Note: Descriptions are shown in the official language in which they were submitted.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-1-
NOVEL DERMATOPHAGOIDES NUCLEIC ACID MOLECULES,
PROTEINS AND USES THEREOF
FIELD OF THE INVENTION
The present invention relates to high molecular weight DeYfnatophagoides
proteins, nucleic acid molecules and therapeutic and diagnostic reagents
derived from
such proteins.
BACKGROUND OF THE INVENTION
Immunoglobulin E (IgE) mediated allergic symptoms afflict many animals.
IgE antibody production in an animal can induce pathogenic IgE responses
including,
for example, atopic disease, asthma and rhinitis. Allergens are proteins or
peptides
characterized by their ability to induce a pathogenic IgE response in
susceptible
individuals.
House dust mite (e.g., Derrnatophagoides fariu.ae and Dernzatophagoides
pteYOrzyssircus; l7er f and Der p, respectively) allergens are major causative
agents
associated With IgE-mediated pathogenesis. Previous investigators have
identified two
major groups of dust mite allergens in humans, group I (Der f I and Der p I,
Mr
25,000) and group 2 (Der f II and Der p II, Mr 14,000); reviewed in Chapman,
et al.,
Allergy, vol. 52, pp.37-379, 1997. Prior investigators have disclosed
nucleotide and/or
annino acid sequences for: Der f I, Der f II, Der p I and Der p II, U.S.
Patent No.
5,552,142, to Thomas et al., issued September 3, 1996, U.S. Patent No.
5,460,977, to
Ando et al., issued October 24, 1995, PCT Patent Publication No. WO 95/28424,
by
Chen et al., published October 26, 1995, U.S. Patent No. 5,433,948, to Thomas
et al.,
issued July 18, 1995, PCT Patent Publication No. WO 93/08279, by Garmen et
al.,
published March 4, 1993, or Chapman, ibid.; Der p III, PCT Patent Publication
No.
WO 95/15976, by Thomas et al., published June 15, 1995; Der p VII, PCT Patent
Publication No. WO 94/20614, by Thomas et al., published September 15, 1994; a
40-
kilodalton (kd) Der f allergen, U.S. Patent No. 5,405,758, to Oka et al.,
issued April
11, 1995, U.S. Patent No. 5,314,991, to Oka et al., issued May 24, 1994; a 70-
kd Des- f
allergen which is a heat shock protein (Hsp70), Aki et al., J. Biochef7z.,
vol. 115, pp.
435-440, 1994; or Noli et al., Vet. Imfzzunol. lyvmmaopath., vol. 52, pp. 147-
157, 1996;
and a 98-kd Der f paramyosin-like allergen, Tsai et al, J. Allergy Clirz.
Imrnuzzol., vol.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-2-
102, pp. 295-303, 1998. None of these published sequences indicates, suggests
or
predicts any of the mite allergic nucleic acid molecules or proteins of the
present
invention, nor the relevance of such proteins as being immunoreactive with IgE
antibodies in canine, feline, or human sera.
Products and processes of the present invention are needed in the art that
provide specific detection and treatment of mite allergy. .
SUMMARY OF THE INVENTION
The present invention relates to novel proteins having molecular weights of
about 60 kilodaltons (kd or kD), 70 kD, or from about 98 kD to about 109 kD.
Such
proteins include at least one epitope of a protein allergen of a mite of the
genus
Dermatophagoides and are designated herein as Def° HMW-map proteins.
Preferred
proteins are Dermatophagoides farinae or Derrnatoplaagoides ptet~onyssius
proteins.
The present invention also provides proteins that are fragments or peptides of
full-
length or mature proteins, as well as antibodies, rnimetopes or muteins of any
of such
proteins. The present invention also provides nucleic acid molecules encoding
any of
such proteins, as well as complements thereof. The present invention also
includes
methods to obtain such proteins, nucleic acid molecules, antibodies, mimetopes
or
muteins, as well as methods to use such compounds in diagnostic or therapeutic
applications. The present invention also relates to reagents comprising non-
proteinaceous epitopes that bind to IgE in mite-allergic dogs and/or cats as
well as to
antibodies raised against such epitopes. The present invention also relates to
therapeutic compositions or assay kits comprising such non-proteinaceous
epitopes, as
well as to methods to identify and/or desensitize an animal susceptible to an
allergic
response to a mite, comprising the use of non-proteinaceous epitopes of the
present
invention.
One embodiment of the present invention is at least one of the following
isolated nucleic acid molecules: (a) a nucleic acid molecule comprising at
least about
150 nucleotides, wherein such a nucleic acid molecule hybridizes, in a
solution
comprising 1X SSC and 0% formamide, at a temperature of about 50°C, to
a nucleic
acid molecule comprising at least one of the following nucleic acid sequences:
SEQ ID
N0:14, SEQ m N0:16, SEQ ID N0:17, SEQ m N0:19, SEQ ID N0:20, SEQ ID
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-3-
N0:22, SEQ m N0:34, SEQ m N0:36, SEQ m N0:37, SEQ ID N0:39, SEQ m
N0:40, SEQ m N0:42, SEQ ID N0:43, SEQ ID N0:45, and a nucleic acid sequence
encoding a protein comprising the amino acid sequence of SEQ ID N0:33 and a
complement thereof; and (b) a nucleic acid molecule comprising a fragment of
any of
the nucleic acid molecules of (a) wherein the fragment comprises at least
about 15
nucleotides. The present invention also includes recombinant molecules,
recombinant
viruses and recombinant cells comprising such nucleic acid sequences as well
as
methods to produce them.
Another embodiment of the present invention is an isolated protein encoded by
at least one of the following nucleic acid molecules: (a) a nucleic acid
molecule
comprising at least about 150 nucleotides, wherein such a nucleic acid
molecule
hybridizes, in a solution comprising 1X SSC and 0% formamide, at a temperature
of
about 50 ° C, to a nucleic acid molecule comprising at least one of the
following
nucleic acid sequences: SEQ m N0:16, SEQ ID N0:19, SEQ ID N0:22, SEQ ID
N0:36, SEQ ID N0:39, SEQ ID N0:42, SEQ ID N0:45, and a complement of a
nucleic acid sequence encoding a protein comprising the amino acid sequence
SEQ ID
N0:33; and (b) a nucleic acid molecule comprising a fragment of any of the
nucleic
acid molecules of (a), wherein the fragment comprises at least about 15
nucleotides.
An isolated protein of the present invention can also be encoded by a nucleic
acid
molecule that hybridizes under stringent hybridization conditions with the
complement
of a nucleic acid molecule that encodes a protein having at least one of the
following
amino acid sequences: SEQ ID NO:1, SEQ ID N0:2, SEQ ID N0:3, SEQ ID N0:4,
SEQ ID N0:5, SEQ m N0:6, SEQ ID N0:7, SEQ ID N0:8, SEQ ID N0:9, SEQ ID
NO:10, SEQ m NO:l 1, SEQ ID N0:12, SEQ ID N0:13, SEQ ID NO:15, SEQ ID
N0:18, SEQ m N0:21, SEQ ID N0:23, SEQ ID N0:24, SEQ ID N0:29, SEQ m
N0:30, SEQ ID N0:31, SEQ ID N0:32, SEQ ID N0:33, SEQ ID N0:35, SEQ ID
N0:38, SEQ ID N0:41, and SEQ m N0:44. The present invention also includes an
antibody that selectively binds to a protein of the present invention as well
as methods
to produce and use such proteins or antibodies.
The present invention also includes a therapeutic composition for treating an
allergic response to a mite. Such a therapeutic composition includes at least
one of the
following desensitizing compounds: (a) an isolated nucleic acid molecule of
the
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-4-
present invention; (b) an isolated mite allergenic protein of the present
invention; (c) a
mimetope of such a mite allergenic protein; (d) a mutein of such a mite
allergenic
protein; (e) an antibody to such a mite allergic protein; and (f) an inhibitor
of binding
of such a mite allergic protein to IgE. Also included is a method to
desensitize a host
animal to an allergic response to a mite. Such a method includes the step of
administering to the animal a therapeutic composition of the present
invention.
One embodiment of the present invention is an assay kit for testing if an
animal
is susceptible to or has an allergic response to a mite. Such a kit includes
an isolated
protein of the present invention and a means for determining if the animal is
susceptible to or has that allergic response. Such a means includes use of
such a
protein to identify animals susceptible to or having allergic responses to
mites. The
present invention also includes a method to identify an animal susceptible to
or having
an allergic response to a mite. Such a method includes the steps of: (a)
contacting an
isolated protein of the present invention with antibodies of an animal; and
(b)
determining immunocomplex formation between the protein and the antibodies,
wherein formation of the immunocomplex indicates that the animal is
susceptible to or
has such an allergic response.
The present invention includes a reagent that comprises a non-proteinaceous
epitope having at least one of the following identifying characteristics: (a)
the epitope
is resistant to ~3-elimination of peptides; (b) the epitope is resistant to
Proteinase-I~
digestion; and (c) the epitope is reactive to a test designed to detect
glycosylated
proteins. Such an epitope binds to at least one of the following antibodies:
canine IgE
from dogs allergic to mites and feline IgE from cats allergic to mites. Also
included is
an isolated antibody that selectively binds such a non-proteinaceous epitope
as well as
derivatives of such an epitope.
The present invention also relates to therapeutic compositions and assay kits
comprising a non-proteinaceous epitope of the present invention, as well as
methods to
identify and/or desensitize an animal susceptible to an allergic response to a
mite,
comprising the use of a non-proteinaceous epitope of the present invention.
BRIEF DESCRIPTION OF THE FIGURES
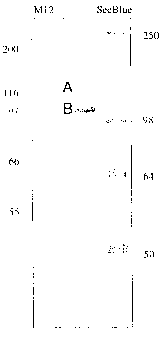
Fig. 1 illustrates high molecular weight Der f proteins resolved by 12% Tris-
Glycine SDS-PAGE.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-5-
Fig. 2 illustrates an about 60 kD Def~ f protein resolved by 14% Tris-Glycine
SDS-PAGE.
DETAILED DESCRIPTION OF THE INVENTION
The present invention provides for isolated proteins having molecular weights
ranging from about 60 kilodaltons (kD) to about 109 kD, that include at least
one
epitope of a protein allergen of a mite of the genus Dermatophagoides, in
particular a
mite of the species Dermatophagoides farinae and/or Derma.tophagoides
pteYOnyssius.
Such proteins are referred to herein as Der HMW-map proteins. The present
invention
further includes methods to isolate and identify nucleic acid molecules
encoding
DeYHMW-map proteins, antibodies directed against Der HMW-map proteins and
inhibitors of Der HMW-map protein activity. As used herein, the term isolated
Der
HMW-map proteins refers to Der HMW-map proteins derived from
Dermatophagoides, and more preferably from Denrnatophagoides far°iT2ae
and/or
Dermatophagoides pteronyssius and, as such, can be obtained from its natural
source
or can be produced using, for example, recombinant nucleic acid technology or
chemical synthesis. Also included in the present invention is the use of this
protein
and antibodies in a method to detect immunoglobulin that specifically binds to
DeY
HMW-map proteins, to treat pathogenesis against mite allergens, and in other
applications, such as those disclosed below. The products and processes of the
present
invention are advantageous because they enable the detection of anti Der HMW-
map
antibodies in fluids of animals and the inhibition of IgE or Des° HMW-
map protein
activity associated with disease.
One embodiment of the present invention is an isolated Derma.toplzagoides
allergenic composition including: (a) a composition produced by a method
comprising: (1) applying soluble proteins of a Dermatophagoides extract to a
gel
filtration column; (2) collecting excluded protein from the gel filtration
column and
applying the excluded protein to an anion exchange column; and (3) eluting
proteins
bound to the anion exchange column with about 0.3 M Tris-HCI, pH 8 to obtain
the
Dermatophagoides allergenic composition; and (b) a composition comprising a
peptide of a protein produced in accordance with step (a), in which the
allergenic
composition is capable of a biological function including binding to IgE,
stimulating a
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-6-
B lymphocyte response and stimulating a T lymphocyte response. Such
Dermatophagoides allergenic composition is also referred to herein as a Der
HMW-
map composition. A suitable gel filtration column includes any gel filtration
column
capable of excluding proteins having a molecular weight between about 50 kD
and
about 150 kD. A preferred gel filtration column includes, but is not limited
to a
Sephacryl S-100 column. A suitable anion exchange column includes any anion
exchange column capable of binding to a protein having a pI of less than about
pI 6. A
preferred anion exchange column includes, but is not limited to a Q-Sepharose
column. As used herein, "stimulating a B lymphocyte response" refers to
increasing a
humoral immune response in an animal that is induced preferentially by a Der
HMW-
map of the present invention and involves the activity of a B lymphocyte in
the
animal. As used herein, "stimulating a T lymphocyte response" refers to
increasing a
cellular immune response in an animal that is induced preferentially by a Der
HMW-
map of the present invention and involves the activity of a T lymphocyte in
the animal.
One embodiment of the present invention is an isolated protein that includes a
Der HMW-map protein. It is to be noted that the term "a" or "an" entity refers
to one
or more of that entity; for example, a protein, a nucleic acid molecule, an
antibody, an
inhibitor, a compound or a therapeutic composition refers to "one or more" or
"at least
one" protein, nucleic acid molecule, antibody, inhibitor, compound or
therapeutic
composition respectively. As such, the terms "a" (or "an"), "one or more" and
"at least
one" can be used interchangeably herein. It is also to be noted that the terms
"comprising", "including", and "having" can be used interchangeably. According
to
the present invention, an isolated, or biologically pure, protein, is a
protein that has
been removed from its natural milieu. As such, "isolated" and "biologically
pure" do
not necessarily reflect the extent to which the protein has been purified. An
isolated
protein of the present invention can be obtained from its natural source, can
be
produced using recombinant DNA technology, or can be produced by chemical
synthesis.
As used herein, a Der HMW-map protein can be a full-length protein or any
homolog of such a protein. As used herein, a protein can be a polypeptide or a
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
peptide, as the terms are used by those of skill in the art. Preferably, a Des
HMW-map
protein comprises at least a portion of a Der HMW-map protein that comprises
at least
one epitope recognized by an IgE antibody (i.e., a protein of the present
invention
binds to an IgE antibody), an antibody on the surface of a B lymphocyte and/or
a T
cell receptor in the presence of a major histocompatability complex (MHC)
molecule
from an animal demonstrating IgE-mediated pathogenesis to a Den HMW-map
protein.
A peptide of the present invention includes a Der- HMW-map protein of the
present invention that is capable of binding to IgE, desensitizing an animal
against
mite allergen, stimulating a B lymphocyte response, and/or stimulating a T
lymphocyte response. Preferably, a peptide of the present invention comprises
a B
lymphocyte epitope or a T lymphocyte epitope. A peptide having a B lymphocyte
epitope can bind to an antibody. A peptide having a T lymphocyte epitope can
bind to
a MHC molecule in such a manner that the peptide can stimulate a T lymphocyte
through a T cell receptor. According to the present invention, a peptide
comprising a
B lymphocyte epitope can be from about 4 residues to about 50 residues in
length,
preferably from about 5 residues to about 20 residues in length. According to
the
present invention, a peptide comprising a T lymphocyte epitope can be from
about 4
residues to about 20 residues in length, preferably from about 8 residues to
about 16
residues in length.
A Der HMW-map protein of the present invention, including a homolog, can
be identified in a straight-forward manner by the protein's ability to induce
an allergic
response to DeY HMW-map protein. Examples of Des HMW-map protein homologs
include Der HMW-map protein in which amino acids have been deleted (e.g., a
truncated version of the protein, such as a peptide), inserted, inverted,
substituted
andlor derivatized (e.g., by glycosylation, phosphorylation, acetylation,
myristoylation, prenylation, palmitoylation, amidation and/or addition of
glycerophosphatidyl inositol) such that the homolog is capable of inducing an
allergic
response to a natural Der HMW-map protein.
Der HMW-map protein homologs can be the result of natural allelic variation
or natural mutation. Der HMW-map protein homologs of the present invention can
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
_g_
also be produced using techniques known in the art including, but not limited
to, direct
modifications to the protein or modifications to the gene encoding the protein
using,
for example, classic or recombinant nucleic acid techniques to effect random
or
targeted mutagenesis.
One embodiment of the present invention is a Der HMW-map gene that
includes the nucleic acid sequence SEQ ID N0:14, SEQ ID N0:16, SEQ ID N0:17,
SEQ ID N0:19, SEQ ID N0:20 SEQ ID N0:22, SEQ ID N0:34, SEQ ID N0:36,
SEQ ID N0:37, SEQ ID N0:39, SEQ ID N0:40, SEQ ID N0:42, SEQ ID N0:43, and
SEQ ID N0:45 as well as the complements of any of these nucleic acid
sequences.
These nucleic acid sequences are further described herein. For example,
nucleic acid
sequence SEQ ID N0:14 represents the deduced sequence of the coding strand of
a
cDNA (complementary DNA) denoted herein as Des HMW-map gene nucleic acid
molecule nDerf981~s2, the production of which is disclosed in the Examples.
Nucleic
acid molecule nDerf981~sz comprises an apparently full-length coding region.
The
complement of SEQ ID N0:14 (represented herein by SEQ ID N0:16) refers to the
nucleic acid sequence of the strand complementary to the strand,having SEQ ID
N0:14, which can easily be determined by those skilled in the art. Likewise, a
nucleic
acid sequence complement of any nucleic acid sequence of the present invention
refers
to the nucleic acid sequence of the nucleic acid strand that is complementary
to (i.e.,
can form a double helix with) the strand for which the sequence is cited. It
should be
noted that since nucleic acid sequencing technology is not entirely error-
free, SEQ ID
N0:14 (as well as other nucleic acid and protein sequences presented herein)
represents an apparent nucleic acid sequence of the nucleic acid molecule
encoding a
Der~ HMW-map protein of the present invention.
In another embodiment, a Der HMW-map gene or nucleic acid molecule can
be an allelic variant that includes a similar but not identical sequence to
SEQ ID
N0:14 or SEQ ID N0:16, or any other Der HMW-map nucleic acid sequence cited
herein. For example, an allelic variant of a Den HMW-map gene including SEQ ID
N0:14 or SEQ ID N0:16, is a gene that occurs at essentially the same locus (or
loci)
in the genome as the gene including SEQ ID N0:14 and SEQ ID N0:16, but which,
due to natural variations caused by, for example, mutation or recombination,
has a
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-9-
similar but not identical sequence. Because natural selection typically
selects against
alterations that affect function, allelic variants (i.e. alleles corresponding
to, or of, cited
nucleic acid sequences) usually encode proteins having similar activity to
that of the
protein encoded by the gene to which they are being compared. Allelic variants
of
genes or nucleic acid molecules can also comprise alterations in the 5' or 3'
untranslated regions of the gene (e.g., in regulatory control regions), or can
involve
alternative splicing of a nascent transcript, thereby bringing alternative
exons into
juxtaposition. Allelic variants are well known to those skilled in the art and
would be
expected to occur naturally within a given dust mite such as
Derm,atop72agoides, since
the respective genomes are diploid, and sexual reproduction will result in the
reassortment of alleles.
In one embodiment of the present invention, an isolated Der HMW-map
protein is encoded by a nucleic acid molecule that hybridizes under stringent
hybridization conditions to a gene encoding a Der HMW-map protein. The minimal
size of a Der HMW-map protein of the present invention is a size sufficient to
be
encoded by a nucleic acid molecule capable of forming a stable hybrid (i.e.,
hybridizing under stringent hybridization conditions) with the complementary
sequence of a nucleic acid molecule encoding the corresponding natural
protein. The
size of a nucleic acid molecule encoding such a protein is dependent on the
nucleic
acid composition and the percent homology between the Der HMW-map nucleic acid
molecule and the complementary nucleic acid sequence. It can easily be
understood
that the extent of homology required to form a stable hybrid under stringent
conditions
can vary depending on whether the homologous sequences are interspersed
throughout
a given nucleic acid molecule or are clustered (i.e., localized) in distinct
regions on a
given nucleic acid molecule.
The minimal size of a nucleic acid molecule capable of forming a stable hybrid
with a gene encoding a Der HMW-map protein is typically at least about 12
nucleotides to about 15 nucleotides in length if the nucleic acid molecule is
GC-rich
and at least about 15 to about 17 bases in length if it is AT-rich. The
minimal size of a
nucleic acid molecule used to encode a Der HMW-map protein homolog of the
present
invention is from about 12 to about 18 nucleotides in length, preferably about
12
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-10-
nucleotides, or about 15 nucleotides, or about 18 nucleotides in length. Thus,
the
minimal size of a Der HMW-map protein homolog of the present invention is from
about 4 to about 6 amino acids in length. There is no limit, other than a
practical limit,
on the maximal size of a nucleic acid molecule encoding a Der HMW-map protein
of
the present invention because a nucleic acid molecule of the present invention
can
include a portion of a gene, an entire gene, or multiple genes. The preferred
size of a
protein encoded by a nucleic acid molecule of the present invention depends on
whether a full-length, fusion, multivalent, or functional portion of such a
protein is
desired. Preferably, the preferred size of a protein encoded by a nucleic acid
molecule
of the present invention is a portion of the protein that induces an immune
response
which is about 30 amino acids, more preferably about 35 amino acids and even
more
preferably about 44 amino acids in length.
Stringent hybridization conditions are determined based on defined physical
properties of the gene to which the nucleic acid molecule is being hybridized,
and can
be defined mathematically. Stringent hybridization conditions are those
experimental
parameters that allow an individual skilled in the art to identify significant
similarities
between heterologous nucleic acid molecules. These conditions are well known
to
those skilled in the art. See, for example, Sambrook, et al., 1989,
Molecular° Clohirzg:
A Laboratory Manual, Cold Spring Harbor Labs Press, and Meinkoth, et al.,
1984,
Arcal. Bioclzern. 138, 267-284. As explained in detail in the cited
references, the
determination of hybridization conditions involves the manipulation of a set
of
variables including the ionic strength (M, in moles/liter), the hybridization
temperature
(°C), the concentration of nucleic acid helix destabilizing agents
(such as formamide),
the average length of the shortest hybrid duplex (n), and the percent G + C
composition of the fragment to which an unknown nucleic acid molecule is being
hybridized. For nucleic acid molecules of at least about 150 nucleotides,
these
variables are inserted into a standard mathematical formula to calculate the
melting
temperature, or Tm, of a given nucleic acid molecule. As defined in the
formula below,
Tm is the temperature at which two complementary nucleic acid molecule strands
will
disassociate, assuming 100% complementarity between the two strands:
Tm 81.5°C + 16.6 log M + 0.41 (%G + C) - 500/n - 0.61
(%formamide).
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-11-
For nucleic acid molecules smaller than about 50 nucleotides, hybrid stability
is
defined by the dissociation temperature (Td), which is defined as the
temperature at
which 50% of the duplexes dissociate. For these smaller molecules, the
stability at a
standard ionic strength is defined by the following equation:
Td=4(G+C)+2,(A+T).
A temperature of 5°C below Td is used to detect hybridization between
perfectly
matched molecules.
Also well known to those skilled in the art is how base-pair mismatch, i.e.
differences between two nucleic acid molecules being compared, including non-
complementarity of bases at a given location, and gaps due to insertion or
deletion of
one or more bases at a given location on either of the nucleic acid molecules
being
compared, will affect Tm or Td for nucleic acid molecules of different sizes.
For
example, Tm decreases about 1°C for each 1% of mismatched base-pairs
for hybrids
greater than about 150 bp, and Td decreases about 5°C for each
mismatched base-pair
for hybrids below about 50 bp. Conditions for hybrids between about 50 and
about
150 base-pairs can be determined empirically and without undue experimentation
using standard laboratory procedures well known to those skilled in the art.
These
simple procedures allow one skilled in the art to set the hybridization
conditions (by
altering, for example, the salt concentration, the formamide concentration or
the
temperature) so that only nucleic acid hybrids with less than a specified %
base-pair
mismatch will hybridize. Stringent hybridization conditions are commonly
understood
by those skilled in the art to be those experimental conditions that will
allow
hybridization between molecules having about 30% or less base-pair mismatch
(i.e.,
about 70% or greater identity). Because one skilled in the art can easily
determine
whether a given nucleic acid molecule to be tested is less than or greater
than about 50
nucleotides, and can therefore choose the appropriate formula for determining
hybridization conditions, he or she can determine whether the nucleic acid
molecule
will hybridize with a given gene under stringent hybridization conditions and
similarly
whether the nucleic acid molecule will hybridize under conditions designed to
allow a
desired amount of base pair mismatch.
Hybridization reactions are often carried out by attaching the nucleic acid
molecule to be hybridized to a solid support such as a membrane, and then
hybridizing
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-12-
with a labeled nucleic acid molecule, typically referred to as a probe,
suspended in a ,
hybridization solution. Examples of common hybridization reaction techniques
include, but are not limited to, the well-known Southern and northern blotting
procedures. Typically, the actual hybridization reaction is done under non-
stringent
conditions, i.e., at a lower temperature and/or a higher salt concentration,
and then
high stringency is achieved by washing the membrane in a solution with a
higher
temperature and/or lower salt concentration in order to achieve the desired
stringency.
For example, if the skilled artisan wished to identify a nucleic acid molecule
that hybridizes under stringent hybridization conditions with a
Dernzatophagoides
farihae and/or Dermatophagoides ptero~zyssius nucleic acid molecule of about
150 by
in length, the following conditions could preferably be used. The average G +
C
content of Dermatophagoides fari~aae and Dermatophagoides pterorayssius DNA is
about 39%. The unknown nucleic acid molecules would be attached to a support
membrane, and the 150 by probe would be labeled, e.g. with a radioactive tag.
The
hybridization reaction could be carried out in a solution comprising 2X SSC
and 0%
formamide, at a temperature of about 37°C (low stringency conditions).
Solutions of
differing concentrations of SSC can be made by one of skill in the art by
diluting a
stock solution of 20X SSC (175.3 gram NaCI and about 88.2 gram sodium citrate
in 1
liter of water, pH 7) to obtain the desired concentration of SSC. In order to
achieve
high stringency hybridization, the skilled artisan would calculate the washing
conditions required to allow up to 30% base-pair mismatch. For example, in a
wash
solution comprising 1X SSC and 0% formamide, the Tm of perfect hybrids would
be
about 80°C:
81.5°C + 16.6 log (.15M) + (0.41 x 39) - (500/150) - (0.61 x 0) =
80.4°C.
Thus, to achieve hybridization with nucleic acid molecules having about 30%
base-
pair mismatch, hybridization washes would be carried out at a temperature of
about
50°C. It is thus within the skill of one in the art to calculate
additional hybridization
temperatures based on the desired percentage base-pair mismatch, formulae and
G/C
content disclosed herein. For example, it is appreciated by one skilled in the
art that as
the nucleic acid molecule to be tested for hybridization against nucleic acid
molecules
of the present invention having sequences specified herein becomes longer than
150
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-13-
nucleotides, the Tm for a hybridization reaction allowing up to 30% base-pair
mismatch will not vary significantly from 50°C.
Furthermore, it is known in the art that there are commercially available
computer programs for determining the degree of similarity between two nucleic
acid
sequences. These computer programs include various known methods to determine
the percentage identity and the number and length of gaps between hybrid
nucleic acid
molecules. Preferred methods to determine the percent identity among amino
acid
sequences and also among nucleic acid sequences include analysis using one or
more
of the commercially available computer programs designed to compare and
analyze
nucleic acid or amino acid sequences. These computer programs include, but are
not
limited to, GCGTM (available from Genetics Computer Group, Madison, WI),
DNAsisTM (available from Hitachi Software, San Bruno, CA) and MacVectorTnn
(available from the Eastman Kodak Company, New Haven, CT). A preferred method
to determine percent identity among amino acid sequences and also among
nucleic
acid sequences includes using the Compare function by maximum matching within
the
program DNAsis Version 2.1 using default parameters.
One embodiment of the present invention includes Der HMW-map proteins.
In one embodiment, Der HMW-map proteins of the present invention include
proteins
that, when submitted to reducing 12% Tris glycine SDS-PAGE; migrate as bands
at a
molecular weight of from about 98 kD to about 109 kD, as shown in Fig. 1. The
bands
in Fig. 1 are obtained when proteins are collected from Dermataphagoides
fari~2ae
mites using the method described in detail in Example 1. Preferably, Der HMW-
map
proteins of the present invention includes proteins having a molecular weight
ranging
from about 90 kD to about 120 kD, and more preferably from about 98 kD to
about
109 kD. Preferred Der HMW-map proteins of the present invention include mapA
and
mapB, the identification of which zs described in the Examples section.
In another embodiment, Der HMW-map proteins of the present invention
include proteins that, when submitted to reducing 14% Tris glycine SDS-PAGE,
migrate as a band at a molecular weight of about 60 kD, as shown in Fig. 2.
The band
in Fig. 2 is obtained when proteins are collected from Def-n2ataphagoides
fariniae mites
using the method described in detail in Example 9. Preferably, Der HMW-map
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-14-
proteins of the present invention includes proteins having a molecular weight
of about
60 kD. Preferred Der HMW-map proteins of the present invention include mapD,
the
identification of which is described in the Examples section.
In another embodiment, a preferred Der HMW-map protein includes a protein
encoded by a nucleic acid molecule which is at least about 50 nucleotides, or
about
150 nucleotides, and which hybridizes under conditions which preferably allow
about
40% or less base pair mismatch, more preferably under conditions which allow
about
35% or less base pair mismatch, more preferably under conditions which allow
about
30% or less base pair mismatch, more preferably under conditions which allow
about
25% or less base pair mismatch, more preferably under conditions which allow
about
20% or less base pair mismatch, more preferably under conditions which allow
about
15% or less base pair mismatch, more preferably under conditions which allow
about
10% or less base pair mismatch and even more preferably under conditions which
allow about 5% or less base pair mismatch with a nucleic acid molecule
selected from
the group consisting of SEQ ID N0:16, SEQ ID N0:19, SEQ ID N0:22, SEQ ID
N0:36, SEQ m NO:39, SEQ ID N0:42, SEQ ID N0:45 and a nucleic acid sequence
encoding a protein comprising the amino acid sequence SEQ ID N0:33 the
complement thereof.
Another embodiment of the present invention includes a Der HMW-map
protein encoded by a nucleic acid molecule selected from the group consisting
of: a
nucleic acid molecule comprising at least about 150 nucleotides, wherein said
nucleic
acid molecule comprising at least about 150 nucleotides hybridizes, in a
solution
comprising 1X SSC and 0% formamide, at a temperature of about 50°C, to
a nucleic
acid sequence selected from the group consisting of SEQ ID N0:16, SEQ ll~
N0:19,
SEQ ID N0:22, SEQ ID N0:36, SEQ ID N0:39, SEQ ID N0:42, SEQ ID N0:45, and
a complement of a nucleic acid sequence encoding a protein comprising the
amino
acid sequence SEQ ID N0:33; and a nucleic acid molecule comprising a fragment
of
any of said nucleic acid molecules comprising at least about 15 nucleotides.
Yet another preferred Der HMW-map protein of the present invention includes
a protein encoded by a nucleic acid molecule which is preferably at least
about 60%
identical, more preferably at least about 65% identical, more preferably at
least about
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-15-
70% identical, more preferably at least about 75% identical, more preferably
at least
about 80% identical, more preferably at least about 85% identical, more
preferably at
least about 90% identical and even more preferably at least about 95%
identical to a
nucleic acid molecule having the nucleic acid sequence SEQ ID N0:14, SEQ ID
N0:17, SEQ ID N0:20, SEQ ID N0:34, SEQ ID N0:37, SEQ ID N0:40, SEQ ID
N0:43, and/or a complement of a nucleic acid sequence encoding a protein
comprising
the amino acid sequence SEQ ID N0:33;, also preferred are fragments of such
proteins. Percent identity as used herein is determined using the Compare
function by
maximum matching within the program DNAsis Version 2.1 using default
parameters.
Additional preferred Der HMW-map proteins of the present invention include
proteins having the amino acid sequence SEQ ID NO:l, SEQ ID N0:2, SEQ ID N0:3,
SEQ ID N0:4, SEQ ID N0:5, SEQ ID N0:6, SEQ ID N0:7, SEQ ID N0:8, SEQ ID
N0:9, SEQ ID N0:10, SEQ ID NO:11, SEQ ID NO:12, SEQ ID N0:13, SEQ ID
N0:15, SEQ ID N0:18, SEQ ID N0:21, SEQ ID N0:23, SEQ ID N0:24, SEQ ID
N0:29, SEQ ID N0:30, SEQ ID N0:31, SEQ ID N0:32, SEQ ID N0:33, SEQ ID
N0:35, SEQ ID NO:38, SEQ ID N0:41, SEQ ID N0:44, and proteins comprising
homologs of a protein having the amino acid sequence SEQ TD NO:l, SEQ ID N0:2,
SEQ ID N0:3, SEQ 117 N0:4, SEQ ZD N0:5, SEQ ID N0:6, SEQ ID N0:7, SEQ ID
N0:8, SEQ ID N0:9, SEQ ID NO:10, SEQ ID N0:11, SEQ ID N0:12, SEQ )D
N0:13, SEQ ID N0:15, SEQ ID N0:18, SEQ ID N0:21, SEQ ID N0:23, SEQ ID
N0:24, SEQ ID N0:29, SEQ ID N0:30, SEQ ID N0:31, SEQ ID N0:32, SEQ 117
N0:33, SEQ ID N0:35, SEQ ID N0:38, SEQ ID N0:41, SEQ ID N0:44 in which
such a homolog comprises at least one epitope that elicits an immune response
against
a protein having an amino acid sequence SEQ ll~ NO:1, SEQ ID N0:2, SEQ ID
N0:3,
SEQ ID N0:4, SEQ ID N0:5, SEQ ID N0:6, SEQ ID N0:7, SEQ ID N0:8, SEQ ID
N0:9, SEQ ID N0:10, SEQ ID NO:l 1, SEQ ID N0:12, SEQ ID N0:13, SEQ D7
N0:15, SEQ ID N0:18, SEQ ID N0:21, SEQ ID N0:23, SEQ ID N0:24, SEQ ID
N0:29, SEQ ID N0:30, SEQ ID N0:31, SEQ ID N0:32, SEQ ID N0:33, SEQ ID
N0:35, SEQ ID N0:38, SEQ ID N0:41, SEQ ID N0:44 Likewise, also preferred are
proteins encoded by nucleic acid molecules encoded by nucleic acid molecules
having
nucleic acid sequence SEQ ID N0:14, SEQ ID N0:17, SEQ ID N0:20, SEQ ID
N0:34, SEQ ID N0:37, SEQ ID N0:40, SEQ ID N0:43 andJor a nucleic acid
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-16-
sequence encoding a protein comprising the amino acid sequence SEQ ID N0:33,
or
by homologs thereof.
A preferred isolated protein of the present invention is a protein encoded by
at
least one of the following nucleic acid molecules: nDerf981~sz, nDerf981~~s,
nDerf981sos~ ~e~98i6zn nDerp981sz~, nDerp9814~0, nDerf60s1o, or allelic
variants of
any of these nucleic acid molecules. Another preferred isolated protein is
encoded by
a nucleic acid molecule having nucleic acid sequence SEQ ID N0:14, SEQ ID
N0:17,
SEQ ID N0:20, SEQ ID N0:34, SEQ ID N0:37, SEQ ID N0:40, SEQ ID N0:43; or
a protein encoded by an allelic variant of any of these listed nucleic acid
molecule.
Translation of SEQ ID N0:14, the coding strand of nDerf981~sz~ Yields a
protein of about 555 amino acids, denoted herein as PDerf98sss, the amino acid
sequence of which is presented in SEQ ID N0:15, assuming a first in-frame
codon
extending from nucleotide 1 to nucleotide 3 of SEQ 117 N0:14. The
complementary
strand of SEQ ID N0:14 is presented herein as SEQ ID N0:16. The amino acid
sequence of PDerf98sss is encoded by the nucleic acid molecule nDerf981~~s,
having a
coding strand denoted SEQ 117 N0:17 and a complementary strand denoted SEQ ID
N0:19. Analysis of SEQ ID N0:15 suggests the presence of a signal peptide
spanning
from about amino acid 1 through about amino acid 19. The proposed mature
protein,
denoted herein as PDerf98s36, contains about 536 amino acids, the sequence of
which
is represented herein as SEQ ID N0:21, and is encoded by a nucleic acid
molecule
refeiTed to herein as nDerf9816os, represented by SEQ ID N0:20, the coding
strand, and
SEQ ID N0:22, the complementary strand.
Translation of SEQ ID NO:34, the coding strand of nDerp981~z~, Yields a
protein of about 509 amino acids, denoted herein as PDerp98so9, the amino acid
sequence of which is presented in SEQ ID NO:35, assuming a first in-frame
codon
extending from nucleotide 14 to nucleotide 16 of SEQ ID N0:34. The
complementary
strand of SEQ ID N0:34 is presented herein as SEQ ID N0:36. The amino acid
sequence of PDerpf98so9 is encoded by the nucleic acid molecule nDerp981sz~,
having a
coding strand denoted SEQ ID NO:37 and a complementary strand denoted SEQ ID
N0:39. Analysis of SEQ ID N0:35 suggests the presence of a signal peptide
spanning
from about amino acid 1 through about amino acid 19. The proposed mature
protein,
denoted herein as PDerp9849o, contains about 490 amino acids, the sequence of
which
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-17-
is represented herein as SEQ ID N0:41, and is encoded by a nucleic acid
molecule
referred to herein as nDerp9814~0, represented by SEQ ID N0:40, the coding
strand,
and SEQ ID N0:42, the complementary strand.
Translation of SEQ ID N0:43, the coding strand of nDerf60sio, a nucleic acid
molecule encoding a portion of the D. fari~ae 60-kD antigen protein yields a
protein
of about 170 amino acids, denoted herein as PDerf601~o, the amino acid
sequence of
which is presented as SEQ ID N0:44, assuming a first in-frame codon extending
from
nucleotide 1 to nucleotide 3 of SEQ ID N0:43. The complementary sequence to
SEQ
ID NO:43 is presented herein as SEQ ID N0:45.
Preferred Der HMW-map proteins of the present invention include proteins
that are at least about 45%, preferably at least about 50%, more preferably at
least
about 55%, even more preferably at least about 60%, even more preferably at
least
about 65%, even more preferably at least about 70%, even more preferably at
least
about 75%, even more preferably at least about 80%, even more preferably at
least
about 85%, even more preferably at least about 90%, and even more preferably
about
95% identical to PDerf98sss. More preferred is a Der HMW-map protein
comprising
PDerf98sss, PDerf98ssG , PDerp98so9, PDerp98ø9o, and/or PDerf601~o; and
proteins
encoded by allelic variants of nucleic acid molecules encoding proteins
PDerf98sss~
PDerf98s3~ , PDerp98so9, PDerp98ø9o, and/or PDerf601~o.
Other preferred Der HMW-map proteins of the present invention include
proteins having amino acid sequences that are at least about 45%, preferably
at least
about 50%, more preferably at least about 55%, even more preferably at least
about
60%, even more preferably at least about 65%, even more preferably at least
about
70%, even more preferably at least about 75%, even more preferably at least
about
80%, even more preferably at least about 85%, even more preferably at least
about
90%, and even more preferably about 95% identical to amino acid sequence SEQ
ID
NO:1, SEQ ID NO:2, SEQ ID N0:3, SEQ ID N0:4, SEQ ID N0:5, SEQ ID N0:6,
SEQ ID N0:7, SEQ ID N0:8, SEQ ID N0:9, SEQ ID NO:10, SEQ ID NO:11, SEQ
ID N0:12, SEQ ID N0:13, SEQ ID N0:15, SEQ ID N0:18, SEQ ID N0:21, SEQ ID
N0:23, SEQ ID N0:24, SEQ ID N0:29, SEQ ID N0:30, SEQ ID N0:31, SEQ ID
N0:32, SEQ ID N0:33, SEQ ID N0:35, SEQ ID N0:38, SEQ ID N0:41, and/or SEQ
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-18-
ID N0:44. More preferred are Der HMW-map proteins comprising amino acid
sequences SEQ ID NO:l, SEQ ID N0:2, SEQ ID N0:3, SEQ ID N0:4, SEQ ID N0:5,
SEQ ID N0:6, SEQ ID N0:7, SEQ ID N0:8, SEQ ID N0:9, SEQ ID N0:10, SEQ ID
NO:l 1, SEQ ID N0:12, SEQ ID N0:13, SEQ ID N0:15, SEQ ID N0:18, SEQ ID
N0:21, SEQ ID N0:23, SEQ ID N0:24, SEQ ID N0:29, SEQ ID N0:30, SEQ ID
NO:31, SEQ ID N0:32, SEQ ID N0:33, SEQ ID N0:35, SEQ ID N0:38, SEQ ID
N0:41, and/or SEQ ID N0:44; and Der HMW-map proteins encoded by allelic
variants of nucleic acid molecules encoding Der HMW-map proteins having amino
acid sequences SEQ ID N0:1, SEQ ID N0:2, SEQ ID N0:3, SEQ ID N0:4, SEQ ID
N0:5, SEQ ID N0:6, SEQ ID N0:7, SEQ ID N0:8, SEQ ID N0:9, SEQ ID NO:10,
SEQ ID NO:l 1, SEQ ID N0:12, SEQ ID N0:13, SEQ ID N0:15, SEQ ID N0:18,
SEQ ID N0:21, SEQ ID N0:23, SEQ ID N0:24, SEQ ID N0:29, SEQ ID NO:30,
SEQ ID N0:31, SEQ ID N0:32, SEQ 117 N0:33, SEQ ID N0:35, SEQ ll~ N0:38,
SEQ ID N0:41, and/or SEQ ID N0:44.
In one embodiment of the present invention, Der HMW-map proteins comprise
amino acid sequence SEQ ID N0:15, SEQ ID N0:35, and/or SEQ ID N0:44
(including, but not limited to, the proteins consisting of amino acid sequence
SEQ ID
N0:15, SEQ ID N0:35, and/or SEQ ID N0:44, fragments thereof, fusion proteins
and
multivalent proteins), and proteins encoded by allelic variants of nucleic
acid
molecules encoding proteins having amino acid sequence SEQ ID N0:15, SEQ ID
N0:35, and/or SEQ ID N0:44.
In one embodiment, a preferred Der HMW-map protein comprises an amino
acid sequence of at least about 35 amino acids in length, preferably at least
about 50
amino acids in length, more preferably at least about 100 amino acids in
length, mode
preferably at least about 200 amino acids in length, even more preferably at
least about
250 amino acids in length. Within this embodiment, a preferred Der HMW-map
protein of the present invention has an amino acid sequence comprising at
least a
portion of SEQ ID N0:15. In another embodiment, a preferred Der HMW-map
protein comprises a full-length protein, i.e., a protein encoded by a full-
length coding
region.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-19-
Additional preferred Der HMW-map proteins of the present invention include
proteins encoded by nucleic acid molecules comprising at least a portion of
nDerf981~sz, nDerf98166s, nDerf9816o8, nDerp9816zv nDerp981sz~, nDerp981a~o,
and
nDerf60slo, as well as Der HMW-map proteins encoded by allelic variants of
such
nucleic acid molecules.
Also preferred are Der HMW-map proteins encoded by nucleic acid molecules
having nucleic acid sequences comprising at least a portion of SEQ ID N0:14,
SEQ
ID N0:17, SEQ ID N0:20, SEQ ID N0:34, SEQ ID N0:37, SEQ ID N0:40 SEQ ID
N0:43 and/or a nucleic acid sequence encoding a protein comprising the amino
acid
sequence SEQ ID N0:33, as well as allelic variants of these nucleic acid
molecules.
In another embodiment, a preferred Der HMW-map protein of the present
invention is encoded by a nucleic acid molecule comprising at least about 12
nucleotides, preferably at least about 16 nucleotides, more preferably at
least about 18
nucleotides, more preferably at least about 20 nucleotides, more preferably at
least
about 25 nucleotides, more preferably at least about 50 nucleotides, more
preferably at
least about 100 nucleotides, more preferably at least about 350 nucleotides,
more
preferably at least about 450 nucleotides, more preferably at least about 500
nucleotides, and even more preferably at least about 800 nucleotides. Within
this
embodiment is a Der HMW-map protein encoded by at least a portion nDerf981~sz~
nDerp9816zv and/or nDerf60slo or by an allelic variant of these nucleic acid
molecules.
In yet another embodiment, a preferred Der HMW-map protein of the present
invention is encoded by a nucleic acid molecule comprising an apparently full-
length
Der HMW-map coding region, i.e., a nucleic acid molecule encoding an
apparently
full-length Der HMW-map protein.
One embodiment of a Der HMW-map protein of the present invention is a
fusion protein that includes a Der HMW-map protein-containing domain attached
to
one or more fusion segments. Suitable fusion segments for use with the present
invention include, but are not limited to, segments that can: enhance a
protein's
stability; act as an immunopotentiator to enhance an immune response against
aDer
HMW-map protein, reduce an IgE response against a Der HMW-map protein; andlor
assist purification of a Def° HMW-map protein (e.g., by affinity
chromatography). A
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-20-
suitable fusion segment can be a domain of any size that has the desired
function (e.g.,
imparts increased stability, imparts increased immunogenicity to a protein,
reduces an
IgE response, and/or simplifies purification of a protein). Fusion segments
can be
joined to amino and/or carboxyl termini of the Der HMW-map protein-containing
domain of the protein and can be susceptible to cleavage in order to enable
straight-
forward recovery of a Der HMW-map protein. Fusion proteins are preferably
produced by culturing a recombinant cell transformed with a fusion nucleic
acid
molecule that encodes a protein including the fusion segment attached to
either the
carboxyl and/or amino terminal end of a Der HMW-map protein-containing domain.
Preferred fusion segments include a metal binding domain (e.g., a poly-
histidine
segment); an immunoglobulin binding domain (e.g., Protein A; Protein G; T
cell; B
cell; Fc receptor or complement protein antibody-binding domains); a sugar
binding
domain (e.g., a maltose binding domain); a "tag" domain (e.g., at least a
portion of -
galactosidase, a strep tag peptide, other domains that can be purified using
compounds
that bind to the, domain, such as monoclonal antibodies); and/or a linker and
enzyme
domain (e.g., alkaline phosphatase domain connected to a Der HMW-map protein
by a
linker). More preferred fusion segments include metal binding domains, such as
a
poly-histidine segment; a maltose binding domain; a strep tag peptide, such as
that
available from Biometra in Tampa, FL; and a phage T7 S 10 peptide.
In another embodiment, a Der HMW-map protein of the present invention also
includes at least one additional protein segment that is capable of
desensitizing an
animal from one or more allergens. Such a multivalent desensitizing protein
can be
produced by culturing a cell transformed with a nucleic acid molecule
comprising two
or more nucleic acid domains joined together in such a manner that the
resulting
nucleic acid molecule is expressed as a multivalent desensitizing compound
containing
at least two desensitizing compounds capable of desensitizing an animal from
allergens.
Examples of multivalent desensitizing compounds include, but are not limited
to, a Der HMW-map protein of the present invention attached to one or more
compounds that desensitize against allergies caused by one or more allergens,
such as
a plant allergen, an animal allergen, a parasite allergen or an ectoparasite
allergen,
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-21-
including, but not limited to: pant allergens from grass, Meadow Fescue, Curly
Dock,
plantain, Mexican Firebush, Lamb's Quarters, pigweed, ragweed, sage, elm,
cocklebur, Box Elder, walnut, cottonwood, ash, birch, cedar, oak, mulberry,
cockroach, Derznatophagoides, Alterrzaria, Aspergillus, Cladospoz~ium,
Fusariuzn,
Helmizzthosporium, Mucor, Pezzicillium, Pullulaz°ia, Rlzizopus and/or
Tz~icoplzytozz;
parasite allergens from helminths; or ectoparasite allergens from arachnids,
insects and
leeches, including fleas, ticks, flies, mosquitos, sand flies, black flies,
horse flies, horn
flies, deer flies, tsetse flies, stable flies, myiasis-causing flies and
biting gnats, ants,
spiders, lice; mites and true bugs.
The present invention also includes mimetopes of a Der HMW-map protein of
the present invention. As used herein, a mimetope of a Der HMW-map protein of
the
present invention refers to any compound that is able to mimic the activity of
such a
Der HMW-map protein (e.g., ability to bind to induce an immune response
against
Der HMW-map protein), often because the mimetope has a structure that mimics
the
Dez~ HMW-map protein. It is to be noted, however, that the mimetope need not
have a
structure similar to a Der HMW-map protein as long as the mimetope
functionally
mimics the protein. Mimetopes can be, but are not limited to: peptides that
have been
modified to decrease their susceptibility to degradation; anti-idiotypic
and/or catalytic
antibodies, or fragments thereof; non-proteinaceous immunogenic portions of an
isolated protein (e.g., carbohydrate structures); synthetic or natural organic
or
inorganic molecules, including nucleic acids; and/or any other peptidomimetic
compounds. Mimetopes of the present invention can be designed using computer-
generated structures of Der HMW-map protein of the present invention.
Mimetopes
can also be obtained by generating random samples of molecules, such as
oligonucleotides, peptides or other organic molecules, and screening such
samples by
affinity chromatography techniques using the corresponding binding partner,
(e.g., an
anti-Der HMW-map protein antibody). A mimetope can also be obtained by, for
example, rational drug design. In a rational drug design procedure, the three-
dimensional structure of a compound of the present invention can be analyzed
by, for
example, nuclear magnetic resonance (NMR) or x-ray crystallography. The three-
dimensional structure can then be used to predict structures of potential
mimetopes by,
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-22-
for example, computer modeling. The predicted mimetope structures can then be
produced by, for example, chemical synthesis, recombinant DNA technology, or
by
isolating a mimetope from a natural source. Specific examples of Der HMW-map
protein mimetopes include anti-idiotypic antibodies, oligonucleotides produced
using
SelexTM technology, peptides identified by random screening of peptide
libraries and
proteins identified by phage display technology. A preferred mimetope is a
peptidomimetic compound that is structurally and/or functionally similar to a
Der
HMW-map protein of the present invention, particularly to an epitope of Der
HMW-
map protein that induces an immune response.
The present invention also includes muteins of a Der~ HMW-map protein of the
present invention. As used herein, a mutein refers to a particular homolog of
a Der
HMW-map protein in which desired amino acid residues have been substituted or
removed. Preferred muteins of the present invention include Der HMW-map
protein
homologs in which amino acid residues have been changed to reduce an
anaphylactic
reaction by an animal when the mutein is administered to the animal in then
apeutic
doses. More preferred muteins of the present invention include Der HMW-map
protein homologs in which one or more cysteine residues of a Der HMW-map
protein
have been replaced or removed. Methods to produce muteins are known to those
of
skill in the art and are disclosed herein. Preferably, a mutein is produced
using
recombinant techniques.
Another embodiment of the present invention is an isolated nucleic acid
molecule comprising a Der HMW-map nucleic acid molecule. The identifying
characteristics of such nucleic acid molecules are heretofore described. A
nucleic acid
molecule of the present invention can include an isolated natural Der HMW-map
gene
or a homolog thereof, the latter of which is described in more detail below. A
nucleic
acid molecule of the present invention can include one or more regulatory
regions,
full-length or partial coding regions, or combinations thereof. The minimal
size of a
nucleic acid molecule of the present invention is a size sufficient to allow
the
formation of a stable hybrid (i.e., hybridization under stringent
hybridization
conditions) with the complementary sequence of another nucleic acid molecule.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-23-
In accordance with the present invention, an isolated nucleic acid molecule is
a
nucleic acid molecule that has been removed from its natural milieu (i.e.,
that has been
subjected to human manipulation) and can include DNA, RNA, or derivatives of
either
DNA or RNA. As such, "isolated" does not reflect the extent to which the
nucleic acid
molecule has been purified. An isolated Der HMW-map nucleic acid molecule of
the
present invention, or a homolog thereof, can be isolated from its natural
source or
produced using recombinant DNA technology (e.g., polymerase chain reaction
(PCR)
amplification or cloning) or chemical synthesis. Isolated Der HMW-map nucleic
acid
molecules, and homologs thereof, can include, for example, natural allelic
variants and
nucleic acid molecules modified by nucleotide insertions, deletions,
substitutions,
and/or inversions in a manner such that the modifications do not substantially
interfere
with the nucleic acid molecule's ability to encode aDer HMW-map protein of the
present invention.
A Der HMW-map nucleic acid molecule homolog can be produced using a
number of methods known to those skilled in the art, see, for example,
Sambrook et
al., 1989, Molecular CloT2ing: A Laboratory Manual, Cold Spring Harbor Labs
Press;
Sambrook et al., ibid. For example, nucleic acid molecules can be modified
using a
variety of techniques including, but not limited to, classic mutagenesis and
recombinant DNA techniques such as site-directed mutagenesis, chemical
treatment,
restriction enzyme cleavage, ligation of nucleic acid fragments, PCR
amplification,
synthesis of oligonucleotide mixtures and ligation of mixture groups to
"build" a
mixture of nucleic acid molecules, and combinations thereof. Nucleic acid
molecule
homologs can be selected by hybridization with a Der HMW-map nucleic acid
molecule or by screening the function of a protein encoded by the nucleic acid
molecule (e.g., ability to elicit an immune response against at least one
epitope of a
Der~ HMW-map protein or to effect Der HMW-map activity).
Allelic variants typically encode proteins having similar activity to that of
the
protein encoded by the gene to which they are being compared. Allelic variants
can
also comprise alterations in the 5' or 3' untranslated regions of the gene
(e.g., in
regulatory control regions). Allelic variants are well known to those skilled
in the art
and would be expected to be found within a given dust mite since the genome is
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-24-
diploid and/or among a group of two or more dust mites. The present invention
also
includes variants due to laboratory manipulation, such as, but not limited to,
variants
produced during polymerase chain reaction amplification.
An isolated nucleic acid molecule of the present invention can include a
nucleic acid sequence that encodes at least one Def- HMW-map protein of the
present
invention, examples of such proteins being disclosed herein. Although the
phrase
"nucleic acid molecule" primarily refers to the physical nucleic acid molecule
and the
phrase "nucleic acid sequence" primarily refers to the sequence of nucleotides
on the
nucleic acid molecule, the two phrases can be used interchangeably, especially
with
respect to a nucleic acid molecule, or a nucleic acid sequence, being capable
of
encoding a Def° HMW-map protein.
A preferred nucleic acid molecule of the present invention, when administered
to an animal, is capable of desensitizing that animal from allergic reactions
caused by
a Der HMW-map allergen. As will be disclosed in more detail below, such a
nucleic
acid molecule can be, or encode, an antisense RNA, a molecule capable of
triple helix
formation, a ribozyme, or other nucleic acid-based drug compound. In
additional
embodiments, a nucleic acid molecule of the present invention can encode a
desensitizing protein (e.g., a Def~ HMW-map protein of the present invention),
the
nucleic acid molecule being delivered to the animal, for example, by direct
injection
(i.e, as a DNA reagent) or in a vehicle such as a recombinant virus reagent or
a
recombinant cell reagent.
One embodiment of the present invention is an isolated nucleic acid molecule
that hybridizes under stringent hybridization conditions with a Der HMW-map
gene.
Stringent hybridization conditions refer to standard hybridization conditions
described
herein. A preferred nucleic acid molecule of the present invention includes an
isolated
nucleic acid molecule that hybridizes under stringent hybridization conditions
with a
gene encoding a protein comprising an amino acid sequence including SEQ ID
NO:1,
SEQ D7 N0:2, SEQ ID N0:3, SEQ ID N0:4, SEQ ID N0:5, SEQ ID N0:6, SEQ ID
N0:7, SEQ ID N0:8, SEQ ID N0:9, SEQ ID NO:10, SEQ ID NO:11, SEQ ID N0:12,
SEQ ID N0:13, SEQ ID N0:15, SEQ ID N0:18, SEQ ID N0:21, SEQ ID N0:23,
SEQ ID N0:24, SEQ ID N0:29, SEQ ID N0:30, SEQ ID N0:31, SEQ ID N0:32,
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-25-
SEQ ID N0:33, SEQ ID N0:35, SEQ ID N0:38, SEQ ID N0:41, and/or SEQ ID
N0:44. A more preferred nucleic acid molecule of the present invention
includes an
isolated nucleic acid molecule that hybridizes under stringent hybridization
conditions
with the complement of a nucleic acid sequence that encodes a protein
comprising an
amino acid sequence including SEQ ID NO:1, SEQ ID N0:2, SEQ ID NO:3, SEQ ID
N0:4, SEQ ID N0:5, SEQ ID N0:6, SEQ ID N0:7, SEQ ID N0:8, SEQ ID N0:9,
SEQ ID N0:10, SEQ m NO:11, SEQ ID N0:12, SEQ ll~ N0:13, SEQ ID N0:15,
SEQ ID N0:18, SEQ ID N0:21, SEQ ID N0:23, SEQ ID N0:24, SEQ ID N0:29,
SEQ ID N0:30, SEQ ID N0:31, SEQ ID N0:32, SEQ ID N0:33, SEQ ID N0:35,
SEQ ID N0:38, SEQ ID N0:41, and/or SEQ 117 N0:44.
A more preferred nucleic acid molecule of the present invention includes an
isolated nucleic acid molecule selected from the group consisting of: a
nucleic acid
molecule comprising at least about 150 nucleotides, wherein said nucleic acid
molecule comprising at least about 150 nucleotides hybridizes, in a solution
comprising 1X SSC and 0% formamide, at a temperature of about 50°C, to
a nucleic
acid sequence selected from the group consisting of SEQ ID N0:14, SEQ ID
N0:16,
SEQ ID N0:17, SEQ ID N0:19, SEQ ID N0:20, SEQ ID N0:22, SEQ m N0:34,
SEQ ID N0:36, SEQ ID N0:37, SEQ ID N0:39, SEQ ID N0:40, SEQ ID N0:42,
SEQ ID N0:43, SEQ ID N0:45 and/or a nucleic acid sequence encoding a protein
comprising the amino acid sequence SEQ ID N0:33 and a complement thereof.
The present invention also includes fragments of any nucleic acid molecule
disclosed herein. According to the present invention, a fragment can include
any
nucleic acid molecule or nucleic acid sequence, the size of which can range
between a
length that is smaller than a sequence identified by a SEQ ID NO of the
present
invention and the minimum size of an oligonucleotide as defined herein. For
example,
the size of a fragment of the present invention can be any size that is less
than about
1752 nucleotides and greater than 11 nucleotides in length.
In one embodiment of the present invention, a preferred DeY HMW-map
nucleic acid molecule includes an isolated nucleic acid molecule which is at
least
about 50 nucleotides, or at least about 150 nucleotides, and which hybridizes
under
conditions which preferably allow about 40% or less base pair mismatch, more
preferably under conditions which allow about 35% or less base pair mismatch,
more
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-26-
preferably under conditions which allow about 30% or less base pair mismatch,
more
preferably under conditions which allow about 25% or less base pair mismatch,
more
preferably under conditions which allow about 20% or less base pair mismatch,
more
preferably under conditions which allow about 15% or less base pair mismatch,
more
preferably under conditions which allow about 10% or less base pair mismatch
and
even more preferably under conditions which allow about 5% or less base pair
mismatch with a nucleic acid molecule selected from the group consisting of
SEQ ID
N0:14, SEQ ID N0:16, SEQ D7 N0:17, SEQ ID N0:19, SEQ ID N0:20, SEQ ID
N0:22, SEQ ID N0:34, SEQ ID N0:36, SEQ ID N0:37, SEQ ID N0:40, SEQ ID
N0:42, SEQ ID N0:43, SEQ ID N0:45, and a nucleic acid sequence encoding a
protein comprising the amino acid sequence SEQ ID N0:33 and a complement
thereof.
Another embodiment of the present invention includes a nucleic acid molecule
comprising at least about 150 base-pairs, wherein the nucleic acid molecule
hybridizes, in a solution comprising 1X SSC and 0% formamide, at a temperature
of
about 50°C, to a nucleic acid sequence selected from the group
consisting of SEQ ID
N0:14, SEQ m N0:16, SEQ ID N0:17, SEQ ID N0:19, SEQ ID N0:20, SEQ ID
N0:22, SEQ m N0:34, SEQ ID N0:36, SEQ m N0:37, SEQ ID N0:39, SEQ ID
N0:40, SEQ ID N0:42, SEQ ID N0:43, SEQ ID N0:45, and/or a nucleic acid
sequence encoding a protein comprising the amino acid sequence SEQ ID N0:33
and
a complement thereof. Additional preferred nucleic acid molecules of the
present
invention include fragments of an isolated nucleic acid molecule comprising at
least
about 150 base-pairs, wherein said nucleic acid molecule hybridizes, in a
solution
comprising 1X SSC and 0% formamide, at a temperature of about 50°C, to
a nucleic
acid sequence selected from the group consisting of SEQ ID N0:14, SEQ ID
N0:16,
SEQ m N0:17, SEQ ID N0:19, SEQ ID N0:20, SEQ ID NO:22, SEQ ID N0:34,
SEQ m N0:36, SEQ ID N0:37, SEQ ID N0:39, SEQ ID NO:40, SEQ ID N0:42,
SEQ ID N0:43, SEQ ID N0:45 and a nucleic acid sequence encoding a protein
comprising the amino acid sequence SEQ ID N0:33 and complement thereof.
Additional preferred Der HMW-map nucleic acid molecules of the present
invention include an isolated nucleic acid molecule which is at least about 50
nucleotides, or at least about 150 nucleotides, comprising a nucleic acid
sequence that
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
_27_
is preferably at least about 60% identical, more preferably at least about 65%
identical,
more preferably at least about 70% identical, more preferably at least about
75%
identical, more preferably at least about 80% identical, more preferably at
least about
85% identical, more preferably at least about 90% identical and even more
preferably
at least about 95% identical to a nucleic acid sequence selected from the
group
consisting of SEQ ID N0:14, SEQ ID N0:16, SEQ ID N0:17, SEQ ID N0:19, SEQ
ID N0:20, SEQ ID N0:22, SEQ ID N0:34, SEQ ID N0:36, SEQ m N0:37, SEQ ID
N0:39, SEQ ID N0:40, SEQ ID N0:42, SEQ ID N0:43, SEQ ID N0:45, and a
nucleic acid sequence encoding a protein comprising the amino acid sequence
SEQ ID
N0:33 and a complement thereof. Also preferred are fragments of any of such
nucleic
acid molecules. Percent identity may be determined using the Compare function
by
maximum matching within the program DNAsis Version 2.1 using default
parameters.
One embodiment of the present invention is a nucleic acid molecule
comprising all or part of nucleic acid molecules nDerf981~s2, nDerf981~6s and
nDerf9816o8, nDerp9816av nDerp981sz~, nDerp981~~0, and/or nDerf60slo, or
allelic
variants of these nucleic acid molecules. Another preferred nucleic acid
molecule of
the present invention includes at least a portion of nucleic acid sequence SEQ
ID
N0:14, SEQ ID N0:16, SEQ ID N0:17, SEQ ID N0:19, SEQ ID N0:20, SEQ ID
N0:22, SEQ ID N0:34, SEQ ID N0:36, SEQ ID N0:37, SEQ ID N0:39, SEQ ID
N0:40, SEQ ID N0:42, SEQ ID N0:43, SEQ ID N0:45 and/or a nucleic acid
sequence encoding a protein comprising the amino acid sequence SEQ ID N0:33,
as
well as allelic variants of nucleic acid molecules having these nucleic acid
sequences
and homologs of nucleic acid molecules having these nucleic acid sequences;
preferably such a homolog encodes or is complementary to a nucleic acid
molecule
that encodes at least one epitope that elicits and an immune response against
a protein
having an amino acid sequence SEQ ID NO:1, SEQ ID N0:2, SEQ ID N0:3, SEQ ID
N0:4, SEQ ID N0:5, SEQ ID N0:6, SEQ ID N0:7, SEQ ID N0:8, SEQ ID N0:9,
SEQ ID NO:10, SEQ ID NO:1 l, SEQ ID N0:12, SEQ ID N0:13, SEQ >D N0:15,
SEQ ID N0:18 , SEQ ID N0:21, SEQ ID N0:23, SEQ ID N0:24, SEQ ID N0:29,
SEQ ID N0:30, SEQ ID N0:31, SEQ ID N0:32, SEQ ID N0:33, SEQ >D N0:35,
SEQ ID N0:38, SEQ ID N0:41, SEQ >D N0:41, and/or SEQ m N0:44. Such nucleic
acid molecules can include nucleotides in addition to those included in the
SEQ ID
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-28-
NOs, such as, but not limited to, a full-length gene, a full-length coding
region, a
nucleic acid molecule encoding a fusion protein, or a nucleic acid molecule
encoding a
multivalent protective compound.
In one embodiment, a Der HMW-map nucleic acid molecule of the present
invention encodes a protein that is at least about 45%, preferably at least
about 50%,
more preferably at least about 55%, even more preferably at least about 60%,
even
more preferably at least about 65%, even more preferably at least about 70%,
even
more preferably at least about 75%, even more preferably at least about 80%,
even
more preferably at least about 85%, even more preferably at least about 90%,
and even
more preferably about 95% identical to PDerf98sss, PDerp98so9, and/or
PDerf601~o.
Even more preferred is a nucleic acid molecule encoding PDerf98sss,
PDerf98s3~,
PDerp98so9, PDerp9849o, and/or PDerf601~o, and/or an allelic variant of such
nucleic
acid molecules.
In another embodiment, a Der HMW-map nucleic acid molecule of the present
invention encodes a protein having an amino acid sequence that is at least
about 45%,
preferably at least about 50%, more preferably at least about 55%, even more
preferably at least about 60%, even more preferably at least about 65%, even
more
preferably at least about 70%, even more preferably at least about 75%, even
more
preferably at least about 80%, even more preferably at least about 85%, even
more
preferably at least about 90%, and even more preferably about 95% identical to
SEQ
ID NO:1, SEQ ID N0:2, SEQ ID N0:3, SEQ ID N0:4, SEQ ID N0:5, SEQ ID N0:6,
SEQ ID N0:7, SEQ ID N0:8, SEQ ID N0:9, SEQ ID NO:10, SEQ ID NO:l l, SEQ
ID N0:12, SEQ ID N0:13, SEQ ID N0:15, SEQ ID N0:18 , SEQ ID N0:21, SEQ ID
N0:23, SEQ 117 N0:24, SEQ ID N0:29, SEQ ID N0:30, SEQ ID N0:31, SEQ ID
N0:32, SEQ ID N0:33, SEQ ID N0:35, SEQ ID N0:38, SEQ ID N0:41, SEQ TD
N0:41, and/or SEQ ID NO:44. The present invention also includes a Den HMW-map
nucleic acid molecule encoding a protein having at least a portion of SEQ ID
NO:1,
SEQ ID N0:2, SEQ ID N0:3, SEQ ID N0:4, SEQ ID N0:5, SEQ ll~ N0:6, SEQ ID
N0:7, SEQ ID N0:8, SEQ ID N0:9, SEQ DJ N0:10, SEQ ID NO:1 l, SEQ ID N0:12,
SEQ ID N0:13, SEQ ID N0:15, SEQ ID N0:18 , SEQ ID NO:21, SEQ ID N0:23,
SEQ D7 N0:24, SEQ m N0:29, SEQ ID N0:30, SEQ ID N0:31, SEQ ID N0:32,
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-29-
SEQ 117 N0:33, SEQ 117 N0:35, SEQ ID N0:38, SEQ ID N0:41, SEQ ID N0:41,
and/or SEQ ID N0:44, as well as allelic variants of a Der HMW-map nucleic acid
molecule encoding a protein having these sequences, including nucleic acid
molecules
that have been modified to accommodate codon usage properties of the cells in
which
such nucleic acid molecules are to be expressed.
In another embodiment, a preferred Def~ HMW-map nucleic acid molecule
encodes a Der HMW-map protein comprising at least about at least about 35
amino
acids in length, preferably at least about 50 amino acids in length, more
preferably at
least about 100 amino acids in length, more preferably at least about 200
amino acids
in length, even more preferably at least about 250 amino acids in length.
Knowing the nucleic acid sequences of certain Der- HMW-map nucleic acid
molecules of the present invention allows one skilled in the art to, for
example, (a)
make copies of those nucleic acid molecules, (b) obtain nucleic acid molecules
including at least a portion of such nucleic acid molecules (e.g., nucleic
acid molecules
including full-length genes, full-length coding regions, regulatory control
sequences,
truncated coding regions), and (c) obtain other Der HMW-map nucleic acid
molecules.
Such nucleic acid molecules can be obtained in a variety of ways including
screening
appropriate expression libraries with antibodies of the present invention;
traditional
cloning techniques using oligonucleotide probes of the present invention to
screen
appropriate libraries; and PCR amplification of appropriate libraries or DNA
using
oligonucleotide primers of the present invention. A preferred library to
screen or from
Which to amplify nucleic acid molecules includes a Dermatophagoides farihae
and/or
Dermatophagoides ptero~yssius library, such as the libraries disclosed herein
in the
Examples. Techniques to clone and amplify genes are disclosed, for example, in
Sambrook et al., ibid.
The present invention also includes nucleic acid molecules that are
oligonucleotides capable of hybridizing, under stringent hybridization
conditions, with
complementary regions of other, preferably longer, nucleic acid molecules of
the
present invention such as those comprising Der HMW-map nucleic acid molecules
or
other Der HMW-map nucleic acid molecules. Oligonucleotides of the present
invention can be RNA, DNA, or derivatives of either. The minimum size of such
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-30-
oligonucleotides is the size required for formation of a stable hybrid between
an
oligonucleotide and a complementary sequence on a nucleic acid molecule of the
present invention. A preferred oligonucleotide of the present invention has a
maximum
size of preferably about 200 nucleotides, more preferably about 150
nucleotides and
even more preferably about 100 nucleotides. The present invention includes
oligonucleotides that can be used as, for example, probes to identify nucleic
acid
molecules.
One embodiment of the present invention includes a recombinant vector, which
includes at least one isolated nucleic acid molecule of the present invention,
inserted
into any vector capable of delivering the nucleic acid molecule into a host
cell. Such a
vector contains heterologous nucleic acid sequences, that is nucleic acid
sequences that
are not naturally found adjacent to nucleic acid molecules of the present
invention and
that preferably are derived from a species other than the species from which
the
nucleic acid molecules) are derived. The vector can be either RNA or DNA,
either
prokaryotic or eukaryotic, and typically is a virus or a plasmid. Recombinant
vectors
can be used in the cloning, sequencing, and/or otherwise manipulation of Den
HMW-
map nucleic acid molecules of the present invention.
One type of recombinant vector, referred to herein as a recombinant molecule,
comprises a nucleic acid molecule of the present invention operatively linked
to an
expression vector. The phrase operatively linked refers to insertion of a
nucleic acid
molecule into an expression vector in a manner such that the molecule is able
to be
expressed when transformed into a host cell. As used herein, an expression
vector is a
DNA or RNA vector that is capable of transforming a host cell and of effecting
expression of a specified nucleic acid molecule. Preferably, the expression
vector is
also capable of replicating within the host cell. Expression vectors can be
either
prokaryotic or eukaryotic, and are typically viruses or plasmids. Expression
vectors of
the present invention include any vectors that function (i.e., direct gene
expression) in
recombinant cells of the present invention, including in bacterial, fungal,
endoparasite,
insect, other animal, and plant cells. Preferred expression vectors of the
present
invention can direct gene expression in bacterial, yeast, insect and mammalian
cells
and more preferably in the cell types disclosed herein.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-31-
In particular, expression vectors of the present invention contain regulatory
sequences such as transcription control sequences, translation control
sequences,
origins of replication, and other regulatory sequences that are compatible
with the
recombinant cell and that control the expression of nucleic acid molecules of
the
present invention. In particular, recombinant molecules of the present
invention
include transcription control sequences. Transcription control sequences are
sequences which control the initiation, elongation, and termination of
transcription.
Particularly important transcription control sequences are those which control
transcription initiation, such as promoter, enhancer, operator and repressor
sequences.
Suitable transcription control sequences include any transcription control
sequence
that can function in at least one of the recombinant cells of the present
invention. A
variety of such transcription control sequences are known to those skilled in
the art.
Preferred transcription control sequences include those which function in
bacterial,
yeast, insect and mammalian cells, such as, but not limited to, tac, lac, try,
trc, oxy-
pro, omp/lpp, rrnB, bacteriophage lambda (such as lambda pL and lambda pR and
fusions that include such promoters), bacteriophage T7, T7la.c, bacteriophage
T3,
bacteriophage SP6, bacteriophage SPO1, metallothionein, alpha-mating factor,
Pichia
alcohol oxidase, alphavirus subgenomic promoters (such as Sindbis virus
subgenomic
promoters), antibiotic resistance gene, baculovirus, Heliothis zea insect
virus, vaccinia
virus, herpesvirus, raccoon poxvirus, other poxvirus, adenovirus,
cytomegalovirus
(such as intermediate early promoters), simian virus 40, retrovirus, actin,
retroviral
long terminal repeat, Rous sarcoma virus, heat shock, phosphate and nitrate
transcription control sequences as well as other sequences capable of
controlling gene
expression in prokaryotic or eukaryotic cells. Additional suitable
transcription control
sequences include tissue-specific promoters and enhancers as well as
lymphokine-
inducible promoters (e.g., promoters inducible by interferons or
interleukins).
Transcription control sequences of the present invention can also include
naturally
occurring transcription control sequences naturally associated with canines or
felines.
Suitable and preferred nucleic acid molecules to include in recombinant
vectors
of the present invention are as disclosed herein. Preferred nucleic acid
molecules to
include in recombinant vectors, and particularly in recombinant molecules,
include
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-32-
nDerf981~sz, nDerf981sss nDerf9816os, nDerp98,~zi, nDerp981sz.,, nDerp981a~o,
and
nDerf60s io.
Recombinant molecules of the present invention may also (a) contain secretory
signals (i.e., signal segment nucleic acid sequences) to enable an expressed
Der
HMW-map protein of the present invention to be secreted from the cell that
produces
the protein and/or (b) contain fusion sequences which lead to the expression
of nucleic
acid molecules of the present invention as fusion proteins. Examples 'of
suitable signal
segments include any signal segment capable of directing the secretion of a
protein of
the present invention. Preferred signal segments include, but are not limited
to, tissue
plasminogen activator (t-PA), interferon, interleukin, growth hormone,
histocompatibility and viral envelope glycoprotein signal segments, as well as
natural
signal segments. Suitable fusion segments encoded by fusion segment nucleic
acids
are disclosed herein. In addition, a nucleic acid molecule of the present
invention can
be joined to a fusion segment that directs the encoded protein to the
proteosome, such
as a ubiquitin fusion segment. Recombinant molecules may also include
intervening
and/or untranslated sequences surrounding and/or within the nucleic acid
sequences of
nucleic acid molecules of the present invention.
Another embodiment of the present invention includes a recombinant cell
comprising a host cell transformed with one or more recombinant molecules of
the
present invention. Transformation of a nucleic acid molecule into a cell can
be
accomplished by any method by which a nucleic acid molecule can be inserted
into the
cell. Transformation techniques include, but are not limited to, transfection,
electroporation, microinjection, lipofection, adsorption, and protoplast
fusion. A
recombinant cell may remain unicellular or may grow into a tissue, organ or a
multicellular organism. Transformed nucleic acid molecules of the present
invention
can remain extrachromosomal or can integrate into one or more sites within a
chromosome of the transformed (i.e., recombinant) cell in such a manner that
their
ability to be expressed is retained. Preferred nucleic acid molecules with
which to
transform a cell include Der HMW-map nucleic acid molecules disclosed herein.
Particularly.preferred nucleic acid molecules with which to transform a cell
include
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-33-
nDerf981~sz, nDerf98166s nDerf9816os, nDerp9816zn nDerp981s2~, nDerp981~~0,
and
nDerf60s lo.
Suitable host cells to transform include any cell that can be transformed with
a
nucleic acid molecule of the present invention. Host cells can be either
untransformed
cells or cells that are already transformed with at least one nucleic acid
molecule (e.g.,
nucleic acid molecules encoding one or more proteins of the present invention
and/or
other proteins useful in the production of multivalent vaccines). Host cells
of the
present invention either can be endogenously (i.e., naturally) capable of
producing Def
HMW-map proteins of the present invention or can be capable of producing such
proteins after being transformed with at least one nucleic acid molecule of
the present
invention. Host cells of the present invention can be any cell capable of
producing at
least one protein of the present invention, and include bacterial, fungal
(including
yeast), other insect, other animal and plant cells. Preferred host cells
include bacterial,
mycobacterial, yeast, parasite, insect and mammalian cells. More preferred
host cells
include Sal~io~zella, Escherichia, Bacillus, Listeria, Saccl2as~o~zyces,
Spodoptera,
Mycobacteria, Trichoplusia, BHK (baby hamster kidney) cells, MDCK cells
(normal
dog kidney cell line for canine herpesvirus cultivation), CRFK cells (normal
cat
kidney cell line for feline herpesvirus cultivation), CV-1 cells (African
monkey kidney
cell line used, for example, to culture raccoon poxvirus), COS (e.g., COS-7)
cells, and
Vero cells. Particularly preferred host cells are Escherichia coli, including
E. coli K-
12 derivatives; Salmonella typl2i; Sa.lf~2o~zella typhimuf°ium,
including attenuated strains
such as UK-1 X3987 and SR-11 x4072; Spodoptera frugiperda; Ti~ichoplusia ni;
BHK
cells; MDCK cells; CRFK cells; CV-1 cells; COS cells; Vero cells; and non-
tumorigenic mouse myoblast G8 cells (e.g., ATCC CRL 1246). Additional
appropriate mammalian cell hosts include other kidney cell lines, other
fibroblast cell
lines (e.g., human, murine or chicken embryo fibroblast cell lines), myeloma
cell lines,
Chinese hamster ovary cells, mouse NIH/3T3 cells, LMTK31 cells and/or HeLa
cells.
A recombinant cell is preferably produced by transforming a host cell with one
or more recombinant molecules, each comprising one or more nucleic acid
molecules
of the present invention operatively linked to an expression vector containing
one or
more transcription control sequences. The phrase operatively linked refers to
insertion
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-34-
of a nucleic acid molecule into an expression vector in a manner such that the
molecule is able to be expressed when transformed into a host cell.
A recombinant molecule of the present invention is a molecule that can include
at least one of any nucleic acid molecule heretofore described operatively
linked to at
least one of any transcription control sequence capable of effectively
regulating
expression of the nucleic acid molecules) in the cell to be transformed,
examples of
which are disclosed herein.
A recombinant cell of the present invention includes any cell transformed with
at least one of any Der HMW-map nucleic acid molecule of the present
invention.
Suitable and preferred Der HMW-map nucleic acid molecules as well as suitable
and
preferred recombinant molecules with which to transform cells are disclosed
herein.
Recombinant DNA technologies can be used to improve expression of
transformed nucleic acid molecules by manipulating, for example, the number of
copies of the nucleic acid molecules within a host cell, the efficiency with
which those
nucleic acid molecules are transcribed, the efficiency with which the
resultant
transcripts are translated, and the efficiency of post-translational
modifications.
Recombinant techniques useful for increasing the expression of nucleic acid
molecules
of the present invention include, but are not limited to, operatively linking
nucleic acid
molecules to high-copy number plasmids, integration of the nucleic acid
molecules
into one or more host cell chromosomes, addition of vector stability sequences
to
plasmids, substitutions or modifications of transcription control signals
(e.g.,
promoters, operators, enhancers), substitutions or modifications of
translational control
signals (e.g., ribosome binding sites, Shine-Dalgarno sequences), modification
of
nucleic acid molecules of the present invention to correspond to the codon
usage of the
host cell, deletion of sequences that destabilize transcripts, and use of
control signals
that temporally separate recombinant cell growth from recombinant enzyme
production during fermentation. The activity of an expressed recombinant
protein of
the present invention may be improved by fragmenting, modifying, or
derivatizing
nucleic acid molecules encoding such a protein.
Isolated Der HMW-map proteins of the present invention can be produced in a
variety of ways, including production and recovery of natural proteins,
production and
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-35-
recovery of recombinant proteins, and chemical synthesis of the proteins. In
one
embodiment, an isolated protein of the present invention is produced by
culturing a
cell capable of expressing the protein under conditions effective to produce
the protein,
and recovering the protein. A preferred cell to culture is a recombinant cell
of the
present invention. Effective culture conditions include, but are not limited
to, effective
media, bioreactor, temperature, pH and oxygen conditions that permit protein
production. An effective medium refers to any medium in which a cell is
cultured to
produce a Der HMW-map protein of the present invention. Such a medium
typically
comprises an aqueous medium having assimilable carbon, nitrogen and phosphate
sources, and appropriate salts, minerals, metals and other nutrients, such as
vitamins.
Cells of the present invention can be cultured in conventional fermentation
bioreactors,
shake flasks, test tubes, microtiter dishes, and petri plates. Culturing can
be carried out
at a temperature, pH and oxygen content appropriate for a recombinant cell.
Such
culturing conditions are within the expertise of one of ordinary skill in the
art.
Depending on the vector and host system used for production, resultant
proteins of the present invention may either remain within the recombinant
cell; be
secreted into the fermentation medium; be secreted into a space between two
cellular
membranes, such as the periplasmic space in E. coli; or be retained on the
outer
surface of a cell or viral membrane. The phrase "recovering the protein", as
well as
similar phrases, refers to collecting the whole fermentation medium containing
the
protein and need not imply additional steps of separation or purification.
Proteins of
the present invention can be purified using a variety of standard protein
purification
techniques, such as, but not limited to, affinity chromatogr aphy, ion
exchange
chromatography, filtration, electrophoresis, hydrophobic interaction
chromatography,
gel filtration chromatography, reverse phase chromatography, concanavalin A
chromatography, chromatofocusing and differential solubilization. Proteins of
the
present invention are preferably retrieved in "substantially pure" form. As
used herein,
"substantially pure" refers to a purity that allows for the effective use of
the protein as
a therapeutic composition or diagnostic. A therapeutic composition for
animals, for
example, should exhibit no substantial toxicity and preferably should be
capable of
desensitizing a treated animal.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-3 6-
The present invention also includes isolated (i.e., removed from their natural
milieu) antibodies that selectively bind to a Des HMW-map protein of the
present
invention or a mimetope thereof (i.e., anti-Der HMW-map protein antibodies).
As
used herein, the term "selectively binds to" a Der HMW-map protein refers to
the
ability of antibodies of the present invention to preferentially bind to
specified proteins
and mimetopes thereof of the present invention. Binding can be measured using
a
variety of methods standard in the art including enzyme immunoassays (e.g.,
ELISA),
immunoblot assays, etc.; see, for example, Sambrook et al., ibid. An anti-Den
HMW-
map protein antibody preferably selectively binds to a portion of a Der HMW-
map
protein that induces an immune response in an animal.
Isolated antibodies of the present invention can include antibodies in a
bodily
fluid (such as, but not limited to, serum), or antibodies that have been
purified to
varying degrees. Antibodies of the present invention can be polyclonal or
monoclonal.
Functional equivalents of such antibodies, such as antibody fragments and
genetically-
engineered antibodies (including single chain antibodies or chimeric
antibodies that
can bind to more than one epitope) are also included in the present invention.
A preferred method to produce antibodies of the present invention includes
(a) administering to an animal an effective amount of a protein, peptide or
mimetope
thereof of the present invention to produce the antibodies and (b) recovering
the
antibodies. In another method, antibodies of the present invention are
produced
recombinantly using techniques as heretofore disclosed to produce Der HMW-map
proteins of the present invention. Antibodies raised against defined proteins
or
mimetopes can be advantageous because such antibodies are not substantially
contaminated with antibodies against other substances that might otherwise
cause
interference in a diagnostic assay or side effects if used in a therapeutic
composition.
Antibodies of the present invention have a variety of potential uses that are
within the scope of the present invention. For example, such antibodies can be
used
(a) as tools to detect mite allergen, in particular Der HMW-map protein; (b)
as tools to
screen expression libraries; and/or (c) to recover desired proteins of the
present
invention from a mixture of proteins and other contaminants. Antibodies of the
present invention can also be used, for example, to inhibit binding of Der HMW-
map
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-37-
protein to IgE that binds specifically to Der HMW-map protein, to prevent
immunocomplex formation, thereby reducing hypersensitivity responses to mite
allergens.
A Der HMW-map protein of the present invention can be included in a
chimeric molecule comprising at least a portion of a Der HMW-map protein that
induces an immune response in an animal and a second molecule that enables the
chimeric molecule to be bound to a substrate in such a manner that the Der HMW-
map
protein portion can bind to IgE in essentially the same manner as a Der HMW-
map
protein that is not bound to a substrate. An example of a suitable second
molecule
includes a portion of an immunoglobulin molecule or another ligand that has a
suitable
binding partner that can be immobilized on a substrate, e.g., biotin and
avidin, or a
metal-binding protein and a metal (e.g., His), or a sugar-binding protein and
a sugar
(e.g., maltose).
A Der HMW-map protein of the present invention can be contained in a
formulation, herein referred to as a Der HMW-map protein formulation. For
example,
a Der HMW-map protein can be combined with a buffer in which the Der HMW-map
protein is solubilized, and/or with a carrier. Suitable buffers and carriers
are known to
those skilled in the art. Examples of suitable buffers include any buffer in
which a Der
HMW-map protein can function to selectively bind to an antibody that
specifically
binds to Der HMW-map protein, such as, but not limited to, phosphate buffered
saline,
water, saline, phosphate buffer, bicarbonate buffer, HEPES buffer (N-2-
hydroxyethylpiperazine-N'-2-ethanesulfonic acid buffered saline), TES buffer
(Tris-
EDTA buffered saline), Tris buffer and TAE buffer (Tris-acetate-EDTA).
Examples
of carriers include, but are not limited to, polymeric matrices, toxoids, and
serum
albumins, such as bovine serum albumin. Carriers can be mixed with Der HMW-map
protein or conjugated (i.e., attached) to Der HMW-map protein in such a manner
as to
not substantially interfere with the ability of the Der HMW-map protein to
selectively
bind to an antibody that specifically binds to Der HMW-map protein.
A Der HMW-map protein of the present invention can be produced by a cell
comprising the Der HMW-map protein. A preferred Der HMW-map protein-bearing
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-38-
cell includes a recombinant cell comprising a nucleic acid molecule encoding a
DeY
HMW-map protein of the present invention.
In addition, a Der HMW-map protein formulation of the present invention can
include not only a Der HMW-map protein but also one or more additional
antigens or
antibodies useful in desensitizing an animal against allergy, or preventing or
treating
mite allergen pathogenesis. As used herein, an antigen refers to any molecule
capable
of being selectively bound by an antibody. As used herein, an allergen refers
to any
.antigen that is capable of stimulating production of antibodies involved in
an allergic
response in an animal. As used herein, selective binding of a first molecule
to a
second molecule refers to the ability of the first molecule to preferentially
bind (e.g.,
having higher affinity higher avidity) to the second molecule when compared to
the
ability of a first molecule to bind to a third molecule. The first molecule
need not
necessarily be the natural ligand of the second molecule. Allergens of the
present
invention are preferably derived from mites, and mite-related allergens
including, but
not limited to, other insect allergens and plant allergens.
In accordance with the present invention, virtually any substance can act as
an
antigen and elicit an antibody response, i.e., can function as an epitope. For
example,
antibodies can be raised in response to carbohydrate epitopes, including
saecharides
and/or polysaccharides that are attached to a protein, a so-called
glycosylated protein.
However, a saccharide and/or polysaccharide may act as an antigen alone,
without a
protein being present. The terminal sugar of a carbohydrate moiety, as well as
internal
sugars can serve as an epitope. Polysaccharide may be present as a branched
chain, in
which case epitopes may comprise sugars that are not contiguous in sequence,
but are
adjacent spatially. Unusual, insect-specific sugars, not normally seen in
mammalian
proteins, may be present on glycoprotein derived from insect nucleic acid
molecules,
and these unusual sugars can comprise an epitope recognized by a mammalian
immune system.
One embodiment of the present invention is a reagent comprising a non-
proteinaceous epitope that is capable of binding to IgE of an animal that is
allergic to
mites, of desensitizing an animal against mite allergen, of stimulating a B
lymphocyte
response, and/or of stimulating a T lymphocyte response. Such an epitope,
referred to
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-39-
herein as a Der NP epitope, can exist as part of a Der HMW-map protein of the
present invention or can be isolated therefrom. Such an epitope exists, for
example, on
a protein contained in the D. farihae HMW-map composition produced in
accordance
with Example 1. A Der NP epitope of the present invention can be isolated from
its
natural source or produced synthetically. Such an epitope can be, but need not
be,
joined to a carrier or other molecule. ADer NP epitope has at least one of the
following identifying characteristics: (a) the epitope is resistant to (3-
elimination of
peptides; (b) the epitope is resistant to Proteinase-K digestion; and (c) the
epitope is
reactive to a test designed to detect glycosylated proteins. A preferred Der
NP epitope
has,all such identifying characteristics. ADer NP epitope can selectively bind
to IgE
of dogs or cats that are allergic to mites. While not being bound by theory,
it is
believed that a Der NP epitope comprises a carbohydrate moiety that apparently
does
not include an N-linked glycan. Identification of the structural
characteristics of such
an epitope can be determined by one skilled in the art. In one embodiment,
there is
provided an isolated antibody that selectively binds to a Def° NP
epitope. The present
invention also includes a derivative of a Der NP epitope, i.e., a compound
that mimics
the activity of such an epitope (e.g. is a Der NP epitope mimetope) and is
capable of
binding to antibody raised against a native (i.e. seen in nature) Der NP
epitope.
A reagent comprising a Der NP epitope of the present invention can be used in
a variety of ways in accordance with the present invention. Such a reagent can
be a
desensitizing compound or a detection reagent to test for mite allergy
susceptibility or
sensitivity. In one embodiment, a therapeutic composition of the present
invention
includes a reagent comprising a Der NP epitope. In another embodiment, an
assay kit
of the present invention includes a reagent comprising a Der NP epitope. One
embodiment of the present invention is a method to identify an animal
susceptible to
or having an allergic response to a mite. Such a method includes the steps of
contacting a reagent comprising a Der NP epitope with antibodies of an animal
and
determining immunocomplex formation between the reagent and the antibodies,
wherein formation of the immunocomplex indicates that the animal is
susceptible to or
has said allergic response. Another embodiment of the present invention is a
method
to desensitize a host animal to an allergic response to a mite. Such a method
includes
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-40-
the step of administering to the animal a therapeutic composition that
includes a
reagent comprising a Der NP epitope as a desensitizing compound.
Another embodiment of the present invention is a Der HMW-map protein
lacking Der NP epitopes. Without being bound by theory, it is believed that
such a
protein would be a better desensitizing compound since such a protein is
expected to
have a reduced ability to bind to IgE. Such a protein can be produced by, for
example,
removing Der NP epitopes from a native Der HMW-map protein or by producing the
protein recombinantly, for example in E. coli.
One embodiment of the present invention is an ih vivo test that is capable of
detecting whether an animal is hypersensitive to Der HMW-map protein. An iiz
vivo
hypersensitivity test of the present invention is particularly useful for
identifying
animals susceptible to or having allergy to mite allergens. A- suitable ih
vivo
hypersensitivity test of the present invention can be, but is not limited to,
a skin test
comprising administering (e.g., intradermally injecting or superficial
scratching) an
effective amount of a formulation containing Der HMW-map protein, or a
mimetope
thereof. Methods to conduct skin tests of the present invention are known to
those of
skill in the art and are briefly disclosed herein.
Suitable formulations to use in an irc vivo skin test include Der HMW-map
protein, homologs of Der HMW-map protein and/or mimetopes of Der HMW-map
protein.
It is understood by one of skill in the art that a suitable amount of Der HMW-
map protein formulation for use in a skin test of the present invention can
vary widely
depending on the allergenicity of the formulation used in the test and on the
site at
which the product is delivered. Suitable amounts of Def° HMW-map
protein
formulation for use in a skin test of the present invention include an amount
capable of
forming reaction, such as a detectable wheal or induration (hardness)
resulting from an
allergic reaction to the formulation. Preferred amounts of Der HMW-map protein
for
use in a skin test of the present invention range from about 1 x 10-8
micrograms (fig) to
about 100 fig, more preferably from about 1 x 10-' ~g to about 10 fig, and
even more
preferably from about 1 x 10-~ ~g to about 1 ~g of Der HMW-map protein. It is
to be
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-41-
appreciated by those of skill in the art that such amounts will vary depending
upon the
allergenicity of the protein being administered.
According to the present invention, Dej- HMW-map protein of the present
invention can be combined with an immunopotentiator (e.g., carriers or
adjuvants of
the present invention as defined in detail below). A novel aspect, however, of
the
present invention is that DeY HMW-map protein of the present invention can
induce a
hypersensitive response in the absence of an immunopotentiator, particularly
in
canines.
A skin test of the present invention further comprises administering a control
solution to an animal. A control solution can include a negative control
solution
and/or a positive control solution. A positive control solution of the present
invention
contains an effective amount of at least one compound known to induce a
hypersensitive response when administered to an animal. A preferred compound
for
use as positive control solution includes, but is not limited to, histamine. A
negative
control solution of the present invention can comprise a solution that is
known not to
induce a hypersensitive response when administered to an animal. As such, a
negative
control solution can comprise a solution having compounds essentially
incapable of
inducing a hypersensitive response or simply a buffer used to prepare the
formulation,
such as saline. An example of a preferred negative control solution is
phenolated
phosphate buffered saline (available from Greer Laboratories, Inc., Lenoir,
NC).
Hypersensitivity of an animal to one or more formulations of the present
invention can be evaluated by measuring reactions (e.g., wheal size,
induration or
hardness; using techniques known to those skilled in the art) resulting from
administration of one or more experimental samples) and control samples) into
an
animal and comparing the reactions to the experimental samples) with reactions
resulting from administration of one or more control solution. Preferred
devices for
intradermal injections include individual syringes. Preferred devices for
scratching
include devices that permit the administration of a number of samples at one
time.
The hypersensitivity of an animal can be evaluated by determining if the
reaction
resulting from administration of a formulation of the present invention is
larger than
the reaction resulting from administration of a negative control, and/or by
determining
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
_q.2_
if the reaction resulting from administration of the formulation is at least
about the
same size as the reaction resulting from administration of a positive control
solution.
As such, if an experimental sample produces a reaction greater than or equal
to the size
of a wheat produced by administration of a positive control sample to an
animal, then
that animal is hypersensitive to the experimental sample. Conversely, if an
experimental sample produces a reaction similar to the reaction produced by
administration of a negative control sample to an animal, then that animal is
not
hypersensitive to the experimental sample.
Preferred wheat sizes for evaluation of the hypersensitivity of an animal
range
from about 16 mm to about 8 mm, more preferably from about 15 mm to about 9
mm,
and even more preferably from about 14 mm to about 10 mm in diameter.
Preferably, the ability or inability of an animal to exhibit an immediate
hypersensitive response to a formulation of the present invention is
determined by
measuring wheat sizes from about 2 minutes to about 30 minutes after
administration
of a sample, more preferably from about 10 minutes to about 25 minutes after
administration of a sample, and even more preferably about 15 minutes after
administration of a sample.
Preferably, the ability or inability of an animal to exhibit a delayed
hypersensitive response to a formulation of the present invention is
determined by
measuring induration andlor erythema from about 18 hours to about 30 hours
after
administration of a sample, more preferably from about 20 hours to about 28
hours
after administration of a sample, and even more preferably at about 24 hours
after
administration of a sample. A delayed hypersensitivity response can also be
measured
using other techniques such as by determining, using techniques known to those
of
skill in the art, the extent of cell infiltrate at the site of administr ation
during the time
periods defined directly above.
In a preferred embodiment, a skin test of the present invention comprises
intradermally injecting into an animal at a given site an effective amount of
a
formulation that includes Def-HMW-map protein, and intradermally injecting an
effective amount of a control solution into the same animal at a different
site. It is
within the scope of one of skill in the art to use devices capable of
delivering multiple
samples simultaneously at a number of sites, preferably enabling concurrent
evaluation
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-43-
of numerous formulations. A preferred Der HMW-map protein for use with a skin
test
includes full-length protein. A preferred positive control sample can be a
sample
comprising histamine. A preferred negative control sample can be a sample
comprising diluent.
Animals suitable and preferred to test for hypersensitivity to Der HMW-map
protein using a skin test of the present invention are disclosed herein.
Particularly
preferred animals to test with a skin test of the present invention include
humans,
canines, felines and equines, with human, canines and felines being even more
preferred. As used herein, canine refers to any member of the dog family,
including
domestic dogs, wild dogs and zoo dogs. Examples of dogs include, but are not
limited
to, domestic dogs, wild dogs, foxes, wolves, jackals and coyotes. As used
herein,
feline refers to any member of the cat family, including domestic cats, wild
cats and
zoo cats. Examples of cats include, but are not limited to, domestic cats,
lions, tigers,
leopards, panthers, cougars, bobcats, lynx, jaguars, cheetahs and servals. As
used
herein, equine refers to any member of the horse family, including horses,
donkeys,
mules and zebras.
One embodiment of the present invention is a method to detect antibodies in
vitro that bind to Der HMW-map protein (referred to herein as anti Der HMW-map
antibody) which includes the steps of: (a) contacting an isolated Der HMW-map
protein with a putative anti-Der HMW-map antibody-containing composition under
conditions suitable for formation of a Der HMW-map protein:antibody complex;
and
(b) detecting the presence of the antibody by detecting the Der HMW-map
protein:antibody complex. Presence of such aDer HMW-map protein:antibody
complex indicates that the animal is producing antibody to a mite allergen.
Preferred
anti-Der HMW-map antibody to detect include antibodies having an IgE or IgG
isotype. Preferred anti-Der HMW-map antibody to detect include feline
antibody,
canine antibody, equine antibody and human antibody, with feline, canine and
human
antibody being particularly preferred.
As used herein, the term "contacting" refers to combining or mixing, in this
case a putative antibody-containing composition with a Der HMW-map protein.
Formation of a complex between a Der HMW-map protein and an antibody refers to
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-44-
the ability of the Der HMW-map protein to selectively bind to the antibody in
order to
form a stable complex that can be measured (i.e., detected). As used herein,
the term
selectively binds to an antibody refers to the ability of a Def~ HMW-map
protein of the
present invention to preferentially bind to an antibody, without being able to
substantially bind to other antibodies that do not specifically bind to Der
HMW-map
protein. Binding between a Der HMW-map protein and an antibody is effected
under
conditions suitable to form a complex; such conditions (e.g., appropriate
concentrations, buffers, temperatures, reaction times) as well as methods to
optimize
such conditions are known to those skilled in the art, and examples are
disclosed
herein. Examples of complex formation conditions are also disclosed in, for
example,
in Sambrook et al., ibid.
As used herein, the term "detecting complex formation" refers to determining
if
any complex is formed, i.e., assaying for the presence (i.e., existence) of a
complex. If
complexes are formed, the amount of complexes formed can, but need not be,
determined. Complex formation, or selective binding, between Der HMW-map
protein and an antibody in the composition can be measured (i.e., detected,
determined) using a variety of methods standard in the art (see, for example,
Sambrook et al. ibid.), examples of which are disclosed herein.
In one embodiment, a putative antibody-containing composition of the present
method includes a biological sample from an animal. A suitable biological
sample
includes, but is not limited to, a bodily fluid composition or a cellular
composition. A
bodily fluid refers to any fluid that can be collected (i.e., obtained) from
an animal,
examples of which include, but are not limited to, blood, serum, plasma,
urine, tears,
aqueous humor, cerebrospinal fluid (CSF), saliva, lymph, nasal secretions,
milk and
feces. Such a composition of the present method can, but need not be,
pretreated to
remove at least some of the non-IgE or non-IgG isotypes of immunoglobulin
and/or
other proteins, such as albumin, present in the fluid. Such removal can
include, but is
not limited to, contacting the bodily fluid with a material, such as the
lectin jacalin or
an antibody that specifically binds to the constant region of an IgA
immunoglobulin
(i.e., anti-IgA isotype antibody), to remove IgA antibodies andlor affinity
purifying
IgE or IgG antibodies from other components of the body fluid by exposing the
fluid
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-45-
to, for example, Concanavalin A or protein G, respectively. In another
embodiment, a
composition includes collected bodily fluid that is pretreated to concentrate
immunoglobulin contained in the fluid. For example, immunoglobulin contained
in a
bodily fluid can be precipitated from other proteins using ammonium sulfate. A
preferred composition of the present method is serum.
In another embodiment, an antibody-containing composition of the present
method includes a cell that produces IgE or IgG. Such a cell can have IgE or
IgG
bound to the surface of the cell and/or can secrete IgE or IgG. An example of
such a
cell includes myeloma cells. IgE or IgG can be bound to the surface of a cell
either
directly to the membrane of the cell or bound to a molecule (e.g., an antigen)
bound to
the surface of the cell.
A complex can be detected in a variety of ways including, but not limited to
use of one or more of the following assays: an enzyme-linked immunoassay, a
radioimmunoassay, a fluorescence immunoassay, a chemiluminescent assay, a
lateral
flow assay, an agglutination assay, a particulate-based assay (e.g., using
particulates
such as, but not limited to, magnetic particles or plastic polymers, such as
latex or
polystyrene beads), an immunoprecipitation assay, a BioCoreTM assay (e.g.,
using
colloidal gold) and an immunoblotting assay (e.g., a western blot). Such
assays are
well known to those skilled in the art. Assays can be used to give qualitative
or
quantitative results depending on how they are used. Some assays, such as
agglutination, particulate separation, and immunoprecipitation, can be
observed
visually (e.g., either by eye or by a machines, such as a densitometer or
spectrophotometer) without the need for a detectable marker.
In other assays, conjugation (i.e., attachment) of a detectable marker to the
Def°
HMW-map protein, to antibody bound to the Der~ HMW-map protein, or to a
reagent
that selectively binds to the Der HMW-map protein or to the antibody bound to
the
Der HMW-map protein (described in more detail below) aids in detecting complex
formation. Examples of detectable markers include, but are not limited to, a
radioactive label, an enzyme, a fluorescent label, a chemiluminescent label, a
chromophoric label or a ligand. A ligand refers to a molecule that binds
selectively to
another molecule. Preferred detectable markers include, but are not limited
to,
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-46-
fluorescein, a radioisotope, a phosphatase (e.g., alkaline phosphatase),
biotin, avidin, a
peroxidase (e.g., horseradish peroxidase) and biotin-related compounds or
avidin-
related compounds (e.g., streptavidin or ImmunoPureO NeutrAvidin available
from
Pierce, Rockford, IL).
In one embodiment, a complex is detected by contacting a putative antibody-
containing composition with a Der HMW-map protein that is conjugated to a
detectable marker. A suitable detectable marker to conjugate to a Der HMW-map
protein includes, but is not limited to, a radioactive label, a fluorescent
label, an
enzyme label, a chemiluminescent label, a chromophoric label or a ligand. A
detectable marker is conjugated to a Der HMW-map protein in such a manner as
not to
block the ability of the Der HMW-map protein to bind to the antibody being
detected.
In another embodiment, a Der HMW-map protein: antibody complex is
detected by contacting a putative antibody-containing composition with a Der
HMW-
map protein and then contacting the complex with an indicator molecule.
Suitable
indicator molecules of the present invention include molecules that can bind
to either
the Der HMW-map protein or to the antibody bound to the Der HMW-map protein.
As such, an indicator molecule can comprise, for example, an antigen and an
antibody,
depending upon which portion of the Der HMW-map protein: antibody complex is
being detected. Preferred indicator molecules that are antibodies include, for
example,
anti-IgE antibodies, anti-IgG antibodies and antibodies that are known bind to
Der
HMW-map protein but bind to a different epitope on Der HMW-map protein than
antibodies identified in the putative antibody-containing composition.
Preferred
lectins include those lectins that bind to high-mannose groups. An indicator
molecule
itself can be attached to a detectable marker of the present invention. For
example, an
antibody can be conjugated to biotin, horseradish peroxidase, alkaline
phosphatase or
fluorescein.
In one preferred embodiment, aDer HMW-map protein: antibody complex is
detected by contacting the complex with an indicator molecule that selectively
binds to
an IgE antibody (referred to herein as an anti-IgE reagent) or an IgG antibody
(referred
to herein as an anti-IgG reagent. Examples of such an anti-IgE or an anti-IgG
antibody
include, but are not limited to, a secondary antibody that is an anti-isotype
antibody
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-47-
(e.g., an antibody that selectively binds to the constant region of an IgE or
an IgG), an
antibody-binding bacterial surface protein (e.g., Protein A or Protein G), an
antibody-
binding cell (e.g., a B cell, a T cell, a natural killer cell, a
polymorphonuclear
leukocyte cell, a monocyte cell or a macrophage cell), an antibody-binding
eukaryotic
cell surface protein (e.g., a Fc receptor), and an antibody-binding complement
protein.
Preferred indicator molecules include, but are not limited to, an anti-feline
IgE
antibody, an anti-feline IgG antibody, an anti-canine IgE antibody, an anti-
canine IgG
antibody, an anti-human IgE antibody, and an anti-human IgG antibody. As used
herein, an anti-IgE or anti-IgG antibody includes not only a complete antibody
but also
any subunit or portion thereof that is capable of selectively binding to an
IgE or IgG
heavy chain constant region. For example, an anti-IgE reagent or anti-IgG
reagent can
include an Fab fragment or a Flab' )Z fragment, both of which are described in
detail in
Janeway et al., in Immm2obiology, the Immune Systen-a i~ Health. and Disease,
Garland
Publishing, Inc., NY, 1996.
In another preferred embodiment, aDef~ HMW-map protein:antibody complex
is detected by contacting the complex with an indicator molecule that
selectively binds
to Der HMW-map protein at a different epitope than the epitope at which an
antibody
in a putative antibody-containing composition binds to Der HMW-map protein.
In one embodiment a complex can be formed and detected in solution. In
another embodiment, a complex can be formed in which one or more members of
the
complex are immobilized on (e.g., coated onto) a substrate. Immobilization
techniques are known to those skilled in the art. Suitable substrate materials
include,
but are not limited to, plastic, glass, gel, celluloid, paper, PVDF (poly-
vinylidene-
fluoride), nylon, nitrocellulose, and particulate materials such as latex,
polystyrene,
nylon, nitrocellulose, agarose and magnetic resin. Suitable shapes for
substrate
material include, but are not limited to, a well (e.g., microtiter dish well),
a plate, a
dipstick, a bead, a lateral flow apparatus, a membrane, a filter, a tube, a
dish, a
celluloid-type matrix, a magnetic particle, and other particulates. A
particularly
preferred substrate comprises an ELISA plate, a dipstick, a radioimmunoassay
plate,
agarose beads, plastic beads, latex beads, immunoblot membranes and immunoblot
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-48-
papers. In one embodiment, a substrate, such as a particulate, can include a
detectable
marker.
A preferred method to detect antibody that binds to Der HMW-map protein is
an immunoabsorbent assay. An immunoabsorbent assay of the present invention
comprises a capture molecule and an indicator molecule. A capture molecule of
the
present invention binds to an IgE or an IgG in such a manner that the IgE or
IgG is
immobilized to a substrate. As such, a capture molecule is preferably
immobilized to a
substrate of the present invention prior to exposure of the capture molecule
to a
putative IgE-containing composition or a putative IgG-containing composition.
An
indicator molecule of the present invention detects the presence of an IgE or
an IgG
bound to a capture molecule. As such, an indicator molecule preferably is not
immobilized to the same substrate as a capture molecule prior to exposure of
the
capture molecule to a putative IgE-containing composition or a putative IgG-
containing composition.
A preferred immunoabsorbent assay method includes a step of either:
(a) immobilizing a Der HMW-map protein on a substrate prior to contacting a
Der
HMW-map protein with a putative IgE-containing composition or a putative IgG-
containing composition to form a Der HMW-map protein -immobilized substrate;
and
(b) binding a putative IgE-containing composition or a putative IgG-containing
composition on a substrate prior to contacting Der HMW-map protein with a
putative
IgE-containing composition or a putative IgG-containing composition, to form a
putative IgE-containing composition-bound substrate or a putative IgG-
containing
composition-bound substrate, respectively. Preferably, the substrate includes
a non-
coated substrate, a Der HMW-map protein -immobilized substrate, an anti-IgE
antibody-immobilized substrate or anti-IgG antibody-immobilized substrate.
Both a capture molecule and an indicator molecule of the present invention are
capable of binding to an IgE, an IgG or Der HMW-map protein. Preferably, a
capture
molecule binds to a different region of an IgE, an IgG or Der HMW-map protein
than
an indicator molecule, thereby allowing a capture molecule to be bound to an
IgE, an
IgG or Der HMW-map protein at the same time as an indicator molecule. The use
of a
reagent as a capture molecule or an indicator molecule depends upon whether
the
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-49-
molecule is immobilized to a substrate when the molecule is exposed to an IgE,
an IgG
or Der HMW-map protein. For example, a Der HMW-map protein of the present
invention is used as a capture molecule when the Der HMW-map protein is bound
on
a substrate. Alternatively, a Der HMW-map protein is used as an indicator
molecule
when the Der HMW-map protein is not bound on a substrate. Suitable molecules
for
use as capture molecules or indicator molecules include, but are not limited
to, a Der
HMW-map protein of the present invention, an anti-IgE antibody reagent or an
anti-
IgG antibody reagent of the present invention.
An immunoabsorbent assay of the present invention can further comprise one
or more layers and/or types of secondary molecules or other binding molecules
capable of detecting the presence of an indicator molecule. For example, an
untagged
(i.e., not conjugated to a detectable marker) secondary antibody that
selectively binds
to an indicator molecule can be bound to a tagged (i.e., conjugated to a
detectable
marker) tertiary antibody that selectively binds to the secondary antibody.
Suitable
secondary antibodies, tertiary antibodies and other secondary or tertiary
molecules can
be selected by those of skill in the art. Preferred secondary molecules of the
present
invention include an antigen, an anti-IgE idiotypic antibody (i.e., an
antibody that
binds to an epitope unique to the anti-IgE antibody), an anti-IgE isotypic
antibody, an
anti-IgG idiotypic antibody (i.e:, an antibody that binds to an epitope unique
to the
anti-IgG antibody), and an anti-IgG isotypic antibody. Preferred tertiary
molecules
can be selected by a skilled artisan based upon the characteristics of the
secondary
molecule. The same strategy is applied for subsequent layers.
In one embodiment, Def° HMW-map protein is used as a capture
molecule by
being immobilized on a substrate, such as a microtiter dish well or a
dipstick. A
biological sample collected from an animal is applied to the substrate and
incubated
under conditions suitable (i.e., sufficient) to allow for Dey- HMW-map
protein:antibody complex formation bound to the substrate (i.e., IgE or IgG in
a
sample binds to Der HMW-map protein immobilized on a substrate). Excess non-
bound material (i.e., material from the biological sample that has not bound
to the Der-
HMW-map protein), if any, is removed from the substrate under conditions that
retain
antigen:antibody complex binding to the substrate. Preferred conditions are
generally
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-50-
disclosed in Sambrook et al., ibid. An indicator molecule that can selectively
bind to
an IgE or an IgG bound to the antigen is added to the substrate and incubated
to allow
formation of a complex between the indicator molecule and the Der HMW-map
protein:antibody complex. Excess indicator molecule is removed, a developing
agent
is added if required, and the substrate is submitted to a detection device for
analysis.
A preferred indicator molecule for this embodiment is an anti-IgG antibody to
detect
IgG antibody bound to Der HMW-map protein or an anti-IgE antibody to detect
IgE
antibody bound to Der HMW-map protein. Preferably the anti-IgG or anti-IgE
antibody are conjugated to biotin, to a fluorescent label or to an enzyme
label.
In one embodiment, an anti-IgE or anti-IgG antibody (e.g., isotype or idiotype
specific antibody) is used as a capture molecule by being immobilized on a
substrate,
such as a microtiter dish well or a dipstick. A biological sample collected
from an
animal is applied to the substrate and incubated under conditions suitable to
allow for
anti-IgE antibody:IgE complex or anti-IgG antibody:IgG complex formation,
respectively, bound to the substrate. Excess non-bound material, if any, is
removed
from the substrate under conditions that retain anti-IgE antibody:IgE complex
or anti-
IgG antibody:IgG complex binding to the substrate. Der HMW-map protein is
added
to the substrate and incubated to allow formation of a complex between the Der
HMW-map protein and the anti-IgE antibody:IgE complex or anti-IgG antibody:IgG
complex. Preferably, the Der HMW-map protein is conjugated to a detectable
marker
(preferably to biotin, an enzyme label or a fluorescent label). Excess Der HMW-
map
protein is removed, a developing agent is added if required, and the substrate
is
submitted to a detection device for analysis.
In one embodiment, an immunoabsorbent assay of the present invention does
not utilize a capture molecule. In this embodiment, a biological sample
collected from
an animal is applied to a substrate, such as a microtiter dish well or a
dipstick, and
incubated under conditions suitable to allow for IgE or IgG binding to the
substrate.
Any IgE or IgG present in the bodily fluid is immobilized on the substrate.
Excess
non-bound material, if any, is removed from the substrate under conditions
that retain
IgE or IgG binding to the substrate. Der HMW-map protein is added to the
substrate
and incubated to allow formation of a complex between the Der HMW-map protein
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-51-
and the IgE or IgG. Preferably, the Der HMW-map protein is conjugated to a
detectable marker (preferably to biotin, an enzyme label or a fluorescent
label). Excess
DeY HMW-map protein is removed, a developing agent is added if required, and
the
substrate is submitted to a detection device for analysis.
Another preferred method to detect IgE or IgG is a lateral flow assay,
examples
of which are disclosed in U.S. Patent No. 5,424,193, issued June 13, 1995, by
Pronovost et al.; U.S. Patent No. 5,415,994, issued May 16, 1995, by Imrich et
al; WO
94/29696, published December 22, 1994, by Miller et al.; and WO 94/01775,
published January 20, 1994, by Pawlak et al. In one embodiment, a biological
sample
is placed in a lateral flow apparatus that includes the following components:
(a) a
support structure defining a flow path; (b) a labeling reagent comprising a
bead
conjugated to Der HMW-map protein, the labeling reagent being impregnated
within
the support structure in a labeling zone; and (c) a capture reagent comprising
an IgE-
binding or an IgG-binding composition. The capture reagent is located
downstream of
the labeling reagent within a capture zone fluidly connected to the labeling
zone in
such a manner that the labeling reagent can flow from the labeling zone into
the
capture zone. The support structure comprises a material that does not impede
the
flow of the beads from the labeling zone to the capture zone. Suitable
materials for
use as a support structure include ionic (i.e:, anionic or cationic) material.
Examples
of such a material include, but are not limited to, nitrocellulose (NC), PVDF,
carboxymethylcellulose (CM). The support structure defines a flow path that is
lateral
and is divided into zones, namely a labeling zone and a capture zone. The
apparatus
can further comprise a sample receiving zone located along the flow path, more
preferably upstream of the labeling reagent. The flow path in the support
structure is
created by contacting a portion of the support structure downstream of the
capture
zone, preferably at the end of the flow path, to an absorbent capable of
absorbing
excess liquid from the labeling and capture zones.
In this embodiment, the biological sample is applied to the sample receiving
zone which includes a portion of the support structure. The labeling zone
receives the
sample from the sample receiving zone which is directed downstream by the flow
path. The labeling zone comprises the labeling reagent that binds to IgE or
IgG, or
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-52-
both. A preferred labeling reagent is Der HMW-map protein conjugated, either
directly or through a linker, to a plastic bead substrate, such as to a latex
bead. The
substrate also includes a detectable marker, preferably a colorimetric marker.
Typically, the labeling reagent is impregnated to the support structure by
drying or
lyophilization. The sample structure also comprises a capture zone downstream
of the
labeling zone. The capture zone receives labeling reagent from the labeling
zone
which is directed downstream by the flow path. The capture zone contains the
capture
reagent, in this case an anti-IgE or anti-IgG antibody, or both, as disclosed
above, that
immobilizes the IgE and/or IgG complexed to the DeY HMW-map protein in the
capture zone. The capture reagent is preferably fixed to the support structure
by
drying or lyophilizing. The labeling reagent accumulates in the capture zone
and the
accumulation is assessed visually or by an optical detection device.
In another embodiment, a lateral flow apparatus used to detect IgE or IgG
includes: (a) a support structure defining a flow path; (b) a labeling reagent
comprising
an anti-IgE or an anti-IgG antibody, or both, as described above, the labeling
reagent
impregnated within the support structure in a labeling zone; and (c) a capture
reagent
comprising Der HMW-map protein, the capture reagent being located downstream
of
the labeling reagent within a capture zone fluidly connected to the labeling
zone in
such a manner that the labeling reagent can flow from the labeling zone into
the
capture zone. The apparatus preferably also includes a sample receiving zone
located
along the flow path, preferably upstream of the labeling reagent. The
apparatus
preferably also includes an absorbent located at the end of the flow path.
An animal hypersensitive to Der HMW-map protein is identified by comparing
the level of immunocomplex formation using samples of body fluid with the
level of
immunocomplex formation using control samples. An immunocomplex refers to a
complex comprising an antibody and Der HMW-map protein (i.e., Der HMW-map
protein:antibody complex). As such, immunocomplexes form using positive
control
samples and do not form using negative control samples. As such, if a body
fluid
sample results in immunocomplex formation greater than or equal to
immunocomplex
formation using a positive control sample, then the animal from which the
fluid was
taken is hypersensitive to the Der HMW-map protein bound to the substrate.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-53-
Conversely, if a body fluid sample results in immunocomplex formation similar
to
immunocomplex formation using a negative control sample, then the animal from
which the fluid was taken is not hypersensitive to the Der HMW-map protein
bound to
the substrate.
It is within the scope of the present invention that two or more different
skin
tests and/or ih vitro tests can be used in combination for diagnostic
purposes. For
example, the immediate hypersensitivity of an animal to Der HMW-map protein
can
be tested using an iya vitro immunoabsorbent test capable of detecting IgE
antibodies
specific for Der HMW-map protein in the animal's bodily fluid. While most
animals
that display delayed hypersensitivity to Der HMW-map protein also display
immediate hypersensitivity to the allergen, a small number of animals that
display
delayed hypersensitivity to an allergen do not display immediate
hypersensitivity to
the allergen. In such cases, following negative results from the IgE-specific
in vitro
test, the delayed hypersensitivity of the animal to Der HMW-map protein can be
tested
using an skin test of the present invention.
The present invention also includes kits to detect antibodies that bind
specifically to Der HMW-map protein based on each of the disclosed detection
methods. One embodiment is a kit to detect Der HMW-map protein-specific
antibodies comprising Der HMW-map protein and a means for detecting an IgE
and/or
an IgG. Suitable means of detection include compounds disclosed herein that
bind to
either the Der HMW-map protein or to an IgE and/or an IgG. A preferred kit of
the
present invention further comprises a detection means including an antibody
capable
of selectively binding to an IgE or IgG disclosed herein and/or a compound
capable of
binding to a detectable marker conjugated to a Der HMW-map protein (e.g.,
avidin,
streptavidin and ImmunoPureO NeutrAvidin when the detectable marker is
biotin).
Another preferred kit of the present invention is an allergen kit comprising
Der
HMW-map protein and an allergen commonly detected in the same environment as
mite allergen. Suitable and preferred mite-related allergens for use with the
present kit
include those mite-related allergens disclosed herein.
A preferred kit of the present invention includes those in which Der HMW-
map protein is immobilized on a substrate. If a kit comprises Der HMW-map
protein
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-54-
and another allergen, the kit can comprise one or more compositions, each
composition comprising one allergen. As such, each allergen can be tested
separately.
A kit can also contain two or more diagnostic reagents for IgE or IgG, or
other
compounds as disclosed herein. Particularly preferred are kits used in a
lateral flow
assay format. It is within the scope of the present invention that a lateral
flow assay kit
can include one or more lateral flow assay apparatuses. Multiple lateral flow
apparatuses can be attached to each other at one end of each apparatus,
thereby
creating a fan-like structure.
Another aspect of the present invention includes treating animals susceptible
to
or having mite allergy, with a Der HMW-map protein formulation of the present
invention. According to the present invention, the term treatment can refer to
the
regulation of a hypersensitive response by an animal to mite allergens.
Regulation can
include, for example, immunomodulation of cells involved in the animal's
hypersensitive response. Immunomodulation can include modulating the activity
of
molecules typically involved in an immune response (e.g., antibodies,
antigens, major
histocompatibility molecules (MHC) and molecules co-reactive with MHC
molecules). In particular, immunomodulation refers to modulation of
antigen:antibody
interactions resulting in inflammatory responses, immunosuppression, and
immunotolerization of cells involved in a hypersensitive response.
Immunosuppression refers to inhibiting an immune response by, for example,
killing
particular cells involved in the immune response. Immunotolerization refers to
inhibiting an immune response by anergizing (i.e., diminishing reactivity of a
T cell to
an antigen) particular cells involved in the immune response.
One embodiment of the present invention is a therapeutic composition that
includes desensitizing compounds capable of inhibiting an immune response to
Def
HMW-map protein of the present invention. Such desensitizing compounds include
blocking compounds, toleragens and/or suppressor compounds. Blocking compounds
comprise compounds capable of modulating antigen:antibody interactions that
can
result in inflammatory responses, toleragens are compounds capable of
immunotolerizing an animal, and suppressor compounds are capable of
immunosuppressing an animal. A desensitizing compound of the present invention
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-55-
can be soluble or membrane-bound. Membrane-bound desensitizing compounds can
be associated with biomembranes, including cells, liposomes, planar membranes
or
micelles. A soluble desensitizing compound of the present invention is useful
for: (1)
inhibiting a Type I hypersensitivity reaction by blocking IgE:antigen mediated
de-
granulation of mast cells; (2) inhibiting a Type III hypersensitivity reaction
by
blocking IgG:antigen complex formation leading to complement destruction of
cells;
and (3) inhibiting a Type IV hypersensitivity reaction by blocking T helper
cell
stimulation of cytokine secretion by macrophages. A membrane-bound
desensitizing
compound of the present invention is useful for: (1) inhibiting a Type II
hypersensitivity reaction by blocking IgG:antigen complex formation on the
surface of
cells leading to complement destruction of cells; (2) inhibiting a Type II
hypersensitivity reaction by blocking IgG regulated signal transduction in
immune
cells; and (3) inhibiting a Type IV hypersensitivity reaction by blocking T
cytotoxic
cell killing of antigen-bearing cells. Examples of desensitizing compounds
include,
but are not limited to, muteins, mimetopes and antibodies of the present
invention, as
well as other inhibitors of the present invention that inhibit binding between
a protein
of the present invention and IgE.
A desensitizing compound of the present invention can also be covalently
linked to a ligand molecule capable of targeting the desensitizing compound to
a
specific cell involved in a hypersensitive response to Def~ HMW-map protein.
Appropriate ligands with which to link a desensitizing compound include, for
example, at least a portion of an immunoglobulin molecule, cytokines, lectins,
heterologous allergens, CD8 molecules or major histocompatibility molecules
(e.g.,
MHC class I or MHC class II molecules). Preferred portions of immunoglobulin
molecules to link to a desensitizing compound include variable regions capable
of
binding to immune cell specific surface molecules and constant regions capable
of
binding to Fc receptors on immune cells, in particular IgE constant regions.
Preferred
CD8 molecules include at least the extracellular functional domains of the a
chain of
CDB. An immune cell refers to a cell involved in an immune response, in
particular,
cells having MHC class I or MHC class II molecules. Preferred immune cells
include
antigen presenting cells, T cells and B cells.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-56-
In one embodiment, a therapeutic composition of the present invention ,
includes Der HMW-map protein of the present invention, a mimetope or mutein
thereof, or a Der HMW-map nucleic acid molecule of the present invention.
Suitable
therapeutic compositions of the present invention for treating mite allergy
include Der
HMW-map protein, a mimetope or mutein thereof, or a Der HMW-map nucleic acid
molecule of the present invention. Preferred therapeutic compositions include:
an
isolated mite allergenic protein encoded a nucleic acid molecule that
hybridizes under
stringent hybridization conditions with the, complement of a nucleic acid
molecule that
encodes an amino acid sequence selected from the group consisting of SEQ ID
NO: l,
SEQ ID NO:2, SEQ ID NO:3, SEQ ID N0:4, SEQ ID N0:5, SEQ ID N0:6, SEQ ID
N0:7, SEQ ID N0:8, SEQ ID N0:9, SEQ ID NO:10, SEQ ID NO:11, SEQ ID NO:12,
SEQ ID N0:13, SEQ ID N0:15, SEQ ID N0:18, SEQ ID N0:21, SEQ ID N0:23,
SEQ ID N0:24, SEQ ID NO:30, SEQ ID N0:31, SEQ ID N0:32, SEQ ID N0:33,
SEQ ID NO:33, SEQ ID NO:35, SEQ ID N0:38, SEQ ID N0:41, and SEQ m N0:44;
a mimetope of the mite allergenic protein; a mutein of the mite allergenic
protein; and
an isolated nucleic acid molecule selected from the group consisting of: a
nucleic acid
molecule comprising at least about 150 nucleotides, wherein said nucleic acid
molecule comprising at least about 150 nucleotides hybridizes, in a solution
comprising 1X SSC and 0% formamide, at a temperature of about 50°C, to
a nucleic
acid sequence selected from the group consisting of SEQ ID N0:14, SEQ ID
N0:16,
SEQ ID NO:17, SEQ ID N0:19, SEQ ID N0:20, SEQ ID N0:22, SEQ ID N0:34,
SEQ ID N0:36, SEQ ID N0:37, SEQ II7 N0:39, SEQ ID N0:40, SEQ ID N0:42,
SEQ ID N0:43, SEQ m N0:45, and a nucleic acid sequence encoding a protein
comprising the amino acid sequence SEQ ID N0:33 and a complement thereof; and
a
nucleic acid molecule comprising a fragment of any of said nucleic acid
molecules
comprising at least about 150 nucleotides. A preferred Der HMW-map mutein
comprises at least a portion of Def~ HMW-map protein, in which a suitable
number of
cysteine residues have been removed or replaced with a non-cysteine residue
such that
the altered Der HMW-map protein is not toxic to an animal (e.g., does not
cause
anaphylaxis).
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-57-
In another embodiment, a therapeutic composition of the present invention
includes a nucleic acid molecule encoding a Der HMW-map protein that can be
administered to an animal in a fashion to enable expression of that nucleic
acid
molecule into a Der HMW-map protein in the animal. Nucleic acid molecules can
be
delivered to an animal in a variety of methods including, but not limited to,
(a)
administering a naked (i.e., not packaged in a viral coat or cellular
membrane) nucleic
acid molecule (e.g., as naked DNA or RNA molecules, such as is taught, for
example
in Wolff et al., 1990, Science 247, 1465-1468) or (b) administering a nucleic
acid
molecule packaged as a recombinant virus or as a recombinant cell (i.e., the
nucleic
acid molecule is delivered by a viral or cellular vehicle).
A naked nucleic acid molecule of the present invention includes a nucleic acid
molecule of the present invention and preferably includes a recombinant
molecule of
the present invention that preferably is replication, or otherwise
amplification,
competent. A naked nucleic acid of the present invention can comprise one or
more
nucleic acid molecules of the present invention in the form of, for example, a
bicistronic recombinant molecule having, for example one or more internal
ribosome
entry sites. Preferred naked nucleic acid molecules include at least a portion
of a viral
genome (i.e., a viral vector). Preferred viral vectors include those based on
alphaviruses, poxviruses, adenoviruses, herpesviruses, picornaviruses, and
retroviruses, with those based on alphaviruses (such as Sindbis or Semliki
virus),
species-specific heipesviruses and species-specific poxviruses being
particularly
preferred. Any suitable transcription control sequence can be used, including
those
disclosed as suitable for protein production. Particularly preferred
transcription
control sequence include cytomegalovirus intermediate early (preferably in
conjunction with Intron-A), Rous Sarcoma Virus long terminal repeat, and
tissue-
specific transcription control sequences, as well as transcription control
sequences
endogenous to viral vectors if viral vectors are used. The incorporation of
"strong"
poly(A) sequences are also preferred.
Naked nucleic acid molecules of the present invention can be administered by a
variety of methods. Suitable delivery methods include, for example,
intramuscular
injection, subcutaneous injection, intradermal injection, intradermal
scarification,
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-58-
particle bombardment, oral application, and nasal application, with
intramuscular
injection, intradermal injection, intradermal scarification and particle
bombardment
being preferred, and intramuscular injection being even more preferred. A
preferred
single dose of a naked DNA molecule ranges from about 1 nanogram (ng) to about
1
milligram (mg), depending on the route of administration and/or method of
delivery,
as can be determined by those skilled in the art. Examples of administration
methods
are disclosed, for example, in U.S. Patent No. 5,204,253, by Bruner, et al.,
issued April
20, 1993, PCT Publication No. WO 95/19799, published July 27, 1995, by McCabe,
and PCT Publication No. WO 95/05853, published March 2, 1995, by Carson, et
al.
Naked DNA molecules of the present invention can be contained in an aqueous
excipient (e.g., phosphate buffered saline) and/or with a carrier (e.g., lipid-
based
vehicles), or it can be bound to microparticles (e.g., gold particles).
A recombinant virus of the present invention includes a recombinant molecule
of the present invention that is packaged in a viral coat and that can be
expressed in an
animal after administration. Preferably, the recombinant molecule is packaging
deficient and/or encodes an attenuated virus. A number of recombinant viruses
can be
a used, including, but not limited to, those based on alphaviruses,
poxviruses,
adenoviruses, herpesviruses, picornaviruses and retroviruses. Preferred
recombinant
viruses are those based on alphaviruses (such as Sindbis virus), raccoon
poxviruses,
species-specific herpesvimses and species-specific poxviruses. An example of
methods to produce and use alphavirus recombinant virus is disclosed in PCT
Publication No. WO 94/17813, by Xiong et al., published August 18, 1994.
When administered to an animal, a recombinant virus of the present invention
infects cells within the recipient animal and directs the production of a
protein or RNA
nucleic acid molecule that is capable of reducing Der HMW-map protein-mediated
biological responses in the animal. For example, a recombinant virus
comprising a
Des HMW-map nucleic acid molecule of the present invention is administered
according to a protocol that results in the animal producing an amount of
protein or
RNA sufficient to reduce Der HMW-map protein-mediated biological responses. A
preferred single dose of a recombinant virus of the present invention is from
about 1 x
104 to about 1 x 10' virus plaque forming units (pfu) per kilogram body weight
of the
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-59-
animal. Administration protocols are similar to those described herein for
protein-
based compositions, with subcutaneous, intramuscular, intranasal and oral
administration routes being preferred.
A recombinant cell vaccine of the present invention includes recombinant cells
of the present invention that express at least one protein of the present
invention.
Preferred recombinant cells for this embodiment include Salfnonella, E. coli,
Listeria,
Mycobacterium, S. frugiperda, yeast, (including Saccharomyces cerevisiae ahd
Pichia
pastoris), BHK, CV-1, myoblast G8, COS (e.g., COS-7), Vero, MDCK and CRFK
recombinant cells. Recombinant cell vaccines of the present invention can be
administered in a variety of ways but have the advantage that they can be
administered
orally, preferably at doses ranging from about 10g to about 1012 cells per
kilogram
body weight. Administration protocols are similar to those described herein
for
protein-based vaccines. Recombinant cell vaccines can comprise whole cells,
cells
stripped of cell walls or cell lysates.
The efficacy of a therapeutic composition of the present invention to
desensitize an animal against mite allergy can be tested in a variety of ways
including,
but not limited to, using in vivo skin test methods disclosed herein,
detection of
cellular immunity activity in the treated animal, or determine levels of IgE
that bind
specifically to a Der HMW-map protein of the present invention. Methods to
determine cellular immunity activity and IgE levels in an animal are known to
those of
skill in the art. In one embodiment, therapeutic compositions can be tested in
animal
models such as dogs, cats, rabbits and mice, and can also be tested in humans.
Such
techniques are known to those skilled in the art.
Preferred nucleic acid molecules to use with a therapeutic composition of the
present invention include any Der HMW-map nucleic acid molecule disclosed
herein,
in particular SEQ ID N0:14, SEQ ID N0:16, SEQ ID N0:17, SEQ m N0:19, SEQ
ID N0:20, SEQ ID N0:22, SEQ ID N0:34, SEQ ID N0:36, SEQ ID N0:37, SEQ ID
N0:39, SEQ ID N0:40, SEQ ID N0:42, SEQ ID N0:43, SEQ ID N0:45 and/or a
nucleic acid sequence encoding a protein comprising the amino acid sequence
SEQ m
N0:33 and a complement thereof.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-60-
A recombinant cell useful in a therapeutic composition of the present
invention
includes recombinant cells of the present invention that comprises Der HMW-map
protein of the present invention. Preferred recombinant cells for this
embodiment
include Sahnoizella, E. coli, Listeria, MycobacteYiu~a, S. frugiper-da, yeast,
(including
Saccharomyces cerevisiae), BHK, CV-1, myoblast G8, COS (e.g., COS-7), Vero,
MDCK and CRFK recombinant cells. A recombinant cell of the present invention
can
be administered in a variety of ways but have the advantage that they can be
administered orally, preferably at doses ranging from about 108 to about 1012
cells per
kilogram body weight. Administration protocols are similar to those described
herein
for protein compositions. Recombinant cells can comprise whole cells, cells
stripped
of cell walls or cell lysates.
One embodiment of the present invention is a method of immunotherapy
comprising administering to an animal an effective amount of a therapeutic
composition comprising a Der HMW-map protein of the present invention.
Suitable
therapeutic compositions and methods of administration are disclosed herein.
According to the present invention, a therapeutic composition and method of
the
present invention can be used to prevent or alleviate symptoms associated with
mite
allergen pathogenesis.
The efficacy of a therapeutic composition of the present invention to effect
an
allergic response to Der HMW-map protein can be tested using standard methods
for
detecting Der HMW-map protein-mediated immunity including, but not limited to,
immediate hypersensitivity, delayed hypersensitivity, antibody-dependent
cellular
cytotoxicity (ADCC), immune complex activity, mitogenic activity, histamine
release
assays and other methods such as those described in Janeway et al., ibid.
The present invention also includes a therapeutic composition comprising one
or more therapeutic compounds of the present invention. Examples of such
therapeutic compounds include, for example, other allergens disclosed herein.
Therapeutic compositions of the present invention can be formulated in an
excipient that the animal to be treated can tolerate. Examples of such
excipients
include water, saline, Ringer's solution, dextrose solution, Hank's solution,
and other
aqueous physiologically balanced salt solutions. Nonaqueous vehicles, such as
fixed
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-61-
oils, sesame oil, ethyl oleate, or triglycerides may also be used. Other
useful
formulations include suspensions containing viscosity enhancing agents, such
as
sodium carboxymethylcellulose, sorbitol, or dextran. Excipients can also
contain
minor amounts of additives, such as substances that enhance isotonicity and
chemical
stability. Examples of buffers include phosphate buffer, bicarbonate buffer
and Tris
buffer, while examples of preservatives include thimerosal, o-cresol, formalin
and
benzyl alcohol. Standard formulations can either be liquid injectables or
solids which
can be taken up in a suitable liquid as a suspension or solution for
injection. Thus, in a
non-liquid formulation, the excipient can comprise dextrose, human serum
albumin,
preservatives, etc., to which sterile water or saline can be added prior to
administration.
In one embodiment of the present invention, a therapeutic composition can
include an adjuvant. Adjuvants are agents that are capable of enhancing the
immune
response of an animal to a specific antigen. Suitable adjuvants include, but
are not
limited to, cytokines, chemokines, and compounds that induce the production of
cytokines and chemokines (e.g., granulocyte macrophage colony stimulating
factor
(GM-CSF), granulocyte colony stimulating factor (G-CSF), macrophage colony
stimulating factor (M-CSF), colony stimulating factor (CSF), Flt-3 ligand,
erythropoietin (EPO), interleukin 2 (IL-2), interleukin-3 (IL-3), interleukin
4 (IL-4),
interleukin 5 (IL-5), interleukin 6 (IL-6), interleukin 7 (IL-7), interleukin
8 (IL-8),
interleukin 10 (IL-10), interleukin 12 (IL-12), interferon gamma, interferon
gamma
inducing factor I (IGIF), transforming growth factor beta, RANTES (regulated
upon
activation, normal T cell expressed and presumably secreted), macrophage
inflammatory proteins (e.g., MIP-1 alpha and MIP-1 beta), and Leishmania
elongation
initiating factor (LEIF); bacterial components (e.g., endotoxins, in
particular
superantigens, exotoxins and cell wall components); aluminum-based salts;
calcium-
based salts; silica; polynucleotides; toxoids; serum proteins, viral coat
proteins; block
copolymer adjuvants (e.g., Hunter's TitermaxTM adjuvant (VaxcelTM, Inc.
Norcross,
GA), Ribi adjuvants (Ribi ImmunoChem Research, Inc., Hamilton, MT); and
saponins
and their derivatives (e.g., Quil A (Superfos Biosector A/S, Denmark). Protein
adjuvants of the present invention can be delivered in the form of the protein
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-62-
themselves or of nucleic acid molecules encoding such proteins using the
methods
described herein.
In one embodiment of the present invention, a therapeutic composition can
include a carrier. Carriers include compounds that increase the half life of a
therapeutic composition in the treated animal. Suitable carriers include, but
are not
limited to, polymeric controlled release vehicles, biodegradable implants,
liposomes,
bacteria, viruses, other cells, oils, esters, and glycols.
One embodiment of the present invention is a controlled release formulation
that is capable of slowly releasing a composition of the present invention
into an
animal. As used herein, a controlled release formulation comprises a
composition of
the present invention in a controlled release vehicle. Suitable controlled
release
vehicles include, but are not limited to, biocompatible polymers, other
polymeric
matrices, capsules, microcapsules, microparticles, bolus preparations, osmotic
pumps,
diffusion devices, liposomes, lipospheres, and transdermal delivery systems.
Other
controlled release formulations of the present invention include liquids that,
upon
administration to an animal, form a solid or a gel in situ. Preferred
controlled release
formulations are biodegradable (i.e., bioerodible).
A preferred controlled release formulation of the present invention is capable
of releasing a therapeutic composition of the present invention into the blood
of an
animal at a constant rate sufficient to attain therapeutic dose levels of the
composition
to reduce mite allergy in the animal. As used herein, mite allergy refers to
cellular
responses that occur when mite allergens contact an animal. For example, IgE
that
specifically binds to mite allergen becomes coupled with Fc epsilon receptor,
resulting
in Fc epsilon receptor-mediated biological response including release of
biological
mediators, such as histamine, prostaglandins and/or proteases; that can
trigger clinical
symptoms of allergy. The therapeutic composition is preferably released over a
period
of time ranging from about 1 to about 12 months. A preferred controlled
release
formulation of the present invention is capable of effecting a treatment
preferably for
at least about 1 month, more preferably for at least about 3 months, even more
preferably for at least about 6 months, even more preferably for at least
about 9
months, and even more preferably for at least about 12 months.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-63-
Therapeutic compositions of the present invention can be sterilized by
conventional methods which do not result in protein degradation (e.g.,
filtration)
and/or lyophilized.
A therapeutic composition of the present invention can be administered to any
animal susceptible to mite allergy as herein described. Acceptable protocols
by which
to administer therapeutic compositions of the present invention in an
effective manner
can vary according to individual dose size, number of doses, frequency of dose
administration, and mode of administration. Determination of such protocols
can be
accomplished by those skilled in the art. An effective dose refers to a dose
capable of
treating an animal against hypersensitivity to mite allergens. Effective doses
can vary
depending upon, for example, the therapeutic composition used and the size and
type
of the recipient animal. Effective doses to immunomodulate an animal against
mite
allergens include doses administered over time that are capable of alleviating
a
hypersensitive response by an animal to mite allergens. For example, a first
tolerizing
dose can comprise an amount of a therapeutic composition of the present
invention
that causes a minimal hypersensitive response when administered to a hyper
sensitive
animal. A second tolerizing dose can comprise a greater amount of the same
therapeutic composition than the first dose. Effective tolerizing doses can
comprise
increasing concentrations of the therapeutic composition necessary to tolerize
an
animal such that the animal does not have a hypersensitive response to
exposure to
mite allergens. An effective dose to desensitize an animal can comprise a
concentration of a therapeutic composition of the present invention sufficient
to block
an animal from having a hypersensitive response to exposure to a mite allergen
present
in the environment of the animal. Effective desensitizing doses can include
repeated
doses having concentrations of a therapeutic composition that cause a minimal
hypersensitive response when administered to a hypersensitive animal.
A suitable single dose is a dose that is capable of treating an animal against
hypersensitivity to mite allergens when administered one or more times over a
suitable
time period. For example, a preferred single dose of a mite allergen, or
mimetope
therapeutic composition is from about 0.5 ng to about 1 g of the therapeutic
composition per kilogram body weight of the animal. Further treatments with
the
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-64-
therapeutic composition can be administered from about 1 day to 1 year after
the
original administration. Further treatments with the therapeutic composition
preferably are administered when the animal is no longer protected from
hypersensitive responses to mite allergens. Particular administration doses
and
schedules can be developed by one of skill in the art based upon the
parameters
discussed above. Modes of administration can include, but are not limited to,
subcutaneous, intradermal, intravenous, nasal, oral, transdermal and
intramuscular
routes.
A therapeutic composition of the present invention can be used in conjunction
with other compounds capable of modifying an animal's hypersensitivity to mite
allergens. For example, an animal can be treated with compounds capable of
modifying the function of a cell involved in a hypersensitive response,
compounds that
reduce allergic reactions, such as by systemic agents or anti-inflammatory
agents (e.g.,
anti-histamines, anti-steroid reagents, anti-inflammatory reagents and
reagents that
drive immunoglobulin heavy chain class switching from IgE to IgG). Suitable
compounds useful for modifying the function of a cell involved in a
hypersensitive
response include, but are not limited to, antihistamines, cromolyn sodium,
theophylline, cyclosporin A, adrenalin, cortisone, compounds capable of
regulating
cellular signal transduction, compounds capable of regulating adenosine 3',5'-
cyclic
phosphate (CAMP) activity, and compounds that block IgE activity, such as
peptides
from IgE or IgE specific Fc receptors, antibodies specific for peptides from
IgE or IgE-
specific Fc receptors, or antibodies capable of blocking binding of IgE to Fc
receptors.
Compositions of the present invention can be administered to any animal
having or susceptible to mite allergen hypersensitivity. Preferred animals to
treat
include mammals and birds, with felines, canines, equines, humans and other
pets,
work andlor economic food animals. Particularly preferred animals to protect
are
felines and canines.
Another aspect of the present invention includes a method for prescribing
treatment for animals susceptible to or having hypersensitivity to mite
allergens, using
a formulation of the present invention. A preferred method for prescribing
treatment
for mite allergen hypersensitivity, for example, comprises: (1) intradermally
injecting
into an animal at one site an effective amount of a formulation containing a
mite
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-65-
allergen of the present invention, or a mimetope thereof (suitable and
preferred
formulations are disclosed herein); (2) intradermally injecting into the
animal at a
second site an effective amount of a control solution; (3) evaluating if the
animal has
mite allergen hypersensitivity by measuring and comparing the wheal size
resulting
from injection of the formulation with the wheat size resulting from injection
of the
control solution; and (4) prescribing a treatment for the mite allergen
hypersensitivity.
An alternative preferred method for prescribing treatment for mite allergen
hypersensitivity comprises: (1) contacting a first portion of a sample of
bodily fluid
obtained from an animal to be tested with an effective amount of a formulation
containing mite allergen, or a mimetope thereof (suitable and preferred
formulations
are disclosed herein) to form a first immunocomplex solution; (2) contacting a
positive
control antibody to form a second immunocomplex solution; (3) evaluating if
the
animal has mite allergen hypersensitivity by measuring and comparing the
amount of
immunocomplex formation in the first and second immunocomplex solutions; and
(4)
prescribing a treatment for the mite allergen hypersensitivity. It is to be
noted that
similar methods can be used to prescribe treatment for allergies using mite
allergen
formulations as disclosed herein.
Another aspect of the present invention includes a method for monitoring
animals susceptible to or having mite allergen hypersensitivity, using a
formulation of
the present invention. In vivo and in vitro tests of the present invention can
be used to
test animals for mite allergen hypersensitivity prior to and following any
treatment for
mite allergen hypersensitivity. A preferred method to monitor treatment of
mite
allergen hypersensitivity (which can also be adapted to monitor treatment of
other
allergies) comprises: (1) intradermally injecting an animal at one site with
an effective
amount of a formulation containing mite allergen, or a mimetope thereof
(suitable and
preferred formulations are disclosed herein); (2) intradermally injecting an
effective
amount of a control solution into the animal at a second site; and (3)
determining if the
animal is desensitized to mite allergens by measuring and comparing the wheat
size
resulting from injection of the formulation with the wheat size resulting from
injection
of the control solution.
An alternative preferred method to monitor treatment of mite allergen
hypersensitivity (which can be adapted to monitor treatments of other
allergies)
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-66-
comprises: (1) contacting a first portion of a sample of bodily fluid obtained
from an
animal to be tested with an effective amount of a formulation containing a
mite
allergen or mimetope thereof (suitable and preferred formulations are
disclosed herein)
to form a first immunocomplex solution; (2) contacting a positive control
antibody to
form a second immunocomplex solution; and (3) determining if the animal is
desensitized to mite allergens by measuring and comparing the amount of
immunocomplex formation in the first and second immunocomplex solutions.
The present invention also includes antibodies capable of selectively binding
to
mite allergen, or mimetope thereof. Such an antibody is herein referred to as
an anti-
mite allergen antibody. As used herein, the term "selectively binds to" refers
to the
ability of such an antibody to preferentially bind to mite allergens and
mimetopes
thereof. In particular, the present invention includes antibodies capable of
selectively
binding to Der HMW-map protein. Binding can be measured using a variety of
methods known to those skilled in the art including immunoblot assays,
immunoprecipitation assays, enzyme immunoassays (e.g., ELISA),
radioimmunoassays, immunofluorescent antibody assays and immunoelectron
microscopy; see, for example, Sambrook et al., ibid.
Antibodies of the present invention can be either polyclonal or monoclonal
antibodies. Antibodies of the present invention include functional equivalents
such as
antibody fragments and genetically-engineered antibodies, including single
chain
antibodies, that are capable of selectively binding to at least one of the
epitopes of the
protein or mimetope used to obtain the antibodies. Preferred antibodies are
raised in
response to Der HMW-map proteins, or mimetopes thereof. More preferred Der
HMW-map protein against which to raise an antibody includes at least a portion
of a
protein having the amino acid sequence SEQ ID NO:1, SEQ ID N0:2, SEQ ID N0:3,
SEQ ID N0:4, SEQ ID N0:5, SEQ ID N0:6, SEQ ID N0:7, SEQ ID N0:8, SEQ ID
N0:9, SEQ ID NO:10, SEQ ID NO:l 1, SEQ ID N0:12, SEQ ID N0:13, SEQ ID
N0:15, SEQ ID N0:18, SEQ ID N0:21, SEQ ID N0:23, SEQ ID N0:24, SEQ ID
N0:30, SEQ ID N0:31, SEQ ID N0:32, SEQ ID N0:33, SEQ ID N0:33, SEQ ID
N0:35, SEQ ID N0:38, SEQ ID N0:41, and/or SEQ ID N0:44, or homologs thereof.
Preferably, an antibody of the present invention has a single site binding
affinity of
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-67-
from about 103M-1 to about 1012M-1 for a Der HMWWmap protein of the present
invention.
A preferred method to produce antibodies of the present invention includes
administering to an animal an effective amount of a Der HMW-map protein or
mimetope thereof to produce the antibody and recovering the antibodies.
Antibodies
raised against defined products or mimetopes can be advantageous because such
antibodies are not substantially contaminated with antibodies against other
substances
that might otherwise cause interference in a diagnostic assay or side effects
if used in a
therapeutic composition.
Antibodies of the present invention have a variety of potential uses that are
within the scope of the present invention. For example, such antibodies can be
used
(a) as vaccines to passively immunize an animal in order to protect the animal
from
mite allergen hypersensitivity, (b) as positive controls in test kits, and/or
(c) as tools to
recover desired mite allergens from a mixture of proteins and other
contaminants.
The following examples are provided for the purposes of illustration and are
not intended to limit the scope of the present invention.
EXAMPLES
It is to be noted that the Examples include a number of molecular biology,
microbiology, immunology and biochemistry techniques considered to be known to
those skilled in the art. Disclosure of such techniques can be found, for
example, in
Sambrook et al., ibid., and related references.
Example 1
This example describes the identification of high molecular weight proteins
that bind to IgE from dogs known to be allergic to mite allergens.
About 5.5 grams (g) of frozen wet Dermataplzagoides farazzae (Derv mites
(available from Bayer Allergy, Spokane, WA) were homogenized in a ground glass
homogenizer, in either about 30 ml of phosphate buffered saline (PBS) or 0.1 M
Tris-HCI, pH ~, each containing complete protease inhibitors (available from
Boehringer Mannheim, Indianapolis, IN) to obtain a Der f crude extract. The
resulting
supernatants were collected and each concentrated in a Centriprep 30
concentrator
(available from Amicon, Beverly, MA) by centrifugation at 16,000 x g for about
30
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-68-
minutes. The concentrated supernatants were applied to separate Sephacryl S-
100
columns (2.7 x 70 cm; available from Pharmacia, Piscataway, NJ) in PBS or 0.1
M
Tris-HCl, pH 8, respectively. The excluded fractions from each column were
pooled.
Fractions were dialyzed against 10 mM Tris-HCI, pH 8, when PBS was used. The
fractions were applied to separate Q-Sepharose columns (2.5 x 5 cm; available
from
Pharmacia). The Q-Sepharose column was pre-equilibrated in 10 mM Tris-HCl, pH
8,
when the fractions containing 0.1 M Tris-HCI, pH 8 were used. Each column was
sequentially eluted with about 45 ml of 10 mM Tris-HCl, pH 8, then 0.1 M Tris-
HCI,
pH 8, then 0.2 M Tris-HCI, pH 8, then 0.3 M Tris-HCI, pH 8, then 0.4 M Tris-
HCI, pH
8 and then 0.5 M Tris-HCI, pH 8. Fractions were collected from each elution
step.
Each fraction was analyzed by western blot for the presence of protein that
bound to
IgE antibodies present in dog sera isolated from dogs known to be allergic to
mite
allergens (referred to herein as mite allergic dog antisera or mite allergic
antisera).
Specifically, proteins contained in the fractions were resolved by
12°70 Tris-glycine
SDS-PAGE and then blotted onto nitrocellulose. The blot was incubated with a
pool
of sera obtained from dogs known to be allergic to mite allergens, diluted
1:20, using
standard buffers. The blot was incubated and then washed using standard
procedures.
The blot was then incubated with the mouse monoclonal anti-dog IgE antibody
DEI38
(1 mg/ml, 1:1000 dilution). The blot was incubated and then washed using
standard
procedures. The blot was then incubated with donkey anti-mouse IgG antibody
conjugated to horseradish peroxidase ( 1:1000 dilution; available from Jackson
Labs,
Maine). The presence of HRP-conjugated antibody bound to the blot was detected
using standard techniques. An about 70-kD protein was identified in the 0.2 M
Tris-HCl, pH 8 fraction, an about 98-kD protein and an about 109-kD protein
were
identified in the 0.3 M Tris-HCI, pH 8 fraction.
The fraction described above that was eluted using 0.3 M Tris-HCI, pH 8 was
concentrated in a Centriprep 30 concentrator and then diluted in 20 mM Na-Ac,
pH
5.6. The diluted fraction was then applied to a PolyCat A HPLC canon exchange
column (available from PoIyLC, Columbia, MD). The column was eluted with about
10 ml of 20 mM Na-Ac, pH 5.6, and then with about 45 ml of a linear gradient
from 0
to 0.5 M NaCl in the 20 mM Na-Ac, pH 5.6 buffer at a flow rate of about
lml/min. '
Fractions were collected from the elution procedure and assayed for the
presence of
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-69-
high molecular weight proteins using the mite allergic antisera and western
blot
protocol described above. Fractions containing the high molecular weight pr
oteins
were pooled. Trifluoroacetic acid (TFA) was added to a concentration of about
0.05%.
The solution was applied to a TSK-Gel TMS-250 Cl reverse phase column
(available
from TosoHaas, Montgomeryville, PA) that had been pre-equilibrated in 80%
solvent
A and 20% solvent B. Solvent A was composed of about 0.05% TFA in water and
solvent B was composed of about 0.05% TFA in 90% acetonitrile in water. The
column was eluted with about 5 ml of 20% solvent B and then with 36 ml of a
linear
gradient of about 20% to about 70% solvent B at 0.6 ml/min. The proteins
eluted from
the column were resolved by 12% Tris-Glycine PAGE. The gel, was stained with
Comassie blue. The stained gel is shown in Fig. 1. Lane 1 contains Mark-12
protein
molecular weight markers (available from Novex, San Diego, CA), lane 2
contains the
protein eluted from the reverse phase column, and lane 3 contains SeeBlueT"~
protein
molecular weight markers (available from Novex). Two major proteins were
identified in the eluant. The molecular weights of the proteins were
determined using
a BioRadTM Multi-AnalystTM/PC Image System (available from BioRad Corp.). The
higher molecular weight protein in lane 2 of Fig. 1 was determined to be about
109
kD, referred to herein as mite allergen protein A (mapA). The lower molecular
weight
protein in lane 2 of Fig. 1 was determined to be about 98 kD, referred to
herein as mite
allergen protein B (mapB). The purity of the combined proteins was greater
than 85%
purity, i.e., less than 15% impurities. This purified eluant is referred to
herein as the
D. fariraae high molecular weight map (HMW-map) composition.
Example 2
This example describes N-terminal sequencing of proteins in the D. fariaae
HMW-map composition.
Proteins contained in the 0.3 M Tris-HCI, pH 8 fraction obtained as described
above in Example 1 were resolved by SDS-PAGE using a 12% Tris-glycine
polyacrylamide-SDS gel, followed by coomasie staining. The proteins were
blotted
onto PVDF, stained with Coomasie R-250 and destained using standard
procedures.
The proteins corresponding to the about 98 kD and about 109 kD bands were
excised
and subjected separately to N-terminal amino acid sequencing using techniques
known
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-70-
to those skilled in the art. A partial N-terminal amino acid sequence of about
14
amino acids was deduced for both proteins and the sequences were determined to
be
identical. The N-terminal amino acid sequence is represented herein as SEQ m
NO:1,
having the amino acid sequence: Ser Ile Lys Arg Asp His Asn Asp Tyr Ser Lys
Asn
Pro Met.
The proteins in the D. fariTaae HMW-map composition were also submitted to
proteolytic cleavage in order to obtain internal amino acid sequence data.
Specifically,
the D. farihae HMW-map composition was cleaved with Endoproteinase Asp-N
(available from Boehringer Mannheim Biochemica, Indianapolis, IN) using
methods
standard in the art. The digested protein was then resolved by HPLC using the
method
described by Stone et al., Enzymatic Digestion of Proteins and HPLC Peptide
Isolation, in A Practical Guide to Protein and Peptide Purification for
Microsequencing, PT Matsudaira ed., Academic Press, San Diego, CA. Twelve
proteolytic fragments were isolated, that are referred to herein as map(1),
map(2),
map(3), map(4), map(5), map(6), map(7), map(8), map(9), map(10), map(11) and
map(12).
The N-terminal partial amino acid sequence of map(1) was determined to be
Asp Tyr Glu Tyr Pro Gly Ser Arg Leu Gly Asn Pro Lys Ala Pro Leu Tyr Lys Arg
Pro,
also denoted SEQ ID N0:2. The N-terminal partial amino acid sequence of map(2)
was determined to be Asp Ile Pro His Pro Thr Asn Ile His Lys Tyr Leu Val Cys
Glu
Ser Val Asn Gly Gly, also denoted SEQ 117 N0:3. The N-terminal partial amino
acid
sequence of map(3) was determined to be Asp Pro Ala Lys Gly Met Ser Pro Pro
Gly
Phe Ile Val Gly Glu Glu Gly Val Leu Ser, also denoted SEQ ID N0:4. The N-
terminal partial amino acid sequence of map(4) was determined to be Asp Glu
Lys
Asn Ser Phe Glu Cys Ile Leu Gly Pro, also denoted SEQ ID N0:5. The N-terminal
partial amino acid sequence of map(5) was determined to be Asp Ala Phe Glu Pro
His
Gly Tyr Leu Leu Thr Ala Ala Val Ser Pro Gly Lys, also denoted SEQ ID N0:6. The
N-terminal partial amino acid sequence of map(6) was determined to be Asp Lys
Gln
Asn Tyr Leu Ala Leu Val Arg Glu Leu Lys, also denoted SEQ ID N0:7. The N-
terminal partial amino acid sequence of map(7) was determined to be Asp Met
Ala Gln
Asn Tyr Lys Tyr Arg Gln Gln Phe Ile Gln Ser Val Leu Asn Asn Gly Ala Thr Arg
Gln,
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-71-
also denoted SEQ ID N0:8. The N-terminal partial amino acid sequence of map(8)
was determined to be Asp Glu Xaa Asn Val Met Xaa Tyr Val Leu Tyr Thr Met His
Tyr Tyr Leu Asn Asn Gly Ala Thr Arg, also denoted SEQ ID N0:9, in which Xaa
represents any amino acid. The N-terminal partial amino acid sequence of
map(9) was
determined to be Asp Lys Leu Val Met Gly Val Pro Phe Tyr Gly Arg Ala Xaa Ser
Ile
Glu, also denoted SEQ ll~ NO:10, in which Xaa represents any amino acid. The N-
terminal partial amino acid sequence of map(10) was determined to be Asp Ile
Pro His
Pro Thr Asn Ile His Lys Tyr Leu Val Cys Glu Ser Val Asn Gly, also denoted SEQ
ID
N0:11. The N-terminal partial amino acid sequence of map(11) was determined to
be
Asp Tyr Ala Lys Asn Pro Lys Arg Ile Val Cys Ile Val Gly Thr Glu Gly Val Leu
Ser,
also denoted SEQ ID N0:12. The N-terminal partial amino acid sequence of
map(12)
was determined to be Asp Pro Ala Lys Gly Met Ser Pro Pro Gly He Ile Val Gly
Glu
Glu Gly Val Leu Ser, also denoted SEQ ID N0:13. Since the amino acid sequences
for map(1), map(2), map(3), map(4), map(5), map(6), map(7), map(8), map(9),
map(10), map(11), map(12), and map(13) were generated from a mixture of mapA
and
mapB proteins, these sequences do not necessarily represent partial sequences
of a
single protein.
Example 3
This example describes the purification of a 70-kD protein that binds to IgE
from dogs known to be allergic to mite allergens.
The fraction described above in Example 1 that was eluted using 0.2 M
Tris-HCI, pH 8 was concentrated in a Centriprep 30 concentrator and then
diluted in
20 mM Na-Ac, pH 5.6. The diluted protein was then applied to a PolyCat A HPLC
cation exchange column. The column was eluted with about 10 ml of 20 mM Na-Ac,
pH 5.6, and then with about 45 ml of a linear gradient from 0 to 0.5 M NaCI in
the 20
mM Na-Ac, pH 5.6 buffer at a flow rate of about 1 ml/min. Fractions were
collected
from the elution procedure and assayed for the presence of 70-kD protein using
the
mite allergic antisera and western blot protocol described above. Fractions
containing
the 70-kD protein were pooled. Trifluoroacetic acid (TFA) was added to a
concentration of about 0.05%. The solution was applied to a TSI~-Gel TMS-250
C1
reverse phase column that had been pre-equilibrated in 80% solvent A and 20%
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
_72_
solvent B. Solvent A was composed of about 0.05% TFA in water and solvent B
was
composed of about 0.05% TFA in 90%o acetonitrile in water. The column was
eluted
with about 3 ml of 20% solvent B and then with 36 ml of a linear gradient of
about
20% to about 70% solvent B at 0.6 ml/min. An about 70-kD protein of >90%
purity
was obtained. The about 70-kD protein is referred to herein as mapC.
N-terminal sequence of a region on an SDS-PAGE corresponding to the 70 kD
protein (mapC) was obtained as described in Example 2. An N-terminal amino
acid
sequence of about 21 amino acids was deduced with an 80% confidence level, and
is
represented herein as SEQ ID N0:33, having the following amino acid sequence:
Gln
Ser Arg Asp Arg Asn Asp Lys Pro Tyr Xaa Ile Val Lys Lys Lys Lys Lys Ala Leu
Asp.
Example 4
This example describes the binding of the D. faz~iyzae HMW-map composition
(i.e., containing mapA and mapB) to canine IgE in dog sera isolated from dogs
known
to be allergic to mite allergens.
Multiple wells of an Immulon II microtiter plate were coated with about 100
nanograms per well (nglwell) of a D. fat ireae HMW-map composition isolated
according to the method described above in Example l, diluted in CBC buffer.
The
plate was incubated overnight at 4°C. Following incubation, the D.
farin.ae HMW-map
composition-containing solution was removed from the plate, and the plate was
blotted
dry. The plate was then blocked using about 200 ,ul/well of 4.0% fetal calf
serum
contained in phosphate buffered saline (PBS) having 0.05% Tween-20 (PBSTFCS)
for about 1 hour at room temperature. The plate was then washed four times
with
0.05% Tween-20 in PBS (PBST) using an automatic washer (available from
Dynatech,
Chantilly, VA). About 100 ,ul/well of a 1:10 dilution in PBSTFCS of serum
samples
isolated from different dogs known to be sensitive to mite allergens in
intradermal skin
tests were added to the plate. A negative control group of sera was also added
to the
plate comprising a combination of sera from six dogs that were raised in a
barrier
facility (available from Harlan Bioproducts, Indianapolis, IN). Some wells did
not
receive dog sera so that background binding levels could be determined. The
plate
was incubated for about 1 hour at room temperature and then washed four times
with
PBST. About 100 ,ul/well of a 1:4000 dilution of 40 ,ug/ml biotinylated human
FcER
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-73-
alpha chain protein (produced as described in Frank et al., WO 98/23964,
published
November 24, 1997) contained in PBSTFCS was added. The plate was incubated for
about 1 hour at room temperature and then washed four times with PBST. About
100
,u! of about 0.25 ,ug/ml streptavidin conjugated to horseradish peroxidase
(available
from Kirkegaard and Perry Laboratories (KPL), Gaithersburg, MD; diluted in
PBST)
was added to each well that received experimental or control samples. The
plates were
then incubated for about 1 hour at room temperature and washed four times with
PBST. About 100 ,u! of TMB substrate (available from KPL), that had been
pre-warmed to room temperature, was added to each well. The plate was then
incubated for about 10 minutes at room temperature and then about 100 ,ul/well
of
Stop Solution (available from KPL) was added. Optical densities (0.D.) of
wells were
read on a Spectramax Microtiter Plate (available from Molecular Devices Inc.)
reader
at 450 nm within 10 minutes of adding the stop solution.
The O.D. readings obtained using the negative control sample and the
background wells were 0 O.D. Sera from 5 of 26 mite allergen sensitive dogs
generated O.D. readings between about 2,000 O.D. and about 3,200 O.D. Sera
from 3
other mite allergen sensitive dogs generated O.D. readings between about 1,000
O.D.
and 2,000 O.D. Sera from 3 other mite allergen sensitive dogs generated O.D.
readings between about 500 O.D. and 1,000 O.D. Sera from 7 other mite allergen
sensitive dogs generated O.D. readings between about 200 O.D. and 500 O.D.
Sera
from 6 other mite allergen sensitive dogs generated O.D. readings less than 50
O.D.
Thus, the results indicate that sera from dogs known to be sensitive to mite
allergens
contain IgE antibodies that bind specifically to the mapA and mapB proteins of
the
present invention.
Example 5
This example describes the binding of the 70-kD D. farihae protein to canine
IgE in dog sera isolated from dogs known to be allergic to mite allergens.
Multiple wells of an Immulon II microtiter plate were coated with about 100
nglwell of 70-kD D. fariTZae protein (referred to herein as mapC) isolated
according to
the method described above in Examples 1 and 3, diluted in CBC buffer. The
plate
was incubated overnight at 4°C. The plate was blocked and washed using
the method
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-74-
described in Example 4. About 100 ,ul/well of a 1:10 dilution in PBSTFCS of
serum
samples isolated from different dogs known to be sensitive to mite allergens
in
intradermal skin tests were added to the plate. Negative control samples were
also
added to the plate comprising SPF serum samples (serum from dogs maintained in
a
barrier facility and therefore never exposed to mite allergens). Some wells
did not
receive dog sera so that background binding levels could be determined. The
plate
was incubated for about 1 hour at room temperature and then washed four times
with
PEST. Biotinylated human FcER alpha chain protein was then added and the
presence
of IgE bound to the plate was detected using the methods described in Example
4.
The O.D. readings obtained using the negative control sample and the
background wells were 0 O.D. Sera from 3 of 26 mite allergen sensitive dogs
generated O.D. readings between about 1,500 O.D. and about 2,700 O.D. Sera
from 5
other mite allergen sensitive dogs generated O.D. readings between about 800
and
about 1,500 O.D. Sera from 4 other mite allergen sensitive dogs generated O.D.
readings between about 500 O.D. and about 800 O.D. Sera from 6 other mite
allergen
sensitive dogs generated O.D. readings between about 200 O.D. and 500 O.D.
Sera
from 8 other mite allergen sensitive dogs generated O.D. readings less than 50
O.D.
Thus, the results indicate that sera from dogs known to be sensitive to mite
allergens
contain IgE antibodies that bind specifically to the mapC protein of the
present
invention.
Example 6
This example describes the binding of mapA, mapB or mapC proteins to feline
IgE in eat sera isolated from cats shown by ira vitro testing to be
hypersensitive to mite
allergens.
Multiple wells of an Tmmulon II microtiter plate were coated with about 100
ng/well of a D. farircae HMW-map composition (isolated according to the method
described above in Example 1) and 70-kD D. farifaae protein (isolated
according to the
method described above in Example 3). Other wells of the plate were coated
with 400
ng/well of whole Derfnatophagoides pteronyssius extract (available from Greer
Laboratories, Inc., Lenoir, NC; concentrated 8-fold prior to use) or whole D.
farifZae
extract (available from Miles, Inc., Elkhart, IN). All samples were diluted in
CBC
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-75-
buffer. The plates were incubated overnight at 4°C. The plates were
blocked and
washed using the method described in Example 4. About 100 ,ul/well of a 1:10
dilution in PBSTFCS of serum samples isolated from different cats known to be
sensitive to mite allergens in i~z vitro allergen testing were added to the
plate. Sera
from seven control cats (#15, #16, #17, #18, #19, #20, and #21), shown not to
be
sensitive by ire vitro test to dust mite allergens, were also tested. Some
wells did not
receive cat sera so that background binding levels could be determined. The
plate was
incubated for about 1 hour at room temperature and then washed four times with
PBST. Biotinylated human FcER alpha chain protein was then added and the
presence of IgE bound to the plate was detected using the methods described in
Example 4.
The results are shown below in Table 1. All values represent O.D. values
times 1,000. HDM refers to cats that are sensitive to house dust mite
allergens (by
serological test, i.e. an ELISA to whole D. fariaae extract).
Table 1.
Cat HDM Whole Der Whole Der mapA and mapBmapC
# p ' f
1 + 54 173 211 400
2 + 437 454 245 352
3 + 96 88 17 36
4 + 35 179 278 758
5 + 123 23 0 0
6 + 2 10 0 0
7 + 84 321 439 445
8 + 125 333 611 599
9 + 2459 2737 1613 507
10 + 17 0 0 0
11 + 146 347 243 586
12 + 31 100 102 223
13 + 56 171 267 292
14 + 121 146 163 185
15 - 0 0 0 8
16 - 0 0 0 0
17 - 0 0 0 0
18 - 0 0 0 0
19 - 0 0 0 0
20 - 0 0 0 0
21 - 23 0 0 0
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-76-
The results indicate that sera from some of the cats known to be sensitive to
mite
allergens contain IgE antibodies that bound specifically to the mapA, mapB or
mapC
proteins of the present invention. In addition, some sera containing IgE that
bound to
the mapA, mapB or mapC proteins also contain IgE antibodies that bound to
whole D.
pterouyssius extract. The control sera did not contain IgE antibodies that
bound to
either the mapA, mapB or mapC proteins of the present invention.
Example 7
This example demonstrates the ability of the D. faf-iozae HMW-map
composition to induce a hypersensitive response in dogs.
To determine whether the D. fariT2ae HMW-map composition described in
Example 1 was capable of inducing an allergic response in animals susceptible
to dust
mite allergic responses, skin tests were performed on dogs that actively
demonstrate
clinical signs for dust mite allergy (referred to herein as atopic dogs).
Normal dogs
include dogs that do not show symptoms of mite allergy but may be susceptible
to a
mite allergic response. Each dog (i.e., 4 normal and 4 atopic dogs) was shaved
in the
lateral thorax/abdominal area and intradermally injected in different sites in
that area
with an about 1:50,000 dilution of D. fariuae crude extract isolated by the
method
described in Example l, with about 2 ,ug of the purified D. fari~za.e HMW-map
composition and/or with control solutions, i.e., saline, as a negative
control, and a
1:1000 dilution of histamine as a positive control. All four normal dogs and
all 4
atopic dogs received D. farihae whole extract. Three of the normal dogs and 2
of the
atopic dogs received the D. faf~is2ae HMW-map composition. All 8 of the dogs
received both the negative and positive control samples. The total volume per
injection was 50 microliters (,u1), with the compositions and controls being
diluted in
saline. The injections were administered as single injections.
All injection sites were objectively measured in millimeters (mm) at 15
minutes and scored either (+) or (-) when compared with the control samples.
The
subjective scoring was performed by Andrew Hillier, D.V.M., at Ohio State
University, Columbus, OH. The results are shown in Table 2:
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
_77_
Table 2.
NdrmalNormal:.Normal: NormalAtopicAtopic AtopieAtopic
Dog'1.I)og.2 Dog Dog Dog Dog Dog Dog
' 3. 4 1 2 3 4
Whole Extract+ + + - + + - -
HMW map + + - n/a + - n/a n/a
Neg. Control- - - - - - - -
Histamine + + + + + + + +
n/a = not applicable
The results indicate that the D. faritZae HMW-map composition was capable of
inducing an immediate hypersensitive response in dogs including atopic dogs.
Thus,
the HMW-map composition is sufficiently allergenic to induce a hypersensitive
response in dogs including atopic dogs.
Table 3 describes the results of the following experiment. IgE to the HMW-
map composition was measured in the serum of three groups of dogs: D. farin.ae
allergic (HDM-AD), atopic (to other allergens) but not HDM allergic (AD), and
naive
dogs using ELISA. These dogs were also tested by intradermal skin test to D.
farii2ae
whole extract and to the HMW-map composition.
Table 3. Skin test and ELISA data for D. farioae whole extract and for
HMW-map composition in D. farihae-allergic, atopic but not
HDM-allergic, and naive dogs.
Dog Clinical Df IDST Df ELXSA:~y~!-mapHMW-map
status 1:50;000 IDST'1u ELTSA
:
1 HDM-AD + 1968 + 2876
2 HDM-AD + 407 - 954
3 HDM-AD + 3921 + 3465
4 HDM-AD + 153 + 198
5 HDM-AD + 1712 + 997
6 HDM-AD + 1833 + 2006
7 HDM-AD + 4200 + 4200
8 HDM-AD + 2851 + 3559
9 HDM-AD + 122 + 209
10 HDM-AD + 1627 + 566
11 HDM-AD + 1185 + 1307
12 HDM-AD + 308 + 101
13 HDM-AD + 341 + 433
14 AD - 1 - 0
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
_78_
Dog Clinical Df IDST Df ELISAHMWmap. HMW-map
status 1:50,000 IDST 1u ELISA
v
15 AD - 8 - 2
16 AD ND 66 ND 87
17 Normal - 24 - 40
18 Normal - 53 ND 369
19 Normal - 37 - 21
20 SPF bea ND 0 ND 0
1e
21~ SPF beagleND I 6 ND 1
I
All dogs that were positive by ELISA for whole D. faf°ihae extract were
also positive
for the HMW-map composition allergen. Of the eight dogs that were ELISA
negative
for whole D. farireae extract, 7 of 8 were also negative for the HMW-map
composition.
Example 8
This example describes the isolation of nucleic acid molecules encoding a Der
HMW-map composition of the present invention.
Der HMW-map composition nucleic acid molecules were identified and
isolated as follows.
A. Preparation of a Dermatophagoides farinae cDNA Library.
A Dermatophagoides fariizae cDNA library was prepared as follows. Total
RNA was extracted from about 2 grams of flash frozen and pulverized house dust
mites, using an acid-guanidinium-phenol-chloroform method similar to that
described
by Chomzynski et al., 1987, A~eal. Biochen2. 162,156-159. Poly A+ selected RNA
was
separated from the total RNA preparation by oligo-dT cellulose chromatography
using
the mRNA Purification Kit ( available from Pharmacia Biotech, Newark, NJ),
according to the method recommended by the manufacturer. A cDNA library was
constructed in lambda-Uni-ZAPTM XR vector (available from Stratagene), using
Stratagene's ZAP-cDNA Synthesis Kit protocol. Approximately 5 ~,g of Poly A~
RNA
was used to produce the Dermatoplzadoides farin.ae cDNA library.
B. Preparation of PCR primers.
Further N-terminal amino acid sequence analysis was performed according to
the methods described above in Example 2. A partial N-terminal amino acid
sequence
of 34 amino acids was deduced and is represented by SEQ ID N0:24, having the
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-79-
amino acid sequence: Ser Ile Lys Arg Asp His Asn Asp Tyr Ser Lys Asn Pro Met
Met
Ile Val Xaa Tyr Tyr Gly Gly Ser Ser Gly Tyr Gln Ser Xaa Lys Arg Xaa Xaa Thr
(wherein "Xaa" represents any amino acid residue). The amino acid sequences of
SEQ
ll~ N0:4 (described above in Example 2) and SEQ ID N0:24 were used to design
synthetic oligonucleotide primers. Sense primer Derfl derived from SEQ m
N0:24,
having the nucleotide sequence 5' AAA CGT GAT CAT AAY. GAT TAY TCN AAR
AAY C 3' (wherein Y represents C or T, R represents A or G, and N represents
A, C,
T or G), designated SEQ ID NO: 25 or sense primer Derf2, derived from SEQ ID
N0:24, having the nucleotide sequence 5' AAA CGT GAT CAT AAY GAT TAY
AGY AAR AAY C 3', designated SEQ ID N0:26, were used in combination with
antisense primer Derf3 deriveel from SEQ ID N0:4, having the nucleotide
sequence 5'
CCT TCT TCA CCN ACR ATC AAN CC 3', denoted SEQ 117 N0:27, or antisense
primer Derf4 derived from SEQ ID NO:4, having the nucleotide sequence 5' CCT
TCT TCA CCN ACR ATG AAN CC 3', denoted SEQ ID N0:28.
The foregoing primers were then used to screen the Der f cDNA library using
standard polymerase chain reaction amplification (PCR) techniques. All
attempts to
identify a cDNA that hybridized to the primers failed.
C. Immunoscreening the D. farihae cDNA library using anti-Der HMW-
mapcomposition antibodies.
Since attempts to isolate a cDNA clone using PCR methods failed, the
inventors screened the D. farihae cDNA library using an antiserum produced as
follows. Protein isolated according to the method described above in Example 1
was
used as a source of antigen to generate rabbit polyclonal antibodies, referred
to herein
as anti-Der HMW-map composition antibodies. The preparation of rabbit
polyclonal
antibodies was carried out using standard techniques.
About 7.5 ml of Escherichia coli (XL1 Blue, O.D.boo=0.5) was incubated with
3.0 x 104 pfu of phage from a Dermatophagoides farircae ZAP-cDNA library ( 1.8
x
109pfu/ml), at 37°C for 15 min and plated in 30 ml Luria-Bertani (LB)
medium agar
plates (150 mm). The plates were incubated at 37°C over night. Each
plate was then
overlaid with an IPTG (10 mM) treated nitrocellulose filter for about 4 hours
at 37°C.
The filters were then removed and washed with Tris buffered saline (pH 7.5)
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-80-
containing 0.1% Tween (TBST), for 5 minutes. The filters were blocked with a
solution of 1% dried pwder milk, 1% BSA, 2% goat serum and 0.15% gelatin,
prepared in TBST, for 2 hours at room temperature. Filters were then incubated
with
the anti-Der HMW-map composition antibodies at a dilution of 1:1000, contained
in
the above blocking solution at 4°C, overnight. The mixture was then
incubated with a
donkey anti-rabbit IgG antibody conjugated to horseradish peroxidase
(available from
Jackson ImmunoResearch, West Grove, PN) for 2 hours at room temperature. All
of
the filters were washed with blocking solution contained in TBST (3 x 15
min/wash)
between each incubation. All of the filters were then treated to a final wash
in Tris
buffered saline (pH 7.5) for 5 minutes at room temperature. Immunocomplexed
plaques were identified by immersing the filters into the developing solution
(TMB
Peroxidase Substrate/TMB Peroxidase Solution/TMB Membrane Enhancer from
I~irkegaard & Perry Laboratories) at 1/1/0.1 volume ratio to produce a color
reaction.
One hundred and twenty three plaques were identified and 50 plaques were
further
plaque purified two more times under the same immunoscreening condition as
described above.
D. PCR screening of purified phage plugs
The phage plugs identified in the foregoing immunoscreening study were then
further analyzed by PCR amplification using the primers described above in
section
8B. DNA from the 50 plaques was amplified using a mixture of the 4 primers
identified by SEQ ll~ NO: 25, SEQ ID N0:26, SEQ ID N0:27 and SEQ ID N0:28.
PCR amplification was conducted using standard techniques. One resulting PCR
amplification product comprised a fragment of about 700 nucleotides. The PCR
product was cloned into the InVitrogen, Corp., TAT"' cloning vector
(procedures
provided by InVitrogen, Corp.) and subjected to DNA sequence analysis using
standard techniques. The phagemid from the purified phage that were determined
to
contain sequences encoded in the 700-by PCR product were rescued and subjected
to
DNA sequence analysis using standard techniques.
A clone was isolated that included about a 1752-nucleotide insert, referred to
herein as nDerf981~sz. Nucleic acid sequence was obtained using standard
techniques
from nDerf981~sz, to yield a Derrnatophagoides farihae nucleic acid molecule
named
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-81-
nDerf981~s~ composed of a coding strand having nucleic acid sequence SEQ ID
N0:14
and a complementary strand having a nucleic acid sequence SEQ ID N0:16.
Translation of SEQ ID N0:14 suggests that nucleic acid molecule nDerf981~sz
encodes
a full-length flea protein of about 555 amino acids, referred to herein as
PDerf98sss~
having amino acid sequence SEQ ID N0:15, assuming an open reading frame in
which the first codon spans from nucleotide 1 through nucleotide 3 of SEQ ID
N0:14
and a stop codon spanning from nucleotide 1666 through nucleotide 1668 of SEQ
ID
N0:14. The amino acid sequence of PDerf98sss is encoded by the nucleic acid
molecule'nDerf98166s, having a coding strand with the nucleic acid sequence
SEQ ID
NO:17 and a complementary strand with the nucleic acid sequence SEQ WN0:19.
PDen98sss , also represented by SEQ ID N0:18, has an estimated molecular
weight of
about 63.2 kD and an estimated pI of about 5.33. Analysis of SEQ ID N0:15
suggests
the presence of a signal peptide spanning from about amino acid 1 through
about
amino acid 19. The proposed mature protein, denoted herein as PDerfs36,
contains
about 536 amino acids, the sequence of which is represented herein as SEQ ID
N0:21,
and is encoded by a nucleic acid molecule referred to herein as nDerf981~os~
represented by SEQ ID N0:20, the coding strand, and SEQ ID N0:22, the
complementary strand. The amino acid sequence of flea PDerf98s36 (i.e. SEQ ID
N0:21) predicts that PDerf98s3s has an estimated molecular weight of 61.2 kD,
and an
estimated pI of about 5.26.
Comparison of amino acid sequence SEQ ID N0:15 with amino acid
sequences reported in GenBank indicates that SEQ ID N0:15 showed the most
homology, i.e., about 42% identity, with a chitinase protein fromAfzopl~eles
gambiae
(GenBank accession number 2654602). Comparison of nucleic acid sequence SEQ ID
N0:17 with nucleic acid sequences reported in GenBank indicates that SEQ ID
N0:17
showed the most homology, i.e., about 58% identity between SEQ ID N0:17 and
Chelo~us ,rp. venom chitinase mRNA (GenBank accession number U10422).
Example 9
This example describes the purification of a 60-kD protein that binds to IgE
from dogs known to be allergic to mite allergens and partial amino acid
sequences
derived from this 60-kD protein.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-82-
A. Purification of a 60 kD protein
D. farihae extract was prepared and fractionated on a Sephacryl S-100 column
according to the methods described above in Example 1. Fractions were
collected
from the Sephacryl S-100 column after the excluded peak (fractions 29 through
35)
and were pooled. The pooled fractions were then diluted 1:1 with 10 mM Tris-
HCI,
pH 8, and applied to a Q-sepharose column and fractions obtained using the
methods
described above in Example 1. The fraction that eluted in 0.4 M Tris-HCl was
concentrated and further purified through a TMS 250 reverse phase HPLC column
using the methods described above in Example 1. The proteins in the fractions
were
resolved by 14°lo Tris-glycine SDS-PAGE using similar methods described
for
resolution of proteins on the 12% gel in Example 1. The stained gel is shown
in Fig.
2. A protein was identified having a molecular weight of about 60 kD (Fig. 2,
lane 4)
of about 90°Io purity that eluted at about 50% B (.05%TA in 90%
acetonitrile). The
molecular weight of the denoted 60-kd protein was estimated to be 56.11 kd
using the
BioRad Multi-Analyst/PC Version 1.1 program and Mark-12 protein molecular
weight
markers. The about 60-kd protein is referred to herein as mapD protein.
B. Partial N-terminal and internal sequence obtained from the 60-kd protein
The eluted protein from Part A, above, was blotted onto PVDF, which was
stained with Coomassie R-250 and destained using standard procedures. The
protein
corresponding to the about 60-kd band was excised and subjected to N-terminal
amino
acid sequencing using techniques known to those skilled in the art. A partial
N-terminal amino acid sequence of about 25 amino acids was deduced for the
protein
and the amino acid sequence, represented herein as SEQ TD N0:23, was
determined to
be: Xaa Leu Glu Pro Lys Thr Val Cys Tyr Tyr Glu Ser Trp Val His His Arg Gln
G1y
Glu Gly Lys Met Asp Pro (wherein Xaa refers to any amino acid).
The protein corresponding to the 60 kd region was also submitted to
proteolytic cleavage in order to obtain internal amino acid sequence data.
Digestion of
the 60-kd protein and reverse-phase chromatography were carried out as
described in
Example 1. Four proteolytic fragments were isolated and sequenced, and are
referred
to herein as map(13), map(14), map(15), and map(16).
The N-terminal partial amino acid sequence of map(13) was determined to be
Gln Tyr Gly Val Thr Gln Ala Val Val Thr Gln ProAla, also denoted SEQ m N0:29.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-83-
The N-terminal partial amino acid sequence of map(14) was determined to be Asp
Glu
Leu Leu Met Lys Ser Gly Pro Gly Pro, also denoted SEQ ID N0:30. The N-terminal
partial amino acid sequence of map(15) was determined to be Asp Met Glu His
Phe
Thr Gln His Lys Gly Asn Ala Lys Ala Met Ile Ala Val Gly Gly Ser Thr Met Ser,
also
denoted SEQ ID N0:31. The N-terminal partial amino acid sequence of map(16)
was
determined to be Asp Ala Asn Glu Glu Ala Arg Ser Gln Leu Pro Glu Thr Ala Met
Val
Leu Ile Lys Ser Gln, denoted SEQ ID N0:32.
Example 10.
This example describes the isolation and sequencing of nucleic acid molecules
encoding a portion of the D. farihae 60 kD (mapD) allergen.
A D. farihae library was prepared as described previously in Example 8. A
degenerate synthetic oligonucleotide primer was designed from the N-terminal
amino
acid sequence deduced for D. farinae 60 kD-protein (SEQ ID N0:23): Primer l, a
sense primer corresponding to amino acid residues from about 3 through about
11 of
SEQ ID N0:23 has the sequence 5' GAACCAAAA CHGTNTGYTA YTAYG 3', also
known as SEQ ID N0:46, where H represents A or C or T, N represents A or C or
G
or T, and Y represents C or T. PCR amplification of fragments from the D.
farinae
library was conducted using standard techniques. A PCR amplification product
was
generated using a combination of SEQ ID N0:46 (primer 1) and the M13 forward
universal primer 5'GTAAAACGACG GCCAGT 3', denoted SEQ ID N0:47.
A second, nested PCR reaction was carried out on the products of the first PCR
reaction. A synthetic oligonucleotide was synthesized that corresponded to a
region
spanning from about amino acid residue 1 through amino acid residue 10 of the
60-kD
protein internal amino acid sequence, SEQ ID N0:31. This primer, primer 2, has
the
nucleic acid sequence 5' GATATGGAAC ATTTYACHCA ACAYAARGG 3',
denoted SEQ ID N0:48, where R represents A or G. A PCR amplification product
was generated using the combination of primer 2, SEQ ID N0:48, and the T7
standard
primer, 5' GTAATACGAC TCACTATAGG GC 3', denoted SEQ ID N0:49. The
resultant PCR product Was subjected to DNA sequence analysis using standard
techniques.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-84-
The PCR product was sequenced and found to contain 510 nucleotides, and is
known as nDerf605~o. The nucleotide sequence of the coding strand of
nDerf60s~o is
represented herein as SEQ m N0:43, and its complement is denoted SEQ ID N0:45.
Translation of SEQ ID N0:43 suggests that nDerf605io encodes a partial D.
faritaae 60-
kD protein of about 170 amino acids, referred to herein as PDerf601~o, with an
amino
acid sequence denoted SEQ ID N0:44, assuming an open reading frame in which
the
first codon spans from about nucleotide 1 through nucleotide 3 of SEQ ID
N0:43, and
the last codon spanning from about nucleotide 508 through about nucleotide 510
of
SEQ ID N0:43. PDerf601~o has an estimated molecular weight of 19.2 kD and an
estimated pI of about 6.51.
Nucleic acid molecule nDerf605io was used as a probe to isolate a nucleic acid
molecule that encodes a protein corresponding to a full-length, or larger
partial D.
faYinae 60-kD protein. Using procedures described previously in Example 8, the
whole D. farinae library was screened with the nucleic acid SEQ ID N0:43
radiolabeled with 32P using standard techniques. Hybridization was done in 6X
SSC,
5X Denhardt's solution, 0.5% SDS, 100 mglml ssDNA, at 55°C, for about
36 hours.
The filters were washed 3 times, for 30 minutes per wash, at 55°C in 2X
SSC, 0.2%
SDS, followed by a final wash of about 30 minutes in 0.2X SSC, 0.2% SDS.
PCR amplification was carried out on the primary phage plugs. Primer 1,
denoted as SEQ ID N0:46, and T7 standard primer, denoted as SEQ ID N0:49, were
used as the primers, and a PCR product was generated. Preliminary sequence
analysis
of this 1.6 kilobase PCR product showed that it represents a nucleic acid
sequence that
contains the complete sequence encoding the PDerf60 full-length protein.
Comparison of PDerf601~o, the amino acid sequence of SEQ ID N0:44, with
amino acid sequences reported in GenBank indicates that PDerf601~o showed the
most
homology, i.e. about 39% identity, with a chitinase protein precursor from
Apha~2odidium album. (GenBank accession number P32470). Nucleic acid sequence
SEQ ID N0:43 showed no significant homology to any of the sequences submitted
to
GenB ank.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-85-
Example 11
This example describes the isolation of nucleic acid molecules encoding
DernzatopTzagoides ptero>zyssius 98 kD allergen protein.
Nucleic acid molecules with high homology to the D. farinae 98 kD allergen
(map B) were isolated from a D. ptero~zyssius cDNA library by hybridization
with a
32-P labeled cDNA encoding the D. fari>zae HMW-map composition.
A D. pteronyssius cDNA library was prepared as follows. Total RNA was
extracted from approximately 2 grams of D. pterorzyssius mites, using an acid-
guanidium-phenol-chloroform method, described by Chomzynski et al., 1987,
Aizal.
Biochem 162: pp 156-159. Poly A+ selected RNA was separated from the total RNA
preparation by oligo-dT cellulose chromatography using the mRNA Purification
Kit
(available from Pharmacia, Newark, NJ), according to the method recommended by
the manufacturer. A whole D. pteronyssius cDNA library was constructed in
lambda-
Uni-ZAPTM XR vector (available from Stratagene, La Jolla, CA), using
Stratagene's
ZAP-cDNA Synthesis Kit protocol. Approximately 5 milligram (mg) of Poly A+
RNA was used to produce the D. pteronyssius cDNA library.
Using a modification of the protocol described in the cDNA Synthesis Kit
(available from Stratagene), the whole D, pterorzyssius cDNA library was
screened,
using duplicate plaque lifts, with a 32P-labeled cDNA encoding the D. farizZae
97 kD
Map B allergen, i.e. SEQ ID N0:17. Hybridization was done in 6X SSC (for
recipe
see Sambrook, et al., ibid.), 5X Denhardt's solution (for recipe see Sambrook,
et al.,
ibid.), 0.5% sodium dodecyl sulfate (SDS) (available from Sigma), and 100
mglml of
single stranded DNA (available from Sigma), at 55°C, for about 36
hours. The filters
were washed 3 times, for about 30 minutes per wash, at 55°C, in 2X SSC,
0.2% SDS,
followed by a final wash of about 30 minutes, at 55°C, in 0.2X SSC,
0.2°lo SDS. A
plaque purifiedclone of the D. pteroyzyssius nucleic acid molecule encoding
the D.
pteroTZyssius 97 kD allergen (map B) was converted into a double stranded
recombinant molecule using the ExAssist TM helper phage and SOLRTM E. coli
according to the ih vivo excision protocol described in the ZAP-cDNA Synthesis
Kit
(all available from Stratagene). The plasmid containing the D. pterotzyssius
clone was
subjected to DNA sequence analysis using standard techniques. DNA sequence
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-86-
analysis, including the determination of molecular weight and isoelectric
point (pI)
was performed using the GCGTM program.
A clone was isolated that included an about 1621-nucleotide insert, which
includes the full-length coding region, referred to herein as nDerp981~2i,
with a coding
strand represented as SEQ ll~ N0:34 and a complementary strand represented as
SEQ
ID N0:36. The apparent start and stop codons span from nucleotide 14 through
nucleotide 16, and from nucleotide 1541 through nucleotide 1543, respectively,
of
SEQ ID N0:34. A putative polyadenylation signal (5' AATAAA 3' ) is located in
a
region spanning from nucleotide 1584 to 1589 of SEQ ID N0:34.
Translation of SEQ ID N0:34 yields a protein of about 509 amino acids,
denoted PDerp985o9, the amino acid sequence of which is presented as SEQ ID
N0:35.
The nucleic acid molecule consisting of the coding region encoding PDerp985o~
is
referred to herein as nDerp981sz~, the nucleic acid sequence of which is
represented as
SEQ ID N0:37 (the coding strand), and SEQ ID NO:39 (the complementary strand).
The amino acid sequence of PDerp985o9, also represented herein as SEQ ID
N0:38,
has an estimated molecular weight of about 58.9 kD and an estimated pI of
about 5.61.
Analysis of PDerp985os suggests the presence of a signal peptide spanning from
about
amino acid 1 through about amino acid 19. The proposed mature protein, denoted
herein as PDerp9849o, contains about 490 amino acids, and is represented
herein as
SEQ ID NO:41. The amino acid sequence of PDerp98~~o predicts the protein to
have
an estimated molecular weight of about 56.8 kD, and an estimated pI of about
5.49, as
well as two asparagine-linked glycosylation sites extending from about amino
acid 115
to about amino acid 117, and extending from about amino acid 240 to amino acid
242,
respectively. The nucleic acid molecule encoding PDerp9849o is known as
nDerp981ø~0,
with a coding strand represented by SEQ ID N0:40 and a complementary strand
represented by SEQ ID N0:42.
A BLAST search was performed as described previously. PDerp985o9, SEQ ID
N0:35, showed the highest homology at the amino acid level with the Mazzduca
sexta
chitinase (SwissProt accession number p36362), with about a 34% identity.
nDerp9816au SEQ ID N0:34, showed the highest homology at the nucleic acid
level to
Chelorzus sp. chitinase (accession number U10422), with about a 49% identity.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-87-
Comparison of cDNA regions corresponding to the coding regions for the D.
faf~iuae
98 kD allergen protein and the cDNA regions corresponding to the coding
regions for
the D. pteroiZyssius 98 kD allergen protein shows an identity of about 84%.
Example 14.
This example demonstrates the binding of the D. farifZae HMW-map
composition to human IgE in human sera isolated from humans known to be
allergic
to mite allergens.
A technique called RAST, or radio-allergo-absorbent test, was used because
the amount of human IgE present in human sera is quite low. RAST was
essentially
performed as described in Aalberse, RC et al., (1981) J. Allef~gy CliiZ
In2mma. 68: pp
356-364. To calculate the unit IU/ml, a standard curve was derived by
performing
RAST with several dilutions of a well-characterized chimeric human/mouse IgE
monoclonal antibody against Derp2, (human IgE/monoclonal anti-Derp2, following
the procedure of Schuurman, et al. (1997) JAllergy Clih ImnZUn.ol. 99: pp 545-
550).
Briefly, 50 ,ug of the HMW-map composition, purified as described in
Example 1, was coupled to 50 mg of CNBr-activated Sepharose 4B (available from
Pharmacia, Piscataway, NJ), according to the manufacturer's protocols. Human
sera
were selected (17 different samples, total) on the basis of a positive RAST
for whole
mite D. fariuae extracts, a positive RAST number is greater than 1 IU/ml). Two
negative (less than 0.3 IU) control sera were also included.
To test each individual serum sample, 0.5 mg of the D. fariJ2ae HMW-map
composition-coupled Sepharose was incubated with 50 ,u1 serum in a total
volume of
300 ,u1 of PBS-T (Phosphate buffered saline with added 0.1% volume/volume
Tween-
20, available from Sigma). Incubation was overnight at 27°C, with
shaking. After
incubation, the coupled Sepharose was washed five times with PBS-T.
Radiolabelled
(ias-Iodine) sheep anti-human IgE, made by standard radioiodination protocols,
(diluted in PBS-T with 4.5% bovine serum and 0.5% sheep serum, v/v) in a total
volume of 750 ,u1, was added and incubated overnight at 27°C. After
incubation, the
coupled Sepharose was washed four times with PBS-T and counted in a gamma-
counter to determine the amount of radiolabeled sheep anti-human IgE bound to
the
HMW-map composition-coupled Sepharose. The results are shown in Table 4.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
_88_
Table 4. Binding of human IgE to HMW-map composition from D. farizzae
Serum number RAST, D: farinae RAST, HMW-map
whole comps'n., IU
extract, IU
1445 > 100 48
1456 >100 42
1458 21.1 0.5
1460 14.1 2.5
1463 37.6 0.1
1464 37.2 2.0
1465 14.5 0.7
1466 89.9 7.7
1468 >100 19.9
1471 31.9 0.8
1491 23.8 1.0
1496 25.3 3.6
1505 5.1 0.2
1523 1.0 <0.1
1529 1.2 0.7
1530 (control)0.2 <0.1
1531 (control)0.1 <0.1
Almost 75% of patients (11 of 15) who showed sensitivity to D. farizzae whole
mite extracts were sensitive to the HMW-map composition antigen, implying that
the
HMW-map composition antigen is a major antigen for D. farirzae sensitive
humans.
Sensitivity to the HMW-map composition was defined as a RAST of greater than
or
equal to 0.5 ICT.
Example 15.
This example demonstrates that the D. farizzae HMW-map composition
described in Example 1 includes a glycoprotein.
About (5.4 ,ug) of a D. farirzae HMW-map composition prepared in accordance
with Example 1 was applied to SDS PAGE and electrophoresis was done according
to
standard techniques. The protein was blotted to a nitrocellulose membrane
according
to standard techniques, and glycoprotein was detected using the DIGT"~ Glycine
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-89-
Detection Kit (available from Boehringer Mannheim, Indianapolis, IN), using
the
manufacturer's protocol. The region corresponding to the HMW-map region showed
a
positive reaction with the kit, indicating that the HMW-map composition
includes a
glycoprotein.
Example 16.
This example shows that the D. faritaae HMW-map composition retains its
character as an allergen even when the amino acid residues are removed, both
by
chemical and enzymatic means. The results suggest that the main epitope(s)
could be
a carbohydrate epitope including a polysaccharide attached to an N-linked or O-
linked
glycosylation site on the HMW-map composition.
A. Protein elimination by chemical means ((3-elimination of proteins)
Twelve ,ug (microgram) of HMW-map composition (purified as described in
Example 1) was dissolved in 100 ,u1 (microliter) of distilled deionized water.
To this
mixture was added 5 ,u1 10 M (molar) NaOH and 3.8 mg (milligram) NaBH4
(available
from Sigma) to give a final concentration of 0.5 M NaOH and 1 M NaBH4. This
reaction mixture was heated at 50°C for 30 minutes, then cooled, and
100 ,u1 acetone
was added. To this mixture, sufficient amount, i.e. approximately 150 ,u1, of
Dowex
50 (H+) (available from Pharmacia) was added to make the solution slightly
acidic.
The Dowex 50 adsorbed and removed the protein, leaving any sugar moieties in
the
supernatant. The mixture was centrifuged in a microcentrifuge and washed three
times
with 100 ,u1 of water. The combined supernatants from the centrifugations were
evaporated to dryness, then washed five times from a methanol:HCl solution
(1000:1
v/v), evaporating to dryness after each wash, to remove salts. The mixture was
dissolved in 100 ,u1 of water, and a portion (20 ,u1) was analyzed by SDS-PAGE
using
standard techniques, and both Coomassie blue and Silver staining were used to
determine the amount of protein in the chemically treated samples. No protein
was
detected by either Coomassie or Silver staining, indicating removal of
protein. Any
sugar moieties on the protein would be unaffected by these conditions.
The remainder of the residue from each sample was subjected to ELISA
analysis as described in Example 4. Briefly, 100 ng of either the (3-
eliminated sample
or of non-(3-eliminated sample of the HMW-map composition was coated onto the
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-90-
Immulon plates, and ELISAs were carried out as described in Example 4 with a
D.
farircae sensitive dog sera pool, a D. farahae sensitive cat sera pool, and
various
individual dog sera that are either D. farihae sensitive or not sensitive (as
measured by
ELISA). The results are shown in Table 5.
Table 5. Reactivity of dog and cat sera to HMW-map composition and to
(3-eliminated HMW-map composition (which is carbohydrate
only)
Sera used .. (3-eliminated HMW-map;untreated I3MW-map
.. comps'n., OD X
OD (carbohydrate 10'3
antigen)
D.farinae dog 1233 1931
pool
D. farihae cat 2837 3115
pool
dog 1621A 15 0
dog 1621C 24 21
dog 1621 S 59 420
dog 1626C 23 214
dog SPF-2 16 0
Results from Table 5 indicate that the (3-eliminated HMW-map composition
sample
still retains the ability to bind IgE from dog and cat sera that is sensitive
to D. fari~2ae
HMW-map composition, indicating that the glycans attached to the protein
constitute a
major epitope of the HMW-map composition allergen protein.
B. Protein Elimination by enzymatic means.
14 ,ug of HMW-map composition (purified as described in Example 1) was
digested with l,ug Endoproteinase K, available from Sigma, to remove the
protein
moiety of the molecule. The digestion reaction took place at 56°C for
24 hours, after
which the endoproteinase in the reaction was heat-denatured in boiling water
for 10
minutes.
A portion of this reaction was analyzed by SDS-PAGE using standard
techniques, and both Coomassie blue and Silver staining were used to detect
the
presence of protein in the enzymatically digested samples. No HMW-map
composition was detected by either Coomassie or Silver staining, indicating
elimination of the HMW-map composition. Any glycan that was attached via a
glycosylation site on the protein would be unaffected by these conditions.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-91-
The remainder of the enzymatically digested reaction was tested by ELISA in
the manner described in Example 4. Briefly, 100 ng of either the proteinase-K-
digested sample or of a non-digested sample of the HMW-map composition was
coated onto Immulon plates, and ELISAs were carried out as described in
Example 4
with various individual dog sera that were either D. fari~zae sensitive or not
sensitive
(as measured by ELISA). The results are shown in Table 6.
Table 6. Reactivity of dog sera to HMW-map composition and to
Endoproteinase-K digested HMW-map composition.
dog D. farir2ae OD, wells chafed OD; wells coated with
# sensitive?1 with Proteinase K
HMW-map, cotnps'n. digested HMW-map :,
,
1 yes 120 122
2 yes 1637 1561
3 yes 858 383
4 yes 914 509
5 yes 277 227
6 yes 2891 2636
7 no 10 11
8 yes 4056 3880
9 yes 1920 1626
10 yes 472 432
11 yes 328 213
12 yes 2913 2530
13 yes 1232 984
14 yes 3153 2355
15 no 6 46
16 yes 860 339
17 yes 2429 750
18 yes 1194 351
19 yes 2655 1443
20 yes 3285 1207
0 21 yes 2636 1240
22 yes 1097 848
23 yes 1621 1408
24 yes 2113 1592
25 yes 1169 408
26 yes 4200 4200
27. yes 4200 4200
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-92-
dog D. farinae OD, wells coated OD, wells coated with
# sensitive?1 with Pxoteinase K
HMW-map comps'n. digested HMW-map
28 yes 3222 2932
29 yes 2468 2118
30 yes 3339 2454
31 no 0 4
' by ELISA in a separate experiment
Results from Table 6 indicate that the proteinase-I~ digested HMW-map
composition
sample still retains the ability to bind IgE from dog and cat sera that is
sensitive to D.
farihae HMW-map composition, suggesting that the glycans attached to the
protein
constitute a major epitope on the HMW-map composition.
Example 17
This example describes attempts to remove N-linked glycans from the HMW-
map composition.
HMW-map composition (2 ,ug), purified as in Example 1, was digested with N-
glycosidase F (available from Boehringer-Mannheim), according to the
manufacturer's
directions. The digestion was analyzed by SDS-PAGE and stained according to
standard protocols. 2 ,ug Fetuin (available from Sigma) was used as a positive
N-
linked glycosylated protein control. Analysis of the SDS-PAGE showed that
there
were no apparent differences in the molecular weights of the intact and
digested map B
protein. The positive control, fetuin, did show a reduction of molecular
weight after
digestion with N-glycosidase F. This result indicates that there are no N-
linked
glycans on the HMW-map composition, or alternatively that there are only small
sized
N-glycans on the HMW-map composition.
Example 18
This example describes the isolation and sequencing of a nucleic acid
molecule encoding the full length Dermatophagoides fariyzae 60 kD allergen.
This nucleic acid molecule was isolated from a Derfvatophagoides farircae
cDNA library by it's ability to hybridize with a 32P-labeled cDNA encoding a
portion
of the Dermatoplzagoides fararcae 60 kD allergen described in Example 10.
A Dermatophagoides faf°inae cDNA library was prepared as follows.
Total
RNA was extracted from approximately 2 grams of D. farir~ae. mites, using an
acid-
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-93-
guanidinium-phenol-chloroform method similar to that described by Chomzynski
et
al., 1987, Anal. Biochern. 162,156-159. Poly A+ selected RNA was separated
from the
total RNA preparation by oligo-dT cellulose chromatography using the mRNA
Purification Kit (available from Pharmacia Biotech, Newark, NJ), according to
the
method recommended by the manufacturer. A whole mite cDNA library was
constructed in lambda-Uni-ZAPS XR vector (available from Stratagene), using
Stratagene's ZAP-cDNA Synthesis Kit protocol. Approximately 5 ~g of Poly A+
RNA
was used to produce the D. farinae cDNA library.
Using a modification of the protocol described in the cDNA Synthesis Kit, the
whole mite cDNA library was screened, using duplicate plaque lifts, with 32P-
labeled
cDNA nDerf605io. Hybridization was done at 6XSSC, 5X Denhardt's solutions, 0.5
%
SDS, 100 mg/ml of ssDNA and, at 52°C, for 18 hr. The filters were
washed 2 times,
for 30 minutes per wash, at 55° C in 2XSSC, 0.2°7o SDS, followed
by a final wash of
30 minutes in the same buffer except using aboutØ2XSSC: A plaque purified
clone
of the nucleic acid molecules encoding the D. farinae 60 kD allergen was
converted
into a double stranded recombinant molecule, herein denoted as nDerf601ass ,
using the
ExAssist~ helper phage and SOLRTM E. coli according to the in vivo excision
protocol described in the ZAP-cDNA Synthesis Kit (available from Stratagene).
Double-stranded plasmid DNA was prepared using an alkaline lysis protocol,
such as
that described in Sambrook et al., ibid.
Example 19
This example describes the sequencing of a D. farinae nucleic acid molecule of
the present invention.
The plasmid containing nDerf6014ss was sequenced by the Sanger dideoxy
chain termination method, using the PRISMS Ready Dye Terminator Cycle
Sequencing Kit with Ampli Taq DNA Polymerase, FS (available from the Perkin-
Elmer Corporation, Norwalk, CT). PCR extensions were done in the GeneAmpTM PCR
System 9600 (available from Perkin-Elmer). Excess dye terminators were removed
from extension products using the CentriflexTM Gel Filtration Cartridge
(available
from Advanced Genetics Technologies Corporation, Gaithersburg, MD) following
the
manufacturer's standard protocol. Samples were resuspended according to ABI
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-94-
protocols and were run on a Perkin-Elmer ABI PRISM TM 377 Automated DNA
Sequencer. DNA sequence analysis, including the compilation of sequences and
the
determination of open reading frames, was performed using the GCGTM program
(available from Genetics Computer Group, Madison, WI). Protein sequence
analysis,
including the determination of molecular weight and isoelectric point (pI) was
performed using the GCG~ program.
An about 1455 nucleotide consensus sequence of the entire nDerf6014ss nucleic
acid molecule was determined; the sequences of the two complementary strands
are
presented as SEQ ID N0:50 (the coding strand) and SEQ ID NO: 52 (the
complementary strand). The nDerf6014ss sequence contains a full length coding
region.
The apparent start and stop codons span nucleotides from 14 through 16 and
from
1400 through1402, respectively, of SEQ ID NO: 50. A putative polyadenylation
signal (5' AATAAA 3' ) is located in a region spanning from about nucleotide
1408-
1413 of SEQ ID NO: 50.
Translation of SEQ ID NO: 50 yields a protein of 462 amino acids, denoted
PDerf6046z, the amino acid sequence of which is presented in SEQ ID NO: 51.
The
nucleic acid molecule consisting of the coding region encoding PDerf60d~z is
referred
to herein as nDerf6013s6, the nucleic acid sequence of which is represented in
SEQ ID
NO: 53 (the coding strand) and SEQ ID NO: 54 (the complementary strand). The
amino acid sequence of PDerf604~z (i.e., SEQ ID NO: 51) predicts that
PDerf604~z has
an estimated molecular weight of about 52.1 kD and an estimated pI of about
5.73.
Analysis of SEQ ID NO: 51 suggests the presence of a signal peptide encoded by
a
stretch of amino acids spanning from amino acid 1 through amino acid 25. The
proposed mature protein, denoted herein as PDerf6043~, contains about 437
amino
acids which is represented herein as SEQ ID NO: 56. The amino acid sequence of
PDerf6043~ (i.e., SEQ ID NO: 56) predicts that PDerf60ø6z has an estimated
molecular
weight of about 50.0 kD, an estimated pI of about 5.61. and one predicted
asparagine-
linked glycosylation site extending from amino acids 313 through 315. The
nucleic
acid molecule encoding the mature protein is denoted SEQ ID NO: 55 and its
reverse
complement is denoted SE ID NO: 57.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-95-
A BLASTp search was performed according to Altschul, et al, (1990), J. Mol.
Biol. 215:403-410; and Altschul, et al, (1997), Nucleic Acids Res. 25:3389-
3402. The
protein search was performed using SEQ ID N0:51, which showed significant
homology to chitinase molecules. The highest scoring match of the homology
search
at the amino acid level was PIR accession number A53918: Cheloi2us sp.
chitinase
precursor, which was about 32°1o identical with SEQ ID N0:51. At the
nucleotide
level, the search was performed using SEQ ID N0:53, which did not show
significant
similarity to any sequences in the database. Sequence analysis was performed
using
the GCG GAP program as described above.
Example 20
This example further describes the characterization of the D. farihae HMW-
map composition (also referred to as Der f 15).
Nucleic acid molecule nDerf981~s2 of Example 8 was inserted into appropriate
expression vectors and expressed in E. coli and P. pastoris. When the
resulting
protein, PDerf98sss was expressed in E. coli or P. pastoris, sensitized dog
sera,
produced as described in Example 4, failed to recognize the recombinant
protein. This
is in contrast to the positive results obtained when the native D. farihae HMW-
map
composition of Example 1 (also referred to as native Der f 15) was used; see
Example 4.
The non-reactivity of the protein expressed in E. coli is consistent with the
results shown in Example 16, in which it was shown that the native HMW
allergens
retain their character as allergens, even after the amino acids are removed.
All of these results together suggest that the main epitope(s) are
carbohydrate
regions of the molecule or some other secondary modification.
The antigenicity of the native Der f 15 protein is not lost after periodate
treatment; generally carbohydrate epitopes are destroyed by periodate except
for those
further substituted with additional groups or those having an unusual sugar
with no
geminal hydroxyl groups.
The native Der f 15 antigen was analyzed for carbohydrate content. A
substantial amount of carbohydrate was found, about 30% by weight.
Specifically,
mannose constituted approximately 2.8% by weight of the antigen; galactose
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-96-
approximately 23.2%; glucose approximately 4.3% (the presence of glucosyl
residue
must be considered tentative as glucose often contaminates glycoprotein
samples); and
HexNAc at detectable levels; further investigation revealed that the HexNAc
were
GIcNAc and GalNAc.
The native Der f 15 protein was treated with base in the presence of NaBH4 and
analyzed by a P-4 sizing chromatography. O-linked oligosaccharides present in
Der f
were found to void the column. This result is consistent with either very
large O-
linked oligosaccharides or the presence of acidic groups on the
oligosaccharides such
as sulfate. Attempts to determine the presence or absence of sulfate more
directly gave
10 ambiguous results.
Der f 15 was treated at pH 4, pH 5, and pH 7 overnight at 37° C. The
resulting
samples were then probed with antibody to the protein or dog serum known to be
reactive with Der f 15. In the samples treated at pH 5 and pH 7, all of the
dog
antiserum epitope was destroyed, but in the samples treated at pH 4, some
activity
15 remained. The anti-Der f 15 antibody shows that the molecular weight of Der
f 15 was
decreased at all pH's with some original material left at pH 4, as though
deglycosylation was occurring. It is not known whether this change was self
catalyzed
by the Der f 15 protein or occurred chemically; while not being bound by
theory, it is
believed that self catalysis was involved since the loss of the epitope
occurred under
such mild conditions.
Example 21
This example describes the binding of several house dust mite (HDM)
allergens to feline IgE in cat serum.
The allergen profile of the IgE response of cats to house dust mites appears
to
be different fiom that of dogs. An examination of the results of IgE testing
on cat sera
submitted to Heska's Veterinary Diagnostic Laboratories (VDL) in January 2000
shows that 40% of all allergen-specific IgE positive cats had anti-HDM IgE.
All the
cats were positive to both D. fari~ae and D. pterofzyssiyaus. Eighty-eight
sera known to
be positive for D. farinae were assayed by ELISA on highly purified
preparations of
Der f 1, Der f 2, Der f 15, and the 60 kD allergen. In this assay, 32% of the
cats were
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
-97-
positive for Der f l, 42% were positive for Der f 2, 68% were positive for Der
f 15,
and 86% were positive for the 60 kD allergen.
While various embodiments of the present invention have been described in
detail, it is apparent that modifications and adaptations of those embodiments
will
occur to those skilled in the art. It is to be expressly understood, however,
that such
modifications and adaptations are within the scope of the present invention;
as set forth
in the following claims.
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
SEQUENCE LISTING
<110> Heska Corporation
MCCall, Catherine A.
Hunter, Shirley Wu
Weber, Eric R.
<120> NOVEL DERMATOPHAGOIDES NUCLEIC ACID MOLECULES, PROTEINS AND USES
THEREOF
<130> AL-2-C4-PCT
<140> not yet assigned
<141> 2001-09-13
<150> 09/662,293
<151> 2000-09-14
<150> 09/292,225
<151> 1999-04-15
<150> 60/098,909
<151> 1998-09-02
<150> 60/085,295
<151> 1998-05-13
<150> 6.0/098,565
<151> 1998-04-17
<150> 09/062,013
<151> 1998-04-17
<160> 57
<170> Patentln version 3.1
<210> 1
<211> 14
<212> PRT
<213> Dermatophagoides farinae
<400> 1
Ser Ile Lys Arg Asp His Asn Asp Tyr Ser Lys Asn Pro Met
1 5 10
<210> 2
<211> 20
<212> PRT
<213> Dermatophagoides farinae
<400> 2
Asp Tyr Glu Tyr Pro Gly Ser Arg Leu Gly Asn Pro Lys Ala Pro Leu
1 5 10 15
Tyr Lys Arg Pro
<210> 3
1
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
<211> 20
<212> PRT
<213> Dermatophagoides farinae
<400> 3
Asp Ile Pro His Pro Thr Asn Ile His Lys Tyr Leu Val Cys Glu Ser
1 5 10 15
Val Asn Gly Gly
<210> 4
<211> 20
<212> PRT
<213> Dermatophagoides farinae
<400> 4
Asp Pro Ala Lys Gly Met Ser Pro Pro Gly Phe Ile Val Gly Glu Glu
1 5 10 15
Gly Val Leu Ser
<210> 5
<211> 12
<212>. PRT
<213> Dermatophagoides farinae
<400> 5
Asp Glu Lys Asn Ser Phe Glu Cys Ile Leu Gly Pro
1 5 10
<210> 6
<211> 18
<212> PRT
<213> Dermatophagoides farinae
<400> 6
Asp Ala Phe Glu Pro His Gly Tyr Leu Leu Thr A1a Ala Val Ser Pro
1 5 10 15
Gly Lys
<210> 7
<211> 13
<212> PRT
<213> Dermatophagoides farinae
<400> 7
Asp Lys Gln Asn Tyr Leu Ala Leu Val Arg Glu Leu Lys
1 5 10
2
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
<210> 8
<211> 24
<212> PRT
<213> Dermatophagoides farinae
<400> 8
Asp Met Ala Gln Asn Tyr Lys Tyr Arg Gln Gln Phe Ile Gln Ser Val
1 5 10 15
Leu Asn Asn Gly Ala Thr Arg Gln
<210> 9
<211> 23
<212> PRT
<213> Dermatophagoides farinae
<220>
<221> MISC_FEATURE
<222> (3). (3)
<223> Xaa = any amino acid at position 3
<220>
<221> MISC_FEATURE
<222> (7). (7)
<223> Xaa = any amino acid at position 7
<400> 9
Asp Glu Xaa Asn Val Met Xaa Tyr Val Leu Tyr Thr Met His Tyr Tyr
1 5 10 15
Leu Asn Asn Gly Ala Thr Arg
<210> 10
<211> 17
<212> PRT
<213> Dermatophagoides farinae
<220>
<221> MISC_FEATURE
<222> (14) .(14)
<223> Xaa = any amino acid at position 14
<400> 10
Asp Lys Leu Val Met Gly Val Pro Phe Tyr Gly Arg Ala Xaa Ser Ile
1 5 10 15
Glu
3
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT. ST25 . tact
<210> 11
<211> 19
<212> PRT
<213> Dermatophagoides farinae
<400> 11
Asp Ile Pro His Pro Thr Asn Ile His Lys Tyr Leu Val Cys Glu Ser
1 5 10 15
Val Asn Gly
<210> 12
<211> 18
<212> PRT
<213> Dermatophagoides farinae
<400> 12
Asp Tyr Ala Lys Asn Pro Lys Arg I1e Val Cys Ile Val Gly Thr Glu
1 5 10 15
Gly Val
<210> 13
<211> 20
<212> PRT
<213> Dermatophagoides farinae
<400> 13
Asp Pro Ala Lys Gly Met Ser Pro Pro Gly Phe Ile Val Gly Glu Glu
1 5 10 15
G1y Val Leu Ser
<210> 14
<211> 1752
<212> DNA
<213> Dermatophagoides farinae
<220>
<221> CDS
<222> (1)..(1665)
<223>
<400> 14
atg aaa acc ata tat gca ata ctt agt att atg gcc tgc att ggc ctt 48
Met Lys Thr Ile Tyr Ala Ile Leu Ser Ile Met Ala Cys Ile Gly Leu
1 5 10 15
atg aat gca tcc atc aaa cga gat cat aat gat tat tcg aaa aat ccg 96
Met Asn Ala Ser Ile Lys Arg Asp His Asn Asp Tyr Ser Lys Asn Pro
4
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
20 25 30
atgaga attgtt tgttatgttgga acatggtcc gtatatcat aaagtt 144
MetArg IleVal CysTyrVa1Gly ThrTrpSer ValTyrHis LysVal
35 40 45
gatcca tacact atcgaagatatt gatccattc aagtgtaca cattta 192
AspPro TyrThr IleGluAspIle AspProPhe LysCysThr HisLeu
50 55 60
atgtat ggtttc getaaaattgat gaatacaaa tacacaatt caagtt 240
MetTyr GlyPhe AlaLysIleAsp GluTyrLys TyrThrIle GlnVal
65 70 75 80
ttcgat ccttac caagatgataac cataactca tgggaaaaa cgtggt 288
PheAsp ProTyr GlnAspAspAsn HisAsnSer TrpGluLys ArgGly
85 90 95
tatgaa cgtttc aacaacttgcga ttgaagaat ccagaatta accacc 336
TyrGlu ArgPhe AsnAsnLeuArg LeuLysAsn ProGluLeu ThrThr
100 105 110
atgatt tcactt ggtggttggtat gaaggctcg gaaaaatat tccgat 384
MetIle SerLeu GlyGlyTrpTyr GluGlySer GluLysTyr SerAsp
115 120 125
atgget gcaaat ccaacatatcgt caacaattc atacaatca gttttg 432
MetAla AlaAsn ,ProThrTyrArg GlnGlnPhe IleGlnSer ValLeu
130 135 140
gacttt ttgcaa gaatacaagttc gacggtcta gatttggat tgggag 480
AspPhe LeuGln GluTyrLysPhe AspGlyLeu AspLeuAsp TrpGlu
145 150 155 160
tatcct ggatct cgattgggtaac ccgaaaatc gataaacaa aactat 528
TyrPro GlySer ArgLeuGlyAsn ProLysIle AspLysGln AsnTyr
165 170 175
ttgget ttggtt agagaacttaaa gacgetttt gaacctcat ggctac 576
LeuAla LeuVal ArgGluLeuLys AspAlaPhe GluProHis GlyTyr
180 185 190
ttgttg actget gcagtatcacca ggtaaagac aaaatcgac cgaget 624
LeuLeu ThrAla AlaValSerPro GlyLysAsp LysIleAsp ArgAla
195 200 205
tatgat atcaaa gaattgaacaaa ttgttcgat tggatgaat gtcatg 672
TyrAsp IleLys GluLeuAsnLys LeuPheAsp TrpMetAsn ValMet
210 215 220
acatat gattac cacggtggatgg gaaaacttt tacggtcac aatget 720
ThrTyr AspTyr HisGlyGlyTrp GluAsnPhe TyrGlyHis AsnAla
225 230 235 240
ccgttg tataaa cgaccagatgaa actgatgag ttgcacact tacttc 768
ProLeu TyrLys ArgProAspGlu ThrAspGlu LeuHisThr TyrPhe
245 250 255
aatgtc aactac accatgCactat tatttgaac aatggtgcc accaga 816
AsnVal AsnTyr ThrMetHisTyr TyrLeuAsn AsnGlyAla ThrArg
260 265 270
gac aaa ttg gta atg ggt gtt cca ttc tat ggc cgt get tgg agc att 864
Asp Lys Leu Val Met Gly Val Pro Phe Tyr Gly Arg Ala Trp Ser Ile
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
275 280 285
gaa gatcgaagc aaactcaaa ctt-ggagat ccagccaaa ggcatgtcg 912
Glu AspArgSer LysLeuLys LeuGlyAsp ProAlaLys GlyMetSer
290 295 300
CCC CCaggtttc atttctggt gaagaaggt gtcctctca tatatagaa 960
Pro ProGlyPhe IleSerGly GluGluGly ValLeuSer TyrIleGlu
~
305 310 315 320
ttg tgtcaattg tttcaaaaa gaagaatgg catatccaa tacgatgaa 1008
Leu CysGlnLeu PheGlnLys GluGluTrp HisIleGln TyrAspGlu
325 330 335
tat tacaatget ccatatggt tacaatgat aaaatctgg gtcggttac 1056
Tyr TyrAsnAla ProTyrGly TyrAsnAsp LysIleTrp ValGlyTyr
340 345 350
gatgatctggcc agtata tcatgcaagttg getttcctg aaagaatta 1104
AspAspLeuAla SerIle SerCysLysLeu AlaPheLeu LysGluLeu
355 360 365
ggcgtttctggt gtcatg gtttggtcattg gaaaatgat gatttcaaa 1152
GlyValSerGly ValMet ValTrpSerLeu GluAsnAsp AspPheLys
370 375 380
ggtcactgcgga ccgaaa aatccattgttg aacaaagtt cataatatg 1200
GlyHisCysGly ProLys AsnProLeuLeu AsnLysVal HisAsnMet
385 390 395 400
attaatggcgat gaaaag aactctttcgaa tgcattttg ggtccaagt 1248
IleAsnGlyAsp GluLys AsnSerPheGlu CysIleLeu GlyProSer
405 410 415
acaacgacacca actcca acgacgacaccc acaaccccg actacaacg 1296
ThrThrThrPro ThrPro ThrThrThrPro ThrThrPro ThrThrThr
420 425 430
ccaacaactcct tctccc accaccccgaca acaacccct tctcccacc 1344
ProThrThrPro SerPro ThrThrProThr ThrThrPro SerProThr
435 440 445
accccgacaaca acccct tctcccaccaca ccgacaaca actccttct 1392
ThrProThrThr ThrPro SerProThrThr ProThrThr ThrProSer
450 455 460
cccaccacacca acacca acaacaccaaca ccagcccct acaacatcg 1440
ProThrThrPro ThrPro ThrThrProThr ProAlaPro ThrThrSer
465 470 475 480
acaccttcgcca accacg accgaacacaca agcgaaaca ccaaaatat 1488
ThrProSerPro ThrThr ThrG1uHisThr SerGluThr ProLysTyr
485 490 495
acaacctatgtc gatgga catcttatcaaa tgttacaag gaaggtgat 1536
ThrThrTyrVal AspGly HisLeuIleLys CysTyrLys GluGlyAsp
500 505 510
atcccacatcca accaat atacacaaatat ttggtctgt gaatttgtt 1584
IleProHisPro ThrAsn IleHisLysTyr LeuValCys GluPheVal
515 520 525
aatggtggctgg tgggtt catattatgccc tgtccaccg ggcactatt 1632
AsnGlyGlyTrp TrpVal HisIleMetPro CysProPro GlyThrIle
6
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
530 535 540
tgg tgt caa gaa aaa ttg act tgt ata ggc gaa taattctgaa aaaaaaattc 1685
Trp Cys Gln Glu Lys Leu Thr Cys Ile Gly Glu
545 550 555
aattaaaatt taaaattcaa tttttaatat gaaaaattca aaaaaaaaaa aaaaaaaaaa 1745
aaaaaaa 1752
<210> 15
<211> 555
<212> PRT
<213> Dermatophagoides farinae
<400> 15
Met Lys Thr Ile Tyr Ala Ile Leu Ser Ile Met Ala Cys Ile Gly Leu
1 5 10 15
Met Asn Ala Ser Ile Lys Arg Asp His Asn Asp Tyr Ser Lys Asn Pro
20 25 30
Met Arg Ile Val Cys Tyr Val Gly Thr Trp Ser Val Tyr His Lys Val
35 40 45
Asp Pro Tyr Thr Ile Glu Asp Ile Asp Pro Phe Lys Cys Thr His Leu
50 55 60
Met Tyr Gly Phe Ala Lys Ile Asp Glu Tyr Lys Tyr Thr Ile Gln Val
65 70 75 80
Phe Asp Pro Tyr Gln Asp Asp Asn His Asn Ser Trp Glu Lys Arg Gly
85 90 95
Tyr Glu Arg Phe Asn Asn Leu Arg Leu Lys Asn Pro Glu Leu Thr Thr
100 105 110
Met Ile Ser Leu Gly Gly Trp Tyr Glu Gly Ser Glu Lys Tyr Ser Asp
115 120 125
Met Ala Ala Asn Pro Thr Tyr Arg Gln Gln Phe Ile Gln Ser Val Leu
130 135 140
Asp Phe Leu Gln Glu Tyr Lys Phe Asp Gly Leu Asp Leu Asp Trp Glu
145 150 155 160
Tyr Pro Gly Ser Arg Leu Gly Asn Pro Lys Ile Asp Lys Gln Asn Tyr
165 170 175
Leu Ala Leu Val Arg Glu Leu Lys Asp Ala Phe Glu Pro His Gly Tyr
180 185 190
7
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Leu Leu Thr Ala Al.a Val Ser Pro Gly Lys Asp Lys I1e Asp Arg Ala
195 200 205
Tyr Asp Ile Lys Glu Leu Asn Lys Leu Phe Asp Trp Met Asn Val Met
210 215 220
Thr Tyr Asp Tyr His Gly Gly Trp Glu Asn Phe Tyr Gly His Asn Ala
225 230 235 240
Pro Leu Tyr Lys Arg Pro Asp Glu Thr Asp Glu Leu His Thr Tyr Phe
245 250 255
Asn Val Asn Tyr Thr Met His Tyr Tyr Leu Asn Asn Gly Ala Thr Arg
260 265 270
Asp Lys Leu Val Met Gly Val Pro Phe Tyr Gly Arg Ala Trp Ser Ile
275 280 285
Glu Asp Arg Ser Lys Leu Lys Leu Gly Asp Pro Ala Lys Gly Met Ser
290 295 300
Pro Pro Gly Phe Ile Ser Gly G1u Glu Gly Val Leu Ser Tyr Ile Glu
305 310 315 320
Leu Cys Gln Leu Phe Gln Lys Glu G1u Trp His Ile Gln Tyr Asp G1u
325 330 335
Tyr Tyr Asn Ala Pro Tyr Gly Tyr Asn Asp Lys Ile Trp Val Gly Tyr
340 345 350
Asp Asp Leu Ala Ser Ile Ser Cys Lys Leu Ala Phe Leu Lys Glu Leu
355 360 365
Gly Val Ser Gly Val Met Val Trp Ser Leu Glu Asn Asp Asp Phe Lys
370 375 380
Gly His.Cys Gly Pro Lys Asn Pro Leu Leu Asn Lys Val His Asn Met
385 390 395 400
Ile Asn Gly Asp Glu Lys Asn Ser Phe Glu Cys Ile Leu Gly Pro Ser
405 410 415
Thr Thr Thr Pro Thr Pro Thr Thr Thr Pro Thr Thr Pro Thr Thr Thr
420 425 430
Pro Thr Thr Pro Ser Pro Thr Thr Pro Thr Thr Thr Pro Ser Pro Thr
435 440 445
8
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Thr Pro Thr Thr Thr Pro Ser Pro Thr Thr Pro Thr Thr Thr Pro Ser
450 455 460
Pro Thr Thr Pro Thr Pro Thr Thr Pro Thr Pro Ala Pro Thr Thr Ser
465 470 475 480
Thr Pro Ser Pro Thr Thr Thr Glu His Thr Ser Glu Thr Pro Lys Tyr
485 490 495
Thr Thr Tyr Val Asp Gly His Leu Ile Lys Cys Tyr Lys Glu Gly Asp
500 505 510
Ile Pro His Pro Thr Asn Ile His Lys Tyr Leu Val Cys Glu Phe Val
515 520 525
Asn Gly Gly Trp Trp Val His Ile Met Pro Cys Pro Pro Gly Thr Ile
530 535 540
Trp Cys Gln Glu Lys Leu Thr Cys Ile Gly Glu
545 550 555
<210>
16
<211>
1752
<212>
DNA
<213>
Dermatophagoides
farinae
<400>
16
ttttttttttttttttttttttttttttgaatttttcatattaaaaattgaattttaaat60
tttaattgaatttttttttcagaattattcgcctatacaagtcaatttttcttgacacca120
aatagtgcccggtggacagggcataatatgaacccaccagccaccattaacaaattcaca180
gaccaaatatttgtgtatattggttggatgtgggatatcaccttccttgtaacatttgat240
aagatgtccatcgacataggttgtatattttggtgtttcgcttgtgtgttcggtcgtggt300
tggcgaaggtgtcgatgttgtaggggctggtgttggtgttgttggtgttggtgtggtggg360
agaaggagttgttgtcggtgtggtgggagaaggggttgttgtcggggtggtgggagaagg420
ggttgttgtcggggtggtgggagaaggagttgttggcgttgtagtcggggttgtgggtgt480
cgtcgttggagttggtgtcgttgtacttggacccaaaatgcattcgaaagagttcttttc540
atcgccattaatcatattatgaactttgttcaacaatggatttttcggtccgcagtgacc600
tttgaaatcatcattttccaatgaccaaaccatgacaccagaaacgcctaattctttcag660
gaaagccaacttgcatgatatactggccagatcatcgtaaccgacccagattttatcatt720
gtaaccatatggagcattgtaatattcatcgtattggatatgccattcttctttttgaaa780
caattgacacaattctatatatgagaggacaccttcttcaccagaaatgaaacctggggg840
9
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
cgacatgcctttggctggatctccaagtttgagtttgcttcgatcttcaatgctccaagc900
acggccatagaatggaacacccattaccaatttgtctctggtggcaccattgttcaaata960
atagtgcatggtgtagttgacattgaagtaagtgtgcaactcatcagtttcatctggtcg1020
tttatacaacggagcattgtgaccgtaaaagttttcccat.ccaccgtggtaatcatatgt1080
catgacattcatccaatcgaacaatttgttcaattctttgatatcataagctcggtcgat1140
tttgtctttacctggtgatactgcagcagtcaacaagtagccatgaggttcaaaagcgtc1200
tttaagttctctaaccaaagccaaatagttttgtttatcgattttcgggttacccaatcg1260
agatccaggatactcccaatccaaatctagaccgtcgaacttgtattcttgcaaaaagtc1320
caaaactgattgtatgaattgttgacgatatgttggatttgcagccatatcggaatattt1380
ttccgagccttcataccaaccaccaagtgaaatcatggtggttaattctggattcttcaa1440
tcgcaagttgttgaaacgttcataaccacgtttttcccatgagttatggttatcatcttg1500
gtaaggatcgaaaacttgaattgtgtatttgtattcatcaattttagogaaaccatacat1560
taaatgtgtacacttgaatggatcaatatcttcgatagtgtatggatcaactttatgata1620
tacggaccatgttccaacataacaaacaattctcatcggatttttcgaataatcattatg1680
atctcgtttgatggatgcattcataaggccaatgcaggccataatactaagtattgcata1740
tatggttttcat 1752
<210> 17
<211> 1665
<212> DNA
<213> Dermatophagoides farinae
<220>
<221> CDS
<222> (1)..(1665)
<223>
<400>
17
atgaaa accatatat gcaata cttagtatt atggcctgcatt ggcctt 48
MetLys ThrIleTyr AlaIle LeuSerIle MetAlaCysIle GlyLeu
1 5 10 15
atgaat gcatccatc aaacga gatcataat gattattcgaaa aatccg 96
MetAsn AlaSerIle LysArg AspHisAsn AspTyrSerLys AsnPro
20 25 30
atgaga attgtttgt tatgtt ggaacatgg tccgtatatcat aaagtt 144
MetArg IleValCys TyrVal G1yThrTrp SerValTyrHis LysVal
35 40 45
gatcca tacactatc gaagat attgatcca ttcaagtgtaca cattta 192
AspPro TyrThrIle GluAsp IleAspPro PheLysCysThr HisLeu
50 55 60
atgtat ggtttcget aaaatt gatgaa,tac aaatacacaatt caagtt 240
MetTyr GlyPheAla LysIle AspGluTyr LysTyrThrIle GlnVal
65 70 75 80
10
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
ttcgatccttac caagatgat aaccataac tcatgggaaaaa cgtggt 288
PheAspProTyr GlnAspAsp AsnHisAsn SerTrpGluLys ArgGly
85 90 95
tatgaacgtttc aacaacttg cgattgaag aatccagaatta accacc 336
TyrGluArgPhe AsnAsnLeu ArgLeuLys AsnProGluLeu ThrThr
100 105 110
atgatttcactt ggtggttgg tatgaaggc tcggaaaaatat tccgat 384
MetIleSerLeu GlyGlyTrp TyrGluGly SerGluLysTyr SerAsp
115 120 125
atggetgcaaat ccaacatat cgtcaacaa ttcatacaatca gttttg 432
MetAlaAlaAsn ProThrTyr ArgGlnGln PheIleGlnSer ValLeu
130 135 140
gactttttgcaa gaatacaag ttcgacggt ctagatttggat tgggag 480
AspPheLeuGln GluTyrLys PheAspGly LeuAspLeuAsp TrpGlu
145 150 155 160
tatcct ggatctcga ttgggt aacccgaaa atcgataaacaa aactat 528
TyrPro GlySerArg LeuGly AsnProLys IleAspLysGln AsnTyr
165 170 175
ttgget ttggttaga gaactt aaagacget tttgaacctcat ggctac 576
LeuAla LeuValArg GluLeu LysAspAla PheGluProHis GlyTyr
180 185 190
ttgttg actgetgca gtatca ccaggtaaa gacaaaatcgac cgaget 624
LeuLeu ThrAlaAla ValSer ProGlyLys AspLysI1eAsp ArgAla
195 200 205
tatgat atcaaagaa ttgaac aaattgttc gattggatgaat gtcatg 672
TyrAsp IleLysGlu LeuAsn LysLeuPhe AspTrpMetAsn ValMet
210 215 220
acatat gattaccac ggtgga tgggaaaac ttttacggtcac aatget 720
ThrTyr AspTyrHis GlyGly TrpGluAsn PheTyrGlyHis AsnAla
225 230 235 240
ccgttg tataaacga ccagat gaaactgat gagttgcacact tacttc 768
ProLeu TyrLysArg ProAsp GluThrAsp GluLeuHisThr TyrPhe
245 250 255
aatgtc aactacacc atgcac tattatttg aacaatggtgcc accaga 816
AsnVal AsnTyrThr MetHis TyrTyrLeu AsnAsnGlyAla ThrArg
260 265 270
gacaaa ttggtaatg ggtgtt ccattctat ggccgtgettgg agcatt 864
AspLys LeuValMet GlyVal ProPheTyr GlyArgAlaTrp SerIle
275 280 285
gaagat cgaagcaaa ctcaaa cttggagat ccagccaaaggc atgtcg 912
GluAsp ArgSerLys LeuLys LeuGlyAsp ProAlaLysGly MetSer
290 295 300
ccccca ggtttcatt tctggt gaagaaggt gtcctctcatat atagaa 960
ProPro GlyPheIle SerGly GluGluGly Va1LeuSerTyr IleGlu
305 310 315 320
ttgtgt caattgttt caaaaa gaagaatgg catatccaatac gatgaa 1008
LeuCys GlnLeuPhe GlnLys GluGluTrp HisIleGlnTyr AspGlu
325 , 330 335
11
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
tattacaatget ccatatggt tacaatgataaa atctgggtc ggttac 1056
TyrTyrAsnAla ProTyrGly TyrAsnAspLys IleTrpVal GlyTyr
340 345 350
gatgatctggcc agtatatca tgcaagttgget ttcctgaaa gaatta 1104
AspAspLeuAla SerIleSer CysLysLeuAla PheLeuLys GluLeu
355 360 365
ggcgtttctggt gtcatggtt tggtcattggaa aatgatgat ttcaaa 1152
GlyValSerGly ValMetVa1 TrpSerLeuGlu AsnAspAsp PheLys
370 375 380
ggtcactgcgga ccgaaaaat ccattgttgaac aaagttcat aatatg 1200
GlyHisCysGly ProLysAsn ProLeuLeuAsn LysValHis AsnMet
385 390 395 400
attaatggcgat gaaaagaac tctttcgaatgc attttgggt ccaagt 1248
Ile,AsnGlyAsp GluLysAsn SerPheGluCys IleLeuGly ProSer
405 410 415
acaacgacacca actccaacg acgacacccaca accccgact acaacg 1296
ThrThrThrPro ThrProThr ThrThrProThr ThrProThr ThrThr
420 425 430
ccaacaactcct tctcccacc accccgacaaca accccttct cccacc 1344
ProThrThrPro SerProThr ThrProThrThr ThrProSer ProThr
435 440 445
accccgacaaca accccttct CCCaCCaCaCCg acaacaact ccttct 1392
ThrProThrThr ThrProSer ProThrThrPro ThrThrThr ProSer
450 455 460
cccaccacacca acaccaaca acaccaacacca gcccctaca acatcg 1440
ProThrThrPro ThrProThr ThrProThrPro AlaProThr ThrSer
465 470 475 480
acaccttcgcca accacgacc gaacacacaagc gaaacacca aaatat 1488
ThrProSerPro ThrThrThr GluHisThrSer GluThrPro LysTyr
485 490 495
acaacctatgtc gatggacat cttatcaaatgt tacaaggaa ggtgat 1536
ThrThrTyrVal AspGlyHis LeuIleLysCys TyrLysGlu GlyAsp
500 505 510
atcccacatcca accaatata cacaaatatttg gtctgtgaa tttgtt 1584
IleProHisPro ThrAsnIle HisLysTyrLeu ValCysGlu PheVal
515 520 525
aatggtggctgg tgggttcat attatgccctgt ccaccgggc actatt 1632
AsnGlyGlyTrp TrpValHis IleMetProCys ProProGly ThrIle
530 535 540
tggtgtcaagaa aaattgact tgtataggcgaa 1665
TrpCysGlnGlu LysLeuThr CysIleGlyG1u
545 550 555
<210> 18
<211> 555
<212> PRT
<213> Dermatophagoides farinae
<400> 18
12
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Met Lys Thr Ile Tyr Ala Ile Leu Ser Ile Met Ala Cys Ile Gly Leu
1 5 10 15
Met Asn Ala Ser Ile Lys Arg Asp His Asn Asp Tyr Ser Lys Asn Pro
20 25 30
Met Arg Ile Val Cys Tyr Val Gly Thr Trp Ser Val Tyr His Lys Val
35 40 45
Asp Pro Tyr Thr Ile Glu Asp Ile Asp Pro Phe Lys Cys Thr His Leu
50 55 60
Met Tyr Gly Phe Ala Lys Ile Asp Glu Tyr Lys Tyr Thr Ile Gln Val
65 70 75 80
Phe Asp Pro Tyr Gln Asp Asp Asn His Asn Ser Trp Glu Lys Arg Gly
85 90 95
Tyr Glu Arg Phe Asn Asn Leu Arg Leu Lys Asn Pro Glu Leu Thr Thr
100 105 110
Met Ile Ser Leu Gly Gly Trp Tyr Glu Gly Ser Glu Lys Tyr Ser Asp
115 120 125
Met Ala Ala Asn Pro Thr Tyr Arg Gln Gln Phe Ile Gln Ser Val Leu
130 135 140
Asp Phe Leu Gln Glu Tyr Lys Phe Asp Gly Leu Asp Leu Asp Trp Glu
145 150 155 160
Tyr Pro Gly Ser Arg Leu Gly Asn Pro Lys Tle Asp Lys Gln Asn Tyr
165 170 175
Leu Ala Leu Val Arg Glu Leu Lys Asp Ala Phe Glu Pro His Gly Tyr
180 185 190
Leu Leu Thr Ala Ala Val Ser Pro Gly Lys Asp Lys Ile Asp Arg A1a
195 200 205
Tyr Asp Ile Lys Glu Leu Asn Lys Leu Phe Asp Trp Met Asn Val Met
210 215 220
Thr Tyr Asp Tyr His Gly Gly Trp Glu Asn Phe Tyr Gly His Asn Ala
225 230 235 240
Pro Leu Tyr Lys Arg Pro Asp Glu Thr Asp Glu Leu His Thr Tyr Phe
245 250 255
13
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT. ST25 . tact
Asn Val Asn Tyr Thr Met His Tyr Tyr Leu Asn Asn Gly Ala Thr Arg
260 265 270
Asp Lys Leu Val Met Gly Val Pro Phe Tyr Gly Arg Ala Trp Ser Ile
275 280 285
Glu Asp Arg Ser Lys Leu Lys Leu Gly Asp Pro Ala Lys Gly Met Ser
290 295 300
Pro Pro Gly Phe Ile Ser Gly Glu Glu Gly Val Leu Ser Tyr Ile Glu
305 310 315 320
Leu Cys Gln Leu Phe Gln Lys Glu Glu Trp His Ile Gln Tyr Asp Glu
325 330 335
Tyr Tyr Asn Ala Pro Tyr Gly Tyr Asn Asp Lys Ile Trp Val Gly Tyr
340 345 350
Asp Asp Leu Ala Ser Ile Ser Cys Lys Leu Ala Phe Leu Lys Glu Leu
355 360 365
Gly Val Ser Gly Val Met Val Trp Ser Leu Glu Asn Asp Asp Phe Lys
370 375 380
Gly His Cys Gly Pro Lys Asn Pro Leu Leu Asn Lys Val His Asn Met
385 390 395 400
Ile Asn Gly Asp Glu Lys Asn Ser Phe Glu Cys Ile Leu Gly Pro Ser
405 410 415
Thr Thr Thr Pro Thr Pro Thr Thr Thr Pro Thr Thr Pro Thr Thr Thr
420 425 430
Pro Thr Thr Pro Ser Pro Thr Thr Pro Thr Thr Thr Pro Ser Pro Thr
435 440 445
Thr Pro Thr Thr Thr Pro Ser Pro Thr Thr Pro Thr Thr Thr Pro Ser
450 455 460
Pro Thr Thr Pro Thr Pro Thr Thr Pro Thr Pro Ala Pro Thr Thr Ser
465 470 475 480
Thr Pro Ser Pro Thr Thr Thr Glu His Thr Ser Glu Thr Pro Lys Tyr
485 490 495
Thr Thr Tyr Val Asp Gly His Leu Ile Lys Cys Tyr Lys Glu Gly Asp
500 505 510
14
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Ile Pro His Pro Thr Asn Ile His Lys Tyr Leu Val Cys Glu Phe Val
515 520 525
Asn Gly Gly Trp Trp Val His Ile Met Pro Cys Pro Pro Gly Thr Ile
530 535 540
Trp Cys Gln Glu Lys Leu Thr Cys Ile Gly Glu
545 550 555
<210>
19
<211>
1665
<212>
DNA
<213>
Dermatophagoides
farinae
<400>
19
ttcgcctatacaagtcaatttttcttgacaccaaatagtgcccggtggacagggcataat60
atgaacccaccagccaccattaacaaattcacagaccaaatatttgtgtatattggttgg120
atgtgggatatcaccttccttgtaacatttgataagatgtccatcgacataggttgtata180
ttttggtgtttcgcttgtgtgttcggtcgtggttggcgaaggtgtcgatgttgtaggggC240
tggtgttggtgttgttggtgttggtgtggtgggagaaggagttgttgtcggtgtggtggg300
agaaggggttgttgtcggggtggtgggagaaggggttgttgtcggggtggtgggagaagg360
agttgttggcgttgtagtcggggttgtgggtgtcgtcgttggagttggtgtcgttgtact420
tggacccaaaatgcattcgaaagagttcttttcatcgccattaatcatattatgaacttt480
gttcaacaatggatttttcggtccgcagtgacctttgaaatcatcattttccaatgacca540
aaccatgacaccagaaacgcctaattctttcaggaaagccaacttgcatgatatactggc600
cagatcatcgtaaccgacccagattttatCattgtaaccatatggagcattgtaatattc660
atcgtattggatatgccattcttctttttgaaacaattgacacaattctatatatgagag720
gacaccttcttcaccagaaatgaaacctgggggcgacatgcctttggctggatctccaag780
tttgagtttgcttcgatcttcaatgctccaagcacggccatagaatggaacacccattac.840
caatttgtctctggtggcaccattgttcaaataatagtgcatggtgtagttgacattgaa900
gtaagtgtgcaactcatcagtttcatctggtcgtttatacaacggagcattgtgaccgta960
aaagttttcccatccaccgtggtaatcatatgtcatgacattcatccaatcgaacaattt1020
gttcaattctttgatatcataagctcggtcgattttgtctttacctggtgatactgcagc1080
agtcaacaagtagccatgaggttcaaaagcgtctttaagttctctaaccaaagccaaata1140
gttttgtttatcgattttcgggttacccaatcgagatccaggatactcccaatccaaatc1200
tagaccgtcgaacttgtattcttgcaaaaagtccaaaactgattgtatgaattgttgacg1260
atatgttggatttgcagccatatcggaatatttttccgagccttcataccaaccaccaag1320
tgaaatcatggtggttaattctggattcttcaatcgcaagttgttgaaacgttcataacc1380
15
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
acgtttttcccatgagttatggttatcatcttggtaaggatcgaaaacttgaattgtgta1440
tttgtattcatcaattttagcgaaaccatacattaaatgtgtacacttgaatggatcaat1500
atcttcgatagtgtatggatcaactttatgatatacggaccatgttccaacataacaaac1560
aattctcatcggatttttcgaataatcattatgatctcgtttgatggatgcattcataag1620
gccaatgcaggccataatactaagtattgcatatatggttttcat 1665
<210> 20
<211> 1608
<212> DNA
<213> Dermatophagoides farinae
<220>
<221> CDS
<222> (1)..(1608)
<223>
<400>
20
tcc atc,aaacga gatcataat gattattcg aaaaatccg atgagaatt 48
Ser IleLysArg AspHisAsn AspTyrSer LysAsnPro MetArgIle
1 5 10 15
gtt tgttatgtt ggaacatgg tccgtatat cataaagtt gatccatac 96
Val CysTyrVal,GlyThrTrp SerValTyr HisLysVal AspProTyr
20 25 30
act atcgaagat attgatcca ttcaagtgt acacattta atgtatggt 144
Thr IleGluAsp IleAspPro PheLysCys ThrHisLeu MetTyrGly
35 ~ 40 45
ttc getaaaatt gatgaatac aaatacaca attcaagtt ttcgatcct 192
Phe AlaLysIle AspGluTyr LysTyrThr IleGlnVal PheAspPro
50 55 60
tac caagatgat aaccataac tcatgggaa aaacgtggt tatgaacgt 240
Tyr GlnAspAsp AsnHisAsn SerTrpGlu LysArgGly TyrGluArg
65 70 75 80
ttc aacaacttg cgattgaag aatccagaa ttaaccacc atgatttca 288
Phe AsnAsnLeu ArgLeuLys AsnProGlu LeuThrThr MetIleSer
85 90 95
ctt ggtggttgg tatgaaggc tcggaaaaa tattccgat atggetgca 336
Leu GlyGlyTrp TyrGluGly SerGluLys TyrSerAsp MetAlaAla
100 105 110
aat ccaacatat cgtcaacaa ttcatacaa tcagttttg gactttttg 384
Asn ProThrTyr ArgGlnGln PheIleGln SerValLeu AspPheLeu
115 120 125
caa gaatacaag ttcgacggt ctagatttg gattgggag tatcctgga 432
Gln GluTyrLys PheAspGly LeuAspLeu AspTrpGlu TyrProGly
130 135 140
tct cgattgggt aacccgaaa atcgataaa caaaactat ttggetttg 480
Ser ArgLeuGly AsnProLys IleAspLys GlnAsnTyr LeuAlaLeu
145 150 155 160
16
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
gttagagaactt aaagacget tttgaacct catggctac ttgttgact 528
ValArgGluLeu LysAspAla PheGluPro .HisGlyTyr LeuLeuThr
165 170 175
getgcagtatca ccaggtaaa gacaaaatc gaccgaget tatgatatc 576
AlaAlaValSer ProGlyLys AspLysIle AspArgAla TyrAspIle
180 185 190
aaagaattgaac aaattgttc gattggatg aatgtcatg acatatgat 624
LysGluLeuAsn LysLeuPhe AspTrpMet AsnValMet ThrTyrAsp
195 200 205
taccacggtgga tgggaaaac ttttacggt cacaatget ccgttgtat 672
TyrHisGlyGly TrpGluAsn PheTyrGly HisAsnAla ProLeuTyr
210 215 220
aaacgaccagat gaaactgat gagttgcac acttacttc aatgtcaac 720
LysArgProAsp GluThrAsp GluLeuHis ThrTyrPhe AsnValAsn
225 230 235 240
tacaccatgcac tattatttg aacaatggt gccaccaga gacaaattg 768
TyrThrMetHis TyrTyrLeu AsnAsnGly AlaThrArg AspLysLeu
245 250 255
gtaatgggtgtt Ccattctat ggccgtget tggagcatt gaagatcga 816
ValMetGlyVal ProPheTyr GlyArgAla TrpSerIle GluAspArg
260 265 270
agcaaactcaaa cttggagat ccagccaaa ggcatgtcg cccccaggt 864
SerLysLeuLys LeuGlyAsp ProAlaLys GlyMetSer ProProGly
275 280 285
ttcatttctggt gaagaaggt gtcctctca tatatagaa ttgtgtcaa 912
PheIleSerGly G1uGluGly ValLeuSer TyrIleGlu LeuCysGln
290 295 300
ttgtttcaaaaa gaagaatgg catatccaa tacgatgaa tattacaat 960
LeuPheGlnLys GluGluTrp HisIleGln TyrAspGlu TyrTyrAsn
305 310 315 320
getccatatggt tacaatgat aaaatctgg gtcggttac gatgatctg 1008
AlaProTyrGly TyrAsnAsp LysIleTrp ValGlyTyr AspAspLeu
325 330 335
gccagtatatca tgcaagttg getttcctg aaagaatta ggcgtttct 1056
AlaSerIleSer CysLysLeu AlaPheLeu LysGluLeu GlyValSer
340 345 350
ggtgtcatggtt tggtcattg gaaaatgat gatttcaaa ggtcaCtgc 1104
GlyValMetVal TrpSerLeu GluAsnAsp AspPheLys GlyHisCys
355 360 365
ggaccgaaaaat ccattgttg aacaaagtt cataatatg attaatggc 1152
GlyProLysAsn ProLeuLeu AsnLysVa1 HisAsnMet IleAsnGly
370 375 380
gatgaaaagaac tctttcgaa tgcattttg ggtccaagt acaacgaca 1200
AspGluLysAsn SerPheGlu CysIleLeu GlyProSer ThrThrThr
385 390 395 400
ccaactccaacg acgacaccc acaaccccg actacaacg ccaacaact 1248
ProThrProThr ThrThrPro ThrThrPro ThrThrThr ProThrThr
405 410 415
17
CA 02420459 2003-02-21
WO PCT/USO1/28730
02/22807
AL-2-C4-PCT.ST25.txt
cct tctCCC aCCaCCCCgaca acaacc ccttctcccacc accccgaca 1296
Pro SerPro ThrThrProThr ThrThr ProSerProThr ThrProThr
420 425 430
aca acccct tctcccaccaca ccgaca acaactccttct cccaccaca 1344
Thr ThrPro SerProThrThr ProThr ThrThrProSer ProThrThr
435 440 445
cca acacca acaacaccaaca ccagcc cctacaacatcg acaccttcg 1392
Pro ThrPro ThrThrProThr ProAla ProThrThrSer ThrProSer
450 455 460
cca accacg accgaacacaca agcgaa acaccaaaatat acaacctat 1440
Pro ThrThr ThrGluHisThr SerGlu ThrProLysTyr ThrThrTyr
465 470 475 480
gtc gatgga catcttatcaaa tgttac aaggaaggtgat atcccacat 1488
Val AspGly HisLeuIleLys CysTyr LysGluGlyAsp IleProHis
485 490 495
cca accaat atacacaaatat ttggtc tgtgaatttgtt aatggtggc 1536
Pro ThrAsn IleHisLysTyr LeuVal CysGluPheVal AsnGlyGly
500 505 510
tgg tgggtt catattatgccc tgtcca ccgggcactatt tggtgtcaa 1584
Trp TrpVal HisI1eMetPro CysPro ProGlyThrIle TrpCysGln
515 520 525
gaa aaattg acttgtataggc gaa 1608
Glu LysLeu ThrCysIleGly Glu
530 535
<210> 21
<211> 53
6
<212> PRT
<213> Dermatophagoides farinae
<400> 21
Ser Ile Lys Arg Asp His Asn Asp Tyr Ser Lys Asn Pro Met Arg I1e
1 5 10 15
Val Cys Tyr Val Gly Thr Trp Ser Val Tyr His Lys Val Asp Pro Tyr
20 25 30
Thr Ile Glu Asp Ile Asp Pro Phe Lys Cys Thr His Leu Met Tyr Gly
35 40 45
Phe Ala Lys Ile Asp Glu Tyr Lys Tyr Thr Ile Gln Val Phe Asp Pro
50 55 60
Tyr Gln Asp Asp Asn His Asn Ser Trp Glu Lys Arg Gly Tyr Glu Arg
65 70 75 80
Phe Asn Asn Leu Arg Leu Lys Asn Pro Glu Leu Thr Thr Met Ile Ser
85 90 95
18
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Leu Gly Gly Trp Tyr Glu Gly Ser Glu Lys Tyr Ser Asp Met Ala Ala
100 105 110
Asn Pro Thr Tyr Arg Gln Gln Phe Ile Gln Ser Va1 Leu Asp Phe Leu
115 120 125
Gln Glu Tyr Lys Phe Asp Gly Leu Asp Leu Asp Trp Glu Tyr Pro Gly
130 135 140
Ser Arg Leu Gly Asn Pro Lys Ile Asp Lys Gln Asn Tyr Leu Ala Leu
145 150 155 160
Val Arg Glu Leu Lys Asp A1a Phe Glu Pro His Gly Tyr Leu Leu Thr
165 170 175
AlaAla Val Ser Pro Gly Lys Asp Lys Ile Asp Arg Ala Tyr Asp Tle
180 185 190
Lys Glu Leu Asn Lys Leu Phe Asp Trp Met Asn Val Met Thr Tyr Asp
195 200 205
Tyr His Gly Gly Trp Glu Asn Phe Tyr Gly His Asn Ala Pro Leu Tyr
210 215 220
Lys Arg Pro Asp Glu Thr Asp Glu Leu His Thr Tyr Phe Asn Val Asn
225 230 235 240
Tyr Thr Met His Tyr Tyr Leu Asn Asn Gly Ala.Thr Arg Asp Lys Leu
245 250 255
Val Met Gly Val Pro Phe Tyr Gly Arg Ala Trp Ser Ile Glu Asp Arg
260 265 270
Ser Lys Leu Lys Leu Gly Asp Pro Ala Lys Gly Met Ser Pro Pro Gly
275 280 285
Phe Ile Ser Gly Glu Glu Gly Val Leu Ser Tyr Ile Glu Leu Cys Gln
290 295 300
Leu Phe Gln Lys Glu Glu Trp His Ile Gln Tyr Asp Glu Tyr Tyr Asn
305 310 315 320
Ala Pro Tyr Gly Tyr Asn Asp Lys Ile Trp Val Gly Tyr Asp Asp Leu
325 330 335
Ala Ser Ile Ser Cys Lys Leu Ala Phe Leu Lys Glu Leu Gly Val Ser
340 345 350
19
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Gly Val Met Val Trp Ser Leu Glu Asn Asp Asp Phe Lys Gly His Cys
355 360 365
Gly Pro Lys Asn Pro Leu Leu Asn Lys Val His Asn Met Ile Asn Gly
370 375 380
Asp Glu Lys Asn Ser Phe Glu Cys Ile Leu Gly Pro Ser Thr Thr Thr
385 390 395 400
Pro Thr Pro Thr Thr Thr Pro Thr Thr Pro Thr Thr Thr Pro Thr Thr
405 410 415
Pro Ser Pro Thr Thr Pro Thr Thr Thr Pro Ser Pro Thr Thr Pro Thr
420 425 430
Thr Thr Pro Ser Pro Thr Thr Pro Thr Thr Thr Pro Ser Pro Thr Thr
435 440 ~ 445
Pro Thr Pro Thr Thr Pro Thr Pro Ala Pro Thr Thr Ser Thr Pro Ser
450 455 460
Pro Thr Thr Thr Glu His Thr Ser Glu Thr Pro Lys Tyr Thr Thr Tyr
465 470 475 480
Val Asp G1y His Leu Ile Lys Cys Tyr Lys Glu Gly Asp I1e Pro His
485 490 495
Pro Thr Asn Ile His Lys Tyr Leu Va1 Cys Glu Phe Val Asn Gly Gly
500 505 510
Trp Trp Val His Ile Met Pro Cys Pro Pro Gly Thr Ile Trp Cys Gln
515 520 525
Glu Lys Leu Thr Cys Ile Gly Glu
530 535
<210>
22
<211>
1608
<212>
DNA
<213>
Dermatophagoides
farinae
<400>
22
ttcgcctatacaagtcaatttttcttgacaccaaatagtgcccggtggacagggcataat60
atgaacccaccagccaccattaacaaattcacagaccaaatatttgtgtatattggttgg120
atgtgggatatcaccttccttgtaacatttgataagatgtccatcgacataggttgtata180
ttttggtgtttcgcttgtgtgttcggtcgtggttggcgaaggtgtcgatgttgtaggggc240
tggtgttggtgttgttggtgttggtgtggtgggagaaggagttgttgtcggtgtggtggg300
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
agaaggggttgttgtcggggtggtgggagaaggggttgttgtcggggtggtgggagaagg360
agttgttggcgttgtagtcggggttgtgggtgtcgtcgttggagttggtgtcgttgtact420
tggacccaaaatgcattcgaaagagttcttttcatcgccattaatcatattatgaacttt480
gttcaacaatggatttttcggtccgcagtgacctttgaaatcatcattttccaatgacca540
aaccatgacaccagaaacgcctaattctttcaggaaagccaacttgcatgatatactggc600
cagatcatcgtaaccgacccagattttatcattgtaaccatatggagcattgtaatattc660
atcgtattggatatgccattcttctttttgaaacaattgacacaattctatatatgagag720
gacaccttcttcaccagaaatgaaacctgggggcgacatgcctttggctggatctccaag780
tttgagtttgcttcgatcttcaatgctccaagcacggccatagaatggaacacccattac840
caatttgtctctggtggcaccattgttcaaataatagtgcatggtgtagttgacattgaa900
gtaagtgtgcaactcatcagtttcatctggtcgtttatacaacggagcattgtgaccgta960
aaagttttcccatccaccgtggtaatcatatgtcatgacattcatccaatcgaacaattt1020
gttcaattctttgatatcataagctcggtcgattttgtctttacctggtgatactgcagc1080
agtcaacaagtagccatgaggttcaaaagcgtctttaagttctctaaccaaagccaaata1140
gttttgtttatcgattttcgggttacccaatcgagatccaggatactcccaatccaaatc1200
tagaccgtcgaacttgtattcttgcaaaaagtccaaaactgattgtatgaattgttgacg1260
atatgttggatttgcagccatatcggaatatttttccgagccttcataccaaccaccaag1320
tgaaatcatggtggttaattctggattcttcaatcgcaagttgttgaaacgttcataacc1380
acgtttttcccatgagttatggttatcatcttggtaaggatcgaaaacttgaattgtgta1440
tttgtattcatcaattttagcgaaaccatacattaaatgtgtacacttgaatggatcaat,1500
atcttcgatagtgtatggatcaactttatgatatacggaccatgttccaacataacaaac1560
aattctcatcggatttttcgaataatcattatgatctcgtttgatgga 1608
<210> 23
<211> 25
<212> PRT
<213> Dermatophagoides farinae
<220>
<221> MISC_FEATURE
<222> (1) . (1)
<223> Xaa --- any amino acid at position 1
<400> 23
Xaa Leu Glu Pro Lys Thr Val Cys Tyr Tyr Glu Ser Trp Val His His
1 5 10 15
Arg Gln Gly Glu Gly Lys Met Asp Pro
20 25
21
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
<210> 24
<211> 33
<212> PRT
<213> Dermatophagoides farinae
<220>
<221> MISC_FEATURE
<222> (18) .(18)
<223> Xaa = any amino acid at position 18
<220>
<221> MISC_FEATURE
<222> (28) .(28)
<223> Xaa = any amino acid at position 28
<220>
<221> MISC_FEATURE
<222> (31) . (31)
<223> Xaa = any amino acid at position 31
<220>
<221> MISC_FEATURE
<222> (32) .(32)
<223> Xaa = any amino acid at position 32
<400> 24
Ser Ile Lys Arg Asp His Asn Asp Tyr Ser Lys Asn Pro Met Met Ile
1 5 10 15
Val Xaa Tyr Gly Gly Ser Ser Gly Tyr Gln Ser Xaa Lys Arg Xaa Xaa
20 25 30
Thr
<210> 25
<211> 31
<212> DNA
<213> Artificial sequence
<220>
<223> Synthetic Primer
<220>
<221> misc_feature
<222> (24) .(24)
<223> n = a, c, t or g at position 24
<400> 25
aaacgtgatc ataaygatta ytcnaaraay c 31
<210> 26
22
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
<211> 31
<212> DNA
<213> Artificial sequence
AL-2-C4-PCT.ST25.txt
<220>
<223> Synthetic Primer
<400> ~ 26
aaacgtgatc ataaygatta yagyaaraay c 31
<210> 27
<211> 23
<212> DNA
<213> Artificial sequence
<220>
<223> Synthetic Primer
<220>
<221> misc_feature
<222> (12) .(12)
<223> n = a, c, t or g at position 12
<220>
<221> misc_feature
<222> (21) .(21)
<223> n = a, c, t or g at position 21
<400> 27
ccttcttcac cnacratcaa ncc 23
<210> 28
<211> 23
<212> DNA
<213> Artificial sequence
<220>
<223> Synthetic Primer
<220>
<221> misc_feature
<222> (12) .(12)
<223> n = a, c, t or g at position 12
<220>
<221> misc-feature
<222> (21) . . (21)
<223> n = a, c, t or g at position 21
<400> 28
ccttcttcac cnacratgaa ncc 23
<210> 29
<211> 13
<212> PRT
<213> Dermatophagoides farinae
23
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
<400> 29
Gln Tyr Gly Val Thr Gln Ala Val Val Thr Gln Pro Ala
1 5 10
<210> 30
<211> 11
<212> PRT
<213> Dermatophagoides farinae
<400> 30
Asp Glu Leu Leu Met Lys Ser Gly Pro Gly Pro
1 5 10
<210> 31
<211> 24
<212> PRT
<213> Dermatophagoides farinae
<400> 31
Asp Met Glu His Phe Thr Gln His Lys Gly Asn Ala Lys A1a Met Ile
1 5 10 15
Ala Val Gly Gly Ser Thr Met Ser
<210> 32
<211> 21
<212> PRT
<213> Dermatophagoides farinae
<400> 32
Asp Ala Asn Glu Glu Ala Arg Ser Gln Leu Pro Glu Thr Ala Met Val
1 5 10 15
Leu Ile Ly's Ser Gln
<210> 33
<211> 21
<212> PRT
<213> Dermatophagoides farinae
<220>
<221> MISC_FEATURE
<222> (11) .(11)
<223> Xaa = any amino acid at position 11
<400> 33
Gln Ser Arg Asp Arg Asn Asp Lys Pro Tyr Xaa I1e Val Lys Lys Lys
1 5 10 15
24
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
Lys Lys Ala Leu Asp
AL-2-C4-PCT.ST25.txt
<210> 34
<211> 1621
<212> DNA
<213> Dermatophagoides farinae
<220>
<221> CDS
<222> (14)..(1540)
<223>
<400>
34
agaacttatg tt 49
aaa gca
atg ttg
aaa ttt
acg tgt
aca ata
t tgg
gcc
Met
Lys
Thr
Thr
Phe
Ala
Leu
Phe
Cys
Ile
Trp
Ala
1 5 1 0
tgcatt ggcttgatg aatgcg gccactaaa cgagatcac aataattat 97
CysIle GlyLeuMet AsnAla AlaThrLys ArgAspHis AsnAsnTyr
15 20 25
tcgaaa aatccaatg cgaatc gtatgttat gttggaaca tggtccgtt 145
SerLys AsnProMet ArgIle ValCysTyr ValGlyThr TrpSerVal
35 40
tatcat aaagttgat ccatac acaattgaa gatattgat cctttcaaa 193
TyrHis LysValAsp ProTyr ThrIleGlu AspIleAsp ProPheLys
45 50 55 60
tgtact catttgatg tatggt tttgetaaa atcgatgaa tacaaatac 241
CysThr HisLeuMet TyrGly PheAlaLys TleAspGlu TyrLysTyr
65 70 75
accatt caagttttt gatcca tttcaagat gataaccat aactcatgg 289
ThrIle GlnValPhe AspPro PheGlnAsp AspAsnHis AsnSerTrp
80 85 90
gaaaaa cacgggtat gaacgt ttcaacaac ttgagattg aagaatcca 337
GluLys HisGlyTyr GluArg PheAsnAsn LeuArgLeu LysAsnPro
95 100 105
gaattg accaccatg atttca ttgggtggt tggtatgaa ggttcagaa 385
GluLeu ThrThrMet 21eSer LeuGlyGly TrpTyrGlu GlySerGlu
110 115 120
aaatat tcggatatg gcagcc aatccaaca tatcgtcag caatttgtt 433
LysTyr SerAspMet AlaAla AsnProThr TyrArgGln GlnPheVal
125 130 135 140
Caatca gttttggac tttttg caagaatac aaattcgat ggcctagat 481
GlnSer ValLeuAsp PheLeu GlnGluTyr LysPheAsp GlyLeuAsp
145 150 155
ttggat tgggaatat cctgga tcacggtta ggcaatcct aaaatcgat 529
LeuAsp TrpGluTyr ProGly SerArgLeu GlyAsnPro LysIleAsp
160 165 170
aaacaa aactattta acatta gttagagaa cttaaagag gcatttgaa 577
LysGln AsnTyrLeu ThrLeu ValArgGlu LeuLysGlu AlaPheGlu
175 180 185
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
cctttcggctac ttgttgact gccgcagta tcacccggtaaa gataaa 625
ProPheGlyTyr LeuLeuThr AlaAlaVal SerProG1yLys AspLys
190 195 200
attgacgtaget tatgagctc aaagaattg aaccaattgttc gattgg 673
IleAspValAla TyrGluLeu LysGluLeu AsnGlnLeuPhe AspTrp
205 210 215 220
atgaatgtcatg acttatgat taccatggc ggatgggaaaat gttttc 721
MetAsnValMet ThrTyrAsp TyrHisGly GlyTrpGluAsn ValPhe
225 230 235
ggccataatget ccgttgtat aaacgaccc gatgaaacggat gaattg 769
G1yHisAsnAla ProLeuTyr LysArgPro AspGluThrAsp GluLeu
240 245 250
cacacttacttc aatgtcaac tacaccatg cactattatttg aacaat 817
HisThrTyrPhe AsnValAsn TyrThrMet HisTyrTyrLeu AsnAsn
255 260 265
ggcgetactcga gacaaactt gttatgggt gttccattctat ggtcgt 865
GlyAlaThr.Arg AspLysLeu ValMetGly ValProPheTyr GlyArg
270 275 280
gettggagcatc gaagatcga agcaaagtc.aaacttggcgat CCggCC 913
AlaTrpSerIle GluAspArg SerLysVal LysLeuGlyAsp ProAla
285 290 295 300
aaaggcatgtct cctcctggt tttattact ggtgaagaaggt gttctc 961
LysGlyMetSer ProProGly PheIleThr GlyGluGluGly Va1Leu
305 310 315
tcatacatcgaa ttgtgtcag ttattccag aaagaagaatgg catatt 1009
SerTyrIleGlu LeuCysGln LeuPheGln LysGluGluTrp HisIle,
320 325 330
caatacgatgaa tattacaat getccatac ggatataatgat aaaatc 1057
GlnTyrAspGlu TyrTyrAsn AlaProTyr GlyTyrAsnAsp LysIle
335 340 345
tgggt~ ggttac gatgatctg getagtata tcatgcaagttg gccttt 1105
TrpVal GlyTyr AspAspLeu AlaSerIle SerCysLysLeu AlaPhe
350 355 360
ctcaaa gaattg ggcgtctct ggcgttatg atatggtcattg gaaaac 1153
LeuLys GluLeu GlyValSer GlyValMet IleTrpSerLeu GluAsn
365 370 375 380
gatgat ttcaaa ggtcattgc ggaccgaaa tatccattgttg aacaaa 1201
AspAsp PheLys GlyHisCys GlyProLys TyrProLeuLeu AsnLys
385 390 395
gttcac aatatg atcaatggt gatgaaaag aactcttacgaa tgtctt 1249
ValHis AsnMet IleAsnGly AspGluLys AsnSerTyrGlu CysLeu
400 405 410
ttgggc ccaagt acaaccaca ccaacacca accaccccgtca actact 1297
LeuGly ProSer ThrThrThr ProThrPro ThrThrProSer ThrThr
415 420 425
tcgact accaca cca.acgcct accaccacc gatagcacaagc gaaaca 1345
SerThr ThrThr ProThrPro ThrThrThr AspSerThrSer GluThr
430 435 440
26
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
ccaaaatacactacg tatattgat ggacatttg attaaatgc tataaa 1393
ProLysTyrThrThr TyrIleAsp GlyHisLeu IleLysCys TyrLys
445 450 455 460
caaggttatcttcca catccaact gatgttcat aaatattta gtttgt 1441
GlnGlyTyrLeuPro HisProThr AspValHis LysTyrLeu ValCys
465 470 475
gaatatattgccaca ccaaacggt ggttggtgg gtacacatt atggat 1489
GluTyrIleAlaThr ProAsnGly GlyTrpTrp ValHisIle MetAsp
480 485 490
tgtccaaaaggaact agatggcac gcaacatta aaaaattgt attcaa 1537
CysProLysGlyThr ArgTrpHis AlaThrLeu LysAsnCys IleGln
495 500 505
gaatgatctgata tatttgtaac taaatgaaat ttaaataaaa
1590
tgttttttgc
Glu
ttatttgaat ccattaaaaa aaaaaaaaaa a 1621
<210> 35
<211> 509
<212> PRT
<213> Dermatophagoides farinae
<400> 35
Met Lys Thr Thr Phe Ala Leu Phe Cys Ile Trp Ala Cys Ile Gly Leu
1 5 10 15
Met Asn Ala Ala Thr Lys Arg Asp His Asn Asn Tyr Ser Lys Asn Pro
20 25 30
Met Arg Ile Val Cys Tyr Val Gly Thr Trp Ser Val Tyr His Lys Val
35 40 45
Asp Pro Tyr Thr Ile Glu Asp Ile Asp Pro Phe Lys Cys Thr His Leu
50 55 60
Met Tyr Gly Phe Ala Lys Ile Asp Glu Tyr Lys Tyr Thr Ile Gln Val
65 70, 75 80
Phe Asp Pro Phe Gln Asp Asp Asn His Asn Ser Trp Glu Lys His G1y
85 90 95
Tyr Glu Arg Phe Asn Asn Leu Arg Leu Lys Asn Pro Glu Leu Thr Thr
100 105 110
Met Ile Ser Leu Gly Gly Trp Tyr Glu Gly Ser Glu Lys Tyr Ser Asp
115 120 125
Met Ala Ala Asn Pro Thr Tyr Arg Gln Gln Phe Val Gln Ser Val Leu
130 ' 135 140
27
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Asp Phe Leu Gln Glu Tyr Lys Phe Asp Gly Leu Asp Leu Asp Trp Glu
145 150 155 160
Tyr Pro Gly Ser Arg Leu Gly Asn Pro Lys Ile Asp Lys Gln Asn Tyr
165 170 175
Leu Thr Leu Val Arg Glu Leu Lys G1u Ala Phe Glu Pro Phe Gly Tyr
180 185 190
Leu Leu Thr Ala Ala Val Ser Pro Gly Lys Asp Lys Ile Asp Val Ala
195 200 205
Tyr G1u Leu Lys Glu Leu Asn Gln Leu Phe Asp Trp Met Asn Val Met
210 215 220
Thr Tyr Asp Tyr His Gly Gly Trp Glu Asn Val Phe Gly His Asn Ala
225 '230 235 240
Pro Leu Tyr Lys Arg Pro Asp Glu Thr Asp Glu Leu His Thr Tyr Phe
245 250 255
Asn Va1 Asn Tyr Thr Met His Tyr Tyr Leu Asn Asn Gly Ala Thr Arg
260 265 270
Asp Lys Leu Val Met Gly Val Pro Phe Tyr Gly Arg Ala Trp Ser Ile
275 280 285
Glu Asp Arg Ser Lys Val Lys Leu Gly Asp Pro Ala Lys Gly Met Ser
290 295 300
Pro Pro Gly Phe Ile Thr Gly Glu Glu Gly Val Leu Ser Tyr Ile Glu
305 310 315 320
Leu Cys Gln Leu Phe Gln Lys Glu Glu Trp His Ile Gln Tyr Asp Glu
325 330 335
Tyr Tyr Asn Ala Pro Tyr Gly Tyr Asn Asp Lys Ile Trp Val Gly Tyr
340 345 350
Asp Asp Leu Ala Ser Ile Ser Cys Lys Leu Ala Phe Leu Lys Glu Leu
355 360 365
Gly Val Ser Gly Val Met Ile Trp Ser Leu Glu Asn Asp Asp Phe Lys
370 375 380
Gly His Cys Gly Pro Lys Tyr Pro Leu Leu Asn Lys Val His Asn Met
385 390 395 400
28
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Ile Asn Gly Asp Glu Lys Asn Ser Tyr Glu Cys Leu Leu Gly Pro Ser
405 410 415
Thr Thr Thr Pro Thr Pro Thr Thr Pro Ser Thr Thr Ser Thr Thr Thr
420 425 430
Pro Thr Pro Thr Thr Thr Asp Ser Thr Ser Glu Thr Pro Lys Tyr Thr
435 440 445
Thr Tyr Ile Asp Gly His Leu Ile Lys Cys Tyr Lys Gln Gly Tyr Leu
450 455 460
Pro His Pro Thr Asp Val His Lys Tyr Leu Val Cys Glu Tyr Ile Ala
465 470 475 480
Thr Pro Asn Gly Gly Trp Trp Val His Ile Met Asp Cys Pro Lys Gly
485 490 495
Thr Arg Trp His Ala Thr Leu Lys Asn Cys Ile Gln Glu
500 505
<210> 36
<211> 1621
<212> DNA
<213> Dermatophagoides farinae
<400> 36
tttttttttt ttttttaatg gattcaaata attttattta aatttcattt agcaaaaaac 60
agttacaaatatatcagatcattcttgaatacaattttttaatgttgcgtgccatctagt120
tccttttggaCaatccataatgtgtacccaccaaccaccgtttggtgtggcaatatattc180
acaaactaaatatttatgaacatcagttggatgtggaagataaccttgtttatagcattt240
aatcaaatgtccatcaatatacgtagtgtattttggtgtttcgcttgtgctatcggtggt300
ggtaggcgttggtgtggtagtcgaagtagttgacggggtggttggtgttggtgtggttgt360
acttgggcccaaaagacattcgtaagagttcttttcatcaCcattgatcatattgtgaac420
tttgttcaacaatggatatttcggtccgcaatgacctttgaaatcatcgttttccaatga480
ccatatcataacgccagagacgcccaattctttgagaaaggccaacttgcatgatatact540
agccagatcatcgtaaccaacccagattttatcattatatccgtatggagcattgtaata600
ttcatcgtattgaatatgccattcttctttctggaataactgacacaattcgatgtatga660
gagaacaccttcttcaccagtaataaaaccaggaggagacatgcctttggccggatcgcc720
aagtttgactttgcttcgatcttcgatgctccaagcacgaccatagaatggaacacccat780
aacaagtttgtctcgagtagcgccattgttcaaataatagtgcatggtgtagttgacatt840
29
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
gaagtaagtgtgcaattcatccgtttcatcgggtcgtttatacaacggagcattatggcc900
gaaaacattttcccatccgccatggtaatcataagtcatgacattcatccaatcgaacaa960
ttggttcaattctttgagctcataagctacgtcaattttatctttaccgggtgatactgc1020
ggcagtcaacaagtagccgaaaggttcaaatgcctctttaagttctctaactaatgttaa1080
atagttttgtttatcgattttaggattgcctaaccgtgatccaggatattcccaatccaa1140
atctaggccatcgaatttgtattcttgcaaaaagtccaaaactgattgaacaaattgctg1200
acgatatgttggattggctgccatatccgaatatttttctgaaccttcataccaaccacc1260
caatgaaatcatggtggtcaattctggattcttcaatctcaagttgttgaaacgttcata1320
cccgtgtttttcccatgagttatggttatcatcttgaaatggatcaaaaacttgaatggt1380
gtatttgtattcatcgattttagcaaaaccatacatcaaatgagtacatttgaaaggatc1440
aatatcttcaattgtgtatggatcaactttatgataaacggaccatgttccaacataaca1500
tacgattcgcattggatttttcgaataattattgtgatctcgtttagtggccgcattcat1560
caagccaatgcaggcccatatacaaaacaatgcaaatgtcgttttcattttcataagttc1620
t 1621
<210>
37
<211>
1527
<212>
DNA
<213>
Dermatophagoides
farinae
<220>
<221> CDS
<222> (1)..(1527)
<223>
<400>
37
atgaaa acgaca tttgcattg ttttgtata tgggcctgcatt ggcttg 48
MetLys ThrThr PheAlaLeu PheCysIle TrpAlaCysIle GlyLeu
1 5 10 15
atgaat gcggcc actaaacga gatcacaat aattattcgaaa aatcca 96
MetAsn AlaAla ThrLys'ArgAspHisAsn AsnTyrSerLys AsnPro
20 25 30
atgcga atcgta tgttatgtt ggaacatgg-tccgtttatcat aaagtt 144
MetArg IleVal CysTyrVal GlyThrTrp SerValTyrHis LysVal
35 40 45
gatcca tacaca attgaagat attgatcct ttcaaatgtact catttg 192
AspPro TyrThr IleGluAsp IleAspPro PheLysCysThr HisLeu
50 55 60
atgtat ggtttt getaaaatc gatgaatac aaatacaccatt caagtt 240
MetTyr GlyPhe AlaLysIle AspGluTyr LysTyrThrIle GlnVal
65 70 75 80
tttgat ccattt caagatgat aaccataac tcatgggaaaaa cacggg 288
PheAsp ProPhe GlnAspAsp AsnHisAsn SerTrpGluLys HisGly
85 90 95
30
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
tatgaacgtttc aacaacttg agattgaagaat ccagaattg accacc 336
TyrGluArgPhe AsnAsnLeu ArgLeuLysAsn ProGluLeu ThrThr
100 105 110
atgatttcattg ggtggttgg tatgaaggttca gaaaaatat tcggat 384
MetIleSerLeu GlyGlyTrp TyrGluGlySer GluLysTyr SerAsp
115 120 125
atggcagccaat ccaacatat cgtcagcaattt gttcaatca gttttg 432
MetAlaAlaAsn ProThrTyr ArgGlnGlnPhe ValGlnSer ValLeu
130 135 140
gactttttgcaa gaatacaaa ttcgatggccta gatttggat tgggaa 480
AspPheLeuGln GluTyrLys PheAspG1yLeu AspLeuAsp TrpGlu
145 150 155 160
tatCctggatca cggttaggc aatcctaaaatc gataaacaa aactat 528
TyrProGlySer ArgLeuGly AsnProLysIle AspLysGln AsnTyr
165 170 175
ttaacattagtt agagaactt aaagaggcattt gaacctttc ggctac 576
LeuThrLeuVal ArgGluLeu LysGluAlaPhe GluProPhe GlyTyr
180 185 190
ttgttgactgcc gcagtatca cccggtaaagat aaaattgac gtaget 624
LeuLeuThrAla AlaValSer ProGlyLysAsp LysT1eAsp ValAla
195 200 205
tatgagctcaaa gaattgaac caattgttcgat tggatgaat gtcatg 672
TyrGluLeuLys GluLeuAsn GlnLeuPheAsp TrpMetAsn ValMet
210 215 220
acttatgattac catggcgga tgggaaaatgtt ttcggccat aatget 720
ThrTyrAspTyr HisGlyGly TrpGluAsnVal PheGlyHis AsnAla
225 230 235 240
ccgttgtataaa cgacccgat gaaacggatgaa ttgcacact taCttc 768
ProLeuTyrLys ArgProAsp GluThrAspGlu LeuHisThr TyrPhe
245 250 255
aatgtcaactac accatgcac tattatttgaac aatggcget actcga 816
AsnValAsnTyr ThrMetHis TyrTyrLeuAsn AsnGlyAla ThrArg
260 265 270
gacaaacttgtt atgggtgtt ccattctatggt cgtgettgg agcatc 864
AspLysLeuVal MetGlyVal ProPheTyrGly ArgAlaTrp SerIle
275 280 285
gaagatcgaagc aaagtcaaa cttggcgatccg gccaaaggc atgtct 912
GluAspArgSer LysValLys LeuGlyAspPro AlaLysGly MetSer
290 295 300
cctcctggtttt attactggt gaagaaggtgtt ctctcatac atcgaa 960
ProProGlyPhe IleThrGly GluGluGlyVal LeuSerTyr IleGlu
305 310 315 320
ttgtgtcagtta ttccagaaa gaagaatggcat attcaatac gatgaa 1008
LeuCysGlnLeu PheGlnLys GluGluTrpHis IleGlnTyr AspGlu
325 330 335
tattacaatget ccatacgga tataatgataaa atctgggtt ggttac 1056
TyrTyrAsnAla ProTyrGly TyrAsnAspLys IleTrpVal GlyTyr
340 345 350
31
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
gatgat ctgget agtatatca tgcaagttggcc tttctcaaa gaattg 1104
AspAsp LeuAla SerIleSer CysLysLeuAla PheLeuLys GluLeu
355 360 36 5
ggcgtc tctggc gttatgata tggtcattggaa aacgatgat ttcaaa 1152
GlyVal SerGly ValMetI1e TrpSerLeuGlu AsnAspAsp PheLys
370 375 380
ggtcat tgcgga ccgaaatat ccattgttgaac aaagttcac aatatg 1200
GlyHis CysGly ProLysTyr ProLeuLeuAsn LysValHis AsnMet
385 390 395 400
atcaat ggtgat gaaaagaac tcttacgaatgt Cttttgggc ccaagt 1248.
IleAsn GlyAsp GluLys.AsnSerTyrGluCys LeuLeuGly ProSer
405 410 415
acaacc acacca acaccaacc accccgtcaact acttcgact accaca 1296
ThrThr ThrPro ThrProThr ThrProSerThr ThrSerThr ThrThr
420 425 430
Ccaacg cctacc accaccgat agcacaagcgaa acaccaaaa tacact 1344
ProThr ProThr ThrThrAsp SerThrSerGlu ThrProLys TyrThr
435 440 445.
acgtatattgat ggacatttg attaaatgc tataaacaaggt tatctt 1392
ThrTyrIleAsp G1yHisLeu IleLysCys TyrLysG1nGly TyrLeu
450 455 460
ccacatccaact gatgttcat aaatattta gtttgtgaatat attgcc 1440
.
ProHisProThr AspValHis LysTyrLeu ValCysGluTyr IleAla
465 470 475 480
acaccaaacggt ggttggtgg gtacacatt atggattgtcca aaagga 1488
ThrProAsnGly GlyTrpTrp ValHisIle MetAspCysPro LysGly
485 490 495
actagatggcac gcaacatta aaaaattgt attcaagaa 1527
ThrArgTrpHis AlaThrLeu LysAsnCys IleGln.Glu
500 505
<210> 38
<211> 509
<212> PRT
<213> Dermatophagoides farinae
<400> 38
Met Lys Thr Thr Phe Ala Leu Phe Cys Ile Trp Ala Cys Ile Gly Leu
1 5 10 15
Met Asn Ala Ala Thr Lys Arg Asp His Asn Asn Tyr Ser Lys Asn Pro
20 25 30
Met Arg Ile Val Cys Tyr Val Gly Thr Trp Ser Val Tyr His Lys Val
35 40 45
Asp Pro Tyr Thr Tle Glu Asp Ile Asp Pro Phe Lys Cys Thr His Leu
50 55 60
32
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Met Tyr Gly Phe Ala Lys Ile Asp Glu Tyr Lys Tyr Thr Ile Gln Val
65 70 75 80
Phe Asp Pro Phe Gln Asp Asp Asn His Asn Ser Trp Glu Lys His Gly
85 90 95
Tyr Glu Arg Phe Asn Asn Leu Arg Leu Lys Asn Pro Glu Leu Thr Thr
100 105 110
Met Ile Ser Leu Gly Gly Trp Tyr Glu Gly Ser Glu Lys Tyr Ser Asp
115 120 125
Met Ala Ala Asn Pro Thr Tyr Arg Gln Gln Phe Val Gln Ser Val Leu
130 135 140
Asp Phe Leu Gln Glu Tyr Lys Phe,Asp Gly Leu Asp Leu Asp Trp Glu
145 150 155 160
Tyr Pro Gly Ser Arg Leu Gly Asn Pro Lys Ile Asp Lys Gln Asn Tyr
165 170 175
Leu_Thr Leu Val Arg Glu Leu Lys Glu Ala Phe Glu Pro Phe Gly Tyr
180 185 190
Leu Leu Thr Ala Ala Val Ser Pro Gly Lys Asp Lys 21e Asp Val Ala
195 200 205
Tyr Glu Leu Lys Glu Leu Asn Gln Leu Phe Asp Trp Met Asn Val Met
210 215 220
Thr Tyr Asp Tyr His Gly Gly Trp Glu Asn Val Phe Gly His Asn Ala
225 230 235 240
Pro Leu Tyr Lys Arg Pro Asp Glu Thr Asp G1u Leu His Thr Tyr Phe
245 250 255
Asn Val Asn Tyr Thr Met His Tyr Tyr Leu Asn Asn Gly Ala Thr Arg
260 265 270
Asp Lys Leu Val Met Gly Val Pro Phe Tyr Gly Arg Ala Trp Ser Ile
275 280 285
Glu Asp Arg Ser Lys Val Lys Leu Gly Asp Pro Ala Lys Gly Met Ser
290 295 300
Pro Pro Gly Phe Ile Thr Gly Glu Glu Gly Val Leu Ser Tyr Ile Glu
305 310 315 320
33
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Leu Cys Gln Leu Phe Gln Lys Glu Glu Trp His Ile Gln Tyr Asp G1u
325 330 335
Tyr Tyr Asn Ala Pro Tyr Gly Tyr Asn Asp Lys Ile Trp Val Gly Tyr
340 345 350
Asp Asp Leu Ala Ser Ile Ser Cys Lys Leu Ala Phe Leu Lys Glu Leu
355 360 365
Gly Val Ser Gly Val Met Ile Trp Ser Leu Glu Asn Asp Asp Phe Lys
370 375 380
Gly His Cys Gly Pro Lys Tyr Pro Leu Leu Asn Lys Val His Asn Met
385 390 395 400
Ile Asn Gly Asp Glu Lys Asn Ser Tyr Glu Cys Leu Leu Gly Pro Ser
405 410 415
Thr Thr Thr Pro Thr Pro Thr Thr Pro Ser Thr Thr Ser Thr Thr Thr
420 425 430
Pro Thr Pro Thr Thr Thr Asp Ser Thr Ser Glu Thr Pro Lys Tyr Thr
435 440 445
Thr Tyr I1e Asp Gly His Leu Ile Lys Cys Tyr Lys Gln Gly Tyx Leu
450 455 460
Pro His Pro Thr Asp Val His Lys Tyr Leu Val Cys Glu Tyr Ile Ala
465 470 475 480
Thr Pro Asn Gly Gly Trp Trp Val His Ile Met Asp Cys Pro Lys Gly
485 490 495
Thr Arg Trp His Ala Thr Leu Lys Asn Cys Ile Gln Glu
500 505
<210>
39
<211>
1527
<212>
DNA
<213>
Dermatophagoides
farinae
<400>
39
ttcttgaatacaattttttaatgttgcgtgccatctagttccttttggacaatccataat60
gtgtacccaccaaccaccgtttggtgtggcaatatattcacaaactaaatatttatgaac120
atcagttggatgtggaagataaccttgtttatagcatttaatcaaatgtccatcaatata180
cgtagtgtattttggtgtttcgcttgtgctatcggtggtggtaggcgttggtgtggtagt240
cgaagtagttgacggggtggttggtgttggtgtggttgtacttgggcccaaaagacattc300
34
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
gtaagagttcttttcatcaccattgatcatattgtgaactttgttcaacaatggatattt360
cggtccgcaatgacctttgaaatcatcgttttccaatgaccatatcataacgocagagac420
gcccaattctttgagaaaggccaacttgcatgatatactagccagatcatcgtaaccaac480
ccagattttatcattatatccgtatggagcattgtaatattcatcgtattgaatatgcca540
ttcttctttctggaataactgacacaattcgatgtatgagagaacaccttcttcaccagt600
aataaaaccaggaggagacatgcctttggccggatcgccaagtttgactttgcttcgatc660
ttcgatgctccaagcacgaccatagaatggaacacccataacaagtttgtctcgagtagc720
gccattgttcaaataatagtgcatggtgtagttgacattgaagtaagtgtgcaattcatc780
cgtttcatcgggtcgtttatacaacggagcattatggccgaaaacattttcccatccgcc840
atggtaatcataagtcatgacattcatccaatcgaacaattggttcaattctttgagctc900
ataagctacgtcaattttatctttaccgggtgatactgcggcagtcaacaagtagccgaa960
aggttcaaatgcctctttaagttctctaactaatgttaaatagttttgtttatcgatttt1020
aggattgcctaaccgtgatccaggatattcccaatccaaatctaggccatcgaatttgta1080
ttcttgcaaaaagtccaaaactgattgaacaaattgctgacgatatgttggattggctgc1140
catatccgaatatttttctgaaccttcataccaacCacccaatgaaatcatggtggtcaa1200 "
ttctggattcttcaatctcaagttgttgaaacgttcatacccgtgtttttcccatgagtt1260
atggttatcatcttgaaatggatcaaaaacttgaatggtgtatttgtattcatcgatttt1320
agcaaaaccatacatcaaatgagtacatttgaaaggatcaatatcttcaattgtgtatgg1380
atcaactttatgataaacggaccatgttccaacataacatacgattcgcattggattttt1440
cgaataattattgtgatctcgtttagtggccgcattcatcaagccaatgcaggcccatat1500
acaaaacaatgcaaatgtcgttttcat 1527
<210>40
<211>1470
<212>DNA
<213>Dermatophagoides farinae
<220>
<221>CDS
<222>(1)..(1470).
<223>
<400> 40
gcc act aaa cga gat cac aat aat tat tcg aaa aat cca atg cga atc 48
A1a Thr Lys Arg Asp His Asn Asn Tyr Ser Lys Asn Pro Met Arg Ile
1 5 10 15
gta tgt tat gtt gga aca tgg tcc gtt tat cat aaa gtt gat cca tac 96
Val Cys Tyr Val Gly Thr Trp Ser Val Tyr His Lys Val Asp Pro Tyr
20 25 30
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
acaattgaagat attgatcct ttcaaatgtact catttgatg tatggt 144
ThrIleGluAsp IleAspPro PheLysCysThr HisLeuMet TyrGly
35 40 45
tttgetaaaatc gatgaatac aaatacaccatt caagttttt gatcca 192
PheAlaLysIle AspGluTyr LysTyrThrIle GlnValPhe AspPro
50 55 60
tttcaagatgat aaccataac tcatgggaaaaa cacgggtat gaacgt 240
PheGlnAspAsp AsnHisAsn SerTrpGluLys HisGlyTyr GluArg
65 70 75 80
ttcaacaacttg agattgaag aatccagaattg accaccatg atttca 288
PheAsnAsnLeu ArgLeuLys AsnProGluLeu ThrThrMet IleSer
85 90 95
ttgggtggttgg tatgaaggt tcagaaaaatat tcggatatg gcagcc 336
LeuGlyGlyTrp TyrG1uGly SerGluLysTyr SerAspMet AlaAla
100 105 110
aatccaacatat cgtcagcaa tttgttcaatca gttttggac tttttg 384
AsnProThrTyr ArgGlnGln PheValGlnSer ValLeuAsp PheLeu
115 120 125
caagaatacaaa ttcgatggc ctagatttggat tgggaatat cctgga 432
GlnGluTyrLys PheAspGly LeuAspLeuAsp TrpGluTyr ProGly
130 135 140
tcacggttaggc aatcctaaa atcgataaacaa aactattta acatta 480
SerArgLeuGly AsnProLys IleAspLysGln AsnTyrLeu ThrLeu
145 150 155 160
gttagagaactt aaagaggca tttgaacctttc ggctacttg ttgact 528
ValArgGluLeu LysGluAla PheGluProPhe GlyTyrLeu LeuThr
165 170 175
gccgcagtatca cccggtaaa gataaaattgac gtagettat gagctc 576
AlaAlaValSer ProGlyLys AspLysIleAsp ValAlaTyr GluLeu
180 185 190
aaagaattgaac caattg.ttcgattggatgaat gtcatgact tatgat 624
LysGluLeuAsn GlnLeuPhe AspTrpMetAsn ValMetThr TyrAsp
195 200 205
taccatggcgga tgggaaaat gttttcggccat aatgetccg ttgtat 672
TyrHisGlyGly TrpGluAsn ValPheGlyHis AsnAlaPro LeuTyr
210 215 220
aaacgacccgat gaaacggat gaattgcacact tacttcaat gtcaac 720
LysArgProAsp GluThrAsp GluLeuHisThr TyrPheAsn ValAsn
225 230 235 240
tacaccatgcac tattatttg aacaatggcget actcgagac aaactt 768
TyrThrMetHis TyrTyrLeu AsnAsnGlyAla ThrArgAsp LysLeu
245 250 255
gttatgggtgtt ccattctat ggtcgtgettgg agcatcgaa gatcga 816
ValMetGlyVal ProPheTyr GlyArgAlaTrp Sex'IleGlu AspArg
260 265 270
agcaaagtcaaa cttggcgat ccggccaaaggc atgtctcct cctggt 864
SerLysValLys LeuGlyAsp ProAlaLysGly MetSerPro ProGly
275 280 285
36
CA 02420459 2003-02-21
WO PCT/USO1/28730
02/22807
AL-2-C4-PCT.ST25. txt
ttt attactggt gaagaaggt gttctctca tacatcgaa ttgtgtcag 912
Phe IleThrGly GluGluGly ValLeuSer TyrIleGlu LeuCysGln
290 295 300
tta ttccagaaa gaagaatgg catattcaa tacgatgaa tattacaat 960
Leu PheGlnLys GluGluTrp HisIleGln TyrAspGlu TyrTyrAsn
305 310 315 320
get ccatacgga tataatgat aaaatctgg gttggttac gatgatctg 1008
Ala ProTyrGly TyrAsnAsp LysIleTrp ValGlyTyr AspAspLeu
325 330 335
get agtatatca tgcaagttg gcctttctc aaagaattg ggcgtctct 1056
Ala SerI1eSer CysLysLeu AlaPheLeu LysGluLeu GlyVa1Ser
340 345 350
ggc gttatgata tggtcattg gaaaacgat gatttcaaa ggtcattgc 1104
Gly ValMetIle TrpSerLeu GluAsnAsp AspPheLys GlyHisCys
355 360 365
gga ccgaaatat ccattgttg aacaaagtt cacaatatg atcaatggt 1152
Gly ProLysTyr ProLeuLeu AsnLysVal HisA'snMet TleAsnGly
370 375 380
gat gaaaagaac tcttacgaa tgtcttttg ggcccaagt acaaccaca 1200
Asp G1uLysAsn SerTyrGlu CysLeuLeu GlyProSer ThrThrThr
385 390 395 400
cca acaccaacc accccgtca actacttcg actaccaca ccaacgcct 1248
Pro ThrProThr ThrProSer ThrThrSer ThrThrThr ProThrPro
405 410 415
acc accaccgat agcacaagc gaaacacca aaatacact acgtatatt 1296
Thr ThrThrAsp SerThrSer GluThrPro LysTyrThr ThrTyrI1e
420 425 430
gat ggacatttg attaaatgc tataaacaa ggttatctt ccacatcca 1344
Asp GlyHisLeu IleLysCys TyrLysGln GlyTyrLeu ProHisPro
435 440 445
act gatgttcat aaatattta gtttgtgaa tatattgcc acaccaaac 1392
Thr AspValHis LysTyrLeu ValCysGlu TyrIleAla ThrProAsn
450 455 460
ggt ggttggtgg gtacacatt atggattgt ccaaaagga actagatgg 1440
Gly GlyTrpTrp ValHisIle MetAspCys ProLysGly ThrArgTrp
465 470 475 480
cac gcaacatta aaaaattgt attcaagaa 1470
His AlaThrLeu LysAsnCys 21eGlnGlu
485 490
<210> 41
<211> 490
<212> PRT
<213> Dermatophagoides farinae
<400> 41
Ala Thr Lys Arg Asp His Asn Asn Tyr Ser Lys Asn Pro Met Arg Ile
1 5 10 15
37
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Val Cys Tyr Val Gly Thr Trp Ser Val Tyr His Lys Val Asp Pro Tyr
20 25 30
Thr Ile~Glu Asp Ile Asp Pro Phe Lys Cys Thr His Leu Met Tyr Gly
35 40 45
Phe Ala Lys Ile Asp Glu Tyr Lys Tyr Thr Tle Gln Val Phe Asp Pro
50 55 60
Phe Gln Asp Asp Asn His Asn Ser Trp Glu Lys His Gly Tyr Glu Arg
65 70 75 80
Phe Asn Asn Leu Arg Leu Lys Asn Pro Glu Leu Thr Thr Met Ile Ser
85 90 95
Leu Gly Gly Trp Tyr Glu Gly Ser Glu Lys Tyr Ser Asp Met Ala Ala
100 105 110
Asn Pro Thr Tyr Arg Gln Gln Phe Val Gln Ser Val Leu Asp Phe Leu
115 120 125
Gln Glu Tyr Lys Phe Asp Gly Leu Asp Leu Asp Trp Glu Tyr Pro Gly
130 135 140
Ser Arg Leu Gly Asn Pro Lys Ile Asp Lys Gln Asn Tyr Leu Thr Leu
145 150 155 160
Val Arg Glu Leu Lys Glu Ala Phe Glu Pro Phe Gly Tyr Leu Leu Thr
165 170 175
Ala Ala Val Ser Pro Gly Lys Asp Lys Ile Asp Val Ala Tyr Glu Leu
l80 185 1.90
Lys Glu Leu Asn Gln Leu Phe Asp Trp Met Asn Val Met Thr Tyr Asp
195 200 205
Tyr His Gly Gly Trp Glu Asn Val Phe Gly His Asn Ala Pro Leu Tyr
210 215 220
Lys Arg Pro Asp Glu Thr Asp Glu Leu His Thr Tyr Phe Asn Val Asn
225 230 235 240
Tyr Thr Met His Tyr Tyr Leu Asn Asn Gly Ala Thr Arg Asp Lys Leu
245 250 255
Val Met Gly Val Pro Phe Tyr Gly Arg Ala Trp Ser Ile Glu Asp Arg
260 265 270
38
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Ser Lys Val Lys Leu Gly Asp Pro Ala Lys Gly Met Ser Pro Pro Gly
275 280 285
Phe Ile Thr Gly Glu Glu Gly Val Leu Ser Tyr Ile Glu Leu Cys Gln
290 295 300
Leu Phe Gln Lys Glu Glu Trp His Ile Gln Tyr Asp Glu Tyr Tyr Asn
305 310 315 320
Ala Pro Tyr Gly Tyr Asn Asp Lys Ile Trp Val Gly Tyr Asp Asp Leu
325 330 335
Ala Ser Ile Ser Cys Lys Leu Ala Phe Leu Lys Glu Leu Gly Va1 Ser
340 345 350
Gly Val Met Ile Trp Ser Leu Glu Asn Asp Asp Phe Lys Gly His Cys
355 360 365
Gly Pro Lys Tyr Pro Leu Leu Asn Lys Val His Asn Met,Ile Asn Gly
370 375 380
Asp Glu Lys Asn Ser Tyr Glu Cys Leu Leu Gly Pro Ser Thr Thr Thr
385 390 395 400
Pro Thr Pro Thr Thr Pro Ser Thr Thr Ser Thr Thr Thr Pro Thr Pro
405 410 415
Thr Thr Thr Asp Ser Thr Ser Glu Thr Pro Lys Tyr Thr Thr Tyr Ile
420 425 430
Asp Gly His Leu Ile Lys Cys Tyr Lys Gln Gly Tyr Leu Pro His Pro
435 440 445
Thr Asp Val His Lys Tyr Leu Val Cys Glu Tyr Ile Ala Thr Pro Asn
450 455 460
Gly Gly Trp Trp Val His Ile Met Asp Cys Pro Lys G1y Thr Arg Trp
465 470 475 480
His Ala Thr Leu Lys Asn Cys Ile Gln Glu
485 ' 490
<210> 42
<211> 1470
<212> DNA
<213> Dermatophagoides farinae
<400> 42
ttcttgaata caatttttta atgttgcgtg ccatctagtt ccttttggac aatccataat 60
39
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
gtgtacccac caaccaccgtttggtgtggcaatatattcacaaactaaatatttatgaac120
atcagttgga tgtggaagataaccttgtttatagcatttaatcaaatgtccatcaatata180
cgtagtgtat tttggtgtttcgcttgtgctatcggtggtggtaggcgttggtgtggtagt240
cgaagtagtt gacggggtggttggtgttggtgtggttgtacttgggcccaaaagacattc300
gtaagagttc ttttcatcaccattgatcatattgtgaactttgttcaacaatggatattt360
cggtccgcaa tgacctttgaaatcatcgttttccaatgaccatatcataacgccagagac420
gcccaattct ttgagaaaggccaacttgcatgatatactagccagatcatcgtaaccaac480
ccagatttta tcattatatccgtatggagcattgtaatattcatcgtattgaatatgcca540
ttcttctttc tggaataactgacacaattcgatgtatgagagaacaccttcttcaccagt.600
aataaaacca ggaggagacatgcctttggccggatcgccaagtttgactttgcttcgatc660
ttcgatgctc caagcacgaccatagaatggaacacccataacaagtttgtctcgagtagc720
gccattgttc aaataatagtgcatggtgtagttgacattgaagtaagtgtgcaattcatc780
cgtttcatcg ggtcgtttatacaacggagcattatggccgaaaacattttcccatccgcc840
atggtaatca taagtcatgacattcatccaatcgaacaattggttcaattctttgagctc900
ataagctacg tcaattttatctttaccgggtgatactgcggcagtcaacaagtagccgaa960
aggttcaaat gcctctttaagttctctaactaatgttaaatagttttgtttatcgatttt1020
aggattgcct aaccgtgatccaggatattcccaatccaaatctaggccatcgaatttgta1080
ttcttgcaaa aagtccaaaactgattgaacaaattgctgacgatatgttggattggctgc1140
catatccgaa tatttttctgaaccttcataccaaccacccaatgaaatcatggtggtcaa1200
ttctggattc ttcaatctcaagttgttgaaacgttcatacccgtgtttttcccatgagtt1260
atggttatca tcttgaaatggatcaaaaacttgaatggtgtatttgtattcatcgatttt1320
agcaaaacca tacatcaaatgagtacatttgaaaggatcaatatcttcaattgtgtatgg1380
atcaacttta tgataaacggaccatgttccaacataacatacgattcgcattggattttt1440
cgaataatta ttgtgatctcgtttagtggc 1470
<210> 43
<211> 510
<212> DNA
<213> Dermatophagoides farinae
<220>
<221> CDS
<222> (1)..(510)
<223>
<400> 43
gat atg gaa cat ttt aca caa cat aag ggc aac gcc aaa gcc atg atc 48
Asp Met G1u His Phe Thr G1n His Lys Gly Asn Ala Lys Ala Met Ile
1 5 10 15
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
gccgtcggtggt tcgactatg tccgatcaattt tccaagact gcagcg 96
AlaValGlyGly SerThrMet SerAspGlnPhe SerLysThr AlaAla
20 25 30
gtagaacattat cgggaaacg tttgttgttagc acagttgat cttatg 144
ValGluHisTyr ArgGluThr PheValValSer ThrValAsp LeuMet
35 40 45
actcgttatggt ttcgatggt gtcatgattgat tggtctggc atgcaa 192
ThrArgTyrGly PheAspGly ValMetIleAsp TrpSerGly MetGln
50 55 60
gccaaagatagt gataatttc attaaattgttg gacaaattc gacgaa 240
AlaLysAspSer AspAsnPhe IleLysLeuLeu AspLysPhe AspGlu
65 70 75 80
aagtttgetcac acctcgttt gtgatgggtgtt accttgccg gcaacg 288
LysPheA1aHis ThrSerPhe ValMetGlyVal ThrLeuPro AlaThr
85 90 95
atcgcatcatac gataactat aacattcctgcc atctccaac tatgtc 336
IleAlaSerTyr AspAsnTyr AsnIleProAla IleSerAsn TyrVal
100 105 110
gattttatgaac gtgcttagt ctggattacact ggatcatgg gcccat 384
AspPheMetAsn ValLeuSer LeuAspTyrThr GlySerTrp AlaHis
115 120 125
acggtcggtcat gettctccg tttcctgaacaa ctcaaaacg ctagaa 432
ThrValGlyHis AlaSerPro PheProGluGln LeuLysThr LeuGlu
130 135 140
gettaccacaaa cgaggcget ccacgtcataag atggtcatg getgta 480
AlaTyrHisLys ArgGlyAla ProArgHisLys MetValMet AlaVal
145 150 155 160
ccattttatgca cgtacctgg attctcgag 510
ProPheTyrAla ArgThrTrp IleLeuGlu
165 170
<210> 44
<211> 170
<212> PRT
<213> Dermatophagoides farinae
<400> 44
Asp Met G1u His Phe Thr Gln His Lys Gly Asn Ala Lys Ala Met Ile
1 5 10 15
Ala Val Gly Gly Ser Thr Met Ser Asp Gln Phe Ser Lys Thr Ala Ala
20 25 30
Val Glu His Tyr Arg Glu Thr Phe Val Val Ser Thr Val Asp Leu Met
35 40 45
Thr Arg Tyr Gly Phe Asp Gly Val Met Ile Asp Trp Ser Gly Met Gln
50 55 60
41
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Ala Lys Asp Ser Asp Asn Phe Ile Lys Leu Leu Asp Lys Phe Asp Glu
65 70 75 80
Lys Phe Ala His Thr Ser Phe Val Met Gly Val Thr Leu Pro Ala Thr
85 90 95
Ile Ala Ser Tyr Asp Asn Tyr Asn Ile Pro Ala Ile Ser Asn Tyr Val
100 105 110
Asp Phe Met Asn Val Leu Ser Leu Asp Tyr Thr Gly Ser Trp Ala His
115 120 125
Thr Val G1y His Ala Ser Pro Phe Pro Glu Gln Leu Lys Thr Leu G1u
130 135 140
Ala Tyr His Lys Arg Gly Ala Pro Arg His Lys Met Va1 Met Ala Val
145 150 155 160
Pro Phe Tyr Ala Arg Thr Trp Ile Leu Glu
165 170
<210> 45
<211> 510
<212> DNA
<213> Dermatophagoicles farinae
<400>
45
ctcgagaatccaggtacgtgcataaaatggtacagccatgaccatcttatgacgtggagc60
gcctcgtttgtggtaagcttctagcgttttgagttgttcaggaaacggagaagcatgacc120
gaccgtatgggcccatgatccagtgtaatccagactaagcacgttcataaaatcgacata180
gttggagatggcaggaatgttatagttatcgtatgatgcgatcgttgccggcaaggtaac240
acccatcacaaacgaggtgtgagcaaacttttcgtcgaatttgtccaacaatttaatgaa300
attatcactatctttggcttgcatgccagaccaatcaatcatgacaccatcgaaaccata360
acgagtcataagatcaactgtgctaacaacaaacgtttcccgataatgttctaccgctgc420
agtcttggaaaattgatcggacatagtcgaaccaccgacggcgatcatggctttggcgtt480
gcccttatgttgtgtaaaatgttccatatc 510
<210> 46
<211> 25
<212> DNA
<213> Artificial sequence
<220>
<223> Synthetic Primer
<220>
<221> misc feature
42
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
<222> (15)..(15)
<223> n = a, c, t, or g at position 15
<400> 46
gaaccaaaaa chgtntgyta ytayg 25
<210> 47
<211> 17
<212> DNA
<213> Artificial sequence
<220>
<223> Synthetic Primer
<400> 47
gtaaaacgac ggccagt 17
<210> 48
<211> 29
<212> DNA
<213> Artificial sequence
<220>
<223> Synthetic Primer
<400> 48
gatatggaac atttyachca acayaargg 29
<210> 49
<211> 22
<212> DNA
<213> Artificial sequence
<220>
<223> Synthetic Primer
<400> 49
gtaatacgac tcactatagg gc 22
<210> 50
<211> 1445
<212> DNA
<213> Dermatophagoides farinae
<220>
<221> CDS
<222> (14)..(1399)
<223>
<400> 50
atcccaaata aaa atg act cga ttc tct ttg act gta ttg gcc gta ctt 49
Met Thr Arg Phe Ser Leu Thr Val Leu Ala Val Leu
1 5 10
gcc get tgt ttc ggt tca aat att cgt ccg aat gtg gca act ttg gaa 97
Ala Ala Cys Phe Gly Ser Asn Ile Arg Pro Asn Val Ala Thr Leu Glu
15 20 25
43
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
cctaaa actgtatgt tactatgaa tcttgggta cattggcgc caaggt 145
ProLys ThrValCys TyrTyrGlu SerTrpVal HisTrpArg GlnGly
30 35 40
gaaggc aaaatggat cccgaagac atagataca tcgttgtgt actcac 193
GluGly LysMetAsp ProGluAsp IleAspThr SerLeuCys ThrHis
45 50 55 60
attgtc tactcttat ttcggcatt gatgetgcc actcatgag attaaa 241
IleVal TyrSerTyr PheGlyIle AspAlaAla ThrHisGlu IleLys
65 70 75
ctattg gatgaatat cttatgaaa gatttacat gacatggaa catttc 289
LeuLeu AspGluTyr LeuMetLys AspLeuHis AspMetGlu HisPhe
80 85 90
acgcag cataagggc aacgccaaa gccatgatc gccgtcggt ggttcg 337
ThrGln HisLysGly AsnAlaLys AlaMetIle AlaValGly GlySer
95 100 105
actatg tccgatcaa ttttccaag actgcagcg gtagaacat tatcgg 385
ThrMet SerAspGln PheSerLys ThrAlaAla ValGluHis TyrArg
110 115 120
gaaacg tttgttgtt agcaca gttgatcttatg actcgttat ggtttc 433
GluThr PheValVal SerThr ValAspLeuMet ThrArgTyr GlyPhe
125 130 135 140
gatggt gtcatgatt gattgg tctggcatgcaa gccaaagat agtgat 481
AspGly ValMetIle AspTrp SerGlyMetGln AlaLysAsp SerAsp
145 150 155
aatttc attaaattg ttggac aaattcgacgaa aagtttget cacacc 529
AsnPhe IleLysLeu LeuAsp LysPheAspGlu LysPheAla HisThr
160 165 170
tcgttt gtgatgggt gttacc ttgccggcaacg atcgcatca tacgat 577
SerPhe ValMetGly ValThr LeuProAlaThr IleAlaSer TyrAsp
175 180 185
aactat aacattcct gccatc tccaactatgtc gattttatg aacgtg 625
AsnTyr AsnIlePro AlaIle SerAsnTyrVal AspPheMet AsnVal
190 195 200
cttagt ctggattac actgga tcatgggcccat acggtcggt catget 673
LeuSer LeuAspTyr ThrGly SerTrpAlaHis ThrValGly HisAla
205 210 215 220
tctccg tttcctgaa caactc aaaacgctagaa gettaccac aaacga 721
SerPro PheProGlu GlnLeu LysThrLeuGlu AlaTyrHis LysArg
225 230 235
ggcget ccacgtcat aagatg gtcatggetgta ccattttat gcacgt 769
GlyAla ProArgHis LysMet ValMetAlaVal ProPheTyr AlaArg
240 245 250
acctgg attctcgag aaaatg aacaaacaggac attggcgat aaaget 817
ThrTrp IleLeuGlu LysMet AsnLysGlnAsp IleGlyAsp LysAla
255 260 265
agtgga ccaggccca cgaggt cagtttacacag actgatggt ttcctt 865
SerGly ProGlyPro ArgGly GlnPheThrGln ThrAspGly PheLeu
270 275 280
44
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
agctacaacgaa ttgtgcgttcag attcaggcc gaaacgaat gcattc 913
SerTyrAsnGlu LeuCysValGln IleGlnAla GluThrAsn AlaPhe
285 29 0 295 300
accattactcgt gatcatgataat accgcaatt tacgetgtc tatgtg 961
ThrIleThrArg AspHisAspAsn ThrAlaIle TyrAlaVal TyrVal
305 310 315
catagcaaccat gcagaatggatc tctttcgaa gaccgacat acactt 1009
HisSerAsnHis AlaGluTrpIle SerPheGlu AspArgHis ThrLeu
320 325 330
ggtgaaaaagca aaaaacataacc caacaagga tatgetgga atgtca 1057
GlyGluLysAla LysAsnIleThr GlnGlnGly TyrAlaGly MetSer
335 340 345
gtctacacattg tccaacgaagat gtgcacggc gtttgtggt gataaa 1105
ValTyrThrLeu SerAsnGluAsp ValHisGly ValCysGly AspLys
350 355 360
aaccctttgttg catgetatccaa tcgaactat tatcatggc gtggta 1153
AsnProLeuLeu HisAlaIleGln SerAsnTyr TyrHisGly ValVal
365 370 375 380
accgaaccgacc gtcgttacactt cctccagtc acacataca acagaa 1201
ThrGluProThr ValValThrLeu ProProVal ThrHisThr ThrGlu
385 390 395
catgtgaccgat ataccaggcgtg tttcattgc catgaagaa ggattc 1249
HisValThrAsp IleProGlyVal ,PheHisCys HisGluGlu GlyPhe
400 405 410
ttccgcgataag acctattgtgcc acatactac gaatgcaaa aaaggc 1297
PheArgAspLys ThrTyrCysAla ThrTyrTyr GluCysLys LysGly
415 420 425
gattttggactg gagaaaaccgtg catcattgt gccaatcac ttacag 1345
AspPheGlyLeu GluLysThrVal HisHisCys AlaAsnHis LeuGln
430 435 440
gcatttgacgaa gtaagtcggaca tgtattgat cataccaaa ataccc 1393
AlaPheAspGlu ,ValSerArgThr CysIleAsp HisThrLys IlePro
445 450 455 460
ggttgttgaatacaaa taaaattaca aaaaaa 1445
atcactttaa
aaaaaaaaaa
GlyCys .
<210> 51
<211> 462
<212> PRT
<213> Dermatophago idesfarinae
<400> 51
Met Thr Arg Phe Ser Leu Thr Val Leu Ala Val Leu Ala Ala Cys Phe
1 5 10 15
Gly Ser Asn Ile Arg Pro Asn Val Ala Thr Leu Glu Pro Lys Thr Val
20 25 30
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Cys Tyr Tyr Glu Ser Trp Val His Trp Arg Gln Gly Glu Gly Lys Met
35 40 45
Asp Pro Glu Asp Ile Asp Thr Ser Leu Cys Thr His Ile Val Tyr Ser
50 55 60
Tyr Phe Gly Ile Asp Ala Ala Thr His Glu Ile Lys Leu Leu Asp Glu
65 70 75 80
Tyr Leu Met Lys Asp Leu His Asp Met Glu His Phe Thr Gln His Lys
85 90 95
Gly Asn Ala Lys Ala Met Tle Ala Val Gly Gly Ser Thr Met Ser Asp
100 105 110
Gln Phe Ser Lys Thr Ala Ala Val Glu His Tyr Arg Glu Thr Phe Val
115 120 l25
Val Ser Thr Va1 Asp Leu Met Thr Arg Tyr Gly Phe Asp Gly Val Met
130 135 140
Ile Asp Trp Ser Gly Met Gln Ala Lys Asp Ser Asp Asn Phe Ile Lys
145 l50 155 160
Leu Leu Asp Lys Phe Asp Glu Lys Phe Ala His Thr Ser Phe Val Met
165 170 175
Gly Val Thr Leu Pro Ala Thr Ile Ala Ser Tyr Asp Asn Tyr Asn Tle
180 185 190
Pro Ala Ile Ser Asn Tyr Val Asp Phe Met Asn Val Leu Ser Leu Asp
195 200 205
Tyr Thr Gly Ser Trp Ala His Thr Val Gly His'Ala Ser Pro Phe Pro
210 215 220
Glu G1n Leu Lys Thr Leu Glu Ala Tyr His Lys Arg Gly Ala Pro Arg
225 230 235 240
His Lys Met Val Met Ala Val Pro Phe Tyr Ala Arg Thr Trp Ile Leu
245 250 255
Glu Lys Met Asn Lys Gln Asp Ile Gly Asp Lys Ala Ser Gly Pro Gly
260 265 270
Pro Arg Gly Gln Phe Thr Gln Thr Asp Gly Phe Leu Ser Tyr Asn Glu
275 280 285
46
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Leu Cys Val Gln Ile Gln Ala Glu Thr Asn Ala Phe Thr Ile Thr Arg
290 295 300
Asp His Asp Asn Thr Ala Ile Tyr Ala Val Tyr Val His Ser Asn His
305 310 315 ' 320
Ala Glu Trp Ile Ser Phe Glu Asp Arg His Thr Leu Gly Glu Lys Ala
325 330 335
Lys Asn Ile Thr Gln Gln Gly Tyr Ala Gly Met Ser Val Tyr Thr Leu
340 345 350
Ser Asn Glu Asp Val His Gly Va1 Cys Gly Asp Lys Asn Pro Leu Leu
355 360 365
His Ala Ile Gln Ser Asn Tyr Tyr His Gly Val Val Thr Glu Pro Thr
370 375 380
Val Val Thr Leu Pro Pro Val Thr His Thr Thr Glu His Val Thr Asp
385 390 395 400
Ile Pro Gly Val Phe His Cys His Glu Glu Gly Phe Phe Arg Asp Lys
405 410 415
Thr Tyr Cys A1a Thr Tyr Tyr Glu Cys Lys Lys Gly Asp Phe Gly Leu
420 425 430
Glu Lys Thr Val His His Cys Ala Asn His Leu Gln Ala Phe Asp Glu
435 440 445
Val Ser Arg Thr Cys Ile Asp His Thr Lys Ile Pro Gly Cys
450 455 460
<210> 52
<211> 1445
<212> DNA
<213> Dermatophagoides farinae
<400> 52
tttttttttt ttttttttaa agtgattgta attttatttg tattcaacaa ccgggtattt 60
tggtatgatc aatacatgtc cgacttactt cgtcaaatgc ctgtaagtga ttggcacaat 120
gatgcacggt tttctccagt ccaaaatcgc cttttttgca ttcgtagtat gtggcacaat 180
aggtcttatc gcggaagaat ccttcttcat ggcaatgaaa cacgcctggt atatcggtca 240
catgttctgt tgtatgtgtg actggaggaa gtgtaacgac ggtcggttcg gttaccacgc 300
catgataata gttcgattgg atagcatgca acaaagggtt tttatcacca caaacgccgt 360
gcacatcttc gttggacaat gtgtagactg acattccagc atatccttgt tgggttatgt 420
47
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
tttttgctttttcaccaagtgtatgtcggtcttcgaaagagatccattctgcatggttgc480
tatgcacatagacagcgtaaattgcggtattatcatgatcacgagtaatggtgaatgcat540
tcgtttcggcctgaatctgaacgcacaattcgttgtagctaaggaaaccatcagtctgtg600
taaactgacctcgtgggcctggtccactagctttatcgccaatgtcctgtttgttcattt660
tctcgagaatccaggtacgtgcataaaatggtacagccatgaccatcttatgacgtggag720
cgcctcgtttgtggtaagcttctagcgttttgagttgttcaggaaacggagaagcatgac780
cgaccgtatgggcccatgatccagtgtaatccagactaagcacgttcataaaatcgacat840
agttggagatggcaggaatgttatagttatcgtatgatgcgatcgttgccggcaaggtaa900
cacccatcacaaacgaggtgtgagcaaacttttcgtcgaatttgtccaacaatttaatga960
aattatcactatctttggcttgcatgccagaccaatcaatcatgacaccatcgaaaccat1020
aacgagtcataagatcaactgtgctaacaacaaacgtttcccgataatgttctaccgctg1080
cagtcttggaaaattgatcggacatagtcgaaccaccgacggcgatcatggctttggcgt1140
tgcccttatgctgcgtgaaatgttccatgtcatgtaaatctttcataagatattcatcca1200
atagtttaatctcatgagtggcagcatcaatgccgaaataagagtagacaatgtgagtac1260
acaacgatgtatctatgtcttcgggatccattttgccttcaccttggcgccaatgtaccc1320
aagattcatagtaacatacagttttaggttccaaagttgccacattcggacgaatatttg1380
aaccgaaacaagcggcaagtacggccaatacagtcaaagagaatcgagtcatttttattt1440
gggat
1445
<210> 53
<211> 1386
<212> DNA
<213> Dermatophagoides farinae
<400>
53
atgactcgattctctttgactgtattggccgtacttgccgcttgtttcggttcaaatatt60
cgtCCgaatgtggcaactttggaacctaaaactgtatgttactatgaatcttgggtacat120
tggcgccaaggtgaaggcaaaatggatcccgaagacatagatacatcgttgtgtactcac180
attgtctactcttatttcggcattgatgctgccactcatgagattaaactattggatgaa240
tatcttatgaaagatttacatgacatggaacatttcacgcagcataagggcaacgccaaa300
gccatgatcgccgtcggtggttcgactatgtccgatcaattttccaagactgcagcggta360
gaacattatcgggaaacgtttgttgttagcacagttgatcttatgactcgttatggtttc420
gatggtgtcatgattgattggtctggcatgcaagccaaagatagtgataatttcattaaa480
ttgttggacaaattcgacgaaaagtttgctcacacctcgtttgtgatgggtgttaccttg540
ccggcaacgatcgcatcatacgataactataacattcctgccatctccaactatgtcgat600
tttatgaacgtgcttagtctggattacactggatcatgggcccatacggtcggtcatgct660
48
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
tctccgtttcctgaacaactcaaaacgctagaagcttaccacaaacgaggcgctccacgt720
cataagatggtcatggctgtaccattttatgcacgtacctggattctcgagaaaatgaac780
aaacaggacattggcgataaagctagtggaccaggcccacgaggtcagtttacacagact840
gatggtttccttagctacaacgaattgtgcgttcagattcaggccgaaacgaatgcattc900
accattactcgtgatcatgataataccgcaatttacgctgtctatgtgcatagcaaccat960
gcagaatggatctctttcgaagaccgacatacacttggtgaaaaagcaaaaaacataacc1020
caacaaggatatgctggaatgtcagtctacacattgtccaacgaagatgtgcacggcgtt1080'
tgtggtgataaaaaccctttgttgcatgctatccaatcgaactattatcatggcgtggta1140
accgaaccgaccgtcgttacacttcctccagtcacacatacaacagaacatgtgaccgat1200
ataccaggcgtgtttcattgccatgaagaaggattcttccgcgataagacctattgtgcc1260
acatactacgaatgcaaaaaaggcgattttggactggagaaaaccgtgcatcattgtgcc1320
aatcacttacaggcatttgacgaagtaagtcggacatgtattgatcataccaaaataccc1380
ggttgt 1386
<210>
54
<211>
1386
<212>
DNA
<213>
Dermatophagoides
farinae
<400>
54
acaaccgggtattttggtatgatcaatacatgtccgacttacttcgtcaaatgcctgtaa60
gtgattggcacaatgatgcacggttttctccagtccaaaatcgccttttttgcattcgta120
gtatgtggcacaataggtcttatcgcggaagaatccttcttcatggcaatgaaacacgcc180
tggtatatcggtcacatgttctgttgtatgtgtgactggaggaagtgtaacgacggtcgg240
ttcggttaccacgccatgataatagttcgattggatagcatgcaacaaagggtttttatc300
accacaaacgccgtgcacatcttcgttggacaatgtgtagactgacattccagcatatcc360
ttgttgggttatgttttttgctttttcaccaagtgtatgtcggtcttcgaaagagatcca420
ttctgcatggttgctatgcacatagacagcgtaaattgcggtattatcatgatcacgagt480
aatggtgaatgcattcgtttcggcctgaatctgaacgcacaattcgttgtagctaaggaa540
accatcagtctgtgtaaactgacctcgtgggcctggtccactagctttatcgccaatgtc600
ctgtttgttcattttctcgagaatccaggtacgtgcataaaatggtacagccatgaccat660
cttatgacgtggagcgcctcgtttgtggtaagcttctagcgttttgagttgttcaggaaa720
cggagaagcatgaccgaccgtatgggcccatgatccagtgtaatccagactaagcacgtt780
cataaaatcgacatagttggagatggcaggaatgttatagttatcgtatgatgcgatcgt840
tgccggcaaggtaacacccatcacaaacgaggtgtgagcaaacttttcgtcgaatttgtc900
49
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
caacaatttaatgaaattatcactatctttggcttgcatgccagaccaatcaatcatgac960
accatcgaaaccataacgagtcataagatcaactgtgctaacaacaaacgtttcccgata1020
atgttctaccgctgcagtcttggaaaattgatcggacatagtcgaaccaccgacggcgat1080
catggctttggcgttgcccttatgctgcgtgaaatgttccatgtcatgtaaatctttcat1140
aagatattcatccaatagtttaatctcatgagtggcagcatcaatgccgaaataagagta1200
gacaatgtgagtacacaacgatgtatctatgtcttcgggatccattttgccttcaccttg1260
gcgccaatgtacccaagattcatagtaacatacagttttaggttccaaagttgccacatt1320
cggacgaatatttgaaccgaaacaagcggcaagtacggccaatacagtcaaagagaatcg1380
agtcat 1386
<210> 55
<211> 1236
<212> DNA
<213> Dermatophagoides
farinae
<220>
<221> CDS
<222> (1)..(1236)
<223>
<400>
55
actttggaacct aaaactgta tgttactat gaatcttgg gtacattgg 48
ThrLeuGluPro LysThrVal CysTyrTyr GluSerTrp ValHisTrp
1 5 10 15
cgccaaggtgaa ggcaaaatg gatcccgaa gacatagat acatcgttg 96
ArgGlnGlyGlu GlyLysMet AspProGlu AspTleAsp ThrSerLeu
20 25 30
tgtactcacatt gtctactct tatttcggc attgatget gccactcat 144
CysThrHisIle ValTyrSer TyrPheGly IleAspAla AlaThrHis
35 40 45
gagattaaacta ttggatgaa tatcttatg aaagattta catgacatg 192
GluIleLysLeu LeuAspGlu TyrLeuMet LysAspLeu HisAspMet
50 55 60
gaacatttcacg cagcataag ggcaacgcc aaagccatg atcgccgtc 240
GluHisPheThr GlnHisLys GlyAsnAla LysAlaMet IleAlaVal
65 70 75 80
ggtggttcgact atgtccgat caattttcc aagactgca gcggtagaa 288
GlyGlySerThr MetSerAsp GlnPheSer LysThrAla AlaValGlu
85 90 95
cattatcgggaa acgtttgtt gttagcaca gttgatctt atgactcgt 336
HisTyrArgGlu ThrPheVal ValSerThr ValAspLeu MetThrArg
100 105 110
tatggtttcgat ggtgtcatg attgattgg tctggcatg caagccaaa 384
TyrGlyPheAsp GlyValMet IleAspTrp SerGlyMet GlnAlaLys
115 120 125
gatagtgataat ttcattaaa ttgttggac aaattcgac gaaaagttt 432
50
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
AspSerAspAsn PheIleLys LeuLeuAsp LysPheAspGlu LysPhe
130 135 140
getcacacctcg tttgtgatg ggtgttacc ttgccggcaacg atcgca 480
AlaHisThrSer PheValMet GlyValThr LeuProAlaThr IleAla
145 150 155 160
tcatacgataac tataacatt cctgccatc tccaactatgtc gatttt 528
SerTyrAspAsn TyrAsnIle ProAlaIle SerAsnTyrVal AspPhe
165 170 175
atgaacgtgctt agtctggat tacactgga tcatgggcccat acggtc 576
MetAsnValLeu SerLeuAsp TyrThrGly SerTrpAlaHis ThrVal
180 185 190
ggtcatgettct ccgtttcct gaacaactc aaaacgctagaa gettac 624
GlyHisAlaSer ProPhePro GluGlnLeu LysThrLeuGlu AlaTyr
195 200 205
cacaaacgaggc getccacgt cataagatg gtcatggetgta ccattt 672
HisLysArgGly AlaProArg HisLysMet ValMetAlaVal ProPhe
210 215 220
tatgcacgtacc tggattctc gagaaaatg aacaaacaggac attggc 720
TyrAlaArgThr TrpIleLeu GluLysMet AsnLysGlnAsp IleGly
225 230 235 240
gataaagetagt ggaccaggc ccacgaggt cagtttacacag actgat 768
AspLysAlaSer GlyProGly ProArgGly GlnPheThrGln ThrAsp
245 250 255
ggtttccttagc tacaacgaa ttgtgcgtt cagattcaggcc gaaacg 816
GlyPheLeuSer TyrAsnGlu LeuCysVal GlnIleGlnAla GluThr
260 265 270
aatgcattcacc attactcgt gatcatgat aataccgcaatt tacget 864
AsnAlaPheThr IleThrArg AspHisAsp AsnThrAlaIle TyrAla
275 280 285
gtctatgtgcat agcaaccat gcagaatgg atctctttcgaa gaccga 912
ValTyrValHis SerAsnHis AlaGluTrp IleSerPheGlu AspArg
290 295 300
catacacttggt gaaaaagca aaaaacata acccaaCaagga tatget 960
HisThrLeuGly GluLysAla LysAsnIle ThrGlnGlnGly TyrAla
305 310 315 320
ggaatgtcagtc tacacattg tccaacgaa gatgtgcacggc gtttgt 1008
GlyMetSerVal TyrThrLeu SerAsnGlu AspValHisGly ValCys
325 330 335
ggtgataaaaac cctttgttg catgetatc caatcgaactat tatcat 1056
GlyAspLysAsn ProLeuLeu HisAlaIle GlnSerAsnTyr TyrHis
340 345 350
ggcgtggtaacc gaaccgacc gtcgttaca cttcctccagtc acacat 1104
GlyValValThr GluProThr ValVa1Thr LeuProProVal ThrHis
355 360 365
acaacagaacat gtgaccgat ataccaggc gtgtttcattgc catgaa 1152
ThrThrGluHis ValThrAsp IleProGly ValPheHisCys HisGlu
370 375 380
gaaggattcttc cgcgataag acctattgt gccacatactac gaatgc 1200
51
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
Glu Gly Phe Phe Ar,g Asp Lys Thr Tyr Cys Ala Thr Tyr Tyr Glu Cys
385 390 395 400
aaa aaa ggC gat ttt gga ctg gag aaa acc gtg cat 1236
Lys Lys Gly Asp Phe Gly Leu Glu Lys Thr Val His
405 410
i
<210> 56
<211> 412
<212> PRT
<213> Dermatophagoides farinae
<400> 56
Thr Leu Glu Pro Lys Thr Val Cys Tyr Tyr G1u Ser Trp Val His Trp
1 5 10 15
Arg Gln Gly Glu Gly Lys Met Asp Pro Glu Asp Ile Asp Thr Ser Leu
20 25 30
Cys Thr His Ile Val Tyr Ser Tyr Phe Gly Ile Asp Ala Ala Thr His
35 40 45
Glu Tle Lys Leu Leu Asp Glu Tyr Leu Met Lys Asp Leu His Asp Met
50 55 60
Glu His Phe Thr Gln His Lys Gly Asn Ala Lys Ala Met Ile Ala Va1
65 70 75 80
Gly Gly Ser Thr Met Ser Asp G1n Phe Ser Lys Thr Ala Ala Val Glu
85 90 95
His Tyr Arg Glu Thr Phe Val Val Ser Thr Val Asp Leu Met Thr Arg
100 105 110
Tyr Gly Phe Asp Gly Val Met Ile Asp Trp Ser Gly Met Gln Ala Lys
115 120 125
Asp Ser Asp Asn Phe Ile Lys Leu Leu Asp Lys Phe Asp Glu Lys Phe
130 135 140
Ala His Thr Ser Phe Val Met Gly Val Thr Leu Pro Ala Thr I1e A1a
145 150 155 160
Ser Tyr Asp Asn Tyr Asn Ile Pro Ala I1e Ser Asn Tyr Val Asp Phe
165 170 175
Met Asn Val Leu Ser Leu Asp Tyr Thr Gly Ser Trp Ala His Thr Val
180 185 190
Gly His Ala Ser Pro Phe Pro Glu Gln Leu Lys Thr Leu Glu Ala Tyr
52
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
195 200 205
His Lys Arg Gly Ala Pro Arg His Lys Met Val Met Ala Val Pro Phe
210 215 220
Tyr Ala Arg Thr Trp Ile Leu Glu Lys Met Asn Lys Gln Asp Ile Gly
225 230 235 240
Asp Lys Ala Ser G1y Pro Gly Pro Arg Gly Gln Phe Thr Gln Thr Asp
245 250 255
Gly Phe Leu Ser Tyr Asn Glu Leu Cys Val Gln Ile Gln Ala Glu Thr
260 265 270
Asn Ala Phe Thr Ile Thr Arg Asp His Asp Asn Thr Ala I1e Tyr Ala
275 280 285
Val Tyr Val His Ser Asn His Ala Glu Trp Ile Ser Phe Glu Asp Arg
290 295 300
His Thr Leu Gly Glu Lys Ala Lys Asn Ile Thr Gln Gln Gly Tyr Ala
305 310 315 320
Gly Met Ser Val Tyr Thr Leu Ser Asn Glu Asp Val His Gly Val Cys
325 330 335
Gly Asp Lys Asn Pro Leu Leu His Ala Ile Gln Ser Asn Tyr Tyr His
340 345 350
G1y Val Val Thr Glu Pro.Thr Val Val Thr Leu Pro Pro Val Thr His
355 360 365
Thr Thr G1u His Val Thr Asp Ile Pro Gly Val Phe His Cys His Glu
370 375 380
Glu Gly Phe Phe Arg Asp Lys Thr Tyr Cys Ala Thr Tyr Tyr Glu Cys
385 390 395 400
Lys Lys Gly Asp Phe Gly Leu Glu Lys Thr Va1 His
405 410
<210> 57
<211> 1236
<212> DNA
<213> Dermatophagoides farinae
<400> 57
atgcacggtt ttctccagtc caaaatcgcc ttttttgcat tcgtagtatg tggcacaata 60
ggtcttatcg cggaagaatc cttcttcatg gcaatgaaac acgcctggta tatcggtcac 120
53
CA 02420459 2003-02-21
WO 02/22807 PCT/USO1/28730
AL-2-C4-PCT.ST25.txt
atgttctgttgtatgtgtgactggaggaagtgtaacgacggtcggttcggttaccacgcc180
atgataatagttcgattggatagcatgcaacaaagggtttttatcaccacaaacgccgtg240
cacatcttcgttggacaatgtgtagactgacattccagcatatccttgttgggttatgtt300
ttttgctttttcaccaagtgtatgtcggtcttcgaaagagatccattctgcatggttgct360
atgcacatagacagcgtaaattgcggtattatcatgatcacgagtaatggtgaatgcatt420
cgtttcggcctgaatctgaacgcacaattcgttgtagctaaggaaaccatcagtctgtgt480
aaactgacctcgtgggcctggtccactagctttatcgccaatgtcctgtttgttcatttt540
ctcgagaatccaggtacgtgcataaaatggtacagccatgaccatcttatgacgtggagc600
gcctcgtttgtggtaagcttctagcgttttgagttgttcaggaaacggagaagcatgacc660
gaccgtatgggcccatgatccagtgtaatccagactaagcacgttcataaaatcgacata720
gttggagatggcaggaatgttatagttatcgtatgatgcgatcgttgccggcaaggtaac780
acccatcacaaacgaggtgtgagcaaacttttcgtcgaatttgtccaacaatttaatgaa840
attatcactatctttggcttgcatgccagaccaatcaatcatgacaccatcgaaaccata900
acgagtcataagatcaactgtgctaacaacaaacgtttcccgataatgttCtaCCgCtgC960
agtcttggaaaattgatcggacatagtcgaaccaccgacggcgatcatggctttggcgtt1020
gcccttatgctgcgtgaaatgttccatgtcatgtaaatctttcataagatattcatccaa1080
tagtttaatctcatgagtggcagcatcaatgccgaaataagagtagacaatgtgagtaca1140
caacgatgtatctatgtcttcgggatccattttgccttcaccttggcgccaatgtaccca1200
agattcatagtaacatacagttttaggttccaaagt 1236
54