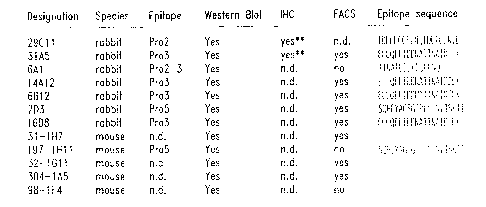Note: Descriptions are shown in the official language in which they were submitted.
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
COMPOSITIONS AND METHODS FOR THE THERAPY. DIAGNOSIS AND
MONITORING OF BREAST CANCER
TECHNICAL FIELD
The present invention relates generally to therapy, diagnosis and
monitoring of cancer, such as breast cancer. The invention is more
specifically related
to specific epitopes of mammaglobin, to antibodies and immune cells that
recognize
such epitopes and to methods for detecting mammaglobin in patient serum. Such
peptides, antibodies and cells may be used in vaccines and pharmaceutical
compositions
for prevention and treatment of breast cancer, and for the diagnosis and
monitoring of
breast cancers.
BACKGROUND OF THE INVENTION
Breast cancer is a significant health problem for women in the United
States and throughout the world. Although advances have been made in detection
and
treatment of the disease, breast cancer remains the second leading cause of
cancer-
related deaths in women, affecting more than 180,000 women in the United
States each
year. For women in North America, the life-time odds of getting breast cancer
are now
one in eight.
No vaccine or other universally successful method for the prevention or
treatment of breast cancer is currently available. Management of the disease
currently
relies on a combination of early diagnosis (through routine breast screening
procedures)
and aggressive treatment, which may include one or more of a variety of
treatments
such as surgery, radiotherapy, chemotherapy and hormone therapy. The course of
treatment for a particular breast cancer is often selected based on a variety
of prognostic
parameters, including an analysis of specific tumor markers. See, e.g., Porter-
Jordan
and Lippman, Breast Cancer 8:73-100, 1994. However, the use of established
markers
often leads to a result that is difficult to interpret, and the high mortality
observed in
breast cancer patients indicates that improvements are needed in the
treatment,
diagnosis and prevention of the disease.
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
In spite of considerable research into therapies and diagnostic methods,
there is a need in the art for improved methods for detecting and treating
breast cancers.
The present invention fulfills these needs and further provides other related
advantages.
SUMMARY OF THE INVENTION
Briefly stated. the present invention provides compositions and methods
for the diagnosis, therapy and monitoring of breast cancer. In one aspect, the
present
invention provides polypeptides comprising at least 7, preferably at least 9
and more
preferably at least 15 consecutive amino acid residues of an epitope of human
mammaglobin, wherein the epitope is selected from the group consisting of
IDELKECFLNQTDETLSNVE (Pro2; SEQ ID NO: I ); TTNAIDELKECFLNQ (Pro2-
3; SEQ ID NO: 2); SQHCYAGSGCPLLENVISKTI (Pros: SEQ ID NO: 3)
EYKELLQEFIDDNATTNAID (peptide SA: SEQ ID NO: 4) and KLLMVLMLA (mgb
1; SEQ ID NO: 5), such that the polypeptides contain no more than 30
consecutive
amino acid residues present within human mammaglobin.
Within other aspects, the present invention provides pharmaceutical
compositions comprising a polypeptide as described above and a physiologically
acceptable carrier.
Within a related aspect of the present invention, vaccines are provided.
Such vaccines comprise a polypeptide described above and an immunostimulant.
Within further aspects, the present invention provides antibodies, such as
monoclonal antibodies, or antigen-binding fragments thereof, that bind to a
mammaglobin epitope as described above, as well as diagnostic kits comprising
such
antibodies.
Also provided are isolated antibodies. or antigen-binding fragments
thereof, that specifically bind to glycosylated mammaglobin, and diagnostic
kits
comprising such antibodies.
The present invention further provides pharmaceutical compositions that
comprise: (a) an antibody or antigen-binding fragment thereof that
specifically binds to
an epitope as described above; and (b) a physiologically acceptable carrier.
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
Within further aspects, the present invention provides methods for
inhibiting the development of breast cancer in a patient. comprising
administering to a
patient a pharmaceutical composition or vaccine as recited above.
Within further aspects. the present provides methods for determining the
presence or absence of breast cancer in a patient, comprising (a) contacting a
biological
sample obtained from a patient with an antibody or antigen-binding fragment
thereof
that specifically binds to an epitope as described above; (b) detecting in the
sample an
amount of polypeptide that binds to the antibody or fragment thereof; and (c)
comparing
the amount of polypeptide with a predetermined cut-off value. Within preferred
embodiments, the antibody is a monoclonal antibody. Step (b) may comprise. for
example, a two-antibody sandwich assay.
The present invention also provides, within other aspects, methods for
monitoring the progression of breast cancer in a patient. Such methods
comprise the
steps of: (a) contacting a biological sample obtained from a patient at a
first point in
time with an antibody or antigen-binding fragment thereof that specifically
binds to an
epitope as described above; (b) detecting in the sample an amount of
polypeptide that
binds to the antibody or fragment thereof; (c) repeating steps (a) and (b)
using a
biological sample obtained from the patient at a subsequent point in time; and
(d)
comparing the amount of polypeptide detected in step (c) with the amount
detected in
step (b).
Within further aspects, the present invention provides methods for
removing tumor cells from a biological sample, comprising contacting a
biological
sample with T cells that specifically react with a mammaglobin epitope
selected from
the group consisting of EYKELLQEFIDDNATTNAID (peptide SA; SEQ ID NO: 4)
and KLLMVLMLA (mgb 1; SEQ ID NO: 5), wherein the step of contacting is
performed under conditions and for a time sufficient to permit the removal of
cells
expressing mammaglobin or a peptide epitope thereof from the sample.
Biological
samples include, for example, blood and fractions thereof.
Methods are further provided. within other aspects, for inhibiting the
development of breast cancer in a patient, comprising administering to a
patient a
biological sample treated as described above.
-,
J
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
The present invention also provides. within further aspects, methods for
stimulating and/or expanding T cells specific for mammaglobin. comprising
contacting
T cells with a peptide comprising the sequence EYKELLQEFIDDNATTNAID (peptide
SA; SEQ ID NO: 4) or KLLMVLMLA (mgb 1; SEQ ID NO: ~). wherein the peptide
comprises at least 7, preferably at least 9 and more preferably at least 15
consecutive
amino acid residues of human mammaglobin, wherein the peptide comprises no
more
than 30 consecutive amino acid residues of human mammaglobin, and wherein the
contact is performed under conditions and for a time sufficient to permit
stimulation
and/or expansion of T cells.
In related aspects, isolated T cell populations are provided, comprising T
cells prepared as described above.
Methods are also provided, within further aspects, for inhibiting the
development of breast cancer in a patient. comprising administering to a
patient an
effective amount of a T cell population as described above.
Within further aspects, methods are provided for inhibiting the
development of breast cancer in a patient, comprising the steps of: (a)
incubating CD4+
and/or CD8+ T cells isolated from a patient with a peptide comprising at least
7,
preferably at least 9 and more preferably at least 15 consecutive amino acid
residues of
human mammaglobin, wherein the peptide comprises no more than 30 consecutive
amino acid residues of human mammaglobin, and wherein the peptide comprises
the
sequence EYKELLQEFIDDNATTNAID (peptide SA; SEQ ID NO: 4) or
KLLMVLMLA (mgb l; SEQ ID NO: 5), such that T cells proliferate; and (b)
administering to the patient an effective amount of the proliferated T cells.
Methods are further provided for inhibiting the development of breast
cancer in a patient, comprising the steps of: (a) incubating CD4+ and/or CD8+
T cells
isolated from a patient with a peptide comprising at least 7, preferably at
least 9 and
more preferably at least 15 consecutive amino acid residues of human
mammaglobin,
wherein the peptide comprises no more than 30 consecutive amino acid residues
of
human mammaglobin, and wherein the peptide comprises the sequence
EYKELLQEFIDDNATTNAID (peptide SA; SEQ ID NO: 4) or KLLMVLMLA (mgb
4
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
l; SEQ ID NO: 5), such that T cells proliferate; (b) cloning at least one
proliferated cell;
and (c) administering to the patient an effective amount of the cloned T
cells.
Still further methods are provided wherein the presence or absence of
breast cancer may be determined in a patient by detecting the level of
mammaglobin
mRNA in sample of whole blood, or a fraction thereof, obtained from the
patient.
wherein epithelial cells have been removed from the sample. For example, such
detection may be achieved by (a) contacting a biological sample obtained from
a patient
with an oligonucleotide that hybridizes to a polynucleotide encoding
mammaglobin or a
complement thereof; (b) detecting in the sample an amount of a polynucleotide
that
hybridizes to the oligonucleotide; and (c) comparing the amount of
polvnucleotide that
hybridizes to the oligonucleotide to a predetermined cut-off value, and
therefrom
determining the presence or absence of breast cancer in the patient.
Within other aspects, the progression of breast cancer may be monitored
in a patient by detecting the level of mammaglobin mRNA in sample of whole
blood, or
a fraction thereof, obtained from the patient, wherein epithelial cells have
been removed
from the sample at two different times. For example, such monitoring may be
achieved
by: (a) contacting a sample obtained from a patient with an oligonucleotide
that
hybridizes to a mammaglobin polynucleotide; (b) detecting in the sample an
amount of
a polynucleotide that hybridizes to the oligonucleotide; (c) repeating steps
(a) and (b)
using a sample obtained from the patient at a subsequent point in time; and
(d)
comparing the amount of polynucleotide detected in step (c) to the amount
detected in
step (b) and therefrom monitoring the progression of the cancer in the
patient.
These and other aspects of the present invention will become apparent
upon reference to the following detailed description and attached drawings.
All
references disclosed herein are hereby incorporated by reference in their
entirety as if
each was incorporated individually.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1A is a summary of representative rabbit and mouse monoclonal
antibodies raised against the human mammaglobin protein. Included is a summary
of
assays in which these anti-mammaglobin monoclonal antibodies have been used to
detect mammaglobin. The epitope binding sequence for each monoclonal antibody
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
(SEQ ID Nos: 11-18) is also listed. Abbreviations are: n.d. = not determined;
FACS =
fluorescence activate cell sorter: IHC = immunohistochemistry.
Figures 1 B-1 C present the CDR sequence for rabbit monoclonal
antibodies 6A1 (SEQ ID NO: 19), 16D8 (SEQ ID Nos: 20-21), 6B12 (SEQ ID NO:
22),
2D3 (SEQ ID NO: 23). 14A12 (SEQ ID NO: 24), 29C11 (SEQ ID NO: 25) and 31A5
(SEQ ID NO: 26).
Figure 2 presents the human mammaglobin amino acid sequence (SEQ
ID NO: 27), along with peptide and recombinant regions used for epitope
mapping
studies. Various peptides (Prol-9 (SEQ ID NO: 27), Pro-20 (SEQ ID NO: 27) and
Glob-2 ( SEQ ID NO: 27)) spanning mammaglobin protein sequence were
synthesized
and used for epitope mapping of the monoclonal antibodies using the ELISA
method.
Each peptide sequence is indicated in bold and underlined. In addition, an N-
terminal
recombinant fragment of mammaglobin (SEQ ID NO: 28) was also used for epitope
mapping studies.
Figures 3A-3D present epitope mapping data for the rabbit and mouse
monoclonal antibodies obtained by the ELISA method. Figure 3A shows the
epitope
binding regions of the mouse monoclonal antibodies. Shaded areas are
considered
positive for the antibody. Epitope binding specificity for the affinity-
purified rabbit
polyclonal 967 is also demonstrated. Figures 3B-3D present epitope mapping
data for
the rabbit monoclonal antibodies 6B12 (Figure 3B), 29C11 (Figure 3C) and 2D3
(Figure 3D) using decreasing concentration of mammaglobin peptides and
recombinant
fragments.
Figures 4A-4B present the results of monoclonal antibody
characterization by FACS analysis. Each monoclonal antibody was used to detect
mammaglobin expression in MDA-MB-415 cells. Samples were fixed in 2%
formaldehyde and permeabilized with 0.5% saponin. MCF-7 cells do not express
mammaglobin and were used as a negative control.
Figure 5 presents Western blot detection of mammaglobin by each
monoclonal antibody. SDS-PAGE was performed on media in which MDA-MB-41 S
cells were grown, MDA-MB-41 S cell lysate and bacterially expressed
recombinant
6
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
mammaglobin, as indicated. Mammaglobin expression was detected with the
indicated
antibody.
Figure 6 is a table showing mammaglobin expression in breast tissue. but
not in other tissues tested. Mammaglobin expression in various tissues was
evaluated
by immunohistochemistry analysis using a combination of 29C 11 and 31 A~
rabbit
monoclonal antibodies.
Figures 7A-7C are graphs illustrating the results of sandwich assays
performed using the indicated rabbit monoclonal antibodies to detect
mammaglobin in
lysates and supernatants of MB415 cells.
Figure 8 is a graph showing the standard curve for a sandwich assay
using the polyclonal anti-967 serum in combination with the monoclonal
antibody 2D3
biotinylated.
Figure 9 is a table showing the results of sandwich assays using the
representative indicated antibodies to detect mammaglobin in patients with and
without
breast cancer.
Figure 10 presents the human mammaglobin amino acid sequence (SEQ
ID NO: 27), with underlined and bold peptide regions (SEQ ID Nos: 29-36) used
for
epitope mapping studies.
Figures 11 A and 11 B are graphs illustrating the recognition of CD4 T
cell lines for mammaglobin and various portions thereof, as indicated. Figure
11 A
shows T cell proliferation of three different CD4 T cell lines in response to
various
proteins and peptides. Figure 11 B shows interferon-y production by the same
cells lines
in response to the same proteins and peptides.
Figure 12 presents the human mammaglobin amino acid sequence (SEQ
ID NO: 27), along with peptide regions (SEQ ID Nos: 37-45) used for CD4+ and T
cell
epitope mapping studies.
Figures 13A-13C are graphs illustrating the recognition of JurkatA2Kb
cells pulsed with mgb-1 by CTL from HLA A2 transgenic mice immunized with mgb
1.
CTL from three different mice were tested at different effectoraarget ratios,
as
indicated. Each figure shows the percent specific lysis of cells that are
(solid circles)
and are not (open circles) pulsed with mgb-1.
7
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
Figures 14A-14C are graphs illustrating the recognition of JurkatA2Kb
cells pulsed with mgb-1 (triangles) or expressing full length mammaglobin
(mammaglobin) by CTL from HLA A2 transgenic mice immunized with mgb 1. CTL
from three different mice were tested at different effectoraarget ratios. as
indicated. In
each figure, the percent specific lysis of cells that do not express mgb-1 or
mammaglobin is represented by circles.
Figure 1 ~ is a histogram showing the tissue distribution for
mammaglobin. Copies of mammaglobin per ng (3-actin are shown for a variety of
normal and tumor tissues, as indicated.
Figure 16 is a graph showing the number of copies of mammaglobin
message in the breast cancer cell line MB415 as a function of the amount of
cells.
Figure 17 is a histogram showing the detection of mammaglobin in
epithelial cells isolated, using the Dynal isolation method, from the
peripheral blood of
patients with metastatic breast cancer compared to similar isolates from
normal blood
samples. Copies of mammaglobin per ng (3-actin are shown for thirty three
metastatic
and 11 normal samples, as indicated.
DETAILED DESCRIPTION OF THE INVENTION
As noted above, the present invention is generally directed to
compositions and methods for the therapy, diagnosis and monitoring of cancer,
such as
breast cancer. The compositions described herein may include mammaglobin
polynucleotides, polypeptides, epitopes or antibodies that specifically
recognize such
epitopes. The present invention is based, in part, on the discovery of certain
specific
epitopes of human mammaglobin, and antibodies that bind such epitopes. The
invention is further based, in part, on the discovery of antibodies that bind
mammaglobin in a glycosylation-sensitive manner. Other methods described
herein
employ techniques for detecting mammaglobin nucleic acid in patient blood, or
fractions thereof. These discoveries, within the context of the present
invention, permit
the generation of antibodies suited for diagnostic purposes, improved
therapies for
breast cancer, as well as diagnostic methods that can be based on the
detection of
mammaglobin RNA in blood permits sensitive diagnosis of breast cancer.
8
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
MAMMAGLOBIN POLYNUCLEOTIDES
The diagnostic methods provided herein generally employ mammaglobin
polynucleotides (e.g., oligonucleotides) as probes or primers to detect the
level of
mammaglobin nucleic acid in a sample obtained from a patient. A mammaglobin
oligonucleotide may encode a portion of a mammaglobin protein (e.g.. at least
15, 30 or
45 consecutive nucleotides). Oligonucleotides complementary to any such
sequences
are also encompassed by the present invention. Polynucleotides may be single-
stranded
(coding or antisense) or double-stranded, and may be DNA (genomic, cDNA or
synthetic) or RNA molecules. RNA molecules include HnRNA molecules. which
contain introns and correspond to a DNA molecule in a one-to-one manner, and
mRNA
molecules, which do not contain introns. Additional coding or non-coding
sequences
may, but need not, be present within a polynucleotide of the present
invention, and a
polynucleotide may, but need not, be linked to other molecules and/or support
materials.
Polynucleotides may comprise a native sequence (i.e.. a portion of
endogenous mammaglobin) or may comprise a variant of such a sequence.
Polynucleotide variants may contain one or more substitutions, additions,
deletions
and/or insertions such that the ability of the polynucleotide to hybridize to
a
mammaglobin polynucleotide under assay conditions is not substantially
diminished.
Preferably, such polynucleotide variants are capable of hybridizing under
moderately
stringent conditions to a naturally occurring DNA sequence encoding a native
mammaglobin (or a complementary sequence). Suitable moderately stringent
conditions include prewashing in a solution of 5 X SSC, 0.5% SDS, 1.0 mM EDTA
(pH
8.0); hybridizing at 50°C-65°C, 5 X SSC, overnight; followed by
washing twice at 65°
C for 20 minutes with each of 2X, O.SX and 0.2X SSC containing 0.1% SDS.
Polynucleotides may be prepared using any of a variety of techniques.
For example, polynucleotides may be amplified from cDNA prepared from cells
expressing mammaglobin, such as breast tumor cells. Such polynucleotides may
be
amplified via polymerase chain reaction (PCR). For this approach, sequence-
specific
primers may be designed based on known mammaglobin sequences, and may be
purchased or synthesized.
9
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
A portion of a coding sequence or a complementary sequence may be
designed as a probe or primer to detect gene expression. Probes may be labeled
by a
variety of reporter groups, such as radionuclides and enzymes, and are
preferably at
least 10 nucleotides in length, more preferably at least 20 nucleotides in
length and still
more preferably at least 30 nucleotides in length. Primers are preferably 22-
30
nucleotides in length.
Any polynucleotide may be further modified to increase stability in vivo.
Possible modifications include. but are not limited to, the addition of
flanking
sequences at the 5' and/or 3' ends; the use of phosphorothioate or 2' O-methyl
rather
than phosphodiesterase linkages in the backbone; and/or the inclusion of
nontraditional
bases such as inosine, queosine and wybutosine, as well as acetyl- methyl-.
thio- and
other modified forms of adenine, cytidine, guanine, thymine and uridine.
Nucleotide sequences as described herein may be joined to a variety of
other nucleotide sequences using established recombinant DNA techniques. For
example, a polynucleotide may be cloned into any of a variety of cloning
vectors,
including plasmids, phagemids, lambda phage derivatives and cosmids. Vectors
of
particular interest include expression vectors, replication vectors, probe
generation
vectors and sequencing vectors. In general, a vector will contain an origin of
replication
functional in at least one organism, convenient restriction endonuclease sites
and one or
more selectable markers. Other elements will depend upon the desired use, and
will be
apparent to those of ordinary skill in the art.
MAMMAGLOBIN EPITOPES AND POLYPEPTIDES
Within the context of the present invention, polypeptides comprise at
least one mammaglobin epitope, or a variant thereof. An "epitope" is a portion
of
mammaglobin to which one or more antibodies within an anti-mammaglobin
antiserum
specifically binds, or with which one or more mammaglobin-specific T cells
specifically reacts, as described herein. An epitope may, but need not, be
specifically
bound by an antibody in a glycosylation-sensitive manner (i.e., the antibody
may bind
to a glycosylated epitope, to a deglycosylated epitope or to both).
Polypeptides
comprising a mammaglobin epitope generally comprise at least 7 consecutive
amino
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
acid residues of human mammaglobin, and preferably 9-30 consecutive amino acid
residues of human mammaglobin. It should be noted that the size of an epitope
may
vary depending on whether the epitope is recognized by CD4' T cells , CD8+ T
cells or
antibodies. In general, however, a 9-amino acid sequence is sufficient for TCL
S recognition. Polypeptides as described herein may be of any length.
Additional
sequences derived from the native protein and/or heterologous sequences may be
present, and such sequences may (but need not) possess further immunogenic or
antigenic properties.
Certain preferred epitopes comprise one of the following sequences, or a
portion thereof that comprises at least 7, preferably at least 9 and more
preferably at
least 15 consecutive amino acid residues of such a sequence:
IDELKECFLNQTDETLSNVE (Pro2; SEQ ID NO: 1);
TTNAIDELKECFLNQ (Pro2-3; SEQ ID NO: 2);
SQHCYAGSGCPLLENVISKTI (Pros; SEQ ID NO: 3);
EYKELLQEFIDDNATTNAID (peptide SA; SEQ ID NO: 4) or
KLLMVLMLA (mgb 1; SEQ ID NO: 5).
Other preferred epitopes comprise a glycosylation site of mammaglobin. Such
epitopes
are particularly useful for the generation of antibodies that specifically
bind to
glycosylated mammaglobin. Two such sites are the N-linked glycosylation sites
asparagine (Asp)-53 (QEFIDDNATTNAI) (SEQ ID NO: 6) and Asp-68
(LKECFLNQTDETL) (SEQ ID NO: 7). Other such sites may be readily identified
using, for example, an antibody library comprising antibodies to different
glycosylation
combinations. The binding of such antibodies to native mammaglobin from breast
carcinoma cell lines may be assayed using conventional ELISA and blotting
techniques.
Established biochemical techniques may also be used to identify other
mammaglobin
glycosylation sites.
As noted above, a polypeptide may comprise a variant of a native
mammaglobin epitope. A "variant," as used herein, differs from a native
epitope in one
or more substitutions, deletions, additions and/or insertions, such that the
ability of the
variant to be bound by an antibody specific for the epitope is not
substantially
diminished. In other words, the ability of a variant to react with epitope-
specific
11
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
antisera or isolated antibodies may be enhanced or unchanged. relative to the
native
protein. or may be diminished by less than 50%, and preferably less than 20%.
relative
to the native protein. Such variants may generally be identified by modifying
an
epitope as provided herein and evaluating the reactivity of the modified
epitope with
epitope-specific antibodies or antisera as described herein. Preferred
variants include
those in which substitutions are made at no more than 20% of the residues in
the
epitope.
Preferably, a variant contains conservative substitutions. A
"conservative substitution" is one in which an amino acid is substituted for
another
amino acid that has similar properties, such that one skilled in the art of
peptide
chemistry would expect the secondary structure and hydropathic nature of the
polypeptide to be substantially unchanged. Amino acid substitutions may
generally be
made on the basis of similarity in polarity, charge, solubility,
hydrophobicity,
hydrophilicity and/or the amphipathic nature of the residues. For example,
negatively
charged amino acids include aspartic acid and glutamic acid; positively
charged amino
acids include lysine and arginine; and amino acids with uncharged polar head
groups
having similar hydrophilicity values include leucine, isoleucine and valine;
glycine and
alanine; asparagine and glutamine; and serine, threonine, phenylalanine and
tyrosine.
Other groups of amino acids that may represent conservative changes include:
(1) ala,
pro, gly, glu, asp, gln, asn, ser, thr; (2) cys, ser, tyr, thr; (3) val, ile,
leu, met, ala, phe;
(4) lys, arg, his; and (5) phe, tyr, trp, his. A variant may also, or
alternatively, contain
nonconservative changes. Variants may also (or alternatively) be modified by,
for
example, the deletion or addition of amino acids that have minimal influence
on the
immunogenicity, secondary structure and hydropathic nature of the polypeptide.
As noted above, polypeptides may comprise a signal (or leader)
sequence at the N-terminal end of the protein, which co-translationally or
post-
translationally directs transfer of the protein. The polypeptide may also be
conjugated
to a linker or other sequence for ease of synthesis, purification or
identification of the
polypeptide (e.g., poly-His), or to enhance binding of the polypeptide to a
solid support.
For example, a polypeptide may be conjugated to an immunoglobulin Fc region.
12
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
Polypeptides may be prepared using any of a variety of well known
techniques. Recombinant polypeptides may be readily prepared from mammaglobin
DNA sequences using any of a variety of expression vectors known to those of
ordinary
skill in the art. Expression may be achieved in any appropriate host cell that
has been
transformed or transfected with an expression vector containing a DNA molecule
that
encodes a recombinant polypeptide. Suitable host cells include prokaryotes,
yeast and
higher eukaryotic cells. Preferably, the host cells employed are E. coli,
yeast or a
mammalian cell line such as COS or CHO. Supernatants from suitable host/vector
systems that secrete recombinant protein or polypeptide into culture media may
be first
concentrated using a commercially available filter. Following concentration,
the
concentrate may be applied to a suitable purification matrix such as an
affinity matrix or
an ion exchange resin. Finally. one or more reverse phase HPLC steps can be
employed
to further purify a recombinant polypeptide.
Polypeptides having fewer than about 100 amino acids, and generally
fewer than about 50 amino acids, may also be generated by synthetic means,
using
techniques well known to those of ordinary skill in the art. For example, such
polypeptides may be synthesized using any of the commercially available solid-
phase
techniques, such as the Merrifield solid-phase synthesis method, where amino
acids are
sequentially added to a growing amino acid chain. See Merrifield, J. Am. Chem.
Soc.
85:2149-2146, 1963. Equipment for automated synthesis of polypeptides is
commercially available from suppliers such as Applied BioSystems, Inc. (Foster
City,
CA), and may be operated according to the manufacturer's instructions.
Within related aspects, polynucleotides that encode a polypeptide as
provided herein are provided. In general, polypeptides and polynucleotides as
described
herein are isolated. An "isolated" polypeptide or polynucleotide is one that
is removed
from its original envirornnent. For example, a naturally-occurring protein is
isolated if
it is separated from some or all of the coexisting materials in the natural
system.
Preferably, such polypeptides are at least about 90% pure, more preferably at
least about
95% pure and most preferably at least about 99% pure. A polynucleotide is
considered
to be isolated if, for example, it is cloned into a vector that is not a part
of the natural
environment.
13
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
ANTIBODIES AND FRAGMENTS THEREOF
The present invention further provides agents, such as antibodies and
antigen-binding fragments thereof, that specifically bind to a mammaglobin
epitope. As
used herein. an antibody, or antigen-binding fragment thereof, is said to
"specifically
bind" to a mammaglobin epitope if it reacts at a detectable level (within, for
example,
an ELISA) with the epitope, and does not react detectable with unrelated
proteins under
similar conditions. Preferred antibodies bind detectably to an epitope of
mammaglobin,
but do not bind detectably to other portions of mammaglobin that do not
overlap with
the epitope (or that overlap by less than five amino acid residues). As used
herein,
"binding" refers to a noncovalent association between two separate molecules
such that
a complex is formed. The ability to bind may be evaluated by. for example,
determining a binding constant for the formation of the complex. The binding
constant
is the value obtained when the concentration of the complex is divided by the
product of
the component concentrations. In general, two compounds are said to "bind," in
the
context of the present invention, when the binding constant for complex
formation
exceeds about 10~ L/mol. The binding constant maybe determined using methods
well
known in the art.
Binding agents may be further capable of differentiating between
patients with and without a cancer, such as breast cancer, using the
representative assays
provided herein. In other words, antibodies or other binding agents that bind
to a
mammaglobin epitope will generate a signal indicating the presence of a cancer
in at
least about 20% of patients with the disease, and will generate a negative
signal
indicating the absence of the disease in at least about 90% of individuals
without the
cancer. To determine whether a binding agent satisfies this requirement,
biological
samples (e.g., blood, sera, urine and/or tumor biopsies) from patients with
and without a
cancer (as determined using standard clinical tests) may be assayed as
described herein
for the presence of polypeptides that bind to the binding agent. It will be
apparent that a
statistically significant number of samples with and without the disease
should be
assayed. Each binding agent should satisfy the above criteria; however, those
of
14
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
ordinary skill in the art will recognize that binding agents may be used in
combination
to improve sensitivity.
Any agent that satisfies the above requirements may be a binding agent.
For example, a binding agent may be a ribosome, with or without a peptide
component.
and RNA molecule or a polypeptide. In a preferred embodiment, a binding agent
is an
antibody or an antigen-binding fragment thereof. Antibodies may be prepared by
any of
a variety of techniques known to those of ordinary skill in the art. See,
e.g., Harlow and
Lane, Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, 1988. In
general, antibodies can be produced by cell culture techniques. including the
generation
of monoclonal antibodies as described herein, or via transfection of antibody
genes into
suitable bacterial or mammalian cell hosts, in order to allow for the
production of
recombinant antibodies. In one technique, an immunogen comprising the
polypeptide is
initially injected into any of a wide variety of mammals (e.g., mice, rats,
rabbits, sheep
or goats). In this step, the polypeptides of this invention may serve as the
immunogen
without modification. Alternatively, particularly for relatively short
polypeptides, a
superior immune response may be elicited if the polypeptide is joined to a
carrier
protein, such as bovine serum albumin or keyhole limpet hemocyanin. The
immunogen
is injected into the animal host, preferably according to a predetermined
schedule
incorporating one or more booster immunizations, and the animals are bled
periodically.
Polyclonal antibodies specific for the polypeptide may then be purified from
such
antisera by, for example, affinity chromatography using the polypeptide
coupled to a
suitable solid support.
Monoclonal antibodies specific for an antigenic polypeptide of interest
may be prepared, for example, using the technique of Kohler and Milstein, Eur.
J.
Immunol. 6:511-519, 1976, and improvements thereto. Briefly, these methods
involve
the preparation of immortal cell lines capable of producing antibodies having
the
desired specificity (i.e., reactivity with the polypeptide of interest). Such
cell lines may
be produced, for example, from spleen cells obtained from an animal immunized
as
described above. The spleen cells are then immortalized by, for example,
fusion with a
myeloma cell fusion partner, preferably one that is syngeneic with the
immunized
animal. A variety of fusion techniques may be employed. For example, the
spleen cells
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
and myeloma cells may be combined with a nonionic detergent for a few minutes
and
then plated at low density on a selective medium that supports the growth of
hybrid
cells, but not myeloma cells. A preferred selection technique uses HAT
(hypoxanthine.
aminopterin, thymidine) selection. After a sufficient time, usually about 1 to
2 weeks,
colonies of hybrids are observed. Single colonies are selected and their
culture
supernatants tested for binding activity against the polypeptide. Hybridomas
having
high reactivity and specificity are preferred.
Monoclonal antibodies may be isolated from the supernatants of growing
hybridoma colonies. In addition, various techniques may be employed to enhance
the
yield, such as injection of the hybridoma cell line into the peritoneal cavity
of a suitable
vertebrate host, such as a mouse. Monoclonal antibodies may then be harvested
from
the ascites fluid or the blood. Contaminants may be removed from the
antibodies by
conventional techniques, such as chromatography, gel filtration,
precipitation, and
extraction. The polypeptides of this invention may be used in the purification
process
in, for example, an affinity chromatography step.
Certain preferred monoclonal antibodies specifically bind to an epitope
sequence recited above (Pro2, Pro2-3, Pros, peptide SA or mgb 1 ). Such
antibodies
include the rabbit antibodies designated 29C 11, 6A 1, 2D3 and 16D8 and the
mouse
antibody designated 197-1H11 herein. Other preferred antibodies bind to other
sequences, such as conformationally dependent sequences. Such antibodies
include
those designated 31-1H7, 32-1611, 304-lA5 and 98-1F4 herein. Other preferred
antibodies bind to a glycosylation site of mammaglobin with an affinity that
is
dependent on glycosylation. For example, certain antibodies specifically bind
to
glycosylated mammaglobin (i.e., require glycosylation of a particular
glycosylation site
for optimal binding). As used herein, an antibody, or antigen binding fragment
thereof,
specifically binds to glycosylated mammaglobin if it binds to a glycosylated
mammaglobin with an affinity that is at least two-fold, preferably at least
five-fold,
greater than the affinity with which it binds deglycosylated mammaglobin
(mammaglobin that is enzymatically deglycosylated, using well known
techniques, so
as to remove substantially all glycosylation). Glycosylation results when
oligosaccharide units are attached to the protein via asparagine (N-linked) or
serine and
16
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
threonine residues (O-linked). Compared to normal cells, protein glycosylation
is often
altered in tumor cells. This difference in protein glycosylation can be
exploited to
provide a tumor-specific antibody for diagnostic purposes (e.g., for the
diagnosis of
breast cancer). This is particularly true for heavily glycosylated proteins,
such as
mammaglobin. Although the predicted molecular weight of mammaglobin is 9.2
kDa,
the mature form of this protein expressed in breast carcinoma cells runs at a
molecular
weight of approximately 18-25kDa. It has been found, within the context of the
present
invention, that the additional molecular weight of mammaglobin is due to the
attachment of oligosaccharides. Thus, roughly one half or more of the
molecular
weight of mammaglobin is due to glycosylation.
Within certain embodiments, the use of antigen-binding fragments of
antibodies may be preferred. Such fragments include Fab fragments. which may
be
prepared using standard techniques. Briefly, immunoglobulins may be purified
from
rabbit serum by affinity chromatography on Protein A bead columns (Harlow and
Lane,
Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, 1988) and
digested
by papain to yield Fab and Fc fragments. The Fab and Fc fragments may be
separated
by affinity chromatography on protein A bead columns.
Monoclonal antibodies of the present invention may be coupled to one or
more therapeutic agents. Suitable agents in this regard include radionuclides,
differentiation inducers, drugs, toxins, and derivatives thereof. Preferred
radionuclides
include 9°Y, ''~I, '25I, 'i'I, '86Re, 'ggRe, ''"At, and 2'2Bi.
Preferred drugs include
methotrexate, and pyrimidine and purine analogs. Preferred differentiation
inducers
include phorbol esters and butyric acid. Preferred toxins include ricin,
abrin, diptheria
toxin, cholera toxin, gelonin, Pseudomonas exotoxin, Shigella toxin, and
pokeweed
antiviral protein.
A therapeutic agent may be coupled (e.g., covalently bonded) to a
suitable monoclonal antibody either directly or indirectly (e.g., via a linker
group). A
direct reaction between an agent and an antibody is possible when each
possesses a
substituent capable of reacting with the other. For example, a nucleophilic
group, such
as an amino or sulfhydryl group, on one may be capable of reacting with a
carbonyl-
17
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
containing group, such as an anhydride or an acid halide, or with an alkyl
group
containing a good leaving group (e.g., a halide) on the other.
Alternatively, it may be desirable to couple a therapeutic agent and an
antibody via a linker group. A linker group can function as a spacer to
distance an
antibody from an agent in order to avoid interference with binding
capabilities. A
linker group can also serve to increase the chemical reactivity of a
substituent on an
agent or an antibody. and thus increase the coupling efficiency. An increase
in
chemical reactivity may also facilitate the use of agents, or functional
groups on agents,
which otherwise would not be possible.
It will be evident to those skilled in the art that a variety of bifunctional
or polyfunctional reagents, both homo- and hetero-functional (such as those
described
in the catalog of the Pierce Chemical Co., Rockford, IL), may be employed as
the linker
group. Coupling may be effected, for example. through amino groups, carboxyl
groups,
sulfhydryl groups or oxidized carbohydrate residues. There are numerous
references
describing such methodology, e.g., U.S. Patent No. 4,671,958, to Rodwell et
al.
Where a therapeutic agent is more potent when free from the antibody
portion of the immunoconjugates of the present invention, it may be desirable
to use a
linker group which is cleavable during or upon internalization into a cell. A
number of
different cleavable linker groups have been described. The mechanisms for the
intracellular release of an agent from these linker groups include cleavage by
reduction
of a disulfide bond (e.gl., U.S. Patent No. 4.489,710, to Spitler), by
irradiation of a
photolabile bond (e.g., U.S. Patent No. 4,625,014, to Senter et al.), by
hydrolysis of
derivatized amino acid side chains (e.g., U.S. Patent No. 4,638,045, to Kohn
et al.), by
serum complement-mediated hydrolysis (e.g., U.S. Patent No. 4,671,958, to
Rodwell
et al.), and acid-catalyzed hydrolysis (e.g., U.S. Patent No. 4,569,789, to
Blattler et al.).
It may be desirable to couple more than one agent to an antibody. In one
embodiment, multiple molecules of an agent are coupled to one antibody
molecule. In
another embodiment, more than one type of agent may be coupled to one
antibody.
Regardless of the particular embodiment, immunoconjugates with more than one
agent
may be prepared in a variety of ways. For example, more than one agent may be
18
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
coupled directly to an antibody molecule, or linkers that provide multiple
sites for
attachment can be used. Alternatively, a carrier can be used.
A carrier may bear the agents in a variety of ways, including covalent
bonding either directly or via a linker group. Suitable carriers include
proteins such as
albumins (e.g., U.S. Patent No. 4,507.234, to Kato et al.), peptides and
polysaccharides
such as aminodextran (e.g., U.S. Patent No. 4,699,784. to Shih et al.). A
carrier may
also bear an agent by noncovalent bonding or by encapsulation, such as within
a
liposome vesicle (e.g., U.S. Patent Nos. 4,429,008 and 4,873.088). Carriers
specific for
radionuclide agents include radiohalogenated small molecules and chelating
compounds. For example, U.S. Patent No. 4,735,792 discloses representative
radiohalogenated small molecules and their synthesis. A radionuclide chelate
may be
formed from chelating compounds that include those containing nitrogen and
sulfur
atoms as the donor atoms for binding the metal, or metal oxide, radionuclide.
For
example, U.S. Patent No. 4,673,562, to Davison et al. discloses representative
chelating
compounds and their synthesis.
A variety of routes of administration for the antibodies and
immunoconjugates may be used. Typically, administration will be intravenous,
intramuscular, subcutaneous or in the bed of a resected tumor. It will be
evident that the
precise dose of the antibody/immunoconjugate will vary depending upon the
antibody
used, the antigen density on the tumor, and the rate of clearance of the
antibody.
T CELLS
Immunotherapeutic compositions may also, or alternatively, comprise T
cells that recognize mammaglobin. Such cells may generally be prepared in
vitro or ex
vivo, using standard procedures. For example, T cells may be isolated from
bone
marrow, peripheral blood, or a fraction of bone marrow or peripheral blood of
a patient,
using a commercially available cell separation system. such as the IsolexTM
System,
available from Nexell Therapeutics, Inc. (Irvine, CA; see also U.S. Patent No.
5,240,856; U.S. Patent No. 5.215,926; WO 89/06280; WO 91/16116 and WO
92/07243). Alternatively, T cells may be derived from related or unrelated
humans,
non-human mammals, cell lines or cultures. Briefly, T cells, which may be
isolated
19
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
from a patient or a related or unrelated donor by routine techniques (such as
by
Ficoll/Hypaque density gradient centrifugation of peripheral blood
lymphocytes), are
incubated with a mammaglobin polypeptide. For example, T cells may be
incubated in
vitro for 2-9 days (typically 4 days) at 37°C with a mammaglobin
polypeptide (e.g., 5 to
25 ~g/ml) or cells synthesizing a comparable amount of mammaglobin
polypeptide. It
may be desirable to incubate a separate aliquot of a T cell sample in the
absence of
mammaglobin polypeptide to serve as a control.
T cells may be stimulated with a mammaglobin polypeptide,
polynucleotide encoding a mammaglobin polypeptide and/or an antigen presenting
cell
(APC) that expresses such a polypeptide. Such stimulation is performed under
conditions and for a time sufficient to permit the generation of T cells that
are specific
for the polypeptide. Preferably, a mammaglobin polypeptide or polynucleotide
is
present within a delivery vehicle, such as a microsphere, to facilitate the
generation of
specific T cells.
T cells are considered to be specific for a mammaglobin polypeptide if
the T cells specifically proliferate, secrete cytokines or kill target cells
coated with the
polypeptide or expressing a gene encoding the polypeptide. T cell specificity
may be
evaluated using any of a variety of standard techniques. For example, within a
chromium release assay or proliferation assay, a stimulation index of more
than two
fold increase in lysis and/or proliferation, compared to negative controls,
indicates T
cell specificity. Such assays may be performed, for example, as described in
Chen et
al., Cancer Res. X4:1065-1070, 1994. Alternatively, detection of the
proliferation of
T cells may be accomplished by a variety of known techniques. For example, T
cell
proliferation can be detected by measuring an increased rate of DNA synthesis
(e.g., by
pulse-labeling cultures of T cells with tritiated thymidine and measuring the
amount of
tritiated thymidine incorporated into DNA). Other ways to detect T cell
proliferation
include measuring increases in interleukin-2 (IL-2) production, Ca2+ flux, or
dye
uptake, such as 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium.
Alternatively,
synthesis of lymphokines (such as interferon-gamma) can be measured or the
relative
number of T cells that can respond to a mammaglobin polypeptide may be
quantified.
Contact with a mammaglobin polypeptide (100 ng/ml - 100 ug/ml, preferably 200
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
ng/ml - 2~ ~g/ml) for 3 - 7 days should result in at least a two fold increase
in
proliferation of the T cells. Contact as described above for 2-3 hours should
result in
activation of the T cells, as measured using standard cytokine assays in which
a two
fold increase in the level of cytokine release (e.g., TNF or IFN-y) is
indicative of T cell
activation (see Coligan et al., Current Protocols in Immunology, vol. 1. Wiley
Interscience (Greene 1998)). Mammaglobin-specific T cells may be expanded
using
standard techniques. Within preferred embodiments, the T cells are derived
from a
patient, a related donor or an unrelated donor, and are administered to the
patient
following stimulation and expansion.
T cells that have been activated in response to a mammaglobin
polypeptide, polynucleotide or polypeptide-expressing APC may be CD4~ and/or
CD8+.
Specific activation of CD4+ or CD8+ T cells may be detected in a variety of
ways.
Methods for detecting specific T cell activation include detecting the
proliferation of
T cells, the production of cytokines (e.g., lymphokines), or the generation of
cytolytic
activity (i. e., generation of cytotoxic T cells specific for mammaglobin).
For CD4+
T cells, a preferred method for detecting specific T cell activation is the
detection of the
proliferation of T cells. For CD8+ T cells, a preferred method for detecting
specific
T cell activation is the detection of the generation of cytolytic activity.
For therapeutic purposes, CD4+ or CD8+ T cells that proliferate in
response to a mammaglobin polypeptide, polynucleotide or APC can be expanded
in
number either in vitro or in vivo. Proliferation of such T cells in vitro may
be
accomplished in a variety of ways. For example, the T cells can be re-exposed
to a
mammaglobin polypeptide (e.g., a short peptide corresponding to an immunogenic
portion of such a polypeptide) with or without the addition of T cell growth
factors,
such as interleukin-2, and/or stimulator cells that synthesize a mammaglobin
polypeptide. The addition of stimulator cells is preferred where generating
CD8+ T cell
responses. T cells can be grown to large numbers in vitro with retention of
specificity
in response to intermittent restimulation with mammaglobin polypeptide.
Briefly, for in
vitro stimulation, lymphocytes may be placed in a vessel with media containing
human
serum, mammoglobin protein or peptide and cytokines such as IL-2, IL-10 and IL-
7.
Cells may be incubated for seven to fourteen days and then restimulated in a
similar
21
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
manner using autologous antigen presenting cells, mammoglobin protein or
peptide and
cytokines. Antigen specific T cells may also be expanded in vita°o
using either antigen
or a mitogen or non-specific stimulator such as oe-CD3 or PHA.
Alternatively, one or more T cells that proliferate in the presence of
mammaglobin polypeptide can be expanded in number by cloning. Methods for
cloning cells are well known in the art, and include limiting dilution.
Responder T cells
may be purified from the peripheral blood of sensitized patients by density
gradient
centrifugation and sheep red cell rosetting and established in culture by
stimulating with
the nominal antigen in the presence of irradiated autologous filler cells. In
order to
generate CD4+ T cell lines, mammaglobin polypeptide is used as the antigenic
stimulus
and APC derived from autologous peripheral blood lymphocytes (PBL) or
lymphoblastoid cell lines (LCL) immortalized by infection with Epstein Barr
virus are
used as antigen presenting cells. In order to generate CD8+ T cell lines,
autologous
antigen-presenting cells transfected with an expression vector that produces
mammaglobin polypeptide may be used as stimulator cells. Established T cell
lines
may be cloned following antigen stimulation by plating stimulated T cells at a
frequency of 0.5 cells per well in 96-well flat-bottom plates with 1 x 106
irradiated PBL
or LCL cells and recombinant interleukin-2 (rIL2) (50 U/ml). Wells with
established
clonal growth may be identified at approximately 2-3 weeks after initial
plating and
restimulated with appropriate antigen in the presence of autologous antigen-
presenting
cells, then subsequently expanded by the addition of low doses of rIL2 (10
U/ml) 2-3
days following antigen stimulation. T cell clones may be maintained in 24-well
plates
by periodic restimulation with antigen and rIL2 approximately every two weeks.
Cloned and/or expanded cells may be administered back to the patient as
described, for
example, by Chang et al., Crit. Rev. Oncol. Hematol. 22:213, 1996.
Within certain embodiments, allogeneic T-cells may be primed (i.e.,
sensitized to mammaglobin) in vivo and/or in vitro. Such priming may be
achieved by
contacting T cells with a mammaglobin polypeptide, a polynucleotide encoding
such a
polypeptide or a cell producing such a polypeptide under conditions and for a
time
sufficient to permit the priming of T cells. In general, T cells are
considered to be
primed if, for example, contact with a mammaglobin polypeptide results in
proliferation
22
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
and/or activation of the T cells, as measured by standard proliferation,
chromium
release and/or cytokine release assays as described herein. A stimulation
index of more
than two fold increase in proliferation or lysis. and more than three fold
increase in the
level of cytokine, compared to negative controls, indicates T-cell
specificity. Cells
primed in vitro may be employed, for example, within a bone marrow
transplantation or
as donor lymphocyte infusion.
PHARMACEUTICAL COMPOSITIONS AND VACCINES
Within certain aspects, polypeptides, polynucleotides, T cells and/or
binding agents described herein may be incorporated into pharmaceutical
compositions
or immunogenic compositions (i.e., vaccines). Alternatively, a pharmaceutical
composition may comprise an antigen-presenting cell (e.g.. a dendritic cell)
transfected
with a mammaglobin polynucleotide such that the antigen presenting cell
expresses a
mammaglobin polypeptide. Pharmaceutical compositions comprise one or more such
compounds and a physiologically acceptable earner. Vaccines may comprise one
or
more such compounds and an immunostimulant. An immunostimulant may be any
substance that enhances or potentiates an immune response (antibody- and/or
cell-
mediated) to an exogenous antigen. Examples of immunostimulants include
adjuvants,
biodegradable microspheres (e.g., polylactic galactide) and liposomes (into
which the
compound is incorporated; see e.g., Fullerton, U.S. Patent No. 4,235,877).
Vaccine
preparation is generally described in, for example, M.F. Powell and M.J.
Newman, eds.,
"Vaccine Design (the subunit and adjuvant approach)," Plenum Press (NY, 1990.
Pharmaceutical compositions and vaccines within the scope of the present
invention
may also contain other compounds, which may be biologically active or
inactive. For
example, one or more immunogenic portions of other tumor antigens may be
present,
either incorporated into a fusion polypeptide or as a separate compound,
within the
composition or vaccine.
A pharmaceutical composition or vaccine may contain DNA encoding
one or more of the polypeptides as described above, such that the polypeptide
is
generated in situ. As noted above, the DNA may be present within any of a
variety of
delivery systems known to those of ordinary skill in the art, including
nucleic acid
23
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
expression systems, bacteria and viral expression systems and mammalian
expression
systems. Numerous gene delivery techniques are well known in the art, such as
those
described by Rolland, Crit. Rev. Therap. Drub Carrier .Systems 1:143-198,
1998, and
references cited therein. Appropriate nucleic acid expression systems contain
the
necessary DNA sequences for expression in the patient (such as a suitable
promoter and
terminating signal). Bacterial delivery systems involve the administration of
a
bacterium (such as Bacillus-Calmette-Guerrin) that expresses an immunogenic
portion
of the polypeptide on its cell surface or secretes such an epitope. In a
preferred
embodiment, the DNA may be introduced using a viral expression system (e.g.,
vaccinia or other pox virus, retrovirus, or adenovirus), which may involve the
use of a
non-pathogenic (defective), replication competent virus. Suitable systems are
disclosed,
for example, in Fisher-Hoeh et al.. Proc. Natl. Acad. Sci. USA 86:317-321,
1989;
Flexner et al., Ann. N.Y. Acad. Sci. 569:86-103, 1989; Flexner et al., Vaccine
8:17-21,
1990; U.S. Patent Nos. 4,603,112, 4,769,330, and 5,017,487; WO 89/01973; U.S.
Patent No. 4,777,127; GB 2,200,651; EP 0,345,242; WO 91/02805; Berkner,
Biotechniques 6:616-627, 1988; Rosenfeld et al., Science 252:431-434, 1991;
Kolls et
al., Proc. Natl. Acad. Sci. USA 91:215-219, 1994; Kass-Eisler et al., Proc.
Natl. Acad
Sci. USA 90:11498-11502, 1993; Guzman et al., Circulation 88:2838-2848, 1993;
and
Guzman et al., Cir. Res. 73:1202-1207, 1993. Techniques for incorporating DNA
into
such expression systems are well known to those of ordinary skill in the art.
The DNA
may also be "naked," as described, for example, in Ulmer et al., Science
29:1745-1749.
1993 and reviewed by Cohen, Science 259:1691-1692, 1993. The uptake of naked
DNA may be increased by coating the DNA onto biodegradable beads, which are
efficiently transported into the cells. It will be apparent that a vaccine may
comprise
both a polynucleotide and a polypeptide component. Such vaccines may provide
for an
enhanced immune response.
It will be apparent that a vaccine may contain pharmaceutically
acceptable salts of the polynucleotides and polypeptides provided herein. Such
salts
may be prepared from pharmaceutically acceptable non-toxic bases, including
organic
bases (e.g., salts of primary, secondary and tertiary amines and basic amino
acids) and
24
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
inorganic bases (e.g.. sodium, potassium, lithium, ammonium. calcium and
magnesium
salts).
While any suitable carrier known to those of ordinary skill in the art may
be employed in the pharmaceutical compositions of this invention. the type of
carrier
will vary depending on the mode of administration. Compositions of the present
invention may be formulated for any appropriate manner of administration,
including
for example, topical, oral, nasal, intravenous, intracranial, intraperitoneal,
subcutaneous
or intramuscular administration. For parenteral administration, such as
subcutaneous
injection, the carrier preferably comprises water, saline, alcohol, a fat, a
wax or a buffer.
For oral administration, any of the above carriers or a solid carrier. such as
mannitol,
lactose, starch, magnesium stearate, sodium saccharine, talcum, cellulose,
glucose,
sucrose, and magnesium carbonate, may be employed. Biodegradable microspheres
(e.g., polylactate polyglycolate) may also be employed as carriers for the
pharmaceutical compositions of this invention. Suitable biodegradable
microspheres
are disclosed, for example, in U.S. Patent Nos.4,897,268; 5,075,109;
5,928,647;
5,811,128; 5,820,883; 5,853,763; 5,814,344 and 5,942,252.
Such compositions may also comprise buffers (e.g., neutral buffered
saline or phosphate buffered saline), carbohydrates (e.g., glucose, mannose,
sucrose or
dextrans), mannitol, proteins, polypeptides or amino acids such as glycine,
antioxidants,
bacteriostats, chelating agents such as EDTA or glutathione, adjuvants (e.g.,
aluminum
hydroxide), solutes that render the formulation isotonic, hypotonic or weakly
hypertonic
with the blood of a recipient, suspending agents, thickening agents and/or
preservatives.
Alternatively, compositions of the present invention may be formulated as a
lyophilizate. Compounds may also be encapsulated within liposomes using well
known
technology.
Any of a variety of immunostimulants may be employed in the vaccines
of this invention. For example, an adjuvant may be included. Most adjuvants
contain a
substance designed to protect the antigen from rapid catabolism, such as
aluminum
hydroxide or mineral oil, and a stimulator of immune responses. such as lipid
A,
Bortadella pertussis or Mycobacterium tuberculosis derived proteins. Suitable
adjuvants are commercially available as, for example, Freund's Incomplete
Adjuvant
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
and Complete Adjuvant (Difco Laboratories. Detroit, MI); Merck Adjuvant 65
(Merck
and Company, Inc., Rahway, NJ); AS-2 (SmithKline Beecham, Philadelphia, PA):
aluminum salts such as aluminum hydroxide gel (alum) or aluminum phosphate;
salts of
calcium, iron or zinc; an insoluble suspension of acylated tyrosine: acylated
sugars;
canonically or anionically derivatized polysaccharides; polyphosphazenes:
biodegradable microspheres; monophosphoryl lipid A and quil A. Cytokines, such
as
GM-CSF or interleukin-2, -7, or -12, may also be used as adjuvants.
Within the vaccines provided herein. the adjuvant composition is
preferably designed to induce an immune response predominantly of the Thl
type.
High levels of Thl-type cytokines (e.g., IFN-y, TNFa, IL-2 and IL-12) tend to
favor the
induction of cell mediated immune responses to an administered antigen. In
contrast,
high levels of Th2-type cytokines (e.g., IL-4, IL-5, IL-6 and IL-10) tend to
favor the
induction of humoral immune responses. Following application of a vaccine as
provided herein, a patient will support an immune response that includes Th 1-
and Th2-
type responses. Within a preferred embodiment, in which a response is
predominantly
Thl-type, the level of Thl-type cytokines will increase to a greater extent
than the level
of Th2-type cytokines. The levels of these cytokines may be readily assessed
using
standard assays. For a review of the families of cytokines, see Mosmann and
Coffman,
Ann. Rev. Immunol. 7:145-173, 1989.
Preferred adjuvants for use in eliciting a predominantly Thl-type
response include, for example, a combination of monophosphoryl lipid A,
preferably 3-
de-O-acylated monophosphoryl lipid A (3D-MPL), together with an aluminum salt.
MPL adjuvants are available from Corixa Corporation (Seattle, WA; see US
Patent Nos.
4,436,727; 4,877,611; 4,866,034 and 4,912,094). CpG-containing
oligonucleotides (in
which the CpG dinucleotide is unmethylated) also induce a predominantly Thl
response. Such oligonucleotides are well known and are described, for example,
in WO
96/02555 and WO 99/33488. Immunostimulatory DNA sequences are also described.
for example, by Sato et al., Science 273:352, 1996. Another preferred adjuvant
is a
saponin, preferably QS21 (Aquila Biopharmaceuticals Inc., Framingham, MA),
which
may be used alone or in combination with other adjuvants. For example, an
enhanced
system involves the combination of a monophosphoryl lipid A and saponin
derivative,
26
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
such as the combination of QS21 and 3D-MPL as described in WO 94/00153. or a
less
reactogenic composition where the QS21 is quenched with cholesterol. as
described in
WO 96/33739. Other preferred formulations comprise an oil-in-water emulsion
and
tocopherol. A particularly potent adjuvant formulation involving QS21, 3D-MPL
and
tocopherol in an oil-in-water emulsion is described in WO 95/17210.
Other preferred adjuvants include Montanide ISA 720 (Seppic. France).
SAF (Chiron, California, United States), ISCOMS (CSL), MF-59 (Chiron), the
SBAS
series of adjuvants (e.g., SBAS-2 or SBAS-4. available from SmithKline
Beecham.
Rixensart. Belgium), Detox (Corixa Corporation; Seattle, WA), RC-529 (Corixa
Corporation: Seattle, WA) and aminoalkyl glucosaminide 4-phosphates (AGPs).
Any vaccine provided herein may be prepared using well known
methods that result in a combination of antigen. immunostimulant and a
suitable carrier
or excipient. The compositions described herein may be administered as part of
a
sustained release formulation (i. e., a formulation such as a capsule, sponge
or gel
(composed of polysaccharides, for example) that effects a slow release of
compound
following administration). Such formulations may generally be prepared using
well
known technology (sec, e.g., Coombes et al., vaccine 1-x:1429-1438, 1996) and
administered by, for example, oral, rectal or subcutaneous implantation, or by
implantation at the desired target site. Sustained-release formulations may
contain a
polypeptide, polynucleotide or antibody dispersed in a carrier matrix and/or
contained
within a reservoir surrounded by a rate controlling membrane.
Carriers for use within such formulations are biocompatible, and may
also be biodegradable; preferably the formulation provides a relatively
constant level of
active component release. Such carriers include microparticles of poly(lactide-
co-
glycolide), as well as polyacrylate, latex, starch, cellulose and dextran.
Other delayed-
release carriers include supramolecular biovectors, which comprise a non-
liquid
hydrophilic core (e.g.. a cross-linked polysaccharide or oligosaccharide) and,
optionally,
an external layer comprising an amphiphilic compound, such as a phospholipid
(see
e.g., U.S. Patent No. 5,151,254 and PCT applications WO 94/20078, WO/94/23701
and
WO 96/06638). The amount of active compound contained within a sustained
release
27
CA 02375049 2001-11-23
WO 00/73338 PCTNS00/14845
formulation depends upon the site of implantation. the rate and expected
duration of
release and the nature of the condition to be treated or prevented.
Any of a variety of delivery vehicles may be employed within
pharmaceutical compositions and vaccines to facilitate production of an
antigen-specific
immune response that targets tumor cells. Delivery vehicles include antigen
presenting
cells (APCs), such as dendritic cells. macrophages, B cells, monocytes and
other cells
that may be engineered to be efficient APCs. Such cells may. but need not, be
genetically modified to increase the capacity for presenting the antigen, to
improve
activation and/or maintenance of the T cell response, to have anti-tumor
effects per se
and/or to be immunologically compatible with the receiver (i. e., matched HLA
haplotype). APCs may generally be isolated from any of a variety of biological
fluids
and organs, including tumor and peritumoral tissues, and may be autologous,
allogeneic, syngeneic or xenogeneic cells.
Certain preferred embodiments of the present invention use dendritic
cells or progenitors thereof as antigen-presenting cells. Dendritic cells are
highly potent
APCs (Banchereau and Steinman, Nature 392:245-251, 1998) and have been shown
to
be effective as a physiological adjuvant for eliciting prophylactic or
therapeutic
antitumor immunity (see Timmerman and Levy, Ann. Rev. Med. 50:507-529, 1999).
In
general, dendritic cells may be identified based on their typical shape
(stellate in situ,
with marked cytoplasmic processes (dendrites) visible in vitro), their ability
to take up,
process and present antigens with high efficiency and their ability to
activate naive T
cell responses. Dendritic cells may, of course, be engineered to express
specific cell-
surface receptors or ligands that are not commonly found on dendritic cells in
vivo or ex
vivo, and such modified dendritic cells are contemplated by the present
invention. As
an alternative to dendritic cells, secreted vesicles antigen-loaded dendritic
cells (called
exosomes) may be used within a vaccine (see Zitvogel et al., Nature Med. 4:594-
600,
1998).
Dendritic cells and progenitors may be obtained from peripheral blood,
bone marrow, tumor-infiltrating cells, peritumoral tissues-infiltrating cells,
lymph
nodes, spleen, skin, umbilical cord blood or any other suitable tissue or
fluid. For
example, dendritic cells may be differentiated ex vivo by adding a combination
of
28
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
cytokines such as GM-CSF, IL-4, IL-13 and/or TNFa to cultures of monocytes
harvested from peripheral blood. Alternatively, CD34 positive cells harvested
from
peripheral blood, umbilical cord blood or bone marrow may be differentiated
into
dendritic cells by adding to the culture medium combinations of GM-CSF, IL-3:
TNFa.
CD40 ligand, LPS, flt3 ligand and/or other compounds) that induce
differentiation,
maturation and proliferation of dendritic cells.
Dendritic cells are conveniently categorized as "immature" and "mature"
cells, which allows a simple way to discriminate between two well
characterized
phenotypes. However, this nomenclature should not be construed to exclude all
possible intermediate stages of differentiation. Immature dendritic cells are
characterized as APC with a high capacity for antigen uptake and processing,
which
correlates with the high expression of Fcy receptor and mannose receptor. The
mature
phenotype is typically characterized by a lower expression of these markers,
but a high
expression of cell surface molecules responsible for T cell activation such as
class I and
class II MHC, adhesion molecules (e.g., CD54 and CD11) and costimulatory
molecules
(e.g., CD40, CD80, CD86 and 4-1BB).
APCs may generally be transfected with a polynucleotide encoding a
mammaglobin protein (or portion or other variant thereof) such that the
mammaglobin
polypeptide, or an immunogenic portion thereof, is expressed on the cell
surface. Such
transfection may take place ex vivo, and a composition or vaccine comprising
such
transfected cells may then be used for therapeutic purposes, as described
herein.
Alternatively, a gene delivery vehicle that targets a dendritic or other
antigen presenting
cell may be administered to a patient, resulting in transfection that occurs
in vivo. In
vivo and ex vivo transfection of dendritic cells, for example, may generally
be
performed using any methods known in the art, such as those described in WO
97/24447, or the gene gun approach described by Mahvi et al., Immunology and
cell
Biology 75:456-460, 1997. Antigen loading of dendritic cells may be achieved
by
incubating dendritic cells or progenitor cells with the mammaglobin
polypeptide, DNA
(naked or within a plasmid vector) or RNA; or with antigen-expressing
recombinant
bacterium or viruses (e.g.. vaccinia, fowlpox, adenovirus or lentivirus
vectors). Prior to
loading, the polypeptide may be covalently conjugated to an immunological
partner that
29
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
provides T cell help (e.g., a carrier molecule). Alternatively, a dendritic
cell may be
pulsed with a non-conjugated immunological partner. separately or in the
presence of
the polypeptide.
Vaccines and pharmaceutical compositions may be presented in unit-
s dose or multi-dose containers. such as sealed ampoules or vials. Such
containers are
preferably hermetically sealed to preserve sterility of the formulation until
use. In
general, formulations may be stored as suspensions, solutions or emulsions in
oily or
aqueous vehicles. Alternatively, a vaccine or pharmaceutical composition may
be
stored in a freeze-dried condition requiring only the addition of a sterile
liquid carrier
immediately prior to use.
CANCER THERAPY
In further aspects of the present invention, the compositions described
herein may be used for immunotherapy of cancer, such as breast cancer. Within
such
methods, pharmaceutical compositions and vaccines are typically administered
to a
patient. As used herein, a "patient" refers to any warm-blooded animal,
preferably a
human. A patient may or may not be afflicted with cancer. Accordingly, the
above
pharmaceutical compositions and vaccines may be used to prevent the
development of a
cancer or to treat a patient afflicted with a cancer. A cancer may be
diagnosed using
criteria generally accepted in the art, including the presence of a malignant
tumor.
Pharmaceutical compositions and vaccines may be administered either prior to
or
following surgical removal of primary tumors and/or treatment such as
administration
of radiotherapy or conventional chemotherapeutic drugs.
Within certain embodiments, immunotherapy may be active
immunotherapy, in which treatment relies on the in vivo stimulation of the
endogenous
host immune system to react against tumors with the administration of immune
response-modifying agents (such as tumor vaccines, bacterial adjuvants and/or
cytokines).
Within other embodiments, immunotherapy may be passive
immunotherapy, in which treatment involves the delivery of agents with
established
tumor-immune reactivity (such as effector cells or antibodies) that can
directly or
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
indirectly mediate antitumor effects and does not necessarily depend on an
intact host
immune system. Examples of effector cells include T lymphocytes (such as CD8-
cytotoxic T lymphocytes and CD4' T-helper tumor-infiltrating lymphocytes).
killer
cells (such as Natural Killer cells and lymphokine-activated killer cells), B
cells and
antigen-presenting cells (such as dendritic cells and macrophages) expressing
a
polypeptide provided herein. T cell receptors and antibody receptors specific
for the
polypeptides recited herein may be cloned, expressed and transferred into
other vectors
or effector cells for adoptive immunotherapy. The polypeptides provided herein
may
also be used to generate antibodies or anti-idiotypic antibodies (as described
above and
in U.S. Patent No. 4,918,164) for passive immunotherapy.
Effector cells may generally be obtained in sufficient quantities for
adoptive immunotherapy by growth in vitro, as described herein. Culture
conditions for
expanding single antigen-specific effector cells to several billion in number
with
retention of antigen recognition in vivo are well known in the art. Such in
vitro culture
conditions typically use intermittent stimulation with antigen, often in the
presence of
cytokines (such as IL-2) and non-dividing feeder cells. As noted above,
immunoreactive polypeptides as provided herein may be used to rapidly expand
antigen-specific T cell cultures in order to generate a sufficient number of
cells for
immunotherapy. In particular, antigen-presenting cells, such as dendritic,
macrophage
or B cells, may be pulsed with immunoreactive polypeptides or transfected with
one or
more polynucleotides using standard techniques well known in the art. For
example,
antigen-presenting cells can be transfected with a polynucleotide having a
promoter
appropriate for increasing expression in a recombinant virus or other
expression system.
Cultured effector cells for use in therapy must be able to grow and distribute
widely,
and to survive long term in vivo. Studies have shown that cultured effector
cells can be
induced to grow in vivo and to survive long term in substantial numbers by
repeated
stimulation with antigen supplemented with IL-2 (sec, for example, Cheever et
al.,
Immunological Reviews 1~ 7:177, 1997).
The polypeptides provided herein may also be used to generate and/or
isolate tumor-reactive T cells, which can then be administered to a patient.
In one such
technique, antigen-specific T cell lines may be generated by in vivo
immunization with
31
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
short peptides corresponding to immunogenic portions of the disclosed
polypeptides.
The resulting antigen-specific CD8- CTL clones may be isolated from the
patient,
expanded using standard tissue culture techniques and returned to the patient.
Within another embodiment, syngeneic or autologous dendritic cells may
be pulsed with peptides corresponding to at least an immunogenic portion of a
polypeptide disclosed herein. The resulting antigen-specific dendritic cells
may either
be transferred into a patient or employed to stimulate T cells to provide
antigen-specific
T cells which may, in turn, be administered to a patient. The use of peptide-
pulsed
dendritic cells to generate antigen-specific T cells and the subsequent use of
such
antigen-specific T cells to eradicate tumors in a murine model has been
demonstrated
by Cheever et al., Immunological Reviews 1 ~ 7:177, 1997.
Alternatively, a vector expressing a polypeptide recited herein may be
introduced into antigen presenting cells taken from a patient and clonally
propagated ex
vivo for transplant back into the same patient. Transfected cells may be
reintroduced
into the patient using any means known in the art, preferably in sterile form
by
intravenous, intracavitary, intraperitoneal or intratumor administration.
Routes and frequency of administration, as well as dosage, will vary
from individual to individual, and may be readily established using standard
techniques.
In general, the pharmaceutical compositions and vaccines may be administered
by
injection (e.g., intracutaneous, intramuscular, intravenous or subcutaneous),
intranasally
(e.g., by aspiration) or orally. Preferably, between 1 and 10 doses may be
administered
over a 52 week period. Preferably, 6 doses are administered, at intervals of 1
month,
and booster vaccinations may be given periodically thereafter. Alternate
protocols may
be appropriate for individual patients. A suitable dose is an amount of a
compound that,
when administered as described above, is capable of promoting an anti-tumor
immune
response, and is at least 10-50% above the basal (i.e., untreated) level. Such
response
can be monitored by measuring the anti-tumor antibodies in a patient or by
vaccine-
dependent generation of cytolytic effector cells capable of killing the
patient's tumor
cells in vitro. Such vaccines should also be capable of causing an immune
response that
leads to an improved clinical outcome (e.g., more frequent remissions,
complete or
partial or longer disease-free survival) in vaccinated patients as compared to
non-
32
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
vaccinated patients. In general. for pharmaceutical compositions and vaccines
comprising one or more polypeptides, the amount of each polypeptide present in
a dose
ranges from about 100 ~g to 5 mg per kg of host. Suitable dose sizes will vary
with the
size of the patient, but will typically range from about 0.1 mL to about 5 mL.
In general, an appropriate dosage and treatment regimen provides the
active compounds) in an amount sufficient to provide therapeutic and/or
prophylactic
benefit. Such a response can be monitored by establishing an improved clinical
outcome (e.g., more frequent remissions, complete or partial, or longer
disease-free
survival) in treated patients as compared to non-treated patients. Increases
in
preexisting immune responses to mammaglobin generally correlate with an
improved
clinical outcome. Such immune responses may be evaluated using standard
proliferation, cytotoxicity or cytokine assays, which may be performed using
samples
obtained from a patient before and after treatment.
1 S METHODS FOR DETECTING CANCER
In general, a cancer may be detected in a patient based on the presence of
one or more mammaglobin epitopes or antibodies thereto in a biological sample
obtained from the patient. In other words, such epitopes may be used as
markers to
indicate the presence or absence of a cancer such as breast cancer. In
general, such an
epitope or antibody should be present at a level that is at least three fold
higher in tumor
tissue than in normal tissue
There are a variety of assay formats known to those of ordinary skill in
the art for using a binding agent to detect polypeptide markers in a sample.
See, e.g.,
Harlow and Lane, Antibodies: A Laboratory Manual, Cold Spring Harbor
Laboratory,
1988. In general, the presence or absence of a cancer in a patient may be
determined by
(a) contacting a biological sample obtained from a patient with a binding
agent; (b)
detecting in the sample a level of polypeptide that binds to the binding
agent; and (c)
comparing the level of polypeptide with a predetermined cut-off value.
In a preferred embodiment, the assay involves the use of binding agent
immobilized on a solid support to bind to and remove the polypeptide from the
remainder of the sample. The bound polypeptide may then be detected using a
-, .,
CA 02375049 2001-11-23
WO 00/73338 PCTNS00/14845
detection reagent that contains a reporter group and specifically binds to the
binding
agent/polypeptide complex. Such detection reagents may comprise, for example,
a
binding agent that specifically binds to the polypeptide or an antibody or
other agent
that specifically binds to the binding agent. such as an anti-immunoglobulin,
protein G,
protein A or a lectin. Alternatively, a competitive assay may be utilized, in
which a
polypeptide is labeled with a reporter group and allowed to bind to the
immobilized
binding agent after incubation of the binding agent with the sample. The
extent to
which components of the sample inhibit the binding of the labeled polypeptide
to the
binding agent is indicative of the reactivity of the sample with the
immobilized binding
agent.
The solid support may be any material known to those of ordinary skill
in the art to which the binding agent may be attached. For example, the solid
support
may be a test well in a microtiter plate or a nitrocellulose or other suitable
membrane.
Alternatively, the support may be a bead or disc, such as glass, fiberglass,
latex or a
plastic material such as polystyrene or polyvinylchloride. The support may
also be a
magnetic particle or a fiber optic sensor, such as those disclosed, for
example, in U.S.
Patent No. 5,359,681. The binding agent may be immobilized on the solid
support
using a variety of techniques known to those of skill in the art, which are
amply
described in the patent and scientific literature. In the context of the
present invention,
the term "immobilization" refers to both noncovalent association, such as
adsorption,
and covalent attachment (which may be a direct linkage between the agent and
functional groups on the support or may be a linkage by way of a cross-linking
agent).
Immobilization by adsorption to a well in a microtiter plate or to a membrane
is
preferred. In such cases, adsorption may be achieved by contacting the binding
agent,
in a suitable buffer, with the solid support for a suitable amount of time.
The contact
time varies with temperature, but is typically between about 1 hour and about
1 day. In
general, contacting a well of a plastic microtiter plate (such as polystyrene
or
polyvinylchloride) with an amount of binding agent ranging from about 10 ng to
about
10 ~tg, and preferably about 100 ng to about 1 fig, is sufficient to
immobilize an
adequate amount of binding agent.
34
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
Covalent attachment of binding agent to a solid support may generally be
achieved by first reacting the support with a bifunctional reagent that will
react with
both the support and a functional group, such as a hydroxyl or amino group. on
the
binding agent. For example, the binding agent may be covalently attached to
supports
having an appropriate polymer coating using benzoquinone or by condensation of
an
aldehyde group on the support with an amine and an active hydrogen on the
binding
partner (see, e.g., Pierce Immunotechnology Catalog and Handbook, 1991. at
A12-A13).
In certain embodiments, the assay is a two-antibody sandwich assay.
This assay may be performed by first contacting an antibody that has been
immobilized
on a solid support, commonly the well of a microtiter plate, with the sample,
such that
polypeptides within the sample are allowed to bind to the immobilized
antibody.
Unbound sample is then removed from the immobilized polypeptide-antibody
complexes and a detection reagent (preferably a second antibody capable of
binding to a
different site on the polypeptide) containing a reporter group is added. The
amount of
detection reagent that remains bound to the solid support is then determined
using a
method appropriate for the specific reporter group.
More specifically, once the antibody is immobilized on the support as
described above, the remaining protein binding sites on the support are
typically
blocked. Any suitable blocking agent known to those of ordinary skill in the
art, such
as bovine serum albumin or Tween 20TM (Sigma Chemical Co., St. Louis, MO). The
immobilized antibody is then incubated with the sample, and polypeptide is
allowed to
bind to the antibody. The sample may be diluted with a suitable diluent, such
as
phosphate-buffered saline (PBS) prior to incubation. In general, an
appropriate contact
time (i.e., incubation time) is a period of time that is sufficient to detect
the presence of
polypeptide within a sample obtained from an individual with breast cancer.
Preferably,
the contact time is sufficient to achieve a level of binding that is at least
about 95% of
that achieved at equilibrium between bound and unbound polypeptide. Those of
ordinary skill in the art will recognize that the time necessary to achieve
equilibrium
may be readily determined by assaying the level of binding that occurs over a
period of
3~
CA 02375049 2001-11-23
WO 00/73338 PCTNS00/14845
time. At room temperature. an incubation time of about 30 minutes is generally
sufficient.
Unbound sample may then be removed by washing the solid support
with an appropriate buffer, such as PBS containing 0.1% Tween 20T". The second
antibody, which contains a reporter group, may then be added to the solid
support.
Preferred reporter groups include those groups recited above.
The detection reagent is then incubated with the immobilized antibody-
polypeptide complex for an amount of time sufficient to detect the bound
polypeptide.
An appropriate amount of time may generally be determined by assaying the
level of
binding that occurs over a period of time. Unbound detection reagent is then
removed
and bound detection reagent is detected using the reporter group. The method
employed for detecting the reporter group depends upon the nature of the
reporter
group. For radioactive groups, scintillation counting or autoradiographic
methods are
generally appropriate. Spectroscopic methods may be used to detect dyes,
luminescent
groups and fluorescent groups. Biotin may be detected using avidin, coupled to
a
different reporter group (commonly a radioactive or fluorescent group or an
enzyme).
Enzyme reporter groups may generally be detected by the addition of substrate
(generally for a specific period of time), followed by spectroscopic or other
analysis of
the reaction products.
To determine the presence or absence of a cancer, such as breast cancer,
the signal detected from the reporter group that remains bound to the solid
support is
generally compared to a signal that corresponds to a predetermined cut-off
value. In
one preferred embodiment, the cut-off value for the detection of a cancer is
the average
mean signal obtained when the immobilized antibody is incubated with samples
from
patients without the cancer. In general, a sample generating a signal that is
three
standard deviations above the predetermined cut-off value is considered
positive for the
cancer. In an alternate preferred embodiment, the cut-off value is determined
using a
Receiver Operator Curve, according to the method of Sackett et al., Clinical
Epidemiology: A Basic Science .for Clinical Medicine, Little Brown and Co.,
1985,
p. 106-7. Briefly, in this embodiment, the cut-off value may be determined
from a plot
of pairs of true positive rates (i.e., sensitivity) and false positive rates
(100%-specificity)
36
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
that correspond to each possible cut-off value for the diagnostic test result.
The cut-off
value on the plot that is the closest to the upper left-hand corner (i.e., the
value that
encloses the largest area) is the most accurate cut-off value, and a sample
generating a
signal that is higher than the cut-off value determined by this method may be
considered
positive. Alternatively, the cut-off value may be shifted to the left along
the plot, to
minimize the false positive rate, or to the right, to minimize the false
negative rate. In
general, a sample generating a signal that is higher than the cut-off value
determined by
this method is considered positive for a cancer.
For certain embodiments (e.g., sandwich assays), quantitative
measurements of antigen may be obtained. Within such embodiments, a standard
curve
may be generated. Signals obtained for antigen levels in particular samples
may then be
compared to the standard curve, to allow quantitation. The cut-off value
within such
assays may be an amount of mammaglobin indicative of the presence of breast
cancer.
In a related embodiment, the assay is performed in a flow-through or
strip test format, wherein the binding agent is immobilized on a membrane,
such as
nitrocellulose. In the flow-through test, polypeptides within the sample bind
to the
immobilized binding agent as the sample passes through the membrane. A second,
labeled binding agent then binds to the binding agent-polypeptide complex as a
solution
containing the second binding agent flows through the membrane. The detection
of
bound second binding agent may then be performed as described above. In the
strip test
format, one end of the membrane to which binding agent is bound is immersed in
a
solution containing the sample. The sample migrates along the membrane through
a
region containing second binding agent and to the area of immobilized binding
agent.
Concentration of second binding agent at the area of immobilized antibody
indicates the
presence of a cancer. Typically, the concentration of second binding agent at
that site
generates a pattern, such as a line, that can be read visually. The absence of
such a
pattern indicates a negative result. In general, the amount of binding agent
immobilized
on the membrane is selected to generate a visually discernible pattern when
the
biological sample contains a level of polypeptide that would be sufficient to
generate a
positive signal in the two-antibody sandwich assay, in the format discussed
above.
Preferred binding agents for use in such assays are antibodies and antigen-
binding
37
CA 02375049 2001-11-23
WO 00/73338 PCTNS00/14845
fragments thereof. Preferably, the amount of antibody immobilized on the
membrane
ranges from about 25 ng to about 1 fig, and more preferably from about 50 ng
to about
500 ng. Such tests can typically be performed with a very small amount of
biological
sample.
Of course, numerous other assay protocols exist that are suitable for use
with the epitopes and binding agents of the present invention. The above
descriptions
are intended to be exemplary only. For example, it will be apparent to those
of ordinary
skill in the art that the above protocols may be readily modified to use
polypeptides as
described herein to detect antibodies that bind to such polypeptides in a
biological
sample. The detection of such mammaglobin epitope-specific antibodies may
correlate
with the presence of a cancer. Other preferred assay protocols include laser
scanning
cytometry (a microscopic technique in which cells are stained with labeled
antibody)
and immunohistochemical detection. Such techniques may generally be performed
according to techniques known in the art. Antibodies as provided herein may
further be
used to facilitate cell identification and sorting in vitro, permitting the
selection of cells
expressing mammaglobin (or varying levels of mammaglobin). Preferably,
antibodies
for use in such methods are linked to a detectable marker. Suitable markers
are well
known in the art and include radionuclides, luminescent groups, fluorescent
groups,
enzymes, dyes, constant immunoglobulin domains and biotin. Within one
preferred
embodiment, an antibody linked to a fluorescent marker, such as fluorescein,
is
contacted with the cells, which are then analyzed by fluorescence activated
cell sorting
(FACS).
In another embodiment, the above polypeptides may be used as markers
for the progression of cancer. In this embodiment, assays as described above
for the
diagnosis of a cancer may be performed over time, and the change in the level
of
reactive polypeptide(s) evaluated. For example, the assays may be performed
every
24-72 hours for a period of 6 months to 1 year, and thereafter performed as
needed. In
general, a cancer is progressing in those patients in whom the level of
polypeptide
detected by the binding agent increases over time. In contrast, the cancer is
not
progressing when the level of reactive polypeptide either remains constant or
decreases
with time.
38
CA 02375049 2001-11-23
WO 00/73338 PCTNS00/14845
Certain irmivo diagnostic assays may be performed directly on a tumor.
One such assay involves contacting tumor cells with a binding agent. The bound
binding agent may then be detected directly or indirectly via a reporter
group. Such
binding agents may also be used in histological applications.
To improve sensitivity, assays as described herein may be combined
with assays to detect other tumor-associated antigens. It will be apparent
that binding
agents specific for different proteins may be combined within a single assay.
The
selection of tumor protein markers may be based on routine experiments to
determine
combinations that results in optimal sensitivity.
By alternative embodiments of the present invention, a cancer may be
detected in a patient based on the presence of mammaglobin polynucleotides in
a
biological sample obtained from the patient. In other words, such
polynucleotides may
be used as markers to indicate the presence or absence of a cancer such as
breast cancer.
In particular, polynucleotide primers and probes may be used to detect the
level of
mRNA encoding mammaglobin, which is indicative of the presence or absence of
breast cancer. In general, the presence of a mammaglobin polynucleotide at a
level that
is at least two fold, preferably at least three fold, higher than in normal
tissue is
indicative of breast cancer.
There are a variety of biological samples that may be used for an assay
provided herein, including various body fluids and tumor samples. Preferred
samples
are blood, and fractions thereof, such as peripheral blood, serum or plasma.
In general,
RNA may be isolated from blood or a fraction thereof using any standard
technique.
Prior to PCR or hybridization analysis, a sample is treated by any
standard technique to remove epithelial cells. It has been found, within the
context of
the present invention, that such treatment improves the sensitivity of the
assay by up to
10 fold. One method for removing epithelial cells employs Dynal's Epithelial
cell
enrichment beads (Dynal, Oslo, Norway), which may be used according to the
manufacturer's instructions. Preferred samples for analysis are patient whole
blood
samples, from which epithelial cells have been removed.
Within certain embodiments, at least two oligonucleotide primers may
be employed in a polymerase chain reaction (PCR) based assay to amplify a
portion of a
39
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
mammaglobin cDNA derived from a biological sample, wherein at least one of the
oligonucleotide primers is specific for (i.e., hybridizes to) a polynucleotide
encoding
mammaglobin. The amplified cDNA is then separated and detected using
techniques
well known in the art, such as gel electrophoresis and autoradiography.
Similarly,
oligonucleotide probes that specifically hybridize to a mammaglobin
polynucleotide
may be used in a hybridization assay to detect mammaglobin expression in a
biological
sample.
To permit hybridization under assay conditions, oligonucleotide primers
and probes should comprise an oligonucleotide sequence that has at least about
60%,
preferably at least about 75% and more preferably at least about 90%, identity
to a
portion of a mammaglobin polynucleotide that is at least 10 nucleotides, and
preferably
at least 20 nucleotides, in length. Oligonucleotide primers and/or probes
which may be
usefully employed in the diagnostic methods described herein preferably are at
least 10-
40 nucleotides in length. Techniques for both PCR based assays and
hybridization
assays are well known in the art (see, for example, Mullis et al., Cold Spring
Harbor
Symp. Quant. Biol. 51:263, 1987; Erlich ed., PCR Technology, Stockton Press,
NY,
1989).
One preferred assay employs RT-PCR, in which PCR is applied in
conjunction with reverse transcription. Typically, RNA is extracted from a
sample
tissue and is reverse transcribed to produce cDNA molecules. PCR amplification
using
at least one specific primer generates a cDNA molecule, which may be separated
and
visualized using, for example, gel electrophoresis. Amplification may be
performed on
samples obtained from biological samples taken from a test patient and an
individual
who is not afflicted with a cancer. The amplification reaction may be
performed on
several dilutions of cDNA spanning two orders of magnitude. A two-fold or
greater
increase in expression in several dilutions of the test patient sample as
compared to the
same dilutions of the non-cancerous sample is typically considered positive.
Yet another amplification technique that may be used within such assays
is real-time PCR (see Gibson et al., Genome Research 6:995-1001, 1996; Heid et
al.,
Genome Research 6:986-994, 1996). Real-time PCR is a technique that evaluates
the
level of PCR product accumulation during amplification, permitting
quantitative
CA 02375049 2001-11-23
WO 00/73338 PCTNS00/14845
evaluation of mRNA levels. Briefly, mRNA is initially extracted from cells of
interest
using standard techniques. Real-time PCR may then be performed, for example,
using a
Perkin Elmer/Applied Biosystems (Foster City, CA) 7700 Prism instrument.
Matching
primers and fluorescent probes may be designed for mammaglobin using, for
example,
the primer express program provided by Perkin Elmer/Applied Biosystems (Foster
City,
CA). Optimal concentrations of primers and probes may be initially determined
by
those of ordinary skill in the art, and control (e.g., ~3-actin) primers and
probes may be
obtained commercially from, for example, Perkin Elmer/Applied Biosystems
(Foster
City, CA). To quantitate the amount of mammaglobin RNA in a sample, a standard
curve may be generated alongside using a plasmid containing a mammaglobin
gene.
Standard dilutions ranging from 10-106 copies of the gene of interest are
generally
sufficient. In addition, a standard curve may be generated for the control
sequence, to
permit standardization of initial RNA content of a tissue sample to the amount
of
control for comparison purposes.
Certain in vivo diagnostic assays may be performed directly on a tumor.
One such assay involves contacting tumor cells with a polynucleotide probe.
Bound
probe may be detected directly or indirectly using a reporter group.
As noted above, to improve sensitivity, multiple breast tumor protein
markers may be assayed within a given sample. For example, a polynucleotide
probe or
primer as described herein may be used concurrently with a probe or primer
designed to
detect a different marker. The selection of breast tumor markers may be based
on
routine experiments to determine combinations that results in optimal
sensitivity.
DIAGNOSTIC KITS
The present invention further provides kits for use within any of the
above diagnostic methods. Such kits typically comprise two or more components
necessary for performing a diagnostic assay. Components may be compounds,
reagents, containers and/or equipment. For example, one container within a kit
may
contain a monoclonal antibody or fragment thereof that specifically binds to a
mammaglobin epitope. Such antibodies or fragments may be provided attached to
a
support material, as described above. One or more additional containers may
enclose
41
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
elements, such as reagents or buffers, to be used in the assay. Such kits may
also. or
alternatively. contain a detection reagent as described above that contains a
reporter
group suitable for direct or indirect detection of antibody binding.
Preferred kits are those designed for use within sandwich assays. Such
kits comprise two or more components for use within such assays. For example,
such a
kit may comprise standards based on recombinant mammaglobin for use in
preparing a
standard curve. Such a kit may comprise one or both antibodies for use within
the assay
(i.e., the capture antibody and/or signal antibody), with or without
additional reagents
for use in detecting mammaglobin binding.
Kits designed to detect the level of mRNA encoding mammaglobin in a
biological sample may comprise at least one oligonucleotide probe or primer,
as
described above. Such an oligonucleotide may be used, for example, within a
PCR or
hybridization assay. Additional components that may be present within such
kits
include a second oligonucleotide and/or a diagnostic reagent or container to
facilitate
the detection of a mammaglobin polynucleotide.
The following Examples are offered by way of illustration and not by
way of limitation.
42
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
EXAMPLES
Example 1
Identification of Mamma~lobin Epitopes and Preparation of Antibodies
This Example illustrates the preparation of anti-mammaglobin antibodies
and epitope mapping.
Rabbits and mice were immunized with full-length human
mammaglobin protein. Mouse monoclonal antibodies were isolated with standard
hybridoma technology. Rabbit monoclonal antibodies were isolated with selected
lymphocyte antibody method (SLAM) technology. In addition to these antibodies,
a
purified polyclonal antibody directed against the C-terminus of mammaglobin
was also
developed following immunization of rabbits with a C-terminal peptide.
Figure 1A illustrates the monoclonal antibodies that were developed for
mammaglobin. For the rabbit monoclonal antibodies the Ig variable regions were
sequenced. The sequence for the variable regions of each rabbit
antimammaglobin
monoclonal antibody is shown in Figures 1 B-1 C.
In order to better define the epitope binding region of each monoclonal
antibody a series of peptides was generated that spans the entire mammaglobin
protein
sequence. The amino acid sequence for mammaglobin is shown in Figure 2, and
the
corresponding peptides are indicated. In addition to the peptides, a short
recombinant
form of mammaglobin was generated by cleavage with protease. 96 well
microtiter
plates (Costar) were coated with either peptide or recombinant antigen at 200
ng/well.
Coating was overnight at 4°C. Plates were then aspirated and blocked
with phosphate
buffered saline containing 1% (w/v) BSA for 2 hours at room temperature, and
then
washed in PBS containing 0.1% Tween 20 (PBST). Purified rabbit antibodies at
different dilutions (1000 to 7.8 ng/ml) in PBST was added to the wells and
incubated
for 30 minutes at room temperature. This was followed by washing 6 times with
PBST
and then incubating with Protein-A HRP conjugated at a 1/20000 dilution for a
fiuther
30 minutes. Plates were washed 6 times in PBST and then incubated with
Tetramethylbenzidine (TMB) substrate for a further 1 S minutes. The reaction
was
43
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
stopped by the addition of 1 N sulfuric acid and plates were read at 450 run
using an
ELISA plate reader.
ELISA with the mouse monoclonal antibodies was performed with
supernatants from tissue culture run neat in the assay.
A summary of the data is shown in Figure 3A. Shaded cells are
considered positive for the antibody. The reactivity of three different
epitopes is shown
in Figures 3B, 3C and 3D, where 2D3 reacts with pros and the N-terminal
recombinant
and 29C 11 reacts weakly with pro2. The epitope binding sites of the
antimammaglobin
antibodies are summarized in Figure 1 A.
Subsequent to epitope mapping, the antibodies were tested by FACS
analysis on a cell line that expresses mammaglobin, MB415 breast carcinoma
cells. In
order to ensure specificity of antibody binding, MCF-7 cells that do not
express
mammaglobin were also tested by FACS analysis under identical conditions.
Cells
were fixed with 4% formaldehyde for 20 min before being washed 2 times. Cells
were
then permeabilized for 10 minutes with PBS containing 0-.1% saponin. 0.5 ~g of
anti-mammaglobin monoclonal antibody was added and cells were incubated at
room
temperature for 30 minutes before being washed 2 times and incubated with a
F1TC-labeled goat anti-rabbit or mouse secondary antibody for 20 minutes.
After being
washed 2 times, cells were analyzed with an Excalibur fluorescent activated
cell sorter.
The results are illustrated in Figure 4A.
Western blot analysis was also used to characterize anti-mammaglobin
monoclonal specificity (Figure 5). SDS-PAGE was performed on 1) media in which
MDA-MB-415 cells were grown, 2) MDA-MB-415 cell lysate and 3) bacterially
expressed recombinant mammaglobin. Protein was transferred to nitrocellulose
and
then Western blotted for the antimammaglobin monoclonal antibodies at an
antibody
concentration of 1 pg/ml. Protein was detected using horse radish peroxidase
(HRP)
conjugated to either a goat anti-mouse monoclonal antibody or to protein A-
Sepharose.
The purified anti-mammaglobin polyclonal antibody recognized bacterial
expressed
recombinant mammaglobin, as well as mammaglobin expressed in and secreted from
MDA-MB-415 breast carcinoma cells. All mouse and rabbit monoclonal antibodies
recognized recombinant bacterial expressed mammaglobin. With the exception of
197-
44
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
1 H 11, all of the mouse monoclonal antibodies recognized mammaglobin secreted
into
the cell media or expressed within the cytoplasm. The rabbit monoclonal
antibodies
14A12, 6B12 and 2D3 recognized recombinant bacterial expressed mammaglobin, as
well as mammaglobin expressed in the cytoplasm and secreted into the media.
Although unable to recognize mammaglobin secreted into the media, rabbit
monoclonal
antibody 6A1 was able to recognize bacterial expressed mammaglobin and
mammaglobin expressed in the cytoplasm of MB415 cells. The inability of
monoclonal
antibodies 197-1H11 and 6A1 to associate with specific forms of mammaglobin
likely
reflect differential posttranslation modifications such as glycosylation
and/or relative
affinity of the antibody to mammaglobin.
In order to determine which tissues express mammaglobin,
immunohistochemistry (IHC) analysis was performed on a diverse range of tissue
sections. Tissue samples were fixed in formalin solution for 24 hours and
embedded in
paraffin before being sliced into 10 micron sections. Tissue sections were
permeabilized
and incubated with anti-mammaglobin antibody for 1 hour. HRP-labeled anti-
mouse or
anti-rabbit antibody was used to visualize mammaglobin immunoreactivity.
Figure 6
summarizes the tissue-specific distribution of mammaglobin protein.
Mammaglobin
was highly expressed in breast tissue but not found in other tissues tested
including
adrenal, cervix, colon, duodenum, gall bladder, ileum, kidney, ovary,
pancreas, parotid
gland, prostate, skeletal muscle, spleen, and testis.
Example 2
Sandwich Immunoassays for Mamma~lobin
This Example illustrates the use of antibodies provided herein for
detection of mammaglobin in serum.
Monoclonal antibodies and the rabbit polyclonal antibody 967 directed
to the C-terminal 16 amino acid peptide of mammaglobin were evaluated in
sandwich
ELISAs for their ability to detect mammaglobin in lysates of MB41 ~ cells,
cell
supernatants of MB415 cells and also in the serum of breast cancer patients.
Antibodies
were paired based on their ability to detect different epitopes. The following
describes
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
some of the sandwich combinations tested. In all assays a standard curve was
constructed by spiking recombinant mammaglobin into male serum.
Mouse/Rabbit Antibody Sandwiches
Assays were designed to capture the mouse monoclonal antibody using a
solid phase of goat anti-mouse IgG as part of the sandwich. 96 well plates
(Costar
Corning) were coated overnight at 4°C with 200ng/well of goat anti
mouse IgG
(Rockland antibodies, Rockland, ME). Plates were washed in Phosphate buffered
saline
(PBS) containing 0.05% Tween 20 (PBST) then blocked with 1% BSA in phosphate
buffered saline(PBS) for 2 hours. Mouse monoclonal supernatants were then
added
(SOuI) at 1:10 dilution in PBS and the plates incubated for a further hour at
room
temperature. Plates were washed six times in PBS Tween 20 (PBST) then blocked
with 1 % normal mouse serum and 1 % normal human serum for 1 hour then washed
again. Samples and standards were applied to the wells and the plate was
incubated for
1 hour at room temperature. The plate was then washed six times in PBS
containing
0.05% Tween. Biotinylated 31A5 or 2D3 were used as the conjugate at lug/ml in
PBS
and 1 % Normal mouse serum. These were incubated for 1 hour at room
temperature
then washed six times in PBST. 501 of a 1:10000 dilution of streptavidin HRP
in PBS
containing 1 % normal mouse serum was added and the plate incubated at room
temperature for 30 minutes at which time the plate was again washed six times.
TMB
(tetramethyl benzidine) substrate (Kirkegaard and Perry) was then added to the
well and
incubated for a further 15 minutes. The reaction was stopped with 1N H~SO4
(1001)
and the signal generated read at 450rnn. A standard curve relating pg
mammaglobin in
the assay was constructed using recombinant mammaglobin spiked into normal
male
serum and samples run in the test were quantitated using this standard curve.
Rabbit/Rabbit antibody sandwiches
Two assays were performed. The first utilized affinity purified rabbit
anti 967 peptide (C-terminal 16 amino acid peptide) as the solid phase and 2D3
rabbit
monoclonal biotinylated as the signal antibody. The second used 2D3 as the
solid phase
antibody and 31A5-biotinylated as the signal antibody. In the first assay the
affinity
46
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
purified polyclonal antibody was coated overnight at 4°C on a 96 well
plate
(200ng/well) in 50 mM carbonate/bicarbonate buffer pH9.~. Plates were washed
in
PBST then blocked for 2 hours at room temperature with 1% BSA in PBS. Serum
(501) was then added to the plate and incubated for 1 hour at room
temperature. Plates
were then washed six times with Phosphate buffered saline containing 0.05%
Tween 20.
Biotinylated 2D3 monoclonal antibody (2ug/ml) in PBS containing 1% nornlal
rabbit
serum was then added and the plate further incubated at room temperature for I
hour.
After again washing six times, 501 of a 1:10000 dilution of streptavidin HRP
in PBS
containing 1 % normal rabbit serum was added and the plate incubated at room
temperature for 30 minutes at which time the plate was again washed six times.
Development of signal with TMB substrate was as described above.
In the second assay 2D3 was coated on 96 well plates at 200ngiwell
(Costar/Corning, Cambridge MA) overnight at 4°C washed in PBST then
blocked for
two hours at room temperature. Serum (501) was then added to the plate and
incubated for 1 hour at room temperature. Plates were then washed six times
with
Phosphate buffered saline containing 0.05% Tween 20. Biotinylated 31A5
monoclonal
antibody (O.S~g/ml) in PBS containing 1% normal rabbit serum was then added
and the
plate further incubated at room temperature for 1 hour. After again washing
six times
the streptavidin-HRP and TMB incubations were performed as described above.
Figures 7A-7C are examples of the sandwiching of the mouse
monoclonal antibodies with 31 A5, 6B 12 or 2D3 biotinylated and the ability to
detect
mammaglobin in MB415 lysates as well as supernatants. In Figure 8 is shown the
linear portion of the standard curve for the polyclonal anti-967 serum in
combination
with the monoclonal 2D3 biotinylated. This curve was used to quantitate
mammaglobin
serum samples of 7 patients with metastatic breast cancer. Of these 5/7 were
positive
for mammaglobin with serum levels in the 1-lOng/ml range. In the same
experiment
9/11 normal samples were negative and below the cut-off. Mammaglobin and
mammaglobin RNA were detectable in these same breast cancer patient samples
using a
sandwich of 2D3 and 29C11 as shogun in Figure 9. For all experiments,
mammaglobin
levels were obtained using a standard curve, and negative and positive
controls were as
expected.
47
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
Example 3
Identification of Anti-oli~osaccharide Antibodies ~ecific for Mamma~lobin
This Example illustrates the preparation of antibodies that specifically
bind mammaglobin in a glycosylation-sensitive manner. including antibodies to
differentially glycosylated sites of mammaglobin expressed in breast cancer
cells.
An antibody library (Glycotech Corp., Rockville MD) that encompasses
20-30 antibodies to different glycosylation combinations is screened using
native
mammaglobin from breast carcinoma cell lines via conventional ELISA and
blotting
techniques. Native mammaglobin is purified from MDA-MB-415 breast carcinoma
cells using standard biochemical purification procedures. Both rabbits and
mice are
immunized with native mammaglobin. SLAM technology is used to develop rabbit
monoclonal antibodies that bind to native mammaglobin but not to mammaglobin
that
has been stripped of oligosaccharides using deglycosylation enzymes. Identical
approaches are used to screen hybridoma supernatants that are generated from
mice
immunized with native mammaglobin.
The glycosylation epitopes for the antibodies generated in rabbits and
mice are mapped using a carbohydrate library (Glycotech Corp.). The
carbohydrate
library consists of a diverse array of oligosaccharide combinations permits
the
definition of carbohydrate epitopes to the antibodies. Upon identification,
isolation and
characterization of anti-oligosaccharide antibodies specific for mammaglobin,
a
sandwich ELISA assay is performed in which mammaglobin is captured from the
sera
of breast cancer patients with an anti-mammaglobin polyclonal or monoclonal
antibody
(which binds an epitope that is the 16 C-terminal amino acids of mammaglobin)
and the
anti-carbohydrate antibody is used for detection. The resulting ELISA assay
provides
sensitive and accurate diagnosis of breast cancer.
48
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
Example 4
Identification of Human CD4 T cell Epitopes for Mamma~lobin
This Example illustrates the generation of CD4 T cells that recognize
mammaglobin.
CD4 T cell responses were generated from PBMC of normal donors
using dendritic cells (DC) pulsed with overlapping 20-mer peptides spanning
the entire
mammaglobin protein sequence. CD4+ T cells were stimulated 3-4 times with DC
pulsed with a mixture of overlapping peptides (10 ~g/mL each) in Iscoves
modified
Dulbecco's Medium (IMDM) containing IL-6 and IL-12 in the primary stimulation,
and
0.5 ng/mL IL-2 and ~ ng/mL IL-7 in all other stimulations. The peptides are
shown in
Figure 10. These lines were subsequently assayed for reactivity with the
priming
peptides or recombinant E. coli-derived mammaglobin. As shown in Figures 11 A
an
11 B, a number of CD4 T cell lines demonstrated reactivity with the priming
peptides as
well as mammaglobin protein. The dominant reactivity of these lines appeared
with
peptide SA (EYKELLQEFIDDNATTNAID; SEQ ID NO: 4), corresponding to amino
acids 41-60 of the mammaglobin sequence. These results indicate that peptide
SA
represents an immunogenic CD4 epitope of mammaglobin.
Example 5
Identification of Human CD8 T cell Epitopes for Mamma~lobin
This Example illustrates the generation of CD8 T cells that recognize
mammaglobin.
HLA A2Kb mice were immunized with 9-mer peptides predicted to bind
HLA A2 (shown in Figure 12). Immunizations were performed subcutaneously in
the
footpad, using 100 ~g of peptide together with 140 pg of hepatitis B virus
core peptide
(a Th peptide) in Freund's incomplete adjuvant. Three weeks post immunization,
spleen cells were removed and cultured in vitro with peptide-pulsed APC to
elicit CTL
lines. CTL lines were subsequently evaluated for recognition of peptide-pulsed
mammaglobin-transduced target cells in a standard chromium release assay. CTL
lines
recognizing peptide pulsed targets were then tested on targets transduced and
stably
49
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
expressing the mammaglobin protein. CTL lines from mice immunized with the 9-
mer
peptide KLLMVLMLA (mgb 1; SEQ ID NO: 5) corresponding to amino acids 2-10 of
mammaglobin were shown to recognize both peptide-pulsed and mammaglobin
transduced targets (Figures 13A-13C and 14A-14C). These data demonstrate that
the 9-
mer peptide KLLMVLMLA (SEQ ID NO: 5) is a naturally processed CTL epitope of
mammaglobin, and is restricted by HLA A2.
Example 6
Detection of Mamma~lobin RNA in Patient Blood Samples
This Example illustrates the use of PCR to detect mammaglobin
expression in blood for the purpose of diagnosing breast cancer.
RNA extraction: RNA was extracted from frozen tumors and normal
tissues and cell lines (MB415) as follows. Tissue samples were homogenized in
Trizol
reagent (Gibco, BRL) at lml/50-100mg of tissue using a homogenizer (Polytron)
and
cells mixed with Trizol reagent at lml 5-10x106 cells. The homogenized samples
were
then incubated at room temperature for 5 minutes followed by the addition of
0.2m1 of
chloroform per lml of Trizol reagent. Sample tubes were capped and vigorously
shaken for 15 seconds followed by a further incubation at room temperature for
2-3
minutes. Samples were centrifuged at 12,OOOg for 15 minutes at 2-8°C,
and the upper
aqueous phase was removed. The RNA preparation was transferred to a new tube
and
precipitated by addition of O.SmI isopropyl alcohol per lml Trizol reagent
used in the
homogenation step. Samples were incubated at room temperature for 10 minutes,
and
then centrifuge for 10 minutes, 12,OOOg at 2-8°C. The supernatant was
removed from
the gel like pellet and the pellet was washed once with 75% ethanol (1 ml/lml
of
Trizol). The sample was mixed and then centrifuged at 7,SOOg for 5 minutes at
2-8°C.
Supernatant was removed and the RNA pellet was briefly dried at room
temperature and
dissolved in RNase free water.
Isolated RNA was treated with DNase to remove any DNA
contamination. The RNA (SO~g) in 751 nuclease free water and first strand
buffer
(Gibco BRL) was incubated with DNaseI (Ambion) in the presence of RNase
inhibitor
RNasin (Promega) at 37°C for 30 minutes. The reaction mix was then
precipitated with
CA 02375049 2001-11-23
WO 00/73338 PCTNS00/14845
phenol/chloroform and centrifuged for ~ minutes in an eppendorf centrifuge
maximum
speed. The top layer was transferred to new tube, to which 201 3M sodium
acetate and
4401 of 100% cold ethanol was added. The mixture was vortexed and spun again
for 5
minutes. Supernatant was discarded and the pellet was washed with 75% cold
ethanol
and centrifuged. The RNA pellet was resuspended in RNase free water at 1-2p
g/ml.
RNA was extracted from whole blood using Dynal's Epithelial cell
enrichment beads and Dynal's mRNA Direct kit (Dynal, Oslo, Norway) according
to the
manufacturer's instructions. RNA extracted via the Dvnal extraction kit was
immediately resuspended in 20m1 of Reverse transcription mix shown below and
reverse transcribed.
Reverse Transcription: cDNA for use in real time PCR tissue panels was
prepared as follows. 25~g of RNA was incubated with 25y1 Oligo dT (Boehringer
Mannheim) (100ng/ml) at 70°C for 10 minutes, and then with 1251 of
diluted reverse
transcriptase buffer (Gibco, BRL containing O.SmM dNTP's 1000 units RNasin,
0.02mM dithiothreitol and Superscipt II (Gibco BRL) at 42°C for 1 hour.
The reaction
mix was then cooled to 4°C for use in real-time PCR or frozen. The
reaction mix for
the epithelial extracted material was 201 of Superscript RT mix (4u1 of Sx
buffer, 2~1
of 0.1 M DTT, 1 ~l 1 OmM dNTP mix, 1 ~1 (200 units) of superscript II and 12~
1 of
RNAse free water. The mix was then incubated at 50°C for 5 minutes
followed by
42°C for 50 minutes then inactivated at 70°C for 15 minutes.
Real Time PCR: Real time PCR analysis was performed on the Perkin
Elmer/Applied Biosystems 7700 Prism instrument. Matching primers and
fluorescent
probes were designed for each of the genes of interest according to the primer
express
program provided by Perkin Elmer/Applied Biosystems (Foster City, CA). Primers
and
probes so produced can be used in the universal thermal cycling program in
Real time
PCR. Initially the primers and probes were titrated to determine the optimal
concentrations using a checkerboard approach. A pool of cDNA from target
tumors
was used in this optimization process. These reagents were then used in Real
Time
PCR at their optimal concentrations. The reaction was performed in 25p1
volumes. In
all cases the final probe concentration was 155nM. dATP, dCTP and dGTP were at
0.2mM and dUTP at 0.4mM. Amplitaq gold and Amperase UNG (Perkin
51
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
Elmer/Applied Biosystems, Foster City CA) were used at 0.625 units and 0.25
units per
reaction. MgCI, was at a final concentration of SmM. Trace amounts of
glycerol,
gelatin and Tween 20 (Sigma Chem Co, St Louis, MO) were added to stabilize the
reaction. Each reaction contained 2~1 of template. B-actin primers and probes
were
obtained from Perkin Elmer/Applied Biosystems (Foster City, CA) and used in a
similar manner to quantitate the presence of B-actin in the samples. The
forward primer
was at 900nM, reverse primer at 300nM.
In order to quantitate the amount of specific RNA in the sample a
standard curve was generated alongside using the plasmid containing the gene
of
interest. Standard curves were generated using the Ct values determined in the
real-
time PCR which are related to the initial cDNA concentration used in the
assay.
Standard dilutions ranging from 10-106 copies of the gene of interest were
used for this
purpose. In addition, a standard curve was generated for (3-actin ranging from
200fg-2000pg. This enabled standardization of initial RNA content of a tissue
sample
to the amount of (3-actin for comparison purposes.
The primers and probes used were as shown in Table 1.
52
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
Table I
Mamma~lobin Primers and Probes
Mammaglobin SEQ ID NO:
Forward PrimerTGCCATAGATGAATTGAAGGAATG 8
Reverse PrimerTGTCATATATTAATTGCATAAACACCTCA 9
Probe TCTTAACCAAACGGATGAAACTCTGAGCAATG 10
The mammaglobin gene sequence contains three exons. Exon one spans
bases 992 through 1110, exon two from 1713 through 1900, and exon three from
3789
through 3974. The start Met is at base 1056 and the stop codon is at base
3725. The
primers and probes used for the quantitative real time PCR are located in exon
2;
however, the reverse primer is divided between exon 2 and exon 3. The primer
placement does not exclude amplification of genomic DNA. All tissue samples
were
DNase treated with Ambion DNase I. These samples were tested for the presence
of
contaminating DNA prior to use. RNA extracted from whole blood using Dynal's
Epithelial cell enrichment beads and Dynal's mRNA Direct kit were not DNase
treated,
but this is a highly specific isolation method for RNA only.
Figure 15 shows the tissue distribution for mammaglobin showing the
high degree of specificity for breast tissue. The skin sample shown to be
positive was
from a breast reduction. Figure 16 shows the copies of mammaglobin message
detectable in the breast cancer cell line MB415 as a function of the amount of
cells
indicating that one cell has 10000 copies. Figure 17 shows the detection of
mammaglobin in epithelial cells isolated, using the Dynal isolation method,
from the
peripheral blood of patients with metastatic breast cancer compared to similar
isolates
from normal blood samples. Thirty three metastatic and 11 normal samples were
tested.
The data indicate that mammaglobin can be detected in the blood of individuals
with
metastatic breast cancer, and that such detection may be used to diagnose the
disease.
From the foregoing it will be appreciated that, although specific
embodiments of the invention have been described herein for purposes of
illustration,
various modifications may be made without deviating from the spirit and scope
of the
invention. Accordingly, the invention is not limited except as by the appended
claims.
53
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
1
SEQUENCE LISTING
<110> Corixa Corporation
Fanger, Gary R.
Foy, Theresa M.
Houghton, Raymond L.
Reed, Steven G.
<120> COMPOSITIONS AND METHODS FOR THE
THERAPY, DIAGNOSIS AND MONITORING OF BREAST CANCER
<130> 210121.479PC
<140> PCT
<141> 2000-05-26
<160> 45
<170> FastSEQ for Windows Version 4.0
<210> 1
<211> 20
<212> PRT
<213> Homo sapien
<400> 1
Ile Asp Glu Leu Lys Glu Cys Phe Leu Asn Gln Thr Asp Glu Thr Leu
1 5 10 15
Ser Asn Val Glu
<210> 2
<211> 15
<212> PRT
<213> Homo sapien
<400> 2
Thr Thr Asn Ala Ile Asp Glu Leu Lys Glu Cys Phe Leu Asn Gln
1 5 10 15
<210> 3
<211> 21
<212> PRT
<213> Homo sapien
<400> 3
Ser Gln His Cys Tyr Ala Gly Ser Gly Cys Pro Leu Leu Glu Asn Val
1 5 10 15
Ile Ser Lys Thr Ile
<210> 4
<211> 20
<212> PRT
<213> Homo sapien
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
2
<400> 4
Glu Tyr Lys Glu Leu Leu Gln Glu Phe Ile Asp Asp Asn Ala Thr Thr
1 5 10 15
Asn Ala Ile Asp
<210> 5
<211> 9
<212> PRT
<213> Homo sapien
<400> 5
Lys Leu Leu Met Val Leu Met Leu Ala
1 5
<210> 6
<211> 13
<212> PRT
<213> Homo sapien
<400> 6
Gln Glu Phe Ile Asp Asp Asn Ala Thr Thr Asn Ala Ile
1 5 10
<210> 7
<211> 13
<212> PRT
<213> Homo sapien
<400> 7
Leu Lys Glu Cys Phe Leu Asn Gln Thr Asp Glu Thr Leu
1 5 10
<210> 8
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> PCR primer
<400> 8
tgccatagat gaattgaagg aatg 24
<210> 9
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> PCR primer
<400> 9
tgtcatatat taattgcata aacacctca 29
<210> 10
<211> 32
<212> DNA
CA 02375049 2001-11-23
WO 00/73338 PCTNS00/14845
-,
<213> Artificial Sequence
<220>
<223> Probe
<400> 10
tcttaaccaa acggatgaaa ctctgagcaa tg 32
<210> 11
<211> 20
<212> PRT
<213> Oryctolagus cuniculus
<400> 11
Ile Asp Glu Leu Lys Glu Cys Phe Leu Asn Gln Thr Asp Glu Thr Leu
1 5 10 15
Ser Asn Val Glu
<210> 12
<211> 20
<212> PRT
<213> Oryctolagus cuniculus
<400> 12
Glu Leu Leu Gln Glu Phe Ile Asp Asp Asn Ala Thr Thr Asn Ala Ile
1 5 10 15
Asp Glu heu Lys
<210> 13
<211> 15
<212> PRT
<213> Oryctolagus cuniculus
<400> 13
Thr Thr Asn Ala Ile Asp Glu Leu Lys Glu Cys Phe Leu Asn Gln
1 5 10 15
<210> 14
<211> 20
<212> PRT
<213> Oryctolagus cuniculus
<400> 14
Glu Leu Leu Gln Glu Phe Ile Asp Asp Asn Ala Thr Thr Asn Ala Ile
1 5 10 15
Asp Glu Leu Lys
<210> 15
<211> 20
<212> PRT
<213> Oryctolagus cuniculus
<400> 15
Glu Leu Leu Gln Glu Phe Ile Asp Asp Asn Ala Thr Thr Asn Ala Ile
1 5 10 15
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
4
Asp Glu Leu Lys
<210> 16
<211> 21
<212> PRT
<213> Oryctolagus cuniculus
<400> 16
Ser Gln His Cys Tyr Ala Gly Ser Gly Cys Pro Leu Leu Glu Asn Val
1 5 10 15
Ile Ser Lys Thr Ile
<210> 17
<211> 20
<212> PRT
<213> Oryctolagus cuniculus
<400> 17
Glu Leu Leu Gln Glu Phe Ile Asp Asp Asn Ala Thr Thr Asn Ala Ile
1 5 10 15
Asp Glu Leu Lys
<210> 18
<211> 21
<212> PRT
<213> Mus musculus
<400> 18
Ser Gln His Cys Tyr Ala Gly Ser Gly Cys Pro Leu Leu Glu Asn Val
1 5 10 15
Ile Ser Lys Thr Ile
<210>
19
<211>
405
<212>
DNA
<213>
Oryctolagus
cuniculus
<400>
19
caccatggagacaggcctgcgctggcttctcctggtcgctgtgctcaaaggtgtccagtg 60
tcagtcgctggaggagtccggcggtcgcctggtaacgcctggaggatccctgacactcac 120
ctgcacagtctctggaatcgacctcagtagctatggagtgggctgggtccgccaggctcc 180
agggaaggggctggaatacatcggaatcattagtaaaattgataacacatactacgcgaa 240
ctgggcgaaaggccgattcaccatctccaaaacctcgtcgaccacggtggatctgaaaat 300
gaccagtctgacaaccgaggacacggccacctatttctgtaccagagggtcttttgatcc 360
ctggggcccaggcaccctggtcaccgtctcctcagggcaacctaa 405
<210>
20
<211>
414
<212>
DNA
<213>
Oryctolagus
cuniculus
<400>
20
caccatggagacaggcctgcgctggcttctcctggtcgctgtgctcaaaggtgtccagtg 60
tcagtcggtggaggagtccgggggtcgcctggtcacgcctgggacacccctgacactcac 120
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
ctgcacagtctctggattctccctcagcagctacgacatgacctgggtccgccaggctcc 180
agggaaggggctggaatggatcggaaccattagtactattggtagcccattttacgcgag 240
ctgggcgagaggccgattcaccatctccaaaacctcgaccacggtggatctgaaaatcac 300
caatccgacaaccgaggacacggccacgtatttttgcggcagatttcggattgctggtga 360
tggtgccttctggggcccaggcacgctggtcaccgtctcctcagggcaacctaa 414
<210>
21
<211>
414
<212>
DNA
<213>
Oryctolagus
cuniculus
<400>
21
caccatggagacaggcctgcgctggcttctcctggtcgctgtgctcaaaggtgtccagtg 60
tcagtcggtggaggagtccgggggtcgcctggtcacgcctaggacacccctgacactcac 120
ctgcacagtctctggattctccctcagcagctacgacatgacctgggtccgccaggctcc 180
agggaaggggctggaatggatcggaaccattagtactattggtagcccattttacgcgac 240
ctgggcgagaggccgattcaccatctccaaaacctcgaccacggtggatctgaaaatcac 300
caatccgacaaccgaggacacggccacgtatttttgcggcagatttcggattgctggtga 360
tggtgccttctggggcccaggcacgctggtcaccgtctcctcagggcaacctaa 414
<210>
22
<211>
414
<212>
DNA
<213>
Oryctolagus
cuniculus
<400>
22
caccatggagacaggcctgcgctggcttctcctggtcgctgtgctcaaaggtgtccggtg 60
tcagtcggtggaggagtccgggggtcgcctggtcacgcctgggacacccctgagattcac 120
ctgcacagtctctggaatcgacctcagcacctacgacatgacctgggtccgccaggctcc 180
agggaagggactggaatggatcggaaccattagtactcttggtacccctttttccgccaa 240
ttgggcgagaggccgattcaccatctccaagacctcgaccacggtggatctgaaaatcgc 300
cagtccgacgaccgaagacactgccacatatttttgtggcagattgcggattgctcatga 360
tggtgccttctggggcccaggcacgctggtcaccgtctcctcagggcaacctaa 414
<210>
23
<211>
422
<212>
DNA
<213>
Oryctolagus
cuniculus
<400>
23
cccatggagacaggcctgcgctggcttctcctggtcgctgtgctcaaaggtgtccagtgt 60
caggagcagctgaaggagtccggaggaggcctggtcacgcctgggacacccctgacactc 120
acctgcacagtgtctggaatcgacctcaatatcgatgcaatgagctgggtccgccaggct 180
ccagggaaggggctggaatggatcggaattattggtactcgtggtggcacatggttcgcg 240
agctgggcgaaaggccgattcaccatctccaaaaccccgaccacagtggatctgaaaatc 300
cccagtccgacaaccgaggacacggccacctatttctgtgccagtatctattctgatagt 360
ggtacttatacgaccttgtggggcccaggcaccccggtcaccgtctcctcagggcaacct 420
as 422
<210> 24
<211> 414
<212> DNA
<213> Oryctolagus cuniculus
<400> 24
caccatggag acaggcctgc gctggcttct cctggtcgct gtgctcaaag gtgtccagtg 60
tcagtcggtg gaggagtccg ggggtcgcct ggtcacgcct gggacacccc tgacactcac 120
ctgcaccgtc tctggattct ccctcagcag cgtcgacatg acctgggtcc gccaggctcc 180
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
6
agggaaggggctggaatggatcggaaccattagtactcgtagtagcacatactacgcgag240
ctgggcgaaaggccgattcaccatctccaaaacctcgaccacggtggatctgaaaatcac300
cagtccgacaaccgaggacacggccacgtatttctgtggcagatttcggattgctggtga360
tggtgccttctggggcccaggcacgctggtcaccgtctcctcagggcaacctaa 414
<210>
25
<211>
412
<212>
DNA
<213> culus
Oryctolagus
cuni
<220>
<221> feature
misc_
<222> .(412)
(1)..
<223>
n = A,T,C
or G
<400>
25
ggaaggctgcgctggcttttcctggtcgctgtgctcagaggtgtccagtgtcagtcgctg60
gaggagtccgggggtngcctggtaacgcctgggacacccctganantcacctgcacagcc120
tttggattttccctcagtagctggtcaatgagctgggtccgccaggctccagggaagggg180
ctggaatggatcggaatgattggtattgttggtagtggcacataatangcgacctgggcg240
aaaggccgattcaccatttccaaaaccttgtgaccacggtcgatttgaaaatgaccagtt300
tgacaaccgaggacacggccacctatttttgtgtcagagggggtagttttanttttgcta360
ccgccttgtggggcccaggcaccctggtcaccgtntcctcagggcaacctas 412
<210>
26
<211>
402
<212>
DNA
<213>
Oryctolagus
cuniculus
<220>
<221> feature
misc
_
<222>
(1).
.(402)
<223>
n = A,T,C
or G
<400>
26
ttgcaggctgcgtggttttcctggtcgctgtgctcaaaggtgtccagtgtcagtcggtgg60
aggagtccgggggtngcctggtaacncctgggacacccctgacanttttttgcaaagtnt120
ttggattttccctcagcagntacganatgacctgggtccgccaggctccagggaaggggc180
tggaatggatnggaaccattagtanttgtggtaatggataatacgcgacctgggcgaaag240
gccgattcaccatttccaaaaccttgaccaccgtggatttgaaaatcaccagtccgacaa300
ccgaggacacggccaagtatttttgtggcagatttcggattgctggtgatggtgcttttg360
gggcccgggcacgctggtcaccgtntcctcagggcaacctas 402
<210>
27
<211>
93
<212>
PRT
<213> sapien
Homo
<400>
27
Met Lys Leu Ala Leu Ser His Cys
Leu Leu Ala Gln
Met Val
Leu Met
1 5 10 15
Tyr Ala Leu Glu Val Ile Lys Thr
Gly Ser Asn Ser
Gly Cys
Pro Leu
20 25 30
Ile Asn Glu Tyr Glu Leu Gln Glu
Pro Gln Lys Leu
Val Ser
Lys Thr
35 40 45
Phe Ile Asn Ala Asp Glu Lys Glu
Asp Asp Ile Leu
Asn Ala
Thr Thr
50 55 60
Cys Phe Thr Leu Asn Val Val Phe
Leu Asn Ser Glu
Gln Thr
Asp Glu
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
7
65 70 75 80
Met Gln Leu Ile Tyr Asp Ser Ser Leu Cys Asp Leu Phe
85 90
<210> 28
<211> 55
<212> PRT
<213> Homo sapien
<400> 28
Gly Ser Gly Met Lys Glu Thr Ala Ala Ala Lys Phe Glu Arg Gln His
1 5 10 15
Met Asp Ser Pro Asp Leu Gly Thr Asp Asp Asp Asp Lys Ala Met Ala
20 25 30
Ile Ser Asp Pro Asn Ser His Cys Tyr Ala Gly Ser Gly Cys Pro Leu
35 40 45
Leu Glu Asn Val Ile Ser Lys
50 55
<210> 29
<211> 13
<212> PRT
<213> Homo sapien
<400> 29
Met Lys Leu Leu Met Val Leu Met Leu Ala Ala Leu Ser
1 5 10
<210> 30
<211> 20
<212> PRT
<213> Homo sapien
<400> 30
Ala Leu Ser Gln His Cys Tyr Ala Gly Ser Gly Cys Pro Leu Leu Glu
1 5 10 15
Asn Val Ile Ser
<210> 31
<211> 20
<212> PRT
<213> Homosapien
<400> 31
Gly Cys Leu Leu Glu Asn Val SerLys Thr Ile Asn Pro
Pro Ile Gln
1 5 10 15
Val Ser Thr
Lys
20
<210> 32
<211> 20
<212> PRT
<213> Homosapien
<400> 32
Lys Thr Asn Pro Gln Val Ser ThrGlu Tyr Lys Glu Leu
Ile Lys Leu
1 5 10 15
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
8
Gln Glu Phe Ile
<210> 33
<211> 20
<212> PRT
<213> Homo sapien
<400> 33
Glu Tyr Lys Glu Leu Leu Gln Glu Phe Ile Asp Asp Asn Ala Thr Thr
1 5 10 15
Asn Ala Ile Asp
<210> 34
<211> 20
<212> PRT
<213> Homo sapien
<400> 34
Asp Asp Asn Ala Thr Thr Asn Ala Ile Asp Glu Leu Lys Glu Cys Phe
1 5 10 15
Leu Asn Gln Thr
<210> 35
<211> 20
<212> PRT
<213> Homo sapien
<400> 35
Glu Leu Lys Glu Cys Phe Leu Asn Gln Thr Asp Glu Thr Leu Ser Asn
1 5 10 15
Val Glu Val Phe
<210> 36
<211> 23
<212> PRT
<213> Homo sapien
<400> 36
Asp Glu Thr Leu Ser Asn Val Glu Val Phe Met Gln Leu Ile Tyr Asp
1 5 10 15
Ser Ser Leu Cys Asp Leu Phe
<210> 37
<211> 9
<212> PRT
<213> Homo sapien
<400> 37
Lys Leu Leu Met Val Leu Met Leu Ala
1 5
<210> 38
<211> 9
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
9
<212> PRT
<213> Homo sapien
<400> 38
Leu Leu Met Val Leu Met Leu Ala Ala
1 5
<210> 39
<211> 9
<212> PRT
<213> Homo sapien
<400> 39
Leu Met Val Leu Met Leu Ala Ala Leu
1 5
<210> 40
<211> 9
<212> PRT
<213> Homo sapien
<400> 40
Phe Leu Asn Gln Thr Asp Glu Thr Leu
1 5
<210> 41
<211> 10
<212> PRT
<213> Homo sapien
<400> 41
Leu Ile Tyr Asp Ser Ser Leu Cys Asp Leu
1 5 10
<210> 42
<211> 10
<212> PRT
<213> Homo sapien
<400> 42
Lys Leu Leu Met Val Leu Met Leu Ala Ala
1 5 10
<210> 43
<211> 10
<212> PRT
<213> Homo sapien
<400> 43
Phe Met Gln Leu Ile Tyr Asp Ser Ser Leu
1 5 10
<210> 44
<211> 10
<212> PRT
<213> Homo sapien
<400> 44
CA 02375049 2001-11-23
WO 00/73338 PCT/US00/14845
Ala Ile Asp Glu Leu Lys Glu Cys Phe Leu
1 5 10
<210> 45
<211> 10
<212> PRT
<213> Homo sapien
<400> 45
Leu Leu Gln Glu Phe Ile Asp Asp Asn Ala
1 5 10