Note: Descriptions are shown in the official language in which they were submitted.
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
NUCLEAR-ENVELOPE AND NUCLEAR-LAMINA BINDING CHIMERAS FOR
MODULATING GENE EXPRESSION
Field of the Invention
The present invention is directed to nucleic acid target-specific chimeric
proteins
comprising a nuclear-envelope and/or nuclear-lamina binding domain and a DNA
binding
domain. These proteins, as well as the nucleic acids encoding those proteins,
can be used in
methods to modulate gene expression and are particularly useful to repress or
down-regulate
expression of selected target genes. The DNA binding domains are preferably
from naturally-
occuring zinc finger proteins (ZFPs) or artificial zinc finger proteins
(AZPs). The invention
also relates to molecular switch systems for gene repression and derepression.
Background of the Invention
Transcriptional repression of genes can be achieved by a variety of
mechanisms. A
classic example is the lac repressor which, when bound to its target sequence
on the lac
operon, prevents RNA polymerise from binding and thereby initiating
transcription. In
eukaryotes, additional mechanisms exist to control gene repression. For
example, genes
found in constitutive heterochromatin are transcriptionally silent.
Heterochromatin is not
positioned randomly and appears to be associated with the nuclear periphery
[Cohen et al.
(2001) Trends Biochem. Sci. 26:41-47], suggesting that bringing genes into
proximity with
heterochromatin or the nuclear periphery may play a role, at least in part, in
gene silencing.
Transcriptional repressors are also found at the nuclear periphery in
eukaryotes. In
some cases, it appears that such proteins are only active as repressors when
localized to the
nuclear periphery. The nuclear periphery of higher eukaryotes (metazoans and
above)
consists of a nuclear envelope (NE) with inner and outer membranes and a
nuclear lamina.
The nuclear lamina resides underneath the inner nuclear membrane and is
composed of
intermediate filaments termed lamins and lamina-associated proteins (LAPs).
Certain LAPs
are also integral membrane proteins of the inner nuclear membrane. A
discussion of the
composition of the nuclear lamina from several different species is provided
in Cohen et al.
Oct-1 is a repressor of the aging-associated collagenase gene. Experimental
evidence
shows that dissociation of Oct-1 from the nuclear periphery induces
collagenase gene
-1-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
expression [Imai et al. (1997) Mol. Biol. Cell 8:2404-2419]. Furthermore, when
the active
form of the retinoblastoma protein (Rb) is associated with the transcription
factor E2F, the
complex co-localizes with lamins A/C at the nuclear periphery in vivo and
represses
transcription [Mancini et al. (1999) Dev. Biol. 215:288-297]. The mouse germ-
cell-less
protein (GCL), also involved in gene repression, [Nili et al. (2001) J. Cell.
Sci. 114:3297-
3307], has been reported to bind LAP2~ at the nuclear lamina (Cohen et al.).
Transcription factors and other DNA binding proteins bind their targets in a
sequence
specific manner to modulate gene expression and thereby activate or repress
expression of the
target gene. Modulation of gene expression can be achieved temporally (e.g.,
at different
times in development or during the cell cycle) and/or spatially (e.g., in
different tissues). In
some instances, it may be desirable to turn off expression of undesired genes
at particular
times or in particular cell types. For example, genes that become associated
with and
activated in oncogenesis may be targets for repression. Since heterochromatin
and genes
localized to the nuclear periphery are known to be silenced, a sequence-
specific method to
bring a gene into association with the nuclear periphery could provide a route
to silence or
down regulate (repress) expression of that target gene. Alternatively, a
method to release
genes from a state of repression (i.e., to derepress or activate those genes)
would also be
valuable.
However, known transcription factors have limited utility --such proteins are
useful to
control genes associated with their natural target sequences or to a limited
set of closely
related target sequences. One way to overcome this drawback is to design and
construct
DNA binding proteins with predetermined sequence specificity, particularly for
unique target
sequences in a large, complex genome. One particular class of proteins shown
amenable to
such manipulation is zinc finger proteins (ZFPs)
ZFPs are well-known DNA-binding proteins that recognize and bind to DNA target
sequences by interaction of the target sequence with particular amino acids in
the alpha helix
of each zinc finger in the ZFP. ZFPs typically contain from three to nine, and
sometimes
more, zinc fingers and there are many classes of ZFPs; for a review, see,
e.g., Laity et al.
(2001) Curr. Opin. Struct. Biol. 11:39-46. The Cys2Hisz class of ZFPs has been
extensively
studied and proved particularly useful in development of a universal
recognition code to
permit the design of artificial zinc finger proteins (AZPs) that bind
predetermined DNA target
-2-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
sequences. See, e.g., Wolfe et al. (2000) Ann. Rev. Biophys. Biomol. Struct.
29:183-212;
Choo et al. (2000) Curr. Opin. Struct. Biol. 10:411-416; Segal et al. (1999)
Proc. Natl. Acad.
Sci. USA 96:2758-2763; Kim et al. (1998) Proc. Natl. Acad. Sci. USA 95:2812-
2817; and
U.S. Serial No.09/911,261 to Takashi Sera, filed July 23, 2001 and entitled
"Zinc Finger
Domain Recognition Code and Uses Thereof."
The availability of AZPs enables the design of proteins that can regulate
target genes
associated with any unique sequence not just known regulatory sequences. When
these AZPs
(or other DNA binding proteins) are combined with one or more protein domains
capable of
associating with the nuclear periphery, a chimeric protein is created which
can be used to bind
a nucleotide sequence associated with a target gene and localize that target
gene to the nuclear
periphery for silencing or down regulation. When the domains of these chimeric
proteins are
rearranged into a molecular switch system, it is possible to provide systems
for either
activation or repression of gene expression.
Summary of the Invention
The present invention relates to nucleic acid target-specific, chimeric
proteins having
one or more first domains capable of specifically binding a nucleotide
sequence associated
with a target gene and having one or more second domains capable of
associating with or
binding to the nuclear periphery. These proteins are useful in regulating gene
expression.
Multiple first and second domains, preferably from one to five additional
domains can also be
present in the chimeric proteins of the invention. The preferred first domain
is an AZP and
the preferred second domain is a GCL protein. In certain embodiments the
chimeric proteins
can include additional domains to facilitate cellular uptake and/or transport
to the nucleus.
Other aspects of the invention provide isolated nucleic acids encoding the
chimeric
proteins of the invention, expression vectors comprising those nucleic acids,
and host cells
transformed (by any method) with the expression vectors. Such host cells can
be used, e.g.,
in a method of preparing the chimeric protein by culturing the host cell for a
time and under
conditions to express the chimeric protein and recovering the chimeric
protein. In addition
the host cells can be used as a source of expression vectors to deliver the
chimeric protein by
gene transfer methods into a cell or an organism. In addition, the invention
provides
pharmaceutical compositions of these chimeric proteins, nucleic acids and
expression vectors.
-3-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
A still further aspect of the invention relates to a method of binding a
target nucleic
acid with chimeric protein of the invention by contacting the target nucleic
acid (having a
nucleotide sequence associated with the target gene) with a chimeric protein
of the invention
in an amount and for a time sufficient for that protein to bind to the target
nucleic acid. In a
preferred embodiment the chimeric protein is introduced into a cell via a
nucleic acid for in
vivo binding. Alternatively, the method provides the chimeric protein can be
used for an in
vitro binding assay.
A yet further aspect of the invention provides a method of repressing or down
regulating expression of a target gene which comprises contacting a nucleic
acid containing a
nucleotide sequence associated with or in sufficient proximity to the target
gene with a
chimeric protein of the invention in an amount and for a time sufficient for
the protein to
decrease the expression level of the target gene relative to an appropriate
control. In certain
embodiments, the chimeric protein is introduced into a cell or an organism as
a protein or as a
nucleic acid encoding the chimeric protein.
In the contemplated method of binding a target nucleic acid or the
contemplated
methods of repressing gene expression, the target gene encodes, or the
targeted nucleotide
sequence site is from or controls, a plant gene, a mammalian gene, an insect
gene, a yeast
gene or is from a virus such as a DNA virus. When the target gene or site is
from a mammal,
it can encode or control a cytokine, an interleukin, an oncogene, an anti-
angiogenesis factor, a
drug resistance gene andlor any other desired target which allows a selected
gene to be
brought into proximity with the nuclear periphery and thereby silenced or down
regulated.
Plant genes of interest include, but are not limited to, genes from tomato,
corn, rice and cereal
plants. Moreover, multiple target genes that share a common nucleotide target
sequence can
be coordinately or simultaneously controlled.
A still further aspect of the invention relates to molecular switch systems
useful for
gene repression. These systems comprise (a) a first fusion protein with a
first domain capable
of specifically binding a nucleotide sequence associated with a target gene,
and a second
domain capable of specifically binding to a first binding moiety of a divalent
ligand, where
the ligand is capable of uptake by a cell, and the first domain and second
domains are
heterologous with respect to each other; and (b) a second fusion comprising a
first domain
capable of associating with the nuclear periphery and a second domain capable
of specifically
-4-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
binding to the second binding moiety of the divalent ligand. The first domain
of the first
fusion protein is the same as the first domain of the chimeric proteins of the
invention; and
the first domain of the second fusion protein is the same as the second domain
of the chimeric
proteins of the invention. The second domains of the two fusion proteins can
be a single
chain variable region (scFv) of an antibody with specificity for its
respective binding moiety
of the divalent ligand.
Other aspects of the invention provide isolated nucleic acids encoding the
fusion
proteins for gene repression of the invention, expression vectors comprising
those nucleic
acids, and host cells transformed (by any method) with the expression vectors.
Such host cells
can be used in a method of preparing the fusion proteins by culturing the host
cell for a time
and under conditions to express the fusion proteins and recovering the fusion
proteins. In
addition the host cells can be used as a source of expression vectors to
deliver the fusion
proteins by gene transfer methods into a cell or an organism. In addition, the
invention
provides pharmaceutical compositions of these fusions proteins, the molecular
switch
systems, nucleic acids and expression vectors.
The molecular switch useful for gene repression can be used in a method of
temporally or spatially repressing expression of a target gene by (a)
contacting a cell or an
organism containing a target nucleic acid having a nucleotide sequence
associated with a
target gene with these molecular switch systems, and (b) contacting the cell
or organism with
the divalent ligand of the molecular switch system at a time or in a location
to allow
formation of a complex between the fusion proteins to thereby repress
expression of the said
target gene by localizing the target gene to the nuclear periphery. The fusion
proteins of this
molecular switch system can be introduced into the cell or organism as
proteins, as one or
more nucleic acids encoding one or more of those proteins, or as a combination
thereof.
Yet another aspect of the invention relates to molecular switch systems useful
for gene
derepression, i.e., activation of repressed genes. These systems comprise (a)
a first fusion
protein comprising a first domain capable of specifically binding a nucleotide
sequence
associated with a target gene, and a second domain capable of specifically
binding to a
binding partner, where the first and second domains are heterologous with
respect to each
other; and (b) a second fusion protein comprising a first domain capable of
associating with
the nuclear periphery and a second domain comprising the binding partner of
the second
-5-
' CA 02472729 2004-07-08
domain of said first fusion protein, wherein said first domain is heterologous
with respect to
said second domain. The first domain of the first fusion protein is the same
as the first
a domain of the chimeric proteins of the invention; and the first domain of
the second fusion
protein is the same as the second domain of the chimeric proteins of the
invention: The
second domain of the first fusion protein can be an S-protein and the second
domain of said
second fusion protein can be an S-tag, or vice-a-versa.
Other aspects of the invention provide isolated nucleic acids encoding these
fusion
proteins for gene derepression of the invention, expression vectors comprising
those nucleic
acids, and host cells transformed (by any method) with the expression vectors:
Such host cells
:3
can be used in a method of preparing the fusion proteins by culturing the host
cell for a time
and under conditions to express the fusion proteins and recovering the fusion
proteins. In
' addition the host cells can be used as a source of expression vectors to
deliver the fusion
proteins by gene transfer methods into a cell or an organism. In addition, the
invention
provides pharmaceutical compositions of these fasions proteins, the molecular
switch
systems, nucleic acids and expression vectors
The molecular switch useful for gene derepression can be used in a method of
temporally or spatially altering expression of a target gene by (a) contacting
a cell or an
organism containing a target nucleic acid having a nucleotide sequence
associated with a
target gene with these molecular switch systems, and (b) contacting the cell
or organism with
a ligand of the molecular switch system at a time or in a location to disrupt
association of the
first and second fusion proteins and thereby derepress expression o~ said
target gene.
by releasing the target gene from its association with the nuclear periphery.
The fusion
_.
v proteins of this molecular switch system can be introduced into the cell or
organism as
v_
proteins, as one or more nucleic acids encoding one or more of those proteins,
or as a
combination thereof.
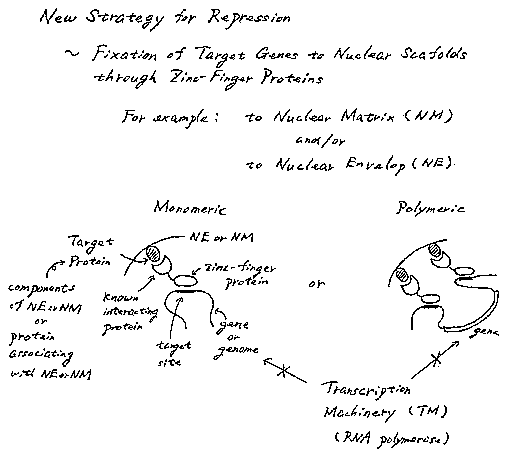
Brief Description of the DraWIIIQS
Fig.l schematically illustrates monomeric and polymeric gene repression using
chimeric proteins of the invention to bring one or more target genes into
proximity to the
nuclear periphery. Cross hatch: target protein in or associated with nuclear
periphery; open:
known-interacting protein; stippled: AZP or ZFP; thick black line: AZP or ZFP
target site;
thin black line: gene or genome; NE: nuclear envelope; NM: nuclear membrane.
-6-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
Detailed Description of the Invention
A. Chimeric Proteins of the Invention
The present invention relates to target-specific, chimeric proteins for
repressing gene
expression by bringing a target gene into proximity to the nuclear periphery
and thereby
silence or down regulate expression of that gene. The chimeric proteins
comprise at least two
heterologous domains: a first domain capable of specifically binding a
nucleotide sequence
associated with the target gene, and a second domain capable of associating
with the nuclear
periphery by binding to or associating with proteins at or in the nuclear
envelope, nuclear
lamina, heterochromatin or any combination of the three. The chimeric proteins
of the
invention are useful in regulating gene expression, particularly to repress or
down regulate
expression of the selected gene. For example, it may be desirable to down
regulate or shut off
genes involved in oncogenesis, cellular proliferation and regeneration,
angiogenesis (when
unwanted blood vessel formation occurs such as in tumors), or in plants at
particular stages of
development or growth. Similarly, the chimeric proteins of the invention can
be used to
down regulate or shut off viral genes.
As used herein, the term "nuclear periphery" includes the nuclear envelope and
the
nuclear lamina. A gene in proximity to the nuclear periphery is physically
adjacent to the
nuclear periphery and, in accordance with the invention, is so positioned by
forming an
association (covalently or non-covalently) with proteins that bind to or form
part of the
nuclear envelope, the nuclear lamina or heterochromatin associated with the
nuclear envelope
or nuclear lamina. For purposes of the present invention, it is not necessary
to determine the
actual physical location of a gene relative to the nuclear periphery, but
rather, one can
measure and use the reduction in gene expression relative to the normal
expression level, or
other control level of expression, to assess whether the gene is at or in
proximity to the
nuclear periphery.
As used herein, the term "chimeric protein" or "chimeric proteins" is used to
denote
that the proteins of the invention are non-naturally occurring proteins. The
chimeric proteins
of the invention are artificial constructs combining a nucleic acid binding
domain and a
domain capable of associating with the nuclear periphery from different
sources, i.e., the two
domains are heterologous with respect to each other. When multiple domains are
present, it
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
is sufficient that only one nucleic acid binding domain be from a source
different from the
domain capable of associating with the nuclear periphery. The sources of the
heterologous
domains can be, independently, from different species, from different strains
of an organism,
from different proteins of a single organism or from artificial proteins
designed to have the
desired activity, provided that none of the combinations are such to produce a
naturally-
occurring protein.
The nucleic acid binding domain of the chimeric protein specifically binds to
a
nucleotide sequence associated with the target gene. The identity and
characteristics of that
domain is determined by the nucleotide sequence desired to be bound by the
chimeric protein.
As used herein, "specifically binds" means, and includes reference to, the
binding or
association of a DNA binding moiety or protein (for example, as a whole
protein, as a
domain, or as present in a chimeric protein of the invention) to a specified
nucleotide
sequence to a detectably greater degree (e.g., at least 1.5-fold over
background) than its
binding to other nucleotide sequences and to the substantial exclusion of
other nucleotide
sequences under a particular set of conditions, such, e.g., as temperature,
ionic strength,
solvent polarity and the like. The gel shift assay, well known in the art, is
one method useful
to assess and verify whether the binding is specific for a particular
nucleotide sequence.
It is possible to control the nature and position of the nucleotide sequence
relative to
the target gene. As used herein, a "target polynucleotide," "nucleotide
sequence associated
with a target gene," or "targeted nucleotide sequence," or other similar
terminology refers to a
portion of a double-stranded polynucleotide, preferably DNA, to which the DNA
binding
domain of the chimeric proteins binds. This targeted nucleotide sequence may
be at any
location, near or within the target gene to be regulated, provided that
location is suitable for
repressing expression of that target gene. For example, the targeted
nucleotide sequence can
be within the coding region, immediately upstream or downstream thereof or it
can be some
distance away (e.g., several hundred nucleotides) if the selected nucleotide
sequence still
allows the gene to be brought into sufficient proximity to the nuclear
periphery to reduce
expression of that gene from its normal or other control level. The targeted
nucleotide
sequence can also be all or part of a known transcriptional control element
for a target gene.
The length of the targeted nucleotide sequence can range from about 6-10
nucleotides
to about 50, 60, 70 or more nucleotides. Examples of suitable nucleotide
sequence lengths
_g_
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
are about 8 to about 30, about 10 to about 25, and about 10 to about 20
nucleotides. A length
of about 16 nucleotides is sufficient to provide a unique target site in the
human genome. The
specificity and affinity of the DNA binding domain, the organism being
targeted and the
nature of the sequence can all be factors in determining the appropriate
length of the targeted
nucleotide sequence. Those of skill in the art can readily determine the
length and identity of
the targeted nucleotide sequence based on such considerations.
The nucleic acid binding domain of the chimeric protein can be a known or
artificial
DNA binding protein or a fragment thereof with DNA binding activity. Examples
of DNA
binding proteins include, but are not limited to, zinc finger proteins (ZFPs),
artificial zinc .
finger proteins (AZPs), the DNA binding moiety of a transcription factor,
nuclear hormone
receptors, homeobox domain proteins such as engrailed or antenopedia, helix-
turn-helix motif
proteins such as lambda repressor and tet repressor, Gal4, TATA binding
protein, helix-loop-
helix motif proteins such as myc and myoD, leucine zipper type proteins such
as fos and jun,
and beta-sheet motif proteins such as met, arc, and mnt repressors, or the DNA
binding
moiety of any of those proteins. Such proteins and moieties are known to those
of skill in the
art.
The preferred DNA binding proteins for the nucleic acid binding domains of the
invention are ZFPs and AZPs. There are many classes of ZFPs, including but not
limited to,
Cys2His2 class (examples, SpIC and Zif 268), Cys6 (example, the Gal4 DNA
binding protein)
and Cys4 (example, estrogen hormone receptor); any of these proteins with the
desired
nucleotide sequence specificity can be used.
By "zinc finger protein", "zinc finger polypeptide," "ZFP," "artificial zinc
finger
protein" or "AZP" is meant a polypeptide having DNA binding domains that are
stabilized by
zinc. The individual DNA binding domains are typically referred to as
"fingers," such that a
ZFP or peptide has at least one finger, more typically two fingers, more
preferably three
fingers, or even more preferably four or five fingers, to at least six or more
fingers. Each
finger binds three or four base pairs of DNA. In the Cyst-His2 class of ZFPs
and AZPs, each
finger is typically an approximately 30 amino acid, zinc-chelating, DNA-
binding moiety
domain. A representative sequence motif for the Cyst-His2 class is -Cys-(X)2_4-
Cys-(X)12-
His-(X)3_5-His, where X is any amino acid (SEQ ID NO: 1). The two invariant
histidine
-9-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
residues and the two invariant cysteine residues bind a zinc cation [see,
e.g., Berg et al.
(1996) Science 271:1081-1085].
In one embodiment of the invention, the chimeric protein has a first domain
which is
an AZP comprising at least one zinc finger, each finger represented by the
formula
-X3-Cys-X2_4-Cys-XS-Z 1-X-Z2-Z3-X2-Z6-His-X3_5-His-X4-, (SEQ )D NO: 2)
and where multiple fingers, when present, are independently covalently joined
to each
other with from 0 to 10 amino acid residues, wherein X is any amino acid, X~
represents the
number of occurrences of X in the polypeptide chain and Z-~, Z2, Z3, and Z6
are determined by
a recognition code shown in Tables 1 and 2 (and as further explained below).
The amino acids represented by X form the framework of a CysZHis2 zinc finger
and
can be a known zinc finger framework, a consensus framework, a framework
obtained by
varying the sequence of any of these frameworks or any artificial framework.
Preferably
known frameworks are used to determine the identities of each X. In certain
embodiments,
the framework for determining X is that from Spl, SpIC or Zif268. In a
preferred
embodiment, the framework has the sequence of SpIC domain 2 (i.e., the middle
finger of
SplC) , which sequence is -Pro-Tyr-Lys-Cys-Pro-Glu-Cys-Gly-Lys-Ser-Phe-Ser-Z-1-
Ser- Z2-
Z3-Leu-Gln- Z6-His-Gln-Arg-Thr-His-Thr-Gly-Glu-Lys- (SEQ >D NO: 3). Such AZPs
are
more fully described in U.S. Serial No. 09/911,261 to Takashi Sera, filed July
23, 2001.
The AZPs of the invention can comprise from 3 to 40 zinc fingers, from 3 to 15
fingers, 3 to 12 fingers, 3 to 9 fingers or 3 to 6 fingers, as well as ZFPs
with 3, 4, 5, 6, 7, 8 or
9 fingers.
The four nucleic acid-contacting residues of the zinc finger, designated as Z-
1, Z2, Z3
and Z6 in the above formula, are primarily responsible for determining
specificity and affinity
of DNA binding. These four amino acid residues may also be referred to as the
base-
contacting amino acids. These four residues occur in the same position
relative to the first
consensus histidine and second consensus cysteine. The first residue is seven
residues to the
N-terminal side of the first consensus histidine and six residues to the C-
terminal side of the
second consensus cysteine. The first residue is also referred to as the "-1
position" and is so
designated because it represents the residue immediately adjacent to the N-
terminus of the a-
helix in the zinc finger (with position 1 thus being the first N-terminal
residue of the a-helix).
The other three amino acids occur at positions two, three and six of the a-
helix, and are
-10-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
referred to as the "2 position", "3 position" and "6 position", respectively.
These four
positions are interchangeably referred to herein as the Z-1, Z2, Z3 and Z6
positions.
The recognition code table provides a method to determine the identify of Z-l,
Z2, Z3
and Z6 for a given nucleotide sequence. In the recognition code table (and for
each 4 base-
pair portion of a nucleotide sequence), the bases are always provided in 5' to
3' order. The
fourth base, however, is always the complement of the fourth base provided in
the target
sequence. For example, if the target sequence is written as ATCC, then it
means a sense
strand target sequence of 5'-ATCC-3' and an antisense strand of 3'-TAGG-5'.
Thus, when
the sense strand sequence ATCC is translated to amino acids from Table 1
below, the first
base of A means there is glutamine at position 6, the second base of T means
there is serine at
position 3 and the third base of C means there is glutamic acid at position -
1. However, with
the fourth base written as C, it means that the complement of C, i.e., G, is
found in the table
and used to identify the amino acid of position 2. In this case, the amino
acid at position two
is serine.
Tables 1 and 2 provide the preferred and alternative recognition code tables
for the
AZPs that are useful in the invention, respectively, in summary format:
Table 1
ls' base 2 base 3' base 4' base
G Arg His Arg Ser
A Gln Asn Gln Asn
T Thr, Tyr, Ser Thr, Met Thr
Leu
C Glu Asp Glu Asp
Position Position Position Position
6 3 -1 2
-11-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
Table 2
ls' base 2" base 3' base 4~ base
G Arg, Lys His, Lys Arg, Lys Ser, Arg
A Gln, Asn Asn, Gln Gln, Asn Asn, Gln
T Thr, Tyr, Ser, Ala, Thr, Met, Thr, Val,
Leu, Val, Leu, Ala
lle, Met Thr lle
C Glu, Asp Asp, Glu Glu, Asp Asp, Glu
Position Position Position Position
6 3 -1 2
In Table 2, the order of amino acids listed in each box represents, from left
to right, the most
preferred to least preferred amino acid at that position.
These recognition code tables can also be described as follows below. The
preferred
recognition code table for the AZPs (equivalent to Table 1) is, for each four
base target
sequence, in 5' to 3' order:
(i) if the first base is G, then Z6 is arginine,
if the first base is A, then Z6 is glutamine,
if the first base is T, then Z6 is threonine, tyrosine or leucine,
if the first base is C, then Z6 is glutamic acid,
(ii) if the second base is G, then Z3 is histidine,
if the second base is A, then Z3 is asparagine,
if the second base is T, then Z3 is serine,
if the second base is C, then Z3 is aspartic acid,
(iii) if the third base is G, then Z-1 is arginine,
if the third base is A, then Z-t is glutamine,
if the third base is T, then Z-1 is threonine or methionine,
if the third base is C, then Z-1 is glutamic acid,
(iv) if the complement of the fourth base is G, then Z2 is serine,
if the complement of the fourth base is A, then Z2 is asparagine,
if the complement of the fourth base is T, then Z2 is threonine, and
if the complement of the fourth base is C, then Z2 is aspartic acid.
-12-
-11-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
In a preferred embodiment of the above recognition code (i.e., the Table 1
recognition code),
if the first base is T, then Z6 is threonine; and if the third base is T, then
Z-~ is threonine
(Table 1 ).
The alternative recognition code table (equivalent to Table 2) can also be
presented as
follows:
(i) if the first base is G, then Z6 is arginine or lysine,
if the first base is A, then Z6 is glutamine or asparagine,
if the first base is T, then Z6 is threonine, tyrosine, leucine, isoleucine
or methionine,
if the first base is C, then Z6 is glutamic acid or aspartic acid,
(ii) if the second base is G, then Z3 is histidine or lysine,
if the second base is A, then Z3 is asparagine or glutamine,
if the second base is T, then Z3 is serine, alanine, valine or threonine
if the second base is C, then Z3 is aspartic acid or glutamic acid,
(iii) if the third base is G, then Z-1 is arginine or lysine,
if the third base is A, then Z-' is glutamine or asparagine,
if the third base is T, then Z-~ is threonine, methionine leucine or
isoleucine,
if the third base is C, then Z-~ is glutamic acid or aspartic acid,
(iv) if the complement of the fourth base is G, then ZZ is serine or
arginine,
if the complement of the fourth base is A, then Z2 is asparagine or
glutamine,
if the complement of the fourth base is T, then Z2 is threonine, valine
or alanine, and
if the complement of the fourth base is C, then Z2 is aspartic acid or
glutamic acid.
To use the recognition code table to design and identify an AZP for a given
nucleotide
sequence, a nucleotide sequence of length 3N+1 base pairs, wherein N is the
number of
overlapping 4 base pair segments in the target, is divided into overlapping 4
base pair
segments, where the fourth base of each segment, up to the N-1 segment, is the
first base of
-13-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
the immediately following segment. The identities of each Z-~, Z2, Z3 and Z6
in the zinc
finger are then determined according to the recognition code table.
Zinc fingers designed in accordance with this invention are either covalently
joined
directly to one another or can be separated by a linker of from 1-10 amino
acids. The linker
amino acids can provide flexibility or some degree of structural rigidity. The
choice of linker
can be, but is not necessarily, dictated by the desired affinity of the ZFP
for its cognate
nucleotide sequence. It is within the skill of the art to test and optimize
various linker
sequences to improve the binding affinity of the AZP for its cognate target
sequence. For
example, one useful arrangement for a six finger ZFP, is to have the first
three zinc fingers be
joined without amino acid linkers, a flexible amino acid linker between the
third and fourth
fingers and the last three fingers joined without amino acid linkers. This
arrangement appears
to allow each three finger group to independently bind its target sequence
while minimizing
steric hindrance for the binding of the other three finger group.
In an embodiment, longer genomic sequences are targeted using mufti-finger
AZPs
linked to other mufti-fingered AZPs using flexible linkers including, but not
limited to,
GGGGS, GGGS and GGS (these sequences can be part of the 1-10 additional amino
acids in
the AZPs; SEQ ID NO: 4, residues 2-5 of SEQ ID NO: 4; and residues 3-5 of SEQ
117 NO: 4,
respectively).
In addition, the nucleic acid binding domain of the chimeric proteins of the
invention
can be designed to bind to non-contiguous nucleotide sequences, either using a
single domain
or multiple domains. For example, the nucleotide sequence bound by a six-
finger AZP can be
a ten base pair sequence (recognized by three fingers) with intervening bases
(that do not
contact the zinc fingers) and a second ten base pair sequence (recognized by
the other three
fingers). The number of intervening bases can vary, such that one can
compensate for this
intervening distance with an appropriately designed amino acid linker between
the two three-
finger parts of the AZP. A range of intervening nucleic acid bases in a target
binding site can
be from 5-100, and preferably from 10-20 or less bases, more preferably 10 or
less, and even
more preferably 6 or less bases. Of course, the linker maintains the reading
frame between
the linked parts of the AZP.
Methods of designing and constructing nucleic acids encoding ZFPs and AZPs by
phage display, random mutagenesis, combinatorial libraries, computer/rational
design,
-14-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
affinity selection, PCR, cloning from cDNA or genomic libraries, synthetic
construction and
the like are known. (see, e.g., U.S. Pat. No. 5,786,538; Wu et al., Proc.
Natl. Acad. Sci. USA
92:344-348 (1995); Jamieson et al., Biochemistry 33:5689- 5695 (1994); Rebar &
Pabo,
Science 263:671-673 (1994); Choo & Klug, Proc. Natl. Acad. Sci. USA 91: 11168-
11172
(1994); Desjarlais et al., Proc. Natl. Acad. Sci. USA 89:7345-5349 (1992);
Desjarlais et al.,
Proc. Natl. Acad. Sci. USA 90:2256-2260 (1993); Desjarlais et al., Proc. Natl.
Acad. Sci.
USA 91:11099-11103; Pomerantz et al., Science 267:93-96 (1995); Pomerantz et
al., Proc.
Natl. Acad. Sci. USA 92:9752-9756 (1995); Liu et al., Proc. Natl. Acad. Sci.
USA 94:5525-
5530 (1997); Griesman & Berg, Science 275:657-661 (1997); and U.S. Serial No.
09/911,261
to Sera filed July 23, 2001 (the Sera application). For example, the Sera
application describes
a modular method of making AZPs that can be adapted to produce combinatorial
libraries of
AZPs. These AZPs can then be used in screening and/or selection assays to
identify AZPs
that bind at or near a target gene. Once such AZPs are known, they can serve
as the first
domain of the chimeric proteins of the invention. Similarly, any AZP (or ZFP)
obtained by a
screening or selection procedure, whether an in vitro or in vivo procedure,
can be used as the
first domain, provided that the AZP (or ZFP) specifically binds to or
associates with a target
gene in the manner contemplated by the invention.
In accordance with the invention, the chimeric proteins of the invention can
have
multiple first, nucleic acid binding domains. Each such domain specifically
binds to a
selected nucleotide sequence. Such sequences can be near one another or
located at some
distance provided that the distance does not prevent the chimeric protein from
being localized
to the nuclear periphery and repressing expression of the associated target
gene or genes.
When one first domain is present, the nucleotide sequence can be at any
location relative to
the intended target gene, provided that binding or association of the chimeric
protein with
both the nucleotide sequence and the nuclear periphery represses gene
expression. Additional
first domains can be added to the chimeric proteins of the invention to
enhance transcriptional
repression. The chimeric proteins have from one to six first domains, from one
to three first
domains, or one first domain.
Examples of other transcriptional repressors include, but are not limited to,
the KRAB
repression domain from the human KOX- I protein (Thiesen et al., New Biologist
2:363-374
(1990); Margolin et al., Proc. Natl. Acad. Sci. U.S.A. 91:4509-4513 (1994);
Pengue et al.,
-15-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
Nucl. Acids Res. 22:2908-2914 (1994); Witzgall et al., Proc. Natl. Acad. Sci.
U.S.A.
91:4514-4518 (1994)). KAP-1, a KRAB co-repressor, can be used with KRAB
(Friedman et
al., Genes Dev. 10:2067-2078 (1996)). KAP- I can also be used alone. Other
transcription
factors and transcription factor domains that act as transcriptional
repressors include MAD
(see, e.g., Sommer et al., J. Biol. Chem. 273:6632-6642 (1998); Gupta et al.,
Oncogene
16:1149- 1159 (1998); Queva et al., Oncogene 16:967-977 (1998); Larsson et
al., Oncogene
:737-748 (1997); Laherty et al., Cell 89:349-356 (1997); and Cultraro et al.,
Mol. Cell. Biol.
17:2353-2359 (19977)); FKHR (forkhead in rhapdosarcoma gene; Ginsberg et al.,
Cancer
Res. 15:3542-3546 (1998); Epstein et al., Mol. Cell. Biol. 18:4118-4130
(1998)); EGR- I
(early growth response gene product- 1; Yan et al., Proc. Natl. Acad. Sci.
U.S.A. 95:8298-
8303 (1998); and Liu et al., Cancer Gene Ther. 5:3-28 (1998)); the ets2
repressor factor
repressor domain (ERD; Sgouras et al., EMBO J. 14:4781- 4793 ((19095)); and
the MAD
smSIN3 interaction domain (SID; Ayer et al., Mol. Cell. Biol. 16:5772-5781
(1996)).
The second domain of the chimeric proteins of the invention is capable of
associating
with the nuclear periphery. This association may be direct or indirect and is
typically
mediated by protein-protein interactions between the second domain and one or
more protein
components of the nuclear envelope, the nuclear lamina, heterochromatin or any
combination
thereof. For example, the second domains) can associate with or bind to (1) a
protein that is
a component of the nuclear lamina or (2) a another protein that associates
with a protein that
is a component of the nuclear lamina. Hence, the second domain can comprise a
nuclear
envelope-binding protein, nuclear lamina-binding protein (alternatively known
as a lamina-
associated polypeptide), a heterochromatin-binding protein, the binding moiety
of any of
these proteins, a protein capable of associating with or binding to any of the
foregoing, or any
combination thereof.
The nuclear envelope- and nuclear lamina-binding proteins (or the appropriate
binding
moiety thereof) are known to or engineered to interact, respectively, with the
nuclear
envelope (particularly the inner nuclear membrane) or the nuclear lamina by
binding directly
or indirectly to those structural components of the nucleus. In some cases,
the second domain
of the chimeric protein may interact with both the inner nuclear membrane and
the nuclear
lamina. Preferred nuclear envelope- and/or nuclear lamina-binding proteins (or
their binding
moiety) include, but are not limited to, lamins (e.g., lamins A, B and C) and
lamina-binding
-16-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
proteins such as LAP 2(3 and the LAP 2(3 interaction region (amino acids 138-
524). [Nili et
al., 2001]. Other preferred proteins for the second domain include the 524-
amino acid mouse
GCL protein [Leatherman et al. (2000) Mech. Dev. 92: 145-153], or any other
GCL protein
such as from Drosophila or any other mammalian species. GCL appears to bind
indirectly to
the nuclear lamina via a lamina-associated protein (LAP). Other useful
proteins (or their
binding moieties) for the second domain include the hyperphosphorylated form
of Rb, Oct-1
and the insulin activator IPF/PDX-1 (which in the presence of low glucose is
localized to the
nuclear membrane). For all second domains, it may be useful to select a domain
that is from
the same species as the target gene. Heterochromatin-binding proteins (or the
moieties
thereof with binding activity) can also be used as second domains in the
chimeric proteins of
the invention. Useful heterochromatin-binding proteins include, but are not
limited to, HP1
and polycomb-group proteins.
In another aspect of the invention, a nuclear localization peptide can be
attached to the
chimeric proteins of the invention to aid in transporting that protein to the
nuclear
compartment. The nuclear localization peptide facilitates the transport of
proteins present in
the cytoplasm into the nucleus. The nuclear localization peptide can be used
alone or on
conjunction with other domains. One example of a nuclear localization peptide
is a peptide
from the SV40 large T antigen having the sequence Pro-Lys-Lys-Lys-Arg-Lys-Val
(SEQ m
NO: 9).
In addition, the chimeric proteins can have a cellular uptake signal attached,
either
alone or in conjunction a nuclear localization peptide, to aid in transport of
the protein into
the cell. Such cellular uptake signals include, but are not limited to,
the minimal Tat protein transduction domain which is residues 47-57 of the
human
immunodeficiency virus Tat protein: YGRKKRRQRRR (SEQ ll7 NO: 5);
residues 43-58 of the Antenapedia (pAntp) homeodomain: Arg-Gln-Ile-Lys-lle-Trp-
Phe-Gln-Asn-Arg-Arg-Met-Lys-Trp-Lys-Lys (SEQ m NO: 10) (Derossi et al., (1994)
J. Biol.
Chem. 269:10444-10450);
residues 267-300 of the herpes simplex virus (HSV) VP22 protein: Asp-Ala-Ala-
Thr-
Ala-Thr-Arg-Gly-Arg-Ser-Ala-Ala-Ser-Arg-Pro-Thr-Glu-Arg-Pro-Arg-Ala-Pro-Ala-
Arg-Ser-
Ala-Ser-Arg-Pro-Arg-Arg-Pro-Val-Glu (SEQ >D NO: 11) (Elliott et al. (1997)
Cell 88:223-
233);
-17-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
various basic peptides with reported cellular uptake signal activity such as
Tyr-Ala-
Arg-Ala-Ala-Ala-Arg-Gln-Ala-Arg-Ala (SEQ )D NO: 12)(Ho et al. (2001) Cancer
Res.
61:474-477), Arg-Arg-Arg-Arg-Arg-Arg-Arg-Arg-Arg (SEQ >D NO: 13) , also known
as R9
(Jin et al. (2001) Free Rad. Biol. Med. 31:1509-1519) and the all D-arginine
form of R9
(Winder et al. (2000) Proc. Natl. Acad. Sci. USA 97:13003-13008); and
the peptides described by the Temsamani group which include the peptides
capable of
carrying substances across the blood brain barrier of WO00/32236, the peptides
capable of
carrying an anti-cancer agent into a cancer cell as described in WO00/32237,
the amphipathic
peptide moieties of the antibiotic peptides of W002/02595, the amphipathic
peptides for
transporting negatively charged substances into cells or cell nuclei as
described in
W002/053583, and the peptide vector moieties of the analgesic molecules of
W002/067994.
The peptides described by Temsamani, include but are not limited, to n-
penetratin
(rqikiwfqnrnnkwkk; all amino acids being in the D form) (SEQ >D NO: 14), pAntp
and active
variants thereof, SynBl (RGGRLSYSRRRFSTSTGR) (SEQ )D NO: 15), L-SynB3
(RRLSYSRRRF) (SEQ >D NO: 16), and D-SynB3 (rrlsysrrrf; all amino acids being
in the D
form) (SEQ m NO: 17).
For ease of purification, monitoring expression, or monitoring cellular and
subcellular
localization, a chimeric protein of the invention can also be expressed as a
fusion protein with
such proteins or protein moieties as maltose binding protein ("MBP"), green
fluorescent
protein (GFP), glutathione S transferase (GST), hexahistidine, c-myc, and the
FLAG epitope
Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys (SEQ m NO: 18).
The chimeric proteins of the invention can be prepared either synthetically or
recombinantly, preferably recombinantly, using any of the multitude of
techniques well-
known in the art. When the proteins are prepared recombinantly, e.g., via a
DNA encoding
the chimeric protein, the codon usage can be optimized for high expression in
the organism in
which that protein is to be expressed. Such organisms include bacteria, fungi,
yeast, animals,
insects and plants. More specifically the organisms, include but are not
limited to, human,
mouse, E. coli, cereal plants, rice, tomato and corn. When the nucleic acid
will be used to
deliver the chimeric protein of the invention, codon usage can also be
optimized for the
eukaryotic organism which will receive the nucleic acid construct.
-18-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
Any suitable method of protein purification known to those of skill in the art
can be
used to purify the chimeric proteins of the invention [see, e.g., Sambrook et
al. (1989)
Molecular Cloning: A Laboratory Manual (2d ed., Cold Spring Harbor Laboratory
Press,
Plainview, New York]. In addition, any suitable host can be used for protein
expression, e.g.,
bacterial cells, insect cells, yeast cells, mammalian cells, plant cells and
the like.
The chimeric proteins of the invention and the nucleic acids encoding same are
used
to repress, down regulate or decrease gene expression of a target gene (as
determined by its
association with a particular nucleotide sequence) in any eukaryotic organism,
including
yeast, animals and plants. The target gene can encode any eukaryotic gene for
which
repression of expression is desired. For example, target genes can encode
cytokines,
interleukins, oncogenes, angiogenesis factors, anti-angiogenesis factors, drug
resistance
proteins, growth factors and/or tumor suppressors. The target gene can also be
a viral gene,
particularly from DNA viruses. The target gene can encode a plant gene.
Preferred sources
of those plant genes are from tomato, corn, rice and cereal plants.
The target genes can be oncogenes, including, but not limited to, myc, jun,
fos, myb,
max, mad, rel, ets, bcl, myb, mos family members and their associated factors
and modifiers.
Oncogenes are described in, for example, Cooper, Oncogenes, 2nd ed., The Jones
and Bartlett
Series in Biology, Boston, MA, Jones and Bartlett Publishers, 1995. The ets
transcription
factors are reviewed in Waslylk et al., Eur. J. Biochem. 211:7-18 (1993). Myc
oncogenes are
reviewed in, for example, Ryan et al., Biochem. J. 314:713-21 (1996). The Jun
and fos
transcription factors are described in, for example, The Fos and Jun Families
of Transcription
Factors, Angel & Herrlich, eds. (1994). The max oncogene is reviewed in Hurlin
et al., Cold
Spring Harb. Symp. Quant. Biol. 59:109- 16. The myb gene family is reviewed in
Kanei-Ishii
et al., Curr. Top. Microbiol. Immunol. 211:89-98 (1996). The mos family is
reviewed in
Yew et al., Curr. Opin. Genet. Dev. 3:19-25 (1993).
The chimeric proteins of the present invention can be used to inhibit the
expression of
a disease-associated genes. In one example, the disease-associated gene is an
oncogene such
as a BCR-ABL fusion oncogene or a ras oncogene, and the DNA binding domain is
designed
to bind to the DNA sequence GCAGAAGCC (SEQ )D NO: 6) and is capable of
inhibiting the
expression of the BCR-ABL fusion oncogene by both targeting to the nuclear
periphery and
by inhibiting expression by binding to a sequence needed in the transcription
process.
-19-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
Transcription factors involved in disease are reviewed in Aso et al., J Clin.
Invest. 97:1561-9
(1996).
B. Methods of Use
Another aspect of the invention relates to a method of repressing or down
regulating
expression of a target gene by localizing the gene to the nuclear periphery.
This method
involves contacting the target nucleic acid containing a nucleotide sequence
associated with
or in sufficient proximity to the target gene with a chimeric protein of the
invention. The
nucleic acid can be present in a cell or in an organism and is preferably
genomic DNA.
However, the nucleic acid can also be extrachromosomal DNA present in the
nucleus. The
nucleotide sequence and target gene are as described hereinabove. The
proximity of the
nucleotide sequence to the target gene is sufficient to allow measurable
repression or down
regulation of the target gene after exposure to a chimeric protein of the
invention.
In accordance with the invention, the chimeric protein can be introduced into
a cell as
a protein or as a nucleic acid encoding that protein. When protein is used,
the chimeric
protein can, optionally, have a cellular-uptake signal andlor a nuclear
localization signal to
facilitate uptake of the protein by the cell and its transport into the
nucleus. The amount of
the chimeric protein needed to repress or down regulate expression of the
target gene can be
readily determined by those of skill in the art. When a nucleic acid such as
RNA or DNA is
used, it can be delivered in any of a variety of forms, including as naked
plasmid or other
DNA, formulated in liposomes, in a viral vector (including RNA viruses and DNA
viruses),
via a pressure injection apparatus such as the PowderjectTM system using RNA
or DNA, or by
any other convenient means. Again, the amount of nucleic acid needed to
repress or down
regulate expression of the target gene can be readily determined by those of
skill in the art
based on the target cell or organism, the delivery formulation and mode and
whether the
nucleic acid is DNA or RNA. Preferably DNA is used.
In accordance with the invention, the chimeric protein binds to the target
nucleic acid.
with a chimeric protein at the nucleotide sequence associated with a target
gene. Assays to
determine whether binding has occurred and the efficiency by which the
repression of the
target gene or protein of interest occurs are known. In brief, in one
embodiment, a reporter
gene such as 3-glucuronidase (GUS), chloramphenicol acetyl transferase (CAT),
(3
-20-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
galactosidase (~i-gal) or green fluorescent protein (GFP) is operably linked
to the target gene
sequence controlling promoter, ligated into a transformation vector, and
transformed into an
animal or plant cell. After introduction of the chimeric protein (whether as a
protein or as a
nucleic acid which is translated to produce the protein) the level of the
reporter gene can be
assessed relative to the appropriate controls. As an alternative, levels of
RNA can be
measured by a Northern blot or other means. This latter method is useful when
reporter
constructs are not practical.
The invention contemplates gene regulation which may be tissue specific or
not,
inducible or not, and which may occur in animal cells, yeast cells, insect
cells, or plant cells
either in culture or in intact plants. Useful repression levels can vary,
depending on how
tightly the target gene is normally regulated, the effects of changes in
regulation, and other
similar factors. Desirably, the change in gene expression is modified by at
least about 1.5-
fold to 2-fold; about 3-fold to 5-fold; about 8- to 10- to 15-fold; or even
more such as 20- to
25- to 30-fold; and even 40-, 50-, 75-, or 100-fold, or more. The degree of
change in gene
expression again varies from system to system
"Organisms" as used are any eukaryotic organism including yeast, animals,
birds,
insects, plants and the like. Animals include, but are not limited to, mammals
(humans,
primates, etc.), commercial or farm animals (fish, chickens, cows, cattle,
pigs, sheep, goats,
turkeys, etc.), research animals (mice, rats, rabbits, etc.) and pets (dogs,
cats, parakeets and
other pet birds, fish, etc.). As contemplated herein, particular animals may
be members of
multiple animal groups.
The chimeric proteins of the present invention (or nucleic acids encoding
those
proteins) can be used, for example, to repress, down regulate or decrease gene
expression,
over a broad range of plant types and plant tissue, preferably the class of
higher plants
amenable to transformation techniques, particularly monocots and dicots.
A "plant" refers to any plant or part of a plant at any stage of development,
including
seeds, suspension cultures, embryos, meristematic regions, callus tissue,
leaves, roots, shoots,
gametophytes, sporophytes, pollen, and microspores, and progeny thereof. Also
included are
cuttings, and cell or tissue cultures. As used in conjunction with the present
invention, the
term "plant tissue" includes, but is not limited to, plant cells, plant organs
(e.g., leafs, stems,
-21-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
roots, meristems) plant seeds, protoplasts, callus, cell cultures, and any
groups of plant cells
organized into structural and/or functional units.
Particularly preferred are monocots such as the species of the Family
Gramineae
including Sorghum bicolor and Zea mays. The isolated nucleic acid and proteins
of the
present invention can also be used in species from the genera: Cucurbita,
Rosa, Vitis, Juglans,
Fragaria, Lotus, Medicago, Onobrychis, Trifolium, Trigonella, Vigna, Citrus,
Linum,
Geranium, Manihot, Daucus, Arabidopsis, Brassica, Raphanus, Sinapis, Atropa,
Capsicum,
Datura, Hyoscyamus, Lycopersicon, Nicotiana, Solanum, Petunia, Digitalis,
Majorana,
Ciahorium, Helianthus, Lactuca, Bromus, Asparagus, Antirrhinum, Heterocallis,
Nemesis,
Pelargonium, Panieum, Pennisetum, Ranunculus, Senecio, Salpiglossis, Cucumis,
Browaalia,
Glycine, Pisum, Phaseolus, Lolium, Oryza, Avena, Hordeum, Secale, and
Triticum.
Preferred plants and plant tissue includes those from corn (Zea mays), canola
(Brassica napus, Brassica rapa ssp.), alfalfa (Medicago sativa), rice (Oryza
sativa). rye (Secale
cereale), sorghum (Sorghum bicolor, Sorghum vulgare), sunflower (Helianthus
annuus),
wheat (Triticum aestivum), soybean (Glycine max), tobacco (Nicotiana tabacum),
potato
(Solanum tuberosum), peanuts (Arachis hypogaea), cotton (Gossypium barbadense,
Gossypium hirsutum), sweet potato Qpomoea batatus), cassava (Manihot
esculenta), coffee
(Cqfea spp.), coconut (Cocos nucijra), pineapple (Ananas comosus), citrus
trees (Citrus spp.),
cocoa (Theobroma cacao), tea (Camellia sinensis), banana (Musa spp.), avocado
(Persea
americana), fig (Ficus casica), guava (Psidium guajava), mango (Mangifera
indica), olive
(Olea europaea), papaya (Carica papaya), cashew (Anacardium occidentale),
macadamia
(Macadamia integr~fblia), almond (Prunus amygdalus), sugar beets (Beta
vulgaris), sugarcane
(Saccharum spp.), duckweed (Lemma spp.), oats, barley, vegetables,
ornamentals, and
conifers.
Preferred vegetables include tomatoes (Lycopersicon esculentum), lettuce
(e.g.,
Lactuca sativa), green beans (Phaseolus vulgaris), lima beans (Phaseolus
limensis), peas
(Lathyrus spp.), and members of the genus Cucumis such as cucumber (C.
sativus),
cantaloupe (C cantalupensis), and musk melon (C. melo).
Preferred ornamentals include azalea (Rhododendron spp.), hydrangea
(Macrophylla
hydrangea), hibiscus (Hibiscus rosasanensis), roses (Rosa spp.), tulips
(Tulipa spp.), daffodils
-22-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
(Narcissus spp.). petunias (Petunia hybrida), carnation (Dianthus
caryophyllus), poinsettia
(Euphorbiapulcherrima), and chrysanthemum.
Conifers that may be employed in practicing the present invention include, for
example, pines such as loblolly pine (Pinus taeda), slash pine (Pinus
elliotii), ponderosa pine
(Pinus ponderosa), lodgepole pine (Pinus contorta), and Monterey pine (Pinus
radiata);
Douglas-fir (Pseudotsuga menziesii); Western hemlock (Isuga canadensis); Sitka
spruce
(Picea glauca); redwood (Sequoia sempervirens); true firs such as silver fir
(Abies amabilis)
and balsam fir (Abies balsamea); and cedars such as Western red cedar (Thuja
plicata) and
Alaska yellow-cedar (Chamaecyparis nootkatensis).
Most preferably, plants and plant tissue of the present invention are crop
plants (for
example, corn, alfalfa, sunflower, canola, soybean, cotton, peanut, sorghum,
wheat, tobacco,
etc.), even more preferably corn and soybean plants, yet more preferably corn
plants.
As used herein, "transgenic plant" or "genetically modified plant" includes
reference
to a plant which comprises within its genome a heterologous polynucleotide
(i.e., a
polynucleotide from a source other than the recipient organism). Generally,
and preferably,
the heterologous polynucleotide is stably integrated within the genome such
that the
polynucleotide is passed on to successive generations. The heterologous
polynucleotide may
be integrated into the genome alone or as part of a recombinant expression
cassette.
"Transgenic" is used herein to include any cell, cell line, callus, tissue,
plant part or plant, the
genotype of which has been altered by the presence of heterologous nucleic
acid including
those transgenics initially so altered as well as those created by sexual
crosses or asexual
propagation from the initial transgenic. The term "transgenic" as used herein
does not
encompass the alteration of the genome (chromosomal or extra-chromosomal) by
conventional plant breeding methods or by naturally occurring events such as
random cross-
fertilization, non- recombinant viral infection, non-recombinant bacterial
transformation, non-
recombinant transposition, or spontaneous mutation.
C. Expression Systems
The present invention also provides recombinant expression cassettes
comprising a
chimeric protein-encoding nucleic acid of the present invention. A nucleic
acid sequence
coding for the desired polynucleotide of the present invention can be used to
construct a
-23-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
recombinant expression cassette which can be introduced into a desired host
cell. A
recombinant expression cassette will typically comprise a polynucleotide of
the present
invention operably linked to transcriptional initiation regulatory sequences
which will direct
the transcription of the polynucleotide in the intended host cell, such as
tissues of a
transformed plant. The expression vectors can be a mammalian expression
vector, an insect
expression vector, a yeast expression vector or a plant expression vector.
When the protein is
being expressed for the purpose of preparing and purifying the protein (which
can then be
used, e.g., in the methods of the invention), the expression vector can be a
bacterial
expression vectors. Expression vectors are well known in the art and can be
readily selected
for the desired purpose.
The elements for transcription include but are not limited to promoters active
in
eukaryotic cells, enhancers, transcription termination signals including
polyadenylation
signals or polyA tracts, elements to facilitate nucleocytoplasmic transport,
and the like.
Suitable transcription termination elements include the SV 40 transcription
termination
region and terminators derived therefrom.
Any mammalian, yeast, bacterial, insect, viral, other eukaryotic expression
vector or
expression cassette can be employed in the present invention and can be
selected from, e.g.,
any of the many commercially available vectors or cassettes, such as pCEP4 or
pRc/RSV
obtained from Invitrogen Corporation (San Diego, Calif.), pXTl, pSGS, pPbac or
pMbac
obtained from Stratagene (La Jolla, Calif.), pPUR or pMAM obtained from
ClonTech (Palo
Alto, Calif.), and pSV(i-gal obtained from Promega Corporation (Madison,
Wis.), or
synthesized either de novo or by adaptation of a publically or commercially
available
eukaryotic expression system.
The individual elements within the expression cassette can be derived from
multiple
sources and may be selected to confer specificity in sites of action or
longevity of the
cassettes in the recipient cell. Such manipulation of the expression cassette
can be done by
any standard molecular biology approach.
Plant expression vectors may include (1) a cloned plant gene under the
transcriptional
control of 5' and 3' regulatory sequences and (2) a dominant selectable
marker. Such plant
expression vectors may also contain, if desired, a promoter regulatory region
(e.g., one
conferring inducible or constitutive, environmentally- or developmentally-
regulated, or cell-
-24-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
or tissue-specific/selective expression), a transcription initiation start
site, a ribosome binding
site, an RNA processing signal, a transcription termination site, and/or a
polyadenylation
signal.
Typical vectors useful for expression of genes in higher plants are well known
in the
art and include vectors derived from the tumor-inducing (Ti) plasmid of
Agrobacterium
tumefaciens described by Rogers et al., Meth. in Enzymol., 153:253-277 (1987).
These
vectors are plant integrating vectors in that on transformation, the vectors
integrate a portion
of vector DNA into the genome of the host plant. Exemplary A. tumefaciens
vectors useful
herein are plasmids pKYLX6 and pKYLX7 of Schardl et al., Gene, 6 1: 1 -11
(1987) and
Berger et al., Proc. Natl. Acad. Sci. U.S.A., 86:8402-8406 (1989). Another
useful vector is
plasmid pBI101.2.
Cell transformation techniques and gene delivery methods (such as those for in
vivo
use to deliver genes) are well known in the art. Any such technique can be
used to deliver a
nucleic acid encoding the chimeric proteins of the invention to a cell or in
vivo to the cells of
a subject, respectively.
The term "expression cassette" as used herein means a DNA sequence capable of
directing expression of a particular nucleotide sequence in an appropriate
host cell,
comprising a promoter operably linked to the nucleotide sequence of interest
which is
operably linked to termination signals. It also typically comprises sequences
required for
proper translation of the nucleotide sequence. The coding region usually codes
for a protein
of interest but may also code for a functional RNA of interest, for example
antisense RNA or
a nontranslated RNA, in the sense or antisense direction. The expression
cassette comprising
the nucleotide sequence of interest may be chimeric, meaning that at least one
of its
components is heterologous with respect to at least one of its other
components. The
expression cassette may also be one which is naturally occurring but has been
obtained in a
recombinant form useful for heterologous expression. Typically, however, the
expression
cassette is heterologous with respect to the host, i.e., the particular DNA
sequence of the
expression cassette does not occur naturally in the host cell and must have
been introduced
into the host cell or an ancestor of the host cell by a transformation event.
The expression of
the nucleotide sequence in the expression cassette may be under the control of
a constitutive
promoter or of an inducible promoter which initiates transcription only when
the host cell is
-25-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
exposed to some particular external stimulus. In the case of a multicellular
organism, such as
a plant, the promoter can also be specific to a particular tissue or organ or
stage of
development.
Various promoters well-known to be useful for driving expression of genes in
animal
cells, such as the viral-derived SV40, CMV immediate early and, RSV promoters
or
eukaryotic derived 3-casein, uteroglobin, 3-actin or tyrosinase promoters. The
particular
promoter is not critical to the invention, unless the object is to obtain
temporal- or tissue-
specific expression. For example, a promoter can be selected which is only
active in the
desired tissue or selected cell type. Examples of tissue-specific promoters
include, but are not
limited to, b' S1- and ~-casein promoters which are specific for mammary
tissue (Platenburg
et al., Trans. Res., 3:99-108 (1994); and Maga et al., Trans. Res., 3:36-42
(1994)); the
phosphoenolpyruvate carboxykinase promoter which is active in liver, kidney,
adipose,
jejunum and mammary tissue (McGrane et al., J. Reprod. Fert., 41:17-23
(1990)); the
tyrosinase promoter which is active in lung and spleen cells, but not testes,
brain, heart, liver
or kidney (Vile et al., Canc. Res., 54:6228-6234 (1994)); the involucerin
promoter which is
only active in differentiating keratinocytes of the squamous epithelia
(Carroll et al., J. Cell
Sci., 103:925-930 (1992)); and the uteroglobin promoter which is active in
lung and
endometrium (Helftenbein et al., Annal. N.Y. Acad. Sci., 622:69-79 (1991)).
Alternatively, cell specific enhancer sequences can be used to control
expression, for
example human neurotropic papovirus JCV enhancer regulates viral transcription
in glial
cells alone (Remenick et al., J. Virol., 65:5641-5646 (1991)). Yet another way
to control
tissue specific expression is to use a hormone responsive element (HRE) to
specify which cell
lineages a promoter will be active in, for example, the MMTV promoter requires
the binding
of a hormone receptor, such as progesterone receptor, to an upstream HRE
before it is
activated (Beato, FASEB J., 5:2044-2051 (1991); and Truss et al., J. Steroid
Biochem. Mol.
Biol., 41:241-248 (1992)).
A plant promoter fragment can be employed which will direct expression of a
polynucleotide of the present invention in all tissues of a regenerated plant.
Such promoters
are referred to herein as "constitutive" promoters and are active under most
environmental
conditions and states of development or cell differentiation. Examples of
constitutive
promoters include the cauliflower mosaic virus (CaMV) 35S transcription
initiation region,
-26-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
the P- or 2'- promoter derived from T-DNA of Agrobacterium tumefaciens, the
ubiquitin I
promoter, the Smas promoter, the cinnamyl alcohol dehydrogenase promoter (U.S.
Patent No.
5,683,439), the Nos promoter, the pEmu promoter, the rubisco promoter, the GRP
1 - 8
promoter, and other transcription initiation regions from various plant genes
known to those
of skill in the art.
Alternatively, the plant promoter can direct expression of a polynucleotide of
the
present invention in a specific tissue or may be otherwise under more precise
environmental
or developmental control. Such promoters are referred to here as "inducible"
promoters.
Environmental conditions that may effect transcription by inducible promoters
include
pathogen attack, anaerobic conditions, or the presence of light. Examples of
inducible
promoters include the AdhI promoter which is inducible by hypoxia or cold
stress, the IIsp70
promoter which is inducible by heat stress, and the PPDK promoter which is
inducible by
light. Examples of promoters under developmental control include promoters
that initiate
transcription only, or preferentially, in certain tissues, such as leaves,
roots, fruit, seeds, or
flowers. An exemplary promoter is the anther specific promoter 5126 (U.S.
Patent Nos.
5,689,049 and 5,689,051). The operation of a promoter may also vary depending
on its
location in the genome. Thus, an inducible promoter may become fully or
partially
constitutive in certain locations.
Both heterologous and non-heterologous (i.e., endogenous) promoters can be
employed to direct expression of the nucleic acids of the present invention.
These promoters
can also be used, for example, in recombinant expression cassettes to drive
expression of
antisense nucleic acids to reduce, increase, or alter concentration and/or
composition of the
proteins of the present invention in a desired tissue. Thus, in some
embodiments, the nucleic
acid construct will comprise a promoter functional in a plant cell, such as in
Zea mays,
operably linked to a polynucleotide of the present invention. Promoters useful
in these
embodiments include the endogenous promoters driving expression of a
polypeptide of the
present invention.
In some embodiments, isolated nucleic acids which serve as promoter or
enhancer
elements can be introduced in the appropriate position (generally upstream) of
a non-
heterologous form of a polynucleotide so as to up or down regulate its
expression. For
example, endogenous promoters can be altered in vivo by mutation, deletion,
and/or
-27-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
substitution (U.S. Patent 5,565,350; PCT/LJS93/03868), or isolated promoters
can be
introduced into a plant cell in the proper orientation and distance from a
gene of the present
invention so as to control the expression of the gene. Gene expression can be
modulated
under conditions suitable for plant growth so as to alter the total
concentration and/or alter the
composition of the polypeptides of the present invention in plant cell.
A variety of promoters will be useful in the invention, particularly to
control the
expression of the chimeric proteins, the choice of which will depend in part
upon the desired
level of protein expression and desired tissue-specific, temporal specific, or
environmental
cue-specific control, if any in a plant cell. Constitutive and tissue specific
promoters are of
particular interest. Such constitutive promoters include, for example, the
core promoter of the
Rsyn7, the core CaMV 35S promoter (Odell et al. (1985) Nature 313:810-812),
rice actin
(McElroy et al. (1990) Plant Ceil 2:163-171); ubiquitin (Christensen et al.
(1989) Plant Mol.
Biol. 12:619-632 and Christensen et al. (1992) Plant Mol. Biol. 18:675-689),
pEMU (Last et
al. (1991) Theor. Appl. Genet. 81:581-588), MAS (Veltenet al. (1984) EMBO J.
3:2723-
2730), and constitutive promoters described in, for example, U.S. Patent Nos.
5,608,149;
5,608,144; 5,604,121; 5,569,597; 5,466,785; 5,399,680; 5,268,463; and 5,
608,142.
Tissue-specific promoters can be utilized to target enhanced expression within
a
particular plant tissue. Tissue-specific promoters include those described by
Yamamoto et al.
(1997) Plant J. 12(2)255-265, Kawamata et al. (1997) Plant Cell Physiol.
38(7):792-803,
Hansen et al. (1997) Mol. Gen Genet. 254(3):337), Russell et al. (1997)
Transgenic Res.
6(2):15 7-168, Rinehart et al. (1996) Plant Physiol. 112(3):1331, Van Camp et
al. (1996)
Plant Physiol. 112(2):525-535, Canevascini et al. (1996) Plant Physiol.
112(2):513-524,
Yamamoto et al. (1994) Plant Cell Physiol. 35(5):773 -778, Lam (1994) Results
Probl. Cell
Differ. 20:181-196, Orozco et al. (1993) Plant Mol. Biod. 23 (6):1129-113 8,
Matsuoka et
ad. (1993) Proc NatL. Acad. Sci. USA 90(20):9586-9590, and Guevara-Garcia et
al. (1993)
Plant J. 4(3):495-505. Such promoters can be modified, if necessary, for weak
expression.
Leaf-specific promoters are known in the art, and include those described in,
for
example, Yamamoto et al. (1997) Plant J. 12(2):255-265, Kwon et al. (1994)
Plant Physiol.
105:357- 67, Yamamoto et al. (1994) Plant Cell Physiol. 35(5):773-778, Gotor
et al. (1993)
Plant J. 3:509-18, Orozco et al. (1993) Plant Mol. Biol. 23(6):1129-1138, and
Matsuoka et
al. (1993) Proc. Natl. Acad. Sci. U.S.A .90(20):9586-9590.
-28-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
Any combination of constitutive or inducible and non-tissue specific or tissue
specific
may be used to control the expression of the chimeric proteins of the
invention. The desired
control may be temporal, developmental or environmentally controlled using the
appropriate
promoter. Environmentally controlled promoters are those that respond to
assault by
pathogen, pathogen toxin, or other external compound (e.g., intentionally
applied small
molecule inducer). An example of a temporal or developmental promoter is a
fruit ripening-
dependent promoter. Particularly preferred are the inducible PRl promoter, the
maize
ubiquin promoter, and ORS.
Methods for identifying promoters with a particular expression pattern, in
terms of,
e.g., tissue type, cell type, stage of development, and/or environmental
conditions, are well
known in the art. See, e.g., The Maize Handbook, Chapters 114-115, Freeling
and Walbot,
Eds., Springer, New York (1994); Corn and Corn Improvement, Pedition, Chapter
6, Sprague
and Dudley, Eds., American Society of Agronomy, Madison, Wisconsin (1988).
Plant transformation protocols as well as protocols for introducing nucleotide
sequences into plants may vary depending on the type of plant or plant cell,
i.e., monocot or
dicot, targeted for transformation. Suitable methods of introducing nucleotide
sequences into
plant cells and subsequent insertion into the plant genome include
microinjection (Crossway
et al. (1986) Biotechniques 4:320-334), electroporation (Riggs et al. (1986)
Proc. Natl. Acad
Sci. USA 83:5602- 5606, Agrobacterium-mediated transformation (Townsend et
al., U.S. Pat
No. 5,563,055), direct gene transfer (Paszkowski et al. (1984) EMBO J. 3:2717-
2722), and
ballistic particle acceleration (see, for example, Sanford et al., U. S.
Patent No. 4,945,050;
Tomes et al. (1995) "Direct DNA Transfer into Intact Plant Cells via
Microprojectile
Bombardment," in Plant Cell Tissue and Oman Culture: Fundamental Methods, ed.
Gamborg and Phillips (Springer-Verlag, Berlin); and McCabe et al. (1988)
Biotechnology
6:923-926). Also see Weissinger et al. (1988) Ann. Rev. Genet. 22:421-477;
Sanford et al.
(1987) Particulate Science and Technology 5:27-37 (onion); Christou et al.
(1988) Plant
Physiol. 87:671- 674 (soybean); McCabe et al. (1988) BioTechnology 6:923-926
(soybean);
Finer and McMullen (199 1) In Vitro Cell Dev. Biol. 2 7P: 175-182 (soybean);
Singh et al.
(1998) Theor. Appl. Genet. 96:319-324 (soybean); Datta et al. (1990)
Biotechnology 8:736-
740 (rice); Klein et al. (1988) Proc. Natl. Acad Sci. USA 85:4305-4309
(maize); Klein et al.
(1988) Biotechnology 6:559-563 (maize); Tomes, U.S. Patent No. 5,240,855;
Buising et al.,
-29-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
U.S. Patent Nos. 5,322, 783 and 5,324,646; Tomes et al. (1995) "Direct DNA
Transfer into
Intact Plant Cells via Microprojectile Bombardment," in Plant Cell, Tissue,
and Orgy
Culture: Fundamental Methods, ed. Gamborg (Springer-Verlag, Berlin) (maize);
Klein et al.
(198 8) Plant Physiol. 91:440-444 (maize); Fromm et al. (1990) Biotechnology
8:833-839
(maize); Hooykaas-Van Slogteren et al. (1984) Nature (London) 311:763-764;
Bowen et al.,
U.S. Patent No. 5,736,369 (cereals); Bytebier et al. (1987) Proc. Natl. Acad
Sci. USA
84:5345-5349 (Liliaceae); De Wet et al. (1985) in The Experimental
Manipulation of Ovule
Tissues, ed. Chapman et al. (Longman, New York), pp. 197-209 (pollen);
Kaeppler~et al.
(1990) Plant Cell Reports 9:415- 418 and Kaeppler et al. (1992) Theor. Appl.
Genet. 84:560-
566 (whisker- mediated transformation); D'Halluin et al. (1992) Plant Cell
4:1495-1505
(electroporation); Li et al. (1993) Plant Cell Reports 12:250-255 and Christou
and Ford
(1995) Annals of Botany 75:407-413 (rice); Osjoda et al: (1996) Nature
Biotechnology
14:745-750 (maize via Agrobacterium tumefaciens); all of which are herein
incorporated by
reference.
The modified plant may be grown into plants by conventional methods. See, for
example, McCormick et al. (1986) Plant Cell. Reports :81-84. These plants may
then be
grown, and either pollinated with the same transformed strain or different
strains, and the
resulting hybrid having the desired phenotypic characteristic identified. Two
or more
generations may be grown to ensure that the subject phenotypic characteristic
is stably
maintained and inherited and then seeds harvested to ensure the desired
phenotype or other
property has been achieved.
D. Molecular Switch stems for Gene Repression
Another aspect of the invention relates to molecular switch systems for
controlling
gene expression, and particularly molecular switch systems for repressing or
down regulating
gene expression using the domains in the chimeric proteins of the inventions.
Such systems
(also called "chemical switches") provide a further tool to manipulate the
timing of or
location where gene expression is being regulated or controlled. Briefly, the
molecular switch
system introduces two fusion proteins, one with the nucleic acid binding
domain and the other
with the nuclear periphery-binding domain, into a cell or organism. These two
fusion
proteins each have a second domain which specifically bind to one or the other
moiety of a
-30-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
divalent ligand. Upon introduction of the divalent ligand into the cell or
organism containing
the two fusion proteins, the ligand acts as a switch to trigger formation of a
complex among
the three entities. This complex is then similar in function to the chimeric
proteins of the
invention since once formed it can bring a target gene into association with
the nuclear
periphery to repress or down regulate gene expression.
An example is a complex formed by a divalent chemical ligand having moieties A
and
B, a first fusion protein encoding an AZP and an antibody specific for moiety
A (or an active
fragment of such antibody) and a second fusion protein encoding the domain
capable of
associating with the nuclear periphery and an antibody specific for moiety B
(or an active
fragment of such antibody). The two fusion proteins can be separately or
coordinately
expressed in the same cell. Upon addition to the cell or organism of the
divalent chemical
that includes moiety A and moiety B linked together, the affinity of each
fusion protein for
either moiety A or moiety B mediates formation of a complex.
Accordingly, the first fusion protein of this aspect of the invention
comprises a first
domain capable of specifically binding a nucleotide sequence associated with a
target gene,
and a second domain capable of specifically binding to a first binding moiety
of a divalent
ligand, said ligand capable of uptake by a cell, wherein the first and second
domains are
heterologous with respect each other. The first domain of the fusion protein
is the same as
the first domain of the chimeric proteins of the invention. For example, this
first domain of
the first fusion protein can be a ZFP, an AZP, a leucine zipper protein, a
helix-turn-helix
protein, a helix-loop-helix protein, a homeobox domain protein, the DNA
binding moiety of
any of those proteins, or any combination thereof.
Likewise, the nucleotide sequence associated with the target gene, and the
target gene
is the same as described for the chimeric proteins of the invention.
The second fusion protein of this aspect of the invention comprises a first
domain
capable of associating with the nuclear periphery and a second domain capable
of specifically
binding to the second binding moiety of the divalent ligand, wherein said
first domain is
heterologous with respect to said second domain. The first domain of these
fusion proteins is
the same as the second domain as the chimeric proteins of the invention. Thus,
the first
domain of said second fusion protein binds the nuclear envelope, the nuclear
lamina,
heterochromatin, or any combination thereof, and is preferably a nuclear
envelope-binding
-31-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
protein, nuclear lamina-binding protein, a heterochromatin-binding protein,
the binding
moiety of any of those proteins, or any combination thereof.
The second domains of the first and second fusion proteins of this molecular
switch
system are each capable of specifically binding to one binding moiety of a
divalent ligand.
The first fusion protein binds to one of the binding moieties (e.g., moiety A)
of the divalent
ligand and the second fusion protein binds to the other binding moiety (e.g.,
moiety B) of the
divalent ligand. In an embodiment, the second domain of each fusion protein
can be a single
chain variable region (scFv) of an antibody with specificity for its
respective binding moiety
of the divalent ligand.
Numerous possibilities exist for moieties A and B. The criteria are that the
moiety is
sufficiently antigenic to allow selection of a antibody specific for that
moiety, and that the
two moieties, linked together, form a compound that can enter and act within a
cell to mediate
formation of the complex. In one embodiment, moiety A can have a structure,
for example,
as depicted below:
H03S
moiety B can have a structure, for example, as depicted below:
OCH3
H
O
OCH3
and moieties A and B can be linked by a linker of any suitable length, having
units such as
those depicted below:
CH2
CH2
Any compound capable of entry into cell and having moieties against which
antibodies can be raised is suitable for the divalent ligand aspect of the
invention. This
-32-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
embodiment of the invention permits sequence-specific localization of the
target gene domain
to the nuclear periphery by allowing a complex to form in the presence of the
divalent ligand.
In the absence of the divalent chemical no tertiary complex is formed.
In a preferred embodiment, a chemical switch is used which is a divalent
chemical
comprising two linked compounds. These compounds may be any compounds to which
antibodies can be raised linked by a short linker, for example, CH2CH2. In one
preferred
embodiment, a single chain antibody (e.g., a single chain F,, (scFv)) binds to
one portion of
the divalent chemical to link it to a nucleic acid binding domain. The other
portion of the
divalent chemical binds to a second single chain antibody, for example a
single chain F,,
(scF"), which recognizes and binds to protein domain capable of associating
with or binding
to the nucelar periphery.
In another embodiment, the second domain of the two fusion proteins can be
mutant
S-tag and S-proteins (described below) which can only bind to each other in
the presence of a
small molecule or chemical. This small molecule thus acts as the divalent
ligand to bring the
two fusion proteins into a single complex that localizes to the nuclear
periphery and leads to
gene repression or down regulation.
This molecular switch system can be used in a method to regulate repression of
a
target gene in a temporally or spatially manner. In particular the method
involves contacting
a cell or organism containing a target nucleic acid having a nucleotide
sequence associated
with a target gene with the molecular switch system of the invention (as
described in this
section), and contacting the cell or organism with the appropriate divalent
ligand at a time or
in a location to allow formation of a complex with the fusion proteins and
thereby repress or
down regulate expression of the target gene by virtue of its localization to
the nuclear
periphery. As with the chimeric proteins, the fusion proteins of the molecular
switch system
can be introduced into the cell or organism as proteins, as one or more
nucleic acids encoding
one or more of the proteins, or as a combination thereof. When a single
nucleic acid is used
to deliver the fusion proteins, expression of each protein can be coordinately
or independently
controlled. Likewise the method is useful with the same target genes as
contemplated for the
methods using the chimeric proteins of the invention.
-33-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
The fusion proteins can be expressed, isolated and purified as described above
for the
chimeric proteins. Likewise they can be introduced into the cells or organism
as described
above for the chimeric proteins.
E. Molecular Switch Systems for Gene Derepression
Molecular switch systems can be provided in another format that allows
controlled
regulation for derepression of a target gene, i.e., activating expression of a
target gene
currently being repressed. In this aspect of the invention, the "switch" is
used to disrupt the
interaction between two fusion proteins (rather than to promote the
interaction as in section
D). Again, these systems (also called "chemical switches") provide another
tool to
manipulate the timing of or location where gene expression is regulated or
controlled.
Briefly, the molecular switch system introduces two fusion proteins, one with
the nucleic acid
binding domain and the other with the nuclear periphery-binding domain, into a
cell or
organism. These two fusion proteins each have second domains which
specifically bind to
each other, e.g., the second domains are binding partners for one another. In
this system,
introduction of the fusion proteins leads to formation of a complex which
localizes to the
nuclear periphery and represses or down regulates expression of the associated
target gene.
When the chemical switch is introduced into the cells or organisms at the
desired time (or in
the particular cell types), it acts to disrupt the complex and release the
state of repression, i.e.,
presence of the chemical switch leads to derepression of the target gene.
Accordingly, the first fusion protein of this aspect of the invention
comprises a first
domain capable of specifically binding a nucleotide sequence associated with a
target gene,
and a second domain capable of specifically binding to the second binding
moiety of the
divalent ligand, wherein said first domain is heterologous with respect to
said second domain.
These fusions proteins are distinct from those described in Section D.
The first domain of the fusion protein is the same as the first domain of the
chimeric
proteins of the invention. For example, this first domain of the first fusion
protein can be a
ZFP, an AZP, a leucine zipper protein, a helix-turn-helix protein, a helix-
loop-helix protein, a
homeobox domain protein, the DNA binding moiety of any of those proteins, or
any
combination thereof. Likewise, the nucleotide sequence associated with the
target gene, and
the target gene is the same as described for the chimeric proteins of the
invention.
-34-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
The second fusion protein of this aspect of the invention comprises a first
domain
capable of associating with the nuclear periphery and a second domain
comprising the
binding partner of the second domain of said first fusion protein, wherein
said first domain is
heterologous with respect to said second domain. The first domain of these
second fusion
proteins is the same as the second domain as the chimeric proteins of the
invention. Thus, the
first domain of said second fusion protein binds the nuclear envelope, the
nuclear lamina,
heterochromatin, or any combination thereof, and is preferably a nuclear
envelope-binding
protein, nuclear lamina-binding protein, a heterochromatin-binding protein,
the binding
moiety of any of those proteins, or any combination thereof.
The second domains of the first and second fusion proteins of this molecular
switch
system are each capable of specifically binding to one another. One example of
second
domains is represented by the S-tag/S-protein system [Kim et al.(1993) Protein
Sci. 3:348-
356]. The S-tag is a short peptide (15 amino acids) and S-protein is a small
protein (104
amino acids) and can be used interchangeably as the second domain for either
of the two
fusion proteins. The affinity of the S-tag and S-protein complex is high
(Kd=1nM). The
chemical switch or ligand is then a molecule which can disrupt the interaction
between the S-
tag and the S-protein. For example, free or conjugated S-tag protein may act
as the chemical
switch.
This molecular switch system can be used in a method to regulate repression of
a
target gene in a temporally or spatially manner. In particular the method
involves contacting
a cell or organism containing a target nucleic acid having a nucleotide
sequence associated
with a target gene with a molecular switch system of the invention (as
described in this
section) and contacting the cell or organism with a ligand at a time or in a
location to disrupt
association of the first and second fusion proteins and thereby derepress
expression of the
target gene. As with the chimeric proteins, the fusion proteins of the
molecular switch system
can be introduced into the cell or organism as proteins, as one or more
nucleic acids encoding
one or more of the proteins, or as a combination thereof. When a single
nucleic acid is used
to deliver the fusion proteins, expression of each protein can be coordinately
or independently
controlled. Likewise the method is useful with the same target genes as
contemplated for the
methods using the chimeric proteins of the invention.
-35-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
These fusion proteins can be also be expressed, isolated and purified as
described
above for the chimeric proteins. Likewise they can be introduced into the
cells or organism as
described above for the chimeric proteins.
F. Pharmaceutical Formulations
Therapeutic formulations of the chimeric proteins, the molecular switch
systems (as
provided in Section D or E), the various fusion proteins (of Section D or E)
or the nucleic
acids encoding any of those proteins or systems of the invention are prepared
for storage by
mixing those entities having the desired degree of purity with optional
physiologically
acceptable carriers, excipients or stabilizers (Remington's Pharmaceutical
Sciences 16th
edition, Osol, A. Ed. (1980)), in the form of lyophilized formulations or
aqueous solutions.
Acceptable carriers, excipients, or stabilizers are nontoxic to recipients at
the dosages and
concentrations employed, and can include buffers such as phosphate, citrate,
and other
organic acids; antioxidants including ascorbic acid and methionine;
preservatives (such as
octadecyldimethylbenzyl ammonium chloride; hexamethonium chloride;
benzalkonium
chloride, benzethonium chloride; phenol, butyl or benzyl alcohol; alkyl
parabens such as
methyl or propyl paraben; catechol; resorcinol; cyclohexanol; 3-pentanol; and
m-cresol); low
molecular weight (less than about 10 residues) polypeptide; proteins, such as
serum albumin,
gelatin, or immunoglobulins; hydrophilic polymers such as
polyvinylpyrrolidone; amino acids
such as glycine, glutamine, asparagine, histidine, arginine, or lysine;
monosaccharides,
disaccharides, and other carbohydrates including glucose, mannose, or
dextrins; chelating
agents such as EDTA; sugars such as sucrose, mannitol, trehalose or sorbitol;
salt-forming
counter-ions such as sodium; metal complexes (e.g., Zn-protein complexes);
andlor non-ionic
surfactants such as TWEENT'~', PLURONICS~ or polyethylene glycol (PEG).
The formulations herein may also contain more than one active compound as
necessary for the particular indication being treated, preferably those with
complementary
activities that do not adversely affect each other. Such molecules are
suitably present in
combination in amounts that are effective for the purpose intended.
The active ingredients may also be entrapped in microcapsule prepared, for
example,
by coacervation techniques or by interfacial polymerization, for example,
hydroxymethylcellulose or gelatin-microcapsule and poly-(methylmethacylate)
microcapsule,
-36-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
respectively, in colloidal drug delivery systems (for example, liposomes,
albumin
microspheres, microemulsions, nano-particles and nanocapsules) or in
macroemulsions. Such
techniques are disclosed in Remington's Pharmaceutical Sciences, 16th edition,
Osol, A. Ed.
(1980).
The formulations to be used for in vivo administration are sterile. While this
can be
readily accomplished by filtration through sterile filtration membranes, other
sterilization
methods can be used provided that the activity of the active ingredients is
not destroyed or
altered.
Sustained-release preparations may be prepared. Suitable examples of sustained-
release preparations include semipermeable matrices of solid hydrophobic
polymers
containing the polypeptide variant, which matrices are in the form of shaped
articles, e.g.,
films, or microcapsule. Examples of sustained-release matrices include
polyesters, hydrogels
(for example, poly(2-hydroxyethyl-methacrylate), or poly(vinylalcohol)),
polylactides (U.S.
Pat. No. 3,773,919), copolymers of L-glutamic acid and y ethyl-L-glutamate,
non-degradable
ethylene-vinyl acetate, degradable lactic acid-glycolic acid copolymers such
as the LUPRON
DEPOTS (injectable microspheres composed of lactic acid-glycolic acid
copolymer and
leuprolide acetate), and poly-D-(-)-3-hydroxybutyric acid. While polymers such
as ethylene-
vinyl acetate and lactic acid-glycolic acid enable release of molecules for
over 100 days,
certain hydrogels release proteins for shorter time periods. Rational
strategies can be devised
for stabilization depending on the mechanism involved. For example, if the
aggregation
mechanism is discovered to be intermolecular S-S bond formation through thio-
disulfide
interchange, stabilization may be achieved by modifying sulfhydryl residues,
lyophilizing
from acidic solutions, controlling moisture content, using appropriate
additives, and
developing specific polymer matrix compositions.
Those of skill in the art can readily determine the amounts of the chimeric
proteins,
the molecular switch systems (as provided in Section D or E), the various
fusion proteins (of
Section D or E) or the nucleic acids encoding any of those proteins or systems
of the
invention to be included in any pharmaceutical composition and the appropriate
dosages for
the contemplated use.
-37-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
Throughout this application, various publications, patents, and patent
applications
have been referred to. The teachings and disclosures of these publications,
patents, and patent
applications in their entireties are hereby incorporated by reference into
this application.
It is to be understood and expected that variations in the principles of
invention herein
disclosed in exemplary embodiments may be made by one skilled in the art and
it is intended
that such modifications, changes, and substitutions are to be included within
the scope of the
present invention.
Example 1
Repression of human VEGF-A
To down regulate the expression of the human vascular endothelial growth
factor A
(VEGF-A) gene, a recombinant construct encoding a chimeric protein (CPl-vegf)
containing
the 524-amino acid mouse GCL protein [Leatherman et al. (2000)] and an AZP
targeted for
the sequence 5'-GTG TGG GTG AGT GAG TGT G-3' (SEQ ID NO: 7) is prepared. A
second construct encoding another chimeric protein (CP2-vegf) is prepared
using the same
mouse GCL protein and an AZP targeted for the sequence 5'-GGG GCT GGG GGC GGT
GTC T-3' (SEQ ID NO: 8). The target nucleotide sequences are from the promoter
of the
human VEGF-A gene [Tischer et al. (1991) J. Biol. Chem. 266:11947-11954]. The
AZPs
have 6 zinc fingers, each with the framework sequence of -Pro-Tyr-Lys-Cys-Pro-
Glu-Cys-
Gly-Lys-Ser-Phe-Ser-Z-'-Ser- Z2- Z3-Leu-Gln- Z6-His-Gln-Arg-Thr-His-Thr-Gly-
Glu-Lys-
(SEQ ID NO: 3); each framework is joined to the next without additional amino
acid
residues. The identities of the residues that determine DNA binding
specificity (Z-', Z2, Z3
and Z6) for CP1-vegf and CP2-vegf are provided in Table 3.
To test for repression activity, the chimeric proteins constructs are co-
transfected into
the human histiocytic lymphoma cell line U-937 with a luciferase gene reporter
plasmid
containing the luciferase gene under control of the human VEGF-A native
promoter. This
luciferase gene reporter plasmid contains nucleotides from -2279 to +1041 of
the VEGF-A
gene upstream of the luciferase gene [Liu et al. (2001) J. Biol. Chem.
276:11323-11334]. For
a positive control, the U-937 cells are transfected with the luciferase gene
reporter plasmid
alone or co-transfected with the luciferase gene reporter plasmid and a
chimeric protein
construct (as a protein or as nucleic acid) of GCL and an AZP (or other DNA
binding
-38-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
domain) for an unrelated target sequence. A decrease in luciferase activity
relative to the
control level indicates that CP1-vegf and CP2-vegf down regulate VEGF-A
promoter activity.
Alternatively, repression activity can be monitored by treating cells with the
CPl-vegf
or CP2-vegf proteins or by transfecting the U-937 cells with a nucleic acid
encoding the CPl-
vegf or CP2-vegf protein, and monitoring the levels of endogenous VEGF-A mRNA
by
Northern blotting techniques.
TABLE 3
Protein Domain/Target Z-1 Z' Z' Z"
Nucleotides
CP1-vegf 1 GTGT Arg Asn Ser Arg
2 TGGG Arg Asp His Thr
3 GTGA Arg Thr Ser Arg
4 AGTG Thr Asp His Gln
5 GAGT Arg Asn Asn Arg
6 TGTG Thr Asp His Thr
CP2-vegf 1 GGGG Arg Asp His Arg
2 GCTG Thr Asp Asp Arg
3 GGGG Arg Asp His Arg
4 GGCG Glu Asp His Arg
5 GGTG Thr Asp His Arg
6 GTCT Glu Asn Ser Arg
-39-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
SEQUENCE LISTING
<110> Sera, Takashi
S <120> Nuclear-Envelope and Nuclear-Lamina Binding Chimeras for
Modulating Gene Expression
<130> 109845-163
<160> is
<170> PatentIn version 3.2
<210> 1
IS <211> 25
<212> PRT
<213> Artificial Sequence
<220>
<223> Zinc finger domain
<220>
<221> MISC FEATURE
ZS <222> (2) . . (5)
<223> Amino acids 2-5 are Xaa wherein Xaa = any amino acid, and up to
two amino acids can be missing.
<220>
<221> MISC FEATURE
<222> (7)..(18)
<223> Xaa can be any amino acid
<220>
3S <221> MISC FEATURE
<222> (20)..(24)
<223> Amino acids 20-24 are Xaa wherein Xaa = any amino acid, and up to
two amino acids can be missing.
<400> 1
Cys Xaa Xaa Xaa Xaa Cys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
1 5 10 15
-1-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
Xaa Xaa His Xaa Xaa Xaa Xaa Xaa His
20 25
<210> 2
<211> 32
<212> PRT
<213> Artificial Sequence
<220>
<223> Zinc finger domain
<220>
<221> MISC FEATURE
<222> (1)..(3)
<223> Xaa can be any amino acid
<220>
<221> MISC FEATURE
<222> (5)..(8)
<223> Amino acids 5-8 are Xaa wherein Xaa = any amino acid, and up to
two amino acids can be missing
<220>
<221> MISC FEATURE
<222> (10)..(14)
<223> Xaa can be any amino acid
<220>
<221> MISC FEATURE
<222> (15)..(15)
<223> Amino acid 15 is Z(-1) wherein Z(-1) = Arg, Lys, Gln, Asn, Thr,
Met, Leu, Ile, Glu or Asp.
<220>
<221> MISC FEATURE
<222> (16)..(16)
<223> Xaa can be any amino acid
<220>
-2-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
<221> MISC FEATURE
<222> (17)..(17)
<223> Amino acid 17 is Z2 wherein Z2 = Ser, Arg, Asn, Gln, Thr, Val,
Ala, Asp or Glu.
<220>
<221> MISC FEATURE
<222> (18)..(18)
<223> Amino acid 18 is Z3 wherein Z3 = His, Lys, Asn, Gln, Ser, Ala,
lO Val, Thr, Asp or Glu..
<220>
<221> MISC FEATURE
<222> (18)..(18)
<223> Amino acid 18 is Z3 wherein Z3 = His, Lys, Asn, Gln, Ser, Ala,
Val, Thr, Asp or Glu.
<220>
<221> MISC FEATURE
<222> (19) . . (20)
<223> Xaa can be any amino acid
<220>
<221> MISC FEATURE
<222> (21) . . (21)
<223> Amino acid 21 is Z6 wherein Z6 = Arg, Lys, Gln, Asn, Thr, Tyr,
Leu, Ile, Met, Glu or Asp.
<220>
3~ <221> MISC FEATURE
<222> (23) . . (27)
<223> Amino acids 5-8 are Xaa wherein Xaa = any amino acid, and up to
two amino acids can be missing
<220>
<221> MISC FEATURE
<222> (29) . . (32)
<223> Xaa can be any amino acid
4O <400> 2
Xaa Xaa Xaa Cys Xaa Xaa Xaa Xaa Cys Xaa Xaa Xaa Xaa Xaa Xaa Xaa
1 5 10 15
-3-
CA 02472729 2004-07-08
10
WO 03/062447 PCT/US03/01529
Xaa Xaa Xaa Xaa Xaa His Xaa Xaa Xaa Xaa Xaa His Xaa Xaa Xaa Xaa
20 25 30
<210> 3
<211> 28
<212> PRT
<213> Artificial Sequence
<220>
<223> Zinc finger domain
IS <220>
<221> MISC FEATURE .
<222> (13)..(13)
<223> Amino acid 13 is Z(-1) wherein Z(-1) = Arg, Lys, Gln, Asn, Thr,
Met, Leu, Ile, Glu or Asp.
<220>
<221> MISC FEATURE
<222> (15)..(15)
<223> Amino acid 15 is Z2 wherein Z2 = Ser, Arg, Asn, Gln, Thr, Val,
Ala, Asp or Glu.
<220>
<221> MISC FEATURE
<222> (16)..(16)
<223> Amino acid 16 is Z3 wherein Z3 = His, Lys, Asn, Gln, Ser, Ala,
Val, Thr, Asp or Glu.
<220>
<221> MISC FEATURE
<222> (19)..(19)
<223> Amino acid 19 is Z6 wherein Z6 = Arg, Lys, Gln, Asn, Thr, Tyr,
Leu, Ile, Met, Glu or Asp.
<400> 3
Pro Tyr Lys Cys Pro Glu Cys Gly Lys Ser Phe Ser Xaa Ser Xaa Xaa
1 5 10 15
-4-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
Leu Gln Xaa His Gln Arg Thr His Thr Gly Glu Lys
20 25
<210> 4
<211> 5
<212> PRT
<213> Artificial Sequence
<220>
<223> peptide
<400> 4
IS Gly Gly Gly Gly Ser
1 5
<210> 5
<211> 11
<212> PRT
<213> Human immunodeficiency virus
<220>
<221> misc feature
<223> HIV Tat protein domain
<400> 5
Tyr Gly Arg Lys Lys Arg Arg Gln Arg Arg Arg
1 5 10
<210> 6
<211> 9
<212> DNA
<213> Human immunodeficiency virus
<220>
<221> misc feature
<223> DNA binding domain
-S-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
<400> 6
gcagaagcc 9
<210> 7
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> DNA target sequence
<400> 7
gtgtgggtga gtgagtgtg 19
<210> 8
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> DNA target sequence
<400> s
ggggctgggg gcggtgtct 19
<210> 9
<211> 7
<212> PRT
<213> Simian virus 40
<220>
<221> misc feature
<223> Peptide from SV40 large T antigen
<400> 9
Pro Lys Lys Lys Arg Lys Val
1 5
-6-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
<210> 10
<211> 16
<212> PRT
<213> Artificial Sequence
<220>
<223> Peptide, residues 43-58 of the Antennapeida homeodomain protein
<400> to
Arg Gln Ile Lys Ile Trp Phe Gln Asn Arg Arg Met Lys Trp Lys Lys
1 5 10 15
<210> 11
<211> 34
<212> PRT
<213> Herpes Simplex Virus
<220>
<221> mist feature
<223> Residues 267-300 of the HSV VP22 protein
<400> 11
Asp Ala Ala Thr Ala Thr Arg Gly Arg Ser Ala Ala Ser Arg Pro Thr
1 5 10 15
Glu Arg Pro Arg Ala Pro Ala Arg Ser Ala Ser Arg Pro Arg Arg Pro
20 25 30
Val Glu
<210> 12
<211> 11
<212> PRT
_7_
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
<213> Artificial Sequence
<220>
<223> Basic peptide with cellular uptake signal acitivty
<400> 12
Tyr Ala Arg Ala Ala Ala Arg Gln Ala Arg Ala
1 5 10
<210> 13
<211> 9
<212> PRT
IS <213> Artificial Sequence
<220>
<223> Basic peptide with cellular uptake signal activity, "R9"
<400> 13
Arg Arg Arg Arg Arg Arg Arg Arg Arg
1 5
<210> 14
<211> 16
<212> PRT
<213> Artificial Sequence
<220>
<223> D-penetratin peptide
<220>
<221> MISC FEATURE
<222> (1) . . (16)
<223> All amino acids are in the D-form.
<400> 14
Arg Gln Ile Lys Ile Trp Phe Gln Asn Arg Arg Met Lys Trp Lys Lys
1 5 10 15
_g_
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
<210> 15
<211> 18
<212> PRT
<213> Artificial Sequence
15
25
<220>
<223> Peptide Syn B1 from Antennapedia homeodomain protein
<400> 15
Arg Gly Gly Arg Leu Ser Tyr Ser Arg Arg Arg Phe Ser Thr Ser Thr
1 5 10 15
Gly Arg
<210> 16
<211> 10
<212> PRT
<213> Artificial Sequence
<220>
<223> L-SynB3 peptide from Antennapedia homeodomain protein
<400> 16
Arg Arg Leu Ser Tyr Ser Arg Arg Arg Phe
1 5 10
<210> 17
<211> to
<212> PRT
<213> Artificial Sequence
<220>
<223> D-SynB3 peptide from Antennapedia homeodomain protein
-9-
CA 02472729 2004-07-08
WO 03/062447 PCT/US03/01529
<220>
<221> MISC FEATURE
<222> (1)..(10)
<223> All amino acids are in the D-form.
<400> 17
Arg Arg Leu Ser Tyr Ser Arg Arg Arg Phe
1 5 10
<210> 18
<211> 8
<212> PRT
IS <213> Artificial Sequence
<220>
<223> Flag Epitope Peptide
<400> 18
Asp Tyr Lys Asp Asp Asp Asp Lys
1 5
-10-