Note: Descriptions are shown in the official language in which they were submitted.
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
INHIBITION OF SMAD3 TO PREVENT FIBROSIS AND IMPROVE WOUND HEALING
Background of the Invention
Field of the Invention
The invention is related to inhibition of Smad3 to prevent fibrosis and
improve wound healing.
Descriution of the Related Art
Both Smad3 and its closely related homologue, Smad2, are intracellular
mediators of TGF-(3 function, acting
as nuclear transcriptional activators (Massague, J. TGF-beta signal
transduction. Annu Rev. Biochem. 67, 753-791
(1998)); (Derynck, R., Zhang Y. & Feng, X. H. Smalls: transcriptional
activators of TGF-beta responses. Ce// 95, 737-
740 (1998)1. Smad2 and Smad3 mediate intracellular signaling from TGF-[3s 1,
2, 3 and activin, each of which has
been implicated as an important factor in the cellular proliferation,
differentiation and migration pivotal to cutaneous
wound healing (Roberts, A. B. TGF-beta: activity and efficacy in animal models
of wound healing. Wound Repair
Regen. 3, 408-418 (1995)); (0'Kane, S. & Ferguson, M. W. J. TGF-beta s and
wound healing. /nt. J. Biochem. CellBiol.
29, 63-78 (1997)). Mice null for Smad3 (Smad3exaiexs mice) survive into
adulthood, unlike Smad2-null mice which do not
survive embryogenesis (Yang, X. et al. Targeted disruption of SMAD3 results in
impaired mucosal immunity and
diminished T cell responsiveness to TGF-beta. EMBO J. 188, 1280-1291 (1999));
(Datto, M. B. et al. Targeted
disruption of Smad3 reveals an essential role in transforming growth factor
beta-mediated signal transduction. Mo/.
CellBiol. 19, 2495-2504 (1999)); (Zhu, Y., Richardson, J. A., Parada, L. F., &
Graff, J. M. Smad3 mutant mice develop
metastatic colorectal cancer. Cell 18, 703-714 (1998)1; (Weinstein, M., Yang,
X., Li, C., Xu, X., & Deng, C. Failure of
extraembryonic membrane formation and mesoderm induction in embryos lacking
the tumor suppressor Smad2. Proc.
NatlAcad Sci. USA 95, 9378-9383 (199811. Here, to identify selective targets
of Smad3 signaling pathways in viva,
we studied its role in cutaneous wound healing using wild-type mice or mice
heterozygous or null for the Smad3 gene
following targeted disruption (Yang, X. et al. Targeted disruption of SMAD3
results in impaired mucosal immunity and
diminished T cell responsiveness to TGF-beta. EMBOJ. 188, 1280-1291 (1999)).
Summary of the Invention
The generation of animals lacking SMAD proteins, which transduce signals from
transforming
growth factor-(3 (TGF-(3), has made it possible to explore the contribution of
the SMAD proteins to TGF-(3 activity in
viva. Here we report that, in contrast to predictions made on the basis of the
ability of exogenous TGF-(3 to improve
wound healing, Smad3-null (Smad3eXaiexa) mice paradoxically showed accelerated
cutaneous wound healing compared
with wild-type mice, characterized by an increased rate of re-
epithelialization and significantly reduced local infiltration
of monocytes. Smad3e'8~exa keratinocytes showed altered patterns of growth and
migration, and Smad3exaiexa
monocytes exhibited a selectively blunted chemotactic response to TGF-(3.
These data are, to our knowledge, the first
to implicate Smad3 in specific pathways of tissue repair and in the modulation
of keratinocyte and monocyte function
in viva
-1-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
Brief Description of the Drawings
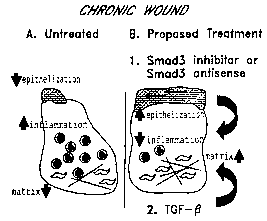
Figure A: Proposed suppression of endogenous Smad3 to improve wound healing.
Chronic wounds
are characterized by delayed re-epithelialization and increased inflammation.
Application of TGF-(3 to these wounds
impairs healing further by inhibiting keratinocyte proliferation and
stimulating monocyte and neutrophil recruitment.
Conversely, treatment of chronic wounds with agents that suppress Smad3
expression would be predicted to stimulate
re-epithelialization, inhibit inflammation, and reduce local levels of TGF-(3.
Subsequence application of exogenous TGF-
[3 to such wounds would stimulate matrix deposition via Smad3-independent
pathways, but would have no impact on
the Smad3-dependent pathways.
Figure 1: Accelerated wound healing in Smad3-null mice is associated with a
reduced monocytic
response. a, Wound areas were determined using image analysis. Results are
means ~s.e.m., n=10 for each time
point and group. 'P < 0.05 compared with wild-type (Student's t-test). d, day.
b, Re-epithelialization was determined
as the percentage of distance migrated by the neo-epidermis compared with the
upper wound width. Results are
means ~ s.e.m., n=10 for each time point and group. 'P < 0.05 compared with
wild-type. S2 HT, Smad2
heterozygotes. c, Cell numbers per unit area were quantified at days 1 and 3
post-wounding. Results are
means~s.e.m., n=10 for each time point and group. 'P < 0.05 compared with wild-
type.
Figure 2: Addition of TGF-(31 to Smad3-~~ wounds has no effect on re-
epithelialization but enhances
matrix production. a, Serum levels of TGF-(31 do not differ significantly
between phenotypes; n=8 for each group.
b, Expression of TGF-(31 is markedly reduced in Smad3-null and heterozygote
wound tissue. Values shown are
expressed relative to pooled total messenger RNA levels; n=9 per group. At day
3, no expression was detected in
wild-type and null tissue. RNase-protection assays showed a decrease in
expression of TGF-(32 and TGF-(33 from days
1-3 post-wounding, with no differences between phenotypes. c, Expression of
TGF-(311 was detectable but reduced in
day-1 wounds of Smad3-null and heterozygote mice. The type-I receptor was
barely detectable in all samples. Values
shown are expressed relative to pooled total mRNA levels; n=9 per group.
Figure 3: Smad3 is required for TGF-(3 induced monocyte chemotaxis and TGF-(3
expression. a,
Smad3-null monocytes showed a significant decrease in chemotaxis to TGF-(31
compared with wild-type cells but a
normal response to the classical chemoattractant fMet-Leu-Phe (fMet). Data
shown are the means~s.e.m. of five
experiments. 'P < 0.01 compared with media alone. b, Impaired upregulation of
TGF-(31 expression by TGF-(3 itself in
Smad3-null monocytes. Data shown are the means~s.e.m. of four experiments. 'P
< 0.01 compared with media
alone. Values shown are expressed relative to total mRNA levels. WT+, HT+ and
Null+ indicate cells treated with
TGF-(3 for 24 h. c, Expression of integrin a5 integrin is upregulated by TGF-
(3 treatment in monocytes of all genotypes.
'P < 0.05. Values are expressed relative to levels of mRNA expressed from the
housekeeping gene HPRT.
Figure 4: Smad3 deletion modulates keratinocyte proliferation and migration.
a, TGF-(31 regulates
its own expression in keratinocytes; this response is absent in Smad3-null
cells. n=20 animals in each group. C,
control medium. 'P < 0.05, treatment versus control. b, TGF-(31 inhibits
growth of wild-type and heterozygote
-2-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
keratinocytes, with a partial response in Smad3-null cells. I3HITdr, tritiated
thymidine. c, Migration of Smad3-null
keratinocytes to TGF-(31 and KGF was significantly reduced compared with wild-
type cells; i'P < 0.01, wild-type
versus Smad3-null mutants and heterozygotes; ~P < 0.01, wild-type versus Smad3-
null cells. The response of null cells
to conditioned medium (CM) was the same as that of the wild-type cells. d, The
expression of integrin oc5 in response
to TGF-(31 was impaired in null keratinocytes, with maintained upregulation of
integrin (3~. ~P < 0.01, treated versus
untreated cells. Syndecan-1 and E-cadherin were weakly expressed in all
samples, with no significant differences
observed between phenotypes or treatments.
Detailed Description of the Preferred Embodiment
Smad3 is a member of the Smad family of cytoplasmic proteins that functions to
mediate signals from TGF-[3
and activin receptors to promoters of target genes in the nucleus. To identify
selective pathways downstream of the
TGF-(3 receptors, we have characterized mice in which the Smad3 gene has been
disrupted by homologous
recombination. Studies in these mice and sibling wild-type mice showed that
the loss of Smad3 is beneficial to normal
wound healing. The data implicate Smad3 in vivo both in the inhibition of re-
epithelialization, with specific effects on
keratinocyte proliferation, and in TGF-[3-mediated chemotaxis of both
monocytes and keratinocytes. Our results
demonstrate that Smad3 mediates in vivo signalling pathways that are
inhibitory to wound healing, as its deletion
leads to enhanced re-epithelialization and contracted wound areas. The data
indicate that the disruption of the Smad3
pathway in vivo, optionally coupled with exogenous TGF signalling through
intact alternative pathways, is to be of
therapeutic benefit in accelerating all aspects of impaired wound healing.
Additionally, we propose Smad3 inhibitors as anti-fibrotic agents that have a
protective effect against
induction of fibrosis. The data indicate that Smad3 null mice are protected
from fibrosis in response to high dose
radiation. Inhibitors of Smad3 are to have clinical application in prevention
of fibrosis, including radiation-induced
fibrosis.
Definitions
The term "isolated" requires that a material be removed from its original
environment (e.g., the natural
environment if it is naturally occurring). For example, a naturally occurring
polynucleotide or polypeptide present in a
living cell is not isolated, but the same polynucleotide or polypeptide,
separated from some or all of the coexisting
materials in the natural system, is isolated.
The term "purified" does not require absolute purity; rather it is intended as
a relative definition, with
reference to the purity of the material in its natural state. Purification of
natural material to at least one order of
magnitude, preferably two or three magnitudes, and more preferably four or
five orders of magnitude is expressly
contemplated.
The term "enriched" means that the concentration of the material is at least
about 2, 5, 10, 100, or 1000
times its natural concentration (for example), advantageously 0.01 % by
weight. Enriched preparations of about 0.5%,
1 %, 5%, 10%, and 20% by weight are also contemplated.
-3-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
The Smad3 Gene
To date, nine vertebrate Smalls have been identified, and these have been
divided into subgroups based on
their functional role in various pathways. Smad1, 5, and SmadB, all mediate
signal transduction from BMPs, while
Smad2 and Smad3 mediate signal transduction from TGF-(3s and activins.
Collectively, these Smalls are known as the
pathway-restricted Smalls and can form homo or heterodimers. Smad4 has been
shown to be a shared hetero-
oligomerization partner to the pathway-restricted Smalls and is known as the
common mediator. The last two
members of the family, Smad6 and 7, act to inhibit the Small signaling
cascades often by forming unproductive dimers
with other Smalls and therefore classified as antagonistic Smalls (Heldin et
al., Nature, 1997, 390, 465-471;
Kretzschmar and Massague, Cui~ Opin. Genet. Dev., 1998, 8,103-111).
The published cDNA sequence of human Smad3 is available as GenBank accession
number 068019 and
provided as SEO ID N0:1. The deduced amino acid sequence is provided as SEO ID
N0:2. The genomic sequence is
also known.
The Smad3 nucleotide sequences of the invention include: (a) the cDNA sequence
given in SEQ ID N0:1; (b)
the nucleotide sequence that encodes the amino acid sequence given in SEO ID
N0:2; (c) any nucleotide sequence that
hybridizes to the complement of the cDNA sequence given in SEO ID N0:1 under
highly stringent conditions, e.g.,
hybridization to filter-bound DNA in 0.5 M NaHP04, 7% sodium dodecyl sulfate
(SDS), 1 mM EDTA at 65° C., and
washing in 0.1× SSCI0.1 % SDS at 68° C. (Ausubel F. M. et al.,
ells., 1989, Current Protocols in Molecular
Biology, Ilol. I, Green Publishing Associates, Inc., and John Wiley & sons,
Inc., New York, at p. 2.10.3) and encodes a
functionally equivalent gene product; and (d) any nucleotide sequence that
hybridizes to the complement of the cDNA
sequence given in SEO ID N0:1 under less stringent conditions, such as
moderately stringent conditions, e.g., washing
in 0.2× SSCI0.1 % SDS at 42° C. (Ausubel et al., 1989, supra),
yet which still encodes a functionally equivalent
gene product. Functional equivalents of Smad3 include naturally occurring
Smad3 present in other species, and mutant
Smad3s whether naturally occurring or engineered. The invention also includes
degenerate variants of sequences (a)
through (d).
The invention also includes nucleic acid molecules, preferably DNA molecules,
that hybridize to, and are
therefore the complements of, the nucleotide sequences (a) through (d), in the
preceding paragraph. Such hybridization
conditions may be highly stringent or less highly stringent, as described
above. In instances wherein the nucleic acid
molecules are deoxyoligonucleotides ("oligos"), highly stringent conditions
may refer, e.g., to washing in 6X
SSCI0.05% sodium pyrophosphate at 37° C. (for 14-base oligosl,
48° C. (for 17-base oligosl, 55° C. (for 20-base
oligos), and 60° C. (for 23-base oligos). These nucleic acid molecules
may encode or act as Smad3 antisense
molecules, useful, for example, in Smad3 gene regulation (for andlor as
antisense primers in amplification reactions of
Smad3 gene nucleic acid sequenced. With respect to Smad3 gene regulation, such
techniques can be used to regulate,
for example, radiation-induced fibrosis andlor cutaneous wound healing.
Further, such sequences may be used as part
of ribozyme andlor triple helix sequences, also useful for Smad3 gene
regulation.
_q._
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
In addition to the Smad3 nucleotide sequences described above, full length
Smad3 cDNA or gene sequences
present in the same species andlor homologs of the Smad3 gene present in other
species can be identified and readily
isolated, without undue experimentation, by molecular biological techniques
well known in the art. The identification of
homologs of Smad3 in related species can be useful for developing animal model
systems more closely related to
humans for purposes of drug discovery. For example, expression libraries of
cDNAs synthesized from mRNA derived
from the organism of interest can be screened using labeled TGF-j3 or activin
receptors (or Smalls involved in forming
dimers with Smad3) derived from that species. Alternatively, such cDNA
libraries, or genomic DNA libraries derived
from the organism of interest can be screened by hybridization using the
nucleotides described herein as hybridization
or amplification probes. Furthermore, genes at other genetic loci within the
genome that encode proteins which have
extensive homology to one or more domains of the Smad3 gene product can also
be identified via similar techniques. In
the case of cDNA libraries, such screening techniques can identify clones
derived from alternatively spliced transcripts
in the same or different species.
Screening can be by filter hybridization, using duplicate filters. The labeled
probe can contain at least 15-30
base pairs of the Smad3 cDNA sequence. The hybridization washing conditions
used should be of a lower stringency
when the cDNA library is derived from an organism different from the type of
organism from which the labeled
sequence was derived. With respect to the cloning of a human Smad3 homolog,
using murine Smad3 probes, for
example, hybridization can, for example, be performed at 65° C.
overnight in Church's buffer (7% SDS, 250 mM
NaHP04, 2,uM EDTA, 1 % BSA). Washes can be done with 2X SSC, 0.1 % SDS at
65°C. and then at 0.1 X SSC, 0.1
SDS at 65°C.
Low stringency conditions are well known to those of skill in the art, and
will vary predictably depending on
the specific organisms from which the library and the labeled sequences are
derived. For guidance regarding such
conditions see, for example, Sambrook et al., 1989, Molecular Cloning, A
Laboratory Manual, Cold Springs Harbor
Press, N.Y.; and Ausubel et al., 1989, Current Protocols in Molecular Biology,
Green Publishing Associates and Wiley
Interscience, N.Y.
Alternatively, the labeled Smad3 nucleotide probe may be used to screen a
genomic library derived from the
organism of interest, again, using appropriately stringent conditions. The
identification and characterization of human
genomic clones is helpful for designing clinical protocols for protecting
against fibrosis and improving wound healing in
human patients. For example, sequences derived from regions adjacent to the
intronlexon boundaries of the human
gene can be used to design primers for use in amplification assays to detect
mutations within the exons, introns, splice
sites (e.g. splice acceptor andlor donor sites), etc.
Further, a Smad3 gene homolog may be isolated from nucleic acid of the
organism of interest by performing
PCR using two degenerate oligonucleotide primer pools designed on the basis of
amino acid sequences within the
Smad3 gene product disclosed herein. The template for the reaction may be cDNA
obtained by reverse transcription of
mRNA prepared from, for example, human or non-human cell lines or tissue known
or suspected to express a Smad3
gene allele.
-5-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
The PCR product may he subcloned and sequenced to ensure that the amplified
sequences represent the
sequences of a Smad3 gene. The PCR fragment may then be used to isolate a full
length cDNA clone by a variety of
methods. For example, the amplified fragment may be labeled and used to screen
a cDNA library, such as a
bacteriophage cDNA library. Alternatively, the labeled fragment may be used to
isolate genomic clones via the
screening of a genomic library.
PCR technology may also be utilized to isolate full length cDNA sequences. For
example, RNA may be
isolated, following standard procedures, from an appropriate cellular or
tissue source (i.e., one known, or suspected, to
express the Smad3gene). A reverse transcription reaction may be performed on
the RNA using an oligonucleotide
primer specific for the most 5' end of the amplified fragment for the priming
of first strand synthesis. The resulting
RNAIDNA hybrid may then be "tailed" with guanines using a standard terminal
transferase reaction, the hybrid may be
digested with RNAase H, and second strand synthesis may then be primed with a
poly-C primer. Thus, cDNA
sequences upstream of the amplified fragment may easily be isolated. For a
review of cloning strategies which may be
used, see e.g., Sambrook et al., 1989, supra.
The Smad3 gene sequences may additionally be used to isolate mutant Smad3 gene
alleles. Such mutant
alleles may be isolated from individuals either known or proposed to have a
genotype which contributes to fibrosis and
or wound healing. Mutant alleles and mutant allele products may then be
utilized in the therapeutic systems described
below. Additionally, such Smad3 gene sequences can be used to detect Smad3
gene regulatory (e.g., promoter or
promotorlenhancer) defects which can affect fibrosis ar wound healing.
A cDNA of a mutant Smad3 gene may be isolated, for example, by using PCR, a
technique which is well
known to those of skill in the art. In this case, the first cDNA strand may be
synthesized by hybridizing an oligo-dT
oligonucleotide to mRNA isolated from tissue known or suspected to be
expressed in an individual putatively carrying
the mutant Smad3 allele, and by extending the new strand with reverse
transcriptase. The second strand of the cDNA
is then synthesized using an oligonucleotide that hybridizes specifically to
the 5' end of the normal gene. Using these
two primers, the product is then amplified via PCR, cloned into a suitable
vector, and subjected to DNA sequence
analysis through methods well known to those of skill in the art. By comparing
the DNA sequence of the mutant
Smad3 allele to that of the normal Smad3 allele, the mutations) responsible
for the loss or alteration of function of the
mutant Smad3 gene product can be ascertained.
Alternatively, a genomic library can be constructed using DNA obtained from an
individual suspected of or
known to carry the mutant Smad3 allele, or a cDNA library can be constructed
using RNA from a tissue known, or
suspected, to express the mutant Smad3 allele. The normal Smad3 gene or any
suitable fragment thereof may then be
labeled and used as a probe to identify the corresponding mutant Smad3 allele
in such libraries. Clones containing the
mutant Smad3 gene sequences may then be purified and subjected to sequence
analysis according to methods well
known to those of skill in the art.
Additionally, an expression library can be constructed utilizing cDNA
synthesized from, for example, RNA
isolated from a tissue known, or suspected, to express a mutant Smad3 allele
in an individual suspected of or known
-6-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
to carry such a mutant allele. In this manner, gene products made by the
putatively mutant tissue may be expressed
and screened using standard antibody screening techniques in conjunction with
antibodies raised against the normal
Smad3 gene product, as described, below, in the sections. (For screening
techniques, see, for example, Harlow, E, and
Lane, eds., 1988, "Antibodies: A Laboratory Manual", Cold Spring Harbor Press,
Cold Spring Harbor.) Additionally,
screening can be accomplished by screening with labeled Smad3 fusion proteins.
In cases where a Smad3 mutation
results in an expressed gene product with altered function (e.g., as a result
of a missense or a frameshift mutation), a
polyclonal set of antibodies to Smad3 are likely to cross-react with the
mutant Smad3 gene product. Library clones
detected via their reaction with such labeled antibodies can be purified and
subjected to sequence analysis according
to methods well known to those of skill in the art.
The invention also encompasses nucleotide sequences that encode mutant Smad3s,
peptide fragments of
Smad3, truncated Smad3s, and Smad3 fusion proteins. These include, but are not
limited to nucleotide sequences
encoding mutant Smad3s described in subsequent sections or peptides
corresponding to a domain of Smad3 or
portions of these domains; truncated Smad3s in which one or two of the domains
is deleted, or a truncated,
nonfunctional Smad3 lacking all or a portion of a domain. Nucleotides encoding
fusion proteins may include but are not
limited to full length Smad3, truncated Smad3 or peptide fragments of Smad3
fused to an unrelated protein or peptide,
such as for example, a transmembrane sequence, which anchors the Smad3 to the
cell membrane; an Ig Fc domain
which increases the stability and half life of the resulting fusion protein in
the bloodstream; or an enzyme, fluorescent
protein, luminescent protein which can be used as a marker.
The invention also encompasses (a) DNA vectors that contain any of the
foregoing Smad3 coding sequences
andlor their complements (i.e., antisense); (b) DNA expression vectors that
contain any of the foregoing Smad3 coding
sequences operatively associated with a regulatory element that directs the
expression of the coding sequences; and
(c) genetically engineered host cells that contain any of the foregoing Smad3
coding sequences operatively associated
with a regulatory element that directs the expression of the coding sequences
in the host cell. As used herein,
regulatory elements include but are not limited to inducible and non-inducible
promoters, enhancers, operators and
other elements known to those skilled in the art that drive and regulate
expression. Such regulatory elements include
but are not limited to the cytomegalovirus hCMV immediate early gene, the
early or late promoters of SV40 adenovirus,
the lac system, the trp system, the TAC system, the TRC system, the major
operator and promoter regions of phage A,
the control regions of fd coat protein, the promoter for 3-phosphoglycerate
kinase, the promoters of acid phosphatase,
and the promoters of the yeasta-mating factors.
Particular polynucleotides are DNA sequences having three sequential
nucleotides, four sequential
nucleotides, five sequential nucleotides, six sequential nucleotides, seven
sequential nucleotides, eight sequential
nucleotides, nine sequential nucleotides, ten sequential nucleotides, eleven
sequential nucleotides, twelve sequential
nucleotides, thirteen sequential nucleotides, fourteen sequential nucleotides,
fifteen sequential nucleotides, sixteen
sequential nucleotides, seventeen sequential nucleotides, eighteen sequential
nucleotides, nineteen sequential
nucleotides, twenty sequential nucleotides, twenty-one, twenty-two, twenty-
three, twenty-four, twenty-five, twenty-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
six, twenty-seven, twenty-eight, twenty-nine, thirty, thirty-one, thirty-two,
thirty-three, thirty-four, thirty-five, thirty-
six, thirty-seven, thirty-eight, thirty-nine, forty, forty-one, forty-two,
forty-three, forty-four, forty-five, forty,-six, forty-
seven, forty-eight, forty-nine, fifty, fifty-one, fifty-two, fifty-three,
fifty-four, fifty-five, fifty-six, fifty-seven, fifty-
eight, fifty-nine, sixty, sixty-one, sixty-two, sixty-three, sixty-four, sixty-
five, sixty-six, sixty-seven, sixty-eight, sixty-
nine, seventy, seventy-one, seventy-two, seventy-three, seventy-four, seventy-
five, seventy-six, seventy-seven,
seventy-eight, seventy-nine, eighty, ninety, one-hundred, two-hundred, or
three-hundred or more sequential nucleotides.
Smad3 Proteins and Polypeptides
Smad3 protein, polypeptides and peptide fragments, mutated, truncated or
deleted forms of Smad3 andlor
Smad3 fusion proteins can be prepared for a variety of uses, including but not
limited to the generation of antibodies,
as reagents for research purposes, or the identification of other cellular
gene products involved in the regulation of
fibrosis and wound healing, as reagents in assays for screening for compounds
that can be used in the prevention of
fibrosis and improvement of wound healing, and as pharmaceutical reagents
useful in protecting against fibrosis and
improving wound healing related to Smad3.
The Smad3 amino acid sequences of the invention include the amino acid
sequence, or the amino acid
sequence encoded by the cDNA or encoded by the gene. Further, Smad3 of other
species are encompassed by the
invention. In fact, any Smad3 encoded by the Smad3 nucleotide sequences
described in the sections above are within
the scope of the invention.
The invention also encompasses proteins that are functionally equivalent to
Smad3 encoded by the
nucleotide sequences described in the above sections, as judged by any of a
number of criteria, including but not
limited to the ability to bind TGF-(3 or activin receptors or Smalls involved
in forming dimers with Smad3, the binding
affinity for these ligands, the resulting biological effect of Smad3 binding,
e.g., signal transduction, a change in cellular
metabolism or change in phenotype when the Smad3 equivalent is present in an
appropriate cell type, or the regulation
of fibrosis or wound healing. Such functionally equivalent Smad3 proteins
include but are not limited to additions or
substitutions of amino acid residues within the amino acid sequence encoded by
the Smad3 nucleotide sequences
described in the sections above, but which result in a silent change, thus
producing a functionally equivalent gene
product. Amino acid substitutions may be made on the basis of similarity in
polarity, charge, solubility, hydrophobicity,
hydrophilicity, andlor the amphipathic nature of the residues involved. For
example, nonpolar (hydrophobic) amino acids
include alanine, leucine, isoleucine, valine, proline, phenylalanine,
tryptophan, and methionine; polar neutral amino acids
include glycine, serine, threonine, cysteine, tyrosine, asparagine, and
glutamine; positively charged (basic) amino acids
include arginine, lysine, and histidine; and negatively charged (acidic) amino
acids include aspartic acid and glutamic
acid. While random mutations can be made to Smad3 DNA (using random
mutagenesis techniques well known to those
skilled in the art) and the resulting mutant Smad3s tested for activity, site-
directed mutations of the Smad3 coding
sequence can be engineered (using site-directed mutagenesis techniques well
known to those skilled in the art) to
generate mutant Smad3s with altered function, e.g., different binding affinity
for TGF-(3 or activin receptors or Smalls
involved in forming dimers with Smad3, andlor different signalling capacity.
_g_
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
For example, identical amino acid residues of a mouse form of Smad3 and the
human Smad3 homolog can be
aligned so that regions of identity are maintained, whereas the variable
residues are altered, e.g., by deletion or
insertion of an amino acid residuels) or by substitution of one or more
different amino acid residues. Conservative
alterations at the variable positions can be engineered in order to produce a
mutant Smad3 that retains function; e.g.,
ligand binding affinity or signal transduction capability or both. Non-
conservative changes can be engineered at these
variable positions to alter function, e.g., ligand binding affinity or signal
transduction capability, or both. Alternatively,
where alteration of function is desired, deletion or non-conservative
alterations of the conserved regions (i.e., identical
amino acids) can be engineered. For example, deletion or non-conservative
alterations (substitutions or insertions) of a
domain can be engineered to produce a mutant Smad3 that binds a ligand but is
signalling-incompetent. Non-
conservative alterations to residues of identical amino acids can be
engineered to produce mutant Smad3s with altered
binding affinity for ligands. The same mutation strategy can also be used to
design mutant Smad3s based on the
alignment of other non-human Smad3s and the human Smad3 homolog by aligning
identical amino acid residues.
Other mutations to the Smad3 coding sequence can be made to generate Smad3s
that are better suited for
expression, scale up, etc. in the host cells chosen. For example, cysteine
residues can be deleted or substituted with
another amino acid in order to eliminate disulfide bridges; N-linked
glycosylation sites. can be altered or eliminated to
achieve, for example, expression of a homogeneous product that is more easily
recovered and purified from yeast hosts
which are known to hyperglycosylate N-linked sites.
Peptides corresponding to one or more domains of Smad3, as well as fusion
proteins in which the full length
Smad3, a Smad3 peptide or truncated Smad3 is fused to an unrelated protein are
also within the scope of the
invention and can be designed on the basis of the Smad3 nucleotide and Smad3
amino acid sequences given in SEQ ID
NOS:1 and 2. Such fusion proteins include but are not limited to IgFc fusions
which stabilize the Smad3 protein or
peptide and prolong half-life in vivo; or fusions to any amino acid sequence
that allows the fusion protein to be
anchored to the cell membrane; or fusions to an enzyme, fluorescent protein,
or luminescent protein which provide a
marker function.
While the Smad3 polypeptides and peptides can be chemically synthesized (e.g.,
see Creighton, 1983,
Proteins: Structures and Molecular Principles, W. H. Freeman & Co., N.Y.),
large polypeptides derived from Smad3 and
the full length Smad3 itself may advantageously be produced by recombinant DNA
technology using techniques well
known in the art for expressing nucleic acid containing Smad3 gene sequences
andlor coding sequences. Such methods
can be used to construct expression vectors containing the Smad3 nucleotide
sequences and appropriate
transcriptional and translational control signals. These methods include, for
example, in vitro recombinant DNA
techniques, synthetic techniques, and in vivo genetic recombination. See, for
example, the techniques described in
Sambrook et al., 1989, supra, and Ausubel et al., 1989, supra. Alternatively,
RNA capable of encoding Smad3
nucleotide sequences may be chemically synthesized using, for example,
synthesizers. See, for example, the techniques
described in "Oligonucleotide Synthesis", 1984, Gait, M. J. ed., IRL Press,
Oxford.
-9- '
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
A variety of host-expression vector systems may be utilized to express the
Smad3 nucleotide sequences of
the invention. Where the Smad3 peptide or polypeptide is soluble, the peptide
or polypeptide can be recovered from the
culture, ie., from the host cell in cases where the Smad3 peptide or
polypeptide is not secreted, and from the culture
media in cases where the Smad3 peptide or polypeptide is secreted by the
cells. However, the expression systems also
encompass engineered host cells that express the Smad3 or functional
equivalents in situ, i.e., anchored in the cell
membrane. Purification or enrichment of the Smad3 from such expression systems
can be accomplished using
appropriate detergents and lipid micelles and methods well known to those
skilled in the art. However, such engineered
host cells themselves may be used in appropriate situations.
The expression systems that may be used for purposes of the invention include
but are not limited to
microorganisms such as bacteria (e.g., E. coli, B. subtilis) transformed with
recombinant bacteriophage DNA, plasmid
DNA or cosmid DNA expression vectors containing Smad3 nucleotide sequences;
yeast (e.g., Saccharomyces, Pichia)
transformed with recombinant yeast expression vectors containing the Smad3
nucleotide sequences; insect cell
systems infected with recombinant virus expression vectdrs (e.g., baculovirus)
containing the Smad3 sequences; plant
cell systems infected with recombinant virus expression vectors (e.g.,
cauliflower mosaic virus, CaMV; tobacco mosaic
virus, TMII) or transformed with recombinant plasmid expression vectors (e.g.,
Ti plasmid) containing Smad3
nucleotide sequences; or mammalian cell systems (e.g., COS, CHO, BHK, 293,
3T3) harboring recombinant expression
constructs containing promoters derived from the genome of mammalian cells
(e.g., metallothionein promoter) or from
mammalian viruses (e.g., the adenovirus late promoter; the vaccinia virus 7.5K
promoter).
In bacterial systems, a number of expression vectors may be advantageously
selected depending upon the
use intended for the Smad3 gene product being expressed. For example, when a
large quantity of such a protein is to
be produced, for the generation of pharmaceutical compositions of Smad3
protein or for raising antibodies to the
Smad3 protein, for example, vectors which direct the expression of high levels
of fusion protein products that are
readily purified may be desirable. Such vectors include, but are not limited,
to the E. coli expression vector pUR278
(Ruther et al., 1983, EMBO J. 2:1791), in which the Smad3 coding sequence may
be ligated individually into the vector
in frame with the IacZ coding region so that a fusion protein is produced; pIN
vectors (Inouye & Inouye, 1985, Nucleic
Acids Res. 13:3101-3109; Uan Heeke & Schuster, 1989, J. Biol. Chem. 264:5503-
5509); and the like. pGEX vectors
may also be used to express foreign polypeptides as fusion proteins with
glutathione S-transferase (GST). In general,
such fusion proteins are soluble and can easily be purified from lysed cells
by adsorption to glutathione-agarose beads
followed by elution in the presence of free glutathione. The PGEX vectors are
designed to include thrombin or factor Xa
protease cleavage sites so that the cloned target gene product can be released
from the GST moiety.
In an insect system, Autographa californica nuclear polyhidrosis virus (AcNPV)
is used as a vector to express
foreign genes. The virus grows in Spodoptera frugiperda cells. The Smad3 gene
coding sequence may be cloned
individually into non-essential regions (for example the polyhedrin gene) of
the virus and placed under control of an
AcNPV promoter (for example the polyhedrin promoter). Successful insertion of
Smad3 gene coding sequence will
result in inactivation of the polyhedrin gene and production of non-occluded
recombinant virus, (i.e., virus lacking the
-10-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
proteinaceous coat coded for by the polyhedrin genet. These recombinant
viruses are then used to infect Spodoptera
frugiperda cells in which the inserted gene is expressed. (E.g., see Smith et
al., 1983, J. Virol. 46: 584; Smith, U.S.
Pat. No. 4,215,0511.
In mammalian host cells, a number of viral-based expression systems may be
utilized. In cases where an
adenovirus is used as an expression vector, the Smad3 nucleotide sequence of
interest may be ligated to an adenovirus
transcriptionltranslation control complex, e.g., the late promoter and
tripartite leader sequence. This chimeric gene may
then he inserted in the adenovirus genome by in vitro or in vivo
recombination. Insertion in a non-essential region of the
viral genome (e.g., region E1 or E3) will result in a recombinant virus that
is viable and capable of expressing the
Smad3 gene product in infected hosts. (E.g., See Logan & Shenk, 1984, Proc.
Natl. Acad. Sci. USA 81:3655-3659).
Specific initiation signals may also be required for efficient translation of
inserted Smad3 nucleotide sequences. These
signals include the ATG initiation colon and adjacent sequences. In cases
where an entire Smad3 gene or cDNA,
including its own initiation colon and adjacent sequences, is inserted into
the appropriate expression vector, no
additional translational control signals may be needed. However, in cases
where only a portion of the Smad3 coding
sequence is inserted, exogenous translational control signals, including,
perhaps, the ATG initiation colon, must be
provided. Furthermore, the initiation colon must be in phase with the reading
frame of the desired coding sequence to
ensure translation of the entire insert. These exogenous translational control
signals and initiation colons can be of a
variety of origins, both natural and synthetic. The efficiency of expression
may be enhanced by the inclusion of
appropriate transcription enhancer elements, transcription terminators, etc.
(See Bittner et al., 1987, Methods in
Enzymol.153:516-5441.
In addition, a host cell strain may be chosen which modulates the expression
of the inserted sequences, or
modifies and processes the gene product in the specific fashion desired. Such
modifications (e.g., glycosylation) and
processing (e.g., cleavage) of protein products may be important for the
function of the protein. Different host cells
have characteristic and specific mechanisms for the post-translational
processing and modification of proteins and
gene products. Appropriate cell lines or host systems can be chosen to ensure
the correct modification and processing
of the foreign protein expressed. To this end, eukaryotic host cells which
possess the cellular machinery for proper
processing of the primary transcript, glycosylation, and phosphorylation of
the gene product may be used. Such
mammalian host cells include but are not limited to CHO, VERO, BHK, HeLa, COS,
MDCK, 293, 3T3, and WI38.
For long-term, high-yield production of recombinant proteins, stable
expression is preferred. For example, cell
lines which stably express the Smad3 sequences described above may be
engineered. Rather than using expression
vectors which contain viral origins of replication, host cells can be
transformed with DNA controlled by appropriate
expression control elements (e.g., promoter, enhancer sequences, transcription
terminators, polyadenylation sites,
etc.), and a selectable marker. Following the introduction of the foreign DNA,
engineered cells may be allowed to grow
for 1-2 days in an enriched media, and then are switched to a selective media.
The selectable marker in the
recombinant plasmid confers resistance to the selection and allows cells to
stably integrate the plasmid into their
chromosomes and grow to form foci which in turn can be cloned and expanded
into cell lines. This method may
-11-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
advantageously be used to engineer cell lines which express the Smad3 gene
product. Such engineered cell lines may
be particularly useful in screening and evaluation of compounds that affect
the endogenous activity of the Smad3 gene
product.
A number of selection systems may be used, including but not limited to the
herpes simplex virus thymidine
kinase (Wigler, et al., 1977, Cell 11:2231, hypoxanthine-guanine
phosphoribosyltransferase (Szybalska & Szybalski,
1962, Proc. Natl. Acad. Sci. USA 48:2026), and adenine
phosphoribosyltransferase (Lowy, et al., 1980, Cell 22:817)
genes can be employed in tk-, hgprt- or aprt- cells, respectively. Also,
antimetabolite resistance can be used as the
basis of selection for the following genes: dhfr, which confers resistance to
methotrexate (Wigler, et al., 1980, Natl.
Acad. Sci. USA 77:3567; 0'Hare, et al., 1981, Proc. Natl. Acad. Sci. USA
78:1527); gpt, which confers resistance to
mycophenolic acid (Mulligan & Berg, 1981, Proc. Natl. Acad. Sci. USA 78:2072);
neo, which confers resistance to the
aminoglycoside G-418 (Colberre-Garapin, et al., 1981, J. Mol. Biol. 150:1);
and hygro, which confers resistance to
hygromycin (Santerre, et al., 1984, Gene 30:147).
Alternatively, any fusion protein may be readily purified by utilizing an
antibody specific for the fusion protein
being expressed. For example, a system described by Janknecht et al. allows
for the ready purification of non-
denatured fusion proteins expressed in human cell lines (Janknecht, et al.,
1991, Proc. Natl. Acad. Sci. USA 88: 8972-
8976). In this system, the gene of interest is subcloned into a vaccinia
recombination plasmid such that the gene's
open reading frame is translationally fused to an amino-terminal tag
consisting of six histidine residues. Extracts from
cells infected with recombinant vaccinia virus are loaded onto
Ni2+.nitriloacetic acid-agarose columns and histidine-
tagged proteins are selectively eluted with imidazole-containing buffers.
The Smad3 gene products can also be expressed in transgenic animals. Animals
of any species, including, but
not limited to, mice, rats, rabbits, guinea pigs, pigs, micro-pigs, goats, and
non-human primates, e.g., baboons,
monkeys, and chimpanzees may be used to generate Smad3 transgenic animals.
Particular polypeptides are amino acid sequences having three sequential
residues, four sequential residues,
five sequential residues, six sequential residues, seven sequential residues,
eight sequential residues, nine sequential
residues, ten sequential residues, eleven sequential residues, twelve
sequential residues, thirteen sequential residues,
fourteen sequential residues, fifteen sequential residues, sixteen sequential
residues, seventeen sequential residues,
eighteen sequential residues, nineteen sequential residues, twenty sequential
residues, twenty-one, twenty-two,
twenty-three, twenty-four, twenty-five, twenty-six, twenty-seven, thirty,
forty, fifty, sixty, seveny, eighty, ninety, or
more sequential residues.
Screening Assays for Compounds that inhibit Smad3 Expression or Activity
The following assays are designed to identify compounds that inhibit Smad3,
compounds that interfere with
the interaction of Smad3 with intracellular proteins, and compounds that
interfere with the interaction of Smad3 with
transmembrane proteins, e.g., TGF-(3 and activin receptors, and compounds
which inhibit the activity of the Smad3
gene or modulate the level of Smad3. Assays may additionally be utilized which
identify compounds which bind to
Smad3 gene regulatory sequences (e.g., promoter sequences) and which may
inhibit Smad3 gene expression. Assays
-12-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
may additionally be utilized to identify compounds which interfere with the
interaction of Smad3 with promoters of
target genes.
The compounds which may be screened in accordance with the invention include,
but are not limited to
peptides, antibodies and fragments thereof, and other organic compounds (e.g.,
peptidomimetics) that bind to Smad3,
or to intracellular proteins that interact with Smad 3, or to transmembrane
proteins that interact with Smad3, and
inhibit the activity triggered by Smad3 or mimic the inhibitors of Smad3; as
well as peptides, antibodies or fragments
thereof, and other organic compounds that mimic the ligands of Smad3 (or a
portion thereof) and bind to and
"neutralize" Smad3.
Such compounds may include, but are not limited to, peptides such as, for
example, soluble peptides,
including but not limited to members of random peptide libraries; (see, e.g.,
Lam, K. S. et al., 1991, Nature 354:82-84;
Houghten, R. et al., 1991, Nature 354:84-86), and combinatorial chemistry-
derived molecular libraries made of D-
andlor L- configuration amino acids, phosphopeptides (including, but not
limited to, members of random or partially
degenerate, directed phosphopeptide libraries; see, e.g., Songyang, Z. et al.,
1993, Cell 72:767-778), antibodies
(including, but not limited to, polyclonal, monoclonal, humanized, anti-
idiotypic, chimeric or single chain antibodies, and
FAb, F(ab')2 and FAb expression library fragments, and epitope-binding
fragments thereofl, and small organic or
inorganic molecules.
Other compounds which can be screened in accordance with the invention include
but are not limited to small
organic molecules that affect the expression of the Smad3 gene or some other
gene balancing the interaction of
intracellular proteins with Smad3 or the interaction of transmembrane proteins
with Smad3 (e.g., by interacting with
the regulatory region or transcription factors involved in gene expression);
or such compounds that affect the activity
of Smad3 or the activity of some other intracellular protein that interacts
with Smad3 or of some other
transmembrane protein that interacts with Smad3 or of promoters of target
genes regulated by Smad3.
Computer modelling and searching technologies permit identification of
compounds, or the improvement of
already identified compounds, that can inhibit Smad3 expression or activity.
Having identified such a compound or
composition, the active sites or regions are identified. Such active sites
might typically be ligand binding sites, such as
the interaction domains of the ligand with Smad3 itself. The active site can
be identified using methods known in the
art including, for example, from the amino acid sequences of peptides, from
the nucleotide sequences of nucleic acids,
or from study of complexes of the relevant compound or composition with its
ligand. In the latter case, chemical or X-
ray crystallographic methods can be used to find the active site by finding
where on the factor the complexed ligand is
found. Next, the three dimensional geometric structure of the active site is
determined. This can be done by known
methods, including X-ray crystallography, which can determine a complete
molecular structure. On the other hand,
solid or liquid phase NMR can be used to determine certain intra-molecular
distances. Any other experimental method
of structure determination can be used to obtain partial or complete geometric
structures. The geometric structures
may be measured with a complexed ligand, natural or artificial, which may
increase the accuracy of the active site
structure determined. Indeed, the Smad interaction domains have been
determined for known inhibitors of Smad3,
-13-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
including the transcriptional repressors TGIF and SIP1, the adenoviral
oncoprotein E1A, and the human oncogenes Ski,
SnoN, and Evi-1 and may serve as the basis for rational drug design.
If an incomplete or insufficiently accurate structure is determined, the
methods of computer based numerical
modelling can be used to complete the structure or improve its accuracy. Any
recognized modelling method may be
used, including parameterized models specific to particular biopolymers such
as proteins or nucleic acids, molecular
dynamics models based on computing molecular motions, statistical mechanics
models based on thermal ensembles, or
combined models. For most types of models, standard molecular force fields,
representing the forces between
constituent atoms and groups, are necessary, and can be selected from force
fields known in physical chemistry. The
incomplete or less accurate experimental structures can serve as constraints
on the complete and more accurate
structures computed by these modeling methods.
Finally, having determined the structure of the active site, either
experimentally, by modeling, or by a
combination, candidate inhibiting compounds can be identified by searching
databases containing compounds along
with information on their molecular structure. Such a search seeks compounds
having structures that match the
determined active site structure and that interact with the groups defining
the active site. Such a search can be
manual, but is preferably computer assisted. The compounds found from this
search are potential Smad3 inhibiting
compounds.
Alternatively, these methods can be used to identify improved inhibiting
compounds from an already known
inhibiting compound or ligand. The composition of the known compound can be
modified and the structural effects of
modification can be determined using the experimental and computer modelling
methods described above applied to the
new composition. The altered structure is then compared to the active site
structure of the compound to determine if
an improved fit or interaction results. In this manner systematic variations
in composition, such as by varying side
groups, can be quickly evaluated to obtain modified inhibiting compounds or
ligands of improved specificity or activity.
Further experimental and computer modeling methods useful to identify
inhibiting compounds will be apparent
to those of skill in the art based upon identification of the active sites of
Smad3, and of intracellular and
transmembrane proteins that interact with Smad3, and of related transduction
and transcription factors, as well as of
promoters of target genes regulated by Smad3.
Examples of molecular modelling systems are the CHARMM and QUANTA programs
(Polygon Corporation,
Waltham, Mass.). CHARMm performs the energy minimization and molecular
dynamics functions. QUANTA performs
the construction, graphic modelling and analysis of molecular structure.
QUANTA allows interactive construction,
modification, visualization, and analysis of the behavior of molecules with
each other.
A number of articles review computer modelling of drugs interactive with
specific-proteins, such as
Rotivinen, et al., 1988, Acta Pharmaceutical Fennica 97:159-166; Ripka, New
Scientist 54-57 (Jun. 16, 1988);
McKinaly and Rossmann, 1989, Annu. Rev. Pharmacol. Toxiciol. 29:111-122; Perry
and Davies, OSAR: Quantitative
Structure-Activity Relationships in Drug Design pp. 189-193 (Alan R. Liss,
Inc. 1989); Lewis and Dean, 1989 Proc. R.
Soc. Land. 236:125-140 and 141-162; and, with respect to a model receptor for
nucleic acid components, Askew, et
-14-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
al., 1989, J. Am. Chem. Soc. 111:1082-1090. Other computer programs that
screen and graphically depict chemicals
are available from companies such as BioDesign, Inc. (Pasadena, Calif.),
Allelix, Inc. (Mississauga, Ontario, Canadal,
and Hypercube, Inc. (Cambridge, Ontariol. Although these are primarily
designed for application to drugs specific to
particular proteins, they can be adapted to design of drugs specific to
regions of DNA or RNA, once that region is
identified.
Although described above with reference to design and generation of compounds
which could alter binding,
one could also screen libraries of known compounds, including natural products
or synthetic chemicals, and biologically
active materials, including proteins, for compounds which are inhibitors of
Smad3.
Compounds identified via assays such as those described herein may be useful,
for example, in elaborating
the biological function of the Smad3 gene product, and for preventing fibrosis
and improving wound healing.
In Vitro Screening Assays for Compounds that Bind to Smad3
In vitro systems may be designed to identify compounds capable of interacting
with (e.g., binding to) Smad3.
Compounds identified may be useful, for example, in inhibiting the activity of
wild type andlor mutant Smad3 gene
products; may be useful in elaborating the biological function of Smad3; may
be utilized in screens for identifying
compounds that disrupt normal Smad3 interactions; or may in themselves disrupt
such interactions.
The principle of the assays used to identify compounds that bind to Smad3
involves preparing a reaction
mixture of Smad3 and the test compound under conditions and for a time
sufficient to allow the two components to
interact and bind, thus forming a complex which can be removed andlor detected
in the reaction mixture. The Smad3
species used can vary depending upon the goal of the screening assay. For
example, where compounds that bind and
inhibit or mimic the inhibitors or mimic the ligands of Smad3 and bind to and
"neutralize" Smad3 are sought, the full
length Smad3 protein, a peptide corresponding to a domain or a fusion protein
containing a Smad3 domain fused to a
protein or polypeptide that affords advantages in the assay system (e.g.,
labeling, isolation of the resulting complex,
etc.) can be utilized.
The screening assays can be conducted in a variety of ways. For example, one
method to conduct such an
assay would involve anchoring the Smad3 protein, polypeptide, peptide or
fusion protein or the test substance onto a
solid phase and detecting Smad3ltest compound complexes anchored on the solid
phase at the end of the reaction. In
one embodiment of such a method, the Smad3 reactant may be anchored onto a
solid surface, and the test compound,
which is not anchored, may be labeled, either directly or indirectly.
In practice, microtiter plates may conveniently be utilized as the solid
phase. The anchored component may
be immobilized by non-covalent or covalent attachments. Non-covalent
attachment may be accomplished by simply
coating the solid surface with a solution of the protein and drying.
Alternatively, an immobilized antibody, preferably a
monoclonal antibody, specific for the protein to be immobilized may be used to
anchor the protein to the solid surface.
The surfaces may be prepared in advance and stored.
In order to conduct the assay, the nonimmobilized component is added to the
coated surface containing the
anchored component. After the reaction is complete, unreacted components are
removed (e.g., by washing) under
-15-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
conditions such that any complexes formed will remain immobilized on the solid
surface. The detection of complexes
anchored on the solid surface can be accomplished in a number of ways. Where
the previously nonimmobilized
component is pre-labeled, the detection of label immobilized on the surface
indicates that complexes were formed.
Where the previously nonimmobilized component is not pre-labeled, an indirect
label can be used to detect complexes
anchored on the surface; e.g., using a labeled antibody specific for the
previously nanimmobilized component (the
antibody, in turn, may be directly labeled or indirectly labeled with a
labeled anti-Ig antibody).
Alternatively, a reaction can be conducted in a liquid phase, the reaction
products separated from unreacted
components, and complexes detected; e.g., using an immobilized antibody
specific for Smad3 protein, polypeptide,
peptide or fusion protein or the test compound to anchor any complexes formed
in solution, and a labeled antibody
specific for the other component of the possible complex to detect anchored
complexes.
Alternatively, cell-based assays can be used to identify compounds that
interact with Smad3. To this end,
cell lines that express Smad3, or cell lines (e.g., COS cells, CHO cells,
fibroblasts, etc.) that have been genetically
engineered to express Smad3 (e.g., by transfection or transduction of Smad3
DNA) can be used. Interaction of the test
compound with, for example, the Smad3 expressed by the host cell can be
determined by comparison or competition
with native ligand.
Assays for Intracellular or Transmembrane Proteins that Interact with the
Smad3
Any method suitable for detecting protein-protein interactions may be employed
for identifying
transmembrane proteins or intracellular proteins that interact with Smad3.
Among the traditional methods which may
be employed are co-immunoprecipitation, crosslinking and co-purification
through gradients or chromatographic
columns of cell lysates or proteins obtained from cell lysates and the Smad3
protein to identify proteins in the lysate
that interact with the Smad3 protein. For these assays, the Smad3 component
used can be a full length Smad3
protein, a peptide corresponding to a domain of Smad3 or a fusion protein
containing a domain of Smad3. Once
isolated, such an intracellular or transmembrane protein can be identified and
can, in turn, be used, in conjunction with
standard techniques, to identify proteins with which it interacts. Far
example, at least a portion of the amino acid
sequence of an intracellular or transmembrane protein which interacts with
Smad3 can be ascertained using
techniques well known to those of skill in the art, such as via the Edman
degradation technique. (See, e.g., Creighton,
1983, "Proteins: Structures and Molecular Principles", W.H. Freeman & Co.,
N.Y., pp.34-49). The amino acid sequence
obtained may be used as a guide for the generation of oligonucleotide mixtures
that can be used to screen for gene
sequences encoding such intracellular and transmembrane proteins. Screening
may be accomplished, for example, by
standard hybridization or PCR techniques. Techniques for the generation of
oligonucleotide mixtures and the screening
are well-known. (See, e.g., Ausubel et al., 1989, Current Protocols in
Molecular Biology, Green Publishing Associates
and Wiley Interscience, N.Y., and PCR Protocols: A Guide to Methods and
Applications, 1990, lnnis, M, et al., eds.
Academic Press, Inc., New York).
Additionally, methods may be employed which result in the simultaneous
identification of genes which
encode the transmembrane or intracellular proteins interacting with Smad3.
These methods include, for example,
-16-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
probing expression, libraries, in a manner similar to the well known technique
of antibody probing of 7~gt11 libraries,
using labeled Smad3 protein, or a Smad3 polypeptide, peptide or fusion
protein, e.g., a Smad3 polypeptide or Smad3
domain fused to a marker (e.g., an enzyme, fluor, luminescent protein, or
dye), or an Ig-Fc domain.
One method which detects protein interactions in vivo, the two-hybrid system,
is described in detail for
illustration only and not by way of limitation. One version of this system has
been described (Chien et al., 1991, Proc.
Natl. Acad. Sci. USA, 88:9578-9582) and is commercially available from
Clontech (Palo Alto, Calif.). The assay
identifies proteins that interact with Smad3, whether physiologically or
pharmacologically.
Briefly, utilizing such a system, plasmids are constructed that encode two
hybrid proteins: one plasmid
consists of nucleotides encoding the DNA-binding domain of a transcription
activator protein fused to a Smad3
nucleotide sequence encoding Smad3, a Smad3 polypeptide, peptide or fusion
protein, and the other plasmid consists
of nucleotides encoding the transcription activator protein's activation
domain fused to a cDNA encoding an unknown
protein which has been recombined into this plasmid as part of a cDNA library.
The DNA-binding domain fusion plasmid
and the cDNA library are transformed into a strain of the yeast Saccharomyces
cerevisiae that contains a reporter
gene whose regulatory region contains the transcription activator's binding
site. Either hybrid protein alone cannot
activate transcription of the reporter gene: the DNA-binding domain hybrid
cannot because it does not provide
activation function and the activation domain hybrid cannot because it cannot
localize to the activator's binding sites.
Interaction of the two hybrid proteins reconstitutes the functional activator
protein and results in expression of the
reporter gene, which is detected by an assay for the reporter gene product.
The two-hybrid system or related methodology may be used to screen activation
domain libraries for proteins
that interact with the "bait" gene product. By way of example, and not by way
of limitation, Smad3 may be used as
the bait gene product. Total genomic or cDNA sequences are fused to the DNA
encoding an activation domain. This
library and a plasmid encoding a hybrid of a bait Smad3 gene product fused to
the DNA-binding domain are co-
transformed into a yeast reporter strain, and the resulting transformants are
screened for those that express the
reporter gene. For example, and not by way of limitation, a bait Smad3 gene
sequence, such as the open reading frame
of Smad3 (or a domain of Smad3), can be cloned into a vector such that it is
translationally fused to the DNA encoding
the DNA-binding domain of the GAL4 protein. These colonies are purified and
the library plasmids responsible for
reporter gene expression are isolated. DNA sequencing is then used to identify
the proteins encoded by the library
plasmids.
A cDNA library of the cell line from which proteins that interact with bait
Smad3 gene product are to be
detected can be made using methods routinely practiced in the art. According
to the particular system described
herein, for example, the cDNA fragments can be inserted into a vector such
that they are translationally fused to the
transcriptional activation domain of GAL4. This library can be co-transformed
along with the bait Smad3 gene-GAL4
fusion plasmid into a yeast strain which contains a IacZ gene driven by a
promoter which contains GAL4 activation
sequence. A cDNA encoded protein, fused to GAL4 transcriptional activation
domain, that interacts with bait Smad3
gene product will reconstitute an active GAL4 protein and thereby drive
expression of the HIS3 gene. Colonies which
-17-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
express HISS can be detected by their growth on petri dishes containing semi-
solid agar based media lacking histidine.
The cDNA can then be purified from these strains, and used to produce and
isolate the bait Smad3 gene-interacting
protein using techniques routinely practiced in the art.
Assays for Compounds that Interfere with Smad311ntracellular or
Smad3lTransmemhrane Macromolecule
Interaction
The macromolecules that interact with Smad3 are referred to, for purposes of
this discussion, as "ligands".
These ligands are likely to be involved in the Smad3 signal transduction
pathway, and therefore, in the role of Smad3
in wound healing and fibrosis. Therefore, it is desirable to identify
compounds that interfere with or disrupt the
interaction of such ligands with Smad3 which may be useful in regulating the
activity of Smad3 and control wound
healing and fibrosis associated with Smad3 activity.
The basic principle of the assay systems used to identify compounds that
interfere with the interaction
between Smad3 and its ligand or ligands involves preparing a reaction mixture
containing the Smad3 protein,
polypeptide, peptide or fusion protein and the ligand under conditions and for
a time sufficient to allow the two to
interact and bind, thus forming a complex. In order to test a compound far
inhibitory activity, the reaction mixture is
prepared in the presence and absence of the test compound. The test compound
may be initially included in the
reaction mixture, or may be added at a time subsequent to the addition of the
Smad3 moiety and its ligand. Control
reaction mixtures are incubated without the test compound or with a placebo.
The formation of any complexes
between the Smad3 moiety and the ligand is then detected. The formation of a
complex in the control reaction, but not
in the reaction mixture containing the test compound, indicates that the
compound interferes with the interaction of
Smad3 and the interactive ligand. Additionally, complex formation within
reaction mixtures containing the test
compound and normal Smad3 protein may also be compared to complex formation
within reaction mixtures containing
the test compound and a mutant Smad3. This comparison may be important in
those cases wherein it is desirable to
identify compounds that disrupt interactions of mutant but not normal Smad3
proteins.
The assay for compounds that interfere with the interaction of Smad3 and
ligands can be conducted in a
heterogeneous or homogeneous format. Heterogeneous assays involve anchoring
either the Smad3 moiety product or
the ligand onto a solid phase and detecting complexes anchored on the solid
phase at the end of the reaction. In
homogeneous assays, the entire reaction is carried out in a liquid phase. In
either approach, the order of addition of
reactants can be varied to obtain different information about the compounds
being tested. Far example, test
compounds that interfere with the interaction by competition can be identified
by conducting the reaction in the
presence of the test substance; i.e., by adding the test substance to the
reaction mixture prior to or simultaneously
with the Smad3 moiety and interactive ligand. Alternatively, test compounds
that disrupt preformed complexes, e.g.
compounds with higher binding constants that displace one of the components
from the complex, can be tested by
adding the test compound to the reaction mixture after complexes have been
formed. The various formats are
described briefly below.
-18-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
In a heterogeneous assay system, either the Smad3 moiety or the interactive
ligand, is anchored onto a solid
surface, while the non-anchored species is labeled, either directly or
indirectly. In practice, microtiter plates are
conveniently utilized. The anchored species may be immobilized by non-covalent
or covalent attachments. Non-covalent
attachment may be accomplished simply by coating the solid surface with a
solution of the Smad3 gene product or
ligand and drying. Alternatively, an immobilized antibody specific for the
species to be anchored may be used to anchor
the species to the solid surface. The surfaces may be prepared in advance and
stored.
In order to conduct the assay, the partner of the immobilized species is
exposed to the coated surface with or
without the test compound. After the reaction is complete, unreacted
components are removed (e.g., by washing) and
any complexes formed will remain immobilized on the solid surface. The
detection of complexes anchored on the solid
surface can be accomplished in a number of ways. Where the non-immobilized
species is pre-labeled, the detection of
label immobilized on the surface indicates that complexes were formed. Where
the non-immobilized species is not pre
labeled, an indirect label can be used to detect complexes anchored on the
surface; e.g., using a labeled antibody
specific for the initially non-immobilized species (the antibody, in turn, may
be directly labeled or indirectly labeled with °
a labeled anti-Ig antibody). Depending upon the order of addition of reaction
components, test compounds which inhibit
complex formation or which disrupt preformed complexes can be detected.
Alternatively, the reaction can be conducted in a liquid phase in the presence
or absence of the test
compound, the reaction products separated from unreacted components, and
complexes detected; e.g., using an
immobilized antibody specific for one of the binding components to anchor any
complexes formed in solution, and a
labeled antibody specific for the other partner to detect anchored complexes.
Again, depending upon the order of
addition of reactants to the liquid phase, test compounds which inhibit
complex or which disrupt preformed complexes
can be identified.
In an alternate embodiment of the invention, a homogeneous assay can be used.
In this approach, a
preformed complex of the Smad3 moiety and the interactive ligand is prepared
in which either the Smad3 or its ligand
is labeled, but the signal generated by the label is quenched due to formation
of the complex (see, e.g., U.S. Pat. No.
4,109,496 by Rubenstein which utilizes this approach for immunoassays). The
addition of a test substance that
competes with and displaces one of the species from the preformed complex will
result in the generation of a signal
above background. In this way, test substances which disrupt Smad311igand
interaction can be identified.
In a particular embodiment, a Smad3 fusion can be prepared for immobilization.
For example, Smad3, or a
peptide fragment, e.g., corresponding to a domain, can be fused to a
glutathione-S-transferase (GST) gene using a
fusion vector, such as pGEX-5X-1, in such a manner that its binding activity
is maintained in the resulting fusion
protein. The interactive ligand can be purified and used to raise a monoclonal
antibody, using methods routinely
practiced in the art. This antibody can be labeled with the radioactive
isotope'z51, for example, by methods routinely
practiced in the art. In a heterogeneous assay, e.g., the GST-Smad3 fusion
protein can be anchored to glutathione-
agarose beads. The interactive ligand can then be added in the presence or
absence of the test compound in a manner
that allows interaction and binding to occur. At the end of the reaction
period, unbound material can be, washed away,
-19-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
and the labeled monoclonal antibody can be added to the system and allowed to
bind to the complexed components.
The interaction between the Smad3 gene product and the interactive ligand can
be detected by measuring the amount
of radioactivity that remains associated with the glutathione-agarose beads. A
successful inhibition of the interaction
by the test compound will result in a decrease in measured radioactivity.
Alternatively, the GST-Smad3 fusion protein and the interactive (igand can be
mixed together in liquid in the
absence of the solid glutathione-agarose beads. The test compound can be added
either during or after the species are
allowed to interact. This mixture can then be added to the glutathione-agarose
beads and unbound material is washed
away. Again the extent of inhibition of the Smad311igand interaction can be
detected by adding the labeled antibody
and measuring the radioactivity associated with the beads.
I0 In another embodiment of the invention, these same techniques can be
employed using peptide fragments
that correspond to the binding domains of Smad3 andlor the interactive ligand
(in cases where the ligand is a protein),
in place of one or both of the full length proteins. Any number of methods
routinely practiced in the art can be used to
identify and isolate the binding sites. These methods include, but are not
limited to, mutagenesis of the gene encoding
one of the proteins and screening for disruption of binding in a co-
immunoprecipitation assay. Compensating mutations
in the gene encoding the second species in the complex can then be selected.
Sequence analysis of the genes encoding
the respective proteins will reveal the mutations that correspond to the
region of the protein involved in interactive
binding. Alternatively, one protein can be anchored to a solid surface using
methods described above, and allowed to
interact with and bind to its labeled ligand, which has been treated with a
proteolytic enzyme, such as trypsin. After
washing, a short, labeled peptide comprising the binding domain may remain
associated with the solid material, which
can be isolated and identified by amino acid sequencing. Also, once the gene
coding for the interactive ligand is
obtained, short gene segments can be engineered to express peptide fragments
of the protein, which can then be
tested for binding activity and purified or synthesized.
For example, and not by way of limitation, a Smad3 gene product can be
anchored to a solid material as
described above, by making a GST-Smad3 fusion protein and allowing it to bind
.to glutathione agarose beads. The
interactive ligand can be labeled with a radioactive isotope, such as 35S, and
cleaved with a proteolytic enzyme such as
trypsin. Cleavage products can then be added to the anchored GST-Smad3 fusion
protein and allowed to bind. After
washing away unbound peptides, labeled bound material, representing the
interactive ligand binding domain, can be
eluted, purified, and analyzed for amino acid sequence by well-known methods.
Peptides so identified can be produced
synthetically or fused to appropriate facilitative proteins using recombinant
DNA technology.
In one embodiment, the "ligand" is Smad4, with which Smad3 heteroligomerizes
upon receptor activation. In
another embodiment, the "ligand" is SARA (Small anchor for receptor
activation), which recruits the cytoplasmic signal
transducer Smad3. In a further embodiment, the "ligand" is the cognate DNA
binding site for Smad3. Small MH2
domains are the locus of Small-dependent transcriptional activation activity,
and are the site of protein-protein
interactions responsible for oligomerization of Small proteins as well as
heteromerization with other transcription
-20-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
factors. Thus, in a further embodiment, the MH2 domain of Smad3 is substituted
for Smad3 itself in the assays
described herein.
Assays for Identification of Compounds that Prevent Fibrosis or Improve Wound
Healing
Compounds including, but not limited to, binding compounds identified via
assay techniques such as those
described in the preceding sections, can be tested for the ability to prevent
fibrosis and improve wound healing. The
assays described above can identify compounds which affect Smad3 activity
(e.g., compounds that bind to Smad3,
inhibit binding of a natural ligand, and either block activation (antagonists)
or mimic inhibitors of activation (agonists),
and compounds that bind to a natural ligand of Smad3 and neutralize ligand
activity); or compounds that affect Smad3
gene activity (by affecting Smad3 gene expression, including molecules, e.g.,
proteins or small organic molecules, that
affect or interfere with splicing events so that expression of the full length
or a truncated form of Smad3 can be
modulated). However, it should be noted that the assays described can also
identify compounds that inhibit Smad3
signal transduction (e.g., compounds which affect upstream or downstream
signalling events). The identification and
use of such compounds which affect another step in the Smad3 signal
transduction pathway in which the Smad3 gene
andlor Smad3 gene product is involved and, by affecting this same pathway may
modulate the effect of Smad3 on
fibrosis and wound healing are within the scope of the invention. Such
compounds can be used as part of a method for
the prevention of fibrosis and improvement of wound healing.
The invention encompasses cell-based and animal model-based assays for the
identification of compounds
exhibiting such an ability to prevent fibrosis and improve wound healing.
Cell-based systems can be used to identify compounds which may act to prevent
fibrosis and improve wound
healing. Such cell systems can include, for example, recombinant or non-
recombinant cells, such as cell lines, which
express the Smad3 gene. For example monocyte cells, keratinocyte cells, or
cell lines derived from monocytes or
keratinocytes can be used.
In utilizing such cell systems, cells may be exposed to a compound suspected
of exhibiting an ability to
protect against fibrosis and improve wound healing, at a sufficient
concentration and for a time sufficient to elicit a
cellular phenotype associated with such a protection against fibrosis and
improvement of wound healing in the exposed
cells, e.g., altered migration and selective chemotactic response to TGF-(3.
After exposure, the cells can be assayed to
measure alterations in the expression of the Smad3 gene, e.g., by assaying
cell lysates for Smad3 mRNA transcripts
(e.g., by Northern analysis) or for Smad3 protein expressed in the cell;
compounds which inhibit expression of the
Smad3 gene are good candidates as therapeutics. Alternatively, the cells are
examined to determine whether one or
more cellular phenotype associated with fibrosis or impaired wound healing has
been altered to resemble a cellular
phenotype associated with protection against fibrosis and improvement of wound
healing. Still further, the expression
andlor activity of components of the signal transduction pathway of which
Smad3 is a part, or the activity of Smad3
signal transduction pathway itself can be assayed.
For example, after exposure, the cell lysates can be assayed for the presence
of host cell proteins, as
compared to lysates derived from unexposed control cells. The ability of a
test compound to inhibit expression of
-21-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
specific Smad3 target genes in these assay systems indicates that the test
compound inhibits sigrial transduction
initiated by Smad3 activation. The cell lysates can be readily assayed using a
Western blot format; i.e., the host cell
proteins are resolved by gel electrophoresis, transferred and probed using a
anti-host cell protein detection antibody
(e.g., an anti-host cell protein detection antibody labeled with a signal
generating compound, such as radiolabel, fluor,
enzyme, etc.). Alternatively, an ELISA format could be used in which a
particular host cell protein is immobilized using
an antibody specific for the target host cell protein, and the presence or
absence of the immobilized host cell protein is
detected using a labeled second antibody. In yet another approach, ion flux,
such as calcium ion flux, can be measured
as an end point for Smad3 stimulated signal transduction. In yet a further
approach, assays for compounds that
interfere with Smad3 binding to its cognate DNA binding site utilize specific
reporter constructs, such as (SBE)4-
luciferase reporter, driven by four repeats of the sequence identified as a
Smad binding element in the Jung promoter.
In addition, animal-based systems for protection against fibrosis and
improvement of wound healing, for
example, may be used to identify compounds capable of protecting against
fibrosis and improving wound healing. Such
animal models may be used as test substrates for the identification of drugs,
pharmaceuticals, therapies and
interventions which may be effective in protecting against fibrosis and
improving wound healing. For example, animal
models may be exposed to a compound, suspected of protecting against fibrosis
or improving wound healing, at a
sufficient concentration and for a time sufficient to elicit a protection
against fibrosis and improvement of wound
healing in the exposed animals. The response of animals to the exposure may be
monitored by assessing
radioprotection or cutaneous wound healing. With regard to intervention, any
treatments which protect against any
aspect of fibrosis or improve any aspect of wound healing should be considered
as candidates for human therapeutic
intervention in protecting against fibrosis and improving wound healing.
Dosages of test agents may be determined by
deriving dose-response curves, as discussed in the sections below.
Inhibition of Smad3 Expression or Smad3 Activity to Prevent Fibrosis or
Improve Wound Healing
Any method which neutralizes Smad3 or inhibits expression of the Smad3 gene
(either transcription or
translation) can be used to protect against fibrosis and improve wound
healing. Such approaches can be used to
reduce the size of wounds, to treat chronic non-healing wounds, to promote
closure in surgical wounds, to speed the
re-epithelialization of wounds, to treat ulcers, e.g., decubitus ulcers,
diabetic ulcers, and venous stasis ulcers, to
improve the growth of autologous skin grafts, and to hasten the recovery of
severe burn patients. Such methods can
also be useful for imparting resistance to fibrosis resulting from chronic
inflammation, e.g., pulmonary fibrosis,
glomerulosclerosis, and cirrhosis, protecting against radiation-induced
fibrosis, supporting members of the armed
forces who might be expected to encounter high dose radiation, permitting dose
escalation of radiation treatment, e.g.,
in cancer patients, and decreasing the accumulation of scar tissue.
For example, the administration of soluble peptides, proteins, fusion
proteins, or antibodies (including anti-
idiotypic antibodies) that bind to and "neutralize" Smad3 can be used to
protect against fibrosis and improve wound
healing. To this end, peptides corresponding to the cytoplasmic domain of the
TGF-[3 or activin receptor (or a domain
of a Smad involved in forming dimers with Smad3) can be utilized.
Alternatively, anti-idiotypic antibodies or Fab
-22-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
fragments of antiidiotypic antibodies that mimic the cytoplasmic domain of the
TGF-(3 or activin receptor (or the
domain of a Smad involved in forming dimers with Smad3) and that neutralize
Smad3 can be used. Such Smad3
peptides, proteins, fusions proteins, antibodies, anti-idiotypic antibodies or
Fabs are administered to a subject in
amounts sufficient to neutralize Smad3 and protect against fibrosis or improve
wound healing.
In one embodiment, the peptides, proteins, fusions proteins, antibodies, anti-
idiotypic antibodies or Fabs are
cell-permeable compounds. In an alternative embodiment, cells are genetically
engineered using recombinant DNA
techniques to introduce the coding sequence for the peptide, protein, fusion
protein, antibody, anti-idiotypic antibody or
Fab into the cell, e.g., by transduction (using viral vectors, such as
retroviruses, adenoviruses, and adeno-associated
viruses) or transfection procedures, including but not limited to the use of
naked DNA or RNA, plasmids, cosmids,
YACs, electroporation, liposomes, etc. The coding sequence can be placed under
the control of a strong constitutive or
inducible promoter, or a tissue-specific promoter, to achieve expression of
the gene product. The engineered cells
which express the gene product can be produced in vitro and introduced into
the patient, e.g., systemically,
intraperitoneally, at the site of cutaneous wound healing, or the cells can be
incorporated into a matrix and implanted
in the body, e.g., genetically engineered cells can be implanted as part of a
skin graft. Alternatively, the engineered
cells which express the gene product can be produced following in vivo gene
therapy approaches.
In a preferred embodiment, monoclonal antibodies are produced in one of three
different ways. They can be
generated as mouse antibodies that are subsequently "humanized" by
recombination with human antibody genes
(Kohler and Milstein, Nature 256,495 (1975); Winter and Harris, Trends
Pharmacol. Sci. 14, 139 (1993); and Queen et
al., Proc. Nat/. Acad Sci. USA 86, 10029 (19891). Alternatively, human
antibodies are raised in nude mice grafted
with human immune cells (Bruggemann and Neuberger, immunol. Today 8, 391
(1996)). Finally antibodies can also be
made by phase display techniques (Huse et al., Science 246, 1275 (1989);
Hoogenboom et al., lmmunotechno%gy 4, 1
(19981; and Rodi and Makowski, Curr. Opin. Biotechnol. 10, 87 (1999)1.
For the production of antibodies, various host animals may be immunized by
injection with Smad3, a Smad3
peptide, functional equivalents or mutants of Smad3. Such host animals may
include but are not limited to rabbits,
mice, and rats, to name but a few. Various adjuvants may be used to increase
the immunological response, depending
on the host species, including but not limited to Freund's (complete and
incomplete), mineral gels such as aluminum
hydroxide, surface active substances such as lysolecithin, pluronic polyols,
polyanions, peptides, oil emulsions, keyhole
limpet hemocyanin, dinitrophenol, and potentially useful human adjuvants such
as BCG (bacille Calmette-Guerin) and
Corynebacterium parvum. Polyclonal antibodies are heterogeneous populations of
antibody molecules derived from the
sera of the immunized animals.
Monoclonal antibodies, which are homogeneous populations of antibodies to a
particular antigen, may be
obtained by any technique which provides for the production of antibody
molecules by continuous cell lines in culture.
These include, but are not limited to, the hybridoma technique of Kohler and
Milstein, (1975, Nature 256:495-497; and
U.S. Pat. No. 4,376,110), the human B-cell hybridoma technique (Kosbor et al.,
1983, Immunology Today 4:72; Cole et
al., 1983, Proc. Natl. Acad. Sci. USA 80:2026-2030), and the EBV-hybridoma
technique (Cole et al., 1985, Monoclonal
-23-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
Antibodies And Cancer Therapy, Alan R. I_iss, Inc., pp. 77-96). Such
antibodies may be of any immunoglobulin class
including IgG, IgM, IgE, IgA, IgD and any subclass thereof. The hybridoma
producing the mAb of this invention may be
cultivated in vitro or in vivo. Production of high titers of mAbs in vivo
makes this the presently preferred method of
production.
In addition, techniques developed for the production of "chimeric antibodies"
(Morrison et al., 1984, Proc.
Natl. Acad. Sci., 81:6851-6855; Neuberger et al., 1984, Nature, 312:604-608;
Takeda et al., 1985, Nature, 314:452
454) by splicing the genes from a mouse antibody molecule of appropriate
antigen specificity together with genes from
a human antibody molecule of appropriate biological activity can be used. A
chimeric antibody is a molecule in which
different portions are derived from different animal species, such as those
having a variable region derived from a
murine mAb and a human immunoglobulin constant region.
Alternatively, techniques described for the production of single chain
antibodies (U.S. Pat. No. 4,946,778;
Bird, 1988, Science 242:423-426; Huston et al., 1988, Proc. Natl. Acad. Sci.
USA 85:5879-5883; and Ward et al.,
1989, Nature 334:544-546) can be adapted to produce single chain antibodies
against Smad3 gene products. Single
chain antibodies are formed by linking the heavy and light chain fragments of
the Fv region via an amino acid bridge,
resulting in a single chain polypeptide.
Antibody fragments which recognize specific epitopes may be generated by known
techniques. For example,
such fragments include but are not limited to: the Flab')2 fragments which can
be produced by pepsin digestion of the
antibody molecule and the Fab fragments which can be generated by reducing the
disulfide bridges of the F(ab')2
fragments. Alternatively, Fab expression libraries may be constructed (Huse et
al., 1989, Science, 246:1275-1281) to
allow rapid and easy identification of monoclonal Fab fragments with the
desired specificity.
Antibodies to ligands of Smad3 can, in turn, be utilized to generate anti-
idiotype antibodies that "mimic"
these ligands, using techniques well known to those skilled in the art. (See,
e.g., Greenspan & Bona, 1993, FASEB J
7(5):437-444; and Nissinoff, 1991, J. Immunol. 147(81:2429-24381. For example
antibodies which bind to the
cytoplasmic domain of the TGF-~i or activin receptor (or the domain of a Smad
involved in forming dimers with Smad3)
and competitively inhibit the binding of Smad3 to the TGF-(3 or activin
receptor (or a Smad involved in forming dimers
with Smad3) can be used to generate anti-idiotypes that "mimic" these ligands
and, therefore, bind and neutralize
Smad3. Such neutralizing anti-idiotypes or Fab fragments of such anti-
idiotypes can be used in therapeutic regimens to
neutralize Smad3 and protect against fibrosis and improve wound healing.
In an alternate embodiment, interventions to prevent fibrosis and improve
wound healing can be designed by
reducing the level of endogenous Smad3 gene expression, e.g., using antisense
or ribozyme approaches to inhibit or
prevent translation of Smad3 mRNA transcripts; triple helix approaches to
inhibit transcription of the Smad3 gene; or
targeted homologous recombination to inactivate or "knock out" the Smad3 gene
or its endogenous promoter. Delivery
techniques could be preferably designed for a systemic approach.
Alternatively, the antisense, ribozyme or DNA
constructs described herein could be administered directly to the site
containing the target cells, e.g., sites of
cutaneous wound healing.
-24-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
Antisense approaches involve the design of oligonucleotides (either DNA or
RNA) that are complementary to
Smad3 mRNA. The antisense oligonucleotides will bind to the complementary
Smad3 mRNA transcripts and prevent
translation. Absolute complementarity, although preferred, is not required. A
sequence "complementary" to a portion of
an RNA, as referred to herein, means a sequence having sufficient
complementarity to be able to hybridize with the
RNA, forming a stable duplex; in the case of double-stranded antisense nucleic
acids, a single strand of the duplex DNA
may thus be tested, or triplex formation may be assayed. The ability to
hybridize will depend on both the degree of
complementarity and the length of the antisense nucleic acid. Generally, the
longer the hybridizing nucleic acid, the
more base mismatches with an RNA it may contain and still form a stable duplex
(or triplex, as the case may be). One
skilled in the art can ascertain a tolerable degree of mismatch by use of
standard procedures to determine the melting
point of the hybridized complex.
Oligonucleotides that are complementary to the 5' end of the message, e.g.,
the 5' untranslated sequence up
to and including the AUG initiation colon, should work most efficiently at
inhibiting translation. However, sequences
complementary to the 3' untranslated sequences of mRNAs have recently shown to
be effective at inhibiting
translation of mRNAs as well. See generally, Wagner, R., 1994, Nature 372:333-
335. Thus, oligonucleotides
complementary to either the 5'- or 3'- non-translated, non-coding regions of
Smad3 could be used in an antisense
approach to inhibit translation of endogenous Smad3 mRNA. Oligonucleotides
complementary to the 5' untranslated
region of the mRNA should include the complement of the AUG start colon.
Antisense oliganucleotides complementary
to mRNA coding regions could also be used in accordance with the invention.
Whether designed to hybridize to the 5'-,
3'- or coding region of Smad3 mRNA, antisense nucleic acids should be at least
six nucleotides in length, and are
preferably oligonucleotides ranging from 6 to about 50 nucleotides in length.
In specific aspects the oligonucleotide is
at least 6 nucleotides, at least 17 nucleotides, at least 25 nucleotides or at
least 50 nucleotides.
Regardless of the choice of target sequence, it is preferred that in vitro
studies are first performed to
quantitate the ability of the antisense oligonucleotide to inhibit gene
expression. It is preferred that these studies
utilize controls that distinguish between antisense gene inhibition and
nonspecific biological effects of oligonucleotides.
It is also preferred that these studies compare levels of the target RNA or
protein with that of an internal control RNA
or protein. Additionally, it is envisioned that results obtained using the
antisense oligonucleotide are compared with
those obtained using a control oligonucleotide. It is preferred that the
control oligonucleotide is of approximately the
same length as the test oligonucleotide and that the nucleotide sequence of
the oligonucleotide differs from the
antisense sequence no more than is necessary to prevent specific hybridization
to the target sequence.
The oligonucleotides can be DNA or RNA or chimeric mixtures or derivatives or
modified versions thereof,
single-stranded or double-stranded. The oligonucleotide can be modified at the
base moiety, sugar moiety, or phosphate
backbone, for example, to improve stability of the molecule, hybridization,
etc. The oligonucleotide may include other
appended groups such as peptides le.g., for targeting host cell receptors in
vivo), agents facilitating transport across
the cell membrane (see, e.g., Letsinger et a1.,1989, Proc. Natl. Acad. Sci.
U.S.A. 86:6553-6556; Lemaitre et al., 1987,
Proc. Natl. Acad. Sci. 84:648-652; PCT Publication No. W088109810, published
Dec. 15, 1988) or other barriers,
-25-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
hybridization-triggered cleavage agents (See, e.g., Krol et al., 1988,
BioTechniques 6:958-976) or intercalating agents
(See, e.g., Zon, 1988, Pharm. Res. 5:539-549). To this end, the
oligonucleotide may be conjugated to another
molecule, e.g., a peptide, hybridization triggered cross-linking agent,
transport agent, hybridization-triggered cleavage
agent, etc.
The antisense oligonucleotide may comprise at least one modified base moiety
which is selected from the
group including but not limited to 5-fluorouracil, 5-bromouracil, 5-
chlorouracil, 5-iodouracil, hypoxanthine, xantine, 4-
acetylcytosine, 5-(carboxyhydroxylmethyl) uracil, 5-carboxymethylaminomethyl-2-
thiouridine, 5-
carboxymethylaminomethyluracil, dihydrouracil, beta-D-galactosylqueosine,
inosine, N6-isopentenyladenine, 1-
methylguanine, 1-methylinosine, 2,2-dimethylguanine, 2-methyladenine, 2-
methylguanine, 3-methylcytosine, 5-
methylcytosine, N6-adenine, 7-methylguanine, 5-methylaminomethyluracil, 5-
methoxyaminomethyl-2-thiouracil, beta-D-
mannosylqueosine, 5'-methoxycarboxymethyluracil, 5-methoxyuracil, 2-methylthio-
N6-isopentenyladenine, uracil-5-
oxyacetic acid (v), wybutoxosine, pseudouracil, queosine, 2-thiocytosine, 5-
methyl-2-thiouracil, 2-thiouracil, 4-
thiouracil, 5-methyluracil, uracil5-oxyacetic acid methylester, uracil-5-
oxyacetic acid (v), 5-methyl-2-thiouracil, 3-(3-
amino-3-N-2-carboxypropyl) uracil, (acp3)w, and 2,6-diaminopurine.
The antisense oligonucleotide may also comprise at least one modified sugar
moiety selected from the group
including but not limited to arabinose, 2-fluoroarabinose, xylulose, and
hexose.
In yet another embodiment, the antisense oligonucleotide comprises at least
one modified phosphate
backbone selected from the group consisting of a phosphorothioate, a
phosphorodithioate, a phosphoramidothioate, a
phosphoramidate, a phosphordiamidate, a methylphosphonate, an alkyl
phosphotriester, and a formacetal or analog
thereof.
Oligonucleotides of the invention may be synthesized by standard methods known
in the art, e.g. by use of an
automated DNA synthesizer (such as are commercially available from Biosearch,
Applied Biosystems, etc.). As
examples, phosphorothioate oligonucleotides may be synthesized by the method
of Stein et al. (1988, Nucl. Acids Res.
16:3209), methylphosphonate oligonucleotides can be prepared by use of
controlled pore glass polymer supports (Satin
et al., 1988, Proc. Natl. Acad. Sci. U.S.A. 85:7448-74511, etc.
The antisense molecules should be delivered to cells which express the Smad3
protein in vivo, e.g., sites of
cutaneous wound healing. A number of methods have been developed for
delivering antisense DNA or RNA to cells;
e.g., antisense molecules can be injected directly into the tissue site, or
modified antisense molecules, designed to
target the desired cells (e.g., antisense linked to peptides or antibodies
that specifically bind receptors or antigens
expressed on the target cell surface] can be administered systemically.
However, it is often difficult to achieve intracellular concentrations of the
antisense sufficient to suppress
translation of endogenous mRNAs. Therefore a preferred approach utilizes a
recombinant DNA construct in which the
antisense oligonucleotide is placed under the control of a strong pol III or
pol II promoter. The use of such a construct
to transfect target cells in the patient will result in the transcription of
sufficient amounts of single stranded RNAs
that will form complementary base pairs with the endogenous Smad3 transcripts
and thereby prevent translation of
-26-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
the Smad3 mRNA. For example, a vector can be introduced in vivo such that it
is taken up by a cell and directs the
transcription of an antisense RNA. Such a vector can remain episomal or become
chromosomally integrated, as long as
it can be transcribed to produce the desired antisense RNA. Such vectors can
be constructed by recombinant DNA
technology methods standard in the art. Vectors can be plasmid, viral, or
others known in the art, used far replication
and expression in mammalian cells. Expression of the sequence encoding the
antisense RNA can be by any promoter
known in the art to act in mammalian, preferably human cells. Such promoters
can be inducible or constitutive. Such
promoters include but are not limited to: the SV40 early promoter region
(Bernoist and Chambon, 1981, Nature
290:304-310), the promoter contained in the 3' long terminal repeat of Rous
sarcoma virus (Yamamoto et al., 1980,
Cell 22:787-7971, the herpes thymidine kinase promoter (Wagner et al., 1981,
Proc. Natl. Acad. Sci. U.S.A. 78:1441-
1445), the regulatory sequences of the metallothionein gene (Brinster et al.,
1982, Nature 296:39-42/, etc. An
epidermal specific promoter may be desireable, such as a keratin based vector
that has its expression induced by a
variety of appropriate stimuli including wounding. Any type of plasmid,
cosmid, YAC or viral vector can be used to
prepare the recombinant DNA construct which can be introduced directly into
the tissue site; e.g., the site of
cutaneous wound healing. Alternatively, viral vectors can be used which
selectively infect the desired tissue; (e.g., for
skin, papillomavirus vectors may be used), in which case administration may be
accomplished by another route (e.g.,
systemicallyl.
Ribozyme molecules-designed to catalytically cleave Smad3 mRNA transcripts can
also be used to prevent
translation of Smad3 mRNA and expression of Smad3. (See, e.g., PCT
International Publication W090111364,
published Oct. 4, 1990; Sarver et al., 1990, Science 247:1222-12251. While
ribozymes that cleave mRNA at site
specific recognition sequences can be used to destroy Smad3 mRNAs, the use of
hammerhead ribozymes is preferred.
Hammerhead ribozymes cleave mRNAs at locations dictated by flanking regions
that form complementary base pairs
with the target mRNA. The sole requirement is that the target mRNA have the
following sequence of two bases: 5'-
UG-3'. The construction and production of hammerhead ribozymes is well known
in the art and is described more fully
in Haseloff and Gerlach, 1988, Nature, 334:585-591. There are a plurality of
potential hammerhead ribozyme cleavage
sites within the nucleotide sequence of human Smad3 cDNA. Preferably the
ribozyme is engineered so that the
cleavage recognition site is located near the 5' end of the Smad3 mRNA; i.e.,
to increase efficiency and minimize the
intracellular accumulation of non-functional mRNA transcripts.
The ribozymes of the present invention also include RNA endoribonucleases
(hereinafter "Cech-type
ribozymes") such as the one which occurs naturally in Tetrahymena Thermophila
(known as the IVS, or L-19 IVS RNA)
and which has been extensively described by Thomas Cech and collaborators
(Zaug, et al., 1984, Science, 224:574
578; Zaug and Cech, 1986, Science, 231:470-475; Zaug, et al., 1986, Nature,
324:429-433; published International
patent-application No. WO 88104300 by University Patents Inc.; Been and Cech,
1986, Cell, 47:207-216). The Cech
type ribozymes have an eight base pair active site which hybridizes to a
target RNA sequence whereafter cleavage of
the target RNA takes place. The invention encompasses those Cech-type
ribozymes which target eight base-pair active
site sequences that are present in Smad3.
-27-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
As in the antisense approach, the ribozymes can be composed of modified
oligonucleotides (e.g. for improved
stability, targeting, etc.) and should be delivered to cells which express
Smad3 in vivo, e.g., sites of cutaneous wound
healing. A preferred method of delivery involves using a DNA construct
"encoding" the ribozyme under the control of a
strong constitutive pol III or pol II promoter, so that transfected cells will
produce sufficient quantities of the ribozyme
to destroy endogenous Smad3 messages and inhibit translation. Because
ribozymes unlike antisense molecules, are
catalytic, a lower intracellular concentration is required for efficiency.
Endogenous Smad3 gene expression can also be reduced by inactivating or
"knocking out" the Smad3 gene
or its promoter using targeted homologous recombination. (E.g., see Smithies
et al., 1985, Nature 317:230-234;
Thomas & Capecchi, 1987, Cell 51:503-512; Thompson et al., 1989 Cell 5:313-
321). For example, a mutant, non-
functional Smad3 protein (or a completely unrelated DNA sequence) flanked by
DNA homologous to the endogenous
Smad3 gene (either the coding regions or regulatory regions of the Smad3 gene)
can be used, with or without a
selectable marker andlor a negative selectable marker, to transfect cells that
express Smad3 in vivo. Insertion of the
DNA construct, via targeted homologous recombination, results in inactivation
of the Smad3 gene. This approach is
acceptable for use in humans provided the recombinant DNA constructs are
directly administered or targeted to the
required site using appropriate viral vectors, e.g., papillomavirus vectors
for in vivo delivery to sites of cutaneous
wound healing, or retrovirus vectors for in vitro transduction of autologous
skin grafts.
Alternatively, endogenous Smad3 gene expression can be reduced by targeting
deoxyribonucleotide
sequences complementary to the regulatory region of the Smad3 gene /i.e., the
Smad3 promoter andlor enhancers) to
form triple helical structures that prevent transcription of the Smad3 gene in
target cells in the body. (See generally,
Helene, C. 1991, Anticancer Drug Des., 6(6):569-84; Helene, C., et al., 1992,
Ann, N.Y. Accad. Sci., 660:27-36; and
Maher, L. J., 1992, Bioassays 14(12):807-15).
In yet another embodiment of the invention, the activity of Smad3 can be
reduced using a "dominant
negative" approach to protect against fibrosis and improve wound healing. To
this end, constructs which encode
defective Smad3 proteins, can be used in gene therapy approaches to diminish
the activity of Smad3 in appropriate
target cells. For example, nucleotide sequences that direct host cell
expression of Smad3 in which a domain or portion
of a domain is deleted or mutated can be introduced into cells at sites of
high-dose radiation exposure or cutaneous
wound healing (by gene therapy methods described above). Alternatively,
targeted homologous recombination can be
utilized to introduce such deletions or mutations into the subject's
endogenous Smad3 gene at sites of high-dose
radiation exposure or cutaneous wound healing. The engineered cells will
express non-functional Smad3 (i.e., a Smad 3
that is capable of binding its natural ligand, but incapable of signal
transduction). Such engineered cells at sites of high-
dose radiation exposure or cutaneous wound healing should demonstrate a
heightened response to TGF-(3, resulting in
protection against fibrosis and improved wound healing.
Pharmaceutical Preparations and Methods of Administration
The compounds that are determined to affect Smad3 gene expression or Smad3
activity can be administered
to a patient at therapeutically effective doses to protect against fibrosis
and improve wound healing. A therapeutically
_28_
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
effective dose refers to that amount of the compound sufficient to result in
protection against fibrosis and
improvement of wound healing. The compounds of the invention are generally
administered to animals, including
humans.
Effective Dose
Toxicity and therapeutic efficacy of such compounds can be determined by
standard pharmaceutical
procedures in cell cultures or experimental animals, e.g., for determining the
LD50 (the dose lethal to 50% of the
population) and the ED50 (the dose therapeutically effective in 50% of the
population). The dose ratio between toxic
and therapeutic effects is the therapeutic index and it can be expressed as
the ratio LD50 IED50. Compounds which
exhibit large therapeutic indices are preferred. While compounds that exhibit
toxic ,side effects may be used, care
should be taken to design a delivery system that targets such compounds to the
site of affected tissue in order to
minimize potential damage to uninfected cells and, thereby, reduce side
effects.
The data obtained from the cell culture assays and animal studies can be used
in formulating a range of
dosage for use in humans. The dosage of such compounds lies preferably within
a range of circulating concentrations
that include the ED50 with little or no toxicity. The dosage may vary within
this range depending upon the dosage form
employed and the route of administration utilized. For any compound used in
the method of the invention, the
therapeutically effective dose can be estimated initially from cell culture
assays. A dose may be formulated in animal
models to achieve a circulating plasma concentration range that includes the
IC50 (i.e., the concentration of the test
compound which achieves a half-maximal inhibition of symptoms) as determined
in cell culture. Such information can
be used to more accurately determine useful doses in humans. Levels in plasma
may be measured, for example, by high
performance liquid chromatography.
It will be appreciated that the actual preferred amounts of active compound in
a specific case will vary
according to the specific compound being utilized, the particular compositions
formulated, the mode of application, and
the particular situs and organism being treated. Dosages for a give host can
be determined using conventional
considerations, e.g., by customary comparison of the differential activities
of the subject compounds and of a known
agent, e.g., by means of an appropriate, conventional pharmacological
protocol.
Formulation and Use
The pharmacologically active compounds of this invention can be processed in
accordance with conventional
methods of galenic pharmacy to produce medicinal agents for administration to
patients, e.g., mammals including
humans.
The compounds of this invention can be employed in admixture with conventional
excipients, i.e.,
pharmaceutically acceptable organic or inorganic carrier substances suitable
for parenteral, enteral (e.g., oral) or
topical application, which do not deleteriously react with the active
compounds. Suitable pharmaceutically acceptable
carriers include but are not limited to water, salt solutions, alcohols, gum
arabic, vegetable oils, benzyl alcohols,
polyethylene glycols, gelatin, carbohydrates such as lactose, amylose or
starch, magnesium stearate, talc, silicic acid,
viscous paraffin, perfume oil, fatty acid monoglycerides and diglycerides,
pentaerythritol fatty acid esters, hydroxy
-29-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
methylcellulose, polyvinyl pyrrolidone, etc. The pharmaceutical preparations
can be sterilized and if desired mixed with
auxiliary agents, e.g., lubricants, preservatives, stabilizers, wetting
agents, emulsifiers, salts for influencing osmotic
pressure, buffers, coloring, flavoring andlor aromatic substances and the like
which do not deleteriously react with the
active compounds. They can also be combined where desired with other active
agents, e.g., vitamins.
For parenteral application, particularly suitable are injectable, sterile
solutions, preferably oily or aqueous
solutions, as well as suspensions, emulsions, or implants, including
suppositories. Ampoules are convenient unit
dosages.
For enteral application, particularly suitable are tablets, dragees, liquids,
drops, suppositories, or capsules. A
syrup, elixir, or the like can be used wherein a sweetened vehicle is
employed.
Sustained or directed release compositions can be formulated, eg., by
inclusion in liposomes or incorporation
into an epidermal patch with a suitable carrier, for example DMSO. It is also
possible to freeze-dry these compounds
and use the lyophilizates obtained, for example, for the preparation of
products for injection.
For topical application, there are employed as non-sprayable forms, viscous to
semi-solid or solid forms
comprising a carrier compatible with topical application and having a dynamic
viscosity preferably greater than water.
Suitable formulations include but are not limited to solutions, suspensions,
emulsions, creams, ointments, powders,
liniments, salves, aerosols, etc., which are, if desired, sterilized or mixed
with auxiliary agents, e.g., preservatives,
stabilizers, wetting agents, buffers or salts for influencing osmotic
pressure, etc. For topical application, also suitable
are sprayable aerosol preparations wherein the active ingredient, preferably
in combination with a solid or liquid inert
carrier material, is packaged in a squeeze bottle or in admixture with a
pressurized volatile, normally gaseous
propellant, e.g., a freon.
The compositions may, if desired, be presented in a pack or dispenser device
which may contain one or more
unit dosage forms containing the active ingredient. The pack may for example
comprise metal or plastic foil, such as a
blister pack. The pack or dispenser device may be accompanied by instructions
for administration.
Smad3 Disruption Leads To Accelerated Wound Healinn.
Following full-thickness incisional wounds (Ashcroft, G.S. et al. Estrogen
accelerates cutaneous wound
healing associated with an increase in TGF-beta 1 levels. Nature Med 3, 1209-
1215 (1997)), the rate of wound
healing was markedly accelerated in healthy Smad3e'siexa mice (Table 1), with
complete re-epithelialization occurring by
day 2 post-wounding in the null mice versus day 5 in the wild-type mice (Fig.
1 b), and with significantly reduced wound
areas (Fig. 1 a) and wound widths visible. Total cell numbers (fibroblasts and
inflammatory cells) were markedly
reduced in the wounds of the Smad3exaiexs mice, with intermediate numbers
present in the heterozygous mice (Fig 1c),
compared with wild-type controls. Giemsa staining of sections in conjunction
with immunostaining for a monocyte
marker indicated that both neutrophils and monocytes were largely absent from
the early wounds of Smad3exeie.a mice.
The wound areas of the Smad3e'siexs mice were significantly smaller than those
of wild-type mice, with reduced
quantities of granulation tissue present at all time points. Wound contraction
occurs through the relative contributions
of re-epithelialization and myofibroblast action, and thus the accelerated re-
epithelialization in the Smad3exsiexs mice,
-3 0-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
andlor increased contractility of wound fibroblasts, presumably contribute to
this phenotype. This observation
corroborates earlier controversial studies indicating that central granulation
tissue may not be critical to wound closure
(Gross, J. et al. On the mechanism of skin wound "contraction": a granulation
tissue "knockout" with a normal
phenotype. Prac. NatiAcad Sci. USA 92, 5982-5986 (1995)).
Table 1 Accelerated
wound healing
after targeted
Smad3 disruption
Phenotype Day 1 Day 2 Day 3 Day 5
Wild-type Inflammation No re- No re- Re-
(+++) EpithelializationEpithelializationEpithelial-ized
Wide wound Granulation Moderate
(++) tissue (++) wound width
Smad3 Inflammation No re- Re- Moderate
Heterozygote(++) epithelializationEpithelialized wound width
Wide wound Granulation
(+) tissue (++)
Smad3 Reduced Re- Re- Narrow
Knockout Inflammation epithelializedEpithelialized wound width
Narrow Reduced
Wound granulation
tissue
Effects Of Exo4enous TGF-Q on the Wound-Healinn Response.
TGF-(3 released from degranulating platelets at wound sites has a broad
spectrum of effects on, and is
secreted by, each of the diverse cell types involved in wound healing.
Specifically, these cells include the keratinocyte,
responsible for reconstruction of the cutaneous barrier, the fibroblast,
responsible for matrix production, and the
monocyte, which infiltrates the wound at an early stage and secretes a vast
array of cell-regulatory cytokines,
including TGF-(3 (Roberts, A. B. TGF-beta: activity and efficacy in animal
models of wound healing. Wound Repair
Regen. 3, 408-418 (1995)); (0'Kane, S. & Ferguson, M. W. J. TGF-beta s and
wound healing. /nt. J. Biochem. CeiiBiol.
29, 63-78 (1997)). As we observed a marked reduction in the number of
monocytes in the wounds of the null mice,
we proposed that part of the healing phenotype was secondary to the reduced
levels of TGF-(3, a potent monocyte
chemoattractant, secreted by these inflammatory cells (Wahl, S.M. et al.
Transforming growth factor type beta
induces monocyte chemotaxis and growth factor production. Proc. Nat/ Acad Sci,
USA 84, 5788-5792 (1987)).
Moreover, depletion of monocytes in animal models leads to a reduced fibrotic
response, consistent with the role of
these cells in TGF-(3 secretion (Leibovich, S.J. & Ross, R. The role of the
macrophage in wound repair. A study with
hydrocortisone and antimacrophage serum. Am. J. PathoL 78, 71-100 (197511;
(McCartney-Francis, N., & Wahl, S.M.
Transforming growth factor beta: a matter of life and death. J, Leuk. BioL 55,
401-109 (1994)). Although TGF-[31 was
present at equivalent levels in the serum of all animals, probably
representing TGF-(31 released from platelet oc-
granules (Fig. 2a), the null mice showed reduced immunostaining for TGF-[3
isoforms in. wound leukocytes and
-31-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
decreased TGF-(31 RNA levels, particularly at day 3 (Fig. 2b), supporting our
hypothesis that a reduction in local TGF-
(31 amounts contribute to the aberrant wound-healing phenotype of these mice.
To address this question, we applied topical TGF-j31 immediately before
wounding. Following treatment
with TGF-j31, inflammatory-cell numbers were increased in the heterozygote but
not in the Smad3e'eiexa wounds,
indicating that Smad3 may be critical for TGF-(3-mediated chemotaxis. Despite
a failure to influence monocyte
recruitment, addition of TGF-(31 to the wounds of the Smad3e"aiexe mice
increased matrix deposition, corroborating
previous studies that showed that monocytes affect matrix deposition
indirectly through the production of TGF-(31
(Pierce, G.F. et al. Transforming growth factor beta reverses the
glucocorticoid-induced wound-healing deficit in rats:
possible regulation in macrophages by platelet-derived growth factor. Proc.
Nat/. Acad Sci. USA 86, 2229-2233
(19891). Exogenous TGF-(31 stimulated matrix deposition, most notably in the
null and heterozygous mice, without
evidence of increasing fibroblast numbers, consistent with the idea that
reduced local levels of TGF-(31 in the
Smad3e'e~exe mice underlie the decreased matrix deposition in these animals.
Moreover, these data indicate that
expression of TGF-(3 receptors in the wounds of the null mice is adequate for
matrix production. (Fig. 2c) The SMAD
signaling pathway may be important for collagen expression, whereas
fibronectin (matrix) synthesis may be induced by
TGF-(3 through a c-Jun (SMAD-independent) pathway (Vindevoghel, L. et al.
SMAD314-dependent transcriptional
activation of the human type VII collagen gene (COL7A1) promoter by
transforming growth factor beta. Proc. Nat/
Acad. Sci. USA 95, 14769-14774 (1998)); (Chen, S.J. et al. Stimulation of type
I collagen transcription in human skin
fibroblasts by TGF-beta: involvement of Smad3. J. Invest. Dermatol. 112, 49-57
(1999)); (Hocevar, B:A., Brown, T.L.
& Howe, P.H. TGF-beta induces fibronectin synthesis through a c-Jun N-terminal
Kinase-dependent, Smad4-
independent pathway. EMBD J, 18, 1345-1356 (1999)). In agreement with this,
our data also implicate a Smad3-
independent pathway in early fibroblast matrix production in vivo.
Mechanisms Underlyinn A Reduced Local Monocyte Influx.
As Smad3 appeared to be potentially important in monocyte function, we focused
on the mechanisms
underlying these observations. If circulating monocytes are to infiltrate the
sites of injurylinflammation, they must first
respond to a local chemoattractant signal and traverse the endothelial
basement membrane. TGF-(3 is a key factor in
this response because, in vivo, femtomolar concentrations of TGF-(3 induce the
most potent known chemoattractant
response by circulating blood monocytes (Wahl, S.M. et al. Transforming growth
factor type beta induces monocyte
chemotaxis and growth factor production. Proc. Nat/ Acad Sci. USA 84, 5788-
5792 (19871); (Wiseman, D.M.,
Polverini, P.J., Kamp, D.W. & Leibovich, S.J. Transforming growth factor-beta
(TGF beta) is chemotactic for human
monocytes and induces their expression of angiogenic activity. Biochem.
Biophys. Res. Commun. 157, 793-800
(1988)). To investigate the mechanisms underlying the observed reduction in
wound monocyte numbers, we
determined the effects of Smad3 deletion on monocyte chemotaxis and on the
expression of TGF-(3-regulated cell-
adhesion molecules potentially important in the traps-endothelial migration
and adhesion of monocytes (Wahl, S.M.,
Allen, J.B., Weeks, B.S., Wong, H.L. & Klotman, P.E. Trandforming growth
factor beta enhances integrin expression
-32-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
and type IV collagenase secretion in human monocytes. Proc. NatlAcad Sci. USA
90, 4577-4581 (1993)). Cultured
Smad3e~eiexe monocytes exhibited significantly reduced specific chemotaxis to
TGF-(31, but migrated normally to the
classical chemoattractant fMet-Leu-Phe (FMLP), a G-protein-mediated response
(Fig. 3a). Smad3exsiexa monocytes also
showed a failure to upregulate TGF-j31 expression in an autocrine fashion
(Fig. 36) despite a TGF-j3 madiated increase
in levels of TGF-(3 receptor II (TGF-(3RII). The data indicate that regulation
of TGF-j31 and its receptor may occur
independently, with Smad3 being involved in induction of TGF-(31 expression
and Smad3-independent pathways (such
as those involving Smad2 or MAP kinase) regulating receptor expression. Smad3-
independent events may also be
involved in TGF-(3-mediated expression of integrins by monocytes (Fig. 3c).
To test the hypothesis that the initial reduction in monocyte numbers observed
in the wounds of the Smad3-
null mice contributed to the wound-healing phenotype, we applied freshly
extracted monocytes from wild-type mice to
Smad3e"$~e's wounds. Direct addition of wild-type monocytes at the time of
wounding has a similar effect to that of
injection of TGF-(3. That is, reduced matrix deposition in the wounds of the
Smad3e"eiexs mice does not reflect
impairment of the ability of Smad3e"8~exa fibroblasts to elaborate matrix
proteins per se, but instead results from the
reduced levels of TGF-(3 in the wounds of the Smad3e'siexe mice (reduced TGF-
(3 levels being themselves a direct result
of the reduced monocytic infiltrate). Injection of neither monocytes nor TGF-
(3 affected re-epithelialization, so these
two effects - matrix deposition and re-epithelialization - can be
distinguished. We suggest that the decrease in
monocyte infiltration is related to a lack of response by Smad3exaiexs
monocytes to an initial TGF-(31 chemotactic
signal, despite retention of the ability to respond in terms of integrin
upregulation. These events subsequently lead to
reduced local levels of TGF-(3, a characteristic that is secondary not only to
reduced cell numbers but also to an
absence of autocrine induction of TGF-(31.
Role of Smad3 in Wound Re-Epithelialization.
As re-epithelialization is critical to optimal wound healing, not only because
of the reformation of a cutaneous
barrier but also because of its role in wound contraction, we further
investigated the effects of Smad3 disruption on
this process. In vitro, the effects of TGF-(3 are paradoxical: integrin-
mediated keratinocyte migration is enhanced
whereas keratinocyte proliferation is inhibited (Zambruno, G. et al,
Transforming growth factor-beta 1 modulates beta
1 and beta 5 integrin receptors and induces the de novo expression of the
alpha v beta 6 heterodimer in normal human
keratinocytes: implications for wound healing. J. Cell Biol. 129, 853-865
(19951. Moreover, studies of the role of
exogenous TGF-(3 on re-epithelialization have generated conflicting results,
depending upon the dosage, kinetics of
administration, and model chosen (Mustoe, T.A., Pierce, G.F., Morishima, C. &
Deuel, T.F. Growth factor-induced
acceleration of tissue repair through direct and inductive activities in a
rabbit dermal ulcer model. J, Clin. Invest. 87,
694-703 (199111; (Hebda, P.A. Stimulatory effects of transforming growth
factor-beta and epidermal growth factor on
epidermal cell outgrowth from porcine skin explant cultures. J. Invest.
Dermatol. 91, 440-445 (1988)). Here, despite
the presence of similar wound widths in the wild-type and heterozygous mice at
day 3, complete re-epithelialization
had occurred in the heterozygous mice by this time point, indicating that TGF-
(3 signaling in vivo in keratinocytes is a
-33-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
Smad3-dependent process that ultimately leads to the inhibition of re-
epithelialization. To evaluate the specificity of
Smad3 in this signaling pathway, we also analyzed the wound-healing phenotype
in Smad2 heterozygotes. Wounds of
these mice heal to produce wound widths and areas that are similar to those
seen in Smad3 heterozygotes and wild-
type mice at day 3 (Fig.1 ); however, in contrast to the Smad3 heterozygotes,
wounds of Smad2 heterozygotes did not
re-epithelialize IFig. 1 b). These results indicate that Smad3 may have
effects on in vivo epithelial biology that are
different to those of Smad2. Although Smad2 and Smad3 occasionally appear to
function interchangeably when
overexpressed in vitro, the unique abilities Smad3 to bind DNA directly and to
interact with oncogenes such Evi-7 and
nuclear receptors such as the vitamin D3 receptor indicate that these two
SMADs may regulate distinct target genes
in vivo (Yanagisawa, K. et al, Induction of apoptosis by Smad3 and down-
regulation of Smad3 expression in response
to TGF-beta in human normal lung epithelial cells. 0ncogene 17, 1743-1747
(1998)); (Dennler, S., Huet, S. & Gauthier,
J.M. A short amino--acid sequence in MH1 domain is responsible for functional
differences between Smad2 and
Smad3. Oncogene 18, 1643-1648 (1999)); (Ulloa, L., Doody, J. & Massague, J.
Inhibition of transforming growth
factor-betaISMAD signaling by the interferon-gammaISTAT pathway. Nature 397,
710-713 (1999)); (Yanagisawa, J.
et al. Convergence of transforming growth factor-beta and vitamin D signaling
pathways on SMAD transcriptional
coactivators. Science 283, 1317-1321 (1999)); (Kurokawa, M. et al. The
oncoprotein Evi-1 represses TGF-beta
signaling by inhibiting Smad3. Nature 2, 92-96 (1998)). This idea is supported
by the striking differences in their
respective null phenotypes (Yang, X. et al. Targeted disruption of SMAD3
results in impaired mucosal immunity and
diminished T cell responsiveness to TGF-beta. EMBO J. 18, 1280-1291 (1999));
(Datto, M. B. et al. Targeted
disruption of Smad3 reveals an essential role in transforming growth factor
beta-mediated signal transduction Mo/.
Cellbiol. 19, 2495-2504 (1999)1; (Zhu, Y., Richardson, J.A., Parada, L.F., &
Graff, J.M. Smad3 mutant mice develop
metastatic colorectal cancer. Cell18, 703-714 (199811; (Weinstein, M., Yang,
X., Li, C., Xu, X., & Deng. C. Failure of
extraembryonic membrane formation and mesoderm induction in embryos lacking
the tumor suppressor Smad2. Proc.
Nat/. Acad Sci USA 95, 9378-9383 (1998)).
To identify the mechanisms underlying the in vivo effects of Smad3 on re-
epithelialization, we tested
whether keratinocyte functions crucial to wound repair, namely migration and
proliferation, were modified by Smad3
disruption. Although expression levels of TGF-(3 receptors in keratinocytes
were independent of the Smad3 genotype,
Smad3e'aiexa keratinocytes lacked the ability to upregulate TGF-(3 expression
in response to TGF-[31 (Fig. 4a). As
Smad3 is involved in the inhibition of cell growth, we reasoned that enhanced
re-epithelialization in the Smad3exeie»e
mice might be secondary to enhanced proliferative capacity (Datto, M. B. et
al. Targeted disruption of Smad3 reveals
an essential role in transforming growth factor beta-mediated signal
transduction Mol, Cell biol. 19, 2495-2504
(1999)). In culture, primary keratinocytes derived from the Smad3-null mice
showed a reduced sensitivity to growth
inhibition by TGF-(3 (Fig. 4b). These findings were paralleled by an increase
in basal keratinocyte proliferation (as
judged by incorporation of bromodeoxyuridine (BrdU)) at the wound edge in the
null cells compared with wild-type cells
(Fig. 4b).1-he results show that high levels of exogenous TGF-(3 can inhibit
the growth of the heterozygous and wild-
type keratinocytes equally. However, we interpret the intermediate result in
terms of re-epithelialization of cutaneous
-34-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
wounds in the heterozygous mice to result from the reduced level of endogenous
TGF-(3 produced (compared with wild-
type levels), as the inflammatory response is still blunted compared with the
wild-type response.
A further aspect of re-epithelialization involves cell migration across matrix
components in response to a
chemoattractant gradient. Smad3exaiexs keratinocytes exhibited reduced
adhesion to matrix and migration towards
TGF-(3 and keratinocyte growth factor (KGF), while maintaining a normal
response towards growth factors present in
conditioned media (Fig. 4c). An increasing number of cytokines and alternative
signaling pathways have been shown to
affect SMAD activity (Ulloa, L., Doody, J. & Massague, J. Inhibition of
transforming growth factor-betaISMAD
signaling by the interferon-gammaISTAT pathway. Nature 397, 710-713 (1999));
(Yanagisawa, J. et al. Convergence
of transforming growth factor-beta and vitamin D signaling pathways on SMAD
transcriptional coactivators. Science
283, 1317-1321 (1999)); (Kurokawa, M. et al. The oncoprotein Evi-1 represses
TGF-beta signaling by inhibiting
Smad3. Nature 2, 92-96 (1998)); (de Caestecker, M.P. et al. Smad2 transducer
common signals from receptor serine-
threonine and tyrosine kinases. Genes Dev. 1Z, 587-592 (1998)), so it is
possible that KGF may mediate some of its
effects on wild-type cells through interplay with the Smad3 signaling pathway.
Because integrins are pivotal in
mediating cell migration, we reasoned that Smad3 may be required for TGF-(3-
induced integrin expression by
keratinocytes. Exogenous TGF-(31 upregulated expression of (3~ integrins but
not of the a,5 subunit in the null cells;
this may represent an underlying mechanism for impaired migration across
fibronectin (Fig. 4d). This effect differs from
that of altered Smad3 signaling in the monocyte, indicating that the effects
of Smad3 disruption on a particular gene
target depend on the cellular context and cannot be generalized. We also
assessed the effect of Smad3 disruption on
cell-adhesion molecules specific to keratinocytes, namely E-cadherin and
syndecan-1. The expression levels of both
were equivalent in all phenotypes, both basally and following TGF-(3
treatment. Thus, in the context of wound healing,
one possible mechanism of enhanced re-epithelialization in the Smad3exsiexs
mice may involve increased keratinocyte
proliferation (compared with wild-type keratinocytes) in the presence of TGF-
(3, coupled with a migratory response
stimulated by growth factors other than TGF-(3 and KGF in a Smad3-independent
process. These data indicate the
importance of the early proliferative response in accelerating in vivo re-
epithelialization, which appears to be inhibited
by a Smad3-dependent pathway.
Smad3 Disruution Leads To Protection Against Radiation-Induced Fibrosis.
Male wildtype or Smad3 null littermates, 6 weeks of age, were exposed to
radiation on the right thigh region.
The left leg served as an internal control. In this initial experiment, mice
were either not radiated, or given 30 or 60 Gy
in a single dose. Mice were killed at 2 weeks and 5 weeks post-radiation.
Tatoo marks 1 cm apart were used to
assess contraction of the skin and a torsion test was used to measure
contractility of the leg. Sections of the skin
and muscle were fixed in neutral buffered formalin for histology.
Analysis of the histology of the skin at 2 weeks post-radiation demonstrated
that the skin of Smad3 null
mice is resistant to the damaging effects of radiation. Comparison of the non-
radiated skin of the left thigh of
wildtype mice and the skin of the right thigh that received 60 Gy radiation
showed a severe hyperplasia of the
-3 5-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
epidermis and hair follicles resulting from this high dose of radiation. In
contrast, there was only the mildest
hyperplasia in the skin of the radiated thigh of the Smad3 null mice, and the
hair follicles looked normal. The area of
compacted connective tissue (scar) had a greater area in the radiated wildtype
compared to the Smad3 null skin. The
inflammatory response was also stronger in the wildtype mice. These data
establish that Smad3 plays an essential
role in the response of epidermalldermal hair follicle cells to radiation
damage and that cells lacking Smad3 are
resistance to radiation-induced injury.
Pictures were taken of the radiated right thighs of littermate wildtype or
Smad3 null male mice 5 weeks
post-exposure to a single 60 Gy dose of radiation. The skin of the wildtype
mice was thickened, contracted (as
measured by the distance of the two tatoo marks) and lacking regrown hair over
the radiated area. In striking
contrast, the skin of the Smad3 null mice had retained normal flexibility,
pigment, and showed regrowth of hair over
the radiated area. These observations support the conclusion that loss of
Smad3 prevents the long-term effects of
high-dose radiation, such as fibrosis, scarring, and alopecia.
Histology was analyzed of the skin of the radiated right thighs of littermate
wildtype or Smad3 null male
mice 5 weeks post-exposure to a single 60 Gy dose of radiation. The two
wildtype and two Smad3 null mice examined
showed a variable response condition. Nevertheless, patterns could be
discerned. On average, the degree of epidermal
hyperplasia was significantly higher in the wildtype mice. Additionally, the
area of mild hyperplasia in the Smad3 null
mice was quite limited, whereas in the wildtype mice the area of epidermal
involvement was quite extensive and
uniform. These observations further support the conclusion that Smad3
disruption leads to protection against
radiation-induced fibrosis.
Example
Wound-healin4 experiments.
Smad3exeiexa mice were generated by targeted disruption of the Smad3 gene by
homologous recombination.
Targeted embryonic-stem-cell clones were injected into germline transmission.
Mice heterozygous for the targeted
disruption were intercrossed to produce homozygous offspring (Yang, X. et al.
Targeted disruption of SMAD3 results in
impaired mucosal immunity and diminished T cell responsiveness to TGF-beta.
EMBO J. 188, 1280-1291 (1999)). 48
4-6-week-old mice (Smad3 wild-type, heterozygotes and null mice) were
anaesthetized with methoxyfluorane, and the
dorsum was shaved and cleaned with alcohol. Four equidistant 1-cm full-
thickness incisional wounds were made
through the skin and panniculus carnosus muscle. For a subset of animals,
before wounding, the area to be incised
was injected subcutaneously with 50,1 of either vehicle (PBS + 4 mM HCI) or
TGF-j31 (1 ~.g), or was left
unmanipulated. Treatments were rotated to ensure no site bias. Wounds were
collected at days 1, 2, 3 and 5 post
wounding and were bisected for histology and immunostaining, or snap-frozen in
liquid nitrogen for RNA analysis. In
addition, ten healthy Smad2 heterozygote mice (aged 4-6 weeks) underwent 1-cm
incisional wounds as described, with
wound excision at day 3 or 5. For analysis of BrdU incorporation, 150 mg kg'
BrdU solution (Sigma) was injected
intraperitoneally 1 h before the mice were killed, and tissues were stained
with monoclonal mouse anti-BrdU antibody
(DAKO). Serum levels of TGF-j31 were measured using a Quantikine kit (R&D
systems).
-3 6-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
Histology, immunocytochemistry and image Analysis.
Histological sections were prepared from wound tissue fixed in 10% buffered
formal saline and embedded in
paraffin. 7-~,m sections were stained with haematoxylin and eosin, Masson's
trichrome or Giemsa, or were subjected
to immunohistochemistry with antibodies to TGF-[31, 2 and 3 (Santa Cruz) or
fibronectin, used at a dilution of 1:20 in
PBS. Image analysis and quantification of cell numbers per unit area, of wound
area (measured below the clot and
above the panniculus muscle) and of re-epithelialization was done using an
Optimas program as described (Ashcroft,
G.S, et al. Estrogen accelerates cutaneous wound healing associated with an
increase in TGF-beta 1 levels. Nature
Med. 3, 1209-1215 (199711.
Culture of bone-marrow monocytes and chemotaxis assay.
Bone marrow was collected from the femurs and tibias of 4-6-week-old male
mice. Mononuclear cells were
isolated using a two-component step gradient (Cardinal Associates Inc., Santa
Fe), and incubated for 4-7 days in
monocyte colony-stimulating factor (10ngmf') as described (Feldman, G. et al.,
STATSA-deficient mice demonstrate a
defect in granulocyte-macrophage colony-stimulating factor-induced
proliferation and gene expression. Blood 90,
1768-1776 (1997)). Chemotaxis of monocytes was stimulated in a 12-well
chemotaxis chamber (Corning Costar
Transwell Plate), in triplicate wells containing 400 ml FMLP (1 ~,M), control
media, or TGF-(3 (1 pgml~'). Monocytes
were resuspended in chemotaxis buffer (Hank's buffer with 0.5% BSA) at a final
concentration of 3 x 105 per 100,1;
1001 was added to the upper chamber, and the monocytes were incubated for 90
min at 37°C in a humidified
atmosphere (5% CO21. Cells that migrated across the membrane (pore size 3 Vim)
were fixed in 40 ~I chemotaxis
fixative (100mM EDTA and 10% formaldehyde in PBS) and counted in 500-~,I
volume using a Coulter counter. For
wound-healing experiments using monocytes, bone-marrow monocytes removed from
wild-type mice were resuspended
in PBS and 0.5x106 cells (or PBS vehicle alone) were injected subcutaneously
at the site to be incised. Immediately
after injection, 1-cm full-thickness incisions were made (as above) and the
wounds excised at day 3 post-wounding.
Keratinocyte adhesionlmigration and uroliferation assays.
Keratinocytes were isolated from the skin of newborn mice from crosses of
Smad3 heterozygote adults by
standard methods ( Dlugosz, A.A., Glick, A.B., Tennenbaum, T., Weinberg, W.C.
& Yuspa, S.H. Isolation and utilization
of epidermal keratinocytes for oncogene research. Methods Enzymol. 254, 3-20
(1995)). Cells were plated in EMEM
medium, 8°l° chelexed fetal bovine serum, 0.2mM CaCl2 with
antibiotics, and then switched to the same media with
0.05 mM CaCl2. For migration assays, cells were trypsinized, washed and
resuspended to 1x10° cells ml~' in serum-
free EMEM. 5x104 cells were added to the upper well of a chemotaxis chamber
(Neuro Probe Inc.); this upper well was
separated from the test medium (which was EMEM, conditioned medium from wild-
type keratinocytes, KGF or TGF-(31
at 1 ng ml~') in the lower chamber by a fibronectinlcollagen-I-coated
membrane. Cells that had migrated through the
membrane after 5 h at 37°C were stained using Diff-Quick and counted
from video images obtained with a Leitz
photomicroscope. Each value represents the average number of cells migrated
from triplicate wells. For proliferation
assays, cells were seeded at 80,000 cells per well in a 24-well tissue-culture
tray and allowed to proliferate for 3
-37-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
days. Porcine TGF-j31 (R&D Systems) was added at varying concentrations for
20h and the wells were pulsed with
1 ~,Ci ['H]thymidine for an extra 4h. Radioactivity incorporated into DNA was
determined by established methods
(Danielpour, D. et al. Immunodetection and quantitation of the two forms of
transforming growth factor-beta (TGF
beta 1 and TGF-beta 2) secreted by cells in culture. J. Cell PhysioL 138, 79-
86 (1989)). Each value represents the
average of triplicate wells.
Exuression of cell-adhesion molecules and TGF-(3 isoforms.
Wound tissue (microdissected to avoid contamination from unwounded adjacent
skin) and normal skin from
the dorsal area were homogenized and total RNA was extracted with trizol. In
addition, total RNA was extracted in a
similar fashion from monocytes and keratinocytes. Reverse transcription with
polymerase chain reaction was done
using the following primers (band intensities were normalized to those of the
keratinocytelmonocyte housekeeping
gene HPRT (hypoxanthine phosphoribosyl transferase); a, integrin, 5'-
CATTTCCGAGTCTGGGCCA (SEO ID N0:3) and
5'-TGGAGGCTTGAGCTGAGCTT (SEO ID N0:4); [3, integrin, 5'-TGTTCAGTGCAGAGCCTTCA
(SEO ID N0:5) and 5'-
CCTCATACTTCGGATTGACC (SE(1 ID N0:6); intercellular adhesion molecule (ICAM),
5'-
TTCAACCCGTGCCAAGCCCACGCT (SEO ID N0:7) and 5'-GCCAGCACCGTGAATGTGATCTCC (SEO ID
N0:8); E-
cadherin, 5'-TCAGCACCCACACACATACA (SEO ID N0:9) and 5'-GCATTTTCTCAGGAAGCAGG
(SEO ID N0:10);
syndecan-1, 5'-GATCCCAAAGCCACTGTGTT (SEQ ID N0:11) and 5'-ACACTGTGGAACCAGCCTTC
(SEO ID N0:12).
In addition, RNase-protection assays were done according to the manufacturer's
instructions (Pharmingen) using
multiprobe templates on 3wg total RNA, and were developed using
phosphorimaging. Band densities were normalized
to those of the keratinocyte monocyte housekeeping gene L32 for both the
cytokine and the receptor templates, using
an image-analysis program (image Quant, Molecular Dynamics). All data were
analyzed by Student's t-test or analysis
of variance.
Although the invention has been described with reference to embodiments and
examples, it should be
understood that various modifications can be made without departing from the
spirit of the invention. Accordingly, the
invention is limited only by the following claims. All references cited herein
are hereby expressly incorporated by
reference.
-38-
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
SEQUENCE LISTING
<110> Government of the United States of America, represented by the
Secretary,
Department of Health and Human Services
Anita B. Roberts
Gillian S. Ashcroft
Angelo Russo
James B. Mitchell
Chu~ia Deng
<120> Inhibition of Smad3 to Prevent Fibrosis
and Improve Wound Healing
<130> NIH193.OOlPCT
<160> 12
<170> FastSEQ for Windows Version 4.0
<210> 1
<211> 2303
<212> DNA
<213> homo Sapiens
<220>
<221> CDS
<222> (67)...(1344)
<400> 1
cccggcgtco cgtcgagccc agccccgccg ggggcgCtcc tcgccgcccg cacgccctcc 60
ccagcc atg tcg tcc atc ctg cct ttc act ccc ccg atc gtg aag cgc 108
Met Ser Ser Tle Leu Pro Phe Thr Pro Pro Ile Val Lys Arg
1 5 10
ctg ctg ggc tgg aag aag ggc gag cag aac ggg cag gag gag aaa tgg 156
Leu Leu Gly Trp Lys Lys Gly Glu Gln Asn Gly Gln Glu Glu Lys Trp
15 20 25 30
tgc gag aag gcg gtc aag agc ctg gtc aag aaa ctc aag aag acg ggg 204
Cys G1u Lys Ala Val Lys Ser Leu Val Lys Lys Leu Lys Lys Thr Gly
35 40 45
cag ctg gac gag ctg gag aag gcc atc acc acg cag aac gtc aac acc 252
Gln Leu Asp Glu Leu Glu Lys Ala Ile Thr Thr Gln Asn Val Asn Thr
50 55 60
aag tgc atc acc atc ccc agg tcc ctg gat ggc cgg ttg cag gtg tcc 300
Lys Cys Ile Thr Ile Pro Arg Ser Leu Asp Gly Arg Leu Gln Val Ser
65 70 75
cat cgg aag ggg ctc cct cat gtc atc tac tgc cgc ctg tgg cga tgg 348
His Arg Lys Gly Leu Pro His Val Ile Tyr Cys Arg Leu Trp Arg Trp
80 85 90
1
SUBSTITUTE SHEET (RULE 26)
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
ccagacctgcac agccaccac gagctgcgg gccatggag ctgtgt gag 396
ProAspLeuHis SerHisHis GluLeuArg AlaMetGlu LeuCys Glu
95 100 105 110
ttcgccttcaat atgaagaag gacgaggtc tgcgtgaat ccctac cac 444
PheAlaPheAsn MetLysLys AspGluVal CysValAsn ProTyr His
1I5 120 125
taccagagagta gagacacca gttctacct cctgtgttg gtgcca cgc 492
TyrGlnArgVal GluThrPro ValLeuPrp ProValLeu ValPro Arg
130 135 140
cacacagagatc ccggccgag ttcccccca ctggacgac tacagc cat 540
HisThrGluIle ProAlaGlu PheProPro LeuAspAsp TyrSer His
145 150 155
tccatccccgaa aacactaac ttccccgca ggcatcgag ccccag agc 588
SerIleProGlu AsnThrAsn PheProAla GlyIleGlu ProGln Ser
160 165 170
aatattcca gagacccca ccccctggc tacctgagt gaagatgga gaa 636
AsnIlePro GluThrPro ProProGly TyrLeuSer GluAspGly Glu
175 180 185 190
accagtgac caccagatg aaccacagc atggacgca ggttctcca aac 684
ThrSerAsp HisGlnMet AsnHisSer MetAspAla GlySerPro Asn
195 200 205
ctatccccg aatccgatg tccccagca cataataac ttggacctg cag 732
LeuSerPro AsnProMet SerProAla HisAsnAsn LeuAspLeu Gln
210 215 220
ccagttacc tactgcgag ccggccttc tggtgctcc atctcctac tac 780
ProValThr TyrCysGlu ProAlaPhe TrpCysSer IleSerTyr Tyr
225 230 235
gagctgaac cagcgcgtc ggggagaca ttccacgcc tcgcagcca tcc 828
GluLeuAsn GlnArgVal GlyGluThr PheHisAla SerGlnPro Ser
240 245 250
atgactgtg gatggcttc accgacccc tccaattcg gagcgcttc tgc 876
MetThrVal AspGlyPhe ThrAspPro SerAsnSer GluArgPhe Cys
255 260 265 270
ctagggctg ctctccaat gtcaacagg aatgcagca gtggagctg aca 924
LeuGlyLeu LeuSerAsn ValAsnArg AsnAlaAla ValGluLeu Thr
275 280 285
cggagacac atcggaaga ggcgtgcgg ctctactac atcggaggg gag 972
ArgArgHis IleGlyArg GlyValArg LeuTyrTyr IleGlyGly Glu
290 295 300
gtcttcgca gagtgcctc agtgacagc getattttt gtccagtct ccc 1020
ValPheAla GluCysLeu SerAspSer AlaIlePhe ValGlnSer Pro
305 310 315
aac tgt aac cag cgc tat ggc tgg cac ccg gcc acc gtc tgc aag atc 1068
2
SUBSTITUTE SHEET iRU't~°26)
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
Asn Cys Asn Gln Arg Tyr Gly Trp His Pro Ala Thr Val Cys Lys Ile
320 325 330
cca cca gga tgc aac ctg aag atc ttc aac aac cag gag ttc get gcc 1116
Pro Pro Gly Cys Asn Leu Lys Ile Phe Asn Asn Gln Glu Phe Ala Ala
335 340 345 350
ctc ctg gcc cag tcg gtc aac cag ggc ttt gag get gtc tac cag ttg 1164
Leu Leu Ala Gln Ser Val Asn Gln Gly Phe Glu Ala Val Tyr Gln Leu
355 360 365
acc cga atg tgc acc atc cgc atg agc ttc gtc aaa ggc tgg gga gcg 1212
Thr Arg Met Cys Thr Ile Arg Met Ser Phe Val Lys Gly Trp Gly Ala
370 375 380
gag tac agg aga cag act gtg acc agt acc ccc tgc tgg att gag ctg 1260
Glu Tyr Arg Arg Gln Thr Val Thr Ser Thr Pro Cys Trp Ile Glu Leu
385 390 395
cac ctg aat ggg cct ttg cag tgg ctt gac aag gtc ctc acc cag atg 1308
His Leu Asn Gly Pro Leu Gln Trp Leu Asp Lys Val Leu Thr Gln Met
400 405 410
ggc tcc cca agc atc cgc tgt tcc agt gtg tct tag agacatcaag 1354
Gly Ser Pro Ser Ile Arg Cys Ser Ser Val Ser
415 420 425
tatggtaggg gagggcaggc ttggggaaaa tggccataca ggaggtggag aaaattggaa 1414
ctctactcaa cccattgttg tcaaggaaga agaaatcttt ctccctcaac tgaaggggtg 1474
cacccacctg ttttctgaaa cacacgagca aacccagagg tggatgttat gaacagctgt 1534
gtctgccaaa cacatttacc ctttggcccc actttgaagg gcaagaaatg gcgtctgctc 1594
tggtggctta agtgagcaga acaggtagta ttacaccacc ggcaccctcc ccccagactc 1654
tttttttgag tgacagcttt ctgggatgtc acagtccaac cagaaacgcc cctctgtcta 1714
ggactgcagt gtggagttca ccttggaagg gcgttctagg taggaagagc ccgcacgatg 1774
cagacctcat gcccagctct ctgacgcttg tgacagtgcc tcttccagtg aacattccca 1834
gcccagcccc gccccgttgt gagctggata gacttgggat ggggagggag ggagttttgt 1894
ctgtctccct cccctctcag aacatactga ttgggaggtg cgtgttcagc agaacctgca 1954
cacaggacag cgggaaaaat cgatgagcgc cacctcttta aaaactcact tacgttgtcc 2014
tttttcactt tgaaaagttg gaaggactgc tgaggcccag tgcatatgca atgtatagtg 2074
tctattatca cattaatctc aaagagattc gaatgacggt aagtgttctc atgaagcagg 2134
aggcccttgt cgtgggatgg catttggtct caggcagcac cacactgggt gcgtctccag 2194
tcatctgtaa gagcttgctc cagattctga tgcatacggc tatattggtt tatgtagtca 2254
gttgcattca ttaaatcaac tttatcatat gctcaaaaaa aaaaaaaag 2303
<210> 2
<211> 425
<212> PRT
<213> homo sapiens
<400> 2
Met Ser Ser Ile Leu Pro Phe Thr Pro Pro Ile Val Lys Arg Leu Leu
1 5 ' 10 15
Gly Trp Lys Lys Gly Glu Gln Asn Gly Gln G1u Glu Lys Trp Cys G1u
20 25 30
Lys Ala Val Lys Ser Leu Val Lys Lys Leu Lys Lys Thr Gly Gln Leu
35 40 45
Asp Glu Leu Glu Lys Ala Ile Thr Thr Gln Asn Val Asn Thr Lys Cys
3
SUBSTITUTE SHEET (RULE 26~'
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
50 55 60
Ile Thr Ile Pro Arg Ser Leu Asp Gly Arg Leu Gln Val Ser His Arg
65 70 75 80
Lys Gly Leu Pro His Val Ile Tyr Cys Arg Leu Trp Arg Trp Pro Asp
85 90 95
Leu His Ser His His Glu Leu Arg Ala Met Glu Leu Cys Glu Phe Ala
100 105 110
Phe Asn Met Lys Lys Asp Glu Va1 Cys Val Asn Pro Tyr His Tyr Gln
115 120 125
Arg Val Glu Thr Pro Val Leu Pro Pro Val Leu Val Pro Arg His Thr
130 135 140
Glu Tle Pro Ala Glu Phe Pro Pro Leu Asp Asp Tyr Ser His Ser Ile
145 150 155 160
Pro Glu Asn Thr Asn Phe Pro Ala Gly Ile Glu Pro Gln Ser Asn Ile
165 7.70 175
Pro Glu Thr Pro Pro Pro Gly Tyr Leu Ser Glu Asp Gly Glu Thr Ser
180 185 190
Asp His Gln Met Asn His Ser Met Asp Ala Gly Ser Pro Asn Leu Ser
195 200 205
Pro Asn Pro Met Ser Pro Ala His Asn Asn Leu Asp Leu Gln Pro Val
210 215 220
Thr Tyr Cys Glu Pro Ala Phe Trp Cys Ser Ile Ser Tyr Tyr Glu Leu
225 230 235 240
Asn Gln Arg Val Gly Glu Thr Phe His Ala Ser Gln Pro Ser Met Thr
245 250 255
Val Asp Gly Phe Thr Asp Pro Ser Asn Ser Glu Arg Phe Cys Leu Gly
260 265 270
Leu Leu Ser Asn Val Asn Arg Asn A1a Ala Val Glu Leu Thr Arg Arg
275 280 285
His Tle Gly Arg Gly Val Arg Leu Tyr Tyr Tle Gly Gly Glu Val Phe
290 295 300
Ala Glu Cys Leu Ser Asp Ser Ala Ile Phe Va1 G1n Ser Pro Asn Cys
305 310 315 320
Asn Gln Arg Tyr Gly Trp His Pro A1a Thr Val Cys Lys Tle Pro Pro
325 330 335
Gly Cys Asn Leu Lys Ile Phe Asn Asn Gln Glu Phe Ala Ala Leu Leu
340 345 350
Ala Gln Ser Val Asn Gln Gly Phe Glu Ala Val Tyr Gln Leu Thr Arg
355 360 365
Met Cys Thr Ile Arg Met Ser Phe Val Lys Gly Trp Gly Ala Glu Tyr
370 375 380
Arg Arg Gln Thr Val Thr Ser Thr Pro Cys Trp Ile Glu Leu His Leu
385 390 395 400
Asn Gly Pro Leu Gln Trp Leu Asp Lys Val Leu Thr Gln Met Gly Ser
405 410 415
Pro Ser Ile Arg Cys Ser Ser Val Ser
420 425
<210> 3
<21l> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
4
SUBSTITUTE SHEET (RULE 26)
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
<400> 3
catttccgag tctgggcca 19
<210> 4
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
<400> 4
tggaggcttg agctgagctt ' 20
<210> 5
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
<400> 5
tgttcagtgc agagccttca 20
<210> 6
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
<400> 6
cctcatactt cggattgacc 20
<210> 7
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
<400> 7
ttcaacccgt gccaagccca cgct 24
<210> 8
<2l1> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
<400> 8
gccagcaccg tgaatgtgat ctcc 24
SUBSTITUTE SHEET (RULE 26)
CA 02410987 2002-11-19
WO 01/89556 PCT/US00/13725
<2l0> 9
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
<400> 9
tcagcaccca cacacataca 20
<210> l0
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
<400> 10
gcattttctc aggaagcagg 2p
<210> 11
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
<400> 11
gatcccaaag ccactgtgtt 20
<210> 12
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> primer
<400> 12
acactgtgga accagccttc 20
6
SUBSTITUTE SHEET (RULE 26)