Note: Descriptions are shown in the official language in which they were submitted.
CA 02412764 2007-06-28
1
STIMULATION OF CELLULAR REGENERATION
AND DIFFERENTIATION IN THE INNER EAR
Field of the Invention
The present invention relates to methods and compositions for stimulating the
formation of inner ear cells, including inner ear sensory. hair cells and
inner ear
support cells.
Background of the Invention
Sensorineuronal hearing loss (SNHL), also called "nerve deafness," is a
significant communication problem that affects tens of millions of people in
the U.S.
alone. Loss of the inner ear sensory hair cells that detect sound is thought
to be a
major cause of this deficit. The anatomy of the inner ear is well known to
those of
ordinary skill in the art (see, e.g., Gray's Anatomy, Revised American Edition
(1977),
pages 859-867). In
brief, the inner ear includes
three sensory portions: the cochlea, which senses sound; the semicircular
canals,
which sense angular acceleration; and the otolithic organs, which sense linear
acceleration. In each of these sensory portions, specialized sensory hair
cells are
arrayed upon one or more layers of inner ear supporting cells. Supporting
cells
underlie, at least partially surround, and physically support sensory hair
cells within
the inner ear. In operation, the sensory hair cells are physically deflected
in response
to sound or motion, and their deflection is transmitted to nerves which send
nerve
impulses to the brain for processing and interpretation.
In mammals, the inner ear is normally incapable of regenerating damaged or
dead inner ear sensory hair cells. Thus, hearing disorders that result from
the death
or deterioration of sensory hair cells typically result in a permanent hearing
impairment. Sensorineuronal hearing loss can be caused by a multitude of
events
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
2
including age-related loss (presbycusis), noise exposure, drug exposure (e.g.,
antibiotics and anti-cancer therapeutics), infections, genetic mutations
(syndromic
and non-syndromic) and autoimmune disease.
Currently, the treatment for acquired sensorineuronal hearing loss involves
the use of external hearing aids and cochlear implants. Both devices have
rather
limited therapeutic potential and more importantly, do not address the problem
of
restoring structure or function to the auditory sensory epithelium.
A more recent approach to the problem of regenerating sensory inner ear hair
cells is disclosed in published international application serial number
PCT/US99/24829 which discloses methods for stimulating the regeneration of
inner
ear cells (including sensory hair cells) that include the step of introducing
into inner
ear cells nucleic acid molecules that encode a transcription factor capable of
stimulating the regeneration of inner ear cells.
The present inventors have discovered that destruction of existing inner ear
sensory hair cells promotes the re-entry of normally quiescent inner ear
supporting
cells (that express reduced levels of one or more cell cycle inhibitor
proteins, or in
which cell cycle protein activity has been reduced) into the cell cycle to
yield
progeny cells that can be induced to form inner ear sensory hair cells, as
disclosed
herein. In some instances, destruction of existing inner ear sensory hair
cells is
sufficient to stimulate underlying and/or surrounding inner ear support cells
to
develop into sensory hair cells. In other instances, efficient regeneration of
sensory
hair cells from support cells requires destruction of existing inner ear
sensory hair
cells in combination with at least one other stimulus, as described herein.
Additionally, the present inventors have discovered that stimulating the
proliferation
of inner ear support cells (with or without stimulating the regeneration of
inner ear
sensory hair cells) improves the auditory function of the inner ear.
Summary of the Invention
The present invention provides methods for stimulating the formation of inner
ear cells, including inner ear sensory hair cells and inner ear support cells.
The
methods of the present invention rely on the unexpected observation that
damaging
and/or killing inner ear cells stimulates the formation of new, inner ear
cells.
In one aspect, the present invention provides methods for stimulating the
formation of inner ear sensory hair cells from inner ear support cells. The
methods
of this aspect of the present invention include the step (a) of damaging one
or more
inner ear sensory hair cells under conditions that promote the formation of
one or
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
3
more new sensory hair cells from one or more support cells that are in contact
with
the damaged sensory hair cell(s). Preferably a plurality of inner ear sensory
hair cells
are formed from a plurality of inner ear support cells. The methods of this
aspect of
the invention optionally include the step (b) of further stimulating the
formation of
one or more inner ear sensory hair cells from inner ear support cells that are
in
contact with the damaged inner ear sensory hair cell. Step (b) can occur
before,
during, after or overlapping with step (a). In one embodiment, the step of
stimulating
the formation of one or more inner ear sensory hair cells from one or more
inner ear
support cells that are in contact with the damaged inner ear sensory hair cell
includes
the steps of stimulating the inner ear support cells to enter the cell cycle,
then
stimulating at least some of the progeny of the inner ear support cells to
differentiate
to form inner ear sensory hair cells.
Inner ear sensory hair cells can be damaged, for example, by contact with an
amount of an ototoxic agent, such as an antibiotic, preferably an
aminoglycoside
antibiotic, that is effective to damage inner ear sensory hair cells. The
ototoxic agent
can be introduced into the inner ear by any art-recognized means, for example
by
injection (such as with a needle and syringe), or through a cannula. In one
embodiment of this aspect of the invention, inner ear sensory hair cells are
sufficiently damaged to cause their death.
In some embodiments of the present invention, damage inflicted on an inner
ear sensory hair cell stimulates the formation of one or more new inner ear
sensory
hair cell from an inner ear support cell that is in contact with the damaged
inner ear
sensory hair cell. In other embodiments, however, damage inflicted on an inner
ear
sensory hair cell is insufficient, by itself, to efficiently stimulate the
formation of one
or more new inner ear sensory hair cells from an inner ear support cell that
is in
contact with the damaged inner ear sensory hair cell. Thus, in one embodiment
of
the methods of this aspect of the present invention, the formation of inner
ear sensory
hair cells from inner ear support cells is stimulated by damaging inner ear
sensory
hair cells and expressing within inner ear support cells (before, during
and/or after
the step of damaging sensory hair cells) a transcription factor capable of
stimulating
inner ear sensory hair cells to form from inner ear support cells. For
example, in one
embodiment of the present invention, a nucleic acid molecule encoding a
transcription factor capable of stimulating inner ear sensory hair cells to
form from
inner ear support cells is introduced into inner ear support cells under
conditions that
enable expression of the transcription factor. Representative examples of
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
4
transcription factors capable of stimulating the formation of inner ear
sensory hair
cells from inner ear support cells include POU4F1, POU4F2, POU4F3, Brn3a,
Brn3b
and Brn3c.
In another embodiment of the methods of this aspect of the present invention,
the formation of inner ear sensory hair cells from inner ear support cells is
stimulated
by damaging inner ear sensory hair cells and inhibiting (before, during and/or
after
the step of damaging the sensory hair cells) the expression of one or more
cell cycle
inhibitors active in inner ear support cells. Inhibitors of cell cycle
inhibitors can be
substances, such as proteins, that act on the cell cycle inhibitor directly or
indirectly
within the cell. By way of representative example, cell cycle inhibitors
active in
inner ear support cells include cyclin-dependent kinase inhibitors, such as
cyclin-
dependent kinase inhibitors of the so-called CIP/KIP family including p21ciP1,
prxipt and p57xip2. For example, the expression of a cell cycle inhibitor
active in
inner ear support cells can be inhibited by introducing into inner ear support
cells an
expression vector that expresses a nucleic acid molecule that hybridizes under
stringent conditions (such as stringency greater than 2xS SC at 55 C) to a
nucleic acid
molecule (such as an mRNA molecule) encoding a cell cycle inhibitor active in
inner
ear support cells.
In addition, various recombinant growth factors such as TGF-alpha, insulin
and IGF-1 can be used to stimulate the formation of inner ear sensory hair
cells from
inner ear support cells. A representative, effective concentration range for
recombinant growth factors utilized in vitro in the practice of the present
invention is
1-1000 ng/ml. More specifically, TGF-alpha is preferably used at an effective
concentration of from 1-100 ng/ml; insulin is preferably used at an effective
concentration of from 100-1000 ng/ml; and IGF-1 is preferably used at an
effective
concentration of from 10-1000 ng/ml. For in vivo applications, a sufficient
amount
of recombinant growth factor would be administered to produce the foregoing
concentrations in vivo.
In preferred embodiments of this aspect of the invention, the formation of
inner ear sensory hair cells from inner ear support cells results in
improvement in the
auditory function of the treated inner ear. Thus, in one aspect, the invention
provides
methods for improving auditory function in an inner ear comprising the steps
of: (a)
damaging a first inner ear sensory hair cell under conditions that promote the
formation of one or more new inner ear sensory hair cells from a support cell
that is
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
in contact with the damaged, first inner ear sensory hair cell; and (b)
measuring an
improvement in auditory function in the inner ear treated in accordance with
step (a).
In another aspect, the present invention provides methods for stimulating the
formation of inner ear support cells. The methods of this aspect of the
invention
5 include
the steps of damaging inner ear support cells under conditions that promote
the formation of new inner ear support cells (for example by cell division of
inner ear
support cells that are in contact with damaged inner ear support cells). In
this aspect
of the invention, the inner ear support cell is damaged, and the formation of
new
inner ear support cells is stimulated, using the same techniques described
herein for
the methods of the present invention that stimulate the formation of inner ear
sensory
hair cells from inner ear support cells. Thus, for example, inner ear support
cells can
be damaged by contact with an amount of an ototoxic agent, such as an
aminoglycoside antibiotic, that is effective to damage inner ear support
cells. Again
by way of example, new inner ear support cell formation can be further
stimulated by
damaging inner ear support cells and expressing (before, during and/or after
damaging inner ear support cells) within inner ear support cells a
transcription factor
(such as POU4F1, POU4F2, POU4F3, Brn3a, Brn3b and Bm3c) capable of
stimulating inner ear support cells to divide and form new inner ear support
cells. In
preferred embodiments of this aspect of the invention, the proliferation of
inner ear
support cells results in improvement in the auditory function of the treated
inner ear.
The methods of the present invention are useful for stimulating the formation
of inner ear cells, such as sensory hair cells and support cells. Further, the
methods
of the present invention are useful to ameliorate the symptoms of a hearing
disorder
in a mammal, such as a human, that is caused by the death or damage of inner
ear
cells. Additionally, the methods of the present invention can be used to
identify
genes and/or proteins that are capable of stimulating the formation of inner
ear
support cells and/or the formation of inner ear sensory hair cells from inner
ear
support cells.
Brief Description of the Drawings
The foregoing aspects and many of the attendant advantages of this invention
will become more readily appreciated as the same become better understood by
reference to the following detailed description, when taken in conjunction
with the
accompanying drawings, wherein:
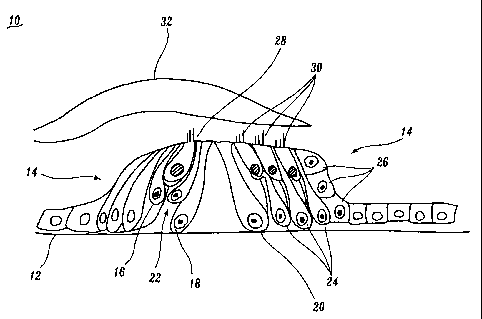
FIGURE 1 shows a cross section of the Organ of Corti.
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
6
FIGURE 2 shows the number of BrdU-labeled, guinea pig JH4 cells
following serum deprivation for 24 hours and a 13rdU pulse for the last 4
hours of the
24 hour period. Cells were counted under fluorescence microscopy. The
combination of lipids and p27kiP1 AS reversed growth arrest to 40% of that
seen with
10% FBS stimulation (p<.0001). (+)
FBS; (-) no FBS; (AS) antisense
oligonucleotide; (lipid) lipofection.
FIGURE 3 shows the ABR threshold of the right ears of mice two weeks after
the inner ears of the mice had been treated with amikacin sulfate.
Abbreviations are:
ABR, auditory brainstem response; dB, decibels; SPL, sound pressure level; Wt,
wild
type; Het, p27 heterozygote; Ko, p27 knock-out; kHZ, kilo Hertz.
FIGURE 4 shows the ABR threshold of the left ears of mice two weeks after
the inner ears of the mice had been treated with amikacin sulfate.
Abbreviations are
the same as those set forth in the description of FIGURE 3.
FIGURE 5 shows the ABR threshold of the right ears of mice four weeks
after the inner ears of the mice had been treated with amikacin sulfate.
Abbreviations
are the same as for FIGURE 3.
FIGURE 6 shows the ABR threshold of the left ears of mice four weeks after
the inner ears of the mice had been treated with amikacin sulfate.
Abbreviations are
the same as for FIGURE 3.
Detailed Description of the Preferred Embodiment
As used herein, the abbreviation "SSC" refers to a buffer used in nucleic acid
hybridization solutions. One liter of the 20X (twenty times concentrate) stock
SSC
buffer solution (pH 7.0) contains 175.3 g sodium chloride and 88.2 g sodium
citrate.
As used herein, the phrase "damaging one or more inner ear sensory hair
cells", or "damaging a first inner ear sensory hair cell", or grammatical
equivalents
thereof, means causing a deleterious change in the structure, biochemistry
and/or
physiology of the damaged, sensory hair cell (including killing the damaged
cell)
compared to an inner ear sensory hair cell that is cultured under
substantially the
same conditions as the damaged cell, but which is not damaged.
As used herein, the phrase "improving auditory function" or "improvement in
auditory function", or grammatical equivalents thereof, means improving, by at
least
10%, the sensitivity to sound of an inner ear by treating the inner ear in
accordance
with the methods of the present invention, or effecting any measurable
improvement
in the sensitivity to sound of an inner ear that is completely unresponsive to
sound
prior to treatment in accordance with the present invention. The sensitivity
to sound
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
7
of the treated inner ear is measured by any art-recognized means (such as the
auditory brainstem response) and compared to the sensitivity to sound of a
control
inner ear that is= not treated in accordance with the present invention and
which is
cultured under substantially the same conditions as the treated inner ear.
As applied to nucleic acid sequence comparisons or amino acid sequence
comparisons herein, the term "sequence homology" (also referred to as
"sequence
identity") is defined as the percentage of amino acid residues or nucleic acid
residues
in a subject amino acid sequence or nucleic acid sequence that are identical
with part
or all of a candidate amino acid sequence or nucleic acid sequence, after
aligning the
sequences and introducing gaps, if necessary, to achieve the maximum percent
homology (identity), and not considering any conservative substitutions as
part of the .
sequence homology. Neither N- or C- terminal extensions nor insertions shall
be
construed as reducing homology., No weight is given to the number or length of
gaps
introduced, if necessary, to achieve the maximum percent homology (identity).
In one aspect, the present invention provides methods for stimulating the
formation of inner ear sensory hair cells from inner ear support cells. The
methods
of this aspect of the present invention include the step (a) of damaging one
or more
inner ear sensory hair cells under conditions that promote the formation of
one or
more new sensory hair cells from one or more support cells that are in contact
with
the damaged sensory hair cell(s). Preferably a plurality of inner ear sensory
hair cells
are formed from a plurality of inner ear support cells. The methods of this
aspect of
the invention optionally include the step (b) of further stimulating the
formation of
one or more inner ear sensory hair cells from inner ear support cells that are
in
contact with the damaged inner ear sensory hair cell. Step (b) can occur
before,
during, after or overlapping with step (a). The methods of this aspect of the
present
invention can be utilized in vivo and in vitro.
The anatomy of the inner ear is well known to those of ordinary skill in the
art (see, e.g., Gray's Anatomy, Revised American Edition (1977), pages 859-
867,
incorporated herein by reference). In particular, the cochlea includes the
Organ of
Corti which is 'primarily responsible for sensing sound. As shown in FIGURE 1,
the
Organ of Corti 10 includes a basilar membrane 12 upon which are located a
variety
of supporting cells 14, including border cells 16, inner pillar cells 18,
outer pillar
cells 20, inner phalangeal cells 22, Dieter's cells 24 and Hensen's cells 26.
Supporting cells 14 support inner hair cells 28 and outer hair cells 30.
Tectorial
membrane 32 is disposed above inner hair cells 28 and outer hair cells 30. The
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
8
present invention is adapted, in one aspect, to stimulate regeneration of
sensory hair
cells 28 and 30 from underlying supporting cells 14. In another aspect, the
present
invention is adapted to stimulate the formation of supporting cells 14.
The present inventors have observed that destruction of existing inner ear
sensory hair cells promotes the re-entry of normally quiescent inner ear
supporting
cells into the cell cycle to yield progeny cells that can be induced to form
inner ear
sensory hair cells as disclosed herein. In some instances, destruction of
existing
inner ear sensory hair cells is sufficient to stimulate underlying and/or
surrounding
inner ear support cells to develop into sensory hair cells. In other
instances, efficient
regeneration of sensory hair cells from support cells requires destruction of
existing
inner ear sensory hair cells in combination with another stimulus, as
described
herein.
In the practice of one aspect of the present invention, inner ear sensory hair
cells are damaged, for example by contact with an amount of an ototoxic agent
that is
effective to damage inner ear sensory hair cells. Representative examples of
ototoxic
agents useful for damaging inner ear sensory hair cells include aminoglycoside
antibiotics (such as, neomycin, gentamycin, streptomycin, kanamycin, amikacin
and
tobramycin). In the practice of the present invention, the foregoing
aminoglycoside
antibiotics are typically used in vitro at an effective concentration in the
range of
from about 0.01 mM-10mM, and in vivo at an effective concentration in the
range of
from about 100 to about 1,000 milligrams per kilogram body weight per day
(mg/kg/d). Additional, representative examples of chemical agents useful for
damaging inner ear sensory hair cells include the following anti-cancer
agents:
cisplatin, carboplatin and methotrexate which are typically used in vitro at
an
effective concentration in the range of from about 0.01-0.1 mM, and in vivo at
an
effective concentration in the range of from about 5 to about 10 mg/kg/d.
Other
useful chemical agents include poly-L-lysine at an effective concentration in
the
range of from about 0.1-1.0 mM in vitro, and magnesium chloride at an
effective
concentration in vitro in the range of from about 5-100 mM.
The ototoxic agent, or agents, can be introduced into the inner ear by any art-
recognized means, for example by injection using a needle and syringe, or by
cochleostomy. Cochleostomy involves puncturing the cochlea and inserting a
catheter through which a chemical agent can be introduced into the cochlea. A
cochleostomy method is disclosed, for example, in Lalwani, A.K. et al.,
Hearing
CA 02412764 2007-06-28
9
Research 114: 139-147 (1997).
In one embodiment of the methods of the present invention, the formation of
inner ear .sensory hair cells from inner ear support cells is stimulated by
damaging
inner ear sensory hair cells and expressing (before, during, and/or after
damaging the
inner ear sensory hair cells) within at least some of. the inner ear support
cells a
transcription factor capable of stimulating the formation of an inner ear
sensory hair
cell from an inner ear support cell. For example, in one embodiment, a nucleic
acid
molecule encoding a transcription factor capable of stimulating the formation
of an
inner ear sensory hair cell is introduced into inner ear support cells under
conditions
that enable expression of the transcription factor.
Transcription factors useful in this aspect of the present invention have the
ability to stimulate regeneration of inner ear sensory hair cells from inner
ear
supporting cells when utilized in the practice of the methods of the present
invention.
Some transcription factors useful in this aspect of the present invention are
required
for the normal development, and/or for the normal functioning, of inner ear
sensory
hair cells.
Representative examples of transcription factors useful in this aspect of the
present invention 'include POU4F1 (Collura, R.G. et al., Nucleic Acids
Research
20(18): 4919-4925 (1992)), POU4F2 (Xiang et al., Neuron 11: 689-701 (1993)),
POU4F3 (Vahava, 0., Science 279(5358): 1950-1954 (1998), Bm3a (also known as
Brn3.0), Bm3b (also known as Brn3.2) and Brn3c (also known as Bm3.1) as
disclosed in Garr= et al., Proc. Nat'l. Acad. Sci. (U.S.A.) 90(22): 10841-
10845
(1993), Xiang, M. et al., Proc. Nat'l. Acad. Sci. (U.S.A.) 93(21): 11950-11955
(1996), Xiang, M. et al., J. Neurosci. 15(7Part 1): 4762-4785 (1995), Erlunan,
L.
et al., Nature 381(6583): 603-606 (1996), Xiang, M. et al., Proc. Nat'l. Acad.
Sci.
(U.S.A.) 94(17): 9445-9450 (1997).
Some transcription factors useful in this aspect of the present
invention possess at least one homeodomain and/or at least one POU-specific
domain, and have a molecular weight in the range of from about 33 kDa to about
37
kDa.
As used herein, the term "homeodomain" means an amino acid sequence that
is at least 50% homologous (such as at least 75% homologous, or at least 90%
homologous) to the homeodomain amino acid sequence set forth in SEQ ID NO: 1.
CA 02412764 2007-06-28
As used herein, the term "POU-specific domain" means an amino acid
sequence that is at least 50% homologous (such as at least 75% homologous, or
at
least 90% homologous) to the POU-specific domain amino acid sequence set forth
in
SEQ ID NO:2.
5 An
example of an algorithm that can be used to determine the percentage
homology between two protein sequences, or between two nucleic acid sequences,
is
the algorithm of Karlin and Altschul (Proc. Natl. Acad. Sci. USA 87:2264-2268
(1990)), modified as in Karlin and Altschul (Proc. NatL Acad. Sci. USA
90:5873-5877 (1993)). Such an algorithm is incorporated into the NBLEST and
10 XBLEST programs of Altschul et al. (.I. MoL Biol. 215:403-410 (1990)).
Presently more preferred inner ear cell transcription factors useful in the
practice of the present invention are POU4F3 transcription factor homologues
(hereinafter referred to as POU4F3 homologues). POU4F3 homologues useful in
the
practice of the present invention are capable of stimulating the regeneration
of inner
ear sensory hair cells from supporting cells and are at least 25% homologous
(such as
at least 50% homologous or at least 75% homologous, or at least 90%
homologous)
to the POU4F3 transcription factor having the amino acid sequence set forth in
SEQ
ID NO:4 and which is encoded by the nucleic acid molecule of SEQ ID NO:3. As
used herein, the term "POU4F3 homologues" includes the POU4F3 protein having
the amino acid sequence set forth in SEQ DD NO:4, which is the presently most
preferred inner ear cell transcription factor useful in the practice of the
present
invention. Representative examples of other POU4F3 homologues useful in the
practice of the present invention are set forth in Xiang, M. et al., .1
Neuroscience, 15
(7): 4762-4785 (1995).
Additional nucleic acid molecules encoding transcription factors useful in the
practice of the present invention can be isolated by using a variety of
cloning
techniques known to those of ordinary skill in the art. For -example, cloned
POU4F3
homologues cDNAs pr genes, or fragments thereof, can be used as hybridization
probes utilizing, for example, the technique of hybridizing radiolabeled
nucleic acid
probes to nucleic acids immobilized on nitrocellulose filters or nylon
membranes as
set forth at pages 9.52 to 9.55 of Molecular Cloning, A Laboratory Manual (2nd
edition), J. Sambrook,. E.F. Fritsch and T. Maniatis eds.
Presently preferred hybridization probes for
identifying additional nucleic .acid molecules encoding POU4F3 homologues are
fragments, of at least 15 nucleotides in length, of the cDNA 'molecule (or its
CA 02412764 2007-06-28
11
complementary sequence) having the nucleic acid sequence set forth in SEQ ID
NO:3, although the complete cDNA molecule having the nucleic acid sequence set
forth in SEQ ID NO:3 is also useful as a hybridization probe for identifying
additional nucleic acid molecules encoding POU4F3 homologue. A presently most
preferred hybridization probe for identifying additional nucleic acid
molecules
encoding POU4F3 homologues is the oligonucleotide having the nucleic acid
sequence 5'-TAG AAG TGC AGG GCA CGC TGC TCA TGG TAT G-3' (SEQ ID
NO:5).
Exemplary high stringency hybridization and wash conditions useful for
identifying (by Southern blotting) additional nucleic acid molecules encoding
POU4F3 homologues are: hybridization at 68 C in 0.25 M Na2HPO4 buffer (pH 7.2)
containing 1 mM Na2EDTA, 20% sodium dodecyl sulfate; washing (three washes of.
twenty minutes each at 65 C) is conducted in 20 mM Na2HPO4 buffer (pH 7.2)
containing 1 mM Na2EDTA, 1% (w/v) sodium dodecyl sulfate.
Exemplary moderate stringency hybridization and wash conditions useful for
identifying (by Southern blotting) additional nucleic acid molecules encoding
P0U4F3 homologues are: hybridization at 45 C in 0.25 M Na2HPO4 buffer (pH 7.2)
containing 1 mM Na2EDTA, 20% sodium dodecyl sulfate; washing is conducted in
5X SSC, containing 0.1% (w/v) sodium dodecyl sulfate, at 55 C to 65 C.
Again, by way of example, nucleic acid molecules encoding transcription
factors useful in the present invention can be isolated by the polymerase
chain
reaction (PCR) described in The Polyrnerase Chain Reaction (K.B. Mullis, F.
Ferre,
R.A. Gibbs, eds), Birkhauser Boston (1994).
Thus,
for example, first strand DNA synthesis can be primed using an oligo(dT)
primer,
and second strand cDNA synthesis can be primed using an oligonucleotide primer
that corresponds to a portion of the 5'-untranslated region of a cDNA molecule
that is
homologous to the target DNA molecule. Subsequent rounds of PCR can be primed
using the second strand cDNA synthesis primer and a primer that corresponds to
a
portion of the 3'-untranslated region of a cDNA molecule that is homologous to
the
target DNA molecule.
By way of non-limiting example, representative PCR reaction conditions for
amplifying nucleic acid molecules encoding transcription factors useful in the
present
invention are as follows. The following reagents are mixed in a tube (on ice)
to form
the PCR reaction mixture: DNA template (e.g., up to 1 lig genomic DNA, or up
to
0.1 lig cDNA), 0.1-0.3 mM dNiTes, 1014.1 10 X PCR buffer (10 X. PCR buffer
CA 02412764 2007-06-28
12
contains 500 mM KC1, 15mM MgC12, 100 rnM Tris-HC1, pH 8.3), 50 pmol of each
PCR primer (PCR primers should preferably be greater than 20 bp in length and
have
a degeneracy of 102 to 103), 2.5 units of Taq DNA polymerase (Perkin Elmer,
Norwalk, CT) and deionized water to a final volume of 50 f.d. The tube
containing
the reaction mixture is placed in a thermocycler and a thennocycler program is
run as
follows. Denaturation at 94 C for 2 minutes, then 30 cycles of: 94 C for 30
seconds,
47 C to 55 C for 30 seconds, and 72 C for 30 seconds to two and a half
minutes.
Preferably, PCR primers will be designed against conserved amino acid
sequence motifs found in some or all of the known target protein sequences.
Examples of conserved amino acid sequence motifs against which PCR primers can
be designed for cloning additional POU4F3 homologues are the POU-specific
domain having the amino acid sequence set forth in SEQ
NO:2, and the
homeodomain having the amino acid sequence set forth in SEQ ID NO:l.
Further, additional nucleic acid molecules encoding transcription factors
useful in the practice of the present invention can also be isolated, for
example, by
utilizing antibodies that = recognize transcription factor proteins. Methods
for
preparing monoclonal and polyclOnal antibodies are well known to those of
ordinary
skill in the art and are set forth, for example, in chapters five and six of
Antibodies A
Laboratory Manual, E. Harlow and D. Lane, Cold Spring Harbor Laboratory
(1988).
By way of non-
limiting example, antibodies were successfully raised against a fusion protein
constructed from the C-terminal end of Bm3 as described in Xiang, M. et al.,
J.
Neuroscience 15(7): 4762-4785 (1995) and Xiang, M. et al., P.N.A.S. (U.S.A.)
94:
9445-9450 (1997).
Nucleic acid molecules that encode transcription factors useful in the
practice
of the present invention can be isolated, for example, by screening expression
libraries. By way of non-limiting example, a cDNA expression library can be
screened using anti-POU4F3 homologue antibodies in order to identify one or
more
clones that encode a POU4F3 homologue protein. DNA expression library
technology is well known to those of ordinary skill in the art. Screening cDNA
expression libraries is fully discussed in Chapter 12 of Sambrook, J.,
Fritsch, E. F.
and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd ed., Cold
Spring Harbor Laboratory, Cold Spring Harbor, NY.
CA 02412764 2007-06-28
13
By way of representative example, antigen useful for raising antibodies for
Screening expression libraries can be prepared in the following manner. A full-
length
cDNA molecule encoding a transcription factor, such as a POU4F3 homologue, (or
a
cDNA molecule that is not full-length, but which includes all of the coding
region)
can be cloned into a plasmid vector, such as a Bluescript plasmid (available
from
Stratagene, Inc., La Jolla, California). The recombinant vector is then
introduced
into an E. colt stain (such as E. coil XL1-Blue, also available from
Stratagene, Inc.)
and the protein encoded by the cDNA is expressed in E. coil and then purified.
For
example, E. coli XL1-Blue harboring a Bluescript vector including a cDNA
molecule
of interest can be grown overnight at 37 C in LB medium containing 100 p.g
ampicillin/ml. A 50 1.1.1 aliquot of the overnight culture can be used to
inoculate 5 ml
of fresh LB medium containing ampicillin, and the culture grown at 37 C with
vigorous agitation to A600 = 0.5 before induction with 1 inM IPTG. After an
additional two hours of growth, the suspension is centrifuged (1000 x g, 15
min,
4 C), the media removed, and the pelleted cells resuspended in 1 ml of cold
buffer
that preferably contains 1 mM EDTA and one or more proteinase inhibitors. The
cells can be disrupted by sonication with a microprobe. The chilled sonicate
is
cleared by centrifugation and the expressed, recombinant protein purified from
the
supernatant by art-recognized protein purification techniques, such as those
disclosed
in Methods in Enzymology, Vol. 182, Guide to Protein Purification, Murray P.
Deutscher, ed (1990).
Methods for preparing monoclonal and polyclonal antibodies are well known
to those of ordinary skill in the art and are set forth, for example, in
chapters five and
six of Antibodies A Laboratory Manual, E. Harlow and D. Lane, Cold Spring
Harbor
Laboratory (1988).
In one representative example, polyclonal antibodies specific for a purified
protein
can be raised in a New Zealand rabbit implanted with a whiffle ball. One tg of
protein is injected at intervals directly into the whiffle ball granuloma. A
representative injection regime is injections (each of 1 p.g protein) at day
1, day 14
and day 35. Granuloma fluid is withdrawn one week prior to the first injection
(preimmune serum), and forty days after the final injection (postinu-nune
serum).
Sequence variants, produced by deletions, substitutions, mutations and/or
insertions, of the transcription factors useful in the practice of the present
invention
can also be used in the methods of the present invention. The amino acid
sequence
variants of the transcription factors useful in the practice of the present
invention may
CA 02412764 2007-06-28
14
be constructed by mutating the DNA sequences that encode the wild-type
transcription factor proteins, such as by using techniques commonly referred
to as
site-directed mutagenesis. Nucleic acid molecules encoding the transcription
factors
useful in the practice of the present invention can be mutated by a variety of
PCR
techniques well known to one of ordinary skill in the art. (See, for example,
the
following publications:
"PCR Strategies", M.A. Innis, D.H. Gelfand and J.J. Sninslcy, eds., 1995,
Academic Press, San Diego, CA (Chapter 14); "PCR Protocols: A Guide to Methods
and Applications", M.A. Innis, D.H. Gelfand, J.J. Sninsky and T.J. White,
eds.,
Academic Press, NY (1990)).
By way of non-limiting example, the two primer system utilized in the
Transformer Site-Directed Mutagenesis kit from Clontech, may be employed for
introducing site-directed mutants into nucleic acid molecules encoding
transcription
factors useful in the practice of the present invention. Following
denaturation of the
target plasmid in this system, two primers are simultaneously annealed to the
plasmid; one of these primers contains the desired site-directed mutation, the
other
contains a mutation at another point in the plasmid resulting in elimination
of a
restriction site. Second strand synthesis is then carried out, tightly linking
these two
mutations, and the resulting plasmids are transformed into a mutS strain of E.
colt.
Plasmid DNA is isolated from the transformed bacteria, restricted with the
relevant
restriction enzyme (thereby linearizing the unmutated plasmids), and then
retransformed into E. colt. This system allows for generation of mutations
directly in
an expression plasmid, without the necessity of subcloning or generation of
single-
stranded phigemids. The tight linkage of the two mutations and the subsequent
linearization of unmutated plasmids results in high mutation efficiency and
allows
minimal screening. Following synthesis of the initial restriction site primer,
this
method requires the use of only one new primer type per mutation site. Rather
than
prepare each positional mutant separately, a set of "designed degenerate"
oligonucleotide primers can be synthesized in order to introduce all of the
desired
mutations at a given site simultaneously. Transformants can be screened by.
sequencing the plasmid DNA through the mutagenized region to identify and sort
mutant clones. Each mutant DNA can then be fully sequenced or restricted and
analyzed by electrophoresis on Mutation Detection Enhancement gel (J.T. Baker)
to
confirm that no other alterations in the sequence have occurred (by _band
shift
comparison to the unmutagenized control).
CA 02412764 2007-06-28
Again, by way of non-limiting example, the two primer system utilized in the
QuikChangeTM Site-Directed Mutagenesis kit froin Stratagene (La Jolla,
California),
may be employed for introducing site-directed mutants into nucleic acid
molecules
encoding transcription factors useful in the practice of the present
invention. Double-
5 stranded plasmid DNA, containing the insert bearing the target mutation
site, is
denatured and mixed with two oligonucleotides complementary to each of the
strands
of the plasmid DNA at the target mutation site. The annealed oligonucleotide
primers are extended using Pfu DNA polymerase, thereby generating a mutated
plasmid containing staggered nicks. After temperature cycling, the unmutated,
10 parental DNA template is digested with restriction enzyme Dpni which
cleaves
methylated or hemimethylated DNA, but which does not cleave unmethylated DNA.
The parental, template DNA is almost always methylated or hemimethylated since
most strains of E. coli, from which the template DNA is obtained, contain the
required methylase activity. The remaining, annealed vector DNA incorporating
the
15 desired mutation(s) is transformed into E. coli.
In the design of a particular site directed mutagenesis experiment, it is
generally desirable to first make a non-conservative substitution (e.g., Ala
for Cys,
His or Glu) and determine if activity is greatly impaired as a consequence. If
the
residue is by this means demonstrated to be important by activity impairment,
or
knockout, then conservative substitutions can be made, such as Asp for Glu to
alter
side chain length, Ser for Cys, or Arg for His. For hydrophobic segments, it
is
largely size that is usefully altered, although aromatics can also be
substituted for
alkyl side chains.
Other site directed mutagenesis techniques may also be employed with
nucleic acid molecules encoding transcription factors useful in the practice
of the
present invention. For example, restriction endonuclease digestion of DNA
followed
by ligation may be used to generate deletion variants of transcription factors
useful in
the practice of the present invention, as described in Section 15.3 of
Sambrook et al.
Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory
Press, New York, NY (1989). A similar strategy
may be used to construct insertion variants, as described in section 15.3 of
Sambrook
et al., supra.
Oligonucleotide-directed mutagenesis may also be employed for preparing
substitution variants of transcription factors useful in the practice of the
present
invention. It may also be used to conveniently prepare the deletion and
insertion
CA 02412764 2007-06-28
16
variants of transcription factors useful in the practice of the present
invention. This
technique is well known in the art as described by Adelman et at. (DNA 2:183
[1983]); Sambrook et al., supra; "Current Protocols in Molecular Biology",
1991,
Wiley (NY), F.T. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.D. Seidman,
IA. Smith and K. Struhl, eds.
Generally, oligonucleotides of at least 25 nucleotides in length are used to
insert, delete or substitute two or more nucleotides in the nucleic acid
molecules
encoding transcription factors useful in the practice of the present
invention. An
optimal oligonucleotide will have 12 to 15 perfectly matched nucleotides on
either
side of the nucleotides coding for the mutation. To mutagenize wild-type
transcription factor proteins useful in the practice of the present invention,
the
oligonucleotide is annealed to the -single-itranded DNA template molecule
under
suitable hybridization conditions. A DNA polymerizing enzyme, usnally the
Klenow
fragment of E. coil DNA polymerase I, is then added. This enzyme uses the
oligonucleotide as a primer to complete the synthesis of the mutation-bearing
strand
of DNA. Thus, a heteroduplex molecule is formed such that one strand of DNA
encodes the wild-type protein inserted in the vector, and the second strand of
DNA
encodes the mutated form- of the protein inserted into the same vector. This
heteroduplex molecule is then transformed into a suitable host cell.
Mutants with more than one amino acid substituted may be generated in one
of several ways. If the amino acids are located close together in the
polypeptide
chain, they may be mutated simultaneously using one oligonucleotide that codes
for
all of the- desired amino acid substitutions. If, however; the amino acids are
located
some distance from each other (separated by more than ten amino acids, for
example)
it is more difficult to generate a single. oligonucleotide that encodes all of
the desired
changes. Instead, one of two alternative methods may be employed. In the first
method, a separate oligonucleotide is generated for each amino acid to be
substituted.
The oligonucleotides are then annealed to the single-stranded template DNA
simultaneously, and the second strand of DNA that is synthesized from the
template
.will encode all of the desired amino acid substitutions. An alternative
method
involves two or more rounds of mutagenesis to produce the desired mutant. The
first
round is as described for the single mutants: wild-type protein DNA is used
for the
template, an oligonucleotide encoding the first desired amino acid
substitution(s) is.
annealed to this template, and the heteroduplex DNA molecule is then
generated.
The second round of mutagenesis utilizes the mutated DNA produced in the first
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
17
round of mutagenesis as the template. Thus, this template already contains one
or
more mutations. The oligonucleotide encoding the additional desired amino acid
substitution(s) is then annealed to this template, and the resulting strand of
DNA now
encodes mutations from both the first and second rounds of mutagenesis. This
resultant DNA can be used as a template in a third round of mutagenesis, and
so on.
Prokaryotes may be used as host cells for the initial cloning steps of
transcription factors useful in the practice of the present invention. They
are
particularly useful for rapid production of large amounts of DNA, for
production of
single-stranded DNA templates used for site-directed mutagenesis, for
screening
many mutants and/or putative inner ear cell transcription factors
simultaneously, and
for DNA sequencing of the mutants generated. Suitable prokaryotic host cells
include E. coli K12 strain 94 (ATCC No. 31,446), E. coli strain W3110 (ATCC
No. 27,325) E. coli X1776 (ATCC No. 31,537), and E. coli B; however many other
strains of E. coli, such as HB101, JM101, NM522, NM538, NM539, and many other
species and genera of prokaryotes including bacilli such as Bacillus subtilis,
other
enterobacteriaceae such as Salmonella typhimurium or Serratia marcesans, and
various Pseudomonas species may all be used as hosts. Prokaryotic host cells
or
other host cells with rigid cell walls are preferably transformed using the
calcium
chloride method as described in section 1.82 of Sambrook et al., supra.
Alternatively, electroporation may be used for transformation of these cells.
Prokaryote transformation techniques are set forth in Dower, W.J., in Genetic
Engineering, Principles and Methods, 12:275-296, Plenum Publishing Corp.,
1990;
Hanahan et al., Meth. EnzyMol., 204:63 (1991).
As will be apparent to those skilled in the art, any.plasmid vectors
containing
replicon and control sequences that are derived from species compatible with
the host
cell may also be used to clone, express and/or manipulate nucleic acid
molecules
encoding transcription factors useful in the practice of the present
invention. The
vector usually has a replication site, marker genes that provide phenotypic
selection
in transformed cells, one or more promoters, and a polylinker region
containing
several restriction sites for insertion of foreign DNA. Plasmids typically
used for
transformation of E. coli include pBR322, pUC18, pUC19, pUCI18, pUC119, and
Bluescript M13, all of which are described in sections 1.12-1.20 of Sambrook
et al.,
supra. However, many other suitable vectors are available as well. These
vectors
contain genes coding for ampicillin and/or tetracycline resistance which
enables cells
transformed with these vectors to grow in the presence of these antibiotics.
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
18
The promoters most commonly used in prokaryotic vectors include the
P-lactamase (penicillinase) and lactose promoter systems (Chang et al. Nature,
375:615 [1978]; Itakura et al., Science, 198:1056 [1977]; Goeddel et al.,
Nature,
281:544 [1979]) and a tryptophan (trp) promoter system (Goeddel et al., NucL
Acids
Res., 8:4057 [1980]; EPO App!. Pub!. No. 36,776), and the alkaline phosphatase
systems. While these are the most commonly used, other microbial promoters
have
been utilized, and details concerning their nucleotide sequences have been
published,
enabling a skilled worker to ligate them functionally into plasmid vectors
(see
Siebenlist et al., Cell, 20:269 [1980]).
The construction of suitable vectors containing DNA encoding replication
sequences, regulatory sequences, phenotypic selection genes and the DNA
encoding
a transcription factor useful in the practice of the present invention are
prepared using
standard recombinant DNA procedures. Isolated plasmids and DNA fragments are
cleaved, tailored, and ligated together in a specific order to generate the
desired
vectors, as is well known in the art (see, for example, Sambrook et al.,
supra).
In another embodiment of the methods of the present invention, the formation
of inner ear sensory hair cells from inner ear support cells is stimulated by
damaging
inner ear sensory hair cells and inhibiting the expression (before, during
and/or after
damaging the inner ear sensory hair cells) of one or more cell cycle
inhibitors active
in inner ear support cells. In this way, inner ear support cells that are in
contact with
damaged sensory hair cells can be stimulated to divide and at least some of
the
resulting progeny form inner ear sensory hair cells. By way of representative
example, cell cycle inhibitors active in inner ear support cells include
cyclin-
dependent kinase inhibitors, such as cyclity-dependent kinase inhibitors of
the so-
called CIP/KIP family including p21Cip1, p27Kip1 and p57xip2.
Specific examples of cell cycle inhibitors active within inner ear support
cells
include: p57KiP2 (Lee et al., Genes Deli. 9(6): 639-649 (1995)(SEQ ID NO:6));
p27KiP1 (Cell 78(1): 59-66 (1994)(SEQ ID NOS:8 and 9)); p2lciP1 (El-Diery et
al.,
Cell 75(4): 817-825 (1993)(SEQ ID NOS:10 and 11)); p19 Ink 4d (Chan et al.,
MoL
Cell. Biol. 15(5): 2682-2688 (1995)(SEQ ID NOS:12 and 13)); p18 Ink 4c (Guan
et al., Genes Dev. 8(24): 2939-2952 (1994)(SEQ ID NOS:14 and 15)); p15 Ink 4b
(Hannon and Beach, 371(6494): 257-261 (1994)(SEQ ID NOS:16 and 17)); and p16
Ink 4a (Serrano, M. et al., Nature 366(6456): 704-707 (1993)(SEQ ID
= NOS:18 and 19)). Nucleic acid molecules that encode cell cycle inhibitors
useful in
the practice of the present invention hybridize to the antisense strands of
any one of
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
19
the nucleic acid molecules set forth in SEQ ID NOS: 6, 8, 10, 12, 14, 16 and
18
under at least one hybridization stringency greater than 2 X SSC at 55 C, such
as
1 X SSC at 60 C, or 0.2 X SSC at 60 C.
Inhibitors of cell cycle inhibitors can be substances, such as proteins, that
act
on the cell cycle inhibitor in an intracellular, direct or indirect manner.
Additionally,
inhibitors of cell cycle inhibitors can be antisense nucleic acid molecules
that are
complementary to all or a portion of a nucleic acid molecule (such as an mRNA
molecule) that encodes a cell cycle inhibitor protein, and that hybridize to
the nucleic
= acid molecule encoding a cell cycle inhibitor protein under stringent
conditions (such
as a stringency greater than 2 X SSC at 55 C, e.g., 1 X SSC at 60 C or 0.2 X
SSC at
60 C).
Any art-recognized method can be used to inhibit cell cycle inhibitor gene
expression in inner ear support cells. For example, the expression of a cell
cycle
inhibitor active in inner ear support cells can be inhibited by introducing
into inner
ear support cells a vector that includes a portion (or all) of a nucleic acid
molecule, in
antisense orientation relative to a promoter sequence, that encodes a cell
cycle
inhibitor active in inner ear support cells.
In general, antisense technology utilizes a DNA sequence that is inverted
relative to its normal orientation for transcription and so expresses an RNA
transcript
that is complementary to a target mRNA molecule expressed within the host cell
(i.e., the RNA transcript of the anti-sense gene can hybridize to the target
mRNA
molecule through Watson-Crick base pairing). An anti-sense gene may be
constructed in a number of different ways provided that it is capable of
interfering
with the expression of a target gene, such as a gene encoding a cell cycle
inhibitor.
The anti-sense gene can be constructed by inverting the coding region (or a
portion
thereof) of the target gene relative to its normal orientation for
transcription to allow
the transcription of its complement, hence the RNAs encoded by the anti-sense
and
sense gene are complementary.
The anti-sense gene generally will be substantially identical to at least a
portion of the target gene or genes. The sequence, however, need not be
perfectly
identical to inhibit expression. Generally, higher homology can be used to
compensate for the use of a shorter anti-sense gene. The minimal identity will
typically be greater than about 65%, but a higher identity might exert a more
effective repression of expression of the endogenous sequences. Substantially
CA 02412764 2007-06-28
greater identity of more than about 80% is preferred, though about 95% to -
absolute
identity would be most preferred.
Furthermore, the anti-sense gene need not have the same intron or exon
pattern as the target gene, and non-coding segments of the target gene may be
equally
5
effective in achieving anti-sense suppression of target gene expression as
coding
segments. Normally, a DNA sequence of at least about 30 or 40 nucleotides
should
be used as the anti-sense gene, although a longer sequence is preferable. The
construct is then introduced into one or more inner ear support cells and the
antisense
strand of RNA is produced.
10
Catalytic RNA molecules or ribozymes can also be used to inhibit expression
of target genes. It is possible to design ribozyme transgenes that encode RNA
ribozymes that specifically pair with a target RNA and cleave the
phosphodiester
backbone at a specific location, thereby functionally inactivating the target
RNA. In
carrying out this cleavage, the ribozyme is not itself altered, and is thus
capable of
15
recycling and cleaving other molecules. The inclusion of ribozyme sequences
within
antisense RNAs confers RNA-cleaving activity upon them, thereby increasing the
activity of the antisense constructs. Tabler et al. (1991, Gene 108:175) have
greatly
simplified the construction of catalytic RNAs by combining the advantages of
the
anti-sense RNA and the ribozyme technologies in a single construct. Smaller
regions
20 of
homology are required for ribozyme catalysis, therefore this can promote the
repression of different members of a large gene family if the cleavage sites
are
conserved.
An additional strategy suitable for suppression of target gene activity
entails
the. sense expression of a mutated or partially deleted form of the protein
encoded by
the target gene according to general criteria for the production of dominant
negative
mutations (Herskowitz I, Nature 329: 219-222 (1987)).
Any art-recognized gene delivery method can be used to introduce a nucleic
acid molecule encoding a transcription factor (or a vector including an
antisense
DNA molecule) into inner ear cells for expression therein, including: direct
injection, electroporation, virus-mediated gene delivery, amino acid-mediated
gene
delivery, biolistic gene delivery, lipofection and heat shock. .Non-viral
methods of
gene delivery into inner ear cells are disclosed in Huang, L., Hung, M-C, and
Wagner, E., Non-Viral Vectors for Gene Therapy, Academic Press, San Diego,
California (1999).
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
21
For example, genes can be introduced into cells in situ, or after removal of
the
cells from the body, by means of viral vectors. For example, retroviruses are
RNA
viruses that have the ability to insert their genes into host cell chromosomes
after
infection. Retroviral vectors have been developed that lack the genes encoding
viral
proteins, but retain the ability to infect cells and insert their genes into
the
chromosomes of the target cell (A.D. Miller, Hum. Gen. Ther.1:5-14 (1990)).
Adenoviral vectors are designed to be administered directly to patients.
Unlike retroviral vectors, adenoviral vectors do not integrate into the
chromosome of
the host cell. Instead, genes introduced into cells using adenoviral vectors
are
maintained in the nucleus as an extrachromosomal element (episome) that
persists for
a limited time period. Adenoviral vectors will infect dividing and non-
dividing cells
in many different tissues in vivo including airway epithelial cells,
endothelial cells,
hepatocytes and various tumors (B.C. Trapnell, Adv Drug Del Rev. 12:185-199
(1993)).
Another viral vector is the herpes simplex virus, a large, double-stranded
DNA virus that has been used in some initial applications to deliver
therapeutic genes
to neurons and could potentially be used to deliver therapeutic genes to some
follus
of brain cancer (D.S. Latchman, MoL Biotechnol. 2:179-95 (1994)). Recombinant
forms of the vaccinia virus can accommodate large inserts and are generated by
homologous recombination. To date, this vector has been used to deliver
interleukins
(ILs), such as human IL-1p and the costimulatory molecules B7-1 and B7-2
(G.R. Peplinski et al., Ann. Surg. Oncol. 2:151-9 (1995); J.W. Hodge et al.,
Cancer
Res. 54:5552-55 (1994)).
Another approach to gene therapy involves the direct introduction of DNA
plasmids into patients. (F.D. Ledley, Hum. Gene Ther. 6:1129-1144 (1995)). The
plasmid DNA is taken up by cells within the body and can direct expression of
recombinant proteins. Typically plasmid DNA is delivered to cells in the form
of
liposomes in which the DNA is associated with one or more lipids, such as
DOTMA
(1,2,-diolcyloxypropy1-3-trimethyl ammonium bromide) and DOPE
(dioleoylphosphatidylethanolamine). Formulations with DOTMA have been shown
to provide expression in pulmonary epithelial cells in animal models (K.L.
Brigham
et al., Am. J. Med. Sci, 298:278-281 (1989); A.B. Canonico etal., Am. J.
Respir. Cell.
MoL Biol. 10:24-29 (1994)).
Additionally, studies have demonstrated that
intramuscular injection of plasmid DNA formulated with 5% PVP (50,000 kDa)
increases the level of reporter gene expression in muscle as much as 200-fold
over
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
22
the levels found with injection of DNA in saline alone (R.J. Mumper et al.,
Pharm.
Res. 13:701-709 (1996); R.J. Mumper et al., Proc. Intern. Symp. Cont Rol.
Bioac.
Mater. 22:325-326 (1995)). Intramuscular administration of plasmid DNA results
in
gene expression that lasts for many months (J.A. Wolff et al., Hum. Mol.
Genet.
1:363-369 (1992); M. Manthorpe et al., Hum. Gene Ther. 4:419-431 (1993);
G. Ascadi et al., New Biol. 3:71-81 (1991), D. Gal etal., Lab. Invest. 68:18-
25
(1993)).
Additionally, uptake and expression of DNA has also been observed after
direct injection of plasmid into the thyroid (M. Sikes et al., Hum. Gene Ther.
5:837-844 (1994)) and synovium (J. Yovandich et al., Hum. Gene Ther. 6:603-610
(1995)). Lower levels of gene expression have been observed after interstitial
injection into liver (M.A. Hickman et al., Hum. Gene Ther. 5:1477-1483
(1994)),
skin (E. Raz et al., Proc. Natl. Acad. Sci. 91:9519-9523 (1994)), instillation
into the
airways (K.B. Meyer et all, Gene Therapy 2:450-460 (1995)), application to the
endothelium (G.D. Chapman et al., Circulation Res. 71:27-33 (1992); R. Riessen
et al., Human Gene Therapy, 4:749-758 (1993)), and after intravenous
administration
(R.M. Conry et al., Cancer Res. 54:1164-1168 (1994)).
Various devices have been developed for enhancing the availability of DNA
to the target cell. A simple approach is to contact the target cell physically
with
catheters or implantable materials containing DNA (G.D. Chapman et al.,
Circulation Res. 71:27-33 (1992)). Another approach is to utilize needle-free,
jet
injection devices which project a column of liquid directly into the target
tissue under
high pressure. (P.A. Furth et al., Anal Biochem. 20:365-368 (1992); (H.L.
Vahlsing
et al., J. Immunol. Meth. 175:11-22 (1994); (F.D. Ledley et al., Cell Biochem.
18A:226 (1994)).
Another device for gene delivery is the "gene gun" or BiolisticTM, a ballistic
device that projects DNA-coated micro-particles directly into the nucleus of
cells
in vivo. Once within the nucleus, the DNA dissolves from the gold or tungsten
microparticle and can be expressed by the target cell. This method has been
used
effectively to transfer genes directly into the skin, liver and muscle (N. S.
Yang et al.,
Proc. Nazi. Acad. Sci. 87:9568-9572 (1990); L. Cheng et al., Proc. Natl. Acad.
Sci.
USA. 90:4455-4459 (1993); R.S. Williams et al., Proc. Natl. Acad. Sci.
88:2726-2730 (1991)).
Cochleostomy involves puncturing the cochlea and inserting a catheter
through which a chemical agent, such as a nucleic acid molecule, can be
introduced
CA 02412764 2007-06-28
23
into the cochlea. A cochleostomy method is disclosed, for example, in Lalwani,
A.K.
et al., Hearing Research 114: 139-147 (1997).
Another approach to targeted gene delivery is the use of molecular
conjugates, which consist of protein or synthetic ligands to which a nucleic
acid- or
DNA-binding agent has been attached for the specific targeting of nucleic
acids to
cells (R.J. Cristiano et al., Proc. Natl. Acad. Sci. USA 90:11548-52 (1993);
B.A.
Bunnell et al., Somat. Call MoL Genet. 18:559-69 (1992); M. Cotten et al.,
Proc.
Natl. Acad. Sci. USA 89:6094-98 (1992)). Once the DNA is coupled to the
molecular conjugate, a protein-DNA complex results. This gene delivery system
has
been shown to be capable of targeted delivery to many cell types through the
use of
different ligands (R.J. Cristiano et al., Proc. Natl. Acad. ScL USA 90:11548-
52
(1993)). For example, the vitamin folate has been used as a ligand to promote
delivery of plasmid DNA into cells that overexpress the folate receptor (e.g.,
ovarian
carcinoma cells) (S. Gottschalk etal., Gene Ther. 1:185-91(1994)). The malaria
circumsporozoite protein has been used for the liver-specific delivery of
genes under
conditions in which ASOR receptor expression on hepatocytes is low, such as in
cirrhosis, diabetes, and hepatocellular carcinoma (Z. Ding et al., J. Biol.
Chem.
270:3667-76 (1995)). The overexpression of receptors for epidermal growth
factor
(EGF) on cancer cells has allowed for specific uptake of EGF/DNA complexes by
lung cancer cells (R. Cristiano et al., Cancer Gene Then 3:4-10 (1996)). The
presently preferred gene delivery method is lipofection.
When the methods of the present invention are utilized in vitro, the whole
inner ear, including the Organ of Corti, is preferably excised and cultured
and
manipulated in a culture vessel. Presently preferred embodiments of an
apparatus
that is useful for culturing inner ears in vitro are disclosed in U.S. Patent
Serial
No: 5,437,998; U.S. Patent Serial No: 5,702,941 and U.S. Patent Serial
No: 5,763,279.
In general, presently preferred embodiments of an apparatus for culturing
inner ears include a gas permeable bioreactor comprising a tubular vessel with
walls
that may be constructed at least partially of a gas permeable material, such
as silicone
rubber. The vessel in one preferred embodiment is constructed such that half
of it is
comprised of gas permeable material and the remaining . portion is made of
nonpermeable material. The gas permeable materials commonly available are
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
24
opaque. Thus, using nonpermeable material for at least part of the bioreactor
may
provide an advantage in allowing visual inspection of the tubular vessel
chamber.
The tubular vessel has closed ends, a substantially horizontal longitudinal
.
central axis, and one or more vessel access ports. The vessel access ports
provide
access to the bioreactor for input of medium and cells, and for removal of old
medium from the tubular vessel. This is easily done through the vessel access
ports
which are also referred to as valves or syringe ports. In the preferred
embodiment,
the vessel access ports are constructed of valves with syringe ports.
Preferably the vessel is rotatable about its horizontal longitudinal central
axis.
A preferred means for rotation is a motor assembly which sits on a mounting
base
and has means for attachment to the tubular vessel. The speed of rotation can
be
adjusted so that the inner ear within the tubular vessel is constantly in
motion, but
rotation of the tubular vessel should not be fast enough to cause significant
turbulence in the aqueous medium within the tubular vessel.
If so desired, the use of gas permeable material in the construction of at
least
part of the tubular vessel wall permits 02 to diffuse through the vessel walls
and into
the cell culture media in the vessel chamber. Correspondingly, CO2 diffuses
through
the walls and out of the vessel. Thus, the use of gas permeable material in
the
construction of at least part of the tubular vessel wall typically overcomes
the need
for air injection into the bioreactor vessel. Air injection into the aqueous
medium
within the bioreactor vessel may be utilized, however, if additional oxygen is
required to culture an inner ear. When an air pump is utilized to inject air
into the
aqueous medium, an air filter is also employed to protect the air pump valves
from
dirt.
An alternative embodiment of the bioreactor useful in the practice of the
present invention is an annular vessel with walls that may be constructed at
least
partially of a gas permeable material. Annular is defined herein to include
annular,
toroidal and other substantially symmetrical ring-like shaped tubular vessels.
The
annular vessel has closed ends and a substantially horizontal longitudinal
central
axis.
In another embodiment, the bioreactor useful in the practice of the present
invention comprises a tubular vessel constructed at least partially of a gas
permeable
material. The vessel has closed ends and a substantially horizontal
longitudinal
central axis around which it rotates. The vessel furthermore has two slidably
interconnected members wherein a first member fits slidably into a second
member,
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
forming a liquid tight seal therebetween and providing a variable volume
tubular
vessel. The bioreactor has means for rotating the tubular vessel about its
substantially horizontal longitudinal central axis. One or more vessel access
ports are
provided for transferring materials into and out of the vessel.
5 In
situations where minimization of contamination is necessary (e.g., AIDS or
human tissue research), disposability of the bioreactor useful in the practice
of the
present invention is a particular advantage. Moreover, the embodiment of the
bioreactor with slidably interconnected members may be adjusted to provide the
exact size bioreactor needed.
10 Presently
preferred, commercially available bioreactors useful in the practice
of the present invention for culturing fluid-filled sensory organs are known
as the
High Aspect Ratio Vessel (HARVTM) and the Cylindrical Cell Culture Vessel
(CCCVTM) and are manufactured by Synthecon, Inc. (8054 El Rio, Houston,
Texas).
NeuralbasalTM media from Gibco BRL (Gibco BRL media are produced by
15 Life
Technologies, Corporate Headquarters, Gaithersburg, MD), which requires the
addition of B27 or N2 media supplement, is the presently preferred culture
medium
for culturing inner ears in vitro. Other culture media can be successfully
used,
however, to culture fluid-filled sensory organs in the practice of the present
invention. Other suitable media include DME, BME and M-199 with fetal calf
20 serum or
horse serum. All of the foregoing media are sold by Gibco -BRL. When
using NeuralbasalTM medium, N2 or B27 supplements play a more significant role
when extended periods of culture (>96 hr) are attempted.
In another aspect, the present invention provides methods for stimulating the
formation of inner ear support cells. The methods of this aspect of the
invention
25 include
the steps of damaging inner ear support cells under conditions that promote
the formation of new inner ear support cells (for example by cell division of
inner ear
support cells that are in contact with damaged inner ear support cells). In
this aspect
of the invention, the inner ear support cell is damaged, and the formation of
new
inner ear support cells is stimulated, using the same techniques described
herein for
the methods of the present invention that stimulate the formation of inner ear
sensory
hair cells from inner ear support cells. Thus, for example, inner ear support
cells can
be damaged by contact with an amount of an ototoxic agent, such as an
aminoglycoside antibiotic, that is effective to damage inner ear support
cells. Again
by way of example, new inner ear support cell formation can be further
stimulated by
damaging inner ear support cells and expressing (before, during and/or after
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
26
damaging inner ear support cells) within inner ear support cells a
transcription factor
(such as POU4F1, POU4F2, POU4F3, Bm3a, Brn3b and Brn3c) capable of
stimulating inner ear support cells to divide and form new inner ear support
cells. In
preferred embodiments of this aspect of the invention, the proliferation of
inner ear
support cells results in improvement in the auditory function of the treated
inner ear.
The following examples merely illustrate the best mode now contemplated
for practicing the invention, but should not be construed to limit the
invention.
Example 1
Overexpressing POU4F3 in immortalized supporting-cell lines in vitro.
POU4F3 is a DNA binding transcription factor exhibiting remarkable
specificity to the hair-cells in the inner ear. Mutations in POU4F3 are known
to
cause developmental failures in mice, and hearing loss in both mice and
humans.
The role of POU4F3 in directing the development of hair-cell precursors is
investigated by transfecting inner ear supporting-cell lines with POU4F3.
To detect the expression of POU4F3 in live cultures, the expression of
Enhanced Green Fluorescent Protein (EGFP), translated from a bicistronic mRNA
which includes both POU4F3 and GFP coding regions, is monitored. Specifically,
a
1250 bp cDNA encoding P0154F3 including 70 bp of 5'UTR and 73 bp of 3' UTR is
directionally cloned into the unique EcoRV restriction site of the pIRES2-EGFP
vector (Clonetech) directly down stream from the human CMV major immediate
early promoter/enhancer. An intervening synthetic intron is cloned downstream
from
the POU4F3 gene to enhance the stability of the mRNA. The internal ribosomal
entry site (IRES) from encephelomyocarditis virus is cloned between the POU4F3
gene and the GFP gene to allow for the translation of GFP and POU4F3 protein
from
the same mRNA. Immediately following the GFP coding region is a poly-
adenylation signal from the bovine growth hormone gene. This expression
cassette is
designed to take advantage of the bicistronic promoter to allow tracking of
transfection and expression of POU4F3 and GFP by visualization of expressed
GFP
under fluorescence microscopy. This second generation GFP vector has a red
shifted
variant of wild-type GFP (Excitation maximum = 488 urn; emission maximum = 507
urn) which has been optimized for brighter fluorescence. To optimize the
identification of cells expressing high levels of the POU4F3 protein, the
pIRES2
vector utilizes a partially disabled IRES sequence (Rees, S. et al.,
BioTechniques
20:102110 (1996)). This attenuated IRES leads to a reduced rate of translation
initiation at the GFP start codon relative to that of the POU4F3 gene.
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
27
The supporting-cell line (UCL) was established from the inner ear vestibular
sensory epithelia of the 112kb-tsA 58 transgenic mouse (Immortamouse).
Utricles
from postnatal day one mice were dissected and the sensory epithelia (hair-
cells and
supporting-cells) isolated after brief thermolysin treatment at 37 C. The
resultant
supporting-cell line was derived after several passages at permissive
conditions
(33 C and yINF) which stimulates rapid cell proliferation. Confluence was
achieved
in 3-4 days. This process resulted in the death of all hair-cells as
determined by ICC
and electron microscopy (EM). The UCLs were further characterized by an
antibody
called ZO-1 (which labels tight junctions that are present in supporting-
cells) and EM
(which showed tight junction complexes, secretory vesicles and luminal surface
microvilli, all characteristic of supporting-cells).
Culture medium for the UCL cell line consisted of DMEM/F12 (Gibco), fetal
bovine serum (10%) and yINF (20 u/ml). Media changes were done 2 to 3 times a
week depending on the growth rate of the cells. Single-cell clones were
developed
using a special seeding method that enabled single cells to become confluent
and
passageable in 3-4 weeks. At non-permissive conditions (37 C or 39 C, no yINF
and
low or no FBS) cell growth arrests.
Given the high transfection efficiencies already observed in the UCLs, these
cells are grown and passaged in defined media that is serum free. Once this is
established, these cells are lipofected with the IRES-GFP-POU4F3 encoding
plasmid. Cultures are monitored for GFP expression over 1-6 DIV. When periods
of
high GFP fluorescence are observed in live cultures, the culture are fixed and
prepared for POU4F3 and calbindin ICC.
Example 2
Overexpression of POU4F3 in lesioned organ of Corti cultures.
Cultures from P7-P14 mice are established and lesioned with 1 mM neomycin
for 2 DIV. The media are removed and the cultures lipofected with pIRES2-GFP-
POU4F3 for 6 hr and recovered in fresh media for 1-6 DIV. Cultures are
aldehyde-
fixed and processed for POU4F3 and calbindin immunocytochemistry. pIRES2-GFP
only lipofected cultures serve as controls. The presence of a triple labeled
cell
(positive for GFP, POU4F3, and calbindin imtnunoreactivity) indicates that
POU4F3
is capable of promoting the adoption of a hair-cell phenotype in the lesioned
organ of
Corti. Further determination of this phenotype is corroborated with antibodies
against other hair-cell selective markers such as myosin 6 and myosin 7a using
polyclonal antibodies derived against these proteins.
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
28
Expression of hair-cell specific markers such as myosin 6 and 7a are observed
in the embryonic mouse organ of Corti at El 6, 2-3 days after the expression
of the
Brn 3.1 transcription factor begins at E13.5.
Example 3
Regeneration of Inner ear hair cells in p271(iP1 -/- mice
Previous reports in p271ciP1 -/- and +/- mice showed qualitative evidence of
supernumerary hair cells (HCs) in both inner hair cell (IHC) and outer hair
cell
(OHC) regions (Chen, P., & Segil, N. Development 126: 1581-1590 (1999);
Lowenheim, H., et al., Proc. Natl. Acad. Sci. USA. 96: 4084-4088 (1999)).
Unfortunately, the significant dysplasia of the surrounding supporting cells
could
very well account for some if not all of these observations. Therefore, the
number of
IHCs and OHCs in the cochlea of p271ciP1 -/-, +/- and +/+ mice was measured.
To
more accurately assess whether there was a true increase in HC number, several
regions from the same cochlea were analyzed. Using a HC specific marker, a
myosin-VIIa antibody, a 20% increase in the number of IHCs in p271(iP1 -1-
cochlea
was observed when compared with that in p271(i11 +/- and +/+ cochlea. However,
there was no statistically significant difference in the overall number of
OHCs
between p2716P1 -/-, +/- and +1+ cochlea, except for a 10% increase in the
number of
OHCs in one analyzed region (Table 1). Table 1 shows the number of hair cells
(n)
in four-week old p27KiP1 +/+, +/-, -/- mice. Counts were made from a 100 ni
length
of sensory epithelium from three different locations along the longitudinal
axis of the
organ of Corti. Distance corresponds to degrees from the apical tip of the
cochlea
s.d.). Comparisons were made with the same hair cell region across +/+, +/-
and -/-. Statistical significance was determined using ANOVA.
Table 1
Genotype Distance IHC OHC
+/+ 90 6 13.0 0.0 42.8 I 2.5
+/- 90 16 13.1 0.7 44.1 2.9
-/- 90 11 16.0 1.9 * 43.5 2.3
+/+ 180 6 13.3 0.8 40.5 5.2
+/- 180 16 13.2 0.8 43.1 2.2
-/- 180 11 17.0 1.3 * 45.9 3.3 *
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
=
29
Table 1
Genotype Distance IHC OHC
+1+ 360 6 13.7 0.8 41.7
5.1
+/- 360 16 13.4 0.5 43.1
2.8
360 11 16.2 1.8 * 41.0
4.6
*p-value <.001
To determine whether auditory hair cells were being produced after the onset
of hearing at postnatal day 10, two-week old p27KiP1 -/-, +/-, +/+ mice (P10-
12)
received three daily systemic injections of bromodeoxyuridine (BrdU;
30mg/kg/s.c),
a nucleotide analog which is incorporated into proliferating cells during S
phase.
Mice were then permitted to recover for two-days or two-weeks without further
injections. BrdU positive HCs were identified with immunocytochemistry using
light and fluorescence microscopy. Cochlea were also labeled with antibodies
against myosin-VI and myosin-VIIa. In two-week old p27KiP1 -/- cochlea that
were
recovered for 2d after the last injection, no BrdU/myosin-VIIa positive cells
were =
observed among the BrdU positive cells. However, in four-week old p2716P1 -/-
cochlea that were recovered for 14 days after the last injection, BrdU/myosin-
VIIa
positive HCs were observed. Most of those double labeled were IHCs. This
qualitative finding is similar to our quantitative assessments of IHCs and
OHCs
numbers. p27KiP1 +/-, +1+ mice were completely devoid of BrdU positive cells
at 2d
and 14d of recovery. These data are summarized in Table 2. Table 2 shows the
number (n) of BrdU labeled cells in the organ of Corti of two- and four-week
old
p27KiP1 +/+, +/-, -/- mice injected with BrdU or BrdU/Amikacin with 2d or 14d
of
recovery. Counts were performed on a 10001.1m length of sensory epithelium
taken
in the apical half of the cochlea ( s.d.). Proliferation in the BrdU groups
were
compared across +/+, +/- and -/-. Proliferation in the BrdU/Amikacin group was
compared with that in the BrdU only group of the same genotype. Statistical
significance was determined using ANOVA.
Table 2
Genotype BrdU + 2d p
BrdU/Amikacin + 2d
=
CA 02412764 2007-06-28
+/+ 10 0.0 0.0 0.0
0.0
+/- 12 0.0 0.0 4.7
7.6
-1- 4 82.3 11.0* 96.3
9.5
Genotype n BrdU + 14d n
BrdU/Amikacin + 14d
+/+ 10 0.0 0.0 10 0.0
0.0
+/- 18 0.0 0.0 10 10.5
12.0*
-/- 6 82.2 22.5* 12 137.4
17.0*
*p-value <.00.1
To ascertain whether auditory HCs could be regenerated, HCs were lesioned
using systemic injections of amikacin sulfate (P7-P12) and then injected with
BrdU
5 (P10-
12). Mice were sacrificed either 2 d or 14 d after the last injection. The
effects
of an amikacin lesioning were at least two-fold. First, in both p27KiP1 -/-
and +/-
mice,. the number of BrdU positive cells increased following amikacin/BrdU vs
BrdU
alone treatment. In p27KiPi +/- cochlea, the numbers of BrdU labeled cells
increased
in the majority of specimens examined, however, not all p27KiPi +/- cochlea
10
displayed BrdU positive cells. Evidence of HC regeneration was confirmed by
sectioning the labeled cochlea. Second, a greater number of BrdU positive HCs
were
observed following amikacin lesioning in p27KiPI -/- cochlea. The majority of
the
BrdU positive HCs appeared in the regions of the cochlea where the amikacin
sulfate
had injured or killed HCs (in the basal half of the cochlea). No BrdU positive
cells
15 were
observed in wt cochlea following amikacin/BrdU or BrdU alone treatment.
These data are summarized in Table 2.
To measure specific protein levels, single cochlear lysates were serially
diluted
and run on polyacrylamide gels. Western blotting showed that myosin-VI and
Vila
levels appeared roughly equal across p27KiP1 -/-, +/- and +1+ cochlea,
although a
20
stronger myosin-Vila band was observed from p27KiP1 -/- cochlea. p27KiP1 +/-
cochlea contained approximately 50% of the prI protein levels found in wt
cochlea, indicating that a reduction of p27KiP1 to 50% of normal can stimulate
supporting cell proliferation and allow some hair cell regeneration to occur
following
amikacin sulfate treatment.
25 The protocol used to measure protein levels in the cochlea is as
follows. A
cochlea is transferred to a tube with 10 4iil of extraction buffer which
contains:HEPES
(25 inM), NP-40Th (0.7%), Aprotintin (1 mM), Leupeptin (1 lig/m1), Pepstatin
CA 02412764 2007-06-28
.31
(10 iiM), phenyhnethylsulfonylfluoride (PMSF) (1 mM), dithiothreitol (DT)
(1 mM), and ethylenediaminetetraacetic acid (EDTA) (2 mM). The cochlea is
homogenized immediately and the tube is placed on ice for about 30 min. Add 5
I
of 4X sample buffer and adjust the salt concentration up to 0.5 M. Heat up the
sample to 90-100 C for 5 min then spin at 13,000 rpm for 10 min and collect
the
supematant. Aliquots of-protein from the supernatant are run on a 15% SDS-PAGE
gel for 50 min at 200 V and the proteins are transferred onto PVDF membrane
for
1 hr at 100 V. The membrane. is blocked with 10% AmershamTm blocking buffer
for 1
hr or overnight. Primary antibody in blocking buffer is applied to the
membrane for
1 hr and the membrane is then washed five times with PBS/Tweeni'm for 5
minutes per
wash. The membrane is probed with goat anti-mouse or goat anti-rabbit-alkaline
phosphatase plus anti-biotin-AP for 1 hr and washed five times with
PBS/TweenTm for
5 minutes per wash.
To determine whether p271ciPI plays a similar role in the peripheral
vestibular
system, the proliferative capacity of the utricle, saccule and cristae of
p271ciP1 +/-, and
+/+ mice was examined. Mice (P7-P12) received systemic injections of amikacin
sulfate (500mg/kg/d/s.c). for six consecutive days that were combined with
injections
of the replication marker, bromodeoxyuridine (BrdU; 30mg/kg/d/s.c) between P10-
P12. Mice were also injected with BrdU alone in a similar fashion. Mice were
then
sacrificed 14 d later and the vestibular sensory organs were fixed, dissected
and
processed for BrdU immunocytochemistry. BrdU positive nuclei were counted from
whole mounts using light microscopy and Nomarski optics. Selected organs were
further processed for cross section analysis.
In p27KiPl. -/- mice that received BrdU only, very low levels of BrdU-labeled
cells were observed in the saccule and utricle. However, after combined
amikacin/BrdU treatment, a 40-fold increase in the number of BrdU positive
cells
was observed in both organs. Approximately one-half of the labeled cells
appeared
as doublets suggesting recent or ongoing cell divisions. Plastic cross
sections
showed that the majority of the BrdU labeled cells were in the basal layer of
the
sensory epithelium, along the basal membrane. BrdU positive HCs were also
observed in both otolithic organs. Most of these regenerated HC appeared as
type
HCs in that they were contacted by a calyx. In p271(iP1 +/- mice that received
BrdU
only, no proliferation was observed in either the saccule or utricle. After
combined
amikacin/BrdU treatment, a very low level of proliferation was induced. In
p271<
+/+ mice, no BrdU positive nuclei in either the saccule or utricle were
observed after
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
32
amikacin/BrdU or BrdU alone. Interestingly, no BrdU positive cells were
observed
in the cristae of any genotype following amikacin/BrdU or BrdU only. These
data
indicate significantly different effects in deleting p27KIP1' among the
various
vestibular sensory organs and between the vestibular sensory organs and the
organ of
Corti. These data are summarized in Table 3. Table 3 shows the numbers of BrdU
labeled cells in the vestibular organs of four-week old p27KiP1 +/+, +/- and -
/- mice
injected with BrdU or BrdU/Amikacin and after a 14 d recovery. Counts were
performed from whole utricular, saccular and cristae sensory epithelium (
s.d.). In
the BrdU group, statistical significance was determined by comparing
proliferation
levels in the same sensory organ across +/+, +/- and -/-. In the BrdU/Amikacin
group, statistical significance was determined by comparing proliferation
levels in
the same sensory organ of the same genotype. Statistical significance was
determined using ANOVA.
Table 3:
utricle (n) genotype BrdU BrdU/Amikacin
8 +/+ 0.0 0.0 0.0 0.0
+/- 0.0 0.0 0.4 0.7
12 -/- 19.7 13.4* 45.5 19.2*
saccule (n) genotype BrdU BrdU/Amikacin
6 +/+ 0.0 0.0 0.0 0.0
16 +/- 0.0 0.0 1.6 1.9
9 -/- 32.7 14.3* 51.8 15.3*
cristae (n) genotype BrdU BrdU/Amikacin
12 +/+ 0.0 0.0 0.0 0.0
=
12 +/- 0.0 0.0 0.0 0.0
12 -/- 0.0 0.0 0.0 0.0
15 *p-value <.001
Selected semi-thin sections were re-embedded in plastic, thin sectioned and
examined under electron microscopy. BrdU positive HCs displayed stereociliary
bundles, cuticular plates, calyceal innervation with evidence of synapse
formation.
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
33
Example 4
Antisense inhibition of p27KiP I expression
To test whether inhibition of the p27KiP1 gene product in p27KiP1 +1+ cochlea
would allow non-mitotic supporting cells to proliferate, wild type organ of
Corti
explants (P7-P10, at age of hearing onset) were treated with p27KiPI antisense
oligonucleotides (ONs). Explant cultures were established by dissecting the
organ of
Corti from the cochlea, removing the tectorial membrane and adhering the organ
of
Corti to a glass slide coated with Cell-tak and maintained at 37 C in a 5% CO2
environment. The explant was then exposed to an ototoxic antibiotic (1 mM
neomycin sulfate) for 48 h which killed >95% of the HCs (Ku, J., et al., ARO
abs.,
21: 672 (1998)). The neomycin containing media was then removed and p27KiP1
antisense oligonucleotides (Ons) (40 nM) were administered using a cationic
lipid
(lipofection) for a period of 24-48 hr. Some of these living cultures were
examined
under fluorescence to detect the presence of FITC conjugated antisense ON.
FITC-positive supporting cells were detected in 18-24 h and increased in
number and fluorescent intensity between 24-48 h. Cultures were aldehyde fixed
and
processed for BrdU immunocytochemistry. BrdU-positive supporting cells
appeared
in most cochlear cultures after 24 h of antisense oligonucleotide (ON)
treatment.
BrdU-positive doublets appeared in antisense ON treated cultures that were
recovered for an additional 24 hr without antisense ONs. This indicated that
successful completion of M phase and subsequent cell division could occur.
Lipofection-only treated cultures contained very low levels of BrdU-labeled
supporting cells.
We observed that administration of p27KiP1 antisense oligonucleotides can
induce supporting cells to proliferate in wild-type cochlea. This observation
is
unique and rather significant. Previous work demonstrating p271(i1)1 antisense
ON
induced proliferation were shown in actively dividing cells that had been
transiently
or reversibly growth arrested (Coats, S., et al, Science, 272: 877-880 (1996);
Dao, M.A., et al, Proc. Natl. Acad. Sci. USA, 95: 13006-13011(1998)). Our
results
are the first demonstration that terminally mitotic cells in a terminally
differentiated
organ can reenter the cell-cycle after inhibition of p27KiP1 gene products.
Again, supporting cell proliferation normally ceases between E12-14 in the
mouse organ of Corti. After this point, supporting cell differentiation
continues
through the second week of postnatal life. At the time of explant, cultures
treated as
described herein have already developed some adultlike morphologic
characteristics.
CA 02412764 2007-06-28
34
These data further support the role for p27KiP1 antisense ON as a potential
means for
inducing hair cell regeneration. In. p27Kipl +I- mice, a reduction to 50% of
normal
protein levels allows some terminally differentiated supporting cells to
overcome the
p271(iP1 blockade and re-enter the cell cycle and proliferate.
The addition of growth factors that induce cell proliferation in other
epithelial
organs, does not promote cell proliferation or hair cell regeneration in the
postnatal
organ of Corti, either in vitro or in vivo. The experiments reported herein
identify
identified a rather ubiquitous and potent cell cycle inhibitor that, when
deleted,
allows the organ of Corti to regenerate some of its hair cells spontaneously.
The
= organ of Corti of mice containing one copy of the gene and 50% of normal
protein
levels are capable of auditory hair cell regeneration.
In addition, organotypic cultures can be established on glass slides (Nunc)
coated with CellTakTm (Collaborative Research) in 100 Is of NeuroB asal media
(Gibco) containing 1 mM neomycin. This treatment kills 95% of the hair cells
and
also facilitates the level of transfection. Cultures are lipofected with
antisense
molecules using commercially available lipofection reagents (e.g, Perfect
Lipofection
Kit; InVitrogen, Inc.). The media also contains BrdU (1011.M) to identify
proliferating cells. In addition, various recombinant growth factors such as
TGF-
alpha (1-100 nM), insulin (10-100 [LM) and IGF-1 (1-100 M) can be used to
increase or drive this proliferative effect.
Example 5
The use of n271(iP1 antisense oligonucleotides to stimulate cell proliferation
in a
guinea Dig fibroblast cell line in vitro
Cultures of cell Hues were established that are responsive to p27KiP1
antisense
oligonucleotides (ON) during the period of serum withdrawal and growth arrest.
These cultures include a guinea pig fibroblast line (JH4). Lipofection of the
16 mer
antisense ON (having the nucleic acid sequence set forth in SEQ ID NO:20)
reversed growth arrest in the .TH4 cell line to over 40% of normal.
Example 6
Stimulation of Supporting Cell Proliferation in Guinea Pig Cochlea Using
p271(iPI
Antisense Molecules
Two- recent, independent studies indicate that antisense ONs can be
successfully delivered through the perilymphatic space and elicit changes in
the
organ of Corti of mature guinea pigs (D'Aldin, C., et al., Mel. Brain Pas.,
55: 151-
"
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
164 (1998); Leblanc, C.R., et al. Hear. Res., 135: 105-112 (1999)). The
presence of
FITC-antisense ON against specific sequences to the mRNA of G1uR2 was seen in
the spiral ganglia, supporting cells, and inner and outer spiral sulcus cells
within 24 h
after the osmotic pump installation. A subsequent selective decrease in
G1uR2/3
5 protein was also observed in situ (D'Aldin et al., 1998, supra).
The expression pattern of p27KiP1 in the mature guinea pig organ of Corti was
found to be similar to that observed in developing and adult mice. Supporting
cell
proliferation in guinea pig cochlea is stimulated as follows:
General anesthesia is induced by inhalation of Isoflurane (5% for the
induction
10 and 2-3% for the maintenance) or by intramuscular injection of ketamine
(50 mg/kg)
and xylazine (9 mg/kg). 1% lidocaine is injected behind the pinna locally. The
animal is placed on a warming pad to keep basal body temperature constant
during
the surgical operation. Respiration and circulation is monitored carefully.
Drugs are
delivered into the guinea pig's inner ear by surgically implanting an infusion
unit
15 which consists of a coiled catheter and an osmotic minipump (Alzet, cat
no. 2002,
0.5 Oh flow rate for up to 14 days). The coil is loaded with the drug and the
osmotic minipump carries a dye. The total volume of drug pumped into the inner
ear
can be monitored by the amount of dye which is pumped into the coil. The drug
is
dissolved in a solution of artificial perilymph which consists of: 137 mM
NaC1,
20 5 mM KC1, 2 mM CaC12, 1 mM MgCl2, 10 mM Hepes, 11 mM Glucose, pH 7.4 and
osmolarity 300mOsm/L.
All surgical procedures are done under a dissection microscope and under
sterile conditions. The mastoid bulla (middle ear space) is exposed via a post-
auricular incision and opened using a 1 mm cutting burr to allow visualization
of the
25 basal turn of the cochlea. A cochleostomy about 1 mm inferior to the
round window
is fashioned using a 0.5 mm diamond paste burr. Observed inner ear perilymph
fluid
leaking from this site confirms correct positioning of the cochleostomy. The
tip of
the infusion unit is inserted into the cochleostomy and the tubing is secured
on the
=
wall of the middle ear by dental cement. The infusion unit is stored in a
30 subcutaneous pocket created behind the neck. The skin incision is closed
in layers
with 2-0 silk. This procedure is performed on both ears to avoid any
subsequent
imbalance or rotational behavior and to reduce the number of animals needed to
complete the experiment. The procedure takes approximately 30 minutes per
side.
Repeat surgery for changing the infusion unit is done under general anesthesia
35 one week after implantation. The procedure is done only through the
initial post-
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
36
auricular skin incision and does not involve re-entry into the middle ear
space. This
procedure takes 5-10 minutes per infusion unit. The post-surgical condition of
the
animal is monitored daily by checking activity, appetite, drinking, feces and
body
weight. The animal is sacrificed if it has lost more than 20% of its body
weight post-
surgery or exhibits severe rotational behavior or head tilt. No operative
discomfort
should occur and this procedure produces only slight postoperative discomfort.
As shown in Table 4 below, normal controls are given a solution of artificial
perilymph for one or two weeks. The hair cell loss group receives gentamycin
sulfate for one week to kill the hair cells with immediate sacrifice and two
week
recovery. Three concentrations are used that should provide a total loss of
HCs as
well as a graded loss of HCs. Cationic liposomes in 5% (w/v) dextrose solution
are
=
delivered for one to two weeks. The purpose of this group is to see if any
damage is
caused by lipofection. Lipofection after hair cell loss involves the
substitution of a
gentamycin containing pump to a lipid containing pump. Lipofection plus
antisense
after hair cell loss involves lipofecting FITC-antisense for another week.
Lipofection
plus antisense plus growth factors after damage are delivered for one week
followed
by the lipofection of antisense and growth factors for another week. Several
groups
involve combining antisense with growth factors, including TGF-alpha, insulin,
and
IGF-1. In all animals, a separate osmotic pump loaded with BrdU is implanted
subcutaneously to allow identification of mitotically active cells
Table 4
Animals Drugs Concentration Delivery time of drugs (d)
(n) (order) (range) Recovery time after (d)
12 GM/L 0.1-10 mg/ml 7
0, 7, 14
12 GM/L/FITC-AS 10-20 nM 7
7,14
12 GM/L/AS 10-20 nM 7
7,14
12 GM/FITZ-AS or AS 10-20 nM 7
7, 14
12 plus TGF-alpha 10-100 ng/ml 7
3,7, 14
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
37
Table 4
Animals Drugs Concentration Delivery time of
drugs (d)
(n) (order) (range) Recovery time after (d)
12 plus Insulin 1-10 ig/m1 7
3, 7, 14
12 plus IGF-1 10-100 ng/ml 7
3, 7, 14
Hair cell damage resulting from gentamycin is checked by myosin-VIIa
immunocytochemistry (a hair cell specific marker) and phalloidin
histochemistry
(F-actin marker) using a whole mount staining technique. The transfection
efficiency
of liposome and p271(iP1 antisense oligonucleotides is assessed by observing
for the
presence of FITC-labeled nuclei under epifluorescence. BrdU
immunohistochemistry is used to determine whether proliferation was induced
with
p271(1P1 antisense ON treatment. Selected specimens are analyzed under
electron
microscopy.
Some experiments are done using a double labeling with BrdU and myosin-
Vila to assess the number of new supporting cells versus the number of total
hair
cells.
Other experiments are done using double labeling with BrdU and vimentin to
assay the number of new supporting cells. Double labeling with BrdU, vimentin
and
myosin, distinguishes between the number of new hair cells compared to the
number
of new supporting cells. The baseline 'for organ of Corti supporting cell
proliferation
and auditory hair cell regeneration in mammals, is zero, making statistical
significance easier to attain with a low number of observed events using one-
way
ANOVAs.
Example 7
Assessment of Auditory Function after Ototoxic Insult and/or p271(iP1
Antisense
Treatment in Guinea Pigs
Auditory Brainstem Responses (ABR) is tested in guinea pigs after ototoxic
insult and/or p27KiP1 antisense treatment. ABR thresholds are compared within
the
same animal over time and across animals from the same or different groups
(pre-
and post-surgical). Significance is determined using one-way analysis of
variance
(ANOVA) for each stimulus frequency and intensity. Differences are considered
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
38
statistically significant for p-values <0.05. Previous studies have shown that
this
operation does not attenuate ABR responses post-operatively.
To record elicit ABRs, guinea pigs are anesthetized with Avertin (0.2 m1/10g
body weight/i.m. using a 1.2% stock solution). The active electrodes are
placed
subcutaneously near the external meatus of the ear (0.1-mm silver wire;
Narishige).
The dural reference electrode is placed in a drilled hole rostral to the
bregma (or an
insert earphone into the ear canal and the sound delivery tube secured to the
pinna
with surgical tape). The ground electrode (Ag/AgC12 pellet) is fixed on the
back.
The sound stimuli is either a broad band click of 100us duration or a 10 ms
tone burst
(1 ms rise/fall time). Guinea pigs are in a sound attenuated chamber (TDT
model
AC-1). The responses are measured and recorded via an Auditory Evoked Response
Workstation (SmartEP; Intelligent Hearing System). Guinea pigs are presented
with
a stimulus intensity series incremented from 20 to 85 dB in 5 dB steps for
both click
and tone burst stimuli. For tone bursts, a stimulus frequency series (1, 2, 4,
8, 16, 32
Hz) at a constant intensity of 50 dB is also used. Stimuli are repeated 5
times/s and a
total of 512 trials will be averaged. Threshold is defined as the lowest
intensity
capable of eliciting a replicable and visually detectable ABR.
Example 8
Improvement in Auditory Function in Amikacin Sulfate-Treated p27 Heterozygote
Mice
Experimental animals were treated the same as the mice described in the
experiments reported in Table 2 herein, except that, in the present example,
there was
an additional recovery time point at four weeks after amikacin treatment.
Auditory
function was measured using the auditory brainstem response (ABR) using
subcutaneous recording electrodes placed on three head points in Isofluorane
anesthetized mice. The sound intensity threshold was determined by presenting
single frequencies as different sound intensities (intensity measured in
decibels). The
higher the tone intensity that is required to elicit the ABR, the higher the
auditory
threshold, i.e., the worse the auditory function. The data set forth in
FIGURES 3-6
shows auditory improvement in five out of eight p27 heterozygotes (p<0.001).
In Table 2 herein, five out of ten p27 heterozygotes showed evidence of inner
cell proliferative regeneration as assessed by BrdU-labeling and morphologic
criteria.
These data showed that the majority of BrdU-labeled cells were supporting
cells, not
hair cells. The improved auditory function in the p27 heterozygotes may,
therefore,
CA 02412764 2007-06-28
39
be due to regeneration of supporting cells either alone, or in combination
with
regeneration of hair cells.
Example 9
The lipofection method of gene delivery and gene expression in the mouse organ
of
Corti culture system.
Lipofecting the organ of Corti utilized cochlear explants obtained from
postnatal
day 7-14 mice grown for a total of up to 8 days in vitro (DIV). Cultures were
grown
in defined culture media composed of Neurobasal Media with B27 supplement
(Gibco). Cultures were exposed to an arninoglycoside antibiotic (1 mM neomycin
sulfate for 48 hrs) which selectively killed the inner ear sensory hair-cells.
Eight
different lipid combinations were then tested from the Perfect Lipofection Kit
(InVitrogen). A bacterial plasmid encoding a betagalactosidase reporter gene
driven
by a CMV immediate/early gene promoter (InVitrogen) was delivered over a 6 hr
period. The cultures were aldehyde-fixed and processed for betagalactosidase
expression using x-gal histochemistry. X-gal labeling appeared in supporting-
cells
(54.3 +/- 15.3 labeled cells per 1000 pm length in the regions of the sensory
epithelium that once contained hair-cells, versus 5-10 labeled cells per 1000
gm
length in tissue that had not been lesioned).
Given the labor involved in detecting beta-galactosidase expression, the size
of the betagalactosidase encoding construct (i.e., 4.1 kbp) and the reduced
compatibility of this = technique with other desired ICC procedures, cultures
are
lipofected with a plasmid encoding Green Fluorescent Protein (GFP; Clonetech).
Detection of GFP requires a standard FITC filter set (excitation maximum 488
rim,
emission maximum 509 rim) and has been successfully transfected into cochlear
hair-
cells, supporting-cells and neurons using an AAV vector system.
Organ of Corti cultures established from P7-P14 Swiss Webster mice derived
from our breeding colony are lipofected using a variety of commercially
available
lipofection reagents (i.e., FuGENE Transfection Reagent; Boehringer-Mannheim).
These efficiences are compared against the transfection efficiences achieved
by the
InVitrogen Kit. The superior lipofection reagent and the superior lipid to DNA
ratio
(3:1, 6:1, 9: 1) are determined by counting the number of GFP-positive cells
within
the organ of Corti along a 1000 pm length taken at the middle of the explant.
Cells
are visualized using a Nikon Tm epifluorescence microscope equipped with a CCD
digital camera that outputs images directly into an imaging software program
where
cell counts are performed.
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
An aminoglycoside antibiotic lesion of the hair-cells is then be combined with
subsequent lipofection. Media containing 1 mM neomycin sulfate (Sigma) is
administered to kill the hair-cells over a 48 period in culture. Unlike
cultures derived
from neonatal mice, the two-week old mouse which has developed auditory
function
5 is more
easily affected by this concentration of neomycin resulting in a greater than
95% loss of hair-cells as determined by calbindin immunoreactivities and
plastic
cross-section analysis. The remaining supporting-cells are. lipofected for 6
hr with a
GFP encoding plasmid. Cultures are rinsed and grown in fresh media for an
additional 1-4 DIV for a total of 3-6 DIV. Cultures are aldehyde-fixed and GFP
is
10
visualized directly under epifluorescence. Several reports have demonstrated a
loss
of GFP fluorescence after aldehyde fixation. This may necessitate the use of a
commercially available GFP antibody (Clonetech) to enhance the fluorescence of
lipofected cells.
Example 10
15 Excision and In Vitro Culture of Mouse Inner Ear
The inner ear of a mouse was excised in the following manner. Postnatal day
7-14 Swiss Webster mice were decapitated and their skulls immersed in 70%
ethanol
for 5 min to disinfect. Under sterile conditions, the skull was cut into
halves along
the mid-sagittal axis and placed into 3 ml of culture media (NeuralbasalTM
Media at
20 pH 7.4;
Gibco) in a 35 mm plastic culture dish (Nalge Nunc International, 2000
North Aurora Road, Naperville, IL 60563). Using surgical forceps, the bony
inner
ear labyrinth was visualized and separated from the temporal bone. The
overlying
connective tissue, stapes bone, facial nerve and stapedial artery were
removed.
Using a fine forcep, a small hole about 2 mm in diameter was made through the
25 apical
turn of the lateral cochlear wall. This surgically created conduit, along with
the patent oval and round windows of the cochlea, permit ready diffusion of
the
culture media into the fluid-filled inner ear.
Typically, an inner ear excised and prepared in the foregoing manner is
transferred to the HARVTM or CCCVTM vessel which contains 50 or 55 ml of
30
NeuralbasalTM Media supplemented with either N2 or B27 media supplement (both
sold by Gibco-BRL, Catalogue number 17504-036), 10 U/ml of penicillin and
.25 [tg/ 1 of fungizone. The B27 supplement is sold as a 50X concentrate which
is
used at a working concentration of 0.5X (e.g., 550 I of 50X B27 stock
solution is
added to 55 ml of NeuralbasalTM Media). The N2 supplement stock solution is
100X
35 and is
used at a working concentration of 1X (e.g., 550 gl of 100X N2 stock solution
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
41
is added to 55 ml of NeuralbasalTM Media). The vessel is then placed in a
tissue
culture incubator at 37 C and in a 95% air/5% CO2 environment. The vessel is
then
rotated at 39 rpm for periods of 24-168 hr. 50% media changes are made every
48 hr. As few as 2 and as many as 12 inner ears have been successfully
cultured in
one vessel.
To lesion the inner ear sensory hair-cells, the inner ear is placed in
*NeuralbasalTm/N2 or B27 media that contain 1 mM neomycin sulfate (Sigma, P.O.
Box 14508, St. Louis, MO 63178) for 24-48 hr. After this culture period, the
media
is completely replaced with media devoid of neomycin.
Example 11
Culture Media
Table 5 shows the composition of NeuralbasalTM medium (1x) sold by Gibco.
All concentrations are working concentrations, i.e., the concentrations of the
components in the medium in which the fluid-filled sensory organ is incubated.
Table 5. NeuralbasalTM media composition
Component mg/liter uM
Inorganic salts
CaC12 (anhydrous) 200 1,800
Fe (NO3)3 9H20 0.1 0.2
KC1 400 5,360
MgC12 (anhydrous) 77.3 812
NaCl 3,000 51,300
NaHCO3 2,200 26,000
NaH2PO4H20 125 900
D-glucose 4,500 25,000
Phenol Red 8.1 23
HEPES 2,600 10,000
Sodium Pyruvate 25 230
Amino Acids
L-alanine 2.0 20
L-arginine HC1 84 400
L-asparagine H20 0.83 5
=
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
42
Component mg/liter M
L-cysteine 1.21 10
=
L-glutamate
Glycine 30 400
L-histidine HC1 H20 42 200
L-isoleucine 105 800
L-lysine HC1 146 5
L-methionine 30 200
L-phenylalanine 66 400
L-proline 7.76 67
L-serine 42 400
L-threonine 95 800
L-trptophan 16 80
L-tyrosine 72 400
L-valine 94 800
D-Ca pantothenate 4 8
Choline chloride 4 28
Folic acid 4 8
i-Inositol 7.2 40
Niacinamide 4 30
Pyridoxal HC1 4 20
Riboflavin 0.4 10
Thiamine HC1 4 10
Vitamin B12 0.34 0.2
The following antibiotics may be added to NeuralbasalTM medium.
Fungizone reagent (amphotericin B, 0.25 g/ml, and sodium desoxycholate,
0.25 gimp which is sold by Gibco-BRL, Catalog number 17504-036. Penicillin G
(10 units/m1) which is sold by Sigma, Catalog number P 3414. Neomycin sulfate
(1 mM), sold by Sigma, Catalog number N 6386. NeuralbasalTM medium may also
be supplemented with L-Glutamine (2mM).
Example 12
Assay for Sensory Epithelium Vitality During Long Term Culture
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
43
In the practice of one aspect of the present invention, the microgravitational
environment provided by the rotation of a culture vessel allows the sensory
epithelium of the inner ear to be maintained for prolonged periods of culture
(>168 hr.) without significant degradation or loss of the sensory hair-cells
or non-
sensory supporting-cells. Data demonstrating the continued vitality of the
sensory
hair cells during prolonged culture were obtained by labeling the sensory
epithelia
with a probe against F-actin (phalloidin-FITC) that labels the surfaces of
sensory and
non-sensory cells, and with a hair-cell specific antibody against calbindin, a
calcium
binding protein. Both labels were detected and photographed under
epifluorescence
microscopy.
Cross-sectional data indicated that the normal cytoarchitecture of the inner
ear
sensory epithelia. are maintained. For example, the Organ of Corti has several
fluid-
filled spaces called the tunnel of Corti and spaces of Nuel that are necessary
for
normal auditory function. These spaces occur between hair-cells and supporting-
cells and are maintained after prolonged periods of culture. In normal
gravitational
environments, (i.e., when the inner ear is floated without rotating the
culture vessel)
the sensory epithelia begin to degenerate. Without rotation, within 24 hr. the
hair-
cells are either completely missing or appear to be undergoing various
endstages of
cell death. After 48 hr., the supporting-cells are completely missing, or are
present
but with the total loss of the tunnel of Corti and spaces of Nuel. Rotating
the vessel
prevents this degradation and maintains normal cytoarchitecture.
While the preferred embodiment of the invention has been illustrated and
described, it will be appreciated that various changes can be made therein
without
departing from the spirit and scope of the invention.
=
CA 02412764 2003-04-30
-1-
SEQUENCE LISTING
<110> Otogene USA, Inc.
Otogene AG
<120> Stimulation of Cellular Regeneration and
Differentiation in the Inner Ear
<130> 356-160
<140> 2,412,764
<141> 2001-07-10
<150> US 09/614,099
<151> 2000-07-11
<160> 20
<170> PatentIn Ver. 2.0
<210> 1
<211> 82
<212> PRT
<213> Mus musculus
<400> 1
Gly Ala Gin Arg Glu Lys Met Asn Lys Pro Glu Leu Phe Asn Gly Gly
1 5 10 15
Glu Lys Lys Arg Lys Arg Thr Ser Ile Ala Ala Pro Glu Lys Arg Ser
20 25 30
Leu Glu Ala Tyr Phe Ala Val Gin Pro Arg Pro Ser Ser Glu Lys Ile
35 40 45
Ala Ala Ile Ala Glu Lys Leu Asp Leu Lys Lys Asn Val Val Arg Val
50 55 60
Trp Phe Cys Asn Gin Arg Gin Lys Gin Trp Arg Asn Lys Phe Ser Ala
65 70 75 80
Thr Tyr
<210> 2
<211> 70
<212> PRT
<213> Mus musculus
<400> 2
Leu Glu Ala Phe Ala Glu Arg Phe Lys Gin Arg Arg Ile Lys Leu Gly
1 5 10 15
Val Thr Gin Ala Asp Val Gly Ser Ala Leu Ala Asn Leu Lys Ile Pro
20 25 30
. = ,
CA 02412764 2002-12-19
VIM) 02/04605 PCT/US01/21793
-2-
Gly val Gly Ser Leu Ser Gin Ser Thr Ile Cys Arg Phe Glu Ser Leu
35 40 45
Thr Leu Ser His Asn Asn Met Ile Ala Leu Lys Pro Ile Leu Gin Ala
50 55 60
Trp Leu Glu Glu Ala Glu
65 70
<210>3
.<211> 1014
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (1)..(1014)
<400> 3
atg atg gcc atg aac tcc aag cag cct ttc ggc atg cac ccg gtg ctg 48
Met Met Ala Met Asn Ser Lys Gin Pro Phe Gly Met His Pro Val Leu
1 5 10 15
caa gaa ccc aaa ttc tcc agt ctg cac tct ggc tee gag gcc atg cgc 96
Gin Glu Pro Lys Phe Ser Ser Leu His Ser Gly Ser Glu Ala Met Arg
20 25 30
cga gte tgt etc cca gee ccg cag ctg cag ggt aat ata ttt gga agc 144
Arg Val Cys Leu Pro Ala Pro Gin Leu Gin Gly Asn Ile Phe Gly Ser
35 40 45
ttt gat gag age ctg ctg gca cgc gee gaa get ctg gcg gcg gtg gat 192
Phe Asp Glu Ser Leu Leu Ala Arg Ala Glu Ala Leu Ala Ala Val Asp
50 55 60
atc gtc ttc cac ggg aag aac cat ccg ttt aag ccc gac gcc ace tac 240
Ile Val Phe His Gly Lys Asn His Pro Phe Lys Pro Asp Ala Thr Tyr
65 70 75 80
cat acc atg age age gtg ccc tgc acg tee act tcg tee ace gtg ccc 288
His Thr Met Ser Ser Val Pro Cys Thr Ser Thr Ser Ser Thr Val Pro
85 90 95
ate tee cac cca get gcg etc ace tea cac cct cac cac gcc gtg cac. 336
Ile Ser His Pro' Ala Ala Leu Thr Ser His Pro His His Ala Val His
100 105 110
cag ggc etc gaa ggc gac ctg ctg gag cac ate tcg ccc acg ctg agt 384
Gin Gly Leu Glu Gly Asp Leu Leu Glu His Ile Ser Pro Thr Leu Ser
llg 120 125
gtg ac ggc ctg ggc get ccg gaa cac tcg gtg atg ccc gca cag atc 432
Val Ser Gly Leu Gly Ala Pro Glu His Ser Val Met Pro Ala Gin Ile
130 135 140
CA 02412764 2003-04-30
-3-
cat cca cac cac ctg ggc gcc atg ggc cac ctg cac cag gcc atg ggc 480
His Pro His His Leu Gly Ala Met Gly His Leu His Gin Ala Met Gly
145 150 155 160
atg agt cac ccg cac acc gtg gcc cct cat agc gcc atg cct gca tgc 528
Met Ser His Pro His Thr Val Ala Pro His Ser Ala Met Pro Ala Cys
165 170 175
ctc agc gac gtg gag tca gac ccg cgc gag ctg gaa gcc ttc gcc gag 576
Leu Ser Asp Val Glu Ser Asp Pro Arg Glu Leu Glu Ala Phe Ala Glu
180 185 190
cgc ttc aag cag cgg cgc atc aag ctg ggg gtg acc cag gcg gac gtg 624
Arg Phe Lys Gin Arg Arg Ile Lys Leu Gly Val Thr Gin Ala Asp Val
195 200 205
ggc gcg gct ctg gct aat ctc aag atc ccc ggc gtg ggc tcg ctg agc 672
Gly Ala Ala Leu Ala Asn Leu Lys Ile Pro Gly Val Gly Ser Leu Ser
210 215 220
caa agc acc atc tgc agg ttc gag tct ctc act ctc tcg cac aac aac 720
Gin Ser Thr Ile Cys Arg Phe Glu Ser Leu Thr Leu Ser His Asn Asn
225 230 235 240
atg atc gct ctc aag ccg gtg ctc cag gcc tgg ttg gag gag gcc gag 768
Met Ile Ala Leu Lys Pro Val Leu Gin Ala Trp Leu Glu Glu Ala Glu
245 250 255
gcc gcc tac cga gag aag aac agc aag cca gag ctc ttc aac ggc agc 816
Ala Ala Tyr Arg Glu Lys Asn Ser Lys Pro Glu Leu Phe Asn Gly Ser
260 265 270
gaa cgg aag cgc aaa cgc acg tcc atc gcg gcg ccg gag aag cgt tca 864
Giu Arg Lys Arg Lys Arg Thr Ser Ile Ala Ala Pro Glu Lys Arg Ser
275 280 285
ctc gag gcc tat ttc gct atc cag cca cgt cct tca tct gag aag atc 912
Leu Glu Ala Tyr Phe Ala Ile Gln Pro Arg Pro Ser Ser Glu Lys Ile
290 295 300
gcg gcc atc gct gag aaa ctg gac ctt aaa aag aac gtg gtg aga gtc 960
Ala Ala Ile Ala Glu Lys Leu Asp Leu Lys Lys Asn Val Val Arg Val
305 310 315 320
tgg ttc tgc aac cag aga cag aaa cag aaa cga atg aag tat tcg gct 1008
Trp Phe Cys Asn Gin Arg Gin Lys Gin Lys Arg Met Lys Tyr Ser Ala
325 330 335
gtc cac 1014
Val His
<210> 4
<211> 338
in=umvitn.**========rn... ,
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
-4-
<212> PRT
<213> Homo sapiens
<400> 4
Met Met Ala Met Asn Ser Lys Gin Pro Phe Gly Met His Pro Val Leu
1 5 10 15
Gin Glu Pro Lys Phe Ser Ser Leu His Ser Gly Ser Glu Ala Met Arg
20 25 30
Arg Val Cys Leu Pro Ala Pro Gin Leu Gin Gly Asn Ile Phe Gly Ser
35 40 45
Phe Asp Glu Ser Leu Leu Ala Arg Ala Glu Ala Leu Ala Ala Val Asp
50 55 60
Ile Val Phe His Gly Lys Asn His Pro Phe Lys Pro Asp Ala Thr Tyr
65 70 75 80
His Thr Met Ser Ser Val Pro Cys Thr Ser Thr Ser Ser Thr Val Pro
85 90 - 95
Ile Ser His Pro Ala Ala Leu Thr Ser His Pro His His Ala Val His
100 105 110
Gin Gly Leu Glu Gly Asp Leu Leu Glu His Ile Ser Pro Thr Leu Ser
115 120 125
Val Ser Gly Leu Gly Ala Pro Glu His Ser Val Met Pro Ala in Ile
130 135 , 140
His Pro His His Leu Gly Ala Met Gly His Leu His Gin Ala Met Gly
145 150 155 160
Met Ser His Pro His Thr Val Ala Pro His Ser Ala Met Pro Ala Cys
165 170 175
Leu Ser Asp Val Glu Ser Asp Pro Arg Glu Leu Glu Ala Phe Ala Glu
180 185 190
Arg Phe Lys Gin Arg Arg Ile Lys Leu Gly Val Thr Gin Ala Asp Val
195 200 205
Gly Ala Ala Leu Ala Asn Leu Lys Ile Pro Gly Val Gly Ser Leu Ser
210 215 220
Gin Ser Thr Ile Cys Arg Phe Glu Ser Leu Thr Leu Ser His Asn Asn
225 230 235 240
Met Ile Ala Leu Lys Pro Val Leu Gin Ala Trp Leu Glu Glu Ala Glu
245 250 255
Ala Ala Tyr Arg Glu Lys Asn Ser Lys Pro Glu Leu Phe Asn Gly Ser
260 265 270
Glu Arg Lys Arg Lys Arg Thr Ser Ile Ala Ala Pro Glu Lys Arg Ser
CA 02412764 2002-12-19
VIM) 02/04605 PCT/US01/21793
-5-
275 280 285
Leu Glu Ala Tyr Phe Ala Ile Gin Pro Arg Pro Ser Ser Glu Lys Ile
290 295 300
Ala Ala Ile Ala Glu Lys Leu Asp Leu Lys Lys Asn Val Val Arg Val
305 310 315 320
Trp Phe Cys Asn Gin Arg Gin Lys Gin Lys Arg Met Lys Tyr Ser Ala
325 330 335
Val His
<210> 5
<211> 31
<212> DNA
<213> Artificial Sequence =
<220>
<223> Description of Artificial Sequence:
oligonucleotide'
<220>
<221> misc_feature
<222> (1)..(31)
<223> Oligonucleotide for identifying POU4F3 homologs
<400> 5
tagaagtgca gggcacgctg ctcatggtat g 31
=
<210> 6
<211> 1355
<212> DNA
<213> Mus musculus
<220>
<221> CDS
<222> (41)..(1084)
<400> 6
ccgcaggagc cgtccatcac caatcagcca gccttcgacc atg ggc atg tcc gac 55
Met Gly Met Ser Asp
, 1 5
gtg tac ctc'cgc agc aga aca gcg atg gaa cgc ttg gcc tcc agc gat 103
Val Tyr Leu Arg Ser Arg Thr Ala Met Glu Arg Leu Ala Ser Sex Asp
10 15 20
acc ttc cca gtg ata gcg cgt agc agc gcc tgc cgc agc ctc ttc ggg 151
Thr Phe Pro Val Ile Ala Arg Ser Ser Ala Cys Arg Ser Leu Phe Gly
25 30 35
cct gta gac cac gag gag ctg ggc cgc gag ctg cgg atg cgc ctg gcc 199
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
-6-
Pro Val Asp His Glu Glu Leu Gly Arg Glu Leu Arg Met Arg Leu Ala
40 45 50
gag ctg aac gcc gag gac cag aac cgc tgg gac ttc aac ttc cag cag 247
Glu Leu Asn Ala Glu Asp Gln Asn Arg Trp Asp Phe Asn Phe Gln Gln
55 60 65
gat gtg cct ctt cga ggc cot ggt cgt ctg cag tgg atg gag gtg gac 295
Asp Val Pro Leu Arg Gly Pro Gly Arg Leu Gln Trp Met Glu Val Asp
70 75 80 85
ago gag tct gtg ccc gcc ttc tac cgc gag acg gtg cag gtg ggg cgc 343
Ser Glu Ser Val Pro Ala Phe Tyr Arg Glu Thr Val Gln Val Gly Arg
90 95 100
tgt cgc ctg cag ctg ggg ccc cgg cca ccc ccg gtg gcc gtg gct gtc 391
Cys Arg Leu Gln Leu Gly Pro Arg Pro Pro Pro Val Ala Val Ala Val
105 110 115
atc ccg cgt tct ggg ccg ccg gct ggc gag gcc ccc gac ggc cta gag 439
Ile Pro Arg Ser Gly Pro Pro Ala Gly Glu Ala Pro Asp Gly Leu Glu
120 125 130
=
gag gcg cct gag cag ccg ccc agc gcc cca gcc tcg gcc gtg gtc gcg 487
Glu Ala Pro Glu Gln Pro Pro Ser Ala Pro Ala Ser Ala Val Val Ala
135 140 145
gac gcc acc cca ccc gcg acc ccg gcc ccg gct tca gat ctg acc tca 535
Asp Ala Thr Pro Pro Ala Thr Pro Ala Pro Ala Ser Asp Leu Thr Ser
150 155 160 165
gac cca att ccg gag gtg acc ctg gtc gcg acc tcc gac ccg act ccg 583
Asp Pro Ile Pro Glu Val Thr Leu Val Ala Thr Ser Asp Pro Thr Pro
170 175 180
gac ccg atc ccg gac gcg aac ccg gac gtg gcg act cgg gac ggc gag 631
Asp Pro Ile Pro Asp Ala Asn Pro Asp Val Ala Thr Arg Asp Gly Glu
185 190 195
gaa cag gtc cct gag cag gtc tct gag cag ggc gag gag tcg ggt gct 679
Glu Gln Val Pro Glu Gln Val Ser Glu Gin Gly Glu Glu Ser Gly Ala
200 205 210
gag ccg ggt gat gag ctg gga act gag ccg gtc tct gag cag ggc gag 727
Glu Pro Gly Asp Glu Leu Gly Thr Glu Pro Val Ser Glu Gln Gly Glu
215 220 225
gag cag ggc gca gag ccg gtc gag gag aag gac gag gag cog gag gag 775
Glu Gln Gly Ala Glu Pro Val Glu Glu Lys Asp Glu Glu Pro Glu Glu
230 235 240 245
gag cag ggc gcg gag ccg gtc gag gag cag ggt gcg gag cog gtc gag 823
Glu Gln Gly Ala Glu Pro Val Glu Glu Gln Gly Ala Glu Pro Val Glu
250 255 260
gag cag aat ggg gag ccg gtc gag gag cag gac gag aat caa gag cag 871
CA 02412764 2003-04-30
Giu Gin Asn Gly Glu Pro Val Glu Glu Gin Asp Glu Asn Gin Glu Gin
265 270 275
cgc ggc cag gag ctg aag gac cag cct ctc tcg ggg att cca gga cgt 919
Arg Gly Gin Glu Leu Lys Asp Gin Pro Leu Ser Gly Ile Pro Gly Arg
280 285 290
cct gca ccc ggg act gct gcg gcc aat gcg aac gac ttc ttc gcc aag 967
Pro Ala Pro Gly Thr Ala Ala Ala Asn Ala Asn Asp Phe Phe Ala Lys
295 300 305
cgc aag aga act gcg cag gag aac aag gcg tcg aac gac gtc cct cca 1015
Arg Lys Arg Thr Ala Gin Glu Asn Lys Ala Ser Asn Asp Val Pro Pro
310 315 320 325
ggg tgt ccc tct cca aac gtg gct cct ggg gtg ggc gcg gtg gag cag 1063
Gly Cys Pro Ser Pro Asn Val Ala Pro Gly Val Gly Ala Val Glu Gin
330 335 340
acc ccg cgc aaa cgt ctg aga tgagttagtt tagaggctaa cggccagaga 1114
Thr Pro Arg Lys Arg Leu Arg
345
gaacttgctg ggcatctggg cagcggacga tggaagaact ctgggcttcg gctgggacct 1174
ttcgttcatg tagcaggaac cggagatggt tgcgtagagc agcccacggt tttgtggaaa 1234
tctgaaaact gtgcaatgta ttgagaacac tctgtaccat gtgcaaggag tacgctggtc 1294
ccaaggtgta aagctttaaa tcatttatgt aaaatgttta atctctactc gctctcagtg 1354
1355
<210> 7
<211> 348
<212> PRT
<213> Mus musculus
<400> 7
Met Gly Met Ser Asp Val Tyr Leu Arg Ser Arg Thr Ala Met Glu Arg
1 5 10 15
Leu Ala Ser Ser Asp Thr Phe Pro Val Ile Ala Arg Ser Ser Ala Cys
20 25 30
Arg Ser Leu Phe Gly Pro Val Asp His Glu Giu Leu Gly Arg Glu Leu
35 40 45
CA 02412764 2002-12-19
VIM) 02/04605
PCT/US01/21793
-8-
Arg Met Arg Leu Ala Glu Leu Asn Ala Glu Asp Gin Asn Arg Trp Asp
50 55 60
Phe Asn Phe Gin Gin Asp Val Pro Leu Arg Gly Pro Gly Arg Leu Gin
65 70 75 80
Trp Met Glu Val Asp Ser Glu Ser Val Pro Ala Phe Tyr Arg Glu Thr
85 90 95
Val Gin Val Gly Arg Cys Arg Leu Gin Leu Gly Pro Arg Pro Pro Pro
100 105 110
Val Ala Val Ala Val Ile Pro Arg Ser Gly Pro Pro Ala Gly Glu Ala
115 120 125
Pro Asp Gly Leu Glu Glu Ala Pro Glu Gin Pro Pro Ser Ala Pro Ala
130 135 140
' Ser Ala Val Val Ala Asp Ala Thr Pro Pro Ala Thr Pro Ala Pro Ala
145 150 155 160
Ser Asp Leu Thr Ser Asp Pro Ile Pro Glu Val Thr Leu Val Ala Thr
165 170 175
Ser Asp Pro Thr Pro Asp Pro Ile Pro Asp Ala Asn Pro Asp Val Ala
180 185 190
Thr Arg Asp Gly Glu Glu Gin Val Pro Glu Gin Val Ser Glu Gin Gly
195 200 205
Glu Glu Ser Gly Ala Glu Pro Gly Asp Glu Leu Gly .Thr Glu Pro Val
210 215 220
Ser Glu Gin Gly Glu Glu Gin Gly Ala Glu Pro Val Glu Glu Lys Asp
225 230 235 240
Glu Glu Pro Glu Glu Glu Gin Gly Ala Glu Pro Val Glu Glu Gin Gly
245 250 255
Ala Glu Pro Val Glu Glu Gin Asn Gly Glu Pro Val Glu Glu Gin Asp
260 265 270
Glu Asn Gin Glu Gln Arg Gly Gin Glu Leu Lys Asp Gin Pro Leu Ser
275 280 285
Gly Ile Pro Gly Arg Pro Ala Pro Gly Thr Ala Ala Ala Asn Ala Asn
290 295 300
Asp Phe Phe Ala Lys Arg Lys Arg Thr Ala Gin Glu Asn Lys Ala Ser
305 310 315 320
Asn Asp Val Pro Pro Gly Cys Pro Ser Pro Asn Val Ala Pro Gly Val
325 330 335
Gly Ala Val Glu Gin Thr Pro Arg Lys Arg Leu Arg
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
-9-
340 345
<210> 8
<211> 597
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (1)..(594)
<400> 8
atg tca aac gtg cga gtg tct aac ggg agc cct agc ctg gag cgg atg 48
Met Ser Asn Val Arg Val Ser Asn Gly Ser Pro Ser Leu Glu Arg Met
1 5 10 15
gac gcc agg cag gcg gag cac ccc aag ccc tcg gcc tgc agg aac ctc 96
Asp Ala Arg Gin Ala Glu His Pro Lys Pro Ser Ala Cys Arg Asn Leu
20 25 30
ttc ggc ccg gtg gac cac gaa gag tta acc cgg gac ttg gag aag cac 144
Phe Gly Pro Val Asp His Glu Glu Leu Thr Arg Asp Leu Glu Lys His
35 40 45
tgc aga gac atg gaa gag gcg agc cag cgc aag tgg aat ttc gat ttt 192
Cys Arg Asp Met Glu Glu Ala Ser Gin Arg Lys Trp Asn Phe Asp Phe
50 55 60
cag aat cac aaa ccc cta gag ggc aag tac gag tgg caa gag gtg gag 240
Gin Asn His Lys Pro Leu Glu Gly Lys Tyr Glu.Trp Gin Glu Val Glu
65 70 75 80
aag ggc agc ttg ccc gag ttc tac tac aga ccc ccg cgg ccc ccc aaa 288
Lys Gly Ser Leu Pro Glu Phe Tyr Tyr Arg Pro Pro Arg Pro Pro Lys
85 90 95
ggt gcc tgc aag gtg ccg gcg cag gag agc cag gat gtc agc ggg agc 336
Gly Ala Cys Lys Val Pro Ala Gin Glu Ser Gin Asp Val Ser Gly Ser
100 105 110
cgc ccg gcg gcg cct tta att ggg gct ccg gct aac tct gag gac acg 384
Arg Pro Ala Ala Pro Leu Ile Gly Ala Pro Ala Asn Ser Glu Asp Thr
115 120 125
cat ttg gtg gac cca aag act gat ccg tcg gac agc cag acg ggg tta 432
His Leu Val Asp Pro Lys Thr Asp Pro Ser Asp Ser Gin Thr Gly Leu
130 135 140
gcg gag caa tgc gca gga ata agg aag cga cct gca acc gac gat tct 480
Ala Glu Gin Cys Ala Gly Ile Arg Lys Arg Pro Ala Thr Asp Asp Ser
145 150 155 160
=
tct act caa aac aaa aga gcc aac aga aca gaa gaa aat gtt tca gac 528
Ser Thr Gin Asn Lys Arg Ala Asn Arg Thr Glu Glu Asn Val Ser Asp
165 170 ' 175
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
-10-
ggt tcc cca aat gcc ggt tot gtg gag cag acg ccc aag aag cct ggc 576
Gly Ser Pro Asn Ala Gly Ser Val Glu Gin Thr Pro Lys Lys Pro Gly
180 185 190
ctc aga aga cgt caa acg taa 597
Leu Arg Arg Arg Gin Thr
195
<210> 9
<211> 198
<212> PRT
<213> Homo sapiens =
<400> 9
Met Ser Asn Val Arg Val Ser Asn Gly Ser Pro Ser Leu Glu Arg Met
1 5 10 15
Asp Ala Arg Gin Ala Glu His Pro Lys Pro Ser Ala Cys Arg Asn Leu
20 25 30
Phe Gly Pro Val Asp His Glu Glu Leu Thr Arg Asp Leu Glu Lys His
35 40 45
Cys Arg Asp Met Glu Glu Ala Ser Gin Arg Lys Trp Asn Phe Asp Phe
50 55 60
Gin Asn His Lys Pro Leu Glu Gly Lys Tyr Glu Trp Gan Glu Val Glu
65 70 75 80
Lys Gly Ser Leu Pro Glu Phe Tyr Tyr Arg Pro Pro Arg Pro Pro Lys
85 90 95 ,
Gly Ala Cys Lys Val Pro Ala Gin Glu Ser Gin Asp Val Ser Gly Ser'
* 100 105 110
Arg Pro Ala Ala Pro Leu Ile Gly Ala Pro Ala Asn Ser Glu Asp Thr
115 120 125
His Leu Val Asp Pro Lys Thr Asp Pro Ser Asp Ser Gin Thr Gly Leu
130 135 140
Ala Glu Gin Cys Ala Gly Ile Arg Lys Arg Pro Ala Thr Asp Asp Ser
145 150 155 160
=
Ser Thr Gin Asn Lys Arg Ala Asn Arg Thr Glu Glu Asn Val Ser Asp
165 170 175
Gly Ser Pro Asn Ala Gly Ser Val Glu Gin Thr Pro Lys Lys Pro Gly
180 185 190
Leu Arg Arg Arg Gin Thr
195
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
-11-
<210> 10
<211> 2121
<212> DNA
<213> Homo sapiens .
<220>
<221> CDS
<222> (76)..(567).
<400> 10
gccgaagtca gttccttgtg gagccggagc tgggcgcgga ttcgccgagg caccgaggca 60
ctcagaggag gcgcc atg tca gaa ccg gct ggg gat gtc cgt cag aac cca 111
Met Ser Glu Pro Ala Gly Asp Val Arg Gin Asn Pro
1 5 10
tgc ggc agc aag gcc tgc cgc cgc ctc ttc ggc cca gtg gac agc gag 159
Cys Gly Ser Lys Ala Cys Arg Arg Leu Phe Gly Pro Val Asp Ser Glu
15 20 25
cag ctg agc cgc gac tgt gat gcg cta atg gcg ggc tgc atc cag gag 207
Gin Leu Ser Arg Asp Cys Asp Ala Leu Met Ala Gly Cys Ile Gin Glu
30 35 40
gcc cgt gag cga tgg aac ttc gac ttt gtc acc gag aca cca ctg gag 255
Ala Arg Glu Arg Trp Asn Phe Asp Phe Val Thr Glu Thr Pro Leu Glu
45 50 55 60
ggt gac ttc gcc tgg gag cgt gtg cgg ggc ctt ggc ctg ccc aag ctc 303
Gly Asp Phe Ala Trp Glu Arg Val Arg Gly Leu Gly Leu Pro Lys Leu
65 70 75
tac ctt ccc acg ggg ccc cgg cga ggc cgg gat gag ttg gga gga ggc 351
Tyr Leu Pro Thr Gly Pro Arg Arg Gly Arg Asp Glu Leu Gly Gly Gly
80 85 90
agg cgg cct ggc acc tca cct gct ctg ctg cag ggg aca gca gag gaa 399
Arg Arg Pro Gly Thr Ser Pro Ala Leu Leu Gin Gly Thr Ala Glu Glu
95 100 105
gac cat gtg gac ctg tca ctg tct tgt acc ctt gtg cct cgc tca ggg 447
Asp His Val Asp Leu Ser Leu Ser Cys Thr Leu Val Pro Arg Ser Gly
110 115 120
gag cag gct gaa ggg tcc cca ggt gga c'ct gga gac tct cag ggt cga 495
Glu Gin Ala Glu Gly Ser Pro Gly Gly Pro Gly Asp Ser Gin Gly Arg
125 130 135 140
aaa cgg cgg cag acc agc atg aca gat ttc tac cac tcc aaa cgc cgg 543
Lys Arg Arg Gin Thr Ser Met Thr Asp Phe Tyr His Ser Lys Arg Arg
145 150 155
ctg atc ttc tcc aag agg aag ccc taatccgccc acaggaagcc tgcagtcctg 597
Leu Ile Phe Ser Lys Arg Lys Pro
160
=
CA 02412764 2003-04-30
-12--
gaagcgcgag ggcctcaaag gcccgctcta catcttctgc cttagtctca gtttgtgtgt 657
cttaattatt atttgtgttt taatttaaac acctcctcat gtacataccc tggccgcccc 717
ctgcccccca gcctctggca ttagaattat ttaaacaaaa actaggcggt tgaatgagag 777
gttcctaaga gtgctgggca tttttatttt atgaaatact atttaaagcc tcctcatccc 837
gtgttctcct tttcctctct cccggaggtt gggtgggccg qcttcatgcc agctacttcc 897
tcctccccac ttgtccgctg ggtggtaccc tctggagggg tgtggctcct tcccatcgct 957
gtcacaggcg gttatgaaat tcaccccctt tcctggacac tcagacctga attctttttc 1017
atttgagaag taaacagatg gcactttgaa ggggcctcac cgagtggggg catcatcaaa 1077
aactttggag tcccctcacc tcctctaagg ttgggcaggg tgaccctgaa gtgagcacag 1137
cctagggctg agctggggac ctggtaccct cctggctctt gatacccccc tctgtcttgt 1197
gaaggcaggg ggaaggtgqg gtactggagc agaccacccc gcctgccctc atggcccctc 1257
tgacctgcac tggggagccc gtctcagtgt tgagcctttt ccctctttgg ctcccctgta 1317
ccttttgagg agccccagct tacccttctt ctccagctgg gctctgcaat tcccctctgc 1377
tgctgtccct cccccttgtc tttcccttca gtaccctctc atgctccagg tggctctgag 1437
gtgcctgtcc cacccccacc cccagctcaa tggactggaa ggggaaggga cacacaagaa 1497
gaagggcacc ctagttctac ctcaggcagc tcaagcagcg accgccccct cctctagctg 1557
tgggggtgag ggtcccatgt ggtggcacag gcccccttga gtggggttat ctctgtgtta 1617
ggggtatatg atgggggagt agatctttct aggagggaga cactggcccc tcaaatcgtc 1677
cagcgacctt cctcatccac cccatccctc cccagttcat tgcactttga ttagcagcgg 1737
aacaaggagt cagacatttt aagatggtgg cagtagaggc tatggacagg gcatgccacg 1797
tgggctcata tggggctggg agtagttgtc tttcctggca ctaacgttga gcccctggag 1857
4.1101110... me W.,.
___________________ No.
CA 02412764 2003-04-30
-13-
gcactgaagt gcttagtgta cttggagtat tggggtctga ccccaaacac cttccagctc 1917
ctgtaacata ctggcctgga ctgttttctc tcggctcccc atgtgtcctg gttcccgttt 1977
ctccacctag actgtaaacc tctcgagggc agggaccaca ccctgtactg ttctgtgtct 2037
ttcacagctc ctcccacaat gctgaatata cagcaggtgc tcaataaatg attcttagtg 2097
actttaaaaa aaaaaaaaaa aaaa 2121
<210> 11
<211> 164
<212> PRT
<213> Homo sapiens
<400> 11
Met Ser Glu Pro Ala Gly Asp Val Arg Gin Asn Pro Cys Gly Ser Lys
1 5 10 15
Ala Cys Arg Arg Leu Phe Gly Pro Val Asp Ser Glu Gin Leu Ser Arg
20 25 30
Asp Cys Asp Ala Leu Met Ala Gly Cys Ile Gin Glu Ala Arg Glu Arg
35 40 45
Trp Asn Phe Asp Phe Val Thr Glu Thr Pro Leu Glu Gly Asp Phe Ala
50 55 60
Trp Glu Arg Val Arg Gly Leu Gly Leu Pro Lys Leu Tyr Leu Pro Thr
65 70 75 80
Gly Pro Arg Arg Gly Arg Asp Glu Leu Gly Gly Gly Arg Arg Pro Gly
85 90 95
Thr Ser Pro Ala Leu Leu Gin Gly Thr Ala Glu Glu Asp His Val Asp
100 305 110
Leu Ser Leu Ser Cys Thr Leu Val Pro Arg Ser Gly Glu Gin Ala Glu
115 120 125
Gly Ser Pro Gly Gly Pro Gly Asp Ser Gin Gly Arg Lys Arg Arg Gin
130 135 140
Thr Ser Met Thr Asp Phe Tyr His Ser Lys Arg Arg Leu Ile Phe Ser
145 150 155 160
Lys Arg Lys Pro
-------
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
-14-
<210> 12
<211> 706
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (1)..(498)
<400> 12
atg ctg ctg gag gag gtt cgc gcc ggc gac cgg ctg agt ggg gcg gcg 48
Met Leu Leu Glu Glu Val Arg Ala Gly Asp Arg Leu Ser Gly Ala Ala
1 5 10 15
gcc cgg ggc gac gtg cag gag gtg cgc cgc ctt ctg cac cgc gag ctg 96
Ala Arg Gly Asp Val Gln Glu Val Arg Arg Leu Leu His Arg Glu Leu
20 25 30
gtg cat ccc gac gcc ctc aac cgc ttc ggc aag acg gcg ctg cag gtc 144
Val His Pro Asp Ala Leu Asn Arg Phe Gly Lys Thr Ala Leu Gln Val
35 40 45
atg atg ttt ggc agc acc gcc atc gcc ctg gag ctg ctg aag caa ggt 192
Met Met Phe Gly Ser Thr Ala Ile Ala Leu Glu Leu Leu Lys Gln Gly
50 55 60
gcc agc ccc aat gtc cag gac acc tcc ggt acc agt cca gtc cat gac 240
Ala Ser Pro Asn Val Gln Asp Thr Ser Gly Thr Ser Pro Val His Asp
65 70 75 80
gca gcc cgc act gga Ãtd ctg gac acc ctg aag gtc cta gtg gag cac 288
Ala Ala Arg Thr Gly Phe Leu Asp Thr Leu Lys ValLeu Val Glu His
85 90 95
ggg gct gat gtc aac gtg cct gat ggc acc ggg gca ctt cca atc cat 336
Gly Ala Asp Val Asn Val Pro Asp Gly Thr Gly Ala Leu Pro Ile His
100 105 110
ctg gca gtt caa gag ggt cac act gct gtg gtc agc ttt ctg gca gct 384
Leu Ala Val Gln Glu Gly His Thr Ala Val Val Ser Phe Leu Ala Ala
115 120 125
gaa tct gat ctc cat cgc agg gac gcc agg ggt ctc aca ccc ttg gag 432
Glu Ser Asp Leu His Arg Arg Asp Ala Arg Gly Leu Thr Pro Leu Glu
130 135 140
ctg gca ctg cag aga ggg gct cag gac ctc gtg gac atc ctg cca ggc 480
Leu Ala Leu Gln Arg Gly Ala Gln Asp Leu Val Asp Ile Leu Pro Gly
145 150 155 160
cac atg gtg gcc ccg ctg tgatctgggg tcaccctctc cagcaagaga 528
His Met Val Ala Pro Leu
165
acccccccgt ggttatgtat cagaagagag gggaagaaac actttctctt cttgtttctc 588
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
-15-
ctgcccactg ctgcagtagg ggaggagcac agtttgtggc ttataggtgt tggttttggg 648
ggtgtgagtg tttgggggac gttctcattt gtttttctca ctccttttgg tgtgttgg 706
<210> 13
<211> 166
<212> PRT
<213> Homo sapiens
<400> 13
Met Leu Leu Glu Glu Val Arg Ala Gly Asp Arg Leu Ser Gly Ala Ala
1 5 10 15
Ala Arg Gly Asp Val Gln Glu Val Arg Arg Leu Leu His Arg Glu Leu
20 25 30
Val His Pro Asp Ala Leu Asn Arg Phe Gly Lys Thr Ala Leu Gln Val
35 40 45
Met Met Phe Gly Ser Thr Ala Ile Ala Leu Glu Leu Leu Lys Gln Gly
50 55 60
Ala Ser Pro Asn Val Gln Asp Thr Ser Gly Thr Ser Pro Val His Asp
65 70 75 80
Ala Ala Arg Thr Gly Phe Leu Asp Thr Leu Lys Val Leu Val Glu His
85 90 95
Gly Ala Asp Val Asn Val Pro Asp Gly Thr Gly Ala Leu Pro Ile His
100 105 110
Leu Ala-Val Gln Glu Gly His Thr Ala Val Val Ser Phe Leu Ala Ala
115 120 125
Glu Ser Asp Leu His Arg Arg Asp Ala Arg Gly Leu Thr Pro Leu Glu
130 135 140
Leu Ala Leu Gln Arg Gly Ala Gln Asp Leu Val Asp Ile Leu Pro Gly
145 150 155 160
His Met Val Ala Pro Leu
165
<210> 14
<211> 600
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (94)..(597)
<400> 14
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
-16-
ccgatgccat catgcagcct ggttaggagc aaaggaaagg ggaaaaagaa aaacgactaa GO
ttcatctttt cctgatcgtc aggaccctaa aga atg gcc gag cct tgg ggg aac 114
Met Ala Glu Pro Trp Gly Asn
1 5
gag ttg gcg tcc gca gct gcc agg ggg gac cta gag caa ctt act agt 162
Glu Leu Ala Ser Ala Ala Ala Arg Gly Asp Leu Glu Gin Leu Thr Ser
15 20
ttg ttg caaaat aat gta aac gtc aat gca caa aat gga ttt gga agg 210
Leu Leu Gln Asn Asn Val Asn Val Asn Ala Gin Asn Gly Phe Gly Arg
25 30 35
act gcg ctg cag gtt atg aaa ctt gga aat ccc gag att gcc agg aga 258
Thr Ala Leu Gin Val Met Lys Leu Gly Asn Pro Glu Ile Ala Arg Arg
40 45 50 55
ctg cta ctt aga ggt gct aat ccc gat ttg aaa gac cga act ggt ttc 306
Leu Leu Leu Arg Gly Ala Asn Pro Asp Leu Lys Asp Arg Thr Gly Phe
60 65 70
gct gtc att cat gat gcg gcc aga gca ggt ttc ctg gac act tta cag 354
Ala Val Ile His Asp Ala Ala Arg Ala Gly Phe Leu Asp Thr Leu Gin
75 80 85
act ttg ctg gag ttt caa gct gat gtt aac atc gag gat aat gaa ggg 402
Thr Leu Leu Glu Phe Gin Ala Asp Val Asn Ile Glu Asp Asn Glu Gly
90 95 100
aac ctg ccc ttg cac ttg gct gcc aaa gaa ggc cac ctc cgg gtg gtg 450
Asn Leu Pro Leu His Leu Ala Ala Lys Glu Gly His Leu Arg Val Val
105 110 115
. gag ttc ctg gtg aag cac acg gcc agc aat gtg ggg cat cgg aac cat 498
Glu Phe Leu Val Lys His Thr Ala Ser Asn Val Gly His Arg Asn His
120 125 130 135
aag ggg gac acc gcc tgt gat ttg gcc agg ctc tat ggg agg aat gag 546
Lys Gly Asp Thr Ala Cys Asp Leu Ala Arg Leu Tyr Gly Arg Asn Glu
140 145 150
gtt gtt agc ctg atg cag gca aac ggg gct ggg gga gcc.aca aat ctt 594
Val Val Ser Leu Met Gin Ala Asn Gly Ala Gly Gly Ala Thr Asn Leu
155160 165
=
caa taa 600
Gin
<210> 15
<211> 168
<212> PRT
<213> Homo sapiens
<400> 15
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
-17-
Met Ala Glu Pro Trp Gly Asn Glu Leu Ala Ser Ala Ala Ala Arg Gly
1 5 10 15
Asp Leu Glu Gln Leu Thr Ser Leu Leu Gln Asn Asn Val Asn Val Asn
20 25 30
Ala Gln Asn Gly Phe Gly Arg Thr Ala Leu Gln Val Met Lys Leu Gly
35 40 45
Asn Pro Glu Ile Ala Arg Arg Leu Leu Leu Arg Gly Ala Asn Pro Asp
50 55 60
Leu Lys Asp Arg Thr Gly Phe Ala Val Ile His Asp Ala Ala Arg Ala
65 70 75 80
Gly Phe Leu Asp Thr Leu Gln Thr Leu Leu Glu Phe Gln Ala Asp Val
85 90 95
Asn Ile Glu Asp Asn Glu Gly Asn Leu Pro Leu His Leu Ala Ala Lys
100 105 110
Glu Gly His Leu Arg Val Val Glu Phe Leu Val Lys His Thr Ala Ser
115 120 125
Asn Val Gly His Arg Asn His Lys Gly Asp Thr Ala Cys Asp Leu Ala
130 135 140
Arg Leu Tyr Gly Arg Asn Glu Val Val Ser Leu Met Gln Ala Asn Gly
145 150 155 160
Ala Gly Gly Ala Thr Asn Leu Gln
165
<210> 16
<211> 837
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (328)..(738)
<400> 16
gaggactccg cgacggtccg caccctgcgg ccagagcggc tttgagctcg gctgcttccg 60
cgctaggcgc tttttcccag aagcaatcca ggcgcgcccg ctggttcttg agcgccagga 120
aaagcccgga gctaacgacc ggccgctcgg cactgcacgg ggccccaagc cgcagaagaa 180
ggacgacggg agggtaatga agctgagccc aggtctccta ggaaggagag agtgcgccgg 240
agcagcgtgg gaaagaaggg aagagtgtcg ttaagtttac ggccaacggt ggattatccg 300
ggccgctgcg cgtctggggg ctgcgga atg cgc gag gag aac aag ggc atg ccc 354
Met Arg Glu Glu Asn Lys Gly Met Pro
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
-18-
1 5
agt ggg ggc ggc agc gat gag ggt ctg gcc acg ccg gcg cgg gga cta 402
Ser Gly Gly Gly Ser Asp Glu Gly Leu Ala Thr Pro Ala Arg Gly Leu
15 20 25
gtg gag aag gtg cga cac tcc tgg gaa gcc ggc gcg gat ccc aac gga 450
Val Glu Lys Val Arg His Ser 'Trp Glu Ala Gly Ala Asp Pro Asn Gly
30 35 40
gtc aac cgt ttc ggg agg cgc gcg atc cag gtc atg atg atg ggc agc 498
Val Asn Arg Phe Gly Arg Arg Ala Ile Gin Val Met Met Met Gly Ser
45 50 55
gcc cgc gtg gcg gag ctg ctg ctg ctc cac ggc gcg gag ccc aac tgc 546
Ala Arg Val Ala Glu Leu Leu Leu Leu His Gly Ala Glu Pro Asn Cys
60 65 70
gca gac cct gcc act ctc acc cga cc4 gtg cat gat gct gcc cgg gag 594
Ala Asp Pro Ala Thr Leu Thr Arg Pro Val His Asp Ala Ala Arg Glu
75 80 85
ggc ttc ctg gac acg ctg gtg gtg ctg cac cgg gcc ggg gcg cgg ctg 642
Gly Phe Leu Asp Thr Leu Val Val Leu His Arg Ala Gly Ala Arg Leu
90 95 100 105
gac gtg cgc gat gcc tgg ggt cgt ctg ccc gtg gac ttg gcc gag gag 690
Asp Val Arg Asp Ala Trp Gly Arg Leu Pro Val Asp Leu Ala Glu Glu
110 115 120
cgg ggc cac cgc gac gtt gca ggg tac ctg cgc aca gcc acg ggg gac 738
Arg Gly His Arg Asp Val Ala Gly Tyr Leu Arg Thr Ala Thr Gly Asp
125 130 135
tgacgccagg ttccccagcc gcccacaacg actttatttt cttacccaat ttcccacccc 798
cacccaccta attcgatgaa ggctgccaac ggggagcgg 837
<210> 17
<211> 137
<212> PRT
<213> Homo sapiens
=
<400> 17
Met Arg Glu Glu Asn Lys Gly Met Pro Ser Gly Gly Gly Ser Asp Glu
1 5 10 15
Gly Leu Ala Thr Pro Ala Arg Gly Leu Val Glu Lys Val Arg His Ser
20 25 30
Trp Glu Ala Gly Ala Asp Pro Asn Gly Val Asn Arg Phe Gly Arg Arg
35 40 45
Ala Ile Gin Val Met Met Met Gly Ser Ala Arg Val Ala Glu Leu Leu
50 55 60
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
-19-
Leu Leu His Gly Ala Glu Pro Asn Cys Ala Asp Pro-Ala Thr Leu Thr
65 70 75 80
Arg Pro Val His Asp Ala Ala Arg Glu Gly Phe Leu Asp Thr Leu Val
85 90 95
Val Leu His Arg Ala Gly Ala Arg Leu Asp Val Arg Asp Ala Trp Gly
100 105. 110
Arg Leu Pro Val Asp Leu' Ala Glu Glu Arg Gly His Arg Asp Val Ala
115 120 125
Gly Tyr Leu Arg Thr Ala Thr Gly Asp
130 135
<210> 18
<211> 987
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (41)..(508)
<400> 18
cggagagggg gagaacagac aacgggcggc ggggagcagc atg gag ccg 9-Cg gcg 55
Met Glu Pro Ala Ala
1 5
ggg agc agc atg gag cct tcg gct gac tgg ctg gcc acg gcc gcg gcc 103
Gly Ser Ser Met Glu Pro Ser Ala Asp Trp Leu Ala Thr Ala Ala Ala
15 20
cgg ggt cgg gta gag gag gtg cgg gcg ctg ctg gag gcg ggg gcg ctg 151
Arg Gly Arg Val Glu Glu Val Arg Ala Leu Leu Glu Ala Gly Ala Leu
25 30 35
ccc aac gca ccg aat agt tac ggt cgg agg ccg atc cag gtc atg atg 199
Pro Asn Ala Pro Asn Ser Tyr Gly Arg Arg Pro Ile Gln Val Met Met
40 45 50
atg ggc agc gcc cga gtg gcg gag ctg ctg ctg ctc cac ggc gcg gag 247
Met Gly Ser Ala Arg Val Ala Glu Leu Leu Leu Leu His Gly Ala Glu
55 60 65
ccc aac tgc gcc gac ccc gcc at ctc acc cga ccc gtg cac gac gct 295
Pro Asn Cys Ala Asp Pro Ala Thr Leu Thr Arg Pro Val His Asp Ala
70 75 80 85
gcc cgg gag ggc ttc ctg gac acg ctg gtg gtg ctg cac cgg gcc ggg 343
Ala Arg Glu Gly Phe Leu Asp Thr Leu Val Val Leu His Arg Ala Gly
90 95 100
gcg cgg ctg gac gtg cgc gat gcc tgg ggc cgt ctg ccc gtg gac ctg 391
CA 02412764 2002-12-19
WO 02/04605 PCT/US01/21793
-20-
Ala Arg Leu Asp Val Arg Asp Ala Trp Gly Arg Leu Pro Val Asp Leu
105 110 115
gct gag gag ctg ggc cat cgc gat gtc gca cgg tac ctg cgc gcg gct 439
Ala Glu Glu Leu Gly His Arg Asp Val Ala Arg Tyr Leu Arg Ala Ala
120 125 130
gcg ggg ggc acc aga ggc agt aac cat gcc cgc ata gat gcc gcg gaa 487
Ala Gly Gly Thr Arg Gly Ser Asn His Ala Arg Ile Asp Ala Ala Glu
135 140 145
ggt ccc tca gac atc ccc gat tgaaagaacc agagaggctc tgagaaacct 538
Gly Pro Ser Asp Ile Pro Asp
150 155
cgggaaactt agatcatcag tcaccgaagg tcctacaggg ccacaactgc ccccgccaca 598
acccaccccg ctttcgtagt tttcatttag aaaatagagc ttttaaaaat gtcctgcctt 658
,ttaacgtaga tataagcctt cccccactac cgtaaatgtc catttatatc attttttata 718
tattcttata aaaatgtaaa aaagaaaaac accgcttctg ccttttcact gtgttggagt 778
tttctggagt gagcactcac gccctaagcg cacattcatg tgggcatttc ttgcgagcct 838
cgcagcctcc ggaagctgtc gacttcatga caagcatttt gtgaactagg gaagctcagg 898
ggggttactg gcttctcttg agtcacactg ctagcaaatg gcagaaccaa agctcaaata 958
aaaataaaat aattttcatt cattcactc 987
<210> 19
<211> 156
<212> PRT
<213> Homo sapiens
<400> 19
Met Glu Pro Ala Ala Gly Ser Ser Met Glu Pro Ser Ala Asp Trp Leu
1 5 10 15
Ala Thr Ala Ala Ala Arg Gly Arg Val Glu Glu Val Arg Ala Leu Leu
20 25 30_
Glu Ala Gly Ala Leu Pro Asn Ala Pro Asn Ser Tyr Gly Arg Arg Pro
35 40 45
Ile Gln Val Met Met Met Gly Ser Ala Arg Val Ala Glu Leu Leu Leu
50 55 60
Leu His Gly Ala Glu Pro Asn Cys Ala Asp Pro Ala Thr Leu Thr Arg
65 70 75 80
Pro Val His Asp Ala Ala Arg Glu Gly Phe Leu Asp Thr Leu Val Val
85 90 95
CA 02412764 2002-12-19
WO 02/04605
PCT/US01/21793
-21-
'Leu His Arg Ala Gly Ala Arg Leu Asp Val Arg Asp Ala TLE, Gly Arg
100 105 110
Leu Pro Val Asp Leu Ala Glu Glu Leu Gly His Arg Asp Val Ala Arg
115 120 125
Tyr Leu Arg Ala Ala Ala Gly Gly Thr Arg Gly-Ser Asn His Ala Arg
130 135 140
Ile Asp Ala Ala Glu Gly Pro Ser Asp Ile Pro Asp
145 150 155
<210> 20
<211> 15
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<220>
<221> misc_feature
<222> (1)..(15)
<223> p27 antisense oligonucleotide (5' to 3'
orientation)
<400> 20
tggctctcct gcgcc 15