Note: Descriptions are shown in the official language in which they were submitted.
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME 1 DE 3
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME OF
NOTE: For additional volumes please contact the Canadian Patent Office.
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
Modifications of HIV Env, Gag, and Pol Enhance Immunogenicity
for Genetic Immunization
Field of the Invention
The present invention relates to the field of molecular biology. The present
invention discloses modified HIV Env, Gag, Pol, and Nef proteins, related
nucleotide sequences, and usage for genetic immunization.
Background of the Invention
Protective immunity against human immunodeficiency virus-1 (HIV-1) is
likely to require recognition of linear and conformation epitopes from
multiple HIV
antigens. Whether these responses can be elicited more effectively by virion-
like
structures or fused CTL epitopes is unknown.
The immune response to HIV infection in long-term non-progressors (Cao Y
et al., 1995, N Engl J Med, 332:201-8; Pantaleo G et al., 1995, N Engl J Med,
332:209-16) and HIV-exposed sex workers suggests that specific viral immunity
may limit infection and the symptoms of disease. No single characteristic yet
correlates with protective immunity, but studies in non-human of primates
suggest
that both humoral and cellular immunity are required for this response.
Depletion of
cytotoxic T cells (CTLs) in chronically-infected macaques enhances viremia
(Jin X
et al.,1999, J Exp Med, 189:991-8; Schmitz JE et al., 1999, Science, 283:857-
60).
In humans, higher CTL responses correlate with lower viral load and
stabilization of
clinical symptoms (Musey L et al., 1997, N Engl J Med, 337:1267-74; Ogg GS et
al., 1998, Science, 279:2103-6). In animal models, passive transfer of
neutralizing
antibodies can also contribute to protection against virus challenge (Burton
DR,
Montefiori DC, 1997, AIDS, 11 Suppi A:S87-S98; Gauduin MC et al., 1997, Nat
Med, 3:1389-93; Moore JP, Ho DD., 1995, AIDS, 9 Suppl A:S117-5136; Muster T
et al., 1994, J Virol, 68:4031-4; Muster T et al., 1993, J Virol, 67:6642-7;
Poignard
P et al., 1996, Immunol Today, 17:239-46; Sattentau QJ, 1996, Curr Opin
Immunol,
8:540-5; Shibata R et al., 1999, Nat Med, 5:204-10). Neutralizing antibody
responses can also be developed in HIV-infected individuals (Burton DR,
Montefiori DC., 1997, AIDS, 11 Suppl A:S87-S98; Poignard P et al., 1996,
Immunol Today, 17:239-46; Sattentau QJ., 1996, Curr Opin Immunol, 8:540-5) and
are associated with lower viral loads in long-term non-progressors (Montefiori
DC et
al., 1996, J Infect pis, 173:60-7). Though this neutralizing antibody response
is
-1-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
uncommon, it is directed largely against the Env protein of the virus (Robey
WG et
al., 1986, Proc Natl Acad Sci U S A, 83:7023-7; Steimer KS et al., 1991,
Science,
254:105-8).
In early human vaccine trials, gp120 protein immunogens have yielded
disappointing results: vaccine-induced antibodies have not been broadly
neutralizing
and have sometimes enhanced infection in vitro (Bolognesi DP, Matthews TJ,
1998,
Nature, 391:638-9; Connor RI et al., 1998, J Virol, 72:1552-76; Haynes BF,
1996,
Lancet, 348:933-7; VanCott TC et at., 1995, J Immunol, 155:4100-10). Monomeric
gp120 loses oligomer-dependent epitopes and does not include sequences in the
ectodomain of the gp41 that become exposed during virus entry (Broder CC et
al.,
1994, Proc Natl Acad Sci USA, 91:11699-703; Moore JP, 1995, Nature, 376:115;
Muster T et at., 1993, J Virol, 67:6642-7). It is assumed that broadly
neutralizing
antibodies bind to native gpl20/gp41 complex on the surface of the virus
rather than
soluble gp120 (Burton DR, Montefiori DC, 1997, AIDS, 11 Suppl A:S87-S98).
The development of a cytotoxic T lymphocyte (CTL) response to viruses is
often crucial to the outcome of infections. Lysis of infected cells prior to
the
production of progeny virions may limit virus burst size (Yang, 0 et al.,
1996, J.
Virol., 70:5799-5806), and HIV specific CD8+ cytotoxic T lymphocytes (CTL)
have
been shown to be important in viral clearance and in the control of initial
HIV-1
spread (Borrow, P et al., 1994, J. Virol., 68:6103-6110; Yang, 0 et at., 1996,
J.
Virol. 70:5799-5806). CTL responses specific to HIV also contribute to
reduction in
viral load during acute and asymptomatic infection (Klein, MR, et at., 1995,
J. Exp.
Med. 181:1365-1372; Moss, PAH et al., 1995, Proc. Natl. Acad. Sci. USA,
92:5773-
5777) and may be involved in protection against the establishment of
persistent HIV
infections (Rowland-Jones, SL et al., 1993, Lancet, 341:860-861; Rowland-
Jones,
SL et.al., 1995, Nat. Med., 1:59-64). High-frequency CTL responses to HIV-1
correlated with low viral load and slow disease progression in chronically
infected
individuals (Musey, L et al., 1997, N. Engl. J. Med., 337:1267-1274; Ogg, GS
et al.,
1998, Science, 279:2103-2106.). More compelling evidence of an antiviral
effect of
CD8+ cells was demonstrated in controlled studies in macaques, in which CD8"'
cells
were depleted in vivo using a monoclonal antibody. The viral loads in these
animals
increased or decreased as the CD8 cells were depleted or reappeared,
respectively
(Jin, X et al., 1999, J. Exp. Med., 189:991-998; Schmitz, JE, et al., 1,999,
Science,
283:857-860). Therefore, induction of a CTL response specific to these
proteins
represents a desirable response in an HIV-1 vaccine.
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
HIV-1 internal structural and enzymatic proteins contain conserved domains
that preserve their functions and thus exhibit less antigenic diversity that
may elicit
more effective CTL responses (Nixon, DF et al., 1988, Nature, 336:484-487.).
Efficient and durable CTL responses require endogenous antigen synthesis and
processing. Current vaccine delivery techniques include immunization with
live,
attenuated viruses, inactivated recombinant virus infection (Letvin, NL, 1998,
Science, 280:1875-1880) or plasmid DNA expression vectors. A major obstacle in
the induction of CTL responses with naked DNA or recombinant virus during
development of an HIV vaccine is that the expression of HIV-1 structural and
enzymatic genes is tightly regulated by the virus itself. The expression of
these
proteins is heavily dependent upon the existence of the Rev-responsive element
(RRE) of HIV-1 in recombinant vectors (Cullen, BR, 1992, Microbiol. Rev.,
56:375-394; Felber, BK et al., 1989, Proc Natl Acad Sci U S A, 86:1495-1499).
Poor expression is caused by the presence of AT rich inhibitory nucleotide
sequences (INS) in the gag, pol and env genes, which inhibit the nuclear
export and
efficient expression of unspliced HIV1 mRNAs. Early studies of DNA vaccination
against HIV in mice required the inclusion of Rev in their expression vectors
(Lu, S
et al., 1995, Virology, 209:147-154; Okuda, K et al., 1995, AIDS Res. Hum.
Retroviruses, 11:933-943; Wang, B et al., 1993, Proc. Natl. Acad. Sci. U. S. A
90:4156-4160), but modification of INS has been shown to facilitate Rev-
independent expression of HIV-1 Gag (Qiu, J-T et al., 1999, J. Virol., 73:9145-
9152; zur Megede, J et al., 2000, J Virol 74:2628-2635), allowing detectable
humoral and CTL responses against this protein (Qiu, J-T et al., 1999, J.
Virol.,
73:9145-9152). These modified HIV-1 Gag genes produced viral-like particles of
the expected density and morphology and induced an immune response to HIV-1
Gag after DNA immunization in mice (zur Megede, J et al., 2000, J Virol,
74:2628-
2635).
Summary of the Invention
Protective immunity against human immunodeficiency virus-I (HIV-1) is
likely to require recognition of linear and conformation epitopes from
multiple HIV
antigens, and whether these responses can be elicited more effectively by
virion-like
structures or fused CTL epitopes was previously unknown. Herein is provided a
modified HIV Env with deletions in the cleavage site, fusogenic domain, and
spacing of heptad repeats I and 2 to expose the core protein for optimal
antigen
-3-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
presentation and recognition. Additionally we provide a Gag-Pol or Gag-Pol-Nef
fusion protein that is a polyprotein designed to maximize epitope
presentation. The
invention extends the mutation in HIV Env, Gag, Pot, and Nef to any HIV Glade
or
strain and to related proteins of other viruses. Different combinations,
different
orders, and different variations on a theme are envisioned, the theme being to
optimize presentation of epitopes that generate broad CTL and antibody
responses.
More particularly, we have investigated the effect of specific mutations in
human immunodeficiency virus type I (HIV-1) envelope (Env) on humoral and
cellular immune responses after DNA vaccination. Mice were injected with
plasmid
expression vectors encoding HIV Env with modifications of conserved
glycosylation
sites or different COON-terminal mutations intended to mimic a fusion
intermediate.
Elimination of conserved glycosylation sites did not substantially enhance
humoral
or CTL immunity. In contrast, a modified gpl40 with deletions in the cleavage
site,
fusogenic domain and spacing of heptad repeats 1 and 2 enhanced humoral
immunity without reducing the efficacy of the CTL response. Because of its
ability
to stimulate the antibody response to native gp160 without affecting cellular
immunity, this modified gpl40 or a related derivative is envisioned to be a
useful
component of an AIDS vaccine.
In addition, we have examined the immune response to HIV-1 Gag and Pot
after plasmid DNA immunization with Rev-independent expression vectors
encoding various forms of these proteins. Immune responses were analyzed after
vaccination with four expression vectors, including Gag alone or Gag-Pol, both
of
which gave rise to virion-like particles (VLPs), compared to Pot alone or a
Gag-Pol
fusion protein that did not form VLPs. The Gag-Pol fusion protein induced the
most
broad and potent CTL responses to Gag and Pot in DNA-vaccinated mice, and this
immunogen also readily elicited an antibody response to HIV-1 Gag and Pot
determinants. Through its ability to induce broad CTL and antibody responses,
this
Gag-Pol fusion protein or a related derivative is envisioned to be a useful
component
of an AIDS vaccine.
-4-
CA 02419822 2009-09-17
Various embodiments of this invention provide a polynucleotide encoding a
mutated HIV Env protein, wherein the polynucleotide comprises a deletion of
nucleic acid
sequences encoding the Env cleavage site, the Env fusion domain, and the
interspace
between the two heptad repeats of Env, and wherein the polynucleotide
optionally
comprises one or more of. (a) a deletion of one or more nucleic acid sequences
encoding
glycosylation sites of Env, (b) a nucleic acid sequence encoding an HIV Gag-
Pol fusion
protein or an HIV Gag-Pol-Nef fusion protein in a continuous open reading
frame, and (c)
a deletion of a nucleic acid sequence encoding a cytoplasmic domain of Env.
Various embodiments of this invention provide a composition comprising a
polynucleotide of this invention and a pharmaceutically acceptable carrier.
Various embodiments of this invention provide a composition comprising at
least
one mutated HIV Env protein encoded by the polynucleotide of this invention
and a
pharmaceutically acceptable carrier.
Various embodiments of this invention provide a polynucleotide of this
invention
for use in amelioration of symptoms of AIDS or infection by HIV, by genetic
immunization.
Various embodiments of this invention provide a mutated Env protein encoded by
the polynucleotide of this invention.
Various embodiments of this invention provide a mutated Env protein encoded by
the polynucleotide of this invention for use in amelioration of the symptoms
of AIDS or
infection by HIV, by protein immunization.
4a
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Brief Description of the Drawings
Figure A. Comparison of GP2 with the structures of viral and cellular
membrane fusion proteins.
(A) Recombinant Ebola Zaire GP2, (B) Recombinant Mo-S5 from the TM
subunit of MoMuLv, (C) Low pH-treated HA2 from influenza virus, (D)
Recombinant, proteolysis-resistant core of HIV-1 gp4l, (E) Recombinant SIV
gp4l,
NMR structure, (F) Recombinant core coiled segments of the SNARES syntaxin 1-
A, synaptobrevin-II, and SNAP-25B. Weissenhorn et al., 1998, Molecular Cell,
2,
605-616.
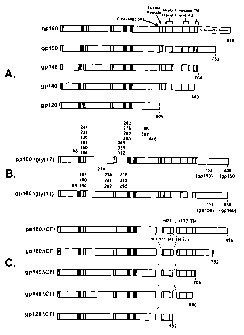
Figure I. Schematic representation of functional domains and mutations in
HIV-1 Env glycoproteins.
Full-length envelope polyprotein, gp160, with the indicated features based on
the amino acid residues of HXB2 is shown (top). Functional domains include the
gpl20/gp4l cleavage site (residues 510/511), the fusion domain (512-527), the
two
heptad repeats (546-579 and 628-655), the transmembrane domain (684-705), and
the cytoplasmic domain (706-856). The mutant forms of the envelope proteins
are
shown below the structure of gp160. COOH deletions were introduced that
terminate the envelope protein at positions 752, 704, or 680 to produce gp150,
gp145, or gp140, respectively. Two internal deletions that removed the
cleavage
site, the fusion domain, and the region between the two heptad repeats were
introduced into gp160, gpl50, gp145, and gp140. A further deletion in the 000H-
terminal region at position 592 removed the second heptad repeat, the
transmembrane domain, and the interspace region to produce gp128ACFI. To
disrupt potential glycosylation sites, asparagine (N) residues at eleven
positions (88,
156, 160, 197, 230, 234, 241, 262, 276, 289, and 295) were replaced with
aspartic
acid (D) residues in both gp160 and gp150. Versions of both gp160 and gp 1 S0
were
created with a total of 17 mutated glycosylation sites by including six
additional N
to D substitutions at positions 332, 339, 356, 386, 392, and 448.
-5-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
Figure II. Comparison of the expression of the HIV-1 gp160 with codon-
optimized gp160.
A. Expression of plasmids encoding Rev-dependent and Rev-independent
codon-modified gp160. Upper panel: expression of Rev-dependent viral gp160
(left) and codon-modified gp160 (right) in transfected 293 cells. Lower panel:
comparable expression of (3-actin in these transfected cells.
B-D. Expression of mutant CXCR4- and CCR5-tropic HIV Env
glycoproteins with COOH-terminal truncations (panels A and B, respectively).
CXCR4-tropic envelope proteins containing mutant glycosylation sites and
mutant
functional domains are shown (panels C and D, respectively). The indicated
proteins were detected by immunoblotting as above. Cell lysates produced by
transfection with vector containing no insert were used as controls (first
lane in each
panel).
Figure III. Cytotoxicity of full-length gp160 is eliminated by deletion of the
COOH-terminal cytoplasmic domain.
Cell rounding and detachment was not observed in control-transfected 293
cells (A), in contrast to full-length gp160 (B) and to a lesser extent in
cells
transfected with gpl50 (C), in contrast to gp145 (D) or gp140 (E).
Figure IV. Expression of soluble gp14OACFI ]HIV-1 envelope variant.
Immunoprecipitation and Western blot analysis of supernatants from the
indicated transfected cells.
Figure V. Antibody response against HIV-1 envelope proteins in DNA
immunized mice.
A. Comparison of the antibody response in mice immunized with gp140
(ACFI) or other Env plasmid expression vectors. Sera were collected 2 weeks
after
the last immunization and used to immunoprecipitate codon-altered gp160 from
lysates of transfected 293 cells. The quantitation of the immunoprecipitated
gpl60
was done as described in Figure 5B. The average of the normalized data has
been
presented as a bar diagram.
B. Antibody responses in mice immunized with different mutant Env
expression vectors. Antisera from immunized mice were diluted in IP buffer and
1
-6-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
l of each diluted serum was used to immunoprecipitate codon-altered HIV-I
gpl60
from lysates of transfected 293 cells as described in Figure 3A. The gels were
scanned and the intensity of the gp160 band was determined by densitometry
using
the program Image Quant and presented relative to the intensity of gpl60
immunoprecipitated with positive control sera (rabbit anti-gpl60), which was
used
to normalize data between experiments. These data are presented graphically to
facilitate comparison among groups.
C. Antibody responses in mice immunized with gp140 or gpl40 (zCFI)
were determined by immunoprecipitation and Western blotting. Animals received
two booster doses (100 g) of the same plasmid, two weeks apart. Sera (1 l)
collected 2 weeks after the last immunization was used to immunoprecipitate
codon-
optimized HIV-1 gp160 from lysates of transfected 293 cells containing 400 g
of
total protein. Each lane corresponds to the sera from an animal immunized with
either the control vector (lanes 1 and 2), CXCR4-tropic gpl40 (lanes 3-6), or
plasmid that expresses gp140 with the indicated mutant functional domains
(lanes 7-
10). A mouse monoclonal antibody to gpl60 (HIV-1 V3 Monoclonal (HIB-V3-13),
NIH AIDS Reagent Program) was used as a positive control (lane 11).
Figure VI. CTL response against HIV-1 envelope proteins in DNA immunized
mice.
The CTL response to CXCR4-tropic Env and indicated deletion mutants is
shown (A). The CTL responses to CXCR4-tropic envelope with glycosylation site
and zCFI mutations are shown (B and C, respectively). Dependence of CTL
activity on CD8 cells was shown by magnetic bead depletion using the indicated
representative immunogens (D). Spleen cells were isolated from immunized mice
two weeks after the final immunization and stimulated in vitro with irradiated
cells
expressing gpl60 with addition of hIL2 (5 U/ml) at day 4. The cytolytic
activity of
the restimulated spleen cells was tested after 7 days against V3 peptide-
pulsed
BC10ME cells. Similar findings were observed with target cells that stably
express
full-length Env.
Figure VII. Schematic representation of HIV-1 Gag-Pol expression constructs.
The protein sequences of Gag (amino acids 1-432) from HXB2 (GenBank
accession number K03455) and Pol (amino acids 3-1003) from NL4-3 (GenBank
-7-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
accession number M19921) were used to create a synthetic version of hGag-Pol
using codons found in human cells. hGag-Po!AFSL\Pr was made by modification of
the frame shift site (FS) and inactivation of protease. For hPol, 432 amino
acids
were deleted from the NH2-terminal region of hGag-Pol and addition of an ATG
codon. hGag was made by deletion of 925 amino acids from the COOH-terminal
region of hGag-Pol. hGag-Pol, hGag-PolLFSAPr, hPol and hGag are expressed
from the pNGVL-3 vector backbone.
Figure VIII. HIV-1 Gag-Pol expression in transfected 293T cells and stably
transfected CT26 and BCIO ME cells.
Cell lysates from 293T cells transfected with pCMV AR8.2 containing viral
Gag-Pol (vGag-Pol) (Naldini, L et al., 1996, Science, 272:263-267), pNGVL-
hGag,
hPol, hGag-PolAFS\Pr and hGag-Pol were separated by 4-15% gradient SDS-
PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting
with
(A) human anti HIV-1-IgG, (B) monoclonal anti-p24, and (C) rabbit anti-RT. (D)
Cell lysates from CT26 and BCIOME cells stably transduced with either hGag or
hPol were analyzed with human anti HIV-1-IgG.
Figure IX. Transmission electron microscopy of HIV-1 immature virus-like
particles (VLP) produced by transfected 293T.
Cells were transfected with pNGVL-hGag 48 hours prior to harvesting and
fixing (magnification 25,000X).
Figure X. Gag or Pol specific CTL response mediated by CD8 positive cells in
immunized mice.
Two weeks after mice were immunized with a control vector, hGag, hPol,
hGag-Po hFSAPr, and hGag-Pol, splenic cells were harvested and sensitized with
naive mouse splenic cells pulsed with Gag or Pol peptides. One week later,
effector
cells were tested for cytolytic activity in a 5-h 51Cr release assay using
S1Cr-labeled
BC10ME target cells that were pulsed for 2 hours with either (A) HIV-1 Gag
peptides, or (B) HIV-1 Pol peptides. (C) CD4+ or CD8+ lymphocytes were
depleted from splenic cells of immunized mice with anti-mouse- CD4+ or CD8+
Dynal beads according to the manufacturer's instructions.
-8-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
Figure XI. Gag or Pol specific CTL response mediated by CD8 positive cells in
immunized mice using stable expressing cell lines as target cells.
Two weeks after immunization in mice, splenic cells were harvested and
sensitized with naive mouse splenic cells pulsed with Gag or Pol peptides. One
week later, effector cells were tested for cytolytic activity in a 5-h 51Cr
release assay
using 51 Cr-labeled BCI OME target cells expressing either (A) HIV-1 Gag or
(B) Pol
protein.
Figure XII. HIV-1 p24 antibody ELISA assays, HIV-1 immunoblotting and
immunoprecipitation Western blotting.
(A) An HIV-1 p24 antibody ELISA assay was performed by coating 96-well
plates with 50 l of purified recombinant HIV-iIIIB p24 antigen at a
concentration of
2 g/ml in PBS buffer, pH 7.4.
(B) HIV-1 immunoblotting of strips containing HIV-1 proteins were
incubated with pooled mouse sera at a dilution of 1:25. Bands were visualized
using
the ECL western blotting detection reagent.
(C) Immunoprecipitation and Western blotting of hPol gene-transfected
293T cell lysates three days after transfection with RIPA buffer. The pooled
mouse
serum was diluted with IP buffer. After adding 10 g of the cell lysate
containing
HIV-1 Pot protein, the reactions were incubated overnight on a rotator at 4 C.
The
next day, 250 l of Protein G and A Sepharose beads (10%V/V in IP buffer) were
added, and the reactions were incubated on a rotator for 2 hours at 4 C. The
reactions were washed 4x with IP buffer, re-suspended with 30 l of IX sample
buffer, and then loaded onto SDS-PAGE. The reactions were transferred to an
Immobilon P membrane, and then incubated with anti HIV-1-IgG. Bands were
visualized using the ECL Western blotting detection reagent.
-9-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
7 10 ^- IN M V' !n ~O I- 00 10 N M d I~ t` 00 O
r-. 'N M U1 ~ v 00 QA .-. N N N N IN N N N N IN M
L4
_Q
~O In ~O O
zN IM d' t-- coq 0~ O N M Imo' V') r' 00 c
N N N N N N 'N N N IN M
Q IQ
Q Q
U U U U
cl
cj
U,U
z o _ n v w 'u Iw Iw w N M M f
cd c In ~~I WI 0' b.UUUUU ~t NN M
U U U "~ a a/ 1 a~ a~ o N M 0 0 v 0 0 v 0 v
U > > ^ i. ti n .-..-~,4..-.
fs k R. C s N IT Ln Li -0 'U 4u Lc CL W Li Li
w M
a1 u -q, Cr,
-UUUUUU ^UUUUUUUU
2 Z Z z z `c 0 vv 0 b ~c n a~ a~ -0 ~T. v v .0 -0 v -0 v -0
rnt`rn 0000 lno 665 Ty1oUUooooo0
P. M V_¾pl M_¾p _M¾C
V ~_[p1. N_¾C ~¾p3 d _V¾p ~¾~I d¾p V d V¾ _V¾ _~¾ ~O D' _d¾ d 'd Iqp d ~¾pY d
7~
bA bn bA bA bA' bq bn bn bn bn' bn `"' bn bn bD b4 bA bA pp.b1) bD bn bn bn bq
bn bq Lf)
In C4 Lr) F~ b4 bn'~ 04 Lf) LY LY Rix fY
to h fA fn N to N fn to to (/1 VA fn to Cn w to w w to to w w w w to w
N N N N N N N N_ N_ N N N N N N N_ N N N N N N_ N N N N N N N N
O O O O 0 O O O O O O O O O O O O O O_ O O O O O O O O
p. P, Cl.I Ar P- P. 0. C1. ca. O. O. Q. O. in. 0. 0.' O. O.
'--d rn 1
-4
H
W x ~ 1
v -0 ,b I -0
U U U U
ww u. w
u. _
v o d '
rs
(Z) 00 In
V' N d
I ~ r
o ~ ~ ~ I~nl I~, I~,
OQ 'x x, x I
U U d~-
Q al~Q' xx k x x
~ b I'U ~ O_ _O ~O I _O O
ro U b~ R d
U' > > >
'd x x U P.' C1. O. CL Cs. ~-. - c0
Ca ~4 p; a CL Iw Iw Iw u, N d v 'ter N
0 O p p U
a U U v v >> > >>>>>>
.c
c c c ~ v b, d b b b b b b b b v 0
a SIG u u N M'v to u u. w w r= u w w u w wI
~I~IUu 0 U U UI UIUIU U U U UUUU UIU UU UIU U
z a 4'~d ci u u Qj- 0v
a vvvv 0bbbb
-0 -0 -0 -0 -0 "
O< c, o r- 0 CD wn In 0 c) to 0 0 0 0 0 C 0 õ 0 0 C
O 0 0 0 0 0 O C)
-tld~d era dIb ~ib ~,d d dd-dl~l~r~r~rf
~n a s' C~. a Ia a a Ia a O.a a a. a. _ pp.. a, a. a. a. _ Ca.O.a a.f~. a.
bn
on 01) bn bn 01) 00 bq on on bn bn on on on on bn on on on on bA bn bn on on
bn on
~ I~ lk x X I C SG k '>C In 'n D ID 24, lc IC4 2 I04 C IC I04
>C C4 V] t, fn Vi N to N !n to V) to V) Vl to V) tZ V) A 1 V) 1 &) t/) VJ N N
tZ V:4 cr"
) A
N N N N N N N iN N Cl N hl N N N N N N N N Ll N N N N N N N N N
O a a a O I O a O O a 'a a a a O a a a O O 'a a a O Ia 'a O O
u I~' c. Y. 0~ r) ~ 2' c' Ire "~ ID Cp ac c~ C, :2- p~ oG :d C! 04 is aC4 C'
rx a: lrc
> > > > I> > I> > > > > > I> > > > > > > > > > '> I> I> > > '> '> >
O. C1 O.. O. O. O. 0. S1 Or C1 02 C1 C].1 C1 C1 O. C].. 02 1s. O. $1. C1. O.
O O O N O O N t - O -- V to O r-- N M O N M d- !n CO 00 SOS 0
0 0 0 0 0 0 0 0 0 0 0 0 0 O ^-- r- N N N N N N N N N N M
-- N M M V Its [~ r- h 00 00 00 00 00 00 DO 00 00 00 00 00 110
CV N N 00 N 00
R] N N N N IN N N N N N N N N N N N N N N N 11 N
CJ N
CL
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
N m 'IT vD AO N 00 C O IN iM - In L.O r 00 rn O Ir. N m V' !n r- DD (T O N
bn 'm IM 'M m M 'M M M m '~- - t t V a) `7 'd' V" Id 'I V1 V'1 1!1 I1n I In Vl
v1 Ilp SAO t0
w I
C7 c1l
CT O N m ~ ~r- 00 Oll O ~'o r, 00 d n 'In IV m1 I In I In I I I~ l0 Lr) z m m
m m m m m m V ~r NT M m
m ('(N
N m V m (N (N N_ N M M N
b b b b b b 'c7 b 'U^ 'c7 'c7 - 'O b 'b
a, u u'u wwwwr,tZrZr,
U IU U U U U U U U U U U U U U E `c -C E E
C~s ~r ~r m - -
~ N m m Cy O O O O O O 0'0 0 0 0 0 0 0 0 wti. ww w~ N M d M
¾ Nõ_~rvd v
~_r_d_-_vv_'_'r'rU U U U ~NmV - N
v o 0 0 fin' ~n l ~n ~n gn' ~n b n' fin' oan gn gn' oan o b b -o -0 ;:- - - -
ry w Ci k V) 1n In in In In 4-1 v, v, v, vz v, !n ~n +n
v n
Nm wwwr~www
w
0~ all ce C4 04 04
U IU U U u 'u C.) U U U U U U U U U U U U
O O O In V'1 V1 In V1 d' Vl V'1 !n 1n Ul
v 4 v O
~r x x x ~x x 'x 'x x~ v d ~r d ~r d d v ~r
y '0. IR' C1' x 0.' R R.' R' R' R' R' L~ R'
A' R' 'Q' '~ '> y > > > > > > >
N N N N U U U U U U U U U U U U U U U N N N N N N N N N N N N N
7 D O
d d d d d d d'd d d d d d d d ^
~ c~' u b b -o,b b,b b b -d b -v b b ro b R' -
> > > '> Id d d d Id d d d d d d 'd 'd 'dd '> > > >> > '> > > > > > >
CL 0. 0. a a0. a 0. 0. a a' 0., 0. a' 0. CL 0. 0. 0. a' 0. 0.'a 0. 0. R~ 0. 0.
x 0. a. P.
rn I
r-+
0
a)
a
w
a)
0
u I I
U
N
N I
E
m I U U U U 'U > > > > > '> > >
gy N ~r -O -c7 'c7 10 "O b ^"C7 "O
is obw~_I
r~ u ~u >I>I>>I>>>>>>>>'yI>> ~c,Imd ~ww'wIw'ri www
U U U U - b b -o v v IU IU U IU U IU U U U U 'U U U
"F3 u 73 't; Z
ft C IC L? Iw [i w f% Ci G r? [? L4 IL4 [i 'O 'b 'b O
O O O O lI'1 V1 V') 'b ~'V7 N. Vl 177 V..i7 V7 V'1 Vl !n
Id d I~r 'U IU U U U U U U IU U 'U'U U U U 'd- 'ar 'ar v'a) a I~r d d d v d 0.
0. 0. 0 o s 0 0 0 ( 0 0 C:0 0 C. 0 in. s a a a a s
tjo enl on lo o N IN ,N 0 0 o 1
t)l) ti) ) In and A n en' bb
vzILnItn d'd d d _vw ~r-Iv'd v 'tv'a L n In nh In V L r)
c~ c cG c4 ¾, c~ cG cx cx 0= a: c 0G ~ 'u R P4 0:
wual,il~n~n~000a))oabn0b000 a to wmwmww
'Zs Ln v, ~ v, v Ln v ~ v, v Ln Ln A, Zi k Zi I S2 `x l x
N N ~N N '~ I~ '~ I~ N N IN N N_ N _N N 'N IN N N_ N_
O O p 'O a, a a a M. a a a R a a a 0. R a 0 0 0 0 0 0 0 0 0 O O O O
dddlddldddddddddd
-o vIb -o v -o -o ro 7v -c7 -o -o -0 -C) -0 icCIcy r a' ne 0L'~' _ cGI~ a' rw'
< < I < < d < < < 1<'d d d < > > > I l '
aaIa a a a al0. a s a a'0. a 0. 0. IM- a a in. a 0. P. a ala a~ala a a a
r n m M M ~o a- v aN- In ~r I'd Iw a)- II1 II (n In IT c) I~ irr) I~ '~
I~
00 00 00 00 00 loo loo 00 loo 00 cc 00 W I00 00 00 00 00 00 cc 00 00 00 00 00
00 00 00 00 00 00 00
ro N N N N N N N N N N N N N N N N N N C11 N N N N N N N N N N CV N N
a I
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
a)
M d Yn t oo a o N m ~ v1 N oo O N mid- Yn ~,D t-- 00 0~ o N Im
b4 140 N N t~ t~ N fs t ` N N N 00 100 00 00 00 00 00 00 00 00 0v Dv rn C~ Q
O M V r tom 00 Cr) O N M I'V' 1tn CO N 00 0A O r-+ N m d' (n ~O r- 00 OA O N M
V
z I~ AO lD l0 N N N N N N N N N N 00 00 00 00 00 00 00 00 00 00 Cl C 0, ON 0~
00
O
Lt
v Q)
CIO cC ~+ x -j `C
d
E L .4 Q N '
w 'IT
z ~ m rd N N N o q o U w U o b U U b
o ovbvv^o U a~ -bb o s' t~ u o o ww
-,,Q U on v n v u Q ~, v
UUUUUUU ~r,Caxoo no's nt a
oov~~ov~o~c v~vd~c~oo u N_~ tonn U
Ln n h V't to In n O 0 O IO O O O O O Rai R; p '~ u
a ~r~r ~r~rd v n VV) ~n V,~o I'D~o
o as bo u U
p p opno~,o~n~u o 0 0~~~'a~ y o ~'~n~nann
Yn Yn to v t ~t V d' d' ~t '0 N 0 rn rn ro Yn Yn Yn Yn Yn
Z 2 Z ztv U U 2 !Q C4 P4 0-0 to to (n fo to N fo y to to N VI to 0 to to M to
to to to to to w to h w to w h
Zs ~~x~s'`kZs Zs ~~~'~~Zs ~i'sZZs ~I~~ s~'~ k~ k~ZS SZ~~~ SZZs
N N N N N N N N N N N N N N N N N N N N N N N N N N N N N N N N
O O O O O 'O -r4 IO O O O O O O IO IO _ O IO _O O _O O O O O O O O O_ O O
t~ t~ C1 C~ (~ - R. C? P R F14 F2 - 01. C~, P4 04 or" 24
> > > > > '> > > > > > > > > > > > > I> > > > > > > I> > > > > > >
.Øi 0. In. 0.a ¾. In. In. 0. 0. a 0. a 0. 0. 0.I 0. a. 0. Cl. 0. 0. a ~a.
in. 0. 0. 0. Im. 0. Cl.
rn I a x
r~ -o'
W x x on
b1
IE
u u
t~.
0
O o' a,' I ~n
o ^I UI
En cn
y N
n C!J
0
X p
Q
1) IN N '=--+
LT.
1ti
O
C~ N N N UJ
I~ 1
O O 1 X '. y~ 'p 'O
cci N 4J
Q^ .~ ,_.,~ 'O L1
Z ct
o U U U U
> > ~ _
'IT 'Cl- m
~l- U ^
1Ci Q1 L
'n M M 'n N a l 1 0. -
N M I fJ-. U. Gx, I p.
N II N N -
> > > > > I> _ a \c a a
b b b -o - 'v o O O t} a, ti pa b d. o v
o
s_>n1 ~I u ~` CG n UIIId~
LZ wu..rs LL, >, E >1 Q O~ oaa
UIUIUIUIU U UI UIab UooCA CaI oo U n oI~ b^an ~'1~I
o v o 0 0 o b. < 4 Id IQ4 IQ 1 u
IU 'N 14 Id U U d d IW W
tntn~ n nIn noloooo 0CD 0o I , X ao pp,aUla) v 0 a~a~
d 1~ Id' 1~ ~' d- cY IU, I~n'Yn IVl v, lD 1~ bq bA bo1 'O cr 'cJ I'U 'O v
a n [¾, ¾ plaaa a a a o,. a. C. ul~c1L U1~ alb ou u u c~i u
bo bo bo bo bn' an an co 1 bn bn an on on co cn an on -. G on =._
US Yn to Yn Yn V) V) f) V) d- V Id' d` d' 'V' IV d' N QJ N u o\ O, Ot V) n Ln
In Ln
f~ CL r~ R ' c4 C~ ' ~ C4 X X 1 SC X ~G x Z z z 00 I 00 I00 a CL fx 04
to to to to to Ito N to Ito to to to to to to to Cn to to V) to y - to v1 to
to to Ito
x~k~~~ sZ sZ k`X x1 k Y~~~IX~ X kIU s~'~~~~
N_ N_ N N N N IN N N N N N N N IN N IN N N N N N N IN N N N 'IN N_ N N IN -om
O O O O_ O IO O 0 O 0 O O O O O O O 0 O 0 O m 0 O 10 IO IO O
C~ IRS f~ 1~ ICC LLB ICS f~ L~ 1~ CC r`C C~ F". - U IG'C F U ICC I2 Ice I~
> > > > > 1> > > > > > > > > > > > > > > > >I- > > kn > > > > > >
a0. ICL a a a1Ca. 0. In. al al M. al o. an.2. a a a are I¾a. aa a a a a
00 10~ I0 IF N M 'c- IO O IO H O
-- N IM O ~ I,- O O O 0 10 ^ N M O S
0 0 M d' 1~ CD 1IN
~~~ NN 0000000 CCCCC C'C' Iooooloo
00 00 00 00 00 00 00 10 N N N N M M V d' Ul \o t` 00 N N N N M M M M M m m
N N IN IN N N N N M M M m m M M M M IM M M M V) V) to In V1 Ul 1n f) In V7 Vl
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
7 tl1 t` W tT O 1r-+ N M It V) l0 !` '00 OA O --N M V IUl 10 h 0 QA O N M to
I'0
C Do 0 10 O O 'O 0 0 0 0 0 0 '^- ^^ N N N N N N
- - - - - - - - - - - - - - - - -
Lc.
O M CD tv 00 U O N M d' In 00 1~ O N 'M 1d' to ~O ['00 O~ O .--N M V) CO
d r- 0 0 0 0 0 10 O O O O '~ -- 'N N N N N 1N N
1~ V COI .C~ I'V"
M M N 't1 V' \
N N NN N M M d' c r
N N N N M M M M d' V' N V M M
N N N N M M > -N 'N N N M M K}' 1
j 'C7 0 'l7 t7 C7 'U 0 C7 '0 '0 U TJ C1 'd c7 c7 70 '0 'C7 'T1 '0 'G 'd Tl 'c7
'l7 0 T1 "U
~ w w w w w w rJ, e~. w w w w w' w w w w w r.=. w w w w w w w w w w'
w ~-,U U U U U UU U UUIU U UU U U U UU U U U UIU U U UU U U
R -0 0 0 0) "C -C "o -C vv 0 C "C "C "C -c) vim, C 'C ,'C v 'o "C b v "C -C "
v
a~ U 0 "1 L v
0. WCA W W 01,10.1 W 0.l 0.1 W W (~ 10. Ga Ga W f~ CY1 CA W 0.7 W 0] Lr1 0.
UUUUUUUUUUUUUUUUUUUUUUUUUUUUUUU
N N N N N N 0 N N C) Q) N 1 N N 9) d) 0 Q) O N 4) N N N Q) d) O N N N
~^ c~a c~c'0'O'0 "0)'0 'b'O "0'0'0 TJ "0)'O'0'0'O'o C'0'o'0 "C7'0'0'0 "U'0'0
'C'O
at c0 cC cC 0) cC c0 0) 0) 0) c0 RS 0) c0 c0 c0 0) 0) 0) 0 cG cV c0 c) c0 0)
0) 0.S c0 0)
UUUU'OO'OIOOO'OIOOO'OOOOOOOOOOOOOOOOOO
O O Cl O O O C O O O C C Cl 'O Cl C n to ~n ~n to V, v-, V) kr) cl
0- r d t :I- -T T V 4 4 4 T '4 4 d' Y d' V d Y -'r d' r
aba aa - aaaaa Q3 QaQaaaaa a - a
n n to to to tn v-) vn n to to to to n to to to In V) n kn 'n tn to to to
c4 c4 c4 c4 04 lm~ 99c4c49 c4cGrx c4C~4 c4
M N to to to w 0 to to to to to to to to to to to N to to w to to N to V) rn
to m w
N N N N_ N N N N N N N N N N IN N N N N N N N N N N N N N N
_O O O O O Cl O C O O 0 0 0 0 0 0 0 0 c -c, 0 0 O O O 0 0 0 O 0 O O
C~ CL~ 0.~ C~ 1 C~ 14 Cl 1 C~ 10. C~ C~ C~ C~ C~ C~ C~ CG
> > > > > > > > > > > > > > > > > >> > > > > > > > > > > > > >
R. C1. r). C). 0). 0- In. 10. 0 P- 0. ri. 0- u. 0. $Z.11 CL a. Cs. R. r). $n.
C). ¾. C1 0- Cl. R. O. R. r1. O.
b I
C
Cl,
G
x x I I
N 9)
y _ _ I
a U U
Li. Cs.
U I
~ v
b b V y
N N N M m M M M N N M M I00 V
IN M M M to
~~"' 10 N N 1 d N rn - N N
a) l' 'CY N N N N M IM N N N N N M M
'Ct
I> "U 'i7 'C7 '0 '0 Ib d - - - - - - - - - - - - - - -
N N N1 N N IN N N 1N cC m Q d p~ 0. CA 0.l 10~ W
0 U. 'Ci. 1f%, u. 1Li [i. ICt, Lz. [i. w Li. 1Cr., Ci ICi. [-L. 1Li, 1Li k.
[?. G. ICi. Cx., fs, 'w G. [_, L+.. w 1Lc, 'Ci,
U) U U U U U U U U 'U U U U lU U U U U U U U U IU U U U U U U U U
UU v 0 0 c~ d10 C'C 0 0v 0v 0v 0 0v,Olo-C drov110bb-ob
0 000000010o1ololoo''n nln n n1nlnlnv n o nln~
1.~ .b It T r i' It - 1! :V V' V V 1d' Vt i In _v n
a a o. a 0. ca. a n. 0- o.. a p.
u a
bn bnbnbAbn~nnr.0 bnanbnltna~~~ ~ol~n~nl~nbn~o
u 1 ~ ~ 1 ~ bn
---
V1 to v^i ti rn in ti y I rn v) rn to I. vi rn v) ,- in v in in Iin cn to ~ t -
ti - rn Ito 'I y
C~ C4 '
to vz I S~ k X I YC ~. ~ ~7 k L.~ 1ZS ~ 1 k k~ k k 1 ~S k ~ k' k ~. k I k Xk'
>C iC X.
N N N Ip O O 0 0 0 0 0 0 0 0 0 0 0 O O O O 0 0 0 0 I O O O O 0 O O
~~UIUUUIUUIUUUUU~C4 UUUUIUUUUIUIUUUUUUU1UU'U
C~ C~ C~ C4 CC C~ CL' CL' C~ C 0L C4~ C C4 C4 C4 C4 C4 r~ C4 CL~ C4 C~ C4 0~
aL cL c~ C~ C~
> > > > > 1> > > > > '> > I> > > > I> > > > > >1> > > > > 1> > > > 1>
C1 0. 0.I c)' ¾' n. "' 0. ¾' 0. 0. in. a. a. 0. a C1ICD 0- a. a. 1a. 0. C), w
Ia 0. CL 0. CL CL CL I oo nvN,vM,~~ntX n n~,o,c CDCI~~
M M M M M M M M M M M M M M M M M M M M M M M I M M M M M 00 M
to to to to to to Iõ V) Ln V) to t/-) V) I to Ito to to to to to v) to V) to
to to Vi to to V) to to I
P. I
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
~" l~ 00 OA O N M 00 ~O l~ CO 1OA O N M d` 00 lp N 00 O, O ~= N M ~' 00 l0
N N N M M 'M M M M M M M iM ~Y d' 'cY d" d' V ~Y d' mil' Vl Vl to 00 00
b0
0 N 00 OC O N M IV V} N 00 Oll O N M ~Y !` 00 Q\ O N M 'ch 00 10
N N N M M M M M M M 1, M V d' d` d' ~f" d' 'V' fit' '~h fit' 00 V'1 00 00
V] ^^
V M M M
N_ N_ N_ 'N M M 'd M 1M d' 'd- N N N_ M M 'ct M
M 'IT M N d'
^ N IN N N M 'M V N N N N 1M
Z > > > >> > >> > > > > > > >> > >> > > > > > > >
b -a b b 1'a b 70 7b -0 b b b 10 'a 'a 70 'a "a b b b "a "a b b b '0 b b b
a. Q Q Q Q Q 1Q Q Q Q IQ IQ Q Q Q Q Q Q Q Q 'Q Q Q Q Q Q IQ Q Q Q
0000DO U)U)0UoIV) DO V)cnCA cn U)vc000DO co &)0cl) 0000000
v ~. w w w w w w w Iw ~. w w w rJ, w w w w w w 'u. 'w k., w w w w w w w
U U U U U U U U U U UUU U U U UUUU U U U U UU U U U U
-ate -a v -a -a -a -a -a -0 -a -a -a -a -a -a v -a :-a -a -a v -a S-0 -a v v -
a v -a
O O O O O O O O O 0 O 0 O O O Vl d V) ~ in V) ~ V) V1 'V1 V) ~ V7 V1 v1 V7
pa',,
b-0 b0 b0 b0 bA b0 bq b0' b0 00 bq 0q b0 b0 b0 bq b0 b0 b0 b0 bq b0 b0 bq b))
b0 b0 b0 b0 bq
kn Vl 00 V) 00 00 00 V) V) V) V, 00 0 0 00 00 00 00 V) V) 00 V) V) 0 V) V) X00
00 00 00
0G 01. 0~
CL C~ C~ C4 C~ C~ C~ C~ ~. C~ CG C~
to to V V V) to to to V) V V ti v) V rn rn V ti to V V rn V to y V w V
N N N N N N N N N_ N N N N N N N N N N N N N N N N N N N N N
O O O O O O O_ O_ 10 O O O O_ O O O O _O O_ O _O O_ _O O O O 10 O O O
> > > > > > > > > > > > > > > > '> > > > > > > > > I> > > >
a. a. a. a.1 a. a. a. a. l a. a. a. a 1 a. a.' a, a. a. a. C). a. a. a. C). a.
a. a. a. a. a. a.
E
a
lW
a
K I I
a.
U
N
Q L
N
CO M 00 A 0 a-
Z 1 'N N _M _M 'ct' M M Imo- 00 00 N N IN M N N d` y Im M N V'
> '> > > N N IN N M M d' --1N N N N M M 'cY ^
b b b > > > > '> > 1> > > > > > > '> > > > > > I> > > > > 'y
~, wwwwlr~.we=,wr~.wwww,wwr~,wlwwwlw~wwwwww
bU,UaIUUUIUUUIUUIUUIUIUUUUIUUUUIUUUUUIUUU
a a b b b v a -a :2, -a -a -a -a 'b -a v -a-a'b -a -a -a -a'a'-a -a =a `a
~lCC0000I0oQ00Q0tnnni nnnInILnInn n~nn
d a- d- tv- Ira- d- 'T V-Id-It a- dt d- a- It TT It va1It atavd-
T I t a bz: 01) OA 0^0 00 bn O0 00 bn 00 0!) 00 00 b0 b1)I 00 0000 00 01) 0i)
G0 00 00 00 01) 00 07) 00 00' 00
UU,UIUUUIUUIUUIUIUIUIUIUUUUUIUUIUUUUIUIUUI~ U1
Q d Q Q Q Q IQ Q Q Q Q Q Q Q Q Q Q Q Q IQ Q ~Q IQ Q Q Q Q Q Q
00 Cn V) Cl) Cn Cn V] V) C) C/3 Cn V) V) Cl) V] C/a V3 V). IU) IV3 V) C/) Cl)
CQ V) CA V) I V] V) V)
n U: v: n )n in n n U n n n~ n U ~ in lU nnn
CG C~ C~ R C~ C~ C4 f~ CC R C~ CG C4 ol CG 0~ C4 C~ CG I0.' L CL 0. CG
V) V V) Vt V) to to V V) V IO to Ito V) V1 V V) V V V to V] V) V I V) V V) V
k 'k k k k lk k k 'k k lk 'k k k k k k k k k lk k k k k k ?~ `3~
N N N N N N N N N N N N N N N N N N IN N N N N N N N N N N
C, Io to o ICD C) o o o 0 o to to o to _) _0 to _o o_ o Io o_ CD to 0 to o_ 'o
Io
CCa~Cdll a 1a _ IddC~Ic~a'a'a:a'a:r~r'c~r a'uu:c~u c~da'c4 u_
> > > '> > > I> > > > > > > > > > > > > 11> > > > > > I> > > I> >
alala a s a a a1a a aja a s a,a,ala a a1ala a a.la al a. a. a.la.
-a
O ^ N M It 00 Co N- 00 O ^ N M ~ '00 ' o t~ 00 Qt 0 ^ N M Imo' Vl CD f~ Oq 101
0 0 0 0 0 0 0 0 0 0 N N N N N N N CJ N N
C C C C C C C C- ~r'3 V3 'n - -n Ul V7 -n to N N N N N N N N IN
la I I I
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
N X00 a\ O N M Icy- U1 ISO r- 00 to O ,-- IN M d' Ul ,~O
L~ loo 1 O N M 'd' U1 'N W d> O N M d' 4l
i
z w -o z v
- to ~'~ d Z -b b Z
~n 'Z Z rL c o~ Z
b b
H H - H -a
cam'
c~ -o
b b ~ _
a u C~ d o
to -
a
to m b
z ~.
z
to a ~G
' ba: bb a b n
CIO
w C- v~ oo
PO u" P. 0. b L7 r 0.
t 0a ova v~~raaa . w w
a, b w w a
0 Ca 5 v V -o w
~,~a~ a~aw ob'n o v
0 ww
w bp > o bi) > :2, - U b
c'F d
u IC7 v1 2 11 C7 C7 C7 vi on on n bn C7 4. on w `c on
[n rn rn N rn cn v) rn v) to [n tC cC 7 cG N cC td tcJ
sZ sZ '~ sZ Z sz yZ o C7 C7 C7 Z C7 C7 Z 'C7
N N N IN N N N_ N N N N N N N N N_ ~N N N
O 10 O O O 10 O_ O O O_ O O_ O O O O ~j O O_ O_ 'O
> > > > > > > > > > > > > > > > > N > N > H >
n a'p-'Q Q u a a a Z a s a oIp a a a-o'o_Z arm' a
U
z
to -d
a 4
an
b ~
_o z Z z z Z
u '
v~~~ :d d ~4
2 o b'H H HI
ra a _ _ o a o
E- I Z
w''a 2 I: o ;b H cC
(1) V
wl ~4 mod' to w w ~ "b v -to ro b `n 0
~I ii d d 'o o -o Iv 0 '~ N'
o 0 0 0 0 0'0 CL o to o.
~laa r' la as i ,~ Z
bb on qn btl bny - w w Q
C7 C71C7 7 a a C7 C71I nIw 0 dI
bn cs
> > > ~ > d ~o ,. CJ I v I~ I
Ix v3 x C7 va ~c ou on ol CID on 4 on o nq
rn y i v~ to vs v, to m - N ' M CO to to N I m ca
k~ 's~~ kZi N XZC7C7C7C7 C7 Z C7 U,
N N N N N N N N N N N N N N N N N N N
o to o_ to o_ to o_ o_ _o o_ o_ to _o 0 0 0 0_ Io I b o
a' I~ w'Ia'IaICC a Ix c'C~4I~ 0d 00 a
> > > H' > > > to
>
o a M. c7. r:'. I CL C1. CL n 0. CL w' a Q. a n. CL a a. C7 a
-d I
O O -- O 0 0 N M 'V' Ul lp CO OL O ^_ N M
p O0i 00 00 O O O O 10 O O O 0 O O O M M IM M
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Detailed Description of the Invention
The present invention provides modifications of HIV Env, Gag, Pol, and Nef
that enhance immunogenicity for genetic immunization. Both HIV and SIV are
genetically related members of the lentivirus genus of the Retrovif-idae
family.
Lentivirus isolates from humans are grouped into one of two types, designated
HIV-
I and HIV-2. A classification scheme recognizes nine subtypes (clades) of HIV-
1
(A through 1) and five subtypes of HIV-2 (A through E). A compendium of HIV
and SIV sequence information is found in a database prepared by Myers et al.,
Los
Alamos, NM: Los Alamos National Laboratory.
Nucleic Acid Molecules
As indicated herein, nucleic acid molecules of the present invention may be
in the form of RNA or in the form of DNA obtained by cloning or produced
synthetically. The DNA may be double-stranded or single-stranded. Single-
stranded
DNA or RNA may be the coding strand, also known as the sense strand, or it may
be
the non-coding strand, also referred to as the anti-sense strand.
By "isolated" nucleic acid molecule(s) is intended a nucleic acid molecule,
DNA or RNA, which has been removed from its native environment. For example,
recombinant DNA molecules contained in a vector are considered isolated for
the
purposes of the present invention. Further examples of isolated DNA molecules
include recombinant DNA molecules maintained in heterologous host cells or
purified (partially or substantially) DNA molecules in solution. Isolated RNA
molecules include in vivo or in vitro RNA transcripts of the DNA molecules of
the
present invention. Isolated nucleic acid molecules according to the present
invention
further include such molecules produced synthetically.
Nucleic acid molecules of the present invention include DNA molecules
comprising an open reading frame (ORF) of a wild-type HIV gene; and DNA
molecules which comprise a sequence substantially different from those
described
above but which, due to the degeneracy of the genetic code, still encode an
ORF of a
wild-type HIV polypeptide. Of course, the genetic code is well known in the
art.
Degenerate variants optimized for human codon usage are preferred.
In another aspect, the invention provides a nucleic acid molecule comprising
a polynucleotide which hybridizes under stringent hybridization conditions to
a
portion of the polynucleotide in a nucleic acid molecule of the invention
described
above. By "stringent hybridization conditions" is intended overnight
incubation at 42
degree C in a solution comprising: 50% fonnamide, 5 times SSC (750 mM NaCl, 75
-16-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
mM trisodium citrate), 50 mM sodium phosphate (pH 7.6), 5 times Denhardt's
solution, 10% dextran sulfate, and 20 g/ml denatured, sheared salmon sperm
DNA,
followed by washing the filters in 0.1 times SSG at about 65 degree C.
By a polynucleotide which hybridizes to a "portion" of a polynucleotide is
intended a polynucleotide (either DNA or RNA) hybridizing to at least about 15
nucleotides (nt), and more preferably at least about 20 nt, still more
preferably at
least about 30 nt, and even more preferably about 30-70 nt of the reference
polynucleotide.
By a portion of a polynucleotide of "at least 20 nt in length," for example,
is
intended 20 or more contiguous nucleotides from the nucleotide sequence of the
reference polynucleotide. Of course, a polynucleotide which hybridizes only to
a
complementary stretch of T (or U) resides, would not be included in a
polynucleotide of the invention used to hybridize to a portion of a nucleic
acid of the
invention, since such a polynucleotide would hybridize to any nucleic acid
molecule
containing a poly T (or U) stretch or the complement thereof (e.g.,
practically any
double-stranded DNA clone).
As indicated herein, nucleic acid molecules of the present invention which
encode an HIV polypeptide may include, but are not limited to those encoding
the
amino acid sequence of the full-length polypeptide, by itself, the coding
sequence
for the full-length polypeptide and additional sequences, such as those
encoding a
leader or secretory sequence, such as a pre-, or pro- or prepro-protein
sequence, the
coding sequence of the full-length polypeptide, with or without the
aforementioned
additional coding sequences, together with additional, non-coding sequences,
including for example, but not limited to introns and non-coding 5' and 3'
sequences,
such as the transcribed, non-translated sequences that play a role in
transcription,
mRNA processing, including splicing and polyadenylation signals, for example,
ribosome binding and stability of mRNA; and additional coding sequence which
codes for additional amino acids, such as those which provide additional
functional i ties.
The present invention further relates to variants of the nucleic acid
molecules
of the present invention, which encode portions, analogs or derivatives of the
HIV
protein. Variants may occur naturally, such as a natural allelic variant. By
an "allelic
variant" is intended one of several alternate forms of a gene occupying a
given locus
on a genome of an organism. Genes II, Lewin, B., ed., John Wiley & Sons, New
-17-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
York (1985). Non-naturally occurring variants may be produced using art-known
mutagenesis techniques.
Such variants include those produced by nucleotide substitutions, deletions
or additions, which may involve one or more nucleotides. The variants may be
altered in coding regions, non-coding regions, or both. Alterations in the
coding
regions may produce conservative or non-conservative amino acid substitutions,
deletions or additions. Especially preferred among these are silent
substitutions,
additions and deletions, which do not alter the properties and activities of
the HIV
polypeptide or portions thereof. Also especially preferred in this regard are
conservative substitutions.
Further embodiments of the invention include nucleic acid molecules
comprising a polynucleotide having a nucleotide sequence at least 95%
identical,
and more preferably at least 96%, 97%, 98% or 99% identical to a nucleotide
sequence encoding a polypeptide having the amino acid sequence of a wild-type
HIV polypeptide or a nucleotide sequence complementary thereto.
By a polynucleotide having a nucleotide sequence at least, for example, 95%
"identical" to a reference nucleotide sequence encoding a HIV polypeptide is
intended that the nucleotide sequence of the polynucleotide is identical to
the
reference sequence except that the polynucleotide sequence may include up to
five
point mutations per each 100 nucleotides of the reference nucleotide sequence
encoding the HIV polypeptide. In other words, to obtain a polynucleotide
having a
nucleotide sequence at least 95% identical to a reference nucleotide sequence,
up to
5% of the nucleotides in the reference sequence may be deleted or substituted
with
another nucleotide, or a number of nucleotides up to 5% of the total
nucleotides in
the reference sequence may be inserted into the reference sequence. These
mutations
of the reference sequence may occur at the 5' or 3' terminal positions of the
reference
nucleotide sequence or anywhere between those terminal positions, interspersed
either individually among nucleotides in the reference sequence or in one or
more
contiguous groups within the reference sequence.
As a practical matter, whether any particular nucleic acid molecule is at
least
95%, 96%, 97%, 98% or 99% identical to the reference nucleotide sequence can
be
determined conventionally using known computer programs such as the Bestfit
program (Wisconsin Sequence Analysis Package, Version 8 for Unix, Genetics
Computer Group, University Research Park, 575 Science Drive, Madison, Wis.
53711). Bestfit uses the local homology algorithm of Smith and Waterman,
-18-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
Advances in Applied Mathematics 2: 482-489 (1981), to find the best segment of
homology between two sequences. When using Bestfit or any other sequence
alignment program to determine whether a particular sequence is, for instance,
95%
identical to a reference sequence according to the present invention, the
parameters
are set, of course, such that the percentage of identity is calculated over
the full
length of the reference nucleotide sequence and that gaps in homology of up to
5%
of the total number of nucleotides in the reference sequence are allowed.
The present application is directed to nucleic acid molecules at least 95%,
96%, 97%, 98% or 99% identical to the nucleic acid sequences shown herein in
the
Sequence Listing which encode a polypeptide having HIV polypeptide activity.
By
"a polypeptide having HIV activity" is intended polypeptides exhibiting HIV
activity in a particular biological assay. For example, Env, Gag, and Pol
protein
activity can be measured for changes in immunological character by an
appropriate
immunological assay.
Of course, due to the degeneracy of the genetic code, one of ordinary skill in
the art will immediately recognize that a large number of the nucleic acid
molecules
having a sequence at least 95%, 96%, 97%, 98%, or 99% identical to a nucleic
acid
sequence shown herein in the Sequence Listing will encode a polypeptide
"having
HIV polypeptide activity." In fact, since degenerate variants of these
nucleotide
sequences all encode the same polypeptide, this will be clear to the skilled
artisan
even without performing the above described comparison assay. It will be
further
recognized in the art that, for such nucleic acid molecules that are not
degenerate
variants, a reasonable number will also encode a polypeptide having HIV
polypeptide activity. This is because the skilled artisan is fully aware of
amino acid
substitutions that are either less likely or not likely to significantly
effect protein
function (e.g., replacing one aliphatic amino acid with a second aliphatic
amino
acid).
For example, guidance concerning how to make phenotypically silent amino
acid substitutions is provided in Bowie, J. U. et al., "Deciphering the
Message in
Protein Sequences: Tolerance to Amino Acid Substitutions," Science 247:1306-
1310
(1990), wherein the authors indicate that proteins are surprisingly tolerant
of amino
acid substitutions.
-19-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Polypeptides and Fragments
The invention further provides a HIV polypeptide having the amino acid
sequence encoded by an open reading frame (ORF) of a wild-type HIV gene, or a
peptide or polypeptide comprising a portion thereof (e.g., gp 120).
It will be recognized in the art that some amino acid sequences of the FIIV
polypeptides can be varied without significant effect of the structure or
function of
the protein. If such differences in sequence are contemplated, it should be
remembered that there will be critical areas on the protein which determine
activity.
Thus, the invention further includes variations of the HIV polypeptide which
show substantial HIV polypeptide activity or which include regions of HIV
protein
such as the protein portions discussed below. Such mutants include deletions,
insertions, inversions, repeats, and type substitutions. As indicated,
guidance
concerning which amino acid changes are likely to be phenotypically silent can
be
found in Bowie, J. U., et al., "Deciphering the Message in Protein Sequences:
Tolerance to Amino Acid Substitutions," Science 247:1306-1310 (1990).
Thus, the fragment, derivative or analog of the polypeptide of the invention
may be (i) one in which one or more of the amino acid residues are substituted
with
a conserved or non-conserved amino acid residue (preferably a conserved amino
acid residue) and such substituted amino acid residue may or may not be one
encoded by the genetic code, or (ii) one in which one or more of the amino
acid
residues includes a substituent group, or (iii) one in which additional amino
acids are
fused to the mature polypeptide, such as an IgG Fe fusion region peptide or
leader or
secretory sequence or a sequence which is employed for purification of the
mature
polypeptide or a proprotein sequence. Such fragments, derivatives and analogs
are
deemed to be within the scope of those skilled in the art from the teachings
herein.
As indicated, changes are preferably of a minor nature, such as conservative
amino acid substitutions that do not significantly affect the folding or
activity of the
protein (see Table A).
-20-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Table A
Conservative Amino Acid Substitutions
Aromatic Phenylalanine
Tryptophan
Tyrosine
Ionizable: Acidic Aspartic Acid
Glutamic Acid
Ionizable: Basic Arginine
Histidine
Lysine
Nonionizable Polar Asparagine
Glutamine
Selenocystine
Serine
Threonine
Nonpolar (Hydrophobic) Alanine
Glycine
Isoleucine
Leucine
Proline
Valine
Sulfur Containing Cysteine
Methionine
Of course, the number of amino acid substitutions a skilled artisan would
make depends on many factors, including those described above. Generally
speaking, the number of amino acid substitutions for any given HIV polypeptide
will not be more than 50, 40, 30, 20, 10, 5 or 3.
Amino acids in the HIV polypeptides of the present invention that are
essential for function can be identified by methods known in the art, such as
site-
directed mutagenesis or alanine-scanning mutagenesis (Cunningham and Wells,
Science 244:1081-1085 (1989)). The latter procedure introduces single alanine
mutations at every residue in the molecule. The resulting mutant molecules are
then
tested for biological activity such as changes in immunological character.
-21-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
The polypeptides of the present invention are conveniently provided in an
isolated form. By "isolated polypeptide" is intended a polypeptide removed
from its
native environment. Thus, a polypeptide produced and/or contained within a
recombinant host cell is considered isolated for purposes of the present
invention.
Also intended as an "isolated polypeptide" are polypeptides that have been
purified, partially or substantially, from a recombinant host cell or a native
source.
For example, a recombinantly produced version of the HIV polypeptide can be
substantially purified by the one-step method described in Smith and Johnson,
Gene
67:31-40 (1988).
The polypeptides of the present invention include a polypeptide comprising a
polypeptide shown herein in the Sequence Listing; as well as polypeptides
which are
at least 95% identical, and more preferably at least 96%, 97%, 98% or 99%
identical
to those described above and also include portions of such polypeptides with
at least
30 amino acids and more preferably at least 50 amino acids.
By a polypeptide having an amino acid sequence at least, for example, 95%
"identical" to a reference amino acid sequence of an HIV polypeptide is
intended
that the amino acid sequence of the polypeptide is identical to the reference
sequence except that the polypeptide sequence may include up to five amino
acid
alterations per each 100 amino acids of the reference amino acid of the HIV
polypeptide. In other words, to obtain a polypeptide having an amino acid
sequence
at least 95% identical to a reference amino acid sequence, up to 5% of the
amino
acid residues in the reference sequence may be deleted or substituted with
another
amino acid, or a number of amino acids up to 5% of the total amino acid
residues in
the reference sequence may be inserted into the reference sequence. These
alterations of the reference sequence may occur at the amino or carboxy
terminal
positions of the reference amino acid sequence or anywhere between those
terminal
positions, interspersed either individually among residues in the reference
sequence
or in one or more contiguous groups within the reference sequence.
As a practical matter, whether any particular polypeptide is at least 95%,
96%, 97%, 98% or 99% identical to, for instance, the amino acid sequence shown
herein in the Sequence Listing can be determined conventionally using known
computer programs such the Bestfit program (Wisconsin Sequence Analysis
Package, Version 8 for Unix, Genetics Computer Group, University Research
Park,
575 Science Drive, Madison, Wis. 53711). When using Bestfit or any other
sequence alignment program to determine whether a particular sequence is, for
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
instance, 95% identical to a reference sequence according to the present
invention,
the parameters are set, of course, such that the percentage of identity is
calculated
over the full length of the reference amino acid sequence and that gaps in
homology
of up to 5% of the total number of amino acid residues in the reference
sequence are
allowed.
The polypeptides of the invention may be produced by any conventional
means. Houghten, R. A. (1985) General method for the rapid solid-phase
synthesis
of large numbers of peptides: specificity of antigen-antibody interaction at
the level
of individual amino acids. Proc. Natl. Acad. Sci. USA 82:5131-5135. This
"Simultaneous Multiple Peptide Synthesis (SMPS)" process is further described
in
U.S. Pat. No. 4,631,211 to Houghten et al. (1986).
The present invention also relates to vectors which include the nucleic acid
molecules of the present invention, host cells which are genetically
engineered with
the recombinant vectors, and the production of HIV polypeptides or fragments
thereof by recombinant techniques.
The polynucleotides may be joined to a vector containing a selectable marker
for propagation in a host. Generally, a plasmid vector is introduced in a
precipitate,
such as a calcium phosphate precipitate, or in a complex with a charged lipid.
If the
vector is a virus, it may be packaged in vitro using an appropriate packaging
cell line
and then transduced into host cells.
The DNA insert should be operatively linked to an appropriate promoter,
such as the phage lambda PL promoter, the E. coli lac, trp and tac promoters,
the
SV40 early and late promoters and promoters of retroviral LTRs, to name a few.
Other suitable promoters will be known to the skilled artisan. The expression
constructs will further contain sites for transcription initiation,
termination and, in
the transcribed region, a ribosome binding site for translation. The coding
portion of
the mature transcripts expressed by the constructs will preferably include a
translation initiating at the beginning and a termination codon (UAA, UGA or
UAG)
appropriately positioned at the end of the polypeptide to be translated.
As indicated, the expression vectors will preferably include at least one
selectable marker. Such markers include dihydrofolate reductase or neomycin
resistance for eukaryotic cell culture and tetracycline or ampicillin
resistance genes
for culturing in E. coli and other bacteria. Representative examples of
appropriate
hosts include, but are not limited to, bacterial cells, such as E. coli,
Streptomyces
and Salmonella typhirnurium cells; fungal cells, such as yeast cells; insect
cells such
-23-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
as Drosophila S2 and Spodoptera Sf9 cells; animal cells such as CHO, COS and
Bowes melanoma cells; and plant cells. Appropriate culture mediums and
conditions
for the above-described host cells are known in the art.
Among vectors preferred for use in bacteria include pQE70, pQE60 and
pQE-9, available from Qiagen; pBS vectors, Phagescript vectors, Bluescript
vectors,
pNH8A, pNHl6a, pNH18A, pNH46A, available from Stratagene; and ptrc99a,
pKK223-3, pKK233-3, pDR540, pRIT5 available from Pharmacia. Among preferred
eukaryotic vectors are pWLNEO, pSV2CAT, pOG44, pXT1 and pSG available from
Stratagene; and pSVK3, pBPV, pMSG and pSVL available from Pharmacia. Other
suitable vectors will be readily apparent to the skilled artisan.
Introduction of the construct into the host cell can be effected by calcium
phosphate transfection, DEAE-dextran mediated transfection, cationic lipid-
mediated transfection, electroporation, transduction, infection or other
methods.
Such methods are described in many standard laboratory manuals, such as Davis
et
al., Basic Methods In Molecular Biology (1986).
The HIV polypeptide can be recovered and purified from recombinant cell
cultures by well-known methods including ammonium sulfate or ethanol
precipitation, acid extraction, anion or cation exchange chromatography,
phosphocellulose chromatography, hydrophobic interaction chromatography,
affinity chromatography, hydroxylapatite chromatography and lectin
chromatography. Most preferably, high performance liquid chromatography
("HPLC") is employed for purification. Polypeptides of the present invention
include naturally purified products, products of chemical synthetic
procedures, and
products produced by recombinant techniques from a prokaryotic or eukaryotic
host,
including, for example, bacterial, yeast, higher plant, insect and mammalian
cells.
Depending upon the host employed in a recombinant production procedure, the
polypeptides of the present invention may be glycosylated or may be non-
glycosylated. In addition, polypeptides of the invention may also include an
initial
modified methionine residue, in some cases as a result of host-mediated
processes.
Pharmaceutical Formulations, Dosages, and Modes of Administration
The compounds of the invention may be administered using techniques well
known to those in the art. Preferably, compounds are formulated and
administered
by genetic immunization. Techniques for formulation and administration may be
found in "Remington's Pharmaceutical Sciences", 18"' ed., 1990, Mack
Publishing
Co., Easton, PA. Suitable routes may include parenteral delivery, such as
-24-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
intramuscular, intradermal, subcutaneous, intramedullary injections, as well
as,
intrathecal, direct intraventricular, intravenous, intraperitoneal,
intranasal, or
intraocular injections, just to name a few. For injection, the compounds of
the
invention may be formulated in aqueous solutions, preferably in
physiologically
compatible buffers such as Hanks' solution, Ringer's solution, or
physiological
saline buffer.
In instances wherein intracellular administration of the compounds of the
invention is preferred, techniques well known to those of ordinary skill in
the art
may be utilized. For example, such compounds may be encapsulated into
liposomes, then administered as described above. Liposomes are spherical lipid
bilayers with aqueous interiors. All molecules present in an aqueous solution
at the
time of liposome formation are incorporated into the aqueous interior. The
liposomal contents are both protected from the external microenvironment and,
because liposomes fuse with cell membranes, are effectively delivered into the
cell
cytoplasm.
Nucleotide sequences of the invention which are to be intracellularly
administered may be expressed in cells of interest, using techniques well
known to
those of skill in the art. For example, expression vectors derived from
viruses such
as retroviruses, adenoviruses, adeno-associated viruses, herpes viruses,
vaccinia
viruses, polio viruses, or sindbis or other RNA viruses, or from plasmids may
be
used for delivery and expression of such nucleotide sequences into the
targeted cell
population. Methods for the construction of such expression vectors are well
known. See, for example, Sambrook et al., 1989, Molecular Cloning, A
Laboratory
Manual, Cold Spring Harbor Press, Cold Spring Harbor, NY, and Ausubel et al.,
1989, Current Protocols In Molecular Biology, Greene Publishing Associates and
Wiley Interscience, NY.
The invention extends to the use of a plasmid for primary immunization
(priming) of a host and the subsequent use of a recombinant virus, such as a
retrovirus, adenovirus, adeno-associated virus, herpes virus, vaccinia virus,
polio
virus, or sindbis or other RNA virus, for boosting said host, and vice versa.
For
example, the host may be immunized (primed) with a plasmid by DNA
immunization and receive a boost with the corresponding viral construct, and
vice
versa. Alternatively, the host may be immunized (primed) with a plasmid by DNA
immunization and receive a boost with not the corresponding viral construct
but a
different viral construct, and vice versa.
-25-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
With respect to HIV Env, Gag, and Pol, protein sequences of the invention
may be used as therapeutics or prophylatics (as subunit vaccines) in the
treatment of
AIDS or HIV infection. A therapeutically effective dose refers to that amount
of the
compound sufficient to result in amelioration of symptoms or a prolongation of
survival in a patient. Toxicity and therapeutic efficacy of such compounds can
be
determined by standard pharmaceutical procedures in cell cultures or
experimental
animals, e.g., for determining the LD50 (the dose lethal to 50% of the
population)
and the ED50 (the dose therapeutically effective in 50% of the population).
The
dose ratio between toxic and therapeutic effects is the therapeutic index and
it can be
expressed as the ratio LD50/ED50. Compounds which exhibit large therapeutic
indices are preferred. The data obtained from cell culture assays and animal
studies
can be used in formulating a range of dosage for use in humans. The dosage of
such
compounds lies preferably within a range of circulating concentrations that
includes
the ED50 with little or no toxicity. The dosage may vary within this range
depending upon the dosage form employed and the route of administration
utilized.
For any compound used in the method of the invention, the therapeutically
effective
dose can be estimated initially from cell culture assays. A dose may be
formulated
in animal models to achieve a circulating plasma concentration range that
includes
the IC50 (e.g., the concentration of the test compound which achieves a half-
maximal inhibition of viral infection relative to the amount of the event in
the
absence of the test compound) as determined in cell culture. Such information
can
be used to more accurately determine useful doses in humans. Levels in plasma
may
be measured, for example, by high performance liquid chromatography (HPLC).
The compounds of the invention may, further, serve the role of a
prophylactic vaccine, wherein the host produces antibodies and/or CTL
responses
against HIV Env, Gag and Pol, which responses then preferably serve to
neutralize
HIV viruses by, for example, inhibiting further HIV infection. Administration
of the
compounds of the invention as a prophylactic vaccine, therefore, would
comprise
administering to a host a concentration of compounds effective in raising an
immune
response which is sufficient to elicit antibody and/or CTL reponses to HIV
Env,
Gag, and Pol, and/or neutralize HIV, by, for example, inhibiting HIV ability
to
infect cells. The exact concentration will depend upon the specific compound
to be
administered, but may be determined by using standard techniques for assaying
the
development of an immune response which are well known to those of ordinary
skill
in the art.
-26-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
The compounds may be formulated with a suitable adjuvant in order to
enhance the immunological response. Such adjuvants may include, but are not
limited to mineral gels such as aluminum hydroxide; surface active substances
such
as lysolecithin, pluronic polyols, polyanions; other peptides; oil emulsions;
and
potentially useful human adjuvants such as BCG and Corynebacterium parvum.
Adjuvants suitable for co-administration in accordance with the present
invention should be ones that are potentially safe, well tolerated and
effective in
people including QS-21, Detox-PC, MPL-SE, MoGM-CSF, TiterMax-G, CRL-
1005, GERBU, TERamide, PSC97B, Adjumer, PG-026, GSK-1, GcMAF, B-
alethine, MPC-026, Adjuvax, CpG ODN, Betafectin, Alum, and MF59 (see Kim et
at., 2000, Vaccine, 18: 597 and references therein).
Other contemplated adjuvants that may be administered include lectins,
growth factors, cytokines and lymphokines such as alpha-interferon, gamma-
interferon, platelet derived growth factor (PDGF), gCSF, gMCSF, TNF, epidermal
growth factor (EGF), IL-1, IL-2, IL-4, IL-6, IL-8, IL-10 and IL-12.
For all such treatments described above, the exact formulation, route of
administration and dosage can be chosen by the individual physician in view of
the
patient's condition. (See e.g., Fingl et al., 1975, in "The Pharmacological
Basis of
Therapeutics", Ch. I p. 1).
It should be noted that the attending physician would know how to and when
to terminate, interrupt, or adjust administration due to toxicity, or to organ
dysfunctions. Conversely, the attending physician would also know to adjust
treatment to higher levels if the clinical response were not adequate
(precluding
toxicity). The magnitude of an administered dose in the management of the
viral
infection of interest will vary with the severity of the condition to be
treated and the
route of administration. The dose and perhaps prime-boost regimen, will also
vary
according to the age, weight, and response of the individual patient. A
program
comparable to that discussed above may be used in veterinary medicine.
The pharmacologically active compounds of this invention can be processed
in accordance with conventional methods of galenic pharmacy to produce
medicinal
agents for administration to patients, e.g., mammals including humans.
The compounds of this invention can be employed in admixture with
conventional excipients, i.e., pharmaceutically acceptable organic or
inorganic
carrier substances suitable for parenteral, enteral (e.g., oral) or topical
application
which do not deleteriously react with the active compounds. Suitable
-27-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
pharmaceutically acceptable carriers include but are not limited to water,
salt
solutions, alcohols, gum arable, vegetable oils, benzyl alcohols, polyethylene
glycols, gelatine, carbohydrates such as lactose, amylose or starch, magnesium
stearate, talc, silicic acid, viscous paraffin, perfume oil, fatty acid
monoglycerides
and diglycerides, pentaerythritol fatty acid esters, hydroxy methylcellulose,
polyvinyl pyrrolidone, etc. The pharmaceutical preparations can be sterilized
and if
desired mixed with auxiliary agents, e.g., lubricants, preservatives,
stabilizers,
wetting agents, emulsifiers, salts for influencing osmotic pressure, buffers,
coloring,
flavoring and/or aromatic substances and the like which do not deleteriously
react
with the active compounds. They can also be combined where desired with other
active agents, e.g., vitamins.
For parenteral application, which includes intramuscular, intraderrnal,
subcutaneous, intranasal, intracapsular, intraspinal, intrasternal, and
intravenous
injection, particularly suitable are injectable, sterile solutions, preferably
oily or
aqueous solutions, as well as suspensions, emulsions, or implants, including
suppositories. Formulations for injection may be presented in unit dosage
form, e.g.,
in ampoules or in multi-dose containers, with an added preservative. The
compositions may take such forms as suspensions, solutions or emulsions in
oily or
aqueous vehicles, and may contain formulatory agents such as suspending,
stabilizing and/or dispersing agents. Alternatively, the active ingredient may
be in
powder form for constitution with a suitable vehicle, e.g., sterile pyrogen-
free water,
before use.
For enteral application, particularly suitable are tablets, dragees, liquids,
drops, suppositories, or capsules. The pharmaceutical compositions may be
prepared
by conventional means with pharmaceutically acceptable excipients such as
binding
agents (e.g., pregelatinised maize starch, polyvinylpyrrolidone or
hydroxypropyl
methylcellulose); fillers (e.g., lactose, microcrystalline cellulose or
calcium
hydrogen phosphate); lubricants (e.g., magnesium stearate, talc or silica);
disintegrants (e.g., potato starch or sodium starch glycolate); or wetting
agents (e.g.,
sodium lauryl sulphate). The tablets may be coated by methods well known in
the
art. Liquid preparations for oral administration may take the form of, for
example,
solutions, syrups or suspensions, or they may be presented as a dry product
for
constitution with water or other suitable vehicle before use. Such liquid
preparations
may be prepared by conventional means with pharmaceutically acceptable
additives
such as suspending agents (e.g., sorbitol syrup, cellulose derivatives or
hydrogenated
-28-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
edible fats); emulsifying agents (e.g., lecithin or acacia); non-aqueous
vehicles (e.g.,
almond oil, oily esters, ethyl alcohol or fractionated vegetable oils); and
preservatives (e.g., methyl or propyl-p-hydroxybenzoates or sorbic acid). The
preparations may also contain buffer salts, flavoring, coloring and sweetening
agents
as appropriate. A syrup, elixir, or the like can be used wherein a sweetened
vehicle
is employed.
Sustained or directed release compositions can be formulated, e.g., liposomes
or those wherein the active compound is protected with differentially
degradable
coatings, e.g., by microencapsulation, multiple coatings, etc. It is also
possible to
freeze dry the new compounds and use the lyophilizates obtained, for example,
for
the preparation of products for injection.
For administration by inhalation, the compounds for use according to the
present invention are conveniently delivered in the form of an aerosol spray
presentation from pressurized packs or a nebulizer, with the use of a suitable
propellant, e.g., dichlorodifluoromethane, trichlorofluoromethane,
dichlorotetrafluoroethane, carbon dioxide or other suitable gas. In the case
of a
pressurized aerosol the dosage unit may be determined by providing a valve to
deliver a metered amount. Capsules and cartridges of e.g. gelatin for use in
an
inhaler or insufflator may be formulated containing a powder mix of the
compound
and a suitable powder base such as lactose or starch.
For topical application, there are employed as non-sprayable forms, viscous
to semi-solid or solid forms comprising a carrier compatible with topical
application
and having a dynamic viscosity preferably greater than water. Suitable
formulations
include but are not limited to solutions, suspensions, emulsions, creams,
ointments,
powders, liniments, salves, aerosols, etc., which are, if desired, sterilized
or mixed
with auxiliary agents, e.g., preservatives, stabilizers, wetting agents,
buffers or salts
for influencing osmotic pressure, etc. For topical application, also suitable
are
sprayable aerosol preparations wherein the active ingredient, preferably in
combination with a solid or liquid inert carrier material, is packaged in a
squeeze
bottle or in admixture with a pressurized volatile, normally gaseous
propellant, e.g.,
a freon.
The compositions may, if desired, be presented in a pack or dispenser device
which may contain one or more unit dosage forms containing the active
ingredient.
The pack may for example comprise metal or plastic foil, such as a blister
pack. The
pack or dispenser device may be accompanied by instructions for
administration.
-29-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Genetic Immunization
Genetic immunization according to the present invention elicits an effective
immune response without the use of infective agents or infective vectors.
Vaccination techniques which usually do produce a CTL response do so through
the
use of an infective agent. A complete, broad based immune response is not
generally
exhibited in individuals immunized with killed, inactivated or subunit
vaccines. The
present invention achieves the full complement of immune responses in a safe
manner without the risks and problems associated with vaccinations that use
infectious agents.
According to the present invention, DNA or RNA that encodes a target
protein is introduced into the cells of an individual where it is expressed,
thus
producing the target protein. The DNA or RNA is linked to regulatory elements
necessary for expression in the cells of the individual. Regulatory elements
for DNA
include a promoter and a polyadenylation signal. In addition, other elements,
such as
a Kozak region, may also be included in the genetic construct.
The genetic constructs of genetic vaccines comprise a nucleotide sequence
that encodes a target protein operably linked to regulatory elements needed
for gene
expression. Accordingly, incorporation of the DNA or RNA molecule into a
living
cell results in the expression of the DNA or RNA encoding the target protein
and
thus, production of the target protein.
When taken up by a cell, the genetic construct which includes the nucleotide
sequence encoding the target protein operably linked to the regulatory
elements may
remain present in the cell as a functioning extrachromosomal molecule or it
may
integrate into the cell's chromosomal DNA. DNA may be introduced into cells
where it remains as separate genetic material in the form of a plasmid.
Alternatively,
linear DNA which can integrate into the chromosome may be introduced into the
cell. When introducing DNA into the cell, reagents which promote DNA
integration
into chromosomes may be added. DNA sequences which are useful to promote
integration may also be included in the DNA molecule. Since integration into
the
chromosomal DNA necessarily requires manipulation of the chromosome, it is
preferred to maintain the DNA construct as a replicating or non-replicating
extrachromosomal molecule. This reduces the risk of damaging the cell by
splicing
into the chromosome without affecting the effectiveness of the vaccine.
Alternatively, RNA may be administered to the cell. It is also contemplated to
-30-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
provide the genetic construct as a linear minichromosome including a
centromere,
telomeres and an origin of replication.
The necessary elements of a genetic construct of a genetic vaccine include a
nucleotide sequence that encodes a target protein and the regulatory elements
necessary for expression of that sequence in the cells of the vaccinated
individual.
The regulatory elements are operably linked to the DNA sequence that encodes
the
target protein to enable expression.
The molecule that encodes a target protein is a protein-encoding molecule
which is translated into protein. Such molecules include DNA or RNA which
comprise a nucleotide sequence that encodes the target protein. These
molecules
may be cDNA, genomic DNA, synthesized DNA or a hybrid thereof or an RNA
molecule such as mRNA. Accordingly, as used herein, the terms "DNA construct",
"genetic construct" and "nucleotide sequence" are meant to refer to both DNA
and
RNA molecules.
The regulatory elements necessary for gene expression of a DNA molecule
include: a promoter, an initiation codon, a stop codon, and a polyadenylation
signal.
In addition, enhancers are often required for gene expression. It is necessary
that
these elements be operable in the vaccinated individual. Moreover, it is
necessary
that these elements be operably linked to the nucleotide sequence that encodes
the
target protein such that the nucleotide sequence can be expressed in the cells
of a
vaccinated individual and thus the target protein can be produced.
Initiation codons and stop codons are generally considered to be part of a
nucleotide sequence that encodes the target protein. However, it is necessary
that
these elements are functional in the vaccinated individual.
Similarly, promoters and polyadenylation signals used must be functional
within the cells of the vaccinated individual.
Examples of promoters useful to practice the present invention, especially in
the production of a genetic vaccine for humans, include but are not limited to
promoters from Simian Virus 40 (SV40), Mouse Mammary Tumor Virus (MMTV)
promoter, Human Immunodeficiency Virus (HIV) such as the HIV Long Terminal
Repeat (LTR) promoter, Moloney virus, ALV, Cytomegalovirus (CMV) such as the
CMV immediate early promoter, Epstein Barr Virus (EBV), Rous Sarcoma Virus
(RSV) as well as promoters from human genes such as human Actin, human
Myosin, human Hemoglobin, human muscle creatine and human metalothionein.
-31-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Examples of polyadenylation signals useful to practice the present invention,
especially in the production of a genetic vaccine for humans, include but are
not
limited to SV40 polyadenylation signals and LTR polyadenylation signals. In
particular, the SV40 polyadenylation signal which is in pCEP4 plasmid
(Invitrogen,
San Diego Calif), referred to as the SV40 polyadenylation signal, can be used.
In addition to the regulatory elements required for DNA expression, other
elements may also be included in the DNA molecule. Such additional elements
include enhancers. The enhancer may be selected from the group including but
not
limited to: human Actin, human Myosin, human Hemoglobin, human muscle
creatine and viral enhancers such as those from CMV, RSV and EBV.
Genetic constructs can be provided with mammalian origin of replication in
order to maintain the construct extrachromosomally and produce multiple copies
of
the construct in the cell. Plasmids pCEP4 and pREP4 from Invitrogen (San
Diego,
Calif.) contain the Epstein Barr virus origin of replication and nuclear
antigen
EBNA-1 coding region which produces high copy episomal replication without
integration.
An additional element may be added which serves as a target for cell
destruction if it is desirable to eliminate cells receiving the genetic
construct for any
reason. A herpes thymidine kinase (tk) gene in an expressible form can be
included
in the genetic construct. When the construct is introduced into the cell, tk
will be
produced. The drug gangcyclovir can be administered to the individual and that
drug
will cause the selective killing of any cell producing tk. Thus, a system can
be
provided which allows for the selective destruction of vaccinated cells.
In order to be a functional genetic construct, the regulatory elements must be
operably linked to the nucleotide sequence that encodes the target protein.
Accordingly, it is necessary for the initiation and termination codons to be
in frame
with the coding sequence.
Open reading frames (ORFs) encoding the protein of interest and another or
other proteins of interest may be introduced into the cell on the same vector
or on
different vectors. ORFs on a vector may be controlled by separate promoters or
by a
single promoter. In the latter arrangement, which gives rise to a
polycistronic
message, the ORFs will be separated by translational stop and start signals.
The
presence of an internal ribosome entry site (IRES) site between these ORFs
permits
the production of the expression product originating from the second ORF of
-32-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
interest, or third, etc. by internal initiation of the translation of the
bicistronic or
polycistronic mRNA.
According to the invention, the genetic vaccine may be administered directly
into the individual to be immunized or ex vivo into removed cells of the
individual
which are reimplanted after administration. By either route, the genetic
material is
introduced into cells which are present in the body of the individual. Routes
of
administration include, but are not limited to, intramuscular,
intraperitoneal,
intradermal, subcutaneous, intravenous, intraarterially, intraoccularly and
oral as
well as transdermally or by inhalation or suppository. Preferred routes of
administration include intramuscular, intraperitoneal, intradermal and
subcutaneous
injection. Genetic constructs may be administered by means including, but not
limited to, traditional syringes, needleless injection devices, or
microprojectile
bombardment gene guns. Alternatively, the genetic vaccine may be introduced by
various means into cells that are removed from the individual. Such means
include,
for example, ex vivo transfection, electroporation, microinjection and
microprojectile bombardment. After the genetic construct is taken up by the
cells,
they are reimplanted into the individual. It is contemplated that otherwise
non-
immunogenic cells that have genetic constructs incorporated therein can be
implanted into the individual even if the vaccinated cells were originally
taken from
another individual.
The genetic vaccines according to the present invention comprise about 1
nanogram to about 1000 micrograms of DNA. In some preferred embodiments, the
vaccines contain about 10 nanograms to about 800 micrograms of DNA. In some
preferred embodiments, the vaccines contain about 0.1 to about 500 micrograms
of
DNA. In some preferred embodiments, the vaccines contain about I to about 350
micrograms of DNA. In some preferred embodiments, the vaccines contain about
25
to about 250 micrograms of DNA. In some preferred embodiments, the vaccines
contain about 100 micrograms DNA.
The genetic vaccines according to the present invention are formulated
according to the mode of administration to be used. One having ordinary skill
in the
art can readily formulate a genetic vaccine that comprises a genetic
construct. In
cases where intramuscular injection is the chosen mode of administration, an
isotonic formulation is preferably used. Generally, additives for isotonicity
can
include sodium chloride, dextrose, mannitol, sorbitol and lactose. In some
cases,
isotonic solutions such as phosphate buffered saline are preferred.
Stabilizers include
-33-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
gelatin and albumin. In some embodiments, a vaso-constriction agent is added
to the
formulation. The pharmaceutical preparations according to the present
invention are
provided sterile and pyrogen free.
Genetic constructs may optionally be formulated with one or more response
enhancing agents such as: compounds which enhance transfection, i.e.
transfecting
agents; compounds which stimulate cell division, i.e. replication agents;
compounds
which stimulate immune cell migration to the site of administration, i.e.
inflammatory agents; compounds which enhance an immune response, i.e.
adjuvants
or compounds having two or more of these activities.
In one embodiment, bupivacaine, a well known and commercially available
pharmaceutical compound, is administered prior to, simultaneously with or
subsequent to the genetic construct. Bupivacaine and the genetic construct may
be
formulated in the same composition. Bupivacaine is particularly useful as a
cell
stimulating agent in view of its many properties and activities when
administered to
tissue. Bupivacaine promotes and facilitates the uptake of genetic material by
the
cell. As such, it is a transfecting agent. Administration of genetic
constructs in
conjunction with bupivacaine facilitates entry of the genetic constructs into
cells.
Bupivacaine is believed to disrupt or otherwise render the cell membrane more
permeable. Cell division and replication is stimulated by bupivacaine.
Accordingly,
bupivacaine acts as a replicating agent. Administration of bupivacaine also
irritates
and damages the tissue. As such, it acts as an inflammatory agent which
elicits
migration and chemotaxis of immune cells to the site of administration. In
addition
to the cells normally present at the site of administration, the cells of the
immune
system which migrate to the site in response to the inflammatory agent can
come
into contact with the administered genetic material and the bupivacaine.
Bupivacaine, acting as a transfection agent, is available to promote uptake of
genetic
material by such cells of the immune system as well.
In addition to bupivacaine, mepivacaine, lidocaine, procains, carbocaine,
methyl bupivacaine, and other similarly acting compounds may be used as
response
enhancing agents. Such agents acts a cell stimulating agents which promote the
uptake of genetic constructs into the cell and stimulate cell replication as
well as
initiate an inflammatory response at the site of administration.
Other contemplated response enhancing agents which may function as
transfecting agents and/or replicating agents and/or inflammatory agents and
which
may be administered include lectins, growth factors, cytokines and lymphokines
-34-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
such as alpha-interferon, gamma-interferon, platelet derived growth factor
(PDGF),
gCSF, gMCSF, TNF, epidermal growth factor (EGF), IL-1, IL-2, IL-4, IL-6, IL-8,
IL-10 and IL-12 as well as collagenase, fibroblast growth factor, estrogen,
dexamethasone, saponins, surface active agents such as immune-stimulating
complexes (ISCOMS), Freund's incomplete adjuvant, LPS analog including
monophosphoryl Lipid A (MPL), muramyl peptides, quinone analogs and vesicles
such as squalene and squalane, hyaluronic acid and hyaluronidase may also be
used
administered in conjunction with the genetic construct. In some embodiments,
combinations of these agents are co-administered in conjunction with the
genetic
construct. In other embodiments, genes encoding these agents are included in
the
same or different genetic construct(s) for co-expression of the agents.
With respect to HIV Env, Gag, and Pol nucleotide sequences of the
invention, particularly through genetic immunization, may be used as
therapeutics or
prophylatics in the treatment of AIDS or HIV infection. A therapeutically
effective
dose refers to that amount of the compound sufficient to result in
amelioration of
symptoms or a prolongation of survival in a patient. Toxicity and therapeutic
efficacy of such compounds can be determined by standard pharmaceutical
procedures in cell cultures or experimental animals, e.g., for determining the
LD50
(the dose lethal to 50% of the population) and the ED50 (the dose
therapeutically
effective in 50% of the population). The dose ratio between toxic and
therapeutic
effects is the therapeutic index and it can be expressed as the ratio
LD50/ED50.
Compounds which exhibit large therapeutic indices are preferred. The data
obtained
from cell culture assays and animal studies can be used in formulating a range
of
dosage for use in humans. The dosage of such compounds lies preferably within
a
range of circulating concentrations that includes the ED50 with little or no
toxicity.
The dosage may vary within this range depending upon the dosage form employed
and the route of administration utilized. For any compound used in the method
of
the invention, the therapeutically effective dose can be estimated initially
from cell
culture assays. A dose may be formulated in animal models to achieve a
circulating
plasma concentration range that includes the (e.g., the concentration of the
test
compound which achieves a half maximal inhibition of viral infection relative
to the
amount of the event in the absence of the test compound) as determined in cell
culture. Such information can be used to more accurately determine useful
doses in
humans. Levels in plasma may be measured, for example, by high performance
liquid chromatography (HPLC).
-35-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
The compounds (for genetic immunization) of the invention may, further,
serve the role of a prophylactic vaccine, wherein the host produces antibodies
and/or
CTL responses against HIV Env, Gag, and Pol which responses then preferably
serve to neutralize HIV viruses by, for example, inhibiting further HIV
infection.
Administration of the compounds of the invention as a prophylactic vaccine,
therefore, would comprise administering to a host a concentration of compounds
effective in raising an immune response which is sufficient to elicit antibody
and/or
CTL reponses to HIV Env, Gag, and Pol and/or neutralize HIV, by, for example,
inhibiting HIV ability to infect cells. The exact concentration will depend
upon the
specific compound to be administered, but may be determined by using standard
techniques for assaying the development of an immune response which are well
known to those of ordinary skill in the art.
Env
To improve the immune response to native gpl60 and to expose the core
protein for optimal antigen presentation and recognition, we have analyzed the
immune response to modified forms of the protein. The role of conserved N-
linked
glycosylation sites has been studied, and analogues of fusion intermediates
have
been developed. Expression vectors with deletions in the cleavage site (C),
the
fusion peptide (F), and the interspace (I) between the two heptad repeats were
termed ACFI. Plasmid DNA vaccination has been a useful technology for the
development and analysis of immunogens. This method of vaccination allows
appropriate post-translational modification, proper intracellular trafficking,
and
antigen presentation. Direct injection of naked DNA either intramuscularly or
intradennally in rodents induces immune responses, and the ability to easily
modify
plasmid expression vectors to express different forms of HIV envelope proteins
enables rapid and systematic testing of vaccine immunogens. In this
disclosure, we
have analyzed the immune response to modified Env candidates expressed in
plasmids with modified codons to improve gene expression. Both antibody and
CTL responses were analyzed after injection of plasmid DNA into muscle. A
modified gp140 DNA with improved ability to elicit antibody and CTL. responses
to
HIV Env has now been identified that is envisioned as a prototype immunogen
that
can elicit broadly neutralizing antibody responses to HIV.
-36-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Exposing the Core Protein of Viral Membrane Fusion Proteins
Described herein are modified HIV envelope proteins that improve the
immune response to native gpl60 and expose the core protein for optimal
antigen
presentation and recognition. Weissenhorn et al., Molecular Cell, 2, 605-616,
1998
proposes a core protein as a model for a fusion intermediate of viral
glycoproteins,
where the glycoproteins are characterized by a central triple stranded coiled
coil
followed by a disulfide-bonded loop that reverses the chain direction and
connects to
an a helix packed antiparallel to the core helices, as, for example, in the
case of
Ebola Zaire GP2, Murine Moloney Leukemia virus (MuMoLv) 55-residue segment
of the TM subunit (Mo-55), low-pH-treated influenza HA2, protease resistant
core
of HIV gp4l, and SIV gp4l (Figure A). Thus, the strategy for improving the
immune response by exposing the protease resistant core of HIV gp4l extends to
other viral membrane fusion proteins that are characterized by a central
triple
stranded coiled coil followed by a disulfide-bonded loop that reverses the
chain
direction and connects to an a helix packed antiparallel to the core helices.
The present approach involves a series of internal mutations designed to
replace the cleavage site (C), the fusion domain (F), and the interspace (I)
between
the two heptad repeats all on a backbone of COOH-terminal truncations to
expose
the core protein of the viral membrane fusion protein Env, based on modified
gpl40
as a prototype immunogen. By replacement is meant deletions, insertions,
and/or
substitutions of amino acid residues. In one embodiment, deletions are meant
(i.e.,
amino acids are deleted to create the ACFI mutations).
In this embodiment, the AC mutation is intended to eliminate proteolysis by
deleting the gpl20/gp41 cleavage site that links the envelope covalently to
the
ectodomain by 100%, 99%, 98%, 97%, 96%, 95%, 94%, 93%, 92%, 91%, 90%,
89%, 88%, 87%, 86%, 85%, 84%, 83%, 82%, 81,%, 80%, 79%, 78%, 77%, 76%,
75%, 74%, 73,%, 72%, 71%, 70%, 69%, 68%, 67%, 66%, 65%, 64%, 63%, 62%,
61%, 60%, 59%, 58%, 57%, 56%, 55%, 54%, 53%, 52%, 51%, 50%, 49%, 48%,
47%,46%,45%,44%,43%,42,41%,40%, 39%, 38%, 37%, 36%, 35%, 34%, 33%,
32%, 31%, 30%, 29%, 28%, 27%, 26%, 25%, 24%, 23%, 22%, 21%, 20%, 19%,
18%,17%,16%,15%,14%,13%,12%,11%,10%, 9%, 8%, 7%, 6%, 5%, 4%, 3%,
2%, or 1%.
In this embodiment, the AF mutation is intended to solubilize the molecule
by deleting the fusion domain by 100%, 99%, 98%, 97%, 96%, 95%, 94%, 93%,
-37-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
92%, 91%, 90%, 89%, 88%, 87%, 86%, 85%, 84%, 83%, 82%, 81,%, 80%, 79%,
78%, 77%, 76%, 75%, 74%, 73,%, 72%, 71%, 70%, 69%, 68%, 67%, 66%, 65%,
64%, 63%, 62%, 61%, 60%, 59%, 58%, 57%, 56%, 55%, 54%, 53%, 52%, 51%,
50%,49%,48%,47%,46%,45%,44%,43%,42, 41%, 40%, 39%, 38%, 37%, 36%,
35%, 34%, 33%, 32%, 31%, 30%, 29%, 28%, 27%, 26%, 25%, 24%, 23%, 22%,
21%, 20%, 19%,18%,17%, 16%,15%,14%, 13%,12%,11%,10%, 9%, 8%, 7%,
6%,5%,4%,3%,2%, or 1%.
In this embodiment, the At mutation is intended to stabilize oligomer
formation by deleting the interspace between the two heptad repeats by 100%,
99%,
98%, 97%, 96%, 95%, 94%, 93%, 92%, 91%, 90%, 89%, 88%, 87%, 86%, 85%,
84%, 83%, 82%, 81,%, 80%, 79%, 78%, 77%, 76%, 75%, 74%, 73,%, 72%, 71%,
70%, 69%, 68%, 67%, 66%, 65%, 64%, 63%, 62%, 61%, 60%, 59%, 58%, 57%,
56%, 55%,54%,53%, 52%, 51%, 50%,49%,48%,47%,46%,45%,44%,43%,42,
41%, 40%, 39%, 38%, 37%, 36%, 35%, 34%, 33%, 32%, 31%, 30%, 29%, 28%,
27%, 26%, 25%, 24%, 23%, 22%, 21%, 20%, 19%, 18%, 17%, 16%, 15%, 14%,
13%,12%,11%,10%,9%,8%,7%,6%,5%,4%,3%,2%, or 1%.
In this embodiment, the COON-terminal truncation is intended to reduce
toxicity by deleting the cytoplasmic domain by 100%, 99%, 98%, 97%, 96%, 95%,
94%, 93%, 92%, 91%, 90%, 89%, 88%, 87%, 86%, 85%, 84%, 83%, 82%, 81,%,
80%, 79%, 78%, 77%, 76%, 75%, 74%, 73,%, 72%, 71%, 70%, 69%, 68%, 67%,
66%, 65%, 64%, 63%, 62%, 61%, 60%, 59%, 58%, 57%, 56%, 55%, 54%, 53%,
52%,51.%,50%,49%,48%,47%,46%,45%,44%,43%,42,41%,40%, 39%,38%,
37%, 36%, 35%, 34%, 33%, 32%, 31%, 30%, 29%, 28%, 27%, 26%, 25%, 24%,
23%,22%,21%,20%,19%,1,8%,17%,1,6%,15%,14%,13%,12%,11%,10%,9%,
8%, 7%,6%,5%,4%,3%,2%, or 1%.
In this embodiment, optionally, the COOH-terminal truncation is extended
so as to solubilize the molecule by deleting the transmembrane domain by 100%
99%, 98%, 97%, 96%, 95%, 94%, 93%, 92%, 91%, 90%, 89%, 88%, 87%, 86%,
85%, 84%, 83%, 82%, 81,%, 80%, 79%, 78%, 77%, 76%, 75%, 74%, 73,%, 72%,
71%, 70%, 69%, 68%, 67%, 66%, 65%, 64%, 63%, 62%, 61%, 60%, 59%, 58%,
57%, 56%, 55%, 54%, 53%, 52%, 51%, 50%, 49%, 48%, 47%, 46%, 45%, 44%,
43%, 42, 41%, 40%, 39%, 38%, 37%, 36%, 35%, 34%, 33%, 32%, 31%, 30%, 29%,
28%, 27%, 26%, 25%, 24%, 23%, 22%, 21%, 20%, 19%, 18%, 17%, 16%, 15%,
14%,13%,12%,11%,10%,9%,8%,7%,6%,5%,4%,3%,2%, or 1%.
-38-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Amino acid substitutions may encompass those of a conserved or non-
conserved nature. Presumably, a non-conserved substitution of a domain would
act
like a deletion of the domain. Conserved amino acid substitutions constitute
switching one or more amino acids with amino acids of similar charge, size,
and/or
hydrophobicity characteristics. Non-conserved amino acid substitutions
constitute
switching one or more amino acids with amino acids of dissimilar charge, size,
and/or hydrophobicity characteristics. The families of amino acids include the
basic
amino acids (lysine, arginine, histidine); the acidic amino acids (aspartic
acid,
glutamic acid); the non-polar amino acids (alanine, valine, leucine,
isoleucine,
proline, phenylalanine, methionine, tryptophan); the uncharged polar amino
acids
(glycine, asparagine, glutamine, cysteine, serine, threonine, tyrosine); and
the
aromatic amino acids (phenylalanine, tryptophan, and tyrosine). One or more
substitutions may be introduced to achieve the AC mutation intended to
eliminate
proteolysis by acting like a deletion of the gpl20/gp41, cleavage site to link
the
envelope covalently to the ectodomain, the AF mutation intended to solubilize
the
molecule by acting like a deletion of the fusion domain, the Al mutation
intended to
stabilize oligomer formation by acting like a deletion of the interspace
between the
two heptad repeats, the COOH-terminal truncation intended to reduce toxicity
by
acting like a deletion of the cytoplasmic domain, and, optionally, the COOH-
terminal truncation extended so as to solubilize the molecule by acting like a
deletion of the transmembrane domain.
Amino acid insertions may constitute single amino acid residues or stretches
of residues. The insertions may be made at the carboxy or amino terminal end
of a
domain, as well as at a position internal to the domain. Such insertions will
generally range from 2 to 15 amino acids in length. One or more insertions may
be
introduced to achieve the AC mutation intended to eliminate proteolysis by
acting
like a deletion of the gp l20/gp4l cleavage site to link the envelope
covalently to the
ectodomain, the AF mutation intended to solubilize the molecule by acting like
a
deletion of the fusion domain, the Al mutation intended to stabilize oligomer
formation by acting like a deletion the interspace between the two heptad
repeats,
the COOH-terminal truncation intended to reduce toxicity by acting like a
deletion
of the cytoplasmic domain, and, optionally, the COOH-terminal truncation
extended
so as to solubilize the molecule by acting like a deletion of the
transmembrane
domain.
-39-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
The nucleic acids of the present invention are optionally DNA, RNA, or
mRNA. Most typically, the nucleic acids are provided by recombinantly making a
DNA, which is expressed in a cell as RNA and/or as mRNA. Given the strategy
for
making the nucleic acids of the present invention, one of skill can construct
a variety
of clones containing functionally equivalent nucleic acids. Cloning
methodologies to
accomplish these ends, and sequencing methods to verify the sequence of
nucleic
acids are well known in the art. Examples of appropriate cloning and
sequencing
techniques, and instructions sufficient to direct persons of skill through
many
cloning exercises are found in Berger and Kimmel, Guide to Molecular Cloning
Techniques, Methods in Enzymology, volume 152, Academic Press, Inc., San
Diego, Calif. (Berger); Sambrook, et al. (1989) Molecular Cloning--A
Laboratory
Manual (2nd ed.) Vol. 1-3, Cold Spring Harbor Laboratory, Cold Spring Harbor
Press, N.Y., (Sambrook); and Current Protocols in Molecular Biology, F. M.
Ausubel, et al., eds., Current Protocols, a joint venture between Greene
Publishing
Associates, Inc. and John Wiley & Sons, Inc., (1994 Supplement) (Ausubel).
Product information from manufacturers of biological reagents and experimental
equipment also provide information useful in known biological methods. Such
manufacturers include the SIGMA chemical company (Saint Louis, Mo.), R&D
systems (Minneapolis, Minn.), Pharmacia LIMB Biotechnology (Piscataway, N.J.),
CLONTECH Laboratories, Inc. (Palo Alto, Calif), Chem Genes Corp., Aldrich
Chemical Company (Milwaukee, Wis.), Glen Research, Inc., GIBCO BRL Life
Technologies, Inc. (Gaithersberg, Md.), Fluka Chemica-Biochemika Analytika
(Fluka Chemie AG, Buchs, Switzerland), Invitrogen, San Diego, Calif, and
Applied
Biosystems (Foster City, Calif), as well as many other commercial sources
known
to one of skill.
The nucleic acid compositions of this invention, whether RNA, cDNA,
mRNA, genomic DNA, or a hybrid of the various combinations, are isolated from
biological sources or synthesized in vitro. The nucleic acids of the present
invention
are present in transformed or transfected whole cells, in transformed or
transfected
cell lysates, or in a partially purified or substantially pure form.
In vitro amplification techniques suitable for amplifying sequences to
provide a nucleic acid or for subsequent analysis, sequencing or subcloning
are
known. Examples of techniques sufficient to direct persons of skill through
such in
vitro amplification methods, including the polymerase chain reaction (PCR) the
-40-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
ligase chain reaction (LCR), Q3-replicase amplification and other RNA
polymerase
mediated techniques (e.g., NASBA) are found in Berger, Sambrook, and Ausubel,
as
well as Mullis, et al., (1987) U.S. Pat. No. 4,683,202; PCR Protocols A Guide
to
Methods and Applications (Innis, et al. eds) Academic Press Inc. San Diego,
Calif.
(1990) (Innis); Arnheim & Levinson (Oct. 1, 1990) C&EN 36-47; The Journal Of
NIH Research (1991) 3:81-94; Kwoh, et al., Proc. Natl. Acad. Sci. USA, 86:1173
(1989); Guatelli, et al., Proc. Natl. Acad. Sci. USA, 87:1874 (1990); Lomell,
et al., J.
Clin. Chem., 35:1826 (1989); Landegren, et al., Science, 241:1077-1080 (1988);
Van Brunt, Biotechnology, 8:291-294 (1990); Wu and Wallace, Gene, 4:560
(1989);
Barringer, et al., Gene, 89:117 (1990), and Sooknanan and Malek,
Biotechnology,
13:563-564 (1995). Improved methods of cloning in vitro amplified nucleic
acids
are described in Wallace, et al., U.S. Pat. No. 5,426,039. Improved methods of
amplifying large nucleic acids (up to 40 kb) are summarized in Cheng, et al.,
Nature,
369:684-685 (1994) and the references therein. One of skill will appreciate
that
essentially any RNA can be converted into a double stranded DNA suitable for
restriction digestion, PCR expansion and sequencing using reverse
transcriptase and
a polymerase. See, Ausubel, Sambrook, Innis, and Berger, all supra.
One of skill will recognize many ways of generating alterations in a given
nucleic acid construct. Such well-known methods include site-directed
mutagenesis,
PCR amplification using degenerate oligonucleotides, exposure of cells
containing
the nucleic acid to mutagenic agents or radiation, chemical synthesis of a
desired
oligonucleotide (e.g., in conjunction with ligation and/or cloning to generate
large
nucleic acids) and other well-known techniques. See, Giliman and Smith, Gene
8:81-97 (1979), Roberts, et al., Nature, 328:731-734 (1987) and Sambrook,
Innis,
Ausubel, Berger, and Mullis (all supra).
Most modifications to nucleic acids are evaluated by routine screening
techniques in suitable assays for the desired characteristic. For instance,
changes in
the immunological character of encoded polypeptides can be detected by an
appropriate immunological assay. For instance, changes in the cellular
immunological character of the polypeptide can be detected by an appropriate
antibody or CTL assay. Modifications of other properties such as nucleic acid
hybridization to a complementary nucleic acid, redox or thermal stability of
encoded
proteins, hydrophobicity, susceptibility to proteolysis, or the tendency to
aggregate
are all assayed according to standard techniques.
-41-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
A wide variety of formats and labels are available and appropriate for
detection of polypeptide sequences. These include analytic biochemical methods
such as spectrophotometry, radiography, electrophoresis, capillary
electrophoresis,
high performance liquid chromatography (HPLC), thin layer chromatography
(TLC), hyperdiffusion chromatography, and the like, and various immunological
methods such as fluid or gel precipitin reactions, immunodiffusion (single or
double), immunoelectrophoresis, radioimmunoassays (RIAs), enzyme-linked
immunosorbent assays (ELISAs), western blot assays, immunofluorescent assays,
and the like. Several commercially available ELISA assays for the detection of
retroviral components, including Env domains, are available, allowing one of
skill to
detect Env in biological samples.
Similarly, the detection of the nucleic acids of the present invention
proceeds
by well known methods such as Southern analysis, northern analysis, gel
electrophoresis, PCR, radiolabeling and scintillation counting, and affinity
chromatography. Many assay formats are appropriate, including those reviewed
in
Tijssen (1993) Laboratory Techniques in biochemistry and molecular biology--
hybridization with nucleic acid probes parts I and II, Elsevier, New York and
Choo
(ed) (1994) Methods In Molecular Biology Volume 33--In Situ Hybridization
Protocols, Humana Press Inc., New Jersey (see also, other books in the Methods
in
Molecular Biology series); see especially, Chapter 21 of Choo (id.) "Detection
of
Virus Nucleic Acids by Radioactive and Nonisotopic in Situ Hybridization".
Finally,
PCR is also routinely used to detect nucleic acids in biological samples (see,
Innis,
supra, for a general description of PCR techniques).
In one preferred embodiment, antibodies are used to detect polypeptide
sequences. Methods of producing polyclonal and monoclonal antibodies are known
to those of skill in the art, and many anti-HIV antibodies are available. See,
e.g.,
Coligan (1991) Current Protocols in Immunology, Wiley/Greene, NY; and Harlow
and Lane (1989) Antibodies: A Laboratory Manual, Cold Spring Harbor Press, NY;
Stites, et al. (eds.) Basic and Clinical Immunology (4th ed.), Lange Medical
Publications, Los Altos, Calif., and references cited therein; Goding (1986)
Monoclonal Antibodies: Principles and Practice (2d ed.), Academic Press, New
York, N.Y.; and Kohler and Milstein, Nature, 256:495-497 (1975). Other
suitable
techniques for antibody preparation include selection of libraries of
recombinant
antibodies in phage or similar vectors. See, Huse, et al., Science, 246:1275-
1281
(1989); and Ward, et al., Nature, 341:544-546 (1989). Specific monoclonal and
-42-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
polyclonal antibodies and antisera will usually bind with a KD of at least
about 0.1
mM, more usually at least about I M, preferably at least about 0.1 M or
better,
and most typically and preferably, 0.01 M or better.
Development of HIV Env Vectors
To develop Env glycoprotein variants that might effectively induce humoral
and cellular immunity, a series of plasmid expression vectors were generated
(Fig.
I). HIV Env is encoded by nucleic acid sequences that contain RNA structures
that
limit gene expression (Andre S et al, 1998, J Virol, 72:1497-503; Qiu JT et
al., 1999,
J Virol, 73:9145-52; Rana TM et al., 1999, Arch Biochem Biophys, 365:175-85;
Roebuck KA, Saifuddin M, 1999, Gene Expr, 8:67-84; Romano G, et al., 1999, J
Cell Biochem, 75:357-68; Schneider R et al., 1997, J Virol, 71:4892-903).
These
vectors were therefore synthesized using codons found in human genes that
allow
these structures to be eliminated without affecting the amino acid sequence.
Full-
length HIV Env (gp160) was highly expressed in the absence of HIV accessory
proteins at levels >10-fold higher than Rev-dependent viral gp 160 in
transfected
293 cell by Western blot analysis (Fig. IIA), and the relevant mutant proteins
were
detected at the expected apparent molecular weights (Fig. ITC-D). As might be
anticipated, gp16O expressed from the synthetic gene was not efficiently
processed
in transfected 293 cells, presumably because over-expression of gp160
saturates the
cellular proteases responsible for cleavage. (Binley JM et al., 2000, J.
Virol, 74:627-
643.) Synthetic HIV gpl60 induced toxicity in transfected cells, with cell
rounding
and detachment evident within 48 hours (Fig. ILIA vs B). This cytotoxicity was
reduced by elimination of the COON-terminal cytoplasmic domain. Env protein
that terminated at amino acid 752 (gpl50) was less cytotoxic than gp160, while
the
shorter proteins (gp145 and gp140) produced little or no effect (Fig. IIIC, D,
and E,
respectively).
To alter Env immunogenicity, two different approaches were explored. First
the effects of glycosylation on cellular and humoral immunity were evaluated
by
analysis of mutants in which conserved N-linked glycosylation sites were
eliminated
by site-directed mutagenesis. Two sets of mutations were introduced into both
gp160 and gp150 (Fig. 1). The first set included eleven potential sites
(zgly11), and
the second set included an additional 6 sites downstream (4glyl7). Expression
studies showed that the glycosylation mutants were efficiently expressed, and
the
-43-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
glycosylation mutant protein was appropriately reduced in size compared to
wild
type gp160 or gp150, consistent with reduced N-linked glycosylation (Fig 11C).
The second approach involved a series of internal deletions designed to
stabilize and expose functional domains of the protein that might be present
in an
extended helical structure prior to the formation of the six-member coiled-
coil
structure in the hairpin intermediate (Chan DC et al., 1997, Cell, 89:263-73;
Weissenhorn W et al., 1997, Nature, 387:426-30). To generate this putative pre-
hairpin structure, the cleavage site was removed to prevent the proteolytic
processing of the envelope and stabilize the protein by linking it covalently
to the
gp4l extracellular and/or transmembrane domain. To reduce toxicity and enhance
stability, the fusion peptide domain was deleted. The heptad repeats in the
envelope
protein are important tertiary structure domains involved in the ability of
the
envelope protein to form trimers (Lu M, Kim PS, 1997, J Biomol Struct Dyn,
15:465-71). The sequence between the heptad repeats was removed to stabilize
the
formation of trimers and eliminate formation of the hairpin intermediate.
These
zCFI deletions were introduced into full-length gp160 and COOH-terminal
truncation mutants. Though cells trasfected with vectors encoding gp l40ACFI,
gpl45ACFI and gp160 readily expressed these proteins (Fig. 11D), only
gpl40ACFI,
which lacks the transmembrane domain, was readily detected in the supernatant
(Fig. IV), indicating that it can give rise to soluble antigen.
Immunogenicity of Env Mutants After DNA Vaccination
The ability of these Env proteins to elicit an immune response was
determined in mice by injection with these plasmid DNA expression vectors.
Antibody responses were monitored by the ability of antisera from injected
mice to
immunoprecipitate wild type gpl60 from cell lysates by SDS-PAGE and Western
blotting (Fig. V). In some cases, antibody reactivity was also confirmed by
immunofluorescence. To quantitate the antibody response, immunoprecipitation
followed by Western blotting with different dilutions of immunized mouse sera
was
tested. The intensity of the gp 160 band was determined by densitometry and
standardized relative to a positive control sera used to normalize data
between
experiments. The approximately linear dose response of gpl60 intensity with
serum
dilution allowed quantification of the anti-gpl60 antibody response in mouse
sera.
Data from immunized mice showed that none of the wild type Env proteins,
neither
the gp 160, gp 150, gp 145, nor gp 140 COOH-terminal truncations generated
-44-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
consistently high antibody responses (Fig. VA). Of these proteins, gp140 was
somewhat more effective than gp160 in generating antibody responses but
remained
low and inconsistent. Immunization with vectors designed to express
glycosylation
deficient envelope proteins also did not improve the humoral response. In
contrast,
the ACFI mutants in the Env truncation vectors substantially increased anti-
gp160
antibody response (Fig. VA; gp 145, gp 140, and gp128). The longer ACFI Env
proteins (gp160 and gp150) did not show comparable enhancement of this
response.
The gp140 (ACFI) provided more consistent and a greater increase in antibody
response than gp128ACFI (Fig VB). In all cases, these vectors that encoded
gpl40ACFI, but not wild type gpl40, induced antibodies that were reactive to
native
gp160 (Fig. VC).
To determine whether these modifications of Env adversely affected CTL
responses, spleen cells from immunized mice were tested for their ability to
lyse
relevant target cells. All mice immunized with codon-altered Env vectors,
including
COOH-terminal deletion mutations (Fig. VIA) or glycosylation mutants (Fig.
VIB),
elicited strong CTL responses directed to cell lines pulsed with HIV Env
peptides.
These findings were also confirmed using stably transfected cells that
expressed
Env. Importantly, gpl40 (ACFI), which elicited increased antibody responses
relative to the comparable wild typ Env, readily induced CTL responses to
native
Env, as did other ACFI mutants (Fig. VIC). Addition of anti-CD8 antibody
inhibited
cytolytic activity, as did depletion using magnetic beads coupled with anti-
CD8
antibody, thus confirming a cytotoxic T cell response to these immunogens
after
genetic immunization (Fig. VID). This response was detectable for at least six
months after immunization.
Glycosylation and ACFI HIV Env Mutants
To develop DNA vaccine candidates for HIV, we developed a series of
synthetic genes designed to express HIV Env mutants in human cells. In the
absence of HIV regulatory proteins, these codon-altered envelope protein genes
expressed well in human cells. Like other DNA vaccines, immunization with
these
vectors elicited strong CTL responses in mice, and antibody responses were not
robust in mice immunized with wild type Env expression vectors. Mutations in
highly conserved N-linked glycosylation sites did not significantly alter
humoral or
cellular immune response to native Env. In contrast, a mutant Env with
deletions in
the cleavage site, fusion domain, and a region between the heptad repeats
elicited a
-45-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
more potent humoral immune response and retained its ability to stimulate Env-
specific CTL.
Recent reports suggest that gp160 forms trimers in vivo and the domain
required for trimer formation resides in the ectodomain of the gp41 (Yang X et
al.,
2000, J Virol, 74:5716-25). Such trimeric forms of HIV envelope protein are
likely
to present different epitopes to the immune system compared to monomeric
gp120.
In addition to the linear epitopes in the envelope, this trimeric structure is
likely to
expose conformational epitopes important for B cell triggering of a relevant
antibody response. In this regard, gp140 (ACFI), which induced the greatest
antibody response, is released in a soluble form (Fig. IVB). In contrast, wild
type
Env did not elicit high titer antibody responses. The toxicity of Env in
mammalian
cells has been seen and could limit both the amount and duration of envelope
protein
expression in vivo that would affect immunogenicity. The envelope is also
heavily
glycosylated, and removal of partial or complete gp120 glycosylation sites has
resulted in higher titers of strain-specific neutralizing antibody responses
to mutant
SIVs in monkeys (Binley JM et al., 1998, AIDS Res Hum Retroviruses, 14 :191-8;
Reitter JN et al., 1998, Nat Med, 4:679-84). Though it seemed reasonable that
deglycosylation would reveal epitopes otherwise masked in the native protein,
we
did not observe enhanced immune reactivity by DNA vaccination using different
glycosylation site mutants, both in gp160 and gp150. This difference with the
previous study is likely due to the fact that DNA vaccination rather than
viral
infection was utilized for immunization. Though glycosylation mutants are
unlikely
to prove helpful with this former method of immunization, we envision that
modification of glycosylation sites will be effective with other vectors or
adjuvants.
HIV-1 Env is proteolytically cleaved by a cellular convertase into gp120 and
gp41 (Eckert DM et al., 1999, Cell, 99:103-15). The gp4l subunit is composed
of
cytoplasmic, transmembrane, and ectodomain segments. The role of the
ectodomain
of the envelope in membrane fusion, particularly its hydrophobic glycine-rich
fusion
peptide, is well established. Two regions with heptad coiled-coil repeats in
the
ectodomain of gp4l are involved in viral fusion (Eckert DM et al., 1999, Cell,
99:103-15; Weissenhorn W et al., 1996, EMBO J, 15:1507-14). Upon fusion, these
two alpha helices, connected via a disulfide-stabilized loop (Gallaher WR et
al.,
1989, AIDS Res Hum Retroviruses, 5:431-40; Kent KA, Robinson J, 1996, AIDS,
10 Suppl A:S107-S114; Sattentau QJ et al., 1995, Virology, 206:713-7),
presumably
-46-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
undergo a transient conformational change to a fusion active state. These
changes
allow the formation of a six-member helical hairpin intermediate structure
that
presumably exposes the fusion peptide at the NH2-terminus of gp4l, allowing
fusion
to the target cell membrane (Binley J, Moore JP,[published erratum appears in
Nature 1997 Sep 11;389(6647):131], 1997, Nature, 387 :346-8; LaCasse R et al.,
1999, Science, 283:357-62). The ACFI mutation was intended to eliminate
cleavage
of gpl40, remove the unstable hydrophobic region and stabilize oligomer
formation.
Though detailed structural data is not yet available on this protein, these
mutations
apparently stabilize Env in a conformation that elicits both humoral and
cellular
immune responses. For example, the neutralizing epitope in the ectodomain of
gp4l
(Muster T et al., 1994, J Virol, 68:4031-4) is present in the series of
deletions and
truncations of the envelope and gpl40 (ACFI) is reactive with the 2F5
neutralizing
monoclonal antibody that binds to this epitope. Importantly, these immunogens
also
induced CTL responses to Env. Though gp 128 (ACFI) induced slightly more
potent
CTL activity, gp140 (ACFI) was better able to elicit such responses, both to
peptide-
pulsed cells and stably transduced target cells. Thus the enhanced humoral
immune
response introduced by this vaccine candidate did not appear to diminish the
CTL
response. Taken together, these results indicate that gpl40ACFI serves as an
improved immunogen that can more effectively elicit an antibody response
against
the envelope by DNA vaccination while preserving its ability to induce a CTL
response.
Gag and Pol
In this disclosure, we have prepared synthetic HIV-1 B Glade Gag and Pol
expression vectors that are based on human (h) codon usage. These vectors
encode
hGag-Pol and its derivatives, hGag, hPol and an hGag-Pol fusion protein. The
synthetic Gag-Pol genes show little nucleotide homology to HIV-1 but are the
same
in protein sequence. The modified Gag-Pol genes were subcloned into a
eukaryotic
plasmid expression vector for expression and DNA immunization studies.
Synthetic
Gag-Pol genes allowed high level Rev-independent expression of HIV-1 Gag-Pol
precursor proteins in human and mouse cell lines and induced significant
cellular
and humoral responses in mice. The Gag-Pol fusion protein induced the broadest
responses to Gag and Pol determinants and thus is envisioned as a prototype
immunogen that maximizes epitope presentation.
-47-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Eliminating The Frame Shift Site To Create Viral Polyproteins
Described herein are HIV Gag-Pol fusion proteins encoded by a continuous
open reading frame so to improve the immune response to native Gag and Pol. In
some viruses, translational frame shifting is exploited during protein
synthesis.
Specific sequences in the RNA are required for the frame shifting. The viral
RNA
sequences cause ribosomal slippage so that viral proteins are produced in non-
equivalent ratios. For example, during translation in HIV, the ribosomes shift
reading frames to synthesize Gag precursor protein and the gag-pol fusion
protein in
a 20:1 ratio. The strategy here is to maximize epitope presentation by
transcribing
an immunogen from a continuous open reading frame by eliminating the frame
shift
site. Thus, the strategy for improving the immune response by the use of a HIV
Gag-Pol fusion protein encoded by a continuous open reading frame extends to
other
viral proteins that are produced in non-equivalent ratios by virtue of
translational
frame shifting.
The present invention involves HIV Gag-Pol fusion proteins encoded by a
single continuous open reading frame due to mutation of the frame shift site.
The
frame shift site is mutated by deletions, insertions, and/or substitutions of
nucleotides to create a single continuous open reading frame. In one
embodiment,
deletions are meant (i.e., nucleotides are deleted to create the same open
reading
frame).
The frame shift site is a mutated frame shift to create a single continuous
open reading frame. For example, a set of similar retroviral gag-pol frame
shift sites
are optionally made for a given fusion protein, for example, by synthesizing
different gag-pol frame shift regions and cloning the sequences appropriately,
or by
site-directed mutagenesis of a given frame shift clone. The efficacy of the
frame
shift sites are assessed by measuring the production of the fusion protein.
The
sequence that shows the highest level of expression is a "optimized" frame
shift
mutation for the set assessed. Alternatively, where a particular level of
expression is
desired, a frame shift site from a particular set of possible frame shift
sites which is
closest to the desired activity level is considered to be "optimized."
Although a full length Gag sequence is preferred for use in the fusion protein
of the present invention, Gag is optionally deleted of subsequences without
negating
a polyepitope response. For example, regions of the matrix protein (p 17),
regions of
the capsid protein (p24), regions of p2, regions of the nucleocapsid protein
(p7),
regions of pl, and regions of p6 can be deleted while preserving the
polyepitope
-48-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
response. Alternatively, regions of the matrix protein (p17), regions of the
capsid
protein (p24), regions of p2, regions of the nucleocapsid protein (p7),
regions of pl,
and regions of p6 can be substituted while preserving the polyepitope response
.
Alternatively, regions of the matrix protein (p17), regions of the capsid
protein
(p24), regions of p2, regions of the nucleocapsid protein (p7), regions of pl,
and
regions of p6 can be interrupted by insertions while preserving the
polyepitope
response. Optionally, regions of the matrix protein (p17), regions of the
capsid
protein (p24), regions of p2, regions of the nucleocapsid protein (p7),
regions of pl,
and regions of p6 can be mutated to inactivate these proteins.
Likewise, a full length Pol sequence is preferred for use in the fusion
protein
of the present invention, and Pol is optionally deleted of subsequences
without
negating a polyepitope response. For example, regions of the protease protein,
regions of the reverse transcriptase protein, and regions of the integrase
protein can
be deleted while preserving the polyepitope response. Alternatively, regions
of the
protease protein, regions of the reverse transcriptase protein, and regions of
the
integrase protein can be substituted while preserving the polyepitope
response.
Alternatively, regions of the protease protein, regions of the reverse
transcriptase
protein, and regions of the integrase protein can be interrupted by insertions
while
preserving the polyepitope response. Optionally, regions of the protease
protein,
regions of the reverse transcriptase protein, and regions of the integrase
protein can
be mutated to inactivate these enzymes.
In other embodiments, the Gag-Pol fusion proteins are co-expressed with
other proteins, either as fusion proteins or separately, preferably with Env
sequence
proteins, such as the modified HIV Env ACFI proteins described herein.
In another aspect, the invention involves chimeric nucleic acid molecules.
The chimeric nucleic acid molecules of the invention typically have a
retroviral gag
nucleic acid sequence and a retroviral pol nucleic acid sequence in the same
open
reading frame due to mutation of the frame shift site. The continuous open
reading
frame encodes a fusion protein such as those described above.
The chimeric nucleic acids of the present invention are optionally DNA,
RNA, or mRNA. Most typically, the chimeric nucleic acids are provided by
recombinantly making a DNA, which is expressed in a cell as RNA and/or as
mRNA. Given the strategy for making the chimeric nucleic acids of the present
invention, one of skill can construct a variety of clones containing
functionally
-49-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
equivalent nucleic acids. Cloning methodologies to accomplish these ends, and
sequencing methods to verify the sequence of nucleic acids are well known in
the
art. Examples of appropriate cloning and sequencing techniques, and
instructions
sufficient to direct persons of skill through many cloning exercises are found
in
Berger and Kimmel, Guide to Molecular Cloning Techniques, Methods in
Enzymology, volume 152, Academic Press, Inc., San Diego, Calif (Berger);
Sambrook, et al. (1989) Molecular Cloning--A Laboratory Manual (2nd ed.) Vol.
1-
3, Cold Spring Harbor Laboratory, Cold Spring Harbor Press, N.Y., (Sambrook);
and Current Protocols in Molecular Biology, F. M. Ausubel, et al., eds.,
Current
Protocols, a joint venture between Greene Publishing Associates, Inc. and John
Wiley & Sons, Inc., (1994 Supplement) (Ausubel). Product information from
manufacturers of biological reagents and experimental equipment also provide
information useful in known biological methods. Such manufacturers include the
SIGMA chemical company (Saint Louis, Mo.), R&D systems (Minneapolis, Minn.),
Pharmacia LKB Biotechnology (Piscataway, N.J.), CLONTECH Laboratories, Inc.
(Palo Alto, Calif.), Chem Genes Corp., Aldrich Chemical Company (Milwaukee,
Wis.), Glen Research, Inc., GIBCO BRL Life Technologies, Inc. (Gaithersberg,
Md.), Fluka Chemica-Biochemika Analytika (Fluka Chemie AG, Buchs,
Switzerland), Invitrogen, San Diego, Calif., and Applied Biosystems (Foster
City,
Calif), as well as many other commercial sources known to one of skill.
The chimeric nucleic acid compositions of this invention, whether RNA,
cDNA, mRNA, genomic DNA, or a hybrid of the various combinations, are isolated
from biological sources or synthesized in vitro. The chimeric nucleic acids of
the
present invention are present in transformed or transfected whole cells, in
transformed or transfected cell lysates, or in a partially purified or
substantially pure
form.
In vitro amplification techniques suitable for amplifying sequences to
provide a nucleic acid or for subsequent analysis, sequencing or subcloning
are
known. Examples of techniques sufficient to direct persons of skill through
such in
vitro amplification methods, including the polymerase chain reaction (PCR) the
ligase chain reaction (LCR), Q(3-replicase amplification and other RNA
polymerase
mediated techniques (e.g., NASBA) are found in Berger, Sambrook, and Ausubel,
as
well as Mullis, et al., (1987) U.S. Pat. No. 4,683,202; PCR Protocols A Guide
to
Methods and Applications (Innis, et al. eds) Academic Press Inc. San Diego,
Calif.
-50-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
(1990) (Innis); Arnheim & Levinson (Oct. 1, 1990) C&EN 36-47; The Journal Of
NIH Research (1991) 3:81-94; Kwoh, et al., Proc. Natl. Acad. Sci. USA, 86:1173
(1989); Guatelli, et al., Proc. Natl. Acad. Sci. USA, 87:1874 (1990); Lomell,
et al., J.
Clin. Chem., 35:1826 (1989); Landegren, et al., Science, 241:1077-1080 (1988);
Van Brunt, Biotechnology, 8:291-294 (1990); Wu and Wallace, Gene, 4:560
(1989);
Barringer, et al., Gene, 89:117 (1990), and Sooknanan and Malek,
Biotechnology,
13:563-564 (1995). Improved methods of cloning in vitro amplified nucleic
acids
are described in Wallace, et al., U.S. Pat. No. 5,426,039. Improved methods of
amplifying large nucleic acids (up to 40 kb) are summarized in Cheng, et al.,
Nature,
369:684-685 (1994) and the references therein. One of skill will appreciate
that
essentially any RNA can be converted into a double stranded DNA suitable for
restriction digestion, PCR expansion and sequencing using reverse
transcriptase and
a polymerase. See, Ausubel, Sambrook, Innis, and Berger, all supra.
One of skill will recognize many ways of generating alterations in a given
nucleic acid construct. Such well-known methods include site-directed
mutagenesis,
PCR amplification using degenerate oligonucleotides, exposure of cells
containing
the nucleic acid to mutagenic agents or radiation, chemical synthesis of a
desired
oligonucleotide (e.g., in conjunction with ligation and/or cloning to generate
large
nucleic acids) and other well-known techniques. See, Giliman and Smith, Gene
8:81-97 (1979), Roberts, et al., Nature, 328:731-734 (1987) and Sambrook,
Innis,
Ausubel, Berger, and Mullis (all supra).
Most modifications to nucleic acids are evaluated by routine screening
techniques in suitable assays for the desired characteristic. For instance,
changes in
the immunological character of encoded polypeptides can be detected by an
appropriate immunological assay. For instance, changes in the cellular
immunological character of the polypeptide can be detected by an appropriate
antibody or CTL assay. Modifications of other properties such as nucleic acid
hybridization to a complementary nucleic acid, redox or thermal stability of
encoded
proteins, hydrophobicity, susceptibility to proteolysis, or the tendency to
aggregate
are all assayed according to standard techniques.
A wide variety of formats and labels are available and appropriate for
detection of fusion protein sequences. These include analytic biochemical
methods
such as spectrophotometry, radiography, electrophoresis, capillary
electrophoresis,
high performance liquid chromatography (HPLC), thin layer chromatography
(TLC), hyperdiffusion chromatography, and the like, and various immunological
-51-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
methods such as fluid or gel precipitin reactions, immunodiffusion (single or
double), immunoelectrophoresis, radioimmunoassays (RIAs), enzyme-linked
immunosorbent assays (ELISAs), western blot assays, immunofluorescent assays,
and the like. Several commercially available ELISA assays for the detection of
retroviral components, including Env domains, are available, allowing one of
skill to
detect Env in biological samples.
Similarly, the detection of the chimeric nucleic acids of the present
invention
proceeds by well known methods such as Southern analysis, northern analysis,
gel
electrophoresis, PCR, radiolabeling and scintillation counting, and affinity
chromatography. Many assay formats are appropriate, including those reviewed
in
Tijssen (1993) Laboratory Techniques in biochemistry and molecular biology--
hybridization with nucleic acid probes parts I and II, Elsevier, New York and
Choo
(ed) (1994) Methods In Molecular Biology Volume 33--In Situ Hybridization
Protocols, Humana Press Inc., New Jersey (see also, other books in the Methods
in
Molecular Biology series); see especially, Chapter 21 of Choo (id.) "Detection
of
Virus Nucleic Acids by Radioactive and Nonisotopic in Situ Hybridization".
Finally,
PCR is also routinely used to detect nucleic acids in biological samples (see,
Innis,
supra, for a general description of PCR techniques).
In one preferred embodiment, antibodies are used to detect polypeptide
sequences. Methods of producing polyclonal and monoclonal antibodies are known
to those of skill in the art, and many anti-HIV antibodies are available. See,
e.g.,
Coligan (1991) Current Protocols in Immunology, Wiley/Greene, NY; and Harlow
and Lane (1989) Antibodies: A Laboratory Manual, Cold Spring Harbor Press, NY;
Stites, et al. (eds.) Basic and Clinical Immunology (4th ed.), Lange Medical
Publications, Los Altos, Calif, and references cited therein; Goding (1986)
Monoclonal Antibodies: Principles and Practice (2d ed.), Academic Press, New
York, N.Y.; and Kohler and Milstein, Nature, 256:495-497 (1975). Other
suitable
techniques for antibody preparation include selection of libraries of
recombinant
antibodies in phage or similar vectors. See, Huse, et al., Science, 246:1275-
1281
(1989); and Ward, et al., Nature, 341:544-546 (1989). Specific monoclonal and
polyclonal antibodies and antisera will usually bind with a KD of at least
about 0.1
mM, more usually at least about I M, preferably at least about 0.1 M or
better,
and most typically and preferably, 0.01 kM or better.
-52-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Expression of Synthetic HIV-1 Gag and Pol Genes
Four synthetic HIV-1 Gag-and/or Pol expression vectors, hGag-Pol, hGag-
PolAFsAPr, hPol and hGag genes were prepared (Fig. VII). To confirm
expression,
the synthetic or viral Gag-Pol genes were transiently transfected into 293T
cells, a
human kidney-derived cell line. When cell lysates were analyzed by
immunoblotting with human anti-HIV-1 IgG (Fig. VIIIA), monoclonal anti-p24
(Fig. VIIIB), and rabbit anti-RT (Fig. VIIIC), Gag p55, Pol p110 and Gag-Pol
p160
precursor proteins were detected in hGag, hPol, and hGag-Pol fusion plasmids
transfected 293T cells, as was expected. Mature virion proteins, p24 and
RTp66,
were detected in the hGag-Pol gene transfected cells (Figs. VIIIA, B and Q.
This
might be a result of the activation of protease inside cells which was itself
a result of
the high-level expression of Gag and Gag-Pol protein (Karacostas, V et al.,
1993,
Virology, 193:661-671). The expression of Gag precursor proteins from codon-
altered vectors was _>10-fold higher than viral Gag-Pol (Fig. VIII),
determined by
quantitative phosphorimaging. The level of accumulated Gag-Pol fusion protein
was 100-fold higher in cells transfected with hGag-Pol compared to viral Gag-
Pol.
Virus-like particles were released from the hGag gene transfected cells (Fig.
IX),
detected by transmission electron microscopy. Though such particles were
observed
at a lower frequency with hGag-Pol, no particles were seen in cells
transfected with
hGag-PoIAFsAPr or hPol vectors. Stable expression of HIV-1 Gag and Pol
proteins
from codon-optimized genes in mouse CT26 and BHIOME cells was also observed
(Fig. VIRD).
Induction of HIV-1 Gag and Pol CTL Responses in Mice by DNA Vaccination
To evaluate the cellular immune response to HIV-1 Gag and Pol proteins,
Balb/C female mice were injected intramuscularly with the eukaryotic
expression
vector plasmids containing the codon-optimized genes. Two weeks after the
final
vaccination, splenocytes were harvested from the immunized mice and sensitized
with either Gag or Pol peptide-pulsed naive mouse splenocytes. One week later,
CTL responses were analyzed using a 5-hour chromium release assay.
CTL responses specific to HIV-1 Gag and/or Pol were first analyzed using
Gag or Pol peptide-pulsed BCIOME cells, or mouse fibrosarcoma cell lines
derived
from B/C-N cells (Collins, JL et al., 1981, J Exp. Med., 153:89-106).
Immunization
with hGag, hGag-PolAFSAPr or hGag-Pol genes induced comparably strong CTL
responses specific to Gag (Fig. XA); however, after immunization with hPol,
hGag-
-53-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
PolAFSAPr or hGag-Pol genes, only the fusion protein, hGag-PoIAFSAPr, and hPol
to a lesser extent, elicited a marked CTL response to Pol (Fig. XB). To
confirm that
the specific killing in the CTL assays was induced by CD8+ cytotoxic T
lymphocytes, CD4+ or CD8+ cells were depleted from sensitized splenocytes by
Dynal beads (Dynal, Inc., Lake Success, NY). Depletion of CD8+ cells abolished
the specific lysis in the hGag-Pol AFSAPr gene-immunized mice, while depletion
of
CD4+ had little effect on lysis (Fig. XC), suggesting that CD8+ lymphocytes
were
responsible for specific cytotoxicity.
The responses were further analyzed and confirmed with the hGag or hPol
gene transduced syngeneic CT26 and BCIOME cell lines. Responses to Gag in the
mice immunized with the hGag, hGag-Po1AFSAPr or hGag-Pol genes were similar
when peptide-pulsed cells were used as targets in the CTL assay (Fig. XIA).
Mice
immunized with the hPol gene generated a specific response to HIV-1 Pol on
BCIOME cell lines stably expressing Pol as target cells (Fig. XIB). The same
results have been observed with CT26 cell lines. These stably transfected cell
lines
were therefore more sensitive as target cells than peptide-pulsed cells in the
Pol CTL
assays.
Antibody Response in the Immunized Mice
Sera from mice immunized with different plasmids was analyzed with a p24
ELISA. hGag immunized mice demonstrated the highest p24 antibody titers (Fig.
XIIA). Unexpectedly, hGag-Pol virus-like particles elicited the lowest levels
of p24
antibody. Similar results were observed by Western blotting with pooled sera
(Fig.
XIIB). The HIV-1 Pol specific antibodies were not detected by a commercially
available Western blotting kit (Fig. XIIB), but antibodies to Pol were
detected in
mice immunized with hPol and hGag-PolAFSAPr with a more sensitive method,
IP/Western blotting (Fig. XIIC). Presumably, this assay is more sensitive and
better
able to detect native conformational epitopes. Though such antibodies were
found
in mice immunized with Pol and Gag-Pol fusion proteins in this assay, minimal
response was detected in the mice immunized with hGag-Pol. Though both
immunogens elicited similar Gag responses, the Gag-Pol fusion protein was
therefore more effective in the stimulation of CTL and antibody responses to
Pol.
HIV Gag-Pol Fusion Proteins
In this disclosure, HIV-1 B-clade Gag and Pol genes were modified to
increase Rev-independent expression of HIV-1 Gag-Pol proteins. This
modification
-54-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
allowed synthesis of HIV Gag and Pol, as well as fusion proteins at levels 10-
to
100-fold higher than the corresponding Rev-dependent viral gene in the absence
of
Rev and RRE elements. These viral proteins were recognized by standard
polyclonal and monoclonal antibodies (Fig. VIII), and immature VLP were
produced and released from 293T cells transfected with hGag (Fig. IX).
The immune response induced by Rev-independent Gag-Pol was directed
against both Gag and Pol determinants. The hGag, hGag-Pol fusion and hGag-Pol
all induced strong CTL responses specific for Gag in mice immunized with
plasmid
DNA, but a significant Pol response was elicited only in the mice immunized
with
the hGag-Po AFSAPr or Pol alone. Because immunization with hGag-Pol gene
failed to induce detectable cellular or humoral responses to HIV-1 Pol
protein, these
findings indicate that the Gag-Pol fusion protein induces a broader range of
responses and allows delivery of an immunogen with a larger number of epitopes
in
a single continuous open reading frame. During viral replication, viral gag-
pol
produces Gag precursor protein and the gag-pol fusion protein by frame
shifting in a
20:1 ratio (Wilson, W et al., 1988, Cell, 55:1159-1169). The deletion of a
frame
shift site in hGag-Po1AFSAPr results in production of only the Gag-Pol fusion
protein. Expression of Gag-Pol proteins alone in human cells is not adequate
to
form releasable viral particles because HIV-1 viral assembly requires Gag
precursor
proteins (Park, J and CD Morrow, 1992, J Virol, 66:6304-6313; Smith, AJ et
al.,
1993, J Virol, 67:2266-2275). The ability of hGag-Po1AFSAPr to elicit strong
Gag
and Pot specific CTL responses in mice may be explained by high level
expression
of Gag-Pol fusion protein and its retention on the inside of cells, which
could create
a new condition not existing during normal viral replication and could provide
sufficient proteins for antigen presentation. Moreover, the mutation in viral
protease
prevents viral protein from intracellular activation and reduced cellular
toxicity.
Overexpression of this polyprotein is also likely to affect its intracellular
localization/transport and is envisioned as improving antigen presentation.
The Pol gene of HIV-1 is the precursor protein for viral protease, reverse
transcriptase and integrase, which are crucial to viral replication (Kohl, NE
et al.,
1988, Proc. Natl. Acad. Sci. USA, 85:4686-4690.), Retroviral extracellular
maturation, resulting from self activation of protease after release produces
mature
RT and IN which have important functions in reverse transcription and
integration
respectively, for HIV-1, HIV-2, and S[V replication. The catalytic cores of
these
-55-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
enzymes have relatively conserved domains in order to preserve their functions
and
thus are envisioned as inducing cross-clade CTL responses. As early as 1988,
CTLs
specific for HIV-1 RT were found in blood samples from HIV-1 infected
individuals
(Hosmalin, A et al., 1990, Proc. Natl. Acad. Sci. USA, 87:2344-2348; Walker,
BD
et al., 1988, Science, 240:64-66.). Relatively strong Gag-specific CTL
responses
have been shown in numerous non-human primate and human studies, using DNA
vaccines or a live recombinant vector containing viral Gag-Pol constructs
(Evans,
TG, 1999, J Infect Dis, 180:290-298; Ferrari, G, 1997, Blood, 90:2406-2416;
Gorse,
GJ et al., 1999, Vaccine, 18:835-849; Seth, A, 1998, Proc. Natl. Acad. Sci.
USA,
95:10112-10116; Seth, A et al., 2000, J Virol, 74:2502-2509), but fewer Pol-
specific
CTL responses have been reported. The detection of significant CTL responses
specific to Pol in our disclosure may be attributed in part to establishment
of stable
Pol expressing cell lines, in which codon alteration and inactivation of FS
and PR in
the Pol gene allow high level expression of the Pol protein without cellular
toxicity.
Though it remains possible that the hGag-Pol, or a combination of hGag and
hPol,
may exert similar effects with appropriate adjuvants or with different prime-
boost
regimens, the Rev-independent Gag-Pol fusion protein stimulates HIV-1 Gag and
Pol specific CTL responses in mice and is envisioned as proving useful in an
AIDS
vaccine.
PLASMID DESCRIPTIONS
Env Plasmids
VRC21 00
pVR10l2x/s R5gp139-Nef (delta) MHC (delta) CD4/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. The envelope
protein gene from pX4gpl6O/h (Nabel lab #1272) was ligated in frame with the
mutant Nef gene from pNefDMHCDCD4/h (Mabel lab #1278) to produce
-56-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
pX4gpl39-NefDMHCDCD4/h. The envelope-Nef fusion protein expressed from
pX4gpl39-NefDMHCDCD4/h contains the first 668 amino acids from the HIV
envelope glycoprotein (gp 139) fused to the entire mutant Nef protein. The
truncated
envelope polyprotein (gp139) contains the entire SU protein and a portion of
the TM
protein including the fusion domain, but lacking the transmembrane domain and
regions important for oligomer formation. The protein sequence of the Nef
protein
from HIV-1 PV22 (GenBank accession number K02083) was used to create a
synthetic version of the Nef gene (Nef/h) using codons optimized for
expression in
human cells. To disrupt the ability of Nef to limit both MHC class I and CD4
expression, point mutations were introduced into the Nef gene from pNef/h
(Nabel
lab #1275). The resulting amino acids substitutions in pNefDMHCDCD4/h are:
P69A, P72A, P75A, P78A, D174A and D175A. X4gp139-NefDMHCDCD4/h is
expressed from the pVRl012x/s (Nabel lab #1267) vector backbone.
VRC2200
pVR1012x/s R5gpl57-Nef (delta) MHC (delta) CD4/h
The protein sequence of the envelope polyprotein (gp l60) from HXB2 (X4-
tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp l 60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, bu t the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (Nabel lab number
1272) were replaced with the corresponding region from the BaL strain of HIV-1
(GeneBank accession number M68893, again using human preferred codons). The
envelope-Nef fusion protein expressed from pR5gpl57-Nef/h contains the first
820
amino acids from the HIV envelope glycoprotein (gp 157) fused to the entire
mutant
Nef protein. The gene for gp157 was ligated in frame with the full-length
mutant
Nef gene from pNefDMHCDCD4/h (Nabel lab #1278) to produce pR5gpl57-
NefDMHCDCD4/h. The protein sequence of the ref protein from HIV-1 PV22
(GenBank accession number K02083) was used to create a synthetic version of
the
Nef gene (Nef/h) using codons optimized for expression in human cells. To
disrupt
the ability of Nef to limit both MHC class I and CD4 expression, point
mutations
-57-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
were introduced into the Nef gene from pNef/h (Nabel lab #1275). The resulting
amino acids substitutions in pNefDMHCDCD4/h are: P69A, P72A, P75A, P78A,
D174A and D175A. R5gp157-NefDMHCDCD4/h is expressed from the
pVR1012x/s (Nabel lab #1267) vector backbone.
VRC2300
pVR1012x/s X4 gp139-NefdeltaMHC deltaCD4/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gpl60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. The envelope
protein gene from pX4gpl6O/h (Nabel lab #1272) was ligated in frame with the
mutant Nef gene from pNefDMHC/h (Nabel lab #1276) to produce pX4gp139-
NefDMHC/h. The envelope-Nef fusion protein expressed from pX4gp139-
NefDMHC/h contains the first 668 amino acids from the HIV envelope
glycoprotein
(gp139) fused to the entire mutant Nef protein. The truncated envelope
polyprotein
(gp139) contains the entire SU protein and a portion of the TM protein
including the
fusion domain, but lacking the transmembrane domain and regions important for
oligomer formation. The protein sequence of the Nef protein from HIV-1 PV22
(GenBank accession number K02083) was used to create a synthetic version of
the
Nef gene (Nef/h) using codons optimized for expression in human cells. To
disrupt
the ability of Nef to limit MHC class I expression, point mutations were
introduced
into the Nef gene from pNef/h (Nabel lab #1275). The resulting amino acids
substitutions in pNefDMHC/h are: P69A, P72A, P75A, and P78A. X4gp139-
NefDMHC/h is expressed from the pVR1012x/s (Nabel lab #1267) vector backbone
VRC2302
pVR1012x/s X4gp130-Nef/h
For the X4gp130/h (VRC2703) portion, the protein sequence of the envelope
polyprotein (gpl60) from HXB2 (X4-tropic, GenBank accession number K03455)
was used to create a synthetic version of the gene (X4gpl60/h) using codons
-58-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
optimized for expression in human cells. The nucleotide sequence X4gpl 60/h
shows
little homology to the HXB2 gene, but the protein encoded is the same with the
following amino acid substitutions: F53L, N94D, K192S, 1215N, A224T, A346D,
and P470L. The full-lenth X4-tropic version of the envelope protein from
pX4gpl60/h (VRC3300) was terminated after the codon for amino acid 602. The
truncated envelope polyprotein contains the entire SU protein and a portion of
the
TM protein including the fusion domain, but lacking the transmembrane domain.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization.
Regions important for oligomer formation may be partially functional. This
X4gp130/h (VRC2703) gene was fused to Nef gene. The Nef protein from HIV-1
PV22 (GenBank accession number K02083) was used to create the viral Nef gene
(pVR1012-Nef)(Nabel Lab #1093. The nucleotide sequence is homologous to the
viral gene, and the protein encoded is the same. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC2400
pVR10l2x/s X4gpl57-NefDMHCDCD4/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp l60/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. The envelope-
Nef fusion protein expressed from pX4gpl57-NefDMHCDCD4/h contains the first
820 amino acids from the HIV envelope glycoprotein (gp157) fused to the entire
mutant Nef protein. The truncated envelope polyprotein (gp157) contains the
entire
SU protein and a portion of the TM protein including the fusion domain, the
transmembrane domain and regions important for oligomer formation. Heptad(H)
1,
Heptad 2 and their interspace(IS) are required for oligomerization. The gene
for
gpl57 was ligated in frame with the full-length mutant Nef gene from
pNefDMHCDCD4/h (VRC3600) to produce pX4gp157-NeIDMHCDCD4/h. The
protein sequence of the Nef protein from HIV-1 PV22 (GenBank accession number
K02083) was used to create a synthetic version of the Nef gene (Neflh) using
codons
optimized for expression in human cells. To disrupt the ability of Nef to
limit both
-59-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
MHC class I and CD4 expression, point mutations were introduced into the Nef
gene from pNef/h (VRC3500). The resulting amino acids substitutions in
pNefDMHCDCD4/h are: P69A, P72A, P75A, P78A, D174A and D175A.
X4gp 160-NefDMHCDCD4/h is expressed from the pVRl 012x/s (VRC2000) vector
backbone.
VRC2700
pVR1012x/s X4gp 140/h
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. The full-lenth
X4-tropic version of the envelope protein from pX4gpl6O/h (VRC3300) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the transmembrane domain. Regions important for oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC2701
pVRl012x/s X4gp140(del F/CL del HIS)/h
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gpl60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. The full-lenth X4-tropic version
of the envelope protein from pX4gpl6O/h (VRC3300) was terminated after the
codon for amino acid 680. The truncated envelope polyprotein contains the
entire
SU protein and a portion of the TM protein including the fusion domain, but
lacking
-60-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
the transmembrane domain. Regions important for oligomer formation may be
partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 503-
536, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2, from
amino acids 593-620, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC2702
pVRl012x/s X4gp128(del F/CL)/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. The full-lenth X4-tropic version
of the envelope protein from pX4gp160/h (VRC3300) was terminated after the
codon for amino acid 592. The truncated envelope polyprotein contains the
entire
SU protein and a portion of the TM protein including the fusion domain, but
lacking
the transmembrane domain. Regions important for oligomer formation may be
partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 503-
536, have been deleted. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC2706
pVR1012x/s X4gp145/h
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. The full-lenth X4-tropic version
of the envelope protein from pX4gp160/h (VRC3300) was terminated after the
codon for amino acid 704. The truncated envelope polyprotein contains the
entire
-61-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
SU protein and a portion of the TM protein including the fusion domain, but
lacking
the transmembrane domain. Regions important for oligomer formation may be
partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC2707
pVR1012x/s X4gp 145 (del F/CL del H IS)Ih
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. The full-lenth X4-tropic version
of the envelope protein from pX4gpl60/h (VRC3300) was terminated after the
codon for amino acid 704. The truncated envelope polyprotein contains the
entire
SU protein and a portion of the TM protein including the fusion domain, but
lacking
the transmembrane domain. Regions important for oligomer formation may be
partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 503-
536, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2, from
amino acids 593-620, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC2800
pVR1012x/s R5gp140/h
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gpl60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gpl 60/h), the region encoding
HIV-1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
-62-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-lenth
R5-
tropic version of the envelope protein gene from pR5gpl60/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the transmembrane domain. Regions important for oligomer
formation may be partially functional. The expression vector backbone is
pVR1012x/s (VRC2000).
-63-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC2801
pVR1012x/s R5gp140(del F/CL del H IS)/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gpl60/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-lenth
R5-
tropic version of the envelope protein gene from pR5gpl60/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the transmembrane domain. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The Fusion and Cleavage
(F/CL)
domains, from amino acids 503-536, have been deleted. The Interspace (IS)
between
Heptad (H) I and 2, from amino acids 593-620, have been deleted. Regions
important for oligomer formation may be partially functional. The expression
vector
backbone is pVR1012x/s (VRC2000).
VRC2804
pVR1012x/s R5gp145/h
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
-64-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
number M68893, again using human preferred codons). The full-lenth R5-tropic
version of the envelope protein gene from pR5gpl6O/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVR1012x/s (VRC2000).
VRC2805
pVR1012x/s R5gp 145 (del F/CL del H IS)/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gpl60Ih), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-lenth R5-tropic
version of the envelope protein gene from pR5gp160/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
Fusion and Cleavage (F/CL) domains, from amino acids 503-536, have been
deleted. The Interspace (IS) between Heptad (H) I and 2, from amino acids 593-
620,
have been deleted. The expression vector backbone is pVR1012x/s (VRC2000).
-65-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 2810
pVR1012 xis R5gp140delCI(delCFI)/h
Constant domain 1 was deleted from gp140delCFl from amino acid 33-127 and was
replaced with a Nhel site.
VRC 2811
pVR1012 xis R5gp l40delC2 (delCFI)/h
Constant domain 2 was deleted from gpl40delCFI from amino acid 199-293, and
was replaced with an Nhel site
VRC2812
pVR 1012 x/s R5gpl40de1C3 (delCFI)/h
Constant domain 3 was deleted from gpl40delCFI from amino acid 333-380, and
was replaced with an Nhel site.
VRC2813
pVRI O12 xis R5gpl40de1C4 (delCFI)/h
Constant domain 4 was deleted from gpl40delCFI from amino acide 419-458, and
was replaced with an Nhel site.
VRC2814
pVR1012 x/s R5gpl40de1C5 (delCFI)/h
Constant domain 5 was deleted from gpl40delCFI from amino acid 472 -498 and
was replaced with an Nhel site.
VRC 2820
pVR1012xis R5gp140(dCFI)/dV1
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
-66-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the VI loop (a.a.129 to 154) and transmembrane domain.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
Fusion and Cleavage (F/CL) domains, from amino acids 503-536, have been
deleted. The Interspace (IS) between Heptad (H) I and 2, from amino acids 593-
620,
have been deleted. Regions important for oligomer formation may be partially
functional. The expression vector backbone is pVR1012x/s (VRC2000).
VRC 2821
pVR1012x/s R5gp140(dCFI)/dV2
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp l 60/h shows little homology to the HXB2 gene, but
the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T723I, and S745T. To produce an
R5-tropic version of the envelope protein (R5gpl60/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl60/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V2 loop (a.a.160 to 193) and transmembrane domain.
-67-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
Fusion and Cleavage (F/CL) domains, from amino acids 503-536, have been
deleted. The Interspace (IS) between Heptad (H) I and 2, from amino acids 593-
620,
have been deleted. Regions important for oligomer formation may be partially
functional. The expression vector backbone is pVRI012x/s (VRC2000).
VRC 2822
pVR1012x/s R5gp140(dCFI)/dV3
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gpl60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gpl60/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V3 loop (a.a.299 to 327) and transmembrane domain.
Heptad (H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization.
The Fusion and Cleavage (F/CL) domains, from amino acids 503-536, have been
deleted. The Interspace (IS) between Heptad (H) I and 2, from amino acids 593-
620,
have been deleted. Regions important for oligomer formation may be partially
functional. The expression vector backbone is pVR1012x/s (VRC2000).
VRC 2823
pVR1012x/s R5gp140(dCFI)/dV4
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
-68-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V4 loop (a.a.386 to 413) and transmembrane domain.
Heptad (H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization.
The Fusion and Cleavage (F/CL) domains, from amino acids 503-536, have been
deleted. The Interspace (IS) between Heptad (H) 1 and 2, from amino acids 593-
620,
have been deleted. Regions important for oligomer formation may be partially
functional. The expression vector backbone is pVR1012x/s (VRC2000).
VRC 2824
pVR1012x/s R5gp140(dCFI)/dV12
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the VI, V2 loops (a.a.129 to 154,and a.a.160 to 193) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
-69-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) I and 2,
from
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pVRl012x/s
(VRC2000).
VRC 2825
pVRI 01 2x/s R5gpl40(dCFI)/dVl3
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gpl60/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the VI, V3 loops (a.a.129 to 154,and a.a.299 to 327) and
transmembrane domain. Heptad (H) 1, Heptad 2 and their Interspace (IS) are
required for oligomerization. The Fusion and Cleavage (F/CL) domains, from
amino
acids 503-536, have been deleted. The Interspace (IS) between Heptad (H) 1 and
2,
from amino acids 593-620, have been deleted. Regions important for oligomer
formation may be partially functional. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC 2826
pVR1012x/s R5gpl40(dCFI)/dV 14
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
-70-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the VI, V4 loops (a.a.129 to 154,and a.a.386 to 413) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) I and 2,
from
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 2827
pVRl012x/s R5gpl40(dCFI)/dV23
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gpl60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gpl60/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
-71-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
domain, but lacking the V2, V3 loops (a.a.160 to 193,and a.a.299 to 413) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2,
from
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 2828
pVR1012x/s R5gp140(dCFI)/dV24
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gpl60/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T723I, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V2, V4 loops (a.a.160 to 193,and a.a.386 to 413) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2,
from
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pVR1012x/s
(VRC2000).
-72-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 2829
pVR1012x/s R5gp140(dCFI)/dV34
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gpl60/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V3, V4 loops (a.a.299 to 327,and a.a.386 to 413) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2,
from
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pVRl012x/s
(VRC2000).
VRC 2830
pVR1012x/s R5gpl40(dCFI)/dV123
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
-73-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl60/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V1,V2,V3 loops (a.a. 129 to 154, a.a.160 to 193,and
a.a.299
to 327) and transmembrane domain. Heptad(H) 1, Heptad 2 and their
Interspace(IS)
are required for oligornerization. The Fusion and Cleavage (F/CL) domains,
from
amino acids 503-536, have been deleted. The Interspace (IS) between Heptad (H)
1
and 2, from amino acids 593-620, have been deleted. Regions important for
oligomer formation may be partially functional. The expression vector backbone
is
pVR1012x/s (VRC2000).
VRC 2831
pVRI 012x/s RSgp 140(dCFI)/dV 124
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl60/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V1,V2,V4 loops (a.a. 129 to 154, a.a.160 to 193,and
a.a.386
to 413) and transmembrane domain. Heptad (H) 1, Heptad 2 and their Interspace
(IS) are required for oligomerization. The Fusion and Cleavage (F/CL) domains,
from amino acids 503-536, have been deleted. The Interspace (IS) between
Heptad
(H) I and 2, from amino acids 593-620, have been deleted. Regions important
for
-74-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
oligomer formation may be partially functional. The expression vector backbone
is
pVR1012x/s (VRC2000).
VRC 2832
pVR1012x/s R5gp I 40(dCFI)/dV 134
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V1,V3,V4 loops (a.a. 129 to 154, a.a.299 to 327,and
a.a.386
to 413) and transmembrane domain. Heptad(H) 1, Heptad 2 and their
Interspace(IS)
are required for oligomerization. The Fusion and Cleavage (F/CL) domains, from
amino acids 503-536, have been deleted. The Interspace (IS) between Heptad (H)
I
and 2, from amino acids 593-620, have been deleted. Regions important for
oligomer formation may be partially functional. The expression vector backbone
is
pVRI012x/s (VRC2000).
VRC 2833
pVR1012x/s R5gpl40(dCFI)/dV234
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
-75-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp 160/h), the region encoding
HIV-1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V2,V3,V4 loops (a.a. 160 to 193, a.a.299 to 327,and
a.a.386
to 413) and transmembrane domain. Heptad(H) 1, Heptad 2 and their
Interspace(IS)
are required for oligomerization. The Fusion and Cleavage (F/CL) domains, from
amino acids 503-536, have been deleted. The Interspace (IS) between Heptad (H)
I
and 2, from amino acids 593-620, have been deleted. Regions important for
oligomer formation may be partially functional. The expression vector backbone
is
pVRl0l2x/s (VRC2000).
VRC 2834
pVRl012x/s R5gp140(dCFI)/dV1234
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V 1,V2,V3,V4 loops (a.a. 129 to 154, a.a. 160 to 193,
a.a.299 to 327,and a.a,386 to 413) and transmembrane domain. Heptad(H) 1,
Heptad
2 and their Interspace(IS) are required for oligomerization. The Fusion and
Cleavage
-76-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
(F/CL) domains, from amino acids 503-536, have been deleted. The Interspace
(IS)
between Heptad (H) 1 and 2, from amino acids 593-620, have been deleted.
Regions
important for oligorner formation may be partially functional. The expression
vector
backbone is pVR1012x/s (VRC2000).
VRC 2835
pAdApt R5gp 140(dCFI)/dV 1
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V1 loop (a.a.129 to 154) and transmembrane domain.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
Fusion and Cleavage (F/CL) domains, from amino acids 503-536, have been
deleted. The Interspace (IS) between Heptad (H) 1 and 2, from amino acids 593-
620,
have been deleted. Regions important for oligomer formation may be partially
functional. The expression vector backbone is pAdApt.
VRC 2836
pAdApt R5gp140(dCFI)/dV2
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
-77-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gpl6O/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V2 loop (a.a.160 to 193) and transmembrane domain.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
Fusion and Cleavage (F/CL) domains, from amino acids 503-536, have been
deleted. The Interspace (IS) between Heptad (H) I and 2, from amino acids 593-
620,
have been deleted. Regions important for oligomer formation may be partially
functional. The expression vector backbone is AdApt.
VRC 2837
pAdApt R5gp140(dCFI)/dV3
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, I215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gpl60/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V3 loop (a.a.299 to 327) and transmembrane domain.
Heptad (H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization.
The Fusion and Cleavage (F/CL) domains, from amino acids 503-536, have been
-78-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
deleted. The Interspace (IS) between Heptad (H) 1 and 2, from amino acids 593-
620,
have been deleted. Regions important for oligomer formation may be partially
functional. The expression vector backbone is pAdApt.
VRC 2838
pAdApt R5gpl40(dCFI)/dV4
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp1.60/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gpl60/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V4 loop (a.a.386 to 413) and transmembrane domain.
Heptad (H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization.
The Fusion and Cleavage (F/CL) domains, from amino acids 503-536, have been
deleted. The Interspace (IS) between Heptad (H) 1 and 2, from amino acids 593-
620,
have been deleted. Regions important for oligomer formation may be partially
functional. The expression vector backbone is pAdApt.
VRC 2839
pAdApt R5gp140(dCFI)/dV12
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
-79-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the VI, V2 loops (a.a.129 to 154,and a.a.160 to 193) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) I and 2,
from
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pAdApt.
VRC 2840
pAdApt R5gp140(dCFI)/dV13
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V1, V3 loops (a.a.129 to 154,and a.a.299 to 327) and
transmembrane domain. Heptad (H) 1, Heptad 2 and their Interspace (IS) are
required for oligomerization. The Fusion and Cleavage (F/CL) domains, from
amino
acids 503-536, have been deleted. The Interspace (IS) between Heptad (H) 1 and
2,
-80-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
from amino acids 593-620, have been deleted. Regions important for oligomer
formation may be partially functional. The expression vector backbone is
pAdApt.
VRC 2841
pAdApt R5gp 140(dCFI)/dV 14
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T723I, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the VI, V4 loops (a.a.129 to 154,and a.a.386 to 413) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) I and 2,
from
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pAdApt.
VRC 2842
pAdApt R5gp l40(dCFI)/dV23
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
-81-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gp160/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V2, V3 loops (a.a.160 to 193,and a.a.299 to 413) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2,
from
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pAdApt.
VRC 2843
pAdApt R5 gp I 40(dCFI)/dV24
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gpl60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gpl60/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl60/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V2, V4 loops (a.a.160 to 193,and a.a.386 to 413) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) I and 2,
from
-82-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pAdApt.
VRC 2844
pAdApt R5gp 140(dCFI)/dV34
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gpl60/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V3, V4 loops (a.a.299 to 327,and a.a.386 to 413) and
transmembrane domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) I and 2,
from
amino acids 593-620, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pAdApt.
VRC 2845
pAdApt R5gp I40(dCFI)/dV 123
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
-83-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
R5-tropic version of the envelope protein (R5gpl60/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the VI,V2,V3 loops (a.a. 129 to 154, a.a.160 to 193,and
a.a.299
to 327) and transmembrane domain. Heptad(H) 1, Heptad 2 and their
Interspace(IS)
are required for oligomerization. The Fusion and Cleavage (F/CL) domains, from
amino acids 503-536, have been deleted. The Interspace (IS) between Heptad (H)
I
and 2, from amino acids 593-620, have been deleted. Regions important for
oligomer formation may be partially functional. The expression vector backbone
is
pAdApt
VRC 2846
pAdApt RSgp 140(dCFI)/dV 124
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp1.60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V1,V2,V4 loops (a.a. 129 to 154, a.a.160 to 193,and
a.a.386
to 413) and transmembrane domain. Heptad (H) 1, Heptad 2 and their Interspace
(IS) are required for oligomerization. The Fusion and Cleavage (F/CL) domains,
from amino acids 503-536, have been deleted. The Interspace (IS) between
Heptad
-84-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
(H) 1 and 2, from amino acids 593-620, have been deleted. Regions important
for
oligomer formation may be partially functional. The expression vector backbone
is
pAdApt.
VRC 2847
pAdApt R5gpl40(dCFI)/dV 134
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the VI,V3,V4 loops (a.a. 129 to 154, a.a.299 to 327,and
a.a.386
to 413) and transmembrane domain. Heptad(H) 1, Heptad 2 and their
Interspace(IS)
are required for oligomerization. The Fusion and Cleavage (F/CL) domains, from
amino acids 503-536, have been deleted. The Interspace (IS) between Heptad (H)
1
and 2, from amino acids 593-620, have been deleted. Regions important for
oligomer formation may be partially functional. The expression vector backbone
is
pAdApt.
VRC 2848
pAdApt R5gp140(dCFI)/dV234
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
-85-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl60/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V2,V3,V4 loops (a.a. 160 to 193, a.a.299 to 327,and
a.a.386
to 413) and transmembrane domain. Heptad(H) 1, Heptad 2 and their
Interspace(IS)
are required for oligomerization. The Fusion and Cleavage (F/CL) domains, from
amino acids 503-536, have been deleted. The Interspace (IS) between Heptad (H)
1
and 2, from amino acids 593-620, have been deleted. Regions important for
oligomer formation may be partially functional. The expression vector backbone
is
pAdApt.
VRC 2849
pAdApt R5gpl 40(dCFI)/dV 1234
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-length
R5-tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 680. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the V1,V2,V3,V4 loops (a.a. 129 to 154, a.a. 160 to 193,
a.a.299 to 327,and a.a.386 to 413) and transmembrane domain. Heptad(H) 1,
Heptad
-86-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
2 and their Interspace(IS) are required for oligomerization. The Fusion and
Cleavage
(F/CL) domains, from amino acids 503-536, have been deleted. The Interspace
(IS)
between Heptad (H) I and 2, from amino acids 593-620, have been deleted.
Regions
important for oligomer formation may be partially functional. The expression
vector
backbone is pAdApt.
VRC2850
pVR1012 x/s R5gpI45delCI (delCFI)/h
Constant domain 1 was deleted from gp145delCFI from amino acid 33-127 and was
replaced with an Nhe I site.
VRC2851
pVR1012 x/s R5gp145de1C2 (delCFI)/h
Constant domain 2 was deleted from gp145dCFI from amino acid 199-293 and was
replaced with an Nhe I site.
VRC2852
pVR1012 x/s R5gp145delC3 (de1CFI)/h
Constant domain 3 was deleted from gp145delCFI from amino acide 333-380 and
was replaced with an Nhe I site.
VRC2853
pVRI012 x/s R5gp145deIC4 (delCFI)/h
Constant domain 4 was deleted from gp145delCF1 from amino acid 419-458 and
was replaced with an Nhe I site.
VRC2854
pVR1012 x/s R5gp145delC5 (delCFI)/h
Constant domain 5 was deleted from gpl45delCFl from amino acid 472-498 and
was replaced with an Nhe I site.
-87-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 2860
pVR1012x/s R5gpl45(dCFI)/h/dV1
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gpl60/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl6O/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
V1 loop (a.a. 129-154) and Fusion and Cleavage (F/CL) domains (a.a. 503-536)
have been deleted. Also, the Interspace (IS) between Heptad (H) I and 2
(a.a.593-
620) have been deleted. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 2861
pVR1012x/s R5gp145(dCFI)/h/dV2
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gpl60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp 160/h (VRC3300) were replaced
with
-88-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl60/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp
145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
V2 loop (a.a. 160-193) and Fusion and Cleavage (F/CL) domains (a.a. 503-536)
have been deleted. Also, the Interspace (IS) between Heptad (H) 1 and 2
(a.a.593-
620) have been deleted. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 2862
pVR1012x/s R5gp145(dCFI)/h/dV3
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gp160/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization.
The V3 loop (a.a. 299-327) and Fusion and Cleavage (F/CL) domains (a.a. 503-
536)
have been deleted. Also, the Interspace (IS) between Heptad (H) 1 and 2
(a.a.593-
620) have been deleted. The expression vector backbone is pVR1012x/s
(VRC2000).
-89-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
VRC 2863
pVR1012x/s R5gp145(dCFI)/h/dV4
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp l60/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gpl60/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl6O/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The V4 loop (a.a. 386-413) and Fusion and Cleavage (F/CL) domains (a.a. 503-
536)
have been deleted. Also, the Interspace (IS) between Heptad (H) 1 and 2
(a.a.593-
620) have been deleted. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 2864
pVRt012x/s R5gp145(dCFI)/hldV12
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp l60/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
-90-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl60/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
VI, V2 loops (a.a. 129-154 and 160-193) and Fusion and Cleavage (F/CL) domains
(a.a. 503-536) have been deleted. Also, the Interspace (IS) between Heptad (H)
1
and 2 (a.a.593-620) have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC 2865
pVR1012x/s R5gp145(dCFI)/h/dV13
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl6O/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The VI, V3 loops (a.a. 129-154 and 299-327) and Fusion and Cleavage (F/CL)
domains (a.a. 503-536) have been deleted. Also, the Interspace (IS) between
Heptad
(H) 1 and 2 (a.a.593-620) have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
-91-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 2866
pVR1012x/s R5gp145(dCFI)/h/dV14
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gp160/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The V1, V4 loops (a.a. 129-154 and 386-413) and Fusion and Cleavage (F/CL)
domains (a.a. 503-536) have been deleted. Also, the Interspace (IS) between
Heptad
(H) 1 and 2 (a.a.593-620) have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC 2867
pVR1012x/s R5gp145(dCFI)/h/dV23
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp 160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp 160/h (VRC3300) were replaced
with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
-92-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gp160/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad(H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The V2, V3 loops (a.a. 160-193 and 299-327) and Fusion and Cleavage (F/CL)
domains (a.a. 503-536) have been deleted. Also, the Interspace (IS) between
Heptad
(H) I and 2 (a.a.593-620) have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC 2868
pVR1012x/s R5gp145(dCFI)/h/dV24
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, I215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl6O/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad(H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The V2, V4 loops (a.a. 160-193 and 386-413) and Fusion and Cleavage (F/CL)
domains (a.a. 503-536) have been deleted. Also, the Interspace (IS) between
Heptad
(H) 1 and 2 (a.a.593-620) have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
-93-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 2869
pVR1012x/s R5gp145(dCFI)/h/dV34
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl6O/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The V3, V4 loops (a.a. 299-327 and 386-413) and Fusion and Cleavage (F/CL)
domains (a.a. 503-536) have been deleted. Also, the Interspace (IS) between
Heptad
(H) 1 and 2 (a.a.593-620) have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC 2870
pVR1012x/s R5gp145(dCFI)/h/dV134
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp 160/h (VRC3300) were replaced
with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
-94-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl6O/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The VI, V3, V4 loops (a.a. 129-154, 299-327 and 386-413) and Fusion and
Cleavage (F/CL) domains (a.a. 503-536) have been deleted. Also, the Interspace
(IS) between Heptad (H) I and 2 (a.a.593-620) have been deleted. The
expression
vector backbone is pVR1012x/s (VRC2000).
VRC 2871
pVRI012x/s R5gp145(dCFI)/h/dV234
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gpl60/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp1.60/h (VRC3300) were replaced
with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl6O/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization.
The V2, V3, V4 loops (a.a. 160-193, 299-327 and 386-413) and Fusion and
Cleavage (F/CL) domains (a.a. 503-536) have been deleted. Also, the Interspace
(IS) between Heptad (H) 1 and 2 (a.a.593-620) have been deleted. The
expression
vector backbone is pVRl012x/s (VRC2000).
-95-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 2872
pVR1012x/s R5gp145(dCFI)dv123/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gpl6O/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The V1, V2, V4 loops (a.a. 129-154, 160-193 and 386-413) and Fusion and
Cleavage (F/CL) domains (a.a. 503-536) have been deleted. Also, the Interspace
(IS) between Heptad (H) I and 2 (a.a.593-620) have been deleted. The
expression
vector backbone is pVR1012x/s (VRC2000).
VRC 2873
pVR1012x/s R5gp145(dCFI)/h/dV124
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
-96-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gp 160/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The VI, V2, V4 loops (a.a. 129-154, 160-193 and 386-413) and Fusion and
Cleavage (F/CL) domains (a.a. 503-536) have been deleted. Also, the Interspace
(IS) between Heptad (H) 1 and 2 (a.a.593-620) have been deleted. The
expression
vector backbone is pVR1012x/s (VRC2000).
VRC 2874
pVR1012x/s R5gp145(dCFI)/h/dV1234
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S,1215N, A224T, A346D, and P470L. To produce an R5-tropic version
of the envelope protein (R5gp160/h), the region encoding HIV-1 envelope
polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-1 (GeneBank accession
number M68893, again using human preferred codons). The full-length R5-tropic
version of the envelope protein gene from pR5gp160/h (VRC3000) was terminated
after the codon for amino acid 704. The truncated envelope polyprotein (gp145)
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, the transmembrane domain, and regions important for oligomer
formation.
Heptad (H) 1, Heptad 2 and their Interspace (IS) are required for
oligomerization.
The V1, V2, V3, V4 loops (a.a. 129-154, 160-193, 299-327 and 386-413) and
Fusion and Cleavage (F/CL) domains (a.a. 503-536) have been deleted. Also, the
Interspace (IS) between Heptad (H) I and 2 (a.a.593-620) have been deleted.
The
expression vector backbone is pVR1012x/s (VRC2000).
-97-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC2900
pVR1012x/s R5gp150/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gpl60/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
1
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). The full-lenth
R5-
tropic version of the envelope protein gene from pR5gpl6O/h (VRC3000) was
terminated after the codon for amino acid 752. The truncated envelope
polyprotein
(gp150) contains the entire SU protein and a portion of the TM protein
including the
fusion domain, the transmembrane domain, and regions important for oligomer
formation. The expression vector backbone is pVR1012x/s (VRC2000).
VRC3000
pVR1012x/s R5gp160/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gpl60/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an
R5-tropic version of the envelope protein (R5gp160/h), the region encoding HIV-
l
envelope polyprotein amino acids 275 to 361 from X4gp160/h (VRC3300) were
replaced with the corresponding region from the BaL strain of HIV-1 (GeneBank
accession number M68893, again using human preferred codons). Full length SU
and TM proteins are expressed from the pVR1012x/s (VRC2000) vector backbone.
-98-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC3200
pVR1012x/s X4gp 150/h
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. The full-lenth
X4-tropic version of the envelope protein gene from pX4gp1.60/h (VRC3300) was
terminated after the codon for amino acid 752. The truncated envelope
polyprotein
(gpl50) contains the entire SU protein and a portion of the TM protein
including the
fusion domain, the transmembrane domain, and regions important for oligomer
formation. The expression vector backbone is pVR1012x/s (VRC2000).
VRC3201
pVR1012x/s X4gp150(del F/CL del H IS)/h
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. The full-lenth
X4-tropic version of the envelope protein gene from pX4gpl,60/h (VRC3300) was
terminated after the codon for amino acid 752. The truncated envelope
polyprotein
(gp150) contains the entire SU protein and a portion of the TM protein
including the
fusion domain, the transmembrane domain, and regions important for oligomer
formation. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 503-
536, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2, from
amino acids 593-620, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
-99-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC3202
pVR1012x/s X4gp150 z\gly/h.
Eukaryotic vector with humanized codons expressing the HIV envelope
glycoprotein gp150 from HXB2, X4 tropic mutated in the Glycosylation sites.
VRC3202 pVR1012x/s X4gp150Dgly/h The protein sequence of the envelope
polyprotein (gp160) from HXB2 (X4- tropic, GenBank accession number K03455)
was used to create a synthetic version of the gene (X4gp160/h) using codons
optimized for expression in human cells. The nucleotide sequence X4gp160/h
shows
little homology to the HXB2 gene, but the protein encoded is the same with the
following amino acid substitutions: F53L, N94D, K192S, 1215N, A224T, A346D,
P470L, T723I, and S745T. To disrupt potential glycoslylation sites in the HIV-
1
envelope proteins, point mutations were introduced into the full-length X4-
tropic
version of the envelope protein gene from pX4gp160/h (VRC3300). The resulting
amino acids substitutions in X4gpl60Dgly/h are: N88D, N156D, N160D, N197E,
N230D, N234D, N241D, N276D, L288V, N289D, S291T, N295D, N332D, N339D,
N356D, N386D, and N448D. The full-length X4-tropic version of the envelope
protein gene from pX4gpl60/h (VRC3300) was terminated after the codon for
amino acid 752. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required
for
oligomerization. Full length SU and TM proteins are expressed from the
pVR1012x/s (VRC2000) vector backbone.
VRC3203
pVR1012x/s X4gp150 AB Agly/h
The protein sequence of the envelope polyprotein (gp160) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The AB
designation means the amino acids from 1-307 (Xbal to EcoRl). The nucleotide
sequence X4gp160/h shows little homology to the HXB2 gene, but the protein
encoded is the same with the following amino acid substitutions: F53L, N94D,
K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To disrupt potential
glycoslylation sites in the HIV-1 envelope proteins, point mutations were
introduced
into the full-lenth X4-tropic version of the envelope protein gene from pX4gp
l 60/h
-100-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
(VRC3300). The resulting amino acids substitutions in X4gpl6ODgly/h are: N88D,
N156D, N160D, N197E, N230D, N234D, N241D, N276D, L288V, N289D, S291T,
and N295D. The full-lenth X4-tropic version of the envelope protein gene from
pX4gpl6O/h (VRC3300) was terminated after the codon for amino acid 752.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligornerization. Full
length SU and TM proteins are expressed from the pVR1012x/s (VRC2000) vector
backbone.
VRC3300
pVR1012x/s X4gp160/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp 160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. Heptad(H) 1,
Heptad 2 and their Interspace(IS) are required for oligomerization. Full
length SU
and TM proteins are expressed from the pVR1012x/s (VRC2000) vector backbone.
VRC3301
pVR1012x/s X4gp160(del F/CL del H IS)/h
The protein sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic,
Gen-Bank accession number K03455) was used to create a synthetic version of
the
gene (X4gp160/h) using colons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, I215N, A224T, A346D, P470L, T7231, and S745T. Full length SU
and TM proteins are Expressed. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The Fusion and Cleavage (F/CL) domains, from
amino
acids 503-536, have been deleted. The Interspace (IS) between Heptad (H) 1 and
2,
from amino acids 593-620, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
-101-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC3400
pVR1012x/s X4gp 1 60Agly/h
The protein sequence of the envelope polyprotein (gp l60) from HXB2 (X4-
tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the
protein encoded is the same with the following amino acid substitutions: F53L,
N94D, K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To disrupt
potential glycoslylation sites in the HIV-1 envelope proteins, point mutations
were
introduced into the full-lenth X4-tropic version of the envelope protein gene
from
pX4gpl6O/h (VRC3300). The resulting amino acids substitutions in X4gpl60Dgly/h
are: N88D, N156D, N160D, N197E, N230D, N234D, N241D, N276D, L288V,
N289D, S291T, N295D, N332D, N339D, N356D, N386D, and N448D. Heptad(H)
1, Heptad 2 and their Interspace(IS) are required for oligomerization. Full
length SU
and TM proteins are expressed from the pVR1012x/s (VRC2000) vector backbone.
VRC3401
pVR1012x/s X4gpI60AB mut Agly/h
The protein sequence of the envelope polyprotein (gp1.60) from HXB2 (X4-
tropic,
GenBank accession number K03455) was used to create a synthetic version of the
gene (X4gp160/h) using codons optimized for expression in human cells. The AB
designation means the amino acids from 1-307 (Xbal to EcoRl). The nucleotide
sequence X4gp160/h shows little homology to the HXB2 gene, but the protein
encoded is the same with the following amino acid substitutions: F53L, N94D,
K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To disrupt potential
glycoslylation sites in the HIV-1 envelope proteins, point mutations were
introduced
into the full-lenth X4-tropic version of the envelope protein gene from
pX4gpl60/h
(VRC3300). The resulting amino acids substitutions in X4gp160Dgly/h are: N88D,
N156D, N I60D, N197E, N230D, N234D, N241D, N276D, L288V, N289D, S291T,
and N295D. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. Full length SU and TM proteins are expressed from the
pVR1012x/s (VRC2000) vector backbone.
-102-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC3500
pVR1012x/s Nef/h
The protein sequence of the Nef protein from HIV-1 PV22 (GenBank accession
number K02083) was used to create a synthetic version of the Nef gene (Nef/h)
using codons optimized for expression in human cells. The nucleotide sequence
Nef/h shows little homology to the viral gene, but the protein encoded is the
same.
Nef/h is expressed from the pVR1012x/s (VRC2000) vector backbone.
VRC3600
pVR1012x/s NefDMHCDCD4/h
The protein sequence of the Nef protein from HIV-1 PV22 (GenBank accession
number K02083) was used to create a synthetic version of the Nef gene (Nef/h)
using codons optimized for expression in human cells. To disrupt the ability
of Nef
to limit both MHC class I and CD4 expression point mutations were introduced
into
the Nef gene from pNef/h (VRC3500). The resulting amino acids substitutions in
pNefDMHCDCD4/h are: P69A, P72A, P75A, P78A, D174A and D175A.
pNefDMHCDCD4/h is expressed from the pVR1012x/s (VRC2000) vector
backbone.
VRC3700
pVR1012x/s NefDCD4/h
The protein sequence of the Nef protein from HIV-1 PV22 (GenBank accession
number K02083) was used to create a synthetic version o f the Nef gene (Nef/h)
using codons optimized for expression in human cells. To disrupt the ability
of Nef
to limit CD4 expression, point mutations were introduced into the Nef gene
from
pNef/h (VRC3500). The resulting amino acids substitutions in pNefDCD/h are:
D174A and D175A. pNefDCD4/h is expressed from the pVR1012x/s (VRC2000)
vector backbone. VRC3700 pVR1012x/s NefDCD4/h The protein sequence of the
Nef protein from HIV-1 PV22 (GenBank accession number K02083) was used to
create a synthetic version o f the Nef gene (Nef/h) using codons optimized for
expression in human cells. To disrupt the ability of Nef to limit CD4
expression,
-103-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
point mutations were introduced into the Nef gene from pNef/h (VRC3500). The
resulting amino acids substitutions in pNefDCD/h are: D174A and D175A.
pNefDCD4/h is expressed from the pVR1012x/s (VRC2000) vector backbone.
VRC3800
pVR1012x/s NefDMHC/h
The protein sequence of the Nef protein from HIV-1 PV22 (GenBank accession
number K02083) was used to create a synthetic version of the Nef gene (Nef/h)
using codons optimized for expression in human cells. To disrupt the ability
of Nef
to limit MHC class I expression, point mutations were introduced into the Nef
gene
from pNef/h (VRC3500). The resulting amino acids substitutions in pNefDMHC/h
are: P69A, P72A, P75A, and P78A. pNefDMHC/h is expressed from the
pVR1012x/s (VRC2000) vector backbone.
VRC5200
pVR1012x/s 89.6Pgp128(del F/CL)/h
The protein sequence of the envelope polyprotein (gpl60) from 89.6P (Dual-
tropic,
GenBank accession number u89134/LOCUS:SIU89134) was used to create a
synthetic version of the gene (89.6Pgpl6O/h) using codons optimized for
expression
in human cells. The nucleotide sequence 89.6Pgp160/h shows little homology to
the
89.6P gene, but the protein encoded is the same. The full-lenth 89.6P, dual-
tropic
version of the envelope protein gene from 89.6P gpl60/h (VRC3000) was
terminated after the codon for amino acid 596. The truncated envelope
polyprotein
contains the entire SU protein and a portion of the TM protein including the
fusion
domain, but lacking the transmembrane domain. The Fusion and Cleavage (F/CL)
domains, from amino acids 508-541, have been deleted. The expression vector
backbone is pVR1012x/s (VRC2000).
VRC5201
pVR1012x/s 89.6Pgpl4O(del F/CL del H IS/h
The protein sequence of the envelope polyprotein (gpl60) from 89.6P (dual-
tropic,
GenBank accession number u89134/locus S1U89134) was used to create a synthetic
-104-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
version of the gene (Dualtropic gpl60/h) using codons optimized for expression
in
human cells. The nucleotide sequence dualtropic gp160/h shows little homology
to
the 89.6P gene, but the protein encoded is the same. The full-lenth 89.6P,
dual-tropic
version of the envelope protein gene was terminated after the codon for amino
acid
683. The truncated envelope polyprotein contains the entire SU protein and a
portion
of the TM protein including the fusion domain, but lacking the transmembrane
domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 508-
541, have been deleted. The Interspace (IS) between Heptad (H) I and 2, from
amino acids 597-625, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC5202
pVR1012x/s 89.6Pgpl45(del F/CL del HIS/h
The protein sequence of the envelope polyprotein (gp1.60) from 89.6P (dual-
tropic,
GenBank accession number u89134/locus SIU89134) was used to create a synthetic
version of the gene (Dualtropic gpl60/h) using codons optimized for expression
in
human cells. The nucleotide sequence dualtropic gp l60/h shows little homology
to
the 89.6P gene, but the protein encoded is the same. The full-lenth 89.6P,
dual-tropic
version of the envelope protein gene was terminated after the codon for amino
acid
709. The truncated envelope polyprotein contains the entire SU protein and a
portion
of the TM protein including the fusion domain, but lacking the transmembrane
domain. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 508-
541, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2, from
amino acids 597-625, have been deleted. Regions important for oligomer
formation
may be partially functional. The expression vector backbone is pVR1012x/s
(VRC2000).
-105-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC5203
pVR1012x/s 89.6Pgp160/h
The protein sequence of the envelope polyprotein (gp160) from 89.6P (Dual-
tropic,
GenBank accession number U89134/LOCUS: SIU89134) was used to create a
synthetic version of the gene (dual tropic gpl60/h) using codons optimized for
expression in human cells. The nucleotide sequence 89.6P gpl60/h shows little
homology to the 89.6P gene, but the protein encoded is the same. Full length
SU and
TM proteins are expressed from the pVR1012x/s (VRC2000) vector backbone
VRC5300
pVR1012x/s R5(clade C)gp140(del F/CL del H IS)/h
The protein sequence of the envelope polyprotein (gp160) from 92br025 (R5-
tropic,
GenBank accession number U52953) was used to create a synthetic version of the
gene (R5gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence R5gp160/h shows little homology to the 92br025 gene, but
the
protein encoded is the same. The full-length R5-tropic version of the envelope
protein was synthesized by Operon under the name: kongene. The Xbal (18nt up-
stream from ATG) to BglII. (1376nt down-stream from ATG) fragment which
contains polylinker at the 5' end, Kozak sequence and ATG was cloned into the
Xbal to BglII sites of VRC2701 pVR1012x/s X4gp140(del F/CL del H IS)/h
backbone. Therefore, the gene is R5 (clade C) gp160/h up to the BglII site
(1376nt
from ATG) and the rest of the gene after BgIII site is VRC2701 pVR1012x/s
X4gp140(del F/CL del H IS)/h. The truncated envelope polyprotein contains the
entire SU protein and a portion of the TM protein including the fusion domain,
but
lacking the transmembrane domain. Regions important for oligomer formation may
be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) I and 2,
from
amino acids 593-620, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000). The full-lenth X4-tropic version of the envelope protein
from pX4gp160/h (VRC3300) was terminated after the codon for amino acid 680.
-106-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC5301
pVR1012x/s R5(clade C)gp145(del F/CL del H IS)/h
The protein sequence of the envelope polyprotein (gpl60) from 92br025 (R5-
tropic,
GenBank accessionnumber U52953) was used to create a synthetic version of the
gene (R5gp160/h) using codons optimized for expression in human cells. The
nucleotide sequence R5gp160/h shows little homology to the 92br025 gene, but
the
protein encoded is the same. The full-length R5-tropic version of the envelope
protein was synthesized by Operon under the name: kongene. The Xbal (18nt up-
stream from ATG) to Bg1Il (1376nt down-stream from ATG) fragment which
contains polylinker at the 5' end, Kozak sequence and ATG was cloned into the
Xbal to BglII sites of VRC2701 pVR1012x/s X4gpl40(del F/CL del H IS)/h
backbone. Therefore, the gene is R5 (clade C) gp160/h up to the BglII site
(1376nt
from ATG) and the rest of the gene after Bg1Il site is VRC2707 pVR1012x/s
X4gp145(del F/CL del H IS)/h. The truncated envelope polyprotein contains the
entire SU protein and a portion of the TM protein including the fusion domain,
but
lacking the transmembrane domain. Regions important for oligomer formation may
be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids
503-536, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2,
from
amino acids 593-620, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000). The full-lenth X4-tropic version of the envelope protein
from pX4gp160/h (VRC3300) was terminated after the codon for amino acid 704.
VRC5303
pVR1012x/s R5gp145CladeC(Brazil) delCFl/h
Authentic Clade C unlike 5301 which is a hybrid between C and B.
VRC 5304
pVR1012x/s R5(clade A)gpl40(del F/CL del H, IS)/h
The protein sequence of the envelope polyprotein (gpl60) from 92rw020 (R5-
tropic,
GenBank accessionnumber U51283) was used to create a synthetic version of the
-107-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
gene (Clade-A gpl45delCFl) using codons optimized for expression in human
cells.
The nucleotide sequence R5gpl45delCFl shows little homology to the 92rw020
gene, but the protein encoded is the same. The Xbal (18nt up-stream from ATG)
to
BamH1 (1837 nt down-stream from ATG) fragment which contains polylinker at the
5' end, Kozak sequence and ATG was cloned into the Xbal to BamHl sites of
pVR1012x/s backbone. The truncated envelope polyprotein contains the entire SU
protein and the TM domain, but lacking the the Fusion domain and Cytoplasmic
domain. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
Fusion and Cleavage (F/CL) domains, from amino acids 486-519, have been
deleted. The Interspace (IS) between Heptad (H) 1 and 2, from amino acids 576-
604,
have been deleted. The expression vector backbone is pVR1012x/s (VRC2000).
VRC 5305
pVR1012x/s R5(clade A)gp145(del F/CL del HIS)/h
The protein sequence of the envelope polyprotein (gpl60) from 92rw020 (R5-
tropic,
GenBank accessionnumber U51283) was used to create a synthetic version of the
gene (Clade-A gpl45delCFl) using codons optimized for expression in human
cells.
The nucleotide sequence R5gpl45delCFI shows little homology to the 92rw020
gene, but the protein encoded is the same. The Xbal (18nt up-stream from ATG)
to
BamHl (1912 nt down-stream from ATG) fragment which contains polylinker at the
5' end, Kozak sequence and ATG was cloned into the Xbal to BamH1 sites of
pVR1012x/s backbone. The truncated envelope polyprotein contains the entire SU
protein and the TM domain, but lacking the the Fusion domain and Cytoplasmic
domain. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
Fusion and Cleavage (F/CL) domains, from amino acids 486-519, have been
deleted. The Interspace (IS) between Heptad (H) 1 and 2, from amino acids 576-
604,
have been deleted. The expression vector backbone is pVR1012x/s (VRC2000).
-108-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 5306
pVR1012x/s R5(clade E)gpl40(del F/CL del HIS)/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 93th966.8
(R5-tropic, GenBank accessionnumber U08456) was used to create a synthetic
version of the gene (Clade-C gpl40delCFt) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl40delCFt shows little homology to
the gene 93th966.8, but the protein encoded is the same. The Xbal (1 8nt up-
stream
from ATG) to BamHI (1856 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein
contains the entire SU protein and the TM domain, but lacking the the Fusion
domain and Cytoplasmic domain. Regions important for oligomer formation may be
partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 497-
530, have been deleted. The Interspace (IS) between Heptad (H) I and 2, from
amino acids 588-613, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC 5307
pVR1012x/s R5(clade E)gp145(del F/CL del H IS)/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 93th966.8
(R5-tropic, GenBank accessionnumber U08456) was used to create a synthetic
version of the gene (Clade-C gpl45delCFt) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 93th966.8, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH1 (1928 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the XbaI to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein
contains the entire SU protein and the TM domain, but lacking the the Fusion
domain and Cytoplasmic domain. Regions important for oligomer formation may be
partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 497-
530, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2, from
-109-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
amino acids 588-613, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC 5308
pVR1012x/s R5(clade C South African)gp140(del F/CL del H IS)/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accessionnumber AF286227) was used to create a synthetic
version of the gene (Glade-C gpl45delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1833 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein
contains the entire SU protein and the TM domain, but lacking the the Fusion
domain and Cytoplasmic domain. Regions important for oligomer formation may be
partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 487-
520, have been deleted. The Interspace (IS) between Heptad (H) 1 and 2, from
amino acids 577-605, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC 5309
pVR1012x/s R5(clade C South African)gp145(del F/CL del H IS)/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accessionnumber AF286227) was used to create a synthetic
version of the gene (Glade-C gpl45delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gp 145delCFl shows little homology
to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein
contains the entire SU protein and the TM domain, but lacking the the Fusion
domain and Cytoplasmic domain. Regions important for oligomer formation may be
-110-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
partially functional. Heptad(H) 1, Heptad 2 and their interspace(IS) are
required for
oligomerization. The Fusion and Cleavage (F/CL) domains, from amino acids 487-
520, have been deleted. The Interspace (IS) between Heptad (H) I and 2, from
amino acids 577-605, have been deleted. The expression vector backbone is
pVR1012x/s (VRC2000).
VRC 5350
pVRC1012(x/s)-gpI 40(dCFt)(Brazil C)/dV 1
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1 loop(a.a.133-148) of the SU protein.
It also
lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the Interspace
(IS)
between Heptad (H) 1 and 2(a.a. 586-612), transmembrane domain, and
cytoplasmic domain in the TM protein. Regions important for oligomer formation
may be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 5351
pVRC1012(x/s)-gp140(dCFI)(Brazil C)/dV12
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH 1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
-111-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V 1,V2 loops (a.a.133-191) of the SU
protein. It
also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the
Interspace
(IS) between Heptad (H) 1 and 2(a.a. 586-612), transmembrane domain, and
cytoplasmic domain in the TM protein. Regions important for oligomer formation
may be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 5352
pVRC1012(x/s)-gp140(dCFI)(Brazil C)/dV123
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gp l40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp140(de1CF1) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamH1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1,V2,V3 loops (a.a.130-191, 330-358) of
the
SU protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-
529),
the Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), transmembrane
domain, and cytoplasmic domain in the TM protein. Regions important for
oligomer formation may be partially functional. Heptad(H) 1, Heptad 2 and
their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC 5353
pVRC 1012(x/s)-gpl40(dCFI)(Brazil C)/dV 1234
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
-112-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(l8nt up-stream from ATG) to BamHI (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1,V2,V3,V4 loops (a.a.130-191, 330-358,
384-
408) of the SU protein. It also lacks the Fusion and Cleavage (F/CL) domains(
a.a.
496-529), the Interspace (IS) between Heptad (H) 1 and 2(a.a. 586-612),
transmembrane domain, and cytoplasmic domain in the TM protein. Regions
important for oligomer formation may be partially functional. Heptad(H) 1,
Heptad
2 and their Interspace(IS) are required for oligomerization. The expression
vector
backbone is pVR1012x/s (VRC2000).
VRC 5354
pVRC1 01 2(x/s)-gp 140(dCFI)(Brazil C)/dV124
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1,V2,V4 loops (a.a. 130-191, 384-408) of
the
SU protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-
529),
the Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), transmembrane
domain, and cytoplasmic domain in the TM protein. Regions important for
oligomer formation may be partially functional. Heptad(H) 1, Heptad 2 and
their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
-113-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 5355
pVRC1012(x/s)-gpl40(dCFI)(Brazil C)/dV13
The protein sequence of the envelope polyprotein (gp160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(1 8nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamH1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V 1,V3 loops (a.a.133-148, 330-358) of
the SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) 1 and 2(a.a. 586-612), transmembrane
domain,
and cytoplasmic domain in the TM protein. Regions important for oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC 5356
pVRC1012(x/s)-gp140(dCF1)(Brazil C)/dV134
The protein sequence of the envelope polyprotein (gp160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gp140delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1,V3,V4 loops (a.a.130-148, 330-358, 384-
408) of the SU protein. It also lacks the Fusion and Cleavage (F/CL) domains(
a.a.
496-529), the Interspace (IS) between Heptad (H) I and 2(a.a. 586-612),
transmembrane domain, and cytoplasmic domain in the TM protein. Regions
important for oligomer formation may be partially functional. Heptad(H) 1,
Heptad
-114-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
2 and their Interspace(IS) are required for oligomerization. The expression
vector
backbone is pVR1012x/s (VRC2000).
VRC 5357
pVRC 101 2(x/s)-gp I 40(dCFI)(Brazil C)/dV14
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V I,V4 loops (a.a. 130-148, 384-408) of
the SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), transmembrane
domain,
and cytoplasmic domain in the TM protein. Regions important for oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC 5358
pVRC1012(x/s)-gpl40(dCF1)(Brazil C)/dV2
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(de]CFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V2 loop(a.a.154-191) of the SU protein.
It also
lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the Interspace
(IS)
-115-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
between Heptad (H) 1 and 2(a.a. 586-612), transmembrane domain, and
cytoplasmic domain in the TM protein. Regions important for oligomer formation
may be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 5359
pVRC1012(x/s)-gpl40(dCFI)(Brazil C)/dV23
The protein sequence of the envelope polyprotein (gp160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR10l2x/s backbone. The truncated envelope
polyprotein contains deletion in the V2,V3 loops (a.a. 154-191, 330-358) of
the SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), transmembrane
domain,
and cytoplasmic domain in the TM protein. Regions important for oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC 5360
pVRC1012(x/s)-gp140(dCFI)(Brazil C)/dV234
The protein sequence of the envelope polyprotein (gp160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH 1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
-116-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V2,V3,V4 loops (a.a.154-191, 330-358, 384-
408) of the SU protein. It also lacks the Fusion and Cleavage (F/CL) domains(
a.a.
496-529), the Interspace (IS) between Heptad (H) 1 and 2(a.a. 586-612),
transmembrane domain, and cytoplasmic domain in the TM protein. Regions
important for oligomer formation may be partially functional. Heptad(H) 1,
Heptad
2 and their Interspace(IS) are required for oligoinerization. The expression
vector
backbone is pVR1012x/s (VRC2000).
VRC 5361
pVRC1012(x/s)-gp140(dCFI)(Brazil C)/dV24
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp l40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V2,V4 loops (a.a. 154-191, 384-408) of
the SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) 1 and 2(a.a. 586-612), transmembrane
domain,
and cytoplasmic domain in the TM protein. Regions important for oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligoinerization. The expression vector
backbone is
pVRI012x/s (VRC2000).
VRC 5362
pVRC1012(x/s)-gpl40(dCFI)(Brazil C)/dV3
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(de]CFI) shows little
-117-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamH 1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V3 loop(a.a.330-358) of the SU protein.
It also
lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the Interspace
(IS)
between Heptad (H) 1 and 2(a.a. 586-612), transmembrane domain, and
cytoplasmic domain in the TM protein. Regions important for oligomer formation
may be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 5363
pVRC1012(x/s)-gpl40(dCFI)(Brazil C)/dV34
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamH1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V3,V4 loops (a.a.330-358, 384-408) of the
SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), transmembrane
domain,
and cytoplasmic domain in the TM protein. Regions important for oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligornerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
-118-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 5364
pVRC1012(x/s)-gpl40(dCFI)(Brazil C)/dV4
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl40delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl40(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamH1 sites of pVR101,2x/s backbone. The truncated envelope
polyprotein contains deletion in the V4 loop(a.a.384-408) of the SU protein.
It also
lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the Interspace
(IS)
between Heptad (H) 1 and 2(a.a. 586-612), transmembrane domain, and
cytoplasmic domain in the TM protein. Regions important for oligomer formation
may be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 5365
PVRC1012(x/s)-gpl45(dCFI)(Brazil C)/dVl
The protein sequence of the envelope polyprotein (gp160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gp145delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the XbaI to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the VI loop(a.a.133-148) of the SU protein.
It also
lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the Interspace
(IS)
between Heptad (H) 1 and 2(a.a. 586-612), and Cytoplasmic domain in the TM
protein. Regions important for oligomer formation may be partially functional.
-119-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVR1012x/s (VRC2000).
VRC 5366
PVRC1012(x/s)-gp145(dCFI)(Brazil C)/dV12
The protein sequence of the envelope polyprotein (gp160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl45de1CFl) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(de1CFl) shows little
homology to the gene 92BR025, but the protein encoded is the same. The XbaI
(18nt up-stream from ATG) to BamHI (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamH1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1,V2 loops (a.a.133-191) of the SU
protein. It
also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the
Interspace
(IS) between Heptad (H) 1 and 2(a.a. 586-612), and Cytoplasmic domain in the
TM
protein. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVRl012x/s (VRC2000).
VRC 5367
PVRC1012(x/s)-gp145(dCFI)(Brazil C)/dV123
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl45delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCFl) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the VI, V2,V3 loops (a.a. 133-191,330-358) of
the
SU protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-
529),
the Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), and Cytoplasmic
-120-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
domain in the TM protein. Regions important for oligomer formation may be
partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC 5368
PVRC1012(x/s)-gp145(dCFI)(Brazil C)/dV1234
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gp145delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1,V2, V3,V4 loops (a.a. 133-191,330-
358,384-
408) of the SU protein. It also lacks the Fusion and Cleavage (F/CL) domains(
a.a.
496-529), the Interspace (IS) between Heptad (H) 1 and 2(a.a. 586-612), and
Cytoplasmic domain in the TM protein. Regions important for oligomer formation
may be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 5369
PVRC1012(x/s)-gpI45(dCFI)(Brazi1 C)/dV124
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl45delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xba1 to BamH1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1, V2,V4 loops (a.a. 133-148,154-191,384-
-121-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
408) of the SU protein. It also lacks the Fusion and Cleavage (F/CL) domains(
a.a.
496-529), the Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), and
Cytoplasmic domain in the TM protein. Regions important for oligomer formation
may be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 5370
PVRC1012(x/s)-gp145(dCFI)(Brazil C)/dV13
The protein sequence of the envelope polyprotein (gp 160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl45delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCF1) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V 1,V3 loops (a.a.133-148, 330-358) of
the SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), and Cytoplasmic
domain
in the TM protein. Regions important for oligomer formation may be partially
functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC 5371
PVRC1012(x/s)-gpI 45(dCFI)(Brazil C)/dV 134
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gp145delCFJ) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(de1CFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH 1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
-122-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1, V3,V4 loops (a.a. 133-148,330-358,384-
408) of the SU protein. It also lacks the Fusion and Cleavage (F/CL) domains(
a.a.
496-529), the Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), and
Cytoplasmic domain in the TM protein. Regions important for oligomer formation
may be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 5372
PVRC1012(x/s)-gp145(dCFI)(Brazil C)/dV14
The protein sequence of the envelope polyprotein (gp160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl45delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCFl) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V1,V4 loops (a.a.133-148, 384-408) of the
SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), and Cytoplasmic
domain
in the TM protein. Regions important for oligomer formation may be partially
functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The expression vector backbone is pVRI012x/s (VRC2000).
VRC 5373
PVRC1012(x/s)-gp145(dCFI)(Brazil C)/dV2
The protein sequence of the envelope polyprotein (gp160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gp145delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl45(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
-123-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
(18nt up-stream from ATG) to BamHI (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V2 loop (a.a.154-191) of the SU protein.
It also
lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the Interspace
(IS)
between Heptad (H) I and 2(a.a. 586-612), and Cytoplasmic domain in the TM
protein. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVR1012x/s (VRC2000).
VRC 5374
PVRC1012(x/s)-gp145(dCFI)(Brazi1 C)/dV23
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gp145deICFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamH1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V2,V3 loops (a.a.154-191,330-358) of the
SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) 1 and 2(a.a. 586-612), and Cytoplasmic
domain
in the TM protein. Regions important for oligomer formation may be partially
functional. Heptad(H) 1, Heptad 2 and their interspace(IS) are required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC 5375
PVRC1012(x/s)-gpl45(dCFI)(Brazil C)/dV234
The protein sequence of the envelope polyprotein (gp 160) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gp145delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gpl45(delCFI) shows little
-124-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xba1 to BamH1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V2, V3,V4 loops (a.a. 154-191,330-358,384-
408) of the SU protein. It also lacks the Fusion and Cleavage (F/CL) domains(
a.a.
496-529), the Interspace (IS) between Heptad (H) 1 and 2(a.a. 586-612), and
Cytoplasmic domain in the TM protein. Regions important for oligomer formation
may be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS)
are
required for oligomerization. The expression vector backbone is pVR1012x/s
(VRC2000).
VRC 5376
PVRC1012(x/s)-gp145(dCFI)(Brazil C)/dV24
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl45delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(de1CFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamH1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V2,V4 loops (a.a.154-191, 384-408) of the
SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), and Cytoplasmic
domain
in the TM protein. Regions important for oligomer formation may be partially
functional. Heptad(H) 1, Heptad 2 and their lnterspace(IS) are required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC 5377
PVRC1012(x/s)-gp145(dCFI)(Brazil C)/dV3
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
-125-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
of the gene (Brazil-C gpl45delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCFl) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V3 loop (a.a.330-358) of the SU protein.
It also
lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the Interspace
(IS)
between Heptad (H) 1 and 2(a.a. 586-612), and Cytoplasmic domain in the TM
protein. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVRI012x/s (VRC2000).
VRC 5378
PVRC1012(x/s)-gp145(dCFI)(Brazi1 C)/dV34
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gpl45delCFl) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCFI) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamH1 (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamHl sites of pVRI012x/s backbone. The truncated envelope
polyprotein contains deletion in the V3,V4 loops (a.a.330-358, 384-408) of the
SU
protein. It also lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529),
the
Interspace (IS) between Heptad (H) I and 2(a.a. 586-612), and Cytoplasmic
domain
in the TM protein. Regions important for oligomer formation may be partially
functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
-126-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC 5379
PVRC1012(x/s)-gpl45(dCFI)(Brazil C)/dV4
The protein sequence of the envelope polyprotein (gpl60) from 92BR025 (R5-
tropic, GenBank accession number U52953) was used to create a synthetic
version
of the gene (Brazil-C gp145delCFI) using codons optimized for expression in
human cells. The nucleotide sequence of Brazil-C gp145(delCFl) shows little
homology to the gene 92BR025, but the protein encoded is the same. The Xbal
(18nt up-stream from ATG) to BamHl (1910 nt down-stream from ATG) fragment
which contains polylinker at the 5' end, Kozak sequence and ATG was cloned
into
the Xbal to BamH1 sites of pVR1012x/s backbone. The truncated envelope
polyprotein contains deletion in the V4 loop (a.a.384-408) of the SU protein.
It also
lacks the Fusion and Cleavage (F/CL) domains( a.a. 496-529), the Interspace
(IS)
between Heptad (H) 1 and 2(a.a. 586-612), and Cytoplasmic domain in the TM
protein. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVR1012x/s (VRC2000).
VRC5500
pVR1012x/s R5(SA-C)gpl4O(dCFI) dVl/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gp14Ode1CFI shows little homology to
the gene 97ZAO12, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH 1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the VI loop (a.a.136-150), the Fusion and Cleavage (F/CL) domains (a.a. 487-
520),
the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605), the
transmembrane
domain and the intracellular region. Regions important for oligomer formation
may
be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
-127-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC5501
pVR1012x/s R5(SA-C )gp140(dCFI) dV12/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gp l40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl40delCFl shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the VI, V2 loops (a.a.136-194), the Fusion and Cleavage (F/CL) domains (a.a.
487-
520), the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605), the
transmembrane domain and the intracellular region. Regions important for
oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC 5502
pVRl0I2x/s R5(SA-C )gp140(dCFI) dV123/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl40delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the VI, V2, V3 loops (a.a. 136-194, 297-325), the Fusion and Cleavage (F/CL)
domains (a.a. 487-520), the Interspace (IS) between Heptad (H) 1 and 2
(a.a.577-
605), the transmembrane domain and the intracellular region. Regions important
for
oligomer formation may be partially functional. Heptad(H) 1, Heptad 2 and
their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR10l2x/s (VRC2000).
-128-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC5503
pVR1012x/s R5(SA-C)gpl40(dCFI)dV 1234/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gp l40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl4OdelCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHI (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V1, V2, V3,V4 loops (a.a.136-194,297-325, 385-399), the Fusion and
Cleavage
(F/CL) domains (a.a. 487-520), the Interspace (IS) between Heptad (H) 1 and 2
(a.a.577-605), the transmembrane domain and the intracellular region. Regions
important for oligomer formation may be partially functional. Heptad(H) 1,
Heptad
2 and their Interspace(IS) are required for oligomerization. The expression
vector
backbone is pVR1012x/s (VRC2000).
VRC5504
pVRI 012x/s R5(SA-C)gp 140(dCFI)dV 124/h
The protein sequence of the envelope polyprotein (gp l40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl40deICFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (I 8nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the XbaI to
BamHI sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V1, V2, V4 loops (a.a. 136-194, 385-399), the Fusion and Cleavage (F/CL)
domains (a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2
(a.a.577-
605), the transmembrane domain and the intracellular region. Regions important
for
oligomer formation may be partially functional. Heptad(H) 1, Heptad 2 and
their
-129-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVRI012x/s (VRC2000).
VRC5505
pVR1012x/s R5(SA-C)gpl40(dCFI)dV 13/h
The protein sequence of the envelope polyprotein (gpl40de]CFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl40delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the VI, V3 loops (a.a.136-150, 297-325), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) 1 and 2 (a.a.577-605),
the
transmembrane domain and the intracellular region. Regions important for
oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC5506
pVR1012x/s R5(SA-C)gpl40(dCFI)dV134/h
The protein sequence of the envelope polyprotein (gpl40delCF1) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl40deICFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the VI, V3, V4 loops (a.a. 136-150, 297-325, 385-399), the Fusion and Cleavage
(F/CL) domains (a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2
(a.a.577-605), the transmembrane domain and the intracellular region. Regions
-130-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
important for oligomer formation may be partially functional. Heptad(H) 1,
Heptad
2 and their Interspace(IS) are required for oligomerization. The expression
vector
backbone is pVR1012x/s (VRC2000).
VRC5507
pVR1012x/s R5(SA-C)gp140(dCFI)dVI4/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl4OdelCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V1, V4 loops (a.a.136-150, 385-399), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605),
the
transmembrane domain and the intracellular region. Regions important for
oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC5508
pVR1012x/s R5(SA-C)gpl4O(dCFI)dV2/h
The protein sequence of the envelope polyprotein (gp140delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl40delGFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (I8nt up-
stream
from ATG) to BamH1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH 1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V2 loop (a.a.156-194), the Fusion and Cleavage (F/CL) domains (a.a. 487-
520),
the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605), the
transmembrane
-131-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
domain and the intracellular region. Regions important for oligomer formation
may
be partially functional. Heptad(H) 1, Heptad 2 and their interspace(IS) are
required
for oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC5509
pVR1012x/s R5(SA-C)gp 140(dCFI)dV23/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl4OdelCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V2, V3 loops (a.a.156-194, 297-325), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605),
the
transmembrane domain and the intracellular region. Regions important for
oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC5510
pVRI012x/s R5(SA-C)gpl4O(dCFI)dV234/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl4OdelCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V2, V3, V4 loops (a.a. 156-194, 297-325, 385-399), the Fusion and Cleavage
(F/CL) domains (a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2
-132-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
(a.a.577-605), the transmembrane domain and the intracellular region. Regions
important for oligomer formation may be partially functional. Heptad(H) 1,
Heptad
2 and their Interspace(IS) are required for oligomerization. The expression
vector
backbone is pVR1012x/s (VRC2000).
VRC5511
pVR1012x/s R5(SA-C)gpl40(dCFI)dV24/h
The protein sequence of the envelope polyprotein (gp140de1CFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl40delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V2, V4 loops (a.a.156-194, 385-399), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605),
the
transmembrane domain and the intracellular region. Regions important for
oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC5512
pVRI012x/s R5 (SA-C)gp I 40(dCFI)dV3/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl4OdelCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V3 loop (a.a.297-325), the Fusion and Cleavage (F/CL) domains (a.a. 487-
520),
-133-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605), the
transmembrane
domain and the intracellular region. Regions important for oligomer formation
may
be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC5513
pVR1012x/s R5 (SA-C)gp 1 40(dCFI)dV34/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gp14OdelCF1) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl4OdelCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V3, V4 loops (a.a. 297-325, 385-399), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) 1 and 2 (a.a.577-605),
the
transmembrane domain and the intracellular region. Regions important for
oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC5514
pVRI012x/s R5(SA-C)gpl40(dCF1)dV4/h
The protein sequence of the envelope polyprotein (gpl40delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gpl40delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl4OdelCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V4 loop (a.a 385-399), the Fusion and Cleavage (F/CL) domains (a.a. 487-
520),
-134-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
the Interspace (IS) between Heptad (H) 1 and 2 (a.a.577-605), the
transmembrane
domain and the intracellular region. Regions important for oligomer formation
may
be partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required
for oligomerization. The expression vector backbone is pVRI012x/s (VRC2000).
VRC5515
pVR1012x/s R5(SA-C)gp145(dCFI)dV 1/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gp145delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delGFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V1 loop (a.a.136-150), the Fusion and Cleavage (F/CL) domains (a.a. 487-
520),
the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605), and the
intracellular
region. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVRl012x/s (VRC2000).
VRC5516
pVR1012x/s R5(SA-C)gp145(dCFI)dVI2/h
The protein sequence of the envelope polyprotein (gpl45de1CFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gp145delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (1.8nt up-
stream
from ATG) to BamH 1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH,l sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V 1,V2 loops (a.a.136-194), the Fusion and Cleavage (F/CL) domains (a.a.
487-
520), the Interspace (IS) between Heptad (H) 1 and 2 (a.a.577-605), and the
-135-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
intracellular region. Regions important for oligomer formation may be
partially
functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC5517
pVR1012x/s R5(SA-C)gpl45(dCFI)dV 123/h
The protein sequence of the envelope polyprotein (gp145delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gpl45delCFl) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45de1CFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V1, V2, V3 loops (a.a.136-194, 297-325), the Fusion and Cleavage (F/CL)
domains (a.a. 487-520), the Interspace (IS) between Heptad (H) 1 and 2
(a.a.577-
605), and the intracellular region. Regions important for oligomer formation
may be
partially functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are
required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC5518
pVR1012x/s R5(SA-C)gp145(dCFI)dV1234/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gpl45delCFl) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH 1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V1,V2,V3 V4 loops (a.a.t36-194, 297-325, 385-399), the Fusion and Cleavage
(F/CL) domains (a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2
(a.a.577-605), and the intracellular region. Regions important for oligomer
-136-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC5519
pVR1012x/s R5(SA-C)gp145(dCFI)dV2/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gp145delCF1) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V2 loop (a.a.156-194), the Fusion and Cleavage (F/CL) domains (a.a. 487-
520),
the Interspace (IS) between Heptad (H) 1 and 2 (a.a.577-605), and the
intracellular
region. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVR1012x/s (VRC2000).
VRC5520
pVR1012x/s R5(SA-C)gpl45(dCF1)dV23/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gpl45delCFl) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V2, V3 loops (a.a.156-194, 297-325), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605),
and
the intracellular region. Regions important for oligomer formation may be
partially
-137-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligornerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC5521
pVR1012x/s R5(SA-C)gp145(dCFI)dV234/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gp 145deICFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFl shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V2, V3 V4 loops (a.a.156-194, 297-325, 385-399), the Fusion and Cleavage
(F/CL) domains (a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2
(a.a.577-605), and the intracellular region. Regions important for oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC5522
pVR1012x/s R5(SA-C)gp145(dCFI)dV24/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl45delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BarHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the XbaI to
BarHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V2, V4 loops (a.a.156-194, 385-399), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) 1 and 2 (a.a.577-605),
and
the intracellular region. Regions important for oligomer formation may be
partially
-138-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC5523
pVR1012x/s R5(SA-C)gp145(dCFI)dV3/h
The protein sequence of the envelope polyprotein (gpl45delCF1) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gp145delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVRl0l2x/s backbone. The truncated envelope polyprotein lacks
the V3 loop (a.a.297-325), the Fusion and Cleavage (F/CL) domains (a.a. 487-
520),
the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605), and the
intracellular
region. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVR1012x/s (VRC2000).
VRC5524
pVRI012x/s R5(SA-C)gp145(dCFI)dV34/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gp145delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHi sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V3, V4 loops (a.a. 297-325, 385-399), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605),
and
the intracellular region. Regions important for oligomer formation may be
partially
-139-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
functional, Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The expression vector backbone is pVRl012x/s (VRC2000).
VRC5525
pVR1012x/s R5(SA-C)gp145(dCFI)dV4/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gpl45delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFl shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVRl0l2x/s backbone. The truncated envelope polyprotein lacks
the V4 loop (a.a.385-399), the Fusion and Cleavage (F/CL) domains (a.a. 487-
520),
the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605), and the
intracellular
region. Regions important for oligomer formation may be partially functional.
Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The
expression vector backbone is pVR1012x/s (VRC2000).
VRC5526
pVR1012x/s R5(SA-C)gpI45(dCFI)dV13/h
The protein sequence of the envelope polyprotein (gp145delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gp l45delCFl) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFl shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamH 1 (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the V1, V3 loops (a.a.136-150, 297-325), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) 1 and 2 (a,a.577-605),
and
the intracellular region. Regions important for oligomer formation may be
partially
-140-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
VRC5527
pVR1012x/s R5(SA-C)gp145(dCFI)dV134/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gp145deICFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHI sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the VI, V3, V4 loops (a.a.136-150, 297-325, 385-399), the Fusion and Cleavage
(F/CL) domains (a.a. 487-520), the Interspace (IS) between Heptad (H) 1 and 2
(a.a.577-605), and the intracellular region. Regions important for oligomer
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVRl0l2x/s (VRC2000).
VRC5528
pVRI012x/s R5(SA-C)gp145(dCFI)dV124/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Glade-C gp145delCFf) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamHl sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the VI, V2, V4 loops (a.a.136-150, 156-194, 385-399), the Fusion and Cleavage
(F/CL) domains (a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2
(a.a.577-605), and the intracellular region. Regions important for oligomer
-141-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
formation may be partially functional. Heptad(H) 1, Heptad 2 and their
Interspace(IS) are required for oligomerization. The expression vector
backbone is
pVR1012x/s (VRC2000).
VRC5529
pVR1012x/s R5(SA-C)gp145(dCFI)dV14/h
The protein sequence of the envelope polyprotein (gpl45delCFI) from 97ZA012
(R5-tropic, GenBank accession number AF286227) was used to create a synthetic
version of the gene (Clade-C gp145delCFI) using codons optimized for
expression
in human cells. The nucleotide sequence R5gpl45delCFI shows little homology to
the gene 97ZA012, but the protein encoded is the same. The Xbal (18nt up-
stream
from ATG) to BamHl (1914 nt down-stream from ATG) fragment which contains
polylinker at the 5' end, Kozak sequence and ATG was cloned into the Xbal to
BamH1 sites of pVR1012x/s backbone. The truncated envelope polyprotein lacks
the VI, V4 loops (a.a.136-150, 385-399), the Fusion and Cleavage (F/CL)
domains
(a.a. 487-520), the Interspace (IS) between Heptad (H) I and 2 (a.a.577-605),
and
the intracellular region. Regions important for oligomer formation may be
partially
functional. Heptad(H) 1, Heptad 2 and their Interspace(IS) are required for
oligomerization. The expression vector backbone is pVR1012x/s (VRC2000).
Gag-Pol Plasmids
VRC3900
pVR1012x/s Gag/h
The protein sequence of the gag polyprotein (Pr55, amino acids 1-432) from
HXB2
(GenBank accession number K03455) was used to create a synthetic version of
the
gag gene using codons optimized for expression in human cells. The nucleotide
sequence of the synthetic gag gene shows little homology to the HXB2 gene, but
the
protein encoded is the same. The synthetic gag gene was ligated in frame with
sequences encoding the pol polyprotein to produce pGag(fs)Pol/h (VRC4200). The
protein sequence of the pol polyprotein (amino acids 3-1003) from NL4-3
(GenBank
accession number M19921) was used to create a synthetic version of the pol
gene
using codons optimized for expression in human cells. To produce a gene that
-142-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
expresses all of the gag proteins, the region encoding pol amino acids 77-1003
were
deleted from Gag(fs)Pol/h to produce Gag/h. This construct also encodes most
of the
protease (98 amino acids) gene encoding amino acids 3-77. Gag/h is expressed
from
the pVR1012x/s vector backbone.
VRC3901
pVR1012x/s SIV Gag/h
The protein sequence of the gag polyprotein (amino acid from 1-550)
SlVmac239(GenBank accession number M33262) was used to create a synthetic
version of the gag gene using codons optimized for expression in human cells.
The
nucleotide sequence of the synthetic gag gene shows little homology to the
SIVMac239 gene, but the protein encoded is the same.
VRC4000
pVR1012x/s Gag-Pol/h
The protein sequence of the gag polyprotein (Pr55, amino acids 1-432) from
HXB2
(GenBank accession number K03455) was used to create a synthetic version of
the
gag gene using codons optimized for expression in human cells. The nucleotide
sequence of the synthetic gag gene shows little homology to the HXB2 gene, but
the
protein encoded is the same. The synthetic gag gene contains all of the mature
Gag
proteins except for the last two that are normally cleaved from the carboxy-
terminus
of the gag polyprotein, pl and p6 (amino acids 433-500). The synthetic gag
gene
was ligated in frame with sequences encoding the pol polyprotein. The protein
sequence of the pol polyprotein (amino acids 3-1003) from NL4-3 (GenBank
accession number M19921) was used to create a synthetic version of the pol
gene
(Pol/h) using codons optimized for expression in human cells. Gag-Pol/h is
expressed from the pVR1012x/s vector backbone.
-143-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
VRC4001
pVR1012x/s SIVGag-Pol/h
Eukaryotic vector with humanized codons expressing the Gag-pol gene of
SIVmac239. The Sal I Xbal fragment of SIV gag(VRC3901) was inserted into Sal
I XbaI of SIV Pol(VRC4101) to create VRC4001.
VRC4100
pVRl012x/s Pol/h
The protein sequence of the pol polyprotein (amino acids 3-1003) from NL4-3
(GenBank accession number M19921) was used to create a synthetic version of
the
pol gene using codons optimized for expression in human cells. To initiate
translation at the beginning of Pol, a methionine codon was added to the 5'-
end of
the synthetic polymerase gene to create the Pol/h gene. Pol/h is expressed
from the
pVR1012x/s (VRC2000) vector backbone.
V RC4101
pVR1012x/s SIV Pol/h
The protein sequence of the pol polyprotein (amino acids 3-1017) from
SIVmac239
(GenBank accession number M19921) was used to create a synthetic version of
the
pol gene using codons optimized for expression in human cells. To initiate
translation at the beginning of Pol, a methionine codon was added to the 5'-
end of
the synthetic polymerase gene to create the Pol/h gene. The Protease (Pr)
mutation
is at pol amino acid 123 and is AAG->GGA or amino acids R->G. Reverse
transcriptase(RT) mutation is at as 352(GAC to CAT D(D to H) -RT) and
Integrase
mutation is at as 788(GAC to GGC(D to A) -IN). Pol/h is expressed from the
pVR1012x/s (VRC2000) vector backbone.
VRC4200
pVR1012x/s Gag(fs)Pol/h
The protein sequence of the gag polyprotein (Pr55, amino acids 1-432) from
HXB2
(GenBank accession number K03455) was used to create a synthetic version of
the
-144-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
gag gene using codons optimized for expression in human cells. The nucleotide
sequence of the synthetic gag gene shows little homology to the HXB2 gene, but
the
protein encoded is the same. The synthetic gag gene contains all of the mature
Gag
proteins except for the last two that are normally cleaved from the carboxy-
tenninus
of the gag polyprotein, pl and p6 (amino acids 433-500). The synthetic gag
gene
was ligated in frame with sequences encoding the pol polyprotein. The protein
sequence of the pol polyprotein (amino acids 3-1003) from NL4-3 (GenBank
accession number M19921) was used to create a synthetic version of the pol
gene
(Pol/h) using codons optimized for expression in human cells. To create the
possibility for translational frameshifting as means to express the gag-pol
polyprotein, the synthetic coding region for the last four amino acids of the
NC
protein through the rest of gag plus an additional 3 amino acids from pol were
replaced with the corresponding viral sequences (nucleotides 2074-2302 on the
HXB2 genome) from NL4-3 (GenBank accession number M19921). The
substitution of viral for synthetic sequences both introduces the sites
required for
frameshifting and restores the ability to express all gag proteins, including
p1 and
p6. Gag(fs)Pol/h is expressed from the pVR1,012x/s vector backbone.
VRC4300
pVR1012 Gag-Pol(d delta RT delta IN)/h
Eukaryotic vector with humanized codons expressing the Gag and the frame
shifted
Pol genes of HIV HXB2 subtype B with deletions in Reverse transcriptase, and
Integrase regions. The protein sequence of the gag polyprotein (Pr55, amino
acidsl-
432) from HXB2 (GenBank accession number K03455) was used to create a
synthetic version of the gag gene using codons optimized for expression in
human
cells. The nucleotide sequence of the synthetic gag gene shows little homology
to
the HXB2 gene, but the protein encoded is the same. The synthetic gag gene
contains all of the mature Gag proteins except for the last two that are
normally
cleaved from the carboxy-terminus of the gag polyprotein, pl and p6 (amino
acids
433-500). The synthetic gag gene was ligated in frame with sequences encoding
the
pol polyprotein. The protein sequence of the pol polyprotein (amino acids 3-
1003)
from NL4-3 (GenBank accession number M19921) was used to create a synthetic
version of the pol gene (Pol/h) using codons optimized for expression in human
cells. To create the possibility for translational frameshifting as means to
express
-145-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
the gag-pol polyprotein, the synthetic coding region for the last four amino
acids of
the NC protein through the rest of gag plus an additional 3 amino acids from
pot
were replaced with the corresponding viral sequences (nucleotides 2074-2302 on
the
HXB2 genorne) from NL4-3 (GenBank accession number M19921). The
substitution of viral for synthetic sequences both introduces the sites
required for
frameshifting and restores the ability to express all gag proteins, including
pl and p6
The Reverse Transcriptase (RT) mutation is at gag-pol amino acid 771 and is
GAC-
>CAC or amino acids D->H. The Integrase (IN) mutation is at gag-pol amino acid
1209 and is ACT->CAT or amino acids D->A. Note: This vector is not in the
pVR1012x/s backbone.
VRC4301
pVR1012x/s-Gag(FS)-Pol-delta RT IN-IRES-R5 gp 15 7-Nef
For the Gag(FS)Pol delta RT delta IN/h portion, VRC4302 pVR1012x/s
Gag(fs)Pol(delta PR deltalh was used for the protein sequence of the gag
polyprotein
(Pr55, amino acids 1-432) from HXB2 (GenBank accession number K03455) was
used to create a synthetic version of the gag gene using codons optimized for
expression in human cells. The nucleotide sequence of the synthetic gag gene
shows
little homology to the HXB2 gene, but the protein encoded is the same. The
synthetic gag gene contains all of the mature Gag proteins except for the last
two
that are normally cleaved from the carboxy- terminus of the gag polyprotein,
p1 and
p6 (amino acids 433-500). The synthetic gag gene was ligated in frame with
sequences encoding the pot polyprotein. The protein sequence of the pot
polyprotein
(amino acids 3-1003) from NL4-3 (GenBank accession number M19921) was used
to create a synthetic version of the pot gene (Pol/h) using codons optimized
for
expression in human cells. To create the possibility for translational
frameshifting as
means to express the gag-pol polyprotein, the synthetic coding region for the
last
four amino acids of the NC protein through the rest of gag plus an additional
3
amino acids from pot were replaced with the corresponding viral sequences
(nucleotides 2074-2302 on the HXB2 genome) from NL4-3 (GenBank accession
number M19921). The substitution of viral for synthetic sequences both
introduces
the sites required for frameshifting and restores the ability to express all
gag
proteins, including p1 and p6. Gag(fs)Pol/h is expressed from the pVR1012x/s
vector backbone. The Reverse Transcriptase (RT) mutation is at gag-pot amino
acid
-146-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
771 and is GAC->CAC or amino acids D->H. The Integrase (IN) mutation is at gag-
pol amino acid 1209 and is ACT->CAT or amino acids D->A. This gene has been
fused to an Internal Ribosomal Entry Site (IRES) and then fused to the R5gp
157-
Nef from VRC2200 pVR1012x/s R5gp 157- NefDMHCDCD4/h in which the protein
sequence of the envelope polyprotein (gpl60) from HXB2 (X4-tropic, GenBank
accession number K03455) was used to create a synthetic version of the gene
(X4gp160/h) using codons optimized for expression in human cells. The
nucleotide
sequence X4gp160/h shows little homology to the HXB2 gene, but the protein
encoded is the same with the following amino acid substitutions: F53L, N94D,
K192S, 1215N, A224T, A346D, P470L, T7231, and S745T. To produce an R5-tropic
version of the envelope protein (R5gp160/h), the region encoding HIV-1
envelope
polyprotein amino acids 275 to 361 from X4gpl60/h (VRC3300) were replaced with
the corresponding region from the BaL strain of HIV-l. (GeneBank accession
number M68893, again using human preferred codons). The envelope-Nef fusion
protein expressed from pR5gpl57-Nef/h contains the first 820 amino acids from
the
HIV envelope glycoprotein (gp157) fused to the entire mutant Nef protein. The
gene
for gp157 was ligated in frame with the full-length mutant Nef gene from
pNefDMHCDCD4/h (VRC3600) to produce pR5gpl57- NefDMHCDCD4/h. The
protein sequence of the Nef protein from HIV-1 PV22 (GenBank accession number
K02083) was used to create a synthetic version of the Nef gene (Nef/h) using
codons
optimized for expression in human cells. To disrupt the ability of Nef to
limit both
MHC class I and CD4 expression, point mutations were introduced into the Nef
gene from pNef/h (VRC3500). The resulting amino acids substitutions in
pNefDMHCDCD4/h are: P69A, P72A, P75A, P78A, D174A and D175A. R5gp157-
NefDMHCDCD4/h is expressed from the pVRl012x/s (VRC2000) vector
backbone.
VRC4302
pVR1012 Gag(delFS)Pol(delta PR delta RT delta IN)/h
The protein sequence of the gag polyprotein (Pr55, amino acids 1-432) from
HXB2
(GenBank accession number K03455) was used to create a synthetic version of
the
gag gene using codons optimized for expression in human cells. The nucleotide
sequence of the synthetic gag gene shows little homology to the HXB2 gene, but
the
protein encoded is the same. The synthetic gag gene contains all of the mature
Gag
-147-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
proteins except for the last two that are nonnally cleaved from the carboxy-
terminus
of the gag polyprotein, p1 and p6 (amino acids 433-500). The synthetic gag
gene
was ligated in frame with sequences encoding the pol polyprotein. The protein
sequence of the pol polyprotein (amino acids 3-1003) from NL4-3 (GenBank
accession number M19921) was used to create a synthetic version of the pol
gene
(Pol/h) using codons optimized for expression in human cells. To create the
possibility for translational frameshifting as means to express the gag-pol
polyprotein, the synthetic coding region for the last four amino acids of the
NC
protein through the rest of gag plus an additional 3 amino acids from pol were
replaced with the corresponding viral sequences (nucleotides 2074-2302 on the
HXB2 genome) from NL4-3 (GenBank accession number M19921). The
substitution of viral for synthetic sequences both introduces the sites
required for
frameshifting and restores the ability to express all gag proteins, including
p1 and
p6. The deleted Frame Shifted (delFS) has 5 T nucleotides deleted between the
Gag
and Pol sequences. Gag(fs)Pol/h is expressed from the pVR1012x/s vector
backbone. The Protease (PR) mutation is at gag-pol amino acid 553 and is AGG-
>GGC or amino acids R->G. The Reverse Transcriptase (RT) mutation is at gag-
pol
amino acid 771 and is GAC->CAC or amino acids D->H. The Integrase (IN)
mutation is at gag- pol amino acid 1209 and is ACT->CAT or amino acids D->A.
Note: This vector is not in the pVR1012x/s backbone.
VRC4303
pVRIO12 SIV Gag(delFS)Pol(delta PR delta RT delta IN)/h
Eukaryotic vector with humanized codons expressing the Gag and the frame-shift-
deleted Pol genes SIVmac239 with deletions in the Protease, Reverse
transcriptase,
and Integrase regions. The 5 of Ts from 3188 to 3192 of SIV gag-pol(VRC4001)
were deleted to create VRC4303.
VRC4304
pVRI 012 Gag (delFS)Pol delta PR delta RT delta IN/h
The protein sequence of the gag polyprotein (Pr55, amino acids 1-432) from HIV-
1
C clade(GenBank accession number U52953) was used to create a synthetic
version
of the gag gene using codons optimized for expression in human cells. The
-148-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
nucleotide sequence of the synthetic gag gene shows little homology to the HIV-
1
gene, but the protein encoded is the same. The synthetic gag gene contains all
of the
mature Gag proteins except for the last two that are normally cleaved from the
carboxy-terminus of the gag polyprotein, pl and p6 (amino acids 433-500). The
synthetic gag gene was ligated in frame with sequences encoding the pol
polyprotein. The protein sequence of the pol polyprotein (amino acids 3-1003)
from
NL4-3 (GenBank accession number M19921) was used to create a synthetic version
of the pol gene (Pol/h) using codons optimized for expression in human cells.
To
create the possibility for translational frameshifting as means to express the
gag-pol
polyprotein, the synthetic coding region for the last four amino acids of the
NC
protein through the rest of gag plus an additional 3 amino acids from pol were
replaced with the corresponding viral sequences (nucleotides 2074-2302 on the
HXB2 genome) from NL4-3 (GenBank accession number M19921). The
substitution of viral for synthetic sequences both introduces the sites
required for
frameshifting and restores the ability to express all gag proteins, including
pl and
p6. The deleted Frame Shifted (delFS) has 5 T nucleotides deleted between the
Gag
and Pol sequences. Gag(fs)Pol/h is expressed from the pVR1012x/s vector
backbone. The Protease (PR) mutation is at gag-pol amino acid 553 and is AGG-
>GGC or amino acids R->G. The Reverse Transcriptase (RT) mutation is at gag-
pol
amino acid 771 and is GAC->CAC or amino acids D->H. The Integrase (IN)
mutation is at gag- pol amino acid 1209 and is ACT->CAT or amino acids D->A.
Note: This vector is not in the pVR1012x/s backbone.
VRC4305
pVR1012 Gag-A(delFS)Pol(delta PR delta RT delta IN)/h
Eukaryotic vector with humanized codons expressing the Gag and the frame
shifted
Pol genes of HIV HIV-IA Glade with deletions in the Protease, Reverse
transcriptase, and Integrase regions. VRC4305 pVRIO12 Gag(-Ade1FS)Pol(delta PR
delta RT delta IN)/h The protein sequence of the gag polyprotein (Pr55, amino
acids
1-432) from HIV-1 A Glade (GenBank accession number AF004885) was used to
create a synthetic version of the gag gene using codons optimized for
expression in
human cells. The nucleotide sequence of the synthetic gag gene shows little
homology to the HIV-1 gene, but the protein encoded is the same. The synthetic
gag
gene contains all of the mature Gag proteins except for the last two that are
normally
-149-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
cleaved from the carboxy-terminus of the gag polyprotein, pl and p6 (amino
acids
433-500). The synthetic gag gene was ligated in frame with sequences encoding
the
pol polyprotein. The protein sequence of the pol polyprotein (amino acids 3-
1003)
from NL4-3 (GenBank accession number M19921) was used to create a synthetic
version of the pol gene (Pol/h) using codons optimized for expression in human
cells. To create the possibility for translational frameshifting as means to
express the
gag-pol polyprotein, the synthetic coding region for the last four amino acids
of the
NC protein through the rest of gag plus an additional 3 amino acids from pol
were
replaced with the corresponding viral sequences (nucleotides 2074-2302 on the
HXB2 genome) from NL4-3 (GenBank accession number M19921). The
substitution of viral for synthetic sequences both introduces the sites
required for
frameshifting and restores the ability to express all gag proteins, including
pl and
p6. The deleted Frame Shifted (delFS) has 5 T nucleotides deleted between the
Gag
and Pol sequences. Gag(fs)Pol/h is expressed from the pVR1012x/s vector
backbone. The Protease (PR) mutation is at gag-pol amino acid 553 and is AGG-
>GGC or amino acids R->G. The Reverse Transcriptase (RT) mutation is at gag-
pol
amino acid 771 and is GAC->CAC or amino acids D->H. The Integrase (IN)
mutation is at gag- pol amino acid 1209 and is ACT->CAT or amino acids D->A.
Note: This vector is not in the pVR1012x/s backbone.
pVRC4306
pVRIO12 Gag(delFS)Pol delta PR delta RT delta IN/Nef/h
The protein sequence of the Gag polyprotein (Pr55, amino acids 1-432) from
HXB2
(GenBank accession number K03455) was used to create a synthetic version of
the
Gag gene using codons optimized for expression in human cells. The nucleotide
sequence of the synthetic Gag gene shows little homology to the HXB2 gene, but
the
protein encoded is the same. The synthetic Gag gene contains all of the mature
Gag
proteins except for the last two that are normally cleaved from the carboxy-
terminus
of the Gag polyprotein, pl and p6 (amino acids 433-500). The synthetic Gag
gene
was ligated in frame with sequences encoding the Pol polyprotein. The protein
sequence of the Pol polyprotein (amino acids 3-1003) from NL4-3 (GenBank
accession number M19921) was used to create a synthetic version of the Pol
gene
(Pol/h) using codons optimized for expression in human cells. To create the
possibility for translational frameshifting as means to express the Gag-Pol
-150-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
polyprotein, the synthetic coding region for the last four amino acids of the
NC
protein through the rest of Gag plus an additional 3 amino acids from Pol were
replaced with the corresponding viral sequences (nucleotides 2074-2302 on the
HXB2 genome) from NL4-3 (GenBank accession number M19921). The
substitution of viral for synthetic sequences both introduces the sites
required for
frameshifting and restores the ability to express all Gag proteins, including
pl and
p6. The deleted Frame Shifted (delFS) has 5 T nucleotides deleted between the
Gag
and Pol sequences. The Protease (PR) mutation is at Gag-Pol amino acid 553 and
is
AGG->GGC or amino acids R->G. The Reverse Transcriptase (RT) mutation is at
Gag-Pol amino acid 771 and is GAC->CAC or amino acids D->H. The Integrase
(IN) mutation is at Gag- Pol amino acid 1209 and is ACT->CAT or amino acids D-
>A. The Nef/h gene was fused to downstream of Pol gene of Gag(delFS)Pol(delta
PR delta RT delta IN). No loss or extra-amino acid was created by the fusion
between Nef and Pol. The ATG of Nef was preserved.The protein sequence of the
Nef protein from HIV-1 PV22 (GenBank accession number K02083) was used to
create a synthetic version of the Nef gene (Nef/h) using codons optimized for
expression in human cells. The nucleotide sequence Nef/h shows little homology
to
the viral gene, but the protein encoded is the same. Note: This vector is not
in the
pVR1012x/s backbone.
VRC 4308
VRC4302-myr
pVR1012 Gag (delFS) Pol deltaPR deltaRT deltalN deltaMyr/h
The myristoylation site was deleted from pVRC4302. The protein sequence of the
gag polyprotein (Pr55, amino acids 1-432) from HXB2 (GenBank accession number
K03455) was used to create a synthetic version of the gag gene using codons
optimized for expression in human cells. The nucleotide sequence of the
synthetic
gag gene shows little homology to the HXB2 gene, but the protein encoded is
the
same. The synthetic gag gene contains all of the mature Gag proteins except
for the
last two that are normally cleaved from the carboxy-terminus of the gag
polyprotein,
p1 and p6 (amino acids 433-500). The synthetic gag gene was ligated in frame
with
sequences encoding the pol polyprotein. The protein sequence of the pol
polyprotein (amino acids 3-1003) from NL4-3 (GenBank accession number
M 19921) was used to create a synthetic version of the pol gene (Pol/h) using
codons
-151-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
optimized for expression in human cells. To create the possibility for
translational
frameshifting as means to express the gag-pol polyprotein, the synthetic
coding
region for the last four amino acids of the NC protein through the rest of gag
plus an
additional 3 amino acids from pol were replaced with the corresponding viral
sequences (nucleotides 2074-2302 on the HXB2 genome) from NL4-3 (GenBank
accession number M19921). The substitution of viral for synthetic sequences
both
introduces the sites required for frameshifting and restores the ability to
express all
gag proteins, including p1 and p6. The deleted Frame Shifted (delFS) has 5 T
nucleotides deleted between the Gag and Pol sequences. Gag(fs)Pol/h is
expressed
from the pVRI012x/s vector backbone. The Protease (PR) mutation is at gag-pol
amino acid 553 and is AGG->GGC or amino acids R->G. The Reverse
Transcriptase (RT) mutation is at gag-pot amino acid 771 and is GAC->CAC or
amino acids D->H. The Integrase (IN) mutation is at gag-pol amino acid 1209
and
is ACT->CAT or amino acids D->A. The myristylation site was deleted. Note:This
vector is not in the pVR1012x/s backbone.
VRC 4309
pVR1012 Gag (delFS) Pol deltaPR deltaRT deltalN delta Myr/Nef/h
The myristoylation site was deleted from pVRC4306. The protein sequence of the
gag polyprotein (Pr55, amino acids 1-432) from HXB2 (GenBank accession number
K03455) was used to create a synthetic version of the gag gene using codons
optimized for expression in human cells. The nucleotide sequence of the
synthetic
gag gene shows little homology to the HXB2 gene, but the protein encoded is
the
same. The synthetic gag gene contains all of the mature Gag proteins except
for the
last two that are normally cleaved from the carboxy-terminus of the gag
polyprotein,
p1 and p6 (amino acids 433-500). The synthetic gag gene was ligated in frame
with
sequences encoding the pol polyprotein. The protein sequence of the pol
polyprotein (amino acids 3-1003) from NL4-3 (GenBank accession number
M19921) was used to create a synthetic version of the pol gene (Pol/h) using
codons
optimized for expression in human cells. To create the possibility for
translational
frameshifting as means to express the gag-pol polyprotein, the synthetic
coding
region for the last four amino acids of the NC protein through the rest of gag
plus an
additional 3 amino acids from pol were replaced with the corresponding viral
sequences (nucleotides 2074-2302 on the HXB2 genome) from NL4-3 (GenBank
-152-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
accession number M19921). The substitution of viral for synthetic sequences
both
introduces the sites required for frameshifting and restores the ability to
express all
gag proteins, including p1 and p6. The deleted Frame Shifted (delFS) has 5 T
nucleotides deleted between the Gag and Pol sequences. Gag(fs)Pol/h is
expressed
from the pVR1012x/s vector backbone. The Protease (PR) mutation is at gag-pol
amino acid 553 and is AGG->GGC or amino acids R->G. The Reverse
Transcriptase (RT) mutation is at gag-pol amino acid 771 and is GAC->CAC or
amino acids D->H. The Integrase (IN) mutation is at gag-pol amino acid 1209
and
is ACT->CAT or amino acids D->A. The stop codon TAG was removed and
synthetic B Glade Nef ( Genbank access number) was fusion to the 3' end of pol
by
PCR. Note: This vector is not in the pVR1012x/s backbone.
VRC 4310
pVR1012 Nef Gag (del fs) (del Myr) Pol (delta PR delta RT delta IN)/h
The protein sequence of the gag polyprotein (Pr55, amino acids 1-432) from
HXB2
(GenBank accession number K03455) was used to create a synthetic version of
the
gag gene using codons optimized for expression in human cells. The nucleotide
sequence of the synthetic gag gene shows little homology to the HXB2 gene, but
the
protein encoded is the same. The synthetic gag gene contains all of the mature
Gag
proteins except for the last two that are normally cleaved from the carboxy-
terminus
of the gag polyprotein, pl and p6 (amino acids 433-500). The synthetic gag
gene
was ligated in frame with sequences encoding the pol polyprotein. The protein
sequence of the pol polyprotein (amino acids 3-1003) from NL4-3 (GenBank
accession number M19921) was used to create a synthetic version of the pol
gene
(Pol/h) using codons optimized for expression in human cells. To create the
possibility for translational frameshifting as means to express the gag-pol
polyprotein, the synthetic coding region for the last four amino acids of the
NC
protein through the rest of gag plus an additional 3 amino acids from pol were
replaced with the corresponding viral sequences (nucleotides 2074-2302 on the
HXB2 genome) from NL4-3 (GenBank accession number M19921). The
substitution of viral for synthetic sequences both introduces the sites
required for
frameshifting and restores the ability to express all gag proteins, including
pl and
p6. The deleted Frame Shifted (delFS) has 5 T nucleotides deleted between the
Gag
and Pol sequences. Gag(fs)Pol/h is expressed from the pVR1012x/s vector
-153-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
backbone. The Protease (PR) mutation is at gag-pol amino acid 553 and is AGG-
>GGC or amino acids R->G. The Reverse Transcriptase (RT) mutation is at gag-
pol
amino acid 771 and is GAC->CAC or amino acids D->H. The Integrase (IN)
mutation is at gag-pol amino acid 1209 and is ACT->CAT or amino acids D->A.
The stop codon TAG of synthetic B Glade Nef ( Genbank access number) gene was
removed and was fused to the 5' end of gene gene by PCR.Note: This vector is
not
in the pVR1012x/s backbone.
VRC 4311
pVR1012 Gag-C(delFS)PoI(delta PR delta RT delta IN) Nef/h
VRC4304 + Clade C Nef
The protein sequence of the gag polyprotein (Pr55, amino acids 1-432) from HIV-
1
C clade(GenBank accession number U52953) was used to create a synthetic
version
of the gag gene using codons optimized for expression in human cells. The
nucleotide sequence of the synthetic gag gene shows little homology to the HIV-
1
gene, but the protein encoded is the same. The synthetic gag gene contains all
of the
mature Gag proteins except for the last two that are normally cleaved from the
carboxy-terminus of the gag polyprotein, p1 and p6 (amino acids 433-500). The
synthetic gag gene was ligated in frame with sequences encoding the pol
polyprotein. The protein sequence of the pol polyprotein (amino acids 3-1003)
from
NL4-3 (GenBank accession number M19921) was used to create a synthetic version
of the pol gene (Pol/h) using codons optimized for expression in human cells.
To
create the possibility for translational frameshifting as means to express the
gag-pol
polyprotein, the synthetic coding region for the last four amino acids of the
NC
protein through the rest of gag plus an additional 3 amino acids from pol were
replaced with the corresponding viral sequences (nucleotides 2074-2302 on the
HXB2 genome) from NL4-3 (GenBank accession number M19921). The
substitution of viral for synthetic sequences both introduces the sites
required for
frameshifting and restores the ability to express all gag proteins, including
pl and
p6. The deleted Frame Shifted (delFS) has 5 T nucleotides deleted between the
Gag
and Pol sequences. Gag(fs)Pol/h is expressed from the pVRI012x/s vector
backbone. The Protease (PR) mutation is at gag-pol amino acid 553 and is AGG-
>GGC or amino acids R->G. The Reverse Transcriptase (RT) mutation is at gag-
pol
amino acid 771 and is GAC->CAC or amino acids D->H. The Integrase (IN)
-154-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
mutation is at gag-pol amino acid 1209 and is ACT->CAT or amino acids D->A.
The stop codon TAG was removed and synthetic C Glade Nef ( Genbank accession
number:U52953) was fusion to the 3' end of pol by PCR. Note:This vector is not
in
the pVR1012x/s backbone.
VRC 4312
Gag (del fs) (del Myr) Nef Pol APRARTAIN/h
VRC-Myr-gag-Dnef-Dpol
(Eukaryotic vector with humanized codons expressing the Gag, truncated Nef,
and
truncated Pol proteins of HIV subtype B .ns.)
This construct was derived from VRC4302. The protein sequence of the gag
polyprotein (Pr55, amino acids 1-432) from HXB2 (GenBank accession number
K03455) was used to create a synthetic version of the gag gene using codons
optimized for expression in human cells. The nucleotide sequence of the
synthetic
gag gene shows little homology to the HXB2 gene, but the protein encoded is
the
same. The synthetic gag gene contains all of the mature Gag proteins except
for the
last two that are normally cleaved from the carboxy-terminus of the gag
polyprotein,
p1 and p6 (amino acids 433-500). The synthetic gag gene was ligated in frame
with
sequences encoding synthetic nef gene that 51 as were deleted from 5'. 77 as
were
deleted from 5' of pol polyprotein, and ligated with 3' of nef in which tag
stop
codon was deleted . The protein sequence of the pol polyprotein (amino acids 3-
78 -
1.003) from NL4-3 (GenBank accession number M19921) was used to create a
synthetic version of the pol gene (Pol/h) using codons optimized for
expression in
human cells. To create the possibility for translational frameshifting as
means to
express the gag-pol polyprotein, the synthetic coding region for the last four
amino
acids of the NC protein through the rest of gag plus an additional 3 amino
acids
from pol were replaced with the corresponding viral sequences (nucleotides
2074-
2302 on the HXB2 genome) from NL4-3 (GenBank accession number M19921).
The substitution of viral for synthetic sequences both introduces the sites
required
for frameshifting and restores the ability to express all gag proteins,
including p1
and p6. The deleted Frame Shifted (delFS) has 5 T nucleotides deleted between
the
Gag and Pol sequences. Gag(fs)Pol/h is expressed from the pVR1012x/s vector
backbone. The Protease (PR) mutation is at gag-pol amino acid 553 and is AGG-
-155-
CA 02419822 2003-02-13
WO 02/32943 PCT/USO1/25721
>GGC or amino acids R->G. The Reverse Transcriptase (RT) mutation is at gag-
pol
amino acid 771 and is GAC->CAC or amino acids D->H. The Integrase (IN)
mutation is at gag-pol amino acid 1209 and is ACT->CAT or amino acids D->A.
Note: This vector is not in the pVR1012x/s backbone.
VRC 4313
pVR1012 Gag Clade A (del fs)Pol(A PR A RT A IN)/h
VRC4305+ Clade A Nef
The protein sequence of the gag polyprotein (Pr55, amino acids 1-432) from HIV-
1
A Glade (GenBank accession number AF004885) was used to create a synthetic
version of the gag gene using codons optimized for expression in human cells.
The
nucleotide sequence of the synthetic gag gene shows little homology to the HIV-
1
gene, but the protein encoded is the same. The synthetic gag gene contains all
of the
mature Gag proteins except for the last two that are normally cleaved from the
carboxy-terminus of the gag polyprotein, pl and p6 (amino acids 433-500). The
synthetic gag gene was ligated in frame with sequences encoding the pol
polyprotein. The protein sequence of the pol polyprotein (amino acids 3-1003)
from
NL4-3 (GenBank accession number M19921) was used to create a synthetic version
of the pol gene (Pol/h) using codons optimized for expression in human cells.
To
create the possibility for translational frameshifting as means to express the
gag-pol
polyprotein, the synthetic coding region for the last four amino acids of the
NC
protein through the rest of gag plus an additional 3 amino acids from pol were
replaced with the corresponding viral sequences (nucleotides 2074-2302 on the
HXB2 genome) from NL4-3 (GenBank accession number M19921). The
substitution of viral for synthetic sequences both introduces the sites
required for
frameshifting and restores the ability to express all gag proteins, including
p1 and
p6. The deleted Frame Shifted (delFS) has 5 T nucleotides deleted between the
Gag
and Pol sequences. Gag(fs)Pol/h is expressed from the pVR1012x/s vector
backbone. The Protease (PR) mutation is at gag-pol amino acid 553 and is AGG-
>GGC or amino acids R->G. The Reverse Transcriptase (RT) mutation is at gag-
pol
amino acid 771 and is GAC->CAC or amino acids D->H. The Integrase (IN)
mutation is at gag-pol amino acid 1209 and is ACT->CAT or amino acids D->A.
The stop codon TAG was removed and synthetic A Glade Nef ( Genbank accession
-156-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
number:AF069670) was fused to the 3' end of pol by PCR. Note: This vector is
not
in the pVR1012x/s backbone.
Examples
Env
Immunogens
Plasmids expressing the CXCR4-tropic HIV-1 HXB2 Env were made
synthetically with sequences designed to disrupt viral RNA structures that
limit
protein expression by using codons typically found in human cells. Briefly,
the
synthetic Env gene of HXB2 (GenBank accession number K03455) was generated
in three fragments by assembling the overlapping synthetic oligonucleotides
using
PCR amplification. To produce a CCR5-tropic version of the HIV-1 envelope, the
region encoding amino acids 275 to 361 of HXB2 (CXCR4-tropic) gp160 was
replaced with CCR5-tropic HIV-1 BaL sequence (GenBank accession number
M68893), which includes the V3 loop. Glycosylation mutants were generated by
site-directed mutagenesis to replace asparagine with glutamic acid residues at
seventeen conserved glycosylation sites between amino acids 88 and 448. To
express truncation mutant Env proteins, stop codons were introduced after
positions
752, 704, 680 or 592 to produce gp150, gp1.45, gpl40, or gp128, respectively.
The
Env protein was further changed by deleting amino acids 503 to 537 and 593 to
619,
which removes the cleavage site sequence, the fusion domain, and a part of the
spacer between the two heptad repeats. The structures of the synthetic HIV
envelope genes are shown (Fig. 1). The cDNAs were cloned in the expression
vector
pNGVL or pVRIO12 under the control of the cytomegalovirus immediate-early
enhancer, promoter, and first intron. The protein sequence was identical to
HXB2
Env except for the following amino acid substitutions: F53L, N94D, K192S,
1215N,
A224T, A346D, P470L, T7231, and S745T.
Expression of Envelope Proteins in Transfected Cells
293 cells (106) were plated in 60mm dishes. Cells were transfected on the
following day with 2 g of plasmid using calcium phosphate (Graham FL, van der
Eb AJ, 1973, Virology, 52:456-67). Cells were harvested 48 hours after
transfection
and lysed in buffer containing 50mM HEPES pH 7.0, 250mM NaCl and 0.5%
NP40. The protein concentration in the lysates was determined using the
Bradford
reagent (Bio-Rad). Proteins (25 g) in lysates were separated by 7.5% SDS-PAGE
-157-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
and transferred to Immobilon-P membrane (Millipore, Bedford, MA). Env was
detected by immunoprecipitation followed by Western blotting using polyclonal
antibody against gp160 (Intracel, Rockville, MD).
Cell Surface Expression of Envelope by FAGS Analysis
293 cells were harvested 48 hours after transfection and washed twice with
phosphate-buffered saline (PBS) containing 1% bovine serum albumin and
incubated for 30 minutes on ice with polyclonal immunoglobulin from an HIV-1
infected patient. The secondary antibody against human IgG conjugated with
FITC
(Jackson Immuno Research) was added, incubated for 30 minutes on ice, washed 3
times with PBS, and analyzed by flow cytometry (FACScan). The median
fluorescent intensity values were derived using Cell Quant software.
DNA Injection in Mice
Six week old, female BALB/c mice were injected intramuscularly with 100
g of purified plasmid DNA suspended in 200 l of normal saline. For each
plasmid DNA, a group of 4 mice was injected three times at intervals of two
weeks.
The mice were bled two weeks after the last injection, sera collected and
stored at
4 C.
Quantitation of the Antibody Response
Immunoprecipitation and Western blotting was used to detect the antibodies
that bind to native envelope proteins. Sera from immunized mice were used to
immunoprecipitate gpl60 from cell lysates of gpl60-transfected 293 cells.
Indicated
dilutions were used to immunoprecipitate gpl60 from the cell lysate (400 g).
Immunocomplexes were separated by 7.5% SDS-PAGE and analyzed by
immunoblotting using the polyclonal antibody against gp160.
Analysis of C'I'E Response
Spleens were removed aseptically and gently homogenized to a single cell
suspension, washed, and resuspended to a final concentration of 5x107
cells/ml.
Cells were incubated for 7 days in presence of IL-2 (10 U/ml) and either
irradiated
peptide-pulsed splenocytes from naive mice or an irradiated stable cell line
expressing the full length gpl60 BC-eni/rev, (Irwin MJ et al., 1994, J Virol,
68:5036-44). Three types of target cells were used: peptide-pulsed P815 cells
(ATCC TIB64), BC10ME cells stably expressing gpl60, and BC10ME cells
(Collins JL, Patek PQ, Cohn M. Tumorigenicity and lysis by natural killers. J
Exp
Ivied, 1981; 153:89-106) pulsed with peptides derived from gp l 60 sequence.
Target
-158-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
cells were labeled with 51Cr for 90 minutes and washed three times with RPMI-
1640
with 10% FBS, 2mM glutamine, 5x 10-5M (3-mercaptoethanol and fungisone,
250units/ml, and resuspended in this media. Cytolytic activity was determined
in
triplicate samples using all different target cell dilutions in a 5-hour 51Cr-
release
assay (Ohno T et al., 1997, Gene Ther, 4:361-6).
Gag and Pol
Development of Synthetic HIV-1 Gag-Pol Expression Vectors
The protein sequences of Gag (amino acids 1-432) from HXB2 (GenBank
accession number K03455) and Pol (amino acids 3-1003) from NL4-3 (GenBank
accession number M19921) were reverse translated by the GCG Package (Genetic
Computer Group, Inc., Madison, WI) using codons expected for human cells. A
226-
bp fragment spanning the frame shift site and the overlapping region of the
two
reading frames from NL4-3 were retained to allow expression of Gag and Gag-Pol
precursor polyproteins in the same construct. 86 oligonucleotides covering
4325
DNA base pairs with 5' Sall and 3' EcoRI sites were purchased from GIBCO Life
Technologies. Each of the oligonucleotides was 75 base-pairs with 25 nt of
overlap.
The codon optimized Gag-Pol gene (hGag-Pol) was assembled by PCR with Pwo
(Boehringer Mannheim) and Turbo Pfu (Stratagene) high fidelity DNA polymerase.
The PCR conditions were optimized with a PCR optimization kit (Stratagene) on
a
gradient Robocycler (Stratagene). Full-length synthetic Gag-Pol gene was
cloned
into the Sal I and blunted Bgl II site of the mammalian expression vector
pNGVL-3
(Xu, Ling et al., 1998, Nature Medicine, 4:37-42) and confirmed by DNA
sequencing. Three additional constructs were derived from the hGag-Pol gene. 5
Thymidines (Ts) in the frame shift site (FS) of the hGag-Pol gene were deleted
(AFS) and the protease was inactivated by replacing AGG in protease to GGC
(R42G) to create hGag-PohhFSAPr (Hung, M, 1998, 1 Virol, 72:4819-4824; Loeb,
DD et al., 1989, Nature 340:397-400). 432 amino acids of the NH2- terminal of
hGag-Pol gene were deleted and an ATG start codon was added to create the hPol
gene. 925 amino acids of the COOH-terminal hGag-Pol were deleted to create the
hGag gene. hGagPolAFS sPr, hPol and hGag genes were expressed in the pNGVL-3
plasmid, derived by insertion of a polylinker into pVRI O12. The plasmid
expressing
viral Gag-Pol, pCMV1 8.2 was a kind gift (Naldini, L et al., 1996, Science,
272:263-
267).
-159-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
Transient Transfection and Analysis of Expression
293T cells were maintained in Dulbecco's modified Eagle medium (DMEM;
GIBCO-BRL), supplemented with 10% fetal bovine serum (FBS). Plasmid DNAs
were purified with double cesium chloride sedimentation gradients.
Approximately
3x106 293T cells were placed in a 10-cm dish one day before transfection. 10
g of
pCMVdR8.2 plasmid (containing the viral gag-pol gene), or 5 g of pVR1012s
(containing the codon altered genes), were used to transfect 293T cells, using
the
calcium phosphate method (Chen, C and H Okayama, 1987, Mol. Cell. Biol, 7:2745-
2752). Three days after transfection, cell lysates were prepared with RIPA
buffer
(Boehringer Mannheim, Indianapolis, IN) and separated by 4-15% gradient SDS-
polyacrylamide gel electrophoresis (PAGE), then transferred onto an Immobilon
P
membrane (Millipore). Membranes were then incubated with anti HIV-1-IgG
(AIDS Research and Reference Reagent Program), monoclonal anti-p24 (ICN), or
rabbit anti-RT (Intracel, Rockville, MD). Bands were visualized using the ECL
Western blotting detection reagent (Amersham Pharmacia Biotech, Piscataway,
NJ),
as described by the manufacturer. Expression levels were determined using a
phosphorimager.
Generation of Stably Transfected Cell Lines
hGag and hPol genes were individually subcloned into the Xho I and EcoRI
sites of a retroviral vector, pPGS-CITE-Neo. Three plasmid systems were used
to
produce recombinant retroviruses containing the hGag or hPol genes (Yang, Z et
al.,
1998, Science, 279:1034-1037). 48 hours after transfection, the supernatants
were
collected to transduce CT26 and BC10ME (Collins, JL, 1981, J Exp. Med, 153:89-
106) which are syngeneio to Balb/C mice, and selected in 0.8 mg/ml of G418 two
days after infection. The positive clones were screened and confirmed by
Western
blotting and maintained in 10% FCS supplemented RPMI (GIBCO-BRL) with
0.5mg/ml G418.
DNA Vaccination of Mice
Female BALB/C mice, 6-8 weeks old, were used for immunogenicity
studies. For DNA vaccination, mice were immunized with 100 l (0.5 g/ml DNA
, 0.9% NaCl) in the quadriceps muscle of each hind leg every two weeks for a
total
of 4 injections.
-160-
CA 02419822 2003-02-13
WO 02/32943 PCT/US01/25721
CTL Assay
Animals were sacrificed after the last immunization and spleens removed
from both naive and immunized mice using aseptic techniques. Splenic
lymphocytes were harvested and the chromium release CTL assay was performed in
triplicate as previously described (Ohno, T. et al., 1997, Gene Ther, 4:361-
366).
The peptides used for sensitizing cells are as follows: Two peptide mixtures
from
the Gag protein P17(88-115) and p24(62-76). Seven peptide mixtures from the
Pol
protein: 1) P66(175-189), 2) P66(179-193), 3) P66(183-197), 4) P66(187-201),
5)
P66(223-237), 6) P66(227-241), 7) P66(367-381).
Measurement of Antibody Responses
Anti p24 ELISA assays
The anti-p24 ELISA assay was performed in Immunlon ninety-six-well
plates (Dynet Technologies Inc., Chantilly, VA). The plates were coated with
50 l
of purified recombinant HIV-111113 p24 antigen (Intracel) at a concentration
of 2
g/ml in PBS buffer, pH 7.4 (GIBCO) with 0.05% sodium azide. Plates were
washed 3x in PBS containing 3% BSA and 0.05% Tween20 (blocking buffer) and
incubated for 2 hours. Mouse sera were serially diluted from 1:100 to 1:12,800
in
blocking buffer, added to the p24-coated plates and incubated overnight at 4
C.
Plates were then washed four times with PBS (0.05% Tween 20), and incubated
with
goat anti-mouse IgG (1:10,000 dilution, Sigma) for 2 hours at room
temperature.
Plates were washed four times, and then pNPP alkaline phosphatase substrate
(75 l;
Sigma, St. Louis, MO) was added to each well. The reaction was stopped after
one
hour by addition of 0.5 N NaOH (25 l). The plates were read on an ELISA
reader
at 405 nm, and titers were calculated at a cutoff optical density of 0.4.
HIV-1 inummoblotting
The strips containing HIV-1 proteins (Immunectics Inc., Cambridge, MA)
were incubated with pooled mouse sera at a dilution of 1:25. Purified human
anti-
HIV IgG (AIDS Research and Reference Reagent Program, Rockville, MD) was
used as a positive control. Bands were visualized using the ECL Western
blotting
detection reagent (Amersham Pharmacia Biotech, Piscataway, NJ).
Im munoprecipitation and Western blotting
Three days after transfection, hPol gene-transfected 293T cell lysates were
prepared with RIPA buffer. The pooled mouse sera were diluted with
immunoprecipitation (1P) buffer (100mM KC1, 2.5mM MgC12, 20mM I-EPES, pH
-161-
CA 02419822 2009-09-17
7.9, 0.1 % NP-40, 1 mM DTT and proteinase inhibitors). After adding 10 .tg of
the
cell lysate containing the HIV-1 Pol protein, the reactions were incubated
overnight
on a rotator at 4 C. 250 l of Protein G and A SepharoseTM beads (10%V/V in IP
buffer) were then added, and the reactions were incubated for 2 hours on a
rotator at
4 C. The beads were washed four times with IP buffer, resuspended in 30 i of
IX
sample buffer and loaded onto SDS-PAGE. After transfer onto an Immobilon P
membrane (Millipore), membranes were incubated with anti HIV-1-IgG (AIDS
Research and Reference Reagent Program). Bands were visualized using the ECL
Western blotting detection reagent (Amersham Pharmacia Biotech).
Prime/Boost Vaccination Strategy
Groups of guinea pigs (4/group) were immunized (primed) with the vectors
listed below, 3 times at 2 week intervals. Two weeks after the third DNA
immunization, blood was collected for immune analysis. 2-3 days after the
blood
collection, the animals received a boost with a corresponding adenoviral
construct,
AdApt (Sullivan, N. et al., 2000, Nature, 408:605-608). Blood was again
collected 2
weeks after the adenoviral boost. Blood was tested for neutralizing antibodies
and
(using an ELISA) the presence of antibody against the envelope. The groups are
summarized below.
Prime Adenoviral Boost
gpl40delCFI (R5) Adv gpl40delCFI(R5)
gpl45delCFI (R5) Adv gp I 45delCFI(R5)
gp145delCFl (R5) Adv gpl40delCFI(R5)
gpl40delCFI (89.6P) Adv gp14OdelCFI(89.6P)
gpI45delCFI (89.6P) Adv gp145delCFI(89.6P)
gpl45delCFI (89.6P) Adv gpl40delCFI(89.6P)
Adv gpI40delCFI (R5) Adv gpl40delCFI(R5)
Adv gpl45delCFI (R5) Adv gpl45delCFI(R5)
Adv gpl45delCFI (R5) Adv gpl40delCFI(R5)
Adv gpl40delCFI (89.6P) Adv gpI40delCF1(89.6P)
Adv gpl45delCFI (89.6P) Adv gp145delCF1(89.6P)
Adv gpl45delCFI (89.6P) Adv gp I 40delCFI(89.6P)
-162-
CA 02419822 2009-09-17
Results
The level of antibody in the sera after adenoviral boost was 8- tolO-fold
higher than the level of antibody present after 3 injections of DNA alone, in
almost
all of the cases. Neutralizing antibodies were detected in the sera of animals
immunized with either gpl45delCFl DNA or gpl40delCFI DNA followed by
Advgp 140de1CFI.
Neutralization was also observed in animals immunized with gpl45de1CFI
DNA followed by Adv gpl45delCFl, and in animals both primed and boosted with
adenovirus.
Optimal neutralization was seen when animals were primed with
gpl45delCFl DNA followed by adenovirus expressing gpl40delCFI in both Glade B
(R5-Bal strain) and 89.6P (Glade B, dual tropic strain).
All neutralizing activity was able to be blocked with the V3 peptide of the
corresponding strains.
While the present invention has been described in some detail for purposes
of clarity and understanding, one skilled in the art will appreciate that
various
changes in form and detail can be made without departing from the true scope
of the
invention.
-163-
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME DE
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 3
NOTE: For additional volumes please contact the Canadian Patent Office.