Note: Descriptions are shown in the official language in which they were submitted.
CA 02388445 2002-05-31
1
GENETIC MARKERS, METABOLIC MARKERS, AND METHODS FOR
EVALUATING PATHOGENICITY OF STRAINS OF E. COLI
BACKGROUND OF THE INVENTION
a) Field of the invention
The present invention is concerned with genetic and metabolic markers and
with methods to identify pathogenic or potentially pathogenic strains of E.
coli.
More particularly, the invention provides nucleotide and amino acid sequences,
antibodies, probes, cells, kits and methods concerning genes expressed mostly
by
pathogenic strains E. coli.
b) Brief description of the prior art
Escherichia coli is a heterogeneous species consisting of both enteric
commensal and pathogenic strains. Different types of E. coli cause different
diseases in a range of hosts, including extra-intestinal and enteric
infections. For
example, enteropathogenic E. coil (EPEC) is the leading cause of severe
infantile
diarrhea in developing countries, and enterohaemorrhagic E. coli (EHEC)
(including the well-known 0157:H7) have recently been shown to be the cause of
bloody diarrhea and hemolytic-uremic syndrome in major food-borne outbreaks in
the United States, Europe, and Asia (CMR 1998, 11:142).
Over the last five years, studies have been published on the E. coli
chromosome. The whole genome sequence of the laboratory strain K-12 MG1655
was published in 1997 (Science 1997, 277:1453). The genome of E, coli 0157: H7
(EHEC strain EDL933) was recently sequenced (Nature 2001, 409:529). Although
comparative analysis of these sequences have resulted in the identification of
virulence genes and the characterization of pathogenicity islands, the
specific
virulence regions associated with the pathogenesis of E, coli causing various
diseases remains to be elucidated.
Recently, some of the present inventors identified in the genome of
S. enferica serovar Typhi, an operon of three genes (deoK operon) regulated by
a
repressor DeoQ and missing in E. coli K12 (J. Bacteriol., 2000, 182:869-873).
In
E. coli strain AL862, sequences similar to the deoK operon have been sequenced
CA 02388445 2002-05-31
2
(GenBankT"" accession Nos. AF286670 and AF286671 ) but no function has been
assigned to these sequences.
Furthermore, although the use of 2-Deoxy-D-ribose by E. coli strains has
been previously described (Br. J. Biomed. Sci., 1995; 52: 173), this property
was
never associated with the pathogenic status of the strains and the genes
encoding
this function were not identified.
In view of the above, there is a need for methods, nucleic acid molecules,
polypeptides, antibodies, vectors and cells useful for the identification of
pathogenic strains of E. coli.
The present invention fulfils this need and also other needs as it will be
apparent to those skilled in the art upon reading the following specification.
SUMMARY OF THE INVENTION
The present inventors have found that a sugar (deoxyribose) that is not
fermented by E. coli K12, is metabolized by a large number of pathogenic
isolates
belonging to various pathotypes. The present inventors have identified the
genes
encoding this function and demonstrated that they are conserved among several
pathogenic strains. The present inventors have also developed genetic and
bacteriological assays to identify deoxyribose-positive E. coli strains.
In general, the invention features an isolated or purified nucleic acid
molecule, such as genomic, cDNA, antisense, DNA, RNA or a synthetic nucleic
acid molecule that encodes or corresponds to a E. coli deoK polypeptide.
According to a first aspect, the invention features isolated or purified
nucleic
acid molecules, polynucleotides, polypeptides, E. coli deoK proteins and
fragment
thereof. Preferred nucleic acid molecules consist of a DNA.
According to another aspect, the invention features a nucleotide probe.
According to another aspect, the invention features a purified antibody. In a
preferred embodiment, the antibody is a monoclonal or a polyclonal antibody
that
specifically binds to a E. coli deoK protein and/or to a fragment thereof.
A further aspect of the invention relate to a method for evaluating
pathogenicity of a strain of E. coli, comprising assaying a metabolic activity
of the
CA 02388445 2002-05-31
3
strain. Preferably the metabolic activity consists of metabolization of 2-
Deoxy-D-
ribose and capacity of the strain to metabolize of 2-Deoxy-D-ribose is
assessed.
In another aspect, the present invention further features a method for
identifying a pathogenic or potentially pathogenic strain of E. coli. In a
related
aspect, the invention relates to a method for determining likelihood of
pathogenicity of a strain of E. coli. In one embodiment, the method comprises
detecting deoxyribokinase enzymatic activity of the strain. Preferably this is
done
by assaying, under suitable culture conditions, the capabilities of the strain
to
metabolize 2-Deoxy-D-ribose. In another embodiment, the method comprises
assaying the E. coli strain for the presence of genes or proteins involved in
metabolization of 2-Deoxy-D-ribose. According to the invention ability of E.
coli
strains to metabolize 2-Deoxy-D-ribose and/or presence of genes or proteins
involved in metabolization of 2-Deoxy-D-ribose is indicative that the strain
of E. coli
is pathogenic or a potentially pathogenic. Of course, both aspects of the
method
may be carried out simultaneously or in parallel.
In another aspect, the present invention further features a method for
identifying a pathogenic or a potentially pathogenic strain of E. coli, the
method
having a level of specificity of at least 30%, 40%, 45%, 46%, 47%, 48%, 49% or
50% for pathogenic E. coli from some clinical isolates. More preferably, the
method detect less than 25%, 20%, 18%, 15% or 10% of commensal E. coli from
healthy peoples.
In another related aspect, the invention features a kit for identifying a
strain
of E. coli or evaluating pathogenicity of a strain of E. coli, the kit
comprising
preferably an antibody or a probe as defined previously.
The present invention also features a method of treatment of E. coli
infections.
One of the greatest advantages of the present invention is that it provides
genetic and proteinic markers, antibodies, probes, kits and methods that can
be
used for identifying pathogenic strains of E. coli and/or for evaluating
pathogenicity
of a strain of E. coli and eventually treat or prevent E. coli infections.
CA 02388445 2002-05-31
4
Other objects and advantages of the present invention will be apparent
upon reading the following non-restrictive description of the preferred
embodiments thereof and from the claims.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1 is a schema illustrating operon deoK in Escherichia coli.
Figure 2 represents nucleic acids and amino acids sequences of deoK
operon in Escherichia coli - strain AL862. Underlined sequence
corresponds to probe A and doubled underlined sequence corresponds to
probe B. Bold nucleotides correspond to primers used in PCR assay to
amplify probes A and B.
Figure 3 represents nucleic acids and amino acids sequences of deoK
operon in Escherichia coli - strain 55989.
Figure 4 represents nucleic acids sequence of Probe A.
Figure 5 represents nucleic acids sequence of Probe B.
DETAILED DESCRIPTION OF THE INVENTION
A) Definitions
Throughout the text, the word "kilobase" is generally abbreviated as "kb",
the words "deoxyribonucleic acid" as "DNA", the words "ribonucleic acid" as
"RNA", the words "complementary DNA" as "cDNA", the words "polymerase chain
reaction" as "PCR", and the words "reverse transcription" as "RT". Nucleotide
sequences are written in the 5' to 3' orientation unless stated otherwise.
In order to provide an even clearer and more consistent understanding of
the specification and the claims, including the scope given herein to such
terms,
the following definitions are provided:
CA 02388445 2002-05-31
Antisense: As used herein in reference to nucleic acids, is meant a nucleic
acid sequence, regardless of length, that is complementary to the coding
strand of
a gene.
Expression: Refers to the process by which gene encoded information is
5 converted into the structures present and operating in the cell. In the case
of
cDNAs, cDNA fragments and genomic DNA fragments, the transcribed nucleic
acid is subsequently translated into a peptide or a protein in order to carry
out its
function if any. By "positioned for expression" is meant that the DNA molecule
is
positioned adjacent to a DNA sequence which directs transcription and
translation
of the sequence (i.e., facilitates the production of, e.g., a deoK
polypeptide, a
recombinant protein or a RNA molecule).
Fragment: Refers to a section of a molecule, such as a protein, a
polypeptide or a nucleic acid, and is meant to refer to any portion of the
amino acid
or nucleotide sequence.
Host: A cell, tissue, organ or organism capable of providing cellular
components for allowing the expression of an exogenous nucleic acid embedded
into a vector. This term is intended to also include hosts which have been
modified
in order to accomplish these functions. Bacteria, fungi, animal (cells,
tissues, or
organisms) and plant (cells, tissues, or organisms) are examples of a host.
Isolated or Purified or Substantially pure: Means altered "by the hand of
man" from its natural state, i.e., if it occurs in nature, it has been changed
or
removed from its original environment, or both. For example, a polynucleotide
or a
protein/peptide naturally present in a living organism is not "isolated", the
same
polynucleotide separated from the coexisting materials of its natural state,
obtained by cloning, amplification and/or chemical synthesis is "isolated" as
the
term is employed herein. Moreover, a polynucleotide or a protein/peptide that
is
introduced into an organism by transformation, genetic manipulation or by any
other recombinant method is "isolated" even if it is still present in said
organism.
Nucleic acid: Any DNA, RNA sequence or molecule having one nucleotide
or more, including nucleotide sequences encoding a complete gene. The term is
intended to encompass all nucleic acids whether occurring naturally or non-
naturally in a particular cell, tissue or organism. This includes DNA and
fragments
CA 02388445 2002-05-31
6
thereof, RNA and fragments thereof, cDNAs and fragments thereof, expressed
sequence tags, artificial sequences including randomized artificial sequences.
Open reading frame ("ORF"): The portion of a cDNA that is translated into
a protein. Typically, an open reading frame starts with an initiator ATG codon
and
ends with a termination codon (TAA, TAG or TGA).
Percent identity and Percent similarity: Used herein in nucleic acid
and/or among amino acid sequences comparisons. Sequence identity is typically
measured using sequence analysis software with the default parameters
specified
therein (e.g., Sequence Analysis Software Package of the Genetics Computer
Group, University of Wisconsin Biotechnology Center, 1710 University Avenue,
Madison, Owl 53705). This software program matches similar sequences by
assigning degrees of homology to various substitutions, deletions, and other
modifications. Conservative substitutions typically include substitutions
within the
following groups: glycine, alanine, valine, isoleucine, leucine; aspartic
acid,
glutamic acid, asparagine, glutamine; serine, threonine; lysine, arginine; and
phenylalanine, tyrosine.
Polypeptide or Protein: Means any chain of more than two amino acids,
regardless of post-translational modification such as glycosylation or
phosphorylation.
Potentially pathogenic: Refers to a strain which has the capacity to be
involved in a pathogenic process. Examples of potentially pathogenic strains
are
extra-intestinal E. coli strains which are distinct from the commensal and
from the
intestinal pathogenic strains.
Specifically binds: Means an antibody that recognizes and binds a protein
or polypeptide but that does not substantially recognize and bind other
molecules
in a sample, e.g., a biological sample, that naturally includes protein.
Substantially the same: Refers to nucleic acid or amino acid sequences
having sequence variation that do not materially affect the nature of the
protein.
With particular reference to nucleic acid sequences, the term "substantially
the
same" is intended to refer to the coding region and to conserved sequences
governing expression, and refers primarily to degenerate codons encoding the
same amino acid, or alternate codons encoding conservative substitute amino
CA 02388445 2002-05-31
7
acids in the encoded polypeptide. With reference to amino acid sequences, the
term "substantially the same" refers generally to conservative substitutions
and/or
variations in regions of the protein that are not involved in determination of
structure or function of the protein. "Substantially the same" encompasses
"degenerate variants" of nucleic acid or amino acid sequences.
Substantially pure polypeptide: Means a polypeptide that has been
separated from the components that naturally accompany it. Typically, the
polypeptide is substantially pure when it is at least 60%, by weight, free
from the
proteins and naturally-occurring organic molecules with which it is naturally
associated. Preferably, the polypeptide is at least 75%, 80%, or 85%, more
preferably at least 90%, 95% or 97% and most preferably at least 99%, by
weight,
pure. A substantially pure polypeptide or protein may be obtained, for
example, by
extraction from a natural source (including but not limited to E. Coh) by
expression
of a recombinant nucleic acid encoding the polypeptide, or by chemically
synthesizing the protein. Purity can be measured by any appropriate method,
e.g.,
by column chromatography, polyacrylamide gel electrophoresis, or HPLC
analysis.
A protein is substantially free of naturally associated components when it is
separated from those contaminants which accompany it in its natural state.
Thus,
a protein which is chemically synthesized or produced in a cellular system
different
from the cell from which it naturally originates will be substantially free
from its
naturally associated components. Accordingly, substantially pure polypeptides
include those derived from eukaryotic organisms but synthesized in E. coli or
other
prokaryotes. By "substantially pure DNA" is meant DNA that is free of the
genes
which, in the naturally-occurring genome of the organism from which the DNA of
the invention is derived, flank the gene. The term therefore includes, for
example,
a recombinant DNA which is incorporated into a vector; into an autonomously
replicating plasmid or virus; or into the genomic DNA of a prokaryote or
eukaryote;
or which exists as a separate molecule (e.g., a cDNA or a genomic or cDNA
fragment produced by PCR or restriction endonuclease digestion) independent of
other sequences. It also includes a recombinant DNA which is part of a hybrid
gene encoding an additional polypeptide sequence.
CA 02388445 2002-05-31
Transformed or Transfected or Transduced or Transgenic cell: Refers
to a cell into which (or into an ancestor of which) has been introduced, by
means
of recombinant DNA techniques, an exogenous DNA molecule encoding a
polypeptide of interest. By "'transformation" is meant any method for
introducing
foreign molecules into a cell. Lipofection, calcium phosphate precipitation,
retroviral delivery, electroporation, and ballistic transformation are just a
few of the
teachings which may be used.
Vector: A self-replicating RNA or DNA molecule which can be used to
transfer an RNA or DNA segment from one organism to another. Vectors are
particularly useful for manipulating genetic constructs and different vectors
may
have properties particularly appropriate to express proteins) in a recipient
during
cloning procedures and may comprise different selectable markers. Bacterial
plasmids are commonly used vectors. Modified viruses such as adenoviruses and
retroviruses are other examples of vectors.
B) General overview of the invention
The present inventors have shown that a sugar (deoxyribose) that is not
fermented by E. coli K12, is metabolized by a large number of pathogenic
isolates
belonging to various pathotypes. The present inventors have also identified
the
genes encoding this function and they demonstrated that they are conserved
among several pathogenic strains. The present inventors have further developed
genetic and bacteriological assays to identify deoxyribose-positive E, coli
strains.
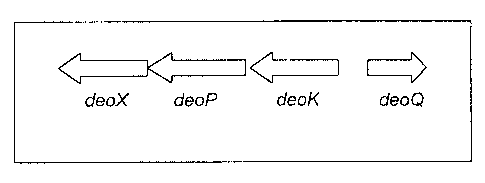
i) Cloning and molecular characterization of deoK operon in E, coli
As it will be described hereinafter in the exemplification section of the
invention, the inventors have discovered, cloned and sequenced the DNA
encoding the deoK operon in two pathogenic strains of E. coli. The DNA
sequences and the predicted amino acid sequence of the encoded proteins are
shown in Figures 2 and 3. Computer analysis revealed four open reading frames
(ORF), deoX, deoP, deoK, and deoQ, which mapped to the same loci as had
similar sequences to the deoX, deoP, deoK, and deoQ genes from the deoK
operon from Salmonella, respectively (See Figure 1 ).
CA 02388445 2002-05-31
9
The function of deoP, deoK, and deoQ is known. These E. coli genes
encode a putative 2-Deoxy-D-ribose permease, a deoxyribokinase and a putative
repressor protein, respectively. Function of deoX remains to be elucidated.
DeoX
gene encodes a protein of 337 amino acids (A.A.) long. In silico analysis
indicates
that the protein has the following features: it has a molecular weight of
about 38
kDa, an isoelectric point of about 5.2; an instability index of about 45.4
(i.e.
Unstable); an aliphatic index of about 79.6; and a grand average of
hydropathicity
(GRAVY) of about -0.136.
ii) deoK homology with other genes and proteins
As shown in Table 1 on the exemplification section, a blast search indicates
that deoK operon in E. coli shares high level of identity with deoK operon in
S. Typhi (about 75 to 80%).
Therefore, the present invention concerns an isolated or purified nucleic acid
molecule (such as DNA) comprising a sequence selected from the group
consisting of
a) sequences provided in part or all of SEQ ID NO: 1 or 6;
b) complements of the sequences provided in part or all of SEQ ID NO: 1 or 6;
c) sequences consisting of at least 20 contiguous residues of a sequence
provided in SEQ ID NO: 1 or 6;
d) sequences that hybridize to part or all of nucleic acids of SEQ ID NO: 1 or
6,
under moderately, preferably high, stringent conditions;
e) sequences having at least 80% identity to part or all of SEQ ID NO: 1 or 6;
f) degenerate variants of a sequence provided in part or all of SEQ ID NO: 1
or 6;
and
g) sequences encoding part or all of polypeptides provided in SEQ ID NO: 2-5
and 7-10.
More preferably, the nucleic acid molecule of the invention comprises a
sequence selected from the group consisting of:
a) a nucleotide sequence having at least 80%, 85%, 90%, 95% or 97% nucleotide
sequence identity with SEQ ID NO: 1 or 6; and
CA 02388445 2002-05-31
b) a nucleotide sequence having at least 80%, 85%, 90%, 95% or 97% nucleotide
sequence identity with a nucleic acid encoding an amino acid sequence of
SEQ ID NO: 2-5 and 7-10.
More preferably, the nucleic acid molecule comprises a sequence
5 substantially the same or having 100% identity with SEQ ID NO: 1 or 6, or a
sequence substantially the same or having 100% identity with nucleic acids
encoding an amino acid sequence of SEQ ID NO: 2-5 and 7-10.
The present invention also concerns isolated or purified nucleic acid
molecules comprising a sequence encoding a E. coli polypeptide involved in
10 metabolization of 2-Deoxy-D-ribose, or degenerate variants thereof, the E.
coli
polypeptide or degenerate variant comprising part or all of SEQ ID N0:2-5 and
7-10.
The present invention also concerns isolated or purified nucleic acid
molecule which hybridizes under moderate, preferably high stringency
conditions
with part or all of any of the nucleic acid molecules of the invention
mentioned
hereinbefore or with part or all of a complementary sequence thereof. The
"hybridizing" nucleic acid could be used as probe or as antisense molecules as
it
will be described hereinafter.
In a related aspect, the present invention concerns an isolated or purified
polypeptide or a protein comprising an amino acid sequence selected from the
group consisting of:
a) sequences encoded by a nucleic acid as defined previously;
b) sequences having at least 80% identity to part or all of any of SEQ ID N0:2-
5
and 7-10;
c) sequences having at least 85% homology to part or all of any of SEQ ID
N0:2-5 and 7-10; and
d) sequence provided in part or all of any of SEQ ID N0:2-5 and 7-10.
More preferably, the polypeptide comprises an amino acid sequence
substantially the same or having 100% identity with any of SEQ ID N0:2-5 and
7-10. Most preferred polypeptides are those having a biological activity that
permit
E. coli to metabolize 2-Deoxy-D-ribose.
CA 02388445 2002-05-31
11
iii) Anti-deoK antibodies
The invention features purified antibodies that specifically bind to a protein
encoded by the E. colt deoK operon. The antibodies of the invention may be
prepared by a variety of methods using the deoK proteins or polypeptides
described above. For example, the deoK polypeptide, or antigenic fragments
thereof, may be administered to an animal in order to induce the production of
polyclonal antibodies. Alternatively, antibodies used as described herein may
be
monoclonal antibodies, which are prepared using hybridoma technology (see,
e.g.,
Hammerling et al., In Monoclonal Antibodies and T-Cell Hybridomas, Elsevier,
NY,
1981 ).
The invention features antibodies that specifically bind E. colt deoK operon
polypeptides, or fragments thereof. In particular, the invention features
"neutralizing" antibodies. By "neutralizing" antibodies is meant antibodies
that
interfere with any of the biological activities of any of the E. colt deoK
operon
polypeptides, particularly the ability of E. colt to metabolize 2-Deoxy-D-
ribose. The
neutralizing antibody may reduce the ability of E. colt deoK proteins to
metabolize
2-Deoxy-D-ribose by, preferably 50%, more preferably by 70%, and most
preferably by 90% or more. Any standard assay of 2-Deoxy-D-ribose
metabolization, including those described herein, may be used to assess
potentially neutralizing antibodies. Once produced, monoclonal and polyclonal
antibodies are preferably tested for specific deoK proteins recognition by
Western
blot, immunoprecipitation analysis or any other suitable method.
In addition to intact monoclonal and polyclonal anti-deoK antibodies, the
invention features various genetically engineered antibodies, humanized
antibodies, and antibody fragments, including F(ab')2, Fab', Fab, Fv and sFv
fragments. Antibodies can be humanized by methods known in the art. Fully
human antibodies, such as those expressed in transgenic animals, are also
features of the invention.
Antibodies that specifically recognize deoK proteins (or fragments deoK),
such as those described herein, are considered useful to the invention. Such
an
antibody may be used in any standard immunodetection method for the detection,
CA 02388445 2002-05-31
12
quantification, and purification of deoK proteins. The antibody may be a
monoclonal or a polyclonal antibody and may be modified for diagnostic
purposes.
The antibodies of the invention may, for example, be used in an immunoassay to
monitor deoK expression levels, to determine the subcellular location of a
deoK or
deoK fragment produced by E. coli, to determine the amount of deoK or fragment
thereof in a biological sample and evaluate the pathogenicity of a strain of
E. coli.
In addition, the antibodies may be coupled to compounds for diagnostic and/or
therapeutic uses such as gold particles, alkaline phosphatase, peroxidase for
imaging and therapy The antibodies may also be labeled (e.g.
immunofluorescence) for easier detection.
iv) Identification of E. coli pathogenic strains
According to the present invention, the ability of the E. coli strain to
metabolize 2-Deoxy-D-ribose and/or the presence of genes or proteins involved
in
metabolization of 2-Deoxy-D-ribose in the E. coli strain is indicative that
this strain
is pathogenic or at least potentially pathogenic.
Therefore, the invention provides a method for evaluating pathogenicity of a
strain of E. coli comprising assaying a metabolic activity of that strain.
Preferably,
the metabolic activity consists of metabolization of 2-Deoxy-D-ribose and the
assessment step consists of growing the strain of a minimal medium comprising
2-Deoxy-D-ribose as a sole source of carbon.
The antibodies described above and probes described hereinafter rnay be
used to monitor deoK protein expression and/or to identify a pathogenic strain
of
E, coli in a biological sample or in a human or an subject. Accordingly, the
invention provides a method for identifying a pathogenic strain of E, coli
and/or for
evaluating likelihood of pathogenicity of a strain of E. coli as compared to a
commensal strain.
According to a first embodiment, the method comprises assaying the E. coli
strain for the presence of genes or proteins involved in metabolization of 2-
Deoxy-
D-ribose. Preferably, oligonucleotides such as probes, or cloned nucleotide
(RNA
or DNA) fragments corresponding to unique portions of genes and proteins from
operon deoK are used to assess deoK proteins cellular levels or detect deoK
CA 02388445 2002-05-31
13
mRNAs (both indicative of E. coli pathogenicity). Such an assessment may also
be
done in vifro using well-known methods (Northern analysis, PCR, quantitative
PCR, microarrays, etc.). The methods of the invention may be carried out by
contacting, in vitro or in vivo, an E, coli isolate or a biological sample
(such as a
urine sample, feces, blood, cerebral spinal fluid, from an individual or an
individual
or an animal suspected of harboring pathogenic E. coli. or an extract thereof,
witty
an anti-deoK antibody or a probe according to the invention, in order to
determine
the presence or evaluate the amount of deoK proteins or gene in the sample or
the
cells therein.
According to a preferred embodiment, the method comprises assessment of
the E, coli strain for the presence of a nucleic acid sequence selected from
the
group consisting of:
a) sequences provided in part or all of SEQ ID NO: 1 or 6;
b) complements of the sequences provided in part or all of SEQ ID NO: 1 or 6;
a) sequences consisting of at least 20 contiguous residues of a sequence
provided in SEQ ID NO: 1 or 6;
b) sequences that hybridize to part or all of nucleic acids of SEQ ID NO: 1 or
6,
under moderately, preferably high, stringent conditions;
c) sequences having at least 80% identity to part or all of SEQ ID NO: 1 or 6;
d) degenerate variants of a sequence provided in part or all of SEQ ID NO: 1
or 6;
and
e) sequences encoding part or all of polypeptides provided in SEQ ID NO: 2-5
and 7-10.
According to another preferred embodiment, the method comprises
assessment of the E. coli strain for the presence of a polypeptide comprising
an
amino acid sequence selected from the group consisting of:
a) sequences encoded by a nucleic acid as defined in claim 7;
b) sequences having at least 80% identity to part or all of any of SEQ ID N0:2-
5
and 7-10;
c) sequences having at least 85% homology to part or all of any of SEQ ID N0:2-
5 and 7-10; and
d) sequence provided in part or all of any of SEQ ID N0:2-5 and 7-10.
CA 02388445 2002-05-31
14
Accordingly, the invention encompasses nucleotide probes comprising a
sequence of at least 15, 20, 25, 30, 40, 50, 75, 100 or more sequential
nucleotides
cf SEQ ID NO: 1 or 6, or of a sequence complementary to SEQ ID NO: 1 or 6.
More preferably, the probe consists of SEQ ID NO: 11 or 12.
Of course, it may be preferable to further assay the presence (or absence)
of other genes/proteins in order to increase sensitivity and/or specificity of
the
method.
According to another embodiment, the method for identifying a pathogenic
strain of E. coli comprises detecting deoxyribokinase enzymatic activity of
the
strain. Preferably this is done by assaying, under suitable culture
conditions, the
capabilities of the strain to metabolize 2-Deoxy-D-ribose. This may be
achieved by
grow'ng in vitro an E. coli isolate or a biological sample suspected of
harboring
pathogenic E. coli on a minimal medium comprising 2-Deoxy-D-ribose as a sole
source of carbon and evaluating bacteria growth and survival in that medium.
Preferably, the minimal medium comprises from about 0.01 % 2-Deoxy-D-ribose
and the bacteria are cultured in the minimal medium for about 24h to about
48h.
Assay kits for determining the amount of deoK genes and proteins in a
sample and/or for identifying a pathogenic strain of E. coli, are also within
the
scope of the present invention. According to one embodiment, such a kit would
preferably comprises anti-deoK antibody(ies) or probes) according to the
invention and other elements) selected such as instructions for using the kit,
assay tubes, enzymes, reagents or reaction buffers}, enzymes}. In another
embodiment, the kit would comprises means for assaying capabilities of a
strain of
E. coli to metabolize 2-Deoxy-D-ribose.
A non-limitative example of use for the methods, kits and probes of the
invention is the detection of pathogenic or potentially pathogenic E. coli
bacteria in
food which may be contaminated by E. coli.
v) Downmodulation of deoK proteins expression
As mentioned previously, expression of proteins of the deoK operon allows
E. coli to metabolize 2-Deoxy-D-ribose. Modulation of deoK may be useful.
CA 02388445 2002-05-31
More particularly downmodulation of deoK proteins could be used to
prevent and/or treat E. coli infections. Therefore, the invention also relates
to
methods for preventing or treating E. Coli infections comprising
downmodulating
expression or biological activity of deoK proteins or genes. This may be
achieved
5 by administering a molecule or compound having such property.
vii) Vectors and Cells
The invention is also directed to a host, such as a genetically modified cell,
comprising any of the nucleic acid sequence according to the invention and
more
10 preferably, a host capable of expressing the peptide/protein encoded by
this
nucleic acid.
The host cell may be any type of cell (a transiently-transfected mammalian
cell line, an isolated primary cell, or a bacterium (such as E. coh). More
preferably
the host is Escherichia coli bacterium and it is selected from the Escherichia
coli
15 bacteria filed on May 14, 2002 at the CNCM under accession numbers I-2867
and
I-2867.
A number of vectors suitable for stable transfection of mammalian cells and
bacteria are available to the public (e.g. plasmids, adenoviruses, adeno-
associated viruses, retroviruses, Herpes Simplex Viruses, Alphaviruses,
Lentiviruses), as are methods for constructing such cell lines. The present
invention encompasses any type of vector comprising any of the nucleic acid
molecule of the invention and more particularly the vectors capable of
directing
expression of the peptide encoded by such nucleic acid in a vector-containing
cell.
The cells of the invention may be particularly useful for diagnostic purposes
and for drug screening (by measuring effect of a compound on expression or
activity levels of deoK genes of proteins for instance).
vii) Synthesis of E. coli deoK proteins and functional derivative thereof
;knowledge of E. coli deoK operon gene sequences open the door to a
series of applications. For instance, the characteristics of the cloned E.
coli deoK
genes sequences may be analyzed by introducing the sequence into various cell
types or using in vitro extracellular systems. The function of E. coli deoK
genes
CA 02388445 2002-05-31
16
may then be examined under different physiological conditions. The deoK cDNA
sequences may be manipulated in studies to understand the expression of the
gene and gene product. Alternatively, cell lines may be produced which
overexpress the gene product allowing purification of deoK proteins for
biochemical characterization, large-scale production, antibody production, and
patient therapy.
For protein expression, eukaryotic and prokaryotic expression systems may
be generated in which the deoK operon gene sequences is introduced into a
plasmid or other vector which is then introduced into living cells. Gonstructs
in
which the deoK cDNA sequences containing the entire open reading frame
inserted in the correct orientation into an expression plasmid may be used for
protein expression. Alternatively, portions of the sequence, including wild-
type or
mutant deoK sequences, may be inserted. Prokaryotic and eukaryotic expression
systems allow various important functional domains of the protein to be
recovered
as fusion proteins and then used for binding, structural and functional
studies and
also for the generation of appropriate antibodies. The deoK DNA sequences may
be altered by using procedures such as restriction enzyme digestion, DNA
polymerase fill-in, exonuclease deletion, terminal deoxynucleotide transferase
extension, ligation of synthetic or cloned DNA sequences and site directed
sequence alteration using specific oligonucleotides together with PCR.
Accordingly, the invention also concerns a method for producing a
polypeptide involved in E. coli metabolization of 2-Deoxy-D-ribose. The method
comprises the steps of: (i) providing a cell transformed with a nucleic acid
sequence encoding the polypeptide positioned for expression in the cell; (ii)
culturing the transformed cell under conditions suitable for expressing the
nucleic
acid; (iii) producing the polypeptide; and optionally, (iv) recovering the
polypeptide
produced.
Once the recombinant protein is expressed, it is isolated by, for example,
affinity chromatography. In one example, an anti-deoK polypeptide antibody,
which
may be produced by the methods described herein, can be attached to a column
and used to isolate the deoK proteins. Lysis and fractionation of deoK-
harboring
CA 02388445 2002-05-31
17
cells prior to affinity chromatography may be performed by standard methods.
Once isolated, the recombinant protein can, if desired, be purified further.
Methods and techniques for expressing recombinant proteins and foreign
sequences in prokaryotes and eukaryotes are well-known in the art and will not
be
described in more detail. One can refer, if necessary to Joseph Sambrook,
David
W. Russell, Joe Sambrook Molecular Cloning: A Laboratory Manual 2.001 Cold
Spring Harbor Laboratory Press. Those skilled in the art of molecular biology
will
understand that a wide variety of expression systems may be used to produce
the
recombinant protein. The precise host cell used is not critical to the
invention. The
deoK proteins may be produced in a prokaryotic host (e.g., E. coh) or in a
eukaryotic host. These cells are publicly available, for example, from the
American
Type Culture Collection, Rockville, MD. The method of transduction and the
choice
of expression vehicle will depend of the host system selected.
Polypeptides of the invention, particularly short deoK fragments, may also
be produced by chemical synthesis. These general techniques of polypeptide
expression and purification can also be used to produce and isolate useful
deoK
fragments or analogs, as described herein.
Skilled artisans will recognize that a deoK polypeptide, or a fragment
thereof (as described herein), may serve for various purposes, in diagnostic
kits
and methods, and for the obtaining of anti-deoK antibodies for instance.
viii) Identification of Molecules that Modulate deoK Proteins Expression
deoK cDNAs may be used to facilitate the identification of molecules that
increase or decrease deoK genes expression. In one approach, candidate
molecules are added, in varying concentration, to the culture medium of cells
expressing deoK mRNA. deoK expression is then measured (or capabilities of the
cell to metabolize 2-Deoxy-D-ribose), for example, by Northern blot analysis
using
a deoK cDNA, or cDNA or RNA fragment, as a hybridization probe. The level of
deoK expression (or cell metabolizing activity) in the presence of the
candidate
molecule is compared to the level of deoK expression (or cell metabolizing
activity)
in the absence of the candidate molecule, all other factors (e.g. cell type
and
culture conditions) being equal.
CA 02388445 2002-05-31
18
Compounds that modulate the level of deoK expression (or cell
metabolizing activity) may be purified, or substantially purified, or may be
one
component of a mixture of compounds such as an extract or supernatant obtained
from cells. In an assay of a mixture of compounds, deoK expression (or cell
metabolizing activity) is tested against progressively smaller subsets of the
compound pool (e.g., produced by standard purification techniques such as HPLC
or FPLC) until a single compound or minimal number of effective compounds is
demonstrated to modulate deoK expression (or cell metabolizing activity).
The effect of candidate molecules on deoK-biological activity may, instead,
be measured at the level of translation by using the general approach
described
above with standard protein detection techniques, such as Western blotting or
immunoprecipitation with a deoK-specific antibody (for example, the anti-deoK
antibody described herein).
Another method for detecting compounds that modulate the activity of deoK
is to screen for compounds that interact physically with a given deoK
polypeptide.
Depending on the nature of the compounds to be tested, the binding interaction
may be measured using methods such as enzyme-linked immunosorbent assays
(ELISA), filter binding assays, FRET assays, scintillation proximity assays,
microscopic visualization, immunostaining of the cells, in situ hybridization,
PCR,
etc.
A molecule that decreases deoK activity is considered particularly useful to
the invention; such a molecule may be used, for example, as a therapeutic to
decrease and/or block proliferation of pathogenic bacteria (see section (v)
hereinbefore).
Molecules that are found, by the methods described above, to effectively
modulate deoK gene expression or polypeptide activity, may be tested further
in
animal models. If they continue to function successfully in an in vivo
setting, they
may be used as therapeutics to prevent or treat bacterial infections.
EXAMPLES
The following examples are illustrative of the wide range of applicability of
the present invention and is not intended to limit its scope. Modifications
and
CA 02388445 2002-05-31
19
variations can be made therein without departing from the spirit and scope of
the
invention. Although any method and material similar or equivalent to those
described herein can be used in the practice for testing of the present
invention,
the preferred methods and materials are described.
EXAMPLE 1: Cloning and expression of deoxyribose-catalyzing genes in
E. coli strains.
Introduction
Escherichia coli is a heterogeneous species consisting of both enteric
commensal and pathogenic strains. Different types of E. coli cause different
diseases in a range of hosts, including extra-intestinal and enteric
infections.
Extra-intestinal infections due to E. coli are common in groups of age and can
involve almost any organ or anatomical site. Typically extra-intestinal
infections
include urinary tract infection (UTI), meningitis (mostly in neonates and
after
neurosurgery), diverse intra-abdominal infections, pneumonia (particularly in
hospitalized and institutionalized patients), intravascular-device infection,
osteomyelitis, and soft-tissue infection, which usually occurs when the tissue
is
compromised. Bacteremia can accompany infection at any of these sites (JID
2000, 181:1753; JID 2001,183:596). In 1999, extra-intestinal pathogenic E.
coli
strains were the most frequently isolated organisms in US patients receiving
antimicrobials (JAMA, 2001, 285: 1565). Bacterial UTI are second in incidence
only to those causing respiratory infections. E. coli accounts for up to 90 %
of all
UTIs in non-hospitalized patients (5th ed. Williams & Wilkins, Baltimore,
Md.1997).
85 to 95 % of uncomplicated cystitis in pre-menopausal women are due to E.
coli
strains; they globally represent 150-300 million cases per year in the world
(Est. $
6 billion dollars direct cost/ year in US) (JID 2001;183:51). In US, there are
at least
250,000 cases of uncomplicated pyelonephritis per year, allowing to 100,000
hospitalizations and an E. coli estimate cost of $ 175 million dollars /year
(JAMA,
2001; 283:1583). E, coli is responsible for one third of all cases of neonatal
meningitis with an incidence rate of 0.1 per 1,000 live births (JAC 1994, 34
(suppl.
A):61). The extra-intestinal E. coli strains are epidemiologically and
phylogenetically distinct from both the commensal and the intestinal
pathogenic
CA 02388445 2002-05-31
strains; they appear to be unable of causing enteric disease, but they can
stably
colonize the host intestinal tract. In contrast, intestinal pathogenic strains
of E. coli
are rarely encountered in the fecal flora of healthy hosts and, instead,
appear to be
essentially obligate pathogens, causing gastroenteritis or colitis when
ingested in
5 sufficient quantities by a naive host. Various pathotypes of E. coli are
responsible
for significant worldwide diarrheal disease (to date, six have been well
characterized). For example, enteropathogenic E. coli (EPEC) are the leading
cause of severe infantile diarrhea in developing countries, and
enterohaemorrhagic E. coli (EHEC) (including the well-known 0157:H7) have
10 recently been shown to be the cause of bloody diarrhea and hemolytic-uremic
syndrome in major food-borne outbreaks in the United States, Europe, and Asia
(CMR 1998, 11:142). Although there is some overlap between certain
diarrhoeagenic pathotypes, with respect to virulence traits, each pathotype
possesses a unique combination of virulence traits that results in a
distinctive
15 pathogenic mechanism. Recent studies have identified other categories of
pathogenic E. coli, such as strains isolated from diarrhoeagenic stools of HIV-
positive patients, and E. coli that were abnormally predominant in early and
chronic ileal lesions of patients with Crohn's disease.
Knowledge of the pathogenic or non-pathogenic status of an isolate may be
20 of use for clinicians for diagnosis, especially in cases of opportunistic
pathogens.
Isolation of an E. coli strain from a clinical specimen does not, by itself,
confer the
designation of pathogenic isolate, since commensal strains of E. coli can
cause
infections (in particular extraintestinal infections) when the host is
compromised.
However, no single virulence factor is limited to (or absolutely required for)
infection at any one given site or for any particular syndrome. Consequently,
multiple phenotypic and genotypic assays are necessary to identify the
pathotype
of clinical isolates. The aim was to identify genes encoding functions that
are
conserved in all pathogenic strains but are absent in commensal E. coli and to
use
these data to develop new diagnostic and therapeutic tools.
Over the last five years, studies have been published on the E. coli
chromosome. The whole genome sequence of the laboratory strain K-12 MG1655
was published in 1997 (Science 1997, 277:1453), and the size of E. coli
CA 02388445 2002-05-31
?_ 1
chromosome was shown to var~~ from 4.5 to 5.5 megabases (Mb) (1A1 1999,
19:230). Comparative restriction mapping among the chromosome of E. coli K-12,
newborn sepsis-associated strain RS218, and uropathogenic strain J96, showed
that the overall gene order is conserved in the three strains, that large
accessory
segments (some carrying virulence genes) are unique to the chromosome of
pathogenic strains, and that some segments are only absent from the
chromosome of pathogenic strains (1A1 1999, 19:230). Comparison of the E. coli
K-
12 genome and those of different pathogenic E. coli allowed us to identify the
major differences. The genome of E. coli 0157: H7 (EHEC strain EDL933) was
recently sequenced (Nature 2001, 409:529). Comparison with the E. coli K-12
reference strain genome confirmed that the two chromosomes share a common
4.1 Mb 'backbone' sequence and lineage-specific segments (specific islands)
were
found throughout both genomes in clusters of up to 88 kilobases. Roughly 26%
of
the EDL933 genome lies completely within these specific islands, and 33% of
these contain genes of unknown function. The Genome Center of Wisconsin is
currently sequencing the genome of the newborn sepsis-associated strain RS218,
the uropathogenic strain CFT073 and three strains belonging to different
pathotypes of diarrhoeagenic E. coli [enterotoxigenic E. coli (ETEC), EPEC,
and
enteroaggregative E. coli (EAEC) (http://genome.wisc.edu)). It will take
probably
several years before information from the comparison of the pathogenic
specific
islands of various pathogenic E. coli isolates becomes available.
Most studies on pathogenic E. coli strains concern the identification of
specific virulence regions associated with the pathogenesis of E. coli causing
various diseases. Virulence genes have been identified, and pathogenicity
islands
have been characterized and sequenced. The first studies that investigated the
relationship between groups of pathogenic and non-pathogenic E. coli strains
were
based on multilocus enzyme electrophoresis analysis (1A1 1997, 65:2685) and
sequencing of housekeeping genes (Nature 2000, 406.64). They suggested that
pathogenic isolates do not have a single evolutionary origin within E. coli
but that
they arose many times and that the high virulence of clones is a recent,
derived
state resulting from the acquisition of virulence genes rather than an
ancestral
condition of primitive E. coli.
CA 02388445 2002-05-31
22
E. coli strains expressing the K1 polysaccharide colonize the large intestine
of newborn infants and are the leading cause of gram-negative septicaemia and
meningitis during the neonatal period. A recent study used signature-tagged
rnutagenesis to identify E. coli K1 genes that are required for colonization
of the
gastrointestinal tract, which is one of the initial steps in the development
of enteric,
urinary and systemic infections caused by E. coli (MM 2000, 37:1293). One of
these genes is absent from the genome of E. coli K-12, although related
sequences have been found in some representative pathogenic strains
(uropathogenic E. coli, EAEC, and EPEC). The sequence of this gene is not
available. These data strongly suggest that common (or strongly related)
sequences that are absent from the genome of commensal E. coli, are present in
all pathogenic E. coli strains.
A comparative analysis of metabolic functions expressed by pathogenic and
commensal strains of E. coli was developed. The inventors showed that a sugar
(deoxyribose) that is not fermented by E. coli K12, is metabolized by a large
number of pathogenic isolates belonging to various pathotypes. The inventors
identified the genes encoding this function and demonstrated that they are
conserved among several pathogenic strains. They have developed genetic and
bacteriological assays to identify deoxyribose-positive E. coli strains.
Materials and Methods
Bacterial strains, cosmids, and culture conditions
E. coli K-12/MG1655 (Blattner et al., 1997, Science 277:1453-1474) was
used as a host for maintaining cosmid clones.
E. coli strains were routinely grown in Luria broth with glucose (10 g of
tryptone, 5 g of yeast extract, and 5 g of NaCI per liter (pH 7.0] or on Luria
agar
plates (containing 1.5 % agar) at 37°C. E. coli-harboring cosmid clones
were
grown with 100 ~g of carbenicillin per ml.
Collections of human commensal and pathogenic E, coli strains were used
in this study. One hundred fifteen E. coli strains were isolated from blood
cultures
from cancer patients. These strains were previously partially characterized
(J. Clin.
Microbiol., 2001, 30:1738; Infect. A Immun., 2000, 68:3983). One hundred E.
coli
CA 02388445 2002-05-31
2'
J
strains were isolated from urine specimen from patients (children and adults)
clinically diagnosed with pyelonephritis. They were previously partially
characterized and were from various geographical origin (France, USA,
Romania).
Thirty six isolates were from urine specimen from patients with cystitis. They
were
isolated in Romania and USA. Twenty five strains were from the stools of
patients
with CD4 lymphocyte counts <400 cells/mm presenting persistent diarrhea.
Eleven
isolates were from diarrhoeagenic stools of children in Brazil. Commensal E.
coli
strains were isolated from normal flora of healthy people in France, Romania,
Senegal (children), and Central African Republic.
Expression of deoxyribose-catalyzing genes by E. coli strains.
The capacity of bacteria to grow on a minimal medium (K5) (J Bacteriol
1971, 108:639) supplemented with 2-Deoxy-D-ribose 0,1 % as sole source of
carbon was tested by inoculating agar plates with a bacterial suspension and
incubating the plates at 37°C for 24 and 48 h. Inoculations of those
plates were
performed with a loop from a 1 ml bacterial suspension (in water) prepared
with a
loop of bacteria grown on LB agar plates.
The fermentation (Methodes de laboratoire pour ('identification des
enterobacteries, 1e Minor et Richard, Institut Pasteur, p 169) of 2-Deoxy-D-
ribose
by E. coli strains was tested as follows: a drop (15 ~I) of an overnight
culture in LB
broth was inoculated in 3 ml of peptone water containing 1,5% (v/v) of
bromothymol blue and 1 % (w/v) of 2-Deoxy-D-ribose in a 12 x 120 mm glass
tube.
The suspension was incubated 24 h at 37°C without shaking.
Activity assay: 2-Deoxy-D-ribose is phosphorylated by deoxyribokinase to
deoxyribose-5 phosphate which is subsequently cleaved to acetaldehyde and
glyceraldehyde-3phosphate by deoxyribose-5P aldolase also called
phosphopentose aldolase. Deoxyribose-5P aldolase activity was determined by
coupling deoxyribose-5P cleavage to NADH oxidation using glycerophosphate
dehydrogenase and triosephosphate isomerase as coupling enzymes. The
reaction medium (0.5 ml final volume) contains 50 mM Tris-HCI (pH 7.4); 0.2 mM
NADH; 9U and 3U of glycerophosphate dehydrogenase and triosephosphate
isomerase respectively. The reaction was started with crude material extract
CA 02388445 2002-05-31
24
followed by 1 mM deoxyribose-5Phosphate, then the absorption decrease at 334
nm was monitored with an EppendortT"" PCP6121 photometer thermostated at
30°C. One unit of deoxyribose-5P aldolase corresponds to 1 mole of
product
formed per minute.
DNA analysis and genetic technigues.
Cosmid libraries were previously constructed from the genomic DNA from
E. coli AL862 isolated from the blood of a cancer patient (1A1, 2001;69:937)
and
from E. coli 55989 isolated from the stools of a patient with persistent
diarrhea
(C. Bernier, P. Gounon, and C. Le Bouguenec, In press, IAI august 2002). Sau3A
restriction fragments (35 to 50 kb) were sized on a sucrose gradient and
ligated to
the BamHl-digested and alkaline phosphatase-treated cosmid vector pHC79
(Collins J, 1979, Methods Enzymol., 68:309-326) DNA . The recombinant cosmids
pILL1272 and pILL1287 resulted from cloning of DNA from AL862 and 55989
strains, respectively.
Recombinant cosmids were routinely isolated by alkaline lysis. The
sequence of the primers to amplify probe A (GenBankT"" AF286671) and probe B
(GenBankT"~ AF286670) were derived from the partial sequence of PAI IA~ss2
(1A1,
2001 69:937, and Erratum in IAI June 2002). The sequences of the primers to
amplify probe A were 5'-ATCAGATGCCTAAAGAAGGAGAAAC-3' and
5'-CAATACTCGGATAAGATGATTGC-3' and the size of the amplicon was 831 by
(see Figure 4; SEQ 1D N0:11). The sequences of the primers to amplify the
probe
B were 5'-GGACGATAATGTGATCGTCTATAAG-3' and 5'-GTGGAAGA
TACTCATCTGCTACACG-3' and the size of the amplicon was 816 by (see
Figure 5; SEQ ID N0:12). The cycling conditions were initial denaturation at
95°C
for 5 min followed by 30 cycles at 95°C for 30 s, 60°C or
65°C (for amplification of
probe A and probe B, respectively) for 30 s, and 72°C for 1 min.
Hybridization.
Bacteria grown for 3 h on nitrocellulose filters were used for colony
hybridization. Hybridization was performed under stringent conditions
(overnight at
65°C), with PCR products labeled with 32P using the MegaprimeT"" DNA
labeling
CA 02388445 2002-05-31
system (Amersham International) as probes. The 100 ml hybridization solution
contained: 2 ml EDTA 0.5M; 20 mg ATP; and 10 ml 20x SSC.
DNA seauencina.
5 Double-stranded DNA was sequenced by Genome Express (France).
Multiple sequence alignments were generated with the CLUSTAL W program.
Statistical analysis
Proportions were compared by using the chi-square test.
Results
Presence of the deoK operon in the pathogenic E. coli isolates.
While a large number of bacteria are able to use the 2'-deoxyribosyl moiety
of 2'-deoxyribonucleosides as carbon and energy sources via the well-known
deo-operon, few organisms as Salmonella are able to use 2-Deoxy-D-ribose
(dRib) as the sole carbon source through deoxyribokinase which catalyses the
ATP-dependant phosphorylation of dRib to dRib-5 phosphate. Recently, the
inventors identified in the genome of S. enterica serovar Typhi, not only the
gene
encoding deoxyribokinase, deoK but a whole operon (deoK operon) of three genes
regulated by a repressor DeoQ (J. Bacteriol., 2000, 182:869-873). Searches in
databanks showed that this operon was fully represented in one Citrobacter
freundii strain and partially present in Agrobacterium tumefaciens,
Rhodobacter
sphaeroides, and the pathogenic E. coli strain AL862 isolated from a blood
culture.
Use of 2-Deoxy-D-ribose by E. coli strains has been previously described (Br.
J.
Biomed. Sci., 1995; 52: 173), however this property was never associated with
the
pathogenic status of the strains and the genes encoding this function were not
identified.
In strain AL862, the sequences similar to the deoK operon corresponded to
ORF3', ORF4, ORFS and ORF 6 of the partial (and not continuous) sequence of a
pathogenicity island (PAI IA~ss2)(GenBankT"" Nos. AF286670 and AF286671). No
function was previously assigned to these sequences. Two probes derived from
this PAI IA~862 region (probes A and B) corresponded to the deoK homologous
CA 02388445 2002-05-31
2C
sequences. Analysis of the distribution of PAI IA~as2 among pathogenic E. coli
isolates strongly suggested that the A and B regions are widely distributed
among
pathogenic strains (1A1, 2001, 69: 937-948; IAI June 2002 Errata).
To confirm the presence of the deoK operon in pathogenic E. coli strains,
the inventors sequenced again the region of PAI IA~asz that previously showed
similarities to the deoK operon of Salmonella. The sequencing was performed on
the recombinant cosmid pILL1272 (see Material and Methods). They identified a
4486-pb linear region displaying similarities to the entire deoK operon of
Salmonella. Computer analysis revealed four open reading frames (ORF), deoX,
deoP, deoK, and deoQ, which mapped to the same loci as had similar sequences
to the deoX, deoP, deoK, and deoQ genes from the deoK operon from Salmonella,
respectively (See Figure 1 ). These results confirmed that the genetic
organization
of the deoK operon from E. coli was similar to that of the deoK operon from
Salmonella.
The detailed sequence analysis of E. coli - strain AL862 is presented in
Figure 2. The deoK operon from E. coli strain AL862 displayed 78 % identity
with
that from Salmonella (4486 bp14517 bp).
The position and sequence (determined here) of the two probes (probe A
and probe B) that were used in the hybridization experiments are indicated in
Figure 2 (single and doubled underline respectively). In both cases, the
sequence
of the primers used in PCR assays are indicated in bold. These primer
sequences
are identical to those previously described and used (IAI, 2001, 69:937-948;
IAI
June 2002 Errata). Probes A and B are PCR products obtained from strain AL862.
To study the degree of conservation of the deoK operon among pathogenic
E. coli isolates, the inventors determined the nucleotide sequence of the deoK
region in E. coli strain 55989 isolated from the stools of a patient with
persistent
diarrhea. This isolate was shown to belong to the EAEC pathotype of pathogenic
intestinal E. coli. A cosmid library from the genomic DNA of strain 55989 was
previously constructed (Bernier et al., In press, IAI August 2002). The
recombinant
cosmid pILL1287 resulted from the screening of the 55989 cosmid library with
both
the probe A and the probe B. The sequence of the chromosomal region from
strain
55989 that carries the deoK operon is presented in Figure 3.
CA 02388445 2002-05-31
27
The deoK operon from E. coli strain AL862 and strain 55989 showed 98%
identity (4486 bp/4489 bp). The degrees of identities of the deo genes from E.
coli
and Salmonella strains are summarized in Table 1.
TABLE 1: Degrees of identities of the deo genes from E, coli and Salmonella
strains
Strains % of identity No. of nucleotides
55989 / AL862 98 % 4489bp/4486bp
55989 / S. Typhi 78 % 4489bp/4517bp
AL862 / S. Typhi 78 % 4486bp/4517bp
Genes % of identity No. of nucleotides
deoX 55989 / AL862 99% 1014bp/1014bp
deoX 55989 / S. Typhi75% 1014bp/1014bp
deoXAL862 / S. Typhi75% 1014bp/1014bp
deoP 55989 I AL862 99% 1317bp/1317bp
deoP 55989 / S. Typhi83% 1317bp/1317bp
deoP AL862 / S. Typhi82% 1317bp/1317bp
deoK 55989 / AL862 99% 921 bp/921 by
deoK 55989 / S, Typhi80% 921 bp/921 by
deoK AL862 / S, Typhi80% 921 bp/921 by
deoQ 55989 / AL862 96% 783bp/783bp
deoQ 55989 / S. Typhi77% 783bp/786bp
deoQ AL862 / S. Typhi76% 783bp1786bp
Expression of the deoK operon in E. coli strains.
The inventors demonstrated the expression of the deoK operon in clinical
isolates 55989 and AL862, as well as in the recombinant strain MG1655 carrying
either the cosmid pILL1272 or the cosmid pILL1287. All these four strains were
able to grow on K5 plates containing 2-Deoxy-D-ribose as a carbon source. The
growth of the strains was evident after 48 h of incubation at 37°C. As
a negative
control, strain MG1655 alone did not grow on such medium. Deoxyribose-5P
CA 02388445 2002-05-31
28
aldolase activity, easier to determine than that of deoxyribokinase, is
reported in
Table 2.
Table 2: Deoxyribose-5P aldolase activity in E. coli strains
Strain Deoxyribose-5P aldolase
+dR -dR
AL862 0.47 Ulmg 0.06 U/mg
55989 0.45 U/mg 0.08 U/mg
K-12 MG1655 (+1272)0.36 U/mg 0.10 U/mg
K-12 MG1655 (+1287)0.24 U/mg 0.10 U/mg
Analysis of the distribution of deoK operon among commensal and pathogenic E.
coli isolates
To determine whether deoK operon sequences were specific for pathogenic
E. coli, the frequency of occurrence of the A and B regions (corresponding to
parts
of deoK and deoX genes, respectively) was investigated. These regions were
amplified from strain AL862 DNA and used as probes to screen by colony
hybridization collections of E. coli isolates. The strains were also tested
for their
ability to use 2-Deoxy-D-ribose as a carbon source.
These collections comprised strains representative of the various
pathotypes of pathogenic E. coli. Archetypal ExPEC (extraintestinal pathogenic
E.
coh~ familiar to investigators in the field include strains CFT073
(pyelonephritis
isolate), 536 (pyelonephritis isolate), J96 (pyelonephritis isolate), RS218
(neonatal
meningitis isolate). Prototype strains of the various diarrheagenic E. coli
pathotypes are also considered: EDL933 (EHEC), EDL1493 (ETEC), E2348/69
EPEC), 042 and JM221 (EAEC), C1845 (diffusely-adherent E. coli (DAEC)). As
shown in Table 3, the results indicated that the deoK operon is carried by
pathogenic strains belonging to various pathotypes of E. coli and associated
with
both extra-intestinal and intestinal infections.
CA 02388445 2002-05-31
29
Table 3: Frequency of occurrence of the A (deoK) and B (deo~ regions in
various E, coli strains
E. coli strains Probe Probe Deoxyribose utilization
A B
CFT073 (pyelonephritis) + + +
536 (pyelonephritis) + + +
J96 (pyelonephritis) - - -
RS218 (meningitis) - - -
EDL933 (EHEC) - - -
EDL 1493 (ETEC) + + +
E2348/69 (EPEC) - - -
042 (EAEC) + + +
JM221 (EAEC) + + +
C1845 (DAEC) - - -
The collections studied also comprised clinical isolates from 115 human
with septicemia (isolated in France), 100 clinical isolates from patients with
pyelonephritis (origin France, USA, Romania), 36 clinical isolates from
patients
with cystitis (origin USA, Romania), 25 EAEC isolated from HIV-positive
patients
with persistent diarrhea (origin Central African Republic and Senegal), 11
EPEC
with a diffuse adherent pattern (DA-EPEC) on epithelial cells isolated from
infants
with diarrhea in Brazil. We also investigated 257 commensal E. coli strains
isolated from normal flora of healthy patients (origin France (36), Romania,
Senegal, Central African Republic). The results are summarized in Table 4.
CA 02388445 2002-05-31
Table 4: Percentage of occurrence of the A (deol~ and B (deo~ regions in
various E, coli clinical isolates
E. coli strains Probe Probe Probe DeoxyriboseProbe A
A + + 2-
A B Probe utilizationDeoxy-D-
B
(level of ribose
significance)utilization
Septicemia 49 48 48 50 46
(n = 115) (p<0.0001
)
Pyelonephritis 50 53 48 50 48
(n = 100) (p<0.0001
)
Cystitis (n = 7 10 7 8 7
36)
(0.2<p<0.4)
Diarrhea (EAEC) 13 13 13 12 12
(n = 25)
(p<0.0001
)
Diarrhea (DA-EPEC)11 11 11 11 11
(n - 11 ) (NA)
Commensal (France)10 11 10 9 8
(n = 36)
Cornmensal ~ NT NT NT 31 NT
(Romania, Senegal,
Central African
Republic) (n =
221 )
NT, not tested; NA, not appiicaoie.
5 The sensitivity of the two DNA probes appeared equivalent: 43%, and 45%
of the strains were positive with the A and B probes, respectively.
A total of 147 isolates (36 commensal strains and 113 pathogenic E. coli )
were tested for both the growth on K5 plates containing 2-Deoxy-D-ribose and
fermentation of this sugar. A 100 % correlation was observed between the two
10 bacteriological tests; all the strains that grew on K5 plates with 2-Deoxy-
D-ribose
showed the ability to ferment the sugar. The 2-Deoxy-D-ribose utilization test
appeared sensitive but, at a small extend, less specific than the genetic
detection
of the deoK operon (53 % of positive strains). Using both molecular and
CA 02388445 2002-05-31
31
bacteriological approaches (probe A and growth on K5 plates with deoxyribose)
a
total of 40.8 % of the strains are positive.
Taking account of all the data, a significant association of the deoK operon
with pyelonephritis- and septicemia-associated isolates, as well as with
diarrhea
associated EAEC isolates was evidenced.
Conclusion
This work confirmed that metabolic characters may be specific of E. coli
strains and that those expressed by pathogenic isolates may be considered as
virulence-associated factors. Utilization of 2-Deoxy-D-ribose by some E. coli
isolates has been previously reported. Here, the inventors identified the
genes
involved in utilization of 2-Deoxy-D-ribose by E. coli strains. These genes
are
organized in an operon (deoK) that is highly related to that previously
identified in
Salmonella enterica strains. Analysis of the sequences adjacent to the deoK in
several E. coli isolates and in Salmonella strongly suggested that E. coli
strains
acquired the deoK operon by horizontal transfer from Salmonella strains. The
inventors demonstrated that the deoK operon is highly conserved among E. coli
strains. From this observation, the inventors defined two probes that were
used to
study the distribution of the deoK operon among collections of commensal and
pathogenic E, coli isolates. Preliminary studies indicated an association of
the
deoK operon with strains belonging to various pathotypes of E. coli including
strains causing pyelonephritis, septicemia, and some type of diarrhea in
children.
If 40 to 50% of strains associated with pyelonephritis, septicemia, and
diarrhea (EAEC and DA-EPEC strains) carry the deoK operon, we also detected it
in 14 to 22 % of commensal isolates. This may be explained by the fact that
commensal strains of E. coil can be potential pathogens when the host is
compromised. It is interesting to note that the deoK operon is less prevalent
in
commensal strains from Romania, Senegal and Central African Republic than in
French commensal strains.
In conclusion, the inventors have identified a metabolic character
significantly associated with some pathogenic E, coil. The inventors have
developed bacteriological and molecular tests to identify strains expressing
this
CA 02388445 2002-05-31
32
character. These tests could be associated with others in a future diagnostic
kit for
the identification of the pathogenic status of an E. coli isolate.
While several embodiments of the invention have been described, it will be
understood that the present invention is capable of further modifications, and
this
application is intended to cover any variations, uses, or adaptations of the
invention, following in general the principles of the invention and including
such
departures from the present disclosure as to came within knowledge or
customary
practice in the art to which the invention pertains, and as may be applied to
the
essential features hereinbefore set forth and falling within the scope of the
invention or the limits of the appended claims.
CA 02388445 2003-07-09
2003-07-08 Listage pour 1e BdB corrige.txt
SEQUENCE LISTING
(1) GENERAL INFORMATION;
(i) APPLICANT:
(A) NAME: Inst:il:ut Pasteur
(B) STREET: 25-:?8 rue' du Docte~ur Roux
(C) CITY: Paris
(E) COUNTRY: France
(F) POSTAL CODE (ZIP): 75724
(ii) TITLE OF INVENTIC)N: Genetic markers, metabolic markers, and
methods for evaluating pathogenicity of
strains of E.coli
(iii) NUMBER OF SEQUENCES: 1?.
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: RobiC
(B) STRE:E:T': 55 St-J'acqueS
(C) CTTY: ~l~ont,real
( D ) STAT ~; . QC
(E) COUNTRY: Canada
(F) ZIP: H2Y 3X<?
{G) TFLEPH~~IVE: 'i19-987-6242
(H) 'fELEc'A.k: 514-895-7874
( v ) COMPU'1 ER RF~ADABLI=, FORM
(A) MhDTUNi TYPE:: ~?:i.sk 3.5" / 1.44 Ml3
(B) COMPU'CER: Tt=,M ?C compatible
{C) (:)J?ERA~C:ING S'~S'fh)M: fC-DOS/MS-DO:,
(D) SOFTWARE: T:~'I' ?,SCI
(vi) CURREN'P APPLICAT.CON DATA:
(A) Al?PLIC:ATION 'JCJMEIER: 2.388.945
(B) r':CLING DATE: :32 May 2002
( 2 ) INFORMATI02J FOR SEQ I L; J() : 1
(i) SEQUEPJCE CHARACTER:CSTICS:
(A) LHNGTH: 4489 nucleotides
(B) "..""PE: nucle:_c: ~:xcid
(ii) MOLECULE TYPE: D~dA
(vi) ORIGICJ~1L SG(JRCE:
(A) ORGANISM: Escherichia coli
(B) STRAIN: 5'i9t39
( xi ) SEQUENCE DESCRI P'I':I C)PJ : SEQ I D NO : l
ggacgataatgtgat:cgtctatangcJgcaacgctatcatagt:cttgtcctg'gcgggtaaa60
aaagcgcgcttaccta ataagcgcgccgctgttcaggccttgagtggttattcaat 120
aacg
tcctgtggtgactgt:aaaagtgcclcdt:ttgcr_gcggtgcaacctgaatcagcgtgccatt 180
acgttgcgcggcaactatacc:ccta:a<7gccgacaggttgcaggtaatgcaaaggcggctac 240
ctgttgctctccgt:t:ataaaggatcc::aagc~~Itgtcac:ataattta<Ittcagcactgtagaa 300
acgagtaacaaacgt.agtgccatc:gggagagatcat~g~-gaaactctggctgatctgtata 360
agcgtccagtttgt.cagcaaaga~:uJac.;aat:t:tctggaJ:cat.aaaattccggttgactcag 920
cgtcgacagagaggcatctcCCtgCdfaatccgttgattaaacgccagccactgagcggt 480
gggattaacatgcgaggcactgat:tcacgcaatct:taat:at.t r_<:gtccgggatattctg 540
gctgaatgtagcat.t.tggtatatat-_qr:ataattcatgtggcacatatattgtagtggcat 600
atctacagaagccactattggttar.Jclc:catcataat:atr_c~aacagtgta3gaggatttgtg 660
aaggaccactgttgcrttgagccac~:~t:aatgatgaccgaaacccattacatactcgtaacg 720
Page:
1
CA 02388445 2003-07-09
2003- 07-08 BdB corrige.txt
Tistage
pour
1e
ccggttaaggcgtaacatatctc:c,gtctaat.accagc~catget:tcatccatcgcggcaca780
ggccatttcaccgtgtagcagat:gagtat:cttccgcagatgggcagccattagccagcaa840
acctgaatgaaaagcaaaacagccataggtctctatcacctctgtcgccggtttaggctg900
gcgaaacatattgcacatggtgac~gccgt:gt.ccat:caaattgc:gcatc:ccaaatcatctg960
ccccatccagggaagaataatcaaaat:gtcc:ac:gactc3tt.tgcaatttt:aagcccctcgac1020
accgctgtcatagcgaaaagacgi:gacagt.aaaatcactattt.:tccagcaagatacgagg1080
tttctcgccaaaaagcgcccgcc<zcaaat:t:aatacgcgtactc.,at:aacggttctcctcag1140
gacgctgtgacttcagcc::3gtgcc;gt:accttacattgc:tttcac.;gccagaagtagactccg1200
acatagacaaagcagagcatagaaaccaggaatgaaagctgtagtgagtggaacatatct1260
gcaatatatccctgaattgccggaaccaccgcggcaccgacaatagcc:ataacaatgact1320
gctcctgccatttctgtat:gt:tegtt:atcaacagtatccagtctttcct:gcatagatcgtc1380
gcccagcaagggccaaacaaaae,act:taccaggacggr_gacat:agaccgcgctgaaactt1440
ggagccagtgcaacatatgccaggaacagcgcccctataacggaatagagaatcaatact1500
ttttccggattaaaacgcgtcataaggat.gtt:ggctataaact:tc~ccaat:aaagaagcag1560
gcaaagctatagac:catg<3ac~tt:t:gaagcatcacgttcgttgatatcgc~~caactccagc1620
gccagacggatggtaaatgaccatactgc.gacctgcatacccacataaaggaactgcgcc1680
acaataccgcgacc~aaagcgcggatt.tca:agccagat.agcgcaigcgt<it<:cattgctgac1740
gggcgtttatagt~acttc~tctgtc;<~~racattacagc~ti:gggaagcgggttaaaaggaac1800
aacaccatgaccacaaccaaatcataatcatatact.tataccTgttcaagggtgttctct1860
g
aacatcagcacctt:aaagt:tgtga,nt:tt:gctcggcgtt:cattcvcggacatc;t:gcttctca1920
aggctttccccctc:ggag<saaaccagatat:t:tgcccaataaaataccagacgcagcacca1980
atcggataaaaggtctgg<.tgatattgagc:cgcaatgtggcat:aggct.tctggaccgatc2040
attgaactgtatgtgttcc~ctgc:agtttcaaggaaactcaggccaatcgc;aatcgcaaaa2100
atagctgcaagga._i::ata<.~tgtactgt:t:gcc:at;atgcga~ggcac~ggaaaaaaagtgtacaa2160
ccaccaatatacagcgtcagccaattaaaattgccaccatat.aactggtctttttaatc2220
g
acaagggatgctggtattgcaatt:aaaaaataacctccataaaatgcgctcagcaccaat2280
gctgaagcaaagtt~ctt;~gcgaaaatacact:ttt:gaattgacttgattaat~atgtcattt2340
aatgcagctgcgc~tcccc:atagc:gggaataaacacgataac~aaaataaactggaacaag2400
ggagtcttattcag.stac<:catr._cggcatctgaatgatgtttt.tatcgttcatagtgcta2460
cctttaactgtgca~~gat<~at:tatt.cgti,taaggttaaaaatt.c,attaaai:t:gttcaata2.520
ctcggataagatg,~ttgccttacct tt gtgacgct:gaaacrcggcaaaa<lagagcggct2580
ccc:t
tttttcaaagcggcttcaacatc.;cccgctttgaacataataatgggaaaagcaaccaata2640
aatgcgtcaccagc:gccac::tagt,utcaa<::agcat:ttactttgaatgcagctaacatgga<a2700
tcctgatcgcgggt~~atcc:ataatgcgcca:t.tttc:gctcatc~gtaacaat:aatattgtt:c2760
agccctttatcaactaacgaacgt:gcggccaaacgaatatgatcataagt:atcaaccgac2820
ataccggttaatatatccagttct<~r.ttcattcgggataaagaaatcacat:ta gcaggca2880
taagacatatcta:a..-
.tcar:gc:aat.g;:~.~:ggagccggatt:taa~aac:acttc::aataccattt2940
ttcttaccaaacti::~atcc7cgtgcat.aaac:.tgtttccac3ttg:3acttccac~tagtaaaacg3000
atcaatttgcattTvttcagatcttctgcagctcgatcgarat.cttccgc~ggaaagaaat3060
ttattcgctccct~::aattatt:aa~atartattgctcgagtt:ggcattaac::~aagatcggt3120
gcaacaccactgc,:ggtacagggr~a::'~~t:tctcaac_-ataagtgqtattaat:t:ccccatgat3180
tcaagattacgaat:agtai.tatccg~caaaaatatcatcacctactttagt:cagcatcagg3240
acttttgaattca<.u~ttacaccgc~gwcac:r.gcttgatt:agcacctttccc:accacatccg3300
attttgaaggcag<~~:gcti:cag<sg-t cctt:.ttt:aggc~atctgattagtgtaagt:a3360
~~ wtc:t
atgagatccaccat~attgc,aaccaataac.tgcaatgt~catttcactacctcttataaac3420
tttcgcataacaat::c~gtat:ttaa:~t;~.~c:att:agcatgt.tact:tttgcatcatttgtgac:t3480
gagatcgcgattac,c:acat:caacc:c~at::gt.Matt taatagactr_ccagtctcatcactc3590
aggccaacactat<~t:aatc:ataagcaacctaacaagattagtgcccaaaactcagcagcc3600
tataccctttcatttcaaagggcycc~gtcgtatagtat.ggr_:.atgaaaac:aatgtttact3660
t
aacgccaaaatgti::atttt:tata:~c:~r_t.cttacggagaga~fiagtl:gatgctaa.acgaagc:a3720
aaaagagcgtatccgacgtttgatggaeactgcttaag:~aaa:.cgacagaatccatttgaa3'780
agacgcagcgcgaai:gctcrgaagt_vct.gtaatga,~tattc:;tcgcgatctccatcagga3840
agatgaacct ctgcc,actcaaccci:.-
ic:.:t:c;ggv:ggcar_attctt:aai:ggtg~~ataaacccgc3900
gccatccatgcca<xt:aatc:c~atga:gi~t:cc<3aaa<iatc;at.ytgatgactt: acctattgc3960
aattctggctgccggaatggttaatgaaaatgat.r_-.tg~,t.ctt:ctttgatGatggccagga4020
gataccactcgtt<_t:aagcatgat:-:c::;;gg~-
~tgcaatc::cctt:caccggc~::t:cagtt:acts4080
acatcgcgtcttt<;i:tgcgtt:gaatca<3aaagcctaatgt:a_~::ag.aatac:t:ttgtggtgg4140
tacgtatcgtgccagaagt.gatgc:tl~tt.tacgatgccagtaactcttcgc:cattagactc4200
tctcaatccgcgaaaaatatttat:ti=c-cgccagcggtgtgcataatcactttggcgtcag42.60
ctggtttaaccctgaagat:cttgcca~t.aagcgt:~iaacxcga':gaaccgtggactacggaa4320
aattttgctcgcccqccacgcgt~:gt!=c:gat_gaag':ggcct:ag.~~cagcrt:cgcaccgat4380
ctctgcatttgacqt_tctgattactcc~atcc~i:ccg~taccggcagattatc_~ttacgcactg4440
ccagaatggttctctt:aaagateat:ta<,acctgat:tcaaa~s;rricg.,~atga 4489
Paste 2
CA 02388445 2003-07-09
2003-07-~~i8 Listage pour 1e BdEj corrige.txt
(2) INFORMATION FOR SEQ TC: NO; 2:
( i ) SEQUE;~IOE CF~ARACTE.',RI STICS
(A) LENGTH: 33~ am:irw acids
(B) 'TYPE: amino aci ci
( D ) '"~JPOLOGY : l ine~a:r
(ii) MOLECJLE TYPE: protein
(xi) SEQUEiVCE DESCRIPTION: SEQ ID N0;2:
Met Ser Thr Arg Ile Asn Le:u Trp Arg Ala Leu Phe Gly G:Lu Lys
10 15
Pro Arg Ile Leu Leu Glu Asn Sex Asp Phe Thr Val Thr Ser Phe
20 25 30
Arg Tyr Asp Se:r Gly Val. GLu Gly Leu Lys Ile A.la Asn Ser Arg
35 90 45
Gly His Leu Ile Ile Leu P:ro 'Prp Met Gly Gln Met. Ile Trp Asp
50 55 60
Ala Gln Phe Asp Gly His GLy Leu Thr Met Cys Asn Met Phe Arg
65 70 '75
Gln Pro Lys Pro Ala 'rhr GLu 'dal Ile Glu Thr Tyz Gly Cys I'he
80 85 90
Ala Phe His Sexy Gly Leu L~.u Al.a Asn Gly C;ys Pra Ser A).a Glu
95 100 105
Asp Thr His Leu Leu His G.y "1.u Met ALa Cys Ala Ala Met Asp
110 115 120
Glu Ala Trp Leu Glu Leu Asp c~l.y Asp Met Leu Arg Leu Asn Arg
125 130 135
Arg Tyr Glu Ty° Val Met G:Ly L?he ~~:Ly His His Tyz :Leu Ala Gln
140 115 150
Pro Thr Val Va.Leu His Ly:per Ser Thr lieu Phe Asp Ile Lys
155 160 165
Met Ala Val Thz: Asn Leu A'a :ver Val Asp Met Pro Leu Gln Tyr
170 1'75 180
Met Cys His Met Asn Tyr A:_a 'tyr Ile Pro t~sn Ala Thr Phe Ser
185 190 195
Gln Asn Ile Pro Asp Glu Ia_e Leu Arg Leu Arg Glu Ser Val Pro
200 205 210
Ser His Val Asn Pro Thr A1_a Gln Trp Leu Ala Phe Asn Gln Arg
215 220 225
Ile Met Gln Gly Glu Ala Seer Leu Ser Thr Leu Ser Gln Pro Glu
230 23ti 2.40
Phe Tyr Asp Pro Glu Ile Val Phe Phe Ala Asp Lys Leu Asp Ala
245 250 255
Tyr Thr Asp Gln Pro Glu Phe Arg Met Ile Ser Pro Asp Gly Thr
Pan°
CA 02388445 2003-07-09
2003-07-08 L:istage pour 1e BdB corric~e.txt
260 265 X70
Thr Phe Val Thr Arg Phe Tyr Ser Ala G1u Leu Asn Tyr Val Thr
275 280 285
Arg Trp Ile Leu Tyr Asn G~y Gl~_z Gln Gln 'Tal Ala A1a Phe Ala
290 2G5 300
Leu Pro Ala Thr_ Cys Arg Pi:o ~;,1u Gly Tyr :~~eu Ala Ala G:Ln Arg
305 310 315
Asn Gly Thr Lea Ile Gln V~s.l. A.L~a Pro G~n Gln 'z: hr Arg Thr Phe
320 325 330
Thr Va1 Thr Thr G:Ly Ile Gl a
335
( 2 ) INFORMATION FOR SEQ I U L~,TO : 3 :
(i) SEQUENCE CHARACT~:RISTICS:
(A) .'..:~_'.NGTI-1: 43 . a:ni.no acids>
(B) '1'~'PE: amino acid
(D) 'TOPOLOGY: l.im~ar
(ii) MOLECC1:~E TYPE: peotr~-~n.
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:3:
Met Asn Asp Ly:> Asn I1e IIe Gl.n Met Pro Asp (:,1y Tyr Leu Asn
1 C? 7. 5
Lys Thr Pro Let:c Phe Gln Phe Ile heu Leu Ser Cys Leu Phe Pro
20 25 30
Leu Trp Gly Cys Ala Ala A.la L~eu Asn Asp IIe l,eu Ile Thr Gl.n
35 40 45
Phe Lys Ser Val Phe Ser Le>u Ser Asn Phe Ala Ser Ala Leu Val
50 55 60
Gln Ser Ala Phe Tyr Gly GLy 'Cyr Phe Leu :Ile Ala Ile Pro Ala
65 70 75
Ser Leu Val IlELys Lys Thr Ser Ty.r Lys Val A_La Ile Leu Ile
80 85 90
Gly Leu Thr Leu Tyr Ile G.~y ~::~l.y Cys Thr L~eu Phe Phe Pro A:La
95 100 7.05
Ser His Met Ala Thr Tyr Thr Met Phe Leu .Ala A'__a :Ile Phe A1a
110 17.5 120
Ile Ala Ile Gly Leu Ser Phe L.eu Glu 'I'hr Ala Ala Asn 'Phr Tyr
125 130 135
Ser Ser Met Ile Gly Pro Lys Ala Tyr Ala Thr I,eu Arg heu Asn
140 145 150
Ile Ser Gln Thr Phe 'ryr Pro Ile G'__y Ala ;Ill.a Sc~~r c;ly Ile Leu
155 160 165
Leu Gly Lys Tyr Leu Val Phe :3er t~Lu Gly ~.:;la Ser heu Glu hys
170 175 180
Page 9
CA 02388445 2003-07-09
2003-07-08 Listage pour 1e HdI3 corrig~.txt
Gln Met Ser Gly Met Asn A;~a Glu Gln Ile His Asn Phe Lys Val
185 190 1.95
Leu Met Leu Glu Asn Thr I:eu Gl~.i Pro 'Pyr Lys Tyr Met Ile Met
200 205 210
Ile Leu Val Va:L Val Met Va.l. Leu Phe Leu Leu Th~~ Ar.g Phe Pro
215 220 225
Thr Cys Lys Val Ala Gln Thr Ser Hips Tyr Lys Ar<I Pro Ser Ala
230 235 240
Met Asp Thr Leu Arg Tyr Leu A:La Arg Asn Pro Arg Phe Arg Arg
245 250 2.55
Gly Ile Val Ala Gln Phe Leu Tyr Val Gly Met Gln Val Ala Val
26U 265 270
Trp Ser Phe Th.r Ile Arg L~~u .Ala Leu G _u Leu G.Ly Asp I:Le Asn
275 280 <'?85
Glu Arg Asp Al,a Ser Asn P~~e Met: Va1 Tyr Ser .P.he Ala Cys Phe
290 295 300
Phe Ile Gly Ly;_: Phe Ile Al.a .Elsru Ile Leu Met T:hr Arg Phe Asn
305 310 315
Pro Glu Lys Va.L Leu Ile L,~_~u 'I'yr Ser Va 1 1~ 1e G.Ly Ala Leu I'he
320 325 330
Leu Ala Tyr Va:1 Ala Leu Ala Prc; Ser Phe Ser A.1G Val Tyr Val
335 340 345
Ala Val Leu Va:l. Ser Val Lf~~u ahe Gly Pro Cys Trp Al.a Thr ILe
350 355 360
Tyr Ala Gly Th:~: Leu As.p T!,.r 'Jal. Asp Asn C~lu ,H:i~. Thr G~.u Met
365 370 375
A1a Gly Ala Va:L Ile Val Mrt Ala .I: 1e Va 1. Gi y .A:1 G Ala Val Val
380 385 390
Pro Ala Ile Gln Gly Tyr I:Le AI_a Asp M~:et E?he Eli. Ser Le~u G1n
395 400 405
Leu Ser Phe Leu Val Ser Met Leu Cys Phe Va1 Tyr Val G1y Val
410 415 420
Tyr Phe Trp Arg Glu Ser Lays Val. Arg Thr Ala Leu Ala Glu Val
425 4?~0 935
Thr Ala Ser
(2) INFORMATION FOR SEQ ID t4(:): 4:
( i ) SEQUEPdC:E CHARACTE~~R:I,:p'I'IC::>
(A) LENGTH : 30Ei <~~rli rio acids
(B) T"pE: ami.no a<:;.d
(D) TOPOLC%GY: l_rn~ar
Page 5
CA 02388445 2003-07-09
2003-07-C)8 Listage pour ie BdB corrige.txt
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIP'.I'ION: SEQ ID N(7:4:
Met Asp Ile Ala Val Ile G__y Ser Asn Met Val Asp Leu Ile Thr
1 Ci 15
Tyr Thr Asn Gln Met Pro Lys Giu Gly Glu Thr Leu Glu Ala Pro
20 2'e 30
Ala Phe Lys Ile Gly Cys Gly G:ly Lys Gly Ala Asru Gln Ala Val
35 90 45
Al.a Ala Ala Lys Leu Asn Seer hys Val Leu Met Ge~.x Thr Lys Val
50 55 60
Gly Asp Asp Ile Phe Ala Asp Asn '('hr Ile Arg Asn Leu G.Lu Ser
65 70 75
Trp Gly Ile As.n Thr Thr 7'yr Val. Glu Lys Val Pro Cys Thr Ser
80 8J 9O
Ser Gly Val Al~a Pro Tle Prze V<31 Asn Ala Asn Ser Ser Asn Ser
95 100 105
Ile Leu Ile Ile Lys Gly Ala .?~sn l.ys Phe Leu Ser Pro Glu Asp
110 1:15 120
Ile Asp Arg Al,a Ala Glu A=;p Leu Lys Lys C:ys Lya Leu Tle Val
125 130 7.35
Leu Gln Leu Glm Val Gln L~=~u. Glu Thr V<~1 Tyr HL> Ala I~.e Glu
140 145 7.50
Phe Gly Lys Lys Asn Gly Ile ~:~l.u Val Leu Leu Asn Pro Al.a Pro
155 160 1.65
Ala Leu Arg Glu Leu Asp M~_~t Ser Tyr Aia Cys Lys Cys Asp Phe
170 1'75 1.80
Phe Ile Pro Asn Glu 'Phr G:Lu Leu Glu Ile Leu Thr Gly Met Ser
185 190 195
Val Asp Thr Tyr Asp His I:Le Arg Leu Ala Ala Arg Ser Leu Val
200 205 2:10
Asp Lys Gly Lets Asn Asn I:le Ile Val Thr Met Ser Glu Lys Gly
215 220 225
Ala Leu Trp Met Thr Arg Asp Gln Glu Va1 His Val Pro Ala Phe
230 23'p 240
Lys Val Asn Ala Val Asp Thr Ser Gly Ala Gly A.>p Ala Pr:e Ile
245 250 255
Gly Cys Phe Sei: His Tyr Tyr_ ~~al Gln Ser G1y Asp Va1 Gl.u A1a
260 265 270
Ala Leu Lys Ly: Ala Ala LEe;a !?7e Ala A.la E'he S~e.r 'Ja1 Thr G1y
275 287 295
Lys Gly Thr Glr~ Ser Ser Tsr~:~ -?ro rer I.le :'~lu C: n P:ze .As;n C~Lu
290 295 300
Paqo
CA 02388445 2003-07-09
2073-07-f78 I,istace pour 1.e Bd3 corrige.txt
Phe Leu Thr Leu Asn G1u
305
(2) INFORMAT2CN FOR SEA TI:) LSO:
(i) SEQUENCE CHARACT1~RIST:fC~:
(A) LENGTH: 261) amino acids
(B) 'TYPE: amino amid
(D) TOPOLOGY: .. inear
(ii) MOLECCJLE 'T'IPE: Li:°<,tvei.n
(xi) SEQUENCE D:F~SCRIP'f'ION: SEQ ID N0:5:
Met Glu Thr Lys Gln Lys lxlu Arg Ile Arg Arg Leu Met Glu Leu
10 75
Leu Lys Lys Thr Asp Arg Ile His I,eu Lys Asp Ala Ala Arg Met
20 25 30
Leu Glu Val Ser Va:1 Met Ttrr Tle Arg Arg Asp Leu His Gln Glu
35 40 45
Asp Glu Pro Leu Pro Leu Thr heu Leu G1y Gly Tyr_ Ile Val Met
50 55 60
Val Asn Lys Pro Ala Pro SEea: Met: Pro Val :Lle His Asp Val Pro
65 7C 75
Lys Asn His Arg Asp Asp L~eu Pro Ile Ala Ile Leu Ala A.La Gly
80 85 90
Met Val Asn Glu Asn Asp Leu Ile Phe Phe Asp Asr:~. Gly G.Ln G1u
95 100 105
Ile Pro Leu Va1 Ile Ser Met I1e Pro Asp Ala Ile: 'Thr Phe Thr
110 115 I20
Gly Ile Cys Tyr Ser His Arg Val Phe Val Al.a Leu Asn Glu Lys
125 130 135
Pro Asn Val Thr Ala Ile L~~~~u Cys Gly Gly Thr Tyr Arg Al.a Arg
140 195 x.50
Ser Asp Ala Phe Tyr Asp Ala Ser Asn Ser Ser Prc? Leu Asp Ser
155 160 7.65
Leu Asn Pro Arg Lys Ile Phe Ile Ser Ala Ser Gly Val His Asn
170 175 180
His Phe Gly Val Ser Trp Ph.e Asn Pro Gl.u Asp Leu Ala Thr Lys
185 190 195
Arg Lys Ala Met Asn Arg G:ly ~eu Arg Lys Ile Leu Leu Ala Arg
200 205 210
His A1a Leu Phe Asp Glu Va1 A1a Ser Ala Ser Leu Ala Pro Ile
215 220 225
Ser Ala Phe Asl:~ Val heu I 1e :per Asp Arg Pro I,eu Pro Al,a Asp
Page
CA 02388445 2003-07-09
2003-07-()8 Listage pc:ur 1e BdB corrige.txt
230 2.35 240
Tyr Val Thr His Cys Gln P.:~ru Gly Se:r Val Lys Ile Ile Thr Pro
245 250 255
Asp Ser Glu Asp Glu
260
(2) INFORMATION FOR SEQ ID NG: 6:
(i) SEQUENCE CHARACT3R.TSTICS:
(A) :LENGTIV: 4486 nucleotides
(B) TYPE: nucl~:i c acs d
(ii) MOLECULE TYPE: D2dA.
(vi) ORIGINAL Sc7URCE:
(A) ORGANISM: f;;c~erichia coi:i
(B) STRAIN: AL8Cs2
(xi) SEQUENCE D°SCRIEe'''1:0N: SEQ IC N0:6:
ggacgataatgtgatcgtcaataagggca:cacgctatc:atagtc~ttgtc:ctggcgggtaaa60
aaaacgcgcttaccttaaa:gataa:gcgcgccgctgttcaggccttgagtggttattcaat120
tcctgtggtgactgtaaa~.gtgcgcgt ctgcc;gt.gcaacctgaatcagcgtgccatt180
tt:c~
acgttgcgcggcaagatac:ccoca ggcc_:gacagqtt.gc:aggl::aatgraaaggcggctac240
ctgttgctctccgttata ggat:cc:vagc:gtgtc<~ca.taa.t~t:tagttcac~::actgtagaa300
~a
acgagtaacaaacgtagtgccats:~gygagagatcatgcgaaactctggctgatctgtata360
agcgtccagtttgtctgc~aagaagacaatttctggatcataaaattccggttgactcag420
cgtcgacagagaggcttct;ccctctc:~:taatccgtt.gattaa:~<:gccagcc:actgagcggt480
gggattaacatgcclaaggc:actc~~~tt:cargcaatctt<zatatt.tcgt<:cc~ggatattct=g540
gctgaatgtagcai:ttggtat:at~3t:c:~ca:ataat.tcatc~i:ggc:.ac~at:ataqtagtggc<~t600
t~:
atctacagaagccagatt~gtaacggcc:atcttaatatcgaacagtgtagaggatttgtg660
aaggaccactgttggctgagccactat.aat:gatgacccaaacccattacatactcgtaacg720
cccgttaaggcgtaacatatctcc~qtctaattccagccatgctetcatc::c:r'.:cgcggcaca'780
ggccatttcaccgi:gtagcagat,tac7tt;~tr.t:t.cr_:acac~at.dr,Iqcagccat:~~<~gccagcaa84
0
acctgaatgaaaagcaaaacagcc:ataggtctctatc:acct~ct:gtcgccdgtataggctg900
gcgaaacatattgcacat<rgtgaagc:cgt.gtccatcaaattgcgcatcccaaatcatctg960
ccccatccagggaagaat:,iat:ca-
a:ct:gt.c:c:acgac:U:a;tt:tgc<:at:tttaactcccctcgac1020
accgctgtcatat c;gaaaaagacgt:ctac<igt:aaaatcactattttccagc<3<3gatacgagg1080
tttctcgccaaaaa:~cgr_c,cgcc_cc:~.iaat:vtaatacgc~<Ltactcvataac:gat.-
.tctcctcag1140
gacgctgtgacttcagccaagtgc~3gtacgtactttgctttcac:gccaga<3gtagactccg1200
acatagacaaagcagagcataga_caccaggaatgaaagctgtagtgagtggaacatatct1260
gcaatatatccct:Iaat~t.ccggnac:cs~ccgc:ggc:acc;gac.aatagcc:at:aacaatga<:t1320
c
gctcctgccattt::tgtat:gttc~;t.t:aitc:aacagtatccar~tc;ttcctgc:at:agatcgtc1:380
gcccagcaagggc~.aaac<3aaac._,ct aggacggcgacat agaccgc:g<agaaactt1440
t<icc:
ggagccagtgcaac.atatgccag~~~aac:agcgcccctataacgcfaatagagaatcaatact1500
ttttccggattaa<iacgccftcat cac~gat~att.ggctat:aaac.t:tgccaai:aaagaagcag1560
gcaaagctataga::::atg~:3agtt tgaagcatcacctttcctttgatatcgcccaactccagc1620
gccagacggatggt:aaatc,tac:catactgc~gac:ctgcat:acc;:acataaaddaactgcgc:c1680
acaataccgcgacg:aaag<:gcggatttctagccagatagcgc<3gcgtatc~cattgctgac1740
gggcgtttatggtg:acttcJtctgt:gccactttacaggttgggaagcgggt:t:aaaaggaac1800
aacaccatgacca~;..aacca~gaat~:at;:,aatcatata<:ttat<~cctgttcaaciggtgttctct1860
aacatcagcaccttaaagi:tgtg:uat,ttgct:cggcgtt:cattc:ctgac:ai:ct:gcttttca1920
aggctttccccctcggagaaaac~~:agatat:ttgcccaataa~~~aaccag<icgcagcacca1980
atcggataaaaggt~~tggc.:tgatatt:gagecgcaatgtgge~ataggctti:tggaccgatc2040
attgaactgtatgt~~ttrc:xct..gc:e.gtaggaaact:caclgccaatcgc:aatcgcaaaa2100
t.i~ca
atagctgcaagga,_~~ata<tt<ttaa~=~c3l::t,c~c:.catatgcgaggcactggaaaaa~<tagtgtacaa21
60
ccaccaatatacag~~gtcaggcc~:attaaaattgcca<:cttat:aactggt:cattttaat:c2220
acaagggatgctggtatt<3caatta.aaaaataacctccataaaatgcgct:ctgcaccaat2280
gctgaagcaaagttacttagcga,~aatacacttttgaattgagtgattaatatgtcattt2390
Paste 8
CA 02388445 2003-07-09
2003-07-C~E3 BdB corr:ige.txt
f.~istage
pour
:1e
aatgcagctgcgcat:ccccatagcg<~gaataaacacgataacaaaataaactggaacaag 2400
ggagtcttattcaciataccr_atcaggcatctgaatgat.gtttttatcgtt;catagtgcta 2460
cctttaactgtgcac~gatgattat:tc~gtataaggttaaaaattc:attaaat:tgttcaat.a2520
ctcggataagatgatagcgtaces=ti:c_cctgtgacgc2:gaaagcggcaaagagagcggct 2580
tttttcaaagcggcatcaacatcacc~c~ctttgaacataataatgggaaaagcaaccaata 2640
aatgcgtcaccagcc~ccac;tagt,:3t:s.~<~aragcatttar;tttgaatgcagctaacatggac,t2700
tcctgatcgcgggt:catcc:ataat:cacc:Lcc:tttttcgct.catggtaacaat;aatattgttc 2760
agccctttatcaar_i~aacgaacgtgcggccaaacgaat.atgatcataagtatcaaccgac 2820
ataccggttaatat.t~tccagttct:cyt~:t:cattcgggataaac~naatcacatt.tgcaggca2880
taagacatatctaactcac:gc:aatc3~~c~qgagccggatttaataacacttc:aataccattt 2990
ttcttaccaaactc~<~atc<~cgtgtt~3aactgtttc~cagttgaact.t:cc:agttgtaaaacg 3000
atcaatttgcatti:.i~ttcagatctt:~rtgcagctcgatcgatat.cttccg<iggaaagaaat 3060
ttattcgctccctt:<~attattaavatact.attgctcgagtt~3gcattaac:aaagatcggt 3120
gcaacaccactgc!::c~gtac::aggg~,~a;attctcaacataagt:ggtattaat:.t:ccccatgat3180
tcgagattacgaatagtattatc_:gcaaaaatatcatcacctactttagti_agcatcagg3290
acttttgaattcaavttac7ccgc~~:g~car_cgcttgattagcacctttccc:accacatccg 3300
attttgaaggcaggtgctt:.ccag,ugt:ttct:ccttc~ttt:aggr,atctgatt:agtgtaagta 3;360
atgagatccacca~t,3ttg<~aacc~uat;aactgcaatgtccattt cactacc:t_cttataaac3420
tttcgcataacaatggtatataaataacattagcatgttacttttgcatcatttgtgact 3480
gagatcgcgatta~;i;:acat:caac~:cgai~gttt.atttaatagac-ttccagtcttatcactc 3540
aggccaacactat::taatc:ataactcaac;ctaacaggat:t:aataccgaaaat~t:cagcagtc3600
tatacccttttcatttcaaagggt:cggtcgtatagtat~ggt.-3ar_taaaac:aatgtttact 3660
aatgccataatgtt.atttttataacattttacggagagagttgatggaaacgaagcaaaa 3720
agagcgtatccgacgtttgat:tg_uaatact.taagaaaaccgac:agaatccatttgaaaga 3780
cgcggcacgaatg,ctgga<igt:tr_c.t:cttaat:gactatt.<:gtagc:vgatct:cc~at=caggaaga3840
tgaacctctgccactgaccctact:gggtggctatattgt:aatggtgcataaacccgcacc 3900
atccatgccagtaatccaggacgt:tccgagaaatcatc:gtgatgactt.acctattgcaat 3960
tctggccgccggaatggttaatgaaaatgat:ctgatca:t:cttt.gataaat~:~gccaggagat 4020
accgctcgttataagcatgatccc:ggatycaatcacc:ttcactggr_atc=gttactcaca 4080
tcgtgtcttt gttgcgttgaatgaaaaacc:taatgtgar_agcaatactttgtggtggtac 4140
gtatcgtgccagaagtgatgc.~tt.i.t:t:.acc:tatgccagt:aact<a.tcgccatt:agactctct4200
caatccgcgaaaaatattt:atttc.ccaccagc;ggtgta~c~atgat:cactttggcgtcagctg 4260
gtttaatcccgaagatcttgccactaagcgtaaagcgatggcccgtggactaaggaaaat 4320
tttgctcgcccgcc:acgc~:atgt.tcgatgaagtagcctctgc<aagcct:cgc~accgctctc4380
tgcatttgat gttctgattagcgagc.gtccgt:t:accctgcagat;.tatgttacgcactgccg 4440
gaatgcttcgtaaagat~,at t. t:cactaaagacgautga 4486
ttacvacctga
(2) INFORMATION FOR SEQ II:7 N0: 7:
(i) SEQUENCE CHARAC'!'i;R.IST.ICS:
(A) LENGTH: 3~'~' domino acic>
(B) TYPE: amine ac:,id
(D) TOPOLOGY: :spear
(ii) MOLECULE TYPE: p:co.ein
(xi) SEQUENCE DESCRIF'iT~~N: SEQ ID NO:7:
Met Ser Thr Arg Ile Asn i_.n_u:: Trp Arg F,~' a f.~eu Fnee G1y Glu L~ys
I. ~ ~. 5
Pro Arg Ile Leu Leu Glu F.:=,n. Ser P.sp P:.e Thr ua!. ~'hr Ser Phe
20 <:~~ 30
Arg Tyr Asp Ser Gly Val ~:l.m Gl~Y~ heu hys I1_e P.la Asn Ser Arg
35 =?(i e5
Gly His Leu Ile Ile Leu Pro Trp Met G~.y Gln Mev Ile Trp Asp
50 5!-i 60
Ala Gln Phe Asp Gly His G..y Leu Thr Met Cys Asn Met Phe Arg
65 70 75
Pave 9
CA 02388445 2003-07-09
2003-07-O8 histage poeir1e BdEcorrige.txt
GlnProLys PrrrAlaThr G ';!alI GJ.Thr'TyxGly CysPhe
1.,a 1e a
80 8'~ 90
AlaPheHis Se:rGlyLeu h_~uAlaAsn G:LyCysPrc:~Ser ValGlu
95 100 ~05
AspThrHis Le~_iLeuHis Gl.yG1L.Met.Ala(:ysAlaAI_aMetAsp
110 115 120
GluAlaTrp Le,zGluLeu A.>p.,1yAsp MetLeuAr_c)Leu AsnGly
125 1 135
30
ArgTyrGlu TyrValMet.Gl_yPheGly HisHi_sTyr:-Leu AlaGln
140 195 150
ProThrVa1 Va:LLeuHis hysSerSer Thr7_~euPheAsp ILeLys
155 160 1.65
MetAlaVal ThrAsnLeu A=_aSerVal AspMetPrc-.Leu GLnTyr
170 175 1.80
MetCysHis MetAsnTyr A:LaTyrIle ProAsnAlaThr PheSer
185 190 195
GlnAsnIle ProAspGlu I:LeL,euArg LeuArgGlmSer ValPro
200 205 210
SerHisVal AsnProThr RiaGl_nTrp LeuA:LaPheAsn GLnArg
215 220 225
IleMetGln GlyGluAla :>c~rLeiaSer ThrLeuSerGln ProGlu
230 235 290
PheTyrAsp ProGluIle Val.PhePhe ALaAspLysLeu AspAla
295 2~,0 255
TyrThrAsp GlnProGlu PlueArgMet IieSerPrc~Asp GlyThr
260 2E>5 270
ThrPheVal ThrArgPhe TyxSerA:laGl.uLeuFsnTyr Va1Thr
275 280 285
ArgTrpIle LeuTyrAsn C:lyG1~~Gl.nGLnValAl..:xA1a PheAla
290 215 300
LeuProAla ThrCysArg k'rc>GluG_y TyrLeuAlaAla G.lnArg
305 31.0 315
AsnGlyThr LeL.IleGln V,a:l.AlaPro GLnGlnThr_Arg ThrPhe
320 325 ;330
ThrValThr Tl-~rGlyIle Ca:Ll.i
335
( 2 ) INFORMATIOI~I FOR. SEQ I 1) I'!0 : 8 :
(i) SEQUE;DICE CHARAC'1'E; I:STICS:
(A) hENGTH: 4 ,t3 amino acids
(B) TYPE: aminas a-3cv.i.d
(D) TOPOLOGY: ' ira<ear
Page '.0
CA 02388445 2003-07-09
2003-07-08 L~istage pour1e BdBcorrige.txt
(ii)MOL ECULE protein
TYPE:
(xi)SEQUEN(:E P':CIOt~T; :8:
DESCRI SEQ
ID
N0
MetAsnAsp LysAsnIle I:Le;7 MetP:r Faspu.LyTyr LE.~uAsn
n o
5 7.0 7.5
LysThrPro LeuPheGln PheI1_eheuLeu L~er.~.~JSLeu PheI~'ro
20 25 30
LeuTrpGly Cy::~AlaAla Ala?~euAsnAsp Il.eLei:Ile TtArGin
35 4~ 45
PheLysSer Va:LPheSer IaE_;a:3erAsnP'~eA1aSerAla LeuVal
50 5.'> 60
GlnSerAla Ph<~TyrGly G 'ryrP:heLeu 7: i~ I ProAla
l 1e La 1e
y
65 70 'S
SerLeuVal IleLysLys T!-~r,:perTyrLys ValAlaI1_eLeu7:1e
80 85 90
GlyLeuThr Le~.rTyrI1e Gl.y~:~'.yCysThr LeuP:ze.~Phe PwoAla
95 100 105
SerHisMet A1.3ThrTyr Tr:r!hetPheLeu AlaA1<~ile PheAla
110 1'15 120
IleAlaIle GlyLeuSer F:hc~_LeuGluThr F~7.aAlaA.snThrTyr
125 1:~0 '_35
SerSerMet IleGlyPro hysAiaTyrAia ThrL~uArg LceuAsn
140 145 ~~50
IleSerGln ThrPheTyr ProI:LGlyAla A7.aS2rGly I:l_eI~eu
e:.
155 1E~0 165
LeuGlyLys Tyr_LeuVa:LFheSerGluG1y GluSerLeu GluLys
1'70 175 180
GlnMetSer GlyMetAsn A7 G:LiaGlnI HisAsrzPhe LysVal
a 1e
185 19C 195
LeuMetLeu GluAsnThr.LeuGluProTyr I~ysTyrMet I:LeMet
200 205 210
IleLeuVal Va:LValMet.Va7.LeuPheLeu LeuThr-Arg PhePro
215 220 'Z25
ThrCysLys Va:LAlaGln ThrSerHisHis -LysArgPro SerAla
230 235 240
MetAspThr LeuArgTyr L,euA.laArgAsn ProAr<(Phe ArgArg
245 250 255
GlyIleVal AlaGlnPhe L,~~uT ValGly MetGlnVal AlaVal
yr
260 265 270
TrpSerPhe Th.rIleArg I,euAlaLeuGlu :LeuGlvrAsp I1eAsn
275 28 0 285
GluArgAsp A1;~SerAsn PheMetVa1Tyr SerPheAla CysPhe
290 2035 300
Page 11
CA 02388445 2003-07-09
2003-07-(:~E3 L~istage pour1e BdBc:orrige.
txt
PheIleGly LysPheIle A_aAsnIleLeu MetThrArg PheAsn
305 310 315
ProGluLys Va~_LeuIle Lea.x'CyrSerV<i:LLleG:LyAla LeuPhe
320 325 330
LeuAlaTyr VaI.Al.aLeu A:LaProSerPhe SerAlaVal TyrVal
335 390 345
AlaValLeu ValSerVal L<:uPheGlyPro CysTrpAla ThrIle
350 355 360
TyrA1aGly ThwLeuAsp Th.r',7a1AspAsn GluIsisThr GluMet
365 370 375
AlaGlyAla Vaa.Il.eVal McaA.laIl.eVal GlyALaAla ValVal
380 385 390
ProAlaIle GlnGlyTyr IleAlaAspMet PheH.isSex LeuGln
395 400 405
LeuSerPhe LeuValSer M._aLeuC:ysPhe ValT Val G7_yVal
yr
410 915 420
TyrPheTrp ArgGluSer LysValArgThr AlaLeuAla GluVal
425 430 435
Thr Ala Ser
(2) INFORMATIOiV FOR SEQ_ IC~ NO: 9:
( i ) SEQUENCE C'?ARACTE:,R1 S'_"I CS
{A) IaGNGTII: 3UFi amino acida
{B) :CYPE: amine ;7cid
(D) TOPOLOGY: lp_r_ear
(ii) MOLECULE T'~PE: pi:r.:tr:in
(xi) SEQUENCE DhSCRIP~"I:C?N: SEQ ID NO:.°.:
Met Asp Ile Ala Val Ile G:y Ser Asn Met 'JaJ. Asp heu Ile Thr
10 :15
Tyr Thr Asn Gln Met Pro L~js Glu Gly Glu 'rrr Leu Glu Aia Pro
20 J 25 30
Ala Phe Lys Ile Gly Cys C:,:Ly G1y hys Gl.y Al.a As~a Gln ALa 'Jal
35 90 45
Ala Ala Ala Lys Leu Asn Scar Lys Va1 Leu Met Leu Thr Lys Val
50 5'60
Gly Asp Asp Ile Phe Ala A:~L:~ Asn Thr T:ie Arg Asu heu Glu Ser
65 70 75
Trp Gly Ile Asn Thr Thr '1'yr Val Glu Lys Val Pro Cys Thr Ser
80 85 90
Ser Gly Val Ala Pro Ile Phe Val Asn Ala Asn Ser Ser Asn Ser
95 1. C)0 1.05
Page 12
CA 02388445 2003-07-09
2003-07 -08I~ist~age pourle BdBcorr:ige.txt
IleLeuIle IleLysGly ALaAsnLysPhe heuSerProGlu Asp
110 115 120
IleAspArg AlaAlaGlu Asp!:.~euhysLys O:yshya,LeuI7_eVal
125 130 1.35
LeuGlnLeu GluVal.Gin LeuGluThrVal 'I'yrH:LsAl.aIl.eGlu
140 195 1.50
PheGlyLys LysAsnGly ILeGluValLeu L~euAsnProAl.aPro
155 160 I_65
AlaLeuArg GlnzLeuAsp Met:SerTyrA:laCysLysCysAsp Phe
1'70 115 180
PheIlePro AsnGlu'rhrGLuLeuGluIle LeuThxGlyMet Ser
185 190 195
ValAspThr Ty:c:AspHis I .?ergLeuAla AlaArc)SerLeu Val
Le
200 205 210
AspLysGly LeuAsnAsn Il.e:L:LeVal.'rhrMetS~~rG1_uLys Gly
215 220 2.25
AlaLeuTrp Met.ThrArg A~;p~:LnGluVal HisVaif'roAl.aPhe
230 235 240
LysValAsn Al._iValAsp ThrSerGlyAl.aGlyAspAlaPhe Ile
245 250 255
GlyCysPhe Ser_HisTyr Ty:rValGlnSer GlyAsiaValG_LuAla
260 265 270
AlaLeuLys LysAlaAla LeuPheAl.aAla hheSerValThr Gly
275 280 285
LysGlyThr GlnSerSer TyrProSerIle GluG1nPheA.snGlu
290 295 300
PheLeuThr LeuAsnGlu
305
(2) INFORMATION FOR SEQ LD N0: 10:
(i) SEQUENCE CIiARACThRIS'PICS:
(A) LENGTH: 30E: amino acids
(B) 'TYPE: amino avid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE C)L,SCRIPT~GN: SEQ ID NC3:10:
Met Asp Ile Ala Val Ile G':y Sez Asn Mc~t Jai Ast~ Leu I1e Thr
1C' 15
Tyr Thr Asn Gl~z Met Pro I~ys G1u GI_y Glu Thr L,e G.I_u A1a Pro
20 25 30
Ala Phe Lys Ile Gly Cys GL.y Gly Lvs G?~y .~lla Pan Ciln Ala Val
35 4C) 95
Page 13
CA 02388445 2003-07-09
2003-07-C3 '~~is"_:age po~:_ir.1e I3clBcorrige.txt
AlaAlaAla Lye;LeuAsn Ser,ysVa.LLeu MetLeu'l LysVal
hr
50 55 60
G1yAspAsp Ile:PheA1 A:>palsnTh.rI7_e~,rgAsnLeu Gl.uSer
a
65 70 7~
TrpGlyIle Asr:ThrThr Tsrr'1<a1.C~:l.uLy,>N'alPrc~:ysThrSer
8 8 9
0 0
SerGlyVal AlaProIle PheValAsnAia AsnSerSer AsnSer
95 i~:?~ 105
IleLeuIle I1<;LysGly A:laAsnLysPhe LeuSerPro GluAsp
110 115 120
IleAspArg AlmAlaGlu A:~p:L,euLysLys C'.ysLysLeu I~.eVal
125 130 135
LeuGlnLeu GlozValGln L. GluThrVal TyrH A7_aI:LeGlu
~u L:;
140 195 150
PheGlyLys LysAsnGly Il.eGluVal.Leu LeuAsr;Pro A1aPro
155 1.60 165
AlaLeuArg GluLeuAsp MeatSerTyrAla CysLysCys AspPhe
170 175 180
PheIlePro AsnGluThr Gl.uLeuGluIle L,eu'rhrGly MetSer
185 190 195
ValAspThr TyrAspHis IleArgL,euAla AlaArc;Sex LeuVal
200 205 210
AspLysGly LeuAsnAsn I'_eIleValThr MetSerGlu LysGly
215 220 225
AlaLeuTrp MetThrArg AspGLnGl.uVal HisValPro AlaPhe
230 235 290
LysValAsn AlaValAsp 7'h~:SerGl.yA:laGlyAspAla P:helle
245 250 255
GlyCysPhe Se:rHisTyr TyrValGl.nSer Gl.yAspVal GluAla
260 265 270
AlaLeuLys LysAlaAla I:E=u.PheAlaAla PheSerVal ThrGly
2.75 280 285
LysGlyThr GlnSerSer TyrPr<>SerI GluGl.nPhe AsnGlu
~e
290 2.95 300
PheLeuThr LeuAsnGlu
305
( 2 ) INFORMATI01\! FOR SEQ I D NC): 11
(i) SEQUEI~ICE CHARACTERISTICS:
(A) LENGTH: 8'~4 nucLeot~.ide,>
(B) TYPE: nucle~:i.c: acid
( i i ) MOLEC'.LILE TYPE : C>NA
Page 14
CA 02388445 2003-07-09
2003-07-C'_'s histage poszr 1e BdB c:orrige.txt
(vi) ORIGINF~L SOURCE: lsc:herichia coli
( xi ) SEQUENCE DESCRI P'l.' LC)N : S EQ I D N0 : 1 1
caatactcggataactatgattgccttacctttccctgtgacgcagaaagcggcaaagagag 60
cggcttttttcaaagcggcttca<ccat:caccgctttgaacat aataatgggaaaagcaac 120
caataaatgcgtc<~c:cagcgccar:t<~cJt:atcaacagcatttactttgaat:gcaggaacat 180
ggacttcctgatc<Jc:gggt:catcc:ai:~~atg<:gcctttttcgctcatggtaacaataatat 2.40
tgttcagccctttat:caact=aaccJaac;gtgcggcc:aaacgaatatgatcat:aagtatca.a300
ccgacataccggttt~atat:ttcc~igi:t_<agt:ttcattcgggataaagaaat: cacatttgc360
aggcataagacatat:ctaactca<:gc:.aatgccggagccggat:ttaataac:acttcaatac 420
catttttcttacca.aactcaatcgcgtggtaaactgt?-tccagttgaactt:ccagttgta480
aaacgatcaattt<Jcattt:tttcrigat:ctr.ctgcagctcgatcgatatctt:ccggggaaa540
gaaatttattcgct:cccttaattati_aatatacta"~tgct,,:,~t:gttggc~c't:taacaaaga600
tcggtgcaacaccacagctggtac,~gc~ggactttctcaacata.agtggtat:taattcccc660
atgattcgagattac:gaatgta':t,~;:~cgcaaaa;~tatca'~c a:~ctact,t:t.agttagc;a720
a
tcaggacttttgaai:tcaactttacJcc:~c:cgc<:ac~~gct..t:gat:tac~cacctt:t:.cccaccac780
atccgattttgaacxgcaggtgctt~:~c~agagtttc;:cctt:ct'Ytaggcatc:t:gat 8:34
( 2 ) INFORMATIOt~I FOR SEQ IC~ 1v0: 12
( i ) SEQUEtVC:E CHARACTL,ft . STI CS
{A) LENGTH: 81~= !m.ic7_e~otides
{B) '1"fPE: nucleic acid
( i i ) MOLECiJ::~E TYPE : Dl>IA
(vi) ORIGIN~~L SOURCE: I_,scher.ichia ca7.i.
{xi) SEQUE19CE DESCRIP'I'I~DiV: SEQ ID N0:7.2:
ggacgataatgtg,_~rcgtc::tata,~q;Jgcaacgctatcatagtcatgtcct:ggcgggtaaa 60
aaaacgcgcttaccttaa<:gata~~~g.~.gcgccgctgttc:aggccttgagt.<~gttattcaat 120
tcctgtggtgact~ataaaagtgc~:~cqtttgctgcggtcJcaa :cvtgaatcac~cgtgccat:t180
acgttgcgcggcaagatac:cc:ct~::ag:~<:cgacaggttgc:aggt:aatgcaaaggcggctac 240
ctgttgctctccgt:tata~~aggatccagcqtgtcacat:aa-tt agttcagc~actgtagaa300
acgagtaacaaac:Jtagtgccat.:gcx:~agagatcatgcgaasc:t~ctggctgatctgtata 360
agcgtccagtttgt~tgct3aagaagac ttct:ggat:c:a!:aar.aattcccJgttgactcag 420
aat
cgtcgacagagag;Jcttct:ccct.~fc:ataatccgttgattaaacgccagcc::actgagcggt 480
gggattaacatgc;fiaagg~actg=u.t caatctt<iatatt:tcgtccgcJgatattctg540
c<~cg
gctgaatgtagcat:ttggn:atat:~stgcat:.aat.tcatgt:ggcac:atatat?:dt:agtggcat600
atctacagaagccagatt<;gttac:ggccatct.taatat=cgaac:agtgtac4aggatttgtg 660
aaggaccactgttc~gctg<igccac~~ataatgat:gaccgaaaccc:attaca'_actcgtaacg '720
cccgttaaggcgtaacata-itctccgtctaattccagrcatgcttcatcc~itcgcggcaca 780
ggccatttcaccgi:gtagc:agat_c~~agtatc:ttc:cac 816
PacJee 1. S