Note: Descriptions are shown in the official language in which they were submitted.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
1
COMPOSITIONS AND METHODS FOR TREATING CELLULAR RESPONSE TO
INJURY AND OTHER PROLIFERATING CELL DISORDERS REGULATED BY
HYALADHERIN AND HYALURONANS
TECHNICAL FIELD
The present invention relates generally to the understanding, diagnosis and .
treatment of a wide variety of diseases, and more specifically, to
compositions and
methods for treating cellular response to injury and/or the abnormal
proliferationof cells.
BACKGROUND OF THE INVENTION
The normal cell in normal tissue is confined to a narrow range of
function and structure regulated by its differentiation state, genetic program
of
metabolism, tissue specialization, by constraints induced by neighboring
cells, the
extracellular matrix and availability of exogenous factors and metabolic
substrates.
Cells are able to handle normal physiological demands (homeostasis) or adapt
to
excessive physiological stresses and pathological stimuli by altering their
steady state to
preserve cell viability and function. If the adaptive responses to stimuli axe
exceeded,
then a sequence of cellular events follow that transform normal cells to
injured or
diseased cells in an attempt to remodel local tissue. This process can lead to
a number
of diseases driven by enhanced cell proliferation, migration and invasion,
production of
matrix metalloproteinases, infiltration of inflammatory cells, tissue
destruction and
dysfunctional tissue remodeling. Regardless of the etiology, such disease
processes are
commonly found in inflammatory diseases (such as arthritis, multiple
sclerosis,
psoriasis, inflammatory bowel diseases, diabetes), proliferative diseases
(such as cancer
and metastases), degenerative diseases (such as osteoarthritis, osteoporosis,
Alzheimer's, Parkinson's) and injuries caused by wounds or burns.
Current medical approaches to treat or prevent such diseases typically
involve the use of reagents that attempt to block mechanisms affecting cell
proliferation, cell migration, or the production of enzymes or growth factors.
However,
because such reagents are not specific to diseased cells and current practices
do not yet
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
2
allow targeting of these reagents specifically to sites of disease, such a
therapeutic
approach is typically toxic to the host if used for any length of time or at
high dosages
as may be required to treat or prevent the disease. This toxicity of current
reagents is a
severe limitation to the efficacy of current medical treatments.
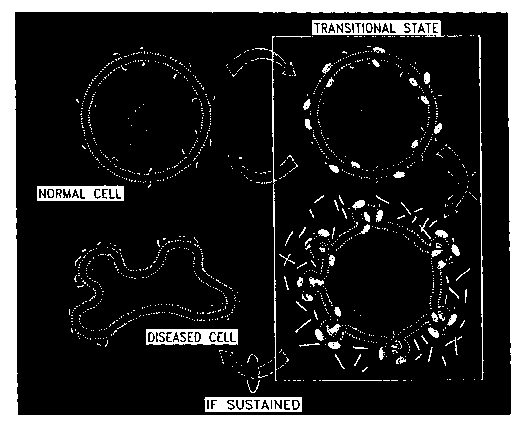
The present invention discloses a sequence of cellular transition states
that are involved in the transformation of normal cells to diseased cells that
is
characteristic to all cell types, and thus, all tissues. Transitory molecules
produced
during the early phases of disease which are responsible for the transition of
cells from
normal to diseased state are described, as well as the use of such molecules
in the
diagnosis, treatment and/or prevention of a wide variety of diseases is
provided.
SUMMARY OF THE INVENTION
Briefly stated, the present invention provides compositions and methods
for treating a tissue disorder associated with a response-to-injury process,
or
proliferating cells in a mammal. The methods include administering to the
mammal an
effective amount of a composition that alters the activity of transition
molecules within
a cell Transition molecules are comprised of hyaladherins, hyalauronans or
molecules
regulated by an amount of intracellular or extracellular hyaladherins or
hyalauronans.
The activity of hyaladherins and hyalauronans are shown to interact with
regulatory
processes associated with a response to injury and/or proliferative/invasive
cell types.
Within one aspect of the present invention, methods are provided for
treating a wide variety of inflammatory or proliferative diseases, comprising
the step of
administering to a warm-blooded animal (e.g., a human, horse, cow, sheep, dog,
cat, or
mouse) a compound to treat the inflammatory or proliferative disease.
A wide variety of compounds may be utilized within this regard,
including for example (a) a polypeptide comprising the amino acid sequence
BX7B
(SEQ ID N0:2S) which binds HA; phage display selected peptides that bind HA
such as
polypeptides comprising P-15 (Sequence ID No. 70), P-16 (Sequence ID No. 26);
P-32
(Sequence ID no. 71); and GAHWQFNALTVR (Sequence ID No. 72); (b) an antibody
which binds any one of domains Dl, D2, D3, D4, or, DS of RHAMM (e.g., an anti-
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
3
TAM antibody); (c) a polypeptide fragment which encodes a D1, D2, D3, D4, or,
D5
domain of RHAMM; and (d) a gene delivery vector which expresses antisense
RHAMM, or, delivers and expresses any one of (a), (b), or (c). In one
embodiment, the
polypeptide can be (a) a first peptide comprised of a hyaluronic acid-binding
domain;
(b) a hyaladherin polypeptide; (c) a second peptide comprised of a domain from
a
hyaladherin polypeptide; (d) a hyaladherin-binding polypeptide; (e) a third
peptide
comprised of a hyaladherin binding domain. Also provided are antibodies which
binds
to a peptide or polypeptide of (a)-(d); and/or vectors (e.g., gene delivery
vectors
described below) that expresses a gene encoding a polypeptide as described
above or
herein. More particular embodiments include methods where a hyaladherin. is
administered comprised of a RHAMM polypeptide. In a particular embodiment, a
first.
peptide is administered comprised of a sequence selected from the group
consisting of
SEQ. ID NO: 1-10. In another embodiment, a second peptide is administered
comprised of a sequence selected from the group consisting of SEQ. ID NO: 11-
20. In
another embodiment, a hyaladherin-binding polypeptide comprised of SEQ. ID NO:
21.
In still other embodiments, an antibody is administered that binds to a
peptide
comprised of SEQ. ID NO: 11-20 or an antibody is administered that binds to a
RHAMM polypeptide. In a more general embodiment, at least one hyaladherin is
administered that is a RHAMM polypeptide.
Within other embodiments, the compound to be delivered is a
polypeptide comprising less than an entire RHAMM polypeptide (i.e., less than
95 kD
or 73kD molecular weight), and containing at least a portion of RHAMM domains
Dl
(e.g., as 1-164 of human RHAMM), D2 (the "leucine zipper" domain, e.g., as 195-
122
of human RHAMM), D3 (the "TAM domain", e.g., as 219-240 of human RHAMM),
D4 (the repeat or "R" domain - e.g., aa. 442-546 of mouse and aa. 442-463 of
human
RHAMM), or, D5 (two HA binding domains - e.g., aa. 721-730 and 742-752 of
mouse
RHAMM, and aa.635-645 and aa. 657-666 RHAMM). Equivalent domains in other
species may be determined by comparison with the human sequence (see, e.g.,
Figure
50; SEQ ID Nos. 47 and 48). Within further embodiments, antibodies are
provided
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
4
which bind to any one of the above RHAMM domains (i.e., domains D1, D2, D2,
D4,
or, DS). Examples include antibodies to SEQ ID Nos. 14, 17, 23, 24, 26 and 27.
A wide variety of inflammatory or proliferative diseases may be treated
with the aforementioned compounds, including for example, inflammatory or
proliferative neurological diseases such as Parkinsons or Alzheimer's disease,
arthritic
diseases (e.g., rheumatoid arthritis and osteoarthritis), diseases associated
with
demylination of the nerve sheath (e.g., multiple sclerosis), inflammatory
dermatosis
(e.g., psoriasis), inflammatory bowel diseases, a variety of wounds (e.g.,
surgical
excisions adhesions, scars, and cheloids, and skin ulcers), stenosis and/or
restenosis (as
well as proliferative. inflammatory responses or fibrotic response associated
with
medical implants, such as hip implants, vascular wraps, catethers, and the
like), cancer
and other malignant diseases, kidney fibrosis, 'inflammatory lung diseases
(e.g.,
emphysema, asthma, and cystic fibrosis), obesity and obesity-related diseases
(including
for example, diabetes), lupus, tissue transplantation (e.g., skin grafts) and
cardiovascular
diseases (e.g., atherosclerosis, and stroke).
Within the context of the present invention, it should be noted that the
above-noted diseases are deemed to be "treated" if the typical disease course
is altered,
slowed, inhibited, or prevented in a statistically significant manner, for at
least one
symptom or sign of the disease.
In related embodiments, the invention provides a composition for
treating a tissue disorder associated with a response-to-injury process or
proliferating
cells in a mammal comprising the compositions described in the aforementioned
methods of treating.
In a different aspect, the invention provides cell cultures which are
comprised of a transition cells, wherein the transition cells include an
activated er~k kinase
signaling activity, a stimulated AP-1 binding activity and at least one
characteristic
selected from the group consisting of: (a) increased podosome formation, (b)
increased
flux of intracellular or extracellular hyalauronans or hyaladherins, (c)
increased expression
of a hyaladherin, (d) an inability to form focal adhesions, (e) increased
metalloproteinase
activity, and (f) increased expression of hyaladherin; wherein measurements of
activity or
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
S
amounts are relative to a normal quiescent cell. In more specific embodiments,
the cell
culture is comprised of cells transfected with a nucleic acid that
overexpresses a
hyaladherin. In a still more specific embodiment, the cell culture
overexpresses a
RHAMM polypeptide. In a related aspect, the invention provides embodiments of
an
animal cell culture wherein the activated erk signaling activity is less than
the activated
er~k signaling activity displayed by normal quiescent cells in response to a
growth factor
stimulation, wherein the growth factor stimulation is selected from the group
consisting
of IL-1 stimulation, PDGF stimulation, and NTF a stimulation.
In still another aspect, the invention provides methods of identifying a
~10 composition for treating a tissue disorder associated with a response-to-
injury process
or proliferating cells in a mammal comprising, (a) treating the cell culture
as described
above with a candidate composition; (b) scoring for the ability of the
candidate
composition to inhibit podosome formation in comparison to an untreated
culture
containing transition cells as described above; and (c) selecting the
composition that
inhibits podosome formation. One embodiment provides a method where scoring
for
the ability to inhibit podosome formation comprises testing the treated cell
culture for a
reduced ability to release a protease that degrades a cellular matrix
component. In a
more particular embodiment of the aforementioned method, the protease is a
fibronectin
degrading protease that releases a CS-1 sequence from a fibronectin matrix,
and the
20' scoring comprises testing the ability of the treated cells to bind an
antibody specific for
the CS-1 sequence.
In a different aspect, the invention provides peptide compositions that
bind a hyalauronan comprising a peptide of the sequence BX7B (SEQ ID N0:28)
wherein B is a basic amino acid and X7 is a sequence of about seven residues
selected
from any amino acid other than an acidic amino acid, wherein the peptide forms
an
alpha helix and each occurrence of B is oriented on the same side of the alpha
helix, and
with the proviso that the peptide does not consist of the sequences
BBXXBBBXXBB,
KQKIKHVVKLK, KLKSQLVKRK, RYPISRPRKR, KNGRYSISR,
RDGTRYVQKGEYR, RRRCGQKKK, RGTRSGSTR, RRRKKIQGRSKR,
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
6
RKSYGKYQGR, KVGKSPPVR, KTFGKMKPR, RIKWSRVSK, KRTMRPTRR,
KVGKSPPVR, or HREARSGKYK (SEQ ID Nos. 29-44 respectively).
In a related aspect, the invention provides peptide compositions that bind
a hyalauronan comprising a peptide of the sequence BX7B (SEQ ID N0:28) wherein
B
is a basic amino acid and X7 is a sequence of seven residues selected from
amino acids
other than acidic amino acids, wherein the peptide satisfies one of the
following sets of
requirements: (a) the amino acid residue at position 4, 5 or 6 is K, H, R, A,
C, G, S, or
T, (e.g., P-peptide (RSHKTRSHH; SEQ ID N0:61) and R-peptide (RGGGRGRRR;
SEQ ID NO:27)); (b) the amino acid residue at position 4 or S is K, H, R, A,
C, S, or T
1~0 and the amino acid residue at position 6 is A, P, F, W, M, I, V, or L; (c)
the amino acid
residues at positions 2 and 6 are I, L, or V, and the amino acid residue at
position 5 is K,
H, R, Q, or N, (e.g., fibromodulin (KVGRKVFSK; SEQ ID N0:64), CD40 receptor
associated factor-1 (KCSVQTLLR; SEQ ID N0:65), probable G protein coupled
receptor (RTHLKHVLR; SEQ ID N0:66), leukoctye derived chemotaxin-2
(KNAINNGVR; SEQ ID N0:67), laminin gamma-2 (KGQINNSIK; SEQ ID N0:68));
(d) the amino acid residues at position 2 and 6 are I, L, or V, the amino acid
residue at
position 4 is N, G, Q, G, S, or T, the amino acid residue at position 5 is R,
N, C, Q, H,
or K, the amino acid residue at position 8 is R, N, C, Q, G, H, K, S, or T,
and the amino
acid residues at positions 3 and 7 are any residue except D, E, P, W, Y, M, or
F; (e) the
amino acid residues at positions 2 and 6 are A, I, L, or V, the amino acid
residue at
position 3 is anything except D, E, P, W, Y, M, or F, the amino acid residue
at position
5 is R, N, Q, H, or K, the amino acid residue at position 7 is H, R, or K,
(e.g., cdc37
(RVRGRAKLR; SEQ ID NO: 69)); or (f) the amino acid residues at positions 2 and
6
are I, L, or V, the amino acid residue at position 3 is any residue except D,
E, P, W, Y,
M, or F, the amino acid residue at position 4 is K, R, or H, the amino acid
residue at
position 5 is R, N, Q, H, or K, and the amino acid residue at position 7 is H,
R, or K.
The invention also provides peptide compositions comprising a peptide that is
a part of
any peptide that meets the requirements of set (b) described above as long as
the peptide
has no acidic residues and also satisfies the requirements of set (b).
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
7
In yet another related aspect, peptide compositions are provided that
binds a hyalauronan or block podosome formation in a cell, wherein the peptide
is
selected from group consisting of: (H4-5)B3 (SEQ. ID. NO: 1); (H4-5)BXBBXB
(SEQ. ID. NO: 2); (H4-5)BXBXBBB (SEQ. ID. NO: 3); (H4-5)BXBBB (SEQ. ID. NO:
4); and (H4-5)BXBB (SEQ. ID. NO: 5). In these embodiments B is lysine (K) or
arginine (R), X is a hydrophobic or neutral amino acid selected from the group
consisting of (I, L,V,Q, S) and H4-5 is sequence of 4 to 5 amino acids
selected to form
an alpha helix. In more particular embodiments of this aspect, the peptide is
selected
from the group consisting of MMTVLKR (SEQ. ID. NO: 6); MMTVLKVKRLR
(SEQ. ID. NO: 7); MMTVLKVKVKRK (SEQ. ID. NO: 8); MMTVLKVRKR (SEQ.
ID. NO: 9); and MMTVLKVRK (SEQ. ID. NO: 10).
In yet a different aspect, the invention provides methods of identifying a
peptide or polypeptide composition for treating a tissue disorder associated
with a
response-to-injury process or proliferating cells in a mammal comprising the
steps of:
(a) selecting a sequence from a database screened for sequences comprising a
peptide of
the sequence BX7B (SEQ ID N0:28) wherein B is a basic amino acid and X7 is a
sequence of about seven residues selected from any amino acid other than an
acidic
amino acid, wherein the peptide forms an alpha helix and each occurrence of B
is
oriented on the same side of the alpha helix, (b) preparing a composition
comprised of
the selected sequence; and (c) testing the prepared composition for the
ability to inhibit
podosome formation according to other methods taught by this invention, and
with the
proviso that the peptide in (a) does not consist of the sequences BBXXBBBXXBB,
KQKIKHVVKLK, KLKSQLVKRK, RYPISRPRKR, KNGRYSISR,
RDGTRYVQKGEYR, RRRCGQKKK, RGTRSGSTR, RRRKKIQGRSKR,
RKSYGKYQGR, KVGKSPPVR, KTFGKMKPR, RIKWSRVSK, KRTMRPTRR,
KVGKSPPVR, or HREARSGKYK (SEQ ID Nos. 29-44 respectively). Within certain
embodiments the peptides provided herein can be less than 250, 200, 150, 100,
75, 50,
or, 25 amino acids in length.
In a related aspect, the invention provides methods of identifying a
peptide or polypeptide composition for treating a tissue disorder associated
with a
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
8
response-to-injury process or proliferating cells in a mammal comprising the
steps of:
(a) forming an expression library comprised of cloned sequences expressed by a
cell
during a transition stage response; (b) screening the expression library for
sequences
encoding a peptide or polypeptide that binds a hyaladherin that is stimulated
in cells
during the transition stage; (c) preparing a peptide or polypeptide encoded by
the
hyaladherin binding sequences; and (d) testing the peptide or polypeptide for
the ability
to affect at least one activity associated with transition stage cells,
wherein the activity
is selected from the group consisting of: increased erk kinase signal
activation,
podosome formation, metalloproteinase expression, flux of intracellular or
extracellular
hyaluronans or hyaladherins, expression of a hyaladherin, and inability to
form focal
adhesions. In one embodiment of this method, the expression vector is a two
hybrid
phage display system, the hyaladherin is RHAMM and the testing is for the
ability to
inhibit podosome formation and inhibition of e~°k kinase signaling
activation.
In a further aspect of the present invention polypeptides are provided
which comprises or consist only of any one of SEQ ID Nos, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54,
55, 56, 57, 58, 59,.
60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, or, 72. When the polypeptide
comprises
additional sequences, the polypeptide may, in certain embodiments, be less
than 50kd or
20kd of molecular weight. In other embodiments, the polypeptide may be less
than 60,
50, 40, 30, or, 20 amino acids in length. Also provided are such polypeptides
that are
purified (e.g., free of cells), sterilized, and/or suitable for
pharmaceuticaluse.
In yet other aspects of the present invention antibodies are provided which
bind to any one of the peptides provided herein, including for example, a
polypeptide
comprising or consisting only of any one of SEQ ID Nos, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54,
55, 56, 57, 58, 59,
60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, or, 72. Also provided are
antibodies such as
noted above, which are purified (e.g., free of cells), sterilized, and/or
suitable for
pharmaceuticaluse (e.g., for use in one of the methods describedherein).
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
9
In still another aspect, the invention provides methods for detecting
hyalauronic acid in a sample comprising the steps of: (a) incubating the
sample with a
hyalauronic acid binding peptide comprising a sequence selected from the group
consisting of SEQ. ID NO: 1-10; and (b) detecting an amount of a complex
formed
between hyalauronic acid and the hyalauronic acid binding peptide. In one
embodiment, the detecting employs an antibody that specifically binds to the
hyalauronic acid binding peptide.
In a related aspect, the invention provides methods of detecting a
molecule that binds to a RHAMM polypeptide in a sample comprising the steps of
(a)
incubating the sample with the RHAMM polypeptide and with a RHAMM-binding
polypeptide comprised of SEQ. ID NO: 21; and (b) detecting an amount of a
complex
formed between the sample and the RHAMM polypeptide by scoring for reduced
binding between the RHAMM polypeptide and the RHAMM-binding polypeptide. In
one embodiment, detecting employs an antibody that specifically binds to the
RHAMM-binding polypeptide.
Yet another embodiment of the invention includes compositions for
treating a tissue disorder associated with a response-to-injuryprocess or
proliferating cells
in a mammal, wherein the composition is comprised of a hyaluronan that binds
to a
hyaladherin that regulates a transition stage cell associated with the
response to injury or
proliferating cell disorder. In a related aspect, the invention provides a
method for treating
a tissue disorder associated with a response-to-injury process or
proliferating cells in a
mammal comprising, administeringto the mammal the aforementionedhyaluronan.
Within further aspects of the present invention, vaccinating agents are
provided for preventing a tissue disorder associated with a response-to-injury
process or
proliferating cells in a mammal wherein the vaccinating agent includes an
antigen
comprised of one of the aforementioned polypeptides, peptides or hyaluronans.
Generally, within this aspect, the vaccinating agent is administered in a
dosage and/or
therapeutic regimen sufficient to generate an immune response against the
selected
antigen, and to prevent the tissue disorder or proliferation of cells.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
These and other aspects of the present invention will become evident
upon reference to the following detailed description and attached drawings. In
addition, various references are set forth herein which describe in more
detail certain
procedures or compositions (e.g., plasmids, etc.) and are therefore
incorporated by
5 reference in their entirety.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1 is a schematic illustration of the impact of the AP-1 pathway on
disease.
Figure 2 is a schematic illustration of cell activation in response to a
10 variety of factors, and this impact on disease pathways.
Figure 3 depicts cells transition from a normal cell to a diseased cell.
Figure 4 shows immunofluorescence micrographs and phosphoprotein
assays which indicate a requirement for cell adhesion for activation of the
ey°kl kinase
signaling.
Figure 5 schematically illustrates the involvement of RHAMM as a
transitional molecule.
Figures 6A and 6B are two blots which show erk activity.
Figures 7A and 7B show the increased expression levels of e-fos, c jun,
junB genes by overexpression of RHAMM.
Figure 8 shows the increased expression levels of c-fos and c-jun gene in
cells overexpressing RHAMM regardless whether grown on PL or FN.
Figure 9A and 9B are northern blots probed with gelatinase B,
stromelysin, timp-l, and GAPDH CDNAS.
Figure 10 is a blot which shows that LR21 cells overexpressing
RHAMMv4 are restricted in the extent to which proinflammatory cytokines can
activate
erk kinase.
Figures 11A and 11B are northern analysis of IL-1 and TNF-alpha
induction of c fos.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
11
Figures 12A and 12B are photographs of LR21 cells, showing, in 12A,
the formation of discrete focal adhesions.
Figures 13 A and 13B are photos, and graphs (respectively), which show
that overexpression of RHAMM results in podosome formation.
Figures 14A, 14B, and 14C are graphs which illustrate the relationship of
RHAM, e~k activity and podosome formation. Figures 14D, 14E, and 14F are
photos
which supplement this data.
Figure 15 A is a bar graph which shows a comparison of RHAMM
expression at a cell surface. Figure 15B provides the sequence of various
RHAMM
peptides (SEQ ID Nos. 14-20).
Figures 16A and 16B are photographs showing cells treated with
peptides. Figures 16C and 16D are bar graphs quantifying the effect.
Figures 17A and 17B are photographs showing podosome formation
under various conditions.
Figure 18 is two photographs showing MDA-231 cells trated with
hyaluronan together with anti-RHAMM antibody.
Figure 19 is a chart that compares rate of locomotion for various cell
lines and various substrates.
Figures 20A and 20B show that anti-RHAMM antibodies inhibit the
ability of MDA231 cells to invade in vitro.
Figures 21A, 21 B and 21 C show RHAMM binding to fibronectin being
blocked by selected antibodies.
Figure 22 is a bar graph which shows that MDA231 cells expressing
RHAMM have a high degree of motility.
Figure 23A and 23B are micrographs which show podosome formation
in various cells.
Figure 24A and 24B are a graph, and a blot, respectively, which show
the effects of exon 4 antibody and LZP on the formation of podosomes.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
12
Figures 25A-D show that both v4 and v5 forms of RHAMM associated
with erkl in vivo and in vitro, but that only the short form stongly activates
the erk
kinase cascade.
Figures 26A, 26B and 26C illustrate that HA binding peptides (SEQ ID
Nos. 28,56-58) including artificial mimics are able to block cell motility.
Figure 27 is a bar graph which shows that treatment of injured cells with
P-peptide (CSTMMSRSHI~TRSHHV- Seq. ID. No. 26) inhibits migration of HFF
cells.
Figure 28 is a bar graph which shows velocity of cells after addition of
peptide aa423-432 (Sequence ID No. 24).
Figure 29 is two bar graphs which, show MMP release from knockout
fibroblasts compared to normal ones (on fibronectin vs. cell culture plastic).
Figure 30 is a bar graph and blots which show that erk phosphorylation
is influenced by RHAMM expression.
Figure 31 is a bar graph which shows knockout fibroblasts have
decreased motility compared to wild-type fibroblasts.
Figures 32a, 32b, 32c and 32d are photographs of bleomycin-induced
lung fibrosis.
Figure 33 is a bar graph which illustrates that a significant increase in
motility of macrophages from both bleomycin and saline-treated animals.
Figure 34 is a bar graph which illustrates the motility of BAL cells four
days after injury in response to administration of RHAMM peptides.
Figure 35 is a bargraph, and a blot which shows the ability of HA
binding peptides to inhibit firbosis.
Figures 36a-f are a series of photographs from a histological analysis of
lung tissue.
Figure 37 is a table which shows. the percentage of cells with various
isoforms of RHAMM, from a variety of RA patients.
Figures 38A-F are a series of photographs of stained RA tissue.
Figure 39 is a graph which shows attenuation of clinical signs of MS
after treatment.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
13
Figures 40A and 40B are bar graphs which show that collagen
production following treatment with P-peptide.
Figure 41 shows that P-peptide reduces infiltration of macrophages into
the site of a wound.
Figure 42 is a bar graph which compares glucosamine activity after
treament of various peptides.
Figure 43A and 43B illustrate expression of RHAMM isoforms from
various cell lines.
Figures 44A (bar graphs) and 44B (a blot) illustrate random cell motility
and matrigel cell invasion utilizing various peptides.
Figure 45 is a graph which shows weight change in transgenic mice.
Figure 46A and 46 B show that RHAMM is most highly expressed in the
most invasive lung cancer cell lines.
Figure 47A and 47B show that RHAMM is most highly expressed in
high grade or invasive astnocytomas .
Figure 48A is a schemata showing domains of various RHAMM
polypeptides required for podosome formation and activation of erk kinase
signaling and
Figure 48B is a protein gel showing that intracellularRHAMMv4 binds to ERK
kinase.
Figure 49 shows (A) a partial amino acid (SEQ ID N0:46) and nucleotide
sequence (SEQ ID N0:45) of a RHAMM binding protein (RABP) isolated using a
phage
two hybrid system; (B) a Northern blot of R.ABP expression in transitional
cells; (C) a
Western blot of transitional cell lysate indicating that RA.BP is a 60 kDa
protein; and (D) a
FACE analysis illustratingthat RABP is present on the cell surface.
Figure 50 depicts the human and murine sequence of RHAMM (SEQ ID
Nos. 47 and 48 respectively).
Figure 51 is a bar graph that depicts the incidence of abnormal blood
glucose levels in NOD mice.
Figure 52 is a bar graph that depicts the incidence of abnormal urine
glucose level in NOD mice.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
14
Figure 53 is a graph that indicates the effect of P-16 peptide on water
consumption in NOD mice.
Figure 54 is a graph that indicates the effect of P-16 peptide on kidney
weight in NOD mice.
DETAILED DESCRIPTION OF THE INVENTION
Definitions
Prior to setting forth the invention, it may be helpful to an understanding
thereof to first set forth definitions of certain terms that will be used
hereinafter.
"Expression vector" and "expression cassette" refers to an assembly
which is capable of directing the expression of a sequence or gene of
interest. The
nucleic acid expression vector must include a promoter which, when
transcribed, is
operably linked to the sequences) or genes) of interest, as well as a
polyadenylation
sequence. Within certain embodiments of the invention, the expression vectors
described herein may be contained within a plasmid construct. In addition to
the
components of the nucleic acid expression vector, the plasmid construct may
also
include a bacterial origin of replication, one or more selectable markers, a
signal which
allows the plasmid construct to exist as single-stranded DNA (e.g., a M13
origin of
replication), a multiple cloning site, and a "manunalian" origin of
replication (e.g., an
SV40 or adenovirus origin of replication).
As used herein, "nucleic acid" or "nucleic acid molecule" refers to any of
deoxyribonucleic acid (DNA), ribonucleic acid (RNA), oligonucleotides,
fragments
generated by the polymerase chain reaction (PCR), and fragments generated by
any of
ligation, scission, endonuclease action, and exonuclease action. Nucleic acids
can be
composed of monomers that are naturally-occurring nucleotides (such as
deoxyribonucleotides and ribonucleotides), or analogs of naturally-occurring
nucleotides (e.g., a-enantiomeric forms of naturally-occurring nucleotides),
or a
combination of both. Modified nucleotides can have modifications in sugar
moieties
and/or in pyrimidine or purine base moieties. Sugar modifications include, for
example,
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
replacement of one or more hydroxyl groups with halogens, alkyl groups,
amines, and
azido groups, or sugars can be functionalized as ethers or esters. Moreover,
the entire
sugar moiety can be replaced with sterically and electronically similar
structures, such
as aza-sugars and carbocyclic sugar analogs. Examples of modifications in a
base
5 moiety include alkylated purines and pyrimidines, acylated purines or
pyrimidines, or
other well-known heterocyclic substitutes. Nucleic acid monomers can be linked
by
phosphodiester bonds or analogs of such linkages. Analogs of phosphodiester
linkages
include phosphorothioate, phosphorodithioate, phosphoroselenoate,
phosphorodiselenoate, phosphoroanilothioate, phosphoranilidate,
phosphoramidate, and-
10 the like. The term "nucleic acid" also includes so-called "peptide nucleic
acids," which
comprise naturally-occurring or modified nucleic acid bases attached to a
polyamide
backbone. Nucleic acids can be either single stranded or double stranded.
An "isolated nucleic acid molecule" is a nucleic acid molecule that is not
integrated in the genomic DNA of an organism. For example, a DNA molecule that
15 encodes a RHAMM binding protein that has been separated from the genomic
DNA of
a eukaryotic cell is an isolated DNA molecule. Another example of an isolated
nucleic
acid molecule is a chemically-synthesized nucleic acid molecule that is not
integrated in
the genome of an organism.
An "isolated polypeptide" is a polypeptide that is essentially free from
contaminating cellular components, such as carbohydrate, lipid, or other
proteinaceous
impurities associated with the polypeptide in nature. That a particular
protein
preparation contains an isolated polypeptide can be shown by the appearance of
a single
band following sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis
of the
protein preparation and Coomassie Brilliant Blue staining of the gel.
"Gene delivery vehicle" refers to a construct which is capable of
delivering, and, within preferred embodiments expressing, one or more genes)
or
sequences) of interest in a host cell. Representative examples of such
vehicles include
viral vectors, nucleic acid expression vectors, naked DNA, and certain
eukaryotic cells
(e.g., producer cells).
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
16
A "riboz~me" is a nucleic acid molecule that contains a catalytic center.
The term includes RNA enzymes, self splicing RNAs, self cleaving RNAs, and
nucleic
acid molecules that perform these catalytic functions. A nucleic acid molecule
that
encodes a ribozyme is termed a "ribozyme gene."
"Treatment" refers to both therapeutic treatment and prophylactic or
preventative measures. Those in need of treatment include those already with
the
disorder as well as those prone to have the disorder or those in which the
disorder is to
be prevented.
"Mammal" for purposes of treatment refers to any animal classified as a
mammal, including humans, domestic and farm animals, and zoo, sports, or pet
animals, such as dogs, horses, cats, cows, etc. Preferably, the mammal herein
is human.
"Antibodies" (Abs) and "immuno~lobulins" (Igs) are glycoproteins
having the same structural characteristics. While antibodies exhibit binding
specificity
to a specific antigen, immunoglobulins include both antibodies and other
antibody-like
molecules for .which antigen specificity has not been defined. Polypeptides of
the latter
kind are, for example, produced at low levels by the lymph system and at
increased
levels by myelomas.
"Native antibodies and immuno~lobulins" are usually heterotetrameric
glycoproteins of about 150,000 daltons, composed of two identical light (L)
chains and
two identical heavy (H) chains. Each light chain is linked to a heavy chain by
one
covalent disulfide bond, while the number of disulfide linkages varies between
the
heavy chains of different immunoglobulin isotypes. Each heavy and light chain
also has
regularly spaced intrachain disulfide bridges, Each heavy chain has at one end
a
variable domain (V<sub>H</sub>) followed by a number of constant domains. Each light
chain
has a variable domain at one end (V<sub>L</sub>) and a constant domain at its other
end; the
constant domain of the light chain is aligned with the first constant domain
of the heavy
chain, and the light chain variable domain is aligned with the variable domain
of the
heavy chain. Particular amino acid residues are believed to form an interface
between
the light- and heavy-chain variable domains (Clothia et al., J. Mol. Biol.
186:651
(195); Novotny and Haber, Pf~oc. Natl. Acad. Sci. U.S.A. 82:4592 (1955)).
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
17
The term "variable" refers to the fact that certain portions of the variable
domains differ extensively in sequence among antibodies and are used in the
binding
and specificity of each particular antibody for its particular antigen.
However, the
variability is not evenly distributed throughout the variable domains of
antibodies. It is
concentrated in three segments called complementarity-determining regions
(CDRs) or
hypervariable regions both in thelight-chain and the heavy-chain variable
domains. The
more highly conserved portions of variable domains are called the framework
(FR). The
variable domains of native heavy and light chains each comprise four FR
regions,
largely adopting a .beta.-sheet configuration, comzected by three CDRs, which
form
loops connecting, and in some cases forming part of, the .beta.-sheet
structure. The
CDRs in each chain are held together in close proximity by the FR regions and,
with the
CDRs from the other chain, contribute to the formation of the antigen-binding
site of
antibodies (see Kabat et al., Sequences of Proteins of Immunological Interest,
Fifth
Edition, National Institute of Health, Bethesda, Md. (1991)). The constant
domains are
not involved directly in binding an antibody to an antigen, but exhibit
various effector
functions, such as participation of the antibody in antibody-dependent
cellular toxicity:
Papain digestion of antibodies produces two identical antigen-binding
fragments, called "Fab" fragments, each with a single antigen-binding site,
and a
residual "Fc" fragment, whose name reflects its ability to crystallize
readily. Pepsin
treatment yields an F(ab')2 fragment that has two antigen-combining sites and
is still
capable of cross-linking antigen.
"Fv" is the minimum antibody fragment which contains a complete antigen-
recognition and -binding site. This region consists of a dimer of one heavy-
and one light-
chain variable domain in tight, non-covalent association. It is in this
configuration that the
three CDRs of each variable domain interact to define an antigen-binding site
on the surface
of the V<sub>H</sub> -V<sub>L</sub> dimer. Collectively, the six CDRs confer antigen-
binding
specificity to the antibody. However, even a single variable domain (or half
of an Fv
comprising only three CDRs specific for an antigen) has the ability to
recognize and bind
antigen, although at a lower affinity than the entire binding site.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
l~
The Fab fragment also contains the constant domain of the light chain
and the first constant domain (CH 1) of the heavy chain. Fab' fragments differ
from Fab
fragments by the addition of a few residues at the carboxy terminus of the
heavy chain
CH 1 domain including one or more cysteines from the antibody hinge region.
Fab'-SH
is the designation herein for Fab' in which the cysteine residues) of the
constant
domains bear a free thiol group. F(ab')2 antibody fragments originally were
produced as
pairs of Fab' fragments which have hinge cysteines between them. Other
chemical
couplings of antibody fragments are also known.
The "light chains" of antibodies (immunoglobulins) from any vertebrate
species can be assigned to one of two clearly distinct types, called kappa
(.kappa.) and
lambda (.lambda.), based on the amino acid sequences of their constant
domains.
Depending on the amino acid sequence of the constant domain of their
heavy chains, immunoglobulins can be assigned to different classes. There are
five
major classes of immunoglobulins: IgA, IgD, IgE, IgG, and IgM, and several of
these
can be further divided into subclasses (isotypes), e.g., IgG<sub>l</sub>, IgG<sub>2</sub>,
IgG<sub>3</sub>,
IgG<sub>4</sub>, IgA<sub>l</sub>, and IgA<sub>2</sub>. The heavy-chain constant domains that
correspond
to the different classes of immunoglobulins are called .alpha., .delta.,
.epsilon.,
.gamma., and µ, respectively. The subunit structures and three-dimensional
configurations of different classes of immunoglobulins are well known.
The term "monoclonal antibody" (mAb) as used herein refers to an
antibody obtained from a population of substantially homogeneous antibodies,
a. e., the
individual antibodies comprising the population axe identical except for
possible
naturally occurring mutations that may be present in minor amounts. Monoclonal
antibodies are highly specific, being directed against a single antigenic
site.
Furthermore, in contrast to conventional (polyclonal) antibody preparations
which
typically include different antibodies directed against different determinants
(epitopes),
each mAb is directed against a single determinant on the antigen. In addition
to their
specificity, the monoclonal antibodies are advantageous in that they can be
synthesized
by hybridoma culture, uncontaminated by other immunoglobulins. Thus, the
modifier
"monoclonal" indicates the character of the antibody as being obtained from a
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
19
substantially homogeneous population of antibodies, and is not to be construed
as
requiring production of the antibody by any particular method. For example,
the
monoclonal antibodies to be used in accordance with the present invention can
be made
by the hybridoma method first described by Kohler and Milstein, Nature 256:495
(1975), or can be made by recombinant DNA methods (Cabilly et al., supra).
The monoclonal antibodies herein specifically include "chimeric"
antibodies (immunoglobulins) in which a portion of the heavy and/or light
chain is
identical with or homologous to corresponding sequences in antibodies derived
from a
particular species or belonging to a particular antibody class or subclass,
while the
remainder of the chains) is identical with or homologous to corresponding
sequences in
antibodies derived from another species or belonging to another antibody class
or
subclass, as well as fragments of such antibodies, so long as they exhibit the
desired
biological activity (Cabilly et al., sups°a; Morrison et al., Proc.
Natl. Acad. Sci. U.S.A.
81:6851 (1984)).
"Humanized" forms of non-human (e.g., murine) antibodies are specific
chimeric immunoglobulins, immunoglobulin chains or fragments thereof (such as
Fv,
Fab, Fab', F(ab')z, or other antigen-binding subsequences of antibodies) which
contain
minimal sequence derived from non-human immunoglobulin. For the most part,
humanized antibodies are human immunoglobulins (recipient antibody) in which
residues from a complementarity-determining region (CDR) of the recipient are
replaced by residues from a CDR of a non-human species (donor antibody) such
as
mouse, rat, or rabbit having the desired specificity, aff pity, and capacity.
In some
instances, Fv framework residues of the human immunoglobulin are replaced by
corresponding non-human residues. Furthermore, humanized antibodies can
comprise
residues which are found neither in the recipient antibody nor in the imported
CDR or
framework sequences. These modifications are made to further refine and
optimize
antibody performance. In general, the humanized antibody will comprise
substantially
all of at least one, and typically two, variable domains, in which all or
substantially all
of the CDR regions correspond to those of a non-human immunoglobulin and all
or
substantially all of the FR regions are those of a human immunoglobulin
consensus
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
sequence. The humanized antibody optimally also will comprise at least a
portion of an
immunoglobulin constant region (Fc), typically that of a human immunoglobulin.
For
further details see Jones et al., Nature 321:522 (1986); Reichmann et al.,
Nature
332:323 (1988); and Presta, Cury~. Op. St~uct. Biol. 2:593 (1992).
5 As noted above, the present invention provides compositions and
methods for treating a tissue disorder associated with a response-to-injury
process or
proliferating cells in a mammal. More specifically, based upon the pathways
and
progression of disease described herein, it is now understood that many
diseases are
related in the sense that transition molecules are involved in the initiation
and
1, 0 progression of disease. Provided in more detail below are: (A) assays for
detecting
molecules suitable for use within the present invention; (B) suitable
candidate
molecules for use within the present invention (whether for assaying, or for
therapeutic
purpose); (C) antibodies (for either assaying, or for therapeutic purpose);
(D) expression
systems for producing and or delivering therapeutic quantities of a desired
polypeptide;
15 (E) methods of treating a wide variety of diseases; and (F) the preparation
of
pharmaceutical compositions, including vaccines.
A. Assays for Detecting Therapeutic Molecules
The present invention provides a number of assays suitable for detecting
therapeutic molecules, which are briefly described herein, as well as in more
detail
20 below in the examples.
For example, within one aspect of the invention methods of identifying a
peptide or polypeptide composition for treating a tissue disorder associated
with a
response-to-injury process, or, the proliferation of cells in a mammal is
provided,
comprising the general steps of: (a) selecting a sequence from a database
screened for
sequences comprising a peptide of the sequence BX7B (SEQ ID N0:28) wherein B
is a
basic amino acid, and X7 is a sequence of about seven residues is selected
from any
amino acid other than an acidic amino acid, wherein the peptide forms an alpha
helix
and each occurrence of B is oriented on the same side of the alpha helix, (b)
preparing a
composition comprised of the selected sequence; and (c) testing the prepared
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
21
composition for the ability to inhibit podosome formation. Within certain
embodiments, the peptide in (a) does not consist of the sequences BBXXBBBXXBB,
KQKIKHVVKLK, KLKSQLVKRK, RYPISRPRKR, KNGRYSISR;
RDGTRYVQKGEYR, RRRCGQKKK, RGTRSGSTR, RRRKKIQGRSKR,
RKSYGKYQGR, KVGKSPPVR, KTFGKMKPR, RIKWSRVSK, KRTMRPTRR,
KVGKSPPVR, or HREARSGKYK (SEQ ID Nos. 29-44 respectively).
In a related aspect, the invention provides methods of identifying a
peptide or polypeptide composition for treating a tissue disorder associated
with a
response-to-injury process or proliferating cells in a mammal comprising the
steps of:
(a) forming an expression library comprised of cloned sequences expressed by a
cell
during a transition stage response; (b) screening the expression library for
sequences
encoding a peptide or polypeptide that binds a hyaladherin that is stimulated
in cells
during the transition stage; (c) preparing a peptide or polypeptide encoded by
the
hyaladherin binding sequences; and (d) testing the peptide or polypeptide for
the ability
to affect at least one activity associated with transition stage cells wherein
the activity is
selected from the group consisting of: increased erk kinase signal activation,
podosome
formation, metalloproteinase expression, flux of intracellular or
extracellular HA or
hyaladherins, expression of a hyaladherin, and inability to form focal
adhesions. In one
embodiment of this method, the expression vector is a two hybrid phage display
system,
the hyaladherin is RHAMM and the testing is for the ability to inhibit
podosome
formation and inhibition of e~k kinase signaling activation. In a related
embodiment of
these methods, the library is a library of nucleic acid molecules, or organic
molecules,
and the library is tested in order to determine its ability to affect one of
the activities set
forth in (d) above. If the test is positive, the library may be deconvoluted,
and screened
until a single molecule is identified.
In still another aspect, the invention provides methods for detecting
hyalauronic acid in a sample comprising the steps of: (a) incubating the
sample with a
hyalauronic acid binding peptide comprising a sequence selected from the group
consisting of SEQ. ID NO: 1-10; and (b) detecting an amount of a complex
formed
between hyalauronic acid and the hyalauronic acid binding peptide. In one
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
22
embodiment, the detecting employs an antibody that specifically binds to the
hyalauronic acid binding peptide.
In a related aspect, the invention provides methods of detecting a
molecule that binds to a RHAMM polypeptide in a sample comprising the steps of
(a)
incubating the sample with the RHAMM polypeptide and with a RHAMM-binding
polypeptide comprised of SE(~. ID NO: 21; and (b) detecting an amount of a
complex
formed between the sample and the RHAMM polypeptide by scoring for reduced
binding between the RHAMM polypeptide and the RHAMM-binding polypeptide. In
one embodiment, this method includes detecting which employs an antibody that
I O specifically binds to the RHAMM-binding polypeptide.
These as well as other methods are described in more detail below in the
examples.
B. Candidate Molecules
Utilizing the assays provided herein, a wide variety of molecules may
be assayed for their ability to treat or prevent a tissue disorder associated
with a
response-to-injury process or proliferating cells. Representative examples
which are
discussed in more detail below include organic molecules, proteins or
peptides, and
nucleic acid molecules.
1. Organic Molecules
Numerous organic molecules may be assayed for their ability to treat or
prevent a tissue disorder associated with a response-to-injuryprocess or
proliferating cells.
For example, within one embodiment of the invention suitable organic
molecules may be selected from either from a chemical library, wherein
chemicals are
assayed individually, or from combinatorial chemical libraries where multiple
compounds are assayed at once, then deconvoluted to determine and isolate the
most
active compounds.
Representative examples of such combinatorial chemical libraries include
those described by Agrafiotis et al., "System and method of automatically
generating
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
23
chemical compounds with desired properties," U.S. Patent No. 5,463,564;
Armstrong,
R.W., "Synthesis of combinatorial arrays of organic compounds through the use
of
multiple component combinatorial array syntheses," WO 95/02566; Baldwin, J.J.
et al.,
"Sulfonamide derivatives and their use," WO 95/24186; Baldwin, J.J. et al.,
"Combinatorial dihydrobenzopyran library," WO 95/30642; Brenner, S., "New kit
for
preparing combinatorial libraries," WO 95/16918; Chenera, B. et al.,
"Preparation of
library of resin-bound aromatic carbocyclic compounds," WO 95/16712; Ellman,
J.A.,
"Solid phase and combinatorial synthesis of benzodiazepine compounds on a
solid
support," U.S. Patent No. 5,288,514; Felder, E. et al., "Novel combinatorial
compound
libraries," WO 95/16209; Lerner, R. et al., "Encoded combinatorial chemical
libraries,"
WO 93/20242; Pavia, M.R. et al., "A method for preparing and selecting
pharmaceutically
useful non-peptide compounds from a structurally diverse universal library,"
WO 95/04277; Summerton, J.E. and D.D. Weller, "Morpholino-subunit
combinatorial
library and method," U.S. Patent No. 5,506,337; Holmes, C., "Methods for the
Solid
Phase Synthesis of Thiazolidinones, Metathiazanones, and Derivatives thereof,"
WO 96/00148; Phillips, G.B. and G.P. Wei, "Solid-phase Synthesis of
Benzimidazoles,"
Tet. Letters 37:4887-90, 1996; Ruhland, B. et al., "Solid-supported
Combinatorial Synthesis
of Structurally Diverse (3-Lactams," .l. Amer. Chem. Soc. 111:253-4, 1996;
Look, G.C. et al.,
"The Identification of Cyclooxygenase-1 Inhibitors from 4-Thiazolidinone
Combinatorial
Libraries," Bioorg and Med. Chem. Letters 6:707-12,1996.
2. Proteins and Peptides
A wide range of proteins and peptides may likewise be assayed for their
ability to treat or prevent a tissue disorder associated with a response-to-
injury process
or proliferating cells.
a. Combinatorial Peptide Libraries
Suitable peptide molecules may be obtained through the screening of
combinatorial peptide libraries. Such libraries may either be prepared by one
of skill in
the art (see e.g., U.S. Patent Nos. 4,528,266 and 4,359,535, and Patent
Cooperation
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
24
Treaty Publication Nos. WO 92/I5679, WO 92/15677, WO 90/07862, WO 90/02809,
or purchased from commercially available sources (e.g., New England Biolabs
Ph.D.TM
Phage Display Peptide Library Kit).
b. Peptide mimetics
Numerous peptide mimetics may also be utilized within the present
invention, including for example peptides such as:
SEQ. ID. NO: 1 (H4-5)B3;
SEQ. ID. NO: 2 (H4-5)BXBBXB;
SEQ. ID. NO: 3 (H4-5)BXBXBBB;
~10 SEQ. ID. NO: 4 (H4-5)BXBBB; and
SEQ. ID. NO: 5 (H4-5)BXBB
where B is either lysine (K) or arginine (R and X is a hydrophobic or neutral
amino
acid (i.e., L,V,Q, S) and H represents a series of amino acids such that an
alpha helix is
formed, as determined by NN-predict EMBL protein analysis. . This need not be
an
amphipathic or coiled coil helix but such would also be suitable. Specific
examples of
sequences fitting these motifs that have been analyzed for effectiveness on
podosome
include the following:
SEQ. ID. NO: 6 MMTVLKR;
SEQ. ID. NO: 7 MMTVLKVKRLR;
SEQ. ID. NO: 8 MMTVLKVKVKRK;
SEQ. ID. NO: 9 MMTVLKVRKR; and
SEQ. ID. NO: 10 MMTVLKVRK.
In addition, the following RHAMM sequences are more highly exposed
on cell surfaces and more effective at blocking podosomes, cell motility and
cell
invasion. These are:
SEQ ID NO: 11 KLQATQKPLTESK, and
SEQ ID NO: 12 VSIEKEKIDEKS.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
Other peptides may likewise be developed based upon the TAM domain
("Transient Activator of Map kinases"). This sequence is
SEQ ID NO: 13 VS(I/L)EKE.
Since this sequence is included in those used to prepare polyclonal
5 antibodies against RHAMM, and because such antibodies blocks cell motility
and
activation of erk by growth factors, TAM domains have been identified as key
sites of
protein-protein interaction that are required for controlling map kinase
pathways. This
in turn regulates the activation of the cell to migrate, proliferate and
remodel
extracellular matrix. Reagents to this sequence will be useful in therapeutic
treatment
10 of the diseases described above.
Antibodies, peptide mimics or antisense technology of these motifs can
be used to disrupt the transient phenotype and for treatment of disease as
described in
more detail below.
c. Antibodies
15 Antibodies, as described in more detail below, may likewise be assayed
for their ability to treat or prevent a tissue disorder associated with a
response-to-injury
process or proliferating cells.
d. Production of proteins
Although various genes (or portions thereof) have been provided herein,
20 it should be understood that within the context of the present invention,
reference to one
or more of these genes includes derivatives of the genes that are
substantially similar to
the genes (and, where appropriate, the proteins (including peptides and
polypeptides)
that are encoded by the genes and their derivatives). As used herein, a
nucleotide
sequence is deemed to be "substantially similar" if: (a) the nucleotide
sequence is
25 derived from the coding region of the above-described genes and includes,
for example,
portions of the sequence or allelic variations of the sequences discussed
above, (b) the
nucleotide sequence is capable of hybridization to nucleotide sequences of the
present
invention under moderate, high or very high stringency (see Sambrook et al.,
Molecular
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
26
Cloning: A Laborato~ y Manual, 2nd ed., Cold Spring Harbor Laboratory Press,
NY,
1989); or (c) the DNA sequences are degenerate as a result of the genetic code
to the
DNA sequences defined in (a) or (b). Further, the nucleic acid molecule
disclosed
herein includes both complementary and non-complementary sequences, provided
the
sequences otherwise meet the criteria set forth herein. Within the context of
the present
invention, high stringency means standard hybridization conditions (e.g.,
SXSSPE,
0.5% SDS at 65°C, or the equivalent). '
The structure of the proteins encoded by the nucleic acid molecules
described herein may be predicted from the primary translation products using
the
hydrophobicity plot function of, for example, P/C Gene or Intelligenetics
Suite
(Intelligenetics, Mountain View, California), or according to the methods
described by
Kyte and Doolittle (J. Mol. Biol. 157:105-132, 1982).
Proteins of the present invention may be prepared in the form of acidic or
basic salts, or in neutral form. In addition, individual amino acid residues
may be modified
by oxidation or reduction. Furthermore, various substitutions, deletions, or
additions may be
made to the amino acid or nucleic acid sequences, the net effect of which is
to retain or
further enhance or decrease the biological activity of the mutant or wild-type
protein.
Moreover, due to degeneracy in the genetic code, for example, there may be
considerable
variation in nucleotide sequences encoding the same amino acid sequence.
Other derivatives of the proteins disclosed herein include conjugates of
the proteins along with other proteins or polypeptides. This may be
accomplished, for
example, by the synthesis of N-terminal or C-terminal fusion proteins which
may be
added to facilitate purification or identification of proteins (see U.S.
Patent No.
4,851,341, see also, Hopp et al., BiolTechvcology 6:1204, 1988.)
Alternatively, fusion
proteins such as Flag/desired protein binding protein be constructed in order
to assist in
the identification, expression, and analysis of the protein.
Proteins of the present invention may be constructed using a wide variety
of techniques described herein. Further, mutations may be introduced at
particular loci
by synthesizing oligonucleotides containing a mutant sequence, flanked by
restriction
sites enabling ligation to fragments of the native sequence. Following
ligation, the
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
27
resulting reconstructed sequence encodes a derivative having the desired amino
acid
insertion, substitution, or deletion.
Alternatively, oligonucleotide-directed site-specific (or segment specific)
mutagenesis procedures may be employed to provide an altered gene having
particular
codons altered according to the substitution, deletion, or insertion required.
Exemplary
methods of making the alterations set forth above are disclosed by Walder et
al. (Gene
42:133, 1986); Bauer et al. (Gene 37:73, 1985); Craik (BioTechniques, January
1985,
12-19); Smith et al. (Genetic Engiueerifzg: Principles and Methods, Plenum
Press,
1981); and Sambrook et al. (supra). Deletion or truncation derivatives of
proteins (e.g.,
a soluble extracellular portion) may also be constructed by utilizing
convenient
restriction endonuclease sites adjacent to the desired deletion. Subsequent to
restriction,
overhangs may be filled in, and the DNA relegated. Exemplary methods of making
the
alterations set forth above are disclosed by Sambrook et al. (Molecular
Cloning: A
Laboratoi y Manual, 2d Ed., Cold Spring Harbor Laboratory Press, 1989).
Mutations which are made in the nucleic acid molecules of the present
invention preferably preserve the reading frame of the coding sequences.
Furthermore,
the mutations will preferably not create complementary regions that could
hybridize to
produce secondary mRNA structures, such as loops or hairpins, that would
adversely
_ affect translation of the mRNA. Although a mutation site may be
predetermined, it is
not necessary that the nature of the mutation per se be predetermined. For
example, in
order to select for optimmn characteristics of mutants at a given site, random
mutagenesis may be conducted at the target codon and the expressed mutants
screened
for indicative biological activity. Alternatively, mutations may be introduced
at
particular loci by synthesizing oligonucleotides containing a mutant sequence,
flanked
by restriction sites enabling legation to fragments of the native sequence.
Following
legation, the resulting reconstructed sequence encodes a derivative having the
desired
amino acid insertion, substitution, or deletion.
Nucleic acid molecules which encode proteins of the present invention
may also be constructed utilizing techniques of PCR mutagenesis, chemical
mutagenesis (Drinkwater and Klinedinst, PNAS 83:3402-3406, 1986), by forced
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
28
nucleotide misincorporation (e.g., Liao and Wise Gene 88:107-11 I, 1990), or
by use of
randomly mutagenized oligonucleotides (Horwitz et al., GenorrZe 3:112-117,
1989).
The present invention also provides for the manipulation and expression
of the above described genes by culturing host cells containing a vector
capable of
expressing the above-described genes. Such vectors or vector constructs
include either
synthetic or cDNA-derived nucleic acid molecules encoding the desired protein,
which
are operably linked to suitable transcriptional or translational regulatory
elements.
Suitable regulatory elements may be derived from a variety of sources,
including
bacterial, fungal, viral, mammalian, insect, or plant genes. Methods for
expressing
genes of interest are describe in more detail above.
Proteins can be isolated by, among other methods, culturing suitable host
and vector systems to produce the recombinant translation products of the
present
invention. Supernatants from such cell lines, or protein inclusions or whole
cells where
the protein is not excreted into the supernatant, can then be treated by a
variety bf
purification procedures in order to isolate the desired proteins. For example,
the
supernatant may be first concentrated using commercially available protein
concentration
filters, such as an Amicon or Millipore Pellicon ultrafiltration unit.
Following
concentration, the concentrate may be applied to a suitable purification
matrix such as, for
example, an anti-protein antibody bound to a suitable support. Alternatively,
anion or
canon exchange resins may be employed in order to purify the protein. As a
further
alternative, one or more reverse-phase high performance liquid chromatography
(RP-
HPLC) steps may be employed to further purify the protein. Other methods of
isolating
the proteins of the present invention are well known in the skill of the art.
A protein is deemed to be "isolated" within the context of the present
invention if no other (undesired) protein is detected pursuant to SDS-PAGE
analysis
followed by Coomassie blue staining. Within other embodiments, the desired
protein
can be isolated such that no other (undesired) protein is detected pursuant to
SDS-
PAGE analysis followed by silver staining.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
29
3. Nucleic Acid Molecules
Within other aspects of the invention, nucleic acid molecules can be
assayed for their ability to treat or prevent a tissue disorder associated
with a response-
to-injury process or proliferating cells. For example, within one embodiment
antisense
S oligonucleotide molecules are provided which specifically inhibit expression
of nucleic
acid sequences which are associated with a response-to injury process or the
proliferation of cells (see generally, Hirashima et al. in Molecular Biology
of RNA: New
Perspectives (M. Inouye and B. S. Dudock, eds., 1987 Academic Press, San
Diego, p.
401); Oligonucleotides: Antisense Inhibitors of Gene Expression (J.S. Cohen,
ed., 1989
MacMillan Press, London); Stein and Cheng, Science 261:1004-1012, 1993; WO
9S/10607; U.S. Patent No. S,3S9,OS1; WO 92/06693; and EP-A2-612844). Briefly,
such molecules are constructed such that they are complementary to, and able
to form
Watson-Crick base pairs with, a region of a transcribed mRNA sequence. The
resultant
double-stranded nucleic acid interferes with subsequent processing of the
mRNA,
1 S thereby preventing protein synthesis.
Within other aspects of the invention, ribozymes are provided which are
capable of inhibiting the expression of sequences which are associated with,
or which
encode proteins or polypeptides that are associated with the disorders
described herein.
As used herein, "ribozymes" are intended to include RNA molecules that contain
anti-
sense sequences for specific recognition, and an RNA-cleaving enzymatic
activity. The
catalytic strand cleaves a specific site in a target RNA at greater than
stoichiometric
concentration. A wide variety of ribozymes may be utilized within the context
of the
present invention, including for example, the hammerhead ribozyme (for
example, as
described by Forster and Symons, Cell 48:211-220, 1987; Haseloff and Gerlach,
Nature
2S 328:596-600, 1988; Walbot and Bruening, Nature 334:196, 1988; Haseloff and
Gerlach, Natur°e 334:S8S, 1988); the hairpin ribozyme (for example, as
described by
Haseloff et al., U.S. Patent No. S,2S4,678, issued October 19, 1993 and Hempel
et al.,
European Patent Publication No. 0 360 257, published March 26, 1990); and
Tetrahyrnena ribosomal RNA-based ribozymes (see Cech et al., U.S. Patent No.
4,987,071). Ribozymes of the present invention typically consist of RNA, but
may also
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
be composed of DNA, nucleic acid analogs (e.g., phosphorothioates), or
chimerics
thereof (e.g., DNA/RNA/RNA).
4. Labels
The gene product or any of the candidate molecules described above and
5 below, may be labeled with a variety of compounds, including for example,
fluorescent
molecules, toxins, and radionuclides. Representative examples of fluorescent
molecules
include fluorescein, Phycobili proteins, such as phycoerythrin, rhodamine,
Texas red and
luciferase. Representative examples of toxins include ricin, abrin diphtheria
toxin, cholera
toxin, gelonin, pokeweed antiviral protein, tritin, Shigella toxin, and
Pseudomonas
10 exotoxin A. Representative examples of radionuclides include Cu-64, Ga-67,
Ga-68, Zr-
89, Ru-97, Tc-99m, Rh-105, Pd-109, Tn-111, I-123, I-125, I-131, Re-186, Re-
188, Au-
198, Au-199, Pb-203, At-211, Pb-212 and Bi-212. In addition, the antibodies
described
herein may also be labeled or conjugated to one partner of a ligand binding
pair.
Representative examples include avidin-biotin, and riboflavin-
riboflavinbinding protein.
15 Methods for conjugating or labeling the molecules described herein with
the representative labels set forth above may be readily accomplished by one
of
ordinary skill in the art (see Trichothecene Antibody Conjugate, U.S. Patent
No.
4,744,981; Antibody Conjugate, U.S. Patent No. 5,106,951; Fluorogenic
Materials and
Labeling Techniques, U.S. Patent No. 4,018,884; Metal Radionuclide Labeled
Proteins
20 for Diagnosis and Therapy, U.S. Patent No. 4,897,255; and Metal
Radionuclide
Chelating Compounds for Improved Chelation Kinetics, U.S. Patent No.
4,988,496; see
also Inman, Methods In Enzyrnology, Vol. , 34, Affinity Techniques, Enzyme
Purification: Part B, Jakoby and Wilchek (eds.), Academic Press, New York, p.
30,
1974; see also Wilchek and Bayer, "The Avidin-Biotin Complex in Bioanalytical
25 Applications," Anal. Biochena. 171:1-32, 1988).
C. Antibodies
Antibodies to the polypeptides, fragments, or peptides described herein
may readily be prepared by one of skill in the art given the disclosure
provided herein.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
31
Within the context of the present invention, the term "antibody" should be
understood to
include monoclonal antibodies, polyclonal antibodies, anti-idiotypic
antibodies, antibody
fragments (e.g., Fab, and F(ab')2, Fv variable regions, or complementarity
determining
regions), whether obtained from animals or humans, generated utilizing
hybridoma
technology, or recombinantly produced. Antibodies are generally accepted as
specific
against an antigen if they bind with a I~d of at least 10-7 M (moles/liter),
and more
preferably, at least 10-8 M, 10-9 M, 10-10 M, 10'11 M, 10-12 M, 10-12 M, 10-13
M, or,
10-14 M. The affinity of a monoclonal antibody or binding partner can be
readily
determined by one of oxdinary skill in the art (see Scatchard, Ann. N. ~ Acad.
Sci. 51:660-
672, 1949). Antibodies of the present invention should also bind to the
desired domain or
peptide with the specificity noted above, and not against randomized peptides.
Briefly, a polyclonal antibody preparation may be readily generated in a
variety of warm-blooded animals such as rabbits, mice, or rats. Typically, an
animal is
immunized with a desired antigen or peptide thereof, which may be conjugated
to a
carrier protein, such as keyhole limpet hemocyanin. Routes of administration
include
intraperitoneal, intramuscular, intraocular, or subcutaneous injections,
usually in an
adjuvant (e.g., Freund's complete or incomplete adjuvant). Particularly
preferred
polyclonal antisera demonstrate binding in an assay that is at least three
times greater
than background.
Monoclonal antibodies may also be readily generated from hybridoma
cell lines using conventional techniques (see U.S. Patent Nos. RE 32,011,
4,902,614,
4,543,439, and 4,411,993; see also Antibodies: A Laboratory Manual, Harlow and
Lane
(eds.), Cold Spring Harbor Laboratory Press, 1988). Briefly, within one
embodiment, a
subject animal such as a rat or mouse is injected with an antigen of interest
or a portion
thereof. The protein may be administered as an emulsion in an adjuvant such as
Freund's complete or incomplete adjuvant in order to increase the immune
response.
Between one and three weeks after the initial immunization the animal is
generally
boosted and may tested for reactivity to the protein utilizing well-known
assays. The
spleen and/or lymph nodes are harvested and immortalized. Various
immortalization
techniques, such as mediated by Epstein-Barr virus or fusion to produce a
hybridoma,
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
32
may be used. In a preferred embodiment, immortalization occurs by fusion with
a
suitable myeloma cell line (e.g., NS-1 (ATCC No. TIB 18), and P3X63 - Ag 8.653
(ATCC No. CRL 1580) to create a hybridoma that secretes monoclonal antibody.
The
preferred fusion partners do not express endogenous antibody genes. Following
fusion,
the cells are cultured in medium containing a reagent that selectively allows
for the
growth of fused spleen and myeloma cells such as HAT (hypoxanthine,
aminopterin,
and thymidine) and are subsequently screened for the presence of antibodies
that are
reactive against the desired antigen of interest. A wide variety of assays may
be
utilized, including for example countercurrent immuno-electrophoresis,
radioimmunoassays, radioimmunoprecipitations, enzyme-linked immunosorbent
assays
(ELISA), dot blot assays, western blots, immunoprecipitation, inhibition or
competition
assays, and sandwich assays (see U.S. Patent Nos. 4,376,110 and 4,486,530; see
also
Antibodies: A Laboratory Manual, Harlow and Lane (eds.), Cold Spring Harbor
Laboratory Press, 1988).
Other techniques may also be utilized to construct monoclonal antibodies
(see Huse et al., Science 246:1275-1281, 1989; Sastry et al., Pf~oc. Natl.
Acad. Sci.
USA 86:5728-5732, 1989; Alting-Mees et al., Strategies in Molecular Biology
3:1-9,
1990; describing recombinant techniques). Briefly, RNA is isolated from a B
cell
population and utilized to create heavy and light chain immunoglobulin cDNA
expression libraries in suitable vectors, such as ~,ImmunoZap(H) and
~,ImmunoZap(L).
These vectors may be screened individually or co-expressed to form Fab
fragments or
antibodies (see Huse et al., supra; Sastry et al., supra). Positive plaques
may
subsequently be converted to a non-lytic plasmid that allows high level
expression of
monoclonal antibody fragments from E. eoli.
Similarly, portions or fragments, such as Fab and Fv fragments, of
antibodies may also be constructed utilizing conventional enzymatic digestion
or
recombinant DNA techniques to yield isolated variable regions of an antibody.
Within
one embodiment, the genes which encode the variable region from a hybridoma
producing a monoclonal antibody of interest are amplified using nucleotide
primers ~or
the variable region. These primers may be synthesized by one of ordinary skill
in the
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
33
art, or may be purchased from commercially available sources (e.g.,
Stratacyte, La Jolla,
CA) Amplification products are inserted into vectors such as ImmunoZAPTM H or
ImmunoZAPTM L (Stratacyte), which are then introduced into E. coli, yeast, or
mammalian-based systems for expression. Utilizing these techniques, large
amounts of
S a single-chain protein containing a fusion of the VH and VL domains may be
produced
(see Bird et al., Science 242:423-426, 1988). In addition, techniques may be
utilized to
change a "murine" antibody to a "human" antibody, without altering the binding
specificity of the antibody. Examples of humanized antibodies include chimeric
or
CDR-grafted antibodies (LT.S. Pat. Nos. 4,816,567 and 5,225,539), antibodies
produced
in genetically-altered mice (see PCT Application No. 93/12227).
One of ordinary skill in the art will appreciate that a variety of alternative
'
techniques for generating antibodies exist. In this regard, the following U.S.
patents
teach a variety of these methodologies and are thus incorporated herein by
reference:
U.S. Patent Nos. 5,840,479; 5,770,380; 5,204.,244; 5,482,856; 5,849,288;
5,780,225;
5,395,750; 5,225,539; 5,110,833; 5,693,762; 5,693,761; 5,693,762; 5,698,435;
and
5,328,834.
Once suitable antibodies have been obtained, they may be isolated or
purified by many techniques well known to those of ordinary skill in the art
(see
Antibodies: A Laboratory Manual, Harlow and Lane (eds.), Cold Spring Harbor
Laboratory Press, 1988). Suitable techniques include peptide or protein
affinity
columns, HPLC (e.g., reversed phase, size exclusion, ion-exchange),
purification on
protein A or protein G columns, or any combination of these techniques.
D. Expression Systems
1. Vectors, host cells and means of expressing and producing protein
Proteins or polypeptides of the present invention may be readily
expressed in a variety of host cells or organisms. For protein production and
purification, proteins are preferably secreted and produced in bacteria, such
as E. coli,
for which many expression vectors have been developed and are available. Other
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
34
suitable host organisms include other bacterial species (e.g., Bacillus, and
eukaryotes,
such as yeast (e.g., Saccharomyces cerevisiae), mammalian cells (e.g., CHO and
COS-
7), plant cells and insect cells (e.g., Sue). Vectors for these hosts are well
known.
Briefly, within one embodiment a DNA sequence encoding a desired
protein or polypeptide is introduced into an expression vector appropriate for
the host.
The sequence is derived from an existing clone or synthesized. A preferred
means of
synthesis is amplification of the gene from cDNA, genomic DNA, or a
recombinant
clone using a set of primers that flank the coding region or the desired
portion of the
protein. Restriction sites are typically incorporated into the primer
sequences and are
chosen with regard to the cloning site of the vector. If necessary,
translational initiation
and termination codons can be engineered into the primer sequences. The
desired
sequence can be codon-optimized for expression in a particular host. For
example, a
secreted form of a desired protein that is expressed in a fungal host, such as
yeast, can
be altered in nucleotide sequence to use codons preferred in yeast. Codon-
optimization
may be accomplished by methods such as splice overlap extension, site-directed
mutagenesis, automated synthesis, and the like.
At minimum, the vector must contain a promoter sequence. Other
regulatory sequences however may also be included. Such sequences include a
transcription termination signal sequence, secretion signal sequence, origin
of
replication, selectable marker, and the like. The regulatory sequences are
operationally
associated with one another to allow transcription or translation.
2. Expression in bacteria
The plasmids used herein for expression of a desired protein or
polypeptide include a promoter designed for expression of the proteins in a
bacterial host.
Suitable promoters are widely available and are well known in the art.
Inducible or
constitutive promoters are preferred. Such promoters for expression in
bacteria include
promoters from the T7 phage and other phages, such as T3, T5, and SP6, aiid
the trp, lpp,
and lac operons. Hybrid promoters (see, U.S. Patent No. 4,551,433), such as
tac and trc,
may also be used. Promoters for expression in eukaryotic cells include the P10
or
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
polyhedron gene promoter of baculovirus/insect cell expression systems (see,
e.g., U.S.
Patent Nos. 5,243,041, 5,242,687, 5,266,317, 4,745,051, and 5,169,784), MMTV
LTR,
RSV LTR, SV40, metallothionein promoter (see, e.g., U.S. Patent No. 4,870,009)
and
other inducible promoters. For expression of the proteins, a promoter is
inserted in
5 operative linkage with the coding region of the desired protein or
polypeptide.
The promoter controlling transcription of the desired protein may be
controlled by a repressor. In some systems, the promoter can be derepressed by
altering
the physiological conditions of the cell, for example, by the addition of a
molecule that
competitively binds the repressor, or by altering the temperature of the
growth media.
10 Preferred repressor proteins include, but are not limited to the E coli
lacI repressor
responsive to IPTG induction, the temperature sensitive ~,cI857 repressor, and
the like.
The E coli lacI repressor is preferred.
In other preferred embodiments, the vector also includes a transcription
terminator sequence. A "transcription terminator region" has either a sequence
that
15 provides a signal that terminates transcription by the polymerase that
recognizes the
selected promoter and/or a signal sequence for polyadenylation.
Preferably, the vector is capable of replication in bacterial cells. Thus,
the vector preferably contains a bacterial origin of replication. Preferred
bacterial
origins of replication include the fl-on and col E1 origins of replication,
especially the
20 on derived from pUC plasmids.
The plasmids also 'preferably include at least one selectable marker that
is functional in the host. A selectable marker gene includes any gene that
confers a
phenotype on the host that allows transformed cells to be identified and
selectively
grown. Suitable selectable marker genes for bacterial hosts include the
ampicillin
25 resistance gene (Ampr), tetracycline resistance gene (Tcr) and the
kanamycin resistance
gene (KanT). Suitable markers for eukaryotes usually require a complementary
deficiency in the host (e.g., thymidine kinase (tk) in tk- hosts). However,
drug markers
are also available (e.g., G418 resistance and hygromycin resistance).
The sequence of nucleotides encoding the desired protein or polypeptide
30 may also include a classical secretion signal, whereby the resulting
peptide is a
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
36
precursor protein processed and secreted. The resulting processed protein may
be
recovered from the periplasmic space or the fermentation medium. Secretion
signals
suitable for use are widely available and are well known in the art (von
Heijne, J. Mol.
Biol. 14:99-I05, 1985). Prokaryotic and eukaryotic secretion signals that are
functional in E. coli (or other host) may be employed. The presently preferred
secretion
signals include, but are not limited to pelB, mates,, extensin and glycine-
rich protein.
One skilled in the art appreciates that there are a wide variety of suitable
vectors for expression in bacterial cells and which are readily obtainable.
Vectors such
as the pET series (Novagen, Madison, WI) and the test and trc series
(Pharmacies,
Uppsala, Sweden) are suitable for expression of a wide variety of proteins. A
suitable
plasmid is ampicillin resistant, has a colEI origin of replication, lacIq
gene, a lac/trp
hybrid promoter in front of the lest Shine-Dalgarno sequence, a hexes-his
coding
sequence that joins to the 3' end of the inserted gene, and an rrnB terminator
sequence.
The choice of a bacterial host for the expression of the desired protein or
polypeptide is dictated in part by the vector. Commercially available vectors
are paired
with suitable hosts. The vector is introduced in bacterial cells by standard
methodology.
Typically, bacterial cells are treated to allow uptake of DNA (for protocols,
see generally,
Ausubel et al., supra; Sambrook et al., supra). Alternatively, the vector may
be
introduced by electroporation,phage infection, or another suitable method.
3. Expression in other organisms
A variety of other organisms are suitable for use in the present invention.
For example, various fungi, including yeasts, molds, and mushrooms, insects,
especially
vectors for diseases and pathogens, and other animals, such as cows, mice,
goats, birds,
aquatic animals (e.g., shrimp, turtles, fish, lobster and other crustaceans),
amphibians
and reptiles and the like, may be transformed with a desired transgene.
The principles that guide vector construction for bacteria and plants, as
discussed above, are applicable to vectors for these organisms. In general,
vectors are
well known and readily available. Briefly, the vector should have at least a
promoter
functional in the host in operative linkage with the desired protein or
polypeptide.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
37
Usually, the vector will also have one or more selectable markers, an origin
of
replication, a polyadenylation signal and transcription terminator.
The sequence of nucleotides encoding the desired protein or polypeptide
may also include a classical secretion signal, whereby the resulting peptide
is a
precursor protein processed and secreted. Suitable secretion signals may be
obtained
from a variety of genes, such as mat-alpha or invertase genes.
4. Transgenic animals
Within related aspects of the present invention, proteins of the present
invention may be expressed in a transgenic animal whose germ cells and somatic
cells
contain a gene which encodes the desired protein and which is operably linked
to a
promoter effective for the expression of the gene. Alternatively, in a similar
manner
transgenic animals may be prepared that lack the desired gene (e.g.,
"knockout" mice).
Such transgenics may be prepared in a variety non-human animals, including
mice, rats,
rabbits, sheep, dogs, goats and pigs (see Hammer et al., Natm°e 315:680-
683, 1985,
Palmiter et al., SciefZCe 222:809-814, 1983, Brinster et al., Py~oc. Natl.
Acad. Sci. USA
$2:4438-4442, 1985, Palmiter and Brinster, Cell 41:343-345, 1985, PCT
Publication
No. WO 99/01164, and U.S. Patent Nos. 5,175,383, 5,087,571, 4,736,866,
5,387,742,
5,347,075, 5,221,778, 5,162,215; 5,545,808; 5,741,957; 4,873,191; 5,780,009;
4,736,866; 5,567,607; 5,633,076 and 5,175,384). Briefly, an expression vector,
including a nucleic acid molecule to be expressed together with appropriately
positioned expression control sequences, is introduced into pronuclei of
fertilized eggs,
for example, by microinjection. Integration of the injected DNA is detected by
blot
analysis of DNA from tissue samples. It is preferred that the introduced DNA
be
incorporated into the germ line of the animal so that it is passed on to the
animal's
progeny. Tissue-specific expression may be achieved through the use of a
tissue-
specific promoter, or through the use of an inducible promoter, such as the
metallothionein gene promoter (Palmiter et al., 1983, ibid), which allows
regulated
expression of the transgene.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
38
E. Gene Delivery Vectors
A wide variety of gene delivery vectors may be utilized to deliver and/or
express a desired gene of interest in host cells. For example, within one
aspect of the
present invention, retroviral gene delivery vehicles may be utilized. Briefly,
retroviral
gene delivery vehicles of the present invention may be readily constructed
from a wide
variety of retroviruses, including for example, B, C, and D type retroviruses
as well as
spumaviruses and lentiviruses (see RNA Tumor Viruses, Second Edition, Cold
Spring
Harbor Laboratory, 1985). Such retroviruses may be readily obtained from
depositories
or collections such as the American Type Culture Collection ("ATCC";
Rockville,
Maryland), or isolated from known sources using commonly available techniques.
Representative examples of retroviral gene delivery vectors are described in
more detail
in EP 0,415,731; PCT Publication Nos. WO 90/07936; WO 91/0285, WO 9311230;
WO 9310218, WO 9403622; WO 9325698; WO 9325234; and U.S. Patent Nos.
5,219,740, 5,716,613, 5,851,529, 5,591,624, 5,716,826, 5,716,832, and
5,817,491.
Other suitable gene delivery vectors can be generated from alphaviruses
(see e.g., U.S. Patent Nos. 5,091,309 and 5,217,879, 5,843,723, and
5,789,245),
recombinant adenoviral vectors (see e.g., U.S. Patent No. 5,872,005), and
numerous
other viruses such as pox viruses, such as canary pox virus or vaccinia virus
(Fisher-
Hoch et al., PNAS 86:317-321, 1989; Flexner et al., Ann. N Y. Aead. Sci.
569:86-103,
1989; Flexner et al., haccine 8:17-21, 1990; U.S. Patent Nos. 4,603,112,
4,769,330 and
5,017,487; WO 89/01973); SV40 (Mulligan et al., Natz~re 277:108-114, 1979);
influenza virus (Luytjes et al., Cell 59:11071113, 1989; McMicheal et al., N.
Eng. J.
Med. 309:13-17, 1983; and Yap et al., Natzzf°e 273:238-239, 1978);
herpes (I~it, Adv.
Exp. Med. Biol. 215:219-236, 1989; U.S. Patent No. 5,288,641); HIV (Poznansky,
J.
Viz°ol. 65:532-536, 1991); measles (EP 0 440,219); Semliki Forest
Virus, and
coronavirus, as well as other viral systems (e.g., EP 0,440,219; WO 92/06693;
U.S.
Patent No. 5,166,057).
In addition to the above viral-based vectors, numerous non-viral gene
delivery vehicles may likewise be utilized within the context of the present
invention.
Representative examples of such gene delivery vehicles include direct delivery
of
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
39
nucleic acid expression vectors or naked DNA alone (see e.g., U.S. Patent Nos.
5,814,482 and 5,580,859), polycation condensed DNA linked or unlinked to
killed
adenovirus (Curiel et al., Hunt. Gene Then. 3:147-154, 1992), DNA ligand
linked to a
ligand (Wu et al., J. ofBiol. Cheyrr 264:16985-16987, 1989), and nucleic acid
containing
liposomes (e.g., WO 95/24929 and WO 95/12387).
F. Compounds
As noted above, a wide variety of compounds may be utilized within this
regard, including for example (a) a polypeptide comprising the amino acid
sequence
BX7B (SEQ ID N0:28) which binds HA; (b) an anti-TAM antibody; (c) a
polypeptide
fragment which encodes a Dl, D2, D3, D4, or, DS domain of RHAMM; and (d) a
gene
delivery vector which expresses antisense RHAMM, or, delivers and expresses
any one
of (a), (b), or (c).
Within one embodiment, the polypeptide BX7B (SEQ ID N0:28)
comprises a polypeptide wherein B is a basic amino acid and X7 is a sequence
of about
seven residues selected from any amino acid other than an acidic amino acid,
wherein
the peptide forms an alpha helix and each occurrence of B is oriented on the
same side
of the alpha helix, and with the proviso that the peptide does not consist of
the
sequences BBXXBBBXXBB, KQKIKHVVKLK, KLKSQLVKRK, RYPISRPRKR,
KNGRYSISR, RDGTRYVQKGEYR, RRRCGQKKK, RGTRSGSTR,
RRRKKIQGRSKR, RKSYGKYQGR, KVGKSPPVR, KTFGKMKPR, RIKWSRVSK,
KRTMRPTRR, KVGKSPPVR, or HREARSGKYK (SEQ ID Nos. 29-44 respectively).
In one embodiment, the polypeptide can be (a) a first peptide comprised of
a hyaluronic acid-binding domain; (b) a hyaladherin polypeptide; (c) a second
peptide
comprised of a domain from a hyaladherin polypeptide; (d) a hyaladherin-
binding
polypeptide; (e) a third peptide comprised of a hyaladherin binding domain.
Also
provided are antibodies which binds to a peptide or polypeptide of (a)-(d);
and/or vectors
(e.g., gene delivery vectors described below) that expresses a gene encoding a
polypeptide
as described above or herein. In a particular embodiment, peptides are
provided
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
comprised of a sequence selected from the group consisting of SEQ. ID NO: 1-
20. In
another embodiment, a hyaladherin-bindingpolypeptide comprised of SEQ. ID NO:
21.
Within particularly preferred embodiments of the invention, the
compound is an antibody. Representative examples of antibodies suitable for
use
5 within the present invention include antibodies to domain D1 of RHAMM amino
acids
1-164 of human RHAMM (including for example: sequences recognizing the murine
D 1 sequence, aa. 97-111 - QERGTQDKRIQDME (SEQ ID N0:21 ); and sequences
recognizing human RHAMM, as 151-164 - LKSKFSENGNQKNL (SEQ ID N0:18));
antibodies to domain D2 of RHAMM - the "leucine zipper" domain of human RHAMM
10 from as 195-222; antibodies which recognize the domain D3 -the TAM domains
of
RHAMM (aa 219-240 of the human RHAMM sequence, including antibodies which
recognize the sequence VSIEKEKIDEK (SEQ ID N0:49)); domain D4 (repeat or "R"
domain - as 442-546 for mouse, and as 442-463 for human) and domain D5 (HA
binding domain, including two domains: as 721-730 and as 742-752 for mouse; as
15 635-645 and as 657-666 for human). In other embodiments, antibodies are
provided
which bind to a polypeptide comprised of SEQ. ID NO: 11-20.
As utilized herein, reference may be made to the human sequence of
RHAMM for identification of the domains. However, the domains can be
identified
and specific antibodies generated for other species, such as, for example,
mouse. Figure
20 50 (SEQ ID Nos. 47 and 48) provides the amino acid sequence of human and
mouse
RHAMM (see PCT publication No. WO 97/38098 and Genbank Accession Nos.
AAC52049 & Q00547). As utilized herein, it should be understood that
antibodies
"bind" to the above sequence if they do so with a Kd of at least 10-7 M
(moles/liter)
(see "antibodies" above).
25 Also provided are polypeptides comprising a fragment of the RHAMM
protein, of less than 95 or 73kD molecular weight. Representative fragments of
polypeptides contain at least all, or a portion of one of domains D1, D2, D3,
D4, or D5,
as set forth above. Within various embodiments, the polypeptides are less than
250,
200, 150, 100, 75, 50, or, 25 amino acids in length.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
41
G. Methods of Treatment
As described in more detail below, a wide variety of diseases share
common disease processes such as local production of cytokines, degradative
enzymes,
reactive oxygen species resulting in increased cell migration and
proliferation and
eventual tissue destruction and cell death. These diseases may be readily
treated or
prevented, by administration of a composition that alters the activity of
transition
molecules within a cell Transition molecules are comprised of hyaladherins,
hyalauronans or molecules regulated by an amount of intracellular or
extracellular
hyaladherins or hyalauronans. The activity of hyaladherins and hyalauronans
are shown
to interact with a regulatory processes associated with a response to injury
and/or
proliferative/invasive cell types.
In one embodiment, methods are provided comprising the general steps
of administering to a mammal an effective amount of a composition comprised of
any
one of (a) a first peptide comprised of a hyaluronic acid-binding domain; (b)
a
hyaladherin polypeptide; (c) a second peptide comprised of a domain from a
hyaladherin polypeptide; (d) a hyaladherin-binding polypeptide; (e) a third
peptide
comprised of a hyaladherin binding domain; (f) an antibody that binds to a
peptide or
polypeptide of (a)-(e); and/or (g) a vector that expresses a gene encoding any
of (a)-(f).
Briefly, cells in a homeostatic environment such as normally occurs within
adult tissues are characterized by a differentiated state that varies with the
tissue, and a
low or contained rate of cell proliferation or motility. This differentiated
state has
typically been viewed to occur as a result of specific gene regulation. As
described in
more detail below, differentiated cells that are functioning within a
physiological,
homeostatic tissue environment are also restricted from expressing genes that
regulate
response-to-injury processes. These processes are regulated by master switch
transcription heterodimers termed AP-1 as illustrated in Figure 1. The three
map kinase
cascades identified in mammalian cells so far include ey~k, jnk and p38 hog
pathways.
These pathways collectively regulate expression of the transcription factors c
fos and c-
jun. Heterodimerization of these two transcription factors results in
formation of AP-1
which controls the expression of genes required for cell migration, cell
proliferation,
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
42
extracellular matrix remodeling and production of growth factors and cytokines
that are
required for amplification and maintenance of the response to injury process.
It is the
deregulated activation of these pathways that leads to the diseased state.
These transcription factors when activated control the expression of a
plethora of molecules required for efficient repair and include proteases such
as
collagenases, various extracellular matrix proteins and molecules that allow
the cell to
respond to cytokines/growth factors by proliferating and migrating
efficiently.
Response-to-injury is a well-defined term referring to the ability of cells to
repair and to
remodel their extracellular environment to promote and ultimately re-establish
the
differentiated state.
As shown in more detail in the Figures, nomal cells undergo a number of
intermediate changes until it becomes a diseased cell involved in chronic
inflammatory
diseases, proliferating diseases and degenerative diseases. In Figures 1, 2,
3, and 5, we
show schematically a model for the key transition steps involved in the
transformation
of normal cell to diseased cells which is applicable to all cell types
surrounded by
matrix and is most likely involved in all diseases including cancers,
inflammatory and
degenerative diseases, wound healing and injury related diseases, inflammatory
implications of host verses graft or devices. Briefly, in the normal tissue,
cells are
quiescent and responsible for normal and controlled tissue remodelling. Normal
cells
express growth factor receptors (represented by small circles) but these are
not grouped
together and the cell is restricted in its ability to respond to pro-
inflammatory cytokines
and growth factors. Rather, the cell remains in a non-proliferative state and
responds to
factors that regulate its state of differentiation (homeostatic responses).
Upon injury,
the cell rapidly releases glycosylphosphatidyl inositol linked proteins (co-
receptors
indicated by triangles) onto the cell surface and releases hyaluronic acid
(represented by
X) and other matrix molecules such as fibronectin (represented by ~) that
allow the cell
to initiate the beginnings of an activated state. The presence of the
coreceptors permits
growth factors to aggregate slightly enhancing their ability to respond to pro-
inflammatory cytokines and growth factors yet at the same time preventing the
full
response that is seen in the fully diseased cell. This regulated ability to
respond to
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
43
growth factors as well as the production of molecules such as hyaluronic acid
and
fibronectin allows the formation of podosomes (represented by small triangular
extensions) that facilitate the localized release of proteases and other
enzymes that
produce fragments of extracellular matrix. These fragments serve to recruit
other cells
to the site of injury including white cells that allow enhancement and
stabilization of the
response to injury. Furthermore, these fragments contribute to the evolution
of
podosomes to focal contacts. This intermediary transitional state is termed
Stage C and
reagents prepared against the molecules regulating podosome structure and
function are
predicted to prevent development of the next state, Stage D which is one that
allows full
responses to pro-inflammatory cytokines and growth factors. In Stage D, growth
factor
and cytokine receptors axe aggregated into structures called focal contacts,
which
contain all the signaling molecules required for activation of multiple
pathways. In this
aggregated state, cells are able to maximally respond to growth factors and
cytokines
and maintenance of this state leads to disease.
Cytokines and other pro-inflammatory mediators are not capable of
stimulating the expression of AP-1 dependent genes involved in cell
proliferation
migration, and tissue destruction. However upon injury or stress, the cells
under go a
series of changes which result in the transformation of normal cells to
diseased cells.
While not being bound by theory, it is proposed herein that cells that are
initially
responding to stress, whether due to heat, chemical, free radical, mechanical
injury or to
mutations of key proteins, react to these insults in a standard pattern. The
initial stage
involves the expression and secretion of matrix molecules involved in edema
and
inflammatory responses (for example hyaluronic acid (HA), collagen type VIII,
osteopontin, tenascin, serglycin, addressin, laminin), as well as expression
of transition
molecules on the cell membrane and surface such as heat shock proteins and
HA-binding proteins (Stage B). It is known that differentiated cells
undergoing
transition respond initially to injury by activating ERK kinase cascades that
regulate at
the least, activation of heat shock protein transcription factors and
potentially other
transition molecules allowing cells to remain viable as Stage B cells. These
cells are
characterized by enhanced production of heat shock proteins that protect the
cell from
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
44
aggregation of key proteins, organelle damage and ultimately apoptosis, as
well as by
increased presence of HA-binding molecules on the cell surface.
Once the cell has entered stage B, the presence of growth factor
concentrations and other molecules at the site of injury will likely determine
whether
the cell now returns to its differentiated state or proceeds to Stage C. The
present
invention provides the unexpected discovery that closely following the initial
responses,
cells enter a transitional stage (defined as Stage C) which is characterized
by (1) the
formation of transient structures called podosomes or invadapodia; (2)
disassembled
actin cytoskeleton (e.g., a paucity of focal adhesion); amd (3) dependence
upon
hyaluronan related molecules and hyaladherins for regulation of signaling
cascades; and
(4) altered control of growth factor initiated signaling. As illustrated in
the Examples,
cells plated onto plastic transiently form podosomes at 12-18 h., but this is
reduced by
24 h. Plating of cells onto fibronectin enhances and stabilizes podosome
formation.
Fibronectin is also necessary for the formation of focal contacts. The
presence of podosomes correlates with cell surface RHAMM expression. Such
cells
importantly exhibit the first stage of release from the restriction of AP-1
activation
exhibited by Stage A cells. Podosomes allow the cell to efficiently release
proteases at
lamellae tips to promote cell invasion into the matrix that will ultimately
initiate
controlled remodelling of its extracellular matrix detected by exposure of the
CS-1
sequence in fibronectin (described in more detail hereafter). This event is
believed to be
required for an ability to maximally respond to growth factors/cytokines and
re-
establishment of tissue homeostasis. Podosomes are also ultimately the sites
of focal
adhesion assembly that ultimately allow a cell to proceed to Stage D. At the
podosome
sites, increased and persistent matrix degradation results in increased
degradation
fragments of matrix molecules such as collagen and CS-1 fibronectin which
suppresses
the expression and levels of cell surface transitions molecules. Podosomes
require
interactions between hyaluronan and hyaladherins as well as interactions
between
hyaladherins and other proteins for their stmctural and functional integrity.
As the
levels of transitions molecules such as HA-binding molecules (e.g., RHAMM,
CD37)
on the cell surface and cytoplasm decrease, there is an increase in the
formation of focal
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
adhesions and Local accumulation of cell surface cytokine receptors,
intracellular
signaling molecules and cytoskeletal components (Stage D). Focal Adhesions
couple
integrins to growth factor/cytokine receptors and allow the cell to enter the
next stage in
the injury response which is characterized by heightened ability to respond to
pro-
s inflammatory cytokines and growth factors. The formation of focal adhesions
removes
restriction of activation of AP-1 dependent genes by cytokines and growth
factors and
results in increased cell migration and proliferation, and tissue destruction.
As described in more detail below, the sustained presence of these cells,
termed Stage D are largely responsible for tissue deterioration following
sustained and
10 escalated response to injury that is characteristic of many inflammatory,
degenerative
and proliferative type of diseases including for instance arthritis, multiple
sclerosis,
psoriasis, inflammatory bowel diseases, restenosis, fibronosis,
atherosclerosis, diabetes,
osteoarthritis, cancers, Alzheimer's, Parkinson's and wound healing.
These transitional stages (Stages B-C) are necessary for all differentiated
15 cells in tissues to activate AP-1 dependent genes and AP-1 dependent
disease processes
such as cell proliferation, migration, invasion and production of matrix
metalloproteinases, therefore, inflammatory, proliferative and degenerative
diseases are
dynamic processes that involve the continual recruitment of differentiated
tissue into the
pathway culminating in the stage D cells. The ability of the cell to acquire a
20 transitional phenotype is absolutely required for it to progress to Stage D
where it
responds to pro-inflammatory cytokines/growth factors. Molecules that regulate
this
transient cellular phase, such as those that either disrupt
hyaluronan/hyaladherin or
hyaladherin/other protein interactions, make excellent therapeutic and
diagnostic taxgets
in a variety of human diseases since these molecules will not be expressed in
most cells
25 and only transiently expressed in diseased tissue. This expression pattern
will provide
tissue specificity and low toxicity to the human body, allowing for chronic
use of
reagents, a requirement for managing many diseases.
Normal cells surrounded by a normal tissues are quiescent and involved
in the turnover of matrix. Furthermore, consistent with the present
disclosure, cells do
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
46
not possess focal adhesions in normal tissue in vivo whereas focal adhesions
have been
observed in diseased tissue.
Transitional stages such as that described above are evident in a wide variety
of disease processes, including for example, Parkinson's, Alzheimer's,
Arthritis and
Osteoporosis. These, as well as other disease processes which involve
transition molecules
that remove AP-1 restriction from normal cells are discussed in more detail
below.
1. Parkinson's
Parkinsonism is a clinical syndrome characterized by a disturbance in
motor functions such as slowness of voluntary movement, diminished facial
expressions,
stooped posture, rigidity and tremor. The disease appears later in life.
Although little is
known on the cause of the disease, there is substantial evidence indicating
that damage to
the nigrostriatal dopaminergic system is central to the disease. The
dopaminergic neurons
- of the substantial nigra project to the striatum in normal brain.
In~Parkinson's disease, the
loss of these neurons results in a decrease in striatal dopamine content and
this is
proportional to the severity of the motor syndrome. Similar to other brain
diseases, there
is an increase in glisosis, which involves the recruitment and activation of
glial cells.
These cells are recruited as part of the repair process, however, destructive
enzymes,
reactive oxygen species, cytokines and pro-inflammatorymediators produced by
activated
glial cells contribute and acerbate the disease.
Thus, within one embodiment methods are provided for treating
inflammatory neurological diseases such as Parkinsons, comprising
administering to a
patient a compound selected from the group consisting of (a) a polypeptide
comprising
the amino acid sequence BX7B (SEQ ID N0:2~) which binds HA; phage display
selected peptides that bind HA such as polypeptides comprising P-15 (Sequence
ID No.
70), P-16 (Sequence ID No. 26); P-32 (Sequence ID no. 71); and GAHWQFNALTVR
(Sequence ID No. 72); (b) an antibody which binds one of domains D1, D2, D3,
D4, or
DS of RHAMM; (c) a peptide of less than 95 kD or 73 kd, comprising all or a
portion of
domains Dl, D2, D3, D4, or, DS of RHAMM; and (d) a gene delivery vector which
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
47
expresses antisense RHAMM, or, delivers and expresses any one of (a), (b), or
(c), such
that the disease is treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
2. Alzheimer Disease
Alzheimer's disease is clinically manifested as insidious impairment of
higher intellectual function with alterations in mood and behavior. Later,
progressive
memory loss and disorientation are observed and eventually, profound
disability and
death. Alzheimer's disease affects a large portion of the increasingly aging
population
with a prevalence as high as 47% of those over 85 years old. The total costs
required for
formal and informal care of AD patients was $67 million in the United States.
Although
there is much variability, average life expectancy is 8-10 years after
dementia onset.
Alzheimer's disease is characterized by the appearance of cerebral
extracellular beta-amyloid deposits as senile plaques, intraneuronal
neurofibrillary
tangles, granulovascular degeneration and amyloid angiopathy. Senile plaques
are
extracellular lesions comprised of degenerating neuronal processes and
abnormal
deposits of beta-amyloid protein. Senile plaques range in size from 20 to 200
~,m in
diameter. Microglia and reactive fibrous astrocytes are enriched in the
periphery of
plaques, suggesting the recruitment of cells to the diseased site. These
plaques are
widely distributed in the cerebral cortex and are considered a critical
process in the
development of the disease.
Neurofibrillary tangles are intraneuronal structures consisting of paired
helical filaments in which the major constituent is a hyperphosphorylated tau
protein, an
axonal protein involved in microtubule assembly, and neurite regeneration and
remodeling. The MAP kinase, ERIC, and ubiquitin are also tightly associated
with these
helical filaments and may be directly involved in the stimulation of the MAP
kinase
pathway. The neurofibrillary tangles represent abnormal organization of
cytoskeletal
elements in neurons of patients with Alzheimer's disease.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
48
Other pathological findings associated with Alzheimer's include
granulovascular degeneration, Hirano bodies, neuronal and synaptic loss, and
beta
amyloid deposition in the wall of small cortical blood vessels. Although some
of these
disease processes are found in normal aging brains, their prevalence is
significantly
lower than in Alzheimer's disease and correlate with the severity of dementia.
Response to neuronal injury is characterized by the activation of glial
cells and the expression of a number of genes that participate in the repair
of damaged
neurons. Some of those products include the beta-amyloid precursor protein and
neurotrophins. The glial cell recruitment and responses may compromise
neuronal
viability by producing cytokines, reactive oxygen species and degradative
enzymes. It
is generally hypothesized that in local neuronal injury, an increased beta-
amyloid
production results in glial cell recruitment and activation which results in
the production
of pro-inflammatory processes and tissue destruction. Thus, it is most likely
the
accumulative effects of a defective repair process that results in neuronal
cell death and
the formation of senile plaques. A potential therapeutic target would include
the
inhibition of glial cell recruitment and activation. This would prevent
exacerbation of
the local inflammation and tissue destruction.
Thus, within one embodiment methods are provided for treating
Alzheimer disease, comprising administering to a patient a compound selected
from the
group consisting of (a) a polypeptide comprising the amino acid sequence BX7B
(SEQ
ID N0:28) which binds HA; phage display selected peptides that bind HA such as
polypeptides comprising P-15 (Sequence ID No. 70), P-16 (Sequence ID No. 26);
P-32
(Sequence ID no. 71); and GAHWQFNALTVR (Sequence ID No. 72); (b) an antibody
which binds one of domains D1, D2, D3, D4, or DS of RHAMM; (c) a peptide of
less
than 95 kD or 73 kd, comprising all or a portion of domains Dl, D2, D3, D4,
or, DS of
RHAMM; and (d) a gene delivery vector which expresses antisense RHAMM, or,
delivers and expresses any one of (a), (b), or (c), such that the disease is
treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
49
3. Arthritis and other inflammatory joint diseases
A number of inflammatory joint diseases have been characterized in
humans based on analysis of signs and symptoms, including for example,
rheumatoid
arthritis, systemic lupus erythomatosus, Reiter's syndrome, psoriatic
arthritis,
ankylosing spondylitis, to name just a few. Briefly, rheumatoid arthritis (RA)
is the
most prevalent type of inflammatory arthritis which occurs in approximately
1.5% of
the population (2). Therefore, the present characterization of human
inflammatory joint
disease is based on findings concerning RA. RA is characterized by synovial
hyperplasia, destruction of articular cartilage and bone, infiltration of
lymphocytes and
macrophages into synovial tissues and accumulation of autoantibody immune
complexes in synovial fluid. However, the contribution of infiltrating
lymphocyte cells
to the disease process is not clear. Cytokines, such as interleukin 1 (IL-1)
and
granulocyte-macrophage colony-stimulation factor (C'~M-CSF), are present in
increased
levels in inflamed joints and play a major role in the production of
metalloproteases
including MMP1 (collagenase), MMP2 (gelatinase) and MMP3 released by synovial
cells which are responsible for the destruction of cartilage. IL-1, together
with tumor
necrosis factor (TNF), also plays a major role in accumulation of lymphocytes
in the
joints. Joint inflammation is mediated by plasma and lipid derived mediators,
prostaglandin E2 and leukotriene B4.
Gradual destruction of articular cartilage is the most debilitating sign of
the disease. Cartilage is a connective tissue which consists of chondrocytes
and
extracellular matrix. Collagens and proteoglycans are the major components of
the
matrix. Chondrocytes are responsible for preservation of the integrity of the
matrix
which mostly depends on the collagenous network, the majority of which
consists of
collagen type II. Proteolytic enzymes that degrade the cartilage components
are
metalloproteases, which are produced by synovial cells, chondrocytes,
neutrophils, and
serine proteases derived from neutrophils. Several factors can induce the
expression of
metalloproteases, the most potent being secretion of IL-1 by macrophages (4).
Tissue
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
inhibitor of metalloproteases (TIMP) is a ubiquitous protein and natural
metalloprotease
inhibitor that is present in R.A synovial fluid in elevated levels. Another
feature of RA
is an increase in bone resorption due to activation of osteoclasts. It has
been shown that
monocyte derived mediators such a IL-1 and TNF, are responsible for the
increase in
5 osteoclastic activity.
In terms of treatment of RA, there are two types of drugs currently used:
anti-inflammatory drugs, including non-steroid or steroid, which alleviate the
inflammatory process only, and disease modifying anti-rheumatoid drugs which
interfere with the disease process. However, the mechanisms of action of these
drugs is
10 mostly unknown.
Osteoarthritis (OA) is a slowly progressive degeneration of the articular
cartilage that manifests in the weight-bearing j oints such as the knees and
hips.
Osteoarthritis, described as "wear and tear" arthritis, is characterized by
narrowing of the
joint owing to the loss of articular cartilage and thickening of the
subchondral bone. At a
15 later stage, inflammation of the synovium may occur, which plays an
important role in the
pathologic process by accelerating the catabolism. All these events lead to
nonfunctional
and painful joint. The prevalence and severity of OA increase with age,
affecting 80% of
the population after 55 years of age with higher frequency in women
(Altmm,1987). The
primary cause of OA remains unclear, joint trauma, obesity, bone
microfractures and
20 aging constitute the risk factors for OA (Altman,1987; Hough et a1.,1989).
Although the mechanisms involved in the pathogenesis of cartilage
destruction in OA are not well-characterized, much evidence suggests that
cytokines
may play an important role. During the progression of OA, cartilage fragments
in the
synovial fluid elicit an inflammatory response (Loyau and Puj o1, 1990;
Pelletier et al.,
25 1991 and 1993). This response results in enhanced protease and cytokine
release and
the production of reactive oxygen species. The cytokines, including IL-1 and
TNFa,
can activate MMP synthesis from chondrocytes and synoviocytes setting off a
cascade
leading to OA (Howell, 1986; Pelletier et al., 1983b). Apart from cytokines,
growth
factors also have significant effects on cartilage remodeling.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
51
Thus, within one embodiment methods are provided for treating arthritis
(e.g., rheumatoid arthritis or osteoarthritis), comprising administering to a
patient a
compound selected from the group consisting of (a) a polypeptide comprising
the amino
acid sequence BX7B (SEQ ID N0:28) which binds HA; phage display selected
peptides
that bind HA such as polypeptides comprising P-15 (Sequence ID No. 70), P-16
(Sequence ID No. 26); P-32 (Sequence ID no. 71); and GAHWQFNALTVR (Sequence
ID No. 72); (b) an antibody which binds one of domains Dl, D2, D3, D4, or D5
of
RHAMM; (c) a peptide of less than 95 kD or 73 kd, comprising all or a portion
of
domains D1, D2, D3, D4, or, D5 of RHAMM; and (d) a gene delivery vector which
expresses antisense RHAMM, or, delivers and expresses any one of (a), (b), or
(c), such
that the disease is treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally. .
4. Osteoporosis
Osteoporosis is a term used to define increased bone porosity of the
skeleton resulting from a reduction in bone mass. This disease affects the
elderly, is
particularly prevalent amongst females, and is sometimes a secondary responses
to
other clinical conditions. Thus, osteoporosis may be primary or secondary, and
depending on numerous parameters, can be localized to a certain bone region or
limb, or
may involve the entire skeleton. Osteoporosis normally refers to the common
primary
forms such as senile and postmenopausal osteoporosis, whereas secondary forms
include endocrine disorders (hyperparathyroidism, hyperthyroidism,
hypothyroidism,
acromegaly, Cushing's syndrome, prolactinaoma, Type I diabetes), neoplasia
(multiple
myeloma, sarcinomatosis, mast cell disease, thyroidlpar~athyf°oid
ademo),
gastrointestinal disorders (malnutrition, malabsorption, hepatic
insufficiency),
osteoarthritis and rheumatoid arthritis, drugs (anticoagulants,
chemotherapeutics,
corticosteroids, lithium), and a number of other non-specific disorders
(immobilization
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
52
or inactivity, pulmonary disease, anemia). Regardless of the etiology, the
critical loss
of bone makes the skeleton vulnerable to fractures and pain. Over 15 million
individuals suffer from primary osteoporosis in the United States and their
direct
medical costs are over $1 billion annually.
The maximum bone mass is achieved during young adulthood. In
normal adults the level of bone mass is determined by genetic factors, diet,
physical
activity and hormonal state. During adult years and aging this bone is turned
over by a
continuous, controlled resorption and formation cycle. In normal individuals,
a small
deficit in bone mass accrues with every bone resorption and formation cycle,
which can
average 0.7% of the total bone mass per year. Although there is no doubt that
an
imbalance in the ~resorption and formation cycle is responsible for
osteoporosis, little is
known on the origins of primary osteoporosis. Most of the focus has been on
age-
related changes, reduced physical activity and hormonal changes (particularly
associated with menopause). It is well established that osteoblasts from 'the
elderly,
which are cells responsible for bone formation, have reduced biosynthetic
potential
relative to osteoblasts from young adults. In addition, peptides (bone
morphogenic
proteins) deposited in the mineralized matrix which stimulate osteoprogenitor
cells and
osteoblastic activity are less effective with aging. Thus, decreased capacity
of bone
formation combined with normal or elevated osteoclastic activity are largely
responsible
for osteoporosis associated with aging and physical inactivity.
Postmenopausal osteoporosis is characterized by a hormonal dependent
accelerated bone loss. Following menopause, the yearly loss of bone mass may
reach
2% of the cortical bone and 9% of the cancellous bone. Estrogen is believed to
play an
important role in the reduction of bone loss. The estrogen effects are thought
to be
mediated by cytokines, which are found elevated in osteoporotic bone. It
appears that
decreased estrogen levels are capable of inducing cytokines such as IL-1,
which are
capable stimulating bone resorption. IL-1 is the most potent stimulator of
osteoclast
recruitment and activity and thought to play an important role in bone
resorption in
post-menopausal osteoporosis. A number of genes that are induced by IL-1
(cathepsin
K, matrix metalloproteinases and COX-2) are elevated in osteoporotic bone and
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
53
produced by osteoblasts and osteoclasts in vitro. Inhibition of osteoclast
recruitment
and activation are key steps in shifting the balance from resorption to bone
formation,
resulting in increased bone mass.
Thus, within one embodiment methods are provided for treating
osteoporosis, comprising administering to a patient a compound selected from
the group
consisting of (a) a polypeptide comprising the amino acid sequence BX7B (SEQ
ID
N0:28) which binds HA; phage display selected peptides that bind HA such as
polypeptides comprising P-15 (Sequence ID No. 70), P-16 (Sequence ID No. 26);
P-32
(Sequence ID no. 71); and GAHWQFNALTVR (Sequence ID No. 72); (b) an antibody
. which binds one of domains D1, D2, D3, D4, or DS of RHAMM; (c) a peptide of
less
than 95 kD or 73 kd, comprising all or a portion of domains D1, D2, D3, D4,
or, DS of
RHAMM; and (d) a gene delivery vector which expresses antisense RHAMM, or,
delivers and expresses any one of (a), (b), or (c), such that the disease is
treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
5. Multiple Sclerosis
Multiple sclerosis is the most common of the demyelinating disorders,
having a prevalence of approximately 1 in 1000 persons in most of the United
States
and Europe. Although the etiology of multiple sclerosis (MS) is unknown,
genetic,
environmental and immunological factors are believed responsible for a
coordinated
attack on myelin. The hallmark lesion in MS is a punched-out area in which the
axon is
surrounded by astrocytic processes. The accompanying inflammatory reaction is
characterized by infiltration of lymphocytes, monocytes and macrophages into
the
parenchyma of the central nervous system (CNS), analogous to the chronic
inflammation in other diseases such as arthritis and psoriasis. Thus, in MS,
there is
increased inflammatory cell activation and infiltration, increased fibrous
astrocyte
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
54
activation, migration and proliferation, increased production of cytokines and
matrix
metalloproteinases, increased demyelination, axonal degeneration and plaque
formation.
Thus, within one embodiment methods are provided for treating multiple
sclerosis, comprising administering to a patient a compound selected from the
group
consisting of (a) a polypeptide comprising the amino acid sequence BX7B (SEQ
ID
N0:28) which binds HA; phage display selected peptides that bind HA such as
polypeptides comprising P-15 (Sequence ID No. 70), P-16 (Sequence ID No. 26);
P-32
(Sequence ID no. 71); and GAHWQFNALTVR (Sequence ID No. 72); (b) an antibody
which binds one of domains D1, D2, D3, D4, or DS of RHAMM; (c) a peptide of
less
than 95 kD or 73 kd, comprising all or a portion of domains D1, D2, D3, D4,
or, DS of
RHAMM; and (d) a gene delivery vector which expresses antisense RHAMM, or,
delivers and expresses any one of (a), (b), or (c), such that the disease is
treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
6. Inflammatory Dermatosis
Inflammatory dermatological diseases, such as psoriasis, are very
common, affecting as many as 1 to 2% of the people in the United States. It is
often
associated with arthritis, myopathy, spondylitic heart disease and AIDS.
Psoriasis is a
chronic inflammatory disease characterized by keratinocyte hyperproliferation
and a
distinct inflammatory pattern that is dependent on the type of psoriasis. The
underlying
pathogenesis involves three predominant and interdependent biologic processes:
inflammation, epidermal hyperproliferation,and altered differentiationwith
parakeratosis.
The homeostasis of the epidermis depends on the balance of growth
regulatory signals, which appear to be altered in psoriasis. The epidermis
serves a number
of important barrier functions against protein and water loss, entry of
microorganisms,
physiochemical trauma including UV. The squamous epithelium undergoes terminal
differentiationresulting in an insoluble cornified envelope providing an
important barner.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
Keratinocyte proliferation takes place in the basal layer and migrate through
the epidermis
where differentiation specific proteins such as involucrin and keratins are
expressed.
Normal epidermis represents a normal balance between kaeratinocyte production
in the
basal layer a.nd corneocyte shedding at the skin surface. Upon wounding or
psoriasis,
5 there are rapid increases in the proliferationof keratinocytes.
Thus, within one embodiment methods are provided for treating
inflammatory dermatosis (e.g., psoriasis), comprising administering to a
patient a
compound selected from the group consisting of (a) a polypeptide comprising
the amino
acid sequence BX7B (SEQ ID N0:28) which binds HA; phage display selected
peptides
10 that bind HA such as polypeptides comprising P-15 (Sequence ID No. 70), P-
16
(Sequence ID No. 26); P-32 (Sequence ID no. 71); and GAHWQFNALTVR (Sequence
ID No. 72); (b) an antibody which binds one of domains D1, D2, D3, D4, or DS
of
RHAMM; (c) a peptide of less than 95 kD or 73 kd, comprising all or a portion
of
domains D1, D2, D3, D4, or, DS of RHAMM; and (d) a gene delivery vector which
15 expresses antisense RHAMM, or, delivers and expresses any one of (a), (b),
or (c), such
that the disease is treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
7. Inflammatory Bowel Diseases
There is overwhelming evidence that genetic and environmental factors
play a role in the development of inflammatory bowel diseases (IDB),
ulcerative colitis
and Crohn's disease. These diseases are chronic relapsing inflammatory
diseases and
share many common features of unknown etiology. Crohn's disease is a
granulomatous
disease that may affect any portion of the gastrointestinal tract from mouth
to anus, but
most often involves the small intestine and colon. Ulcerative colitis is a non-
granulomatosis disease limited to the colon. These diseases affect
approximately 3 to 6
people per 100,000, but the incidence can vary markedly between populations.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
56
The clinical manifestations, biochemistry and pathology of IDB
demonstrate that infiltration and activation of inflammatory cells, increased
local
mucosal responses, overproduction of cytokines and destructive enzymes are
associated
with the disease process ultimately leading to tissue injury. It is not known
whether the
immune system infiltrates the intestine in response to luminal or mucosal
antigens or
that local insult or disease results in the expression of adhesion molecules
and
chemoattractant cytokines that induce the infiltration of inflarmnatory cells
resulting in
the immune mediated tissue injury. Regardless of the etiology, there are
similarities
between the disease processes in IDB and other chronic inflammatory diseases.
Similar to other inflammatory diseases, there are very high levels of pro-
inflammatory cytokines (IL-1, IL-6, IL-8 and TNF), as well as anti-
inflammatory
cytokines (IL-4, IL-10 and IL-11) in IBD biopsies. In IBD, there is a
disturbed balance
between the levels of pro-inflammatory cytokines and anti-inflammatory
cytokines that
favors the former. The expression of IL-1, IL-6, IL-8 is increased in
inflammatory
lesions of patients with IDB (p3 82-4). These cytokines are produced by
infiltrating
inflammatory cells and local epithelial cells and fibroblasts. It is thought
that these
imbalances result in increased expression of genes such as adhesion molecules,
matrix
metalloproteinases and inflammatory mediators that are involved in cell
migration and
proliferation, and tissue destruction. Current therapeutic strategies aim at
inhibiting IL
1 and TNF activity.
Thus, within one embodiment methods are provided for treating
inflammatory bowel disease, comprising administering to a patient a compound
selected
from the group consisting of (a) a polypeptide comprising the amino acid
sequence
BX7B (SEQ ID N0:28) which binds HA; phage display selected peptides that bind
HA
such as polypeptides comprising P-15 (Sequence ID No. 70), P-16 (Sequence ID
No.
26); P-32 (Sequence ID no. 71); and GAHWQFNALTVR (Sequence ID No. 72); (b) an
antibody which binds one of domains D 1, D2, D3, D4, or DS of RHAMM; (c) a
peptide
of less than 95 kD or 73 kd, comprising all or a portion of domains D1, D2,
D3, D4, or,
DS of RHAMM; and (d) a gene delivery vector which expresses antisense RHAMM,
or,
delivers and expresses any one. of (a), (b), or (c), such that the disease is
treated.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
57
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
8. Other Inflammatory Diseases
As described above, there are several classes of molecules and disease
processes that are common to all chronic inflammatory diseases. These include
increased expression of adhesion molecules, cytokines and matrix
metalloproteinases,
increased cell proliferation and migration, increased inflammatory cell
activation and
infiltration, increased angiogenesis, and increased tissue destruction and
dysfunctional
matrix remodeling. These disease processes are tightly regulated in normal
differentiated cells and require the activation of AP-1 transcription factors
and AP-1
dependent genes. Since the restriction of AP-1 activation in normal cells can
be
reversed in a controlled fashion by transition molecules (such , as RHAMM),
the
inhibition of expression, activity and signaling of transition molecules will
be useful
therapeutically for not only the diseases described above, but also for other
inflammatory diseases such as diabetes mellitus; restenosis; atherosclerosis;
systemic
lupus erythematosus; emphysema; AIDS; chronic endometriosis; pulmonary,
myocardial and hepatic fibrosis; inflammatory polyradiculoneuropathy; chronic
cystitis;
acute mastitis; cholecystitis; gastritis; nephritis; hepatitis; bronchial
asthma; vasculitis;
chronic bronchitis; kidney fibrosis, pericarditis and myocarditis;
pancreatitis;
peritonitis; prostatitis; septic shock; periodentitis, thyroiditis;
retinopathy.
. Thus, within one embodiment methods are provided for treating the
above described treating diseases (e.g., lupus, diabetes mellitus, or, kidney
fibrosis),
comprising administering to a patient a compound selected from the group
consisting of
(a) a polypeptide comprising the amino acid sequence BX7B (SEQ ID N0:28) which
binds HA; phage display selected peptides that bind HA such as polypeptides
comprising -15 (Sequence ID No. 70), P-16 (Sequence ID No. 26); P-32 (Sequence
ID
no. 71); and GAHWQFNALTVR (Sequence ID No. 72); (b) an antibody which binds
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
58
one of domains D1, D2, D3, D4, or DS of RHAMM; ~(c) a peptide of less than 95
kD or
73 kd, comprising all or a portion of domains Dl, D2, D3, D4, or, DS of RHAMM;
and
(d) a gene delivery vector which expresses antisense RHAMM, or, delivers and
expresses any one of (a), (b), or (c), such that the disease is treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
9. Wound Healing and Responses to Injury
Wound healing responses to injury involve a complex series of cellular
and inflammatory processes resulting in the deposition of connective tissues
and its
remodeling into abnormal tissue or scarring. The underlying mechanisms of
wounding
or injury responses involve the induction of an acute inflammation, production
of
cytokines and growth factors, regeneration of parenchymal cells, migration,
proliferation and differential of parenchymal and connective tissue cells,
synthesis of
extracellular matrix proteins, angiogenesis and fibrosis, and remodeling of
connective
tissues. In addition, these healing processes are common in a variety of
clinical areas
such as scarring from surgical incisions, wounds or various derma inflammatory
diseases, restenosis following angioplasty, vascular grafts, stroke and
surgical
adhesions. Interference of processes that induce abnormal tissue deposition
and
remodeling will enhance an orderly wound or injury repair resulting in the
development
of normal functional tissue. Since transition molecules, such as RHAMM,
regulate a
number of the diseased processes in wound healing and the transformation of
normal to
diseased cells, it is likely that agents which inhibit the function of
transition molecules
would be useful therapeutically for the treatment of restenosis following
angioplasty,
vascular grafting, ballooning or any other type of injury to the vascular
system, stroke,
surgical incisions, burns, wounds, inflammatory skin diseases, and surgical
adhesions.
The simplest form of wound repair or healing is observed following a
clean surgical incision. The incision causes a limited amount of tissue
disruption,
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
59
which results in responses by epithelial cells and connective tissue cells, as
well as
infiltration of inflammatory cells. Immediately following the incision, the
incision
space is bathed with blood, containing fibrin and blood cells that clots and
leads to the
formation of a scab that covers the wound. The initial process involves the
response of
local basal cells in the production of cytokines and other pro-inflammatory
mediators,
and infiltration of neutrophils. The basal cells become mitotic and produce
matrix,
resulting in the thickening of the epidermis. This is followed by the
migration of the
epithelial cells along the cut margins and depositing basement membrane
underneath
the scab. The neutrophils are replace by macrophages and granulation tissue is
progressively laid down containing collagen fibrils vertically oriented rather
than
oriented in fashion that would enhance bridging the incision space. Epithelial
cell
proliferation and migration continues, as well as tissue thickening.
Neovascularization
reaches maximal levels and the surface cells differentiate and produce normal
epidermal
architecture. The last stages of incision healing involve the disappearance of
all
inflammatory cells, edema and increased vascularization, as well as
accumulation of
normal collagen fibrils and strengthening of tissue.
In cases where there are more extensive surface wounds such as burns,
abscess formation, inflammatory ulceritis, the reparative process is also more
extensive.
The larger tissue defects have greater cell loss, more fibrin and more
inflammation,
increased amounts of granulation tissue and wound contraction involving
myofibroblasts.
Regardless of the wound, the mechanisms of responsible for the processes of
healing
described above are similar. Wound healing is ultimately regulated by growth
factors and
cytokines that balance matrix synthesis and degradation locally. Collagen
synthesis is a
key component of wound healing and provides the tensile strength required
closing of the
incision. The type of collagen produced is dependent on the tissue repaired,
and changes
in the type of collagen may lead to dysfunction tissue. Collagen synthesis is
stimulated
early in tissue repair by factors such PDGF, FGF, and TGF. On the other hand,
degradation of collagen fibrils and other matrix molecules are also important.
The
degradative enzymes involved during wound healing include matrix
metalloproteinases,
neutrophil elastase, cathepsin G, kinins, plasmin and other enzymes.
Inflammatory and
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
local cells produce these enzymes. Degradation may aid in the remodeling of
the
connective tissue repair. If the inflammatory destructive processes are
suppressed, then it
is more likely to achieve a more rapid formation of the connective tissues and
decrease the
accumulation of scar tissue.
5 One type of wound healing occurs in surgical adhesions. Briefly,
surgical adhesion formation is characterized by abnormal adherence and scar
formation
between two adjacent tissues that occur most often following surgery.
Adhesions are a
major cause of surgical therapy and can result in bowel or urethral
obstruction. Surgical
adhesions are thought to be an inflammatory response to surgical trauma. Local
tissues
10 and inflammatory cells produce and secrete pro-inflammatory cytokines which
increase
vascular permeability, inflammatory cell infiltration, cellular migration and
proliferation, and the laying down of matrix between just-neighboring tissues.
The
accumulation of fibroblasts results in the accumulation of matrix and eventual
adhesion
of the two tissues. In theory, any agent that inhibits the inflammatory
response and
15 tissue remodeling would prevent the formation of surgical adhesions,
particularly if
these agents can be administered locally.
Thus, within one embodiment methods are provided for treating the
afore-mentioned diseases associated with wounds / wound healing, comprising
administering to a patient a compound selected from the group consisting of
(a) a
20 polypeptide comprising the amino acid sequence BX7B (SEQ ID N0:28) which
binds
HA; phage display selected peptides that bind HA such as polypeptides
comprising P-
15 (Sequence ID No. 70), P-16 (Sequence ID No. 26); P-32 (Sequence ID no. 71);
and
GAHWQFNALTVR (Sequence ID No. 72); (b) an antibody which binds one of
domains D1, D2, D3, D4, or DS of RHAMM; (c) a peptide of less than 95 kD or 73
kd,
25 comprising all or a portion of domains D1, D2, D3, D4, or, DS of RHAMM; and
(d) a
gene delivery vector which expresses antisense RHAMM, or, delivers and
expresses
any one of (a), (b), or (c), such that the disease is treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
30 intramuscularly, and orally.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
61
10. Restenosis/Stenosis following Therapeutic Interventions in Vascular
Disease: Angioplasty, Stent Insertion and Vascular Replacement/Grafts
Vascular diseases such as atherosclerosis are a leading cause of death and
disability in the developed world. Several therapeutic interventions have been
developed
to treat vascular diseases such as atherectomy, balloon angioplasty, insertion
of stems, and
insertion of arterial and venous grafts. For example, over 300,000 procedures
of
percutaneous transluminol coronary angioplasty are performed in the United
States per
year. Although these interventions are less costly and less invasive to the
patient, there
are a number of morphological changes and disease states produced in response
to injury
that are introduced by these new modes of therapy, namely restenosis.
Restenosis is characterized by thickening of the blood vessel wall in
response to injury that progresses until full occlusion of the vessel. Despite
the
significant advance made in these therapies, chronic restenosis of the dilated
lesions occur
in 30 to 50 % of the cases, remaining a serious and frequent problem.
Fu~thermof-e,
eventually sterzosis occuf°s in virtually all g~°afted vessels.
Restenosis has been suggested
to represent an exaggerated healing response to local injury, in which smooth
muscle cells
in the media migrate to and proliferate in the intima. Local production of
cytokines and
growth factors by local cells and inflammatory cells results in abnormal
matrix deposition
and remodeling. There are number of underlying mechanisms which can play a
role in
the induction of this disease. An injury to the endothelial cell layer will
expose blood
vessel layers to serum components and platelets, initiating a wound healing
process.
Factors released locally lead to increased cell proliferation and increased
expression of
matrix metalloproteinases required for cell proliferation and migration. These
cells
accumulate in the intima and form lesions that eventually block the vessel.
Utilizing the
therapeutic compositions provided herein, blocking the activation of smooth
muscle cells
and inhibition of their migration and proliferation in response to injury can
be utilized to
therapeuticallytreat stenosis and restenosis.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
62
Thus, within one embodiment methods are provided for inflammatory /
proliferative diseases associated with surgical procedures or intervention
(e.g.,
restenosis, stenosis, medical implants and the like), comprising administering
to a
patient a compound selected from the group consisting of (a) a polypeptide
comprising
the amino acid sequence BX7B (SEQ ID N0:28) which binds HA; phage display
selected peptides that bind HA such as polypeptides comprising -15 (Sequence
ID No.
70), P-16 (Sequence ID No. 26); P-32 (Sequence ID no. 71); and GAHWQFNALTVR
(Sequence ID No. 72); (b) an antibody which binds one of domains D1, D2, D3,
D4, or
DS of RHAMM; (c) a peptide of less than 95 kD or 73 kd, comprising all or a
portion of
domains D1, D2, D3, D4, or, DS of RHAMM; and (d) a gene delivery vector which
expresses antisense RHAMM, or, delivers and expresses any one of (a), (b), or
(c), such
that the disease is treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally. In addition, within certain embodiments the
compounds
described herein may be administered by balloon catheter, or, delivered from a
stmt
which is adapted to release the desired compound.
11. Atherosclerosis and Related Diseases: Myocardial Infarction and Stroke
Cardiovascular disease is a serious problem and accounts for 44% of the
mortality in the USA. Atherosclerotic cardiovascular disease is generalized
process that
involves the brain, heart and peripheral arteries. Atherosclerosis is
characterized by
intimal thickening caused by the accumulation of cells, infiltration of
inflammatory
cells, lipids, and connective tissues that can lead to cardiac and cerebral
infarction (such
as heat attack and stroke). Although the role of injurious stimuli is not
known, the
responses of the endothelial cells and the adaptive changes within the intima
are critical
in vascular remodeling leading to atherosclerotic plaques. Endothelia cells,
monocytes
and smooth muscle cells express biologically active molecules such as adhesion
molecules, cytokines, coagulation and fibrinolytic factors, metalloproteinases
and
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
63
vasoactive substances that contribute to atherogenesis and thrombosis. It is
thought that
atherosclerotic lesions develop by (1) invasion of artery wall by inflammatory
cells,
particularly monocytes; (2) smooth muscle cell migration, proliferation, and
synthesis
of matrix molecules; (3) intracellular lipoprotein uptake and lipid
accumulation. Briefly,
inflammatory cytokines induce the production of adhesion molecules resulting
in
inflammatory cell infiltration and responses. Activated smooth muscle cells
migrate in
response to local injury and produce large amounts of matrix and express
lipoprotein
scavenger receptors and can become involved in a generalized immune reaction.
Occlusion of the artery leads to a series of clinical complications such as
myocardial
infarction and stroke. Prevention of inflammatory cell infiltration,
production of matrix
metalloproteinases, cell proliferation and migration will reduce smooth muscle
cell and
matrix accumulation, and inhibit vessel occlusion.
Thus, within one embodiment methods are provided for treating the
above-noted atherosclerotic diseases, comprising administering to a patient a
compound
selected from the group consisting of (a) a polypeptide comprising the amino
acid .
sequence BX7B. (SEQ ID N0:28) which binds HA; phage display selected peptides
that
bind HA such as polypeptides comprising P-15 (Sequence ID No. 70), P-16
(Sequence
ID No. 26); P-32 (Sequence ID no. 71); and GAHWQFNALTVR (Sequence ID No.
72); (b) an antibody which binds one of domains D1, D2, D3, D4, or DS of
RHAMM;
(c) a peptide of less than 95 kD or 73 kd, comprising all or a portion of
domains D1,
D2, D3, D4, or, DS of RHAMM; and (d) a gene delivery vector which expresses
antisense RHAMM, or, delivers and expresses any one of (a), (b), or (c), such
that the
disease is treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
64
12. Tissue Transplantation
The increasing use of transplantation for bone marrow, renal, pulmonary,
cardiovascular and hepatic disorders has generated a series of clinical
complications. In
addition, with recent advances in tissue engineering, there is considerable
potential that
skin, cartilage, bone and many other tissues will be transplanted in the
future. In many
cases transplantation is the only form of treatment. For example, lung
transplant is the
only effective treatment of terminal lung diseases such as idiopathic
pulmonary fibrosis,
primary pulmonary hypertension, emphysema, and cystic fibrosis. The same is
true for
specific renal, hepatic and heart diseases. There are three major
complications in the
transplantation of organs: ( 1 ) host versus graft disease; (2) non-
immunological damage;
and (3) infection. Acute and chronic rejection is a significant problem where
the host
immune system invades the donor organ. This inflammatory response and
mononuclear
cell infiltrates are treated with immunosuppressive drugs with some success.
However,
these drugs can be very toxic and result in other clinical complications. The
non-
immunological damage from preservation injury results in inflammation and
tissue
damage. The role of infection can be treated with antibiotics. The disease
processes
involved in organ rejection are similar to other inflammatory diseases.
Disease intervention with devices has increased significantly over the
past decade. These include the use of devices fox hip and knee replacements,
cardiovascular stems, esophageal stems, vascular wraps, bone grafts, venous
and arterial
grafts, many others. A common problem with the use of these devices is an
inflammatory reaction to particles produced from the device or loosening of
the device
or injury caused by the local application of the device. It would seem likely
that
systemic or local application of the inflammatory response and local tissue
reaction to
the devices would inhibit this problem.
Thus, within one embodiment methods are provided for treating patients
undergoing tissue or cell transplation, comprising administering to a patient
a
compound selected from the group consisting of (a) a polypeptide comprising
the amino
acid sequence BX7B (SEQ ID N0:28) which binds HA; phage display selected
peptides
that bind HA such such as polypeptides comprising P-15 (Sequence ID No. 70), P-
16
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
(Sequence ID No. 26); P-32 (Sequence ID no. 71); and GAHWQFNALTVR (Sequence
ID No. 72); (b) an antibody which binds one of domains D1, D2, D3, D4, or DS
of
RHAMM; (c) a peptide of less than 95 kD or 73 kd, comprising all or a portion
of
domains D1, D2, D3, D4, or, DS of RHAMM; and (d) a gene delivery vector which
5 expresses antisense RHAMM, or, delivers and expresses any one of (a), (b),
or (c), such
that the disease is treated.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
13. Cancer and Metastases
Cancer is a generic term representing a collection of diseases arising
from mutations of key molecules that regulate cell proliferation, invasion,
and
metastasis. A representative cancer for which the key mutations are known is
exemplified by colorectal cancer. This cancer originates as a benign growth as
a result
of a mutation in a gene termed APC. Mutation of three additional molecules are
required for this benign growth to progress to a rapidly proliferating and
invasive tumor.
A plethora of mutations arises within the tumor as it progresses and these
enhance the
ability of the mutant tumor cells to attract normal endothelial cells to
migrate into the
growing tumor and form new blood vessels, a process known as angiogenesis. As
angiogenesis proceeds and as mutations affecting the ability of tumors to
respond to
growth factors accumulate, subsets of tumor cells develop the capacity to
invade blood
vessels as well as lymphatics and to metastasize.
The ability of tumor cells to metastasize involves deregulation via
overproduction or mutation of genes that allow cells to invade out of the
tissue of
origin, survive in a contact-independent manner, escape immune recognition,
lodge at a
distant site, then invade to a suitable place within the new tissue and grow
there. The
molecules that are commonly involved in tumor initiation, progression and
metastasis
include adhesion molecules, growth factor receptors, factors regulating the
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
66
cytoskeleton, master switches regulating cell cycle, proliferation repressor
genes,
proteases and transcription factors.
Although our understanding of master switches, proliferation repressors,
growth factors and proteases is quite well developed and pre-clinical and
clinical
approaches to targeting these molecules, particularly proteases have been
developed,
very little is known about the molecular characteristics of the invasive
phenotype. The
invasive tumor phenotype is predicted to be similar to the transitional
phenotype noted
for the above diseases and to be characterized by a propensity to form
invadapodia or
podosomes to release proteases and to express transition molecules that permit
and
prepare a cell to invade, move, and ultimately respond to growth factors and
cytokines
in a focal adhesion-dependent manner. It is likely that molecules required for
generating this phenotype are also expressed transiently in tumor cells since
they may
be only temporarily required and permanent expression would not necessarily be
advantageous. Thus, it is predicted that transitional molecules defining an
invasive
phenotype would appear in a subpopulation of tumor cells in a given tumor. A
transient
nature is likely one reason that markers of invasive phenotype have been so
elusive to
define. However, the ability of most tumors to kill is directly related to
their capacity to
invade and ultimately to metastasize. Therefore, identification of transient
molecules is
key for diagnosis, prognosis, adjuvant treatment or therapeutic treatment of a
variety of
cancers including: head and neck tumors (lip, oral cavity, auropharynx,
nasopharynx,
hypopharynx, larynx, glottis, supraglottis, subglottis, maxillary sinus, major
salivary
gland, lung, esophageal, gastric, colorectal cancer, anal, pancreatic liver,
gall bladder,
extrahepatic bile duct cancer, breast cancer, gynecologic cancers (cervix,
endometrium,
ovary, cancer of the uterine body, vaginal, vulvar, gestational
trophoboblastic),
testicular, urinary tract (renal, urinary bladder, penile, urethral,
prostatic) neurologic,
endocrine skin (basal cell and squamous cell melanoma) sarcomas, blood
(leukemia,
lymphoma) childhood neoplasm's (leukemia, lymphoma, neuroblastoma, Wilms'
tumor
rhabdomyosarcoma, Ewing's sarcoma, retinoblastoma) mediastinum, thymic germ
cell,
retroperitoneal, cardiovascular tumors, mastocytosis, carcinosarcomas, adenoid
cystic
carcinoma, dental tumors olfactory, neuroblastoma, paraganglioma.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
67
With regard to transitional molecules involved in proliferative cancers,
the present invention shows that RHAMM is highly overexpressed in subsets of
cells in
primary breast cancer tissue and this overexpression is prognostic of lymph
node
metastasis and poor outcome. Furthermore, RHAMM is shown to regulate ERK
activation, a key player in AP-1 activation. ERIC is also shown to regulate
cell
locomotion, a key behavior required for cell invasion into lymph nodes and is
required
for the invasion of tumor cells both iyz vitro and in transgenic models of
breast cancer.
Furthermore, CD44 is required for efficient signaling through her2/neu, an
oncogene
strongly implicated in regulating lymph node metastasis of breast cancer
cells. Finally,
HA promotes the expression of podosomes in invasive cancer cells and podosome
formation is one important characteristic of the transitional phenotype. In
addition, and
consistent with this observation, HA promotes the invasion of these cells into
collagen
gels in vitro.
Thus, within one embodiment methods are provided for treating cancer
and other metaseses, comprising administering to a patient a compound selected
from
the group consisting of (a) a polypeptide comprising the amino acid sequence
BX7B
(SEQ ID N0:2$) which binds HA; phage display selected peptides that bind HA
such as
polypeptides comprising -15 (Sequence ID No. 70), P-16 (Sequence ID No. 26); P-
32
(Sequence ID no. 71); and GAHWQFNALTVR (Sequence ID No. 72); (b) an antibody
which binds one of domains Dl, D2, D3, D4, or DS of RHAMM; (c) a peptide of
less
than 95 kD or 73 kd, comprising all or a portion of domains D1, D2, D3, D4,
or, DS of
RHAMM; and (d) a gene delivery vector which expresses antisense RHAMM, or,
delivers and expresses any one of (a), (b), or (c), such that the disease is
treated.
The' polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
68
14. Chronic and Acute respiratory distress syndrome:
Due to injury of the lung such as occurs in premature birth and consequent
positive pressure breathing measures as well as in adults following accidents
or
chemotherapy, the lung is injured and macrophages and neutrophils accumulate
within the
lung eventually destroying type II aveolar cells that produce surfactant
proteins required
for maintenance of positive pressure following lung expansion. As a result,
lungs are
poorly functional and patients become cyanotic and breathe rapidly. This
syndrome ends
in death. Clinical indications characterized by lung inflammation include
emphysema,
asthma, cystic fibrosis, new-born lung disease involving chronic respiratory
distress
syndrome, and the acute respiratory distress syndrome that affects accident
victims. Local
inflammatory responses that recruit macrophages into the lung result in
destruction of
alveolar type II cells, which make the surfactant responsible for normal lung
inflation.
The infiltration of macrophages and abnormal local tissue responses result in
further tissue
destruction and disease. This pathological sequence results in improper
lung~expansion.
As described in more detail herein, reagents that inhibit transitional
proteins prevent
massive accumulation of white cells that result in this syndrome and prevent
the
development of a surfactant deficit in the lung.
Thus, within one embodiment methods are provided for treating chronic
and acute distress syndromes, comprising administering to a patient a compound
selected from the group consisting of (a) a polypeptide comprising the amino
acid
sequence BX7B (SEQ ID N0:28) which binds HA; phage display selected peptides
that
bind HA such as polypeptides comprising -15 (Sequence ID No. 70), P-16
(Sequence
ID No. 26); P-32 (Sequence ID no. 71); and GAHWQFNALTVR (Sequence ID No.
72); (b) an antibody which binds one of domains Dl, D2, D3, D4, or DS of
RHAMM;
(c) a peptide of less than 95 kD or 73 kd, comprising all or a portion of
domains D1,
D2, D3, D4, or, DS of RHAMM; and (d) a gene delivery vector which expresses
antisense RHAMM, or, delivers and expresses any one of (a), (b), or (c), such
that the
disease is treated.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
69
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
15. Diabetes Mellitus
Diabetes mellitus is a group of diseases characterized by high glucose
resulting from defects in insulin secretion, insulin action, or both. Diabetes
mellitus can
be associated with serious complications such as heart disease, stroke, kidney
disease,
nervous system disease, blindness and complications in pregnancy.
Type I diabetes mellitus, also referred to as insulin dependent diabetes
mellitus (IDDM), develops most often in children and young adults over a short
period
of time. About 30-40% of diabetic children eventually develop nephropathy.
Type II
diabetes mellitus usually develops in adults. Risk factors include obesity and
family
history of diabetes. The symptoms usually develop gradually and are not as
noticeable
as in Type I diabetes.
Type I diabetes mellitus is an autoimmune disorder, the onset of which
results from a well characterized insulitis. During this condition the
inflammatory cells
are apparently specifically directed against the insulin producing beta cells
of the
pancreatic islets. The destruction of pancreatic beta cells by invading
leukocytes result
in deterioration of the insulin-dependent homeostasis.
The inflammatory cascade is a complex process that involves triggering
of the immunological response, release of chemokines, cytokines and a toxic
agents by
the activated cells, up-regulation of cell surface adhesion molecules and
transendothelial
cell migration. Although the triggering mechanism of IDDM remains elusive, it
is clear
that the entire process depends on the migration of inflammatory cells into
the
pancreatic islets and their interaction with matrix.
Within one embodiment methods are provided for treating or preventing
diabetes mellitus, comprising administering to a patient a compound selected
from the
group consisting of (a) a polypeptide comprising the amino acid sequence BX7B
(SECT
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
ID N0:28) which binds HA; phage display selected peptides that bind HA such as
polypeptides comprising P-15 (Sequence ID No. 70), P-16 (Sequence ID No. 26);
P-32
(Sequence ID no. 71); and GAHWQFNALTVR (Sequence ID No. 72); (b) an antibody
which binds one of domains D1, D2, D3, D4, or DS of RHAMM; (c) a peptide of
less
5 than 95 kD or 73 kd, comprising all or a portion of domains D1, D2, D3, D4,
or, DS of
RHAMM; and (d) a gene delivery vector which expresses antisense RHAMM, or,
delivers and expresses any one of (a), (b), or (c), such that the disease is
treated. Within
certain embodiments of the invention, any of the above-described compounds may
be
administered prior, during, or subsequent to islet cell transplantation.
Within other
10 related embodiments, the above-described compounds may be utilized to treat
related
diseases, including for example, obesity.
The polypeptides, antibodies, or, vectors may be delivered to the patient
by a variety of routes, including for example, systemically, intravenously,
intramuscularly, and orally.
PHARMACEUTICAL COMPOSITIONS
As noted above, the present invention also provides a variety of
pharmaceutical compositions, comprising one of the above-described molecules
with a
pharmaceutically or physiologically acceptable carrier, excipients or
diluents.
Generally, such carriers should be nontoxic to recipients at the dosages and
concentrations employed. Ordinarily, the preparation of such compositions
entails
combining the therapeutic agent with buffers, antioxidants such as ascorbic
acid, low
molecular weight (less than about 10 residues) polypeptides, proteins, amino
acids,
carbohydrates including glucose, sucrose or dextrins, chelating agents such as
EDTA,
glutathione and other stabilizers and excipients. Neutral buffered saline or
saline mixed
with nonspecific serum albumin are exemplary appropriate diluents.
In addition, the pharmaceutical compositions of the present invention
may be prepared for administration by a variety of different routes (e.g.,
systemically,
orally, rectally, intravenously, intramuscularly, ocularly, or, topically).
Further within
other embodiments the compounds or compositions provided herein may be admixed
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
71
with other carriers (e.g., polymers), and implanted on or contained within
devices which
are designed to release such compounds. Within further embodiments, the
compounds
may be delivered under radioscopic or other visual guidance to a desired site
(e.g.,
outside the lumen of a desired vessel, or outside of an organ, or, tissue to
be treated).
As should be readily evident, the compounds or compositions of the
present invention should be administered sufficient to have the desired
therapeutic
outcome. As an example, it is generally desirable to administer between a
total of 1 ng
of the desired compound, and up to 80 mg/kg. Within certain embodiments, the
dosage
will be adjusted for the therapeutic regimen desired (e.g., from 1 ug/kg to 1
mg/kg).
Within other embodiments the dosage for local administration may range from 1
to 100
ug/ml (2.Sng/kg to 80 mg/kg), and for systemic administration from 1 ng/kg to
10
mg/kg. Further, the dosage can be adjusted based upon the desired route of
treatment,
e.g., a smaller dose may be given if applied locally or topically, whereas a
larger dose
may be given if the compound is administered systemically. Further, the dosage
may
vary with the desired regimen (e.g., daily, weekly, or monthly).
In addition, pharmaceutical compositions of the present invention may be
placed within containers, along with packaging material which provides
instructions
regarding the use of such pharmaceutical compositions. Generally, such
instructions will
include a tangible expression describing the reagent concentration, as well as
within
certain embodiments, relative amounts of excipient ingredients or diluents
(e.g., water,
saline or PB S) which may be necessary to reconstitute the pharmaceutical
composition.
The following examples are offered by way of illustration, and not by
way of limitation.
EXAMPLES
EXAMPLE 1
REQUIREMENT FOR FOCAL ADHESIONS FOR MAXIMAL ACTIVATION OF
ERKKINASE IN RESPONSE TO GROWTH FACTORS
In disease or injury, mediators such as cytokines, growth factors and
genetic mutations activate a myriad of responses leading in increased
expression of AP-1
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
72
dependent genes (Figure 1 ). These genes are required for cell proliferation,
migration,
inflammation, tissue destruction and abnormal tissue remodeling. The
activation of the
AP-1 pathway occurs through the activation of the MAP kinase. The present
invention
discloses that in normal cells the activation of the AP-1 pathway by cytokines
and other
mediators is restricted and thus genes involved in disease cannot be induced
significantly.
Further this restriction is a result of the lack of ERK-1 activation in normal
cells (Figure
2). Normal cells must undergo a series of transitional stages to form a
diseased state cell
containing focal adhesions and is then responsive to inflammatory mediators.
Transition
stage cells provided by the present invention constitutively form podosomes
and are
unable to establish focal adhesions. Sustained formation of podosomes leads to
the
formation of focal adhesions and results in a diseased state (Figure 3). The
present
invention further discloses a requirement for focal adhesions for maximal
activation of erk
kinase in response to growth factors and cytokines. Cellular response-to-
injury processes
including growth factor mediated responses which lead to cellular
proliferation, migration, '
production of destructive enzymes and abnormal tissue remodeling are
characterized by a
maximal activation of the e~°k kinase signaling pathway. To demonstrate
that this
response requires the presence of focal adhesions, the response to IL-1
induction of erk
kinase signaling was measured in cells grown under conditions permitting or
preventing
the formation of focal adhesions.
Cells were either plated without serum on culture dishes precoated at
4°C
overnight with 25 ~,g/ml fibronectin which permits formation of focal
adhesions or with
100 ~,g/ml poly-1-Lysine which prevents formation of focal adhesions.
Formation of
focal contacts was detected by positive immunofluorescence of the marker
protein,
vinculin. Activation of erk kinase signaling in comparison to other MAP kinase
signaling pathways regulated by growth factors was estimated by detection of
proteins
phsophoryalted by components of the differing signaling cascades.
Phosphorylation of
myelin basic protein (MBP) is an indicator of erk kinase signaling,
phsophoryaltion of
GST-c jun is an indicator of jnk signaling, and phsophoryaltion of GST-ATF2 is
an
indicator of p3 ~ kinase signaling cascade. Results of this analysis is shown
in Figure 4.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
73
More specifically, Figure 4B shows that cells plated onto fibronectin
(FN) are able to form focal contacts as detected by positive
immunofluorescence for the
marker protein vinculin. Cells that are maintained on a non-physiological yet
adhesive
substratum poly-L-lysine (PL), attach but do not form focal contacts (4A).
Figure 4C
shows that normal quiescent phase cells plated onto fibronectin substrata
which make
focal contacts are able to activate the erk kinase cascade as indicated by the
phosphorylation of myelin basic protein (MBP) in response to the cytokine IL-1
(lane
2). These same cells plated onto poly-L-lysine do not make focal contacts and
are
unable to activate ef~k as detected by MBP phosphorylation (lanel). However,
cells
plated onto fibronectin ("FN") or in suspension are equally able to activate
the other
map kinases, jnk or p38 (lanes l and 2). Figure 4C also shows, that normal
cells in the
absence of focal adhesions, when plated onto fibronectin or grown in
suspension (SP),
are restricted in their ability to activate erk in response to IL-1 in
comparison to disease
cells containing focal adhesions, but able to activate the other MAP kinases,
jnk and p38
(lanes 3 and 4). These results indicate that responsiveness of the ef°k
kinase cascade is
restricted in transition stage cells but that the erk kinase cascade becomes
maximally
active when focal contacts are made as occurs upon entry of cells into a post-
transitional
stage that is fully responsive to growth factor stimulation, as indicated in
Figure 3.
Northern analysis was used to further demonstrate IL-1 induction of the
AP-1 transcriptional activator, c fos, by cells able to form focal contacts.
IL-1(3 was
added to cells grown either on FN or PL, then RNA was isolated and analyzed by
Northern blotting for levels of c fos mRNA. Figure 4D shows a Northern
analyses of
cells plated on fibronectin or PL and incubated with 20 ng/ml of IL-1 (3. 20
ng/ml IL-1 (3
was able to induce c fos expression in cells grown on FN (cells with focal
adhesions)
but not in cells grown on PL (in the absence of focal adhesions). Blots were
first
probed for c fos mRNA expression, stripped, and then reprobed with control
radiolabeled GAPDH cDNA to assess equality of RNA loading.
Figure 4E shows that the level of AP-1 activated in response to IL-1
induction requires the ability to make focal adhesions (cells grown on PL
which are
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
74
unable to form focal adhesions have reduced levels of AP-1 induction relative
to cells
grown on fibronectin).
More specifically, to further demonstrate a requirement of focal
adhesions for full IL-1 induction, the amount of the transcriptional factor AP-
1 binding
induced in response to IL-1 stimulation was analyzed. The level of DNA binding
to an
AP-1 oligonucleotide was measured in nuclear extracts from cells either grown
on
fibronectin or poly-1-Lysine coated dishes in medium without serum. Briefly,
tissue
culture dishes were precoated with 25 ~,g/ml fibronectin or 100 p,g/ml poly-1-
Lysine as
before and washed twice with PBS before use. Cells were then incubated under
starving condition for 6 h, media were removed and fresh serum-free medium
containing IL-1 (20 ng/ml) was added to the cells for 4 h.
For the preparation of nuclear extracts, cells were washed twice with
PBS (phosphate-buffered saline) and lysed with 1 ml buffer 1 (10 mM Tris-Cl,
pH 7.5,
10 mM NaCI, 3 mM MgClz, 0.5% Nonidet P-40, 0.5 mM pheny°lmethylsulfonyl
fluoride (PMSF). Cells were scraped into an eppendorf tube and put on ice for
10 min.
The nuclei were collected after centrifugation at 5000 rpm for 10 min. Nuclear
proteins
were prepared by resuspending the nuclei in buffer 2 (20 mM Hepes, pH7.9, 5 mM
MgClz, 0.2 mM EDTA, 1 mM DTT, 300 mM NaGI, 20% glycerol, 0.5 mM PMSF),
after centrifugation at 14,000 rpm for 10 min, supernatant was harvested.
Double-
stranded AP-1 oligonucleotide (Santa Cruz Biotech, Inc) was end-labeled with
[y-32P]
ATP (DuPont NEN) using T4 polynucleotide kinase (Pharmacia). Labeled probe was
separated from free nucleotide through a Sephadex G-50 mini-spin column
(Pharmacia).
DNA-protein binding was performed by mixing 10 p,g of nuclear extract with 3'-
P-
labeled double-stranded AP-1 consensus oligonucleotide in a total volume of 20
~,1
containing 20 mM Hepes, pH 7.9, 1 mM MgClz, 4% Ficoll, 0.5 mM DTT, 50 mM KCI,
1 mM EDTA, 2 ~,g poly(dI~dC) and 1 mg/ml BSA for 45 min on ice. The DNA
protein
complex was separated on a 4% native polyacrylamide gel using O.SX Tris-borate-
EDTA buffer at 150 V. Gels were then dried and autoradiographed.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
EXAMPLE 2
RHAMM OVEREXPRESSION IS ASSOCIATED WITH INCREASED ERK KINASE ACTIVATION
AND AP-1 ACTIVATION.
As noted above, expression of transitional molecules such as RHAMM
5 results in the initiation of cell transformation from a normal state to a
diseased state.
RHAMM is believed to play a role in the initial activation of ERK pathway,
thus
removing the ERK restriction found in normal cells. This activation leads to
the
expression of c-fos and c jun resulting in the AP-1 activation and induction
of AP-1
dependent genes involved in many of the disease processes associated with
10 inflammatory, degenerative and proliferative diseases (Figure 5).
Cells that overexpress a hyaladherin such as RHAMM in response to
stress or during proliferation exhibit elevated activation of erk kinase
signaling activity
as shown in Figure 6. E~k kinase activation is stimulated directly by
overexpression of
a hyaladherin such as RHAMM. Briefly, the cell line LR21 was constructed by
15 transfecting normal quiescent parental lOTl/2 cells with a vector
expressing a
RHAMMv4 polypeptide. Cells that overexpress RHAMM show increased erk
activation as indicated by phosphoryation of the MAP kinase activated myelin
basic
protein (MBP), p44 ERK1 and p42 ERK2, and by increased AP-1 binding activity.
Figure 6A illustrates that MAP kinase activity in quiescent lOTl/2 cells
20 is reduced relative to the levels present in RHAMM transfected LR21 cells.
Cells were
growth in DMEM with 10% FBS, cell monolayers were washed three times with PBS
and total cellular extracts were prepared in a buffer containing 25 mM Hepes,
pH 7.7,
100 mM NaCI, 2 mM MgCl2, 0.2 mM EDTA, 0.5% Triton X-100, 0.5 mM DTT, 20
mM (3-glycerophosphate, 0.1 mM sodium orthovanadate, 0.5 ~,ghnl leupeptin, 100
25 q,glml PMSF. Cellular lysates of 100 ~g total protein were incubated with
anti-ERK2
antibody conjugated agarose (ERK(C-14), Santa Cruz Biotech., Inc), immuno-
complexes were washed twice with the above lysis buffer and twice with kinase
buffer
(20 mM Hepes, pH 7.7, 10 mM MgCl2, 2 mM MnCIZ, 2 mM DTT and 25 ~,M ATP).
ERK2 activity was determined by in vitro kinase assay using 2 ~,g substrate
MBP and 1
30 ~Ci ['y-3zP] ATP in 20 ~,1 of kinase buffer. After incubation at
30°C for 20 min, the
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
76
reactions were terminated with Laemmli buffer, proteins were separated by SDS-
PAGE
and the gels were dried and autoradiographed.
The amount of ERK2 and phosphorylated MAPK was detected from the
total extracts by western blot using an ECL chemiluminescence system. In
brief,
lysates of 25 ~,g total protein were resolved by 10% SDS-PAGE and transferred
onto
nitrocellulose membrane (BioBlot, Costar) using Trans-Blot° Semi-Dry
Electrophoretic
Transfer Cell (BioRad) with a transfer-blotting buffer containing 20 mM Tris,
150 mM
glycine, 0.01% SDS and 20% methanol. The filters were blocked for non-fat skim
milk
in TBS-T (20 mM Tris, pH 7.5, 150 mM NaCI and 0.1% Tween 20) at 4°C
overnight.
The membranes were then probed with phospho-specific anti-p44/p42 MAP kinase
antibody (New England BioLabs, Inc.) by incubation at room temperature for 1.5
h.
After washing three times with TBS-T for 30 min, blots were incubated with
horseradish peroxidase conjugated anti-rabbit antibodies (NEB) for 1 h. The
filters
were washed three times for 30 min and visualized on X-ray film using . the
chemiluminescence detection method (NEB).
Figure 6B illustrates that AP-1 DNA binding activity is stimulated in
LR21 cells relative to parental lOTl/2 cell. Parental lOTl/2 cell and LR21
cells were
grown in DMEM with 10% FBS. Cells were then starved in the medium without
serum
for 8 h. Cells were washed twice with PBS (phosphate-buffered saline) and
lysed with
1 ml buffer 1 (10 mM Tris-Cl, pH7.5, 10 mM NaCI, 3 mM MgCl2, 0.5% Nonidet P-
40,
0.5 mM phenylmethylsulfonyl fluride [PMSF]). Cells were scraped into an
eppendorf
tube and put on ice for 10 min. The nuclei were collected after centrifugation
at 5000
rpm for 10 min. Nuclear proteins were prepared by resuspending the nuclei in
buffer 2
(20 mM Hepes, pH7.9, 5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 300 mM NaCI, 20%
glycerol, 0.5 mM PMSF), after centrifugation at 14,000 rpm for 10 min the
supernatant
was harvested as nuclear extract. Double-stranded AP-1 oligonucleotide (Santa
Cruz
Biotech, Inc.) was end-labeled with ['y-3zP] ATP (DuPont NEN) using T4
polynucleotide kinase (Pharmacia). Labeled probe was separated from free
nucleotide
through a Sephadex G-50 mini-spin column (Pharmacia). DNA-protein binding was
performed by mixing 10 ~g of nuclear extract with 3'-P-labeled double-stranded
AP-1
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
77
consensus oligonucleotide in a total volume of 20 ~1 containing 20 mM Hepes,
pH 7.9,
1 mM MgClz, 4% Ficoll, 0.5 mM DTT, 50 mM KCI, 1 mM EDTA, 2 ~g poly(dI~dC)
and 1 mg/ml BSA for 45 min on ice. The DNA protein complex was separated on a
4%
native polyacrylaminde gel using O.SX Tris-borate-EDTA buffer at 150 V. Gels
were
then dried and autoradiographed.
EXAMPLE 3
OVEREXPRESSION OF RHAMM ACTIVATES EXPRESSION OF C-FOS AND C-JUN, AND
MATRIX METLLOPROTEINASES ASSOCIATED WITH RESPONSE-TO-INJURY PROCESSES
Expression of the transcription factors c fos, c jun, jun B are associated
with response to injury processes in mammalian tissues. Northern analysis was
used to
show that of c fos, c jun, and jun B but not jun D expression are stimulated
by
overexpression of the transition stage hyaladherin, RHAMM.
Briefly, cells were'grown in DMEM with 10% FBS and W ere starved in
the absence of serum for 6 h. Cells were washed twice with PBS and total RNA
was
isolated by guanidine isothiocyanate method. In concise, cells were Iysed in 4
ml
solution D (5.3 M guanidine isothiocyanate, 30 mM sodium citrate, 0.7% N-
laurylsarcosine, 0.72% 2-mercaptoethanol). To each sample, added 4 ml of acid
phenol,
1 ml of chloroform and 0.45 ml of 2 M sodium acetate. The solution was mixed
well
and centrifuged at 7000 rpm fox 30 min, the aqueous phase was collected and
precipitated with an equal volume of 2-propanol. Pelleted RNA was dissolved in
50 ~l
diethyl pyrocarbonate (DEPC) treated water. The RNA was second extracted with
0.4
ml of TRIzoI reagent (GibcoBRL) with the addition of 0.1 ml chloroform. After
vigorous mixing and centrifugation, the RNA supernatant was precipitated with
2-
propanol and washed in 75% ethanol. Finally, the RNA was dissolved the DEPC-
treated water. The expression level of c fos, c jury, jun B and jun D were
probed with a
rat c fos and a human c jun probe, respectively. The blots were stripped and
re-probed
with rat GAPDH cDNA as internal standard. The results, as shown in Figure 7,
show
that expression of c fos, c juu, juh B but not juh D is stimulated in LR21
cells that
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
78
overexpress RHAMM. In addition, LR21 cells which overexpress RHAMM
constitutively form podosomes and form few focal adhesions (data not shown).
C fos and c jun expression are stimulated in LR21 cells in comparison to
parental lOTl/2 whether or not they are grown on fibronectin (Figure 8).
Briefly, cells
were grown in DMEM with 10% FBS and cell monolayers were trypsinized. Cells
were washed and plated on fibronectin and poly-1-Lysine coated dishes in the
medium
without serum. These tissue culture dishes were precoated with 25 ~.g/ml
fibronectin or
100 ~g/ml poly-1-Lysine at 4°C for overnight and washed twice with PBS
before use.
Cells were then incubated under this starving condition for 6 hr, total RNAs
were
extracted and hybridized as previously described. The levels of c fos and c
jun were
determined by hybridization with a rat c fos cDNA and a human c jun cDNA. The
blot
was stripped and re-probed with rat GAPDH cDNA as internal control. These
results,
as shown in Figure 8, further illustrate that a cell culture overexpressing a
transition
molecules such as RHAMM exhibits an activated signaling phenotype
characteristics of
transition stage cells.
Another characteristic of transition cells is an increase in the expression of
matrix metalloproteinases. This increase in metalloproteinases expression is
exhibited in
cells that overexpress RHAMM and these cells show reduced expression of matrix
metalloproteinase inhibitors. The association of increased RHAMM and
metalloproteinases activity is demonstrated by Northern analysis of RHAMM and
matrix
metalloproteinasemRNA levels in 102T1/2 and LR21 cell lines as illustrated in
Figure 9.
Briefly, cells were grown in DMEM with 10% FBS and were starved for 6
hours in the medium without serum. Cells were washed twice with PBS and total
RNA
was isolated by guanidine isothiocyanate method. In concise, cells were lysed
in 4 ml
solution D (5.3 M guanidine isothiocyanate, 30 mM sodium citrate, 0.7% N-
laurylsarcosine, 0.72% 2-mercaptoethanol). To each sample, added 4 ml of acid
phenol, l
ml of chloroform and 0.45 ml of 2 M sodium acetate. The solution was mixed
well and
centrifuged at 7000 rpm for 30 min, the aqueous phase was collected and
precipitated with
an equal volume of 2-propanol. Pelleted RNA was dissolved in 50 ~l diethyl
pyrocarbonate (DEPC) treated water. The RNA was second extracted with 0.4 ml
of
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
79
TRIzoI reagent (GibcoBRL) with the addition of 0.1 ml chloroform. After
vigorous
mixing and centrifugation, the RNA supernatant was precipitated with 2-
propanol and
washed in 75% ethanol. Finally, the RNA was dissolved the DEPC-treatedwater.
Denatured RNA samples of 20 ~g were separated in 1% agarose gel
containing 2.2 M formaldehyde, transferred to a Zeta probe membrane (BioRad),
cross
linked with an ultraviolet cross-linker (Strategene). The membrane was
prehybridized
in 0.35 M phosphate buffer containing 1% BSA, 7% SDS and 30% formamide for 5-6
h
at 55°C. The expression level of RHAMM, gelatinase B, and stromelysin
were detected
by hybridizing, the membrane with a 32P-labeled cDNA of a mouse full length
RHAMMv2. After washing the membrane in O.SXSSC and 0.5% SDS at 55°C
for 1.5
h, the membrane was autoradiographed. The blot was subsequent stripped and re-
probed with a mouse gelatinase B cDNA, a human stromelysin cDNA, or a rat
GAPDH
cDNA as internal standard.
In addition to showing increased expression of metalloproteinases, cells
that overexpress RHAMM also show decreased expression of inhibitors of
metalloproteinase such as timp-1. Northern analysis of tissue inhibitor of
matrix
metalloproteinase (timp-1 ) expression in LR21 cell shows a reduced level in
comparison to normal quiescent cells as illustrated in Figure 9. LR21 cells
which
overexpress RHAMMv4 show decreased expression of time-1, which normally blocks
activity of metalloproteinases.
EXAMPLE 4
OVEREXPRESSION OF RHAMM RESTRICTS THE EXTENT TO WHICH CYTOIfINES
AND GROWTH FACTORS ACTIVATE ERK SIGNALING PATHWAYS
As previously mentioned, the overexpression of RHAMM produces a
transition cell phenotype that only partially activates er~k signaling
pathways. This is
further illustrated in Figures 10 and 11 which show that signaling molecules
ordinarily
fully activated by growth factor induction are restricted, or partially
activated by
overexpression of RHAMM.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
Figure 10 shows a phosphoprotein activity analysis that directly illustrates
that cells that overexpress RHAMM have elevated erk activation of MAP kinases
but that
this activation is restricted relative to the level of activity observed in
normal cells induced
by a growth factor (PDGF). The Figure shows both phosphoylation of e~k
molecules and
5 er~k2 dependentphosphorylationof MBP molecules.
Briefly, cells were grown in DMEM with 10% FBS, and cells were
starved for 6 h in the medium without serum. Cells were then stimulated with
PDGF
(25 ng/ml) for 30 and 60 min. Cell monolayers were washed three times with PBS
and
total cellular extracts were prepared in a buffer containing 25 mM Hepes, pH
7.7, 100
10 mM NaCI, 2 mM MgCl2, 0.2 mM EDTA, 0.5% Triton X-100, 0.5 mM DTT, 20 mM
(3-glycerophosphate, 0.1 mM sodium orthovanadate, 0.5 ~,g/ml leupeptin, 100
~g/ml
PMSF. Cellular lysates of 100 ~g total protein were incubated with anti-ERK2
antibody conjugated agarose (ERK(C-14), Santa Cruz Biotech., Inc.), immuno-
complexes were washed twice with the above lysis buffer and twice by kinase
buffer
15 (20 mM Hepes, pH 7.7, 10 mM MgCh, 2 mM MnClz, 2 mM DTT and 25 ~M ATP).
ERK2 activity was determined by in vitro kinase assay using 2 ~.g substrate
MBP and 1
~.Ci [y-3zP] ATP in 20 ~,1 of kinase buffer. After incubation at 30°C
for 20 min, the
reactions were terminated with Laemmli buffer, and proteins were separated by
SDS-
PAGE, gels were dried and autoradiography.
20 The amount of ERI~2 and phosphorylated MAPK were detected from the
total extracts by western blot analysis and ECL chemiluminescence system.
Lysates of
25 ~,g total protein were resolved by 10% SDS-PAGE and transferred onto
nitrocellulose membrane (BioBlot, Costar) using Trans-Blot° Semi-Dry
Electrophoretic
Transfer Cell (BioRad) with a transfer-blotting buffer containing 20 mM Tris,
150 mM
25 glycine, 0.01% SDS and 20% methanol. The filters were blocked for non-fat
skim milk
in TBS-T (20 mM Tris, pH 7.5, 150 mM NaCI and 0.1% Tween 20) at 4°C
overnight.
The membranes were then probed with phospho-specific anti-p44/p42 MAP kinase
antibody (New England BioLabs, Inc) by incubation at room temperature for 1.5
h.
After washing three times with TBS-T for 30 min, blots were incubated with
30 horseradish peroxidase conjugated anti-rabbit antibodies (NEB) for 1 h. The
filters
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
81
were washed three times for 30 min and visualized on X-ray film with
chemiluminescence detection method (NEB).
As shown in Figure 10, the results of this analysis shows that LR21 cells
overexpressing RHAMMv4 are restricted in the extent to which proinflammatory
S cytokines/growth factors (e.g. PDGF) can activate er~k kinase.
Figure 11A shows a Northern analysis of IL-1 induction of c fos
expression in lOTl/2 and LR21 cell lines. Cells were grown in DMEM with 10%
FBS
and starved for 6 hours in the medium without serum. Cells were then
stimulated with
IL-1 (20 ng/ml) for 30 min and 60 min. Total RNAs were extracted and
hybridized as
described above. The level of c fos was measured by hybridization with a rat c
fos
cDNA. The blot was stripped and re-probed with rat GAPDH cDNA as internal
control. The results show that expression of c fos in response to IL-1 and TNF
is
restricted in LR21 cells.
Figure 11B shows a Northern analysis of TNF-a induction of c fos
1S expression in lOTl/2 and LR21 cell lines. Cells were grown in DMEM with 10%
FBS
and starved for 6 h in the medium without serum. Cells were then stimulated
with
TNF-a (30 ng/ml) for 30 min and 60 min. Total RNAs were extracted and
hybridized
as described above. The level of c fos was measured by hybridization with a
rat c fos
cDNA. Again it can be seen that the expression of c fos in response to a
injury response
growth factor i.e., TNF-a, is restricted in LR21 cells.
EXAMPLE S
RHAMM OVEREXPRESSION PREVENTS FOCAL ADHESION
FORMATION AND INDUCES CONSTITUTIVE
2S PODOSOME PRODUCTION
A key feature of cells over-expressing RHAMM, LR21, prepared as
described above is that they do not form focal adhesions. Figure 12 A shows
that the parent
cell line, lOTl/2, form very discreet focal adhesions, as demonstrated with
anti-vinculin
staining. In contrast LR21 cells do not form focal adhesions (Figure 12B).
This inhibition
of focal adhesion formation may be responsible for the lack of response of
these cells to
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
82
cytokines such as IL-1 and TNF. It would appear that as long as cells are
expressing
RHAMM they do not form focal adhesions and remain unresponsive to cytokines.
In addition, lOTl/2 cells, the parent cell line when plated form small
numbers of podosomes immediately following plating as shown in Figure 13. By
12 to
24 hours, there is little formation of podosomes and there is now the
formation of focal
adhesions in these cells. In contrast to lOTl/2 cells, LR21 cells that over-
express
RHAMM form podosomes constitutively. The level of podosome formation is higher
and continuous in cells over-expressing RHAMM. These data indicate that RHAMM
overexpression is required for podosome formation in cells immediately
following
injury or sustained disease conditions.
EXAMPLE 6
BLOCKAGE OF ERK ACTIVITY INHIBITS THE FORMATION OF P~DOSOMES
AND MIGRATION OF CELLS TOWARD WOUNDS PROMOTED BY OVERE3~PRESSION
RHAMM
To further illustrate the relationship between RHAMM, erk activity and
podosome formation in transient stage dells, podosome formation is shown to be
inhibited by inhibitors of erk activity. Figure 14 shows that enhanced
podosome
formation resulting from RHAMMv4 overexpression, is blocked by inhibitors of
erk
kinase which also blocks cell migration into wound sites. Overexpression of
RHAMMv4 results in a sustained high production of podosomes, detected by the
marker protein cortactin (A). Inhibition of erk kinase by PD09058 reduces the
number
of podosomes (B) as does mutation of intracellular RHAMMv4 (C) so that er~k
does not
bind to RHAMM. Overexpression of RHAMMv4 enhances cell migration into wounds
(D) compared to the parent lOTl/2 fibroblast line that produces little RHAMMv4
(E).
The addition of PD09058 blocks wound repair of RHAMMv4 overexpressing cells
(F).
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
83
EXAMPLE 7
RHAMM IS TRANSIENTLY DETECTED ON THE SURFACE OF CELLS
AND IS REQUIRED FOR PODOSOME FORMATION AND CELL MOTILITY:
METHOD OF DETECTING TRANSIENT CELLS
Exon 3 and 4 of RHAMM provide peptides and antibodies thereto which
are useful for detecting RHAMM expression and demonstrating that RHAMM is
associated with podosomes. Figure 15A shows a comparison of the expression of
RHAMM at the cell surface using anti-exon 4 RHAMM antibody or antibody "R3.8"
raised against a whole RHAMM polypeptide. The chart summarizes the results of
FACS analysis of ire vitro growth of invasive MDA231 cells in comparison to
MCF-7
human breast cancer cells which are non invasive cells. The results show that
cell
surface RHAMM is present in larger amounts on MDA-231 cells than on MCF-7
cells.
Figure 15B shows the amino acid sequences of rnurine and human
MAMM peptides including: a peptide from marine exon 3 (SEQ. ID NO: 14); a
smaller
peptide used to raise anti-exon 3 antibodies (SEQ. ID NO: 15); a peptide from
marine
exon 4 (SEQ. ID NO: 16); a smaller peptide used to raise anti-exon 4
antibodies (SEQ. ID
NO: 17), a human RHAMM peptide from exon 5 (SEQ. ID NO: 18); a hiunan RHAMM
peptide homologous to marine exon 3 (SEQ. TD NO: 19); and a marine RHAMM
peptide
homologous to human exon 5 (SEQ. ID NO: 20). The human exon 4 homologue is
identical to the marine sequence used to raise anti-exon 4 antibodies. The C
residues
shown in parenthesis were added during synthesis of the peptides.
Figure 16 shows that cells treated by administering peptides mimicking
exon 3, i.e., SEQ. ID NO: 15 (peptide 1, panel A) block the motility of
invasive cells
relative to a scrambled peptide (peptide 2, panel B). This effect is
quantified in the
graph shown in 16C and is highly significant (P<0.001, student's T test).
Figure 16D
shows that administration of antibodies against peptides mimicking exon 4
(i.e., antibodies to SEQ. ID NO: 16) also inhibit the motility of invasive
cells.
Figure 17, panel A are micrographs that show that podosome formation
is enhanced on the perimeter of LR21 cells that overexpress RHAMM in
comparison to
control lOTll2 cells. Panel B shows that administration of exon 4, i. e.,
peptide SEQ. ID
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
84
NO: 16 (peptide 1) blocks the formation of podosomes while scrambled exon 4
peptide
(peptide 2) does not. Podsomome formation was visualized using either
fluorescent
cortactin or CAS, the latter being a particularly useful marker fox podosomes
as
illustrated by the micrographs in Figure 17B.
In Figure 18, MDA-231 cells were treated with hyaluronan together with
anti-RHAMM antibody (exon 4 antibody). The antibody blocked the formation of
podosomes as detected by cortactin staining.
Figure 19 is a chart that quantitatively shows that anti-RHAMM
antibodies block the rapid motility characteristic of MDA231 human breast
cancer cells
but have only a small effect on the less rapid motility of cells of the benign
MCF
human breast cancer cell line.
Figure 20 further shows that anti-RHAMM antibodies inhibit the ability
of MDA231 cells to invade i~c vitf°o. The chart in 20A illustrates the
invasiveness of a
variety of cell lines while 20B shows the ability of a variety of RHAMM
antibodies to
reduce invasiveness of MDA231 cells.
Figure 21 shows that RHAMM binds to fibronectin but is blocked by
antibody to exon 3 indicating that exon 3 contains a fibronectin binding
domain. Figure
21 further illustrates that RHAMM binds to the CS-1 fragment of fibornectin
and not to
the RGDS sequence which was previously considered to be a critical sequence
for
fibronectin signaling of matrix protein degradation. Panel A shows that RHAMM
binds
to fibronectin as detected by an ELISA. Panel B shows that exon 3 of RHAMM
binds
to fibronectin but not through the RGDS region but rather through the CS-1
region.
Panel C shows that peptides mimicking exon 3 are able to block the binding of
intact
RHAMM to fibronectin, providing a rationale for why peptides block cell
locomotion
and podosome formation.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
EXAMPLE 8
ERK KINASE INVOLVED IN CELLULAR MOTILITY
Elevated erk activity is associated with, and required for, rapid cell
motility characteristic of proliferative or invasive cells such as the breast
cancer cell line
5 MDA231 which express high levels of RHAMM.
The relationship between erk activation and cell motility is illustrated in
Figure 22 which shows that when cells overexpress RHAMM such as in the case of
MDA231 cells, the level of cell motility is high. When er~k kinase activity is
inhibited by
treatment with a MEK inhibitor (PD09058), that inhibition strongly reduces
cellular
10 locomotion and blocks cell invasion. Figure 22 further shows that MDA231
cells
expressing a mutant version of RHAMM (HA mutant) have reduced mobility. The
relationship between RHAMM expression and cell motility is further established
by
treating MDA231 cells with anti RHAMM antibodies, resulting in reduced cell
mobility.
15 EXAMPLE 9
OVEREXPRESSION OF RHAMM PROMOTES PODOSOME FORMATION
Podosomes are transient structures at lamellae tips that are required for
the efficient release of the MMPs. Together with other proteinases, MMPs
initiate
extracellular matrix remodeling. This initial remodeling of matrix attracts
white cells to
20 the site of injury, providing additional source of pro-inflammatory
cytokines and
growth factors that are responsible for the amplification of the response-to-
injury. This
experiment shows that transient RHAMM overexpression will alter podosome
formation in transfected lOTI/2 cells.
Briefly, RHAMMv4 cDNA was tagged with HA and transfected into
25 lOTl/2 cells. IOTl/2 cells were cultured to 40-50% confluence and
transfected with 10
~,g of RHAMMv4 CDNA tagged with HA in 60 ~l of superFect reagent. After five
hours of incubation, monolayers were washed twice with PBS and the transfected
cells
were cultured an additional 48 hours with growth medium supplemented withl 0%
FBS. The cells were harvested in RIPA buffer and RHAMM expression was detected
30 by Western analysis. Only the transfectants that expressed similar level of
(2-3 fold
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
86
higher than parenthal cells) were used for assays. HAv4tag cells were selected
and used
in this experiment. For the experimental purposes transfected cells were
plated on the
fibronectin (FN) substrate at the SO% density. To visualize the presence of
Hav4tag
cDNA cells were stained at different time points: 1/2h, 2h, 6h, and 24 h
respectively
S with monoclonal antibody against HA. Additionally, cells were stained with
the
monoclonal Ab against HA (dil.l:SO) 1h at room temperature. Cells were then
washed
with 1%BSA in PBS and incubated with x4 antibody. X4 antibody was detected by
Texas-red (1:100). Cells were incubated 1h at room temperature in Texas red.
Some
cells were plated onto RITC-labeled fibronectin in order to detect the ability
of cells to
digest this extracellular matrix protein providing an assessment of the
functional
capability of the podosomes. A clearing of fluorescence indicates that cells
have
released collagenases that are able to digest fibronectin.
In both experimental paradigms, results were examined under the
confocal microscope. .
1S As shown on Figure 23A, at the 2h point 100% of plated cells formed
podosomes. Additionally, v4 tagged RHAMM cDNA was found in perinuclear region
as well as in the podosomes. Evidence that podosomes made by RHAMM transfected
cells are releasing proteases is provided in Figure 23B which shows the
clearing of
FITC-fibronectin underneath the plated cells. The dark area indicates that
fibronectin
has been proteolyzed and released from the cell substratum. Based upon this
experiment it is evident that overexpression of RHAMM promotes podosome
formation
in lOTl/2 cells.
EXAMPLE 10
2S ANTIBODIES AGAINST TAM DOMAINS AND LEUCINE ZIPPER INHIBIT PODOSOME
FORMATION
The objective of this experiment was to investigate whether RHAMM
induces podosome formation in a system where RHAMM surface sites were blocked
with a exon4 (TAM) A antibody. Leucine zipper peptide (LZP) was also tested
for its
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
87
capability to compete for RHAMM surface sites with fibronectin, since this is
the
binding site for fibronectin.
Briefly, LR21 cells were plated in DMEM with 10% serum at 70-80%
density and allow to grow for 8h. Cells were then washed twice with PBS. After
being
washed cells were incubated in cell dissociation medium to detach from the
plates. Cell
dissociation medium was harvested and centrifuged at 1000 rpm's for 31/2 min.
Then,
cells were plated at fibronectin-coated coverslips at 50% density in DMEM
supplemented with 10% FBS. Cells were allow to grow for up to 9h. Plates were
then
divided into 4 groups and treated in the following manner: control group was
treated
with 50 ~g/ml of BSA in DMEM supplemented with 10% FBS; second group was
treated with 50 ~,g/ml of v4 antibody; third group was treated with 100 mg/ml
of LZP
and the fourth group was treated with combination of v4 antibody and LZP at
the same
concentrations as they were used in separate treatments. Cells were kept with
the
'
proteins for 30 min and they were fixed with 3% paraformaldehyde. Cells were
stained
with cortactin (di1.1:100) for 1h. Subsequently, cells were washed with 1% BSA
in
PBS and stained with Texas-red mouse IgG (dil. 1:100). Staining of the cells
was:
examined by confocal microscope.
LR21 cells were plated onto fibronectin substrata as outlined above for
8-12 hrs in serum free medium in the presence of IgG alone or anti-TAM
antibody
("exon 4"). The supernatant culture medium was collected at that time and
concentrated on an amicon filter that retains proteins of over 20 kDa. The
retentate was
suspended in loading buffer without mercaptoethanol or SDS-PAGE and run on a
polyacrylamide gel impregnanted with gelatin. The gel was then incubated in
PBS
containing Mg++ and Ca++ buffer to permit collagenase activity at 37C for
several
hours. The gel was washed and stained with Coomassie Blue. Cleared areas
indicate
that collagenases released into the supernatant medium by LR21 cells is
active.
As shown on Figure 24A, v4 antibody added to the medium competed
for the RHAMM binding sites with fibronectin which resulted in reduced
podosome
formation by those cells up to 25% compared to the BSA-treated control. LZP
and
combination of LZP and v4 antibody didn't result in any changes in podosome
number.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
$g
The reason for this result could be the fairly high concentration of LZ
peptide used in
this experiment.
Based upon these results it is evident that RHAMM on the cell surface is
required for the efficient podosome formation. Addition of v4 antibodies
competed
S with fibronectin for the RHAMM binding sites which resulted in lower
podosome
formation by LR21 cells. The antibody also blocked release of collagenase
(Figure
24B), consistent with its blocking podosome formation.
EXAMPLE 11
1 O RHAMM V4 AND FULL LENGTH RHAMMVS INTERACT WITH ERK 1 KINASE
The most common murine RHAMM RNA transcript encodes a 9S kDa
protein (referred to as "vS"). In addition, a shorter form of RHAMM may exist
encoding a 73 kDa protein (v4), which lacks 163 N-terminal amino acids found
in the
longer RHAMM form. The objective of this experiment was to determine which
form
1 S of RHAMM associates with erk and which particular domain of RHAMM is
responsible for this interaction.
A. I~ vitro binding competition assays.
Purified GST-RHAMM proteins were released from GST with trombin
and RHAMM was coupled to Amino Link plus coupling gel (Pierce). After several
20 washes with PBS, RHAMM-coupled beads were incubated with purified erkl His-
6
tagged fusion proteins in binding buffer for 1h at 4°C on Nutator
rotor. After several
washes with cold binding buffer, the beads were boiled for 2 min in cold
loading
buffer, then proteins were separated on SDS-PAGE and transfer to
nitrocellulose blots
for western analysis. Anti-erkl antibody (K23) was used to detect this kinase
on
2S western blots. For competition assays, 1 ~,g of purified erkl His-6 fusion
protein was
incubated with 10 ~,g of soluble RHAMM protein for 1h at room temperature,
then
incubated with beads-RHAMM for an additional 1h. For peptide competition
assays, 1
~g of erkl His-6 fusion protein was incubated with beads-RHAMM for another 1h
on a
Rotator. Three different peptides were used in competition binding assay, D4:
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
89
QEKYNDTAQSLRDVTAQLESV (SEQ ID NO:50), D5:
KQKIKHVVKLKDENSQLKSEVSKLRSQLVKRK (SEQ ID NO:51), and P-16
peptide: CSTMMSRSHKTRSHHV (SEQ ID N0:26).
B. Immunoprecipitation.
Parental IOTl/2 cells and transfected cell lines were plated at 50%
confluence for 6-24 h and washed two times with cold PBS and lysed in a lysis
buffer,
containing leupeptin (lmg/ml), aprotinin (0.2 TIU/ml) and dichloroisocoumarin
(200
yM). The lysates were centrifuged and equal amounts of protein (300-400 fig)
from
each sample were added to 2 ~,g of anti-RHAMM antibody (R3.2), and anti-erk-1
(K23)
antibody. After 1h of incubation at 4°C on a Nutator rotor, 50 ~g of a
50% solution of
protein G Sepharose was added and incubated at 4°C for an additional
1h, then washed
four times with lysis buffer.
C. Western analysis.
Cells were plated at 50% confluence and grown for 6-24 h. Then,
monolayers were washed with cold PBS, lysed in RIPA buffer and subjected to
SDS-
PAGE. Separated proteins were transferred onto nitrocellulose membranes
(BioRad)
using a Transfer buffer. Non-specific binding sites were blocked with 5%
defatted milk
in Tris buffer. RHAMMv4 antibody was prepared against following sequence:
VSIEKEKIDEK (SEQ ID NO:50). RHAMMvS antibody was prepared against
following sequence: QERGTQDKRIQDME (SEQ ID NO:21). Membranes were
washed three times with TBST, then incubated with horseradish peroxidase-
conjugated
goat anti-rabbit IgG (1;10000) for 30 min at room temperature. Bound antibody
was
visualized by chemiluminescence (ECL). The densitometry was performed with a
Multi-Analyst program (Bio-Rad). To determine antibody specificity, anti-RHAMM
antibodies were incubated with beads-linked with RHAMM protein (leg
antibody/20
~l beads) for 1h at 4°C on a Rotator, and then centrifuged for 5 min.
The supernatant
was used to probe membranes.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
Results are shown in Figure 25. Briefly, Figure 25B shows erkl binding
to v4 and v5 obtained in vitro, whereas Figure 25C shows similar results in
vivo. The
Bottom Western blot represents total cellular erk kinase and densitometery was
calculated as the ratio of total cellular erk versus erk associated with
RHAMM. Both
5 RHAMMvS and v4 associated with erkl kinase. RHAMMv4 more strongly activates
erk kinase than RHAMMvS (Figure 25D), which contains all of the domains of v4
but
in addition N-terminal sequence that negatively regulates the functions of the
activating
D2-5 domains (Figure 25B). The presence of mutations in both DS or D4 domains
or in
competition assays the presence of both D4 and DS peptides, reduced erkl
binding fo
10 v4 by 90% (Figure 25C), suggesting key roles of those domains in binding.
Thus, in summary both forms of RHAMM (v4 and v5) associate with erkl
in vivo and in vitro but only the short form strongly activates the erk kinase
cascade. The
hyaluronan binding domains (See Figures 25A, DS) and a repeated sequence (D4)
are
required for binding of erkl to RHAMM. However, both D3 (encoding the TAM
15 domain) and D2 (encoding the leucine zipper) are required for activation of
erk kinase
although they are not involved in the binding of erkl to RHAMM.
EXAMPLE 12
HA BINDING DOMAINS OF RHAMM PEPTIDES AND ANTIBODIES TI-IERETO
2O FOR AFFECTING A RESPONSE-TO-INJURY PROCESS
Figure 26 illustrates that HA binding peptides including artificial mimics
of hyaluronan binding domains of RHAMM are able to block cell motility in
podosome
forming cells while scrambled peptides do not. Figure 26A provides the
sequence of
several artificial HA binding peptides of the formula BX7B (SEQ ID N0:28)
discussed
25 above. Panel B shows that each of these peptides are able to block cell
motility when
administered to cells. Panel C shows that an HA binding peptide according to
one of the
general structures provided in SEQ ID NO: 1-5, and more particularly, having
one of the
structures provided in SEQ. ID NOS. 6-10 is even more effective in blocking
cell motility
and that a scrambled version of this peptide is not.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
91
EXAMPLE 13
HA-BINDING PEPTIDE MIMETIC (P-16) AND RHAMM SEQUENCE PEPTIDE (423-432 AA)
INHIBIT MIGRATION OF HUMAN FIBROBLAST
Wound healing is the response to injury. By day three after the
wounding, fibroblasts appear in the fibronectin - fibrin framework and
initiate collagen
synthesis. Fibroblast proliferate in response to growth factors present on the
wound site
and this complex series of cellular and inflammatory processes resulting in
deposition
of connective tissues and its remodeling into the scar tissue. The
fibroproliferative
response is accompanied with wound contraction and fibrosis due to the
presence of
myofibroblasts and to the enhanced production of collagen. In adult humans,
the
extracellular matrix is remodeled to sustain and direct the cellular changes
and to restore
the tissue integrity. Such exuberant healing responses often lead to tissue
fibrosis and
contraction commonly referred to us as scarring. Fibrosis of adult human
tissue is a
serious clinical problem that results in malfunction of tissue due to, for
example:
formation of intraabdominal adhesions, cirrhosis of liver, failure of
anastomoses as well
as adhesions following injury.
In animal models of skin wounding, expression of an active (73 kDa)
RHAMM form is transiently increased early after injury and this elevated
expression
occurs in most cell types present in the wound site. A specific domain within
RHAMM (DS) that is responsible for interactions of hyaluronan with cell
surface
RHAMM and erkl binding to intracellular RHAMM was identified and utilized to
develop a peptide mimetic reagent (p-16), which blocks function of cell
surface
RHAMM. Another RHAMM sequence consisted of 9 AA (423-432) which was also
tested in the following experiment.
The objectives of following experiment were to test the abilities of two
RHAMM synthetic peptides to inhibit migration of human fibroblasts. One
experimental model tested a 16 amino acid RHAMM peptide mimetic (P-peptide) to
inhibit migration of Human Foreskin Fibroblast (HFF) through the wound gap.
Another peptide consisted of 9 AAs (RHAMM sequence, 423-432 AA) was also
tested
in regards of cell locomotion of human fibroblasts.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
92
Experiment A.
Human fibroblasts were seeded at 5 X 105 cells/well in 6 well plates
using a-MEM supplemented with glucose and 10% FBS. After being 6 hours in the
culture (80-90% confluency), cells were injured with the single edge cell
scraper (one
injury/dish). Cells were washed twice with PBS and treated with two different
concentrations of P-peptide (10 p,g and 100 ~.g) for 15 h. Untreated cells
served as
control. Following 15 hours of incubation, images were taken using a SX
modulation
objective (leis, Germany) attached to the Zeiss Axiovert 100 inverted
microscope
equipped with Hoffman Modulation contrast optical filters (Greenvale, NY). The
number of migrated cells in each image was counted choosing the ~70% of the
middle
of each injury. Statistically significant (P<0.05) differences between means
were
assessed by the unpaired Student's t-test method, using Microsoft Excel '97
software.
Experiment B.
To quantify the effect of RHAMM sequence (423-432 AA) to alter
velocity of cell locomotion human fibroblasts were seeded on T-12.5
fibronectin coated
flasks using a-MEM supplemented with glucose and 10% FBS. 2.5 x 10~ cells were
seeded and cells incubated for 4 hrs at 37°C. After incubation time
cells were treated
with increasing concentrations of RHAMM sequence peptide (423-432 AA, 0.1,
1.0,
5.0, 10 and 50 ng/ml) and cell locomotion was monitored over the period of 16
hrs on a
37 °C using lOX modulation objective (Zeiss, Germany) attached to a
Zeiss Axiovert
100 inverted microscope equipped with Hoffman Modulation contrast optical
filters
(Greenvale, NY). Cell images were captured with a CCD video camera module
attached to a Hamamatsu CCD camera controller. Motility was assessed using
Northern Exposure 2.9 image analysis software (Empix Imaging, Mississauga,
Ontario).
Nuclear displacement of 7 -10 cells was measured and data were subjected to
statistical
analysis. Statistically significant (P<0.05) differences between means were
assessed by
the unpaired Student's T-test method, performed using Microsoft Excel "97
software.
Results are shown in Figures 27 and 28. Briefly, Figure 27 shows that
treatment of injured cells with 100 pg/ml of P-peptide inhibited migration of
HFF cells
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
93
approximately 4 fold compared to control cells (P<0.01). Lower concentration
(10
~g/ml) of P-peptide didn't have any effect. As shown in Figure 28, different
concentrations of RHAMM sequence (423-432 AA) progressively inhibited
migration
of human fibroblasts up to 40% .
Both treatments were successful in inhibition of cell migration ih
vitf°o.
These important data suggests potential implementation of the both P-peptide
and
RHAMM sequence 423-432 AA peptide in prevention of tissue contraction and
fibrosis
and ultimately prevention of abnormal tissue remodeling and scaring.
EXAMPLE 14
FIBROBLASTS FROM RHAMM KNOCKOUT MOUSE PRODUCE TWO TIMES
LESS MMP'S THAN WILD TYPE
MMP expression is involved in a wide variety of inflammatory diseases
and cancers. This experiment investigates whether fibroblasts which are
obtained from
RHAMM knockout mice have altered MMP production.
Briefly, embryonic stem cells (ES) were transfected with antisense
cDNA that recombined with the RHAMM gene, resulting in recombination and a
genetic deletion of the RHAMM gene. The ES cells were injected into mouse
blastocysts, and placed into pseudo-pregnant mice. Mice from the resultant
litters were
crossed and examined for the presence of a genetic deletion, in order to
determine germ
line transmission. Founders were identified and homozygotes obtained.
Embryos from normal and knockout mice were taken out at the 13"' day
of their intrauterine development. Tissue was cut and tripsinized in the
incubator at 37
°C for 10 min. Cell suspension was pipeted up and down several times in
order to
release fibroblasts from the tissue. Then, fibroblasts were plated on the
Petri dishes
(one embryo per one Petri dish). Cells were grown for 2 days before first
passage was
done. Five passages were done before actual experiment was performed.
For experimental purposes, fibroblasts were plated on 6-well dishes, normal
ones and fibronectin coated, both at the 70% confluency. Cells were grown in
DMEM
3 0 medium for 2 h. After 2 h, medium was changed to DMEM without serum and
cells kept
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
94
in starvation medium for 24 h. Then, medium was taken out and the amount of
gelatinase
released into medium measured by zymografic analysis. Briefly, DMEM medium
taken out
from plates was run on overnight in a cold room (+4 °C) on 10%
acrylamide gel containing
lmg/ml of gelatin. Then, gel was washed in TritonX-100 for 1h and subsequently
incubated
S in a bufFer on 37°C for 24 h in order to develop zymogram. Then, gel
was stained with
commassi blue whereas areas with MMPs were left unstained. Intensities of
unstained
bands were measured and presented as relative numbers.
Results are shown in Figure 29. Briefly, MMP release from knockout
fibroblasts is approximately 2.S times lower compared to normal ones.
Thus, it is evident that RHAMM expression regulates production of
MMPs. Absence of RHAMM in knockout fibroblasts resulted in maxked decrease in
MMP production.
EXAMPLE 1 S
1 S RHAMM INFLUENCES ERK PHOSPHORYLATION UPON PDGF TREATMENT IN MOUSE
PRIMARY FIBROBLASTS
The purpose of this experiment was to investigate if erk phosphorylation
is decreased in primary fibroblasts of the RHAMM knockout mouse.
Briefly, mouse normal and RHAMM knockout fibroblasts are plated in
DMEM medium and starved overnight. Medium was changed and two different
concentrations of PDGF added. After 10 min cells were lysed in RIPA buffer.
Western
blot analysis was done and proteins separated by SDS-PAGE. Bands were
visualized
by phospho-specific erk antibody. Subsequently, blot was stripped and reprobed
with
erk antibody.
2S Results are shown in Figure 30. Briefly, erk ,phosphorylation is
influenced by RHAMM expression: knockout mice exhibited at least two folds
lower
phosphorylation of erkl isoform compared to wild type. PDGF concentration of
lng/ml
produced the largest decrease (2.3 fold) in erk phosphorylation (Figure 30).
Thus,
deficiency in RHAMM expression in knockout fibroblasts down regulates the
capability
of PDGF to activate erk pathway.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
EXAMPLE 16
FIBROBLASTS ISOLATED FROM RHAMM KNOCKOUT MOUSE LOCOMOTE AT
SIGNIFICANTLY LOWER RATE THAN WILD TYPE FIBROBLASTS
5 This experiment investigates the impact of lacking RHAMM expression
on cell migration in mouse knockout fibroblasts.
Briefly, knockout and wild type mouse fibroblasts were plated in 100
mm Petry dishes (normal or gelatin and fibronectin coated) and grown in normal
DMEM medium. Cells were plated sparsely and left for 2h to attach, spread and
start
10 to migrate. Two hours after plating, cells were checked for migration and
pictures were
taken from the same spot every 15 min. The images were overlaid and cell
migration
analyzed by measurement of the migration distance.
Results are shown in Figure 31. Briefly, knockout fibroblasts have
decreased cell motility compared to wild type by two folds. Combination of
gelatin
15 and fibronectin coating seem to potentiate slower migration of mouse RHAMM
knockout fibroblasts. Thus, it is evident that mouse RHAMM knockout
fibroblasts
migrate at the slower rate compared to wild type cells. Attenuation of cell
migration is
between 2 to 4 fold.
20 EXAMPLE 17
ADMINISTRATION OF RHAMM PEPTIDES OR ANTIBODIES INHIBIT RESPONSE-TO-INJURY
PROCESSES ASSOCIATED WITH MACROPHAGES IN INJURED LUNG TISSUE
Clinical diseases characterized by lung inflammation include
emphysema, asthma, cystic fibrosis, new-born lung disease involving chronic
25 respiratory distress syndrome, and the acute respiratory distress syndrome
that affects
accident victims. Local inflammatory responses that recruit macrophages into
the lung
result in destruction of alveolar type II cells, which make the surfactant
responsible for
normal lung inflation. The infiltration of macrophages and abnormal local
tissue
responses result in further tissue destruction and disease. This pathological
sequence
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
96
results in improper lung expansion. A common response to injury in mammalian
tissue
is increased motility of macrophages and macrophage accumulation near a wound
site.
As described in more detail below, the RHAMM peptide mimetics
prevent these pathological events, particularly recruitment and activation of
macrophages, from occurring following bleomycin-induced lung fibrosis (Figure
32).
In particular, the infiltration of inflammatory cells and local responses such
as fibrosis is
completely absent with treatment. Yet, no toxicity was observed in the animals
even at
very high concentrations. Therefore, these reagents should be effective for
treating lung
diseases that involve recruiting macrophages and inflammatory cells, as well
as fibrosis.
This experiment shows that macrophage responses are inhibited by
administration of antisera to RHAMM peptides. More specifically, Figure 33
illustrates
that a significant increase occurs in the motility of macrophages from both
bleomycin
and saline-treated animals at four days after intratracheal instillation
(*p<0.01 versus
control; p < 0.01 versus saline and control). Normal rabbit IgG had no effect
on
macrophage motility, but anti-RHAMM peptide as antiserum inhibited macrophage
motility from both saline - (# p < 0.01 versus saline) and bleomycin-treated
(p < 0.01
versus bleomycin, saline and bleomycin + normal IgG) animals to levels
observed in
macrophages from untreated healthy control animals. Values represent mean and
standard errors of five animals studied for each condition with mean
velocities
calculated.
Figure 34 illustrates motility of BAL cells four days after injury in
response to administration of RHAMM peptides. Macrophages from bleomycin-
treated
animals showed increased motilities as compared to those from control and
saline
animals (*p < 0.001). Animals pretreated with Scrambled Peptide A showed the
same
motility as macrophages obtained from animals injured with bleomycin. However,
macrophages from animals treated with Peptide A prior to bleomycin-induced
injury
showed significantly lower cell locomotion than either bleomycin-injured or
scrambled
peptide treated controls (# p < 0.001). Values represent mean and standard
error with
three animals studied for each condition and at least 20 cells tracked per
animal studied.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
97
Based on these findings, peptide mimetic and antibody formulations can
be utilized in the treatment of a variety of "response to injury" indications,
including for
example, emphysema, asthma and the chronic respiratory distress syndrome
associated
with newborn lung disease.
EXAMPLE 18
ADMINISTRATION OF HA BINDING PEPTIDES INHIBIT RESPONSE-TO-INJURY PROCESSES
ASSOCIATED WITH FIBROSIS IN INJURED LUNG TISSUE
Increased N-acetyl-(3 -glucosaminidase activity is a known marker for
fibrosis in lung tissue. Figure 35A shows in vivo effects of a HA-binding
(Peptide A)
on N-acetyl-(3 -glucosaminidase activity of BAL cells obtained 7 days after
injury.
Briefly, bleomycin injury results in an increased glucosaminidase
activity (* p < 0.01 versus controls and saline animals). Scrambled Peptide A
had no
effect on the glucosaminidase activity whereas Peptide A significantly
decreased
glucosaminidase activity (# p<0.05 versus bleomycin alone and bleomycin +
scrambled
peptide A). Values represent mean and standard error with five animals studied
for
each condition.
Figure 35B illustrates that mRNA of collagen type la in lungs harvested
4 days after injury is reduced in response to administration of HA binding
Peptide A.
Collagen type la is common indicator of fibrosis in lung tissue injury models
as used
throughout this invention. An increase in collagen type 1 a mRNA was observed
by 4
days after injury in control tissue, however, this increase was completely
inhibited by
administration of HA binding Peptide A, whereas scrambled Peptide A had no
effect on
the mRNA expression levels for this collagen. The data shown are
representative of
three independent experiments.
The ability of HA binding peptides to inhibit fibrosis is further illustrated
by histological analysis. Figure 36 are micrographs from a histological
analysis of lung
tissue treated with and without HA binding Peptide A after bleomycin injury.
Panels
(a-c) show tissue after treatment with saline alone, and panels (d-f) show
tissues injured
3 0 by bleomycin treatment. Panel (d) shows a fibrotic morphology . in the
presence of
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
98
bleomycin alone while panel (e) shows the morphology in the presence of
bleomycin
and scrambled RHAMM peptide. In contrast, panel (f) shows that injection of
bleomycin-treated animals with the sense RHAMM HA binding Peptide A results in
a
normal lung architecture despite the injury caused by bleomycin.
EXAMPLE 19
EXPRESSION OF RHAMM IN DIFFERENT CELLS PRESENT IN SYNOVIAL FLUIDS ISOLATED
FROM RA PATIENTS
This experiment determines which cell type from the synovial fluids of
RA patients express RHAMM isoforms.
Briefly, samples of synovial fluids from different RA patients were
centrifuged at 1600 rpm's for 10 min and pellets resuspended in 2-5 ml of
Blocking
r
buffer (BB, 1% human serum albumin in HBSS). After counting, 106 to 2.5x106
cells
per ml, were taken into each tube. Cells were washed once with lml of BB and
the
pellets resuspended in 100 ~,l of BB. First antibody was added (dil. 1:100)
and samples
incubated for 30 min on ice. Along with the first antibody 20 ~l of specific
markers for
certain cell type present in the synovial fluid were added, as well. Rabbit
IgG was used
as a control. Samples were washed twice with 1 ml of BB. After washing,
secondary
antibody was added (FITC, dil. 5:100) and cells kept 30 min on ice. Again,
samples
were washed twice, each time with 1 ml of BB and fixed with 0.3 ml of 0.5%
paraformaldehyde. Immunofluorescence was determined by flowcytometre.
Results are shown on Figure 37. Briefly, the majority of cells present in
synovial fluid are neutrophils. Macrophage/monocyte cells are present as 5-10%
of cells
and T cells are also present as a minority. Macrophage/monocyte cells
exhibited the
highest RHAMM expression. In some cases the number of exon4-positive cells was
as
high as 99.8%. A similar pattern was observed in neutrophil populations but
the
percentage of positively labeled cells was between 54.6% and 99.3%. T cells
also express
RHAMM isoforms, although to a lesser extent compared to the other two cell
types.
In summary, all tested RA patients expressed RHAMM on the surface of
cells present in synovial fluid. The most abundant cell type is neutrophils.
In all tested
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
99
patients more than 50% of neutrophil cell population was X4-positive.
Significant
number of macrophages expressing x4 was uncovered: in all tested RA patients
more
than 75% of macrophage/monocyte population was labeled x4 positive.
EXAMPLE 20
RHAMMX4 AND RHAMM R3.8 ARE PRESENT IN THE SYNOVIUM TISSUE SECTIONS
FROM RA PATIENTS
Rheumatoid arthritis is the most prevalent type of inflammatory arthritis,
affectingl.5% of the human population. RA is characterized by synovial
hyperplasia,
destruction of articular cartilage and bone and macrophage infiltration into
synovial
joints. Cytokines like IL-1 are present in increased levels and they play a
major role in
production of MMPs, such as collagenase and gelatinase.
In order to investigate if there is any RHAMM expressed in the
synovium tissue of RA patients, immunohistochemistry was done. Briefly, pannus
formed from synovium tissue was isolated and embedded in wax. Three microns
tissue
sections were obtained and slides were heated on 58°C for 30 min. To
deparafinized
slides the following procedure was done: tissue sections were washed in xylene
three
times each four minutes. After washing in hylene, slides were washed in 100%
ethanol
two times each three minutes. Additionally sections were washed in 96% ethanol
the
same amount of time. Slides were then incubated in dH20 two times each three
minutes
and once in PBS. Tissue on the slides was then marked with barrier-pen. The
activity
of endogenous peroxidase was blocked with 0.3% of hydrogen peroxide for 10
min.
Slides were washed with dH20 two times each 3 minutes and with PBS two times
each
5 minutes. Unspecific binding was blocked with 1% bovine serum albumin (BSA)
in
PBS at 37°C for 30 minutes. Different dilutions of RHAMMv4 and
RHAMM R3.8
antibodies were made: 1:100, 1:50, 1:25) in 1% BSA-PBS and incubated with
tissue
samples overnight at +4°C. Two tissue sections served as controls and
they were
incubated with either rabbit IgG (at the same dilution as the antibodies) or
with vehicle
which was 1%BSA PBS, without primary antibody. After incubation with primary
antibodies, slides were washed with PBS three times, 10 minutes each.
Consequently,
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
100
biotinylated antirabbit IgG was added and slides kept at room temperature for
1 hour
(di1.1:200 in BSA-PBS). Slides were again washed with PBS three times each 10
minutes. Additionally, Avidin-biotin ABC reagent was premixed and incubated
with
slides at room temperature for one hour. Slides were washed with PBS three
times each
time 5 minutes. After washing, DAB solution was premixed and incubated with
slides
for 5 minutes at room temperature. Samples were washed with dHzO three times
each
time 5 minutes and counterstained with hematoxylene for 1-2 minutes. Samples
were
washed with regular water and dehydrated. For dehydration similar procedure
was done
as for deparafinization only this time steps were done backwards. Slides were
mounted
and left to dry overnight.
Results are shown in Figure 38. Briefly, synovium tissue isolated from
joints of RA patient was positively stained (brawn staining) with RHAMM exon4
(pictures A and B) and RHAMM R3.8 (pictures C and D). Areas of synovial lining
. cells are enriched in RHAMM staining which are most likely macrophage cell
type,
although other cell types in the RA synovium also express RHAMM (pictures A,
B, C
and D). Controls BSA (picture E) and rabbit IgG (picture F) are unstained.
Hence, it is evident that RHAMM is present in high levels in human
arthritic joints.
EXAMPLE 21
RHAMM PEPTIDE MIMETIC INHIBITS PROGRESSION IN EXISTING
MULTIPLE SCLEROSIS (MS) MODEL
Multiple sclerosis (MS) is a major human neurological disease in North
America and Western Europe. Although the mechanism by which demyelination
takes
place in MS is not fully understood, it appears that the persistence of high
levels of
improperly assembled myelin which is prone to destruction is a leading cause
for on set
of the disease. Creation of ND 4 model of transgenic mice (Mastronardi et. al.
J.
Neurosci Res (93) Vol. 36 pp. 315-324) provides useful tool for investigation
of the
possible mechanism involving destabilization of the myelin membranes and
appearance
of distinctive features of MS disease.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
101
The purpose of this experiment was to attenuate clinical signs of
demyelination in MS by inhibition of function of the cells involved in
pathological
processes.
Briefly, transgenic mice (ND 4) bearing 70 copies of the transgene for
DM20, a myelin proteolipid protein, were utilized for assignment of scores
based upon
clinical signs of demyelination. Clinical signs which were assessed included
general
shaking, seizures, head jerk, hind-limb and tail shiver, unsteadiness, wobbly
gait and
limp tail. Within each sign score between 0 - 4 was given: where zero score
means
absent and score 4 means constant and uncontrollable appearance of the sign of
the
disease. Experimental groups of mice were divided into 4; each group contained
5
animals: one normal, one ND 4 mouse untreated and three ND 4 mice treated with
RHAMM mimetic - P-peptide. Animals were treated three times per week with 10
mg/kg of P-peptide intraperitonealy. Peptide was resuspended in 300 ~,1 of
PBS.
Results are shown in Figure 39. Briefly, treatment of ND 4 mice with P-
peptide showed significant attenuation of clinical signs of MS symptoms from 3
to 6
months of age (Figure 39). Applied in a fairly high dosage (10 mg/kg), the
peptide
exhibited 2 fold inhibition of disease symptoms, without observing
toxicological or
lethal effects on animals.
EXAMPLE 22
SCAR REDUCTION: P-PEPTIDE REDUCES COLLAGEN I AND III EXPRESSION IN
EXCISIONAL MODEL OF RAT SKIN
Wound-healing responses to injury involve a complex series of cellular
and inflammatory processes resulting in deposition of connective tissues and
its
remodeling into the scar tissue. The fibroproliferative response is
accompanied by
wound contraction and fibrosis due to the presence of myofibroblasts and to
the
enhanced production of collagen. In adult humans, the extracellular matrix is
remodeled to sustain and direct the cellular changes and to restore tissue
integrity. Such
exuberant healing responses often lead to tissue fibrosis and contraction,
commonly
referred to as scarring. Fibrosis of adult human tissue is a serious clinical
problem that
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
102
results in malfunction of tissue due to intraabdominal adhesions, cirrhosis of
liver,
failure of anastomoses as well as adhesions following surgery.
A fibrotic wound response contrasts with repair of fetal skin wound
wounds which exhibit reduced leukocyte infiltration, reduced fibroplasia and
altered
extracellular matrix remodeling resulting in a non-scared appearance of the
healed
wound. Additionally, hyaluronan accumulation is sustained in fetal skin while
its
accumulation is only transient in wounded adult skin.
This experiment tests the ability of 16 amino acid RHAMM peptide
mimetic (P-peptide) to reduce tissue fibrosis in a rat punch biopsy model of
skin repair.
A. Animal Model.
Three-month old female Sprague-Dawley (200-250 g) rats were
anesthetized with Somnitol (1 ml/kg) and subjected to 4 mm full-thickness
dorsal punch
biopsies. Series of the P-peptide concentrations (1 ng-20 mg) were mixed into
a diluted
bovine/1% collagen (type I) suspension and applied once only per biopsy punch
at the
time of wounding. A SOqI of the peptide/collagen solution was applied to the
punch
biopsy wound and allowed to polymerize over several hours. Collagen was used
as
vehicle to stimulate inflammation and fibrosis as rat skin normally shows
minimal
fibrosis. Collagen alone (control wounds) does not influence the rate of
healing when
compared to phosphate buffer saline. A twenty four hours after dorsal punch
biopsies,
animals were anesthetized with Isofluorane inhalant with oxygen and nitrous
oxide and
the experimental and control wounds (collagen alone) were excised. Samples
were
flash frozen in liquid nitrogen for RNA extraction.
B. RNA extraction.
Frozen wounds were homogenized in 1 ml of Trizol (Gibco, BRL) until
completely homogenous. After being homogenized, samples were incubated at room
temperature for 5 min and 200 ~,I of chloroform was added. Tubes were tightly
capped
and shaken vigorously for ~ 15 sec. Then, samples were incubated at room
temperature for
2-3 min. After incubation, samples were centrifuged at 11200 rpm's for 15 min,
at
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
103
2 - 8°C. Upper aqueous phase was transferred to another tube, carefully
not to disturb
interphase or organic phase of extract solution. After transfer,10 ~,g of T-
RNA was added
into the tube along with 0.5 ml of isopropyl alcohol. Samples were incubated
at room
temperature for 10 min. Subsequently, they were centrifuged in picofuge at
11200 rpm's
for 10 min at 4 - 8°C. After being centrifuged, supernatant was removed
and pellet
washed once in 75% ethanol. Samples were vortexed for 15 sec and spun in
picofuge for
5 minutes on 8800 rpm's at 4 - 8°C. Remaining ethanol solution was
carefully removed
and RNA pellet allowed to air dry. Pellet was dissolved in DEPC HZO
(Diethylpyrocarbonate). Concentration of RNA was determined by
spectrophotometer.
RNA was aliquoted into 20 ~g portions and stored in -70°C freezer until
required.
C. RT-PCR Analysis.
Frozen wound samples (50-100 mg tissue) were homogenized in 1 ml of
Trizol reagent and RNA was isolated according to standard Trizol Reagent.
Protocol.
For the synthesis of oligo-dT-primed cDNA, 2~g of total RNA, 1 ~,g of
oligo(dT)
primers and Moloney Murine Leukemia Virus Reverse Transcriptase (Gibco Brl #
28025-013) were used. Following 1 h incubation at 37°C, the reaction
was stopped by
heating samples at 95°C for 5 min and 2 q1 of RT reaction mixture was
used for PCR.
PCR amplification was performed with platinum Taq DNA polymerase (Gibco BRL
#10966-018) and specific primers for collagen I and III were used: 5' CGA TGT
CGC
TAT CCA GCT GA (SEQ ID N0:52) for collagen I and the following primer 5' ATC
AGT CAG CCA TCT ACC ACC (SEQ ID N0:53) was used for collagen type III.
Thermal cycles for collagen type I and III were as follows: denaturation at
94°C,
annealing at 60°C and polymerization at 72°C for 20 cycles. In
addition, a set of
primers of a common housekeeping gene B-actin, were run in parallel on 1.5%
agarose
gel as a loading standard.
Results are shown in Figure 40. Briefly, collagen production, which is a
marker for fibrosis, was assessed by semiquantitative RT-PCR analysis of
collagen type
I and III mRNA within the wound site. Levels of collagen type I and III mRNA
following P-peptide (lng/ml-20 mg/ml) application are shown in Figure 40.
Treatment
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
104
of wound sites with P-peptide reduced levels of collagen type I and III
measured at 24 h
post wounding.
EXAMPLE 23
S SCAR REDUCTION: ED-1 EXPRESSION IS REDUCED BY P-PEPTIDE TREATMENT IN
EXCISIONAL MODEL OF RAT SKIN
Fibrosis of adult human tissues is a serious clinical problem that results
in malfunction of tissue due to keloids, hypertrophic scars, anatomonic
strictures,
intraabdominal adhesions, cirrhosis of the liver, neurologic deficits
following injury to
the spinal cord, valvular heart disease, burned-injured joints as well as
failure of
anastomoses and adhesions following surgery.
The P-peptide was assessed for its effect on the course of wound repair
by measuring macrophage infiltration into the wound through the measurement of
ED-
' 1 expression, a marker for macrophages and fibroblasts.
1 S A. Animal Model.
Three-month old female Sprague-Dawley (200-2S0 g) rats were
anesthetized with Somnitoh (1 ml/kg) and subjected to 4 mm full-thickness
dorsal punch
biopsies. Series of the P-peptide concentrations (lng - 20 mg) were mixed into
a
diluted bovine/1% collagen (type I) suspension and applied once only per
biopsy punch
at the time of wounding. A SO ~,1 of the peptide/collagen solution was applied
to the
punch biopsy wound and allowed to polymerize over several hours. Collagen was
used
as vehicle to stimulate inflammation and fibrosis as rat skin normally shows
minimal
fibrosis. Seven days after dorsal punch biopsies, animals were anesthetized
with
Isofluorane inhalant with oxygen and nitrous oxide and the experimental and
control
2S wounds (collagen alone) were excised. Samples were flash frozen in liquid
nitrogen for
RNA extraction.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
105
B. RNA extraction.
Frozen wounds were homogenized in 1 ml of Trizol (Gibco, BRL) until
completely homogenous. After homogenization, samples were incubated at room
temperature for 5 min and 200 ~,l of chloroform was added. Tubes were tightly
capped
and shaken vigorously for 15 seconds. Then, samples were incubated at room
temperature for 2-3 min. After incubation, samples were centrifuged at 11200
rpm's for
min, at 2 - 8°C. Upper aqueous phase was transferred to another RNAse
free tube,
carefully not disturbing interphase or organic phase of extract solution.
After transfer, 10
~g of T-RNA was added into the tube together with 0.5 ml of isopropyl alcohol.
Samples
10 were incubated at room temperature for 10 min. Subsequently, they were
centrifuged in
picofuge at 11200 rpm's for 10 min at 4 - 8°C. After being centrifuged,
supernatant was
removed and pellet washed once in 75% ethanol. Samples were vortexed for 15
sec and
spun in picofuge for 5 minutes on 8800 rpm's between 4-8°C. Remaining
ethanol
solution was carefully removed and RNA pellet allowed to air dry. Pellet was
dissolved
15 in DEPCHZO). Concentration of RNA was determined by spectrophotometer. RNA
was
aliquoted into 20 q,g portions and stored in -70°C freezer until
required.
C. RT-PCR Analysis.
Frozen wound samples (50-100 mg tissue) were homogenized in 1 ml of
Trizol reagent and RNA was isolated. For the synthesis of oligo-dT-primed
cDNA, 2
~g of total RNA, 1 ~.g of oligo(dT) primers and Moloney Murine Leukemia Virus
Reverse Transcriptase (Gibco Brl # 28025-013) were used. Following 1 h
incubation at
37°C, the reaction was stopped by heating samples at 95°C for 5
min and 2 ~,1 of RT
reaction mixture was used for PCR. PCR amplification was performed with
platinum
Taq DNA polymerase (Gibco BRL #10966-018) and specific primers that used for
ED-
1 is: for ED-1 - 5' CGA TGG CAG GAC AGT AGT CGC (SEQ ~ID N0:54) and/or 5'
AAG GCT GCT GTT GAA AGG ACG (SEQ ID NO:55).
Thermal cycles for ED-1 was as follows: denaturation at 94°C,
annealing at 59°C and polymerization at 72 °C for 28 and 29
cycles. In addition, a set
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
106
of primers of a common housekeeping gene B-actin, were run in parallel on 1.5%
agarose gel as a loading standard.
Results are shown in Figure 41. Briefly, RT-PCR analysis of mRNA
isolated from the wound site treated with P-peptide (1 ng/ml to 100 ug/ml)
showed
down regulation of ED-1 expression at 7 days after injury in comparison to the
untreated wounds.
Thus, P-peptide reduces infiltration of macrophages into the site of the
wound.
EXAMPLE 24
RHAMM HA BINDING PEPTIDES INHIBIT MACROPHAGE
INFILTRATION FOLLOWING SKIN WOUNDING
Several key processes are involved in excisional wounding healing and
scarring. These include local inflammation and infiltration of macrophages and
neutrophils. The objective of this study was to determine whether different HA
binding
peptides inhibit macrophage infiltration.
The excisional wound healing rat model used and the method of local
application of peptides was similar to that described in example 23. Tissue
biopsies
were removed and assayed for Glucosiminidase activity, a biological marker for
macrophages.
As shown in Figure 42, the data demonstrate that HA-binding peptides
A (RGGGRGRRR; SEQ ID N0:27), B (RGGGRGGRR; SEQ ID NO:56), C
(RGGGRGGGR; SEQ ID N0:57) inhibited the infiltration of macrophages in wounded
biopsies, whereas peptide D (RGGGGGGGR; SEQ ID N0:58) wvhich has a similar
sequence but does not bind HA does not inhibit macrophage infiltration in
wounding.
In addition the scrambled peptide A did not have any effect on macrophage
levels.
In conclusion these data demonstrate that HA-binding peptides inhibit
macrophage motility and infiltration in wounding, and thus have potential to
promote
wound healing and reduce scarring.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
107
EXAMPLE 25
RHAMM REGULATES PROSTATE CANCER PROGRESSION
This experiment investigates whether functional expression of the HA
receptor RHAMM is required for enhancement of CaP cell motility and invasion
in vitf°o.
Briefly, Dunning CaP cell lines (AT-1, MatLyLu) were grown in DMEM
medium supplemented with 10% FBS at 37°C in a humidified atmosphere
containing 5%
C02. All cell lines were passaged every 3-4 days upon reaching confluency.
A. Immunofluorescence.
Cells were seeded sparsely on glass coverslip and incubated in growth
media for 24 h. cells were then fixed with 3% paraformaldehyde and
permeabilized
with 0.2% triton X-100. RHAMM was visualized by indirect immunofluorescence
using a polyclonal antibody to the C-terminus (tram 2.3, 1:100) and Texas red
conjugated donkey anti-rabbit antibody (1:100). Images are obtained using a
Zeiss laser
scamling confocal microscope.
B. Western blotting.
Cells were also grown to 50-60% confluency were lysed using RIPA
buffer. Equal amounts of total cell protein were loaded onto a 10% SDS-PAGE
gel.
RHAMM was probed using a polyclonal antibody to the C-terminus (tram, 1:1000)
and HRP-conjugated goat anti-rabbit antibody (1:5000). RHAMM was visualized by
chemiluminescence.
C. Cell motility.
Cell were seeded sparsely and grown in 25 cm2 flasks overnight. Serum-
free medium was used for the experiments. Random cell motility of cells
untreated, or
treated with either RHAMM polyclonal antibody (Re4) or peptide mimicking the
HA-
binding domain over two hours was visualized by videomicroscopy. Cell motility
tracks were analyzed using a Northern Exposure software. Statistical analysis
was
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
108
performed on 100 cells per field and statistical significance was determined
using
unpaired Student t-test.
D. Cell invasion.
Cell were grown to confluency in growth media, detached, and equal
number of cells were seeded in 24-well Matrigel invasion chambers. Cells were
left
untreated with RHAMM peptide and allowed to invade for 24 h. For statistical
analysis, 5
high power fields(400X) were counted for the number of cells that invaded
through the
membrane. Statistical significance was determined using unpaired Student t-
test.
E. MMP activity.
Cells were grown to confluency in growth media, detached and equal
number of cells were seeded in 6-.well plates uncoated or coated with 50%
Matrigel in
media. Cells were allowed to adhere for 1h to the substrate, and then treated
with the
peptide mimicking the HA-binding domain of RHAMM (100 ~g/ml) for 24 h in serum-
free media. The activity of MMP secreted into the media was determined by
zymography using 8% SDS-PAGE.
Results are shown in Figure 43. Briefly, Figure 43A shows the Western
blot analysis using a RHAMM polyclonal antibody detecting progressively
increasing
expression of 54 kDa RHAMM isoform in proportion to motility/invasivity: the
highly
motile/invasive subline, Fb2 > the weakly motile/invasive parental line, MC2 >
the
nontumotigenic parental NbE epithelial line. Figure 43B shows RHAMM
localization
to a sites of cell extension and to podosomes of invasive CaP cells. Open
arrows point
to sites of cell protrusion, whereas closed arrows point to circular
structures known as
podosomes or invadopodia. Left panel Figure 44A shows that RHAMM regulates
Dunning CaP cell line motility and invasion, whereas right panel of Figure 44A
showed
that MaTLyLu cells treated with a RHAMM peptide showed a .significant
reduction
(p<0.025) of about 20% in invasive potential as determined using Matrigel irz
vitro
invasion chambers. However no effect of peptide was observed upon treatment of
the
AT-1 cells. Figure 44B shows that secretion of MMP was higher in AT-1 cells
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
109
compared to MatLyLu cells when grown on plastic. Matrigel did not reduce MMP
activity in AT-1 in MatLyLu. When RHAMM blocking peptide was added, MMP
activity was suppressed.
Thus, RHAMM is preferentially expressed in more motile/invasive and
metastatic CaP cells. Blocking RHAMM function significantly and preferentially
reduces motility, invasion, and MMP activity in highly metastatic CaP cells.
EXAMPLE 26
INFLUENCE OF ntlAMM PEPTIDE MIMETIC ON WEIGHT
1 O GAIN IN MURINE MODEL OF SLE
F1 (NZB/W) mice, hybrids of New Zealand Black (NZB) and New
Zealand White (NZW) mice, a.re a murine model of SLE (Systemic Lupus
Erythematosus). These mice develop spontaneously autoantibodies to DNA and
other
cell components. Female mice develop a more rapid disease course than males,
'with
death from renal failure occurring by 8-10 months of age in females and 18-20
months
of age in males. Females of 8 weeks of age are free of overt symptoms of
disease, with
gradual development of autoantibodies, glomerulonephritis, proteinuria, renal
failure
and death. The renal disease is likely secondary to the immune dysfunction.
In addition to progressive renal inflammatory disease, these mice show
increase in body weight of 20% - 30%, which is manifested by increased
accumulation
of body fat. These lupus mice also have elevated triglycerides, similar to
that seen in
human SLE patients. A number of studies in murine SLE model have shown that
dietary manipulations and restrictions have an effect on the development on
this life
shortening autoimmune disease. Several lines of evidence have supported a link
between adipose tissue and immunocompetent cells. For example, in obesity,
excess
adiposity is linked to impaired immune function. Studies in rodents with
genetic
abnormality of leptin and leptin receptors, which result in obesity, revealed
obesity
related changes in macrophage phagocytosis and the production of
proinflammatory
cytokines. These data identify an important link between obesity and
regulation of
inflammatory and immune responses.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
110
This experiment assesses the effect of the P-peptide on body fat
accumulation in murine SLE model.
Briefly, female NZB/WFl were obtained from Jackson Laboratories at 6
weeks of age and housed locally for 2 weeks prior to initiation of the
studies. The study
design comprised of four groups of 10 female NZBIWFl mice; one control and
three
experimental groups. The control group of mice were not treated with 16 amino
acid
RHAMM peptide mimetic (P-peptide). First group of mice were given P-peptide (5
mg/kg), three times a week via the IP route. The treatment started at 8 weeks
of age and
continued up to 28 weeks of age. The animals in the other two experimental
groups
were started on the P-peptide (5 mg/kg) at 16 and 24 weeks of age to determine
whether
interference with P-peptide can arrest or reverse active weight gain. The
treatment in
these animals also continued up to 28 weeks. The animals were assessed for
weight
gain during the development of the disease at weekly intervals.
Results are shown in Figure 45. Briefly, the control group of rilice
showed a trend of increase in body weight of approximately 5 g per month. The
total
average increase after 20 weeks was 13 g. The group of mice that was treated
from 8
weeks of age showed significant reduction in weight gain in comparison with
the
control group. The average increase of body weight in this group was 2 g per
month,
whereas total accumulation of weight was 6 g after 20 weeks. Weight gain in
mice in the
other two experimental groups was identical to the control group until the
initiation of
treatment. The body weigh in these mice showed decrease within the first week
of the
treatment, with the trend of further decrease toward the levels observed in
animals that
were treated at early stage of the disease (Figure 45). The treatment with P-
peptide did
not effect the weight gain in NOD mice, which served as a control for this
experiment.
In summary, the weight gain in mice that were treated at the early stage of
disease (8 weeks) was similar to the weight gain in normal mice. Mice that
were treated
at later stages of disease showed not only arrest but reverse of weight gain
that was similar
to early stage treated mice. Thus, the P-peptide can be utilized as a
therapeutic agent in
the treatment of obesity and obesity related diseases (e.g., diabetes and
cardiovascular
disease), as well as for diseases such as kidney fibrosis and lupus (SLE).
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
111
EXAMPLE 27
CORRELATION BETWEEN RHAMM LEVELS AND CANCER CELL INVASIVENESS
This experiment assesses the relationship between RHAMM expression
and aggressiveness of cancer cell lines. RT-PCR was conducted as described in
the
attached Wang et al., (Clinical Cancer Research, 4:567-576, 1998). Western
blot
analyses was conducted as described above.
Results of these experiments are shown in Figure 46. Briefly, the levels
of erk kinase correlated significantly with the levels of RHAMM expression
(r=.21,
p<0.007, Students "t" test). The cell lines H125, Hl 57 and H226 are less
invasive and
aggressive than the H460 and MGH 7. As shown in the Figures, RHAMM expression
is highest in the latter two cell lines. Of the two cell lines, the H460 is
more invasive in
matrigel assays than the MGH7 cell lines.
Based upon this experiment it is evident that the highest level of
RHAMM expression is observed in the most invasive lung cancer cell lines.
EXAMPLE 28
CORRELATION BETWEEN ASTROCYTOMA CELL
METASTASES AND RHAMM EXPRESSION
Invasive astrocytoma cell lines (U87MG and U343MG-A), astrocytoma
biopsies from patients, cervical tumor cell lines (HeLa) were extracted for
mRNA and
analyzed for the presence of RHAMM using northern blots and RT-PCR as
described
by Sambrook et al. Results are shown in Figure 47. Briefly, astrocytoma cell
lines
express approximately equal amounts of RHAMM, as detected by Northern blot
analysis. RT-PCR analysis show that RHAMM is most highly expressed in high
grade
or invasive astrocytomas (Figure 47B)
These results support that RHAMM is involved in the tumor invasion step
of neoplastic progression and this is consistent with its ability to regulate
podosome
formation, structures that permit release of collagenases that are required
for cell invasion.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
112
EXAMPLE 29
SCREENING FOR PROTEINS THAT REGULATE HA TRANSPORT IN A TRANSITIONAL CELL
A RHAMM induced cDNA expression library is obtained from mRNA
populations extracted from RHAMM transfected cells maintained in serum free
medium
for 24 h. These culture conditions allow uptake of HA into the cell cytoplasm
but will
not allow HA uptake into the cytoplasm of non-transfected cells unless a HA
transport
protein is expressed. The cDNA library is used to infect COS or CHO cells
which are
then exposed to Texas red-labeled HA in the presence of cytochalasin D which
inhibits
endocytic uptake of HA. Under these conditions cells would not ordinarily take
up HA
into the cytoplasm, hence, HA uptake will depend on the expression of a HA
transporter. Infected cells are briefly exposed to streptomyces hyaluronidase
to remove
any Texas red labeled HA coating the outside of the cell and then cells are
sorted for
positive fluorescence with FACS.
Cells that are positive are cloned and rescreened for HA uptake.
' Transfected genes encoding an HA transporter are then retrieved by RT-PCR of
mRNA
and sequenced. These genes are then transfected into lOTl/2 cells which do not
take up
HA into the cytoplasm unless they are exposed to phorbol ester to activate
protein
kinase C. These cells are in turn assessed for uptake of Texas red-labeled HA
into the
cytoplasm and scored for altered growth factor sensitivity by techniques
previously
described herein.
The cDNA encoding a HA transporter is then cloned into an appropriate
expression vector that will permit expression and isolation of the transporter
protein.
Appropriate vectors and expression systems are well known in the art.
Antibodies are
then be prepared against this protein. In addition, peptide regions
instrumental in taking
up HA (i.e., an HA binding domains) are identified and peptides that mimic
these sites
are prepared for assessment of the ability to compete with HA transport or
otherwise
impact signaling pathways, podosome formation and/ cell motility which
characterize
transition stage cells.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
113
EXAMPLE 30
IDENTIFICATION OF RHAMM BINDING PROTEINS OR OTHER TRANSITION STAGE
MOLECULES BY USE OF RHAMM OVEREXPRESSING CELLS CULTURES
To identify proteins that are transiently regulated with RHAMM to
control cell activation, cDNA expression libraries obtained from the "CHIP"
differential
screen described above are used to establish libraries expressing transition
molecules
that is capable of binding to a hyaladherin or other transition stage
molecule. Several
techniques are known in the art for identifying an expressed binding partner.
These
include a two hybrid phage display system and a two hybrid yeast expression
system.
The two hybrid expression system is used to screen for peptides or
polypeptides that
bind to RHAMM or other transition molecule, and the ability to actually bind
the
transition molecule is further characterized using a far Western assay system.
Specific
binding regions of the RHAMM binding partners can be further identified using
the
functional regions of RHAMM exons and the regions of RHAMM known to be
involved in the transient phenotype through the ability to activate erk
kinases as
provided for example, in Figure 48. Antibodies may be made to the identified
binding
protein and assessed for the ability to affect cell motility or e~°k
signaling cascades
according as previously described.
One such protein herein designated as RABP for RHAMM Associated
Binding Protein has been identified using this method by using a phage display
library
mentioned above to bind to the peptide regions of exon 4 described as SEQ. ID
NO: 17.
A partial polypeptide and nucleic acid sequence for RABP is provided as SEQ.
ID NO:
47 and 46. Antibody against this protein has been prepared and shov~nl to be
effective in
inhibiting RHAMM activated podosome formation and signaling in RHAMM
overexpressing cells. Figure 49 shows the sequence for this novel RHAMM
binding
protein which was determined to be a 60 kDa protein that binds to exon 4 of
RHAMM,
and which is transiently present on the cell surface. Panel A shows the
partial sequence
of RABP isolated by a two hybrid screen using exon 4 of RHAMM. Panel B shows a
Northern blot of RABP in transitional cells. Panel C shows a Western blot of
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
114
transitional cell lysate indicating that RABP occurs within a 60 kDa protein.
Panel D is
a FACE analysis showing that RABP is present on the cell surface.
Other proteins regulating transition stage cells can be identified using
cell cultures characterized by the transition stage phenotypes described for
LR21 cells
provided herein. Briefly, transitional cell cultures that overexpress RHAMM at
precisely the levels required for enhancing podosome formation are grown to
subconfluence (50-60%) in 10% FBS then released from their substratum using a
0.15
median, PBS, non-enzymatic disassociation medium. These cells are maintained
in
suspension in defined medium for 30 minutes and then plated for 24 hrs, at
5x15
cells/ml on plasma fibronectin coated dishes which promotes podosome positive,
transitional phenotype. PolyA mRNA is isolated from the transitional cells and
a
differential screen is conducted using a cell line that is plated onto plastic
dishes so that
podosomes are not produced. RNA is placed into CHIPS for differential
screening and
genes associated with the transition phenotype are identified using CHIP
protein
technology. Positive cDNA's are sequenced, cDNA libraries are screened and
RACE
technology is used to obtain a full length cDNA.
The CHIP will contain cDNA's encoding proteins involved in the
transition stage phenotype that do not necessarily bind to HA but are
nevertheless
involved in regulating this stage. Hence, this method for obtaining
transitional mRNA
is useful for identifying other important dominant acting proteins involved
with the
transitional stage of response to injury processes. The CHIP screen can be
used for
proteins that bind to important podosome proteins such as CAS and cortactin,
and full
length sequences of these can be obtained. The function of such sequences may
be
analyzed for their effect on podosome formation by transient transfection. The
entire
differentially screened mRNA can be used to transiently transfect cells to
determine
whether they can induce podosome formation, using CAS or cortactin to detect
podosomes as described above. Particular sequences are identified by
increasingly
restricting the number of mRNAs included in a transfection group until
ultimately
restricting the size of the group to single mRNAs encoding single genes
affecting the
transition stage phenotype.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
115
EXAMPLE 31
IDENTIFICATION OF HYALADHERINS BY SEARCHING DATABASES
FOR HYALURONAN BINDING MOTIFS
In addition to specific peptides such as those described in SEQ. ID NOS:
1-10 that represent hyaladherins which bind to hyalauronic acid, a variant of
additional
polypeptides may be identified, generated and tested fox use within the
methods
described herein. All such binding motifs are characterized by the presence of
general
amino acid motifs including staggered basic residues. These motifs can be more
generally described as BX7B (SEQ ID N0:28) where B is any basic amino acid and
X7
is any amino acid sequence of about seven residues but usually including at
least one
hydrophobic amino acids or an additional basic amino acid. Most importantly
however,
none of the intervening X amino acids should be acidic, as acidic amino acids
appear to
interfere with binding to hyaluronan, a negatively charged polymer. Peptides
which are
specifically excluded from this motif include: BBXXBBBXXBB, KQKIKHVVKLK,
KLKSQLVKRK, RYPISRPRKR, KNGRYSISR, RDGTRYVQKGEYR,
RRRCGQKKK, RGTRSGSTR, RRRKKIQGRSKR, RKSYGKYQGR, KVGKSPPVR,
KTFGKMKPR, RIKWSRVSK, KRTMRPTRR, KVGKSPPVR, or HREARSGKYK
(SEQ ID Nos. 29-44 respectively). These excluded peptides do not bind HA with
the
'20 same high affznity as peptides of the present invention which require are
peptides that
form an alpha helix. All motifs that bind to hyaluronan also preferably form
strong
alpha helices as predicted in secondary structure protein analysis programs
which
further show that hyaluronan binding motifs contain at least two basic amino
acids
aggregating along one plane of the helix.
Using this information, a search of the data bases for previously
undetected hyaladherins can be made, searching first for the aforementioned
motif then
coupling this with analysis of the structural requirements again using protein
prediction
programs such as for example are available on the Internet (e.g., GCG).
Additional
sequence candidates can be found by searching appropriate databases of the
technical
literature such as Medline, Biosis, Chemical Abstracts and the like. Searches
can then
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
116
be made to determine which of the newly identified hyaladherins have
previously been
associated with disease and with the expression of podosomes. Those that are
present at
the cell surface can then be screened for their potential therapeutic use.
EXAMPLE 32
SCREENING FOR INHIBITORS OF PODOSOME FORMATION
The present invention provides for novel cell lines that overexpress
RHAMM and that produce enhanced formation of podosomes. These cell lines may
be
utilized to screen for inhibitors of podosome formation. Concomitant with the
formation of podosomes and development of a transient phenotype, cells release
proteases that result in degradation of fibronectin, revealing a previously
sequestered
CS-1 sequence. Antibodies to this CS-1 sequence have been prepared. The
presently
provided RHAMM overexpressing -cell lines are coated on microsphe~e beads in
- conjunction with plasma fibronectin to form an assay mixture which is
incubated at
37°C for 2-3 h. The aforementioned CS antibody conjugated to a
fluorochrome is
added to this mixture causing a fluorescence response indicative of
fibronectin
degradation which is in turn indicative of the formation of functional
podosomes.
Candidate inhibitors of podosome formation are identified by the ability to
reduce
fluorescence in this assay and these candidates may be screened using any of
several
high through-put screening systems known in the art.
Inhibitors to be screened include, but are not limited to antibodies, HA
binding peptides/polypeptides and RHAMM binding peptides/polypeptides
associated
with regulation of the novel transitional stage cells as provided in this
invention. In
addition, upon identification of functional portions of newly discovered
transitional
proteins using the methods described herein, candidate inhibitors comprised of
small
chemicals or peptide mimics of these functional portions can be synthesized
according
to methods known in the art. One set of peptide mimics includes for example,
HA
binding mimics.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
117
EXAMPLE 33
HUMANIZED ANTIBODIES THAT INHIBIT TRANSITIONAL MOLECULE FUNCTION
Humanized antibodies raised against transitional molecules identified
above (e.g., hyaladherins, RHAMM binding partners, transitional proteins) can
be
screened for their inhibition of specific cell signaling pathways involved in
cell
transition (inhibition of ef~k kinase activity, AP-1 activity, MMP expression
or specific
transition states of the cell (e.g., podosome formation, cell migration, cell
proliferation)
in fluorescent screening assays. Cell lines over expressing specific
transition molecules
will activate ERIC kinases, c fos expression, AP-1 activity, MMP expression,
and
increased transitional states of the cell such as podosome formation resulting
increased
cell migration and proliferation.
Humanized antibodies to identified transition molecules such as RHAMM
can be screened for inhibition of the aforementioned cell signaling pathways,
gene
expression, podosome formation, and/or cell motility and proliferation. These
studies will
identify potent antibodies which inhibit the transition of normal cells to
diseased cells
which can be utilized clinically in humans for the treatment and diagnosis of
disease.
EXAMPLE 34
COMPLEMENTARY PEPTIDES AND PEPTIDE MIMICS
2O THAT INTERFERE WITH TRANSITION MOLECULE FUNCTION
A variety of candidate peptides affecting transition state cells can be
detected and/or screened using the methods provided by the present invention.
Candidate peptides include peptides generated from transition molecules
provided in the
present disclosure (such as the RHAMM peptides), peptides of the transition
stage
which may further be identified using the aforementioned methods, peptides
that bind
strongly to active regions of transition molecules, or peptides that compete
with binding
of transition molecules of specific ligands generated by standard synthetic
processes.
Each of these can also be screened for effects on the particular features of
transition
cells disclosed herein including effects on specific signaling pathways (e.g.,
ERIC
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
118
activity, AP-1 activity,) gene expression (e.g., c fos and MMP expression),
podosome
formation, cell motility and proliferation.
The structure of peptides effective in inhibiting transition molecule
induced processes can be determined by several methods including standard
structure
function analyses of a proteins shown to inhibit podosome formation andlor by
using
the above screening methods for analyzing peptide sequences encoded by a gene
shown
to be involved in podosome formation. Complementary peptides and peptide
mimics
can be designed based upon functional peptide motifs, particularly when an
inhibitory
peptide motiff is small (e.g., 10 amino acids or less). Such peptides and
their mimics
would be candidate molecules for therapeutic treatment of a variety of disease
states
dependent upon entry and passage of cells through the transition stage
phenotype taught
by the present invention. Candidate molecules would be tested for efficacy by
assay in
animal models of disease or using cultured cells expressing a transitional
stage cells.
Similar studies may be performed to screen small molecules for their
inhibition of transition molecule function and the progression of cells from
the normal
to diseased state as described in the present invention.
EXAMPLE 35
DIAGNOSTICS METHOD FOR DETECTING HA, HYALADHERINS AND INJURED CELLS
1. Detection of Intracellular and Plasma HA
Serum and tissue levels of HA are valuable diagnostic markers of
arthritis and neoplasia. Thus, levels of HA in the serum are currently used to
follow the
course of osteoarthritis response to steroid therapy. Further, HA accumulation
within
colorectal and breast cancers is prognostic of a poor outcome. Because HA
levels are
enhanced following most forms of tissue injury, other conditions including
restenosis,
MS, Alzheimer's, stroke, myocardial infarction, sports injuries, burns and
other
inflammatory diseases would benefit from methods of detecting HA. In addition,
HA
increase in plasma is associated with a variety of other diseases,
particularly rheumatoid
arthritis and in tumors such as mesetheliomas and Wilm's tumors. Therefore
testing of
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
119
HA levels in serum or in biopsy tissue will be useful, alone or in combination
with other
disease markers for determination of a variety of disease conditions.
Currently, HA is routinely detected using fragments of HA binding
proteins such as the 60 kDa fragment of aggrecan or link protein. The
procedures for
purifying these proteins is laborious and results are inconsistent making it
difficult to
routinely assess HA as a diagnostic parameter. The sensitivity of this
technique is 5 pg
HA in serum using ELISA assays.
The present invention provides a method of similar sensitivity but which
is cheaper and more reliable. The method is based on using HA binding partners
discovered using the techniques described above for detecting RHAMM binding
partners. Using a phage display library to bind to biotinylated HA permitted
identification of five particular species of HA binding motifs described in
SEQ. ID
NOS: 6-10. This was accomplished by isolating phage that attached to HA which
were
further isolated, rescreened twice and recloned. The clones were then bound to
biotinylated HA-sepharose beads and only those phage that could be released
with
unlabeled HA were retrieved, recloned and sequenced. Five clones comprising
the
sequences identified above were detected. These sequences all bind to HA and
are
useful for detecting HA in serum and tissues in an assay described below.
An assay was developed based upon HA binding to these newly
discovered peptide sequences. Synthetic peptides comprising these sequences
were
synthesized with a linker arm of glycine-glycine-cysteineto which I~LH was
covalently
linked using EDAC. One to 200 ~,g of any one of these peptides were coated
onto the
surface of ELISA plates in phosphate buffered saline for 1 h at room
temperature. Plates
were washed in PBS and then coated with 1 ~g/ml of HA and washed with PBS and
0.1%
triton. Texas red labeled peptide was then applied to the coated plate for 1
h. Serum
samples and HA standard solutions were then applied to the plates and left on
a mixer for
2 h. The plates were then washed and read in a fluorescentELISA plate reader
at 545 um.
The amount of HA in the samples was determined by comparison to the HA
standards.
This assay has a similar sensitivity to previously described assays using
aggrecan but is more reproducible due to the standardization possible using
peptide
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
120
synthesis. This contrast to the more variable results obtained using assays
based on
preparation of purified aggrecan for which a reproducible reagent capable of
binding to
HA is difficult to make.
These newly discovered peptides are also useful for detecting HA present
in tissue (e.g., biopsy tissue). In one example, frozen or paraffin embedded
tissue sections
are incubated with biotinylated peptides for 1 h in sections that have been
either exposed
to hyaluronidase, used as a control, or to buffer alone. Sections are washed
then
developed with horseradish peroxidase labeled streptavidin and sections are
then
examined for a positive reaction indicated by brown staining. This procedure
can be
readily adapted for use in a kit as can the ELISA assay for detecting HA in
plasma.
2. Development of an Assay for Detection of Soluble Hyaladherins
The above mentioned HA binding peptides are also useful in an assay for
soluble hyaladherins. In this regard, an important aspect of the present
disclosure is that
the transition phenotype plays a heretofore undisclosed role in many disease
processes
such as inflammatory .diseases, cancers, degenerative diseases and wound
healing. In
each case HA will be shed during the podosome stage of a cell that typifies
the
transitional phenotype. Therefore, the presence of a transitional phenotype
during the
early stages of disease establishment may be detected by assaying for the
presence of
hyalauronan or hyaladherins present in serum.
An assay for hyaladherins can be provided using the small peptides that
bind to HA as described herein before. In one example, these peptides can be
synthesized with an additional cysteine at the carboxy terminus. The peptides
are then
covalently linked to sepharose as per standard procedures. The sepharose beads
are
incubated with biotinylated HA for one hour, then washed. Tlie beads
containing
biotinylated HA are then incubated for 1-2 hours at room temperature with an
aliquot of
sample serum. Hyaladherins that axe present within the serum will compete with
the
peptide bound to sepharose for the biotinylated HA and therefore the amount of
biotin
label remaining with the sepharose beads will be inversely proportional to the
amount
of hyaladherins present in the serum sample.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
121
An alternative to the general hyaladherins assay mentioned above is a
specific hyaladherins assay for selected hyaladherins observed to increase
during a
particular disease or cellular response as may be detected using the screening
methods
provided in the foregoing Examples. In this specific assay, monoclonal
antibodies are
prepared against the selected hyaladherins observed to increase during disease
as
detected by these screening methods. The monoclonal antibodies are used in a
standard
ELISA assay where antibodies are coated onto the ELISA well, serum is added to
this
coating, washed and a second layer of anti-hyaladherin will be layered on top.
The top
layer of antibody is detected using a fluorochrome labeled secondary antibody
and the
amount of label quantified in an ELISA plate reader.
The presently described assays based upon use of HA binding peptides
hyaladherins and antibodies thereto are readily adaptable for detecting other
components associated with the transitional state such as HA transporters or
other
proteins which may be detected using the aforementioned screening systems.
EXAMPLE 36
RHAMM REGULATES PROSTATE CANCER PROGRESSION
This experiment investigates whether functional expression of the HA
receptor RHAMM is required for enhancement of CaP cell motility and invasion
in vitro.
Briefly, Dunning CaP cell lines (AT-1, MatLyLu) were grown in DMEM
medium supplemented with 10% FBS at 37°C in a humidified atmosphere
containing 5%
C02. All cell lines were passaged every 3-4 days upon reaching confluency.
A. Immunofluorescence.
Cells were seeded sparsely on glass coverslip and incubated in growth
media for 24 h. cells were then fixed with 3% paraformaldehyde and
permeabilized
with 0.2% triton X-100. RHAMM was visualized by indirect immunofluorescence
using a polyclonal antibody to the C-terminus (tram 2.3, 1:100) and Texas red
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
122
conjugated donkey anti-rabbit antibody (1:100). Images are obtained using a
Zeiss laser
scanning confocal microscope.
B. Western blotting.
Cells were also grown to 50-60% confluency were lysed using RIPA
buffer. Equal amounts of total cell protein were loaded onto a 10% SDS-PAGE
gel.
RHAMM was probed using a polyclonal antibody to the C-terminus (tram, 1:1000)
and HRP-conjugated goat anti-rabbit antibody (1:5000). RHAMM was visualized by
chemiluminescence.
C. Cell motility.
Cell were seeded sparsely and grown in 25 cm2 flasks overnight. Serum-free
medium was used for the experiments. Random cell motility of cells untreated,
or treated
with oither RHAMM polyclonal antibody (Re4) or peptide mimicking the HA-
binding
domain over two hours was visualized by videomicroscopy. Cell motility tracks
were
analyzed using a Northern Exposure software. Statistical analysis was
performed on 100
cells per field and statistical significance was determined using unpaired
Student t-test.
D. Cell invasion.
Cell were grown to confluency in growth media, detached, and equal
number of cells were seeded in 24-well Matrigel invasion chambers. Cells were
left
untreated with RHAMM peptide and allowed to invade for 24 h. For statistical
analysis, 5
high-power fields(400X) were counted for the number of cells that invaded
through the
membrane. Statistical significance was determined using unpaired Student t-
test.
E. MMP activity.
Cells were grown to confluency in growth media, detached and equal
number of cells were seeded in 6-well plates uncoated or coated with 50%
Matrigel in
media. Cells were allowed to adhere for 1h to the substrate, and then treated
with the
peptide mimicking the HA-binding domain of RHAMM ( 100 p,g/ml) for 24 h in
serum-
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
123
free media. The activity of MMP secreted into the media was determined by
zymography using 8% SDS-PAGE.
Thus, RHAMM is preferentially expressed in more motile/invasive and
metastatic CaP cells. Blocking RHAMM function significantly and preferentially
reduces motility, invasion, and MMP activity in highly metastatic CaP cells.
EXAMPLE 37
TREATMENT AND/OR PREVENTION OF DIABETES MELLITUS
The purpose of these experiments was to evaluate the RHAMM (P-16)
peptide on the treatment of diabetes in the non-obese diabetic (NOD) mouse
model.
NOD mouse is a model of human type I diabetes mellitus, which is characterized
by a
cell-mediated autoimmune process resulting in spontaneous diabetes (see, e.g.,
Zhao et
al. Lithium (1991), 2(4)227-34; see also, The Jackson Laboratory). Studies
have shown
that the major populations of cells infiltrating the islets of Langerhans in
the early stage
of insulitis in NOD mice are T cells and macrophages.
There are different colonies of NOD mice and there can be some
variability between the colonies. The colony used develops the disease between
15-20
weeks of age and there is a 70-80% incidence of diabetes mellitus. The mice
treated
were divided into two groups of 10 animals; the first group being treated with
P-16
peptide and the other group comprising of the control group, which was treated
with
saline. Once the NOD mice were 5 weeks old, the P-16 peptide was injected
three
times a week interperitoneally at a dose of Smg/kg for 23 weeks. The untreated
mice
and five mice from the treated group were sacrificed at 28 weeks of age. The
remaining
five mice from the treated group were taken off the peptide treatment at 28
weeks of age
and were assessed for the disease after 16 weeks.
As shown in figure 51, the incidence of diabetes measured by blood
glucose level in untreated NOD mice was 70% whereas the incidence in the
treated
mice was 20%. The untreated mice also had a higher incidence of abnormal urine
glucose level, 80%, compared to 0% in the treated mice (figure 52). Further,
when
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
124
examining water consumption associated with diabetes, water consumption
increased
significantly in untreated animals with the onset of diabetes around week 12
to 13
(figure 53). In contrast, the water consumption did not change in animals
treated with P-
16. These data clearly demonstrate that P-16 peptide inhibited the incidence
of diabetes.
The treated mice that had the treatment stopped at 28 weeks have not
developed any signs of the disease after 16 weeks. They looked healthy and did
not
show presence of polydipsia or urinary glucose.
In NOD mice, there was an increase in kidney weight due to renal
hypertrophy that is associated with the onset and progression of diabetic
symptoms. As
shown in figure 54, treatment with the P-16 completely inhibited the increase
in kidney
weight, presumably by inhibiting glomerulosclerosis.
The histological analysis of pancreatic tissue showed that treated mice
had more intact pancreatic islets than the untreated animals and significantly
smaller
inflammation of the islets with inflammatory cells.
Presented results clearly show that RHAMM (P-16) peptide administration
potently prevents the development of diabetes and associated complications in
the NOD
model of Type I diabetes mellitus in the absence of any toxicity. The diabetes-
sparing
effect is probably due to the inhibition of the destruction of beta cells in
the pancreatic
islets. The effectiveness of RHAMM (P-16) peptide administration to induce
long-term
inhibition of disease was demonstrated by the negative results of urinary
glucose and
polydipsiaof 16 weeks post-peptidetreatmentNOD mice.
These results indicate that RHAMM and its major ligand HA associate
functionally with autoimmune insulitis leading to IDDM, and that by using
specific
RHAMM peptides they can serve as potential therapeutic targets.
These findings also show that the RHAMM peptides, peptide mimetics,
antibodies and potential HA binding peptides can be used as an effective
method for
preventing and/or treating diabetes mellitus by interfering with the
penetration of the
inflammatory cells into the islets and destructive invasion of the islets.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
125
From the foregoing, it will be appreciated that, although specific
embodiments of the invention have been described herein for purposes of
illustration,
various modifications may be made without deviating from the spirit and scope
of the
invention.
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
1
SEQUENCE LISTING
<1l0> Transition Therapeutics
Turley, Eva A.
Cruz, Tony F.
<120> COMPOSITIONS AND METHODS FOR TREATING
CELLULAR RESPONSE TO INJURY AND OTHER PROLIFERATING CELL
DISORDERS REGULATED BY HYALADHERIN AND HYALURONANS
<130> 910130.40101PC
<140> PCT
<141> 2000-10-05
<160> 72
<170> FastSEQ for Windows Version 4.0
<210> 1
<211> 8
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<221> VARIANT
<222> (1)...(5)
<223> Xaa = any amino acid
<221> HELIX
<222> (1)...(5)
<223> Alpha-helix
<221> VARIANT
<222> (6) ... (8)
<223> Xaa = Lysine or Arginine
<400> 1
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
1 5
<210> 2
<211> 11
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<221> VARIANT
<222> (l)...(5)
<223> Xaa = any amino acid
<221> HELIX
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
<222> (1)...(5)
<223> Alpha-helix
<221> VARIANT
<222> (6) ... (6)
<223> Xaa = Lysine or Arginine
<221> VARIANT
<222> (7) .. . (7)
<223> Xaa = Hydrophobic or neutral amino acid consisting
of I, L, V,Q,S
<221> VARIANT
<222> (8)...(9)
<223> Xaa = Lysine or Arginine
<221> VARIANT
<222> (10)...(10)
<223> Xaa = Hydrophobic or neutral amino acid consisting
of I, L, V, Q, S
<221> VARIANT
<222> (11)...(11)
<223> Xaa = Lysine or Arginine
<400> 2
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
1 5 10
<210> 3
<211> 12
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<221> VARIANT
<222> (1)...(5)
<223> Xaa = any amino acid
<221> HELIX
<222> (1)...(5)
<223> Alpha-helix
<221> VARIANT
<222>~(6)...(6)
<223> Xaa = Lysine or Arginine
<221> VARIANT
<222> (7)...(7)
<223> Xaa = Hydrophobic or neutral amino acid consisting
of I,L,V,Q,S
<221> VARIANT
<222> (8)...(8)
<223> Xaa = Lysinse or Arginine
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
3
<221> VARIANT
<222> (9)...(9)
<223> Xaa = Hydrophobic or neutral amino acid consisting
of I, L, V, Q, S
<221> VARIANT
<222> (10)...(12)
<223> Xaa = Lysine or Arginine
<400> 3
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
1 5 10
<210> 4
<21l> 10
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<221> VARIANT
<222> (1)...(5) _
<223> Xaa = any amino acid
<221> HELIX
<222> (1)...(5)
<223> Alpha-helix
<221> VARIANT
<222> (6)...(6)
<223> Xaa = Lysine or Arginine
<221> VARIANT
<222> (7)...(7)
<223> Xaa = Hydrophobic or neutral amino acid consisting
of I, L, V, Q, S
<221> VARIANT
<222> (8)...(10)
<223> Xaa = Lysine or Arginine
<400> 4
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
1 5 10
<210> 5
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<221> VARIANT
<222> (1)...(5)
<223> Xaa = Any amino acid
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
4
<221> HELIX
<222> (1)...(5)
<223> Alpha-helix
<221> VARIANT
<222> (6)...(6)
<223> Xaa = Lysine or Arginine
<221> VARIANT
<222> (7)...(7)
<223> Xaa = Hydrophobic or neutral amino acid consisting
of I, L, V,Q,S
<221> VARIANT
<222> (8)...(9)
<223> Xaa = Lysine or Arginine
<400> 5
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
1 5
<210> 6
<2I1> 7
<212> PRT
<223> Artificial Sequence
<220> ~ .
<223> Peptide that binds a hyalauronan -
<221> HELIX
<222> (1)...(5)
<223> Alpha-helix
<400> 6
Met Met Thr Val Leu Lys Arg
1 5
<210> 7
<211> 11
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<221> HELIX
<222> (1)...(5)
<223> Alpha-helix
<400> 7
Met Met Thr Val Leu Lys Val Lys Arg Leu Arg
1 5 10
<210> 8
<211> 12
<212> PRT
<213> Artificial Sequence
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
<220>
<223> Peptide that binds a hyalauronan
<221> HELIX
<222> (1)...(5)
<223> Alpha-helix
<400> S
Met Met Thr Val Leu Lys Val Lys Val Lys Arg Lys
1 5 10
<210> 9
<211> l0
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<221> HELIX
<222> (1)...(5)
<223> Alpha-helix
<400> 9
Met Met Thr Val Leu Lys Val Arg Lys Arg
1 ~ 5 10
<2l0> 10
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<221> HELIX
<222> (1)...(5)
<223> Alpha-helix
<400> 10
Met Met Thr Val Leu Lys Val Arg Lys
1 5
<210> 11
<211> 13
<212> PRT
<213> Homo sapiens
<400> 11
Lys Leu Gln Ala Thr Gln Lys Pro Leu Thr Glu Ser Lys
1 5 10
<210> 12
<211> 12
<212> PRT
<213> Homo Sapiens
<400> 12
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
6
Val Ser Ile G1u Lys Glu Lys Ile Asp Glu Lys Ser
1 5 10
<210> 13
<211> 6
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide developed based upon the TAM domain
(Transient Activator of MAP kinases)
<221> VARIANT
<222> (3) ... (3)
<223> Xaa = Any amino acid
<400> 13
Val Ser Xaa Lys Glu Lys
1 5
<210> 14
<211> 23
<212> PRT
<213> Mus musculus
<400> 14
Lys Leu Gln Ala Thr Gln Lys Asp Leu Thr Glu Ser Lys Gly Lys Ile
1 5 10 15
Val Gln Leu Glu Gly Lys Leu
<210> 15
<211> 14
<212> PRT
<213> Mus musculus
<400> l5
Lys Leu Gln A1a Thr Gln Lys Asp Leu Thr Glu Ser Lys Gly
1 5 10
<210> 16
<211> 25
<212> PRT
<213> Mus musculus
<400> 16
Val Ser Ile Glu Lys Glu Lys Ile Asp Glu Lys Cys Glu Thr G1u Lys
1 5 10 15
Leu Leu Glu Tyr Ile Gln Glu Ile Ser
20 25
<210> 17
<211> 12
<212> PRT
<213> Mus musculus
<400> 17
Val Ser Ile Glu Lys Glu Lys I1e Asp Glu Lys Cys
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
1 5 10
<210> 18
<211> 14
<212> PRT
<213> Homo sapien
<400> 18
Leu Lys Ser Lys Phe Ser Glu Asn Gly Asn Gln Lys Asn Leu
1 5 10
<210> 19
<211> 14
<212> PRT
<213> Homo Sapiens
<400> 19
Lys Leu Gln Val Thr Gln Arg Ser Leu Glu Glu Gln Lys Gly
1 5 10
<210> 20
<211> 14
<212> PRT
<213> Mus musculus
<400> 20
Leu Lys Ala Lys Phe Ser G1u Asp Gly His Gln Lys Asn Met
l 5 l0
<2l0> 21
<211> 14
<212> PRT
<213> Mus musculus
<400> 21
Gln Glu Arg Gly Thr Gln Asp Lys Arg Ile Gln Asp Met Glu
1 5 10
<210> 22
<211> 21
<212> PRT
<213> Homo sapien
<400> 22
Gly Thr Leu Lys Leu Asp Lys Leu Gly Ser Gln Ala Asp Thr Gly Gln
1 5 10 15
Lys Glu Leu Lys Gln
<210> 23
<211> 20
<212> PRT
<213> Homo sapien
<400> 23
Glu Ser Thr Asn Gln Glu Tyr Ala Arg Met Val Gln Asp Leu Gln Asn
1 5 10 15
Arg Ser Thr Leu
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
g
<210> 24
<211> 11
<212> PRT
<213> Homo sapien
<400> 24
Lys Leu Arg Ser Gln Leu Val Lys Arg Lys Gln
1 5 10
<210> 25
<211> 11
<212> PRT
<213> Artificial Sequence
<220>
<223> Scrambled hyalauron binding peptide
<400> 25
Arg Gln Lys Val Leu Lys Arg Gln Leu Lys Ser
1 5 l0
<210> 26
<211> 16
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<400> 26
Cys Ser Thr Met Met Ser Arg Ser His Lys Thr Arg Ser His His Val
1 5 10 15
<210> 27
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<400> 27
Arg Gly Gly Gly Arg Gly Arg Arg Arg
1 5
<210> 28
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<221> VARIANT
<222> (1)...(1)
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
9
<223> Xaa = Any basic amino acid
<221>VARIANT
<222>(2)...(8)
<223>Xaa = Any amino other an acidic
acid than amino
acid
<221>HELIX
<222>(2)...(8)
<223>Alpha-helix
<221>VARIANT
<222>(9)...(9)
<223>Xaa = Any basic acid
amino
<400>28
Xaa Xaa Xaa
Xaa
Xaa
Xaa
Xaa
Xaa
Xaa
1 5
<210>29
<211>11
<212>PRT
<213>Artificial Sequence
<220>
<223>Peptide compositionhat bindshyalauronan
t a
<221>VARIANT
<222>(1)...(2)
<223>Xaa = any basic acid
amino
<22l>VARIANT
<222>(3)...(4)
<223>Xaa = any amino other an acidic
acid than amino
acid
<221>VARIANT
<222>(5)...(7)
<223>Xaa = any basic acid
amino
<221>VARIANT
<222>(8)...(9)
<223>Xaa = any amino other an acidic
acid than amino
acid
<221>VARIANT
<222>(10)...(11)
<223>Xaa = any basic acid
amino
<400>29
Xaa Xaa Xaa
Xaa Xaa Xaa
Xaa
Xaa
Xaa
Xaa
Xaa
1 5 10
<210>30
<211>11
<212>PRT
<213>Artificial Sequence
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
1~
<220>
<223> Peptide composition that binds a hyalauronan
<400> 30
Lys Gln Lys Ile Lys His Val Val Lys Leu Lys
1 5 l0
<2l0> 31
<211> 10
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 31
Lys Leu Lys Ser Gln Leu Val Lys Arg Lys
1 5 10
<210> 32
<2l1> 10
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 32
Arg Tyr Pro Ile Ser Arg Pro Arg Lys Arg
1 5 10
<210> 33
<21l> 9
<212> PRT
<2l3> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 33
Lys Asn Gly Arg Tyr Ser Ile Ser Arg
1 5
<210> 34
<211> 13
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 34
Arg Asp Gly Thr Arg Tyr Val Gln Lys Gly Glu Tyr Arg
1 5 10
<210> 35
<211> 9
<212> PRT
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
11
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 35
Arg Arg Arg Cys Gly Gln Lys Lys Lys
1 5
<210> 36
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 36
Arg Gly Thr Arg Ser Gly Ser Thr Arg
1 5
<210> 37
<21l> 12
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
~400> 37
Arg Arg Arg Lys Lys Ile Gln Gly Arg Ser Lys Arg
1 5 10
<210> 38
<211> 10
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 38
Arg Lys Ser Tyr Gly Lys Tyr G1n Gly Arg
1 5 10
<210> 39
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 39
Lys Val Gly Lys Ser Pro Pro Val Arg
l 5
<210> 40
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
12
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 40
Lys Thr Phe Gly Lys Met Lys Pro Arg
1 5
<210> 41
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 41
Arg I1e Lys Trp Ser Arg Val Ser Lys
1 5
<210> 42
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 42
Lys Arg Thr Met Arg Pro Thr Arg Arg
1 5
<210> 43
<21l> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 43
Lys Val Gly Lys Ser Pro Pro Val Arg
1 5
<210> 44
<211> 10
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide composition that binds a hyalauronan
<400> 44
His Arg Glu Ala Arg Ser Gly Lys Tyr Lys
1 5 10
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
13
<210> 45
<211> 588
<212> DNA
<213> Homo sapien
<220>
<221> CDS
<222> (1)...(486)
<400> 45
gaa ttc gcg gcg gcg tcg acc aac aag ccc cct get gtt tcc ccg ggg 48
Glu Phe Ala Ala Ala Ser Thr Asn Lys Pro Pro Ala Val Ser Pro Gly
1 5 10 15
gtg gtc tcc cca acc ttt gaa ctt aca aat ctt cta aat cat cct gac 96
Val Val Ser Pro Thr Phe Glu Leu Thr Asn Leu Leu Asn His Pro Asp
20 25 30
cat tat gta gaa aca gag aac att cag cat ctc aca gac ccg get cta 144
His Tyr Val Glu Thr Glu Asn Ile Gln His Leu Thr Asp Pro Ala Leu
35 40 45
gca cat gtg gat aga ata agc caa gcc cgg aaa ctg agt atg gga tct 192
Ala His Val Asp Arg Ile Ser Gln Ala Arg Lys Leu Ser Met Gly Ser
50 55 60
gat gat get gcc tac aca caa get ctg ctg gtg cac cag aag gcc agg 240
Asp Asp Ala A1a Tyr Thr Gln Ala Leu Leu Val His Gln Lys Ala Arg
65 70 75 80
atggaa cggcttcaaaga gagctcgag atgcaaaag aaaaagctggat 288
MetGlu ArgLeuGlnArg GluLeuGlu MetGlnLys LysLysLeuAsp
85 90 95
aaactc aaatctgaggtc aatgagatg gaaaataat ctaactcgaagg 336
LysLeu LysSerGluVal AsnGluMet GluAsnAsn LeuThrArgArg
100 105 110
cgcctg aagagatcaaat tccatttcc cagataccg tcactcgaagaa 384
ArgLeu LysArgSerAsn SerI1eSer GlnIlePro SerLeuGluGlu
115 120 125
atgcag cagttgagaagt tgtaataga caactccag attgacattgac 432
MetGln GlnLeuArgSer CysAsnArg GlnLeuGln IleAspIleAsp
130 135 140
tttgac tgcttaaccaaa gaaattgca tctttttca agcccgaggacc 480
PheAsp CysLeuThrLys GluT1eAla SerPheSer SerProArgThr
l45 150 155 160
acattt taaccccagc caatattgga tttgtaggcc
536
gctattcata
acttttatga
ThrPhe
ctgtgccacc aaaacccaaa gatcaaaggt ccaccatcaa aggtcgacgc gg 588
<210> 46
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
14
<211> 162
<212> PRT
<213> Homo sapien
<400> 46
Glu Phe Ala Ala Ala 5er Thr Asn Lys Pro Pro Ala Val Ser Pro Gly
1 5 10 15
Val Val Ser Pro Thr Phe Glu Leu Thr Asn Leu Leu Asn His Pro Asp
20 25 30
His Tyr Val Glu Thr Glu Asn Ile Gln His Leu Thr Asp Pro Ala Leu
35 40 45
Ala His Val Asp Arg Ile Ser Gln Ala Arg Lys Leu Ser Met Gly Ser
50 55 60
Asp Asp Ala Ala Tyr Thr Gln Ala Leu Leu Val His Gln Lys Ala Arg
65 70 75 80
Met Glu Arg Leu Gln Arg Glu Leu Glu Met Gln Lys Lys Lys Leu Asp
85 90 95
Lys Leu Lys Ser Glu Val Asn Glu Met Glu Asn Asn Leu Thr Arg Arg
100 105 110
Arg Leu Lys Arg Ser Asn Ser Ile Ser Gln Ile Pro Ser Leu Glu Glu
115 120 125
Met Gln Gln Leu Arg Ser Cys Asn Arg Gln Leu Gln Ile Asp Ile Asp
130 135 140
Phe Asp Cys Leu Thr Lys Glu Ile Ala Ser Phe Ser Ser Pro Arg Thr
145 150 155 160
Thr Phe
<210> 47
<211> 725
<212> PRT
<213> Homo sapiens
<400> 47
Met Ser Phe Pro Lys Ala Pro Leu Lys Arg Phe Asn Asp Pro Ser Gly
1 5 10 15
Cys Ala Pro Ser Pro Gly Ala Tyr Asp Val Lys Thr Leu Glu Val Leu
20 25 30
Lys Gly Pro Val Ser Phe Gln Lys Ser Gln Arg Phe Lys G1n G1n Lys
35 40 45
Glu Ser Lys Gln Asn Leu Asn Val Asp Lys Asp Thr Thr Leu Pro Ala
50 55 60
Ser Ala Arg Lys Val Lys Ser Ser Glu Ser Lys Lys Glu Ser Gln Lys
65 70 75 80
Asn Asp Lys Asp Leu Lys Ile Leu Glu Lys Glu Ile Arg Val Leu Leu
85 90 95
Gln Glu Arg Gly Ala Gln Asp Arg Arg Ile Gln Asp Leu Glu Thr Glu
100 105 110
Leu Glu Lys Met Glu Ala Arg Leu Asn Ala Ala Leu Arg Glu Lys Thr
115 120 125
Ser Leu Ser Ala Asn Asn Ala Thr Leu Glu Lys Gln Leu Ile Glu Leu
130 135 140
Thr Arg Thr Asn Glu Leu Leu Lys Ser Lys Phe Ser G1u Asn Gly Asn
145 150 155 160
Gln Lys Asn Leu Arg Ile Leu Ser Leu Glu Leu Met Lys Leu Arg Asn
165 170 175
Lys Arg Glu Thr Lys Met Arg Gly Met Met Ala Lys Gln Glu Gly Met
180 185 190
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
Glu Met Lys Leu Gln Val Thr Gln Arg Ser Leu Glu Glu Ser Gln Gly
195 200 205
Lys Ile Ala Gln Leu Glu Gly Lys Leu Val Ser Ile Glu Lys Glu Lys
210 215 220
Ile Asp Glu Lys Ser Glu Thr Glu Lys Leu Leu Glu Tyr Ile Glu Glu
225 230 235 240
Ile Ser Cys Ala Ser Asp Gln Val Glu Lys Tyr Lys Leu Asp Ile Ala
245 250 255
Gln Leu Glu Glu Asn Leu Lys Glu Lys Asn Asp Glu Ile Leu Ser Leu
260 265 270
Lys Gln Ser Leu Glu Glu Asn Ile Val Ile Leu Ser Lys Gln Val Glu
275 280 285
Asp Leu Asn Val Lys Cys Gln Leu Leu Glu Lys Glu Lys Glu Asp His
290 295 300
Val Asn Arg Asn Arg Glu His Asn Glu Asn Leu Asn Ala Glu Met Gln
305 310 315 320
Asn Leu Lys Gln Lys Phe Ile Leu Glu Gln Gln Glu His Glu Lys Leu
325 330 335
Gln Gln Lys Glu Leu Gln Ile Asp Ser Leu Leu Gln Gln Glu Lys Glu
340 345 350
Leu Ser Ser Ser Leu His G1n Lys Leu Cys Ser Phe Gln Glu Glu Met
355 360 365
Val Lys Glu Lys Asn Leu Phe Glu Glu Glu Leu Lys Gln Thr Leu Asp
370 375 380
Glu Leu Asp Lys Leu Gln Gln Lys Glu Glu Gln Ala Glu Arg Leu Val
385 390 395 400
Lys G1n Leu Glu Glu Glu Ala Lys Ser Arg Ala Glu G1u Leu Lys Leu
405 410 415
Leu Glu Glu Lys Leu Lys Gly Lys Glu Ala Glu Leu Glu Lys Ser Ser
420 425 430
Ala Ala His Thr Gln Ala Thr Leu Leu Leu G1n Glu Lys Tyr Asp Ser
435 440 445
Met Val Gln Ser Leu Glu Asp Val Thr Ala Gln Phe Glu Ser Tyr Lys
450 455 460
Ala Leu Thr Ala Ser Glu Ile Glu Asp Leu Lys Leu Glu Asn Ser Ser
465 470 475 480
Leu Gln Glu Lys Ala Ala Lys Ala Gly Lys Asn Ala Glu Asp Val Gln
485 490 495
His Gln Ile Leu Ala Thr Glu Ser Ser Asn Gln Glu Tyr Va1 Arg Met
500 505 510
Leu Leu Asp Leu Gln Thr Lys Ser Ala Leu Lys Glu Thr Glu Ile Lys
515 520 525
Glu Ile Thr Val Ser Phe Leu Gln Lys Ile Thr Asp Leu Gln Asn Gln
530 535 540
Leu Lys Gln Gln Glu Glu Asp Phe Arg Lys Gln Leu Glu Asp Glu Glu _
545 550 555 560
Gly Arg Lys Ala Glu Lys Glu Asn Thr Thr Ala Glu Leu Thr Glu Glu
565 570 575
Ile Asn Lys Trp Arg Leu Leu Tyr Glu Glu Leu Tyr Asn Lys Thr Lys
580 585 590
Pro Phe Gln Leu Gln Leu Asp Ala Phe Glu Val Glu Lys Gln Ala Leu
595 600 605
Leu Asn Glu His Gly Ala Ala Gln Glu Gln Leu Asn Lys Ile Arg Asp
610 615 620
Ser Tyr Ala Lys Leu Leu Gly His Gln Asn Leu Lys Gln Lys Ile Lys
625 630 635 640
His Val Val Lys Leu Lys Asp Glu Asn Ser Gln Leu Lys Ser Glu Val
645 650 655
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
16
Ser Lys Leu Arg Cys Gln Leu Ala Lys Lys Lys Gln Ser Glu Thr Lys
660 665 670
Leu Gln Glu Glu Leu Asn Lys Val Leu Gly Ile Lys His Phe Asp Pro
675 680 685
Ser Lys Ala Phe His His Glu Ser Lys Glu Asn Phe Ala Leu Lys Thr
690 695 700
Pro Leu Lys Glu Gly Asn Thr Asn Cys Tyr Arg Ala Pro Met Glu Cys
705 710 7l5 720
Gln Glu Ser Trp Lys
725
<210> 48
<211> 631
<212> PRT
<213> Mus musculus
<400> 48
Met Arg Ala Leu Ser Leu Glu Leu Met Lys Leu Arg Asn Lys Arg Glu
1 5 10 15
Thr Lys Met Arg Ser Met Met Val Lys Gln Glu Gly Met Glu Leu Lys
20 25 30
Leu Gln Ala Thr Gln Lys Asp Leu Thr Glu Ser Lys Gly Lys Ile Val
35 40 45
Gln Leu Glu Gly Lys Leu Val Ser Ile Glu Lys Glu Lys I1e Asp Glu
50 55 60
Lys Cys Glu Thr Glu Lys Leu Leu Glu Tyr Ile Gln Glu Ile Ser Cys
65 70 75 80
Ala Ser Asp Gln Val Glu Lys Cys Lys Val Asp Ile Ala Gln Leu flu
85 90 95
Glu Asp Leu Lys Glu Lys Asp Arg Glu I1e Leu Ser Leu Lys Gln Ser
100 105 110
Leu Glu G1u Asn Ile Thr Phe Ser Lys Gln Ile Glu Asp Leu Thr Val
115 120 125
Lys Cys Gln Leu Leu Glu Thr Glu Arg Asp Asn Leu Val Ser Lys Asp
130 135 140
Arg Glu Arg Ala Glu Thr Leu Ser Ala Glu Met Gln Ile Leu Thr Glu
145 150 155 160
Arg Leu Ala Leu Glu Arg Gln Glu Tyr Glu Lys Leu G1n Gln Lys G1u
165 170 175
Leu Gln Ser Gln Ser Leu Leu Gln Gln Glu Lys Glu Leu Ser Ala Arg
180 185 190
Leu Gln Gln Gln Leu Cys Ser Phe Gln Glu Glu Met Thr Ser Glu Lys
195 200 205
Asn Val Phe Lys Glu Glu Leu Lys Leu Ala Leu A1a Glu Leu Asp Ala
210 215 220
Val Gln Gln Lys Glu Glu Gln Ser Glu Arg Leu Val Lys Gln Leu Glu
225 230 235 240
Glu Glu Arg Lys Ser Thr Ala Glu Gln Leu Thr Arg Leu Asp Asn Leu
245 250 255
Leu Arg Glu Lys Glu Val Glu Leu Glu Lys His Ile Ala Ala His Ala
260 265 270
Gln Ala Ile Leu Ile A1a Gln Glu Lys Tyr Asn Asp Thr Ala Gln Ser
275 280 285
Leu Arg Asp Val Thr Ala Gln Leu Glu Ser Val Gln Glu Lys Tyr Asn
290 295 300
Asp Thr Ala Gln Ser Leu Arg Asp Val Thr Ala Gln Leu Glu Ser Glu
305 310 315 320
Gln Glu Lys Tyr Asn Asp Thr Ala Gln Ser Leu Arg Asp Val Thr Ala
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
1~
325 330 335
Gln Leu Glu Ser Glu Gln Glu Lys Tyr Asn Asp Thr Ala Gln Ser Leu
340 345 350
Arg Asp Val Thr Ala Gln Leu Glu Ser Val Gln Glu Lys Tyr Asn Asp
355 360 365
Thr Ala Gln Ser Leu Arg Asp Val Ser Ala Gln Leu Glu Ser Tyr Lys
370 375 380
Ser Ser Thr Leu Lys Glu Ile Glu Asp Leu Lys Leu Glu Asn Leu Thr
385 390 395 400
Leu Gln Glu Lys Val Ala Met Ala Glu Lys Ser Val Glu Asp Val Gln
405 410 415
Gln Gln Ile Leu Thr Ala Glu Ser Thr Asn Gln Glu Tyr Ala Arg Met
420 425 430
Va1 Gln Asp Leu Gln Asn Arg Ser Thr Leu Lys Glu Glu Glu Ile Lys
435 440 445
Glu Ile Thr Ser Ser Phe Leu Glu Lys Ile Thr Asp Leu Lys Asn Gln
450 455 460
Leu Arg Gln Gln Asp Glu Asp Phe Arg Lys Gln Leu Glu Glu Lys Gly
465 470 475 480
Lys Arg Thr Ala Glu Lys Glu Asn Val Met Thr Glu Leu Thr Met Glu
485 490 495
21e Asn Lys Trp Arg Leu Leu Tyr Glu Glu Leu Tyr Glu Lys Thr Lys
500 505 510
Pro Phe Gln Gln Gln Leu Asp Ala Phe Glu Ala Glu Lys Gln Ala Leu
515 520 525
Leu Asn Glu His Gly Ala Thr Gln Glu Gln Leu Asn Lys Ile Arg Asp
530 535 540
Ser Tyr Ala Gln Leu Leu Gly His G1n Asn Leu Lys Gln Lys Ile Lys
545 550 555 560
His Val Val Lys Leu Lys Asp Glu Asn Ser Gln Leu Lys Ser Glu Val
565 570 575
Ser Lys Leu Arg Ser Gln Leu Val Lys Arg Lys Gln Asn Glu Leu Arg
580 585 590
Leu Gln Gly Glu Leu Asp Lys Ala Leu Gly Ile Arg His Phe Asp Pro
595 600 605
Ser Lys Ala Phe Cys His Ala Ser Lys Glu Asn Phe Thr Pro Leu Lys
610 615 620
Glu Gly Asn Pro Asn Cys Cys
625 630
<210> 49
<211> 11
<212> PRT
<213> Homo sapien
<400> 49
Val Ser Ile Glu Lys G1u Lys Ile Asp Glu Lys
1 5 10
<210> 50
<211> 21
<212> PRT
<213> Unknown
<220>
<223> Peptide used in competition binding assay
<400> 50
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
18
Gln Glu Lys Tyr Asn Asp Thr Ala Gln Ser Leu Arg Asp Val Thr Ala
1 5 10 25
Gln Leu Glu Ser Val
<2l0> 51
<211> 32
<212> PRT
<213> Unknown
<220>
<223> Peptide used in competition binding assay
<400> 51
Lys Gln Lys Ile Lys His Val Val Lys Leu Lys Asp Glu Asn Ser Gln
1 5 10 l5
Leu Lys Ser Glu Val Ser Lys Leu Arg Ser G1n Leu Val Lys Arg Lys
20 25 30
<210> 52
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Primer for PCR amplication of collagen I
<400> 52
cgatgtcgct atccagctga 20
<210> 53
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Primer for PCR amplication of collagen III
<400> 53
atcagtcagc catctaccac c 21
<210> 54
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Primer for PCR amplication of ED-1
<400> 54
tggcaggaca gtagtcgc ~ 1g
<210> 55
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
19
<223> Primer for PCR amplication of ED-1
<400> 55
aaggctgctg ttgaaaggac g 21
<210> 56
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<400> 56
Arg Gly Gly Gly Arg Gly Gly Arg Arg
1 5
<210> 57
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<400> 57
Arg Gly Gly Gly Arg Gly Gly Gly Arg
1 5
<210> 58
<211> 9
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<400> 58
Arg Gly Gly Gly Gly G1y G1y Gly Arg
l 5
<210> 59
<211> 9
<212> PRT
<213> Homo sapien
<400> 59
Lys Leu Arg Ser Gln Leu Val Lys Arg
1 5
<210> 60
<211> 9
<212> PRT
<213> Homo sapien
<400> 60
Lys Gln Lys Ile Lys His Val Val Lys
1 5
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
<210> 61
<211> 9
<212> PRT
<213> Homo sapien
<400> 61
Arg Ser His Lys Thr Arg Ser His His
1 5
<210> 62
<211> 7
<212> PRT
<2l3> Homo sapien
<400> 62
Arg Pro His Phe His Lys Arg
1 5
<210> 63
<211> 11
<2l2> PRT
<213> Homo sapien
<400> 63
Arg Lys Ile Gln Lys His Lys Thr Ile Pro Lys
l 5 ~ 10
<210> 64
<211> 9
<212> PRT
<213> Homo Sapiens
<400> 64
Lys Val Gly Arg Lys Val Phe Ser Lys
1 5
<210> 65
<211> 9
<212> PRT
<213> Homo Sapiens
<400> 65
Lys Cys Ser Val Gln Thr Leu Leu Arg
1 5
<2l0> 66
<211> 9
<212> PRT
<213> Homo Sapiens
<400> 66
Arg Thr His Leu Lys His Val Leu Arg
1 5
<210> 67
<211> 9
<212> PRT
CA 02448483 2003-11-24
WO 02/28415 PCT/IB00/01534
21
<2l3> Homo sapiens
<400> 67
Lys Asn Ala Ile Asn Asn Gly Val Arg
l 5
<210> 68
<211> 9
<212> PRT
<213> Homo sapiens
<400> 68
Lys Gly Gln Ile Asn Asn Ser Ile Lys
l 5
<210> 69
<211> 9
<2l2> PRT
<213> Homo sapiens
<400> 69
Arg Val Arg Gly Arg Ala Lys Leu Arg
1 5
<210> 70
<211> 15
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<400> 70
Ser Thr Met Met Ser Arg Ser His Lys Thr Arg Ser His His Val
1 5 10 15
<210> 71
<211> 32
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide that binds a hyalauronan
<400> 71
Cys Ser Thr Met Met Ser Arg Ser His Lys Thr Arg Ser His His Val
1 5 10 15
Cys Ser Thr Met Met Ser Arg Ser His Lys Thr Arg Ser His His Val
20 25 30
<210> 72
<211> 12
<212> PRT
<213> Homo sapien
<400> 72
Gly Ala His Trp Gln Phe Asn Ala Leu Thr Val Arg
1 5 10