Note: Descriptions are shown in the official language in which they were submitted.
^
CA 02390309 2002-05-07
DESCRIPTION
METHOD FOR SYNTHESIZING THE NUCLEIC ACID
Technical Field
The present invention relates to a method of synthesizing
nucleic acid composed of a specific nucleotide sequence, which
is useful as a method of amplifying nucleic acid.
Background Art
An analysis method based on complementarity of a nucleic
acid nucleotide sequence can analyze genetic traits directly.
Accordingly, this analysis is a very powerful means for
identification of genetic diseases, canceration,
microorganisms etc. Further, a gene itself is the object of
detection, and thus time-consuming and cumbersome procedures
such as in culture can be omitted in some cases.
Nevertheless, the detection of a target gene present in
a very small amount in a sample is not easy in general so that
amplification of a target gene itself or its detection signal
is necessary. As a method of amplifying a target gene, the PCR
(polymerase chain reaction) method is known (Science, 230,
1350-1354, 1985). Currently, the PCR method is the most popular
method as a technique of amplifying nucleic acid in vitro. This
method was established firmly as an excellent detection method
by virtue of high sensitivity based on the effect of exponential
1
CA 02390309 2002-05-07
amplification. Further, since the amplification product can
be recovered as DNA, this method is applied widely as an important
tool supporting genetic engineering techniques such as gene
cloning and structural determination. In the PCR method,
however, there are the following noted problems: a special
temperature controller is necessary for practice; the
exponential progress of the amplification reaction causes a
problem in quantification; and samples and reaction solutions
are easily contaminated from the outside to permit nucleic acid
mixed in error to function as a template.
As genomic information is accumulated, analysis of single
nucleotide polymorphism (SNPs) comes to attract attention.
Detection of SNPs by means of PCR is feasible by designing a
primer such that its nucleotide sequence contains SNPs. That
is, whether a nucleotide sequence complementary to the primer
is present or not can be inferred by determining whether a reaction
product is present or not. However, once a complementary chain
is synthesized in error in PCR by any chance, this product
functions as a template in subsequent reaction, thus causing
an erroneous result. In practice, it is said that strict control
of PCR is difficult with the difference of only one base given
at the terminal of the primer. Accordingly, it is necessary
to improve specificity in order to apply PCR to detection of
SNPs.
On one hand, a method of synthesizing nucleic acid by a
2
1
CA 02390309 2002-05-07
ligase is also practically used. The LCR method (ligase chain
reaction, Laffler TG; Garrino JJ; Marshall RL; Ann. Biol. Clin.
(Paris), 51:9, 821-6, 1993) is based on the reaction in which
two adjacent probes are hybridized with a target sequence and
ligated to each other by a ligase. The two probes could not
be ligated in the absence of the target nucleotide sequence,
and thus the presence of the ligated product is indicative of
the target nucleotide sequence. Because the LCR method also
requires control of temperature f or separation of a complementary
chain from a template, there arises the same problem as in the
PCR method. For LCR, there is also a report on a method of
improving specificity by adding the step of providing a gap
between adjacent probes and filling the gap by a DNA polymerase.
However, what can be expected in this modified method is
specificity only, and there still remains a problem in that
control of temperature is required. Furthermore, use of the
additional enzyme leads to an increase in cost.
A method called the SDA method (strand displacement
amplification) [Proc. Natl. Acad. Sci. USA, 89, 392-396, 1992]
[Nucleic Acid. Res., 20, 1691-1696, 1992] is also known as a
method of amplifying DNA having a sequence complementary to a
target sequence as a template. In the SDA method, a special
DNA polymerase is used to synthesize a complementary chain
starting from a primer complementary to the 3' -side of a certain
nucleotide sequence while displacing a double-stranded chain
3
CA 02390309 2002-05-07
if any at the 5'-side of the sequence. In the present
specification, the simple expression "5'-side" or "3'-side"
refers to that of a chain serving as a template. Because a
double-stranded chain at the 5'-side is displaced by a newly
synthesized complementary chain, this technique is called the
SDA method. The temperature-changing step essential in the PCR
method can be eliminated in the SDA method by previously inserting
a restriction enzyme recognition sequence into an annealed
sequence as a primer. That is, a nick generated by a restriction
enzyme gives a 3'-OH group acting as the origin of synthesis
of complementary chain, and the previously synthesized
complementary chain is released as a single-stranded chain by
strand displacement synthesis and then utilized again as a
template for subsequent synthesis of complementary chain. In
this manner, the complicated control of temperature essential
in the PCR method is not required in the SDA method.
In the SDA method, however, the restriction enzyme
generating a nick should be used in addition to the strand
displacement-type DNA polymerase. This requirement for the
additional enzyme is a major cause for higher cost. Further,
because the restriction enzyme is to be used not for cleavage
of both double-stranded chains but for introduction of a nick
(that is, cleavage of only one of the chains) , a dNTP derivative
such as a-thio dNTP should be used as a substrate for synthesis
to render the other chain resistant to digestion with the enzyme.
4
CA 02390309 2002-05-07
Accordingly, the amplification product by SDA has a different
structure from that of natural nucleic acid, and there is a limit
to cleavage with restriction enzymes or application of the
amplification product to gene cloning. In this respect too,
there is a major cause for higher cost. In addition, when the
SDA method is applied to an unknown sequence, there is the
possibility that the same nucleotide sequence as the restriction
enzyme recognition sequence used for introducing a nick may be
present in a region to be synthesized. In this case, it is
possible that a complete complementary chain is prevented from
being synthesized.
NASBA (nucleic acid sequence-based amplification, also
called the TMA/transcription mediated amplification method) is
known as a method of amplifying nucleic acid wherein the
complicated control of temperature is not necessary. NASBA is
a reaction system wherein DNA is synthesized by DNA polymerase
in the presence of target RNA as a template with a probe having
a T7 promoter added thereto, and the product is formed with a
second probe into a double-stranded chain, followed by
transcription by T7 RNA polymerase with the formed
double-stranded chain as a template to amplify a large amount
of RNA (Nature, 350, 91-92, 1991). NASBA requires some heat
denaturation steps until double-stranded DNA is completed, but
the subsequent transcriptional reaction by T7 RNA polymerase
proceeds under isothermal conditions. However, a combination
CA 02390309 2002-05-07
of plural enzymes such as reverse transcriptase, RNase H, DNA
polymerase and T7 RNA polymerase is essential, and this is
unfavorable for cost similarly to SDA. Further, because it is
complicated to set up conditions for a plurality of enzyme
reaction, this method is hardly widespread as a general
analytical method. In the known reactions of amplification of
nucleic acid, there remain problems such as complicated control
of temperature and the necessity for plural enzymes as described
above
For these known reactions of synthesizing nucleic acid,
there are few reports on an attempt for further improving the
efficiency of synthesis of nucleic acid without sacrificing
specificity or cost. For example, in a method called RCA
(rolling-circle amplification), it was shown that
single-stranded DNA having a series of nucleotide sequences
complementary to a padlock probe can be synthesized continuously
in the presence of a target nucleotide sequence (Paul M. Lizardi
et al., Nature Genetics, 19, 225-232, July, 1998). In RCA, a
padlock probe having a special structure wherein each of the
5'- and 3'-terminals of a single oligonucleotide constitutes
an adjacent probe in LCR is utilized. Then, the continuous
reaction of synthesizing complementary chain with the padlock
probe as a template which was ligated and cyclized in the presence
of a target nucleotide sequence is triggered by combination with
a polymerase catalyzing the strand displacement-type reaction
6
CA 02390309 2002-05-07
of synthesizing complementary chain. Single-stranded nucleic
acid having a structure of a series of regions each consisting
of the same nucleotide sequence is thus formed. A primer is
further annealed to this single-stranded nucleic acid to
synthesize its complementary chain and a high degree of
amplification is thus realized. However, there still remains
the problem of the necessity fora plurality of enzymes. Further,
triggering of synthesis of complementary chain depends on the
reaction of ligating two adjacent regions, and its specificity
is basically the same as in LCR.
For the object of supplying 3' -OH, there is a known method
in which a nucleotide sequence is provided at the 3'-terminal
with a sequence complementary thereto and a hair pin loop is
formed at the terminal (Gene, 71, 29-40, 1988) . Synthesis of
complementary chain with a target sequence itself as a template
starts at the hairpin loop to form single-stranded nucleic acid
composed of the complementary nucleotide sequence. For example,
a structure in which annealing occurs in the same chain at the
terminal to which a complementary nucleotide sequence has been
linked is realized in PCT/FR95/00891. In this method, however,
the step in which the terminal cancels base pairing with the
complementary chain and base pairing is constituted again in
the same chain is essential. It is estimated that this step
proceeds depending on a subtle equilibrium state at the terminal
of mutually complementary nucleotide sequences involving base
7
CA 02390309 2002-05-07
pairing. That is, an equilibrium state maintained between base
pairing with a complementary chain and base pairing in the same
chain is utilized and the only chain annealing to the nucleotide
sequence in the same chain serves as the origin of synthesis
of a complementary chain. Accordingly, it is considered that
strict reaction conditions should be set to achieve high reaction
efficiency. Further, in this prior art, the primer itself forms
a loop structure. Accordingly, once a primer dimer is formed,
amplification reaction is automatically initiated regardless
of whether a target nucleotide sequence is present or not, and
an unspecific synthetic product is thus formed. This can be
a serious problem. Further, formation of the primer dimer and
subsequent consumption of the primer by unspecific synthetic
reaction lead to a reduction in the amplification efficiency
of the desired reaction.
Besides, there is a report that a region not serving as
a template for DNA polymerase was utilized to realize a
3'-terminal structure annealing to the same chain (EP713922).
This report also has the same problem as in PCT/FR95/00891 supra
in respect of the utilization of dynamic equilibrium at the
terminal or the possibility of unspecific synthetic reaction
due to formation of a dimer primer. Further, a special region
not serving as a template for DNA polymerase should be prepared
as a primer.
Further, in various signal amplification reactions to
8
CA 02390309 2002-05-07
which the principle of NASBA described above is applied, an
oligonucleotide having a hairpin structure at the terminal
thereof is often utilized to supply a double-stranded promoter
region (JP-A5-211873). However, these techniques are not those
permitting successive supply of 3'-OH for synthesis of a
complementary chain. Further, a hairpin loop structure having
a 3'-terminal annealed in the same chain is utilized for the
purpose of obtaining a DNA template transcribed by RNA polymerase
is utilized in JP-A 10-510161 (W096/17079). In this method,
the template is amplified by using transcription into RNA and
reverse transcription from RNA to DNA. In this method, however,
the reaction system cannot be constituted without a combination
of a plurality of enzymes.
Disclosure of the Invention
The object of the present invention is to provide a method
of synthesizing nucleic acid based on a novel principle. Amore
specific object is to provide a method capable of realizing the
synthesis of nucleic acid depending on sequence efficiently at
low costs. That is, an object of the present invention is to
provide a method capable of achieving the synthesis and
amplification of nucleic acid by a single enzyme even under
isothermal reaction conditions. Another object of the present
invention is to provide a method of synthesizing nucleic acid
which can realize high specificity difficult to achieve in the
9
s
CA 02390309 2002-05-07
known reaction principle of nucleic acid synthesis, as well as
a method of amplifying nucleic acid by applying said synthetic
method.
The present inventors focused their attention on the fact
that the utilization of a polymerase catalyzing strand
displacement-type synthesis of complementary chain is useful
for nucleic acid synthesis not depending on complicated control
of temperature. Such a DNA polymerase is an enzyme utilized
in SDA and RCA. However, even if such an enzyme is used, another
enzyme reaction is always required for supplying 3'-OH as the
origin of synthesis in the known means based on primers, such
as SDA.
Under these circumstances, the present inventors examined
supply of 3'-OH from a completely different viewpoint from the
known approach. As a result, the present inventors found that
by utilizing an oligonucleotide having a special structure, 3'-OH
can be supplied without any additional enzyme reaction, thereby
completing the present invention. That is, the present
invention relates to a method of synthesizing nucleic acid, a
method of amplifying nucleic acid by applying said method of
synthesizing nucleic acid and a novel oligonucleotide enabling
said methods, as follows:
CA 02390309 2002-05-07
1. A method of synthesizing nucleic acid having complementary
nucleotide sequences linked alternately in a single-stranded
chain, comprising:
a) the step of giving nucleic acid which is provided at the
3'-terminal thereof with a region Fl capable of annealing to
a part Fic in the same chain and which upon annealing of the
region F1 to Fic, is capable of forming a loop containing a region
F2c capable of base pairing,
b) the step of performing synthesis of a complementary chain
wherein the 3'-terminal of Fl having annealed to Fic serves as
the origin of synthesis,
c) the step of annealing, to a region F2c, of an oligonucleotide
provided with the 3'-terminal thereof with F2 consisting of a
sequence complementary to the region F2c,followed by synthesis,
with said oligonucleotide as the origin of synthesis, of a
complementary chain by a polymerase catalyzing the strand
displacement reaction of synthesizing a complementary chain to
displace the complementary chain synthesized in step b), and
d) the step of annealing, to the complementary chain displaced
in step c) to be ready for base pairing, of a polynucleotide
provided at the 3'-terminal thereof with a sequence complementary
to an arbitrary region in said chain synthesized in step c),
followed by synthesis, with said 3' -terminal as the origin of
synthesis, of a complementary chain by a polymerase catalyzing
the strand displacement reaction of synthesizing a complementary
11
CA 02390309 2002-05-07
chain to displace the complementary chain synthesized in step
c) .
2. The method according to item 1, wherein in step d) , the origin
of synthesis is a region R1 present at the 3'-terminal in the
same chain and capable of annealing to a region Ric, and a loop
containing the region R2c capable of base pairing is formed by
annealing R1 to Rlc.
3. An oligonucleotide composed of at least two regions X2 and
Xlc below, and Xic is linked to the 5'-side of X2,
X2: a region having a nucleotide sequence complementary to an
arbitrary region X2c in nucleic acid having a specific nucleotide
sequence, and
Xlc: a region having substantially the same nucleotide sequence
as in a region Xlc located at the 5' -side of the region X2c in
nucleic acid having a specific nucleotide sequence.
4. The method according to item 1, wherein the nucleic acid in
step a) is second nucleic acid provided by the following steps:
i) the step of annealing, to a region F2c in nucleic acid serving
as a template, of a region F2 in the oligonucleotide described
in item 3 wherein the region X2 is a region F2 and the region
X1c is a region Fic,
ii) the step of synthesizing first nucleic acid having a
nucleotide sequence complementary to the template wherein F2
in the oligonucleotide serves as the origin of synthesis,
iii) the step of rendering an arbitrary region in the first nucleic
12
CA 02390309 2002-05-07
acid synthesized in step ii) ready for base pairing, and
iv) the step of annealing an oligonucleotide having a nucleotide
sequence complementary to the region made ready for base pairing
in the first nucleic acid in step iii) , followed by synthesizing
second nucleic acid with said oligonucleotide as the origin of
synthesis and rendering Fl at the 3' -terminal thereof ready for
base pairing.
5. The method according to item 4, wherein the region enabling
base pairing in step iii) is R2c, and the oligonucleotide in
step iv) is the oligonucleotide described in item 3 wherein the
region X2c is a region R2c and the region Xlc is a region Ric.
6. The method according to item 4 or 5, wherein the step of
rendering base pairing ready in steps iii) and iv) is conducted
by the strand displacement synthesis of complementary chain by
a polymerase catalyzing the strand displacement reaction of
synthesizing complementary chain wherein an outer primer
annealing to the 3'-side of F2c in the template and an outer
primer annealing to the 3'-side of the region used as the origin
of synthesis in step iv) for the first nucleic acid serve as
the origin of synthesis.
7. The method according to item 6, wherein the melting temperature
of each oligonucleotide and its complementary region in the
template used in the reaction is in the following relationship
under the same stringency:
(outer primer/region at the 3'-side in the template) 5 (F2c/F2
13
CA 02390309 2002-05-07
and R2c/R2) S (Flc/F1 and Rlc/R1).
B. The method according to any one of items 4 to 7, wherein the
nucleic acid serving as the template is RNA, and the synthesis
of complementary chain in step ii) is conducted by an enzyme
having a reverse transcriptase activity.
9. A method of amplifying nucleic acid having complementary
nucleotide sequences linked alternately in a single-stranded
chain by repeatedly conducting the following steps:
A) the step of providing a template which is provided at the
3'- and 5'-terminals thereof with a region consisting of a
nucleotide sequence complementary to each terminal region in
the same chain and which upon annealing of these mutually
complementary nucleotide sequences, forms a loop capable of base
pairing therebetween,
B) the step of performing the synthesis of complementary chain
wherein the 31-terminal of said template annealed to the same
chain serves as the origin of synthesis,
C) the step of annealing, to the loop portion, of an
oligonucleotide provided at the 3'-terminal thereof with a
complementary nucleotide sequence to a loop which among said
loops, is located at the 3'-terminal site, f ollowed by synthesis,
with the oligonucleotide as the origin of synthesis, of a
complementary chain by a polymerase catalyzing the strand
displacement reaction of synthesizing a complementary chain to
displace the complementary chain synthesized in step B) to make
14
CA 02390309 2002-05-07
the 3'-terminal thereof ready for base pairing, and
D) the step wherein the chain with the 3'-terminal made ready
for base pairing in step C) serves as a new template.
10. The method according to item 9, wherein the oligonucleotide
in step C) is provided at the 5' -terminal thereof with a nucleotide
sequence complementary to the 3'-terminal serving as the origin
of synthesis in step B).
11. The method according to item 10, further comprising the step
where a complementary chain synthesized with the oligonucleotide
in step C) as the origin of synthesis is used as a template in
step A).
12. The method according to item 9, wherein the template in step
A) is synthesized by the method described in item 5.
13. The method according to item 1 or 9, wherein the strand
displacement reaction of synthesizing complementary chain is
carried out in the presence of a melting temperature regulator.
14. The method according to item 13, wherein the melting
temperature regulator is betaine.
15. The method according to item 14, wherein 0.2 to 3. 0 M betaine
is allowed to be present in the reaction solution.
16. Amethod of detecting a target nucleotide sequence in a sample,
which comprises performing an amplification method described
in any one of items 9 to 15 and observing whether an amplification
reaction product is generated or not.
17. The method according to item 16, wherein a probe containing
CA 02390309 2002-05-07
a nucleotide sequence complementary to the loop is added to the
amplification reaction product and hybridization therebetween
is observed.
18. The method according to item 17, wherein the probe is labeled
on particles and aggregation reaction occurring upon
hybridization is observed.
19. The method according to item 16, wherein an amplification
method described in any one of items 9 to 15 is conducted in
the presence of a detector for nucleic acid, and whether an
amplification reaction product is generated or not is observed
on the basis of a change in the signal of the detector.
20. A method of detecting a mutation in a target nucleotide
sequence by the detection method described in item 16, wherein
a. mutation in a nucleotide sequence as the subject of
amplification prevents synthesis of any one of complementary
chains constituting the amplification method.
21. A kit for synthesis of nucleic acid having complementary
chains alternately linked in a single-stranded chain, comprising
the following elements:
i) the oligonucleotide described in item 3 wherein the region
F2c in nucleic acid as a template is X2c, and Flc located at
the 5'-side of F2c is Xlc;
ii) an oligonucleotide containing a nucleotide sequence
complementary to an arbitrary region in a complementary chain
synthesized with the oligonucleotide in (i) as a primer;
16
CA 02390309 2002-05-07
iii) an oligonucleotide having a nucleotide sequence
complementary to a region F3c located at the 3-side of the region
F2c in the nucleic acid serving as a template;
iv) a DNA polymerase catalyzing the strand displacement-type
reaction of synthesizing complementary chain; and
v) a nucleotide serving as a substrate for the element iv).
22. The kit according to item 21, wherein the oligonucleotide
in ii) is the oligonucleotide described in item 3 wherein an
arbitrary region R2c in a complementary chain synthesized with
the oligonucleotide in i) as the origin of synthesis is X2c,
and Rlc located at the 5' of R2c is Xlc.
23. The kit according to item 22, further comprising:
vi) an oligonucleotide having a nucleotide sequence
complementary to a region Ric located at the 3'-side of the
arbitrary R2c in a complementary chain synthesized with the
oligonucleotide in i) as the origin of synthesis.
24. A kit for detection of a target nucleotide sequence,
comprising a detector for detection of a product of nucleic acid
synthetic reaction additionally in a kit described in any one
of items 21 to 23.
The nucleic acid having complementary nucleotide
sequences linked alternately in a single-stranded chain as the
object of synthesis in the present invention means nucleic acid
having mutually complementary nucleotide sequences linked side
by side in a single-stranded chain. Further, in the present
17
CA 02390309 2002-05-07
invention, it should contain a nucleotide sequence for forming
a loop between the complementary chains. In the present
invention, this sequence is called the loop-forming sequence.
The nucleic acid synthesized by the present invention is composed
substantially of mutually complementary chains linked via the
loop-forming sequence. In general, a strand not separated into
2 or more molecules upon dissociation of base pairing is called
a single-stranded chain regardless of whether it partially
involves base pairing or not. The complementary nucleotide
sequence can form base pairing in the same chain. An
intramolecular base-paired product, which can be obtained by
permitting the nucleic acid having complementary nucleotide
sequences linked alternately in a single-stranded chain
according to the present invention to be base-paired in the same
chain, gives a region constituting an apparently double-stranded
chain and a loop not involving base pairing.
That is, the nucleic acid having complementary nucleotide
sequences linked alternately in a single-stranded chain
according to the present invention contains complementary
nucleotide sequences capable of annealing in the same chain,
and its annealed product can be defined as single-stranded
nucleic acid constituting a loop not involving base pairing at
a bent hinged portion. A nucleotide having a nucleotidesequence
complementary thereto can anneal to the loop not involving base
pairing. The loop-forming sequence can be an arbitrary
18
CA 02390309 2002-05-07
nucleotide sequence. The loop-forming sequence is capable of
base pairing so as to initiate the synthesis of a complementary
chain for displacement, and is provided preferably with a
sequence distinguishable from a nucleotide sequence located in
the other region in order to achieve specific annealing. For
example, in a preferred embodiment, the loop-forming sequence
contains substantially the same nucleotide sequence as a region
F2c (or R2c) located at the 3'-side of a region (i.e. Flc or
Ric) derived from nucleic acid as a template and annealed in
the same chain.
In the present invention, substantially the same
nucleotide sequence is defined as follows. That is, when a
complementary chain synthesized with a certain sequence as a
template anneals to a target nucleotide sequence to give the
origin of synthesizing a complementary chain, this certain
sequence is substantially the same as the target nucleotide
sequence. For example, substantially the same sequence as F2
includes not only absolutely the same nucleotide sequence as
F2 but also a nucleotide sequence capable of functioning as a
template giving a nucleotide sequence capable of annealing to
F2 and acting as the origin of synthesizing complementary chain.
The term "anneal" in the present invention means formation of
a double-stranded structure of nucleic acid through base pairing
based on the law of Watson-Crick. Accordingly, even if a nucleic
acid chain constituting base pairing is a single-stranded chain,
19
CA 02390309 2002-05-07
annealing occurs if intramolecular complementary nucleotide
sequences are base-paired. In the present invention, annealing
and hybridization have the same meaning in that the nucleic acid
constitutes a double-stranded structure through base pairing.
The number of pairs of complementary nucleotide sequences
constituting the nucleic acid according to the present invention
is at least 1. According to a desired mode of the present
invention, it may be 2 or more. In this case, there is
theoretically no upper limit to the number of pairs of
complementary nucleotide sequences constituting the nucleic
acid. When the nucleic acid as the synthetic product of the
present invention is constituted of plural sets of complementary
nucleotide sequences, this nucleic acid is composed of repeated
identical nucleotide sequences.
The nucleic acid having complementary nucleotide
sequences linked alternately in a single-stranded chain
synthesized by the present invention may not have the same
structure as naturally occurring nucleic acid. It is known that
if a nucleotide derivative is used as a substrate when nucleic
acid is synthesized by the action of a DNA polymerase, a nucleic
acid derivative can be synthesized. The nucleotide derivative
used includes nucleotides labeled with a radioisotope or
nucleotide derivatives labeled with a binding ligand such as
biotin or digoxin. These nucleotide derivatives can be used
to label nucleic acid derivatives as the product. Alternatively,
CA 02390309 2002-05-07
if fluorescent nucleotides are used as a substrate, the nucleic
acid as the product can be a fluorescent derivative. Further,
this product may be either DNA or RNA. Which one is formed is
determined by a combination of the structure of a primer, the
type of a substrate for polymerization and polymerization
reagents for carrying out polymerization of nucleic acid.
Synthesis of the nucleic acid having the structure
described above can be initiated by use of a DNA polymerase having
the strand displacement activity and nucleic acid which is
provided at the 3'-terminal thereof with a region Fl capable
of annealing to a part Flc in the same chain and which upon
annealing of the region F1 to Fic, is capable of forming a loop
containing a region F2c capable of base pairing. There are many
reports on the reaction of synthesizing complementary chain
wherein a hairpin loop is formed and a sample sequence itself
is used as a template, while in the present invention the portion
of the hairpin loop is provided with a region capable of base
pairing, and there is a novel feature on utilization of this
region-in synthesizing complementary chain. By use of this
region as the origin of synthesis, a complementary chain
previously synthesized with a sample sequence itself as a
template is displaced. Then, a region Rlc (arbitrary region)
located at the 3' -terminal of the displaced chain is in a state
ready for base-pairing. A region having a complementary
sequence to this Ric is annealed thereto, resulting in formation
21
CA 02390309 2002-05-07
of the nucleic acid (2 molecules) having a nucleotide sequence
extending from Fl to Ric and its complementary chain linked
alternately via the loop-forming sequence. In the present
invention, the arbitrary region such as R1c above can be selected
arbitrarily provided that it can be annealed to a polynucleotide
having a nucleotide sequence complementary to that region, and
that a complementary chain synthesized with the polynucleotide
as the origin of synthesis has necessary functions for the present
invention.
In the present invention, the term "nucleic acid" is used.
The nucleic acid in the present invention generally includes
both DNA and RNA. However, nucleic acid whose nucleotide is
replaced by an artificial derivative or modified nucleic acid
from natural DNA or RNA is also included in the nucleic acid
of the present invention insofar as it functions as a template
for synthesis of complementary chain. The nucleic acid of the
present invention is generally contained in a biological sample.
The biological sample includes animal, plant or microbial tissues,
cells, cultures and excretions, or extracts therefrom. The
biological sample of the present invention includes
intracellular parasitic genomic DNA or RNA such as virus or
mycoplasma. The nucleic acid of the present invention may be
derived from nucleic acid contained in said biological sample.
For example, cDNA synthesized from mRNA, or nucleic acid
amplified on the basis of nucleic acid derived from the biological
22
CA 02390309 2002-05-07
sample, is a typical example of the nucleic acid of the present
invention.
The nucleic acid characteristic of the present invention
which is provided at the 3'-terminal thereof with a region F1
capable of annealing to a part Flc in the same chain and which
upon annealing of the region Fl to Flc, is capable of forming
a loop containing a region F2c capable of base pairing can be
obtained in various methods. In the most preferable embodiment,
the reaction of synthesizing complementary chain utilizing an
oligonucleotide having the following structure can be used to
give the structure.
That is, the useful oligonucleotide in the present
invention consists of at least two regions X2 and Xlc below wherein
Xlc is ligated to the 5'-side of X2.
X2: a region having a nucleotide sequence complementary to a
region X2c in nucleic acid having aspecific nucleotide sequence.
Xlc: a region having substantially the same nucleotide sequence
as a region Xlc located at the 5' -side of the region X2c in nucleic
acid having a specific nucleotide sequence.
Here, the nucleic acid having a specific nucleotide
sequence by which the structure of the oligonucleotide of the
invention is determined refers to nucleic acid serving as a
template when the oligonucleotide of the present invention is
used as a primer. In the case of detection of nucleic acid based
on the synthetic method of the present invention, the nucleic
23
CA 02390309 2002-05-07
acid having a specific nucleotide sequence is a detection target
or nucleic acid derived from the detection target. The nucleic
acid having a specific nucleotide sequence refers to nucleic
acid wherein at least a part of the nucleotide sequence is revealed
or predictable. The part of the nucleotide sequence revealed
is the region X2c and the regionXlc located at the 5' -side thereof .
It can be supposed that these 2 regions are contiguous or located
apart from each other. By the relative positional relationship
of the two, the state of a loop formed upon self-annealing of
nucleic acid as the product is determined. The distance between
the two is preferably not very apart from each other in order
that nucleic acid as the product is subjected to self-annealing
preferentially over intermolecular annealing. Accordingly,
the positional relationship of the two is preferably that they
are contiguous via a distance of usually 0 to 100 bases. However,
in the formation of a loop by self-annealing described below,
there can be the case where it would be disadvantageous for
formation of a loop in a desired state that the two are too close
to each other. In the loop, there is a need for a structure
for annealing of a new oligonucleotide and for readily initiating
the strand-displacement reaction of synthesizing a
complementary chain with said oligonucleotide as the origin of
synthesis. More preferably, the distance between the region
X2c and the region Xlc located at the 5' -side of X2c is designed
to be 0 to 100 bases, more desirably 10 to 70 bases. This numerical
24
CA 02390309 2008-12-17
value shows a length excluding Xlc and X2. The number of bases
constituting the part of a loop is that of this length plus a
region corresponding to X2.
Both the terms "same" and "complementary" used for
characterization of the nucleotide sequence constituting the
oligonucleotide based on the present invention do not mean being
absolutely the same or absolutely complementary. That is, the
same sequence as a certain sequence includes sequences
complementary to nucleotide sequences capable of annealing to
a certain sequence. On the other hand, the complementary
sequence means a sequence capable of annealing under stringent
conditions to provide a 3'-terminal serving as the origin of
synthesis of complementary chain.
Usually, the regions X2 and Xlc constituting the
oligonucleotide of the present invention for the nucleic acid
having a specific nucleotide sequence are located contiguously
without being overlapped. If there is a common part in both
the nucleotide sequences, the two can be partially overlaid.
Because X2 should function as a primer, it should always be a
31-terminal. On the other hand, Xlc should give the function
of a,primer as described below to the 3'-terminal of a complementary
chain synthesized with the nucleic acid as a template, and thus
it shall be arranged at the 5'-terminal. The complementary chain
obtained with this oligonucleotide as the origin of synthesis
serves as a template for synthesis of complementary chain in
CA 02390309 2002-05-07
the reverse direction in the next step, and finally the part
of the oligonucleotide of the present invention is copied as
a template into a complementary chain. The 3'-terminal
generated by copying has the nucleotide sequence X1, which
anneals to X1c in the same chain to form a loop.
In the present invention, the oligonucleotide means the
one that satisfies the 2 requirements, that is, it must be able
to form complementary base pairing and give an -OH group serving
as the origin of synthesis of complementary chain at the
3'-terminal. Accordingly, its backbone is not necessarily
limited to the one via phosphodiester linkages. For example,
it may be composed of a phosphothioate derivative having S in
place of 0 as a backbone or a peptide nucleic acid based on peptide
linkages. The bases may be those capable of complementary base
pairing. In the nature, there are 5 bases, that is, A, C, T,
G and U, but the base can be an analogue such as bromodeoxyuridine.
The oligonucleotide used in the present invention functions
preferably not only as the origin of synthesis but also as a
template for synthesis of complementary chain. The term
polynucleotide in the present invention includes
oligonucleotides. The term "polynucleotide" is used in the case
where the chain length is not limited, while the term
"oligonucleotide" is used to refer to a nucleotide polymer having
a relatively short chain length.
The oligonucleotide according to the present invention
26
^
CA 02390309 2002-05-07
has such a chain length as to be capable of base pairing with
a complementary chain and to maintain necessary specificity under
the given environment in the various reactions of synthesizing
nucleic acid described below. Specifically, it is composed of
to 200 base pairs, more preferably 10 to 50 base pairs. The
chain length of a primer recognizing the known polymerase
catalyzing the sequence-dependent nucleic acid synthetic
reaction is at least about 5 bases, so the chain length of the
annealing part should be longer than that. In addition, a length
of 10 bases or more is desired statistically in order to expect
specificity as the nucleotide sequence. On the other hand,
preparation of a too long. nucleotide sequence by chemical
synthesis is difficult, and thus the chain length described above
is exemplified as a desired range. The chain length exemplified
here refers to the chain length of a part annealing to a
complementary chain. As described below, the oligonucleotide
according to the present invention can anneal finally to at least
2 regions individually. Accordingly, it should be understood
that the chain length exemplified here is the chain length of
each region constituting the oligonucleotide.
Further, the oligonucleotide according to the present
invention can be labeled with a known labeling. substance. The
labeling substance includes binding ligands such as digoxin and
biotin, enzymes, fluorescent substances and luminescent
substances, and radioisotopes. The techniques of replacing a
27
CA 02390309 2002-05-07
base constituting an oligonucleotide by a fluorescent analogue
are also known (W095/05391, Proc. Natl. Acad. Sci. USA, 91,
6644-6648, 1994).
Other oligonucleot ides according to the present invention
can also have been bound to a solid phase. Alternatively, an
arbitrary part of the oligonucleotide may be labeled with a
binding ligand such as biotin, and it can be immobilized
indirectly via a binding partner such as immobilized avidin.
When the immobilized oligonucleotide is used as the origin of
synthesis, nucleic acid as the synthetic reaction product is
captured by the solid phase, thus facilitating its separation.
The separated product can be detected by a nucleic acid-specific
indicator or by hybridization with a labeling probe. The target
nucleic acid fragments can also be recovered by digesting the
product with arbitrary restriction enzymes.
The term "template" used in the present invention means
nucleic acid serving as a template for synthesizing a
complementary chain. A complementary chain having a nucleotide
sequence complementary to the template has a meaning as a chain
corresponding to the template, but the relationship between the
two is merely relative. That is, a chain synthesized as the
complementary chain can function again as a template. That is,
the complementary chain can become a template.
The oligonucleotide useful in the present invention is
not limited to the 2 regions described above and can contain
28
^
CA 02390309 2002-05-07
an additional region. While X2 and Xlc are arranged at the 3' -
and 5'-terminals respectively, an arbitrary sequence can be
interposed therebetween. For example, it can be a restriction
enzyme recognition site, a promoter recognized by RNA polymerase,
or DNA coding for ribozyme. By using it as a restriction enzyme
recognition sequence, the nucleic acid having a complementary
sequence alternately linked in a single-stranded chain as the
synthetic product of the present invention can be cleaved into
double-stranded nucleic acids of the same length. By arranging
a promoter sequence recognized by RNA polymerase, the synthetic
product of the present invention serves as the template to permit
further transcription into RNA. By further arranging DNA coding
for ribozyme, a system where the transcriptional product is
self-cleaved is realized. These additional nucleotide
sequences are those functioning after formed into a
double-stranded chain. Accordingly, when the single-stranded
nucleic acid according to the present invention has formed a
loop, these sequences do not function. They do not function
until the nucleic acid is elongated and annealed in the absence
of a loop to a chain having a complementary nucleotide sequence.
When a promoter is combined with the oligonucleotide based
on the present invention in such a direction as to permit
transcription of the synthesized region, the reaction product
based on the present invention where the same nucleotide sequence
is repeated realizes a highly efficient transcriptional system.
29
CA 02390309 2002-05-07
By combining this system with a suitable expression system,
translation into a protein is also feasible. That is, the system
can be utilized for transcription and translation into protein
in bacteria or animal cells or in vitro.
The oligonucleotide of the present invention having the
structure described above can be chemically synthesized.
Alternatively, natural nucleic acid may be cleaved with e.g.
restriction enzymes and modified so as to be composed of, or
ligated into, the nucleotide sequence described above.
The basic principle of the reaction for performing
synthesis by utilizing the useful oligonucleotide described
above in combination with DNA polymerase having the strand
displacement activity in the reaction of synthesizing nucleic
acid according to the present invention is describedby reference
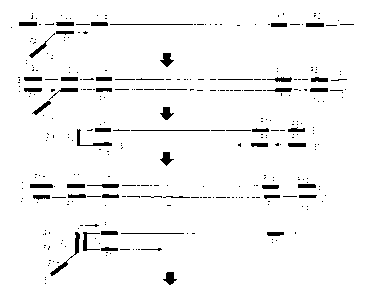
to Figs. 5 to 6. The oligonucleotide described above (FA in
Fig. 5) anneals at X2 (corresponding to F2) to nucleic acid as
a template, to provide the origin of synthesis of complementary
chain. In Fig. 5, a complementary chain synthesized from FA
as the origin of synthesis is displaced by synthesis of
complementary chain (described below) from an outer primer (F3),
to form a single-stranded chain (Fig. 5-A). When synthesis of
complementary chain to the resulting complementary chain is
further conducted, the 3'-terminal of nucleic acid synthesized
as complementary chain in Fig. 5-A has a nucleotide sequence
complementary to the oligonucleotide of the present invention.
CA 02390309 2002-05-07
That is, because the 5' -terminal of the oligonucleotide of the
present invention has the same sequence as a region Xlc
(corresponding to Fic) , the 3'-terminal of the nucleic acid thus
synthesized has a complementary sequence Xl (Fl). Fig. 5 shows
that the complementary chain synthesized from R1 as the origin
of synthesis is displaced by synthesis of complementary chain
by primer R3 as the origin of synthesis. Once the 3'-terminal
portion is made ready for base pairing by this displacement,
X1 (Fl) at the 3' -terminal anneals to Xic (F1c) in the same chain,
and elongation reaction with itself as a template proceeds (Fig.
5-B). Then, X2c (F2c) located at the 3'-terminal thereof is
left as a loop not involving base pairing. X2 (F2) in the
oligonucleotide according to the present invention anneals to
this loop, and a complementary chain is synthesized with said
oligonucleotide as the origin of synthesis (Fig. 5-B) . A product
of complementary chain synthetic reaction with the previously
synthesized product as a template is displaced by the strand
displacement reaction so that it is made ready for base pairing.
By the basic constitution using one kind of oligonucleotide
according to the present invention and an arbitrary reverse
primer capable of conducting nucleic acid synthesis where a
complementary chain synthesized with said oligonucleotide as
a primer is used as a template, a plurality of nucleic acid
synthetic products as shown in Fig. 6 can be obtained. As can
be seen from Fig. 6, (D) is the desired nucleic acid product
31
CA 02390309 2002-05-07
of the invention having complementary nucleotide sequence
alternately linked in a single-stranded chain. Once converted
into a single-stranded chain by treatment such as heat
denaturation, the other product (E) serves again as a template
for forming (D) . If the product (D) as nucleic acid in the form
of a double-stranded chain is converted into a single-stranded
chain by heat denaturation, annealing occurs within the same
chain at high probability without forming the original
double-stranded chain. This is because a complementary chain
having the same melting temperature (Tm) undergoes
intramolecular reaction preferentially over intermolecular
reaction. Each single-stranded chain derived from the product
(D) annealed in the same chain is annealed in the same chain
and returned to the state of (B), and each chain further gives
one molecule of (D) and (E) respectively. By repeating these
steps, it is possible to successively synthesize the nucleic
acid having complementary nucleotide sequences linked
alternately in a single-stranded chain. The template and the
product formed in 1 cycle are increased exponentially, thus
making the reaction very efficient.
To realize the state of Fig. 5(A), the initially
synthesized complementary chain should, in at least the portion
to which the reverse primer anneals, should be ready for base
pairing. This step can be achieved by an arbitrary method. That
is, an outer primer (F3), which anneals to the first template
32
CA 02390309 2002-05-07
at a region F3c at the 3'-side of the region F2c to which the
oligonucleotide of the present invention anneals, is separately
prepared. If this outer primer is used as the origin synthesis
to synthesize a complementary chain by a polymerase catalyzing
the strand displacement-type synthesis of complementary chain,
the complementary chain synthesized from the F2c as the origin
of synthesis in the invention is displaced, and as a result the
region Ric to be annealed by R1 is made ready for base pairing
(Fig. 5). By utilization of the strand displacement reaction,
the reaction up to now can proceed under isothermal conditions.
When an outer primer is used, synthesis from the outer
primer (F3) should be initiated after synthesis from F2c. In
the most simple method, the concentration of the inner primer
is made higher than the concentration of the outer primer.
Specifically, the primers are used at usually 2- to 50-fold,
preferably 4- to 10-fold different concentrations, whereby the
reaction can proceed as expected. Further, the melting
temperature (Tm) of the outer primer is set to be lower than
the Tm of the X1 (corresponding to Fl and Rl) in the inner primer
whereby the timing of synthesis can be controlled. That is,
(outer primer F3 . F3c) S F2c/F2) <- (Fic/F1) or (outer
primer/region at the 3'-side in the template). <_ (X2c : X2) 5
(Xlc : Xl) . Here, the reason for (F2c/F2) 5 (Fic/F1) is for
annealing between Flc/Fi prior to annealing of F2 to the loop.
The annealing between Fic/Fl is an intramolecular reaction and
33
CA 02390309 2002-05-07
may thus proceed preferentially at high probability. However,
it is meaningful to consider Tm in order to give more desired
reaction conditions. As a matter of course, similar conditions
should be considered even in the design of a reverse primer.
By using such a relationship, statistically ideal reaction
conditions can be achieved. If other conditions are fixed,
melting temperature (Tm) can be theoretically calculated by a
combination of the length of an annealing complementary chain
and bases constituting base-pairing. Accordingly, those
skilled in the art can derive preferable conditions on the basis
of the disclosure of this specification.
Further, the phenomenon called contiguous stacking can
also be applied for controlling timing of annealing of the outer
primer. Contiguous stacking is a phenomenon in which an
oligonucleotide not capable of annealing independently is made
capable of annealing upon being contiguous to the part of a
double-stranded chain (Chiara Borghesi-Nicoletti et al., Bio
Techniques, 12, 474-477 (1992)) . That is, the outer primer is
designed so as to be contiguous to F2c (X2c) and not to be able
to anneal independently. By doing so, annealing of the outer
primer does not occur until F2c (X2c) anneals, and thus the
annealing of F2c (X2c) occurs preferentially. On the basis of
this principle, the Examples show setting of the nucleotide
sequence of an oligonucleotide necessary as a primer for a series
of reactions. This step can also be achieved by denaturation
34
CA 02390309 2002-05-07
under heating or with a DNA helicase.
If the template nucleic acid having F2c (X2c) is RNA, the
state of Fig. 5- (A) can also be realized by a different method.
For example, if this RNA chain is decomposed, R1 is made ready
for base pairing. That is, F2 is annealed to F2c in RNA and
a complementary chain is synthesized as DNA by a reverse
transcriptase. The RNA serving as a template is decomposed by
alkali denaturation or by enzymatic treatment with a ribonuclease
acting on RNA in a double-stranded chain of DNA/RNA whereby the
DNA synthesized from F2 is formed into a single-stranded chain.
For the enzyme selectively decomposing RNA in a double-stranded
chain of DNA/RNA, the ribonuclease activity of RNase H or some
reverse transcriptases can be utilized. In this manner, the
reverse primer can be annealed to Ric made capable of base pairing.
Accordingly, the outer primer for rendering Ric ready for base
pairing becomes unnecessary.
Alternatively, the strand displacement activity of
reverse transcriptase can be utilized for the strand displacement
by an outer primer as described above. In this case, a reaction
system can be constituted by a reverse transcriptase only. That
is, using RNA as a template, it is made possible by a reverse
transcriptase to synthesize a complementary chain from F2
annealing to F2c in the template and to synthesize a complementary
chain from the outer primer F3 as the origin of synthesis annealing
to F3c located at the 3' -side of F2c and to simultaneously displace
CA 02390309 2002-05-07
the previously synthesized complementary chain. When the
reverse transcriptase performs the reaction of synthesizing a
complementary chain with DNA as the template, all the reactions
of synthesizing complementary chains including the synthesis
of a complementary chain with R1 as the origin of synthesis
annealing to Rlc in the displaced complementary chain as the
template, the synthesis of a complementary chain with R3 as the
origin of synthesis annealing to Ric located at the 3' -side of
Rlc and the simultaneous displacement reaction, proceed by the
reverse transcriptase. If it is not possible to expect that
the reverse transcriptase exhibits the DNA/RNA strand
displacement activity under given reaction c.onditions, a DNA
polymerase having the strand displacement activity described
above may be combined. The mode of obtaining a first
single-stranded nucleic acid with RNA as a template as described
above constitutes a preferable mode of the present invention.
On the other hand, if a DNA polymerase such as Bca DNA polymerase
having both strand displacement activity and reverse
transcriptase activity is used, not only synthesis of a first
single-stranded nucleic acid from RNA but also subsequent
reaction with DNA as a template can proceed similarly by the
same enzyme.
The reaction system described above brings about various
variations inherent in the present invention by utilization of
the reverse primer having a specific structure. The most
36
CA 02390309 2002-05-07
effective variation is described below. That is, the
oligonucleotide constituted as described in [5] is used as the
reverse primer in the most advantageous mode of the present
invention. The oligonucleotide in [5] is an oligonucleotide
wherein arbitrary regions R2c and Ric in a complementary chain
synthesized with F2 as a primer are X2c and Xlc respectively.
By use of such a reverse primer, a series of reactions for forming
a loop and f or synthesizing and displacing a complementary chain
from this loop occur in both the sense and antisense chains
(forward side and reverse side). As a result, the reaction
efficiency for synthesis of the nucleic acid having complementary
nucleotide sequences linked alternately in a single-stranded
chain according to the present invention is greatly improved
while a series of these reactions are feasible under isothermal
conditions. Hereinafter, this mode is described in more detail
by reference to Figs. 1 to 3 where this mode is summarized.
In the following mode, 2 kinds of oligonucleotides based
on the present invention are prepared. For explanation, these
are designated FA and RA. The regions constituting FA and RA
are as follows:
X2 Xlc
FA F2 Flc
RA R2 R1c
Here, F2 is a complementary nucleotide sequence to a region
F2c in nucleic acid as the template. R2 is a nucleotide sequence
37
CA 02390309 2002-05-07
complementary to an arbitrary region R2c contained in a
complementary chain synthesized with F2 as a primer. Flc and
R1c are arbitrary nucleotide sequences located downward from
F2c and R2c respectively. The distance between F2 and R2 may
be arbitrary. Even if its length is about 1 kbp, sufficient
synthesis is feasible under suitable conditions, though
depending on the synthetic ability of DNA polymerase to perform
the synthesis of a complementary chain. Specifically, when Bst
DNA polymerase is used, the desired product is certainly
synthesized if the distance between F2 and R2c is 800 bp,
preferably 500 bp or less. In PCR involving temperature cycle,
the reduction in the enzyme activity by the stress of temperature
change is considered to reduce the efficiency of synthesis of
a long nucleotide sequence. In a preferable mode of the present
invention, the temperature cycle in the step of amplifying
nucleic acid is not required, and thus the synthesis and
amplification of an even long nucleotide sequence can be
certainly achieved.
First, F2 in FA is annealed to nucleic acid as a template
and used as the origin of synthesis of a complementary chain.
The subsequent reaction steps until Fig. 1(4) are the same as
in the previously described basic mode (Fig. 5) in the present
invention. The sequence annealed as F3 in Fig. 1 (2) is the outer
primer described above. A DNA polymerase for conducting the
strand displacement-type synthesis of a complementary chain with
38
CA 02390309 2002-05-07
this primer as the origin of synthesis is used so that the
complementary chain synthesized from FA is displaced and made
ready for base pairing.
When R2c is made ready for base pairing in (4), RA as a
reverse primer anneals thereto in the combination of R2c/R2.
Synthesis of a complementary chain with this site as the origin
of synthesis proceeds until the chain reaches Flc at the
5'-terminal of FA. Following this reaction of synthesizing a
complementary chain, the outer primer R3 for displacement anneals
thereto to synthesize a complementary chain, during which strand
displacement also proceeds so that the complementary chain
synthesized from RA as the origin of synthesis is displaced.
In the complementary chain thus displaced, RA is located at the
5'-side thereof and a sequence complementary to FA is located
at the 3'-terminal thereof.
At the 3' -side of the single-stranded nucleic acid thus
displaced, there is a sequence F1 complementary to Flc in the
same chain. Fl rapidly anneals to Flc in the same molecule to
initiate synthesis of a complementary chain. When the
3'-terminal (Fl) anneals to Flc in the same chain, a loop
containing F2c is formed. As is also evident from Fig. 2- (7) ,
the part of this loop remains ready for base pairing. The
oligonucleotide FA of the invention having a nucleotide sequence
complementary to F2c anneals to the part of this loop and acts
as the origin of synthesis of a complementary chain (7).
39
CA 02390309 2002-05-07
Synthesis of a complementary chain from the loop proceeds while
the reaction product in the previously initiated complementary
chain synthesis from Fl is displaced. As a result, the
complementary chain synthesized with itself as the template is
made ready for base pairing again at the 3'-terminal. This
3'-terminal is provided with a region R1 capable of annealing
to Rlc in the same chain, and the two are annealed preferentially
due to the rapid intramolecular reaction. The same reaction
as the above-described reaction starting from the 3'-terminal
synthesized with FA as a template proceeds in this region as
well. As a result, the nucleic acid having complementary
nucleotide sequences linked alternately in the same
single-stranded chain according to the present invention is
continued to be extended from R1 as the starting point at the
3'-terminal by successive synthesis of a complementary chain
and subsequent displacement thereof. Because R2c is always
contained in the loop formed by intramolecular annealing of the
3'-terminal R1, the oligonucleotide (RA) provided with R2 anneals
to the loop at the 3'-terminal in the subsequent reaction.
When attention is paid to nucleic acid synthesized as
complementary chain from the oligonucleotide annealing to the
loop in the single-stranded nucleic acid elongated with itself
as the template, synthesis of the nucleic acid having
complementary nucleotide sequences linked alternately in the
same single-stranded chain according to the present invention
CA 02390309 2002-05-07
also proceeds here. That is, synthesis of a complementary chain
from the loop is completed when it reached RA in e . g. Fig. 2- (7) .
Then, when the nucleic acid displaced by this nucleic acid
synthesis initiates synthesis of complementary chain (Fig.
3- (8) ) , the reaction reaches the loop which was once the origin
of synthesis, and displacement is initiated again. In this
manner, the nucleic acid initiated to be synthesized from the
loop is also displaced, and as a result, the 3' -terminal R1 capable
of annealing in the same chain is obtained (Fig. 3- (10)) . This
3'-terminal Ri anneals to Rlc in the same chain to initiate
synthesis of complementary chain. This reaction is the same
as in Fig. 2- (7) except that F is used in place of R. Accordingly,
the structure shown in Fig. 3- (10) can function as a new nucleic
acid which continues self-elongation and new nucleic acid
formation.
The reaction of synthesizing nucleic acid, initiated from
the nucleic acid shown in Fig. 3-(10), causes elongation from
the 3'-terminal Fl as the origin of synthesis, as opposed to
the reaction described above. That is, in the present invention,
as one nucleic acid is elongated, the reaction of continuing
to supply a new nucleic acid initiating elongation separately
proceeds. Further, as the chain is elongated, a plurality of
loop-forming sequences are brought about not only at the terminal
but also in the same chain. When these loop-forming sequences
are made ready for base pairing by the strand displacement
41
CA 02390309 2002-05-07
synthetic reaction, an oligonucleotide anneals thereto to serve
as a base for the reaction of forming a new nucleic acid. Further
efficient amplification is achieved by the synthetic reaction
starting not only at the terminal but also in the chain. The
oligonucleotide RA based on the present invention is combined
as the reverse primer as described above whereby elongation and
subsequent formation of a new nucleic acid occur. Further, in
the present invention, this newly formed nucleic acid itself
is elongated and brings about subsequent formation of a new
nucleic acid. A series of these reactions continue
theoretically permanently to achieve very efficient
amplification of nucleic acid. In addition, the reaction in
the present invention can be conducted under isothermal
conditions.
The reaction products thus accumulated possess a structure
having a nucleotide sequence between Fl and Rl and its
complementary sequence linked alternately therein. However,
both the terminals of the repeating unit have a region consisting
of the successive nucleotide sequences F2-F1 (F2c-F1c) and R2-R1
(R2c-Rlc). For example, in Fig. 3-(9), the sequences
(R2-F2c) - (F1-R2c) - (Rl-Flc) - (F2-R2c) are linked in this order
from the 5'-side. This is because the amplification reaction
based on the present invention proceeds on the principle that
the reaction is initiated from F2 (or R2) with an oligonucleotide
as the origin of synthesis and then a complementary chain is
42
CA 02390309 2002-05-07
elongated by the synthetic reaction from Fl (or Ri) with the
3'-terminal as the origin of synthesis.
Here, in the most preferable mode, oligonucleotides FA
and RA according to the present invention were used as
ol igonucleot ides annealing to the part of a loop. However, even
if these oligonucleotide having a limited structure are not used,
the amplification reaction according to the present invention
can be carried out by use of an oligonucleotide capable of
initiating the synthesis of a complementary chain from the loop.
That is, the elongating 3'-terminal, once displaced by a
complementary chain synthesized from the loop, gives the part
of a loop again. Because the nucleic acid having complementary
nucleotide sequences linked alternately in a single-stranded
chain is always used as a template in the complementary chain
synthesis starting from the loop, it is evident that the nucleic
acid desired in the present invention can be synthesized.
However, the nucleic acid thus synthesized performs synthesis
of a complementary chain by forming a loop after displacement,
but there is no 3'-terminal available for subsequent formation
of a loop, and thus it cannot function as a new template.
Accordingly, the product in this case, unlike nucleic acid
initiated to be synthesized by FA or RA, cannot be expected to
be exponentially amplified. From this reason, an
oligonucleotide having the structure of FA or RA is useful for
highly efficient synthesis of nucleic acid based on the present
43
CA 02390309 2002-05-07
invention.
A series of these reactions proceed by adding the following
components to single-stranded nucleic acid as a template and
then incubating the mixture at such a temperature that the
nucleotide sequence constituting FA and RA can form stable base
pairing with its complementary nucleotide sequence while the
enzyme activity can be maintained.
= 4 kinds of oligonucleotides:
FA,
RA,
outer primer F3, and
outer primer R3,
= DNA polymerase for performing the strand displacement-type
synthesis of complementary chain,
= an oligonucleotide serving as a substrate for DNA polymerase.
Accordingly, temperature cycle such as in PCR is not
necessary. The stable base pairing referred to herein means
a state in which at least a part of an oligonucleotide present
in the reaction system can give the origin of synthesis of
complementary chain. For example, the desired condition for
bringing about stable base pairing is to set lower than melting
temperature (Tm) . Generally, melting temperature (Tm) is
regarded as the temperature at which 50 % of nucleic acids having
mutually complementary nucleotide sequences are base-paired.
Setting at melting temperature (Tm) or less is not an essential
44
CA 02390309 2002-05-07
condition in the present invention, but is one of the reaction
conditions to be considered for attaining high efficiency of
synthesis. If nucleic acid to be used as a template is a
double-stranded chain, the nucleic acid should, in at least a
region to which the oligonucleotide anneals, be made ready for
base pairing. For this, heat denaturation is generally
conducted, and this may be conducted only once as pretreatment
before the reaction is initiated.
This reaction is conducted in the presence of a buffer
giving suitable pH to the enzyme reaction, salts necessary for
annealing or for maintaining the catalytic activity of the enzyme,
a protective agent for the enzyme, and as necessary a regulator
for melting temperature (Tm). As the buffer, e.g. Tris-HC1
having a buffering action in a neutral to weakly alkaline range
is used. The pH is adjusted depending on the DNA polymerase
used. As the salts, KC1, NaCl, (NH4) 2SO4 etc. are suitably added
to maintain the activity of the enzyme and to regulate the melting
temperature (Tm) of nucleic acid. The protective agent for the
enzyme makes use of bovine serum albumin or sugars. Further,
dimethyl sulfoxide (DMSO) or formamide is generally used as the
regulator for melting temperature (Tm) . By use of the regulator
for melting temperature (Tm), annealing of the oligonucleotide
can be regulated under limited temperature conditions. Further,
betaine (N,N,N-trimethylglycine) or a tetraalkyl ammonium salt
is also effective for improving the efficiency of strand
CA 02390309 2002-05-07
displacement by virtue of its isostabilization. By adding
betaine in an amount of 0.2 to 3.0 M, preferably 0.5 to 1.5 M
to the reaction solution, its promoting action on the nucleic
acid amplification of the present invention can be expected.
Because these regulators for melting temperature act for lowering
melting temperature, those conditions giving suitable
stringency and reactivity are empirically determined in
consideration of the concentration of salts, reaction
temperature etc.
An important feature in the present invention is that a
series of reactions do not proceed unless the positional
relationship of a plurality of regions is maintained. By this
feature, unspecific synthetic reaction accompanied by
unspecific synthesis of complementary chain is effectively
prevented. That is, even if a certain unspecific reaction occurs,
the possibility for the product to serve as a starting material
in the subsequent amplification step is minimized. Further,
the regulation of the progress of reactions by many regions brings
about the possibility that a detection system capable of strict
identification of the desired product in analogous nucleotide
sequences can be arbitrarily constituted.
This feature can be utilized for detection of mutations
in a gene. In the mode of the invention where the outer primer
is used, 4 primers, that is, 2 outer primers and 2 primers
consisting of the oligonucleotides of the present invention,
46
CA 02390309 2002-05-07
are used. That is, unless the 6 regions contained in the 4
oligonucleotides work as designed, the synthetic reaction of
the present invention do not proceed. In particular, the
sequences of the 3'-terminal of each oligonucleotide as the
origin of synthesis of complementary chain and of the 5' -terminal
of the X1c region where the complementary chain serves as the
origin of synthesis are important. Hence, these important
sequences is designed so as to correspond to a mutation to be
detected, and the synthetic reaction product of the present
invention is observed whereby the presence or absence of a
mutation such as base deletion or insertion, or genetic
polymorphism such as SNPs can be comprehensively analyzed.
Specifically, bases estimated to have a mutation or polymorphism
are designed so as to correspond to the vicinity of the 3' -terminal
of an oligonucleotide as the origin of synthesis of complementary
chain, or of 5' -terminal thereof when a complementary chain is
the origin of synthesis. If a mismatch is present at the
3'-terminal as the origin of synthesis of complementary chain
or in its vicinity, the reaction of synthesizing a complementary
chain to nucleic acid is significantly inhibited. In thepresent
invention, a high degree of amplification reaction is not
achieved unless the structure of the terminals of a product in
the initial reaction brings about repeated reactions.
Accordingly, even if erroneous synthesis occurs, complementary
chain synthesis constituting amplification reaction is always
47
CA 02390309 2002-05-07
interrupted in some of the steps, and thus a high degree of
amplification reaction does not occur in the presence of a
mismatch. As a result, the mismatch effectively inhibits
amplification reaction, and an accurate result is finally brought
about. That is, it can be said that the amplification reaction
of nucleic acid based on the present invention has a highly
completed mechanism for checking the nucleotide sequence.
These features are an advantage hardly expectable in e. g. the
PCR method where amplification reaction is performed in mere
2 regions.
The region Xlc characterizing the oligonucleotide used
in the present invention can serve as the origin of synthesis
after a complementary sequence is synthesized, and this
complementary sequence anneals to the sequence Xl in the same
newly synthesized chain whereby synthetic reaction with itself
as a template proceeds. Therefore, even if the so-called primer
dimer which is often problematic in the prior art is formed,
this oligonucleotide does not form a loop. Accordingly,
unspecific amplification attributable to the primer dimer cannot
occur theoretically, and thus the present oligonucleotide
contributes to an improvement in the specificity of the reaction.
Further, according to the present invention, the outer
primers shown as F3 (Fig. 1- (2) ) or R3 (Fig. 2- (5) ) are combined
whereby a series of the reactions described above can be conducted
under isothermal conditions. That is, the present invention
48
CA 02390309 2002-05-07
provides a method of amplifying nucleic acid having complementary
sequences linked alternately in a single-stranded chain, which
comprises the steps shown in item 9 above. In this method,
temperature conditions where stable annealing occurs between
F2c/F2, between R2c/R2, between Fic/Fl, and between Ric/R1 are
selected, and preferably F3c/F3 and R3c/R3 are set up to be
annealed by the phenomenon of contiguous stacking facilitated
by annealing of F2c/F2 and R2c/R2, respectively.
In the present invention, the terms "synthesis" and
"amplification" of nucleic acid are used. The synthesis of
nucleic acid in the present invention means the elongation of
nucleic acid from an oligonucleotide serving as the origin of
synthesis. If not only this synthesis but also the formation
of other nucleic acid and the elongation reaction of this formed
nucleic acid occur continuously, a series of these reactions
is comprehensively called amplification.
The single-stranded nucleic acid which is provided at the
3'-terminal thereof with a region Fl capable of annealing to
a part Flc in the same chain and which upon annealing of the
region Fl to Fic in the same chain, is capable of forming a loop
containing a region F2c capable of base pairing is an important
element of the present invention. Such a single-stranded
nucleic acid can also be supplied on the following principle.
That is, the synthesis of a complementary chain is allowed to
proceed on the basis of a primer having the following structure.
49
CA 02390309 2002-05-07
5' - [region Xl annealing to region Xlc located in primer] - [loop
forming sequence ready for base pairing] -(region Xlc] - [region
having a sequence complementary to a template]-3'
As the region having a sequence complementary to a template,
two nucleotide sequences, that is, a nucleotide sequence (primer
FA) complementary to Fl and a nucleotide sequence (primer RA)
complementary to Ric, are prepared. The nucleotide sequence
constituting nucleic acid to be synthesized contains a nucleotide
sequence extending from the region F1 to the region Rlc and a
nucleotide sequence extending from the region R1 having a
nucleotide sequence complementary to this nucleotide sequence
to the region Fic. Xlc and X1 capable of annealing in the inside
of the primer can be arbitrary sequences. However, in a region
between primers FA and RF, the sequence of the region Xlc/X1
is made preferably different.
First, the synthesis of a complementary chain by the primer
FA from the region Fl in template nucleic acid is conducted.
Then, the region Rlc in the synthesized complementary chain is
made ready for base pairing, to which the other primer is annealed
to form the origin of synthesis of complementary chain. The
3'-terminal of the complementary chain synthesized in this step
has a nucleotide sequence complementary to the primer FA
constituting the 5'-terminal of the initially synthesized chain,
so it has been provided at the 3' -terminal thereof with the region
X1 which anneals to the region Xlc in the same chain to form
CA 02390309 2002-05-07
a loop. The characteristic 3'-terminal structure according to
the present invention is thus provided, and the subsequent
reaction constitutes the reaction system shown previously as
the most preferable mode. The oligonucleotide annealing to the
portion of the loop is provided at the 3' -terminal thereof with
the region X2 complementary to the region X2c located in the
loop and at the 5'-terminal thereof with the region X1. In the
previous reaction system, primers FA and RA were used to
synthesize a chain complementary to template nucleic acid thereby
giving a loop structure to the 3' -terminal of the nucleic acid.
In this method, the terminal structure characteristic of the
present invention is provided by the short primers. In this
mode, on the other hand, the whole of a nucleotide sequence
constituting a loop is provided as a primer, and synthesis of
this longer primer is necessary.
If a nucleotide sequence containing restriction enzyme
recognition regions is used as a reverse primer, a different
mode according to the present invention can be constituted. The
reaction with a reverse primer containing a restriction enzyme
recognition sequence is specifically described by reference to
Fig. 6. When Fig. 6-(D) is completed, a nick is generated by
a restriction enzyme corresponding to a restriction enzyme
recognition site in the reverse primer. The strand
displacement-type reaction of synthesizing complementary chain
is initiated from this nick as the origin of synthesis. Because
51
CA 02390309 2002-05-07
the reverse primers are located at both the terminals of a
double-stranded nucleic acid constituting (D), the reaction of
synthesizing complementary chain is also initiated from both
the terminals. Though basically based on the SDA method
described as the prior art, the nucleotide sequence serving as
a template has a structure having complementary nucleotide
sequences alternately linked according to the present invention
so that the nucleic acid synthetic system unique to the present
invention is constituted. A part serving as a complementary
chain of the reverse primer to be nicked should be designed to
incorporate a dNTP derivative such that it is rendered nuclease
resistance to prevent cleavage of the double-stranded chain by
the restriction enzyme.
It is also possible to insert a promoter for RNA polymerase
into the reverse primer. Transcription from both the terminals
in Fig. 6- (D) is performed by a RNA polymerase recognizing this
promoter in this case too similar to the previous mode where
the SDA method was applied.
The nucleic acid synthesized in the present invention is
a single-stranded chain but is composed of partial complementary
sequences, and thus the majority of these sequences are
base-paired. By use of this feature, the synthesized product
can be detected. By carrying out the method of synthesizing
nucleic acid according to the present invention in the presence
of a fluorescent pigment as a double-stranded chain-specific
52
CA 02390309 2002-05-07
intercalater such as ethidiumbromide, SYBR Green I or Pico Green,
the increased density of fluorescence is observed as the product
is increased. By monitoring it, it is possible to trace the
real-time synthetic reaction in a closed system. Application
of this type of detection system to the PCR method is also
considered, but it is deemed that there are many problems because
the signal from the product cannot be distinguished from signals
from primer dimers etc. However, when this system is applied
to this invention, the possibility of increasing unspecific base
pairing is very low, and thus high sensitivity and low noises
can be simultaneously expected. Similar to use of the
double-stranded chain-specific intercalater, the transfer of
fluorescent energy can be utilized for a method of realizing
a detection system in a uniform system.
The method of synthesizing nucleic acid according to the
present invention is supported by the DNA polymerase catalyzing
the strand displacement-type reaction for synthesis of
complementary chain. During the reaction described above, a
reaction step not necessarily requiring the strand
displacement-type polymerase is also contained. However, for
simplification of a constitutional reagent and in an economical
viewpoint, it is advantageous to use one kind of DNA polymerase.
As this kind of DNA polymerase, the following enzymes are known.
Further, various mutants of these enzymes can be utilized in
the present invention .insofar as they have both the
53
^
CA 02390309 2002-05-07
sequence-dependent activity for synthesis of complementary
chain and the strand displacement activity. The mutants
referred to herein include those having only a structure bringing
about the catalytic activity required of the enzyme or those
with modifications to catalytic activity, stability or
thermostability by e.g. mutations in amino acids.
Bst DNA polymerase
Bca (exo-)DNA polymerase
DNA polymerase I Klenow fragment
Vent DNA polymerase
Vent (exo-)DNA polymerase (Vent DNA polymerase deficient in
exonuclease activity)
Deep Vent DNA polymerase
Deep Vent(exo-)DNA polymerase (Deep Vent DNA polymerase
deficient in exonuclease activity)
029 phage DNA polymerase
MS-2 phage DNA polymerase
Z-Taq DNA polymerase (Takara Shuzo Co., Ltd.)
KOD DNA polymerase (Toyobo Co., Ltd.)
Among these enzymes, Bst DNA polymerase and Bca (exo-)
DNA polymerase are particularly desired enzymes because they
have a certain degree of thermostability and high catalytic
activity. The reaction of this invention can be carried
isothermally in a preferred embodiment, but because of the
adjustment of melting temperature (Tm) etc., it is not always
54
CA 02390309 2002-05-07
possible to utilize temperature conditions desired for the
stability of the enzyme. Accordingly, it is one of the desired
conditions that the enzyme is thermostable. Although the
isothermal reaction is feasible, heat denaturation may be
conducted to provide nucleic acid as a first template, and in
this respect too, utilization of a thermostable enzyme broadens
selection of assay protocol.
Vent (exo-) DNA polymerase is an enzyme having both strand
displacement activity and a high degree of thermostability. It
is known that the complementary chain synthetic reaction
involving strand displacement by DNA polymerase is promoted by
adding a single strand binding protein (Paul M. Lizardi et al.,
Nature Genetics, 19, 225-232, July, 1998). This action is
applied to the present invention, and by adding the single strand
binding protein, the effect of promoting the synthesis of
complementary chain can be expected. For example, T4 gene 32
is effective as a single strand binding protein for Vent (exo-)
DNA polymerase.
For DNA polymerase free of 3'-5' exonuclease activity,
there is a known phenomenon where the synthesis of complementary
chain does not stop at the 5' -terminal of a template, resulting
in generation ofa one-base protrusion. In the present invention,
this phenomenon is not preferable because when synthesis of the
complementary chain reaches the terminal, the sequence of the
3'-terminal leads to initiation of next synthesis of
CA 02390309 2002-05-07
complementary chain. However, because a base "A" is added at
high probability to the 3'-terminal by the DNA polymerase, the
sequence may be selected such that the synthesis from the
3'-terminal starts at "A", so that there is no problem if an
additional base is added erroneously by dATP. Further, even
if the 3'-terminal is protruded during synthesis of complementary
chain, the 3'-a5' exonuclease activity can be utilized for
digesting the protrusion to make it blunt-ended. For example,
since natural Vent DNA polymerase has this activity, this enzyme
may be used as a mixture with Vent (exo-) DNA polymerase in order
to solve this problem.
Various reagents necessary for the method of synthesizing
or amplifying nucleic acid according to the present invention
maybe previously packaged and provided as a kit. Specifically,
a kit is provided for the present invention, comprising various
kinds of oligonucleotides necessary as primers for synthesis
of complementary chain and as outer primers for displacement,
dNTP as a substrate for synthesis of complementary chain, a DNA
polymerase for carrying out the strand displacement-type
synthesis of complementary chain, a buffer giving suitable
conditions to the enzyme reaction, and as necessary regents
necessary for detection of synthetic reaction products. In
particular, the addition of reagents is necessary during the
reaction in a preferable mode of the present invention, and thus
the reagents necessary for one reaction are supplied after
56
CA 02390309 2002-05-07
pipetted into reaction vessel, whereby the reaction can be
initiated by adding only a sample. By constituting a system
in which the reaction product can be detected in situ in a reaction
vessel by utilizing a luminescent signal or a fluorescent signal,
it is not necessary to open and shut the vessel after reaction.
This is very desirable for prevention of contamination.
The nucleic acid having complementary nucleotide
sequences alternately linked in a single-stranded chain,
synthesized according to the present invention, has e.g. the
following usefulness. The first feature is use of an advantage
resulting from the special structure having complementary
sequences in one molecule.. This feature can be expected to
facilitate detection. That is, there is a known system for
detecting nucleic acid wherein its signal is varied depending
on base pairing with a complementary nucleotide sequence. For
example, by combination with the method of utilizing a
double-stranded chain-specific intercalater as a detector as
described above, a detection system making full use of the
characteristics of the synthetic product of the present invention
can be realized. If the synthetic reaction product of the present
invention is once heat-denatured in said detection system and
returned to the original temperature, intramolecular annealing
occurs preferentially thus permitting complementary sequences
to be rapidly base-paired. If there are unspecific reaction
products, they have not complementary sequences in the molecule
57
CA 02390309 2002-05-07
so that after separated by heat denaturation into 2 or more
molecules, they cannot immediately be returned to the original
double-stranded chain. By providing the step of heat
denaturation before detection, noises accompanying the
unspecific reaction can be reduced. If the DNA polymerase not
resistant to heat is used, the step of heat denaturation has
the meaning of termination of the reaction and is thus
advantageous for the control of reaction temperature.
The second feature is to always form a loop capable of
base pairing. The structure of a loop capable of base pairing
is shown in Fig. 4. As can be seen from Fig. 4, the loop is
composed of the nucleotide sequence F2c (X2c) which can be
annealed by the primer and a nucleotide sequence intervening
between F2c-Flc(Xlc). The sequence between F2c-Flc (or between
X2c-Xlc in a more universal form) is a nucleotide derived sequence
derived from the template. Accordingly, if a probe having a
complementary nucleotide sequence is hybridized with this region,
template-specific detection is feasible. In addition, this
region is always ready for base pairing, and therefore, heat
denaturation prior to hybridization is not necessary. The
nucleotide sequence constituting a loop in the amplification
reaction product in the present invention may have an arbitrary
length. Accordingly, if hybridization with a probe is desired,
a region to be annealed by the primer and a region to be hybridized
by the probe are arranged separately to prevent their competition,
58
^
CA 02390309 2002-05-07
whereby ideal reaction conditions can be constituted.
According to a preferable mode of the present invention,
a large number of loops capable of base pairing are given in
a single strand of nucleic acid. This means that a large number
of probes can be hybridized with one molecule of nucleic acid
to permit highly sensitive detection. It is thus possible to
realize not only the improvement of sensitivity but also a method
of detecting nucleic acid based on a special reaction principle
such as aggregation. For example, a probe immobilized onto fine
particles such as polystyrene latex is added to the reaction
product of the present invention, the aggregation of latex
particles is observed as the hybridization of the product with
the probe proceeds. Highly sensitive and quantitative
observation is feasible by optically measuring the strength of
the aggregation. Because the aggregation can also be observed
with the naked eyes, a reaction system not using an optical
measuring device can also be constituted.
Further, the reaction product of the present invention
permitting many labels to be bound thereto per nucleic acid
molecule enables chromatographic detection. In the field of
immunoassay, an analytical method (immunochromatography) using
a chromatographic medium utilizing a visually detectable label
is used practically. This method is based on the principle that
an analyte is sandwiched between an antibody immobilized on a
chromatographic medium and a labeled antibody, and the unreacted
59
CA 02390309 2002-05-07
labeled component is washed away. The reaction product of the
present invention makes this principle applicable to analysis
of nucleic acid. That is, a labeled probe toward the part of
a loop is prepared and immobilized onto a chromatographic medium
to prepare a capturing probe for trapping thereby permitting
analysis in the chromatographic medium. As the capturing probe,
a sequence complementary to the part of the loop can be utilized.
Since the reaction product of the present invention has a large
number of loops, the product binds to a large number of labeled
probes to give a visually recognizable signal.
The reaction product according to the present invention
always giving a region as a loop capable of base pairing enables
a wide variety of other detection systems. For example, a
detection system utilizing surface plasmon resonance using an
immobilized probe for this loop portion is feasible. Further,
if a probe for the loop portion is labeled with a double-stranded
chain-specific intercalater, more sensitive fluorescent
analysis can be conducted. Alternatively, it is also possible
to positively utilize the ability of the nucleic acid synthesized
by the present invention to form a loop capable of base pairing
at both the 3' - and 5'-sides. For example, one loop is designed
to have a common nucleotide sequence between a normal type and
an abnormal type, while the other loop is designed to generate
a difference therebetween. It is possible to constitute a
characteristic analytic system in which the presence of a gene
CA 02390309 2002-05-07
is confirmed by the probe for the common portion while the presence
of an abnormality is confirmed in the other region. Because
the reaction of synthesizing nucleic acid according to the
present invention can also proceed isothermally, it is a very
important advantage that real-time analysis can be effected by
a general fluorescent photometer. Heretofore, the structure
of nucleic acid to be annealed in the same chain is known. However,
the nucleic acid having complementary nucleotide sequences
linked alternately in a single-stranded chain obtained by the
present invention is novel in that it contains a large number
of loops capable of base pairing with other oligonucleotides.
On the other hand, a large number of loops themselves given
by the reaction product according to the present invention can
be used as probes. .For example, in a DNA chip, probes should
be accumulated at high density in a limited area. In present
technology, however, the number of oligonucleotides which can
be immobilized in a certain area is limited. Hence, by use of
the reaction product of the present invention, a large number
of probes capable of annealing can be immobilized at high density.
That is, the reaction product according to the present invention
maybe immobilized as probes on a DNA chip. After amplification,
the reaction product may be immobilized by any techniques known
in the art, or the immobilized oligonucleotide is utilized as
the oligonucleotide in the amplification reaction of the present
invention, resulting in generating the immobilized reaction
61
CA 02390309 2002-05-07
product. By use of the probe thus immobilized, a large number
of sample DNAs can be hybridized in a limited area, and as a
result, high signals can be expected.
Brief Description of the Drawings
Fig. 1 is an illustration of a part (1) to (4) of the reaction
principle in a preferable mode of the present invention.
Fig. 2 is an illustration of a part (5) to (7) of the reaction
principle in a preferable mode of the present invention.
Fig. 3 is an illustration of a part (8) to (10) of the
reaction principle in a preferable mode of the present invention.
Fig. 4 is an illustration of the structure of a loop formed
by the single-stranded nucleic acid according to the present
invention.
Fig. 5 is an illustration of a part (A) to (B) in a basic
mode of the present invention.
Fig. 6 is an illustration of a part (C) to (D) in a basic
mode of the present invention.
Fig. 7 is a drawing showing the positional relationship
of each nucleotide sequence constituting an oligonucleotide in
the target nucleotide sequence of M13mp18.
Fig. 8 is a photograph showing the result of agarose
electrophoresis of a product obtained by the method of
synthesizing single-stranded nucleic acid with M13mp18 as a
template according to the present invention.
62
CA 02390309 2002-05-07
Lane 1: XIV size marker
Lane 2: 1 fmol M13mp18 dsDNA
Lane 3: No target
Fig. 9 is a photograph showing the result of agarose gel
electrophoresis of a restriction enzyme-digested product
obtained in Example 1 by the nucleic acid synthetic reaction
according to the present invention.
Lane 1: XIV size marker
Lane 2: BamHI digest of the purified product
Lane 3: PvuII digest of the purified product
Lane 4: HindlIl digest of the purified product
Fig. 10 is a photograph showing the result of agarose gel
electrophoresis of a product obtained by the method of
synthesizing single-stranded nucleic acid according to the
present invention using M13mp18 as a template in the presence
of betaine. 0, 0.5, 1 and 2 indicate the concentration (M) of
betaine added to the reaction solution. N indicates the negative
control, and -21 indicates the concentration 10-21 mol of template
DNA.
Fig. 11 is a drawing showing the positional relationship
of each nucleotide sequence constituting an oligonucleotide in
a target nucleotide sequence derived from HVB.
Fig. 12 is a photograph showing the result of agarose gel
electrophoresis of a product obtained by the method of
synthesizing single-stranded nucleic acid according to the
63
CA 02390309 2002-05-07
present invention wherein HBV-M13mp18 integrated in M13mp18 was
used as a template.
Lane 1: XIV size marker
Lane 2: 1 fmol HBV-M13mp18 dsDNA
Lane 3: No target
Fig. 13 is a photograph showing the result of gel
electrophoresis of an alkali-denatured product obtained by the
method of synthesizing single-stranded nucleic acid according
to the present invention.
Lane 1: HindlIl-digested fragment from lambda-phage
Lane 2: The reaction product in Example 1.
Lane 3: The reaction product in Example 3.
Fig. 14 is a photograph showing the result of agarose gel
electrophoresis of a product obtained by the method of
synthesizing single-stranded nucleic acid according to the
present invention wherein the concentration of Ml3mpl8 as a
target was varied. The upper and lower photographs show the
result of the reaction for 1 and 3 hours respectively.
Lane 1: M13mp18 dsDNA 1x10-15 mol/tube
Lane 2: Ml3mpl8 dsDNA 1x10-16 mol/tube
Lane 3: Ml3mpl8 dsDNA 1x10-17 mol/tube
Lane 4: M13mpl8 dsDNA 1x10-18 mol/tube
Lane 5: M13mpl8 dsDNA 1x10-19 mol/tube
Lane 6: M13mpl8 dsDNA 1x10-20 mol/tube
Lane 7: M13mpl8 dsDNA 1x10-21 mol/tube
64
CA 02390309 2002-05-07
Lane 8: Ml3mp18 dsDNA 1x10-22 mol/tube
Lane 9: No target
Lane 10: XIV size marker
Fig. 15 is a drawing showing the position of a mutation
and the positional relationship of each region toward a target
nucleotide sequence (target) . Underlined guanine is replaced
by adenine in the mutant.
Fig. 16 is a photograph showing the result of agarose gel
electrophoresis of a product according to the amplification
reaction of the present invention.
M: 100 bp ladder (New England Biolabs)
N: No template (purified water)
WT: 1 fmol wild-type template M13mp18
MT: 1 fmol mutant template Ml3mp18FM
Fig. 17 is a drawing showing the positional relationship
of each nucleotide sequence constituting an oligonucleotide in
a nucleotide sequence coding for target mRNA.
Fig. 18 is a photograph showing the result of agarose
electrophoresis of a product obtained by the method of
synthesizing single-stranded nucleic acid according to the
present invention using mRNA as a target.
Best Mode for Carrying Out the Invention
Example 1. Amplification of a region in M13mp18
The method of synthesizing the nucleic acid having
CA 02390309 2002-05-07
complementary chains alternately linked in a single-stranded
chain according to the present invention was attempted using
M13mp18 as a template. Four kinds of primers, that is, M13FA,
M13RA, M13F3, and M13R3, were used in the experiment. M13F3
and M13R3 were outer primers for displacing the first nucleic
acid obtained respectively with M13FA and M13RA as the origin
of synthesis. Because the outer primers are primers serving
as the origin of synthesis of complementary chain after synthesis
with M13FA (or M13RA) , these were designed to anneal to a region
contiguous to M13FA (or M13RA) by use of the phenomenon of
contiguous stacking. Further, the concentrations of these
primers were set high such that annealing of M13FA (or M13RA)
occurred preferentially.
The nucleotide sequence constituting each primer is as
shown in the Sequence Listing. The structural characteristics
of the primers are summarized below. Further, the positional
relationship of each region toward the target nucleotide sequence
(target) is shown in Fig. 7.
Primer Region at the 5'-side/region at the 3'-side
M13FA The same as region Flc in complementary chain
synthesized by M13FA/complementary to region F2c in M13mp18
M13RA The same as region Ric in complementary chain
synthesized by M13RA/complementary to region R2c in
complementary chain synthesized by M13FA
M13F3 Complementary to F3c contiguous to the 3' -side of
66
CA 02390309 2002-05-07
region F2c in M13mp18
M13R3 Complementary to R3c contiguous to the 3'-side of
region F2c in complementary chain synthesized by M13FA
By such primers, nucleic acid wherein a region extending
from Flc to Ric in M13mp18, and its complementary nucleotide
sequence, are alternately linked via a loop-forming sequence
containing F2c in a single-stranded chain, is synthesized. The
composition of a reaction solution for the method of synthesizing
nucleic acid by these primers according to the present invention
is shown below.
Composition of the reaction solution (in 25 L)
20 mM Tris-HC1 pH8.8
mM KC1
10 mm (NH4) 2SO4
6 mM MgSO4
0.1 % Triton X-100
5% dimethyl sulfoxide (DMSO)
0.4 mM dNTP
Primers:
800 nM M13FA/SEQ ID NO:1
800 nM M13RA/SEQ ID NO:2
200 nM M13F3/SEQ ID NO:3
200 nM M13R3/SEQ ID NO:4
Target: M13mp18 dsDNA/SEQ ID NO:5
Reaction: The above reaction solution was heated at 95
67
^
CA 02390309 2002-05-07
C for 5 minutes, and the target was denatured into a
single-stranded chain. The reaction solution was transferred
to ice-cold water, and 4 U of Bst DNA polymerase (NEW ENGLAND
Biolabs) was added thereto and the mixture was reacted at 65
C for 1 hour. After reaction, the reaction was terminated at
80 C for 10 minutes and transferred again to ice-cold water.
Confirmation of the reaction: 1 l loading buffer was
added to 5 l of the above reaction solution, and the sample
was electrophoresed for 1 hour at 80 mV on 2 % agarose gel (0.5 %
TBE). As a molecular-weight marker, XIV (100 bp ladder,
Boehringer Mannheim) was used. The gel after electrophoresis
was stained with SYBR Green I (Molecular Probes, Inc.) to confirm
the nucleic acid. The results are shown in Fig. 8. The
respective lanes correspond to the following samples.
1. XIV size marker.
2. 1 fmol M13mpl8 dsDNA.
3. No target.
In lane 3, no band was confirmed except that the unreacted
primers were stained. In lane 2 in the presence of the target,
the products were confirmed as a low size band ladder, as smeared
staining at high size and as a band hardly electrophoresed in
the gel. Among the low-size bands, bands in the vicinity of
2 90 by and 4 50 by agree with the products estimated in the synthetic
reaction of this invention, that is, double-stranded chains of
SEQ ID NOS:11 and 12 (corresponding to double-stranded chains
68
CA 02390309 2002-05-07
formed as shown in Figs. 2- (7) and 2- (10) ) and a single-stranded
chain of SEQ ID NO: 13 (corresponding to the long single-stranded
chain in Fig. 3- (9) ) , and it was thus confirmed that the reaction
proceeds as expected. It was estimated that the electrophoresis
results of the smeared pattern at high size and the band not
electrophoresed were brought about because this reaction was
basically a continuous reaction to permit varying molecular
weights of the reaction product and further because the product
has a complicated structure having a partially single-stranded
chain and a double-stranded complex.
Example 2. Confirmation of the reaction products by digestion
with restriction enzymes
For the purpose of clarifying the structure of the nucleic
acid having complementary nucleotide sequences linked
alternately in a single-stranded chain obtained in Example 1
according to the present invention, the digestion of the products
with restriction enzymes was conducted. If fragments are
theoretically generated by digestion thereof with restriction
enzymes and simultaneously the smear pattern at high size and
the band not electrophoresed as observed in Example 1 disappear,
then it can be estimated that any of these products are the nucleic
acid having complementary sequences linked alternately in a
single-stranded chain synthesized according to the present
invention.
The reaction solution (200 l) from 8 tubes in Example
69
CA 02390309 2002-05-07
1 was pooled and purified by treatment with phenol and
precipitation with ethanol. The resulting precipitates were
recovered and dissolved again in 200 l TE buffer, and 10 l
aliquot was digested at 37 C for 2 hours with restriction enzymes
BamHI, PvuII, and Hindlil respectively. The digest was
electrophoresed for 1 hour at 80 mV on 2 % agarose gel (0.5 %TBE) .
As a molecular marker, Super Ladder-Low (100 bp ladder) (Gensura
Laboratories, Inc.) was used. The gel after electrophoresis
was stained with SYBR Green I (Molecular Probes, Inc.) to confirm
the nucleic acid. The results are shown in Fig. 9. The
respective lanes correspond to the following samples.
1. XIV size marker
2. BamHI digest of the purified product
3. PvuII digest of the purified product
4. HindlIl digest of the purified product
It is estimated that nucleotide sequences constituting
relatively short amplification products are those of SEQ ID
NOS:13, 14, 15 and 16. From these nucleotide sequences, the
estimated size of each fragment digested with the restriction
enzymes is as shown in Table 1. "L" in the table indicates that
its position in electrophoresis is not established because L
is a fragment containing a loop (single-stranded chain).
Table 1. Restriction enzyme-digested fragments of the
amplification products according to the present invention
CA 02390309 2002-05-07
SEQID NO BamHI PvuII Hindlll
13 177 +L 56 +L 147 +L
14 15+101 +L - 142 +L
15 171+101 +L 56 +L 147+161 +L
16 11+101+230+L 237 +L 142+170 +L
Summary 101,177,230 56,237 142,147,161,170
(11,15; not confirmed)
Because almost all bands before digestion were changed
into estimated bands, it was confirmed that the object reaction
products were amplified. Further, it was also shown that there
were no or less unspecific products.
Example 3. Promotion of amplification reaction by addition of
betaine
An experiment for examining the effect of betaine
(N,N,N-trimethylglycine, Sigma) added to the amplification
reaction solution on the amplification reaction of nucleic acid
was conducted. Synthesis of the nucleic acid having
complementary chains alternately linked in a single-stranded
chain according to the present invention was conducted using
M13mp18 as a template similarly in Example 1 in the presence
of betaine at various concentrations. The primers used in the
experiment were identical to those used in Example 1. The amount
of the template DNA was 10-21 mol (Ml3mp18) and water was used
as the negative control. Betaine was added at concentrations
of 0, 0.5, 1 and 2 M to the reaction solution. The composition
of the reaction solution is shown below.
Composition of the reaction solution (in 25 L)
71
CA 02390309 2008-12-17
20 mM Tris-HC1 pH8.8
4 mM MgSO4
0.4 mM dNTPs
mM KC1
10 MM (NH4) 2SO4
0.1% Triton` X-100
Primers:
800 nM M13FA/SEQ ID NO:1
800 nM M13RA/SEQ ID NO:2
200 nM M13F3/SEQ ID NO:3
200 nM M13R3/SE.Q ID NO:4
Target: M13mp18 dsDNA/SEQ ID NO:5
The polymerise, reaction conditions, and conditions for
electrophoresis after the reaction were identical to those
described in Example 1.
The results are shown in Fig.. 10. In the reaction in the
presence of betaine at a concentration of 0.5 or 1.0 M, the amount
of the amplification product was increased. Further, if its
concentration was increased to 2.0 M, no amplification was
observed. It was thus shown that the amplification reaction
was promoted in the -presence of betaine at a suitable
concentration. The estimated reason that the amount of the
amplification product was decreased when the concentration of
betaine was 2 M was that Tm was lowered too much.
Example 4. Amplification of HBV gene sequence
72
CA 02390309 2002-05-07
The method of synthesizing nucleic acid according to the
present invention was attempted wherein M13mp18 dsDNA having
a partial sequence of HBV gene integrated therein was used as
a template. Four kinds of primers, HB65FA (SEQ ID NO: 6) , HB65RA
(SEQ ID NO: 7) , HBF3 (SEQ ID NO: 8) and HBR3 (SEQ ID NO: 9) , were
used in the experiment. HBF3 and HBR3 were outer primers for
displacement of the first nucleic acid obtained respectively
with HB65FA and HB65RA as the origin of synthesis. Because the
outer primers are primers serving as the origin of synthesis
of complementary chain after synthesis with HB65FA (or HB65RA),
these were designed to anneal to a region contiguous to HB65FA
(or HB65RA) by use of the phenomenon of contiguous stacking.
Further, the concentrations of these primers were set high such
that annealing of HB65FA (or HB65RA) occurred preferentially.
The target sequence (430 bp) in this example, derived from HBV
integrated in M13mp18, is shown in SEQ ID NO:10.
The nucleotide sequence constituting each primer is shown
in the Sequence Listing. The structural feature of each primer
is summarized below. Further, the positional relationship of
each region toward the target nucleotide sequence (target) is
shown in Fig. 11.
Primer Region at the 5'-side/region at the 3'-side
HB65FA The same as region Flc in complementary chain
synthesized by HB65FA/complementary to region F2c in HBV-M13mpl8
HB65RA The same as region Rlc in complementary chain
73
CA 02390309 2002-05-07
synthesized by HB65RA/complementary to region R2c in
complementary chain synthesized by HB65FA
HBF3 Complementary to F3c contiguous to the 3'-side of
region F2c in HBV-M13mp18
HBR3 Complementary to R3c contiguous to the 3'-side of
region F2c in complementary chain synthesized by HB65FA
By such primers, nucleic acid wherein a region extending
from Flc to Rlc in M13mp18 (HBV-M13mp18) having a partial sequence
of HBV gene integrated therein, and its complementary nucleotide
sequence, are alternately linked via a loop-forming sequence
containing F2c in a single-stranded chain, is synthesized. The
reaction was conducted under the same conditions as in Example
1 except that the primers described above were used, and the
reaction solution was analyzed by agarose electrophoresis. The
results are shown in Fig. 12. The respective lanes correspond
to the following samples.
1. XIV size marker
2. 1 fmol HBV-M13mp18 dsDNA.
3. No target
Similar to Example 1, the products were confirmed only
in the presence of the target as a low size band ladder, as smeared
staining at high size and as a band hardly electrophoresed in
the gel (lane 2) . Among the low-size bands, bands in the vicinity
of 310 bp and 480 bp agree with the products estimated in the
synthetic reaction of this invention, that is, double-stranded
74
CA 02390309 2002-05-07
chains of SEQ ID NOS:17 and 18, and it was thus confirmed that
the reaction proceeds as expected. As described in the results
in Example 1, it was estimated that the smeared pattern at high
size and the band not electrophoresed were caused by the structure
of the synthetic product characteristic of the present invention.
From this experiment, it was confirmed that the present invention
can be practiced even if a different sequence (target) is used
for amplification.
Example 5. Confirmation of the sizes of the synthetic reaction
products
To confirm the structure of the nucleic acid synthesized
according to the present invention, its length was analyzed by
electrophoresis under alkali-denaturing conditions. 1 1
alkaline loading buffer was added to 5 l of each reaction solution
in the presence of the target in Example 1 or 4 and electrophoresed
at 50 mA in 0.7 % agarose gel (50 mM NaOH, 1 mM EDTA) for 14
hours. As the molecular-weight size marker, Hindlll-digested
lambda-phage fragments were used. The gel after
electrophoresis was neutralized with 1 M Tris, pH 8 and stained
with SYBR Green I (Molecular Probes, Inc.) to confirm the nucleic
acid. The results are shown in Fig. 13. The respective lanes
correspond to the following samples.
1. HindlIl-digested fragments from lambda-phage.
2. The reaction product in Example 1.
3. The reaction product in Example 4.
CA 02390309 2002-05-07
When the reaction product was electrophoresed under
alkali-denaturing conditions, its size in a single-stranded
state could be confirmed. It was confirmed that the sizes of
the major products in both Example 1 (lane 2) and Example 4 (lane
3) were within 2 kbase. Further, it was revealed that the product
according to the present invention had been extended to have
a size of at least 6 kbase or more within the range capable of
confirmation by this analysis. In addition, it was confirmed
again that bands not electrophoresed under non-denaturing
conditions in Examples 1 and 4 were separated in a denatured
state into individual single-stranded chains of smaller size.
Example 6. Confirmation of amplification depending on the
concentration of a target in the amplification of a region in
M-13mp13
The influence of a varying concentration of a target on
the method of synthesizing nucleic acid according to the present
invention was observed. The method of synthesizing nucleic acid
according to the present invention was carried out under the
same conditions as in Example 1 except that the amount of M13mp18
dsDNA as the target was 0 to 1. fmol and the reaction time was
1 hour or 3 hours. Similar to Example 1, the sample was
electrophoresed in 2 % agarose gel (0.5 % TBE) and stained with
SYBR Green I (Molecular Probes, Inc.) to confirm the nucleic
acid. As a molecular size marker, XIV (100 by ladder, Boehringer
Mannheim) was used. The results are shown in Fig. 14 (upper:
76
CA 02390309 2002-05-07
1-hour reaction, below: 3-hour reaction) The respective lanes
correspond to the following samples:
1.. M13mp18 dsDNA 1x10-15 mol/tube.
2. M13mp18 dsDNA 1x10-16 mol/tube.
3. M13mp18 dsDNA 1x10-17 mol/tube.
4. M13mp18 dsDNA 1x10-18 mol/tube.
5. M13mp18 dsDNA 1x10-19 mol/tube.
6. M13mp18 dsDNA 1x10-2 mol/tube.
7. M13mp18 dsDNA 1x10-21 mol/tube.
8. M13mp18 dsDNA 1x10-22 mol/tube.
9. No target.
10. XIV size marker.
A common band among the respective lanes appears in a lower
part in the electrophoretic profile and shows the unreacted
stained primers. Regardless of the reaction time, no
amplification product is observed in the absence of the target.
A staining pattern, depending on the concentration of the target,
of the amplification product was obtained only in the presence
of the target. Further, the amplification product could be
confirmed at lower concentration as the reaction time was
increased.
Example 7. Detection of a point mutation
(1) Preparation of M13mpl8FM (mutant)
The target DNA used was M13mp18 (wild-type) and M13mpl8FM
(mutant) . For the construction of the mutant M13mp18FM, LA PCRTM
77
CA 02390309 2002-05-07
in vitro Mutagenesis Kit (Takara Shuzo Co., Ltd.) was used to
replace one nucleotide for mutation. Thereafter, the sequence
was confirmed by sequencing. The sequence of the F1 region is
shown below:
Wild-type: CCGGGGATCCTCTAGAGTCG (SEQ ID NO:19)
Mutant: CCGGGGATCCTCTAGAGTCA..(SEQ ID NO:20)
(2) Design of primers
The FA primers used for the wild-type and the mutant were
provided at the 5'-terminal of the Flc region thereof with
different nucleotide sequences, respectively. The location of
the mutation and the positional relationship of each region
toward the target nucleotide sequence (target) are shown in Fig.
15.
(3) Amplification reaction
An experiment was conducted to examine whether
template-specific amplification reaction occurs using a
combination of specific primers shown below by use of M13mp18
(wild-type) and M13mp18FM (mutant) as primers.
Primer set for wild-type amplification: FAd4, RAd4, F3, R3
Primer set for mutant amplification: FAMd4, RAd4, F3, R3
The nucleotide sequence of each primer is as follows:
FAd4 : CGACTCTAGAGGATCCCCGGTTTTTGTTGTGTGGAATTGTGAGCGGAT (SEQ
ID NO:21)
FAMd4: TGACTCTAGAGGATCCCCGGTTTTTGTTGTGTGGAATTGTGAGCGGAT (SEQ
ID NO:22)
78
CA 02390309 2002-05-07
RAd4 : CGTCGTGACTGGGAAAACCCTTTTTGTGCGGGCCTCTTCGCTATTAC (SEQ
ID NO:23)
F3: ACTTTATGCTTCCGGCTCGTA (SEQ ID NO:24)
R3: GTTGGGAAGGGCGATCG (SEQ ID NO:25)
(4) Detection of the point mutation in M13mp18
The composition of the reaction solution is as follows:
Final concentration
D2W 3.75 L
lOX Thermo pol buffer(NEB) 2.5 L 20 mM Tris-HC1 pH B.8
mM KC1
10 mM (NH4) 2SO4
6 mM MgSO4
0.1 % TritonX-100
2.5 mM dNTP 4 L 400 }.LM
100 MM MgSO4 0.5 L
4 M Betaine 6.25 L 1 M
M13FAd4 primer (10 pmol/ L) or
M13FAMd4 primer (10 pmol/ L) 2 L 800 nM
M13RAd4 primer (10 pmol/ L) 2 .LL 800 nM
M13F3 primer (10 pmol/ L) 0.5 L 200 nM
M13R3 primer (10 pmol/ L) 0.5 L 200 nM
Total amount 22 L
1 fmol (2 l) of the target M13mp18 or M13mp18 FM was added
to the reaction solution and heated at 95 C for 5 minutes whereby
the target was denatured into a single-stranded chain. The
79
CA 02390309 2002-05-07
reaction solution was transferred to ice-cold water, and 1 l
(8 U) of Bst DNApolymerase (NEW ENGLAND Biolab) was added thereto
and reacted for 1 hour at 68 C or 68.5 C. After reaction, the
reaction was terminated at 80 C for 10 minutes, and the reaction
solution was transferred again to ice-cold water.
As shown in Fig. 16, when FAd4 for wild type was used as
the FA primer, effective amplification was observed only in the
presence of the wild-type template. On the other hand, when
FAMd4 for mutant was used as the FA primer, effective
amplification was observed only in the presence of the wild-type
[sic.] template.
From the results described above, it was shown that the
point mutation could be detected efficiently by use of the
amplification reaction of the present invention.
Example 8. Amplification reaction of mRNA as a target
The method of synthesizing nucleic acid according to the
present invention was attempted using mRNA as the target nucleic
acid. To prepare the target mRNA, prostate cancer cell line
LNCaP cells (ATCC No. CRL-1740) expressing prostate specific
antigen (PSA) were mixed with chronic myeloid leukemia cell line
K562 cells (ATCC No. CCL-243) as non-expressing cells at 1 :
106 to 100 : 106, followed by extraction of the total RNA by
use of an RNeasy Mini kit from Qiagen (Germany). Four kinds
of primers, that is, PSAFA, PSARA, PSAF3 and PSAR3, were used
in the experiment. PSAF3 and PSAR3 are outer primers for
CA 02390309 2002-05-07
displacing the first nucleic acid obtained respectively with
PSAFA and PSARA as the origin of synthesis. Further, the
concentrations of these primers were set high such that annealing
of PSAFA (or PSARA) occurred preferentially. The nucleotide
sequences constituting the respective primers are as follows.
Primer:
PSAFA: TGTTCCTGATGCAGTGGGCAGCTTTAGTCTGCGGCGGTGTTCTG (SEQ ID
NO: 26)
PSARA: TGCTGGGTCGGCACAGCCTGAAGCTGACCTGAAATACCTGGCCTG (SEQ ID
NO: 27)
PSAF3: TGCTTGTGGCCTCTCGTG (SEQ ID NO: 28)
PSAR3: GGGTGTGGGAAGCTGTG (SEQ ID NO: 29)
The structural features of the primers are summarized below.
Further, the positional relationship of each primer toward the
DNA nucleotide sequence coding for the target mRNA and
recognition sites of restriction enzyme Sau3AI are shown in Fig.
17.
Primer Region at the 5'-side/region at the 3'-side
PSAFA The same as region Flc in complementary chain
synthesized by PSAFA/complementary to region F2c in the target
nucleotide sequence
PSARA The same as region Rlc in complementary chain
synthesized by PSARA/complementary to region R2c in
complementary chain synthesized by PSAFA
PSAF3 Complementary to F3c contiguous to the 3'-side of
81
fi
CA 02390309 2002-05-07
region F2c in the target nucleotide sequence
PSAR3 Complementary to Ric contiguous to the 3'-side of
region R2c in complementary chain synthesized by PSAFA
The composition of a reaction solution for the method of
synthesizing nucleic acid according to the present invention
is as follows:
Composition of the reaction solution (in 25 L)
20 mM Tris-HC1 pH 8.8
4 MM MgSO4
0.4 mM dNTPs
mM KC1
10 mm (NH4) 2SO4
0.1 % Triton X-100
0.8 M betaine
5 mM DTT
1600 nM PSAFA & PSARA primer
200 nM PSAF3 & PSAR3 primer
8 U Bst DNA polymerase
100 U Rever Tra Ace (Toyobo Co., Ltd., Japan)
5 g total RNA
All ingredients were mixed on ice. In this experiment,
mRNA (single-stranded chain) is used as a target, and thus the
step of making single-stranded chain by heat denaturation is
not necessary. The reaction was conducted at 65 C for 45 minutes,
and the reaction was terminated at 85 C for 5 minutes. After
82
CA 02390309 2002-05-07
reaction, 5 l of the reaction solution was electrophoresed in
2 % agarose and detected by SYBR Green I.
The results are shown in Table 18. The respective lanes
correspond to the following samples.
Lane Bst RT Number of LNCaP cells/106 K562 cells
1 - + 0
2 - + 10
3 + - 0
4 + - 10
+ + 0
6 + + 1
7 + + 10
8 Sau3AI digest of 1 L aliquot of the reaction solution in
lane 6
9 Sau3AI digest of 1 .tL aliquot of the reaction solution in
lane 7
Size maker, 100 bp ladder (New England Biolabs)
In the absence of either Bst DNA polymerase or Rever Tra
Ace, no amplification product could be obtained (lanes 1 to 4) .
In the presence of both the enzymes, an amplification product
was detected (lanes 5 to 7) if RNA derived from LNCaP was present.
RNA extracted from one LNCaP cell/one million K562 cells could
be detected (lane 6) . When the amplification product was
digested at the restriction enzyme site Sau3AI located in the
inside of the target, the product was digested into a fragment
83
CA 02390309 2002-05-07
of estimated size (lanes 8 and 9).
From the results described above, it was confirmed that
the desired reaction product can be obtained in the method of
synthesizing nucleic acid according to the present invention
even if RNA is used as a target.
Industrial Applicability
According to the novel oligonucleotide according to the
present invention and the method of synthesizing nucleic acid
by using said oligonucleotide, there is provided a method of
synthesizing nucleic acid having complementary nucleotide
sequences linked alternately in a s ingle-stranded chain, without
requiring any complicated control of temperature. A
complementary chain synthesized with the oligonucleotide as a
primer based on the present invention serves as the origin of
synthesizing a new complementary chain with the 3' -terminal of
said synthesized chain as a template. This is accompanied by
formation of a loop causing annealing of a new primer, and a
product of the reaction of synthesizing complementary chain with
the previously synthesized chain as a template is displaced again
by synthesis of complementary chain from the loop and made ready
for base pairing. The thus obtained nucleic acid synthesized
with itself as a template is combined with e. g. a known nucleic
acid synthesizing method such as SDA, to contribute the
improvement of efficiency of nucleic acid synthesis.
84
CA 02390309 2002-05-07
According to an additional preferred mode of the present
invention, there is provided a novel method of synthesizing
nucleic acid, which achieves the improvement of efficiency of
the known method of synthesizing nucleic acid, does not require
complicated control of temperature, can be expected to attain
high efficiency of amplification and can achieve high specificity.
That is, the oligonucleotide based on the present invention is
applied to a template chain and its complementary chain whereby
nucleic acid having complementary sequences linked alternately
in a single-stranded chain can be successively synthesized.
This reaction continues theoretically until the starting
materials necessary for synthesis are exhausted, during which
new nucleic acid initiated to be synthesized from the loop
continues tobe formed. The elongation from the oligonucleotide
having annealed to the loop performs strand displacement for
supplying 3' -OH for elongation of long single-stranded nucleic
acid (that is, nucleic acid having plural pairs of complementary
chains linked therein). On the other hand, the 3'-OH of the
long single-stranded chain performs the reaction of synthesizing
complementary chain with itself as a template whereby its
elongation is achieved, during which a new complementary chain
whose synthesis is initiated from the loop is displaced. Such
an amplification reaction step proceeds under isothermal
conditions while maintaining high specificity.
The oligonucleotides in the present invention can, when
CA 02390309 2002-05-07
two contiguous regions are arranged as designed, function as
primers for the reaction of synthesizing nucleic acid according
to the present invention. This contributes significantly to
the preservation of specificity. By comparison with e.g. PCR
where unspecific amplification reaction is initiated by
unspecific missannealing regardless of the intended positional
relationship of 2 primers, it can be easily explained that high
specificity can be expected in the present invention. This
feature can be utilized to detect SNPs highly sensitively and
accurately.
The characterizing feature of the present invention lies
in that such reaction can be easily achieved by a very simple
constitution of reagents. For example, the oligonucleotide
according to the present invention has a special structure, but
this is a matter of selection of nucleotide sequence, and it
is a simple oligonucleotide as substance. Further, in a
preferred mode, the reaction can proceed by only a DNA polymerase
catalyzing the strand displacement-type reaction of
synthesizing complementary chain. Further, if the present
invention is carried out with RNA as a template, a DNA polymerase
such as Bca DNA polymerase having both reverse transcriptase
activity and strand displacement-type DNA polymerase activity
is used so that all enzyme reactions can be conducted by the
single enzyme. The reaction principle of realizing a high degree
of nucleic acid amplification reaction by such simple enzyme
86
CA 02390309 2002-05-07
reaction is not known. Even for the application of the present
invention to a known nucleic acid synthesizing reaction such
as SDA, no additional enzyme is necessary for their combination,
and such a simple combination with the oligonucleotide based
on the present invention can be applied to various reaction
systems. Accordingly, it can be said that the method of
synthesizing nucleic acid according to the present invention
is also advantageous in respect of cost.
As described above, the method of synthesizing nucleic
acid according to the present invention and the oligonucleotide
therefor provide a new principle of simultaneously solving a
plurality of difficult problems such as operativeness
(temperature control is not necessary), improvement of
efficiency of synthesis, economization, and high specificity.
87
CA 02390309 2002-10-28
SEQUENCE LISTING
<110> Eiken Kagaku Kabushiki. Kaisha
<120> Method for Synthesizing The Nucleic Acid.
<130> 12871-4
<140> CA 2,390,309
<141> 2000-03-28
<150> PCT/JP99/06213
<151> 1999-11-08
<160> 29
<170> Patentln Ver. 2.0
<210> 1
<211> 52
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequense
<400> 1
cgactctaga ggatccccgg gtactttttg ttgtgtggaa ttgtgagcgg at 52
<210> 2
<211> 51
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequense
<400> 2
acaacgtcgt gactgggaaa accctttttg tgcgggcctc ttcgctatta c 51
<210> 3
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificiall.y
synthesized primer sequense
<400> 3
actttatgct tccggctcgt a 21
<210> 4
<211> 17
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
88
CA 02390309 2002-10-28
synthesized primer sequense
<400> 4
gttgggaagg gcgatcg 17
<210> 5
<211> 600
<212> DNA
<213> Bacteriophage M13mpl8
<400> 5
gcgcccaata cgcaaaccgc ctctccccgc gcgttggccg attcattaat gcagctggca 60
cgacaggttt cccgactgga aagcgggcag tgagcgcaac gcaattaatg tgagttagct 120
cactcattag gcaccccagg ctttacactt tatgcttccg gctcgtatgt tgtgtggaat 180
tgtgagcgga taacaatttc acacaggaaa cagctatgac catgattacq aattcgagct 240
cggtacccgg ggatcctcta gagtcgacct gcaggcatgc aagcttggca ctggccgtcg 300
ttttacaacg tcgtgactgg gaaaaccctg gcgttaccca acttaatcgc cttgcagcac 360
atcccccttt cgccagctgg cgtaatagcg aagdj.ggcccg caccgat_cgc cctt:cccaac 420
agttgcgcag cctgaatggc gaatggcgct t:tgcctggtt tccggcacca gaagcggtgc 480
cggaaagctg gctggagtgc gatcttcctg aggc;cgatac ggtcgtcgtc ccctcaaact 540
ggcagatgca cggttacgat gcgcccatct acaccaacgt aacctatccc attacggtca 600
<210> 6
<211> 63
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 6
ctcttccaaa agtaaggcag gaaatgtgaa accagatcgt aatttggaag acccagcatc 60
cag 63
<210> 7
<211> 43
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 7
gtggattcgc actcctcccg ctgatcggga cctgcctcgt cgt 43
<210> 8
<211> 16
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequeence:Artificially
synthesized primer sequence
<400> 8
gccacctggg tgggaa 16
<210> 9
<211> 22
89
CA 02390309 2002-10-28
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artif.iciaily
synthesized primer sequence
<400> 9
ggcgagggag ttcttcttct ag 22
<210> 10
<211> 430
<212> DNA
<213> Hepatitis B virus
<400> 10
ctccttgaca ccgcctctgc tctgtatcgg gaggccttag agtctccgga acattgttca 60
cctcaccata cagcactcag gcaagctatt ctgtgttggg gtgagttaat_ gaatctggcc 120
acctgggtgg gaagtaattt ggaagaccca gcatccaggg aattagtagt cagctatgtc 180
aatgttaata tgggcctaaa aatcagacaa ctactgtggt ttcacatttc ctgccttact 240
tttggaagag aaactgtttt ggagtatttg gtatc:ttttg gagtgtggat tcgcactcct 300
cccgcttaca gaccaccaaa tgcccctatc ttatcaacac ttccggaaac tactgttgtt 360
agacgacgag gcaggtcccc tagaagaaga act-ccctcgc ctcgcagacg aaggtctcaa 420
tcgccgcgtc 430
<210> 11
<211> 293
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized sequence
<400> 11
acaacgtcgt gactgggaaa accctttttg tgcgggcctc t.tcgctatta cgccagctgg 60
cgaaaggggg atgtgctgca aggcgattaa gttgggtaac gccagggttt tcccagtcac 1.20
gacgttgtaa aacgacggcc agtgccaagc ttgcatgcct gcaggtcgac tctagaggat 180
ccccgggtac cgagctcgaa ttcgtaatca tggtcatagc tgtttcctgt gtgaaattgt 240
tatccgctca caattccaca caacaaaaag tacccgggga t.cctctagag tcg 293
<210> 12
<211> 293
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized sequence
<400> 12
cgactctaga ggatccccgg gtactttttg ttgtgtggaa ttgtgagcgg ataacaattt 60
cacacaggaa acagctatga ccatgattac gaattcgagc tcggtacccg gggatcctct 120
agagtcgacc tgcaggcatg caagctt.ggc actggccgtc gttttacaac gtcgtgactg 180
ggaaaaccct ggcgttaccc aacttaatcg ccttgcagca catccccctt tcgccagctg 240
gcgtaatagc gaagaggccc gcacaaaaag ggttttcc:ca. gtcacgacgt tgt 293
<210> 13
<211> 459
<212> DNA
<213> Artificial Sequence
CA 02390309 2002-10-28
<220>
<223> Description of Artificial Sequence:Artificially
synthesized sequence
<400> 13
acaacgtcgt gactgggaaa accctttttg tgcgggcctc ttcgctatta cgccagctgg 60
cgaaaggggg atgtgctgca aggcgattaa gttgggtaac gccagggttt: tcccagtcac 120
gacgttgtaa aacgacggcc agtgccaagc ttgcatgcct gcaggtcgac tctagaggat 180
ccccgggtac cgagctcgaa ttcataatca tggtaatagc tgtttcctgt gtgaaattgt 240
tatccgctca caattccaca caacaaaaag tacccgggga tcctctagag tcgacctgca 300
ggcatgcaag cttggcactg gccgtcgttt tacaacgtcg tgactgggaa aaccctggcg 360
ttacccaact taatcgcctt gcagcacatc cccctttcgc cagctggcgt aatagcgaag 420
aggcccgcac aaaaagggtt ttcccagtca cgacgttg't 459
<210> 14
<211> 458
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized sequence
<400> 14
cgactctaga ggatccccgg gtactttttg ttgtgtggaa ttgtgagcgg ataacaattt 60
cacacaggaa acagctatga ccat:gattac gaattcgagc tcggtacccg gggatcctct 120
agagtcgacc tgcaggcatg caagcttggc actggccgtc gttttacaac gtcgtgactg 180
ggaaaaccct ggcgttaccc aact:taatcg ccttgcagca catccccctt tcgccagctg 240
gcgtaatagc gaagaggccc gcacaaaaag ggttttccca gtcacgacgt tgtaaaacga 300
cggccagtgc caagcttgca tgcctgcagg tcgactctag aggatccccg ggtaccgagc 360
tcgaattcgt aatcatggtc atagctgttt cctgtgtgaa attgttatcc gctcacaatt 420
ccacacaaca aaaagtaccc ggggatcctc tagagtcg 458
<210> 15
<211> 790
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized sequence
<400> 15
acaacgtcgt gactgggaaa accctttttg tgcgggcctc ttcgctatta cgccagctgg 60
cgaaaggggg atgtgctgca aggcgattaa gttgggtaac gccagggttt tcccagtcac 120
gacgttgtaa aacgacggcc agtgccaagc ttgcatgcct gcaggtcgac tctagaggat 180
ccccgggtac cgagctcgaa ttcgtaatca tggtcatagc tgtttcctgt: gtgaaattgt 240
tatccgctca caattccaca caacaaaaag tacccgggga tcctctagag tcgacctgca 300
ggcatgcaag cttggcactg gccgtcgttt tacaacgtcg tgactgggaa aaccctggcg 360
ttacccaact taatcgcctt gcagcacatc cccct.ttcgc: cagctggcgt aatagcgaag 420
aggcccgcac aaaaagggtt ttcc:cagt::ca cgacgttgta aaacgacggc cagtgccaag 480
cttgcatgcc tgcaggtcga ctctagagga tccccgggta ctttttgttg tgtggaattg 540
tgagcggata acaatttcac acaggaaaca gctctgacca tgattacgaa ttcgagctcg 600
gtacccgggg atcctctaga gtcgacctgc aggcatgcaa gcttggcact ggccgtcgtt 660
ttacaacgtc gtgactggga aaaccctggc gttacccaac ttaatcgcct tgcagcacat 720
ccccctttcg ccagctggcg taatagcgaa gaggcccgca caaaaagggt: tttcccagtc 780
acgacgttgt 790
<210> 16
<211> 789
91
CA 02390309 2002-10-28
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized sequence
<400> 16
cgactctaga ggatccccgg gtactttttg ttgtgtggaa ttgtgagcgg ataacaattt 60
cacacaggaa acagctatga ccatgattac gaat.tcgagc tcggtacccg gggatcctct 120
agagtcgacc tgcaggcatg caagcttggc actcggccgtc gttttacaac gtcgtgactg 180
ggaaaaccct ggcgttaccc aact:taat.cg ccttgcagca catccccctt. tcgccagctg 240
gcgtaatagc gaagaggccc gcacaaaaag ggttttccca gtcacgacgt tgtaaaacga 300
cggccagtgc caagcttgca tgcctgcagg tcgactctag aggatccccg ggtaccgagc 360
tcgaattcgt aatcatggtc atagctgttt cctgtgtgaa attgttatcc gctcacaatt 420
ccacacaaca aaaagtaccc ggggatcctc tagagtcgac ctgcaggcat gcaagcttgg 480
cactggccgt cgttttacaa cgtcgtgact gggaaaaccc tttttgtgcq ggcctcttcg 540
ctattacgcc agctggcgaa agggggatgt gctgcaaggc gattaagttg ggtaacgcca 600
gggttttccc agtcacgacg ttgt:aaaacg acggccagtg ccaagcttgc atgcctgcag 660
gtcgactcta gaggatcccc gggtaccgag ctcgaattcg taatcatggt: catagctgtt 720
tcctgtgtga aattgttatc cgct:cacaat t.ccacacaac aaaaagtacc cggggatcct 780
ctagagtcg 789
<210> 17
<211> 310
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized sequence
<400> 17
gtggattcgc actcctcccg ctgatcggga cctgcctcgt cgtctaacaa cagtagtttc 60
cggaagtgtt gataagatag gggcatttgg tggtctgtaa gcgggaggag tgcgaatcca 120
cactccaaaa gataccaaat actccaaaac agtrt:ctctt ccaaaagtaa ggcaggaaat 180
gtgaaaccac aatagttgtc tgatttttag gcccatatta acattgacat agctgactac 240
taattccctg gatgctgggt cttccaaatt acgat:ctggt ttcacatttc ctgccttact 300
tttggaagag 310
<210> 18
<211> 465
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Ar.tif.icially
synthesized sequence
<400> 18
gtggattcgc actcctcccg ctgatcggga cctgcctcgt cgtctaacaa cagtagtttc 60
cggaagtgtt gataagatag gggcatttgg tggtctgtaa gcgggaggag tgcgaatcca 120
cactccaaaa gataccaaat actccaaaac agttttctctt ccaaaagtaa ggcaggaaat 180
gtgaaaccac aatagttgtc tgatttttag gcccatatta acattgacat agctgactac 240
taattccctg gatgctgggt cttccaaatt acgatctggt t.tcacatttc ctgccttact 300
tttggaagag aaactgtttt ggagtatttg gtatcttttg gagtgtggat tcgcactcct 360
cccgcttaca gaccaccaaa tgcccctatc ttatt.caacac ttccggaaac tactgttgtt 420
agacgacgag gcaggtcccg atcagcggga ggagtgcgaa tccac 465
<210> 19
<211> 20
92
CA 02390309 2002-10-28
<212> DNA
<213> M13mp18
<400> 19
ccggggatcc tctagagtcg 20
<210> 20
<211> 20
<212> DNA
<213> M13mp18 mutant
<400> 20
ccggggatcc tctagagtca 20
<210> 21
<211> 48
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 21
cgactctaga ggatccccgg tttttgttgt gtggaattgt gagcggat 48
<210> 22
<211> 48
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 22
tgactctaga ggatccccgg tttttgttgt gtggaattgt gagcggat 48
<210> 23
<211> 47
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 23
cgtcgtgact gggaaaaccc tttttgttcg ggcctcttcg ctattac 47
<210> 24
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 24
actttatgct tccggctcgt a 21
93
CA 02390309 2002-10-28
<210> 25
<211> 17
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequerce:Artificially
synthesized primer sequence
<400> 25
gttgggaagg gcgatcg 17
<210> 26
<211> 44
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 26
tgttcctgat gcagtgggca gctttagtct gcggcggtgt tctg 44
<210> 27
<211> 45
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 27
tgctgggtcg gcacagcctg aagctgacct gaaatacctg gcctg 45
<210> 28
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 28
tgcttgtggc ctctcgtg 18
<210> 29
<211> 17
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Artificially
synthesized primer sequence
<400> 29
gggtgtggga agctgtg 17
94