Note: Descriptions are shown in the official language in which they were submitted.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 1
THE IDENTIFICATION AND USE OF EFFECTORS AND ALLOSTERIC
MOLECULES FOR THE ALTERATION OF GENE EXPRESSION
Field of the Invention
The present invention relates to the use of identified effectors to alter the
catalytic activity of polynucleotides and to methods of use of these
components in
the alteration of gene expression.
Background of the Invention
The following is a brief description of enzymatic nucleic acid molecules.
This summary is not meant to be all-inclusive but is provided only for a
better
understanding of the invention that follows. This summary is not an admission
that all of the work described below is prior art to the claimed invention.
Enzymatic nucleic acid molecules (e.g., ribozymes) are nucleic acids
capable of catalyzing one or more of a variety of reactions, including the
ability to
2 0 cleave, ligate or splice either themselves or other separate nucleic acid
molecules
in a nucleotide base sequence-specific manner. In general, enzymatic nucleic
acids act by first binding to a target nucleic acid. Such binding occurs
through a
target-binding portion of the enzymatic nucleic acid molecule which is in
close
proximity to the enzymatic portion of the molecule that acts, for example, to
2 5 cleave the target. Thus, the enzymatic nucleic acid first recognizes and
then binds
a target through complementary base-pairing, and once bound to the correct
site,
acts enzymatically upon the target. In one use, the enzymatic activity may
involve a cleavage reaction. Strategic cleavage of a target RNA, for example,
will
destroy that RNA's ability to direct synthesis of an encoded protein. After an
3 0 enzymatic nucleic acid has bound and cleaved its target, it is released
from that
molecule to search for another target and can repeatedly bind and cleave new
targets. If the nucleic acid is cis-acting (e.g., self-cleaving, self-
interacting,
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 2 -
autolytic), then the activity is eliminated with the destruction of the
nucleic acid.
If the nucleic acid is trans-acting (cleaves or interacts with a another
nucleic
molecule or sequence), then the activity need not be eliminated with a single
reaction.
The use of ribozymes has been proposed to treat diseases or genetic
disorders by cleaving a target RNA, such as a viral RNA or a messenger RNA
transcribed from a gene that should be turned off such as in cancer. These
techniques have been described as an alternative to the blockage of the RNA
transcript by the use of antisense sequences. Because of the enzymatic nature
of
certain ribozymes, a single ribozyme molecule may be used to cleave many
molecules of target RNA, and therefore, therapeutic activity is achieved with
relatively lower concentrations of material than required in an antisense
treatment.
Because of their sequence specificity, trans-cleaving enzymatic nucleic
acid molecules are studied and described as therapeutic agents to treat human
disease. The enzymatic nucleic acid molecules can be designed or selected to
cleave specific RNA targets within the background of cellular RNA. Such a
cleavage event renders the mRNA nonfunctional and abrogates protein production
from that RNA. In this manner, the synthesis of a protein associated with a
disease state can be selectively inhibited. While there have been several
2 0 descriptions of the use of ribozymes as therapeutic agents (Cotton,
TIBTECH,
8:174-178, 1990; Usman & McSwiggen, Anrz. Rep. Med. Chem. 30:285-294,
1995; Couture and Stinchcomb, TIG, 12(12):510-515, 1996; Christoffersen and
Marr, J. Med. Chem. 38, 2023-2037, 1995; Gibson and Shillitoe, Molecular
Biotechnology, 7:125-137, 1997; Persidis, Nature Biotechnology, 15:921-922,
2 5 1997; and Jaeger, Current Opinion in Structural Biology, 7:324-335, 1997)
there
have been relatively few studies of the use of traps- or cis-acting enzymatic
nucleic acid molecules to alter gene expression (Chuat and Galibert,
Biochemical
and Biophysical Research Communications, 162(3):1025-1029, 1989; Innovir
U.S. Patent No. 5,741,679) or the control of gene expression by means of
3 0 translation inhibition using small molecule RNA interactions (Werstuck and
Green, Science, 282:296-298, 1998).
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 3 -
Researchers have demonstrated the remarkable diversity of catalytic
functions which RNA molecules can perform besides the cleavage of other RNA
molecules. See for example, Robertson and Joyce, Nature 344:467, 1990;
Ellington and Szostak, Nature 346:818, 1990; Piccirilli, et al., Science
256:1420,
1992; Noller, et al., Science 256:1416, 1992; Ellington and Szostak, Nature
355:850, 1992; Bock, et al., Nature 355:564, 1992; Beaudry and Joyce, Science
257:635, 1992; and Oliphent, et al., Mol. Cell. Biol. 9:2944, 1989. RNA
molecules with a given function, e.g., catalytic or ligand-binding, can be
selected
from a complex mixture of random molecules in what has been referred to as "in
vitro genetics" (Szostak, TIBS 19:89, 1992) or "in vitro evolution". In brief,
a
large pool of RNA molecules bearing random and defined sequences is
synthesized and that complex mixture, for example, approximately 10'5
individual
sequences, is subjected to a selection and enrichment process. For example, by
repeated cycles of affinity chromatography and polymerase chain reaction (PCR)
amplification of the molecules bound to a ligand on the affinity column,
Ellington
and Szostak (1990) estimated that 1 in 10'° RNA molecules folded in
such a way
as to bind a given ligand. DNA molecules with such ligand-binding behavior
have been isolated (Ellington and Szostak, 1992, supra; Bock et al., 1992,
supra).
2 0 The control or regulation of gene expression is a highly desired objective
in the fields of protein production, diagnostics, transgenics, cell therapy
and gene
therapy. A variety of expression control systems have been described as means
to
transcriptionally control the expression of a transgene in a recipient host
cell.
Control means or gene switches include, but are not limited to, the following
2 5 systems.
Rapalogs may be used to dimerize chimeric proteins which contain a small
molecule-binding domain and a domain capable of initiating a biological
process,
such as a DNA-binding protein or transcriptional activation protein (as
described
in WO 9641865 (PCT/US96/099486); WO 9731898 (PCT/LTS97/03137) and WO
3 0 9731899 (PCT/LTS95/03157)). The dimerization of the proteins can be used
to
initiate transcription of the transgene.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 4 -
An alternative regulation technology uses a method of storing proteins,
expressed from the gene of interest, inside the cell as an aggregate or
cluster. The
gene of interest is expressed as a fusion protein that includes a conditional
aggregation domain which results in the retention of the aggregated protein in
the
endoplasmic reticulum. The stored proteins are stable and inactive inside the
cell.
The proteins can be released, however, by administering a drug (e.g., small
molecule ligand) that removes the conditional aggregation domain and thereby
specifically breaks apart the aggregates or clusters so that the proteins may
be
secreted from the cell. See Science 287:816-817 and 826-830, 2000.
Mifepristone (RU486) is used as a progesterone antagonist. The binding
of a modified progesterone receptor ligand-binding domain to the progesterone
antagonist activates transcription by forming a dimer of two transcription
factors
which then pass into the nucleus to bind DNA. The ligand binding domain is
modified to eliminate the ability of the receptor to bind to the natural
ligand. The
modified steroid hormone receptor system is further described in U.S. Patent
No. 5,364,791; WO 9640911 and WO 9710337.
Yet another control system uses ecdysone (a fruit fly steroid hormone)
which binds to and activates an ecdysone receptor (cytoplasmic receptor). The
receptor then translocates to the nucleus to bind a specific DNA response
element
2 0 (promoter from ecdysone-responsive gene). The ecdysone receptor includes a
transactivation domain/DNA-binding domain/ligand-binding domain to initiate
transcription. The ecdysone system is further described in U.S. Patent
No. 5,514,578; WO 9738117; WO 9637609 and WO 9303162.
Another control means uses a positive tetracycline-controllable
2 5 transactivator. This system involves a mutated tet repressor protein DNA-
binding
domain (mutated tet R - 4 amino acid changes which resulted in a reverse
tetracycline-regulated transactivator protein, i.e., it binds to a tet
operator in the
presence of tetracycline) linked to a polypeptide which activates
transcription.
Such systems are described in U.S. Patent Numbers 5,464,758; 5,650,298 and
3 0 5,654,168.
CA 02401654 2002-08-28
WO 01/64956 PCT/US01/06615
- 5 -
Knockout and transgenic animals are well known to those skilled in the
art. Swanson et al., Annu. Rep. Med. Chem., 29:265-274, 1994; Fassler et al.,
Int.
Arch. Allergy Immunol., 106:323-334, 1995; Polites, H. G., Int. J. Exp.
Pathol.,
77(6):257-262, 1996; Harris and Foord, Pharmacogenomics, 1(4):433-443, 2000.
A knockout animal has been genetically altered to disrupt the expression of a
targeted gene, resulting in the elimination of the target gene product.
Knockout
animals are widely used to demonstrate the function of a protein of interest.
In
particular, the elimination of the expression of the targeted gene in the
knockout
animal can indicate the effect of inhibiting the protein product of the gene.
One
limitation with the technology is that targeted gene disruption can cause
developmental defects which, although not indicative of the effect of target
gene
inhibition in an adult animal, result in embryonic lethality. Thus, certain
disruptions of gene function cannot be studied in a viable animal. Another
limitation of current knockout technology is the effect of developmental
compensation on targeted gene disruption. In the course of development, other
related gene products may compensate for the lost function of the disrupted
gene
and, thus, obscure its function in the adult animal. These well known
limitations
of the technology can significantly restrict the utility of targeted gene
disruption in
knockout animals. In the case of transgenic animals, an additional copy of the
2 0 gene of interest is introduced into the organism and results in the over
expression
of the gene product. Protein over expression during development can cause
defects or lead to compensation for or inhibition of over expression. These
problems can obscure the effects of transgene over expression and limit the
ability
to interpret the biological effects of target gene over expression. The
ability to
2 5 create a conditional knockout animal is particularly important and
relevant to
overcome these limitations.
The references cited above are distinct from the presently claimed
invention since they do not disclose and/or contemplate the identification of
3 0 effectors and their use in the control of gene expression as provided by
the instant
invention. Nor do they involve allosteric control modules of the present
invention.
CA 02401654 2002-08-28
WO 01/64956 PCT/USOI/06615
- 6 -
DESCRIPTION OF THE DRAWINGS
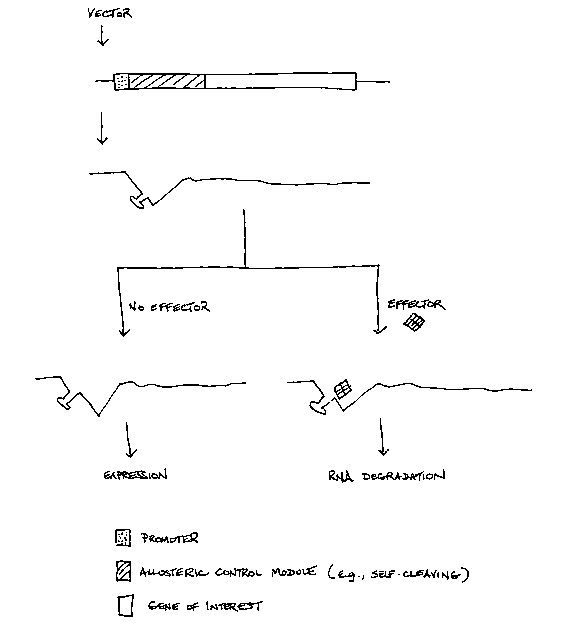
Figure I is a diagrammatic representation of the present invention
involving an allosteric control module containing a self-cleaving RNA domain,
the activity of which is inhibited by interacting with an effector, thereby
resulting
in the translation of the mRNA. It will be appreciated by those skilled in the
art
that the allosteric control module may be placed 5' or 3' of the gene of
interest.
Figure 2 is a diagrammatic representation of the present invention
involving an allosteric control module containing a self-cleaving RNA domain,
the activity of which is initiated by interacting with an effector. The
cleaving of
the allosteric control module results in the degradation of the mRNA and
inhibition of translation. In the absence of the effector, translation occurs.
Figure 3 is a diagrammatic representation involving an allosteric control
module containing a self-splicing RNA domain, the activity of which is
initiated
by interacting with an effector. In this embodiment of the present invention,
the
2 0 presence of effector provides for the activation of the allosteric control
module
such that splicing of the precursor mRNA occurs to result in the formation of
a
translatable mRNA.
Figure 4 is a diagrammatic representation involving an allosteric control
2 5 module containing a self-splicing RNA domain in an engineered intron. In
the
depicted embodiment, the presence of effector provides for the activation of
the
allosteric control module such that splicing of the precursor mRNA occurs to
result in the formation of a translatable open reading frame.
3 0 Figure 5 is a diagrammatic representation of the present invention
involving an allosteric control module containing a self-splicing RNA domain.
In
CA 02401654 2002-08-28
WO 01!64956 PCT/USO1/06615
this embodiment, the activity of the allosteric control module is inhibited by
interacting with an effector.
Figure 6 is a diagrammatic representation of the present invention
involving an allosteric control module containing a self-splicing RNA domain
inserted in a region of an intron necessary for spliceosome assembly. In this
embodiment, the activity of the allosteric control module is inhibited in the
presence of effector, resulting in the expression of the gene of interest.
Figure 7 depicts the secondary structure of RNA template 1 comprising a
stem-loop structure of the theophylline-binding aptamer which is connected
through a contiguous stretch of randomized nucleotides to a hammerhead
ribozyme. The molecular structure of theophylline is also shown (SEQ ID NO:
6).
Figure 8 depicts the secondary structure of the RNA sequence TA-50,
selected after seven cycles.
Figure 9 depicts the self-cleaving ability of TA-50 in the presence of
theophylline.
2 0 SUMMARY OF THE INVENTION
The means to identify a useful effector molecule is described as is the use
of the identified effector to evolve a cognate aptamer. The construction of an
allosteric control module is described in which a catalytic RNA forms a part
of or
2 5 is linked to the effector-binding RNA domain or aptamer, thereby placing
the
activity of the catalytic RNA under the control of the effector and requiring
the
presence of the effector for activation or inactivation. RNA molecules are
constructed in which at least one portion is capable of binding an effector
and
another portion is a catalytic RNA. The present invention involves both the
3 0 evolution of RNA sequences which bind the effector and a selection process
in
which the allosteric control modules are identified by their catalytic
function in
the presence and absence of the effector. In this manner, regulatable
catalytic
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
_ g _
RNAs may be selected for use in cleaving, splicing or ligating a target RNA in
the
presence of an effector, or in cleaving, splicing or ligating a target RNA in
the
absence of an effector.
These methods of effector selection and the construction of allosteric
control modules are useful in altering the expression of a target RNA molecule
in
a controlled fashion. It is particularly useful when the target RNA molecule
is
formed in or delivered to the cell in combination with the allosteric control
module.
DETAILED DESCRIPTION OF THE INVENTION
Disadvantages associated with previously known control constructs and
their uses include potential toxicities in in vivo use, including the
transcriptional
activation or repression of endogenous genes, the activation or inhibition of
proteins or cellular processes that the small molecule entity normally
regulates, or
the induction of an immune response towards the foreign proteinaceous gene
products of the control system. In addition, there may be size constraints
upon the
2 0 control coding sequences that can be delivered together with the gene of
interest
by means of certain viral and non-viral vectors. Thus, there is still a need
to
develop alternative methods and materials for the controlled expression of a
gene
of interest. Furthermore, the development of a means of controlling gene
expression wherein that means may also be varied by the individual components
2 5 used would contribute significantly to any strategy of gene therapy as
well as to
the production of therapeutic proteins. The present invention uniquely solves
these problems.
The expression of a specific gene can be altered at any step in the process
of producing an active protein. The modulation of total protein activity may
occur
3 0 via transcriptional, transcript-processing, translational or post-
translational
mechanisms. Transcription may be modulated by altering the rate of
transcriptional initiation or the progression of RNA polymerase. Transcript-
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 9 -
processing may be influenced by circumstances such as the pattern of RNA
splicing, the rate of mRNA transport to the cytoplasm, or mRNA stability. The
present invention primarily concerns the identification and use of effector
molecules and the creation of allosteric control module molecules which act
together to alter the expression of a target gene (for example, altering the
in vivo
concentration of a target protein by altering RNA processing.) The present
invention provides an identification process for suitable effectors for use in
the
control of gene expression, which process has not been previously described.
Descriptions of various effector-controlled RNA molecules are available.
See for example, Chuat and Galibert, Biochemical and Biophysical Research
Communications, 162(3):1025-1029, 1989; Ellington et al., Nature 355:850-852,
1992; Porta et al., BiolTechnology 13:161-164, 1995; and Soukup and Breaker,
Proc. Natl. Acad. Sci. USA, 96:3584-3589, 1999. The identification and in
vitro
selection processes, however, have been designed for and directed to the use
of
ribozymes to control the expression of deleterious genes (such as HIV, HCV,
CMV, VEGF and TNF) or for biosensors to detect specific ligands.
Aminoglycoside antibiotics are among the most studied molecules which react
with RNAs (von Ahsen et al., Nature 353:368-370, 1991; von Ahsen et al., J.
Mol. Biol. 226:935-941, 1992; Murray and Arnold, Biochem. J. 317:855-860,
2 0 1996; Werstuck and Green, Science 282:296-298, 1998). Such previously
described molecules, however, react directly with naturally occurnng
ribozymes.
The present invention describes the novel identification of effectors which
are
used to evolve aptamers for use in expression regulation constructs.
Alternatively, Werstuck and Green (Science, 282:296-298, 1998)
2 5 described the use of effectors and aptamers to regulate translation. In
that
research they described the use of multiple aptamers and dyes as effectors to
repress the translation of a reporter gene. The method did not involve the use
of a
catalytic RNA or allosteric control module of the present invention. While
other
disclosures relate to the use of regulated gene expression using a ligand,
ligand
3 0 binding sequence and catalytic RNA (Innovir Laboratories Inc.; expression
control systems and nucleic acid constructs are described in U.S. Patent
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 10 -
No. 5,741,679 and U.S. Patent No. 5,834,186), the descriptions have not
included
a process for the selection of suitable effectors.
The present inventors are not aware of any prior reports of a systematic
process for the identification of effectors, their use to select and evolve
non-
naturally occurnng aptamers for the construction of allosteric control
modules,
and the combined use of effector and allosteric control module for the
alteration
of gene expression. Such a process is an object of the present invention. The
procedures described herein also serve to produce molecules not previously
envisioned in the regulation of gene expression. The present invention
provides a
means for determining whether a "non-therapeutic" molecule can specifically
modulate the expression of a gene of interest, and the ultimate clinical use
of such
an effector molecule provides an advantage over the use of previous
biologicals,
or drugs which have a therapeutic function.
Methods and compositions are described for the controlled expression of
targeted RNA molecules by means of an allosteric control module. The activity
of the allosteric control module is altered through the presence, absence or
amount
of a pre-identified effector. In one embodiment, the allosteric control module
is
active in the presence of an effector, in another embodiment the allosteric
control
module is inactive in the presence of an effector.
The following definitions are used in the description of the present
invention.
An "allosteric control module" as used herein refers to a non-naturally
2 5 occurring RNA composed of at least two domains, one a receptor for a pre-
identified ligand and the other a catalytic domain. The receptor domain or
effector-binding domain may also be referred to as an aptamer, and the ligand
to
which it binds is referred to as an effector. The catalytic domain is a RNA
which
is capable of interacting with a target RNA. Such activity may include
cleaving,
3 0 splicing, or ligating the target RNA.
The allosteric regulation of gene expression refers to the alteration of the
expression of a gene, preferably a transgene in gene therapy, by means of the
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 11 -
interaction of the effector and allosteric control module. The effectors of
the
present invention bind to a domain of the allosteric control module and alter
the
activity of the module's catalytic domain. Without being bound by any
particular
theory, it is believed that the influence of the effector upon the activity of
the
catalytic domain is brought about by a change in the conformation of the
module
which change is induced by the binding of the effector and the effector-
binding
domain or aptamer. The combination of the aptamer and catalytic RNA domain is
selected such that the conformational change may either preclude formation of
an
active catalytic domain or induce the formation of an active catalytic domain.
For
example, the effector-induced conformational change may be selected to cause
the
inhibition of or a reduction in the activity of the catalytic domain due to
steric
interference between the aptamer and the catalytic domain tertiary structures.
Alternatively, the effector-induced conformational change may be selected to
cause the initiation of or an increase in the activity of the catalytic
domain.
Therefore, the term "activation" or "activated" is used herein to refer to
either the
initiation of or an increase in or enhancement of catalytic activity.
It will be appreciated by those skilled in the art that the domains of the
allosteric control module may be non-overlapping or partially overlapping such
that one or more domains are encoded in part by the same polynucleotide. Thus,
2 0 the domains are primarily distinguished by their function rather than by
their
sequence. In addition, the domains may be separately prepared and then joined
to
form the allosteric control module, or the allosteric control module may be
prepared as a single polynucleotide having both aptamer and catalytic domains.
In contrast to previously described ribozymes and ribozyme-like
2 5 molecules, the allosteric control module of the present invention does not
have
true "enzymatic" activity. In a preferred construction of the DNA of the
present
invention, the allosteric control module acts on an intramolecular basis and
is only
required to provide one reaction rather than multiple reactions with multiple
RNA
or DNA molecules. In addition, the allosteric regulation of the catalytic
function
3 0 differs from that of inhibitors which block the catalytic sites of
structures such as
a ribozyme. In the present invention, the effector binds to an aptamer which
is a
site located apart from the active site, and its influence on the activity of
the
CA 02401654 2002-08-28
WO 01/64956 PCTNSO1/06615
- 12 -
polynucleotide is thought to be brought about by changes in polynucleotide
conformation which result from the interaction of the aptamer and effector. It
will
also be appreciated that the position of the allosteric control module in the
DNA
constructs of the present invention may vary. All that is required is that the
allosteric control module be positioned such that altering the activity of the
allosteric control module by means of an effector will result in the
alteration of
expression of the gene of interest.
"Allosteric" or "allostery" as used herein refers to the alteration of the
activity of a molecule by the interaction of an effector with that molecule.
The
effector interacts a domain distinct from the molecule's catalytic domain and
that
interaction causes a change in the activity of the molecule. The typical
activity
involved in the constructs of the present invention may be referred to as
catalytic
activity, which is further described herein.
An "aptamer" or "effector-binding domain" as used herein refers to a
polynucleotide that binds to an effector. The polynucleotide typically
comprises
at least 20 nucleotides and may comprise at least 300 nucleotides. The
aptamers
of the present invention may comprise naturally occurring polynucleotides,
which
are not chemically modified. Preferably, the aptamer comprises a synthetic or
non-naturally occurring polynucleotide. In addition, the aptamer is preferably
a
2 0 ribonucleic acid (RNA) selected by in vitro evolution to interact with a
previously
identified effector. The in vitro evolution of aptamers begins with a pool of
RNA
molecules created by chemical and/or enzymatic synthesis. Typically, the
desired
aptamer is selected based upon its ability to interact (e.g., recognize and
bind)
with an effector. Thus, in a preferred embodiment, the effector is a
predetermined
2 5 molecule which is used to select and further evolve a suitable aptamer.
Alternatively, the aptamer may be constructed and libraries of molecules
screened
to identify and select a suitable effector.
For use in the control of gene expression in gene therapy techniques, the
aptamer is not an isolated and purified chemical entity. Instead, the aptamer
is
3 0 encoded by a DNA which is delivered to a cell, and the aptamer becomes a
portion of the mRNA transcribed from that DNA in the host cell. In addition,
the
effectors of the present invention do not modify a biological activity of an
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 13 -
aptamer because the aptamers of the present invention have no inherent
physiological activity in the recipient cell.
There is no set number of bases required for the interaction (e.g.,
hybridization or binding) of an aptamer to an effector. In general the aptamer
will
contain 20 to 300 nucleotides, selected as described herein for binding to a
specific effector. The small size of the molecule (typically 200 nucleotides
or
less, preferably between 20-30 nucleotides in length) is advantageous for cell
delivery as compared with conventional expression regulation constructs which
are much larger in size. Although referred to herein as a "random" sequence,
it is
understood that the RNA is random only as originally used in the evolution
process. The product of the evolution process, i.e., the aptamer(s), is not a
random sequence. It is a specific sequence which binds with a high degree of
affinity and specificity to a defined effector.
The specific aptamers of the allosteric control modules described herein
are not limiting in the invention. Those skilled in the art will recognize
that all
that is important in an aptamer of the present invention is that it
selectively and
specifically interact with a suitable effector, and that it have the ability
to alter the
catalytic activity of the allosteric control module when the aptamer has
interacted
with the effector. Multiple aptamers (as well as multiple allosteric control
2 0 modules) may also be used so that multiple effectors and even multiple
different
effectors may be used to react with allosteric control modules and thereby
alter
gene expression by altering the precursor mRNA.
The term "domain" as used herein refers to a polynucleotide which
provides a selected activity or function to the allosteric control modules of
the
2 5 present invention.
An "effector" as used herein refers to a molecule which interacts with an
aptamer. Binding may be the result of the interaction and may include, but is
not
limited to, hydrogen binding, hydrophobic interactions, intercollations, etc.
Suitable molecules for use as effectors of the present invention include, but
are
3 0 not limited to, organic or inorganic molecules, peptides, polypeptides,
proteins,
oligonucleotides, polynucleotides, nucleic acids, naturally occurnng
metabolites
and biological effectors, lipids, carbohydrates (polysaccharides, sugar),
fatty
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 14 -
acids, and polymers. The preferred effector molecules of the present invention
are
distinguished from those described in the art in that the present effectors
have
either no pharmalogical effect or are used at concentrations wherein a
pharmalogical effect either is not observed or is negligible.
"Suitability" of the effector for use with the allosteric control module may
include the following characteristics. (1) The effector has little or no
pharmalogical effect in the dosage range used in altering gene expression or
has a
negligible pharmalogical effect. This refers to either the lack of any
alteration in
function of the structure or process of a cell in which the effector acts
(other than
the allosteric control module) or the occurrence of a insignificant alteration
in
function of the structure or process of a cell in which the effector acts
(other than
the allosteric control module). In other words, if the effector may be used as
a
pharmaceutical agent that has an effect on a structure or process other than
the
allosteric control module, then that effect does not cause any harm to the
patient
not in need of treatment by that pharmaceutical agent. (2) The effector will
undergo biodistribution to that cell or tissue which will contain the
allosteric
control module. (3) The effector has the ability to pass to subcellular
structures,
i.e., to the allosteric control module in the nucleus of cells transformed for
regulated gene expression. This may be by means of intracellular diffusion or
2 0 transport, and most preferably, by intranuclear diffusion or transport.
(4) The
effector has the ability to interact with an aptamer wherein the interaction
occurs
with high specificity and a high affinity.
Additional considerations for the identification of a suitable effector may
further include the following. (1) If the effector is a molecule which has a
dose
2 5 effect relationship, then the molecule typically is used at a daily
maximum dose
which is less than the usual daily minimum dose of the molecule when used for
an
approved indication. Preferably such an effector is used at a dosage level
which is
below 25% of the effective dose (ED25) of the molecule when used as a
pharmaceutical agent. It will be appreciated by those skilled in the art,
however,
3 0 that such a preference is based on a case-by-case evaluation of the agent.
For
example, in the case of an effector which is an antiviral agent, the effector
could
be used at any dosing range in the absence of the virus. More preferably the
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 15 -
dosage level is below the ED10 for the molecule. Most preferably, if the
effector
is a molecule which has a dose-effect relationship, then the molecule is used
as an
effector at a dose which is below the lowest effective dose (threshold dose)
of the
molecule when used as a pharmaceutical agent. (2) The use of a pharmaceutical
agent related to the effector is not contraindicated in patients having the
condition
to be treated by the regulated gene. (3) There are no significant side effects
or
adverse reactions produced by the effector itself, and no significant adverse
reaction is due to an overdose of the effector itself. Typically, non-
significant
effects would include, for example, events such as headache, dizziness,
lightheadedness, sedation, nausea, vomiting, rash, constipation, diarrhea,
abdominal pain, euphoria, dysphoria, fatigue, arthralgia which can be
controlled
by dose adjustment or other common intervention or which occurs in less than
five percent of the population receiving the effector. (4) There are no known
contraindications in pregnancy, heart disease, or hypersensitivity to related
agents.
In certain preferred embodiments of the present invention, the effector is a
small molecule. The nature of the effector can be chosen to be exogenously
supplied, such as some non-toxic molecule or drug which readily enters at
least
the cells containing the targeted RNA. Alternatively, an entirely endogenous
system can be used in which the controlling effector is some endogenous
2 0 metabolite or macromolecule which is directly or indirectly related to the
pathology to be corrected or the gene to be expressed or the molecule to be
produced. For example, a protein encoded by the target RNA could be the
effector. The construct may be designed such that the activity of the
allosteric
control module is dependent on binding to the expressed protein and as the
level
2 5 of protein increases the activity of the module increases to cause a
decrease in
expression. As the level of target RNA falls due to alteration (e.g.,
cleavage) by
the allosteric control module, the concentration of the protein (as ligand or
effector) also falls. When the concentration falls below that at which the
regulatable RNA molecules are all occupied, the rate of alteration will begin
to
3 0 fall off. By selecting for differing aptamer-effector affinities, the
appropriate
level of regulation of allosteric-mediated destruction of the target RNA can
be
achieved for any given situation.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 16 -
The "pharmalogical activity" of a molecule is used to refer to the activity
of that molecule as a drug or medication.
A "database" as used herein refers to any compilation of information on
potential effectors, such as small molecules, containing information
concerning
the suitability of the effector for use in controlling gene expression.
The "catalytic activity" of the allosteric control module refers to the
activity of the "catalytic domain" or "catalytic sequence" which is a nucleic
acid
which acts on a target nucleic acid in a desirable manner. Examples of
possible
actions include, but are not limited to binding of the target, reacting with
the target
in a way which modifies/alters the target as by cleavage, splicing or ligation
or the
functional activity of the target, or facilitating the reaction between the
target and
another molecule. In preferred embodiments of the present invention, the
catalytic activity is a self-cleaving activity, a ligase activity, or a
splicing activity.
Such activities are often associated with ribozymes. Ribozymes, including
ribozyme-like molecules and portions of such molecules, may be used to form
the
catalytic domain of the allosteric control module of the present invention. It
will
be appreciated by those skilled in the art that it is primarily the
catalytically active
portion of the naturally occurring ribozyme or non-naturally occurring
ribozyme-
like molecules that is used in the allosteric control modules of the present
2 0 invention, but that additional domains also may be used. For example, if
the
allosteric control module is self-cleaving, then in addition to an aptamer and
a
catalytic domain, the module will further include a substrate domain. It will
also
be appreciated that for the purposes of the present invention, the "catalytic
activity" of the allosteric control module merely refers to the alteration or
2 5 modification of an interaction with a target RNA. The catalytic domain may
be
designed such that it may or may not be consumed in the process, and
therefore,
the domain is not required to be a true "catalyst".
Ribozymes which may be useful to the present invention in the preparation
of catalytic domains include, but are not limited to, molecules in the classes
of
3 0 hammerhead, axehead, hairpin, hepatitis delta virus, neurospora, self-
splicing
introns (group I or group II), ligases, phosphatases and polymerases. Each
class
of ribozyme cleaves a different sequence of nucleotides using distinct
mechanisms
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 17 -
of action. Moreover, each class is further distinguished based on how many
nucleotide bases are essential for catalytic activity and to the extent the
intended
target sequence and the ribozyme can be manipulated to alter specificity.
Thus,
naturally occurring ribozymes may be used as a basis to create non-naturally
occurnng (totally unique or modified) ribozyme-like structures. For
simplicity,
the term ribozyme may be used in place of catalytic domain in the present
invention, but it will be appreciated that the catalytic domain need not
contain all
of a naturally occurnng ribozyme structure to be used in the present
invention,
and it may involve a synthetic ribozyme-like structure.
The term "RNA ligase" or "ligase domain" as used herein refers to a
polynucleotide capable of catalyzing (altering the occurrence, velocity and/or
rate
of) a ligation reaction (the joining together of two polynucleotides) in a
nucleotide
base sequence-specific manner. As with other catalytic domains, the ligase
domain may act in cis, i.e., as an intramolecular reaction, or in traps, i.e.,
as an
intermolecular reaction when the gene of interest, the expression of which is
to be
regulated, is provided to the cell as a separate DNA construct. The RNA ligase
may have complementarity in a substrate binding region to a specified target
polynucleotide, and also has a catalytic activity to specifically join RNA in
that
target. By "complementarity" is meant a nucleic acid that can base pair (e.g.,
2 0 form hydrogen bond(s)) with other RNA by either traditional Watson-Crick
or
other non-traditional types (for example, Hoogsteen type) of base-paired
interactions. This complementarity functions to provide sufficient
hybridization
of the ligase domain to the target RNA to allow the reaction to occur.
Complementarity of 100% is preferred, but complementarity as low as 50-75%
2 5 may also be useful. The nucleic acids may be modified at the base, sugar,
and/or
phosphate groups. The specific ligase domains of the allosteric RNA ligase
polynucleotides are not limiting in the invention, and those skilled in the
art will
recognize that all that is important in a ligase domain of this invention is
that it
has a specific substrate binding site which is complementary to one or more of
the
3 0 target polynucleotide regions, and that it include a site within or
surrounding that
substrate binding site which imparts a ligase activity to the ligase domain.
It will
be appreciated by those skilled in the art that catalytic domains having
cleaving or
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 18 -
splicing activities may also involve complementarity in a substrate binding
domain to a specified target polynucleotide in order to specifically interact
the
catalytic RNA and the desired target.
The term "gene of interest" or "desired gene" as used herein refers to a
gene providing a physiologically relevant benefit to the cell or organism,
such as
encoding a therapeutically relevant molecule for which expression is
controlled by
means of the effector and allosteric control module. For example, a
therapeutic
gene of interest is a gene that corrects or compensates for an underlying
protein
deficit or, alternately, that is capable of down-regulating a particular gene.
Moreover, a gene of interest can be a gene that mediates cell killing, for
instance,
in gene therapy for the treatment of cancer. The term "transgene" as used
herein
refers to a gene, such as the gene of interest, which is transferred to a
cell.
The term "messenger RNA" (mRNA) is used to refer to a polynucleotide
which transfers information from DNA to the protein-forming system of the
cell.
The term "precursor mRNA" or "pre-mRNA" is used to refer to a polynucleotide
which is directly transcribed from the coding DNA strand, may contain an
intron
or introns, and may or may not be capped with an inverted methylated guanosine
nucleotide.
The term "non-natural" polynucleotide as used herein refers to a
2 0 polynucleotide sequence or construct that does not occur in nature. The
preferred
allosteric polynucleotides of the present invention do not occur in nature.
The
term "isolated polynucleotide" refers to a nucleic acid molecule of the
invention
that is free from at least one contaminating nucleic acid molecule with which
it is
naturally associated, and preferably substantially free from any other
2 5 contaminating mammalian nucleic acid molecules which would interfere with
its
use in protein production or its therapeutic or diagnostic use.
The term "splice recognition region" as used herein refers to a sequence in
the precursor RNA which serves as a splice donor, splice acceptor or
spliceosome
binding site. The splice donor is typically a site at the 3' end of the exon
(located
3 0 at the 5' end of the intron to be removed), and the splice acceptor refers
to a site at
the 5' end of the adjacent exon to be ligated (the 3' end of the intron to be
removed). "Spliceosome" refers to a large, multicomponent complex of cellular
CA 02401654 2002-08-28
WO 01/64956 PCT/US01/06615
- 19 -
protein and RNA that binds to and directs the processing of the pre-RNA into
mRNA by cleavage, the removal of introns, and the ligation of exons.
The term "intron" is used to refer to a section of RNA occurring in a
transcribed portion of a gene which is included in a precursor mRNA but which
is
then excised during processing of the transcribed RNA before translation
occurs.
Therefore, intron sequences are not found in mRNA nor translated into protein.
The term "exon" is used to refer to a portion of a gene that is represented
in the transcript of the gene and that survives processing of the RNA in the
cell to
become part of a mRNA. Exons generally encode three distinct functional
regions of the RNA transcript. The first region, located at the 5' end which
is not
translated into protein, is called the 5'-untranslated region (5'-UTR). The 5'-
UTR
signals the beginning of RNA transcription and contains sequences that direct
the
mRNA to the ribosomes and cause the mRNA to be bound by ribosomes so that
protein synthesis can occur. The second region is known as an open reading
frame and contains the information that can be translated into the amino acid
sequence of the protein or function as a bioactive RNA. The third region,
located
at the 3' end, contains the signals for the termination of translation and for
the
addition of a polyadenylation tail (poly(A)) and is not translated into
protein (i.e.,
the 3'-UTR). In particular, the 3'-UTR can provide mRNA stability. The
2 0 intron/exon boundary will be that portion in a particular gene where an
intron
section connects to an exon section. The terms "TATA box" and "CAP site" are
used as they are recognized in the art.
The term "vector" is used to refer to any molecule (e.g., nucleic acid,
plasmid, virus, small molecule, liposome, Garner molecule, etc.) used to
transfer
2 5 coding information to a host cell.
The terms "control sequences" and "control elements" are used to refer
collectively to non-coding regulatory sequences including, but not limited to,
promoters, polyadenylation signals, transcription termination sequences,
upstream
regulatory domains, origins of replication, internal ribosome entry sites,
3 0 enhancers, and the like, which are operably linked to the DNA encoding a
gene of
interest to provide for the transcription and translation of the coding
sequence in a
recipient cell. Not all of these control elements need always be present so
long as
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 20 -
the sequence encoding the gene of interest is capable of being transcribed and
translated in an appropriate host cell in accordance with the expression
regulatory
means of the present invention.
A "promoter" is used to refer to a DNA sequence that directs the binding
of RNA polymerase and thereby promotes RNA synthesis. An "origin of
replication" is a sequence on a vector or host cell chromosome that renders
extragenomic elements (e.g., viruses or plasmids) capable of replicating
independently of the host cell genome. "Enhancers" are cis-acting elements of
DNA that stimulate or inhibit transcription of adjacent genes. An enhancer
that
inhibits transcription is also termed a "silencer". Enhancers differ from DNA-
binding sites for sequence-specific DNA binding proteins found only in the
promoter (which are also termed "promoter elements") in that enhancers can
function in either orientation, and over distances of up to several kilobase
pairs,
even from a position downstream of a transcribed region.
The term "operably linked" is used to refer to an arrangement of elements
wherein the elements so described are configured or assembled so as to perform
their usual function. Thus, a control sequence operably linked to a coding
sequence is capable of effecting the replication, transcription and/or
translation of
the coding sequence. For example, a coding sequence is operably linked to a
2 0 promoter when the promoter is capable of directing transcription of that
coding
sequence. The control sequence need not be contiguous with the coding
sequence, so long as it functions correctly. Thus, for example, intervening
untranslated yet transcribed sequences can be present between a promoter
sequence and the coding sequence and the promoter sequence can still be
2 5 considered "operably linked" to the coding sequence.
The term "co-element" as used herein refers to a separate molecule or a
separate domain in the precursor mRNA which interacts with or complexes with
the catalytic domains and/or aptamers of the allosteric control module and/or
the
effector to yield a catalytic complex. For example, the co-element may
complete
3 0 a missing portion of the allosteric control module so that it becomes
catalytically
active. Such co-elements include components as described in WO 9808974.
Alternative co-elements may include the bridge elements described by Soukup
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 21 -
and Breaker (Proc. Natl. Acad. Sci. USA 96:3584-3589, 1999) to construct
ligand
responsive allosteric ribozymes.
The term "gene transfer" or "gene delivery" is used to refer to methods
and/or systems for reliably inserting a particular polynucleotide (e.g., DNA)
into a
target cell. Gene transfer may take place in vivo (e.g., adeno-associated
virus
gene therapy) or ex vivo (e.g., as with extracellular modification of cells
with a
retrovirus followed by transfer or implantation of the transformed cells into
the
host). Such methods may result in the integration of the transferred genetic
material into the genome of target cells or the transferred genetic material
may
function independently of the host cell genome.
The terms "reduction", "inactivation", "inhibition", "initiation" and
"decrease" in expression as used herein means that the level of translation of
a
target mRNA is reduced below that observed in the absence of the regulation
means of the present invention. Thus, the inhibition of gene expression may
range from a decrease in the translation of the target mRNA to the complete
inactivation or inability of the transcript to be used to express the gene of
interest.
The terms "enhancement", "activation", "induction" or "increase" in
expression as used herein means that the level of translation of a target mRNA
is
increased above that observed in the absence of the regulation means of the
2 0 present invention. Thus, induction of expression may range from the
initiation of
the translation of the target mRNA to an increase in the translation of the
target
mRNA.
The phrase "specifically controlling the expression of the gene of interest"
as used herein means altering the expression of the gene of interest without
2 5 altering the expression of other genes in the cell in a way which would
cause an
adverse effect on (a) an organism containing the cell in the case where the
cell is
within the organism or (b) the growth or the culturing of the cell, in the
case
where the cell is being grown or cultured to make a product where the amount
of
product produced is associated with expression of the gene of interest.
CA 02401654 2002-08-28
WO 01/64956 PCT/USOi/06615
- 22 -
Alteration of Gene Expression
The production of a recombinant protein or peptide can be affected by the
efficiency with which DNA (or an episomal nucleic acid) is transcribed into
mRNA. Conventional control systems seek to affect the transcription event. The
production of a recombinant protein or peptide, however, can also be affected
by
the efficiency with which precursor mRNA is modified to form the mRNA which
is translated into protein. The novel constructs and methods of the present
invention advantageously provide for the regulated expression of a gene of
interest, such as a therapeutic protein, by altering the process of pre-mRNA
processing.
The allosteric control modules of the present invention contain catalytic
and effector-binding domains that are specifically selected such that the
interaction of the effector-binding domain or aptamer with an effector alters
the
activity of the allosteric control modules. For example, the interaction of
the
effector and aptamer may result in a conformational change in the allosteric
control module. Depending upon the selection of the allosteric control
modules,
the conformational change can result in either an increase or a decrease in
the
activity of the catalytic domain of the module. This in turn affects whether
or not
2 0 the precursor mRNA is appropriately modified to form a mRNA capable of
being
translated into a protein of interest. While not limiting to the present
invention, a
conformational change may be caused by the binding energy derived from the
effector-aptamer interaction which is used to shift the thermodynamic balance
between two possible confirmations of the allosteric control module. Depending
2 5 upon the format design, as described in greater detail in the different
embodiments, the allosteric control module is either activated or inactivated
by
the effector.
The present invention includes an expression regulation format involving
an allosterically activated self-cleaving RNA as the catalytic domain of the
3 0 allosteric control module. In one aspect of this embodiment, the
allosteric control
module may be encoded in a mRNA with the gene of interest. For example, the
DNA sequence is designed to encode a self-cleaving site that separates the cap
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 23 -
and message sequences. As described in further detail herein, the aptamer and
catalytic domain may be selected such that in the absence of an effector, the
catalytic domain is active and that activity results in the cleavage of the
pre-
mRNA or mRNA. Thus, the gene of interest is not expressed because the mRNA
can not be translated. In the presence of the effector, the catalytic domain
is
inhibited and thus inactive or unable to act upon the RNA substrate. As a
result,
the mRNA is not cleaved. Thus, the gene of interest is expressed because the
RNA can be appropriately processed and translated. A preferred embodiment of
the present invention involves constructs which provide gene expression in the
presence of the effector.
In an alternative embodiment involving an allosterically activated self-
cleaving RNA, the aptamer and catalytic domain may be selected such that in
the
presence of an effector, the catalytic domain is active and that activity
results in
the cleavage of the mRNA. Thus, the gene of interest is not expressed because
the mRNA can not be translated. In the absence of the effector, the catalytic
domain is inactive or unable to act upon the RNA substrate. As a result, the
mRNA is not cleaved and the gene of interest is expressed. Thus, in the
absence
of effector the gene is expressed.
In yet another embodiment the catalytic domain may include an
2 0 engineered intron which contains a self-cleaving site. The aptamer and
effector
are selected such that the domain does not cleave if effector is absent. As a
result,
the mRNA is not recognized as an active molecule and is not translated.
The present invention also includes an expression regulation format
involving an allosterically activated splicing nucleic acid as the catalytic
domain
2 5 having cleaving and ligase activities. In one aspect of this embodiment,
the
allosteric control module is inserted into an exon of the gene of interest.
This
results in the formation of a "engineered intron" (eI) as depicted in Figure
3. In
the absence of the effector, the allosteric control module is inactive, thus
leaving
the engineered intron in place and resulting in an altered open reading frame
that
3 0 leads to the inhibition or reduction of protein expression. In the
presence of the
effector, the allosteric control module is active. This activity results in
the
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 24 -
removal of the engineered intron and the ligation of the exon to form a
productive
open reading frame that leads to the activation or increase in protein
expression.
The presence of the eI in the pre-mRNA can result in the inhibition of
gene expression by a variety of mechanisms that can act individually or
jointly.
Such mechanisms include, but are not limited to 1) the eI may be designed to
contain several stop codons which would arrest translation; 2) the eI could
code
for nonsense amino acids resulting in a protein with multiple inactivating
mutations; and 3) the presence of the eI could activate the mRNA surveillance
system native to cells that would sequester or destroy the altered pre-mRNA.
It
will be further appreciated that the format may involve an engineered intron
designed such that in the presence of effector the allosteric control module
is
inactive and in the absence of the effector the module is active.
The present invention further includes an expression regulation format
involving an inhibitable allosteric self-splicing intron. In this embodiment,
the
allosteric control module is inserted into an intron in a region necessary for
spliceosome assembly such that the action of the self-splicing intron results
in the
removal of a nucleotide sequence necessary for normal splicing. This
embodiment is depicted in Figure 6. In the absence of effector, the allosteric
control module is active, thus leading to the removal of the vital splice
recognition
2 0 site. The removal of the splice recognition site results in an altered
open reading
frame that leads to the inhibition or reduction of protein expression. In the
presence of effector, the allosteric control module is inactive and, as a
result,
normal splicing occurs at the splice recognition sites, the correct open
reading
frame is formed, and the protein is expressed.
Effector Selection
A fundamental aspect of the present invention is the identification of an
effector for use with an allosteric control module to alter the expression of
a gene.
Associated with such a use is the use of the effector to evolve a cognate
aptamer
3 0 which will form a part of the allosteric control module. The present
invention
provides a novel, systematic method for identifying an effector and generating
an
interactive aptamer (or aptamers). The method involves the following steps.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 25 -
First, the desired characteristics for the effector are selected. These
desired
characteristics are selected from one or more of the following attributes
which are
useful in identifying a potential effector suitable for use in protein
expression and
gene therapy techniques:
a) has at least 1 % bioavailability;
b) is biodistributed to tissue containing an allosteric control module;
c) has the ability to pass to the nucleus of the cell;
d) exhibits either no drug interactions or manageable drug interactions;
e) exhibits either no toxicity or acceptable toxicity at the dosage range
used;
f) exhibits either no side effects or acceptable side effects at the dosage
range
used;
g) exhibits either no pharmacological effect at the dosage range used in
regulating transgene expression or a negligible pharmacological effect; and
h) possess physical properties suitable for the in vitro evolution of an
aptamer
(desirable physical properties may include a planar molecule which has a rigid
structure.)
These characteristics indicate that the effector is suitable for aptamer
generation, human consumption and use with an allosteric control module for
the
regulation of gene expression. Information on these characteristics may be
2 0 obtained by accessing one or more databases containing data on the
selected
characteristics. Databases containing the relevant information include, but
are not
limited to, Investigational Drugs database (IDdb, Current Drugs; Current Drugs
Ltd., Philadelphia, Pennsylvania), Drug Data Report (MDDR, MDL Information
Systems Inc., San Leandro, California; Prous Science Publishers, Barcelona,
2 5 Spain), World Drug Index (WDI, Derwent Information, Alexandria, Virginia)
and
Derwent Drug File, R&D Insight (Adis International Inc., Langhorne,
Pennsylvania), R&D Focus (IMS HEALTH, IMSworld Publications Ltd.,
London, England), Pharmaprojects (PJB Publications, Surrey, United Kingdom),
MEDLINE (The National Library of Medicine) and EMBASE (Elsevier Science,
3 0 B.V). A set of effectors having the selected characteristics is then
identified.
"Bioavailability" as used herein refers to the ability of the effector to
reach
its intended site of action after administration. The effector will have a
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 26 -
bioavailability of at least 1%. Preferably, the effector will have a
bioavailability
of at least 5%, and more preferably the effector will have a bioavailability
of at
least 10%. A preferred effector will also be bioavailable upon oral delivery,
but it
will be appreciated that the route of delivery is not limiting to the present
invention. The effector may be administered parenterally, e.g., by injection
intravenously, intraperitoneally, intramuscularly, intrathecally or
subcutaneously.
The selected effector may be formulated as a composition for oral
administration
(including sublingual and buccal), pulmonary administration (intranasal and
inhalation), topical administration, transdermal administration, and rectal
administration. Delivery may involve a single dose schedule or a multiple dose
schedule.
Using this method, suitable effectors may be identified from a variety of
molecules including, but not limited to, small organic molecules, peptides,
polypeptides, proteins, oligonucleotides, polynucleotides, nucleic acids,
naturally
occurring metabolites and biological effectors, lipids, carbohydrates
(polysaccharides, sugar), fatty acids, and polymers. In a preferred embodiment
of
the present invention, the effector is a small molecule.
It will be appreciated that any database, or combination of databases, may
2 0 be used in the performance of the present invention. Suitable databases of
potential effectors will include molecules such as:
a) marketed drugs with stereoselectivity for an isomer that comprises the
pharmaceutically active component and another isomer with little or no
pharmacological activity (the latter being the possible effector of interest);
2 5 b) known drug metabolites having little or no activity;
c) nuclear receptor targeted molecules (for example, Vitamin D, retinoic acid,
steroids);
d) drug candidates which entered clinical trials, but the trials were
discontinued
due to a relative lack of efficacy;
3 0 e) drugs that were removed from the market because of lack of therapeutic
efficacy;
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 27 -
f) drugs that are efficacious but which are not marketed because of low
relative
benefit;
g) drugs designed as antiviral/anti-infectives, for use in patients not
affected by
the targeted virus or infectious agent;
h) well characterized food additives;
i) generic drugs with well known mechanisms of action; and
j) drugs that were displaced from the market or clinical trials by best in
class
molecules.
The identified effectors may then be used to generate and select aptamers
to the effectors in the set by means of in vitro evolution. The combinations
of
effectors and aptamers, as well as effectors and allosteric control modules,
may
then be evaluated to identify those molecules best suited for the control of
gene
expression.
Beyond allosteric control module binding, efficacy and cell penetration,
effectors of interest will undergo a variety of additional tests as part of
the effector
suitability and selection process and as part of the regulatory approval
process.
Such testing includes those tests typically conducted as part of the
development
and regulatory approval of a pharmaceutical product, including evaluations of
pharmacokinetic properties, pharmacodynamic properties, pharmaceutic
2 0 properties, toxicology, mutagenicity, reproductive toxicity and the like.
Pharmacokinetic evaluations may include analyses of an effector's
absorption, distribution, metabolism and excretion profiles in in vitro cell
systems,
animals, animal disease models, normal humans and patients. The absorption
profile includes the rate of absorption, the maximum plasma concentration
2 5 achieved, the effect of formulation modifications, the effect of different
salt and
crystal forms, the effect of food and other medications on absorption and the
like.
The distribution profile includes the determination of the location and
concentration of the effector in the various tissues and fluids of the body,
protein
binding and the like. The metabolism profile includes the determination of the
3 0 mechanism by which the effector is metabolized, such as by enzymes of the
liver
or kidney, a determination of the structure and activity of the metabolites
produced, the effect of the effector and metabolites on the metabolism of
other
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 28 -
drugs, the effect of food and other drugs on the metabolism of the effector
and the
like. The excretion profile includes the determination of the mechanism and
distribution of excretion, such as through the bile or kidney, the clearance
rate, the
half-life (amount of time needed to clear fifty percent of the plasma level of
the
administered effector), accumulation, and the like of the effector and its
metabolites.
Pharmacodynamic evaluations involve an analysis of an effector's
physiological activity. Such an analysis may include an assessment of the
duration of activity, dosing regiment and formulation effects on activity,
therapeutic threshold (minimum effector plasma concentration needed for
activity) and mode of administration effects.
Pharmaceutic evaluations will involve an analysis of an effector's physical
properties with respect to formulating the effector. Effector physical
properties of
interest include solubility, chemical stability, such as the effect of
temperature,
moisture and light, crystal form (solid), salt form (solid or in solution) and
the
like, solution stability, effect on solution pH, crystal vs. amorphous solid
vs. oil
vs. liquid, crystal density, etc. Also the effector formulation properties are
evaluated. These properties include, but are not limited to, stability, effect
of the
crystal form and size on absorption, effect of the salt form on absorption,
solid
2 0 compressibility and malleability, solid flowability, uniformity of crystal
size,
compatibility with other formulation ingredients, packing density, blend and
content uniformity of each formulation.
Toxicology involves determining the toxic and other side effect profile of
the effector, its metabolites and its formulation, generally initially in
animals and
2 5 later in humans, to ascertain the potential risks involved in
administering the
effector. Analyses may include an evaluation of undesirable effects on the
central
nervous system, cardiovascular system, pulmonary system, gastrointestinal
system, renal system, hepatic system, genitourinary system, hematopoietic
system, immunologic system and dermal system. The analyses may include
3 0 determining toxic dose, maximum tolerable dose, agonistic or antagonistic
activity against other enzymes, receptors, binding proteins and the like,
carcinogenicity, immunogenicity, and the like. Means and methods of
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 29 -
toxicological analysis are well known in the art. Descriptions include those
found
in Principles and Methods of Toxicology (Third Edition, 1994, Ed. A. Wallace
Hayes, Raven Press, NY) and Toxicology: The Basic Science of Poisons (by
Cassarett and Doull, Fifth Edition, 1996, Ed. Curtis D. Klaassen, McGraw-Hill,
NY).
Effector development may also include the study and evaluation of one or
more of these attributes in special populations such as pediatrics and
geriatrics. In
addition, the analyses may also require the evaluation of the effects of
gender and
ethnicity.
As mentioned above, the final selection of a suitable effector will include
testing similar to that performed for any new molecular entity (NME) proposed
for human use. A variety of screening strategies may be used. Until a few
years
ago, it was not possible to predict the absorption and metabolism
characteristics of
NMEs without conducting appropriate whole animal in vivo studies. However,
recent advances in the understanding of the molecular biology and functional
specificity of metabolic enzymes and absorption and transport mechanisms have
provided a mechanistic basis for gathering absorption and metabolism data
utilizing "humanized" in vitro systems. The development and availability of
these
2 0 humanized in vitro systems coupled with advances in analytical
instrumentation
are speeding the development process. It is increasingly possible to conduct
high-
throughput pharmacokinetic screening of new molecules. The following
description will highlight the in vitro and in vivo methods and techniques
that are
being applied.
In vitro methods
Absorption assays
The ideal drug candidate should possess good metabolism and absorption
characteristics. An early prediction of the oral bioavailability of a series
of
3 0 compounds provides invaluable information regarding the structure activity
relationship (SAR). An essential part of selecting compounds with good
systemic
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 30 -
bioavailability involves an accurate prediction of absorption through the gut.
One
of the most successfully applied techniques for prediction of absorption in
man is
the 21-day Caco-2 model. Caco-2 cells are of human origin, derived from human
colon cancer cells. When cultured on porous membranes these cells
spontaneously differentiate into highly functionalized monolayers that are
similar
in characteristics to the small intestinal enterocytes (Pinto et al., Biol.
Cell 47:323-
330, 1983; Hidalgo et al., Gastroenterology 96:736-749, 1989.) Data gathered
over the years show this model to be a good predictor of in vivo human
intestinal
absorption (Artursson et al., Biochem. Biophys. Res. Comm. 175:880-885, 1991.)
Though this model provides reasonable estimates of in vivo absorption, its use
as a
high-throughput screening tool has been challenged by some investigators. The
labor-intensive and time-consuming nature of conducting studies with these
cells
has prompted some researchers to look for viable alternatives such as the fast-
growing Madin-Darby Canine Kidney (MDCK) cells (Irvine et al., J. Pharm. Sci.
88(1):28-33, 1999) or the three-day Caco-2 culture model (Chong et al., Pharm.
Res. 14 (12):1835-1837, 1997) while others have attempted to automate the Caco-
2 absorption assessment methodology (Garberg et al., Pharm. Res. 16(3):441-
445,
1999.) Irvine et al. utilized a large number of compounds to evaluate the use
of
MDCK cells as an alternative to Caco-2 cells for estimating membrane
2 0 permeability. Overall, a good correlation (r- =0.79) was observed for
apparent
permeability (PaP~) values between the Caco-2 and MDCK cells. Based on their
findings these authors suggest MDCK cells to be a practical permeability
screening tool for increasing throughput in the early discovery phase. The
three-
day Caco-2 model for studying the permeability of a compound has also been
2 5 investigated. These three-day cultures provide reasonable Pa~~ values for
transcellularly transported compounds. The monolayers from three-day cultures,
however, are about 4- to 6-fold leakier than the traditional 21 day cultured
cells,
and the predictive nature of these cells for compounds that are transported
paracellularly or by efflux and carrier-mediated mechanisms is not as robust
(Yee
3 0 S, Day W: Applications of Caco-2 Cells in Drug Discovery and Development
In
Handbook of Drug Metabolism. Woolf, T (Eds), Marcel Dekker, Inc. New York:
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 31 -
508-519.) Garberg and colleagues developed an automated set-up for assessment
of in vitro permeability to increase the throughput capacity during the
screening
phase utilizing the traditional Caco-2 model. This automation was accomplished
by the incorporation of a liquid handling system that performed all necessary
pipetting steps during the course of the experiment. Based on the results
obtained,
these authors suggested that utilizing an automatic sample processor can
substantially increase the capacity of in vitro absorption screening. Another
strategy to accelerate the absorption screening throughput has been
investigated
(Taylor et al., Pharm. Res. 14(5):572-577, 1997.) These researchers conducted
absorption experiments using the Caco-2 model with mixtures of
physicochemically diverse compounds and were able to successfully select
potential orally bioavailable compounds. The pooling of samples obtained from
experiments conducted with a single compound or a mixture of compounds has
also been investigated utilizing a mass spectrometer as the analytical tool
(McCarthy et al., Pharm. Res. 13(9):S242, 1996).
New higher throughput methods are also being developed to screen NMEs
for physicochemical properties such as solubility that could influence NME
absorption (Tarbit et al., Curr. Opin. Chem. Biol. 2:411-416, 1998.) Others
are
developing methods to determine if NMEs are substrates of various intestinal
2 0 transporters (e.g., p-glycoprotein). Permeability data, coupled with
physiochemical and transport data, should enhance the ability to predict
absorption in the future and will lead to faster selection of lead candidates
with
desirable absorption characteristics.
2 5 Metabolism assays
First-pass metabolism by the intestine or liver is another major
determinant of in vivo oral bioavailability. Metabolic reactions can be
broadly
classified into Phase I and Phase II reactions. Phase I reactions create a
functional
group on the molecule so that it can either be further conjugated by phase II
3 0 enzymes or excreted upon modification. Most of the oxidative metabolic
reactions are carned out by the cytochrome P450 (CYP) enzyme system, a
CA 02401654 2002-08-28
WO 01/64956 PCTNSO1/06615
- 32 -
superfamily of heme-containing enzymes predominantly found in the liver. CYP
catalyzed metabolism can have significant effects on the overall dispositional
characteristics of a molecule which could result in a short half-life, low
bioavailability, non-linear kinetics, drug-drug interactions, toxicity, lack
of
efficacy, and intersubject variability. These defects alone or in combination
are
the leading cause of failure of drugs in development or in some instances have
led
to withdrawal of a marketed product.
To address these issues metabolism studies are instituted early in the
program. High-throughput in vitro screening methods have been developed for
predicting the metabolic stability of an NME, and the potential for drug-drug
interactions. The traditional test tube and water bath incubation method for
metabolic stability testing has been replaced with the 96-well plate technique
that
is more amenable to automation. The incubations are typically conducted
utilizing pooled human or animal liver microsomes or S-9 fraction, and the
reaction is carried out at 37~C on a reaction block. Following termination of
the
reaction, the plate is placed directly on the high pressure liquid
chromatography
(HPLC) autosampler coupled to a mass spectrometer. The use of human
hepatocytes in a 96-well format has also been proposed (Li A., RAPS, New
Orleans, LA, 1999.) Sample pooling from a 96-well plate system along with
2 0 liquid chromatography mass spectrometry (LC/MS) for separation and
detection
and automated data acquisition has also been used to increase the throughput
in
the generation of metabolic stability data (Michelson et al., LC-MS Analysis
of
the Metabolic Stability of Human Liver Microsome Samples Prepared in a 96-
well Format. RAPS Pharmsci. 1(4):S-300, 1999.)
After data acquisition, an estimate of in vitro half-life (t,2) or the
apparent
clearance (CIa~P) is made based on the rate of depletion of the parent
compound
(Obach et al., Journal of Pharmacol. Exp. Ther. 283(1):46-58, 1999.)
Predictions
of in vivo bioavailability can also be made using the Cl~~~ for the species in
question. Rank ordering of the screened compounds based on the ChpP or the
3 0 predicted in vivo bioavailability can then be performed. These results can
be
generated for a large number of compounds in a rapid turnaround manner and
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 33 -
provide structure activity relationship insights for improving pharmacokinetic
(PK) characteristics by the means of structural modifications.
The prediction of drug-drug interactions is also an integral part of drug
development programs. Traditionally, cytochrome P450 mediated drug-drug
interaction studies have been conducted utilizing human liver microsomes, and
the analysis of samples is accomplished using HPLC separation techniques with
UV, or fluorescence detection. Complete chromatographic separation is quite
time-consuming and labor-intensive and is therefore not appealing for use in a
high-throughput screening setting. Recently, Yin and colleagues (ISSX
Proceedings 15:87, 1999) described a rapid throughput method for the
determination of CYP isoform activity to evaluate the inhibitory potential of
NMEs. This method utilizes human liver microsomes and is performed on a 96-
well platform with solid phase extraction and pooling of samples for high-
pressure liquid chromatography coupled to mass spectrometry (LC/MS/MS)
quantification. Crespi and coworkers (Curr. Opin. Chem. Biol. 2(1):15-19,
1999)
have also described microtiter plate-based fluorometric assays for activities
of
CYP1A2, CYP2C9, CYP2C19, CYP2D6, and CYP3A4, the five major human
drug metabolizing CYP enzymes. Radiometric assays to obtain data from in vitro
studies for various CYP enzymes have also been reported (Rodrigues et al.,
Drug
2 0 Met. Dis. 24:126-136, 1996; Rodrigues et al., Anal. Biochem. 219:309-320,
1994;
and Riley RJ, Howbrook D: J. Pharmac. Toxicol. Meth. 38:189-193, 1998.)
Moody et al. (Xenobiotica, 9(1):53-75, 1999) recently reported the development
and application of a fully automated method for the analysis of catalytic
activities
of major CYP enzymes. This procedure is based on fluorometric and radiometric
2 5 assays and is performed by a robotic sample processor.
The methods described above could successfully provide a rapid
estimation of ICso values and thus an initial determination of the inhibitory
capability of the NME. Further characterization of the inhibitory potential of
NMEs may be pursued using classical approaches that utilize established CYP
3 0 isoform specific probe substrates (Newton et al., Drug Met. Dis. 23(1):154-
158,
1995).
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 34 -
Early identification of induction based drug-drug interactions is another
important issue. Previously, induction assays required the use of primary
cultures
of human hepatocytes that meant heavy reliance of such studies on the
availability
of human tissue. Recent knowledge of the molecular basis for the induction of
CYP3A4 via the PXR nuclear receptor has provided new means for exploring
induction based drug-drug interactions in a rapid and relatively inexpensive
fashion (Lehmann et al., J. Clin. Invest. 102(5):1016-1023, 1998.
High-throughput screening for metabolic stability and drug-drug
interactions generates valuable data that can be exploited to develop "ideal"
molecules. In order to maximize the benefit of existing information, the data
should be organized in a format that is easily searchable and retrievable. To
this
end, several drug metabolism databases are now commercially available (Erhardt
P: Drug Metabolism Data: Past and Present Status, Med. Chem. Res. 8(7/8):400-
421, 1998.) A "knowledge-based" database with systematic organization of
literature reports on drug interactions is also being developed by Professor
Rene
Levy's group at the University of Washington, Seattle (Levy RH et al.,
Metabolic
drug interactions, www.a~ptechsys.com/drug_/.) This object-oriented database
design allows the user to retrieve information regarding the involvement of
particular CYP isoforms in the metabolism of a drug, and also includes
2 0 information on other relevant PK parameters to allow an in vitro-in vivo
correlation. The parallel use of these databases along with the data generated
from high throughput screening studies will speed up the nomination and
development process of lead candidates.
2 5 In vivo pharmacokinetic assays
Despite successes in the development of in vitro absorption and
metabolism assays, in vivo PK studies still play an important role in drug
development (Smith DA, van de Waterbeemd H: Pharmacokinetics and
metabolism in early discovery, Curr. Opin. Chem. Biol. 3:373-378, 1999.) To
3 0 obtain the PK parameter values of an NME, the NME is dosed both
intravenously
and orally to an animal, blood is sampled at various time points and then
analyzed
CA 02401654 2002-08-28
WO 01/64956 PCT/US01/06615
- 35 -
by HPLC or gas chromatography (GC) for the compound of interest. The PK
parameters (e.g., clearance, volume of distribution, elimination half-life,
and oral
bioavailability) which describe the absorption and disposition of the NME in a
whole animal are calculated from the serum concentration-time profiles.
Feedback on the structure-PK relationship is important in directing research
and
synthetic efforts and information on in vivo exposures is useful in designing
in
vivo pharmacology/efficacy studies. In vivo PK screening may also be necessary
when in vitro metabolism assays are poorly predictive of in vivo kinetics
(e.g.,
when the compound is eliminated predominantly by renal or biliary mechanisms).
Increasingly, pharmaceutical scientists are generating in vivo PK data for use
in
developing and validating models to predict in vivo kinetics from in vitro
data and
for use in in-silico modeling (Tarbit et al., Curr. Opin. Chem. Biol. 2:411-
416,
1998; Bayliss et al., Curr. Opin. Drug. Dis. Dev. 2(1), 20-25, 1999). In
recent
years, efforts to increase throughput in PK evaluation of NMEs have focused on
mixture dosing and sample pooling to minimize bioanalytical workload. HPLC
with mass spectrometric detection (LC/MS and LC/MS/MS) has greatly enhanced
the ability to monitor more than one compound in a matrix.
Mixture Dosing
2 0 Mixture dosing (also referred to as N-in-One, cassette, or cocktail
dosing)
involves the simultaneous administration of two or more NMEs to the same
animal and the assay of the plasma samples by LC/MS. PK parameter values are
determined from the resulting concentration-time profiles. Berman et al. (J.
Med.
Chem. 40(6):827-829, 1997; and Olah et al., Rapid Commun. Mass Spectrom
2 5 11:17-23, 1997) reported the successful application of the mixture dosing
approach to speed candidate selection during drug discovery. Since then,
several
publications have reported on the PK of two to 22 compounds dosed
simultaneously to mice, rats, dogs, and monkeys by intravenous and oral routes
(Bayliss et al., Curr. Opin. Drug. Dis. Dev. 2(1), 20-25, 1999; Allen et al.,
Pharm.
3 0 Res. 15(1):93-97, 1998; Frick et al., Pharm. Sci. Tech. Today. 1(1):12-18,
1998;
and Gao et al., J. Chromatogr. A828:141-148, 1998). The reports have
CA 02401654 2002-08-28
WO 01/64956 PCT/US01/06615
- 36 -
demonstrated good agreement between PK parameter values obtained from
mixture dosing compared to those obtained from traditional approaches. Mixture
dosing offers significant cost- and time-savings because fewer animals are
studied, less compound is required, and fewer samples are generated compared
to
traditional methods. An additional advantage of mixture dosing is the ability
to
examine tissue distribution (e.g., brain-blood ratios), urinary excretion
patterns
(Frick et al., Pharm. Sci. Tech. Today. 1(1):12-18, 1998) and protein binding
(Allen et al., Pharm. Res. 15(1):93-97, 1998) of multiple NMEs along with the
PK studies. Disadvantages of N-in-One dosing include difficulties in
developing
formulations for dosing, the possibility of increased adverse events in the
animal,
and method development issues (e.g., the need for increased sensitivity
because of
lower doses, avoidance of molecular weight redundancies between metabolites
and analytes of interest). There is also a concern that drug-drug interactions
(e.g.,
inhibition of metabolism or other routes of elimination of one compound by
another) may alter the PK of NMEs. Compound-compound interactions have
occurred during oral mixture dosing and during intravenous dosing when a
potent
cytochrome P450 inhibitor was introduced into a mixture (Olah et al., Rapid
Commun. Mass Spectrom. 11:17-23, 1997). The interactions were identified by
inclusion of a reference compound in all mixture dosing sessions. A list of
factors
2 0 to consider when designing mixture dosing experiments to minimize the
chance of
obtaining false data and a statistical analysis describing the probability
that an
NME may experience a compound-compound interaction as a function of mixture
size are provided in the review by Frick and colleagues (Pharm. Sci. Tech.
Today
1(1):12-18, 1998). Future advances in mixture dosing will likely focus on the
2 5 automation of various steps such as dose solution preparation, MS tuning
and data
reduction and on the development of robust, flexible chromatography methods
(Bayliss et al., Curr. Opin. Drug Dis. Dev. 2(1), 20-25, 1999; Gao et al., J.
Chromatogr. A828:141-148, 1998). Future work should also be focused on
defining and minimizing the PK risks (e.g., compound-compound interactions)
3 0 associated with the method and the use of PK data for the development of
structure-PK relationships (see for example, Shaffer et al., J Pharm. Sci.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 37 -
88(3):313-318, 1999 suggesting that structure-pharmacokinetic relationships
can
be derived for a set of chemical analogs from data generated using a mixture
dosing technique.)
Pooling of samples from pharmacokinetic studies to increase bioanalytical
throughput.
Several labs have successfully used a plasma-pooling approach to increase
bioanalytical and PK throughput (Olah et al., Rapid Commun. Mass Spectrom.
11:17-23, 1997; and Hop et al., J. Pharm. Sci. 87(7):901-903, 1998). In the
pooling approach, multiple animals are dosed with individual NMEs, and
aliquots
of the samples from common time points are pooled and analyzed by LC/MS.
The advantage of pooling is that there are fewer samples to analyze and the
possibility of in vivo compound-compound interactions is eliminated. The
pooling of samples, however, can be very time-consuming and detection limits
may be compromised because of sample dilution.
Hop and colleagues (J. Pharm. Sci. 87(7):901-903, 1998) revived a
pooling technique that has been used to obtain PK parameters in pediatric
patients. For each animal, aliquots of plasma from each time point were pooled
in
proportion to the time interval it covered to yield just one sample that had a
2 0 concentration proportional to the area under the curve (AUC). The major
advantage of this approach is a significant reduction in the number of samples
and
bioanalytical workload. The disadvantages are that information about the
concentration-time course (Cmax, Tmax, half-life) is no longer available and
pooling is very tedious. The unused portion of the plasma sample for promising
2 5 compounds can always be reanalyzed to obtain the full PK profile and
automation
would facilitate pooling. The researchers have applied this technique to the
analysis of NMEs for a discovery program where AUC after oral dosing was the
main concern. The technique could also be used to determine clearance and
bioavailability of compounds.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 38 -
In Vitro Evolution
For purposes of the present invention, in vitro evolution strategies are
typically used to evolve an aptamer for an identified effector. The selected
and
specific aptamer-effector reaction is a useful tool for in vivo applications:
e.g., it
allows the engineering of constructs which are not naturally found in the
cell, and
therefore, are not expected to adversely affect normal cell function.
Several in vitro evolution (selection) strategies (Orgel, Proc. R. Soc.
London, B 205: 435, 1979) have been used to evolve new nucleic acid catalysts
capable of catalyzing a variety of reactions, such as the cleavage and
ligation of
RNA and DNA (Joyce, Gene, 82:83-87, 1989; Beaudry et al., Science 257:635-
641, 1992; Joyce, Scientific American, 267:90-97, 1992; Breaker et al.,
TIBTECH
12:268, 1994; Bartel et al., Science 261:1411-1418, 1993; Szostak, TIBS 17:89-
93, 1993; Kaufmann et al. U.S. Patent No. 5,814,476; Kumar et al., FASEB J.,
9:1183, 1995; Breaker, Curr. Op. Biotech., 7:442, 1996; and Berzal-Herranz et
al., Genes & Develop. 6:129, 1992), and new nucleic acids which act as
aptamers,
such as an ATP aptamer, HIV Rev aptamer, HIV Tat aptamer and others
including oligonucleotide effectors (see Ellington and Szostak, Nature 346:818-
822, 1990; Famulok and Szostak, Angew. Chem. Int. Ed. Engl. 31:979-988, 1992;
Ellington, Current Biology, 4(5):427-429, 1990; Porta and Lizardi,
2 0 Bioll'echnology 131:161-164, 1995; Tang and Breaker, Chemistry & Biology,
4(6):453-459, 1997; Tang and Breaker, RNA, 3:914-925, 1997; Werstuck and
Green, Science, 282:296-298, 1998; Tang and Breaker, Nucleic Acids Research
26(18):4214-4221, 1998; Soukup and Breaker, Proc. Natl. Acad. Sci. USA,
96:3584-3589, 1999; and Robertson and Ellington, Nature Biotechnology 17:62-
2 5 66, 1999). In addition to these, further descriptions of in vitro
evolution processes
are provided in Innovir U.S. Patent No. 5,741,679 and U.S. Patent No.
5,834,186,
WO 9843993 (PCT/LTS98/06231, Yale University), WO 9827104
(PCT/US97/24158, Yale University) and U.S. Patent No. 5,817,785 (NeXstar
Pharmaceuticals).
3 0 In a basic form, the process may involve the following steps.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 39 -
1) A candidate mixture of nucleic acids of differing sequence is prepared.
The candidate mixture typically includes nucleic acids having regions of fixed
sequences (i.e., each of the members of the candidate mixture contains the
same
sequences in the same location) and regions of randomized sequences. The fixed
sequence regions may be used for a variety of reasons, including: (a) to
assist in
the amplification steps described below, (b) to mimic a sequence known to bind
to
the effector, or (c) to enhance the concentration of a given structural
arrangement
of the nucleic acids in the candidate mixture. The randomized sequences can be
totally randomized (i.e., the probability of finding a base at any position
being one
in four) or only partially randomized (e.g., the probability of finding a base
at any
location can be selected at any level between 0 and 100 percent).
2) The candidate mixture is contacted with the selected effector under
conditions favorable for an interaction between the effector and nucleic acids
of
the candidate mixture. Under these circumstances, the interaction between the
effector and the nucleic acids can be considered as forming aptamer-effector
pairs
between the effector and those nucleic acids having the strongest affinity for
the
effector.
3) The nucleic acids with the highest affinity for the effector are
partitioned from those nucleic acids with a lesser affinity to the effector.
Because
2 0 only an extremely small number of different molecules (and possibly only
one
molecule) corresponding to the highest affinity nucleic acid sequences exist
in the
candidate mixture, it may be desirable to set the partitioning criteria so
that a
significant amount of the nucleic acids in the initial candidate mixture
(approximately 5-50%) are retained during partitioning.
2 5 4) Those nucleic acids selected during partitioning as having relatively
higher affinity to the effector are then amplified to create a new candidate
mixture
that is enriched in nucleic acids having a relatively higher affinity for the
effector.
5) By repeating these partitioning and amplifying steps, the newly formed
candidate mixture contains fewer and fewer unique sequences, and the average
3 0 degree of affinity of the nucleic acids to the effector will generally
increase. With
repeated cycles, the process yields a candidate mixture containing one or more
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 40 -
unique nucleic acids representing those nucleic acids from the original
candidate
mixture having the highest affinity to the effector.
There is no known set number of bases required for the interaction of an
aptamer and an effector. A variety of nucleic acid primary, secondary and
tertiary
structures are known to exist. The structures or motifs that have been shown
most
commonly to be involved in non-Watson-Crick type interactions are referred to
as
hairpin loops, symmetric and asymmetric bulges, psuedoknots and myriad
combinations of the same. Research suggests that they can be formed in a
nucleic
acid sequence of less than 30 nucleotides. For this reason, it is often
preferred
that in vitro evolution procedures with contiguous randomized segments be
initiated with nucleic acid sequences containing a randomized segment of about
to about 50 nucleotides. Thus, the aptamer will typically contain about 20 to
about 50 nucleotides, or more preferably about 20 to about 30 nucleotides,
15 evolved for binding to a specific effector. The small size of the molecule
is
advantageous for cell delivery as compared with conventional expression
regulation constructs which are much larger in size.
It will be appreciated by persons skilled in the art that in vitro evolution
techniques may be used to create and identify separate aptamers which may then
2 0 be used as modular units with catalytic structures for the construction of
a variety
of different allosteric control modules. Alternatively, the allosteric control
modules may be evolved as a single unit with separate regions or domains
including an aptamer or effector-binding domain, a catalytic domain, and in
some
embodiments additional domains including, but not limited to, a target RNA
2 5 recognition domain and a substrate domain.
It will also be appreciated that for any given aptamer, large combinatorial
libraries of effectors (e.g., organic compounds, peptides, small molecules,
etc.)
produced by chemical synthesis can be evaluated for aptamer binding. Phage
display libraries may also be screened for the presence of an effector to bind
a
3 0 selected aptamer. Thus, in another embodiment of the present invention the
aptamer may be identified and the effector selected through a screening
process.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 41 -
Catalytic Domain
The other main component of the allosteric control module is the catalytic
domain. As described above, in vitro evolution may also be used to evolve new
nucleic acid catalysts capable of catalyzing a variety of reactions, such as
cleavage, ligation and splicing. In addition, the catalytic domain is a highly
specific construct, with the specificity of activity depending not only on the
base-
pairing mechanism of binding, but also on the mechanism by which the molecule
affects the expression of the RNA to which it binds. For example, if
inhibition is
caused by cleavage of the RNA target, specificity is defined as the ratio of
the rate
of cleavage of the targeted RNA over the rate of cleavage of non-targeted RNA.
Gene of interest
The present invention contemplates, in one embodiment, the provision of a
desired gene that encodes a protein that is defective or missing from a target
cell
genome in a patient. The present invention also contemplates a method of
treating
a patient suffering from a disease state by providing the patient with human
cells
genetically engineered to encode a required protein. Yet another embodiment is
the delivery of a gene to correct a genetic defect. In each of these
embodiments of
2 0 the present invention, the gene of interest is delivered to the recipient
cell with an
allosteric control module which has been designed to alter the expression of
the
gene in response to the presence or absence of an identified effector.
Exemplary uses of constructs of the present invention include gene therapy
for hereditary diseases. These diseases include, but are not limited to:
familial
2 5 hypercholesterolemia or type II hyperlipidemias (LDL receptor), familial
lipoprotein lipase deficiency or type I hyperlipidemias (lipoprotein lipase),
phenylketonuria (phenylalanine hydroxylase), urea cycle deficiency (ornithine
transcarbamylase), von Gierke's disease (e.g., glycogen storage disease, type
I;
glucose-6-phosphotases), alpha 1-antitrypsin deficiency (alpha 1-antitrypsin),
3 0 cystic fibrosis (cystic fibrosis transmembrane conductant regulator), von
Willebrand's disease and hemophilia A (Factor VIII), hemophilia B (Factor IX),
sickle cell anemia (beta globin), beta thalassemias (beta globin), alpha
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 42 -
thalassemias (alpha globin), hereditary sperocytosis (spectrin), severe
combined
immune deficiency (adenosine deaminase), Duchenne muscular dystrophy
(dystrophin minigene), Lesch-Nyhan syndrome (hypoxanthine guanine
phosphoribosyl transferase), Gaucher's disease (beta-glucocerebrosidase),
Nieman-Pick disease (sphingomyelinase), Tay-Sachs disease (lysosomal
hexosaminidase), and maple syrup urine disease (branched-chain keto acid
dehydrogenase).
Suitable transgenes for use in the present invention further include, but are
not limited to, those encoding proteins such as: nerve growth factor (NGF),
ciliary neurotrophic factor (CNTF), brain-derived neurotrophic factor (BDNF),
neurotrophins 3 and 4/5 (NT-3 and 4/5), glial cell derived neurotrophic factor
(GDNF), transforming growth factors (TGF), and acidic and basic fibroblast
growth factor (aFGF and bFGF); sequences encoding tyrosine hydroxylase (TH)
and aromatic amino acid decarboxylase (AADC); sequences encoding superoxide
dismutase (SOD 1 or 2), catalase and glutathione peroxidase; sequences
encoding
interferons, lymphokines, cytokines and antagonists thereof such as tumor
necrosis factor (TNF), CD4 specific antibodies, and TNF or CD4 receptors;
sequences encoding GABA receptor isoforms, the GABA synthesizing enzyme
glutamic acid decarboxylase (GAD), calcium dependent potassium channels or
2 0 ATP-sensitive potassium channels; and sequences encoding thymidine kinase,
angiostatin, dopamine, blood clotting factors, erythropoietin, the colony
stimulating factors G-, GM- and M-CSF, tissue plasminogen activator, human or
animal growth hormones, IGF-1, insulin, KGF, leptin, MGDF, multiple drug
resistance, osteoprotogerin, VEGF, VEGF-ra, alpha-interferon, beta-interferon,
2 5 consensus-interferon, IFN-gamma, IL-12, IL-lra, IL-2, IL-4, and TNFbp.
Other genes of interest contemplated by the invention encode pathogens
for use as vaccines. Exemplary genes include, but are not limited to, those
encoding: HIV-1 and HIV-2 (sequences other than rev and gp160 sequences);
human T-lymphotrophic virus types I and II; respiratory syncytial virus;
3 0 parainfluenza virus types 1-4; measles virus; mumps virus; rubella virus;
polio
viruses; influenza viruses; non-human influenza viruses (avian, equine,
porcine);
hepatitis virus types A, B, C, D and E; rotavirus; Norwalk virus;
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 43 -
cytomegaloviruses; Epstein-Barr virus; herpes simplex virus types 1 and 2;
varicella-zoster virus; human herpes virus type 6; hantavirus; adenoviruses;
hepatitis viruses, rabies, foot and mouth disease virus, chlamydia pneumoniae;
chlamydia trachomatis; mycoplasma pneumoniae; mycobacterium tuberculosis;
atypical mycobacteria; feline leukemia virus; feline immunodeficiency virus;
bovine immunodeficiency virus; equine infectious anemia virus; caprine
arthritis
encephalitis virus; visna virus, infectious mononucleosis; roseola; pneumonia
and
adult respiratory distress syndrome; upper and lower respiratory tract
infections;
conjunctivitis; upper and lower respiratory tract infections; genital tract
infections;
and pneumonia and encephalitis in sheep. The vaccine vectors may be used to
generate intracellular immunity if the gene product is cytoplasmic (e.g., if
the
gene product prevents integration or replication of a virus). Alternatively,
extracellular/systemic immunity may be generated if the gene product is
expressed on the surface of the cell or is secreted.
A host (especially a human host) may be immunized against a polypeptide
of a disease-causing organism by administering to the host an immunity-
inducing
amount of a vector of the present invention which encodes the polypeptide.
Immunization of a human host with a vector of the invention typically involves
administration by inoculation of an immunity-inducing dose of the virus by the
2 0 parenteral route (e.g., by intravenous, intramuscular or subcutaneous
injection), by
surface scarification or by inoculation into a body cavity. Typically, one or
several inoculations of between about 1000 and about 10,000,000 infectious
units
each, as measured in susceptible human or nonhuman primate cell lines, are
sufficient to effect immunization of a human host.
2 5 Additional uses of the materials and methods described herein include, but
are not limited to:
1. increasing the expression of monoclonal antibodies by hybridoma cells,
i.e., cell lines resulting from the fusion of a B-lymphocytes with a myeloma
cell
lines. Monoclonal antibodies could be produced by growing the hybridoma in
3 0 tissue culture or in vivo;
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 44 -
2. increasing plant derived products as used in fragrances and perfumes,
flavoring compounds or sweeteners e.g. the basic proteins from Thaumatococcus
danielli, insecticides, anti-fungal compounds or pesticides;
3. increasing the expression of a polypeptide which is associated with the
rate limiting step in bioleaching of metals such as uranium, copper, silver,
manganese etc. For example, as carned out by the organism Thiobacillus sp.,
algae or fungi;
4. increasing the expression of a polypeptide which is associated with the
rate limiting step in removal of nitrogen or phosphate or toxic waste minerals
from water, e.g., as carried out by Nitrobacter sp. or Acinetobacter sp.;
5. increasing the expression of a polypeptide which is associated with the
rate limiting step in stimulating methane production from biological waste,
typically from the methanogenic micro-organisms archaebacteria;
6. increasing the expression of a polypeptide which is associated with the
rate limiting step in the biodegradation of marine oil spills (e.g., aliphatic
hydrocarbons, halogenated aliphatics, halogenated aromatics). In one instance,
biodegradation is effected by the conversion of petroleum products to
emulsified
fatty acids. Bacteria useful in this invention include, but are not restricted
to,
Archromobacter, Arthrobacter, Flavobacterium, Nocardia, Pseudomonas (e.g.
2 0 Pseudomonas oleovorans) and Cytophaga. Yeast useful in this invention
include,
but are not restricted to, Candida (e.g., Candida tropicalis), Rhodotorula,
and
Trichosporon;
7. increasing the expression of a polypeptide which is associated with the
rate limiting step in biodegradation of lignin;
2 5 8. increasing the expression of a polypeptide which is associated with the
rate limiting step in the biotransformation of steroids and sterols, e.g., by
Rhizopus sp., Saccharomyces, Corynebacterium sp.; D sorbitol to L sorbose by
Acetobacter suboxydans; racemic mixtures; prochiral substrates; terpenoids;
alicyclic and heteroalicylic compounds; antibiotics; aromatic and heterocyclic
3 0 structures including phthalic acid esters, lignosulfonates, surfactants
and dyes;
naphthyridines by Penicillium sp.; polynuclear aromatic hydrocarbons;
aliphatic
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 45 -
hydrocarbons; amino acid and peptides; glucose to fructose; glucose to
gluconic
acid; raffinose to sucrose and galactose; lactose; and sucrose.
9. increasing the expression of a polypeptide which is associated with the
rate limiting step in the production of commercially important enzymes from
micro-organisms, e.g., lactase from Aspergillus oryzae, Escherichia coli,
Bacillus
stearothermophilus;
10. increasing the expression of a polypeptide which is associated with
the rate limiting step in the growth of Saccharomyces on molasses; the growth
of
Candida on spent sulphite liquor; the growth of yeast on higher n-alkanes; the
growth of bacteria on higher n-alkanes; the growth of bacteria or yeast on
methane or methanol;
11. increasing the expression of a polypeptide which is associated with
the rate limiting step in the assimilation of atmospheric nitrogen by e.g.,
Azotobacteria sp., Rhizobium sp., or Cyanobacteria;
12. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of insecticides from Bacillus sp.,
e.g.,
Bacillus thuringiensis;
13. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of insecticides from entomogenous
fungi
2 0 such as Deuteromycetes, e.g., Verticillium lecanii and Hirsutella
thompsonii;
14. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of ethanol from cellulosic materials,
starch
crops, sugar cane, fodder beats, or molasses by, for example, Saccharomyces
cerevisiae, S. uvarum, Schizosaccharomyces pombe or Kluyveromyces sp.;
2 5 15. increasing the expression of a polypeptide which is associated with
the rate limiting step in acetic acid production from ethanol by Acetobacter
sp. or
Gluconobacter sp., the rate limiting step in the lactic acid production by the
family
of Lactobacillaceae, the rate limiting step in the citric acid production by
Candida
sp. or Aspergillus niger using e.g., molasses or starch, the rate limiting
step in
3 0 gluconic acid production by e.g., Pseudomonas sp., Gluconobacter sp., and
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 46 -
Acetobacter sp., or the rate limiting step in the production of amino acids by
bacteria or fungi;
16. increasing the expression of a enzyme in a cell, which enzyme
catalyzes the resolution of racemic mixtures of amino acids;
17. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of extracellular polysaccharides,
e.g., by
Corynebacterium sp., Pseudomonas sp., or Erwinia tahitica. Other examples
include the production of scleroglycan from the fungus Sclerotium sr.,
pullulan
from Aureobasidium pullulans, curdlan from Alcaligeans faecalis, and dextrans
from Streptobacterium sp. or Streptocucus sp. Other examples include anionic
polysacharides from Arthrobacter viscosus, bacterial alginates from
Azotobacter
vinelandii, and xanthan from Xanthomonas campestris;
18. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of an antifungal compound (e.g.,
Griseofulvin) and penicillins from Penicillium sps.;
19. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of antibiotics by fungi, e.g.,
polyether
antibiotics, chloramphenicol, ansamycines, tetracyclines, macrolides,
aminoglycosides, clavans, cephalosporins, cephamycins from Streptomyces sp.;
2 0 20. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of antitumor substances, e.g.,
actinomycin
D, anthracyclines, and bleomycin from Streptomyces sp.;
21. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of nucleic acids, nucleotides and
related
2 5 compounds, e.g., 5' inosinate (IMP), 5' guanylate (GMP), cAMP by e.g.,
Brevibacterium ammoniagenes;
22. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of vitamins, e.g., vitamin B 12 by
Pseudomonas denitrificans, Propionibacterium shermanii, or Rhodopseudomonas
3 0 protamicus;
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 47 -
23. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of riboflavin by Ashbya gossypii or
Bacillus subtilis.;
24. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of ergosterol by yeast, e.g.,
Saccharomyces
cerevisiae;
25. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of ergot alkaloids by Claviceps sp.;
26. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of secondary metabolites useful for
selected therapeutic uses in human medicine, e.g., cyclosporin from
Trichoderma
polysporum;
27. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of secondary products from plant cell
cultures, e.g., cinnamic acid derivatives in Coleus blume and shikonins from
Lithospermum erythrophizon;
28. increasing the expression of a polypeptide which is associated with
the rate limiting step in the production of wine and beer by Saccharomyces
sp.;
29. increasing the expression of a polypeptide which is associated with
2 0 the rate limiting step in the production of yogurt or cheese by
Staphylococcus sp.,
Lactobacillus sp. and Propionibacterium sp.;
30. increasing the expression of a polypeptide which is associated with
the rate limiting step in the fermentation of cocoa from Theobroma cacao by
fungi
and bacteria; and
2 5 31. increasing the expression of a polypeptide which is associated with
the rate limiting step in the fermentation of coffee beans from Coffea sp. by
fungi
and bacteria.
The gene of interest, whose expression is associated with a defined
physiological or pathological effect within a multicellular organism, may also
be a
3 0 plant gene. The plant gene may encode an agronomically important trait.
Examples of agronomically important traits may include, but are not limited
to,
germination, sprouting, flowering, fruit ripening, salt tolerance, herbicide
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 48 -
resistance, pesticide resistance, fungicide resistance, temperature
resistance, and
growth.
Additionally, in the practice of the invention the gene of interest may be a
protozoan gene. Examples of protozoans may include, but are not limited to, a
selection from the group consisting of Trypanosoma, Plasmodium, Leishmania,
Giardia, Entamoeba, Toxoplasma, Babesia, and Cryptosporidiosis. Moreover, the
gene of interest whose expression is associated with a defined physiological
or
pathological effect within a multicellular organism, may be a helminth gene.
Clearly, the present invention has commercial applications both in the case
where the polypeptide itself is commercially or therapeutically important, and
in
the case where expression of the polypeptide mediates the production of a
molecule which is commercially or therapeutically important.
A further use of the allosteric control modules of the present invention is
in the creation of conditional knockout and transgenic animals. In conditional
knockouts, the targeted gene product is expressed normally in the genetically
altered animal and expression is inhibited only in the presence of an
effector. In
applying the effectors described herein and the allosteric control modules
evolved
by the methods described herein to conditional knockouts, one desires to
evolve
allosteric self-cleaving RNAs that are activated by a small molecule effector
2 0 identified by the inventive methods. In this case, a DNA construct is
created
which codes for the gene product of interest that has been altered to place an
activatable self-cleaving allosteric control module comprising a catalytic
domain,
in a preferred embodiment, a ribozyme, in an intron or untranslated region of
the
gene. The choice of insertion site is made through empirical investigation so
that
2 5 the gene product is expressed at normal or near normal levels in the
absence of
effector, but is completely or nearly completely inhibited in the presence of
effector.
Thus, insertion of an allosteric ontrol module of the present invention in
place of the native gene through site specific genetic recombination methods
in
3 0 embryonic stem (ES) cells and subsequent micro-injection of the altered ES
cells
into blastocysts allows for the creation of an altered organism that normally
or
near normally expresses the gene of interest during development of the
genetically
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 49 -
altered organism. This allows for the gene product to be present during
development, thus eliminating problems of embryonic lethality or developmental
compensation. Subsequently, adult animals can be treated with the effector
molecule that activates the cis-acting catalytic domain, resulting in
degradation of
the RNA coding for the targeted gene product.
Alternatively, one could evolve an inhibitable self-splicing intron that
processes itself normally in the absence of effector. In the presence of
effector,
the activity of the self-splicing intron would be inhibited and the intron
would
remain in the mRNA resulting in inhibition of gene expression. In either case
of
activation or inhibition, direct assessment of the biological effects of
inhibiting
expression of the targeted gene product in the adult animal is achieved.
In the case of transgene over expression, one desires to evolve a cis-acting
inhibitable self cleaving ribozyme or an activatable self splicing intron in a
very
similar manner to that described for the application of RMST in a gene therapy
approach for human therapeutic applications. In this particular application,
the
insertion of the altered gene product that contains the cis-acting inhibitable
self-
cleaving allosteric control module or activatable self-splicing intron could
be
accomplished through standard methods employed for the creation of transgenic
animals. Swanson et al., Annu. Rep. Med. Chem., 29:265-274, 1994; Polites,
Int.
J. Exp. Pathol., 77(6):257-262, 1996. Alternatively, the altered gene
construct
could be introduced to adult animals via viral or naked DNA transfer methods,
akin to those being contemplated for gene therapy applications. In either
case,
over expression of the gene of interest would normally be inhibited due the
insertion of the desired allosteric control modules as described herein. The
2 5 subsequent dosing of the animal with the effector would then result in the
over
expression of the gene product for assessment of functional outcomes.
Cell therapy or ex vivo gene therapy, e.g., implantation of cells containing
the DNA constructs of the present invention, is also contemplated. This
3 0 embodiment would involve implanting cells containing the DNA constructs by
which the expression of the gene of interest is then regulated. In order to
minimize a potential immunological reaction, it is preferred that the cells be
of
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 50 -
human origin and produce a human gene of interest. It is envisioned, however,
that the vectors may be used to modify heterologous donor cells and xenogeneic
cells, as well as autologous cells, for delivery or implantation.
In some cases, vectors may be delivered through implanting into patients
certain cells that have been genetically engineered, using methods such as
those
described herein, to express and secrete the polypeptides, fragments,
variants, or
derivatives. Such cells may be animal or human cells, and may be derived from
the patient's own tissue (autologous) or from another source, either human
(allogeneic) or non-human (xenogeneic). Optionally, the cells may be
immortalized. In order to further decrease the chance of an immunological
response, the cells may be encapsulated to avoid the infiltration of
surrounding
tissues. The encapsulation materials are typically biocompatible, semi-
permeable
polymeric enclosures or membranes that allow release of the protein products)
but prevent destruction of the cells by the patient's immune system or by
other
detrimental factors from the surrounding tissues.
Techniques for the encapsulation of living cells are known in the art, and
the preparation of the encapsulated cells and their implantation in patients
may be
accomplished without undue experimentation. For example, Baetge et al. (WO
9505452; PCT/US94/09299) describe membrane capsules containing genetically
2 0 engineered cells for the effective delivery of biologically active
molecules. The
capsules are biocompatible and are easily retrievable. The capsules
encapsulate
cells transfected with recombinant DNA molecules comprising DNA sequences
coding for biologically active molecules operatively linked to promoters that
are
not subject to down regulation in vivo upon implantation into a mammalian
host.
2 5 The devices provide for the delivery of the molecules from living cells to
specific
sites within a recipient. In addition, see U.S. Patent Numbers 4,892,538,
5,011,472, and 5,106,627. A system for encapsulating living cells is described
in
PCT Application WO 9110425 of Aebischer et al. See also, PCT Application
WO 9110470 of Aebischer et al., Winn et al., Exper. Neurol. 113:322-329, 1991,
3 0 Aebischer et al., Exper. Neurol. 111:269-275, 1991; and Tresco et al.,
ASAIO
38:17-23, 1992.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 51 -
Promoter
Those skilled in the art will appreciate that it may also be beneficial to
provide a promoter with the gene of interest as well as one more additional
operational control sequences. The choice of promoter or other operational
control sequences, however, is not limiting to the selection of effectors, the
evolution of aptamers or the construction and use of the allosteric control
modules
of the present invention.
Promoter regions vary in length and sequence, and can further encompass
one or more DNA-binding sites for sequence-specific DNA binding proteins,
and/or an enhancer or silencer. The present invention may employ, for example,
a
CMV promoter or a PS promoter. Such promoters, as well as mutations thereof,
are well known and have been described in the art (see, e.g., Hennighausen et
al.,
EMBO J. 5:1367-1371, 1986; Lehner et al., J. Clin. Microbiol. 29:2494-2502,
1991; Lang et al., Nucleic Acids Res. 20:3287-95, 1992; Srivastava et al., J.
Virol.
45:555-564, 1983; and Green et al., J. Virol. 36:79-92, 1980). Other
promoters,
however, can also be employed, such as the Ad2 or Ad5 major late promoter and
tripartite leader, the Rous sarcoma virus (RSV) long terminal repeat, and
other
constitutive promoters, as have been described in the literature. For
instance, the
herpes thymidine kinase promoter (Wagner et al., Proc. Natl. Acad. Sci. USA.,
78:144-145, 1981), the regulatory sequences of the metallothionine gene
(Brinster
et al., Nature, 296:39-42, 1982) promoter elements from yeast or other fungi,
such
as the Gal 4 promoter, the alcohol dehydrogenase promoter, the phosphoglycerol
kinase promoter, and the alkaline phosphatase promoter, can be employed.
Similarly, promoters isolated from the genome of mammalian cells or from
viruses that grow in these cells (e.g., Ad, SV40, CMV, and the like) can be
used.
Delivery
The DNA constructs described herein can be incorporated into a variety of
vectors for introduction into cells. Suitable vectors include, but are not
restricted
3 0 to, naked DNA, plasmid DNA vectors, viral DNA vectors (such as adenovirus
or
adeno-associated virus vectors), viral RNA vectors (such as retroviral or
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 52 -
alphavirus vectors) (for a review see Couture and Stinchcomb, 1996, supra) and
non-viral vectors (such as DNA complexed with cationic lipids or packaged
within liposomes). It will be appreciated by those skilled in the art that an
expression vector will also include a) a transcription initiation region; b) a
transcription termination region, and c) expression control sequences. It will
also
be appreciated that the DNA constructs and vectors may be produced by joining
separately produced components. For example, the promoter, aptamer and
catalytic domain may be separately manufactured by chemical synthesis or
recombinant DNA/RNA technology and then joined.
Vectors containing the DNA constructs of the present invention may be
delivered to cells by a variety of plasmid and non-viral delivery methods
known to
those familiar with the art, including, but not restricted to, liposome-
mediated
transfer or lipofection, by incorporation into other delivery vehicles, such
as
hydrogels, cyclodextrins, biodegradable nanocapsules, and bioadhesive
microspheres, naked DNA delivery (direct injection or direct uptake), receptor-
mediated transfer (ligand-DNA complex), electroporation, calcium phosphate
mediated transformation, microinjection, osmotic shock, microparticle
bombardment (e.g., gene gun), bio-chip materials and combinations of the
above.
Delivery materials and methods may also involve the use of components
2 0 including, but not limited to, inducible promoters, tissue-specific
enhancer-
promoters, DNA sequences designed for site-specific integration, DNA sequences
capable of providing a selective advantage over the parent cell, labels to
identify
transformed cells, negative selection systems (safety measures), cell-specific
binding agents (for cell targeting), cell-specific internalization factors,
2 5 transcription factors to enhance expression by a vector as well as methods
of
vector manufacture. Such additional methods and materials for the practice of
gene therapy techniques are described in U.S. Patent No. 4,970,154
electroporation techniques; WO 9640958 nuclear ligands; U.S. Patent No.
5,679,559 concerning a lipoprotein-containing system for gene delivery; U.S.
3 0 Patent No. 5,676,954 involving liposome carriers; U.S. Patent No.
5,593,875
concerning methods for calcium phosphate transfection; and U.S. Patent
No. 4,945,050 wherein biologically active particles are propelled at cells at
a
CA 02401654 2002-08-28
WO 01/64956 PCT/US01/06615
- 53 -
speed whereby the particles penetrate the surface of the cells and become
incorporated into the interior of the cells.
A typical use of the constructs of the present invention involves the
transfer of a vector to a recipient cell. The recipient cell or host cell is
typically a
prokaryotic cell in protein production techniques and is preferably a
eukaryotic
cell in gene therapy techniques. The eukaryotic host cell can be modified
either in
vitro or in vivo. According to the invention, "contacting" of cells with the
vectors
of the present invention can be by any means by which the vectors will be
introduced into the cell. In one preferred embodiment, viral vectors will be
introduced by infection using the natural capability of the virus to enter
cells (e.g.,
the capability of adenovirus to enter cells via receptor-mediated
endocytosis).
The viral and plasmid vectors, however, can be introduced by any suitable
means.
Suitable viral vectors include, but are not limited to, retrovirus,
adenovirus, herpes simplex virus, lentivirus, hepatitis virus, parvovirus,
papovavirus, poxvirus, alphavirus, coronavirus, rhabdovirus, paramyxovirus,
and
papilloma virus vectors. U.S. Patent No. 5,672,344 describes an in vivo viral-
mediated gene transfer system involving a recombinant neurotrophic HSV-1
vector. U.S. Patent No. 5,399,346 provides examples of a process for providing
a
patient with a therapeutic protein by the delivery of human cells which have
been
2 0 treated in vitro to insert a DNA segment encoding a therapeutic protein.
Additional methods and materials for the practice of gene therapy techniques
are
described in U.S. Patent No. 5,631,236 involving adenoviral vectors; U.S.
Patent
No. 5,672,510 involving retroviral vectors; and U.S. Patent No. 5,635,399
involving retroviral vectors expressing cytokines.
2 5 Preferably, the vectors persist in the cells to which they are delivered.
Alternatively, some vectors may be used that provide for transient expression
of
the DNA constructs. Such vectors might be repeatedly administered as
necessary.
Compositions and Administration
3 0 Host cells may be transformed with the vectors of the present invention
either in vivo or in vitro. If in vitro, the desired target cell type may be
removed
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 54 -
from the subject, recombinantly modified by vector delivery, and reintroduced
into the subject. The modified cells can be screened for those cells harboring
the
gene of interest, using conventional techniques such as Southern blots and/or
PCR. Such modified cells may be transplanted directly into the subject or may
be
placed in a device which is implanted in the subject. It is also envisioned
that the
ex vivo modified cells may include non-human cell lines for direct or indirect
implantation.
If delivered irZ vivo, the vector may be formulated into a pharmaceutical
composition. The vector may be administered parenterally, e.g., by injection
intravenously, intraperitoneally, intramuscularly, intrathecally or
subcutaneously.
Additional vector formulations suitable for other modes of administration
include
oral (including sublingual and buccal) and pulmonary (intranasal and
inhalation)
formulations, topical and transdermal formulations, and suppositories.
Delivery
may involve a single dose schedule or a multiple dose schedule.
For example, the intramuscular injection of a vector of the present
invention, such as a rAAV particles containing the DNA construct, may provide
efficient transduction of postmitotic muscle fibers and prolonged transgene
expression. According to the invention, this is accomplished without
significant
inflammation or activation of immunity to the transgene product. Muscle is
also
2 0 particularly well suited for the production of secreted therapeutic
protein, such as
factor IX or apolipoprotein (Apo) E, among other genes of interest.
When administered by injection, it will be appreciated by those skilled in
the art that the vectors of the present invention are typically suspended in a
biologically compatible solution or pharmaceutically acceptable delivery
vehicle.
2 5 One such suitable and common vehicle is sterile saline. Other aqueous and
non-
aqueous isotonic sterile injection solutions and aqueous and non-aqueous
sterile
suspensions may also be employed and are well known as suitable
pharmaceutically acceptable carriers in the art.
In one embodiment of the present invention the vectors contain a gene
3 0 encoding a therapeutic protein. The vectors are administered in sufficient
amounts to provide sufficient levels of expression of the selected protein
such that
a therapeutic benefit may be obtained without undue adverse effects and with
CA 02401654 2002-08-28
WO 01/64956 PCT/US01/06615
- 55 -
medically acceptable physiological effects which can be determined by those
skilled in the medical arts. Dosages of the vector will depend primarily on
factors
such as the condition being treated, the selected transgene, the age, weight
and
health of the patient, and thus, may vary among patients. For example, a
therapeutically effective dose of the vector of the present invention may be
in the
range of from about 1 to about 50 ml of saline solution containing
concentrations
of from about 1 x 10$ to 1 x 10" particles/ml rAAV virions containing the DNA
constructs of the present invention. A more preferred human dosage may be
about 1-20 ml saline solution at the above concentrations. The levels of
expression of the selected gene can be monitored to determine the selection,
adjustment or frequency of administration. Administration of the vector might
then be repeated as needed.
When practiced in vivo, any suitable organs or tissues or component cells
can be targeted for vector delivery. Preferably, the organs/tissues/cells
employed
are of the circulatory system (i.e., heart, blood vessels or blood),
respiratory
system (i.e., nose, pharynx, larynx, trachea, bronchi, bronchioles, lungs),
gastrointestinal system (i.e., mouth, pharynx, esophagus, stomach, intestines,
salivary glands, pancreas, liver, gallbladder), urinary system (i.e., kidneys,
ureters,
urinary bladder, urethra), nervous system (i.e., brain and spinal cord, and
special
2 0 sense organs such as the eye) and integumentary system (i.e., skin). Even
more
preferably the cells being targeted are selected from the group consisting of
heart,
blood vessel, lung, liver, gallbladder, urinary bladder, and eye cells.
Accordingly, the present invention also provides a method of obtaining
stable gene expression in a host, or modulating gene expression in a host,
which
2 5 comprises administering the vectors of the present invention using any of
the
aforementioned formulations and routes of administration or alternative routes
known to those skilled in the art and appropriate for a particular
application. The
"effective amount" of the vector or pharmaceutical composition is such as to
produce the desired effect in a host which can be monitored using several end-
3 0 points known to those skilled in the art. For example, effective nucleic
acid
transfer to a host cell could be monitored in terms of a therapeutic effect
(e.g.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 56 -
alleviation of some symptom associated with the disease or syndrome being
treated), or by further evidence of the transferred gene or coding sequence or
its
expression within the host (e.g., using the polymerase chain reaction,
Northern or
Southern hybridizations, or transcription assays to detect the nucleic acid in
host
cells, or using immunoblot analysis, antibody-mediated detection, or
particularized assays to detect protein or polypeptide encoded by the
transferred
nucleic acid, or impacted in level or function due to such transfer). One such
particularized assay includes an assay for expression of a reporter or marker
gene.
While it will be appreciated that the constructs of the present invention
may be used with any vector system, a preferred system involving the use of
rAAV particles is described as an exemplary system for the following
descriptions
of pharmaceutical compositions. Such compositions may comprise a
therapeutically effective amount of an rAAV particle product in admixture with
a
pharmaceutically acceptable agent such as a pharmaceutically acceptable
carrier.
An exemplary carrier material may be water for injection, preferably
supplemented with other materials common in solutions for administration to
mammals. Typically, an rAAV particle therapeutic compound will be
administered in the form of a composition comprising particles in conjunction
with one or more physiologically acceptable agents. Neutral buffered saline or
2 0 saline mixed with serum albumin are exemplary appropriate Garners. Other
standard pharmaceutically acceptable agents may be included as desired. For
example, other compositions may comprise a buffer or preservative.
The rAAV particle pharmaceutical compositions typically include a
therapeutically or prophylactically effective amount of rAAV particles in
2 5 admixture with one or more pharmaceutically and physiologically acceptable
formulation agents selected for suitability with the mode of administration.
Suitable formulation materials or pharmaceutically acceptable agents include,
but
are not limited to, antioxidants, preservatives, diluting agents, suspending
agents,
solvents, fillers, bulking agents, buffers, delivery vehicles, and diluents.
For
3 0 example, a suitable vehicle may be water for injection, physiological
saline
solution, or artificial cerebrospinal fluid, possibly supplemented with other
materials common in compositions for parenteral administration. Neutral
CA 02401654 2002-08-28
WO 01/64956 PCT/US01/06615
- 57 -
buffered saline or saline mixed with serum albumin are further exemplary
vehicles. The term "pharmaceutically acceptable carrier" or "physiologically
acceptable Garner" as used herein refers to a formulation agents) suitable for
accomplishing or enhancing the delivery of the rAVV particles as a
pharmaceutical composition.
The primary solvent in a composition may be either aqueous or non-
aqueous in nature. In addition, the vehicle may contain other formulation
materials for modifying or maintaining the pH, osmolarity, viscosity, clarity,
color, sterility, stability, rate of dissolution, or odor of the formulation.
Similarly,
the composition may contain additional formulation materials for modifying or
maintaining the rate of release of rAVV particles, or for promoting the
absorption
or penetration of rAVV particles.
When systemically administered, the therapeutic compositions for use in
this invention may be in the form of a pyrogen-free, parenterally acceptable
aqueous solution. The preparation of such pharmaceutically acceptable
solutions,
with due regard to pH, isotonicity, stability and the like, is within the
skill of the
art.
Therapeutic formulations of rAVV particles compositions useful for
practicing the present invention may be prepared for storage by mixing the
2 0 selected composition having the desired degree of purity with optional
physiologically acceptable stabilizers (Remington's Pharmaceutical Sciences,
18th Edition, A.R. Gennaro, ed., Mack Publishing Company, 1990). Acceptable
stabilizers preferably are nontoxic to recipients and are preferably inert at
the
dosages and concentrations employed, and preferably include buffers such as
2 5 phosphate, citrate, or other organic acids; antioxidants such as ascorbic
acid; low
molecular weight polypeptides; proteins, such as serum albumin, gelatin, or
immunoglobulins; hydrophilic polymers such as polyvinylpyrrolidone; amino
acids such as glycine, glutamine, asparagine, arginine or lysine;
monosaccharides,
disaccharides, and other carbohydrates including glucose, mannose, or
dextrins;
3 0 chelating agents such as EDTA; sugar alcohols such as mannitol or
sorbitol; salt-
forming counterions such as sodium; and/or nonionic surfactants such as Tween,
pluronics or polyethylene glycol (PEG).
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 58 -
The optimal pharmaceutical formulation will be determined by one skilled
in the art depending upon the intended route of administration, delivery
format
and desired dosage. See for example, Remington's Pharmaceutical Sciences, 18th
Ed. (Mack Publishing Co., Easton, PA 18042 pages 1435-1712, 1990.)
An effective amount of an rAVV particle composition to be employed
therapeutically will depend, for example, upon the therapeutic objectives such
as
the indication for which the gene delivered by the rAVV particle is being
used,
the route of administration, and the condition of the patient. Accordingly, it
may
be necessary for the therapist to titer the dosage and modify the route of
administration as required to obtain the optimal therapeutic effect.
Typically, a
clinician will administer the composition until a transgene dosage is reached
that
achieves the desired effect. The composition may therefore be administered as
a
single dose, or as two or more doses (which may or may not contain the same
amount of rAAV particles) over time, or as a continuous infusion via
implantation
device or catheter.
As further studies are conducted, information will emerge regarding
appropriate dosage levels for the treatment of various conditions in various
patients, and the ordinary skilled worker, considering the therapeutic
context, the
type of disorder under treatment, the age and general health of the recipient,
will
2 0 be able to ascertain proper dosing.
A particularly suitable vehicle for parenteral injection is sterile distilled
water in which the rAVV particle composition is formulated as a sterile,
isotonic
solution, properly preserved. Yet another preparation may involve the
formulation of rAVV particles with an agent, such as injectable microspheres,
2 5 bio-erodible particles or beads, or liposomes, that provides for the
controlled or
sustained release of the product which may then be delivered as a depot
injection.
The preparations of the present invention may include other components,
for example parenterally acceptable preservatives, tonicity agents,
cosolvents,
wetting agents, complexing agents, buffering agents, antimicrobials,
antioxidants
3 0 and surfactants, as are well known in the art. For example, suitable
tonicity
enhancing agents include alkali metal halides (preferably sodium or potassium
chloride), mannitol, sorbitol and the like. Suitable preservatives include,
but are
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 59 -
not limited to, benzalkonium chloride, thimerosal, phenethyl alcohol,
methylparaben, propylparaben, chlorhexidine, sorbic acid and the like.
Hydrogen
peroxide may also be used as preservative. Suitable cosolvents are for example
glycerin, propylene glycol and polyethylene glycol. Suitable complexing agents
are for example caffeine, polyvinylpyrrolidone, beta-cyclodextrin or
hydroxypropyl-beta-cyclodextrin. Suitable surfactants or wetting agents
include
sorbitan esters, polysorbates such as polysorbate 80, tromethamine, lecithin,
cholesterol, tyloxapal and the like. The buffers can be conventional buffers
such
as borate, citrate, phosphate, bicarbonate, or Tris-HCI.
The formulation components are present in concentrations that are
acceptable to the site of administration. For example, buffers are used to
maintain
the composition at physiological pH or at slightly lower pH, typically within
a pH
range of from about 5 to about 8.
A pharmaceutical composition may be formulated for inhalation. For
example, rAVV particles may be formulated as a dry powder for inhalation.
Alternatively, rAVV particle inhalation solutions may be formulated in a
liquefied
propellant for aerosol delivery. In yet another formulation, solutions may be
nebulized.
Additional rAVV particle formulations will be evident to those skilled in
2 0 the art, including formulations involving rAVV particles in combination
with one
or more other therapeutic agents. Techniques for formulating a variety of
other
sustained- or controlled-delivery means, such as liposome carriers, bio-
erodible
microparticles or porous beads and depot injections, are also known to those
skilled in the art.
2 5 Regardless of the manner of administration, the specific dose may be
calculated by the delivery of a gene that produces a therapeutic effect
according to
body weight, body surface area or organ size. Further refinement of the
calculations necessary to determine the appropriate dosage for treatment
involving
each of the above mentioned formulations is routinely made by those of
ordinary
3 0 skill in the art and is within the ambit of tasks routinely performed by
them.
Appropriate dosages may be ascertained through use of appropriate dose-
response
data.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 60 -
The route of administration of the composition is in accord with known
methods, e.g., inhalation, injection or infusion by intravenous,
intraperitoneal,
intracerebral (intraparenchymal), intracerebroventricular, intramuscular,
intraocular, intraarterial, or intralesional routes, or by sustained release
systems
which may optionally involve the use of a catheter.
In this regard, the cell to be transformed will depend on the purpose for
gene transfer; for example, the disease state being treated. For example, the
rAAV particle can be used to deliver selected nucleotide sequences into any
nucleated cell including stem, progenitor and erythroid cells; as well as any
of the
various white blood cells such as lymphocytes, neutrophils, eosinophils,
basophils, monocytes; tissue specific cells, such as those derived from lung,
heart,
kidney, liver, spleen, pancreatic tissue, connective tissue, bone tissue
including
osteocytes, gangliocytes, epithelial and endothelial cells, ependymal cells,
reticuloendothelial cells, dendritic and neural cells, skeletal muscle,
cardiac
muscle and smooth muscle cells, and the like. It is further envisioned that
the
constructs of the present invention are useful in the delivery of a gene of
interest
to tumor cells and pathogen infected cells. AAV has been reported to infect
all
established cell lines thus far examined.
It will also be appreciated that the same dosage calculations and
2 0 considerations of routes of administration and pharmaceutical formulations
are
applicable to the effector used to alter the expression of the gene of
interest.
EXAMPLES
The following non-limiting examples further illustrate the identification
and selection methods as well as the synthesis and use of exemplary constructs
of
the present invention. Those of ordinary skill will recognize that these are
non-
limiting examples and that the present invention discloses a means to provide
a
3 0 broad array of other effectors, allosteric control modules and vectors
which can be
used as described herein for the regulation of gene expression.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 61 -
Example 1
Effector Identification
A method for identifying an effector for use in the evolution of aptamers
and the alteration of gene expression involves the following steps. The first
step
is the selection a set of desired characteristics for an effector, wherein the
desired
characteristics include one or more of the following:
a) at least 1 7o bioavailability;
b) biodistribution to tissue containing an allosteric
control module;
c) the ability to pass to the nucleus of the cell;
d) either no drug interactions or manageable drug interactions;
e) either no toxicity or acceptable toxicity at the
dosage range used;
f) either no side effects or acceptable side effects
at the dosage range used;
g) either no pharmacological effect at the dosage range
used in regulating
transgene expression or a negligible pharmacological
effect; and
h) physical properties suitable for the in vitro evolution
of an aptamer.
The selected
characteristics
indicate
that
the effector
is suitable
for aptamer
generation, human consumption and use with an allosteric control module for
the
2 0 alteration of gene expression. Second, one or more databases containing
information on the selected effector characteristics is accessed to evaluate
these
attributes and a set of effectors having the selected characteristics is
identified.
The effectors may then be used to generate aptamers by means of in vitro
evolution.
Physical properties which will further aid in the identification of a suitable
effector for the in vitro evolution of an aptamer include, but are not limited
to the
following attributes. S
~ sufficient structural and functional group complexity to provide interactive
3 0 sites for the identification of high affinity aptamers; preferably the
molecules will be rigid, planar entities
CA 02401654 2002-08-28
WO 01/64956 PCT/USOi/06615
- 62 -
~ solubility in an aqueous solution at a level which allows molecular
evolution techniques to be performed
~ appropriate charge, i.e., lack of an excessive number of ionizable groups
that would lead to unfavorable interactions with nucleic acid ligands
Thus, highly flexible, lipophilic molecules are not favored.
For purposes of the present invention, manageable drug-drug interactions
are interactions that can be avoided by avoiding concomitant use of the drugs
or
reduced through a combination of monitoring and dose adjustments. For purposes
of the present invention, acceptable toxicity at the dosage range used
includes the
use of the effector at a dose which is not toxic or which has reversible
toxicity or
is acceptable in view of desired treatment. For purposes of the present
invention,
acceptable side effects at the dosage range used includes side effects which
abate
or end with continued use and those which are managed by other means (for
example, prevented or suppressed by the use of other drugs, diet, etc. )
Using databases, such as those described above, the following molecules
were identified for evaluation as potential effectors:
1) Atevirdine / nonnucleoside Pharmacia & Upjohn
reverse
transcriptase inhibitor
Development Status Phase III; discontinued / more
potent
successor
Bioavailability: Oral; well absorbed orally,
30-60 minutes
to peak serum
Biodistribution: Widely distributed, crosses
blood-brain
barrier and placenta; maximum
concentration of drug in serum
(oral
delivery): 7.3 uM
Intracellular Localization: Intracellular target (HIV RT)
Toxicity / Drug Interactions: Well tolerated (tested up to
1600 mg oral
I, delivery); no effects on vital
signs, ECG
CA 02401654 2002-08-28
WO 01/64956 PCT/USOI/06615
- 63 -
or lab tests; buffering interferes
with
absorption; rash in some symptomatic
HIV patients
Other Pharmacology: Viral target, no other known
activity
Target Localization: Intracellular (HIV RT)
Functionality for Aptamer: Hydrophobic, basic, large surface
Synthesis / Scale Up: Produced in mufti-ton quantities
Further described in: W091/9849 1991, priority US
457483
EtNH
H ~ N
N ~-~ N
Me0
Structure:
2) Talviraline / nonnucleoside Bayer/Hoechst Marion/Glaxo-Wellcome
reverse (BAY-10-8979, HBY-097)
transcriptase inhibitor
Development Status: Phase II; discontinued / more
potent
successor
Bioavailability: Oral absorption (375-3000 mg/day
in
Phase II)
Biodistribution: Wide
Intracellular Localization: Intracellular Target (HIV RT)
Toxicity: Well tolerated (rash in two
patients who
received dosing three times
daily)
Other Pharmacology: Viral target, no other known
activity
Target Localization: Intracellular (HIV RT)
Functionality for Aptamer: Hydrophobic, basic
Synthesis / Scale Up: Produced in sufficient quantities
for 180
patient Phase II clinical trial
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 64 -
O OPr-i
Me0 / N
S SMe
N S
H
Structure:
3) Quinolones/ DNA topoisomerasemultiple sources
inhibitors
Development Status: Phase II, III, active
Bioavailability: Oral
Biodistribution: Wide / muscle
Toxicity: Acceptable / phototoxicity
Other Pharmacology: Antibacterial
Target Localization: Intracellular / intrabacterial
Name(s): Fandofloxacin, DW-116
F
Met ~ C02 H
HC1
Structure:
Development status: Dong Wa, Phase II complete, development
ongoing
Criteria:
Bioavailability: Orally absorbed
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 65 -
Biodistribution: Good organ distribution including muscle
Intracellular Localization: Intracellular, intrabacterial
Toxicity / Drug Interactions: Well tolerated, v. low phototoxicity
Other Pharmacological Activity: Anti-infective, anti-microbial
Functionality for Aptamer: Basic, hydrophobic
Synthesis / Scale Up: Sufficient for Phase II in Europe and Korea
Further description: U.S. 5,496,947
Name(s): Bay-y-3118
C02H
Structure: ~ Hcl
Development status: Bayer, Phase II 1993, patent on more potent
analogs
Criteria:
Bioavailability: Orally absorbed (Tmax 1.3 Hrs, Half life
11.4 Hrs
Intracellular Localization: Intracellular, intrabacterial
Toxicity / Drug Interactions: No abnormal findings in humans
Other Pharmacological Activity: Anti-infective, anti-microbial
Functionality for Aptamer: Basic, hydrophobic
2 0 Synthesis / Scale Up: Phase II supplied
Further description: EP 520240
4) Vitamin D3 analogs Leo
Development Status: Phase II, III, active
Bioavailability: Low Oral (Form. Diff.) /transdermal
CA 02401654 2002-08-28
WO 01/64956 PCT/USOI/06615
- 66 -
Biodistribution: Highly potent/ side effects
common
Toxicity: Hypercalcemia
Other Pharmacology: Vitamin D receptor
Target Localization: Intracellular (nuclear receptor)
5) MCC-555 / peroxisome proliferationJohnson & Johnson / Mitsubishi
activated receptor
Development Status: Phase II; active / diabetes
Bioavailability: Oral
Biodistribution: Muscle and adipose
Toxicity: Liver toxicity (controversial)
Other Pharmacology: Peroxisome proliferation activated
receptor / hypoglycaemia
Target Localization: Nuclear
6) GW-2570 / peroxisome proliferationGlaxo
activated receptor
Development Status: Phase II; active / diabetes
Bioavailability: Oral
Biodistribution: Muscle and adipose
Toxicity: Unknown
Other Pharmacology: Peroxisome proliferation activated
receptor / hypoglycaemia
Target Localization: Nuclear; high stereoselectivity
for active
component
7) Triclabendazole Novartis
Development Status: Phase II; third world, veterinary
(Fasinex)
Bioavailability: Oral
Biodistribution: Liver / metabolism important
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 67 -
Toxicity: Low / none in humans
Other Pharmacology: antihelmintic / flukecide
Target Localization: Unknown
Structure
C / N SMe
Cl
1 Cl
Currently, the preferred effectors include the nonnucleoside reverse
transcriptase inhibitors. These compounds have good oral bioavailability, are
nontoxic and have no known activity other than against the viral target.
Example 2
In Vitro Evolution
An exemplary in vitro evolution strategy is described as follows. A
random pool of nucleic acids is synthesized wherein, each member contains two
portions: a) one portion consists of a region with a defined (known)
nucleotide
sequence; b) the second portion consists of a region with a degenerate
(random)
sequence. The known nucleotide sequences may provide several advantages/uses.
For example, a certain nucleotide sequence may be known or expected to bind to
a given effector. Alternatively, the known sequence may facilitate or provide
for
2 0 complementary DNA (cDNA) synthesis and PCR amplification of molecules
selected for their effector binding. In yet another aspect, the sequences may
be
used to introduce a restriction endonuclease site for the purpose of cloning.
The
random sequence can be created to be completely random (each of the four
nucleotides represented at every position within the random region) or the
2 5 degeneracy can be partial (Beaudry and Joyce, 1992 supra) and involve the
use of
rational design. Sequence variation in test nucleic acids can be introduced or
CA 02401654 2002-08-28
WO 01/64956 PCT/US01/06615
- 68 -
increased by mutagenesis before or during the selection/amplification
iterations.
This random library of nucleic acids is incubated under conditions that ensure
folding of the nucleic acids into conformations that facilitate the desired
activity
(e.g., effector binding and catalysis). Following incubation, nucleic acids
are
converted into complementary DNA (if the starting pool of nucleic acids is
RNA).
Nucleic acids with desired trait may be separated or partitioned from the
rest of the population of nucleic acids by a variety of methods. For example,
a
filter-binding assay can be used to separate the fraction that binds the
desired
effector from those that do not. The fraction of the population that is bound
by
the effector (for example) may be the population that is desired (active
pool).
Typically, the binding of the effector and the RNA is assessed by applying the
RNA pool or mixture to an affinity matrix containing the effector with which
the
aptamer will specifically bind or react. The non-binding RNA species are
removed or washed away, and the specifically-binding species are eluted from
the
effector for further use in the evolution process. A new piece of DNA
(containing
new oligonucleotide primer binding sites for PCR and restriction sites for
cloning)
may be introduced to the termini of molecules in the active pool (to reduce
the
chances of contamination from previous cycles of selection) to facilitate PCR
amplification and subsequent cycles (if necessary) of evolution.
2 0 Amplification is preferably performed by means of reverse transcription of
the eluted species into DNA followed by polymerise chain reaction. The result
of
the amplification process is the production of a large number of the selected
RNA-encoding DNA molecules. The final pool of nucleic acids with the desired
trait (i.e., aptamer(s) which bind to the effector) may be cloned into a
plasmid
2 5 vector and transformed into bacterial hosts. Recombinant plasmids can then
be
isolated from transformed bacteria, and the identity of clones can be
determined
using DNA sequencing techniques.
Multiple cycles of selection and amplification should result in the selective
enrichment of RNA species that bind to the effector tightly and specifically.
3 0 Cycles of selection and amplification are repeated until a desired goal is
achieved.
With the current techniques and materials, the cycles are typically repeated
five or
more times. In the most general case, the evolutionary steps of selection and
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 69 -
amplification may be continued until no significant improvement in binding
strength of RNA to effector is achieved upon repetition of a cycle.
In some cases, however, it is not necessarily desirable to repeat the
iterative steps of in vitro evolution until a single RNA is identified. The
RNA
pool may include a family of nucleic acid structures or motifs that have a
number
of conserved sequences and a number of sequences which can be substituted or
added without significantly effecting the affinity of the nucleic acid
molecules to
the effector. By terminating the process prior to the identification of a
single
aptamer, it is possible to determine the sequence of a number of suitable
RNAs.
To further improve specificity, a negative selection process can also be
used. A negative selection procedure may be employed before, during or after
the
in vitro evolution process. The negative selection provides the ability to
discriminate between closely related but different effectors. Thus, negative
selection can be introduced to identify aptamers that have a high specificity
for an
effector but do not recognize either other members of the effector's family or
other structurally similar molecules. For example, in the evolution of an
aptamer
for theophylline, caffeine may be used to counter-select and eliminate those
aptamers which would cross-react with the structurally similar molecule
caffeine.
One post-evolution method would be to perform a negative selection on a
2 0 pool that has already been evolved against the desired effector. The
process
would involve the use of either an effector family member or a molecule
structurally similar to the desired effector as the negative selection target.
The
selected population is passed over an affinity column containing the negative
selection target and those nucleic acids which bind to the negative selection
target
2 5 are removed from the selected pool. Alternatively, the selected population
may be
passed over an affinity column containing the desired effector, and the pool
is
then challenged by the addition of a negative selection target. Preferably,
this
process would also involve the performance of two to three negative selections
using the negative selection target and a late-round, highly evolved pool that
was
3 0 evolved using the effector. The binding of certain sequences to the
negative
selection target would be used to subtract those sequences from the evolved
pool.
This method allows one to quickly eliminate from several hundred to several
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 70 -
thousand nucleic acid sequences that demonstrate a high affinity for both the
effector and molecules having similar structural characteristics.
It will be appreciated that "separating" or "partitioning" may include any
process for separating the selected RNAs from the remainder of the unreacted
RNA candidate mixture. Separation can be accomplished by various methods
known in the art. Filter binding, size separation, affinity chromatography,
liquid-
liquid partitioning, filtration, gel shift, density gradient centrifugation
are all
examples of suitable partitioning methods. Separation may also be accomplished
by means of the presence of a PCR priming site that remains on the
catalytically
inactive RNA. Equilibrium partitioning methods can also be used. The simple
partitioning methods include any method for separating a solid from a liquid,
such
as, centrifugation with and without oils, membrane separations and simply
washing. The RNAs may also be specifically eluted from the effector with a
specific antibody or ligand. The choice of partitioning method will depend on
properties of the effector and the RNA and can be made according to principles
and properties known to those of ordinary skill in the art.
The amplification process may be any process or combination of process
steps that increases the amount or number of copies of a molecule or class of
2 0 molecules. In preferred embodiments, amplification occurs after members of
the
test mixture have been partitioned, and it is the facilitating nucleic acid
associated
with a desirable product that is amplified. For example, the amplification of
RNA
molecules can be carried out by a sequence of three reactions: the use of
reverse
transcription to make cDNA copies of selected RNAs, the use of the polymerise
2 5 chain reaction to increase the copy number of each cDNA, and the
transcription of
the cDNA copies to obtain RNA molecules having the same sequences as the
selected RNAs. Any reaction or combination of reactions known in the art can
be
used as appropriate, including direct DNA replication, direct RNA
amplification
and the like, as will be recognized by those skilled in the art. The
amplification
3 0 method should result in the proportions of the amplified mixture being
essentially
representative of the proportions of different sequences in the mixture prior
to
amplification.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 71 -
Example 3
Allosteric Control Module
The allosteric control module contains a catalytic domain and an aptamer,
evolved as described above, that is selected such that the interaction of the
aptamer with an effector alters the activity of the allosteric control module.
The
interaction of the effector and aptamer may result in an alteration of the
catalytic
activity of the catalytic domain. Depending upon the selection of the
components,
the alteration can result in either an increase or a decrease in the activity
of the
catalytic domain of the module.
Ribozymes, ribozyme-like molecules and portions of such molecules are
used to form the catalytic domains of the present invention. Ribozymes which
are
useful in the present invention include, but are not limited to, molecules in
the
classes of hammerhead, axehead, hairpin, hepatitis delta virus, neurospora,
self-
splicing introns (including group I and group II), newt satellite ribozymes,
Tetrahymena ribozymes, ligases, peptide ligases, phosphatases and polymerases.
The nucleic acids of these molecules may be used or the molecules may be used
2 0 as the starting point for the production of ribozyme-like, synthetic, non-
naturally
occurring sequences.
The allosteric control module may include additional components or
domains including a substrate domain (for example, in the case of self-
cleaving
catalytic domains) or a recognition domain (to aid in the recognition of the
site at
2 5 which catalytic activity is to be directed). The allosteric control module
may also
be designed such that the catalytic domain and aptamer are joined by a
structural
bridge, wherein the interaction of the aptamer and effector results in an
alteration
of the bridge which in turn results in an alteration of catalytic activity of
the
catalytic domain (see Soukup and Breaker, Proc. Nat. Acad. Sci. USA, 96:3584-
3 0 3589, 1999).
CA 02401654 2002-08-28
WO 01/64956 PCT/US01/06615
- 72 -
The allosteric control module is then tested in vitro and/or in vivo. The
optimal constructs in an effector-activated expression control system will
provide
the maximum inhibition of expression in the absence of effector and the
maximum enhancement of expression in the presence of effector. In an effector-
inactivated expression control system, the optimal constructs will provide the
maximum inhibition of expression in the presence of effector and the maximum
enhancement of expression in the absence of effector.
While not being limiting to the present invention, the catalytic domain
may be synthesized by procedures for normal chemical synthesis of RNA as
described in Usman et al., J. Am. Chem. Soc. 109:7845, 1987; Scaringe et al.,
Nucleic Acids Res. 18:5433, 1990; and Wincott et al., Nucleic Acids Res.
23:2677-2684, 1995. The details will not be repeated here, but such procedures
may involve the use of common nucleic acid protecting and coupling groups,
such
as dimethoxytrityl at the 5'-end, and phosphoramidites at the 3'-end.
In addition, the catalytic activity of the molecules can be optimized as
described by Draper et al., PCT W093/23569, and Sullivan et al., PCT
W094/02595; Ohkawa et al., Nucleic Acids Symp. Ser. 27:15-6, 1992; Taira et
al.,
Nucleic Acids Res. 19:5125-30, 1991; Ventura et al., Nucleic Acids Res.
21:3249-55, 1993; and Chowrira et al., J. Biol. Chem. 269:25856, 1994. The
2 0 details will not be repeated here, but include altering the length of the
ribozyme
binding arms, or chemically synthesizing ribozymes with modifications (base,
sugar and/or phosphate) that prevent their degradation by serum ribonucleases
and/or enhance their enzymatic activity (see e.g., Eckstein et al.,
International
Publication No. WO 9207065; Perrault et al., Nature 344, 565, 1990; Pieken et
al., Science 253, 314, 1991; Usman and Cedergren, Trends in Biochem. Sci. 17,
334, 1992; Usman et al., International Publication No. WO 9315187; and Rossi
et
al., International Publication No. WO 9103162; and Sproat, U.S. Patent No.
5,334,711; all of these describe various chemical modifications that can be
made
to the base, phosphate and/or sugar moieties of enzymatic RNA molecules).
3 0 Modifications which enhance their efficacy in cells, and removal of bases
from
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 73 -
stem loop structures to shorten RNA synthesis times and reduce chemical
requirements may be used.
Example 4
Aptamer Evolution and Selection
One embodiment of the present invention involves effector identification
in accordance with Example 1 together with an evolution and selection of an
aptamer. The aptamer evolution first involves preparing a pool or mixture of
random sequence single-stranded RNA (ssRNA) with constant regions that are
necessary for reverse transcription and PCR amplifications. Typically the
individual ssRNAs contain at least 20 nucleotides, but fewer nucleotides may
be
used. The mixture of ssRNA is then contacted with an effector. Next, the RNAs
which bind to the effector are separated from the remainder of RNAs in the
pool
which do not bind to the effector. Those separated RNAs are amplified to form
DNA, and the amplified DNA is used to form an enriched mixture of RNAs
which bind to the effector. The effector-recognition, partitioning and
amplification steps are performed for one or more cycles as needed to identify
one
2 0 or more of the RNAs as an aptamer(s) which best bind the effector.
The evolved aptamer or aptamers are then used in an allosteric control
module. The selection of the aptamer for use in an allosteric control module
is
performed by first linking the aptamer to a catalytic RNA to form an
allosteric
control module; and then identifying those allosteric control modules in which
the
2 5 interaction of the effector and aptamer alters the activity of the
catalytic RNA.
This analysis may be performed in vivo or in vitro.
Example 5
In Vitro Aptamer Evolution and Selection for Theophylline
This example describes the evolution of an allosteric regulatable
hammerhead ribozyme comprising an aptamer specific for theophylline.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 74 -
Theophylline was chosen as an effector for the reason, among others, that it
has
been approved for use by the FDA since 1940 and its safety and toxicity
profiles
are acceptable and well-known. Jenison et al. previously characterized a
theophylline-binding aptamer, Science 263:1425-1429, 1994, to which
Zimmerman et al., assigned its secondary structure. Nature Struct. Biol. 4:644-
649, 1997. The two RNA strands that connect the hammerhead catalytic domain
to the theophylline aptamer domain, the so-called "communication module,"
contains only six nucleotides and was completely randomized. The complexity of
this library is 1.7 x 106 individual molecules.
Selection was initiated with 2 nmoles of synthetic single-stranded DNA
Template 1 (SEQ ID NO: 1; Fig. 1) which was made into its double stranded form
by the polymerise chain reaction (PCR).
DNA Template 1
5' GGGAGAGGGA TCCAGCTGAC GANNNNNNAA
TACCAGCCGA ,~1AGGCCCTTG GCAGGNNNNN
NGAAACGCCT TCGGCGTCCT GGAT 3'
Five rounds of PCR were carried out in 500 pL reaction volume containing 50
mM KCI, 10 mM Tris-HCl (pH 9.0 at 25 °C), 0.01% TritonX-100, 2.5 mM
2 0 EDTA, 0.25 mM dNTP each, and 2 nmoles of the two primers, Forward T7-11:
5' GCTTAATACGACTCA CTATAGGGAGAGGGATCCAGC 3' (SEQ m NO:
2)
and Reverse T7-11:
5' GAAGTTCTAATC CAGGACGCCGAAGGCGTTT 3' (SEQ ID NO: 3).
2 5 Parameters for each PCR cycle were 30 sec. at 93 °C, 30 sec. at 55
°C, and
1 min at 72 °C. Resulting double-stranded DNA containing a priming site
for T7
RNA polymerise template was concentrated by ethanol precipitation.
Approximately 25% of DNA was used for in vitro transcription carried out at 37
°C overnight in 500 ~,L reaction volume containing 40 mM Tris-HCl (pH
8.0 at 25
3 0 °C), 20 mM MgCh, 2 mM Spermidine (Sigma, St. Louis, MO), 0.01 %
Triton X-
100, 5 mM DTT, 400 U of T7 RNA polymerise and 4 mM NTP each. DNA
template in the post-transcription mixture was removed by a brief DNAse I
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 75 -
treatment (20 U/500 ~L) at 37 °C for 15 min. The ribozyme library
containing the
random sequence region in the Stem-II was isolated by running transcribed RNA
on an 8% polyacrylamide gel under denaturing conditions (denaturing PAGE).
Ribozymes that did not cleave in response to Theophylline were removed
by incubating the RNA in a ribozyme cleavage buffer (RZCL buffer: 50 mM Tris-
HCl (pH 7.5 at 25 °C) and 10 mM MgCl2) at 25 °C, punctuated at
30 min intervals
by incubation at 85 °C for 30 sec. This negative selection was carned
out for 5
cycles of thermal punctuation. The RNA population resistant to ligand-
independent self-cleavage was isolated by denaturing PAGE. A positive
selection
of the ligand-independent cleavage resistant RNA population was carried out by
incubating this RNA in RZCL buffer containing 200 ~,M Theophylline at 25
°C
for 5 min to facilitate Theophylline-dependent cleavage of ribozymes. The
resulting population of 5' fragments upon self-cleavage was isolated by
denaturing PAGE. The 5'-fragment RNA population was subsequently used as
the template for reverse transcription in 30 ~L reaction volume containing 50
mM
KCI, 50 mM Tris-HCl (pH 8.0 at 25 °C), 5 mM MgCl2, 5 mM DTT, 1 mM
dNTP
each, 10 U of avian myeloblastosis virus reverse transcriptase, and 500 pmoles
of
Reverse T7-11 primer at 42 °C for 30 min. The resulting cDNA was used
as the
template for PCR to obtain the template to generate RNA for the next round of
in
2 0 vitro selection.
We carried out seven cycles of selections, during which the concentration
of Theophylline and MgCIZ in the positive selection step was gradually
decreased
from 200- 20 mM and 10 - 2 mM, respectively. After seven cycles, the enriched
population exhibited theophylline-dependent self-cleavage activity. The
resultant
2 5 PCR products were purified by gel filtration and ethanol precipitation.
Purified
PCR products (0.1 pmoles) were cloned into pT7Blue-3 Vector (Novagen ,
Madison, WI) in EcoRV site with Perfectly Blunt Cloning Kit (Novagen)
according to the protocol supplied by the manufacturer. The ligation mixture
was
transformed into NovaBlue Singles Competent Cells (Novagen) using standard
3 0 protocols. Plasmids in the transformants were isolated and sequenced with
U 19
vector-based primer (SEQ ID NO: 4) (5' GTTTTCCCAGTCACGACGT 3') and
subjected to further analysis.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 76 -
An example of an individual ribozyme sequence, AT-50, that responds to
Theophylline for cleavage is illustrated in Fig. 8 and has the following
sequence
(SEQ >D NO:S):
5' GGGAGAGGGA UCCAGCUGAC GAGLTACUGAA
UACCAGCCGA AAGGCCCULTG GCAGGUUUGG
UGAAACGCCU UCGGCGUCCU GGAUUAGAAC UUC 3'
The self-cleaving ability of ribozyme TA-50 is strongly dependent on the
presence of theophylline (Fig. 9). The highest cleavage activity of TA-50
ribozyme was observed with theophylline concentration between 10-50 ~,M.
Since this concentration of theophylline is within the therapeutically
achievable
range, it is possible to envision an in vivo application of TA-50 or another
ribozyme with similar performance characteristics to modulate gene expression
in
response to theophylline intake.
Example 6
Effector Inactivation of an Allosteric Control Module
2 0 The following procedure produces allosteric control modules having a
catalytic activity which is inactivated or inhibited in the presence of the
identified
effector. Effector identification is performed in accordance with Example 1
followed by the evolution and selection of an aptamer which involves the steps
of:
a) preparing a pool of random sequence ssRNA wherein each ssRNA comprises
2 5 an aptamer, a proposed catalytic domain and one or more constant regions
suitable for reverse transcription and PCR amplification;
b) identifying those RNAs which have catalytic activity;
c) amplifying the catalytically active RNAs to form coding DNA molecules;
d) transcribing the amplified DNA to form an enriched mixture of catalytically
3 0 active RNA;
e) contacting the mixture with an effector;
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
77 _
f) selecting those RNAs which bind to the effector but which do not retain
catalytic activity upon binding the effector;
g) amplifying the selected RNAs to form coding DNA molecules;
h) transcribing the amplified DNA to form an enriched mixture of allosteric
control modules having a catalytic activity which is inactivated or inhibited
in
the presence of the effector; and
i) performing steps b) through h) for one or more cycles as needed to identify
one or more allosteric control modules which recognize, bind and interact with
the effector and which are inactivated or inhibited by effector binding when
the effector and the selected allosteric control module are used in the
modulation of gene expression.
In an alternative embodiment, the evolution of the allosteric control
module, involves the steps of:
a) preparing a pool of random sequence ssRNA wherein each ssRNA comprises
an aptamer, a proposed catalytic domain and one or more constant regions
suitable for reverse transcription and PCR amplification;
b) contacting the mixture with an effector;
c) selecting those RNAs which bind to the effector but which do not
demonstrate
2 0 catalytic activity upon binding the effector;
d) amplifying the selected RNAs to form DNA molecules;
e) transcribing the amplified DNA to form a RNA mixture;
f) selecting those RNA as one or more allosteric control modules which
demonstrate catalytic activity in the absence of the effector;
2 5 g) amplifying the selected RNAs;
h) transcribing the amplified RNA to form an enriched mixture of allosteric
control modules having a catalytic activity which is inactivated or inhibited
in
the presence of the effector; and
i) performing steps b) through h) for one or more cycles as needed to identify
3 0 one or more allosteric control modules which recognize, bind and interact
with
the effector and which are inactivated or inhibited by effector binding when
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
_ 78 _
the effector and the selected allosteric control module are used in the
modulation of gene expression.
In one embodiment the "catalytic activity" is a self-cleaving activity and
the self-cleaving allosteric control module is used for the inhibition or
reduction
the expression of a gene of interest in the absence of the effector. In
further
embodiments, the catalytic activity involves ligase activity or splicing
activity.
Example 7
Effector Activation of an Allosteric Control Module
The following procedure produces allosteric control modules having a
catalytic activity which is activated in the presence of the identified
effector.
Effector identification is performed in accordance with Example 1 followed by
the evolution and selection of an aptamer which involves the steps of:
a) preparing a mixture of random sequence ssRNA wherein each ssRNA
comprises an aptamer, a proposed catalytic domain and one or more constant
regions suitable for reverse transcription and PCR amplification;
b) identifying those RNAs which do not demonstrate catalytic activity in the
2 0 absence of effector;
c) amplifying the identified RNAs to form coding DNA molecules;
d) transcribing the amplified DNA to form an enriched mixture of RNA;
e) contacting the mixture with an effector;
f) identifying those RNAs which bind to the effector and demonstrate catalytic
2 5 activity upon binding the effector;
g) amplifying the identified RNAs to form coding DNA molecules;
h) transcribing the amplified DNA to form an enriched mixture of allosteric
control modules having a catalytic activity which is activated in the presence
of effector; and
3 0 i) performing steps b) through h) for one or more cycles as needed to
identify
one or more allosteric control modules which recognize, bind and interact with
the effector and which are activated or enhanced by effector binding when the
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 79 -
effector and the selected allosteric control module are used in the modulation
of gene expression.
In an alternative embodiment, the evolution of the allosteric control
module, involves the steps of:
a) preparing a pool of random sequence ssRNA wherein each ssRNA comprises
an aptamer, a proposed catalytic domain and one or more constant regions
suitable for reverse transcription and PCR amplification;
b) contacting the pool with an effector;
c) identifying those RNAs which bind to the effector and demonstrate catalytic
activity while bound to the effector;
d) amplifying the identified RNAs to form coding DNA molecules;
e) transcribing the amplified DNA to form an enriched mixture of RNA having a
catalytic activity in the presence of effector;
f) selecting those RNA which are catalytically inactive in the absence of
effector;
g) amplifying the selected RNAs to form coding DNA molecules;
h) transcribing the amplified DNA to form an enriched mixture of allosteric
control modules having a catalytic activity which is activated in the presence
of
effector; and
2 0 i) performing steps b) through h) for one or more cycles as needed to
identify one
or more allosteric control modules which recognize, bind and interact with the
effector and which are activated or enhanced by effector binding when the
effector and the selected allosteric control module are used in the modulation
of gene expression.
In one embodiment the "catalytic activity" is a self-cleaving activity and
the self-cleaving of the allosteric control module results in the formation of
a
functional mRNA encoding a gene of interest. In further embodiments, the
catalytic activity involves ligase activity or splicing activity.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 80 -
Example 8
Alternate Effector Selection Procedure
An alternate method of selecting an effector, involves the steps of:
a) providing an allosteric control module suitable for use in the modulation
of
gene expression;
b) contacting the allosteric control module with one or more effectors; and
c) determining whether or not the interaction of the allosteric control module
and
an effector results in an alteration of the catalytic activity of the
allosteric
control module.
Another method of determining whether a molecule not previously known
to be an effector may be used in combination with an allosteric control module
to
specifically alter the expression of a gene of interest involves the steps of:
(a) contacting a sample which contains a predefined number of eucaryotic cells
with the molecule to be tested, each cell comprising a DNA construct
encoding,
i) an allosteric control module, and
ii) a reporter gene that produces a detectable signal, coupled to, and under
the
2 0 control of, a promoter,
under conditions wherein the molecule if capable of acting as a modulator of
the gene of interest, causes a detectable signal to be produced by the
reporter
gene;
(b) quantitatively determining the amount of the signal produced in (a);
2 5 (c) comparing the amount of signal determined in (b) with the amount of
signal
produced and detected in the absence of any molecule being tested or with the
amount of signal produced and detected upon contacting the sample in (a)
with other molecules, thereby identifying the test molecule as an effector
which causes a change in the amount of detectable signal produced by the
3 0 reporter gene, and thereby determining whether the test molecule
specifically
alters expression of the gene of interest.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 81 -
EXAMPLE 9
Inducible Gene Expression
The Figures are schematics of several specific embodiments of this
technology to regulate expression of genes. The system's utility can be
extended
beyond direct medical applications into more basic research applications,
diagnostic applications and environmental testing applications.
In one embodiment, one can select for an effector-responsive allosteric
control module and gene of interest to be introduced into the cell by means of
a
viral or nonviral vector. Where the allosteric control module involves a self-
cleaving catalytic domain, the interaction of the effector and allosteric
control
module results in the expression of the gene of interest. In the absence of
the
effector, the mRNA is untranslatable. Upon removal of effector from the system
the allosteric control module should return to the active conformation and the
expression levels decrease due to the presence of untranslatable mRNA.
In an alternative embodiment, one can select for an effector-responsive
allosteric control module and gene of interest to be introduced into the cell
by
means of a viral or nonviral vector. Where the allosteric control module
involves
2 0 a self-cleaving catalytic domain, the interaction of the effector and
allosteric
control module may be designed to result in the inactivation of the mRNA and
the
non-expression of the gene of interest in the presence of effector. In the
absence
of the effector, the mRNA is translatable.
In another embodiment, one can select for an effector-responsive allosteric
2 5 control module and gene of interest to be introduced into the cell by
means of a
viral or nonviral vector. Where the allosteric control module involves a self-
splicing catalytic domain, the interaction of the effector and allosteric
control
module results in the expression of the gene of interest. In the absence of
the
effector, the mRNA is untranslatable.
3 0 In an alternative embodiment, one can select for an effector-responsive
allosteric control module and gene of interest to be introduced into the cell
by
means of a viral or nonviral vector. Where the allosteric control module
involves
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 82 -
a self-splicing catalytic domain, the interaction of the effector and
allosteric
control module may be designed to result in the inactivation of the mRNA and
the
non-expression of the gene of interest. In the absence of the effector, the
mRNA
is translatable.
It will be appreciated that the DNA constructs may include a regulatable
allosteric control module (e.g., self-cleaving format) placed under the
control of a
constitutive promoter. The same construct may include the coding region for
the
gene of interest, a stop codon and a poly(A) tail. The introduction of the
vector
into a cell will result in the production of mRNA encoding both the allosteric
control module and gene. If the allosteric control module is active in the
absence
of effector, then the mRNA will be cleaved by the catalytic domain to yield
pieces
available for exonucleolytic attack; that is, mRNA will be inactivated. When
the
effector is administered and enters the cells, the allosteric control module
becomes
inactive and the mRNA will be translated normally. Hence, gene expression will
be under the inducible control of the allosteric control module.
All of the references cited herein, including patents, patent applications,
2 0 and publications, are hereby incorporated in their entireties by
reference.
While this invention has been described with an emphasis upon preferred
embodiments, it will be apparent to those of ordinary skill in the art that
variations
in the preferred embodiments can he prepared and used and that the invention
can
2 5 be practiced otherwise than as specifically described herein. The present
invention is intended to include such variations and alternative practices.
Accordingly, this invention includes all modifications encompassed within the
spirit and scope of the invention as defined by the following claims.
CA 02401654 2002-08-28
WO 01/64956 PCT/USO1/06615
- 1 -
SEQUENCE LISTING
<110> AMGEN INC.
MARSHALL, WILLIAM
KHVOROVA, ANASTASIA
JAYASENA, SUMEDHA
<120> THE IDENTIFICATION AND USE OF EFFECTORS AND ALLOSTERIC MOLECULES FOR
THE ALTERATION OF GENE EXPRESSION
<130> A-655
<140> 09/729,258
<141> 2000-11-28
<150> 60/186,240
<151> 2000-03-01
<160> 5
<170> PatentIn version 3.0
<210> 1
<211> 84
<212> DNA
<213> synthetic construct
<220>
<221> misc_feature
<223> N = any nucleotide
<400> 1
gggagaggga tccagctgac gannnnnnaa taccagccga aaggcccttg gcaggnnnnn 60
ngaaacgcct tcggcgtcct ggat 84
<210> 2
<211> 36
<212> DNA
<213> synthetic construct
<400> 2
gcttaatacg actcactata gggagaggga tccagc 36
<210> 3
<211> 31
<212> DNA
<213> synthetic construct
<400> 3
gaagttctaa tccaggacgc cgaaggcgtt t 31
<210> 4
<211> 19
<212> DNA
CA 02401654 2002-08-28
WO 01/64956 - 2 - PCT/USO1/06615
<213> synthetic construct
<400> 4
gttttcccag tcacgacgt 19
<210> 5
<211> 93
<212> DNA
<213> synthetic construct
<400> 5
gggagaggga uccagcugac gaguacugaa uaccagccga aaggcccuug gcagguuugg 60
ugaaacgccu ucggcguccu ggauuagaac uuc 93
<210> 6
<211> 93
<212> DNA
<213> synthetic construct
<220>
<221> misc_feature
<223> N = any nucleotide
<400> 6
gggagaggga uccagcugac gannnnnnaa uaccagccga aaggcccuug gcaggnnnnn 60
ngaaacgccu ucggcguccu ggauuagaac uuc 93