data on the plants in the
composition (plant-
related data), marker infoririation obtained after exposure of a biosystem to
the herbal
composition including a dose dependent study, and a database of BioResponse
data obtained
after exposure of a biosystein to the herbal composition. The data in any
particular HBR Array
can be statistically analyzed~in'eitlier.2- or 3-dimensional space.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-19-
HBR~Arrays may be ~desigmated as batch HBR Arrays and standardized HBR Arrays.
Batch HBR Arrays are aiTays of data associated with specific batches of an
herbal
composition. Standardized HBR Arrays axe arrays of data associated with a
standardized
herbal composition. ~ ' ' ,
Major Data Set. As used herein,~the term °'major data set" refers to
the data set which
acts as the baseline set of dat'a~by Which various other sets of data are
compared or otherwise
analyzed for the same or different herbal compositions. Generally, the major
data set is created
using biotechnological techniques to ascertain some genetic or protein aspect
of the herbal
compositions. Thus, the major data set will usually, but not always, be based
on a genomic or
proteomic set of data. For example, nucleic acid microarray results could be
the major data set
which is used to compare toother, dependent or minor data sets.
Minor 'or Dependent data Set. 'As used herein, the "minor data set" or
"dependent
data set" refers to one or:rizore~data sets which are used for comparing to
the major data set.
Generally, but riot always,y the minor data set will consist of information on
an herbal
composition which are collected by more traditional methods. For example, the
minor, or
dependent, data set may consist of a collection of plant-related data obtained
by more
conventional.means: Example's' 'of plant-related' data include, but axe not
limited to, the
genus/species of the herbs) iri the herbal composition, the particular plant
parts of the herbs)
in the composition and the geographic location where the herbs) were located.
Another
example of a minor data set might consist of a set of biological responses of
a cell, tissue,
organ or organism after treatment with' one or more different amounts of the
herbal
,~
composition. Exarilples.of suich=biological data or a whole organism may
include, but are not
limited to, cell toxicity studi~es;~.'eilzyme treatirient studies, growth
rates, weight gain or loss,
changes in motor skills and changes in mental abilities.
Herb. Technically speaking an herb is 'a~small, non-woody (i.e., fleshy
stemmed),
annual or perennial seed-b~earing.plant in which all the aerial parts die back
at the end of each
growing season. Herbs are:valued for their medicinal, savory or aromatic
qualities. As the
word is more generally used: and: as tie word is used herein, an "herb" refers
to any plant or
plant part which has a food supplement, medicinal, drug, therapeutic or life-
enhancing use.
Thus, as used herein, an k~erli°is.not limited to the botancal
definition of an herb but rather to
any botanical, plant or plant part used for such purposes, including any plant
or plant part of
any plant species or subspecies of the IVIetaphyta kingdom, including herbs,
shrubs, subshrubs,
and trees. Plant parts used.-iri'herbalcompositions include, but are not
limited to, seeds, leaves,
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-20-
stems, twigs, branches, buds;' flo~?vers, bulbs, corms, tubers, rhizomes,
runners, roots, fruits,
cones, bernes, cambium and' bark.
. , , .''
I~erbal Composition.' ,A's used herein, an "herbal composition" refers to any
. . . .:
composition which includes herbs, herbal plants or herbal plant parts. Thus,
as used herein, an
herbal composition is any herbal preparation, including herbal food
supplements, herbal
medicines, herbal drugs and medical foods. Examples of herbal compositions
include, but are
not limited to, the following components: a whole plant or a plant part of a
single plant species;
whole plants or plant parts 'of multiple plant species; multiple components
derived from a
single plant species; multiple components derived from multiple plant species;
or any
combination of these various. components. For a thorough review of various
herbal
compositions, see, for example,~Kee.Chang'Huang, The Pharmacology of Chinese
Herbs, CRC
Press (1993), herein iricorporat'ed',iri,its'entirety. Representative examples
of various herbal
compositions are provided 'iiz'the'following paragraphs.
-',
Herbal compositions 'vc~hicli include the bark of the willow tree have been
used to treat
fever since the mid-eighteenth century in England. The active ingredient in
willow bark is a
bitter glycoside called saliciri;which on hydrolysis yields glucose and
salicylic alcohol.
Aspirin (acetylsalicylic'acid)'a~id aspirin-like~drugs (e.g., ibuprofen), all
ofwhich are often
called nonsteroidal antiiriflammatory-drugs (NSAIDs), are frequently used to
treat pain, fever,
and inflammation: Meadbwsweet~ is another herb that contains salicylates.
Treatment of
arthritic and arthritic-like syiiiptoxris with willow bark or meadowsweet
requires the
consumption of large quantities of herbalteas made from these plants. The
entire Populus
species (i.e., poplar tree's arid shrubs) also contains salicylate precursors
and poplar-buds have
been used in antiinflamniatoiy,~ antipyretic and analgesic medications.
U.S. Patents Have been:issued for herbal compositions used for the treatment
of various
diseases and other health-rebated problems afflicting humans and animals. For
example, U.S.
Patent No. 5,417,979 discloses a composition comprising a mixture of herbs,
including species
of Stephahia and GlycyYxhiza; as vaell, as their extracts, which is used as an
appetite stimulant
and for the treatment of pain: ~ ~-F-ierbal compositions which include
GlycyYrhiza uralensis have
been found useful for treat'img eczema;~psoriasis, pruritis and inflammatory
reactions of the
skin (U.S. Patent No. 5466;452). U.S. Patent No. 5,595,743 discloses various
herbal
compositions which include licorice extract (Glycyrrhiza) and siegesbeckia,
sophora, stemona
and tetrandra herbs used ~fo~~the treatment of various mammalian diseases,
including
~,,.: ..
inflammation and rheumatoid; arthritis. Ocular inflammation can be treated
with a
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-21-
pharmaceutical composition containing the plant alkaloid tetrandrine (U.S.
Patent No.
5,627,195). ' ' . ,
U.S. Patent No. 5,683,697 discloses a pharmaceutical composition having anti
inflammatory, anti-fever,~expectorant or anti-tussive action, wherein the
composition includes
plant parts from the, species Melia, Arzgepica, Dendrobium, Impatiens, Citrus,
LoYarethus,
° :..
Celosia, Cyhahchum and Glehhaa. , An herbal composition which includes
extracts of the roots,
rhizomes, andlor vegetation ofAlphinia, Smilax, Tinospora, Tribulus, Withauia
and Zihgiber
has been found to reduce or~alleviate the symptoms associated with rheumatoid
arthritis,
osteoarthritis, reactive arthritis and for reducing the production of
proinflammatory cytokines
(U.S. Patent No. 5,683,698)., ,
..
Herbal compositions are available in many forms, including capsules, tablets,
or coated
tablets; pellets; extracts ontinctures; powders; fresh or dried plants or
plant parts; prepared
teas; juices; creams and o.intinents; essential oils;.or, as combinations of
any of these forms.
Herbal medicines are adniiriist'ered by any one of various methods, including
orally, rectally,
paxenterally, enterally, trarisdermally, intravenously, via feeding tubes, and
topically.
Herbal compositions.emcompassed by the present invention include herbal
compositions whichwalso contain non-Herbal components. Examples of such non-
herbal
components include, but are-riot.lmiited.to, whole insects and insect parts,
worms, animal or
insect feces, natural or petroleuiYi'oils, carbonate of ammonia, salt of
tartar, liquor, water,
glycerin, steroids, pharmaceuticals, vitamins, nutrient extracts, whey, salts,
and gelatin.
For oral administration, the herbal compositions disclosed may take the form
of, for
example, tablets or capsules prepared by conventional means in admixture with
generally
acceptable excipierlts such-~as bindimg.agents (e:g., pregelatinised maize
starch,
polyvinylpyrrolidone or hydroxypropyl methylcellulose); fillers (e.g.,
lactose, microcrystalline
cellulose or~calciuin phospliale); lubricants (e.g., magnesium stearate, talc
or silica);
disintegrarits (e.g.; potato starch,or sodium starch glycolate); or wetting
agents (e.g., sodium
lauryl sulphate); glidant~s;'arti~'icial~,amd natural flavors and sweeteners;
artificial or natural
colors and dyes; and solubilizers.~ The herbal compositions may be
additionally formulated to
release the active agents in a'tiizie-release manner as is known in the art
and as discussed in
U.S. Patent Nos. 4,690,825 !amd'S,~O'55,300. The tablets may be coated by
methods well known
in the art.
Liquid preparations fox oral administration may take the form of, for example,
solutions, syrups, suspensions~or slurries (such~as the liquid nutritional
supplements described
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-22-
in Mulchandani et al., 1992 LT.S..Patent No. 5,108,767), or they may be
presented as a dry
product for reconstitution with water or other suitable vehicles before use.
Liquid preparations
of folic acid, and other vitamins and minerals may come in the form of a
liquid nutritional
supplement specifically designed for ESRD patients. Such liquid preparations
may be
prepared by conventional rrieans with pharmaceutically acceptable additives
such as
suspending agents (e.g:, sorbitol syrup, methyl cellulose or hydrogenated
edible fats);
.,
emulsifying agents (e.g., alecithin or.acacia); non-aqueous vehicles (e.g.,
almond oil, oily esters
or ethyl alcohol); .preservatives (e.g., methyl or propyl p-hydroxybenzoates
or sorbic acid); and
-~ ,
artificial or natural colors and/or sweeteners.
For topical administration, herbal components may be combined in admixture
with at
least one other ingredient constituting an acceptable, Garner, diluent or
excipient in order to
provide,a composition, such;as'a,cre~arii, gel, solid, paste, salve, powder,
lotion, liquid, aerosol
treatment, or the like, wc~hich is rriost.suitable for topical application.
Sterile distilled water
alone and simple cream, ointrizent. and gel bases may be employed as carriers
of the herbal
components. Preservatives and buffers may also be added. The formulation may
be applied to
a sterile dressing, biodegradable, absorbable patches or dressings for topical
application, or to
slow release implant systeW s with' a high initial release decaying to slow
release.
For a more complete~oveiview and discussion of herbal-based compositions see
Earl
Mindell, Earl Mindell's Herb°Bible. Simon & Schuster (1992);
Cul~eper°s Complete Herbal,
W. Foulsham & Co., Ltd. .(,originally published in the mid 1600's); and,
Rodale°s Illustrated
Enc,~pedia of Herbs; ~Rodal'e vPress (1987).
Standardized;Herbal'.Composition. ' As used herein, a "standardized herbal
composition" or, a "charactdrized~herlial composition" refers to a particular
herbal composition
wluch is chosen as the standardwlierbal composition for evaluating batch
herbal compositions
which have the same, similar'br different components as the components of the
standardized
herbal composition. Sometimes herein al'so.referred to as the "master herbal
composition."
Standardized herbal' ~cornpositions: are generally herbal compositions which
have been well
characterized,and which~d 'emonstrate the desired biological responses in a
particular
biosystem. Standardi~ed~herbal compositions are'usually standardized by
chemical tests well
known to one skilledun the'ant~and are properly stored for long term usage and
reference. The
standardized herbal composition is used to establish a standardized HBR Array
based on
observations and measurenierits for. the plants (i:e., plant-related data),
markers and
BioResponses so as to characterize the~herbal composition.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-23-
Batch Herbal Composition. ~ As used herein, a "batch herbal composition"
refers to
any test herbal composition. which is used to establish a HBR Array based on
observations and
measurements for the plants and markers so as to characterize the herbal
composition.
Sometimes herein also referred to as a ".test" or "batch" herbal composition.
Observations and
measurements of Biolzesp~orises riiay,or may not be included. The herbal
compositions used to
establish the standardized~herbal composition may also be referred to as
"batch herbal
compositions" until designated.as "standardized herbal compositions."
Batch. ,As used herein,. a "batch" refers to a particular quantity of an
herbal
composition which can.be identif ed as to some particular attribute so as to
distinguish it from
any other particular quantity'of that same herbal composition. For example,
one batch of an
herbal composition may differ from: another batch of that same herbal
composition in that one
.. .
of the batches was hai-veste'd:'at~ a~different time, or in a different
geographical location than the
other batch. Other.differences that distinguish particular batches may
include, but are not
limited to, the following:' 1:) the particular plant part used (e.g., the root
of an herb was used in
one batch while the leaves of that same herb were used in a different batch);
2) the post-harvest
treatment ofthe individual' herbs~~or herbal composition (e.g., one batch may
be processed with
distilled water while a differentbatch may be processed with Hydrogen Chloride
to simulate
the acidity of the human.stomach); arid; 3) the relative proportions of the
individual herbs in an
herbal composition (e.g., one batch may have equal parts by weight or volume
of three
different herbs while another batch has proportionally more of one herb than
the other two).
Biosysterri.' As used lierein~ a "biosystem" refers to any biological entity
fox which
biological responses may lie obs,etved, or measured. Thus, a biosystem
includes, but is not
limited to, any cell, fiissueorgan, whole organism or in vitro assay.
Biological Activity'.' As fused herein, the °'biological activity" of
an herb refers to the
specific biological effect~p~eculiai to an.herbal composition on a given
biosystem.
Plant-Related Data: '.As used herein, "Plant-related data" refers to the data
collected
on the herbal composition including; but not limited to, data about the
plants, their growing
conditions and the handling ofvtlie plants during and after harvesting. The
plant-related data
also includes the relative ~iroportioris of the~components in an herbal
compositions, wherein the
components may be different plant parfs~ different plant species, other non-
plant ingredients
(e.g., insect parts, chemical~drugs) or ariy combinations of these variables.
Plant-related data whicli~inay'be gathered for an herbal composition includes,
but is not
limited to, the following:'1) the plaint species (and, if available, the
specific plant variety,
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-24-
cultivar, clone, line, etc.) and specific plant parts used in the composition;
2) the geographic
origin of the herbs, including the longitude/latitude and elevation; 3) the
growth conditions of
the herbs, including fertilizer:types and amounts, amounts and times of
rainfall and irrigation,
average microEinsteins received per day, pesticide usage, including
herbicides, insecticides,
miticides and fungicides, acid tillage methods; 4) methods and conditions used
for processing
the herbs, including age/maturity of the herbs, soaking times, drying times,
extraction methods
... , . ,
and grinding methods; and 5) storing methods and conditions for the herbal
components and
the final herbal composition.
Additionally, the standardized herbal composition may be analyzed chemically.
Chemical characterization may be accomplished by any chemical analysis method
generally
known by one skilled in,the'art.y~.Examples of applicable chemical analyses
include, but are not
. ,
limited to, HPLC, TLC; chemical~fingerprinting, mass spectrophotometer
analyses and gas
chromatography.
Cell Banking System. .As used herein, a "cell banking system" includes a
Master Cell
Bank (MCB) and a Working'.Cell Bank (WCB) of cells. The use of a cell banking
system
minimizes cell variability,for herbal medicine testing, and is used for all
types of cells in
nucleic acid microarray studies. . .
Bioinformatics: As~used herein, "bioinformatics" refers to the use and
organization of
information of biological interest. Bioinformatics covers, among other things,
the following:
(1) data acquisition and analysis; (2) database development; (3) integration
and links; and (4)
fiuther analysis of the,resul'~ing database. Nearly all bioinformatics
resources were developed
as public domain freeware until the. early.1990s, and much is still available
free over the
Internet. Some companies.have~developed proprietary databases or analytical
software.
Genomic or Genomlcs. As used herein, the term "genomics" refers to the study
of
genes and their function. ~ Genomics 'emphasizes the integration of basic and
applied research
in comparative gene mapping;,:molecular cloning, large-scale restriction
mapping, and DNA
sequencing and computatidnal~analysis. 'Genetic information is extracted using
fundamental
techniques, such as DNA; sequencing, protein sequencing and PCR.
,.
Gene function is deterriiirled (1) by analyzing the effects of DNA mutations
in genes on
normal development.and health of the cell, tissue, organ or organism; (2) by
analyzing a
variety of signals encoded'viri~tlie DNA sequence; and (3) by studying the
proteins produced by
a gene or system of related:genes'~
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-25-
Proteomic or Proteoriiics. ~As~ used herein, the term "proteomics", also
called
.: .. , . , ; t;: , . :~
"proteome research" or "pheriome", refers to the quantitative protein
expression pattern of a
genome under defined conditions. As used generally, proteomics refers to
methods of high
throughput, automated arialysis~using protein biochemistry.
Conducting proteorrie~iesearch in addition.to genome research is necessary for
a
r. .
number of reasons. First,ahe level of gene expression does not necessarily
represent the
j' .
amount of active protein in~a cell. Also, the gene sequence does not describe
post-tranlsational
modifications which are essential for the function and activity of a protein.
In addition, the
genome itself does not describe~the dynamic cell processes which alter the
protein level either
up or down.
Proteome programs seek to characterize all the proteins in a cell, identifying
at least
part of their amino acid sequence of ~an isolated protein. In general, the
proteins are first
separatedwsing 2D gels or,HPLC arid then the peptides or proteins are
sequenced using high
throughput mass spectrometiy.yUsing~a computer, the output of the mass
spectrometry can be
analyzed so as to' link a gerie,~amd ;the particular protein for which it
codes. This overall process
is sometimes referred ~to has '"functional genomics". A number of commercial
ventures now
offer proteomic~services (e.g,,'Pharm~aceutical ProteomicsTM, The
ProteinChipTM System from
Ciphergen BiosystemPerS,ep~ive Biosystems):
For general information on proteome research, see, for example, J.S. Fruton,
1999,
Proteins, Enzymes. Genes:.The Interplay of Chemistry and Biology, Yale Univ.
Pr.; Wilkins et
al., 1997, Proteome Research: New Frontiers in Functional Genomics (Principles
and Practicel,
Springer: Verlag; A.J. .Liuk~ f999, 2-D Proteome Analysis Protocals (Methods
in Molecular
Biolo~v. ~I12, HumariarPr:.T~amp et al:, 1999, Proteome and Protein AnalXsis,
Springer
Verlag. , ~ , ..'. , . . . .
Signal Transduetion:vAs' used herein, "signal transduction", also known as
cellular
signal transductiori, refers to the pathways through which cells receive
external signals and
transmit, amplify and direct theim internally. Signaling pathways require
intercoxmnunicating
chains of proteins that. transiriit ~the'signal in a stepwise fashion. Protein
kinases often
participate in this cascade of peactions, since many signal transductions
involve receiving an
. :.. .
extracellular chemical signal,~t?vhich triggers~the phosphorylation of
cytoplasmic proteins to
amplify the signal. .. . , .. ..
Post-translatiorial~Modification. As used herein, "post-translational
modification" is
a blanket,term used to cover the alterations that:~happen to a protein after
it has been
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-26-
synthesized as a primary polypeptide. ~ Such post-translational modifications
include, but are
not limited to, glycosylatiori,.removal of the N-terminal methionine (or N-
formyl methionine),
signal peptide removal, aceiylation, foimylation, amino acid modifications,
internal cleavage
of peptide chains to release smaller proteins or peptides, phosphorylation,
and modification of
methionine.
Array or Microarray.. As 'used herein, ail "array" or "microarray" refers to a
grid
system which has each position or probe cell occupied by a defined nucleic
acid fragment. The
arrays themselves are sometimes referred to as "chips", "biochips", "DNA
chips" or "gene
chips". High-density nucleic acid illicroarrays often have thousands of probe
cells in a variety
of grid styles.
,., .
Once the array is fabricated,' DNA or protein molecules derived from a
biosystem are
added and some form.of cllerriistry:occurs between the DNA or protein
molecules and the array
to give some recognition pattern that is particular to that array and
biosystem.
Autoradiography of radiolabeled batches is a traditional detection strategy,
but other options
are available, including fluorescence, coloririletry, and electronic signal
transduction.
Markers. As used herein, the term "markers" refers to any biological-based
measurement or observatioil~for a particular herbal compositionthat is
characteristic of a
particular biosystem~ whichvis being exposed~to a particular batch of an
herbal composition.
The term "marker" encorilpasses~ both qualitative and ,qualitative
measurements and
observations of a biosystein: ~ The~riiarker database constitutes a data set
which characterizes
gene expression patterns in.response to~herbal therapies, wherein the patterns
show which
genes are turned on; off up; or down in response to specific herbal
compositions. Thus,
"markers" refers to any'biologically-based measurement or observation whose up-
and down-
or temporal regulations; or qualitative or quantitative changes of expression
levels in a
biosystem are used to characterize ~di'fferential biological responses of a
biosystem to an herbal
composition.
The particular batch' of an herbal composition to which the biosystem is
exposed may
be an unknown herbal cofripositiori, a'known herbal composition, or a
standardized herbal
composition. Examples of markers useful in accomplishing the present invention
include, but
are not limited to, molecular~iiiarkers, cytogenetic markers, biochemical
markers or
macromolecular markers.' Macroinolecular markers include, but are not limited
to, enzymes,
polypeptides, peptides; sugars; antifodies, DNA, RNA, proteins (both
translational proteins
and post-translational proteins),.'nucleic'acids, polysaccharides.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-27-
Any marker that satisfies the definition of "marker" herein is appropriate for
conducting
the present invention. The term "markers" includes related, alternative terms,
such as
.. ~~w " .., °~r~ ~ w .. .. '
biomarker or genetic marker or gene marker. There may be one or more primary
markers along with secondary markers; or a hierarchy of markers for achieving
the purposes of
increasing the discriminatimg~power of a HBR array. Thus, selected molecular
markers may be
combined with various other molecular, cytogenetic, biochemical or
macromolecular markers
to enable an even more accurate, extended HBR Array.
A molecular marker comprisesyone or more microscopic molecules from one or
more
classes of molecular compounds, .such as DNA, RNA, cDNA, nucleic acid
fragments, proteins,
protein fragments, lipids; .fatty 'acids, carbohydrates, and glycoproteins.
The establishment;; generation and use of applicable molecular markers are
well known
to one skilled in the art. Examples of particularly useful technologies for
the characterization
of molecular markers include' differential display, reverse transcriptase
polymerase chain
~; _ ~,.; . .
reactions (RT-PCR)~, large=scale sequencing of expressed sequence tags (ESTs),
serial analysis
of gene expression (SAGE); Western immunoblot or 2D, 3D study of proteins, and
microarray
.,
technology. One skilled in he 'art.:of molecular marker technology is familiar
with the methods
and uses of such technology';('see; e.g:,'Bernard R. Glick and Jack J.
Pasternak, Molecular
Biotechnolo~v, Principles and Applications of Recombinant DNA. Second Edition,
ASM Press
(1998); Mathew R. Walker arid Ralph Rapley, Route Mays in Gene Teclmolo~v,
Blackwell
Science (1997); Roe et al:, DIsTA Isolatiori~ arid Seduencing, John Wiley &
Sons (1996)James
D. Watsori et al.; Recombinant DNA.' Second Edition, Scientific American Books
(1992)).
DNA, RNA and p'roteirr isolation and sequencing methods are well known to
those
skilled in the wart. Exarilples' of such well known techniques can be found in
Molecular
Cloning;: A Laboratory Maritiaf2rid Edition, Sambrook et al., Cold Spring
Harbor N.Y.
(1989); Hanspeter.Saluz and J.vP..~Jost, A LaboratorX Guide to Genomic
Seduencin~ The
Direct Seduencing~ ofNativeIIJricloned'IDNA (,Biomethods Vol 1), Birkhauser
(1988); and B.
Roe et al., DNA Isolation and Se~uencins, Wiley (1996). Examples of
conventional molecular
biology techniques includes liti't are not limited to, in vitro ligation,
restriction endonuclease
digestion, PCR, cellular'traris'formation, hybridization, electrophoresis, DNA
sequencing, cell
culture, and the like. Specific kits and tools available commercially fox use
in the present
invention include, but are not limited to; those useful for RNA isolation, PCR
cDNA library
construction, retroviral expression libraries, vectors, gene expression
analyses, protein
antibody purification; cy'totoXicily assays, protein expression and
purification, and high-
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
_28_
throughput plasmid purification (see, e.g., CLONTECHnidues product catalog,
XIII(3), 1-32
(1998) or www.clontech.com; AtlasTM cDNA Expression Assays product catalog
(1998);
SI MA~ product catalog (1997)). ~ .
For discussions, methodologies and applications of oligonucleotide arrays,
microarrays,
DNA chips or biochips,.see, for,example, U.S. Patent Numbers.5,445,934,
5,605,662,
5,631,134, 5,736,257, 5,74'1,644, 5,744,305, 5,795,714; Schena et al.,
Parallel human genome
analysis: Microarray-based expression monitoring of 1000 genes, Proc. Natl.
Acad. Sci. USA
,; ,.,. , , .
93, 10614-10619 (1996); Del2isi etfal., Exploring the Metabolic and Genetic
Control of Gene
Expression on a Genomic Scale, Science 278, 680-686 (1997); Wodicka, et al.,
Genome-wide
Expression Monitoring in,Saccharofnyces ce~evisiae, Nature Biotechnoloav 15,
1359-1367
(1997); Pardee, Complete Genome Expression Monitoring: The Human Race, Nature
Biotechnoloav 15, 1343-134.4 (1997); Schafer et al., DNA variation and the
Future of Human
Genetics, Nature Biotechnolo~v 16,.3.3-39 (1998); DeRisi et al., Use of a cDNA
Microarray to
Analyze Gene Expre'ssiorl~ Patterns in Human Cancer, Nature Genetics 14, 457-
460 (1996);
1 S Heller et al., Discovery and Analysis of Inflammatory Disease-Related
Genes Using cDNA
Microarrays, Proc. Natl. Acad.'.~Sci. USA 94, 2150-2155 (1997); Marshall et
al., DNA Chips:
An Array of Possibilities,.Nature Biotechnolo~v 16, 27-31 (1998); Schena et
al., Microarrays:
Biotechnology's DiscoveryPlatform~for,Functional Genomics, Tibtech 16, 301-306
(1998);
Ramsay, DNA Chips: State-of the-art, Nature Biotechnoloay 16, 40-44 (1998);
Chee et al.,
Accessing Genetic Information with.High-Density DNA Arrays, cience 274, 610-
614 (1996);
and Chen et al., Profilimg.Expression.Patterns and Isolating Differentially
Expressed Genes by
cDNA Microarray System vvith'.Coloriinetry Detection, Genomics S0, 1-12
(1998); P. Andrew
Outinen et al., Character'iz_atio'n of the stress-inducing effects of
homocysteine, Biochem. J.
. y
332, 213-221 (.1998); and Gelbert et al., Will genetics really revolutionize
the drug discovery
process, Curr Opin Biotechilol~8(6), 669-674'(1997).
Other, more specific,, references applicable to the instant invention include,
but are not
limited to, .those addressing-. the .expression technologies, such as ESTs
(see, e.g., Michael R.
Fannon, Gene expressibri~iri normal and disease states - identification of
therapeutic targets,
TIBTECH 14, 294-298 (1996)); the generation of protein profiles (see, e.g.,
Robinson et al., A
Tyrosine Kinase Profile o~1'rostate' Carcinoma, Proc. Natl. Acad. Sci. USA 93,
5958-5962
(1996)); chemical and spectroscopic methods for identifying components of
herbal
compositions (Kojima et ~al.°, ~Saponiris from Gliricidia sepium,
PhytochemistrX 48(5), 885-888
(1998)); the determination of furictiorial antigens (see, e.g., Aris Persidis,
Functional
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-29-
antigenics, Nature Biotechnology 16, 305-307 (1998)); HPLCs (see, e.g., Milton
T. W. Hearn
(Editor), HPLC of ProteinsYPe~ties~ and Pol~cleotides: Contemporary Topics and
Applications (Analytical Technidues in Clinical Chemistry and Laboratory
Manual), VCH
Pub. (1991); electrophoresis (see e:g., Westermeier et al., Electrophoresis in
Practice: A Guide
to Methods and Applications of DNA and Protein Separations, John Wiley & Sons
(1997));
~.. ..7.. , . ,
and cross-reactivity marker assays (see, e.g., Irving Millinan et al.,
Woodchuck Hepatitis
Virus: Experimental Infection and Natural Occurrence, Hepatolog,~ 4(5):817-823
(1984)). The
use of structural genomics for solving the structures of all the proteins
encoded for in
completed genorries, wherein the.methodology includes high-throughput direct
structure
determinations and computational methods, is discussed by Terry Gaasterland,
Structural
genomics: Bioinformatics in the driver's seat, Nature Biotechnoloav 16, 625-
627. For
bioinformatics methodologies, see, for example, Andreas Baxevanis (Editor),
Bioinformatics:
A Practical Guide to the AnalXsis of Genes and Proteins, John Wiley & Sons
(1998) and Luke
..
Alphey, DNA Seduenciny.,, From Experimental Methods to Bioinformatics
(Introduction to
Biotechnidues Series,',i;~Sprimger Verlag (1997).
Cytogenetic parameters include, but are not limited to, karyotype analyses
(e.g.,
relative chromosome lengths,; centromere positions, presence or absence of
secondary
constrictions), ideograms; (i.'e:; a' diagrammatic representation of the
karyotype of an organism),
the behavior of chromosomes during mitosis and meiosis, chromosome staining
and banding
patterns, DNA-protein interactions (also known as nuclease protection assays),
neutron
scattering studies, rolling circles (A.M: Diegelrilan and E.T. Fool, Nucleic
Acids Res
26(13):3235-324.'1 (1998); Backert et al:, Mol. Cell. Biol. 16(11):6285-6294
(1996); Skaliter et
al., J. Viol. 70(2):11'32-1136 (.1996); A. Fire and S.Q. Xu, Proc. Natl. Acad.
Sci. LTSA
92(10):4641-4645 (1995)),,and~auforadiogr~phy of whole nuclei following
incubation with
radiolabelled riboriucleatides:' . '
Biochemical pararrieters include, but are not limited to, specific pathway
analyses, such
as signal transduction, protein: 'synthesis and transport, RNA transcription,
cholesterol synthesis
and degradation, glucogenesis and glycolysis.
Fingerprinting. As 'used herein, the term °'fingerprinting" as used
herein refers to the
means of making a characteristio.profile of a substance, particularly an herb,
in order to
identify it. The term "fingerprint" as used herein refers to the display of
the result of the
particular means employed. for the fingerprinting.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-30-
Examples of the various types of fingerprinting means include, but are not
limited to,
DNA fingerprinting, protein 'fiiigeiprinting, chemical fingerprinting and
footprinting.
DNA fingerprinting; or profiling, refers to a way of making a unique pattern
from the
DNA of particular biological.~source (e.g., a particular plant, plant species,
genus of plant, plant
part or plant tissue): 'The DNA~fingeiprint, or profile, can be used to
distinguish that particular
biological source from a'differerlt biological source: The patterns obtained
by analyzing a
batch using microarrays;'olig~onucletide arrays, DNA chips or biochips axe
also referred to as
.'fingerprints". ' .. .
Protein fingerprinting refers, to generating a pattern of proteins in a cell,
tissue, organ or
organism, such as a plant,. whicli provides a completely characteristic
"fingerprint" of that cell,
tissue, organ or organism at that' time.
Chemical fingerprinting'refers to the' analysis of the low molecular weight
chemicals in
~ ~ ~v:F'.. . '
a cell and the resulting pa,.ttern~ used,to identify a cell, tissue, organ or
organism, such as a plant.
The analysis is usually dene using Gas Chromatography (GC), HPLC or mass
spectrometry.
Footprinting refers to' a method of finding how two molecules stick together.
In the
case of DNA, a protein is bound'to a labeled piece of DNA, and then the DNA is
broken down,
by enzymes or by chemical~attack. This process produces a "ladder'" of
fragments of all sizes.
Where the DNA is protected by the Bound protein it is degraded less, and so
the "ladder"
appears fainter. Footprinting;is a,cornmon technique for homing in on where
the proteins that
regulate gene activity actually'vbind to the DNA.
The means, 'or methods,;used, ~o accomplish each type of fingerprinting are
described in
detail elsewhere herein. . ; ' ~ '
SioResponses:~ As:iised herein, a'"BioResponse" refers to any observation or
~,
measurement of a biologicah'res~ onse of la biosystem following exposure to an
herbal
composition. Sometimes hereimalso referred to as a "biological effect." A
BioResponse is a
qualitative or quantitative data'pomt ;for the biological activity of a
particular herbal
composition: . Biol~esponse'°data includes~both dosage and temporal
information, wherein such
information is well known'to~ one skilled in the art of measuring responses of
biosystems to
various treatments. Thus,~BioResp~onse data includes information on the
specific biological
response of a specific biosysfein to 'a specific dosage of herbal composition
administered in a
particular manner for a~speci~c'period of.time.
BioResponses inclucie,.b-ut are not limited to, physiological responses,
morphological
responses, cognitive responses; motivational responses, autonomic responses
and post-
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-31-
translational modification's, ;such as signal transduction measurements. Many
herbal
compositions demonstrate' more than one BioResponse (see, e.g., Kee Chang
Huang, The
Pharmacology of Chinese Herbs, CRC'Press (1993)). Some particular BioResponses
may be
included in more than one of the delineated groups or have aspects or
components of the
response that encompass iriore than one group. BioResponses applicable to the
instant
invention are well known to one skilled in the art. The following references
are representative
of the state of art iwthe field: .Kee .Chang~Huang, The Pharmacology of
Chinese Herbs, CRC
Press (1993); Earl Miridell, Earl Mindell's Herb Bible, Simon & Schuster
(1992); Goodman &
Gilinan's The Pharmacological~B'asis of Therapeutics~Ninth Edition, Joel G.
Hardman, et. al.
(eds.), McGraw Hill, Health Professions Division (1996); P. J. Bentley,
Elements of
nharmacology.TA primer on:diu ag ction, Cambridge University Press (1981); P.
T. Marshall
and G. IVI. Hughes, Ph, s~~ iology of mammals and other vertebrates, Second
Edition, Cambridge
University 'Press (1980); Report of~tlie Committee on Infectious Diseases,
American Academy
of Pediatrics (1991); nut S~chmidt-Nielsen, Animal Ph, siolow: Adaptation and
Envirornnent,
5th Edition, Cambridge Univefsity' Press (1997); Elain N. Marieb, Human
Anatomy &
Physiology, Addison-Wessley Pub: Co. (1997); William F. Ganong, Review of
Medical
PhXsiologv~l8th Edl, Appletori ~ Lange (1997); Arthur C. Guyton and John E.
Hall,
Textbook of Medical Phs sy iologX, W'. B. Saunders Co. (1995).
A "physiological response" refers to any characteristic related to the
physiology, or
functioning, of a biosystem,v.Physiological responses on a cellular, tissue or
organ level
include, but are not limited ~to, temperature, blood flow, rate, pulse rate,
oxygen concentration,
b'ioelectric potential; pH value, cholesterol levels, infection state (e.g.,
viral, bacterial) and ion
flux. Physiological responses'ori a~wholeorganism basis include
gastrointestinal functioning
(e.g., ulcers, upset stomach; 'indigestion, heartburn), reproductive tract
functioning (e.g.,
physiologically-based iiilpotence;; uterine cramping, menstrual cramps),
excretory functions
(e.g., urinary tract problems, kidney' ailments, diarrhea, constipation),
blood circulation (e.g.,
hypertension, heart disorders); oXygenconsumption, skeletal health (e.g.,
osteoporosis),
condition of the cartilage and connective~tissues (e.g., joint pain and
inflammation),
locomotion, eyesight (e.g.; riiyopia, bluidness), muscle tone (e.g. wasting
syndrome, muscle
strains), presence or absence~of pain, epidermal and dermal health (e.g., skin
irritation, itching,
skin wounds), functioning of the endocrine system, caxdiac functioning,
nervous coordination,
head-related health (e.g.,~he~daches, dizziness), age (e.g., life span,
longevity) and respiration
(e.g., congestion, respiratory ailrilents).
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-32-
A "morphological response°' refers to any characteristic related to the
morphology, or
the form and structure, of a~liiosystem following exposure to an herbal
composition.
Morphological responses, regardless of the type of biosystem, include, but are
not limited to,
size, weight, height, width; color, 'degree of inflammation, general
appearance (e.g.,
opaqueness, transparericy;.paleriess), degree of wetness or dryness, presence
or absence of
cancerous growths, and the presence or lack of parasites or pests (e.g., mice,
lice, fleas).
Morphological responses ;on avwhole organism basis~include, but are not
limited to, the amount
and location of hair growth (e.g:;v hirsutism, baldness), presence or absence
of wrinkles, type
and degree of nail and skin growth, .degree of blot clotting, presence or
absence of sores or
wounds, and presence or absence of hemorrhoids.
A "cognitive response" refers to any characteristic related to the
cognitive,'or mental
state, of a biosystem folloWimg exposure to an Herbal composition. Cognitive
responses
include, but are not limited ~toperceivirig~ recognizing, conceiving, judging,
memory,
reasoning and imagining: ~ ~ ,~ . - ~ v
A '°motivatiorial response°''refers to any"characteristic
related to the motivation, or
induces action, of'a biosyste~rii following exposure to an herbal composition.
Motivational
responses include, but are not limited, to, emotion (e.g., cheerfulness),
desire, learned drive,
particular physiological needs, (e.g:; appetite, sexual drive) or similar
impulses that act as
incitements to action (e.g., stamina; sex drive).
,.,
An "autonomic response"'refers to any characteristic related to autonomic
responses of
a biosystem following exposure to ,an~lierbal composition. Autonomic responses
are related to
the autonomic nervous systerxi of the biosystem. Examples of autonomic
responses include,
but art not limited to,Linvoluntary~fiinctio~ung~(e.g., nervousness, panic
attacks), or
physiological needs (e.g., respir~.'tion, cardiac~rhythm, hormone release,
immune responses,
insomnia, narcolepsy). ; ~ '
BioResponses~of~cells; tissues; organs and whole organisms treated with
various herbal
compositions or herbal, cornporlents are well known in the herbal arts. For
example, the herbal
compositions Sairei-to.(T-J-1:1,4), alismatis~rhizoma (Japanese name'Takusha')
and hoelen
~,,
(Japanese name 'Bukuryoit') yvere, each found toinhibit the synthesis and
expression of
endothelin-1 in rats (Hattori e't aZ., Sairei-to may inhibit the synthesis of
endothelin-1 in
nephritic glomeruli,'ISTippon Jinzo~Gakkai Shi 39(2), 121-128 (1997)).
Interleukin (IL,)-1 alpha
production was significantly'promoted~by treatment of cultured human epidermal
keratinocytes
with the Herbal medicine Sho=saiko-to (Matsmnoto et al., Enhancement of
interleukin-1 alpha
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-33-
mediated autocrine growthrof ;cultured human keratinocytes by sho-saiko-to,
Jpn J. Pharmacol
73(4), 333-336 (1997). Add~iig'Sho-saiko-to to a culture of peripheral blood
mononuclear
cells obtained from healthy volunteers resulted in a dose-dependent increase
in the production
of granulocyte colony=stiriiula'ting~factor (G-CSF) (Yamashiki et al., Herbal
medicine "sho-
saiko-to" induces in vitro graiiulocyte colony-stimulating factor production
on peripheral blood
mononuclear cells, J Clin'I~;ab Immunol 37(2), 83-90 (1992)). These
researchers concluded
that the administration of Sho-saiko-to may be useful for the treatment of
chronic liver disease,
malignant diseases and acute,infectious diseases where G-CSF is efficacious.
Plasminogen
activator inhibitor type l ,(PAI-1)-specific mRNA expression decreased and
tissue-type
plasminogen activator~(t-P~)-specific mRNA increased after treatment of human
umbilical
vein endothelial cells (HUVECs) with the saponin astragaloside IV (AS-IV)
purified from the
Chinese herb Astragalus meiiibrarlaceus' (Zhang et al., Regulation of the
fibrinolytic potential
of cultured human umbilicalv~invendothelial cells: astragaloside IV down
regulates
plasminogen activator inhibitor=1 arid~'up regulates tissue-type plasminogen
activator
expressibn, J Vasc Res 34(4)',273-280'(1997)). One component out of four
components
isolated from the roots o'f Panax ginseng was found to be a potent inducer of
IL-8 production
by human monocytes and TI3P-1 cells,,'and this induction was accompanied by
increased IL-8
mRNA expression (Sonoda et izl.; Stimulation of interleukin-8 production by
acidic
polysaccharides from the root of panax ginseng, Immunopharmacolosv 38(3), 287-
294
(1998)). By flow cytometric analysis, the expression of Fc gamma 11/111
receptors and
complement receptor 3 ~ (CR3) on macrophages were found to be increased by
treatment with
the Kampo=herbal medicine~.Toki-shakuyalcusan (TSS) (Cyong, New BRM from kampo-
herbal
medicine, Ni~pon Yakuri~aku'Zasshi 110 Suppl 1, 87P-92P (1997)). Using
computer image
analysis, Chen et al. (Image~arialysis for intercellular adhesion molecule-1
expression in
l~~RI/lpr mice: effects of Chinese herb medicine, Chung Hua I Hsueh Tsa Chih
75(4), 204-206
(1995)) found that the distribution intensity of intercellular adhesion
molecule-1 (ICAM-1),
immunoglobulins and C3.were significantly decreased in MRL/lpr mice after
treatment with
the Chinese herb stragaliri. western blot analysis showed that tetradrine,
isolated from a
natural Chinese herbal medicine, inhibited signal-induced NF-kappa B
activation in rat
~ alveolar macrophages CChem et al:, Tetrandrine inhibits signal-induced NF-
kappa B activation
in rat alveolar macrophages; Bioclieril BiophXs°Res Common 231(1), 99-
102 (1997)).
Algorltlini. ~ As used Herein, an "algorithm" refers to a step-by-step problem-
solving
procedure, especially an established, recursive computational procedure with a
finite number
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-34-
of steps. Appropriate algorithms for two- and three-dimensional analyses of
the plant-related,
marker arid BioResponse dafa~sets~ are well known to one skilled in the
computational arts.
Such algorithms are useful in constructing the Herbal BioResponse Arrays of
the present
invention. For general information on algorithiris, see, for example, Jerrod
H. Zar,
Biostatistical Analwsis, second edition, Prentice Hall (1984); Robert A.
Schowengerdt,
Technidues for image processing and classification in remote sensing, Academic
Press (1983);
Steven Gold et al., New Algorithms for 2D and 3D Point Matching: Pose
Estimation and
Correspondence, Pattern Recog-nition, 31(8):1019-1031 (1998); Berc Rustem,
Algorithms for
Nonlinear Prog_rammin~ and lVlultinle-Objiective Decisions, Wiley-hlterscience
Series in
Systems and Optixilization;,~Johm. Whey & Sons (1998); Jeffrey H. Kingston,
Algorithms and
Data Structures' Design Correctness Analysis International Computer Science
series,
Addison-Wesley Pub. Co. ,(1997); .Steven S~. Skiena, The Algorithm Desi,~m
Manual, Springer
Verlag (1997); and Marcel F~''NeutsAl~;orithm Probability A Collection of
Problems
-(Stochastic Modeling), Cliapmam'& Hall (1995). For information more specific
to the
application of algorithms togenetic-based data, see, for example, Dan
Gusfield, Alg-oritluns on
Strings-. Trees, and Seduences: Computer Science and Computational Biology,
Cambridge
University Press (1997); lVlelanie Mitchell, An Introduction to Genetic
Algorithms (Complex
Adaptive Svstemsl, MIT Press' (1996); David E. Goldberg, Genetic Algorithms in
Search
Optimization and Machine Learning, Addison-Wessley Pub. Co. (1989); Zbigniew
Michalewicz, Genetic Alg_orithms~+ Data Structures = Evolution Programs,
Springer Verlag
(1996); Andre G. Uitterlirideri'arid JamVijg, Two-Dimensional DNA Typing: A
Parallel
Approach to Genome Anal°~, Ellis Horwood Series in Molecular Biology,
Ellis Horwood
Ltd. (1994); 'and Pierre.Baldi. amd Soien Brunak, Bioinformatics: The Machine
Learning
Approach Adaptive Com~utationvand Machine Learnin~l, MIT Press (1998).
Combinatorial Cheiiiistry: As used herein, "combinatorial chemistry" refers to
the
numerous technologies used to' create hundreds or thousands of chemical
compounds, wherein
each of the chemical compounds' differ for one or more features, such as their
shape, charge,
and/or hydrophobic characteristics: Combinatorial chemistry can be utilized to
generate
compounds which are cheniic'al vxiiations of herbs or herbal components. Such
compounds
can be evaluated using 'the methods .of the present invention.
Basic combinatorial. chemistry concepts are well known to one of ordinary
skill in the
chemical arts and can also' be found in Nicholas K. Terrett, Combinatorial
Chemistr;~(Oxford
Chemistry. Masters), Oxford Univ: Press (1998); Anthony W. Czarnik and Sheila
Hobbs
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
~35-
Dewitt (Editors), A Practicah~Guide to Combinatorial ChemistrX, Amer. Chemical
Society
(1997); Stephen R. Wilsori,,(Editor) and Anthony W. Czarnik (Contributor),
Combinatorial
Chemistry: Synthesis and Application, John Wiley & Sons (1997); Eric M. Gordon
and James
F. Kerwin (Editors), ombinatorial Chemistry and Molecular Diversi in
Drug_Discovery,
Wiley-Liss (1998); Sluriuel~Cabilly.(Editor),~Combinatorial Peptide Library
Protocols
(Methods in Molecular Biolo~vl, Human Press (1997); John P. Devlin, High
Throushput
Screeuns, Marcel Dekker~(1998);'Larry Gold and Joseph Alper, Keeping pace with
genomics
through combinatorial chemistry;~Nature Biotechnolo~v 15, 297 (1997); Aris
Persidis,
Combinatorial chemistry,.Nature Biotechnolosv' 16, 691-693 (1998).
' v . . ~ EXAMPLES
Example 1. ~Estalilishing 'a Standardized HBR Array for Selected Herbal
,..;
,,.,:
Compositions. . , , ~ ';, ', :.
' _ 6 v ~ ,
The basic scheme for establishing a Standardized HBR Array is provided in
Figure 1.
Definitions of each com~onerit~ of the schematic are provided above.
Following selection~o~,aWherbal composition of interest, data is collected for
various
traits associated with the~herb'al'composition, including, but not limited to
plant-related
characteristics.and inarker'.and.Bio.Response information.
Plant-related data.irichtdes, but is not limited to, the plant species,
specific plant parts,
geographic origin of the plarits'in the herbal composition, the growth
conditions of the plants,
the processing methods used to~prepare the herbal components, storage methods
and
conditions, and various chemical analyses of the herbal composition. Marker
information
includes qualitative and quantitative 'data for markers collected after
exposure of a biosystem to
the herbal compost. Applicable iriarl~ers include, but are not limited to,
molecular markers,
cytogenetic markers bi'ocheinical',markers and macromolecular markers.
BioResponse
information includes qualitative' ~ancT quantitative data for biological
responses collected after
exposure of a biosystem.to,the lierb~al composition:
Each type of data (e:'g.'chemical, marker, BioResponse) cari be obtained using
one or
more assays'on the same;'similar, substantially similar, or different batches
of the herbal
composition of interest. Suclivdi'fferent assays can be conducted at the same
or different times.
In addition, data can be collected for the same or~different markers at the
same or different
times. Similarly; BioResp~onse' data can be collected for the same or
different biological
responses at the same or~different times. ~ Thus, collection of the data for
the HBR Array is
either collected at one time'or bollected on an on-going basis. Where a
biosystem is exposed
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-36-
to an herbal composition so 'as to collect data; information is recorded on
the administered
dosages of the herbal composition as well as treatment times. BioResponse data
may also
consist of post-translational modifications, such as measurements of signal
transduction.
.,. :~ .E;.
After.collection of tW0 or more types of data (e.g., data for two or more
markers and a
BioResponse; data for plant-related traits and' data for a BioResponse), the
data is analyzed
,; , :; ,, ,. , , , . ~
using algorithms so as to~'create~2- and/or 3-dimensional Herbal BioResponse
Arrays.
Various statistical parameters may be calculated for the HBR Array and may
become
" ~ h~ . ~~'~ ,
part of the HBR Array data set: .These statistical parameters may include, but
are not limited
V. 1
to, means, standard deviations, correlation or match (or mismatch) matrices,
ratios, regression
coefficients, and transformed values (e.g., arcsin percentage transformations
of the raw data).
Thus, the HBR Array.may consist of the raw data as well as certain
calculations, distributions,
graphical presentations and' other data rrianipulations associated with the
raw data. Particular
examples'of such~informatiowinclude, but are not limited to, digital images,
scatter graphs,
cluster analyses and large Iscaley gene. expression profiles for marker data.
The total accumulated data and resultant analyses constitute a standardized
HBR Array
for the particular herbal composition used to establish the HBR Array data
set. Due to the
:1,~ :..,
iterative nature of the process'used to establish and maintain an HBR Array
for an herbal
' . . ,.: fw~
composition, such arrays can be viewed as either static at any one point in
time or dynamic
over time. ~ '
The resulting analyses can identify subsets of the standardized HBR Arrays
which axe
correlated (positively or negatively) or associated (i.e., showing a general
trend) with one or
more specific biological activities of any particular herbal composition.
Example 2. Establisliingva Batch HBR Array for. Batch Herbal Compositions.
The basic scheme for establishing a HBR Array for a batch of an herbal
composition is
provided in Figure 2. ~ Definitions ,of~each component of the schematic are
provided above.
The procedure for establishing such an array is the same as that set forth
immediately above
for the standardized'HBR~'Ai-~ay:' ~~ ,
Generally, the arribunt~of data collected for a batch HBR Array will be less
than that
collected to establish''a ~staridardized HBR Array. However, data collected
for a batch herbal
composition may be added to an,established HBR Array or used to establish a
new
standardized HBR Array.: v ~ ~, .. ' ,
Generally, the only data collected for a batch herbal composition is that data
which has
been found to be highlycorrelated or associated with the desired biological
activities of the
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-37-
herbal composition being:tested. '.For example, if it has been determined that
a particular subset
of plant-related and marker data, is highly correlated to a desired biological
activity of a
particular herbal composition,(based on the standardized HBR Array data and
analyses
discussed above), it is only necessary to test the batch herbal composition
for that subset of
traits in order to determine whether or not the batch has the desired
biological activity. By
comparing the data obtained-for that subset of traits obtained from the batch
(i.e., the batch
HBR Array) with the standardised HBR Array.for that particular herbal
composition, one
skilled in the art can deterinine whether or not that particular batch has the
desired biological
aCtlvlty. . " ,,
. Example 3. Establishing and Using a Major Data Set.
The basic scheme for,establishing and using a major data set for an herbal
composition
is provided in Figure 3.. 'Defiri~tioiis, of each component of the schematic
axe provided above.
The first step, is .the establishment of a major data set for a selected
herbal composition
or batch herbal coiTipositibn' This, is accomplished by exposing a biosystem
to the herbal
composition and collecting the resultant marker information which will
constitute the major
data set. In most, but not instances, the major data set will consist of
genomics and/or
proteomics data in'the form~o~an axr'ay, such as an array obtained with a DNA
biochip.
Next, the majof data set is' analyzed to see if differential
expression/results have been
obtained for the tested herbal composition. Differential expression/results
are necessary in
order to generate meariingful~algorithms in the next step. Examples of such
differential
expression/results include, but axe not limited to, indications that certain
genes are up- or
down-regulated in respoMSe to exposure so the herbal composition or that the
levels of certain
proteins have been iricreasedvor decreased iii response to the exposure.
If no meariingful~ot''usefixl differential expression/results are obtained,
then it is
necessary to repeat the exposufe.and marker collection step. If it is believed
that experimental
error lead to the lack of a adequate~~esult the first time then the
exposure/data collection step
can be repeated with all of the variables the same~as the first time (e.g.,
same biosystem, same
marker set, same experirrierival protocol; etc.): I~owever, it may be
necessary to vary the
biosystem sampling (e.g.'; type ~of cell utilized, stage of cell growth), use
a different marker set
and/or change the experirriental protocol in order to get differential
expression/result..
Example 4. Using HBI~ Array Information.
The,HBR Array iriforination discussed herein can be used for many different
purposes
including, but not limited to, 'the following: '1) evaluating the components
of an herbal
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-3 8-
composition; 2) predicting the BioResponse of an herbal composition; 3)
determining which
marker information is most highly correlated with a particular BioResponse of
an herbal
composition; 3) determinrig what data set of information (i.e., plant-related
data, marker data,
and BioResponse data) is iiiost..correlated with a particular BioResporise of
an herbal compost;
4) determining which type ofbiosystem is best for evaluating the biological
activity of an
herbal composition; 5) adjusting or changing the components of a herbal
composition so that
the I~BR Array of that herbal composition corresponds to a standardized HBR
Array for the
same or substantially the same herbal composition; 6) adjusting or changing
the components of
an herbal composition so that the herbal composition will have the desired
biological activity;
7) measuring the relatedness of different herbal compositions; 8) creating and
updating
standardized HBR Arrays; 9)Aidentifying specific components (e.g., plant
parts, proteins,
molecules) which retain the 'desired biological activity of an herbal
composition; 10)
determining which compomerits~ of aWherbal composition can be eliminated while
maintaining
or improving the desired ~fiological activity of the herbal composition; 11)
identifying one or
more previously unknown biological activities for an herbal composition; 12)
aiding in the
design of therapeutics which include Herbal and non-herbal components, such as
chemically-
synthesized drugs or pharmaceuticals and 13) utilizing the HBR Array
information to
complement combinatorial oheinistiy rizethods of designing therapeutics. Each
of these
embodiments of the present invention can be accomplished by one skilled in the
applicable art
4
using the methods and tools provided herein.
Exaaazple 5. Qu~ali'ty G~ntrol.
The HBR Array ethnology o~f the present invention is used to correlate or to
determine
a substantial equivalence'of a~'specific batch of an herbal composition
(single herb or multiple
,,:.
herbs of a~ formula) to ~a st'aaidardized, or master, batch of a same or
substantial similar herbal
composition. The HBR Arrays utilized in this process include the'acceptable
range of
quantitative variation for ~each~°o~f the biological effects (i.e.,
BioResponse), and possibly a
global score composed of weighted values assigned to each of the biological
effects, which
may consist of markers from multiple biochemical pathways of a biosystem.
"Data mining" refers.to a'process used to determine or select which subset of
biological
effects is the rninimuni number of biological effects required in any specific
I~BR Array. The
information for data mining results from exposing a biosystem (e.g., a cell
line) in a dose
dependent manner to a standaxdized~herbal composition to establish a
standardized HBR
Array. This standardized I~'BR Art~ay can then be compared to various HBR
Arrays
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-39-
established for test herbal, compositions. These test herbal compositions
include, but are not
limited to, different batches prepared at different dates; different batches
prepared from raw
herbs collected at different~times; and different batches prepared from raw
herbs collected at
different locations: . . ' ~ '
Example 6. Improving an Herbal Composition or Identifying New Uses for an
Herbal Composition. , '
HBR Arrays are ,generated by exposingFbiosystems to either extracts from
individual
herbs of a formula, or to extracts from the whole formula, and examining the
biological effects
of the extracts. The observed biological effects can be from multiple
biochemical pathways of
a biosystem _and/or from multiple.tissues of an animal, wherein various
markers are evaluated
for their corresponding qualitative ancUor quantitative changes. The resulting
HBR Arrays can
be compared to novel~HBR Arrays~or.to similar HBR Arrays from different herbal
,.
compositions or herbal ~coriipositions.prepared by different processes. This
procedure is useful
for selecting a given set of biological effects and the minimum number of
markers required to
predict that a given batch herbal composition has the given set of biological
effects.
In order to construct:HBR'Axrays, one skilled in the art utilizes various data
mining
tools including, but are not limited o; statistical analyses, artificial
intelligence, and database
research on neural work: The statistical methods of choice include, but are
not limited to, basic
exploratory data analysis ~ (EDA), graphic EDA (such as bushing) and
multivariate exploratory
techniques (e.g., cluster analysis, discriminating factor analyses, stepwise
linear on non-linear
regression,classification,tree) ~see~~'e.g., STATISTICATM, software packages
from StatSoft,
Tulsa, OK 74104; Tel: 918-749-1119; Fax: 918-749-2217; www.statsoft.com).
Data mining tools are usedrto explore large amounts of HBR Array data in
search of
constructing an HBR Array and~corisistent pattern within, between or among
various HBR
Arrays. The procedure consists of exploration, construction of an HBR array,
and validation.
This procedure is typically.xepeated iteratively until a robust HBR Array, or
standardized HBR
Array, is identified. , ° ~ ~ '
Example '7. Establishing a Standardized HBR Array for Ginseng Recipes.
For the purposes of this exairiple,~standard ginseng is chosen to be Panax
Ginsefzg C.A.
Meyer 6115 grown neither in Manchuria or in Korea. The climate for growth is
between -10 to
+I O°C with an annual rainfall ~of 50-I00 cni (see Huang in The
Pharmacology of Chinese
Herbs, (1993) pp 2I-45, CRC'.Press, $oca Raton, FL, fully incorporated by
reference). Ginseng
batches will first be characterized ~by geographic origin, species, plant part
(e.g., rhizome, root,
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-40-
leaf skin, seed, bud and flower); growth conditions, processing methods and
storage conditions
both before and after processing. Verification of chemical content for these
batches will be
performed by qualitative HPLC analysis for determination of ginsenoside
saponins (e.g., Ro,
Ral, Ra2, Rbl, Rb2, Rb3, Rc, Rgl, Rg2, Rd, Re, Rf, Rhl, Rh2, NG-R2 and Z-Rl),
including
S TLC qualitative analysis for lipophilic constituents (see, Elkin et al.,
Chumg Kuo Yao Li
Hsueh Pao (1993) 14: 97-100 and Yoshikawa et al., Yaku~:aku Zasshi (1993) 113:
460-467).
The saponin content of different herbs should be between 2.1 and 20.6% (by
weight)
depending on the species (see Table 1). These data will then be stored,
preferably in the
memory of a computer processor, for further manipulation.
Table 1. Saponin Content of Different Ginseng Herbs.*
Species ~ I Total saponins (% by weight)
Pahax gihseng C.A. Meyer .. , , , 2.1-4.4%
Panax quiquefolius ' ~ 4.9%
Pahax fZOtogihsehg and Panax japoi2ica ~ 13.6-20.6%
Pahaxjapohica ear. iriajor °-' ' ' I 9.34%
*from Huang~in The Pharmacology~of Chinese Herbs, (1993) page 29, CRC Press,
Boca Raton, FL.
Expressionbiomarke~s for standard ginseng (i.e., G11S) include the following:
IL-8,
IL-2, GM-CSF, NficB, ICAM-1,, interferon gamma, choline acetyl transferase,
trk A, nerve
1S growth factor (Kim et al:,tPlantaMed.(19~98) 64: 110-115; Sonoda et al.,
Immunopharmacolo~v (1998)'38: 287-294;.Baum et al., Eur J Appl Phvsiol (1997)
76: 16S-
169; Iwangawa et al., Free.Radic.Biol.Med (1998) 24: 1256-1268; Rhind et al.,
Eur J Anal
Ph siol (1996) 74: 348-360). Alternatively, for a broader batch size, the
400,000
oligonucleotide group/1.6 cma chip of Affymetrix can be used (U.S. Pat.
No.S,SS6,7S2). The
expression biomarkers fo"r standard ginseng will be prepared by nucleic acid
microarray
technology using eitherphotolithography, mechanical microspotting or ink jet
application (see
Schena et al., TIBTECIi (1998),16: 301-306). Selected sets of cells will be
contacted with
standard ginseng for varyiiig periods of time, under varying conditions to
generate multiple
microarray sets. The microarray set's~will then lie analyzed by hybridization-
based expression
2S monitoring of biochemical e~~tracts via deduction of steady state mRNA
levels from
fluorescence intensity at each.position on the microarrays (Schena et al.,
cience (1995) 270:
467-470; Sohena et al., Proc IVatl Acad Sci USA' (1996) 93: 10614-10619;
Lockhart et al. Nat
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-41-
Biotechnol (1996) 14: 167.5-1680;.DeRisi et al., Nat Genet (1996) 14: 457-460;
Heller et al.,
Proc Natl Acad Sci USA (1997),94: 2150-2155). The array data sets are then
input into
algorithms to generate statistical expression biomarker values for standard
ginseng.
Biochemical biomarkers for standard ginseng include quantitative analysis for
increases in
cycloheximide sensitive. [3H]-Ieucine incorporation proportional to protein
synthesis and [3H]-
thymidine incorporation;reflective of mitosis. (see Yamamoto et al.,
Arzneimittelforschun~
(1977) 27: 1169-1173). For biochemical biomarkers,.bone marrow cells will be
contacted with
standard ginseng for varying time periods under varying conditions in the
presence of [3H]-
thymidine (for DNA syntliesis) or in the presence and absence of cycloheximide
and [3H]-
leucine (for protein synthesis) to :perform multiple quantitative analysis of
biochemical
biomarkers (i.e., BBM sets).'.The BBM sets are then input into algorithms to
generate statistical
biochemical biomarker values. for. standard ginseng. Statistical data will
then be stored,
preferably in the memory of a computer processor, for further manipulation.
Biological resporise,'ofa biosystem (i.e., BioResponses) will be determined
using cells
and whole animals. For cells,.' ginseng batches will be exposed to specific
cell types, including,
but not limited to~ fibroblasts, macrophages, monocytes, PMNL, LAK cells, B16-
F10
melanoma cells, THP-1 cells and hippocampal neurons at a concentration of 0.5
mg/m1 to 100
mg/ml. For animal treatments,' 0.5-100 mg/kg of ginseng herbal extract will be
administered
orally, by intraperitorieal irijectiomor subcutaneous injection.
To determine a biological response of a biosystem to standardized ginseng,
human
ovarian cancer cells willrbe inoculated into nude mice, which results in the
formation of
palpable tumors. After tumor' formation the mice will be treated by co-
administration of cis-
diamminecichloroplatinum arid ~staiidard. ginseng. Mice will be examined for
tumor growth
inhibition, increase iii sv=vival time and lowered adverse side-effects on
hematocrit values and
body weight (Nakata~et'al.;~ J~h J Cancer Res (1998) 89:733-740). The assay
will be repeated
using various concentrations "of stamdard ginseng to generate measures of
central tendency,
dispersion and variability for.eacli variable.
The data collected will then~be subj ected ~to multidimensional analysis to
generate
multivariant normal distribution sets as a means of determining a baseline
correlation between
biological activity and standard ginseng (see Zar, J. H., in Biostatistical
Anal~~sis. 2nd ed.
(I984), pp 328-360, ~PreriticeTIaII, Eriglewood Cliffs, NJ). A second
independent
determination of a Iiiologicah response.of a biosystem to standard ginseng
will be the effect of
standard ginseng ori~physicalvperforinance during exercise. Rats will be
treated for 4 days with
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-42-
standard ginseng at various" concentrations (between 0.5-100 mg/kg/day) and
animals will be
tested for increasedplasma free~fatty acid level and maintenance of glucose
level during
exercise at approximately'70% V02max (see Wang et al., Planta Med (1998) 64130-
133).
The data generated will be:/collected and then subjected to multidimensional
analysis to
generate multivariant~ribrmal distribution sets as a means of.detertizining a
baseline correlation
between biological activity and standard ginseng (see Zax, J. H., in
Biostatistical Anal, sis, 2nd
ed. (1984), pp 328-360, Preritice Hall,~Englewood Cliffs, NJ, Herein, fully
incorporated by
reference). The distribution sets for each BioResponse are then put~into
algorithms to generate
statistical values fox standard ginseng: Statistical data will then be stored,
preferably in a
memory of a computer processor, for further manipulation.
Each of these steps (a. e., chemical analysis, generation of biomarker
information and
determination of responsess of a biosystem) is reiterated to generate a large
database of
statistical values. These values are compiled and input into an algorithm to
generate 2- and 3-
dimensional Herbal Response' Arrays (HBR Array) for standardized ginseng.
Through
reiteration, the resitltirig arrays (a.,e:~'Standardized Arrays) display the
highest correlation
between composition(iric'ludirig'~growth conditions), biomarker information
and biological
response for standardized ,gin5erig.', ,By determining two or more known
associated variables
for composition and biomarkar information values via display on an HBR Array
for a test
batch, the values for biological response variables can be predicted for the
test batch by
comparing test values'agairist Standardized HBR Array values for standardized
ginseng. The
resulting prediction will be used. to evaluate the quality of a given ginseng
batch without
necessitating the use.of amobserve~.biological-response of a biosystem (see
Example 2).
Example 8. Evaluation of a Selected. Herbal Composition of Ginseng Using a
Subset of Variables Correlated with a Specific Biological Response.
To evaluate tf~e quality.~ofa~ est batch herbal composition, data is first
collected
concerning the plant-related,parameters for the~herbs in the selected herbal
composition (e.g.,
plant species, plant parts, ge'ographic'origin, growth conditions, processing
methods and
storage conditions). The selected herbal composition is then manipulated such
that chemical
analysis can be performed t'o' determine the chemical content of the herb (see
Elkin et al.,
Chumg Kuo Yao Li I=Isueli~Pab (1993)=14: 97-100 and Yoshikawa et al.,
Yaku~;aku Zasshi
(1993) 113: 460-467). Previously obtained ginseng data has demonstrated a
strong correlation
between oxygen consumption during aerobic exercise performance and the
presence of a
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-43-
subset of saponin components; especially Rg1 and Rbl (Wang et al., Planta Med
(1998) 64:
130-133).
The test batch,is then exposed to test cells including, but not limited to,
fibroblasts,
macrophages, monocytes; PMNL, LAK cells, B16-F10 melanoma cells, THP-1 cells
and
hippocampalneurons at a concentration of 0.5 mg/m1 to 100 mg/ml to determine
expression
biomarker values. . mRNA is isolated from exposed cells which is subsequently
manipulated to
serve as a substrate for hybridization-based expression moutoring of
biochemical extracts
using microarrays comprising IL-8,'IL-2 and Interferon gamma cDNA (Schena et
al., Science
(1995) 270: 467-470; Schena et al.; Proc Natl Acad Sci USA (1996) 93: 10614-
10619;
Lockhart et al., Nat Biotechnol (1996) 14: 1675-1680; DeRisi et al., Nat enet
(1996) 14: 457-
460; Heller et al., Proc Natl~Acad S~ci USA (1997) 94: 2150-2155). Previously
obtained
ginseng data has demoristraYed a~strong correlation between oxygen consumption
during
aerobic exercise performanceand the induction of the expression biomarkers IL-
8, IL-2 and
Interferon gamma in test cellsv(Venkatramawet al.; Med Sci Sports Exerc (1997)
29: 333-344
and Wang et al., Planta Med (1,998).64: 130-133). For biochemical biomarkers,
rat bone
marrow cells will then be exliosed to the test batch and assayed for [3HJ-
thymidine
incorporation reflective of~mitosis. Previously obtained ginseng data has
demonstrated that
Rbl and Rgl show a strong correlation with DNA synthesis in rat bone marrow
cells
(Yamamoto et al., Arzneiriiittelforschuns (1'978) 28: 2238-2241).
After reiterative~aiialysis,'°data from each assay will be input into
an algorithm to
generate a test HBR array~-for the;selected herbal composition based on the
enumerated plant-
related data, including chemical analyses, and data concerning the subset of
biomarkers. The
quality of a test batch wVill lie deterrizined by comparing test HBR and
standard ginseng
Standardized HBR Array variables directed toward analysis of the above
observations and
subsets, wherein the demoris~ration bf the induction of IL-2, IL-8 and INF
gamma mRNA i~a.
vitro and an increase in [3H],=thyinidirie. incorporation in rat bone marrow
cells (including data
collected on growth conditions, origin, and verification of the saponins Rgl
and Rbl) is
predictive cif an equivalent Bioltesponse effect of the test batch on oxygen
consumption as that
exhibited by standard ginseng. Based~on this procedure it can be determined
Whether or not
the test batch is of a similar~'or different quality than that of the standard
for the given
biological response or biological response of interest.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
=44-
Example 9. Establishing a Standardized HBR Array for Huang Ling (HL)
,:
Recipes. ' ' . , .
f . ~,,~t...~.W
For the purposes of this example, standard huang ling (HL) is chosen to be
Coptis
chiyaesis France, from southwest Asia, wherein growth conditions are well
known to one
skilled in the art (see Huang in The~Pharmacolog"~! of Chinese Herbs, (1993),
pp 69 and 287-
288, CRC Press, Boca Raton, FL).:Dried rhizomes of Coptis c7Zinesis France
will be verified
for chemical content by quantitative chemical analysis for determination
arsenic, berberine,
. ;v
caeruleic acid, columbamine,scopsine, coptine, coptiside-I, coptiside-II,
coptisine, coreximine,
epiberberine, ferulic acid, ,greemlandicine, isocoptisine, lumicaerulic acid,
magnoflorine,
oxybererine, thalifendine, umbellatine; urbeune, worenine, palmatine,
jatrorrhizine and
.. . , ..
colubamine (see also Zhu M.~ Chung Yao Tung Pao (1984) 9: 63-64). Content of
the alkaloid
berberine of different herbs ~sliould be between 7-9% (by weight). These data
will be stored,
preferably in the memory,of a computer processor, for further manipulation.
Expression biomarkersrfor standard HL include the following: NfxB; bcl-2
analog, Al;
zinc forger protein, A20; °II;-2'receptor; cell,cycle probes; c-Ki-
ras2; growth regulators probes
and glucocorticoid receptor,dependent.apoptosis probes (see Chi et al., Life
Sci (1994) 54:
2099-2107; Yang et al:; ~NaiiW n Schrriiedebergs Arch Pharmacol (1996) 354:
102-108; Miura
et al., Biochem Pharniacol:(1'997) 53; Chang K.S., J Formos Med Assoc (1991)
90: 10-14).
Alternatively; for a broader batch size, the 400,000 oligonucleotide group/1.6
cm2 chip of
Affymetrix can be used ~U.S:'Pat. No.5,556,752). The expression biomarkers for
standard HL
will be prepared by microairay technology as described in Example 1, including
analysis and
statistical data generation..Biocheinical biomarkers for standard HL include
increase in
glucocorticoid receptor and'inhibitiomof alpha-fetoprotein secretion in HL
exposed HepG2
cells (see Chi et al., Life-Sci;(1994) 54: 2099-2107). BBM sets are generated
and analyzed as
described in Example l: ~ Statistical data will then be stored, preferably in
the memory of a
computer processor, for'fiirther'mariipulation.
Biological responseof:'a:biosystem will be determined using cells and whole
animals.
Batches of the selected herbal'coriiposition will be exposed to specific cell
types, including but
not limited to, human HepCi2 ~hepatoma cells, human embryonal carcinoma cells
and
thymocytes at concentrations°froin.0:1-100mg/ml. For animal treatments
O.lmg-2g/kg of
Coptic herbal composit'ion.(i.e.;'HL) will be administered orally, by
intraperitoneal injection or
subcutaneous inj ection. Towdetermine~ a biological response of a biosystem to
standardized HL,
;., ,
human embryonal carciriorna~clone, NT2/D1 is exposed to various concentrations
of standard
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-45-
HL and cells will be exariliiied for differentiation into cells with neuronal-
like cell morphology
(Chang K.S,, J Formos Med Assoc (1991) 90: 10-14). The assay will be repeated
to generate
measures and analysis will be performed as described for ginseng in Example 1.
A second
independent determinatiori'of a biological response of a biosystem to standard
HL will be the
effect of standard HL on diarrhea due to enteroytoXigenic EscheYichia coli
(ETEC). Patients
with active diarrhea due to~ET.EC will~be treated with various concentrations
of HL (e.g.,
2g/kg) and stool volumes will be determined (see, e.g., Rabbani G.H., Dan Med
Bull (1996)
43: 173-185). The assay:will be repeated to generate measures and analysis
will be performed
as described for ginseng imExarriple 1. The distribution sets for each
biological system are
n,.: ~, ,~
then put into algorithms to generate statistical values for standard HL.
Statistical data will then
be stored, preferably in the memory of a computer processor, for further
manipulation.
Lastly, as in Exayple~'l, the tees are reiterated to generate HBR arrays for
standardized HL, wherein~the r~esultirig HBR arrays will then be used to
predict biological
activity and evaluate'batch qii~~ity. Using~this method, a Standardized HBR
Array can be
generated~and updated periodically. '
Example 10: Evahtation of a.Selected Herbal Composition of Huang Ling Using a
Subset of Variables Correlated vwith a Specific Biological Response.
To evaluate the quality of a selected test batch of an herbal composition of
Huang
Ling, data is first collected concerning the plant-related characteristics
(e.g., plant species,
plant parts, geographic origin, growth conditions, processing methods and
storage conditions).
The herbal composition.is~'then manipulated such that chemical analysis can be
performed to
determine the chemical aopferit of the composition (see also Zhu M., Chung Yao
Tun,g Pao
(1984) 9: 63-64). ~ . ,,
Previously obtained HL data has demonstrated terminal differentiation of human
embryonal carcinoma clones into neuronal-like cells is strongly correlated
with the presence of
berberine (see Chang K.S:, .f.Formos Med Assoc (1991) 90: 10-14). The test
batch is then
exposed to test cells inclufing liuman ~embryonal, carcinoma clone, NT2/D 1 at
a concentration
starting at a non-toxic coiicentratiori-(determiriation of which is within the
skill of the ordinary
artisan). mRNA is isolated fxom exposed cells which is subsequently
manipulated to serve as
substrate for hybridization~~based expression monitoring of biochemical
extracts using
microarrays comprising~TL=2 receptor and NficB; (see Chi et al., Life ci
(1994) 54: 2099-
2107; Yang et al., Naunyri Sclimiedebergs Arch Pharmacol (1996) 354: 102-108;
Miura et al.,
Biochem Pharmacol (1997)' S3~; .Chang K.S., J Formos Med Assoc (1991) 90: 10-
14; U.S. Pat.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
46-
No.5,556,752), and which~can be used to determine down regulation of c-Ki-ras2
gene
expression in said.cells. ~Previou'sly obtained HL data has demonstrated
terminal
. , r,..
differentiation of human emliryomal carcinoma clones into neuronal-like cells
is strongly
I
correlated with induction~of riiitogen probes and down regulation of c-Ki-ras2
gene expression
(see Chang K.S., J Formos IVIed~Assoc (1991) 90: 10-I4).
For biochemical markers; HepG2 cells are exposed to the test composition and
cells are
assayed for increase in glucocorticoid receptor and inhibition of alpha-
fetoprotein secretion
. -. a
(see Chi et al., Life ci (1994) 542099-2107). Previously obtained HL data has
demonstrated
that inhibition of glucocorticoid induced apoptosis is strongly correlated
with berberine-type
alkaloids (see IVIiura et al.;Bi~chem Pharmacol (1997) 53: 1315-1322). After
reiterative
. ~,. . . .
analysis, data from each assay will be input into an algorithm to generate a
test HBR array
based on the enumerated observational data, chemical data and data concerning
the subset of
biomarkers.
The quality of a.test batchwill be determined by comparing test HBR and
standard HL
HBR Array variables directed tovcrar~d analysis of the above observations and
subsets, wherein
the demonstration of tlie.induction of IL-2 receptor and NficB, the down
regulation of c-Ki-
ras2 gene expression, an'increase in.glucocorticoid receptor and inhibition of
alpha-fetoprotein
secretion for HepG2 cells (to ~inoluding data collected on growth conditions,
origin, and
verification of berberine alkaloid) is predictive of an equivalent BioResponse
effect of the test
batch on terminal differentiation of human embryonal carcinoma clones into
neuronal-like
cells and inhibition of dexamethasone induced apoptosis as that exhibited by
standard HL.
Based on this procedure it aari be deterrriined whether or not the test batch
is of a similar or
different quality than that.~ofvthe:standard.
Example 11. Evaluation of Xiao Chai Hu Tang (sho-saiko-to) Using Two
Bioassays. ~ , .
To evaluate the,.quali'ty'of three sources of Xiao Chai Hu Tang, two bioassays
were
used: 1) cell growth inhibition"and 2) hepatitis B virus secretion from
infected cells. The Xiao
Chai I3u Tang composition is made from a mixture of 6-7 herbal plants (Radix
Bupeuri,
Rhizoma Pinelliae, RhiaqinavZiyagiber°i's, Radix Scutellariae,~F~uctus
Ziziphi, Codonopsis
Pilosula, Radix Ginseng a~hd Radix Glycy~rhizae, see Table 2 for relative
amounts, by weight).
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-47-
Table B. Composition of ~iao Chef fIu Tang.
Source ~ . Plant
Species
Radix RhizontrtRhizontaRadix FnrctusCodonopsisRadix' Radix
, .
bupleuripinelliaezingiberisscutellariae'~ziziphipilosulaginsengglycyrrhizae
. * .
.
Relative
Amount
by Weight
Singapore1 1 ; ~ ~ . ' 0.375----- 0.3750.375
,'' ~ 0.375
'
,'0.375
~
,, ,._-:,.
Korea 1 0.717 ,,.. 0.492 ---- 0.429 ---- 0.288
Ø51
'
Taiwan 1 0.25 0:3750.375 0.25 ---- 0.3750.375
The three "recipe's"°~ originate iri either Singapore, Korea or Taiwan.
Batches were
evaluated for toxicity aiidvfor the ability to inhibit hepatitis B virus as
detected by DNA
quantitation or detection~ofhepatitis~B surface antigen (HbsAg) (see Dong et
al., Proc Natl
Aced Sci USA (1991) ~88:~r8495=8499).
Briefly, one grarri of preparation was added with 10 ml of water. The mixture
was
boiled for 30 minutes. 'The supernatant was collected after centrifugation and
filtered through
a 0.22 ~.m filter. Two cell,types were used: a) 2.2.15 cells which secrete
hepatitis B virons
(kindly provided by Professor~G.;Ace; see 'Ace et al. Proc Natl Acad Sci USA
(1987) 84: 1005-
1009) and b) HepG2 cell's (ATCC~cat # HB-8065). One to fifty dilutions were
used for each
assay. The cell growth inhibition assay was performed for 72 hours. All other
procedures
were performed as described"byDong''et al., Proc Natl~Acad Sci Z~ (1991) 88:
8495-8499.
The results of the assays ,using.tfie. three batches is displayed in Table 3.
Based on these data,
the Taiwan source would'lie selected as a standard herbal composition because
of its low
..**, ,. . . ..-
toxicity combined with its"e~fectivemess in reducing secretion HbsAG (which is
proportional to
viral release) by W ore 'thari-half ~ ~ '
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-48-
Table 3. Bioassay of Xiao .Chaff Hu Tang (Sho-saiko-to).
Source Cell Growth.Inhibition Hepatitis
(%) B Virus
. (secreted)
% Inhibition
HepG2 , 2.2.15 DNA HbsAG
Singapore 73 100 65 38
Korea 13 ' 60 ~ 20 42
Taiwan 0 ' ~ ~ 42 0 47
The data presented in:Tables 2 and 3 for the Taiwan herbal composition
constitute the
initial data for the standardized HBR Array for this herbal composition.
Therefore, this data
set would initially include the source of the herbal composition, the plant
species and relative
amounts of each the herbal, composition, and two BioResponses (i.e., cell
growth inhibition
and hepatitis B virus secret'ion~ froiri infected cells).
Using the procedurES setforth in the schematic of Figure 1 and in Examples l
and 3,
additional data can be collected on,,plant-related data, markers and
BioResponses for the
standard herbal compositiori.~ This .additional data is added to the initial
standardized HBR
Array to generate an expanded ~stai~dardized HBR Array. Appropriate analyses
of the resulting
database can be conducted as set forth in the detailed description and the
examples in order to
ascertain the subset of variables ~hicli is most highly correlated or
associated with the
BioResponse of interest. 'Batch HB,R Arrays may be determined using the
methods depicted in
Figure 2 and in the-procedures jo'f Examples 2 and 4.
The'resultant batch HBR Array. can be compared to the standardized HBR Array
so as
to predict the BioResporis'~e ofvthe'batch herbal compositions.
Example 12: Herbal Preparation
The standardized protocol 'for the herbal extract preparation was as follows:
The
ingredients of herbal raw material's~with proper ratios were placed in a
jacketed reactor and
extracted with water at an eleyated'constant temperature with mixing. The
solid was separated
from the liquid with a_120-mesh screen. The resultant filtrate was collected
and then
concentrated by evaporating''the water under reduced pressure. The
concentrated liquor was
spray dried at elevated temperature~to yield granulated powder. This bulk
substance was then
formulated into the desired'ddsage form.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-49-
Example 13. Evaluation of Huang Qing Tang
Huang Qing Tang (HQT)'is' an ancient Chinese botanical formula composed of
four distinct
herbs: Scutellariae (scute), . Glycyrrhizae (licorice), Paeo~rie lact~o~a
pallus (white peo~ey
root), ahd Fructus zizip7ia'(date): (Table 4). This herbal formula has been
long used in Asia to
treat a variety of gastrointestinal ailments since 3b0 AD.
Table 4: Herbal Ingredients of TCM Formula HQT
Scientific Common NaineTraditional Use
Name
Scutellariae Scute BaicalUsed to reduce capillary permeability:
Radix to reduce
Skullcap inflammation: to treat enteritis
xoot and dysentery: incxease the
secretion of bile to treat jaundice:
to relieve muscle
spasms to treat coughing: to expel
parasites.
Glycyrrhizae Licorice Used to~moisten the Lungs and stop
~ Root coughs: to relax spasm
Radix . . and stop pain: to moderate the
action of herbs; to reduce
(Gancao) ~ ~ ~ ' ~ fire and release toxins.
,
Fructus ZiziphiDate ~ ~ Has diuretic and strengthening
~ effects.
Paeohie lactifloraWhite Peony Used to suppress and soothe pain;
' to soothe ligaments and
pallus radix Root .'', . .purify the blood.
;'.:_ '
" , ,
Biolosical and EnzXrne Assays_ ; .
. , Table.5. Batch Properties (I3QT)
Property Batch A ~ Batch B Batch C
~
Origin Taiwan; Sun-Ten~ Taiwan, Sun-TenTaiwan, Sun-Ten
Preparation Standard Standard Boiled 30 min.
method .,,s.~ ~
~
Plant part Root ; , Root -
,
Eriefly, one. gram of, each :batch of Huang Qing Tang (HQT) was added with 10
ml of
water (1 mg/ml).. The ~riixture,was treated~as outlined in Table 5. The
supernatant was
collected after centrifugation and filtered through a 0.22 ~,m filter. Two
cell types were used to
test for biological effects of 'each batch of HQT: a) Jurkat T cells (ATCC cat
#TIB-152) and b)
HepG2 cells (ATCC cat # I3B'-X065-). One to fifty dilutions were used for each
assay. Frozen
cells (107/m1) were quickly.thawed~in a~water bath at 37 °C. The cells
were then diluted in 10
ml of pre-warmed media (see Life Technologies, Inc., Catalogue and Reference
Guide, 1998-
1999, Cell Culture section) followed;by centrifugation at 1500 rpm for 5 min.
The supernatant
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-SO-
was then discarded and the cells were cultured in 100 ml media at 37
°C, S% CC~Z. After 2
days, the cells were counted (approximately 8 x 105/m1, total 100 ml).
Batches were also evaluated for the ability to inhibit hepatitis B virus as
detected by
DNA quantitation (see Dong et al., Proc Natl Acad Sci USA (1991) 88: 8495-
8499). Briefly,
S one gram of preparation was-'added with 10~m1 of water. The mixture was
boiled for 30
minutes. The supernatant, was collected after centrifugation and filtered
through a 0.22 ~,m
filter. HepG2.2.1 S cells which secrete hepatitis B virons (kindly provided by
Professor G.
Ace; see Ace et al. Proc Natl Acad Sci~USA (1987) 84: 1005-1009) were used in
this assay.
One to fifty dilutions were used for each assay. The cell growth inhibition
assay was
performed for 72 hours. All other procedures were performed as described by
Dong et al.,
Proc Natl Acad Sci USA (199I) 88: 8495-8499.
(3-glucuronidase.was assayed as HQT is known for its anti-diarrhea properties.
Different HQT extracts were added ~to triplicate wells of a 96-well plate
which contained
0.lmM phenolphthalein glucuronidate, 70 mM Tris-HCl (pH 6.8) and 0.8 ng of
dialyzed (3-
glucuronidase (from E. Coli; purchased from SigmaTM) to a final volume of 80
~,1. After 2 hr'
incubation. at 37°C, the reactions~were terminated with 200 ~.1 of
stopping solution which
contained 0.2.M Glycine and~0.2 M NaCI (pH 10.4), and the OD was monitored
with a kinetic
microplate reader at S40 nm. ,
The results of the assays using the three batches are displayed in Table 6.
Based on
these data, HQT sources A and B have relatively low toxicities combined with
higher
. , .
inhibitory activity relative to batch HQT C (i. e.~ approximately S fold
greater toxicity toward
HepG2 cells and 3.3 fold less. inhibitory activity against (3-glucuronidase
than either HQT A or
B, see Table 6).
2S
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-51-
Table ~6. Biological Assay of Three Preparations of HQT*
' E. Coli ~ HepG2 Jurkat HBV$
(3=Glucuronidase DNA
HQT A : 0.6 , . 1.50 0.76 None
HQT B . ,~ ' 0.7 . . , 1.6 0.81 ND
HQT C ' ~:2.2 .: , 0.32 ND ND
*Values $, % of Control
represent ICso
values. ' ~ . '
ND, not ,
determined. ~ ' ,
Evaluation of HOT Effects on Protein Expression
. ~.y'., . ... : . ~ ~ .
HepG2 cells.(l~ x 106),~were seeded in 25 em2 flasks in 3.0 ml of RPMI-1640
medium
(see Life Technologies,, Irio':,~Catalogiie and~Reference Guide, 1998-1999,
Cell Culture section)
24 hr before the drug addition. . The~cells were treated with or without
herbal medicine, where
the former is added at two final.concentrations of 0.2 mg/ml or 4 mg/ml,
respectively, and
incubated at 37°C for 24 hours': The medium was removed and the cells
were washed twice
... .. ~.: .
with cold PBS. The cells were harvested into 1 ml of PBS and centrifuged at
10,000 rpm for 2
minutes, extracted on ice with a buffer containing 50 mM Tris-Cl (pH 7.5), 0.2
mM PMSF and
10% glycerol, followed by,three.freeze-thaw cycles. Potassium chloride was
added to the cell
lysate at a final concentration of 0':15' M prior to centrifugation. The
protein concentration was
determined and the. cell extract was electrophoresed according to the method
of Laemmli
(Ncztu~e (1970) 227;680-685). Western.blots were performed by standard
techniques known in
the art, see for example S ~ainbrook, et al (1989).. The antibodies used were
directed to the
following proteins:.Topo °Z; Sfat'(20Z07); Cyclin Bl; MAPK (Ab2) and Nm
23 H1.
Figure 4 demonstxat~es~tlia~ the higher concentrations of HQT A or HQT B
differentially
effects the expression of c~cliri Bl,polypeptide.
HPLC Analysis :. . ,.._, ,. , . , . , . ,
..- . . ,.. , . .~ ~ ..,_.
The herbal batches~v~ere analyzed by HPLC with a Beckman ODS UltrasphereTM
column (5 micron particles, 4.6 mm X 25 cm) and detected with an UV
spectrophotometer
(Perkin Eliner). The vvavelerigths for UV' detection were monitored at 280 nm
and 340 run. The
mobile phase was pumped~at ~1'rril/min'and consisted of Solvent A: H20 and
Solvent B: 20%
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-S2-
MeOH with the following gradient: 1) the solvent was 100% solvent A for the
first S minutes;
2) the sblvent composition vvas changed to 10% solvent A / 90% solvent B for
the next 10
minutes; and 3) the solvent was changed to 10% solvent A / 90% solvent B for
the next 40
minutes. This was followed~liy..the'addition of 100 % solvent A for S minutes.
The HPLC
markers are baicalin aridy~li'aicaleiii. '.
Mass Spectrometry
The.herbal extract'vvas analyzed by MarinerTM ESI-TOF Mass Spectrometry (MS)
from
PE Biosystems. Control tracings were generated using baicalein and baicalin,
two known
active ingredients in HQT: ' ' ' ' ' '
HQT samples in water aild~acid treated batches were been analyzed by HPLC and
Mass
Spectrometry. While.watef treated HQT batches A and B had distinct HPLC and MS
tracings,
acid treated batches gave alinbst identical patterns (data not shown).
Algorithm y - ~ . ' 't : : . , .
The data collect'ed~°forin part of the multidimensional analysis used
to generate
1S multivariant normal distribiitiom sets as a~means of determining a baseline
correlation between
biological activity and standard HQT chemical (HPLC and Mass Spec), arid
origin/growth
characteristics: ~ . ~~ '
Example 14. Indiw7ic~ual components
A. Licorice. ' ~ ; ~ ,
Evaluation of Glvcvr~hizae Radix ~corice~
Licorice is useful for moistening the lungs and reducing coughs, helps to
relax spasm
~.Y '.. , '
and pain. The properties of the licorice batches used in this example are
presented in Table 7.
..~;, ~:f~_'.',;
Talole'7. Batch Properties (licorice)
Property . .~atch~.A:; .Batch Batch C Batch D
, :y~ B
Plant NameGlycyrrhizaev.'~'Glycyri~hizaeGlycyrrhizaeGlycyrrhizae
:': '
. Radix : . Radix Radix
Radix ' ,
.
Origin Imier MongoliaInner MongoliaU.S., Kin U.S., Kin
Man Man
' ~ Herb Center Herb Center
PrepaxationStandard Standard Boiled 30 Warm H20,
, - min. 30
method min.
Plant partRoot , ' Root _ - -
~
ZS
Biological
and
Enz~!me
Assa,~,
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-53-
To assay the quality, of herbal sources, each herbal extract supernatant was
assayed and
the analysis was repeated three: times. For a given sample to be assayed, 1
gram of herbal
powder was dissolved in 10 ml of ~0° C deionized water (neutral pH) in
a polypropylene tube.
The tube was then incubated as outlined in Table 7, then centrifuged to obtain
the supernatant.
Batches of licorice were tested against either HepG2 cells (ATCC cat # HB-
8065) or Jurkat T
cells (ATCC cat #TIB-152) or.both. Cells.were cultured for 24 hours as
described above.
Batches were also evaluated for the ability to inhibit hepatitis B virus as
detected by
DNA quantitation (see Dong et al.,,Proc Natl Acad Sci USA (1991) 88: 8495-
8499). Briefly,
one gram of preparation.was added with 10 ml of water. The mixture was boiled
for 30
minutes. The supernatant was collected after centrifugation and filtered
through a 0.22 p,m
filter.2.2.15 cells which secrete hepatitis B virons (kindly provided by
Professor G. Ace; see
Ace et al. Proc Natl Acad Sci USA (1987) 84: 1005-1009) were used in this
assay. One to fifty
dilutions were used for ~each'~assay.,~ .The~cell growth inhibition assay was
performed for 72
hours. All other procedures v'vere~performed as described by Dong et al., Proc
Natl Acad Sci
SSA (1991) 88: 8495=8499:' °"
Again, (3-glucuronidase was assayed. Different'licorice extracts were added to
triplicate
wells of a 96-well plate which contained O.lmM phenolphthalein glucuronidate,
70 mM Tris-
. . .
HCl (pH 6.8) and 0.8 ng of dialyzed beta-glucuronidase (from E.Coli, purchased
from Sigma)
to a final volume~of 80 ~,l and'assayed as above.
The results of the assays using the two batches is displayed in Table 8. Based
on these
data, licorice batch A was much more toxic to Jurkat cells than batches B
(approximately 9
fold) and a more effective inhibitor of (3-glucuronidase (see Table 8).
'Table 8. Biologica~.Assay of Four Preparations of Licorice*
y . E: Coli'. , IiepG2 Jurkat HBV$
(3-Glucuronidase DNA
Licorice A . -' 1.1 ' ~ ~ , 1.07 0.41 None
Licorice B ~ , : ND '' ND 3:6 ND
Licorice C , . , r 2.1 . ND ND ND
Licorice D . .:ND "~ ND - >2.0 53.8
*Values represent ICSO , $, % of Control
values. r,
ND, not determined.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-54-
Expression Assay
In order to assay gene expression, Jurkat T cells were treated with herbal
extract as
follows: Jurkat cells (107/m1) °were quickly thawed in a water bath at
37 °C. The cells were
.. , .~..
then diluted in 10 ml of pre-warnied media (see Life Technologies, Inc.,
Catalogue and
Reference Guide, 1998-1999; Cell Culture section) followed by centrifugation
at 1500 rpm for
5 min. The supernatant was.then'discarded and the cells were cultured in 100
ml media at 37
°C, 5% CO2. After 2 days~,'the cells. were counted (approximately 8 x
105/m1, total 100 ml).
The herbal extract solution was prepared as outlined above (e.g., 2 g of an
herbal
powder to obtain 20 nil of sterile 'solution (0.1 g/ml). The cells were
divided into 3 flasks at a
density of 2.5 x 105/m1, 1~OO m1/eacli flask. Assays were carried out with
control (no extract),
and 10 ml of extract at I O mg/ml,_ and 1 mg/ml. Again, toxicity results were
used to determine
the "high" and "low"' concentrations for any given extract. After extract
addition, cell cultures
were incubated for X24 hours' under conditions as outlined above. The cells
were counted and
subsequently collected im Sa~uril centrifuge tubes. The resulting cell pellet
was treated with an
RNA isolation means to extract~ri~RNA (see, for example, Sambrook et al., 1989
at pages 7.3-
7.39). . ,
Microarrav
Microarray printirigFwas carried out as follows:
Human gene olones'yvere obtained from the IMAGE Consortium libraries through
its
distributors and comprise'geries from various tissues. Most clones have been
partially
sequenced and the sequericesv vveie-available as expressed sequence tags in
the dbEST database
of GenEank. Clones were cultured and,amplified using commercially available
primers prior to
application on nylon meriibraries ~(Chen et al., Genomics (1998) 51:313-324).
Approximately
10 ng of each amplified target~:was applied on a positively charged nylon
membrane using a PG
(personal computer) controlledvarraying system. The arraying system allows
high density
spotting and is capable of depositing 31,000 spots on a piece of nylon
membrane measuring 18
by 27 mm using a 24-pin arraying tool.
cI~NA probe and Membrane Hybridization
Two microgram ofyeacli rnRNA sample (mRNA was isolated as outlined above) was
_ ., s
labeled with biotin and~or~ digoXigeiiin using random primed reverse
transcription. The labeled
samples were treated'with alkali and the resulting labeled nucleic acids were
precipitated prior
to use in hybridization: Membrane ~liybridization and washing were carried out
using the
labeled probes as disclosed iri~.Clien~et al. (1998). To detect the spots on
the membrane in dual
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-55-
color mode (i. e., both biotin and digoxigenin), j3-galactosidase-conjugated
streptavidin (Strept-
Gal) and alkaline phosphafase-conjugated ~digoxigenin antibody (anti-Dig-AP)
were employed.
After color development, image digitization using an imaging means was
employed (e.g., a
flatbed scanner or digital .camera). Quantitative measurements were determined
by computer
. :~.~~., ..
analysis which uses a program that measures the integrated density of the
primary color
...v . ;',.
components of each spot, performs regression analysis of the integrated
density data anal
locates statistical outliers asdifferentially expressed genes.
Gene expression data for samples 1, 2 and licorice fST117)
Extract 1, 2 and 6 corresponding to extract of Cordyceps sinensis, Po~ia cocos
(ST
027) and licorice, respectively, ,were assayed by the following method:
Batches were evaluated
for toxicity using Jurkat T°c.ells..
The eXtracts were' prepared as outlined in Example 6. The cells were divided
into 24
well culture plates by addiiig~'l:,~ml~of Jurkat~cells at a density of 5 x
1OS/ml. Assays were
carried out with control (no~ ~Xtract)and 5 concentrations of extracts as
described (see Table
9). The high and low concentrations for the cell culture assays were varied
between 10 mg/ml
and 0.05 mg/ml (i. e., mg dry ~?veight of herbal extract per ml) depending on
the toxicity of the
extract to cells. For certain sariiples the toxicities at 10 mg/ml were such
that "high" and °'low"
concentrations were adjusted ~downv~ard, nevertheless, at least one order of
magnitude between
extremes was maintained.v'For'exaxnple; for licorice (ST117) the "high" was
0.5 mg/ml and the
"low" was 0.05 mg/ml (see,Table.:9). After extract addition, cell cultures
were incubated for 24
hours under conditions vas. outlined in Example 6. The cells were counted and
the resulting data
tabulated to demonstrate extract toxicity. The resulting data is shown in
Table 9.
30
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-56-
Table 9.
Survival
Cell Number
at Different
Concentration
of Herbal
Extract
Solution
. experiment
concentration
no.x105m1 . S 2 1 O.S mg/ml High conc Low
10 mg/mlmg/ml'mglmlNo drug conc.
mg/ml
(mg/ml) (mg/ml)
.
1 Cordyceps 8.4 11.9 11.5 9.2 9.0 12.4 10 1
sinensis ',
mycelium
~
2 ST027 4.2 ~ 10 7.5 10 10 10 1
8.7
,
3 STO-44 ' x':5.98.4 9.9 9.4 S 0.5
-
4 STOSl - - 1.7 S.4 0.5 O.OS
S ST093 - - .1.9 3.8 4.4 O.S O.OS
.
,.
6 STI17 . . ~ 4.6 S.8 O.S O.OS
1'.6.~3.6
7 ST123 3.4 ' 8 9.3 7.8 5 O.S
6.4'
~
8 ST128 3.5 .' ~ 7.7 8.3 S O.S
v .7.7.7.9
9 STI34 2.9' ' 11.2 9.6 9.8 5 O.S
~ ~6.1,..,.
"
ST237 - . 2.5 6.6 8.7 1 0.1
Note: original cell number is SX10'/ml and the number to 1OX10'/ml after 24h
incubation. "-°'
describes all dead cells:
5 Protocol: : ,.. ' .
1. Add 1 ml of Sx105/inl Jurkat cells into 24 well culture plates.
2. Prepare 12 kinds of herbal~,extract solutions and sterilize.
3. Test 5 concentration per s'ample~ 1,0 mg/ml, 5 mg/ml, 2 mg/ml, 1 mg/ml, 0.5
mg/ml
4. Culture the cells for 24.'hvirl 37C with 5% C02 incubator.
10 ' '. . '
In each analysis; 144X96 genes (i.e., 13,824 genes) were analyzed (data not
shown)
and about 100 genes showed ~sigriificamt differences in comparison with that
of control (Table
10). Some of the,genes were'up-regulated and others were down-regulated. The
magnitude of
the difference with the control. sometirn.es varied depending on the relative
amount of the
herbal composition to.which~ the particular cells' were exposed. Numbers under
C1 (control
treatment) and H or L (herbs) represent intensities of mRNA expressed after
subtraction of
background (Table 10). The gene designation is encoded in Array AD, which can
be traced to
a specific GenBanlc clone.' The level of expression was determined by H or L
divided by C.
Only a fraction of 13,824 genes iri each herb treated samples showed
significant changes,
namely, up, down or unchanged (see Table 10).
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-57-
n n n n n n n n n n n n n n n n n n n n n a
a\ a, J v J J J J ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~
0o r0 O N N W ~ ~P O O N N W .p v~ Cn CW1 ~p ...~ .p
O ~ ~ O ~--' O ~ O ~ D O ~-~ O ~ ~ .r O O O O ~C
G~ O O ~ O W ~ O W N N 0o N ~I ~ N W co 0o vp w
.? w1 ~ .? ~--~ N N oo tw0 J N ~D c~ N v0 ~ w W w1 O
V V V V V V V V V V V V V V V V V V V V V d
o ~ ~ ~ ~ ~ ~ x ~ ~ ~ coo
~ x
s ~ ~ '~ ~y cio v~ ~° ~ t7
~_ ~' r
o ~ ° o '~ o tT~ ~. o ~,
o ~ ~~ ~. ~, ~
a ~ Wa ~. °° ~~ x'°
a a ~~ ~ o rx r~
o ~ Q ~ ~ ~r ~~. x x
0 o k ~ ~ ~ r
~o a~ c~ z
Zo N° b o
°
' ~ '~ ~°° ~ O b
o. ~ ~ ~ H °
N a. (~'p z "~ r o ,-' t~
a o ~ .. Z
a.
,~a ~ ~
''fir' ~ o
~ n o
w o, ~ o w .p ~ a, o~ ~ oo w wo 00 00 0o v, ~
N V O .ice 41 ~D .f~ C1 -P 01 cn O 00 vp tJ~ O v0 V~ O v0 cm
N i0 0o w ~D oo G1 W O N ~ N 0o c~ N C1 :p. W G1 cn N
.P N 01 W d1 01 00 N W 1 ~--~ W W 01 W .P 0o W 0o t!1 ~D
k
x .~
'''wv,vN,w°oJa~oJO°o~oc»o~o~ooo x
J.PtNnOOo~OWONc,NnWO~I~JCm~NN~~~~c~m
<'
o~ r ,~
_ _ ,r ~, ~ o
c~ GN1 ~ W 0o N J O~o ~ O ~ ~ 00 V O~o N ~ J ~ O w1 x
W i.r N W W W v1 O i-. ~ J sD C ~ 0o s1 O~ sl :-. N ...-~ ..r
N w ~-.~ cn V~ W vp ~D w ~ a1 cf ~O ~l v~ N V~ w o~ c.n
~ N
i
~O W ~. ~ 00 N w1 09 W J .? W V7 V~ V~ 00
O N W N O V~ ~ C1 00 .-~ 01 w1 c.n O1 .p ~l O~ W ~ -P O~
N 00 w oo c~ s0 Ov ~ N o0 ~ -P oo ~ C s0 G1 N 00 00 s1 '~
N v0 J .p w1 ~ N ~ oo W .-. .p ..... .p J 00 c.n 01 W
w r
N ~ N ~r Q1 -P Ov ~ 0o a1 OW N w1 Q\ N N LO W CJ ~,.
0o O N s1 v0 0o Q1 O w1 ~ s0 ~ ~O s0 O w1 cw0 W o0 ~"
N iT 01 v1 0o .-. s0 N s1 ~ J~ 0o O ~ ~l O c.n vW1 0o O
CJ~ -P V~ 01 J Vt W ~r cJW1 -P .~ ~O ~ ~-' 00 ~ -P .? W W
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-58-
x x
W ~-..N 01.pV~W ~. .~N ~ W .AW N
a ~
1 01O ~ ~ N ...-..V ooW .p cfw o000v~w OvO~'O ~-h
v0 W ~ N ~DW tn~ W ~ ~P V~W N ~ O~J .-.p
N
c~ 01 W ~ ~ N 0oN W o0 00w1t W v W 01N ~ p"'
1 1 ,n
U4r ~'
-Ptn W ~ W 01wt~ t w1 01~ tnt w1-PW 0oW ~ O
x
O ~ O s1N V~O N w oo.p 01N ~DvDcrp N v0O ~
a1c~ ~ ~.-..V~~ O ~ ~ 00W ~ N ~DW W s10ow 0100
,pc,nJ ~-...--.-PN ~O~O~lJ v~ J V w 010oN cryw N
oc o W c o ~ ~ ~.. o c c o ~-r.c c ~ o x o
. . . .
V~0o N -PCJD J :-r "-.,.r ~-' y l O O ~
O ~
01N N tm~ ~,tn0oN O w ~ ~ J ~ O ~ G w0c~
010o N O oo.-.01~ ~OW 0o-P O W N tnO w1s1J -P b c~
~OW Q100V~O 0100W 00N w1
V W W O
01N .J~O V .-.W ,r'pW -P0o N O ~ N ~ c.~ W
O n
-PW N ~--~N 01N 01N W W w1 ~ 00J c.l~N ~ W ~D'O
No w o ~ ~-. ~ ~-.0 0 0 _ ,r,-. o o ,
~ W ~
w10 O ~ 1 ~ ~ ~ W N O ~ ~ W N ~ p V O _
0 0 O
N
Ooo J c,nO .p.p~ c~c.nc.nO w vpoow ..r~,h...ppvp ('
tno0 0ow1w1~O01W O1W O ~ ~ Oow1N w1~ V -PW
W'-'ooO1v~V~~ N O ~ W P W O
01v0 J V 00v000v D ooc~- 01O w 1 N N O~cry~- '-,~'' t?.
N 00~l~D~--'-PV~~Ooo
N O ~ 0 0 0 0 ~--~O ~ O O O ~ 0 0 0 0 0 '~~ ~
~ W 0oN J v1s0J o0C 01 C1s1N O tnOvV sD00 O ~'
Oo0 .P~ N OoO N ~ V ~ N W W V ~ 01N -A.p.p
~1~ r
D. 00O ~ N ~Ow ~-.O oo.r o00oO w G W.~ ~lc.n
WO D1O ~ W
. tnO~ .pW N ~--'N O O Oow1O ~ON W
w1w1 W d1~ ~ W ~..0oN w1~ N C w1v001~ W O O~
t!~N ~ ~OO\.-~~ W W t!1 v0 W W .-..-.W 00W N ~
W
N ''.''
o o v,o o ~-.. 0 0 0 0 0 0 0 0 -.o r..,~
~l 01N W C1, C ~ 'OO ~ t~C1OoJ ~ N W C d1
,_ n
~'c~ p1O 00N w 0 ~ s001 W 01~ ~1OWO 01~.00
-
C1c,r~~l00O v0P c~O O 0oW ~DW W W w1'OW 1 00 -r
ON v0N c~~ ~ .pV Cw0 w W 01W ~-00w W J Cn
~ N W J ~ ~ ~ ~ J N ~ ~ W ~ ~ ~
~
O O J J . c.n J n
1 O P
~ ~r
o a o c.~ c o c c c o 0 0 0 0 0 0 o x
0 v1O t,r~lVN ootnC1v1 ~ J~ooc.nw O v~~ W
s1
,r~O 01N N 01N ~ w O O -P G~ooO W c.n.pV O N
w1 OoW W W O C ~ ~ W W 'O~ ~ .p00W N N Oo
00-P 0o~--O V~00~--c.rWO cm0o t~N Oow1W ~p
N _ _ ~ ~ J ~ ~ N ~ _ J ~
-PN . O . c~ O 0 W W W J w1W
P o P O 1
O O O ~-' ~ O O ~-~ O O ~ O O O O ~ O
~wo'o~oJ~°°,°~v~",o~v~",°o~ov~,aJ,o~ooo°v~",
0o W J ~O N Cn ~ 01 01 v0 w N ~l w1 -P .1~ ~ W O O J
~O W N O Oo N 0o c.l~ ~..~ V~ 01 O 01 v0 O 0o w1 00 ~O 00 .P
N O Qv ~ ~i 0o tn W .... J cn .p 0o w1 ~I v0 01 VW1 v0 N
~Dw1W01~--~CJW1.7~~Nt~~--~~1W T~~~ ~t.»
~-r rr
V1 (J1 i-r i--. n--.. .-.. ~.. .~. rr ~... i..r V1 O ~J1 ,.r
try N O~ N 01 w1 ~ W w1 w1 ~l ~ oo J ~ ~ ~1 00 ~-~ .r O O
~w0 .P ~ w v0 cJwl N N ~ oo .~ N v~ 00 N 00 O w
~D O .p w1 ~O W Qv tn N ~--~ pp N ~ J ~--~ cn N w1 O W c~
N O Ov VW D W1 O 01 W 01 W 00 O1 W ~, ~.... p .1~ ~D
w1 ~O ~O ~O N N N N 01 cm .p cm W w1 ~ oo V~ ~ 00 N O C7
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-59-
n n n n n n n n n n n n n n n n n n n n n n n n n
.h .p .yes .~ .t~ .~ .~ ~, v, ~, ~n v, ~, v, v, ~, v, ~, v, v, ~, a, a\ o, c,
W W W rP cn 01 s1 0 ~ ~ N W W ~ tn pv 01 J 0o r0 ~0 ~-~ N N N
O O O ~ ~ O O O ~ W--~ O O
,..,. W W O O
OO~OC.n WO__W WN~..W~001000~0"'r.r~. .-.
V ~ V OV V V V V V V V V ~V OV OV V ~ V V V V OV V V
ao ~~ob ~ xa ~ x~ ~ ~a~ a
w d a- ~ ~' o
~.rJ~ ~ ~ coo
o ~ ~z~ z~~ ~ ~°c~ x
xr~ aza ~ W~ ~ x
o az
H ~ ~~ ~, ~x
.d
° a:
n d o ~ tn o r~ x ~.. o
C~J ., ~ c ~ o
7~ ~ cu ° ~, ~ C~ "' ~y °°
O b a. ~ ~ ~ ~-~3 'J ° ~ o
0
~ ~'~ a .'_' ~ a°a x .~ ~.
0
n ~ ~ ..r a
c~ ~
O o ~
~O G~.. .
W ~ Q CD
U4
a
n
~l 00 CW 1 N W W ~ 01 0o V~ J w1 41 ~ 0o ~G O
N 01 tm N Gv -P 0o O 00 N v0 J O .-~ Cv .-. J O W v0 y1 ~ a\
O~oO~l~~c~~WO~oc.~nN~~tNn00~J~N~wN~w
N ~ c,l~ W W .? .-. ~, ~O Vm--~ W O ~ Vwl v0
-~ W 00 -P W Cn ~.-~ O O~ ~ W N N G1 O~ O tn ~O ~
O ~ O W ~''~ sG C1 ~--~ 0o ~O cm N C1 ~ 0o N ~ sD ~O a1
.P Oo N C1 G1 tJW O O wI O .-~ 0o O O 00 00 01 w1 O~ W 0o J N O ~l
W G1 O O N N ~O O O N -P N -P ~ N ~ .-PP ~ ~ J ~ W W
w N oo IJ ~ .p :p N ~. G 'p' W W :p O c~ Cv W 0o ~1 ~ ~ sD
00 W W w1 W ~p t!~ ~ Vt O~ ~ W O 00 J 00 W ~---~ W W CJ~ ~ p1 O 00
N .r ~... J c," W .p w w J w w ~ tn W w Ow0 N ~ ~ ~ 00
~1 00 ~--~ c.l~ CW C ~ .~ 00 \O N 00 W W W Ov VW O .1~ w ~l 41 N W ~-'
c.n ~' cT .p 00 00 ~''~ W O W v0 -P .P ~ 01 00 ~ vt ~ O J 0o O c!v W
~C .P 00 W 01 ~--~ 00 -P ~D W (J~ 00 wI w1 J 01 01 ~-' W ~-' .-' 01 .-' V~ ~O
--" ~D .p 01 ~ ~ W 1 ~ G~ N W N -P ~l 01
N N ~ .-r 0o W V .P G1 C!~ ~ 00 OWD G1 a1 N Oo w1 W 0O O
00 ~1 ~ V~ tJ~ ~ ~ W O ~ tl~ N .~ O 00 00 t!1 ~ 00 O ~G V~
w ~ 0o W v0 v0 ~ O ~ W V~ N O W N v0 v0 ~O N J 01 v~ .p 0o cn
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-59/1-
"-' W W V~ W N 01 N W J J V~ c.n 00 N 01 Oo W
W N ~ J ..-. O ~D O 0o N W J N ~ N O 01 D1 W tn W N
~. s0 00 ~ tV 0o w J N W N N C1 ~ s1 00 -P sl W N ~ s0 0o J
J v0 N cn G1 Q1 01 ~O cn 01 ~~ -P O ~ O .P J W 0o v0 N v~ -P ~ J
c.n N ~ O W ~ V~ O ~ O J O ~O N O O C!i 01 W Q1 .p 0~1 J .(~
s0 N O 0o N s1 ~''~ O O1 ~ W N cwl s0 -P N crW''~ """ Cw0 0o J '"''
'0 00 01 Q1 v0 ~ N ~ .p 01 cn .p w .P W J o0 N .p N 0o N J ~ 00
O O O O ~ ~ O O O O -~ O O O ~
O O O
N W v N O o0 O s0N ~ sDN 01 .P~ O\v1 00
-P01N tn00-PN ~ C.nN W N.N O 000op1.P Oo
W 01N ~ .pv0N v0 O ~ v w1v0N N oov0O J
~ ooc.no00oW .~ 01 N O G1 0101v0 0oO ~ a1
0001~O~D~ON J cf 01J .-~ O J W N N O O J
0001N tntw0 01O W O .p~.O -P W tn.p J 0001W O
O O O O ~--'O O '--~O N O ~ ~ O O O O O
w -PW ~DO s0v~'~O ~ ~ a1 -~ O G WD O ~''~01J s1
N ~ J V O ~ O 01c,nJ v0N w -AN 00 0101O tn 00
N J N V~c.noo~.O N J O .P W o0N N c,r~01~.w Q1
CJ~~ 0000J W 00N O N ~ J ~ Q1O ~' .P-P~ w ~O
J O v0W W W O N N N W O tn .-.00~--~cnO1.~J v0
N ooJ J 0101.--J w oo.pV~O W W O v W N 01N O W
1
O O O -~N ~ ~ V~P ~ O O O O ~ N O O ~ O
W N N O ~-.:....i.r~ V~ '-'.' i-.. ' '_' ....
~ ~ J J
0oO O .PW O W .rO ~ o ~ ~ V O ~ N N N . W v0O
1 O p 1
0 ~ V _ J W ~ ~
0 0 0 - W O ~ ~ ~ J ~ O W ~ ~ ~ 0 ~ 1 0 ~ J
1 o P O 0 0
tn.p~O~OW ~OJ O ~ O ~ 000o O 0001O ~ W J O ~ N
J N .pW 0oD1J v0-P W 0oJ 0o O~ v wpN W ~-.N N Cm--
O O O .-r~ ~-.~--~ .-~N ~. O O O O N O O O
lr.>N o000:p Q1 ~O~ s0 N s0N ~ ~ .J~00Vt
J .pW 1 cl~-Pt! V~ 00N ~ J ~OW 00 01W 00..r~ C!~
J 0100r oo.pa1 ~ tn01~D W w1N ~ N 00O N W J
O 'O01O ~ .PO W ~ C100 N V~J J J ~ 000oN V~
O cnJ N N O w ~.J w 0 ~ 00Cn N -Pv0v0J o0
-PW ~ W G1W 00O V W c~J O 01 G~cm00 0oW 0oW .p01
D
O O O ~ O ~ O .P ~ ~ O O O O ~ ~--~O O O
O ~ ~D~ ~DVto0.p '~N ~ 00 J J ~ J J 00N J
.?.PO V v0O W O N O o0O 0o ~"O W ~ 0oW 01~ N
J ~ON ~DcryO O J ~ 01w N N N w O ~GN W .J~O r
0ot_n~.O ~ J 01J v0 ~O.-~O o0 00N t~ J O w W W V~
J try..rW ~,c~,~.-.N Q1 w v0O -P W .pJ N .?cm00V,00
01~ ~ ~D00J V~ ~ ~ J O t~ OWD ~ ~..r,.rpy-..pN
O O ~ --~O ~ N O O O ~ O O O ~--~ O ~ O
N w "''i ~ W ~ J
-P . t : o ~ J W ~O01~O
n O
O ooc.n~--~V~cfO ~ V~ N 01v0 J ~ V~v~ N N v0J J
N
01a1O N -P-~J W a1 N ~ c.n N 00W J J ~.O ~ N J
J cmN .p.O ~ 01O wl~ 00~ .-..N N .p W ~ O N V~
W O1N ~Dv00oN W v~ J v0J N 01 000ov0 01000ov0J 01
.h ~ Cn v~ t~ t~ cn 01 N N ~--~ N w W 01 c,n .-~ N N N
0o N v0 N ~ ~-~ N ~ N N 0o N ~ 0o N W O -P -P ~ N ~ W N .J~
00 O N 01 ~ ~' 01 O v0 .p Q1 ~ Q1 J 00 ~ ~O J Cn O ~ O Q1 00
_O~_DO_~~cON~-~~NN~_OOWOCnv~NooN_OWO_ W
w0 ~ O ~ ~ ~ owo O N ~ c.n 001 ~ J O WD w O oo O~o N
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-59/2-
N N N N N N N N N N N N N W W W W W W W w W w
NwW~~cnvylrlOOOO~DvDOONN~t~0o0o~D~00N
O O ~' O O !-~ ~' O ~' O ~ O O OO ~ O ~-' ~ O O O O ~ --~ O
~-w0 N ~1 v0 O N ~ ~ W O w tn w O W .-r ~ ~D Oc v0 O ~ O v0
W v0 01 ~ 00 01 01 w1 v0 cl~ .p Cl~ Oo W N ~ Oo W a1 ~ ~l ~l 00 O ~D
V V V V V V V V V V V V V V V V V V V V V V V V V
o a~ ~ ~ o a
o ~ ~ ~ ~ o
-C ~ x "'~ ø, ~. m x ~ ~r
O ,.~ O ~ ~ ~ t-' ~ x- w ua
H ~s C~ c~ ~, o a ~
C7 ° ~ o ~ r». cr ~ ~ °or
~--~ v~ x ~. rr ~ ~-.
~_ w ~ ~ ~.
a d ~. . w a ~.. ~. .~
a ro ~ ~' t7~ ~o o ~ o.
O O ~. o ~ Y ~~ o~
t~ ~Z ~ ~ x
w
(.a~ ~ o~~, ~ b '~~,' "00
x x~
0
.O~' ~ ~s. o Y ~ ~c
'°
a ~ ~ (gyp f~D
0
O
O
n
d1 N ~ ~ ~l 01 N ~1 w1 N 41 .1~ O ~--' w
a1 c~ cn w ~. 01 N 01 ~ O v~ O N 0o .-~ w1 r-. ..-. vp ~ p
"'' ~. ,-. yp N '"'' s0 C ~ ~ J -P ~ ~ s0 N W tI~ 0o -P N N
0o W 0o N O~ ~O W Q1 O O~ O ~D ~l ~D W ~..-. 00 W tn ~ .-~ 01 a1 O c!i
.J~ v~ N -P N N w cm ~ w v~ N oo N w
vp ~.-~ 00 V~ oo N ~ vo s1 01 O N P oo N ~ N Ov ~ O w
s0 ~O :~ ~ ~ W C Vt \C ~ 01 W ~''~ i-..y0 ~O V~ C1 ~O G W w1
W w w0 W 0o W W O W O N 01 N ~ V~ .-. 'p N ~ .p 01 ~ O W
.(~ ~ ~ N v~ oo O~ .-.. J ~--' N oo N ~--~ w
,r .... O p~ N W v0 v0 ~ G1 tn C1 vG :P Vo s0 w tn N 01
W N O :'~ "'y .? oo N o0 01 ~ O w ~ ~ O G~1 N N ~
N c!~ 00 '--' 00 ~-~ 00 Q1 O G1 O 01 O W 01 01
N w W o vw0 01 J .P ~ W .P c.n ~ O~ N W W N o0 N W W
N 0o .P O C1 ~ J 01 00 ~--~ s1 C1 v0 s1 N ~D w O N N O G1 w1 o0
y1 00 ~ N ~1 O N N ~ ~ 00 ~-r W W a 01 ~''~ 00 ~ O N W 01
01 00 01 01 ~-' Wl ~O w1 c!~ 00 tn G1 .? ~-' N Oo .p 00 0o w1 ~--
.p ~. ~ p~ w v~ .p N w v~ w w w N ~ w ~..r N .-~. c,n
v0 .p N C1 J 00 O ~ ~ O ~ G~ W w ~ ~ P O~ N w_1 01 v~ 0o N
c,Wn~JONo~~O-PO~ICVnJ~D.NPc~IW O ~ WO~o-P~O~OWO~oC1
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-59/3-
0o w W N N -P d1 N .p ~--~ N ~-~ N w 00 .L~
-P -P O O ~ ~ -P O w1 ~.O 4\ CO ~.1 ~1 J ~O a1 N N N O
W N cm Vi o0 00 ~ :.. O O O VW n -P O .-~ sD :..~ N . ~,a c~, w
w.1 ~-w0 w1 ~ 00 01 CT O N O ~ Ln r1 N N Q~ W v0 W W oo cW O GT
N ~ ~ O~ -A t~ W W W vi ~-~ W -is O~ N w W ~-~ .P .1~
O 00 O 00 O~ ~ O O O cl~ 00 00 a1 ~.1 ~--~ N .P V~ ~p 00 .p v0 w1 00
O s1 V Oo J 0o cn O :"' t~ ~D -P Cn c.r~ ~ cry s0 ~ N o0 tn i.a G1 ~P .1~
-P. w w O~ ~l w w ..r .-. .p J ~.1 ~ N v0 v0 ~U ~~ v0 N ~ O1 ~.l w1 w
OO O O ~ O O ~--~O O O O O O O O O O O O
0ow N s0O ~ tncryc.n s0 01 .4~ooG~.1~C tnootryN O
Ow1~ ~--'-P.00~ N O -P -1~.w .P~OO ~ ~ ~--~00v~~.1
oo01N O 0o'"rw W ~ ~ ~ ~ ~ ~ W ~ J w ~ W ~ O
NN W N tnr O G o o
w o O O O
v~~ V~ V~N 0oO'w0 vD v0 0o w c.nO O~W -P~ W w ..-...p
Noov~ '10oW ~ ~1 ~O tn 0o N -P~ ~.DN 01~ 0o-PN
OO ~ O O O O O O ~-' O O ~ O O O N ~ O
Q1N s0 s00ov1O~cros1 -P s0c,WO ~ ~ 0oO Ov~ N
_ _ _
W ~ ~ ~ O ~ J N ~ W ~ ~ ~ ~ N ~
t o O c o N 01 . w1
n o 1 m O ?
0001~.O0otnN O C w1 ~.l ~.ON ~ ~O~ N ~Gw W cii
-P01.p 0ov0v0~ 00 N ~1 ~ ~.1~ .P~ w W O ~ N
Vt~Ov N ~DN w700 ~ .p,O ~ 00O~J W O w V~.p~D
O ~ O N ~ ~ P O O N O N O N O O O 01N 0o N
4~V ~ s1W -PcnJ ~ J W N N w -A~ -PN J J ~.1 O
~.101cm 0 ~ ~.1~.1w cry ~.lN .?0ow1w 0 0oN 0ov~t!~
NO a1 tnV~-P00N wl N N W W 01t.rwp~DtmV~01~- O~
OO ~ Q1W W W 00 w O W 1 00~O00~ ~OW ~.100..r V1
l O
0 J ~ _ ~ ~ W W W ~ _ ~ J
0v0O N 0 W W J ~-' 0 ,.000 l
1 O 1 0 r 0 l1
N N ~ ~ O O ~ O O '--'O ~ O -rO O O ~ N ~--
OJ i.~O VosD~Dcm -P ~ N .~N VrO N O~J ~ C100 ~l
J~ w W w W ~ ~ W .? 00 w1W .-w0~.1~DO O v00o O
v0J J Q~01~ O t!~O O ~O W O C~N G1~ W ~ W -P
-PN 00 ~.lOvw 0oc~ W ~....0o W ~.1~ ~ ~ O a N
d1J w d100~lw 01 - 00 w1 ~1O
a1~ 0o c!~01W .P~.1~ ~O N w J Q1J 0ow ~ N d1~.1 w
OO O ~ ~ O O ~ O ~ O O O O O O O N O
a10ocn N N ~ t~cJ~c~ O :~ a101W s1~ J 00V -P.t~ W
~Ow1c~ O1CvJ~~.1O d1 ~ O vDO 01v0~ 0000~ ~--~N w
O~Dv0 O O W O ~ ~ W .P O~tnw .p~ cryV~~ O 0o -P
~O~ oo .pvDN O W ~. 01 ~O
,....,.-w1w ~ .p~pO ~ ~ N
w~ .p ooN O~oocm w N O N N N cm-PN N V~-PO~
vDN 01 N W V~N ~D W ~ W 1 0ov0~OVvooW N 00v0 tn
OW N ~--~~ O O ~ O O O O G7~-'O O O ~ ~ N -''
c~.~p~cO O s0OoJ oo vo sD s1 to""~s1O ~ J ~1v,a1N O~
NcmW ..7N w .-.p~ ,.-..J .-..-,:~00.~V~-POoC10001 O
.P~ N ~.lw7w1~--~oo-,....w p ooW cmt~'pv~01~.1v0~D O
NO 0o N 0oW --Q\ W .? -P O vG~.~DooW w w N v0
.t~7~--~ooO~.--c~.-.w N .P ootnooc,nO O N ~ ~--v wO
NN v0 -P.V,000000 c.n .P -~ 0001w1~--wl~ O~Qvo0w N
tn ~ ~ tis J -P N a1 ~ ~ ~ 0o W oo WI ~ ~ N o0 00 N N o0 00
GO O1 tn ~. N J 0o -P -P c.n .r 0o N N O cn N w ~ ~ w W .P O~
Co .-wp w1 w_1 0o w1 01 01 w1 ~. ~.l N W N .P c,ml ~.G C1 01 ~.1 W
-OPJO~1~~.PW-~P.NN~~o~oC~t~n~O~~~t~nW ~cwmN
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-59/4-
n n n n n n n n n n n n n n n n n n n n n n n n n
O O ~ ~~ ...., ,-, ..... ~... .r ..r ~. ~..._. ..r .-. ..r ~. ..r ..r ... ,_,
~--. N N N N
~Or000~-r~~.rNNWrP~VyljTslVl~ICOOO~DO00N
n--. r.a V..r n-.... Q ~. .... ,r ~. n... O ..r .~. r-. r... p O r-. Q ,~,
~~... ,~. r-.
W O_ W c.n O W ~_ .P ~ ~_ O ~ O O a1 v0 O 0o O ~D O O N O
~V ~ V ~ V V V V V V V V V ~ V V V V V V V V V ~
0 o zz o ~ zz~~x~ x~W
.° o o as o ° aa'
x ~ ~ p ~ ~ ~y p ~ ~~~ ~d
cr w ~
z
b ~ ~ o ~. ~, ~ ~' ~ ~ ~ ~ z ~xC
7~ N ~cs ~ Z ~ rn ~ ~ 0 0 o CJ ~
a ~
x N '° ~ '~, n x ~ crn ~~-3
a.o~', ~ dc~ ~' ~x
p" ~~ ~D C~ p
a ~. N ~ ~ ,~~' a c~D O '.~ ~ C/~ rn
O a' N W r ~ C/~
O. o tv .b a
x o ~ ~' ~ 0
ci ..fl ao cr ~ ~ O
o ~ ~ w o ~n ~ C~C"
0
n Q. c~ ',. 'LS
~ ° O
H a~ ~ ~ ~ ~
Z w ~ ø. w
~~ ~'zoa
v~ w1 w0 ~ ~1 N .p N w0 tn W ~ N N o0
~O W -P O 0o O O 01 O w1 w1 O~ O N ~ -P w1 ~ N ~1 W
G1 W ~ N W C s1 C1 ~. C1 ~ s1 s1 V~ C1 :p s1 ~ O C
W N cm ~ oo w1 -P .P Oo -P N w1 ~O O N N O w1 v0 two N w1
.p ~.. 0o N m--. c~ ~. W W .-y v1 N ~ ~-.. N th
n-. ~ ~. W .P .P t!t 01 W .P -P ~-~' O W lJ~ ~--~ 'p r-. tJt N w1
s1 tD W C1 V~ .P N C ~ 00 0o N a1 ~D N N ~~ ~ i,.~ N .-. :.r pp
W t!~ -P W 00 ~ ~D ~ N w1 w1 p~ W O N w1 ~ O ~1 W 00 C1 w1 vG ~
00 ~ \p ~ 00 ~ ~1 Q1 W ~ O -P W ~ ~ W
00 w1 W O ~ CJwI O~ C~ ~.w' 00 O ~ W ~ O~ O O ~1 W W
s0 ~--~ sD sp r~ .cW o0 W .-. ~D N ~. 00 ~ a1 N ~.~ O s1 W
N 01 J .P 0o vG 00 .p ~ N o0 00 ~l w1 O .J~ ~ O C1 ~l v0 O W 01 00
t!»1 .p 00 ~.~r W 00 w1 V~ W CJ~ -P W Q1 W W V~ W w1 CJ~
t!»1 w1 W C!i 01 O J 01 ~ 0o t~ a1 00 .p v0 w1 W W 1 N ~ -P 01 O
W c.a cn ~ Ov Oo -P J ~ N J 01 W O :~ J J v0 ~ W O O~ ~D 0o O
N W 1 00 ~D N ~ 0o t~ CT w1 N 01 W v0 w1 01 V~ Ov ~ ~ 0o w1 .p
W 00 N ~l N 00 ~ V~ W W N O1 ~.-' 01 N N ~-' W ~
cn w O~ .p oo cn ~7 a1 O N tn oo N oo ~ ~D ~ N O w1 o0 0o v0 N ~
cW a G1 0o Oo 0o N 'O O W ~''~ ~ s0 N 0o W W 0o O ~ s0 01 ~ s0 im
01 ~--~ oo c.w1 ~l N ~l ~. .P 01 tn o0 00 ,p two w1 ~-~ 0o w1 w1 ~l w1 W
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-59/5-
-P 01 W t~ ~ W 01 01 N v~ W N
V»O Q1 .p J ~--~ O ~O cl~ O O Vt 0o O 01. 0 W W ~ V ~-.-. O J 0~1
W .N~ o ~ W ~ ~ ~~,, W N C V~ N Oo 01 00 ~ 01 W .~ N J ~ N W
J w1 W N N ~D w1 cm W .~ V V c,n W to ~ W
01 ~l -P 01 N 0o tn Cn cn N .-~ ~ pp ~-. Cn ~ W W W W
O ~-- O tm oo .p w1 tn 01 N N O N 01 ~ W .p s0 01 W ~..,.. 'p c~ W v~
C G~ ~ -P v0 ~ N 01 sG ~ N ~ ~ G~ ~ ~ W 00 :p, N N O 00 00 .p
W 00 W --~ r-.. ...r pp 'D N V -P W W W 00 -P VW 1 W \p V1 W
N""'"-' "-'~-' O O O ~ O O ~ ~ O O O O O O
'"~
~tn ~ C J W 0oC~ s1tJV V W W C1000001 00C1
' '
'
-Oo ~-~O .PN w1 W ~ O O o 00 w1 V O G10o N ~D
1
''rO ~ O N N ai~ N _ W C1 ""'ppOOGI .-~cr~
w~ N . O ~O W OO ~ Wl N P
p I
D O 01 -P-~N N ~O .pc,rW N J 0 W ~ C1W .
o
~..p
v000 ~1W c.nW ~ c.n0001 .pcr1 O v0 ~O oo W vp
Q101 ~ W
N..r~-~.~-.~--m..r ..rO O .-r,r
~ O O -~ O O O O O ~ O
00.p O N - o ~ s ~
P 0 p O ~ O sI00 ~ s1 00 ~ 01 vi N
NV1 N
N ~ 00\pVt\pW W _
l w W O ~ ~ V
ww1 w N c~V V ,_,O 'p 0 w1 ~ ~ t ~ ~ W
1 n
, o
N ~ N W N N 0oO w1O w1 O N Cnc CJ . o
n
N V . W1w ~DooO w1 V c~ Q1 .-., m...w
N
d100 W 00 W V w .PW ~ON .1~G1 ~ W G~ W r ~ .p
1
r O 00 0100
W ~'~-'~ O ~--~O N O N N ~ O N ~ N N N w N O
~ h ~ ~ .P.oosGoo~ ooC .~ .p'pJ i W
.
v0 o v0c.r~~O~ ~ 0o ~.- N ~ ~ o0 ~ ~Ow1W ~. ..O~o
o ~ j v0 O
w1N ~ oo~lN O N O W G1G1.Pw1 N .p c.nV t!~V~-1~~
0100 ~D~O~ N 'Oc~O c!~W N t!w1 G1 0o w1 C1Oo~-N l P
Oo~l 0o~ W W p~V~01N N 'ON N 'O O~ ~1 ~ N ~1. w -
~O W N
00v0 N W W I V w1w V N 00v0N Gv o0 w1 wIv0N 0
1 J
p W O ,_,O
. ' O ~-... .-. p ~-.O
O W '-00~ ~pi-.~ '~'~-.i~.~W O v1 O ~ C~~ 0o N
s0
VWO V1O ~ N Q1C!~V1tltW v0v0.P ~ ~O 00N ~D0
1 N W
~O a ~ W ~ tnO ~ O~ O N c~00 ~1 V 0o Q
- c 1 00~lw1 O W
P m -Pv I ~ W 01N ~
.?~O00 N O
v0N ~100~lV 01vDW o0 N ~--~01O ~ N ~
0 C~-PW ~ J O
'O01.1~-P.1~\ON 01 V1r-.~ .....i--~(~ 0
W
00W ~ W
N~-.N ~ O ~-..O O
_ _ O N ~ O O O O ~-O
J N W 00J cmW p t!~
~ o V W ~ ~ ~
W~ ~ -Po 0 O N N ~ V~ G~ ~lO O w1 O cm
0
w1.p01N 01~ N tn0oW p~ N 01 ~ N N W tn 0oJ
W~ O W o0Cn~DO .~O c~a1.pcryc,mV~ o0 0001p 1 ~
. w ~--O
C,!1 N ~ W 1 O ~ O ~l O ~ ~ OO CI l W ~Op p W
. . ~
wN 01.rV 0oN ~ ~--~~ .p~DooN 00 ~G 0o W . ,
N
00C~ N N
Ww.-.(JO N V~O ~ O r-.m .-. ~7 O ~O ,...,.i-.~....y .-...rO
J~~ \G00s1C1~DW C W i.rpp~.,.rpp -P O W s1N G1 N
01~O -Pwltw0 a1v0~O.P O N W C v0 .p.W J c~N N c!~N
~D0o O ~G0oW C1O O N ~ G~01cn O~ O W tn01w1N w C
_ _ _ v
~N aiJ ~ N ~ ~ ~ ~ V ~
G O O N t,N - OO~ J O N ~ O O
l o o n P O o
Nw 0 C~00~.oov0Oo-P.V~N ~1W 0o ~D w1 C~Cn011~
. G1Q1
V7 Q1 tJ~ (!~ Ch W ~.... c~ W tJ~ W cJ~ t!~ V1 N (,h V1 W CJ~
~ O .-~ .-. .p W .-. -P N J~. W ~ .P O .? V~ N .? ~ ~. .P 01 G1 0o G1
OVOO~o-P W ~OO~ W C.~nOGNI~~J~~~NW W~~~JOwo
O W -P N c.n Gv N w1 0o O ~l ~ a1 V~ N V»1 N W 00 00 N v0 01 Q1
O O~ N W w1 G1 O W O ~--~ N V»I ~ ~ W -P ? G1 N w1 00 N 01 01
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-59/6-
H n n n n n n n n n n n n n n n n n n n n
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
-.~ N w w ~p gyp. c~ a1 0~ a~ o~ J J o0 00 00 00 0o VD
.-. ~ ;.m-.... Q ~. n--. ,.r Q Q ,.r ~~. ~. ,r ,~. ~ ~.-. err... ~i--. i....~
O
.-~ tn W ~ ~D ~ O W O W O O W O ~ V~ O N W W o0
~-. 0o 01 w0 O Ov ~ 01 ~ ~ N c.n ~l ~ cn W N N 0o V~
o~ V V V V V V V V V V V V V V V V V V V V
n O ~ C~ls ~ ~ ~ ~ ~ C~J~ O
~ rn ~ ~: ~ ~ yn tn
O
n
i~. ~ ~. cro. ,~ ~' c~
c.
cu ~ ~. o x ~. ~ ~ o
~ o
p, ~ ~
a. o x ~ ~ o 0
o v, ,..,. ~~ d
O w
o. N x o~
cu
C7 ~ ~
~ cL n
0 o y
0
a
a.
0
., n '~'Wr
y o C7
n
o a.
N W N c~ ~ W W W G1 W G1 .p, o0
O ~ .p N 01 cr»O .p v0 ~ ~. w ~.-. 'p N w1 ~ O
O~ W oo v0 C J O ~ oo ~ v? ~ '~ cry C N cry :r
0o CWD w1 0o ~ w O O w N N w1 0o v~ 0o ~ N w1 .P
v~ w N ~ oo N cn W w .-.. .p N N O o0
w .p W o0 00 w .p ~ N O~ N w ~ O c.n w N a1 00
W 0o C1 tn t~.~ 0o G~ s1 s0 C C1 N C1 C ~ N o0 ~ 00
V7 .? W J W ~-.. N i-.. O CW W 0o W w1 i-' w1 ~--' .? ~ W
w1 N 01 N ~ W N ~ N 0o -P O 01 vD
w w ~-~ ~-~ ~--~ p~ -P N 0o ~D 0o N ~. ~l W v0 C1
N ~O N N N v0 C1 N v~ 0o C v1 00 :p J C1 cry N
G1 tn c.Im.-.~ ~O .P ~--' O O .-wl ~ ~.... c./ W 'G .P c.l~ W N
01 c.wl tn ~ W 01 ~-' .p ~l N G1 v0 w1
N w ~1 N ~O 01 ~1 00 ~t O ~ W ~1 ~O w O W o0
cn ~ .P OO ~''~ s1 :W O s1 N W OO OO Q O Cn N
01 .p ~ W ~ .yes 0o O O c,n ~ oo N -P w1 cm N Ow1 ?
c~ -P N N w1 N tn W .P O V~ W 01 W o0 00 w1
01 V1 00 W .p n~r 00 W ~ p1 ~ pp ~ ~ 00 (J1 00 n--W 1
tn 01 00 ~. O v0 00 v0 ~ C :p 00 W C~ ~O G1 :p, N
00 01 .p 00 -P .P N O W G1 -P 00 W ~-~ p1 ~ .p W 01 -P
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-59/7-
cn J cr, c~ ~ cr»o v, ~ oo w J tn J
N O V~ W .p C1 P c,lyD W N y0 00 y0 N C1 v0 a1
00 01 ~ N -P G1 00 0o O J N V 00 v0 ~ 0o ~ O
-" V C1 01 N W W O O V»1 N v0 C~ O~ Op Op cW p t,h
~ V1 C1 .p 00 tn 01 W Vt N O~ c.rWD ~ 00 C1 00
..~ t,m .~ N s1 v0 w oo O ;P O~ ~ 01 N
wCWlt,~nOINWOO~W N0~1~W0~1c.~ntmW
-~f-. O O w...O ,-..-
.-~
0oC1C10o cryCnW Ov s0c~J :p O oos1cm.1~
wtW V O w1w D C~ O w1O V~ N .pN N W O
~O wt0o V~N v0 cW . y1G1O 01W crO N o0
NO N ~ O J W O 0o W W ~ CvO v0~D.P
1
O00J . W O W ~ N O 00N V~W 00O V w
1
N01~ 01 ~ C101 ~ N vDW cm .pW 01N ~ Cn
.-.h.-.O O r-.~-. O O ~ ~-.~ ~-.m -..~--~ ..r
O J N ~ ~ ~ ~ ~ 00s0v~ i-.N ~ Cn N
N ~ W ~ ~ J "'v0
w1,-.C101 N w O V N O ~ ~ ~ C1O
WN cryV ~C~O.p W ~Oc~Vt~l w1N .pvCV 01
c.nw w1 O O N w w V~try~O O~ocO cr,N -P
w1V~00~ W m - 'O ~ 01.pN v001~ON 01vD
N N N i--~N i--~00 H...~--.~pN i--~w..~ ~-.r-.i-.
O.pJ N G1v000 w1 v1-PC W .PW "''W s1C
t
OJ _ ~ O ~ ~ N ~ ~''~~ ~D N O ~O.~C1O
00 ~O ~1 ~DO 01~1N W
~N ~ ~ _ O ~ N a'~t~'.P N ' 0oN c.nN
0l
o O c N v W v W ~ ~ J '"'G1.P
o o r~
No0w1Gv 00.1~~-- cry cry O100~O
O O m - r-..,..,W i.-......,m-. i.-~O ~.."..,,.rO
.~s0C1 v1~D~--~c~.os0~G
W N N N O ~
.- ~O CnN W O wlv0m --~c.nG1
Nw 01N O w ~l ~l O~O N tn ~1O w Cn-PO~
?w1c~N N v0O ~ N 0o~ ~O 1~w .~Cri w
WC~'CO v~O .iceJ O ~OO~O W ~lO W V O
N.poow ~l~ C~ oo w ~...p~~D Gv~ N ~ v O
f-.~-.i--.N ~... W r..f.-.c~ N .r~ ~.. p
-'~DC1W W ~''~""' vN :pW C '~ t,lWV C ~ ""~s0
vDO N ~ N c~C1 ~l 00w...,r
~O.p.N oo ~1~QC~ ~l w N O .p cmw .pW w o0
~OO N 01 O w1~ Oo -PN ~OO G1~ O~N w p
ON .IW W 0oO w1 N W ~ N .p,COOoO~O O~
1
J W -PW o0 V W ~-'01 ~--'0oW
1
...N ~.-.~--m-..w ~. J ~ ~ t~N N ..... .-..r
Vt~ J 00 ~.c!~C1 W ~ W i....N i~""'':y~.'"'G1O
..,-.W O -PO vW l~ W ~..rtn~ 00W OvN O W
N O oo w1~ O C1 ~ oot w1 OvG1w O tmw
W.1~O 00 N v0w1 G1 J 'OW N ~--~.p01~lN W
w1tnO 00 ~--~O 00 cJ~ O CJ~01O cr1 O N ~
!rW ~O~--~00C1~ ~D 0100rr00 .pCvc!~W ~ J
W~~P.~.NWONOW0~1WWONON~~WO
O V ~ N .p ~ c~ O G1 O O 01 ~O O~ ~ O 0o C~ N W
~O N W N c~ w ~O 01 O O~ J ~1 ~O 00 ~ 01 tn .P -P tn
N 00 N 0o ~O. O w1 w1 ~O C1 O vC w1 00 ~.... O 0o N o0
-' V J ~ c,n ~O ~C N .p V~ O O O O .1W1 O w1 O
SUBSTITUTE SHEET (RULE 26)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-60-
In this manner, we are able to correlate specific gene expression with the
exposure of a
cell to no, low (L) or high (H) amounts of an herbal composition. Many of the
genes identified
in this way code for proteins important in known metabolic or biochemical
pathways. Many of
these proteins have direct and indirect effects on certain physiological,
morphological and
psychological parameters: Thus, this method permits the association of a
particular genetic
fingerprint of an herbal composition with its array biological effects. Such
associations can be
used to profile or characterize an herbal composition for the purposes of
Quality Control and
Quality Assurance and evaluating pharmacological or toxicological properties.
The role of
primary and secondary herbs in an herbal formula can also be assessed by this
approach.
HPLC Anal.~is
The herbal batches were analyzed by HPLC with a Beckman ODS LTltrasphereTM
column (5 micron particles, 4.6 mm X' 25 cm) and detected with an W
spectrophotometer
(I'erl~in Elmer). The wavelengths for UV detection were monitored at 280 nm
and 340 nm. The
mobile phase was pumped at 1'ml/min and consisted of Solvent A: HZO and
Solvent B: 20%
MeOH with the following gradient: 1) the solvent was 100% solvent A for the
first 5 minutes;
2) the solvent composition Was changed to 10% solvent A l 90% solvent B for
the next 10
minutes; and 3) the solvent'was changed to 10% solvent A / 90% solvent B for
the next 40
minutes. This was followed by the addition of 100 % solvent A for 5 minutes.
The HPLC
marker is glycyrrhizin. '
A1 og rithm
The data collected form part of the multidimensional analysis used to generate
multivariant normal distribution sets as a means of determining a baseline
correlation between
biological activity and standard licorice molecular, chemical (HPLC and Mass
Spec), and
origin/growth characteristics.
B. Scute
Evaluation of Radix~Scutellaf~iae (Scutel
Scute has been found to be useful in reducing capillary permeability and
inflammation.
It can also be used treat eri~eritis''and dysentery, increases the secretion
of bile to treat j aundice;
to relieve muscle spasms; to~ treat coughizig and to expel parasites. The
properties of the scute
batches used in this example are presented in Table 11.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-61-
-Table 11. Batch Properties (Scute)
Property Batch A Batch B Batch C Batch D
.
Plant NameScutellariaeScutellariaeScutellariaeScutellariae
~ radix radix radix
radix
Origin Sanxi Province.U.S., Kin U.S., Kin U.S., Boston
Man Herb Man
Center Herb Center
PreparationStandard Boiled, 30 Warm H20, Soiled , 2
method : , min 30 hours
. . min.
Plant partRoot - - -
Biological and Enz~tme Assavs
Briefly, one gram of each preparation of scute extract was added with 10 ml of
water (1
rng/ml). The mixture was treated as Outlined in Table 11. The supernatant was
collected after
centrifugation and filtered through a 0:22 p,m filter. Batches of scute were
tested against either
HepG2 cells (ATCC cat #~HB-8065) or Jurkat T cells (ATCC cat #TIB-152) or
both. One to
fifty dilutions were used for each assay. Cells were cultured for 24 hours as
described above.
Batches were also evaluated for the ability to inhibit hepatitis B virus as
detected by
DNA quantitation (see Dong et al.; Proc Natl Acad Sci USA (1991) 88: 8495-
8499). Briefly,
one gram of preparation was added with 10 ml of water. The mixture was treated
as outlined
in Table 11. The supernatant was collected after centrifugation and filtered
through a 0.22 ~m
filter.2.2.15 cells which secrete hepatitis B virons (kindly provided by
Professor G. Ace; see
Ace et al. Proc Natl Acad S'ci~USA'~~(1987) 84: 1005-1009) were used in this
assay. One to fifty
dilutions were used for each assay_ ' The cell growth inhibition assay was
performed for 72
hours. All other procedures were performed as' described by Dong et al., Proc
Natl Acad Sci
LTSA (1991) 88: 8495-8499:
For (3-glucuronidase, different scute extracts were added to triplicate wells
of a 96-well
plate which contained O.lmM phenolphthalein glucuronidate, 70 mM Tris-HCl (pH
6.8) and
0.8 ng of dialyzed [3-glucuroriidase (from E. Coli, purchased from Sigma) to a
final volume of
80 ~tl. After 2 hr incubation.at 37°C,~the reactions were terminated
with 200 ~.1 of stopping
solution which contained 0.2 M Glycine and 0.2 M NaCI (pH 10.4), and the OD
was
monitored with a kinetic microplate reader at 540 nm.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-62-
The results of the assays using the
three batches is displayed in Table
12.
Table 12. Biological Assay of Four arations
Prep of Scute*
. . "'E. Coli HepG2 Jurkat HBV$ DNA
(3-Glucuronidase
Scute A ~..,1.5,. 0.33 0.45 None
Scute B ~ ' ; ' l., g ~,. ; ,' , ND ND
. ND
Scute C ' ~ ' . - "Ø3 ' ~ ND ND ND
Scute D ND " 0.65 ND 27.5
*Values represent $, % of Control
ICS° values.
ND, not determined. ' e.
Evaluation of Scute Effects orl~Protein Expression
HepG2 cells (1 x' 10~) were seeded in 25 cm2 flasks in 3.0 ml of RPMI-1640
medium
(see Life Technologies, Inc.,~Gatalogue and Reference Guide, 1998-1999, Cell
Culture section)
24 hr before the extract' addition. The cells were treated with or without
herbal medicine,
where the former is added at.two final concentrations of 0.2 mg/ml or 4 mg/ml,
respectively,
and~incubated at 37°C for 24~hours: The medium was removed and the
cells were washed
twice with cold PBS. The cells were harvested into 1 ml of PBS and centrifuged
at 10,000 rpm
for 2 minutes, extracted on ice with a buffer containing 50 mM Tris-Cl (pH
7.5), 0.2 mM
PMSF and 10% glycerol, 'followed by three freeze-thaw cycles. Potassium
chloride was added
to the cell lysate at a final concentration of 0.15 M prior to centrifugation.
The protein
concentration was detertTiirieei'arid,the cell extract was electrophoresed
according to the method
of Laemmli U.K. (Nature (1970 227:680-685): Western blots were performed by
standard
techniques knowr'i in the art, see for example Sambrook, et al (1989). The
antibodies used
were directed to the folloWing'proteins: Topo I; Stat (20707); Cyclin B1; MAPK
(l-~b2) and
Nm 23 H1.
Figure 4 demonstrates that scute batches A and B do not differentially affect
the
expression of the polypeptides~resolved on Western blots.
HPLC Anal, ' . . . .
The herbal batches wefeyanalyzed by'HPLC with a Beckman ODS UltrasphereT'~ ,
column (5 micron particles, ~.~6' ri~xri X 25 cm) and detected with an UV
spectrophotometer
(Perkin Elmer). The wavelengths for U~ detection were monitored at 280 nm and
340 nm. The
mobile phase~was pumped~at'1 iril/rriin and consisted of Solvent A: HaO and
Solvent B: 20%
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-63-
lVIeOH with the following grac~iemt: 1) the solvent was 100% solvent A for the
first 5 minutes;
2) the solvent composition was changed to 10% solvent A / 90% solvent B for
the next 10
minutes; and 3) the solvent was changed to 10% solvent A / 90% solvent B for
the next 40
minutes. This was followed by the addition of 100 % solvent A for 5 minutes.
The HPLC
markers are baicalin and baicalein. .
Scute batches in water and acid treated samples were analyzed by HPLC. Water
and
acid treated batches were virtually indistinguishable.
Al o
The data collected form part of the multidimensional analysis used to generate
multivariant normal distribution sets as a means of determining a baseline
correlation between
biological activity and~standard scute chemical (HPLC), and origin/growth
characteristics.
C. White Peon, Root'''' ' I ~ .
Evaluation of Paeorcie lacti of°a callus radix (Peony)
Peony is used to suppress ~amd soothe pain. It is also known to soothe
ligaments and
purify the blood. The properties of the peony batches used in this example are
presented in
Table 13.
,' ~ , ~,able~ l3. Batch Properties (Peony)
Property ' ~ 'Batch A Batch B
Plant Name Paeoiiie lact~o~aPaeonie lact~ora pallus
pallus
Origin Anwey,~'rovince iJ.S., Boston
"
Preparation Standard'" Boiled 2 hours.
method :~';
Plant part . Root ' ' . . Root
Biological and Enzyme Assays
Briefly, one gram of each preparation of scute extract was added with 10 ml of
water (1
mg/ml). The mixture was treated .as outlined. in Table 13. The supernatant was
collected after
centrifugation and filteredvthrough a 0.22 ~,m filter. Batches of peony were
tested against either
HepG2 cells (ATCC cat # HB-8065) or Jurkat T cells (ATCC cat #TIB-152) or
both. One to
fifty dilutions were used fore each assay.' Cells 'were cultured for 24 hours
as described above.
Batches were also .evaluated for the ability to inhibit hepatitis B virus as
detected by
I~NA quantitation (see Dong.~t~al:,°,Proc Natl Acad Sci USA (1991) 88:
8495-8499).
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-64-
..
Briefly, one gram of preparation was added with 10 ml of water. The mixture
was
treated as outlined in Table 13. ~~The supernatant was collected after
centrifugation and filtered
through a 0.22 ~.m filter: , 2.2,:15 cells~which secrete hepatitis B virons
(kindly provided by
Professor G. Ace; see Ace,et al. Proc Natl Acad Sci USA (1987) 84: 1005-1009)
were used in
this assay. One to fifty dilutioris were used for each assay. The cell growth
inhibition assay
was performed fox 72 hours~:~ All~other procedures were performed as described
by Dong et czl.,
Proc Natl Acad Sci USA (1991) 88: 8495-8499.
Different peony extracts were added to triplicate wells of a 96-well plate
which
contained O.lmM phenolphthalein glucuronidate, 70 mM Tris-HCl (pH 6.8) and 0.8
ng of
dialyzed beta-glucuronidase (from E. eoli, purchased from Sigma) to a final
volume of 80 p,1.
After 2 hr incubation at 37°C.,-the reactions were terminated with 200
~.l of stopping solution
which contained 0.21VI Glycine :aiid 0.2 M NaCI (pH 10.4), and the OD was
monitored with. a
kinetic microplate reader at: 540 nm. Results are shown in Table 14.
Table 14. Biological Assay of Two Preparations of Peony*
''; : E. Cola , IIepG2 Jurkat I3B~l~
(3=Glucuronidase
Peony A . 2.8 ~ >1.5 1.1 None
Peony B >2.5 ' ND ND ND
*values represent$, % of Control
ICS
values. . '
ND, not determined.,
HPLC Analysis -
The herbal batches were analyzed by HPL,C with a Beckman ODS Ultrasphere
column
(5 micron particles, 4.6°mm X 25 cin) and detected with an UiT
spectrophotometer (Perkin
lElmer). The wavelengths for UV detection were monitored at 280 nm and 340 nm.
The mobile
phase was pumped at 1 ml/rinri arid~consisted~ of Solvent A: Ha0 and Solvent
B: 20% MeOH
with the following gradient::l).thevsolvent was 100% solvent A for the first 5
minutes; 2) the
.. . . . :. ..,. .
solvent composition was changed to 10% solvent A / 90% solvent B for the next
10 minutes;
and 3) the solvent was changed to 10% solvent A / 90% solvent B for the next
40 minutes.
This was followed by the addition~of ~l~0 % solvent A for 5 minutes. HPLC
marker is
paeoniflorin.
Peony batches were analyzed~by HPLC as shown in Figure 5.
A1 o_g rithm_
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-65-
The data collected foim part-of the multidimensional analysis used to generate
multivariant normal distribution sets;as a means of determining a baseline
correlation between
biological activity and standard peony chemical (HPI,C), and origin/growth
characteristics.
D. Date
Evaluation of Zizi~hi F~uctics (Date)
Date has been used fondiuretic properties and strengthening effects. The
properties of
the date batches used in this example~are presented in Table 15.
,. . .
Table 15. Batch Properties (Date)
. . . ,.. .
Property ~ Satch 'A~~. ~ Batch B Batch C
~
Plant Name Ziziphi F,ructus.~Ziaiphi FructusZiziphi Fructus
Origin Hebe'i Province.U.S., Kin Man U.S., Kin Man
Herb Herb
Center Center
Preparation Standard',. Boiled, 30 Warm HZO, 30
method . , ~ min min.
Plant part Fruit . ~ - -
, ~
Biological and Enz~nne Assays
Briefly, one gram of each batch of scute extract was added with 10 ml of water
(1
mg/rill). The mixture waswtreated as~:outlined in Table 15. The supernatant
was collected after
centrifugation and filtered through a 0.22 ~,m filter. Batches of date were
tested against either
HepG2 cells (ATCC cat#;HB-8065) or~Jurkat T cells (ATCC cat #TIB-152) or both.
One to
fifty dilutions were used,for-each, assay. Cells were cultured for 24 hours as
described above.
Batches were also evaluated for the ability to inhibit hepatitis B virus as
detected by
DNA quantitation (see Doug et~al., Proc Natl Acad Sci USA (1991) 88: 8495-
8499). Briefly,
one gram of preparation was added with.l0 ml of water. The mixture was treated
as outlined in
Table 15. The supernatant was collected after centrifugation and filtered
through a 0.22 ~,m
filter. HepG2.2.15 cells which secrete hepatitis B virons (kindly provided by
Professor G.
Ace; see Ace et al. Proc N~,tl Acad Sci USA (1987) 84: 1005-1009) were used in
this assay.
One to fifty dilutions were used for each assay. The cell growth inhibition
assay was
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-66-
performed for 72 hours. ~ All other procedures were performed as described by
Dong et al., .
Proc Natl Acad Sci USA (1991) 888495-8499.
Different peony eXtzacts were added to triplicate wells of a 96-well plate
which
contained O.lmM phenolphthalein.glucuronidate, 70. mM Tris-HCl (pH 6.8) and
0.8 ng of
dialyzed beta-glucuronidase (from E. Coli, purchased from Sigma) to a final
volume of 80 ~.1.
After 2 hr incubatiomat~37°C, the. reactions were terminated with 200
~,1 of stopping solution
which contained 0.2 M Glycirie arid 0.2 M NaCI (pH 10.4), and the OD was
monitored with a
kinetic microplate reader at 540 iim. Results are shown in Table 16.
Table 16. Biological Assay of Three Preparations of Date*
E: Coli HepG2 Jurkat HBV$ DNA
(3-Glucuronidase
Date A ~1~:2 ~ 1.5 v 5.1 None
. . . ',
Date B ~ ND >2.0 ND 52.3
Date C r 2.5 ~ ND ND ND
*Values represent$, % of Comtrol
.
ICS values. . ' ,
ND, not determined.;, ,
I0 HPLC Anal, sis
The herbal batches.were analyzed by HPLC with a Beckman ODS Ultrasphere column
(5 micron particles, 4.6 mm X.25 cm) and detected with an UV spectrophotometer
(Perkin
Elmer). The wavelengths fb~ UV detection were monitored at 280 nm and 340 nm.
The mobile
phase was pumped at l~ml/min.and consist of Solvent A: H20 and Solvent B: 20%
MeOH with
the following gradient: 1) the solvent was 100% solvent A for the first 5
minutes; 2) the
solvent composition was changed to f0% solvent A / 90% solvent B for the next
10 minutes;
and 3) the solvent was changed to 10% solvent A / 90% solvent B for the next
40 minutes.
This was followed by the addition of.100 %~ solvent A for 5 minutes. HPLC
markers for date
are chelidonic acid and cAlVIl':~ '
Date batches samples were ~.nalyzed by HPLC as shown in Figure 6.
Al og n
The data collected form part of the multidimensional analysis used to generate
multivariant normal distribution sets as a means of determining a baseline
correlation between
biological activity and standard peony chemical ~(HI'LC), and origin/growth
characteristics.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-67-
Example 15. Characterizing Herbal Medicines b~~lVucleic Acid Microarra~
Anal~ s~-is. .
Introduction.
The rapid development of nucleic acid microarray technology has led to an
explosion
of gene expression data, (Larder, 1999, Duggan et al., 1999). Four
characteristics of the gene
expression account for the great value of using nucleic acid microarrays to
study the gene
expression profiles. (i) Nucleic acid microarray makes it easier to measure
the transcripts of
thousands of genes at once. (ii) Close association between the function of a
gene product and
its expression pattern makes gene function predictable. (iii) Cells respond to
the micro-
environmental changes by changing the expression level of specific genes. (iv)
The sets of
genes expressed in a cell. determine,what the cell is derived of, what
biochemical and
regulatory systemsare involved; and so on (Brown and Botstein, 1999). By using
a microarray
system, the above features can be studied in an ensemble manner.
The expression of any. desired number of genes can be detected using the
nucleic acid
microarray technology: 'For' example, up to about 20;000 genes may be placed
on a single
array. i~Ve have developed a nucleic acid microarray with colorimetric
detection system
(Microarray/CD) (Chen ~et al., 1998). Gene expression profiles of different
cell lines were
studied using microarray filter'rnembranes (2.7 cm x 1..8 cm) with about
10,000 cDNA
representing approximately 10;000: distinct human transcripts. The sensitivity
and detection
limits of the microarray/CD 'system have been characterized and are comparable
to the system
with radioactive detectiori'~r'tfie system with laser induced fluorescence
detection (Bertucci et
al.; 1999). ~f; ;.; ~, , . , ,
As previously descriliec~', cellular gene expression profiles portray the
origin, the
present differentiation of ~tlie cell; and the cellular responses to external
stimulants. In other
words, the gene expressiori~.profiles reveal the, state of the cell and
microarray is a perfect tool
for the rendering purpbse. 'In'tlie present 'studies, we apply the
microarray/CD system to
characterize cellular responses ~to external stimulants, in this case, the
Chinese herbal
medicines. Conversely, we also based on the stimulated gene expression
profiles to classify
different herbal mediciries:e'
Figure 7 is a flowchart depicting a general method that may be used for
establishing an
expression response data~set for~cells treated with an herbal composition. The
method
comprises the steps of
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-68-
(a) Determine the' ICSO concentration of~an herbal composition by incubating
various
concentrations of the herbal medicine in mammalian cell cultures and identify
the
concentration that leaves 50% of survival cells after a predetermined time.
(b) Incubate the mammalian cell 'cultures with herbal extracts of various
fractions of
;. .
ICSO concentrations:
(c) Harvest and count the~cultured cells after a predetermined culture time.
(d) Immediately lyse the cells after they are removed from the incubator and
extract
mRNA from cell lysate.
(e) Label the mRNA byreverse transcription reaction to turn mRNA into labeled
cDNA.
(f) Mix the labeled cIDNA with control cDNA of plant origin and perform
hybridization
to a microarray of mammalian gene probes.
(g) Measure expression ;level of genes by 'analyzing digitized images of the
microarray
hybridization results:
(h) Perform data pre=processing' to select data for statistical analysis.
(r) Acquired'expression data generated by microarray experiments of an herbal
composition with various concentrations.
(j) Data pre-processing to select the genes with statistical significance in
cells treated
with different concentrations of the herbal medicine.
(k) Categorize expression profiles into clusters by statistical methods such
as the self
organizing-map algorithm:
(1) Deduce the characteristic expression profiles for the herbal medicine
based on the
expression profile clusters.
Figure 8 is a flowchart demonstrating how data sets of expression data for
various batches of
the herbal compositiow are integrated to make an expression profile database
for the particular
herbal composition. The expression profile database then becomes part of the
HBR Array.
HBR,Arrays contaiiiirig expression profiles may also be used to identify an
unknown
herbal composition. Figure 9 is a'flowchart depicting a general method for
identifying an
unknown herbal composition, the method comprising the steps of
(a) Construct an HBR Array containing characteristic expression profiles for
an herbal
medicine or a collection of expression profiles. of various herbal medicines
by the
..
aforementioned steps. '
(b) Obtain the characteristic expression prof 1e data set of the unknown
herbal
composition.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-69-
(c) Compare the HBR Array~~coiitaining the characteristic expression profile
induced by
the said unknov~m herbal composition with a standardized HBR Array containing
expression data by~algorithms such as the Hamming distance algorithm.
(d) Score possible alignments to identify the most probable herbal composition
whose
characteristic expression profiles are archived in the said HBR Array
Scoring possible alignments of HBR Arrays containing expression profiles may
be performed
using hierarchical cluster analysis of the Hamming distance matrix. ~Jse of
hierarchical cluster
analysis for the Hamming distance matrix is well known in the art.
The gene expression profiles may also be incorporated into the standardized
HBR Array.
As has been already discussed, the standardized HBR Array containing such gene
expression
profiles induced by an herbal composition can be used for studying the
pharmacological
~.
mechanisms of the herbal; composition, for discovering new application of the
herbal
composition, and for designing optimized formulation of a complex herbal
preparation. As can
be seen from the flowchart.of Figure I0, 'the method may be generally outlined
as comprising
the steps of
(a) Construct a data set coritairiing the characteristic gene expression
profiles for an
herbal composition:'
(b) Score each gene li"'y the consistency of its expression profiles in the
data set using
known statistical liarameters, such as the coefficient of variation.
(c) Based on the statistical scoring, gene expression profiles for an herbal
medicine are
selected to be incorporated into the standardized HBR Array.
HBR Arrays containing-gene~expression profiles may also be used to identify
signature
gene expression profiles Yinduced; b~y individual chemical constituents in an
herbal composition
consisting of complex chemical 'constituents, as outlined in the flowchart of
Figure 11. The
method comprises the steps. of
(a) Construct a HBO Array. containing characteristic gene expression profiles
for an
herbal compositidn~by'~the aforementioned steps.
(b) Determine the cornpositiori of chemical constituents in an herbal medicine
by high
performance liquid cl~om~.tography (HPLC) or liquid chromatography mass
spectrometrsr (LC-MASS).'
(c) Repeat the step (1i) for'varibits batches of herbal medicine preparations.
(d) Score the correlat'ioti'coefficients between the expression levels of each
gene with the
amount of individual chemical constituent in an herbal preparation.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-70-
(e) The signature gene expression profiles for individual chemical constituent
are selected
with a Pearson correlation coefficient exceeding 0.99 or smaller than -0.99.
Any herbal composition can then be characterized through the use of gene
expression
profiles generated through the use of nucleic acid microarrays. Moreover, one
can choose any
number of genes that "are differentially expressed to be included in the data
set represented the
gene expression profiles. !For example; one may choose about 10 genes, about
100 genes,
r ".
about 500 genes, about"1000 genes, about 1500 genes, abut 2000 genes, about
2500 genes or
more, or any number in between.
The prescription of the" Chinese herbal medicine Scute and Licorice
combination
(Huang Chin Tang) stops diarrhea, relieves spasms and clears fever. The
ingredients of Huang
Chin Tang are Scute, .Peony, "Licorice and Jujube. This recipe has been used
for more than
1000 years but the clieriiical and biomedical studies oy the prescription have
not been carried
out until recent decades.;Iri~this study we used the nucleic acid microarray
technology to study
the gene expression profiles'~of'.h'erba'1 medicines treated cells. Our aims
are to demonstrate the
feasibility of using the rriicro'array/CD system for classification of
different herbal
compositions or different'prepaxations and to find the predictor genes (marker
genes) for the
Huang Chin~Tang prescription. The lomg-term goals are to find the correlation
of the
biochemical 'ingredierits'in ~eacli herbal composition with the gene
expression profiles of
various treated cells arid to decipher the molecular pharmacological
mechanisms of the
Chinese herbal medicines ima~rational fashion.
Materials and Methodse ~ ~ " ~ '
1. I)e~ei~pnnent of;'a'vcell banking systegn. ,
Purpose: ' Microarray,~ystem is a sensitive detection method to monitor gene
expression
patterns of:cells"It' is necessary "to build a Cell Banking System with a
Master
Cell Bank (MCB) '.and a Working Cell Bank (WCB) to minimize cell variability
for herbal medicine testing. .
Scope: The Cell Baxik System is used for all types of cells in microarray
studies.
Apparatus: " C02 Air-Jacketed~Incubator (NUAIRETM DH autoflow)
Centrifuge (KI~BOTA 2100)
Freezingwial~:(Coi'ning Costar, Cat. #430659)
Tissue culture flask 750 ml (Falcon, Cat. #3045)
Tissue culture dish "150x25 mm (Falcon, Cat. #3025)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-71-
Cell: Jurkat T cell~~from~Dr: Alexandra Ho
Reagents: RPMI Medium 1640 (GIBCO BRL, Cat. #31800-014)
Dimethyl Sulphoxide (DMSO) (Sigma, Cat. #D-2650)
Fetal Bovine Serum (HyClone, Cat. #5H30070.03, Lot#AGL7258)
2-mercaptoethanol (GIBCO BRL, Cat. #21985-023, SxlO-Z M)
Media: I. Culture medium: 90% RPMI + 10% fetal bovine serum + 2-
mercaptoethanol (5x10-5 M)
II. Freezing medium: 90% RPMI 1640 + 10% DMS~
Procedure:
A. Master Cell Bank
..
1. Follow the standard sterile procedure of cell culture.
2. Seed Jurkat T cells'in culture medium in flask at 37°C with 5% COZ
incubator.
3. After incubation for two days, count the cell number and spread the cells
to two flasks.
Note. Cell density 1s kept a°~~out 5x104- 2x10 per ml.
4. Culture and count the cell riuinber until~the number reaches 2x107.
5. Collect the cells iri'S0'ml centrifuge tubes and spin at 1300 rpm (300~g)
for 5 min.
6. ' Discard the supernatant and re-suspend the cell pellet with chilled
freezing media. The
cell number in eachJvial is about fx106 per ml.
7. Slowly freeze the cells:by the following temperature profile: -20°C
for 2 hr,
-80°C for 24 hr and then place the cells in liquid N2 storage. Store a
total of 20 frozen
vials in MCB. , . ',
B. T~Yorkihg Cell Bank. ' . ' t .
1. Retrieve one vial of ~ell~ from MCB in liquid N2 tank and quickly thaw at
37°C water
bath. ' ; y '
2. Transfer the cells into ,1,0 ml of warm culture medium.
3. Spin down the cells at 1300 rpm ~(300Xg) for 5 min. Discard the
supernatant. Culture
the cells with 20 ml 'medium in a flask. '
4. Sub-culture the cells to~ 2 flasks. ~ .
5. Seed 5x107 cells with 500 ml culture medium in each flask with stirring for
a total of 2
flasks. Culture the'cells~for 2 days.
6. Culture until the cell density reaches 1x1061rn1 arid a total volume of 1
L.
7. Prepare freezing media by adding 100 ml of fetal bovine serum and 10 ml of
DMSO.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
.72_
8. Centrifuge, discard the supernatant and re-suspend the cells to 110 ml of
freezing
medium. . . '
9. Dispense 1 'rnl to every freezing vial (10 million cells per vial) for a
total of 100 vials.
Slowly freeze the cells by;the temperature profile described above.
2. Determination
of growth
inhibition
concentration
of herbal
extract in
cell
cultures.
Purpose: Most drugs are toxic to cells. This experiment is
designed to examine the
toxicity of herbal extracts in Jurkat T cells and
to determine the growth
inhibition concentration of herbal extracts that keeps
the cells alive.
Scope: This assay'can be used in all kinds of herbal extracts
to examine the toxicity.
Apparatus: COz Air-Jacketed,Incubator (NUAIRETM DH autoflow)
Counting chamber (Hemacytometer, Reichert, USA)
Microscope (Zeiss~ .A~ciovert 100)
Cell: Jurkat T cell
Reagents: RPMI Medium 1640 (GIBCO BRL, Cat. #31800-014)
Fetal Bovirle~:Serum'(HyClone, Cat. #5H30070.03, Lot
#AGL7258)
2-mercaptoetlianol (GIBCO BRL, Cat. #21985-023, SxlO-2
M)
Culture media: 90% RPMI + 10% fetal bovine serum +
2-mercaptoethanol
(5x10-S M) , _ ~ ~ '
Disposable sterile syringe filters. (0.2 m, Corning,
Cat. #21052-25)
herbal
extracts:.:
. ,
1. Cordyceps Sinensis Mycelium.'
2. ST 024:
3. ST 044:
4. ST 051:
5. ST 093:
6. ST 117:
7. ST 123:
8. ST 128e
9. ST 134:
10. ST 237:
11. PH~.'906-303503: Complex~mix composed
of 4, 6, 7, 10
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
_73_
12. P~IY906-284003: Complex mix composed of 4, 6, 7, 10
Procedure: ,
A. Herbal Extract Prenarataoh
1. Dissolve 1 gram of herbal powder in 10 ml of 80 °C deionized water
(neutral pIi) in a
polypropylene tube.
2. Incubate the tube at'80 °C water bath for 30 minutes with gentle
shaking then
centrifuge at 4000 rpril (1500~g) for 5 min to obtain the supernatant.
3. Centrifuge at 11000 rpm (14000~g) for 10 min to collect the supernatant.
4. ZJsing the disposable sterile syringe filter to filter the supernatant.
B. Cell Survival Test
1. Culture Jurkat T cells as described above.
2. Dispense 1 ml of Sx105Ym1 cells per well to 24-well culture plates.
3. Prepare 12 kinds of herbal extract solutions. The extract solutions must be
freshly
prepared and used~iinmediately.
4. Add 100, 50, 20,10, 5 ,~,1 of each herbal extract solution into the 24 well
culture plates
to get the five different concentrations: r10, 5, 2, l, 0.5 mg/ml.
5. Culture the cells for 24 h at 37°C W an incubator filled with 5%
C02.
6. Count the number of cells per well. Mix 10 ~,1 of cell solution with 10 ~,l
of Trypan
blue dye and load irito~~cell counting chamber.
7. Count the four major square areas to calculate the cell number.
(number of cells in 4 areas)/4 x,104 x dilution factor = number of cells per
ml
3. Profiling gene~expressiori patterns of Jurkat T cells treated with herbal
... :.. ,
extracts.
Purpose: Profile the gene expression patterns of Jurkat T cells treated with
herbal
extracts. A high-density nucleic acid microarray with colorimetric detection
system is used:
Apparatus: Heat block (Boekel, lVIode1 110002)
Spectrophotometer (Beckman, D~T640)
Centrifuge.(KIIBOTA 1910)'
Water bath (SI;M.~AMINCO, Model 800)
I~ybridization incubator (YI~I DER OIi-800)
Heat sealer'(TISH-300, TEW)
Reagents: RNAzaITM B (Tel=Test, Cat. #CS-104)
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-74-
Oligotex rriRNA Midi Kit (Qiagen, Hilden, Germany)
Hybridization Bags,(GIBCO BRL, Cat. #18278-010)
EasiSeal (Hybaid,'Cat: #HBOSSSEZlE)
Glass slides (Matsunami, 52214, Japan)
Aerosol Resistant Tips (ART'tip) (molecular BIO-Products, Cat. #2139)
Random hexamer primer (GIBCO BRL, Cat. #48190-Ol 1)
Reverse transcriptase and 5~ buffer (GIBCO BRL, Cat. #18064-014)
RNase inhibitor (GIBCO BRL, Cat. #10777-019)
Biotin-16-dUTP (Boehringer Mannheim, Cat. #1093070)
Dig-11-dUTP (Boehringer Mannheim, Cat. #1558706,)
,~
Blocking powder for hybridization (Boehringer Mannheim, Cat. #1096176)
Bovine.seriim albumin, (Sigma, Cat. #A2153)
., ., ,
20X SSC (Arizr~sco, Cat.#09185-2-20XPTMSL)
SDS (Merck,~Cat.#1'13760)
Dextran Sulfatev(Sigma,~ Cat. #D6001)
Streptavidin=(3'=galactosidase (GIBCO BRL, Cat. #19536-010)
..
Anti-digoxigeriirl-AP Fab fragments (Boehringer Mannheim, Cat. #1093274,)
X-gal (GIBCO BRL, Cat: #15520-018)
Malefic acid (Sigma; Cat. #M1125)
~ N-lauroylsarcosiri (Sigma, Cat. #L5777)
Fast red TR/AS~MX substrate kit~(PIERCE, Cat. #34034)
Polyethylene glycol (Sigma, Cat.#P2139)
Reagent Preparation:
1~ hybridization buffer (4X SSC, 0.1% N-lauroylsarcosine, 0.02% SDS, 1% BM
bl~cking reagent) ~ ~ ~ ~ - . ' ,
20X SSC ~ ~ ' , - ~16 m
I % N-lauroylsarcosine 8 ml
IO% SDS '. , . ~ 160 ~.tl
BM blocl~ing powder ' ' . ' 0.8 g
Ha0 ." . 51"ml
total 80 ml
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-75-
Heat to 65°C to dissolve the powder then store at -20°C.
50% PEG-8000 (Polyethylene Glycol)
PEG-8000 ~ , 10 g. - .
H20 up ~to ' 20 ml
Heat to 65°C to dissolve then autoclave..Aliquot and store at -
20°C.
10~ TBS (100 mM Txis, 1:SM'NaCI, pH 7.4)
Tris base ~ 12.1 g '
Na.CI ' 87.6 g
H20 up to 1000 ml'
120 mM X-gal
X-gal' 100 ing.' ' ~: ;,,
DMF 2 ml '
Store at -20°C. , ' '
X-gal Substrate Buffer {lmlVI MgCl2, 3 mM K3Fe(CN)6, 3 mM K4Fe(CN)6 in 1X TBS
buffer) . S00 ml
lOX TBS/pH7.4 , ' ' S0 ml
Potassium Ferrocyanide 633.5 mg
Potassium Ferncyariitle 493.9 mg
MgCla ~ , 101.6 rng
Filter and store at -20C.
BM Blocking Dilution Buffer/pH7.5
(0.1 M malefic acid, 0.15
M NaCI)
'
' ~ .' 100 ml
1M Maleic~Acid
SM NaCI ' ~ ~ ' 30 ml
Solid NaOH ' '7.5 'g
H20 ~ ~ up to 1000 ml
10% Blocking lteagent~ 100 ml
Blocking Powder ' 10 g
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-76-
Blocking Dilution Buffer (no tween 20) 100 ml
Heat to 70°C then autoclave: Store ~at 4°C.
20% Dextran Sulfate''
S Dextran Sulfate ~ v ° 2 g
Ha0 ~- up ao ~ 8 ml
Autoclave then store at -20°C.
DEPC-treated water 800 ml
Diethyl Pyrocarbonate (store at 4°C) 400 ~l
H20 , 800 ml
Put in 37°C shaking water bath for 4 hxs and then in 37°C warm
room for overnight. Autoclave
the solution for 4S minutes or-2S eininutes each for two times.
Procedure:
1 S A. P~e~at~atioh of'Jurkat 'T Cells
1. Thaw one vial of Jurkat T cells. Transfer to 10 ml growth medium. The
number of vials
to thaw depends on the 'number of test to perform. In general, one vial of
cells is needed
for 2 herbal extract tests.
2. Re-suspend the cells in SO ml of culture medium.
3. Incubate the cells f~r one day. Add lSOvml of culture medium and divide
into two
flasks with 100 ml each.
4. Culture the cells for 3 days.
S. Change the medium end distribute 4 flasks with 100 ml of medium each.
6. Culture the cells for ~2 'days. '
2S 7. Count the cell number. Collect the cells and spin down. Ize-suspend the
cell pellet with
culture medium~to,Sx1.05 cells per ml and 100 ml per flask.
8. Culture the cells for 3.hrs before adding the herbal extract. '
~'. ~Ie~bal extract treatment''
1. Prepare the herbal extract (see herbal extract preparation).
2. According to the growth inhibition concentration of herbal extract
determined by the
cell survival experiments, c~.lculate the'SO% growth inhibition concentration
of each
1 herbal extract. .
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
_77_
3. Define the 50% growth inhibition concentration as H. Treat the cell with
serial dilution
of herbal extracts with the following concentrations: H, H/2.5, H/5, H/10,
H/20.
4. Culture the cell for 24.~hrs.
5. Collect cells and count the number of cells. Centrifuge to obtain cell
pellet.
6. Wash the cell pellet with 1 XPBS once.
7. Discard the supernatant. The cell pellet is ready for total RNA isolation.
C. Isolation of total R A , ~ .
1. Add 1 ml of RNAzoIT~ B per 10' cells. Homogenize the cell pellet but do not
vortex.
2. Add 0.1 ml chloroform per ml of homogenate, cover the samples tightly,
shade
vigorously for 1 min°,(do not vortex). Place on ice for 15 min.
3. Centrifuge at 12000. rpm (13500~g) at 4°C for 15 min.
4. After centrifugation, tliehoriiogenate develops two phases: a lower blue
phenol-
chloroform phase and a colorless upper aqueous phase. DNA and proteins are in
the
interphase and the organic phase. Transfer the aqueous phase to a new tube,
add an
equal volume of isopiropanol' and store the samples at -80°C. Note. The
range of
isopropanol additioi~~'ir'~rorn 0.7 tb 1 volume of the aqueous phase solution.
5. Keep the samples at'=~0°C',uritil use: Let the sample completely
thaw before
centrifuging and mix 2 to 3 times by inverting the tube. Centrifuge samples
for 15 min
at 13000 rpm' (15000~g). ~ '
6. Remove the~supernatant'and wash the RNA pellet once with 1 ml of 75%
ethanol.
Centrifuge for 3'rriiii at' 13~OOO ipm (15000Xg) and at 4°C.
7. Discard the~supernataiit. I~ry the pellet under vacuum for 1 min. Note. Do
not Iet the
RNA pellet diy coriipletely.rIt will greatly decrease its solubility.
8. Dissolve the RNA pellet iri X50-100 p,1 of diethylpyrocarbonate (DEPC) -
treated water
by pipetting. Note. ~If tlie'pellet is hard to dissolve, incubating the pellet
for 10 - 15 min
at 60°C may help': 5:, .,, ; : , , , , ~~ a
v':
9. Measure absorbance ~~.~ 260'iun ~(Aiso) and 280nm (A28o) with a
spectrophotometer.
Concentration analysis:' OD26o x 40' ng/p,l X dilution factor = total RNA
(ng/~,l).
D. Isolation ofRolv A~+ mRNA fYOm total RNA p~
1. Determine the amount of starting RNA and the appropriate volume of Buffer
OBB and
Oligotex Suspension ~solutiori to be added in the RNA solution according to
the Table
17.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
_78_
Table 17. Buffer amounts for Oligotex mRNA Spin-Column Protocol.
Total RNA Add RNase-free Buffer Oligotex Prep
vaster to: (~,l)OBB Suspension size
(~,l) (~,1)
20 ~,g 100 100 6 Mini
0.25 mg 250 . . ~ 250 . 15 Mini
0.25-0.5 mg 500 500 30 Midi
0.5-0.75 mg 500 500 45 Midi
0.75-1 mg 500 : 500 55 Midi
The following procedures are based on using 500 ~,g total RNA as an example.
2. Add 500 ~.1 of 2X Binding Buffer and 30 ~,l Oligotex Suspension to the
total RNA
sample. Mix the contents thoroughly by flipping the tube.
3. Incubate the sample for 10 min at 70°C.
4. Incubate for. 20 rriin, at room.temperature.
5. Centrifuge for 2 min -at maximum speed (14000 to 18000Xg) and aspirate the
supernatant. ._, ;.. ~ .
6. Re-suspend the pellet in 400 ~,1 of Wash Buffer OW2 and transfer onto spin
column
and centrifuge the spin column for '1 min.
7. Wash with 400 ~1 of ~OW2 and centrifuge as above.
8. Add 20 ~,l of preheated (70°C) Elution Buffer onto the column and re-
suspend the
resin. Close the microcentrifuge tube.
i..., ,
9. Put the spin column with 1.5 ml microcentrifuge tube at 70°C for 3
min.
10. Centrifuge the c~oluniii at maximum speed for 2 min at room temperature.
11. Elute again. (repeat step 8 to ~1~0 to get better yield) .
E. cDNA Labelih~
1. Mix 2~,g mRNA,~ ~1, ~.l of control plants mRNA for single color label
(Hat22: 1 X 109,
Rbcl: 5 X 1 O8, Ga4: 1 X 108, Rca: 5 X 107, Asal : 1 X 107, Atps: 5 X 1 O6
molecule/~,1), 6 ~,1 of
. , ,. .F;: . . . . .
~50 mM random hexamer and DEPC-H20 to 28.88 ~,1 final volume. For dual-color
mode, use 2 ~.g of ~mRNA each ~in Biotin or Dig labeling and individual
addition of
control plants mRNA~: .1~. biotin labeling: Hat22: 1 X 1 O8, Rbcl: 5 X 107,
Ga4: 2 X 107, Rca:
1 X 107, Asal : ~1 X 107;' Atps:. fX 107, Hat4: 1 X 107/x.1. 2. Dig labeling:
Hat22: 1 X 107, Rbcl:
..
1 X 107, Ga4: 1 X 107 Rca: 1 X.107, Asal : 1X 108, Atps: SX 107, Hat4: 2X
107/x,1.
2. Denature for IO min at 70°C, then chill quickly in ice for 5 min.
3. Add 10 ~,l of 5 X first strand buffer, 5 ~.1 of 0.1 M DTT, 1 ~,l of 25 mM
dATP, dCTP,
dGTP mixture 1 ~.1 of 2 mM dTTP , 2 ~,l of 1 mM biotin-16-dUTP, or Dig-11-dUTP
(1
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-79-
mM), 0.63 ~,l of 40 ~ IJ/~1- RNAsin and 1.5 p,1 of Superscript II (reverse
transcriptase,
GIBCO BRL)(200'L1/~.l).
4. Mix well and incubate for 10 min at 25°C, then for 90 min at
42°C.
5. Stop the reaction for 5 min at 94°C.
6. Add 5.5 ~.l of 3 M NaOH for 30 min at 50°C.
7. Add 5.5 ~,1 of 3 M, CH3COOH for 30 min at 50°C.
8. Precipitate the labeled cDNA by adding 34 ~,1 of water, 50 ~.1 of 7.5 M
anunonia
acetate, 10 ~,g of linear polyacrylamide as carrier and 380 ~.1 of absolute
alcohol.
9. Incubate the sample for 30 min at -80°C. Centrifuge at 13000 rpm for
15 min.
10. Wash the pellet with 1 ml of 70% ethanol and centrifuge at 13000rpm for 5
min.
11. Dissolve the pellet in 36 ~,1 of autoclaved HZO. For dual color, combine
two labeled
cDNA together.
F. Arrav Hvbridization
1. The filter membrane carrying the 9600 EST PCR products is pre-hybridized in
5 ml of
lx hybridization buffer,~(4X SSC, 0.1% N-lauroylsarcosine, 0.02% SDS, 1% BM
blocking reagent (Boehringer Mannheim)), and 50 ~.g/ml salmon sperm DNA (GIBCO
BRL) at 63°C for 1.5 hours. Note. You can prepare 80 ml of 1 ~
hybridization buffer
and store it at -20°C: Thaw the buffer at 60°C before use.
2. Stick one side of adhesive EasiSeal~ square to a clean glass slide and
place the pre-
hybridized membrane in the center of the square with the spots facing up.
3. Mix the probe with 2 ~,I of poly-d(A)10 (10 ~.gl~.l) and 2 ~,1 of human Cot-
1 DNA (10
~,g/~.1) (GIBCO BRL) and'40 ~.1 of 2~ hybridization buffer to 80 ~.1 final
volume.
4. Denature the probe iizixture at 95°C for 5 min and then cool on ice.
5. Seal the filter membrane with the probe solution in the hybridization bag.
6. Incubate at 95°C for 5~inin~and then at 63°C for 12-16 h
(overnight).
,. .
7. Wash the filter membrane twice with 5 ml of 2X SSC, 0.1% SDS for 5 min at
room
temperature. ' ',-
8. Wash three times for l5~inin each with 5 ml of 0.1 X SSC, 0.1% SDS at
63°C.
9. Block the filter membrane with 5 ml of 1% BM blocking reagent containing 2%
dextran sulfate at roorri temperature for 1 h.
10. Incubate with 5, ml~' mixture containing 700X diluted Streptavidin-(3-
galactosidase
(1.38U/ml, enzyme activity)(GIBCO BRL), 10000 diluted anti-Digoxigenin-
alkaline
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-80-
phosphatase (0.07SLT/rizl,.,enzyme activity)(Boehringer Mannheim), 4%
polyethylene
glycol 8000 (Sigma); and .0:3% BSA in 1 X TBS buffer at room temperature for 2
hours.
Note. This formula,is~for dhal-color mode. For single color mode, anti-Dig-AP
is not
needed and the incubation time can be reduced to 1 hour.
S 11. Wash with 1 X TBS buffer three times for 5 minutes each.
12. Freshly prepare X=gale, substrate solution (1.2 mM X-gal; 1mM MgCl2, 3 mM
K3Fe(CN)6, 3 mM K4Fe(CN)6 in 1 X TBS buffer) by mixing SO p.1 of 120 mM X-Gal
and S ml of X-Gal substrate buffer. Immerse the filter membrane in the X-gal
substrate
solution for 4S min at 37°C with gentle shaking.
13. Wash with lx TBS. ~ . - .
14. Dual color development: stain the membrane with S ml of Fast red
TR/naphthol AS-
MXsubstrate(Pierce;, Rdckford, IL) at room temperature for 30 minutes with
gentle
shaking.
1S. Wash with dexonized water. Stop the reaction with 1X PBS containing 20 mM
EDTA
for 20 min. ~ ' '
16. Air dry the filter membrane: ,
Results. ~ . . ; . ,~ .
1. Determine the growth irihiliition concentrations of herbal extracts in cell
cultures.
Each herbal extract has~different cellular toxicity, thus it is necessary to
determine the
growth inhibition concentration of every herbal medicine before treating the
cells. Five serial
dilutions of herbal extrac'tv(lO;vS, 2; ~1, O.S mg/ml) were added to Sx105/ml
cultured cells and
incubated fo'r 24 hours in an incubator at 37°C with S% C02. The
numbers of survival cells at
different concentrations~of herbal 'extracts are shown in Table. 18.
2S
3S
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-81-
Table 18. Survival Cell Number at Different Concentrations of Herbal Extracts
experimental
concentration
no.x105/ml 10 5 2 1 0.5 No High Low cone.
rng/mlmg/mlmg/mlmg/mlmg/mldrugcone. (mg/ml)
(mg/ml)
1 Cordyceps sinensis8.4' 11:911.59.2 9.0 12.410 1
mycelium
2 ST024 ' 4.2 ' 10 7.5 10 10 10 1
8.7
3 ST044 .5.98.4 9.9 9.4 5 0.5
~
4 STO51 ~ ' - - 1.7 5.4 0.5 0.05
S ST093 - - 1.9 3.8 4.4 0.5 0.05
6 ST117 - 1.6 3.6 4.6 5.8 0.5 0.05
7 ST123 3.4 6.4 8 9.3 7.8 S 0.5
8 ST128 3.5 7.7 7.9 7.7 8.3 5 0.5
9 ST134 2.9 6.1 11.29.6 9.8 5 0.5
ST237 - - 2.5 6.6 8.7 1 0.1
11 PHY906-303503 4.5 4.9 8.2 1 0.1
12 PHY906-284003 - = 3.8 5.7 7.6 1 0.1
~ '
Note: The original 10
cell number /ml.
was 5x The
number
increased
to
10x10
/ml
after
24h
incubation. "-" indicates all. dead cells..
5 ,
The cell number without herbal extract addition doubled after 24 hours
incubation. On
the other hand, the number,of survival cells varies with different herbal
medicine treatments.
We chose the 50% growth inhibition concentration (ICSO) as the high
concentration and one-
tenth of it as the low coricentratiori. In order to maintain consistence of
the Jurkat T cell line, a
;.
10 cell banking system was established. In the cell bank, a total of 100 vials
of cells (10 million
cells per vial) were. frozen in .a.-150°C freezer.
B. Molecular classification of herbal medicines by nucleic acid microarray
analysis. ~ '
Analysis of 3 single-element herbal medicines: Three single-element herbal
medicines,
Cordyceps Siriensis Mycelium (CSM), ST024, and ST117 were used to treat the
cell cultures
as described in the methods"section. Gene expression measurements were
performed by using
microarrays of 13824 cD~tA.,fragmerlts each representing a distinct human
transcript. For the
data analyses, gene spots of l~gh~ data quality were selected. The selection
was based on signal
to background ratio greatervtliaw2.5 or the coefficient of variation (CV) of
spot area smaller
than 10%. All the data sets were normalized with the control cells, which
received no herbal
treatments. The spot intensity'wa's rounded up to 10 for those intensities
that were less than 10.
Based on the selectioncriteria; a total of 492 genes with differential
expression ratio greater
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-82-
than 1.5 were ~selecteii foreclusteranalysis. These data pre-processing
procedures were
performed by the prograrii ~"D'ataExtract" and "Ratio2" developed in-house.
These 492 genes'were'clu'ster analyzed by the average-linkage method. The
distance
between genes is used as~the linear correlation coefficient or resemblance
coefficient. The
cluster analysis programs; Cluster and TreeView, were based on hierarchical
clustering method
and were written by Dr. Michael Eisen of Stanford University (Eisen, 1999,
Eisen et al., 1998).
The results are shown in Figure 1.2. From Figure 12C, one can clearly identify
that three
different herbal medicines ~of high and low concentrations are each clustered
together. For
instance, CSM-L is closer.to:CSM-I3 and less.similar to ST024 or ST117 in the
clustering tree.
A different clustering algorithm, the self organizing map algorithm, which is
based on non-
hierarchical method, yields 'thesame results (data not shown). From the
clustering results
. ~; ,
shown in Figures ~12A &' ~1~3A~vseveral' features are noted. (1) 4 genes were
up regulated by
ST117 treatment but down~regul'ated by other herbal treatments (Figure 12B).
(2) 34 genes
were down regulated by CSM~~treatment but up regulated by others (Figure 13B).
(3) 2 genes
were up regulated by all the three 'herbal treatments, one is Malic enzyme 2
and the other one is
an anonymous gene (clorie'IlD: 328351) (Figure 13C). (4) 12 genes were highly
induced by the
,~;
high concentration treatmerit~arid less induced by the low concentration
treatment in all the
three herbal medicines (Fig.r 13D). ' ' '
Analysis of 2 ~re~arations'of mufti-element herbal medicines. Two batches of
1=Iuang
Chin Tang, PHY906-303503(#1.1) & PHY906-284003 (#12), each with low and high
concentrations were used to treat the cell cultures in three independent
experiments. The gene
expression profiles were ~acqi~ired with microarrays of 9600 non-redundant
cDNA elements.
After the data pre-processing procedures as described above about 5000 genes
were selected
x;. , ,
for the subsequent data analysis.TThere were 3 repeats for each herbal
treatment. For data
analysis, we use a modified methof;based on the one reported by Slonim et al.
(Slonim, 1999).
The following algorithm is designed to search for the candidate marker genes
that have high
differential expression ratios ~liut low deviation in the three repeats. We
designate a P(i) value
to account for the gene' i with 'tlie aforementioned features.
P(i) = square root of~(E(~m:-p.~)2)~-(6~+Esm)
. r.
: Mean expression ~levels.iri three repeat experiments for herbal treated
cells (p,m) or
untreated control, cells (~,~): ~'. , ~ .
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-83-
cr : Standard deviation of the expression levels in three repeat experiments
for herbal
treated cells (6m) or untreated control cells (6~).
We.calculated the P(i) value for each gene and selected 500 genes with the
highest
..
scores as candidate genes; for cluster analysis (Figure 14). The values of
each gene were
averaged over the:3 repeats: As shown in Figure 14B, two different
concentrations of #12 are
clustered together (12-H c~ 12-L). The 1>igher concentration of the #11
preparation is closer to
the #12 preparation cluster than the~lower concentration of #11 preparation.
However, all these
clusters have sirnilax resemblance coefficient (distance between clusters)
compared with the
tree shown in Figure 12. These results suggest that the gene expression
profiles of #11 and #12
preparations ofHuang ChinTang are similar. The results are justified based on
the fact that
these two preparations are' based on'the same herbal medicine mix.
Several features are rioted in the expression profiles illustrated in Figures
14A & 15.
The averaged gene expression levels are shown in Figure 15A The Boxl encloses
genes that
were down regulated in #11'~-L treated cells but up-regulated in others. These
genes include 2
tRNA synthetase (isoleucirie'aii~ methion), RNA polymerase II polypeptide B
(Clone ID
42020), KIAA0212 gener(Clorle ~~D 310497, containing ATP/GTP-binding site
motif A), and
KTAA0577 (Clone'ID,29263,vATP-dependent RNA helicase). It is interesting to
note that 3 out
of the 6 genes were involved~'iii tlie~RNA replication. Box2 encloses the
genes that were up
regulated by all the #1 l arid #12 treatments. Box3 encloses the genes which
showed no
response by #11-L treatment but were down regulated by the others. Box4
encloses the genes
that were highly repre'ssed'by low concentration herbal treatment but were
less repressed by
high concentration herbal treatrrierit. Finally, in Boxl and Box3, the
expression profiles of #11
treated cells are different~from the profiles generated by the other 3
treatments. This result is
consistent with the fincting'tiepicted in Figure 14B. '
Combining the dataysets of the gene eXpression profiles of the 3 single-
elements and the
', , ,
2 preparations of mufti-elernerit herbal medicines together, a couple features
are noted as an
illustration. The KIAA0212.gene ~(Clorie ID 310497, containing ATP/GTP-binding
site motif
A) was highly induced by all the'high concentration herbal treatments except
that it was only
mildly induced by the #11-Iand-the CSM treatments. Two genes, an anonymous
gene, (Clone
ID 510908) and Proteasoriie chain 7 precursor (Clone ~ 70088) were highly up-
regulated on
all the treatments except~dowxi-regulated by the CSM treatment.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-84-
We next wdrked~ori the ciux to 'cluster analyze the gene expression profiles
of the 5
.,.
different types of herbal.medicine treatments. The data pre-processing
procedures were
performed as aforementioned and~500 genes were selected for cluster analysis.
A hierarchical
clustering was performed by the prograrri "Cluster'° described above.
The hierarchical tree was
cut at the position where the 'range of distance between clusters is the
greatest (Romesburg,
1989) and the result is shown ire Figure 16A. The 3 single-element herbal
medicines, CSM,
ST024, and ST117 are clustered together. Out of the 2 different batches of the
mufti-element
herbal medicine, #12-H, #12-L and #11H are clustered together and the #11-L
stands by itself.
The result suggests that higher similarity exists between the #11 and #12 as
compared with that
of CSM, ST024, and ST117: In order to better classify the different herbal
medicines, the data
analysis algorithm was improved.by~standardizing all the data sets so that the
expression level
of each gene across thevdifferent data sets has zero-mean and unit=variance
(Tavazoie et al.,
1999; Chen et al., 1999). ThYS yields the transformed variables:
Y
1 s x; _ (xi-pX)~ a~X
mean expression levels in the' data set
6X: standard,deviation of the expression levels in the data set
x; : un-transformed gene expression level
~; : transformed gene ~eXpression level
After staridardizingtthe 'dataYset;, #1 l and #12 are clustered together as
shown in Figure
.,
16B. The ST024 and ST1'l7~are clustered together and the CSM is in an
independent cluster.
Furthermore, the clustering also 'suggests that CSM is more similar to #11 and
#12 than to the
ST024 and ST117. Another clustering algorithm, self organizing maps, was
performed with
the same standardized data sets and yields the same result as the hierarchical
clustering (Figure
16C).
Class predictors for discriminating #11 and #12 herbal treated expression
a~rofiles- The
above cluster analyses for.the #11 and #12 show that they are similar and
further classification
is difficult by the hierarchical~clusteririg or self organizing maps methods
with the data set
containing the 500 genes of the highest P(i) values. We then modified the
algorithm to select
genes with larger expression ratio difference between #1 l and #12 herbal
treated cells, but
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-85-
smaller variation in the two Herbal treated cells. The T(i) value is defined
to score this feature
as following: ~ ~ ~ ~ .. . '
T(i) _ log('~,yl) -= log(Wa) ~ (611'612)
p,: Mean expression ratios din three time experiments for #11 treated cells
(p.11) or #12
r.
treated cells (~12) v ' ~~ '
o' : Standard deviation of'the expression ratios for #11 treated cells (611)
or #12 treated
cells (~11)
We calculated the T,(i), ealue for each gene and selected 50 genes with the
highest
.~ a
scores as class predictors~(Figure 17): 18 genes were up-regulated by the #l
l~treatment and
down-regulated by'the #l2.treatment. The rest of the genes were up-regulated
by the #12
treatment and down-regulated by the #11 treatment. We then used these class
predictors to
classify two test herbal preparations based on a modified method described by
Ciolub et al.,
1999. . . ,
Two different,batches of°Huang Chin Tang preparations, PHY010401
(#16) ~
PHY010402 (#17) were obtained from Sun Ten Pharmaceutical Co. and were used
for the
class prediction test. The.'gene expression profiles of #16 and #17
preparations were
normalized with the expression profile of the untreated control cells and
standardized with. the
class predictors. Each predictor g; votes for either #11 or #12 herbal
preparation depending on
whether its expression level X; .is closer~to #11 or #12. The vote for each
gene is given by v; _
~xi - (I~m+~.c)~2I~ where ; '
~.: Mean expression'ratio: iri thiee~ repeat experiments for #11 (x,11) or #12
herbal treated
cells (~,lz)
The average votes~~Tl1 and~Vy2 were collected from the predictor genes
correlated with
the predictor on #11 and #~12,~respectively. The prediction strength (PS)
reflects the margin of
victory and it was defined as PS = (Vll-~la)~ (Vll+Vlz). If the PS was greater
than 0, it
indicated that the herbal preparation was more similar to #11 and less similar
to #12. The
results obtained from the analyses on,#16 and #17 indicated that #16-H was
similar to #11
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-86-
(PS=0.1) and #16-L, #17-H; arid #I7-L were similar to #12 (PS=-0.29, -0.21 and
-0.2,
respectively). Based on the information of #16 and #17 preparations, this test
failed to
correctly identify #16-L asmore similar to #11.
Discussion. ' ~. . . .
Characteristic ec~ ne. expression profiles for herbal medicines treated cells.
The predictor
genes are.selected from the differentially expressed genes in two herbal
treated cells. These
genes represent the cellular responses to the herbal medicine treatments. In
this study, we have
identified some interesting genes based on their responses across different
herbal treatments
(Figures 13, 15, and 17). These genes are valuable assets in studying the
signaling pathways of
cells in response to the herbal stimulation and in deciphering the molecular
pharmacological
mechanism of herbal medicines.
Classification bf herbal rrieelicines by nucleic acid microarra,~analvsis. In
surrnnary, a
two-step classification proced~e ~is proposed. An initial classification
procedure based on the
standardized data sets and the clustering algorithms is performed and followed
by a final
classification procedure with'the class predictors. All the procedures can be
integrated in a
computer program. In these preliW inary studies, all the genes have the same
contribution for
classification. When the data sets are large enough, the weight for each gene
(or predictor) can
be acquired from the linear correlation coefficient (Golub et al., 1,999, Chen
et al., 1999).
The #11-L failed to be clustered with #11-H by significant association. A
similar
preparation of #11, the #I6 preparation yielded the same results. It was
interesting to discover
that no matter what clustering algorithms were applied, the #11 and the #16
preparations did
not yield the expected result's. Even with independent experiments, the
results remained the
same. The reason behind the~~failure will be investigated with detailed
information of #11 and
#16 preparations.
~uality control and analysis in microarra,~ystem. The quality of acquired
microarray
data and the 'choice of the statistical analysis methods are both important
factors for achieving
meaningful results. We have recognized that the variation among arrays
contributes to errors in
measuring gene expression levels. Based on the data in this report, we have
found that for
every herbal preparation, the high concentration treated expression profiles
always cluster with
its lower concentrationcounterpart (Figures 16B and 16C) and we could classify
the STI 17,
ST024., CSM and Huang Chin~'Tang with two different clustering methods. In the
past three
' months, the array quality has'been~improved to have less than 7% CV. We have
also set up a
standard procedure for assessing the:quality of every batch of arrays
fabricated in the lab. All
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
_87-
these experimental findimgs:and improvements in the microarray technologies
suggest that
classification and characterization of the Chinese herbal medicines by
microarray system are
feasible.
References for Exam~le~l5: v
Bertucci-F; Bernard-K; Loriod-B:; Chang-YC; Granjeaud-S; Birnbaum-D; Nguyen-C;
Peck-K;
Jordan-BR (1999) Sensitivity issues. imDNA array-based expression measurements
and
performance of nylon microarrays'for small samples Human Mol. Genetics 8(9):
1715-1722.
Bittner-L, Trent-J, Meltaer-P~(1999) Data analysis and integration: of steps
and arrows. Nature
Genet. 22, 213-215
. ..
Brown-PO; Botstein-D, (1999j' Exploring the new world of the genome with DNA
microarrays.
Nature genetics 21 (1) suppleirient; 33-37.
Chen-JJ; Wu-R; Yang=P~;~ Fluang-JY;v Sher-YP; Han-MH; Kao-WC; Lee-PJ; Chiu-TF;
Chang-
F; Chu-YW; Wu-CW; Peck-~~(1~998)~expression patterns and isolating
differentially expressed
genes by cDNA microarray,systern'with colorimetry detection. Genomics. 51: 313-
24.
Chen-Y, Bittner-M, Dougherty-ER (1999) Issues associated with microarray data
analysis and
integration. Information supplementary to article by Michael Bittner, Jeffrey
Trent and Paul
Meltzer (Nature Genet. 22; 213-215).
Duggan-DJ; Bittner-1VI; Clieii=YlVleltzer-P Trent -JM (1999) Expression
profiling using
b,
cDNA microarrays.~Natur~'gerietics. 21 (1) supplements 10-14.
Eisen-M (1999) Cluster and'~Treeview manual.~(rana.stanford.edu/software)
. ..., , s.R .
~:, .r, y,, ,. .. . .
Eisen-M, Spellman-PT, Brbwn=PO, Botstein-D. (1998) Cluster analysis and
display of gene-
wide expression patterms. P'roc. IsTatl. Acad. Sci. LTSA 99:14863-14868
' . ~ '
Golub-TR, Slonim=DK; 7Camayo-P, Hoard-C, Gaasenbeek-M, Mesirov-JP, Coller-I~,
Loh-ML,
Downing-JR, Caligiuri-MA,~B~loomfield-CD, and Lander-ES (1999) Molecular
classification
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
_88_
of cancer: class discovery' arid: class prediction by gene expression
monitoring. Science ~ct 15:
531-537 . . ,
Larder-ES (1999) Array of~hbpe. Nature'genetics 21 (1) supplement, 3-4.
.:
Romesburg-HC (1989) Cluster analysis for researchers. Chapter 16: How to make
classifications. P203-216, I~rieger Publishing Co. Malabar, Florida, LTSA
Slonim-DK, Tamayo-P, Mesirov-JP,.Golub-TR, Larder-ES (1999) Class prediction
and
discovery using gene expression data. ~(www.genome.wi.mit.edu/MPR)
Tavazole-S, Hughes-JDCampbell=MJ, Cho-RJ, Church-GM. (1999) Systemic
determination
of genetic network architecture: Nature genetic 22: 281-285
Example 16. Identify characteristic gene expression profiles induced by an
herbal
medicine . , , , ..
As stated in Example 15; the prescription of the Chinese herbal medicine Scute
and
Licorice combination (Huang Chin 'Tang) stops diarrhea, relieves spasms and
clears fever. The
ingredients of Huang Chiri 'Tang are Scute, Peony, Licorice and Jujube. In
this study we used
the nucleic acid microaxray technology to study the gene expression profiles
induced by herbal
medicines in mammalian cells: To investigate the characteristic expression
profiles induced by
the Huang Chin Tang, Jurkat'.fi cells~were treated with 5 batches of Huang
Chin Tang
(PHY01040; #16, PH3~'010402; #17, PHY03061; #18, PHY03062; #19 and PHY02231;
#20
obtained from Surf Ten Pharmaceutical Co.) by 5 concentrations (1/2, 1/2.5,
1/5, 1/10, and 1/20
of ICso)
Nucleic Acid microarray with a two color detection method was employed to
measure the
expression profiles. The mRNA extracted from herbal treated cells was labeled
with
digoxigenin and the mRNA extracted from untreated cells was labeled with
biotin. Arrays of
9600 features were erriployed arid the procedures described by Chen et al.
(Genomics, 51, 313-
324, 1998) were adopted in the''experiments. For data pre-processing, only
array spots of high
data quality were selected: rThe selection was based on signal to background
ratio greater than
2.5 and 1.5X differential eXpressiori ratio. Ey these criteria, 1081 genes
were selected for
further statistical analysis:~Non-hierarchical cluster analysis programs such
as the Genecluster
program developed in Massachusetts Institute of Technology (Tamayo et al.,
1999) was
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-89-
employed to categorize the expression profiles. The Genecluster program is
based on self
organizing map (SOM) principle. A°6X4 SOM clustering of expression
profiles are shown in
Figure 18A. The ol.etails of geneexpression profiles for the selected clusters
are shown in
Figure 18B. In these clusters, °clusters 3 and 20 (labeled c3 and c20)
were selected for that the
gene expression levels increase with higher herbal concentrations. Similarly,
clusters 5 and 9
for that the gene expression levels decrease with higher herbal
concentrations. Cluster 23
collects genes whose expression levels were up regulated compared with that in
untreated cells
and cluster 0 for down regulated genes. These expression profile clusters are
further condensed
into two major groups A & ~B. Group A collects genes up regulated by herbal
treatment and
Group B collects.genes down regulated by herbal treatment. The expression
profiles in Group
A & B form the basis of a characteristic expression data set for an herbal
preparation. The
same procedures were repeated for 5 different batches of Huang Chin Tang, and
952 genes
were selected to establish tliercliaracteristic expression profile database of
the Huang Chin
Tang. ' T
As shown in Figure 1.9,'bythe aforementioned procedures, a gene can be
categorized as
Group A, B or none (non-A and non-B) and its expression profile can be
represented by l, -l,
and 0 respectively. The niimlier.of different gene expression profiles between
batch #1 and
batch #2 are 3 in Group°A.(Gene 6,'7, and 8) and 2 in Group B (Gene 15
&16). By the same
principle, the number of different expression profiles between batch #1 and #3
are 10 in Group
A and B and the number isvl,l' between batch #2 and batch #3. These numbers
indicate that
batch #1 and #2 are more. similar than batch #3. This principle was applied to
classify 5
different hatches of herbal, preparation's. The following algorithm is
designed to calculate the
distance between a pair ofiieilial preparation batches, i and j.
d;; = E ~(X;, X;) ~~ ~(Harriming distance)
The gene X in i batch of preparation.is assigned to Group A, B or none
if x; ~ x; ; s(x;~"x;) =,1, ~ ; ..
if x; = X; ; s~x;,.xj ~= o. , .
We calculated all the d;;~ value between pairs of herbal preparations for
cluster analysis.
The analysis programs, Kitscli'Gluster was based on hierarchical clustering
principle and was
written by Dr. 3oseph Felsenstein of Washington University
(http://evolution.genetics.washirigton.edu/phylip.html). From the Hamming
distance table
(Figure 20), one can clearly ieleritify that the shortest distance lie between
batch #17 and batch
#18 and that batch #17 is siriiilar to #18. Batch #16 also similar with. batch
#17 and #18 but
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-90-
batch #19 is dissimilar to~.the rest~of~batches. The results were confirmed by
HPLC analyses as
described below. . ' .'
By HPLC, the chemical 'composition of the 5 batches of herbal preparations was
analyzed.
Four major peaks (BG, B; Gly, and Pf) in the chromatograms were selected for
statistical
analyses. Two additional.para'rneters, BG+B and BGB, were included in plotting
the 6-
coordinate radar graph as shown in Figure. 21. The distance on each coordinate
is the integrated
intensity of that particular.cliemical constituent in the chromatogram. In
general, #16, #1~7 and
#18 were similar in their constituent content (liaicalin and baicalein) of
Scutellariae Radix,
which are within 33:55-36:08; while the amount of the same constituents are
higher in #19 and
#20 (42.49 and 44.96, respectively). The resemblance of #16, #17, and #18 can
be seen from
the coincident radar plots in Figure 21B.
. ;,...
In order to identify the ~unl~nown herbal medicine based on the characteristic
expression
profile database established ~as described as above, Jurkat T cells were
treated with a tester
sample #17 in 5 concentrations to set up the characteristic expression data
set for the tester.
The Hamming. distances between the tester and each of the data sets (#16, #17,
#18, #19 and
#20) in the characteristic expre~ssiori;database were calculated and the
scores are: #16: 502,
#17: 405, #18: 402; #19:'699, arid #20:531. These data show that the tester is
most similar to
#17 having a lowest. Ha ~rrirning distance score of 405. The example
demonstrates that this
invention teaches a method'to'identify'unknown herbal medicine based on the
gene expression
profiles induced by the herbal~riiedicine iri mammalian cells. The identity of
the unknown
herbal medicine can be inferred by aligning the characteristic expression
profiles with a
collection of characteristicveXpression profiles of herbal medicines in an HBR
Array.
Based on the characte~istic~expression database, marker genes and signature
expression
profiles can be deduced fort'air herbal medicine for studying its
pharmacological mechanisms
and for optimizing the formulation of aAcomplex herbal preparation. For this
example, 5
different batches of IIuang'Clun Taiig'preparatioris (#16, #17, #18, #19 and
#20) were obtained
from Sun Ten 1'laarmaceuticalvCo.~ and a characteristic expression profile
database was
constructed based ~on aforementioned procedures. For each gene, the
consistency of expression
profiles in the database wa's "scored by the coefficient of variation (CV
value):
CV = ~ / (E ~,; /n)
p.;: Mean expression ratios for.#i treated cells.
n : Number of the data set, i1 --~5 in this case.
. .
6 : Standard deviation of tie .expression ratios for #16, #17, #18, #19 and
#20.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-91-
Since the CV reflects the variation of data, the marker genes for an herbal
medicine were
selected based on the CV'score. The top SO genes with the minimum CV scores
were selected.
Figure 22 shows 2S marker, genes :with up regulated signature profiles and 2S
marker genes
S with down regulated signature.profiles for Huang Chin Tang.
The characteristic expression profile database can be used to infer the
expression profiles
of individual chemical constituents in a~ mixture as complex as an herbal
medicine if the
amount of the chemical constituents can be semi-quantitatively determined. In
this example,
the chemical composition of.an herbal medicine is determined by high
performance liquid
chromatography. The integrated intensities of 4 chemical constituents in five
batches of Huang
Chin Tang preparation were quantified by HPLC analysis. The gene expression
ratios for each
batch of herbal preparation were calculated by taking the median of the
expression ratios
induced by S concentrations 'of herbal preparation. The correlation between a
constituent and a
gene expression profile was quantified by the~Pearson correlation coefficient.
The Pearson
1S correlation coefficient forvgeme x~.and the constituent y is:
R = (1/n) E(x;-~,X)( Ysvl~v)'/ sX ay , i= 1 to n
n : Number of the herbal preparation,, n = S in this case.
~,X: Mean expression ratios in ,five herbal preparation for gene x.
~,y: Mean integrated intensity in five batches of herbal preparation for
constituent y.
x;: Gene expression ratios in #i herbal preparation for gene x.
y;: Integrated intensity.in #i hezbal preparation for constituent y
6: Standard deviation of the expression ratios (ax) or integrated intensities
(6Y) for five herbal
preparations. . '
2S Several genes whose expression levels highly correlated (with ~R~>0.99)
with the amount of
. ;,: ,~
chemical constituents in Huar~g Chin Tang were identified for each
constituent. For example,
the R value between the gene (clonelD: 67185) and Glycyrrhizin was 0.998
(Figure 23A). One
the other hand, the gene ~(clomelD:.344720) whose expression levels increase
with the decrease
of VVogonin(WG) has an R value of -0.997 (Figure 23B). In addition to the
above two
examples, 191 and 170 genes were highly correlated with individual
constituents with R value
> 0.9 and R value < -0.9, respectively. For instance, 17 and 18 genes were
positively and
negatively, respectively, correlated with Albiflorin (Af) (Figure 24). This
example teaches a
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-92-
method to profile gene expressiomfor individual constituents in a mixture
without isolating
them to perform the expression' analyses one constituent by another.
References for Example 16::
US Patent documents , ,
Stoughton-Roland and Friend-SH. USP#~ 5965352: Methods for identifying
pathways of drug
action ~ ~ . , ,
Brown-PO and Shalon-TD.LJSP#5807522: Methods for fabricating rnicroarrays of
biological
samples '
~ ~...
Lockhart-DJ, Brown-EL, Wong;,GG,°Chee-MS, and Gingeras-TR. USP#6040138:
Expression
monitoring by hybridization to.'liigli~density oligonucleotide arrays
Ladunga-I. USP#5987390:~1Vlethods.and systems for identification of protein
classes
. . ~ .' . ~ . . '
Mahant-S, Shivaling-5,~-'.Vivek-G. USP#5951711: Method and device for
determining
hamming distance between two ~iulti=bit digital words
Fo~eign.Patent documents
Brown-P~ and Shalon-TID~,EP#913485A1: Method and apparatus for fabricating
microarrays
of biological samples
. - .. : .:.~., . . ..
y , . ° , ,°, ~~
Other Publications
Bertucci-F; Bernard-K; Loriod-B; Chang-YC; Granjeaud-S; Birnbaum-D; Nguyen-C;
Peck-K;
Jordan-BR (1999) Sensitivityl;issues in DNA array-based expression
measurements and
performance of nylon microairays for.small samples Human Mol. Genet. 8(9):
1715-1722.
.. , ~.
Bittner-L, Trent-J; Meltzer-P,(1999) Data analysis and integration: of steps
and arrows. Nature
Genet. 22, 213-215
.
Brown-PO; .Botsteiri-D (1999)rExploring the new world of the genome with DNA
microarrays.
Nature genet. 21 (1) supplement, 33-37.
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-93-
Chen-JJ; Wu-R; Yang-PC; Huang-JY; Sher-YP; Han-MH; Kao-WC; Lee-PJ; Chiu-TF;
Chang-
F; Chu-YW; Wu-CW;~Peck-K (1998) expression patterns and isolating
differentially expressed
genes by cDNA microarray system with colorimetry detection. Genomics. 51: 313-
24.
,.
Chen-Y, Bittner-M, Douglierty-ER,(1999) Issues associated with microarray data
analysis and
integration. Information supplementary to article by Michael Bittner, Jeffrey
Trent and Paul
Meltzer, Nature Genet. 22~ 213-215.
Duggan-DJ; Bittner-M; Cherl-Y; Meltzar-P Trent -JM (1999) Expression profiling
using
cDNA microarrays. Nature genet. 21 (1) supplement, 10-14.
Eisen-M (1999) Cluster and ~'reeviev~ manual.
(ftp:l/rana.stanford.edu/software)
Eisen-M, Spellman-PT, Brovv~i-PO, Botsteiri-D. (1998) Cluster analysis and
display of gene-
wide expression patterns. Proc: Natl. Acad. Sci. USA 99:14863-14868
Golub-TR, Slonim-DK, Tamay~-P, Huard-C, Gaasenbeek-M, Mesirov-JP, Coller-H,
Loh-ML,
Downing-JR, Caligiuri-MA; Bloomfield-CD, and Lander-ES (1999) Molecular
classification
of cancer: class discovery and class prediction by gene expression monitoring.
Science Oct 15:
531-537 . . ; ~ .
Lander-ES (1999) Array of hope. Nature genet: 21 (1) supplement, 3-4.
Tamayo-P, Slonim-D, Merirov-J, Zhu-Q, Kitareewan-S, Dmitrovsky-E, Lander-ES,
and
Golub-TR (1999) Iiiterpretiii'g~patterris of gene expression with self
organizing maps: Methods
and application'to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA
99:2907-2912
. i 6 .
Romesburg-HC (1989) Cluster analysis for researchers. Chapter 16: How to make
classifications. P203-216; Krieger Publishing Co. Malabar, Florida, USA
Scherf U, Ross-DT, Walthaxiz-M, .Smith=LH, Lee-JK, Tanabe-L, Kohn-KW, Reinhold-
WC,
Myers-TG, Andrews-DT, Scudiero-DA, Eisen-MB, Sausville-EA, Pommier-Y, Botstein-
D,
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-94-
Brown-PO, Weinstein-JN (2000) A gene expression database for the molecular
pharmacology
of caucer. Nature genet. 24: 236=44: .
Slonirn-DK, Tamayo-P, Mesirov-JP, Golub-TR, Lander-ES (1999) Class prediction
and
discovery using gene expression data: (http://www.genome.wi.mit.edu/MPR)
Tavazole-S, Hughes=JD, 'Campbell-MT, Cho-RJ, Church-GM. (1999) Systemic
determination
of genetic network architecture. Nature genet. 22: 281-285
Example 17. Identification of the biores~onses and the signature gene
herlbal composition.
To further investigate the expression profiles induced by the.Huang Chin Tang,
Jurkat T
cells were treated with Huang hiii Tamg (PHY906#2 obtained from Sun Ten
Pharmaceutical
Co. Taiwan) by 5 concentrations (1/20, 1/10, 1/5, 1/2.5, and 1 of ICSO).
Nucleic acid
microarray with dual-color detection was employed to measure the expression
profiles. The
mRNA extracted from herbal'treated and untreated cells were labeled with
Biotin-16-dUTP
and Dig-11-dUTP9 respectively.~A control~group was established by using mRNA
extracted
from untreated cells and labeled the mRNA with biotin and dig respectively in
equal
proportions. Five sample groups with different concentrations of herbal
treatment and one
control group were employed for the study by the procedures described by Chen
et al. (1998)
with minor modifications. For data pre-processing, only array spots of high
data quality were
selected: The selection criteria,were spots with signal to background ratio
greater than 2.5-fold
and more than 1.5-fold in differential expression ratio: By these criteria,
1044 genes were
.. ~ v ~~,.;.
selected for further statistical: analysis. The gene expression profiles of
the control group were
highly correlated with only 4.8 genes listed as statistical outliers that lie
beyond the 2-fold
differential expression range (Figure 25A). For the sample group, many
differentially
expressed genes are evident 'as illustrated in Figure 25B. The number of genes
whose
differential expression 'ratio greater than 2-fold 'increases with
concentration of herbal
treatment as shown in Figure 25C. These results~prove that the identified
differential
expressed genes are truly iritiuc~ed by Huang Chin Tang treatments.
To identify the genes that were specifically induced by PHY906 (signature
genes for
PHY906), the expression profiles were clustered by a non-hierarchical cluster
analysis
programs "GeneCluster" developeci'by Tamayo et al., 1999. The computer program
is based
on self organizing map (SOM) principle and the clusters of expression profiles
are shown in
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-95-
Figure 26. The X-axis represents the°herbal concentration from low to
high and the Y-axis is
the gene-expression ratio~,=~Th~ aignature genes were selected from the
expression profiles
which exhibit dosage response~to the PHY906#2. The induced and repressed genes
were
selected from cluster 3 &°4 and cluster 18 &~ 19, respectively. In
order to identify signature
genes for PHY906, another:b~atcli~.of Huang Chin Tang, PHY906#3, with the same
formula and
manufacturing process were performed as described for PHY906#2. The induced
and
repressed genes commonly foundin both batches are shown in Figure 27.
Score similarity of bioresponses by self organizing map (SOlVn
To differentiate.herbal 'medicines of similar compositions, a scoring method
is developed
and the score S represents the'difference in bioresponses of a biosystem to
two different herbal
compositions.
S = E P,~W~, '~
Where Pty is the number' of the common genes induced both by herbal prep. A
and herbal
prep. B iri cluster i and in cluster j. For example, the SOM clustering
results for the expression
profiles of both batches of P ~.HY906'are shown in Figure 28A. In cluster C13
and C14, 17 and
genes share the same 'eXpressiom profiles for both batches of PHY906,
respectively. In
addition, 10 genes whose'eXpres'siori profiles induced by PHY906#2 are
clustered in C13 but
are clustered in C14 for.PH~'906#3. Therefore, P~313=17, P1a14= 25, and
PI314=lO. A weighing
factor, Wl~, describes the d"istance'between the cluster i and j to indicate
the similarity of the
20 two expression profile clusters. In the case of C13 and C14, these 10 genes
have similarly
response to PHY906#2 and PHY90;6#3 '(Figure 28B). The weighing factor is
defined as:
Wl~ =1- EZ~ /Max(El~), where E~~ ~is the Euclidean distance between the
cluster i and j and
the value is normalized by Elf /MaX(E1~). When i = j, W=~ is 1. The number
decreases as cluster
i and cluster j become more~different (Figure 28C).
25 Classification of 5 batches of herbal medicines
To test how well the valiowle riiethod performs in classifying 5 batches of
similar herbal
preparations, Jurkat 'I' °cells v~e~eweated with 5 batches of Huang
Chin Tang (PHY01040; #16,
.. ~°, , . . .
PHY010402; #17, .PHY0306~1;',#18, PHY03062; #19 and PHY02231; #20 obtained
from Sun
Ten Pharmaceutical Co.) liy 5 concentrations (1, 1/2.5, 1/5, 1/10, and 1120 of
ICSO). The S=~
scores were calculated between pairs of herbal preparations in cluster
analysis (Figure 29). The
analysis programs, Kitsch Cluster was based on hierarchical clustering
principle and was
written by Dr. Joseph Fel~en'stein of ~7Vashington University
(http://evolution.genetics:washington.edu/phylip.htrnl). The S scores
(distance) are tabulated
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-96-
(Figure 29A), one can clearly identify.that the shortest distance lie between
batch #17 and
batch #18 and that batch#:17 is.similar to #18. Batch #16 also similar with
batch #17 and #18
but batch #19 is dissimilar to the rest of batches. .The results were
confirmed by HPLC
analyses.
Characterize an unknown herbal medicine based on the expression profiles
To identify an unknown herbal medicine based on the characteristic expression
profile
database established as described asabove, Jurkat T cells were treated with a
tester sample #17
in 5 concentrations to set up the characteristic expression data set for the
tester. The S score
between the tester and eacli~of the data sets (#16, #17, #18, #19 and #20) in
the characteristic
expression.database were calculated and the S scores are: #I6: 0.78, #17:
0.85, #18: 0.84, #19:
0.77, and #20: 0.79.These data show that the tester is most similar to #17
having a higher S
score of 0.84. The example deindiistrates that one can apply the method to
identify an
unl~own herbal medicine based~on the gene expression profiles induced by the
herbal
medicine in mammalian cells. The identity of the.unknown herbal medicine can
be inferred by
aligning the characteristic expression profiles with a collection of
characteristic expression
profiles of herbal~medicinesvin'an.HBR Array.
The property of an'herb can be described by four natures and five flavors (in
Chinese
Herbal Phramaceuticals; Ed. Zheng Hua Yen, People's Health publications,
Beijing, China,
1997; Book of Ben Cao Gan IVIu by Shi Zeng Li, Ming Dynasty, China). Each of
the four
herbs in PHY906 may relate;to_another set of herbs with similar property (see
Table 19). Or
herbs with similar propeity~~rriay'exhibit similar bioresponse. HBR Arrays may
be used to
determine or measure the :relatedness in terms of the property of herbs. Such
information may
be useful in creating a new Herbal formulation.
30
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-97-
7Cable 19: Herbs with PHY906 Properties
Herb Properties Drugs with similar properties
, -
Scutellariae Bitter tasting,Coptidis Rhizoma, Phellodendri
Radix . Cortex, Gentianae Radix,
cold natured~Gardeniae Fructus
Paeoniae RadixBitter and Canarii Fructus (sweet and sour
sour ' ~ tasting, moderate
tasting, natured), Potulacae Herba (sour
mildly., tasting, cold natured),
' .
cold in nature:. Fraxini. Cortex( bitter tasting,
cold natured), Sophorae
., Flos (bitter tasting, mild cold
in nature), Bletillae Tuber
(bitter, sweet and harsh in taste,
and mildly cold in
nature).
Glycyrrhizae Sweet tasting,Lycii Fructus, Polygonati Officinalis
Rhizoma, Polygonati
Radix moderate Rlaizoma
natured
Zizyphi FructusSweet tasting.Sacchartim Granorum, Juglandis
. and' Semen
warm'natured
Properties:
Four natures-cold; hot; .warm, cool. '
Five flavors-acrid, bitter., sweet;sour, bland.
Example 18. Evaluation ~of an herbal medicines by HBR Array.
As stated in Example'16','the component herbs of Huang Chin Tang are Scute,
Peony,
Licorice and Jujube. .The geiie,expression profiles induced by five batches of
Huang Chin
Tang in mammalian cells were cliaracterized. A standard formula for Huang Chin
Tang can be
defined and characterized with animal studies or with clinical studies. For
example, the #17
was used as the standard formula for. Huang Chin Tang based on the quality
control and other
standards set up by Sun.Ten'Pha'rniaceutical Co. The bioresponses of #17 were
used to build
the HBR Array for Huang~ Chin Tang. The marker genes in the HBR Array were
selected to
evaluate other preparations "of Huang Chiri Tang composition. A tester Huang
Chin Tang may
contain the same herb compositions but the component herbs may be grown under
various
environmental characteristics: Comparing the bioresponses of the tester with
the marker genes
of standardized HBR Array.~the biological activities of the tester were
evaluated.
Furthermore, the inarker'genes whose expression levels are lv.ghly correlated
(with
~R~>0.99) with the dosage of corriponent herbs in Huang Chin Tang (as stated
in Example 16
and Figure 25) are selected, for' evaluation purpose. The tester Huang Chin
Tang can be
evaluated by comparirig'the specific bioresponses or expression levels of the
selected set of
marker genes with the HBR AiTay. 'Tf'the expression levels or bioresponses of
tlae selected
marker genes are~beyond'the~acceptable variation region, the amount or
characteristics of the
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-98-
component herbs 'are adjusted or modified to meet the acceptable variation.
The process is
repeated until the bioresponses induced by the revised herbal composition are
within the
acceptable variation range by comparing with the standard HBR Array.
Example 19. Pi-edicting~biological,- activit~and therapeutic applications of
an
herbal composition. ~ " ~ ~'
According to the identified marker genes for PHY 906 (Figure 27), these genes
can be
used to predict the biological activities of the herbal composition. For
example, the following
underlined marker genes of PHY906 have been reported to involve in the
following biological
activities and therapeutic effects. ~ The only effective drug against ALL is
to inhibit the
aspara ine svnthetase due to increased cellular apoptosis (Handy et al.,
1998). Long-acting
drug somatostatin analogs are applied in the treatment of neurofibroma for
their tumor growth
inhibitory effect because they induce antiproliferative action mediated by the
inhibition of
G6PD, transketolase, or both (Boros et al., 1998). Ephrin-A1 is a new melanoma
growth
factor and is highly expressed during melanoma progression (Easty et al.,
1999). Mito,~en-
activated protein kinase (1VIAPKI family members have been recently reported
to have
opposing effects on apoptosis (Dabrowski et al., 2000). The expressions of
asparagine
synthetase, transketolase,~ ephrim-A1'- and MAPK are repressed with the higher
concentration of
PHY906 treatments. 'The dovsin=regulation of these genes are involved in cell
apoptosis. The
expression of the enzyme argininosuccinate synthetase, cathepsin G and
chemokirie RANTES
are highly induced in inflammatory mechanism. By the PHY906 treatment the
inflammatory
involved genes are suppressed: These literature reports provide a basis for
predicting the
biological activities or therapeutic.effects of an herbal composition.
References for Exaiiilile 19.
Handy-P; Periclou~AP; Avramis-VI (1998) The synergism of 6-mercapopurine plus
cytosine arabinoside~follo~ued by PEG-asparaginase in human leukemia cell
lines
(CCRF/CEM/0 and CCRF/CEM/ara-C/7A) 'is due to increased cellular apoptosis.
Anticancer
Research 18: 727-737-
Boros~LG; Brandes-JL;~~'u'suf FI; Cascante-M; Williams-RD; Schirmer-WJ (1998)
Inhibition of the oxidative and' nonoxidative pentose phosphate pathways by
somatostatin: a
possible mechanism of antiturilor action. Medical Hypotheses 50: 501-506
Easty-DJ; Hill-SP; Hsu-MY; Fallowfield-ME; Florenes-VA; Herlyn-M; Benett-DC
CA 02373708 2002-O1-15
WO 01/66803 PCT/USO1/07608
-99-
(1999) Up-regulation of ephrin=Al.during melanoma progression. Tnt. J. Cancer
84:494-501
Dabrowski-A; Tribilla-I; Dabrowska MI; Wereszczynska-SU; Gabryelewicz-A (2000)
Activation of mitogen-activated protein kinases in different models of
pancreatic acinar cell
damage. Z-Gastroentero1..38: 469-4:81
The foregoing detailed description has been given for clearness of
understanding only and
no unnecessary limitations should be understood therefrom as modifications
will be obvious to
those skilled in the art. ~ : ~ ..
While the invention has been described in connection with specific embodiments
thereof,
it will be understood. thatf if is capable of further modifications and this
application is intended
to cover any variations,yu~es; ;or adaptations of the invention following, in
general, the
principles of the invention and including such departures from the present
disclosure as come
within known or custorriary'practice:v,~ithin the art to which the invention
pertains and as may
be applied to the 'essential features hereinbefore set forth and as follows in
the scope of the
appended claims.