Note: Descriptions are shown in the official language in which they were submitted.
..
CA 02379165 2002-02-06
Chimeric proteins
The present invention concerns recombinant chimeric proteins which have a
nucleic
acid synthesis activity, complexes containing these proteins, nucleic acids
encoding
these proteins, vectors and cells containing these proteins, an antibody
directed
against them, applications of these proteins, kits containing them as well as
methods
for the elongation, amplification, reverse transcription, sequencing and
labelling of
nucleic acids.
Many proteins are not present in the cell as monomers but as part of a
functional
multimeric complex. Examples of such complexes have been described in almost
all
fields of cell biology (e.g. transcription, translation, replication,
cytoskeleton, signal
transduction, mRNA processing).
The interaction or binding of a protein with or to another protein which is
referred
to in the following as the donor protein or acceptor protein is effected by
certain
amino acids or amino acid sequences which, in the folded protein, are usually
on the
surface of the protein and are responsible for the specific binding or
interaction to or
with the partner. These amino acids or such an amino acid sequence is referred
to
herein in the following as the "interaction-effecting sequence" or interaction-
mediating sequence. Persons skilled in the art know that the amino acids
involved in
the formation of an interaction are not necessarily directly adjacent to one
another in
the primary amino acid sequence but rather that positions within the amino
acid
sequence of the protein or peptide that are conserved to a greater or lesser
degree are
responsible for the interaction. If reference is made in the following to the
determination of a sequence that mediates an interaction or effects an
interaction,
this is intended to mean both aforementioned aspects. In this connection donor
proteins are understood as those proteins which interact with another protein
and
optionally bind another protein. Likewise acceptor proteins are understood as
those
proteins which interact with another protein and may bind another protein.
There are a number of methods for determining the interaction-effecting
sequence
of a protein-protein interaction site. These include (i) the determination of
the three-
-2-
dimensional structure of a complex of donor and/or acceptor protein by X-ray
structural analysis or (ii) NMR methods. Another method is (iii) to
reconstitute a
complex binding in vitro using recombinant proteins and to determine the
sequences) which effect the interaction by specifically changing the donor or
acceptor protein. Such specific changes include the mutation of individual
amino
acids, for example to form alanine (alanine scanning) or the deletion of
sequences in
the protein. If the change or deletion of an amino acid or sequence leads to
the loss
of the interaction or binding activity then it is a part of the sequence
effecting the
interaction and conversely if the change or deletion of an amino acid or
sequence
does not lead to the loss of the interaction or binding activity then it is
not part of
the interaction-effecting sequence. The interaction-effecting sequence can be
defined in this manner.
Another method for determining the interaction-effecting sequence of a protein
is
based on the use of a two-hybrid system also abbreviated in the following as
"Y2H". Y2H systems are based on the expression of a protein having a
detectable
activity (such as the enzyme dihydrofolate reductase) as two non-covalently
linked
parts. This protein is inactive when the two parts are not in spatial vicinity
to one
another. The two proteins to be examined which are known to interact with one
another and thus can form a complex i.e. donor protein and acceptor protein,
or
which are to be investigated in this regard, are each fused with one of the
two parts
of the protein having the detectable activity (such as dihydrofolate
reductase) to
form a fusion protein and expressed, which results in the formation of two
fusion
proteins. If the fused donor protein binds the fused acceptor protein, the two
halves
of the detectable protein (such as dihydrofolate reductase) come into spatial
proximity. This restores the activity of the protein which can then be
detected.
Suitable proteins having detectable activity are enzymes (dihydrofolate
reductase,
beta-galactosidase), signal transduction proteins (Cdc25 from Saccharomyces
cerevisiae) or transcription activators (Gal4, LexA-VP16). The determination
of the
binding region using the two hybrid system is based on the same considerations
as
with the in vitro reconstitution of binding: If the change or deletion of an
amino acid
or sequence leads to the loss of binding activity, then it is part of the
interaction-
effecting sequence and conversely if the change or deletion of a sequence does
not
lead to the loss of binding activity then it is not part of the interaction-
effecting
sequence.
CA 02379165 2002-02-06
. .. _3_
In addition the interaction-effecting sequence can be defined by examining the
interaction of numerous fragments of the protein and determining which parts
of the
protein are always present in the interacting fragments. This region which is
always
present is the interaction-effecting sequence. In order to identify the amino
acids in
an interaction-effecting sequence that are essential for the interaction or
binding, it
is possible to change individual amino acids by targeted mutations. A loss or
increase in the binding activity indicates that these positions are directly
involved in
the interaction.
The complexes that are particularly preferred for in vitro applications
include the
thermostable complexes of prokaryotic and eukaryotic replication apparatuses
which often contain polymerases as an important enzymatic activity.
As already mentioned the interaction between proteins in vivo plays a major
role for
example in the replication of nucleic acids in biological systems. Thus highly
processive replication mechanisms are known which are, on the one hand,
cellular
mechanisms and, on the other hand, the replication mechanisms which occur in
the
bacteriophages T4 and T7.
The replication apparatus comprises many components. These include among
others
a) proteins having polymerase activity, b) proteins which are involved in the
formation of a clamp structure, the function of the clamp structure being,
among
others, to bind a polymerase activity to its template and to stabilize the
binding and
thus to change the dissociation constant of the complex of polymerase and
nucleic
acid accordingly, c) proteins which load the clamp onto the template, d)
proteins
which stabilize the template and optionally e) proteins which guide the
polymerase
onto the template.
Proteins having polymerase activity are understood herein in particular as
those
proteins which are able to bind one or several nucleotides or nucleosides to a
nucleotide or nucleoside or polynucleotide or polynucleoside. In each of the
above
cases these can be ribonucleotides/ribonucleosides or deoxynucleotides/deoxy-
nucleosides or polymers thereof. Hence these proteins include, apart from DNA
polymerases, also RNA polymerases irrespective of whether a template is
required
CA 02379165 2002-02-06
CA 02379165 2002-02-06
-4-
or not required for the polymerization reaction of the protein. These proteins
having
polymerase activity are thus also proteins having nucleic acid synthesis
activity.
These also include the elongation proteins known as such in the prior art. An
elongation protein as used herein is also understood as a protein or complex
having
polymerase activity that has at least one or more of the following properties:
Uses
RNA as a template, uses DNA as a template, synthesizes RNA, synthesizes DNA,
exonuclease activity in the 5'-3' direction or exonuclease activity in the 3'-
5'
direction, strand displacement activity and processivity or non-processivity.
DNA polymerases belong to a group of enzymes which use single-stranded DNA as
a template for the synthesis of a complementary DNA strand. These enzymes play
a
major role in nucleic acid metabolism including the processes of DNA
replication,
repair and recombination. DNA polymerases have been identified in all cellular
organisms from bacterial to human cells, in many viruses as well as in
bacteriophages (Kornberg, A. & Baker, T.A. (1991) DNA Replication WH
Freeman, New York, NY). The archaebacteria and eubacteria are usually combined
to form the prokaryote group which are organisms without a real cell nucleus
in
contrast to the eukaryotes which are organisms with a real cell nucleus. A
common
feature of many polymerases from the diverse organisms is often a similarity
of the
amino acid sequence and a similarity of structure (Wang, J., Sattar, A.K.M.A.;
Wang, C.C., Karam, J.D., Konigsberg, W.H. & Steitz, T.A. (1997) Crystal
Structure
of pol a family replication DNA polymerase from bacteriophage RB69.Ce11 89,
1087-1099). Organisms such as humans have numerous DNA-dependent
polymerases which are, however, not all responsible for DNA replication but
some
also carry out DNA repair. Replicative DNA polymerases are usually composed in
vivo of protein complexes with several units which replicate the chromosomes
of
the cellular organisms and viruses. A general property of these replicating
polymerases is in general a high processivity which means their ability to
polymerise thousands of nucleotides without dissociating from the DNA template
(Kornberg, A. & Baker, T.A. ( 1991 ) DNA Replication, WH Freeman, New York,
NY).
DNA polymerases are characterized, among others, by two properties, their
elongation rate i.e. the number of nucleotides which they can incorporate per
second
into a growing DNA strand and their dissociation constant. If the polymerase
_5_
dissociates again from the strand after each step of incorporating one
nucleotide into
the growing chain (i.e. one elongation step occurs per binding event), then
the
processivity has the value 1 and the polymerase is not processive. If the
polymerase
remains connected to the strand for repeated nucleic acid incorporations, then
the
elongation or replication modus and thus also the polymerase is referred to as
processive and can reach a value of several thousand (see also: Methods in
Enzymology Volume 262, DNA replication, edited by J.L. Campbell, Academic
Press 1995, pp. 270-280).
The proteins mentioned under b) form structures which are either open or
closed,
for example circular or semi-circular structures. Such structures can be
formed by
one or several species of proteins. One of the said protein species may have a
polymerase activity.
The proteins responsible for the formation of these structures are referred to
in the
following as "sliding clamp proteins" or "clamp proteins" provided they have
no
polymerase activity.
It is known that the replication apparatus in archaea is similar to the
eukaryotic
replication apparatus although the genome organisation in eukaryotes and
archaea is
completely different and the cellular structure of the eubacteria is similar
to that of
the archaea (Edgell, D.R. and Doolittle, W.F. (1997), Archaea and the origins)
of
DNA replication proteins, Cell 89, 995-998).
The sliding clamp is frequently bound to an elongation protein via one or
several
other proteins, in other words it is coupled to the elongation protein. Such a
coupling protein is referred to herein in the following as a coupling protein
whereby
the coupling may take place via a plurality of coupling proteins.
The three-dimensional structure of various sliding clamp proteins has already
been
determined. The overall structure of these sliding clamps is very similar; the
pictures of the circular total protein structure of PCNA, of the (3 subunit
and gp45
rings are superimposable when laid on top of one another (Kelman, Z. &
O'Donnel,
M. (1995) Structural and functional similarities of prokaryotic and eukaryotic
CA 02379165 2002-02-06
CA 02379165 2002-02-06
~- -6_
sliding clamps. Nucleic Acids Res. 23, 3613-3620). Each ring has comparable
dimensions and a central opening which is large enough to encircle duplex DNA
i.e.
a DNA double strand composed of the two complementary DNA strands.
The sliding clamp cannot position itself in vivo around the DNA but must be
clamped around the DNA. In prokaryotes and eukaryotes such a protein complex
is
composed of numerous subunits. The protein complex recognises the 3'-end of
the
primer in the "primer-template duplex" which is to be elongated to form a
longer
double strand by incorporating nucleotides and positions the sliding clamp
around
the DNA.
In the case of the bacteriophage T7 the same object i.e. a processive DNA
synthesis,
as defined below, is achieved by means of a protein complex with a different
structure. The phage expresses its own catalytic polymerase, T7 polymerase,
the
gene product of gene 5 which binds to a protein from the host Escherichia coli
i.e.
thioredoxin and enables a highly processive DNA replication as a replicase. In
this
case there is also clamp formation but this clamp does not have the same
structure
as for example in the case of the eukaryotic PCNA.
It is often necessary, as for example in the case of human polymerase 8, for
proteins
(coupling proteins) to make the connection between the catalytically active
part of
the polymerase and a processivity factor. A processivity factor is a compound
or a
molecule which influences the processivity of a polymerase and preferably
increases
it. Sliding clamp proteins are examples of processivity factors. In humans
this
coupling protein is a small subunit of the 8 polymerase (Zhang, S.-J., Zeng,
X.-R.,
Zhang, P., Toomey, N.L., Chuang, R.Y., Chang, L.-S., and Lee, M.Y.W.T. (1994).
A conserved region in the amino terminus of DNA polymerase 8 is involved in
proliferating cell nuclear antigen binding, J. Biol. Chem. 270, 7988-7992).
However, in the case of T7 polymerase the processivity factor binds the
catalytic
unit of the polymerase directly without involvement of a~coupling protein.
In vitro applications of proteins having nucleic acid synthesis activity such
as
polymerases or elongation proteins are widespread in the prior art e.g. for
the
polymerase chain reaction (PCR), nucleic acid sequencing or reverse
transcription.
CA 02379165 2002-02-06
- -7_
For most in vitro applications such as PCR or sequencing processes,
processivity is
a desired property that, however, the thermostable enzymes of the prior art
that are
used in these reactions only have to a slight extent.
The US patents 4,683,195, 4,800,195 and 4,683,202 describe the application of
such
thermostable DNA polymerases in the polymerase chain reaction (PCR). In PCR
DNA is newly synthesized using primers, templates (also referred to as
matrices),
nucleotides, a DNA polymerase, an appropriate buffer and suitable reaction
conditions. A thermostable polymerase which survives the cyclic thermal
melting of
the DNA strands is preferably used for this PCR. Thus Taq DNA polymerase is
often used (LJS patent 4,965,188). However, the processivity of Taq DNA
polymerase is relatively low compared to that of T7 polymerase.
DNA polymerases are also used for DNA sequence determination (Sanger et al.,
Proc. Natl. Acad. Sci., USA 74:5463-5467 (1997)). One of the polymerases that
can
be used for this is for example the Taq polymerase mentioned above (US patent
5,075,216) or the polymerase from Thermotoga neapolitana (WO 96/10640) or
other thermostable polymerases. Recent methods couple the exponential
amplification and sequencing of a DNA fragment in one step so that it is
possible to
directly sequence genomic DNA. One of the methods, the so-called DEXAS method
(Nucleic Acids Res. 1997 May 15:25(10):2032-2034 Direct DNA sequence
determination from total genomic DNA. Kilger C, Paabo S, Biol Chem. 1997 Feb;
378(2):99-105 Direct exponential amplification and sequencing (DEXAS) of
genomic DNA. Kilger C, Paabo S and DE 19653439.9 and DE 19653494.1 ), uses a
polymerase with a reduced ability to discriminate against dideoxynucleotides
(ddNTPs) compared to deoxynucleotides (dNTPs) as well as a reaction buffer,
two
primers which are preferably not present in equimolar amounts and the above-
mentioned nucleotides in order to then obtain a complete, sequence-specific
DNA
ladder of a fragment in several cycles which is flanked by the primers. A
further
development of this method comprises the use of a polymerase mixture in which
one of the two polymerases discriminates between ddNTPs and dNTPs whereas the
second has a reduced discrimination ability (Nucleic Acids Res. 1997 May 1 S;
25(10):2032-2034 Direct DNA sequence determination from total genomic DNA.
Kilger C, Paabo S).
_8_
DNA polymerises are also used for the reverse transcription of RNA into DNA.
In
this case RNA serves as a template and the polymerise synthesizes a
complementary DNA strand. The thermostable DNA polymerise from the organism
Thermus thermusphilis (Tth) (US patent 5,322,770) is for example used in this
case.
Some polymerises may also have a "proof reading" activity i.e. a 3'-5'
exonuclease
activity. This property is particularly desirable when the product to be
synthesized
should be produced with a low rate of nucleotide incorporation errors. The
polymerises from the organism Pyrococcus wosei are an example of this.
The majority of the elongation proteins mentioned above that are used in the
aforementioned applications are not actually replication enzymes in vivo but
are
mostly enzymes which are assumed to be involved in DNA repair which is why
their processivity is relatively small. In addition each organism has many
polymerises that have numerous properties.
As mentioned above such elongation proteins have for example the following
properties: Use of RNA as a template, use of DNA as a template, synthesis of
RNA,
synthesis of DNA, exonuclease activity in the 5'-3' direction and exonuclease
activity in the 3'-5' direction, strand displacement activity, processivity or
non-
processivity or thermostability or thermosensitivity. However, in vivo it is
frequently the case that a protein complex combines one or several of these
properties. In particular replication complexes are often present in vivo
whose
processivity is increased, as described above, by the presence of a sliding
clamp
protein.
In addition to the aforementioned components of replication apparatuses, other
DNA-modifying proteins are also often found in vivo in large complexes with
other
proteins. It is often the case in such complexes that a DNA-modifying activity
such
as terminal transcriptase activity from a first protein is combined with one
or several
activities introduced into the complex by an additional protein such as
exonuclease
activity. DNA-modifying activity is understood herein as any enzymatic
activity
which leads to a chemical, physical or structural change of an initial nucleic
acid. It
is also sometimes the case that a DNA-modifying activity only occurs when
there is
CA 02379165 2002-02-06
CA 02379165 2002-02-06
-9-
an interaction with at least one further protein. It is also possible that a
DNA-
modifying activity is reduced or increased by interaction with at least one
further
protein. Hence in vivo a protein complex is often formed which for example
carries
the sum of the individual activities or whose activity is improved compared to
the
individual activity.
The above-mentioned shows that in vivo complexes which contain a nucleic acid
synthesis activity often have additional desirable properties for technical,
i.e. in
vitro, applications which go beyond the actual nucleic acid synthesis activity
and are
contributed by other components forming the complex.
A direct technical application of such in vivo complexes e.g. for DNA
sequencing,
performing a polymerase chain reaction or introducing labels into nucleic
acids has
previously been unsuccessful for a number of reasons. One reason was and is
the
lack of knowledge of all factors or individual components that are involved in
forming the complex of interest. Another reason is that the complex comprising
several components also has one or several undesired properties in addition to
the
desired properties.
An alternative approach for the in vitro use of in vivo complexes is to
combine
components of different origins but having the desired property and in this
manner
to form the complex having the desired properties. Such a property can for
example
be a higher processivity. A practical hurdle to this approach was that the
individual
components forming the complex which would have to interact with one another
did not do this or only very poorly and hence it was not possible to give the
complex
the desired properties.
Hence the object of the present invention is to provide proteins having a
nucleic
acid synthesis activity which have an increased processivity.
A further object of the present invention is to provide a method which allows
the
construction of such proteins.
CA 02379165 2002-02-06
' - l~-
Finally it is an object of the invention to provide methods for the
amplification,
especially by PCR, and sequencing of nucleic acids which allow longer nucleic
acids to be amplified and/or sequenced.
The object is achieved according to the invention by a recombinant chimeric
protein
comprising
a) a first domain with nucleic acid synthesis activity and
b) an interaction-mediating sequence whereby
the interaction-mediating sequence results in the formation of a complex of
nucleic
acid synthesis activity and sliding clamp protein and the complex is different
from
the complex that the nucleic acid synthesis activity and/or the sliding clamp
protein
form with their natural interaction partner(s).
In a further aspect of the invention the object is achieved by a chimeric
protein
comprising a part having nucleic acid synthesis activity whereby this part is
derived
from a part having nucleic acid synthesis activity and a basic protein
containing an
interaction-mediating component, and at least one interaction-mediating
component
wherein the interaction-mediating component is different from the interaction-
mediating component of the basic protein.
In one embodiment the protein contains the interaction-mediating component of
the
basic protein.
In a further aspect the object is achieved by a chimeric protein comprising a
part
having a nucleic acid synthesis activity, the part being derived from a basic
protein
which comprises a part having nucleic acid synthesis activity but no
interaction-
mediating part, and an interaction-mediating part.
In the chimeric proteins according to the invention provision can be made for
the
interaction-mediating part to mediate binding between the nucleic acid
synthesis
CA 02379165 2002-02-06
-11-
activity and a factor which influences the synthesis rate of the nucleic acid
synthesis
activity. In a preferred embodiment the factor is a sliding clamp protein.
In one embodiment the nucleic acid synthesis activity is intended to comprise
a
consensus peptide sequence, the sequence being selected from the group
comprising
the sequences SEQ ID NO.: l, 2 and 3.
In a further embodiment the sliding clamp protein can comprise a consensus
peptide
sequence, the sequence being selected from the group comprising the sequences
SEQ >D NO.: 4, 5, 6 and 7.
In yet a further embodiment the interaction-mediating sequence can comprise a
consensus peptide sequence which is selected from the group comprising the
sequences SEQ >D NO.: 8, 9, 10, 11 and 12. In a particularly preferred
embodiment
the interaction-mediating sequence comprises a consensus peptide sequence
according to SEQ ID NO.: 8.
In a further embodiment the interaction-mediating sequence is at the C-
terminal end
of the sequence carrying the nucleic acid synthesis activity.
In a further embodiment a linker is located between the interaction-mediating
sequence and the sequence carrying the nucleic acid activity.
Furthermore in one embodiment of the chimeric proteins according to the
invention
the recombinant chimeric proteins can be thermostable.
In one embodiment of the chimeric proteins according to the invention the
protein
can have a DNA polymerase activity. It is particularly preferred that the
proteins
have a 3'-5' exonuclease activity.
In a further embodiment of the chimeric proteins according to the invention
the
protein has an RNA polymerase activity.
-12-
In a further embodiment of the chimeric proteins according to the invention
the
protein has a reverse transcriptase activity.
In yet a further preferred embodiment of the chimeric proteins according to
the
invention the incorporation rate of dNTPs and ddNTPs by the nucleic acid
synthesis
activity differs by a factor of less than 5.
In a further aspect of the invention the object is achieved by a complex
comprising
a) a recombinant chimeric protein according to the invention and
b) a sliding clamp protein.
In a preferred embodiment of the complex according to the invention the
sliding
clamp protein contains a consensus peptide sequence which is selected from the
group comprising the sequences according to SEQ >D NO.: 4, 5, 6 and 7.
In a further embodiment the complex additionally contains a nucleic acid.
In a further aspect the object is achieved by a nucleic acid which codes for a
chimeric protein according to the invention and especially for a recombinant
protein.
In yet a further aspect the object is achieved by a vector containing the
nucleic acid
according to the invention. In a preferred embodiment the vector is an
expression
vector.
In a further aspect the object is achieved by a cell which contains the vector
according to the invention.
The object is also achieved by the use of the chimeric protein according to
the
invention to elongate nucleic acids.
CA 02379165 2002-02-06
. -13-
The object is also achieved by the use of the chimeric protein according to
the
invention to amplify nucleic acids.
The object is also achieved by the use of the chimeric protein according to
the
invention for the reverse transcription of RNA into DNA.
The object is also achieved by the use of the chimeric protein according to
the
invention to sequence nucleic acids and in particular DNA.
In yet a further aspect the object is achieved by a kit and in particular a
reagent kit
for the elongation and/or amplification and/or reverse transcription and/or
sequencing and/or labelling of nucleic acids which comprises in one or several
containers:
a) a chimeric protein according to the invention which is preferably a
recombinant
protein and/or
b) a complex according to the invention and
c) preferably optionally at least one primer, buffer, nucleotides, cofactors
and/or
pyrophosphatase.
In a preferred embodiment of the kit it contains deoxynucleotides or/and
derivatives
thereof in addition to the substances a) and/or b) to amplify nucleic acids.
In a further embodiment the kit contains a DNA polymerase having 3'-5'
exonuclease activity.
1n another embodiment the kit contains substances according to a) and/or b)
which
have reverse transcriptase activity and preferably deoxynucleotides and/or
derivatives thereof for reverse transcription.
The kit according to the invention can contain dideoxynucleotides or/and
derivatives thereof for sequencing in addition to deoxynucleotides or
ribonucleotides or/and derivatives thereof.
CA 02379165 2002-02-06
CA 02379165 2002-02-06
- 14-
In a further aspect the object is achieved by a method for the template-
dependent
elongation of nucleic acids in which the nucleic acid to be elongated or at
least one
strand thereof is provided with at least one primer under hybridization
conditions
whereby the primers are sufficiently complementary to a part of or to a
flanking
region of the nucleic acid to be elongated and the primer is elongated by a
polymerase in the presence of nucleotides wherein a chimeric protein according
to
the invention is used as the polymerase and preferably a sliding clamp protein
is
present in the reaction.
In yet a further aspect the object is achieved by a method for the
amplification of a
nucleic acid in which at least two primers are added to the nucleic acid to be
amplified under hybridization conditions, each of the two primers being
complementary to a part of or to a flanking region of the nucleic acid to be
amplified and the primers are elongated by a polymerase in the presence of
nucleotides wherein a chimeric protein according to the invention and in
particular a
recombinant chimeric protein is used as the polymerase and a sliding clamp
protein
is preferably added to the reaction.
In a preferred embodiment a polymerase chain reaction is carned out.
In an especially preferred embodiment the reaction mixture contains two DNA
polymerases of which at least one has a 3'-5' exonuclease activity and the 3'-
5'
exonuclease activity is either added to the reaction mixture by way of the
chimeric
protein or by an additional polymerase.
In yet a further embodiment two chimeric proteins are present and in
particular
recombinant chimeric proteins wherein one of the proteins has a DNA polymerase
activity and the other has a 3'-5' exonuclease activity.
In the method according to the invention for template-dependent elongation,
nucleic
acids can be sequenced in one embodiment starting with a primer which is
complementary to a region neighbouring the nucleic acid to be sequenced, and a
template-dependent elongation or reverse transcription is carried out using
CA 02379165 2002-02-06
- IS -
deoxynucleotides and dideoxynucleotides or derivatives thereof according to
the
method of Singer.
In a further embodiment of the method according to the invention for template-
dependent elongation at least one label can be introduced during the
elongation of
the nucleic acids.
In a particularly preferred embodiment an agent is used which is selected from
the
group comprising labelled primers, labelled deoxynucleotides and derivatives
thereof, labelled dideoxynucleotides and derivatives thereof and labelled
ribonucleotides and derivatives thereof.
In yet a further aspect the object is achieved by a method for labelling
nucleic acids
by generating single breaks in phosphodiester bonds of the nucleic acid chain
and
replacing a nucleotide at the breakage points by a labelled nucleotide with
the aid of
a polymerise wherein a chimeric protein according to the invention and in
particular
a recombinant chimeric protein is used as the polymerise.
In yet a further aspect the object is achieved by a method for producing a
chirneric
protein which comprises a base sequence and a heterologous interaction-
effecting
sequence and binds to an interaction partner or such a binding is strengthened
as a
result of the interaction-effecting sequence, wherein
a) an interaction system comprising a protein referred to as donor protein and
a
protein referred to as acceptor protein is used to determine which sequence of
the
donor protein or acceptor protein effects the interaction between the two
interaction
partners and
b) the interaction-effecting sequence is introduced into a recipient protein
that is
different from the donor protein and acceptor protein and contains the base
sequence.
CA 02379165 2002-02-06
- - 16-
In one embodiment the donor protein and acceptor protein form a complex which
binds the nucleic acid.
In a further embodiment the donor protein and the acceptor protein form a
complex
which has an activity that is selected from the group comprising polymerase
activity, DNA binding activity, RNA binding activity, 5'-3' exonuclease
activity, 3'-
5' exonuclease activity and ligase activity.
In a preferred embodiment the donor protein is selected from the group
comprising
elongation protein, sliding clamp proteins, sliding clamp loader protein and
coupling proteins.
In a further embodiment the acceptor protein is selected from the group
comprising
elongation protein, sliding clamp proteins, sliding clamp loader protein and
coupling proteins.
In yet a further embodiment the recipient protein is selected from the group
comprising elongation protein, sliding clamp proteins, sliding clamp loader
protein
and coupling proteins.
In one embodiment of the method according to the invention step a) can be
repeated
several times and a consensus sequence can be determined from the interaction-
effecting sequences determined in this manner which represents an interaction-
effecting sequence and introduced in step b) as the interaction-effecting
sequence
into a recipient protein that is different from the donor protein and acceptor
protein
and contains a base sequence.
In a further aspect the object is achieved by a chimeric protein which is
obtainable
by the method according to the invention. In a preferred embodiment the base
sequence is a part of the amino acid sequence of a protein that is selected
from the
group comprising elongation proteins, sliding clamp proteins, sliding clamp
loader
proteins and coupling proteins.
CA 02379165 2002-02-06
- 17-
Finally the object is also achieved by an in vitro complex for the template-
dependent elongation of nucleic acids comprising a sliding clamp protein and
an
elongation protein wherein at least one of the proteins is a chimeric protein
according to the invention. In this connection an embodiment is particularly
preferred in which the complex is thermostable.
In connection with the present invention any of the chimeric proteins
according to
the invention can be a recombinant chimeric protein.
The object is also achieved according to the invention by a recombinant
chimeric
protein comprising a) a first domain having nucleic acid synthesis activity
and b) an
interaction-mediating sequence characterized in that the interaction-mediating
sequence results in the formation of a complex of nucleic acid synthesis
activity and
sliding clamp protein, the complex being different from the complex that the
nucleic
acid synthesis activity and/or the sliding clamp protein form with their
natural
interaction partner(s).
Hence the recombinant chimeric protein according to the invention enables a
nucleic acid synthesis activity to be bound to a sliding clamp protein that
cannot
bind the nucleic acid synthesis activity as it for example occurs naturally.
Hence the
natural interaction partner is a partner that can be present bound to the
nucleic acid
synthesis activity under normal physiological conditions within an organism.
The object is achieved according to the invention by a chimeric protein
comprising
a part having nucleic acid synthesis activity, the part being derived from a
part
having a nucleic acid synthesis activity and a basic protein containing an
interaction-mediating part, and at least one interaction-mediating part the
interaction-mediating part being different from the interaction-mediating part
of the
basic protein, and by a chimeric protein comprising a part having nucleic acid
synthesis activity, the part being derived from a basic protein which
comprises a
part having nucleic acid synthesis activity but no interaction-mediating part
and an
interaction-mediating part.
CA 02379165 2002-02-06
-18-
According to the invention the object is additionally achieved by a method for
the
template-dependent elongation of nucleic acids wherein the nucleic acid to be
elongated or at least a strand thereof is provided with at least one primer
under
hybridization conditions, the primer being sufficiently complementary to a
part of
the nucleic acid to be elongated or to a flanking region thereof and a primer
elongation by a polymerase is carned out in the presence of nucleotides
characterized in that a recombinant chimeric protein according to the
invention is
used as the polymerase and that a sliding clamp protein is preferably present
in the
reaction.
In the sense of the present invention "recombinant" is for example understood
to
mean that the chimeric protein is for example produced by genetic engineering
(see
"Gentechnologie, Rompp Basislexikon Chemie, Georg Thieme Verlag" 1998) or for
example when it is chemically synthesized.
Other embodiments arise from the subclaims.
The basis of the invention is the surprising discovery that starting with a
protein
having nucleic acid synthesis activity referred to herein in the following as
the basic
protein, it is possible to increase the processivity of this protein or more
exactly the
nucleic acid synthesis activity by interacting it with a factor such as a
sliding clamp
protein that increases the processivity whereby the basic protein as such
cannot
interact with this factor or if it can interact with this factor, it is not
associated with
an increase in processivity. This is achieved by providing the protein
carrying the
nucleic acid synthesis activity with a group of several amino acids, typically
in the
form of a consecutive amino acid sequence, which is referred to herein as the
interaction-mediating or interaction-effecting sequence, and only as a result
of
which is an interaction possible between the said protein and a factor
increasing the
processivity. This overcomes the aforementioned problem of the prior art i.e.
the
lack of compatibility of the desired individual components of a complex
carrying
nucleic acid synthesis activity.
CA 02379165 2002-02-06
' - 19-
Without wanting to be limited thereto, this results in at least the following
three
approaches relating to the formation of such a chimeric protein carrying a
nucleic
acid synthesis activity.
The basic protein which forms the basis for the chimeric protein can be
present in
two basic forms. In the first form the basic protein only contains the nucleic
acid
synthesis activity but no sequence (also referred to herein as domain) which,
in
particular in vivo, can or could mediate an interaction between the nucleic
acid
synthesis activity and a processivity factor and especially not in such a
manner that
the interaction would lead to an increase in the processivity.
In a second form the basic protein contains a nucleic acid synthesis activity
and
additionally a sequence which, in particular in vivo, can or could promote an
interaction between the nucleic acid synthesis activity and a processivity
factor and
especially in such a manner that the interaction increases the processivity.
The chimeric protein according to the invention can on the basis of the
various
forms of the basic protein be present in different fundamental embodiments.
(i) a first embodiment of the chimeric protein provides that the first form of
the basic protein is used and that a sequence is attached thereto which
mediates an
interaction with a factor that increases the processivity of the nucleic acid
synthesis
activity. This attachment is typically carried out such that the interaction-
mediating
sequence adjoins the sequence of the nucleic acid synthesis activity and is
optionally
separated by a linker. Hence in this first embodiment of the chimeric protein
the
basic protein is supplemented by an interaction-mediating sequence.
(ii) In a second embodiment of the chimeric protein the second form of the
basic protein is used. In this case the inherent sequence of the basic protein
which
mediates an interaction with a factor that increases the processivity of the
nucleic
acid synthesis activity is replaced by another sequence mediating an
interaction with
a factor increasing the processivity of the nucleic acid synthesis activity.
This other
sequence can be derived from another gene of the same organism, from the same
gene of another organism or from another gene of another organism and is thus
in
CA 02379165 2002-02-06
-20-
every case of a different origin. Consequently such a construction enables the
nucleic acid synthesis activity of the basic protein to interact for the first
time or to
an increased extent with a factor that increases the processivity of the
nucleic acid
synthesis activity.
(iii) In a third embodiment of the chimeric protein the second form of the
basic protein is used. In this case the inherent sequence of the basic protein
which
mediates an interaction with a factor that increases the processivity of the
nucleic
acid synthesis activity is supplemented by another sequence mediating an
interaction with a factor increasing the processivity of the nucleic acid
synthesis
activity. This other sequence can be derived from another gene of the same
organism, from the same gene of another organism or from another gene of
another
organism and is thus in every case of a different origin. Consequently such a
construction enables the nucleic acid synthesis activity of the basic protein
to
interact for the first time or to an increased extent with a factor that
increases the
processivity of the nucleic acid synthesis activity.
For all embodiments of the chimeric protein mentioned above it can be said
that the
(amino acid) sequence of such a chimeric protein typically differs from the
sequence
of the basic protein. A further, but not obligatory, consequence can be that
the
complex that is formed from the chimeric protein having nucleic acid synthesis
activity and the factor increasing the processivity is different from the
complex of
basic protein and factor increasing the processivity.
All proteins, polymerases and elongation proteins mentioned in the description
are
suitable as basic proteins, to the disclosure of which reference is herewith
made.
The same applies to all other components in particular to those of the
replication
apparatus such as processivity factors, sliding clamp proteins and interaction-
mediating or interaction-effecting sequences. Finally the definitions given in
the
introduction also apply to this part of the disclosure.
Although the nucleic acid synthesis activity can be derived from many
organisms it
is preferred that it is for example derived from the organism Carboxythermus
hydrogenoformans (European Patent Application EP 0 834 569 Al) or one of the
CA 02379165 2002-02-06
-21 -
organisms such as e.g. Thermus aquaticus, Thermus caldophilus, Thermus
chliarophilus, Thermus filiformis, Thermus Jlavus, Thermus oshimai, Thermus
ruber, Thermus scotoductus, Thermus silvanus, Thermus species ZOS, Thermus
species sp. 17, Thermus thermusphilus, Therotoga maritima, Therotoga
neapolitana, Thermosipho africanus, Anaerocellum thermophilum, Bacillus
caldotenax or Bacillus stearothermophilus.
If a sequence is mentioned herein, this usually refers to an amino acid
sequence.
Nucleic acid sequences are usually referred to directly as nucleic acid
sequences.
The various nucleic acids coding for the recombinant chimeric proteins
according to
the invention can be easily determined by persons skilled in the field by
means of
the genetic code and subsequently synthesized. Likewise suitable vectors for
cloning
and expressing the recombinant chimeric proteins according to the invention
and
methods for their preferably recombinant production are also known to persons
skilled in the art (see for example Maniatis et al.; supra)
Methods for producing antibodies, including monoclonal antibodies, which are
directed against the recombinant chimeric proteins according to the invention
are
also known to persons skilled in the art.
The recombinant chimeric protein according to the invention can be used to
elongate nucleic acids e.g. for the polymerase chain reaction, DNA sequencing,
to
label nucleic acids and for other reactions which include the in vitro
synthesis of
nucleic acids.
Hence a further subject matter of the present invention is a method for
template-
dependent elongation in which the nucleic acid to be elongated or at least a
strand
thereof is provided with at least one primer under hybridization conditions,
the
primer being sufficiently complementary to a part of or to a flanking region
of the
nucleic acid to be elongated and a primer elongation is carried out by a
polymerase
in the presence of nucleotides in which a recombinant chimeric protein
according to
the invention is used as the polymerase and additionally a sliding clamp
protein is
present in the reaction or in the reaction mixture in a preferred embodiment.
CA 02379165 2002-02-06
-22-
Methods for the template-dependent elongation of nucleic acids in which the
elongation is initiated by a primer which has been hybridized to the template
nucleic
acid and has a free 3'-OH end available for the elongation are known to a
person
skilled in the art. In particular a polymerase chain reaction is carned out
for the
amplification. A double-stranded DNA sequence is usually used as the starting
material of which it is intended to amplify a certain target region. Two
primers are
used for this which are complementary to the regions flanking the target
sequence
on each of the partial strands of the DNA double strand. However, in order to
hybridize primers the DNA double strands are firstly denatured and in
particular
thermally melted. After the primer hybridization, an elongation is carried out
by
means of the polymerase, it is subsequently again denatured to separate the
newly
formed DNA strands from the template strands whereupon the nucleic acid
strands
formed in the first step are also available as a template together with the
original
template strands for a further elongation cycle, these are each again
hybridized with
primers and a new elongation takes place. This procedure is carried out in
cycles
with a thermal denaturation as an intermediate step.
The recombinant chimeric protein according to the invention can also be used
for
reverse transcription in which case either the protein according to the
invention
itself has reverse transcriptase activity or a suitable enzyme is additionally
added
which has reverse transcriptase activity irrespective of whether the
thermostable in
vitro complex has an inherent reverse transcriptase activity.
A recombinant chimeric protein according to the invention is also used for the
reverse transcription of RNA into DNA which is preferred according to the
invention in which case the nucleic acid synthesis activity of the protein has
an
inherent reverse transcriptase activity. This reverse transcriptase activity
can either
be the only polymerase activity or it can also be present together with an
existing 5'-
3' DNA polymerase activity. A preferred embodiment according to the invention
of
the recombinant chimeric protein contains the elongation protein derived from
the
organism Carboxydothermus hydrogenformans as disclosed in EP-A 0 834 569.
A further preferred use of the recombinant chimeric protein according to the
invention is to sequence nucleic acids starting with at least one primer which
is
CA 02379165 2002-02-06
-23-
sufficiently complementary to a part of the nucleic acid to be sequenced in
which
again a template-dependent elongation is carried out or, in the case of RNA
sequencing, a reverse transcription using deoxynucleotides and
dideoxynucleotides
is carned out according to the method of Sanger. Within the framework of this
preferred embodiment the respective derivatives described above are also
suitable as
deoxynucleotide and dideoxynucleotides. In particular it is preferable for the
method according to the invention for elongating nucleic acids that the
nucleic acids
that are formed to be labelled. For this purpose it is possible to use
labelled primers
and/or labelled deoxynucleotides or/and labelled dideoxynucleotides and/or
labelled
ribo-nucleotides or appropriate derivatives thereof as already described
above.
A further subject matter of the present invention is a method for labelling
nucleic
acids by inserting individual breaks in phosphodiester bonds of the nucleic
acid
chain and replacing a nucleotide at the breaks by a labelled nucleotide with
the aid
of a polymerase in which a thermostable in vitro complex according to the
invention
is used as the polymerase.
Such a method which is generally referred to as nick translation enables a
simple
labelling of nucleic acids. All aforementioned labelled ribonucleotides or
deoxyribo-nucleotides or derivatives thereof are suitable for this provided
they are
accepted by the polymerase as a substrate.
The invention is further elucidated by the following examples and figures from
which additional advantages and embodiments ensue.
Fig. 1 shows four sequence alignments of different elongation protein
domains;
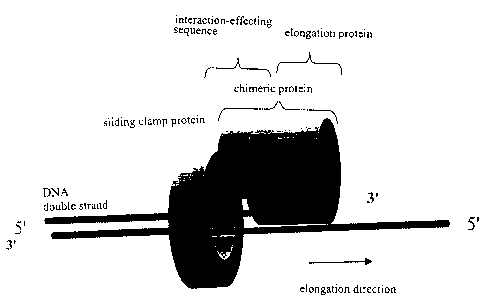
Fig.2 shows a schematic representation of the recombinant chimeric
protein according to the invention;
Fig. 3 shows four consensus sequences of sliding clamp proteins;
Fig. 4 shows a table of the results of a yeast two-hybrid system;
Fig. 5, SA-C show alignments of several conserved regions of the sequence that
is
responsible for the interaction;
CA 02379165 2002-02-06
- -24-
Fig. 6 A shows the entire sequence of one embodiment of a recombinant
chimeric protein according to the invention;
Fig. 6 B shows a diagram of the basic construction of a chimeric protein
according to the invention;
Fig. 7 shows the result of a polymerase chain reaction that was carried out
using a chimeric protein according to the invention;
Fig. 8 A shows all fragments of Afn497 that can interact with the sliding
clamp protein of Archaeoglobus fulgidus (Aft7335);
Fig. 8 B shows an alignment of C-terminal sequences of various genes from
Archaeglobus fulgidus;
Fig. 9 shows the result of a yeast two-hybrid experiment which shows the
interaction between an elongation protein and a sliding clamp protein
and
Fi. 10 shows the result of the amplification of genomic DNA using a
recombinant chimeric protein according to the invention.
Example 1: Use of the yeast two-hybrid system to determine the amino acids
that are important for the interaction between replication factors and sliding
clamp protein
The yeast two-hybrid system (Fields S., Song O., Nature 1989 Jul 20; 340
(6230):245-6) is used to determine the binding region of sliding clamp
proteins
(referred to herein in the following as SCP) and replication factors (referred
to
herein in the following as RF). The genes for SCPs and RFs are expressed in
the
vectors pGBT9 and pGAD424 (CLONTECH Laboratories, Inc.) to form fusion
proteins having the DNA binding domains or the activation domains of GAL4:
DNA is purified by known methods from the organism Archaeoglobus fulgidus
(DSM No. 4304). The microorganisms were cultured by the DSM ("Deutsche
Sammlung fur Mikroorganismen"). Then the open reading frame of the genes are
amplified by means of PCR from genomic DNA of Archaeoglobus fulgidus. The
DNA obtained in this manner is cloned into the vectors pGBT9 and pGAD424.
Other vectors that are used in the two-hybrid system are also suitable for the
method
described in the following (these for example include pAD-GAL4-2.1, pBD-GAL4,
pBD-GAL4 Cam, pCMV-AD, pCMV-BD, pMyr, pSos, pACT2, pAS2-1, pHISi,
CA 02379165 2002-02-06
-25-
pLexA, pM, pHISi-1, pB42AD, pVPl6, pGADlO, pGBKT7, pLacZi, p8op-lacZ,
pGAD GH, pGilda, pAD GL, pGADT7, pGBDU, pDBLeu, pPC86, pDBTrp, which
are obtainable from CLONTECH Laboratories, Inc.). In addition modified clones
are cloned into the same vectors. The modified clones are deletion mutations
and
mutations affecting individual amino acids or several amino acids of the SCP
and/or
RF. Afterwards the ability of the modified clones to interact with one another
in the
two-hybrid system is measured. This enables domains or even individual amino
acid
residues to be determined that are important or essential for the interaction.
There are a number of methods for introducing deletions into a gene. One
method
for generating deletions is to prepare DNA fragments which contain the genes
for
SCP or RF. These fragments are obtained from a suitable vector either by PCR
or by
restriction digestion. In addition genomic DNA is used from the organism from
which the two proteins are derived. These various DNA fragments and the
genomic
DNA are reduced to small pieces by ultrasonic treatment and fragments which
have
lengths between the total length of the genes and about 100 bases are purified
by
preparative agarose gel electrophoresis. The ends of the fragments are filled
in by
treatment with a suitable enzyme (Klenow, Pwo-polymerase or others) or
digested
such that both strands have the same length and are thus blunt. These
fragments are
now incorporated by ligation into a pGAD424 and pGBT9 vector cleaved with SmaI
(or otherwise linearized and made blunt) or into other suitable vectors. This
results
in banks for numerous subfragments of the genes for SCP and RF which are each
in
both vectors of the two-hybrid system. The DNA of these banks is replicated
and
purified after transformation in Escherichia coli. In the next step the two
banks are
transformed into a suitable haploid yeast strain for the two-hybrid system
such that
the fusions of SCP or RF with the GAL4 DNA binding domains in one strain are
present as a pairing type which is different from the fusion of RF or SCP and
the
GAL4 activation domain. Diploid cells in which the plasmids from the two banks
are present in the same cell are now generated by pairing the two strains.
Suitable
strains are PJ69-4 (James P, Halladay J, Craig EA, Genetics 1996 Dec; 144(4):
1425-36) or any other strain having a suitable genotype for the two-hybrid
system.
Cells in which the reporter genes are activated are isolated and the plasmid
DNA is
isolated therefrom by preparation or the inserts are specifically amplified by
PCR.
The sequences of the fusion fragments are determined by DNA sequence analysis.
The binding region (or the binding regions) is (are) determined by determining
those
CA 02379165 2002-02-06
- -26-
regions in all clones (or which are always found in certain groups of clones)
in
which an interaction of the two fusion proteins comprising RF and SCP occurs.
This
determination is carned out four times for each of the genes: once with the
bank in
the vector having the activation domain (preferentially: pGAD424) and once
with
the bank in the vector having the DNA binding domain (preferentially: pGTB9)
and
once with the total length clone of the binding partner and once with the gene
bank
of the fragments.
The described experiments enable minimum binding regions to be defined that
can
be utilized to construct recombinant chimeric proteins having an affinity for
SCP.
Amino acid mutations are introduced for a more detailed characterization of
the
binding region and their effect on the binding characteristics is tested. This
further
narrows down the region and determines the positions in the protein involved
in the
binding. In a parallel preparation the positions involved in the binding are
also
determined by introducing random mutations in the binding region and analysing
the effect on binding. In both cases it is determined whether the mutations
destabilize the protein and thus have an indirect effect or whether they have
no
influence on the stability of the protein and thus have a direct effect.
The experimental procedures that are necessary for this are described in
Maniatis et
al., (Molecular Cloning(2"'~ edition, 3 volume set): A laboratory Manual, Cold
Spring Harbor Laboratory Press, N.Y. (1989)), in Ausubel et al (Current
Protocols
in Molecular Biology, John Wiley and Sons (1988)), in Abelson, J.N., and
Simon,
M.I. (editors) 1991 (Methods in Enzymology, volume 194, Guide to Yeast
Genetics
and Molecular Biology, Academic press) or in Adams, A. Gottschling, D.E.
Kaiser,
CA, and Steams, T., 1997 (Methods in Yeast Genetics, Cold Spring Harbor
Laboratory Press). The handling of the two-hybrid system was carried out
according
to the instructions of the Clontech Company (yeast protocols handbook, PT3024-
1).
Example 2: Further methods for mapping the binding region of SCP and RF
SCP and RF were obtained as recombinant proteins. In each case one of the two
proteins was immobilized on a support and then the other protein was added in
an
unbound form in order to allow binding to the immobilized partner. Free i.e.
non-
_ _27_
bound material was washed out and the bound protein was eluted for example by
denaturation. The amount of bound protein is a measure for the binding (e.g.
used
by Anderson D, Koch CA, Grey L, Ellis C. Moran MF, Pawson T, Science 1990
Nov. 16;250(4983): 979-82).
The analysis of deletion mutants of the proteins and mutants having a modified
amino acid sequence allowed the mapping of the binding region. Deletions can
be
generated by proteolytic digestion or by the recombinant expression of deleted
genes. In the same manner the binding region can be determined by co-immuno
precipitation from cell extracts which contain the deleted forms of the
proteins.
Competition of binding by peptides can give information on the sequence of the
binding region.
A further approach for mapping the binding region is to prepare peptides and
to
measure the binding of the peptides to the other protein in each case. The
sequence
of the peptides can be random (Songyang Z, Prog Biophys Mol Biol. 1999; 71 (3-
4):359-72) or be based on the amino acid sequence of the protein whose binding
region is to be identified (peptide scans, Brix J, Rudiger S, Bukau B,
Schneider-
Mergener J, Pfanner N, J. Biol. Chem. 1999 Jun 4;274(23):16522-30). Such
peptides can be prepared by chemical synthesis or by other methods such as
phage
display and be used for binding. An example is shown in fig. 3. In this case
alignments of various consensus sequences were determined which are suitable
for
the preparation of such peptides.
A further method is to prepare antibodies against epitopes of the protein
whose
binding region is to be identified. If the antibody inhibits the binding, the
epitope of
the antibody overlaps with the binding region (e.g. Fumagalli S, Totty NF,
Hsuan JJ,
Courtneidge SA, Nature 1994 Apr 28;368(6474):871-4).
Finally another method for determining or mapping the binding region
(interaction-
mediating sequence) is to elucidate the fine structure of the complex using
the
methods of X-ray structural analysis, nuclear magnetic resonance or electron
microscopy.
CA 02379165 2002-02-06
- - 28 -
Example 3: Study of the interaction of proteins from Archaeoglobus fulgidus
using the yeast two-hybrid system (Y2H)
The coding regions of genes from Archaeoglobus fulgidus whose gene products
can
be used in vitro as an interaction-effecting sequence and/or nucleic acid
synthesis
activity were amplified by means of PCR, cloned into the vectors pGBT9
(vertical
columns of fig. 9) and pGAD424 (horizontal rows of fig. 9) and expressed as
hybrid
proteins by gap repair in yeast PJ69-4a (for pGAD424) and PJ69-4alpha (for
pGBT9). A positive control was also amplified by PCR, cloned into the vector
pGBT9 and pGAD424 (see also horizontal rows of fig. 9) and expressed by gap
repair in the yeast strain PJ69-4a (for pGAD424) and PJ69-4alpha (for pGBT9)
as
hybrid proteins. Diploid cells containing the two vectors were generated by
pairing.
The expression of three independent reporters (HIS3, ADE2 and MEL 1 ) was
measured. The expression of the HIS3 and ADE2 gene leads to a histidine or
adenine prototrophy of the cells. Transcription of the MEL 1 gene leads to the
production of the beta-galactosidase enzyme the activity of which can for
example
be detected by a colour reaction. In the experiment shown in fig. 9 the cells
that
grow in a histidine- and adenine-deficient medium are those which carry both
vectors and in which additionally the expression products of these two vectors
bind
to one another. The interaction of the fusion proteins results in a
reconstitution of a
functional transcription factor that initiates the transcription of the
reporter genes.
The production of the proteins coded on the reporter genes abolishes the
histidine
and adenine auxotrophy. This is due to the fact that the binding of the
expression
products initiates a transcription which makes it possible for the cells to
grow.
All positive clones in this experiment were also positive with respect to the
expression of the MEL 1 gene. The handling of the two-hybrid system is carried
out
according to the instructions of the Clontech Company (yeast protocol
handbook,
PT3024-1 ).
Example 4: Determination of the interaction-effecting sequence of Af0497
In order to determine the interaction-effecting sequence on the protein AfU497
(an
elongation protein) which results in the interaction with the sliding clamp
protein,
CA 02379165 2002-02-06
~- -29-
the DNA of the gene for AfZ7497 was firstly obtained by restriction of a
suitable
vector which contained an elongation protein which contained an interaction-
effecting sequence. This DNA was then fragmented by ultrasound and ligated
into
the two vectors pGAD424 and pGBDU. This resulted in banks of various fragments
of the gene in the two vectors. Subsequently the yeast two-hybrid system was
used
to determine which of these fragments can result in an interaction with
AfU335. For
this purpose both banks were transformed in suitable yeast strains (pGAD424:
PJ69-4a, PGBDU: PJ69-4alpha). Diploid cells which contain the banks (AfU497)
as
well as the sliding clamp protein in a suitable vector for use in the two-
hybrid
system were obtained by pairing the yeast strains. All clones that activated
the HIS2
reporter (see above) were isolated on plates without histidine and the inserts
were
selectively amplified by PCR of the yeast colony. The sequence of the inserts
was
determined and their position on the Afl7497 gene was determined. The result
is
shown diagrammatically in figure 8A: All found clones with the exception of
one
clone comprised the carboxyterminal end of the elongation protein which
contains
one or several interaction-effecting sequences. The smallest clone was
composed of
S 1 amino acids.
In order to determine whether the interaction-effecting sequence of the
homologous
protein of another organism is also present at the carboxyterminal end of the
protein, the experiment was repeated using the polymerase protein and the
sliding
clamp protein from Pyrococcus horikoshi. For this purpose banks of the large
subunit of the polymerase from P. horikosii were prepared and tested for
interaction
with the sliding clamp protein from P. horikosii by the method described
above.
Again interacting fragments contained the carboxyterminal end of the
elongation
protein.
In order to check whether the interaction between the carboxyterminal end of
the
protein AfU497 and the sliding clamp protein is specific, it was tested
whether the
carboxyterminal protein fragment of AfiJ497 can also interact with other
proteins
from Archaeglobus fulgidus and other organisms. In no case was it possible to
measure an interaction which indicates that the binding is very specific for
the
sliding clamp protein.
CA 02379165 2002-02-06
CA 02379165 2002-02-06
-30-
Thus the polymerase Taq does not bind to the SCP (PCNA from Archaeoglobus
fulgidus) and no interaction whatsoever was measured in the yeast two-hybrid
system. In order to test whether the carboxyterminal end of the protein AfU497
can
result in an interaction with another protein to which it is fused, a fusion
protein
consisting of Taq and the S 1 carboxytenninal amino acids of AfU497 was
prepared.
This protein was tested in the yeast two-hybrid system for interaction: As
shown in
fig. 4 the grafting of the interaction-effecting sequence of AfU497 to Taq
results in a
specific interaction of Tag with the sliding clamp protein Af1~335. The
results of the
corresponding Y2H experiments are shown in fig. 4.
This shows that the property of this fragment (the carboxyterminal end of the
elongation protein Afl7497) to result in an interaction with the sliding clamp
protein
AfU335 can be grafted onto or transferred to another protein and in particular
to
another polymerase in order to result there in an interaction with the sliding
clamp
protein which is actually specific for Af0497.
An interaction with PCNA was measured with six proteins (see also fig. 5 and
fig.
5B) from A. fulgidus. This shows that all these proteins contain similar
interaction-
effecting sequences that mediate the interaction with PCNA. Thus for example
the
polymerase delta large subunit (Af 0497, TREMBL number: 029753), the
polymerase delta small subunit (Af 1790, TREMBL number: 028484), DP2 (Af
1722, TREMBL number: 028552), RPA2 (replication factor A), RFC2 (replication
factor C) and PCNA (Af 0335, TREMBL number: 029912).
Such interaction-effecting sequences are contained in the last SO amino acids
of the
protein Afl7497. An examination of the proteins that interact with PCNA showed
that all contain a motif that is located just before the carboxyterminal end
of the
amino acid sequence. Fig. 8B shows a list of the related sequences in which
the
conserved region is marked with a black bar.
This motif is also conserved in other organisms and genes. Fig. 5 shows the
result of
the search for such interaction-effecting sequences as well as the consensus
sequences generated therefrom.
-31 -
Example 5: Increasing the efficiency of a PCR by using PCNA as the SCP and
a chimeric elongation protein according to the invention
This example shows the influence of PCNA on the efficiency of a PCR reaction.
In
this PCR reaction a 463 by fragment was amplified from plasmid DNA. The Taq
fusion protein was used as the polymerase as also shown in fig. 6A. The
reaction
conditions for the PCR were as follows: 0.4 mM of each dNTP (pH 8.3) and 20
pmol of each primer in one reaction. A first primer (SEQ ID NO.: 13) having
the
sequence 5'-AGGGCGTGGTGCGGAGGGCGGT-3' and a second primer (SEQ 1D
NO.: 14) having the sequence 5'-TCGAGCGGCCGCCCGGGCAGGT-3' were
used.
It turned out that the fusion of a 50 amino acid domain to the C-terminus of
the
native Taq DNA polymerase had no adverse effect on the polymerase properties
since the Taq fusion protein per se already yields a product in a PCR
reaction. This
domain can hence be used to mediate interaction between the sliding clamp
protein
PCNA from Archaeoglobus fulgidus and the Taq fusion protein and increase the
efficiency of the PCR reaction due to its property as a processivity factor
which is
reflected in a considerably higher yield of PCR product. The result is also
shown in
fig. 7.
Example 6: Yeast two-hybrid system for detecting the interaction between the
elongation protein Af 0497 of Archaeoglobus fulgidus and the sliding clamp
protein Af 0335 of Archaeoglobus fulgidus
Fig. 9 shows the results of a Y2H experiment in which row A contains cells
which
carry the empty pGAD424 vector (Clontech, Palo Alto, USA) such that a
transcription activation domain is expressed, row B contains cells that carry
the
pGAD424 vector from which the Sacharomyces cerevesiae gene CDC48 is
expressed as a fusion protein with the transcription activation domain; row C
contains cells which carry the pGAD424 vector from which the sliding clamp
gene
from Archaeoglobus fulgidus is expressed as a fusion protein with the
transcription
activation domain; row D contains no cells and raw E contains cells which
carry the
CA 02379165 2002-02-06
-32-
pGAD424 vector from which the elongation protein gene from Archaeoglobus
fulgidus is expressed as a fusion protein with the transcription activation
domain.
Column 1 contains cells which carry the empty pGBT9 vector (Clontech, Palo
Alto,
USA), column 2 contains cells which carry the pGBT9 vector from which the
Saccharomyces cerevisiae gene UFD3 is expressed as a fusion protein with the
DNA binding domain; column 3 contains cells which carry the pGBT9 vector from
which the sliding clamp protein from Archaeoglobus fulgidus is expressed as a
fusion protein with the DNA binding domain; column 4 contains cells which
carry
the pGBT9 vector from which the coupling protein from Archaeoglobus fulgidus
is
expressed as a fusion protein with the DNA binding domain and column 5
contains
cells which carry the pGBT9 vector from which the elongation protein from
Archaeoglobus fulgidus is expressed as a fusion protein with the DNA binding
domain.
Example 7: Polymerase chain reaction of genomic DNA using a chimeric
elongation protein
Example 7 shows the influence of the sliding clamp protein on the efficiency
of a PCR
reaction on long DNA fragments using the recombinant chimeric protein
according to
the invention. In this PCR reaction a 4954 by fragment was amplified from
human
genomic DNA. The recombinant chimeric protein according to the invention was
used
as the polymerase. The conditions correspond to the standard conditions of a
PCR
reaction: pH 8.3, 0.4 mM of each pNTP and 20 pmol of each primer in one
reaction. A
first primer (SEQ ID NO.: 15): S-AGGAACAACATATGACGCACTCT-3) and a
second primer (SEQ ID NO.: 16): (5'-TAGGTGGCCTGCAGTAATGTTAG-3') were
used.
It turned out that only the recombinant chimeric protein according to the
invention
having the sequence shown in fig. 6A allowed the generation of an unequivocal
and
specific PCR product under stimulation by the sliding clamp protein PCNA
whereas
the same reaction mixture containing native Taq DNA polymerase yielded no
defined product. This can be explained by the interaction of the sliding clamp
protein with the recombinant chimeric protein according to the invention
leading to
CA 02379165 2002-02-06
. _ _33_
less dissociation of the recombinant chimeric protein according to the
invention
from the target DNA.
The figures which have been partially already referred to in the examples are
described in more detail in the following.
All sequence alignments that are disclosed in the following figures were
determined
using the BLAST algorithm according to Altschul, S.F., Gish, W. Miller, W.,
Myers, E.W., and Lipman, D.J., J. Mol. Biol. 215, 403-410 (1990).
Fig. 1:
Fig. 1 shows sequence alignments of a total of four different elongation
protein
domains from various organisms i.e. for the elongation protein 1 from humans,
Archaeglobus fulgidus, Methanococcus thermoautotrophicusm, PHBT (Pyrococcus
horikoshii) and Methanococcus janashii. In the figures there is often a
consensus
sequence directly under the aligilment in which the variable positions are
indicated
by a single symbol. The amino acids in square brackets represent those amino
acids
expressed in the single letter code which can be present at the special
position.
The amino acids are named according to the standard IUPAC single letter
nomenclature and listed according to the prosite pattern description standard.
The
following amino acids are often grouped together:
G, A, V, L, I, M, P, F or W (amino acids with non-polar side chains), also
often written as "$" directly under the sequence
S, T, N, Q, Y or C (amino acids with uncharged polar side chains),
K, R, H, D or E (amino acids with charged and polar side chains) also often
written as "&" directly under the sequence
in addition X in the sequences or the sequence protocols denotes an arbitrary
amino acid or insertion or deletion.
Fig. I shows four different consensus sequences for different regions of
elongation
proteins whereby for example the elongation protein 3 can be frequently found
in
CA 02379165 2002-02-06
CA 02379165 2002-02-06
' ' -34-
eubacteria, the elongation protein 4 also includes the Taq polymerase and can
often
be found in Pol I type polymerases. Hence the elongation protein 3 exhibits an
alignment with a conserved region of the elongation protein from eubacteria
and the
consensus sequences derived therefrom. The following genes are shown:
DP3A ECOLI: DNA Pol III, alpha subunit, Escherichia coli, BB0579: DNA Pol III,
alpha subunit, Borrelia burgdorferi, DP3A_HELPY: DNA Pol III, alpha subunit,
Helicobacter pylori AASO: Aquifex aeolicus, section 50 and DP3A SALTY: DNA
Pol III, alpha subunit, Salmonella typhimurium).
Fig. 2:
Fig. 2 shows a diagram of a possible form of the recombinant chimeric protein
according to the invention in which the sliding clamp, which is the factor
increasing
the synthesis activity of the polymerase, binds to the elongation protein
carrying the
nucleic acid synthesis activity via the domain containing the interaction-
effecting
sequence.
Fig. 3:
Fig. 3 shows four sliding clamp consensus sequences.
The following genes are shown for the sliding clamp re ion l: human PCNA as
well as orthologues thereto from Archaeoglobus fulgidus, from Methanococcus
janashii, from Pyrococcus horikoschii and from Methanococcus
thermoautothrophicus.
Re '~ shows an alignment of a second conserved region of the sliding clamp
from eukaryotes and Archae and the consensus sequences derived therefrom. The
alignment was established using a second region of the sliding clamps that
were
already used above for region 1.
Re ig on 3 shows an alignment of a conserved region of the sliding clamp from
eubacteria and the consensus sequences derived therefrom. The following genes
are
- 35 -
shown: AAPOL3B, DP3B ECOLI, S.TYPHIM, DP3B PROMI, DP3B PSEPU
and DP3B STRCO (AAPOL3B: Aquifex Aeolicus section 93: DP3B ECOLI:
DNA Pol III, beta chain, Escherichia coli, S.TYPHIM: DNA Pol III, beta chain,
Salmonella typhimurium, P3B PROMI: DNA Pol III, beta chain, Proteus mirabilis
DP3B PSEPU: DNA Pol III, beta chain, Pseudomonas putida DP3B STRCO:
DNA Pol III, beta chain, Streptomyces coelicolor).
Re ion 4 shows an alignment of a second conserved region of the sliding clamp
from eubacteria (see organisms in region 3) and the consensus sequences
derived
therefrom.
Fig. 4.
Fig. 4 shows the interaction of chimeric proteins in the yeast two-hybrid
system.
The result shown in fig. 4 proves that the property that promotes an
interaction with
the sliding clamp protein Af1~335 can be transferred to another protein and in
particular to another polymerase where it results in an interaction with the
sliding
clamp protein. It is shown that the SCP binds itself and that the SCP binds a
Taq
fusion protein which contains an interaction-effecting sequence from
Archaeoglobus fulgidus.
Fig. 5:
Fig. 5 shows three alignments of conserved regions of an interaction-effecting
sequence from various organisms and for various genes:
Sequence 1 (polymerase gene, Af~497 homologue): Archaeoglobus fulgidus,
Pyrodictium occultum, Aeropyrum pernix, Pyrococcus glycovorans, Pyrococcus
furiosus, Thermococcus gorgonarius, Pyrococcus abyssi, Pyrococcus horikoshii,
Thermococcus litoralis, Thermococcus fumicolans and Methanococcus jannaschii.
CA 02379165 2002-02-06
. . -36-
Sequence 2 (polymerase gene, Af1722 homologue), Archaeoglobus fulgidus,
Methanococcus jannaschii, Pyrococcus furiosus, Methanobacterium thermoauto-
trophicum, Pyrococcus horikoshii and Pyrococcus abyssi and
Sequence 3 (Af, 1347 homologue), Archaeoglobus fulgidus, Pyrococcus abyssi,
Pyrococcus horikoshii, Methanobacterium thermoautotrophicum and Aeropyrum
pernix.
The interaction-effecting sequences can be introduced into a chimeric protein
according to the invention in order to bind a SCP.
Fig. 5B:
Fig. 5B shows two alignments of conserved regions of an interaction-promoting
sequence from various organisms and for various genes:
Sequence 4 (RNAse gene, Af 0621 homologue): Archaeoglobus fulgidus,
Pyrococcus abyssi, Pyrococcus horikoshii and Arabidopsis thaliana and
Sequence 5 (polymerase gene): Archaeoglobus fulgidus, Pyrococcus abyssi, Metha-
nococcus thermoautotrophicus, Methanococcus janashii, Mus musculus and Homo
sapiens.
The interaction-effecting sequences can be introduced into a chimeric protein
according to the invention in order to bind a SCP.
Fig. SC:
Fig. SC shows a list of characters that are used to label one or several
different
amino acids in which the amino acid or the property of the group is listed
under
"class" and the symbol used is listed under "key" and the amino acids in the
group
are listed under "residue".
CA 02379165 2002-02-06
_37_
Fig. 6A:
Fig. 6A shows the total sequence of a preferred embodiment of the recombinant
chimeric protein according to the invention. In this case the elongation
protein and
thus the nucleic acid synthesis activity was derived from Thermus aquaticus
and the'
interaction-effecting sequence was derived from Archaeoglobus fulgidus.
Fig. 6B:
Fig. 6B shows a graphical overview of the construction of a preferred
embodiment
of the recombinant chimeric protein according to the invention. In this case
the
elongation protein is from Thermus aquaticus and the interaction-effecting
sequence
is from Archaeoglobus fulgidus.
Fig. 7:
Fig. 7 shows the use of a recombinant chimeric protein according to the
invention in
PCR; its sequence is shown in fig. 6A. In all lanes of this 1 % agarose gel
one band
can be seen having a size of 463 by for the amplificate of a PCR reaction
initiated
with plasmid DNA (pCR 2.1 vector including a 463 by fragment; Invitrogen, 9704
CH Groningen; Netherlands) which was carried out under the following
conditions:
(95°C 5'denaturation; 30 x {95°C 30" denaturation; SS°C
30" hybridization; 72°C
40"elongation}; 72°C 7 additional elongations). 20 p1 of a 50 p.1
reaction mixture
was applied to the gel. Lane 7 shows a size standard.
In lanes 2 (0.3 pg PCNA) and 3 (2.4 pg PCNA) it can be seen that the intensity
of
the bands increase compared to lane 1 (PCR reaction without PCNA) after adding
increasing amounts of the sliding clamp protein (PCNA) as a stimulating agent
at a
constant Mg2+ ion concentration (2.5 mM). An additional increase in the
intensity of
the bands and hence an increased yield in the PCR reaction can be achieved at
a
higher Mgz+ ion concentration (3.5 mM) cf lane 4 (without PCNA) and lanes 5
(0.3 ug PCNA) and 6 (2.4 pg PCNA) with increasing amounts of PCNA.
CA 02379165 2002-02-06
i . _38_
Fig. 8A:
Diagram of all fragments of AfD497 (see above) that interact with the sliding
clamp
protein from Archaeoglobus fulgidus (Afn335). The arrows labelled "B"
represent
clones that were found as a fusion with the DNA binding domain, the arrows
labelled "A" represent clones that were found as a fusion with the activation
domain.
Fig. 8B:
Fig. 8B shows an alignment of the C-terminal sequences of the following genes
Af
0497-grUE, of 1195 Rfc, Af 0264 Rad2-Fenl, Af 0621 RNAseH, Acheaoglobus
fulgidus.
Fig. 9:
Fig. 9 shows the result of a yeast two-hybrid test and proves a binding
between the
yeast proteins ufd3 and cdc48 (2B; positive control), between the elongation
protein
and the sliding clamp protein (3E; and in the reverse orientation SC), between
the
sliding clamp protein and the sliding clamp protein (3C) and between the
coupling
protein and the sliding clamp protein (4C). The fusion proteins are expressed
from
the vectors pGBT9 (vector) in column 1, pGBT9::ufd3 (positive control) in 2,
pGBT9::PCNA in 3, pGBT9::klUE in 4, pGBT9::grUE in 5, pGAD424 (vector) in
row A, pGAD424::cdc48 in B (positive control), pGAD424::PCNA in C, empty in
D and pGAD::grUE in E.
Interactions of proteins from Archaeoglobus fulgidus were demonstrated with
the
Y2H system. The coding regions of genes from Archaeoglobus fulgidus whose gene
products can be used in vitro were amplified in PCR, cloned into the vectors
pGBT9
(vertical columns of fig. 27) and pGAD424 (horizontal rows of fig. 9) and
expressed as hybrid proteins by gap repair in yeast PJ69-4a (for pGAD424) and
PJ69-4alpha (for pGBT9). A positive control was also amplified by PCR, cloned
into the vectors pGBT9 (see also vertical columns of fig. 9) and pGAD424 (see
also
CA 02379165 2002-02-06
-39-
horizontal rows of fig. 9) and expressed as hybrid proteins by gap repair in
the yeast
strain PJ69-4a (for pGAD424) and PJ69-4alpha (for pGBT9). Diploid cells
containing the two vectors were generated by pairing according to the grid
shown in
fig. 9. The expression of three independent reporters (HIS3, ADE2 and MELD was
measured. Fig. 9 shows the growth on medium without histidine and adenine. The
cells which grow in this experiment are those that carry both vectors and
where
additionally the expression products of these two vectors bind to one another.
The
binding of the expression products initiates transcription which leads to the
abolition of the histidine and adenine auxotrophy.
All positive clones in this experiment were also positive with respect to the
expression of the MEL 1 gene. The handling of the two-hybrid system was carned
out according to the instructions of the Clontech Company (yeast protocol
handbook, TP3024-1).
Fig. 10:
In all lanes of this 1 % agarose gel with the exception of lane 5 which shows
a size
standard, 20 p1 of a SO ~I preparation of the amplificate of a PCR reaction
has been
applied to the gel where the PCR was carried out on human genomic DNA having a
size of 4954 base pairs (Roche Diagnostics, D-68305 Mannheim) under the
following conditions: 95°C 5'denaturation; 35 x {95°C 30"
denaturation; 47°C 30"
hybridization; 72°C 12'elongation}; 72°C 10 additional
elongations. In lane 3
(2.4 ~g PCNA) it can be seen that after adding increasing amounts of the
sliding
clamp protein (PCNA from Archaeoglobus fulgidus) to the recombinant chimeric
protein complexes according to the invention as a stimulating agent one band
is
obtained at a constant Mgz+ ion concentration (6 mM) compared to lane 1 (PCR
reaction without PCNA) and lane 2 (0.3 pg PCNA), whereas it was not possible
to
generate a defined product in the PCR reaction at the same MgZ+ ion
concentration
with Taq DNA polymerase (lane 4).
The features of the invention disclosed in the present description and in the
claims
can be important individually as well as in any combination for the
realization of the
invention in its various embodiments.
CA 02379165 2002-02-06