Note: Descriptions are shown in the official language in which they were submitted.
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
HUMAN MITOCHONDRIAL DNA POLYMORPHISMS, HAPLOGROUPS,
ASSOCIATIONS WITH PHYSIOLOGICAL CONDITIONS, AND GENOTYPING
A BRAYS
CROSS-REFERENCE TO RELATED APPLICATIONS
This application claims priority to U.S. Patent Applications Serial No.
60/316333
filed August 30, 2001 and Serial No. 60/380,546 filed May 13, 2002, and to
Canadian Patent
Application No. 2,356,536 filed on August 31, 2001, which are, hereby
incorporated in their
entirety by reference to the extent not inconsistent with the disclosure
herein.
STATEMENT REGARDING FEDERALLY SPONSORED RESEARCH
This invention was made in part with funding from the United States Government
(NI~-I grants AG13154, HL64017, NS21328, and NS37167). The United States
Government
may have certain rights therein.
BACKGROUND OF THE INVENTION
Human mitochondria) DNA (mtDNA) is maternally inherited. Mutations accumulate
sequentially in radiating lineages creating branches on the human evolutionary
tree. Using
sequences of mtDNA, human populations are divisible evolutionarily into
haplogroups
(Wallace, D.C. et al. (1999) Gezze 238:211-230; Ingman M. et al., (2000)
Nature 408:708-
713; Maca-Meyer, N. (August 200I) .BioMed Central 2:13; T. G. Schurr et al.,
(1999)
American Jozzrnal of Physical Anthropology 108:1-39; and V. Macaulay et al.,
(1999)
American Journal of Human Genetics 64:232-249). Related haplogroups can be
combined
into macro-haplogroups. Haplogroups can be subdivided into subhaplogroups. The
complete
Cambridge mitochondria) DNA sequence may be found at MITOMAP,
http://www.gen.emory.edu/cgi-gin/MITOMAP, Genbank accession no. J01415, and is
provided in SEQ ID N0:2. Also see Andrews et al. (1999), "Reanalysis and
Revision of the
Cambridge Reference Sequence for Human Mitochondria) DNA," Nature Genetics 23
:147.
Publications on the subject of mitochondria) biology include: Scheffler, LE.
(1999)
Mitochondria, Wiley-Liss, NY; Lestienne P Ed.; Mitochondria) Diseases: Models
and
Methods, Springer-Verlag, Berlin; Methods in EnzYmology (2000) 322:Section V
Mitochondria and Apoptosis, Academic Press, CA; Mitochondria and Cell Death
(1999)
Princeton University Press, NJ; Papa S, Ferruciio G, and Tager J Eds.;
Frontiers of Cellular
1
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Bioenergetics~ Molecular Biolog~Biochemistry, and Physiopatholo~y, Kluwer
Academic /
Plenum Publishers, NY; Lemasters, J. and Nieminen, A. (2001) Mitochondria in
Pathogenesis, Kluwer Academic / Plenum Publishers, NY; MITOMAP,
http://www.gen.emory.edu/cgi-gin/MITOMAP; Wallace, D.C. (2001) "A
mitochondria)
paradigm for degenerative diseases and ageing" Novartis Foundation Symposium
235:247-
266; Wallace, D.C. (1997) "Mitochondria) DNA in Aging and Disease" Scientific
American
August 277:40-47; Wallace, D.C. et al., (1998) "Mitochondria) biology,
degenerative
diseases and aging," BioFactors 7:187-190; Heddi, A. et al., (1999)
"Coordinate Induction of
Energy Gene Expression in Tissues of Mitochondria) Disease Patients" JBC
274:22968-
22976; Wallace, D.C. (1999) "Mitochondria) Diseases in Man and Mouse" Science
283:1482-
1488; Saraste, M. (1999) "Oxidative Phosphorylation at the fin de siecle"
Science 283:1488-
1493; Kokoszka et. al. (2001) "Increased mitochondria) oxidative stress in the
Sod2 (+/-)
mouse results in the age-related decline of mitochondria) function culminating
in increased
apoptosis" PNAS 98:2278-2283; Wallace, D.C. (2001) Mental Retardation and
Developmental Disabilities 7:158-166; Wallace, D.C. (2001) Am. J. Med. Gen.
106:71-93;
Wei, Y-H et al. (2001) Chinese Medical Journal (Taipei) 64:259-270; and
Wallace, D.C.
(2001) EuroMit 5 Abstract.
Certain mitochondria) mutations have been associated with physiological
conditions ,
(U.S. Patent 6,280,966 issued on August 28, 2001; U.S. Patent 6,140,067 issued
on October
31, 2000; U.S. Patent 5,670,320; U.S. Patent 5,296,349; U.S. Patent
5,1.85,244; U.S. Patent
5,494,794; Wallace, D.C. (1999) Science 283:1482-1488; Brown, M.D. et al.
(2001)
American Society for Human Genetics Poster #2332; Brown, M.D. et al., (2001)
Human
Genet. 109:33-39; and Brown, M.D. et al. (January 2002) Human Genet. 110:130-
138),
Wallaee, D.C. et al. (1999) Gene 238:211-230 describes analysis of LHON
mutants.
Grossman, L.I. et al. (2001) Molecular Phylogenetics and Evolution 18(1):26-
36, describes
changes in the biochemical machinery for aerobic energy metabolism. Kalman, B.
et al.
(1999) Acta Neurol. Scand. 99(1):16-25 describes mitochondria) mutations and
multiple
sclerosis (MS). Wei, Y.H. et al. (2001) Chinese Medical Journal 64:259-270
describes
recent results in support of the mitochondria) theory of aging.
Ivanova, R. et al. (1998) Geronotology 44:349 describes mitochondria)
haplotypes
and longevity in a French population. Tana.ka, M. et aI. (I998) Lancet 351:185-
186 describes
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
longevity and haplogroups in a Japanese population. De Benedictis, G. et al.
(1999) FASEB
13:1532-1536 describes haplogroups and longevity in an Italian population.
Rose, G. et al.
(2001) European Journal of Human Genetics 9:701-707 describes haplogroup J in
centenarians. Ross, O.A. et al. (2001) Experimental Gerontology 36(7):1161-
1178 describes
haplotypes and longevity in an Irish population.
Haplogroup T has been associated with reduced sperm motility in European males
(E.
Ruiz-Pesini et al., [2000] American Journal of Human Genetics 67:682-696), the
tRNAG~° np
4336 variant in haplogroup H is associated with late-onset Alzheimer Disease (
J. M.
Shoffner et al., [1993] Genomics 17:171-184).
Taylor, R.W. (1997) J. of Bioeraergetics arad Biomembranes 29(2):195-205
describes
methods for treating mitochondria) disease. Collombet, J. and Coutelle, C.
(1998) Molecular
Medicine Today 4(1):1-8 describes gene therapy for mitochondria) disorders,
including using
cell fusion to introduce healthy mitochondria. Owen, R. and Flotte, T.R.
(2001) Antioxidants
and Redox Signaling 3(3):451-460 discuss approaches and limitations to gene
therapy for
mitochondria) diseases.
Human mitochondria) DNA sequence variation, except that which has been
associated
with particular diseases, has not been associated with specific phenotypic
conditions, has
been considered neutral, and has been used to reconstruct human phylogenies
(Henry Gee,
"Statistical Cloud over African Eden," (13 February 1992) Nature 355:f83;
Marcia Barinaga,
"'African Eve Backers Beat a Retreat," (7 February 1992). Science, 255:687; S.
Blair Hedges
et al., "Human Origins and Analysis of Mitochondria) DNA Sequences," (7
February 1992)
Science, 255:737-739; Allan C. Wilson and Rebecca L. Cann, "The Recent African
Genesis
of Humans," (April 1992) Scientific American, 68). The average number of base
pair
differences between two human mitochondria) genomes is estimated to be from
9.5 to 66
(Zeviani M. et al. (1998) "Reviews in molecular medicine: Mitochondria)
disorders,"
Medicine 77:59-72).
The D-loop is the most variable region in the mitochondria) genome, and the
most
polymorphic nucleotide sites within this loop are concentrated in two
'hypervariable
segments', HVS-I and HVS-II (Wilkinson-Herbots, H.M. et al., (1996) "Site 73
in
hypervariable region 1I of the human mitochondria) genome and the origin of
European
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
populations," Ann Hum Genet 60:499-508). Population-specific, neutral mtDNA
variants
have been identified by surveying mtDNA restriction site variants or by
sequencing
hypervariable segments in the displacement loop. Restriction analysis using
fourteen
restriction endonucleases allowed screening of 1S-20% of the mtDNA sequence
for
variations (Chen Y.S. et al., (1995) "Analysis of mtDNA variation in African
populations
reveals the most ancient of all human continent-specific haplogroups," Am JHum
Genet
57:133-149). The large majority of mtDNA sequence data published to date are
limited to
HVS-I. Bandelt, H.J. et al., (1995) "Mitochondria) portraits of human
populations using
median networks" Genetics 141:743-753).
The coding and classification system that has been used for mtDNA haplogroups
refers primarily to the information provided by RFLPs and the hypervariable
segments of the
control region. (Torroni, A. et al. (1996) "Classification of European mtDNAs
from an
analysis of three European populations," Genetics 144:1835-1850 and Richards
MB et al.,
(1998) "Phylogeography of mitochondria) DNA in western Europe," Ann Hum Genet
62:241-
260.)
Methods are known for testing the likelihood of neutrality of mutations
(Tajima, F.
(1989) Genetics 123:585-S9S; Fu, Y. and Li, W. (1993} Genetics 133:693-709;
Li, W, et al.
(1985) Mol. Biol. Evol. 2(2):150-174; and Nei, M. and Gojobori, T. (1986) Mol.
Biol. Evol.
3(5):418-426). All of the methods in these publications are used to compare
datasets taken
from separate groups. None of these methods are used to analyze a dataset not
containing
data representing an outgroup.
Wise, C.A. et al. (1998) Genetics 148:409-421, describes neutrality analysis
of the
human mitochondria) NADH Dehydrogenase Subunit 2 gene, when compared to the
NADH
Dehydrogenase Subunit 2 gene from chimpanzees. Templeton, A.R. (1996) Genetics
144:1263-1270, describes neutrality analysis of the human mitochondria)
Cytochrome
Oxidase II (COXII) gene when compared to the COXII gene in hominoid primates.
Messier,
W. and Stewart, C. (I997) Nature 385:151-154 describes neutrality analysis of
primate
lysozymes. Endo, T. et al. (1996) Mol. Biol. Evol. 13(5):685-690 describes
large-scale
neutrality analysis of sequences from DDBJ, EMBL, and GenBank databases.
Hughes, A.L.
and Nei, M. (1988) Nature 335:167-170 describes neutrality analysis of MHC
Class I loci.
Nachman, M.W. (1996) Genetics 142:953-963 describes neutrality analysis of the
human
4
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
mitochondrial NADH Dehydrogenase subunit 3 (NADH3) gene, when compared to the
NADH Dehydrogenase subunit 3 gene from chimpanzees. Nachman, M.W. et al.
(1994)
Proc. Natl. Acad. Sci. USA 76:5269-5273 describes neutrality analysis of the
mitochondrial
NADH dehydrogenase subunit 3 gene in 3 strains of mouse. Rand, D.M. et al.
(1994)
Genetics 138:741-756; Ballard, J.W.O. and Kreitman, M. (1994) Genetics 138:757-
772; and
Kaneko, M.Ir. et al. (1993) Genet. Res. 61:195-204, describe neutrality
analysis for
mitochondria) NADH dehydrogenase subunit 5, Cytochrome b, and ATPase6 in
strains of
Drosophila.
In the above-mentioned publications, neutrality testing, including Ka/KS
analysis, has
not been applied for the purpose of identifying disease-associated mutations.
Populations for
neutrality testing analysis were identified by observation of normal
phenbtypic variation.
Neutrality testing has been performed to determine whether a gene is under
selection. None
of these publications describe neutrality analysis with the purpose of
identifying phenotype-
associated mutations, and no suspected phenotype-associated mutations were
identified.
US Patent 6,228,586 (issued May 8, 2001) and US Patent 6,280,953 (issued
August
28, 2001) describe methods for identifying polynucleotide and polypeptide
sequences in
human and/or non-human primates, which may be associated with a physiological
condition.
The methods employ comparison of human and non-human primate sequences using
statistical methods. U.S. Patent 6,274,319 (issued August 14, 2001) describes
I~.a/KS methods
for identifying polynucleotide and polypeptide sequences that may be
associated with
commercially or aesthetically relevant traits in domesticated plants or
animals. The methods
employ comparison of homologous genes from the domesticated organism and its
wild
ancestor to identify evolutionarily significant changes. In the above-
mentioned publications,
neutrality testing, including KalKS analysis, is only applied to
interspecific, nat intraspecific,
comparisons, and only genes from the nuclear genome, not from organelle
genomes, are
analyzed.
Methods for constructing peptide and nucleotide libraries are well known to
the art,
e.g. as described in U.S. Patents 6,156,511 and 6,130,092. Sequencing methods
are also
known to the art, e.g., as described in U.S. Patent 6,087,095. Arrays of
nucleic acid have
been used for sequencing and for identifying exceptional alleles including
disease-associated
alleles. Nucleic acid arrays have been described, e.g., in patent nos.: U.S.
5,837,832, U.S.
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
5,807,522, U.S. 6,007,987, U.S. 6,110,426, WO 99/05324, 99/05591, WO 00/58516,
WO
95/11995, WO 95135505A1, WO 99/42813, JP10503841T2, GR3030430T3, ES2134481T3,
EP804731B1, DE69509925C0, CA2192095AA, AU2862995A1, AU709276B2, AT180570,
EP 1066506, and AU 2780499. Computational methods are useful for analyzing
hybridization results, e.g., as described in PCT Publication WO 99105574, and
U.S. Patents
5,754,524; 6228,575; 5,593,839; and 5,856,101. Methods for screening for
disease markers
are also known to the art, e.g. as described in U.S. Patents 6,228,586;
6,160,104; 6,083,698;
6,268,398; 6,228,578; and 6,265,174.
The development of microarray technologies has stemmed from the desire to
examine
very large numbers of nucleic acid probe sequences simultaneously, in an
effort to obtain
information about genetic mutations, gene expression or nucleic acid
sequences. Microarray
technologies are intimately connected with the Human Genome Proj ect, which
has
development of rapid methods of nucleic acid sequencing and genome analysis as
key
objectives (E. Marshall, (1995) Science 268:1270), as well as elucidation of
sequence-
function relationships (M. Schena et al., (1996) Proc. Nat'1. Acad. ~'ci. USA,
93:10614).
Microarray hybridization of PCR-amplified fragments to allele-specific
oligonucleotide
(ASO) probes is widely used in large-scale single nucleotide polymorphism
(SNP)
genotyping (Huber M. et al. (2002) Analytical Bioche~raistsy 303:25-33 and
Southern, E.M.
(1996) Trends Genet. 12:110-115).
The Affymetrix GeneChip HuSNPTM Array enables whole-genome surveys by
simultaneously tracking nearly 1,500 genetic variations, known as single
nucleotide
polymorphisms (SNPs), dispersed throughout the genome. The HuSNP Affymetrix
Array is
being used for familial linkage studies that aim to map inherited disease or
drug .
susceptibilities as well as for tracking de ~vvo genetic alterations. For
genotyping, arrays rely
on multiple probes to interrogate individual nucleotides in a sequence. The
identity of a
target base can be deduced using four identical probes that vary only in the
target position,
each containing one of the four possible bases. Alternatively, the presence of
a consensus
sequence can be tested using one or two probes representing specific alleles.
To genotype
heterozygous or genetically mixed samples, arrays with many probes can be
created to
provide redundant information.
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Arrays, also called DNA microarrays or DNA chips, are fabricated by high-speed
robotics, generally on glass but sometimes on nylon substrates, for which
probes (Phimister,
B. (1999) Nature Genetics 21s:1-60) with known identity are used to determine
complementary binding. An experiment with a single DNA chip can provide
researchers
information on thousands of genes simultaneously. There are several steps in
the design and
implementation of a DNA array experiment. Many strategies have been
investigated at each
of these steps: 1) DNA types; 2) Chip fabrication; 3) Sample preparation; 4)
Assay; 5)
Readout; and 6) Software (informatics).
There are two major application forms for the array technology: 1)
Determination of
expression level (abundance) of genes; and 2) Identification of sequence (gene
/ gene
mutation). There appear to be two variants of the array technology, in terms
of intellectual
pxoperty, of arrayed DNA sequence with known identity: Format I consists of
probe cDNA
(5005,000 bases long) immobilized to a solid surface such as glass using robot
spotting and
exposed to a set of targets either separately or in a mixture. This method,
"traditionally"
called DNA microarray, is widely considered as having been developed at
Stanford
University. (R. Ekins and F.W. Chu "Microarrays: their origins and
applications," [1999)
Trends in Bioteclanology, 17:217-218). Format IC consists of an array of
oligonucleotide
(2080-mer oligos) or peptide nucleic acid (PNA) probes synthesized either in
situ (on-chip)
or by conventional synthesis followed by on-chip immobilization. The array is
exposed to
labeled sample DNA, hybridized, and the identity/abundance of complementary
sequences is
determined. This method, "historically" called DNA chips, was developed at
Affymetrix,
Inc., which sells its photolithographically fabricated products under the
GeneChip~
trademark. Many companies are manufacturing oligonucleotide-based chips using
alternative
in-situ synthesis or depositioning technologies.
Probes' on arrays can be hybridized with fluorescently-labeled target
polynucleotides
and the hybridized array can be scanned by means of scanning fluorescence
microscopy. The
fluorescence patterns are then analyzed by an algorithm that determines the
extent of
mismatch content, identifies polymorphisms, and provides some general
sequencing
information (M. Chee et al., [1996] Science 274:610). Selectivity is afforded
in this system
by low stringency washes to rinse away non-selectively adsorbed materials.
Subsequent
analysis of relative binding signals from array elements determines where base-
pair
mismatches may exist. This method then relies on conventional chemical methods
to
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
maximize stringency, and automated pattern recognition processing is used to
discriminate
between fully complementary and partially complementary binding.
Devices such as standard nucleic acid microarrays or gene chips, require data
processing algorithms and the use of sample redundancy (i.e., many of the same
types of
array elements for statistically significant data interpretation and avoidance
of anomalies) to
provide semi-quantitative analysis of polymorphisms or levels of mismatch
between the
target sequence and sequences immobilized on the device surface.
Labels appropriate for array analysis are known in the art. Examples are the
two-
color fluorescent systems, such as Cy3/Cy5 and Cy3.5/Cy5.5 phosphoramidites
(Glen
Research, Sterling-Virginia). Patents covering cyanine dyes include: U.S.
6,114,350 (Sept.
5, 2000); U.S. 6,197,956 (March 6, 2001); U.S. 6,204,389 (March 20, 2001) and
U.S.
6,224,644 (May l, 2001). Array printers and readers axe available in the art.
A process of using arrays is described in Grigorenko, E.V. ed., (2002) DNA
Arrays:
Technologies and Experimental Strategies, CRC Press, NY; Vrana, K.E. et al.,
(May 2001)
Microarrays and Related Technologies: Miniaturization and Acceleration of
Genornics
Research, CHI, Upper Falls, MA; and Branca, M.A. et al., (February 2002) DNA
Microarray
Informatics: Key Technological Trends and Commercial Opportunities, CHI, Upper
Falls,
MA.
All publications referred to herein axe incorporated by reference to the
extent not
inconsistent herewith. The mention of a publication in this Background Section
does not
constitute an admission that it is prior art.
SUMMARY OF INVENTION
The high mitochondria) DNA mutation rate of human mitochondria) DNA has been
thought to result in the accumulation of a wide range of neutral, population-
specific base
substitutions in mtDNA. These have accumulated sequentially along radiating
maternal
lineages that have diverged approximately on the same time scale as human
populations have
colonized different geographical regions of the world.
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
About 76% of all African mtDNAs fall into haplogroup L, defined by an HpaI
restriction site gain at by 3592. 77% of Asian mtDNAs are encompassed within a
super-
haplogroup defined by a DdeI site gain at by 10394 and an AIuI site gain at by
10397.
Essentially all native American mtDNAs fall into four haplogroups, A-D.
Haplogroup A is
defined by a HaeIII site gain at by 663, B by a 9 by deletion between by 8271
to by 8281, C
by a HincII site loss at by 13259, and D defined by an AluI site loss at by
5176. Ten
haplogroups encompass almost all mtDNAs in European populations. The ten-mtDNA
haplogroups of Europeans can be surveyed by using a combination of data from
RFLP
analysis of the coding region and sequencing of the hypervariable segment I.
About 99% of
European mtDNAs fall into one of ten haplogroups: H, I, J, K, M, T, U, V, W or
X.
This invention provides human mtDNA polymorphisms that are diagnostic of all
the
major human haplogroups and methods of diagnosing those haplogroups and
selected sub-
haplogroups.
This invention also provides methods for identifying evolutionarily
significant
mitochondrial DNA genes, nucleotide alleles, and amino acid alleles.
Evolutionarily
significant genes and alleles are identified using one or two populations of a
single species.
The process of identifying evolutionarily significant nucleotide alleles
involves identifying
evolutionarily significant genes and then evolutionarily significant
nucleotide alleles in those
genes, and identifying evolutionarily significant amino acid alleles involves
identifying
amino acids encoded by all nonsynonymous alleles. Synonymous codings of the
nucleotide
alleles encoding evolutionarily significant amino acid alleles of this
invention are equivalent
to the evolutionarily significant amino acid alleles disclosed herein and axe
included within
the scope of this invention. Synonymous codings include alleles at neighboring
nucleotide
loci that are within the same codon.
This invention also provides methods for associating haplogroups and
evolutionarily
significant nucleotide and amino acid alleles with predispositions to
physiological conditions.
Methods for diagnosing predisposition to LHON, and methods for diagnosing
increased
likelihood of developing blindness, centenaria, and increased longevity that
are not dependent
on the geographical location of the individual being diagnosed are provided
herein.
Diagnosis of an individual with a predisposition to an energy metabolism-
related
physiological condition is dependent on the geographic region of the
individual.
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Physiological conditions diagnosable by the methods of this invention include
healthy
conditions and pathological conditions. Physiological conditions that are
associated with
haplogroups and with alleles provided by this invention include energetic
imbalance,
metabolic disease, abnormal energy metabolism, abnormal temperature
regulation, abnormal
oxidative phosphorylation, abnormal electron transport, obesity, amount of
body fat, diabetes,
hypertension, and cardiovascular disease.
Molecules having sequences provided by this invention are provided in
libraries and
on genotyping arrays. This invention provides methods of making and using the
genotyping
arrays of this invention. The arrays of this invention are useful far
determining the presence
and absence of nucleotide alleles of this invention, for determining a
haplogroup, and for
diagnosis.
This invention also provides machine-readable storage devices and program
devices
for storing data and programmed methods for diagnosing haplogroups and
physiological
conditions.
The arrays of this invention are useful for determining the presence and
absence of
nucleotide alleles of this invention, for determining a haplogroup, and for
diagnosis. This
invention also provides machine-readable storage devices and program devices
for storing
data and programmed methods for diagnosing haplogroups and physiological
conditions.
BRIEF DESCRIPTION OF THE FIGURES
FIG. 1 shows a consensus neighbor joining tree of 104 human mtDNA complete
sequences and two primate sequences. Numbers correspond to bootstrap values (%
of 500
total bootstrap replicates) (Felsenstein, J. (1993) PHYLIP (Phylogeny
Inference Package)
3.53c. Distributed by author, Department of Genetics, University of
Washington, Seattle,
WA). Maximum Likelihood (ML) and UPGMA yielded consistent branching orders
with
respect to continent-specific mtDNA haplogroups. Sequences: Il-53: Genbank
AF346963-
AF347015 (4); E21U: Genbank X93334, AlLla: Genbank D38112, cam revise: Genbank
NC 001807 corrected according to ( R. M. Andrews et al., Nature Genetics 23,
147 (1999));
the rest are 48 sequences generated in this invention using an ABI 377.
Specific mutations in
patient samples that have been implicated in disease were excluded from this
analysis, as well
as gaps and deletions, with the exception of the 9 by deletion (nucleotide
position (np) 8272
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
to 8281). Haplogroups A, B, C, D, and X were drawn from both Eurasia and the
Americas.
Haplogroup names are designated with capital letters. P. paniscus and P.
troglodytes mtDNA
sequences were used as outgroups. Haplogroups LO and Ll encompass previously
assigned
Lla and Llb mtDNAs, respectively (Y. S. Chen et al., American Journal ofHuman
Genetics 66, 1362-1383 (2000)).
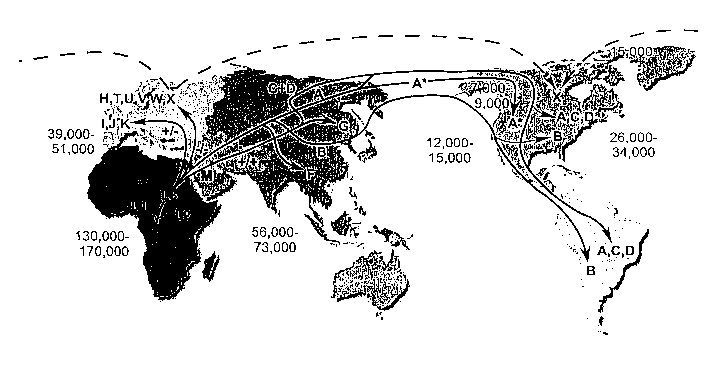
FIG. 2 shows the migrations of human haplogroups around the world. +/-,
+/+,.or -/-
equals Dde I 10394 and Alu I 10397. * equals Rsa I 16329. The mutation rate is
2.2-2.9%
per million years. Time estimates are YBP (years before present).
FIG. 3 shows a cladogram listing nucleotide alleles describing 21 major human
haplogroups, 21 sub-haplogroups, and several macro-haplogroups. The groups on
the left are
described by the alleles to their right. A vertical bar designates that each
group to the left of
the bar has all of the alleles to the right of the bar.
FIG. 4 shows the selective constraint (k~ values) of mtDNA protein genes with
comparisons among mammalian species. Statistical significance (P < 0.05) was
determined
using ANOVA, t-tests or the Tukey-Kramer Multiple Comparisons tests. Most
programs
used are from DNAsp ( J. Rozas and R. Rozas, (1999) Bioinforrnatics 15:174-5).
DNA
sequence divergence was analyzed using the DIVERGE program (Wisconsin Package
Version 10.0, Genetics Computer Group (GCG), Madison, WI). For all thirteen
mtDNA
genes, data is shown for human, human compared to P. troglodytes, human
compared to P.
paniscus, and nine species of primates. For only ATP6 and ATPB, data is also
shown for
fourteen species of mammals.
DETAILED DESCRIPTION OF THE INVENTION
Table 1 shows human mitochondria) nucleotide alleles, which have been
associated
with physiological conditions. In Table 1, columns three (nucleotide locus),
five
(physiological condition nucleotide allele), and column two (physiological
condition) make
up the set of Human Mitochondria) Nucleotide Alleles Known to be Associated
with
Physiological Conditions.
11
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
TABLE 11
Human Mitochondria) Alleles Known to be Associated with Physiolo~ical
Conditions
Physiological Physiological
Cambridge Cambridge
ene hysiological ConditionucleotideNucleotideConditionA~no Condition
Locus Allele NucleotideAcid Anuno
Allele Allele Acid
Allele
MTND1*MELAS 3308 T C M T
MTND1*NIDDM; LHON; PEO 3316 G A A T
MTND1*LHON 3394 T C Y H
MTND1*NIDDM 3394 T C Y H
MTND1*ADPD 3397 A G M V
MTND *LHON 3460 G A A T
1
MTND1*LHON 3496 G T A S
MTND1*LHON 3497 C T A V
MTND1*LHON 4136 A G - Y C
MTND *LHON 4160 T C L P
1
MTND1*LHON 4216 T C Y H
MTND2*LHON 4917 A G D N
MTND2*LHON 5244 G A G S
MTND2*AD 5460 G A A T
MTND2*AD - 5460 G T A S
MTCO1*M o lobinuria, Exercise5920 G A W Ter
Intolerance
MTCO1*Multisystem Disorder 6930 G A G Ter
MTCO1*LHON 7444 G A Ter K
MTC02*Mitochondrial Ence 7587 T ' C M T
halom o ath
MTC02*MM 7671 T A M K
MTC02*Multisystem Disorder 7896 G A W Ter
MTC02*Lactic Acidosis 8042 AT 2 nt de) M Ter
(AT)
MTATP6*NARP 8993 T G L R
MTATP6*NARP / Lei h Disease 8993 T C L P
MTATP6*LHON 9101 T C I T
MTATP6*FBSN l Lei h Disease 9176 T C L P
MTATP6*Lei h Disease 9176 T G L R
MTC03*LHON 9438 G A G S
MTC03*Lei h-like 9537 C C ins Q frameshifr
MTC03*LHON 9738 G T A S
MTC03*LHON 9804 G A A T
MTC03*Mitochondrial Ence 9952 G A W Ter
halo athy
MTC03*PEM; MELAS 9957 T C F L
MTND3*ESOC 10191 T C S P
MTND4*MELAS 11084 A G T A
MTND4*LHON 11778 G A R H
MTND4*Exercise Intolerance 11832 G A W Ter
MTND4*DM 12026 A G I V
MTNDS*MELAS 13513 G A D N
' (MITOMAP: A Human Mitochondria) Genome Database. Center for Molecular
Medicine,
Emory University, Atlanta, GA, USA. httw//www yen emory edu/mitomab,html,
2001).
12
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
MTNDS*MELAS 13514 A G D G
MTNDS*LHON-like 13528 A G T A
MTNDS*LHON 13708 G A A T
MTNDS*LHON 13730 G A G E
MTND6*MELAS 14453 G A A V
MTND6*LDYT 14459 G A A V
MTND6*LHON 14484 T C M V
MTND6*LHON 14495 A G L S
MTND6*LHON 14568 C T G S
14787 TTAA 4 nt I frameshift
MTCYB*PD l MELAS de)
(TTAA)
MTCYB*MM 15059 G A G Ter
MTCYB*Exercise Intolerance 15150 G A W Ter
MTCYB*Exercise Intolerance 15197 T C S P
MTCYB*Mitochondrial Ence 15242 G A G Ter
halomyo ath
MTCYB*LHON 15257 G A D N
MTCYB*Exercise Intolerance 15615 G A G D
MTCYB*MM 15762 G A G E
MTCYB*LHON 15812 G A V M
fi 1)et~nitions:
LHON Leber Hereditary Optic Neuropathy
MM Mitochondria) Myopathy
AD Alzheimer's Disease
LIMM Lethal Infantile Mitochondria) Myopathy
ADPD Alzheimer's Disease and Parkinson's Disease
MMC Maternal Myopathy and Cardiomyopathy
NARP Neurogenic muscle weakness, Ataxia, and Retinitis Pigmentosa; alternate
phenotype at this locus is
reported as Leigh Disease
FICP Fatal Infantile Cardiomyopathy Plus a MELAS-associated cardiomyopathy
MELAS Mitochondria) Encephalomyopathy, Lactic Acidosis, and Stroke-like
episodes LDYT Leber's
hereditary optic neuropathy and DYsTonia
MERRF Myoclonic Epilepsy and Ragged Red Muscle Fibers
MHCM Maternally inherited Hypertrophic CardioMyopathy
CPEO Chronic Progressive External Ophthalinoplegia
KSS Kearns Sayre Syndrome
DM Diabetes Mellitus
DMDF Diabetes Mellitus + DeaFness
CIPO Chronic Intestinal Pseudoobstruction with myopathy and Ophthalinoplegia
DEAF Maternally inherited DEAFness or aminoglycoside-induced DEAFness
PEM Progressive encephalopathy
SNHL SensoriNeural Hearing Loss
Thirteen protein-coding mitochondria) genes are known (MitoMap,
http://www.gen.emory.edu/cgi-bin/MITOMAP).
13
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Table 2
Protein-coding Human MtDNA Genes
Gene Map Locusa AbbreviationLocation
', NADH dehydrogenaseMTND1 ND1 3307-4262
1
II NADH dehydrogenaseMTND2 ND2 4470-5511
2
I NADH dehydrogenaseMTND3 ND3 10059-10404
3
NADH dehydrogenase MTND4L ND4L 10470-10766
4L
NADH dehydrogenase MTND4 ND4 10760-12137
4
NADH dehydrogenase MTNDS ND5 12337-14148
I
NADH dehydrogenase MTND6 ND6 14149-14673
6
Cytochrome b MTCYB Cytb 14747-15887
Cytochrome c oxidaseMTCO1 COI 5904-7445
I
Cytochrome c oxidaseMTC02 COII 7586-8269
II
Cytochrome c oxidaseMTC03 COIII 9207-9990
III
ATP synthase 6 MTATP6 ATP6 8527-9207
ATP synthase 8 MTATP8 ATP8 8366-8572
a,b As defined on MitoMap, http://www.gen.emory.edu/cgi-bin/MITOMAP, which is
numbered relative to the Cambridge Sequence (Genbank accession no. J01415 and
Andrews"
et al. (1999), A Reanalysis and Revision of the Cambridge Reference Sequence
for Human
Mitochondrial DNA, Nature Genetics 23:147.
Codon usage for mtDNA diffexs slightly from the universal code. For example,
UGA
codes for tryptophan instead of termination, AUA codes for methionine instead
of
isoleucine, and AGA and AGG are terminators instead of coding for arginine.
As used herein "printing" refers to the process of creating an array of
nucleic acids on
known positions of a solid substrate. The arrays of this invention can be
printed by spotting,
e.g., applying arrays of probes to a solid substrate, or to the synthesis of
probes in place on a
solid substrate. As used herein "glass slide" refers to a small piece of glass
of the same
dimensions as a standard microscope slide. As used herein, "prepared
substrate" refers to a
substrate that is prepared with a substance capable of serving as an
attachment medium for
attaching the probes to the substrate, such as poly Lysine. As used herein,
"sample" refers to
a composition containing human mitochondrial DNA that can be genotyped. As
used herein,
14
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
"quantitative hybridization" refers to hybridization performed under
appropriate conditions
and using appropriate materials such that the sequence of one nucleotide
allele (a single
nucleotide polymorphism) can be determined, such as by hybridization of a
molecule
containing that allele to two or more probes, each containing different
alleles at that
nucleotide locus, all as is known in the art.
As used herein, "physiological condition" includes diseased conditions,
healthy
conditions, and cosmetic conditions. Diseased conditions include, but are not
limited to,
metabolic diseases such as diabetes, hypertension, and cardiovascular disease.
Healthy
conditions include, but are not limited to, traits such as increased
longevity. Physiological
conditions include cosmetic conditions. Cosmetic conditions include, but are
not limited to,
traits such as amount of body fat. Physiological conditions can change health
status in
different contexts, such as fox the same organism in a different environment.
Such different
environments for humans are different cultural environments or different
climatic contexts
such as are found on different continents.
As used herein, "neutrality analysis" refers to analysis to determine the
neutrality of
one or more nucleotide alleles and/or the gene containing the alleles) using
at least two
alleles of a sequence. Commonly, the alleles in a sequence to be analyzed are
divided into
two groups, synonymous and nonsynonymous. Codon usage tables showing which
codons .
encode which amino acids are used in this analysis. Codon usage tables for
many organisms
and genomes are available in the art. If a gene is determined to not be
neutral, the gene is
determined to have had selection pressure applied to it during evolution, and
to be
evolutionarily significant. The alleles that change amino acids in the gene
(nonsynonymous)
are then determined to be non-neutral and evolutionarily significant.
As used herein, "Ka/KS" refers to a ratio of the proportion of nonsynonymous
differences to the proportion of synonymous differences in a DNA sequence
analysis, as is
known to the art. The proportion of nonsynonymous differences is the number of
nonsynonymous nucleotide substitutions in a sequence per site at which a
nonsynonymous
substitution could occur. The proportion of synonymous differences is the
number of
synonymous nucleotide substitutions in a sequence per site at which synonymous
substitutions could occur. Alternatively, instead of only including the number
of sites in the
denominator of each proportion, the number of alternative substitutions that
could occur at
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
each site are also included. Either definition may be used as long as similar
definitions are
used for both Ka and KS in an analysis. K~ is KalKS.
As used herein "nonsynonymous" refers to mutations that result in changes to
the
encoded amino acid. As used herein, "synonymous" refers to mutations that do
not result in
changes to the encoded amino acids.
As used herein, "haplogroup" refers to radiating lineages on the human
evolutionary
tree, as is known in the art. As used herein, "macro-haplogroup" refers to a
group of
evolutionarily related haplogroups. As used herein, "sub-haplogroup" refers to
an
evolutionarily related subset of a haplogroup. An individual's haplotype is
the haplogroup to
which he belongs.
As used herein, "extended longevity" or "extended lifespan" refers to living
longer
than the average expected lifespan for the population to which one belongs. As
used herein,
"centenaria" refers to an extended lifespan that is at least 100 years.
As used herein, "abnormal energy metabolism" in an individual who is non-
native to
the geographical region in which he lives refers to energy metabolism that
differs from that of
the population that is native to where the individual lives. As used herein,
"abnormal
temperature regulation" in such an individual refers to temperature regulation
that differs
from that of the population that is native to where he lives. As used herein,
"abnormal
oxidative phosphorylation" in such an individual refers to oxidative
phosphorylation that
differs from that of the population that is native to where he lives. As used
herein, "abnormal
electron transport" in such an individual refers to electron transport that
differs from that of
the population that is native to where he lives. As used herein "metabolic
disease" of such an
individual refers to metabolism that differs from that of the population that
is native to where
he lives. As used herein, "energetic imbalance" of such an individual refers
to a balance of
energy generation or use that differs from that of the population that is
native to where he
lives. As used herein, "obesity" of such an individual refers to a body weight
that, for the
height of the individual, is 20% higher than the average body weight that is
recommended for
the population native to where the individual lives. As used herein, "amount
of body fat" of
such an individual refers to a low or high percentage of body fat relative to
What is
recommended for the population that is native to where he lives.
16
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
As used herein, an isolated nucleic acid is a nucleic acid outside of the
context in
which it is found in nature. The term covers, for example: (a) a DNA which has
the sequence
of part of a naturally-occurring genomic DNA molecule but is not flanked by
both of the
coding or noncoding sequences that flank that part of the molecule in the
genome of the
organism in which it naturally occurs; (b) a nucleic acid incorporated into a
vector or into the
genomic DNA of a prokaryote or eukaryote in a manner such that the resulting
molecule is
not identical to any naturally-occurnng vector or genomic DNA; (c) a separate
molecule such
as a cDNA, a genomic fragment, a fragment produced by polymerase chain
reaction (PCR),
or a restriction fragment; and (d) a recombinant nucleotide sequence that is
part of a hybrid
gene, i.e., a gene encoding a fusion protein, or a modified gene having a
sequence not found
in nature.
As used herein, "nucleotide locus" refers to a nucleotide position of the
human
mitochondrial genome. The Cambridge sequence SEQ )lD N0:2 is used as a
reference
sequence, and the positions of the mitochondria) genome referred to herein are
assigned
relative to that sequence. As used herein, "loci" refers to more than one
locus. As used
herein, "nucleotide allele" refers to a single nucleotide at a selected
nucleotide locus from a
selected sequence when different bases occur naturally at that locus in
different individuals.
The nucleotide allele information is provided herein as the nucleotide locus
number and the
base that is at that locus, such as 3796C, which means that at human
mitochondria) position
3796 in the Cambridge sequence, there is a cytosine (C). As used herein,
"amino acid allele"
refers to the amino acid that is at a selected amino acid location in the
human mitochondria)
genome when different amino acids occur naturally at that location in
different individuals.
There are thirteen protein-coding genes in the human mitochondria. For each
gene, the
encoded protein consists of amino acids that are numbered starting at one. ND)
304 H,
means that there is a histidine at amino acid 304 in the NDl protein. Amino
acids are
encoded by codons. As used herein, "codon" refers to the group of three
nucleotides that
encode an amino acid in a protein, as is known in the art. An amino acid
allele can be
referred to by one or more of the nucleotide loci that code for it. For
example, ntl 15884 P
means that thexe is a proline (P) encoded by the codon containing nucleotide
locus 15884.
As used herein, "evolutionarily significant gene" refers to a gene that has
statistically
significantly more nonsynonymaus nucleotide changes, when compared to the
corresponding
17
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
gene in another individual, than would be expected by chance. As used herein,
"evolutionarily significant nucleotide allele" refers to a nucleotide allele
that is located in a
gene that has been determined to be evolutionarily significant using that
nucleotide allele, or
an equivalent nucleotide allele in a corresponding gene in another individual.
As used herein,
"intraspecific" means within one species. As used herein, "subpopulation"
refers to a
population within a larger population. A subpopulation can be as small as one
individual. As
used herein, "geographic region" refers to a geographic area in which a
statistically
significant number of individuals have the same haplotype. As used herein,
being "native" to
a geographic region refers to having the haplotype associated with that
geographic region.
The haplotype associated with a geographic region is that which originated in
the region or of
many individuals who settled historically in the region with respect to human
evolution.
As used herein, "target" or "target sample" refers to the collection of
nucleic acids
used as a sample for array analysis. The target is interrogated by the probes
of the array. A
"target" or "target sample" may be a mixture of several samples that are
combined. For
example, an experimental target sample may be combined with a differently
labeled control
target sample and hybridized to an array, the combined samples being referred
to as the
"target" interrogated by the probes of the array during that experiment. As
used herein,
"interrogated" means tested. Probes, targets, and hybridization conditions are
chosen such
that the probes are capable of interrogating the target, i.e., of hybridizing
to complementary .
sequences in the target sample.
As used hexein, "increased likelihood of developing blindness" refers to a
higher than
normal probability of losing the ability to see normally andlor of losing the
ability to see
normally at a younger age.
All sequences defined herein are meant to encompass the complementary strand
as
well as double-stranded polynucleotides comprising the given sequence.
This invention provides a list of human mtDNA polymorphisms found in all the
major
human haplogroups. Example 1 summarizes data from sequencing over 100 human
mtDNA
genomes that are representative of the major human haplogroups around the
world. The
summary includes over 900 point mutations and one nine-base pair deletion.
Table 3, Human
MtDNA Nucleotide Alleles, lists the alleles identified in 103 such sequences
in the third
18
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
column, the corresponding alleles of the Cambridge mtDNA sequence in the
second column
and the nucleotide loci (position in the Cambridge sequence), in the first
column. Table 3
lists the set of human mtDNA nucleotide alleles that occur naturally in
different haplogroups.
Table 3 does not include alleles previously known to be associated with
disease (i.e., does not
include the alleles of Table 1). The nucleotide alleles listed in column three
of Table 3,
together with the corresponding nucleotide loci in column one, make up the set
of non-
Cambridge human mtDNA nucleotide alleles. Table 4 lists the nucleotide alleles
identified
by the inventors hereof in 48 human mtDNA genomes in column three, and the
corresponding Cambridge alleles in column two. Columns one and three of Table
4 make up
the set of non-Cambridge human mtDNA nucleotide alleles in 48 genomes.
The nucleotide alleles listed in Table 3, including the Cambridge nucleotide
alleles,
being naturally occurnng, are useful for identifying alleles that are
associated with abnormal
physiological conditions. These nucleotide alleles can be ignored during
analysis steps when
performing methods for identifying novel alleles associated with selected
physiological
conditions.
As described below, certain alleles of Table 3 are useful for identifying
physiological
conditions related to energy metabolism such as energetic imbalance, metabolic
disease,
abnormal energy metabolism, abnormal temperature regulation, abnormal
oxidative
phosphorylation, abnormal electron transport, obesity, amount of body fat,
diabetes,
hypertension, and cardiovascular disease when the affected individuals have
the abnormal
physiological condition because they are in a geographical region that is not
native for their
haplogroup.
The nucleotide alleles listed in Table 3, including the Cambridge nucleotide
alleles,
are also useful for identifying mtDNA sequences associated with and diagnostic
of human
haplogroups. Example 2 summarizes phylogenetic analyses of the sequence data
of the 103
individuals and the Cambridge sequence along with two chimpanzee mtDNA
sequences. The
results are shown in FIG. 1 in a cladogram. Calculations of the time since the
most recent
common ancestor (MRCA) are shown in Table 5. The 104 individuals were chosen
from
known haplogroups, and the corresponding haplogroups are labeled on the
figure.
Combining the sequence data of the 104 individuals with FIG. l and the
geographic regions
native to human haplogroups, as is known in the art, results in FIG.2 (Example
3), which
19
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
tracks human mtDNA migrations. Analysis of several mtDNA genomic sequences
representing each haplogroup demonstrated which alleles are segregating within
a haplogroup
as well as which alleles are present in every individual within one or more
haplogroups. The
alleles that are present in every individual within each haplogroup are shown
in FIG. 3
(Example 4). On the left, sub-haplogroups and haplogroups are listed.
Macrohaplogroups
are shown in parentheses. Nucleotide loci and alleles that are present in all
the members of
each group (sub-haplo or haplo) are listed. A vertical bar designates that all
of the alleles to
the right are present in all of the haplogroups and/or sub-haplogroups to the
left. FIG. 3 is
drawn as a cladogram. For example, FIG. 3 demonstrates that the
macrohaplogroup (R)
individuals all contain 127050 and 162230, and no other individuals are known
to have these
alleles, therefore macro-haplogroup (R) can be diagnosed by identifying in a
sample
containing mtDNA, the presence of either 127050 or 162230. Similarly, macro-
haplogroup
(N) can be diagnosed by identifying the presence of 8701A, 9540T, or 10873T.
Analysis of the data in FIG. 3 demonstrated sets of alleles useful for
diagnosing the
haplogroups (Example 5). These alleles are listed by haplogroup in Tables 6
and 7, and by
sub-haplogroup in Tables 8 and 9. A set of alleles useful for diagnosing all
of the
haplogroups and sub-haplogroups in FIG. 3 is listed in Table 10. Table 10
lists the nucleotide
loci in column one and the nucleotide alleles useful for diagnosing
haplogroups in column
two. Table 10 contains some alleles from the Cambridge sequence. There are
many
equivalent methods for diagnosing the haplogroups. Methods for diagnosing
haplogroups
that require testing only one or a few loci are listed in Example 5. The
presence of only one
particular allele i's usually sufficient for diagnosing a haplogroup, however,
often it is not
known which locus needs to be tested. By determining the allele at each
nucleotide locus
listed in Table 10, the haplogroup of an unknown sample can be diagnosed.
Alternatively,
macro-haplogroups can be diagnosed or excluded first, thereby decreasing the
number of loci
that need to be tested to distinguish between the remaining, possible
haplogroups. Alleles
useful for diagnosing macro-haplogroups by methods that require testing only
one or a few
loci are included in Table 11. Further analysis of the data provided by this
invention will
demonstrate which sets of alleles identify additional sub-haplogroups and
additional macro-
haplogroups.
Diagnosing the haplogroup of a sample is useful in criminal investigations and
forensic analyses. Identifying a sample as belonging to a particular
haplogroup, and knowing
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
which alleles have not been associated with a selected physiological condition
and context,
are useful when identifying novel alleles associated with a selected
physiological condition,
as described above and in Example 6. Diagnosing the haplogroup of a sample is
also useful
for identifying a novel allele associated with a selected physiological
condition when the
novel allele causes the physiological condition only in the genetic context of
a particular
haplogroup, as shown in Example 6. In example 6, the list of alleles
associated with
haplogroups found in Russia was used in the sequence analysis of two Russian
LHON
families. By eliminating alleles listed in Table 3, two novel mutations were
identified that
are associated with LHON. These new complex I mutations, 3635A and 4640C, are
useful
for diagnosing a predisposition to Leber Hereditary Optic Neuropathy (LHON).
Example 7 demonstrates the identification of a new primary LHON mutation,
10663C, in complex I, that appears to cause a predisposition to LHON only when
associated
with haplogroup J. Haplogroup J is defined by a nonsynonymous difference that
is useful
for diagnosing haplogroup J, 458T in NDS. This invention provides a method of
diagnosing
a person with a predisposition to LHON and/or to developing early onset
blindness by
identifying, in a sample containing mtDNA from the person, the nucleotide
allele, or a
synonymous nucleotide allele of 10663C and also identifying alleles diagnostic
of
haplogroup J, such as 458T in NDS. Because NDS 458T is a missense mutation in
all
haplogroup J individuals, this particular mutation may be directly involved in
causing LHON. -~
ND 1 304H is another missense mutation that is present in all haplogroup J
individuals, and
may also be directly involved in causing LHON. 458T is also present in
haplogroup T
individuals. Haplogroup J is also associated with a predisposition to
centenaria and an
extended lifespan. NDS 458T and NDl 304H may also be directly involved in
causing the
predisposition to centenaria and extended lifespan.
Example 8 demonstrates the importance of demographic factors in
intercontinental
mtDNA sequence radiation. Haplogroups are combined and separated into various
populations for statistical analyses.
Previously in the art, it has been thought that polymorphisms in human mtDNA,
such
as the nucleotide alleles listed in Table 3, were neutral in all contexts and
could not be
associated with physiological conditions. It has been thought that differences
in human
21
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
mtDNA diversity associated with inter-continental migrations were due to
random genetic
drift (e.g. founder effects followed by rapid population expansion). In this
invention, the
biological and clinical significance of these human mtDNA polymorphisms are
disclosed.
The neutrality of the nucleotide alleles listed in Table 3 was tested using
neutrality analysis
(Examples 9-12).
Some of the nucleotide loci in Table 3 are located in the mitochondria)
protein-coding
genes (Table 2). Of those loci, some of the identified nucleotide alleles
alter the protein
encoded by the codon in which the nucleotide locus resides. This is determined
using the
mitochondria) codon usage table, as is known in the art. Nucleotide alleles
that change an
amino acid are called missense mutations, missense polymorphisms, or
nonsynomymous
differences. Missense polymorphisms alter the protein sequence relative to a
compared
sequence, but they still may be neutral because they do not affect the
function of the encoded
protein. Without performing biochemical studies on the affected proteins,
statistical analyses
can be performed to determine whether a polymorphism is neutral, whether
evolution
imposed selection on the encoding allele, and whether that selection is
positive. This
invention provides results of the statistical analyses of the polymorphisms in
Table 3 and
provides a list of which alleles are not neutral, and therefore evolutionarily
significant.
Neutrality testing of nucleotide alleles first requires neutrality testing of
the genes
containing those nucleotide alleles. Neutrality testing of one or more genes
by comparing
two sets of allelic genes from two intraspecific populations was performed, as
described in
Example 9. Haplogroups were combined to make populations for the comparison.
In
example 9, nucleotide alleles from the entire coding region of the mtDNA
genome,
representing haplogroups native to a geographic region, were combined to make
a first
population and first set of sequences. Nucleotide alleles of the entire coding
region of the
mtDNA genome, from haplogroups native to a different geographic region, were
combined to
make the second population and the second set of sequences. Nucleotide alleles
were divided
into those encoding synonymous and non-synonymous differences. The ratio of
Ka/KS for
each gene, separated by the population containing the allele, is shown in
Table 12. Neutrality
testing of genes by comparing one set of at least two nucleotide alleles of at
least one gene
from one population of one species was performed in Example 10. In Example 10,
sequences
of the entire coding region of the mtDNA genome, of haplogroups in all
geographic regions
on earth, were combined to make one population and set of sequences for
analysis. FIG 4
22
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
shows the results of the comparison of one set of sequences from one
population of only one
species, 104 human sequences. Example 11 includes comparisons of sets of
sequences
between two populations, human vs. P. paniscus, human vs. P. troglodytes,
human vs. eight
other primate species, and human vs. thirteen mammalian species.
To identify an evolutionarily significant gene, two sets of nucleotide
sequences, each
set from a different population, are compared to each other. Nucleotide
sequences
representing parts of genes or one or more whole genes are useful. The sets of
sequences are
compared to each other by neutrality analysis. Differences in the sequences
from each set are
determined to be synonymous or nonsynonymous differences. The proportion of
nonsynonymous differences is compared to the proportion of synonymous
differences
(Ka/KS). The results of the analysis are compiled in a data set and the data
set is analyzed, as
is known in the art, to identify one or more evolutionarily significant genes.
When the
nonsynonymous differences occur significantly more often than is expected by
chance than
the synonymous differences, the gene or part of the gene is determined to be
evolutionarily
significant. When the synonymous differences occur significantly more often
than is
expected by chance than the nonsynonymous differences, the gene or part of the
gene is
determined to be conserved. When the ratio is as expected by chance, then
there is no
evidence of selection or evolutionary significance.
To identify an evolutionarily significant gene, only one set of nucleotide
sequences
(from only one population) may also be analyzed, e.g., the nucleotide
sequences
representative of humans living on one continent. When only one set of
sequences is
analyzed, the set must contain at least two corresponding nucleotide alleles
(i.e., there must
be sequence polymorphism). Corresponding sequences are sequences of the same
gene or
gene part from at least two individuals. The sequences from different
individuals within the
population must contain polymorphisms with respect to each other. Differences
in the
sequences relative to each other are determined to be synonymous or
nonsynonymous.
Neutrality analysis is performed to generate a data set. The data set is
analyzed to identify an
evolutionarily significant gene. If an analysis determines that none of the
analyzed genes are
evolutionarily significant, the set of nucleotide sequences can be increased,
such as by
increasing the size of the population from which the sequences are derived, to
determine if
one or more genes are evolutionarily significant in the enlarged population.
23
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Example 12 is similar to example 9 except that the data is further analyzed by
manipulating I~a/KS to K~. Examples 9-12 demonstrate that all but one mtDNA
gene are not
neutral and therefore are evolutionarily significant. Genes are determined to
not be neutral
by statistical significance tests known in the axt. Some genes are only
evolutionarily
significant when comparing selected populations. For example, ND4 was
demonstrated to be
significant when comparing Native American sequences to African sequences and
when
comparing all human sequences to each other, but not when comparing European
to African
sequences. ND4L is the only mtDNA gene not shown to be evolutionarily
significant by the
current analyses. ND4L might be demonstrated to be evolutionarily significant
by the
methods of this invention using one or more different populations or using
only part of the
gene sequence. In examples 9-12, the entire sequence of each gene was used for
analysis,
however portions of genes are also useful in the methods of this invention.
The statistical
significance tests prevent too small a gene portion from being used to
determine non-
neutrality.
After identifying evolutionarily significant genes, evolutionarily significant
nucleotide
alleles can be identified. To identify an evolutionarily significant
nucleotide allele, the steps
for identifying an evolutionarily significant gene, using one or two
populations, are
performed with the addition of a step of analyzing the sequence data set to
determine an
evolutionarily significant nucleotide allele. An evolutionarily significant
nucleotide allele is
part of a sequence incoding an allelic amino acid in an evolutionarily
significant gene or part
of a gene. Examples 13 and 14 demonstrate identification of evolutionary
significant
nucleotide alleles and evolutionarily significant amino acid alleles in the
evolutionarily
significant genes identified in Examples 9-12. Evolutionarily significant
amino acid alleles
are the amino acids encoded by the codons containing evolutionarily
significant nucleotide
alleles. In these examples, nucleotides at loci not listed in Table 3 axe
identical to the
Cambridge sequence so that the entire codon containing an evolutionarily
significant
nucleotide allele and the amino acid encoded by that codon can be determined.
All
nucleotide alleles that are part of a codon encoding the same amino acid as an
evolutionarily
significant amino acid allele identified herein, or identified by methods of
this invention, are
also evolutionarily significant and are intended to be within the scope of
this invention. An
evolutionarily significant amino acid allele may include more than one
nucleotide allele, such
as at two neighboring nucleotide loci. Evolutionarily significant nucleotide
alleles and
evolutionarily significant amino acid alleles in human mitochondria)
sequences, identified by
24
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
the methods of this invention, are listed in Table 14. In column one, Table 14
lists the gene
containing the alleles, column two indicates the locus of the nucleotide
allele, column three
lists the Cambridge nucleotide allele at that nucleotide locus, column four
lists a non-
Cambridge allele of this invention, column five lists the amino acid encoded
by the codon
containing the Cambridge nucleotide allele (when other Cambridge nucleotides
are present at
the other nucleotide loci of the codon), and column six lists the amino acid
encoded by the
codon containing the non-Cambridge allele (when Cambridge nucleotides are
present at the
other nucleotide loci of the codon). Columns two, three, and four make the set
of
evolutionarily significant human mitochondria) nucleotide alleles. Columns
two, five, and
six make the set of evolutionarily significant human mitochondria) amino acid
alleles. Table
14 designates the nucleotide locus of the listed alleles. For the amino acid
alleles listed in
columns five and six, the relevant loci are all three nucleotide loci in the
encoding codon
containing the nucleotide locus listed in column two.
To identify an evolutionarily significant amino acid allele, the steps for
identifying an
evolutionarily significant gene, using one or two populations, are performed
with the addition
of two steps: 1) analyzing the data set to determine an evolutionarily
significant nucleotide
allele; and 2) determining the encoded amino acid allele. An evolutionarily
significant amino
acid allele is a different amino acid, representing a nonsynonymous
difference, relative to the
corresponding amino acid allele against which it was compared, wherein the
gene has been
determined to be evolutionarily significant in the corresponding one or more
populations.
In this invention it is demonstrated that amino acid substitution mutations
(nonsynonymous differences) are much more common in human mtDNAs than would be
expected by chance, and that most of them are evolutionarily significant. This
invention
demonstrates that these alleles have become fixed by selection. The
mitochondria) genes
encode proteins that are responsible for generating energy and for generating
heat to maintain
body temperature. As humans migrated to different parts of the world, they
encountered
changes in diet and climate. The high mutation rate of mtDNA and the central
role of
mitochondria) proteins in cellular energetics make the mtDNA an ideal system
for permitting
rapid mammalian adaptation to varying climatic and dietary conditions. The
increased amino
acid sequence variability that has been found among human mtDNA genes is due
to the fact
that natural selection favored mtDNA alleles that altered the coupling
efficiency between the
electron transport chain (ETC) and ATP synthesis, determined by the
mitochondria) inner
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
membrane proton gradient (~'If). The coupling efficiency between the ETC and
ATP
synthesis is mediated to a considerable extent by the proton channel of the
ATP synthase,
which is composed of the mtDNA-encoded ATP6 protein and the nuclear DNA-
encoded
ATP9 protein. Mutations in the ATP6 gene, which create a more leaky ATP
synthase proton
channel, reduced ATP production but increased heat production for each calorie
consumed.
Such a change in energy balance was beneficial in a temperate or arctic
climate, but
deleterious in a tropical climate. Humans acquiring mtDNA alleles enabling
better
adaptation to the encountered changes in diet and climate experienced a higher
genetic fitness
and those alleles were selected for. In particular, these alleles were
established genetically
because they had an adaptive advantage as humans moved from the African
tropics into the
EurAsian temperate zone and on into the arctic (FIG 2). The lack of
recombination of the
maternally inherited mtDNAs favored the rapid segregation, expression and
adaptive
selection of advantageous mtDNA alleles. The apparent non-randomness of the
differences
in non-synonymous versus synonymous mtDNA variation between continents
demonstrates
that selection also influenced inter-continental colonization. Random genetic
hitchhiking,
such as in the synonymous alleles, then resulted in identifiable continent-
specific
haplogroups.
Modern mtDNA variation has been shaped by adaptation as our ancestors moved
into
different environmental conditions. Variants that are advantageous in one
climatic and
dietary environment are maladaptive when individuals locate to a different
environment. The
methods of this invention associate mtDNA nucleotide alleles with haplogroups
and combine
this data with native haplogroup geographic regions as is known in the art, to
diagnose
individuals as having predispositions to late-onset clinical disorders such as
obesity, diabetes,
hypertension, and cardiovascular disease when those individuals live in
climatic arid dietary
environments that are disadvantageous with respect to their mtDNA alleles.
When humans
having regional mtDNA alleles move into a different thermal andlor dietary
environment
from the one in which the alleles were selected, they are energetically
unbalanced with their
environment, and as a result are predisposed to having metabolic diseases such
as diabetes,
hypertension, cardiovascular disease, and other diseases known to the art to
be associated
with metabolism and mitochondria) functions. The above-mentioned late-onset
clinical
disorders are rapidly becoming epidemic around the world in members of our
globally mobile
society. This invention provides a method of diagnosing a human with a
predisposition to a
physiological condition such as, but not limited to, energetic imbalance,
metabolic disease,
26
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
abnormal energy metabolism, abnormal temperature regulation, abnormal
oxidative
phosphorylation, abnormal electron transport, obesity, amount of body fat,
diabetes,
hypertension, and cardiovascular disease. The method involves testing a sample
containing
mitochondria) nucleic acid from an individual in a geographic region to
determine the
haplogroup of the sample and therefore of the individual, comparing the
haplogroup of the
individual to the set of haplogroups known to be native to that geographic
region, and
diagnosing the individual human with a predisposition to the above-mentioned
conditions if
the haplogroup of the individual is not in the set of haplogroups native to
that geographic
region. This invention enables treatment of one of the above-mentioned
conditions that is
diagnosed by the above-mentioned method, comprising relocating the diagnosed
human to a
geographic region that is of similar climate as the regions) native to the
human's haplogroup
and/or changing the diagnosed human's diet to more closely match the diet
historically
available in the regions) native to the human's haplogroup.
The above-described method for diagnosing a predisposition to a physiological
condition is also useful for associating an amino acid allele with the
physiological condition.
The evolutionarily significant amino acid alleles present in the haplogroup of
the diagnosed
individual and not in the haplogroups native to the individual's geographic
location are
associated with the physiological condition by the methods of this invention.
Amino acid
alleles, and the corresponding nucleotide alleles, useful for diagnosing
haplogroups, and the
haplogroup they are useful for diagnosing, are listed in Table 15. The amino
acid alleles and
corresponding nucleotide alleles listed in Table 15, and synonymously coding
nucleotide
alleles, are associated with the above-mentioned physiological conditions.
Table 15 lists the
set of amino acid alleles useful for diagnosing haplogroups. Column one of
Table 15 lists the
gene, column two lists the nucleotide locus, column three lists the useful
nucleotide allele,
column four lists the useful amino acid allele encoded by the useful
nucleotide allele when
Cambridge nucleotides are present at the other nucleotide loci of the encoding
codon, and
column five lists the haplogroups or sub-haplogroups, in parentheses, that
contain the
corresponding alleles. The amino acid alleles (column four) can be identified
by the codon
containing the nucleotide locus (column two). For example, the proline in the
ND 1 gene is
identified as ntl 3796 P, where ntl signifies the codon containing the
nucleotide locus (ntl)
3796. When an individual of one of the haplogroups listed in column five of
Table 15 is
diagnosed with one of the above-mentioned physiological conditions by the
above-mentioned
method, the physiological condition is associated with the presence of one of
the alleles listed
27
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
in Table 15. When the haplogroup of the individual is haplogroup G, the amino
acid allele
likely to have caused the physiological condition is ntl 4833 A. When the
haplogroup of the
individual is haplogroup T, the amino acid allele is selected from the group
consisting of ntl
14917 D, ntl 8701 T, and ntl 15452 I. When the haplogroup is haplogroup W, the
amino acid
allele is selected from the group consisting of ntl 5046 I, ntl 5460 T, ntl
8701 T, and ntl
15884 P. When the haplogroup is haplogroup D, the amino acid allele is
selected from the
group consisting of ntl S 178 M and ntl 8414 F. When the haplogroup is
haplogroup L0, the
amino acid allele is selected from the group consisting of ntl 5442 L, ntl
7146 A, ntl 9402 P,
ntl 13105 V, and ntl 13276 V. When the haplogroup is haplogroup L1, the amino
acid allele
is selected from the group consisting of ntl 7146 A, ntl 7389 H, ntl 13105 V,
ntl 13789 H, and
ntl 14178 V. When the haplogroup is haplogroup C the amino acid allele is
selected from the
group consisting of ntl 8584 T and ntl 14318 S. When the haplogroup is
selected from the
group consisting of haplogroups A, I, X, B, F, Y, and U the amino acid allele
is ntl 8701 T.
When the haplogroup is haplogroup J the amino acid allele is selected from the
group
consisting of ntl 8701 T, ntl 13708 T, and ntl 15452 I. When the haplogroup is
haplogroup
selected from the group consisting of haplogroups V and H, the amino acid
allele is selected
from the group consisting of ntl 8701 T and ntl 14766 T.
Evolutionarily significant nucleotide and amino acid alleles also exist in
nuclear-
encoded ATP9 that are useful for diagnosing predisposition to an energy
metabolism-related ..
physiological condition such as energetic imbalance, metabolic disease,
abnormal energy
metabolism, abnormal temperature regulation, abnormal oxidative
phosphorylation, abnormal
electron transport, obesity, centenaria, diabetes, hypertension, and
cardiovascular disease.
These alleles may be identified by methods of this invention.
The evolutionarily significant amino acid alleles and corresponding nucleotide
alleles
are candidates for alleles causing a physiological condition for which a
predisposition is
diagnosable by the methods of this invention. The evolutionarily significant
amino acid and
nucleotide alleles identified by the methods of this invention (Table 19) are
useful for gene
therapy and mitochondria) replacement therapy to neat the corresponding
physiological
conditions. The evolutionarily significant genes, amino acid alleles, and
nucleotide alleles
identified by the methods of this invention are useful for identifying targets
for traditional
therapy, and for designing corresponding therapeutic agents. The
evolutionarily significant
28
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
genes and amino acid and nucleotide changes identified by the methods of this
invention are
useful for generating animal models of the corresponding human physiological
conditions.
As is known to the art, individuals may contain more than one mitochondria)
DNA
allele at any given nucleotide locus. One cell contains many mitochondria, and
one cell or
different cells within one organism may contain genetically different
mitochondria.
Heteroplasmy is the occurrence of more than one type of mitochondria in an
individual or
sample. Varying degrees of heteroplasmy are associated with varying degrees of
the
physiological conditions described herein. Heteroplasmy may be identified by
means known
to the art, and the severity of the physiological condition associated with
specific nucleotide
alleles is expected to vary with the percentage of such associated alleles
within the individual.
The methods of this invention are used to analyze the human mitochondria)
genome
in the listed examples, but the methods are also useful for analyzing other
genomes and other
species. The methods of this invention are useful for identifying
evolutionarily significant
protein-coding genes and the correspondingly encoded mutations in other
genomes in
addition to mitochondria) genomes, such as in nuclear and chloroplast genomes.
Using
human haplogroups as populations (FIG 1), the methods of this invention are
useful for
identifying evolutionarily significant protein-coding genes and the
corresponding
evolutionarily significant alleles in human nuclear genes. The methods of this
invention are
also useful for identifying evolutionarily significant protein-coding genes
and the
corresponding alleles in many species. .For example, the methods of this
invention are
applicable to varieties of beef or dairy cattle, or pig lines. Corn lines are
divisible by
phenotypic and/or molecular markers into heterotic groups that are useful
populations in the
methods of this invention. Using corn heterotic groups as populations, the
methods of this
invention axe useful for identifying evolutionarily significant protein-coding
genes and the
corresponding mutations in the nuclear, chloroplast, and mitochondria) genomes
of corn.
This invention provides isolated nucleic acid molecules containing novel
nucleotide
alleles of this invention in libraries. The libraries contain at least two
such molecules.
Preferably the molecules have unique sequences. The molecules typically have a
length from
about 7 to about 30 nucleotides. "About" as used herein means within about 10%
(e.g.,
"about 30 nucleotides" means 27-33 nucleotides). However, the molecules may be
longer,
such as about 50 nucleotides long. A library of this invention contains at
least two isolated
29
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
nucleic acid molecules each containing at least one non-Cambridge nucleotide
allele of this
invention. A library of this invention may contain at least ten, twenty-five,
fifty, 100, 500 or
more isolated nucleic acid molecules, at least one of which contains a
nucleotide allele of this
invention. A library of this invention may contain molecules having at least
two to all of the
nucleotide alleles of this invention, including synonymous codings of
evolutionarily
significant amino acid alleles. The nucleotide alleles of this invention are
defined by a
nucleotide locus, the nucleotide location in the human mitochondria) genome,
and by the A G
C T (or LT) nucleotide. An isolated nucleic acid molecule, in a library of
this invention, can
be identified as containing a nucleotide allele of this invention, because the
nucleotide allele
of this invention is bounded on at least one side by its context in the
mitochondria) genome.
Statistically, to be unique in the human mitochondria) genome, such a molecule
would need
to be at least about seven nucleotides long. Statistically, to be unique in
the total human
genome, including the mitochondria) genome, such a molecule would need to be
at least
about fifteen nucleotides long. Examples of isolated nucleic acid molecules of
this invention
are molecules containing the following nucleotide alleles: 1) Cambridge
alleles at human
mtDNA nucleotide loci 168-170, non-Cambridge alleles at locus 171A, and
Cambridge
alleles at human mtDNA nucleotide loci 172-174; and 2) Cambridge alleles at
11940-11946,
non-Cambridge alleles at 119476, and Cambridge alleles at 11948-11954. An
isolated
nucleic acid molecule of this invention may contain more than one nucleotide
allele of this
invention. The nucleotide allele of this invention may be at any position in
the isolated
nucleic acid molecule. Often it is useful to have the relevant nucleotide
allele in the center of
the isolated nucleic acid molecule or on the 3' end of the molecule. Isolated
nucleic acid
molecules of this invention are useful for interrogating, determining the
presence or absence
of, a nucleotide allele at the corresponding nucleotide locus in the
mitochondria) genome in a
sample containing mitochondria) nucleic acid from a human, using any method
known in the
art. Methods for determining the presence of absence of the nucleotide allele
include allele-
specific PCR and nucleic acid array hybridization or sequencing.
The alleles and libraries of this invention are useful for designing probes
for nucleic
acid arrays. This invention provides nucleic acid arrays having two or more
nucleic acid
molecules or spots (each spot comprising a plurality of substantially
identical isolated nucleic
acid molecules), each molecule having the sequence of an allele of this
invention. The
molecules on the arrays of this invention are usually about 7 to about 30
nucleotides long.
The arrays are useful for detecting the presence or absence of alleles. Arrays
of this
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
invention are also useful for sequencing human mtDNA. Alleles may be selected
from sets
of nucleotide alleles including human mtDNA nucleotide alleles, non-Cambridge
human
mtDNA nucleotide alleles, human mtDNA nucleotide alleles in 48 genomes and the
Cambridge sequence, non-Cambridge human mtDNA nucleotide alleles in 48
genomes,
nucleotide alleles useful for diagnosing human haplogroups and macro-
haplogroups,
nucleotide alleles useful for diagnosing human haplogroups, and evolutionarily
significant
human mitochondrial nucleotide alleles as listed in the various Tables and
portions of tables
hereof. Arrays of this invention may contain molecules capable of
interrogating all of the
alleles in one of the above-mentioned sets of alleles. A genotyping array
useful for detecting
sequence polymorphisms, such as are provided by this invention, are similar to
Affymetrix
(Santa Clara, CA, USA) genotyping arrays containing a Perfect Match probe (PM)
and a
corresponding Mismatch probe (MM). A PM probe could comprise a non-Cambridge
allele
at a selected nucleotide locus and the corresponding MM probe could comprise
the
corresponding Cambridge allele at the selected nucleotide locus. Arrays of
this invention
include sequencing arrays for human mtDNA.
As used herein, "array" refers to an ordered set of isolated nucleic acid
molecules or
spots consisting of pluralities of substantially identical isolated nucleic
acid molecules.
Preferably the molecules are attached to a substrate. The spots or molecules
are ordered so
that the location of each (on the substrate) is known and the identity of each
is known.
Arrays on a microscale can be called microarrays. Microarays on solid
substrates, such as
glass or other ceramic slides, can be called gene chips or chips.
Arrays are preferably printed on solid substrates. Before printing, substrates
such as
glass slides are prepared to provide a surface useful for binding, as is known
to the art.
Arrays may be printed using any printing techniques and machines known in the
art. Printing
involves placing the probes on the substrate, attaching the probes to the
substrate, and
blocking the substrate to prevent non-specific hybridization. Spots are
printed at known
locations. Arrays may be printed on glass microscope slides. Alternatively,
probes may be
synthesized in known positions on prepared solid substrates (Affymetrix, Santa
Clara, CA,
USA).
Arrays of this invention may contain as few as two spots, or more than about
ten
spots, more than about twenty-five spots, more than about one hundred spots,
more than
31
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
about 1000 spots, more than about 65,000 spots, or up to about several hundred
thousand
spots.
Using microarrays may require amplification of target sequences (generation of
multiple copies of the same sequence) of sequences of interest, such as by PCR
or reverse
transcription. As the nucleic acid is copied, it is tagged with a fluorescent
label that emits
light like a light bulb. The labeled nucleic acid is introduced to the
microarray and allowed to
react for a period of time. This nucleic acid sticks to, or hybridizes, with
the probes on the
array when the probe is sufficiently complementary to the labeled, amplified,
sample nucleic
acid. The extra nucleic acid is washed off of the array, leaving behind only
the nucleic acid
that has bound to the probes. By obtaining an image of the array with a
fluorescent scanner
and using software to analyze the hybridized array image, it can be determined
if, and to what
extent, genes are switched on and off, or whether or not sequences are
present, by comparing
fluorescent intensities at specific locations on the array. The intensity of
the signal indicates
to what extent a sequence is present. In expression arrays, high fluorescent
signals indicate
that many copies of a gene are present in a sample, and lower fluorescent
signal shows a gene
is less active. By selecting appropriate hybridization conditions and probes,
this technique is
useful for detecting single nucleotide polymorphisms (SNPs) and for
sequencing. Methods
of designing and using microarrays are continuously being improved (Relogio,
A. et al.
(2002) Nuc. Acids. Res. 30(11):e51; Iwasaki, H et al. (2002) DNA Res. 9(2):59-
62; and
Lindroos, K. et al. (2002) Nuc. Acids. Res. 30(14):E70).
Arrays of this invention may be made by any array synthesis methods known in
the
art such as spotting technology or solid phase synthesis. Preferably the
arrays of this
invention are synthesized by solid phase synthesis using a combination of
photolithography
and combinatorial chemistry. Some of the key elements of probe selection and
array design
are common to the production of all arrays. Strategies to optimize probe
hybridization, for
example, are invariably included in the process of probe selection.
Hybridization under
particular pH, salt, and temperature conditions can be optimized by taking
into account
melting temperatures and by using empirical rules that correlate with desired
hybridization
behaviors. Computer models may be used for predicting the intensity and
concentration-
dependence of probe hybridization.
32
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Detecting a particular polymorphism can be accomplished using two probes. One
probe is designed to be perfectly complementary to a target sequence, and a
partner probe is
generated that is identical except for a single base mismatch in its center.
In the Affymetrix
system, these probe pairs are called the Perfect Match probe (PM) and the
Mismatch probe
(MM). They allow for the quantitation and subtraction of signals caused by non-
specific
cross-hybridization. The difference in hybridization signals between the
partners, as well as
their intensity ratios, serve as indicators of specific target abundance, and
consequently of the
sequence.
Arrays can rely on multiple probes to interrogate individual nucleotides in a
sequence.
The identity of a target base can be deduced using four identical probes that
vary only in the
target position, each containing one of the four possible bases.
Alternatively, the presence of
a consensus sequence can be tested using one or two probes representing
specific alleles. To
genotype heterozygous or genetically mixed samples, arrays with many probes
can be created
to provide redundant information, resulting in unequivocal genotyping.
Probes fixed on solid substrates and targets (nucleotide sequences in the
sample) are
combined in a hybridization buffer solution and held at an appropriate
temperature until
annealing occurs. Thereafter, the substrate is washed free of extraneous
materials, leaving
the nucleic acids on the target bound to the fixed probe molecules allowing
for detection and
quantitation by methods known in the art such as by autoradiograph, liquid
scintillation
counting, andlor fluorescence. As improvements axe made in hybridization and
detection
techniques, they can be readily applied by one of ordinary skill in the art.
As is well known
in the art, if the probe molecules and target molecules hybridize by forming a
strong non-
covalent bond between the two molecules, it can be reasonably assumed that the
probe and
target nucleic acid are essentially identical, or almost completely
complementary if the
annealing and washing steps are carned out under conditions of high
stringency. The
detectable label provides a means for determining whether hybridization has
occurred.
When using oligonucleotides or polynucleotides as hybridization probes, the
probes
may be labeled. In arrays of this invention, the target may instead be labeled
by means
known to the art. Target may be labeled with radioactive or non-radioactive
labels. Targets
preferably contain fluorescent labels.
33
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Various degrees of stringency of hybridization can be employed. The more
stringent
the conditions are, the greater the complementarity that is required for
duplex formation.
Stringency can be controlled by temperature, probe concentration, probe
length, ionic
strength, time, and the like. Hybridization experiments are often conducted
under moderate
to high stringency conditions by techniques well know in the art, as
described, for example in
Keller, G.H., and M.M. Manak (1987) DNA Probes, Stockton Press, New York, NY.,
pp.
169-170, hereby incorporated by reference. However, sequencing arrays
typically use lower
hybridization stringencies, as is known in the art.
Moderate to high stringency conditions for hybridization are known to the art.
An
example of high stringency conditions for a blot are hybridizing at 68°
C in SX SSC/SX
Denhardt's solution/0.1% SDS, and washing in 0.2X SSC/0.1% SDS at room
temperature.
An example of conditions of moderate stringency are hybridizing at 68°
C in SX SSC/SX
Denhardt's solution/0.1% SDS and washing at 42° C in 3X SSC. The
parameters of
temperature and salt concentration can be varied to achieve the desired level
of sequence
identity between probe and target nucleic acid. See, e.g., Sambrook et al.
(1989) vide infra or
Ausubel et al. (1995) Current Protocols in Molecular Biolo~y, John Wiley &
Sons, NY, NY,
for further guidance on hybridization conditions.
The melting temperature is described by the following formula (Beltz, G.A. et
al.,
[1983] Methods ofEnzymolo~y, R. Wu, L. Grossman and K. Moldave (Eds.] Academic
Press, New York 100:266-285).
Tm=8l.So C + 16.6 Log[Na+]+0.41(+G+C)-0.61(%formamide)-600/length of duplex in
base
pairs.
Washes can typically be carried out as follows: twice at room temperature for
15
minutes in 1X SSPE, 0.1% SDS (low stringency wash), and once at TM-20o C for
15
minutes in 0.2X SSPE, 0.1% SDS (moderate stringency wash).
Nucleic acid useful in this invention can be created by Polymerase Chain
Reaction
(PCR) amplification. PCR products can be confirmed by agarose gel
electrophoresis. PCR
is a repetitive, enzymatic, primed synthesis of a nucleic acid sequence. This
procedure is
well known and commonly used by those skilled in this art (see Mullis, U.S.
Patent Nos.
34
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
4,683,195, 4,683,202, and 4,800,159; Saiki et al. [1985] Science 230:1350-
1354). PCR is
used to enzymatically amplify a DNA fragment of interest that is flanked by
two
oligonucleotide primers that hybridize to opposite strands of the target
sequence. The
primers are oriented with the 3' ends pointing towards each other. Repeated
cycles of heat
denaturation of the template, annealing of the primers to their complementary
sequences, and
extension of the annealed primers with a DNA polymerase result in the
amplification of the
segment defined by the 5' ends of the PCR primers. Since the extension product
of each
primer can serve as a template for the other primer, each cycle essentially
doubles the amount
of DNA template produced in the previous cycle. This results in the
exponential
accumulation of the specific target fragment, up to several million-fold in a
few hours. By
using a thermostable DNA polymerase such as the Taq polymerase, which is
isolated from
the thermophilic bacterium Thermus aquaticus, the amplification process can be
completely
automated. Other enzymes that can be used are known to those skilled in the
art.
Polynucleotide sequences of the present invention can be truncated andlor
mutated
such that certain of the resulting fragments and/or mutants of the original
full-length
sequence can retain the desired characteristics of the full-length sequence. A
wide variety of
restriction enzymes that are suitable for generating fragments from larger
nucleic acid
molecules are well known. In addition, it is well known that Ba131 exonuclease
can be
conveniently used for time-controlled limited digestion of DNA. See, for
example, Maniatis
(1982) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory,
New
York, pages 135-139, incorporated herein by reference. See also Wei et al.
(1983) J. Biol.
Chem. 258:13006-13512. By use of Ba131 exonuclease (commonly referred to as
"erase-a-
base" procedures), the ordinarily skilled artisan can remove nucleotides from
either or both
ends ofthe subject nucleic acids to generate a wide spectrum of fragments that
are
functionally equivalent to the subject nucleotide sequences. One of ordinary
skill in the art
can, in this manner, generate hundreds of fragments of controlled, varying
lengths from
locations all along the original molecule. The ordinarily skilled artisan can
routinely test or
screen the generated fragments for their characteristics and determine the
utility of the
fragments as taught herein. It is also well known that the mutant sequences
can be easily
produced with site-directed mutagenesis. See, for example, Larionov, O.A. and
Nikiforov,
V.G. (1982) Genetika 18(3):349-59; and Shortle, D. et al., (1981) Aranu. Rev.
Genet. 15:265-
94, both incorporated herein by reference. The skilled artisan can routinely
produce deletion-
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
insertion-, or substitution-type mutations and identify those resulting
mutants that contain
the desired characteristics of wild-type sequences, or fragments thereof.
Percent sequence identity of two nucleic acids. may be determined using the
algorithm
of Karlin and Altschul (1990) Proc. Natl. Acad. Sci. USA 87:2264-2268,
modified as in
Karlin and Altschul (1993) Proc. Natl. Acad. Sci. USA 90:5873-5877. Such an
algorithm is
incorporated into the NBLAST and XBLAST programs of Altschul et al. (1990) J.
Mol. Biol.
215:402-410. BLAST nucleotide searches are performed with the NBLAST program,
score
=100, wordlength = 12, to obtain nucleotide sequences with the desired percent
sequence
identity. To obtain gapped alignments for comparison purposes, Gapped BLAST is
used as
described in Altschul et al. (1997) Nucl. Acids. Res. 25:3389-3402. When
utilizing BLAST
and Gapped BLAST programs, the default parameters of the respective programs
(NBLAST
and XBLAST) are used. See http://www.ncbi.nih.gov.
Standard techniques for cloning, DNA isolation, amplification and
purification, for
enzymatic reactions involving DNA ligase, DNA polymerase, restriction
endonucleases and
the like, and various separation techniques useful herein are those known and
commonly
employed by those skilled in the art. A number of standard techniques are
described in
Sambrook et al. (1989) Molecular Cloning, Second Edition, Cold Spring Harbor
Laboratory,
Plainview, New York; Maniatis et al. (1982) Molecular Cloning, Cold Spring
Harbor
Laboratory, Plainview, New York; Wu (ed.) (1993) Meth. Enzymol. 218, Part I;
Wu (ed.)
(1979) Meth. EnzynZOl. 68; Wu et al. (eds.) (1983) Meth. Efazymol. 100 and
101; Grossman
and Moldave (eds.) Meth. Enzymol. 65; Miller (ed.) (1972) Experiments in
Molecular
Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York; Old and
Primrose
(1981) Princit~les of Gene Manipulation, University of California Press,
Berkeley; Schleif
and Wensink (1982) Practical Methods in Molecular Biology; Glover (Ed.) (1985)
DNA
Cloning Vol. I and II, IRL Press, Oxford, UK; Hames and Higgins (Eds.) (1985
Nucleic
Acid Hybridization, IEZL Press, Oxford, UK; Setlow and Hollaender (1979)
Genetic
E~ineeringLPrinciples and Methods, Vols. 1-4, Plenum Press, New York; and
Ausubel et al.
(1992) Current.Protocols in Molecular Biolo.~y, Greene/Wiley, New York, NY.
Abbreviations and nomenclature, where employed, are deemed standard in the
field and
commonly used in professional journals such as those cited herein.
36
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
This invention provides machine-readable storage devices and program storage
devices having data and methods for diagnosing haplogroups and physiological
conditions.
One program storage device provided by this invention contains the program
steps: a)
determining the haplogroup of a sample from an individual using nucleotide
sequence data
from nucleic acid in the sample; b) associating the haplogroup with
information identifying
the geographic region of the individual; c) comparing the haplogroup and
geographic region
of the sample to the set of haplogroups native to the geographic region of the
individual; and
d) diagnosing the individual with a predisposition to an energy metabolism-
related
physiological condition if the haplogroup of the individual is not within the
set of
haplogroups native to the geographic region of the individual; all said
program steps being
encoded in machine readable form, and all said information encoded in machine
readable
form. This invention also provides a data set, encoded in machine-readable
form, containing
nucleotide alleles listed in Table 19, with each allele associated with
encoded information
identifying a physiological condition in humans. These physiological
conditions are energy-
metabolism-related conditions including energetic imbalance, metabolic
disease, abnormal
energy metabolism, abnormal temperature regulation, abnormal oxidative
phosphorylation,
abnormal electron transport, obesity, amount of body fat, diabetes,
hypertension, and
cardiovascular disease. This storage device may also contain information
associating each
allele with one or more native geographic regions. A program storage device
provided by
this invention contains input means for inputting the haplogroup of an
individual and the
geographic region of that individual, and contains information associating
alleles with native
geographic regions, and program steps for diagnosing the individual with a
predisposition to
a physiological condition. A storage device containing a data set in machine
readable form
provided by this invention may include encoded information comprising amino
acid alleles
listed in Table 19, with each allele associated with a physiological condition
in humans.
It will be appreciated by those of ordinary skill in the art that populations,
subpopulations, organelles, and amino acid and nucleotide sequence comparison
methods,
neutrality test methods, nucleotide sequencing methods, codons, samples,
sample collection
techniques, sample preparation techniques, probes, probe generation
techniques, genes
involved in mitochondria) biology, hybridization techniques, array printing
techniques,
physiological conditions, cell lines, mutant strains, organisms, tissues,
solid substrates,
machine-readable storage devices, program devices, and methods of data
analyses other than
those specifically disclosed herein are available in the art and can be
employed in the practice
37
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
of this invention. All art-known functional equivalents are intended to be
encompassed
within the scope of this invention.
The following examples are provided for illustrative purposes, and are not
intended to
limit the scope of the invention as claimed here, Any variations in the
compositions and
methods exemplified that occur to the skilled artisan are intended to fall
within the scope of
the present invention.
EXAMPLES
Example 1
This invention provides human mtDNA polymorphisms found in all the major human
haplogroups. Table 3 shows naturally occurnng nucleotide alleles identified in
the complete
mtDNA sequences of 103 individuals, as compared to the mtDNA Cambridge
sequence. All
nucleotide sequences not listed are identical to the Cambridge sequence.
Nucleotide alleles
previously known to be associated with disease conditions, such as those
listed in Table 1, are
not listed in Table 3. Some deletion or rearrangement polymorphisms have also
been
excluded. All polymorphisms listed are nucleotide substitutions except for a
nine-adenine
nucleotide deletion at positions 8271-8279.
38
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Table 227 A G
3
Human NA tide es
MtD NucleoAllel
228 G A
non- 235 A G
nucleotideCambridgeCambridge 236 T C
locus allelesalleles
247 G A
64 C T
250 T C
72 T C
252 T C
73 A G
263 A G
89 T C
291 A G
93 A G
295 C T
95 A C
297 A G
114 C T
316 G A
143 G A
317 C A
146 T C
317 C G
150 C T
320 C T
151 C T
325 C T
152 T C
340 C T
153 A G
357 A G
171 G A
373 A G
180 T C
400 T G
182 C T
408 T A
183 A G
418 C T
185 G A
456 C T
185 G T
462 C T
186 C A
465 C T
189 A C
467 C T
189 A G
471 T C
194 C T
480 T C
195 T A
482 T C
195 T C
489 T C
198 C T
493 A G
199 T C
499 G A
200 A G
508 A G
204 T C
593 T C
207 G A
597 C T
208 T C
663 A G
210 A G
678 T C
212 T C
680 T C
215 A G
709 G A
217 T C
710 T C
225 G A
39
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
721 T C 2245 A C
750 A G 2245 A G
769 G A 2263 C A
825 T A 2308 A G
827 A G 2332 C T
850 T C 2352 T C
921 T C 2358 A G
930 G A 2380 C T
961 T C 2416 T C
961 T G 2483 T C
1018 G A 2581 A G
1041 A G 2639 C T
1048 C T 2650 C T
1119 T C 2706 A G
1189 T C 2755 A G
1243 T C 2758 G A
1290 C T 2768 A G
1382 A C 2789 C T
1406 T C 2792 A G
1415 G A 2834 C T
1420 T C 2836 C A
1438 A G 2857 T C
1442 G A 2863 T C
1503 G A 2885 T C
1598 G A 3010 G A
1700 T C 3083 T C
1703 C T 3197 T C
1706 C T 3200 T A
1709 G A 3202 T C
1715 C T 3204 C T
1719 G A 3206 C T
1736 A G 3221 A G
1738 T C 3290 T C
1780 T C 3308 T C
1811 A G 3316 G A
1888 G A 3372 T C
1927 G A 3394 T C
2000 C T 3438 G A
2060 A G 3450 C T
2092 C T 3480 A ~ G
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
3505 A G 4203 A G
3513 C T 4216 T C
3516 C A 4221 C T
3516 C G 4225 A G
3547 A G 4232 T C
3549 C T 4248 T C
3552 T A 4312 C T
3552 T C 4336 T C
3565 A G 4370 T C
3594 C T 4388 A G
3644 T C 4454 T A
3666 G A 4491 G A
3693 G A 4506 A G
3699 C T 4508 C T
3720 .A G 4512 G A
3756 A G 4529 A C
3796 A G 4529 A T
3796 A T 4541 G A
3796 A C 4580 G A
3808 A G 4586 T C
3816 A G 4596 G A
3834 G A 4646 T C
3843 A G 4655 G A
3847 T C 4688 T C
3866 T C 4695 T C
3918 G A 4715 A G
3921 C A 4742 T C
3927 A G 4767 A G
3970 C T 4769 A G
3981 A G 4820 G A
4025 C T 4824 A G
4040 C T 4833 A G
4044 A G 4841 G A
4048 G A 4883 C T
4086 C T 4907 T C
4104 A G 4917 A G
4117 T C 4960 C T
4122 A G 4977 T C
4123 A G 4994 A G
4158 A G 5004 T C
I
41
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
5027 C T 5821 G A
5036 A G 5826 T C
5043 G T 5843 A G
5046 G A 5951 A G
5063 T C 5984 A G
5096 T C 5987 G T
5108 T C 6026 G A
5147 G A 6029 C T
5153 A G 6045 G T
5178 C A 6071 T C
5231 G A 6077 C T
523? G A 6104 C T
5255 C T 6150 G A
5262 G A 6152 T C
5263 C T 6164 C T
5285 A G 6167 T C
5300 G T 6182 G A
5330 C A 6185 T C
5331 C A 6185 T C
5390 A G 6221 T C
5393 T C 6227 T C
5417 G A 6253 T C
5426 T C 625? G A
5442 T C 6324 G A
5460 G A 6366 G A
5465 T C 6371 C T
5471 G A 6392 T C
~
5492 T C 6473 C T
5495 T C 6491 C A
5580 T C 6524 T C
5581 A G 6548 C T
5601 C T 6587 C T
5603 C T 6607 T' C
5606 C T 6680 T C
5633 C T 6713 G T
5655 T C 6719 T C
5711 A G 6734 G A
5773 G A 6752 A G
5811 A _ G 6770 A G
5814 T C ~ 6776 T C
42
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
6815 T C 7675 C T
6827 T C 7693 C T
6875 C A 7694 C T
6938 C T 7697 G A
6962 G A 7744 T C
6989 A G 7765 A G
7028 C T 7768 A G
7052 A G 7771 A G
7055 A G 7858 C T
7058 T A 7861 T C
7076 A G 7864 C T
7146 A G 7867 C T
7154 A G 7933 A G
7175 T C 7948 C T
7196 C A 7999 T C
7202 A G 8014 A G
7226 G A 8020 G A
7256 C T 8027 G A
7257 A G 8080 C T
7271 A G 8087 T C
7274 C T 8113 C A
7319 T C 8142 C T
7337 G A 8149 A G
7347 G A 8152 G A
7389 T C 8155 G A
7403 A G 8185 T C
7424 A G 8200 T C
7444 G A 8206 G A
7476 C T 8248 A G
7493 C T 8251 G A
7521 G A 8260 T C
7561 T C 8269 G A
7571 A G , 8271-8279A DEL
7600 G A 8286 T C
7624 T A 8292 G A
7645 T C 8298 T C
7648 C T 8344 A G
7660 T C 8387 G A
7664 G A 8389 A G
7673 A G 8392 G A
--
43
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
8404 T C 8943 C T
8414 C T 8962 A G
8428 C T 8994 G A
8448 T C 9042 C T
8460 A G 9053 G A
8468 C T 9055 G A
8472 C T 9072 A G
8473 T C 9077 T G
8485 G A 9090 T C
8545 G A 9093 A C
8553 C T 9103 T C
8563 A G 9114 A G
8566 A G 9120 A G
8577 A G 9123 G A
8584 G A 9123 G A
8618 T C 9136 A G
8655 C T 9151 A G
8697 G A 9156 A G
8701 A G 9174 T C
8703 C T 9221 A G
8705 T C 9237 G A
8709 C T 9242 A G
8721 A G 9248 C T
8733 T C 9263 A G
8764 G A 9272 C T
8781 C A 9296 C T
8784 A G 9311 T C
8790 G A 9325 T C
8793 T C 9335 C T
8794 C T 9347 A G
8805 A G 9355 A G
8836 A G 9356 C T
8838 G A 9377 A G
8856 G A 9402 A C
8860 A G 9449 C T
8875 T C 9456 A G
8877 T C 9477 G A
8911 T C 9509 T C
8913 A G 953 C T
6
8928 T C I _ T C
_
9540
~
44
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
9545 A G 10358 A G
9548 G A 10370 T C
9554 G A 10398 A G
9559 C G 10400 C T
9575 G A 10410 T C
9591 G A 10414 G T
9599 C T 10427 G A
9632 A G 10463 T C
9647 T C 10499 A G
9667 A G 10505 T C
9682 T C 10550 A G
9698 T C 10586 G A
9755 G A 10589 G A
9818 C T 10609 T C
9822 C A 10637 C T
9824 T A 10640 T C
9911 C T 10646 G A
9932 G A 10659 C T
9950 T C 10664 C T
9957 T C 10667 T C
9966 G A 10688 G A
9977 T C 10736 C T
10034 T C 10790 T C
10086 A G 10792 A G
10086 A C 10793 C T
10115 T C 10804 A G
10118 T C 10810 T C
10142 C T 10819 A G
10151 A G 10828 T C
10152 G C 10873 T C
101.72 G A 10876 A G
10182 G C 10894 C T
10197 G A 10915 T C
10238 T C 10920 C T
10253 T C 10939 C T
10256 T C 10966 T C
10310 G A 10984 C G
10313 A G 11002 A G
10321 T C 11016 G A
10325 G A 11017 T C
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
11023 A G 11947 A G
11078 A G 11959 A G
11092 A G 11963 G A
11147 T C 11969 G A
11150 G A 12007 G A
11167 A G 12049 C T
11172 A G 12070 G A
11176 G A 12083 T G
11177 C T 12121 T C
11215 C T 12134 T C
11251 A G 12153 C T
11257 C T 12172 A G
11296 C T 12175 T C
11299 T C 12234 A G
11332 C T 12236 G A
11362 A G 12239 C T
11365 T C 12248 A G
11377 G A 12308 A G
11467 A G 12346 C T
11476 C T 12358 A G
11536 C T 12361 A G
11590 A G 12372 G A
11611 G A 12373 A G
11641 A G 12397 A G
11653 A G 12406 G A
11654 A G 12414 T C
11674 C T 12477 T C
11701 T C 12501 G A
11719 G A 12507 A G
11722 T C 12519 T C
11767 C T 12528 G A
11812 A G 12540 A G
11854 T C 12612 A G
11884 A G 12630 G A
11887 G A 12633 C T
11893 A G 12635 T C
11899 T C 12669 C T
11909 A G 12672 A G
11914 G A 12693 ~ A G
11944 T C 12705 C T
46
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
12720 A G 13512 A G
12738 T C 13563 A G
12768 A G 13590 G A
12771 G A 13594 A G
12810 A G 13602 T C
12822 A G 13611 A G
12850 A G 13617 T C
12879 T C 13641 T C
12882 C T 13650 C T
12930 A T 13651 A G
12940 G A 13660 A G
12948 A G 13708 G A
12967 A C 13722 A G
12972 A G 13734 T C
12999 A G 13759 G A
13020 T C 13780 A G
13059 C T 13789 T C
13068 A G 13803 A G
13101 A C 13812 T C
13104 A G 13818 T C
13105 A G 13819 T C
13135 G A 13827 A G
13143 T C 13880 C A
13145 G A 13886 T C
13149 A G 13914 C A
13194 G A 13924 C T
13197 C T 13927 A T
13212 C T 13928 G C
13221 A G 13958 G C
13263 A G 13965 T C
13276 A G 13966 A G
13281 T C 13980 G A
13368 G A 14000 T A
13440 C G 14016 G A
13477 G A 14020 T C
13485 A G 14022 A G
13494 C T ' 14025 T C
13500 T C 14034 T C
13500 T G 14059 A G
13506 C I T ~ 14070 A T
47
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
14070 A G 14668 C T
14088 T C 14693 A G
14094 T C 14766 C T
14097 C T 14769 A G
14118 A G 14783 T C
14128 A G 14793 A G
14148 A G 14798 T C
14152 A G 14812 C T
14167 C T 14836 A G
14178 T C 14861 G A
14182 T C 14862 C T
14200 T C 14905 G A
14203 A G 14911 C T
14209 A G 14971 T C
14212 T C 14974 C G
14215 T C 14979 T C
14221 T C 15016 C T
14233 A G 15034 A G
14272 C G 15043 G A
14284 C T 15110 G A
14308 T C 15113 A G
14311 T C 15115 T C
14318 T C 15136 C T
14319 T C 15172 G A
14371 T C 15204 T C
14374 T C 15217 G A
14384 G C 15218 A C
14455 C T 15229 T C
14459 G A 15238 C G
14470 T C 15244 A G
14484 T C 15257 G A
14488 T C 15261 G A
14502 T C 15301 G A
14560 G A~ 15317 G A
14566 A G 15318 C T
14569 G A 15323 G A
14571 T A 15326 A G
14580 A G 15346 G A
14587 A G 15358 A G
14605 A G 15431 G A
48
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
15442 A G 15968 T C
15452 C A 16017 T C
15466 G A 16038 A G
15470 T C 16051 A G
15487 A T 16069 C T
15497 G A 16071 C T
15514 T C 16075 T C
15519 T C 16086 T C
15535 C T 16093 T C
15607 A G 16108 C T
15626 C T 16111 C T
15629 T C 16114 C A
15646 C T 16124 T C
15661 C T 16126 T s C
15663 T C 16129 G A
15670 T C 16129 G C
15724 A G 16140 T C
15731 G A 16144 T C
15746 A G 16145 G A
15766 A G 16147 C T
15784 T C 16148 C T
15793 C T 16153 G A
15803 G A 16162 A G
15806 G A 16163 A C
15812 G A 16166 A C
15824 A G 16167 C T
15833 C T 16168 C T
15849 C T 16169 C T
15884 G C 16171 A G
15900 T C 16172 T C
15904 C T 16175 A G
15907 A G 16176 C T
15924 A G 16182 A C
15927 G A 16183 A C
15928 G A 16184 C T
-15930 G A 16185 C T
15932 T C 16186 C T
15939 C T 16187 C T
15941 T C 16188 C A
5942 T C I 16188 C G
49
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
16189 T C 16278 C T
16192 C T 16284 A G
16193 C T 16286 C G
16207 A G 16287 C T
16209 T C 16288 T C
16212 A G 16290 C T
16213 G A 16291 C T
16214 C T 16292 C T
16217 T C 16293 A G
16219 A G 16294 C T
16223 C T 16296 C T
16224 T C 16298 T C
16227 A G 16304 T C
16229 T C 16309 A G
16230 A G 16311 T C
16231 T C 16316 A G
16232 C T 16317 A T
16234 C T 16318 A T
16235 A G 16319 G A
16239 C T 16320 C T
16241 A G 16324 T C
16242 C T 16325 T C
16243 T C 16326 A G
16245 C T 16327 C T
16247 A G 16343 A G
16249 T C 16344 C T
16254 A C 16354 C T
16255 G A 16355 C T
16256 C T 16356 T C
16257 C T 16357 T C
16258 A G 16360 C T
16260 C T 16362 T C
16261 C T 16366 C T
16264 C T 16368 T C
16265 A C 16390 G A
16266 C T 16391 G A
16268 C T 16399 A G
16270 C T 16438 G A
16_271 T C 16439 C A
16274 G A 16483 G A
I
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
16519 T C
16527 C T
51
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Table 4 lists the nucleotide alleles identified in 48 mitochondria) genomes as
compared to the Cambridge sequence.
Table 4
Human MtDNA Nucleotide Alleles in 48 Genomes
52
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
non- 316 G A
l C
tid brid
nuc am Cambridge
eo ge alleles 317 C G
e ,alleles
locus
64 C T 320 C T
72 T C 325 C T
73 A G 340 C T
89 T C 357 A G
93 A G 400 T G
95 A C 418 C T
114 C T 456 C T
146 T C 462 C T
150 C T 467 C T
151 C T 482 T C
152 T C 489 T C
153 A G 493 _ G
A
171 G A 499 G A
180 T C 508 A G
182 C T 597 C T
185 G A 663 A G
185 G T 680 T C
186 C A 709 G A
189 A C 710 T C
194 C T 750 A G
195 T C 769 G A
198 C T 825 T A
199 T C 827 A G
200 A G 921 T C
204 T C 930 G A
207 G A 961 T C
210 A G 961 T G
217 T C 1018 G A
225 G A 1048 C T
227 A G 1189 T C
228 G A 1243 T C
235 A G 1290 C T
236 T C 1406 T C
247 G A 1415 G A
250 T C 1438 A G
263 A G 1442 G A
295 C T 1598 G A
297 A G 1700 T C
53
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
1703 C T 3516 C G
1706 C T 3547 A G
1709 G A 3552 T A
1715 C T 3552 T C
1719 G A 3565 A G
1736 A G 3594 C T
1738 T C 3644 T C
1780 T C 3666 G A
1811 A G 3693 G A
1888 G A 3720 A G
2092 C T 3756 A G
2245 A C 3796 A G
2245 A G 3796 A T
2308 A G 3796 A C
2332 C T 3808 A G
2352 T C 3816 A G
2358 A G 3834 G A
2416 T C 3843 A G
2581 A G 3847 T C
2639 C T 3866 T C
2706 A G 3921 C A
2758 G A 3970 C T
2768 A G 3981 A G
2789 C T 4025 C T
2834 C T 4040 C T
2857 T C 4044 A G
2885 T C 4086 C T
3010 G A 4104 A G
3083 T C 4122 A G
3197 T C 4123 A G
3200 T A 4158 A G
3202 T C 4216 T C
3221 A G 4221 C T
3308 T C 4225 A G
3316 G A 4232 T C
3394 T C 4248 T C
3450 C T 4312 C T
3480 A G 4336 T C
3505 A G 4370 T C
3516 C A 4454 T A
54
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
4529 A C 5601 C T
4529 A T 5603 C T
4580 G A 5606 C T
4586 T C 5633 C T
4596 G A 5711 A G
4646 T C 5773 G A
4715 A G 5814 T C
4767 A G 5951 A G
4769 A G 5984 A G
4820 G A 6026 G A
4824 A G 6029 C T
4833 A G 6045 C T
4841 G A 6071 T C
4883 C T 6152 T C
4907 T C 6185 T C
4917 A G 6221 T C
4960 C T 6227 T C
4977 T C 6257 G A
5027 C T 6371 C T
5036 A G 6392 T C
5043 G T 6473 C T
5046 G A 6491 C A
5096 T C 6607 T C
5108 T C 6680 T C
5147 G A 6713 C T
5153 A G 6734 G A
5178 C A 6752 A G
5231 G A 6776 T C
5300 C T 6815 T C
5331 C A 6827 T C
5390 A G 6962 G A
5393 T C 6989 A G
5417 G A 7028 C T
5426 T C . 7052 A G
5442 T C 7055 A G
5460 G A 7146 A G
5465 T C 7154 A G
5471 G A 7175 T C
5495 T C 7196 C A
5581 A G 7256 C T
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
7271 A G 8428 C T
7274 C T 8448 T C
~
7389 T C 8460 A G
7424 A G 8468 C T
7476 C T 8472 C T
7521 G A 8545 G A
7561 T C 8553 C T
7600 G A 8563 A G
7624 T A 8566 A G
7664 G A 8584 G A
7694 C T 8618 T C
7765 A G 8655 C T
7771 A G 8697 G A
7864 C T 8701 A ' G
7867 C T 8705 T C
7933 A G 8709 C T
7999 T C 8721 A G
8027 G A 8790 G A
8080 C T 8794 C T
8087 T C 8836 A G
8113 C A 8856 G A
8142 C T 8860 A G
8149 A G 8875 T C
8152 G A 8913 A G
8155 G A 8962 A G
8185 T C 8994 G A
8200 T C 9042 C T
8206 G A 9053 G A
8248 A G 9055 G A
8251 G A 9072 A G
8260 T C 9077 T C
8269 G A 9090 T C
8271-8279A DEL 9093 A C
8286 T C 9114 A G
8298 T C 9120 A G
8344 A G 9123 G A
8387 G A 9151 A G
8389 A G 9221 A G
8392 G A 9237 G A
8414 C I~ 9325 T C
56
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
9335 C T 10463 T C
9347 A G 10550 A G
9355 A G 10586 G A
9377 A G 10589 G A
9402 A C 10609 T C
9449 C T 10637 C T
9456 A G 10646 G .A
9477 G A 10659 C T
9540 T C 10664 C T
9545 A G 10688 G A
9548 G A 10790 T C
9559 C G 10810 T C
9575 G A 10828 T C
9632 A G 10873 ~ T C
9682 T C 10876 A G
9698 T C 10915 T C
9755 G A 10966 T C
9818 C T 10984 C G
9822 C A 11002 A G
9911 C T 11078 A G
9932 G A 11092 A G
9950 T C 11147 T C
9957 T C 11167 A G
9966 G A 11172 A G
10034 T C 11176 G A
10086 A G 11177 C T
10086 A C 11215 C T
10115 T C 11251 A G
10151 A G 11257 C T
10152 G C 11299 T C
10172 G A 11332 C T
10182 G C 11362 A G
10238 T C 11377 G A
10256 T C 11467 A G
10310 G A 11476 C T
10321 T C 11536 C T
10325 G A 11590 A G
10398 A G 11641 A G
10400 C T 11674 C T
10414 G T 11719 G A
57
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
11767 C T 12967 A C
11812 A G ~ 12972 A G
11854 T C 13020 T C
11899 T C 13068 A G
11914 G A 13101 A C
11944 T C 13104 A G
11947 A G 13105 A G
11969 G A 13194 G A
12007 G A 13263 A G
12083 T G 13276 A G
12121 T C 13368 G A
12172 A G 13440 C G
12234 A G 13485 A G
12236 G A 13494 C T
12308 A G 13500 T G
12358 A G 13506 C T
12361 A G 13512 A G
12372 G A 13563 A G
12373 A G 13590 G ' A
12397 A G 13617 T C
12406 G A 13650 C T
12414 T C 13708 G A
12501 G A 13734 T C
12507 A G 13759 G A
12519 T C 13780 A G
12528 G A 13789 T C
12540 A G 13803 A G
12612 A G 13812 T C
12633 C T 13827 A G
12669 C T 13880 C A
12672 A G 13886 T C
12693 A G 13914 C A
12705 C T 13924 C T
12720 A G 13928 G C
12738 T C 13958 G C
12810 A G 13966 A G
12822 A G 14000 T A
12882 C T 14016 G A
12930 A T 14034 T C
12948 A G 14059 A G
58
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
14070 A G 15136 C T
14088 T C 15172 G A
14118 A G 15204 T C
14128 A G 15217 G A
14148 A G 15218 A C
14167 C T 15238 C G
14178 T C 15257 G A
14200 T C 15261 G A
14203 A G 15301 G A
14215 T C 15317 G A
14221 T C 15318 C T
14233 A G 15323 G A
14272 C G 15326 A G
w14284 C T 15431 G A
14308 T C 15442 A G
14318 T C 15452 C A
14374 T C 15466 G A
14459 G A 15487 A T
14470 T C 15497 G A
14484 T C 15519 T C
14488 T C 15535 C T
14502 T C 15607 A G
14560 G A 15661 C T
14566 A G 15724 A G
14569 G A 15766 A G
14668 C T 15784 T C
14693 A G 15793 C T
14766 C T 15806 G A
14783 T C 15812 G A
14?93 A G 15824 A G
14798 T C 15833 C T
14836 A G 15849 C T
14861 G A 15884 G C
14905 G A 15900 T C
14911 C T 15904 C T
14974 C G 15907 A G
15034 A G 15924 A G
15043 G A 15928 G A
15110 G A 15930 G A
15115 T C 15939 C T
59
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
15941 T C 16224 T C
15968 T C 16227 A G
16017 T C 16229 T C
16051 A G 16230 A G
16069 C T 16231 T C
16086 T C 16232 C T
16093 T C 16234 C T
16108 C T 16235 A G
16111 C T 16239 C T
16114 C A 16243 T C
16124 T C 16245 C T
16126 T C 16249 T C
16129 G A 16254 A C
16129 G C 16255 G ' A
16145 G A 16256 C T
16148 C T 16258 A G
16153 G A 16260 C T
16162 A G 16261 C T
16163 A C 16264 C T
16167 C T 16265 A C
16168 C T 16266 C T
16172 T C 16270 C T
16176 C G 16274 G A
16182 A C 16278 C T
16183 A C 16284 A G
16184 C T 16290 C T
16185 C T 16291 C T
16186 C T 16292 C T
16187 C T 16293 A G
16188 C A ' 16294 C T
16188 C G 16296 C T
16189 T C 16298 T C
16192 C T 16304 T C
16193 C T 16309 A G
16212 A G 16311 T C
16213 G A 16317 A T
16214 C T 16318 A T
16217 T C 16319 G A
16219 A G 16320 C T
-
16223 C T 16325 T C
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
16327 C T
16355 C T
16356 T C
16360 C T
163 T C
62
16366 C T
16368 T C
16390 G A
16391 G A
16399 A G
16519 T C
61
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Example 2
The mtDNA sequences of Example 1 were chosen because they represent all of the
major haplogroup lineages in humans. Analysis of these sequences has
reaffirmed that all
human mtDNAs belong to a single maternal tree, rooted in Africa (R. L. Cann et
al., Nature
325:31-36 (1987); M. J. Johnson et al., (1983) .journal ofMolecaclar Evolution
19:255-271;
D. C. Wallace et al., "Global Mitochondrial DNA Variation and the Origin of
Native
Americans" in The Origin of Humankind, M. Aloisi, B. Battaglia, E. Carafoli,
G. A. Danieli,
Eds., Venice (IOS Press, 2000); M. Ingman et al., (2000) Nature 408:708-13;
and D. C.
Vijallace et al., (1999) Gene 238:211-230). A cladogram of these mtDNA
sequences is
shown in FIG 1. Haplogroups are designated on branches of the tree. A
calibration of the
sequence evolution rate for the coding regions of the mtDNA, based on a human-
chimpanzee
divergence time of 6.5 million years ago (MYA) ( M. Goodman et al., (1998) Mol
Phylogenet. Evol. 9:585-98), has permitted an estimate of the time to the most
recent
common ancestor (MRCA) of the human mtDNA phylogeny at 200,000 years before
present (YBP), and an estimate of the time of the MRCA for each major
haplogroup (Table
5).
Table 5
Coalescence dates for haplogroups*
62
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Sample Time to MRCA s.e. Time to MRCA s.e.
Haplogroup sizes (x 10~ mutations per (X 103 yeaxs) b
n ) a
chimp+human 1 + 104 818.05 0.75 6,500
humans 104 24.88 0.90 198 19
LO 8 17.92 1.87 142 17
Ll 9 17.81 1.77 142 17
L2 7 11.57 1.30 91.9 11.8
N 50 8.090.53 64.35.8
A 4 4.06 0.92 32.3 7.6
R 37 7:,660.51 60.95.5
HV 15 3.61 0.73 28.76.1
H 11 2.400.40 19.1 3.4
V 3 1.71 0.60 13.64.8
JT 7 6.29 0.74 50.0 6.7
J 4 4.330.87 34.47.2
T 3 1.400.55 11.14.4
U 4 6.510.66 51.76.2
M 22 8.150.74 64.87.1
CZ 10 5.91 0.87 47.0 -4- 7.6
C 9 3.560.65 2_8.35.5
_ _
D 6 4.190.67 33.35.7
- _
G 3 ~ 3~.7 7.g
4.75 0.93
* The high probability of reverse mutations in the control region led us to
calculate the times
to the MRCAs using the entire mtDNA, excluding the control region (np 577-
16023).
a Based on this value we estimated the average sequence evolution rate as
(1.26 ~ 0.08) x 10-
g per nucleotide per year, using the HKY85 model ( M. Hasegawa et al., (1985)
JMoI. Evol.
22:160-74 (1985)).
b Standard errors calculated from the inverse hessian at the maximum of the
likelihood do not
include any uncertainty in the calibration point, and were calculated using
the delta method.
The coalescence times of the various haplogroups may well be underestimated
because of
their small sample size.
Example 3
Inter-Continental Founder Events
The most striking feature of the mtDNA tree is the remarkable reduction in the
number of mtDNA lineages that are associated with the transition from one
continent to
another. For example, when humans moved to Eurasia from Africa, the number of
mitochondrial lineages was reduced from dozens to two lineages. While
northeastern Africa
encompasses the entire range of African mtDNA variation from the exclusively
African
haplogroups LO- L2 to the progenitors of the European and Asian mtDNA
lineages, only two
African mtDNA lineages, macro-haplogroups M and N, which arose about 65,000
YBP, left
63
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Africa to colonize Eurasia. Moreover, the times of the MRCAs of macro-
haplogroups M and
N as well as sub-macro-haplogroup R are similar, suggesting rapid population
expansion
associated with the colonization of Eurasia.
Similarly, when humans later moved from Central Asia to the Americas, the
number
of lineages was again reduced from dozens to about five. There is great mtDNA
diversity in
Asia, yet this diversity is substantially reduced in Siberia, and only five
mtDNA haplogroups
(A, B, C, D, and X), which arose in Asia about 28,000-34,000 YBP, successfully
crossed the
Bering land bridge to occupy the Americas. Human mtDNA haplogroup migrations
are
depicted in FIG 2.
Example 4
Further analysis demonstrated which alleles are descriptive of the major
haplogroups,
selected sub-haplogroups, and selected macro-haplogroups. The mtDNA nucleotide
positions and the relevant alleles are shown in FIG 3. The data is arranged as
a cladogram,
such that a group on the left contains all of the alleles to its right. A
vertical bar designates
that the alleles to the right of the bar are present in all of the groups to
the left of the bar. The
haplogroup data in FIG. 3 is summarized in Tables 6 and 7. The sub-haplogroup
data is
summarized in Tables 8 and 9. Each group contains the alleles listed below it.
64
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Table 6
LO L1 L2 L3 C D E G Z
1048T 23520 325T 23520 35520 4883T 162276 4833 110786
G
3516A 37960 6800 86180 47156 5178A 82000 16185T
4312T 59516 24160 100860 7196A 8414T 160170 162240
45860 59846 27586 10398A 8584A 14668T 16129A 16260T
54420 60710 41586 108196 95456 15487T
61850 90726 8206A 142120 13263 163620 163620
G
8113A 10586A 92216 161240 143180
8251A 128106 119440 16278T 162980 162980
93476 134856 138036 163620 16327T
94020 3666A 139580 4890 4890 4890 4890 4890
9818T 70556 16278T 10400T 10400T 10400T 10400T 10400T
10589A 73890 163906 147830 147830 147830 147830 147830
10664T 137890 15043A 15043A 15043A 15043A 15043A
109150 141786 15301A 15301A 15301A 15301A 15301A 15301A 15301A
12007A
132766
13506T
825A 825A
2758A 2758A
28850 28850
71466 71466
8468T 8468T
8655T 8655T -
10688A 10688A
108100 108100
131056 131056
769A 769A
1018A 1018A
3594T 3594T
41046 41046
7256T 7256T
7521A 7521A
13650T 13650T
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
C7HC7U E-~U r~, Ud dt7dE-~ C_7dU U U E-
N MM M 10O~dV d M00V'fh DO
-aMM ~Do0000oh h ~Ot0OO N~ b ~n~N O N ~O ~
00~ON ~ -,~ oo..-.~~ VM G1~DCvN .~N rt-~ h N O~to0
..-Nd-~D10vDCOO\~UOm ~h u~~D N ~ vW N ~Dt'~~nO
E~r.~.,~ ...-~-.~. oo~. ~ O
. ., r~ r .'-.,~r, ~.,-,.~,-. ,-,,~000,,~
U ~ N'U ~ M d M
H UN O ~ ~D~ ~1 H
N
_ O N O h
~ ~
hN ~tN -M~ N ~~O N ~ ~v O
'1
--n.~.r rr,-.np~p~.~
HC7C7d C7C7C7t7U UU C7U U f-~H E-aC7UU U E-
U UU d Nh ooN d~Oh ~--~O~NO OW avO~ oo v~ dE-~
t
M on M _ M
h ~Doo~nMvOOh O hO V1N ho0~N ~th-~ wtvoO N O h
C''DV1Met'MM ~ OO~O.-a.-r,-iNN N NM MM Mh N Od'OO
~Dh O .-,r..~NN M d'v7~OW O~Dl0~O~O~O~G~O~D~ON ~Oh~nO
M <fh 01~.~..r..r.-n.--n~ ,..,.-i.~~ ,--i.-a~ .r,--m--~,-r.~.-n,-100O~~
'~U ~ ~ h M dE-~m
_ O N ~O h
O N d
h
xN h N O~ 00
d~ oUo o~,.Uo U'U d-~H
v .~ E c
U 00 0,~.-.h ~ c r,
c0QvN h O N wO h
'
N V1W \O,.-n~ l~N Od 00
M N ~Oh~ O
h V~ -,h.-n-, .-n-r00Q1~
N
VU1M dE1M
M NN M ~O O N -iO h
M O~~ N N
Mv0~Dv0 h N Od'00
5"~h o0~ -,~ N ~Oh~nO
. .-
~ d M
O O ' M -H h
O N O
t~N Oeto0
N ~
N ~Oh~nO
.-,
~ r,oop~.-.
h M dH M
O N -~O h
'
" h N Od'00
N ~GhV1O
~'
,-.,r,ooOv..,
d C7U o oU,o'~o
a vor,h ~h
.~N ~ .~N
,~h 'Odr~D~O Od'00
~ O
M .
U d dd d~ ~ N
U dM ~ o.-<r~rooa.
O ON O ~-N
i~ d
N N.-,h v70000.-r,-.
H ~~ a
Q~MN O~
N O.~M
U1O~Dv0
i-.yt .~.-.,~
~O ~
~~ d
00Ov-r
.
66
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
d
a, o
'b
M M
MO v1
a~
U UE"'~U d
~O~I'CON ~'
00NI~~O O
MO _
~O
a,~rr~ ~-r
U d
coN o
M~p~p h
a~ ~
U C7U d
N O~N
W .-n.-r O
Mm O
aN -n.-n ,--n
' ~ ~ ~ oQ dH ~H do
E-~Uv U C~Q ~ett~a,o d oo~rWo .-.m
o 00~o.~
'~N OOv~ NO N01N MM 01rO~O~nN~O
NN o0M ~Yl~N NW O ~Ov) ~OOV7.-,N V1M
aM ~O~ N N0001~ ~~ l~~M d'h l~
C7 U C7d C7~ oho~oa dE'~c7E'~dHo
o w o 00~ ,-,d-t~o,o d ov ~~o...,w
o
N~ _ ~N N~ tNp~~ ~ _~ ~N hM
d' O
a'd' N No001.-~.r...a,., I~~--nM CYl~l~.~
o '~ ~ a ~ ~~ a
~ ~~ ~ d ~ ~O
N00 ~ hO NO1N MM Qv.-~OvOh NM
M ~hl~N N~ tpv0v~ ~OOW .-N V1
N No0Ow--~,--~.-..~ h ~M ~l~l~-~
U C7C7U C7~ o~ d C7U ~oUo d U C7E-~E~odoo ~ dE-~C7E"'~do
'
Nyp.~d'.-nNo0~-.io0l0V1O~00l~ do0V1~D00~1~ ~--~pQ,c0~hV~O.-W
n
.GO yn00I~l~V100~h~O~no0I~~ Nv1oOd'~Ov1~Oo0.~O~.-~OvO~ N~D
'r'~~ OvO~O OO NM v0OM M~h Nh o0~d W0O O M~OOV'1~N V1M
1-~M ~V1~OOv.-~~.-1M hl~~~ 00N N l~QO00.-i..r..~l~~M d'l~I~~
d C7U UU d U C7E~E-~ohoUo~ df-~C7E-~dHo
'~"'U ~o~no~oo doo~,~oo~noo..,od ood~<rto',~,
Gy" ,o '~ ,O OtnNv0
oot~
.~ ~OV1c0l~ ' Vn oodO WO o0.-~Ov-v ~N ~M
l0OM MV Nl~00~et~OO O M~OOV1
aN M l~I~.-r,--n CON N l~0000.~.-~.-n[v.-rM d'l~l~
d dE~d (~U UU C7~~ ~r ~~ d U C7E-~E-~odoo ~ dE-~C7E'~do
NM .-io0~ONN ~1N l~00~D~O C~O ~r00v1~Oo0W00,-.Od o0~h'V'v0 ~!1
..,v1d ~ ~~i'00O V ~WO O\O N~nv~v'1oDd'~O~n~Dao.--nQv.-~OvOv'1Nv0
.-~NO V7MV ~~tIy74O ON MM Nt~00~'cT~OO O M~OO~n~N Y1M
a0000-~M d'V~~O01O~.--m--i.-~.,.-~.~00N N l~o~00.--m--~.-~[~~c't'Vl~I~
U HE-~d E-iU UU C7~Hv~~ ~~ d U C7E~Hodoo ~ dE-~C7E-~do
"~~Do000~ONN ~nN l~00~ .-.O t~O da0~n~o0oWoo.-~Od a0~h~h~D~v~
00,-~d'~ ~et00O ct~WO avO N~n~nV~00Vv0t!1~Ooo'..nOv,-1~ OV1Nv0
adue'U~ m dM'~ \O~ U ~O O~ .-M,~ oN0N N l~~ 0~0~ o ~1t~0OrNnd'~ ~.-M~
67
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
E-~U HU Ud dC7d E-a t7d UU U H
M ~OO~d CJd M00V1h 00d'U ~N ~O~ M dE~M
M~Oo000c0hh l~~OOO N OWO v7v1NO N -~O h
M
~D.~~~ ~ .-W1d'M OWO ~ N.~Nd'-~I~N OV 00
~ 0~
c~c~ Ud dc~d H c~d UU U H
d ~d 0 V7h 00~'U r"N ~OV1M dH M
NM 00t~IwM0 OO N O~~OV~v'1NO N ~O h
M 0~D
00N oor"O~d'M O~~OO~N~ N~t~h N Od'00
-k~et o0O~~DOM d'V1TWON ~V'1~ON ~Ol~~nO
E-I.-aN .~d'00~.~~.~~ .--y-~~ .~.~~ 00Ov
U U d H
v - E-~r
Ot~00'ch~DOh w , ~O ,
O N h
.--~O .~N ~tMM h N O~Yeo
T"~M~t~O~ .-iNN N ~Ol~~!1O
~.-r'rH .-~~..r .~~ 00O~
H
~N ~ M M
0 l~o V N dE"'~h
O 'I ~
O ~
00 etMM O o
N 1 ~ ~' o
O
~1~O~O .-~NN N O Iv
1
M h CON ~ M dh M
N N ~ ~~ O
O -~
~., r.~ c0\
UN ~ ~ ~N ~ M df-~c-Hn
l0M V1 ~OOh O N -~O h
VM_M d'MM h N Od'
NN ~ ~
V~ ~ -a-i.1 00O~
...
UC7 ~ ~N ~!1M dE"'~M
O OM ~ N _O
~~ N d'M OV
~ O
rte--nh ~O ~ NN N LOIV1
Mh .-. .-.,-,.-. ,-..-,coa
~ Oh O N ~O h
~ N ~ ~~ O
W O~ NN
'
r.~--~.., r,~.-~ .-,.-~opOv
7.
d
~ O~ O N O ~
~U
d~ ~ ~ 00N V7M dE"'~M
~N ~ ~DOh O N ~O h
~N M d'MM h N O~tO~
N ~ ohO~
68
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Example 5
Further analysis of the data in FIG. 3 demonstrated sets of nucleotide alleles
useful for
diagnosing the haplogroups. A set of nucleotide alleles useful for diagnosing
all of the
haplogroups and sub-haplogroups in FIG. 3 is listed in Table 10. There are
many equivalent
methods for diagnosing the haplogroups. Examples of methods requiring testing
only or a
few loci follow. Alleles are identified in human samples containing mtDNA.
Haplogroup LO
can be diagnosed by identifying 45860, 9818T, or 8113A. Haplogroup Ll can be
diagnosed
by identifying 825A, 2758A, 28850, 71466, 8468T, 8655T, 10688A, 108100, or
131056.
Haplogroup L2 can be diagnosed by identifying 24160, 27586, 8206A, 92216,
119440, or
163906. Haplogroup L3 can be diagnosed by identifying 108196, I4212C, 86I80,
100860,
163620, 10398A, or 161240. Haplogroup C can be diagnosed by identifying 35520,
47156,
7196A, 8584A, 95456, 132636, 143180, or 16327T. Haplogroup D can be diagnosed
by
identifying 4883T, 5178A, 8414T, 14668T, or 15487T. Haplogroup E can be
diagnosed by
identifying I6227G. Haplogroup G can be diagnosed by identifying 48336, 82000,
or
160170. Haplogroup Z can be diagnosed by identifying 110786, 16185T, or
16260T.
Haplogroup A can be diagnosed by identifying 6636, 16290T, or 16319A.
Haplogroup I can
be diagnosed by identifying 4529T, 100340, or 16391A. Haplogroup W can be
diagnosed by
identifying 2040, 207A, 12430, 5046A, 5460A, 8994A, 119476, 158840, or 16292T.
Haplogroup X can be diagnosed by identifying 1719A, 35166, 6221 C, or 144700.
Haplogroup F can be diagnosed by identifying 12406A or 163040. Haplogroup Y
can be
diagnosed by identifying 79336, 8392A, 162310, or 16266T. Haplogroup U can be
diagnosed by identifying 31970, 46460, 77686, 9055A, 11332T, 131046, 140706,
159076,
160516, 161290, 161720, 162196, 162490, 16270T, 16311T, 16318T, 163436, or
163560.
Haplogroup J can be diagnosed by identifying 295T, I2612G, I3708A, or 16069T.
Haplogroup T can be diagnosed by identifying 118126, 12633T, 142336, 161630,
16186T,
1888A, 49176, 8697A, 104630, 13368A, 14905A, 156076, 15928A, or 16294T.
Haplogroup V can be diagnosed by identifying 720, 4580A, or 15904T. Haplogroup
H can
be diagnosed by identifying 2706A or 70280. Diagnosis of haplogroup B is more
complicated, requiring three steps. Haplogroup B can be diagnosed by
identifying 161890;
and by identifying the absence of 1719A, 35166, 62210, 144700, or 16278T; and
by
identifying the absence of 1888A, 42160, 49176, 8697A, 104630, I I251G,
114676,
123086, 12372A, 12633T, 131046, 13368A, 140706, 14905A, 15452A, 156076,
15928A,
161260, 16I630, 16186T, 162490, or 16294T.
69
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Tabl e 10 8392 A - T14668~
.
Nucleotidelleles ful 8414 14905 A
A Use
T
for Dia~nosin~Human 8468 15301
T A
l u
H s
a~ p
o ~ro
72 8584 A 15452 A
C
204 8618 C 15487 T
C
207 8655 .1. 15607 G
A
295 8697 A 15884 C
T
663 8994 A 15904 T
G
825 9055 A 15907 G
A
1243 9221 G 15928 A
C
1719 9545 G 16017 C
A
1888 9818 T 16051 G
A
2416 10034 C 16069 T
C
2706 10086 C 16124 C
A
2758 10398 A 16126 C
A
2758 10463 C 16129 C
G
2885 10688 A 16163 C
C
3197 10810 C 16172 C
C
3516 10819 G 16185 T
G
3552 11078 G 16186 T
C
4216 11251 G 16219 G
C
4529 11332 T 16227 G
T
4580 11467 G 16231 C
A
4586 11812 G 16249 C
C
4646 11944 C 16260 T
C
4715 11947 G 16266 T
G
4833 12308 G 16270 T
G
4883 12372 A 16278 T
T
4917 12406 A 16290 T
G
5046 12612 G 16292 T
A
5178 12633 T 16294 T
A
5460 13104 G 16304 C
A
6221 13105 G 16311 T
C
7028 13263 G 16318 T
C
7146 13368 A 16319 A
G
7196 13708 A 16327 T
A
7768 14070 G 16343 G
G
7933 14212 C 16356 C
G
8113 A 14233 G 16362 C
8200 14318 C 16390 G
C
8206 A 14470 C 16391 A
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Additional alleles are included in Table I1. These alleles are useful for
designing
equivalent methods, to those described above, for diagnosing the haplogroups.
Alleles in
Table 11 are useful for designing efficient methods for diagnosing macro-
haplogroups. The
data in Tables 10 and 11 and FIG 3 are also useful for identifying sub-
haplogroups. This
invention provides a method for diagnosing sub-haplogroup L1 al by identifying
in a human
sample, one of the nucleotide alleles selected from the group consisting of
45860 and 9818T.
This invention provides a method for diagnosing sub-haplogroup Lla2 by
identifying in a
human sample, one of the nucleotide alleles selected from the group consisting
of 8113A and
82S 1A. This invention provides a method for diagnosing sub-haplogroup Llbl by
identifying in a human sample, the nucleotide allele 23520 and one of the
nucleotide alleles
selected from the group consisting of 3666A, 70SSG, 73890, 137890, and 141780.
This
invention provides a method for diagnosing sub-haplogroup Llb2 by identifying
in a human
sample, one of the nucleotide alleles selected from the group consisting of
37960, S9S1G,
59846, 6071 C, 90726, 10586A, 128106, and 134856. This invention provides a
method for
diagnosing sub-haplogroup L2a by identifying in a human sample the nucleotide
allele
138036. This invention provides a method for diagnosing sub-haplogroup L2b by
identifying in a human sample the nucleotide allele 4I586. This invention
provides a
method for diagnosing sub-haplogroup L2c by identifying in a human sample, one
of the
nucleotide alleles selected from the group consisting of 325T, 6800, and
139580. This
invention provides a method for diagnosing sub-haplogroup L3a by identifying
in a human
sample, one of the nucleotide alleles selected from the group consisting of
232S0, 108196,
and 142120. This invention provides a method for diagnosing sub-haplogroup L3b
by
identifying in a human sample the nucleotide allele 86180. This invention
provides a method
for diagnosing sub-haplogroup L3c by identifying in a human sample the
nucleotide allele
100860. This invention provides a method for diagnosing sub-haplogroup L3d by
identifying in a human sample the nucleotide allele 10398A. This invention
provides a
method for diagnosing sub-haplogroup Uk by identifying in a human sample, one
of the
nucleotide alleles selected from the group consisting of9055A and 16311T. This
invention
provides a method for diagnosing sub-haplogroup U7 by identifying in a human
sample the
nucleotide allele 16318T. This invention provides a method for diagnosing sub-
haplogroup
U6 by identifying in a human sample, one of the nucleotide alleles selected
from the group
consisting of 161720 and 162196. This invention provides a method fox
diagnosing sub-
haplogroup US by identifying in a human sample, one of the nucleotide alleles
selected from
the group consisting of 31970, 77686, and 16270T. This invention provides a
method for
71
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
diagnosing sub-haplogroup U4 by identifying in a human sample, one of the
nucleotide
alleles selected from the group consisting of 46460, 11332T, 163560. This
invention
provides a method for diagnosing sub-haplogroup U3 by identifying in a human
sample the
nucleotide allele 163436. This invention provides a method for diagnosing sub-
haplogroup
U2 by identifying in a human sample, one of the nucleotide alleles selected
from the group
consisting of 159076, 160516, and 161290. This invention provides a method for
diagnosing sub-haplogroup Ul by identifying in a human sample, one of the
nucleotide
alleles selected from the group consisting of 131446, 140706, 161890, and
162490. This
invention provides a method fox diagnosing sub-haplogroup T* by identifying in
a human
sample, one of the nucleotide alleles selected from the group consisting of
118126 and
142336. This invention provides a method for diagnosing sub-haplogroup T 1 by
identifying
in a human sample, one of the nucleotide alleles selected from the group
consisting of
12633T, 161630, and 16186T.
72
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Tabl e 11 4646 C 9402
Nucleotidellelesful 4715 9540 .
A Use .
G 1
for Dia~nosin~ n
Huma
G 4833 9545
Hap and o-
H lo~roupsMacr
Haplo rg~ou~s 4883 T 9818
72 C 4917 G 10034 C
73 A 5046 A 10086
204 C 5178 A 10398
207 A 5442 C 10400 .1.
295 T 5460 A 10463 C
325 T 5951 G 10586
489 C 5984 G 10589
663 G 6071 C 10664
680 C 6185 C 10688
769 A 6221 C 10810
825 A 7028 C 10819
1018 A 7055 G 10873
1048 T 7146 G 10915
1243 C 7196 A 11078
1719 A 7256 .1. 11251
1888 A 7389 C 11332
2352 C 7521 A 11467
2416 C 7768 G 11719
2706 A 7933 G 11812
2758 A 8113 A 11944 C
2758 G 8200 C 11947
2885 C 8206 A ..12007
3197 C 8251 A 12308
3516 A 8392 A 12372
3516 G 8414 T 12406
3552 C 8468 .~ 12612
3594 T 8584 A 12633
3666 A 8618 C 12705
3796 C 8655 T 12810
4104 G 8697 A 13104
4158 G 8701 A 13105
4216 C 8994 A 13263
4312 T 9055 A 13276
4529 Z, 9072 G 13368
4580 A 9221 G 13485
4586 C 9347 G 13506
73
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
13650 .1, 16231 C
13708 A 16249 C
13789 C 16260
13803 G 16266
13958 C 16270
14070 G 16278
14178 C 16290
14212 C 16292
14233 G 16294
14318 C 16298
14470 C 16304
14668 T 16311
14766 C 16318
14783 C . 16319
14905 A 16327
15043 A 16343
15301 A 16356 C
15452 A 16362
15487 T 16390
15607 G 16391
15884
15904
15907
15928
16017 C
16051 G
16069
16124
16126 C
16129
16129 C
16163
16172 C
16185
16186
16189 C
16219 G
16223 C
16224 C
16227 G
74
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
An equivalent method fox diagnosing a haplogroup is diagnosing haplogroup LO
by
identifying the presence of one of 825A, 2758A, 288S0, 71466, 8468T, 8655T,
10688A,
108100, or 131056; and identifying the absence of one of 3666A, 70556, 73890,
137890,
or 141780. Other equivalent methods can be derived from the data in FIG 3, and
are within
the scope of this invention.
Example 6
Lebers Hereditary Optic Neuropathy (LHON) is a form of blindness caused by
mitochondrial DNA (mfiDNA) mutations. Four mutations, 3460A, 11778A, 144840,
and
14459A, account for over 90% of LHON worldwide and are designated "primary"
mutations.
Primary mutations strongly predispose carriers to LHON, are not found in
controls, are all in
Complex I genes, and do not co-occur with each other. It has been demonstrated
that the
11778A and 144840 mutations occurred more frequently than expected in
association with
European mtDNA haplogroup J (found in 9% of European-derived mtDNAs),
suggesting a
synergistic interaction among mtDNA mutations increased the probability of
disease
expression. Sequence analysis of two Russian LHON families without primary
LHON
mutations, including removal of nucleotide alleles listed in Table 3,
demonstrated two new
complex I mutations, 3635A and 46400. Venous blood samples were obtained from
the
family members. Genomic DNA was isolated from the buffy coat blood fraction
using
Chelex 100 (fetus, Emberyville, CA, USA). mtDNA was amplified by PCR in 2-3kb
fragments, purified on Centricon 100 columns, and cycle-sequenced using BigDye
Terminators (ABI/Perkin Elmer fetus) and an ABI Prism 377 automated DNA
sequencer.
The mutations were confirmed using mutation-specific restriction enzyme
digestion
following mismatched-primer PCR amplification of white blood cell mtDNA (Brown
M.D, et
al., (1995) Human Mutat. 6:311-325).
Example 7
A new primary LHON mtDNA mutation, 106630, affecting a Complex I gene was
homoplasmic in 3 Caucasian LHON families, all of which belonged to haplogroup
J. These 3
families were the only haplogroup J-associated LHON families (out of 17) that
did not harbor
a known, primary LHON mutation. Comprehensive phylogenetic analysis of
haplogroup J
using complete mtDNA sequences demonstrated that the 106630 variant has arisen
3
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
independent times on this background. This mutation was not present in over
200 non-
haplogroup J European controls, 74 haplogroup J patient and control mtDNAs, or
36 putative
LHON patients without primary mutations. A partial Complex I defect was found
in 10663 C-
containing lymphoblast and cybrid mitochondria. Thus, the 106630 mutation has
occurred
three independent times, each time on haplogroup J and only in LHON patients
without a
known LHON mutation. This makes the 106630 mutation unique among all
pathogenic
mtDNA mutations in that it appears to require the genetic background provided
by
haplogroup J for expression. These results provide further evidence for the
predisposing role
of haplogroup J and for the paradigm of "mild" mtDNA mutations interacting in
an additive
way to precipitate disease expression. Europeans with the mild ND6 np 14484
and ND3 np
10663 Leber's Hereditary Optic Neuropathy (LHON) missense mutations are more
prone to
blindness if they also possess the mtDNA haplogroup J.
Example 8
To assess the importance of demographic factors in inter-continental mtDNA
sequence radiation, deviations from the standard neutral model were tested for
in the
distribution of mtDNA sequence variants using the Tajima's D and Fu and Li D*
tests ( Y. X.
Fu, W. H. Li, (1993) Genetics 133:693-709. and F. Tajima, (1989) Genetics 123,
585-95).
The standard neutral model of population genetics assumes a random-mating
population of
constant size, with all mutations uniquely arising and selectively neutral.
The continental
frequency distribution of pairwise mtDNA sequence differences was calculated
to test for
rapid population expansion using the method of A. R. Rogers, H. Harpending,
(1992) Mol.
Biol. Evol. 9:552-569.
For the African mtDNA sequences (ra = 32), the results did not significantly
deviate
from the standard neutral model, and the frequency distribution of pairwise
sequence
difference counts was broad and ragged. Both of these results are consistent
with the model
that the African population has been relatively stable for a long time. By
contrast, the non-
African mtDNAs (n = 72) showed a highly significant deviation from neutrality
(Tajima's D
= -2.43, P < 0.01; Fu and Li D* _ -5.09, P < 0.02), as well as a bell-shaped
frequency
distribution of pairwise sequence differences. Thus, these results are
consistent with
population expansions having distorted the frequency distribution (L.
Excoffier, J. Mol. Evol.
30:125-39 (1990) and D.A. Merriwether et al. (1991) J. Mol. Evol 33:543-555).
76
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
To better define the regional distribution of these demographic influences,
the
Eurasian samples were divided into European and Asian plus Native American.
Analysis of
all European mtDNAs also revealed significant deviations from the standard
neutral model
(Tajima's D = -2.19, P < 0.01; Fu and Li D* _ -3.31, P < 0.02). The
distribution of pairwise
sequence differences for the European mtDNAs revealed two sharp peaks, hinting
at two
major expansion phases. The most recent of these peaks was lost when
haplogroup H and V
mtDNAs were deleted from the sample. Hence, haplogroup H, which represents 40%
of
modern European mtDNAs ( A. Torroni et al., American Journal of Human Genetics
62,
1137-1152 (1998)) and has a MRCA of 19,000 YBP, came to predominate in Europe
relatively recently.
Analysis of the aggregated Asian and Native American mtDNAs (n = 41) also
revealed significant deviations from the standard neutral model (Tajima's D = -
2.28, P <
0.01, Fu and Li D* _ -4.31; P < 0.02) as well as revealing a broad, bell-
shaped distribution of
pairwise differences consistent with rapid population expansion.
When the Asian-Native American haplogroups A, B, C, D and X mtDNAs (n = 26)
were analyzed separately, they also showed significant deviation from
neutrality for the Fu
and Li D* test (D* _ -2.65, P < 0.05), although not for the Tajima's D test (D
=-1.60, ns).
Their distribution of pairwise sequence differences was also strongly uni-
modal, indicating
that the population expanded as people moved through Siberia and Beringia and
into the
Americas.
Example 9
Variable Replacement Mutation Rates in Human mtDNA Genes
To determine if selection was an important factor in causing the sudden shifts
in
mtDNA sequence variation between continents, the number of non-synonymous to
synonymous base substitutions was analyzed for all 13 mtDNA protein genes of
those
haplogroups which contributed to the colonization of each of the major
continental spaces:
African, European, and Native American. For example, for the "Native Americans
" the
mtDNAs from the Asian-Native American haplogroups A, B, C, D and X were
combined.
The Asian-Native American mtDNAs from the haplogroups were combined because
random
mutations accumulate in founder populations and those mtDNAs which prove
advantageous
in new environments are enriched. Hence, the founding mutations of the
haplogroup are
77
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
important in the continental success of the lineage. We then tested for
possible selective
effects during the colonization of each continent by comparing the ratio of
non-synonymous
versus synonymous nucleotide substitutions for each mtDNA gene. An increase in
the non-
synonynous to synonymous mutation ratio suggests that selection has favored
the propagation
of a functionally altered protein.
The comparison of the ratio of nonsynonymous to synonymous mutations, counting
each change only once, revealed great variation between continents for several
genes (Table
12). Marked increases in the accumulation of non-synonymous mutations were
seen for ND3
in Africans, Cytb and COIII in Europeans, and ATP6 in Native Americans. The
number of
non-synonymous and synonymous mutations for each gene was also compared
between the
different continents by computing the P value using a Two-tailed Fisher Exact
Test. This
revealed significant differences between Africans and both Europeans and
Native Americans
for COIII, between Africans and Native Americans for ATP6, and between
Africans and
Europeans for the sum of all mtDNA genes (Table I2). Hence, this analysis
supports the
hypothesis that selection has played a role in shaping continental mtDNA
protein variation.
Table I2*
Number Two-Tail
Gene of FET
Polymorphic P-value
Sites
_
African European Native Afr Afr Eur
American
N- Syn RatioSyn N- RatioSyn N- Ratiovs vs vs
syn syn syn Eur Am Am
NDl 10 17 0.59 5 5 1.00 4 4 1.00 0.710.69 1.00
ND2 9 22 0.41 4 9 0.44 3 7 0.43 1.001.00 1.00
ND3 6 2 3.00 1 3 0.33 1 4 0.25 0.220.10 I.00
ND4L 0 7 0.00 0 1 0.00 1 4 0.25 1.000.42 1.00
ND4 4 3S 0.11 2 13 0.15 3 12 0.25 1.000.38 I.00
NDS 15 31 0.48 8 20 0.40 2 14 0.14 0.800.19 0.28
ND6 2 14 0.14 1 6 0.17 3 5 0.60 1.000.29 0.57
Cytb 11 19 0.58 14 9 1.56 5 12 0.42 0.100.75 0.60
COI 7 30 0.23 0 9 0.00 0 13 0.00 0.320.17 1.00
COII 3 19 0.16 0 4 0.00 2 6 0.33 1.000.59 0.52
COIII 1 13 0.08 6 5 1.20 7 10 0.70 0.02O. 0.70
OS
ATP6 3 15 0.20 5 6 0.83 7 5 1.40 0.20O. 0.68
OS
ATP8 2 3 0.67 2 0 1 3 0.33 0.431.00 0.40
~ Total73 227 0.32 48 90 0.53 39 99 0.39 0.030.41 0.30
~ ~ ~ ~ ~ ~ ~ i ~ ~ ~
78
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
* Replacement versus synonymous mutation numbers of mtDNA genes. Rplmt =
replacement mutations, ratio = rplmt/silent. FET = Fisher Exact Test. Afr =
Africa, Eur =
Europe, Am = Native American. The ratios of polymorphic sites in bold-italics
highlight
some of the higher values observed. Those in bold-italics under Two-Tailed FET
indicate
comparisons that are significant at the 0.05 level.
Example 10
Since the above analysis counts each mutation only once, irrespective of its
frequency
within the haplogroup, it under-emphasizes the importance of nodal mutations
and over-
emphasizes the importance of terminal private polymorphisms. As an alternative
to this
approach, we calculated the corrected non-synonymous (Ka) and synonymous (KS)
mutation
frequencies and then determined the relative selective constraints acting on
that gene by
calculating the kc value { kc= - ln(Ka/KS)}. A high kc value is indicative of
high protein
sequence conservation and low amino acid variation, while a low value is
indicative of low
protein conservation and high amino acid variation (N. Neckelinann et al.,
(1987) Proc. Natl.
Acad. Sci. USA 84:7580-7584).
The k~ values for each human mtDNA gene were compared across the total global
collection of human mtDNA sequences (Figure 4). The ATP6 gene was the least
conserved
gene in the human mtDNA, though previously it had been shown to be relatively
highly
conserved in inter-specific comparisons (N. Neckelmann et al., (1987) Proc.
Natl. Acad. Sci.
USA 84:7580-7584).
Example 11
The higher inter-specific conservation of ATP6 was confirmed by comparing the
k~
values of human versus chimpanzee (Pan troglodytes) and bonobo (Pan paniscus);
human
versus eight primate species (baboon, Borneo and Sumatran orangutan, gibbon,
gorilla,
lowland gorilla, bonobo, and chimpanzee); and human versus 13 diverse
mammalian species
(bovine, mouse, cat, dog, pig, rat, rhinoceros, horse, gibbon, gorilla ,
orangutan, bonobo,
chimpanzee) (Figure 3). Thus, while ATP6 is highly conserved between species,
it is very
poorly conserved within humans. These results are consistent with the reduced
infra-specific
versus inter-specific conservation observed for other genes (C. A. Wise et
al., (1998)
Genetics 148:409-21), and with the hypothesis that mitochondrial protein
variation is
accelerated in humans and other primates, as seen in cytochrome c oxidase
genes (L. I.
Grossman et al., (2001) Mol. Phylogeraet. Evol. 18:26-36).
79
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Example 12
To further investigate the possibility that individual mtDNA protein genes
differ in
their selective constraints in different human continental populations, kC
values for all 13
mtDNA protein genes from each set of continental haplogroups were calculated:
African,
European, and the Native American. The cumulative selective pressure that
separated the
mtDNAs of pairs of continents by pair-wise comparison of the k~ values was
calculated for
the genes of each mtDNA (Table 13). Comparison of mtDNA protein k~ values in
Europeans
versus Africans revealed that three genes (ND 1, cytb and COIF had
significantly lower
sequence conservation in Europeans. A comparison of the k~ values of Native
American
vexsus African mtDNA genes revealed six genes (ND4, ND6, COII, COTII, ATP6 and
ATPB)
that had significantly lower sequence conservation in Native Americans.
Finally, comparison
of the k~ values of Africans versus Europeans or Native Americans revealed
four mtDNA
genes (ND3, NDS, cytb, and C0~ had significantly lower sequence conservation
in Africans.
The greatest differences in k~ values were seen for the comparisons of COIII
and ATP6
between Africans and Native Americans and for C~III between African and
Europeans
(Table 13).
Table 13*
GENES African European T-test Native AmericanT-test
sequences sequences P value sequences P value
(n=32) (n=31) {A,B,C,D,X~
(n=26)
ND1 2.08 1.18 0.27 1.90 P<0.0001 2.07 1.92 NS
~2 1.72 1.07 1.57 1.85 NS 1.81 1.1 I NS
ND3 0.51 1.87 0.91 2.32 NS 1.70 +_ 1.32 P<0.01
ND4L * * NS 2.41 3.83
ND4 3.49 1.34 3.39 2.23 NS 2.20 1.19 P<0.001
NDS 1.78 0.71 2.20 1.20 NS 3.63 3.56 P<0.01
ND6 2.51 1.19 3.13 3.99 NS 1.15 1.52 P<0.001
Cytb 1.89 0.96 0.34 1.51 P<0.0001 2.46 1.15 P<0.05
COI 2.37 0.95 3.85 3.93 P<0.05 * *
COII 2.73 1.32 * * 1.74 2.12 P<0.05
CO>II 4.65 3.94 0.94 2.08 P<0.0001 2.1 I I.26 P<0.01
ATP6 2.31 1.28 1.48 2.28 NS -0.14 I .34 P<0.0001
ATPB 2.62 1.89 * * 1.25 1.94 P<0.01
* Estimates of coefficients of selective constraint (k~) stratified by gene
and region. k~ values
and standard deviations calculated for African, European and Asian-American
haplogroups
A,B,C,D and X mtDNA protein-coding genes. * indicates that k~ values could not
be
calculated, since either KS or Ka were 0. Haplogroup X is represented only by
the Native-
American sequence, the European X sequence being excluded.
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Taken together, these data show that different selective forces have acted on
individual mtDNA genes as humans colonized different continents. Moreover, the
observed
differences in mtDNA protein sequence correlate with the climatic transitions
that humans
would have experienced as they migrated out of tropical and sub-tropical
Africa and into
temperate Eurasia and arctic Siberia and Beringia. The mtDNA genes that showed
the
highest amino acid sequence variation between continents were COIII and ATP6.
Example 13
The nucleotide alleles in Table 3 residing in evolutionarily significant genes
identified
in Examples 9-12 were analyzed for evolutionary significance. Evolutionarily
significant
alleles reside in evolutionarily significant genes and cause amino acid
changes. A list of the
evolutionarily significant nucleotide alleles in ND1, ND2, ND3, ND4, NDS, ND6,
Cytb,
COI, COIL COIII, ATP6, and ATP8 appear in Table 14. The Cambridge nucleotide
alleles in
Table 14 are evolutionarily significant. These amino acid alleles, including
the Cambridge
alleles, are evolutionarily significant. The locations of the amino acid
alleles are identified by
the location of the nucleotide allele listed in Table 3. Other evolutionarily
significant
nucleotide alleles not listed in Table 14, include alleles at neighboring
nucleotide loci that are
within the same codon and code for the same amino acids that are listed in
Table 14.
Table 14
Evolutionarily Significant Human Mitochondria) Nucleotide and Amino Acid
Alleles
Non-
CambridgeCambridgeNon-
enomeambridgeNucl. Amino Cambridge
ene LocationNucleotideAlleleAcid AA
Allele
ND 3308 T C M T
1
ND1 3316 G A A T
NDi 3394 T C Y H
ND1 3505 A G T A
ND1 3547 A G I V
ND1 3565 A G T A
ND1 3644 T C V A
ND1 3796 A T T A
ND1 3796 A G T S
ND1 3796 A C T P
ND1 3808 A G T A
ND1 3866 T C I T
ND 4025 C T T M
1
ND 4040 C T T M
1
81
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
ND 4048 G A D N
1
ND 4123 A G I V
1
ND1 4216 T C Y H
ND 4225 A G M V
1
ND1 4232 T C I T
ND2 4491 G A V I
ND2 4506 A G I V
ND2 4512 G A A T
ND2 4596 G A V I
ND2 4695 T C V I
ND2 4767 A G M V
ND2 4824 A G T A
ND2 4833 A G T A
ND2 4917 A G N D
ND2 4960 C T A G
ND2 5043 G T A S
ND2 5046 G A V I
ND2 5178 C A L M
ND2 5262 G A A T
ND2 5263 C T A V
ND2 5331 C A L I
ND2 5442 T C F L
ND2 5460 G A A T
COI 6150 G A V I
COI 6253 T C M T
COI 6324 G A A T
COI 6366 G A V I
COI 6607 T C F S
COI ? A G T A
146
COI 725? A G I V
COI ?347 G A V I
'COI7389 T C Y H
COI 7444 G A TER K
COII7664 G A A T
COII7673 A G I V
COII7697 G A V I
COII8027 G A A T
COII8142 C T A V
ATPB8387 G A V M
ATP88414 C T L F
ATP88448 T G M T
ATP88460 A G N S
ATPB8472 C T P L
ATP88553 C T S L
ATP68545 G A A T
ATP68563 A G T A
ATP68566 A G I V
ATP68584 G A A T
ATP68618 T C I T
82
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
ATP68701 A G T A
ATP68705 T C M T
ATP68764 G A A T
ATP68794 C T H Y
ATP68836 A G M V
ATP68860 A G T A
ATP68875 T C F L
ATP68962 A G T A
ATP69053 G A S N
ATP69055 G A A T
ATP69077 T C I T
ATP69103 T C F L
ATP69136 A G I V
ATP69151 A G I V
COIII9237 G A V M
COIII9325 T C M T
COIII9355 A G N S
COIII9456 A G I V
COIII9402 A C T P
COIII9477 G A V I
COIII9559 C G P R
COIII9591 G A V I
COIII9667 A G N S
COIII9682 T C M T
COIII9822 C A L I
COIII9957 T C F L
COIII9966 G A V I
ND3 10086A G N D
ND3 10086A C N H
ND3 10152G C E Q
ND3 10182G C D H
ND3 10197G A A T
ND3 10321T _ V A
C
ND3 10398A G T A
ND4L10609T C C R
ND4 10816A T K N
ND4 10920C T P L
ND4 11016G A S N
ND4 11078A G I V
ND4 11150G A A T
ND4 11172A G N S
ND4 11177C T P S
ND4 11654A G T A
ND4 11909A G T A
ND4 11963G A V I
ND4 11969, G A A T
ND4 12083T G S A
ND4 12134T C S P
ND5 12346C T H Y
83
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
ND5 12358A G T A
ND5 12361A G T A
ND5 12373A G T A
NDS 12397A G T A
ND5 12406T A V I
ND5 12635T C I T
ND5 12850A G I V
ND5 12940G A A T
ND5 12967A C T P
ND5 13104A G I V
ND5 13105A G I V
ND5 13135G A A T
ND5 13145G A S N
ND5 13276A ' G M V
NDS 13477G A A T
ND5 13651A G T A
ND5 13660A G N D
ND5 13708G A A T
NDS 13759G A A T
ND5 13780A G I V
ND5 13789T C Y H
ND5 13819T C F L
ND5 13880C A S Y
ND5 13886T C L P
ND5 13924C T P S
ND5 13927A T S C
ND5 13928G C S T
ND5 13958G C G A
ND5 13966A G T A
ND5 14000T A L Q
ND5 14059A G I V
ND5 14128A G T A
ND6 14178T C I V
ND6 14272C G L F
ND6 14318T C N S
ND6 14319T C N D
ND6 14384G C A G
ND6 14459G A A V
ND6 14484T C M V
ND6 14502T C I V
ND6 14571T A S C
C 14766C T T I
C 14?69A G N S
C 14793A G H R
C 14798T C F L
C 14861G A A T
C 14862C T A V
B
C 14979T C I T
C 15110G A A T
84
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
C 15113A G T A
C 15204T C I T
B
C 15218A C T P
C 15218A G T A
B
C 15238C G I M
C 15257G A D N
C 15261G A S N
B
C 15317G A A T
C 15318C T A V
B
C 15323G A A T
C 15326A G T A
C 15431G A A T
B
C 15452C A L I
B
C 15497G A G S
B
CytB 15519T C L P
CytB 15663T C I T
C 15731G A A T
C 15746A G I V
CytB 15803G A V M
C 15806G A A T
CytB 15812G A V M
C 15824A G T A
C 15849C T T I
CytB 15884G C A P
A subset of the alleles in Table 14 that are associated with predispositions
to physiological
conditions using the methods of this invention is listed in Table 15.
Table 15
Amino Acid Alleles Associated with Physiological Conditions in this Invention
Nucleotide Amino Acid Haplogroups
Genome Alleles Alleles Diagnosable
ene Location Useful for Useful for by Alleles
Diagnosing Diagnosing
Ha to ou s Ha to ou s
ND 1 3796 C P (Llb2)
ND2 4833 G A G
ND2 4917 G D T
ND2 5046 A I
ND2 5178 A M D
ND2 5442 C L LO
ND2 5460 A T W
COI 7146 G A L0, L1
COI 7389 C H Ll
ATPB 8414 T F D
ATP6 8584 A T C
ATP6 8618 C T (L3b)
A, I, W, X,
ATP6 8701 A T B, F, Y, U,
J, T,
V, H
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
ATP6 9055 A T (~)
COIII 9402 C P LO
ND3 10086 C H (L3c)
ND3 10398 A T (L3d)
ND4 11078 G V
ND5 12406 A I
ND5 13104 G V (U1)
ND5 13105 G V L0, L1
ND5 13276 G V LO
NDS 13708 A T J
NDS 13789 C H L1
ND5 13958 C A (L2c)
ND6 14178 C V L1
ND6 14318 C S C
CytB 14766 C T V, H
C B 15452 A I J, T
C 15884 C P W
Example 14
Continent-Specific Afnino Acid Substitutions in ATP6
To further investigate the biological significance of the human continent-
specific
ATP6 amino acid substitutions, the amino acid conservation for each variable
human position
using 39 animal species mtDNAs (12 primates, 22 other mammals, four non-
mammalian
vertebrates, and Drosophila) was analyzed. This revealed that many of the ATP6
substitutions that are associated with particular mtDNA haplogroups alter
evolutionarily
conserved, and hence potentially functionally important, amino acids.
A threonine to alanine substitution at codon 59 (T59A, nucleotide location
8701-
8703) in ATP6 separates the mtDNAs of macro-haplogroup N from the rest of the
World.
The polar threonine at position 59 is conserved in all great apes and some old-
world
monkeys.
Among the haplogroups of macro-haplogroup M, the related Siberian-Native
American haplogroups C and Z are delineated by an A20T (nucleotide location
8584-8586)
variant. A non-polar amino acid found in this position occurs in all animal
species except for
Macaca, Papio, Balaenoptera and Drosophila.
86
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
Among the haplogroups of macro-haplogroup N, the non-R lineage Nlb harbors two
distinctive amino acid substitutions: M104V (nucleotide location 8836-8838)
and T146A.
(nucleotide location 8962-8964) The methionine at position 104 is conserved in
all mammals,
and the threonine at position 146 is conserved throughout all animal mtDNAs.
Moreover, the
T146A substitution is within the same transmembrane a-helix as the pathogenic
mutation
L156R that alters the coupling efficiency of the ATP synthase and causes the
NARP and
Leigh syndromes (I. Trounce, S. Neill, D. C. Wallace, Proceedings of the
National Academy
of Sciences of the United States of America 91, 8334-8338 (1994)).
Also in macro-haplogroup N, haplogroup A mtDNAs harbor a H90Y (nucleotide
location 8794-8796) amino acid substitution. The histidine in this position is
conserved in all
placental mammals except Porago, Cebus and Loxodonta and occurs within a
highly
conserved region. Furthermore, among the heterogeneous group of mtDNAs
carrying the
tRNALYs-COII 9 by deletion and arbitrarily assigned to haplogroup B, one mtDNA
harbored a
F193L (nucleotide location 9103-9105) substitution. This position is conserved
in all
mammals except Pongo, Papio, Cebus and Erinaceus.
Since each of the mtDNA sequences used in this comparison of different species
is
derived from only one or two individuals, it is possible that the rare deviant
cases are due to
the accumulation of environmentally adaptive mutations in those species that
parallel those in
humans. Thus, the above ATP6 amino acid polymorphisms have the characteristics
expected
for evolutionarily adaptive mutations.
Table 16
NucleotideNucleotideWIPO
Locus Alleles code
64 CT
72 TC
73 AG r
89 TC
93 AG r
95 AC m
114 CT
143 GA r
146 TC
150 CT
151 CT
152 TC
153 AG r
87
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
171 GA r 471 TC
180 TC 480 TC
182 CT 482 TC
x 83 AG r 489 TC
i8S GAT d 493 AG r
285 GAT d 499 GA r
18b CA m 508 AG r
i89 ACG v 593 TC
i$9 ACG v S97 CT
194 CT 663 AG r
19S TAC h 678 TC
i9S TAC h b80 TC y
198 CT y 709 GA r
i99 TC '1i0 TC
200 AG x 723 TC
204 TC y 750 AG r
207 GA x 769 GA r
208 TC ~ 82S TA w
210 AG r 827 AG r
212 TC 8S0 TC
21S AG r 921 TC
217 TC 930 GA r
225 GA r 961 TCG b
227 AG r 961 TCG b
228 GA r 1018 GA z
23S AG r 1041 AG r
236 TC y 1048 CT
247 GA r 1119 TC
250 TC 1189 TC
2S2 TC y 1243 TC
263 AG r 1290 CT y
291 AG r 1382 AC m
29S CT y 1406 TC
297 AG r 1415 GA r
316 GA r 1420 TC
31'7 CAG v 143$ AG r
317 CAG v 1442 GA r
320 CT 1503 GA r
32S CT 1598 GA r
340 CT 1700 TC
3S7 AG r 1703 CT
373 AG r 1706 CT
400 TG k 1709 GA r
408 TA w 1715 CT
418 CT 1719 GA r
4S6 CT I73b AG r
462 CT 173$ TC
465 CT 1780 TC
467 CT 181 i AG r
88
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
1888 GA r 3547 AG r
1927 GA r 3549 CT
2000 CT 3552 TCA h
2060 AG r 3552 TCA h
2092 CT 3565 AG r
2245 ACG v 3594 CT
2245 ACG v 3644 TC
2263 CA m 3666 GA r
2308 AG r 3693 GA r
2332 CT 3699 CT
2352 TC 3720 AG r
2358 AG r 3756 AG r
2380 CT 3796 AGTC n
2416 TC 3796 ACGT n
2483 TC 3808 AG r
2581 AG r 3816 AG r
2639 CT 3834 GA r
2650 CT y 3843 AG r
2706 AG r 3847 TC y
2755 AG r 3866 TC
2758 GA r 3918 GA r
2768 AG r 3921 CA m
2789 CT 3927 AG r
2792 AG r 3970 CT
2834 CT y ' 3981 AG r
2836 CA m 4025 CT
2857 TC 4040 CT
2863 TC y 4044 AG r
2885 TC 4048 GA r
3010 GA r 4086 CT y
3083 TC y 4104 AG r
3197 TC 4117 TC
3200 TA w 4122 AG r
3202 TC , 4123 AG r
3204 CT 4158 AG r
3206 CT 4203 AG r
3221 AG r 4216 TC
3290 TC 4221 CT
3308 TC y 4225 AG r
3316 GA r 4232 TC
3372 TC 4248 TC
3394 TC y 4312 CT
3438 GA r 4336 TC
3450 CT 4370 TC
3480 AG r 4388 AG r
3505 AG r 4454 TA w
3513 CT 4491 GA r
3516 CGA v 4506 AG r
3516 CGA v ' 4508 CT
89
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
4512 GA r 5442 TC
4529 ATC h 5460 GA r
4529 ATC h 5465 TC
4541 GA r 5471 GA r
4580 GA r 5492 TC
4586 TC 5495 TC
4596 GA r 5580 TC
4646 TC 5581 AG r
4655 GA r 5601 CT
4688 TC 5603 CT
4695 TC 5606 CT
4715 AG r 5633 CT y
4742 TC 5655 TC y
4767 AG r 5711 AG r
4769 AG r 5773 GA r
4820 GA r 5811 AG r
4824 AG r 5814 TC
4833 AG r 5821 GA r
4841 GA r 5826 TC
4883 CT ' 5843 AG r
4907 TC y 5951 AG r
4917 AG r 5984 AG r
4960 CT 5987 CT
4977 TC y 6026 GA r
4994 AG r 6D29 CT y
5004 TC 6045 CT
5027 CT y 6071 TC
5036 AG r 6077 CT
5043 GT k 6104 CT
5046 GA r 6150 GA r
5063 TC 6152 TC
5096 TC 6164 CT
5108 TC y 6167 TC
5147 GA r ' 6182 GA r
5153 AG r 6185 TC
5178 CA m 6221 TC
5231 GA r 6227 TC
5237 GA r 6253 TC
5255 CT 6257 GA r
5262 GA r 6324 GA r
5263 CT 6366 GA r
5285 AG r 6371 CT
5300 CT 6392 TC
5330 CA m 6473 CT
5331 CA m 6491 CA m
5390 AG r 6524 TC
5393 TC 6548 CT
5417 GA r 6587 CT
5426 TC 6607 TC
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
6680 TC 7694 CT
6713 CT 7697 GA r
6719 TC y 7744 TC
6734 GA r 7765 AG r
6752 AG r 7768 AG r
6770 AG r 7771 AG r
6776 TC 7858 CT y
6815 TC 7861 TC
6827 TC 7864 CT
6875 CA m 7867 CT y
6938 CT 7933 AG r
6962 GA r 7948 CT
6989 AG r 7999 TC y
7028 CT 8014 AG r
7052 AG r 8020 GA r
7055 AG r 8027 GA r
7058 TA w 8080 ' CT
7076 AG r 8087 TC
7146 AG r 8113 CA m
7154 AG r 8142 CT
7175 TC y 8149 AG r
7196 CA m 8152 GA r
7202 AG r 8155 GA r
7226 GA r 8185 TC
7256 CT 8200 TC y
7257 AG r 8206 GA r
7271 AG r 8248 AG r
7274 CT 8251 GA r
7319 TC 8260 TC
7337 GA r 8269 GA r
7347 GA r 8271-8279 accccctctJ-
7389 TC 8286 TC
7403 AG r 8292 GA r
7424 AG r 8298 TC
7444 GA r 8344 AG r
7476 CT y 8387 GA r
7493 CT 8389 AG r
7521 GA r 8392 GA r
7561 TC 8404 TC
7571 AG r 8414 CT
7600 GA r 8428 CT
7624 TA w 8448 TC
7645 TC 8460 AG r
7648 CT 8468 CT
7660 TC 8472 CT y
7664 GA r 8473 TC
7673 AG r 8485 GA r
7675 CT 8545 GA r
7693 CT 8553 CT
91
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
8563 AG x 9242 ~ AG r
8566 AG r 9248 CT
8577 AG r 9263 AG r
8584 GA r 9272 CT
8618 TC 9296 CT
8655 CT 9311 TC
8697 GA r 9325 TC
8701 AG r 9335 CT
8703 GT 9347 AG r
8705 TC y 9355 AG r
8709 CT 9356 GT
8?21 AG r 9377 AG r
8'733 TC 9402 AG m
8764 GA r 9449 CT
8781 CA m 9456 AG r
8784 AG r 9477 GA r
8790 GA x 9509 ~ TC
8793 TC 9536 GT
8'794 CT 9540 TC
8805 AG r 9545 AG r
8836 AG r 9548 GA r
8838 GA x 9554 GA r
8856 GA r 9559 CG s
8860 AG r 9575 GA r
8875 TC 9591 GA x
$877 TC 9599 CT
8911 TC 9632 AG r
8913 AG r 9647 TC
8928 TC 9667 AG r
8943 CT 9682 TC
8962 AG r 9698 TC
8994 GA r 9755 GA r
9042 C,"F 9818 CT
9053 GA r , 9822 CA m
9055 GA r 9824 TA w
9072 AG r 9911 CT
9077 TC 9932 GA r
9090 TC 9950 TC
9093 AG m 99x7 TC
9103 TC 9966 GA r
9114 AG r 9977 TC
9120 AG r 30034 TC
9123 GA r 10086 ~ ACG v
9136 AG r 10086 ACG v
9151 AG r x0115 TC
9156 AG r 10118 TC
91?4 TC 10142 CT
9221 AG r 10151 AG r
9237 GA r 10152 GC s
92
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
10172 GA r IIOi7 TC
10182 GC s 11023 AG r
10197 GA x 1,1078 AG r
10238 TC 11092 AG r
10253 TC 11147 TG
10256 TC 11150 GA r
10310 GA r 11167 AG r
10313 AG z 11172 AG r
10321 TC 11176 GA r
10325 GA r III77 CT
10358 AG r 11215 GT
10370 TC 11251 AG r
10398 AG r 11257 CT
10400 CT 11296 CT
10410 TC 11299 TG y
10414 GT k 11332 CT y
10427 GA r Ii362 AG r
10463 TC y 11365 TC
10499 AG r 1137? GA r
10505 TC 11467 AG r
10550 AG r ~ 11476 CT
10586 GA r 11536 CT y
10589 GA z ' 11590 AG r
10609 TC 11611 GA r
10637 CT y 11641 AG ~ r
10640 TC 11653 AG ~ r
10646 GA r 11654 AG r
10659 GT 11674 CT
10664 CT 11701 TC
10667 TC 11719 GA r
10688 GA r 11722 TG
10736 CT 11767 CT
10790 TG 11812 AG z
10792 AG r 11854 TC
10793 CT 11884 AG r
10804 AG r 11887 GA r
10810 TC 11893 AG r
10819 AG r 11899 TC
10828 TC 11909 AG r
10873 TC 11914 GA r
10876 AG r 11944 TC
10894 CT 11947 AG r
10915 TC 11959 AG r
10920 CT 11963 GA r
10939 CT 11969 GA r
10966 TC 12007 GA z
10984 CG s 12049 CT
11002 AG r 12070 GA z'
I10I6 GA r 12083 TG k
93
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
12121 TC 13068 AG r
12134 TC 13201 AC m
12153 CT 13104 AG r
12172 AG r 13105 AG r
12175 TC 13135 GA r
12234 AG r 13143 TC
12236 GA r 13145 GA x
12239 CT 13149 AG x
12248 AG r 13194 GA r
12308 AG r 13197 CT y
12346 CT 13212 CT
12358 AG r x3221 AG r
12361 AG r 13263 AG r
12372 GA r 13276 AG r
12373 AG r 13281 TC
12397 AG r 13368 _ GA r
12406 GA r 13440 CG s
12414 TC 13477 GA r
12477 TC 13485 AG r
12501 GA r 13494 CT
12507 AG r 13500 TCG b
12519 TC y 13500 TCG b
12528 GA r 13506 CT
12540 AG r I3S12 AG r
12612 AG r 13563 AG r
12630 GA r 13590 GA r
12633 CT y 13594 AG r
12635 TC 13602 TC y
12669 CT 13611 AG r
12672 AG r 13617 TC
12693 AG r 13641 TC
12705 CT 33650 CT
12720 AG r 13651 AG x
12738 TC ~ 13660 AG r
12768 AG r ' 13708 GA r
12771 GA r 13722 AG r
12810 AG r 13734 TC
12822 AG r 13TS9 GA r
12850 AG r 13780 AG r
12879 TC 13789 TC
12882 CT 13803 AG r
12930 AT w 13812 TC y
12940 GA r 13818 TC
12948 AG r 13819 TC
12967 AC m 33827 AG r
12972 AG r 13880 CA rn
12999 AG r 13886 TC
13020 TC 13914 CA m
13059 CT y 13924 CT
94
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
13927 AT w 14569 GA x
13928 GC s 14571 TA w
13958 GC s 14580 AG r
13965 TC 14587 AG r
_ AG r 14605 AG r
13966
13980 GA r 14668 CT
_ TA w 14693 AG x
14000
14016 GA r 14766 CT
14020 TC 14769 AG x
14022 AG r 14783 TC
14025 TC 14793 AG x
14034 TC 14798 TC
14059 AG r 14812 CT
14070 AGT d ~ 14836 AG x
14070 AGT d 14861 GA x
14088 TC 14862 CT
14094 TC 14905 GA r
_ CT y 14911 CT y
14097
14118 AG r 14971 TC
14128 AG r 14974 CG s
14148 AG r 14979 TC
14152 AG r 15016 CT
14167 CT 15034 AG r
14178 TC 15043 GA r
14182 TC 15110 GA r
14200 TC 15113 AG r
14203 AG r 15115 TC y
14209 AG r 15136 CT y
14212 TC 15172 GA r
14215 TC y 15204 TC
142 TC 15217 GA r
21
_ AG r 15218 AC m
_
142_33
14272 CG s 15229 TC
14284 CT 15238 CG s
14308 TC 15244 AG r
14311 TC 15257 GA r
14318 TC 15261 GA r
14319 TC 15301 GA r
14371 TC 15317 GA r
14374 TC 15318 CT
14384 GC s 15323 GA r
14455 CT y 15326 AG r
14459 GA r 15346 GA r
14470 TC 15358 AG r
14484 TC y 15431 GA r
14488 TC 15442 AG r
14502 TC 15452 CA m
14560 GA r 15466 GA r
14566 AG r 15470 TC
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
15487 AT w 16126 TC
15497 _ ~ 16129 GCA v
GA
15514 TC 16129 GCA v
15519 TG I614t~ TC
15535 CT 16144 TC
15607 AG r 16145 GA r
15626 CT 16147 CT
15629 TC 16148 CT
15646 CT 16153 GA r
15661 CT 16162 AG r
15663 TC 16163 AC m
15670 TC 16166 AC m
15724 AG r 16167 CT
15731 GA r 16168 CT
15746 AG r 16169 CT y
15766 AG r 16171 AG r
15784 TC 16172 . 7C
15793 CT 161'75 AG r
158Q3 GA r 16176 CT
15846 GA r 16182 AC m
15812 GA r 16183 AC m
15824 AG r 16184 Cfi y
15833 CT 16185 CT
15849 CT 16186 CT
15884 GC s 16187 CT
15900 TG 16188 CAG v
15904 CT 16188 CAG v
15907 AG r 16189 TC
15924 AG r 16192 CT
15927 GA r 16193 CT
15928 GA r 16207 AG r
15930 GA r 16209 TC
15932 TC 16212 AG r
15939 CT 16213 GA r
15941 TC 16214 CT
15942 TC 16217 TC
15968 TC 16219 AG r
16017 TC 16223 CT
16038 AG x 16224 TC
~
16051 AG r 16227 AG r
''
16069 CT x6229 TC
16Ur71 CT 16230 AG r
16075 TC 16231 TC
16086 TC 16232 CT
16093 TC m 16234 CT r
16148 CT 16235 AG
16111 CT 16239 CT x
16214 CA 16241 AG
16124 TC 16242 CT
96
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
16243 TC 16309 AG r
16245 CT 16311 _
TC
16247 AG r 16316 AG r
16249 TC 16317 AT w
_ AC m 16318 AT w
16254
16255 GA r 16319 GA r
16256 CT 16320 CT
16257 CT 16324 TC
16258 AG r 16325 TC
16260 CT y 16326 AG r
16261 CT 16327 CT
16264 CT 16343 AG r
16265 AC m 16344 CT
26266 CT 16354 CT
16268 CT 16355 CT
16270 CT 16356 TC
16271 TC 16357 TC
16274 GA r 16360 CT
16278 CT 16362 TC y
16284 AG r 16366 CT
16286 CG s 16368 TC
16287 CT 16390 GA r
16288 TC 16391 GA r
16290 CT 16399 AG r
16291 CT 16438 GA r
16292 CT 16439 CA m
16293 AG r 16483 GA r
16294 CT 16519 TC
16296 CT 16527 CT
16298 TC
16304 TC
REFERENCE TO SEQUENCE LISTINGS
SEQ DJ N0:1 is a theoretical human mtDNA genome sequence containing the
nucleotide alleles of this invention as listed in Table 3.
SEQ ID N0:2 is the human mtDNA reference sequence called the Cambridge
Sequence (GenBank Accession No. J01415).
97
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
1/10
SEQ ID NO:1
1 gatcacaggt ctatcaccct attaaccact cacgggagct ctccatgcat ttggtatttt
61 cgtytggggg gyrtgcacgc gatagcatyg cgrgmcgctg gagccggagc accytatgtc
121 gcagtatctg tctttgattc ctrccycaty yyrttattta tcgcacctac rttcaataty
181 ayrgdmgavc atayhtayyr aagygtrytr aytartyaat gcttrtrrga cataryaata
241 acaattraay gyctgcacag ccrctttcca cacagacatc ataacaaaaa rtttycrcca
301 aaccccccct cccccrvtty tggcyacagc acttaaacay atctctgcca aaccccraaa
361 acaaagaacc ctracaccag cctaaccaga tttcaaattk tatctttwgg cggtatgyac
421 ttttaacagt caccccccaa ctaacacatt attttyccct cycaytycca yactactaay
481 cycatcaaya carcccccrc ccatcctrcc cagcacacac acaccgctgc taaccccata
541 ccccgaacca accaaacccc aaagacaccc cccacagttt atgtagctta ccycctyaaa
601 gcaatacact gaaaatgttt agacgggctc acatcacccc ataaacaaat aggtttggtc
661 ctrgcctttc tattagcycy tagtaagatt acacatgcaa gcatccccry tccagtgagt
721 ycaccctcta aatcaccacg atcaaaaggr acaagcatca agcacgcarc aatgcagctc
781 aaaacgctta gcctagccac acccccacgg gaaacagcag tgatwarcct ttagcaataa
841 acgaaagtty aactaagcta tactaacccc agggttggtc aatttcgtgc cagccaccgc
901 ggtcacacga ttaacccaag ycaatagaar ccggcgtaaa gagtgtttta gatcaccccc
961 bccccaataa agctaaaact cacctgagtt gtaaaaaact ccagttgaca caaaatarac
1021 tacgaaagtg gctttaacat rtctgaayac acaatagcta agacccaaac tgggattaga
1081 taccecacta tgcttagccc taaacctcaa cagttaaayc aacaaaactg ctcgccagaa
7.141 cactacgagc cacagcttaa aactcaaagg acctggcggt gcttcatayc cctctagagg
1201 agcctgttct gtaatcgata aaccccgatc aacctcacca ccycttgctc agcctatata
1261 ccgccatctt cagcaaaccc tgatgaaggy tacaaagtaa gcgcaagtac ccacgtaaag
7.321 acgttaggtc aaggtgtagc ccatgaggtg gcaagaaatg ggctacattt tctaccccag
7.381 amaactacga tagcccttat gaaacytaag ggtcraaggy ggatttagca gtaaactrag
7.441 artagagtgc ttagttgaac agggccctga agcgcgtaca caccgcccgt caccctcctc
1501 aartatactt caaaggacat ttaactaaaa cccctacgca tttatataga ggagacaagt
7.561 cgtaacatgg taagtgtact ggaaagtgca cttggacraa ccagagtgta gcttaacaca
1621 aagcacccaa cttacactta ggagatttca acttaacttg accgctctga gctaaaccta
1,681 gccccaaacc cactccaccy taytaycara caacyttarc caaaccattt acccarayaa
1741 agtataggcg atagaaattg aaacctggcg caatagatay agtaccgcaa gggaaagatg
1801 aaaaattata rccaagcata atatagcaag gactaacccc tataccttct gcataatgaa
1861 ttaactagaa ataactttgc aaggagarcc aaagctaaga cccccgaaac cagacgagct
1921 acctaaraac agctaaaaga gcacacccgt ctatgtagca aaatagtggg aagatttata
1981 ggtagaggcg acaaacctay cgagcctggt gatagctggt tgtccaagat agaatcttag
2041 ttcaacttta aatttgcccr cagaaccctc taaatcccct tgtaaattta aytgttagtc
2101 caaagaggaa cagctctttg gacactagga aaaaaccttg tagagagagt aaaaaattta
2161 acacccatag taggcctaaa agcagccacc aattaagaaa gcgttcaagc tcaacaccca
2221 ctacctaaaa aatcccaaac atatvactga actcctcaca ccmaattgga ccaatctatc
2281 accctataga agaactaatg ttagtatrag taacatgaaa acattctcct cygcataagc
2341 ctgcgtcaga tyaaaacrct gaactgacaa ttaacagccy aatatctaca atcaaccaac
2401 aagtcattat tacccycact gtcaacccaa cacaggcatg ctcataagga aaggttaaaa
2461 aaagtaaaag gaactcggca aaycttaccc cgcctgttta ccaaaaacat cacctctagc
2521 atcaccagta ttagaggcac cgcctgccca gtgacacatg tttaacggcc gcggtaccct
2581 raccgtgcaa aggtagcata atcacttgtt ccttaaatag ggacctgtat gaatggctyc
2641 acgagggtty agctgtctct tacttttaac cagtgaaatt gacctgcccg tgaagaggcg
2701 ggcatracac agcaagacga gaagacccta tggagcttta atttattaat gcaarcarta
2761 ectaacarac ccacaggtcc taaactacya arcctgcatt aaaaatttcg gttggggcga
2821 cctcggagca gaaycmaacc tccgagcagt acatgcyaag acytcaccag tcaaagcgaa
2881 ctacyatact caattgatcc aataacttga ccaacggaac aagttaccct agggataaca
2942 gcgcaatcct attctagagt ccatatcaac aatagggttt acgacctcga tgttggatca
3001 ggacatcccr atggtgcagc cgctattaaa ggttcgtttg ttcaacgatt aaagtcctac
3061 gtgatctgag ttcagaccgg agyaatccag gtcggtttct atcta-cttc aaattcctcc
3121 ctgtacgaaa ggacaagaga aataaggcct acttcacaaa gcgccttccc ccgtaaatga
3181 tatcatctca acttagyatw ayaycyacac ccacccaaga rcagggtttg ttaagatggc
3241 agagcccggt aatcgcataa aacttaaaac tttacagtca gaggttcaay tcctcttctt
3301 aacaacayac ccatgrccaa cctcctactc ctcattgtac ccattctaat cgcaatggca
3361 ttcctaatgc tyaccgaacg aaaaattcta ggcyatatac aactacgcaa aggccccaac
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
2/10
3421 gttgtaggcc cctacggrct actacaaccy ttcgctgacg ccataaaact cttcaccaar
3481 gagcccctaa aacccgccac atctrccatc acyctvtaca tcaccgcccc gaccttrgct
3541 ctcaccatyg chcttctact atgarccccc ctccccatac ccaaccccct ggtyaacctc
3601 aacctaggcc tcctatttat tctagccacc tctagcctag ccgyttactc aatcctctga
3661 tcaggrtgag catcaaactc aaactacgcc ctratcggyg cactgcgagc agtagcccar
3721 acaatctcat atgaagtcac cctagccatc attctrctat caacattact aataagtggc
3781 tcctttaacc tctccnccct tatcacarca caagarcacc tctgattact cctrccatca
3841 tgrcccytgg ccataatatg atttayctcc acactagcag agaccaaccg aacccccttc
3901 gaccttgccg aaggggartc mgaactrgtc tcaggcttca acatcgaata cgccgcaggc
3961 cccttcgccy tattcttcat rgccgaatac acaaacatta ttataataaa caccctcacc
4021 actayaatct tcctaggaay aacrtatrac gcactctccc ctgaactcta cacaacatat
4081 tttgtyacca agaccctact tctracctcc ctgttcytat grrttcgaac agcatacccc
4141 cgattccgct acgaccarct catacacctc ctatgaaaaa acttcctacc actcacccta
4201 gcrttactta tatgayatgt ytccrtaccc aytacaatct ccagcatycc ccctcaaacc
4261 taagaaatat gtctgataaa agagttactt tgatagagta aataatagga gyttaaaccc
4321 ccttatttct aggacyatga gaatcgaacc catccctgag aatccaaaay tctccgtgcc
4381 acctatcrca ccccatccta aagtaaggtc agctaaataa gctatcgggc ccataccccg
4441 aaaatgttgg ttawaccctt cccgtactaa ttaatcccct ggcccaaccc rtcatctact
4501 ctaccrtytt trcaggcaca ctcatcachg cgctaagctc rcactgattt tttacctgag
4561 taggcctaga aataaacatr ctagcyttta ttccarttct aaccaaaaaa ataaaccctc
4621 gttccacaga agctgccatc aagtayttcc tcacrcaagc aaccgcatcc ataatccttc
4681 taatagcyat cctcytcaac aatatactct ccggrcaatg aaccataacc aatactacca
4741 aycaatactc atcattaata atcatartrg ctatagcaat aaaactagga atagccccct
4801 ttcacttctg agtcccagar gttrcccaag gcrcccctct racatccggc ctgcttcttc
4861 tcacatgaca aaaactagcc ccyatctcaa tcatatacca aatctcyccc tcactaracg
4921 taagccttct cctcactctc tcaatcttat ccatcatagy aggcagttga ggtggaytaa
4981 accaAaccca gctrcgcaaa atcytagcat actcctcaat tacccayata ggatgrataa
5041 takcarttct accgtacaac ccyaacataa ccattcttaa tttaactatt tatatyatcc
5101 taactacyac cgcattccta ctactcaact taaactccag caccacracc ctrctactat
5161 ctcgcacctg aaacaagmta acatgactaa cacccttaat tccatccacc ctcctctccc
5221 taggaggcct rcccccrcta accggctttt tgccyaaatg grycattatc gaagaattca
5281 caaaraacaa tagcctcaty atccccacca tcatagccac catcaccctm mttaacctct
5341 acttctacct acgcctaatc tactccacct caatcacact actccccatr tcyaacaacg
5401 taaaaataaa atgacarttt gaacayacaa aacccacccc aytcctcccc acactcatcr
5461 ccctyaccac rctactccta cctatctccc cyttyatact aataatctta tagaaattta
5521 ggttaaatac agaccaagag ccttcaaagc cctcagtaag ttgcaatact taatttctgy
5581 racagctaag gactgcaaaa ycycaytctg catcaactga acgcaaatca gcyactttaa
5641 ttaagctaag ccctyactag accaatggga cttaaaccca caaacactta gttaacagct
5701 aagcacccta rtcaactggc ttcaatctac ttctcccgcc gccgggaaaa aaggcgggag
5761 aagccccggc agrtttgaag ctgcttcttc gaatttgcaa ttcaatatga raaycacctc
5821 rgagcyggta aaaagaggcc tarcccctgt ctttagattt acagtccaat gcttcactca
5881 gccattttac ctcaccccca ctgatgttcg ccgaccgttg actattctct acaaaccaca
5941 aagacattgg racactatac ctattattcg gcgcatgagc tggrgtycta ggcacagctc
6001 taagcctcct tattcgagcc gagctrggyc agccaggcaa ccttytaggt aacgaccaca
6061 tctacaacgt yatcgtyaca gcccatgcat ttgtaataat cttyttcata gtaataccca
6121 tcataatcgg aggctttggc aactgactar tycccctaat aatyggygcc cccgatatgg
6181 crttyccccg cataaacaac ataagcttct gactcttacc yccctcyctc ctactcctgc
6241 tcgcatctgc tayagtrgag gccggagcag gaacaggttg aacagtctac cctcccttag
6301 cagggaacta ctcccaccct ggarcctccg tagacctaac catcttctcc ttacacctag
6361 caggtrtctc ytctatctta ggggccatca ayttcatcac aacaattatc aatataaaac
6421 cccctgccat aacccaatac caaacgcccc tcttcgtctg atccgtccta atyacagcag
6481 tcctacttct mctatctctc ccagtcctag ctgctggcat cacyatacta ctaacagacc
6541 gcaacctyaa caccaccttc ttcgaccccg ccggaggagg agacccyatt ctataccaac
6601 acctatyctg atttttcggt caccctgaag tttatattct tatcctacca ggcttcggaa
6661 taatctccca tattgtaacy tactactccg gaaaaaaaga accatttgga tayataggya
6721 tggtctgagc tatratatca attggcttcc trgggtttat cgtgtgagcr caccayatat
6781 ttacagtagg aatagacgta gacacacgag catayttcac ctccgcyacc ataatcatcg
6841 ctatccccac cggcgtcaaa gtatttagct gactmgccac actccacgga agcaatatga
6901 aatgatctgc tgcagtgctc tgagccctag gattcatytt tcttttcacc gtaggtggcc
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
3/10
6961 tractggcat tgtattagca aactcatcrc tagacatcgt actacacgac acgtactacg
7021 ttgtagcyca cttccactat gtcctatcaa trggrgcwgt atttgccatc ataggrggct
7081 tcattcactg atttccccta ttctcaggct acaccctaga ccaaacctac gccaaaatcc
7141 atttcrctat catrttcatc ggcgtaaatc taacyttctt cccacaacac tttctmggcc
7201 trtccggaat gccccgacgt tactcrgact accccgatgc atacaccaca tgaaayrtcc
7261 tatcatctgt rggytcattc atttctctaa cagcagtaat attaataatt ttcatgatyt
7321 gagaagcctt cgcttcraag cgaaaartcc taatagtaga agaaccctcc ataaacctgg
7381 agtgactaya tggatgcccc ccrccctacc acacattcga agarcccgta tacataaaat
7441 ctaracaaaa aaggaaggaa tcgaaccccc caaagytggt ttcaagccaa ccycatggcc
7501 tccatgactt tttcaaaaag rtattagaaa aaccatttca taactttgtc aaagttaaat
7561 yataggctaa rtcctatata tcttaatggc acatgcagcr caagtaggtc tacaagacgc
7621 tacwtcccct atcatagaag agctyatyac ctttcatgay cacrccctca tartyatttt
7681 ccttatctgc ttyytartcc tgtatgccct tttcctaaca ctcacaacaa aactaactaa
7741 tacyaacatc tcagacgctc aggaratrga raccgtctga actatcctgc ccgccatcat
7801 cctagtcctc atcgccctcc catccctacg catcctttac ataacagacg aggtcaayga
7861 yccytcyctt accatcaaat caattggcca ccaatggtac tgaacctacg agtacaccga
7921 ctacggcgga ctratcttca actcctayat acttccccca ttattcctag aaccaggcga
7981 cctgcgactc cttgacgtyg acaatcgagt agtrctcccr attgaarccc ccattcgtat
8041 aataattaca tcacaagacg tcttgcactc atgagctgty cccacaytag gcttaaaaac
8101 agatgcaatt ccmggacgtc taaaccaaac cactttcacc gytacacgrc crggrgtata
8161 ctacggtcaa tgctctgaaa tctgyggagc aaaccacagy ttcatrccca tcgtcctaga
8221 attaattccc ctaaaaatct ttgaaatrgg rcccgtatty accctatarc accccctcta
8281 cccccyctag arcccacygt aaagctaact tagcattaac cttttaagtt aaagattaag
8341 agarccaaca cctctttaca gtgaaatgcc ccaactaaat actaccrtrt grcccaccat
8401 aatyaccccc ataytcctta cactattyct catcacccaa ctaaaaayat taaacacaar
8461 ctaccacyta cyyccctcac caaarcccat aaaaataaaa aattataaca aaccctgaga
8521 accaaaatga acgaaaatct gttcrcttca ttyattgccc ccrcartcct aggcctrccc
8581 gccrcagtac tgatcattct atttccccct ctattgaycc ccacctccaa atatctcatc
8641 aacaaccgac taatyaccac ccaacaatga ctaatcaaac taacctcaaa acaaatrata
8701 rcyayacaya acactaaagg rcgaacctga tcycttatac tagtatcctt aatcattttt
8761 attrccacaa ctaacctcct mggrctcctr ccyyactcat ttacrccaac cacccaacta
8821 tctataaacc tagccrtrgc catcccctta tgagcrggcr cagtgattat aggcytycgc
8881 tctaagatta aaaatgccct agcccacttc ytrccacaag gcacaccyac accccttatc
8941 ccyatactag ttattatcga arccatcagc ctactcattc aaccaatagc cctrgccgta
9001 cgcctaaccg ctaacattac tgcaggccac ctactcatgc ayctaattgg aarcrccacc
9061 ctagcaatat craccaytaa ccttccctcy acmcttatca tcytcacaat tctrattctr
9121 ctractatcc tagaartcgc tgtcgcctta rtccargcct acgttttcac actyctagta
9181 agcctctacc tgcacgacaa cacataatga cccaccaatc rcatgcctat catatartaa
9241 arcccagycc atgaccccta acrggggccc tytcagccct cctaatgacc tccggyctag
9301 ccatgtgatt ycacttccac tccayaacgc tcctyatact aggcctrcta accaryacac
9361 taaccatata ccaatgrtgg cgcgatgtaa cacgagaaag cmcataccaa ggccaccaca
9421 caccacctgt ccaaaaaggc cttcgatayg ggatartcct atttattacc tcagaarttt
9481 ttttcttcgc aggatttttc tgagccttyt accactccag cctagcccct accccycaay
9541 taggrggrca ctgrccccsa acaggcatca ccccrctaaa tcccctagaa rtcccactyc
9601 taaacacatc cgtattactc gcatcaggag trtcaatcac ctgagcycac catagtctaa
9661 tagaaarcaa ccgaaaccaa ayaattcaag cactgctyat tacaatttta ctgggtctct
9721 attttaccct cctacaagcc tcagagtact tcgartctcc cttcaccatt tccgacggca
9781 tctacggctc aacatttttt gtagccacag gcttccaygg amtwcacgtc attattggct
9841 caactttcct cactatctgc ttcatccgcc aactaatatt tcactttaca tccaaacatc
9901 actttggctt ygaagccgcc gcctgatact grcattttgt agatgtggty tgactayttc
9961 tgtatrtctc catctaytga tgagggtctt actcttttag tataaatagt accgttaact
10021 tccaattaac tagytttgac aacattcaaa aaagagtaat aaacttcgcc ttaattttaa
10081 taatcvacac cctcctagcc ttactactaa taatyatyac attttgacta ccacaactca
10141 ayggctacat rsaaaaatcc accccttacg artgcggctt csaccctata tcccccrccc
10201 gcgtcccttt ctccataaaa ttcttcttag tagctatyac cttcttatta ttygayctag
10261 aaattgccct ccttttaccc ctaccatgag ccctacaaac aactaacctr ccrctaatag
10321 ytatrtcatc cctcttatta atcatcatcc tagccctrag tctggcctay gagtgactac
10381 aaaaaggatt agactgarcy gaattggtay ataktttaaa caaaacraat gatttcgact
10441 cattaaatta tgataatcat atytaccaaa tgcccctcat ttacataaat attatactrg
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
4/10
10501 cattyaccat ctcacttcta ggaatactag tatatcgctc acacctcatr tcctccctac
10561 tatgcctaga aggaataata ctatcrctrt tcattatagc tactctcaya accctcaaca
10621 cccactccct cttagcyaay attgtrccta ttgccatayt agtyttygcc gcctgcgaag
10681 cagcggtrgg cctagcccta ctagtctcaa tctccaacac atatggccta gactaygtac
10741 ataacctaaa cctactccaa tgctaaaact aatcgtccca acaattatay trytaccact
10801 gacrtgacty tccaaaaarc acataatytg aatcaacaca accacccaca gcctaattat
10861 tagcatcatc ccyctrctat tttttaacca aatyaacaac aacctattta gctgytcccy
10921 aaccttttcc tccgacccyc taacaacccc cctcctaata ctaacyacct gactcctacc
10981 cctsacaatc atggcaagcc arcgccactt atccarygaa ccrctatcac gaaaaaaact
11041 ctacctctct atactaatct ccctacaaat ctccttartt ataacattca crgccacaga
11101 actaatcata ttttatatct tcttcgaaac cacacttatc cccaccytgr ctatcatcac
11161 ccgatgrggc arccarycag aacgcctgaa cgcaggcaca tacttcctat tctayaccct
11221 agtaggctcc cttcccctac tcatcgcact ratttayact cacaacaccc taggctcact
11281 aaacattcta ctactyacyc tcactgccca agaactatca aactcctgag cyaaCaactt
11341 aatatgacta gcttacacaa trgcytttat agtaaarata cctctttacg gactccactt
11401 atgactccct aaagcccatg tcgaagcccc catcgctggg tcaatagtac ttgccgcagt
11461 actcttraaa ctaggyggct atggtataat acgcctcaca ctcattctca accccctgac
11521 aaaacacata gcctayccct tccttgtact atccctatga ggcataatta taacaagctc
11581 catctgcctr cgacaaacag acctaaaatc rctcattgca tactcttcaa tcagccacat
11641 rgccctcgta gtrrcagcca ttctcatcca aacyccctga agcttcaccg gcgcagtcat
11701 yctcataatc gcccacggrc tyacatcctc attactattc tgcctagcaa actcaaacta
11761 cgaacgyact cacagtcgca tcataatcct ctctcaagga cttcaaactc trctcccact
11821 aatagctttt tgatgacttc tagcaagcct cgcyaacctc gccttacccc ccactattaa
11881 cctrctrgga garctctcyg tgctagtarc cacrttctcc tgatcaaata tcactctcct
11941 actyacrgga ctcaacatrc tartcacarc cctatactcc ctctacatat ttaccacaac
12001 acaatgrggc tcactcaccc accacattaa caacataaaa ccctcattya cacgagaaaa
12061 caccctcatr ttcatacacc takcccccat tctcctccta tccctcaacc ccgacatcat
12121 yaccgggttt tccycttgta aatatagttt aaycaaaaca tcagattgtg artcygacaa
12181 cagaggctta cgacccctta tttaccgaga aagctcacaa gaactgctaa ctcrtrccyc
12241 catgtctrac aacatggctt tctcaacttt taaaggataa cagctatcca ttggtcttag
12301 gccccaaraa ttttggtgca actccaaata aaagtaataa ccatgyacac tactatarcc
12361 rccctaaccc trrcttccct aattcccccc atccttrcca ccctcrttaa cccyaacaaa
12421 aaaaactcat acccccatta tgtaaaatcc attgtcgcat ccacctttat tatcagyctc
12481 ttccccacaa caatattcat rtgcctrgac caagaagtya ttatctcraa ctgacactgr ~~
12541 gccacaaccc aaacaaccca gctctcccta agcttcaaac tagactactt ctccataata
12601 ttcatccctg trgcattgtt cgttacatgr tcyaycatag aattctcact gtgatatata
12661 aactcagayc craacattaa tcagttcttc aartatctac tcatyttcct aattaccatr
12721 ctaatcttag ttaccgcyaa caacctattc caactgttca tcggctgrga rggcgtagga
12781 attatatcct tcttgctcat cagttgatgr tacgcccgag crgatgccaa cacagcagcc
12841 attcaagcar tcctatacaa ccgtatcggc gatatcggyt tyatcctcgc cttagcatga
12901 tttatcctac actccaactc atgagacccw caacaaatar cccttctraa cgctaatcca
12961 agcctcmccc crctactagg cctcctccta gcagcagcrg gcaaatcagc ccaattaggy
13021 ctccacccct gactcccctc agccatagaa ggccccacyc cagtctcrgc cctactccac
13081 tcaagcacta tagttgtagc mggrrtcttc ttactcatcc gcttccaccc cctarcagaa
13141 aayarcccrc taatccaaac tctaacacta tgcttaggcg ctatcaccac tctrttygca
13201 gcagtctgcg cycttacaca raatgacatc aaaaaaatcg tagccttctc cacttcaagt
13261 carctaggac tcatartagt yacaatcggc atcaaccaac cacacctagc attcctgcac
13321 atctgtaccc acgccttctt caaagccata ctatttatgt gctccggrtc catcatccac
13381 aaccttaaca atgaacaaga tattcgaaaa ataggaggac tactcaaaac catacctcts
13441 acttcaacct ccctcaccat tggcagccta gcattarcag gaatrccttt cctyacaggb
13501 ttctaytcca argaccacat catcgaaacc gcaaacatat catacacaaa cgcctgagcc
13561 ctrtctatta ctctcatcgc tacctccctr acargcgcct ayagcactcg rataatyctt
13621 ctcaccctaa caggtcaacc ycgcttcccy rcccttactr acattaacga aaataacccc
13681 accctactaa accccattaa acgcctgrca gccggaagcc trttcgcagg attyctcatt
13741 actaacaaca tttcccccrc atcccccttc caaacaacar tccccctcya cctaaaactc
13801 acrgccctcg cygtcacyyt cctaggrctt ctaacagccc tagacctcaa ctacctaacc
13861 aacaaactta aaataaaatm cccacyatgc acattttatt tctccaacat actmggattc
13921 tacyctwsca tcacacaccg cacaatcccc tatctagscc ttctyrcgag ccaaaacctr
13981 cccctactcc tcctagaccw aacctgacta gaaaarctay trccyaaaac aatytcacag
5/10
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
5/10
14041 caccaaatct ccacctccrt catcacctcd acccaaaaag gcataatyaa actytayttc
14101 ctctctttct tcttcccrct catcctarcc ctactcctaa tcacatarcc trttcccccg
14161 agcaatytca attacaayat ayacaccaac aaacaatgty carccagtra cyacyactaa
14221 ycaacgccca tartcataca aagcccccgc accaatagga tcctcccgaa tsaaccctga
7.4287, cccytctcct tcataaatta ttcagctycc yacactayya aagtttacca caaccaccac
14341 cccatcatac tctttcaccc acagCacCaa yccyacctcc atcsctaacc ccactaaaac
14401 actcaccaag acctcaaccc ctgaccccca tgcctcagga tactcctcaa tagcyatcrc
14461 tgtagtatay ccaaagacaa ccaycatycc ccctaaataa aytaaaaaaa ctattaaacc
14521 catataacct cccccaaaat tcagaataat aacacacccr accacrccrc waacaatcar
14581 tactaarccc ccataaatag gagarggctt agaagaaaac cccacaaacc ccattactaa
14641 acccacactc aacagaaaca aagcatayat cattattctc gcacggacta carccacgac
14701 caatgatatg aaaaaccatc gttgtatttc aactacaaga acaccaatga ccccaatacg
14761 caaaaytarc cccctaataa aaytaattaa ccrctcaytc atcgacctcc cyaccccatc
14821, caacatctcc gcatgrtgaa acttcggctc actccttggc ryctgcctga tcctccaaat
14881 caccacagga ctattcctag ccatrcacta ytcaccagac gcctcaaccg ccttttcatc
14941 aatcgcccac atcactcgag acgtaaatta yggstgaayc atccgctacc ttcacgccaa
15001 tggcgcctca atattyttta tctgcctctt cctrcacatc ggrcgaggcc tatattacgg
15061 atcatttctc tactcagaaa cctgaaacat cggcattatc ctcctgcttr carcyatagc
15127. aacagccttc ataggytatg tcctcccgtg aggccaaata tcattctgag grgccacagt
15181 aattacaaac ttactatccg ccaycccata cattggrmca gacctagtyc aatgaatstg
15241 aggrggctac tcagtaraca rtcccaccct cacacgattc tttacctttc acttcatctt
15301 rcccttcatt attgcarycc tarcarcact ccacctccta ttcttrcacg aaacgggrtc
15361 aaacaacccc ctaggaatca cctcccattc cgataaaatc accttccacc cttactacac
15421 aatcaaagac rccctcggct trcttctctt cmttctctcc ttaatracay taacactatt
15481 ctcaccwgac ctcctargcg acccagacaa ttayacccya gccaacccct taaayacccc
15541 tccccacatc aagcccgaat gatatttcct attcgcctac acaattctcc gatccgtccc
15601 taacaarcta ggaggcgtcc ttgccytayt actatccatc ctcatyctag caataatccc
15667. yaycctccay atatccaaac aacaaagcat aatatttcgc ccactaagcc aatcacttta
15721 ttgrctccta rccgcagacc tcctcrttct aacctgaatc ggaggrcaac cagtaagcta
15781 cccytttacc atyattggac aartarcatc crtactatac ttcrcaacaa tcytaatcct
15841 aataccaayt atctccctaa ttgaaaacaa aatactcaaa tggscctgtc cttgtagtay
15901 aaaytartac accagtcttg taarccrrar aygaaaacyt yyttccaagg acaaatcaga
15967. gaaaaagyct ttaactccac cattagcacc caaagctaag attctaattt aaactaytct
16021 ctgttctttc atggggargc agatttgggt rccacccaag tattgactya yccaycaaca
16081 accgcyatgt atytcgtaca ttactgcyag ycamcatgaa tatygyacvg taccataaay
16141 actyrayyac ctrtagtaca trmaamyyya ryccryatca ammyyyyvyc cyyatgctta
16201 caagcargya crryaaycra ccyycarcyr yyayrcatya ryygyarcyc caamryyrcy
16261 yctymycyay yagratayca acarasyyay yyrycytyaa cagyacatrg yacatrwwry
26321 catyyrycgt acatagcaca ttryagtcaa atcyyyycty gycccyaygg atgacccccc
16381 tcagataggr rtcccttgrc caccatcctc cgtgaaatca atatcccgca caagagtrmt
16441 actctcctcg ctccgggccc ataacacttg ggggtagcta aartgaactg tatccgacat
16501 ctggttccta cttcagggyc ataaagycta aatagcccac acgttcccct taaataagac
16561 atcacgatg .
SEQ ID N0:2
Human Mitochondrial DNA Revised Cambridge Reference Sequence
Note: the sequence below is a modified version of the 2001 Revised Cambridge
Reference Sequence (GenBank #NC_001807, a derivation of #J01415). Corrections
have been made and annotated per the reanalyses by Andrews et al (1999).
Rare polymorphisms are shown with flanking green nucleotides. Corrected
sequencing errors are shown with flanking red nucleotides. Clicking on the
corrected nucleotides will return to the annotation. A summary table of the
Andrews et a1 revisions is available online.
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
6/10
This sequence differs from GenBank #N0_001807 in that:
HISTORICAL NUCLEOTIDE NUMBERS ARE MAINTAINED. 3106de1 is maintained in the
sequence as a gap.
Rare polymorphism 7SOA is retained, as in both the original Anderson et al
(1981) paper & in the sequence reanalysis by Andrews et al.
This Revised Cambridge Reference Sequence (RCRS) has eighteen annotated
nucleotides.
Seven are rare polymorphisms: Nucleotides 263A, 3110-3150, 750A, 1438A,
4769A, 8860A, and 15326A axe considered to be rare polymorphisms. See summary
table. Rare polymorphisms are shown with flanking green nucleotides.
Eleven are error corrections: Nucleotides 3106deI, 3423T, 4985A, 95590,
113350, 7.37020, 14I99T, i42720, 143650, 143680, 247660 are corrections of the
original Cambridge sequence. The errors in the original Cambridge_sequence
have been attributed to sequencing errors (8 instances) and to the inclusion.
of bovine (2 instances) or HeLa (1 instance) DNA. See summary table.
Corrected sequencing errors are shown with flanking red nucleotides.
1 gatcacaggt ctatcaccct attaaccact cacgggagct ctccatgcat ttggtatttt
61 cgtctggggg gtatgcacgc gatagcattg cgagacgctg gagccggagc accctatgtc
121 gcagtatctg tctttgattc ctgcctcatc ctattattta tcgcacctac gttcaatatt
181 acaggcgaac atacttacta aagtgtgtta attaattaat gcttgtagga cataataata
241 acaattgaat gtctgcacag ccactttcca cacagacatc ataacaaaaa atttccacca
301 aaccccccct cccccgcttc tggccacagc acttaaacac atctctgcca aaccecaaaa
361 acaaagaacc ctaacaccag cctaaccaga tttcaaattt tatcttttgg cggtatgcac
421 ttttaacagt caccccccaa ctaacacatt attttcccct cccactccca tactactaat
481 ctcatcaata caacccccgc ccatcctacc cagcacacac acaccgctgc taaccccata
541 ccccgaacca accaaacccc aaagacaccc cccacagttt atgtagctta cctcctcaaa
601 gcaatacact gaaaatgttt agacgggctc acatcacccc ataaacaaat aggtttggtc
661 ctagcctttc tattagctct tagtaagatt acacatgcaa gcatccccgt tccagtgagt
721 tcaccctcta aatcaccacg atcaaaagga acaagcatca agcacgcagc aatgcagctc
781 aaaacgctta gcctagccac acccccacgg gaaacagcag tgattaacct ttagcaataa
841 acgaaagttt aactaagcta tactaacccc agggttggtc aatttcgtgc cagccaccgc '~
901 ggtcacacga ttaacccaag tcaatagaag ccggcgtaaa gagtgtttta gatcaccccc
961 tccccaataa agctaaaact cacctgagtt gtaaaaaact ccagttgaca caaaatagac
1021 tacgaaagtg gctttaacat atctgaacac acaatagcta agacccaaac tgggattaga
1081 taccccacta tgcttagccc taaacctcaa cagttaaatc aacaaaactg ctcgccagaa
1141 cactacgagc cacagcttaa aactcaaagg acctggcggt gcttcatatc cctctagagg
1201 agcctgttct gtaatcgata aaccccgatc aacctcacca cctcttgctc agcctatata
1261 ccgccatctt cagcaaaccc tgatgaaggc tacaaagtaa gcgcaagtac ccacgtaaag
1321 acgttaggtc aaggtgtagc ccatgaggtg gcaagaaatg ggctacattt tctaccccag
1381 aaaactacga tagcccttat gaaacttaag ggtcgaaggt ggatttagca gtaaactaag
1441 agtagagtgc ttagttgaac agggccctga agcgcgtaca caccgcccgt caccctcctc
1501 aagtatactt caaaggacat ttaactaaaa cccctacgca tttatataga ggagacaagt
1561 cgtaacatgg taagtgtact ggaaagtgca cttggacgaa ccagagtgta gcttaacaca
1621 aagcacccaa cttacactta ggagatttca acttaacttg accgctctga gctaaaccta
1681 gccccaaacc cactccacct tactaccaga caaccttagc caaaccattt acccaaataa
1741 agtataggcg atagaaattg aaacctggcg caatagatat agtaccgcaa gggaaagatg
1801 aaaaattata accaagcata atatagcaag gactaacccc tataccttct gcataatgaa
1861 ttaactagaa ataactttgc aaggagagcc aaagctaaga cccccgaaac cagacgagct
1921 acctaagaac agctaaaaga gcacacccgt ctatgtagca aaatagtggg aagatttata
1981 ggtagaggcg acaaacctac cgagcctggt gatagctggt tgtccaagat agaatcttag
2041 ttcaacttta~aatttgccca cagaaccctc taaatcccct tgtaaattta actgttagtc
2107. caaagaggaa cagctctttg gacactagga aaaaaccttg tagagagagt aaaaaattta
2161 acacccatag taggcctaaa agcagccacc aattaagaaa gcgttcaagc tcaacaccca
2221 ctacctaaaa aatcccaaac atataactga actcctcaca cccaattgga ccaatctatc
2281 accctataga agaactaatg ttagtataag taacatgaaa acattctcct ccgcataagc
2341 ctgcgtcaga ttaaaacact gaactgacaa ttaacagccc aatatctaca atcaaccaac
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
7/10
2401 aagtcattat taccctcact gtcaacccaa cacaggcatg ctcataagga aaggttaaaa
2461 aaagtaaaag gaactcggca aatcttaccc cgcctgttta ccaaaaacat cacctctagc
2521 atcaccagta ttagaggcac cgcctgccca gtgacacatg tttaacggcc gcggtaccct
2581 aaccgtgcaa aggtagcata atcacttgtt ccttaaatag ggacctgtat gaatggctcc
2641 acgagggttc agctgtctct tacttttaac cagtgaaatt gacctgcccg tgaagaggcg
2701 ggcataacac agcaagacga gaagacccta tggagcttta atttattaat gcaaacagta
2761 cctaacaaac ccacaggtcc taaactacca aacctgcatt aaaaatttcg gttggggcga
2821 cctcggagca gaacccaacc tccgagcagt acatgctaag acttcaccag tcaaagcgaa
2881 ctactatact caattgatcc aataacttga ccaacggaac aagttaccct agggataaca
2941 gcgcaatcct attctagagt ccatatcaac aatagggttt acgacctcga tgttggatca
3001 ggacatcccg atggtgcagc cgctattaaa ggttcgtttg ttcaacgatt aaagtcctac
3061 gtgatctgag ttcagaccgg agtaatccag gtcggtttct atcta-cttc aaattcctcc
3121 ctgtacgaaa ggacaagaga aataaggcct acttcacaaa gcgccttccc ccgtaaatga
3181 tatcatctca acttagtatt atacccacac ccacccaaga acagggtttg ttaagatggc
3241 agagcccggt aatcgcataa aacttaaaac tttacagtca gaggttcaat tcctcttctt
3301 aacaacatac ccatggccaa cctcctactc ctcattgtac ccattctaat cgcaatggca
3361 ttcctaatgc ttaccgaacg aaaaattcta ggctatatac aactacgcaa aggccccaac
3421 gttgtaggcc cctacgggct actacaaccc ttcgctgacg ccataaaact cttcaccaaa
3481 gagcccctaa aacccgccac atctaccatc accctctaca tcaccgcccc gaccttagct
3541 ctcaccatcg ctcttctact atgaaccccc ctccccatac ccaaccccct ggtcaacctc
3601 aacctaggcc tcctatttat tctagccacc tctagcctag ccgtttactc aatcctctga
3661 tcagggtgag catcaaactc aaactacgcc ctgatcggcg cactgcgagc agtagcccaa
3721 acaatctcat atgaagtcac cctagccatc attctactat caacattact aataagtggc
3781 tcctttaacc tctccaccct tatcacaaca caagaacacc tctgattact cctgccatca
3841 tgacccttgg ccataatatg atttatctcc acactagcag agaccaaccg aacccccttc
3901 gaccttgccg aaggggagtc cgaactagtc tcaggcttca acatcgaata cgccgcaggc_
3961 cccttcgccc tattcttcat agccgaatac acaaacatta ttataataaa caccctcacc
4021 actacaatct tcctaggaac aacatatgac gcactctccc ctgaactcta cacaacatat
4081 tttgtcacca agaccctact tctaacctcc ctgttcttat gaattcgaac agcatacccc
4141 cgattccgct acgaccaact catacacctc ctatgaaaaa acttcctacc actcacccta
4201 gcattactta tatgatatgt ctccataccc attacaatct ccagcattcc ccctcaaacc
4261 taagaaatat gtctgataaa agagttactt tgatagagta aataatagga gcttaaaccc
4321 ccttatttct aggactatga gaatcgaacc catccctgag aatccaaaat tctccgtgcc
4381 acctatcaca ccccatccta aagtaaggtc agctaaataa gctatcgggc ccataccccg-
4441 aaaatgttgg ttataccctt cccgtactaa ttaatcccct ggcccaaccc gtcatctact
4501 ctaccatctt tgcaggcaca ctcatcacag cgctaagctc gcactgattt tttacctgag
4561 taggcctaga aataaacatg ctagctttta ttccagttct aaccaaaaaa ataaaccctc
4621 gttccacaga agctgccatc aagtatttcc tcacgcaagc aaccgcatcc ataatccttc
4681 taatagctat cctcttcaac aatatactct ccggacaatg aaccataacc aatactacca
4741 atcaatactc atcattaata atcataatag ctatagcaat aaaactagga atagccccct
4801 ttcacttctg agtcccagag gttacccaag gcacccctct gacatccggc ctgcttcttc
4861 tcacatgaca aaaactagcc cccatctcaa tcatatacca aatctctccc tcactaaacg
4921 taagccttct cctcactctc tcaatcttat ccatcatagc aggcagttga ggtggattaa
4981 accaAaccca gctacgcaaa atcttagcat actcctcaat tacccacata ggatgaataa
5041 tagcagttct accgtacaac cctaacataa ccattcttaa tttaactatt tatattatcc
5101 taactactac cgcattccta ctactcaact taaactccag caccacgacc ctactactat
5161 ctcgcacctg aaacaagcta acatgactaa cacccttaat tccatccacc ctcctctccc
5221 taggaggcct gcccccgcta accggctttt tgcccaaatg ggccattatc gaagaattca
5281 caaaaaacaa tagcctcatc atccccacca tcatagccac catcaccctc cttaacctct
5341 acttctacct acgcctaatc tactccacct caatcacact actccccata tctaacaacg
5401 taaaaataaa atgacagttt gaacatacaa aacccacccc attcctcccc acactcatcg
5461 cccttaccac gctactccta cctatctccc cttttatact aataatctta tagaaattta
5521 ggttaaatac agaccaagag ccttcaaagc cctcagtaag ttgcaatact taatttctgt
5581 aacagctaag gactgcaaaa ccccactctg catcaactga acgcaaatca gccactttaa
5641 ttaagctaag cccttactag accaatggga cttaaaccca caaacactta gttaacagct
5701 aagcacccta atcaactggc ttcaatctac ttctcccgcc gccgggaaaa aaggcgggag
5761 aagccccggc aggtttgaag ctgcttcttc gaatttgcaa ttcaatatga aaatcacctc
5821 ggagctggta aaaagaggcc taacccctgt ctttagattt acagtccaat gcttcactca
5881 gccattttac ctcaccccca ctgatgttcg ccgaccgttg actattctct acaaaccaca
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
s/lo
5941 aagacattgg aacactatac ctattattcg gcgcatgagc tggagtccta ggcacagctc
6001 taagcctcct tattcgagcc gagctgggcc agccaggcaa ccttctaggt aacgaccaca
6061 tctacaacgt tatcgtcaca gcccatgcat ttgtaataat cttcttcata gtaataccca
6121 tcataatcgg aggctttggc aactgactag ttcccctaat aatcggtgcc cccgatatgg
6181 cgtttccccg cataaacaac ataagcttct gactcttacc tccctctctc etactcctgc
6241 tcgcatctgc tatagtggag gccggagcag gaacaggttg aacagtctac cctcccttag
6301 cagggaacta ctcccaccct ggagcctccg tagacctaac catcttctcc ttacacctag
6361 caggtgtctc ctctatctta ggggccatca atttcatcac aacaattatc aatataaaac
6421 cccctgccat aacccaatac caaacgcccc tcttcgtctg atccgtccta atcacagcag
6481 tcctacttct cctatctctc ccagtcctag ctgctggcat cactatacta ctaacagacc
6541 gcaacctcaa caccaccttc ttcgaccccg ccggaggagg agaccccatt ctataccaac
6601 acctattctg atttttcggt caccctgaag tttatattct tatcctacca ggcttcggaa
6661 taatctccca tattgtaact tactactccg gaaaaaaaga accatttgga tacataggta
6721 tggtctgagc tatgatatca attggcttcc tagggtttat cgtgtgagca caccatatat
6781 ttacagtagg aatagacgta gacacacgag catatttcac ctccgctacc ataatcatcg
6841 ctatccccac cggcgtcaaa gtatttagct gactcgccac actccacgga agcaatatga
6901 aatgatctgc tgcagtgctc tgagccctag gattcatctt tcttttcacc gtaggtggcc
6961 tgactggcat tgtattagca aactcatcac tagacatcgt actacacgac acgtactacg
7021 ttgtagccca cttccactat gtcctatcaa taggagctgt atttgccatc ataggaggct
7081 tcattcactg atttccccta ttctcaggct acaccctaga ccaaacctac gccaaaatcc
7141 atttcactat catattcatc ggcgtaaatc taactttctt cccacaacac tttctcggcc
7201 tatccggaat gccccgacgt tactcggact accccgatgc atacaccaca tgaaacatcc
7261 tatcatctgt aggctcattc atttctctaa cagcagtaat attaataatt ttcatgattt
7321 gagaagcctt cgcttcgaag cgaaaagtcc taatagtaga agaaccctcc ataaacctgg
7381 agtgactata tggatgcccc ccaccctacc acacattcga agaacccgta tacataaaat
7441 ctagacaaaa aaggaaggaa tcgaaccccc caaagctggt ttcaagccaa ccccatggcc
7501 tccatgactt tttcaaaaag gtattagaaa aaccatttca taactttgtc aaagttaaat
7561 tataggctaa atcctatata tcttaatggc acatgcagcg caagtaggtc tacaagacgc
7621 tacttcccct atcatagaag agcttatcac ctttcatgat cacgccctca taatcatttt
7681 ccttatctgc ttcctagtcc tgtatgccct tttcctaaca ctcacaacaa aactaactaa
7741 tactaacatc tcagacgctc aggaaataga aaccgtctga actatcctgc ccgccatcat
7801 cctagtcctc atcgccctcc catccctacg catcctttac ataacagacg aggtcaacga
7861 tccctccctt accatcaaat caattggcca ccaatggtac tgaacctacg agtacaccga
7921 ctacggcgga ctaatcttca actcctacat acttccccca ttattcctag aaccaggcga ~~
7981 cctgcgactc cttgacgttg acaatcgagt agtactcccg attgaagccc ccattcgtat
8041 aataattaca tcacaagacg tcttgcactc atgagctgtc cccacattag gcttaaaaac
8101 agatgcaatt cccggacgtc taaaccaaac cactttcacc gctacacgac cgggggtata
8-161 ctacggtcaa tgctctgaaa tctgtggagc aaaccacagt ttcatgccca tcgtcctaga
8221 attaattccc _ctaaaaatct ttgaaatagg gcccgtattt accctatagc accccctcta
8281 ccccctctag agcccactgt aaagctaact tagcattaac cttttaagtt aaagattaag
8341 agaaccaaca cctctttaca gtgaaatgcc ccaactaaat actaccgtat ggcccaccat
8401 aattaccccc atactcctta cactattcct catcacccaa ctaaaaatat taaacacaaa
8461 ctaccaccta cctccctcac caaagcccat aaaaataaaa aattataaca aaccctgaga
8521 accaaaatga acgaaaatct gttcgcttca ttcattgccc cc'acaatcct aggcctaccc
8581 gccgcagtac tgatcattct atttccccct ctattgatcc ccacctccaa atatctcatc
8641 aacaaccgac taatcaccac ccaacaatga ctaatcaaac taacctcaaa acaaatgata
8701 accatacaca acactaaagg acgaacctga tctcttatac tagtatcctt aatcattttt
8761 attgccacaa ctaacctcct cggactcctg cctcactcat ttacaccaac cacccaacta
8821 tctataaacc tagccatggc catcccctta tgagcgggcA cagtgattat aggctttcgc
8881 tctaagatta aaaatgccct agcccacttc ttaccacaag gcacacctac accccttatc
8941 cccatactag ttattatcga aaccatcagc ctactcattc aaccaatagc cctggccgta
9001 cgcctaaccg ctaacattac tgcaggccac ctactcatgc acctaattgg aagcgccacc
9061 ctagcaatat caaccattaa ccttccctct acacttatca tcttcacaat tctaattcta
9121 ctgactatcc tagaaatcgc tgtcgcctta atccaagcct acgttttcac acttctagta
9181 agcctctacc tgcacgacaa cacataatga cccaccaatc acatgcctat catatagtaa
9241 aacccagccc atgaccccta acaggggccc tctcagccct cctaatgacc tccggcctag
9301 ccatgtgatt tcacttccac tccataacgc tcctcatact aggcctacta accaacacac
9361 taaccatata ccaatgatgg cgcgatgtaa cacgagaaag cacataccaa ggccaccaca
9421 caccacctgt ccaaaaaggc cttcgatacg ggataatcct atttattacc tcagaagttt
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
9/10
9481 ttttcttcgc aggatttttc tgagcctttt accactccag cctagcccct accccccaat
9541 taggagggca ctggccccCa acaggcatca ccccgctaaa tcccctagaa gtcccactcc
9601 taaacacatc cgtattactc gcatcaggag tatcaatcac ctgagctcac catagtctaa
9661 tagaaaacaa ccgaaaccaa ataattcaag cactgcttat tacaatttta ctgggtctct
9721 attttaccct cctacaagcc tcagagtact tcgagtctcc cttcaccatt tccgacggca
9781 tctacggctc aacatttttt gtagccacag gcttccacgg acttcacgtc attattggct
9841 caactttcct cactatctgc ttcatccgcc aactaatatt tcactttaca tccaaacatc
9901 actttggctt cgaagccgcc gcctgatact ggcattttgt agatgtggtt tgactatttc
9961 tgtatgtctc catctattga tgagggtctt actcttttag tataaatagt accgttaact
10021 tccaattaac tagttttgac aacattcaaa aaagagtaat aaacttcgcc ttaattttaa
10081 taatcaacac cctcctagcc ttactactaa taattattac attttgacta ccacaactca
10141 acggctacat agaaaaatcc accccttacg agtgcggctt cgaccctata tcccccgccc
10201 gcgtcccttt ctccataaaa ttcttcttag tagctattac cttcttatta tttgatctag
10261 aaattgccct ccttttaccc ctaccatgag ccctacaaac aactaacctg ccactaatag
10321 ttatgtcatc cctcttatta atcatcatcc tagccctaag tctggcctat gagtgactac
10381 aaaaaggatt agactgaacc gaattggtat atagtttaaa caaaacgaat gatttcgact
10441 cattaaatta tgataatcat atttaccaaa tgcccctcat ttacataaat attatactag
10501 catttaccat ctcacttcta ggaatactag tatatcgctc acacctcata tcctccctac
10561 tatgcctaga aggaataata ctatcgctgt tcattatagc tactctcata accctcaaca
10621 cccactccct cttagccaat attgtgccta ttgccatact agtctttgcc gcctgcgaag
10681 cagcggtggg cctagcccta ctagtctcaa tctccaacac atatggccta gactacgtac
10741 ataacctaaa cctactccaa tgctaaaact aatcgtccca acaattatat tactaceact
10801 gacatgactt tccaaaaaac acataatttg aatcaacaca accacccaca gcctaattat
10861 tagcatcatc cctctactat tttttaacca aatcaacaac aacctattta gctgttcccc
10921 aaccttttcc tccgaccccc taacaacccc cctcctaata ctaactacct gactcctacc
10981 cctcacaatc atggcaagcc aacgccactt atccagtgaa ccactatcac gaaaaaaact
11041 ctacctctct atactaatct ccctacaaat ctccttaatt ataacattca cagccacaga
11101 actaatcata ttttatatct tcttcgaaac cacacttatc cccaccttgg ctatcatcac
11161 ccgatgaggc aaccagccag aacgcctgaa cgcaggcaca tacttcctat tctacaccct
11221 agtaggctcc cttcccctac tcatcgcact aatttacact cacaacaccc taggctcact
11281 aaacattcta ctactcactc tcactgccca agaactatca aactcctgag ccaacaactt
11341 aatatgacta gcttacacaa tagcttttat agtaaagata cctctttacg gactccactt
11401 atgactccct aaagcccatg tcgaagcccc catcgctggg tcaatagtac ttgccgcagt
11461 actcttaaaa ctaggcggct atggtataat acgcctcaca ctcattctca accccctgac -~
11521 aaaacacata gcctacccct tccttgtact atccctatga ggcataatta taacaagctc
11581 catctgccta cgacaaacag acctaaaatc gctcattgca tactcttcaa tcagccacat
11641 agccctcgta gtaacagcca ttctcatcca aaccccctga agcttcaccg gcgcagtcat
11701 tctcataatc gcccacgggc ttacatcctc attactattc tgcctagcaa actcaaacta
11761 cgaacgcact cacagtcgca tcataatcct ctctcaagga cttcaaactc tactcccact
11821 aatagctttt tgatgacttc tagcaagcct cgctaacctc gccttacccc ccactattaa
11881 cctactggga gaactctctg tgctagtaac cacgttctcc tgatcaaata tcactctcct
11941 acttacagga ctcaacatac tagtcacagc cctatactcc ctctacatat ttaccacaac
12001 acaatggggc tcactcaccc accacattaa caacataaaa ccctcattca cacgagaaaa
12061 caccctcatg ttcatacacc tatcccccat tctcctccta tccctcaacc ccgacatcat
12121 taccgggttt tcctcttgta aatatagttt aaccaaaaca tcagattgtg aatctgacaa
12181 cagaggctta cgacccctta tttaccgaga aagctcacaa gaactgctaa ctcatgcccc
12241 catgtctaac aacatggctt tctcaacttt taaaggataa cagctatcca ttggtcttag
12301 gccccaaaaa ttttggtgca actccaaata aaagtaataa ccatgcacac tactataacc
12361 accctaaccc tgacttccct aattcccccc atccttacca ccctcgttaa ccctaacaaa
12421 aaaaactcat acccccatta tgtaaaatcc attgtcgcat ccacctttat tatcagtctc
12481 ttccccacaa caatattcat gtgcctagac caagaagtta ttatctcgaa ctgacactga
12541 gccacaaccc aaacaaccca gctctcccta agcttcaaac tagactactt ctccataata
12601 ttcatccctg tagcattgtt cgttacatgg tccatcatag aattctcact gtgatatata
12661 aactcagacc caaacattaa tcagttcttc aaatatctac tcatcttcct aattaccata
12721 ctaatcttag ttaccgctaa caacctattc caactgttca tcggctgaga gggcgtagga
12781 attatatcct tcttgctcat cagttgatga tacgcccgag cagatgccaa cacagcagcc
12841 attcaagcaa tcctatacaa ccgtatcggc gatatcggtt tcatcctcgc cttagcatga
12901 tttatcctac actccaactc atgagaccca caacaaatag cccttctaaa cgctaatcca
12961 agcctcaccc cactactagg cctcctccta gcagcagcag gcaaatcagc ccaattaggt
CA 02459127 2004-02-27
WO 03/018775 PCT/US02/28471
10/10
13021 ctccacccct gactcccctc agccatagaa ggccccaccc cagtctcagc cctactccac
13081 tcaagcacta tagttgtagc aggaatcttc ttactcatcc gcttccaccc cctagcagaa
13141 aatagcccac taatccaaac tctaacacta tgcttaggcg ctatcaccac tctgttcgca
13201 gcagtctgcg cccttacaca aaatgacatc aaaaaaatcg tagccttctc cacttcaagt
13261 caactaggac tcataatagt tacaatcggc atcaaccaac cacacctagc attcctgcac
13321 atctgtaccc acgccttctt caaagccata ctatttatgt gctccgggtc catcatccac
13381 aaccttaaca atgaacaaga tattcgaaaa ataggaggac tactcaaaac catacctctc
13441 acttcaacct ccctcaccat tggcagccta gcattagcag gaataccttt cctcacaggt
13501 ttctactcca aagaccacat catcgaaacc gcaaacatat catacacaaa cgcctgagcc
13561 ctatctatta ctctcatcgc tacctccctg acaagcgcct atagcactcg aataattctt
13621 ctcaccctaa caggtcaacc tcgcttcccc acccttacta acattaacga aaataacccc
13681 accctactaa accccattaa acgcctggca gccggaagcc tattcgcagg atttctcatt
13741 actaacaaca tttcccccgc atcccccttc caaacaacaa tccccctcta cctaaaactc
13801 acagccctcg ctgtcacttt cctaggactt ctaacagccc tagacctcaa ctacctaacc
13861 aacaaactta aaataaaatc cccactatgc acattttatt tctccaacat actcggattc
13921 taccctagca tcacacaccg cacaatcccc tatctaggcc ttcttacgag ccaaaacctg
13981 cccctactcc tcctagacct aacctgacta gaaaagctat tacctaaaac aatttcacag
14041 caccaaatct ccacctccat catcacctca acccaaaaag gcataattaa actttacttc
14101 ctctctttct tcttcccact catcctaacc ctactcctaa tcacataacc tattcccccg
14161 agcaatctca attacaatat atacaccaac aaacaatgtt caaccagtaa ctactactaa
14221 tcaacgccca taatcataca aagcccccgc accaatagga tcctcccgaa tCaaccctga
14281 cccctctcct tcataaatta ttcagcttcc tacactatta aagtttacca caaccaccac
14341 cccatcatac tctttcaccc acagcaccaa tcctacctcc atcgctaacc ccactaaaac
14401 actcaccaag acctcaaccc ctgaccccca tgcctcagga tactcctcaa tagccatcgc
14461 tgtagtatat ccaaagacaa ccatcattcc ccctaaataa attaaaaaaa ctattaaacc
14521 catataacct cccccaaaat tcagaataat aacacacccg accacaccgc taacaatcaa
14581 tactaaaccc ccataaatag gagaaggctt agaagaaaac cccacaaacc ccattactaa
14641 acccacactc aacagaaaca aagcatacat cattattctc gcacggacta caaccacgac
14701 caatgatatg aaaaaccatc gttgtatttc aactacaaga acaccaatga ccccaatacg
14761 caaaactaac cccctaataa aattaattaa ccactcattc atcgacctcc ccaccccatc
14821 caacatctcc gcatgatgaa acttcggctc actccttggc gcctgcctga tcctccaaat
14881 caccacagga ctattcctag ccatgcacta ctcaccagac gcctcaaccg ccttttcatc
14941 aatcgcccac atcactcgag acgtaaatta tggctgaatc atccgctacc ttcacgccaa
15001 tggcgcctca atattcttta tctgcctctt cctacacatc gggcgaggcc tatattacgg ~~
15061 atcatttctc tactcagaaa cctgaaacat cggcattatc ctcctgcttg caactatagc
15121 aacagccttc ataggctatg tcctcccgtg aggccaaata tcattctgag gggccacagt
15181 aattacaaac ttactatccg ccatcccata cattgggaca gacctagttc aatgaatctg
15241 aggaggctac tcagtagaca gtcccaccct cacacgattc tttacctttc acttcatctt
15301 gcccttcatt attgcagccc tagcaacact ccacctccta ttcttgcacg aaacgggatc
15361 aaacaacccc ctaggaatca cctcccattc cgataaaatc accttccacc cttactacac
15421 aatcaaagac gccctcggct tacttctctt ccttctctcc ttaatgacat taacactatt
15481 ctcaccagac ctcctaggcg acccagacaa ttatacccta gccaacccct taaacacccc
15541 tccccacatc aagcccgaat gatatttcct attcgcctac acaattctcc gatccgtccc
15601 taacaaacta ggaggcgtcc ttgccctatt actatccatc ctcatcctag caataatccc
15661 catcctccat atatccaaac aacaaagcat aatatttcgc ccactaagcc aatcacttta
15721 ttgactccta gccgcagacc tcctcattct aacctgaatc ggaggacaac cagtaagcta
15?81 cccttttacc atcattggac aagtagcatc cgtactatac ttcacaacaa tcctaatcct
15841 aataccaact atctccctaa ttgaaaacaa aatactcaaa tgggcctgtc cttgtagtat
15901 aaactaatac accagtcttg taaaccggag atgaaaacct ttttccaagg acaaatcaga
15961 gaaaaagtct ttaactccac cattagcacc caaagctaag attctaattt aaactattct
16021 ctgttctttc atggggaagc agatttgggt accacccaag tattgactca cccatcaaca
16081 accgctatgt atttcgtaca ttactgccag ccaccatgaa tattgtacgg taccataaat
16141 acttgaccac ctgtagtaca taaaaaccca atccacatca aaaccccctc cccatgctta
16201 caagcaagta cagcaatcaa ccctcaacta tcacacatca actgcaactc caaagccacc
16261 cctcacccac taggatacca acaaacctac ccacccttaa cagtacatag tacataaagc
16321 catttaccgt acatagcaca ttacagtcaa atcccttctc gtccccatgg atgacccccc
16381 tcagataggg gtcccttgac caccatcctc cgtgaaatca atatcccgca caagagtgct
16441 actctcctcg ctccgggccc ataacacttg ggggtagcta aagtgaactg tatccgacat
16501 ctggttccta cttcagggtc ataaagccta aatagcccac acgttcccct taaataagac
16561 atcacgatg