Note: Descriptions are shown in the official language in which they were submitted.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
DESCRIPTION
METHOD AND REAGENT FOR THE INHIBITION OF GRID
Background Of The Invention
This invention claims priority from Jarvis et al., USSN (60/181,594), filed
February
24, 2000, entitled "METHOD AND REAGENT FOR THE INHIBITION OF GRID". This
application is hereby incorporated by reference herein in its entirety
including the
drawings.
The present invention concerns compounds, compositions, and methods for the
study, diagnosis, and treatment of conditions and diseases related to the
expression of the
T-cell co-stimulatory adapter protein GRID (Grb2-related with Insert Domain).
The following is a brief description of the current understanding of GRID. The
discussion is not meant to be complete and is provided only for understanding
the
invention that follows. The summary is not an admission that any of the work
described
below is prior art to the claimed invention.
One of the emerging paradigms for signal transduction in lymphocytes is that
receptors and other signaling molecules do not operate in isolation, but
through the
recruitment of a complex of other proteins (Pawson and Scott, 1997; Science,
278, 2075;
Rudd, 1999, Cell, 96, 5). These other proteins sexve to amplify and diversify
the signal into
a number of biochemical cascades. The archetypal adapter protein is Grb2,
which serves to
regulate downstream pathways such as Ras activation and Ca2+ mobilization
(Lowenstein
et al., 1992, Cell, 70, 431), and is ultimately responsible for modulating
gene expression
required for proliferation and differentiation. Grb2 is recruited to LAT and
SLP-76 which
are downstream targets in the signaling cascade initiated by ligation of the T-
cell receptor
by MHC-antigen. These functions are mediated by specialized domains which bind
specific motifs and include the phosphotyrosine binding SH2 (Src homology)
domain and
SH3 domain which are associated with proline-rich PXXP motifs. Grb2, whose
sole
function appears to be the formation of bridges between other proteins, is
entirely
comprised of such domains having an SH3-SH2-SH3 structure (Peterson et al.,
1998, CzsYf-.
Opin. Imnaunol., 10, 337; Koretzky, 1997, Immunol Today, 18, 401).
A novel member of the Grb2 family of adapter proteins termed GRID (Grb2-
related
with Insert Domain) has recently been identified (Asada et al, 1999, J. Exp.
Med., 189,
1383; Liu et al., 1999, Cunr. Biol., 9, 67; Liu et al., 1998, Oncogene, 17,
3073; Law et al.,
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
2
1999, J. Exp. 'Med., 189, 1243; Qiu et al., 1998, Biochem. Biophys. Res.
Comrnun., 253,
443; Bourette et al., 1998, Ernbo. J., 17, 7273). GRID is recruited to the T
cell co-
stimulatory receptor CD28 upon activation of this receptor by cross-linking
antibodies.
Although GRID shares significant similarity at the protein level with Grb2,
possessing an
SH3-SH2-SH3 domain structure, GRID also contains a unique proline-glutamine
rich
domain situated between the SH2 and C-terminal SH3 domain. The association of
GRID
with activated CD28 is absolutely dependent upon the integrity of the SH2
domain and
phosphorylation of residue Y173 in the cytoplasmic tail of CD28. Although GRID
has
been shown to associate with other T cell signaling proteins including SLP-76
and LAT
(Asada et al., supra; Liu et al., supra; Law et al., supra), it's role in T
cell signaling
pathways is not well defined.
Tari et al.,, 1999, Oncogene, 18(6), 1325-1332, describe the antisense
inhibition of
Grb2 in breast cancer cells in order to investigate the role of Grb2 in the
proliferation of
breast cancer cells. The resulting Grb2 inhibition led to MAP kinase
inactivation in EGFR
but not in ErbB2 expressing breast cancer cells.
Tari et al., 1998, J. Liposorne Res., 8(2), 251-264, describe P-ethoxy
antisense
oligonucleotides targeting Bcr-Abl, Grb2, Crkl, and Bcl-2 mRNA. Delivery of
these
antisense oligonucleotides via liposome transfection results in the inhibition
of
corresponding proteins, thereby inducing growth inhibition in leukemia and
lymphoma cell
lines.
Lopez-Berestein et al., 1998, International PCT publication No. WO 98/01547,
describe inhibition of chronic myelogenous leukemic cell growth by liposomal-
antisense
oligodeoxynucleotides targeting Grb2 and Crkl.
Tari et al., 1997, Biochenz. Biophys. Res. Comrnun., 235(2), 383-388, describe
the
antisense-based inhibition of Grb2 and Crkl proteins results in growth
inhbition of
Philadelphia chromosome positive leukemic cells.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
3
Summary Of The Invention
The invention features novel nucleic acid-based techniques [e.g., enzymatic
nucleic
acid molecules (for example, ribozymes or DNAzymes), antisense nucleic acids,
2-SA
antisense chimeras, triplex DNA, antisense nucleic acids containing RNA
cleaving
chemical groups] and methods for their use to modulate the expression of GRID
(Grb2-
related with Insert Domain).
The description below of the various aspects and embodiments is provided with
reference to the exemplary gene GRID. However, the various aspects and
embodiments
are also directed to other genes which express GRID-like adapter proteins
involved in T-
cell co-activation. Those additional genes can be analyzed for target sites
using the
methods described for GRID. Thus, the inhibition and the effects of such
inhibition of the
other genes can be performed as described herein.
In a preferred embodiment, the invention features the use of one or more of
the
nucleic acid-based techniques independently or in combination to inhibit the
expression of
the genes encoding GRID. For example, the nucleic acid-based techniques of the
present
invention can be used to inhibit the expression of GRID gene sequences found
at GenBank
Accession NOS. AJ011736, NM 004810, Y18051, AF121002, AF042380, AF129476,
AF090456).
In another preferred embodiment, the invention features the use of an
enzymatic
nucleic acid molecule, preferably in the hammerhead, NCH (Inozyme), G-cleaver,
amberzyme, zinzyme andlor DNAzyme motif, to inhibit the expression of GRID
gene.
By "inhibit" it is meant that the activity of GRID or level of GRID RNAs or
equivalent RNAs encoding one or more protein subunits of GRID or GRID-like
proteins is
xeduced below that observed in the absence of the nucleic acid molecules of
the invention.
In one embodiment, the inhibition with enzymatic nucleic acid molecule
preferably is
below that level observed in the presence of an enzymatically inactive or
attenuated
molecule that is able to bind to the same site on the target RNA, but is
unable to cleave that
RNA. In another embodiment, inhibition with antisense oligonucleotides is
preferably
below that level observed in the presence of, for example, an oligonucleotide
with
scrambled sequence or with mismatches. In another embodiment, inhibition of
GRID or
GRID-like genes with the nucleic acid molecule of the instant invention is
greater than in
the presence of the nucleic acid molecule than in its absence.
By "enzymatic nucleic acid molecule" it is meant a nucleic acid molecule which
has
complementarity in a substrate-binding region to a specified gene target, and
also has an
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
4
enzymatic activity which is active to specifically cleave target RNA. That is,
the
enzymatic nucleic acid molecule is able to intermolecularly cleave RNA and
thereby
inactivate a target RNA molecule. These complementary regions allow sufficient
hybridization of the enzymatic nucleic acid molecule to the target RNA and
thus permit
cleavage. One hundred percent complementarity is preferred, but
complementarity as low
as 50-75% can also be useful in this invention (see for example Werner and
Uhlenbeclc,
1995, Nucleic Acids Research, 23, 2092-2096; Hammann et al., 1999, Antisense
and
Nucleic Acid Df~ug Dev., 9, 25-31). The nucleic acids can be modified at the
base, sugar,
and/or phosphate groups. The term enzymatic nucleic acid is used
interchangeably with
phrases such as ribozymes, catalytic RNA? enzymatic RNA, catalytic DNA,
aptazyme or
aptamer-binding ribozyme, regulatable ribozyme, catalytic oligonucleotides,
nucleozyme,
DNAzyme, RNA enzyme, endoribonuclease, endonuclease, minizyme, leadzyme,
oligozyme or DNA enzyme. All of these terminologies describe nucleic acid
molecules
with enzymatic activity. The specific enzymatic nucleic acid molecules
described in the
instant application are not limiting in the invention and those skilled in the
art will
recognize that all that is important in an enzymatic nucleic acid molecule of
this invention
is that it has a specific substrate binding site which is complementary to one
or more of the
target nucleic acid regions, and that it have nucleotide sequences within or
surrounding that
substrate binding site which impart a nucleic acid cleaving and/or ligation
activity to the
molecule (Cech et al., U.S. Patent No. 4,987,071; Cech et al., 1988, 260 JAMA
3030).
By "nucleic acid molecule" as used herein is meant a molecule having
nucleotides.
The nucleic acid can be single, double, or multiple stranded and may comprise
modified or
unmodified nucleotides or non-nucleotides or various mixtures and combinations
thereof.
By "enzymatic portion" or "catalytic domain" is meant that portion or region
of the
enzymatic nucleic acid molecule essential for cleavage of a nucleic acid
substrate (for
example, see Figures 1-5).
By "substrate binding arm" or "substrate binding domain" is meant that portion
or
region of a enzymatic nucleic acid which is able to interact, for example, via
complementarity (i.e., able to base-pair with), with a portion of its
substrate. Preferably,
such complernentarity is 100%, but can be less if desired. For example, as few
as 10 bases
out of 14 can be base-paired (see for example Werner and Uhlenbeck, 1995,
Nucleic Acids
Research, 23, 2092-2096; Hammann et al., 1999, Antisense and Nucleic Acid Drug
Dev.,
9, 25-31). Examples of such anus are shown generally in Figures 1-5. That is,
these arms
contain sequences within an enzymatic nucleic acid which are intended to bring
enzymatic
nucleic acid and target RNA together through complementary base-pairing
interactions.
The enzymatic nucleic acid of the invention can have binding arms that are
contiguous or
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
non-contiguous and can be of varying lengths. The length of the binding arms)
are
preferably greater than or equal to four nucleotides and of sufficient length
to stably
interact with the target RNA. Preferably, the binding arms) are 12-100
nucleotides in
length. More preferably, the binding arms are 14-24 nucleotides in length
(see, for
5 example, Werner and Uhlenbeclc, supra; Hamman et al., supYa; Hampel et al.,
EP0360257;
Berzal-Herrance et al., 1993, EMBO J., 12, 2567-73). If two binding arms are
chosen, the
design is such that the length of the binding arms are symmetrical (i. e.,
each of the binding
arms is of the same length; e.g., five and five nucleotides, or six and six
nucleotides, or
seven and seven nucleotides long) or asymmetrical (i. e., the binding arms are
of different
length; e.g., six and three nucleotides; three and six nucleotides long; four
and five
nucleotides long; four and six nucleotides long; four and seven nucleotides
long; and the
like).
By "Inozyme" or "NCH" motif is meant, an enzymatic nucleic acid molecule
comprising a motif as is generally described as NCH Rz in Figure 2. Inozymes
possess
endonuclease activity to cleave RNA substrates having a cleavage triplet NCH/,
where N is
a nucleotide, C is cytidine and H is adenosine, uridine or cytidine, and /
represents the
cleavage site. H is used interchangeably with X. Inozymes can also possess
endonuclease
activity to cleave RNA substrates having a cleavage triplet NCNI, where N is a
nucleotide,
C is cytidine, and / represents the cleavage site. "I" in Figure 2 represents
an Inosine
nucleotide, preferably a ribo-Inosine or xylo-Inosine nucleoside.
By "G-cleaver" motif is meant, an enzymatic nucleic acid molecule comprising a
motif as is generally described as G-cleaver in Figure 2. G-cleavers possess
endonuclease
activity to cleave RNA substrates having a cleavage triplet NYN/, where N is a
nucleotide,
Y is uridine or cytidine and / represents the cleavage site. G-cleavers may be
chemically
modified as is generally shown in Figure 2.
By "amberzyme" motif is meant, an enzymatic nucleic acid molecule comprising a
motif as is generally described in Figure 3. Amberzymes possess endonuclease
activity to
cleave RNA substrates having a cleavage triplet NG/N, where N is a nucleotide,
G is
guanosine, and / represents the cleavage site. Amberzymes can be chemically
modified to
increase nuclease stability through substitutions as are generally shown in
Figure 3. In
addition, differing nucleoside and/or non-nucleoside linkers can be used to
substitute the
5'-gaaa-3' loops shown in the figure. Amberzymes represent a non-limiting
example of an
enzymatic nucleic acid molecule that does not require a ribonucleotide (2'-OH)
group
within its ovcm nucleic acid sequence for activity.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
6
By "zinzyme" motif is meant, an enzymatic nucleic acid molecule comprising a
motif as is generally described in Figure 4. Zinzymes possess endonuclease
activity to
cleave RNA substrates having a cleavage triplet including but not limited to
YG/Y, where
Y is uridine or cytidine, and G is guanosine and / represents the cleavage
site. Zinzymes
can be chemically modified to increase nuclease stability through
substitutions as are
generally shown in Figure 4, including substituting 2'-O-methyl guanosine
nucleotides for
guanosine nucleotides. In addition, differing nucleotide andlor non-nucleotide
linkers can
be used to substitute the 5'-gaaa-2' loop shown in the figure. Zinzymes
represent a non
limiting example of an enzymatic nucleic acid molecule that does not require a
ribonucleotide (2'-OH) group within its own nucleic acid sequence for
activity.
By 'DNAzyme' is meant, an enzymatic nucleic acid molecule that does not
require
the presence of a 2'-OH group fox its activity. In particular embodiments the
enzymatic
nucleic acid molecule can have an attached linkers) or other attached or
associated groups,
moieties, or chains containing one or more nucleotides with 2'-OH groups.
DNAzymes can
be synthesized chemically or expressed endogenously in vivo, by means of a
single
stranded DNA vector or equivalent thereof. An example of a DNAzyme is shown in
Figure 5 and is generally reviewed in Usman et al., International PCT
Publication No. WO
95/11304; Chartrand et al., 1995, NAR 23, 4092; Breaker et al., 1995, Chem.
Bio. 2, 655;
Santoro et al., 1997, PNAS 94, 4262; Breaker, 1999, Natu~°e
Biotechnology, 17, 422-423;
and Santoro et. al., 2000, J. Am. Cl2em. Soc., 122, 2433-39. Additional
DNAzyme motifs
can be selected for using techniques similar to those described in these
references, and
hence, are within the scope of the present invention.
By "sufficient length" is meant an oligonucleotide of greater than or equal to
3
nucleotides that is of a length great enough to provide the intended function
under the
expected condition. For example, for binding arms of enzymatic nucleic acid
"sufficient
length" means that the binding arm sequence is long enough to provide stable
binding to a
target site under the expected binding conditions. Preferably, the binding
arms are not so
long as to prevent useful turnover.
By "stably interact" is meant interaction of the oligonucleotides with target
nucleic
acid (e.g., by forming hydrogen bonds with complementary nucleotides in the
target under
physiological conditions) that is sufficient to the intended purpose (e.g.,
cleavage of target
RNA by an enzyme).
By "equivalent" RNA to GRID is meant to include those naturally occurring RNA
molecules having homology (partial or complete) to GRID proteins or encoding
for
proteins with similar function as GRID in various organisms, including human,
rodent,
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
7
primate, rabbit, pig, protozoans, fungi, plants, and other microorganisms and
parasites.
The equivalent RNA sequence also includes in addition to the coding region,
regions such
as 5'-untranslated region, 3'-untranslated region, introns, intron-exon
junction and the like.
By "homology" is meant the nucleotide sequence of two or more nucleic acid
molecules is partially or completely identical.
By "antisense nucleic acid", it is meant a non-enzymatic nucleic acid molecule
that
binds to target RNA by means of RNA-RNA or RNA-DNA or RNA-PNA (protein nucleic
acid; Egholm et al., 1993 Nature 365, 566) interactions and alters the
activity of the target
RNA (for a review, see Stein and Cheng, 1993 Scierace 261, 1004 and Woolf et
al., US
patent No. 5,849,902). Typically, antisense molecules are complementary to a
target
sequence along a single contiguous sequence of the antisense molecule.
However, in
certain embodiments, an antisense molecule can bind to substrate such that the
substrate
molecule forms a loop, and/or an antisense molecule can bind such that the
antisense
molecule forms a loop. Thus, the antisense molecule can be complementary to
two (or
even more) non-contiguous substrate sequences or two (or even more) non-
contiguous
sequence portions of an antisense molecule can be complementary to a target
sequence or
both. For a review of current antisense strategies, see Schmajuk et al., 1999,
J. Biol.
Chem., 274, 21783-21789, Delihas et al., 1997, Nature, 15, 751-753, Stein et
al., 1997,
Antiserase N. A. Drug Dev., 7, 151, Crooke, 2000, Methods Enzyrraol., 313, 3-
45; Croolce,
1998, Biotech. Genet. Eng. Rev., 15, 121-157, Crooke, 1997, Ad. Pharrnacol.,
40, 1-49. In
addition, antisense DNA can be used to target RNA by means of DNA-RNA
interactions,
thereby activating RNase H, which digests the target RNA in the duplex. The
antisense
oligonucleotides can comprise one or more RNAse H activating region, which is
capable of
activating RNAse H cleavage of a target RNA. Antisense DNA can be synthesized
chemically or expressed .via the use of a single stranded DNA expression
vector or
equivalent thereof.
By "RNase H activating region" is meant a region (generally greater than or
equal to
4-25 nucleotides in length, preferably from 5-11 nucleotides in length) of a
nucleic acid
molecule capable of binding to a target RNA to form a non-covalent complex
that is
recognized by cellular RNase H enzyme (see for example Arrow et al., US
5,849,902;
Arrow et al., US 5,989,912). The RNase H enzyme binds to the nucleic acid
molecule-
target RNA complex and cleaves the target RNA sequence. The RNase H activating
region
comprises, for example, phosphodiester, phosphorothioate (preferably at least
four of the
nucleotides are phosphorothiote substitutions; morepreferably, 4-11 of the
nucleotides are
phosphorothiote substitutions); phosphorodithioate, 5'-thiophosphate, or
methylphosphonate backbone chemistry or a combination thereof. In addition to
one or
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
8
more backbone chemistries described above, the RNase H activating region can
also
comprise a variety of sugar chemistries. For example, the RNase H activating
region can
comprise deoxyribose, arabino, fluoroarabino or a combination thereof,
nucleotide sugar
chemistry. Those skilled in the art will recognize that the foregoing are non-
limiting
examples and that any combination of phosphate, sugar and base chemistry of a
nucleic
acid that supports the activity of RNase H enzyme is within the scope of the
definition of
the RNase H activating region and the instant invention.
By "2-SA antisense chimera" is meant an antisense oligonucleotide containing a
5'-
phosphorylated 2'-5'-linked adenylate residue. These chimeras bind to target
RNA in a
sequence-specific manner and activate a cellular 2-SA-dependent ribonuclease
which, in
turn, cleaves the target RNA (Torrence et al., 1993 P>"oc. Natl. Acad. Sci.
USA 90, 1300;
Silverman et al., 2000, Methods Enzyrnol., 313, 522-533; Player and Torrence,
1998,
Plzas~rnacol. They., 78, 55-113).
By "triplex forming oligonucleotides" is meant an oligonucleotide that can
bind to a
double-stranded DNA in a sequence-specific manner to form a triple-strand
helix.
Formation of such triple helix structure has been shown to inhibit
transcription of the
targeted gene (Duval-Valentin et al., 1992 Pt~oc. Natl. Acad. Sci. USA 89,
504; Fox, 2000,
Cum°. Med. Chenz., 7, 17-37; Praseuth et. al., 2000, Biochim. Biophys.
Acta, 1489, 181-
206).
By "gene" it is meant a nucleic acid that encodes RNA, for example, nucleic
acid
sequences including but not limited to structural genes encoding a
polypeptide.
"Complementarity" refers to the ability of a nucleic acid to form hydrogen
bonds)
with another RNA sequence by either traditional Watson-Cxick or other non-
traditional
types. In reference to the nucleic molecules of the present invention, the
binding free
energy for a nucleic acid molecule with its taxget or complementary sequence
is sufficient
to allow the relevant function of the nucleic acid to proceed, e.g., enzymatic
nucleic acid
cleavage, antisense or triple helix inhibition. Determination of binding free
energies fox
nucleic acid molecules is well known in the art (see, e.g., Turner et al.,
1987, CSH Syrnp.
Quant. Biol. LII pp.123-133; Frier et al., 1986, PYOG. Nat. Acad. Sci. USA
83:9373-9377;
Turner et al., 1987, J. Am. Chezn. Soc. 109:3783-3785). A percent
complementarity
indicates the percentage of contiguous residues in a nucleic acid molecule
which can form
hydrogen bonds (e.g., Watson-Criclc base pairing) with a second nucleic acid
sequence
(e.g., 5, 6, 7, 8, 9, 10 out of 10 being 50%, 60%, 70%, 80%, 90%, and 100%
complementary). "Perfectly complementary" means that all the contiguous
residues of a
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
9
nucleic acid sequence will hydrogen bond with the same number of contiguous
residues in
a second nucleic acid sequence.
By "RNA" is meant a molecule comprising at least one ribonucleotide residue.
By
"ribonucleotide" or "2'-OH" is meant a nucleotide with a hydroxyl group at the
2' position
of a (3-D-ribo-furanose moiety.
By "decoy RNA" is meant a RNA molecule that mimics the natural binding domain
for a ligand. The decoy RNA therefore competes with natural binding target for
the binding
of a specific ligand. For example, it has been shown that over-expression of
HIV trans-
activation response (TAR) RNA can act as a "decoy" and efficiently binds HIV
tat protein,
thereby preventing it from binding to TAR sequences encoded in the HIV RNA
(Sullenger
et al., 1990, Cell, 63, 601-608). This is but a specific example and those in
the art will
recognize that other embodiments can be readily generated using techniques
generally
known in the art.
Several varieties of naturally occurring enzymatic RNAs are known presently.
Each
can catalyze the hydrolysis of RNA phosphodiester bonds in traps (and thus can
cleave
other RNA molecules) under physiological conditions. Table I summarizes some
of the
characteristics of these ribozymes. In general, enzymatic nucleic acids act by
first binding
to a target RNA. Such binding occurs through the target binding portion of a
enzymatic
nucleic acid which is held in close proximity to an enzymatic portion of the
molecule that
acts to cleave the target RNA. Thus, the enzymatic nucleic acid first
recognizes and then
binds a target RNA through complementary base-pairing, and once bound to the
correct
site, acts enzymatically to cut the target RNA. Strategic cleavage of such a
target RNA
will destroy its ability to direct synthesis of an encoded protein. After an
enzymatic
nucleic acid has bound and cleaved its RNA target, it is released from that
RNA to search
for another target and can repeatedly bind and cleave new targets. Thus, a
single ribozyme
molecule is able to cleave many molecules of target RNA. In addition, the
ribozyme is a
highly specific inhibitor of gene expression, with the specificity of
inhibition depending
not only on the base-pairing mechanism of binding to the target RNA, but also
on the
mechanism of target RNA cleavage. Single mismatches, or base-substitutions,
near the site
of cleavage can completely eliminate catalytic activity of a ribozyme.
The enzymatic nucleic acid molecule that cleave the specified sites in GRID-
specific
RNAs represent a novel therapeutic approach to treat a variety of pathologic
indications,
including but not limited to tissue/graft rejection and leukemia.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
In one of the preferred embodiments of the inventions described herein, the
enzymatic nucleic acid molecule is formed in a hammerhead or hairpin motif,
but can also
be formed in the motif of a hepatitis delta virus, group I intron, group II
intron or RNase P
RNA (in association with an RNA guide sequence), Neurospora VS RNA, DNAzymes,
5 NCH cleaving motifs, or G-cleavers. Examples of such hammerhead motifs are
described
by Dreyfus, supra, Rossi et al., 1992, AIDS Research and Human Retroviruses 8,
183.
Examples of hairpin motifs are described by Hampel et al., EP0360257, Hampel
and Tritz,
1989 Biochenaistry 28, 4929, Feldstein et al., 1989, Gene 82, 53, Haseloff and
Gerlach,
1989, Gene, 82, 43, Hampel et al., 1990 Nucleic Acids Res. 18, 299; and
Chowrira &
10 McSwiggen, US. Patent No. 5,631,359. The hepatitis delta virus motif is
described by
Perrotta and Been, 1992 Biochemistry 31, 16. The RNase P motif is described by
Guerrier-
Takada et al., 1983 Cell 35, 849; Forster and Altman, 1990, Science 249, 783;
and Li and
Altman, 1996, Nucleic Acids Res. 24, 835. The Neurospora VS RNA ribozyme motif
is
described by Collins (Saville and Collins, 1990 Cell, 61, 685-696; Saville and
Collins,
1991 Proc. Natl. Acad. Sci. USA 88, 8826-8830; Collins and Olive, 1993
Biochemistry 32,
2795-2799; and Guo and Collins, 1995, EMBO. J. 14, 363). Group II introns are
described
by Griffin et al., 1995, Chena. Biol. 2, 761; Michels and Pyle, 1995,
Biochemistry 34, 2965;
and Pyle et al., International PCT Publication No. WO 96/22689. The Group I
intron is
described by Cech et al., U.S. Patent 4,987,071. DNAzymes are described by
Usman et
al., International PCT Publication No. WO 95/11304; Chartrand et al., 1995,
NAR 23,
4092; Breaker et al., 1995, Claem. Bio. 2, 655; and Santoro et al., 1997, PNAS
94, 4262.
NCH cleaving motifs are described in Ludwig & Sproat, International PCT
Publication No.
WO 98/58058; and G-cleavers are described in Kore et al., 1998, Nucleic Acids
Research
26, 4116-4120 and Eckstein et al., International PCT Publication No. WO
99116871.
Additional motifs include the Aptazyme (Breaker et al., WO 98/43993),
Amberzyme
(Class I motif; Figure 3; Beigelman et al., International PCT publication No.
WO
99/55857) and Zinzyme (Beigelman et al., International PCT publication No. WO
99/55857), all these references are incorporated by reference herein in their
totalities,
including drawings and can also be used in the present invention. These
specific motifs are
not limiting in the invention and those skilled in the art will recognize that
all that is
important in an enzymatic nucleic acid molecule of this invention is that it
has a specific
substrate binding site which is complementary to one or more of the target
gene RNA
regions, and that it have nucleotide sequences within or surrounding that
substrate binding
site which impart an RNA cleaving activity to the molecule (Cech et al., U.S.
Patent No.
4,987,071).
In preferred embodiments of the present invention, a nucleic acid molecule of
the
instant invention can be between 13 and 100 nucleotides in length. Exemplary
enzymatic
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
11
nucleic acid molecules of the invention are shown in Tables III-VIII and X.
For example,
enzymatic nucleic acid molecules of the invention are preferably between 15
and 50
nucleotides in length, more preferably between 25 and 40 nucleotides in
length, e.g., 34,
36, or 38 nucleotides in length (for example see Jarvis et al., 1996, J. Biol.
ClZem., 271,
29107-29112). Exemplary DNAzymes of the invention are preferably between 15
and 40
nucleotides in length, more preferably between 25 and 35 nucleotides in
length, e.g., 29,
30, 31, or 32 nucleotides in length (see for example Santoro et al., 1998,
Bioche~raistry, 37,
13330-13342; Chartrand et al., 1995, Nucleic Acids Research, 23, 4092-4096 and
Cairns et
al., 2000, Aratisense & Nucleic Acid Drug Dev., 10, 323-332). Exemplary
antisense
molecules of the invention are preferably between 15 and 75 nucleotides in
length, more
preferably between 20 and 35 nucleotides in length, e.g., 25, 26, 27, or 28
nucleotides in
length (see for example Woolf et al., 1992, PNAS., 89, 7305-7309; Milner et
al., 1997,
Nature Bioteclanology, 15, 537-541). Exemplary triplex forming oligonucleotide
molecules of the invention are preferably between 10 and 40 nucleotides in
length, more
preferably between 12 and 25 nucleotides in length, e.g., 18, 19, 20, or 21
nucleotides in
length (see for example Maher et al., 1990, Bioclae~aistry, 29, 8820-8826;
Strobel and
Dervan, 1990, Science, 249, 73-75). Those skilled in the art will recognize
that all that is
required is for the nucleic acid molecule to be of length and conformation
sufficient and
suitable for the nucleic acid molecule to catalyze a reaction contemplated
herein. The
length of the nucleic acid molecules of the instant invention are not limiting
within the
general limits stated.
Preferably, a nucleic acid molecule that down regulates the replication of
GRID or
GRID-like gene comprises between 12 and 100 bases complementary to a GRID or
GRID-
lilce RNA. Even more preferably, a nucleic acid molecule ~ that down regulates
the
replication of GRID or GRID-like gene comprises between 14 and 24 bases
complementary to a GRID or GRID-lilce RNA.
In a preferred embodiment, the invention provides a method for producing a
class of
nucleic acid-based gene inhibiting agents which exhibit a high degree of
specificity for the
RNA of a desired target. For example, the enzymatic nucleic acid molecule is
preferably
targeted to a highly conserved sequence region of target RNAs encoding GRID or
GRID
lilce proteins such that specific treatment of a disease or condition can be
provided with
either one or several nucleic acid molecules of the invention. Such nucleic
acid molecules
can be delivered exogenously to specific tissue or cellular targets as
required.
Alternatively, the nucleic acid molecules (e.g., ribozymes and antisense) can
be expressed
from DNA andlor RNA vectors that are delivered to target cells.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
12
In a preferred embodiment, the invention features the use of nucleic acid-
based
inhibitors of the invention to specifically target genes that share homology
with the GRID
gene. For example, the invention describes the use of nucleic acid-based
inhibitors to
target the Grb2 (GenBank accession No. NM 002086) and GRAP (GenBank accession
No.
h1M-006613) genes.
As used in herein "cell" is used in its usual biological sense and does not
refer to an
entire multicellular organism. The cell can be present in an organism which
includes
humans but is preferably a non-human multicellular organism, e.g., birds,
plants and
mammals such as cows, sheep, apes, monkeys, swine, dogs, and cats. The cell
can be
prokaryotic (e.g., bacterial cell) or eukaryotic (e.g., mammalian or plant
cell).
By "GRID proteins" is meant, a protein or a mutant protein derivative thereof,
comprising an adapter-protein type of association to the activated CD28 co-
stimulatory
receptor, and to other signaling proteins including but not limited to SLP-76
and LAT.
By "highly conserved sequence region" is meant a nucleotide sequence of one or
more regions in a target gene that does not vary significantly from one
generation to the
other or from one biological system to the other.
The nucleic acid-based inhibitors of GRID expression are useful for the
prevention
and/or treatment of diseases and conditions that are related to or will
respond to the levels
of GRID in a cell or tissue, alone or in combination with other therapies. For
example, the
nucleic acid-based inhibitors of GRID expressions are useful for the
prevention and/or
treatment of tissue/graft rejection and cancer, such as leukemia, among other
conditions.
By "related" is meant that the reduction of GRID expression (specifically GRID
gene) RNA levels and thus reduction in the level of the respective protein
will relieve, to
some extent, the symptoms of the disease or condition.
In a preferred embodiment, the invention features the use of nucleic acid-
based
inhibitors of the invention to specifically target regions of GRID gene that
are not
homologous to Grb2 gene. Specifically, the invention describes the use of
nucleic acid-
based inhibitors to target sequences that are unique to GRID gene.
The nucleic acid-based inhibitors of the invention are added directly, or can
be
complexed with cationic lipids, packaged within liposomes, or otherwise
delivered to
target cells or tissues using well-lrnown methods described herein and
generally lrnown in
the art. The nucleic acid or nucleic acid complexes can be locally
administered to relevant
tissues ex vivo, or in vivo through injection, infusion pump or stmt, with or
without their
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
13
incorporation in biopolymers. In preferred embodiments, the enzymatic nucleic
acid
inhibitors comprise sequences, which are complementary to the substrate
sequences in
Tables III to X. Examples of such enzymatic nucleic acid molecules also are
shown in
Tables III to VIII and X. Examples of such enzymatic nucleic acid molecules
consist
essentially of sequences defined in these Tables.
In yet another embodiment, the invention features antisense nucleic acid
molecules
and 2-SA chimera including sequences complementary to the substrate sequences
shown in
Tables III to X. Such nucleic acid molecules can include sequences as shown
for the
binding arms of the enzymatic nucleic acid molecules in Tables III to VIII and
X and
sequences shown as GeneBlocTM sequences in Table X. Similarly, triplex
molecules can
be provided targeted to the corresponding DNA target regions, and containing
the DNA
equivalent of a target sequence or a sequence complementary to the specified
target
(substrate) sequence. Typically, antisense molecules are complementary to a
target
sequence along a single contiguous sequence of the antisense molecule.
However, in
certain embodiments, an antisense molecule can bind to substrate such that the
substrate
molecule forms a loop, andlor an antisense molecule can bind such that the
antisense
molecule forms a loop. Thus, the antisense molecule can be complementary to
two (or
even more) non-contiguous substrate sequences or two (or even more) non-
contiguous
sequence portions of an antisense molecule can be complementary to a target
sequence or
both.
By "consists essentially of is meant that the active nucleic acid molecule of
the
invention, for example, an enzymatic nucleic acid molecule, contains an
enzymatic center
or core equivalent to those in the examples and binding arms able to bind RNA
such that
cleavage at the target site occurs. Other sequences can be present which do
not interfere
with such cleavage. Thus, a core region can, for example, include one or more
loop, stem-
loop structure, or linker which does not prevent enzymatic activity. Thus, the
underlined
regions in the sequences in Tables III and IV can be such a loop, stem-loop,
nucleotide
linker, and/or non-nucleotide linker and can be represented generally as
sequence "X". For
example, a core sequence for a hammerhead enzymatic nucleic acid can comprise
a
conserved sequence, such as 5'-CUGAUGAG-3' and 5'-CGAA-3' connected by a
sequence X, where X is 5'-GCCGUUAGGC-3' (SEQ ID NO 2236) or any other stem II
region known in the art or a nucleotide and/or non-nucleotide linker.
Similarly, for other
nucleic acid molecules of the instant invention, such as Inozyme, G-cleaver,
amberzyme,
zinzyme, DNAzyme, antisense, 2-SA antisense, triplex forming nucleic acid, and
decoy
nucleic acids, other sequences or non-nucleotide linkers may be present that
do not
interfere with the function of the nucleic acid molecule.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
14
Sequence X can be a linker of >_ 2 nucleotides in length, preferably 3, 4, 5,
6, 7, 8, 9,
10, 15, 20, 26, 30, where the nucleotides can preferably be internally base-
paired to form a
stem of preferably >_ 2 base pairs. Alternatively or in addition, sequence X
can be a non-
nucleotide linker. In yet another embodiment, the nucleotide linker X can be a
nucleic acid
aptamer, such as an ATP aptamer, HIV Rev aptamer (RRE), HIV Tat aptamer (TAR)
and
others (for a review see Gold et al., 1995, Anhu. Rev. Biochem., 64, 763; and
Szostalc &
Ellington, 1993, in The RNA World, ed. Gesteland and Atkins, pp. 511, CSH
Laboratory
Press). A "nucleic acid aptamer" as used herein is meant to indicate a nucleic
acid sequence
capable of interacting with a ligand. The ligand can be any natural or a
synthetic molecule,
including but not limited to a resin, metabolites, nucleosides, nucleotides,
drugs, toxins,
transition state analogs, peptides, lipids, proteins, amino acids, nucleic
acid molecules,
hormones, carbohydrates, receptors, cells, viruses, bacteria and others.
In yet another embodiment, the non-nucleotide linker X is as defined herein.
The
term "non-nucleotide linker" as used herein include either abasic nucleotide,
polyether,
polyamine, polyamide, peptide, carbohydrate, lipid, or polyhydrocarbon
compounds.
Specific examples include those described by Seela and Kaiser, Nucleic Acids
Res. 1990,
18:6353 and Nucleic Acids Res. 1987, 15:3113; Cload and Schepartz, J. Aru.
Chem. Soc.
1991, 113:6324; Richardson and Schepartz, J. Am. Chem. Soc. 1991, 113:5109; Ma
et al.,
Nucleic Acids Res. 1993, 21:2585 and Bioclaemist~y 1993, 32:1751; Durand et
al., Nucleic
Acids Res. 1990, 18:6353; McCurdy et al., Nucleosides & Nucleotides 1991,
10:287;
Jschke et al., Tetrahedron Lett. 1993, 34:301; Ono et al., Biochemistry 1991,
30:9914;
Arnold et al., International Publication No. WO 89/02439; Usman et al.,
International
Publication No. WO 95/06731; Dudycz et al., International Publication No. WO
95/11910
and Ferentz and Verdine, J. Arn. Chem. Soc. 1991, 113:4000, all hereby
incorporated by
reference herein. The term "non-nucleotide" further refers to any group or
compound
which can be incorporated into a nucleic acid chain in the place of one or
more nucleotide
units, including either sugar and/or phosphate substitutions and allows the
remaining bases
to exhibit their enzymatic activity. The group or compound can be abasic in
that it does
not contain a commonly recognized nucleotide base, such as adenosine, guanine,
cytosine,
uracil or thymine. Thus, in a preferred embodiment, the invention features an
enzymatic
nucleic acid molecule having one or more non-nucleotide moieties and having
enzymatic
activity to cleave an RNA or DNA molecule.
In another aspect of the invention, ribozymes or antisense molecules that
interact
with target RNA molecules and inhibit GRID activity (e.g., inhibit GRID gene)
are
expressed from transcription units inserted into DNA or RNA vectors. The
recombinant
vectors are preferably DNA plasmids or viral vectors. Ribozyme or antisense
expressing
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
viral vectors can be constructed based on, but not limited to, adeno-
associated virus,
retrovirus, adenovirus, or alphavirus. Preferably, the recombinant vectors
capable of
expressing the ribozymes or antisense are delivered as described above, and
persist in
target cells. Alternatively, viral vectors can be used that provide for
transient expression of
5 ribozymes or antisense. Such vectors can be repeatedly administered as
necessary. Once
expressed, the ribozymes or antisense bind to the target RNA and inhibit its
function or
expression. Delivery of ribozyme or antisense expressing vectors can be
systemic, such as
by intravenous or intramuscular administration, by administration to target
cells ex-planted
from the patient followed by reintroduction into the patient, or by any other
means that
10 would allow for introduction into the desired target cell. Antisense DNA
can be expressed
endogenously via the use of a single stranded DNA intracellular expression
vector.
By "vectors" is meant any nucleic acid- and/or viral-based technique used to
deliver
a desired nucleic acid.
By "patient" is meant an organism, which is a donor or recipient of explanted
cells or
15 the cells themselves. "Patient" also refers to an organism to which the
nucleic acid
molecules of the invention can be administered. Preferably, a patient is a
mammal or
mammalian cells. More preferably, a patient is a human or human cells.
By "enhanced enzymatic activity" is meant to include activity measured in
cells
and/or in vivo where the activity is a reflection of both the catalytic
activity and the
stability of the nucleic acid molecules of the invention. In this invention,
the product of
these properties can be increased in vivo compared to an all RNA enzymatic
nucleic acid
or all DNA enzyme. In some cases, the individual catalytic activity or
stability of the
nucleic acid molecule can be decreased (i.e., less than ten-fold), but the
overall activity of
the nucleic acid molecule is enhanced in vivo.
The nucleic acid molecules of the instant invention, individually, or in
combination
or in conjunction with other drugs, can be used to treat diseases or
conditions discussed
above. For example, to treat a disease or condition associated with the levels
of GRID, the
patient can be treated, or other appropriate cells can be treated, as is
evident to those skilled
in the art, individually or in combination with one or more drugs under
conditions suitable
for the treatment.
In a further embodiment, the described molecules, such as antisense or
ribozymes,
can be used in combination with other known treatments to treat conditions or
diseases
discussed above. For example, the described molecules can be used in
combination with
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
16
one or more known therapeutic agents to treat tissue/graft rejection, leukemia
and/or other
disease states or conditions which respond to the modulation of GRID
expression.
In another preferred embodiment, the invention features nucleic acid-based
inhibitors
(e.g., enzymatic nucleic acid molecules (ribozymes), antisense nucleic acids,
2-SA
antisense chimeras, triplex DNA, antisense nucleic acids containing RNA
cleaving
chemical groups) and methods for their use to down regulate or inhibit the
expression of
genes (e.g., GRID) related to the progression and/or maintenance of
tissue/graft rejection,
leukemia and/or other disease states or conditions which respond to the
modulation of
GRID expression.
In another aspect, the invention provides mammalian cells containing one or
more
nucleic acid molecules and/or expression vectors of this invention. The one or
more
nucleic acid molecules can independently be targeted to the same or different
sites.
By "comprising" is meant including, but not limited to, whatever follows the
word
"comprising". Thus, use of the term "comprising" indicates that the listed
elements are
required or mandatory, but that other elements are optional and may or may not
be present.
By "consisting of is meant including, and limited to, whatever follows the
phrase
"consisting of'. Thus, the phrase "consisting of indicates that the listed
elements are
required or mandatory, and that no other elements may be present.
Other features and advantages of the invention will be apparent from the
following
description of the preferred embodiments thereof, and from the claims.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
17
Description Of The Preferred Embodiments
First the drawings will be described briefly.
Drawings
Figure 1 shows the secondary structure model for seven different classes of
enzymatic nucleic acid molecules. Arrow indicates the site of cleavage. -------
-- indicate
the target sequence. Lines interspersed with dots are meant to indicate
tertiary interactions.
- is meant to indicate base-paired interaction. Group I Intron: P1-P9.0
represent various
stem-loop structures (Cech et al., 1994, Nature Struc. Bio., 1, 273). RNase P
(M1RNA):
EGS represents external guide sequence (Forster et al., 1990, ~'cietace, 249,
783; Pace et al.,
1990, J. Biol. Chena., 265, 3587). Group II Intron: 5'SS means 5' splice site;
3'SS means
3'-splice site; IBS means intron binding site; EBS means exon, binding site
(Pyle et al.,
1994, Biochemistry, 33, 2716). VS RNA: I-VI are meant to indicate six stem-
loop
structures; shaded regions are meant to indicate tertiary interaction
(Collins, International
PCT Publication No. WO 96119577). HDV Ribozyme: : I-IV are meant to indicate
four
stem-loop structures (Been et al., US Patent No. 5,625,047). Hammerhead
Ribozyme:
I-III are meant to indicate three stem-loop structures; stems I-III can be of
any length and
can be symmetrical or asymmetrical (Unman et al., 1996, Cur. Op.
Sty°uct. Bio., 1, 527).
Hairpin Ribozyme: Helix 1, 4 and 5 can be of any length; Helix 2 is between 3
and 8
base-pairs long; Y is a pyrimidine; Helix 2 (H2) is provided with a least 4
base pairs (i.e.,
n is 1, 2, 3 or 4) and helix 5. can be optionally provided of length 2 or more
bases
(preferably 3 - 20 bases, i.e., m is from 1 - 20 or more). Helix 2 and helix 5
can be
covalently linked by one or more bases (i.e., r is >_ 1 base). Helix 1, 4 or 5
can also be
extended by 2 or more base pairs (e.g., 4 - 20 base pairs) to stabilize the
ribozyme
structure, and preferably is a protein binding site. In each instance, each N
and N'
independently is any normal or modified base and each dash represents a
potential base-
pairing interaction. These nucleotides can be modified at the sugar, base or
phosphate.
Complete base-pairing is not required in the helices, but is preferred. Helix
1 and 4 can be
of any size (i.e., o and p is each independently from 0 to any number, e.g.,
20) as long as
some base-pairing is maintained. Essential bases are shown as specific bases
in the
structure, but those in the art will recognize that one or more can be
modified chemically
(abasic, base, sugar and/or phosphate modifications) or replaced with another
base without
significant effect. Helix 4 can be formed from two separate molecules, i.e.,
without a
connecting loop. The connecting loop when present can be a ribonucleotide with
or
without modifications to its base, sugar or phosphate. "q" _> is 2 bases. The
connecting
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
18
loop can also be replaced with a non-nucleotide linker molecule. H refers to
bases A, U, or
C. Y refers to pyrimidine bases. " " refers to a covalent bond. (Burke et al.,
1996,
Nucleic Acids & Mol. Biol., 10, 129; Chowrira et al., US Patent No.
5,631,359).
Figure 2 shows examples of chemically stabilized ribozyme motifs. HH Rz,
represents hammerhead ribozyme motif (Usman et al., 1996, GuYY. Op.
Stf°uct. Bio., l,
527); NCH Rz represents the NCH ribozyme motif (Ludwig & Sproat, International
PCT
Publication No. WO 98/58058); G-Cleaver, represents G-cleaver ribozyme motif
(Kore et
al., 1998, Nucleic Acids Reseaf~ch 26, 4116-4120). N or n, represent
independently a
nucleotide which can be same or different and have complementarity to each
other; rI,
represents ribo-Inosine nucleotide; arrow indicates the site of cleavage
within the target.
Position 4 of the HH Rz and the NCH Rz is shown as having 2'-C-ally!
modification, but
those skilled in the art will recognize that this position can be modified
with other
modifications well !mown in the art, so long as such modifications do not
significantly
inhibit the activity of the ribozyme.
Figure 3 shows an example of the Amberzyme ribozyme motif that is chemically
stabilized (see, for example, Beigelman et al., International PCT publication
No. WO
99155857, incorporated by reference herein; also referred to as Class I
Motif). The
Amberzyme motif is a class of enzymatic nucleic molecules that do not require
the
presence of a ribonucleotide (2'-OH) group for its activity.
Figure 4 shows an example of the Zinzyme A ribozyme motif that is chemically
stabilized (Beigelman et al., International PCT publication No. WO 99/55857,
incorporated by reference herein; also referred to as Class A or Class II
Motif). The
Zinzyme motif is a class of enzymatic nucleic molecules that do not require
the presence of
a ribonucleotide (2'-OH) group for its activity.
Figure 5 shows an example of a DNAzyme motif described by Santoro et al.,
1997,
PNAS, 94, 4262.
Figure 6 shows a graph of optimization of GeneBloc concentration. A
fluoresceinated randomized antisense GeneBloc (fGB) was used as a marker for
uptake
using a axed concentration of lipid. Cells were either untreated (A) or
treated continuously
for 24hrs with 10-200nM antisense GeneBloc (B-F). Following treatment, cells
were
analyzed by flow cytometry. Gate M1 represents either untransfected cells or
cells
refractory to transfection. Gate M2 represents the transfected cells.
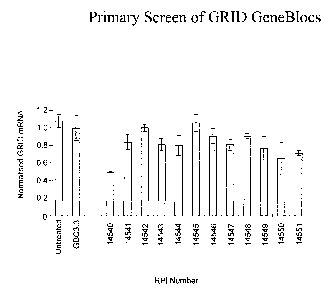
Figure 7 shows a bar graph of a primary screen of twelve GRID GeneBlocs.
Taqman
mRNA assay was used to quantify the level of GRID transcript in Jurkat cells
treated
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
19
continuously for 24 hours with 100nM antisense GeneBloc and S.O~.gml-'
cationic lipid.
For comparison, all data was normalized to the level of (3-actin. Error bars
represent the
standard error of the mean of triplicate points.
Figure 8 shows a graph demonstrating that flow cytometric sorting of
transfected
cells improves antisense GeneBloc mediated inhibition of GRID mRNA expression.
Jurlcat
cells were treated continuously for 24 and 72 hours with GB 14540 (75nM) or
control
GeneBloc GBC3.3 (75nM) spiked with 25nM fluorescent randomized GeneBloc (A) to
facilitate the identiftcation of transfected cells. After transfection, the
10% most and least
fluorescent cells (gates M2 and M1 respectively) were sorted on a FACStar
Plus. Post-sort
low transfecting (B) and high transfecting (C) fractions were re-analyzed fox
purity.
Histograms A-D are representative of results obtained in all experiments and
were taken
from cells treated for 72 hours. The GRID mRNA content of all samples was
quantified by
Taqman RNA assay and normalized to the (3-actin content. For the purposes of
inter-
experiment comparison, all GB 14540 values were also normalized to the
appropriate
, control GBC3.3 value. (D) Normalized GRID mRNA levels in pre-sort samples;
(E)
Normalized GRID mRNA levels in the post-sort low transfecting fraction; (F)
Normalized
GRID mRNA levels in the post-sort high transfecting fraction. Error bars
represent the
range of duplicate points.
Figure 9 shows a graph representing the phenotypic analysis of antisense
GeneBloc
treated Jurlcat cells following activation with anti-CD3 and anti-CD28 anti-
sera. Jurkat
cells were treated continuously for 72 hours with the anti-GRID reagent GB
14540 (A, C)
and the mismatch control reagent GB 17477 (B, D), activated for 22 hours (C,
D) and
stained for the surface activation marker CD69. Unactivated samples are shown
in (A, B).
Mechanism of action of Nucleic Acid Molecules of the Invention
Antisense: Antisense molecules can be modified or unmodified RNA, DNA, or
mixed polymer oligonucleotides which primarily function by specifically
binding to
matching sequences resulting in inhibition of peptide synthesis (Wu-Pong, Nov
1994,
BioPharrn, 20-33). The antisense oligonucleotide binds to target RNA by Watson
Criclc
base-pairing and blocks gene expression by preventing ribosomal translation of
the bound
sequences either by steric blocking or by activating RNase H enzyme. Antisense
molecules can also alter protein synthesis by interfering with RNA processing
or transport
from the nucleus into the cytoplasm (Mukhopadhyay & Roth, 1996, Crit. Rev. in
Oncogenesis 7, 151-190).
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
In addition, binding of single stranded DNA to RNA can result in nuclease
degradation of the heteroduplex (Wu-Pong, supra; Crooke, supr°a). To
date, the only
backbone modified DNA chemistry known to act as substrates for RNase H are
phosphorothioates, phosphorodithioates, and borontrifluoridates. Recently it
has been
5 reported that 2'-arabino and 2'-fluoro arabino- containing oligos can also
activate RNase H
activity.
A number of antisense molecules have been described that utilize novel
configurations of chemically modified nucleotides, secondary structure, and/or
RNase H
substrate domains (Woolf et al., International PCT Publication No. WO
98113526;
10 Thompson et al., International PCT Publication No. WO 99/54459; Hartmann et
al., USSN
60/101,174 which was filed on September 21, 1998) all of these are
incorporated by
reference herein in their entirety.
In addition, antisense deoxyoligoribonucleotides can be used to taxget RNA by
means of DNA-RNA interactions, thereby activating RNase H, which digests the
target
15 RNA in the duplex. Antisense DNA can be expressed endogenously in vivo via
the use of
a single stranded DNA intracellular expression vector or equivalents and
variations thereof.
Triplex Formint~~ Oli~onucleotides (TFO): Single stranded DNA can be designed
to
bind to genomic DNA in a sequence specific manner. TFOs are comprised of
pyrimidine-
rich oligonucleotides which bind DNA helices through Hoogsteen Base-pairing
(Wu-Pong,
20 supra). The resulting triple helix composed of the DNA sense,.DNA
antisense, and TFO
disrupts RNA synthesis by RNA polymerase. The TFO mechanism can result in gene
expression or cell death since binding may be irreversible (Mulchopadhyay &
Roth, supra).
2-SA Antisense Chimera: The 2-SA system is an interferon mediated mechanism
for
RNA degradation found in higher vertebrates (Mitra et al., 1996, Proc Nat Acad
Sci USA
93, 6780-6785). Two types of enzymes, 2-SA synthetase and RNase L, are
required for
RNA cleavage. The 2-SA synthetases require double stranded RNA to form 2'-5'
oligoadenylates (2-SA). 2-SA then acts as an allosteric effector for utilizing
RNase L
which has the ability to cleave single stranded RNA. The ability to form 2-SA
structures
with double stranded RNA makes this system particularly useful for inhibition
of viral
replication.
(2'-5') oligoadenylate structures can be covalently linked to antisense
molecules to
form chimeric oligonucleotides capable of RNA cleavage (Torrence, supra).
These
molecules putatively bind and activate a 2-SA dependent RNase, the
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
21
oligonucleotide/enzyme complex then binds to a target RNA molecule which can
then be
cleaved by the RNase enzyme.
Enzymatic Nucleic Acid: Several varieties of naturally occurring enzymatic
RNAs
are presently known. In addition, several in vitro selection (evolution)
strategies (Orgel,
1979, Proc. R. Soc. London, B 205, 435) have been used to evolve new nucleic
acid
catalysts capable of catalyzing cleavage and ligation of phosphodiester
linkages (Joyce,
1989, Gene, 82, 83-87; Beaudry et al., 1992, Scieface 257, 635-641; Joyce,
1992, Scientific
American 267, 90-97; Breaker et al., 1994, TIBTECH 12, 268; Bartel et
a1.,1993, Science
261:1411-1418; Szostak, 1993, TIBS 17, 89-93; Kumar et al., 1995, FASEB J., 9,
1183;
Breaker, 1996, Curr. Op. Biotech., 7, 442; Santoro et al., 1997, Proc. Natl.
Acad. Sci., 94,
4262; Tang et al., 1997, RNA 3, 914; Nakamaye & Eckstein, 1994, supra; Long &
Uhlenbeclc, 1994, supra; Ishizalca et al., 1995, supra; Vaish et al., 1997,
Biochemistry 36,
6495; all of these are incorporated by reference herein). Each can catalyze a
series of
reactions including the hydrolysis of phosphodiester bonds in tr°ans
(and thus can cleave
other RNA molecules) under physiological conditions.
Nucleic acid molecules of this invention can block to some extent GRID protein
expression and can be used to treat disease or diagnose disease associated
with levels of
GRID.
The enzymatic nature of an enzymatic nucleic acid has significant advantages,
such
as the concentration of enzymatic nucleic acid necessary to affect a
therapeutic treatment is
lower. This advantage reflects the ability of the enzymatic nucleic acid to
act
enzymatically. Thus, a single enzymatic nucleic acid molecule is able to
cleave many
molecules of target RNA. In addition, the enzymatic nucleic acid is a highly
specific
inhibitor, with the specificity of inhibition depending not only on the base-
pairing
mechanism of binding to the target RNA, but also on the mechanism of target
RNA
cleavage. Single mismatches, or base-substitutions, near the site of cleavage
can be chosen
to completely eliminate catalytic activity of an enzymatic nucleic acid
molecule.
Nucleic acid molecules having an endonuclease enzymatic activity are able to
repeatedly cleave other separate RNA molecules in a nucleotide base sequence-
specific
manner. Such enzymatic nucleic acid molecules can be targeted to virtually any
RNA
transcript and achieve efficient cleavage in vitro (Zaug et al., 324, Nature
429 1986 ;
Uhlenbeck, 1987 Nature 328, 596; Kim et al., 84 Proc. Natl. Acad. Sci. LISA
8788, 1987;
Dreyfus, 1988, Eitasteita Quart. J. Bio. Med., 6, 92; Iiaseloff and Gerlach,
334 Nature 585,
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
22
1988; Cech, 260 JAMA 3030, 1988; and Jefferies et al., 17 Nucleic Acids
Researcla 1371,
1989; Santoro et al., 1997 supra).
Because of their sequence specificity, traps-cleaving enzymatic nucleic acid
molecules show promise as therapeutic agents for human disease (iJsman &
McSwiggen,
1995 Atan. Rep. Med. Ghena. 30, 285-294; Christoffersen and Marr, 1995 J. Med.
Chena.
38, 2023-2037). Enzymatic nucleic acid molecules can be designed to cleave
specific RNA
targets within the background of cellular RNA. Such a cleavage event renders
the RNA
non-functional and abrogates protein expression from that RNA. In this manner,
synthesis
of a protein associated with a disease state can be selectively inhibited
(Warashina et al.,
1999, Chemistry and Biology, 6, 237-250).
The nucleic acid molecules of the instant invention are also referred to as
GeneBloc
reagents, which are essentially nucleic acid molecules (e.g., ribozymes,
antisense) capable
of down-regulating gene expression.
GeneBlocs are modified oligonucleotides, including ribozymes and modified
antisense oligonucleotides, that bind to and target specific mRNA molecules.
Because
GeneBlocs can be designed to target any specific mRNA, their potential
applications are
quite broad. Traditional antisense approaches have often relied heavily on the
use of
phosphorothioate modifications to enhance stability in biological samples,
leading to a
myriad of specificity problems stemming from non-specific protein binding and
general
cytotoxicity (Stein, 1995, Nature Medicine, 1, 1119). In contrast, GeneBlocs
contain a
number of modifications that confer nuclease resistance while making minimal
use of
phosphorothioate linkages, which reduces toxicity, increases binding affinity,
and
minimizes non-specific effects compared with traditional antisense
oligonucleotides.
Similar reagents have recently been utilized successfully in various cell
culture systems
(Vassar, et al., 1999, Science, 286, 735) and in vivo (Jarvis et al.,
manuscript in
preparation). In addition, novel cationic lipids can be utilized to enhance
cellular uptake in
the presence of serum. Since ribozymes and antisense oligonucleotides regulate
gene
expression at the RNA level, the ability to maintain a steady-state dose of
GeneBloc over
several days is important for target protein and phenotypic analysis. The
advances in
resistance to nuclease degradation and prolonged activity in vitro have
supported the use of
GeneBlocs in target validation applications.
Target sites
Targets for useful ribozymes and antisense nucleic acids can be determined as
disclosed in Draper et al., WO 93/23569; Sullivan et al., WO 93/23057;
Thompson et al.,
WO 94/02595; Draper et al., WO 95/04818; McSwiggen et al., US Patent No.
5,525,468.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
23
All of these publications are hereby incorporated by reference herein in their
totality.
Other examples include the following PCT applications, which concern
inactivation of
expression of disease-related genes: WO 95/23225, WO 95/13380, WO 94/02595,
all of
which are incorporated by reference herein. Rather than repeat the guidance
provided in
those documents here, specific examples of such methods are provided herein,
not limiting
to those in the art. Ribozymes and antisense to such targets are designed as
described in
those applications and synthesized to be tested in vitro and in vivo, as also
described. The
sequences of human GRID RNAs were screened for optimal enzymatic nucleic acid
and
antisense target sites using a computer-folding algorithm. Antisense,
hammerhead,
DNAzyme, NCH, amberzyme, zinzyme, or G-Cleaver. ribozyme binding/cleavage
sites
were identified. These sites are shown in Tables III to VIII and X (all
sequences are 5' to
3' in the tables; underlined regions can be any sequence or linker X as
previously defined
herein, the actual sequence is not relevant here). The nucleotide base
position is noted in
the Tables as that site to be cleaved by the designated type of enzymatic
nucleic acid
molecule. While human sequences can be screened and enzymatic nucleic acid
molecule
and/or antisense thereafter designed, as discussed in Stinchcomb et al., WO
95/23225,
mouse targeted ribozymes are also useful to test efficacy of action of the
enzymatic nucleic
acid molecule and/or antisense prior to testing in humans.
Antisense, hammerhead, DNAzyme, NCH, amberzyme, zinzyme or G-Cleaver
ribozyme binding/cleavage sites were identified. The nucleic acid molecules
were
individually analyzed by computer folding (Jaeger et al., 1989 Proc. Natl.
Acad. Sci. USA,
86, 7706) to assess whether the sequences fold into the appropriate secondary
structure.
Those nucleic acid molecules with unfavorable intramolecular interactions,
such as
between the binding arms and the catalytic core, were eliminated from
consideration.
Varying binding arm lengths can be chosen to optimize activity.
Antisense, hammerhead, DNAzyme, NCH, amberzyme, zinzyme or G-Cleaver
ribozyme binding/cleavage sites were identified and were designed to anneal to
various
sites in the RNA target. The binding arms are complementary to the target site
sequences
described above. The nucleic acid molecules were chemically synthesized. The
method of
synthesis used follows the procedure for normal DNA/RNA synthesis as described
below
and in Usman et al., 1987 J. Ana. Chem. Soc., 109, 7845; Scaringe et al., 1990
Nucleic
Acids Res., 18, 5433; Wincott et al., 1995 Nucleic Acids Res. 23, 2677-2684;
and
Caruthers et al., 1992, Methods in Enzymology 211,3-19.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
24
Synthesis of Nucleic acid Molecules
Synthesis of nucleic acids greater than 100 nucleotides in length is difficult
using
automated methods, and the therapeutic cost of such molecules is prohibitive.
In this
invention, small nucleic acid motifs ("small refers to nucleic acid motifs no
more than 100
nucleotides in length, preferably no more than 80 nucleotides in length, and
most
preferably no more than 50 nucleotides in length; e.g., antisense
oligonucleotides,
hammerhead or the NCH ribozymes) are preferably used for exogenous delivery.
The
simple structure of these molecules increases the ability of the nucleic acid
to invade
targeted regions of RNA structure. Exemplary molecules of the instant
invention are
chemically synthesized, and others can be similarly synthesized.
Oligonucleotides (e.g.; antisense GeneBlocs) are synthesized using protocols
known
in the art as described in Caruthers et al., 1992, Methods in Enzyfnology 211,
3-19,
Thompson et al., W ternational PCT Publication No. WO 99154459, Wincott et
al., 1995,
Nucleic Acids Res. 23, 2677-2684, Wincott et al., 1997, Methods Mol. Bio., 74,
59,
Brennan et al., 1998, Biotechnol Bioeng., 61, 33-45, and Brennan, US patent
No.
6,001,311. All of these references are incorporated herein by reference. The
synthesis of
oligonucleotides makes use of common nucleic acid protecting and coupling
groups, such
as dimethoxytrityl at the 5'-end, and phosphoramidites at the 3'-end. In a non-
limiting
example, small scale syntheses are conducted on a 394 Applied Biosystems, Inc.
synthesizer using a 0.2 pmol scale protocol with a 2.5 min coupling step for
2'-O-
methylated nucleotides and a 45 sec coupling step for 2'-deoxy nucleotides.
Table IT
outlines the amounts and the contact times of the reagents used in the
synthesis cycle.
Alternatively, syntheses at the 0.2 ~mol scale can be performed on a 96-well
plate
synthesizer, such as the instrument produced by Protogene (Palo Alto, CA) with
minimal
modification to the cycle. A 33-fold excess (60 pL of 0.11 M = 6.6 ~mol) of 2'-
O-methyl
phosphoramidite and a 105-fold excess of S-ethyl tetrazole (60 ~L of 0.25 M =
15 ~cmol)
can be used in each coupling cycle of 2'-O-methyl residues relative to polymer-
bound 5'-
hydroxyl. A 22-fold excess (40 ~,L of 0.11 M = 4.4 ~.mol) of deoxy
phosphoramidite and a
70-fold excess of S-ethyl tetrazole (40 ~L of 0.25 M = 10 ~.mol) can be used
in each
coupling cycle of deoxy residues relative to polymer-bound 5'-hydroxyl.
Average
coupling yields on the 394 Applied Biosystems, Inc. synthesizer, determined by
colorimetric quantitation of the trityl fractions, are typically 97.5-99%.
Other
oligonucleotide synthesis reagents for the 394 Applied Biosystems, Inc.
synthesizer
include; detritylation solution is 3% TCA in methylene chloride (ABI); capping
is
performed with 16% N methyl imidazole in THF (ABI) and 10% acetic
anhydride/10%
2,6-lutidine in THF (ABI); and oxidation solution is 16.9 mM I2, 49 mM
pyridine, 9%
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
water in THF (PERSEPTIVET~. Burdick & Jackson Synthesis Grade acetonitrile is
used
directly from the reagent bottle. S-Ethyltetrazole solution (0.25 M in
acetonitrile) is made
up from the solid obtained from American International Chemical, Inc.
Alternately, for the
introduction of phosphorothioate linkages, Beaucage reagent (3H-1,2-
Benzodithiol-3-one
5 1,1-dioxide, 0.05 M in acetonitrile) is used.
Deprotection of the antisense oligonucleotides is performed as follows: the
polymer-
bound trityl-on oligoribonucleotide is transferred to a 4 mL glass screw top
vial and
suspended in a solution of 40% aq. methylamine (1 mL) at 65 °C for 10
min. After cooling
to -20 °C, the supernatant is removed from the polymer support. The
support is washed
10 three times with 1.0 mL of EtOH:MeCN:H20/3:1:1, vortexed and the
supernatant is then
added to the first supernatant. The combined supernatants, containing the
oligoribonucleotide, axe dried to a white powder.
The method of synthesis used for normal RNA including certain enzymatic
nucleic
acid molecules follows the procedure as described in Usman et al., 1987, J.
Arn. Chern.
15 Soc., 109, 7845; Scaringe et al., 1990, Nucleic Acids Res., 18, 5433;
Wincott et al., 1995,
Nucleic Acids Res. 23, 2677-2684 and Wincott et al., 1997, Methods Mol. Bio.,
74, 59, and
makes use of common.nucleic acid protecting and coupling groups, such as
dimethoxytrityl
at the 5'-end, and phosphoramidites at the 3'-end. In a non-limiting example,
small scale
syntheses are conducted on a 394 Applied Biosystems, Inc. synthesizer using a
0.2 p,mol
20 scale protocol with a 7.5 min coupling step for alkylsilyl protected
nucleotides and a 2.5
min coupling step for 2'-O-methylated nucleotides. Table II outlines the
amounts and the
contact times of the reagents used in the synthesis cycle. Alternatively,
syntheses at the 0.2
~mol scale can be done on a 96-well plate synthesizer, such as the instrument
produced by
Protogene (Palo Alto, CA) with minimal modification to the cycle. A 33-fold
excess (60
25 ~L of 0.11 M = 6.6 ~mol) of 2'-O-methyl phosphoramidite and a 75-fold
excess of S-ethyl
tetrazole (60 ~,L of 0.25 M = 15 ~mol) can be used in each coupling cycle of
2'-O-methyl
residues relative to polymer-bound 5'-hydroxyl. A 66-fold excess (120 ~,L of
0.11 M =
13.2 ~,mol) of allcylsilyl (ribo) protected phosphoramidite and a 150-fold
excess of S-ethyl
tetrazole (120 ~.L of 0.25 M = 30 wmol) can be used in each coupling cycle of
ribo residues
relative to polymer-bound 5'-hydroxyl. Average coupling yields on the 394
Applied
Biosystems, Inc. synthesizer, determined by colorimetric quantitation of the
trityl fractions,
are typically 97.5-99%. Other oligonucleotide synthesis reagents for the 394
Applied
Biosystems, Inc. synthesizer include; detritylation solution is 3% TCA in
methylene
chloride (ABI); capping is performed with 16% N methyl imidazole in THF (ABI)
and
10% acetic anhydride/10% 2,6-lutidine in THF (ABI); oxidation solution is 16.9
mM I2,
49 mM pyridine, 9% water in THF (PERSEPTIVETM). Burdick & Jackson Synthesis
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
26
Grade acetonitrile is used directly from the reagent bottle. S-Ethyltetrazole
solution (0.25
M in acetonitrile) is made up from the solid obtained from American
International
Chemical, Inc. Alternately, for the introduction of phosphorothioate linkages,
Beaucage
reagent (3H-1,2-Benzodithiol-3-one 1,1-dioxide0.05 M in acetonitrile) is used.
Deprotection of the RNA is performed using either a two-pot or one-pot
protocol.
For the two-pot protocol, the polymer-bound trityl-on oligoribonucleotide is
transferred to
a 4 mL glass screw top vial and suspended in a solution of 40% aq. methylamine
(1 mL) at
65 °C for 10 min. After cooling to -20 °C, the supernatant is
removed from the polymer
support. The support is washed three times with 1.0 mL of EtOH:MeCN:H20/3:1:1,
vortexed and the supernatant is then added to the first supernatant. The
combined
supernatants, containing the oligoribonucleotide, are dried to a white powder.
The base
deprotected oligoribonucleotide is resuspended in anhydrous TEA/HF/NMP
solution (300
~L of a solution of 1.5 mL N-methylpyrrolidinone, 750 ~L TEA and 1 mL TEA~3HF
to
provide a 1.4 M HF concentration) and heated to 65 °C. After 1.5 h, the
oligomer is
quenched with 1.5 M NH4HC03.
Alternatively, for the one-pot protocol, the polymer-bound trityl-on
oligoribonucleotide is transferred to a 4 mL glass screw top vial and
suspended in a
solution of 33% ethanolic methylamine/DMSO: 1/1 (0.8 mL) at 65 °C for
15 min. The
vial is brought to r.t. TEA~3HF (0.1 mL) is added and the vial is heated at 65
°C for 15
min. The sample is cooled at -20 °C and then quenched with 1.5 M
NH4HC03.
For purification of the trityl-on oligomers, the quenched NH4HC03 solution is
loaded onto a C-18 containing cartridge that had been prewashed with
acetonitrile followed
by 50 mM TEAA. After washing the loaded cartridge with water, the RNA is
detritylated
with 0.5% TFA for 13 min. The cartridge is then washed again with water, salt
exchanged
with 1 M NaCI and washed with water again. The oligonucleotide is then eluted
with 30%
acetonitrile.
Inactive hammerhead ribozymes or binding attenuated control (BAC)
oligonucleotides) are synthesized by substituting a U for GS and a U for A14
(numbering
from Hertel, K. J., et al., 1992, Nucleic Acids Res_, 20, 3252). Similarly,
one or more
nucleotide substitutions can be introduced in other enzymatic nucleic acid
molecules to
inactivate the molecule and such molecules can serve as a negative control.
The average stepwise coupling yields are typically >98% (Wincott et al., 1995
Nucleic Acids Res. 23, 2677-2684). Those of ordinary skill in the art will
recognize that
the scale of synthesis can be adapted to be larger or smaller than the
examples described
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
27
above including but not limited to 96-well format, all that is important is
the ratio of
chemicals used in the reaction.
Alternatively, the nucleic acid molecules of the present invention can be
synthesized
separately and joined together post-synthetically, for example by Iigation
(Moore et al.,
1992, Science 256, 9923; Draper et al., International PCT publication No. WO
93/23569;
Shabarova et al., 1991, Nucleic Acids Research 19, 4247; Bellon et al., 1997,
Nucleosides
& Nucleotides, 16, 951; Bellon et al., 1997, Biocofzjugate Claena. 8, 204).
The nucleic acid molecules of the present invention are modified extensively
to
enhance stability by modification with nuclease resistant groups, for example,
2'-amino, 2'-
C-allyl, 2'-flouro, 2'-O-methyl, 2'-H (for a review see Usman and Cedergren,
1992, TIBS
17, 34; Usman et al., 1994, Nucleic Acids Syynp. See. 31, 163). Ribozymes are
purified by
gel electrophoresis using general methods or are purified by high pressure
liquid
chromatography (HPLC; See Wincott et al., supra, the totality of which is
hereby
incorporated herein by reference) and are re-suspended in water.
The sequences of the ribozymes and antisense constructs that are chemically
synthesized, useful in this study, are shown in Tables III to X. Those in the
art will
recognize that these sequences are representative only of many more such
sequences where
the enzymatic portion of the ribozyme (all but the binding arms) is altered to
affect activity.
The ribozyme and antisense construct sequences listed in Tables III to X can
be formed of
ribonucleotides or other nucleotides or non-nucleotides. Such ribozymes with
enzymatic
activity are equivalent to the ribozymes described specifically in the Tables.
Optimizing Activity of the nucleic acid molecule of the invention.
Chemically synthesizing nucleic acid molecules with modifications (base, sugar
and/or phosphate) that prevent their degradation by serum ribonucleases can
increase their
potency (see e.g., Eclcstein et al., International Publication No. WO
92/07065; Perrault et
al., 1990 Natuf°e 344, 565; Pieken et al., 1991, Science 253, 314;
Usman and Cedergren,
1992, Trends ih BioclZeyn. Sci. 17, 334; Usman et al., International
Publication No.
WO 93/15187; Rossi et al., International Publication No. WO 91/03162; Sproat,
US Patent
No. 5,334,711; and Burgin et al., supra; all of these describe various
chemical
modifications that can be made to the base, phosphate and/or sugar moieties of
the nucleic
acid molecules described herein). All these references are incorporated by
reference
herein. Modifications which enhance their efficacy in cells, and removal of
bases from
nucleic acid molecules to shorten oligonucleotide synthesis times and reduce
chemical
requirements are preferably desired.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
28
There are several examples in the art describing sugar, base and phosphate
modifications that can be introduced into nucleic acid molecules with
significant
enhancement in their nuclease stability and efficacy. For example,
oligonucleotides are
modified to enhance stability and/or enhance biological activity by
modification with
nuclease resistant groups, for example, 2'-amino, 2'-C-allyl, 2'-flouro, 2'-O-
methyl, 2'-H,
nucleotide base modifications (for a review see Usman and Cedergren, 1992,
TIBS. 17, 34;
Usman et al., 1994, Nucleic Acids Synap. Ser. 31, 163; Burgin et al., 1996,
Biochemistry ,
35, 14090). Sugar modifications of nucleic acid molecules have been
extensively
described in the art (see Eckstein et al., InteYnatiorzal Publication PCT No.
WO 92/07065;
Perrault et al. Nature, 1990, 344, 565-568; Pieken et al. Science, 1991, 253,
314-317;
Usman and Cedergren, Treads in Biochem. Sci. , 1992, 17, 334-339; Usman et al.
Internrational Publication PCT No. WO 93/15187; Sproat, US Patent No.
5,334,711 and
Beigelman et al., 1995, J. Biol. Claem., 270, 25702; Beigelman et al.,
International PCT
publication No. WO 97/26270; Beigelman et al., US Patent No. 5,716,824; Usman
et al.,
US patent No. 5,627,053; Woolf et al., International PCT Publication No. WO
98/13526;
Thompson et al., USSN 60/082,404 which was filed on April 20, 1998; Karpeislcy
et al.,
1998, Tet>"alaedr~on Lett., 39, 1131; Earnshaw and Gait, 1998,
Biopolyrner°s (Nucleic acid
Sciences), 48, 39-55; Verma and Eckstein, 1998, Arrnu. Rev. Biochern., 67, 99-
134; and
Burlina et al., 1997, Bioorg. Med. Claem., 5, 1999-2010; all of the references
are hereby
incorporated by reference herein in their totalities). Such publications
describe general
methods and strategies to determine the location of incorporation of sugar,
base and/or
phosphate modifications and the like into ribozymes without inhibiting
catalysis. In view
of such teachings, similar modifications can be used as described herein to
modify the
nucleic acid molecules of the instant invention.
While chemical modification of oligonucleotide internucleotide linkages with
phosphorothioate, phosphorothioate, and/or 5'-methylphosphonate linkages
improves
stability, too many of these modifications may cause some toxicity. Therefore,
when
designing nucleic acid molecules the amount of these internucleotide linkages
should be
minimized. The reduction in the concentration of these linkages should lower
toxicity
resulting in increased efficacy and higher specificity of these molecules.
Use of the nucleic acid-based molecules of the invention can lead to improved
treatment of the disease progression by affording the possibility of
combination therapies
(e.g., multiple antisense or enzymatic nucleic acid molecules targeted to
different genes,
nucleic acid molecules coupled with known small molecule inhibitors, or
intermittent
treatment with combinations of molecules (including different motifs) and/or
other
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
29
chemical or biological molecules). The treatment of patients with nucleic acid
molecules
can also include combinations of different types of nucleic acid molecules.
Therapeutic nucleic acid molecules (e.g., enzymatic nucleic acid molecules and
antisense nucleic acid molecules) delivered exogenously should preferably be
stable within
cells until translation of the target RNA has been inhibited long enough to
reduce the levels
of the undesirable protein. This period of time varies between hours to days
depending
upon the disease state. The nucleic acid molecules should be resistant to
nucleases in order
to function as effective intracellular therapeutic agents when delivered
exogenously.
Improvements in the chemical synthesis of nucleic acid molecules described in
the instant
invention and in the art (see, e.g., Wincott et al., 1995, Nucleic Acids Res.,
23:2677;
Carruthers, et al., 1992, Methods in Enzymology, 211:3-19, each incorporated
by reference
herein) have expanded the ability to modify nucleic acid molecules by
introducing
nucleotide modifications to enhance their nuclease stability as described
above.
In yet another preferred embodiment, nucleic acid catalysts having chemical
modifications which maintain or enhance enzymatic activity are provided. Such
nucleic
acid is also generally more resistant to nucleases than unmodified nucleic
acid. Thus, in a
cell and/or in vivo the activity may not be significantly lowered. As
exemplified herein
such ribozymes are useful in a cell and/or in vivo even if activity over all
is reduced 10 fold
(Burgin et al., 1996, Biochemistry, 35, 14090). Such ribozymes herein are said
to
"maintain" the enzymatic activity of an all RNA ribozyme.
In another aspect the nucleic acid molecules comprise a 5' and/or a 3'- cap
structure.
By "cap structure" is meant chemical modifications, which have been
incorporated
at either terminus of the oligonucleotide (see, for example, Wincott et al.,
WO 97/26270,
incorporated by reference herein). These terminal modifications protect the
nucleic acid
molecule from exonuclease degradation, and can help in delivery and/or
localization within
a cell. The cap can be present at the 5'-terminus (5'-cap) or at the 3'-
terminus (3'-cap) or
can be present on both termini. In non-limiting examples, the 5'-cap is
selected from the
group consisting of inverted abasic residue (moiety), 4',5'-methylene
nucleotide; 1-(beta-D-
erythrofuranosyl) nucleotide, 4'-thin nucleotide, carbocyclic nucleotide; 1,5-
anhydrohexitol
nucleotide; L-nucleotides; alpha-nucleotides; modified base nucleotide;
phosphorodithioate
linkage; threo-pentofuranosyl nucleotide; acyclic 3',4'-seco nucleotide;
acyclic 3,4-
dihydroxybutyl nucleotide; acyclic 3,5-dihydroxypentyl nucleotide, 3'-3'-
inverted
nucleotide moiety; 3'-3'-inverted abasic moiety; 3'-2'-inverted nucleotide
moiety; 3'-2'-
inverted abasic moiety; 1,4-butanediol phosphate; 3'-phosphoramidate;
hexylphosphate;
aminohexyl phosphate; 3'-phosphate; 3'-phosphorothioate; phosphorodithioate;
or bridging
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
or non-bridging methylphosphonate moiety (for more details see Wincott et al.,
International PCT publication No. WO 97/26270, incorporated by reference
herein).
Suitable 3'-caps include 4',5'-methylene nucleotide; 1-(beta-D-
erythrofuranosyl)
nucleotide; 4'-thio nucleotide, carbocyclic nucleotide; 5'-amino-alkyl
phosphate; 1,3-
5 diamino-2-propyl phosphate, 3-aminopropyl phosphate; 6-aminohexyl phosphate;
1,2-
aminododecyl phosphate; hydroxypropyl phosphate; 1,5-anhydrohexitol
nucleotide; L-
nucleotide; alpha-nucleotide; modified base nucleotide; phosphorodithioate;
tlaYeo-
pentofuranosyl nucleotide; acyclic 3',4'-seco nucleotide; 3,4-dihydroxybutyl
nucleotide;
3,5-dihydroxypentyl nucleotide, 5'-5'-inverted nucleotide moiety; 5'-5'-
inverted abasic
10 moiety; 5'-phosphoramidate; 5'-phosphorothioate; 1,4-butanediol phosphate;
5'-amino;
bridging and/or non-bridging 5'-phosphoramidate, phosphorothioate and/or
phosphorodithioate, bridging or non bridging methylphosphonate and 5'-mercapto
moieties
(for more details, see Beaucage and Iyer, 1993, Tetrahedron 49, 1925;
incorporated by
reference herein).
15 By the term "non-nucleotide" is meant any group or compound which can be
incorporated into a nucleic acid chain in the place of one or more nucleotide
units,
including either sugar and/or phosphate substitutions, and allows the
remaining bases to
exhibit their enzymatic activity. The group or compound is abasic in that it
does not
contain a commonly recognized nucleotide base, such as adenosine, guanine,
cytosine,
20 uracil or thymine.
An "alkyl" group refers to a saturated aliphatic hydrocarbon, including
straight-chain,
branched-chain, and cyclic alkyl groups. Preferably, the alkyl group has 1 to
12 carbons.
More preferably it is a lower alkyl of from 1 to 7 carbons, more preferably 1
to 4 carbons.
The alkyl group can be substituted or unsubstituted. When substituted the
substituted
25 groups) is preferably, hydroxyl, cyano, alkoxy, =O, =S, N02 or N(CH3)2,
amino, or SH.
The term also includes alkenyl groups which are unsaturated hydrocarbon groups
containing at least one carbon-carbon double bond, including straight-chain,
branched-
chain, and cyclic groups. Preferably, the alkenyl group has 1 to 12 carbons.
More
preferably it is a lower alkenyl of from 1 to 7 carbons, more preferably 1 to
4 carbons. The
30 allcenyl group can be substituted or unsubstituted. When substituted the
substituted
groups) is preferably, hydroxyl, cyano, alkoxy, =O, =S, N02, halogen, N(CH3)2,
amino,
or SH. The term "alkyl" also includes alkynyl groups which have an unsaturated
hydrocarbon group containing at least one carbon-carbon triple bond, including
straight-
chain, branched-chain, and cyclic groups. Preferably, the alkynyl group has 1
to 12
carbons. More preferably it is a lower allcynyl of from 1 to 7 carbons, more
preferably 1 to
4 carbons. The alkynyl group can be substituted or unsubstituted. When
substituted the
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
31
substituted groups) is preferably, hydroxyl, cyano, alkoxy, =O, =S, N02 or
N(CH3)2,
amino or SH.
Such alkyl groups can also include aryl, alkylaryl, carbocyclic aryl,
heterocyclic
aryl, amide and ester groups. An "aryl" group refers to an aromatic group
which has at
least one ring having a conjugated ~ electron system and includes carbocyclic
aryl,
heterocyclic aryl and biaryl groups, all of which can be optionally
substituted. The
preferred substituent(s) of aryl groups are halogen, trihalomethyl, hydroxyl,
SH, OH,
cyano, alkoxy, alkyl, alkenyl, alkynyl, and amino groups. An "allcylaryl"
group refers to an
alkyl group (as described above) covalently joined to an aryl group (as
described above).
Carbocyclic aryl groups are groups wherein the ring atoms on the aromatic ring
are all
carbon atoms. The carbon atoms are optionally substituted. Heterocyclic aryl
groups are
groups having from 1 to 3 heteroatoms as ring atoms in the aromatic ring and
the
remainder of the ring atoms are carbon atoms. Suitable heteroatoms include
oxygen,
sulfur, and nitrogen, and include furanyl, thienyl, pyridyl, pyrrolyl, N-lower
alkyl pyrrolo,
pyrimidyl, pyrazinyl, imidazolyl and the like, all optionally substituted. An
"amide" refers
to an -C(O)-NH-R, where R is either alkyl, aryl, alkylaryl or hydrogen. An
"ester" refers to
an -C(O)-OR', where R is either alkyl, aryl, allcylaryl or hydrogen.
By "nucleotide" is meant a heterocyclic nitrogenous base in N-glycosidic
linkage
with a phosphorylated sugar. Nucleotides are recognized in the art to include
natural bases
(standard), and modified bases well known in the art. Such bases are generally
located at
the 1' position of a nucleotide sugar moiety. Nucleotides generally comprise a
base, sugar
and a phosphate group. The nucleotides can be unmodified or modified at the
sugar,
phosphate and/or base moiety, (also referred to interchangeably as nucleotide
analogs,
modified nucleotides, non-natural nucleotides, non-standard nucleotides and
other; see for
example, Usman and McSwiggen, supra; Eckstein et al., International PCT
Publication No.
WO 92/07065; Usman et al., International PCT Publication No. WO 93/15187;
Uhlman &
Peyman, supra all are hereby incorporated by reference herein). There are
several
examples of modified nucleic acid bases known in the art as summarized by
Limbach et
al., 1994, Nucleic Acids Res. 22, 2183. Some'of the non-limiting examples of
chemically
modified and other natural nucleic acid bases that can be introduced into
nucleic acids
include, inosine, purine, pyridin-4-one, pyridin-2-one, phenyl, pseudouracil,
2, 4, 6-
trimethoxy benzene, 3-methyl uracil, dihydrouridine, naphthyl, aminophenyl,
5-alkylcytidines (e.g., 5-methylcytidine), 5-alkyluridines (e.g.,
ribothymidine),
5-halouridine (e.g., 5-bromouridine) or 6-azapyrimidines or 6-alkylpyrimidines
(e.g. 6-
methyluridine), propyne, quesosine, 2-thiouridine, 4-thiouridine, wybutosine,
wybutoxosine, 4-acetylcytidine, 5-(carboxyhydroxymethyl)uridine, 5'-
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
32
carboxymethylaminomethyl-2-thiouridine, 5-carboxymethylaminomethyluridine,
beta-D-
galactosylqueosine, 1-methyladenosine, 1-methylinosine, 2,2-dimethylguanosine,
3-
methylcytidine, 2-methyladenosine, 2-methylguanosine, N6-methyladenosine, 7-
methylguanosine, 5-methoxyaminomethyl-2-thiouridine, 5-
methylaminomethyluridine, 5-
methylcarbonylmethyluridine, 5-methyloxyuridine, 5-methyl-2-thiouridine, 2-
methylthio-
N6-isopentenyladenosine, beta-D-mannosylqueosine, uridine-5-oxyacetic acid, 2-
thiocytidine, threonine derivatives and others (Burgin et al., 1996,
Biochemistry, 35,
14090; Uhlman & Peyman, supra).
By "modified bases" in this aspect is meant nucleotide bases other than
adenine,
guanine, cytosine and uracil at 1' position or their equivalents; such bases
can be used at
any position, for example, within the catalytic core of an enzymatic nucleic
acid molecule
and/or in the substrate-binding regions of the nucleic acid molecule.
By "nucleoside" is meant a heterocyclic nitrogenous base in N-glycosidic
linkage
with a sugar. Nucleosides are recognized in the art to include natural bases
(standard), and
modified bases well known in the art. Such bases are generally located at the
1' position of
a nucleoside sugar moiety. Nucleosides generally comprise a base and sugar
group. The
nucleosides can be unmodified or modified at the sugar, and/or base moiety,
(also referred
to interchangeably as nucleoside analogs, modified nucleosides, non-natural
nucleosides,
non-standard nucleosides and other; see for example, Usman and McSwiggen,
supra;
Eclcstein et al., International PCT Publication No. WO 92/07065; Usman et al.,
International PCT Publication No. WO 93/15187; Uhlman & Peyman, supra all are
hereby
incorporated by reference herein). There are several examples of modified
nucleic acid
bases known in the art as summarized by Limbach et al., 1994, Nucleic Acids
Res. 22,
2183. Some of the non-limiting examples of chemically modified and other
natural nucleic
acid bases that can be introduced into nucleic acids include, inosine, purine,
pyridin-4-one,
pyridin-2-one, phenyl, pseudouracil, 2, 4, 6-trimethoxy benzene, 3-methyl
uracil,
dihydrouridine, naphthyl, aminophenyl, 5-alkylcytidines (e.g., 5-
methylcytidine),
5-allcyluridines (e.g., ribothymidine), 5-halouridine (e.g., 5-bromouridine)
or
6-azapyrimidines or 6-alkylpyrimidines (e.g. 6-methyluridine), propyne,
quesosine, 2-
thiouridine, 4-thiouridine, wybutosine, wybutoxosine, 4-acetylcytidine, 5-
(carboxyhydroxymethyl)uridine, 5'-carboxymethylaminomethyl-2-thiouridine, 5-
carboxymethylaminomethyluridine, beta-D-galactosylqueosine, 1-methyladenosine,
1-
methylinosine, 2,2-dimethylguanosine, 3-methylcytidine, 2-methyladenosine, 2-
methylguanosine, N6-methyladenosine, 7-methylguanosine, 5-methoxyaminomethyl-2-
thiouridine, 5-methylaminomethyluridine, 5-methylcarbonylmethyluridine, 5-
methyloxyuridine, 5-methyl-2-thiouridine, 2-methylthio-N6-
isopentenyladenosine, beta-D-
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
33
mannosylqueosine, uridine-5-oxyacetic acid, 2-thiocytidine, threonine
derivatives and
others (Burgin et al., 1996, Biochemistry, 35, 14090; Uhlman & Peyman, supra).
By "modified bases" in this aspect is meant nucleoside bases other than
adenine,
guanine, cytosine and uracil at 1' position or their equivalents; such bases
can be used at
any position, for example, within the catalytic core of an enzymatic nucleic
acid molecule
and/or in the substrate-binding regions of the nucleic acid molecule.
In a preferred embodiment, the invention features modified ribozymes with
phosphate backbone modifications comprising one or more phosphorothioate,
phosphorodithioate, methylphosphonate, morpholino, amidate carbamate,
carboxymethyl,
acetamidate, polyamide, sulfonate, sulfonamide, sulfamate, formacetal,
thioformacetal,
and/or alkylsilyl, substitutions. For a review of oligonucleotide backbone
modifications
see Hunziker and Leumann, 1995, Nucleic Acid Analogues: Synthesis and
PropeYties, in
Modey~n Synthetic Methods, VCH, 331-417, and Mesmaeker et al., 1994, Novel
Backbone
Replacements fos~ Oligohucleotides, in Carbohydrate Modificatiofas iya
Antisehse ReseaYCh,
ACS, 24-39. These references are hereby incorporated by reference herein.
By "abasic" is meant sugar moieties lacking a base or having other chemical
groups
in place of a base at the 1' position, (for more details, see Wincott et al.,
International PCT
publication No. WO 97/26270).
By "unmodified nucleoside" is meant one of the bases adenine, cytosine,
guanine,
thymine, uracil joined to the 1' carbon of (3-D-ribo-furanose.
By "modified nucleoside" is meant any nucleotide base which contains a
modification in the chemical structure of an unmodified nucleotide base, sugar
and/or
phosphate.
In connection with 2'-modified nucleotides as described for the present
invention,
by "amino" is meant 2'-NHz or 2'-O- NH2, which can be modified or unmodified.
Such
modified groups are described, for example, in Eckstein et al., U.S. Patent
5,672,695 and
Matulic-Adamic et al., WO 98/28317, respectively, which are both incorporated
by
reference herein in their entireties.
Various modifications to nucleic acid (e.g., antisense and ribozyme) structure
can be
made to enhance the utility of these molecules. For example, modifications can
enhance
shelf life, half life in vita°o, stability, and ease of introduction of
such oligonucleotides to
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
34
the target site, e.g., to enhance penetration of cellular membranes, and
confer the ability to
recognize and bind to targeted cells.
Use of these molecules can lead to better treatment of the disease progression
by
affording the possibility of combination therapies (e.g., multiple ribozymes
targeted to
different genes, ribozymes coupled with known small molecule inhibitors, or
intermittent
treatment with combinations of ribozymes (including different ribozyme motifs)
andlor
other chemical or biological molecules). The treatment of patients with
nucleic acid
molecules can also include combinations of different types of nucleic acid
molecules.
Therapies can be devised which include a mixture of ribozymes (including
different
ribozyrne motifs), antisense and/or 2-SA chimera molecules to one or more
targets to
alleviate symptoms of a disease.
Administration of Nucleic Acid Molecules
Methods for the delivery of nucleic acid molecules are described in Akhtar et
al.,
1992, Treyads Gell Bio., 2, 139; and Delivery Strategies for Ahtisefase
Oligonucleotide
Tlaerapeutics, ed. Alchtar, 1995 which are both incorporated herein by
reference. Sullivan
et al., PCT WO 94/02595, further describes the general methods for delivery of
enzymatic
RNA molecules. These protocols can be utilized for the delivery of virtually
any nucleic
acid molecule. Nucleic acid molecules can may be administered to cells by a
variety of
methods known to those familiar to the art, including, but not restricted to,
encapsulation in
liposomes, by iontophoresis, or by incorporation into other vehicles, such as
hydrogels,
cyclodextrins, biodegradable nanocapsules, and bioadhesive microspheres. For
some
indications, nucleic acid molecules can be directly delivered ex vivo to cells
or tissues with
or without the aforementioned vehicles. Alternatively, the nucleic
acid/vehicle
combination can be locally delivered by direct injection or by use of a
catheter, infusion
pump or stmt. Other routes of delivery include, but are not limited to,
intravascular,
intramuscular, subcutaneous or joint injection, aerosol inhalation, oral
(tablet or pill form),
topical, systemic, ocular, intraperitoneal and/or intrathecal delivery. More
detailed
descriptions of nucleic acid delivery and administration are provided in
Sullivan et al.,
supra, Draper et al., PCT W093/23569, Beigelman et al., PCT W099/05094, and
Klimuk
et al., PCT W099/04819 all of which have been incorporated by reference
herein.
The molecules of the instant invention can be used as pharmaceutical agents.
Pharmaceutical agents prevent, inhibit the occurrence, or treat (i.e.,
alleviate a symptom to
some extent, preferably all of the symptoms) of a disease state in a patient.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
The negatively charged polynucleotides of the invention can be administered
(e.g.,
RNA, DNA or protein) and introduced into a patient by any standard means, with
or
without stabilizers, buffers, and the like, to form a pharmaceutical
composition. When it is
desired to use a liposome delivery mechanism, standard protocols for formation
of
5 liposomes can be followed as described in the art. The compositions of the
present
invention can also be formulated and used as tablets, capsules or elixirs for
oral
administration; suppositories for rectal administration; sterile solutions;
suspensions for
injectable administration; and other compositions known in the art.
The present invention also includes pharmaceutically acceptable formulations
of the
10 compounds described. These formulations include salts of the above
compounds, e.g., acid
addition salts, including salts of hydrochloric, hydrobromic, acetic acid, and
benzene
sulfonic acid.
A pharmacological composition or formulation refers to a composition or
formulation in a form suitable for administration, e.g., systemic
administration, into a cell
15 ox patient, preferably a human. Suitable forms, in part, depend upon the
use or the route of
entry, for example oral, transdermal, or by injection. Such forms should not
prevent the
composition or formulation from reaching a target cell (i.e., a cell to which
the negatively
charged polymer is desired to be delivered to). For example, pharmacological
compositions injected into the blood stream should be soluble. Other factors
are known in
20 the art, and include considerations such as toxicity and forms which
prevent the
composition or formulation from exerting its effect.
By "systemic administration" is meant in vivo systemic absorption or
accumulation
of drugs in the blood stream followed by distribution throughout the entire
body.
Administration routes that lead to systemic absorption include, without
limitations:
25 intravenous, subcutaneous, intraperitoneal, inhalation, oral,
intrapulmonary and
intramuscular. Each of these administration routes exposes the desired
negatively charged
polymers, e.g., nucleic acids, to an accessible diseased tissue. The rate of
entry of a drug
into the circulation has been shown to be a function of molecular weight or
size. The use
of a liposome or other drug carrier comprising the compounds of the instant
invention can
30 potentially localize the drug, for example, in certain tissue types, such
as the tissues of the
reticular endothelial system (RES). A liposome formulation that can facilitate
the
association of drug with the surface of cells, such as, lymphocytes and
macrophages is also
useful. This approach can provide enhanced delivery of the drug to target
cells by taking
advantage of the specificity of macrophage and lymphocyte immune recognition
of
35 abnormal cells, such as cancer cells.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
36
By pharmaceutically acceptable formulation is meant, a composition or
formulation
that allows for the effective distribution of the nucleic acid molecules of
the instant
invention in the physical location most suitable for their desired activity.
Non-limiting
examples of agents suitable for formulation with the nucleic acid molecules of
the instant
invention include: P-glycoprotein inhibitors (such as Pluronic P85) which can
enhance
entry of drugs into the CNS (Jolliet-Riant and Tillement, 1999, Fundam. Clin.
Pharmacol.,
13, 16-26); biodegradable polymers, such as poly (DL-lactide-coglycolide)
microspheres
for sustained release delivery after intracerebral implantation (Emerich, DF
et al, 1999,
Cell Transplant, 8, 47-58) Alkermes, Inc. Cambridge, MA; and loaded
nanoparticles, such
as those made of polybutylcyanoacrylate, which can deliver drugs across the
blood brain
barrier and can alter neuronal uptake mechanisms (Pj°og
Neuropsychopharnzacol Biol
Psyclaiat~-y, 23, 941-949, 1999). Other non-limiting examples of delivery
strategies for the
nucleic acid molecules of the instant invention include material described in
Boado et al.,
1998, J. PharnZ. Sci., 87, 1308-1315; Tyler et al., 1999, FEBS Lett., 421, 280-
284;
Pardridge et al., 1995, PNAS USA., 92, 5592-5596; Boado, 1995, Adv.
Dr~ugDelivery Rev.,
15, 73-107; Aldrian-Herrada et al., 1998, Nucleic Acids Res., 26, 4910-4916;
and Tyler et
al., 1999, PNAS USA., 96, 7053-7058.
The invention also features the use of the composition comprising surface-
modified
liposomes containing poly (ethylene glycol) lipids (PEG-modified, or long-
circulating
Iiposomes or stealth liposomes). These formulations offer a method for
increasing the
accumulation of drugs in target tissues. This class of drug carriers resists
opsonization and
elimination by the mononuclear phagocytic system (MPS or RES), thereby
enabling longer
blood circulation times and enhanced tissue exposure for the encapsulated drug
(Lasic et
al. Chern. Rev. 1995, 95, 2601-2627; Ishiwata et al., Cl2em. Pharm. Bull.
1995, 43, 1005-
1011). All incorporated by reference herein. Such liposomes have been shown to
accumulate selectively in tumors, presumably by extravasation and capture in
the
neovascularized target tissues (Lasic et al., Scierace 1995, 267, 1275-1276;
Oku et a1.,1995,
Biochim. Biophys. Acta, 1238, 86-90). All incorporated by reference herein.
The long-
circulating liposomes enhance the pharmacokinetics and pharmacodynamics of DNA
and
RNA, particularly compared to conventional cationic liposomes which are lrnown
to
accumulate in tissues of the MPS (Liu et al., J. Biol. Claem. 1995, 42, 24864-
24870; Choi
et al., International PCT Publication No. WO 96110391; Ansell et al.,
International PCT
Publication No. WO 96/10390; Holland et al., International PCT Publication No.
WO
96/10392; all of which are incorporated by reference herein). Long-circulating
liposomes
are also likely to protect drugs from nuclease degradation to a greater extent
compared to
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
37
cationic liposomes, based on their ability to avoid accumulation in
metabolically
aggressive MPS tissues such as the liver and spleen.
The present invention also includes compositions prepared for storage or
administration which include a pharmaceutically effective amount of the
desired
compounds in a pharmaceutically acceptable carrier or diluent. Acceptable
carriers or
diluents for therapeutic use are well known in the pharmaceutical art, and are
described, for
example, in Renzington's Pharmaceutical Sciences, Mack Publishing Co. (A.R.
Gennaro
edit. 1985) hereby incorporated by reference herein. For example,
preservatives,
stabilizers, dyes and flavoring agents may be provided. These include sodium
benzoate,
sorbic acid and esters ofp-hydroxybenzoic acid. In addition, antioxidants and
suspending
agents can be used.
A pharmaceutically effective dose is that dose required to prevent, inhibit
the
occurrence, or treat (alleviate a symptom to some extent, preferably all of
the symptoms) of
a disease state. The pharmaceutically effective dose depends on the type of
disease, the
composition used, the route of administration, the type of mammal being
treated, the
physical characteristics of the specific mammal under consideration,
concurrent
medication, and other factors which those skilled in the medical arts will
recognize.
Generally, an amount between 0.1 mglkg and 100 mg/kg body weight/day of active
ingredients is administered dependent upon potency of the negatively charged
polymer.
The nucleic acid molecules of the present invention can also be administered
to a
patient in combination with other therapeutic compounds to increase the
overall therapeutic
effect. The use of multiple compounds to treat an indication may increase the
beneficial
effects while reducing the presence of side effects.
Alternatively, certain of the nucleic acid molecules of the instant invention
can be
expressed within cells from eukaryotic promoters (e.g., Izant and Weintraub,
1985,
Science, 229, 345; McGarry and Lindquist, 1986, Proc. Natl. Acad. Sci., USA
83, 399;
Scanlon et al., 1991, Ps°oc. Natl. Acad. Sci. USA, 88, 10591-5; Kashani-
Sabet et al., 1992,
Antisense Res. Dev., 2, 3-15; Dropulic et al., 1992, J. Tirol., 66, 1432-41;
Weerasinghe et
al., 1991, J. ViYOI., 65, 5531-4; Ojwang et al., 1992, Pf~oc. Natl. Acad. Sci.
USA, '89,
10802-6; Chen et al., 1992, Nucleic Acids Res., 20, 4581-9; Sarver et al.,
1990 Science,
247, 1222-1225; Thompson et al., 1995, Nucleic Acids Res., 23, 2259; Good et
al., 1997,
Gene Therapy, 4, 45; all of the references are hereby incorporated in their
totality by
reference herein). Those skilled in the art realize that any nucleic acid can
be expressed in
eukaryotic cells from the appropriate DNA/RNA vector. The activity of such
nucleic acids
can be augmented by their release from the primary transcript by a ribozyme
(Draper et al.,
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
38
PCT WO 93/23569, and Sullivan et al., PCT WO 94/02595; Ohkawa et al., 1992,
Nucleic
Acids Syrnp. Ser., 27, 15-6; Taira et al., 1991, Nucleic Acids Res., 19, 5125-
30; Ventura et
al., 1993, Nucleic Acids Res., 21, 3249-55; Chowrira et al., 1994, J. Biol.
Chern., 269,
25856; all of these references are hereby incorporated in their totalities by
reference
herein).
In another aspect of the invention, RNA molecules of the present invention are
preferably expressed from transcription units (see, for example, Couture et
al., 1996, TIG.,
12, 510) inserted into DNA or RNA vectors. The recombinant vectors are
preferably DNA
plasmids or viral vectors. Ribozyme expressing viral vectors can be
constructed based on,
but not limited to, adeno-associated virus, retrovirus, adenovirus, or
alphavirus. Preferably,
the recombinant vectors capable of expressing the nucleic acid molecules are
delivered as
described above, and persist in target cells. Alternatively, viral vectors can
be used that
provide for transient expression of nucleic acid molecules. Such vectors can
be repeatedly
administered as necessary. Once expressed, the nucleic acid molecule binds to
the target
mRNA. Delivery of nucleic acid molecule expressing vectors can be systemic,
such as by
intravenous or infra-muscular administration, by administration to target
cells ex-planted
from the patient followed by reintroduction into the patient, or by any other
means that
allow for introduction into the desired target cell (for a review, see Couture
et al., 1996,
TIG., 12, 510).
In one aspect, the invention features an expression vector comprising a
nucleic acid
sequence encoding at least one of the nucleic acid molecules disclosed in the
instant
invention. The nucleic acid sequence encoding the nucleic acid molecule of the
instant
invention is operable linked in a manner Which allows expression of that
nucleic acid
molecule.
In another aspect, the invention features an expression vector comprising: a)
a
transcription initiation region (e.g., eukaryotic pol I, II or III initiation
region); b) a
transcription termination region (e.g., eukaryotic pol I, II or III
termination region); c) a
nucleic acid sequence encoding at least one of the nucleic acid catalyst of
the instant
invention; and wherein said sequence is operably linked to said initiation
region and said
termination region, in a manner which allows expression and/or delivery of
said nucleic
acid molecule. The vector can optionally include an open reading frame (ORF)
for a
protein operably linked on the 5' side or the 3'-side of the sequence encoding
the nucleic
acid catalyst of the invention; and/or an intron (intervening sequences).
Transcription of the nucleic acid molecule sequences are driven from a
promoter for
eukaryotic RNA polymerase I (pol I), RNA polymerase II (pol II), or RNA
polymerase III
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
39
(pol III), Transcripts from pol II or pol III promoters are expressed at high
levels in all
cells; the levels of a given pol II promoter in a given cell type depends on
the nature of the
gene regulatory sequences (enhancers, silencers, etc.) present nearby.
Prokaryotic RNA
polymerase promoters also can be used, providing that the prokaryotic RNA
polymerase
enzyme is expressed in the appropriate cells (Elroy-Stein and Moss, 1990,
Pnoc. Natl.
Acad. Sci. U S A, 87, 6743-7; Gao and Huang 1993, Nucleic Acids Res.., 21,
2867-72;
Lieber et al., 1993, Metlzods Erazyznol., 217, 47-66; Zhou et al., 1990, Mol.
Cell. Biol., 10,
4529-37). All of these references are incorporated by reference herein.
Several investigators have demonstrated that nucleic acid molecules, such as
ribozymes expressed from such promoters can function in mammalian cells (e.g.
Kashani-
Sabet et al., 1992, Antisense Res. Dev., 2, 3-15; Ojwang et al., 1992, Proc.
Natl. Acad.
Sci. U S A, 89, 10802-6; Chen et al., 1992, Nucleic Acids Res., 20, 4581-9; Yu
et al.,
1993, Pz°oc. Natl.. Acad. Sci. U S A, 90, 6340-4; L'Huillier et al.,
1992, EMBO J., 1 l,
4411-8; Lisziewicz et al., 1993, Proc. Natl. Acad. Sci. U. S. A, 90, 8000-4;
Thompson et
al., 1995, Nucleic Acids Res., 23, 2259; and Sullenger & Cech, 1993, Science,
262, 1566).
More specifically, transcription units such as the ones derived from genes
encoding U6
small nuclear (snRNA), transfer RNA (tRNA) and adenovirus VA RNA are useful in
generating high concentrations of desired RNA molecules such as ribozymes in
cells
(Thompson et al., sup>"a; Couture and Stinchcomb, 1996, supra; Noonberg et
al., 1994,
Nucleic Acid Res., 22, 2830; Noonberg et al., US Patent No. 5,624,803; Good et
al., 1997,
Gene Ther., 4, 45; and Beigelman et al., International PCT Publication No. WO
96/18736;
all of these publications are incorporated by reference herein. The above
ribozyme
transcription units can be incorporated into a variety of vectors for
introduction into
mammalian cells, including but not restricted to, plasmid DNA vectors, viral
DNA vectors
(such as adenovirus or adeno-associated virus vectors), or viral RNA vectors
(such as
retroviral or alphavirus vectors) (for a review, see Couture and Stinchcomb,
1996, supra).
In yet another aspect, the invention features an expression vector comprising
a
nucleic acid sequence encoding at least one of the nucleic acid molecules of
the invention,
in a manner which allows expression of that nucleic acid molecule. The
expression vector
comprises in one embodiment; a) a transcription initiation region; b) a
transcription
termination region; c) a nucleic acid sequence encoding at least one said
nucleic acid
molecule; and wherein said sequence is operably linked to said initiation
region and said
termination region, in a manner which allows expression and/or delivery of
said nucleic
acid molecule.
In another preferred embodiment, the expression vector comprises: a) a
transcription
initiation region; b) a transcription termination region; c) an open reading
frame; d) a
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
nucleic acid sequence encoding at least one said nucleic acid molecule,
wherein said
sequence is operably linked to the 3'-end of said open reading frame; and
wherein said
sequence is operably linked to said initiation region, said open reading frame
and said
termination region, in a manner which allows expression and/or delivery of
said nucleic
5 acid molecule.
In yet another embodiment the expression vector comprises: a) a transcription
initiation region; b) a transcription termination region; c) an intron; d) a
nucleic acid
sequence encoding at least one said nucleic acid molecule; and wherein said
sequence is
operably linked to said initiation region, said intron and said termination
region, in a
10 manner which allows expression and/or delivery of said nucleic acid
molecule.
In another embodiment, the expression vector comprises: a) a transcription
initiation
region; b) a transcription termination region; c) an ~intron; d) an open
reading frame; e) a
nucleic acid sequence encoding at least one said nucleic acid molecule,
wherein said
sequence is operably linked to the 3'-end of said open reading frame; and
wherein said
15 sequence is operably linked to said initiation region, said intron, said
open reading frame
and said termination region, in a manner which allows expression and/or
delivery of said
nucleic acid molecule.
Examples.
The following are non-limiting examples showing the selection, isolation,
synthesis
20 and activity of nucleic acids of the instant invention.
The following examples demonstrate the selection and design of Antisense,
hammerhead, DNAzyme, NCH, Amberzyme, Zinzyme, or G-Cleaver enzymatic nucleic
acid molecules and binding/cleavage sites within GRID RNA.
Nucleic acid inhibition of GRID target RNA
25 The use of GeneBlocs to modulate the activity of GRID, a putative component
of co-
stimulatory signaling in T cells, is herein described. An array of GeneBlocs
were designed
and screened for their ability to reduce GRID mRNA levels whilst leaving
transcripts from
the closely related genes Grb2 and GRAD unaffected. A series of experiments
were
conducted to optimize delivery of GeneBlocs to the Jurlcat T cell line. Using
these
30 conditions, applicant has demonstrated the efficacy of these reagents at
both the mRNA
and protein level. Anti-CD3/CD28 triggering of Jurkat cells pre-treated with
the anti-GRID
GeneBloc results in an impairment of CD69 up-regulation consistent with an
important
role for GRID in transducing the co-stimulatory signal.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
41
Example 1: Identification of Potential Targ-et Sites in Human GRID RNA
The sequence of human GRID were screened for accessible sites using a computer-
folding algorithm. Regions of the RNA were identified that do not form
secondary folding
structures. These regions contain potential ribozyme andlor antisense
binding/cleavage
sites. The sequences of these binding/cleavage sites are shown in Tables III-
X.
Example 2: Selection of Enzymatic Nucleic Acid Cleavage Sites in Human GRID
RNA
Enzymatic nucleic acid target sites are chosen by analyzing sequences of Human
GR>I7 (for example, GenBank accession numbers: AJ011736 and Y18051) and
prioritizing
the sites on the basis of folding. Enzymatic nucleic acids are designed that
bind each target
and are individually analyzed by computer folding (Christoffersen et al., 1994
J. Mol.
Struc. Tlaeochena, 311, 273; Jaeger et al., 1989, Proc. Natl. Acad. Sci. USA,
86, 7706) to
assess whether the enzymatic nucleic acid sequences fold into the appropriate
secondary
structure. Those enzymatic nucleic acids with unfavorable intramolecular
interactions
between the binding arms and the catalytic core are eliminated from
consideration. As
noted below, varying binding arm lengths can be chosen to optimize activity.
Generally, at
least 5 bases on each arm are able to bind to, or otherwise interact with, the
target RNA.
Example 3: Chemical Synthesis and Purification of Enzymatic nucleic acids and
Antisense
for Efficient Cleavage and/or blocking of GRID RNA
Enzymatic nucleic acids and antisense constructs are designed to anneal to
various
sites in the RNA message. The binding arms of the enzymatic nucleic acids are
complementary to the target site sequences described above, while the
antisense constructs
are fully complimentary to the target site sequences described above. The
enzymatic
nucleic acids and antisense constructs were chemically synthesized. The method
of
synthesis used followed the procedure for normal RNA or DNA synthesis as
described
above and in Usman et al., (1987 J. Arn. Chern. Soc., 109, 7845), Scaringe et
al., (1990
Nucleic Acids Res., 18, 5433) and Wincott et al., supra, and made use of
common nucleic
acid protecting and coupling groups, such as dimethoxytrityl at the 5'-end,
and
phosphoramidites at the 3'-end. The average stepwise coupling yields were
typically
>98%.
Enzymatic nucleic acids and antisense constructs also can be synthesized from
DNA
templates using bacteriophage T7 RNA polymerase (Milligan and Uhlenbeck, 1989,
Methods Enzyrnol. 180, 51). Enzymatic nucleic acid and antisense constructs
are purified
by gel electrophoresis using general methods or are purified by high pressure
liquid
chromatography (HPLC; see Wincott et al., supra; the totality of which is
hereby
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
42
incorporated herein by reference) and are resuspended in water. The sequences
of the
chemically synthesized enzymatic nucleic acid and antisense constructs used in
this study
are shown below in Table III-X.
Example 4: Enzymatic nucleic acid Cleavage of GRID RNA Target in vitro
Enzymatic nucleic acids targeted to the human GRID RNA are designed and
synthesized as described above. These enzymatic nucleic acids can be tested
for cleavage
activity in vitro, for example, using the following procedure. The target
sequences and the
nucleotide location within the GRID RNA are given in Tables III-X.
Cleavage Reactions: Full-length or partially full-length, internally-labeled
target
RNA for enzymatic nucleic acid cleavage assay is prepared by ita vitro
transcription in the
presence of [a-32p] CTP, passed over a G 50 Sephadex~ column by spin
chromatography
and used as substrate RNA without further purification. Alternately,
substrates are 5'-32p-
end labeled using T4 polynucleotide lcinase enzyme. Assays are performed by
pre-
wanning a 2X concentration of purified enzymatic nucleic acid in enzymatic
nucleic acid
cleavage buffer (50 mM Tris-HCI, pH 7.5 at 37°C, 10 mM MgCl2) and the
cleavage
reaction was initiated by adding the 2X enzymatic nucleic acid mix to an equal
volume of
substrate RNA (maximum of 1-5 nM) that was also pre-warmed in cleavage buffer.
As an
initial screen, assays are carried out for 1 hour at 37°C using a final
concentration of either
40 nM or 1 mM ribozyme, i. e., enzymatic nucleic acid excess. The reaction is
quenched by
the addition of an equal volume of 95% formamide, 20 mM EDTA, 0.05%
bromophenol
blue and 0.05% xylene cyanol after which the sample is heated to 95°C
for 2 minutes,
quick chilled and loaded onto a denaturing polyacrylamide gel. Substrate RNA
and the
specific RNA cleavage products generated by enzymatic nucleic acid cleavage
are
visualized on an autoradiograph of the gel. The percentage of cleavage is
determined by
Phosphor Imager~ quantitation of bands representing the intact substrate and
the cleavage
products.
Example 5: Nucleic acid inhibition of GRID i~ vivo
Antisense nucleic acid molecules (GeneBlocs) targeted to the human GRID RNA
are
designed and synthesized as described above. These nucleic acid molecules can
be tested
for cleavage activity in vivo, for example, using the following procedure. The
target
sequences and the nucleotide location within the GRID RNA are given in Tables
III-X.
GRID shares 60.3% and 57.3% homology at the nucleotide level with the closely
related adapter proteins Grb2 and GRAP. In order to discriminate between human
GRID
and other Grb2 family members, twelve GeneBlocs (see Methods for details)
targeting
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
43
human GRID (GenBank accession number Y18051) were designed, each containing a
minimum of six mismatches versus human Grb2 (M96995) and human GRAD (U52518).
In order to determine the optimal site for GeneBloc binding and inhibition of
the target
mRNA, the efficacy of the GeneBlocs was tested on Jurkat cells. A Taqman RNA
assay
was used to quantify the level of GRID transcript in cells treated
continuously for 24hrs.
The efficacy of the twelve GeneBlocs, normalized to the levels of a house-
keeping gene ((3-
actin), is shown in Figure 7. The GeneBloc targeting site 152 (GeneBloc 14540)
was the
most efficacious, reducing GRID mRNA levels by up to 55% when compared with a
randomized control GeneBloc (GBC3.3). To confirm that these effects were
target specific,
a four base-pair mismatch GeneBloc (GB 17477) was synthesized. GRID mRNA
expression was unaffected in cells treated with the mismatch control GeneBloc
compared
to untreated cells.
Efficacy of the anti-GRID GeneBloc (GB 14540) in Jurkat cells
From the primary screen (Figure 7), the optimal GeneBloc, GB 14540, suppressed
GRID mRNA levels by up to 55%. However, this represents the inhibition in a
bulk
population of cells, some of which are refractory to transfection (see Figure
6D-F). To
investigate the correlation between dose and efficacy, GB 14540 was spiked
with 25% fGB.
Based on mixture experiments with active GeneBlocs in other systems, it was
not expected
that the presence of the fluorescent GeneBloc would interfere With anti-GRID
activity of
GB 14540. Thus, the most highly fluorescent cells represent the population of
cells
transfected with the highest concentration of active GeneBloc ('high
transfecting'), whilst
the cells that appear to be refractory to transfection should contain a
significantly lower
concentration active GeneBloc ('low transfecting').
Following transfection of a GB14540:fGB mixture, the high transfecting cells
(Figure 8A, Gate M2, the 10% most fluorescent cells) and the low transfecting
cells
(Figure 8A, Gate Ml, the 10% least fluorescent cells) were purified by FACS
sorting. Re-
analysis of the sorted cell populations confirmed greater than 95% purity
(Figure 8B-C).
Taqman RNA analysis of the treated cells pre- and post-sort (Figure 8D-F)
shows that
although GB 14540 inhibition of GRID mRNA expression in an unsorted population
is
variable between experiments (0-30°l0, Figure 8D), the level of
inhibition is significantly
increased to 45-63% in the 'high transfecting' fraction (Figure 8F). In
contrast, GRID
mRNA levels in the 'low transfecting' fraction was similar to that of cells
treated with
control GBC3.3 (Figure 8E). These data suggest that the degree of GRID mRNA
inhibition is dependent on the dose of GeneBloc delivered to the cells.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
44
To identify the optimal time-point for inhibition of GRID mRNA levels, samples
were sorted as described above at 24 and 72 hours following continuous
transfection.
Analysis of pre- and post-sort samples at these time-points revealed that in
pre-sort
samples, inhibition of GRID transcript occurred within 24 hours and did not
significantly
increase throughout the time-course of the experiment (Figure 8D). In the
'high
transfecting' fractions, reduction of GRID transcript was ~45% at 24 hours and
increased
only fractionally at the 72 hour time-point (50-65%, Figure 8F). This suggests
that
GB 14540 reduced GRID mRNA levels rapidly following transfection and that
inhibition
was sustained in the continued presence of GB 14540.
Analysis of GRID protein levels in GB 14540 treated cells
To determine whether the reduction in GRID transcript levels was associated
with a
loss of GRID protein, the level of GRID protein in cells treated continuously
with active
GeneBloc reagent GB 14540 and the mismatch control GB 17477 was assessed. When
delivered continuously for 72 hours, GB 14540 caused a substantial reduction
in GRID
protein levels as determined by the intensity of the GRID specific band whilst
at earlier
time-points (24 and 48 hrs) no reduction in protein was observed. Cells
treated with the
mismatch control GB 17477 showed GRID levels comparable to the untreated
sample.
Cells treated continuously with GB 14540 for periods up to 144 hours showed no
further
reduction in GRID protein levels, suggesting that the effect of the GeneBloc
was maximal
and sustained from 72 hours onwards. Whilst the effects of the anti-GRID
GeneBloc on
mRNA levels are seen at 24 hours, the reduction in GRID protein is delayed a
further 48
hours indicating that GRID protein may have a relatively long half life.
The GeneBlocs were designed to target and discriminate GRID from the closely
related adapter proteins Grb2 and GRAP. GB 14540 contains 6 and 7 mismatches
respectively when aligned with the human Gxb2 and GRAD sequences. Due to the
presence
of these mismatches, GB 14540 was not expected to inhibit Grb2 mRNA
expression. The
Western blots used for the GRID assay were stripped and re-probed using an
anti-Grb2
antibody. No difference in Grb2 protein levels was observed between the
untreated sample
and cells treated with either GB 14540 or the mismatch control reagent GB
17477,
confirming that the GB 14540 was specific for GRID.
Phenotypic effects of the anti-GRID GeneBloc on T cell activation
GRID is a novel member of the Grb2 family of adapter proteins. A role for GRID
in
T cell signaling has been postulated due to its association with known T cell
signaling
proteins [Law, 1999 #3296][Asada, 1999 #3243][Liu, 1999 #3245] and more
recently the
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
T cell co-stimulatory receptor CD28 following activation by cross-linking
antibodies (Ellis
et al.). To further elucidate the role of GRID in T cell co-stimulatory
pathways, applicant
studied the expression of early surface activation marker CD69 (Jung et al.,
1988, Cellulaf°
Immuzzology, 117, 352, Lanier et al., 1988, .I. Exp. Med., 167, 1572)
following activation of
5 Jurkat cells treated with GB 14540 and GB 17477. Jurkat cells were activated
by cross-
linking anti-CD3 and anti-CD28 monoclonal antibodies using a sub-maximal
stimulus to
increase the sensitivity of the assay. In cells treated with the mismatch
control GeneBloc,
GB 17477, 5.7% stained CD69 positive following activation compared with 0.7%
CD69
positive in unactivated cells (Figure 9D vs. 9B). In cells treated with the
anti-GRID
10 reagent GB 14540, there was a marked reduction in the proportion of
activated cells, with
only 1.3% staining positive for CD69 (Figure 9C). Expression of CD69 in the
unactivated
sample remained unaltered at 0.6% (Figure 9A). As the activation stimulus was
increased,
the relative difference between the cells treated with GB 14540 and GB 17477
decreased
even though the proportion of cells staining positive for CD69 increased. This
can be
15 attributed to the combination of residual GRID protein and supra-maximal
activation
stimulus. The latter component is particularly relevant to T cell activation
since the
dependency on co-stimulation is reduced as the strength of the CD3 signal
increases
(Geppert and Lipslcy, 1988, J. Clin. Invest., 81, 1497, Geppert and Lipslcy,
1987, Journal
oflmmunology, 138, 1660).
20 Taken together, these data suggest that the phenotypic effects described
above can be
attributed to GRID and not the closely related adapter protein Grb2. The
inhibitory effects
of GB 14540 on CD69 expression support a role for GRID in T cell co-
stimulatory
signaling.
Example 6: Delivery of GeneBloc reagents to Jurkat cells
25 As in many mammalian cell culture systems (Marcusson et al., 1998, Nuc.
Acids,
Res. 26, 2016), a cationic lipid was found to be necessary to facilitate
cellular uptake of
oligonucleotide. In preliminary experiments using a fluoresceinated randomized
GeneBloc
as a marker for uptake, a lipid concentration of 2.5-5.0 q,gml-' was found to
be optimal.
Although some cells are readily transfected by the GeneBloc, a sub-population
of cells
30 remained refractory to transfection (see Gate M2 vs. M1 in Figures 6D-6F).
In order to
minimize the refractory population, the concentration of GeneBloc was varied
between 10-
200nM. Transfection frequencies of up to 75% (as determined by fraction of
cells in Gate
M2) were observed in the 50-100nM range of GeneBloc concentration. At lower
concentrations (10-25nM), the transfection frequency dropped off very steeply
whilst at
35 higher concentrations, no further enhancement of transfection was observed.
Cationic
lipids however are not essential for the use of oligonucleotides in vivo (see
McGraw et al.,
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
46
1997, A~zti-Cancer Ds°ug Design, 12, 315-326; Henry et al., 1997, Ahti-
Cafacer Drug
Design, 12, 409-420).
Example 7: Flow Cytometry
Cultures were harvested, washed once and re-suspended in PBS containing 2%
FCS.
Cells were stained with a human anti-CD69 PE-conjugated antibody (Caltag)
using an
IgG2a PE-conjugate as an isotype control (Becton Dickinson). Cells were
analyzed on a
Becton Dickinson FACScan using CellQuest software. Cells were sorted on the
basis of
fluorescence in the FL1 channel using a Becton Diclcinson FACStar Plus. In
order to
compare the efficiency of GeneBloc uptake using different transfection
conditions, a
coefficient of transfection was calculated by multiplying the proportion of
control
GeneBloc (as a fraction of total GeneBloc) and the transfection frequency.
Example 8: Protein Studies
Actively growing Jurkat cells (0.1-1.0 x 106) were harvested, washed once in
PBS
and re-suspended in 25p.1 PBS. Cells were lysed by the addition of an equal
volume of ice-
cold 2x RIPA buffer (2% NP40, 1.0% sodium deoxycholate, 0.2% SDS in PBS with
2x
protease and phosphatase inhibitors). Following a 30 minute incubation on ice,
cell debris
was removed by centrifugation and the supernatant denatured at 100°C
for 5 minutes
following the addition of an equal volume of 2x SDS protein sample buffer.
Prior to
separation by SDS-PAGE electrophoresis, protein content was normalized using a
Coomassie~ Plus-200 protein assay reagent (Pierce). For Western blotting, SDS-
PAGE
gels were transferred to PVDF membrane (Millipore). Antisera specific for GRID
(rabbit
polyclonal courtesy of Claire Ashman, GlaxoWellcome), p85 sub-unit of PI-3-
kinase (#06-
195, Upstate Biotechnology) and Grb2 (sc-255, Santa Cruz) were used as primary
antibodies with an anti-rabbit HRP conjugate as the secondary antibody. Bound
antibody
was visualized using the SuperSignal~ West Dura chemiluminescent reagent. For
re-
probing, chemiluminescent substrate and bound antibody were removed with TBST
(TBS
+ 0.5% Tween-20) and ImmunoPure~ IgG Elution Buffer (Pierce) respectively.
Example 9: Cell Culture
Human Jurlcat cell lines E6.1 and J6 were maintained at 37°C in 5% COZ
in flasks in
RPMI 1641 (+ 25mM HEPES) supplemented with 10% fetal calf serum and glutamine.
Cells were passaged at a density of 1 x 106 cells ml-'. GeneBlocs were
delivered to the
cells using a modified centrifugation-based transfection protocol (Verma et
al., 1998,
BioTeclaniques, 25, 46). Cells were grown to a density of 1 x 106 cells m1',
harvested by
centrifugation and re-suspended in fresh media at 0.75 x 106 cells ml-'.
GeneBloc at lOX
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
47
final concentration and cationic lipid (25p,gml-') at lOX anal concentration
were prepared
separately in RPMI media (no FCS or glutamine), mixed 1:1 and incubated at
37°C for 30
minutes. 1.6m1 aliquots of the cell suspension was dispensed into a 6-well
tissue-culture
treated plate and 0.4m1 of the GeneBloc:lipid mixture added drop-wise. The
GeneBloc:lipid solution was evenly distributed by gentle agitation. Following
centrifugation at 1000rpm for 60 minutes at room temperature, the 6-well
plates were
incubated for 24-72 hours at 37°C.
Example 10: Real-time quantitative PCR (Taqman)
Human GRID oligonucleotide Taqman probe 6FAM-(5'-
ACTCCAGTTTCCCAAATGGTTTCACGAA-3') (SEQ ID NO 2237) -TAMRA and
human actin Taqman probe JOE-(5'-TCGAGCACGGCATCGTCACCAA-3') (SEQ ID
NO 2238) -TAMRA were purchased from PE Applied Biosystems. GRID primers
(forward, 5'-AGGATATGTGCCCAAGAATTTCATA-3') (SEQ ID NO 2239) and
reverse, (5'-TGCCTGGTGTCGAGAGAGG-3') (SEQ ID NO 2240) and actin primers
(forward, 5'-GCATGGGTCAGAAGGATTCCTAT-3') (SEQ ID NO 2241) and reverse,
(5'-TGTAGAAGGTGTGGTGCCAGATT-3') (SEQ ID NO 2242) were purchased from
Life Technologies. The Taqman probes were labeled with a reporter dye (FAM or
JOE) at
the 5' termini and a quencher dye (TAMRA) at their 3' termini. A combination
RT-PCR
and Taqman PCR was performed for each sample in triplicate on an ABI PRISM
7700
Sequence Detection System using the following program: 48°C for 30
minutes, 95°C for
10 minutes and then 40 cycles of 95°C for 15 seconds and 60°C
fox 1 minute. The reaction
was performed in a total volume of 40p.1 with each tube containing 10U RNase
inhibitor
(Promega), 1.25U Amplitaq Gold (PE Biosystems), 100nM of the GRID and Actin
primers, 100nM GRID FAM Taqman probe, 100nM Actin JOE Taqman probe and 10U
MuLV reverse transcriptase. PCR Buffer (PE Biosystems #4304441) and dNTPs (PE
Biosystems #N808-0261) were added according to the manufacturer's guidelines.
A
standard curve was generated using serially diluted purified RNA (300, 100, 33
and 1 lng)
prepared from untreated Jurkat cells.
Example 11: RNA isolation
Total RNA was isolated from Jurkat J6 or Jurkat E6.1 cells using the 96-well
RNeasy
lcit (Qiagen) and a minor modification of their protocol. 901 of RLT buffer
was added to
each sample, followed by an equal volume of 70% ethanol. Samples were mixed
and
transferred to a RNeasy-96-plate. A vacuum was applied for 15-60sec until the
wells were
dry. 80,1 of lx DNase solution was added (40mM Tris-HCl pH 7.5, lOmM MgCl2,
lOmM
CaClz, lOmM NaCl, 1.2U/~,l RNase-free DNase I). Following incubation at room
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
48
temperature for 15 minutes, lml of Buffer RW1 was added and incubated for a
further 5
minutes. The buffer was removed by applying a vacuum. The wells were washed
once in
lml of RPE. A second lml aliquot of Buffer RPE was added and the RNeasy-96-
plate
centrifuged at 6000 rpm for 10 minutes. The RNA was eluted by the addition of
100m1 of
RNase-free water. Following incubation at room temperature for 1 minute, the
RNA was
recovered by centrifugation at 6000rpm for 4 minutes and stored at -
70°C.
Indications
Particular conditions and disease states that can be associated with GRID
expression
modulation include, but are not limited to. tissue/graft rejection and cancer,
such as
leukemia.
The present body of knowledge in GRID research indicates the need for methods
to
assay GRID activity and for compounds that can regulate GRID expression for
research,
diagnostic, and therapeutic use.
Radiation, chemotherapeutic treatments, and Cyclosporin are non-limiting
examples of
compounds and/or methods that can be combined with or used in conjunction with
the
nucleic acid molecules (e.g. ribozymes and antisense molecules) of the instant
invention.
Those slcilled in the art will recognize that other drug compounds and
therapies can be
similarly be readily combined with the nucleic acid molecules of the instant
invention (e.g.
ribozymes and antisense molecules) are hence within the scope of the instant
invention.
Diagnostic uses
The nucleic acid molecules of this invention (e.g., ribozyrnes) can be used as
diagnostic tools to examine genetic drift and mutations within diseased cells
or to detect
the presence of GRID RNA in a cell. The close relationship between ribozyme
activity and
the structure of the target RNA allows the detection of mutations in any
region of the
molecule which alters the base-pairing and three-dimensional structure of the
target RNA.
By using multiple ribozymes described in this invention, one can map
nucleotide changes
which are important to RNA structure and function in vitro, as well as in
cells and tissues.
Cleavage of target RNAs with ribozymes can be used to inhibit gene expression
and define
the role (essentially) of specified gene products in the progression of
disease. In this
manner, other genetic targets can be defined as important mediators of the
disease. These
experiments can lead to better treatment of the disease progression by
affording the
possibility of combinational therapies (e.g., multiple ribozymes targeted to
different genes,
ribozymes coupled with known small molecule inhibitors, or intermittent
treatment with
combinations of ribozymes and/or other chemical or biological molecules).
Other ira vitro
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
49
uses of ribozymes of this invention include detection of the presence of mRNAs
associated
with GRID-related condition. Such RNA is detected by determining the presence
of a
cleavage product after treatment with a ribozyme using standard methodology.
In a specific example, ribozymes which can cleave only wild-type or mutant
forms of
the target RNA are used for the assay. The first ribozyme is used to identify
wild-type
RNA present in the sample and the second ribozyme is used to identify mutant
RNA in the
sample. As reaction controls, synthetic substrates of both wild-type and
mutant RNA are
cleaved by both ribozymes to demonstrate the relative ribozyme efficiencies in
the
reactions and the absence of cleavage of the "non-targeted" RNA species. The
cleavage
products from the synthetic substrates also serve to generate size markers for
the analysis
of wild-type and mutant RNAs in the sample population. Thus, each analysis can
require
two ribozymes, two substrates and one unknown sample, which are combined into
six
reactions. The presence of cleavage products is determined using an RNAse
protection
assay so that full-length and cleavage fragments of each RNA can be analyzed
in one lane
of a polyacrylamide gel. It is not absolutely required to quantify the results
to gain insight
into the expression of mutant RNAs and putative risk of the desired phenotypic
changes in
target cells. The expression of mRNA whose protein product is implicated in
the
development of the phenotype (i.e., GRID) is adequate to establish rislc. If
probes of
comparable specific activity are used for both transcripts, then a qualitative
comparison of
RNA levels is adequate and will decrease the cost of the initial diagnosis.
Higher mutant
form to wild-type ratios are correlated with higher risk whether RNA levels
are compared
qualitatively or quantitatively.
Additional Uses
Potential usefulness of sequence-specific enzymatic nucleic acid molecules of
the
instant invention have many of the same applications for the study of RNA that
DNA
restriction endonucleases have for the study of DNA (Nathans et al., 1975
Aran. Rev.
Biochena. 44:273). For example, the pattern of restriction fragments can be
used to
establish sequence relationships between two related RNAs, and large RNAs can
be
specifically cleaved to fragments of a size more useful for study. The ability
to engineer
sequence specificity of the enzymatic nucleic acid molecule is ideal for
cleavage of RNAs
of unknown sequence. Applicant describes the use of nucleic acid molecules to
down-
regulate gene expression of target genes in bacterial, microbial, fungal,
viral, and
eulcaryotic systems including plant, or mammalian cells.
All patents and publications mentioned in the specification are indicative of
the levels
of skill of those slcilled in the art to which the invention pertains. All
references cited in
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
this disclosure are incorporated by reference to the same extent as if each
reference had
been incorporated by reference in its entirety individually.
One skilled in the art would readily appreciate that the present invention is
well
adapted to carry out the objects and obtain the ends and advantages mentioned,
as well as
5 those inherent therein. The methods and compositions described herein as
presently
representative of preferred embodiments are exemplary and are not intended as
limitations
on the scope of the invention. Changes therein and other uses which are
encompassed
within the spirit of the invention, are defined by the scope of the claims.
It will be readily apparent to one skilled in the art that varying
substitutions and
10 modifications can be made to the invention disclosed herein without
departing from the
scope and spirit of the invention. Thus, such additional embodiments are
within the scope
of the present invention and the following claims.
The invention illustratively described herein suitably may be practiced in the
absence
of any element or elements, limitation or limitations which is not
specifically disclosed
15 herein. Thus, for example, in each instance herein any of the terms
"comprising",
"consisting essentially of and "consisting of may be replaced with either of
the other two
terms. The terms and expressions which have been employed are used as terms of
description and not of limitation, and there is no intention that in the use
of such terms and
expressions of excluding any equivalents of the features shown and described
or portions
20 thereof, but it is recognized that various modifications are possible
within the scope of the
invention claimed. Thus, it should be understood that although the present
invention has
been specifically disclosed by preferred embodiments, optional features,
modification and
variation of the concepts herein disclosed may be resorted to by those skilled
in the art, and
that such modifications and variations are considered to be within the scope
of this
25 invention as defined by the description and the appended claims.
In addition, where features or aspects of the invention are described in terms
of
Markush groups or other grouping of alternatives, those skilled in the art
will recognize
that the invention is also thereby described in terms of any individual member
or subgroup
of members of the Marlcush group or other group.
30 Other embodiments are within the following claims.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
51
TABLE I
Characteristics of naturally occurring ribozymes
Group I Introns
~ Size: 150 to >1000 nucleotides.
~ Requires a U in the target sequence immediately 5' of the cleavage site.
~ Binds 4-6 nucleotides at the 5'-side of the cleavage site.
~ Reaction mechanism: attack by the 3'-OH of guanosine to generate
cleavage products with 3'-OH and 5'-guanosine.
~ Additional protein. cofactors required in some cases to help folding,and
maintenance of the active structure.
~ Over 300 known members of this class. Found as an intervening
sequence in TetrahynZesia therrnophila rRNA, fungal mitochondria,
chloroplasts, phage T4, blue-green algae, and others.
~ Major structural features largely established through phylogenetic
comparisons, mutagenesis, and biochemical studies [;ii].
~ Complete kinetic framework established for one ribozyme (ii ~x ~ ~~i~
~ Studies of ribozyme folding and substrate docking underway (~i ~~ixi,iX].
~ Chemical modification investigation of important residues well
established (~,Xi].
~ The small (4-6 nt) binding site may make this ribozyme too non-specific
for targeted RNA cleavage, however, the Tetrahymena group 1 intron
has been used to repair a "defective" beta-galactosidase message by the
ligation of new beta-galactosidase sequences onto the defective message
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
52
RNAse P RNA (M1 RNA)
~ Size: 290 to 400 nucleotides.
~ RNA portion of a ubiquitous ribonucleoprotein enzyme.
~ Cleaves tRNA precursors to form mature tRNA [Xiii]
~ Reaction mechanism: possible attack by M2+-OH to generate cleavage
products with 3'-OH and 5'-phosphate.
~ RNAse P is found throughout the prokaryotes and eukaryotes. The
RNA subunit has been sequenced from bacteria, yeast, rodents, and
primates.
~ Recruitment of endogenous RNAse P . for therapeutic applications is
possible through hybridization of an External Guide Sequence (EGS) to
the target RNA [xi ~x~]
~ Important phosphate and 2' OH contacts recently identified [X~ ~XVii~
Group II Introns
~ Size: >1000 nucleotides.
~ Trans cleavage of target RNAs recently demonstrated [X~ii ~XiX~.
~ Sequence requirements not fully determined.
~ Reaction mechanism: 2'-OH of an internal adenosine generates cleavage
products with 3'-OH and a "lariat" RNA containing a 3'-5' and a 2'-5'
br anch point.
~ Only natural ribozyme with demonstrated participation in DNA
cleavage [X ;xXi] in addition to RNA cleavage and ligation.
~ Major structural features largely established through phylogenetic
comparisons [Xxii].
~ Important 2' OH contacts beginning to be identified [XXiii]
~ Kinetic framework under development [XXiV]
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
53
Neurospora VS RNA
~ Size: 144 nucleotides.
~ Trans cleavage of hairpin target RNAs recently demonstrated [XX~]
~ Sequence requirements not fully determined.
~ Reaction mechanism: attack by 2'-OH 5' to the scissile bond to generate
cleavage products with 2',3'-cyclic phosphate and 5'-OH ends.
~ Binding sites and structural requirements not fully determined.
~ Only 1 known member of this class. Found in Neurospora VS RNA.
Hammerhead Ribozyme
(see text for references)
~ Size: ~13 to 40 nucleotides.
~ Requires the target sequence UH immediately 5' of the cleavage site.
~ Binds a variable number nucleotides on both sides of the cleavage site.
~ Reaction mechanism: attack by 2'-OH 5' to the scissile bond to genes ate
cleavage products with 2',3'-cyclic phosphate and 5'-OH ends.
~ 14 known members of this class. Found in a number of plant pathogens
(virusoids) that use RNA as the infectious agent.
~ Essenfiial structural features largely defined, including 2 crystal
structures [xxv ~XX~ii'
~ Minimal ligation activity demonstrated (for engineering through irc
vitro selection) [XX~iii]
~ Complete kinetic framework established for two or more ribozymes
~ Chemical modification investigation of important residues well
established [XXX~.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
54
Hairpin Ribozyme
~ Size: ~50 nucleotides.
~ Requires the target sequence GUC immediately 3' of the cleavage site.
~ Binds 4-6 nucleotides at the 5'-side of the cleavage site and a variable
number to the 3'-side of the cleavage site.
~ Reaction mechanism: attack by 2'-OH 5' to the scissile bond to generate
cleavage products with 2',3'-cyclic phosphate and 5'-OH ends.
~ 3 known members of this class. Found in three plant pathogen (satellite
RNAs of the tobacco ringspot vixus, arabis mosaic virus and chicory
yellow mottle virus) which uses RNA as the infectious agent.
~ Essential structural features largely defined (XXXi XXxii XXXiii XXXi~]
~ Ligation activity (in addition to cleavage activity) makes ribozyme
amenable to engineering through in vitro selection (XXX~~
~ Complete kinetic framework established for one ribozyme (XXX~i]
~ Chemical modification investigation of important residues begun
(xxxvii xxxviii,.
Hepatitis Delta Virus (HDV) Ribozyme
~ Size: ~60 nucleotides.
~ Trans cleavage of target RNAs demonstrated (XXXiX~.
~ Binding sites and structural requirements not fully determined,
although no sequences 5' of cleavage site are required. Folded
ribozyme contains a pseudoknot structure (xl~.
~ Reaction mechanism: attack by 2'-OH 5' to the scissile bond to generate
cleavage products with 2',3'-cyclic phosphate and 5'-OH ends.
~ Only 2 known members of this class. Found in human HDV.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
XiiCircular form of HDV isXlii active and shows increased nuclease
stability [xlill~
' . Michel, Francois; Westhof, Eric. Slippery substrates. Nat. Struct. Biol.
(1994),1(1), 5-
7.
ii . Lisacek, Frederique; Diaz, Yolande; Michel, Francois. Automatic
identification of
group I intron cores in genomic DNA sequences. J. Mol. Biol. (1994),
235(4),1206-17.
"' . Herschlag, Daniel; Cech, Thomas R.. Catalysis of RNA cleavage by the
Tetrahymena
thermophila ribozyme.1. Kinetic description of the reaction of an RNA
substrate
complementary to the active site. Biochemistry (1990), 29(44),10159-71.
'° . Herschlag, Daniel; Cech, Thomas R.. Catalysis of RNA cleavage by
the Tetrahymena
thermophila ribozyme. 2. Kinetic description of the reaction of an RNA
substrate that forms
a mismatch at the active site. Biochemistry (1990), 29(44),10172-80.
Knitt, Deborah S.; Herschlag, Daniel. pH Dependencies of the Tetrahymena
Ribozyme Reveal an Unconventional Origin of an Appaxent pKa. Biochemistry
(1996), 35(5),
1560-70.
°' . Bevilacqua, Philip C.; Sugimoto, Naoki; Turner, Douglas H.. A
mechanistic
framework for the second step of splicing catalyzed by the Tetrahymena
ribozyme.
Biochemistry (1996), 35(2), 648-58.
°" . Li, Yi; Bevilacqua, Philip C.; Mathews, David; Turner, Douglas H..
Thermodynamic
and activation parameters for binding of a pyrene-labeled substrate by the
Tetrahymena
ribozyme: docking is not diffusion-controlled and is driven by a favorable
entropy change.
Biochemistry (1995), 34(44),14394-9.
°"' . Banerjee, Aloke Raj; Turner, Douglas H.. The time dependence of
chemical
modification reveals slow steps in the foldW g of a group I ribozyme.
Biochemistry (1995),
34(19), 6504-12.
'x . Zarrinkar, Patxick P.; Williamson, James R.. The P9.1-P9.2 peripheral
extension helps
guide folding of the Tetrahymena ribozyme. Nucleic Acids Res. (1996), 24(5),
854-8.
X . Strobel, Scott A.; Cech, Thomas R.. Minor groove recognition of the
conserved
G.cntdot.U pair at the Tetrahymena ribozyme reaction site. Science
(Washington, D. C.)
(1995), 267(5198), 675-9.
X' . Strobel, Scott A.; Cech, Thomas R.. Exocyclic Amine of the Conserved
G.cntdot.U
Paix at the Cleavage Site of the Tetrahymena Ribozyme Contributes to 5'-Splice
Site Selection
and Transition State Stabilization. Biochemistry (1996), 35(4),1201-11.
x". Sullenger, Bruce A.; Cech, Thomas R.. Ribozyme-mediated repair of
defective
mRNA by targeted traps-splicing. Nature (London) (1994), 371(6498), 619-22.
X"'. Robertson, H.D.; Altman, S.; Smith, J.D. J. Biol. Chem., 247, 5243-5251
(1972).
X'°. Forster, Anthony C.; Altman, Sidney. External guide sequences for
an RNA enzyme.
Science (Washington, D. C.,1883-) (1990), 249(4970), 783-6.
x°. yuan, Y.; Hwang, E. S.; Altman, S. Targeted cleavage of mRNA by
human RNase P.
Proc. Natl. Acad. Sci. USA (1992) 89, 8006-10.
x°' . Harris, Michael E.; Pace, Norman R.. Identification of phosphates
involved in
catalysis by the ribozyme RNase P RNA. RNA (1995),1(2), 210-18.
X°" . Pan, Tao; Loria, Andrew; Zhong, Kun. Probing of tertiary
interactions in RNA: 2'-
hydroxyl-base contacts between the RNase P RNA and pxe-tRNA. Proc. Natl. Acad.
Sci. U. S.
A. (1995), 92(26),12510-14.
X°"' . Pyle, Anna Marie; Green, Justin B.. Building a Kinetic Framework
for Group II Intron
Ribozyme Activity: Quantitation of Interdomain Binding and Reaction Rate.
Biochemistry
(1994), 33(9), 2716-25.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
56
XtX . Michels, William J. Jr.; Pyle, Anna Marie. Conversion of a Group II
Intron into a New
Multiple-Turnover Ribozyme that Selectively Cleaves Oligonucleotides:
Elucidation of
Reaction Mechanism and Strucfure/Function Relationships. Biochemistry (1995),
34(9), 2965-
77.
xX . Zimmerly, Steven; Guo, Huatao; Eskes, Robert; Yang, Jian; Perlman, Philip
S.;
Lambowitz, Alan M.. A group II intron RNA is a catalytic component of a DNA
endonuclease involved in intron mobility. Cell (Cambridge, Mass.) (1995),
83(4), 529-38.
XXa . Griffin, Edmund A., Jr.; Qin, Zhifeng; Michels, Williams J., Jr.; Pyle,
Anna Marie.
Group II intron ribozymes that cleave DNA and RNA linkages with similar
efficiency, and
lack contacts with substrate 2'-hydroxyl groups. Chem. Biol. (1995), 2(11),
761-70.
xxsa . Michel, Francois; Ferat, Jean Luc. Structure and activities of group II
introns. Annu.
Rev. Biochem. (1995), 64, 435-61.
x%"' . Abramovitz, Dana L.; Friedman, Richard A.; Pyle, Anna Marie. Catalytic
role of 2'-
hydroxyl groups within a group II intron active site. Science (Washington, D.
C.) (1996),
271 (5254),1410-13.
Xxa . Daniels, Danette L.; Michels, William J., Jr.; Pyle, Anna Marie. Two
competing
pathways for self-splicing by group II introns: a quantitative analysis of in
vitro reaction
rates and products. J. Mol. Biol. (1996), 256(1), 31-49.
XX° . Guo, Hans C. T.; Collies, Richard A.. Efficient trans-cleavage of
a stem-loop RNA
substrate by a ribozyme derived from Neurospora VS RNA. EMBO J. (1995),14(2),
368-76.
xX°i , Scott, W.G., Finch, J.T., Aaron,K. The crystal structure of an
all RNA hammerhead
ribozyme:Aproposed mechanism for RNA catalytic cleavage. Cell, (1995), 81, 991-
1002.
Xx°" . McICay, Structure and function of the hammerhead ribozyme: an
unfinished story.
RNA, (1996), 2, 395-403.
Xx°"' . Long, D., Uhlenbeck, O., Hertel, K. Ligation with hammerhead
ribozymes. US Patent
No. 5,633,133.
xXiX . Hertel, K.J., Herschlag, D., Uhlenbeck, O. A kinetic and thermodynamic
framework
for the hammerhead ribozyme reaction. Biochemistry, (1994) 33, 3374-
3385.Beigelman, L., et
al., Chemical modifications of hammerhead ribozymes. J. Biol. Chem., (1995)
270, 25702-
25708.
xxx . Beigelman, L., et al., Chemical modifications of hammerhead ribozymes.
J. Biol.
Chem., (1995) 270, 25702-25708.
XxXt , Hampel, Arnold; Tritz, Richard; Hicks, Margaret; Cruz, Phillip.
'Hairpin' catalytic
RNA model: evidence for helixes and sequence requirement for substrate RNA.
Nucleic
Acids Res. {1990),18(2), 299-304.
xXxa , Chowrira, Bharat M.; Berzal-Herranz, Alfredo; Burke, John M.. Novel
guanosine
requirement for catalysis by the hairpin ribozyme. Nature (London) (1991),
354(6351), 320-2.
Xxx"' . Berzal-Herranz, Alfredo; Joseph, Simpson; Chowrira, Bharat M.;
Butcher, Samuel E.;
Burke, John M.. Essential nucleotide sequences and secondary structure
elements of the
hairpin ribozyme. EMBO J. (1993),12(6), 2567-73.
XxX» , Joseph, Simpson; Berzal-Herranz, Alfredo; Chowrira, Bharat M.; Butcher,
Samuel E..
Substrate selection rules for the hairpin ribozyme determined by in vitro
selection, mutation,
and analysis of mismatched substrates. Genes Dev. (1993), 7(1),130-8.
xxx° , Berzal-Herranz, Alfredo; Joseph, Simpson; Burke, John M.. In
vitro selection of
active hairpW ribozymes by sequential RNA-catalyzed cleavage and ligation
reactions.
Genes Dev. (1992), 6(1),129-34.
xxx~' . Hegg, Lisa A.; Fedor, Martha J.. Kinetics and Thermodynamics of
Intermolecular
Catalysis by Hairpin Ribozymes. Biochemistry (1995), 34(48),15813-28.
xXx°" . Grasby, Jane A.; Mersmann, Karin; Singh, Mohinder; Gait,
Michael J.. Purine
Functional Groups in Essential Residues of the Hairpin Ribozyme Required fox
Catalytic
Cleavage of RNA. Biochemistry (1995), 34(12), 4068-76.
xXX°'.' , Schmidt, Sabine; Beigelman, Leorud; Karpeisky, Alexander;
Usman, Nassim;
Sorensen, Ulrik S,; Gait, Michael J.. Base and sugar requirements for RNA
cleavage of
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
57
essential nucleoside residues in internal loop B of the hairpin ribozyme:
implications for
secondary structure. Nucleic Acids Res. (1996), 24(4), 573-81.
XxxiX . Perrotta, Anne T.; Been, Michael D.. Cleavage of oligoribonucleotides
by a ribozyme
derived from the hepatitis .delta. virus RNA sequence. Biochemistry (1992),
31(1),16-21.
%t . Perrotta, Anne T.; Been, Michael D.. A pseudoknot-like structure required
for
efficient self-cleavage of hepatitis delta virus RNA. Nature (London) (1991),
350(6317), 434-6.
xn
Xn~
x<<" . Puttaraju, M.; Perrotta, Arule T.; Been, Michael D.. A circular trans-
acting hepatitis
delta virus ribozyme. Nucleic Acids Res. (1993), 21(18), 4253-8.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
58
Table II;
A. 2.5 umnl Synthesis Cycle Agl 39d Instrument
Reagent EquivalentsAmount Wait Time* Wait Time* Wait Time*RNA
DNA 2'-O-methyl
Phosphoramidites6.5 163 NL 45 sec 2.5 min 7.5 min
S-Ethyl 23.8 238 NL 45 sec 2.5 min 7.5 min
Tetrazole
Acetic 100 233 NL 5 sec 5 sec 5 sec
Anhydride
N-Methyl 186 233 NL 5 sec 5 sec 5 sec
Imidazole
TCA 176 2.3 mL 21 sec 21 sec 21 sec
Iodine 11.2 1.7 mL 45 sec 45 sec 45 sec
Beaucage 12.9 645 NL 100 sec 300 sec 300 sec
AcetonitrileNA 6.67 NA NA NA
mL
B. 0.2 umol Synthesis Cycle ABI 394 Instrument
Reagent EquivalentsAmount Wait Time* Wait Time* Wait Time*RNA
DNA 2'-O-methyl
Phosphoramidites15 31 NL 45 sec 233 sec 465 sec
S-Ethyl 38.7 31 NL 45 sec 233 min 465 sec
Tetrazole
Acetic 655 124 NL 5 sec 5 sec 5 sec
Anhydride
N-Methyl 1245 124 IIL 5 sec 5 sec 5 sec
Imidazole
TCA 700 732 [~L 10 sec 10 sec 10 sec
Iodine 20.6 244 NL 15 sec 15 sec 15 sec
Beaucage 7.7 232 NL 100 sec 300 sec 300 sec
AcetonitrileNA 2.64 NA NA NA
mL
C. 0.2 umol Synthesis Cycle 96 well Instrument
Reagent Equivalents:DNAIAmount: DNA/2'-O-Wait Time* Wait Time*Wait Time*
2'-O-methyIIRibomethyl/Ribo DNA 2'-O Ribo
methyl
Phosphoramidites22/33/66 40/60/120 60 sec 180 sec 360sec
NL
S-Ethyl 70/105/21040/60/120 60 sec 180 min 360 sec
Tetrazole NL
Acetic 265/265/26550150/50 NL 10 sec 10 sec 10 sec
Anhydride
N-Methyl 5021502/50250/50/50 NL 10 sec 10 sec 10 sec
Imidazole
TCA 23S/475/475250/500/500 15 sec 15 sec 15 sec
NL
Iodine 6.8/6,.8/6.880/80/80 NL 30 sec 30 sec 30 sec
Beaucage 34/51/51 80/1201120 100 sec 200 sec 200 sec
AcetonitrileNA 1150/1150/1150NA NA NA
uL
Wait time does not include contact time during delivery.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
59
Table III: Human GRID Hammerhead Ribozyme and Substrate Sequence
Pos Substrate Seq Ribozyme Seq
ID ID
13 GGCACAGUU AAUGGAUC1 GAUCCAUUCUGAUGAGGCCGUUAGGCCGAA ACUGUGCC906
14 GCACAGUUA AUGGAUCU2 AGAUCCAUCUGAUGAGGCCGUUAGGCCGAA AACUGUGC907
21' UAAUGGAUC UGUAAACU3 AGUUUACACUGAUGAGGCCGUUAGGCCGAA AUCCAUUA908
25 GGAUCUGUA AACUUGCA4 UGCAAGW GCCGUUAGGCCGAA ACAGAUCC909
CUGAUGAG
30 UGUAAACUU GCACCCUC5 GAGGGUGCCUGAUGAGGCCGUUAGGCCGAA AGUUUACA910
38 UGCACCCUC UUUCAGAG6 CUCUGAAA GCCGUUAGGCCGAA AGGGUGCA911
CUGAUGAG
40 CACCCUCUU UCAGAGUG7 CACUCUGACUGAUGAGGCCGUUAGGCCGAA AGAGGGUG912
41' ACCCUCUUU CAGAGUGG8 CCACUCUGCUGAUGAGGCCGUUAGGCCGAA AAGAGGGU913
42 CCCUCUUUC AGAGUGGU9 ACCACUCUCUGAUGAGGCCGUUAGGCCGAA AAAGAGGG914
51 AGAGUGGUA CAUGGAAG10 CUUCCAUGCUGAUGAGGCCGUUAGGCCGAA ACCACUCU915
76 AAGUGGAUC CAUACUCU11 AGAGUAUGCUGAUGAGGCCGUUAGGCCGAA AUCCACUU916
80 GGAUCCAUA CUCUGAAA12 UUUCAGAGCUGAUGAGGCCGUUAGGCCGAA AUGGAUCC917
83 UCCAUACUC UGAAAUGC13 GCAUUUCACUGAUGAGGCCGUUAGGCCGAA AGUAUGGA918
95 AAUGCAGUA ACUCUGAU14 AUCAGAGUCUGAUGAGGCCGUUAGGCCGAA ACUGCAUU919
99 CAGUAACUC UGAUGCUUl5 AAGCAUCACUGAUGAGGCCGUUAGGCCGAA AGUUACUG920
107 CUGAUGCUU GAAUUUGU16 ACAAAUUCCUGAUGAGGCCGUUAGGCCGAA AGCAUCAG921
112 GCUUGAAUU UGUUCUCCl7 GGAGAACACUGAUGAGGCCGUUAGGCCGAA AUUCAAGC922
113 CUUGAAUUU GUUCUCCC18 GGGAGAACCUGAUGAGGCCGUUAGGCCGAA AAUUCAAG923
116 GAAUUUGUU CUCCCUUC19 GAAGGGAGCUGAUGAGGCCGUUAGGCCGAA ACAAAUUC924
117 AAUUUGUUC UCCCUUCU20 AGAAGGGACUGAUGAGGCCGUUAGGCCGAA AACAAAUU925
119 UUUGUUCUC CCUUCUUG2l CAAGAAGGCUGAUGAGGCCGUUAGGCCGAA AGAACAAA926
123 UUCUCCCUU CUUGCCAG22 CUGGCAAGCUGAUGAGGCCGUUAGGCCGAA AGGGAGAA927
124 UCUCCCUUC UUGCCAGA23 UCUGGCAA GCCGUUAGGCCGAA AAGGGAGA928
CUGAUGAG
126 UCCCUUCUU GCCAGAAA24 UUUCUGGCCUGAUGAGGCCGUUAGGCCGAA AGAAGGGA929
139 GAAAGGAUU CUAAUAAC25 GUUAUUAGCUGAUGAGGCCGUUAGGCCGAA AUCCUUUC930
140 AAAGGAUUC UAAUAACU26 AGUUAUUACUGAUGAGGCCGUUAGGCCGAA AAUCCUUU931
142 AGGAUUCUA AUAACUCG27 CGAGUUAUCUGAUGAGGCCGUUAGGCCGAA AGAAUCCU932
145 AUUCUAAUA ACUCGGUG28 CACCGAGUCUGAUGAGGCCGUUAGGCCGAA AUUAGAAU933
149 UAAUAACUC GGUGUCAA29 UUGACACCCUGAUGAGGCCGUUAGGCCGAA AGUUAUUA934
155 CUCGGUGUC AAAGCCAA30 UUGGCUUUCUGAUGAGGCCGUUAGGCCGAA ACACCGAG935
169 CAAGACAUA AACUCAAU31 AUUGAGUUCUGAUGAGGCCGUUAGGCCGAA AUGUCUUG936
174 CAUAAACUC AAUCUCUU32 AAGAGAUUCUGAUGAGGCCGUUAGGCCGAA AGUUUAUG937
178 AACUCAAUC UCUUCUCU33 AGAGAAGACUGAUGAGGCCGUUAGGCCGAA AUUGAGUU938
180 CUCAAUCUC UUCUCUUC34 GAAGAGAA GCCGUUAGGCCGAA AGAUUGAG939
CUGAUGAG
182 CAAUCUCUU CUCUUCCA35 UGGAAGAGCUGAUGAGGCCGUUAGGCCGAA AGAGAUUG940
183 AAUCUCUUC UCUUCCAA36 UUGGAAGACUGAUGAGGCCGUUAGGCCGAA AAGAGAUU941
185 UCUCUUCUC UUCCAAAA37 UUUUGGAA GCCGUUAGGCCGAA AGAAGAGA942
CUGAUGAG
187 UCUUCUCUU CCAAAAGC38 GCUUUUGGCUGAUGAGGCCGUUAGGCCGAA AGAGAAGA943
188 CUUCUCUUC CAAAAGCU39 AGCUUUUGCUGAUGAGGCCGUUAGGCCGAA AAGAGAAG944
197 CAAAAGCUU CACGUUAC40 GUAACGUGCUGAUGAGGCCGUUAGGCCGAA AGCUUUUG945
198 AAAAGCUUC ACGUUACA41 UGUAACGUCUGAUGAGGCCGUUAGGCCGAA AAGCUUUU946
203 CUUCACGUU ACAGCAUG42 CAUGCUGUCUGAUGAGGCCGUUAGGCCGAA ACGUGAAG947
204 UUCACGUUA CAGCAUGG43 CCAUGCUGCUGAUGAGGCCGUUAGGCCGAA AACGUGAA948
220 GAAGCUGUU GCCAAGUU44 AACUUGGCCUGAUGAGGCCGUUAGGCCGAA ACAGCUUC949
228 UGCCAAGUU UGAUUUCA45 UGAAAUCACUGAUGAGGCCGUUAGGCCGAA ACUUGGCA950
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
229 GCCAAGUUU GAUUUCAC46 GUGAAAUCCUGAUGAGGCCGUUAGGCCGAA AACWGGC951
233 AGUUUGAUU UCACUGCU47 AGCAGUGACUGAUGAGGCCGUUAGGCCGAA AUCAAACU952
234 GUUUGAUUU CACUGCUU48 AAGCAGUGCUGAUGAGGCCGUUAGGCCGAA AAUCAAAC953
235 WUGAUUUC ACUGCWC49 GAAGCAGUCUGAUGAGGCCGUUAGGCCGAA AAAUCAAA954
242 UCACUGCUU CAGGUGAG50 CUCACCUGCUGAUGAGGCCGUUAGGCCGAA AGCAGUGA955
243 CACUGCUUC AGGUGAGG51 CCUCACCUCUGAUGAGGCCGUUAGGCCGAA AAGCAGUG956
264 ACUGAGCUU UCACACUG52 CAGUGUGACUGAUGAGGCCGUUAGGCCGAA AGCUCAGU957
265 CUGAGCUUU CACACUGG53 CCAGUGUGCUGAUGAGGCCGUUAGGCCGAA AAGCUCAG958
266 UGAGCUUUC ACACUGGA54 UCCAGUGUCUGAUGAGGCCGUUAGGCCGAA AAAGCUCA959
280 GGAGAUGUU UUGAAGAU55 AUCUUCAA GCCGUUAGGCCGAA ACAUCUCC960
CUGAUGAG
281 GAGAUGUUU UGAAGAUU56 AAUCUUCACUGAUGAGGCCGUUAGGCCGAA AACAUCUC961
282 AGAUGUUUU GAAGAUUU57 AAAUCUUCCUGAUGAGGCCGUUAGGCCGAA AAACAUCU962
289 UUGAAGAUU UUAAGUAA58 UUACUUAA GCCGUUAGGCCGAA AUCUUCAA963
CUGAUGAG
290 UGAAGAUUU UAAGUAAC59 GUUACUUACUGAUGAGGCCGUUAGGCCGAA AAUCUUCA964
291 GAAGAUUUU AAGUAACC60 GGUUACUUCUGAUGAGGCCGUUAGGCCGAA AAAUCUUC965
292 AAGAUUUUA AGUAACCA61 UGGUUACUCUGAUGAGGCCGUUAGGCCGAA AAAAUCUU966
296 UUUUAAGUA ACCAAGAG62 CUCUUGGUCUGAUGAGGCCGUUAGGCCGAA ACUUAAAA967
312 GGAGUGGUU UAAGGCGG63 CCGCCUUACUGAUGAGGCCGUUAGGCCGAA ACCACUCC968
313 GAGUGGUUU AAGGCGGA64 UCCGCCUUCUGAUGAGGCCGUUAGGCCGAA AACCACUC969
314 AGUGGUUUA AGGCGGAG65 CUCCGCCUCUGAUGAGGCCGUUAGGCCGAA AAACCACU970
325 GCGGAGCUU GGGAGCCA66 UGGCUCCCCUGAUGAGGCCGUUAGGCCGAA AGCUCCGC971
342 GGAAGGAUP UGUGCCCA67 UGGGCACACUGAUGAGGCCGUUAGGCCGAA AUCCUUCC972
356 CCAAGAAUU UCAUAGAC68 GUCUAUGACUGAUGAGGCCGUUAGGCCGAA AUUCUUGG973
357 CAAGAAUUU CAUAGACA69 UGUCUAUGCUGAUGAGGCCGUUAGGCCGAA AAUUCUUG974
358 AAGAAUUUC AUAGACAU70 AUGUCUAUCUGAUGAGGCCGUUAGGCCGAA AAAUUCUU975
361 AAUUUCAUA GACAUCCA71 UGGAUGUCCUGAUGAGGCCGUUAGGCCGAA AUGAAAUU976
367 AUAGACAUC CAGUUUCC72 GGAAACUGCUGAUGAGGCCGUUAGGCCGAA AUGUCUAU977
372 CAUCCAGUU UCCCAAAU73 AUUUGGGACUGAUGAGGCCGUUAGGCCGAA ACUGGAUG978
373 AUCCAGUUU CCCAAAUG74 CAUUUGGGCUGAUGAGGCCGUUAGGCCGAA AACUGGAU979
374 UCCAGUUUC CCAAAUGG75 CCAUUUGGCUGAUGAGGCCGUUAGGCCGAA AAACUGGA980
384 CAAAUGGUU UCACGAAG76 CUUCGUGACUGAUGAGGCCGUUAGGCCGAA ACCAUUUG981
385 AAAUGGUUU CACGAAGG77 CCUUCGUGCUGAUGAGGCCGUUAGGCCGAA AACCAUUU982
386 AAUGGUUUC ACGAAGGC78 GCCUUCGUCUGAUGAGGCCGUUAGGCCGAA AAACCAUU983
397 GAAGGCCUC UCUCGACA79 UGUCGAGACUGAUGAGGCCGUUAGGCCGAA AGGCCUUC984
399 AGGCCUCUC UCGACACC80 GGUGUCGACUGAUGAGGCCGUUAGGCCGAA AGAGGCCU985
401 GCCUCUCUC GACACCAG81 CUGGUGUCCUGAUGAGGCCGUUAGGCCGAA AGAGAGGC986
420 AGAGAACUU ACUCAUGG82 CCAUGAGUCUGAUGAGGCCGUUAGGCCGAA AGUUCUCU987
421 GAGAACUUA CUCAUGGG83 CCCAUGAGCUGAUGAGGCCGUUAGGCCGAA AAGUUCUC988
424 AACUUACUC AUGGGCAA84 UUGCCCAUCUGAUGAGGCCGUUAGGCCGAA AGUAAGUU989
439 AAGGAGGUU GGCUUCUU85 AAGAAGCCCUGAUGAGGCCGUUAGGCCGAA ACCUCCUU990
444 GGUUGGCUU CUUCAUCA86 UGAUGAAGCUGAUGAGGCCGUUAGGCCGAA AGCCAACC991
445 GUUGGCUUC UUCAUCAU87 AUGAUGAA GCCGUUAGGCCGAA AAGCCAAC992
CUGAUGAG
447 UGGCUUCUU CAUCAUCC88 GGAUGAUGCUGAUGAGGCCGUUAGGCCGAA AGAAGCCA993
448 GGCUUCUUC AUCAUCCG89 CGGAUGAUCUGAUGAGGCCGUUAGGCCGAA AAGAAGCC994
451 UUCUUCAUC AUCCGGGC90 GCCCGGAUCUGAUGAGGCCGUUAGGCCGAA AUGAAGAA995
454 UUCAUCAUC CGGGCCAG91 CUGGCCCGCUGAUGAGGCCGUUAGGCCGAA AUGAUGAA996
471 CCAGAGCUC CCCAGGGG92 CCCCUGGGCUGAUGAGGCCGUUAGGCCGAA AGCUCUGG997
483 AGGGGACUU CUCCAUCU93 AGAUGGAGCUGAUGAGGCCGUUAGGCCGAA AGUCCCCU998
484 GGGGACUUC UCCAUCUC94 GAGAUGGACUGAUGAGGCCGUUAGGCCGAA AAGUCCCC999
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
61
486 GGACUUCUC CAUCUCUG95 CAGAGAUGCUGAUGAGGCCGUUAGGCCGAA AGAAGUCC1000
490 UUCUCCAUC UCUGUCAG96 CUGACAGACUGAUGAGGCCGUUAGGCCGAA AUGGAGAA1001
492 CUCCAUCUC UGUCAGGC97 GCCUGACACUGAUGAGGCCGUUAGGCCGAA AGAUGGAG1002
496 AUCUCUGUC AGGCAUGA98 UCAUGCCUCUGAUGAGGCCGUUAGGCCGAA ACAGAGAU1003
514 GAUGACGUU CAACACUU99 AAGUGUUGCUGAUGAGGCCGUUAGGCCGAA ACGUCAUC1004
Sl5 AUGACGUUC AACACUUC100 GAAGUGUUCUGAUGAGGCCGUUAGGCCGAA AACGUCAU1005
522 UCAACACUU CAAGGUCA101 UGACCUUGCUGAUGAGGCCGUUAGGCCGAA AGUGUUGA1006
523 CAACACUUC AAGGUCAU102 AUGACCUUCUGAUGAGGCCGUUAGGCCGAA AAGUGUUG1007
529 UUCAAGGUC AUGCGAGA103 UCUCGCAUCUGAUGAGGCCGUUAGGCCGAA ACCUUGAA1008
548 ACAAGGGUA AUUACUUU104 AAAGUAAUCUGAUGAGGCCGUUAGGCCGAA ACCCUUGU1009
551 AGGGUAAUU ACUUUCUG105 CAGAAAGUCUGAUGAGGCCGUUAGGCCGAA AUUACCCU1010
552 GGGUAAUUA CUUUCUGU106 ACAGAAAGCUGAUGAGGCCGUUAGGCCGAA AAUUACCC1011
555 UAAUUACUU UCUGUGGA107 UCCACAGACUGAUGAGGCCGUUAGGCCGAA AGUAAUUA1012
556 AAUUACUUU CUGUGGAC108 GUCCACAGCUGAUGAGGCCGUUAGGCCGAA AAGUAAUU1013
557 AUUACUUUC UGUGGACU109 AGUCCACACUGAUGAGGCCGUUAGGCCGAA AAAGUAAU1014
573 UGAGAAGUU UCCAUCCC110 GGGAUGGACUGAUGAGGCCGUUAGGCCGAA ACUUCUCA1015
574 GAGAAGUUU CCAUCCCU111 AGGGAUGGCUGAUGAGGCCGUUAGGCCGAA AACUUCUC1016
575 AGAAGUUUC CAUCCCUA112 UAGGGAUGCUGAUGAGGCCGUUAGGCCGAA AAACUUCU1017
579 GUUUCCAUC CCUAAAUA113 UAUUUAGGCUGAUGAGGCCGUUAGGCCGAA AUGGAAAC1018
583 CCAUCCCUA AAUAAGCU114 AGCUUAUUCUGAUGAGGCCGUUAGGCCGAA AGGGAUGG1019
587 CCCUAAAUA AGCUGGUA115 UACCAGCUCUGAUGAGGCCGUUAGGCCGAA AUUUAGGG1020
595 AAGCUGGUA GACUACUA116 UAGUAGUCCUGAUGAGGCCGUUAGGCCGAA ACCAGCUU1021
600 GGUAGACUA CUACAGGA117 UCCUGUAGCUGAUGAGGCCGUUAGGCCGAA AGUCUACC1022
603 AGACUACUA CAGGACAA118 UUGUCCUGCUGAUGAGGCCGUUAGGCCGAA AGUAGUCU1023
614 GGACAAAUU CCAUCUCC119 GGAGAUGGCUGAUGAGGCCGUUAGGCCGAA AUUUGUCC1024
615 GACAAAUUC CAUCUCCA120 UGGAGAUGCUGAUGAGGCCGUUAGGCCGAA AAUUUGUC1025
619 AAUUCCAUC UCCAGACA121 UGUCUGGACUGAUGAGGCCGUUAGGCCGAA AUGGAAUU1026
621 UUCCAUCUC CAGACAGA122 UCUGUCUGCUGAUGAGGCCGUUAGGCCGAA AGAUGGAA1027
637 AAGCAGAUC UUCCUUAG123 CUAAGGAA GCCGUUAGGCCGAA AUCUGCUU1028
CUGAUGAG
639 GCAGAUCUU CCUUAGAG124 CUCUAAGGCUGAUGAGGCCGUUAGGCCGAA AGAUCUGC1029
640 CAGAUCUUC CUUAGAGA125 UCUCUAAGCUGAUGAGGCCGUUAGGCCGAA AAGAUCUG1030
643 AUCUUCCUU AGAGACAG126 CUGUCUCUCUGAUGAGGCCGUUAGGCCGAA AGGAAGAU1031
644 UCUUCCUUA GAGACAGA127 UCUGUCUCCUGAUGAGGCCGUUAGGCCGAA AAGGAAGA1032
671 ACCAGGGUC ACCGGGGC128 GCCCCGGUCUGAUGAGGCCGUUAGGCCGAA ACCCUGGU1033
699 CCGGAGGUC CCAGGGAG129 CUCCCUGGCUGAUGAGGCCGUUAGGCCGAA ACCUCCGG1034
718 CCACACCUC AGUGGGGC130 GCCCCACUCUGAUGAGGCCGUUAGGCCGAA AGGUGUGG1035
742 GAAGAAAUC CGACCUUC131 GAAGGUCGCUGAUGAGGCCGUUAGGCCGAA AUUUCUUC1036
749 UCCGACCUU CGAUGAAC132 GUUCAUCGCUGAUGAGGCCGUUAGGCCGAA AGGUCGGA1037
750 CCGACCUUC GAUGAACC133 GGUUCAUCCUGAUGAGGCCGUUAGGCCGAA AAGGUCGG1038
768 GAAGCUGUC GGAUCACC134 GGUGAUCCCUGAUGAGGCCGUUAGGCCGAA ACAGCUUC1039
773 UGUCGGAUC ACCCCCCG135 CGGGGGGUCUGAUGAGGCCGUUAGGCCGAA AUCCGACA1040
787 CCGACCCUU CCCCUGCA136 UGCAGGGGCUGAUGAGGCCGUUAGGCCGAA AGGGUCGG1041
788 CGACCCUUC CCCUGCAG137 CUGCAGGGCUGAUGAGGCCGUUAGGCCGAA AAGGGUCG1042
821 CACAGCCUC CGCAAUAU138 AUAUUGCGCUGAUGAGGCCGUUAGGCCGAA AGGCUGUG1043
828 UCCGCAAUA UGCCCCAG139 CUGGGGCACUGAUGAGGCCGUUAGGCCGAA AUUGCGGA1044
873 GCAGCGAUA UCUGCAGC140 GCUGCAGACUGAUGAGGCCGUUAGGCCGAA AUCGCUGC1045
875 AGCGAUAUC UGCAGCAC141 GUGCUGCACUGAUGAGGCCGUUAGGCCGAA AUAUCGCU1046
890 ACCACCAUU UCCACCAG142 CUGGUGGACUGAUGAGGCCGUUAGGCCGAA AUGGUGGU1047
891 CCACCAUUU CCACCAGG143 CCUGGUGGCUGAUGAGGCCGUUAGGCCGAA AAUGGUGG1048
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
62
892 CACCAUUUC CACCAGGA144 UCCUGGUGCUGAUGAGGCCGUUAGGCCGAA AAAUGGUG1049
919 GGCAGCCUU GACAUAAA145 UUUAUGUCCUGAUGAGGCCGUUAGGCCGAA AGGCUGCC1050
925 CUUGACAUA AAUGAUGG146 CCAUCAUUCUGAUGAGGCCGUUAGGCCGAA AUGUCAAG1051
938 AUGGGCAUU GUGGCACC147 GGUGCCACCUGAUGAGGCCGUUAGGCCGAA AUGCCCAU1052
951 CACCGGCUU GGGCAGUG148 CACUGCCCCUGAUGAGGCCGUUAGGCCGAA AGCCGGUG1053
976 GCGGCCCUC AUGCAUCG149 CGAUGCAUCUGAUGAGGCCGUUAGGCCGAA AGGGCCGC1054
983 UCAUGCAUC GGAGACAC150 GUGUCUCCCUGAUGAGGCCGUUAGGCCGAA AUGCAUGA1055
1009 GUGCAGCUC CAGGCGGC151 GCCGCCUGCUGAUGAGGCCGUUAGGCCGAA AGCUGCAC1056
1047 GGCGCUGUA UGACUUUG152 CAAAGUCACUGAUGAGGCCGUUAGGCCGAA ACAGCGCC1057
1053 GUAUGACUU UGAGGCCC153 GGGCCUCACUGAUGAGGCCGUUAGGCCGAA AGUCAUAC1058
1054 UAUGACUUU GAGGCCCU154 AGGGCCUCCUGAUGAGGCCGUUAGGCCGAA AAGUCAUA1059
1083 GCUGGGGUU CCACAGCG155 CGCUGUGGCUGAUGAGGCCGUUAGGCCGAA ACCCCAGC1060
1084 CUGGGGUUC CACAGCGG156 CCGCUGUGCUGAUGAGGCCGUUAGGCCGAA AACCCCAG1061
1108 GUGGAGGUC CUGGAUAG157 CUAUCCAGCUGAUGAGGCCGUUAGGCCGAA ACCUCCAC1062
1115 UCCUGGAUA GCUCCAAC158 GUUGGAGCCUGAUGAGGCCGUUAGGCCGAA AUCCAGGA1063
1119 GGAUAGCUC CAACCCAU159 AUGGGUUGCUGAUGAGGCCGUUAGGCCGAA AGCUAUCC1064
1128 CAACCCAUC CUGGUGGA160 UCCACCAGCUGAUGAGGCCGUUAGGCCGAA AUGGGUUG1065
1165 CUGGGCCUC UUCCCUGC161 GCAGGGAACUGAUGAGGCCGUUAGGCCGAA AGGCCCAG1066
1167 GGGCCUCUU CCCUGCCA162 UGGCAGGGCUGAUGAGGCCGUUAGGCCGAA AGAGGCCC1067
1168 GGCCUCUUC CCUGCCAA163 UUGGCAGGCUGAUGAGGCCGUUAGGCCGAA AAGAGGCC1068
1179 UGCCAACUA CGUGGCAC164 GUGCCACGCUGAUGAGGCCGUUAGGCCGAA AGUUGGCA1069
1200 GACCCGAUA AACUCUUC165 GAAGAGUUCUGAUGAGGCCGUUAGGCCGAA AUCGGGUC1070
1205 GAUAAACUC UUCAGGGG166 CCCCUGAACUGAUGAGGCCGUUAGGCCGAA AGUUUAUC1071
1207 UAAACUCUU CAGGGGAC167 GUCCCCUGCUGAUGAGGCCGUUAGGCCGAA AGAGUUUA1072
1208 AAACUCUUC AGGGGACA168 UGUCCCCUCUGAUGAGGCCGUUAGGCCGAA AAGAGUUU1073
1223 CAGAAGCUU UUUGUCUG169 CAGACAAA GCCGUUAGGCCGAA AGCUUCUG1074
CUGAUGAG
1224 AGAAGCUUU UUGUCUGG170 CCAGACAA GCCGUUAGGCCGAA AAGCUUCU1075
CUGAUGAG
1225 GAAGCUUUU UGUCUGGA171 UCCAGACACUGAUGAGGCCGUUAGGCCGAA AAAGCUUC1076
1226 AAGCUUUUU GUCUGGAG172 CUCCAGACCUGAUGAGGCCGUUAGGCCGAA AAAAGCUU1077
1229 CUUUUUGUC UGGAGCUG173 CAGCUCCACUGAUGAGGCCGUUAGGCCGAA ACAAAAAG1078
1274 GCUGGACUC CAUGACUA174 UAGUCAUGCUGAUGAGGCCGUUAGGCCGAA AGUCCAGC1079
1282 CCAUGACUA UAUAUACA175 UGUAUAUACUGAUGAGGCCGUUAGGCCGAA AGUCAUGG1080
1284 AUGACUAUA UAUACAUA176 UAUGUAUACUGAUGAGGCCGUUAGGCCGAA AUAGUCAU1081
1286 GACUAUAUA UACAUACA177 UGUAUGUACUGAUGAGGCCGUUAGGCCGAA AUAUAGUC1082
1288 CUAUAUAUA CAUACAUC178 GAUGUAUGCUGAUGAGGCCGUUAGGCCGAA AUAUAUAG1083
1292 AUAUACAUA CAUCUAUC179 GAUAGAUGCUGAUGAGGCCGUUAGGCCGAA AUGUAUAU1084
Input Sequence = HSA011736. Cut Site = UH/.
Stem Length = 8 . Core Sequence = CUGAUGAG GCCGUUAGGC CGAA
HSA011736 (Homo sapiens mRNA for growth factor receptor binding protein
(GRBLG) ; 1303 bp)
Underlined region can be any X sequence or linker as defined herein.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
63
Table IV: Human GRID NCH Ribozyme and Substrate Sequence
Pos Substrate Se Riboz Se
ID me ID
GGAGGCACA GUUAAUGG180 CCAUUAACCUGAUGAGGCCGUUAGGCCGAA IUGCCUCC1085
22 AAUGGAUCU GUAAACUU181 AAGUUUACCUGAUGAGGCCGUUAGGCCGAA IAUCCAUU1086
29 CUGUAAACU UGCACCCU182 AGGGUGCACUGAUGAGGCCGUUAGGCCGAA IUUUACAG1087
33 AAACUUGCA CCCUCUUU183 AAAGAGGGCUGAUGAGGCCGUUAGGCCGAA ICAAGUUU1088
35 ACUUGCACC CUCUUUCA184 UGAAAGAGCUGAUGAGGCCGUUAGGCCGAA IUGCAAGU1089
36 CUUGCACCC UCUUUCAG185 CUGAAAGACUGAUGAGGCCGUUAGGCCGAA IGUGCAAG1090
37 UUGCACCCU CUUUCAGA186 UCUGAAAGCUGAUGAGGCCGUUAGGCCGAA IGGUGCAA1091
39 GCACCCUCU UUCAGAGU187 ACUCUGAA GCCGUUAGGCCGA~ IAGGGUGC1092
CUGAUGAG
43 CCUCUUUCA GAGUGGUA188 UACCACUCCUGAUGAGGCCGUUAGGCCGA1~' IAAAGAGG1093
53 AGUGGUACA UGGAAGAC189 GUCUUCCACUGAUGAGGCCGUUAGGCCGAA IUACCACU1094
62 UGGAAGACA GCACAAAG190 CUUUGUGCCUGAUGAGGCCGUUAGGCCGAA IUCUUCCA1095
65 AAGACAGCA CAAAGUGG191 CCACUUUGCUGAUGAGGCCGUUAGGCCGAA ICUGUCUU1096
67 GACAGCACA AAGUGGAU192 AUCCACUUCUGAUGAGGCCGUUAGGCCGAA IUGCUGUC1097
77 AGUGGAUCC AUACUCUG193 CAGAGUAUCUGAUGAGGCCGUUAGGCCGAA IAUCCACU1098
78 GUGGAUCCA UACUCUGA194 UCAGAGUACUGAUGAGGCCGUUAGGCCGAA IGAUCCAC1099
82 AUCCAUACU CUGAAAUG195 CAUUUCAGCUGAUGAGGCCGUUAGGCCGAA IUAUGGAU1100
84 CCAUACUCU GAAAUGCA196 UGCAUUUCCUGAUGAGGCCGUUAGGCCGAA TAGUAUGG1101
92 UGAAAUGCA GUAACUCU197 AGAGUUACCUGAUGAGGCCGUUAGGCCGAA ICAUUUCA1102
98 GCAGUAACU CUGAUGCU198 AGCAUCAGCUGAUGAGGCCGUUAGGCCGAA IUUACUGC1103
100 AGUAACUCU GAUGCUUG199 CAAGCAUCCUGAUGAGGCCGUUAGGCCGAA IAGUUACU1104
106 UCUGAUGCU UGAAUUUG200 CAAAUUCACUGAUGAGGCCGUUAGGCCGAA ICAUCAGA1105
118 AUUUGUUCU CCCUUCUU201 AAGAAGGGCUGAUGAGGCCGUUAGGCCGAA IAACAAAU1106
120 UUGUUCUCC CUUCUUGC202 GCAAGAAGCUGAUGAGGCCGUUAGGCCGAA IAGAACAA1107
121 UGUUCUCCC UUCUUGCC203 GGCAAGAACUGAUGAGGCCGUUAGGCCGAA IGAGAACA1108
122 GUUCUCCCU UCUUGCCA204 UGGCAAGACUGAUGAGGCCGUUAGGCCGAA IGGAGAAC1109
125 CUCCCUUCU UGCCAGAA205 UUCUGGCACUGAUGAGGCCGUUAGGCCGAA IAAGGGAG1110
129 CUUCUUGCC AGAAAGGA206 UCCUUUCUCUGAUGAGGCCGUUAGGCCGAA ICAAGAAG1111
130 UUCUUGCCA GAAAGGAU207 AUCCUUUCCUGAUGAGGCCGUUAGGCCGAA IGCAAGAA1112
141 AAGGAUUCU AAUAACUC208 GAGUUAUUCUGAUGAGGCCGUUAGGCCGAA IAAUCCUU1113
148 CUAAUAACU CGGUGUCA209 UGACACCGCUGAUGAGGCCGUUAGGCCGAA IUUAUUAG1114
156 UCGGUGUCA AAGCCAAG210 CUUGGCUUCUGAUGAGGCCGUUAGGCCGAA IACACCGA1115
161 GUCAAAGCC AAGACAUA211 UAUGUCUUCUGAUGAGGCCGUUAGGCCGAA ICUUUGAC1116
162 UCAAAGCCA AGACAUAA212 UUAUGUCUCUGAUGAGGCCGUUAGGCCGAA IGCUUUGA1117
167 GCCAAGACA UAAACUCA213 UGAGUUUACUGAUGAGGCCGUUAGGCCGAA IUCUUGGC1118
173 ACAUAAACU CAAUCUCU214 AGAGAUUG~ GCCGUUAGGCCGAA IUUUAUGU1119
CUGAUGAG
175 AUAAACUCA AUCUCUUC215 GAAGAGAUCUGAUGAGGCCGUUAGGCCGAA IAGUUUAU1120
179 ACUCAAUCU CUUCUCUU216 AAGAGAAGCUGAUGAGGCCGUUAGGCCGAA IAUUGAGU1121
181 UCAAUCUCU UCUCUUCC217 GGAAGAGACUGAUGAGGCCGUUAGGCCGAA IAGAUUGA1122
184 AUCUCUUCU CUUCCAAA218 UUUGGAAGCUGAUGAGGCCGUUAGGCCGAA IAAGAGAU1123
186 CUCUUCUCU UCCAAAAG219 CUUUUGGACUGAUGAGGCCGUUAGGCCGAA IAGAAGAG1124
189 UUCUCUUCC AAAAGCUU220 AAGCUUUUCUGAUGAGGCCGUUAGGCCGAA IAAGAGAA1125
,
190 UCUCUUCCA AAAGCUUC221 GAAGCUUUCUGAUGAGGCCGUUAGGCCGAA IGAAGAGA1126
196 CCAAAAGCU UCACGUUA222 UAACGUGACUGAUGAGGCCGUUAGGCCGAA ICUUUUGG1127
199 AAAGCUUCA CGUUACAG223 CUGUAACGCUGAUGAGGCCGUUAGGCCGAA IAAGCUUU1128
206 CACGUUACA GCAUGGAA224 UUCCAUGCCUGAUGAGGCCGUUAGGCCGAA IUAACGUG1129
209 GUUACAGCA UGGAAGCU225 AGCUUCCACUGAUGAGGCCGUUAGGCCGAA ICUGUAAC1130
217 AUGGAAGCU GUUGCCAA226 UUGGCAACCUGAUGAGGCCGUUAGGCCGAA ICUUCCAU1131
223 GCUGUUGCC AAGUUUGA227 UCAAACUUCUGAUGAGGCCGUUAGGCCGAA TCAACAGC1132
224 CUGUUGCCA AGUUUGAU228 AUCAAACUCUGAUGAGGCCGUUAGGCCGAA IGCAACAG1133
236 UUGAUUUCA CUGCUUCA229 UGAAGCAGCUGAUGAGGCCGUUAGGCCGAA IAAAUCAA1134
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
64
238 GAUUUCACU GCUUCAGG230 CCUGAAGCCUGAUGAGGCCGUUAGGCCGAA IUGAAAUC1135
241 UUCACUGCU UCAGGUGA231 UCACCUGACUGAUGAGGCCGUUAGGCCGAA ICAGUGAA1136
244 ACUGCUUCA GGUGAGGA232 UCCUCACCCUGAUGAGGCCGUUAGGCCGAA IAAGCAGU1137
258 GGAUGAACU GAGCUUUC233 GAAAGCUCCUGAUGAGGCCGUUAGGCCGAA IUUCAUCC1138
263 AACUGAGCU UUCACACU234 AGUGUGAA GCCGUUAGGCCGAA ICUCAGW1139
CUGAUGAG
267 GAGCUUUCA CACUGGAG235 CUCCAGUGCUGAUGAGGCCGUUAGGCCGAA IAAAGCUC1140
269 GCUUUCACA CUGGAGAU236 AUCUCCAGCUGAUGAGGCCGUUAGGCCGAA IUGAAAGC1141
271 UUUCACACU GGAGAUGU237 ACAUCUCCCUGAUGAGGCCGUUAGGCCGAA IUGUGAAA1142
299 UAAGUAACC AAGAGGAG238 CUCCUCUUCUGAUGAGGCCGUUAGGCCGAA IUUACUUA1143
300 AAGUAACCA AGAGGAGU239 ACUCCUCUCUGAUGAGGCCGUUAGGCCGAA IGUUACUU1144
324 GGCGGAGCU UGGGAGCC240 GGCUCCCACUGAUGAGGCCGUUAGGCCGAA ICUCCGCC1145
332 UUGGGAGCC AGGAAGGA241 UCCUUCCUCUGAUGAGGCCGUUAGGCCGAA ICUCCCAA1146
333 UGGGAGCCA GGAAGGAU242 AUCCUUCCCUGAUGAGGCCGUUAGGCCGAA IGCUCCCA1147
348 AUAUGUGCC CAAGAAUU243 AAUUCUUGCUGAUGAGGCCGUUAGGCCGAA ICACAUAU1148
349 UAUGUGCCC AAGAAUUU244 AAAUUCUUCUGAUGAGGCCGUUAGGCCGAA IGCACAUA1149
350 AUGUGCCCA AGAAUUUC245 GAAAUUCUCUGAUGAGGCCGUUAGGCCGAA IGGCACAU1150
359 AGAAUUUCA UAGACAUC246 GAUGUCUACUGAUGAGGCCGUUAGGCCGAA IAAAUUCU1151
365 UCAUAGACA UCCAGUUU247 AAACUGGACUGAUGAGGCCGUUAGGCCGAA IUCUAUGA1152
368 UAGACAUCC AGUUUCCC248 GGGAAACUCUGAUGAGGCCGUUAGGCCGAA IAUGUCUA1153
369 AGACAUCCA GUUUCCCA249 UGGGAAACCUGAUGAGGCCGUUAGGCCGAA IGAUGUCU1154
375 CCAGUUUCC CAAAUGGU250 ACCAUUUGCUGAUGAGGCCGUUAGGCCGAA IAAACUGG1155
376 CAGUUUCCC AAAUGGUU251 AACCAUUUCUGAUGAGGCCGUUAGGCCGAA IGAAACUG1156
377 AGUUUCCCA AAUGGUUU252 AAACCAUUCUGAUGAGGCCGUUAGGCCGAA IGGAAACU1157
387 AUGGUUUCA CGAAGGCC253 GGCCUUCGCUGAUGAGGCCGUUAGGCCGAA IAAACCAU1158
395 ACGAAGGCC UCUCUCGA254 UCGAGAGACUGAUGAGGCCGUUAGGCCGAA ICCUUCGU1159
396 CGAAGGCCU CUCUCGAC255 GUCGAGAGCUGAUGAGGCCGUUAGGCCGAA IGCCUUCG1160
398 AAGGCCUCU CUCGACAC256 GUGUCGAGCUGAUGAGGCCGUUAGGCCGAA IAGGCCUU1161
400 GGCCUCUCU CGACACCA257 UGGUGUCGCUGAUGAGGCCGUUAGGCCGAA IAGAGGCC1162
405 CUCUCGACA CCAGGCAG258 CUGCCUGGCUGAUGAGGCCGUUAGGCCGAA IUCGAGAG1163
407 CUCGACACC AGGCAGAG259 CUCUGCCUCUGAUGAGGCCGUUAGGCCGAA IUGUCGAG1164
408 UCGACACCA GGCAGAGA260 UCUCUGCCCUGAUGAGGCCGUUAGGCCGAA IGUGUCGA1165
412 CACCAGGCA GAGAACUU261 AAGUUCUCCUGAUGAGGCCGUUAGGCCGAA ICCUGGUG1166
419 CAGAGAACU UACUCAUG262 CAUGAGUACUGAUGAGGCCGUUAGGCCGAA IUUCUCUG1167
423 GAACUUACU CAUGGGCA263 UGCCCAUGCUGAUGAGGCCGUUAGGCCGAA IUAAGUUC1168
425 ACUUACUCA UGGGCAAG264 CUUGCCCACUGAUGAGGCCGUUAGGCCGAA IAGUAAGU1169
431 UCAUGGGCA AGGAGGUU265 AACCUCCUCUGAUGAGGCCGUUAGGCCGAA ICCCAUGA1170
443 AGGUUGGCU UCUUCAUC266 GAUGAAGACUGAUGAGGCCGUUAGGCCGAA ICCAACCU1171
446 UUGGCUUCU UCAUCAUC267 GAUGAUGACUGAUGAGGCCGUUAGGCCGAA IAAGCCAA1172
449 GCUUCUUCA UCAUCCGG268 CCGGAUGACUGAUGAGGCCGUUAGGCCGAA TAAGAAGC1173
452 UCUUCAUCA UCCGGGCC269 GGCCCGGACUGAUGAGGCCGUUAGGCCGAA IAUGAAGA1174
455 UCAUCAUCC GGGCCAGC270 GCUGGCCCCUGAUGAGGCCGUUAGGCCGAA IAUGAUGA1175
460 AUCCGGGCC AGCCAGAG271 CUCUGGCUCUGAUGAGGCCGUUAGGCCGAA ICCCGGAU1176
461 UCCGGGCCA GCCAGAGC272 GCUCUGGCCUGAUGAGGCCGUUAGGCCGAA IGCCCGGA1177
464 GGGCCAGCC AGAGCUCC273 GGAGCUCUCUGAUGAGGCCGUUAGGCCGAA ICUGGCCC1178
465 GGCCAGCCA GAGCUCCC274 GGGAGCUCCUGAUGAGGCCGUUAGGCCGAA IGCUGGCC1179
470 GCCAGAGCU CCCCAGGG275 CCCUGGGGCUGAUGAGGCCGUUAGGCCGAA ICUCUGGC1180
472 CAGAGCUCC CCAGGGGA276 UCCCCUGGCUGAUGAGGCCGUUAGGCCGAA IAGCUCUG1181
473 AGAGCUCCC CAGGGGAC277 GUCCCCUGCUGAUGAGGCCGUUAGGCCGAA IGAGCUCU1182
474 GAGCUCCCC AGGGGACU278 AGUCCCCUCUGAUGAGGCCGUUAGGCCGAA IGGAGCUC1183
475 AGCUCCCCA GGGGACUU279 AAGUCCCCCUGAUGAGGCCGUUAGGCCGAA IGGGAGCU1184
482 CAGGGGACU UCUCCAUC280 GAUGGAGACUGAUGAGGCCGUUAGGCCGAA IUCCCCUG1185
485 GGGACUUCU CCAUCUCU281 AGAGAUGGCUGAUGAGGCCGUUAGGCCGAA IAAGUCCC1186
487 GACUUCUCC AUCUCUGU282 ACAGAGAUCUGAUGAGGCCGUUAGGCCGAA IAGAAGUC1187
488 ACUUCUCCA UCUCUGUC283 GACAGAGACUGAUGAGGCCGUUAGGCCGAA IGAGAAGU1188
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
491 UCUCCAUCUCUGUCAGG284 CCUGACAGCUGAUGAGGCCGUUAGGCCGAA IAUGGAGA1189
493 UCCAUCUCUGUCAGGCA285 UGCCUGACCUGAUGAGGCCGUUAGGCCGAA IAGAUGGA1190
497 UCUCUGUCAGGCAUGAG286 CUCAUGCCCUGAUGAGGCCGUUAGGCCGAA IACAGAGA1191
501 UGUCAGGCAUGAGGAUG287 CAUCCUCACUGAUGAGGCCGUUAGGCCGAA ICCUGACA1192
516 UGACGUUCAACACUUCA288 UGAAGUGUCUGAUGAGGCCGUUAGGCCGAA TAACGUCA1193
519 CGUUCAACACUUCAAGG289 CCUUGAAGCUGAUGAGGCCGUUAGGCCGAA TUUGAACG1194
521 UUCAACACUUCAAGGUC290 GACCUUGACUGAUGAGGCCGUUAGGCCGAA TUGUUGAA1195
524 AACACUUCAAGGUCAUG291 CAUGACCUCUGAUGAGGCCGUUAGGCCGAA TAAGUGUU1196
530 UCAAGGUCAUGCGAGAC292 GUCUCGCACUGAUGAGGCCGUUAGGCCGAA IACCUUGA1197
539 UGCGAGACAACAAGGGU293 ACCCUUGUCUGAUGAGGCCGUUAGGCCGAA IUCUCGCA1198
542 GAGACAACAAGGGUAAU294 AUUACCCUCUGAUGAGGCCGUUAGGCCGAA 2UUGUCUC1199
554 GUAAUUACUUUCUGUGG295 CCACAGAA GCCGUUAGGCCGAA TUAAUUAC1200
CUGAUGAG
558 UUACUUUCUGUGGACUG296 CAGUCCACCUGAUGAGGCCGUUAGGCCGAA IAAAGUAA1201
565 CUGUGGACUGAGAAGUU297 AACUUCUCCUGAUGAGGCCGUUAGGCCGAA TUCCACAG1202
576 GAAGUUUCCAUCCCUAA298 UUAGGGAUCUGAUGAGGCCGUUAGGCCGAA 2AAACUUC1203
577 AAGUUUCCAUCCCUAAA299 UUUAGGGACUGAUGAGGCCGUUAGGCCGAA IGAAACUU1204
580 UUUCCAUCCCUAAAUAA300 UUAUUUAGCUGAUGAGGCCGUUAGGCCGAA TAUGGAAA1205
581 UUCCAUCCCUAAAUAAG301 CUUAUUUACUGAUGAGGCCGUUAGGCCGAA TGAUGGAA1206
582 UCCAUCCCUAAAUAAGC302 GCUUAUUUCUGAUGAGGCCGUUAGGCCGAA TGGAUGGA1207
591 AAAUAAGCUGGUAGACU303 AGUCUACCCUGAUGAGGCCGUUAGGCCGAA ICUUAUUU1208
599 UGGUAGACUACUACAGG304 CCUGUAGUCUGAUGAGGCCGUUAGGCCGAA IUCUACCA1209
602 UAGACUACUACAGGACA305 UGUCCUGUCUGAUGAGGCCGUUAGGCCGAA IUAGUCUA1210
605 ACUACUACAGGACAAAU306 AUUUGUCCCUGAUGAGGCCGUUAGGCCGAA IUAGUAGU1211
610 UACAGGACA 307 AUGGAAUUCUGAUGAGGCCGUUAGGCCGAA IUCCUGUA1212
AAUUCCAU
616 ACAAAUUCCAUCUCCAG308 CUGGAGAUCUGAUGAGGCCGUUAGGCCGAA TAAUUUGU1213
617 CAAAUUCCAUCUCCAGA309 UCUGGAGACUGAUGAGGCCGUUAGGCCGAA IGAAUUUG1214
620 AUUCCAUCUCCAGACAG310 CUGUCUGGCUGAUGAGGCCGUUAGGCCGAA TAUGGAAU1215
622 UCCAUCUCCAGACAGAA311 UUCUGUCUCUGAUGAGGCCGUUAGGCCGAA IAGAUGGA1216
623 CCAUCUCCAGACAGAAG312 CUUCUGUCCUGAUGAGGCCGUUAGGCCGAA TGAGAUGG1217
627 CUCCAGACAGAAGCAGA313 UCUGCUUCCUGAUGAGGCCGUUAGGCCGAA TUCUGGAG1218
633 ACAGAAGCAGAUCUUCC314 GGAAGAUCCUGAUGAGGCCGUUAGGCCGAA ICUUCUGU1219
638 AGCAGAUCUUCCUUAGA315 UCUAAGGACUGAUGAGGCCGUUAGGCCGAA TAUCUGCU1220
641 AGAUCUUCCUUAGAGAC316 GUCUCUAA GCCGUUAGGCCGAA TAAGAUCU1221
CUGAUGAG
642 GAUCUUCCUUAGAGACA317 UGUCUCUACUGAUGAGGCCGUUAGGCCGAA IGAAGAUC1222
650 UUAGAGACAGAACCCGA318 UCGGGUUCCUGAUGAGGCCGUUAGGCCGAA IUCUCUAA1223
655 GACAGAACCCGAGAAGA319 UCUUCUCGCUGAUGAGGCCGUUAGGCCGAA IUUCUGUC1224
656 ACAGAACCCGAGAAGAC320 GUCUUCUCCUGAUGAGGCCGUUAGGCCGAA IGUUCUGU1225
665 GAGAAGACCAGGGUCAC321 GUGACCCUCUGAUGAGGCCGUUAGGCCGAA IUCUUCUC1226
666 AGAAGACCAGGGUCACC322 GGUGACCCCUGAUGAGGCCGUUAGGCCGAA IGUCUUCU1227
672 CCAGGGUCACCGGGGCA323 UGCCCCGGCUGAUGAGGCCGUUAGGCCGAA TACCCUGG1228
674 AGGGUCACCGGGGCAAC324 GUUGCCCCCUGAUGAGGCCGUUAGGCCGAA TUGACCCU1229
680 ACCGGGGCAACAGCCUG325 CAGGCUGUCUGAUGAGGCCGUUAGGCCGAA TCCCCGGU1230
683 GGGGCAACAGCCUGGAC326 GUCCAGGCCUGAUGAGGCCGUUAGGCCGAA TUUGCCCC1231
686 GCAACAGCCUGGACCGG327 CCGGUCCACUGAUGAGGCCGUUAGGCCGAA ICUGUUGC1232
687 CAACAGCCUGGACCGGA328 UCCGGUCCCUGAUGAGGCCGUUAGGCCGAA IGCUGUUG1233
692 GCCUGGACCGGAGGUCC329 GGACCUCCCUGAUGAGGCCGUUAGGCCGAA TUCCAGGC1234
700 CGGAGGUCCCAGGGAGG330 CCUCCCUGCUGAUGAGGCCGUUAGGCCGAA TACCUCCG1235
701 GGAGGUCCCAGGGAGGC331 GCCUCCCUCUGAUGAGGCCGUUAGGCCGAA TGACCUCC1236
702 GAGGUCCCAGGGAGGCC332 GGCCUCCCCUGAUGAGGCCGUUAGGCCGAA TGGACCUC1237
710 AGGGAGGCCCACACCUC333 GAGGUGUGCUGAUGAGGCCGUUAGGCCGAA ICCUCCCU1238
711 GGGAGGCCCACACCUCA334 UGAGGUGUCUGAUGAGGCCGUUAGGCCGAA IGCCUCCC1239
712 GGAGGCCCACACCUCAG335 CUGAGGUGCUGAUGAGGCCGUUAGGCCGAA IGGCCUCC1240
714 AGGCCCACACCUCAGUG336 CACUGAGGCUGAUGAGGCCGUUAGGCCGAA TUGGGCCU1241
716 GCCCACACCUCAGUGGG337 CCCACUGACUGAUGAGGCCGUUAGGCCGAA IUGUGGGC1242
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
66
717 CCCACACCU CAGUGGGG338 CCCCACUGCUGAUGAGGCCGUUAGGCCGAA IGUGUGGG1243
719 CACACCUCA GUGGGGCU339 AGCCCCACCUGAUGAGGCCGUUAGGCCGAA IAGGUGUG1244
727 AGUGGGGCU GUGGGAGA340 UCUCCCACCUGAUGAGGCCGUUAGGCCGAA ICCCCACU1245
743 AAGAAAUCC GACCUUCG341 CGAAGGUCCUGAUGAGGCCGUUAGGCCGAA IAUUUCUU124'6
747 AAUCCGACC UUCGAUGA342 UCAUCGAA GCCGUUAGGCCGAA IUCGGAUU1247
CUGAUGAG
748 AUCCGACCU UCGAUGAA343 UUCAUCGACUGAUGAGGCCGUUAGGCCGAA IGUCGGAU1248
758 CGAUGAACC GGAAGCUG344 CAGCUUCCCUGAUGAGGCCGUUAGGCCGAA IUUCAUCG1249
765 CCGGAAGCU GUCGGAUC345 GAUCCGACCUGAUGAGGCCGUUAGGCCGAA TCUUCCGG1250
774 GUCGGAUCA CCCCCCGA346 UCGGGGGGCUGAUGAGGCCGUUAGGCCGAA 2AUCCGAC1251
776 CGGAUCACC CCCCGACC347 GGUCGGGGCUGAUGAGGCCGUUAGGCCGAA TUGAUCCG1252
777 GGAUCACCC CCCGACCC348 GGGUCGGGCUGAUGAGGCCGUUAGGCCGAA IGUGAUCC1253
778 GAUCACCCC CCGACCCU349 AGGGUCGGCUGAUGAGGCCGUUAGGCCGAA IGGUGAUC1254
779 AUCACCCCC CGACCCUU350 AAGGGUCGCUGAUGAGGCCGUUAGGCCGAA IGGGUGAU1255
780 UCACCCCCC GACCCUUC351 GAAGGGUCCUGAUGAGGCCGUUAGGCCGAA IGGGGUGA1256
784 CCCCCGACC CUUCCCCU352 AGGGGAAGCUGAUGAGGCCGUUAGGCCGAA IUCGGGGG1257
785 CCCCGACCC UUCCCCUG353 CAGGGGAACUGAUGAGGCCGUUAGGCCGAA IGUCGGGG1258
786 CCCGACCCU UCCCCUGC354 GCAGGGGACUGAUGAGGCCGUUAGGCCGAA IGGUCGGG1259
789 GACCCUUCC CCUGCAGC355 GCUGCAGGCUGAUGAGGCCGUUAGGCCGAA IAAGGGUC1260
790 ACCCUUCCC CUGCAGCA356 UGCUGCAGCUGAUGAGGCCGUUAGGCCGAA IGAAGGGU1261
791 CCCUUCCCC UGCAGCAG357 CUGCUGCACUGAUGAGGCCGUUAGGCCGAA IGGAAGGG1262
792 CCUUCCCCU GCAGCAGC358 GCUGCUGCCUGAUGAGGCCGUUAGGCCGAA IGGGAAGG1263
795 UCCCCUGCA GCAGCACC359 GGUGCUGCCUGAUGAGGCCGUUAGGCCGAA ICAGGGGA1264
798 CCUGCAGCA GCACCAGC360 GCUGGUGCCUGAUGAGGCCGUUAGGCCGAA ICUGCAGG1265
801 GCAGCAGCA CCAGCACC361 GGUGCUGGCUGAUGAGGCCGUUAGGCCGAA ICUGCUGC1266
803 AGCAGCACC AGCACCAG362 CUGGUGCUCUGAUGAGGCCGUUAGGCCGAA IUGCUGCU1267
804 GCAGCACCA GCACCAGC363 GCUGGUGCCUGAUGAGGCCGUUAGGCCGAA IGUGCUGC1268
807 GCACCAGCA CCAGCCAC364 GUGGCUGGCUGAUGAGGCCGUUAGGCCGAA ICUGGUGC1269
809 ACCAGCACC AGCCACAG365 CUGUGGCUCUGAUGAGGCCGUUAGGCCGAA IUGCUGGU1270
810 CCAGCACCA GCCACAGC366 GCUGUGGCCUGAUGAGGCCGUUAGGCCGAA IGUGCUGG1271
813 GCACCAGCC ACAGCCUC367 GAGGCUGUCUGAUGAGGCCGUUAGGCCGAA ICUGGUGC1272
814 CACCAGCCA CAGCCUCC368 GGAGGCUGCUGAUGAGGCCGUUAGGCCGAA IGCUGGUG1273
816 CCAGCCACA GCCUCCGC369 GCGGAGGCCUGAUGAGGCCGUUAGGCCGAA IUGGCUGG1274
819 GCCACAGCC UCCGCAAU370 AUUGCGGACUGAUGAGGCCGUUAGGCCGAA ICUGUGGC1275
820 CCACAGCCU CCGCAAUA371 UAUUGCGGCUGAUGAGGCCGUUAGGCCGAA IGCUGUGG1276
822 ACAGCCUCC GCAAUAUG372 CAUAUUGCCUGAUGAGGCCGUUAGGCCGAA IAGGCUGU1277
825 GCCUCCGCA AUAUGCCC373 GGGCAUAUCUGAUGAGGCCGUUAGGCCGAA ICGGAGGC1278
832 CAAUAUGCC CCAGCGCC374 GGCGCUGGCUGAUGAGGCCGUUAGGCCGAA ICAUAUUG1279
833 AAUAUGCCC CAGCGCCC375 GGGCGCUGCUGAUGAGGCCGUUAGGCCGAA IGCAUAUU1280
834 AUAUGCCCC AGCGCCCC376 GGGGCGCUCUGAUGAGGCCGUUAGGCCGAA IGGCAUAU1281
835 UAUGCCCCA GCGCCCCA377 UGGGGCGCCUGAUGAGGCCGUUAGGCCGAA IGGGCAUA1282
840 CCCAGCGCC CCAGCAGC378 GCUGCUGGCUGAUGAGGCCGUUAGGCCGAA ICGCUGGG1283
841 CCAGCGCCC CAGCAGCU379 AGCUGCUGCUGAUGAGGCCGUUAGGCCGAA IGCGCUGG1284
842 CAGCGCCCC AGCAGCUG380 CAGCUGCUCUGAUGAGGCCGUUAGGCCGAA IGGCGCUG1285
843 AGCGCCCCA GCAGCUGC381 GCAGCUGCCUGAUGAGGCCGUUAGGCCGAA IGGGCGCU1286
846 GCCCCAGCA GCUGCAGC382 GCUGCAGCCUGAUGAGGCCGUUAGGCCGAA ICUGGGGC1287
849 CCAGCAGCU GCAGCAGC383 GCUGCUGCCUGAUGAGGCCGUUAGGCCGAA ICUGCUGG1288
852 GCAGCUGCA GCAGCCCC384 GGGGCUGCCUGAUGAGGCCGUUAGGCCGAA ICAGCUGC1289
855 GCUGCAGCA GCCCCCAC385 GUGGGGGCCUGAUGAGGCCGUUAGGCCGAA ICUGCAGC1290
858 GCAGCAGCC CCCACAGC386 GCUGUGGGCUGAUGAGGCCGUUAGGCCGAA ICUGCUGC1291
859 CAGCAGCCC CCACAGCA387 UGCUGUGGCUGAUGAGGCCGUUAGGCCGAA TGCUGCUG1292
860 AGCAGCCCC CACAGCAG388 CUGCUGUGCUGAUGAGGCCGUUAGGCCGAA IGGCUGCU1293
861 GCAGCCCCC ACAGCAGC389 GCUGCUGUCUGAUGAGGCCGUUAGGCCGAA IGGGCUGC1294
862 CAGCCCCCA CAGCAGCG390 CGCUGCUGCUGAUGAGGCCGUUAGGCCGAA IGGGGCUG1295
L ~CCCCCACA GCAGCGAU391 AUCGCUGCCUGAUGAGGCCGUUAGGCCGAA IUGGGGGC1296
864
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
67
867 CCCACAGCA GCGAUAUC392 GAUAUCGCCUGAUGAGGCCGUUAGGCCGAA ICUGUGGG1297
876 GCGAUAUCU GCAGCACC393 GGUGCUGCCUGAUGAGGCCGUUAGGCCGAA IAUAUCGC1298
879 AUAUCUGCA GCACCACC394 GGUGGUGCCUGAUGAGGCCGUUAGGCCGAA ICAGAUAU1299
882 UCUGCAGCA CCACCAW395 AAUGGUGGCUGAUGAGGCCGUUAGGCCGAA ICUGCAGA1300
884 UGCAGCACC ACCAUUUC396 GAAAUGGUCUGAUGAGGCCGUUAGGCCGAA IUGCUGCA1301
885 GCAGCACCA CCAUUUCC397 GGAAAUGGCUGAUGAGGCCGUUAGGCCGAA IGUGCUGC1302
887 AGCACCACC AUUUCCAC398 GUGGAAAUCUGAUGAGGCCGUUAGGCCGAA IUGGUGCU1303
888 GCACCACCA UUUCCACC399 GGUGGAAA GCCGUUAGGCCGAA IGUGGUGC1304
CUGAUGAG
893 ACCAUUUCC ACCAGGAA400 UUCCUGGUCUGAUGAGGCCGUUAGGCCGAA IAAAUGGU1305
894 CCAUUUCCA CCAGGAAC401 GUUCCUGGCUGAUGAGGCCGUUAGGCCGAA IGAAAUGG1306
896 AUUUCCACC AGGAACGC402 GCGUUCCUCUGAUGAGGCCGUUAGGCCGAA IUGGAAAU1307
897 UUUCCACCA GGAACGCC403 GGCGUUCCCUGAUGAGGCCGUUAGGCCGAA IGUGGAAA1308
905 AGGAACGCC GAGGAGGC404 GCCUCCUCCUGAUGAGGCCGUUAGGCCGAA ICGUUCCU1309
914 GAGGAGGCA GCCUUGAC405 GUCAAGGCCUGAUGAGGCCGUUAGGCCGAA ICCUCCUC1310
917 GAGGCAGCC UUGACAUA406 UAUGUCAA GCCGUUAGGCCGAA ICUGCCUC1311
CUGAUGAG
918 AGGCAGCCU UGACAUAA407 UUAUGUCACUGAUGAGGCCGUUAGGCCGAA IGCUGCCU1312
923 GCCUUGACA UAAAUGAU408 AUCAUUUACUGAUGAGGCCGUUAGGCCGAA IUCAAGGC1313
936 UGAUGGGCA UUGUGGCA409 UGCCACAA GCCGUUAGGCCGAA ICCCAUCA1314
CUGAUGAG
944 AUUGUGGCA CCGGCUUG410 CAAGCCGGCUGAUGAGGCCGUUAGGCCGAA ICCACAAU1315
946 UGUGGCACC GGCUUGGG411 CCCAAGCCCUGAUGAGGCCGUUAGGCCGAA IUGCCACA1316
950 GCACCGGCU UGGGCAGU412 ACUGCCCACUGAUGAGGCCGUUAGGCCGAA ICCGGUGC1317
956 GCUUGGGCA GUGAAAUG413 CAUUUCACCUGAUGAGGCCGUUAGGCCGAA ICCCAAGC1318
973 AAUGCGGCC CUCAUGCA414 UGCAUGAGCUGAUGAGGCCGUUAGGCCGAA ICCGCAUU1319
974 AUGCGGCCC UCAUGCAU415 AUGCAUGACUGAUGAGGCCGUUAGGCCGAA IGCCGCAU1320
975 UGCGGCCCU CAUGCAUC416 GAUGCAUGCUGAUGAGGCCGUUAGGCCGAA IGGCCGCA1321
977 CGGCCCUCA UGCAUCGG417 CCGAUGCACUGAUGAGGCCGUUAGGCCGAA IAGGGCCG1322
981 CCUCAUGCA UCGGAGAC418 GUCUCCGACUGAUGAGGCCGUUAGGCCGAA ICAUGAGG1323
990 UCGGAGACA CACAGACC419 GGUCUGUGCUGAUGAGGCCGUUAGGCCGAA IUCUCCGA1324
992 GGAGACACA CAGACCCA420 UGGGUCUGCUGAUGAGGCCGUUAGGCCGAA IUGUCUCC1325
994 AGACACACA GACCCAGU421 ACUGGGUCCUGAUGAGGCCGUUAGGCCGAA IUGUGUCU1326
998 ACACAGACC CAGUGCAG422 CUGCACUGCUGAUGAGGCCGUUAGGCCGAA IUCUGUGU1327
999 CACAGACCC AGUGCAGC423 GCUGCACUCUGAUGAGGCCGUUAGGCCGAA IGUCUGUG1328
1000 ACAGACCCA GUGCAGCU424 AGCUGCACCUGAUGAGGCCGUUAGGCCGAA IGGUCUGU1329
1005 CCCAGUGCA GCUCCAGG425 CCUGGAGCCUGAUGAGGCCGUUAGGCCGAA ICACUGGG1330
1008 AGUGCAGCU CCAGGCGG426 CCGCCUGGCUGAUGAGGCCGUUAGGCCGAA ICUGCACU1331
1010 UGCAGCUCC AGGCGGCA427 UGCCGCCUCUGAUGAGGCCGUUAGGCCGAA IAGCUGCA1332
1011 GCAGCUCCA GGCGGCAG428 CUGCCGCCCUGAUGAGGCCGUUAGGCCGAA IGAGCUGC1333
1018 CAGGCGGCA GGGCGAGU429 ACUCGCCCCUGAUGAGGCCGUUAGGCCGAA ICCGCCUG1334
1036 CGGUGGGCC CGGGCGCU430 AGCGCCCGCUGAUGAGGCCGUUAGGCCGAA ICCCACCG1335
1037 GGUGGGCCC GGGCGCUG431 CAGCGCCCCUGAUGAGGCCGUUAGGCCGAA IGCCCACC1336
1044 CCGGGCGCU GUAUGACU432 AGUCAUACCUGAUGAGGCCGUUAGGCCGAA ICGCCCGG1337
1052 UGUAUGACU UUGAGGCC433 GGCCUCAA GCCGUUAGGCCGAA IUCAUACA1338
CUGAUGAG
1060 UUUGAGGCC CUGGAGGA434 UCCUCCAGCUGAUGAGGCCGUUAGGCCGAA ICCUCAAA1339
1061 UUGAGGCCC UGGAGGAU435 AUCCUCCACUGAUGAGGCCGUUAGGCCGAA IGCCUCAA1340
1062 UGAGGCCCU GGAGGAUG436 CAUCCUCCCUGAUGAGGCCGUUAGGCCGAA IGGCCUCA1341
1077 UGACGAGCU GGGGUUCC437 GGAACCCCCUGAUGAGGCCGUUAGGCCGAA ICUCGUCA1342
1085 UGGGGUUCC ACAGCGGG438 CCCGCUGUCUGAUGAGGCCGUUAGGCCGAA IAACCCCA1343
1086 GGGGUUCCA CAGCGGGG439 CCCCGCUGCUGAUGAGGCCGUUAGGCCGAA IGAACCCC1344
1088 GGUUCCACA GCGGGGAG440 CUCCCCGCCUGAUGAGGCCGUUAGGCCGAA IUGGAACC1345
1109 UGGAGGUCC UGGAUAGC441 GCUAUCCACUGAUGAGGCCGUUAGGCCGAA IACCUCCA1346
1110 GGAGGUCCU GGAUAGCU442 AGCUAUCCCUGAUGAGGCCGUUAGGCCGAA IGACCUCC1347
1118 UGGAUAGCU CCAACCCA443 UGGGUUGGCUGAUGAGGCCGUUAGGCCGAA ICUAUCCA1348
1120 GAUAGCUCC AACCCAUC444 GAUGGGUUCUGAUGAGGCCGUUAGGCCGAA IAGCUAUC1349
1121 AUAGCUCCA ACCCAUCC445 GGAUGGGUCUGAUGAGGCCGUUAGGCCGAA IGAGCUAU1350
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
68
1124 GCUCCAACC CAUCCUGG446 CCAGGAUGCUGAUGAGGCCGUUAGGCCGAA IUUGGAGC1351
1125 CUCCAACCC AUCCUGGU447 ACCAGGAUCUGAUGAGGCCGUUAGGCCGAA IGUUGGAG1352
1126 UCCAACCCA UCCUGGUG445 CACCAGGACUGAUGAGGCCGUUAGGCCGAA IGGUUGGA1353
1129 AACCCAUCC UGGUGGAC449 GUCCACCACUGAUGAGGCCGUUAGGCCGAA IAUGGGUU1354
1130 ACCCAUCCU GGUGGACC450 GGUCCACCCUGAUGAGGCCGUUAGGCCGAA IGAUGGGU1355
1138 UGGUGGACC GGCCGCCU451 AGGCGGCCCUGAUGAGGCCGUUAGGCCGAA IUCCACCA1356
1142 GGACCGGCC GCCUGCAC452 GUGCAGGCCUGAUGAGGCCGUUAGGCCGAA ICCGGUCC1357
1145 CCGGCCGCC UGCACAAC453 GUUGUGCACUGAUGAGGCCGUUAGGCCGAA ICGGCCGG1358
1146 CGGCCGCCU GCACAACA454 UGUUGUGCCUGAUGAGGCCGUUAGGCCGAA IGCGGCCG1359
1149 CCGCCUGCA CAACAAGC455 GCUUGUUGCUGAUGAGGCCGUUAGGCCGAA ICAGGCGG1360
1151 GCCUGCACA ACAAGCUG456 CAGCUUGUCUGAUGAGGCCGUUAGGCCGAA IUGCAGGC1361
1154 UGCACAACA AGCUGGGC457 GCCCAGCUCUGAUGAGGCCGUUAGGCCGAA IUUGUGCA1362
1158 CAACAAGCU GGGCCUCU458 AGAGGCCCCUGAUGAGGCCGUUAGGCCGAA ICUUGUUG1363
1163 AGCUGGGCC UCUUCCCU459 AGGGAAGACUGAUGAGGCCGUUAGGCCGAA ICCCAGCU1364
1164 GCUGGGCCU CUUCCCUG460 CAGGGAAGCUGAUGAGGCCGUUAGGCCGAA IGCCCAGC1365
1166 UGGGCCUCU UCCCUGCC461 GGCAGGGACUGAUGAGGCCGUUAGGCCGAA IAGGCCCA1366
1169 GCCUCUUCC CUGCCAAC462 GUUGGCAGCUGAUGAGGCCGUUAGGCCGAA IAAGAGGC1367
1170 CCUCUUCCC UGCCAACU463 AGUUGGCACUGAUGAGGCCGUUAGGCCGAA IGAAGAGG1368
1171 CUCUUCCCU GCCAACUA464 UAGUUGGCCUGAUGAGGCCGUUAGGCCGAA IGGAAGAG1369
1174 UUCCCUGCC AACUACGU465 ACGUAGUUCUGAUGAGGCCGUUAGGCCGAA ICAGGGAA1370
1175 UCCCUGCCA ACUACGUG466 CACGUAGUCUGAUGAGGCCGUUAGGCCGAA IGCAGGGA1371
1178 CUGCCAACU ACGUGGCA467 UGCCACGUCUGAUGAGGCCGUUAGGCCGAA IUUGGCAG1372
1186 UACGUGGCA CCCAUGAC468 GUCAUGGGCUGAUGAGGCCGUUAGGCCGAA TCCACGUA1373
1188 CGUGGCACC CAUGACCC469 GGGUCAUGCUGAUGAGGCCGUUAGGCCGAA IUGCCACG1374
1189 GUGGCACCC AUGACCCG470 CGGGUCAUCUGAUGAGGCCGUUAGGCCGAA IGUGCCAC1375
1190 UGGCACCCA UGACCCGA471 UCGGGUCACUGAUGAGGCCGUUAGGCCGAA IGGUGCCA1376
1195 CCCAUGACC CGAUAAAC472 GUUUAUCGCUGAUGAGGCCGUUAGGCCGAA IUCAUGGG1377
1196 CCAUGACCC GAUAAACU473 AGUUUAUCCUGAUGAGGCCGUUAGGCCGAA IGUCAUGG1378
1204 CGAUAAACU CUUCAGGG474 CCCUGAAGCUGAUGAGGCCGUUAGGCCGAA IUUUAUCG1379
1206 AUAAACUCU UCAGGGGA475 UCCCCUGACUGAUGAGGCCGUUAGGCCGAA IAGUUUAU1380
1209 AACUCUUCA GGGGACAG476 CUGUCCCCCUGAUGAGGCCGUUAGGCCGAA IAAGAGUU1381
1216 CAGGGGACA GAAGCWU477 AAAGCUUCCUGAUGAGGCCGUUAGGCCGAA IUCCCCUG1382
1222 ACAGAAGCU UUUUGUCU478 AGACAAAA GCCGUUAGGCCGAA ICUUCUGU1383
CUGAUGAG
1230 UUUUUGUCU GGAGCUGC479 GCAGCUCCCUGAUGAGGCCGUUAGGCCGAA IACAAAAA1384
1236 UCUGGAGCU GCCCACAA480 UUGUGGGCCUGAUGAGGCCGUUAGGCCGAA ICUCCAGA1385
1239 GGAGCUGCC CACAAGAA481 UUCUUGUGCUGAUGAGGCCGUUAGGCCGAA ICAGCUCC1386
1240 GAGCUGCCC ACAAGAAA482 UUUCUUGUCUGAUGAGGCCGUUAGGCCGAA IGCAGCUC1387
1241 AGCUGCCCA CAAGAAAG483 CUUUCUUGCUGAUGAGGCCGUUAGGCCGAA IGGCAGCU1388
1243 CUGCCCACA AGAAAGAG484 CUCUUUCUCUGAUGAGGCCGUUAGGCCGAA IUGGGCAG1389
1255 AAGAGGGCA AGGAAAAA485 UUUUUCCUCUGAUGAGGCCGUUAGGCCGAA ICCCUCUU1390
1268 AAAAAGGCU GGACUCCA486 UGGAGUCCCUGAUGAGGCCGUUAGGCCGAA ICCUUUUU1391
1273 GGCUGGACU CCAUGACU487 AGUCAUGGCUGAUGAGGCCGUUAGGCCGAA IUCCAGCC1392
1275 CUGGACUCC AUGACUAU488 AUAGUCAUCUGAUGAGGCCGUUAGGCCGAA IAGUCCAG1393
1276 UGGACUCCA UGACUAUA489 UAUAGUCACUGAUGAGGCCGUUAGGCCGAA IGAGUCCA1394
1281 UCCAUGACU AUAUAUAC490 GUAUAUAUCUGAUGAGGCCGUUAGGCCGAA IUCAUGGA1395
1290 AUAUAUACA UACAUCUA491 UAGAUGUACUGAUGAGGCCGUUAGGCCGAA IUAUAUAU1396
1294 AUACAUACA UCUAUCUA492 UAGAUAGACUGAUGAGGCCGUUAGGCCGAA IUAUGUAU1397
I
Input Sequence = HSA011736. Cut Site = CH/.
Stem Length = 8 . Core Sequence = CUGAUGAG GCCGUUAGGC CGAA
HSA011736 (Homo Sapiens mRNA for growth factor receptor binding protein
(GRBLG); 1303 bp)
Underlined region can be any X sequence or linker as defined herein.
I = Inosine
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
69
Table V: Human GRID G-cleaver Ribozyme and Substrate Sequence
Pos Substrate Se Riboz me Se
ID ID
31 GUAAACUUG CACCCUCU493 AGAGGGUGUGAUGGCAUGCACUAUGCGCG 1398
AAGUUUAC
85 CAUACUCUG AAAUGCAG494 CUGCAUUUUGAUGGCAUGCACUAUGCGCGAGAGUAUG1399
90 UCUGAAAUG CAGUAACU495 AGUUACUGUGAUGGCAUGCACUAUGCGCGAUUUCAGA1400
101 GUAACUCUG AUGCUUGA496 UCAAGCAUUGAUGGCAUGCACUAUGCGCGAGAGUUAC1401
104 ACUCUGAUG CUUGAAUU497 AAUUCAAGUGAUGGCAUGCACUAUGCGCGAUCAGAGU1402
108 UGAUGCUUG AAUUUGUU498 AACAAAUUUGAUGGCAUGCACUAUGCGCG 1403
AAGCAUCA
127 CCCUUCUUG CCAGAAAG499 CUUUCUGGUGAUGGCAUGCACUAUGCGCG 1404
AAGAAGGG
221 AAGCUGUUG CCAAGUUU500 AAACUUGGUGAUGGCAUGCACUAUGCGCGAACAGCUU1405
230 CCAAGUUUG AUUUCACU501 AGUGAAAUUGAUGGCAUGCACUAUGCGCG 1406
AAACUUGG
239 AUUUCACUG CUUCAGGU502 ACCUGAAGUGAUGGCAUGCACUAUGCGCGAGUGAAAU1407
248 CUUCAGGUG AGGAUGAA503 UUCAUCCUUGAUGGCAUGCACUAUGCGCGACCUGAAG1408
254 GUGAGGAUG 504 GCUCAGUUUGAUGGCAUGCACUAUGCGCGAUCCUCAC1409
AACUGAGC
259 GAUGAACUG AGCUUUCA505 UGAAAGCUUGAUGGCAUGCACUAUGCGCGAGUUCAUC1410
283 GAUGUUUUG AAGAUUUU506 AAAAUCUUUGAUGGCAUGCACUAUGCGCG 1411
AAAACAUC
346 GGAUAUGUG CCCAAGAA507 UUCUUGGGUGAUGGCAUGCACUAUGCGCGACAUAUCC1412
389 GGUUUCACG AAGGCCUC508 GAGGCCUUUGAUGGCAUGCACUAUGCGCGGUGAAACC1413
402 CCUCUCUCG ACACCAGG509 CCUGGUGUUGAUGGCAUGCACUAUGCGCGGAGAGAGG1414
503 UCAGGCAUG AGGAUGAC510 GUCAUCCUUGAUGGCAUGCACUAUGCGCGAUGCCUGA1415
509 AUGAGGAUG ACGUUCAA511 UUGAACGUUGAUGGCAUGCACUAUGCGCGAUCCUCAU1416
532 AAGGUCAUG CGAGACAA512 UUGUCUCGUGAUGGCAUGCACUAUGCGCGAUGACCUU1417
534 GGUCAUGCG AGACAACA513 UGUUGUCUUGAUGGCAUGCACUAUGCGCGGCAUGACC1418
566 UGUGGACUG AGAAGUUU514 AAACUUCUUGAUGGCAUGCACUAUGCGCGAGUCCACA1419
657 CAGAACCCG AGAAGACC515 GGUCUUCUUGAUGGCAUGCACUAUGCGCGGGGUUCUG1420
744 AGAAAUCCG ACCUUCGA516 UCGAAGGUUGAUGGCAUGCACUAUGCGCGGGAUUUCU1421
751 CGACCUUCG AUGAACCG517 CGGUUCAUUGAUGGCAUGCACUAUGCGCGGAAGGUCG1422
754 CCUUCGAUG AACCGGAA518 UUCCGGUUUGAUGGCAUGCACUAUGCGCGAUCGAAGG1423
781 CACCCCCCG ACCCUUCC519 GGAAGGGUUGAUGGCAUGCACUAUGCGCGGGGGGGUG1424
793 CUUCCCCUG CAGCAGCA520 UGCUGCUGUGAUGGCAUGCACUAUGCGCGAGGGGAAG1425
823 CAGCCUCCG CAAUAUGC521 GCAUAUUGUGAUGGCAUGCACUAUGCGCGGGAGGCUG1426
830 CGCAAUAUG CCCCAGCG522 CGCUGGGGUGAUGGCAUGCACUAUGCGCGAUAUUGCG1427
838 GCCCCAGCG CCCCAGCA523 UGCUGGGGUGAUGGCAUGCACUAUGCGCGGCUGGGGC1428
850 CAGCAGCUG CAGCAGCC524 GGCUGCUGUGAUGGCAUGCACUAUGCGCGAGCUGCUG1429
870 ACAGCAGCG AUAUCUGC525 GCAGAUAUUGAUGGCAUGCACUAUGCGCGGCUGCUGU1430
877 CGAUAUCUG CAGCACCA526 UGGUGCUGUGAUGGCAUGCACUAUGCGCGAGAUAUCG1431
903 CCAGGAACG CCGAGGAG527 CUCCUCGGUGAUGGCAUGCACUAUGCGCGGUUCCUGG1432
906 GGAACGCCG AGGAGGCA528 UGCCUCCUUGAUGGCAUGCACUAUGCGCG'GGCGUUCC1433
920 GCAGCCUUG ACAUAAAU529 AUUUAUGUUGAUGGCAUGCACUAUGCGCG 1434
AAGGCUGC
929 ACAUAAAUG AUGGGCAU530 AUGCCCAUUGAUGGCAUGCACUAUGCGCGAUUUAUGU1435
959 UGGGCAGUG AAAUGAAU531 AUUCAUUUUGAUGGCAUGCACUAUGCGCGACUGCCCA1436
964 AGUGAAAUG AAUGCGGC532 GCCGCAUUUGAUGGCAUGCACUAUGCGCGAUUUCACU1437
968 AAAUGAAUG CGGCCCUC533 GAGGGCCGUGAUGGCAUGCACUAUGCGCGAUUCAUUU1438
979 GCCCUCAUG CAUCGGAG534 CUCCGAUGUGAUGGCAUGCACUAUGCGCGAUGAGGGC1439
1003GACCCAGUG CAGCUCCA535 UGGAGCUGUGAUGGCAUGCACUAUGCGCGACUGGGUC1440
1023GGCAGGGCG AGUGCGGU536 ACCGCACUUGAUGGCAUGCACUAUGCGCGGCCCUGCC1441
1027GGGCGAGUG CGGUGGGC537 GCCCACCGUGAUGGCAUGCACUAUGCGCGACUCGCCC1442
1042GCCCGGGCG CUGUAUGA538 UCAUACAGUGAUGGCAUGCACUAUGCGCGGCCCGGGC1443
1049CGCUGUAUG ACUUUGAG539 CUCAAAGUUGAUGGCAUGCACUAUGCGCGAUACAGCG1444
1055AUGACUUUG AGGCCCUG540 CAGGGCCUUGAUGGCAUGCACUAUGCGCG 1445
AAAGUCAU
1070UGGAGGAUG ACGAGCUG541 CAGCUCGUUGAUGGCAUGCACUAUGCGCGAUCCUCCA1446
1073AGGAUGACG AGCUGGGG542. CCCCAGCUUGAUGGCAUGCACUAUGCGCGGUCAUCCU1447
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
1143GACCGGCCG CCUGCACA543 UGUGCAGGUGAUGGCAUGCACUAUGCGCGGGCCGGUC1448
1147GGCCGCCUG CACAACAA544 WGWGUG UGAUGGCAUGCACUAUGCGCGAGGCGGCC1449
1172UCUUCCCUG CCAACUAC545 GUAGUUGGUGAUGGCAUGCACUAUGCGCGAGGGAAGA1450
1192GCACCCAUG ACCCGAUA546 UAUCGGGUUGAUGGCAUGCACUAUGCGCGAUGGGUGC1451
1197CAUGACCCG AUAAACUC547 GAGUUUAUUGAUGGCAUGCACUAUGCGCGGGGUCAUG1452
1237CUGGAGCUG CCCACAAG548 CUUGUGGGUGAUGGCAUGCACUAUGCGCGAGCUCCAG1453
1278GACUCCAUG ACUAUAUA549 UAUAUAGUUGAUGGCAUGCACUAUGCGCGAUGGAGUC1454
Input Sequence = HSA011736. Cut Site = YG/M or UG/U.
Stem Length = 8. Core Sequence = UGAUG GCAUGCACUAUGC GCG
HSA011736 (Homo Sapiens mRNA for growth factor receptor binding protein
(GRBLG); 1303 bp)
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
71
Table VI: Human GRID Zlnzyme and Substrate Sequence
Pos Substrate Se Zinz me Se
ID ID
11 GAGGCACAG 550 UCCAUUAA UGUGCCUC1455
UUAAUGGA GCCGAAAGGCGAGUCAAGGUCU
23 AUGGAUCUG 551 CAAGUUUAGCCGAAAGGCGAGUCAAGGUCUAGAUCCAU1456
UAAACUUG
31 GUAAACUUG 493 AGAGGGUGGCCGAAAGGCGAGUCAAGGUCU 1457
CACCCUCU AAGUUUAC
46 CUUUCAGAG 552 AUGUACCAGCCGAAAGGCGAGUCAAGGUCUUCUGAAAG1458
UGGUACAU
49 UCAGAGUGG 553 UCCAUGUAGCCGAAAGGCGAGUCAAGGUCUCACUCUGA1459
UACAUGGA
63 GGAAGACAG 554 ACUUUGUGGCCGAAAGGCGAGUCAAGGUCUUGUCWCC1460
CACAAAGU
70 AGCACAAA UGGAUCCA555 UGGAUCCAGCCGAAAGGCGAGUCAAGGUCUUUUGUGCU1461
G
90 UCUGAAAUG 495 AGUUACUGGCCGAAAGGCGAGUCAAGGUCUAUUUCAGA1462
CAGUAACU
93 GAAAUGCAG 556 CAGAGUUAGCCGAAAGGCGAGUCAAGGUCUUGCAUUUC1463
UAACUCUG
104 ACUCUGAUG 497 AAUUCAAGGCCGAAAGGCGAGUCAAGGUCUAUCAGAGU1464
CUUGAAUU
114 UUGAAUUUG 557 AGGGAGAA 1465
UUCUCCCU GCCGAAAGGCGAGUCAAGGUCU
AAAUUCAA
127 CCCUUCUUG 499 CUUUCUGGGCCGAAAGGCGAGUCAAGGUCU 1466
CCAGAAAG AAGAAGGG
151 AUAACUCGG 558 CUUUGACAGCCGAAAGGCGAGUCAAGGUCUCGAGUUAU1467
UGUCAAAG
153 AACUCGGUG 559 GGCUUUGAGCCGAAAGGCGAGUCAAGGUCUACCGAGUU1468
UCAAAGCC
159 GUGUCAAA CCAAGACA560 UGUCUUGGGCCGAAAGGCGAGUCAAGGUCUUUUGACAC1469
G
194 UUCCAAAA CUUCACGU561 ACGUGAAGGCCGAAAGGCGAGUCAAGGUCUUUUUGGAA1470
G
201 AGCUUCACG 562 UGCUGUAA GUGAAGCU1477.
UUACAGCA GCCGAAAGGCGAGUCAAGGUCU
207 ACGUUACAG 563 CUUCCAUGGCCGAAAGGCGAGUCAAGGUCUUGUAACGU1472
CAUGGAAG
215 GCAUGGAAG 564 GGCAACAGGCCGAAAGGCGAGUCAAGGUCUUUCCAUGC1473
CUGUUGCC
218 UGGAAGCUG 565 CUUGGCAA AGCUUCCA1474
UUGCCAAG GCCGAAAGGCGAGUCAAGGUCU
221 AAGCUGUUG 500 AAACUUGGGCCGAAAGGCGAGUCAAGGUCU 1475
CCAAGUUU AACAGCUU
226 GUUGCCAAG 566 AAAUCAAAGCCGAAAGGCGAGUCAAGGUCUUUGGCAAC1476
UUUGAUUU
239 AUUUCACUG 502 ACCUGAAGGCCGAAAGGCGAGUCAAGGUCUAGUGAAAU1477
CUUCAGGU
246 UGCUUCAGG 567 CAUCCUCAGCCGAAAGGCGAGUCAAGGUCUCUGAAGCA1478
UGAGGAUG
261 UGAACUGAG 568 UGUGAAAGGCCGAAAGGCGAGUCAAGGUCUUCAGUUCA1479
CUUUCACA
278 CUGGAGAUG 569 CUUCAAAAGCCGAAAGGCGAGUCAAGGUCUAUCUCCAG1480
UUUUGAAG
294 GAUUUUAA UAACCAAG570 CUUGGUUAGCCGAAAGGCGAGUCAAGGUCUUUAAAAUC1481
G
307 CAAGAGGAG 571 UUAAACCAGCCGAAAGGCGAGUCAAGGUCUUCCUCUUG1482
UGGUUUAA
310 GAGGAGUGG 572 GCCUUAAA CACUCCUC1483
UUUAAGGC GCCGAAAGGCGAGUCAAGGUCU
317 GGUUUAAGG 573 AAGCUCCGGCCGAAAGGCGAGUCAAGGUCUCUUAAACC1484
CGGAGCUU
322 AAGGCGGAG 574 CUCCCAAGGCCGAAAGGCGAGUCAAGGUCUUCCGCCUU1485
CUUGGGAG
330 GCUUGGGAG 575 CUUCCUGGGCCGAAAGGCGAGUCAAGGUCUUCCCAAGC1486
CCAGGAAG
344 AAGGAUAUG 576 CUUGGGCAGCCGAAAGGCGAGUCAAGGUCUAUAUCCUU7.487
UGCCCAAG
346 GGAUAUGUG 507 UUCUUGGGGCCGAAAGGCGAGUCAAGGUCUACAUAUCC1488
CCCAAGAA
370 GACAUCCAG 577 UUGGGAAAGCCGAAAGGCGAGUCAAGGUCUUGGAUGUC1489
UUUCCCAA
382 CCCAAAUGG 578 UCGUGAAAGCCGAAAGGCGAGUCAAGGUCUCAUUUGGG1490
UUUCACGA
393 UCACGAAGG 579 GAGAGAGGGCCGAAAGGCGAGUCAAGGUCUCUUCGUGA1491
CCUCUCUC
410 GACACCAGG 580 GUUCUCUGGCCGAAAGGCGAGUCAAGGUCUCUGGUGUC1492
CAGAGAAC
429 ACUCAUGGG 581 CCUCCUUGGCCGAAAGGCGAGUCAAGGUCUCCAUGAGU1493
CAAGGAGG
437 GCAAGGAGG 582 GAAGCCAA CUCCUUGC1494
UUGGCUUC GCCGAAAGGCGAGUCAAGGUCU
441 GGAGGUUGG 583 UGAAGAAGGCCGAAAGGCGAGUCAAGGUCUCAACCUCC1495
CUUCUUCA
458 UCAUCCGGG 584 CUGGCUGGGCCGAAAGGCGAGUCAAGGUCUCCGGAUGA1496
CCAGCCAG
462 CCGGGCCAG 585 AGCUCUGGGCCGAAAGGCGAGUCAAGGUCUUGGCCCGG1497
CCAGAGCU
468 CAGCCAGAG 586 CUGGGGAGGCCGAAAGGCGAGUCAAGGUCUUCUGGCUG1498
CUCCCCAG
494 CCAUCUCUG 587 AUGCCUGAGCCGAAAGGCGAGUCAAGGUCUAGAGAUGG1499
UCAGGCAU
499 UCUGUCAGG 588 UCCUCAUGGCCGAAAGGCGAGUCAAGGUCUCUGACAGA1500
CAUGAGGA
512 AGGAUGACG 589 GUGUUGAA GUCAUCCU1501
UUCAACAC GCCGAAAGGCGAGUCAAGGUCU
527 ACUUCAAGG 590 UCGCAUGAGCCGAAAGGCGAGUCAAGGUCUCUUGAAGU1502
UCAUGCGA
532 AAGGUCAUG 512 UUGUCUCGGCCGAAAGGCGAGUCAAGGUCUAUGACCUU1503
CGAGACAA
546 CAACAAGGG 591 AGUAAUUAGCCGAAAGGCGAGUCAAGGUCUCCUUGUUG1504
UAAUUACU
559 UACUUUCUG 592 UCAGUCCAGCCGAAAGGCGAGUCAAGGUCUAGAAAGUA1505
UGGACUGA
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
72
571 ACUGAGAA UUUCCAUC593 GAUGGAAA UUCUCAGU1506
G GCCGAAAGGCGAGUCAAGGUCU
589 CUAAAUAA CUGGUAGAS94 UCUACCAGGCCGAAAGGCGAGUCAAGGUCUWAUUUAG 1507
G
593 AUAAGCUGG S95 GUAGUCUAGCCGAAAGGCGAGUCAAGGUCUCAGCUUAU1508
UAGACUAC
631 AGACAGAA CAGAUCUU596 AAGAUCUGGCCGAAAGGCGAGUCAAGGUCUUUCUGUCU1509
G
669 AGACCAGGG 597 CCCGGUGAGCCGAAAGGCGAGUCAAGGUCUCCUGGUCU1510
UCACCGGG
678 UCACCGGGG 598 GGCUGUUGGCCGAAAGGCGAGUCAAGGUCUCCCGGUGA1511
CAACAGCC
684 GGGCAACAG S99 GGUCCAGGGCCGAAAGGCGAGUCAAGGUCUUGUUGCCC1512
CCUGGACC
697 GACCGGAGG 600 CCCUGGGAGCCGAAAGGCGAGUCAAGGUCUCUCCGGUC1513
UCCCAGGG
708 CCAGGGAGG 601 GGUGUGGGGCCGAAAGGCGAGUCAAGGUCUCUCCCUGG1514
CCCACACC
720 ACACCUCAG 602 CAGCCCCAGCCGAAAGGCGAGUCAAGGUCUUGAGGUGU1515
UGGGGCUG
725 UCAGUGGGG 603 UCCCACAGGCCGAAAGGCGAGUCAAGGUCUCCCACUGA1516
CUGUGGGA
728 GUGGGGCUG 604 UUCUCCCAGCCGAAAGGCGAGUCAAGGUCUAGCCCCAC1517
UGGGAGAA
763 AACCGGAA CUGUCGGA605 UCCGACAGGCCGAAAGGCGAGUCAAGGUCUUUCCGGUU1518
G
766 CGGAAGCUG 606 UGAUCCGAGCCGAAAGGCGAGUCAAGGUCUAGCUUCCG1519
UCGGAUCA
793 CUUCCCCUG 520 UGCUGCUGGCCGAAAGGCGAGUCAAGGUCUAGGGGAAG1520
CAGCAGCA
796 CCCCUGCAG 607 UGGUGCUGGCCGAAAGGCGAGUCAAGGUCUUGCAGGGG1521
CAGCACCA
799 CUGCAGCAG 608 UGCUGGUGGCCGAAAGGCGAGUCAAGGUCUUGCUGCAG1522
CACCAGCA
805 CAGCACCAG 609 GGCUGGUGGCCGAAAGGCGAGUCAAGGUCUUGGUGCUG1523
CACCAGCC
811 CAGCACCAG 610 GGCUGUGGGCCGAAAGGCGAGUCAAGGUCUUGGUGCUG1524
CCACAGCC
817 CAGCCACAG 611 UGCGGAGGGCCGAAAGGCGAGUCAAGGUCUUGUGGCUG1525
CCUCCGCA
823 CAGCCUCCG 521 GCAUAUUGGCCGAAAGGCGAGUCAAGGUCUGGAGGCUG1526
CAAUAUGC
830 CGCAAUAUG 522 CGCUGGGGGCCGAAAGGCGAGUCAAGGUCUAUAUUGCG1527
CCCCAGCG
836 AUGCCCCAG 612 CUGGGGCGGCCGAAAGGCGAGUCAAGGUCUUGGGGCAU1528
CGCCCCAG
838 GCCCCAGCG 523 UGCUGGGGGCCGAAAGGCGAGUCAAGGUCUGCUGGGGC1529
CCCCAGCA
844 GCGCCCCAG 613 UGCAGCUGGCCGAAAGGCGAGUCAAGGUCUUGGGGCGC1530
CAGCUGCA
847 CCCCAGCAG 614 UGCUGCAGGCCGAAAGGCGAGUCAAGGUCUUGCUGGGG1531
CUGCAGCA
850 CAGCAGCUG 524 GGCUGCUGGCCGAAAGGCGAGUCAAGGUCUAGCUGCUG1532
CAGCAGCC
853 CAGCUGCAG 615 GGGGGCUGGCCGAAAGGCGAGUCAAGGUCUUGCAGCUG1533
CAGCCCCC
856 CUGCAGCAG 616 UGUGGGGGGCCGAAAGGCGAGUCAAGGUCUUGCUGCAG1534
CCCCCACA
865 CCCCCACAG 617 UAUCGCUGGCCGAAAGGCGAGUCAAGGUCUUGUGGGGG1535
CAGCGAUA
868 CCACAGCAG 618 AGAUAUCGGCCGAAAGGCGAGUCAAGGUCUUGCUGUGG1536
CGAUAUCU
877 CGAUAUCUG 526 UGGUGCUGGCCGAAAGGCGAGUCAAGGUCUAGAUAUCG1537
CAGCACCA
880 UAUCUGCAG 619 UGGUGGUGGCCGAAAGGCGAGUCAAGGUCUUGCAGAUA1538
CACCACCA
903 CCAGGAACG 527 CUCCUCGGGCCGAAAGGCGAGUCAAGGUCUGUUCCUGG1539
CCGAGGAG
912 CCGAGGAGG 620 CAAGGCUGGCCGAAAGGCGAGUCAAGGUCUCUCCUCGG1540
CAGCCUUG
915 AGGAGGCAG 621 UGUCAAGGGCCGAAAGGCGAGUCAAGGUCUUGCCUCCU1541
CCUUGACA
934 AAUGAUGGG 622 CCACAAUGGCCGAAAGGCGAGUCAAGGUCUCCAUCAUU1542
CAUUGUGG
939 UGGGCAUUG 623 CGGUGCCAGCCGAAAGGCGAGUCAAGGUCU 1543
UGGCACCG AAUGCCCA
942 GCAUUGUGG 624 AGCCGGUGGCCGAAAGGCGAGUCAAGGUCUCACAAUGC1544
CACCGGCU
948 UGGCACCGG 625 UGCCCAAGGCCGAAAGGCGAGUCAAGGUCUCGGUGCCA1545
CUUGGGCA
954 CGGCUUGGG 626 UUUCACUGGCCGAAAGGCGAGUCAAGGUCUCCAAGCCG1546
CAGUGAAA
957 CUUGGGCAG 627 UCAUUUCAGCCGAAAGGCGAGUCAAGGUCUUGCCCAAG1547
UGAAAUGA
968 AAAUGAAUG 533 GAGGGCCGGCCGAAAGGCGAGUCAAGGUCUAUUCAUUU1548
CGGCCCUC
971 UGAAUGCGG 628 CAUGAGGGGCCGAAAGGCGAGUCAAGGUCUCGCAUUCA1549
CCCUCAUG
979 GCCCUCAUG 534 CUCCGAUGGCCGAAAGGCGAGUCAAGGUCUAUGAGGGC1550
CAUCGGAG
1001 CAGACCCAG 629 GAGCUGCAGCCGAAAGGCGAGUCAAGGUCUUGGGUCUG1551
UGCAGCUC
1003 GACCCAGUG 535 UGGAGCUGGCCGAAAGGCGAGUCAAGGUCUACUGGGUC1552
CAGCUCCA
1006 CCAGUGCAG 630 GCCUGGAGGCCGAAAGGCGAGUCAAGGUCUUGCACUGG1553
CUCCAGGC
1013 AGCUCCAGG 631 CCCUGCCGGCCGAAAGGCGAGUCAAGGUCUCUGGAGCU1554
CGGCAGGG
1016 UCCAGGCGG 632 UCGCCCUGGCCGAAAGGCGAGUCAAGGUCUCGCCUGGA1555
CAGGGCGA
1021 GCGGCAGGG 633 CGCACUCGGCCGAAAGGCGAGUCAAGGUCUCCUGCCGC1556
CGAGUGCG
1025 CAGGGCGAG 634 CCACCGCAGCCGAAAGGCGAGUCAAGGUCUUCGCCCUG1557
UGCGGUGG
1027 GGGCGAGUG 537 GCCCACCGGCCGAAAGGCGAGUCAAGGUCUACUCGCCC1558
CGGUGGGC
1030 CGAGUGCGG 635 CGGGCCCAGCCGAAAGGCGAGUCAAGGUCUCGCACUCG1559
I UGGGCCCG
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
73
1034 UGCGGUGGG 636 CGCCCGGGGCCGAAAGGCGAGUCAAGGUCUCCACCGCA1560
CCCGGGCG
1040 GGGCCCGGG 637 AUACAGCGGCCGAAAGGCGAGUCAAGGUCUCCGGGCCC1561
CGCUGUAU
1042 GCCCGGGCG 538 UCAUACAGGCCGAAAGGCGAGUCAAGGUCUGCCCGGGC1562
CUGUAUGA
1045 CGGGCGCUG 638 AAGUCAUAGCCGAAAGGCGAGUCAAGGUCUAGCGCCCG1563
UAUGACUU
1058 ACUUUGAGG 639 CUCCAGGGGCCGAAAGGCGAGUCAAGGUCUCUCAAAGU1564
CCCUGGAG
1075 GAUGACGAG 640 AACCCCAGGCCGAAAGGCGAGUCAAGGUCUUCGUCAUC1565
CUGGGGUU
1081 GAGCUGGGG 641 CUGUGGAA CCCAGCUC1566
UUCCACAG GCCGAAAGGCGAGUCAAGGUCU
1089 GUUCCACAG 642 CCUCCCCGGCCGAAAGGCGAGUCAAGGUCUUGUGGAAC1567
CGGGGAGG
1097 GCGGGGAGG 643 CUCCACCAGCCGAAAGGCGAGUCAAGGUCUCUCCCCGC1568
UGGUGGAG
1100 GGGAGGUGG 644 GACCUCCAGCCGAAAGGCGAGUCAAGGUCUCACCUCCC1569
UGGAGGUC
1106 UGGUGGAGG 645 AUCCAGGAGCCGAAAGGCGAGUCAAGGUCUCUCCACCA1570
UCCUGGAU
1116 CCUGGAUAG 646 GGUUGGAGGCCGAAAGGCGAGUCAAGGUCUUAUCCAGG1571
CUCCAACC
1132 CCAUCCUGG 647 CCGGUCCAGCCGAAAGGCGAGUCAAGGUCUCAGGAUGG1572
UGGACCGG
1140 GUGGACCGG 648 GCAGGCGGGCCGAAAGGCGAGUCAAGGUCUCGGUCCAC1573
CCGCCUGC
1143 GACCGGCCG 543 UGUGCAGGGCCGAAAGGCGAGUCAAGGUCUGGCCGGUC1574
CCUGCACA
1147 GGCCGCCUG 544 UUGUUGUGGCCGAAAGGCGAGUCAAGGUCUAGGCGGCC1575
CACAACAA
1156 CACAACAA CUGGGCCU649 AGGCCCAGGCCGAAAGGCGAGUCAAGGUCUUUGUUGUG1576
G
1161 CAAGCUGGG 650 GGAAGAGGGCCGAAAGGCGAGUCAAGGUCUCCAGCUUG1577
CCUCUUCC
1172 UCUUCCCUG 545 GUAGUUGGGCCGAAAGGCGAGUCAAGGUCUAGGGAAGA1578
CCAACUAC
1181 CCAACUACG 651 GGGUGCCAGCCGAAAGGCGAGUCAAGGUCUGUAGUUGG1579
UGGCACCC
1184 ACUACGUGG 652 CAUGGGUGGCCGAAAGGCGAGUCAAGGUCUCACGUAGU1580
CACCCAUG
1220 GGACAGAA CUUUUUGU653 ACAAAAAGGCCGAAAGGCGAGUCAAGGUCUUUCUGUCC1581
G
1227 AGCUUUUUG 654 GCUCCAGAGCCGAAAGGCGAGUCAAGGUCU 1582
UCUGGAGC AAAAAGCU
1234 UGUCUGGAG 655 GUGGGCAGGCCGAAAGGCGAGUCAAGGUCUUCCAGACA1583
CUGCCCAC
1237 CUGGAGCUG 548 CUUGUGGGGCCGAAAGGCGAGUCAAGGUCUAGCUCCAG1584
CCCACAAG
1253 GAAAGAGGG 656 UUUCCUUGGCCGAAAGGCGAGUCAAGGUCUCCUCUUUC1585
CAAGGAAA
1266 GAAAAAAGG 657 GAGUCCAGGCCGAAAGGCGAGUCAAGGUCUCUUUUUUC1586
I CUGGACUC
I
Input Sequence = HSA011736. Cut Site = G/Y
Stem Length = 8 . Core Sequence = GCcgaaagGCGaGuCaaGGuCu
HSA011736 (Homo sapiens mRNA for growth factor receptor binding protein
(GRBLG); 1303 bp)
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
74
Table VII: Human GRID DNAzyme and Substrate Sequence
Pos Substrate Se DNAz me Se ID
ID
11 GAGGCACAG 550 TCCATTAA TGTGCCTC1587
UUAAUGGA GGCTAGCTACAACGA
15 CACAGUUAA 658 CAGATCCAGGCTAGCTACAACGATAACTGTG1588
UGGAUCUG
19 GUUAAUGGA 659 TTTACAGAGGCTAGCTACAACGACCATTAAC1589
UCUGUAAA
23 AUGGAUCUG 551 CAAGTTTAGGCTAGCTACAACGAAGATCCAT1590
UAAACUUG
27 AUCUGUAA CUUGCACC660 GGTGCAAGGGCTAGCTACAACGATTACAGAT1591
A
31 GUAAACUUG 493 AGAGGGTGGGCTAGCTACAACGA 1592
CACCCUCU AAGTTTAC
33 AAACUUGCA 183 AAAGAGGGGGCTAGCTACAACGAGCAAGTTT1593
CCCUCUUU
46 CUUUCAGAG 552 ATGTACCAGGCTAGCTACAACGATCTGAAAG1594
UGGUACAU
49 UCAGAGUGG 553 TCCATGTAGGCTAGCTACAACGACACTCTGA1595
UACAUGGA
51 AGAGUGGUA 10 CTTCCATGGGCTAGCTACAACGAACCACTCT1596
CAUGGAAG
53 AGUGGUACA 189 GTCTTCCAGGCTAGCTACAACGAGTACCACT1597
UGGAAGAC
60 CAUGGAAGA 661 TTGTGCTGGGCTAGCTACAACGACTTCCATG1598
CAGCACAA
63 GGAAGACAG 554 ACTTTGTGGGCTAGCTACAACGATGTCTTCC1599
CACAAAGU
65 AAGACAGCA 7.91 CCACTTTGGGCTAGCTACAACGAGCTGTCTT1600
CAAAGUGG
70 AGCACAAA UGGAUCCA555 TGGATCCAGGCTAGCTACAACGATTTGTGCT1601
G
74 CAAAGUGGA 662 AGTATGGAGGCTAGCTACAACGACCACTTTG1602
UCCAUACU
78 GUGGAUCCA 194 TCAGAGTAGGCTAGCTACAACGAGGATCCAC1603
UACUCUGA
80 GGAUCCAUA 12 TTTCAGAGGGCTAGCTACAACGAATGGATCC1604
CUCUGAAA
88 ACUCUGAAA 663 TTACTGCAGGCTAGCTACAACGATTCAGAGT1605
UGCAGUAA
90 UCUGAAAUG 495 AGTTACTGGGCTAGCTACAACGAATTTCAGA1606
CAGUAACU
93 GAAAUGCAG 556 CAGAGTTAGGCTAGCTACAACGATGCATTTC1607
UAACUCUG
96 AUGCAGUAA 664 CATCAGAGGGCTAGCTACAACGATACTGCAT1608
CUCUGAUG
102 UAACUCUGA 665 TTCAAGCAGGCTAGCTACAACGACAGAGTTA1609
UGCUUGAA
104 ACUCUGAUG 497 AATTCAAGGGCTAGCTACAACGAATCAGAGT1610
CUUGAAUU
110 AUGCUUGAA 666 AGAACAAA TCAAGCAT1611
UUUGUUCU GGCTAGCTACAACGA
114 UUGAAUUUG 557 AGGGAGAAGGCTAGCTACAACGA 1612
UUCUCCCU AAATTCAA
127 CCCUUCUUG 499 CTTTCTGGGGCTAGCTACAACGA 1613
CCAGAAAG AAGAAGGG
137 CAGAAAGGA 667 TATTAGAA CCTTTCTG1614
UUCUAAUA GGCTAGCTACAACGA
143 GGAUUCUAA 668 CCGAGTTAGGCTAGCTACAACGATAGAATCC1615
UAACUCGG
146 UUCUAAUAA 669 ACACCGAGGGCTAGCTACAACGATATTAGAA1616
CUCGGUGU
151 AUAACUCGG 558 CTTTGACAGGCTAGCTACAACGACGAGTTAT1617
UGUCAAAG
153 AACUCGGUG 559 GGCTTTGAGGCTAGCTACAACGAACCGAGTT1618
UCAAAGCC
159 GUGUCAAA CCAAGACA560 TGTCTTGGGGCTAGCTACAACGATTTGACAC1619
G
165 AAGCCAAGA 670 AGTTTATGGGCTAGCTACAACGACTTGGCTT1620
CAUAAACU
167 GCCAAGACA 213 TGAGTTTAGGCTAGCTACAACGAGTCTTGGC1621
UAAACUCA
171 AGACAUAA CUCAAUCU671 AGATTGAGGGCTAGCTACAACGATTATGTCT1622
A
176 UAAACUCAA 672 AGAAGAGAGGCTAGCTACAACGATGAGTTTA1623
UCUCUUCU
194 UUCCAAAA CUUCACGU561 ACGTGAAGGGCTAGCTACAACGATTTTGGAA1624
G
199 AAAGCUUCA 223 CTGTAACGGGCTAGCTACAACGAGAAGCTTT1625
CGUUACAG
201 AGCUUCACG 562 TGCTGTAA GTGAAGCT1626
UUACAGCA GGCTAGCTACAACGA
204 UUCACGUUA 43 CCATGCTGGGCTAGCTACAACGA 1627
CAGCAUGG AACGTGAA
207 ACGUUACAG 563 CTTCCATGGGCTAGCTACAACGATGTAACGT1628
CAUGGAAG
209 GUUACAGCA 225 AGCTTCCAGGCTAGCTACAACGAGCTGTAAC1629
UGGAAGCU
215 GCAUGGAAG 564 GGCAACAGGGCTAGCTACAACGATTCCATGC1630
CUGUUGCC
218 UGGAAGCUG 565 CTTGGCAAGGCTAGCTACAACGAAGCTTCCA1631
UUGCCAAG
221 AAGCUGUUG 500 AAACTTGGGGCTAGCTACAACGA 1632
CCAAGUUU AACAGCTT
226 GUUGCCAA UUUGAUUU566 AAATCAAA TTGGCAAC1633
G GGCTAGCTACAACGA
231 CAAGUUUGA 673 CAGTGAAA CAAACTTG1634
UUUCACUG GGCTAGCTACAACGA
236 UUGAUUUCA 229 TGAAGCAGGGCTAGCTACAACGAGAAATCAA1635
CUGCUUCA
239 AUUUCACUG 502 ACCTGAAGGGCTAGCTACAACGAAGTGAAAT1636
CUUCAGGU
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
246 UGCUUCAGG UGAGGAUG567 CATCCTCAGGCTAGCTACAACGACTGAAGCA1637
252 AGGUGAGGA UGAACUGA674 TCAGTTCAGGCTAGCTACAACGACCTCACCT1638
256 GAGGAUGAA CUGAGCUU675 AAGCTCAGGGCTAGCTACAACGATCATCCTC1639
261 UGAACUGAG CUUUCACA568 TGTGAAAGGGCTAGCTACAACGATCAGTTCA1640
267 GAGCUUUCA CACUGGAG235 CTCCAGTGGGCTAGCTACAACGAGAAAGCTC1641
269 GCUUUCACA CUGGAGAU236 ATCTCCAGGGCTAGCTACAACGAGTGAAAGC1642
276 CACUGGAGA UGUUUUGA676 TCAAAACAGGCTAGCTACAACGACTCCAGTG1643
278 CUGGAGAUG UUUUGAAG569 CTTCAAAAGGCTAGCTACAACGAATCTCCAG1644
287 UUUUGAAGA UUUUAAGU677 ACTTAAAA CTTCAAAA1645
GGCTAGCTACAACGA
294 GAUUUUAA UAACCAAG570 CTTGGTTAGGCTAGCTACAACGATTAAAATC1646
G
297 UUUAAGUAA CCAAGAGG678 CCTCTTGGGGCTAGCTACAACGATACTTAAA1647
307 CAAGAGGAG UGGUUUAA571 TTAAACCAGGCTAGCTACAACGATCCTCTTG1648
310 GAGGAGUGG UUUAAGGC572 GCCTTAAA CACTCCTC1649
GGCTAGCTACAACGA
317 GGUUUAAGG CGGAGCUU573 AAGCTCCGGGCTAGCTACAACGACTTAAACC1650
322 AAGGCGGAG CUUGGGAG574 CTCCCAAGGGCTAGCTACAACGATCCGCCTT1651
330 GCUUGGGAG CCAGGAAG575 CTTCCTGGGGCTAGCTACAACGATCCCAAGC1652
340 CAGGAAGGA UAUGUGCC679 GGCACATAGGCTAGCTACAACGACCTTCCTG1653
342 GGAAGGAUA UGUGCCCA67 TGGGCACAGGCTAGCTACAACGAATCCTTCC1654
344 AAGGAUAUG UGCCCAAG576 CTTGGGCAGGCTAGCTACAACGAATATCCTT1655
346 GGAUAUGUG CCCAAGAA507 TTCTTGGGGGCTAGCTACAACGAACATATCC1656
354 GCCCAAGAA UUUCAUAG680 CTATGAAA TCTTGGGC1657
GGCTAGCTACAACGA
359 AGAAUUUCA UAGACAUC246 GATGTCTAGGCTAGCTACAACGAGAAATTCT1658
363 UUUCAUAGA CAUCCAGU681 ACTGGATGGGCTAGCTACAACGACTATGAAA1659
365 UCAUAGACA UCCAGUUU247 AAACTGGAGGCTAGCTACAACGAGTCTATGA1660
370 GACAUCCAG UUUCCCAA577 TTGGGAAAGGCTAGCTACAACGATGGATGTC1661
379 UUUCCCAA UGGUUUCA682 TGAAACCAGGCTAGCTACAACGATTGGGAAA1662
A
382 CCCAAAUGG UUUCACGA578 TCGTGAAAGGCTAGCTACAACGACATTTGGG1663
387 AUGGUUUCA CGAAGGCC253 GGCCTTCGGGCTAGCTACAACGAGAAACCAT1664
393 UCACGAAGG CCUCUCUC579 GAGAGAGGGGCTAGCTACAACGACTTCGTGA1665
403 CUCUCUCGA CACCAGGC683 GCCTGGTGGGCTAGCTACAACGACGAGAGAG1666
405 CUCUCGACA CCAGGCAG258 CTGCCTGGGGCTAGCTACAACGAGTCGAGAG1667
410 GACACCAGG CAGAGAAC580 GTTCTCTGGGCTAGCTACAACGACTGGTGTC1668
417 GGCAGAGAA CUUACUCA684 TGAGTAAGGGCTAGCTACAACGATCTCTGCC1669
421 GAGAACUUA CUCAUGGG83 CCCATGAGGGCTAGCTACAACGA 1670
AAGTTCTC
425 ACUUACUCA UGGGCAAG264 CTTGCCCAGGCTAGCTACAACGAGAGTAAGT1671
429 ACUCAUGGG CAAGGAGG581 CCTCCTTGGGCTAGCTACAACGACCATGAGT1672
437 GCAAGGAGG UUGGCUUC582 GAAGCCAA CTCCTTGC1673
GGCTAGCTACAACGA
441 GGAGGUUGG CUUCUUCA583 TGAAGAAGGGCTAGCTACAACGACAACCTCC1674
449 GCUUCUUCA UCAUCCGG268 CCGGATGAGGCTAGCTACAACGAGAAGAAGC1675
452 UCUUCAUCA UCCGGGCC269 GGCCCGGAGGCTAGCTACAACGAGATGAAGA1676
458 UCAUCCGGG CCAGCCAG584 CTGGCTGGGGCTAGCTACAACGACCGGATGA1677
462 CCGGGCCAG CCAGAGCU585 AGCTCTGGGGCTAGCTACAACGATGGCCCGG1678
468 CAGCCAGAG CUCCCCAG586 CTGGGGAGGGCTAGCTACAACGATCTGGCTG1679
480 CCCAGGGGA CUUCUCCA685 TGGAGAAGGGCTAGCTACAACGACCCCTGGG1680
488 ACUUCUCCA UCUCUGUC283 GACAGAGAGGCTAGCTACAACGAGGAGAAGT1681
494 CCAUCUCUG UCAGGCAU587 ATGCCTGAGGCTAGCTACAACGAAGAGATGG1682
499 UCUGUCAGG CAUGAGGA588 TCCTCATGGGCTAGCTACAACGACTGACAGA1683
501 UGUCAGGCA UGAGGAUG287 CATCCTCAGGCTAGCTACAACGAGCCTGACA1684
507 GCAUGAGGA UGACGUUC686 GAACGTCAGGCTAGCTACAACGACCTCATGC1685
510 UGAGGAUGA CGUUCAAC687 GTTGAACGGGCTAGCTACAACGACATCCTCA1686
512 AGGAUGACG UUCAACAC589 GTGTTGAAGGCTAGCTACAACGAGTCATCCT1687
517 GACGUUCAA CACUUCAA688 TTGAAGTGGGCTAGCTACAACGATGAACGTC1688
519 CGUUCAACA CUUCAAGG289 CCTTGAAGGGCTAGCTACAACGAGTTGAACG1689
527 ACUUCAAGG UCAUGCGA590 TCGCATGAGGCTAGCTACAACGACTTGAAGT1690
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
76
530 UCAAGGUCA 292 GTCTCGCAGGCTAGCTACAACGAGACCTTGA1691
UGCGAGAC
532 AAGGUCAUG 512 TTGTCTCGGGCTAGCTACAACGAATGACCTT1692
CGAGACAA
537 CAUGCGAGA 689 CCTTGTTGGGCTAGCTACAACGACTCGCATG1693
CAACAAGG
540 GCGAGACAA 690 TACCCTTGGGCTAGCTACAACGATGTCTCGC1694
CAAGGGUA
546 CAACAAGGG 591 AGTAATTAGGCTAGCTACAACGACCTTGTTG1695
UAAUUACU
549 CAAGGGUAA 691 GAAAGTAA TACCCTTG1696
UUACUUUC GGCTAGCTACAACGA
552 GGGUAAUUA 106 ACAGAAAGGGCTAGCTACAACGA 1697
CUUUCUGU AATTACCC
559 UACUUUCUG 592 TCAGTCCAGGCTAGCTACAACGAAGAAAGTA1698
UGGACUGA
563 UUCUGUGGA 692 CTTCTCAGGGCTAGCTACAACGACCACAGAA1699
CUGAGAAG
571 ACUGAGAA UUUCCAUC593 GATGGAAAGGCTAGCTACAACGATTCTCAGT1700
G
577 AAGUUUCCA 299 TTTAGGGAGGCTAGCTACAACGAGGAAACTT1701
UCCCUAAA
585 AUCCCUAAA 693 CCAGCTTAGGCTAGCTACAACGATTAGGGAT1702
UAAGCUGG
589 CUAAAUAA CUGGUAGA594 TCTACCAGGGCTAGCTACAACGATTATTTAG1703
G
593 AUAAGCUGG 595 GTAGTCTAGGCTAGCTACAACGACAGCTTAT1704
UAGACUAC
597 GCUGGUAGA 694 TGTAGTAGGGCTAGCTACAACGACTACCAGC1705
CUACUACA
600 GGUAGACUA 117 TCCTGTAGGGCTAGCTACAACGAAGTCTACC1706
CUACAGGA
603 AGACUACUA 118 TTGTCCTGGGCTAGCTACAACGAAGTAGTCT1707
CAGGACAA
608 ACUACAGGA 695 GGAATTTGGGCTAGCTACAACGACCTGTAGT1708
CAAAUUCC
612 CAGGACAAA 696 AGATGGAA TTGTCCTG1709
UUCCAUCU GGCTAGCTACAACGA
617 CAAAUUCCA 309 TCTGGAGAGGCTAGCTACAACGAGGAATTTG1710
UCUCCAGA
625 AUCUCCAGA 697 TGCTTCTGGGCTAGCTACAACGACTGGAGAT1711
CAGAAGCA
631 AGACAGAAG 596 AAGATCTGGGCTAGCTACAACGATTCTGTCT1712
CAGAUCUU
635 AGAAGCAGA 698 AAGGAAGAGGCTAGCTACAACGACTGCTTCT1713
UCUUCCUU
648 CCUUAGAGA 699 GGGTTCTGGGCTAGCTACAACGACTCTAAGG1714
CAGAACCC
653 GAGACAGAA 700 TTCTCGGGGGCTAGCTACAACGATCTGTCTC1715
CCCGAGAA
663 CCGAGAAGA 701 GACCCTGGGGCTAGCTACAACGACTTCTCGG1716
CCAGGGUC
669 AGACCAGGG 597 CCCGGTGAGGCTAGCTACAACGACCTGGTCT1717
UCACCGGG
672 CCAGGGUCA 323 TGCCCCGGGGCTAGCTACAACGAGACCCTGG1718
CCGGGGCA
678 UCACCGGGG 598 GGCTGTTGGGCTAGCTACAACGACCCGGTGA1719
CAACAGCC
681 CCGGGGCAA 702 CCAGGCTGGGCTAGCTACAACGATGCCCCGG1720
CAGCCUGG
684 GGGCAACAG 599 GGTCCAGGGGCTAGCTACAACGATGTTGCCC1721
CCUGGACC
690 CAGCCUGGA 703 ACCTCCGGGGCTAGCTACAACGACCAGGCTG1722
CCGGAGGU
697 GACCGGAGG 600 CCCTGGGAGGCTAGCTACAACGACTCCGGTC1723
UCCCAGGG
708 CCAGGGAGG 601 GGTGTGGGGGCTAGCTACAACGACTCCCTGG1724
CCCACACC
712 GGAGGCCCA 335 CTGAGGTGGGCTAGCTACAACGAGGGCCTCC1725
CACCUCAG
714 AGGCCCACA 336 CACTGAGGGGCTAGCTACAACGAGTGGGCCT1726
CCUCAGUG
720 ACACCUCAG 602 CAGCCCCAGGCTAGCTACAACGATGAGGTGT1727
UGGGGCUG
725 UCAGUGGGG 603 TCCCACAGGGCTAGCTACAACGACCCACTGA1728
CUGUGGGA
728 GUGGGGCUG 604 TTCTCCCAGGCTAGCTACAACGAAGCCCCAC1729
UGGGAGAA
740 GAGAAGAA UCCGACCU704 AGGTCGGAGGCTAGCTACAACGATTCTTCTC1730
A
745 GAAAUCCGA 705 ATCGAAGGGGCTAGCTACAACGACGGATTTC1731
CCUUCGAU
752 GACCUUCGA 706 CCGGTTCAGGCTAGCTACAACGACGAAGGTC1732
UGAACCGG
756 UUCGAUGAA 707 GCTTCCGGGGCTAGCTACAACGATCATCGAA1733
CCGGAAGC
763 AACCGGAAG 605 TCCGACAGGGCTAGCTACAACGATTCCGGTT1734
CUGUCGGA
766 CGGAAGCUG 606 TGATCCGAGGCTAGCTACAACGAAGCTTCCG1735
UCGGAUCA
771 GCUGUCGGA 708 GGGGGTGAGGCTAGCTACAACGACCGACAGC1736
UCACCCCC
774 GUCGGAUCA 346 TCGGGGGGGGCTAGCTACAACGAGATCCGAC1737
CCCCCCGA
782 ACCCCCCGA 709 GGGAAGGGGGCTAGCTACAACGACGGGGGGT1738
CCCUUCCC
793 CUUCCCCUG 520 TGCTGCTGGGCTAGCTACAACGAAGGGGAAG1739
CAGCAGCA
796 CCCCUGCAG 607 TGGTGCTGGGCTAGCTACAACGATGCAGGGG1740
CAGCACCA
799 CUGCAGCAG 608 TGCTGGTGGGCTAGCTACAACGATGCTGCAG1741
CACCAGCA
801 GCAGCAGCA 361 GGTGCTGGGGCTAGCTACAACGAGCTGCTGC1742
CCAGCACC
805 CAGCACCAG 609 GGCTGGTGGGCTAGCTACAACGATGGTGCTG1743
CACCAGCC
807 I GCACCAGCA 364 GTGGCTGGGGCTAGCTACAACGAGCTGGTGC1744
CCAGCCAC
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
77
811 CAGCACCAG 610 GGCTGTGGGGCTAGCTACAACGATGGTGCTG1745
CCACAGCC
814 CACCAGCCA 368 GGAGGCTGGGCTAGCTACAACGAGGCTGGTG1746
CAGCCUCC
817 CAGCCACAG 611 TGCGGAGGGGCTAGCTACAACGATGTGGCTG1747
CCUCCGCA
823 CAGCCUCCG 521 GCATATTGGGCTAGCTACAACGAGGAGGCTG1748
CAAUAUGC
826 CCUCCGCAA 710 GGGGCATAGGCTAGCTACAACGATGCGGAGG1749
UAUGCCCC
828 UCCGCAAUA 139 CTGGGGCAGGCTAGCTACAACGAATTGCGGA1750
UGCCCCAG
830 CGCAAUAUG 522 CGCTGGGGGGCTAGCTACAACGAATATTGCG1751
CCCCAGCG
836 AUGCCCCAG 612 CTGGGGCGGGCTAGCTACAACGATGGGGCAT1752
CGCCCCAG
838 GCCCCAGCG 523 TGCTGGGGGGCTAGCTACAACGAGCTGGGGC1753
CCCCAGCA
844 GCGCCCCAG 613 TGCAGCTGGGCTAGCTACAACGATGGGGCGC1754
CAGCUGCA
847 CCCCAGCAG 614 TGCTGCAGGGCTAGCTACAACGATGCTGGGG1755
CUGCAGCA
850 CAGCAGCUG 524 GGCTGCTGGGCTAGCTACAACGAAGCTGCTG1756
CAGCAGCC
853 CAGCUGCAG 615 GGGGGCTGGGCTAGCTACAACGATGCAGCTG1757
CAGCCCCC
856 CUGCAGCAG 616 TGTGGGGGGGCTAGCTACAACGATGCTGCAG1758
CCCCCACA
862 CAGCCCCCA 390 CGCTGCTGGGCTAGCTACAACGAGGGGGCTG1759
CAGCAGCG
865 CCCCCACAG 617 TATCGCTGGGCTAGCTACAACGATGTGGGGG1760
CAGCGAUA
868 CCACAGCAG 618 AGATATCGGGCTAGCTACAACGATGCTGTGG1761
CGAUAUCU
871 CAGCAGCGA 711 TGCAGATAGGCTAGCTACAACGACGCTGCTG1762
UAUCUGCA
873 GCAGCGAUA 140 GCTGCAGAGGCTAGCTACAACGAATCGCTGC1763
UCUGCAGC
877 CGAUAUCUG 526 TGGTGCTGGGCTAGCTACAACGAAGATATCG1764
CAGCACCA
880 UAUCUGCAG 619 TGGTGGTGGGCTAGCTACAACGATGCAGATA1765
CACCACCA
882 UCUGCAGCA 395 AATGGTGGGGCTAGCTACAACGAGCTGCAGA1766
CCACCAUU
885 GCAGCACCA 397 GGAAATGGGGCTAGCTACAACGAGGTGCTGC1767
CCAUUUCC
888 GCACCACCA 399 GGTGGAAA GGTGGTGC1768
UUUCCACC GGCTAGCTACAACGA
894 CCAUUUCCA 401 GTTCCTGGGGCTAGCTACAACGAGGAAATGG1769
CCAGGAAC
901 CACCAGGAA 712 CCTCGGCGGGCTAGCTACAACGATCCTGGTG1770
CGCCGAGG
903 CCAGGAACG 527 CTCCTCGGGGCTAGCTACAACGAGTTCCTGG1771
CCGAGGAG
912 CCGAGGAGG 620 CAAGGCTGGGCTAGCTACAACGACTCCTCGG1772
CAGCCUUG
915 AGGAGGCAG 621 TGTCAAGGGGCTAGCTACAACGATGCCTCCT1773
CCUUGACA
921 CAGCCUUGA 713 CATTTATGGGCTAGCTACAACGACAAGGCTG1774
CAUAAAUG
923 GCCUUGACA 408 ATCATTTAGGCTAGCTACAACGAGTCAAGGC1775
UAAAUGAU
927 UGACAUAA UGAUGGGC714 GCCCATCAGGCTAGCTACAACGATTATGTCA1776
A
930 CAUAAAUGA 715 AATGCCCAGGCTAGCTACAACGACATTTATG1777
UGGGCAUU
934 AAUGAUGGG 622 CCACAATGGGCTAGCTACAACGACCATCATT1778
CAUUGUGG
936 UGAUGGGCA 409 TGCCACAA GCCCATCA1779
UUGUGGCA GGCTAGCTACAACGA
939 UGGGCAUUG 623 CGGTGCCAGGCTAGCTACAACGAAATGCCCA1780
UGGCACCG
942 GCAUUGUGG 624 AGCCGGTGGGCTAGCTACAACGACACAATGC1781
CACCGGCU
944 AUUGUGGCA 410 CAAGCCGGGGCTAGCTACAACGAGCCACAAT1782
CCGGCUUG
948 UGGCACCGG 625 TGCCCAAGGGCTAGCTACAACGACGGTGCCA1783
CUUGGGCA
954 CGGCUUGGG 626 TTTCACTGGGCTAGCTACAACGACCAAGCCG1784
CAGUGAAA
957 CUUGGGCAG 627 TCATTTCAGGCTAGCTACAACGATGCCCAAG1785
UGAAAUGA
962 GCAGUGAA UGAAUGCG716 CGCATTCAGGCTAGCTACAACGATTCACTGC1786
A
966 UGAAAUGAA 717 GGGCCGCAGGCTAGCTACAACGATCATTTCA1787
UGCGGCCC
968 AAAUGAAUG 533 GAGGGCCGGGCTAGCTACAACGAATTCATTT1788
CGGCCCUC
971 UGAAUGCGG 628 CATGAGGGGGCTAGCTACAACGACGCATTCA1789
CCCUCAUG
977 CGGCCCUCA 417 CCGATGCAGGCTAGCTACAACGAGAGGGCCG1790
UGCAUCGG
979 GCCCUCAUG 534 CTCCGATGGGCTAGCTACAACGAATGAGGGC1791
CAUCGGAG
981 CCUCAUGCA 418 GTCTCCGAGGCTAGCTACAACGAGCATGAGG1792
UCGGAGAC
988 CAUCGGAGA 718 TCTGTGTGGGCTAGCTACAACGACTCCGATG1793
CACACAGA
990 UCGGAGACA 419 GGTCTGTGGGCTAGCTACAACGAGTCTCCGA1794
CACAGACC
992 GGAGACACA 420 TGGGTCTGGGCTAGCTACAACGAGTGTCTCC1795
CAGACCCA
996 ACACACAGA 719 GCACTGGGGGCTAGCTACAACGACTGTGTGT1796
CCCAGUGC
1001 CAGACCCAG 629 GAGCTGCAGGCTAGCTACAACGATGGGTCTG1797
UGCAGCUC
1003 GACCCAGUG 535 TGGAGCTGGGCTAGCTACAACGAACTGGGTC1798
CAGCUCCA
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
78
1006 CCAGUGCAG CUCCAGGC630 GCCTGGAGGGCTAGCTACAACGATGCACTGG1799
1013 AGCUCCAGG CGGCAGGG631 CCCTGCCGGGCTAGCTACAACGACTGGAGCT1800
1016 UCCAGGCGG CAGGGCGA632 TCGCCCTGGGCTAGCTACAACGACGCCTGGA1801
1021 GCGGCAGGG CGAGUGCG633 CGCACTCGGGCTAGCTACAACGACCTGCCGC1802
1025 CAGGGCGAG UGCGGUGG634 CCACCGCAGGCTAGCTACAACGATCGCCCTG1803
1027 GGGCGAGUG CGGUGGGC537 GCCCACCGGGCTAGCTACAACGAACTCGCCC1804
1030 CGAGUGCGG UGGGCCCG635 CGGGCCCAGGCTAGCTACAACGACGCACTCG1805
1034 UGCGGUGGG CCCGGGCG636 CGCCCGGGGGCTAGCTACAACGACCACCGCA1806
1040 GGGCCCGGG CGCUGUAU637 ATACAGCGGGCTAGCTACAACGACCGGGCCC1807
1042 GCCCGGGCG CUGUAUGA538 TCATACAGGGCTAGCTACAACGAGCCCGGGC1808
1045 CGGGCGCUG UAUGACUU638 AAGTCATAGGCTAGCTACAACGAAGCGCCCG1809
1047 GGCGCUGUA UGACUUUG152 CAAAGTCAGGCTAGCTACAACGAACAGCGCC1810
1050 GCUGUAUGA CUUUGAGG720 CCTCAAAGGGCTAGCTACAACGACATACAGC1811
1058 ACUUUGAGG CCCUGGAG639 CTCCAGGGGGCTAGCTACAACGACTCAAAGT1812
1068 CCUGGAGGA UGACGAGC721 GCTCGTCAGGCTAGCTACAACGACCTCCAGG187.3
1071 GGAGGAUGA CGAGCUGG722 CCAGCTCGGGCTAGCTACAACGACATCCTCC1814
1075 GAUGACGAG CUGGGGUU640 AACCCCAGGGCTAGCTACAACGATCGTCATC1815
1081 GAGCUGGGG UUCCACAG641 CTGTGGAA CCCAGCTC1816
GGCTAGCTACAACGA
1086 GGGGUUCCA CAGCGGGG439 CCCCGCTGGGCTAGCTACAACGAGGAACCCC1817
1089 GUUCCACAG CGGGGAGG642 CCTCCCCGGGCTAGCTACAACGATGTGGAAC1818
1097 GCGGGGAGG UGGUGGAG643 CTCCACCAGGCTAGCTACAACGACTCCCCGC1819
1100 GGGAGGUGG UGGAGGUC644 GACCTCCAGGCTAGCTACAACGACACCTCCC1820
1106 UGGUGGAGG UCCUGGAU645 ATCCAGGAGGCTAGCTACAACGACTCCACCA1821
1113 GGUCCUGGA UAGCUCCA723 TGGAGCTAGGCTAGCTACAACGACCAGGACC1822
1116 CCUGGAUAG CUCCAACC646 GGTTGGAGGGCTAGCTACAACGATATCCAGG1823
1122 UAGCUCCAA CCCAUCCU724 AGGATGGGGGCTAGCTACAACGATGGAGCTA1824
1126 UCCAACCCA UCCUGGUG448 CACCAGGAGGCTAGCTACAACGAGGGTTGGA1825
1132 CCAUCCUGG UGGACCGG647 CCGGTCCAGGCTAGCTACAACGACAGGATGG1826
1136 CCUGGUGGA CCGGCCGC725 GCGGCCGGGGCTAGCTACAACGACCACCAGG1827
1140 GUGGACCGG CCGCCUGC648 GCAGGCGGGGCTAGCTACAACGACGGTCCAC1828
1143 GACCGGCCG CCUGCACA543 TGTGCAGGGGCTAGCTACAACGAGGCCGGTC1829
1147 GGCCGCCUG CACAACAA544 TTGTTGTGGGCTAGCTACAACGAAGGCGGCC1830
1149 CCGCCUGCA CAACAAGC455 GCTTGTTGGGCTAGCTACAACGAGCAGGCGG1831
1152 CCUGCACAA CAAGCUGG726 CCAGCTTGGGCTAGCTACAACGATGTGCAGG1832
1156 CACAACAAG CUGGGCCU649 AGGCCCAGGGCTAGCTACAACGATTGTTGTG1833
1161 CAAGCUGGG CCUCUUCC650 GGAAGAGGGGCTAGCTACAACGACCAGCTTG1834
1172 UCUUCCCUG CCAACUAC545 GTAGTTGGGGCTAGCTACAACGAAGGGAAGA1835
1176 CCCUGCCAA CUACGUGG727 CCACGTAGGGCTAGCTACAACGATGGCAGGG1836
1179 UGCCAACUA CGUGGCAC164 GTGCCACGGGCTAGCTACAACGAAGTTGGCA1837
1181 CCAACUACG UGGCACCC651 GGGTGCCAGGCTAGCTACAACGAGTAGTTGG1838
1184 ACUACGUGG CACCCAUG652 CATGGGTGGGCTAGCTACAACGACACGTAGT1839
1186 UACGUGGCA CCCAUGAC468 GTCATGGGGGCTAGCTACAACGAGCCACGTA1840
1190 UGGCACCCA UGACCCGA471 TCGGGTCAGGCTAGCTACAACGAGGGTGCCA1841
1193 CACCCAUGA CCCGAUAA728 TTATCGGGGGCTAGCTACAACGACATGGGTG1842
1198 AUGACCCGA UAAACUCU729 AGAGTTTAGGCTAGCTACAACGACGGGTCAT1843
1202 CCCGAUAA CUCUUCAG730 CTGAAGAGGGCTAGCTACAACGATTATCGGG1844
A
1214 UUCAGGGGA CAGAAGCU731 AGCTTCTGGGCTAGCTACAACGACCCCTGAA1845
1220 GGACAGAA CUUUUUGU653 ACAAAAAGGGCTAGCTACAACGATTCTGTCC1846
G
1227 AGCUUUUUG UCUGGAGC654 GCTCCAGAGGCTAGCTACAACGAAAAAAGCT1847
1234 UGUCUGGAG CUGCCCAC655 GTGGGCAGGGCTAGCTACAACGATCCAGACA1848
1237 CUGGAGCUG CCCACAAG548 CTTGTGGGGGCTAGCTACAACGAAGCTCCAG1849
1241 AGCUGCCCA CAAGAAAG483 CTTTCTTGGGCTAGCTACAACGAGGGCAGCT1850
1253 GAAAGAGGG CAAGGAAA656 TTTCCTTGGGCTAGCTACAACGACCTCTTTC1851
1266 GAAAAAAGG CUGGACUC657 I GAGTCCAGGGCTAGCTACAACGACTTTTTTC1852
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
79
1271 AAGGCUGGA CUCCAUGA732 TCATGGAGGGCTAGCTACAACGACCAGCCTT1853
1276 UGGACUCCA UGACUAUA489 TATAGTCAGGCTAGCTACAACGAGGAGTCCA1854
1279 ACUCCAUGA CUAUAUAU733 ATATATAGGGCTAGCTACAACGACATGGAGT1855
1282 CCAUGACUA UAUAUACA175 TGTATATAGGCTAGCTACAACGAAGTCATGG1856
1284 AUGACUAUA UAUACAUA176 TATGTATAGGCTAGCTACAACGAATAGTCAT1857
1286 GACUAUAUA UACAUACA177 TGTATGTAGGCTAGCTACAACGAATATAGTC1858
1288 CUAUAUAUA CAUACAUC178 GATGTATGGGCTAGCTACAACGAATATATAG1859
1290 AUAUAUACA UACAUCUA491 TAGATGTAGGCTAGCTACAACGAGTATATAT1860
1292 AUAUACAUA CAUCUAUC179 GATAGATGGGCTAGCTACAACGAATGTATAT1861
1294 AUACAUACA UCUAUCUA492 TAGATAGAGGCTAGCTACAACGAGTATGTAT1862
Input Sequence = HSA011736. Cut Site = R/Y
Stem Length = 8 . Core Sequence = GGCTAGCTACAACGA
HSA011736 (Homo Sapiens mRNA for growth factor receptor binding protein
(GRBLG); 1303 bp)
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
M<HLf1lDr OD01O r1N M~HLn10r N01O o-1N MV~l7710r aD01Or1N
~
tniovoiotovovor rr rr r rr rr m mm mm m mo00om rno~o~
mm m aom aom m mm mm o mo
o o mm m mm aom m moomm mo~m
y -Irirav-1rirara~-Ir1r1ri.-In-Ir1~-Iririr1r1r-Irir-Irir1rir1r-Ir1riri
UL7~ ',~U L7L7 r.~r.~U",~U "~'C7"aL7r.~UU ~r.~ ChFCUC7 p ",~
U r.CFCFC ~ U'U UL7U U',~C7p L7pFCC7U ~ C7C7U'C7 FC~
UU U 7~ 7 ' " ' "' ' " '
'
~ ~ C C aU r-C~ a L7C7ar$~Ca~ rCa U C7~ C7.~FC~ C7
U U ",~'Ur~pr.~p p ~ U p',~L7r.~ ~U p r.C
C7 P~ ~7 U "' L7U U C7~ C7~ FCL7rU ~ ~U U'a~' 7
~'~ C
".~".~' FCL7 ~ 'a,, pU ",~p UFC~ Ur.~-C7 C7C7',~p C7U L
C7p FCC7 U'U U UL7<;C~ C7hU r.~L7p L7(7U C7'J'aFCr-CU
FC~ 'a C7U
'aFCU r.C~ pp FCUrCU" p p~ UFCFC~r FC ~" t~U U'U C
~ C ~'
, . . r-
C7C7C7ChC7C7C7L7C7C7C7C7C7i7C7C7C7L7C7C7C7L7C7C7C7~hU C7t7c9
C7C7L7t7C7L7C7LJL7C7C7C7L7L7U L7U'L7C7U C7C9C7C9U'ChL7C7C7C7
C7C7C7U'C7C7C9C7C7C7L7C7L7L7L9L7C7U'C7C9C7C7C7C7C'JC7CJC7C'JU'
UU U UU UU U UU UU U UU UU U UU UU U UU UU UU U
UU U UU UU U UU UU U UU UU U UU UU U UU UU UU U
".~'a'a"a'a'a~ 'a"~''a'a''a".~~".~".~p 'a~p ~".~p ".-~''a'.-~'~ ".-~'"'ap
L7C7C7L7C7C7C7C7ChC7C7C7C7C7C7C7C'JC7C7C7C7C7C7U'C7C7L7L7C7C7
UU U UU UU U UU UU U UU UU U UU UU U UU UU UU U
'.~~".~'ap "~~".~pp "~''a"a'a'~'a".~'a".~'~'".~"ap 'a~ "~p '~
UU U UU UU U UU UU U UU UU U UU UU U UU UU UU U
C7t7C7C7ChC7C'JC7C7t9C7U C7C7C7C7LhC7c7C7C7C7CJC7C9C7C7C7C7c7
t7CJC7LJC7C7U'C7C7C7C7C7C7UC7C7U'C7UC7U'C7C7L7C7C7L7C7ChC7
C N
UU U UU UU U UU UU U UU UU U UU UU U UU UU UU U
d ' ' '
"
.-~~ 'a~"ap"~~ ".~~ 'a~ ~ "~~ ".~~ 'a~".~p'a".~~"J".~".~~'a~
UU U UU UU U UU UU U UU UU U UU UU U UU UU UU U
N ~ UU U UU UU U UU UU U UU UU U UU UU U UU UU UU U
UU U UU UU U UU UU U UU UU U UU UU U UU UU UU U
UU U UU UU U UU UU U UU UU U UU UU U UU UU UU U
L7C7C7c9C7C7C7C7C7C7C7C7C7C7C7C7LhL7C7C7i7CJC7U'C7ChC7ChC7i7
.Q L7U'L7U'U'C7ChL7C7U'C7C7U'C7I7C7C7L7C7C7L7C7C7C9L7C7U U'C7U'
UC7C7C7C7U'C7C7C7C7U'C7C7L7C7U'C7C7C7U C7U'C7C7C7C7ChC7U'C7
U'C7U'CJC7U'C7U'CJC7U'L7L7U'L7U'C7U'L7L7C7C7L7U'U'C7C7C7C7C7
'aFC~~ UU r-C~~ ~~U'~ U~ FC~ ',~ ~ U U Ur.CaC
~
..h.a'C7 C7R'U '.7C7 Up C7UR'C7 U U C7"J Ua'"J
".~C7FC~C7Ur.~U'pU ".~'U pC7C7r.CFCC7C7U r.CUU ~U'FCC7
a'a'U L7C7Uh p R'',~C7C7 R'L7"~U ~ R' U' U
~ ~ 'J'a",>A',~ '~.
U~ ~ ~C~'J"~" U Up U' U CU7C '' 7 C -~' '
~~' ~~ ' "
'
'' " U' .. U p 7U ' 'r-.J C r-CU ~~ U ~~ ~p .~,~U
77 7' ' ' ' " '
~ '
.. ~ a L~ U Lp a a. C7U A Up FCU R .~U U
,
L
OdWf1r1M ~ON r MQO01O ~ If1r1Nd~u1~01Drm r a1M 'W11110m a1
NM M If101Mu1M L>1M M~ If1lf1cNV~0101t11T 01O1U101c!d~C~V~~tIf1
y 1r1r r m~ rInr Inr rr Inu1r rd~~ru1v~<r~ru1d~r rr rinu1
a
p r~r~~ ~n~ u~ ~ r~~ ur~~ r~r~uu ~ c~r~~~ ~ ~~ ~~ c7u
U~ U FCFCC7C7r.CU'U U'U"',..Wr$U 'aC7 U ~ ~ U
C
CC "a "a~U 'aC7U r-CFC Ur-C'aU ~ U'J~C7U ~~ !~ C7
'J7 h UU ' ~ "~ UU p ' " ' "
'p ~
~
. L Ur~r.,r. ~ U r.~L9 pa .oU U U,~U
U p U U'"..-~'U r-CC7r.~C7 r.~U Up "aUU U'p 'JC7C7 U 'U
~
'a FCC~7LU7"~.~FC~ ~U ~ CU'J~ p FC ~ ~~ p U r-C~ ,C~7
~ ~ ~
~ ~I7rC~U <;C.'~U''aC7~r-CU pC7FC U ~FCU~ p U C7rcL7~ "
~'
.
~ C7L7U C?C7UL7C7t7C7C7L7C7C7C7C7C9ChC7C7C7C7C7L7C7C7C7C7C7C7
'~ U~ ~ U~ UU~'C~"U~~ ~ U C~7~U ~ UU ~ ~ ~U ~ "U,~'U C~'J
7 ~ ~ ~
V~ a'~ ~ "JU (7A',. C7U ~' ~' C7.=~ L7p C7 ~ UU ~ Up C7
" a' ' ' '
Up r$ JU C7r~r.~UU C7U U ~U aC7 aC7C7 U U
Chp p C7 'a r-CC7'aFC".~' FCU ~r$C7 U",~'~ p r$C7
C7U'p U' U U r~C7'ar.~~ U ~ 'a ''ar~C7U UFCa U
_ UU ~~ UU ~ U U U U U ~ ~ ~ U ~U ~
' ~C U' 'a" Cr-CC FC r-C ~ C7C FC ',~ FC
a 7 7 7
d
_ t-1dm w r.-tumo o,-1M
y -1r m Mr1crtoN a1t11l0~1M ON Mu1O M
p OO r1NM MM V7U7V)
.y -Ir-Ir1NM cftV~~ V~V1l1111110rr ro0O101~ li -I-1l
rr v rr rimlrir1r1
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
81
M ~H U1 ~O r CO o1 O ,-i N M W N l0 r R~ O1 O r1 N M cN tfi t0 r Go 01 O ri N
M V~ u1 l0
O1 01 O1 01 01 01 01 O O O O O O O O O O r1 r1 r1 r1 ,-~ r1 r1 r1 r1 r-~ N N N
N N N N
M N CO N m m m 01 ~1 01 a1 01 O~ 01 01 01 01 01 01 O1 a1 ~1 01 O1 Q1 O1 01 01
01 01 01 01 Q1 01
ri ri ri ra ra r-1 ri r1 rd ri ri ra ri r-I r-1 r~ r-1 r-1 r1 ~-I r-W -I rW i
r-1 n-1 ri r~ r-I r-1 ri r-1 n-1 ri
U p p r.~ '',.~ U r.~ U C7 'a U' r-C C7 ~ C9 U U r.~ r.~ ',~ C7 U U U U p L7 p
U
r-C ~ ~ U U' 'a C7 C7 U ~ C7 FC U C7 p r-C "a U ~ C7 C7 r.C 'a 'a FC ~ C7 U
',~
U ',~ C7 C7 U L7 'a p U U ~ "a ~ U C7 ~ p U U r.~ ",~ C7 ~ "~' U r.~ ~ ',~ U'
C7 ~ 'a U
r.~ U ~ ~ ~ !~ U r-C p L7 U 'a ~ ~ C7 U U "a U ~ "a C7 L7 U U ~ p ~ U U U
C7 C7 U C7 U p FC U' U C7 ',~ U ~ U "a C7 Ch p FC 'a C7 ~ p U ~
C7 U' 'a U' p U U U U' 'a C7 U r.C U U p r.~ p C7 U U U C7 U U 'J U
~ "~ C7 "a FC 'J C7 'a U' C7 p U U ~ p L7 U C7 r-C U 'a 'a ~ ~ ,a U 'a r.~ U
'a C7 ".~ r.C U ~ FC ~ 'a ~ FC ~ U r.C "~' U FC r.C 'a sC U ~
C7 C7 C7 U' L7 Ch. C7 C7 C7 C7 U' C7 C7 C7 C7 C7 C7 L7 C7 C7 U' U' L7 C7 C7 C7
C7 L9 L7 L7 C7 C9 C? C7
C9 C7 C7 L7 L7 C9 C7 L7 C7 C7 C7 L7 L7 U' U L7 C7 C7 L7 C7 C7 C7 C7 C7 L7 C7
C7 C7 C7 C7 t7 C7 Ch L7
U C7 C7 C7 C7 C7 U U U C7 C7 L7 L7 L7 U U C9 L7 L7 C7 C7 U' C7 C~ U U U C7 C7
L7 C7 C7 C7 C7
U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U
U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U
C7 C7 L7 U' C7 C7 L7 C7 Ch U L7 C7 C7 C7 C7 C7 C7 C7 U' C7 L7 C7 C7 C7 C7 L7
Ch UW 7 C7 ~7 Ch C7 C7
U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U
".~p'a~~~".~'a'a~~"a".~".~p'a'a'a~~'a'a".~'a'a'a~~~~"a~'a'a
U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U
C7 C7 C7 L7 L7 L7 C7 C7 U' C7 C7 U' Ch C7 C7 C7 C7 C7 C7 U C7 C7 L7 C7 C7 C7
C7 L7 U U' L'J U' C'J L7
C7 U C'J C7 L7 C7 C7 L7 C7 L7 U C7 C7 C7 C7 C7 C7 C7 C9 U G7 C7 C7 C7 C7 C7 CJ
L7 U' La U C9 U U
U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U
U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U
U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U
U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U U
C7 U' C7 C7 C7 U' U' C7 C7 C7 C7 C7 t') C7 L9 C7 C7 C4 C7 C4 Ch C7 C7 C7 C7 C9
C7 C7 C4 U' U' U' C7 C7
C7 U' C7 C7 C7 U' L7 C7 C9 C7 U' C7 C7 C7 L9 C7 C7 C7 C7 U' C7 U' C9 C7 C7 L9
U C7 U' C7 C7 C7 C7 C7
a' ~
C7 U C7 L5 C7 L7 L7 C7 U' C7 C7 L7 U' C7 C7 C7 C7 L7 C7 C7 C7 C7 Lh U' C7 C7
L7 C7 C7 C7 C7 L7 C7 Ch
C7 C7 U U L7 C7 C7 C7 L7 U C7 L7 L7 L7 U L7 U' U C7 C7 C7 C7 L7 C7 C7 C7 L7 C7
C7 L7 C7 C7 C7 C7
C7 ',~ U' ~ C7 U "~' U' L7 p C7 U r.C '',.~ U '',.~ D "a C7 U p p ''..~' ''.~
FC 'a U p FC U
C7 C7 ~ 'a p r.~ U' r.C U U ,.'~ r.C p U "a U r.C ~ U ".~ U U
'a"a 'Jr~pUUUp~ ~UpUr~UC7C7 UpU U ~UUr$U
".~ r-C U' L7 U U L7 ~ U' p U ~ C7 ".~ U p U ~ FC b r-C 'a U' "..W C U
U ',~ p U C7 r-C C9 U ',~ C7 ".~ U U r-C ~ U ~ L7 a,' U ~ U C7 U U U ~ ",~ p
'a ~ U' U ",.~ r~ U U 'J p U U 'a U U' ',~ p U "J p ''.~ r.~ U p U
~C~.7~"UaU~~tU7U~~~r.~~U~~UC7~~~~UU~UUU~~~UCU7
O r r1 N M o0 01 V~ l17 O l0 r1 N O r M rI N W LO a0 M d~ N ~1 ~O 10 O r !D ~
,-/ O N
lD d~ ~O lD lD ~ d~ l0 ~O O lD O O If7 10 O t11 lf7 O O l0 Ln Ln V1 lD O Lf7 r
If1 V7 Ll1 r l0 r
m r tm in r r mm un mo r mn r r mmn r r r mn r m r r r u~ r u-,
~C U "J r-C C7 ~ ~ U U' 'a 'a 'a L7 'a U' U r.C FC ".~ C7 C7 U' C7 C7 p C7 U
U ~ U' U ~ U' p U U L7 r.~ ',~ ~ p U U U ~ ~ ~ Cg ~ U
r.C U U' 'a C7 C7 ~ U' L7 FC U' U FC 'a rC C7 ~ C7 C7
C7 r-C ~G U' U ~ p U U' r-C U ~ L7 C7 "a C7 'J U ~ C7 'J U' "J U C7 U' U'
Up U C7 C7 U p U C7 ~ U rC C7 r.C L7 p p ~ ',~ p FC U r.C !~ U'
U~~~ U"a'UJ "~~~~'UJC~'JCU9FC~UC7 r~C~'J'a ~ LU7FCL~7UU'
U ~ U p U C7 ~ U p U 'a r-C U C7 ~ r.C C7 r-C ~ FC U t7 FC FC ~ FC 'a r.C C7
FC 'a L7 'J
C7 C7 C7 L7 L7 L7 C7 C7 C7 U' C7 C9 L7 L7 C7 L'J C7 C7 C7 Ch C7 C7 C7 C7 U' L7
C7 L7 C7 C7 C7 L7 C7 C7
U rC ~ C7 b 'a ~ Lh 'a r.C C7 p ".~ r.C '~7 L7 r~ p r-C L7 r-C 'a L7
FC U FC 'a ~ U ~ ~ ~ U U r$ C7 C7 ~ r.C U L7 U p C7 FC C7 r.C C7 L7 p
U U r.~ U r.C L7 U' U' U A,' p U C7 'a U' C7 ~ ~ U C7 C7 L9 ".~ U C7 U' r.~ L7
U U ~ C7 U Ch p U C7 U p r.~ U' p U' ~ U U r.~ "J r~ p p U ~ r~ C7 r.C
U' UU~~Uiya'~L,~7'.U7 "~~C7 "aR'CU'JC~7"U~' C~Ja'R'UU'',7~~ U L7 U' C7
C.~7 ~FCFC~~L7'.7~L~'JUFCU~U'aUi~'JC~'J'a ~FCUC~'JC~7C~7 FC ~(~'J
01 cN V~ r1 r r1 N u1 m v-I ~O O o1 V7 t0 co O r1 cN O~ ri N M LO o~ M ~O V~ N
V~ LO r 01 O
1f1 10 01 O O ,-I r1 r1 ~-I N N M M d~ ~ W ~7 V1 V1 to ~ r r r r N OD 01 O O O
O O r1
r1 ~-1 ri N N N N N N N N N N N N N N N N N N N N N N N N N M M M M M M
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
82
ro001O r1N M d~l11l0ra001O r1NM ~U1torm 01O c-INM cHU11Dro0a1O
NN NM MM M MM M M MH ~ W n L
M ~ d W crV W cHcHWn N t!1tnNtnLnInIntnt0
010101D\0101d10101010~O~01~101010101~1O101Q1O101O~O~0101O~o~a10101O
~
r1rir-Iv-Irir1riv-1r1wiririririrar10-1rW-Ir1riririr1-Ir-1r-1r-Ir~rar-1rW-1ri
U ~ ~U'U UU U UU U'"..~U Up Ur.~U'U~ r$U'C7U",~'C7~ r.~~',~r.~U
UU U U pU'U UW7U'~ U r pChL9U L7C7U ~' ' p
~ ~
. U , G~ U U
U ~ UU ".~U~ U ".~ChpU p U~ C7L7C9 C7"~'r.~'JC7L7Up C7r.~U U C7
.~ U'.7U C7 "aUI7UU ry'C7 'U'p ' U'C7U''7 UC7r U'U U "
~ 7 7 C ~
~U L7U L9 U U L7U U".~',~L7L7.'a , Ur~',~...L7r.C. pU L7,
' ~ FC ~ L7" C7U U' p
'
UU UU U C7C7U rCFCC7p L7 C7U ~ChC7C7U pC7'Jr.~L9 U
'a"~UC7U~ ~ UU C7'.-~b ~C7U ".7a'U' a'"~"~'.~~'U'"~C9U',7R'U".7~ U
" ' '
pU C7U p U U~ p UC~Ur$r.C,~p ,~r.~U U''aUU''aUp ",~r.~U Up U p
C7L7L7C7C7C7U C7C7L7C'JC9C7ChC7C7C7C7ChC7C7C7C7i7C7C7L7t7C7C7C7c7C7C7
C7C7C7C7L7ChC9t7C7C7UC7C7C7U Ch7 7C77 7 7
C C C CU CC7C7UC7C7C7G C7C7C7t7
L7C7C9C7UL9C7L7C7C7C7C7C7C7C7C7L7C7L7C7GU C7C7C9C7C7L7C7C7C7L7C7c7
UU UU UU U UU U UU UU U UU UU U UU UU U UU UU U UU U U
UU UU UU U UU U UU UU U UU UU U UU UU U UU UU U UU U U
~'a~"~'a~ ~ "~~ ~ 'a~ ~'a'~~~ "p "a'a"J"J''J'a~~ "' " "a~ "
" " " " " " ~ " ~7 ~' "
. . . . . . . . ., .- . a
CJC7U'C7U'C7C7L7C7C7C7C7C7C7C7C7C7L7C7C7C7C7C7L7C7i7C7U'L7C7L7C7L7C7
UU UU UU U UU U UU UU U UU UU U UU UU U UU UU U UU U U
"a~ ~"~'~~ C~~~ ~ ~b ~~ ~ ~~ ~b "a~~ ~~ ~ ~~ ~~ " ~" ~ C
" " " " " ~ '
. . . . ~ a'. a . -~
UU UU UU U UU U UU UU U UU UU U UU UU U UU UU U UU U U
C7C7C7ChC7C7CJC7C7C7C7t7C7C7C7ChL7C7C7c7C7C7C7C7C9GC7L7U C7C7C7L7C7
L7C7C9C7C7L')C7C7C7U'C7C7L7C7C7C9U C7C7L7t7C7L7C7C9C7C7L7C7U'C7C7C7U'
UU UU UU U UU U UU UU U UU UU U UU UU U UU UU U UU U U
".-~~ ".~'a~".~"~"a"..~'".~~rJ"J~ "a'a'J"~"J"a"J"~~'~"J'a~ "~'".~'J~".~'",~'a
UU UU UU U UU U UU UU U UU UU U UU UU U UU UU U UU U U
UU UU UU U UU U UU UU U UU UU U UU UU U UU UU U UU U U
UU UU UU U UU U UU UU U UU UU U UU U U U
U U UU U UU U U
UU UU UU U UU U UU UU U UU UU U UU UU U UU UU U UU U U
t7t')C'JC7t7U'U'(')L7C7ChC7C7U'C7L7CJU'C7t7C7C9C7C7C7CJC7ChC7C7C7C7C7C7
C7C7C7C7UC7C7L7C7C7C7C7C7U 7 L77 i77 7 77 7 7 h
C L C L CL CC~L LC7UC7C7C7LhC7C7
C7C7C7C77C77 C7C77 77 79 ' '
C C L CC LC C7UC7C7C7C7CJC7L7C7U ChC7UC7C7C7U U C7
C7U L7C77C77 UC77 7 79
L C C CC9CC C9UC7C7G C7C7G L7C7U ChC7C7C7C7C7C7U C7
UC7Up U'U U pC7U U ",~FGU' ",~ U ',~U Ch',~U L7".~ U U U'U 'aU
C7U pU U ~ UL7p ~'ar.~U U'~L7~ ~ pU'U'C9L7pU ~U C7 ',~U
~ ~
UU UC7 p U U''a".~Ur$pC7U' ".~ ~ UU'r.~p p Up U'C7''..o U U
U'J ~ UU U'C7U U Up r.~L~'a r.~C7 U'Ur.CU'U'U ".~',~ p U U
UC ~U UU77UUp p ~U ~~ U ~ U~ ~'U'~ ~~ U 'UU ~~ U ''U~ ~
U7 " ' '
. C . p C7L L L C U p Jrr~~~U ,.~ ,~
UU UU U U ~ ~U UU ~ ~7 pU J J~ 9U ~ ~~ ~ U UU U
U J p ~
~
".~ C C C U' ".~ C
7 7 h
~M NM w<riwoinr o~m ovor ~N rM m ood~mown o~orm ov,-io ~ N
~or ~oior~o~oior ~ ~o~orr o rr rr r or r
o r wr rr r mm m ao
rm rr inr r rinr rr ru n rr i.nr m nr mn r mr rr r mr r r
'a'JC7C7C7U'L7FCC7r.C"aC7UU' "aC7 L7r.CU'JUL7 U~ 'Jr.CC7C7C7U
U"~U'U'FC~ C7U' ~ FC".~C7 r-CFC~U (7~U pC7~ p UC7r.CL7C7U'
C7U C7C7U ~ C7~ FC'aU''a~ h UU U~ U Up UC 7 ~ C' 7 '
r r. F C p r-U C FC U U
r.CU'~~ U'U U r.~L7U'r.~"JC7U ~ 'JU UU r.~UU 'aU r.~U'U ".~~ U'C7 p C7
" '
'
C7r.CUp L7C7U UC7L7C7r.~pU ,~U U C7a UU L7r.~ ',~ C7C7'aU'
U'L7L7U FCL7UFC C7"aFCU U ~FC ~ 'JU'U "aFCFCC7~ UU ~ C7C7
UC7rL~ L7~ C7U ~ r pC7U U ~b U UU U h h ~
~ ~
. e . r.~C C U r.CL7
C7U C7C UC7C7CU L7~L7r'aU ~ ' ~ ~C7U 7 U ~'
C ~
r- F - FC,L7 ~CL r-C U C7UL7FCC7
C7C7C7C7C7C7U C7C7C7C7C7C7C7C7t7L7C7C7C7C7C7C7C7C7C7C7C7G L7C7C7C7L7
C7UL7r-C~ C7L7FCr.CC7 U''a".~ FCFC'aU'U C7U FCC7r.CFC'aC7U' U'sC
C7U L7S~'JC7C7U r.C~ FCC7 ",~'U ",.~'r-C~ 'aU r~U C7r.C"JC7 U'
~ ~
'a C7C7C7U b 'JC7U UC7 p 'JUr.~U UC7 U U UC7r~U r.~pU ~ C7
L7UU'U DL7L7UC7L7r.~r.~UU p~ ~ U C7",~'r.CUL7U~ U r.~L7U
7C ' " U" ~C U ' "
'
C7FCC7U~ r$Cr.C9U ~ J ~U ~ U U FCFCUU .~UC7C7
C7~ 'a L7U'KCU' C7r.CU r.~L7r.~U'',~UU U U r.~",~'~ UU C7r.~U pC7C7U
"JGU"'~~ ~U CG7C~'lC7CU7CU'JC7U~ ~ L~7FCC~'Jp U CG7"~"~'U U C~7r U~ ~ ~ "GG'
~ ~ C
,. .- . FC J C
J
lDr o1O Ntor aoO ~ t110701VnlDNN Oc-IN ~N MN T OM R1r aD01M cr10
ir1-IN NN N NM M MM M~ d~11l0rm o0W~ 01O O -il ~N N
x
r v . vr NM M M
MM MM MM M MM M MM MM M MM MM M MM Mdoc~~W V~~ ~ ~cr~
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
83
r1N M'W1f1lDr m 01O r1N M V~Lf7l0r mO~O n-iN MV~VIl0r m01O ~N M d~
tovoioiovoiotototor rr r rr rr rr m mm mm m mm mm m avo~m ov
rno,o~m o~o~m m rnm o~m m o~a~rnm mo~o~mo~mm o~o~o~o~a~ata~ofa~at
U"~'UL7"ar-CU'U C7U C7r.~C7L7C7r.Cr.C'aU ",~'pC7"~'~ U FCU "aC7~CLh r.CaC
L7U U',7ry'C7U'U''.7U'a'U'C7U'A',C7C7UU FI,'U',7C9 U C7p C7 ".-~ ~ U U
",.~U pFCC7'aU C7Ur.~U'C7U'".~C7FC'aUU'U UU' U r-C~C7p C7~ r.CU
pp UU'p r.CU ',~C7L7L7C7C7r.Cr.CUU L7".~p ~~ ~U L7rL'a'aC7 C7U ',~
UU U~ ~ L7U U C7C9U'U'" L7U CU ' U C 7 '
~' W
r. . r. ,r.~ F Cr.~p Up C7~ ~ Ur~U C7
UU rCU'C7U'C7'aC9C7~ U r.C~ C7U'~U U UC7 C7r$U'U' rCU p rC
" ~ ~
L7C9UC7C7UU'.~U U C7C7',~',~Up ',~p~ ",.~U U",.~ U U'Ur~C7U
U UL7U Up p ",~'JUU U r.~p Ur.~"aU r.~L7'aUr.~C7~p UU r.~r.~U r-Cp
C7C7UC7CJL7Chc7CJL7t7C7C7U'C7L7C7L7U C7C7C7C7C7C7L7C7U'C7L7C7C7L7C7
C9C7L7U C9C7c~C7L7L7C7C9C7C7C7UL~C7C7C7C7C7C7C7U C7C7UL7L7C7c~ChL7
L7C7C7C~C7C7U'L7C7C7C7U L7C7L9C7C7C7U'C7U'C7L7U C7C7L7C7U'C7L7C7C7L7
UU UU U UU U UU UU U UU UU UU U UU UU U UU UU U UU U U
UU UU U UU U UU UU U UU UU UU U UU UU U UU UU U UU U U
'a'aP".~p ~"a~ ~'a~~ 'a~~ C~'a~'a~ 'a'a~"a'a'a~ ~~ ~ 'a".~'a'a
U'L7C7U'C7U'ChC7L7C7C7Cht7C7U'L7L7C7U'C7C7(7C7U'C7C7C7C7C7L')C7C7C7L7
UU UU U UU U UU UU U UU UU UU U UU UU U UU UU U UU U U
"a.-h"~~ ".~"a"~'a"a~'J'"arh-'.7".~",.7'a'nJ~J"~"arh"~"a~"a"a"a.7'~-7"~~.7.-h
~ R'
UU UU U UU U UU UU U UU UU UU U UU UU U UU UU U UU U U
L7C UC7c7C7C7ChC7C7c7C7C7C~L7C7c7C7C7C7L7C7L7C7C7UC7UC7C7C7L7ChL7
C7C7C7C7C7C7C7C9C7(7C7C7C7C7C7C7C7C7LhL7C7C7t7C7C9C9L7C7U'C7C7t7C7U
UU UU U UU U UU UU U UU UU UU U UU UU U UU UU U UU U U
"a'a~ ".~''a'~-
7".7~"a'a'a".~"J'a".~",7~"a".~~".~'J"a".~'a".~~".~"..~'a".~'J".~
UU UU U UU U UU UU U UU UU UU U UU UU U UU UU U UU U U
UU UU U UU U UU UU U UU UU UU U UU UU U UU UU U UU U U
UU UU U UU U UU UU U UU UU UU U UU UU U UU UU U UU U U
~~ ".~C.~".~~~ "a~~ ".~~ ~ ".~~ ~".~~C~".~~"a~~ "a".~~ ~"a~ b~ C~~
UU UU U UU U UU UU U UU UU UU U UU UU U UU UU U UU U U
U'C7C7C7L7C7U C7UC7C7C7C7C7C7C7C7C7C7L7C7L7C7L7C7ChC7C7L7C7t7C7C7L7
L~L7C7U C7C7C7C7L7C9C7C~C7C7C7L7L7I7C7C7C7C7C7L7C7ChC7~C7C7C9U C7C7
C7C777 7 7 h 7h 7 ) 7 '
CC C LC7L Cc CC7U CC7CC7C7ChL7L7ChC7C7C7ChC7C7C7L7C7C7U C~
C7C7CJC7U'C7L7C7C7U'C7C7U UC7U'C7C9C7L7C7U'C7L7U'U'C7U'U'L7C7L9C7C9
U U'U U C7C7".~'L7U UU "a~U U'"..~'Up ',~ U ~C7"'~".~U Ur.~r.~U"~".~
U C7C7C7U r.CU Up U'U'"aU p~ C7~r.~C7U U U'U ~ U ",..~U'U
U L7U'".~a L7C7U pC7~ p',~rCU r$U U C7C7'ap ".~'a~ ''..~'~ U C7r.C".~U
U C7C7p UU FCC7p C7 UFCU'.~Ub ~ U ~ ~ ~ ~
~ ~ "ap'~C~r~ ~ C U',~U
U ~ ~ U U UU U ~ U~ ~ h ~~ 9 U
~ U
(7C ~C7L ~ ~CC 'a ',~ C7 ~ C U C7
7 7 ' 9 ' ~ ~ ' '~ ' " 'J '
' '
C7C7~L7J U~ C7UC7r-CU C7FCU aC7r$ UU a a U~ C7FC~ a U'
NM Md~tn~tftt0tor m01O rr1mO NM r141~ ON M t!7torr1N m01~ O
mm mm m mm m mm mm 01m01m~ 01OW-Im01O~,~,-~O~O~O~O~01p~p~r-~O
tnr tnr r tntnr tnr rr r tnr tntnrr tntnr tntntnrr rtntnrr tnm
UU r.CU r.~U'b U C7U '~,~'U U C~C7FCU ".~ UU'r.C r.CU'FCU",~r.C~ U
UU U r.CU U r.C U",.~U ~C7C7r.~Lh~ ~ r.CU C7~ U p FCU C7C7~ ~ U
'aC7U UL7U U~ U p Ur.~L7C7Ut7U UC7UU ~p r.C'Jr~C7 t7
UU pr.~C7U~ U UU ~~ U U'Chr.C',~r.~U ".~~~ ~~ ~ U~ ~~ U ~~ Ch
' '
L7~ UU rCC7C7a UFCU!~ U~ C7FCC7 ~
C7p ~U U FCr.~U UU'r.CU ~ r.~r.~pC7pL7L7UU r.~~ r~ p L7U".~' L7
pU C7U UU C7pL7L7r.CU UU r.CC7FC,'~~U"..~p UU'L7UC7p C7r.~U Ch
pC7UC7C7UU r.CUC7C7C7r.C"JC7Ur-CC7r-CFC'aL7'aU FCFCL7C7'a"JL7~ FC
C7C7C7U'LJC7C7C7U'L7LJC7C7U'L7ChC7C7C7L7C7U'C7C7C9C7C7C7C7t7C7C7U'U'
i7 C7U U'L7KCFCFCsCU'C7L7pr.CU'~ r.CC7".~U C7p U r.C C7U'~ pC7'ar.C
U 7U U t7U $ 7 UU 7 '~ ' '
C7C7~' U CU U R'U rC7C '' ~r~L7r.~r-Cr-~U ~~ C UU ~C7U Up U U
~ U U Ury'U'77 UU "~U'C7C7 U "JU " 'U'
~'7
C7C7C7. p UU'C7UU UU rL., ~C7r.C",~'C9'a Up , i7 U~ , ,p C7U
r.C UL9 ~ r.~ ~ ~ U
r.~U'U r~pC7r.~UU UU U pp U'C7Ur.~r.Cr-C ~U'U C7 U U L7
~ ~ ~U U U ~ ~ ~ ~
C L~ p UU U r.~U pU U Up UU LC p C~ U U'Ur.C ~ ~ ~ C C
7 7 77 'J ~ 'J7
C7rgU'U r~pU C7UC7Up U UU p",.~'r.~C7r.Cs~U r.~ C9pU'r.~U ".~Up p p
ro rlior mN vomvorm ovwm ovM tnioovNtorN w ~<rtwo o~,-1N w m
MV~d~If1If7II11010~Or rr r 01O1O1O OO O r1N NM M M~ WW If7t0l01Dl0
V~d~d'V~V~1'~ d~cNd'V~~ d~V~W
~NV1t11tIWflt11t11Ntf1It1tf11f1Int11t11If7Lf1V1LO
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
84
Inb r~ m or~NM tru1iorm rnor1NM ~rInb ro00~o,-~NM <rtnb r ao
0101o1ov01Oo Oo O oO oO O r-I,-I,-ir1r1,-ir1,-.~ri,-1NN NN N NN N N
o~rnalrno~00 00 0 00 00 0 0o ao 00 0 00 0 00 00 0 00 0 0
rir1~-1~-io-INN NN N NN NN N NN NN NN N NN N NN NN N NN N N
".~C7 p b U'r~Chr.~p U'U'r.~r.Ct9UC7U'',.7pU U Ur.~U ".~p U'U'U U'J'U U
C7r~~~ U r.Cp ",~C7U ".~ U'p ",~ (7~p UU U r.~C7U pL7C9r.~U 'aU U r.~
'a~'aU''aC9r$L9''~U~ C9U U L7 U pU r~C7p U L7~ r~U ".~C7U r.~L7
U~ ~p rCC7~ U'~ C7~U' p ~ U'U U'JC7~ C7'aU'C7''.~U UU C7C7r.CC7C7
pp U U r.C".WCU p ~U' U C7'J".~C7U'C7"aC7C7 UU'U4~L7UU'C7L7
Ur~UC7U pL7U'"~U U ".~"aL7C7U LhC7p'aC7U'U ~ L7C7''aC7U UC7C7"J
'a~ U'r-C~=CC7'aL7C7"aC7~ UC7L7U',~C7p UC7L7UU C7C7FCC7U'U 'aC7b U
"" " "' "" ' '
t~D r.CU p pU .~,~,..~aJ .~.-~U'a~ aU UC7U UU p r.~U t9U ",~Up U U
L7C7U'C7C7C7C'JLhC7C7C7C7U'C7L7C7C7C7U C7C7L7C7C9C7C7C7C7C7C7C7C7ChC7
C7C7U'C7U'C7C7C7C7C9C7L7C9U'C9C7C7t7U'C9C7U'C7L9L7C9CUC7U'C7C7L7L7C7
C7C7L7C7U C7c9ChC7C7C7C7ChC7C7C7C7C7C7C7C7C7C7C7C7C7CJC7C7L9UC7t7C7
UU UU U UU UU U UU UU U UU UU UU U UU U UU UU U UU U U
UU UU U UU UU U UU UU U UU UU UU U UU U UU UU U UU U U
L7L7L7L7t7L7U'LhC7L7U'C7U'L7L7L9LhC7L7C7L7L7C7C7c7UL7C')C7C7Cht4C7C7
UU UU U UU UU U UU UU U UU UU UU U UU U UU UU U UU U U
"J~ 'a~ 'a"a~ "a~ ".~~".~".~".~"a'a"~''a"a'a".~~ 'a~ ".~'a".~'a".~~ ".~p
UU UU U UU UU U UU UU U UU UU UU U UU U UU UU U UU U U
C'JL7C7L7C'JC7L7U'L7C7C7L7U'C7U'C7C7C7C')C'JC7C7C7L7C7C7C7U'C7U'C')L7C')C7
C7C7C7L7c~C7C9C7t7C7L7C7C7C7C7t~C7UC7c7C7U C7U CJUL7C7C7C7C)C7C7U
UU UU U UU UU U UU UU U UU UU U U
U U U U U UU U UU U U
".~".~'a~ 'a~ "a".~~ ".~~ "J~ 'a"a"~~'a~~ ".~~".~~ ~"~.-~~'a'.~"a~ 'a
UU UU U UU UU U UU UU U UU UU UU U UU U UU UU U UU U U
UU UU U UU UU U UU UU U UU UU UU U UU U UU UU U UU U U
UU UU U UU UU U UU UU U UU UU UU U UU U UU UU U UU U U
UU UU U UU UU U UU U U UU U UU U UU U UU UU U UU U
U U U
C9C7C7C7c7C7C7L7C7C7C7C7C7C7C7C7C7C7L7C7C7C7C7L7C7ChC7L7C7C7L7C7C7C7
~ ~a'
C7L7ChC7L7U'C7C7U'C7UC7C7C7L7C7L7C7C7C7C7C7C7C9C7C7L7C7C7C7U'C7U L7
L7U C7L7C7L9C7C7U'C7C7C7C9C7C7C7C7U'L7U'ChL7C7U'C7C7C7C7C7C9L7C7U'C7
C7Ur.G'aU",.~'a~ L7"a'J'a p 'Jp UU R'U U UC7U'U".~U"aU ~U U 'a
r.~r.~p C7pC7U' ".~r.~U U U ~U'Ur.~C7U U L7'JU''aU'".~U r.CU'U p U
U pU r.~C7p 'aU U U''a~C7p UU'b,'C7'JU C7"a'ar.~U'U'UU U'U''aU U
iU'JFC''.-~U~'C~.7 ''UJp ~ ~ UCU'JU C~'JU "U'JL~9tU.'lUp''~~'C~'J'U.-~'U UU
~C~'JtU.9~U U'UU'
U ~"~~ ~~ U' U ~77 ' ~ "UU UU '~'~ U ~ bU U'U U UU'U
J " 7 ' ~ '
U'' ', C7 U'C7a L..L7~ C .~R'UU U' U L7L7C UU LC U UL U C7
7 ~C7 A' R'"a U 7 U U U7 C77 J.7 J '
U' U' U'' C7
~
. . , . , a
Md~r-)InN MV~Nl0l0rN 01O InrIN MW rN b rCO01N01Or!N OM c<'Ll7
01o1O01O OO OO 01Oo Or1r1<ir-Ir1r101rir1r10101e-I,-INN N ON N N
tnV7NIPa0ma0mofina0m a0m !n00a00~m Ln0000OIn117Om a0m N bN opaD
Ur.Cr.~U U 'aU U"a ",~ UL7U U'p C7U'U'r~C7UU U C7C7Ur.~L7U'U r~U
'aC7'~ ~ p" 7~ ~ U~ C U 7 7 U C 7U
~ ~
. r. , Cr. Ur- r.~C UC C7 r-C U r~C7UU L7(7U U r.~
FCr-CUp p p U'U ULh U'r.~UL7UU C9~ U rCC7r.~C7r.~UU ~ C9U U U
~
U',~r$U U r.~'J".~r.~ U U'UC7r-CU U ~ Ur.~C9U'Ch(~U U r~C7U U
UU'L7rC~ ~ C7U ~ ~U C7U r~r.CUr.CUU U C7UC7C7p U UL7C7U
~ ~
"aU'r$U'p U r~U'C7Ur.~r.~U ChU pU r.~C7U ~ p UU C7L7U Ur.CL7U'
pp pC U CU U C "7 U U p U C7U U U 7 "
~
F r- r-aC C7 C7 C9 ~ r.~C . UC7r.~C7
U 7~ C U'C ~~ U C ~ 7 C ' 7U ''
L r. r FCr.rC FC C7C p7 U C U C7FCC7r-CC7.~L7C7r-C
ChC7C7C7U ChL7U'C7U UU C7C7C7C7L7L7L7C7C7L7C7L7C7C7.C7CL7L7C7C7C7L7
bU'r.C~C7a'r.C FC~ r.CFCU r-C~ r.CC7C7U U'C7C7r.CbC7UL7rCC7r.CC7L7
Up " U~ UU U' 'U ' C 'U U t7U'U U~ UU 7 '
~'
C7p UU . r.~U Ur.~U'ha UFCU UC7Ur.UaCU UC7~ UU r.CU ( ~U r.GU
' L7 Cp FC U U UU r.~ C9C7U U r~
C.~7 r~U "UaU~ U~ U r~U ~~ r~U~U'L~'Jr.~U~ U ~U U ~C7C~9~U'U U'aU U
~~ ~ ~
U",~bp U FCU pU r-CC7~ UL7C7~U ~U C~7C7L~7pU C7 U ~
C7"ar.C~U C7FC~ UrCFC ~
~FCU C7Lr-CU'L7
FCU ~r-Cr.~U"~'Up r.~UU pp U C7U L7 r.~C7C7C7"aC7 7
~m NM b br wm ,--iwInr~ r avN rm mIwo rao~ movMw o rM w In
ra0010101OO NN M MW d~If7In1I7b bb tor r rr CO00CO01o~o~p~O O O
1f1InInu1If1bb b1010bb bb b 101010b 101Dtob10b l0b lDb ~Otor r r
.
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
01O r1N M WU1~Or N 01O r1N M d'u1~Or ODo~O r1N M ~HL(1~Dro001O r1N
NM MM M MM MM M Md~V'd'tHWH doV~~H<HL(1L~!f7Lf1t11!f71f11nIf1N\O1ptD
OO OO O OO OO O OO OO O OO OO O OO OO O OO O OO OO O O
NN NN N NN NN N NN NN N NN NN N NN NN N NN N NN NN N N
C7L7~p L7C7FCUU U UL7a,"a L7L7U 'a~ U'~"JU'U'U'U'U'C7C4U'U'D U U
C7C7C7L7t7r.CC7r.CU U UFCUU U C7p r.~',~U pU p C7r.~p p 1~p Ur.~U'C7
C7'7"'~9 ' L7'7UU U 'U U' ' ' U ' '~ '
7
, ,C ry , U a , ~A C7U UU U L7UU U UU C7U U U
'aU C7rCC7".~'U UU C7r.Cr-CU C7 , ~C7 C7r.CU'Cht7C7U'C7U'C7 C7U'C7
UU C7C~" UC UC7~ UU '~ 7 U'U U ~ CU ' '
~ ~ '
. F r , L a r UC7FC~a ~ C7C7 C7C7C7
UU FC'JU FCU Ur~U r$U Ur.~ Up LhU U Ur.~C7L7U UC7U'~'r~rC7'JL7
" ~
U',7C7U R'UU U'U a'UU ',7C7~ ",.~'C7Ch',~C7a'L7U'C7L7C7L7t7..U'.C7U C7
U' 'a
~U pr.CU UU aFCU U" pU'L7rCL7Up rCC7U C7rC'a~'a''a"C7rC' U''
' ~ ~
, , . a a
L7C7C7C7C7L7C9C)C7C7L7C7L~C7C7C7C7C7C7C7C7C7C7L7C7C7C7C7C7L7U.C7C7L7
C'JL'JC'Jt')C7C'JC'JCht')t')C7C9C7t')U'C9U'U'C9U'U'L7U'C9C9C7t7t4C9C7t7U'C9C9
C7C7t7C7C7C7C7UL7C7c7C7ChC7C7C7L7U'L7C7UC7UC7C7L7L7C7C7C9C7U L7C7
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
~'~''J"a'a"~'~ "a"a"a~"a"a"a"a"ap 'a'.~"ap"ab"aC~p".~~ C~",.~".~',7
i7L7C7L9C7CJC7C9L9L7C7U'U'C7C7U'C7C7L7C7L7C7L7U'C7L7C7U'C7C7ChC7C7L7
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
'a~ ". 'a'a'a"a". 'a~ ~~ ". ". ~ ~ ". 'J~ ".~'a"a'a"~'a~".~p "a
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
~ a'~
U'C7U'C7C7C77 ChL77 C77 77 7 77 77 ' '
C C L CC L CC CL C7UC7C7C7L7L7C7C7C7C7UC7L7C7
UL7C7L7t7C7C7C7U L7C7C7C7L7C7C7C7C7C7t9C7C7UC7U C7C7C7C7C7UC7C7C9
UU UU U U UU U UU UU U U
U U U U U UU UU U U U UU UU U U
a'~''J"~''a"a".~~ ".~".~"~".~"~'~"a"a".a"a"a~ 'J'".~'~.~'".~~ "a".~"a".~"a'a
"~''J~~ "~"J"a'a~ ~ ". ~"a'a~'a'a"a~ ~"a'a"a'a~". C~"J'"a~ ".~"~
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
~".~~ ~~ ~'a~ "a~ p~ ".~~".~~~ ~ ".~~ 'a~ ".~~".~~ ".~D C~~ p ~
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
L9L7ChC7L7L9L7C7C7U'C7L7U'C7C7L7C7C7U'C9C9L7C7U'C7U'C7U'C7I7C7L7U C7
~ R'
C7C7C9L7U C7C9U'U C7L7U'C7L7C7U'C7C7U L7U'C7C9C7C7C7C7C7L7C7L7C7ChC7
C7L7C7L7U'C7U C7L9C9C7U C7C7C7UC7UU U UL9C7C7C7C7C'JL7CJC7C7L7U L7
UC7r.~U U UC7~U U p 'J",~',~ U ".~L7r.CU!~pC7U'C7ChU'C7U'U'C7C7C7
U'7 UU 7 U U~ ~ ' ' ' '
C U CrC U p C7r.C brCC7ar.CL7a p pp C7L7 L7U U a
L7U'UU U'FCU U'aU 'a ~C7U ~ UU U rZ'lC7U U U'C7~'r C7L7' U
~ " ~
C7",~UC7a'UR'UU ',~"a a'~ '',~C7U t7a'U .'.7C7C7L7C7L7,.-a'C7C7U C7
'aC7Ur-CU r-CU ~ p U~ C7 ~ UC7r.CC7~ U'C7 "..~'',~~',~L7C7".~'JC7L7FC
~ p p C7 ",~
C7p L7U r.CUU U U ".~ U'L7C7U~ UU ~ C7C7~U C7UU U Ur.~UC7U U
UU ~ ~ ~ U
C7Ch U U~ ~',~p pC ~p U rCC"~'p L7C7C7'ap pC7C7~C7U p p
7 7
1Dr1Nr OD01M CHO ,-1NM V~~Or mtt110t11l0rOD01O r a001O r1r1NN M M
NO ON N NO OM M MM Mr1r1r1M MO O MM r1N O OO r1r1N NriN ri
Wl0~DO m Ml01Dm W ma0Nu7Lnll1CONl010a0N V1N 10~DlOl~1DN LOl0N l0
UU U'L7C7U'r.C FC~ U r~r.~U'~"aUr.~r.CUU Ur.~r-~r.~U U FCU C7C7r.~
CU 7p 7 i97 7 p 7C7 7 U
'
r. ~ L C C C U C pC7U U UU U UU U UC7Ur.CU U
Ur.~Ut7p U'U't7 C7~~ UU U Chp C7C9p UU ",~U'U C7C7U'C7p C7U U'U'
U C7~ C7pC7~C ~ U' Up ~ C7U pU r-CFCU pFCFCFCFCFCUFC~U r.C",~
Ur.~L7U ",~L7",~C7C7 "a~ UL9Up L7Ur.~UU U UU U U',~UU U U
~
UU C7C7U ~U'C7r.CC7 C7 U C7U L7L7C7pU UC7U'UU r.C'a UU U U'
UU C7U'' U" 7L7~ U' U p ~ " " U~ U~
~ ~
U , C r J U r.~, r.r-Cr.~r.~U UC7U r~
U'U "' 7 'U p7 7 ~ ~7 ~ ' C U
~
.U L U C L r~ r ~ L U ,~C7r.r.~ U UU U UU UU U U
C7C7L7L7C7UC7C7U C7C9C7UC7C7U'C7C7C7C7C7U'C7C7L7t7ChC7C7L7L7C7C'JC7
r.CL7r-C".~C7U'L7".~"aC7U'FC~U U '.7U C7~ ~ UC7Up r.CFC~ FCFCU ",.WC U FC
C7C UL9p C9C7UL7p C9C7 U p ~U ~U UU U U U '
F r~U U U U ~U ~U U U
UU ~ U U ~U ~ U U ~
CC UU ~ 'Jp CU " CU'~ U U~ L7 ~C7UU p ~~ ~ ~U U U U
7_7 C 9 ,~'J" ' ~ ~ ~
'
FCL7Up U r~C7UL7U ~,.~U~ U ~L7 U U~ UU U UU U UU U U U
Ur.~r~U p Ur.CC7C7C7UChU' r.~''..~'JL7U L7C,~7U U',~U C7C7U'U'C7UChU L7
U ~ U ~U U UU U ~ UU UU U UU U U
U ~ U p CL C C ~~ UC ~~ U ~FC U U~ C C
.'JJ7 7 7 7
rc0ON M W.f1NO r1N~ rcpr1~01OM 10O~O r1M ~O01u1r1rM O1Do0cN
OO NN N NN NM M MM M'WIf1VIu1l0l0l0l0r a0O~0101O ,-I,-IN MM M <H
rr rr r rr rr r rr rr r rr rr r rr rr r r~ o00om oom m m
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
86
mm ior wm o,-~N <nm vor movor-iN m<rumo r aom o ,~N c~d m vo
vovovotoiovoiorr r rr rr r rr wm m mm mm m aom ovova~o~m a~o~
00 00 0 00 00 0 00 00 0 00 00 0 00 00 0 00 0 00 00 0 0
NN NN N NN NN N NN NN N NN NN N NN NN N NN N NN NN N N
L7C7C7U'C7L7'aC7~ ~ r.CU'U~ ",.~UC7pU p r.C~ ~FCU UU r$'aChChL7FC'a
U'~ bC L7C7U'Up 'U'U 7 '7 7 ''
r, U C UC UC7C U U C7r.CU UU U U U
C7U UU L7pp pr.~(7C7"apU'U C9U Up "J~p rU Ch"U U C9U U~ U r
~ ~' ~
C7U'C7U'C7C7U R'ChU'pU ',7U L7U'.7'~.~U A',(7a',.U '.7,U C7UU L7U U .
~ 7 77 t7t~U U7 CU U ~' ' U
C~
UU UU ~ U~ FC~ V C CC U C ~ r. C7~ U ~ U~ ~U U
~ UU U
C7C7C7(7L7U'U C7C7L ' L p '~ C7~ p 'aKCU ~ U U ~
"' "" ' " '' 7 'J ' ' '
' "
C7 U Up r-CFC
~R a.~.7.~L7R.~~ UU C7L7U ,7U ,7 R aU U
U'C7UC7L7C7C7C7L9U'C7t7t7ChC7L7C~C7L7C7C7C7L7C7C7L7L7C7C7C7C7C7ChC7
C7C7C7t')C7LhC7C7U'L7L7C7C7L7C7L7C9C7C7C9C7U'C7C7ChC7C7U'C7C7C7C7ChU'
C7C7t7U C7C7L7C7U L7i7C7L7C7C7C9ChC7C7i7C7C7C7L9Cht7C7U C7L7C7V C7C7
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
'a~ 'a~ 'a'a'a".~~ ".~~'a"ab '.-~'',7",.7',7".~".~".~~ ".~~ ".~~'.~'a".~~ ".~p
UC7C7ChC7C7C7L7C~C7Ui7C7C7C7C7ChL7C7C7L7C7C7L7U'C7C7V c7t7C7L7C7C7
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
L)C9C7L9C7i9L7U'C7L7C7C7CJC'JL7C7L9L7U'C7VC'JC7C7i7C7U'C7C7L7U'C7c7C'J
UL7L7C7C'JUC'JUC7U C7C7C7C7C7C7C7C7U C7Z7C7C7L7U C7U C7C7C7L7C7C7C7
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
~~ "a~~ D"a~ ~~ ~b ~ "a",a". "a~".~",.~"a~ "a"J"a"a~ ~"a
C 'a~ ",.~b~ ~A "a~"a~~ ',~'"J~ ~~ ~ 'a",.~~~ "a~~ ~ 'a~ ~"a
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
'.~~ "a~ 'a~'a"a"J"a~'a~'a~ 'a~ 'a"a'a"a'a"a'a'a".~'~'a~'a~"J'~ 'a
UU UU U UU UU U UU UU U UU UU U UU UU U UU U UU UU U U
C7L7C7C9C7C7L7C7ChC9C7C7LhL7L7C7C7L7C7chC7L7C7U C7C7C7U'C7c7C7C7C7C7
C7C7L7C~C7C7C7C7C9C7L7L7C7C7C7C9C7C7C7L7L7C7C7V C7C7C7U L7L7C7V L7U
C7C'JC7C7C7L7L7I7C7ChC7ChU'C7LJL7U C7C7ChC7C7LJC7U'C7U'LhC7C7C7C7t7C7
C7U'C7C9C9C7L7U'L7C7U'U U'L7U'L7L7L7U'LhU'ClU'L7L7L7C~U'C7C7C7C7U'C7
FC~ 'aCU'Jp U~ p"UaU ~CVhU'.~U U'~ CV7~U'r-CUL7pU ~ ",.~LU7V UC7'aU
UU UU'U ",~".~UU' U'U UU U ",.~U "aU C7",~ U 'aC7 C7p U",~~ r~
C7C7U'LhC7FCFCU'C7U'Up ''..~U C7UC7~a'U ~L7C7U'~ ~ "aU r.~ r~U
'.7W 7LhU pU'pb U C7U UC7p U'L7Up U ~~ U~ C7UU U Ur.CU~ U C7
UU C9~ 'ar.~r.~C9U'i9C7U Up U L7 "J~ U' U ~C7U UU U FCU pr.~'',~'U
U U~ ~ ~~ U U U V V U U
C L " r-CC7 ',~ p( C "a~ ~CFC U C7r-CC7~ ~ ~p ~ U'
7 'J a 9 7
ehW L(110r COIPl00101Or o0r1N c~'1O r1Q1O cHLONM 10d~r I1100a110r r-1N
r1N r1r1rir1N Nr1f~'1cHN NV~d'V~N NN M V~d~NN V~NV~N V'd~NN f~'1('~1
10Inl0tp1Dl0N In~OOJODInInW ODN10lDInInODOD10l0N 10CO1DCOCO101DLhN
r.~U Ur.~r.~',~'U r.~A,'U'r.CChr.CU U U'r.C",~p ',~C7ChC7U pU r.~r.C r.Cp U
UU UU p UL7UU U C7r.~UC7U U r.~C7p C7U C9UC7U C7 C7 (7
~
C7C7UFC~C".~'aUU U UL7C7FCLh ~~ U L7".~U C7U'U'U'"a 'a C7
r~~ UU L7r-CU r~~ U'UC7C7U R'UU C7 C7~'JL9r-CU U'L7C9L7p C7 C7U
' ' '''' ' ' ' ' ' '
UU UU U ,~~ UU U L7FC~C7U UU J,.~U FCa ~U U U~ U ~U a~ a U
U'C7L7U C7r~CC7U UC7C7L7U'FCC7~~ C9Ur C7r U Ur U' "a
C C C C
br~r~U ~ .~ r.Cr.~~ ~U C7r.CU'Ur.CUU ~ C7. r.~C7. r$U ~ I7. r.~U'
L7 U U U
UU UU U Ur-~UU C7 U r.~C7r.~C7U Ur.Cr~C7C7Up (gUU'U C7L7Up
L7L7L7U L7UL7L7U L7C7U'L7L7C'JL7L7L7L7t7L7L7C'JL7L7L7L7C7C7t7L7ChC9Ch
r.C!~r.CFC~ ~U pr.C~CL7U UFCLh~C7r.Cp '.~pU'U'~ ",~C7U U'~C7Lh~ p ',~
UU UU U UC7UU U FC UC7r.~L7r.~Up r.~",~U' (7~U U U'U C7
C7L7C7C7rCC7FC'~C7U U~ C7U C7C7L9C7U ~ C7r 'a~ p U'r U U~ 'a7
~' C C
. . r - C r-C
FCr.~pr~U r.~U r.~",~'r-CUU'UU U r$L7ChU 'JC7FCU ~ U ~ ChU pC9U
U U U U ~U U~'~ ~ aV U ~V UU U U
C7U'C7 R'~'a'"~ d, C7 7 C.a'a' L C.G C Ch" C7"
L 7 'J 7 7 ~ ~
UU UU U U~ U"~~ ~V ~ U UU C7U U ~ ~C7C7U~ C7UU C7'aU'L7
~ ' L7U '' ~ ' ' ' 7C7b ' " '
7
, R RU , . a U C aU UU J R
rO M101f1mO ro ODay~710OJ01r1N InO 01N(~'1C~41<-INr o0NM Wr 01d~
d~u7If7Lf7101Dr rCO0101O OO O r1r!r1N N MM ('~M s~WW W InInIl7LnN ~p
a0N mm N a0c0mW O a001O101Ov01010101010101~1T 010101O10101010101a1
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
87
rm o~o ~ NM d~rntoroorno ,-~NM m vorm o~o .-1NM ~rumo rm avo
010101O O OO OO O OO Or1r1r1r1n-1,--~,-Ir1~-I~-.~N N NN NN N NN NM
OO Or1r1rir1n-Ir1r1r1r1r1v-Ir1r1riv-Ir1v-ir1r1r-1r1rW-Iv-Ir1rir1v-iv-1riw-I
NN NN N NN NN N NN NN N NN NN N NN NN N NN NN N NN NN
r.CU t7".~U UL7U C7(7"~t7r.C'JU UU C7UU C7'aU r.CU r.~U U C7C7'JU
r.~UC7p r.~U'p"Jp U'p Ur.~L7UU U'U (~UL7UU r.~U~ UU U'UU r.~p
pU pC7~ Up L7U U'~U ChU'C7UU'UL7U UU ".~r.~U C7U UU C7UC7UC7
U ~C7U L7r.Cpp C7UC7FCC7"~'C7U U~ U C7".~UU C7UU UL7U'Ur.Cb
FCr.CC7'aC7U'L7C7FCr~U'~ U UU U'U U UU r.~C7U UU C~C7U C7U i7
'J~ UC7"~'r~U ~L9p UC7U'U U UU''aU C7pr.CUU U r.CL7L7C7U U
" '" ' ' ' " ' "
'
JU U~ ~ C7U UC9U C7C7aU U L7a UU U UU UU r.CUC7(7U U C7,~ U
r-C(hUr.~C7U"..~p~ r.C"a',~UC7U ".~U UC7".~r.CU'Ur.~U UC7UU C7FCr.C p
C7C7C7U C7C7C7ChL7C7U'C7C7L7C7C7C7C7C7C7C7C7(7C7C7L7C7UC7U C7C7C7C7
C9C7UC7t7C7C7C9C7L7C7U C7C7C7UC7UC7C7C7L9C7L7C7U.C7C7C7C7C7C7C7C7
UL9UC7U C7U UC7U C7U UU C7UC7UC7C7C7U C7C7C7C7C7C7C7C7U'C7C7C7
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
~".~'a". "a~ ". ~ ~~ ~ 'J~ 'a~ p "a'a'a~J~ 'a~ 'a~ ". '. "a
L7L7C7C7C7C7L7ChC7C7C7C7C7C7L7C7c7C7L7C7c7C7t7C7L7C7U'C7C7C7C7C7ChL7
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
"a~ "a~ p ~".~~".~"a~".~~"a~ '.~~ 'a".-~"a~".~b"a~ "a~ ~"~"a'a~ ".~~
a'~ ~
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
C7C7C9C7C7C7C9C7L7C7C7C7C7ChC7C7C7C7C7C7C7L7t7C7C7L7L7t7L7L7C7C7C7Ch
C7C7C7C7C7C'JC7ChU C7L9C7C7C7L9C7C7L7C7C7C7C7C7L7C7L7C7C7C7C7LaC7C7C7
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
'a~ 'a~ "J-"~~ "J~ "a~".~'a~ "~~"~b~ 'a"J'a"J'a"~"J~ 'J~ ".~~"~
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
"J~ 'a'a'a"~''J~~ 'a~'a~'a~ ".7~ 'J~ '. "J'~'"J~ ". ". "J~~ 'a
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
C7C7C7L)C7C7C9C7L7C7C7C7C7C7C7UC7t7C7C7C~C7C7U C7UC7UC7U C7U C9C7
C7U C7C7L7C7C7UC7C7C7C7C7C7C7C'JL7L7CJL7C7C7C7L7C7ChC7C7C7C7L7C7C7U'
C7C9L7L7C7L7C7C7t7ChC7C7C7L7C7U'L7C7C7C9c7C7L7C7L7C7L7L7L7L7C7z7c7U
C7U U'C7U "a"',..~'ar~C7C7U U'U U'UU L7"ar.CU'U ~U U U'U UU'C7r.~p ",.7U
UU'C7" ' U7 7U ' ~7 U7 " U7 '
~ ~ ~
UC7U', , pC CC7, r.L UL a U'C UU U U~ UU (7ChU C7U r.CpC7UU
FCU p U' U C7U 'aU U pr.CC7UU UC7U'U'U'UC7U r.~ UU'
C7C7r.~U'~ C7C7L7".~C7L9U L7U U Up UU U r.~U UC7C7UU C7r.~r.~U C7C7
~
C7r.~C7U U'"a'J'aU FC~L7",.~U U 'aU r.CU'U UU U'C7U UU'~U ",~"..~ C7r.~
L7C7".7U ',~U'C7UC7C7Up UU U'U~ UU r.~UU'C7U U Ur.~U~ r.CCU C7U
7
~~ UU p ~U U~ U U UU U UU U U UU UU U U ~~ U ~U U
~
C L U''.U' ( L a' U''J C ry"' (., 'J
7 7 ~ 7 7 C 7 a ~
Mo m<r~-fNM <romn om ,-Wo N rcoMiow rowno ,-WoN Mr m aoavow
M1f1NM InLf1InU1N M MIf7Mu1M InV7MM M MV1M~ l0M~O~DM M MM W~O
InOD~DLOm M004D10InlD00lDm lD0000~ON 10Lnm 1DCDN ~DODo0101n~DInLnN
U,.hU'C7~'UC7C7U a'UC7C7L7~'U'U U'.h.C7UU C7L7U U'"~'R'"~'R'pU'C7FC
~ ~ "' U h7 7U 7 ~ ' ' ' ' ' '
'~ "'
UU r~7 r.C U '~" U CC CC7L ..C7UC7U UU UC7U UU ,~FCC7~r.CpU
C U'U Ch~ C7 U C7p C7C7'aC7U UU'C9C7'aU'ap UC7UU'
Up UC7U r.~r~r.~C7p r.~U r.CC7C7FCL7pU U'C7C7UU C9C7U pC7r.~~p U'.~
UU 'aU ~ UU UFCU UU'UU'L7L7r.~U'C7C9''7L7U'U U L7U'Up p L7 UU
'U U" 7 C U UC97~ 7 U7 7 U ' '
U ,.W r$r.U C7 Cr.C C r.C~ U C7L C9 U UU C7U U a
U'U Ur~r$C7U UC7r.C".-~U C7U r.~U'U L7C7C7C7~ C7L7U UC7UC7'ar~U C.U7U
'' "
UL7UU C7r.~r$r.Cp U UC7UC7U UU U~ p UC7,~C7C7UC7C7U U pr.Cr.~Ch
C7C7C7L7C7ChU C7C9C7L7L7C7C7C7UC7L7C7C7C7C7C7C7U C7C7L7C7C7L7C7C7C7
pU C7'aU U'r-CFC~C'aFC~=CL7U C7FCC7U'U FC'.~U C7p LhC7U U'L7U ~J'a",~FC
C7Ur.Cp UC7UU L7UU r.~C7U Ur.~C9C7U'U'C7UU'p C7U UC7C7U~ ~L7
p C7U ~ p7 CU ~ 7U UL7L7ChU ~7 U ~p L7C7' ~U UU
r. C r. r.C L r. U C7C7p p
Ch~ ',~p U r.~U UU U ~'aUFCL7C7C7UL7L7L7C73~U U'L7L7UU L7UU'U
U 7 Up C U 'U ~U C U7 ' UC C7C7 7 'U ''
"
C r.~F U ,. F C Ur.~C7 F U C7C U U Ua
L7 U p ChFCUU'U FCC7UU U C7U C7U U'U'C7~"J'U'UU'C9C7U C7U U'U
U U ~U ~ UU U U U U UU U ~ ~~ pU U
'.L7 FCL77 C FC ',7d,C U'C LU' FCC '.C i L7U FCC9
7 C 7 'J 9 9 7 7'J 7
00O ~-101c~Inr tn~ M ~N MIn1001O ~M N r41ON M 'W~ ~O N LCIO1Inr
10r rr o7c0p7p~O O Or1r1r1r1r-1N NN N NN MM M MM M'~'W~dWnIn
O O OO OO O OO OO O OO OO O OO OO O OO OO
r1r~ri'iv-Iririv-Iririr1ri~-ir1riririr-1rlr1riwiririr-Ir-I
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
88
~-IN MW vor oom o r1N M<rW or mm o .-IN MW vor aoo~o .-iN M<r
MM MM M MM MM d'~d'V'W W d'W cHV~!ntnIn!f1!n!n!n!nInin1Dt0l010to
rir1rir-i'-Ir-1r1r1r1rar1,-1r1<iririr1r-Ir1,-Ir-ir1r1rir1riririririv-Irir-Ir-I
NN NN N NN NN N NN NN N NN NN N NN NN N NN NN N NN NN
bU pU C7r.C",.~UU p C7U U~ U'L7',~UU U UU pU r.~U",~L7C7C7p~ UU
U''aUU'C7UU ~p C7Up C7L7'aU'UL7U Up ' '
"
,.. U~ U JU C7U C7r.CLhUr.~
U UC7C7UU r.CU'U pU C7p t7p ChU U UU UU U UU rL7p L7t9rU
C ~
U L7U'FCp'aUU p UC7L7p C7'aU UU U ~U r.CU FCUFC.'ar.~C7r~.U
U U
C7C7~ U U~ ~'aU C7r.CC7L7p UU'UU p Ur.~U~ U r.~L7U~ U'r.~U U"J
UC7U'U U UU C7U U'r.CU ".~p U C7U UU U UU UU U L7C7"..~U'C7UU pC7
'JL7r.~U p ",.~p UL7r.~UU C7U C7UU Up U r.~U r.~U ~ L7~ r.~U'r.CUr.~C7C7
Ur~U".~'U r~U'pr~U UU pL7U UU 'aU FCUr~U'aU r.~U ",~r.CU r.CU C7U
C7ChC7ChC7U'C7C7C7C7L7C7c')C7C7C7L7C7C7C7C7C7C7ChC7C7L7C7C7L7C7C7C7C9
U'C7C7C7C7L7LhL7U C~C7U'C7U'C7L7C7U'C7C9U'L7C7i~C7C7C7C7C7CJC7C7C7C7
UC7C7C7U L7C7UC7U C7L7C7~7L7C7C7Z7C7U C7L7C7U C7C7CJC7C7C7C7L7L~C7
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
C9C7L9C7C7C7c7C7c7t7UC7c7C7i7C7L7C7ChC7C7C7L9C7C7C7i7C7U'C7C7C7C7Ch
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
''~"J'a'a~ 'a~ p"~''J"a'a"J'J"~'a".~b"~'"J'a'a"a'a".~'a"aC~"~'a~'.~p".~'
~~ R'
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
7C7L9h C77L777 h 7 h7 ' ' '
L L L LC C CC9tL C7UC7t7C7U C7U C7i7C7C7C L7L7C7C7L7C7C7
C7C7C7C7C7C7L7C7L')ChC7C7C9C7L7C7C7L7L7C7C7C~C7C7C7C7J L7C7U L7L7C7L7
Ch
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
".~~ ~"aA ".~~ "a~ ".~~".~~~ ~ b~ p~ ~ C~~ C~~ ~ ~~ ".~".~
'~'~ ".~b ".~~~ ~~ C~"..~".~~".~~ ".~~ ".~",~'"J~~ "a~ "a~~ pp p ".~".~~'.~
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
'a'a"'~.-~".~~ ".~~ 'J~ 'J~'J~'a"~'"J~ 'a"a'aC-~''a"a'a~ "a".-~'"a
UU UU U UU UU U UU UU U UU UU U UU UU U UU UU U UU UU
t7L7C7C7C7C7U L7U C7ChC7ChC7C7UC7C7C7U C7L7C7L7C7L7C7UC7C~C7L7C7C7
~ ~ R'
C7C7L9C7L7L7C7ChL7C7U'C7C7L7C7C9L7C7C7L7C7C7LhC7L7C7L9C7C7C9U'C7C7L7
C9L7C7C7C7C7C7C7C7C7L7C7CJC7C7L9C7C7C7C7L7C7U'C7U'i7L7C7C7CJU'C7CJC7
C7U pU p p'JU'U U U~ C7U U Up Ur.~U ~U pU r-CU"..~C7U FCU',~UC7
C7p U~ C 7U ~U U UU " " "' '
~
r.C r. ~ U .U r.CU r.CUJ Ur~L7pr.~FCr.~U ,~U UC9
C7U Ua'U UC7UU ~ C7UU ",~UU UU U UU UC7C7R',.7C7U U U'L7U'U
'
U 'U p p~ U~ U'C7U",~U Ur.CUr.CU pU r.~L7r~pU L7U p L7U UC7
U" ,~' L7UU U 7 7" UU U U ' ' ' " ' '
~ C~ ~ '
. r., C C. r.~ r.CU p Ua,Lh~t,U UU .~p U UU UL7
Ur-~UChU U'U UC7U'pU'pU r.~UU UU U UC7U'U U L7FC~L7U'UC7C7
pU pU p r.CU L7~ C7p Ur.~U Ur.CU'JU r.CL7~U ",..~r.~C7U'C7U L7L9r.~U
" ' ~' ' '' ' ' ' '
'
U.~L7a U UU a C7aU UU U r.~U ,~U r$C7r.~Ua a UL7UU U C7U UC9
aw vor m ,-iN om o r1~-INN M <rm toM r ~roorno u~,~N voM r cri.wom
Mvovoiovo~<rmo r rd~a~r r rr r~nr ~r roow ooao<rm ~ mooaod~
vom mm m mw om m ooiovom oomaomvom voao0om iocom iom ~ mcomo
Chr-CUr-CC7U'C7~FCU ~U'C7C7C7DU'r.CU'"'.~U'aC7r$'aUU UC7C7UC7C7U
h C' " U ' " "
~ '
FCC r.U r-C.L7 U r.C FCC7~ C7L7~ C7r-CC7aU ,~C7FC~U UU C7UU pC7
L7~ C7U ChUC7U'U U r.CU r~U''aC7C7L7U'C7C9U UL9U'U'a U U C7U U''.~'
C9R''aa'U C7t7C7"JU Ury't7C7C7..hC7'.7C7a'C7"..~U"aC7C7U ~a'U C7U'UU
~U'FCL7~CFC'aC7p ".~UU C7rCC7L7~ U''aC7r.CC7~U ".~r-CC7UL7r.~UC7C9U
UChL7'JU'C7U C7C7p ~U C7C9~ U'C7C7U'U'C7C7IhU U ",.~~ UC7U'UU UC7
U~ C7C ' U7 "C7Lh L7C77 C 7' ' "
~
F a C .- C FCC7~Ch~ C7F CJ U FCJ .~~ L7FCU UU
UU'r.CL7FC~Cr.CUL7C7C7 UC7L7L7r.CCh"aC7pC7FCC7p L7r~UC7p C7r-CC7U
C7C7C7C7C9UC7L7L7L7C7C7L7C7C7C7C7C7CJi7C7C7C7C7C7C7t7L7C7C7C7C7C7Ch
U''aU'r.CU'pU r-C'Jt7U'U'r-CU U'C7C7r.CC7p C7~ t7FCC7"JC7FC'aU'~U'UL7
~CU pC7~ FC~ L7U p C7L7UC7U C7C7C7FCL7~U'~U'r.CUp pU ~ C7'aUU
ChU UU'ChC7C7UL7U "aC7~r.~U'UC7C7C7C9L7U'U'C7C7UU r.CU U L7C7r.~U
U U',~t7C7'aR'~ C7U".~'UU ~ C7U C7U'r-CL7'aC7",.~'C7(~U C7'aU pL7C7r-C
C7UU "aFCFCChChFCL7U Ur.CU r.~C7C7L7C7r.CU'pC7~ C7~ C7r.C~ U'aC7U'
U'C7U U U'U'"~..~U L7r.~C7',~U ~ Ur.CUU'C9L7U'L7L7C7C9L7".~U r~UU ~C7
Ury'U'U U C7U'A,'A,'U C7a'pU U a'U L7U C7C7d,C7'.7U'a'C7UU U pU C7"J
FCC7FCC7U ~FCU'C7~ UC7C7"JU UFCFCC7C7U'U'r-CC7".~C7r-CUU U FC'aC7L7
a0M V~lDr OM U1O 41Or101r1N M~ ~r O1ON MN 10r1N ~Or1N t~L(101O
!Ii~Ol0l0l0rr rr r OJ07OJ01010101Q10101OO OO O rir1r1M M MM MW
OO OO O OO OO O OO OO O OO OO O r1w1rir1rIr1r1r1r1~-I~-I~ ~,-.~
riririr-1ririr1c-Ir-1r1n-Ir~lriv-Iriri~-1riw-Ir1r-Irir1r-1r~rir1rir1r~1r-
1ririr1
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
89
mtoro0ovo,-~N mm Wo r o0ovoH Nc~'~~nmo r o0ovo~ N c~~ uwo r m
voiovoio~ rr r rr rr r rr oom mm m mo~m mm mrnm avm mo~ovm
Hr-iHH H HrtH r-tH rtH r-tHr1HH Hr-tr(rtH H HH HH H r1H Hn-1H H
NN NN N NN N NN NN N NN NN NN N NN N NN NN N NN NN N N
UU Ch~ p C7r-CChpp UL7"aC7~ C7".~Up r.~C7Up "J"..7U Up UU 'J
'aU ".~ C7'aC7U'pC7U'p C7r.~C7 U UU ~U r.~C7L7'aU pU U
C7U'U'U'~ ~ p C7FC'aFCFCC7 U 'JU'~ FCU C7"JU~ ~ UU ~
C7C7''.~",~p U ",..~r.~",.7C7U C7 L7U t7 UC7U L7p 'J "JUU p U
UU ~~ U L7L7C7",~U'C7p ~U''JU ',7 U FCFCp 'aU p~ U UC7~~ U U
' " '
UC7C7U U FCC7FCz7U C7C7 C7p U.~U r.~UU U C7D pU p C7 U C7
L7C~".~U'FCUU''JUr.C~C9L7pU UC7".~~ U'r.CU C7'a~ Ub U ~ ~',~C7r.C
C7r.~pr.~U U~ L7r.~U r.~C7"aUU Up ~ ~ Up r.~D'a~U U ~U "aU r.CU
C7C7C7U'L7C7U C7C7C7U'L7C7VC7C7U'C7C7C7C7U'L9L7C7L7ChChC7C9L7C7C7C7
C7C7U'L9C7C9C7C7C7U'C7C7C7C7C7C7C7C7C7C7C7C9C7C9C7Vt7C7U'U'UC7C7L7
L7L7UL7U C7C7C7L7U C7C7U C7C9C7C9C9C7L7L7L7C7C7C7Ut7C7UC7C7L7L7L7
UU UU U UU U UU UU U UU UU UU U UU U UU UU U UU UU U U
UU UU U UU U UU UU U UU UU UU U UU U UU UU U UU UU U U
"a~ 'a'a~ ".~"J-"a"J"~''a~ "a'a~ ".~'a"~'".~~ "a'a"~'a"a'a'JD "a".~'a"~'"a'a
U'L7C7L7C7C7C7U L7L7L7C7L7C7C7C7C7L7L7U'C7L7C7C7C7L7C9C7C7L7C7C7L7C7
UU UU U UU U UU UU U UU UU UU U UU U UU UU U UU UU U U
~'a~"J'a~'.~~ "a".~~".~~ ~"J~"a'a~ "a~"a'a'a".~p'J".~".~"a".~".~p ",~
UU U U UU U UU UU UU U U U
U U U U U UU UU U UU UU U U
C7C7C7C9Z7L7L7C7C7C7C7L7U C7L7L7C7U'C7C7C7C7U C7L7C7C7L7UL7C7C7C7C'J
C7C~JUC~L7C7C7ChC7U C7C7U i7C7C7C7C7C7C7C7C7C7C7C7UC7C7C7C7C7C7C7C7
UU UU U UU U UU UU U UU UU UU U UU U UU UU U UU UU U U
".~p 'a".:~~ 'a'a'a'a~ 'a".~'~ "a~ p".~p".~'J'a'a'a'.~'a~'a".~".~~ "a".-
~''a".a
~b ~~ ".~~~ ~ p".~b~ ~ ~C~~"a".~~ ".~~~ ".~~p ".~~ ~ ''a
UU UU U UU U UU UU U UU UU UU U UU U UU UU U UU UU U U
UU UU U UU U UU UU U UU UU UU U UU U UU UU U UU UU U U
UU UU U UU U UU UU U UU UU UU U UU U UU UU U UU UU U U
~'.-~'~~ ~ ~~ ~ 'a~ b"J'. ~ ~~ ~~ ~ ~~ ~ ~"a~~ "a~
UU UU U UU U UU UU U UU UU UU U UU U UU UU U UU UU U U
L7C7C7U'C7U'U'U'C7C7C7L7C7C7L7U'C7C7L7C7C7L7C7U'L7L7C7C9U'C7C7C7U'C7
C7U C7C7C7C7C7C7C7L7C7C7CJC7C7C7C7C7C7C7C7C7C7C7ChC7C7C7ChC7C7C7C7C7
C7C7C7C7CJC7C7U C7C7UC7L7C7C7C7C7C7C7U C7U C7L7C7C7'CaC7C7U C7C7C7C7
C7C7C7U U U'U'r.CUC7p"JU UU p~ ~r.~U pC7U'~p UU L7U UC7U 'J
C7",~r-CU L9L7U'U C7p C7FCU U',~C7 L7".~'U~ C7 U UC7 C7r-C'JC7
A',U'UC7L7a'"aU ",~'C7C7'.7U "aU','7U A',U C9U L9'.~U C7p ~ ~ R'U C7a'
U",.~UC7r.CC7"J'L7C7C7L7p p C7p UC7 U C7FCC7~ UU ~p U UU r.CC7
L7p U~ V''~FCC~'JUU''J'aL~'J~ Up U ~ UC7C7'aC7~U U ~ U",~'L7C7
'aU'C7Ch ~ U ~ ~ ~
U'aC7pr-CFCr.CU 'a ~ U f7L"ap U",~Up Cr.C".~r$
~ ' " ~ ' 7 ' ~ 'J
p r.C U C7C7C7r.~U ,~C7p U C7 r.~U t9C7C7U UU J~ UU ~C7r.~U
fnVn01r m ON H 01N l0r O HN Md'Md'Ln1O~700rof01O 10HN Mr d~1f1
cNd~WOJN Ind~If1mN ~d~0101O~a~T Lf1Lf70101If1~ 01O101O Lf1OO OLf7O O
U7l11l0N ofl0Lf1l0Nl0tn!f147NN Wm l0l00007l0lf10700N01t0Q1T d1lOO101
rC "a U UU U pU'r.~U aC L7U",.~',~U U UU U'C7L7r.~ C7C7"...~U p C7
U~ U~ ~ Ur.CU r.C"a',~p C7 C7p C7C7U UFC~ L7 C7 ~ C7Up r.Cp
r.~U UU ~"'..~U Ur$r.~U r.~C7 r$C7UU C7 C7U' r.~U U r$
U U'',7U ~U r.CUU L7 U r.~C7~ C7',7U'U U r.~U C7C7 U'r.CU U
C7 C7U '.7U U UU U ry'Ua'C7U pL7U "aU a'U'C7 L7 U'C7'.7U
".~'U C7U U p U'r.CU U U'r.CU r.CU' 'aC7UU'U C7U pL9U ",~
~ ~ ~ ~
U pC7C)U C7Ur.~U C7C7r.~U ~U r.~U'"aU U' ~ U',~~ U
~
UU UC7U'UU p U'U r.~r.~(7U'L7r.~ Up U'r.CU U r~C7C9U U' U'U C7r.~
C7U'C7C7C7U'C7C7C7L7C7C7C7C7C7L7C7C7C7C7C7L7C7C7C7C7C7C7L7C9U'L7C7L7
U",~ b ChU'".~U ",~C7"~U r~C7U'C7FC "aL7FC".-~ FCL7C7 L7 C7''aC7
~ ' ~
UU U ".~U'U r~U''ar.~U U r.~U'C7U ~ U 'aC7U C7~ C7 U 'J
7 U " ' 7 U ' ' 'C ~ ' UC7C7U~ ' ~ U~
CU L7U JU J UC U a Ua UF C7 J U r. C7U
U'C7~~ C7UU U r.CU UFCb ~U FCL7r-C C7~".~r-CFCi7 L7T.7U C7U'
UU ~ L9~ ~ pr.~Ui7U ",..~UU'U p i7U C7U~ ~ C9U' U'
U U ~ ~ U
r.~C7~ U ~U U U Ur.~U ~U pL7C~7C7~ "U'".~'UU ~L7 r~i U'
.~ 7
L7L7U~ FCUp U ~r-CC7U ~ U".~UFCC7FCD Cue'".~U C7r-C ~CL7C7r-CU'L7
r~r voovo .-tN H md~Nr o .-tN mr or ,-iN~rr mo~,--~N m rm yo ovo
V'V~Lf)If1l0l0r CO0707O~avH riv-IHo-iNN M MM M Wd~tIiIf)IfiLItu1l0l0t0r
HH Hv-In-1Hv-In-I'-1H HH N NN NN NN N NN N NN NN N NN NN N N
HH H~-IH HH H HH HH H HH 0-1v-1riH r1rir-ir1riH rir1H n-In-1riH H H
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
0
0
0
N
U
C7
Ca
1~.
U
Ch N
U O
U H
W
L9
U
C7 ~
U ~
.4
O
1~
U
U
N
ra ?a
U
O
U
U
U
4-1
U
U
b x
~ 1J
U 3
~ ~ O
II CU7
+O.~ CU7 y1
'' I O
m a ,N
U ~
N
to N
m cn
0 0
~ U
x m
U' II co O R~
E ~
U U II x M
U N .~ M
U b' 07 vo
N Fi M ..
U1 N r
a ~I ca
G ~ ~
H ~n x--
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
91
_ _
(Oh.a0O O r NM d'~ c0f~O N M d'~ COI~a~07O
O OO O r t- rV'd"ct~td"d'd'd'V'hLnIn
O OO O O O OO O O O OIntnLn~ LnIntf~tnIfscInLI7
tn
~'d'd'fl'Wit'd'd'd'd'd''d'd'
rr r r r rr r !-r rr r r r rr r r r rr r
H
C~ N Md'l (Of~00O O t-'N Mst'tn(OI~.00O O N M~tLf~
n
y r r _ r rr N N N NN N N N NN M M M MM M
N NN N N N NN N N N NN N N N NN N N N NN N
N NN N N N NN N N N NN N N N NN N N N NN N
t ~cnfn~ fnNN m tea)fnmm CG00m COOL1m CLm pp
n m~ m 5 U ~O U O O UU f0U ~ ~m U 7 ~ Um
~
U ~U O U U (67 U m f0OU ~ U O UU ~ j U O7(6O
U U U U U C
~7fO fn fntnU fpN U ~ f6nm U U (0U
U ~U ~ O LnUU (6U U ~~ ~ m ~~ U U O
U U~ O > > UO U a7L~OU U ~ 7 OO U O U Lnm
U U 07~
U UV ~ D1O UU U m ' ~N U U U m01a)U ~ f0O
t0U U m m mU ~ (0i~~InM U tp!nU fn(nU fn!l7N
U C~N ~ N N ~--~U U U I-~C7v~~ ~ ~nN~nv~
N U
~C~ U U QU U j ~ ~~ ~ ~ ~ ~~ ~ ~ v UN ~
G~ c~ U I U ~ I-I cn ~
,o a r~a tic~c~a,~~ ~ c~~~,
NN N fntnNfnN N N NQ U Q U I-(~U U U F-U
y Fq Q UQ E-7 U U I U U
N U C U ~ U - N~ v~N N ~nN ~n~ u~~I-U'
'~ '~ a U (9 C9!-Q U U
i~ Q UU'U U I-NQN N ~'~ ~fnN tnU Ntnt9N tnNfnt~
~ tn
y N t~tnN N U1Na a ~ U Q
~-~-a c~c~U
tpNN c~fnf~NfnfpN !Of!7H-U Q I1 U U' U U
I-UQ U I-I-UU C~~9C9U~nm ~nQ ---cnU cnU~nN
Q N v~~n c~ N
Q) U UU'Nj U U C~U U'N~ Q U!pV N ~ ~ N N
~ ~ ~ ~~ ~ I-N ~U J cp fOfnt~a f~U
~ a ~- ~c~~ U
tia, ~ c~c~~a a ~-U UO IBf0Q cL~07U a ~-U p~
~ a U p7 U U
~.1 07f0N U p7p7mU U U U p~U U p7U ~~ O U U OU O
U Up~U ~ ~ O7U U O U p~U U ~ U U ~ U n7m
U U~ U7 7 (007 ~ LA ~~ U
U O O ~ ~ ~ U (SSUm
~ 7 O~ ~ ~ O UU (CfU O p~U (6a7U UU 05U ~ Ut9
r UN~t~tpN l~mU tAt~fONQIN O ~ OCn(0N U Um
m U U U ~~ O U ~ m m mf6
N UU ~ fn~ 7 p77 p~
U
~ ~O CA~7NcO~ a~~ l4
N WU ~ aj5 ~CAm m f0a1
O N M d'4n(Oh 00O O rO r N M d'In(Oh 00OO r
O OO O O O OO O O O O O O OO O O O Or r
N NN N N N NN N N N NN N N N NN N N N NN N
N NN N N N N N
N N N NN N N N NN N N N NN N
a U Q ~U U U ~ U= U U'a UQ ~ U U U~ U
U ~U a U U ~~ U U'U ~U ~ U a UU > > U C~U ~
C~U U UCJQ ~J~ ~U ~ U U (~(9U C9Q U
a UQ Q Q a d QU U C~~ ~JU U
U'U' U V V U' j U ~U C9Q (,UU C7Q Q
' 9
C~C~U C9U U U U C9UC7C~U V'Q> U C9C9
U >> C9 U C~U'(9t~UU > > C9UU U U'U
~ U
y C9~Q ~ U U Q> > Q C)C9C7~ Q ~ UU Q ~ = a C
.7
C~C~U C9(~U UC9U Q ~ C!7C7C~U C9UU U C~U
U U
c~ Q C~~ ~ Q Q UU U U U C~Q U ~ U Qd U'C9U U
UU C7> > Q
~ Q Q~ U U U C~> > Q U ~Q Q U ' CJ~
~ U U' ' '
S r" (~ C9U Q ~C9 Q Q U~ U U Ud ~ C9U Qa U
CC ,~ ~ C~~ C~Q U C~C~U U Q U~ C~~ U'QU C~C~U U
p ~ Q= C9U U ~Q Q Q U Q~ ~ ~9UU ~ Q Q QV Q
U ~ U
C7Q U C~C9~C~C~U Q QU C9UU ~ U U UQ Q
~ ~ ~ U U U > > U ~ U U ~ ~U
h~ U ~ ~ Q C9U ~ U ~ Q
l
Q U U C7U (~U Q Q~9(a~ Q UU C~U C~UQ Q
a
Q C7= U d C9U C~U U UQ U ~ U QQ (9U (gUU U N
' U U U
U ~C~> CgU ~ U C~Q(~~ C7~ U U U ~ C~C~Q
C7QU U U U ~Q C7U ~ UU Q C~U UU ~ Q U U
Q UQ U a a QU C9U U C9Q U Q U QQ Q U t7UU C~
y C~~C~Q U U U'Q Q C~U ~(9~ C9Q UU C9Q Q UU ~
~' N
~ 'ct~COCOO LnrM f~.N CO07d.~ t0COd7Inr M r NC400
N V'~ M r N In~ ~ ~ M ON d'tnM rN In~ r ~O CO
M Oc-r r r rM ~ ~ n'MM O c-r rr r
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
M
O
O
O
G
N
l~
O
<r
G1
'~
O
N
U
N
~1
O
l~
U
W
O
O
O
O
U m
C
O
's.. E
~
U m
.
.'~
C
In
N = ,-~
c0
.~I
t~
x m
.C O a a
CV w
.~,
N v m
I I .R
0 ~ ~'
O
ti x
m
N II
M
U o
N ~ ~
~
.u ~
a
~ a
o
I I C w
I
O ~'
m
W
~ cn H C
00 7
~
x
--
92
CA 02397813 2002-07-16
WO 01/62911 PCT/USO1/05957
M
O
O
O
V
93
ro~ rtrom
a~v a~a~va~va~m~ 'va~a~v va~a~a~a~
~~~ ~~
x~ xxx ~E ~~ E~ ~~ ~a , . a E>;
' ~
~1. f-IS-IS-Iy''~r5r,fir.~5rJr,~5v,firNNN NN ,?, ,7r'v
Sd N N
~ N N NN NN NN f.1~1~1S-1~1NN NN N
00 0o Op bG p~ Nvv a7N r.~,~r.~r.Cr.~
~~ G-~-~,-,~-~.~~,~.o~.~zz zz z
roHH HH HN NN NN ~~~ ~~ Aq pA p
xx xxx
or NMd'In(OI~00Wo N Md'InCO1~0007Or NM d'
a00~Na0a0Ne0a0c0of d) ~
O ~ O)mW~ O)(3)OO OO O
NN NNN NN NN NN NN NN NNN NN MM MM M
d NN NNN NN NN NN NN NN NNN NN NN NN N
WP4P7WW
Ub1b1U
O1b7UU
bl~~ ~U
WW P9WW f-0W Wx Pap~WP7FOP7b'1U~ UU
b7t7707~U
U U ~~ drt
blb1td blN a1b1~p OltPU~ ' ~ f ~W WW W
'
p ~
trb1~dUb1 ~ U ~~ b1rtO7~dC7CJC7C7C7~~ ~~ (d
~ UtJ1~U C7L7C7U'C7U~ ~b'107
FCFC~FC~ HH HH H01Ub1b7tTUUU UU Ub1~~ rd
,~ ~ rtS~ ~~ ~ U ~ d
.n~ ~ ~~ ~~ b~~ ~~ Ub7tbit
V b b d
llL31lb1 ~ J1b7b1
C7U,t7UrU,U,U,U,U,U,U UU UU UUU UU t37U ~~ U
V UU UUU UU UU UU UU UU ~
~yb7tf7b'1b1b'1b1b'107131b'7~ rtU rtrt U rt UU UU U
b7blbltJlCJ7b1blb7blb'1~ (17t blb1rdN~ b7rtS
d t7WT1(T7 Ul
U7
mm romm mm mm ~a
b~ ~ NiN ~rt
C~ ~~ ~~~ ~~ ~~ ~b~77 b1b r UU UU U
U '1 d
r,UU UUU UU UU U~ ~~ ~~ UUU UU HH HH H
UU UUU UU UU UU UU UU ~~~ ~~ UU UU U
'~''b~fi0~tr~bib3bitr~b3b1~ b1~ a1bi L7C7C7c7C7
i~ ~ b3btb7b1bib1bia7b5b3~ dd i~ UUU UU EH HH H
rttt rtdUUU UU UU UU U
tdb d
td
rttdN ~dN UUU UU C7C7C7C7L7
'iceN FCr.CFCr.Cr.CFCFCFCFCFC~ ~b~bibi~p~ ~~ L7C7UC7L?
' ' ' o UU UU rtSNtd~dtd1rtdN~dbt
I~ W ~a ~a~ ap pp ~
UU UUU UU UU U
i.i brtSrtrdtdUbiNN b1
~tdftS~U b7(dUtdU~ tIf(dL31bl~1231b1blbltd(db1(J1bl
~tntr~~ Ntr~trU~ blrdb1~ tnOltrb7b7rdN (Ob7p
pN ~db1rtStdof~dN ~U Nb7U
~~ (J1 U
UN tdU~ b'1tJi~rd01b ' '~ ~
Ub1b7trb1rtSN ~rdtd ~b b1~ L7C7C7U[
1
~L7'~U~ rtSU SU bU ~~ b1b1U~~ ~U
n l
~
V
~b1 U
CC3 U(dU C31b1
rd~ NOl
.,H
r1N v-INO riO riO~OO O0101c0MMN Nr10101mc0L~
01a1OOriv-1N NN M01OO rIN 01Or1NM ~a1OriN
Wo~01~1010101410101N O101~101ODav01O~O10~a0O1O101
MM MMM MM MM MM MM MM MMM MM MM MM M
NN NNN NN NN NN NN NN NNN NN NN NN N
V ~
_
10l~a001O r-IN M~ ~f110l~0001O ,-INM V~N ~Oh 07N 01
NInL~Intob10~D1010101010l0l~L~L~!~L~I~1~L~l~tD1~
GINN NNN NN NN NN NN NN NNN NN NN NN N
d
NN NNN NN NN NN NN NN NNN NN NN NN N
C/~ U U
W
pU ~C7r-C"aC7bC7UC7Ur-CUU r.~",.~rC''.~'U ~~ UFCU'
' ' "
r.Cp UUU UU Ur.CUt7C7U L7U C7aU'UU C7.oU C7
" '~'
' N
L7U UUU ~p ~,~pp U Ur.CFCUr.~C7p ~~ pU r.~~
C7 'aC7r.CUU ~ U~ ~U UU C7~U r.CU' UU U
".-~C7U ~p C7U r.~ p pU r.C C7C7U'C7U ",.~",~U-s-.
~ ~ ~ ~
"a~C Up C(~C7~ ~~ UU C7r-C~ FCC Up p~ U
7 h 'J
pU U"aU I7U r~p "aC7C7C7C7C7L7C7L7U'C7r-Cr-C~L7FC~ C
~ ~ 'a"a'JUU UU UFC'aFCFC~ CC~ ~' UU' C 7
C7 ~~U ~C7C7'aL7U U'U UU FFU FCa C7p ~F CN
UU C7 U U
r r-C~ UC7p L9C7r.CC7r-CC7U r.CC7UU'~ C7C7C7U 'aU pN ~L-
~ UU "aU'U ~ ~C7 U Up Up r.~L9C7C7",~r-Cp U~ U
r~p UI7(~U~ Ur.C C7pr.~UU Ur.C~ ~L7C7U r.~'aII II
47
UU 'JC7U 'U'UU U'UU L7" r~UU C7C7C7 U U
7 ~ o
L7FCFCFCU ,U CC7 FCFCFCFC. 7UU C7C FC C C3
U FC
F C F F -.
U U
II
L L
N MM mtnN00~ MO [~O d)Md0hO N (OO MN N
~ ofc-O hO rM Nr,LI3I~ N O N Or o0O hh O
rM'chrM d'COr rM o~00~td'COrr r rM V'
E'~ O J ~
~'