Note: Descriptions are shown in the official language in which they were submitted.
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
METHODS FOR GENOTYPING BY HYBRIDIZATION ANALYSIS
RELATED APPLICATIONS
This application claims priority from United States Application Numbers
60/193,042, filed on March .29, 2000, 60/252,551, filed on November 21, 2000,
and
60/252,747, filed on November 22, 2000.
TECHNICAL FIELD
This invention relates generally to determining the genotype of organisms
by hybridization analysis and, more specifically, to establishing the
relatedness of
individual organisms within a species.
BACKGROUND OF THE INVENTION
A genotype is the genetic constitution of an individual or group. Variations
m genotype are essential for commercial breeding programs, diagnostics,
monitoring
genetic-based diseases, following spread of pathogens, determining parentage,
and the like.
While determining the nucleic ' acid, sequence of genomic DNA- is one way to
unambiguously establish a genotype of an individual, it is not currently
practicable to
accomplish, especially in organisms with complex genomes.
Genotypes can be more readily described in terms of genetic markers. A
genetic_marker identifies a specific region or locus in the genome.. Thus, the
more genetic
markers, the finer defined is the genotype. A genetic marker becomes
particularly useful
when it is allelic between organisms because it then may serve to
unambiguously identify
an individual.
Many different flavors of genetic markers have been described and
exploited, but all are based upon genomic sequence. Examples of methods to
define
genetic markexs include restriction fragment length polymorphism (RFLP)
analysis
(Botstein et aL, Am J Hum Genet 32: 314, 1980); single-sequence repeats (SSR)
analysis
(Weber and May, Am JHum Genet 44: 388, 1989; US Patent No 5874215); rapid-
amplified
1
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
polymorphic DNA (RAPD); amplified fragment length polymorphism (AFLP) (Vos et
al.,
Nucleic Acids Res 23: 4407, 1995); S' nuclease amplifications (LTS Patent No.
5,962,233);
nucleic acid indexing (US Patent No 5,994,068; Guilfoyle, et al., Nucl Acids
Res, 25; 1854,
1997; Unrau and Degau., Gene 145: 163, 1994; US Patent No 5508169) arbitrarily-
primed
nucleic acid amplification (US Patent No. 5,413,909; US Patent No 5861245);
restriction
enzyme amplification display system (READS) (U.S. Patent No. 5712126; Prashar
and
Weissman, Proc Natl Acad Sci USA 93: 659, 1996); consensus sequence primed
polymerase chain reaction (CP-PCR) (LJS Patent No 5437975); hybridization-
based genetic
amplification (W0.98/0721); and the like.
10. ' All of these genotyping methods ~ suffer from the laborious requirement
to
analyze only a single organism at a time. A further burden in some of these
methods is the
need for pre-identification of a poiymoxphism before analysis of other
individuals (LTS
_,
Patent No: 6,100,030). Still others of these methods depend upon expensive
materials and
time-intensive gel electrophoresis, resulting in a low-throughput.
Furthermore, these
methods that base identity on size suffer from additional difficulties in
precisely correlating
bands on gels with alleles. One method has attempted to overcome many of these
restrictions by performing analysis by hybridization to nucleic acids
immobilized on solid
state surfaces (CTS Patent No: 6,100,030). In this technique however, a
genotype of an
organism is not established. Rather, the analysis yields information regarding
a pre
determined polymorphism.
The ability to assign a comprehensive genotype for an individual without
requiring sequence information, existing knowledge of polymorphisms, or having
to
compare lengths is paramount to the mass of genetic information necessary for
breeding,
disease analysis, and so forth. Such systems and analyses also demands a high-
throughput
for optimal and maximal benefit.
The present invention discloses methods and compositions for performing
high throughput genotype determinations by basing analyses on hybridization of
unselected
nucleic acids to genomic nucleic acids immobilized to solid state materials,
and further
provides other related advantages.
2
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
SUNINIARY OF THE INVENTION
The present invention relates to methods and compositions for determining
and relating genotypes of organisms. Within one aspect of the present
invention, a nucleic
acid molecule that contains a polymorphism is identified. Two organisms are
selected, one
may be referred to as a reference organism and the other may be referred to as
the tester
organism. Nucleic acids from each of these organisms are separately amplified.
Amplified
material from the ~ tester organism is cloned ~ or otherwise separated (by
e.g., gel
electrophoresis, HPLC), and individual clones or separated amplified material
is placed
into an addressable array. The amplified material from the reference organism,
which
contains a detectable label is hybridized to the array. Clones on the array
that do not
evidence detectable hybridization are thus identified as containing a
polymorphism.
In a second aspect, the genotype of an organism is determined. . In this
method, nucleic acids from two or more organisms are pooled and used to
generate a first
diversity panel. In one embodiment, the diversity panel is generated by
amplification. In
1 S other embodiments, the diversity panel is generated by restriction enzyme
digestion, a
combination of amplification and restriction ~ digestion, or other means that
creates a
reproducible pattern. The first diversity panel is then separated on the basis
of sequence or
molecular weight, e.g., by~cloning, gel electrophoresis, HPLC, or the like,
and individual
elements of the diversity panel, e.g., clones, are placed into an addressable
array. Nucleic
acids from another organism, which may be one of the organisms in the initial
pool, the
selected organism, is used to generate a second diversity panel.
In one aspect, the polymorphisms detected are caused by insertion elements,
such as transposons. The diversity panels are generated by amplification, and
in some
embodiments amplification in conjunction with restriction enzyme digestion and
ligation of
adapters. Amplification is performed with a primer pair in which one of the
primers
anneals to a sequence found in a family of insertion elements.
In certain embodiments, the first and second diversity panels are generated
by the same technique and using the same primers, enzymes, , or methods. In
other
3
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
embodiments, the techniques differ, and in yet other embodiments, the
techniques are the
same but the primers or enzymes used to generate the two diversity panels are
different.
Tn a preferred embodiment, the second diversity panel contains a detectable
label, such as a fluorochrome, chemiluminescent molecule, radiolabel, enzyme,
ligand, and
the like. - . '
The array is then hybridized with the second diversity panel. A pattern of
hybridization to the array is established. The genotype of the selected
organism is thus
determined. Briefly, the more elements of the array that hybridize with the
diversity panel
of the selected organism, the more related the selected organism is to the
organisms
constituting the array. By generating a diversity panel from each of the
organisms in the
pool and hybridizing them individually to the array, the genotypes and the
relatedness of all
the organisms can be determined. .
In a third aspect of this invention, a first diversity panel is generated and
placed onto an array as described for the second aspect. The array will thus
comprise the
genomes of two or more organisms. A second diversity panel is generated from a
selected
organism, that may or may not be represented in the first diversity panel. _
The second
diversity panel is hybridized to the array, and a pattern of hybridization is
detected. The
genotype of the selected organism is established.
In one embodiment, a third, fourth, and so on diversity panels are generated
from individual organisms and mixed with the second diversity panel. In this
embodiment,
the second, third, and so on diversity panels contain a detectable label, and
each diversity
panel contains a label distinguishable - from the others. The more labels ~
that can be
distinguished, the more diversity panels that can be mixed together. In
certain
embodiments, the labels are fluorochromes or mass-spectometry tags. The
mixture of
diversity panels is hybridized to the array, and a pattern of hybridization
with each diversity
panel is detected. The genotypes of the selected organisms are thus determined
from the
patterns of hybridization.
In a preferred embodiment, genomic nucleic acids from two or more
organisms are digested with a restriction enzyme. The restriction enzyme may
be an
4
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
enzyme sensitive to methylation. In such a case, the polymorphisms detected
are
modifications (methylation) of bases. In one embodiment, fragments are
selected on the
basis of size to comprise a pool of fragments in a desired size range. The
digested
fragments are cloned into a vector and placed into an addressable array on a
solid surface,
. such as ~ a glass slide. Another organism whose genotype is to be determined
(called here
organism X), and which may or may not be the same organism as one in the first
group, is
digested with. the same restriction enzyme: These restriction fragments are
amplified.
Typically, adapter sequences are ligated to the fragments and also used as
primers for
amplification. The amplified fragments are also labeled with one of the labels
described
below. Labeled fragments are hybridized to the addressable array,
nonhybridized
fragments are washed off, and the array is then analyzed for the label. In
this way a pattern
of hybridization is obtained. That pattern is the genotype of the organism X.
In this
example, when an element in the array hybridizes, it indicates that the
organisms share
sequence similarity for that fragment. When an element in the array does not
hybridize, it
indicates a polymorphism. In this particular example, the polymorphism is
analogous to.a
restriction fragment length polymorphism and arises because the restriction
fragment in
organism X is too long to be amplified or too short to hybridize. -
In still .other aspects, kits and arrays are provided that comprise diversity
panels for genotyping.
These and other aspects of the present invention will become evident upon
reference to the following detailed description and attached drawings. In
addition, various
references are set forth below which describe in more detail certain
procedures or
compositions (e.g., plasmids, etc.), and are therefore incorporated by
.reference in their
- entirety.
BRIEF DESCRIPTION OF THE DRAWITTGS
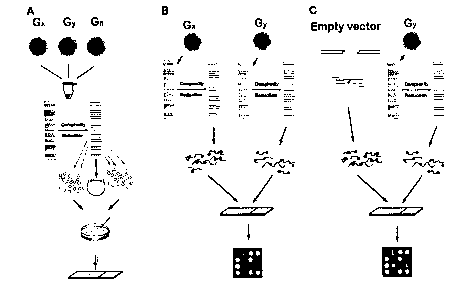
Figures 1A, 1B, and 1C present a schematic representation of various
- embodiments of the present invention. (A) Generation of a diversity panel.
Genomic
DNAs of various organisms to be studied are pooled together. The DNA is cut
with a
5
CA 02404472 2002-09-26
WO 01/73119 PCT/IBU1/00833
chosen restriction enzyme and ligated to adapters. The complexity of the sub-
genomic
profile is reduced in this case by amplification using primers with selective
overhangs. The
amplified sub-genomic fragments are cloned. Clones are selected and inserts
are amplified,
purified and arrayed onto the solid support. (B) Contrasting two samples using
diversity
array technology. Two genomic samples are converted to diversity panels
comprising sub-
genomic samples. Each sub-genomic sample is labeled with a green or red
fluorescent dye,
mixed and hybridized to the diversity array. The ratio of green/red signal
intensity is
measured at each array element. Significant differences in the signal ratio
indicate array
elements (and the relevant fragment of the genome) for which the two samples
differ. (C)
Genetic fingerprinting. The DNA sample for analysis is converted to a sub-
genomic sample
and labeled with green fluorescent dye. Fragments of the cloning vector common
to alI
elements of the array are labeled with red fluorescence and hybridized to the
diversity
panels together with the sub-genomic sample. The ratio of signal intensity is
measured at
each array feature. The ratios across the diversity array provide genetic
fingerprint
information for the sample analyzed.
Figures 2A and 2B show differences between fingerprints of two rice
cultivars, IR64 and Millin. (A) Synthetic array image of 96 spots printed 4
times from an
EcoRI-generated diversity panel. The rice cultivars IR64 and Millin are
labeled with Cy3-
green and Cy5-red respectively. (B) Histogram of green to red normalized
signal intensity
ratios shows tri-modal distribution. The majority of the array features show
signal intensity
ratios are around 1 indicating equal hybridization intensity for Millin and
IR64. The green
and red "tails" are seen at signal intensity ratios above 2.9 indicate
features of the diversity
panel that differentiate IR64 and Millin DNA.
Figures 3A and 3B. Two clones (F4 and F8), representing two polymorphic
features on the EcoRI diversity panel are used as molecular probes. Four
varieties of rice
are analyzed simultaneously, Iane 1, Bala; lane 2, Millin; lane 3, IR64, lane
4, IR20. (A)
Hybridization of labeled F4 and F8 probes to Southern blots of EcoRI-digested
genomic
DNA. (B) Hybridization of labeled F4 and F8 probes to Southern blots of
diversity panels
of sub-genomic samples generated from genomic DNA samples.
6
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
Figure 4 shows the result of hybridization of monomorphic clone F 11 to
EcoRI-digested genomic DNA from strains Millin, Bala, IR20, and IR64.
Figures SA and SB show hybridization of Cy3-labeled IR20 diversity panel
and Cy5-labeled Millin diversity panel (Fig SA) and Cy3-labeled IR64 diversity
panel and
Cy5-labeled Millin diversity panel (Fig SB) to duplicate addressable arrays of
a mixture of
diversity panels.
Figures 6A and 6B show hybridization of Cy3-labeled IR20 diversity panel
and Cy5-labeled Millin diversity panel. (Fig 6A) and Cy3-labeled IR20
diversity panel and
Cy5-labeled vector DNA (Fig 6B) to duplicate addressable arrays of_ a mixture
of diversity
panels.
Figures 7A, 7B, aid ~ 7C show cumulative distribution functions for non-
polymorphic fragments (A), polymorphic fragments (B), and a reference fragment
(C). (A)
Cumulative distribution function of log transformed normalized signal ratios
for 4 different
non-polymorphic spots across 18 different slides. Classification as non-
polymorphic is
1 S based on the monomodal distribution of the ratios ,across the 18 slides.
(B) Cumulative
distribution. function of log transformed normalized signal ratios for 4
different
polymorphic spots across 18 different slides. Classification as polymorphic is
based on a
clear bimodal distribution across the I8 slides. The algorithm calculates the
best value for
separation of the high (value of 1) and low (value of 0) clusters shown as a
cross on the
, curves. (C)~A cumulative distribution function of the normalized log
intensity values of a
'reference fragment (TOPO) across 18 slides adjusted to have equal medians.
Each slide has
2304 spots (384 spots printed 6 times). One curve is shown in red; it is the
result of a
technical problem in a single experiment.
Figure 8 presents a histogram of unique and replicate features from the MspI
diversity'panel. Clones are considered to be replicates if they have the same
apparent gel
mobility and the same polymorphism patterns among the rice cultivars analyzed.
A total of
50 polymorphic spots are analyzed here. The red bars indicate the actual
numbers of spots
found in each category; the blue bars indicate the expected total number. of
spots in the
diversity panel in each category by extrapolation from 50 to 384 spots in the
panel.
7
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
Figure 9 shows dendrograms generated from Mspl (A) and PstI (B) diversity
panels.
Figure 10 presents the results of a reconstruction experiment using mixed
(rice and several microorganisms) diversity panels. A Millin diversity panel
is labeled with
red fluorescent dye and an Enterobacte~ spiked Millin diversity panel is
labeled with green
fluorescent dye. The image and histogram are created using the Pathways
program. (A)
The left half of the array (mostly yellow features) represents rice MspI
diversity array. The
right half of the array contains features from MspI diversity panels from
seven bacterial
species and one 'from yeast. The green spots in the right part of the array
correspond to the
elements of the panel developed from the Enterobacter DNA source. (B)
Histogram of the
signal ratios for the array presented at (A). The EnteYObacter spike is
detected as the green
peak seen at the left edge of the distribution.
Figure 11 presents the result of a diversity array containing DNA from 3
barley cultivars' (Steptoe, Morex, Harrington) and a wild barley Hordeum
spontdneum
hybridized with Cy3-labeled Morex diversity panel and Cy5-labeled Steptoe
diversity
panel.
Figure 12 presents the result of a mouse cDNA diversity array hybridized
with Cy3-labeled C57B1/6 diversity panel and Cy5 labeled NOD K diversity
panel.
Figure 13 presents the result of a rice diversity array hybridized with Cy5
labeled callus-diversity panel and a Cy5-labeled seedling root diversity panel
(upper array) .
an a Cy5-labeled callus diversity panel and a Cy3-labeled immature embryo
diversity
panel:
Figure 14 presents the result of a Southern hybridization of various clones
identified as differentially methylated in fertilized ovary an stigma to DNA
prepared from
12 different diversity panels.
DETAILED DESCRIPTION OF THE INVENTION
Prior to setting forth the invention, it may be helpful to an understanding
thereof to set forth definitions of certain terms that will be used
hereinafter.
8
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
As used herein, an "addressable array" or an "array" means a workspace in
which nucleic acid molecules are positioned in discrete locations, which can
be either
physically or, temporally defined, such that each location is uniquely
identifiable.
Typically, the workspace is a solid substrate in which the ~ locations are an
identifiable
pattern or at regular intervals. Examples of substrates suitable for this
invention include,
but are not limited to, glass slides, silicon chips, or light fiber optic
tubes.
As used herein, a "fingerprint" cbmprises a distinct pattern of nucleic acid
molecules that is a characteristic of the genotype of the organism that the
nucleic acids are
prepared from. The patterns can be generated by a variety of techniques, such
as restriction
enzyme. digestion, amplification, a combination of enzyme digestion and
amplification, or
other method. Fingerprints can reveal sequence differences between nucleic
acid samples
and can be used to establish a genotype of an organism or groups of organisms.
Generally,
fingerprints are used to analyze and compare DNA from different species or
different
individuals of the same species. The differences that are detected are called
i5 polymorphisms, if pre-existing in the population, individual, or gene pool,
or mutations, if
exogenously or spontaneously induced of newly emergent. The precise names
given to the
differences, however, does not change the outcomes.
A "diversity panel" as used herein refers to nucleic acid fragments prepared
from organismal nucleic acids (e.g., genomic DNA) by a method that can reveal
polymorphisms or mutations (e.g., sequence differences)' between samples. When
a
diversity panel is applied to an array, it is called herein a "diversity
array."
As used herein, "organism" refers to an individual entity or a uniform set of
individuals (e.g., species, strain, etc.).
As used herein, "polymorphism" and "mutation"'mean a difference in DNA
sequences among individuals. Differences .include, - without limitation,
changes,
modifications (e.g., methylation, bromination, amination, and the like),
insertions, and
deletions or combinations of these differences and may involve one or more
bases.
9
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
A. PREPARATION OF ARRAYS
The present invention provides addressable arrays, also referred to herein as
arrays, comprising diversity panels of nucleic acid molecules, in which the
molecules on
the array are addressable or uniquely identifiable in some fashion. In the
present invention,
these diversity panels are generated from nucleic acid samples isolated from
multiple
organisms. ' A diversity panel refers to nucleic acid-fragments prepared from
organismal
nucleic acids by a method that can reveal sequence differences between nucleic
acid
samples. As taught-herein, a variety of methods may be used to generate
diversity arrays.
Subsequent to the generation of the diversity panel, the nucleic acid
products of the diversity .panel are separated for application in a uniquely
addressable
format, generally onto . a substrate, hereinafter called an axray or an
addressable array.
Separation may be achieved on the basis of physical parameters, e.g., length,
molecular
weight, or by genetic methods, e.g., cloning.
As exemplified herein, the separated diversity panel is then delivered onto a
substrate to create an addressable array. In the currently most widely used
type of array,
nucleic acid molecules are deposited or synthesized on a glass or silicon
wafer in an
ordered array. Other types of arrays can also be used, such as those that
comprise nucleic
acid molecules immobilized on microspheres that are uniquely encoded and
randomly
deposited in wells of a chemically-etched optical imaging fiber. The codes on
the beads or
particles permit positional registration of beads of a particular sensor type
after assembly.
Thus, the addressing is accomplished by the unique coding signature of each
microsphere.
(see, e.g., U.S. Patent No. 5,814,524; U.S. Patent No. 5,320,814; WO 98/50782;
WO
99/18434; WO 99/45357.)
1. Source of nucleic acids
In the context of the present invention, nucleic acids for generating
diversity
panels are isolated from a variety of organisms. Exemplary organisms include
viruses
.(e.g., HIV and other lentiviruses, papilloma viruses, cytomegalovirus (Clue,
retroviruses,
hepadnaviruses, etc.); bacteria (e.g., enterobacteria, rhizhobia, Hemoplzilus,
etc.); plants,
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
including commercially important crops and weedy plants; fizngi, animals,
including
parasites (e.g., malaria, Giaf-dia, etc.), food animals, rare or endangered
species (e.g.,
condors, Tasmanian devils, spotted owl, etc.); and humans. Briefly, any
organism for
which it is desirable to assess its genotype is a suitable candidate.
The cellular source of the nucleic acids for generating diversity panels may
be genomic DNA, genomic RNA, such as for retroviruses, organelle DNA, such as
mitochondrial DNA, mRNA or cDNA, and the like. Methods for isolation and
preparation
of nucleic acid molecules are well known (see, e.g., Ausebel et al. "Current
Protocols in
Molecular Biology" Greene Publishing, 2000). The nucleic acid molecules used
to
generate diversity panels may furthermore be a mixture of two or more of these
types of
nucleic acids: In some embodiments, the source of the nucleic acids may be
from multiple
organisms or specific sub-fractions of an organism. For example, a soil sample
may
contain a variety of bacterial species, animals, protozoa, plant parts and the
like. When
using mRNA (or cDNAs) as a diversity panel; it may be desirable to choose a
particular
cell type or time as the source of RNA. The choice of the cellular source
depends in part
upon the complexity of the organism, for example a multicellular versus
unicellular
organism, and the intended use of the fingerprint analysis.
2. Methods for generating diversity panels
As discussed above, generating a diversity panel entails using a method that
can reveal sequence differences between nucleic acid samples. Then by
determining and
comparing the fingerprints of different DNA samples, the genetic relatedness
of the
organisms may be established.
There is a large variety of methods for generating diversity panels that are
suitable within the context of this invention. Some of the more popular
methods are
exemplified herein. Other methods will be known to those of skill in the art.
Briefly, the
methods taught herein include both amplification methods and non-amplification
methods.
These two types of methods can also be used in combination (see Examples). As
discussed
above, at Ieast two diversity panels are generated. For purposes of
exemplification: one
. a. .
11
CA 02404472 2002-09-26
WO 0'?3119 PCT/IBO1/00833
panel is arrayed onto a solid substrate and hybridized to the other panel
which is in liquid
phase and is the panel being fingerprinted. Either the same method or
different methods
may be used to generate the two or more diversity panels. While it is not
necessary to
reduce the complexity of the nucleic acids when generating diversity panels
for this
invention, at times it may be desirable to do so. Many of the methods
described herein will
result in a diversity panel with reduced complexity compared to the starting
nucleic acids.
Furthermore, the diversity panel that is being fingerprinted can be a subset
or a superset of
the diversity panel that is arrayed. In preferred embodiments,-the probing
diversity panel is
a superset of the arrayed diversity panel.
a. Amplificatio~z methods
A wide variety of amplification methods may be used to generate diversity
panels. Such methods include adapter-mediated amplification (U.5. Patent No.
5,710,000);
U.S. Patent No. 5,728,524, AFLP (U.5. Patent No. 6,100,030) and other indexing
methods
(CT.S. Patent No. 5,994,068; U.S. Patent No. 5,858,656; U.S. Patent No.
5,508,169),
arbitrarily-primed polymerise chain reaction (U.S. Patent No. 5,487,985; U.S.
Patent No.
5,413,909; U.S. Patent No. 5,126,239; U.S. Patent No. 5,861,245; U.S. Patent
No.
-' 5,126,239); restriction endonuclease amplification and display of cDNAs
(U.5. Patent No.
5,712,126) and other differential display methods (U.5. Patent No. 5,262,311;
U.S. Patent
No. 5,580,726); random-amplified polymozphic DNA (RAPD) (U:5. Patent No.
55,665,572; Williams et al., Nucleic Acids Res 18:' 6531-6535, 1990); simple-
sequence
repeat amplifications (U.5. Patent No. 5,874,215; U.S. Patent No. 5,576,180);
consensus-
sequence primed polymerise chain reaction (CP-PCR) method (U.5. Patent No.
5,437,975); ligation chain reaction and the like.
As discussed above, it is not necessary to reduce the complexity of .the
starting nucleic' acids when generating a diversity panel. However, many of
the methods
cited above are designed to reduce complexity. In one of the more commonly
used
methods (AFLP), complexity is reduced by digesting the DNA with a restriction
enzyme,
ligating adapters to the fragments, and then amplifying the fragments using a
primer that
12
CA 02404472 2002-09-26
WO 01/73119 PCT/IBU1/00833
corresponds to the adapter and restriction site sequences and contains one or
more bases at
the 3' end of the primer. If the primer has one extra base, on average, only
1/16 of the
fragments will amplify (only 1 in 4 fragments will have a complement to the
extra base at
one end of the fragment and 1 in 4 will.have a complement at the other end of
the
fragment). In AFLP and many of the other amplification methods, the choice of
primers
will determine, at least in part, the fraction of the genome that is
represented in the diversity
panel. For example, more extra bases at the 3' end of the primer or primers
used for
amplification will result in a smaller fraction of the genome that will be
amplified. Other
parameters that can be altered to control the fraction of the represented
genome include the
DNA polymerase used, such as whether the enzyme can synthesize long stretches
of
nucleic acids, amplification reaction conditions, such as cycling times and
temperatures,
amount or type of cofactor in the reaction and the Iike. These and other
parameters are
. known to those in the art and are widely used to affect the outcome of
amplifications.
In certain embodiments, regions comprising insertion elements are
amplified. Insertion elements are common in some organisms, . may be mobile or
immobilized, and many groups of such elements have been described. For
example,
transposable elements in plants (e.g., Ac, Ds, miniature inverted-repeat
transposable
elements (MITE) elements), insects (e.g., Drosophila P, gypsy), fungi (e.g.,
impala
element, Scooter), animals (e.g., Tigger, mariner-like elements, B2 elements,
long-
interspersed elements (LINE)), bacteria, and the like; are well known and
characterized.
Amplification of these regions such that polymorphisms are revealed may be
achieved with
several different methods and primer pairs. For exemplary purposes, two
methods are
briefly described herein. In one method, a suitable primer pair comprises a
primer that
anneals to sequences that are conserved in the chosen family of insertion
elements and the
second primer anneals to genomic sequences flai~ldng one side of the
insertion. . The
sequence of the second primer may be chosen arbitrarily, such as for the
arbitrarily-primed
PCR methods cited above. Alternatively, the sequence can comprise (ordered
from the 3'
end) five (or more) arbitrarily chosen bases optionally linked to several or
more bases in
which all four bases are represented at each position followed by a defined
sequence of at
13
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/U0833
least 11 bases (e.g., at least 12 bases, at least 13 bases, at least 14 bases,
and so on). The
first round of amplification uses this primer pair and to obtain a greater
degree of
specificity, subsequent rounds of amplification use the first primer and for
the second
primer use the defined sequence. Variations oi~ primer sequences, such as
incorporating a
restriction site and the like, and variation on methodology, such as
performing nested PCR,
are well known and commonly employed by those skilled in the art.
In another method, the nucleic . acid molecules are digested first with a
restriction enzyme, preferably one that does not cut within the insertion
element. Adapters
are ligated to the fragments, and the fragments are amplified with.a primer
pair in which
the first primer anneals to sequences that are conserved in the chosen family
of insertion
elements and the second primer anneals to the adapter sequence. As for the
methods above,
variations on sequences and methodology can be employed within the context of
this
invention.
b. Non-amplification methods
- Alternatively, methods that do not rely upon amplification may be used to
generate diversity panels. In the simplest form, restriction digestion using
enzymes that
recognize at least a six base sequence 'containing one or more degenerate
bases, enzymes
that cut infrequently, enzymes that cut DNA both 5' and 3' of the recognition
sequence,
enzymes that are sensitive or insensitive to methylation, or the. like may be
used. Other
methods include primer-directed synthesis of DNA and the like.
Furthermore, amplification methods may also be combined with non-
amplification methods. In an exemplary embodiment, fragments are , generated
by
restriction enzyme digestion and ligated with an adapter sequence. These
ligated fragments
are then amplified with primers comprising the adapter sequence. Other
exemplary
embodiments are presented above.
3. Separation of diversity panel products
As presented above, the discrete nucleotide sequences of the diversity panel
are preferably separated prior to applying them to the array. In contrast, the
discrete
14
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
sequences of the diversity panel that are used to probe the array are
preferably not
separated. Separation may be achieved by any of a variety of methods. Such
methods are
known in the art and include, but are not limited to, cloning, gel
electrophoresis,
chromatography, e.g., I~PLC, and dilution.
As an exemplary method, the diversity panel products are cloned into a
suitable vector. Techniques for cloning are well known in~the art (see e.g.,
Ausubel et al.
Current Protocols in Molecular Biology, Greene Publishing, 1999). Briefly, if
the products
do not already have ends that are compatible for ligation into a chosen vector
prepared by
restriction digestion, the products need to be prepared. Typically, the
products will either
be digested with one or more appropriate restriction enzymes or treated with a
DNA
polymerase (e.g., E. toll DNA pol 1) in the presence of all four dNTPs to
produce blunt
ends. The diversity panel products are then ligated to the cloning vector.
Generally, the
cloning vector is one that will replicate in bacteria. Many such vectors are
commercially
available (New England Biolabs, MA USA; Invitrogen, CA, USA; etc.) and include
, pBluescript, pET series vectors, pUC series vectors, and the like. Following
ligation, the
recombinants are transformed into a bacterial host, typically E. toll, and
transformed
bacteria are selected for or screened for.
Alternatively, the diversity panel products may be separated . by gel
electrophoresis, including capillary electrophoresis. Apparatuses for
capillary
electrophoresis are, commercially available (e.g., Hewlett-Packard; CA USA;
SpectruMedix, PA, USA). 'In general, separation by electrophoresis
fractionates the nucleic
acids by length, to an approximation. The separated diversity panel products
are collected
by means known in the art and transferred to the array substrate.
Other types of chromatography can also be employed. Such technologies
include HPLC (high-performance liquid chromatography) and matched ion
polynucleotide
chromatography (Transgenomic, Inc. USA; U.S. Patent No. 5,986,085; U.S. Patent
No.
5,997,742).
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
Another technique for separation, although Iess efficient that the other
methods, is dilution of the diversity panel sample to a point where the sample
drop to be
applied to the array contains a discrete nucleotide molecule.
4. Application of diversit~panels into an addressable array
Many types of materials, such as silicon wafers,' borosilicafe slides,
microtiter plates, nitrocellulose, or nylon membranes, may be used to form
solid supports
for the' array, However, in practice, silicon ~ wafers (readily available from
the
semiconductor industry) and borosilicate slides (e.g., microscope slides) are
presently the
preferred materials to serve as the solid support.
In certain embodiments, the nucleic acid molecule can be directly bound to
the solid support or bound through a linker arm, which is typically positioned
between the
nucleic acid sequence and the solid support. A linker arm that increases the
distance
between the nucleic acid molecule and the substrate can increase hybridization
efficiency.
There are a number of ways to position a linker arm. In one common approach,
the solid
support is coated with a polymeric layer that provides linker arms with a lot
of reactive
ends/sites. A common example of this type is. glass slides coated with
polylysine (see, U.S.
Patent No. 5667976), which are commercially available: Alternatively, the
linker arm may
be synthesized as part of or conjugated to the nucleic acid molecule, and then
this complex
is bonded to the solid support. For example, one approach takes advantage of
the
20~ extremely high affinity biotin-streptavidin interaction. The streptavidin
biotinylated
reaction is stable enough to withstand stringent washing conditions and is
sufficiently
stable that it is not cleaved by laser pulses used in some detection systems,
such as matrix-
assisted laser desorption/ionization time of flight (MALDI-TOF) mass
spectrometry.
Therefore, streptavidin may be covalently attached to a solid support, and the
nucleic acid
molecule is labeled with a biotin group (or vice versa). The biotinylated
nucleic acid
molecule effectively sticks wherever it is placed on the streptavidin-covered
support
surface. In one version of this method, an amino-coated silicon wafer is
reacted with the
n-hydroxysuccinimido-ester bf biotin and complexed with streptavidin.
Biotinylated
16
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
oligonucleotides are bound to the surface at a concentration of about 20 finol
DNA per
~2.
Alternatively, ' one may directly bind DNA to the support using
carbodiimides, for example. In one such method, the support is coated with
hydrazide
groups, then treated with carbodiimide. Carboxy-modified nucleic acid
molecules are then
coupled to the treated support. Epoxide-based chemistries are also being
employed with
amine modified oligonucleotides. Other chemistries for coupling nucleic acid
molecules to
solid substrates are known to those of skill in the art.
The nucleic acid molecules must be delivered to the substrate material.
Because of the miniaturization of the arrays, delivery techniques must be
capable of
positioning very small amounts of liquids (e.g., less than 1~ nanoliter) in
very small regions
(e.g., 100 p,m diameter dots), very close to one another (e.g., 250 ~,m
separation) and
-amenable to automation. Several techniques and apparatus are available to
achieve such
delivery. Among these are mechanical mechanisms (e.g., arrayers from
1S GeneticMicroSystems, MA, USA) and ink jet technology. Very fine pipets may
also be
used.
Other formats are also suitable within the context of this invention. For
ex~nple, a 96-well format with fixation of the nucleic :acids to a
nitrocellulose or nylon
membrane may also be employed.
After the nucleic acid molecules have been bound to the solid support, it is
often essential to block reactive sites on the solid support that are not
consumed in binding
to the nucleic acid molecule. Otheriwise, the probes will, to some extent,
bind directly to
the solid support itself, giving rise to so-called non-specific binding. Non-
specific binding
can defeat the ability to detect low levels of specific binding. A variety of
effective
blocking agents (e.g.; milk powder, serum alburriin or other proteins with
free amine
groups, polyvinylpyrrolidine) can be used and others are known to those
skilled in the art
(see, for example U.S. Patent No. 5994065). The choice depends at least in
part upon the
binding chemistry. -
17
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
B. METHODS FOR LABELING AND DETECTING NUCLEIC ACID PROBES
As discussed above, the nucleic acid molecules of the diversity panel that
are used to probe the array are preferably directly detectable. Generally, a
detectable
molecule, also referred to herein as a label, will be incorporated or added to
the diversity
S panel nucleic acid sequences. Many types of molecules can be used within the
context of
this invention. Such molecules include, but are not limited to, fluorochromes,
chemiluminescent molecules, chromogenic molecules, radioactive molecules, mass
spectometry tags, proteins, and the like. Other labels will be readily
apparent to one skilled
in the art. Indirect detection can also be used within the context of
this~invention. Proteins
and other molecules are available that will bind to double-stranded DNA but
not to single-
stranded DNA. Thus, hybridization can be measured.
To maximize the use of the arrays, diversity panels that are used as probes
may be mixed prior to hybridization as long as each diversity panel can be
distinguished.
Although there are various- means to distinguish nucleic acids, in the
simplest form, the
products of each diversity panel in the mixture comprises a different
detectable molecule.
The number of diversity panels that can then be mixed and applied to the array
at a single
time is dependent on the number of distinguishable detectable molecules.
In one embodiment of this invention, diversity panel products are labeled
with fluorochromes. A plethora of fluorochromes are commercially available or
can be
chemically synthesized. An extensive list of suitable fluorochromes,
procedures for using
them and detecting them is available in "Handbook of Fluorescent Probes and
Research
Chemicals" (T~ Ed. Molecular Probes, Inc., Eugene, OR, USA, (www.probes.com)).
In an alternative embodiment, the nucleic acid molecules are directly or
~indirectly~ coupled to an enzyme. Following hybridization, a chromogenic
substrate is
applied and the colored product is detected by a camera, such as a charge-
coupled camera.
Examples of such enzymes include alkaline phosphatase, horseradish peroxidase
and the
like. The invention also provides methods of labeling nucleic acid molecules
with
cleavable mass spectrometry tags (CMST) (see for example, U.S..Patent No:
60279890).
After an assay is complete, and the uniquely CMST-labeled probes are
distributed across
18
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
the array, a laser beam is sequentially directed to each member of the array.
The light from
the laser beam both _ cleaves the unique tag from the tag-nucleic acid
molecule conjugate
and volatilizes it. The volatilized tag is directed into a mass spectrometer.
Based on the
mass spectrum of the tag and knowledge of how the tagged.nucleotides were
prepared, one
can unambiguously identify the nucleic acid molecules to which the tag was
attached (see,
e.g., W09905319).
The nucleic acids can be labeled readily by any of a variety of techniques.
When the diversity panel is generated by amplification, the nucleic acids can
be labeled
during the reaction by incorporation of a labeled dNTP or use of labeled
amplification
primer. If the amplification primers include a promoter for an RNA polymerase,
a post-
reaction labeling can be achieved by synthesizing RNA in the presence of
labeled NTPs.
Amplified fragments that were unlabeled during amplification or unamplified
nucleic acid
molecules can be labeled by one of a number of end labeling techniques or by a
transcription method, such as nick-translation; random-primed DNA synthesis.
Details of
these methods are well known to one of skill in the art and are set out in
methodology
books (e.g., Ausubel et al., supra). Other types of labeling reactions are
performed by
denaturation of the nucleic acid molecules in the presence of a DNA-binding
molecule,
such as RecA, and subsequent hybridization under conditions that favor the
formation of a
stable RecA-incorporated DNA complex. ,
C. HYBRIDIZATION TO ARRAYS
The invention provides hybridization of a diversity panel to a diversity
array, which is an addressable array containing products of diversity panels.
Typically, stringent hybridization and washing conditions are used for
nucleic acid molecules over about 500 bp. Stringent hybridization conditions
include a
salution~ comprising about 1 M Na+ at 25° to 30°C below the Tm;
e.g., 5 x SSPE, 0.5%
SDS, at 65°C; see, Ausubel, et al., Current Ps-otocols in Molecular
Biology, Greene
Publishing, 1995; Sambrook et al., Molecular Cloning: A LaboYatoYy Manual,
Cold Spring
Harbor Press, 1989j. Tm is dependent on both the G+C content and the
concentration of
19
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
Na+. A forinula to calculate the Tm of nucleic acid molecules greater than
about 500 by is
Tm= 81.5 + 0.41(%(G+C)) - logs°[Na+J. Washing conditions are generally
performed at
least at equivalent stringency conditions as the hybridization. If the
background levels are
high, washing may be performed at higher stringency, such as around
15°C below the Tm.
Low stringency hybridizations are performed ~at conditions approximately
40°C below Tm, and are used. for short fragments; e.g., less than
about_500 bp. For
fragments between about 100 and 500 bp, the Tm decreases about 1.5°C
for every fewer 50
by than 500. For very small fragments, e.g., less than about 50 bp, a formula
for
calculating Tm is 2°C for each AT pair and 4°C for each GC pair.
Very high stringency
hybridizations are performed at conditions approximately 10°C below Tm.
Hybridization conditions are tailored to the length and GC content of the
oligonucleotide. Suitable hybridization conditions may be found in Sambrook et
al., supra,
Ausubel et al., supra, and fiuthermore hybridization solutions may contain
additives such
as tetramethylammonium chloride or other chaotropic reagents or hybotropic
reagents to
increase specificity of hybridization (see for example, PCT/LTS97/17413).
D. DETECTION AND ANALYSIS OF HYBRIDIZATION PRODUCTS
1. Detection
Hybridization may be detected in a vaiiety of ways and with a variety of
equipment. In general, the methods may be categorized as those that rely upon
detectable
molecules incorporated into the diversity panels and those that rely upon
measurable
properties of double-stranded nucleic acids (i.e.~ hybridized nucleic acids)
that distinguish
them from single-stranded nucleic acids (i.e., unhybridized nucleic acids).
The latter
category of methods includes intercalation of dyes, such as ethidium bromide,
into double-
stranded nucleic acids, differential absorbance properties of double and
single stranded
nucleic acids, binding of proteins that preferentially bind double-stranded
nucleic acids,
and the like.
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
In preferred methods, the diversity panels applied to the addressable arrays
are labeled with a detectable molecule. Examples of labels are discussed
above. Following
hybridization, some means of detecting a successful reaction must be
addressed. The
means of detection depend on the type of label used. For example, if a
radioactive label is
used, autoradiography or storage phosphor screens (PhosphorImager) are common
methods
of detection. Other systems, including chemiluminesoent and fluorescent labels
in
conjunction with autoradiography, charge-coupled cameras or confocal
microscopy, are
part of an arsenal of detection systems. .
An alternative detection system that can be used with radioactive,
fluorescent or chemiluminescent labels is a ~CCD integrated silicon wafer. In
this system, a
charge-coupled device (CCD), designed to detect high energy beta particles or
photons, is
placed in direct contact with a silicon support for an array. Upon binding of
the sample to
the immobilized nucleic acids, a radioisotope decay product or photon is
generated.
Electron-hole pairs are generated in the silicon and then electrons are
collected by the CCD.
An alternative detection system for fluorescent molecules is a lens based
camera detecting one or more fluorescent labels. As mentioned above, these
cameras
include epifluorescent microscopes, confocal microscopes, and charge-coupled
cameras. In
the fluorescent systems, a laser excites a fluorescent label, the emitted
light is collected
through a bandpass filter, and the signal is detected by a photomultiplier
tube that has
electronics for counting photons.
Other labels are also amenable to use with either a lens-based camera or a
CCD. For example, chemiluminescent labels or chromogenic substrates can be
detected
with a lens-based charge-coupled camera.
In some embodiments, the label is a cleavable mass-spectrometry tag. Such
labels are then detected using a mass-spectrometer. Many detection ~ systems
are
commercially available (e.g., Affyrnetrix, Santa Clara, CA). One skilled in
the art is able to
choose au appropriate detection means and equipment for the label used.
21
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
2. Anal, '
A genotype of an organism is determined by the pattern of hybridization.
Patterns can be expressed as presence or absence of hybridization, the degree
of
hybridization, or some combination of these. The simplest analysis is
performed by
determining the presence or absence of hybridization. When the complexity of
the genome
of the organism to be genotyped is greater than the complexity of the
genome(s),
represented on the . array, the absence of hybridization conclusively
signifies a
polymorphism. When the complexity is less than on the array, the absence of
hybridization
can signify either a polymorphism or a lack of representation of those
sequences in the
probing diversity panel. The presence of hybridization, however, does not
necessarily
signify the absence of a polymorphism under either scenario. As described in
more detail
below, the pattern of hybridization is informative.
When the presence or absence of signal is assayed, each addressable area is
queried for hybridization using a method appropriate to the label. For
example, when
fluorescent labels are used, such as Cy3 and CyS, both green and red signals
are assayed.
When positive and negative controls are included on the array, signals are
compared to the
controls and each addressable area is assigned a value, e.g., 1 for detectable
hybridization
and 0 for no detectable hybridization. In general, a value of 1 is assigned
for detection over
a threshold level and 0 assigned for detection under a threshold level. It
will be appreciated
by those skilled in the art that detection of polymorphisms is based primarily
on finding a
binary distribution of signal values for any particular array feature when
hybridized with
multiple diversity panels.. Preferably, the panels are the same as those used
to create the
diversity array (see Example 5). In case a diversity panel is generated from a
heterozygote .
for a polymorphism, one will then detect a trimodal distribution. In such a
case two
threshold values are calculated, the: first threshold separates the "0"
cluster (lack of
hybridization) from the "0/1" cluster (heterozygote) and the second threshold
separates the
"Oh" cluster from the "1" cluster (hybridization present). Conventional
statistical methods
may be used to determine the threshold levels.
22
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
The genotype of the organism may then be expressed as a value for each
addressable area. As an exemplary aid to understanding, if the addressable
array is a 96-
spot format (a grid of 8 rows (A-G) x 12 columns (1-12)), and the value for
hybridization is
1 and no detectable hybridization is 0, then visualization of a hypothetical
genotype from
one such grid may look like:
1 2 3 4 5 6 7 8 9 10 11 12
A 0 0 0 1 0 1 1 1 0 1 0 0
~~
B 1 1 0 1 0 0 1 I 0 1 0 1
C 0 1 1- 1 1 1 0 0 0 1 0 1
D 0 0 0 0 0 1 1 1 0 1 0 1
E I 1 I 0 1 0 1 0 0 0 1 I.
F I 1 1 1 1 1 0 1 0 1~ 0 0
G 0 0 0 1 1 1 0 1 0 1 0 1
H 1 1 0 0 0 1 0 0 1 1 0 1
In a similar fashion, if the extent of hybridization is to be measured, then
relative values are assigned to each addressable location. The relative values
will generally
be normalized to controls.
All data can be collected into database formats to facilitate comparisons as
well as perform further analyses, such as construction of genotype trees.
E. USES FOR GENOTYPING BY HYBRIDIZATION
As discussed and elaborated upon herein, genotyping by hybridization
facilitates many different genetic studies, such as breeding of animals or
plants, trait
selection, introgression of traits, genetic disease diagnosis, forensic
analysis, viral family
detection, -genomic mapping, determining origin of germplasm, establishing
relatedness of
germplasm, and the like.
23
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
1. Genotyping / detection of polymorphisms
As described above, this invention provides methods and compositions for
establishing the genotype of an organism. Within the context of this
invention, the
genotype is expressed as presence/absence or extent of hybridization to
individual nucleic
acid molecules from two or more organisms. Until this invention, genotypes
have been
expressed in such ways as complete nucleotide sequence, explicit restriction
fragment or
amplified fragment lengths, a collection of genetic traits and the like. The
present
invention now allows genotypes to be written as it were by hybridization
profiles.
One exemplary application of such genotyping is the determination of a
number of individuals or strains within a species. In the world of plants,
samples of a plant
species are collected from around the world. .Nucleic acids are extracted from
these
individuals. Genotypes of each individual are determined using the methods
taught herein.
Comparisons of the genotypes can reveal the relatedness of the individuals.
Briefly, . the
closer the patterns of hybridization, the more related the individuals. In
this way, for
example gene flow can be documented. .
In other systems, the gene flow or relatedness of viruses can be tracked. In .
this regard, the genotype of HIV infections is becoming crucial for predicting
disease
progression, selecting effective therapies, and the like. Other viruses or
parasites, such as
trypanosomes, that display extensive genotypic variation are useful candidates
for the
present invention.
Breeding programs for both plants and animals will benefit from this
invention: For example, when there is a small population of rare animals that
are being
bred, it is believed. important to interbreed unrelated individuals.
Similarly, for plant
breeding, it would'-be advantageous to characterize at the molecular level the
diversity
available .to the plant breeder, so that he can choose the most appropriate
individuals to
work on before embarking on an extensive crossing and selection program. Most
current
means of determining relatedness are cumbersome, laborious and yield- limited
information.
In contrast, the present invention allows high throughput and yields extensive
information.
24
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
The present invention also provides methods for identifying polymorphisms.
As discussed above, a polymorphism is identified by amplifying nucleic acids
from two
different organisms, preparing diversity panels from the two organisms,
placing the
diversity .panel from one into an addressable array and hybridizing the array
With the
diversity panel of the other organism. When the fraction of the genome
represented in the
diversity panel on the array is the same or less than the fraction of the
genome represented
in the diversity panel in solution, a polymorphism is identified by the
absence of
hybridization. In a preferred embodiment, the arrayed diversity panel is
cloned first and
individual cloned molecules are placed into the array. When a polymorphism is
identified
it is then straightforward to isolate the clone and thus, the polymorphism.
The nature of the
polymorphism can be fiuther characterized by sequence analysis, restriction
site analysis,
heteroduplexing, and the like.
This approach can be applied to the identification of a polymorphism
genetically linked to
a phenotypic trait: the strategy commonly known as Bulk Segregant Analysis can
benefit
from, the present invention. Classically, a large number of individuals are
scored for a
particular trait or phenotype and each individual is placed in one of two
possible categories.
The DNA of individuals in each category is pooled and interrogated to identify
markers
specifically present in one of the two categories. A clear advantage of the
present invention
to perform this analysis is its parallel nature: in a single experiment, a
large number of
markers will be interrogated simultaneously. The chance of detecting a
polymorphic
marker distinguishing between the two categories is therefore higher.
2. Isolation of polymorphism / transgenic plants
The nucleic acid molecule comprising a detected polymorphism is isolated
using techniques known in the art. The nucleic acid molecule may be cloned in
an
appropriate vector if not already cloned. In turn, the clone may be mapped on
the genome
using conventional techniques or mapped to a collection of BAC or YAC clones.
The
nucleotide sequence may be determined as well.
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
In certain embodiments, the polymorphic nucleic acid molecule may be used
to transform a host cell, either a plant or animal. Methods to make transgenic
plants are
known in the art. Depending upon the nature of the transgenic sequence it may
be
desirable to operatively link the sequence to a promoter that are active in
plants. Such
promoters may be constitutive, such as the 35S CaMV promoter, tissue-
dependent,- such as
those active only in root tissues, stage-dependent, such as those active
during
embryogenesis, or the like. Examples of promoters are readily found in public
databases
(e.g., GenBank).
3. Following_polymorphisms through intro~ression / back-crossing
Introgression of specific alleles is a goal frequently pursued in plant
breeding as well as laboratory animal breeding programs. The end product is a
plant or
animal nearly identical to the desired parent except for a specific region of
the genome that
is contributed by another individual. For example, the advent of mice strains
with identical
backgrounds but differing at the Major Histocompatihility Complex locus was
instrumental
in understanding the effect of MHC differences on organ transplantation. in
crop
development, a desirable trait, such as disease resistance, may be identified
in a plant, but is
generally introgressed into elite varieties that are better suited to the
local environment, soil
and climate or to consumer preferences than- the original plant. The
introgression is usually
performed by repeated backcrosses of the new individual with the elite parent.
During the
20~ ~ introgression of the genes that account for the traits, means to follow
that trait are
necessary. In some cases, the trait may not be assayable in the field except
under defined
conditions (e.g., challenge with the pathogen). It is advantageous, however,
to have a
marker for the gene i.e. a polymorphism genetically linked to the desired
trait, which can
then be assayed to identify suitable plants for the breeding program. In order
to accelerate
the speed. of the introgression process ,it is also important to monitor that
the rest of the
genome is as similar as possible to the elite parent. In that regard, the
determination of a
genotype encompassing a Large number of markers in parallel, provided by the
present
invention is a distinct advantage.The present invention provides the means to
follow
26
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
specific markers linked to a desirable traits, as well as genome-wide markers
measuring the
extent of reconversion of the genome, and allows for high throughput
screening.
4. Gonstructin~~ a genetic map' discoyering-important genes through
association studies
The present invention provides the means to build rapidly a genetic map,
'even for organism for which little or no molecular data is available. Once
the genotype of
two individuals is determined according to the invention, the progeny arising
from a cross
between these individuals can be genotyped in a similar manner. Each
individual from the
progeny is genotyped. Commonly used softwares such as Mapmaker will then
extract from
the individual genotypes the co-segregation ratio between markers and
calculate a lir~lcage
map of the markers. Determining rapidly the map position of a large number of
markers
allows breeders or geneticists to associate phenotypic data (such as
qualitative or
quantitative traits) with genetic data (such as molecular markers linked to
the trait, markers
for Quantitative Trait Loci and the like) and molecular data (such as DNA
sequence
associated with the markers, surrounding the markers, comprised between the
markers, arid
. the like). Association studies as described above are an important component
of gene
discovery and gene function identification in the agricultural as well as the
medical field.
With the rapid progress in genomics for an increasing number of plants and
animals (for
example the availability of the complete sequence of the human genome and-the
genome of
Arabidopsis thaliana), this approach to gene discovery will become
increasingly
productive.
5. Varietal identification
The fingerprint of an individual as determined by the present invention can
be used to identify the individual unambiguously. Due to the parallel analysis
of a large
number of markers provided by the present invention, the identification is
highly reliable
and the fingerprinting process has a high throughput and a low cost. This
reliable and cheap
method for identifying plant or animal varieties,is useful in a large range of
activities: it
will facilitate the detection by plant or .animal breeders of unlawful copying
of their
27
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
registered varieties and it will facilitate quality control of identity
preserved crops.
Fingerprinting as provided by the present invention can also be used to
identify the
genotype of grains delivered by a producer, for example for the purpose of
collecting
- royalties on the production of specific varieties.
. ~ -
The following examples are offered by way of illustration, and not by way
of limitation.
EXAMPLES
EXAMPLE 1
1 O GENERATING A DIVERSITY PANEL FROM RICE GENOMIC DNA
Representative samples of rice germplasm are identified for genotyping. The
samples are chosen solely for demonstration purposes and are chosen on the
basis of other
knowledge for being a diverse set of genotypes. This is done, mostly through
analyzing
dendrograms based on sequence and/or molecular marker polymorphism in order to
pick up
15 members of separate groupings. Also representative genotypes can be
identified as
representatives of separate clusters if the results (like Principal Component
Analysis) or
clustering algorithms are available. Alternatively representative genotypes
can be identified
through single pass sequencing of rapidly evolving segment of the genome
followed by
similarity/dissimilarity analysis. DNAs from a sampling of genotypes
representing genetic
20 diversity of rice species (usually 10-15) are used to generate DNA
diversity panels through
a number of techniques, one of which is exemplified below.
In this Example, diversity panels are ~ generated from genomic DNA
prepared from 9 rice cultivars: Azucena, IR20, IR64, Italica, Karolina,
Labelle, L203,
Millin and Nipponbare. Three different restriction endonuclease (Table 1)
digestions of the
25 DNAs generate fragments, which are~ligated with adapters, amplified and
cloned. In this
specific embodiment, primers for amplification are chosen such that the
resulting products
28
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
comprise a subset of the restriction fragments. With this method, complexity
of the
genome is reduced by 100 to 1000-fold compared to total genomic samples.
Genomic DNA is extracted from young seedlings (Murray and Thompson
Nucleic Acid Res. 8: 4321=4326 (1980)). About S ng of DNA from each cultivar
is digested
S ' at 37°C for 1 .hour with 2 units of restriction enzyme in a volume
of 8 p,1. Following
digestion, 2 p,1 of ligase mixture is added, and the reaction is incubated at
37°C for 3 hours.
Ligase mixture comprises 0.2 p,i T4 ligase (NewEngland Biolabs, MA), 0.2 p.1
lOxligase
buffer, 0.1.1 100xBSA (NewEngland Biolabs, MA), 0.2 ~zl SO mM ATP, 1.2 ~,l
MilliQ
(MQ) Hz0 and 0.1 ~l of enzyme-specific adapter (Table 1) at SO pmoU~I for MspI-
specific
adapter and S pmol/~.1 for EcoRI- and PstI-specific adapters.
After ligation, the mixture is diluted to S00 p,1 with MQ H20 and 2 ~.l is
used as a template in a SO p,1 amplification reaction with 2 units of RedTaqTM
polymerase
(Sigma Chemicals, St Louis, MO, USA) and one of the primers (1.S p,1 at
SO.nglp,l) listed
in Table 1. After incubation at 95°C for 3 min the reactions are cycled
30 times: at 94°C
1 S for 30 sec, 60°C for 4S sec and 72°C for 1 min. A final
extension cycle is performed at
72°C for 8 min.
Table 1
' Primer sequence + selective
bases
Restrictio.
Adapter sequence
at
3'
end
of
primer
n enzyme
~
_
GACTGCGTACCAATTC-XXX
(SEQ
ID
No
3)
ECORI CTCGTAGACTGCGTACC (SEQ ID No
1)
CATCGTACGCATGGTTAA (SEQ ID No2)XXX
=
AAG;
AGT,
ACG,
ATG
GATGGATCCAGTGCAG-X (SEQ
PstI CACGATGGATCCAGTGCA (SEQ ID No ID
4) No
6)
GACGTGCTACCTAGGTC (SEQ ID No5)
X
=
T
GTAGACTGCGATGCGG-XX
(SEQ
MSpI GACTGTAGACTGCGATG (SEQ ID No iD
7) No
9)
ACATCTGACGCTACGC (SEQ ID No
B) ',
XX=
TG
29
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
The amplified fragments are ligated into PCR2.1-TOPO vector using the
TOPOTM cloning kit and transformed into heat-shock competent E. coli strain
TOP10F'
{Invitrogen, Carlsbad, CA) according to the manufacturer's insiructions.~
Briefly, amplified
products may be purified to reduce adapter and primer contamination. The
ligation
mixture, which contains approximately 2 p.1 of amplified products, is
incubated for 5 min
and terminated. About 2 - 2.5 ~.l of the ligation reaction is used to
transform E. toll.
Approximately 20-50 p,1 of the transformed E. toll is plated on L plates
containing
ampicillin: for selection and X-gal for blue/white visualization to identify
recombinant
plasmids. Approximately 1000-2000 recombinants are typically isolated. This
number
represents a similar complexity as the diversity panels that are used for
detecting
polymorphisms.
Individual colonies that contain recombinant plasmids (white colonies) are.
transferred by toothpick into 20 p,I of 10% glycerol. From each glycerol
sample, a 5 ~1
aliquot is transferred to 45 p,1 of RedTaqTM amplification master mix
containing 1 S pmols
each of Forward and Reverse M13 primers and 1.5 units of RedTaqTM polymerise.
The
reactions are incubated in microtiter plates for 5 min at 95°C followed
by 30 cycles of 30
sec at 94°C, 30 sec at 54°C and 1 minute at 72°C
(ThermowellTM 96 well plate Model M,
Costar, Corning N~ and 1 cycle of 72°C for 5 min, followed by a hold
at 4°C.
Following amplification the products are precipitated with one vol of
isopropanol {100 p,1) at room temperature. The plate is then centrifuged at
3200 rpm for 20
min at 4°C. All the liquid is removed, and the pellet is washed quickly
with 100 ~.1 of 70%
EtOH. The plate is then f1u-ther centrifuged for 10 min at 4°C. The
EtOH is removed, and
the plate is air dried.. The pellet is resuspended at a concentration of about
20 ng/p,l in MQ
water, 3x SSC, or lx SSC, 0.01% sarcosyl.
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
EXAMPLE 2
PREPARATION OF A DIVERSTTY ARRAY
The amplified DNA inserts are transferred into 384-well plates (Genetix)~
and arrayed using a microarrayer (e.g., 417 microarrayer;.Affymetrix, Palo
Alto, CA) onto
PolysineT"" microscope slides (MenzelGlazer, Germany) or in-house polylysine-
coated
microscope slides. Arrays are made with six replicates per fragment. The
average center to
center spot spacing is 250 ~,m.
At least I day after arraying, slides are processed by Hydration in IX SSC,
quick drying, blocking for 15 min in a solution of NaBrH4/PBS (prepared by
dissolving 1 g
NaBrH4 in 300 ml PBS, pH 7.0) (see also
http://www.microarrays.org/protocols.html,
Protocol for Post Processing Microarrays; June 2000, except that the succinate
anhydride
~pyrolidone is replaced with NaBrH4 in PBS as the blocking solution). Slides
are then
dipped in boiling water for 30 sec to denature the DNA and followed by a 10
sec dip in
100% EtOH. Slides are dried by centrifugation at 1000 rpm in a slide rack on
microtiter
plate carriers for 1 minute. I .
EXAMPLE 3
DETERMINATION OF FINGERPRINTS BY HYBR,mIZATION OF A
LABELED DIVERSITY PANEL TO AN ARRAY
For hybridization to a microarray prepared as taught in Example 3, a
' 20 diversity panel of one or more specific genotypes is generated and
labeled with a
' fluorescent dye. In a single hybridization experiment, a number of genotypes
can be
compared, the number being equal to number of labels that can be unequivocally
detected
and resolved. For example, an Affymetrix 418 scanner is equipped with "green"
and '.'red"
lasers, allowing for simultaneous analysis of. two different samples.
Genomic DNA (200 ng-2 ~,g/pl) is cut with EcoRI and ligated to EcoRI
adapters (1.5 p.1 of 5 pmoles/p.l) using and excess of T4 ligase (40 units)
for 3 hours at
room temperature. For this step, 200 ng of DNA is sufficient, but 1 ~,g of DNA
provides
31
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
sufficient material for a number of hybridizations. Following ligation, the
mixture is
purified on a Qiagen column.
An amplification reaction contains 2.5 units RedTaqTM (Sigma, St. Louis,
MO USA), 1-5 p,1 of ligated genomic DNA from above, 10 p,1 lOx buffer (10x
buffer
contains 500. mM KCl, 1 M Tris-HCl (pH 8.8), 0.1% Triton X-100, 15 mM MgClz),
10 ~1
of 2mM dNTPs, and 1 ~.1 of 20 pmol/~.1 primers. Because the DNA fragments are
ligated
with an adapter a single primer identical to one strand of the adapter is used
with one or
more additional bases added to the 5' end. In some experiments a mix of
primers is used
that are identical to one strand of the adapter but have one or more
additional bases at the 3'
end of the primer. Such a mix serves to limit the complexity of the resulting
fingerprint.
Amplification conditions are 1 cycle of 95°C for 2 rnin, 30 cycles
of 94°C
for 30 sec, 54°C for 30 sec, 72°C for 1.1 min, 1 cycle of
72°C for 5 min and hold at 4°C.
Amplification products are purified using Qiagen Quick PCRTM purification
columns to
remove the dNTPs, which-otherwise will affect the labeling steps.
' Amplified material is labeled by incorporating dUTP-Cy3 or dUTP-Cy5
using a random priming method. In this method, up to ~11 ~,1 of DNA in MQ
water is
mixed with 2 p,1 of E. toll DNA Pol I and 1 ~,l of 3 p,g/~,l hexanucleotides
in 10 mM Tris-
HCl (pH7.5), 5 mlVl MgCl2 and 7.5 mM dithiothreitol, . The mixture is boiled
for 2 min
and snap-cooled on ice for 5 min. The following ingredients are then added: 2
~,l of 2 mM
each dATP, dGTP, dCTP~and 90 p,M dTTP, 1-2 p,1 of dUTP-Cy3 or dUTP-CyS, and 1
~,1 (5
units) E. toll pol I (large fragment). The reaction mixture is incubated for 3
hours at 37°C
and terminated by the addition of 50 mM EDTA. The two labeling reactions (Cy3
and
Cy5) are pooled and purified together using Qiagen columns according to the
manufacturer's recommendations, except that one extra wash using 0.5 ml wash
buffer is
performed. The labeled nucleic acid molecules are eluted in ~30 ~,1 of water.
Alternatively, amplified material can be labeled using a Deca-random-prime
DNA labeling kit from Fermentas (Vilnius). When this kit is used, minor
deviations from
the manufacturer's instructions are used, specifically the reaction volume is
reduced to 5 p1,
32
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
the time increased to 1 hour and 0.4 p.1 of 1 mM Cy3-dUTP or Cy5-dUTP is used
instead of
3zP-dNTPs. probes are not purified for hybridization.
Prior to hybridization, 5 p,1 of the labeled material is mixed with 2 p,1 of
20
mg/ml herring sperm DNA which is dissolved in Express HybridizationTM buffer
(Clontech, Palo Alto, CA, USA), and the mixture is denatured at 96°C
for ~ 3 min. The
denatured probes are mixed with 10 to 15 p,1 of ExpressHyb hybridization
solution, pipetted
directly onto the microarray surface and covered with a glass cover slip (24
mm x 24 mm
Mediglass, Australia). Slides are then quickly placed into a homemade
humidification
chamber in a 65°C water bath for overnight hybridization.
- After hybridization, the coverslips are removed, and the slides are rinsed
in
lxSSC with 0.1% SDS for 5 min; lxSSC for 2 min; 0.2 xSSC for 2 min; and 0.02 x
SSC
for 20' sec; all solutions are at room temperature. Slides are quickly dried
by centrifugation
at 1000 rpm in a slide rack on microtiter plate carriers for 1 min.
The intensity of fluorescence at each spot is measured by scanning the slide
with an array reader (for example Affymetrix 418 microarray scanner).
Fluorescence is
read using scanner settings appropriate for the fluorescent dyes used in
labeling reaction.
For example: for Cy3 dye, the green laser is set to PMT 60 and laser power at
100%, and
for Cy5 the red laser is set to PMT 90 and laser power at.100%. Scanning
conditions are
adjusted if necessary.
~ Identification of polymorphic clones may be made by visual inspection of a
graphic file representing an overlay of scanning results for two genotypes to
be compared.
An overlay can be a result of single hybridization or, alternatively scans
from independent
hybridizations can be overlayed. Polymorphic clones may be identified as those
hybridizing to only one of the two samples compared. Numerous statistical
methods are
available to facilitate conversion of signal intensities into binary
(presence/absence)
characters. Large populations of genotypes can be analyzed in pairs to develop
similarity/dissimilarity measures matrix for the whole population.
In certain experiments, spot signal intensities are analyzed by Scanalyse ver.
2.44 (Stanford University) as well as GenePix Pro v. 3 (Axon Instruments) and
GMS
33
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
Pathways (Affymetrix v. Beta). The outputs of these image analysis programs
are further
analyzed using a program developed for Mathcad v. 8.
Representative examples of outputs can be seen in Figures 5 and 6.
EXAMPLE 4
COMPARISON OF TWO SAMPLES ON A SINGLE ARRAY
In this example, fingerprints of rice cultivars are determined by
hybridization of labeled diversity panels to a diversity array comprising a
diversity panels
generated from a mixture of 9 rice genomes. A schematic of this type of
experiment are
compared on a single array as exemplified in Fig 1B.
The diversity panels are generated using 9 cultivars of rice (3 indica and 6
japonica types). Several panels are constructed using the pair-wise
combination of
restriction enzymes and primers described in Table 1. The resulting fragments
in the
diversity panel range from 0.3 to 2.4 kb with an average insert size of around
1 kb. In
analysis of the fingerprints from hybridization data, an array feature or
element is scored
when the signal is at least 3 times the level of local background for the
vector control
(TOPO). At least 90% of array elements are scored for the panels analyzed in
these
examples. Furthermore, this value is reached without purification of
amplification
products.
Fingerprints for four rice cultivars (Bala, Millin, IR64 and IR20) are
determined by hybridization of a diversity panel from each cultivar to the
EcoRI-generated
diversity panel of the 9 mixed rice genomes. Pairs of the rice cultivars
(e.g., Millin and
IR64; Bala and IR20) are labeled with two different dyes for ease of
detection. A
comparison between Millin (sub-genomic sample labeled with Cy5 dye) and IR64
(sub-
genomic sample labeled with Cy3 dye) shows a high level of variation in signal
intensity
(brightness of array features) and Cy3/Cy5 signal ratios among array elements
(Fig 2A).
Furthermore, a histogram showing green to red channel normalized signal
intensity ratios
(Fig. 2B) shows a tri-modal distribution. The majority of the array features
cluster around
34
CA 02404472 2002-09-26
WO 01/73119 PCT/IB01/00833
a ratio of 1, indicating equal signal intensity for Millin and IR64 samples
(monomorphic
features). The red and green "tails" represent the groups of "polymorphic"
spots.
Several DNA fragments identified in this analysis as potentially
polymorphic between Millin and TR64 (with the red/green ratio above 2.5) are
used as
probes on genomic and sub-genomic Southern blots (Fig. 3). These candidate
clones are
labeled and hybridized to blots of diversity panels and genomic DNA from four
rice
genotypes analyzed most extensively through microarray hybridization method.
Genomic
DNA (2 ~.g) is cut with EcoRI, resolved in 0.8% agarose gel and transferred to
nylon
membranes (Fig 3A). Diversity panels are prepared as described above, resolved
using
1.5% agarose gel and transferred to positively charged nylon membrane
(Boehringer
Mannheim) (Fig 3B). Amplified inserts from the same clone DNA that is arrayed
are
labeled with 32P using the large fragment (Klenow) of E. toll DNA pol I. The
radioactive
labeled probes are hybridized with blots of diversity panels and EcoRI-
digested genomic
DNA.
l 5 Fig 3 (left panels) shows the results of hybridization of candidate clone
F4,
which is polymorphic by fingerprint analysis when a diversity panel of Millin
is tested
against diversity panels of Bala, IR20 and IR64. Thus, in Fig 3B, F4
hybridizes strongly
with Millin diversity panel (lane~2), whereas F4 does not detectably hybridize
to Bala (lane
1), IR64 (lane 3) and IR20 (lane 4) diversity panels. Hybridization of clone
F4 to genomic
DNA (Fig. 3A), which was digested with EcoRI, the same enzyme as used to
generate the
diversity panel, reveals a fragment about 1.6 kb in Millin DNA (lane 2) and a
2.3 kb
fragment in the remaining DNAs. This restriction fragment size difference
accounts for the
presence (Millin) or absence (remaining three genotypes) of a signal apparent
on the
diversity panel Southern (Fig. 3B). In this example a Restriction Fragment
Length
Polymorphism (RFLP) in genomic DNA was converted to the presence/absence
polymorphism in sub-genomic samples that can be identified in a highly
parallel fashion
using the DNA microarray platform.
A second candidate polymorphic fragment, clone F8, also shows
polymorphism on Southern analysis. In this case, a smaller EcoRI fragment (1.3
kb) is
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
detected in Millin and Bala DNA (lanes 1 and 2), whereas both IR20 and IR64
DNA
display a 1.5 kb fragment (lanes 3 and 4) (Fig. 3A). However, while on the
genomic
Southern the band intensities are similar, in the diversity panel Southern,
the hybridization
strength to IR64 and IR20 are much weaker compared to the Millin and Bala
bands. The
difference in the abundance of specific amplified material in the diversity
panel translates
into easily detectable polymorphism in microarray experiment when Millin is
contrasted
with IR20 or IR64. In this case, an RFLP is converted to a quantitative
polymorphism
detected by signal intensity differences between Millin and IR64 sub-genomic
samples on
the array.
i0 One additional clone, F11, is characterized in this example. F11 scores as
monomorphic when analyzed against four rice cultivars, i.e., approximately
equal signal
intensity is observed for this clone when the array containing it is-probed
against any of the
four labeled diversity panels. F11 is also tested as a probe against a
Southern blot of
diversity panels from these genotypes. Fig. 4 shows clearly that similar size -
(and
abundance) products hybridize with the F11 probe in all four genotypes.
This EcoRI-generated diversity panel is also used to determine the minimal
amount of DNA required for generation of reproducible diversity panels. Four
different
amounts of adapter ligation products, from 0.2 ng to 12.5 ng, are used for
amplification of
four genotypes (Bala, Millin, IR64 and IR20) and hybridization results are
analyzed for
polyrizorphisms. All genotypes are scored reproducibly as either present (1)
or absent (0)
for 14 elements identified as polymorphic at the four DNA amount levels (data
not, shown).
EXAMPLE 5
IDENTIFICATION OF ARRAY ELEMENTS AS POLYMORPHIC OR NON-POLYMORPHIC
In order to identify the elements of the array that represent polymorphic
DNA fragments all nine rice cultivars used for Diversity panel generation are
analyzed on
duplicate slides as described in these Examples. The spot intensities
normalization, data
transformation (to obtain near log-normal distribution) and polymorphic spot
detection are
36
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
achieved using the Mathcad 8Ø The program calculates the value (marked as
"x" on each
curve) best ~ separating the two clusters of low and high signal ratios,
respectively, and
classifies each sample analyzed at the particular polymorphic feature as
either. 0 (low value
cluster) or 1 (high value cluster). A table of binary scores is created,
automatically for all
the samples and the polymorphic array. Typical distributions of normalized
ratios of signal
intensities (the signal for MspI sub-genomic sample labeled with Cy3 divided
by the signal
for Topo vector control labeled with Cy5) for four examples of non-
polymorphic' (Fig 7A)
and polymorphic (Fig 7B) spots are presented. For all non- polymorphic spots
the ratios of
signal intensities show a monomodal distribution across 18 slides (9 cultivars
x 2 slides per
cultivar). The polymorphic spots (Fig. 7B) show a, clear bimodal distribution
for the log
transformed signal ratios.
In Fig 7A it is apparent that the range of ratios is larger for spots with an
average ratio value below zero (in which the signal from the sub-genomic
sample is weaker
than the Topo control signal). Distribution of the ratios for alI 384 features
of the MspI
IS panel for the same set of 18 slides (Fig. 7C) shows more variation between
slides at lower
values (especially below -0.2). The presence of a different number of
"positive" spots
among genotypes tested is likely to be one of the sources of the between slide
variation.
However, since the proportion of the polymorphic spots is relatively low this
result most
likely indicates that array features that hybridize weakly to the sub-genomic
sample
(around 30% of the total number) are more influenced by the noise in our
system compared
to the more strongly hybridizing ones.
The number of array features found as polymorphic among nine rice
cultivars is 50 (14.5% of scored spots) for the MspI diversity panel. Apart
from providing
an estimate of polymorphism level detectable by this system, identification of
polymorphic
features allows assessment of the level of redundancy among them. DNA
fragments
representing array elements displaying the same pattern of polymorphism (same
binary
scoring) among the nine rice cultivars are resolved on an. agarose gel. DNA
fragments with
the same apparent mobility are scored as repeats (Fig 8). The analysis
revealed that 50
polymorphic spots represented 28 unique clones of which most (20) had just one
copy in
37
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
the MspI panel of almost 400 clones. Based on the average MspI fragment size
(under 1
kb), the rice genome size of 430 Mb, and 256-fold complexity reduction due to
the
amplification primers used having two selective bases (1/16 x 1/16), over 1000
unique
fragments are expected in the MspI diversity panel, even if less than 50% of
the fragments
amplified efficiently. The presence of mostly unique clones among polymorphic
spots
evidences that this invention can analyze fairly complex samples.
Analysis of diversity panels roughly 16 times more complex (using an
amplification primer with a single selective base) indicates that through
minor modification
of the assay sensitivity (e.g. spotted DNA concentration, diversity panel
labeling and
scanning efficiency, etc.) a comprehensive genome scan can be achieved using
the present
invention.
EXAMPLE 6
USING DIVERSITY ARRAYS TO DETERMINE THE RELATEDNESS OF GENOMES
The binary scoring table for the 28 unique polymorphic features is used to
calculate the distances between the cultivars. A distance table is used to
produce
dendrograms showing the relatedness of the cultivars. Binary scoring tables of
28 unique
features from MspI and 28 from PstI are clustered by Cluster program (Stanford
University) using similarity metric setting of correlation uncentered and
presented by
treeview . (Stanford -University). Differentiation among the cultivars
analyzed and
separation between japonica and indica types is apparent in both dendrograms.
Figure 9A
shows the separation between indica and japonica rice cultivar classes based
on fingerprints
established from using the MspI diversity panel. Similar results are found
using the PstI-
generated diversity panel (Fig 9B).
38
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
EXAMPLE 7
POLYMORPHISMS ARE INHERITED IN A MENDELIAN FASHION
In order to verify that the polymorphisms detected by this system behaved
as Mendelian markers doubled haploid (DH) lines developed from the cross
between IR64
and Azucena (REF) are used for genetic mapping. All 40 polymorphisms
segregating in
the DH lines population are successfully mapped on the microsatellite-based
framework
without any apparent clustering of the new markers.
EXAMPLE 8
FINGERPRINTING USING A COMPLEX MIXTURE OF DIVERSITY PANELS
In this example, complex DNA samples are analyzed to demonstrate that
minor amounts of a genome are detectable. DNA fragments from diversity panels
developed from 8 species are arrayed on the same slide., The mix included rice
and 7
species of micro organisms. This composite panel is then used as a target for
hybridization
with a diversity panel comprising sub-genomic samples from rice with or
without a DNA
admixture from microorganisms. In one example, the diversity panel from rice
cultivar
Millin, which is labeled with Cy5 dye, is hybridized to the composite panel
together with a
mixture (at 10:1 DNA ratio) of Millin and Ehterobacter sp (closest
Buttiauxella agrestis)
(Sproer, C. et al., 1999) diversity panels labeled with Cy3 dye (Fig. 10). The
left part of
the panel (Fig. 10A), containing rice-derived features, shows mostly yellow
spots, .
indicating a similar level_of hybridization signal for the "pure" Millin
diversity panel as for
the Millin mixed with Enterobacter diversity panels. This observation is
confirmed by the
histogram of signal ratio distribution (Fig. 10B) indicating a lack of rice
derived features
with ratio larger than 2.5. At the same time, there is. a clear pattern of
strongly "green"
features (ratios larger than 2.5) located exclusively to the addresses of the
Enterobacter-
, derived features. There is no significant signal detected at other
microorganism-derived
spots on the composite panel, even with closely related species as determined
by 16S
sequence homology analysis.
39
CA 02404472 2002-09-26
WO O1J73119 PCT/IBO1/00833
EXAMPLE 9
EFFICIENT DETECTION OF DNA POLYMORPHISMS IN THE BARLEY GENOME
In this example, diversity arrays are used to identify polymorphisms in the
barley genome, which is more than 10 fold larger than the rice genome. Barley
diversity
panels are generated using DNA from 3 barley cultivars: Steptoe, Morex,
Harrington, and
from Hordeuna spohtaneum (wild barley) accession OSU15. Diversity panels are
constructed according to the Examples above, except that the restriction
enzyme PstI is
used to generate panels having complexities 100 to 1000 fold less than total
genomic
samples (below). Varying complexities of panels are achieved by the choice of
primers
used in amplification. Fragments from the panels are cloned, and inserts are
individually
amplified from bacterial colonies before arraying on glass slides.
Genomic .DNA is extracted from seedlings ~of various cultivars. Genomic
DNA (SO..ng) is digested at 37°C for-1 hour with 2 units of PstI
restriction enzyme in a
volume of 8 p,1. After digestion, 2 pI of ligase mixture is added. Ligase
mixture consists of
0.2 p1 T4 ligase (New England Biolabs, USA), 0.2 p,1 lOx ligase buffer, 0.1
p,1 100x BSA
(New England Biolabs, USA), 0.2 p,1 SOmM ATP, 1.2 p.1 MilliQ (MQ) Hz0 and 0.1
~,1 (5
pmoles) of PstI adapter:
5'-CACGATGGATCCAGTGCA-3' (SEQ ID No: 10) and
5'-CTGGATCCATCGTGCAG-3' (SEQ ID No: 1 I).
After ligation for 3 hours at 37°C,-the mixture is diluted to 500 ~,l
with MQ
H20. 2 p1 of the diluted ligated DNA is used as template from amplification in
a 50 p,1
reaction using 2 units of RedTaqTM polymerase (Sigma, USA). The sequence of
the
amplification primers are either GATGGATCCAGTGCAG (SEQ ID No: 12) or
GATGGATCCAGTGCAG-X (SEQ ID No: 13) where X is A, C, G or T. Single primer for
SEQ ID No: 12 or a combination of primers of SEQ ID No: 13 are used in
amplification to
achieve various levels of complexity reduction. Amplification parameters are 1
cycle at
95°C for 3 min, 30 cycles at 94°C for 30 sec, 60°C for 45
sec, 72°C for 1 min, followed by
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
1 cycle at 72°C for 8 rilin. The amplification products are cloned,
amplified and arrayed
according to methods in Examples 1 and 2.
Diversity panels are prepared as above from cultivars Morex and Steptoe.
The amplification primer used has the sequence 5'-GATGGATCCAGTGCAG-3' (SEQ ID
No: 14). The amplification products are labeled with fluorescent dyes (Cy3 for
the Morex
diversity panel and Cy5 for the Steptoe diversity panel) and the hybridized to
slides
containing the PstI diversity panels from above. Hybridization, washing, image
capture
and analysis is done.according to methods described in Examples 3 and 4. Fig
11 shows a
fragment of the array with polymorphic array features indicated. Depending on
the PCR
primer used the frequency of polymorphic array features detected between Morex
and
Steptoe varied from 10-15%.
EXAMPLE 10
DETECTING POLYMORPHISMS IN THE MOUSE GENOME USING CDNA DTVERSITY ARRAYS
:In this example, diversity arrays prepared from cDNA are used for
genotyping analysis. For any organism, cDNA or EST sequences may be used as .a
diversity panel that can be arrayed and used to establish genotypes.
As an example of this approach, a cDNA library from multiple mouse
strains and tissues is arrayed on glass slides (>5000 independent cDNA clones
per slide).
Arraying and slide processing is done as in Example 2. Diversity panels for
probing the
cDNA are prepared according to. the ~ methods taught in the examples from ~
tyvo mouse
strains, strain C57B1/6 and strain NOD K. Briefly, 0.1 microgram of genomic
DNA is
digested by MspI restriction endonuclease, an adapter with an MspI-compatible
end is
ligated to the restriction fragments, and the fragments are amplified using an
adapter-
specific primer. Amplification products are labeled using fluorescent dyes
(Cy3 and Cy5)
and hybridized to the cDNA diversity arrays using Quick HybT"" buffer
(Clonetech).
41
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
Hybridization, washing, image capture anal analysis is carried out as
described in Examples
4-6.
When a diversity panel generated from strain C57B1/6 that is labeled with
Cy3 is hybridized along with a similarly generated representation from strain
NOD K that
is labeled.with Cy5 to the cDNA diversity array, about 25% (1410/5472) of cDNA
array
features show detectable signal. Among these with detectable signal 144 (10.2
%) have a
ratio of Cy3/Cy5 signal that is >_ 3.0 or _< 0.33, indicating polymorphic
regions. Because
the arrayed nucleic acids are cDNAs, these polymorphic features are markers
not only for a
specific area of the genome but also for a specific gene. A section of the
cDNA diversity
array showing non-polymorphic and polymorphic features is presented in Fig 12.
EXAMPLE 11
DIVERSITY ARRAY ANALYSIS USING CDNA ARRAYS AND DIVERSTTY PANELS GENERATED
WITHOUT AMPLIFICATION
In this example, the diversity panel is generated from genomic DNA by a
method that does not utilize amplification. Instead, the DNA is digested with
a restriction
enzyme and a range of lengths of the restriction fragments are chosen and
isolated. The
panel is then labeled with fluorescent dye and hybridized along with a
similarly prepared
diversity panel from a second sample to a diversity array comprising a large
collection of
cDNAs.
As in the example above, mouse cDNA diversity arrays are prepared using
4000 cDNA clones. Diversity panels are created from two mouse inbred strains,
NOD K
and C57B1/6 by MspI digestion of 10 ~,g of total genomic DNA. Digested DNA is
electrophoresed in a 2.0 % agarose gel, and a section of the gel containing
fragments from
300 by to 700 by is isolated. The DNA is extracted from the agarose and
purified using a
gel extraction kit (Qiagen). The purified DNA is labeled with Cy3 (strain
C57B1/~6) or Cy5
dye (strain NOD K), respectively, using a method described in Example 3.
Hybridization,
42
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
washing and image analysis is done using techniques described in Examples 4
and 5.
Polymorphic array features are identified as those with Cy3/Cy5 signal ratio
>_ 3.0 or <_
0.33. In this particular contrast 9% of the array features are identified-as
polymorphic.
EXAMPLE 12
S DETECTING POLYMORPHISMS DUE TO TRANSPOSON INSERTIONS IN RICE
Diversity array technology is also suitable for detecting polymorphisms
resulting from insertions in the genome. Since transposable elements are among
the
primary source of this type of DNA polymorphism, amplification of transposons
is used as
a method of generating diversity panels for probing rice diversity arrays.
This example
presents polymorphisms due to the transposon, called Stowaway (Bureau et al.,
Proc Natl
Acad Sci USA 93: 8524-8529, 1996), which is a member of the MITE (Miniature
Inverted
Repeat Transposable Elements) class of mobile elements. Diversity arrays are
generated by
amplifying sequences that direct adjoin the Stowaway VII subfamily of MITE
transposable
elements in the rice genome, cloning the amplification products and applying
the cloned
inserts to an array as described in Examples above.
First, genomic DNA of four rice cultivars: Azucena, IR64, Millin,
Nipponbare (500 ng in total, 125 ng mixed from each cultivar) is digested with
MseI
restriction enzyme, and MseI adapters (shown below) are ligated to the
restriction
fragments. Amplification is carried out using the Internal Primer Right.and/or
Left (below)
and MseI adapter Primer 1. After 25 cycles of amplification 1 p1 of amplified
product is
used as a template for another round of amplification using Inverted Repeat
Primer and
MseI adapter Primerl. Amplification products from this reaction are cloned
using a
TopoTM cloning kit: The clone inserts are amplified, purified and arrayed on
glass slides as
in Example 2, resulting in a diversity array comprising 384 clones ready for
polymorphism
detection. The slides are processed as described in Example 3.
43
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
Primer name Primer sequence SEQ ID No
Internal Stowaway VII 5'-ACCGTGTCGCTGTCCTAAAC-3' 15
Right Primer
Internal Stowaway VII 5.'-ATATTCCCAAGGTTTGACTT-3'16
Le$ Primer
Inverted Repeat Primer 5'-CTTTACGAGTATGGAGGGAG-3' 17
MseI adapter Primer 1 5'-CTCGTAGACTGCGTACC-3' 18
MseI adapter Primer 2 5'-TACTCAGGACTCAT-3' 19
Diversity panels are generated from each cultivar separately using the
method above and are labeled with a fluorescent dye (Cy3 for Azucena and Cy5
for IR64).
Labeled panels are hybridized to the diversity arrays and washed. Fluorescent
images are
captured using GMS 4I8 Scanner (Affyrnetrix, CA USA) and analyzed using the
methods
described above. Based on other experimental data, about 17% of the features
axe expected
to be polymorphic.
EXAMPLE 13
ANALYSIS OF POLYMORPHISMS IN RICE USING DIVERSITY PANELS GENERATED BY SEMI-
1 O RANDOM AMPLIFICATION
As an alternative, diversity panels can be generated without the need for a
restriction digestion and adapter ligation step. This offers the possibility
of a complete
automation of this invention. In this example, diversity panels are generated
by a semi-
random, two-step amplification protocol (ST-PCR; Chun et al., Yeast IS: 233-
40, 1997).
ST-PCR requires only genomic DNA and two pairs of amplification primers used
in two
successive amplification reactions.
Genomic DNA (300 ng total) from two rice cultivars, Azucena and IR64, is
used as a template for amplification using two primers: Internal Stowaway VII
Right
Primer ~ (see table above for sequence) and ST-PCRl d Primer (5'-
GGCCACGCGTCGACTAGTACNjoTCGAG-3') (SEQ 117 No: 20). Amplification is
44
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
performed using 0.5 unit Red TaqTM polymerise (Sigma) and using a hot start
program in
which the polymerise is added after the first step of the program. The program
uses the
following steps: (1) 95°C for 3 min; 80°C for 2 min; (2)
94°C for 30 s; (3).42°C for 30 s
and -1.0°C for each subsequent cycle; (4) 72°C for 3 min; (5)
repeat steps 2-4 five times;
(6) 94°C for 30s; (7) 65°C for 30s; (8) 72°C for 3 min;
(9) repeat steps 6-8 for 24 more
times; (10) hold at 4°C.
After completion of this first amplification program, the product is diluted
1:4 with water and 1 ~l is removed for a second amplification. In the second
amplification
reaction, Inverted Repeat Primer (see Table above) and ST-PCR2 primer (5'-
GGCCACGCGTCGACTAGTAC-3' SEQ ID No: 21) are used in the following program:
35 cycles of 94°C for 30 sec; 65°C for 30s; 72°C for 3
min; followed by a hold at 4°C. The
amplification products are cloned. 'Diversity Panels are scaled up as
described in Example 1
and diversity arrays are prepared as in Example 2.
Diversity panels are generated from each cultivar separately using the
method above and are labeled with fluorescent dye (Cy3 for Azucena and Cy5 for
IR64).
Labeled panels are hybridized to diversity arrays slides and washed.
Fluorescent images
are captured .using GMS 418 Scanner, and images are analyzed using the methods
described above. Based on other experimental data, about 17% of the features
are expected
to be polymorphic.
EXAMPLE 14
APPLICATION OF DIVERSITY ARRAY TECHNOLOGY TO DETERMINE DNA METHYLATION
PATTERNS IN RICE.
Analysis of the cytosine methylation status at the CpG .dinucleotide anal
CpXpG trinucleotide within the sequence CCGG. in the rice, genome is performed
using
diversity array technology. For the analysis of developmental variation in
methylation
status among rice tissues, 11 tissues of rice cultivar Millin are collected.
These tissues are:
- ' 45
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
(1) 4-week old seedling leaves, (2) 4-week old seedling roots, (3) mature
pollen and anther,
(4) immature pollen and anther, (5) fertilized ovary and stigma, (6)
unfertilized ovary and
stigma, (7) mature embryo, (8) immature embryo, (9) immature endosperm, (,10)
flag
leaves and (I 1) 3-week callus. - .
Genomic DNA is isolated from these tissues and a mixed sample of DNA is
completely digested with MspI or HpaII, both methylation sensitive. Diversity
panels from
MspI-digested and HpaIl -digested DNA are prepared using the methods described
iri
Example 1 (using MspI adapter and MspI primer .sequences presented in, Table
~. The
diversity panels are scaled up as described in Example 1 and diversity arrays
are prepared .
as in Example 2.
Diversity panels from the various tissues are labeled with either Cy3 or Cy5
and hybridized to the diversity array as described in Examples 4 and 5.
Differentially
methylated regions in DNA between two tissues are identified as array features
that have
Cy3/Cy5 signal ratio >_ 3.0 or 5 0.33. A portion of a diversity array from
two. such
comparisons are presented in Fig. 13 showing clear differences in
hybridization signal for a
number of array features among the tissues compared
Differences in methylation patterns among the tissues analyzed are also
identified through comparison of normalized ratios of signal intensity for a
specific tissue.
The signal is normalized to the signal obtained from hybridization with
labeled TOPO
vector sequence. Statistical methods described herein are used to identify the
features with
developmentally regulated pattern of cytosine methylation. A number of tissue
specific
CpG methylation patterns at CCGG sites are confirmed by Southern analysis in
which
DNA from the diversity panels are hybridized with labeled insert from a clone
identified as
differentially methylated in fertilized ovary and stigma. One such example is
presented in
Fig 14. The absence of hybridization in lane 5 confines the low value of
hybridization
obtained from the normalized data (Figure 14). In addition, DNA sequences are
determined for 20 of the tissue methylation polymorphic fragments. One of the
fragments
has high sequence identity with the rice chloroplast genome and the rest of
the fragments
are derived from the nuclear genome.
46
CA 02404472 2002-09-26
WO 01/73119 PCT/IBO1/00833
From the foregoing it will be appreciated that, although specific
embodiments of the invention have been described herein for purposes of
illustration,
various modifications may be made without deviating from the spirit and scope
of the
invention. Accordingly, the invention is not limited except as by the appended
claims.
47