Note: Descriptions are shown in the official language in which they were submitted.
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
COMPOSITIONS AND METHODS FOR
ARRAY-BASED GENOMIC NUCLEIC ACID ANALYSIS
OF BIOLOGICAL MOLECULES
TECHNICAL FIELD
The present invention claims a closely related family of compounds,
devices, and methods relating to techniques for immobilizing biological
molecules, e.g.,
nucleic acids, to a solid support for the purpose of conducting scientific
investigation or
routine testing upon the bound molecule (e.g., nucleic acid) samples in areas
such as
genome-wide genetic mapping and gene expression studies, protein interaction
studies,
o peptide interaction studies and small molecule interactions with larger
macromolecules.
BACKGROUND
A large percentage of investigation in the biochemical arts is directed to
studies involving nucleic acids, particularly deoxyribonucleic acid, or DNA.
DNA,is a
water-soluble compound, that if left in solution (i.e., a water-based
solution), is likely to
15 degrade, through hydrolysis, and so forth. Obviously this frustrates any
investigation
involving DNA, and so therefore, accurate and reliable study involving DNA
requires a
method or device to ensure the integrity of DNA. To facilitate the study of
DNA, it is
often desirable to affix or immobilize the DNA on a solid surface, such as a
smooth sheet
of glass. Fixed in place in this manner, the DNA can be readily manipulated
(i.e., reacted
2o with other substances). If DNA is envisioned as a long strand, then
immobilizing DNA
means fixing one end of the strand to the solid support so that the remainder
of the strand
is unmodified and free to undergo fixrther reaction depending upon the
particular study.
Indeed, this is a widely used method to conduct laboratory studies involving
DNA.
Perhaps the major problem associated with immobilizing DNA on a solid
2s support is exactly how to do it without altering the DNA (other than that
relatively small
portion that is actually bound to the solid support). This is a very difficult
problem
because whatever solid support is used must be essentially inert. That is, it
must not react
with the DNA, other than simply to immobilize it upon the solid support. Glass
is a
particularly suitable solid support, because it is inexpensive, and highly
inert. At present,
3o the current orthodoxy is that the solid support (e.g., a glass surface)
must first be primed
or derivatized so that it can bind one end of the DNA to the surface. Numerous
techniques exist to do this.
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Unfortunately, derivatizing the otherwise inert surface of glass creates
problems that could confound the results of the laboratory study involving
DNA. One
problem is that derivatizing the glass surface creates a net positive
electrostatic charge on
the glass surface. Since DNA is (net) negatively charged, other DNA (or DNA
used later
in the study but not deliberately affixed to the glass surface) is prone to
stick (by non-
specific electrostatic attraction) to the glass surface. In other words, DNA
"probes" which
are single (rather than double) strands of DNA are often contacted with an
array of DNA
single strands affixed to a solid support. Since the probe has a known
nucleotide sequence
and since a particular single strand of DNA will bind preferentially to a
complementary
o strand, the particular immobilized strand to which the probe reacts reveals
the nucleotide
sequence of the previously unknown immobilized strand. Yet simple experiments
of this
type (pxobe studies) are severely confounded by electrostatic sticking of the
probe to the
derivatized (hence electrostatically charged) glass surface. For instance, the
probe is
often radiolabeled so that its presence can be detected by an ordinary
radiation detector.
Thus, the location of the probe on the glass surface, as evidenced by the
detector, reveals
the chemical identity or sequence of the immobilized DNA strand at that
particular
location on the glass surface (which is known and designated in advance). Yet
the
radiation detector is unable to distinguish between probe that is chemically
bound to a
complementary strand of DNA affixed to the solid support, and probe that is
simply
2o electrostatically stuck to the glass surface (but not to a DNA strand).
Second, derivatized surfaces result in what shall be known as "spreading."
Spreading occurs because the solid support surface becomes hydrophilic upon
derivatization. As a result, when the DNA (desired to be immobilized upon the
solid
support) is contacted with the surface of the solid support, it spreads,
rather than
remaining in a discrete "spot," which it should ideally do, since whether the
radioactive
probe is detected in one spot or another determines whether the scientist
infers that the
probe reacted with this or that immobilized DNA. Spreading is a major
constraint on
array density (i.e., the number of different nucleic acid samples that can be
arranged on a
single solid support). Hence, any means to curtail spreading, and so increase
array
3o density, is lughly desirable.
One very common substance used to prepare a glass surface to receive a
nucleic acid sample is poly-L-lysine. See, e.g., DeRisi (1996) 14 Nature
Genetics 457;
Shalon (1996) 6 Genome Res. 639; and Schena (1995) 270 Science 467. Other
types of
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
pre-derivatized glass supports are commercially available (e.g., sialylated
microscope
slides). See, e.g., Schena (1996) 93 Proc. Natl. Acad. Sci. USA 10614.
Numerous other surface coatings have been disclosed. See, e.g., U.S. Pat.
No. 5,630,932, discloses a coating for a probe (platinum) tip for use in
scanning tunneling
microscopy; numerous means are disclosed for coating the surface, notably,
Si(OCH3)CH2I. U.S. Pat. No. 5,610,287, discloses coating a solid support with
a salt or
cationic detergent to non-covalently bond nucleic acids to the support. U.S.
Pat. No.
5,024,933, discloses coating a solid support with an isolate of naturally
occurring mussel
adhesive protein. U.S. Pat. No. 4,937,188, discloses covalently bonding an
enzyme to a
1o solid support via molecular chain which acts as a substrate for the enzyme.
U.S. Pat. No.
4,818,681, discloses coating a solid support with a nucleoside phosphate
thxough the
heterocyclic moiety of the nucleoside; the nucleic acid is then immobilized
upon the solid
support by enzymatic coupling. U.S. Pat. No. 4,806,631, discloses activating a
nylon solid
support by partially solvolyzing the amine groups (e.g., by treating with an
alkylating
~ 5 group) on the nylon surface.
Another approach to this problem involves derivatizing both the solid
support and the nucleic acid sought to be immobilized. See, e.g., U.S. Pat.
No. 5,641,630,
discloses coating a solid support with a complexing agent that binds to
another
complexing agent to which the nucleic acid sought to be bound is likewise
bound. U.S.
2o Pat. No. 5,554,744, discloses contacting a solid support with
diisopropylcarbodiimide and
an acid catalyst and a succinylated nucleoside to immobilize the nucleoside.
U.S. Pat. No.
5,5I4,785, discloses coating a solid support with, preferably, primary and
secondary
amines, followed by activation of the nucleic acid using cyanuric chloride.
U.S. Pat. No.
5,215,882, discloses modifying the nucleic acid sought to be immobilized with
a primary
25 amine or equivalent, followed by reaction of the modified nucleic acid with
the solid
support (the support must have free aldehyde groups) in the presence of a
reducing agent.
Finally, a third approach to the problem of immobilizing nucleic acids to
solid support material involves creating a novel solid support. See, e.g.,
U.S. Pat. Nos.
5,055,429, 5,008,220, 4,963,436, 4,826,790, and 4,826,789, disclose solid
support
3o material made from aluminosilicate material.
Due to the aforementioned shortcomings of derivatizing the (entire) glass
surface prior to affixing the nucleic acid samples, several methods have been
developed
which involve synthesizing the nucleic acid samples directly to the solid
support. See,
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
e.g., Hacia (1996) 14 Nature Genetics 441 (1996); Lockhart (1996) 14 Nature
Biotechnology 1675 (1996); Maskos (1992) 20 Nucleic Acids Res. 1679 (1992).
To reiterate: at present, the prevailing view in the biochemical arts is that,
in order to effectively immobilize nucleic acids onto solid surfaces, the
solid support must
first be derivatized, or made chemically labile, so that the nucleic acid can
then be reacted
with solid support. In addition, epoxides are known mutagens; that is, they
are known to
damage nucleic acids, particularly DNA.
SUMMARY
This invention provides compositions and methods for affixing biological
o molecules to solid supports. It demonstrates that any biological molecule
can be modified
and affixed to an unmodified solid support. A skilled artisan will recognize
the
significance of first modifying a molecule to enhance its binding affinity by
appropriate
modifications; thus, this modified molecule can be immobilized to an
unmodified solid
surface to generate a fully functional array of molecules for a spectrum of
specific
~ 5 applications.
In one aspect, the invention provides any biological molecule, e.g., DNA
and nucleic acids more generally, that are modified such that they readily
adhere to an
unmodified or underivatized glass surface. In particular, in one aspect of the
invention
epoxide-modified nucleic acids, particularly DNA, are readily affixed to an
unmodified
2o solid support.
The invention provides a modified biological molecule comprising a
biological molecule modified by reaction with a compound having the formula:
Rl-X-R~,
wherein Rl is a cyclic ether group or an amino group, R2 is an alkoxysilane
group and X
2s is a moiety chemically suitable for linking the cyclic ether group or the
amino group to
the alkoxysilane group. In one aspect, the Rl cyclic ether is a compound
comprising an
epoxide group, such as an ethylene oxide, or equivalent. In alternative
aspects, the cyclic
ether is an oxirane group, or equivalent, or a compound comprising an aromatic
hydrocarbon epoxide group.
3o In one aspect, the Rl group reacts with the biological molecule such that
the modified biological molecule is linked to the compound through Rl group.
The
Iinlcage, or association, of the Rl group to the biological molecule can be
such that the Rl
group is covalently or non-covalently bound to the biological molecule.
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
In alternative aspects, the biological molecule comprises a nucleic acid
(e.g., a oligonucleotide), a lipid, a polysaccharide, a polypeptide (e.g., a
peptide), or an
analog or a mimetic thereof, or a combination thereof. The nucleic acid can
comprise a
DNA (e.g., a genomic DNA or a cDNA), an RNA (e.g., an mRNA, rRNA, and the
like)
or an analog or a mimetic thereof or a combination thereof. The nucleic acid
can further
comprise a telomeric structure or a chromatin structure.
In one aspect, the nucleic acid is attached to the compound by the Rl
group, i.e., the nucleic acid reacts with the Rl group at its 5' end.
In one aspect, the cyclic ether is an epoxide group and the alkoxysilane is
-Si(OCH3)3, -Si(OC2 HS)3, -Si(OCH3)H2, -Si(OCH3)(CH3) 2, Or -Sz(OCH) 3) 2
CH3. In one aspect, the cyclic ether is an epoxide group and the compound is 3-
glycidoxypropyltrimethoxysilane.
In one aspect, the Rl group is a primary amino group. In one aspect, the
Rl group is an amino group and the alkoxysilane is selected from the group
consisting of
-Si(OCH3)3, -Si(OC~ HS)3 and
Ri
- Si - R2 ,
2o R3
wherein R1, R2 and R3 are selected from the group consisting of -H,
-CH3, -OCH3, and -OC2 H3, and provided that at least one of Rr, R2 or R3 is
either
-OCH3 or -OC2 H3.
In one aspect, the Rl group is an amino group and the compound is 3-
aminopropyltriethoxysilane.
The invention provides an aa.-ticle of manufacture comprising an arrayed
plurality of biological molecules covalently bound to a surface, wherein,
before
attachment to the surface, the biological molecules are modified by reaction
with a
3o compound having the formula: Rl - X - R2 , wherein Rl is a cyclic ether
group or an
amino group, R2 is an alkoxysilane group and X is a moiety chemically suitable
for
linking the cyclic ether group or the amino group to the alkoxysilane group,
and upon
attachment to the surface the modified biological molecules are covalently
bound to the
surface; wherein each biological molecule is attached to the surface on at
least one
discrete and known location to form a cluster of substantially identical
biological
molecules. In alternative aspects of the article of manufacture the surface is
a glass, a
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
mica, a quartz, or a metal oxide surface. The metal oxide surface can be an
alumina (Ala
03), a titania (Ti02), a Sn02, a Ru02, or a Pt02, or an equivalent thereof.
The surface of
the article of manufacture can comprise a polystyrene, a polyester, a
polycarbonate, a
polyethylene, a polypropylene or a nylon.
In one aspect, the modified biological molecules are covalently bound to
the surface via the R2 group.
On one aspect of the article of manufacture, the biological molecules can
comprise a nucleic acid, a lipid, a polypeptide, a polysaccharide, or an
analog or a
mimetic thereof, or a combination thereof. In alternative aspects, the
biological
1 o molecules are derived from a virus, a bacteria, a yeast, a plant, an
insect, a mammal, such
as a human or a mouse. The biological molecules can comprise nucleic acids or
analogs
or mimetics thereof. The nucleic acids can comprise DNA, RNA or analogs or
mimetics
thereof or a combination thereof. The nucleic acids can be oligonucleotides.
In one aspect of the article of manufacture, the nucleic acids react with the
Rl group at their 5' end.
In one aspect, the nucleic acids immobilized on the article of manufacture
can comprise a plurality of fragments of a genomic nucleic acid. The
biological
molecule, e.g., a genomic nucleic acid or RNA, can be derived from a normal
cell or an
abnormal cell, such as a cell suspected of having a chromosomal defect or
abnormality,
2o e.g., a cancer or tumor cell. In alternative aspects, the genomic DNA is
derived from a
virus, a bacteria, a yeast, a plant, an insect, a mammal, such as a human or a
mouse.
In one aspect of the article of manufacture, the fragments of nucleic acid,
e.g., genomic nucleic acid, further comprise a cloning vehicle. The cloning
vehicle can
comprise a bacterial artificial chromosome (BAC). In alternative aspects, the
cloning
2s vehicle comprises a plasmid, a cosmid, a bacteriophage P1-derived vector
(PAC), a yeast
artificial chromosome (YAC) or a mammalian artificial chromosome (MAC).
In one aspect of the article of manufacture, the nucleic acid comprises a
plurality of CpG island tags.
In one aspect of the article of manufacture, the fragments of genomic
3o nucleic acid comprise sequences representing at least one substantially
complete
chromosome or at least one defined section of a chromosome. In one aspect,
each
genomic nucleic acid fragment has been mapped to a known location on a
chromosome.
In alternative aspects, the nucleic acid, e.g., the genomic nucleic acid
fragments, have a
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
size no more than about 1.5 megabase, no more than about 1.2 megabase, no more
than
about 1.0 megabase, and, no more than about 0.75 megabase in size.
In alternative aspects of the article of manufacture, each cluster of
substantially identical biological molecules consists of between about 5 and
about 400, or,
between about 10 and about 200, or, between about 50 and 100, substantially
identical
copies of a biological molecule. The surface can consist of less than about
800, about
600, about 500, about 400, about 300, about 200 or about 100 clusters per
square
centimeter.
In alternative aspects of the article of manufacture, each cluster of
o substantially identical biological molecules is about 100 microns, about 50
microns, about
25 microns, about 15 microns or about 10 microns in diameter or smaller.
The invention provides an article of manufacture (e.g., array or biochip)
comprising an array of cloned genomic nucleic acid fragments representing a
defined
subsection of or a substantially complete chromosome, wherein, before
attachment to the
~ 5 surface, the cloned fragments are modified by reaction with a compound
having the
formula:
Rl - X - R2 , wherein R~ is an epoxide group, R2 is an alkoxysilane group and
X is a
moiety chemically suitable for linking the epoxide group and the alkoxysilane
group, and
the modif ed cloned fragments are covalently bound to the surface; wherein
each array-
2o bound cloned fragment has been mapped to a known location on a chromosome.
The invention provides a kit comprising an article of manufacture of the
invention, as described herein, and printed matter, wherein the printed matter
comprises
instructions on hybridizing a sample of nucleic acid to an array-bound nucleic
acid.
The invention provides a method for identifying a specific binding partner,
25 comprising: (a) providing an article of manufacture comprising an arrayed
plurality of
biological molecules covalently bound to a surface, wherein, before attachment
to the
surface, the biological molecules are modified by reaction with a compound
having the
formula: Rl - X - Ra, wherein Ri is an cyclic ether group or an amino group,
RZ is an
alkoxysilane group and X is a moiety chemically suitable for linlcing the
cyclic ether
3o group or the amino group to the alkoxysilane group, and upon attachment the
modified
biological molecules are covalently bound to the surface, wherein each
biological
molecule is attached to the surface on at least one discrete and known
location to form a
cluster of identical biological molecules; (b) providing a sample of
biological molecules;
(c) contacting the sample of step (b) with the array-bound biological
molecules as set
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
forth in step (a) under conditions permissive for specific binding of a
molecule in the
sample of step (b) to an array-bound biological molecule; and, (d) screening
for specific
binding of a molecule in the sample of step (b) to an array-bound biological
molecule,
thereby identifying a specific binding partner. In one aspect, the method
further
comprises at least one wash step between the contacting of step (c) and the
screening of
step (d).
The invention provides a method for generating a molecular profile of a
nucleic acid sample, comprising the following steps: (a) providing an article
of
manufacture comprising an array of biological molecules, wherein, before
attachment to
1o the surface, the biological molecules are modified by reaction with a
compound having
the formula: Rl - X - R2, wherein Rl is a cyclic ether group, R2 is an
alkoxysilane
group and X is a moiety chemically suitable for linking the cyclic ether group
and the
alkoxysilane group, and the modified biological molecules are covalently bound
to the
surface; (b) providing a sample comprising a nucleic acid; and (c) contacting
the
~ 5 nucleic acid with the array-bound biological molecules as set forth in
step (a) under
conditions, allowing binding of the sample nucleic acid to the array-bound
biological
molecules, and detecting binding of the sample nucleic acid to the array-bound
biological
molecules, thereby generating a molecular profile of the sample nucleic acid.
In one aspect of this method, the array-bound biological molecules
2o comprise a nucleic acid, such as a DNA corresponding to, or derived from, a
genomic
DNA, or, a message. The binding can comprise hybridization of the sample
nucleic acid
to the array-bound nucleic acid. The array-bound nucleic acid can represent a
section of
at least one a chromosome or at least one substantially complete chromosome.
The
chromosome can be a viral, a bacterial, a yeast, a plant, an insect, or a
mammalian, such
25 as a human or a mouse, chromosome. In one aspect, the array-bound nucleic
acids have
been mapped to a known location on a chromosome.
In one aspect of this method, the molecular profile is a comparative
genomic hybridization (CGH). The molecular profile can comprise detection of a
genomic DNA amplification, a genomic DNA deletion, or a genomic DNA insertion.
so The molecular profile can comprise detection of a point mutation.
In one aspect of this method, the molecular profile is the identification of a
single or multiple point mutations, such as a single-nucleotide polymorphism
(SNP). In
one aspect of this method, the detection of a point mutation can further
comprise use of a
primer extension assay.
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
In one aspect of this method, the modified nucleic acid of the invention, or
the array-bound nucleic acids, comprise on or more CpG island tags. In one
aspect, the
molecular profile is generated by a differential methylation hybridization
(DMH)
reaction. The sample nucleic acids can comprise genomic DNA digested with at
least one
methylation-sensitive restriction endonuclease and the molecular profile
comprises
detection and mapping of hypermethylated regions of the genome. The
methylation-
sensitive restriction endonuclease can be selected from the group consisting
of NotI,
SmaI, SacII, EagI, MspI, HpaII and BssHII.
In one aspect of this method, the molecular profile comprises detection of
o transcriptionally active regions of a genome. In one aspect, the sample of
nucleic acid
can be derived from a nuclear run-off assay; this sample can be modified by
the methods
of the invention, and, in one aspect, these modified nucleic acids are
immobilized onto an
array as set forth in the invention.
In one aspect of this method, the molecular profile comprises an analysis
15 of a chromatin structure. The modified nucleic acids of the invention, or
the array-bound
biological molecule, can comprise a chromatin structure.
In one aspect of this method, the molecular profile comprises an analysis
of a telomeric structure. The molecular profile of a telomeric structure can
comprise an
analysis of telomeric erosion or telomeric addition. The modified nucleic
acids of the
2o invention, or the array-bound biological molecule (e.g., nucleic acid), can
comprise one
or more telomere structures.
The invention provides a method for making a modified biological
molecule comprising (a) providing a biological molecule; (b) providing a
compound
having the formula: Rl - X - R2, wherein Rl is a cyclic ether group or an
amino group,
25 R2 is an alkoxysilane group and X is a moiety chemically suitable for
linking the cyclic
ether group or the amino group to the alkoxysilane group; and (c) reacting the
biological
molecule with the compound, thereby modifying the biological molecule with the
compound.
The invention provides a method for making an article of manufacture
30 (e.g., array or biochip) comprising an arrayed plurality of biological
molecules covalently
bound to a surface comprising (a) providing a biological molecule; (b)
providing a
compound having the formula: Rl - X - R2, wherein Rl is a cyclic ether group
or an
amino group, R2 is an alkoxysilane group and X is a moiety chemically suitable
for
linking the cyclic ether group or the amino group to the alkoxysilane group;
(c)
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
providing a surface comprising hydroxyl groups; (d) reacting the biological
molecule
with the compound, thereby modifying the biological molecule with the
compound; and,
(e) depositing a plurality of modified biological molecules on the surface as
discrete
clusters, wherein a modified biological molecule is attached to the surface on
at least one
s discrete and known location to form at least one cluster of substantially
identical
biological molecules; the array comprises at least two, a plurality of,
clusters.
In one aspect of the method for making an article of manufacture, the
compound used to modify the biological molecule (e.g., a nucleic acid) is a 3-
glycidoxy-
propyltrimethoxysilane. In one aspect, the biological molecule is a nucleic
acid and the
1 o reaction with the 3-glycidoxypropyltrimethoxysilane is at a basic pH,
thereby generating
a modified nucleic acid. In this reaction, the pH can be above about pH 9.5. A
3-
glycidoxy-propyltrimethoxysilane modified nucleic acid can be deposited on an
underivatized glass surface at about a neutral pH.
In another aspect of the method for making an article of manufacture, the
~ 5 compound used to modify the biological molecule (e.g., a nucleic acid) is
a 3-
aminopropyltriethoxysilane. In one aspect, the biological molecule is a
nucleic acid and
the reaction with the 3-aminopropyltriethoxysilane is at about a neutral pH.
This reaction
can take place in the presence of sodium bisulfate, or equivalent. This
modified nucleic
acid can be deposited on an underivatized glass surface.
2o The modified biological molecules, such as a modified nucleic acid, of the
invention, will adhere to a solid surface to allow subsequent biochemical
investigations.
Thus, in one aspect of the present invention, a modified biological molecule,
such as a
modified nucleic acid, comprises a biological molecule (e.g., a nucleic acid)
covalently
bound to moiety containing two crucial functional groups: a cyclic ether group
and an
2s alkoxysilane group. In accordance with other aspects of the present
invention, methods
for preparing the aforementioned modified biological molecules (e.g., nucleic
acids) are
claimed.
In another aspect, the invention provides a high-density microarray
comprising a glass or other inert surface. This array, or "biochip," can be
made by
3o printing numerous highly discrete modified biological molecule (e.g., DNA)
sample
spots, or "clusters," upon the surface.
In another aspect, the invention provides a modified biological molecule
(e.g., a nucleic acid) prepared from a biological molecule (e.g., a nucleic
acid) and a
halogenated silane, or equivalent.
to
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
In another aspect, the invention provides a modified nucleic acid prepared
by reaction of a biological molecule (e.g., a nucleic acid) with a brominated
moiety,
followed by reaction with an aminated silane.
In another aspect, the invention provides a device that allows printing of
the aforementioned high-density microaxrays.
In another aspect, the invention provides modified silanes that allow the
skilled artisan to modulate the electrostatic properties of the solid surface
to optimize
sample density and detection sensitivity.
The present invention possesses numerous advantages over the prior art.
1 o Many of the advantages derive from the fact that the solid surface, which
is can be
ordinary glass, remains highly chemically inert. Thus, the previously
mentioned
problems of probe (or other reactant) sticking to the glass as well as
"spreading" are
entirely eliminated. The ultimate result is, among other things, far higher
detection
sensitivity compared with state-of the-art derivatized solid support.
In addition, the biological molecule (e.g., a nucleic acid) to be
immobilized upon the solid support is readily derivatized. The reaction of the
epoxide
derivatives of the invention is simple to execute; it occurs under mild
conditions, reaction
rates are quick, and equilibrium is highly favorable. Moreover, the epoxide-
modified
biological molecules (e.g., nucleic acids) of the present invention are
essentially
2o permanently stable; thus they can be prepared (derivatized) and stored for
later use
(reaction with a non-derivatized surface).
The details of one or more aspects of the invention are set forth in the
accompanying drawings and the description below. Other features, objects, and
advantages of the invention will be apparent from the description and
drawings, and from
the claims.
All publications, GenBank Accession references (sequences), ATCC
Deposits, patents and patent applications cited herein are hereby expressly
incorporated
by reference for all purposes.
DESCRIPTION OF DRAWINGS
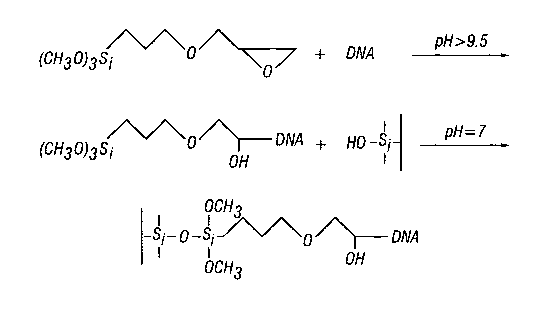
3o FIG. 1 depicts a coupling reaction of nucleic acid (in this instance DNA)
with 3-glycidoxypropyltrimethoxysilane, followed by the reaction of the newly
modified
11
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
DNA and the solid support (in this instance a glass surface). The final
reaction product,
the immobilized DNA, is shown at bottom.
FIG. 2 depicts a coupling reaction of nucleic acid (in this instance DNA)
with 3-aminoproplytriethoxysilane followed by the reaction of the newly
modified DNA
and the solid support (in this instance a glass surface). The final reaction
product, the
immobilized DNA, is shown at bottom.
FIG. 3 depicts a device for making a high-density microarray; both a top
(FIG. 3A) and a side view (FIG. 3B) are shown.
FIG. 4 depicts the silanization of nucleic acid through alkylation of
1 o halogen-containing silane compounds.
FIG. 5a depicts the first step in the silanization of nucleic acid using
amine-containing silane compounds. In this case, the reaction occurs
preferentially at the
guanine base at neutral and slightly basic pH.
FIG. 5b depicts the first step in the silanization of nucleic acid using
~5 amine-containing silane compounds. In this case, the reaction occurs
preferentially at the
cytosine base at more basic pH.
FIG. 5c depicts the second and final step in the silanization of nucleic acid
using amine-containing silane compounds.
Figure 6 is a schematic representation of one aspect of the present
2o invention showing silane linkers by hydrophobic linkers.
Figure 7 is a schematic representation of an exemplary reaction wherein a
biological molecule, a polypeptide, is modified, or "activated," by a method
comprising
use of succinimidyl acetylthiopropionate (SATP) to introduce an active
sulfhydryl
functional group, as described in detail in Example 9, below.
25 Drawings are not necessary to scale. Certain features of the invention may
be exaggerated in scale or shown in schematic form in the interest of clarity
and
conciseness. Like reference symbols in the various drawings indicate like
elements.
DETAILED DESCRIPTION
The invention provides modified biological molecules, such as
3o polypeptides and nucleic acids, and articles of manufacture comprising
arrays, with these
modified biological molecules immobilized to the array surface. The invention
also
provides methods for making and using these compositions.
12
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
One aspect of the invention is chemical modification of the biological
molecule (e.g., nucleic acid) sought to be immobilized. This chemically
modified nucleic
acid is then readily reacted to a solid support such as a glass surface,
rendering the
biological molecule (e.g., nucleic acid) immobilized. Again, this is in direct
contradiction
to the prior art, which teaches modification of the solid support, rather than
the nucleic
acid itself.
The modified the biological molecules (e.g., nucleic acids) of the present
invention readily adhere to a variety of solid surfaces having reactive
functional groups,
e.g., hydroxyl groups. These include, though are not limited to: quartz glass,
mica,
1 o alumina (A1203), titanic (Ti02), SnOa, RuO2, Pt02, plastics such as the
following polymer
materials, polystyrene, polyester, polycarbonate, polyethylene, polypropylene,
and nylon
as well as numerous semi-conductive surfaces, such as numerous other metal
oxide
surfaces and equivalents.
In one family of aspects, the chemically modified biological molecules
15 (e.g., nucleic acids) of the present invention are so modified with
compounds having two
crucial functionalities: a ring ether and an alkoxysilane group. The
biological molecule
(e.g., nucleic acid) reacts with the ring ether, then the newly modified
biological
molecules (e.g., nucleic acids) are contacted with the otherwise inert surface
(e.g., glass),
where the alkoxysilane group reacts with a hydroxyl-containing (e.g., hydroxyl
2o derivatized) surface, e.g., Si--OH groups on the glass surface.
In another distinct family of aspects, the chemically modified biological
molecules (e.g., nucleic acids) of the present invention are so modified with
compounds
having two crucial functionalities: an amino group and an alkoxysilane group.
The
biological molecules (e.g., nucleic acid) react with the amino group, then the
newly
25 modified biological molecules (e.g., nucleic acids) axe contacted with the
otherwise inert
(e.g., glass) surface, where the alkoxysilane group reacts with a hydroxyl-
containing (e.g.,
hydroxyl derivatized) surface, e.g., Si--OH groups on the glass surface.
In yet another distinct family of aspects, the biological molecules (e.g.,
nucleic acids) are modified by reaction with halogenated silane compounds.
3o In another set of aspects, the biological molecules (e.g., nucleic acids)
are
derivatized by a two-step process involving a final reaction with amine-
containing silanes
and brominated nucleic acids.
13
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Other aspects are directed to preparing and optimizing high-density
microarrays utilizing the modified biological molecules (e.g., nucleic acids)
of the other
aspects of the present invention.
Further aspects include compositions and methods of making and using
that comprise any biological molecule or combinations of biological molecules.
One
skilled in the art realizes that nucleic acids, e.g., DNA or RNA, are only one
of many
biological compositions, e.g., biological polymers, that can be modified by
the methods
of the invention and used in the methods of the invention. A polymer refers to
a molecule
that has joined prefabricated units, e.g., monomers or compositions that can
be of limited
o diversity, linked together, usually by identical mechanisms, e.g., a
cellulose is a polymer
is simple sugars or polysaccharides. Exemplary biological molecules include
but are not
limited to DNA, RNA, protein, peptides, lipids, saccharides, polysaccharides
and
mimetics and analogs thereof. Thus, a skilled artisan recognizes that any
biological
molecule, including those having a structure found in nature or a synthetic
structure,
~ s including polymers, can be modified by the methods of the invention and
affixed to a
solid surface similar to the modified nucleic acids of the invention.
Another aspect of the invention is the modification of biological
molecules. One type of modification is chemical cross-linking. It is well
known in the
art that bifunctional "crosslinking" reagents contain two reactive groups,
thus providing a
2o means of covalently crosslinking two target groups. The reactive groups in
a chemical
crosslinking reagent typically belong to the classes of functional groups,
e.g.,
succinimidyl esters, maleimides and iodoacetamides. Bifunctional crosslinking
reagents
can be divided in homobifunctional, heterobifunctional and zero-length
bifunctional
crosslinking reagents. In homobifunctional crosslinking reagents, the reactive
groups are
25 identical. These reagents couple like functional groups, e.g., two thiols,
two amines, two
acids or two alcohols, and are predominantly used to form intramolecular
crosslinlcs. In
heterobifunctional crosslinking reagents, the reactive groups have dissimilar
chemistry,
allowing the formation of crosslinks between unlike functional groups. The
"zero-length"
crosslinlcing reagent forms a chemical bond between two groups without itself
being
3o incorporated into the product. For example, water-soluble cardodiimide
(EDAC) is used
to couple carboxylic acids to amines.
In addition to the traditional bifunctional crosslinking reagents, a
noncovalent interaction between two molecules that has very slow dissociation
lcinetics
can also function as a crosslink. For example, reactive derivatives of
phospholipids can
14
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
be used to link the liposomes or cell membranes to antibodies or enzymes.
Biotinylation
and haptenylation reagents can also be thought of as heterobifimctional
crosslinlcing
reagents because they comprise a chemically reactive group as well as a biotin
or a hapten
moiety that binds with high affinity to avidin or an anti-hapten antibody,
respectively.
s In contrast to chemical crosslinking reagents, photoreactive crosslinlcing
reagents are available. The general scheme involves photoreactive crosslinking
reagents
that contain a chemically reactive group as well as a photoreactive group.
These
crosslinkers are first chemically reacted with one molecule and then this
modified
molecule is coupled to a second molecule using UV illumination. Depending on
the
1 o reactive properties of the chemical and photoreactive groups, these
crosslinkers can be
used to couple like or unlike functional groups.
Other aspects are directed to preparing and optimizing high-density
microarrays utilizing the modified molecules of the invention.
DEFINITIONS
~ 5 Unless defined otherwise, all technical and scientific terms used herein
have the meaning commonly understood by a person skilled in the art to which
this
invention belongs. As used herein, the following terms have the meanings
ascribed to
them unless specified otherwise.
The term "nucleic acid" as used herein refers to a deoxyribonucleotide
20 (DNA) or ribonucleotide (RNA) in either single- or double-stranded form.
The term
encompasses nucleic acids containing known analogues of natural nucleotides.
The term
encompasses mixed oligonucleotides comprising an RNA portion bearing 2'-O-
alkyl
substituents conjugated to a DNA portion via a phosphodiester linkage, see,
e.g., U.S.
Patent No. 5,013,830. The term also encompasses nucleic-acid-like structures
with
25 synthetic backbones. DNA backbone analogues provided by the invention
include
phosphodiester, phosphorothioate, phosphorodithioate, methylphosphonate,
phosphoramidate, alkyl phosphotriester, sulfamate, 3'-thioacetal,
methylene(methylimino),
3'-N-carbamate, morpholino carbamate, and peptide nucleic acids (PNAs); see
Oligonucleotides and Analogues, a Practical Approach, edited by F. Eckstein,
IRL Press at
so Oxford University Press (1991); Antisense Strategies, Annals of the New
York Academy of
Sciences, Volume 600, Eds. Baserga and Denhardt (NYAS 1992); Milligan (1993)
J. Med.
Chem. 36:1923-1937; Antisense Research and Applications (1993, CRC Press).
PNAs
contain non-ionic backbones, such as N-(2-aminoethyl) glycine units.
Phosphorothioate
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
linkages are described, e.g., by U.S. Patent Nos. 6,031,092; 6,001,982;
5,684,148; see also,
WO 97/03211; WO 96/39154; Mata (1997) Toxicol. Appl. Pharmacol. 144:189-197.
Other
synthetic baclcbones encompassed by the term include methyl-phosphonate
linkages or
alternating methylphosphonate and phosphodiester linkages (see, e.g., U.S.
Patent No.
s 5,962,674; Strauss-Soukup (1997) Biochemistry 36:8692-8698), and
benzylphosphonate
linkages (see, e.g., U.S. Patent No. 5,532,226; Samstag (1996) Antisense
Nucleic Acid
Drug Dev 6:153-156). The term nucleic acid is used interchangeably with gene,
DNA,
RNA, cDNA, mRNA, oligonucleotide primer, probe and amplification product.
The terms "polypeptide," "protein," and "peptide" include compositions of
o the invention that also include "analogs," or "conservative variants" and
"mimetics" or
"peptidomimetics" with structures and activity that substantially correspond
to the
polypeptide from which the variant was derived, as discussed in detail, below.
The term "small molecule" means any synthetic small molecule, such as
an organic molecule or a synthetic molecule, such as those generated by
combinatorial
15 chemistry methodologies. These small molecules can be synthesized using a
variety of
procedures and methodologies, which are well described in the scientific and
patent
literature, e.g., Organic Syntheses Collective Volumes, Gilman et al. (Eds)
John Wiley &
Sons, Inc., NY; Venuti (1989) Pha~m Res. 6:867-873. Synthesis of small
molecules, as
with all other procedures associated with this invention, can be practiced in
conjunction
2o with any method or protocol known in the art. For example, preparation and
screening of
combinatorial chemical libraries are well known, see, e.g., U.S. Patent Nos.
6,096,496;
6,075,166; 6,054,047; 6,004,617; 5,985,356; 5,980,839; 5,917,185; 5,767,238.
The terms "array" or "microarray" or "biochip" or "chip" as used herein is
an article of manufacture, a device, comprising a plurality of immobilized
target elements,
25 each target element comprising a "cluster" or "biosite" or defined area
comprising a
biological molecule (e.g., a nucleic acid molecule or polypeptide, such as an
antibody)
immobilized to a solid surface, as discussed in farther detail, below.
The term "sample of nucleic acid targets" or "sample of nucleic acid" as
used herein refers to a sample comprising DNA or RNA, or nucleic acid
representative of
3o DNA or RNA isolated from a natural source, in a form suitable for
hybridization (e.g., as
a soluble aqueous solution) to another nucleic acid or polypeptide or
combination thereof
(e.g., immobilized probes). The nucleic acid may be isolated, cloned or
amplified; it may
be, e.g., genomic DNA, mRNA, or cDNA from substantially an entire genome,
substantially all or part of a particular chromosome, or selected sequences
(e.g. particular
16
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
promoters, genes, amplification or restriction fragments, cDNA, etc.), The
nucleic acid
sample may be extracted from particular cells or tissues. The cell or tissue
sample from
which the nucleic acid sample is prepared is typically taken from a patient
suspected of
having a genetic defect or a genetically-linked pathology or condition, e.g.,
a cancer,
associated with genomic nucleic acid base substitutions, amplifications,
deletions and/or
translocations. Methods of isolating cell and tissue samples are well known to
those of
skill in the art and include, but are not limited to, aspirations, tissue
sections, needle
biopsies, and the like. Frequently the sample will be a "clinical sample"
which is a
sample derived from a patient, including sections of tissues such as frozen
sections or
1 o paraffin sections talcen for histological purposes. The sample can also be
derived from
supernatants (of cells) or the cells themselves from cell cultures, cells from
tissue culture
and other media in which it may be desirable to detect chromosomal
abnormalities or
determine amplicon copy number. In some cases, the nucleic acids may be
amplified
using standard techniques such as PCR, prior to the hybridization. In
alternative aspects,
the target nucleic acid may be unlabeled, or labeled (as, e.g., described
herein) so that its
binding to the probe (e.g., oligonucleotide, or clone, immobilized on an
array) can be
detected. The probe an be produced from and collectively can be representative
of a
source of nucleic acids from one or more particular (pre-selected) portions
of, e.g., a
collection of polymerase chain reaction (PCR) amplification products,
substantially an
2o entire chromosome or a chromosome fragment, or substantially an entire
genome, e.g., as
a collection of clones, e.g., BACs, PACs, YACs, and the like (see below). The
probe or
genomic nucleic acid sample may be processed in some manner, e.g., by blocking
or
removal of repetitive nucleic acids or by enrichment with selected nucleic
acids.
Generating and Manipulating Nucleic Acids
2s The invention provides modified nucleic acids, and articles of manufacture
comprising arrays that include modified nucleic acid compositions and methods
for
making and using these arrays. The nucleic acid is modified by reaction with a
compound having the formula: Rl - X - R2 , where Rl is a cyclic ether group or
an
amino group, R2 is an alkoxysilane group and X is a moiety chemically suitable
for
30 linking the cyclic ether group or the amino group to the alkoxysilane
group. The
modified nucleic acid or the immobilized nucleic acid on the array can be
representative
of genomic DNA, including defined parts of, or entire, chromosomes, or entire
genomes.
In several aspects, the arrays and methods of the invention are used in
comparative
1~
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
genomic hybridization (CGH) reactions, including CGH reactions on arrays (see,
e.g.,
U.S. Patent Nos. 5,830,645; 5,976,790), see discussion below. The invention
can be
practiced in conjunction with any method or protocol or device known in the
art, which
are well described in the scientific and patent literature.
Ge~e~al Techniques
The nucleic acids used to practice this invention, whether RNA, cDNA,
genomic DNA, vectors, viruses or hybrids thereof, may be isolated from a
variety of
sources, genetically engineered, amplified, and/or expressed/ generated
recombinantly
(recombinant polypeptides can be modified or immobilized to arrays in
accordance with
o the invention). Any recombinant expression system can be used, including
bacterial,
mammalian, yeast, insect or plant cell expression systems.
Alternatively, these nucleic acids can be synthesized i~ vitr°o by
well-
known chemical synthesis techniques, as described in, e.g., Carruthers (1982)
Cold
Spring Harbor Symp. Quant. Biol. 47:411-418; Adams (1983) J. Am. Chem. Soc.
105:661; Belousov (1997) Nucleic Acids Res. 25:3440-3444; Frenkel (1995) Free
Radic.
Biol. Med. 19:373-380; Blommers (1994) Biochemistry 33:7886-7896; Narang
(1979)
Meth. Enzymol. 68:90; Brown (1979) Meth. Enzymol. 68:109; Beaucage (1981)
Tetra.
Lett. 22:1859; U.S. Patent No. 4,458,066. Double stranded DNA fragments may
then be
obtained either by synthesizing the complementary strand and annealing the
strands
2o together under appropriate conditions, or by adding the complementary
strand using DNA
polymerase with a primer sequence.
Techniques for the manipulation of nucleic acids, such as, e.g., subcloning,
labeling probes (e.g., random-primer labeling using I~l.enow polymerase, nick
translation,
amplification), sequencing, hybridization and the like are well described in
the scientific
and patent literature, sea, e.g., Sambrook, ed., MOLECULAR CLONING: A
LABORATORY
MANUAL (2ND ED.), Vols. 1-3, Cold Spring Harbor Laboratory, (1989); CURRENT
PROTOCOLS IN MOLECULAR BIOLOGY, Ausubel, ed. John Wiley ~ Sons, Inc., New York
(1997); LABORATORY TECHNIQUES IN BIOCHEMISTRY AND MOLECULAR BIOLOGY:
HYBRIDIZATION WITH NUCLEIC ACID PROBES, Part I. Theory and Nucleic Acid
3o Preparation, Tijssen, ed. Elsevier, N.Y. (1993).
Another useful means of obtaining and manipulating nucleic acids used in
the compositions and methods of the invention is to clone from genomic
samples, and, if
necessary, screen and re-clone inserts isolated (or amplified) from, e.g.,
genomic clones
is
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
or cDNA clones or other sources of complete genomic DNA. Sources of genomic
nucleic
acid used in the methods and compositions of the invention include genomic or
cDNA
libraries contained in, or comprised entirely of, e.g., mammalian artificial
chromosomes
(see, e.g., Ascenzioni (1997) Cancer Lett. 118:135-142; U.S. PatentNos.
5,721,118;
6,025,155) (including human artificial chromosomes, see, e.g., Warburton
(1997) Nature
386:553-555; Roush (1997) Science 276:38-39; Rosenfeld (1997) Nat. Genet.
15:333-
335); yeast artificial chromosomes (YAC); bacterial artificial chromosomes
(BAC); P1
artificial chromosomes (see, e.g., Woon (1998) Genomics 50:306-316; Boren
(1996)
Genome Res. 6:1123-1130); PACs (a bacteriophage P1-derived vector, see, e.g.,
Ioannou
o (1994) Nature Genet. 6:84-89; Reid (1997) Genomics 43:366-375; Nothwang
(1997)
Genomics 41:370-378; Kern (1997) Biotechniques 23:120-124); cosmids, plasmids
or
cDNAs.
Amplification ofNucleicAcids
Amplification using oligonucleotide primers can be used to generate
95 nucleic acids used in the compositions and methods of the invention, to
detect or measure
levels of test or control samples hybridized to an array, and the like. The
skilled artisan
can select and design suitable oligonucleotide amplification primers.
Amplification
methods are also well known in the art, and include, e.g., polymerase chain
reaction, PCR
(PCR PROTOCOLS, A GUIDE TO METHODS AND APPLICATIONS, ed. Innis,
2o Academic Press, N.Y. (1990) and PCR STRATEGIES (1995), ed. Innis, Academic
Press,
Inc., N.Y., ligase chain reaction (LCR) (see, e.g., Wu (1989) Genomics 4:560;
Landegren
(1988) Science 241:1077; Barringer (1990) Gene 89:117); transcription
amplification
(see, e.g., Kwoh (1989) Proc. Natl. Acad. Sci. USA 86:1173); and, self
sustained
sequence replication (see, e.g., Guatelli (1990) Proc. Natl. Acad. Sci. USA
87:1874); Q
25 Beta replicase amplification (see, e.g., Smith (1997) J. Clin. Microbiol.
35:1477-1491),
automated Q-beta replicase amplification assay (see, e.g., Burg (1996) Mol.
Cell. Probes
10:257-271) and other RNA polymerase mediated techniques (e.g., NASBA,
Cangene,
Mississauga, Ontario); see also Berger (1987) Methods Enzymol. 152:307-316;
Sambrook; Ausubel; U.S. Patent Nos. 4,683,195 and 4,683,202; Sooknanan (1995)
3o Biotechnology 13:563-564.
Polypeptides
The invention is directed to modified polypeptides and articles of
manufacture comprising arrays with immobilized polypeptides, peptides and
19
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
peptidomimetics. The polypeptide is modified by reaction with a compound
having the
formula: Rl - X - R2 , where Rl is a cyclic ether group or an amino group, R2
is an
allcoxysilane group and X is a moiety chemically suitable for linking the
cyclic ether
group or the amino group to the alkoxysilane group. As noted above, the terms
"polypeptide," "protein," and "peptide," used to practice the invention,
include
compositions of the invention that also include "analogs," or "conservative
variants" and
"mimetics" or "peptidomimetics." The terms "mimetic" and "peptidomimetic"
refer to a
synthetic chemical compounds. The mimetic can be either entirely composed of
synthetic, non-natural analogues of amino acids, or, is a chimeric molecule of
partly
o natural peptide amino acids and partly non-natural analogs of amino acids.
The mimetic
can also incorporate any amount of natural amino acid conservative
substitutions as long
as such substitutions also do not substantially alter the mimetics' structure
andlor activity.
Polypeptide mimetic compositions can contain any combination of non-natural
structural
components, which are typically from three structural groups: a) residue
linlcage groups
other than the natural amide bond ("peptide bond") linkages; b) non-natural
residues in
place of naturally occurring amino acid residues; or c) residues which induce
secondary
structural mimicry, i.e., to induce or stabilize a secondary structure, e.g.,
a beta turn,
gamma turn, beta sheet, alpha helix conformation, and the like. A polypeptide
can be
characterized as a mimetic when all or some of its residues axe joined by
chemical means
other than natural peptide bonds. Individual peptidomimetic residues can be
joined by
peptide bonds, other chemical bonds or coupling means, such as, e.g.,
glutaraldehyde, N-
hydroxysuccinimide esters, bifunctional maleimides, N,N'-
dicyclohexylcarbodiimide
(DCC) or N,N'-diisopropylcarbodiimide (DIC). Linking groups that can be an
alternative to the traditional amide bond ("peptide bond") linkages include,
e.g.,
lcetomethylene (e.g., -C(=O)-CH2- for -C(=O)-NH-), aminomethylene (CH2-NH),
ethylene, olefin (CH=CH), ether (CHZ-O), thioether (CH2-S), tetrazole (CN4-),
thiazole,
retroamide, thioamide, or ester (see, e.g., Spatola (1983) in Chemistry and
Biochemistry
of Amino Acids, Peptides and Proteins, Vol. 7, pp 267-357, "Peptide Backbone
Modifications," Marcell Dekker, NY). A polypeptide can also be characterized
as a
3o mimetic by containing all or some non-natural residues in place of
naturally occurring
amino acid residues; non-natural residues are well described in the scientific
and patent
literature. The skilled artisan will recognize that individual synthetic
residues and
polypeptides incorporating mimetics can be synthesized using a variety of
procedures and
methodologies, which are well described in the scientific and patent
literature, e.g.,
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Organic Syntheses Collective Volumes, Gilman, et al., supra. Polypeptides
incorporating
mimetics can also be made using solid phase synthetic procedures, as
described, e.g., by
U.S. Pat. No. 5,422,426. Peptides and peptide mimetics can. also be
synthesized using
combinatorial methodologies. Various techniques for generation of peptide and
peptidomimetic libraries are well known, and include, e.g., multipin, tea bag,
and
split-couple-mix techniques; see, e.g., al-Obeidi (1998) Mol. Biotechnol.
9:205-223;
Hruby (1997) Curr. Opin. Chem. Biol. 1:114-119; Ostergaaxd (1997) Mol. Divers.
3:17-27; Ostresh (1996) Methods Enzymol. 267:220-234. Modified polypeptide and
peptides can be further produced by chemical modification methods, see, e.g.,
Belousov
(1997) Nucleic Acids Res. 25:3440-3444; Frenkel (1995) Free Radic. Biol. Med.
19:373-380; Blommers (1994) Biochemistry 33:7886-7896. These peptides can also
be
synthesized, whole or in part, using chemical methods well known in the art
(see e.g.,
Caruthers (1980) Nucleic Acids Res. Symp. Ser. 215-223; Horn (1980) Nucleic
Acids
Res. Symp. Ser. 225-232; Banga, A.I~., Therapeutic Peptides and Proteins,
Formulation,
~5 Processing and Delivery Systems (1995) Technomic Publishing Co., Lancaster,
PA.
Peptide synthesis can be performed using various solid-phase techniques (see
e.g.,
Roberge (1995) Science 269:202; Merrifield (1997) Methods Enzymol. 289:3-13)
and
automated synthesis may be used.
Arrays, or "BioChips"
2o The invention provides "arrays" or "microarrays" or "biochips" or "chip"
comprising the modified biological molecules of the invention, including the
modified
nucleic acids and polypeptides of the invention. Arrays axe generically a
plurality of
target elements immobilized onto the surface of the array as defined
"clusters," or
"biosites," each target element comprising a one or more biological molecules
(e.g.,
25 nucleic acids or polypeptides) immobilized a solid surface for association
(e.g., specific
binding or hybridization) to a sample. The immobilized nucleic acids can
contain
sequences from specific messages (e.g., as cDNA libraries) or genes (e.g.,
genomic
libraries), including a human genome. Other target elements can contain
reference
sequences and the like. The biological molecules of the arrays may be arranged
on the
3o solid surface at different sizes and different densities. The densities of
the biological
molecules in a cluster and the number of clusters on the array will depend
upon a number
of factors, such as the nature of the label, the solid support, and the like.
Each cluster/
biosite may comprise substantially the same biological molecule (e.g., nucleic
acid or
21
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
polypeptide), or, a mixture of biological molecules (e.g., nucleic acids of
different lengths
and/or sequences). Thus, for example, a cluster/ biosite may contain more than
one copy
of a cloned piece of DNA, and each copy may be broken into fragments of
different
lengths. The surface onto which the modified biological molecules of the
invention are
irmnobilized can include nitrocellulose, glass, quartz, fused silica, plastics
and the like, as
discussed further, below. The compositions and methods of the invention can
incorporate
in whole or in part designs of arrays, and associated components and methods,
as
described, e.g., inU.S. PatentNos. 6,197,503; 6,174,684; 6,156,501; 6,093,370;
6,087,112; 6,087,103; 6,087,102; 6,083,697; 6,080,585; 6,054,270; 6,048,695;
6,045,996;
~0 6,022,963; 6,013,440; 5,959,098; 5,856,174; 5,843,655; 5,837,832;
5,770,456; 5,723,320;
5,700,637; 5,695,940; 5,556,752; 5,143,854; see also, e.g., WO 99/51773; WO
99/09217;
WO 97/46313; WO 96/17958; WO 89/10977; see also, e.g., Johnston (1998) Curr.
Biol.
8:8171-8174; Schummer (1997) Biotechniques 23:1087-1092; Kern (1997)
Biotechniques 23:120-124; Solinas-Toldo (1997) Genes, Chromosomes & Cancer
20:399-407; Bowtell (1999) Nature Genetics Supp. 21:25-32; Epstein (2000)
Current
Opinion in Biotech. 11:36-41; Mendoza (1999 Biotechniques 27: 778-788; Lueking
(1999) Anal. Biochem. 270:103-111; Davies (1999) Biotechniques 27:1258-1261.
Substrate Surfaces
The articles of manufacture of the invention comprising arrays can have
2o substrate surfaces of a rigid, semi-rigid or flexible material. The
substrate surface can be
flat or planar, be shaped as wells, raised regions, etched trenches, pores,
beads, filaments,
or the like. Substrates can be of any material upon which a "capture probe"
can be
directly or indirectly bound. For example, suitable materials can include
paper, glass
(see, e.g., U.S. Patent No. 5,843,767), ceramics, quartz or other crystalline
substrates (e.g.
2s gallium arsenide), metals, metalloids, polacryloylmorpholide, various
plastics and plastic
copolymers, NylonTM, TeflonTM, polyethylene, polypropylene, poly(4-
methylbutene),
polystyrene, polystyrene/ latex, polymethacrylate, polyethylene
terephthalate), rayon,
nylon, polyvinyl butyrate), polyvinylidene difluoride (PVDF) (see, e.g., U.S.
Patent No.
6,024,872), silicones (see, e.g., U.S. Patent No. 6,096,817), polyformaldehyde
(see, e.g.,
3o U.S. PatentNos. 4,355,153; 4,652,613), cellulose (see, e.g., U.S. Patent
No. 5,068,269),
cellulose acetate (see, e.g., U.S. Patent No. 6,048,457), nitrocellulose,
various membranes
and gels (e.g., silica aerogels, see, e.g., U.S. Patent No. 5,795,557),
paramagnetic or
superparamagnetic microparticles (see, e.g., U.S. Patent No. 5,939,261) and
the like.
22
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Reactive functional groups can be, e.g., hydroxyl, carboxyl, amino groups or
the like.
Silane (e.g., mono- and dihydroxyalkylsilanes, aminoalkyltxialkoxysilanes, 3-
aminopropyl-triethoxysilane, 3-aminopropyltximethoxysilane) can provide a
hydroxyl
functional group for reaction with an amine functional group.
Generating Molecular Profiles of Sample Nucleic Acids
The invention provides compositions and methods for generating a
molecular profile of a nucleic acid sample, such as a sample of genomic DNA or
a cDNA
library. The invention provides articles of manufacture and methods for
contacting array-
bound nucleic acids with a sample containing nucleic acids and detecting the
binding of
1 o the sample nucleic acids to the array, thereby generating a molecular
profile of the sample
nucleic acid. In alternative aspects of the methods of the invention, the
molecular profile
can be a comparative genomic hybridization (CGH) reaction; detection of a
genomic
DNA amplification, a genomic DNA deletion, or a genomic DNA insertion;
detection of
a point mutation, such as identification of a single-nucleotide polymorphism
(SNP);
differential methylation hybridization (DMH), where the array-bound nucleic
acids axe
CpG island tags; detection of transcriptionally active regions of a genome
(using, e.g.,
nuclear run-off assays); analysis of a chromatin structure; and analysis of a
telomeric
structure (such as telomeric erosion or telomeric addition). All of these
procedures are
well known in the art, and any molecular biology procedure or analysis, can be
performed
2o using the modified biological molecules or arrays of the invention.
Comparative genomic hybridizatio~e (CGH)
In one aspect, the arrays and methods of the invention are used in
comparative genomic hybridization (CGH) reactions. CGH is a molecular
cytogenetics
approach that can be used to detect regions in a genome undergoing
quantitative changes,
i.e. gains or losses of copy numbers. Analysis of genomes of tumor cells can
detect a
region or regions of anomaly under going gains and/or losses. Differential
expression of
hundreds of genes can be analyzed using a cDNA array, thus facilitating
characterization
of gene expression in normal and diseased tissues. Generating a molecular
profile of a
nucleic acid sample by comparative genomic hybridization using methods and
arrays of
3o the invention can be practiced with methods and compositions known in the
art, see, e.g.,
U.S. PatentNos. 6,197,501; 6,159,685; 5,976,790; 5,965,362; 5,856,097;
5,830,645;
5,721,098; 5,665,549; 5,635,351; and, Diago (2001) American J. of Pathol.
23
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
May;lSB(S):1623-1631; Theillet (2001) Bull. Cancer 88:261-268; Werner (2001)
Pharmacogenomics 2:25-36; Jain (2000) Pharmacogenomics 1:289-307.
Detection of single-nucleotide polymoyphisnZS (SNPs)
In one aspect, the arrays and methods of the invention are used to detect
point mutations, such as single-nucleotide polymorphisms (SNPs). Arrays can be
used
for high-throughput genotyping approaches for pharmacogenomics, where numerous
individuals are studied with thousands of SNP markers. SNP mapping has
accelerated
complex disease gene localization; detection of multiple SNPs associated with
a disease
in a relatively small linkage disequilibrium region can narrow the linkage
region for that
1 o disease, and, identification of susceptibility genes will enable a better
understanding of
the mechanisms of the disease processes and will facilitate the discovery of
new and more
efficacious medicines. Generating a molecular profile of a nucleic acid sample
by the
analysis and detection of SNPs using methods and arrays of the invention can
be
practiced with methods and compositions known in the art, see, e.g., U.S.
Patent Nos.
~5 6,221,592; 6,110,709; 6,074,831; 6,015,888; and, Kwok (2000)
Pharmacogenomics 1:95-
100; Riley (2000) Pharmacogenomics 1:39-47; Kokoris (2000) Mol. Diagn. 5:329-
340;
Shi (2001) Clin. Chem. 47:164-172; Fan (2000) Genome Res. 10:853-860; Ianonne
(2000) Cytometry 39:131-140; Cai (2000) Genomics.66:135-143; Chen (2000)
Genome
Res. 10:549-SS7; Syvanen (1999) Hum. Mutat. 13:1-10; Pastinen (1997) Genome
Res.
20 7:606-614.
Differential methylation hybridization (DMH)
The arrays and methods of the invention are used in differential
methylation hybridization (DMH), including, for example, CpG island analysis.
In one
aspect, the array-bound nucleic acids comprise CpG island tags. In one aspect,
the
25 methods and arrays of the invention are used to identify, analyze and map
hypermethylated or hypomethylated regions of the genome. In one aspect, the
sample
nucleic acids can comprise genomic DNA digested with at least one methylation-
sensitive
restriction endonuclease and the molecular profile comprises detection and
mapping of
hypermethylated (or hypomethylated) regions of the genome. Any methylation-
sensitive
3o restriction endonuclease or equivalent endonuclease enzyme can be used,
including, for
example, NotI, SmaI, SacII, EagI, MspI, HpaII, Sau3AI and BssHII. In one
aspect of the
methods of the invention, both a methylation-sensitive enzyme and its
methylation
insensitive isoschizomer is used; see, e.g., Robinson (2000) Chromosome Res.
8:635-643;
24
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
described use of the methylation-sensitive enzyme HpaII and its methylation
insensitive
isoschizomer MspI. Windhofer (2000) Curr. Genet. 37:194-199, described
digestion of
genomic DNA with the methylation-sensitive endonuclease Sau3AI and the
methylation-
insensitive endonuclease NdeII. See also, e.g., Muller (2001) J. Biol. Chem.
276:14271-
14278; Memisoglu (2000) J. Bacteriol. I82:2I04-2112; Roth (2000) Biol. Chem.
381:269-272.
DNA methylation, or the covalent addition of a methyl group to cytosine
within the context of the CpG dinucleotide, has profound effects on the
mammalian
genome. These effects include transcriptional repression via inhibition of
transcription
1 o factor binding or the recruitment of methyl-binding proteins and their
associated
chromatin remodeling factors, X chromosome inactivation, imprinting and the
suppression of parasitic DNA sequences. DNA methylation is also essential for
proper
embryonic development, DNA repair and genome stability. For example, DNA
demethylation influence on chromosome stability is modulated by a sequence-
specific
~5 chromatin structure (the invention also provides modified biological
molecules a.nd arrays
comprising chromatin structures) (see, e.g., Vilain (2000) Cytogenet. Cell.
Genet. 90:93-
101).
Normal methylation patterns axe frequently disrupted in tumor cells with
global hypomethylation accompanying region-specific hypermethylation. When
these
2o hypermethylation events occur within the promoter of a tumor suppressor
gene, they will
silence the gene and provide the cell with a growth advantage in a manner akin
to
deletions or mutations. For example, the Rb tumor suppressor pathway is
frequently
disrupted by methylation-dependent silencing of the pl6INI~4A gene and
stimulation of
Rb degradation by a proteosomal subunit (see, e.g., Buendia (2000) Semin.
Cancer Biol.
2s 10:185-200). Reversal of abnormalities in DNA methylation may therefore
restore the
tumor-suppressive function of these genes and provide a novel approach to
cancer therapy
(see, e.g., Santini (2001) Ann. Intern. Med. 3;134(7):573-586. The
transcriptional
silencing of selected genes by DNA methylation plays a crucial role in the
development
and progression of human gastrointestinal malignancies (see, e.g., Toyota
(2000) J.
3o Gastroenterol. 35:727-734). Generating a molecular profile of a nucleic
acid sample by
the analysis of differential methylation and CpG islands using methods and
arrays of the
invention can be practiced with methods and compositions known in the art,
see, also,
U.S. PatentNos. 6,214,556; 6,180,344; 5,851,762; and, W00127317, WO9928498;
W00044934; and W01999DE03747 19991119.
2s
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Analysis of telomeric sty~uctu~e
The arrays and methods of the invention are used in the analysis of a
telomeric structure, such as telomeric erosion or telomeric addition. The
maintenance of
telomeres, which are specialized nucleoprotein structures, is essential for
chromosome
stability. Without new synthesis of telomeres at chromosome ends the
chromosomes
shorten with progressive cell division. This eventually triggers either
replicative
senescence or apoptosis when telomere length becomes critically short. The
regulation of
telomerase activity in human cells plays a significant role in the development
of cancer
(telomerase is the enzyme that synthesizes the telomere ends of linear
eukaryotic
1o chromosomes). Telomerase is tightly repressed in the vast majority of
normal human
somatic cells, but becomes activated during cellular immortalization and in
cancers.
Thus, telomerase assays are useful for cancer detection and diagnosis (see,
e.g., Hahn
(2001) Ann Med 33:123-129; Meyerson (2000) J. Clin. Oncol. 18:2626-2634;
Meyerson
(1998) Toxicol. Lett. 102-103:41-5). Using the array-based telomeric
structures of the
1 s invention will accelerate understanding of telomerase biology and lead to
clinically
relevant telomerase-based therapies. Generating a molecular profile of a
nucleic acid
sample by the analysis of telomeric structures using methods and arrays of the
invention
can be practiced with methods and compositions known in the art, see, e.g.,
U.S. Patent
Nos. 6,221,590; 6,221,584; 6,022,709; 6,007,989; 6,004,939; 5,972,605;
5,871,926;
20 5,834,193; 5,830,644; 5,695,932; 5,645,986.
Analysis of ch~omatih stf~ucture
The arrays and methods of the invention are used in the analysis of
chromatin structure, including chromatin condensation, chromatin
decondensation,
lustone phosphorylation, histone acylation, and the like (see, e.g., Guo
(2000) Cancer
2s Res. 60:5667-5672; Mahlknecht (2000) Mol. Med. 6:623-644). Chromatin
structure
remodeling occurs in certain cancers (see, e.g., Giamarchi (2000) Adv. Exp.
Med. Biol.
480:155-161). Chromatin structure affects nuclear processes that utilize DNA
as a
substrate, e.g., transcription, replication, DNA repair, and DNA organization
within the
nucleus. Chromatin structure analysis is useful in fertility assessment; for
example,
3o sperm with decondensed chromatin are infertile. DNA damage in patients with
untreated
cancer can be measured using a sperm chromatin structure assay (see, e.g.,
Kobayashi
(2001) Fertil. Steril. 75:469-475). Generating a molecular profile of a
nucleic acid
sample by the analysis of chromatin structure using the methods and arrays of
the
26
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
invention can be practiced with methods and compositions known in the art,
see, e.g.,
U.S. Patent Nos. 6,204,064; 6,187,749; 6,097,485; 5,972,608; 5,919,621;
5,470,709; and,
Dreyer (2000) Anal. Cell Pathol. 20:141-150; Hong (2001) Acta Cytol. 45:163-
168;
Evenson (1991) Reprod. Toxicol. 5:115-125.
s Nuclear gun-off assay
The arrays and methods of the invention are used in the detection and
analysis of transcriptionally active regions of nucleic acid, e.g.,
transcriptionally active
regions of a genome. In one aspect, a sample of nucleic acid can be derived
from a
nuclear run-off assay and detected and analyzed by the compositions and
methods of the
o invention. In another aspect, nuclear run-off samples are modified by the
methods of the
invention. In one aspect, these modified nucleic acids are immobilized onto an
array as
set forth in the invention.
Generating a molecular profile of a nucleic acid sample by the detection
and analysis of transcriptionally active regions of nucleic acid by, e.g.,
nuclear run-off
~5 assays, using the methods and arrays of the invention can be practiced with
methods and
compositions known in the art, see, e.g., U.S. Patent Nos. 6,200,960;
6,184,032;
6,175,060; 6,159,751; 6,022,694; 5,994,523; and, Delany (2001) Methods Mol.
Biol.
151:321-333; Srivastava (1998) Methods Mol. Biol. 86:201-207; Greene (1994) J.
Biochem. Biophys. Methods 29:179-187; Srivastava (1994) Methods Mol. Biol.
31:281-
20 288.
It will be readily apparent to one skilled in the art that various
substitutions
and modifications may be made to the invention disclosed herein without
departing from
the scope and spirit of the invention. It is understood that the examples and
aspects
25 described herein are for illustrative purposes only and that various
modifications or
changes in light thereof will be suggested to persons skilled in the art and
are to be
included within the spirit and purview of this application and scope of the
appended
claims.
3o EXAMPLES
The following example is offered to illustrate, but not to limit the claimed
invention.
2~
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Example 1: Preparation of Modified Nucleic Acid Using 3-glycidoxypropyl-
trimethoxysilane
The following example describes making and using one aspect of modified
nucleic acid of the present invention. The purpose of the chemical
modification is to
enable the nucleic acid to be readily affixed to an underivatized solid
surface. In this
example, the nucleic acid--preferably DNA--is modified by reaction with 3-
glycidoxypropyl-trimethoxysilane (GPTS), according to FIG. 1. GPTS has in fact
been
previously used to derivatize a glass surface upon which (unmodified) DNA
samples are
1o then contacted and immobilized. Yet the use of GPTS is for the opposite
purpose: to
modify the DNA for subsequent attachment to an underivatized glass surface,
has not
been previously disclosed nor suggested. Moreover, GPTS--since it contains an
epoxide
group--is known to damage DNA i~c vivo. For these reasons, ifs use to
derivatize DNA is
actually discouraged by the prior art.
~ 5 Schematically, affixing the nucleic acid to the solid support consists
essentially of two steps. In the first, the nucleic acid reacts with the
epoxide end of the
GPTS molecule; in the second step, the glass surface reacts with the other
end, or the
silane end of the GPTS-modified nucleic acid, thereby affixing the nucleic
acid onto an
underivatized glass surface. The entire reaction is rapid, is characterized by
a favorable
2o equilibrium, and occurs under very mild conditions using a minimum of
inexpensive
reagents. Though there quite obviously are numerous ways to carry out either
step of the
reaction, the preferred method is shown in this and the following example.
As depicted in FIG. 1, a chemical compound having a cyclic or ring ether
and an alkoxysilane--in this instance ethylene oxide and trimethyloxysilane,
respectively-
25 -comprise the two ends of the compound; the two ends are connected by a
four-carbon
ether linkage. The compound shown is 3-glycidoxypropyltrimethoxysilane or
GPTS. In
the first step, DNA is reacted with GPTS at basic pH, preferably above 9.5, to
form the
modified DNA. The modified DNA is then reacted with an underivatized glass (or
other
silanol-containing) surface at neutral pH, thus immobilizing the DNA onto the
glass
so surface. In the first step, the ring ether functionality reacts with the
DNA. Again, the
ring ether need not be ethylene oxide, as it is in GPTS, although the small
ring is
preferred to increase reactivity of the ether functionality, which is
relatively unreactive.
The first reaction, leading to the derivatized DNA, is a ring-opening
reaction likely involving carbon 5 of the ribose ring of the DNA. This
derivatized DNA is
2s
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
unusually stable and can be stored for long periods of time prior to actual
use. The second
reaction, immobilizing the derivatized DNA onto the glass surface, is a simple
substitution reaction creating an Si--O--Si linkage in the glass surface, and
removing one
of the allcoxy groups from the GPTS molecule.
Example 2: Preparation of Modified Nucleic Acid Using 3-aminopropyl-
triethoxysilane
The following example describes making and using another aspect of
modified nucleic acids of the present invention. The purpose of the chemical
modification is to enable the nucleic acid to be readily affixed to an
underivatized solid
o surface. In this example, the nucleic acid, preferably DNA, is modified by
reaction with
3-aminopropyl-trimethoxysilane, according to FIG. 2. As in example 1, affixing
the
nucleic acid to the solid support consists essentially of two steps. In the
first, the nucleic
acid reacts with the amino end of the 3-aminopropyltrimethoxysilane molecule;
in the
second step, the glass surface reacts with the other end, or the silane end of
the 3-
~ 5 aminopropyltrimethoxysilane-modified nucleic acid, thereby affixing the
nucleic acid
onto an underivatized glass surface.
As in example 1, the entire reaction is rapid, is characterized by a
favorable equilibrium, and occurs under very mild conditions using a minimum
of
inexpensive reagents. Though there quite obviously are numerous ways to carry
out
2o either step of the reaction, the preferred method is shown in this and the
following
example.
As depicted in FIG. 2, a chemical compound having an amino group and
an alkoxysilane--in this instance --NH2 and triethyloxysilane, respectively-
comprise the
two ends of the compound; the two ends are connected by a propyl linkage. The
25 compound shown is 3-aminopropyltriethoxysilane. In the first step, DNA is
reacted with
3-aminopropyltriethoxysilane at neutral pH in the presence of sodium
bisulfite, or
equivalent.
The first reaction, leading to the derivatized DNA, is transamination
reaction of the cytosine residues on nucleic acids. The second reaction, as in
Example 1,
3o involving immobilizing the derivatized DNA onto the glass surface, is a
simple
substitution reaction. It creates an Si--O--Si linkage in the glass surface
and removes one
of the alkoxy groups from the GPTS molecule.
29
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Examule 3: Preparation of a High Density Microarray using Modified Nucleic
Acid
The following example describes malting a high-density microarray of the
invention.
Once the modified nucleic acids of the present invention, such as those
described in Examples 1 and 2, are prepared, they can then be exploited in a
variety of
ways, including, e.g., to make a high density array. Again, these modified
nucleic acids
(particularly DNA) can be immobilized onto a glass surface simply by
contacting the
1 o modified DNA onto the underivatized surface. The significance of this is,
among other
things, that spreading (migration of the DNA sought to be immobilized from the
desired
location) and non-specific probe sticking (caused by derivatization of the
glass surface
which creates a net positive electrostatic charge upon the surface which
attracts the net
negatively charged DNA) are essentially eliminated.
~ 5 These advantages allow the creation of extraordinarily high-density
microarrays, which is highly desirable. For instance, due to the elimination
of spreading,
and the effective elimination of probe sticking, a single small glass surface
can contain
virtually thousands of DNA samples to be tested, each of which is microscopic
in size, all
immobilized upon a single glass surface. Indeed, one can construct a
microarray
2o consisting of multiple single sample spots smaller than 50 microns placed
upon a glass
surface.
A high-density microarray consisting of multiple DNA samples of this
type is also easily constructed in accordance with the present invention. The
modified
DNA can be prepared (for instance, in accordance with Examples l and 2) well
in
25 advance of actual use. These chemically modified DNA samples are analogous
to "DNA
chips" that can then be readily "imprinted" upon an unaltered glass sheet in,
for instance,
grid fashion. FIG. 3 illustrates one aspect of a device for preparing such a
high-density
microarray using the DNA chips of the present invention. In one preferred
aspect, the
device is made from a plurality of inexpensive commercially available
capillary
so micropipets, preferably 10 cm micropipets, although other sizes will, of
course, work. As
depicted in FIG. 3 each 10 cm micropipet is pulled to make a taper at one end.
They are
arranged in a hexagonal close-packed array, bounded by a square frame. The
micropipets
can be glued to one another to form a stable unit within the frame. The
tapered ends
(FIG. 3B) are cut off and polished to optical flatness.
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
To prepare the microarray, the tips of the device are dipped into a multi-
well container that contains the (chemically modified in accordance with the
present
invention) DNA samples to be tested, and whose wells are aligned with the
micropipets
of the device. Upon contact of the tips into the wells, a small portion of
each DNA sample
is deposited into the micropipet corresponding to the particular well by
simple capillary
action. The size of the spot can be carefully controlled by the size of the
tapered end.
Using this device and the DNA chips of the present invention, thousands of
samples can
be arrayed in a narrow area, simultaneously and without the need for expensive
robotics.
Indeed, the method (comprising the DNA chips and pipet device) of the present
invention
1 o has been shown to be even more efficient than methods using high-speed
spotting robots.
Finally, the compounds, methods and devices of the present invention are
readily
incorporated into a pre-packaged kit for commercial sale.
The high-density microarray of the present invention can also be readily
incorporated into the microarray systems of the art, such as those disclosed
in the art
~ 5 section above. For instance, fluorescent ih situ hybridization (FISH) and
the method
described in Shalon (1996) 6 Genome Res. 639 (1996), describing a microarray
system is
presented for analyzing DNA samples that involves making microarrays of DNA
samples
on glass substrates, probing them by hybridization with complex fluorescent-
labeled
probes, and using a laser-scanning microscope to detect the fluorescent
signals
2o representing hybridization. Similarly, Sargent, et al. (U.S. Pat. No.
5,601,982) discloses a
method and apparatus for determining the sequence of polynucleotides involving
scanning the nucleic acids by scanning tunneling microscopy.
One skilled in the art recognizes that this invention is not limited to using
only nucleic acids. Other biological molecules, such as biopolymers, e.g.,
DNA, RNA,
25 proteins, peptides or polypeptides, and polysaccharides, can be directly
activated using
the methods of the invention, such as bifunctional silane compounds or other
crosslinking
reagents, resulting in an immobilized biologic molecule, e.g., a biopolymer,
to a solid
surface. This invention demonstrates that the target molecules to be arrayed
(i.e.,
immobilized) are first modified so that they have gained a binding affinity
for solid
3o surfaces without losing their probing (e.g., hybridization) abilities.
Because modification
is a separate process, virtually any biological molecule can be modified and
arrayed
(immobilized). Thus, a skilled artisan realizes that this invention is not
limited to nucleic
acids, but can also be used for a spectrum of biological molecules.
31
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Examule 4: Preparation of Modified Nucleic Acids
The following example demonstrates methods for preparing modified
biological molecules of the invention by describing the modification of
nucleic acids.
Tlsitag Halogehated Silahes: This example describes another form of modified
nucleic acid of the present Invention. Again, the purpose of the chemical
modification
disclosed and claimed here is to enable to nucleic acid to be readily affixed
to an
underivatized solid surface, e.g., ordinary quartz glass. According to Fig. 4,
a modified
nucleic acid in accordance with the present invention is prepared by reacting
unmodified
nucleic acid under near neutral pH with suitable silane compounds. The "X" in
FIG. 4
o can refer to any halide, preferably Cl, Br, or I; Rl, Ra and R3, can be the
same or different,
including, --OCH3, and --OCaHS. In particularly, preferred aspects, the
halogenated
silane depicted to the left of the arrow in Fig. 4 is 8-
bromocytltrichlorosilane, 8-bromo-
cytltrimethoxysilane, 4-chlorobutylmethyldichlorosilane, and 3-
iodopropyltrimethoxysilane.
The conversion depicted in Fig. 4 was performed as follows. The
halogenated silane was dissolved in dimethylformamide (DMF) at a concentration
of
about 30 mM. Next, 3 ~g to 10 ~,g of nucleic acid was dissolved in 100 u1 of
0.01 M
phosphate buffer (pH 7.0). Then 1 to 3 ~.g of 30 mM halogenated silane was
added, the
solution is then mixed well, and allowed to react at about 37°C for
about 3 hours
(alternatively, it can be reacted at ambient temperature overnight). After
reaction, the
desired product -- the modified nucleic acid -- is purified by ethanol
precipitation; then
the modified nucleic acid is dissolved in water.
Example 5: Preparation of Arrays and Controlling Spot (Cluster) Density and
Size
The following example demonstrates an exemplary method for
manufacturing the arrays, or "biochips" of the invention.
As discussed throughout the present application, one particular advantage
of the present invention is that it allows the investigator to prepare
unusually high-density
microarrays to conduct nucleic acid studies. This example is best understood
in relation
3o to example 3, which disclosed the preparation of a high-density microarray
in accordance
with the present invention. This example discloses enhanced methods for
controlling the
size and density of the individual nucleic acid "spots" or "clusters" on the
solid supports,
in accordance with the present invention.
32
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Small "spot" or "cluster" size, in relation to high-density microarrays,
allows higher sample density (i.e., more samples per unit area) and superior
detection
sensitivity (because the signals are less diffuse). In the conventional solid
support
systems, the skilled artisan faces a crucial dilemma. An ordinary clean quartz
glass
surface--of the type used in the experiments described here--is very
hydrophilic. Thus,
nucleic acid samples will naturally tend to spread out when placed on the
glass surface.
Again, this is undesirable. To mitigate spreading, the skilled artisan can
treat the surface
to make it more hydrophobic--e.g., either pretreating the surface with a
hydrophobic
agent, or simply by dehydrating the surface. Naturally, either of these
options makes the
o glass surface less reactive towards silane-modified nucleic acids.
In a family of aspects of the present invention discussed in this example,
the skilled artisan is spared this dilemma. More specifically, spreading can
be eliminated
yet the reactivity of the surface towards the modified nucleic acids can be
maintained
through the use of another type of silanes of the present invention. For
instance, one quite
general aspect of these silanes after hydrolysis contains an Si(OH)3 at each
end, linked by
a hydrophobic group. See FIG. 6. Any of a variety of hydrophobic linkers can
be used.
Particularly preferred aspects include: 1,6-Bis-trichlorosilyhexane, 1,8-Bis-
trichlorosilyloctane, 1,6-Bis-trimethoxysilyhexane, and 1,4 Bis-
trimethoxysilylethylbenzene. Thus, according to these aspects of the present
invention,
one end of the silane attaches to the surface, and the other end remains
reactive to the
modified biological molecule, e.g., nucleic acid. The hydrophobic linker
confers
hydrophobicity to the surface. Thus, the skilled artisan can readily see how
the
electrostatic properties of the surface (hydrophobic versus hydrophilic) can
be readily
modulated--e.g., the chain length of the linker can be adjusted to control
hydrophobicity,
2s and the surface reactivity can be controlled by adjusting the amount of
silane contacted
with the surface.
33
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
To prepare the solid supports in accordance with this aspect of the present
invention, the glass surface was cleaned by slowly boiling in 3 M HCl for
about 2 hrs in a
fume hood. Next, the surfaces were rinsed with deionized water then kept in
0.1 M HCl
until ready for use. When ready for use, the surfaces were rinsed with doubly
distilled
deionized water to remove any extant acid, then rinsed in absolute ethanol.
Next, the
surfaces were immediately transferred to an ethanol solution containing 0.0005
% to
0.002 % of the bi-functional silanes of this aspect of the invention. The
surfaces were
then treated at room temperature for about 48 hours. The surfaces were then
rinsed with
ethanol and air dried. Finally, the glass surfaces were stored in a dust-free
environment
o until ready for use.
Example 6: Preparation of Modified Nucleic Acids Using Amine-Containing
Silane Compounds
The following example describes another form of modified nucleic acid of
the present invention. In this family of aspects, the modified nucleic acid is
prepared by
~ 5 reacting pristine nucleic acids with an amine-contaiung silane.
Heuristically, the
derivatization of nucleic acid with amine-containing silanes is comprised of
two steps: (1)
the halogenation (or bromination, as shown) of the nucleic acid (FIG. 5a, Sb);
and (2) the
derivatization of the halogenated nucleic acid (FIG. 5c). As depicted in FIG.
5a, Sb, the
reaction can occur in the presence of N-bromosuccinimide under mild pH
conditions;
2o varying either of these reaction variables allows the skilled biochemist to
control the
reaction rate. Also as evidenced by FIG. 5a, Sb, the reaction normally occurs
at the
guanine or cytosine base depending upon the pH--i.e., neutral to slightly
basic pH favors
reaction at the guanine residue, more basic pH favors reaction at the cytosine
residue.'
Slightly different reaction protocols are preferably used depending upon
25 whether the nucleic acid is DNA or RNA. For DNA, 5 ~,g of DNA was dissolved
in 100
~1 of 0.1 M NaHC03, to reach a pH of about 9.5. This solution is kept on ice
for about 5
minutes. Contemporaneously, a fresh N-bromosuccinimide solution at
concentration of
about 10 mM was prepared and also chilled on ice. Next, 1 ~.1 of the N-
bromosuccinimide solution is added to the DNA solution; the solution was then
stirred
3o vigorously (to vortex). The reaction was then allowed to proceed on ice for
about 15
minutes. Next, 10 ~ul of 0.5 M aminosilane solution at pH about 9.5 to about
12, was
added to the bromine-activated DNA solution; this new mixture was allowed to
react at
65°C for about 2 hours. Finally, the silane-modified DNA was purified
by methods well
34
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
known in the art; preferably, it is purified by ethanol precipitation, or
equivalent
procedures.
For RNA, a similar, though slightly different protocol was used: 5 ~,g of
RNA was dissolved in 100 ~.1 of 0.1 M phosphate buffer, to reach a pH of about
7.5. This
solution is kept on ice for about 5 minutes. Contemporaneously, a fresh N-
bromosuccinimide solution at concentration of about 10 mM was prepared and
also
chilled on ice. Next, 1 ~,1 of the N-bromosuccinimide solution is added to the
DNA
solution; the solution was then stirred vigorously (to vortex). The reaction
was then
allowed to proceed on ice for about 15 minutes. Next, 10 ~,l of 1 M
aminosilane solution
1 o at pH about 8.0, was added to the bromine-activated DNA solution; this new
mixture was
allowed to react at 45°C for about 2 hours. Finally, the silane-
modified DNA was purified
by methods well known in the art; preferably, it is purified by ethanol
precipitation, or
equivalent procedures.
In these aspects the following silanes are available for these reasons:
R1
H2N - X - Si - OR ,
R2
R can be -CH3, or -C2 H5;
Rl can be H, -CH3, -CZHS,-OCH3, or -OC2H5;
R2 can be H, -CH3, -C2H5,-OCH3, or -OC2H5;
Further any other amino silane compound after hydrolysis that takes the
following form is useful:
R1
HaN - X - Si - OH ,
R2
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
Example 7: Preparation of Modified Biological Molecules (Biopolymers) Using 3-
glycidoxypropyltrimethoxysilane
This example describes methods to modify biological molecules, e.g.,
nucleic acids, using bifunctional silane compounds.
The purpose of the chemical modification is to enable sample (the
biological molecule) to be readily affixed to an underivatized solid surface.
In this
example, a biopolymer is modified by reaction with 3-
glycidoxypropyltrimethoxysilane
(GPTS).
1 o Schematically, affixing the biopolymer to a solid surface consists
essentially of two steps. In the first, the biopolymer reacts with the epoxide
end of the
GPTS molecule; in the second step, the glass surface reacts with the other
end, or the
silane end of the GPTS-modified biopolymer, thereby affixing the biopolymer
onto an
underivatized surface.
15 A skilled artisan recognizes that a variety of bifunctional crosslinking
reagents could be used in the present invention (see above). Crosslinking
reagents and
the conditions required for their use are well known in the art, thus, one
skilled in the art
would be able to extrapolate the information provided by this application and
utilize
specific crosslinking reagents and conditions to obtain a specific modified
biopolymer.
2o Example 8: Preparation of Modified Small Molecules
This example describes methods to modify biological molecules which
may not be described as biopolymers. As described above, any biological
molecule can
be incorporated into the compositions and methods of the invention. These non-
biopolymers are first crosslinked to epoxide silane-activated biopolymers,
e.g.,
25 biopolymers activated according to Example 7. The crosslinking of these non-
biopolymers, which are typically small molecules, increases the size and
stability of the
molecule. Once the non-biopolymer is crosslinked to an activated biopolymer,
e.g.,
polyethylene glycol (PEG) or DNA, these crosslinked molecules can be
immobilized on a
solid surface by direct deposition and curing under proper conditions.
3o Example 9: Silanization of Amine-Containing Biopolymers such as Proteins
and
Polypeptides (e.g., Antibodies), Polysaccharides and Lipids
This example describes methods to modify amine-containing biological
molecules by silanization. Such biological molecules include peptides and
polypeptides
(e.g., antibodies), lipids and polysaccharides, in addition to nucleic acids
compxising
36
CA 02446050 2003-10-29
WO 02/092615 PCT/USO1/15446
amine groups. Nucleic acids comprising amine groups include, e.g., nucleotide
compositions containing aminooxy moieties, as described in U. S. Patent No.
6,127,533.
Biopolymers are effectively silanized and arrayed onto glass surfaces.
Biopolymers are first treated with 2-iminothiolane (commonly known as "Traut's
Reagent") or N-succinimidyl S-acetylthioacetate (SATA) or succinimidyl
acetylthiopropionate (SATP) to introduce an active sulfhydryl functional
group. The
activated biopolymers are silanized by reacting with the epoxide silane
compound as
described previously under mild conditions. A typical reaction is depicted in
Figure 7;
this example uses SATP and an epoxide silane comprising --Si(OCH3)3.
To Antibodies are silanized by various methods. One such method is to first
dissolve an antibody in 0.1 M sodium phosphate buffer (pH 7.3) with 50 mM NaCI
and
mM EDTA at a concentration of about 1 to 3 mg/ml. Then, add 5 ~,1 of 100 mM
SATA or SATP in a DMSO solution to 1 ml antibody solution and react at room
temperature (RT) overnight. Next, add 100 ~.M of 1 M hydroxylamine
hydrochloride and
react at RT for one hour. After the RT activation, add 10 ~.M of 0.2 M 3-
glycidoxypropyltrimethoxysilane (epoxide silane) and react at RT for about 5
hours.
Upon completion of all reactions, antibodies are purified by gel filtration on
a Sephadex
G25 column, or equivalent. The modified antibody is fixed on a glass surface
by direct
deposition.
2o All patents, publications mentioned in this specification are indicative of
the level of those skilled in the art to which the invention pertains. All
patents,
publications hexein are incorporated by reference to the same extent as if
each individual
publication was specifically and individually indicated to be incorporated by
reference.
One skilled in the art will readily appreciate that the present invention is
well adapted to carry out the objects and obtain the ends and advantages
mentioned as
well as those inherent therein. The chemically modified nucleic acids, their
attachment to
solid support, along with the sequences, methods, procedures, assays,
molecules, devices
and specific compounds described herein are presently representative of the
preferred
aspects are exemplary and are not intended as limitations on the scope of the
invention.
3o Changes therein and other uses will occur to those skilled in the art which
are
encompassed within the spirit of the invention and are defined by the scope of
the claims.
37