Note: Descriptions are shown in the official language in which they were submitted.
CA 02398229 2010-01-20
30754-30
1
IMPROVED DNA VACCINES FOR PRODUCTION-TYPE ANIMALS
The present invention relates to improved DNA
vaccines for farm animals, in particular bovines and
porcines.
The use of deoxyribonucleic acid (DNA)
molecules for vaccination has been known since the
beginning of the 1990s (Wolf et al. Science 1990. 247.
1465-1468). This vaccination technique induces cellular
and humoral immunity after in vivo transfection of
cells of the subject to be vaccinated with DNA or RNA
molecules encoding immunologically active proteins.
A DNA vaccine is composed of at least one
plasmid which may be expressed by the cellular
machinery of the subject to be vaccinated and of a
pharmaceutically acceptable vehicle or excipient. The
nucleotide sequence of this plasmid encodes, inter
alia, one or more immunogens, such as proteins or
glycoproteins capable of inducing, in the subject to be
vaccinated, a cellular immune response (mobilization of
the T lymphocytes) and a humoral immune response
(stimulation of the production of antibodies
specifically directed against the immunogen) (Davis
H.L. Current Opinion Biotech. 1997. 8. 635-640).
All the immunogens derived from a pathogen are
not antigens which are naturally sufficiently effective
for inducing an optimum protective immune response in
the animal to be vaccinated. It is therefore necessary
to improve the immune response.
Various routes of administration of the DNA
vaccine have been proposed (intraperitoneal,
intravenous, intramuscular, subcutaneous, intradermal,
mucosal, and the like). Various means of administration
have also been proposed, in particular gold particles
coated with DNA and projected so as to penetrate into
the cells of the skin of the subject to be vaccinated
(Tang et al. Nature 1992. 356. 152-154) and the liquid
jet injectors which make it possible to transfect both
skin cells and cells of the underlying tissues (Furth
et al. Analytical Bioch. 1992. 205. 365-368).
CA 02398229 2010-01-20
30754-30
2
Chemical compounds have been used for the in
vitro transfection of DNA:
A/ - cationic lipids.
The cationic lipids are themselves divided into
four subgroups.
1) The cationic lipids containing quaternary
ammonium salts, such as for example DOTMA (dioleoyl-
oxypropyltrimethylammonium, produced by Gibco under the
name Lipofectine), DOTAP (trimethyl-2, 3-(octadec-
9-eneoyloxy)-1-propaneammonium; Gregoriadis et al. FEBS
Letters 1997. 402. 107-110), DMRIE (N-(2-hydroxyethyl)-
N,N-dimethyl-2,3-bis(tetradecyloxy)-1-propaneammonium;
WO-A-9634109), DLRIE (N-(2-hydroxyethyl)-N,N-dimethyl-
2,3-bis(dodecyloxy)-1-propaneammonium; Feigner et al.
Ann. N Y Acad. Sci. 1995. 772. 126-139).
These cationic lipids containing quaternary
ammonium salts may be combined or otherwise with an
additional neutral lipid, such as DOPC (dioleoyl-
phosphatidylcholine) or DOPE (dioleoylphosphatidyl-
ethanolamine) (J.P. Behr, Bioconjugate Chemistry 1994.
5. 382-389).
2) The lipoamines, such as for example DOGS
(dioctadecylamidoglycylspermine, produced by Promega
under the name Transfectam; Abdallah et al. Biol. Cell.
1995. 85. 1-7), DC-Chol (dimethylamino ethane- carbamoyl-
cholesterol; Gao and Huang, Biochem. Biophys. Res.
Commun. 1991. 179. 280-285), BGSC (bis-guanidine-
spermidine-cholesterol), BGTC (bis-guanidine-tren-
cholesterol) (Vigneron et al. Proc. Natl. Acad. Sci.
USA 1996. 93. 9682-9686).
3) The cationic lipids containing quaternary
ammonium salts and lipoamines, such as for example
DOSPA (N,N-dimethyl-N-(2-(sperminecarboxamido)ethyl)-
2,3-bis(dioleoyloxy)-1-propaneimidium pentahydro-
chloride, marketed by Gibco under the name
LipofectAmine ; Hawley-Nelson et al. Focus 1993. 15.
73-79), GAP-DLRIE (N-(3-aminopropyl)-N,N-dimethyl-
2,3-bis(dodecyloxy)-1-propaneammonium; Wheeler et al.
CA 02398229 2010-01-20
30754-30
3
Proc. Natl. Acad. Sci. USA 1996. 93. 11454-11459;
Norman et al. Vaccine 1997. 1S. 801-803).
4) The lipids containing amidine salts, such as
for example ADPDE, ADODE (Ruysschaert et al. Biochem.
Biophys. Res. Commun. 1994. 203. 1622-1628).
B/ - the polymers, such as for example
SuperFect (molecules of activated dendrimers, produced
by Qiagen; Xu et al. Mol. Genet. Metab. 1998. 64. 193-
197), and
C/ - the biochemical agents, such as for
example toxins, in particular cholera toxins.
Some of these compounds have also been used in
the formulation of DNA vaccines with more than
mitigated results. Knowledge in the field of in vitro
transfection is not transposable to DNA vaccination
where the final objective is to ensure a protective
immune reaction. Negative effects on the induction of
an effective immune protection have even been observed
with compounds known to promote transfection in vitro.
Some formulation chemical compounds are toxic at high
doses for the transfected cells.
In the work by Etchart (Etchart et al. J. Gen.
Virol. 1997. 78. 1577-1580), the use of DOTAP did not
have an adjuvant effect during the administration of
the DNA vaccine by the intranasal route, whereas it had
an adjuvant effect by the oral route. DOTAP has also
been used in DNA vaccines encoding the influenza virus
hemagglutinin (HA) on the mouse model which were
administered by the intranasal route (Ban et al.
Vaccine 1997. 15. 811-813), but the addition of DOTAP
inhibited the immune response. The use of DC-Chol or of
DOTAP/DOPE in DNA vaccines encoding the hepatitis B
virus surface protein (S) on the mouse model which were
administered by the intramuscular route made it
possible to increase the antibody response, whereas the
use of Lipofectine (or DOTMA) did not increase this
response (Gregoriadis et al. FEES Letters 1997. 402.
107-110). DC-Chol/DOPE has also been used in DNA
vaccines against the human immunodeficiency virus (HIV,
CA 02398229 2010-01-20
30754-30
4
Env protein) on the mouse model, whose administration
by the intramuscular route induced a more effective
immune response, whereas the administration by the
subcutaneous or intradermal route did not increase it
(Ishii et al. AIDS Res, Hum. Retro. 1997. 13. 1421-
1428).
The addition of certain cytokines, in
particular interleukins or interferons, can make it
possible to enhance the immune response induced in
particular by DNA vaccines. Each cytokine triggers a
reaction which is specific to it and orients the immune
response to a greater or lesser degree towards a
cellular response or towards a humoral response
(Pasquini et al. Immunol. Cell.. Biol. 1997. 75. 397-
401; Kim et al. J. Interferon Cytokine Res. 1999. 19.
77-84). The adjuvant effects of a cytokine obtained
from a given species are not necessarily the same if
the immune context varies, in particular if this
cytokine is administered to another species, therefore
in a heterologous immune system. The addition of
cytokine may also have no adjuvant effect, or may even
result in a reversal of the effect sought, that is to
say a reduction or an inhibition of the immune
response. Thus, a DNA vaccine encoding a single chain
of an immunoglobulin fused with GM-CSF does not
increase the immune response, whereas direct
administration of this fusion protein to mice is
effective, in the sane way as is the administration of
a fusion protein consisting of Fv and of the cytokine
IL-Lbeta or the administration of a DNA vaccine
encoding the latter fusion protein (Hakim et al. J.
Immun_ol. 1996. 157. 5503-5511). The use of plasmids co-
expressing the cytokine IL-2 and the hepatitis B virus
envelope protein in a fused or nonfused conformation
results in an increase in the humoral and cellular
immune responses (Chow et al. J. Virol. 1997. 71. 169-
78). However, the use of a bicistronic plasmid encoding
the human acquired immunodeficiency virus (HIV-1)
glycoprotein gp120 and the cytokine IL-2 induced a
CA 02398229 2010-01-20
30754-30
lower specific anti-gp120 immune response than that
obtained by the use of a monocistronic plasmid encoding
only gpl20 (Barouch et al. J. Immunol 1998. 161. 1875-
1882). The co-injection, into mice, of two expression
vectors, one coding for the rabies virus G
glycoprotein, the other for murine GM-CSF stimulates
the activity of the B and T lymphocytes, whereas the
co-injection with a plasmid encoding gamma-interferon
(in place of murine GM-CSF) results in a decrease in
the immune response (Xiang et al. Immunity 1995. 2.
129-135).
Certain modifications in the antigens, such as
deletions of part of the nucleotide sequence encoding
the antigen, insertions of a DNA fragment into the
nucleotide sequence encoding the antigen or into non-
translated regions upstream or downstream, can also
enhance the efficacy of DNA vaccines, in particular by
enhancing the level of expression of the antigen or its
presentation.
However, in practice, manipulations on the
n_ucleetide sequence encoding the antigen may bring
about a reduction or loss of the initial immunological
activity. Thus, the deletion of the transmembrane
domain from the gene encoding the rabies virus G
antigen reduced the level of protection induced in the
mouse model after administration by the intramuscular
route of a DNA vaccine encoding this modified antigen
(Xiang et al. Virol. 1995. 209. 569). The deletion of
the transmembrane domain from the gene encoding the
bovine herpesvirus (BHV) gD glycoprotein did not make
it possible to increase the antibody response and
induced only a partial protection in bovines vaccinated
by the intramuscular route (van Drunen Little-van den
Hurk et al. J_ Gen. Virol. 1998. 79. 831-839). The
humoral and cellular immune responses and the
protection conferred are identical in guinea pigs
challenged after having been immunized with the aid of
either a DNA vaccine encoding the Ebola virus GP
glycoprotein, or of a DNA vaccine encoding this GP
CA 02398229 2010-01-20
30754-30
6
glycoprotein but in a secreted form (Xu et a1. Nature
Medicine 1998. 4. 37-42).
The insertion of the signal sequence of the
human tissue plasminogen activator (tPA) into the gene
encoding the malaria Pf332 antigen did not make it
possible to increase the antibody response in mice
vaccinated by the intramuscular route (Haddad et al.
FEMS 1997. 18. 193-202). The addition, in phase, of a
tPA sequence to the gene encoding the murine rotavirus
VP7 antigen also did not make it possible to increase
the antibody response in mice vaccinated by the
intradermal route, whereas the fusion protein
consisting of the VP4 antigen and tPA allowed this
increase, but without inducing an effective protection
(Choi et al. Virology 1998. 250. 230-240).
The modifications carried out on the nucleotide
sequence of one antigen cannot in general be directly
transposed to another antigen, because antigens do not
always have the same structural arrangements.
The applicant has as objective the enhancement
of the efficacy of DNA vaccination. Its objective is in
particular to obtain a better immune response and in
particular an effective protection in farm animals, in
particular bovines and porcines, by DNA vaccinations.
The applicant has as objective the production
of improved DNA vaccines which induce an effective and
protective immune response against the bovine
herpesvirus type 1 (BHV-1) also called infectious
bovine rhinotrachitis (IBR), the bovine respiratory
syncitial virus (BRSV), the mucosal disease virus or
bovine pestivirus type 1 or type 2 (bovine viral
diarrhea virus or BVDV-1 and BVDV-2), the parainfluenza
virus type 3 (bPI-3) in bovines.
The applicant has as objective the production
of improved DNA vaccines which induce an effective and
protective immune response comprising at least one
valency selected from the group consisting of porcine
herpesvirus or Aujeszky's disease (pseudorabies virus
or PRV), the porcine reproductive respiratory syndrome
CA 02398229 2010-01-20
30754-30
7
virus (or PRRSV), the swine influenza virus (or SIV),
the conventional hog cholera virus (or HCV),
parvoviruses in porcines.
The applicant also has as objective the
production of improved DNA vaccines which make it
possible to obtain effective and protective immune
protection in bovines, comprising at least one valency
selected from the group consisting of the BHV-1, BRSV,
BVDV, bPI-3 and rabies viruses.
The subject of the invention is improved DNA
vaccines which make it possible to obtain effective
protection against at least one pathogen which infects
farm, in particular bovines and porcines. The DNA
vaccine is improved: either by its formulation, or by
the addition of GM-CSF, or by the optimization of the
antigen(s), or by combinations of these solutions.
Preferably, the DNA vaccine is improved by its
formulation, and optionally either by the addition of
GM-CSF, or by the optimization of the antigen(s), or
finally by the addition of GM-CSF and by the
optimization of the antigen(s).
By definition, the DNA vaccine comprises, as
active ingredient, a plasmid encoding and expressing a
gene or gene fragment e.g. epitope. The term plasmid
covers a DNA transcription unit comprising a
polynucleotide sequence comprising the sequence of the
gene to be expressed and the elements necessary for its
expression in vivo. The circular plasmid form,
supercoiled or otherwise, is preferred. The linear form
also falls within the scope of this invention.
Each plasmid comprises a promoter capable of
ensuring, in the host cells, the expression of the gene
inserted under its control. It is in general a strong
eukaryotic promoter and in particular a cytomegalovirus
early promoter CMV-IE, of human or murine origin, or
optionally of other origin such as rat or guinea pig.
More generally, the promoter is either of viral origin
or of cellular origin. As a viral promoter other than
CMV-IE, there may be mentioned the SV40 virus early or
CA 02398229 2010-01-20
30754-30
8
late promoter or the Rous Sarcoma virus LTR promoter.
It may also be a promoter the virus from which the gene
is derived, for example the promoter specific to the
gene. As cellular promoter, there may be mentioned the
promoter of a cytoskeleton gene, such as for example
the desmin promoter, or alternatively the actin
promoter. When several genes are present in the same
plasmid, they may be provided in the same transcription
unit or in several different units.
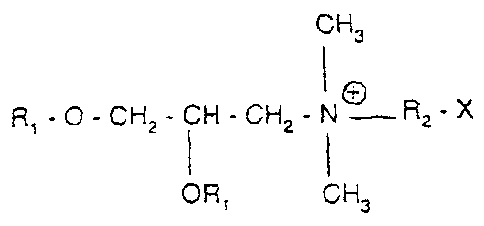
According to a first mode, the DNA vaccines
according to the invention are formulated by adding, as
adjuvant, cationic lipids containing a cfaaternary
ammonium salt of formula:
CH3
I +
R,-O-CH2-CH-CH2-N R2-X
I
OR1 CH3
in which R1 is a saturated or unsaturated
linear aliphatic radical having 12 to 18 carbon atoms,
R2 is another aliphatic radical containing 2 or 3
carbon atoms, and X a hydroxyl or amine group.
Preferably, this is DMRIE (N-(2-hydroxyethyl)-
N,N-dimethyl-2,3-bis(tetradecyloxy)-1-propanammonium
WO-A-9634109), preferably combined with a neutral
lipid, in particular, DOPE (dioleoyl-phosphatidyl-
ethanolamine), to form DMRIE-DOPE.
The subject of the present invention is
therefore a DNA vaccine against at least one pathogen
affecting farm animals, in particular bovines or
porcines, comprising at least one plasmid containing at
least one nucleotide sequence encoding an immunogen of
a pathogen of the animal species considered, under
conditions allowing the in vivo expression of this
sequence, and a cationic lipid containing a quaternary
ammonium salt, in particular DMRIE, preferably combined
with DOPE.
CA 02398229 2011-03-21
51440-160
8a
In one aspect, the present invention relates to a DNA vaccine comprising a
plasmid
comprising a polynucleic acid coding for an immunogen of bovine herpes virus
type 1
(BHV-1), wherein the BHV-1 immunogen is glycoprotein B (gB), glycoprotein C
(gC), or
glycoprotein D (gD), and the elements necessary for its in vivo expression, N-
(2-
hydroxyethyl)-N,N-dimethyl-2,3-bis (tetradecyloxy)-1-propanammonium (DMRIE)
and
dioleoyl-phosphatidyl-ethanolamine (DOPE).
CA 02398229 2010-01-20
30754-30
9
Preferably, the recombinant vector is mixed
with this adjuvant immediately before use and it is
preferable, before its administration to the animal, to
allow the mixture thus prepared to form a complex, for
example for a period ranging from 10 to 60 minutes, in
particular of the order of 30 minutes.
When DOPE is present, the DMRIE:DOPE molar
ratio preferably ranges from 95:5 to 5:95, and is more
particularly 1:1.
The plasmid:DMRIE or DMRIE-DOPE adjuvant weight
ratio may range in particular from 50:1 to 1:10, in
particular from 10:1 to 1:5, preferably from 1:1 to
1:2.
According to a second mode, GM-CSF (granulocyte
macrophage-colony stimulating factor; Clark S.C. et al.
Science 1987. 230. 1229; Grant S.M. et al. Drugs 1992.
53. 516) is added to the vaccines according to the
invention; this may be carried out by incorporating
GM-CSF protein directly into the vaccinal composition
or preferably by inserting the nucleotide sequence
encoding GM-CSF into an expression vector under
conditions allowing its expression in vivo. As
expression vector, the use of a plasmid, e.g. the
plasmid containing the nucleotide sequence encoding the
antigen(s) of interest or another plasmid, is
preferred. The choice of GM-CSF is preferably made
according to the animal species to be vaccinated; thus,
for bovines, bovine GM-CSF is used; for pigs, it is
porcine GM-CSF.
According to a third mode, the nucleotide
sequence(s) encoding the immunogen are in an optimized
form. Optimization is understood to mean any
modification of the nucleotide sequence, in particular
which manifests itself at least by a higher level of
expression of this nucleotide sequence, and/or by an
increase in the stability of the messenger RNA encoding
this antigen, and/or by the triggered secretion of this
antigen into the extracellular medium, and having as
CA 02398229 2010-01-20
30754-30
direct or indirect consequence an increase in the
immune response induced.
In the present invention, the optimization of
the antigen of interest preferably consists in the
deletion of the fragment of the nucleotide sequence
encoding the transmembrane domain of the antigen of
interest (deletion is understood to mean the complete
deletion or a partial deletion sufficient for the
transmembrane domain to no longer, or no longer
substantially, be functional), and/or in the addition,
in frame, of a nucleotide sequence encoding the tPA
(Montgomery et al. Cell. Mol. Biol. 1997. 43. 285-292;
Farris et al. Mol. Biol. Med 1986. 3. 279-292) signal,
and/or in the insertion of a stabilizing intron
upstream of the gene to be expressed. The deletion of
the DNA fragment encoding the transmembrane domain of
the antigen of interest promotes the secretion, into
the extracellular medium, of the antigens thus
truncated and thus increases the possibilities of their
coming into contact with the cells of the immune
system. The insertion of the nucleotide sequence
encoding the tPA signal facilitates the translatability
of the messenger RNA to which the tPA signal is joined,
and thus increases the level of expression of this
messenger RNA and therefore the production of antigens.
The tPA signal also plays a role in the secretion of
the antigen synthesized.
Other nucleotide sequences encoding signal
peptides may be used, in particular those for the
signal peptide of melittin obtained from bees
(Sisk W.P. et al., 1994, J. Viral., 68, 766-775).
The insertion of a stabilizing intron into the
gene encoding the antigen of interest avoids the
aberrant splicings of its messenger RNA and maintains
the physical integrity of the latter.
Preferably, the tPA signal is of human origin-
The nucleotide sequence of the human tPA signal is
accessible from the GenBank database under the
accession number NM 000930. Preferably, the intron is
CA 02398229 2010-01-20
30754-30
11
intron II of the rabbit beta-globin gene (van Ooyen et
al. Science 1979. 206. 337-344), whose nucleotide
sequence is accessible from the GenBank database under
the accession number V00882 and designated by a
reference under intror_ No. 2.
The subject of the present invention is an
improved DNA vaccine capable of inducing an effective
and protective immune response in bovines against
infectious bovine rhinotrachitis (IBR).
The virus responsible for infectious bovine
rhinotrachitis is a bovine herpesvirus type 1 (BHV-1),
a member of the Alphaherpesvirinae family (Babiuk L.A.
et al., 1996, Vet. Microbiol., 53, 31-42). Nucleotide
sequences encoding the glycoproteins gB, gC and gD are
known and are accessible from the GenBank database
under the accession number AJ004801.
According to the invention, the DNA vaccine
against IBR is preferably improved by its formulation
with an adjuvant according to the invention, in
particular DMRIE, preferably DMRIE-DOPE. Optionally,
this may be'combined either with the addition of bovine
GM-CSF (Maliszewski et al., Molec. Immunol., 1988, 25,
843-850), or the optimization of at least one IBR
antigen, or finally the addition of bovine GM-CSF and
the optimization of at least one IBR antigen.
A nucleotide sequence encoding bovine GM-CSF is
accessible from the GenBank database under the
accession number U22385.
The addition of bovine GM-CSF may be carried
out by the incorporation of the bovine GM-CSF
polypeptide into the vaccinal composition or preferably
by the insertion of the nucleotide sequence encoding
the bovine GM-CSF into an in vivo expression vector,
preferably a plasmid. Preferably, the nucleotide
sequence encoding bovine GM-CSF is inserted into a
second expression plasmid (e.g. pLF1032 Example 13),
different from that (or those) into which the gene(s)
encoding the IER antigen(s) is(are) inserted.
CA 02398229 2010-01-20
30754-30
12
The optimization of the antigens derived from
IBR is carried out by substitution, by a "signal"
sequence, in particular that of the tPA signal of human
origin (GenBank accession number NM - 000930), of the
sequence of the signal peptide of the glycoprotein gB
and/or of the glycoprotein gC and/or of the
glycoprotein gD, and/or by the deletion of the DNA
fragment encoding the transmembrane domain of gB and/or
of gC and/or of gD. The deletion of the DNA fragment
encoding the transmembrane domain of one of these
glycoproteins is preferably accompanied by the
contiguous C-terminal part (cytoplasmic portion of the
glycoprotein). The DNA vaccine against IBR according to
the invention can therefore encode and express a single
optimized IBR antigen (gB, gC or gD) or two of them or
all three, that is to say optimized gB, optimized gC
and optimized gD.
Nucleotide sequences encoding the BHV-1
antigens which can be used in the present invention and
various constructs of expression vectors are given in
the accompanying examples and in FR-A1-2751229, in
particular in Examples 7 and 8, and in Figures 3 and 4.
Preferably, according to the invention, the DNA
vaccine against BI--V-1 is formulated with DMRIE-DOPE,
and is composed of an expression plasmid (e.g. pPB281,
Example 3.1.2) encoding the BHV-1 gB antigen optimized
by the deletion of the fragment of the nucleotide
sequence encoding the transmembrane domain and the
contiguous C-terminal part, of a second expression
plasmid (e.g. pPB292, Example 3.2.2) encoding the BF-V-1
gC antigen optimized by the deletion of the fragment of
the nucleotide sequence encoding the transmembrane
domain and the contiguous C-terminal part, and of a
third expression plasmid (e.g. pPB284, Example 3.3.2)
encoding the BIV-1 gD antigen optimized by the deletion
of the fragment of the nucleotide sequence encoding the
transmembrane domain and the contiguous C-terminal
part.
CA 02398229 2010-01-20
30754-30
13
In general, and not only for BHV-1, the
C-terminal part contiguous to the sequence encoding the
transmembrane domain may be conserved. It is however
often easier to delete it at the same time as the
sequence encoding the transmembrane domain.
The subject of the present invention is also an
improved DNA vaccine capable of inducing an effective
and protective immune response in bovines against the
bovine respiratory syncitial virus (BRSV).
The BRSV virus is a Paramyxovirus, also a
member of the Paramyxoviridae family (Baker et al.,
Vet. Clin. North Am. Food Anim. Pratt., 1997, 13,
425-454). Nucleotide sequences encoding the F protein
and the G glycoprotein are known and accessible from
the GenBank database respectively under the accession
number Y17970 and U33539.
The DNA vaccine against BRSV is preferably
formulated with an adjuvant according to the invention,
in particular DMRIE, preferably D1`,MIE-DOPE. This may be
optionally combined with either the addition of bovine
GM-CSF, or the optimization of at least one BRSV
antigen, or finally the addition of bovine GM-CSF and
the optimization of at least one BRSV antigen.
The addition of bovine GM-CSF may be carried
out as is described for BHV-1.
The optimization of the antigens derived from
BRSV is carried out by substitution, by a "signal"
sequence, in particular that of the tPA of human
origin, of the signal sequence of the F protein of BRSV
and/or of the G envelope glycoprotein of BRSV, and/or
by the deletion of the DNA fragment encoding the
transmembrane domain of F and/or of G. The deletion of
the DNA fragment encoding the transmembrane domain of
one of these proteins is preferably accompanied by the
contiguous C-terminal part. The DNA vaccine against
BRSV according to the invention can therefore encode
and express a single optimized BRSV antigen (F or G) or
both (F and G).
CA 02398229 2010-01-20
30754-30
14
Nucleotide sequences encoding the BRSV antigens
which can be used in the present invention and various
expression vector constructs are given in the
accompanying examples and in FR-Al-2751229, in
particular in Examples 9 and 10, and in Figures 5 and
6.
Preferably, according to the invention, the DNA
vaccine against BRSV is formulated with DMRIE-DOPE, and
is composed of an expression plasmid (e.g. pSB114
Example 4.1.3) encoding the F antigen of BRSV optimized
by the insertion of the signal sequence of the human
tPA in place of the signal sequence of F, by the
deletion of the fragment of the nucleotide sequence of
F encoding the transmembrane domain and the contiguous
C-terminal part, and of a second expression plasmid
(e.g. pSB110 Example 4.2.2) encoding the G antigen of
BRSV optimized by the insertion of the signal sequence
of the human tPA in place of the signal sequence of G,
by the deletion of the fragment of the nucleotide
sequence encoding the transmembrane domain of G and the
contiguous C-terminal part.
The subject of the present invention is also an
improved DNA vaccine capable of inducing an effective
and protective immune response in bovines against the
BVDV virus.
The BVDV virus is a pestivirus of the
Flaviviridae family. It is universally distributed in
bovine populations and manifests itself by fetal
malformations, abortions or clinical respiratory
(mucosal disease) and enteric (bovine viral diarrhea)
symptoms.
The BVDV viruses are distinguishable by the
seriousness of the clinical signs and two groups have
been formed, the BVDVs type 1 (inapparent or mild
clinical signs) and those of type 2 (acute clinical
signs, hemorrhage, high morbidity, high mortality)
(Dean H.J. and Leyh R., 1999, Vaccine, 17, 1117-1124).
CA 02398229 2010-01-20
30754-30
When a BVDV virus type is not clearly
specified, this virus is understood to be type 1 or
type 2.
The BVDV virus is an enveloped single-stranded
RNA virus composed of a single gene encoding a
polyprotein which, after cleavage, gives several well-
individualized proteins, in particular the E0 protein
(gp48) and the E2 protein (gp53) (Vassilev V.B. et al.,
1997, J. Virol., 71, 471-478).
Nucleotide sequences encoding the E0-E2
polyproteins are known and accessible from the GenBank
database under the accession number M96687 for BVDV-1
and AF145967 for BVDV-2.
The DNA vaccine against BVDV is preferably
formulated with an adjuvant according to the invention,
in particular DMRIE, preferably DMRIE-DOPE. This may be
optionally combined with either the addition of bovine
GM-CSF, or the optimization of at least one BVDV
antigen, or finally the addition of bovine GM-CSF and
the optimization of at least one BVDV antigen.
The addition of bovine GM-CSF may be carried
out as is described for BHV-1.
The optimization of the antigens derived from
BVDV is carried out by the addition of a "signal"
sequence, in particular that of the tPA of human
origin, upstream of the nucleotide sequence encoding
the EO protein of BVDV and/or of the E2 protein of
BVDV, and/or by the deletion of the DNA fragment
encoding the transmembrane domain of E2, and/or by the
insertion of an intron, in particular intron II of the
rabbit beta-globin gene upstream of the nucleotide
sequence encoding EO and/or E2. The DNA vaccine against
BVDV according to the invention may therefore encode
and express a single optimized BVDV antigen (EO or E2)
or both (EO and E2).
Nucleotide sequences encoding the BVDV antigens
which can be used in the present invention and various
constructs of expression vectors are given in the
CA 02398229 2010-01-20
30754-30
16
accompanying examples and in FR-A1-2751229, in
particular in Example 13, and in Figure 9.
Preferably, according to the invention, the DNA
vaccine against BVDV is formulated with DMRIE-DOPE, and
is composed of an expression plasmid (e.g. pLF1029
Example 5.1.2, pLF1031 Example 6.2.2) encoding the EO
antigen of BVDV optimized by the insertion of the
signal sequence of the human tPA upstream of EO and by
the insertion of intron II of the rabbit beta-globin
gene upstream of E0, and of a second expression plasmid
(e.g. pLF1021 Example 5.2.2, pLF1023 Example 6.1.2)
encoding the E2 antigen of BVDV optimized by the
insertion of the signal sequence of the human tPA
upstream of E2, by the deletion of the fragment of the
nucleotide sequence encoding the transmembrane domain
of E2 and the contiguous C-terminal part and by the
insertion of intron II of the rabbit beta-globin gene
upstream of E2.
A mixture of plasmids can be advantageously
produced. The mixture may comprise at least two
expression plasmids, each expressing a different
immunogen (EO or E2) and/or obtained from a different
type of BVDV (BVDV-1 or BVDV-2). In particular, a
mixture made of four plasmids expressing BVDV-1 E0,
BVDV-1 E2, BVDV-2 EO and BVDV-2 E2.
The subject of the present invention is also an
improved DNA vaccine capable of inducing an effective
and protective immune response in bovines against the
parainfluenza virus type 3 (bPI-3).
The bPI-3 virus is a Paramyxovirus, also a
member of the Paramyxoviridae family (Tsai et al.,
Infect. Imsmun., 1975, 11, 783-803).
Nucleotide sequences encoding the hemagglutinin
and neuraminidase proteins (HN) and the fusion protein
(F) of bPI-3 are known and accessible from the GenBank
database under the accession number U31671.
The DNA vaccine against bPI-3 is preferably
formulated with an adjuvant according to the invention,
in particular DMRIE, preferably DN RIE-DOPE. This may be
CA 02398229 2010-01-20
30754-30
17
optionally combined with either the addition of bovine
GM-CSF, or the optimization of at least one bPI-3
antigen, of finally the addition of bovine GM-CSF and
the optimization of at least one bPI-3 antigen.
The addition of bovine GM-CSF may be carried
out as is described for BR-J-1.
The optimization of the antigens derived from
bPI-3 is carried out by substitution, by a "signal"
sequence, in particular that of the tPA of human
origin, of the signal sequence of hemagglutinin-
neura:nir_idase MN,) of bPI-3 and/or of the fusion
protein (F) of bPI-3, and/or by the deletion of the DNA
fragment encoding the transmembrane domain of HN and/or
of F, and/or by the insertion of an intron, in
particular of intron II of the rabbit beta-globin gene
upstream of the nucleotide sequence encoding HN and/or
F. The deletion of the DNA fragment encoding the
transmembrane domain of one of these proteins is
preferably accompanied by the contiguous C-terminal
part. The DNA vaccine against bPI-3 according to the
invention may therefore encode and express a single
optimized PI-3 antigen (EM or F) or both (HM and F).
Nucleotide sequences encoding the bPI-3
antigens which can be used in the present invention and
various expression vector constructs are given in the
accompanying examples and in FR-AI-2751229, in
particular in Examples 14 and 15, and in Figures 10 and
Z1.
Preferably, according to the invention, the DNA
vaccine against bPI-3 is formulated with DMRIE-DOPE,
and is composed of an expression plasmid (e.g. pLF1025
Example 7.1.2) encoding the HN antigen of bPI-3
optimized by the insertion of the signal sequence of
the human tPA in place of the signal sequence of P2.4, by
the deletion of the fragment of the nucleotide sequence
of encoding the transmembrane domain and the
contiguous C-terminal part and by the insertion of
intron II of the rabbit beta-globin gene upstream of
HN, and of a second expression plasmid (e.g. pLF1027
CA 02398229 2010-01-20
30754-30
18
Example 7.2.2) encoding the F antigen of bPI-3
optimized by the insertion of the signal sequence of
the human tPA in place of the signal sequence of F, by
the deletion of the fragment of the nucleotide sequence
encoding the transmembrane domain of F and the
contiguous C-terminal part and by the insertion of
intron II of the rabbit beta-globin gene upstream of F.
The subject of the present invention is an
improved DNA vaccine capable of inducing an effective
and protective immune response in pigs against porcine
herpesvirus (PRV).
The PRV virus is a member of the
Alphaherpesvirirzae family, this virus is responsible
for Aujeszky's disease (Sawitzky D., Arch. Virol.
Suppl., 1997, 13, 201-206).
Nucleotide secruences encoding the glycoproteins
gB, gC and gD are known and accessible from the GenBank
database under the accession number M17321, AF158090,
AF086702.
The DNA vaccine against PRV is preferably
formulated with an adjuvant according to the invention,
in particular DMRIE, preferably DNLRIE-DOPE. This may be
optionally combined with either the addition of porcine
GM-CSF (Inumaru S. and Takamatsu H., Immunol. Cell.
Biol., 1995, 73, 474-476), or the optimization of at
least one PRV antigen, or finally the addition of
porcine GM-CSF and the optimization of at least one PRV
antigen.
The addition of porcine GM-CSF may be carried
out by the incorporation of the porcine GM-CSF
poly-peptide into the vaccine composition or by the
insertion of a nucleotide sequence encoding the porcine
GM-CSF (e.g. accessible from the GenBank database under
the accession number D21074) into an in vivo expression
vector, preferably a plasmid. Preferably, the
nucleotide sequence encoding porcine GM-CSF is inserted
into a second expression plasmid (e.g. pLF1033
Example 14), different from that (or those) into which
CA 02398229 2010-01-20
30754-30
19
the gene(s) encoding the PRV antigen(s) is (are)
inserted.
The optimization of the antigens derived from
PRV is carried out by substitution, by a "signal"
sequence, in particular that of the tPA signal of human
origin (GenBank access-Lon number NM - 000930), of the
sequence of the signal peptide of the glycoprotein gB
and/or of the glycoprotein gC and/or of the
glycoprotein gD, and/or_ by the deletion of the DNA
fragment encoding the transmembrane domain of gB and/or
of gC and/or of gD_ The deletion of the DNA fragment
encoding the transmembrane domain of one of these
glycoproteins is preferably accompanied by the
contiguous C-terminal part. The DNA vaccine against PRV
according to the invention may therefore encode and
express a single optimized PRV antigen (gB, gC or gD)
or two of them or the three, that is to say optimized
gB, optimized gC and optimized gD.
Nucleotide sequences encoding the PRV antigens
which can be used in the present invention and various
expression vector constructs are given in the
accompanying examples and in FR-A1-2751224, in
particular in Examples 8 and 9 and in Figures 3 and 5.
Preferably, according to the invention, the DNA
vaccine against PRV is formulated with DMRIE-DOPE, and
is composed of an expression plasmid (e.g. pSB102
Example 8.1.2) encoding the gB antigen of PRV optimized
by the deletion of the fragement of the nucleotide
sequence encoding the transmembrane domain and of the
contiguous C-terminal part, of a second expression
plasmid (e.g. pSB104 Example 8.2.2) encoding the gC
antigen of PRV optimized by the deletion of the
fragment of the nucleotide sequence encoding the
transmembrane domain and of the contiguous C-terminal
part, and of a third expression plasmid (e.g. pSBl06
Example 8.3.2) encoding the gD antigen of PRV optimized
by the deletion of the fragment of the nucleotide
sequence encoding the transmembrane domain and of the
contiguous C-terminal part.
CA 02398229 2010-01-20
30754-30
The subject of the present invention is an
improved DNA vaccine capable of inducing an effective
and protective immune response in pigs against the
porcine reproductive respiratory syndrome virus
(PRRSV).
The PRRSV virus is an Arterivirus, a member of
the Arteriviridae family, (Murtaugh et al., Arch.
Virol., 1995, 140, 1451-1460).
Nucleotide sequences encoding the proteins
encoded by the open reading frames ORF3, ORF5 and ORF6
are known and accessible from the GenBank database
under the accession number U87392.
The DNA vaccine against PRRSV is preferably
formulated with an adjuvant according to the invention,
in particular DMRIE, preferably DMRIE-DOPE. This may be
optionally combined with either the addition of porcine
GM-CSF, or the optimization of at least one PRRSV
antigen, or finally the addition of porcine GM-CSF and
the optimization of at least one PRRSV antigen.
The addition of porcine GM-CSF may be carried
out as is described for PRV.
The optimization of the antigens derived from
PRRSV is carried out by substitution, by a "signal"
sequence, in particular that of the tPA signal of human
origin (GenBank accession number NM - 000930), of the
sequence of the signal peptide of the protein encoded
by the open reading frame 3 (ORF3, gp45 or large
envelope glycoprotein) and/or of the glycoprotein ORFS
(gp25 or envelope glycoprotein E) and/or of the
glycoprotein ORF6 (gp18 or membrane protein), and/or by
the deletion of the DNA_ fragment encoding the
transmembrane domain of ORF3 and/or ORFS and/or ORF6.
The deletion of the DNA fragment encoding the
transmembrane domain of one of these glycoproteins is
preferably accompanied by the contiguous C-terminal
part. The DNA vaccine against PRRSV according to the
invention may therefore encode and express a single
optimized PRRSV antigen (ORF3, ORF5 or ORF6) or two of
CA 02398229 2010-01-20
30754-30
21
them or the three, that is to say optimized ORF3,
optimized ORFS and optimized ORF6.
Nucleotide sequences encoding the PRRSV
antigens which can be used in the present invention and
various expression vector constructs are given in the
accompanying examples and in FR-A1-2751224, in
particular in Examples 14 to 17 and in Figures 14 to
17.
Preferably, according to the invention, the DNA
vaccine against PRRSV is formulated with DNERIE-DOPE,
and is composed of an expression plasmid (e.g. pLF1009
Example 9.1.1, pLF1015 Example 10.1.1) encoding the
ORF3 antigen of PRRSV, of a second expression plasmid
(e.g. pLF1012 Example 9.2.2, pLF1018 Example 10.2.2)
encoding the ORF5 antigen of PRRSV optimized by
substitution of the signal sequence of ORF5 by the
human tPA signal peptide sequence and by the deletion
of the fragment of the nucleotide sequence encoding the
transmembrane domain and the contiguous C-terminal
part, and of a third expression plasmid (e.g. pLF1014
Example 9.3.2, pLF1016 Example 10.3.2) encoding the
ORF6 antigen of PRRSV optimized by the substitution of
the signal sequence of ORF6 by the human tPA signal
peptide sequence and by the deletion of the fragment of
the nucleotide sequence encoding the transmembrane
domain and the contiguous C-terminal part.
A mixture of plasmids may be advantageously
produced. The mixture may comprise at least two
expression plasmids, each expressing a different
iri unogen (ORF3, ORFS or ORF6) and/or obtained from a
different strain of PRRSV (e.g. European strain, for
example Lelystad, American strain ATCC VR-2332). In
particular, a mixture made of six plasmids expressing
PRRSV Lelystad ORF3, PRRSV Lelystad ORF5, PRRSV
Lelystad ORF6, PRRSV VR-2332 ORF3, PRRSV VR-2332 ORFS
and PRRSV VR-2332 ORF6.
The subject of the present invention is also an
improved DNA vaccine capable of inducing an effective
CA 02398229 2010-01-20
30754-30
22
and protective immune response in porcines against the
swine influenza virus (SIV).
The SIV virus is an influenza virus group A, a
member of the Orthomyxoviridae family (Marphy B.R. and
Webster R.G., Virology, Second Edition, edited by
B.N. Fields, D.M. Knipe et a?., Raven Press Ltd., New
York 1990).
Nucleotide sequences encoding the hemagglutinin
(HA) and neuraminidase (NA) proteins of the SIV H1N1
and H3N2 strains are known and accessible from the
GenBank database under the accession number K00992,
U86145, U07146, AF153238.
The DNA vaccine against SIV is preferably
formulated with an adjuvant according to the invention,
in particular DMI IE, preferably DMRIE-DOPE. This may be
optionally combined with either the addition of porcine
GM-CSF, or the optimization of at least one SIV
antigen, or finally the addition of porcine GM-CSF and
the optimization of at least one SIV antigen.
The addition of porcine GM-CSF may be carried
out as is described for PRV.
The optimization of the antigens derived from
SIV is carried out by substitution, by a "signal"
sequence, in particular that of the tPA of human
origin, of the signal sequence of SIV hemagglutinin
(FHA) and/or of the SIV neuraminidase (NA) protein,
and/or by the deletion of the DNA fragment encoding the
transmembrane domain of HA and/or of NA, and/or by the
insertion of an intron, in particular of intron 11 of
the rabbit beta-globin gene upstream of the nucleotide
sequence encoding HA and/or NA. The deletion of the DNA
fragment encoding the transmembrane domain of one of
these proteins is preferably accompanied by the
contiguous C-terminal part. The DNA vaccine against SIV
according to the invention may therefore encode and
express a single optimized SIV antigen (F?A or NA) or
both (HA and NA).
Nucleotide sequences encoding SIV antigens
which can be used in the present invention and various
CA 02398229 2010-01-20
30754-30
23
expression vector constructs are given in the
accompanying examples and in FR-A1-2751224, in
particular in Examples 10 and 11, and in Figures 7 and
9 for SIV strain H1N1, and in Examples 12 and 13, and
in Figures 11 and 13 for SIV strain H3N2.
Preferably, according to the invention, the DNA
vaccine against SIV is formulated with DMRIE-DOPE, and
is composed of an expression plasmid (e.g. pLF1002
Example 11.1.2, pLF1006 Example 12.1.2) encoding the HA
antigen of SIV optimized by the insertion of the signal
sequence of the human tPA in place of the signal
sequence of HA, by the deletion of the fragment of the
nucleotide sequence of HA encoding the transmembrane
domain and the contiguous C-terminal part, and by the
insertion of intron II of the rabbit beta-globin gene
upstream of HA, and of a second expression plasmid
(e.g. pLF1004 Example 11.2.2, pLF1008 Example 12.2.2)
encoding the NA antigen of SIV optimized by the
insertion of the signal sequence of the human tPA in
place of thelsignal sequence of NA, by the deletion of
the fragment of the nucleotide sequence encoding the
transmembrane domain of NA and the contiguous
C-terminal part, and by the insertion of intron II of
the rabbit beta-globin gene upstream of NA.
A mixture of plasmids may be advantageously
produced. The mixture may comprise at least two
expression plasmids, each expressing a different
immunogen (HFA or NA) and/or derived from a different
SIV strain (e.g. H1N1 or H3N2). In particular, a
mixture made of four plasmids expressing SIV H1N1 HA,
SIV H1N1 NA, SIV H3N2 HA and SIV H3N2 NA.
Although the invention is described in relation
to specific DNA vaccines, the invention and in
particular the use of the adjuvants according to the
invention also applies to DNA vaccines directed against
other pathogens of these animal species.
In the same line of thought, the vaccines
according to the invention may be, for an animal
species, combined with one another and/or with DNA
CA 02398229 2010-01-20
30754-30
24
vaccines directed against other pathogens of the same
species.
These other pathogens may be in particular the
rabies virus, hog cholera virus and porcine
parvoviruses.
An immunogenic preparation or an improved DNA
vaccine according to the invention against the rabies
virus comprises in particular a plasmid encoding the
unmodified G glycoprotein of the rabies virus and
DMRIE-DOPE and optionally the addition of GM-CSF.
An improved immunogenic preparation or DNA
vaccine according to the invention against the porcine
parvovirus comprises in particular a plasmid encoding
an antigen derived from the porcine parvovirus (e.g.
the VP2 protein, Example 18 and Figure 18 of
FR-A1-2751224) and DMRIE-DOPE and optionally the
addition of porcine GM-CSF (e.g. pLF1Q33, Example 14).
An improved immunogenic preparation or DNA
vaccine according to the invention against the hog
cholera virus (HCV) comprises in particular a plasmid
encoding an antigen derived from HCV (e.g. the El
protein, Example 19 and Figure 19 of or the E2 protein,
Example 20 and Figure 20 of the same document) and
DMRIE-DOPE and optionally porcine GM-CSF (e.g. pLF1033,
Example 14).
Thus, the subject of the present invention is
also improved multivalent DNA vaccines which make it
possible to obtain an effective protection in bovines
against at least two bovine pathogens selected from the
group consisting of the BHV-1, BRSV, BVDV, bPI-3 and
rabies viruses.
The subject of the present invention is also
improved multivalent DNA vaccines which make it
possible to obtain effective protection in pigs against
at least two porcine pathogens selected from the group
consisting of the PRV virus, PRRSV virus, SIV virus,
hog cholera virus (or HCV), and porcine parvoviruses.
The multivalent DNA vaccines may be improved by
their formulation with an adjuvant according to the
CA 02398229 2010-01-20
30754-30
invention, in particular with DMRIE, preferably with
DMRIE-DOPE. This may be optionally combined either with
the addition of GM-CSF as previously described, or with
the optimization of at least one antigen of interest as
previously described, or finally by the addition of
GM-CSF and the optimization of at least one antigen of
interest.
The improved multivalent DNA vaccines according
to the invention are composed of one or more expression
plasmids, such that these vaccines lead to the in vivo
expression of at least one immunogen of a first
pathogen and of at least one immunogen of at least one
other pathogen, infecting the same animal species. At
least one of these immunogens is preferably selected
from the members of the following group:
- F of BRSV, G of BRSV, gB of BHV-l, gC of
BH'V-1, gD of BHV-l, EO of BVDV-1, E2 of BVDV-1, EO of
BVDV-2, E2 of BVDV-2, F of bPI-3 and HN of bPI-3 f or
bovines, and
- gB of PRV, gC of PRV, gD of PRV, ORF3 of
PRRSV strain Lelystad, ORFS of PRRSV strain Lelystad,
ORF6 of PRRSV strainLelystad,' ORF3 of PRRSSV strain
VR-2332, ORFS of PRRSV strain VR-2332, ORF6 of PRRSV
strain VR-2332, HA of SIV strain H1N1, NA of SIV strain
H1N1, HA of SIV strain H3N2 and NA of SIV strain H3N2
for porcines.
The improved monovalent or multivalent DNA
vaccines according to the invention may also be
combined with at least one conventional vaccine
(inactivated, attenuated live, subunit) or recombinant
vaccine using an in vivo expression vector (e.g.
poxvirus, adenovirus, herpesvirus) directed against at
least one different pathogen infecting the same
species.
Persons skilled in the art may refer to
FR-Al-2751229 for the methods for constructing the
plasmids containing these bovine valencies, to
FR-Al-2751224 for the porcine valencies.
CA 02398229 2010-01-20
30754-30
26
The subject of the present invention is also a
method of vaccinating farm animals, in particular
bovines or porcines. This vaccination method comprises
the administration of one of the monovalent or
multivalent improved DNA vaccines as described above.
These vaccination methods concern gestating females for
the passive transfer of immunity or young animals or
adults. This vaccination method comprises the
administration of one or more doses of the improved DNA
vaccine.
The quantity of DNA used in the vaccines
according to the present invention is between about
Vg and about 1000 big, and preferably between about
50 pg and about 500 pg, for a given plasmid. Persons
skilled in the art possess the competence necessary to
precisely define the effective dose of DNA to be used
for each vaccination protocol.
The dose volumes may be preferably between 0.2
and 5 ml, preferably between 1 and 3 ml.
The improved DNA vaccines according to the
invention may be administered, in the context of this
vaccination method, by various routes of administration
proposed in the prior art for polynucleotide
vaccination and by means of known techniques of
administration.
According to a preferred mode of the invention,
the methods of vaccination comprise the administration
of the improved DNA vaccines according to the invention
by the intramuscular route, the subcutaneous route or
with the aid of an injector without needle by the
intradermal route.
The invention will now be described in greater
detail with the aid of embodiments taken as nonlimiting
examples and referring to the drawings, in which:
Figure No. 1: plasmid pVR1012
Figure No. 2: plasmid pAB110
Sequence listing:
SEQ ID NO 1: oligonucleotide PB326
CA 02398229 2010-01-20
30754-30
27
SEQ ID NO 2: oligonucleotide PB329
SEQ ID NO 3: oligonucleotide SB090
SEQ ID NO 4: oligonucleotide SB091
SEQ ID NO 5: oligonucleotide LF001
SEQ ID NO 6: oligonucleotide LF002
SEQ ID NO 7: oligonucleotide PE234
SEQ ID NO 8: oligonucleotide PB235
SEQ ID NO 9: oligonucleotide PB511
SEQ ID NO 10: oligonuc leotide PB512
SEQ ID NO 11: oligonucleotide SB221
SEQ ID NO 12: oligonucleotide SB222
SEQ ID NO 13: oligonucleotide PB507
SEQ ID NO 14: oligonucleotide PBS08
SEQ ID NO 15: oligonucleotide PB513
SEQ ID NO 16: oligonucleotide PB514
SEQ ID NO 17: oligonucleotide S3223
SEQ ID NO 18: oligonucleotide SB224
SEQ ID NO 19: oligonucleotide PB497
SEQ ID NO 20: oligonucleotide PB498
SEQ ID NO 21: oligonucleotide SB225
SEQ ID NO 22: oligonucleotide SB226
SEQ ID NO 23: oligonucleotide SB210
SEQ ID NO 24: oligonucleotide SB211
SEQ ID NO 25: oligonucleotide SB212
SEQ ID NO 26: oligonucleotide SB220
SEQ ID NO 27: oligonucleotide SB213
SEQ ID NO 28: oligonucleotide SB214
SEQ ID NO 29: oligonucleotide SB215
SEQ ID NO 30: oligonucleotide SB216
SEQ ID, NO 31: oligonucleotide LF050
SEQ ID NO 32: oligonucleotide LF051
SEQ ID NO 33: oligonucleotide LF052
SEQ ID NO 34: oligonucleotide LF053
SEQ ID NO 35: oligonucleotide LF039
SEQ ID NO 36: oligonucleotide LF040
SEQ ID NO 37: oligonucleotide LF041
SEQ ID NO 38: oligonucleotide LF042
SEQ ID NO 39: oligonucleotide LF043
SEQ ID NO 40: oligonucleotide LF044
CA 02398229 2010-01-20
30754-30
28
SEQ ID NO 41: oligonucleotide LF045
SEQ ID NO 42: oligonucleotide LF046
SEQ ID NO 43: oligonucleotide LF064
SEQ ID NO 44: oligonucleotide LF065
SEQ ID NO 45: oligonucleotide LF066
SEQ ID NO 46: oligonucleotide LF067
SEQ ID NO 47: oligonucleotide LF047
SEQ ID NO 48: oligonucleotide LF048
SEQ ID NO 49: oligonucleotide LF058
SEQ ID NO 50: oligonucleotide LF059
SEQ ID NO 51: oligonucleotide LF06C
SEQ ID NO 52: oligonucleotide LF061
SEQ ID NO 53: oligonucleotide LF062
SEQ ID NO 54: oligonucleotide LF063
SEQ ID NO 55: oligonucleotide SB201
SEQ ID NO 56: oligonucleotide SB202
SEQ ID NO 57: oligonucleotide SB203
SEQ ID NO 58: oligonucleotide SB217
SEQ ID NO 59: oligonucleotide SB204
SEQ ID NO 60: oligonucleotide SB205
SEQ ID NO 61: oligonucleotide SB206
SEQ ID NO 62: oligonucleotide SB218
SEQ ID NO 63: oligonucleotide SB207
SEQ ID NO 64: oligonucleotide SB208
SEQ ID NO 65: oligonucleotide SB209
SEQ ID NO 66: oligonucleotide SB219
SEQ ID NO 67: oligonucleotide LF027
SEQ ID NO 68: oligonucleotide LF028
SEQ ID NO 69: oligonucleotide LF019
SEQ ID NO 70: oligonucleotide LF020
SEQ ID NO 71: oligonucleotide LF021
SEQ ID NO 72: oligonucleotide LF022
SEQ ID NO 73: oligonucleotide LF023
SEQ ID NO 74: oligonucleotide LF024
SEQ ID NO 75: oligonucleotide LF025
SEQ ID NO 76: oligonucleotide LF026
SEQ ID NO 77: oligonucleotide LF037
SEQ ID NO 78: oligonucleotide LF038
SEQ ID NO 79: oligonucleotide LF029
CA 02398229 2010-01-20
30754-30
29
SEQ ID NO 80: oligonucleotide LF030
SEQ ID NO 81: oligonucleotide LF031
SEQ ID NO 82: oligonucleotide LF032
SEQ ID NO 83: oligonucleotide LF033
SEQ ID NO 84: oligonucleotide LF034
SEQ ID NO 85: oligonucleotide LF035
SEQ ID NO 86: oligonucleotide LF036
SEQ ID NO 87: oligonucleotide LF003
SEQ ID NO 88: oligonucleotide LF004
SEQ ID NO 89: oligonucleotide LF005
SEQ ID NO 90: oligonucleotide LF006
SEQ ID NO 91: oligonucleotide LF007
SEQ ID NO 92: oligonucleotide LF008
SEQ ID NO 93: oligonucleotide LF009
SEQ ID NO 94: oligonucleotide LFO10
SEQ ID NO 95: oligonucleotide LF011
SEQ ID NO 96: oligonucleotide LF012
SEQ ID NO 97: oligonucleotide LF013
SEQ ID NO 98: oligonucleotide LF014
SEQ ID NO 99: oligonucleotide LF015
SEQ ID NO 100: oligonucleotide LF016
SEQ ID NO 101: oligonucleotide LF017
SEQ ID NO 102: oligonucleotide LF018
SEQ ID NO 103: oligonucleotide LF054
SEQ ID NO 104: oligonucleotide LF055
SEQ ID NO 105: oligonucleotide LF056
SEQ ID NO 106: oligonuclectide LF057
EXAMPLES:
For each of the pathogens considered, each gene
encoding the principal antigens (native form and
modified form) was the subject of a particular
construction in a eukaryotic expression plasmid. The
secreted forms of the antigens were obtained by
deletion of the fragments of genes encoding the
transmembrane and cytoplasmic domains. In all cases,
the transmembrane domains of the proteins were
identified on the basis of the hydropathy profiles (on
MacVector 6.5) of the corresponding protein sequences.
CA 02398229 2010-01-20
30754-30
Example 1: Molecular biology methods
1.1 Extraction of viral genomic DNA
Viral suspensions were treated with proteinase
K (100 mg/ml final) in the presence of sodium dodecyl
sulphate (SDS) (0.5% final) for 2 hours at 370C. The
viral DNA was then extracted with the aid of a
phenol /chloroform mixture, and then precipitated with
two volumes of absolute ethanol at -20 C for 16 hours
and then centrifuged at 10,000 g for 15 minutes at 4 C.
The DNA pellets were dried, and then taken up in a
minimum volume of sterile ultrapure water.
1.2 Isolation of viral genomic RNA
The ger_omic RNA of each virus was extracted
using the "guanidinium th_iocyanate/phenol-chloroform"
technique described by P. Chomczynski and N. Sacchi
(Anal. Biochem.. 1987. 162. 156-159).
1.3 Molecular biology techniques
All the constructions of plasmids were carried
out using the standard molecular biology techniques
described by Sambrook et al. (Molecular Cloning: A
Laboratory Manual. 2nd Edition. Cold Spring Harbor
Laboratory, Cold Spring Harbor, New York, 1989). All
the restriction fragments used for the present
invention were isolated with the aid of the "Geneclean"
kit (BIO101 Inc., La Jolla, CA). For all the
constructs, the cloned DNA fragments, as well as the
junctions with the expression vector, were sequenced by
the Sanger method (Sambrook et al., 1989).
1.4 PCR and RT-PCR
The oligonucleotides specific to the genes or gene
fragments cloned were synthesized, some of them
containing, in some cases, at their 5' end, restriction
sites facilitating the cloning of the amplified
fragments. The reverse transcription (RT) reactions and
CA 02398229 2010-01-20
30754-30
31
the polymerase chain reaction (PCR) were carried out
according to standard techniques (Sambrook et al.,
1989),
1.5 Large-scale purification of plasmids
The production, on the scale of about ten mg,
of purified plasmids entering into the vaccinal
compositions was carried out by the caesium chloride-
ethidium bromide gradient method (Sambrook et al_,
1989).
Example 2: Basic plasmid constructs
The eukaryotic expression plasmid pVR1020 (C.J.
Luke et al. Jj. of Infectious Diseases, 1997, 175, 95-
97), derived from the plasmid pVR1012 (Figure No. 1,
Figure 1 and Example 7 of WO-A-9803199), contains the
coding phase of the signal sequence of the human tissue
plasminogen activator (tPA).
A plasmid pVR1020 is modified by BamHI-Bg111
digestion and insertion of a sequence containing
several cloning sites (BamHI, Notl, EcoRI, Xbal, PmII,
PstI, BglII) and resulting from the pairing of the
following oligonucleotides:
P3326 (40 mer) (SEQ ID NO 1)
5' GATCTGCAGCACGTGTCTAGAGGATATCGAATTCGCGGCC 3' and
PB329 (40 mer) (SEQ ID NO 2)
5' GATCCGCGGCCGCGAATTCGATATCCTCTAGACACGTGCT 3'.
The vector thus obtained, having a size of
about 5105 base pairs (or bp), is called pA3110 (Figure
No. 2).
Intron II of the rabbit P-globin gene is cloned
into the vector pCRII (Invitrogen, Carlsbad, CA, USA)
after production of the corresponding DNA fragment by
PCR with the aid of the following cligonucleotides:
SB090 (20 mer) (SEQ ID NO 3)
5' TTGGGGACCCTTGATTGTTC 3' and
SE091 (21 mer) (SEQ ID NO 4)
5' CTGTAGGAAAAAGAAGAAGGC 31
CA 02398229 2010-01-20
30754-30
32
using as template the genomic DNA of rabbit peripheral
blood cells. The resulting plasmid is designated
pNS050,
The expression plasmid pAB110 is modified by
introducing the sequence of intron II of the rabbit
globin gene into the Sall site situated upstream of the
ATG of the signal peptide of tissue plasminogen
activator (tPA). The sequence of intron II of the
rabbit globin gene is amplified by polymerase chain
reaction (PCR) from the plasmid pNS050 using the
following oligonucleotide pair:
LF001 (30 mer) (SEQ ID NO 5)
5' CTCCATGTCGACTTGGGGACCCTTGATTGT 3' and
LF002 (30 mer) (SEQ ID NO 6)
5' CTCCATGTCGACCTGTAGGAAP.AAGAAGAA 3'
The PCR product (573 base pairs or bp) is
digested with Sall and cloned into the plasmid pAB110
previously 'Linearized with Sall, to generate the
plasmid pLF999 of about 5678 bp.
Example 3: Plasmids encoding the various forms of the
bovine herpesvirus type 1 (BEV-1) antigens
Fragments of viral DNA containing the gE, gC
and gD genes of the B901 strain of BHV-1 are isolated
by digesting the viral genome with various restriction
enzymes, by separating them by agarose gel
electrophoresis and by analysing them by Southern
blotting with the aid of probes corresponding to
fragments of the gB, gC and gD genes of the ST strain
of BHV-1 (Leung-Tack P. et al., Virology, 1994, 199,
409-421). The BHV-1 Colorado strain (Cooper] (ATCC
number VR-864) can also be used. The fragments thus
identified are cloned into the vector pBluescript SK+
(Stratager_e, La Jolla, CA, USA) and are at the origin
of the clonings of the three genes into the expression
vector pVR1012.
3.1 Plasmids encoding the various forms of BEV-1 gB
CA 02398229 2010-01-20
30754-30
33
3.1.1 pPB280: gB gene (native form) cloned into the
vector pVR1012
Two Xhol-Xhol fragments containing the 5' and
3' portions of the BHV-1 gB gene are identified by
Southern blotting and cloned into the vector
pBluescript SK+ (Stratagene, La Jolla, CA, USA)
previously digested with Xhol. The plasmids thus
obtained are designated pPB128 and pPB'17 respectively.
The plasmid pPBl28, containing the 5' fragment
of the gB gene, is digested with NotI and Xhol,
generating a fragment of 1708 bp (fragment A).
The plasmid pPB117, containing the 3' portion
of the gB gene, is digested with Xhol and Stul,
generating a fragment of 1345 bp. The latter fragment
is cloned into the vector pBluescript KS+ (Stratagene,
La Jolla, CA, USA) previously digested with EcoRV and
Xhol. The resulting plasmid is called pPB279. The
plasmid pPB279 is then digested with Xho! and BamHI,
generating a DNA fragment of 1413 bp (fragment B).
Fragments A and B are then cloned into a vector
pBluescript KS+ digested with Not! and BamHI,
generating plasmid pPB278 (about 6063 bp) and allowing
the reconstitution of the BaV-i gB gene.
The vector pPB278 then serves as template
during a PCR reaction carried out with the following
oligonucleotides:
PB234 (30 mer) (SEQ ID NO 7)
5' TTGTCGACATGGCCGCTCGCGGCGGTGCTG 3' and
PB23S (21 mer) (SEQ ID NO 8)
5' GCAGGGCAGCGGCTAGCGCGG 3'.
The PCR product (146 bp) is then digested with
the restriction enzymes Sall and NheI.
The plasmid pPB278 is digested with Nhel and
BamHI. The fragment of 2728 bp thus obtained and the
PCR fragment previously digested are ligated into the
vector pVR1012 (Example 2) previously digested with
Sall and Ba'uHI, thus generating the plasmid pPB280,
having a size of about 7742 bp.
CA 02398229 2010-01-20
30754-30
34
The BHV-1 gB gene encodes a protein of 933
amino acids.
3.1.2 pPB281: gB gene (.[TM-CterI form) cloned into the
vector pVR1012
The truncated form (deleted for its
transmembrane (TM) and carboxy-terminal (Cter) domains)
of the BHV-1 gB gene is obtained by ligating into the
plasmid pVR1012 (Example 2) predigested with Sall and
BamHI, both a fragment having a size of 2234 bp
obtained after digestion with Sall-PvuII of the plasmid
pPB280 (Example 3.1.1) and a fragment of 56 bp obtained
by pairing of the following oligonucleotides:
PE511 (52 mer) (SEQ ID NO 9)
5' CTGCACGAGCTCCGGTTCTACGACATTGACCGCGTGGTCAAGACGGACTGAG
3' and
PB512 (57 mer) (SEQ ID NO 10)
5' GATCCTCAGTCCGTCTTGACCACGCGGTCAATGTCGTAGAACCGGAGCTCGT
GCAG 3'.
The plasmid thus generated has a size of about
7154 bp and is called pPB281. The truncated gB gene of
BHV-1 encodes a protein of 759 amino acids.
3.1.3 pSB115: gB gene (tPA A[TM-Cter] form) cloned into
the vector pAB110
The tPA A[TM-Cter) form of the BHV-1 gB gene is
amplified by PCR from the template pPB281 (Example
3.1.2) and with the aid of the following primers:
SB221 (39 mer) (SEQ ID NO 11)
5' AAAATTTCGATATCCGCCGCGGGGCGACCGGCGACAACG 3' and
SB222 (33 mer) (SEQ ID NO 12)
5' GGAAGATCTTCAGTCCGTCTTGACCACGCGGTC 3'
The amplification product (2088 bp) is digested
with the enzymes EcoRV and Bglll and cloned into the
vector pAB110 (Example 2) previously digested with
EcoRV and BglII, generating the plasmid pSB115, having
a size of about 7154 bn.
CA 02398229 2010-01-20
30754-30
The tPA A[TM-Cter) form of the gB gene encodes
a glycoprotein of 729 amino acids, containing the
extracellular domain of the BHV-1 gB glycoprotein.
3.2. Plasmids encoding the various forms of BBV-1 gC
3.2.1 pPB264: gC gene (native form) cloned into the
vector pVR1012
A BamHi-Hindlll fragment of 3.5 kb containing
the complete BH-V-1 gC gene is identifed by Southern
blotting and cloned into the vector pEluescript SK+.
The plasmid thus obtained is called pPB287.
The plasmid pPB287 is then digested with Ncol-
BssSI. A digestion fragment having a size of 1492 bp is
obtained. It is ligated with a synthetic DNA fragment
obtained by the pairing of the following
oligonucelotides:
PB507 (37 mer) (SEQ ID NO 13)
5' TCGTGCCTGCGGCGCAAGGCCCGGGCGCGCCTGTAGT 3' and
PB508 (37 mer) (SEQ ID NO 14)
5' CTAGACTACAGGCGCGCCCGGGCCTTGCGCCGCAGGC 3',
into the plasmid pLitmus 28 (New England Biolabs, Inc.,
Beverly, MA, USA) predigested with NcoI and Xbal,
generating the intermediate plasmid pPB290.
The fragment of 1554 bp derived from the
digestion of pPB290 with PstI and Xbal is cloned into
the vector pVR1012 (Example 2) previously digested with
Pstl and XbaI, thus generating the plasmid pPB264,
having a size of about 6427 bp. The BFV-1 gC gene
encodes a protein of 508 amino acids.
3.2.2 pPB292: gC gene (A[TM-Cterj form) cloned into the
vector pVR1012
The truncated form of the BHV-1 gC gene is
obtained by ligating the following three DNA fragments
into the vector pVR1012 (Example 2) previously digested
with PstI and Xbal:
(a) a fragment of 1035 bp derived from the
digestion of pPB264 (Example 3.2.1) with PstI and Xhol,
CA 02398229 2010-01-20
30754-30
36
(b) a fragment of 350 bp derived from the
digestion of pPB264 with XhoI and BanI and
(c) a synthetic fragment of 43 bp resulting
from the pairing of the oligonucleotides PB513 and
PB514.
These oligonucleotides are the following:
PB513 (43 mer) (SEQ ID NO 15)
5' GCACCGCTGCCCGAGTTCTCCGCGACCGCCACGTACGACTAGT 3' and
PE514 (43 mer) (SEQ ID NO 16)
5' CTAGACTAGTCGTACGTGGCGGTCGCGGAGAACTCGGGCAGCG 3'.
The plasmid having a size of about 6305 bp thus
obtained is called pPB292. The truncated gC gene of
BHV-1 encodes a protein of 466 amino acids.
3.2.3 pSB116: gC gene (tPA 0[TM-Cter2 form) cloned into
the vector pAB110
The tPA A[TM-Cter] form of the BHV-1 gC gene is
amplified by PCR from the template pPB292 (Example
3.2.2) and with the aid of the following primers:
SB223 (39 mer) (SEQ ID NO 17)
5' AAAATTTCGATATCCCGGCGGGGGCTCGCCGAGGAGGCG 3' and
SB224 (32 mer) (SEQ ID NO 18)
5' GGAAGATCTCTAGTCGTACGTGGCGGTCGCGG 3'
The amplification, product (1362 bp) is digested
with the enzymes EcoRV and BglII and cloned into the
vector pAB110 (Example 2) previously digested with
EcoRV and Bglli, generating the plasmid pSB116, having
a size of about E404 bp.
The tPA A [ TM-Cter] form of the gC gene encodes
a glycoprotein of 479 amino acids, containing the
extracellular domain of the B}W-1 gC glycoprotein.
3.3 Plasmids encoding the various forms of BHV-1 gD
3.3.1 pPB148: gD gene (native form) cloned into the
vector pVR1012
A Xhoi-XhoI fragment of 5 kb containing the
BHV-1 gD gene is identified by Southern blotting and
CA 02398229 2010-01-20
30754-30
37
cloned into the vector pEluescript SK+ predigested with
XhoI, generating the plasmid pPB147.
A fragment of 325 bp derived from the digestion
of pPB147 with NdeI and BsrB2 and a fragment of 943 bp
derived from the digestion of pP3147 with Ndel and Styl
are then ligated into the vector pVR1012 (Example 2)
predigested with EcoRV and Xbal, thus generating the
plasmid pPBl48, having a size of about 6171 bp. The
BHV-1 gD gene encodes a protein of 417 amino acids.
3.3.2 pPB284: gD gene (A[TM-Cterj form) cloned into the
vector pVR1012
The truncated gD gene of BHV-1 is obtained from
a fragment obtained after PCR amplification carried out
on the genomic DNA of the B901 strain of the BHV-1
virus previously digested with PstI and Xbal and with
the aid of the following primer pair:
P3497 (33 mer) (SEQ ID NO 19)
5' TTTCTGCAGATGCAAGGGCCGACATTGGCCGTG 3' and
P3498 (31 mer) (SEQ ID NO 20)
5' TTTCTAGATTAGGGCGTAGCGGGGGCGGGCG 3'.
This PCR fragment is then cloned into the
plasm-,d pVR1012 (Example 2) previously digested with
Pstg. and Thal, generating the plasmid pPB284, having a
size of about 5943 bp. The truncated gD gene of BHV-1
encodes a protein of 355 amino acids.
3.3.3 pSB117: gD gene (tPA A[TM-Cter1 form) cloned into
the vector pAB110
The tPA A[TM-Cter] form of the BHV-1 gD gene is
amplified by PCR from the pPB284 template (Exa-nple
3.3.2) and with the aid of the following primers:
SB225 (39 mer) (SEQ ID NO 21)
5' AA.AATTTCGATATCCCCCGCGCCGCGGGTGACGGTATAC 3' and
SB226 (33 mer) (SEQ ID NO 22)
S' GGAAGATCTTTAGGGCGTAGCGGGGGCGGGCGG 3'.
The amplification product (1029 bp) is digested
with the enzymes EcoRV and BglII and cloned into the
vector pABi10 (Example 2) previously digested with
CA 02398229 2010-01-20
30754-30
38
EcoRV and BgiII, generating the plasmid pSB117, having
a size of about 6071 bp.
The tPA A[TM-Cter] form of the gD gene encodes
a glycoprotein of 368 amino acids, containing the
extracellular domain of the BHV-1 gD glycoprotein.
Example 4: Plasmids encoding the various forms of the
bovine respiratory sencitial virus (BRSV) antigens
The genes encoding the F and G antigens of the
BRSV virus are obtained by RT-PCR from the viral RNA of
the Snook strain (Thomas et al. Research in Vet.
Science, 1982, 33, 170-182) . The BRSV A 51908 strain
(ATCC number VR-794) may also be used.
4.1 Plasmids encoding the various forms of BRSV-F
4.1.1 pSB107: F gene (native form) cloned into the
vector pVR1012
The F gene of the Snook strain of BRSV is
amplified by RT-PCR using the viral RNA as template and
with the aid of the following primers:
SB210 (34 mer) (SEQ ID NO 23)
5' AAATTTTCTGCAGATGGCGACAACAGCCATGAGG 3' and
SB211 (35 mer) (SEQ ID NO 24)
5' TThAGGP_TCCTCATTTACTAAAGGAAAGATTGTTG 3'.
The amplification product, having a size of
1739 bp, is digested with the enzymes PstI and BamHI
and cloned into the vector pVR1012 (Example 2)
previously digested with Pstl and BamHI, thus
generating the plasmid pSB107, having a size of about
6583 bp.
The F gene of the BRSV virus encodes a protein
of 574 amino acids.
4.1.2 pSBl08: F gene (i(TM-Cterj form) cloned into the
vector pVRIO12
The truncated form of the F gene of the Snook
strain of BPSV is amplified by RT-PCR using the viral
CA 02398229 2010-01-20
30754-30
39
RNA as template and with the aid of the following
primers:
SB210 (SEQ ID NO 23) and
SB212 (39 mer) (SEQ ID NO 25)
5' AATTTTGGATCCTCATGTGGTGGATTTTCCTACATCTAC 31.
The amplification product (1581 bp) is digested
with the enzymes Pstl and BamHI and cloned into the
vector pVR1012 (Example 2) previously digested with
Pstl and BamFI, generating the plasmid pSB108, having a
size of about 6430 bp.
The truncated form of the F gene encodes a
glycoprotein of 523 amino acids, containing the
extracellular domain of the BRSV F glycoprotein.
4.1.3 pSB114: F gene (tPA A(TM-Cter] form) cloned into
the vector pAB110
The tPA A[TM-Ct:er] form of the F gene of the
BRSV Snook strain is amplified by RT-PCR using the
viral RNA as template and with the aid of the following
primers:
SB212 (SEQ ID NO 25) and
SB220 (38 mer) (SEQ ID NO 26)
5' AA,.AATTCACGTGAACATAACAGAAGA.ATTTTATCAP.TC 3'.
The amplification product (1516 bp) is digested
with the enzymes Pm1I and BglII and cloned into the
vector pAB110 (Example 2) previously digested with Pm1I
and BglII, generating the plasmid pSB114, having a size
of about 6572 bp.
The tPA tl[TM-Cterl form of the F gene encodes a
glycoprotein of 535 amino acids, containing the
extracellular domain of the BRSV F glycoprotein.
4.2 Plasmids encoding the various forms of BRSV-G
In the case of the BRSV G protein (type II
glycoprotein), the signal sequence and the
transmembrane sequence are indistinguishable, requiring
the addition of a signal sequence upstream of the
sequence corresponding to the extracellular domain
during the deletion of the transmembrane domain.
CA 02398229 2010-01-20
30754-30
The plasmid pAB110 (Example 2) is used for the
construction of the plasmids containing the truncated
forms of the gene encoding the BRSV G protein.
4.2.1 pSB109: G gene (native form) cloned into the
vector pVR1012
The G gene of the BRSV Snook strain is
amplified by RT-PCR using the viral RNA as template and
with the aid of the following primers:
58213 (32 mer) (SEQ ID NO 27)
5' ACGCGTCGACATGTCC_AACCATACCCATCATC 3' and
SB214 (38 mer) (SEQ ID NO 28)
S' TTAAA TCTAGATTAGATCTGTGTAGTTGATTGATTTG 3'.
The amplification product (784 bp) is digested
with enzymes Sail and Xbal and cloned into the vector
pVR1012 (Example 2) previously digested with Sall and
XbaI, generating the plasmid pSB109, having a size of
about 5661 bp.
The BRSV G gene encodes a glycoprotein of 257
amino acids.
4.2.2 pSB110: G gene (tPA M[TM-Cter] form) cloned into
the vector pAB110
The truncated form of the G gene of the BRSV
Snook strain is amplified by RT-PCR using the viral RNA
as template and with the aid of the following primers:
SB215 (33 mer) (SEQ ID NO 29)
5' TTTTAAGGATCCGCTAAAGCCAAGCCCACATCC 3' and
SB216 (33 mer) (SEQ ID NO 30)
5' TTAAAATCTAGATTAGATCTGTGTAGTTGATTG 3'.
The amplification product (666 bp) is digested
with the enzymes BamHI and Xbal and cloned into the
vector pAB110 (Example 2) previously digested with
BarII and Xbai, generating the plasmid pSB110, having a
size of about 5660 bp.
The tPA d[TM-Cter] form of the BRSV virus G
gene encodes a glycoprotein of 218 amino acids,
containing the extracellular domain of the G
CA 02398229 2010-01-20
30754-30
41
glycoprotein, but preceded by the signal sequence of
the tissue plasminogen activator.
Example 5: Plasmids encoding the various forms of the
bovine viral diarrhea virus type 1 (BVD-1) antigens
The genes encoding the EO (glycoprotein of
48 kDa or gp48) and E2 (gp53) antigens of the type 1
BVDV viruses are obtained by RT-PCR from the viral RNA
of the Osloss strain (L. De Moerlooze et a1. J. Gen.
Virol. 1993, 74, 1433-1438; A. Renard et al., DNA,
1985, 4, 439-438; A. Renard et al. Ann. Rech. Vet.,
1987, 18, 121-125). The NADL (ATCC VR-534) or New York
(ATCC VR-524) strains may also be used.
5.1 Plasmids encoding the various forms of EO of the
BVDV type 1 Osloss strain
5.1.1 pLF1028: EQ gene (native form) cloned into the
vector pVR1O12
The complementary DNA (cDNA) of the EO gene of
the Osloss strain is synthesized from the corresponding
viral RNA with the aid of the primer LFO51 and
amplified by the PCR reaction with the aid of the
following oligonucleotide pair:
LF050 (36 mer) (SEQ ID NO 31)
5' CATACCGTCGACATGAAGAAACTAGAGAAAGCCCTG 3' and
LF051 (40 mer) (SEQ ID NO 32)
5' CATACCGGATCCTCAGGCTGCATATGCCCCAAACCATGTC 3'.
The DNA fragment of about 765 by obtained by
digesting the PCR product with Sall and BamEl is
ligated with a fragment of 4866 bp resulting from the
digestion of pVR1012 (Example 2) with Sall and Barn= in
order to generate the plasmid pLF1O28 (about 5636 bp).
The E0 gene of BVDV-1 strain Osloss encodes a protein
of 252 amino acids.
An ATG codon is introduced into the sequence of
the oligonucleotide LF050 so as to allow the initiation
of the translation of the corresponding recombinant EO
polypeptide.
CA 02398229 2010-01-20
30754-30
42
5.1.2 pLF1029: EO gene, (5-globin tPA-EO) form cloned
into the vector pLF999.
The EO gene is synthesized by a PCR reaction
from the pLF1028 template (Example 5.1.1) and with the
aid of the following oligonucleotide pair:
LF052 (39 mer) (SEQ ID NO 33)
5' CATGACGCGGCCGCTATGPAGAAACTAGAGAAAGCCCTG 3' and
LF053 (40 mer) (SEQ ID NO 34)
5' CATGACAGATCTTTAGGCTGCATATGCCCCAAACCATGTC 3'.
The DNA fragment of about 770 bp obtained by
digesting the PCR product with NotI and Bglil is
ligated with a fragment of 5642 bp resulting from the
digestion of pLF999 (Example 2) with NotI and BglII in
order to generate the plasmid pLF1029 (about 6417 bp).
The EO gene of BVDV-1 strain Osloss thus
modified ([3-globin tPA-EO) encodes a protein of 283
amino acids.
5.2 Plasmids encoding the various forms of E2 of the
BVDV type 1 Osloss strain
5.2.1 pLF1020: E2 gene (native form) cloned into the
vector pVR1O12
The cDNA of the E2 gene of the Osloss strain is
synthesized from the corresponding viral RNA with the
aid of the primer LF040 and amplified by a PCR reaction
with the aid of the following oligonucleotide pair:
LF039 (33 mer) (SEQ ID NO 35)
5' CATGACGTCGACATGACGACTACTGCATTCCTG 3' and
LF040 (36 mer) (SEQ ID NO 36)
5' CP_TGACAGATCTTCAACGTCCCGAGGTCATTTGTTC 3'.
The DNA fragment of 1235 bp obtained by
digesting the PCR product with Sall and Bg1II is
ligated with a fragment of 4860 bp resulting from the
digestion of pVR1012 (Example 2) with Sa1I and Bg1II in
order to generate the plasmid pLF1020 (about 6100 pb).
The E2 gene of BVDV-1 strain Osloss encodes a
protein of 409 amino acids.
CA 02398229 2010-01-20
30754-30
43
An ATG codon is introduced into the sequence of
the oligonucleotide LF039 so as to allow the initiation
of the translation of the corresponding recombinant E2
polypeptide.
5.2.2 pLF1021: E2 gene, ((3-globin tPA-E2 atTM+Cter])
form cloned into the vector pLF999.
The E2 gene deleted for its transmembrane and
carboxy-terminal domains is synthesized by a PCR
reaction from the pLF1020 template (Example 5.2.1) and
with the aid of the following oligonucleotide pair:
LF041 (36 mer) (SEQ ID NO 37)
5' CATGACGCGGCCGCTATGACGACTACTGCATTCCTG 3' and
LF042 (35 mer) (SEQ ID NO 38)
5' CATGACAGATCTCAAGCGA.AGTAATCCCGGTGGTG 3.
The DNA fragment of 1132 bp obtained by
digesting the PCR product with Notl and Bglil is
ligated with a fragment of 5642 bp resulting from the
digestion of pLF999 (Example 2) with Notl and BglII in
order to generate the plasmid pLF1021 (about 6779 bp).
The E2 gene of BVDV-1 strain Osloss thus
modified (5-globin tPA-E2 A[TM+Cter]) encodes a protein
of 404 amino acids.
Example 6: Plasmids encoding the various forms of the
bovine viral diarrhea virus type 2 (BVDV-2) antigens
The genes encoding the E2 antigen (gp53) of the
B`VTDV type 2 viruses are obtained by RT-PCR from the
viral RNA of the strain 890 (J.F. Ridpath and S.R.
Bolin, Virology, 1995, 212, 36-46). The strain Q140 can
also be used and may be obtained from the Quebec
Ministry of Agriculture, Fisheries and Food, Armand-
Frappier Institute (P. Tijssen at al., Virology, 1996,
217, 356-361). The strains 1373 and 296 may also be
used (J.F. Ridpath, BVDV Research Project, National
Animal Disease Center, 2300 Dayton Avenue, Ames, USA).
6.1 Plasmids encoding the various forms of E2 of the
type 2 - 890 strain
CA 02398229 2010-01-20
30754-30
44
6.1.1. pLF1022: E2 gene (native form) cloned into the
vector pVR1012
The cDNA of the E2 gene of the strain 890 is
synthesized from the corresponding viral RNA with the
aid of the primer LF044 amplified by a PCR reaction
with the aid of the following oligonucleotide pair:
LF043 (36 mer) (SEQ ID NO 39)
5' ACTGTATCTAGAATGACCACCACAGCTTTCCTAATC 3' and
LF044 (39 mer) (SEQ ID NO 40)
5' ACTGTAAGATCTTTA_AGTATTCACTCCAGCACCCATAGC 3'.
The DNA fragment of about 1240 bp obtained by
digesting the PCR product with XbaI and BglII is
ligated with a fragment of 4891 bp resulting from the
digestion of pVR1012 (Example 2) with Xbal and BglII in
order to generate the plasmid pLF1022 (about 6136 bp).
The E2 gene of BVDV-2 strain 890 encodes a
protein of 410 amino acids.
An ATG codon is introduced into the sequence of
the oligonucleotide LF043 so as to allow the initiation
of the translation of the corresponding recombinant E2
polypeptide.
6.1.2 pLF1023: E2 gene, (13-globin tPA-E2 A[TM+Cter])
form, cloned into the vector pLF999
The E2 gene deleted for its transmembrane and
carboxy-terminal domains is synthesized by a PCR
reaction from the pLF1022 template (Example 6.2.1) and
with the aid of the following oligonucleotide pair:
LF045 (41 mer) (SEQ ID NO 41)
5' CATGACGCGGCCGCCCTATGACCACCACAGCTTTCCTAATC 3' and
LF046 (36 mer) (SEQ ID NO 42)
5' CATGACAGATCTTTATATGAACTCTGAGAAGTAGTC 3'.
The DNA fragment of about 1140 bp obtained by
digesting the PCR product with NotI and BglII is
ligated with a fragment. of 5642 bp resulting from the
digestion of pLF999 (Example 2) with NotI and BglII in
order to generate the plasmid pLF1023 (about 6787 bp).
CA 02398229 2010-01-20
30754-30
The E2 gene of B'STDV-2 strain 890 thus modified
(j3-globin tPA-E2 A[TM+Cter)) encodes a protein of 405
amino acids.
6.2 Plasmids encoding the various forms of EO of the
type 2 - 890 strain
6.2.1 pLF 1030: EO gene (native form) cloned into the
vector pVR1O12
The cDNA of the E0 gene of the 890 strain is
synthesized from the corresponding viral RNA with the
aid of the LF065 primer and amplified by a PCR reaction
with the aid of the following oligonuclectide pair:
LF064 (39 mer) (SEQ ID NO 43)
S' CATACCGTCGACATGAG_AAAGAAATTGGAGAAGGCACTG 3' and
LF065 (39 me-r) (SEQ ID NO 44)
5' CATACCGGATCCTCATGCTC-CATGAGCACCAAACCATGC 3'.
The DNA fragment of about 768 bp obtained by
digesting the PCR product with Sall and BamHI is
ligated with a fragment of 4866 bp resulting from the
digestion of pVR1012 (Example 2) with Sall and BamHI in
order to generate the plasmid pLF1030 (about 5639 bp).
The EO gene of BVDV-2 strain 890 encodes a protein of
253 amino acids.
An ATG codon is introduced into the sequence of
the oligonucleotide LF064 so as to allow the initiation
of the translation of the corresponding recombinant FO
polypeptide.
6.2.2 pLF1031: EO gene, (P-globin tPA-EO) form, cloned
into the vector pLF999.
The EO gene is synthesized by a PCR reaction
from the pLF1030 template (Example 6.2.1.) and with the
aid of the following oligonucleotide pair:
LF066 (42 mer) (SEQ ID NO 45)
S' CATGACGCGGCCGCTATGAG;~AGAAATTGGAGAAGGCACTG 3' and
LF067 (39 mer) (SEQ ID NO 46)
5' CATACCAGATCTTCATGCTGCATGAGCACCAA_ACCATGC 3'.
CA 02398229 2010-01-20
30754-30
46
The DNA fragment of about 770 bp obtained by
digesting the PCR product with Notl and BglII is
ligated with a fragment of 5642 bp resulting from the
digestion of pLF999 (Example 2) with NotI and BglII in
order to generate the pla.smid pLF1031 (about 6417 bp).
The EO gene of BVDV-2 strain 890 thus modified
((3-globin tPA-EO) encodes a protein of 283 amino acids.
Example 7: Plasmids encoding the various forms of the
bovine parainfluenza virus type 3 (bPI-3) antigens
The genes encoding the hemagglutinin-
neuraminidase (HN) and fusion (F) antigens of the bPI-3
virus are obtained by RT-PCR from the viral RNA of the
Reisinger SF-4 strain (accessible from ATCC under the
number VR-281) .
7.1 Plasmids encoding the various forms of HN of the
bPI-3 SF-4 strain
7.1.1 pLF1024: HN gene (native form) cloned into the
vector pVR1012
The cDNA of the 1 gene of the SF-4 strain is
synthesized from the corresponding viral RNA with the
aid of the primer LF048 and amplified by a PCR reaction
with the aid of the following oligonucleotide pair:
LF047 (39 mer) (SEQ ID NO 47)
5' CATATCGTCGACATGGAATATTGGAPACACACAAACAGC 3' and
LF048 (38 mer) (SEQ ID NO 48)
5' CATGACGATATCTAGCTGCAGTTTTTCGGAACTTCTGT 3'.
The DNA fragment of 1726 bp obtained by
digesting the PCR product with SaII and EcoRV is
ligated with a fragment of 4896 bp resulting from the
digestion of pVR1012 (Example 2) with Sall and EcoRV in
order to generate the plasmid pLF1024 (about 6619 bp).
The bPI-3 HN gene encodes a protein of 572
amino acids.
7.1.2 pLF1025: HN gene, (3-globin tPA-E2 A[TM]) form,
cloned into the vector pLF999
CA 02398229 2010-01-20
30754-30
47
The HN gene deleted for its transmembrane
domain is synthesized by a PCR reaction from the
pLF1024 template (Example 7.1.1) with the aid of the
following oligonucleotide pair:
LF058 (33 mer) (SEQ ID NO 49)
5' CATACTGCGGCCGCTTTPA'TTCAAGAGAACAAT 3' and
LF059 (35 mer) (SEQ ID NO 50)
5' CATATCGATATCTAGCTGCAGTTTTTCGGAACTTC 31.
The DNA fragment of 1566 bp obtained by
digesting the PCR product with Notl and EcoRV is
ligated with a fragment of 5663 bp resulting from the
digestion of pLF999 (Example 2) with Not? and EcoRV in
order to generate the plasmid pLF1025 (about 7229 bp).
The bPI-3 HN gene thus modified (0-globin tPA-
E2 A[TM)) encodes a protein of 548 amino acids.
7.2 Plasmids encoding the various forms of F of the
bPI-3 SP-4 strain
7.2.1 pLF1026: F gene (native form) cloned into the
vector pVR1012
The cDNA of the F gene of strain SF-4 is
synthesized from the corresponding viral RNA with the
aid of the primer LF061 and amplified by a PCR reaction
with the aid of the following oligonucleotide pair:
LFO660 (36 mer) (SEQ ID NO 51)
5' CATATCGTCGACATGATCATCACAAACACAATCATA 3' and
LF061 (36 mer) (SEQ ID NO 52)
5' CATCACCAGATCTTATTGTCTATTTGTCAGTATATA 3'.
The DNA fragment of 1628 bp obtained by
digesting the PCR product with Sall and BglII is
ligated with a fragment of 4860 bp resulting from the
digestion of pVR1012 (Example 2) with SalI and Bg1II in
order to generate the plasmid pLF1026 (about 6488 bp).
The bPI-3 F gene encodes a protein of 550 amino
acids.
7.2.2 pLF1027: F gene, (P-globin tPA-F AtTM+Cter])
form, cloned into the vector pLF999
CA 02398229 2010-01-20
30754-30
48
The F gene deleted for its transmembrane and C-
terminal domains is synthesized by a PCR reaction from
the pLR1026 template (Example 7.2.1) and with the aid
of the following oligonucleotide pair:
LF062 (42 mer) (SEQ ID NO 53)
5' CATACTGCGGCCGCTCA..AATAGACATAACAAAACTGCAACGT 3' and
LF063 (41 mer) (SEQ ID NO 54)
5' CATATCGATATCTATGCACTAGATTGATACCAACTTCCAAC 3'.
The DNA fragment of 1434 bp obtained by
digesting the PCR product with NotI and EcoRV is
ligated with a fragment of 5663 bp resulting from the
digestion of pLF999 (Example 2) with NotI and EcoRV in
order to generate the plasmid pLF1027 (about 7097 bp).
The bPI-3 F gene thus modified (3-globin tPA-F
A(TM+Cter]) encodes a protein of 504 amino acids.
Example 8: Plasmids encoding the various forms of the
pseudorabies virus (PRV) antigens
The genes encoding the PRV glycoproteins gB, gC
and gD are obtained by PCR from the viral DNA of the
NIA3 strain (M. Riviere et al. J. Virol. 66, 3424-3434;
A. Baskerville et al. The Veterinary Bulletin, 1973, 43
No. 9). Mutants of the PRV NIA3 strain may also be used
and are described in US-A-4, 680, 176 and deposited with
the Collection Nationale de Cultures de Microorganismes
(CNCM), Institut Pasteur, Paris, France, under the
references 1-351 and 1-352.
8.1. Plasmids encoding the various forms of PRV-gB
8.1.1. pSB101: gB gene (native form) cloned into the
vector pVR1012
The gB gene of the PRV NIA3 strain is amplified
by PCR using the viral DNA as template and with the aid
of the following primers:
SB201 (36 mer) (SEQ ID NO S5)
5' TTTTAAGATATCATGCCCGCTGGTGGCGGTCTTTGG 3' and
SB202 (39 mer) (SEQ ID NO 56)
5' TTTTAAGGATCCCTACAGGGCGTCGGGGTCCTCGCTCTC 3'.
CA 02398229 2010-01-20
30754-30
49
The amplification product (2766 bp) is digested
with the enzymes EcoRV and B--mHI and cloned into the
vector pVR1012 (Example 2) previously digested with
EcoRV and BamHI, generating the plasmid pSB101, having
a size of about 7631 bp.
The PRV gB gene encodes a glycoprotein of 913
amino acids.
8.1.2 pSB102: gB gene (MTM-Cter] form) cloned into the
vector pVR1012
The truncated form of the gB gene of the PRV
NIA3 strain is amplified by PCR using the viral DNA as
template and with the aid of the following primers:
SB201 (SEQ ID NO 55) and
SB203 (39 mer) (SEQ ID NO 57)
5' TTTTAP.GGATCCCTAGTGGTCCACCTTGACCACGCGGTC 3'.
The amplification product (2262 bp) is digested
with the enzymes EcoRV and BamHI and cloned into the
vector pVR1012 (Example 2) previously digested with
EccRV and BamHI, generating the plasmid pSE102, having
a size of about 7142 bp.
The truncated form WTM-Cter]) of the gB gene
encodes a glycoprotein of 750 amino acids, containing
the extracellular domain of the PRV gB glycoprotein.
8.1.3 pNS009: g8 gene (tPA AfTM-Cter] form) cloned into
the vector pAB110
The tPA A[TM-Cterl form of the gB gene of the
PRV NIA-3 strain is amplified by PCR from the template
pSB101 (Example 8.1.1) and with the aid of the
following primers:
SB203 (SEQ ID NO 57) and
SB217 (39 mer) (SEQ ID NO 58)
5' AAA<ATTTCGATATCCACCTCGGCCTCGCCGACGCCCGGG 3'.
The amplification product (2088 bp) is digested
with the enzymes EcoRV and BglII and cloned into the
vector pAB110 (Example: 2) previously digested with
EcoRV and Bg1Il, generating the plasmid pNS009, having
a size of about 7127 bp.
CA 02398229 2010-01-20
30754-30
The tPA A[TM-Cter] form of the gB gene encodes
a glycoprotein of 720 amino acids, containing the
extracellular domain of the PRV gB glycoprotein.
8.2 Plasmids encoding the various forms of PRV-gC
8.2.1 pSB103: gC gene (native form) cloned into the
vector pVR1012
The gC gene of the PRV NIA3 strain is amplified
by PCR using the viral DNA as template and with the aid
of the following primers:
SB204 (36 mer) (SEQ ID NO 59)
S' TTTTAAGATATCATGGCCTCGCTCGCGCGTGCGATG 3' and
SB205 (37 mer) (SEQ ID NO 60)
5' TT-TTAAAGATCTTTAAGGCCCCGCCTGGCGGTAGTAG 3'.
The amplification product (1452 bp) is digested
with the enzymes EcoRV and BglII and cloned into the
vector pVR1012 (Example 2) previously digested with
EcoRV and BgiII, generating the plasmid pSB103, having
a size of about 6323 bp.
The PRV gC gene encodes a glycoprotein of 479
amino acids.
8.2.2 pSB104: gC gene (d[TM-Cter] form) cloned into the
vector pVR1012
The truncated form of the gC gene of the PRV
NIA3 strain is amplified by PCR using the viral DNA as
template and with the aid of the following primers:
SB204 (SEQ ID NO 59) and
S32 0 6 (36 mer) (SEQ ID NO 61)
5' TTTTAAAGATCTTTAGGGGGAGGCGTCGTAGCGCTG 3'.
The amplification product (1332 bp) is digested
with the enzymes EcoRV and BglII and cloned into the
vector pVR1012 (Example 2) previously digested with
EcoRV and BglII, generating the plasmid pSB104, having
a size of about 6206 bp.
The truncated form (A[TM-Cter]) of the gC gene
encodes a glycoprotein of 440 amino acids, containing
the extracellular domain of the PRV gC glycoprotein.
CA 02398229 2010-01-20
30754-30
51
8.2.3 pNS0i2: gC gene (tPA A(TM-Cter] form) cloned into
the vector pAB110
The tPA A[TM-Ct.er] form of the gC gene of the
PRV NIA3 strain is amplified by PCR from the template
pSB103 (Example 8.2.1) and with the aid of the
following primers:
SB206 (SEQ ID NO 61) and
SB218 (39 mer) (SEQ ID NO 62)
5' AAAATTTCGATATCCACGGCGCTCGGCACGACGCCCAAC 3'.
The amplification product (1270 bp) is digested
with the enzymes EcoRV and Bg1II and cloned into the
vector pAB110 (Example 2) previously digested with
EcoRV and BglII, generating the plasmid pNS012, having
a size of about 6311 bp.
The tPA A[TM-Cter] form of the gC gene encodes
a glycoprotein of 448 amino acids, containing the
extracellular domain of the PRV gC glycoprotein.
8.3 Plasmids encoding the various forms of PRV-gD
8.3.1 pSB105: gD gene (native form) cloned into the
vector pVR1012
The gD gene of the PRV NIA3 strain is amplified
by PCR using the viral DNA as template and with the aid
of the following primers:
53207 (36 mer) (SEQ ID NO 63)
5' AATTTTGATATCATGCTGCTCGCAGCGCTATTGGCG 3' and
SB208 (36 mer) (SEQ ID NO 64)
5' AATTTTGGATCCCTACGGACCGGGCTGCGCTTTTAG 3'.
The amplification product (1227 bp) is digested
with the enzymes EcoRV and BamHI and cloned into the
vector pVR1012 (Example 2) previously digested with
EcoRV and BamF-I, generating the plasmid pSBl05, having
a size of about 6104 bp.
The PRV gD gene encodes a glycoprotein of 404
amino acids.
CA 02398229 2010-01-20
30754-30
52
8.3.2 pSB106: gD gene (A[TM-Cter] form) cloned into the
vector pVR1012
The truncated form of the gD gene of the PRV
NIA3 strain is amplified by PCR using the viral DNA as
template and with the aid of the following primers:
SB207 (SEQ ID NO 63) and
SE209 (40 mer) (SEQ ID NO 65)
5' P.AATTTTGGATCCCTAGCGGTGGCGCGAGACGCCCGGCGC 3'
The amplification product (1077 bp) is digested
with the enzymes EcoRV and BamHI and cloned into the
vector pVR1012 (Example 2) previously digested with
EcoRV and BamHI, generating the plasmid pSB106 having a
size of about 5957 bp.
The truncated form (A[TM-Cter]) of the gD gene
encodes a glycoprotein of 355 amino acids, containing
the extracellular domain of the PRV gD glycoprotein.
8.3.3 pPB238: gD gene (tPA A[TM-Cter] form) cloned into
the vector pAB110
The tPA A[TM-Cter] form of the gD gene of the
PRV NIA3 strain is amplified by PCR from the template
pSB105 (Example 8.3.1) and with the aid of the
following primers:
SE209 (SEQ ID NO 65) and
SB219 (39 mer) (SEQ ID NO 66)
5' AAAATTTCGATATCCACCT'I'CCCCCCGCCCGCGTACCCG 31.
The amplification product (1015 bp) is digested
with the enzymes EcoRV and BamHI and cloned into the
vector pAB110 (Example 2) previously digested with
EcoRV and BgIII, generating the plasmid pPB238, having
a size of about 6056 bn.
The tPA A[TM-Cter] form of the gD gene encodes
the glycoprotein of 363 amino acids, containing the
extracellular domain of the PRV gD glycoprotein.
Example 9: Plasmids encoding the various forms of the
porcine reproductive respiratory syndrome virus
(PRRSV), strain Lelystad, antigens
CA 02398229 2010-01-20
30754-30
53
The genes encoding the PRRSV ORF3, ORF5 and
ORF6 proteins are obtained by RT-PCR from the viral RNA
of the Lelystad strain (J. Meulenberg et al. Virology,
1993, 19, 62-72; WO-A-92-21375) , deposited June 5, 1991
with the Collection Nationale de Cultures de
Micrcorganismes (CNCM), Institut Pasteur, Paris,
France, under the reference 1-1102.
9.1 Plasmids encoding the various forms of the PRRSV
Lelystad strain ORF3
9.1.1 pLF1009: ORF3 gene (native form) cloned into the
vector pVR1012
The cDNA of the ORF3 gene of the Lelystad
strain is synthesized from the corresponding viral RNA
with the aid of the primer LF028 and amplified by a PCR
reaction with the aid of the following oligonucleotide
pair:
LF027 (30 mer) (SEQ ID NO 67)
5' CACTACGATATCATGGCTCATCAGTGTGCA 3' and
LF028 (30 mer) (SEQ ID NO 68)
51 CACTACAGATCTTTATCGTGATGTACTGGG 3'.
The DNA fragment of 802 bp obtained by
digesting the PCR product with EcoRV and BgliII is
ligated with a fragment of 4879 bp resulting from the
digestion of pVR1012 (Example 2) with Ec0RV and BgiIII
in order to generate the plasmid pLF1009 having a size
of about 5681 bp.
The PRRSV Lelystad ORF3 gene encodes a protein
of 265 amino acids.
9.2 Plasmids encoding the various forms of the PRRSV
Lelystad strain ORF5
9.2.1 pLF1011: ORF5 gene (native form) cloned into the
vector pVR1012
The cDNA of the ORFS gene of the Lelystad
strain is synthesized from the corresponding viral RNA
with the aid of the primer LF020 and amplified by a PCR
CA 02398229 2010-01-20
30754-30
54
reaction with the aid of the following oligonucleotide
pair:
LF019 (30 mer) (SEQ ID NO 69)
5' CTCACCGTCGACATGAGATGTTCTCACAAA 3' and
LF020 (30 mer) (SEQ ID NO 70)
S' CTCACCTCTAGACTAGGCCTCCCATTGCTC 3'.
The DIVA fragment of 802 bp obtained by
digesting the PCR product with Sall and Xbal is ligated
with a fragment of 4879 bp resulting from the digestion
of pvR1012 (Example 2) with Sall and Xbal in order to
generate the plasmid pLF1011 having a size of about
5681 bp.
The PRRSV Lelystad ORFS gene encodes a protein
of 201 amino acids.
9.2.2 pLF1012: ORFS gene (truncated form) cloned into
the vector pAB110
The ORF5 gene deleted for its transmemorane and
carboxy-terminal domains is synthesized by a PCR
reaction from the template pLF1011 (Example 9.2.1) with
the aid of the following oligonucleotide pair:
LF021 (30 mer) (SEQ ID NO 71)
5' CACCTCGGATCCTTTGCCGATGGCAACGGC 3' and
LF022 (33 mer) (SEQ ID NO 72)
5' CACCTCGGATCCTTAGACTTCGGCTTTGCCCAA 3'.
The DNA fragment of 432 bp obtained by
digesting the PCR product with BamHI is ligated with a
fragment of 5105 bp resulting from the digestion of
pAB110 (Example 2) with BamHI in order to generate the
plasmid pLF1012 having a size of about 5537 bp.
The PRRSV Lelystad ORFS gene thus modified (tPA
,&[TM+Cter)) encodes a protein of 168 amino acids.
9.3 Plasmids encoding the various forms of the PRRSV
Lelystad strain ORF6
9.3.1 pLF1013: ORF6 gene (native form) cloned into the
vector pVR1012
CA 02398229 2010-01-20
30754-30
The cDNA of the OR-F6 gene of the Lelystad
strain is synthesized from the corresponding viral RNA
with the aid of the primer LF024 and amplified by a PCR
reaction with the aid of the following oligonucleotide
pair:
LF023 (30 mer) (SEQ ID NO 73)
5' CACTCAGTCGACATGGGAGGCCTAGACGAT 3' and
LF024 (30 mer) (SEQ ID NO 74)
5' CACTCATCTAGATTACCGGCCATACTTGAC 3'.
The DNA fragment of 528 by obtained by
digesting the PCR product with Sall and XbaI is ligated
with the fragment of 4881 bp resulting in the digestion
of pVR1012 (Example 2) with Sall and XbalL in order to
generate the plasmid pLF1013 having a size of about
5409 bp.
The PRRSV Lelystad ORF6 gene encodes a protein
of 173 amino acids.
9.3.2 pLF1014: ORF6 gene (truncated form) cloned into
the vector pAB110
The ORF6 gene deleted for its transmembrane and
carboxy-terminal domains is synthesized by a PCR
reaction from the template pLF1013 (Example 9.3.1) with
the aid of the following oligonucleotide pair:
LF025 (30 mer) (SEQ ID NO 75)
5' CACTACGGATCCGTGTCACGCGGCCGACTC 3' and
LF026 (33 mer) (SEQ ID NO 76)
5' CACTACGGATCCTTAAACAGCTCGTTTGCCGCC 3'.
The DNA fragment of 390 bp obtained by
digesting the PCR product with BamHI is ligated with a
fragment of 5105 by resulting from the digestion of
pAB110 (Example 2) with Bamhl in order to generate the
plasmid pLF1014 having a size of about 5495 bp.
The PP.RSV Lelystad ORF6 gene thus modified
(tPA d[TM-Cter]) encodes a protein of 154 amino acids.
Example 10: Plasmids encoding the various forms of the
porcine reproductive respiratory syndrome virus
(PR.RSV), American strain ATCC VR-2332, antigens
CA 02398229 2010-01-20
30754-30
56
The genes encoding the PRRSV virus ORF3, ORF5
and ORF6 proteins are obtained by RT-PCR from the viral
RNA of the American strain (M. Murtaugh et al. Arch
Virol. 1995, 140, 1451-1460), deposited with the ATCC
under the number VR-2332.
10.1 Plasmids encoding the various forms of PRRSV
VR-2332 strain ORF3
10.1.1 pLF1015: ORF3 gene (native form) cloned into the
vector pVR1012
The cDNA of the ORF3 gene of the VR-2332 strain
is synthesized from the corresponding viral RNA with
the aid of the primer LF038 and amplified by a PCR
reaction with the aid of the following oligonucleotide
pair
LF037 (30 mer) (SEQ ID NO 77)
5' CACTACGATATCATGGTTAATAGCTGTACA 3' and
LF038 (30 mer) (SEQ ID NO 78)
5' CACTACTCTAGACTATCGCCGTACGGCACT 3'_
The DNA fragment of 769 bp obtained by
digesting the PCR product with EcoRV and Xbal is
ligated with a fragment of 4900 bp resulting from the
digestion of pVR1012 (Example 2) with EcoRV and BgIII
in order to generate the plasmid pLF1015 having a size
of about 5669 bp.
The PRRSV strain VR-2332 ORF3 gene encodes a
protein of 254 amino acids.
10.2 Plasmids encoding the various forms of the PRRSV
VR-2332 strain ORPS
10.2.1 pLF1017: ORFS gene (native form) cloned into the
vector pVR1012
The cDNA of the OFFS gene of the VR-2332 strain
is synthesized from the corresponding viral RNA with
the aid of the primer LF030 and amplified by a PCR
reaction with the aid of the following oligonucleotide
pair:
CA 02398229 2010-01-20
30754-30
57
LF029 (30 mer) (SEQ ID 110 79)
S' CACTACGATATCATGTTGGAGAAATGCTTG 3' and
LF030 (30 mer) (SEQ ID NO 80)
5' CACTACAGATCTCTAAGGACGACCCCATTG 3'.
The DNA fragrr,ent of 607 by obtained by
digesting the PCR product with EcoRJ and BglII is
ligated with a fragment of 4879 bp resulting from the
digestion of pVR1012 (Example 2) with EcoRV and BglII
in order to generate the plasmid pLF1017 having a size
of about 5486 bp.
The PRRSV strain VR-2332 ORF5 gene encodes a
protein of 200 amino acids.
10.2.2 pLF1018: ORF5 gene (truncated form) cloned into
the vector pABI10
The ORF5 gene deleted for its transmembrane and
carboxy-terminal domains is synthesized by a PCR
reaction from the template pLF1017 (Example 10.2.1)
with the aid of the following oligonucleotide pair:
LF031 (33 mer) (SEQ ID DNO 81)
5' CACTACGGATCCGCCAGCAACGACAGCAGCTCC 3' and
LF032 (33 mer) (SEQ ID NO 82)
5' CACTACGGATCCTTAGACCTCAACTTTGCCCCT 3'.
The DNA fragment of 426 by obtained by
digesting the PCR product with BanHI is ligated with a
fragment of 5105 bp resulting from the digestion of
pAB110 (Example 2) with BarnHI in order to generate the
plasmid pLF1018 having a size of about 5531 bp.
The PRRSV strain VR-2332 ORFS gene thus
modified (tPA A[TM+Cter}} encodes a protein of 166
amino acids.
10.3 Plasmids encoding the various forms of the PRRSV
VR-2332 strain ORF6
10.3.1 pLF1019: ORF6 gene (native form) cloned into the
vector pVR1012
The cDNA of the ORF6 gene of the VR-2332 strain
is synthesized from the corresponding viral RNA with
CA 02398229 2010-01-20
30754-30
58
the aid of the primer LF034 and amplified by a PCR
reaction with the aid of the following oligonucleotide
pair:
LF033 (33 mer) (SEQ ID NO 83)
5' CACATCCTGCAGATGGGGTCGTCCTTAGATGAC 3' and
LF034 (30 mer) (SEQ ID NO 84)
5' CACATCTCTAGATTATTTGGCATATTTGAC 3'.
The DNA fragment of 527 bp obtained by
digesting the PCR product with Pstl and Xbal is ligated
with a fragment of 4871 bp resulting from the digestion
of pVR1012 (Example 2) with Pstl and XhaI in order to
generate the plasmid pLF1019 having a size of about
5398 bp.
The PRRSV strain VR-2332 ORF6 gene encodes a
protein of 174 amino acids.
10.3.2 pLF1016: ORF6 gene (truncated form) cloned into
the vector pAB110
The ORF6 gene deleted for its transmembrane and
carboxy-terminal domains is synthesized by a PCR
reaction from the template pLF1019 (Example 10.3.1)
with the aid of the following oligonucleotide pair:
LF035 (30 mer) (SEQ ID NO 85)
5' CACTACGGATCCGTGAGTCGCGGCCGACTG 3' and
LF036 (33 mer) (SEQ ID NO 86)
5' CACTACGGATCCTTP_ALACAGCTTTTCTGCCACC 3'.
The DNA fragment of 390 bp obtained by
digesting the PCR product with BamHI is ligated with a
fragment of 5105 bp resulting from the digestion of
pABilO (Example 2) with BamHI in order to generate the
plasmid pLF1016 having a size of about 5459 bp.
The PRRSV strain VR-2332 ORF6 gene thus
modified (tPA A(TM+Cter]) encodes a protein of 154
amino acids.
Example 11: Plasmids encoding the various forms of the
swine influenza virus (SIV), strain H1N1, antigens
The genes encoding the hemagglutinin (HA) and
neuraminidase (NA) antigens of the swine influenza
CA 02398229 2010-01-20
30754-30
59
virus type HIM are obtained by RT-PCR from the viral
RNA of the "Sint" H1N1 strain. Strains are available from
the Virology Research Center, Armand-Frapnier
Institute, University of Quebec, Laval, Canada (D-S.
Agora et al., Virus Genes, 1997, 14, 251-254). See also
G.W. Both et al., Proc. Natl. Acad_ Sci. USA, 1983, 80,
6996-7000.
11.1 Plasmids encoding the various forms of SIV H1N1
strain HA
11.1.1 pLF1001: HA gene (native form) cloned into the
vector pVR1012
The cDNA of the HA gene of the HIN1 strain is
synthesized from the corresponding viral RNA with the
aid of the primer LF004 and amplified by a PCR reaction
with the aid of the following oligonucleotide pair:
LF003 (30 mer) (SEQ ID NO 87)
' CTCCATGATATCATGGAP_GCA.a.AACTP.TTC 3 ' and
LFOO4 (30 mer) (SEQ ID NO 88)
5' CTCCATCAGATCTTAAATGCATATTCTGCA 3'.
The DNA fragment of 1705 by obtained by
digesting the PCR product with EcoRV and BgliI is
ligated with the fragment of 4879 by resulting from the
digestion of pVR1012 (Example 2) with EcoRV and BgliI
in order to generate the plasm-';_d pLF1001 having a size
of about 6584 bp.
The SIV H1NI HA gene encodes a protein of 566
amino acids.
11.1.2 pLF1002: HA gene (modified form) cloned into the
vector pLF999
The F-:A gene deleted for its transme'nbrane and
carboxy-terninal domains is synthesized by a PCR
reaction from the template pLF1001 (Example 11.1.1)
with the aid of the following oligonucleotide pair:
LF005 (30 mer) (SEQ ID NO 89)
5' TCCGCGGCCGCACATGCTAACAATTCCACA 3' and
LF006 (32 mer) (SEQ ID NO 90)
5' TCCGCGGCCGCTTACATTGATTCTAGTTTCAC 3'.
CA 02398229 2010-01-20
30754-30
The DNA fragment of 1515 bp obtained by
digesting the PCR product with Notl is ligated with a
fragment of 5678 bp resulting from the digestion of
pLF999 (Example 2) with Notl in order to generate the
plasmid pLF1002 having a size of 7193 bp.
The SIV H1N1 HA gene thus modified (intron II
of the rabit (3-globin gene, tPA, A(TM+Cter)) encodes a
protein of 530 amino acids.
11.2 Plasmids encoding the various forms of the Sly
R1N1 strain NA
11.2.1 pLF1003: NA gene (native form) cloned into the
vector pVR1012
The cDNA of the NA gene of the H1N1 strain is
synthesized from the corresponding viral RNA with the
aid of the primer LF008 and amplified by a PCR reaction
with the aid of the following dligonucleotide pair:
LF007 (30 mer) (SEQ ID NO 91)
5' CACCTGGTCGACATGAATCCAAATCAGAAG 3' and
LF008 (30 mer) (SEQ ID NO 92)
S' CACCTGTCTAGACTACTTGTCAATGGTGkA 3'.
The DNA fragment of 1416 bp obtained by
digesting the PCR product with Sall and XbaI is ligated
with a fragment of 4881 bp resulting from the digestion
of pVR1012 (Example 2) with Sail and Xbal in order to
generate the plasmid pLF1003 having a size of about
6297 bp.
The SIV H1N1 NA gene encodes a protein of 469
amino acids.
11.2.2 pLF1004: NA gene (modified form) cloned into the
vector pLF999
The NA gene deleted for its transmembrane and
carboxy-terminal domains is synthesized by a PCR
reaction from the template pLF1003 with the aid of the
following oligonucleotide pair:
LF009 (31 mer) (SEQ ID NO 93)
S' CP_CTACC-?AATTCAC_ AATTGGGPATCAAA T 3' and
CA 02398229 2010-01-20
30754-30
61
LF010 (30 mer) (SEQ ID NO 94)
5' AATTTGTGAATTCGCGGCCGCGGATCCGGT 3'.
The DNA fragment of 1207 bp obtained by
digesting the PCR product with EcoRI is ligated with a
fragment of 5678 bp resulting from the digestion of
pLF999 (Example 2) with EcoRl in order to generate the
piasmid pLF1004 having a size of about 6885 bp.
The S_IV HiN1 NA gene thus modified (intron II
of the rabbit (3-globin gene, tPA, [i('TM+Cter]) encodes a
protein of 431 amino acids.
Example 12: Plasmids encoding the various forms of the
swine influenza virus (SIV), strain H3N2, antigens
The genes encoding the HA and NA antigens of
the type H3N2 swine influenza virus are obtained by RT-
PCR from the viral RNA of the "Cotes du Nord 1987"
(cdn87) strain referenced by the World Health
Organization (WHO) and available from the National
Influenza Reference Center, Virology Laboratory, 8
avenue Rockfeller, 69008 Lyon, France.
12.1 Plasmids encoding the various forms of the SIV
H3N2 strain HA
12.1.1 pLF1005: HA gene (native form) cloned into the
vector pVR1012
The cDNA of the HA gene of the H3N2 strain is
synthesized from the corresponding viral RNA with the
aid of the primer LF012 and amplified by a PCR reaction
with the aid of the following oligonucleotide pair:
LF011 (30 mer) (SEQ ID NO 95)
5' CTGCACGTCGACATGAAGACTGTCA_TTGCC 3' and
LF012 (24 mer) (SEQ ID NO 96)
S' GATATCTCAGATGCAAP.TGTTGCA 3'.
The DNA fragment of 1709 by obtained by
digesting the PCR product with EcoRV and Sail is
ligated with a fragment of 4893 bp resulting from the
digestion of pVR1012 (Example 2) with EcoRV and Sall in
CA 02398229 2010-01-20
30754-30
62
order to generate the plasmid pLF1005 having a size of
about 6602 bp.
The SIV H3N2 HA gene encodes a protein of 566
amino acids.
12.1.2 pLF1006: HA gene (modified form) cloned into the
vector pLF999
The HA gene deleted for its transmembrane and
carboxy-terminal domains is synthesized by a PCR
reaction from the template pLF1005 (Example 12.1.1)
with the aid of the following oligonucleotide pair:
LF013 (33 mer) (SEQ ID NO 97)
5' CACCGCGGATCCCTTCCAG_AAAATGGCAGCACA 3' and
LF014 (33 mer) (SEQ ID NO 98)
5' CACCGCGGATCCTTAGTCTTTGTATCCCGACTT 31.
The DNA fragment of 1542 bp obtained by
digesting the PCR product with BainHI is ligated with a
fragment of 5678 by resulting from the digestion of
pLF999 (Example 2) with BamHI in order to generate the
plasmid pLF1006 having a size of about 7220 bp.
The SIV 1-13N2 HA gene thus modified (intron II
of the rabbit 0-globin gene, tPA, A [TM+Cter]) encodes a
protein of 538 amino acids.
12.2 Plasmids encoding the various forms of the SIV
H3N2 strain NA
12.2.1 pLF1007: NA gene (native form) cloned into the
vector pVR1012
The cDNA of the NA gene of the H3N2 strain is
synthesized from the corresponding viral RNA with the
aid of the primer LF016 and amplified by a PCR reaction
with the aid of the following oligonucleotide pair:
LF015 (30 mer) (SEQ ID NO 99)
5' CACTCAGATATCATGAATCCAAAGCAAAAG 3' and
LF016 (30 mer) (SEQ ID NO 100)
5' CACTCATCTAGATTATATAGGCATGAGATC 3'.
The DNA fragment of 1414 bp obtained by
digesting the PCR product with EcoRV and Xoai is
CA 02398229 2010-01-20
30754-30
63
ligated with a fragment of 4900 bp resulting from the
digestion of pVR1012 (Example 2) with EcoRV and XbaI in
order to generate the plasmid pLF1007 having a size of
about 6314 bp.
The SIV H3N2 NA gene encodes a protein of 469
amino acids.
12.2.2 pLF1008: NA gene (modified form) cloned into the
vector pLF999
The NA gene deleted for its transmembrane and
carboxy-terminal domains is synthesized by a PCR
reaction from the template pLF1005 (Example 12.2.1)
with the aid of the following oligonucleotide pair:
LF017 (33 mer) (SEQ ID NO 101)
5' CACTACGGATCCTTCAAGCAATATGAGTGCGAC 3' and
LF018 (33 mer) (SEQ ID NO 102)
5' CACTACGGP..TCCTTATGAAGTCCACCATACTCT 3'.
The DNA fragment of 1221 bp obtained by
digesting the PCR product with BamIll is ligated with a
fragment of 5678 bp resulting from the digestion of
pLF999 (Example 2) with BamHI in order to generate the
plasmid pLF100S having a size of about 6899 bp.
The SIV H3N2 NA gene thus modified (intron 11
of the rabbit (3-globin gene, tPA, A[TM+Cter)} encodes a
protein of 431 amino acids.
Example 13: Plasmid encoding bovine GM-CSF
The cDNA of the bovine GM-CSF gene is
synthesized from the cellular RNA of bovine blood
mononucleated cells with the aid of the primer LF065
and amplified by a PCR reaction with the aid of the
following oligonucleotide pair:
LF054 (36 mer) (SEQ ID NO 103)
5' CATATCGTCGACATGTGGCTGCAGAACCTGCTTCTC 3' and
LF055 (34 mer) (SEQ ID NO 104)
5' CATGACCAGATCTTCACTTCTGGGCTGGTTCCCA 3'.
The DNA fragment of 437 bp obtained by
digesting the PCR product with Sall and BglII is
ligated with a fragment of 4860 bp resulting from the
CA 02398229 2010-01-20
30754-30
64
digestion of pVR1012 (Example 2) with Sall and BglII in
order to generate the plasmid pLF1032 (about 5297 bp).
The bovine GM-CSF gene encodes a protein. of 143 amino
acids.
Example 14: Plasmid encoding porcine GM-CSF
The cDNA of the porcine GM-CSF gene is
synthesized from the cellular RNA of porcine blood
mononucleated cells with the aid of the primer LF067
and amplified by a PCR. reaction with the aid of the
following oligonucleotide pair:
LF056 (36 mer) (SEQ ID NO 105)
5' CATATCGTCGACATGTGGCTGCAGAACCTGCTTCTC 3' and
LF057 (37 mer) (SEQ ID NO 106)
5' CATGACC AGATCTTCACTTCTGGGCTGGTTCCCAGCA 3 ' .
The DNA fragment of 440 bp obtained by
digesting the PCR product with Sall and Bg1II is
ligated with a fragment: of 4860 bp resulting from the
digestion of pVI~1012 (Example 2) with Sall and BglII in
order to generate the plasmid pLF1033 (about 5300 bp).
The porcine GM-CSF gene encodes a protein of 144 amino
acids.
Example 15: Formulation of the vaccinal plasmids
The DNA solution containing one or more
plasmids according to Examples 3 to 14 is concentrated
by ethanolic precipitation as described in Sambrook et
aI. (1989). The DNA. pellet is taken up in a 0.9% NaCl
solution so as to obtain a concentration of 1 mg/ml. A
0.75 x2M DMRIE-DOPE solution is prepared by taking up a
lyophilisate of DMRIE-DOPE with an appropriate volume
of sterile H20.
The formation of the plasmid DNA-lipid
complexes is achieved by diluting, in equal parts, the
0.75 rr~1 DMRIE-DOPE solution with the DNA solution at
mg/ml in 0.9% NaCl. The DNA solution is gradually
introduced, with the aid of a seamed 26G needle, along
the wall of the vial containing the cationic lipid
solution so as to avoid the formation of foam. Gentle
CA 02398229 2010-01-20
30754-30
shaking is carried out as soon as the two solutions
have been mixed. A composition comprising 0.375 mM of
DMRIE-DOPE and 500 }.;.g/ml of plasmid is finally
obtained.
it is desirable for all the solutions used to
be at room temperature for all the operations described
above. The DNA/DMRIE-DOPE complex formation is allowed
to take place at room temperature for 30 minutes before
immunizing the animals.
Example 16: Immunization of bovines against BHV-1
12 bovines are randomized into 3 groups of 4 S.
Group 1 constitutes the control animal group.
A mixture of vaccinal plasmids pPB281 (encoding
B :V-1 gB in a A [TM-Ctei: ] form, Example 3.1.2), pPB292
(encoding BHV-1 gC in a O[TM-Cterl form, Example 3.2.2)
and pPB284 (encoding BNV-1 gD in a A[TM-Cterl form,
Example 3.3.2) is administered to the animals of Group
2.
The same mixture as that in Group 2, but
formulated with DMRIE-DOPE as is described in Example
15, is administered to the animals of Group 3.
An injection of 10 ml, by the intramuscular
route, is performed on. each bovine with the aid of
syringes equipped with needle, and is repeated 21 days
later. The total mass of each plasmid used during each
immunization is 1500 )3.g.
Persons skilled in the art possess the
necessary competence to adjust the volume or the
concentration according to the plasmid dose required.
Monitoring of the serological response induced
by the two mixtures of vaccine plasmids expressing the
BHV-1 gB, gC and gD antigens is carried out over a
period of 35 days after the first vaccination.
The results are presented in the table which
follows:
CA 02398229 2010-01-20
30754-30
66
Plasmids Formulation Artige:ls Dose SN at D28 SN at D35
control --- --- --- 0.2 +/- 0.0 0.2 +/-0.0
pPB281 --- gB d[TM-Cter1 1500 pg 1.0 +!- 0.5 1.2 +/- 0.8
pPB292 gC A[TM-Cter] 1500 pg
pPB294 gD A((TM-Cter1 1500 ug
pPB281 DMRIE-DOPE gB A[TM-Cter] 1500 pg 2.1 +/- 0.6 2.7 +/- 0.6
pP5292 gC A[TM-Cter] 1500 ug
pPB294 gD i[TM-Cter] 1500 ug
Example 17: Immunization of pigs against PRV
15 pigs, about 7 weeks old, are randomized into
3 groups of 5 animals.
Group 1 constitutes the control animal group.
A mixture of vaccinal plasmids pNS009 (encoding
PRV gB in a tPA A[TM-Cter] form, Example 8.1.3), pNS012
(encoding PRV gC in a tPA A[TM-Cter] form, Example
8.2.3) and pPB238 (encoding PRV gD in a tPA A[TM-Cter]
form, Example 8.3.3) is administered to the animals of
Group 2.
The same mixture as that in Group 3 but
formulated with DMRIE-DOPE as is described in Example
15 is administered to the animals of Group 4 so as to
obtain a final DM-2IE-DOPE concentration of 0.0535 msg.
350 g of each plasmid necessary for these
vaccination protocols are mixed in a final volume of
14 ml.
An injection of 2 ml, by the intramuscular
route, is performed with the aid of syringes equipped
with needle on each pig, and is repeated 21 days later.
The pigs are challenged at D35 by nasal
administration of 2 mi of a solution of PRV strain NIA3
challenge virus in an amount of 1 ml per nostril and
having a titre of 10 '.76 CCID50 per ml.
wl
CA 02398229 2010-01-20
30754-30
67
Monitoring of the weight (in kg) of each animal
is carried out over a period of 42 days after the first
vaccination.
AG7 (in percentage) corresponds to the
difference between the mean percentages of weight
growth of a group over a period of 7 days starting on
the day of the challenge and expressed relative to one
dav.
The results are presented in the table which
follows:
CA 02398229 2010-01-20
30754-30
68
a N
Ls
o t` rn
r1 ~r c~
1 I
+ + +
N
Q O --I N
C
N l0 l0
O i1 N Cl N
z
r N CO !I1
I) I i I
In + + +
r''1 M M CO
L[l In M
N N N
tQ
Q bll Gl ~ZS
t!?
O O O O O O O
In LI) In In Lfl In
M M M M M M
CL, G 04 a w 0
1) 11 SJ J..) J-) 41
N a) 4) a) a) Cl)
4.1 u y ii Li L.i
1 U U U U U U
1 F E+ E~ E-1 E- E-
C
a a a a a a
W U A oa U 0
C
o
0
~ I 1
I
.y t H
0
o
~ i
G\ N CO 0) N CO
0 --4 C) -4 C) e--1 M
~t
u O O N C) O N
MMCQMMM
O 0' 04 R 04 04 04
04
CA 02398229 2010-01-20
30754-30
69
it should be clearly understood that the invention
defined by the appended claims is not limited to the
specific embodiments indicated in the description
above, but encompasses the variants which depart
neither from the scope nor the spirit of the present
i nvent i or_.