Note: Descriptions are shown in the official language in which they were submitted.
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
DESCRIPTION
METHODS AND COMPOSITIONS RELATING TO A CARDIAC-SPECIFIC
NUCLEAR REGULATORY FACTOR
BACKGROUND OF THE INVENTION
The government owns rights in the application pursuant to NIH Grant Nos.
P01 HL49953 and HL63926.
1. Field of the Invention
The present invention relates generally to the fields of developmental biology
and molecular biology. More particularly, it concerns proteins involved in the
regulation of cardiomyocyte cell growth and development.
2'. Description of Related Art
The leading cause of morbidity and mortality in industrialized countries is
heart disease, particularly heart disease that is associated with myocardial
infarction.
Myocardial infarction results in the loss of cardiomyocytes. Cardiomyocytes
are post-
mitotic cells and generally do not regenerate after birth. Furthermore, it has
been
discovered that they respond to mitotic signals by cell hypertrophy (Kodama et
al.,
1997; Pan et al., 1997) rather than by cell hyperplasia. The loss of
cardiomyocytes
leads to regional contractile dysfunction. In addition, the necrotized
cardiomyocytes
in the infarcted regions in the ventricular tissues are progressively replaced
by
fibroblasts to form scar tissue.
Recently, fetal cardiomyocytes transplanted in heart scar tissue limited scar
expansion and prevented postinfarction heart failure (Leor et al., 1996).
Although the
transplantation of fetal cardiomyocytes is a proposed treatment of heart
failure, it
remains impractical in the clinical setting, in part because of the difficulty
of obtaining
fetal heart donor tissue. Thus, it is desirable to develop a cardiomyogenic
cell line
that could be used to facilitate the understanding of cardiomyocyte
development and
to facilitate the treatment of heart diseases, such as those associated with
loss of
cardiomyocytes.
1
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Although it is known that the loss of post-mitotic cardiomyocytes results in
increased morbidity and mortality, very little is known about the genes that
are
involved in heart development. It is known that transcription factors such as
d-
HAND, e-HAND (Srivastava et al., 1995), MEF-2C (Edmondson et al. 1994; Lin et
al. 1997), Nkx2.5/Csx, GATA4, and TEF-1 play important roles in cardiac
development (Harvey, 1996), but the lack of a model for cardiomyocyte
differentiation has hindered the understanding of the interactions of these
genes.
A recent report revealed that murine marrow stromal cells that are treated
with
5-azacyidine, a cytosine analog capable of altering expression of certain
genes that
may regulate differentiation, results in a cell line that differentiates into
cardiomyocytes in vitro (Makino et al., 1999). This cardiomyogenic cell line
demonstrated several phenotypic characteristics that are specific to
cardiomyocytes,
e.g., adjoining cells via intercalated discs, forming myotubes, and beating
spontaneously. In addition, the expression of cardiomyocyte specific genes,
such as
horneobox gene N1~2.5, alpha-myosin heavy chain and atrial natriuretic factor,
also
are considered characteristic.
Although the proposed transplantation of fetal cardiomyocytes and
cardiomyogenic cell lines are possible treatments, it is preferable to
discover a
treatment that eliminates any donor/species problems. Thus, identifying new
regulators of cardiomyocyte growth and differentiation is an important goal in
the
search for therapeutics to treat myocardial tissue damage.
SUMMARY OF THE INVENTION
The present invention provides polypeptides capable of modulating cell
phenotype, particularly phenotypic characteristics of cardiomyocyte cells, and
polynucleotides encoding such polypeptides. In particular, pxovided herein is
a family
of peptides, known as myocardins, that share certain sequence homology and
functional activities, as described herein. In one aspect, the polypeptides of
the
present invention comprise mycardin peptides and biologically active fragments
thereof. In another aspect, the present invention provides isolated
polynucleotides
2
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
encoding a myocardin peptide including fragments thereof. Exemplary
biologically
active fragments of myocardin polypeptides are also provided herein.
In a further aspect, there are provided expression cassettes comprising
polynucleotides encoding the polypeptides of the present invention.
Preferably, such
expression cassettes further comprise one or more regulatory sequences
operably
linked to said polynucleotide, capable of enhancing or otherwise modulating
transcription and/or translation of said polynucleotide in a target cell, for
example a
mammalian cell. By way of illustration, in one embodiment, an expression
cassette
comprising a polynucleotide encoding a myocardin polypeptide operably linked
to a
promoter is provided. The promoter may be an inducible promoter or a
constitutive
promoter. The promoter may be heterologous to the myocardin coding sequence. ,
Further, the promoter may be a ubiquitous promoter, for example a
cytomeglovirus
(CMV) promoter, rous sarcoma virus (RSV) promoter or human elongation factor
(e.g., hEF-la) promoter, or it may be active only in certain tissues/cells for
example a .
fibroblast specific promoter (e.g., an alpha collagen promoters) or a muscle-
specific
promoter (e.g., a myosin light chain-2 promoter or a a-myosin heavy chain).
The
regulatory sequence of the expression cassette may further comprise a
polyadenylation signal. The expression cassette may be a viral expression
construct,
for ,example, a retroviral vector, an adenoviral vector, an adeno-associated
viral
vector, a vaccina viral vector, a herpesviral vector, a polyoma viral
construct,
lentiviral vector or a Sindbis viral vector. The expression cassette may
further
comprise a second polynucleotide encoding a second polypeptide. The second..
polypeptide may be, for example, a cardiac transcription factor.
In another aspect of the present invention, there is provided an isolated
nucleic
acid segment comprising at least 1 S contiguous nucleotides of SEQ ID NO: 1,
SEQ ID
NO: 2S, SEQ TD NO: 27 or SEQ ID NO: 29. Also provided is an isolated nucleic
acid
segment of SEQ ID NO: 1, SEQ ID NO: 2S, SEQ ID NO: 27 or SEQ ID NO: 29
comprising 1 S-2000 nucleotides in length. In a related aspect, there is
provided a
peptide of 8-SO residues comprising at least 8-12 consecutive residues of SEQ
ID NO:
2, SEQ ID NO: 26, SEQ ID NO: 28 or SEQ ID NO: 30. In another related aspect,
there are provided antibodies, which may be produced by a hybridoma cell, that
bind
immunologically to a polypeptide comprising SEQ ID NO: 2, SEQ ID NO: 26, SEQ
3
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
ID NO: 28 or SEQ ID NO: 30, or an antigenic fragment thereof. The antibodies
may
be monoclonal or polyclonal antisera.
In a further aspect of the present invention, as described further below,
myocardin peptides from different species are provided. By way of
illustration,
murine mycardin 1 (e.g., SEQ ID NOS: 2 and 30), human myocardin 1 (e.g.,. SEQ
ID
NOS: 26 and 28), human myocardin 2 (SEQ ID NO: 4) and human myocardin 3 (SEQ
117 NO: 6) are described. These myocardin peptides share localized regions of
high
amino acid sequence homology, particularly in the carboxyl-terminal
transcription
activation domain, and, particularly with respect to myocardin l and 2, in
glutamine
(Q) rich domains.
In still a further aspect of the invention, there is provided a transformed
host
cell comprising a polynucleotide encoding a myocardin polypeptide and a
promoter
heterologous to the myocardin-encoding polynucleotide which promoter directs
the
expression of the myocardin polypeptide. The host cell may be prokaryotic or
eukaryotic. In a related aspect of the invention, there is provided a method
of using
the transformed host cell and culturing it under conditions suitable for the
expression
of the myocardin polypeptide. W yet another aspect, there is provided a fusion
protein
comprising a myocardin protein or peptide fused to a second protein or
peptide.
As discussed above, heart disease, especially that resulting in a heart
attack,
typically results in significant cardiac dysfunction. This dysfunction can be
the result
of the activities of cells, especially non-cardiomyocyte cells, in the region
of disease
of the heart. In a further aspect of the present invention, compositions and
methods
are provided that alleviate the deleterious activities of such non-
cardiomyocyte target
cells on the functioning of the heart by modulating the phenotype of said
target cells.
In preferred embodiments, the compositions and methods not only alleviate the
deleterious activities of the target cell population but stimulate the target
cells to
engage in one or more functions typical of cardiomyocytes thereby improving
myocardial functioning in the diseased region. By way of illustration,
fibroblast cells
typically are recruited to form scar tissue in areas of myocardium where
cardiomyocyte necrosis has occurred (for example, as the result of myocardial
infarction) thereby resulting in permanent, regional cardiac dysfunction.
Introduction
4
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
of a composition in accordance herewith into such fibroblasts can prevent
those cells
from engaging in such deleterious activity and, in preferred embodiments, can
actually
stimulate the fibroblasts to engage in one or more functions phenotypical of
cardiomyocytes (for example, spontaneous beating, formation of microtubules or
adjoining to neighboring cells via intercalated discs, and expression of
cardiomyocyte
specific genes, such as homeobox gene Nkx2.5, alpha-myosin heavy chain and
atrial
natriuretic factor) thereby assisting heart function. Advantageously,
introduction of
such compositions in accordance herewith may additionally improve the
functioning
of existing cardiomyocytes by, for example, inducing hypertrophy therein.
Thus, the
present compositions may serve the dual roles of stimulating fibroblast cells
to engage
in functions) phenotypic of cardiomyocytes and stimulating hypertrophy in
existing
cardiomyocytes.
In yet a further and related aspect of the present invention, there is
provided a
method of converting a non-cardiomyocyte target cell, such as a cardiac
fibroblast into
a cardiac myocyte-like cell comprising introducing into the target cell an
expression
cassette. The expression cassette comprises a polynucleotide encoding a
myocardin
polypeptide as well as one or more regulatory sequences, for example, a
promoter
with or without enhancer sequences, which regulatory sequences are active in
the
target cell and direct the expression of the polypeptide. The method may
further
comprise measuring cardiac and muscle cell lineage markers. In another aspect,
the
expression cassette may further comprise one or more additional
polynucleotides
encoding one or more polypeptides. By way of illustration, a second
polypeptide may
be a cardiac transcription factor, for example, GATA4. In a related aspect,
expression
of the additional polynucleotides may be under the control of the same
regulatory
sequences as the first polynucleotide or may be separately controlled by
additional
regulatory sequences.
In another aspect of the present invention, the method further comprises
introducing one or more additional expression cassettes into target cells
separately
from introduction of the myocardin expression cassette. By way of
illustration, a
second expression cassette comprising a polynucleotide encoding a second
polypeptide and including a second promoter able to direct expression of the
second
polypeptide in the target cells may be delivered to the target cell using a
separate gene
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
delivering means from that used to introduce the myocardin expression
cassette.
Thus, for example, a first gene delivery vector comprising a myocardin
expression
cassette may be delivered simultaneously or contemporaneously with a second
gene
delivery vector comprising a second expression cassette. If desired,
polypeptide
expression may be measured, for example, by measuring transcription by RNA
hybridization, RT-PCR or Western analysis.
In yet another aspect, there is provided a method of generating a
cardiomyocyte comprising introducing into a cardiac fibroblast an expression
cassette.
The expression cassette comprising, for example, a polynucleotide encoding a
myocardin polypeptide operatively Linked to a promoter capable of directing
expression of the polypeptide. The promoter may be heterologous to the coding
sequence and may be a ubiquitous (e.g., CMV) or a specific promoter (e.g., an
alpha
collagen promoter). The expression cassette may be introduced into the
fibroblast.by
any of a variety of means known to those of skill in the art. By way of
illustration,
Lipid-based vectors (e.g., liposomes), viral vectors (e.g., retroviral
vectors, vaccine
viral vectors, herpesviral vectors, polyoma viral constructs, lentiviral
vectors or
Sindbis viral vectors), or other macromolecular complexes capable of mediating
delivery of the polynucleotide to the fibroblast or other target cell, may be
employed.
In a further aspect the gene delivery vector may be modified, for example by
means
lcnown to those of skill in the art, to target one or more specific cell
types. The
expression cassette may also comprise a selectable marker, e.g., an
immunologic
marker. The expression cassette may further comprise a second polynucleotide
encoding a second polypeptide, such as the GATA4 cardiac transcription factor.
Such
a second polynucleotide may be under control of a second promoter or the same
promoter as the first polynucleotide. Alternatively, an internal ribosomal
entry site
(IRES) may be employed between the two transgenes to permit expression of the
second transgene.
In a further aspect of the present invention, there is provided a method of
stimulating cardiac tissue regeneration comprising inhibiting the function of
myocardin in a post-mitotic cardiomyocyte. Inhibiting may comprise providing
antisense nucleic acid that inhibits transcription or translation of a
myocardin mRNA.
6
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
The antisense nucleic acid may be provided by introducing an expression
cassette
encoding myocardin antisense RNA.
In still another aspect, there is provided a non-human transgenic animal,
e.g., a
mouse, comprising an expression cassette. The expression cassette comprises a
.
polynucleotide encoding a myocardin peptide or protein and a promoter operably
linked thereto which promoter may be heterologous to the myocardin peptide or
protein encoding region. The promoter may be a constitutive or an inducible
promoter. The expression cassette may further comprise selectable marker(s).
In a
related aspect of the present invention, the non-human transgenic animal may
comprise a defective germ-line myocardin allele or two defective germ-line
myocardin alleles.
In a further aspect of the invention, there is provided a method of treating a
heart disease, such as cardiomyopathy (for example, myocardial infarction or
hypertension). The method comprises administering to an animal suffering from
a
heart disease an expression cassette, which may comprise a polynucleotide
encoding a
myocardin peptide or protein and a promoter operable in eukaryotic cells. The
promoter may be a tissue-specific promoter. The expression cassette may be
comprised within a viral expression vector, for example, a retroviral vector,
an
adenoviral vector, an adeno-associated viral vector, a vaccina viral vector, '
a
herpesviral vector, a polyoma viral construct, a lentiviral vector or a
Sindbis viral
vector or witlun a non-viral vector, for example a lipid-based vector. In a
related
aspect, the method may comprise providing to an animal suffering therefrom a
rnyocardin antisense nucleic acid.
In another related aspect, there is provided a method of alleviating one or
more
symptoms of a heart disease comprising inhibiting the function of myocardin in
post-
mitotic cardiomyocytes in the subject. Another method of alleviating one or
more
symptoms of a heart disease, for example in a subject with heart failure,
comprises
increasing the level of myocardin in fibroblasts to generate cardiomyocytes in
the
subj ect.
An additional aspect of the present invention is to provide compositions and
methods for the identification of downstream target genes of myocardin
polypeptides.
7
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
A gene delivery vector, for example an adenoviral vector, can be employed to
deliver
a myocardin gene to isolated cardiomyocytes thereby permitting over-expression
of
the myocardin polypeptide. Differences in gene profiling between control
(i.e., non-
transfected) cardiomyocytes and ~ transfected (i.e., myocardin-overexpressing)
cardiomyocytes can then be assessed by standard methods, such as differential
display
and microarray (e.g., gene chip) technology. Genes that are activated by
myocardin in
cardiomyocytes can subsequently be evaluated as potential therapeutics, for
example,
using bioinformatics techniques. In yet another aspect of the present
invention, there
is provided a method of screening for a candidate ~ substance for an effect on
myocardin regulation of cardiomyocyte development comprising: (a) providing
myocardin and GATA to a cell; (b) admixing myocardin and GATA in the presence
of the candidate substance; and (c) measuring the effect of the candidate
substance on
the expression of a cardiac lineage maxker, wherein a difference in the
expression of
the cardiac lineage marker, as compared to an untreated cell, indicates that
the
candidate substance effects myocardin regulation of cardiomyocyte development.
Exemplary cells include fibroblast and cardiomyocytes, which may be located
in an animal. The modulator may incxease or decrease the expression of the
cardiac
lineage marker. The cardiac lineage marker may be Nkx2.5. The measuring of the
expression of the cardiac lineage marker may comprise RNA hybridization, RT-
PCR,
immunologic detection, ELISA or immunohisotchemistry, for example.
In still yet another aspect of the invention, there is provided a method of
screening for a modulator of myocardin expression comprising: (a) providing a
cell
that expresses a myocardin polypeptide; (b) contacting the myocardin
polypeptide
with a candidate substance; and (c) measuring the expression of myocardin,
wherein a
difference in myocardin expression, indicates that the candidate substance is
a
modulator of myocardin expression. The modulator may be a pharmaceutical
composition. The modulator may enhance or inhibit myocardin expression.
In another aspect of the invention, there is provided a method of screening a
candidate substance for myocardin binding activity comprising: (a) providing a
myocardin polypeptide; (b) contacting the myocardin polypeptide with the
candidate
substance; and (c) determining the binding of the candidate substance to the
8
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
myocardin polypeptide. The assay may be performed in a cell free system, a
cell or in
vivo. The candidate substance may be an inhibitor or an enhancer of myocardin.
BRIEF DESCRIPTION OF THE DRAWINGS
The following drawings form part of the present specification and are included
to further demonstrate certain aspects of the present invention. The invention
may be
better understood by reference to one or more of these drawings in combination
with
the detailed description of specific embodiments presented herein.
FIG. 1 Schematic diagram of the events of cardiac development. Cardiac
precursors from the cardiac crescent (left) migrate to the midline of the
embryo to
form the linear heart tube, which undergoes rightward looping and eventual
formation
of the mature four-chambered heart. Different populations of cardiac precursor
cells
fated to form the aortic sac (AS), conotruncus (CT), right ventricle (RV),
left ventricle
(LV), and atria (A) are shown. . A schematic diagram of the structure of the
mouse
myocardin 1 gene is also shown. Based on human genomic sequence in public
databases, we have determined that the human gene maps to chromosome 17 and
encompasses 170 kb.
FIG. 2 Amino acid and nucleotide sequence of N-tenninally truncated
myocardin 1. A nucleotide sequence encoding an N-terminally truncated
myocardin 1
and the corresponding amino acid sequence are shown.
FIG. 3A-C Expression pattern of myocardin 1 during early heart
development. FIG. 3A: Expression of myocardin lwas determined by whole-mount
to mouse embryos at E7.75. Myocardin 1 transcripts can be seen localized to
the
cardiac crescent. FIG. 3B: Expression of myocardin 1 was determined by section
to
mouse embryos at E8Ø Transcripts are present throughout the heart tube in a
transverse section. FIG. 3C: Expression of myocardin 1 was determined by in
situ
hybridizations to mouse embryos at E12.5. Transcripts are seen throughout the
developing heart in a sagittal section.
FIG. 4 Expression pattern of myocardin 1 in adult mouse tissues. The
expression of myocardin 1 transcripts in adult mouse tissues was analyzed by
9
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Northern blot. Transcripts are detected only in the heart. Size markers are
shown to
the left.
FIG. 5 Nuclear localization of myocardin 1 protein. Cos cells were
transiently transfected with an expression vector encoding myocardin 1 with a
Flag-
epitope tag. The subcellular location of myocardin 1 protein was determined by
immunostainng with anti-Flag antibody. The myocardin 1 protein is
substantially
localized to the nucleus. The inset in the lower right corner shows an
enlargement of
a single cell, with strong myocardin 1 staining in the nucleus, but excluded
from the
nucleoli.
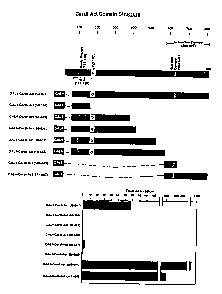
FIG. 6 Structure of myocardin 1 and mapping of transcription activation
domains. A schematic diagram of myocardin 1 is shown at the top. The nuclear
localization sequence (NLS) is located between residues f17 and 126, within a
basic
region. A glutamine-rich (Q) domain is located between residues 159-192. The
transcription activation domain is located at the carboxyl-terminus. Portions
of
myocardin 1 were fused to the DNA binding domain of yeast GAL4 and tested in
transfected Cos cells for transcriptional activity against a GAL4-dependent
luciferase
reporter. Relative transcriptional activities of different myocardin 1
fragments are
shown at the bottom. The carboxyl-terminus is an extremely potent
transcription
activation domain, able to activate, the reporter over 1000-fold, to a level
comparable
to that of the powerful viral coactivator VP16 (not shown).
FIG. 7A-D Trans-activation of the SM22 promoter by myocardin 1. Cos cells
were transiently transfected with a luciferase reporter gene containing the
1.4 kb
SM22 promoter and expression vectors encoding myocardin 1 and SRF, as
indicated.
Forty eight hr later, cells were harvested and luciferase activity was
assayed. FIG. 7A:
shows activity of the wild-type SM22 promoter, which is transactivated about
100-
fold by myocardin 1. FIG. 7B: shows activity of the SM22 promoter with a
mutation
in the distal CArG box (CArG-far). This promoter is also activated by
myocardin 1,
but not to the same extent as the wild-type promoter. FIG. 7C: shows activity
of the
SM22 promoter with a mutation in the proximal CArG box (CArG-near). This
promoter has lost almost all responsiveness to myocardin 1, as has the
promoter with
both CArG boxes mutated (FIG. 7D).
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
FIG. 8 Myocardin 1 and MEF2C cooperatively activate the MLC2V
promoter. Cos cells were transiently transfected with a luciferase reporter
gene
containing the MLC2V promoter and expression vectors encoding myocardin 1 and
MEF2C, as indicated. Forty eight hours later, cells were harvested and
luciferase
activity was assayed. The results show that myocardin 1 and MEF2C
synergistically
activate MLC2V transcription.
FIG. 9 GATA4 represses myocardin 1 activation of the ANF promoter.
HeLa cells and Cos cells were transiently transfected with a luciferase
reporter gene
containing the ANF promoter and expression vectors encoding myocardin 1 and
GATA4 (1 = I00 ng anf luc; 2 = 100 ng anf luc, 100 ng myocardin; 3 = 100 ng
anf
luc, 10 ng GATA4; 4 = 100 ng anf luc, 20 ng GATA4; 5 = 100 ng anf luc, 50 ng
GATA4; 6 = 100 ng anf luc, I00 ng GATA4; 7 = 100 ng anf luc, 100 ng myocardin,
ng GATA4; 8 =100 ng anf luc, 100 ng myocardin, 20-ng GATA4; 9 = 100 ng anf
luc, 100 ng myocardin, 50 ng GATA4; 10 =100 ng anf luc, 100 ng myocardin, 100
ng
GATA4). Forty eight hours later, cells were harvested and luciferase activity
was
assayed. The results show that activation of ANF transcription by myocardin 1
is
repressed in the presence of GATA4.
FIG. 10 Myocardin 1 and Nkx2.5 cooperatively activate the a-MHC
promoter. HeLa cells were transiently transfected with a luciferase reporter
gene
containing the a-MHC promoter and expression vectors encoding myocardin 1 and
Nkx2.5, as indicated. Forty eight hr later, cells were harvested and
luciferase activity
was assayed. The results show that myocardin 1 and Nkx2.5 synergistically
activate
a-MHC transcription.
FIG. 11 Overexpression of myocardin induces serial assembly of sarcomeres
in cardiomyoc~tes. Cardiornyocytes were infected with adenoviruses expressing
either myocardin(Ad-myocardin) or 13-galactosidase(Ad-LacZ), serum deprived,
and
immunostained with anti-sarcomeric-a-actininat antibody 24 hour post-
infection.
FIG. 12 Overexpression of myocardin induces ANF expression in
cardiomyocytes. Cardiomyocytes were infected with adenoviruses expressing
either
myocardin (Ad-myocardin) or l3-galactosidase (Ad-LacZ), serum deprived, and
11
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
immunostained with anti-ANF antibody 24 hour post-infection. Images were
captured
at two different magnifications (x20, x100).
FIG. 13 GATA4 and mvocardin 1 activate of the NKX2.5 promoter. HeLa
cells and Cos cells were transiently transfected with a luciferase reporter
gene
containing the NKX2.5 promoter and expression vectors encoding myocardin I and
GATA4 (1 = 100 ng nl~-luc; 2 = 100 ng nkxf luc, 500 ng myocardin; 3 = 100 ng
nkx-
luc, 10 ng GATA4; 4 = 100 ng nkx-luc, 20 ng GATA4; 5 = 100 ng nkx-luc, 50 ng
GATA4; 6 = 100 ng nkx-luc, 100 ng GATA4; 7 = 100 ng nkx-luc, 500 ng myocardin,
ng GATA4; 8 = 100 ng nkx-luc, 500 ng myocardin, 20 ng GATA4; 9 = 100 ng
nkx-luc, 500 ng myocardin, 50 ng GATA4; 10 = 100 ng nkx-luc, 500 ng myocardin,
100 ng GATA4). Forty eight hours later, cells were harvested and luciferase
activity
was assayed. The results show activation of NKX2.5 transcription by myocardin
1
and GATA4.
DESCRIPTION OF ILLUSTRATIVE EMBODIMENTS
Heart disease is the number one cause of death and hospitalization in the
industrialized world, due in large part to the irreversible nature of the
damage
sustained by the heart following heart attack and other acquired and
congenital
diseases. At present, the only means of repairing a damaged heart is through
complicated surgery and/or heart transplant, which have obvious financial and
medical shortcomings for the patient. The possibility of regenerating cardiac
muscle
cells within the intact human heart following damage represents one of the
most
important challenges in cardiovascular medicine. Perhaps the greatest chance
for
success in this area is to identify "master" control genes for cardiac
development and
to use these genes to reprogram non-muscle cells to a cardiac muscle cell
fate.
A major barrier to cardiac regeneration is the inability of postnatal
cardiomyocytes to divide. In principle, cardiac repair could be achieved
through a
release from this block to cell cycle progression. However, such an approach,
which
might involve introduction of oncogenes or other powerful stimulators of cell
proliferation into the heart, has an obvious downside unless such regulators
were
12
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
somehow cardiac-specific and could be prevented from inducing uncontrolled
proliferation of other cell types. Because about 40% of the cells in the
myocardium
are fibroblasts, an alternate approach would be to reprogram these cells to a
cardiomyocyte fate at sites of cardiac damage through targeted delivery of
cardiac
master control genes.
The loss of cardiomyocytes leads to reduced contractile function of the heart
resulting in increased morbidity and mortality. The present inventors now
report the
discovery of novel cardiac-specific factors, referred to herein as myocardins.
One
such myocardin, myocardin 1, is expressed in cardiac and smooth muscle.
Moreover,
myocardin 1 is first expressed as early as the linear heart tube stage,
embryonic day 8
(E8) in the mouse. This expression is restricted to the heart and to a subset
of vascular
smooth muscle cells throughout embryogenesis to adulthood. The subcellular
distribution of myocardin 1 is localized in the nucleus. Moreover, expression
of
myocardin 1 in transfected cells appears to result in growth arrest of the
cells.
To determine the functions of myocardin 1, the inventors transfected
myocardin 1 expression plasmids into fibroblasts (Cos and HeLa cells) along
with
expression plasmids for the cardiac transcription factor GATA4. The cells were
transiently transfected using FuGENE 6 (Boehringer-Mannheim), according to
manufacturer's instructions. Briefly, 0.1 ~g of expression plasmid encoding
myocardin 1 or the other indicated cardiac transcription factors, along with
the
indicated luciferase plasmids, were mixed with 3 q1 of FuGENE 6 and added to
cells
in six-well plates. Cells were harvested 48 hr later and luciferase activity
was
determined in cell extracts. In all transfections, the amount of DNA per well
was kept
constant by adding the corresponding vector. CMV-lacZ, which contains the lacZ
gene under control of the constitutive cytomegalovirus promoter, was included
in all
transfections as an internal control to normalize for variations in
transfection
efficiency. The results demonstrated that myocardin 1, plus GATA4,
transactivates
regulatory sequences for the cardiac specif c homeobox Nkx2.5, which is the
earliest
marker for the cardiac lineage in vertebrates. These results indicate that
myocardin 1
plays an important role in regulating cardiomyocyte development.
13
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Based upon the functional activity of the marine myocardin 1 and having its
complete cDNA sequence, the inventors have been able to identify other
myocardins,
which they have characterized. Initial searches of DNA sequence databases with
myocardin 1 sequence revealed a number of related sequences. Most of these
sequences are short sequences (for example, ESTs) that share homology to only
small
regions of myocardin 1. None of the sequences located have been identified as
encoding proteins having any particular function, much less any faction
related to cell
regulation, particularly cardiac cell regulation. However, using these
techniques in
combination with the information obtained previously regarding the marine
myocardin, the inventors have identified two sequences that share significant
homology with myocaxdin 1. These appear to be partial sequences from two
additional myocardin genes. cDNA clones for these two related genes, now
designated myocardin 2 and myocardin 3, have been obtained. A comparison of
the
three myocardin species identified has revealed localized regions of high
amino acid
homology between the proteins, particularly in the carboxyl-terminal
transcription
activation domain. By Northern analysis, it was shown that that myocardin 2 is
ubiquitous, and that myocardin 3 appears restricted to heart and liver. These
factors
may be dimerization partners for myocardin 1, and/or may serve analogous
functions
to myocardin 1 in the heart and/or other tissues.
Using similar techniques and information about the marine myocardin 1, the
inventors have also been able locate the genomic sequence of the human homolog
for
myocardin 1 within a particular segment of chromosome 17 (Accession No.
AC005358) and to determine the location of its exons and introns, enabling
identification of the human cDNA sequence. The best EST match fox myocardin 1
is
Accession No. AI607474, for myocardin 2 is Accession No. BE311634, and for
myocardin 3 is Accession No. AW500597.
The discovery of proteins that function to regulate cardiomyocyte growth and
differentiation is important, both for advancing the basic understanding of
heart
development and to provide novel targets for the development of drugs and/or
biotechnological methods to treat cardiac disease, for example by stimulating
the
growth and differentiation of cells into cardiomyocytes after a patient has
suffered
14
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
tissue damage as a result of cardiomyopathy. Since myocardin appears to act as
an
early cardiac inducing factor with the capacity to induce cardiomyocyte
development;
and has the potential to reprogram cardiac fibroblasts, which constitute 40%
of the
cell types in. the heart, to a cardiomyocyte type fate, it may be used in a
variety of
ways to directly treat cardiac disease and to develop additional treatments
for cardiac
disease. Further, because myocardin also appears to induce hypertrophy in
cardiomyocytes, its overexpression may provide an additional benefit in the
treatment
of heart disease by, for example, improving the functioning of dysfunctional
or
malfactional cardiomyocytes.
I. Nucleic Acids
In one aspect, the present invention provides nucleic acid sequences encoding
cardiac cell regulatory factors designated myocardins. In a further aspect the
coding
sequence (as well as substantial non-coding protions) of a novel, N-terminally
tnmcated cardiac-specific factor, designated herein as myocardin 1, is
provided (SEQ
ID NOS 1 and 25). In yet another aspect of the present invention, provided
herein are
nucleic acids encoding mouse and human myocardin l, SEQ ID NOS: 29 and 27;
respectively. The present invention is not limited in scope to any specific
nucleic acid
sequences disclosed herein as one of ordinary skill in the art could, using
these nucleic
acid sequences, readily identify related homologs, including, for example,
homologs
present in any of various species (e.g., rat, rabbit, dog, monkey, gibbon,
chimp, ape,
baboon, cow, pig, horse, sheep, cat and other species).
As discussed below, a "myocardin nucleic acid sequence" may contain a
variety of different bases and yet still produce a myocardin polypeptide
according to
the present invention. Such polypeptides will generally be functionally
equivalent to,
and/or structurally indistinguishable, from the human, mouse and other genes
disclosed herein. Additionally, nucleic acid sequences encoding fragments of
myocardin are provided herein. For example, fragments having increased
activity
(e.g., the carboxy terminal fragments described in FIG. 6) as compared with
the full-
length myoeardin polypeptide are described. Similarly, it will be readily
recognized
that fragments may be employed as probes, for example in the isolation of
homologous sequences. Thus, as will be apparent to those of skill in the art,
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
fragments of the myocardin-encoding nucleic acid sequences as well as homologs
thereof are likewise contemplated herein.
Similarly, any reference to a nucleic acid should be read as encompassing
vectors and host cells containing that nucleic acid and, in some cases,
capable of
expressing the product of that nucleic acid. In addition to therapeutic
considerations,
cells expressing nucleic acids of the present invention may prove useful in
the context
of screening for agents that induce, repress, inhibit, augment, interfere
with, block,
abrogate, stimulate or enhance the function ofmyocardin.
A. Nucleic Acids Encoding Myocardin
Nucleic acids according to the present invention may encode an entire
myocardin gene, a domain of myocardin, or any other fragment of myocardin as
set
forth herein. The nucleic acid may be derived from genomic DNA, i.e., cloned
directly from the genome of a particular organism. In preferred embodiments,
however, the nucleic acid comprises complementary DNA (cDNA). Also
contemplated is a cDNA plus a natural intron or an intron derived from another
gene;
such engineered molecules are sometime referred to as "mini-genes." At a
minimum,
these and other nucleic acids of the present invention may be used as
molecular
weight standards in, for example, gel electrophoresis.
The term "cDNA" is intended to refer to DNA prepared using messenger RNA
(mRNA) as template. The advantage' of using a cDNA, as opposed to genomic DNA
or DNA polymerized from a genomic, non- or partially-processed RNA template,
is
that the cDNA primarily contains coding sequences of the corresponding
protein.
There may be times when the full or partial genomic sequence is preferred,
such as
where the non-coding regions are required for optimal expression or where non-
coding regions such as introns are to be targeted in an antisense strategy.
It also is contemplated that a given myocardin polynucleotide may be
represented by natural or synthetic variants that have slightly different
nucleic acid
sequences but, nonetheless, encode the same or homologous protein (see Table 1
below).
I6
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
As used in this application, the term "a polynucleotide encoding a
polypeptide"
refers to a nucleic acid molecule that is isolated free of total cellular
nucleic acid,
including for example, a synthetic polynucleotide. In exemplary embodiments,
the
invention concerns a nucleic acid sequence essentially as set forth in SEQ ID
NO: 1,
SEQ 1D NO: 25, SEQ ID NO: 27 or SEQ ID NO: 29. The term "comprises SEQ TD
NO: 1 or 27" means that the nucleic acid sequence substantially corresponds to
a
portion of the aforementioned SEQ ID NO: 1 or 27 and likewise for other SEQ ID
NOS providing nucleic acid sequences. The term "functionally equivalent codon"
is
used herein to refer to codons that encode the same amino acid, such as the
six codons
for arginine or serine (Table I, below), and also refers to codons that encode
biologically equivalent amino acids, as discussed in the following pages.
TABLE 1
Amino Acids Codons
Alariine Ala A GCA GCC GCG GCU
Cysteine Cys C UGC UGU
.
Aspartic Asp D GAC GAU
acid
Glutamic Glu E GAA GAG
acid
PhenylalaninePhe F UUC UUU
Glycine Gly G GGA GGC GGG GGU
Histidine His H CAC CAU
Isoleucine Ile I AUA AUC AUU
Lysine Lys K AAA AAG
Leucine Leu L UUA UUG CUA CUC CUG CUU
Methionine Met M AUG
Asparagine Asn N AAC AAU
Proline Pro P CCA CCC CCG CCU
Glutamine Gln Q CAA CAG
Arginine Arg R AGA AGG CGA CGC CGG CGU
Serine Ser S AGC UCA UCC UCG UCU
AGU
Threonine Thr T ACA ACC ACG ACU
Valine Val V GUA GUC GUG GUU
Tryptophan Trp W UGG
Tyrosine Tyr Y UAC UAU
Allowing for the degeneracy of the genetic code, sequences that have at least
about 50%, usually at least about 60%, more usually about 70%, most usually
about
80%, preferably at least about 90% and most preferably about 95% of
nucleotides that
are identical to the nucleotides of a sequence set forth herein, for example
SEQ ID
17
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
NO: 1 or 27 are contemplated. Sequences that are essentially the same as those
set
forth in SEQ ID NO: 1 or 27 also may be functionally defined as sequences that
are
capable of hybridizing to a nucleic acid segment containing the complement of
SEQ
ID NO: 1 or 27 under standard conditions and likewise for other nucleotide
sequences
set forth herein.
The DNA segments of the present invention include those encoding biologically
functional equivalent myocardin proteins, peptides and fragments thereof, as
described
elsewhere herein. Such sequences may arise as a consequence of codon
redundancy
and/or amino acid functional equivalency that are known to those of skill in
the art. For
example, polynucleotides encoding myocardin peptides analogous to the
exemplary
myocardin protein of SEQ m NO: 2 or 28 are likewise contemplated herein. As
discussed fiu-ther below, and as lmown to those of skill in the art, various
amino acid
substitutions, deletions and/or additions may be made to a known amino acid
sequence
without adversely affecting the fimction andlor usefulness thereof.
Alternatively,
functionally equivalent proteins or peptides may be created via the
application of
recombinant DNA technology, in which changes in the protein structure may be
engineered, based on considerations of the properties of the amino acids being
exchanged. Changes designed by man may be introduced through the application
of site-
directed mutagenesis techniques or may be introduced randomly and screened
later for
the desired function, as described below.
B. Oligonucleotide Probes and Primers
Naturally, the present invention also encompasses DNA segments that are
complementary, or essentially complementary, to the sequences set forth
herein, for
example in SEQ m NO:1. Nucleic acid sequences that are "complementary" are
those
that are capable of base-pairing according to-the standard Watson-Crick
complementary
rules. As used herein, the terms "complementary sequences" and "essentially
complementary sequences" means nucleic acid sequences that are substantially
complementary to, as rnay be assessed by the same nucleotide comparison set
forth
above, or are able to hybridize to a nucleic acid segment of one or more
suequences set
forth herein, for example SEQ m NO:1 or 27, under relatively stringent
conditions such
as those described herein. Such sequences may encode an entire myocardin
protein or
peptide or functional or non-functional fragments thereof.
18
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
The hybridizing segments may be short oligonucleotides. Sequences of 17 bases
long should occur only once in the human genome and, therefore, suffice to
specify a
unique target sequence. Although shorter oligomers are easier to make and
increase in
vivo accessibility, numerous other factors are involved in determining the
specificity of
hybridization. Both binding affinity and sequence specificity of an
oligonucleotide to its
complementary target increases with increasing length. It is contemplated that
exemplary oligonucleotides of 8, 9, 10, 11, 12, 13, 14, 1S, 16, 17, 18, 19,
20, 2S, 30, 35,
40, 4S, 50, 5S, 60, 65, 70, 75, 80, 85, 90, 95, 100 or more base pairs will be
used,
although others are contemplated. Longer polynucleotides encoding 250, 500,
750,
1000, 1250, 1500, 2000, 2500, 3000 or 4000 bases and longer are contemplated
as well.
Such oligonucleotides will find use, for example, as probes in Southern and
Northern
blots and as primers in amplification reactions.
Suitable hybridization conditions will be well known to those of skill in the
art.
In certain applications, for example, substitution of amino acids by site-
directed
mutagenesis, it is appreciated that lower stringency conditions are required.
Under these
conditions, hybridization may occur even though the sequences of probe and
target
strand are not perfectly complementary, but are mismatched at one or more
positions.
Conditions may be rendered less stringent by increasing salt concentration and
decreasing temperature. For example, a medimn stringency condition could be
provided
by about 0.1 to 0.25 M NaCI at temperatures of about 37°C to about
55°C, while a low
stringency condition could be provided by about 0.15 M to about 0.9 M salt, at
temperatures ranging from about 20°C to about SS°C. Thus,
hybridization conditions
can be readily manipulated, and thus will generally be a method of choice
depending on
the desired results.
In other embodiments, hybridization may be aclueved under conditions of, for
example, 50 mM Tris-HCl (pH 8.3), 75 mm KCI, 3 mM MgCl2, 10 mM dithiothreitol,
at
temperatures between approximately 20°C to about 37°C. Other
hybridization
conditions utilized could include approximately 10 mM Tris-HCl (pH 8.3), SO mM
KCI,
1.5 p,M MgCl2, at temperatures ranging from approximately 40°C to about
72°C.
Formamide and SDS also may be used to alter the hybridization conditions.
19
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
One method of using probes and primers of the present invention is in the
search
for genes related to myocardin proteins and peptides, including for example,
myocardin
proteins from other species: Normally, the target DNA will be a genomic or
cDNA
library, although screening may involve analysis of RNA molecules. By varying
the
stringency of hybridization, and the region of the probe, different degrees of
homology
may be discovered.
Another way of exploiting probes and primers of the present invention is in
site-directed, or site-specific mutagenesis. Site-specific mutagenesis is a
techtuque
useful in the preparation of individual peptides, or biologically functional
equivalent
proteins or peptides, through specific mutagenesis of the underlying DNA. The
technique further provides a ready ability to prepare and test sequence
variants,
incorporating one or more of the foregoing considerations, by introducing one
or more
nucleotide sequence changes into the DNA. Site-specific mutagenesis allows the
production of mutants through the use of specific oligonucleotide sequences
which
encode the DNA sequence of the desired mutation, as well as a sufficient
number of
adjacent nucleotides, to provide a primer sequence of sufficient size and
sequence
complexity to .form a stable duplex on both sides of the deletion junction
being
traversed. Typically, a primer of about 17 to 25 nucleotides in length is
preferred.
The technique typically employs a bacteriophage vector that exists in both a
single stranded and double-stranded form. Typical vectors useful in site-
directed
mutagenesis include vectors such as the M13 phage. These phage vectors are
commercially available and their use is generally well known to those skilled
in the
art. Double-stranded plasmids are also routinely employed in site-directed
mutagenesis, which eliminates the step of transferring the gene of interest
from a
phage to a plasmid.
In general, site-directed mutagenesis is performed by f rst obtaining a single-
stranded vector, or melting of two strands of a double-stranded vector which
includes
within its sequence a DNA sequence encoding the desired protein. An
oligonucleotide
primer bearing the desired mutated sequence is synthetically prepared. This
primer is
then annealed with the single-stranded DNA preparation, taking into account
the
degree of mismatch when selecting hybridization conditions, and subjected to
DNA
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
polymerizing enzymes such as E. coli polymerase I Klenow fragment, in order to
complete the synthesis of the mutation-bearing strand. Thus, a heteroduplex is
formed
wherein one strand encodes the original non-mutated sequence and the second
strand
bears the desired mutation. This heteroduplex vector is then used to transform
appropriate cells, such as E. coli cells, and clones are selected that include
recombinant vectors bearing the mutated sequence arrangement.
The preparation of sequence variants of the selected gene using site-directed
mutagenesis is provided as a means of producing potentially useful species and
is not
meant to be limiting, as there are other ways in which sequence variants of
genes may
be obtained. For example, recombinant vectors encoding the desired gene may be
treated with mutagenic agents, such as hydroxylamine, to obtain sequence
variants.
C. Antisense Constructs
Antisense methodology takes advantage of the fact that nucleic acids tend to
pair with "complementary" sequences. By complementary, it is meant that
polynucleotides are those which are capable of base-pairing according to the
standard
Watson-Crick complementarity rules. That is, the larger purines will base pair
with
the smaller pyrimidines to form combinations of guanine paired with cytosine
(G:C)
and adenine paired with either thymine (A:T) in the case of DNA, or adenine
paired
with uracil (A:T~ in the case of RNA. Inclusion of less common bases such as
inosine, 5-methylcytosine, 6-methyladenine, hypoxanthine and others in
hybridizing
sequences does not interfere with pairing.
Targeting double-stranded (ds) DNA with polynucleotides leads to triple-helix
formation; targeting RNA will lead to double-helix formation. Antisense
polynucleotides, when introduced into a target cell, specifically bind to
their target
polynucleotide and interfere with transcription, RNA processing, transport,
translation
and/or stability. Antisense RNA constructs, or DNA encoding such antisense
RNA's,
may be employed to inhibit gene transcription or translation or both within a
host cell,
either ifs vitro or in vivo, such as within a host animal, including a human
subject.
Antisense constructs may be designed to bind to the promoter and/or other
control regions, exons, introns or even exon-intron boundaries of a gene. It
is
21
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
contemplated that the most. effective antisense constructs will include
regions
complementary to intron/exon splice junctions. Thus, it is proposed that a
preferred
embodiment includes an antisense construct with complementarity to regions
within
50-200 bases of an intron-exon splice junction. It has been observed that some
exon
sequences can be included in the construct without seriously affecting the
target
selectivity thereof. The amount of exonic material included will vary
depending on
the particular exon and intron sequences used. One can readily test whether
too much
exon DNA is included simply by testing the constructs if2 vitf°o to
determine whether
normal cellular function is affected or whether the expression of related
genes having
complementary sequences is affected.
As stated above, "complementary" or "antisense" means polynucleotide
sequences that are substantially complementary over their entire length and
have very
few base mismatches. For example, sequences of fifteen bases in length may be
termed complementary when they have complementary nucleotides at thirteen or.
fourteen positions. Naturally, sequences which are completely c~rnplementary
will be
sequences which are entirely complementary throughout their entire length and
have
no base mismatches. Other sequences with lower degrees of homology also are
contemplated. For example, an antisense construct which has limited regions of
lugh
homology, but also contains a non-homologous region (e.g., ribozyme; see
below)
could be designed. These molecules, though having less than 50% homology,
would
bind to target sequences under appropriate conditions.
It may be advantageous to combine portions of genomic DNA with cDNA or
synthetic sequences to generate specific constructs. For example, where an
intron is
desired in the ultimate construct, a genomic clone will need to be used. The
cDNA or
a synthesized polynucleotide may provide more convenient restriction sites for
the
remaining portion of the construct and, therefore, would be used for the rest
of the
sequence.
D. Ribozymes
Although proteins traditionally have been used for catalysis of nucleic acids,
another class of macromolecules has emerged as useful in this endeavor.
Ribozymes
are RNA-protein complexes that cleave nucleic acids in a site-specific
fashion.
22
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ribozymes have specific catalytic domains that possess endonuclease activity
(Kim
and Cook, 1987; Gerlach et al., 1987; Forster and Symons, 1987). For example,
a
large number of ribozymes accelerate phosphoester transfer reactions with a
high
degree of specificity, often cleaving only one of several phosphoesters in an
oligonucleotide substrate (Cook et al., 1981; Michel and Westhof, 1990;
Reinhold-
Hurek and Shub, 1992). This specificity has been attributed to the requirement
that
the substrate bind via specific base-pairing interactions to the internal
guide sequence
("IGS") of the ribozyme prior to chemical reaction.
Ribozyme catalysis has primarily been observed as part of sequence-specific
cleavage/ligation reactions involving nucleic acids (Joyce, 1989; Cook et al.,
1981).
For example, U.S. Patent 5;354,855 reports that certain ribozymes can act as
endonucleases with a sequence specificity greater than that of known
ribonucleases
and approaching that of the DNA restriction enzymes. Thus, sequence-specific
ribozyme-mediated inhibition of gene expression may be particularly suited to
therapeutic applications (Scanlon et al., 1991; Sarver et al., 1990).
Recently, it was
reported that ribozymes elicited genetic changes in some cells lines to which
they
were applied; the altered genes included the oncogenes H-ras, c-fos and genes
of HIV.
Most of this work involved the modification of a target rnRNA, based on a
specific
mutant codon that is cleaved by a specific ribozyme.
E. Vectors for Cloning, Gene Transfer and Expression
Within certain embodiments expression vectors are employed to express a
myocardin polypeptide product, which can then be purified and, for example, be
used to
vaccinate animals to generate antisera or monoclonal antibody with which
further studies
may be conducted. In other embodiments, the expression vectors are used in
gene
therapy. Expression requires that appropriate signals be provided in the
vectors,
including, for example, various regulatory elements, such as
enhancers/promoters
from viral and/or mammalian sources that are involved in driving expression of
the
genes of interest in host cells. Elements designed to optimize messenger RNA
stability and translatability in host cells also can be used. The conditions
for the use
of a number of dominant dmg selection markers for establishing permanent,
stable
cell clones expressing the products are also provided, as is an element that
links
expression of the drug selection markers to expression of the polypeptide.
23
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
(i) Regulatory Elements
Throughout this application, the term "expression construct" or "expression
cassette" is meant to include any type of genetic construct containing a
nucleic acid
coding for a gene product in which part or all of the nucleic acid encoding
sequence is
capable of being transcribed. The transcript may be translated into a protein
or
polypeptide, but it need not be. In certain embodiments, expression includes
both-
transcription of a gene and translation of mRNA into a gene product. In other
embodiments, expression only includes transcription of the nucleic acid
encoding a
gene of interest.
As used herein, regulatory elements (or sequences) are nucleotide sequences
that enhance or otherwise modulate transcription and/or translation or that
stabilize
transcription and/or translation products. Thus, for example, promoters
operably
linked to a coding sequence of an expression construct enhance transcription
of that
coding sequence and polyadenylation sequences operably linked to a coding
sequence.
modulate polyadenylation of the gene transcript. Exemplary regulatory
sequences can
include, without limitation, promoters enhancers, introns, termination
sequences,
polyadenylation sequences, stabilization sequences and the like.
In certain embodiments, the nucleic acid encoding a gene product is operably
linked and under transcriptional control of a promoter. A "promoter" refers to
a DNA
sequence recognized by the synthetic machinery of the cell, or introduced
synthetic
machinery, required to initiate the specific transcription of a gene. The
phrase "under
transcriptional control" means that the promoter is in the correct location
and
orientation in relation to the nucleic acid to control RNA polymerase
initiation and
expression of the gene.
The term promoter will be used here to refer to a group of transcriptional
control modules that are clustered around the initiation site for RNA
polymerase II.
Much of the thinking about how promoters are organized derives from analyses
of
several viral promoters, including those for the HSV thymidine kinase (tk) and
SV40
early transcription units. These studies, augmented by more recent work, have
shown
that promoters are typically composed of discrete functional modules, each
consisting
24
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
of approximately 7-20 by of DNA, and containing one or more recognition sites
for
transcriptional activator or repressor proteins.
At least one module in each promoter functions to position the start site for
RNA synthesis. The best known example of this is the TATA .box, but in some
promoters lacking a TATA box, such as the promoter for the mammalian terminal
deoxynucleotidyl transferase gene and the promoter for the SV40 late genes, a
discrete
element overlying the start site itself helps to fix the place of initiation.
Additional promoter elements regulate the frequency of transcriptional
initiation. Typically, these axe located in the region 30-110 by upstream of
the start
site, although a number of promoters have recently been shown to contain
functional
elements downstream of the start site as well. The spacing between promoter
elements frequently is flexible, so that promoter function is preserved when
elements
are inverted or moved relative to one another. In the tk promoter, the spacing
between
promoter elements can be increased to SO by apart before activity begins to
decline.
Depending on the promoter, it appeaxs that individual elements can function
either co-
operatively or independently to activate transcription.
In certain embodiments, the human cytomegalovirus (CMV) immediate early
gene promoter, the SV40 early promoter, the Rous sarcoma virus (RSV) long
terminal
repeat, a human elongation factor (hEF) promoter, rat insulin promoter or
glyceraldehyde-3-phosphate dehydrogenase promoter can be used to obtain high-
level
expression of the coding sequence of interest. The use of other viral or
mammalian
cellular or bacterial phage promoters which are well known in the art to
achieve
expression of a coding sequence of interest is contemplated as well, provided
that the
levels of expression are sufficient fox a given purpose.
By employing a promoter with well-known properties, the level and pattern of
expression of the protein of interest following transfection or transformation
can be
optimized. By way of illustration, a ubiquitous, strong (i.e., high activity)
promoter
may be employed to provide abundant gene expression in a group of host cells,
or a
tissue-specific promoter may be employed to target gene expression to one or
more
specific cell types. Further, selection of a promoter that is regulated in
response to
specific physiologic signals can permit inducible expression of the gene
product.
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Tables 2 and 3 list several regulatory elements that may be employed, in the
context
of the present invention, to regulate the expression of the gene of interest.
This list is
not intended to be exhaustive of all the possible elements involved in the
promotion of
gene expression but, merely, to be exemplary thereof.
Enhancers are genetic elements that increase transcription from a promoter
located at a distant position on the same molecule of DNA. Enhancers are
organized
much like promoters. That is, they are composed of many individual elements,
each
of which binds to one or more transcriptional proteins.
The basic distinction between enhancers and promoters is operational. An
enhancer region as a whole is typically able to stimulate transcription at a
distance;:.
this need not be true of a promoter region or its component elements. On the
other
hand, a promoter typically has one or more elements that direct initiation of
RNA
synthesis at a particular site and in a particular . orientation, whereas
enhancers.'
generally lack these specificities. Promoters and enllancers are often
overlapping and
contiguous, often seeming to have a very similar modular organization.
Tables 2 and 3, provided below, list several regulatory elements that may be
employed, in the context of the present invention, to regulate the expression
of the .
gene of interest. This list is not intended to be exhaustive of all the
possible elements
involved in the promotion of gene expression but, merely, to be exemplary
thereof.
Other promoter/enhancer combinations (see, e.g., the Eukaryotic Promoter Data
Base
EPDB) could also be used to drive expression of the gene. Eukaryotic cells can
suppout cytoplasmic transcription from certain bacterial promoters if the
appropriate
bacterial polymerase is provided, either as part of the delivery complex or as
an
additional genetic expression construct.
26
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
TABLE 2
Promoter and/or
Enhancer
Promoter/Enhancer References
Immunoglobulin HeavyBanerji et al., 1983; Gilles et al.,
Chain 1983; Grosschedl et
al., 1985; Atchinson et al., 1986, 1987;
Imler et al.,
1987; Weinberger et al., 1984; Kiledjian
et al., 1988;
Porton et al.; 1990
Immunoglobulin LightQueen et al., 1983; Picard et al., 1984
Chain
T-Cell Receptor Luria et al., 1987; Winoto et al., 1989;
Redondo et al.;
1990
HLA DQ a and/or Sullivan et al., 1987
DQ (3
(3-Interferon Goodbourn et al., 1986; Fujita et al.,
1987; Goodbourn ;
et al., 1988
Interleukin-2 Greene et al., 1989
Interleukin-2 ReceptorGreene et al., 1989; Lin et al., 1990
MHC Class II 5 Koch et al., 1989
MHC Class II HLA-DRaSherman et al., 1989
(3-Actin Kawamoto et al., 1988; Ng et al.; 1989
Muscle Creatine Jaynes et al., 1988; Horlick et al.,
Kinase 1989; Johnson et al.,
(MCK) 1989
Human Elongation Uetsuki, et al., 1989; Wakabayashi-Ito,
Factor-lA (hEF-lA et al., 1994
or
hEF-1 a)
Prealbumin Costa et al., 1988
(Transthyretin)
Elastase I Ornitz et al., 1987
Metallothionein Karin et al., 1987; Culotta et al.,
(MTII) 1989
Collagenase Pinkert et al., 1987; Angel et al.,
1987a
Albumin Pinkert et al., 1987; Tronche et al.,
1989, 1990
a-Fetoprotein Godbout et al., 1988; Campere et al.,
1989
t-Globin Bodine et al., 1987; Perez-Stable et
al., 1990
[3-Globin Trudel et al., 1987
c-fos Cohen et al., 1987
27
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
TABLE 2
Promoter and/or
Enhancer
Promoter/Enhancer References
c-HA-sas Triesman, 1986; Deschamps et al., 1985
Insulin Edlund et al., 1985
Neural Cell AdhesionHirsh et al., 1990
Molecule (NCAM)
al-Antitrypain Latimer et al., 1990
H2B (TH2B) Histone Hwang et al., 1990
Mouse and/or Type Ripe et al., 1989
I
Collagen
Glucose-Regulated Chang et al., 1989
Proteins (GRP94
and
GRP78) .
Rat Growth Hormone Larsen et al., 1986 .
Human Serum AmyloidEdbrooke et al., 1989
A
(SAA)
Troponin I (TN I) Yutzey et al., 1989
Platelet-Derived Pech et al., 1989
Growth
Factor (PDGF)
Duchenne Muscular Klamut et al., 1990
Dystrophy
SV40 Banerji et al., 1981; Moreau et al.,
1981; Sleigh et al.,
1985; Firak et al., 1986; Herr et al.,
1986; Imbra et al.,
1986; Kadesch et al., 1986; Wang et
al., 1986; Ondek
et al., 1987; Kuhl et al., 1987; Schaffner
et al., 1988
Polyoma Swartzendruber et al., 1975; Vasseur
et al., 1980;
Katinka et al., 1980, 1981; Tyndell
et al., 1981;
Dandolo et al., 1983; de Villiers et
al., 1984; Hen et al.,
1986; Satake et al., 1988; Campbell
and/or Villarreal,
1988
Retroviruses Kriegler et al., 1982, 1983; Levinson
et al., 1982;
Kriegler et al., 1983, 1984a, b, 1988;
Bosze et al.,
1986; Miksicek et al., 1986; Celander
et al., 1987;
Thiesen et al., 1988; Celander et al.,
1988; Choi et al.,
1988; Reisman et al., 1989
28
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
TABLE 2
Promoter and/or
Enhancer
Promoter/Enhancer References
Papilloma Virus Campo et al., 1983; Lusky et al., 1983;
Spandidos
and/or Wilkie, 1983; Spalholz et al.,
1985; Lusky et al.,
1986; Cripe et al., 1987; Gloss et al.,
1987; Hirochika
et al., 1987; Stephens et al., 1987;
Glue et al., 1988
Hepatitis B Virus Bulla et al., 1986; Jameel et al., 1986;
Shaul et al.,
1987; Spandau et al., 1988; Vannice
et al., 1988
Human Muesing et al., 1987; Hauber et al.,
1988; Jakobovits
Immunodeficiency et al., 1988; Feng et al., 1988; Takebe
Virus et al., 1988;
Rosen et al., 1988; Berkhout et al.,
1989; Laspia et al.,
1989; Sharp et al., 1989; Braddock et
al., 1989
Cytomegalovirus Weber et al., 1984; Boshart et al.,
(CMV) 1985; Foecking
et al., 1986
Rous sarcoma virus Gorman, et al., 1982; Guzman, et al.,
1993
(RSV)
Gibbon Ape LeukemiaHolbrook et al., 1987; Quinrl et al.,
1989
VIrLIS
TABLE 3
Inducible Elements
Element Inducer References
MT II Phorbol Ester (TFA) Palmiter et al., 1982;
Haslinger
Heavy metals et al., 1985; Searle
et al., 1985;
Stuart et al., 1985;
Imagawa
et al., 1987, I~arin
et al., 1987;
Angel et al., 1987b;
McNeall
et al., 1989
MMTV (mouse Glucocorticoids Huang et al., 1981;
Lee et al.,
mammary tumor 1981; Majors et al.,
1983;
virus) Chandler et al., 1983;
Lee
et al., 1984; Ponta
et al., 1985;
Sakai et al., 1988
(3-Interferon poly(rI)x Tavernier et al., 1983
poly(rc)
Adenovirus 5 ElA Imperiale et al., 1984
E2
Collagenase Phorbol Ester (TPA) Angel et al., 1987a
Stromelysin Phorbol Ester (TPA) Angel et al., 1987b
SV40 Phorbol Ester (TPA) .Angel et al., 1987b
29
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
TAELE 3
Inducible Elements
Element Inducer References
Murine MX Gene Interferon, NewcastleHug et al., 1988
Disease Virus
GRP7S Gene A23187 Resendez et al., 1988
cx-2-MacroglobulinIL-6 Kunz et al., 1989
Vimentin Serum Rittling et al., 1989
MHC Class I Interferon Blanar et al., 1989
Gene
H-2,Kb
HSP70 , EIA, SV40 Large T Taylor et al., 1989,
Antigen 1990a,
1990b
Proliferin Phorbol Ester-TPA Mordacq et al., 1989
Tumor Necrosis PMA Hensel et al., 1989
Factor
Thyroid StimulatingThyroid Hormone Chatterjee et al.,
Hormone a Gene 1989
In one aspect, tissue-specific promoters, e.g., cardiac-specific and/or
fibroblast-specific promoters, are of particular interest. By way of
illustration,
cardiac-specific promoters include the myosin light chain-2 promoter (Franz et
al.,
1994; Kelly et al., 1995), the alpha actin promoter (Moss et al., 1996), the
troponin 1
promoter (Bhavsar et°al., 1996); the Na~/Ca2+ exchanger promoter
(Barnes et al.,
1997), the dystrophin promoter (Kimura et al., 1997), the creatine kinase
promoter
(Ritchie, M.E., 1996), the alpha? integrin promoter (Ziober & Kramer, 1996),
the
brain natriuretic peptide promoter (LaPointe et al., 1996) and the alpha B-
crystalliWsmall heat shock protein promoter (Gopal-Srivastava, R., 1995),
alpha
myosin heavy chain promoter (Yamauchi-Takihara et al., 1989) and the ANF
promoter (LaPointe et al., 1988).
Where a cDNA insert is employed, one will typically desire to include a
polyadenylation signal to effect proper polyadenylation of the gene
transcript. The
nature of the polyadenylation signal is not believed to be crucial to the
successful
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
practice of the invention, and any such sequence may be employed such as human
growth hormone and SV40 polyadenylation signals. Also contemplated as an
element
of the expression cassette is a terminator. These elements can serve to
enhance
message levels and to minimize read through from the cassette into other
sequences.
(ii) Selectable Markers
In certain embodiments of the invention, in which cells contain nucleic acid
constructs of the present invention, a cell may be identified in vitro or in
vivo by
including a marker in the expression construct. Such markers would confer an
identifiable change to the cell permitting easy identification of cells
containing the
expression construct. Usually the inclusion of a drug selection marker aids in
cloning
and in the selection of transformants, for example, genes that confer
resistance to
neomycin, puromycin, hygromycin, DHFR, GPT, zeocin and histidinol are useful
selectable markers. Alternatively, enzymes such as herpes simplex virus
thymidine
kinase (tk) or chloramphenicol acetyltransferase (CAT) may be employed.
Immunologic markers also can be employed. The selectable marker employed is
not
believed to be important, so long as it is capable of being expressed
simultaneously
with the nucleic acid encoding a gene product. Further examples of selectable
markers are well known to one of skill in the art.
(iii) Multigene Constructs and IRES
In certain embodiments of the invention, the use of internal ribosome entry
site
(IRES) elements are used to create multigene, or polycistronic, messages. IRES
elements is believed to allow bypassing of the ribosome scanning model of 5'
methylated Cap dependent translation and facilitate translation at internal
sites
(Pelletier and Sonenberg, 1988). By way of illustration, IRES elements from
two
members of the picanovirus family (polio and encephalomyocarditis) have been
described (Pelletier and Sonenberg, 1988), as well an IRES from a marmnalian
message (Macejak and Sarnow, 1991). IRES elements can be linked to
heterologous
open reading frames: Multiple open reading frames can be transcribed together,
each
separated by an IRES, creating polycistronic messages. By virtue of the TRES
element, each open reading frame is accessible to ribosomes for efficient
translation.
31
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Multiple genes can be efficiently expressed using a single promoter/enhancer
to
transcribe a single message.
Any heterologous open reading frame can be linked to IRES elements. This
includes genes for secreted proteins, rnulti-subunit proteins, encoded by
independent
genes, intracellular or membrane-bound proteins and selectable markers. In
this way,
expression of several proteins can be simultaneously engineered into a cell
with a
single construct and a single selectable marker.
(iv) Polyadenylation Signals
In expression, one will typically include a polyadenylation signal to effect
proper polyadenylation of the transcript. The nature of the polyadenylation
signal is
not believed to be crucial to the successful practice of the invention, and
any of a .
number of such sequences may be employed. Exemplary embodiments include the
SV40 polyadenylation signal, the bovine growth hormone polyadenylation signal
and
others which are convenient and/or known to function well in various target
cells.
Also contemplated as an element of the expression cassette is a
transcriptional
termination site. These elements can serve to enhance message levels and/or to
minimize read through from the cassette into other sequences.
(v) Vectors
The term "vector" is used to refer to carnet molecules with which a nucleic
acid sequence can be associated for introduction into a cell. The nucleic acid
sequence can be "exogenous," (e.g., foreign to the cell into which it is
introduced) or
"endogenous" (e.g., the same as a sequence in the cell into which it is
introduced.
Exemplary vectors include plasmids, cosmids, viruses (bacteriophage, animal
viruses,
and plant viruses), and artificial chromosomes (e.g., YACs), lipid-based
vectors (e.g.,
liposomes) and other macromolecular complexes capable of mediating delivery of
a
polynucleotide to a host cell. One of skill in the art would be well equipped
to
construct a vector through standard techniques, for example standard
recombinant
techniques such as described in Sambrook et al., 1989 and Ausubel et al.,
1994, both
incorporated herein by reference.
32
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
A large number of viral and non-viral vectors (including lipid-based and other
synthetic delivery systems known in the art) can likewise be employed to
deliver
polynucleotides of the present invention. Such vectors may be modified, as
known to
those of skill in the art, to confer or enhance cell specificity. By way of
illustration,
the surface of viral vectors may be modified such that they preferentially or
exclusively bind to and/or infect a particular target cell population.
As used herein, the term "expression vector" refers to a vector containing a
nucleic acid sequence coding for at least part of a gene product capable of
being
transcribed. In some cases, the transcription products) are then translated
into a
protein, polypeptide, or peptide. In other cases, these sequences are not
translated, for
example, in the production of antisense molecules or ribozymes. Expression
vectors
can contain a variety of "control sequences," which refer to nucleic acid
sequences
that regulate the transcription and possibly translation of an operably linked
coding
sequence in a particular host organism. In addition to control sequences that
govern
transcription and translation, vectors and expression vectors may contain
nucleic acid
sequences that serve other functions as well for example as described infra.
(vi) Host Gells
As used herein, the terms "cell," "cell line," and "cell culture" may be used
interchangeably. These terms also include their progeny, which is any and all
subsequent generations. It is understood that all progeny may not be identical
due to
deliberate or inadvertent mutations.' In the context of expressing a
heterologous
nucleic acid sequence, "host cell" refers to a prokaryotic or eukaryotic cell,
and. it
includes any transformable organisms that is capable of replicating a vector
and/or
expressing a heterologous gene encoded by a vector. A host cell can, and has
been,
used as a recipient for vectors. A host cell may be "transfected" or
"transformed,"
which refers to a process by which exogenous nucleic acid is transferred or
introduced
into the host cell. A transformed cell includes the primary subject cell and
its
progeny.
Some vectors may employ control sequences that allow it to be replicated
and/or expressed in both prokaryotic and eukaryotic cells. One of skill in the
art
would further understand conditions under which to incubate such host cells to
33
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
maintain them and to permit replication of a vector. Also understood and known
are
techniques and conditions that would allow large-scale production of vectors,
as well.
as production of the nucleic acids encoded by vectors and their cognate
polypeptides, .
proteins, or peptides.
(vii) Expression Systems
Numerous expression systems exist that comprise at least a part or all of the
compositions discussed above. Prokaryote- and/or eukaryote-based systems can
be
employed for use with the present invention to produce nucleic acid sequences,
or
their cognate polypeptides, proteins and peptides. Many such systems are
commercially and widely available.
The insect cell/baculovirus system can produce a High level of protein
expression of a heterologous nucleic acid segment, such as described in U.S.
Patent
5,871,986, 4,879,236, both herein incorporated by reference, and which can be
bought, for example, under the name MAXBAC~ 2.0 from INVITROGENO and
BACPACI~TM baculovirus expression system from CLONTECH~.
Other examples of expression systems include STRATAGENE~'s
COMPLETE CONTROLT"" Inducible Mammalian Expression System, which involves
a synthetic ecdysone-inducible receptor, or its pET Expression System, an E.
coli
expression system. Another example of an inducible expression system is
available
from INVITROGENC~, which carries the T-REXTM (tetracycline-regulated
expression) System, an inducible mammalian expression system that uses the
full-.
length CMV promoter. INVITROGEN~ also provides a yeast expression system
called the Pichia methanolica Expression System, which is designed for high-
level
production of recombinant proteins in the methylotrophic yeast Pichia
methanolica.
One of skill in the art would know how to express a vector, such as an
expression
construct, to produce a nucleic acid sequence or its cognate polypeptide,
protein, or
peptide.
(viii) Gene Delivery Means
There are a number of ways in which a gene of interest, for example within an
expression vector, may be introduced into cells. In certain embodiments of the
34
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
invention, the gene delivery means comprises a virus or engineered construct
derived
from a viral genome. The ability of certain viruses to enter cells for example
via
receptor-mediated endocytosis and to express viral genes stably and
efficiently have
made them attractive candidates for the transfer of foreign genes into
mammalian cells
(Ridgeway, 1988; Nicolas and Rubenstein, 1988; Baichwal and Sugden, 1986;
Temin,
1986). The first vimses used as gene vectors were DNA viruses including the
papovaviruses (simian virus 40, bovine papilloma virus, and polyoma)
(Ridgeway,
1988; Baichwal and Sugden, 1986) and adenoviruses (Ridgeway, 1988; Baichwal
and
Sugden, 1986). Although these viral vectors generally have a relatively fixed
capacity
for foreign DNA can accornlnodate up to 5-10 kb of foreign DNA and many
different
viral vectors can be readily introduced into a variety of different cells and
animals
(see, e.g., Nicolas and Rubenstein, 1988; Temin, 1986). Where viral vectors
are
employed to deliver the gene or genes of interest, it is generally preferred
that they be
replication-defective.
One of the preferred methods for in wivo gene delivery involves the use of an
adenovirus expression vector. "Adenovirus expression vector" is meant to
include
those constructs containing adenovims sequences sufficient to (a) support
packaging
of the construct and (b) to express polynucleotide that has been cloned
therein. In this
context, expression does not require that the gene product be synthesized.
An adenivorus expression vector comprises a genetically engineered form of
adenovirus. Knowledge of the genetic organization of adenovirus, a linear,
double-
stranded DNA virus, allows substitution of large pieces of adenoviral DNA with
foreign sequences (typically up to about 7 kB (Grunhaus and Horwitz, 1992)).
Modified adenoviral and other viral vectors have also been constructed to
provide for
increased packaging capacity and are likewise contemplated herein. In contrast
to
retrovirus, the adenoviral infection of host cells does not generally result
in
chromosomal integration. Also, adenoviruses are structurally stable, and no
genome
rearrangement has been detected after extensive amplification. Adenovirus can
infect
various lineages of cells regardless of their cell cycle stage. So far,
adenoviral
infection appears to be linked only to mild disease such as acute respiratory
disease in
humans.
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Adenovirus is particularly suitable for use as a gene transfer vector because
of
its mid-sized genome, ease of manipulation, high titer, wide target cell range
and high
infectivity. In the case of adenovirus serotype 5 (Ad5), for example, both
ends of the
viral genome contain 100-200 base pair inverted repeats (ITRs), which are cis
elements necessary for viral DNA replication and packaging. The early (E) and
late
(L) regions of the genome contain different transcription units that are
divided by the
onset of viral DNA replication. The E1 region (ElA and ElB) encodes proteins
responsible for the regulation of transcription of the viral genome and a few
cellular
genes. The expression of the E2 region (E2A and E2B) results in the synthesis
of the
proteins for viral DNA replication. These proteins are involved in DNA
replication,
late gene expression and host cell shut-off (Reran, 1990). The products of the
late
genes, including the majority of the viral capsid proteins, axe expressed only
after
significant processing of a single primary transcript issued by the major late
promoter
(MLP). The MLP (located at 16.8 m.u.) is particularly efficient during the
late phase
of infection, and the mRNA's issued from this promoter possess a S'-tripartite
leader
(TPL) sequence which makes them preferred mRNA's for translation.
In one system, recombinant adenovirus is generated from homologous
recombination between shuttle vector and provirus vector. Due to the possible
recombination between two proviral vectors, wild-type adenovirus may be
generated
from this process. Therefore, it is important to minimize this possibility by
reducing
or eliminating adnoviral sequence overlaps within the system and/or to isolate
a single
clone of virus from an individual plaque and examine its genomic structure.
Generation and propagation of replication-deficient adenovirus vectors depend
on a unique helper cell line, such as the human 293 cell line, which was
transformed
from human embryonic kidney cells by Adenovirus type 5 DNA fragments to
constitutively expresses El proteins (Graham et al., 1977). Since the E3
region is
dispensable from the adenovirus genome (Jones and Shenk, 1978), the current
adenovirus vectors, with the help of 293 cells, generally carry foreign DNA in
either
the E1, the E3 or both regions (Graham and Prevec, 1991). In nature,
adenovirus can
package approximately 105% of the wild-type genome (Ghosh-Choudhury et al.,
1987), providing capacity for about 2 extra kb of DNA. Combined with the
approximately 5.5 kb of DNA that is replaceable in the E 1 and E3 regions, up
to about
36
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
7.5 kb of foreign DNA may be packaged in an adenovirus. Additionally, modified
adenoviral vectors are now available which have an even greater capacity to
carry
foreign DNA.
Helper cell lines may be derived from human. cells such as human embryonic
kidney cells, muscle cells, hematopoietic cells or other human embryonic
mesenchymal or epithelial cells. Alternatively, the helper cells may be
derived from
the cells of other mammalian species that are permissive for human adenovirus.
Such
cells include, e.g., Vero cells or other monkey embryonic mesenchymal or
epithelial
cells. As stated above, a preferred helper cell line is 293.
Racher et al. (1995) disclosed improved methods for culturing 293 cells and
propagating adenovirus: In one format, natural cell aggregates are grown by
inoculating individual cells into 1 liter siliconized spinner flasks (Techne,
Cambridge,
UK) containing 100-200 ml of medium. Following stirnng at 40 rpm, the cell
viability is estimated with trypan blue. In another format, Fibra-Cel
microcarriers
(Bibby Sterlin, Stone, UK) (5 g/1) are employed as follows. A cell inoculum,
resuspended in 5 ml of medium, is added to the carrier (50 ml) in a 250: ml
Erlenmeyer flask and left stationary, with occasional agitation, for 1 to 4 h.
The
medium is then replaced with 50 ml of fresh medium and shaking initiated. For
virus
production, cells are allowed to grow to about 80% confluence, after which
time the
medium is replaced (to 25% of the final volume) and adenovirus added at an MOI
of
0.05. Cultures are left stationary overnight, following which the volume is
increased
to 100% and shaking commenced for another 72 h.
Other than the preference that the adenovirus vector be replication-defective,
or at least conditionally defective, the nature of the adenovirus vector is
not believed
to be critical to the successful practice of the invention. The adenovirus may
be
selected from any of the 42 different known serotypes or subgroups A-F.
Adenovirus
serotype 5 of subgroup C is a preferred starting material for obtaining a
conditional
replication-defective adenovirus vector for use in the present invention. This
is, in
part, because Adenovirus type 5 is a human adenovirus about which a great deal
of
biochemical and genetic information is known, and it has historically been
used for
most constructions employing adenovirus as a vector. Additionally, various
37
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
modifications can be made to. adenovirus to facilitate cell targeting of the
expression
cassette and/or otherwise modify vector interaction with the host cell. By way
of
illustration, it is known that primary fibroblasts generally express low
levels of the
high-affinity Coxsackie virus and Adenovirus receptor (CAR), which receptor
facilitates transduction of host cells by the adenoviral vector. However, it
is also
known that adenoviral vectors can be modified, for example by altering the
adenovirus fiber, to improve binding to other cell-surface receptors where CAR
receptors are limited (see, e.g. Hidaka et al., 1999).
As stated above, a preferred adenoviral vector according to the present
invention lacks an adenovirus E1 region and thus, is replication defective.
Typically,
it is most convenient to introduce the polynucleotide encoding the gene of
interest at
the position from which the E1-coding sequences have been removed. However,
the
position of insertion of the construct within the adenovirus sequences is not
critical to
the invention. Further, other adenoviral sequences may be deleted and/or
inactivated
in addition to or in lieu of the El region. For example, the E2 and E4 regions
are both
necessary for adenoviral replication and thus may be modified to render an
adenovirus
vector replication-defective, in which case a helper cell Iine or helper virus
complex
may employed to provide such deleted/inactivated genes isz t~as2,r. The
polynucleotide
encoding the gene of interest may alternatively be inserted in lieu of a
deleted E3
region, such as in E3 replacement vectors as described by Karlsson et al.
(1986), or in
the E4 region where a helper cell line or helper virus complements an E4.
defect.
Other modifications are known to those of skill in the art and are likewise
contemplated herein.
Adenovirus is easy to grow and manipulate and exhibits broad host range in
vitro and in vivo. This group of viruses can be obtained in high titers, e.g.,
109-lOlz
plaque-forming units per ml, and they are highly infective. The life cycle of
adenovirus does not require integration into the host cell genome. The foreign
genes
delivered by adenovirus vectors are episomal and, therefore, have low
genotoxicity to
host cells. No side effects have been reported in studies of vaccination with
wild-type
adenovirus (Couch et al., 1963; Top et al., 1971), demonstrating their safety
and
therapeutic potential as in vivo gene transfer vectors.
38
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Adenovirus vectors have been used in eukaryotic gene expression (Levrero et
al., 1991; Gomez-Foix et al., 1992) and vaccine development (Grunhaus and
Horwitz,
1992; Graham and Prevec, 1991). Animal studies initially suggested that
recombinant
adenovirus could be useful for gene therapy (see, e.g., Stratford-Perricaudet
and
Perricaudet, 1991; Stratford-Perricaudet et al:, 1990; Rich et al., 1993).
Studies in
administering recombinant adenovirus to different tissues include
administration via
intracoronary catheter into one or more coronary arteries of the heart
(Hammond, et
al.), U.S. Patents 5,792,453 and 6,100,242), trachea instillation (Rosenfeld
et al:,
1991; Rosenfeld et al., 1992), muscle injection (Ragot et al., 1993),
peripheral
intravenous injections (Herz and Gerard, 1993) and stereotactic inoculation
into the
brain (Le Gal La Salle et al., 1993).
The retroviruses are a group of single-stranded RNA viruses characterized by
an ability to convert their RNA to double-stranded DNA in infected cells by a
process
of reverse-transcription (Coffin, 1990). The resulting DNA then stably
integrates into
cellular chromosomes as a provirus and directs synthesis of viral proteins.
The
integration results in the retention of the viral gene sequences in the
recipient cell and
its descendants. The retroviral genome contains three genes, gag, pol, and env
that
code for capsid proteins, polymerase enzyme, and envelope components,
respectively.
A sequence found upstream from the gag gene contains a signal for packaging of
the
genome into virions. Two long terminal repeat (LTR) sequences are present at
the 5'
and 3' ends of the viral genome. These contain strong promoter and enhancer
sequences and are also required for integration in the host cell genome
(Coffin, 1990).
In order to construct a retroviral vector, a nucleic acid encoding a gene of
interest is inserted into the viral genome in the place of certain viral
sequences to
produce a virus that is replication-defective. In order to produce virions, a
packaging
cell line containing the gag, pol, and env genes but without the LTR and
packaging
components is generally employed (Mann et al., 1983). When a recombinant
plasmid
containing a cDNA, together with the retroviral LTR and packaging sequences is
introduced into this cell line (by calcium phosphate precipitation for
example), the
packaging sequence allows the RNA transcript of the recombinant plasmid to be
packaged into viral particles, which are then secreted into the culture media
(Nicolas
and Rubenstein, 1988; Temin, 1986; Main et al., 1983). The media containing
the
39
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
recombinant retroviruses is then collected, optionally concentrated, and used
for gene
transfer. Retroviral vectors are able to infect a broad variety of cell types.
However,
integration and stable expression require the division of host cells (Paskind
et al.,
1975).
A novel approach designed to allow specific targeting of retrovinxs vectors
was recently developed based on the chemical modification of a retrovirus by
the
chemical addition of lactose residues to the viral envelope. This modification
could
permit the specific infection of hepatocytes via sialoglycoprotein receptors.
A different approach to targeting of recombinant retrovinises was designed in
which biotinylated antibodies against a retroviral envelope protein and
against a
specific cell receptor were used. The antibodies were coupled via the biotin
components by using streptavidin (Roux et al., 1989). Using antibodies against
major
histocompatibility complex class I and class II antigens, they demonstrated
the
infection of a variety of human cells that bore those surface antigens with an
ecotropic
virus iTZ vitro (Roux et al., 1989).
There are certain limitations to the use of retrovirus. For example,
retrovirus
vectors usually integrate into random sites in the cell genome. This can lead
to
insertional rnutagenesis through the interruption of host genes or through the
insertion
of viral regulatory sequences that can interfere with the function of flanking
genes
(Varmus et al., 1981).' Another concern with the use of defective retrovinis
vectors is
the potential appearance of wild-type replication-competent virus in the
packaging
cells. This can result from recombination events in which the intact-sequence
from
the recombinant virus inserts upstream from the gag, pol, env sequence
integrated in
the host cell genome. However, new packaging cell lines are now available that
should greatly decrease the likelihood of recombination (Markowitz et al.,
1988;
Hersdorffer et al., 1990).
Other viral vectors may be employed as expression constructs in the present
invention. Vectors derived from viruses such as vaccinia virus (Ridgeway,
1988;
Baichwal and Sugden, 1986; Coupar et al., 1988) adeno-associated virus (AAV)
(Ridgeway, 1988; Baichwal and Sugden, 1986; Hermonat and Muzycska, 1984) and
herpesviruses may be employed. They offer several attractive features fox
various
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
mammalian cells (Friedmann, 1989; Ridgeway; 1988; Baichwal and Sugden, 1986;
Coupar et al., 1988; Norwich et al., 1990).
With the recent recognition of defective hepatitis B viruses, new insight was
gained into the structure-function relationship of different viral sequences.
Ih vitro
studies showed that the virus could retain the ability for helper-dependent
packaging
and reverse transcription despite the deletion of up to 80% of its genome
(Norwich et
al., 1990). This suggested that large portions of the genorne could be
replaced with
foreign genetic material. The hepatotropism and persistence (integration) were
particularly attractive properties for liver-directed gene transfer. Chang et
al., recently
introduced the chloramphenicol acetyltransferase (CAT) gene into duck
hepatitis B
virus genome in the place of the polymerise, surface, and pre-surface coding
sequences. It was co-transfected with wild-type virus into an avian hepatoma
cell
line. Culture media containing high titers of the recombinant virus were used
to infect
primary duckling hepatocytes. Stable CAT gene expression was detected for at
least
24 days after transfection (Chang et al., 1991).
In order to effect expression of sense or antisense gene constructs, the
expression construct is delivered into a cell. This delivery may be
accomplished ih
vitro, as in laboratory procedures for transforming cells lines, or ih vivo or
ex vivo, as
in the treatment of certain disease states. One mechanism for delivery is via
viral
infection where the expression construct is encapsidated in an infectious
viral particle.
Non-viral methods for the transfer of expression constructs into mammalian
cells can also be used in the context of the present invention. These include
calcium
phosphate precipitation (Graham and Van Der Eb, 1973; Chen and Okayama, 1987;
Rippe et al., 1990) DEAF-dextrin (Gopal, 1985), electroporation (Tur-Kaspa et
al.,
1986; Potter et al., 1984), direct microinjection (Harland and Weintraub,
1985), DNA-
loaded liposomes (Nicolau and Sene, 1982; Fraley et al., 1979) and
lipofectamine-
DNA complexes, cell sonication (Fechheimer et al., 1987), gene bombardment
using
high velocity microprojectiles (Yang et al., 1990), and receptor-mediated
transfection
(Wu and Wu, 1987; Wu and Wu, 1988). Some of these techniques may be
successfully adapted for in vivo or ex vivo use.
41
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Once the expression construct has been delivered into the cell the nucleic
acid
encoding the gene of interest may be positioned and expressed at different
sites. In
certain embodiments, the nucleic acid encoding the gene may be stably
integrated into
the genome of the cell. This integration may be in the cognate location and
orientation via homologous recombination (gene replacement) or it may be
integrated
in a random, non-specific location (gene augmentation). In yet further
embodiments,
the nucleic acid may be stably maintained in the cell as a separate, episomal
segment
of DNA. Such nucleic acid segments or "episomes" encode sequences sufficient
to
permit maintenance and replication independent of or in synchronization with
the host
cell cycle. How the expression construct is delivered to a cell and where in
the cell .
the nucleic acid remains is dependent on the type of expression construct
employed.
In yet another embodiment of the invention, the expression construct may
simply consist of naked recombinant DNA or plasmids. Transfer of the construct
may
be performed by any of the methods mentioned above which physically or
chemically
permeabilize the cell membrane. This is particularly applicable for transfer
in vitro
but it may be applied to in vivo use as well. Dubensky et al. (1984)
successfully
injected polyomavirus DNA in the form of calcium phosphate precipitates into
liver
and spleen of adult and newborn mice demonstrating active viral replication
and acute
infection. Benvenisty and Neshif (1986) also demonstrated that direct
intraperitoneal
injection of calcium phosphate-precipitated plasmids results in expression of
the
transfected genes. It is envisioned that DNA encoding a gene of interest may
also be
transferred in a similar manner in vivo and express the gene product.
In still another embodiment of the invention, a naked DNA expression
construct may be transferred into cells using particle bombardment. This
method
depends on the ability to accelerate DNA-coated microprojectiles to a high
velocity
allowing them to pierce cell membranes and enter cells without killing them
(Klein et
al., 1987). Several devices for accelerating small particles have been
developed. One
such device relies on a high voltage discharge to generate an electrical
current, which
in turn provides the motive force (Yang et al., 1990). The microprojectiles
used have
consisted of biologically inert substances such as tungsten or gold beads.
42
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Selected organs including the liver, skin, and muscle tissue of rats and mice
have been bombarded iri vivo (Yang et al., 1990; Zelenin et al., 1991). This
may
require surgical exposure of the tissue or cells, to eliminate any intervening
tissue
between the gun and the target organ, i.e., ex vivo treatment. Again, DNA
encoding a
particular gene may be delivered via this method and still be incorporated by
the
presentinvention.
In a further embodiment of the invention, the expression construct may be
complexed with one or more lipid components and/or entrapped in a liposome.
Liposomes are vesicular structures characterized by a phospholipid bilayer
membrane
and an inner aqueous medium. Multilamellar liposomes have multiple lipid
layers
separated by aqueous medium. They form spontaneously when phospholipids are
suspended in an excess of aqueous solution. The lipid components undergo self
rearrangement before the formation of closed structures and entrap water and
dissolved solutes between the lipid bilayers (Ghosh and Bachhawat, 1991). Also
contemplated are lipofectamine-DNA complexes.
Liposome-mediated nucleic acid delivery and expression of foreign DNA in
vitro has been very successful. Wong et al., (1980) demonstrated the
feasibility of
liposome-mediated delivery and expression of foreign DNA in cultured chick
embryo,
HeLa and hepatoma cells. Nicolau et al., (1987) accomplished successful
liposome-
mediated gene transfer in rats after intravenous injection.
In certain embodiments of the invention, the liposome may be complexed with
a hemagglutinating virus (HVJ). This has been shown to facilitate fusion with
the cell
membrane and promote cell entry of liposome-encapsulated DNA (Kaneda et al.,
1989). In other embodiments, the liposome may be complexed or employed in
conjunction with nuclear non-histone chromosomal proteins (HMG-1) (Kato et
al.,
1991 ). In yet further embodiments, the liposome may be complexed or employed
in
conjunction with both HVJ and HMG-1. In that such expression constructs have
been
successfully employed in transfer and expression of nucleic acid in vitro and
in vivo,
then they are applicable for the present invention. Where a bacterial promoter
is
employed in the DNA construct, it also will be desirable to include within the
liposome an appropriate bacterial polymerase.
43
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Other expression constructs which can be employed to deliver a nucleic acid
encoding a particular gene into cells are receptor-mediated delivery vehicles.
These
take advantage of the selective uptake of macromolecules by receptor-mediated
endocytosis in most eukaryotic cells. Because of the cell type-specific
distribution of
various receptors, the delivery can be highly specific (Wu and Wu, 1993).
Receptor-mediated gene targeting vehicles generally consist of two
components: a cell receptor-specific ligand and a DNA-binding agent. Several
ligands have been used for receptor-mediated gene transfer. The most
extensively
characterized ligands are asialoorosomucoid (ASOR) (Wu and Wu, 1987) and
transferrin (Wagner et al., 1990). Recently, a synthetic neoglycoprotein,
which
recognizes the same receptor as ASOR, has been used as a gene delivery vehicle
(Ferkol et al., 1993; Perales et al., 1994) and epidermal growth factor (EGF)
has also
been used to deliver genes to squamous carcinoma cells (Myers, EPO 0273085).
In other embodiments, the delivery vehicle may comprise a ligand and a
liposome. For example, Nicolau et al., (1987) employed lactosyl-ceramide, a
galactose-terminal asialganglioside, incorporated into liposomes and observed
an
increase in the uptake of the insulin gene by hepatocytes. Thus, it is
feasible that a
nucleic acid encoding a particular gene also may be specifically delivered
into a cell
type by any number of receptor-ligand systems with or without liposomes. For
example, epidermal growth factor (EGF) may be used as the receptor for
mediated
delivery of a nucleic acid into cells that exhibit upregulation of EGF
receptor.
Mannose can be used to target the mannose receptor on liver cells. Also,
antibodies to
CDS (CLL), CD22 (lymphoma), CD25 (T-cell leukemia) and MAA (melanoma) can
similarly be used as targeting moieties.
In certain embodiments, gene transfer may more easily be performed under ex
vivo conditions. Ex vivo gene therapy refers to the isolation of cells from an
animal,
the delivery of a nucleic acid into the cells in vitro, and then the return of
the modified
cells back into an animal. This may involve the surgical removal of
tissue/organs
from an animal or the primary culture of cells and tissues.
44
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
II. Myocardin Peptides and Polypeptides
The present invention also provides exemplary myocardin protein/polypeptide
sequences. For example, SEQ ID NOS:2, 26, 28 and 30 provide amino acid
sequences for myocardins of SEQ ID NOS:1, 25, 27 and 29, respectively. In
addition
to entire myocardin molecules, the present invention also relates to fragments
of the
polypeptides that may or may not retain the various functions described below.
By
way of illustration, N-terminally truncated myocardin 1 polypeptides from
mouse and
human (SEQ ID NOS: 2 and 26, respectively) are provided, which polypeptides
retain
the various functions described below. Fragments, including the N-terminus of
the
molecule may be generated by genetic engineering of translation stop sites
within the
coding region (discussed below). Alternatively, treatment of the polypeptides
with
proteolytic enzymes, known as proteases, can produces a variety of N-terminal,
C-
terminal and internal fragments. These fragments may be purified according to
known methods, such as pr.;cipitation (e.g., ammonium sulfate), HPLC, ion
exchange
chromatography, affinity chromatography (including immunoaffinity
chromatography) or various size separations (sedimentation, gel
electrophoresis, gel
filtration).
A. Structural and Functional Aspects
Myocardin 1 from human and mouse, shown in SEQ ID N0:28 and SEQ ID
N0:30 are 935 and 938 residues, respectively. In human myocardin 1, the
nuclear
localization sequence (NLS) is located between residues 245 and 254, within a
basic
region at residues 243-260. A glutamine-rich (Q) domain is located between
residues
287-320. The SAP domain is found at residues 380-414. The transcription
activation
domain is located at the carboxyl-terminus at 670 to 935.
In general, myocardins are cell regulatory proteins/peptides that function to
modulate cell phenotype. In particular, myoeardin can be used to reduce the
deleterious effects of non-cardiomyocytes on injured myocardium and/or to
stimulate
non-cardiomyoeytes to perform one or more functions typical of cardiomyocytes,
thereby enhancing cardiac function. By way of illustration, myocardin 1 is a
novel
cardiac-specific regulatory protein capable of modulating the phenotype of
target cells
within the heart, such as fibroblasts. Overexpression of myocardin 1 in
fibroblasts is
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
sufficient to activate expression of a variety of cardiac promoters, including
a-myosin
heavy chain, atrial natriuretic factor, Nkx2.5 and SM22. In combination with
GATA4, myocardin 1 transactivates regulatory sequences in the cardiac specific
homeobox Nkx2.5 gene. In addition myocardin 1 is a potent inhibitor of cell
proliferation, demonstrated by a reduced number of transfected cells
expressing
myocardin 1 compared to those expressing a control marker gene. Further,
though
inhibitory of cell proliferation, myocardin appears to stimulate cardiomyocyte
hypertrophy. These results may provide an explanation for the post-mitotic
feautures
of the cardiomyocytes.
B. Variants of Myocardin
Amino acid sequence variants of a myocardin polypeptide can be substitutional,
insertional or deletion variants. Deletion variants lack one or more residues
of the native
protein which are not essential for function or immunogenic activity, and are
exemplified
by the variants lacking a transmembrane sequence described above. Another
common
type of deletion variant is one lacking secretory signal sequences or signal
sequences
directing a protein to bind to a particular part of a cell. Insertional
mutants typically
involve the addition of material at a non-terminal point in the polypeptide.
This may
include the insertion of an immunoreactive epitope or simply a single residue.
Terminal
additions, called fusion proteins, are discussed below.
Substitutional variants typically contain the exchange of one amino acid for
another at one or more sites within the protein, and may be designed to
modulate one or
more properties of the polypeptide, such as stability against proteolytic
cleavage, without
the loss of other functions or properties. Substitutions of this kind
preferably are
conservative, that is, one amino acid is replaced with one of similar shape
and charge.
Conservative substitutions are well known in the art and include, for example,
the
changes of alanine to serine; arginine to lysine; asparagine to glutamine or
histidine;
aspartate to glutamate; cysteine to serine; glutamine to asparagine; glutamate
to
aspartate; glycine to proline; histidine to asparagine or glutamine;
isoleucine to leucine or
valine; leucine to valine or isoleucine; lysine to arginine; methionine to
leucine or
isoleucine; phenylalanine to tyrosine, leucine or methionine; serine to
threonine;
46
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
threonine to serine; tryptophan to tyrosine; tyrosine to tryptophan or
phenylalanine; and
valine to isoleucine or leucine.
The following is a discussion based upon changing of the amino acids of a
protein to create an equivalent, or even an improved, second-generation
molecule. For
example, certain amino acids may be substituted for other amino acids in a
protein
structure without appreciable loss of interactive binding capacity with
structures such as,
for example, antigen-binding regions of antibodies or binding sites on
substrate
molecules. Since it is the interactive capacity and nature of a protein that
defines that
protein's biological functional activity, certain amino acid substitutions can
be made in a
protein sequence, and its underlying DNA coding sequence, and nevertheless
obtain a
protein with like properties. It is thus contemplated by the inventors that
various changes
may be made in the DNA sequences of genes without appreciable loss of their
biological
utility or activity, as discussed below. Table 1 shows the codons that encode
particular
amino acids.
In making such changes, the hydropathic index of amino acids may be
considered. The importance of the hydropathic amino acid index in conferring
interactive
biologic function on a protein is generally understood in the art (Kyle and
Doolittle,
1982). It is accepted that the relative hydropathic character of the amino
acid
contributes to the secondary structure of the resultant protein, which in turn
defines
the interaction of the protein with other molecules, for example, enzymes,
substrates,
receptors, DNA, antibodies, antigens, and the like. .
Each amino acid has been assigned a hydropathic index on the basis of their
hydrophobicity and charge characteristics (Kyle and Doolittle, 1982), these
are:
isoleucine (+4.5); valine (+4.2); leucine (+3.8); phenylalanine (+2.8);
cysteine/cystine
(+2.5); methionine (+1.9); alanine (+1.8); glycine (-0.4); threonine (-0.7);
serine (-
0.8); tryptophan (-0.9); tyrosine (-1.3); proline (-1.6); histidine (-3.2);
glutamate (-
3.5); glutamine (-3.5); aspartate (-3.5); asparagine (-3.5); lysine (-3.9);
and arginine (-
4.5).
It is known in the art that certain amino acids may be substituted by other
amino acids having a similar hydropathic index or score and still result in a
protein
with similar biological activity, i.e., still obtain a biological functionally
equivalent
47
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
protein. In making such changes, the substitution of amino acids whose
hydropathic
indices are within ~2 is preferred, those which are within +1 are particularly
preferred,
and those within +0.5 are even more particularly preferred.
It is also understood. in the art that the substitution of like amino acids
can be
made effectively on the basis of hydrophilicity. U.S. Patent 4,554,101,
incorporated
herein by reference, states that the greatest local average hydrophilicity of
a protein, as
governed by the hydrophilicity of its adjacent amino acids, correlates with a
biological
property of the protein. As detailed in U.S. Patent 4,554,101, the following
hydrophilicity values have been assigned to amino acid residues: arginine
(+3.0);
lysine (+3.0); aspartate (+3.0 ~ 1); glutamate (+3.0 ~ 1); serine (+0.3);
asparagine
(+0.2); glutamine (+0.2); glycine (0); threonine (-0.4); proline (-0.5 ~ 1);
alanine
(-0.5); histidine *-0.5); cysteine (-1.0); methionine (-1.3); valine (-1.5);
leucine (-1.8);
isoleucine (-1.8); tyrosine (-2.3); phenylalanine (-2.5); tryptophan (-3.4).
It is understood that an amino acid can be substituted for another having a
similar hydrophilicity value and still obtain a biologically equivalent and
immunologically equivalent protein. :fn such changes, the substitution of
amino acids
whose hydrophilicity values are within ~2 is preferred, those that are within
~1 are
particularly preferred, and those within X0.5 are even more particularly
preferred.
As outlined above, amino acid substitutions are generally based on the
relative
similarity of the amino acid side-chain substituents, for example, their
hydrophobicity,
hydrophilicity, charge, size, and the like. Exemplary substitutions that take
various of
the foregoing characteristics into consideration are well known to those of
skill in the
art and include: arginine and lysine; glutamate and aspartate; serine and
threonine;
glutamine and asparagine; and valine, leucine and isoleucine.
Another embodiment for the preparation of polypeptides according to the
invention is the use of peptide mimetics. Mimetics are peptide-containing
molecules
that mimic elements of protein secondary structure (Johnson et al, 1993). The
underlying rationale behind the use of peptide mimetics is that the peptide
backbone
of proteins exists chiefly to orient amino acid side chains in such a way as
to facilitate
molecular interactions, such as those of antibody and antigen. A peptide
mimetic is
expected to permit molecular interactions similar to the natural molecule.
These
48
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
principles may be used, in conjunction with the principles outline above, to
engineer
second generation molecules having many. of the natural properties of
myocardin, but
with altered and even improved characteristics.
C. Domain Switching
Domain switching involves the generation of chimeric molecules using
different but, in this case, related polypeptides. By comparing various
myocardin
proteins, one can make predictions as to the functionally significant regions
of these
molecules. It is possible, then, to switch related domains of these molecules
in an
effort to determine the criticality of these regions to myocardin function.
These
molecules may have additional value in that these "chimeras" can be
distinguished
from natural molecules, while possibly providing the same function. In
particular, it
is contemplated that one will create chimeras between myocardins, for example,
between myocardin 1 & myocardin 2, myocardinl & myocardin 3, myocardin 2 &
myocardin 3, and/or myocardin 1, myocardin 2 & myocardin 3.
D. Fusion Proteins
A specialized kind of insertional variant is the fusion protein. This molecule
generally has all or a substantial portion of the native molecule linked, at
the N- or C-
terminus, to all or a portion of a second polypeptide. For example, fusions
typically
employ leader sequences from other species to permit the recombinant
expression of a
protein in a heterologous host. Another useful fusion includes the addition of
a
immunologically active domain, such as an antibody epitope, to facilitate
purification of
the fusion protein. Inclusion of a cleavage site at or near the fusion
junction will
facilitate removal of the extraneous polypeptide after purification. Other
useful fusions
include linking of functional domains, such as active sites from enzymes,
glycosylation
domains, cellular targeting signals or transmembrane regions.
49
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
E. Purification of Proteins
It may be desirable to purify myocardin or variants thereof. Protein
purification techniques are well known to those of skill in the art. These
techniques
involve, at one level, the crude fractionation of the cellular milieu to
polypeptide and
non-polypeptide fractions. Having separated the polypeptide from other
proteins, the
polypeptide of interest may be further purified using chromatographic and
electrophoretic techniques to achieve partial or complete purification (or
purification
to homogeneity). Analytical methods particularly suited to the preparation of
a pure
peptide are ion-exchange chromatography, exclusion chromatography;
polyacrylamide gel electrophoresis; isoelectric focusing. A particularly
efficient
method of purifying peptides is fast protein liquid chromatography or even
HPLC.
Certain aspects of the present invention concern the purification, and in
particular embodiments, the substantial purification, of an encoded protein or
peptide.
The term "purified protein or peptide" as used herein, is intended to refer to
a
composition, isolatable from other components, wherein the protein or peptide
is
purified to any degree relative to its naturally-obtainable state. A purified
protein or
peptide therefore also refers to a protein or peptide, free from the
environment in
which it may naturally occur.
Generally, "purified" will refer to a protein or peptide composition that has
been subjected to fractionation to remove various other components, and which
composition substantially retains its expressed biological activity. Where the
term
"substantially purified" is used, this designation will refer to a composition
in which
the protein or peptide forms the major component of the composition, such as
constituting about 50%, about 60%, about 70%, about 80%, about 90%, about 95%
or
more of the proteins in the composition.
Various methods for quantifying the degree of purification of the protein or
peptide will be known to those of skill in the art in light of the present
disclosure.
These include, for example, determining the specific activity of an active
fraction, or
assessing the amount of polypeptides within a fraction by SDS/PAGE analysis. A
preferred method for assessing the purity of a fraction is to calculate the
specific
activity of the fraction, to compare it to the specific activity of the
initial extract, and
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
to thus calculate the degree of purity, herein assessed by a "-fold
purification
number." The actual units used to represent the amount of activity will, of
course, be
dependent upon the particular assay technique chosen to follow the
purification and
whether or not the expressed protein or peptide exhibits a detectable
activity.
Various techniques suitable for use in protein purification will be well known
to those of skill in the art. These include, for example, precipitation with
ammonium
sulphate, PEG, antibodies and the like or by heat denaturation, followed by
centrifugation; chromatography steps such as ion exchange, gel filtration,
reverse
phase, hydroxylapatite and affinity chromatography; isoelectric focusing; gel
electrophoresis; and combinations of such and other techniques. As is
generally
known in the art, it is believed that the order of conducting the various
purification
steps may be changed, or that certain steps may be omitted, and still result
in a
suitable method for the preparation of a substantially purified protein or
peptide.
There is no general requirement that the protein or peptide always be provided
in their most purified state. Indeed, it is contemplated that less
substantially purified
products will have utility in certain embodiments. Partial purification may be
accomplished by using .fewer purification steps in combination, or by
utilizing
different forms of the same general purification scheme. For example, it is
appreciated that a canon-exchange column chromatography performed utilizing an
HPLC apparatus will generally result in a greater "-fold" purification than
the same
technique utilizing a low pressure chromatography system. Methods exhibiting a
lower degree of relative purification may have advantages in total recovery of
protein
product, or in maintaining the activity of an expressed protein.
It is known that the migration of a polypeptide can vary, sometimes
significantly, with different conditions of SDSlPAGE (Capaldi et al., 1977).
It will
therefore be appreciated that under differing electrophoresis conditions, the
apparent
molecular weights of purified or partially purified expression products may
vary.
High Performance Liquid Chromatography (HPLC) is characterized by a very
rapid separation with extraordinary resolution of peaks. This is achieved by
the use of
very fine particles and high pressure to maintain an adequate flow rate.
Separation
can be accomplished in a matter of minutes, or at most an hour. Moreover, only
a
51
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
very small volume of the sample is needed because the particles are so small
and
close-packed that the void volume is a very small fraction of the bed volume.
Also,
the concentration of the sample need not be very great because the bands are
so
narrow that there is very little dilution of the sample.
Gel chromatography, or molecular sieve chromatography, is a special type of
partition chromatography that is based on molecular size. The theory behind
gel
chromatography is that the column, which is prepared with tiny particles of an
inert
substance that contain small pores, separates larger molecules from smaller
molecules
as they pass through or around the pores, depending on their size. As long as
the
material of which the particles are made does not adsorb the molecules, the
sole factor
determining rate of flow is the size. Hence, molecules are eluted from the
column in
decreasing size, so long as the shape is relatively constant. Gel
chromatography is
unsurpassed for separating molecules of different size because separation is
independent of other factors such as pH, ionic strength, temperature, etc.
There also is
virtually no adsorption, less zone spreading and the elution volume is related
in a
simple matter to molecular weight.
Affinity Chromatography is a chromatographic procedure that relies on the
specific affinity between a substance to be isolated and a molecule that it
can
specifically bind to. This is a receptor-ligand type interaction. The column
material is
synthesized by covalently coupling one of the binding partners to an insoluble
matrix.
The column material is then able to specifically adsorb the substance from the
solution. Elution occurs by changing the conditions to those in which binding
will not
occur (alter pH, ionic strength, temperature, etc.).
A particular type of affinity chromatography useful in the purification of
carbohydrate containing compounds is lectin affinity chromatography. Lectins
are a
class of substances that bind to a variety of polysaccharides and
glycoproteins.
Lectins are usually coupled to agarose by cyanogen bromide. Conconavalin A
coupled to Sepharose was the first material of this sort to be used and has
been widely
used in the isolation of polysaccharides and glycoproteins other lectins that
have been
include lentil lectin, wheat germ agglutinin which has been useful in the
purification
of N-acetyl glucosaminyl residues and Helix pomatia lectin. Lectins themselves
are
52
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
purified using affinity chromatography with carbohydrate ligands. Lactose has
been
used to purify lectins from castor bean and peanuts; maltose has been useful
in
extracting lectins from lentils and jack bean; N-acetyl-D galactosamine is
used for
purifying lectins from soybean; N-acetyl glucosaminyl binds to lectins from
wheat
germ; D-galactosamine has been used in obtaining lectins from clams and L-
fucose
will bind to lectins from lotus.
The matrix should be a substance that itself does not adsorb molecules to any
significant extent and that has a broad range of chemical, physical and
thermal
stability. The ligand should be coupled in such a way as to not affect its
binding
properties. The ligand should also provide relatively tight binding. And it
should be
possible to elute the substance without destroying the sample or the ligand.
One of
the most common forms of affinity chromatography is immunoaffmity
chromatography. The generation of antibodies that would be suitable for use in
accord with the present invention is discussed below.
F. Synthetic Peptides
The present invention also includes smaller myocardin-related peptides for use
in various embodiments of the present invention. Because of their relatively
small
size, the peptides of the invention can also be synthesized in solution or on
a solid
support in accordance with conventional techniques. Various automatic
synthesizers
are commercially available and can be used in accordance with known protocols.
See,
for example, Stewart and Young (1984); Tam et al. (1983); Mernfield (1986);
and
Barany and Mernfield (1979), each incorporated herein by reference. Short
peptide
sequences, or libraries of overlapping peptides, usually from about 6 up to
about 35 to
50 amino acids, which correspond to the selected regions described herein, can
be
readily synthesized and then screened in screening assays designed to identify
reactive
peptides. Alternatively, recombinant DNA technology may be employed wherein a
nucleotide sequence which encodes a peptide of the invention is inserted into
an
expression vector, transformed or transfected into an appropriate host cell
and
cultivated under conditions suitable for expression.
53
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
G. Antigen Compositions
The present invention also provides for the use of myocardin proteins or
peptides as antigens for the immunization of animals relating to the
production of
antibodies. It is envisioned that myocardin or portions thereof, will be
coupled,
bonded, bound, conjugated or chemically-linked to one or more agents via
linkers,
polylinkers or derivatized amino acids. This may be performed such that a
bispecific
or multivalent composition or vaccine is produced. It is further envisioned
that the
methods used in the preparation of these compositions will be familiar to
those of skill
in the art and should be suitable for administration to animals, i.e.,
pharmaceutically
acceptable. Preferred agents are the carriers are keyhole limpet hemocyannin
(KLH)
or bovine serum albumin (BSA).
III. Generating Antibodies Reactive With Myocardin
In another aspect, the present invention contemplates an antibody that is
immunoreactive with a myocardin molecule of the present invention, or any
portion
thereof. An antibody can be a polyclonal or a monoclonal antibody. In a
preferred
embodiment, an antibody is a monoclonal antibody. Means for preparing and
characterizing antibodies are well known in the art (see, e.g., Harlow and
Lane,
1988).
Briefly, a polyclonal antibody is prepared by immunizing an animal with an
immunogen comprising a polypeptide of the present invention and collecting
antisera
from that immunized animal. A wide range of animal species can be used for the
production of antisera. Typically an animal used for production of anti-
antisera is a
non-human animal including rabbits, mice, rats, hamsters, pigs or horses.
Because of
the relatively large blood volume of rabbits, a rabbit is a preferred choice
for
production of polyclonal antibodies.
Antibodies, both polyclonal and monoclonal, specific for isoforms of antigen
may be prepared using conventional immunization techniques, as will be
generally
known to those of skill in the art. A composition containing antigenic
epitopes of the
compounds of the present invention can be used to immunize one or more
experimental animals, such as a rabbit or mouse, which will then proceed to
produce
specific antibodies against the compounds of the present invention. Polyclonal
54
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
antisera may be obtained, after allowing time for antibody generation, simply
by
bleeding the animal and preparing serum samples from the whole blood.
It is proposed that the monoclonal antibodies of the present invention will
find
useful application in standard immunochemical procedures, such as ELISA and
Western blot methods and in immunohistochemical procedures such as tissue
staining,
as well as in other procedures which may utilize antibodies specific to
myocardin-
related antigen epitopes. Additionally, it is proposed that monoclonal
antibodies
specific to the particular myocardin of different species may be utilized in
other useful
applications
In general, both polyclonal and monoclonal antibodies against myocardin may
be used in a variety of embodiments. For example, they may be employed in
antibody
cloning protocols to obtain cDNAs or genes encoding other myocardin. They may
also be used in inhibition studies to analyze the effects of myocardin-related
peptides
in cells or animals. Myocardin antibodies will also be useful in
immunolocalization
studies to analyze the distribution of myocardins during various cellular
events, for
example, to determine the cellular or tissue-specific distribution of
myocardin
polypeptides at different points in the cell cycle. A particularly useful
application of
such antibodies is in purifying native or recombinant myocardin, for example,
using
an antibody affinity column. The operation of such immunological techniques
will be
known to those of skill in the art in light of the present disclosure.
Means for preparing and characterizing antibodies are well known in the art
(see, e.g., Harlow and Lane, 1988; incorporated herein by reference). More
specific
examples of monoclonal antibody preparation are given in the examples below.
As is well known in the art, a given composition may vary in its
immunogenicity. It is often necessary therefore to boost the host immune
system, as
may be achieved by coupling a peptide or polypeptide immunogen to a carrier.
Exemplary and preferred Garners are keyhole limpet hemocyanin (KLH) and bovine
serum albumin (BSA). Other albumins such as ovalbumin, mouse serum albumin or
rabbit serum albumin can also be used as carriers. Means for conjugating a
polypeptide to a carrier protein are well known in the art and include
glutaraldehyde,
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
m-maleimidobencoyl-N-hydroxysuccinimide ester, carbodiimide and bis-biazotized
benzidine.
As also is well known in the art, the immunogenicity of a particular
immunogen composition can be enhanced by the use of non-specific stimulators
of the
immune response, known as adjuvants. Exemplary and preferred adjuvants include
complete Freund's adjuvant (a non-specific stimulator of the immune response
containing killed Mycobacterium tuberculosis), incomplete Freund's adjuvants
and
aluminum hydroxide adjuvant.
The amount of immunogen composition used in the production of polyclonal
antibodies varies upon the nature of the immunogen as well as the animal used
for
immunization. A variety of routes can be used to administer the immunogen
(subcutaneous, intramuscular, intradermal, intravenous and intraperitoneal).
The
production of polyclonal antibodies may be monitored by sampling blood of the
immunized animal at various points following immunization. A second, booster,
injection may also be given. The process of boosting and titering is repeated
until a
suitable titer is achieved. When a desired level of immunogenicity is
obtained, the
immunized animal can be bled and the serum isolated and stored, and/or the
animal
can be used to generate mAbs.
MAbs may be readily prepared through use of well-known techniques, such as
those exemplified in U.S. Patent 4,196,265, incorporated herein by reference.
Typically, this technique involves immunizing a suitable animal with a
selected
immunogen composition, e.g., a purified or partially purified MCIP protein,
polypeptide or peptide or cell expressing high levels of MCIP. The immunizing
composition is administered in a manner effective to stimulate antibody
producing
cells. Rodents such as mice and rats are preferred animals, however, the use
of rabbit,
sheep frog cells is also possible. The use of rats may provide certain
advantages
(Goding, 1986), but mice are preferred, with the BALB/c mouse being most
preferred
as this is most routinely used and generally gives a higher percentage of
stable
fusions.
56
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
IV. Immunologic Analysis
The use of antibodies of the present invention, in an ELISA assay is
contemplated. For example, anti-myocardin antibodies are immobilized onto a
selected surface, preferably a surface exhibiting a protein affinity such as
the wells of
a polystyrene microtiter plate. After washing to remove incompletely adsorbed
material, it is desirable to bind or coat the assay plate wells with a non-
specific protein
that is known to be antigenically neutral with regard to the test antisera
such as bovine
serum albumin (BSA), casein or solutions of powdered milk. This allows for
blocking
of non-specific adsorption sites on the immobilizing surface and thus reduces
the
background caused by non-specific binding of antigen onto the surface.
After binding of antibody to the well, coating with a non-reactive material to
reduce background, and washing to remove unbound material, the immobilizing
surface is contacted with the sample to be tested in a manner conducive to
immune
complex (antigen/antibody) formation.
Following formation of specific immunocomplexes between the test sample
and the bound antibody, and subsequent washing, the occurrence and even amount
of
immunocomplex formation may be determined by subjecting same to a second
antibody having specificity for myocardin or a fragment thereof that differs
from the
first antibody. Appropriate conditions preferably include diluting the sample
with
diluents such as BSA, bovine gamma globulin (BGG) and phosphate buffered
saline
(PBS)/Tweeri . These added agents also tend to assist in the reduction of
nonspecific
background. The layered antisera is then allowed to incubate for from about 2
to
about 4 hr, at temperatures preferably on the order of about 25° to
about 27°C.
Following incubation, the antisera-contacted surface is washed so as to remove
non-
immunocomplexed material. A preferred washing procedure includes washing with
a
solution such as PBS/Tween~, or borate buffer.
To provide a detecting means, the second antibody will preferably have an
associated enzyme that will generate a color development upon incubating with
an
appropriate chromogenic substrate. Thus, for example, one will desire to
contact and
incubate the second antibody-bound surface with a urease or peroxidase-
conjugated
anti-human IgG for a period of time and under conditions which favor the
57
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
development of immunocomplex formation (e.g., incubation for 2 hr at room
temperature in a PBS-containing solution such as PBS/Tween°).
After incubation with the second enzyme-tagged antibody, and subsequent to
washing to remove unbound material, the amount of label is quantified by
incubation
with a chromogenic substrate such as urea and bromocresol purple or 2,2'-azino-
di-(3-
ethyl-benzthiazoline)-6-sulfonic acid (ABTS) and HZOz, in the case of
peroxidase as
the enzyme label. Quantitation is then achieved by measuring the degree of
color
generation, e.g., using a visible spectrum spectrophotometer.
The preceding format may be altered by first binding the sample to the assay
plate. Then, primary antibody is incubated with the assay plate, followed by
detecting
of bound primary antibody using a labeled second antibody with specificity for
the
primary antibody.
The antibody compositions of the present invention will find great use in
immunoblot or Western blot analysis. The antibodies may be used as high-
affinity
primary reagents for the identification of proteins immobilized onto a solid
support
matrix; such as nitrocellulose, nylon or combinations thereof. In conjunction
with
immunoprecipitation, followed by gel electrophoresis, these may be used as a
single
step reagent for use in detecting antigens against which secondary reagents
used in the
detection of the antigen cause an adverse background. Immunologically-based
detection methods for use in conjunction with Western blotting include
enzymatically-
radiolabel-, or fluorescently-tagged secondary antibodies against the toxin
moiety are
considered to be of particular use in this regard.
V. Cardiomyocyte Regeneration
The present invention also involves, in another embodiment, the treatment of
the loss of cardiomycoytes, for example due to myocardial infarction. In
particular,
myocardin plays a role in cardiac myocyte development. Thus, increasing levels
of
myocardin in non-cardiomyocyte target cells can be used to modulate the
phenotype
of such target cells such that it includes one or more functions of
cardiomyocytes,
whereas decreasing the levels of myocardin activity in cardiomyocytes can be
used to
reduce or inhibit one or more cardiomyocyte functionalities and promote cell
growth.
58
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
One of the therapeutic embodiments contemplated by the present inventors is
the intervention, at the molecular level, in the events involved in
cardiomyocyte
development. Specifically, the present inventors intend to provide, to a non-
cardiomyocyte target cell, for example a cardiac fibroblast cell, an
expression
construct capable of providing myocardin to that cell. The lengthy discussions
of
gene delivery means, expression vectors and the genetic elements employed
therein
are incorporated into this section by reference. Exemplary gene delivery
vectors are
viral vectors such as adenovirus, adeno-associated virus, herpesvirus,
vaccinia virus,
lentivirus and retrovirus, as well as lipid-based vectors.
A. Gene Therapy
One skilled in the art recognizes that various methods of DNA delivery may be
employed to deliver the polynucleotides of the present invention to specific
cells for
gene therapy. Further, in the context of gene therapy, a skilled artisan is
cognizant
that the vector to be utilized will generally contain the gene of interest
operatively
linked to a promoter. One skilled in the art also recognizes that, in certain
instances,
other sequences such as a 5' and/or 3'-UTR regulatory sequences are useful in
expressing the gene of interest.
Where appropriate, the gene therapy vectors can be formulated into
preparations in solid, semisolid, liquid or gaseous forms in the ways known in
the art
for their respective route of administration. Means known in the art can be
utilized to
prevent release and absorption of the composition until it reaches the target
organ or
to ensure timed release of the composition. Alternatively or additionally, the
composition may be targeted by the delivery itself, for example by
intracoronary
delivery to target the heart (see e.g. U.S. Patents 5,792,453 and 6,100,242,
hereby
incorporated by reference in their entirety). A pharmaceutically acceptable
form
should be employed which does not deactivate the compositions of the present
invention. In pharmaceutical dosage forms, the compositions can be used alone
or in
appropriate association, as well as in combination, with other
pharmaceutically active
compounds. Preferably, a sufficient amount of vector containing the
therapeutice
nucleic acid sequence is administered to provide a pharmacologically effective
dose of
the gene product, for example to alleviate symptoms associated with the
disease being
treated.
59
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
One skilled in the art recognizes that other methods o.f delivery may likewise
be utilized to administer an expression cassette into a cell. Examples
include: (1)
methods utilizing physical means, such as electroporation (electricity), a
gene gun
(physical force) or applying large volumes of a liquid (pressure); and (2)
methods
wherein said expression cassette is complexed with another entity, such as a
lipid-
based vector (e.g., a liposome), an aggregated protein or a transporter
molecule.
Certain of these embodiments are primarily suitable for ex vivo applications.
The actual dose and schedule can vary, for example, depending on whether the
compositions are administered in combination with other pharmaceutical
compositions, or depending on inter-individual differences in
pharmacokinetics, drug
disposition, and metabolism. Similarly, amounts to be administered can vary in
in
vitro applications, for example depending on the particular cell line utilized
(e.g.,
based on the variable number and/or type of vector receptors present on the
cell
surface, or the ability of the particular vector employed for gene transfer to
replicate in
that cell line). Furthermore, the amount of vector to be added per cell will
likely vary
with the length and stability of the therapeutic gene inserted in the vector,
as well as
the nature of the sequence itself. Thus, vector amount is particularly a
parameter
which is preferably determined empirically and can be altered due to factors
not
inherent to the present invention (for instance, the cost associated with
synthesis).
One skilled in the art can easily make adjustments to dose in accordance with
the
exigencies of the particular situation.
Those of skill in the art are well aware of how to apply gene delivery to in
vivo
situations. By way of illustration, for viral vectors, one generally will
prepare a viral
vector stock. Depending on the type of virus utilized and the titer
attainable, one will
generally deliver 1 X 104, 1 X 105, 1 X 10~, 1 X 10', 1 X 10g, 1 X 109, 1 X
101°, 1 X
1011, 1 X 1012 to 103 infectious particles to the patient. Similar figures may
be
extrapolated for lipid-based or other non-viral formulations by comparing
relative
uptake efficiencies. Formulation as a pharmaceutically acceptable composition
is
discussed further below. Various routes are contemplated, but local provision
to the
heart, preferably by the method of Hammond, et al., supra and intra-arterial
or
intravenous administration are preferred.
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
In another embodiment, it is contemplated that blocking myocardin activity
may result in stimulation of the cardiomycytes to divide. This may be
accomplished
in one of several ways. First, one may provide an analog of myocardin's target
that
binds and inhibits myocardin function, effectively creating a "suicide
substrate" for
myocardin. This approach also could be exploited using a mimetic (see above).
Second, one could use a similar peptide target, with an additional domain
actually
capable of cleaving myocardin. Third, one could provide a non-functional
myocardin
analog that is capable of competing with myocardin peptide. And fourth,
antisense or
ribozyme techniques could also be used to inhibit the expression of myocardin.
B. Combined Therapy
In another embodiment, it is envisioned to use myocardin in combination with
other therapeutic modalities. For example, it is known that myocardin
interacts with
other transcription factors. Thus, the present invention further contemplates
the
provision of myocardin in conjunction with one or more transcription factors,
and in
particular, one or more cardiac transcription factors. Examples of cardiac
transcription factors include, but are not limited to, GATA4, serum response
factor
(SRF) and Nkx2.5.
In other embodiments, in addition to the therapies described above, one may
also provide to the patient more "standard" pharmaceutical cardiac therapies.
Examples of standard therapies include, without limitation, so-called "beta
Mockers",
anti-hypertensives, cardiotonics, anti-thrombotics, vasodilators, hormone
antagonists,
endothelin antagonists, calcium channel blockers, phosphodiesterase
inhibitors,
angiotensin type 2 antagonists and cytokine blockers/inhibitors.
Combinations may be achieved by contacting cardiac cells with a single
composition or pharmacological formulation that includes both agents, or by
contacting the cell with two distinct compositions or formulations, at the
same time,
wherein one composition includes the expression construct and the other
includes the
agent. Alternatively, gene therapy may precede or follow administration of the
other
agent by intervals ranging from minutes to weeks. In embodiments where the
other
agent and expression construct are applied separately to the cell, one would
generally
ensure that a significant period of time did not expire between the time of
each
61
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
delivery, such that the agent and expression construct would still be able to
exert an
advantageously combined effect on the cell. In such instances, it is
contemplated that
one would typically contact the cell with both modalities within about 12-24
hours of
each other and, more preferably, within about 6-12 hours of each other, with a
delay
time of only about 12 hours being most preferred. In some situations, it may
be
desirable to extend the time period for treatment significantly, however,
where several
days (2, 3, 4, 5, 6 or 7) to several weeks (1, 2, 3, 4, 5, 6, 7 or 8) lapse
between the
respective administrations.
It also is conceivable that more than one administration of either a myocardin
gene, or the other agent will be desired. In this regard, various combinations
may be
employed. By way of illustration, where myocardin is "A" and the other agent
or
cardiac transcription factor is "B," the following permutations based on 3 and
4 total
administrations are exemplary:
ABlA BIAS BB/A A/AB B/A/A ABB BBB/A BBlAB
A/A/BB AB/AB ABBlA BB/A/A B/A/B/A B/A/AB BBB/A
A/A/AB B/A/A/A AB/A/A A/A/B/A ABBB B/A/BB BBlAB
Other combinations are likewise contemplated.
VII. Drug Formulations and Routes for Administration to Patients
Where clinical applications are contemplated, pharmaceutical compositions
will be prepared - e.g., expression vectors, virus stocks and drugs - in a
form
appropriate for the intended application. Generally, this will entail
preparing
compositions that are essentially free of pyrogens, as well as other
impurities that
could be harmful to humans or animals.
One will generally desire to employ appropriate salts and buffers to render
delivery vectors stable and allow for uptake by target cells. Buffers also
will be
employed when recombinant cells are introduced into a patient. Aqueous
compositions of the present invention comprise an effective amount of the
vector or
cells, dissolved or dispersed in a pharmaceutically acceptable carrier or
aqueous
medium. The phrase "pharmaceutically or pharmacologically acceptable" refer to
62
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
molecular entities and compositions that do not produce adverse, allergic, or
other
untoward reactions when administered to an animal or a human. As used herein,
"pharmaceutically acceptable carrier" includes solvents, buffers, solutions,
dispersion
media, coatings, antibacterial and antifungal agents, isotonic and absorption
delaying
agents and the like acceptable for use in formulating pharmaceuticals, such as
pharmaceuticals suitable for administration to humans. The use of such media
and
agents for pharmaceutically active substances is well know in the art. Except
insofar
as any conventional media or agent is incompatible with the active ingredients
of the
present invention, its use in therapeutic compositions is contemplated.
Supplementary
active ingredients also can be incorporated into the compositions, provided
they do
not inactivate the vectors or cells of the compositions.
The active compositions of the present invention may include classic
pharmaceutical preparations. Administration of these compositions according to
the
present invention may be via any common route so long as the target tissue is
available via that route. This includes oral, nasal, buccal, rectal, vaginal
or topical.
Alternatively, administration may be by orthotopic, intradermal, subcutaneous,
intramuscular, intraperitoneal or intravenous injection. Such compositions
would
normally be administered as pharmaceutically acceptable compositions, as
described
supra.
The active compounds may also be administered parenterally or
intraperitoneally. By way of illustration, solutions of the active compounds
as free
base or pharmacologically acceptable salts can be prepared in water suitably
mixed
with a surfactant, such as hydroxypropylcellulose. Dispersions can also be
prepared
in glycerol, liquid polyethylene glycols, and mixtures thereof and in oils.
Under
ordinary conditions of storage and use, these preparations generally contain a
preservative to prevent the growth of microorganisms.
The pharmaceutical forms suitable for injectable use include, for example,
sterile aqueous solutions or dispersions and sterile powders for the
extemporaneous
preparation of sterile injectable solutions or dispersions. Generally, these
preparations
are sterile and fluid to the extent that easy syringability exists.
Preparations should be
stable under the conditions of manufacture and storage and should be preserved
63
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
against the contaminating action of microorganisms, such as bacteria and
fungi.
Appropriate solvents or dispersion media may contain, for example, water,
ethanol,
polyol (for example, glycerol, propylene glycol, and liquid polyethylene
glycol, and
the like), suitable mixtures thereof, and vegetable oils. The proper fluidity
can be
maintained, for example, by the use of a coating, such as lecithin, by the
maintenance
of the required particle size in the case of dispersion and by the use of
surfactants.
The prevention of the action of microorganisms can be brought about by various
antibacterial an antifungal agents, for example, parabens, chlorobutanol,
phenol,
sorbic acid, thimerosal, and the like. In many cases, it will be preferable to
include
isotonic agents, for example, sugars or sodium chloride. Prolonged absorption
of the
injectable compositions can be brought about by the use in the compositions of
agents
delaying absorption, for example, aluminum monostearate and gelatin.
Sterile injectable solutions may be prepared by incorporating the active
compounds in an appropriate amount into a solvent along with any other
ingredients
(for example as enumerated above) as desired, followed by filtered
sterilization.
Generally, dispersions are prepared by incorporating the various sterilized
active
ingredients into a sterile vehicle which contains the basic dispersion medium
and the
desired other ingredients, e.g., as enumerated above. In the case of sterile
powders for
the preparation of sterile injectable solutions, the preferred methods of
preparation
include vacuum-drying and freeze-drying techniques which yield a powder of the
active ingredients) plus any additional desired ingredient from a previously
sterile-
filtered solution thereof.
For oral administration the polypeptides of the present invention generally
may be incorporated with excipients and used in the form of non-ingestible
mouthwashes and dentifrices. A mouthwash may be prepared incorporating the
active
ingredient in the required amount in an appropriate solvent, such as a sodium
borate
solution (Dobell's Solution). Alternatively, the active ingredient may be
incorporated
into an antiseptic wash containing sodium borate, glycerin and potassium
bicarbonate.
The active ingredient may also be dispersed in dentifrices, including: gels,
pastes,
powders and slurnes. The active ingredient may be added in a therapeutically
effective amount to a paste dentifrice that may include water, binders,
abrasives,
flavoring agents, foaming agents, and humectants.
64
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
The compositions of the present invention generally may be formulated in a
neutral or salt form. Pharmaceutically-acceptable salts include, for example,
acid
addition salts (formed with the free amino groups of the protein) derived from
inorganic acids (e.g., hydrochloric or phosphoric acids, or from organic acids
(e.g.,
acetic, oxalic, tartaric, mandelic, and the like. Salts formed with the free
carboxyl
groups of the protein can also be derived from inorganic bases (e.g., sodium,
potassium, ammonium, calcium, or ferric hydroxides) or from organic bases
(e.g.,
isopropylamine, trimethylamine, histidine, procaine and the like.
Upon formulation, solutions are preferably administered in a manner
compatible with the dosage formulation and in such amount as is
therapeutically
effective. The formulations may easily be administered in a variety of dosage
forms
such as injectable solutions, drug release capsules and the like. For
parenteral
administration in an aqueous solution, for example, the solution generally is
suitably
buffered and the liquid diluent first rendered isotonic for example with
sufficient
saline or glucose. Such aqueous solutions may be used, for example, for
intravenous,
intramuscular, subcutaneous and intraperitoneal administration. Preferably,
sterile
aqueous media are employed as is known to those of skill in the art,
particularly in
light of the present disclosure. By way of illustration, a single dose may be
dissolved
in 1 ml of isotonic NaCI solution and either added to 1000 ml of
hypodermoclysis
fluid or injected at the proposed site of infusion, (see for example,
"Remington's
Pharmaceutical Sciences" 15th Edition, pages 1035-1038 and 1570-1580). Some
variation in dosage will necessarily occur depending on the condition of the
subject
being treated. The person responsible for administration will, in any event,
determine
the appropriate dose for the individual subject. Moreover, for human
administration,
preparations should meet sterility, pyrogenicity, general safety and purity
standards as
required by FDA Office of Biologics standards.
VIII. Methods of Making Transgenic Mice
A particular embodiment of the present invention provides transgenic animals
that contain myocardin-related constructs. Transgenic. animals expressing
myocardin,
recombinant cell lines derived from such animals, and transgenic embryos may
be
useful in determining the exact role that myocardin plays in the development
and
differentiation of cardiomyocytes. Furthermore, this transgenic animal may
provide
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
an insight into heart development. The use o.f constitutively expressed
myocardins
provides a model for over- or unregulated expression. Also, transgenic animals
which
are "knocked out" for myocardin, in one or both alleles are contemplated.
In a general aspect, a transgenic animal is produced by the integration of a
given transgene into the genome in a manner that permits the expression of the
transgene. Methods for producing transgenic animals are generally described by
Wagner and Hoppe (U.S. Patent 4,873,191; which is incorporated herein by
reference), Brinster et al., 1985; which is incorporated herein by reference
in its
entirety) and in Hogan et al. (1994).
Typically, a gene flanked by genomic sequences is transferred by
microinjection into a fertilized egg. The microinjected eggs are implanted
into a host
female, and the progeny are screened for the expression of the transgene.
Transgenic
animals may be produced from the fertilized eggs from a number of animals
including, but not limited to reptiles, amphibians, birds, mammals, and fish.
DNA clones for microinjection can be prepared by any means known in the
art. For example, DNA clones for microinjection can be cleaved with enzymes
appropriate for removing the bacterial plasmid sequences, and the DNA
fragments
electrophoresed on 1 % agarose gels in TBE buffer, using standard techniques.
The
DNA bands are visualized by staining with ethidium bromide, and the band
containing
the expression sequences is excised. The excised band is then placed in
dialysis bags
containing 0.3 M sodium acetate, pH 7Ø DNA is electroeluted into the
dialysis bags,
extracted with a 1:l phenol:chloroform solution and precipitated by two
volumes of
ethanol. The DNA is redissolved in 1 ml of low salt buffer (0.2 M NaCI, 20 mM
Tris,pH 7.4, and 1 mM EDTA) and purified on an Elutip-DTM column. The column
is
first primed with 3 ml of high salt buffer (1 M NaCI, 20 mM Tris, pH 7.4, and
1 mM
EDTA) followed by washing with 5 ml of low salt buffer. The DNA solutions are
passed through the column three times to bind DNA to the column matrix. After
one
wash with 3 ml of low salt buffer, the DNA is eluted with 0.4 ml high salt
buffer and
precipitated by two volumes of ethanol. DNA concentrations are measured by
absorption at 260 nm in a UV spectrophotometer. For microinjection, DNA
concentrations are adjusted to 3 p,g/ml in 5 mM Tris, pH 7.4 and 0.1 mM EDTA.
66
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Other methods for purification of DNA for microinjection are described in
Hogan et
al. (1986), in Palmiter et al. (1982); and in Sambrook et al. (1989).
In an exemplary microinjection procedure, female mice six weeks of age are
induced to superovulate with a S IU injection (0.1 cc, ip) of pregnant mare
serum
gonadotropin (PMSG; Sigma) followed 48 hours later by a 5 ILT injection (0.1
cc, ip)
of human chorionic gonadotropin (hCG; Sigma). Females are placed with males
immediately after hCG injection. Twenty-one hours after hCG injection, the
mated
females are sacrificed by COZ asphyxiation or cervical dislocation and embryos
are
recovered from excised oviducts and placed in Dulbecco's phosphate buffered
saline
with 0.5% bovine serum albumin (BSA; Sigma). Surrounding cumulus cells are
removed with hyaluronidase (1 mg/ml). Pronuclear embryos are then washed and
placed in Earle's balanced salt solution containing 0.5 % BSA (EBSS) in a
37.5°C
incubator with a humidified atmosphere at 5% C02, 95% air until the time of
injection. Embryos can be implanted at the two-cell stage.
Randomly cycling adult female mice are paired with vasectomized males.
C57BL/6 or Swiss mice or other comparable strains can be used for this
purpose.
Recipient females are mated at the same time as donor females. At the time of
embryo transfer, the recipient females are anesthetized with an
intraperitoneal
injection of 0.015 ml of 2.5 % avertin per gram of body weight. The oviducts
are
exposed by a single midline dorsal incision. An incision is then made through
the
body wall directly over the oviduct. The ovarian bursa is then torn with
watchmakers
forceps. Embryos to be transferred are placed in DPBS (Dulbecco's phosphate
buffered saline) and in the tip of a transfer pipet (about 10 to 12 embryos).
The pipet
tip is inserted into the infundibulum and the embryos transferred. After the
transfer,
the incision is closed by two sutures.
67
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
IX. Screening Assays
The present invention also contemplates the screening of compounds for
various abilities to interact with and/or affect myocardin expression or
function.
Particularly preferred compounds will be those useful in inhibiting or
promoting the
actions of myocardin in regulating the development and differentiation of
cardiomyocytes. In the screening assays of the present invention, the
candidate
substance may first be screened for basic biochemical activity -- e.g.,
binding to a
target molecule -- and then tested for its ability to inhibit modulate
activity, at the
cellular, tissue or whole animal level.
A. Modulators and Assay Formats
i) Assay Formats
The present invention provides methods of screening for modulators of
myocardin expression and binding to other proteins or nucleic acids. In one
embodiment, the present invention is directed to a method of:
(a) providing a myocardin polypeptide;
(b) contacting the myocai-din polypeptide with the candidate substance;
and
(c) determining the binding of the candidate substance to myocardin
polypeptide.
In yet another embodiment, the assay looks not at binding, but at myocardin
function. Such methods would comprise, for example:
(a) providing myocardin and DATA to a cell;
(b) admixing myocardin and GATA in the presence of a candidate
modulator; and
(c) measuring the effect of the candidate substance on the expression of a
cardiac cell gene product.
68
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
A related assay that examines the interaction of myocardin and GATA would
comprise:
(a) providing myocardin and GATA to a cell;
(b) admixing myocardin and GATA in the presence of a candidate
substance; and
(c) measuring the effect of the candidate substance on the interaction of
myocardin and GATA.
Both of the preceding assays could be performed substituting SRF or Nkx2.5 for
GATA.
In still yet other embodiments, one would look at the effect of a candidate
substance on the expression of myocardin. This can be done by examining mRNA
expression, although alterations in mRNA stability and translation would not
be
accounted for. A more direct way of assessing expression is by directly
examining
protein levels, for example, through Western blot or ELISA.
ii) Inhibitors and Activators
An inhibitor according to the present invention may be one which exerts an
inhibitory effect on the expression or function of myocardin. By the same
token, an
activator according to the present invention may be one which exerts a
stimulatory
effect on the expression or function of myocardin.
69
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
iii) Candidate Substances
As used herein, the term "candidate substance" refers to any molecule that may
potentially modulate myocardin expression or function. The candidate substance
may
be a protein or fragment thereof, a small molecule inhibitor, or even a
nucleic acid
molecule. It may prove to be the case that the most useful pharmacological
compounds will be compounds that are structurally related to compounds which
interact naturally with myocardin. Creating and examining the action of such
molecules is known as "rational drug design," and include making predictions
relating
to the structure of target molecules.
The goal of rational drug design is to produce structural analogs of
biologically active polypeptides or target compounds. By creating such
analogs, it is
possible to fashion drugs which are more active or stable than the natural
molecules,
which have different susceptibility to alteration or which may affect the
function of
various other molecules. In one approach, one would generate a three-
dimensional
structure for a molecule like myocardin, and then design a molecule for its
ability to
interact with myocardin. Alternatively, one could design a partially
functional
fragment of myocardin (binding, but no activity), thereby creating a
competitive
inhibitor. This could be accomplished by x-ray crystallography, computer
modeling
or by a combination of both approaches.
It also is possible to use antibodies to ascertain the structure of a target
compound or inhibitor. In principle, this approach yields a pharmacore upon
which
subsequent drug design can be based. It is possible to bypass protein
crystallography
altogether by generating anti-idiotypic antibodies to a functional,
pharmacologically
active antibody. As a mirror image of a mirror image, the binding site of anti-
idiotype
would be expected to be an analog of the original antigen. The anti-idiotype
could
then be used to identify and isolate peptides from banks of chemically- or
biologically-produced peptides. Selected peptides would then serve as the
pharmacore. Anti-idiotypes may be generated using the methods described herein
for
producing antibodies, using an antibody as the antigen.
On the other hand, one may simply acquire, from various commercial sources,
small molecule libraries that are believed to meet the basic criteria for
useful drugs in
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
an effort to "brute force" the identification of useful compounds. Screening
of such
libraries, including combinatorially generated libraries (e.g., peptide
libraries), is a
rapid and efficient way to screen large number of related (and unrelated)
compounds
for activity. Combinatorial approaches also lend themselves to rapid evolution
of
potential drugs by the creation of second, third and fourth generation
compounds
modeled of active, but otherwise undesirable compounds.
Candidate compounds may include fragments or parts of naturally-occurring
compounds or may be found as active combinations of known compounds which are
otherwise inactive. It is proposed that compounds isolated from natural
sources, such
as animals, bacteria, fungi, plant sources, including leaves and bark, and
marine
samples may be assayed as candidates for the presence of potentially useful
pharmaceutical agents. It will be understood that the pharmaceutical agents to
be
screened could also be derived or synthesized from chemical compositions or
man-
made compounds. Thus, it is understood that the candidate substance identified
by the
present invention may be polypeptide, polynucleotide, small molecule
inhibitors or
any other compounds that may be designed through rational drug design starting
from
known inhibitors of hypertrophic response.
Other suitable inhibitors include antisense molecules, ribozymes, and
antibodies (including single chain antibodies).
It will, of course, be understood that the screening methods of the present
invention are useful in themselves notwithstanding the fact that effective
candidates
may not be found. The invention provides methods for screening for such
candidates,
not solely methods of finding them.
B. In Vitro Assays
A quick, inexpensive and easy assay to run is a binding assay. Binding of a
molecule to a target may, in and of itself, be inhibitory, due to steric,
allosteric or
charge-charge interactions. This can be performed in solution or on a solid
phase and
can be utilized as a first round screen to rapidly eliminate certain compounds
before
moving into more sophisticated screening assays. In one embodiment of this
kind, the
71
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
screening of compounds that bind to a myocardin molecule or fragment thereof
is
provided
The target may be either free in solution, fixed to a support, expressed in or
on
the surface of a cell. Either the target or the. compound may be labeled,
thereby
permitting determining of binding. In another embodiment, the assay may
measure
the inhibition of binding of a target to a natural or artificial substrate or
binding
partner (such as myocardin). Competitive binding assays can be performed in
which
one of the agents (myocardin for example) is labeled. Usually, the target will
be the
labeled species, decreasing the chance that the labeling will interfere with
the binding
moiety's function. One may measure the amount of free label versus bound label
to
determine binding or inhibition of binding.
A technique for high throughput screening of compounds is described in WO
84/03564. Large numbers of small peptide test compounds are synthesized on a
solid
substrate, such as plastic pins or some other surface. The peptide test
compounds are
reacted with, for example, myocardin and washed. Bound pelypeptide is detected
by
various methods.
Purified target, such as myocardin, can be coated directly onto plates for use
in
the aforementioned drug screening techniques. However, non-neutralizing
antibodies
to the polypeptide can be used to immobilize the polypeptide to a solid phase.
Also,
fusion proteins containing a reactive region (preferably a terminal region)
may be
used to link an active region (e.g., the C-terminus of myocardin) to a solid
phase.
C. In Cyto Assays
Various cell lines that express myocardin can be utilized for screening of
candidate substances. For example, cells containing myocardin with an
engineered
indicators can be used to study various functional attributes of candidate
compounds.
In such assays, the compound would be formulated appropriately, given its
biochemical nature, and contacted with a target cell.
Depending on the assay, culture may be required. As discussed above, the cell
may then be examined by virtue of a number of different physiologic assays
(growth,
size, Ca++ effects). Alternatively, molecular analysis may be performed in
which the
72
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
function of myocardin and related pathways may be explored. This involves
assays
such as those for protein expression, enzyme function, substrate utilization,
mRNA
expression (including differential display of whole cell or polyA RNA) and
others.
D. In Yivo Assays
The present invention particularly contemplates the use of various animal
models. Transgenic animals may be created with constructs that permit
myocardin
expression and activity to be controlled and monitored. The generation of
these
animals has been described elsewhere in this document.
Treatment of these animals with test compounds will involve the
administration of the compound, in an appropriate form, to the animal.
Administration will be by any route the could be utilized for clinical or non-
clinical
purposes, including but not limited to oral, nasal, buccal, or even topical.
Alternatively, administration may be by intratracheal instillation, bronchial
instillation, intradermal, subcutaneous, intramuscular, intraperitoneal or
intravenous
injection. Specifically contemplated are systemic intravenous injection,
regional
administration via blood or lymph supply.
E. Production of Inhibitors
In an extension of any of the previously described screening assays, the
present invention also provide for methods of producing inhibitors. The
methods
comprising any of the preceding screening steps followed by an additional step
of
"producing the candidate substance identified as a modulator of ' the screened
activity.
X. Examples
The following examples are included to further illustrate various aspects of
the
invention. It should be appreciated by those of skill in the art that the
techniques
disclosed in the examples which follow represent techniques and/or
compositions
discovered by the inventor to function well in the practice of the invention,
and thus
can be considered to constitute preferred modes for its practice. However,
those of
skill in the art should, in light of the present disclosure, appreciate that
many changes
can be made in the specific embodiments which are disclosed and still obtain a
like or
similar result without departing from the spirit and scope of the invention.
73
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
EXAMPLE 1
Expression Pattern of Myocardin 1 During Early Heart Development
The heart is the first organ to form during mammalian development. Cardiac
muscle cells originate from a region of the embryo known as the cardiac
crescent and
develop into a primitive heart tube along the midline of the embryo (FIG. 1).
Because
this is the only region of the embryo that can give rise to cardiac muscle
cells, it
would be expected to express a unique set of genes responsible for
cardiogenesis. By
identifying the genes that are uniquely expressed in this region, master
control genes
for heart formation can be identified. Subsequent embryonic events of looping,
chamber maturation and alignment with the vascular system give rise to the
mature
four-chambered heart (Olson et al., 1996 and Fishman et al., 1997).
Expression of myocardin 1 was determined by whole-mount (FIG. 3A) or
section (FIG. 3B and FIG. 3C) in situ hybridizations to mouse embryos at E7.75
(FIG.
3A), E8.0 (FIG. 3B), and E12.5 (FIG. 3C).
In situ hybridization to cellular RNA was performed using standard techniques
well known in the art, e.g., fluorescence in situ hybridization (FISH).
Briefly, the
samples were fixed for the appropriate time and dehydrated through a graded
ethanol
series. The samples were then impregnated in paraffin wax and cast into
blocks. The
samples were sectioned on a microtome. A specific labeled probe was prepared.
The
probe can be labeled with biotin or digoxigenin or with a fluorochrome-tagged
deoxynucleotide. Next, the probe was hybrized to the sample. Hybridization
conditions may vary with the different labeled probes. After the
hybridization,
samples were washed for 15 min in 37°C 50% formamide/2 x SSC, 15 min in
37°C 2
x SSC and 15 min in room temperature 1 x SSC. The slides were equilibrated for
5
min in 4 x SSC at room temperature. The slides were drained and allowed to air
dray.
Next, a detection solution was added. After a 45 min incubation in the
detection
solution, the slides were washed. A counterstain of DAPI or propidium iodide
staining solution was added to the slide. The slide was viewed using a
fluorescence
microscope.
74
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
The results in FIG. 3 illustrate the expression pattern of myocardin 1 during
early heart development. In FIG. 3A, myocardin 1 transcripts can be seen
localized to
the cardiac crescent. In FIG. 3B, transcripts are present throughout the heart
tube in a
transverse section. In FIG. 3C, transcripts are seen throughout the developing
heart in
a sagittal section.
As might be expected given its role in embryonic development, myocardin has
also been shown to be expressed in a subset of embryonic vascular and visceral
smooth muscle cells. At E13.5, myocardin expression was evident within smooth
muscle cells lining the walls of the esophagus and aortic arch arteries, as
well as the
pulmonary outflow tract. Expression in these smooth muscle cell types was
still
apparent, but was diminished, by E15.5. Myocardin expression was also detected
in
smooth muscle cells within the lung and gut, as well as in head mesenchyme,
which
may serve as a source of smooth muscle precursors. Myocardin was not expressed
at
detectable levels in skeletal muscle.
EXAMPLE 2
Expression Pattern of Myocardin 1 in Adult 'Mouse Tissues
The expression of myocardin 1 transcripts in adult mouse tissues was analyzed
by Northern blot, utilizing techniques well known in the art. RNA was isolated
from
adult mouse heart, brain, spleen, lung, liver, skeletal muscle, kidney and
testis
according to standard RNA isolation procedures, e.g.,
phenol/chloroform/isoamyl
alcohol (RNAzoI, Life Technologies, Inc.) or guanidine thiocyanate.
Briefly, fractionated RNA was transferred from an agarose gel to a membrane
by upward capillary action. The transferred RNA is cross-linked to the
membrane.
Next, a radiolabeled probe (DNA or RNA) was hybridized to the membrane in a
formamide solution. After hybridization, autoradiography was performed to
detect
the transcripts.
The results in FIG. 4 show that the transcripts were detected only in the
heart.
A RNA molecular marker illustrates the size of the transcripts.
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
EXAMPLE 3
Nuclear Localization of Myocardin 1 Protein
Cos cells were transiently transfected with an expression vector encoding
myocardin 1 with a Flag-epitope tag. Transient transfection assays were
performed
using standard methods, such as LifofectAMINE plus (Life Technologies, Inc.),
calcium phosphate or electroporation. Briefly, the cells were plated 12 hr
before
transfection in tissue culture dishes. They were transfected with a total of
about 0.5-
1.0 ~g of plasmid DNA. The subcellular location of myocardin 1 protein was
determined by immunostaining with anti-Flag antibody.
All myocardin 1 protein is localized to the nucleus as illustrated in FIG. 5.
The inset in the lower right corner shows an enlargement of a single cell,
with strong
myocardin 1 staining in the nucleus, but excluded from the nucleoli.
EXAMPLE 4
Structure of Myocardin 1 and Mapping of Transcription Activation Domains
Portions of myocardin 1 were fused to the DNA binding domain of yeast
GAL4 and tested in transfected Cos cells for transcriptional activity against
a GAL4-
dependent luciferase reporter. The relative transcriptional activities of
different
myocardin 1 fragments are show in FIG. 6.
The nuclear localization sequence (NLS) is located between residues 245 and
254, within a basic region at residues 243-260. A glutamine-rich (Q) domain is
located between residues 287-320. The SAP domain is found at residues 380-414.
The transcription activation domain is located at the carboxyl-terminus at 670
to 935.
The carboxyl-terminus is an extremely potent transcription activation domain,
able to
activate the reporter over 1000-fold, to a level comparable to that of the
powerful viral
coactivator VP16 (not shown).
As might be expected, given its apparent influence on transcription, myocardin
contains an SAP domain (named for nuclear scaffold attachment factors A and
B), as
found in a variety of proteins that affect not only transcription but also
nuclear
architecture. The SAP domain is a 35 amino acid motif containing two predicted
amphipathic helices separated by an intervening region with an invariant
glycine
76
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
residue. Functional aspects of the SAP domain were examined by introducing
proline
mutations into helix-1 or -2. These mutations had only a modest effect on the
ability
of myocardin to transactivate the SM22 promoter (which transactivtation is
discussed
further below). Similarly, the deletion of the linker region between the two
helices of
the SAP domain, shown previously to be required for DNA binding by SAF-A, had
little effect on SM22 activation, but eliminated ANF activation (discussed
below).
EXAMPLE 5
Traps-Activation of the SM22 Promoter by Myocardin
Cos cells were transiently transfected with a luciferase reporter gene
containing the 1.4 kb SM22 promoter and expression vectors encoding myocardin
1
and SRF. Briefly, the cells were, plated 12 hr before transfection in tissue
culture
dishes. They were transfected with plasmid DNA. Forty eight hr after
transfection,
the cells were harvested. Luciferase assays of whole cell extracts were
conducted by
standard methods well known in the art.
FIG. 7A shows activity of the wild-type SM22 promoter, which was
transactivated about 100-fold by myocardin. FIG. 7B shows activity of the SM22
promoter with a mutation in the distal CArG box (CArG-far). This promoter was
also
activated by myocardin, but not to the same extent as the wild-type promoter.
FIG.
7C shows activity of the SM22 promoter with a mutation in the proximal CArG
box
(CArG-near). This promoter has lost almost all responsiveness to myocardin, as
has
the promoter with both the CArG boxes mutated, (FIG. 7D). Both myocardin 1 and
N-
terminally truncated myocardin 1 have demonstrated similar activities in these
assays.
Additional studies have further demonstrated myocardin's potency as a
transactivator and its preferential action via CArG boxes. By way of example,
myocardin's ability to transactivate reporter genes containing four tandem
copies of
SM22 CArG-near or the c fos SRE linked to the E 1 b promoter was tested and
compared to SRF. These reporters were transactivated several hundred-fold by
myocardin, whereas SRF was only able to activate expression by 8-fold.
77
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
EXAMPLE 6
Myocardin 1 and MEF2C Cooperatively Activate the MLC2V Promoter
Cos cells were transiently transfected with a luciferase reporter gene
containing the MLC2V promoter and expression vectors encoding myocardin 1 and
MEF2C, as indicated. Forty eight hr later, cells were harvested and luciferase
activity
was assayed. The results in FIG. 8 show that myocardin 1 and MEF2C
synergistically
activate MLC2V transcription. Both myocardin 1 and N-terminally truncated
myocardin 1 have demonstrated similar activities in these assays.
EXAMPLE 7
Interactions of Myocardin 1 and GATA4
A. GATA4 Represses Myocardin Activation of the ANF Promoter
HeLa cells and Cos cells were transiently transfected with a luciferase
reporter
gene containing the ANF promoter and expression vectors encoding myocardin 1
and
GAT A4, as indicated. Forty eight hr later, cells were harvested and
luciferase activity
was assayed. The results in FIG. 9 show that GATA4 represses myocardin 1
activation of ANF transcription.
B. Myocardin 1 and GATA4 Cooperatively Activate the Nkx2.5 Promoter
HeLa cells (and/or Cos cells) were transiently transfected with a luciferase
reporter gene containing the Nkx2.5 promoter and expression vectors encoding
myocardin 1 and GATA4. Approximately forty eight hr later, cells were
harvested
and luciferase activity was assayed. The results demonstrated that myocardin 1
and
GATA4 cooperatively activated Nkx2.5 transcription. Both myocardin 1 and N-
ternlinally truncated myocardin 1 have demonstrated similar activities in
these
GATA4 interaction assays. FIG.13.
78
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
EXAMPLE 8
Myocardin 1 and Nkx2.5 Cooperatively Activate the a-MHC Promoter
HeLa cells were transiently transfected with a luciferase reporter gene
containing the a-MHC promoter and expression vectors encoding myocardin 1 and
Nkx2.5. Forty eight hr later; cells were harvested and luciferase activity was
assayed.
The results in FIG. 10 show that myocardin 1 and Nkx2.5 synergistically
activate a-
MHC transcription. EXAMPLE 9Myocardin Forms a Complex with SRFTo
further examine the mechanism for CArG box-dependent transcriptional
activation,
the inventors tested whether myocardin translated in vitro could bind to the
CArG
boxes from the SM22 promoter. S1RF' bound to both CArG boxes, but no binding
of
myocardin to either CArG box was detectable in gel mobility shift assays.
However,
myocardin in the presence of SRF gave rise to a prominent ternary complex with
the
CArG box sequence. This ternary complex was supershifted by antibodies against
SRF or FLAG-tagged myocardin. The total amount of SRF DNA binding was
comparable in the presence and absence of myocardin, suggesting that
association of
SIRf with myocardin does not alter the affinity of SRF fro the CArG box.
Myocardin
and SRF also formed a ternary complex with the c fos and ANF CArG boxes, the
intensity of which correlated directly with the relative binding of SRF, to
the site. The
lack of obvious homology in the flanking sequences of these different CArG
boxes
suggests that myocardin associates directly with SRF and does not depend on
specific
DNA sequences for ternary complex formation.
The region of myocardin required for ternary complex formation with SRF
was determined using myocardin deletion mutants. Deletion of the amino-
terminal
276 amino acid abolished association with SRF, as did larger amino-terminal
deletions. In contrast, deletions from near the middle of the protein to the
carboxyl
terminus did not affect SRF interaction. Deletion of the Q-rich domain or the
basic
regions also abolished ternary complex formation, whereas mutation of the SAP
domain did not. These findings are consistent with the interpretation that the
amino
terminus of myocardin confers transcriptional specificity by mediating
association
with SRF, whereas the carboxyl terminus activates transcription.
79
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
To determine whether myocardin interacts with the DNA binding or
transcription activation domain of SRF, the inventors performed gel mobility
shift
assays with an SRF deletion mutant encompassing the MADS domain, but lacking
the
amino and carboxyl termini. This SRF mutation (SRF 100-300) bound the CArG box
sequence and formed a ternary complex with myocardin.
Association of myocardin and SRF was also readily detectable in
coimmunoprecipitation assays of epitope-tagged proteins. Interaction was
dependent
on the amino-terminal regions of myocardin.. The core MADS domain of SRF
(residues 133-266) was also necessary and sufficient to mediate association
with
myocardin in coimmunoprecipitation assays.
Without wishing to be bound by any particular theory, these results suggest
that myocardin interacts with SRF to form a stable ternary complex which may
be an
aspect of the mechanism of action of myocardin as a transcription activator.
Both
myocardin 1 and N-terminally truncated myocardin 1 have demonstrated similar
activities in this regard.
iVlyocardin has also been shown to be sensitive to the level of SRF, such that
at
low concentrations of SRF expression plasmid; myocardin and SRF
synergistically
activated SM22 transcription, whereas at higher concentrations of SRF,
transcriptional
activation by myocardin was reduced. Inhibition of myocardin-dependent
transcription by excess SRF could be relieved by increasing the amount of
myocardin.
Thus, the ratio of SRF to myocardin appears relevant for transcriptional
activation by
myocardin, such that exceeding an optimal ratio with an excess of SRF can
result in
attenuation of myocardin activity.
EXAMPLE 10
Inhibition of Cardiomyocyte Differentiation in Xenopus Embryos by Dominant
Negative Myocardin
Further confirming the role of myocardin in cardiomyocyte differentiation,
mRNA from a dominant negative myocardin mutant was injected into Xenopus
embryos. A dramatic reduction in the expression of transcripts for cardiac a-
actin and
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
a-tropomyosin was observed. The effects on cardiac differentiation were highly
specific as demonstrated by the normal overall appearance of the embryo. Also
observed was a dose-dependent reduction in expression of cardiac markers, such
that
approximately 90% of injected embryos exhibited a reduction or complete
elimination
of cardiac gene expression.
EXAMPLE 11
Overexpression of Myocardin Induces Hypertrophy in Cardiomyocytes
The inventors have investigated how myocardin affects the growth and/or
differentiation of cardiomyocytes by overexpressing myocardin in
cardiomyocytes
using adenoviral delivering system. Cardiomyocyte cultures were prepared by
dissociation of 1-day-old neonatal rat hearts and were plated differentially
to remove
fibroblasts. Cells were plated on glass coverslips coated with 4 pg/cm2
laminin in 4:1
Dulbecco's modified Eagle's medium (DMEM):199 medium with 10% horse serum
and 5% fetal calf serum at a density of 5 x 104 cells/cmZ. Eighteen hours
after plating,
cells were changed into serum-free media and infected with adenoviruses
expressing
either myocardin or 13-galactosidase (as a control) at a multiplicity of
infection (m.o.i.)
of 100.
For immunofluorescence, cells were fixed in 3.7% formaldehyde on ice for 30
min, permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS)
and
blocked with 5% serum in PBS for 1 hour at room temperature. Cells were
incubated
with monoclonal anti-a-actinin (sarcomeric) or anti-ANF (atrial natriuretic
factor)
antibodies at a dilution of 1:200 in blocking buffer for 1 hour at
37°C, washed and
incubated with fluorescein-conjugated horse anti-mouse IgG antibody at a
dilution of
1:200 in blocking buffer for 1 hour at 37°C. Following secondary
antibody
incubation, cells were washed with PBS.
The results are shown in FIGS. 11 and 12. Overexpression of myocardin in
neonatal cardiomyocytes induces assembly of sacomeres and expression of atrium
natriuretic factor (ANF), markers of cardiac hypertrophy.
81
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
References
The following references, to the extent that they provide exemplary procedural
or other details supplementary to those set forth herein, are specifically
incorporated
herein by reference.
EPO Patent 0 273 085
EPO Patent 0 320 308
U.S. Patent 5,354,855
U.S. Patent 4,196,265
U.S. Patent 4,554,101
U.S. Patent 4,683,195
U.S. Patent 4,683,202
U.S. Patent 4,800,159
U.S. Patent 4,873,191
U.S. Patent 4,883,750
U.S. Patent 5,279,721
U.S. Patent 4,879,236
U.S. Patent 5,871,986
U.S. Patent 5,792,453
U.S. Patent 6,100,242
WO 84/03564
WO 90/07641
Aramburu et al., Mol. Cell, 1:627-637, 1998.
Aramburu et al., Science, 285:2129-2133, 1999.
Ausubel et al., In: Current Protocols in Molecular Biology, John, Wiley &
Sons, Inc.,
1994.
Baichwal and Sugden, In: Gene Transfer, Kucherlapati R, ed., New York, Plenum
Press,
pp. 117-148, 1986.
Barany and Merrifield, The Peptides, Gross and Meienhofer, eds., Academic
Press,
New York, pp. 1-284, 1979.
Barnes et al., J. Biol. Chem., 272(17):11510-7, 1997.
Beals et al., Genes Dev., 11:824-834, 1997.
Benvenisty and Neshif, Proc. Nat'l Acad. Sci. USA 83(24):9551-9555, 1986.
82
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Bhavsar et al., Genomics, 35(1):11-23, 1996.
Brinster et al., Proc. Nat'l Acad. Sci. USA, 82: 4438-4442, 1985.
Capaldi et al., Biochem. Biophys. Res. Comm., 76:425, 197.7
Chang et al., 14:124A, 1991. ,
Chen and Okayama, 7:2745-2752, 1987.
Chin et al., Genes Dev., 12:2499-2509, 1998.
Clipstone and Crabtree, Nature, 357:695-697, 1992.
Coffin, Retroviridae and Their Replication. In: Virology, Fields et al., eds.,
Raven Press,
New York, pp. 1437-1500, 1990.
Cook et al., Cell, 27:487-496, 1981.
Couch et al., Am. Rev. Resp. Dis., 88:394-403, 1963.
Coupar et al., Gene, 68:1-10, 1988.
Crabtree, Cell, 96:611-614, 1990.
Ding et al., Circ. Res., 84:729-734, 1999.
Dubensky et al., Proc. Nat'l Acad. Sci. USA, 81:7529-7533, 1984.
Dunn et al., J. Biol. Chem., 274:21908-21912, 1999.
Edmondson et ccl., Development, 120:1251-1263, 1994.
Epstein, In The Metabolic and Molecular Bases of Inherited Disease: in Down
Syndrome (Trisomy 21), Scriver, Beaudet, Valle, (eds), 7th Ed., Vol. 1 , pp.
749-794, McGraw-Hill, Inc., New York, 1995.
Fechheimer et al., Proc. Nat'l Acad. Sci. USA, 84:8463-8467, 1987.
Ferkol et al., FASEB J., 7:1081-1091, 1993.
Fishman and Olson, Cell 91, 153- 156, 1997.
Forster & Symons, Cell, 49:211-220, 1987.
Fraley et al., Proc. Nat'1 Acad. Sci. USA, 76:3348-3352, 1979.
Franz et al., Cardioscience, 5(4):235-43, 1994.
Freifelder, Physical Biochemistry Applications to Biochemistry and Molecular
Biology, 2nd ed. Wm. Freeman and Co., New York, NY, 1982.
Friedmann, "Progress toward human gene therapy", Science, 244:1275-1281, 1989.
Fuentes et al., Genomics, 44:358-361, 1997.
Fuentes et al., Hum. Mol. Genet., 4:1935-1944, 1995.
Gerlach et al., Nature (London), 328:802-805, 1987.
83
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ghosh and Bachhawat, Targeting of Liposomes to Hepatocytes. In: Liver
Diseases,
Targeted Diagnosis and Therapy Using Specific Receptors and Ligands. Wu et
al., eds., Marcel Dekker, New York, pp. 87-104, 1991.
Ghosh-Choudhury et al., EMBO J., 6:1733-1739, 1987.
Goding, 1986, In: Monoclonal Antibodies: Principles and Practice, 2d ed.,
Academic
Press, Orlando, Fla., pp. 60-61, and 71-74, 1986.
Gomez-Foix et al., J. Biol. Chem., 267:25129-25134, 1992.
Gopal, Mol. Cell Biol., 5:1188-1190, 1985.
Gopal-Srivastava et al., Mol. Cell. Biol., 15(12):7081-90, 1995.
Gorman et al., Proc Nat'l Acad. Sci. (USA), 1982; 79(22):6777-81, 1982.
Graef et al., Nature, 401:703-8, 1999.
Graham and Prevec, In: Methods in Molecular Biology: Gene Transfer and
Expression Protocol, E.J. Murray, ed., Humana Press, Clifton, NJ, 7:109-128,
1991.
Graham and van der Eb, Virology, 52:456-467, 1973.
Graham et. al., .l. Gen. Virol., 36(1):59-74, 1977.
Grayson et al., J. Cell. Biochem., 70:366-375, 1998.
Grayson et al., Mol. Cell. Biol., 15:1870-1878, 1995.
Grunhaus and Horwitz, Seminar in Virology, 3:237-252, 1992.
Guzman et al., Circulation, 88(6):2838-48, 1993.
Hammond et al., Clin. Res., 42:123A, 1994.
Harland and Weintraub, J. Cell Biol., 101:1094-1099, 1985.
Harlow and Lane, Antibodies: A Laboratory manual, Cold Spring Harbor
Laboratory,
1988.
Harvey, Dev. Biol. 178:203-216, 1996.
Hermonat and Muzycska, Proc. Nat'l Acad. Sci. USA, 81:6466-6470, 1984.
Hersdorffer et al., DNA Cell Biol., 9:713-723, 1990.
Herz and Gerard, Proc. Nat'I Acad. Sci. USA, 90:2812-2816, 1993.
Hidaka et. al., J. Clin. Invest. 103(4):549-587, 1999.
Ho et al., J. Exp. Med., 184:101-112, 1996.
Ho et al., Cell, 85:973-983, 1996.
Ho et al., J. Biol. Chem., 270:19898-19907, 1995.
Hoey et al., Immunity, 2:461-472, 1995.
84
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Hogan et al., In: Manipulating the Mouse Embryo: A Laboratory Manual, 2nd
ed.,Cold Spring Harbor Laboratory Press, 1994.
Horwich et al., J. Yirol., 64:642-650, 1990.
Innis et al., PCR Protocols, Academic Press, Inc., San Diego CA, 1990.
Johnson et al., Peptide Turn Mimetics" IN: Biotechnology And Pharmacy, Pezzuto
et
al., eds., Chapman and Hall, New York, 1993.
Jones and Shenk, Cell, 13:181-188, 1978.
Joyce, Nature, 338:217-244, 1989.
Kaneda et al., Science, 243:375-378, 1989.
Karlsson et al., EMBO J., 5:2377-2385, 1986.
Kashishian et al., J. Biol. Chem., 273:27412-27419, 1998.
Kato et cal., J. Biol. Chem., 266:3361-3364, 1991.
Kelly et al., J. Cell Biol., 129(2):383-96, 1995.
Kim & Cook, Proc. Nat'l Acad. Sci. USA, 84:8788-8792, 1987.
Kimura et al, Dev. Growth Differ. 39(3):257-65, 1997.
Kingsbury and Cunningham, Yeast Genetics and Molecular Meeting, Abstract , p.
98,
Genetics Society of America, Bethesda, MD, 1998.
Klauck et al., Science, 271:1589-1592, 1996.
Klee et al., J. Biol. Chem., 273:13367-13370, 1998.
Klein et al., Nature, 327:70-73, 1987.
Kodama, et al., Cir. Res. 81:656-663, 1997.
Kyte and Doolittle, J. Mol. Biol., 157(1):105-132, 1982.
Lai et al., J. Biol. Chem., 273:18325-18331, 1998.
LaPointe et al., Hypertension 27(3 Pt 2):715-22, 1996.
Le Gal La Salle et al., Science, 259:988-990, 1993.
Leor et al., Circulation 94(Suppl. II):332-336, 1996.
Levrero et al., Gene, 101:195-202, 1991.
Lin et. al., Science, 276:1404-1407, 1997.
Liu et al., EMBO J., 16:143-153, 1997.
Loh et al., J. Biol. Chem., 271:10884-10891, 1996.
Luo et al., Proc. Nat'l Acad. Sci. USA, 93:8907-8912, 1996.
Macejak and Sarnow, Nature, 353:90-94, 1991.
Makino et. al., J. Clin. Invest., 103:697-705, 1999.
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Mann et al., Cell, 33:153-159, 1983.
Mao and Wiedmann, J. Biol. Chem., 274:31102-31107, 1999.
Mao et al., Science, 286:785-790, 1999.
Marban et al., Proc. Nat'1 Acad. Sci. USA, 84:6005-6009, 1987.
Markowitz et al., J. Virol., 62:1120-1124, 1988.
Masuda et al., Mol. Cell. Biol., 17:2066-2075, 1997.
Masuda et al., Mol. Cell. Biol., 15:2697-2706, 1995.
Merrifield, Science, 232: 341-347, 1986.
Mesaeli et al., J. Cell Biol., 144:857-868, 1999.
Michel & Westhof, J. Mol. Biol., 216:585-610, 1990.
Miyazaki et al., J. Biol. Chem., 271:14567-14571, 1996.
Molkentin et al., Cell, 93:215-228, 1998.
Moss et al., J. Biol. Chem., 271 (49):31688-94, 1996.
Mulligan, Science, 260:926-932, 1993.
Musaro et al., Nature, 400:581-585, 1999.
Naya et al., Development, 126:2045-2052, 1999.
Nicolas and Rubinstein, In: Vectors: A survey of molecular cloning vectors and
their
uses, Rodriguez and Denhardt, eds., Stoneham: Butterworth, pp. 494-513,
1988.
Nicolau and Sene, Biochim. Biophys. Acta, 721:185-190, 1982.
Nicolau et al., Methods Enzymol., 149:157-176, 1987.
O'Keefe et al., Nature, 357:692-694, 1992.
Olson, E.N. 1990. Genes & Dev. 4, 1454-1461.
Olson and Srivastava, D. 1996. Science 272, 671-676.
Palmiter et al., Nature, 300:611, 1982.
Pan, et al., Circ. Res. 81:611-617, 1997.
Parks et al., Anal. Biochem., 216:413-417, 1994.
Paskind et al., Virology, 67:242-248, 1975.
Pelletier and Sonenberg, Nature, 334:320-325, 1988.
Perales et al., Proc. Nat'1 Acad. Sci. 91:4086-4090, 1994.
Pignon et al., Hum. Mutat., 3: 126-132, 1994.
Potter et. al., Proc. Nat'l Acad. Sci. USA, 81:7161-7165, 1984.
Racher et al., Biotechnology Techniques, 9:169-174, 1995.
86
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ragot et al., Nature, 361:647-650, 1993.
Rao et al., Annu. Rev. Immzmol., 15:707-747, 1997.
Reinhold-Hurek & Shub, Nature, 357:173-176, 1992.
Remington's Pharmaceutical Science", 15'h Ed., pg. 1035-1038 and 1570-1580.
Renan, Radiother. Oncol., 19:197-218, 1990.
Rich et al., Hum. Gene Ther., 4:461-476, 1993.
Ridgeway, Mammalian Expression Vectors, In: Vectors: A Survey of Molecular
Cloning
Vectors and Their Uses, Rodriguez et al., eds., Stoneham: Butterworth, pp. 467-
492, 1988.
Rippe et. al., 10:689-695, 1990.
Ritchie, M. E., J. Biol. Chem. 271(41):25485-25491, 1996.
Rosenfeld et al., Cell, 68:143-155, 1992.
Rosenfeld et al., Science, 252:431-434, 1991.
Roux et al., Proc. Nat'l Acad. Sci. USA, 86:9079-9083, 1989.
Sambrook et. al., In: Molecular Cloning.' A Laboratory Manual, 2d Ed., Cold
Spring
Harbor Laboratory Press, Cold Spring Harbor, NY, 1989.
Sarver et al, Science, 247:1222-1225, 1990.
Scanlon et. al., Proc. Nat'l Acad. Sci. USA, 88:10591-10595, 1991.
Schiaffino and Reggiani, Physiol. Rev., 76:371-423, 1996.
Seidman and Seidman, Basic Res. Cardiol., 93:'13-16, 1998.
Semsarian et al., Nature, 400:576-581_, 1999.
Shibasaki et al., Nature, 382:370-373, 1996.
Shub, Cell, 71 (2):183-186, 1992.
Sigal et al., J. Exp. Med., 173:619-628, 1991.
Srivastava et. al., Cell, 56:607-617, 1995.
Stewart and Young, Solid Phase Peptide Synthesis, 2d. ed., Pierce Chemical
Co.,
1984.
Stratford-Perncaudet and Perricaudet, In: Human Gene Transfer, O. Cohen-
Haguenauer
et al., eds., John Libbey Eurotext, France, pp. 51-61, 1991.
Stratford-Perricaudet et al., Hum. Gene. Ther., 1:241-256, 1990.
Sun et al., Immunity, 8:703-71 l, 1998.
Sussman et al., Science, 281:1690-1693, 1998.
Tam et al., J. Am. Chem. Soc., 105:6442, 1983.
87
WO 02/060946 PCT/USO1/50606
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Temin, In: Gene Transfer, Kucherlapati R, ed., New York, Plenum Press, pp. 149-
188,
1986.
Timmerman et al., Nature, 383:837-840, 1996.
Top et. al., J. Infect. Dis., 124:155-160, 1971.
Tur-Kaspa et al., Mol. Cell Biol., 6:716-718, 1986.
Uetsuki et al., J. Biol. Chem., 264(10):5791-5798, 1989.
Varmus et al., Cell, 25:23-36, 1981.
Wagner et al., Proc. Nat'l Acad. Sci. USA 87(9):3410-3414, 1990.
Wakabayashi-Ito, et al., J. Biol. Chem., 269(47):29831-29837, 1994.
Wang et al., Science, 284:339-343, 1999.
Wong et al., Gene, 10:87-94, 1980.
Wu and Wu, Adv. Drug Delivery Rev., 12:1 ~9-167, 1993.
Wu and Wu, Biochemistry, 27:887-892, 1988.
Wu and Wu, J. Biol. Chem., 262:4429-4432, 1987.
Wu et al., Genomics, 4:560, 1989.
Yamauchi-Takihara, et. al., Proc. Nat'1 Acad. Sci. USA 86(10):3504-3508, 1989.
Yang et al., Proc. Nat'l Acad. Sci. USA, 87:9568-9572, 1990.
Yang et al., Mol. Cell. .Biol., 17:5236-5243, 1997.
Youn et al., Science, 286:790-793, 1999.
Zelenin et al., FEBSLett., 280:94-96, 1991.
Zhang et al., Circ. Res., 84:722-728, 1999.
Zhuo et al., Proc. Nat'l Acad. Sci. USA, 96:4650-4655, 1999.
Ziober and Kramer, J. Biol. Chem., 271(37):22915-22, 1996.
88
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
SEQUENCE LISTING
<110> OLSON, ERIC N.
WANG, DA-ZHI
<120> METHODS AND COMPOSITIONS RELATING TO A CARDIAC-SPECIFIC
NUCLEAR REGULATORY FACTOR
<130> UTSD:695US
<140> UNKNOWN
<141> 2001-12-20
<150> 60/257,761
<151> 2000-12-21
<160> 30
<170> PatentIn Ver. 2.1
<210> 1
<211> 4959
<212> DNA
<213> Mus musculus
<220>
<221> CDS
<222> (641)..(3061)
<400> 1
ggaattcggc acgaggccac cctcagagga ggagggtcct gcctgctggg agttaattag 60
cctcgcgagc ggcgaggggg gaggcgccag ttttctgggg acactggcgg ccactgtgcg 120
tcctcctacc caagggagct ccccaagagt tggatgaatt ctgggttgtt agctgctgtc 180
ctctgggctc ccgggagcca gtttctggtg gaaagcgggg cgcctggcca acgaccagcg 240
gcttgctgag actcaccatg acactcctgg ggtctgaaca ctctttgctg attagaagga 300
agttccgatc agtcttacag ttacggcttc aacagagaag gacccaggag cagctggcta 360
accaaggctt aataccgcca ctgaaaggtc caactgaatt ccatgacccg agaaaacaat 420
tggatagtgc caagactgaa gattccctga ggcgcaaggg cagaaacagg tccgaccgtg 480
ccagcctggt tactatgcac attct~ccaag cctccacggc agaaaggtcc attccaactg 540
ctcagatgaa gctcaaaaga gcccgccttg cagatgacct caatgagaag atcgctctcc 600
gccaaggccc ttggaactgg tggagaagaa cattctgccg atg gat tct tcc gtg 655
Met Asp Ser Ser Val
1 5
aaa gag get ata aaa ggt act gag gtg agc ctc tcc aag gca gca gat 703
1
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Lys Glu Ala Ile Lys Gly Thr Glu Val Ser Leu Ser Lys Ala Ala Asp
15 20
gca ttc gcc ttt gag gat gac agc agt aga gat ggg ctc tct cca gat 751
Ala Phe Ala Phe Glu Asp Asp Ser Ser Arg Asp Gly Leu Ser Pro Asp
25 30 35
cag get agg agc gag gac ccc cag ggc tct aca gga tcc acc cca gac 799
Gln Ala Arg Ser Glu Asp Pro Gln Gly Ser Thr Gly Ser Thr Pro Asp
40 45 50
atc aaa tcc act gag get cct ctg gac aca atc cag gat ctc act cct 847
Ile Lys Ser Thr Glu Ala Pro Leu Asp Thr Ile Gln Asp Leu Thr Pro
55 60 65
ggc tca gaa agt gac aag aat gat gca gcc tcc cag cca ggc aac cag 895
Gly Ser Glu Ser Asp Lys Asn Asp Ala Ala Ser Gln Pro Gly Asn Gln
70 75 80 85
tca gac cct ggg aag cag gtt ctc ggc ccc ctc agc acc ccg att cct 943
Ser Asp Pro Gly Lys Gln Val Leu Gly Pro Leu Ser Thr Pro Ile Pro
90 95 100
gtg cac act get gta aag tcc aag tct ttg ggt gac agt aag aac cgc 991
Val His Thr Ala Val Lys Ser Lys Ser Leu Gly Asp Ser Lys Asn Arg
105 110 115
cac aaa aag ccc aaa gac ccc aaa cca aag gtg aag aag ctc aaa tac 1039
His Lys Lys Pro Lys Asp Pro Lys Pro Lys Val Lys Lys Leu Lys Tyr
120 125 130
cat cag tac atc ccc cca gac cag aag gca gag aag tct ccc cca ccc 1087
His Gln Tyr Ile Pro Pro Asp Gln Lys Ala Glu Lys Ser Pro Pro Pro
135 140 145
atg gac tct gcc tat gcc cgg ctg ctc cag caa cag cag cta ttc ctg 1135
Met Asp Ser Ala Tyr Ala Arg Leu Leu Gln Gln Gln Gln Leu Phe Leu
150 155 160 165
cag cta cag atc ctc agc cag cag cag caa cag cag cag caa cag cag 1183
Gln Leu Gln Ile Leu Ser Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln
170 175 180
cag cag caa cag cag cag cag cag cag cag cag cgg ttc agc tac cct 1231
Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Arg Phe Ser Tyr Pro
185 190 195
ggg atg cac caa aca cac ctc aaa gaa cca aat gaa cag atg gcc aga 1279
Gly Met His Gln Thr His Leu Lys Glu Pro Asn Glu Gln Met Ala Arg
200 205 210
aat ccg aat cct tct tca aca cca ctg agc aat acc cct cta tcc cct 1327
Asn Pro Asn Pro Ser Ser Thr Pro Leu Ser Asn Thr Pro Leu Ser Pro
215 220 225
2
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
gtc aaa aat agc att tct gga caa act ggt gtt tct tct ctc aaa cca 1375
Val Lys Asn Ser Ile Ser Gly Gln Thr Gly Val Ser Ser Leu Lys Pro
230 235 240 245
ggc ccc ctc cca ccc aac ctg gat gat ctc aag gtg tca gag tta aga 1423
Gly Pro Leu Pro Pro Asn Leu Asp Asp Leu Lys Val Ser Glu Leu Arg
250 255 260
caa cag ctt cga atc cgg ggc ttg cca gtg tca ggc acc aag aca gcg 1471
Gln Gln Leu Arg Ile Arg Gly Leu Pro Val Ser Gly Thr Lys Thr Ala
265 270 275
ctg gtg gac cgg ctt cgt ccc ttc cag gat tgt get ggc aac cct gtg 1519
Leu Val Asp Arg Leu Arg Pro Phe Gln Asp Cys Ala Gly Asn Pro Val
280 285 290
ccc aac ttt ggg gac atc aca act gtc acc ttt cct gtc acg ccc aac 1567
Pro Asn Phe Gly Asp Ile Thr Thr Val Thr Phe Pro Val Thr Pro Asn
295 300 305
acc ttg ccc agt tat cag tcc tcc ccg aca ggc ttc tac cac ttt ggc 1615
Thr Leu Pro Ser Tyr Gln Ser Ser Pro Thr Gly Phe Tyr His Phe Gly
310 315 320 325
agc aca agc tcc agc cca ccc atc tcc ccc gcc tca tct gac ttg tcc 1663
Ser Thr Ser Ser Ser Pro Pro Ile Ser Pro Ala Ser Ser Asp Leu Ser
330 335 340
get gca ggg tcc ctg cca gac acc ttc acc gat gcg tca cct ggc ttc 1711
Ala Ala Gly Ser Leu Pro Asp Thr Phe Thr Asp Ala Ser Pro Gly Phe
345 350 355
ggc ctg cac gca tct ccg gtg ccc gcc tgc acg gac gag agt ctg ctg 1759
Gly Leu His Ala Ser Pro Val Pro Ala Cys Thr Asp Glu Ser Leu Leu
360 365 370
agc agc ctg aat ggg ggc tcg ggc ccc tcc gag cct gat ggg cta gac 1807
Ser Ser Leu Asn Gly Gly Ser Gly Pro Ser Glu Pro Asp Gly Leu Asp
375 380 385
tct gag aag gac aag atg ctg gtg gag aag cag aaa gtg atc aac cag 1855
Ser Glu Lys Asp Lys Met Leu Val Glu Lys Gln Lys Val Ile Asn Gln
390 395 400 405
ctc acc tgg aag ctg cgg caa gag cag cgg cag gtg gaa gag ctg aga 1903
Leu Thr Trp Lys Leu Arg Gln Glu Gln Arg Gln Val Glu Glu Leu Arg
410 415 420
atg caa ctg cag aag cag aag agc agc tgc agc gac cag aag cca ctg 1951
Met Gln Leu Gln Lys Gln Lys Ser Ser Cys Ser Asp Gln Lys Pro Leu
425 430 435
ccc ttc ttg gcc acc acc atc aaa cag gaa gat gtc tcc agc tgc ccc 1999
Pro Phe Leu Ala Thr Thr Ile Lys Gln Glu Asp Val Ser Ser Cys Pro
440 445 450
3
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
ttc gca ccc cag cag gcg tct ggg aag gga cag ggc cac agc tct gac 2047
Phe Ala Pro Gln Gln Ala Ser Gly Lys Gly Gln Gly His Ser Ser Asp
455 460 465
agt ccc cct ccg get tgt gag acg get cag ctg ctg cct cac tgt gtg 2095
Ser Pro Pro Pro Ala Cys Glu Thr Ala Gln Leu Leu Pro His Cys Val
470 475 480 485
gag tcc tca ggt caa acc cat gta ctc tcg tcc acg ttt ctc agc ccc 2143
Glu Ser Ser Gly Gln Thr His Val Leu Ser Ser Thr Phe Leu Ser Pro
490 495 500
cag tgc tcc cct cag cac tcg ccc ctg ggg ggc ctg aag agc ccg cag 2191
Gln Cys Ser Pro Gln His Ser Pro Leu Gly Gly Leu Lys Ser Pro Gln
505 510 515
cac atc agc ctg cct cca tca ccc aac aac cat tac ttc ctg get tcc 2239
His Ile Ser Leu Pro Pro Ser Pro Asn Asn His Tyr Phe Leu Ala Ser
520 525 530
tct tcg gga get cag aga gag aac cat ggg gtc tct tca ccc agc agc 2287
Ser Ser Gly Ala Gln Arg Glu Asn His Gly Val Ser Ser Pro Ser Ser
535 540 545
agc caa ggg tgc gca cag atg act ggt tta caa tct tct gac aag gtg 2335
Ser Gln Gly Cys Ala Gln Met Thr Gly Leu Gln Ser Ser Asp Lys Val
550 555 560 565
ggg cca acg ttt tca att cca tcc cca act ttt tct aag tca agt tca 2383
Gly Pro Thr Phe Ser Ile Pro Ser Pro Thr Phe Ser Lys Ser Ser Ser
570 575 580
gca gtt tca gat atc acc cag ccc cca tcc tat gaa gat gca gtg aag 2431
Ala Val Ser Asp Ile Thr Gln Pro Pro Ser Tyr Glu Asp Ala Val Lys
585 590 595
cag caa atg act cgg agt cag cag atg gac gaa ctc ctg gat gtc ctc 2479
Gln Gln Met Thr Arg Ser Gln Gln Met Asp Glu Leu Leu Asp Val Leu
600 605 610
att gaa agt gga gaa atg cca gcc gat gcc agg gaa gat cat tca tgt 2527
Ile Glu Ser Gly Glu Met Pro Ala Asp Ala Arg Glu Asp His Ser Cys
615 620 625
ctt cag aaa att cca aag atc cct ggg tcc tcc tgc agc cca act gcc 2575
Leu Gln Lys Ile Pro Lys Ile Pro Gly Ser Ser Cys Ser Pro Thr Ala
630 635 640 645
atc ccc ccg aag ccc tcg get tcc ttt gag cag gca tct tcg gga ggc 2623
Ile Pro Pro Lys Pro Ser Ala Ser Phe Glu Gln Ala Ser Ser Gly Gly
650 655 660
cag atg gcc ttc gat cac tac gcc aac gac agt gac gaa cac ctg gaa 2671
Gln Met Ala Phe Asp His Tyr Ala Asn Asp Ser Asp Glu His Leu Glu
4
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
665 670 675
gtc tta ttg aat tct cac agc ccc atc gga aag gtg agc gat gtt acc 2719
Val Leu Leu Asn Ser His Ser Pro Ile Gly Lys Val Ser Asp Val Thr
680 685 690
ctc ctc aaa atc gga agc gag gag cct cct ttt gac agc atc atg gat 2767
Leu Leu Lys Ile Gly Ser Glu Glu Pro Pro Phe Asp Ser Ile Met Asp
695 700 705
ggc ttc cca ggg aag get gcg gaa gat ctc ttc agt get cac gag ctc 2815
Gly Phe Pro Gly Lys Ala Ala Glu Asp Leu Phe Ser Ala His Glu Leu
710 715 720 725
ttg cct ggg ccc ctc tcc ccg atg cat gca cag ttg tca cct cct tct 2863
Leu Pro Gly Pro Leu Ser Pro Met His Ala Gln Leu Ser Pro Pro Ser
730 735 740
gtg gac agc agt ggt ctg cag ctg agc ttc acg gaa tct cct tgg gaa 2911
Val Asp Ser Ser Gly Leu Gln Leu Ser Phe Thr Glu Ser Pro Trp Glu
745 750 755
aca atg gaa tgg ctg gac ctc act cca cct agt tcc acg cca ggc ttc 2959
Thr Met Glu Trp Leu Asp Leu Thr Pro Pro Ser Ser Thr Pro Gly Phe
760 765 770
agc aac ctt acc tcc agt ggg ccc agc att ttc aac atc gat ttt ctg 3007
Ser Asn Leu Thr Ser Ser Gly Pro Ser Ile Phe Asn Ile Asp Phe Leu
775 780 785
gat gtt aca gat ctt aat ctg aat tcc cct atg gat ctc cac tta cag 3055
Asp Val Thr Asp Leu Asn Leu Asn Ser Pro Met Asp Leu His Leu Gln
790 795 800 805
cag tgg taaacacccg aggtacaaga gctacgagag ctcagtggga attcaatgga 3111
Gln Trp
ggaaagcacg ataccggaaa tgtgtgttcc aaaagatgaa ggggggaaaa tggggaggga 3171
aaaaaaaaaa cagcaacgga ggtttttgtg acaactaacc agaacaaaca gaagtcagct 3231
attaaaatat gtctaaatgt aatatctacc agcattcagt aactgttaat aacttcagtg 3291
atgcattcaa aaatgtgctt tgtcagaata agaatgccaa aaatgttttt tcgctgcctt 3351
atctcatacc agtttttttg ggtttttttt tgtttgtttg ttttttggtt tttttttttt 3411
tgtgtgtgtt gttatttggt tttctttttg cccacagttt gtctcaggca atactgggac 3471
ataggctgac cccattagct tttgttatga atttactaaa ctttctgtgg aaggagaaca 3531
gagcctctgc cgcgggtgtg gggaagccat cctgtgcttg aggcagcaca cgtgtgtcca 3591
tcatcatcag tcagaagagc agggcctgtc tcacccaatc gagtccttaa gacagaataa 3651
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
tcagaatggt cagagggaca gaccaatcaa ttcccaggaa agcaaaagtg actcaatgtc 3711
ccttgactcc caaatggtcc cactggactg gtgatcactg gtgacaacta actagctttg 3771
tccagagaat ccacccagaa cacggtgctt tttagccagt agtccacctc tatgtgcatc 3831
agcaatgcat agcaggtgag aacttgaatc acagaaactt catgccatgg atggagactc 3891
ctgaggcgct caaatactac tacctctagt tccaaagact agagctagat gatcagaaag 3951
gcaactggag gcccagggag ccgtactggg acaagttaga attagagaac gatgtcattt 4011
aacattccga gaaagaaata accatgaatt gctattacag gagtaacaca cagggccagc 4071
ttcttttttc ttctttttta tttttctttt cttattgtga gcagagggaa ttcacctcag 4131
ttcatctttc tctcagtact tttctttcaa gatatcaatc ctttatgact cttttgcttt 4191
taattctctc tctctctctc tctctctctc tctctctttc tctcaaagga gaggtttcag 4251
ttctaacaag ctaccatagt cctattaaag ccattttttt ttttagaata ttaaaagtcc 4311
aaactctctt gccaaactct ttcttcacat gcgcattggc tgaaaacaga atttacaaga 4371
atttctttag gaagaaactg gggatgtggc ccattggtca caaagttttt ttgtttgttt 4431
ttgtttttgt ttcaattctt gtttgattta tggacaatct ttggtttgta ttgctctgga 4491
gaaattggaa atcattgcag agtgaagata aatcagggca ccatgtatag tagagaatgt 4551
ttcagtagtt ttccaaacga gaacacaatt gcacactgta aacaacagga gtgtgaagga 4611
ccacagtctt gaggagttct tgttgccctg tgtttggtga aggcgttggg gaccgaggaa 4671
gacaacatac agtttggcca aggctctcag aggcttgctg tggcgccaat tcaagtatta 4731
caatgttgca tgctgtagaa agtagctgtt gctgttgttt tgttttgttt taatttaagt 4791
caccaaggca ctgttttatt cttttgtaaa aaaaaaaaaa gttcactgtg cacttataga 4851
gaaaataatc aacaatgttg tgaatttttg agaagacttt tttttttttg ataaaccaaa 4911
gatttagaaa tcattccatt gtcaacttgt aaaaaaaaaa aaaaaaaa 4959
<210> 2
<211> 807
<212> PRT
<213> Mus musculus
<400> 2
Met Asp Ser Ser Val Lys Glu Ala Ile Lys Gly Thr Glu Val Ser Leu
1 5 10 15
Ser Lys Ala Ala Asp Ala Phe Ala Phe Glu Asp Asp Ser Ser Arg Asp
6
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
20 25 30
Gly Leu Ser Pro Asp Gln Ala Arg Ser Glu Asp Pro Gln Gly Ser Thr
35 40 45
Gly Ser Thr Pro Asp,Ile Lys Ser Thr Glu Ala Pro Leu Asp Thr Ile
50 55 60
Gln Asp Leu Thr Pro Gly Ser Glu Ser Asp Lys Asn Asp Ala Ala Ser
65 70 75 80
Gln Pro Gly Asn Gln Ser Asp Pro Gly Lys Gln Val Leu Gly Pro Leu
85 90 95
Ser Thr Pro Ile Pro Val His Thr Ala Val Lys Ser Lys Ser Leu Gly
100 105 110
Asp Ser Lys Asn Arg His Lys Lys Pro Lys Asp Pro Lys Pro Lys Val
115 120 125
Lys Lys Leu Lys Tyr His Gln Tyr Ile Pro Pro Asp Gln Lys Ala Glu
130 135 140
Lys Ser Pro Pro Pro Met Asp Ser Ala Tyr Ala Arg Leu Leu Gln Gln
145 150 155 160
Gln Gln Leu Phe Leu Gln Leu Gln Ile Leu Ser Gln Gln Gln Gln Gln
165 170 175
Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln
180 185 190
Arg Phe Ser Tyr Pro Gly Met His Gln Thr His Leu Lys Glu Pro Asn
195 200 205
Glu Gln Met Ala Arg Asn Pro Asn Pro Ser Ser Thr Pro Leu Ser Asn
210 215 220
Thr Pro Leu Ser Pro Val Lys Asn Ser Ile Ser Gly Gln Thr Gly Val
225 230 235 240
Ser Ser Leu Lys Pro Gly Pro Leu Pro Pro Asn Leu Asp Asp Leu Lys
245 250 255
Val Ser Glu Leu Arg Gln Gln Leu Arg Ile Arg Gly Leu Pro Val Ser
260 265 270
Gly Thr Lys Thr Ala Leu Val Asp Arg Leu Arg Pro Phe Gln Asp Cys
275 280 285
Ala Gly Asn Pro Val Pro Asn Phe Gly Asp Ile Thr Thr Val Thr Phe
290 295 300
Pro Val Thr Pro Asn Thr Leu Pro Ser Tyr Gln Ser Ser Pro Thr Gly
305 310 315 320
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Phe Tyr His Phe Gly Ser Thr Ser Ser Ser Pro Pro Ile Ser Pro Ala
325 330 335
Ser Ser Asp Leu Ser Ala Ala Gly Ser Leu Pro Asp Thr Phe Thr Asp
340 345 350
Ala Ser Pro Gly Phe Gly Leu His Ala Ser Pro Val Pro Ala Cys Thr
355 360 365
Asp Glu Ser Leu Leu Ser Ser Leu Asn Gly Gly Ser Gly Pro Ser Glu
370 375 380
Pro Asp Gly Leu Asp Ser Glu Lys Asp Lys Met Leu Val Glu Lys Gln
385 390 395 400
Lys Val Ile Asn Gln Leu Thr Trp Lys Leu Arg Gln Glu Gln Arg Gln
405 410 415
Val Glu Glu Leu Arg Met Gln Leu Gln Lys Gln Lys Ser Ser Cys Ser
420 425 430
Asp Gln Lys Pro Leu Pro Phe Leu Ala Thr Thr Ile Lys Gln Glu Asp
435 440 445
Val Ser Ser Cys Pro Phe Ala Pro Gln Gln Ala Ser Gly Lys Gly Gln
450 455 460
Gly His Ser Ser Asp Ser Pro Pro Pro Ala Cys Glu Thr Ala Gln Leu
465 470 475 480
Leu Pro His Cys Val Glu Ser Ser Gly Gln Thr His Val Leu Ser Ser
485 490 495
Thr Phe Leu Ser Pro Gln Cys Ser Pro Gln His Ser Pro Leu Gly Gly
500 505 510
Leu Lys Ser Pro Gln His Ile Ser Leu Pro Pro Ser Pro Asn Asn His
515 520 525
Tyr Phe Leu Ala Ser Ser Ser Gly Ala Gln Arg Glu Asn His Gly Val
530 535 540
Ser Ser Pro Ser Ser Ser Gln Gly Cys Ala Gln Met Thr Gly Leu Gln
545 550 555 560
Ser Ser Asp Lys Val Gly Pro Thr Phe Ser Ile Pro Ser Pro Thr Phe
565 570 575
Ser Lys Ser Ser Ser Ala Val Ser Asp Ile Thr Gln Pro Pro Ser Tyr
580 585 590
Glu Asp Ala Val Lys Gln Gln Met Thr Arg Ser Gln Gln Met Asp Glu
595 600 605
g
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Leu Leu Asp Val Leu Ile Glu Ser Gly Glu Met Pro Ala Asp Ala Arg
610 615 620
Glu Asp His Ser Cys Leu Gln Lys Ile Pro Lys Ile Pro Gly Ser Ser
625 630 635 640
Cys Ser Pro Thr Ala Ile Pro Pro Lys Pro Ser Ala Ser Phe Glu Gln
645 650 655
Ala Ser Ser Gly Gly Gln Met Ala Phe Asp His Tyr Ala Asn Asp Ser
660 665 670
Asp Glu His Leu Glu Val Leu Leu Asn Ser His Ser Pro Ile Gly Lys
675 680 685
Val Ser Asp Val Thr Leu Leu Lys Ile Gly Ser Glu Glu Pro Pro Phe
690 695 700
Asp Ser Ile Met Asp Gly Phe Pro Gly Lys Ala Ala Glu Asp Leu Phe
705 710 715 720
Ser Ala His Glu Leu Leu Pro Gly Pro Leu Ser Pro Met His Ala Gln
725 730 735
Leu Ser Pro Pro Ser Val Asp Ser Ser Gly Leu Gln Leu Ser Phe Thr
740 745 750
Glu Ser Pro Trp Glu Thr Met Glu Trp Leu Asp Leu Thr Pro Pro Ser
755 760 765
Ser Thr Pro Gly Phe Ser Asn Leu Thr Ser Ser Gly Pro Ser Ile Phe
770 775 780
Asn Ile Asp Phe Leu Asp Val Thr Asp Leu Asn Leu Asn Ser Pro Met
785 790 795 800
Asp Leu His Leu Gln Gln Trp
805
<210> 3
<211> 3907
<212 > DNA
<213> Mus musculus
<220>
<221> CDS
<222> (5)..(2083)
<400> 3
ccaa ggg atc atg ccg cct ttg aaa agt cca gcc gca ttt cat gag cag 49
Gly Ile Met Pro Pro Leu Lys Ser Pro Ala Ala Phe His Glu Gln
1 5 10 15
9
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
aga agg agc ttg gag cgg gcc agg aca gag gac tat ctc aaa cgg aag 97
Arg Arg Ser Leu Glu Arg Ala Arg Thr Glu Asp Tyr Leu Lys Arg Lys
20 25 30
att cgt tcc cgg ccg gag aga tcg gag ctg gtc agg atg cac att ttg 145
Ile Arg Ser Arg Pro Glu Arg Ser Glu Leu Val Arg Met, His Ile Leu
35 40 45
gaa gag acc tcg get gag cca tcc ctc cag gcc aag cag ctg aag ctg 193
Glu Glu Thr Ser Ala Glu Pro Ser Leu Gln Ala Lys Gln Leu Lys Leu
50 55 60
aag aga gcc aga cta gcc gat gac ctc aat gag aag att gca cag agg 241
Lys Arg Ala Arg Leu Ala Asp Asp Leu Asn Glu Lys Ile Ala Gln Arg
65 70 75
cct ggc ccc atg gag ctg gtg gag aag aac atc ctt cct gtt gag tcc 289
Pro Gly Pro Met Glu Leu Val Glu Lys Asn Ile Leu Pro Val Glu Ser
80 85 90 95
agc ctg aag gaa gcc atc att gtg ggc cag gtg aac tat ccc aaa gta 337
Ser Leu Lys Glu Ala Ile Ile Val Gly Gln Val Asn Tyr Pro Lys Val
100 105 110
gca gac agc tct tcc ttc gat gag gac agc agc gat gcc tta tcc ccc 385
Ala Asp Ser Ser Ser Phe Asp Glu Asp Ser Ser Asp Ala Leu Ser Pro
115 120 125
gag cag cct gcc agc cat gag tcc cag ggt tct gtg ccg tca ccc ctg 433
Glu Gln Pro Ala Ser His Glu Ser Gln Gly Ser Val Pro Ser Pro Leu
130 135 140
gag gcc cga gtc agc gaa cca ctg ctc agt gcc acc tct gca tcc ccc 481
Glu Ala Arg Val Ser Glu Pro Leu Leu Ser Ala Thr Ser Ala Ser Pro
145 150 155
acc cag gtt gtg tct caa ctt ccg atg ggc cgg gat tcc aga gaa atg 529
Thr Gln Val Val Ser Gln Leu Pro Met Gly Arg Asp Ser Arg Glu Met
160 165 170 175
ctt ttc ctg gca gag cag cct cct ctg cct ccc cca cct ctg ctg cct 577
Leu Phe Leu Ala Glu Gln Pro Pro Leu Pro Pro Pro Pro Leu Leu Pro
180 185 190
ccc agc ctc acc aat gga acc act atc ccc act gcc aag tcc acc ccc 625
Pro Ser Leu Thr Asn Gly Thr Thr Ile Pro Thr Ala Lys Ser Thr Pro
195 200 205
aca ctc att aag caa agc caa ccc aag tct gcc agt gag aag tca cag 673
Thr Leu Ile Lys Gln Ser Gln Pro Lys Ser Ala Ser Glu Lys Ser Gln
210 215 220
cgc agc aag aag gcc aag gag ctg aag cca aag gtg aag aag ctc aag 721
Arg Ser Lys Lys Ala Lys Glu Leu Lys Pro Lys Val Lys Lys Leu Lys
225 230 235
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
tac cac cag tac atc ccc ccg gac cag aag cag gac agg ggg gca ccc 769
Tyr His Gln Tyr Ile Pro Pro Asp Gln Lys Gln Asp Arg Gly Ala Pro
240 245 250 255
ccc atg gac tca tcc tac gcc aag atc ctg cag cag cag cag ctc ttc 817
Pro Met Asp Ser Ser Tyr Ala Lys Ile Leu Gln Gln Gln Gln Leu Phe
260 265 270
ctc cag ctg cag atc ctc aac cag cag cag cag cag cac cac aac tac 865
Leu Gln Leu Gln Ile Leu Asn Gln Gln Gln Gln Gln His His Asn Tyr
275 280 285
cag gcc atc ctg cct gcc ccg cca aag tca gca ggc gag gcc ctg gga 913
Gln Ala Ile Leu Pro Ala Pro Pro Lys Ser Ala Gly Glu Ala Leu Gly
290 295 300
agc agc ggg acc ccc cca gta cgc agc ctc tcc act acc aat agc agc 961
Ser Ser Gly Thr Pro Pro Val Arg Ser Leu Ser Thr Thr Asn Ser Ser
305 310 315
tcc agc tcg ggc gcc cct ggg ccc tgt ggg ctg gca cgt cag aac agc 1009
Ser Ser Ser Gly Ala Pro Gly Pro Cys Gly Leu Ala Arg Gln Asn Ser
320 325 330 335
acc tca ctg act ggc aag ccg gga gcc ctg ccg gcc aac ctg gac gac 1057
Thr Ser Leu Thr Gly Lys Pro Gly Ala Leu Pro Ala Asn Leu Asp Asp
340 345 350
atg aag gtg gca gag ctg aag cag gag ctg aag ttg cga tca ctg cct 1105
Met Lys Val Ala Glu Leu Lys Gln Glu Leu Lys Leu Arg Ser Leu Pro
355 360 365
gtc tcg ggc acc aaa act gag ctg att gag cgc ctt cga gcc tat caa 1153
Val Ser Gly Thr Lys Thr Glu Leu Ile Glu Arg Leu Arg Ala Tyr Gln
370 375 380
gac caa atc agc cct gtg cca gga gcc ccc aag gcc cct gcc gcc acc 1201
Asp Gln Ile Ser Pro Val Pro Gly Ala Pro Lys Ala Pro Ala Ala Thr
385 390 395
tct atc ctg cac aag get ggc gag gtg gtg gta gcc ttc cca gcg gcc 1249
Ser Ile Leu His Lys Ala Gly Glu Val Val Val Ala Phe Pro Ala Ala
400 405 410 415
cgg ctg agc acg ggg cca gcc ctg gtg gca gca ggc ctg get cca get 1297
Arg Leu Ser Thr Gly Pro Ala Leu Val Ala Ala Gly Leu Ala Pro Ala
420 425 430
gag gtg gtg gtg gcc acg gtg gcc agc agt ggg gtg gtg aag ttt ggc 1345
Glu Val Val Val Ala Thr Val Ala Ser Ser Gly Val Val Lys Phe Gly
435 440 445
agc acg ggc tcc acg ccc ccc gtg tct ccc acc ccc tcg gag cgc tca 1393
Ser Thr Gly Ser Thr Pro Pro Val Ser Pro Thr Pro Ser Glu Arg Ser
I1
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
450 455 460
ctg ctc agc acg ggc gat gaa aac tcc acc ccc ggg gac acc ttt ggt 1441
Leu Leu Ser Thr Gly Asp Glu Asn Ser Thr Pro Gly Asp Thr Phe Gly
465 470 475
gag atg gtg aca tca cct ctg acg cag ctg acc ctg cag gcc tcg cca 1489
Glu Met Val Thr Ser Pro Leu Thr Gln Leu Thr Leu Gln Ala Ser Pro
480 485 490 495
ctg cag atc ctc gtg aag gag gag ggc ccc cgg gcc ggg tcc tgt tgc 1537
Leu Gln Ile Leu Val Lys Glu Glu Gly Pro Arg Ala Gly Ser Cys Cys
500 505 510
ctg agc cct ggg ggg cgg gcg gag cta gag ggg cgc gac aag gac cag 1585
Leu Ser Pro Gly Gly Arg Ala Glu Leu Glu Gly Arg Asp Lys Asp Gln
515 520 525
atg ctg cag gag aaa gac aag cag atc gag gcg ctg acg cgc atg ctc 1633
Met Leu Gln Glu Lys Asp Lys Gln Ile Glu Ala Leu Thr Arg Met Leu
530 535 540
cgg cag aag cag cag ctg gtg gag cgg ctc aag ctg cag ctg gag cag 1681
Arg Gln Lys Gln Gln Leu Val Glu Arg Leu Lys Leu Gln Leu Glu Gln
545 550 555
gag aag cga gcc cag cag ccc gcc ccc gcc ccc gcc ccc ctc ggc acc 1729
Glu Lys Arg Ala Gln Gln Pro Ala Pro Ala Pro Ala Pro Leu Gly Thr
560 565 570 575
ccc gtg aag cag gag aac agc ttc tcc agc tgc cag ctg agc cag cag 1777
Pro Val Lys Gln Glu Asn Ser Phe Ser Ser Cys Gln Leu Ser Gln Gln
580 585 590
ccc ctg ggc ccc get cac cca ttc aac ccc agc ctg gcg gcc cca gcc 1825
Pro Leu Gly Pro Ala His Pro Phe Asn Pro Ser Leu Ala Ala Pro Ala
595 600 605
acc aac cac ata gac cct tgt get gtg gcc ccg ggg ccc ccg tcc gtg 1873
Thr Asn His Ile Asp Pro Cys Ala Val Ala Pro Gly Pro Pro Ser Val
610 615 620
gtg gtg aag cag gaa gcc ttg cag cct gag ccc gag ccg gtc ccc gcc 1921
Val Val Lys Gln Glu Ala Leu Gln Pro Glu Pro Glu Pro Val Pro Ala
625 630 635
ccc cag ttg ctt ctg ggg cct cag ggc ccc agc ctc atc aag ggg gtt 1969
Pro Gln Leu Leu Leu Gly Pro Gln Gly Pro Ser Leu Ile Lys Gly Val
640 645 650 655
gca cct ccc acc ctc atc acc gac tcc aca ggg acc cac ctt gtc ctc 2017
Ala Pro Pro Thr Leu Ile Thr Asp Ser Thr Gly Thr His Leu Val Leu
660 665 670
acc gtg acc aat aag aat gca gac agc cct ggc ctg tcc agt ggg agc 2065
12
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Thr Val Thr Asn Lys Asn Ala Asp Ser Pro Gly Leu Ser Ser Gly Ser
675 680 685
ccc cag cag ccc tcg tcc cagcctggct ctccagcgcc tgccccctct 2113
Pro Gln Gln Pro Ser Ser
690
gcccagatgg acctggagca cccactgcag cccctctttg ggacccccac ttctctgctg 2173
aagaaggaac cacctggcta tgaggaagcc atgagccagc agcccaaaca gcaggaaaat 2233
ggttcctcaa gccagcagat ggacgacctg tttgacattc tcattcagag cggagaaatt 2293
tcagcagatt tcaaggagcc gccatccctg ccagggaagg agaagccatc cccgaagaca 2353
gtctgtgggt cccccctggc agcacagcca tcaccttctg ctgagctccc ccaggctgcc 2413
ccacctcctc caggctcacc ctccctccct ggacgcctgg aggacttcct ggagagcagc 2473
acggggctgc ccctgctgac cagtgggcat gacgggccag agcccctttc cctcattgac 2533
gacctccata gccagatgct gagcagcact gccatcctgg accacccccc gtcacccatg 2593
gacacctcgg aattgcactt tgttcctgag cccagcagca ccatgggcct ggacctggct 2653
gatggccacc tggacagcat ggactggctg gagctgtcgt caggtggtcc cgtgctgagc 2713
ctagcccccc tcagcaccac agcccccagc ctcttctcca cagacttcct cgatggccat 2773
gatttgcagc tgcactggga ttcctgcttg tagctctctg gctcaagacg gggtggggaa 2833
ggggctggga gccagggtac tccaatgcgt ggctctcctg cgtgattcgg cctctccaca 2893
tggttgtgag tcttgacaat cacagcccct gctttttccc ttccctggga ggctagaaca 2953
gagaagccct tactcctggt tcagtgccac gcagggcaga ggagagcagc tgtcaagaag 3013
cagccctggc tctcacgctg gggttttgga cacacggtca gggtcagggc catttcagct 3073
tgacctcctt ttttgaggtc agggggcact gtctgtctgg ctacaatttg gctaaggtag 3133
gtgaagcctg gccaggcggg aggcttctct tctgacccag ggctgagaca ggttaagggg 3193
tgaatctcct tcctttctct ccctgctttg ctgtgaaggg agaaattagc ctgggcctct 3253
accccctatt ccctgtgtct gccaacccca ggatcccagg gctccctgcc attttagtgt 3313
cttggtgtag tgtaaccatt tagtggttgg tggcaacaat tttatgtaca ggtgtatata 3373
cctctatatt atatatcgac atacatatat atttttgggg gggggcggac aggagatggg 3433
tgcaactccc tcccatccta ctctcacaga agggcctgga tgcaaggtta cccttgagct 3493
gtgtgccaca gtctggtgcc cagtctggca tgcagctacc caggcccacc catcacgtgt 3553
13
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
gattgacatg taggtaccct gccacggcct atgccccacc tgccctgctt cctggctcct 3613
tatcagtgcc atgagggcag aggtgctacc tggccttcct gccaggagct ctccacccac 3673
tcacattccg tccccgccgc ctcactgcag ccagcgtggt cctaggacag gaggagcttc 3733
gggcccagct tcaccctgcg gtggggctga ggggtggcca tctcctgccc tggggccact 3793
ggcttcacat tctgggctga ctcatagggg agtaggggtg gagtcaccaa aaccagtgct 3853
gggacaaaga tggggaaggt gtgtgaactt tttaaaataa acacaaaaac acag 3907
<210> 4
<211> 693
<212> PRT
<213> Mus musculus
<400> 4
Gly Ile Met Pro Pro Leu Lys Ser Pro Ala Ala Phe His Glu Gln Arg
1 5 10 15
Arg Ser Leu Glu Arg Ala Arg Thr Glu Asp Tyr Leu Lys Arg Lys Ile
20 25 30
Arg Ser Arg Pro Glu Arg Ser Glu Leu Val Arg Met His Ile Leu Glu
35 40 45
Glu Thr Ser Ala Glu Pro Ser Leu Gln Ala Lys Gln Leu Lys Leu Lys
50 55 60
Arg Ala Arg Leu Ala Asp Asp Leu Asn Glu Lys Ile Ala Gln Arg Pro
65 70 75 80
Gly Pro Met Glu Leu Val Glu Lys Asn Ile Leu Pro Val Glu Ser Ser
85 90 95
Leu Lys Glu Ala Ile Ile Val Gly Gln Val Asn Tyr Pro Lys Val Ala
100 105 110
Asp Ser Ser Ser Phe Asp Glu Asp Ser Ser Asp Ala Leu Ser Pro Glu
115 120 125
Gln Pro Ala Ser His Glu Ser Gln Gly Ser Val Pro Ser Pro Leu Glu
130 135 140
Ala Arg Val Ser Glu Pro Leu Leu Ser Ala Thr Ser Ala Ser Pro Thr
145 150 155 160
Gln Val Val Ser Gln Leu Pro Met Gly Arg Asp Ser Arg Glu Met Leu
165 170 175
Phe Leu Ala Glu Gln Pro Pro Leu Pro Pro Pro Pro Leu Leu Pro Pro
180 185 190
14
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ser Leu Thr Asn Gly Thr Thr Ile Pro Thr Ala Lys Ser Thr Pro Thr
195 200 205
Leu Ile Lys Gln Ser Gln Pro Lys Ser Ala Ser Glu Lys Ser Gln Arg
210 215 220
Ser Lys Lys Ala Lys Glu Leu Lys Pro Lys Val Lys Lys Leu Lys Tyr
225 230 235 240
His Gln Tyr Ile Pro Pro Asp Gln Lys Gln Asp Arg Gly Ala Pro Pro
245 250 255
Met Asp Ser Ser Tyr Ala Lys Ile Leu Gln Gln Gln Gln Leu Phe Leu
260 265 270
Gln Leu Gln Ile Leu Asn Gln Gln Gln Gln Gln His His Asn Tyr Gln
275 280 285
Ala Ile Leu Pro Ala Pro Pro Lys Ser Ala Gly Glu Ala Leu Gly Ser
290 295 300
Ser Gly Thr Pro Pro Val Arg Ser Leu Ser Thr Thr Asn Ser Ser Ser
305 310 315 320
Ser Ser Gly Ala Pro Gly Pro Cys Gly Leu Ala Arg Gln Asn Ser Thr
325 330 335
Ser Leu Thr Gly Lys Pro Gly Ala Leu Pro Ala Asn Leu Asp Asp Met
340 345 350
Lys Val Ala Glu Leu Lys Gln Glu Leu Lys Leu Arg Ser Leu Pro Val
355 360 365
Ser Gly Thr Lys Thr Glu Leu Ile Glu Arg Leu Arg Ala Tyr Gln Asp
370 375 380
Gln Ile Ser Pro Val Pro Gly Ala Pro Lys Ala Pro Ala Ala Thr Ser
385 390 395 400
Ile Leu His Lys Ala Gly Glu Val Val Val Ala Phe Pro Ala Ala Arg
405 410 415
Leu Ser Thr Gly Pro Ala Leu Val Ala Ala Gly Leu Ala Pro Ala Glu
420 425 430
Val Val Val Ala Thr Val Ala Ser Ser Gly Val Val Lys Phe Gly Ser
435 440 445
Thr Gly Ser Thr Pro Pro Val Ser Pro Thr Pro Ser Glu Arg Ser Leu
450 455 460
Leu Ser Thr Gly Asp Glu Asn Ser Thr Pro Gly Asp Thr Phe Gly Glu
465 470 475 480
Met Val Thr Ser Pro Leu Thr Gln Leu Thr Leu Gln Ala Ser Pro Leu
IS
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
485 490 495
Gln Ile Leu Val Lys Glu Glu Gly Pro Arg Ala Gly Ser Cys Cys Leu
500 505 510
Ser Pro Gly Gly Arg Ala,Glu Leu Glu Gly Arg Asp Lys Asp Gln Met
515 520 525
Leu Gln Glu Lys Asp Lys Gln Ile Glu Ala Leu Thr Arg Met Leu Arg
530 535 540
Gln Lys Gln Gln Leu Val Glu Arg Leu Lys Leu Gln Leu Glu Gln Glu
545 550 555 560
Lys Arg Ala Gln Gln Pro Ala Pro Ala Pro Ala Pro Leu Gly Thr Pro
565 570 575
Val Lys Gln Glu Asn Ser Phe Ser Ser Cys Gln Leu Ser Gln Gln Pro
580 585 590
Leu Gly Pro Ala His Pro Phe Asn Pro Ser Leu Ala Ala Pro Ala Thr
595 600 605
Asn His Ile Asp Pro Cys Ala Val Ala Pro Gly Pro Pro Ser Val Val
610 615 620
Val Lys Gln Glu Ala Leu Gln Pro Glu Pro Glu Pro Val Pro Ala Pro
625 630 635. 640
Gln Leu Leu Leu Gly Pro Gln Gly Pro Ser Leu Ile Lys Gly Val Ala
645 650 655
Pro Pro Thr Leu Ile Thr Asp Ser Thr Gly Thr His Leu Val Leu Thr
660 665 670
Val Thr Asn Lys Asn Ala Asp Ser Pro Gly Leu Ser Ser Gly Ser Pro
675 680 685
Gln Gln Pro Ser Ser
690
<210> 5
<211> 35
<212> PRT
<213> Mus musculus
<400> 5
Leu Glu Lys Met Lys Val Ser Asp Leu Lys Gln His Leu Lys Arg Arg
1 5 10 15
Asn Leu Pro Val Ser Gly Pro Lys Pro His Leu Ile Glu Arg Leu Lys
20 25 30
16
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Pro Tyr Leu
<210> 6
<211> 6459
<212> DNA
<213> Mus musculus
<220>
<221> CDS
<222> (1)..(1053)
<400> 6
cag act tca cca caa gca gga atg cag act cag cct cag ata gca act 48
Gln Thr Ser Pro Gln Ala Gly Met Gln Thr Gln Pro Gln Ile Ala Thr
1 5 10 15
get gca caa ata cca act get gcc ttg gcc tca ggc ttg gcc cca act 96
Ala Ala Gln Ile Pro Thr Ala Ala Leu Ala Ser Gly Leu Ala Pro Thr
20 25 30
gta cct cag aca caa gac acg ttc ccg cag cat gtg ctc agt cag cct 144
Val Pro Gln Thr Gln Asp Thr Phe Pro Gln His Val Leu Ser Gln Pro
35 40 45
caa caa gtc aga aag gtt ttc aca aac tca gca tca tca aat aca gtt 192
Gln Gln Val Arg Lys Val Phe Thr Asn Ser Ala Ser Ser Asn Thr Val
50 55 60
ctt cca tat cag aga cat cct gcc cca get gtc cag cag ccc ttt atc 240
Leu Pro Tyr Gln Arg His Pro Ala Pro Ala Val Gln Gln Pro Phe Ile
65 70 75 80
aat aag gcc tcc aac agt gtt ctt caa tcc aga aat get ccg ctt cca 288
Asn Lys Ala Ser Asn Ser Val Leu Gln Ser Arg Asn Ala Pro Leu Pro
85 90 95
tcc ctg caa aat gga cct aac aca ccc aac aag cct agt tca ccc ccg 336
Ser Leu Gln Asn Gly Pro Asn Thr Pro Asn Lys Pro Ser Ser Pro Pro
100 105 110
cca ccc cag caa ttt gtc gtc cag cac tct cta ttt ggg agt cca gtc 384
Pro Pro Gln Gln Phe Val Val Gln His Ser Leu Phe Gly Ser Pro Val
115 120 125
gcc aag aca aaa gat ccc ccc cgc tat gag gag gcc atc aag cag aca 432
Ala Lys Thr Lys Asp Pro Pro Arg Tyr Glu Glu Ala Ile Lys Gln Thr
130 135 140
cgc agc aca cag gcc cct ctg cca gag att tcc aac get cac agt cag 480
Arg Ser Thr Gln Ala Pro Leu Pro Glu Ile Ser Asn Ala His Ser Gln
145 150 155 160
cag atg gat gac ctc ttt gat atc ctc att aag agt gga gag atc tcc 528
17
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Gln Met Asp Asp Leu Phe Asp Ile Leu Ile Lys Ser Gly Glu Ile Ser
165 170 175
ctc ccc ata aaa gaa gaa cct tct cct att tcc aaa atg aga cca gtg 576
Leu Pro Ile Lys Glu Glu Pro Ser Pro Ile Ser Lys Met Arg Pro Val
180 185 190
aca gcc agc atc acc aca atg cca gtg aat aca gtg gtg tcc cgg cca 624
Thr Ala Ser Ile Thr Thr Met Pro Val Asn Thr Val Val Ser Arg Pro
195 200 205
cca ccc caa gtc caa atg gca cca cct gta tct tta gaa cct atg ggc 672
Pro Pro Gln Val Gln Met Ala Pro Pro Val Ser Leu Glu Pro Met Gly
210 215 220
agt tta tct gcc agc tta gag aac caa cta gaa get ttc ttg gat gga 720
Ser Leu Ser Ala Ser Leu Glu Asn Gln Leu Glu Ala Phe Leu Asp Gly
225 230 235 240
act tta ccc tca gcc aat gaa att cct cca cta caa agc agc agt gaa 768
Thr Leu Pro Ser Ala Asn Glu Ile Pro Pro Leu Gln Ser Ser Ser Glu
245 250 255
gac aga gag ccc ttc tct ctg atc gag gac ctc cag aat gat ctg ctg 816
Asp Arg Glu Pro Phe Ser Leu Ile Glu Asp Leu Gln Asn Asp Leu Leu
260 265 270
agt cac tca ggt atg ctg gac cat tca cac tca ccc atg gag act tcc 864
Ser His Ser Gly Met Leu Asp His Ser His Ser Pro Met Glu Thr Ser
275 280 285
gag acc cag ttt get gca ggt act ccc tgt ctg tct ctc gac ctg tca 912
Glu Thr Gln Phe Ala Ala Gly Thr Pro Cys Leu Ser Leu Asp Leu Ser
290 295 300
gac tca aac ttg gac aac atg gag tgg ttg gac att acc atg ccc aac 960
Asp Ser Asn Leu Asp Asn Met Glu Trp Leu Asp Ile Thr Met Pro Asn
305 310 315 320
tcc tct tca gga ctc act cct ctc agc acc acc gcg ccg agc atg ttc 1008
Ser Ser Ser Gly Leu Thr Pro Leu Ser Thr Thr Ala Pro Ser Met Phe
325 330 335
tct get gac ttt cta gac cca cag gac cta ccg ctg cca tgg gac 1053
Ser Ala Asp Phe Leu Asp Pro Gln Asp Leu Pro Leu Pro Trp Asp
340 345 350
taacgtcaca gatttctttt ctgagagttg atgaggttta agaacatgaa gattctaaaa 1113
ggtcagtttt tagagataga tctatagttg cattgttgca atcaaaatat gttgtcacag 1173
aaagaatagg tggaaggtca tagcctggaa cccaagtttg aaaacatttc attgtgttca 1233
gtagtgaatt tctacagttt aacatagcac agggccttct gaaaatcgca cttgtcaaag 1293
18
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
acgactcatc tatttctcca gacttcagta aagaatgaaa agtaccttta gataaaaaca 1353
aagaagagta atatatgcag cacagtgacg ttaggattct ggtaattaac tacatttaaa 1413
tctctggtca ctttaagacc ctaaataaaa ggcagacagc tccactcaaa aactaaggct 1473
gatgtgaggg aggtgagagg tcactgcact tggtacttcc tagagacggc cggagccagg 1533
cccaagacac agcagaggtc aaaggcagtg gagagccttg gccagttcag tgacagcttc 1593
tggctgaaga cttttggttc tattcagaaa cctgtgtcgt ttttttggtg ttgtagttta 1653
ttttgtgatt ttttggaatc gtactttata tttccaaatt taaatttaaa tgcaagatct 1713
ttcaacataa acagaagata cctacaaaat actgtcagaa gtccaggtat actgataaca 1773
ctgaaaattc tattagcaac cttctgggtt ggttagattg attttaatgt atatattaga 1833
catttgtatg tatgctctga cattgtgatt tgtacagcct accaaccaat ctaactaaaa 1893
ttacatatat aatctgtaaa catacatacg agactgtaac actaaacatg tcggatggct 1953
ggggtaagga aatgggtatc caaggtccta cttttttaat agctcgaata tttctagagt 2013
acttgagcca catgtatttc tgtatttaaa gaattgctga ctaactttca ggtaaccaga 2073
cccatctcaa agaaccaaga aaaggcttta gcatgaaata tctttctgag ctggcgagtt 2133
agaagaatgg aaggtaaggg gaaggtctgt catctaccca ggacattccc atgatgagta 2193
caggtcagat tgtgccacaa ggtgggcctc cacgtccctg ccctggccct ctttcttctg 2253
tcactaaccc tggttatcat tttacaggct tgtaactgga tattctacca gagctctcac 2313
tatattgtca agcctaagat tgaaaaactg gaggctttat tagtgttttt atatagaaaa 2373
cagttattac atatgtgtta agtcatttct taagaatttt ctaaaatgcc aactatcaca 2433
ggattatttc aagctagtca ttgaggtata tgacaaaatg taaataacaa aaaactgaaa 2493
tctaccaaaa gagcatggag atttttctta aataataata ttgtgctctc cacctcaccc 2553
ttgtgtaaac cctcaggtca ggttggttcc cctgggtcac aaaatggtaa atgttccata 2613
ctgacatgcc ggaggcagcc tgacccgtta tttggaaaga atttgtgaat tatttctggg 2673
tttgtgtttt gcctaagagc tgatcttatt tctgatttgt gtgtgtgtgg tttataaatt 2733
ttactacgtg taacaaaagt ctgcttttcc agcatgagca cactggagcc ctctgctcac 2793
tggcacttcc tgactcaaag gcactaacag tgctcatgtg ccttccagac ggttcaaggc 2853
agaggccact gtgctcagtg tagtctgtga tgaatagttt aagtgttcag aaattggtaa 2913
accaacacac gggatacaat aactctctgt taaccagaac accgtcattt gaccaagccc 2973
1~
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
tacaagagtt taccacacta gatgttctgt gaatcctcga gtcagttcca ttgagagggg 3033
cctgtctcca gggccaggct atttacttgg gaaagtattt ttcttacagt tcttggccac 3093
agtttttttg ctgagggttt ctgtctgggc ctatcaggtc catacttaga ccctgagcat 3153
cttcttcatt cagatttgaa tggcttatta aattagcacc aaatatcagt gggactgtag 3213
aaggtaaccg aacactagta ccattgactc tcgttgaata actttatatt tccataatcc 3273
tgaattgtgt agatagtttt ctagctctcg ggtcccttta ttgcttttta aaatagggtt 3333
Egaaacatgc cacaggaagg ttgctctcca aaaatacaca gtgcagtgca agaaaatgct 3393
atctcattgg cctactctcc tatgaattgc taaagtgccc acttcacata tgtgtttaaa 3453
cctttataaa ccagtatttc acttttaaaa agacaaggca tctctaccca ttaactctgc 3513
aagccactcc acttgcacca ttccgcttga ccctcctctc tcctggcttg ggtcaccagc 3573
caggcacctg tgacacgagt gctgctctcc aggatctcca ctacatgttc caggttggag 3633
' tgaggacgcg ctatgtgctc acactcatgt gacatgacca aagatgatac tctgtaaaca 3693
aggcccttct gaccggactc agtgcgtgtg atggtgagtg caatgaggaa gggtggatat 3753
tgactaaaga ctggttgttg ttgttgttta ggttagtgtc acagccatta cagcacaagt 3813
caaagtcgcc agttgaattt tcattatgca cgtgtgtggt ttaagcagtg gtactgttgt 3873
atatcatatt gtaaagtatc atactgccaa gaaaataact cctagaaagg cattatctca 3933
catccccatc ttactttcct acatgttctc aaaacacagt agtaccagcc ttacctttct 3993
gctgtatgta gatagtcaga tcatcctact gggggtggga tgaatttaaa agttatacca 4053
aaatttctgt aaagtctttg aaagtctagg agtaggtggt catcttgtac atttcagcaa 4113
agcatccact aggaaacctc ataggacagt gttagtggtt cacgttctag tgtttttcct 4173
gaaatgtgca atctactgta tagtattgcc acataattgt acatagatgt attctgaatt 4233
tgtggaattt cttgcttaga tactattgtg tttgtttcat atgaatattt ttgtaattct 4293
aaaagagatc ttatttaaat ttccttttta aaagccgcat ggttctgtga tccatgtaac 4353
tgacactttt tggctttcag tgctgtttag aaacagtggc aaaggcaggg tggtgctgcc 4413
tgcaagctgc tgcctatgga aggcaaagtc atagtaatga gatagcacct ctgaactgtg 4473
cagtcagcat accctgagtg atggctcagg ggcgcactac ctattttgtc accagagctg 4533
actcaggctt cctggaccct caaccccaga tcattccagg cagcatagct ttcttcacag 4593
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
tcctttcaga attcccaggc tgaaatcagc catagcagtt gacaaaacag ctatccccac 4653
aagtgatgag atagtccctt tactgtcctc aaatggactt ggccttagag acgtggtaaa 4713
gcactttggc agggtttaaa atatttgtga gaagcccaca tttcagtata catcctcact 4773
ggcttccatg cacccacctg acggtagcct cacagaagtc ctggctgtca ctcaggtggg 4833
gagctcatgg tgccgctggg gactttttag agaaatgtaa agagaatagc tattcagtgt 4893
ctactagcag agcaacatgt gtcaatttaa ccaaattcac aaataaccct ccatttttca 4953
atatctgcta ctgtaaacat gaatattgaa tactgacaag agaataccca tacaaatcgt 5013
cccaccgccc tagaggccac agaattagcc caaaattatc aaagaacatt aactggaagg 5073
tcaggttttt caaggaacat agcttacaaa tgcatcagtg tgtatctgga gagcatccta 5133
actgcatttc aactcatctt ttaagtgatt tcagtcaaaa ctggaaaaca actaagatgt 5193
agtaattttt ttttcctggt tcaaaccttc aataacttgc tcattcagca gtctctctga 5253
gctacatttt tatttgtaaa gtgactctgt ctgcatggca atgagcaggt gcgtgttttt 5313
tccacattcg ccctttcttg cagtatccag ggaaacacat cattacaaag ggtttctacc 5373
tgaaatcttt catggaaggc ctacaattcg aaaagctgca catgtttaca gaagagctct 5433
taccctccat gcaaacactt tgctctgtgg tgtcacagct ttgtgacaat aagatggcaa 5493
tctcggatct acaaggtgct gtcgggaatc aaataaaata tgttatcaga gatatcatca 5553
catctatagt gtttaacaga gctttatgcc aactactaag acaaagcttt aacaaagttt 5613
atagaatact gaaactcgta acaattacct ctctacgtga tgctgtaagg aatcttgcta 5673
atttggtagg aagaggaagc atttaggaaa gggtttagta tctaccaaaa gtacttgacc 5733
tcaagtaacc aatagtaatg caaacttgct ttaaaggaac caaaggcatt gccaagtatt 5793
tgccaaaagg aggcactttt tatttaaaat ttgagactaa tgagatctca aaaatcagcc 5853
ccaaaaaggt attatccatt taaggttata attttcacta agatgtagat atttctctta 5913
ttgttttcat gtaaaatagt atagagttgt tttgttggtt taagagtaac attcagtagt 5973
aatacaaagt tttttttcta tgtagaaatt agttttcttt tcttgcttgc aatagaaatg 6033
caatgtgata ggtgtttctc ttcttatttt cattgtcaca ttatgtctta gcatctcact 6093
ttatgaaaaa aaggagaaag ataccaatct acagagccct gcttgttgaa gcactagttt 6153
aatcaacaaa aaattatggc aatcgggggt ccattcatct attgccttta tgttgttttt 6213
taaaagaaaa accatgatgc ctttgtattt gctgtttgca cccctgaaat caattccata 6273
21
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
tcatgtttga atgccataca ttttgcacat gtactgtaca taagtaatgc atactgtatt 6333
tttatatgtg tgcacattta tcatcagatc ttttgtacat agtggcagta ttgtagctga 6393
tcgggaaatg tttgatatct cagcaatttt gcatttttgt gtctcaaata aaagacattt 6453
tgatgt 6459
<210> 7
<211> 351
<212> PRT
<213> Mus musculus
<400> 7
G1n Thr Ser Pro Gln Ala Gly Met Gln Thr Gln Pro Gln Ile Ala Thr
1 5 10 15
Ala Ala Gln Ile Pro Thr Ala Ala Leu Ala Ser Gly Leu Ala Pro Thr
20 25 30
Val Pro Gln Thr Gln Asp Thr Phe Pro Gln His Val Leu Ser Gln Pro
35 40 45
Gln Gln Val Arg Lys Val Phe Thr Asn Ser Ala Ser Ser Asn Thr Val
50 55 60
Leu Pro Tyr Gln Arg His Pro Ala Pro Ala Val Gln Gln Pro Phe Ile
65 70 75 80
Asn Lys Ala Ser Asn Ser Val Leu Gln Ser Arg Asn Ala Pro Leu Pro
85 90 95
Ser Leu Gln Asn Gly Pro Asn Thr Pro Asn Lys Pro Ser Ser Pro Pro
100 105 110
Pro Pro Gln Gln Phe Val Val Gln His Ser Leu Phe Gly Ser Pro Val
115 120 125
Ala Lys Thr Lys Asp Pro Pro Arg Tyr Glu Glu Ala Ile Lys Gln Thr
130 135 140
Arg Ser Thr Gln Ala Pro Leu Pro Glu Ile Ser Asn Ala His Ser Gln
145 150 155 160
Gln Met Asp Asp Leu Phe Asp Ile Leu Ile Lys Ser Gly Glu Ile Ser
165 170 175
Leu Pro Ile Lys Glu Glu Pro Ser Pro Ile Ser Lys Met Arg Pro Val
180 185 190
Thr Ala Ser Ile Thr Thr Met Pro Val Asn Thr Val Val Ser Arg Pro
195 200 205
22
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Pro Pro Gln Val Gln Met Ala Pro Pro Val Ser Leu Glu Pro Met Gly
210 215 220
Ser Leu S°_r Ala Ser Leu Glu Asn Gln Leu Glu Ala Phe Leu Asp Gly
225 230 235 240
Thr Leu Pro Ser Ala Asn Glu Ile Pro Pro Leu Gln Ser Ser Ser Glu
245 250 255
Asp Arg Glu Pro Phe Ser Leu Ile Glu Asp Leu Gln Asn Asp Leu Leu
260 265 270
Ser His Ser Gly Met Leu Asp His Ser His Ser Pro Met Glu Thr Ser
275 280 285
Glu Thr Gln Phe Ala Ala Gly Thr Pro Cys Leu Ser Leu Asp Leu Ser
290 295 300
Asp Ser Asn Leu Asp Asn Met Glu Trp Leu Asp Ile Thr Met Pro Asn
305 310 315 320
Ser Ser Ser Gly Leu Thr Pro Leu Ser Thr Thr Ala Pro Ser Met Phe
325 330 335
Ser Ala Asp Phe Leu Asp Pro Gln Asp Leu Pro Leu Pro Trp Asp
340 345 350
<210> 8
<211> 35
<212> PRT
<213> Mus musculus
<400> 8
Val Thr Lys Met Lys Val Ala Asp Leu Lys Arg Glu Leu Lys Leu Arg
1 5 10 15
Gly Leu Ala Val Asn Gly Asn Lys Thr Glu Leu Gln Asp Arg Leu Gln
20 25 30
Thr Ala Leu
<210> 9
<211> 35
<212> PRT
<213> Mus musculus
<400> 9
Leu Asp Asp Met Lys Val Ala Glu Leu Lys Gln Glu Leu Lys Leu Arg
1 5 10 15
Ser Leu Pro Val Ser Gly Thr Lys Thr Glu Leu Ile Glu Arg Leu Arg
23
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
20 25 30
Ala Tyr Gln
<210> 10
<211> 35
<212> PRT
<213> Mus musculus
<400> 10
Val Lys Lys Leu Lys Val Ser Glu Leu Lys Glu Glu Leu Lys Lys Arg
1 5 10 15
Arg Leu Ser Asp Lys Gly Leu Lys Ala Asp Leu Met Glu Arg Leu Gln
20 25 30
Ala Ala Leu
<210> 11
<211> 35
<212> PRT
<213> Mus musculus
<400> 11
Val Lys Lys Leu Lys Val Ser Glu Leu Lys Glu Glu Leu Lys Lys Arg
1 5 10 15
Arg Leu Ser Asp Lys Gly Leu Lys Ala Glu Leu Met Glu Arg Leu Gln
20 25 30
Ala Ala Leu
<210> 12
<211> 35
<212> PRT
<213> Mus musculus
<400> 12
Val Lys Lys Leu Lys Val Ser Glu Leu Lys Glu Glu Leu Lys Lys Arg
1 5 10 15
Arg Leu Ser Asp Lys Gly Leu Lys Ala Asp Leu Met Asp Arg Leu Gln
20 25 30
Ala Ala Leu
<210> 13
24
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
<211> 35
<212> PRT
<213> Mus musculus
<400> 13
Val Lys Lys Leu Lys Val Ser Glu Leu Lys Glu Glu Leu Lys Lys Arg
1 5 10 15
Arg Leu Ser Asp Lys Gly Leu Lys Ala Asp Leu Met Asp Arg Leu Gln
20 25 30
Ala Ala Leu
<210> 14
<211> 35
<212> PRT
<213> Mus musculus
<400> 14
Met Glu Gln Leu Lys Val Leu Glu Leu Lys Gln Ile Cys Lys Ser Leu
1 5 10 15
Asp Leu Ser Ile Thr Gly Lys Lys Ala Val Leu Gln Asp Arg Ile Lys
20 25 30
Gln Phe Leu
<210> 15
<211> 35
<212> PRT
<213> Mus musculus
<400> 15
Val Lys Lys Leu Lys Val Ser Glu Leu Lys Glu Glu Leu Lys Lys Arg
1 5 10 15
Arg Leu Ser Asp Lys Gly Leu Lys Ala Asp Leu Met Asp Arg Leu Gln
20 25 30
Ala Ala Leu
<210> 16
<211> 35
<212> PRT
<213> Mus musculus
<400> 16
Val Lys Lys Leu Lys Val Ser Glu Leu Lys Glu Glu Leu Lys Lys Arg
1 5 10 15
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Arg Leu Ser Asp Lys Gly Leu Lys Ala Asp Leu Met Asp Arg Leu Gln
20 25 30
Ala Ala Leu
<210> 17
<211> 35
<212> PRT
<213> Mus musculus
<400> 17
Leu Gln Ala Leu Arg Val Thr Asp Leu Lys Ala Ala Leu Glu Gln Arg
1 5 10 15
Gly Leu Ala Lys Ser Gly Gln Lys Ser Ala Leu Val Lys Arg Leu Lys
20 25 30
Gly Ala Leu
<210> 18
<211> 35
<212> PRT
<213> Mus musculus
<400> 18
Leu Gln Ala Leu Arg Val Thr Asp Leu Lys Ala Ala Leu Glu Gln Arg
1 5 10 15
Gly Leu Ala Lys Ser Gly Gln Lys Ser Ala Leu Val Lys Arg Leu Lys
20 25 30
Gly Ala Leu
<210> 19
<211> 35
<212> PRT
<213> Mus musculus
<400> 19
Leu Ser Glu Leu Arg Val Ile Asp Leu Arg Ala Glu Leu Lys Lys Arg
1 5 10 15
Asn Leu Asp Thr Gly Gly Asn Lys Ser Val Leu Met Glu Arg Leu Lys
20 25 30
Lys Ala Val
26
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
<210> 20
<211> 35
<212> PRT
<213> Mus musculus
<400> 20
Leu Ser Asp Leu Arg Val Ile Asp Leu Arg Ala Glu Leu Arg Lys Arg
1 5 10 15
Asn Val Asp Ser Ser Gly Asn Lys Ser Val Leu Met Glu Arg Leu Lys
20 25 30
Lys Ala Ile
<210> 21
<211> 35
<212> PRT
<213> Mus musculus
<400> 21
Val Arg Arg Leu Lys Val Asn Glu Leu Arg Glu Glu Leu Gln Arg Arg
1 5 10 15
Gly Leu Asp Thr Arg Gly Leu Lys Thr Glu Leu Ala Glu Arg Leu Gln
20 25 30
Ala Ala Leu
<210> 22
<211> 35
<212> PRT
<213> Mus musculus
<400> 22
Ile Lys Ala Leu Lys Val Ser Gln Leu Lys Asp Ile Leu Arg Asp Arg
1 5 10 15
Gly Leu Arg Val Ser Gly Lys Lys Ala Asp Leu Leu Asp Asn Leu Thr
20 25 30
Asn Tyr Val
<210> 23
<211> 35
<212> PRT
<213> Mus musculus
<400> 23
27
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ala Asn Lys Leu Lys Val Asp Glu Leu Arg Leu Lys Leu Ala Glu Arg
1 5 10 15
Gly Leu Ser Thr Thr Gly Val Lys Ala Val Leu Val Glu Arg Leu Glu
20 25 30
Glu Ala Ile
<210> 24
<211> 3907
<212> PRT
<213> Mus musculus
<400> 24
Cys Cys Ala Ala Gly Gly Gly Ala Thr Cys Ala Thr Gly Cys Cys Gly
1 5 10 15
Cys Cys Thr Thr Thr Gly Ala Ala Ala Ala Gly Thr Cys Cys Ala Gly
20 25 30
Cys Cys Gly Cys Ala Thr Thr Thr Cys Ala Thr Gly Ala Gly Cys Ala
35 40 45
Gly Ala Gly Ala Ala Gly Gly Ala Gly Cys Thr Thr Gly Gly Ala Gly
50 ~ 55 60
Cys Gly Gly Gly Cys Cys Ala Gly Gly Ala Cys Ala Gly Ala Gly Gly
65 70 75 80
Ala Cys Thr Ala Thr Cys Thr Cys Ala Ala Ala Cys Gly Gly Ala Ala
85 90 95
Gly Ala Thr Thr Cys Gly Thr Thr Cys Cys Cys Gly Gly Cys Cys Gly
100 105 110
Gly Ala Gly Ala Gly Ala Thr Cys Gly Gly Ala Gly Cys Thr Gly Gly
115 120 125
Thr Cys Ala Gly Gly Ala Thr Gly Cys Ala Cys Ala Thr Thr Thr Thr
130 135 140
Gly Gly Ala Ala Gly Ala Gly Ala Cys Cys Thr Cys Gly Gly Cys Thr
145 150 155 160
Gly Ala Gly Cys Cys Ala Thr Cys Cys Cys Thr~Cys Cys Ala Gly Gly
165 170 175
Cys Cys Ala Ala Gly Cys Ala Gly Cys Thr Gly Ala Ala Gly Cys Thr
180 185 190
Gly Ala Ala Gly Ala Gly Ala Gly Cys Cys Ala Gly Ala Cys Thr Ala
195 200 205
28
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Gly Cys Cys Gly Ala Thr Gly Ala Cys Cys Thr Cys Ala Ala Thr Gly
210 215 220
Ala Gly Ala Ala Gly Ala Thr Thr Gly Cys Ala Cys Ala Gly Ala Gly
225 230 235 240
Gly Cys Cys Thr Gly Gly Cys Cys Cys Cys Ala Thr Gly Gly Ala Gly
245 250 255
Cys Thr Gly Gly Thr Gly Gly Ala Gly Ala Ala Gly Ala Ala Cys Ala
260 265 270
Thr Cys Cys Thr Thr Cys Cys Thr Gly Thr Thr Gly Ala Gly Thr Cys
275 280 285
Cys Ala Gly Cys Cys Thr Gly Ala Ala Gly Gly Ala Ala Gly Cys Cys
290 295 300
Ala Thr Cys Ala Thr Thr Gly Thr Gly Gly Gly Cys Cys Ala Gly Gly
305 310 315 320
Thr Gly Ala Ala Cys Thr Ala Thr Cys Cys Cys Ala Ala Ala Gly Thr
325 330 335
Ala Gly Cys A.la Gly Ala Cys Ala Gly Cys Thr Cys Thr Thr Cys Cys
340 345 350
.Thr Thr Cys Gly Ala Thr Gly Ala Gly Gly Ala Cys Ala Gly Cys Ala
355 360 365
Gly Cys Gly Ala Thr Gly Cys Cys Thr Thr Ala Thr Cys Cys Cys Cys
370 375 380
Cys Gly Ala Gly Cys Ala Gly Cys Cys Thr Gly Cys Cys Ala Gly Cys
385 390 395 400
Cys Ala Thr Gly Ala Gly Thr Cys Cys Cys Ala Gly Gly Gly Thr Thr
405 410 415
Cys Thr Gly Thr Gly Cys Cys Gly Thr Cys Ala Cys Cys Cys Cys Thr
420 425 430
Gly Gly Ala Gly Gly Cys Cys Cys Gly Ala Gly Thr Cys Ala Gly Cys
435 440 445
Gly Ala Ala Cys Cys Ala Cys Thr Gly Cys Thr Cys Ala Gly Thr Gly
450 455 460
Cys Cys Ala Cys Cys Thr Cys Thr Gly Cys Ala Thr Cys Cys Cys Cys
465 470 475 480
Cys Ala Cys Cys Cys Ala Gly Gly Thr Thr Gly Thr Gly Thr Cys Thr
485 490 495
Cys Ala Ala Cys Thr Thr Cys Cys Gly Ala Thr Gly Gly Gly Cys Cys
29
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
500 505 510
Gly Gly Gly Ala Thr Thr Cys Cys Ala Gly Ala Gly Ala Ala Ala Thr
515 520 525
Gly Cys Thr Thr Thr Thr Cys Cys Thr Gly Gly Cys Ala Gly Ala Gly
530 535 540
Cys Ala Gly Cys Cys Thr Cys Cys Thr Cys Thr Gly Cys Cys Thr Cys
545 550 555 560
Cys Cys Cys Cys Ala Cys Cys Thr Cys Thr Gly Cys Thr Gly Cys Cys
565 570 575
Thr Cys Cys Cys Ala Gly Cys Cys Thr Cys Ala Cys Cys Ala Ala Thr
580 585 590
Gly Gly Ala Ala Cys Cys Ala Cys Thr Ala Thr Cys Cys Cys Cys Ala
595 600 605
Cys Thr Gly Cys Cys Ala Ala Gly Thr Cys Cys Ala Cys Cys Cys Cys
610 615 620
Cys Ala Cys Ala Cys Thr Cys Ala Thr Thr Ala Ala Gly Cys Ala Ala
625 630 635 640
Ala Gly Cys Cys Ala Ala Cys Cys Cys Ala Ala Gly Thr Cys Thr Gly
645 650 655
Cys Cys Ala Gly Thr Gly Ala Gly Ala Ala Gly Thr Cys Ala Cys Ala
660 665 670
Gly Cys Gly Cys Ala Gly Cys Ala Ala Gly Ala Ala Gly Gly Cys Cys
675 680 685
Ala Ala Gly Gly Ala Gly Cys Thr Gly Ala Ala Gly Cys Cys Ala Ala
690 695 700
Ala Gly Gly Thr Gly Ala Ala Gly Ala Ala Gly Cys Thr Cys Ala Ala
705 710 715 720
Gly Thr Ala Cys Cys Ala Cys Cys Ala Gly Thr Ala Cys Ala Thr Cys
725 730 735
Cys Cys Cys Cys Cys Gly Gly Ala Cys Cys Ala Gly Ala Ala Gly Cys
740 745 750
Ala Gly Gly Ala Cys Ala Gly Gly Gly Gly Gly Gly Cys Ala Cys Cys
755 760 765
Cys Cys Cys Cys Ala Thr Gly Gly Ala Cys Thr Cys Ala Thr Cys Cys
770 775 780
Thr Ala Cys Gly Cys Cys Ala Ala Gly Ala Thr Cys Cys Thr Gly Cys
785 790 795 800
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ala Gly Cys Ala Gly Cys Ala Gly Cys Ala Gly Cys Thr Cys Thr Thr
805 810 815
Cys Cys Thr Cys Cys Ala Gly Cys Thr Gly Cys Ala Gly Ala Thr Cys
820 825 830
Cys Thr Cys Ala Ala Cys Cys Ala Gly Cys Ala Gly Cys Ala Gly Cys
835 840 845
Ala Gly Cys Ala Gly Cys Ala Cys Cys Ala Cys Ala Ala Cys Thr Ala
850 855 860
Cys Cys Ala Gly Gly Cys Cys Ala Thr Cys Cys Thr Gly Cys Cys Thr
865 870 875 880
Gly Cys Cys Cys Cys Gly Cys Cys Ala Ala Ala Gly Thr Cys Ala Gly
885 890 895
Cys Ala Gly Gly Cys Gly Ala Gly Gly Cys Cys Cys Thr Gly Gly Gly
900 905 910
AJ_a Ala Gly Cys Ala Gly Cys Gly Gly Gly Ala Cys Cys Cys Cys Cys
915 920 925
Cys Cys Ala Gly Thr Ala Cys Gly Cys Ala Gly Cys Cys Thr Cys Thr
930 935 940
Cys Cys Ala Cys Thr Ala Cys Cys Ala Ala Thr Ala Gly Cys Ala Gly
945 950 955 960
Cys Thr Cys Cys Ala Gly Cys Thr Cys Gly Gly Gly Cys Gly Cys Cys
965 970 975
Cys Cys Thr Gly Gly Gly Cys Cys Cys Thr Gly Thr Gly Gly Gly Cys
980 985 990
Thr Gly Gly Cys Ala Cys Gly Thr Cys Ala Gly Ala Ala Cys Ala Gly
995 1000 1005
Cys Ala Cys Cys Thr Cys Ala Cys Thr Gly Ala Cys Thr Gly Gly Cys
1010 1015 1020
Ala Ala Gly Cys Cys Gly Gly Gly Ala Gly Cys Cys Cys Thr Gly Cys
1025 1030 1035 1040
Cys Gly Gly Cys Cys Ala Ala Cys Cys Thr Gly Gly Ala Cys Gly Ala
1045 1050 1055
Cys Ala Thr Gly Ala Ala Gly Gly Thr Gly Gly Cys Ala Gly Ala Gly
1060 1065 1070
Cys Thr Gly Ala Ala Gly Cys Ala Gly Gly Ala Gly Cys Thr Gly Ala
1075 1080 1085
31
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ala Gly Thr Thr Gly Cys Gly Ala Thr Cys Ala Cys Thr Gly Cys Cys
1090 1095 1100
Thr Gly Thr Cys Thr Cys Gly Gly Gly Cys Ala Cys Cys Ala Ala Ala
1105 1110 1115 1120
Ala Cys Thr Gly Ala Gly Cys Thr Gly Ala Thr Thr Gly Ala Gly Cys
1125 1130 1135
Gly Cys Cys Thr Thr Cys Gly Ala Gly Cys Cys Thr Ala Thr Cys Ala
1140 1145 1150
Ala Gly Ala Cys Cys Ala Ala Ala Thr Cys Ala Gly Cys Cys Cys Thr
1155 1160 1165
Gly Thr Gly Cys Cys Ala Gly Gly Ala Gly Cys Cys Cys Cys Cys Ala
1170 1175 1180
Ala Gly Gly Cys Cys Cys Cys Thr Gly Cys Cys Gly Cys Cys Ala Cys
1185 1190 1195 1200
Cys Thr Cys Thr Ala Thr Cys Cys Thr Gly Cys Ala Cys Ala Ala Gly
1205 1210 1215
Gly Cys Thr Gly Gly Cys Gly Ala Gly Gly Thr Gly Gly Thr Gly Gly
1220 1225 1230
Thr Ala Gly Cys Cys Thr Thr Cys Cys Cys Ala Gly Cys Gly Gly Cys
1235 1240 1245
Cys Cys Gly Gly Cys Thr Gly Ala Gly Cys Ala Cys Gly Gly Gly Gly
1250 1255 1260
Cys Cys Ala Gly Cys Cys Cys Thr Gly Gly Thr Gly Gly Cys Ala Gly
1265 1270 1275 1280
Cys Ala Gly Gly Cys Cys Thr Gly Gly Cys Thr Cys Cys Ala Gly Cys
1285 1290 1295
Thr Gly Ala Gly Gly Thr Gly Gly Thr Gly Gly Thr Gly Gly Cys Cys
1300 1305 1310
Ala Cys Gly Gly Thr Gly Gly Cys Cys Ala Gly Cys Ala Gly Thr Gly
1315 1320 1325
Gly Gly Gly Thr Gly Gly Thr Gly Ala Ala Gly Thr Thr Thr Gly Gly
1330 1335 1340
Cys Ala Gly Cys Ala Cys Gly Gly Gly Cys Thr Cys Cys Ala Cys Gly
1345 1350 1355 1360
Cys Cys Cys Cys Cys Cys Gly Thr Gly Thr Cys Thr Cys Cys Cys Ala
1365 1370 1375
Cys Cys Cys Cys Cys Thr Cys Gly Gly Ala Gly Cys Gly Cys Thr Cys
32
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
1380 1385 1390
Ala Cys Thr Gly Cys Thr Cys Ala Gly Cys Ala Cys Gly Gly Gly Cys
1395 1400 1405
Gly Ala Thr Gly Ala.Ala Ala Ala Cys Thr Cys Cys Ala Cys Cys Cys
1410 1415 1420
Cys Cys Gly Gly Gly Gly Ala Cys Ala Cys Cys Thr Thr Thr Gly Gly
1425 1430 1435 1440
Thr Gly Ala Gly Ala Thr Gly Gly Thr Gly Ala Cys Ala Thr Cys Ala
1445 1450 1455
Cys Cys Thr Cys Thr Gly Ala Cys Gly Cys Ala Gly Cys Thr Gly Ala
1460 1465 1470
Cys Cys Cys Thr Gly Cys Ala Gly Gly Cys Cys Thr Cys Gly Cys Cys
1475 1480 1485
Ala Cys Thr Gly Cys Ala Gly Ala Thr Cys Cys Thr Cys Gly Thr Gly
1490 1495 1500
Ala Ala Gly Gly Ala Gly Gly Ala Gly Gly Gly Cys Cys Cys Cys Cys
1505 1510 1515 1520
Gly Gly Gly Cys Cys Gly Gly Gly Thr Cys Cys Thr Gly Thr Thr Gly
1525 1530 1535
Cys Cys Thr Gly Ala Gly Cys Cys Cys Thr Gly Gly Gly Gly Gly Gly
1540 1545 1550
Cys Gly Gly Gly Cys Gly Gly Ala Gly Cys Thr Ala Gly Ala Gly Gly
1555 1560 1565
Gly Gly Cys Gly Cys Gly Ala Cys Ala Ala Gly Gly Ala Cys Cys Ala
1570 1575 1580
Gly Ala Thr Gly Cys Thr Gly Cys Ala Gly Gly Ala Gly Ala Ala Ala
1585 1590 1595 1600
Gly Ala Cys Ala Ala Gly Cys Ala Gly Ala Thr Cys Gly Ala Gly Gly
1605 1610 1615
Cys Gly Cys Thr Gly Ala Cys Gly Cys Gly Cys Ala Thr Gly Cys Thr
1620 1625 1630
Cys Cys Gly Gly Cys Ala Gly Ala Ala Gly Cys Ala Gly Cys Ala Gly
1635 1640 1645
Cys Thr Gly Gly Thr Gly Gly Ala Gly Cys Gly Gly Cys Thr Cys Ala
1650 1655 1660
Ala Gly Cys Thr Gly Cys Ala Gly Cys Thr Gly Gly Ala Gly Cys Ala
1665 1670 1675 1680
33
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Gly Gly Ala Gly Ala Ala Gly Cys Gly Ala Gly Cys Cys Cys Ala Gly
1685 1690 1695
Cys Ala Gly Cys Cys Cys Gly Cys Cys Cys Cys Cys Gly Cys Cys Cys
1700 1705 1710
Cys Cys Gly Cys Cys Cys Cys Cys Cys Thr Cys Gly Gly Cys Ala Cys
1715 1720 1725
Cys Cys Cys Cys Gly Thr Gly Ala Ala Gly Cys Ala Gly Gly Ala Gly
1730 1735 1740
Ala Ala Cys Ala Gly Cys Thr Thr Cys Thr Cys Cys Ala Gly Cys Thr
1745 1750 1755 1760
Gly Cys Cys Ala Gly Cys Thr Gly Ala Gly Cys Cys Ala Gly Cys Ala
1765 1770 1775
Gly Cys Cys Cys Cys Thr Gly Gly Gly Cys Cys Cys Cys Gly Cys Thr
1780 1785 1790
Cys Ala Cys Cys Cys Ala Thr Thr Cys Ala Ala Cys Cys Cys Cys Ala
1795 1800 1805
Gly Cys Cys Thr Gly Gly Cys Gly Gly Cys Cys Cys Cys Ala Gly Cys
1810 1815 1820
Cys Ala Cys Cys Ala Ala Cys Cys Ala Cys Ala Thr Ala Gly Ala Cys
1825 1830 1835 1840
Cys Cys Thr Thr Gly Thr Gly Cys Thr Gly Thr Gly Gly Cys Cys Cys
1845 1850 1855
Cys Gly Gly Gly Gly Cys Cys Cys Cys Cys Gly Thr Cys Cys Gly Thr
1860 1865 1870
Gly Gly Thr Gly Gly Thr Gly Ala Ala Gly Cys Ala Gly Gly Ala Ala
1875 1880 1885
Gly Cys Cys Thr Thr Gly Cys Ala Gly Cys Cys Thr Gly Ala Gly Cys
1890 1895 1900
Cys Cys Gly Ala Gly Cys Cys Gly Gly Thr Cys Cys Cys Cys Gly Cys
1905 1910 1915 1920
Cys Cys Cys Cys Cys Ala Gly Thr Thr Gly Cys Thr Thr Cys Thr Gly
1925 1930 1935
Gly Gly Gly Cys Cys Thr Cys Ala Gly Gly Gly Cys Cys Cys Cys Ala
1940 1945 1950
Gly Cys Cys Thr Cys Ala Thr Cys Ala Ala Gly Gly Gly Gly Gly Thr
1955 1960 1965
34
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Thr Gly Cys Ala Cys Cys Thr Cys Cys Cys Ala Cys Cys Cys Thr Cys
1970 1975 1980
Ala Thr Cys Ala Cys Cys Gly Ala Cys Thr Cys Cys Ala Cys Ala Gly
1985 1990 1995 2000
Gly Gly Ala Cys Cys Cys Ala Cys Cys Thr Thr Gly Thr Cys Cys Thr
2005 2010 2015
Cys Ala Cys Cys Gly Thr Gly Ala Cys Cys Ala Ala Thr Ala Ala Gly
2020 2025 2030
Ala Ala Thr Gly Cys Ala Gly Ala Cys Ala Gly Cys Cys Cys Thr Gly
2035 2040 2045
Gly Cys Cys Thr Gly Thr Cys Cys Ala Gly Thr Gly Gly Gly Ala Gly
2050 2055 2060
Cys Cys Cys Cys Cys Ala Gly Cys Ala Gly Cys Cys Cys Thr Cys Gly
2065 2070 2075 2080
Thr Cys Cys Cys Ala Gly Cys Cys Thr Gly Gly Cys Thr Cys Thr Cys
2085 2090 2095
Cys Ala Gly Cys Gly Cys Cys Thr Gly Cys Cys Cys Cys Cys Thr Cys
2100 2105 2110
Thr Gly Cys Cys Cys Ala Gly Ala Thr Gly Gly Ala Cys Cys Thr Gly
2115 2120 2125
Gly Ala Gly Cys A1a Cys Cys Cys Ala Cys Thr Gly Cys Ala Gly Cys
2130 2135 2140
Cys Cys Cys Thr Cys Thr Thr Thr Gly Gly Gly Ala Cys Cys Cys Cys
2145 2150 2155 2160
Cys Ala Cys Thr Thr Cys Thr Cys Thr Gly Cys Thr Gly Ala Ala Gly
2165 2170 2175
Ala Ala Gly Gly Ala Ala Cys Cys Ala Cys Cys Thr Gly Gly Cys Thr
2180 2185 2190
Ala Thr Gly Ala Gly Gly Ala Ala Gly Cys Cys Ala Thr Gly Ala Gly
2195 2200 2205
Cys Cys Ala Gly Cys Ala Gly Cys Cys Cys Ala Ala Ala Cys Ala Gly
2210 2215 2220
Cys Ala Gly Gly Ala Ala Ala Ala Thr Gly Gly Thr Thr Cys Cys Thr
2225 2230 2235 2240
Cys Ala Ala Gly Cys Cys Ala Gly Cys Ala Gly Ala Thr Gly Gly Ala
2245 2250 2255
Cys Gly Ala Cys Cys Thr Gly Thr Thr Thr Gly Ala Cys Ala Thr Thr
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
2260 2265 2270
Cys Thr Cys Ala Thr Thr Cys Ala Gly Ala Gly Cys Gly Gly Ala Gly
2275 2280 2285
Ala Ala Ala Thr Thr Thr Cys Ala Gly Cys Ala Gly Ala Thr Thr Thr
2290 2295 2300
Cys Ala Ala Gly Gly Ala Gly Cys Cys Gly Cys Cys Ala Thr Cys Cys
2305 2310 2315 2320
Cys Thr Gly Cys Cys Ala Gly Gly Gly Ala Ala Gly Gly Ala Gly Ala
2325 2330 2335
Ala Gly Cys Cys Ala Thr Cys Cys Cys Cys Gly Ala Ala Gly Ala Cys
2340 2345 2350
Ala Gly Thr Cys Thr Gly Thr Gly Gly Gly Thr Cys Cys Cys Cys Cys
2355 2360 2365
Cys Thr Gly Gly Cys Ala Gly Cys Ala Cys Ala Gly Cys Cys Ala Thr
2370 2375 2380
Cys Ala Cys Cys Thr Thr Cys Thr Gly Cys Thr Gly Ala Gly Cys Thr
2385 2390 2395 2400
Cys Cys Cys Cys Cys Ala Gly Gly Cys Thr Gly Cys Cys Cys Cys Ala
2405 2410 2415
Cys Cys Thr Cys Cys Thr Cys Cys Ala Gly Gly Cys Thr Cys Ala Cys
2420 2425 2430
Cys Cys Thr Cys Cys Cys Thr Cys Cys Cys Thr Gly Gly Ala Cys Gly
2435 2440 2445
Cys Cys Thr Gly Gly Ala Gly Gly Ala Cys Thr Thr Cys Cys Thr Gly
2450 2455 2460
Gly Ala Gly Ala Gly Cys Ala Gly Cys Ala Cys Gly Gly Gly Gly Cys
2465 2470 2475 2480
Thr Gly Cys Cys Cys Cys Thr Gly Cys Thr Gly Ala Cys Cys Ala Gly
2485 2490 2495
Thr Gly Gly Gly Cys Ala Thr Gly Ala Cys Gly Gly Gly Cys Cys Ala
2500 2505 2510
Gly Ala Gly Cys Cys Cys Cys Thr Thr Thr Cys Cys Cys Thr Cys Ala
2515 2520 2525
Thr Thr Gly Ala Cys Gly Ala Cys Cys Thr Cys Cys Ala Thr Ala Gly
2530 2535 2540
Cys Cys Ala Gly Ala Thr Gly Cys Thr Gly Ala Gly Cys Ala Gly Cys
2545 2550 2555 2560
36
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ala Cys Thr Gly Cys Cys Ala Thr Cys Cys Thr Gly Gly Ala Cys Cys
2565 2570 2575
Ala Cys Cys Cys Cys Cys Cys Gly Thr Cys Ala Cys Cys Cys Ala Thr
2580 2585 2590
Gly Gly Ala Cys Ala Cys Cys Thr Cys Gly Gly Ala Ala Thr Thr Gly
2595 2600 2605
Cys Ala Cys Thr Thr Thr Gly Thr Thr Cys Cys Thr Gly Ala Gly Cys
2610 2615 2620
Cys Cys Ala Gly Cys Ala Gly Cys Ala Cys Cys Ala Thr Gly Gly Gly
2625 2630 2635 2640
Cys Cys Thr Gly Gly Ala Cys Cys Thr Gly Gly Cys Thr Gly Ala Thr
2645 2650 2655
Gly Gly Cys Cys Ala Cys Cys Thr Gly Gly Ala Cys Ala Gly Cys Ala
2660 2665 2670
Thr Gly Gly Ala Cys Thr Gly Gly Cys Thr Gly Gly Ala Gly Cys Thr
2675 2680 2685
Gly Thr Cys Gly Thr Cys Ala Gly Gly Thr Gly Gly Thr Cys Cys Cys
2690 2695 2700
Gly Thr Gly Cys Thr Gly Ala Gly Cys Cys Thr Ala Gly Cys Cys Cys
2705 2710 2715 2720
Cys Cys Cys Thr Cys Ala Gly Cys Ala Cys Cys Ala Cys Ala Gly Cys
2725 2730 2735
Cys Cys Cys Cys Ala Gly Cys Cys Thr Cys Thr Thr Cys Thr Cys Cys
2740 2745 2750
Ala Cys Ala Gly Ala Cys Thr Thr Cys Cys Thr Cys Gly Ala Thr Gly
2755 2760 2765
Gly Cys Cys Ala Thr Gly Ala Thr Thr Thr Gly Cys Ala Gly Cys Thr
2770 2775 2780
Gly Cys Ala Cys Thr Gly Gly Gly Ala Thr Thr Cys Cys Thr Gly Cys
2785 2790 2795 2800
Thr Thr Gly Thr Ala Gly Cys Thr Cys Thr Cys Thr Gly Gly Cys Thr
2805 2810 2815
Cys Ala Ala Gly Ala Cys Gly Gly Gly Gly Thr Gly Gly Gly Gly Ala
2820 2825 2830
Ala Gly Gly Gly Gly Cys Thr Gly Gly Gly Ala Gly Cys Cys Ala Gly
2835 2840 2845
37
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Gly Gly Thr Ala Cys Thr Cys Cys Ala Ala Thr Gly Cys Gly Thr Gly
2850 2855 2860
Gly Cys Thr Cys Thr Cys Cys Thr Gly Cys Gly Thr Gly Ala Thr Thr
2865 2870 2875 2880
Cys Gly Gly Cys Cys Thr Cys Thr Cys Cys Ala Cys Ala Thr Gly Gly
2885 2890 2895
Thr Thr Gly Thr Gly Ala Gly Thr Cys Thr Thr Gly Ala Cys Ala Ala
2900 2905 2910
Thr Cys Ala Cys Ala Gly Cys Cys Cys Cys Thr Gly Cys Thr Thr Thr
2915 2920 2925
Thr Thr Cys Cys Cys Thr Thr Cys Cys Cys Thr Gly Gly Gly Ala Gly
2930 2935 2940
Gly Cys Thr Ala Gly Ala Ala Cys Ala Gly Ala Gly Ala Ala Gly Cys
2945 2950 2955 2960
Cys Cys Thr Thr Ala Cys Thr Cys Cys Thr Gly Gly Thr Thr Cys Ala
2965 2970 2975
Gly Thr Gly Cys Cys Ala Cys Gly Cys Ala Gly Gly Gly Cys Ala Gly
2980 2985 2990
Ala Gly Gly Ala Gly Ala Gly Cys Ala Gly Cys Thr Gly Thr Cys Ala
2995 3000 3005
Ala Gly Ala Ala Gly Cys Ala Gly Cys Cys Cys Thr Gly Gly Cys Thr
3010 3015 3020
Cys Thr Cys Ala Cys Gly Cys Thr Gly Gly Gly Gly Thr Thr Thr Thr
3025 3030 3035 3040
Gly Gly Ala Cys Ala Cys Ala Cys Gly Gly Thr Cys Ala Gly Gly Gly
3045 3050 3055
Thr Cys Ala Gly Gly Gly Cys Cys Ala Thr Thr Thr Cys Ala Gly Cys
3060 3065 3070
Thr Thr Gly Ala Cys Cys Thr Cys Cys Thr Thr Thr Thr Thr Thr Gly
3075 3080 3085
Ala Gly Gly Thr Cys Ala Gly Gly Gly Gly Gly Cys Ala Cys Thr Gly
3090 3095 3100
Thr Cys Thr Gly Thr Cys Thr Gly Gly Cys Thr Ala Cys Ala Ala Thr
3105 3110 3115 3120
Thr Thr Gly Gly Cys Thr Ala Ala Gly Gly Thr Ala Gly Gly Thr Gly
3125 3130 3135
Ala Ala Gly Cys Cys Thr Gly Gly Cys Cys Ala Gly Gly Cys Gly Gly
38
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
3140 3145 3150
Gly Ala Gly Gly Cys Thr Thr Cys Thr Cys Thr Thr Cys Thr Gly Ala
3155 3160 3165
Cys Cys Cys Ala Gly Gly Gly Cys Thr Gly Ala Gly Ala Cys Ala Gly
3170 3175 3180
Gly Thr Thr Ala Ala Gly Gly Gly Gly Thr Gly Ala Ala Thr Cys Thr
3185 3190 3195 3200
Cys Cys Thr Thr Cys Cys Thr Thr Thr Cys Thr Cys Thr Cys Cys Cys
3205 3210 3215
Thr Gly Cys Thr Thr Thr Gly Cys Thr Gly Thr Gly Ala Ala Gly Gly
3220 3225 3230
Gly Ala Gly Ala Ala Ala Thr Thr Ala Gly Cys Cys Thr Gly Gly Gly
3235 3240 3245
Cys Cys Thr Cys Thr Ala Cys Cys Cys Cys Cys Thr Ala Thr Thr Cys
3250 3255 3260
Cys Cys Thr Gly Thr Gly Thr Cys Thr Gly Cys Cys Ala Ala Cys Cys
3265 3270 3275 3280
Cys Cys Ala Gly Gly Ala Thr Cys Cys Cys Ala Gly Gly Gly Cys Thr
3285 3290 3295
Cys Cys Cys Thr Gly Cys Cys Ala Thr Thr Thr Thr Ala Gly Thr Gly
3300 3305 3310
Thr Cys Thr Thr Gly Gly Thr Gly Thr Ala Gly Thr Gly Thr Ala Ala
3315 3320 3325
Cys Cys Ala Thr Thr Thr Ala Gly Thr Gly Gly Thr Thr Gly Gly Thr
3330 3335 3340
Gly Gly Cys Ala Ala Cys Ala Ala Thr Thr Thr Thr Ala Thr Gly Thr
3345 3350 3355 3360
Ala Cys Ala Gly Gly Thr Gly Thr Ala Thr Ala Thr Ala Cys Cys Thr
3365 3370 3375
Cys Thr Ala Thr Ala Thr Thr Ala Thr Ala Thr Ala Thr Cys Gly Ala
3380 3385 3390
Cys Ala Thr Ala Cys Ala Thr Ala Thr Ala Thr Ala Thr Thr Thr Thr
3395 3400 3405
Thr Gly Gly Gly Gly Gly Gly Gly Gly Gly Cys Gly Gly Ala Cys Ala
3410 3415 3420
Gly Gly Ala Gly Ala Thr Gly Gly Gly Thr Gly Cys Ala Ala Cys Thr
3425 3430 3435 3440
39
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Cys Cys Cys Thr Cys Cys Cys Ala Thr Cys Cys Thr Ala Cys Thr Cys
3445 3450 3455
Thr Cys Ala Cys Ala Gly Ala Ala Gly Gly Gly Cys Cys Thr Gly Gly
3460 3465 3470
Ala Thr Gly Cys Ala Ala Gly Gly Thr Thr Ala Cys Cys Cys Thr Thr
3475 3480 3485
Gly Ala Gly Cys Thr Gly Thr Gly Thr Gly Cys Cys Ala Cys Ala Gly
3490 3495 3500
Thr Cys Thr Gly Gly Thr Gly Cys Cys Cys Ala Gly Thr Cys Thr Gly
3505 3510 3515 3520
Gly Cys Ala Thr Gly Cys Ala Gly Cys Thr Ala Cys Cys Cys Ala Gly
3525 3530 3535
Gly Cys Cys Cys Ala Cys Cys Cys Ala Thr Cys Ala Cys Gly Thr Gly
3540 3545 3550
Thr Gly Ala Thr Thr Gly Ala Cys Ala Thr Gly Thr Ala Gly Gly Thr
3555 3560 3565
Ala Cys Cys Cys Thr Gly Cys Cys Ala Cys Gly Gly Cys Cys Thr Ala
3570 3575 3580
Thr Gly Cys Cys Cys Cys Ala Cys Cys Thr Gly Cys Cys Cys Thr Gly
3585 3590 3595 3600
Cys Thr Thr Cys Cys Thr Gly Gly Cys Thr Cys Cys Thr Thr Ala Thr
3605 3610 3615
Cys Ala Gly Thr Gly Cys Cys Ala Thr Gly Ala Gly Gly Gly Cys Ala
3620 3625 3630
Gly Ala Gly Gly Thr Gly Cys Thr Ala Cys Cys Thr Gly Gly Cys Cys
3635 3640 3645
Thr Thr Cys Cys Thr Gly Cys Cys Ala Gly Gly Ala Gly Cys Thr Cys
3650 3655 3660
Thr Cys Cys Ala Cys Cys Cys Ala Cys Thr Cys Ala Cys Ala Thr Thr
3665 3670 3675 3680
Cys Cys Gly Thr Cys Cys Cys Cys Gly Cys Cys Gly Cys Cys Thr Cys
3685 3690 3695
Ala Cys Thr Gly Cys Ala Gly Cys Cys Ala Gly Cys Gly Thr Gly Gly
3700 3705 3710
Thr Cys Cys Thr Ala Gly Gly Ala Cys Ala Gly Gly Ala Gly Gly Ala
3715 3720 3725
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Gly Cys Thr Thr Cys Gly Gly Gly Cys Cys Cys Ala Gly Cys Thr Thr
3730 3735 3740
Cys Ala Cys Cys Cys Thr Gly Cys Gly Gly Thr Gly Gly Gly Gly Cys
3745 3750 3755 3760
Thr Gly Ala Gly Gly Gly Gly Thr Gly Gly Cys Cys Ala Thr Cys Thr
3765 3770 3775
Cys Cys Thr Gly Cys Cys Cys Thr Gly Gly Gly Gly Cys Cys Ala Cys
3780 3785 3790
Thr Gly Gly Cys Thr Thr Cys Ala Cys Ala Thr Thr Cys Thr Gly Gly
3795 3800 3805
Gly Cys Thr Gly Ala Cys Thr Cys Ala Thr Ala Gly Gly Gly Gly Ala
3810 3815 3820
Gly Thr Ala Gly Gly Gly Gly Thr Gly Gly Ala Gly Thr Cys Ala Cys
3825 3830 3835 3840
Cys Ala Ala Ala Ala Cys Cys Ala Gly Thr Gly Cys Thr Gly Gly Gly
3845 3850 3855
Ala Cys Ala Ala Ala Gly Ala Thr Gly Gly Gly Gly Ala Ala Gly Gly
3860 3865 3870
Thr Gly Thr Gly Thr Gly Ala Ala Cys Thr Thr Thr Thr Thr Ala Ala
3875 3880 3885
Ala Ala Thr Ala Ala Ala Cys Ala Cys Ala Ala Ala Ala Ala Cys Ala
3890 3895 3900
Cys Ala Gly
3905
<210> 25
<211> 2424
<212> DNA
<213> Homo Sapiens
<220>
<221> CDS
<222> (1)..(2421)
<400> 25
atg gat tct tcc gtg aaa gag get ata aaa ggt act gag gtg agc ctc 48
Met Asp Ser Ser Val Lys Glu Ala Ile Lys Gly Thr Glu Val Ser Leu
1 5 10 15
tcc aag gca g,ca gat gca ttc gcc ttt gag gat gac agc agt aga gat 96
Ser Lys Ala Ala Asp Ala Phe Ala Phe Glu Asp Asp Ser Ser Arg Asp
20 25 30
41
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
ggg ctc tct cca gat cag get agg agc gag gac ccc cag ggc tct aca 144
Gly Leu Ser Pro Asp Gln Ala Arg Ser Glu Asp Pro Gln Gly Ser Thr
35 40 45
gga tcc acc cca gac atc aaa tcc act gag get cct ctg gac aca atc 192
Gly Ser Thr Pro Asp Ile Lys Ser Thr Glu Ala Pro Leu Asp Thr Ile.
50 55 60
cag gat ctc act cct ggc tca gaa agt gac aag aat gat gca gcc tcc 240
Gln Asp Leu Thr Pro Gly Ser Glu Ser Asp Lys Asn Asp Ala Ala Ser
65 70 75 80
cag cca ggc aac cag tca gac cct ggg aag cag gtt ctc ggc ccc ctc 288
Gln Pro Gly Asn Gln Ser Asp Pro Gly Lys Gln Val Leu Gly Pro Leu
85 90 95
agc acc ccg att cct gtg cac act get gta aag tcc aag tct ttg ggt 336
Ser Thr Pro Ile Pro Val His Thr Ala Val Lys Ser Lys Ser Leu Gly
100 105 110
gac agt aag aac cgc cac aaa aag ccc aaa gac ccc aaa cca aag gtg 384
Asp Ser Lys Asn Arg His Lys Lys Pro Lys Asp Pro Lys Pro Lys Val
115 120 125
aag aag ctc aaa tac cat cag tac atc ccc cca gac cag aag gca gag 432
Lys Lys Leu Lys Tyr His Gln Tyr Ile Pro Pro Asp Gln Lys Ala Glu
130 135 140
aac tct ccc cca ccc atg gac tct gcc tat gcc cgg ctc ctc cag caa 480
Asn Ser Pro Pro Pro Met Asp Ser Ala Tyr Ala Arg Leu Leu Gln Gln
145 150 155 160
cag cag cta ttc ctg cag cta cag atc ctc agc cag cag cag caa cag 528
Gln Gln Leu Phe Leu Gln Leu Gln Ile Leu Ser Gln Gln Gln Gln Gln
165 170 175
cag cag caa cag cag cag cag caa cag cag cag cag cag cag cag cag 576
Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln
180 185 190
cgg ttc agc tac cct ggg atg cac caa aca cac ctc aaa gaa cca aat 624
Arg Phe Ser Tyr Pro Gly Met His Gln Thr His Leu Lys Glu Pro Asn
195 200 205
gaa cag atg gcc aga aat ccg aat cct tct tca aca cca ctg agc aat 672
Glu Gln Met Ala Arg Asn Pro Asn Pro Ser Ser Thr Pro Leu Ser Asn
210 215 220
acc cct cta tcc cct gtc aaa aat agv att tct gga caa act ggt gtt 720
Thr Pro Leu Ser Pro Val Lys Asn Xaa Ile Ser Gly Gln Thr Gly Val
225 230 235 240
tct tct ctc aaa cca ggc ccc ctc cca ccc aac ctg gat gat ctc aag 768
Ser Ser Leu Lys Pro Gly Pro Leu Pro Pro Asn Leu Asp Asp Leu Lys
245 250 255
42
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
gtg tca gag tta aga caa cag ctt cga atc cgg ggc ttg cca gtg tca 816
Val Ser Glu Leu Arg Gln Gln Leu Arg Ile Arg Gly Leu Pro Val Ser
260 265 270
ggc acc aag aca gcg ctg gtg gac cgg ctt cgt ccc ttc cag gat tgt 864
Gly Thr Lys Thr Ala Leu Val Asp Arg Leu Arg Pro Phe Gln Asp Cys
275 280 285
get ggc aac cct gtg ccc aac ttt ggg gac atc aca act gtc acc ttt 912
Ala Gly Asn Pro Val Pro Asn Phe Gly Asp Ile Thr Thr Val Thr Phe
290 295 300
cct gtc acg ccc aac acc ttg ccc agt tat cag tcc tcc ccg aca ggc 960
Pro Val Thr Pro Asn Thr Leu Pro Ser Tyr Gln Ser Ser Pro Thr Gly
305 310 315 320
ttc tac cac ttt ggc agc aca agc tcc agc cca ccc atc tcc ccc gcc 1008
Phe Tyr His Phe Gly Ser Thr Ser Ser Ser Pro Pro Ile Ser Pro Ala
325 330 335
tca tct gac ttg tcc get gca ggg tcc ctg cca gac acc ttc acc gat 1056
Ser Ser Asp Leu Ser Ala Ala Gly Ser Leu Pro Asp Thr Phe Thr Asp
340 345 350
gcg tca cct ggc ttc ggc ctg cac gca tct ccg gtg ccc gcc tgc acg 1104
Ala Ser Pro Gly Phe Gly Leu His Ala Ser Pro Val Pro Ala Cys Thr
355 360 365
gac gag agt ctg ctg agc agc ctg aat ggg ggc tcg ggc ccc tcc gag 1152
Asp Glu Ser Leu Leu Ser Ser Leu Asn Gly Gly Ser Gly Pro Ser Glu
370 375 380
cct gat ggg cta gac tct gag aag gac aag atg ctg gtg gag aag cag 1200
Pro Asp Gly Leu Asp Ser Glu Lys Asp Lys Met Leu Val Glu Lys Gln
385 390 395 400
aaa gtg atc aac cag ctc acc tgg aag ctg cgg caa gag cag cgg cag 1248
Lys Val Ile Asn Gln Leu Thr Trp Lys Leu Arg Gln Glu Gln Arg Gln
405 410 415
gtg gaa gag ctg aga atg caa ctg cag aag cag aag agc agc tgc agc 1296
Val Glu Glu Leu Arg Met Gln Leu Gln Lys Gln Lys Ser Ser Cys Ser
420 425 430
gac cag aag cca ctc ccc ttc ttg gcc acc acc atc aaa cag gaa gat 1344
Asp Gln Lys Pro Leu Pro Phe Leu Ala Thr Thr Ile Lys Gln Glu Asp
435 440 445
gtc tcc agc tgc ccc ttc gca ccc cag cag gcg tct ggg aag gga cag 1392
Val Ser-Ser Cys Pro Phe Ala Pro Gln Gln Ala Ser Gly Lys Gly Gln
450 455 460
ggc cac agc tct gac agt ccc cct ccg get tgt gag acg get cag ctg 1440
Gly His Ser Ser Asp Ser Pro Pro Pro Ala Cys Glu Thr Ala Gln Leu
43
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
465 470 475 480
ctg cct cac tgt gtg gag tcc tca ggt caa acc cat gta ctc tcg tcc 1488
Leu Pro His Cys Val Glu Ser Ser Gly Gln Thr His Val Leu Ser Ser
485 490 495
acg ttt ctc agc ccc cag tgc tcc cct cag cac tcg ccc cgt ggg ggc 1536
Thr Phe Leu Ser Pro Gln Cys Ser Pro Gln His Ser Pro Arg Gly Gly
500 505 510
ctg aag agc ccg cag cac atc agc ctg cct cca tca ccc aac aac cat 1584
Leu Lys Ser Pro Gln His Ile Ser Leu Pro Pro Ser Pro Asn Asn His
515 520 525
tac ttc ctg get tcc tct tcg gga get cag aga gag aac cat ggg gtc 1632
Tyr Phe Leu Ala Ser Ser Ser Gly Ala Gln Arg Glu Asn His Gly Val
530 535 540
tct tca ccc agc agc agc caa ggg tgc gca cag atg act ggt tta caa 1680
Ser Ser Pro Ser Ser Ser Gln Gly Cys Ala Gln Met Thr Gly Leu Gln
545 550 555' 560
tct tct gac aag gtg ggg cca acg ttt tca att cca tcc cca act ttt 1728
Ser Ser Asp Lys Val Gly Pro Thr Phe Ser Ile Pro Ser Pro Thr Phe
565 570 575
tct aag tca agt tca gca gtt tca gat atc acc cag ccc cca tcc tat 1776
Ser Lys Ser Ser Ser Ala Val Ser Asp Ile Thr Gln Pro Pro Ser Tyr
580 585 590
gaa gat gca gtg aag cag caa atg act cgg agt cag cag atg gac gaa 1824
Glu Asp Ala Val Lys Gln Gln Met Thr Arg Ser Gln Gln Met Asp Glu
595 600 605
ctc ctg gat gtc ctc att gaa agt gga gaa atg cca gcc gat gcc agg 1872
Leu Leu Asp Val Leu Ile Glu Ser Gly Glu Met Pro Ala Asp Ala Arg
610 615 620
gaa gat cat tca tgt ctt cag aaa att cca aag atc cct ggg tcc tcc 1920
Glu Asp His Ser Cys Leu Gln Lys Ile Pro Lys Ile Pro Gly Ser Ser
625 630 635 640
tgc agc cca act gcc atc ccc ccg aag ccc tcg get tcc ttt gag cag 1968
Cys Ser Pro Thr Ala Ile Pro Pro Lys Pro Ser Ala Ser Phe Glu Gln
645 650 655
gca tct tcg gga ggc cag atg gcc ttc gat cac tac gca aac gac agt 2016
Ala Ser Ser Gly Gly Gln Met Ala Phe Asp His Tyr Ala Asn Asp Ser
660 665 670
gac gaa cac ctg gaa gtc tta ttg aat tct cac agc ccc atc gga aag 2064
Asp Glu His Leu Glu Val Leu Leu Asn Ser His Ser Pro Ile Gly Lys
675 680 685
gtg agc gat gtt acc ctc ctc aaa atc gga agc gag gag cct cct ttt 2112
44
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Val Ser Asp Val Thr Leu Leu Lys Ile Gly Ser Glu Glu Pro Pro Phe
690 695 700
gac agc atc atg gat ggc ttc cca ggg aag get gcg gaa gat ctc ttc 2160
Asp Ser Ile Met Asp Gly Phe Pro Gly Lys Ala Ala Glu Asp Leu Phe
705 710 715 720
agt get cac gag ctc ttg cct ggg ccc ctc tcc ccg atg cat gca cag 2208
Ser Ala His Glu Leu Leu Pro Gly Pro Leu Ser Pro Met His Ala Gln
725 730 735
ttg tca cct cct tct gtg gac agc agt ggt ctg cag ctg agc tta ccg 2256
Leu Ser Pro Pro Ser Val Asp Ser Ser Gly Leu Gln Leu Ser Leu Pro
740 745 750
gaa tct cct tgg gaa aca atg gaa tgg ctg gac ctc act cca cct agt 2304
Glu Ser Pro Trp Glu Thr Met Glu Trp Leu Asp Leu Thr Pro Pro Ser
755 760 765
tcc acg cca ggc ttc agc aac ctt acc tcc agt ggg ccc agc att ttc 2352
Ser Thr Pro Gly Phe Ser Asn Leu Thr Ser Ser Gly Pro Ser Ile Phe
770 775 780
aac atc gat ttt ctg gat gtt aca gat ctt aat ctg aat tcc cct atg 2400
Asn Ile Asp Phe Leu Asp Val Thr Asp Leu Asn Leu Asn Ser Pro Met
785 790 795 800
gat ctc cac tta cag cag tgg taa 2424
Asp Leu His Leu Gln Gln Trp
805
<210> 26
<211> 807
<212> PRT
<213> Homo sapiens
<400> 26
Met Asp Ser Ser Val Lys Glu Ala Ile Lys Gly Thr Glu Val Ser Leu
1 5 10 15
Ser Lys Ala Ala Asp Ala Phe Ala Phe Glu Asp Asp Ser Ser Arg Asp
20 25 30
Gly Leu Ser Pro Asp Gln Ala Arg Ser Glu Asp Pro Gln Gly Ser Thr
35 40 45
Gly Ser Thr Pro Asp Ile Lys Ser Thr Glu Ala Pro Leu Asp Thr Ile
50 55 60
Gln Asp Leu Thr Pro Gly Ser Glu Ser Asp Lys Asn Asp Ala Ala Ser
65 70 75 80
Gln Pro Gly Asn Gln Ser Asp Pro Gly Lys Gln Val Leu Gly Pro Leu
85 90 95
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ser Thr Pro Ile Pro Val His Thr Ala Val Lys Ser Lys Ser Leu Gly
100 105 110
Asp Ser Lys Asn Arg His Lys Lys Pro Lys Asp Pro Lys Pro Lys Val
115 120 125
Lys Lys Leu Lys Tyr His Gln Tyr Ile Pro Pro Asp Gln Lys Ala Glu
130 135 140
Asn Ser Pro Pro Pro Met Asp Ser Ala Tyr Ala Arg Leu Leu Gln Gln
145 150 155 160
Gln Gln Leu Phe Leu Gln Leu Gln Ile Leu Ser Gln Gln Gln Gln Gln
165 170 175
Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln
180 185 190
Arg Phe Ser Tyr Pro Gly Met His Gln Thr His Leu Lys Glu Pro Asn
195 200 205
Glu Gln Met Ala Arg Asn Pro Asn Pro Ser Ser Thr Pro Leu Ser Asn
210 215 220
Thr Pro Leu Ser Pro Val Lys Asn Xaa Ile Ser Gly Gln Thr Gly Val
225 230 235 240
Ser Ser Leu Lys Pro Gly Pro Leu Pro Pro Asn Leu Asp Asp Leu Lys
245 250 255
Val Ser Glu Leu Arg Gln Gln Leu Arg Ile Arg Gly Leu Pro Val Ser
260 265 270
Gly Thr Lys Thr Ala Leu Val Asp Arg Leu Arg Pro Phe Gln Asp Cys
275 280 285
Ala Gly Asn Pro Val Pro Asn Phe Gly Asp Ile Thr Thr Val Thr Phe
290 295 300
Pro Val Thr Pro Asn Thr Leu Pro Ser Tyr Gln Ser Ser Pro Thr Gly
305 310 315 320
Phe Tyr His Phe Gly Ser Thr Ser Ser Ser Pro Pro Ile Ser Pro Ala
325 330 335
Ser Ser Asp Leu Ser Ala Ala Gly Ser Leu Pro Asp Thr Phe Thr Asp
340 345 350
Ala Ser Pro Gly Phe Gly Leu His Ala Ser Pro Val Pro Ala Cys Thr
355 360 365
Asp Glu Ser Leu Leu Ser Ser Leu Asn Gly Gly Ser Gly Pro Ser Glu
370 375 380
46
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Pro Asp Gly Leu Asp Ser Glu Lys Asp Lys Met Leu Val Glu Lys Gln
385 390 395 400
Lys Val Ile Asn Gln Leu Thr Trp Lys Leu Arg Gln Glu Gln Arg Gln
405 410 415
Val Glu Glu Leu Arg Met Gln Leu Gln Lys Gln Lys Ser Ser Cys Ser
420 425 430
Asp Gln Lys Pro Leu Pro Phe Leu Ala Thr Thr Ile Lys Gln Glu Asp
435 440 445
Val Ser Ser Cys Pro Phe Ala Pro Gln Gln Ala Ser Gly Lys Gly Gln
450 455 460
Gly His Ser Ser Asp Ser Pro Pro Pro Ala Cys Glu Thr Ala Gln Leu
465 470 475 480
Leu Pro His Cys Val Glu Ser Ser Gly Gln Thr His Val Leu Ser Ser
485 490 495
Thr Phe Leu Ser Pro Gln Cys Ser Pro Gln His Ser Pro Arg Gly Gly
500 505 510
Leu Lys Ser Pro Gln His Ile Ser Leu Pro Pro Ser Pro Asn Asn His
515 520 525
Tyr Phe Leu Ala Ser Ser Ser Gly Ala Gln Arg Glu Asn His Gly Val
530 535 540
Ser Ser Pro Ser Ser Ser Gln Gly Cys Ala Gln Met Thr Gly Leu Gln
545 550 555 560
Ser Ser Asp Lys Val Gly Pro Thr Phe Ser Ile Pro Ser Pro Thr Phe
565 570 575
Ser Lys Ser Ser Ser Ala Val Ser Asp Ile Thr Gln Pro Pro Ser Tyr
580 585 590
Glu Asp Ala Val Lys Gln Gln Met Thr Arg Ser Gln Gln Met Asp Glu
595 600 605
Leu Leu Asp Val Leu Ile Glu Ser Gly Glu Met Pro Ala Asp Ala Arg
610 615 620
Glu Asp His Ser Cys Leu Gln Lys Ile Pro Lys Ile Pro Gly Ser Ser
625 630 635 640
Cys Ser Pro Thr Ala Ile Pro Pro Lys Pro Ser Ala Ser Phe Glu Gln
645 650 655
Ala Ser Ser Gly Gly Gln Met Ala Phe Asp His Tyr Ala Asn Asp Ser
660 665 670
Asp Glu His Leu Glu Val Leu Leu Asn Ser His Ser Pro Ile Gly Lys
47
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
675 680 685
Val Ser Asp Val Thr Leu Leu Lys Ile Gly Ser Glu Glu Pro Pro Phe
690 695 700
Asp Ser Ile Met Asp Gly Phe Pro Gly,Lys Ala Ala Glu Asp Leu Phe
7U5 710 715 720
Ser Ala His Glu Leu Leu Pro Gly Pro Leu Ser Pro Met His Ala Gln
725 730 735
Leu Ser Pro Pro Ser Val Asp Ser Ser Gly Leu Gln Leu Ser Leu Pro
740 745 750
Glu Ser Pro Trp Glu Thr Met Glu Trp Leu Asp Leu Thr Pro Pro Ser
755 760 765
Ser Thr Pro Gly Phe Ser Asn Leu Thr Ser Ser Gly Pro Ser Ile Phe
770 775 780
Asn Ile Asp Phe Leu Asp Val Thr Asp Leu Asn Leu Asn Ser Pro Met
785 790 795 800
Asp Leu His Leu Gln Gln Trp
805
<210> 27
<211> 3063
<212> DNA
<213> Homo sapiens
<400> 27
gacgtcgcat gctcccggcc gccatggcgg ccgcgggaat tcgattgact cctggagccc 60
gtcagtatcg gcggaattcg cggccgcgtc gacctggctg ccactgtact cctacccagg 120
ggagctcacg gagagttgga tgaattctgg gttgttagct gcggtcagct gggctcccgg 180
gagcctgttg ctggtggaga acagggggcg cctggccaag ggaccagcgg cttgctgaga 240
ctcaacatga cactcctggg gtctgagcat tccttgctga ttaggagcaa gttcagatca 300
gttttacagt taagacttca acaaagaagg acccaggaac aactggctaa ccaaggcata 360
ataccaccac tgaaacgtcc agctgaattc catgagcaaa gaaaacattt ggatagtgac 420
aaggctaaaa attccctgaa gcgcaaagcc agaaacaggt gcaacagtgc cgacttggtt 480
aatatgcaca tactccaagc ttccactgca gagaggtcca ttccaactgc tcagatgaag 540
ctgaaaagag cccgactcgc cgatgatctc aatgaaaaaa ttgctctacg accagggcca 600
ctggagctgg tggaaaaaaa cattcttcct gtggattctg ctgtgaaaga ggccataaaa 660
ggtaaccagg tgagtttctc caaatccacg gatgcttttg cctttgaaga ggacagcagc 720
agcgatgggc tttctccgga tcagactcga agtgaagacc cccaaaactc agcgggatcc 780
ccgccagacg ctaaagcctc agatacccct tcgacaggtt ctctggggac aaaccaggat 840
cttgcttctg gctcagaaaa tgacagaaat gactcagcct cacagcccag ccaccagtca 900
gatgcgggga agcaggggct tggccccccc agcaccccca tagccgtgca tgctgctgta 960
aagtccaaat ccttgggtga cagtaagaac cgccacaaaa agcccaagga ccccaagcca 1020
aaggtgaaga agcttaaata tcaccagtac attcccccag accagaaggc agagaagtcc 1080
cctccaccta tggactcagc ctacgctcgg ctgctccagc aacagcagct gttcctgcag 1140
ctccaaatcc tcagccagca gcagcagcag cagcaacacc gattcagcta cctagggatg 1200
caccaagctc agcttaagga accaaatgaa cagatggtca gaaatccaaa ctcttcttca 1260
48
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
acgccactga gcaatacccc cttgtctcct gtcaaaaaca gtttttctgg acaaactggt 1320
gtctcttctt tcaaaccagg cccactccca cctaacctgg atgatctgaa ggtctctgaa 1380
ttaagacaac agcttcgaat tcggggcttg cctgtgtcag gcaccaaaac ggctctcatg 1440
gaccggcttc gacccttcca ggactgctct ggcaacccag tgccgaactt tggggatata 1500
acgactgtca cttttcctgt cacacccaac acgctgccca attaccagtc ttcctcttct 1560
accagtgccc tgtccaacgg cttctaccac tttggcagca ccagctccag ccccccgatc 1620
tccccagcct cctctgacct gtcagtcgct gggtccctgc cggacacctt caatgatgcc 1680
tccccctcct tcggcctgca cccgtcccca gtccacgtgt gcacggagga aagtctcatg 1740
agcagcctga atgggggctc tgttccttct gagctggatg ggctggactc cgagaaggac 1800
aagatgctgg tggagaagca gaaggtgatc aatgaactca cctggaaact ccagcaagag 1860
cagaggcagg tggaggagct gaggatgcag cttcagaagc agaaaaggaa taactgttca 1920
gagaagaagc cgctgccttt cctggctgcc tccatcaagc aggaagaggc tgtctccagc 1980
tgtccttttg catcccaagt acctgtgaaa agacaaagca gcagctcaga gtgtcaccca 2040
ccggcttgtg aagctgctca actccagcct cttggaaatg ctcattgtgt ggagtcctca 2100
gatcaaacca atgtactttc ttccacattt ctcagccccc agtgttcccc tcagcattca 2160
ccgctggggg ctgtgaaaag cccacagcac atcagtttgc ccccatcacc caacaaccct 2220
cactttctgc cctcatcctc cggggcccag ggagaagggc acagggtctc ctcgcccatc 2280
agcagccagg tgtgcactgc acagatggct ggtttacact cttctgataa ggtggggcca 2340
aagttttcaa ttccatcccc aactttttct aagtcaagtt cagcaatttc agaggtaaca 2400
cagcctccat cctatgaaga tgccgtaaag cagcaaatga cccggagtca gcagatggat 2460
gaactcctgg acgtgcttat tgaaagcgga gaaatgccag cagacgctag agaggatcac 2520
tcatgtcttc aaaaagtccc aaagataccc agatcttccc gaagtccaac tgctgtcctc 2580
accaagccct cggcttcctt tgaacaagcc tcttcaggca gccagatccc ctttgatccc 2640
tatgccaccg acagtgatga gcatcttgaa gtcttattaa attcccagag ccccctagga 2700
aagatgagtg atgtcaccct tctaaaaatt gggagcgaag agcctcactt tgatgggata 2760
atggatggat tctctgggaa ggctgcagaa gacctcttca atgcacatga gatcttgcca 2820
ggccccctct ctccaatgca gacacagttt tcaccctctt ctgtggacag caatgggctg 2880
cagttaagct tcactgaatc tccctgggaa accatggagt ggctggacct cactccgcca 2940
aattccacac caggctttag cgccctcacc accagcagcc ccagcatctt caacatcgat 3000
ttcctggatg tcactgatct caatttgaat tcttccatgg accttcactt gcagcagtgg 3060
tag 3063
<210> 28
<211> 938
<212> PRT
<213> Homo Sapiens
<400> 28
Met Thr Leu Leu Gly Ser Glu His Ser Leu Leu Ile Arg Ser Lys Phe
1 5 10 15
Arg Ser Val Leu Gln Leu Arg Leu Gln Gln Arg Arg Thr Gln Glu Gln
20 25 30
Leu Ala Asn Gln Gly Ile Ile Pro Pro Leu Lys Arg Pro Ala Glu Phe
35 40 45
His Glu Gln Arg Lys His Leu Asp Ser Asp Lys Ala Lys Asn Ser Leu
50 55 60
Lys Arg Lys Ala Arg Asn Arg Cys Asn Ser Ala Asp Leu Val Asn Met
65 70 75 80
His Ile Leu Gln Ala Ser Thr Ala Glu Arg Ser Ile Pro Thr Ala Gln
49
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
85 90 95
Met Lys Leu Lys Arg Ala Arg Leu Ala Asp Asp Leu Asn Glu Lys Ile
100 105 110
Ala Leu Arg,Pro Gly Pro Leu Glu Leu Val Glu Lys Asn Ile Leu Pro
115 120 125
Val Asp Ser Ala Val Lys Glu Ala Ile Lys Gly Asn Gln Val Ser Phe
130 135 140
Ser Lys Ser Thr Asp Ala Phe Ala Phe Glu Glu Asp Ser Ser Ser Asp
145 150 155 160
Gly Leu Ser Pro Asp Gln Thr Arg Ser Glu Asp Pro Gln Asn Ser Ala
165 170 175
Gly Ser Pro Pro Asp Ala Lys Ala Ser Asp Thr Pro Ser Thr Gly Ser
180 185 190
Leu Gly Thr Asn Gln Asp Leu Ala Ser Gly Ser Glu Asn Asp Arg Asn
195 200 205
Asp Ser Ala Ser Gln Pro Ser His Gln Ser Asp Ala Gly Lys Gln Gly
210 215 220
Leu Gly Pro Pro Ser Thr Pro Ile Ala Val His Ala Ala Val Lys Ser
225 230 235 240
Lys Ser Leu Gly Asp Ser Lys Asn Arg His Lys Lys Pro Lys Asp Pro
245 250 255
Lys Pro Lys Val Lys Lys Leu Lys Tyr His Gln Tyr Ile Pro Pro Asp
260 265 270
Gln Lys Ala Glu Lys Ser Pro Pro Pro Met Asp Ser Ala Tyr Ala Arg
275 280 285
Leu Leu Gln Gln Gln Gln Leu Phe Leu Gln Leu Gln Ile Leu Ser Gln
290 295 300
Gln Gln Gln Gln Gln Gln His Arg Phe Ser Tyr Leu Gly Met His Gln
305 310 315 320
Ala Gln Leu Lys Glu Pro Asn Glu Gln Met Val Arg Asn Pro Asn Ser
325 330 335
Ser Ser Thr Pro Leu Ser Asn Thr Pro Leu Ser Pro Val Lys Asn Ser
340 345 350
Phe Ser Gly Gln Thr Gly Val Ser Ser Phe Lys Pro Gly Pro Leu Pro
355 360 365
Pro Asn Leu Asp Asp Leu Lys Val Ser Glu Leu Arg Gln Gln Leu Arg
370 375 380
$~
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ile Arg Gly Leu Pro Val Ser Gly Thr Lys Thr Ala Leu Met Asp Arg
385 390 395 400
Leu Arg Pro Phe Gln Asp Cys Ser Gly Asn Pro Val Pro Asn Phe Gly
405 410 415
Asp Ile Thr Thr Val Thr Phe Pro Val Thr Pro Asn Thr Leu Pro Asn
420 425 430
Tyr Gln Ser Ser Ser Ser Thr Ser Ala Leu Ser Asn Gly Phe Tyr His
435 440 445
Phe Gly Ser Thr Ser Ser Ser Pro Pro Ile Ser Pro Ala Ser Ser Asp
450 455 460
Leu Ser Val Ala Gly Ser Leu Pro Asp Thr Phe Asn Asp Ala Ser Pro
465 470 475 480
Ser Phe Gly Leu His Pro Ser Pro Val His Val Cys Thr Glu Glu Ser
485 490 495
Leu Met Ser Ser Leu Asn Gly Gly Ser Val Pro Ser Glu Leu Asp Gly
500 505 510
Leu Asp Ser Glu Lys Asp Lys Met Leu Val Glu Lys Gln Lys Val Ile
515 520 525
Asn Glu Leu Thr Trp Lys Leu Gln Gln Glu Gln Arg Gln Val Glu Glu
530 535 540
Leu Arg Met Gln Leu Gln Lys Gln Lys Arg Asn Asn Cys Ser Glu Lys
545 550 555 560
Lys Pro Leu Pro Phe Leu Ala Ala Ser Ile Lys Gln Glu Glu Ala Val
565 570 575
Ser Ser Cys Pro Phe Ala Ser Gln Val Pro Val Lys Arg Gln Ser Ser
580 585 590
Ser Ser Glu Cys His Pro Pro Ala Cys Glu Ala Ala Gln Leu Gln Pro
595 600 605
Leu Gly Asn Ala His Cys Val Glu Ser Ser Asp Gln Thr Asn Val Leu
610 615 620
Ser Ser Thr Phe Leu Ser Pro Gln Cys Ser Pro Gln His Ser Pro Leu
625 630 635 640
Gly Ala Val Lys Ser Pro Gln His Ile Ser Leu Pro Pro Ser Pro Asn
645 650 655
Asn Pro His Phe Leu Pro Ser Ser Ser Gly Ala Gln Gly Glu Gly His
660 665 670
$1
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Arg Val Ser Ser Pro Ile Ser Ser Gln Val Cys Thr Ala Gln Met Ala
675 680 685
Gly Leu His Ser Ser Asp Lys Val Gly Pro Lys Phe Ser Ile Pro Ser
690 695 700
Pro Thr Phe Ser Lys Ser Ser Ser Ala Ile Ser Glu Val Thr Gln Pro
705 710 715 720
Pro Ser Tyr Glu Asp Ala Val Lys Gln Gln Met Thr Arg Ser Gln Gln
725 730 735
Met Asp Glu Leu Leu Asp Val Leu Ile Glu Ser Gly Glu Met Pro Ala
740 745 750
Asp Ala Arg Glu Asp His Ser Cys Leu Gln Lys Val Pro Lys Ile Pro
755 760 765
Arg Ser Ser Arg Ser Pro Thr Ala Val Leu Thr Lys Pro Ser Ala Ser
770 775 780
Phe Glu Gln Ala Ser Ser Gly Ser Gln Ile Pro Phe Asp Pro Tyr Ala
785' 790 795 800
Thr Asp Ser Asp Glu His Leu Glu Val Leu Leu Asn Ser Gln Ser Pro
805 810 815
Leu Gly Lys Met Ser Asp Val Thr Leu Leu Lys Ile Gly Ser Glu Glu
820 825 830
Pro His Phe Asp Gly Ile Met Asp Gly Phe Ser Gly Lys Ala Ala Glu
835 840 845
Asp Leu Phe Asn Ala His Glu Ile Leu Pro Gly Pro Leu Ser Pro Met
850 855 860
Gln Thr Gln Phe Ser Pro Ser Ser Val Asp Ser Asn Gly Leu Gln Leu
865 870 875 880
Ser Phe Thr Glu Ser Pro Trp Glu Thr Met Glu Trp Leu Asp Leu Thr
885 890 895
Pro Pro Asn Ser Thr Pro Gly Phe Ser Ala Leu Thr Thr Ser Ser Pro
900 905 910
Ser Ile Phe Asn Ile Asp Phe Leu Asp Val Thr Asp Leu Asn Leu Asn
915 920 925
Ser Ser Met Asp Leu His Leu Gln Gln Trp
930 935
<210> 29
<211> 4960
<212> DNA
52
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
<213> Mus musculus
<400> 29
ggaattcggc acgaggccac cctcagagga ggagggtcct gcctgctggg agttaattag 60
cctcgcgagc ggcgaggggg gaggcgccag ttttctgggg acactggcgg ccactgtgcg 120
tcctcctacc caagggagct ccccaagagt tggatgaatt ctgggttgtt agctgctgtc 180
ctctgggctc ccgggagcca gtttctggtg gaaagcgggg cgcctggcca acgaccagcg 240
gcttgctgag actcaccatg acactcctgg ggtctgaaca ctctttgctg attagaagga 300
agttccgatc agtcttacag ttacggcttc aacagagaag gacccaggag cagctggcta 360
accaaggctt aataccgcca ctgaaaggtc caactgaatt ccatgacccg agaaaacaat 420
tggatagtgc caagactgaa gattccctga ggcgcaaggg cagaaacagg tccgaccgtg 480
ccagcctggt tactatgcac attctccaag cctccacggc agaaaggtcc attccaactg 540
ctcagatgaa gctcaaaaga gcccgccttg cagatgacct caatgagaag atcgctctcc 600
gccaagggcc cttggaactg gtggagaaga acattctgcc gatggattct tccgtgaaag 660
aggctataaa aggtactgag gtgagcctct ccaaggcagc agatgcattc gcctttgagg 720
atgacagcag tagagatggg ctctctccag atcaggctag gagcgaggac ccccagggct 780
ctacaggatc caccccagac atcaaatcca ctgaggctcc tctggacaca atccaggatc 840
tcactcctgg ctcagaaagt gacaagaatg atgcagcctc ccagccaggc aaccagtcag 900
accctgggaa gcaggttctc ggccccctca gcaccccgat tcctgtgcac actgctgtaa 960
agtccaagtc tttgggtgac agtaagaacc gccacaaaaa gcccaaagac cccaaaccaa 1020
aggtgaagaa gctcaaatac catcagtaca tccccccaga ccagaaggca gagaagtctc 1080
ccccacccat ggactctgcc tatgcccggc tgctccagca acagcagcta ttcctgcagc 1140
tacagatcct cagccagcag cagcaacagc agcagcaaca gcagcagcag caacagcagc 1200
agcagcagca gcagcagcgg ttcagctacc ctgggatgca ccaaacacac ctcaaagaac 1260
caaatgaaca gatggccaga aatccgaatc cttcttcaac accactgagc aatacccctc 1320
tatcccctgt caaaaatagc atttctggac aaactggtgt ttcttctctc aaaccaggcc 1380
ccctcccacc caacctggat gatctcaagg tgtcagagtt aagacaacag cttcgaatcc 1440
ggggcttgcc agtgtcaggc accaagacag cgctggtgga ccggcttcgt cccttccagg 1500
attgtgctgg caaccctgtg cccaactttg gggacatcac aactgtcacc tttcctgtca 1560
cgcccaacac cttgcccagt tatcagtcct ccccgacagg cttctaccac tttggcagca 1620
caagctccag cccacccatc tcccccgcct catctgactt gtccgctgca gggtccctgc 1680
cagacacctt caccgatgcg tcacctggct tcggcctgca cgcatctccg gtgcccgcct 1740
gcacggacga gagtctgctg agcagcctga atgggggctc gggcccctcc gagcctgatg 1800
ggctagactc tgagaaggac aagatgctgg tggagaagca gaaagtgatc aaccagctca 1860
cctggaagct gcggcaagag cagcggcagg tggaagagct gagaatgcaa ctgcagaagc 1920
agaagagcag ctgcagcgac cagaagccac tgcccttctt ggccaccacc atcaaacagg 1980
aagatgtctc cagctgcccc ttcgcacccc agcaggcgtc tgggaaggga cagggccaca 2040
gctctgacag tccccctccg gcttgtgaga cggctcagct gctgcctcac tgtgtggagt 2100
cctcaggtca aacccatgta ctctcgtcca cgtttctcag cccccagtgc tcccctcagc 2160
actcgcccct ggggggcctg aagagcccgc agcacatcag cctgcctcca tcacccaaca 2220
accattactt cctggcttcc tcttcgggag ctcagagaga gaaccatggg gtctcttcac 2280
ccagcagcag ccaagggtgc gcacagatga ctggtttaca atcttctgac aaggtggggc 2340
caacgttttc aattccatcc ccaacttttt ctaagtcaag ttcagcagtt tcagatatca 2400
cccagccccc atcctatgaa gatgcagtga agcagcaaat gactcggagt cagcagatgg 2460
acgaactcct ggatgtcctc attgaaagtg gagaaatgcc agccgatgcc agggaagatc 2520
attcatgtct tcagaaaatt ccaaagatcc ctgggtcctc ctgcagccca actgccatcc 2580
ccccgaagcc ctcggcttcc tttgagcagg catcttcggg aggccagatg gccttcgatc 2640
actacgccaa cgacagtgac gaacacctgg aagtcttatt gaattctcac agccccatcg 2700
gaaaggtgag cgatgttacc ctcctcaaaa tcggaagcga ggagcctcct tttgacagca 2760
tcatggatgg cttcccaggg aaggctgcgg aagatctctt cagtgctcac gagctcttgc 2820
ctgggcccct ctccccgatg catgcacagt tgtcacctcc ttctgtggac agcagtggtc 2880
tgcagctgag cttcacggaa tctccttggg aaacaatgga atggctggac ctcactccac 2940
ctagttccac gccaggcttc agcaacctta cctccagtgg gcccagcatt ttcaacatcg 3000
attttctgga tgttacagat cttaatctga attcccctat ggatctccac ttacagcagt 3060
ggtaaacacc cgaggtacaa gagctacgag agctcagtgg gaattcaatg gaggaaagca 3120
53
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
cgataccgga aatgtgtgtt ccaaaagatg aaggggggaa aatggggagg gaaaaaaaaa 3180
aacagcaacg gaggtttttg tgacaactaa ccagaacaaa cagaagtcag ctattaaaat 3240
atgtctaaat gtaatatcta ccagcattca gtaactgtta ataacttcag tgatgcattc 3300
aaaaatgtgc tttgtcagaa taagaatgcc aaaaatgttt tttcgctgcc ttatctcata 3360
ccagtttttt tgggtttttt tttgtttgtt tgttttttgg tttttttttt tttgtgtgtg 3420
ttgttatttg gttttctttt tgcccacagt ttgtctcagg caatactggg acataggctg 3480
accccattag cttttgttat gaatttacta aactttctgt ggaaggagaa cagagcctct 3540
gccgcgggtg tggggaagcc atcctgtgct tgaggcagca cacgtgtgtc catcatcatc 3600
agtcagaaga gcagggcctg tctcacccaa tcgagtcctt aagacagaat aatcagaatg 3660
gtcagaggga cagaccaatc aattcccagg aaagcaaaag tgactcaatg tcccttgact 3720
cccaaatggt cccactggac tggtgatcac tggtgacaac taactagctt tgtccagaga 3780
atccacccag aacacggtgc tttttagcca gtagtccacc tctatgtgca tcagcaatgc 3840
atagcaggtg agaacttgaa tcacagaaac ttcatgccat ggatggagac tcctgaggcg 3900
ctcaaatact actacctcta gttccaaaga ctagagctag atgatcagaa aggcaactgg 3960
aggcccaggg agccgtactg ggacaagtta gaattagaga acgatgtcat ttaacattcc 4020
gagaaagaaa taaccatgaa ttgctattac aggagtaaca cacagggcca gcttcttttt 4080
tcttcttttt tatttttctt ttcttattgt gagcagaggg aattcacctc agttcatctt 4140
tctctcagta cttttctttc aagatatcaa tcctttatga ctcttttgct tttaattctc 4200
tctctctctc tctctctctc tctctctctt tctctcaaag gagaggtttc agttctaaca 4260
agctaccata gtcctattaa agccattttt ttttttagaa tattaaaagt ccaaactctc 4320
ttgccaaact ctttcttcac atgcgcattg gctgaaaaca gaatttacaa gaatttcttt 4380
aggaagaaac tggggatgtg gcccattggt cacaaagttt ttttgtttgt ttttgttttt 4440
gtttcaattc ttgtttgatt tatggacaat ctttggtttg tattgctctg gagaaattgg 4500
aaatcattgc agagtgaaga taaatcaggg caccatgtat agtagagaat gtttcagtag 4560
ttttccaaac gagaacacaa ttgcacactg taaacaacag gagtgtgaag gaccacagtc 4620
ttgaggagtt cttgttgccc tgtgtttggt gaaggcgttg gggaccgagg aagacaacat 4680
acagtttggc caaggctctc agaggcttgc tgtggcgcca attcaagtat tacaatgttg 4740
catgctgtag aaagtagctg ttgctgttgt tttgttttgt tttaatttaa gtcaccaagg 4800
cactgtttta ttcttttgta aaaaaaaaaa aagttcactg tgcacttata gagaaaataa 4860
tcaacaatgt tgtgaatttt tgagaagact tttttttttt tgataaacca aagatttaga 4920
aatcattcca ttgtcaactt gtaaaaaaaa aaaaaaaaaa 4960
<210> 30
<211> 935
<212> PRT
<213> Mus musculus
<400> 30
Met Thr Leu Leu Gly Ser Glu His Ser Leu Leu Ile Arg Arg Lys Phe
1 5 10 15
Arg Ser Val Leu Gln Leu Arg Leu Gln Gln Arg Arg Thr Gln Glu Gln
20 25 30
Leu Ala Asn Gln Gly Leu Ile Pro Pro Leu Lys Gly Pro Thr Glu Phe
35 40 45
His Asp Pro Arg Lys Gln Leu Asp Ser Ala Lys Thr Glu Asp Ser Leu
50 55 60
Arg Arg Lys Gly Arg Asn Arg Ser Asp Arg Ala Ser Leu Val Thr Met
65 70 75 80
His Ile Leu Gln Ala Ser Thr Ala Glu Arg Ser Ile Pro Thr Ala Gln
54
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
85 90 95
Met Lys Leu Lys Arg Ala Arg Leu Ala Asp Asp Leu Asn Glu Lys Ile
100 105 110
Ala Leu Arg Gln Gly Pro Leu Glu Leu Val Glu Lys Asn Ile Leu Pro
115 120 125
Met Asp Ser Ser Val Lys Glu Ala Ile Lys Gly Thr Glu Val Ser Leu
130 135 140
Ser Lys Ala Ala Asp Ala Phe Ala Phe Glu Asp Asp Ser Ser Arg Asp
145 150 155 160
Gly Leu Ser Pro Asp Gln Ala Arg Ser Glu Asp Pro Gln Gly Ser Thr
165 170 175
Gly Ser Thr Pro Asp Ile Lys Ser Thr Glu Ala Pro Leu Asp Thr Ile
180 185 190
Gln Asp Leu Thr Pro Gly Ser Glu Ser Asp Lys Asn Asp Ala Ala Ser
195 200 205
Gln Pro Gly Asn Gln Ser Asp Pro Gly Lys Gln Val Leu Gly Pro Leu
210 215 220
Ser Thr Pro Ile Pro Val His Thr Ala Val Lys Ser Lys Ser Leu Gly
225 230 235 240
Asp Ser Lys Asn Arg His Lys Lys Pro Lys Asp Pro Lys Pro Lys Val
245 250 255
Lys Lys Leu Lys Tyr His Gln Tyr Ile Pro Pro Asp Gln Lys Ala Glu
260 265 270
Lys Ser Pro Pro Pro Met Asp Ser Ala Tyr Ala Arg Leu Leu Gln Gln
275 280 285
Gln Gln Leu Phe Leu Gln Leu Gln Ile Leu Ser Gln Gln Gln Gln Gln
290 295 300
Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln Gln
305 310 315 320
Arg Phe Ser Tyr Pro Gly Met His Gln Thr His Leu Lys Glu Pro Asn
325 330 335
Glu Gln Met Ala Arg Asn Pro Asn Pro Ser Ser Thr Pro Leu Ser Asn
340 345 350
Thr Pro Leu Ser Pro Val Lys Asn Ser Ile Ser Gly Gln Thr Gly Val
355 360 365
Ser Ser Leu Lys Pro Gly Pro Leu Pro Pro Asn Leu Asp Asp Leu Lys
370 375 380
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Val Ser Glu Leu Arg Gln Gln Leu Arg Ile Arg Gly Leu Pro Val Ser
385 390 395 400
Gly Thr Lys Thr Ala Leu Val Asp Arg Leu Arg Pro Phe Gln Asp Cys
405 410 415
Ala Gly Asn Pro Val Pro Asn Phe Gly Asp Ile Thr Thr Val Thr Phe
420 425 430
Pro Val Thr Pro Asn Thr Leu Pro Ser Tyr Gln Ser Ser Pro Thr Gly
435 440 445
Phe Tyr His Phe Gly Ser Thr Ser Ser Ser Pro Pro Ile Ser Pro Ala
450 455 460
Ser Ser Asp Leu Ser Ala Ala Gly Ser Leu Pro Asp Thr Phe Thr Asp
465 470 475 480
Ala Ser Pro Gly Phe Gly Leu His Ala Ser Pro Val Pro Ala Cys Thr
485 490 495
Asp Glu Ser Leu Leu Ser Ser Leu Asn Gly Gly Ser Gly Pro Ser Glu
500 505 510
Pro Asp Gly Leu Asp Ser Glu Lys Asp Lys Met Leu Val Glu Lys Gln
515 520 525
Lys Val Ile Asn Gln Leu Thr Trp Lys Leu Arg Gln Glu Gln Arg Gln
530 535 540
Val Glu Glu Leu Arg Met Gln Leu Gln Lys Gln Lys Ser Ser Cys Ser
545 550 555 560
Asp Gln Lys Pro Leu Pro Phe Leu Ala Thr Thr Ile Lys Gln Glu Asp
565 570 575
Val Ser Ser Cys Pro Phe Ala Pro Gln Gln Ala Ser Gly Lys Gly Gln
580 585 590
Gly His Ser Ser Asp Ser Pro Pro Pro Ala Cys Glu Thr Ala Gln Leu
595 600 605
Leu Pro His Cys Val Glu Ser Ser Gly Gln Thr His Val Leu Ser Ser
610 615 620
Thr Phe Leu Ser Pro Gln Cys Ser Pro Gln His Ser Pro Leu Gly Gly
625 630 635 640
Leu Lys Ser Pro Gln His Ile Ser Leu Pro Pro Ser Pro Asn Asn His
645 650 655
Tyr Phe Leu Ala Ser Ser Ser Gly Ala Gln Arg Glu Asn His Gly Val
660 665 670
56
CA 02432278 2003-06-20
WO 02/060946 PCT/USO1/50606
Ser Ser Pro Ser Ser Ser=~n Gly Cys Ala Gln Met Thr Gly Leu Gln
675 680 685
Ser Ser Asp Lys Val Gly Pro Thr Phe Ser Ile Pro Ser Pro Thr Phe
690 695 700
Ser Lys Ser Ser Ser Ala Val Ser Asp Ile Thr Gln Pro Pro Ser Tyr
705 710 715 720
Glu Asp Ala Val Lys Gln Gln Met Thr Arg Ser Gln Gln Met Asp Glu
725 730 735
Leu Leu Asp Val Leu Ile Glu Ser Gly Glu Met Pro Ala Asp Ala Arg
740 745 750
Glu Asp His Ser Cys Leu Gln Lys Ile Pro Lys Ile Pro Gly Ser Ser
755 760 765
Cys Ser Pro Thr Ala Ile Pro Pro Lys Pro Ser Ala Ser Phe Glu Gln
770 775 780
Ala Ser Ser Gly Gly Gln Met Ala Phe Asp His Tyr Ala Asn Asp Ser
785 790 795 800
Asp Glu His Leu Glu Val Leu Leu Asn Ser His Ser Pro Ile Gly Lys
805 810 815
Val Ser Asp Val Thr Leu Leu Lys Ile Gly Ser Glu Glu Pro Pro Phe
820 825 830
Asp Ser Ile Met Asp Gly Phe Pro Gly Lys Ala Ala Glu Asp Leu Phe
835 840 845
Ser Ala His Glu Leu Leu Pro Gly Pro Leu Ser Pro Met His Ala Gln
850 855 860
Leu Ser Pro Pro Ser Val Asp Ser Ser Gly Leu Gln Leu Ser Phe Thr
865 870 875 880
Glu Ser Pro Trp Glu Thr Met Glu Trp Leu Asp Leu Thr Pro Pro Ser
885 890 895
Ser Thr Pro Gly Phe Ser Asn Leu Thr Ser Ser Gly Pro Ser Ile Phe
900 905 910
Asn Ile Asp Phe Leu Asp Val Thr Asp Leu Asn Leu Asn Ser Pro Met
915 920 925
Asp Leu His Leu Gln Gln Trp
930 935
$7