Note: Descriptions are shown in the official language in which they were submitted.
WO 01/07638 CA 02380238 2002-O1-25 PCT/US00/40495
-1-
HOMOGENEOUS FLUORESCENCE METHOD FOR ASSAYING
STRUCTURAL MODIFICATIONS OF BIOMOLECULES
PRIORITY CLAIM
This application claims the benefit of the filing date of United States
Provisional
Patent Application Serial Number 60/145,755, filed July 27, 1999, for
"HOMOGENEOUS FLUORESCENCE METHOD FOR ASSAYING STRUCTURAL
MODIFICATIONS OF BIOMOLECULES".
TECHNICAL FIELD
The invention relates to methods useful for the homogenous assay of covalent
modifications to biomolecules which utilize double-labeled biomolecular
substrates.
Also described are methods of preparing and characterizing the double-labeled
biomolecular substrates and methods of using the inventive assay for high-
throughput
screening, for diagnostic and therapeutic applications, and for discovering
substrates of
novel enzymes.
BACKGROUND
The healthy development and function of eukaryotic organisms depends upon
the proper regulation of structural modifications of various biomolecules (the
phrases
"covalent modification" and "structural modification" are used interchangeably
herein).
It is believed that virtually all intracellular biochemical processes in
eukaryotes are
regulated in some fashion by the covalent modification of biomolecules, such
as
proteins or peptides. However, where an intracellular process responsible for
the
covalent modification of a particular type of biomolecule is somehow
dysfunctional,
several disease states can result.
For example, protein kinases represent one of the largest superfamilies of
enzymes in eukaryotic organisms, with an estimated 1-3% of the human genome
coding
for various protein kinases. Protein kinases catalyze the transfer of
phosphate from
ATP to specific amino acids in proteins, and phosphorylation of proteins is
known to be
the most widespread mechanism for reversible covalent modification of protein
structure and function. The dysfunctional regulation of protein
phosphorylation is
WO 01/07638 CA 02380238 2002-O1-25 PCT/USOO140495
-2-
believed to result in several diseases, such as, for example, diabetes,
cancer, and many
forms of heart disease.
As can be appreciated, the ability to assay the activity of the various
intracellular
processes responsible for the covalent modification of particular biomolecules
is
essential in order to gain an understanding of the potential roles such
processes play in
normal cells and various disease states. Assay techniques which detect and
quantify
various types of covalent modifications of particular biomolecules would also
facilitate
the development of diagnostic and therapeutic technologies relating to disease
states
resulting from dysfunctional modification processes. An assay technique
ideally suited
for these purposes would be sensitive and continuous, would allow both in
vitro and in
vivo assays, would be efficient and economical, and would enable high-density,
high-
throughput screening.
A variety of methods are currently used to assay the covalent modification of
biomolecules. Most of these methods, however, are relatively inefficient or
uneconomical in that they require the use of radioactive labels, mufti-
component assay
systems, and/or mufti-step procedures. Moreover, most existing methods of
assaying
the structural modification of biomolecules are discontinuous and, therefore,
necessitate
the sampling of the reaction at specific times in order to determine enzymatic
activity.
Unlike most known methods, U.S. Patents 5,776,720 to Jaffe et al. (July 7,
1998), 5,770,691 to Fields et al. (June 23, 1998), 5,763,181 to Han et al.
(June 9, 1998),
5,733,719 to Jaffe et al. (March 31, 1998), and 5,698,411 to Lucas et al.
(December 16,
1997) teach various methods for assaying enzyme-mediated cleavage of
biomolecules,
such as peptides or nucleic acids, which do not require radioactive labels and
which are
continuous and homogenous. However, the usefulness of even these methods is
limited
to the assay of enzyme-mediated cleavage reactions, and such reactions
constitute only
one subset of the many processes by which biomolecules are structurally
modified
within the cell.
Homogenous assay methods which detect the presence of antibodies, nucleic
acids, or protein kinase activity through the use of fluorogenic tracer
molecules are also
known. For example, PCT International Publication No. W0/03429 ("the WO/03429
publication") teaches homogenous assay techniques which utilize fluorogenic
tracer
molecules that exhibit a change in fluorescence upon association with their
target
WO X1/07638 CA 02380238 2002-O1-25 PCT/US00/40495
-3-
antibody or nucleic acid sequence. Though the WO/03429 publication provides no
solution to the challenge of developing methods or compositions for monitoring
or
detecting covalent biomolecular modifications, a protein kinase assay
technique making
use of the technology disclosed in the WO/03429 application was recently
reported in
an article authored by Geoghegan et al. ("Geoghegan et al.") (See, Geoghegan
et al.,
Bioconjugate Chemistry, 11:71-77 (2000)).
The assay technique disclosed in Geoghegan et al. is relatively complicated
and
expensive, however, and the technique unsuitable for in vivo application. The
assay
technique disclosed in Geoghegan et al. uses double-labeled tyrosine kinase
substrate
peptides and a polypeptide corresponding to an SH2 domain. The SH2 domain
binds
the double-labeled tyrosine kinase substrate peptides when those substrate
peptides are
phosphorylated, and binding of the SH2 domain to the phosphorylated double-
labeled
tyrosine kinase substrate peptides causes the two labels included in each of
the substrate
peptides to dissociate, resulting in a change in fluorescence. Thus, the assay
technique
disclosed in Geoghegan et al. requires the interaction of two expensive
components
(i.e., the interaction of double-labeled tyrosine kinase substrate peptides
with SH2
domain peptides), resulting in a technique that is relatively complex and
expensive.
Moreover, due to its use of a mufti-component system, the assay technique of
Geoghegan et al. is not suitable for performing in vivo assays.
Homogenous fluorescent protein kinase assay methods utilizing various Green
Fluorescent Protein ("GFP") molecules have also been recently reported. For
instance,
an article written by Nagai et al. teaches a method for assaying protein
kinase activity
using a Kinase-Inducible Domain construct containing two GFP groups (See,
Nagai et
al., Nature Biotechnology, 18:313-316 (2000)). The assay method taught by
Nagai et
al., however, depends upon phosphorylation-dependent changes in the
fluorescence
resonance energy transfer ("FRET") among the two GFP groups to detect kinase
activity, and because such changes in FRET are small, the assay technique of
Nagai et
al. does not provide a homogenous assay having a desired level of sensitivity.
In
addition, U.S. Patent 5,912,137 ("the '137 Patent") teaches a protein kinase
assay
utilizing modified GFP molecules as assay substrates. However, because the
assays
taught in the '137 patent can only be carried out using modified GFP
substrates, the
potential applications of such assays are limited.
WO 01/07638 cA 02380238 2002-0l-25 PCT/US00/40495
-4-
Because of the deficiencies existing in the known assay methods, it would be
an
improvement in the art to develop a homogenous assay method which not only
enables
the continuous, real-time assay of covalent biomolecular modifications, such
as
phosphorylation, sulfation, glycation, glycosylation, carboxylation,
myristoylation,
farnesylation, ubiquitination, and biotinylation, but which also provides
sensitive and
economical assays that are simple to carry out in vitro and in vivo and are
adaptable to a
wide variety of applications.
DISCLOSURE OF INVENTION
The present invention includes substrates and methods useful for the assay of
covalent modification of biomolecules. In contrast to assay methods already in
use, due
to the nature of the double-labeled molecular substrates described herein, the
assay
methods of the present invention are sensitive and homogeneous and do not
require the
use of radioisotopes. The assay methods herein disclosed are also relatively
simple and
economical, adaptable to a wide variety of applications, easily used in vitro
and in
living cells, and allow continuous, real-time monitoring of structural
modifications to
biomolecules. Moreover, the assay methods of the present invention are useful
for
detecting and quantifying a wide range of covalent biomolecular modifications
which
do not result in the cleavage of the biomolecule. Thus, the present invention
offers
significant advantages in terms of simplicity, efficiency, and scope when
compared to
presently used methods for detecting covalent biomolecular modifications.
The substrates of the present invention are double-labeled biomolecular
substrates (the phrases "double-labeled biomolecular substrate" and "double-
labeled
substrate" are used interchangeably herein). The double-labeled substrates of
the
present invention include a core molecular backbone covalently labeled at two
positions
with a first fluorescent dye and a second fluorescent dye or with a first
fluorescent dye
and a second non-fluorescent dye (for convenience, the term "dye" is used
herein to
describe a chromophoric or fluorophoric moiety). The core molecular backbone
of a
double-labeled substrate according to the present invention may include a
protein or
peptide sequence, a nucleotide sequence, a sugar, a lipid, a receptor, or a
biopolymer.
As used in the context of the present invention, the term "biopolymer"
includes any
molecule that is a covalent combination of amino acids, nucleic acids, sugars,
lipids, or
W~ 01/07638 CA 02380238 2002-O1-25 pCT/US00/40495
-5-
other small molecules of biological origin. The core molecular backbone may
also
include a substrate determinant specific for a particular process of covalent
biomolecular modification. Furthermore, each double-labeled substrate of the
present
invention is constructed and labeled so that when the double-labeled substrate
is in its
unmodified state, the first and second labeling dyes associate, or stack, to
form an
intramolecular dimer. When the first and second labeling dyes form an
intramolecular
dimer, the fluorescence of one or both of the dyes is quenched, thereby
quenching the
fluorescence of the double-labeled substrate. Upon catalytic or non-catalytic
covalent
modification of the double-labeled substrate, however, the first and second
labeling
dyes dissociate, and the fluorescence of the covalently modified double-
labeled
substrate changes markedly. Therefore, because only the modified double-
labeled
substrate exhibits a change in fluorescence, no need exists to separate the
modified
double-labeled substrate from the unmodified double-labeled substrate in order
to
accurately assay the extent to which an amount of double-labeled substrate has
been
covalently modified. As a result, the invention enables homogenous and
continuous
assay methods which are simple and economical, and which may be employed both
in
vitro and in living cells.
It should be noted that the quenching phenomenon underlying the substrates and
fluorescence assay methods of the present invention is not fluorescence
resonance
energy transfer ("FRET"). Unlike FRET, the quenching phenomenon underlying the
substrates and fluorescence assay methods of the present invention involve
ground state
interaction of two dyes. Advantageously, because the quenching phenomenon
described herein involves ground state interactions which result in changes in
the
absorbance spectra of the two dyes included in a double-labeled substrate
according to
the present invention, double-labeled substrates of the present invention may
be used
for homogenous absorbance-based assays detecting various types of structural
modifications of biomolecules through modification-dependent changes in the
absorbance spectra of the double-labeled substrates.
The assay methods of the present invention are versatile. Because the core
molecular backbone of double-labeled substrates of the present invention can
be
constructed to include substrate determinants for a wide range of
intracellular
processes, the assay methods of the present invention are applicable to a
broad range of
w0 01/07638 CA 02380238 2002-O1-25 PCT/US00/40495
-6-
covalent biomolecular modifications. For example, the core molecular backbone
of a
double-labeled substrate may include a protein kinase substrate determinant.
Such a
double-labeled substrate could then be used to assay the activity of a
particular protein
kinase or a particular class of protein kinases. However, this is but one
example.
Double-labeled substrates of the present invention could be constructed in
order to
assay numerous other modification reactions, such as sulfation, glycation,
glycosylation, carboxylation, myristoylation, farnesylation, ubiquitination,
and
biotinylation, by which biomolecules are structurally modified.
Various other methods are also included within the scope of the present
invention. For example, methods of producing the assays of the present
invention and
of producing and characterizing the double-labeled biomolecular substrates of
the
present invention are described herein. Also described herein are methods for
using the
inventive assay procedure in high-throughput screening, methods for monitoring
the
activity of the intracellular processes by which biomolecules are covalently
modified,
methods for diagnostic and therapeutic applications of the inventive
substrates and
assay procedures, and methods for discovering substrates for novel enzymes
involved in
the covalent modification of particular biomolecules.
BRIEF DESCRIPTION OF DRAWINGS
FIG. 1 provides a schematic illustration of a first embodiment of a double-
labeled biomolecular substrate of the present invention in an unmodified
state.
FIG. 2 is a schematic illustration of a first embodiment of a double-labeled
biomolecular substrate of the present invention in a structurally modified
state.
FIG. 3 schematically depicts the modification-dependent transition between
intramolecular dimer and intramolecular monomer states of a double-labeled
biomolecular substrate according to the present invention.
FIG. 4 is a schematic illustration of a second embodiment of a double-labeled
biomolecular substrate of the present invention in a structurally modified
state.
FIG. 5 illustrates the absorbance spectrum of double-labeled substrate
including
a fluorescein and a rhodamine label before and after phosphorylation by PKA.
FIG. 6 illustrates the fluorescent emission of the fluorescein label of the
double-
labeled substrate of FIG. 5 before and after phosphorylation by PKA.
WO 01/07638 CA 02380238 2002-O1-25 PCT/US00/40495
FIG. 7 illustrates the fluorescent emission of the rhodamine label of the
double-
labeled substrate of FIG. 5 before and after phosphorylation by PKA.
FIG. 8 illustrates an absorbance spectrum of double-labeled substrate
including
two rhodamine labels before and after phosphorylation by PKA.
FIG. 9 is a chart of the fluorescence the rhodamine labels of the double-
labeled
substrate of FIG. 8 before and after phosphorylation by PKA.
FIG. 10. schematically illustrates a double-labeled substrate including a
covalently attached enhancer, the double-labeled substrate transitioning
between an
unphosphorylated and a phosphorylated state upon phosphorylation by PKA.
BEST MODES FOR CARRYING OUT THE INVENTION
A first embodiment of a double-labeled biomolecular substrate according to the
present invention is schematically illustrated in its unmodified state in FIG.
1. The
double-labeled substrate 10 includes a core molecular backbone 20 containing a
substrate determinant 30 which facilitates the modification of the double-
labeled
substrate, a fluorescent dye 40, and a quenching dye 50. As is illustrated in
FIG. 1, the
double-labeled substrate 10 is constructed so that, in the substrate's
unmodified state,
the fluorescent dye 40 and the quenching dye 50 associate, or stack, to forth
an
intramolecular dye dimer 55. The formation of an intramolecular dye dimer 55
results
in the quenching of the fluorescent dye 40 by the quenching dye 50. Thus, in
its
unmodified state, the fluorescence of the double-labeled substrate 10 is
markedly
reduced.
FIG. 2 provides a schematic illustration of the double-labeled substrate 10 of
FIG. 1 after the double-labeled substrate 10 has been structurally modified
by, for
example, a protein kinase. As is illustrated in FIG. 2, the protein kinase
adds a
phosphate group 60 to an amino acid residue of the substrate determinant 30
included in
the core molecular backbone 20 of the double-labeled substrate 10 (protein
kinases
most commonly phosphorylate hydroxyl amino acids, such as serine, threonine,
and
tyrosine). Phosphorylation of the double-labeled substrate 10 results in the
dissociation
of the fluorescent dye 40 and the quenching dye 50 and, thus, the dissociation
of the
intramolecular dye dimer 55, which, in turn, results in a marked increase in
the
fluorescence of the double-labeled substrate because the quenching dye 50 no
longer
W~ 01/07638 CA 02380238 2002-O1-25 PCT/US00/40495
_g_
quenches the fluorescence of the fluorescent dye 40. The double-labeled
substrate 10
only exhibits a change in fluorescence upon covalent modification, in this
case
phosphorylation. Thus, the extent to which an amount of double-labeled
substrate of
the present invention is structurally modified can be continually assayed
without
separation steps merely by quantifying various changes in substrate
fluorescence.
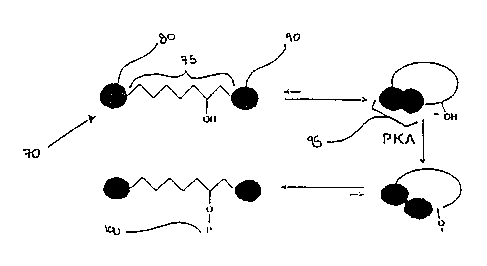
FIG. 3 schematically illustrates the modification dependent transition between
intramolecular dimer and intramolecular monomer of the two dyes 80, 90 of a
double-
labeled substrate 70 of the present invention using a peptide based on the
Kinase-
Inducible Domain (KID) of the Cyclic AMP Response Element Binding Protein
(CREB) and Protein Kinase A ("PKA") as an illustrative system. CREB is a well-
known substrate for PKA and other protein kinases, and FIG. 3 illustrates a
double-
labeled substrate 70 of the present invention which includes the KID sequence
of CREB
within the core molecular backbone 75, and two dyes 80, 90 conjugated to the
KID
sequence. In the double-labeled substrate's 70 unphosphorylated state, the two
dyes 80, 90 form an intramolecular dye dimer 95 and the fluorescence of the
fluorescent
dye 80 is quenched. When the double-labeled substrate is reacted with PKA
however, a
phosphate group, or "P" 100, is introduced in the double-labeled substrate 70,
and the
intramolecular dye dimer 95 dissociates, resulting in an increase in the
fluorescence of
the flourescent dye 80.
Though the double-labeled substrate of the present invention and the
modification-dependent transition of double-labeled substrates of the present
invention
have thus far been described in the context of protein kinase activity, such
descriptions
are illustrative only and do not limit the scope of the present invention. The
double-
labeled substrates of the present invention are useful for assaying a broad
range of
structural modifications to various biomolecules and can be specifically
constructed for
the assay of numerous processes of covalent biomolecular modification. The
core
molecular backbone of double-labeled substrates of the present invention may
be
constructed to include protein or peptide sequences, nucleotide sequences,
sugars,
lipids, receptor molecules, biopolymers, or virtually any other biomolecule
which may
serve as a substrate in one or more intracellular processes of covalent
biomolecular
modification. Thus, double-labeled substrates of the present invention can be
constructed for the assay of numerous catalytic and non-catalytic processes of
covalent
WO 01/07638 CA 02380238 2002-O1-25 pCT/US00/40495
-9-
biomolecular modification. By way of example, double-labeled substrates
according to
the present invention could be constructed to exhibit a change in fluorescence
upon
sulfation, glycation, glycosylation, carboxylation, myristoylation,
farnesylation,
ubiquitination, biotinylation, or other modification reactions.
Although not intending to be bound by a particular theory of the invention,
the
following explanations might explain the excellent results of the invention.
For
instance, the dissociation of intramolecular dye dimer formed by the two dyes
could be
the result of one or more different mechanisms. The simplest mechanism of
intramolecular dye dimer dissociation would be through steric and/or
electrostatic
effects resulting from the introduction of a functional group into the double-
labeled
substrate in close proximity to the intramolecular dye dimer. Another possible
mechanism for the dissociation of the intramolecular dye dimer is through a
modification-dependent conformational change in the double-labeled substrate.
Yet
another possible mechanism for the dissociation of the intramolecular dye
dimer is
through a conformational change in the double-labeled substrate brought about
by the
modification-dependent binding of a second molecule to the double-labeled
substrate.
These second molecules could be considered as fluorescence enhancers. For
example,
where the double-labeled substrate is modified by the addition of a phosphate
group to
a tyrosyl residue in a peptide substrate, the enhancer molecule might be a
phosphotyrosine-specific antibody or a SH2-domain-containing protein with a
high
affinity for phosphotyrosine. As is illustrated in FIG. 10, an enhancer
molecule 200
could be covalently combined with the double-labeled substrate 10 to form a
single
chimeric molecule.
A second embodiment of the double-labeled substrate of the present invention
is
schematically illustrated in FIG. 4. The double-labeled substrate 110 of FIG.
4 is
illustrated in its modified state for ease of description and includes a core
molecular
backbone 120, a substrate determinant 125 within the core molecular backbone
120, a
first spacer segment 130 and a second spacer segment 140. The first spacer
segment 130 is included at a first terminus 135 of the core molecular backbone
120, and
the second spacer segment 140 is included at a second terminus 145 of the core
molecular backbone 120. The first or second spacer segments 130, 140 may be
included in the double-labeled substrate 110 to provide a region of
flexibility between
WO 01/07638 CA 02380238 2002-O1-25 PCT/US00/40495
-10-
the core molecular backbone 120 and one or both of the dyes 150, 160 also
included in
the double-labeled substrate 110. By providing a region or regions of
flexibility
between the core molecular backbone 120 and one or both the dyes 150, 160, the
spacer
segments 130, 140 may ease the formation of an intramolecular dye dimer, and
thus
facilitate more complete quenching of double-labeled substrate 110, resulting
in a more
sensitive assay. The first or second spacer segments 130, 140 may also be
included in
the double-labeled substrate 110 in order to facilitate the use of a
particular dye which
cannot be conjugated to one of the amino acid residues included in the core
peptide
sequence 120.
The embodiment illustrated in FIG. 4 highlights that the exact construction of
double- labeled biomolecular substrates of this invention will vary. The
construction of
a double-labeled substrate may vary not only by including one or more spacer
segments
which may facilitate a more sensitive assay or enable the use of different
dyes, but the
construction may vary to utilize biomolecular substrates corresponding to
different
processes of covalent biomolecular modification. For example, within the
family of
protein kinases, double-labeled substrates of the present invention could be
constructed
using core molecular backbones which include, among others, the following
amino acid
sequences: Arg-Arg-Arg-Val-Thr-Ser-Ala-Ala-Arg-Arg-Ser (SEQ. ID. NO.: 9), a
substrate peptide for Protein Kinase A and Protein Kinase C (See, e.g., PCT
International Application WO Patent Document 98/09169); Phe-Arg-Arg-Leu-Ser-
Ile-
Ser-Thr (SEQ. ID. NO.: 1) and Pro-Leu-Ser-Arg-Thr-Leu-Ser-Val-Ser-Ser (SEQ.
ID.
NO.: 2), substrate peptides for Ca2+/calmodulin-dependent protein kinase II
(See, e.g.,
PCT International Application WO Patent Document 98/09169; Pearson et al.,
Journal
ofBiological Chemistry, 260(27), 14471-76 (1985)); Phe-Leu-Thr-Glu-Tyr-Val-Ala-
Thr-Arg-Trp-Tyr-Arg-Ala-Pro-Glu (SEQ. ID. NO.: 3), a substrate peptide for
mitogen-
activated protein kinase kinase (See, Rossomondo et al., Proceedings of the
National
Academy of Science USA, 89, 5221-25 (June 1992)); or Arg-Arg-Asp-Ile-Tyr-Glu-
Thr-
Asp-Tyr-Tyr-Arg-Lys (SEQ. ID. NO.: 4), a substrate peptide for insulin
receptor
protein-tyrosine kinase (See, Dickens et al., Biochemical and Biophysical
Research
Communications, 174(2), 772-84 (1991)). These examples, however, illustrate
only a
few of the potential substrate determinants which may be included in double-
labeled
substrates of the present invention. These examples do not reflect the myriad
of other
WO 01/07638 CA 02380238 2002-O1-25 PCT/US00/40495
-10-
the core molecular backbone 120 and one or both of the dyes 150, 160 also
included in
the double-labeled substrate 110. By providing a region or regions of
flexibility
between the core molecular backbone 120 and one or both the dyes 150, 160, the
spacer
segments 130, 140 may ease the formation of an intramolecular dye dimer, and
thus
facilitate more complete quenching of double-labeled substrate 110, resulting
in a more
sensitive assay. The first or second spacer segments 130, 140 may also be
included in
the double-labeled substrate 110 in order to facilitate the use of a
particular dye which
cannot be conjugated to one of the amino acid residues included in the core
peptide
sequence 120.
The embodiment illustrated in FIG. 4 highlights that the exact construction of
double- labeled biomolecular substrates of this invention will vary. The
construction of
a double-labeled substrate may vary not only by including one or more spacer
segments
which may facilitate a more sensitive assay or enable the use of different
dyes, but the
construction may vary to utilize biomolecular substrates corresponding to
different
processes of covalent biomolecular modification. For example, within the
family of
protein kinases, double-labeled substrates of the present invention could be
constructed
using core molecular backbones which include, among others, the following
amino acid
sequences: Arg-Arg-Arg-Val-Thr-Ser-Ala-Ala-Arg-Arg-Ser (SEQ. ID. NO.: 9), a
substrate peptide for Protein Kinase A and Protein Kinase C (See, e.g., PCT
International Application WO Patent Document 98/09169); Phe-Arg-Arg-Leu-Ser-
Ile-
Ser-Thr (SEQ. ID. NO.: 1) and Pro-Leu-Ser-Arg-Thr-Leu-Ser-Val-Ser-Ser (SEQ.
ID.
NO.: 2), substrate peptides for Ca2+/calmodulin-dependent protein kinase II
(See, e.g.,
PCT International Application WO Patent Document 98/09169; Pearson et al.,
Journal
ofBiological Chemistry, 260(27), 14471-76 (1985)); Phe-Leu-Thr-Glu-Tyr-Val-Ala-
Thr-Arg-Trp-Tyr-Arg-Ala-Pro-Glu (SEQ. ID. NO.: 3), a substrate peptide for
mitogen-
activated protein kinase kinase (See, Rossomondo et al., Proceedings of the
National
Academy of Science USA, 89, 5221-25 (June 1992)); or Arg-Arg-Asp-Ile-Tyr-Glu-
Thr-
Asp-Tyr-Tyr-Arg-Lys (SEQ. ID. NO.: 4), a substrate peptide for insulin
receptor
protein-tyrosine kinase (See, Dickens et al., Biochemical and Biophysical
Research
Communications, 174(2), 772-84 (1991)). These examples, however, illustrate
only a
few of the potential substrate determinants which may be included in double-
labeled
substrates of the present invention. These examples do not reflect the myriad
of other
WO 01/07638 CA 02380238 2002-0l-25 PCT/US00/40495
-11-
biomolecular substrates which may be included in the core molecular backbone
of
double-labeled substrates of the present invention. Again, the double-labeled
substrates of the present invention have broad application and can be tailor-
made for
use with any one of many enzymatically catalyzed or non-catalyzed
intracellular
processes by which biomolecules are covalently modified.
It is also possible to create double-labeled substrates according to the
present
invention using a variety of dyes and combinations of dyes. For example, the
dyes may
be conventional fluorescent dyes, such as fluorescein, rhodamine, cyanine,
Oregon
Green, Texas Red, Lucifer Yellow, BODIPY, rhodol, coumarin, pyrene, eosin,
erythrosin, napthalene, pyridyloxazole, anthrancene, fluorescamine, acridine,
benzofuran, anthranilic acid, aminobenzoic acid, N-methylisatoic acid,
isoluminol,
bezoxadiazole, carboxybenzoyl-quinoline-carboxyaldehyde, salicylate, bimane,
or
phenathroline, or the dyes may be non-conventional fluorescent dyes, such as a
Yellow
Fluorescent Protein (YFP) or a Green Fluorescent Protein (GFP) (obtainable
from
CLONTECH Laboratories, 1020 East Meadow Circle, Palo Alto, CA 94303). In
addition, the publication entitled "Handbook of Fluorescent Probes and
Research
Chemicals," by Richard P. Haugland, which serves as a catalog for Molecular
Probes,
Inc., of Eugene, Oregon, sets forth additional fluorescent dyes that may be
used in
constructing the double-labeled substrates of the present invention. However,
the dyes
listed here, as well as those described within in the Handbook of Fluorescent
Probes
and Research Chemicals, are provided for illustrative purposes only and do not
comprise a comprehensive list of the dyes usable in the context of the present
invention.
A double-labeled substrate according to the present invention may include a
non-fluorescent dye and a fluorescent dye, or, alternatively, a double labeled
substrate
according to the present invention may be constructed using two fluorescent
dyes.
However, it should be noted that the structure of the GFP and related protein
molecules
might not be able to stack and quench in the same manner as conventional dyes.
As a
result, where double-labeled substrates according to the present invention are
constructed using one of the various GFP molecules, it may be necessary to
include
only one GFP molecule in the combination of two dyes covalently attached to
the
double-labeled substrate ( See, U.S. Patents 5,958,713, 5,925,558, and
5,912,137).
Nevertheless, the combination, nature and location of the two dyes included in
a
WO 01/07638 CA 02380238 2002-0l-25 PCT/US00/40495
-12-
double-labeled substrate of the present invention is of relatively little
import, provided
that the two dyes stack to form a quenched intramolecular dye dimer when the
double-
labeled substrate is unmodified, the two dyes dissociate upon structural
modification of
the double-labeled substrate, and the dissociation of the two dyes upon
structural
modification results in a change in the double-labeled substrate's
fluorescence or
absorbance characteristics. Thus, the construction of a double-labeled
substrate
according to the present invention is variable, and, depending on the
application, the
double-labeled substrate of the present invention may include one or more of
many
biomolecular substrates and any suitable combination of two labels.
Also included within the scope of the present invention are methods of using
the
double- labeled biomolecular substrates of the present invention. For example,
methods of using the double-labeled substrates herein described for the assay
of protein
kinase activity in vitro and in living cells fall within the scope of the
present invention,
as do methods of using the same double-labeled substrates for high-throughput
screening. The double-labeled substrates are also useful for diagnostic and
therapeutic
applications and for methods which facilitate the discovery of substrates,
activators, and
inhibitors for novel protein kinases. Significantly, most efforts to discover
drugs
affecting protein kinase activity are presently aimed at screening for
possible protein
kinase activators and inhibitors.
A preferred method according to the present invention for assaying structural
modification of biomolecules in vitro is homogenous, comparatively simple, and
includes the steps of providing a double-labeled substrate as herein
described, including
the double-labeled substrate in a sample, and quantifying any resultant change
in
fluorescence or absorbance resulting from the structural modification of the
double-
labeled substrate. Because only double-labeled substrate which is structurally
modified
exhibits a change in fluorescence or absorbance, this method requires no
separation of
the unmodified double-labeled substrate from the modified double-labeled
substrate
before the modification of the double-labeled substrate can be accurately
assayed.
Further, the assay methods of the present invention require no special
reagents other
than the double-labeled substrate, and the measurement of changes in
fluorescence or
absorbance of the double-labeled substrate can be easily achieved using a
variety of
well known instruments, such as, for example, known spectrometers, 96-well and
384-
WO 01/07638 CA 02380238 2002-O1-25 PCT/US00/40495
-13-
well microtiter plate readers, other multichannel readers, and micro-array
instruments.
Therefore, the methods of assaying covalent biomolecular modifications
according to
the present invention provide advantages over currently used assays in terms
of
simplicity, throughput, versatility, and economy.
Because the method already described requires no separation steps, it can be
easily modified in order to assay processes of covalent biomolecular
modification in
living cells. A preferred method for assaying biomolecular structural
modification in
living cells includes providing a double-labeled substrate of the present
invention,
introducing the double-labeled substrate into living cells using techniques
well known
in the art, such as microinjection, pinocytosis, or facilitated uptake, and
quantifying any
change in fluorescence or absorbance resulting from the structural
modification of the
double-labeled substrate using well known instruments, such as, for example,
known
spectrometers, fluorescence microscopes, plate readers, cell counters, and
cell sorters.
Again, this method requires no separation steps (although they may be used),
and, thus,
allows for the continuous, real-time assay of the processes resulting in
structural
modification of biomolecules in living cells. Monitoring of biomolecular
structural
modification activities in living cells could be used for purposes of basic
research, drug
discovery, diagnosis of disease states, or efficacy of therapy following
targeted drug
treatment.
Also included within the scope of the invention are methods for the assay of
covalent biomolecular modifications performed in vitro and in living cells
which
simultaneously monitor different processes of biomolecular structural
modification by
utilizing various double-labeled substrates of the present invention, each
double-labeled
substrate being designed to specifically assay the activity of a different
process by
which biomolecules are modified. Such a method is similar to those already
detailed,
except that, in order to accurately simultaneously monitor the activity of
multiple
processes of covalent biomolecular modification, each of the different double-
labeled
substrates must be designed with unique and distinguishable spectral
properties.
Because they enable the simultaneous and continuous monitoring of multiple
processes
by which biomolecules are structurally modified, such assay methods will
likely hasten
the discovery of exactly which processes of biomolecular structural
modification are
associated with specific disease states.
W~ X1/07638 CA 02380238 2002-O1-25 PCT/US00/40495
-14-
The double-labeled substrates of the present invention can also be used in
methods facilitating the discovery of drugs which target intracellular
processes of
covalent biomolecular modification. Such a preferred method would include the
steps
of providing a sample containing the modifying enzymes) to be targeted,
introducing
into the sample a drug designed to target a particular intracellular process
of covalent
biomolecular modification, introducing into the sample a double-labeled
substrate
specific for the targeted modification process, and quantifying any change in
fluorescence or absorbance resulting from the structural modification of the
double-
labeled substrate using well known instruments. Since the covalent
modification of
biomolecules represents one of the major mechanisms by which intracellular
signaling
occurs, many processes facilitating such modifications are likely to be
important drug
targets, and methods of drug discovery facilitated by the double-labeled
peptides of the
present invention are of particular importance.
Yet another aspect of the present invention is a method of using the double-
labeled substrates of the present invention to identify substrates for novel
modifying
enzymes. The human genome is estimated to contain thousands of different
enzymes
responsible for the intracellular modification of biomolecules, many of which
are likely
to be critically involved in disease processes. Protein kinases represent but
one
superfamily of such enzymes, yet there are currently hundreds of putative
protein
kinases in sequence databases, such as GenBank and "EST" databases, whose
function
and regulation are entirely unknown. These putative protein kinases can be
identified
by their homology to known protein kinases and can be cloned and expressed as
proteins, but their enzymatic properties cannot be studied without an
appropriate
peptide or protein substrate. A preferred embodiment of a method of
identifying
peptide substrates for these protein kinases or other putative enzymes with
unknown
enzymatic properties involves constructing (e. g., synthesizing) combinatorial
libraries
of double-labeled substrates according to the present invention with core
molecular
backbones constructed with randomized amino acid (in the case of peptides) and
nucleotide (in the case of DNA and RNA, etc.) sequences, systematically
introducing
individual double-labeled substrates from the combinatorial libraries into a
sample
containing the novel protein kinase or other enzyme of unknown activity, and
quantifying any change in substrate fluorescence or absorbance which results
from
WO 01/07638 CA 02380238 2002-0l-25 PCT/US00/40495
-15-
covalent modification of the double-labeled substrate. Structural modification
of
specific double-labeled substrates in the combinatorial library by the novel
enzyme of
interest would result in a change in their spectral properties, which would
permit these
biomolecules to be identified by standard methods. Once substrates are
identified for a
novel enzyme, they can be used to characterize the activity and properties of
the enzyme
in vitro and in living cells. Further, such substrates would also facilitate
drug
development for the novel enzyme of interest using the methods described
herein (e.g.,
to identify specific inhibitors or activators of a newly discovered protein
kinase).
The present invention is further directed to kits which utilize the double-
labeled
substrates and methods described herein to detect and/or quantify covalent
biomolecular modification. A preferred embodiment of such a kit would include
a
container, one or more different double-labeled substrates of the present
invention
contained within the container, and instructions for use. The kits may also
include, for
convenience, buffers and other reagents necessary to carry out the assay, and
samples of
enzyme for calibration purposes. The reagents included with the kits can be
varied
depending on the application and in order to optimize the sensitivity of the
assay.
A further aspect of the invention is the use of double-labeled substrates to
detect
protein kinase activities and other modification reactions in living cells. A
preferred
embodiment of a method of detecting protein kinase activity in living cells
involves
constructing double-labeled protein kinase substrates as probes which are
sufficiently
cell permeable, or capable of cell permeability with inducement measures well
known
to one skilled in the art, and according to the present invention, with core
molecular
backbones constructed of various peptide sequences. In this situation, the
protein
kinase activity would result in a covalent structural modification of the
double-labeled
substrate, leading to a change in fluorescence or absorbance and in situ
detection of
kinase activity using instruments well known in the art, such as, for example,
known
spectrometers, fluorescence microscopes, plate readers, cell counters, and
cell sorters.
The invention is further described with the aid of the following illustrative
example.
WO 01/07638 CA 02380238 2002-O1-25 pCT/US00/40495
-16-
Example I
A double-labeled protein kinase substrate can be designed, synthesized,
characterized, and used to assay the activity of PKA and other protein
kinases.
The core molecular backbone of the substrate is the synthetic peptide sequence
Asp-Ser-Gln-Arg-Arg-Arg-Glu-Ile-Leu-Ser-Arg-Arg-Pro-Ser-Tyr-Arg-Arg-Ile-Leu-
Asn-Asp-Leu-Cys-Gly (SEQ. ID. NO.: S). This synthetic peptide sequence is
based on
the native sequence for KID, which is Arg-Arg-Pro-Ser-Tyr-Arg-Lys-Ile-Leu-Asn-
Asp-
Leu (SEQ. ID. NO.: 6). To arrive at the peptide sequence of the core molecular
backbone, the Lys residue of the native KID sequence was replaced by an Arg
residue
to facilitate site specific labeling of the peptide's a-amino group. Replacing
the Lys
residue of the native KID sequence resulted in the synthetic KID sequence Arg-
Arg-
Pro-Ser-Tyr-Arg-Arg-Ile-Leu-Asn-Asp-Leu (SEQ. ID. NO.: 7), which represents
the
sequence generally referred to herein as "the KID sequence." Next, a Cys
residue was
added to the C-terminus of the synthetic KID sequence to allow labeling of the
molecular backbone with a dye through the sulfhydryl group in the cysteine
residue, and
a Gly residue was added at the terminal Cys residue to facilitate peptide
synthesis.
Finally, the additional peptide sequence Asp-Ser-Gln-Arg-Arg-Arg-Glu-Ile-Leu-
Ser
(SEQ. ID. NO.: 8) was added at the Arg residue of the N-terminus of the KID
sequence
to give the final peptide more helical structure.
The synthetic peptide sequence of the core molecular backbone of the substrate
was synthesized on a benzhydrylamine resin using conventional (tBOC) solid
phase
peptide synthetic chemistry. See, e.g. Barany and Merrifield in The Peptides,
Analysis,
Synthesis, Biology, Vol. 2, E. Gross and J. Meienhofer, eds., (Acad. Press,
New York,
1980), Glass. D.B., Methods Enzymol., 99, 119-139 (1983). After synthesis, the
peptide
sequence was cleaved from the resin with anhydrous HF using standard protocols
which
yield a crude side-chain deprotected peptide with an amide C-terminus, and the
synthetic peptide sequence was purified to homogeneity by HPLC using a C4
reverse-
phase column. The mass of the purified peptide sequence was confirmed by mass
spectrometry. The synthetic polypeptide sequence of SEQ. ID. NO.: 5 was
selected as
an exemplary core molecular backbone for a double-labeled substrate of the
present
invention not only because the KID sequence embedded in the synthetic
polypeptide is
known to contain the specificity determinants of several protein kinases,
including
WO 01/07638 CA 02380238 2002-O1-25 PCT/US00/40495
-17-
PKA, but also because this exemplary synthetic peptide sequence has proven to
undergo a phosphorylation-dependent change in conformation.
After purification, the core molecular backbone was conjugated with two dyes
to form a double-labeled substrate. First, the synthetic peptide sequence of
the core
molecular backbone was conjugated with tetramethylrhodamine-5-maleimide. The
maleimide on the dye reacts with the cysteine residue at the C-terminus in the
KID
region, and the maleimide group serves as the link between the sulfhydryl
group on the
cysteine and the rhodamine group. The single-labeled substrate was then
conjugated at
the N-terminus with either ~-carboxyfluorescein, succinimidyl ester or 5-
carboxytetramethylrhodamine, succinimidyl ester. The succinimidyl ester group
reacts
with the amino group at the N-terminus of each peptide to form a carboxamide
bond
with the dye. Following HPLC purification on a C4 reversed-phase column, the
double-labeled substrate was subjected to analysis by mass spectrometry
analysis, UV
absorbance spectrophotometry, and fluorescence spectrophotometry.
After such analyses, the double-labeled substrate was phosphorylated with PKA.
The phosphate acceptor amino acid in this double-labeled substrate is the
serine residue
found within the embedded KID sequence.
As illustrated in FIG. 5 through FIG. 9, phosphorylation of double-labeled
substrates prepared as herein described results in detectable changes in the
absorbance
and fluorescence characteristics of the dyes included in the double-labeled
substrates.
FIG. 5. illustrates the absorbance peaks for fluorescein and rhodamine before
and after phosphorylation of a double-labeled substrate having a fluorescein
dye and a
rhodamine dye molecule conjugated thereto. The unphosphorylated substrate
exhibits
an absorbance maximum for fluorescein at SOOnm and an absorbance maximum for
rhodamine at 552 nm. However, after phosphorylation, the two absorbance peaks
shift
to 498 nm and 548 nm for fluorescein and rhodamine, respectively.
FIG. 6 and 7 illustrate the even more dramatic phosphorylation dependent
changes in the fluorescence characteristics a double-Labeled substrate having
a
fluorescein and a rhodamine molecule conjugated thereto. As can be seen in
FIG. 6, the
fluorescence of the fluorescein label increased 340% after phosphorylation,
and FIG. 7
illustrates that phosphorylation of the double-labeled substrate caused a 35%
increase in
the fluorescence of the rhodamine label.
WO 01/07638 CA 02380238 2002-0l-25 PCT/US00/40495
-18-
As mentioned, the molecular backbone described herein may also be labeled
with, among other combinations, two rhodamine dyes instead of a rhodamine dye
and a
fluorescein dye, and FIG. 8 and FIG. 9 illustrate the phosphorylation-
dependent changes
in the optical properties of such a double-labeled substrate. As can be seen
in FIG. 8, in
its unphosphorylated state, the double-labeled substrate exhibited two
absorbance
maxima. The larger peak is at 520 nm, while the smaller peak is at 552 nm.
After
phosphorylation, the peak at 520 nm decreases in size while the peak at 552 nm
shifts
to 550 nm and increases in size. Moreover, as can be appreciated from FIG. 9,
the
fluorescence of rhodamine increases 69% after phosphorylation.
The results illustrated in FIG. 5 through FIG. 9, therefore, show that the
fluorescence of at least one label conjugated to the double-labeled substrates
is
quenched when the substrates are found in their unphosphorylated state. These
results
indicate that when the double-labeled substrates are not phosphorylated, the
two dyes
included in each double-labeled substrate stack on each other to form an
intramolecular
1 ~ dye dimer, resulting in the reduction of the fluorescence of at least one
of the dyes
included in the dye dimers. Stacking is also indicated by the observation that
the UV
absorbance spectrum of the unphosphorylated double-labeled substrate differs
markedly
from the spectra of the same substrate after phosphorylation.
As is also apparent by reference to results illustrated in FIG. 5 through FIG.
9,
phosphorylation of the double-labeled substrates results in an increase in the
intensity of
the fluorescent emission peak of at least one dye conjugated to the double-
labeled
substrates. This indicates that phosphorylation of the double-labeled
substrate causes a
dissociation of the intramolecular dimer. In each instance, there was a large
increase in
fluorescence intensity of at least one of the dyes conjugated to the double-
labeled
substrate, thereby providing a high signal-to-noise ratio. Sensitivity is also
excellent
with changes in dye emission intensity being observable at low nanomolar
concentrations of peptide in a standard spectrofluorometer. The favorable
sensitivity
and signal-to-noise ratio indicate the double-labeled substrate will be useful
for
monitoring protein kinase activity in a variety of applications.
The procedures and methods described herein can be employed to prepare and
use double-labeled protein kinase substrates for assaying most any other
protein kinase.
For example, the KID sequence included in the core molecular backbone
described
W~ 01/07638 CA 02380238 2002-O1-25 PCT/US00/40495
-19-
herein may be modified to contain an appropriate consensus sequence for a
given
protein kinase determinant. Such consensus sequences can be found in the
literature for
many common kinases such as PKA, PKC, CaM kinase II, etc. (cf., Songyang et
al.
Current Biol. 4:479, 1994). Moreover, the double-labeled substrate of the
present
invention can be prepared for assaying most any other intracellular processes
leading to
the structural modification of protein or other biomolecules. Thus, even
though the
present invention has been herein described in terms of certain preferred
embodiments
and specific examples, such descriptions are illustrative only and do not
limit the scope
of the present invention. The scope of the present invention is to be defined
by the
appended claims.